Analysis of scRNA data from Montoro et al.
Jason Willwerscheid
10/19/2018
Last updated: 2019-02-25
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180714)The command
set.seed(20180714)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 4693348
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: data/.DS_Store Ignored: docs/.DS_Store Ignored: docs/figure/.DS_Store Untracked files: Untracked: analysis/gd_notes.Rmd Untracked: analysis/trachea3.Rmd Untracked: code/count_sim.R Untracked: code/pathways.R Untracked: code/trachea3.R Untracked: data/lowrank/ Untracked: data/tmp14.rds Untracked: data/tmpdata.rds Untracked: data/tmplfsr.rds Untracked: data/trachea3/ Untracked: docs/figure/count_notes.Rmd/ Untracked: docs/figure/trachea3.Rmd/ Unstaged changes: Deleted: data/before_bad.Rdata
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | f2b86c1 | Jason Willwerscheid | 2018-12-05 | Build site. |
| Rmd | a89c222 | Jason Willwerscheid | 2018-12-05 | wflow_publish(“analysis/trachea.Rmd”) |
| html | 8327d75 | Jason Willwerscheid | 2018-10-23 | Build site. |
| Rmd | 28891d1 | Jason Willwerscheid | 2018-10-23 | workflowr::wflow_publish(“analysis/trachea.Rmd”) |
Introduction and Code
I fit 30 factors to the drop-seq dataset discussed in Montoro et al. I removed all genes with zero total counts and did a log1p transform of the counts. I used nonnegative priors for gene loadings and normal-mixture priors for cell loadings.
# Add 30 factors with rough backfits after every 5 factors.
fl <- flashier(data, greedy.Kmax = 30, var.type = 1,
prior.type = c("nonnegative", "normal.mix"),
ash.param = list(optmethod = "mixSQP"),
backfit.every = 5, final.backfit = TRUE,
backfit.order = "montaigne", warmstart.backfits = FALSE)
# Refine by backfitting.
fl <- flashier(data, flash.init = fl, backfit = "only",
backfit.order = "dropout", backfit.maxiter = 200)The following code is used to produce the boxplots and tables below.
suppressMessages({
library(ggplot2)
library(topGO)
library(org.Mm.eg.db)
})
#> Warning: package 'ggplot2' was built under R version 3.4.4
fl <- readRDS("~/Downloads/DropSeq30_backfit.rds")
# Remove large data object to free up memory.
fl$fit$Y <- NULL
# Data frame containing cell type and loadings for each factor.
PSdf <- data.frame(fl$loadings$normalized.loadings[[2]])
cell.names <- rownames(fl$loadings$normalized.loadings[[2]])
cell.types <- as.factor(sapply(strsplit(cell.names, "_"), `[`, 3))
levels(cell.types)
#> [1] "Basal" "Ciliated" "Club" "Goblet"
#> [5] "Ionocyte" "Neuroendocrine" "Tuft"
levels(cell.types) <- c("Bas", "Cil", "Clb", "Gob", "Ion", "Nec", "Tft")
PSdf$cell.type <- cell.types
# Need to select signficant genes for topGO.
# Scale gene loadings by scale constant * maximum cell loading.
s <- fl$loadings$scale.constant *
apply(abs(fl$loadings$normalized.loadings[[2]]), 2, max)
gene.loadings <- fl$loadings$normalized.loadings[[1]]
gene.loadings <- gene.loadings * rep(s, each = nrow(gene.loadings))
# Get a pseudo-t statistic by dividing by the residual SE.
gene.t <- gene.loadings * sqrt(fl$fit$tau)
# Convert to a p-value.
gene.p <- 2 * (1 - pnorm(abs(gene.t)))
# Select significant genes using Benjamini-Hochberg.
BH <- function(k, alpha = 0.01) {
pvals <- gene.p[, k]
selected <- rep(0, length(pvals))
names(selected) <- names(pvals)
n <- length(pvals)
sorted.pvals <- sort(pvals)
BH <- sorted.pvals < alpha / (n - 0:(n - 1))
cutoff <- min(which(!BH))
selected[pvals < sorted.pvals[cutoff]] <- 1
return(selected)
}
gene.sig <- sapply(1:ncol(gene.p), BH)
# How many significant genes per factor are there?
colSums(gene.sig)
#> [1] 286 512 98 74 163 183 131 198 168 541 364 365 104 558 535 499 31
#> [18] 504 406 436 3 183 0 96 448 326 361 404 45 156
# Omit factors with PVE below a certain (empirically determined) threshold.
kset <- (1:ncol(gene.p))[fl$pve > 0.0001 & colSums(gene.sig) > 9]
# I also want to see which factors contain genes mentioned in the paper.
paper.genes <- data.frame(
gene = c("Nfia", "Ascl1", "Ascl2", "Ascl3", "Foxq1", "Cdhr3", "Rgs13",
"Muc5b", "Notch2", "Il13ra1", "Krt4", "Krt13", "Krt8", "Ecm1",
"S100a11", "Cldn3", "Lgals3", "Anxa1", "Il25", "Tslp", "Alox5ap",
"Ptprc", "Pou2f3", "Gfi1b", "Spib", "Sox9", "Gp2", "Tff1",
"Tff2", "Lman1l", "P2rx4", "Muc5ac", "Dcpp1", "Dcpp2", "Dcpp3",
"Atp6v1c2", "Atp6v0d2", "Cftr", "Aqp3", "Krt5", "Dapl1", "Hspa1a",
"Trp63", "Scgb1a1", "Krt15", "Cyp2f2", "Lypd2", "Cbr2", "Foxj1",
"Ccdc153", "Ccdc113", "Mlf1", "Lztfl1", "Chga", "Dclk1"),
type = c("Club", "NEC", "Tuft", "Ion", "Goblet", "Cil", "Tuft",
"Club", "Club", "Club", "Hill", "Hill", "Basal", "Hill",
"Hill", "Hill", "Hill", "Hill", "Tuft", "Tuft", "Tuft2",
"Tuft2", "Tuft1", "Tuft2", "Tuft2", "Tuft2", "Goblet", "Gob1",
"Gob1", "Gob1", "Gob1", "Gob1", "Gob2", "Gob2", "Gob2",
"Ion", "Ion", "Ion", "BM", "BM", "BM", "BM",
"BM", "ClbM", "ClbM", "ClbM", "ClbM", "ClbM", "CilM",
"CilM", "CilM", "CilM", "CilM", "NECM", "TftM"))
paper.genes$gene <- as.character(paper.genes$gene)
levels(paper.genes$type) <- c("Basal", "Basal (Marker)", "Ciliated",
"Ciliated (Marker)", "Club (Marker)", "Club",
"Goblet-1", "Goblet-2", "Goblet", "Hillock",
"Ionocyte", "NEC", "NEC (Marker)",
"Tuft (Marker)", "Tuft", "Tuft-1", "Tuft-2")
# Set up topGOdata object.
GO.list <- as.factor(gene.sig[, 1])
GOdata <- new("topGOdata", ontology = "BP", allGenes = GO.list,
annot = annFUN.org, mapping = "org.Mm.eg", ID = "symbol")
#>
#> Building most specific GOs .....
#> ( 11286 GO terms found. )
#>
#> Build GO DAG topology ..........
#> ( 15215 GO terms and 35696 relations. )
#>
#> Annotating nodes ...............
#> ( 15591 genes annotated to the GO terms. )# Loop over kset.
for (k in kset) {
cat(paste0("## Factor ", k, " (PVE: ",
formatC(fl$pve[k], format = "f", digits = 4), ")\n"))
fctr <- paste0("X", k)
plot(ggplot(PSdf, aes_string(x = "cell.type", y = fctr)) +
geom_boxplot(outlier.shape = NA, color = "red") +
geom_jitter(position = position_jitter(0.2), cex = 0.1) +
labs(x = "Cell Type", y = paste("Factor", k, "value")))
cat("\n")
mentions <- which(gene.sig[paper.genes$gene, k] == 1)
if (length(mentions) > 0) {
mentions.df <- paper.genes[mentions, ]
mentions.df$t.val <- gene.t[names(mentions), k]
cat("Significant genes that are also mentioned in the paper:\n")
print(knitr::kable(mentions.df))
cat("\n")
}
cat("Gene Ontology terms:\n")
GOdata@allScores <- as.factor(gene.sig[, k])
result <- suppressMessages(
runTest(GOdata, algorithm = "classic", statistic = "fisher")
)
allRes <- GenTable(GOdata, classic = result, topNodes = 10)
print(knitr::kable(allRes))
cat("\n")
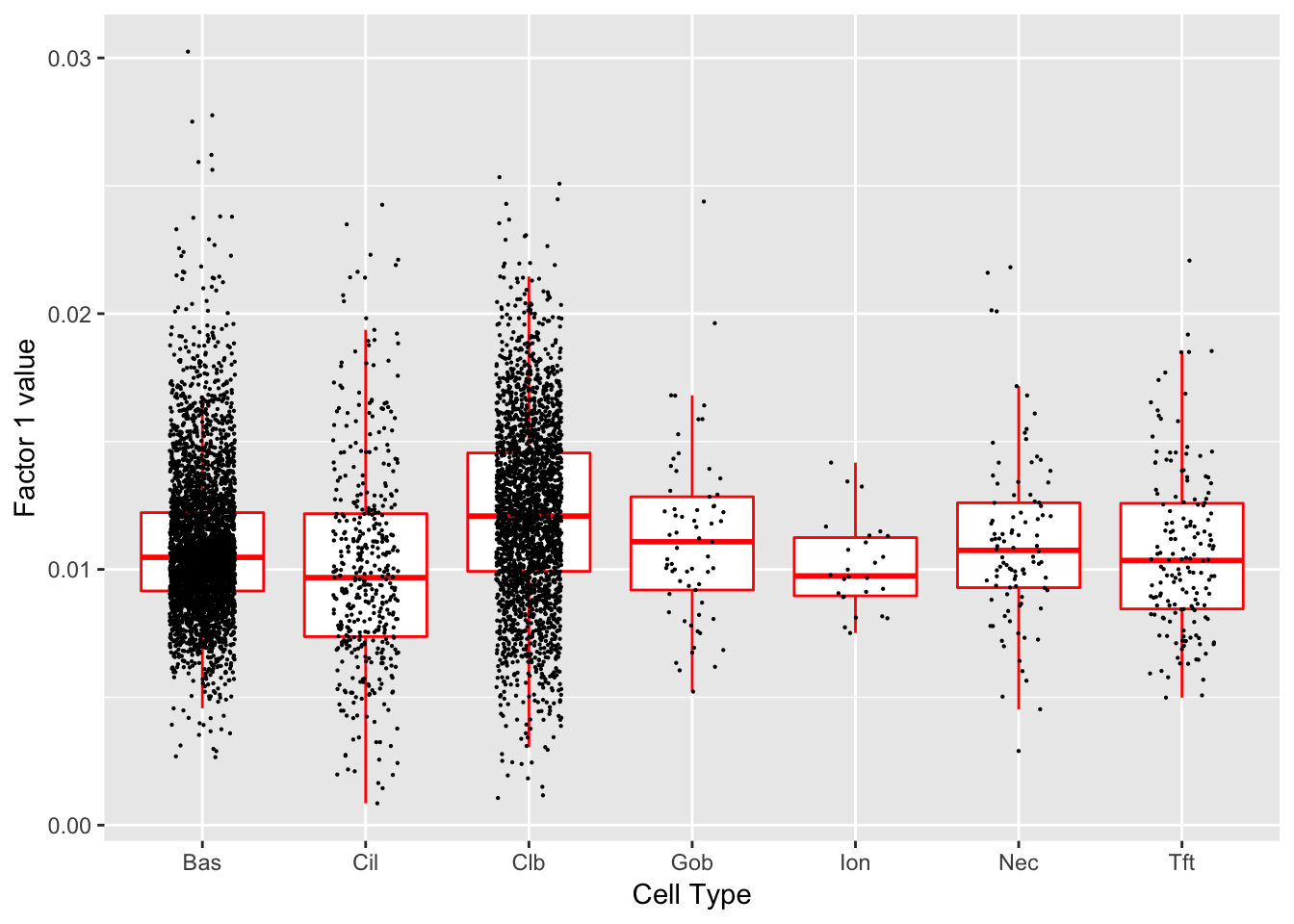
}Factor 1 (PVE: 0.5246)
Expand here to see past versions of factors2-1.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 13 | Krt8 | Basal | 7.242263 |
| 15 | S100a11 | Hillock | 17.425570 |
| 16 | Cldn3 | Hillock | 7.228331 |
| 17 | Lgals3 | Hillock | 10.231025 |
| 18 | Anxa1 | Hillock | 11.827277 |
| 39 | Aqp3 | Basal (Marker) | 7.943683 |
| 42 | Hspa1a | Basal (Marker) | 5.491426 |
| 44 | Scgb1a1 | Club (Marker) | 9.262051 |
| 45 | Krt15 | Club (Marker) | 11.714101 |
| 46 | Cyp2f2 | Club (Marker) | 27.970810 |
| 47 | Lypd2 | Club (Marker) | 11.379180 |
| 48 | Cbr2 | Club (Marker) | 16.672470 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0006518 | peptide metabolic process | 682 | 54 | 11.20 | 1.6e-22 |
| GO:0006412 | translation | 539 | 46 | 8.85 | 1.7e-20 |
| GO:0043043 | peptide biosynthetic process | 560 | 46 | 9.20 | 8.0e-20 |
| GO:0043603 | cellular amide metabolic process | 802 | 54 | 13.17 | 2.8e-19 |
| GO:0043604 | amide biosynthetic process | 626 | 46 | 10.28 | 6.6e-18 |
| GO:1901566 | organonitrogen compound biosynthetic pro… | 1386 | 68 | 22.76 | 6.4e-17 |
| GO:0002181 | cytoplasmic translation | 60 | 14 | 0.99 | 6.4e-13 |
| GO:0046034 | ATP metabolic process | 191 | 21 | 3.14 | 6.1e-12 |
| GO:0009205 | purine ribonucleoside triphosphate metab… | 213 | 21 | 3.50 | 4.9e-11 |
| GO:0009126 | purine nucleoside monophosphate metaboli… | 216 | 21 | 3.55 | 6.4e-11 |
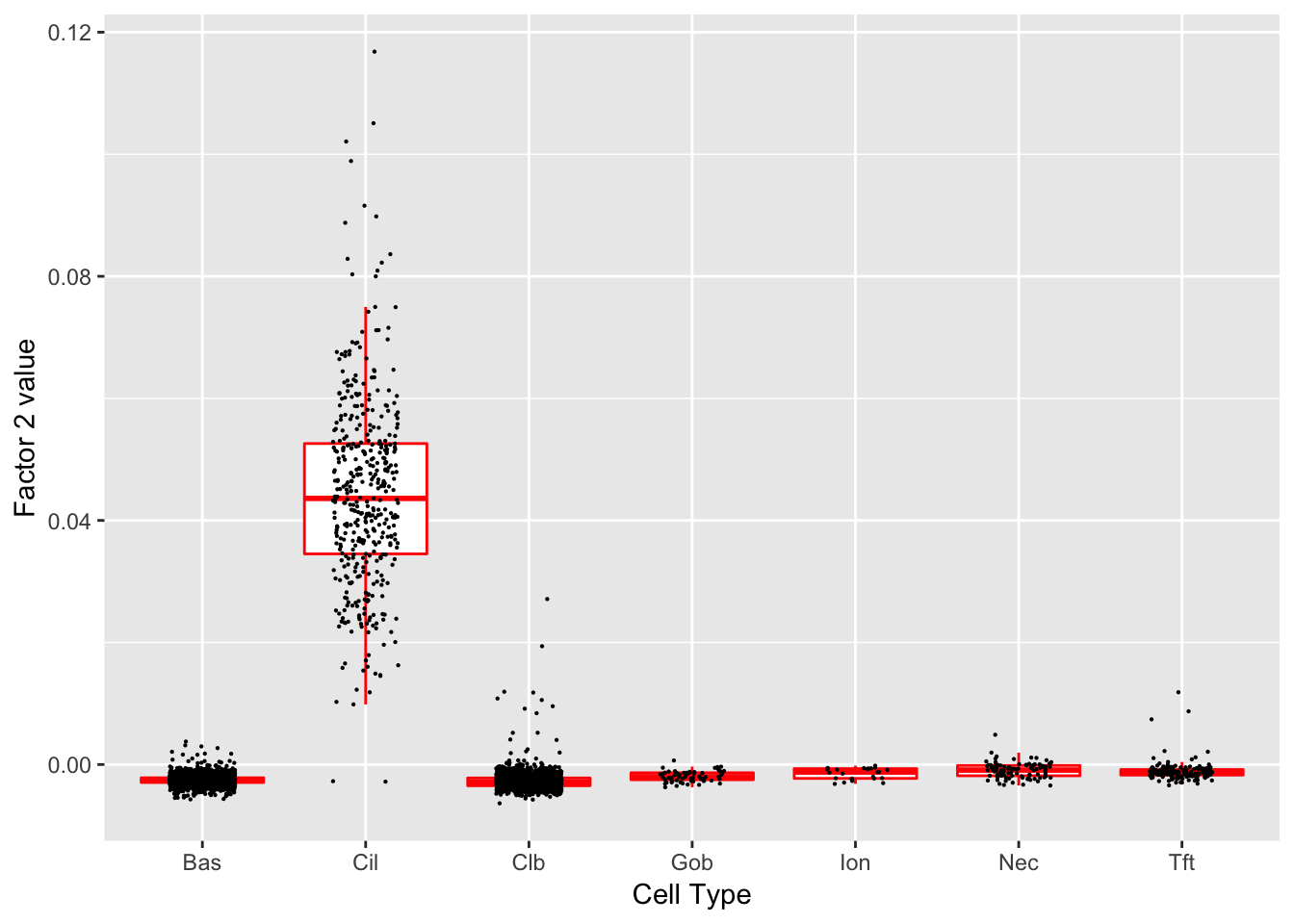
Factor 2 (PVE: 0.0189)
Expand here to see past versions of factors2-2.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 6 | Cdhr3 | Ciliated | 14.432419 |
| 13 | Krt8 | Basal | 7.915175 |
| 15 | S100a11 | Hillock | 8.134064 |
| 16 | Cldn3 | Hillock | 8.365458 |
| 17 | Lgals3 | Hillock | 6.710951 |
| 18 | Anxa1 | Hillock | 8.197110 |
| 42 | Hspa1a | Basal (Marker) | 5.422251 |
| 46 | Cyp2f2 | Club (Marker) | 9.230342 |
| 48 | Cbr2 | Club (Marker) | 6.684108 |
| 49 | Foxj1 | Ciliated (Marker) | 14.103074 |
| 50 | Ccdc153 | Ciliated (Marker) | 33.342495 |
| 51 | Ccdc113 | Ciliated (Marker) | 20.003698 |
| 52 | Mlf1 | Ciliated (Marker) | 20.536544 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0003341 | cilium movement | 68 | 38 | 1.92 | < 1e-30 |
| GO:0044782 | cilium organization | 283 | 65 | 8.00 | < 1e-30 |
| GO:0060271 | cilium assembly | 265 | 60 | 7.50 | < 1e-30 |
| GO:0007018 | microtubule-based movement | 242 | 53 | 6.85 | < 1e-30 |
| GO:0035082 | axoneme assembly | 56 | 29 | 1.58 | 1.8e-30 |
| GO:0120031 | plasma membrane bounded cell projection … | 432 | 65 | 12.22 | 3.6e-29 |
| GO:0030031 | cell projection assembly | 439 | 65 | 12.42 | 9.3e-29 |
| GO:0001578 | microtubule bundle formation | 84 | 30 | 2.38 | 1.8e-25 |
| GO:0007017 | microtubule-based process | 630 | 73 | 17.82 | 1.9e-25 |
| GO:0070925 | organelle assembly | 650 | 68 | 18.39 | 3.5e-21 |
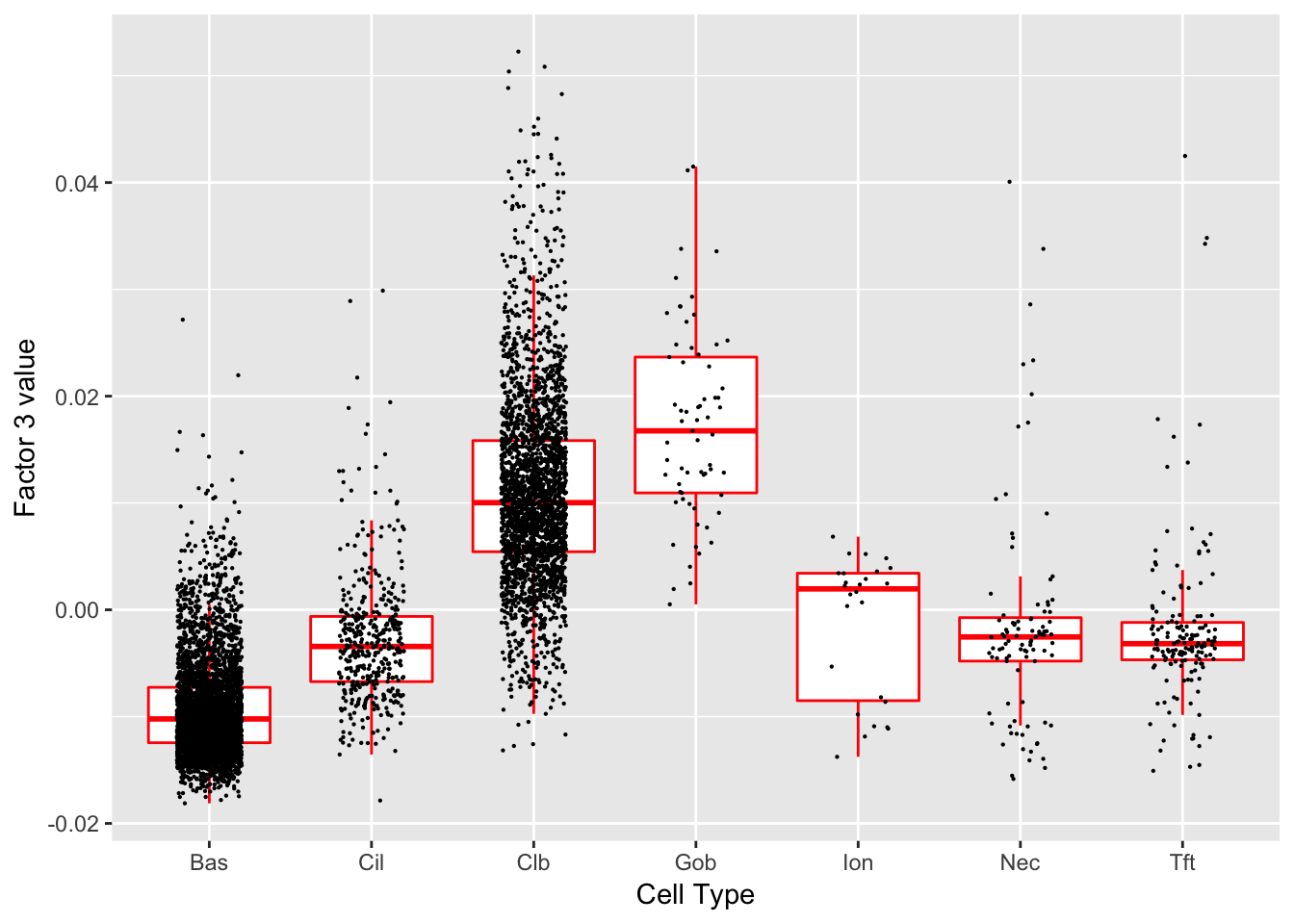
Factor 3 (PVE: 0.0387)
Expand here to see past versions of factors2-3.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 13 | Krt8 | Basal | 5.067801 |
| 16 | Cldn3 | Hillock | 6.660415 |
| 44 | Scgb1a1 | Club (Marker) | 6.303162 |
| 46 | Cyp2f2 | Club (Marker) | 7.803280 |
| 47 | Lypd2 | Club (Marker) | 5.961594 |
| 48 | Cbr2 | Club (Marker) | 5.262580 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0043618 | regulation of transcription from RNA pol… | 44 | 6 | 0.24 | 1.4e-07 |
| GO:0043620 | regulation of DNA-templated transcriptio… | 48 | 6 | 0.26 | 2.4e-07 |
| GO:0035914 | skeletal muscle cell differentiation | 59 | 6 | 0.33 | 8.4e-07 |
| GO:0050896 | response to stimulus | 6040 | 55 | 33.32 | 1.9e-06 |
| GO:0019730 | antimicrobial humoral response | 41 | 5 | 0.23 | 2.9e-06 |
| GO:0007519 | skeletal muscle tissue development | 139 | 7 | 0.77 | 1.1e-05 |
| GO:0009888 | tissue development | 1492 | 22 | 8.23 | 1.4e-05 |
| GO:0060538 | skeletal muscle organ development | 144 | 7 | 0.79 | 1.4e-05 |
| GO:0035821 | modification of morphology or physiology… | 96 | 6 | 0.53 | 1.5e-05 |
| GO:0046686 | response to cadmium ion | 28 | 4 | 0.15 | 1.6e-05 |
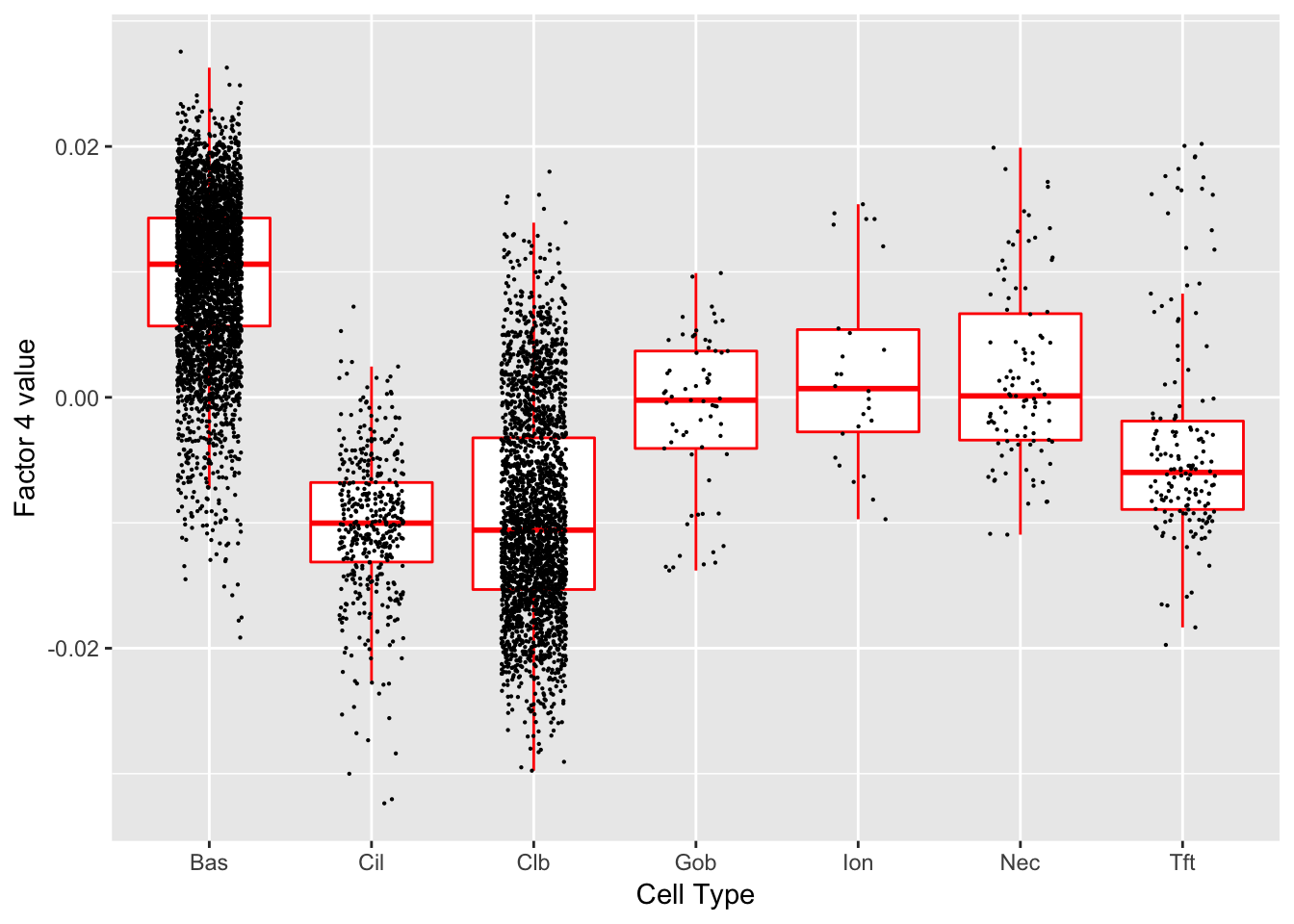
Factor 4 (PVE: 0.0581)
Expand here to see past versions of factors2-4.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 39 | Aqp3 | Basal (Marker) | 5.969027 |
| 42 | Hspa1a | Basal (Marker) | 6.596314 |
| 46 | Cyp2f2 | Club (Marker) | 6.806845 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0043618 | regulation of transcription from RNA pol… | 44 | 4 | 0.17 | 2.6e-05 |
| GO:0010941 | regulation of cell death | 1472 | 17 | 5.76 | 3.3e-05 |
| GO:0043620 | regulation of DNA-templated transcriptio… | 48 | 4 | 0.19 | 3.6e-05 |
| GO:0000028 | ribosomal small subunit assembly | 19 | 3 | 0.07 | 5.3e-05 |
| GO:0035914 | skeletal muscle cell differentiation | 59 | 4 | 0.23 | 8.2e-05 |
| GO:0009612 | response to mechanical stimulus | 119 | 5 | 0.47 | 0.00010 |
| GO:0043604 | amide biosynthetic process | 626 | 10 | 2.45 | 0.00014 |
| GO:0021761 | limbic system development | 70 | 4 | 0.27 | 0.00016 |
| GO:0050896 | response to stimulus | 6040 | 38 | 23.63 | 0.00016 |
| GO:0007519 | skeletal muscle tissue development | 139 | 5 | 0.54 | 0.00021 |
Factor 5 (PVE: 0.0016)
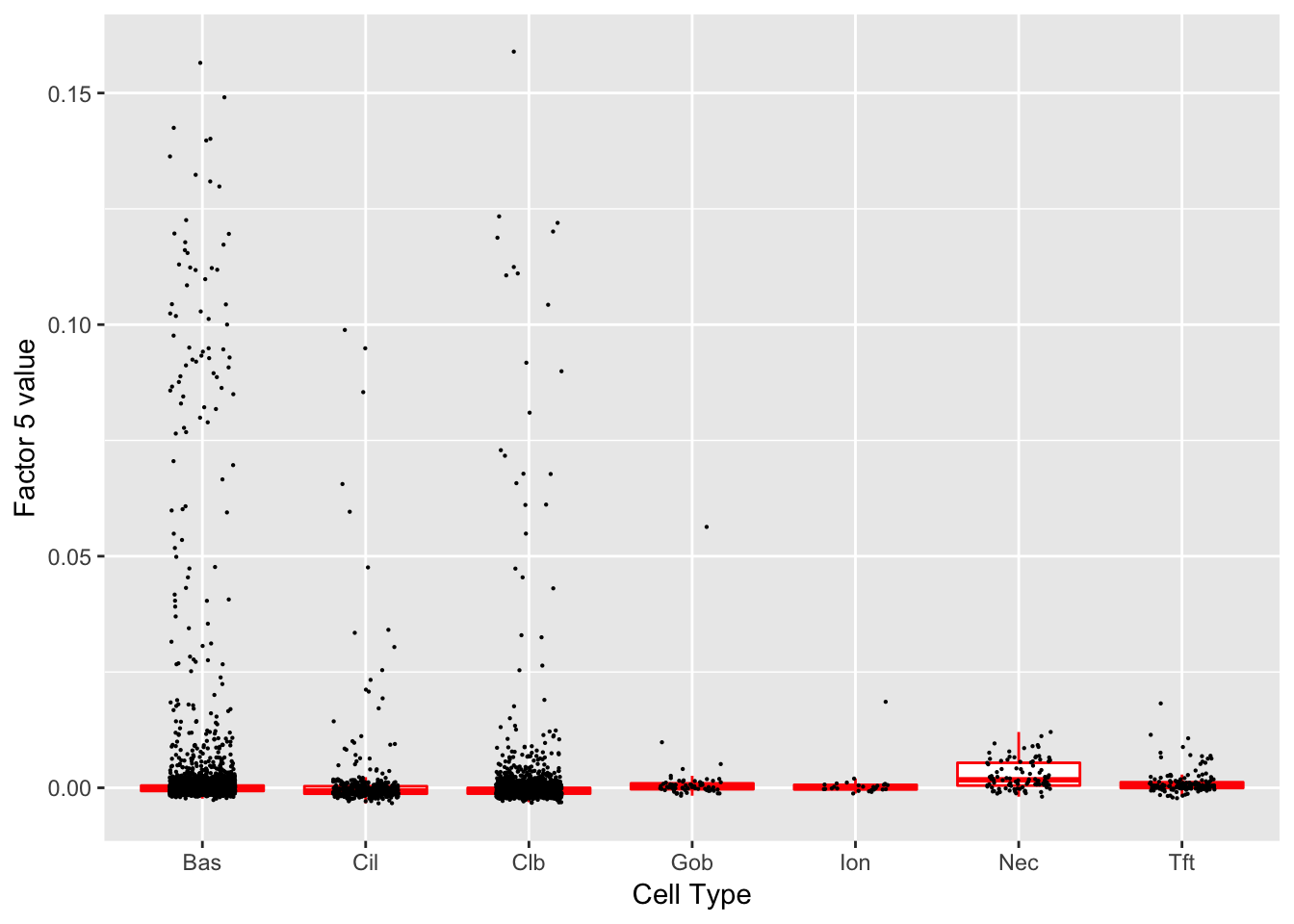
Expand here to see past versions of factors2-5.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0051301 | cell division | 512 | 60 | 4.56 | < 1e-30 |
| GO:0007049 | cell cycle | 1426 | 80 | 12.71 | < 1e-30 |
| GO:1903047 | mitotic cell cycle process | 553 | 56 | 4.93 | < 1e-30 |
| GO:0022402 | cell cycle process | 912 | 66 | 8.13 | < 1e-30 |
| GO:0000278 | mitotic cell cycle | 714 | 60 | 6.37 | < 1e-30 |
| GO:0007059 | chromosome segregation | 275 | 43 | 2.45 | < 1e-30 |
| GO:0140014 | mitotic nuclear division | 222 | 33 | 1.98 | < 1e-30 |
| GO:0000280 | nuclear division | 341 | 38 | 3.04 | < 1e-30 |
| GO:0098813 | nuclear chromosome segregation | 212 | 32 | 1.89 | 1.5e-30 |
| GO:0048285 | organelle fission | 386 | 38 | 3.44 | 3.0e-29 |
Factor 6 (PVE: 0.0076)
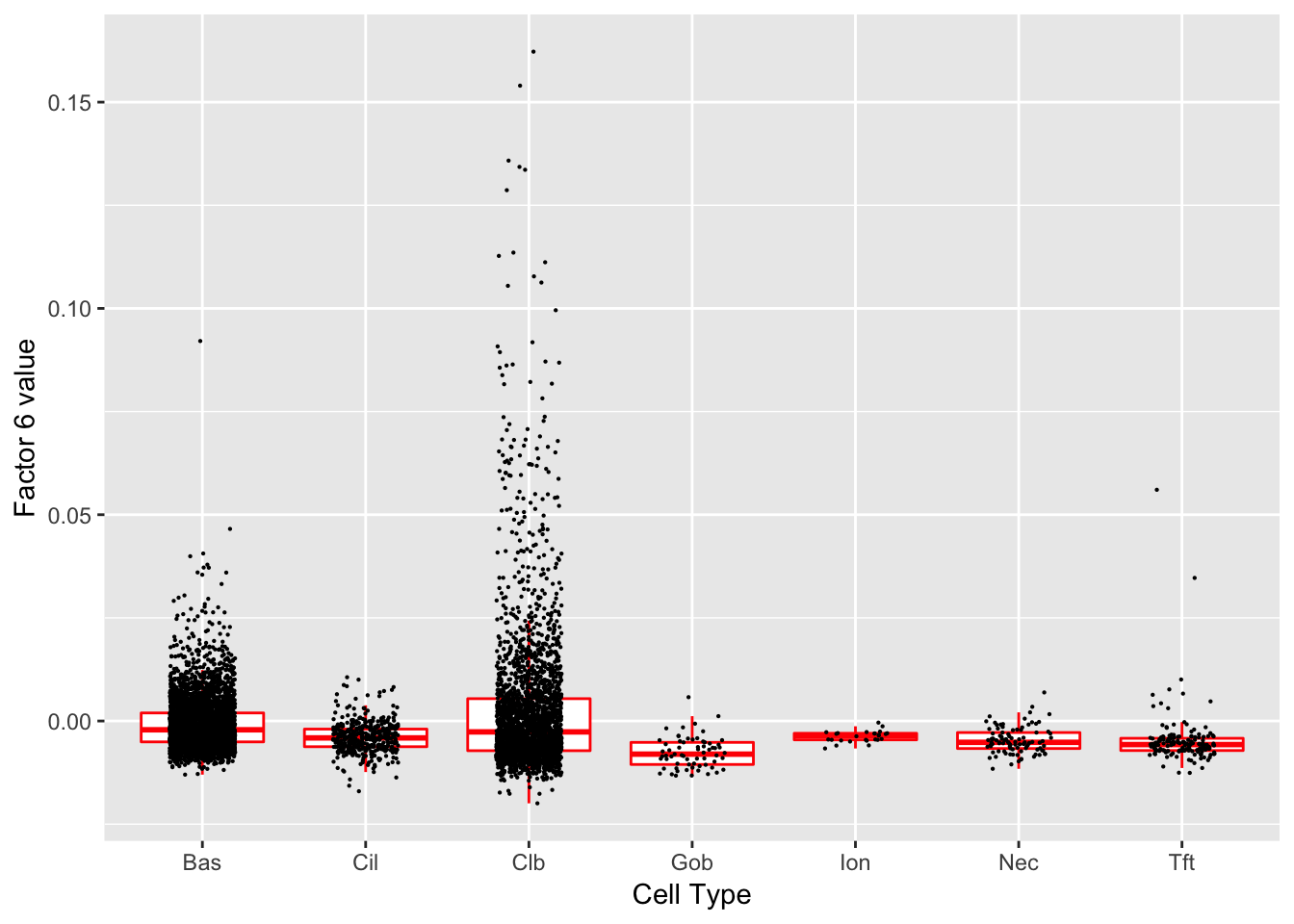
Expand here to see past versions of factors2-6.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 11 | Krt4 | Hillock | 14.016113 |
| 12 | Krt13 | Hillock | 15.089416 |
| 13 | Krt8 | Basal | 7.489441 |
| 14 | Ecm1 | Hillock | 16.277826 |
| 15 | S100a11 | Hillock | 12.758571 |
| 16 | Cldn3 | Hillock | 9.570576 |
| 17 | Lgals3 | Hillock | 16.290688 |
| 18 | Anxa1 | Hillock | 17.406668 |
| 39 | Aqp3 | Basal (Marker) | 10.049004 |
| 45 | Krt15 | Club (Marker) | 10.647146 |
| 46 | Cyp2f2 | Club (Marker) | 9.785778 |
| 48 | Cbr2 | Club (Marker) | 5.267489 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0031579 | membrane raft organization | 22 | 6 | 0.23 | 8.3e-08 |
| GO:0031424 | keratinization | 24 | 6 | 0.25 | 1.5e-07 |
| GO:0007162 | negative regulation of cell adhesion | 224 | 13 | 2.37 | 7.6e-07 |
| GO:0001765 | membrane raft assembly | 10 | 4 | 0.11 | 2.4e-06 |
| GO:0022610 | biological adhesion | 1075 | 29 | 11.38 | 2.7e-06 |
| GO:0022408 | negative regulation of cell-cell adhesio… | 146 | 10 | 1.55 | 3.4e-06 |
| GO:0098609 | cell-cell adhesion | 594 | 20 | 6.29 | 4.8e-06 |
| GO:0071709 | membrane assembly | 25 | 5 | 0.26 | 5.6e-06 |
| GO:0007155 | cell adhesion | 1064 | 28 | 11.26 | 6.6e-06 |
| GO:0030216 | keratinocyte differentiation | 97 | 8 | 1.03 | 8.6e-06 |
Factor 7 (PVE: 0.0185)
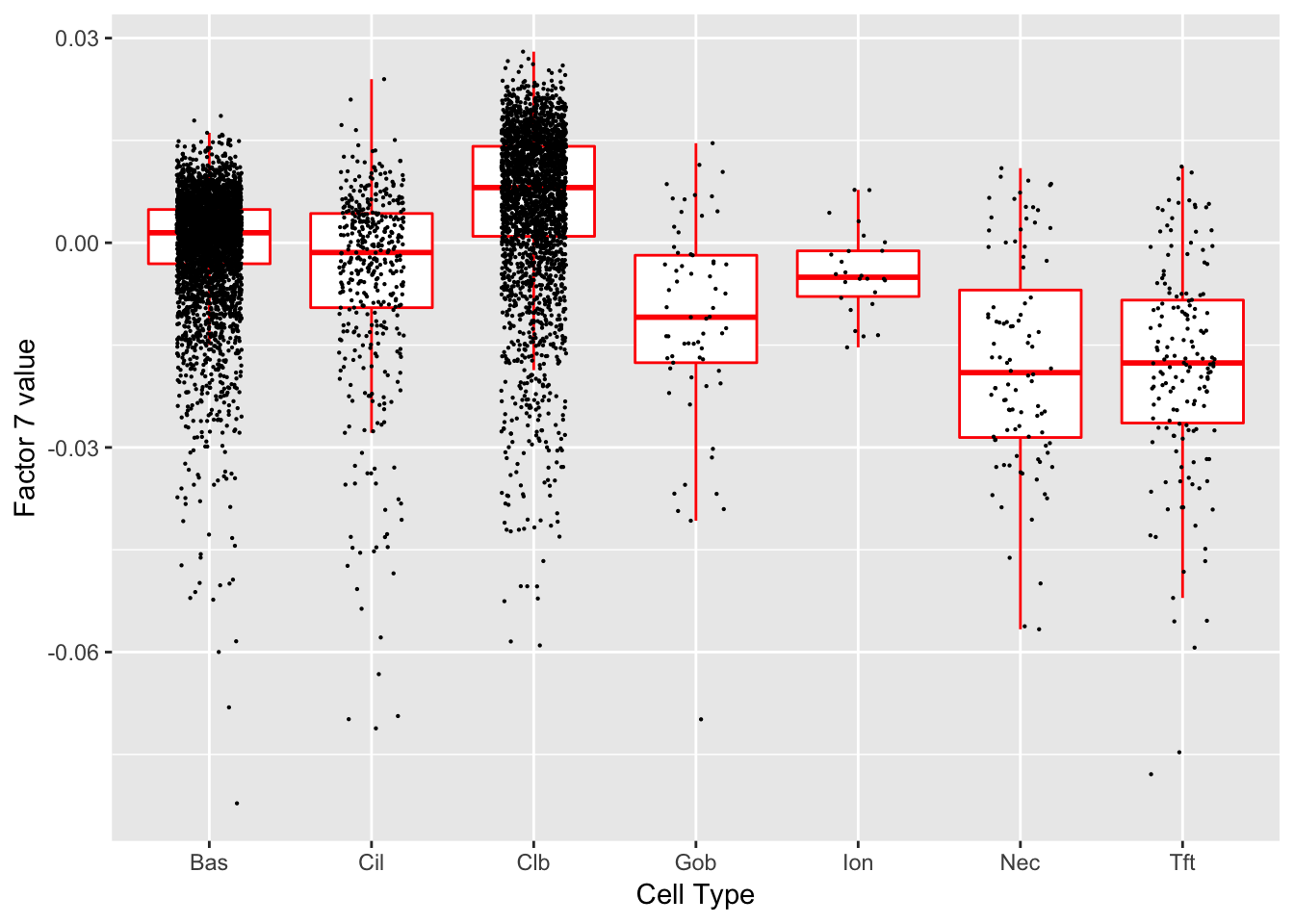
Expand here to see past versions of factors2-7.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 15 | S100a11 | Hillock | 10.231936 |
| 16 | Cldn3 | Hillock | 6.118989 |
| 17 | Lgals3 | Hillock | 6.959122 |
| 18 | Anxa1 | Hillock | 7.969162 |
| 44 | Scgb1a1 | Club (Marker) | 9.736032 |
| 45 | Krt15 | Club (Marker) | 7.180916 |
| 46 | Cyp2f2 | Club (Marker) | 20.148326 |
| 47 | Lypd2 | Club (Marker) | 8.531190 |
| 48 | Cbr2 | Club (Marker) | 11.853278 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0006518 | peptide metabolic process | 682 | 19 | 4.86 | 3.1e-07 |
| GO:0019730 | antimicrobial humoral response | 41 | 6 | 0.29 | 4.2e-07 |
| GO:0006412 | translation | 539 | 16 | 3.84 | 1.3e-06 |
| GO:0043043 | peptide biosynthetic process | 560 | 16 | 3.99 | 2.1e-06 |
| GO:0043603 | cellular amide metabolic process | 802 | 19 | 5.71 | 3.5e-06 |
| GO:0002181 | cytoplasmic translation | 60 | 6 | 0.43 | 4.2e-06 |
| GO:0000028 | ribosomal small subunit assembly | 19 | 4 | 0.14 | 8.7e-06 |
| GO:0043604 | amide biosynthetic process | 626 | 16 | 4.46 | 8.9e-06 |
| GO:0042221 | response to chemical | 2842 | 39 | 20.23 | 1.6e-05 |
| GO:0006880 | intracellular sequestering of iron ion | 2 | 2 | 0.01 | 5.0e-05 |
Factor 8 (PVE: 0.0028)
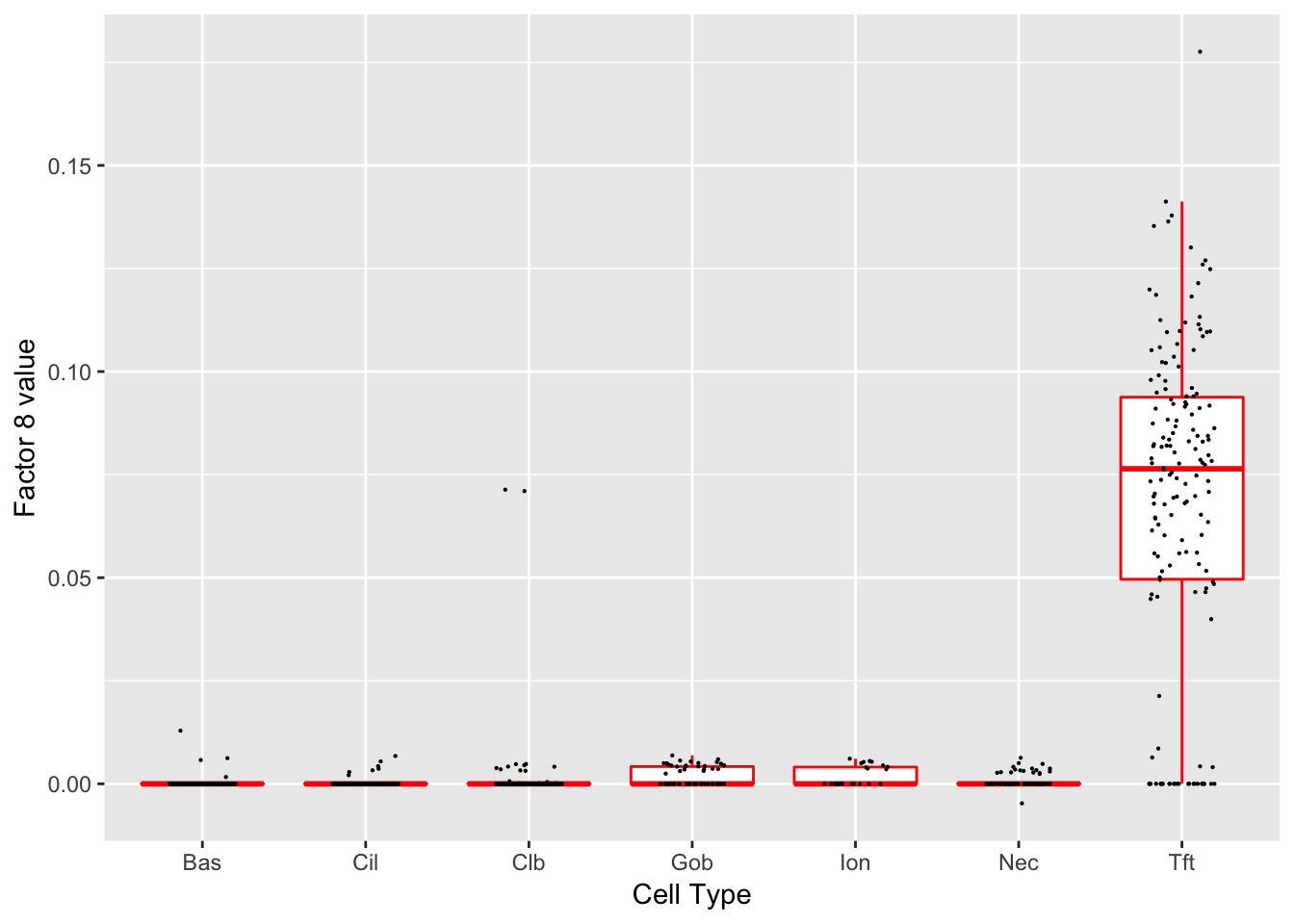
Expand here to see past versions of factors2-8.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 7 | Rgs13 | Tuft | 24.955241 |
| 13 | Krt8 | Basal | 7.649654 |
| 18 | Anxa1 | Hillock | 5.991935 |
| 19 | Il25 | Tuft | 8.547482 |
| 21 | Alox5ap | Tuft-2 | 16.380566 |
| 23 | Pou2f3 | Tuft-1 | 8.797474 |
| 25 | Spib | Tuft-2 | 13.945543 |
| 26 | Sox9 | Tuft-2 | 9.185370 |
| 44 | Scgb1a1 | Club (Marker) | 5.237961 |
| 55 | Dclk1 | Tuft (Marker) | 15.476724 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0007186 | G-protein coupled receptor signaling pat… | 536 | 19 | 6.02 | 9.6e-06 |
| GO:0065007 | biological regulation | 8828 | 126 | 99.09 | 1.8e-05 |
| GO:0021979 | hypothalamus cell differentiation | 6 | 3 | 0.07 | 2.7e-05 |
| GO:0050896 | response to stimulus | 6040 | 94 | 67.80 | 3.8e-05 |
| GO:0006518 | peptide metabolic process | 682 | 20 | 7.66 | 8.2e-05 |
| GO:0043603 | cellular amide metabolic process | 802 | 22 | 9.00 | 9.5e-05 |
| GO:0019370 | leukotriene biosynthetic process | 9 | 3 | 0.10 | 0.00011 |
| GO:0006880 | intracellular sequestering of iron ion | 2 | 2 | 0.02 | 0.00013 |
| GO:0097577 | sequestering of iron ion | 2 | 2 | 0.02 | 0.00013 |
| GO:0098609 | cell-cell adhesion | 594 | 18 | 6.67 | 0.00013 |
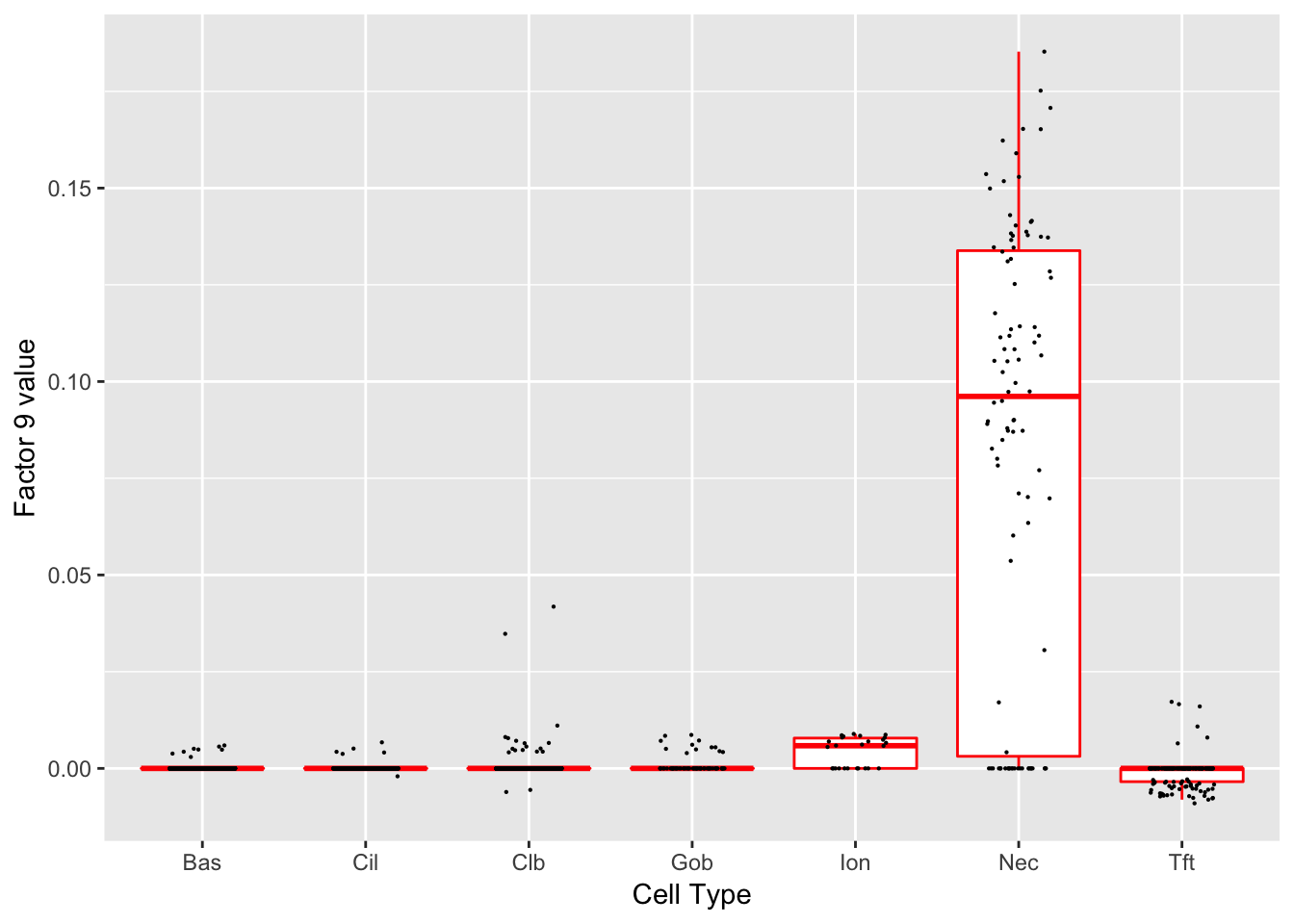
Factor 9 (PVE: 0.0012)
Expand here to see past versions of factors2-9.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 2 | Ascl1 | NEC | 14.57633 |
| 54 | Chga | NEC (Marker) | 45.57047 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0050877 | nervous system process | 698 | 28 | 6.72 | 1.1e-10 |
| GO:0003008 | system process | 1209 | 37 | 11.63 | 1.7e-10 |
| GO:0007610 | behavior | 526 | 24 | 5.06 | 2.1e-10 |
| GO:0099504 | synaptic vesicle cycle | 113 | 11 | 1.09 | 1.1e-08 |
| GO:0007268 | chemical synaptic transmission | 471 | 20 | 4.53 | 2.6e-08 |
| GO:0098916 | anterograde trans-synaptic signaling | 471 | 20 | 4.53 | 2.6e-08 |
| GO:0099537 | trans-synaptic signaling | 472 | 20 | 4.54 | 2.7e-08 |
| GO:0099536 | synaptic signaling | 473 | 20 | 4.55 | 2.8e-08 |
| GO:0050804 | modulation of chemical synaptic transmis… | 300 | 16 | 2.89 | 3.2e-08 |
| GO:0006812 | cation transport | 840 | 26 | 8.08 | 1.1e-07 |
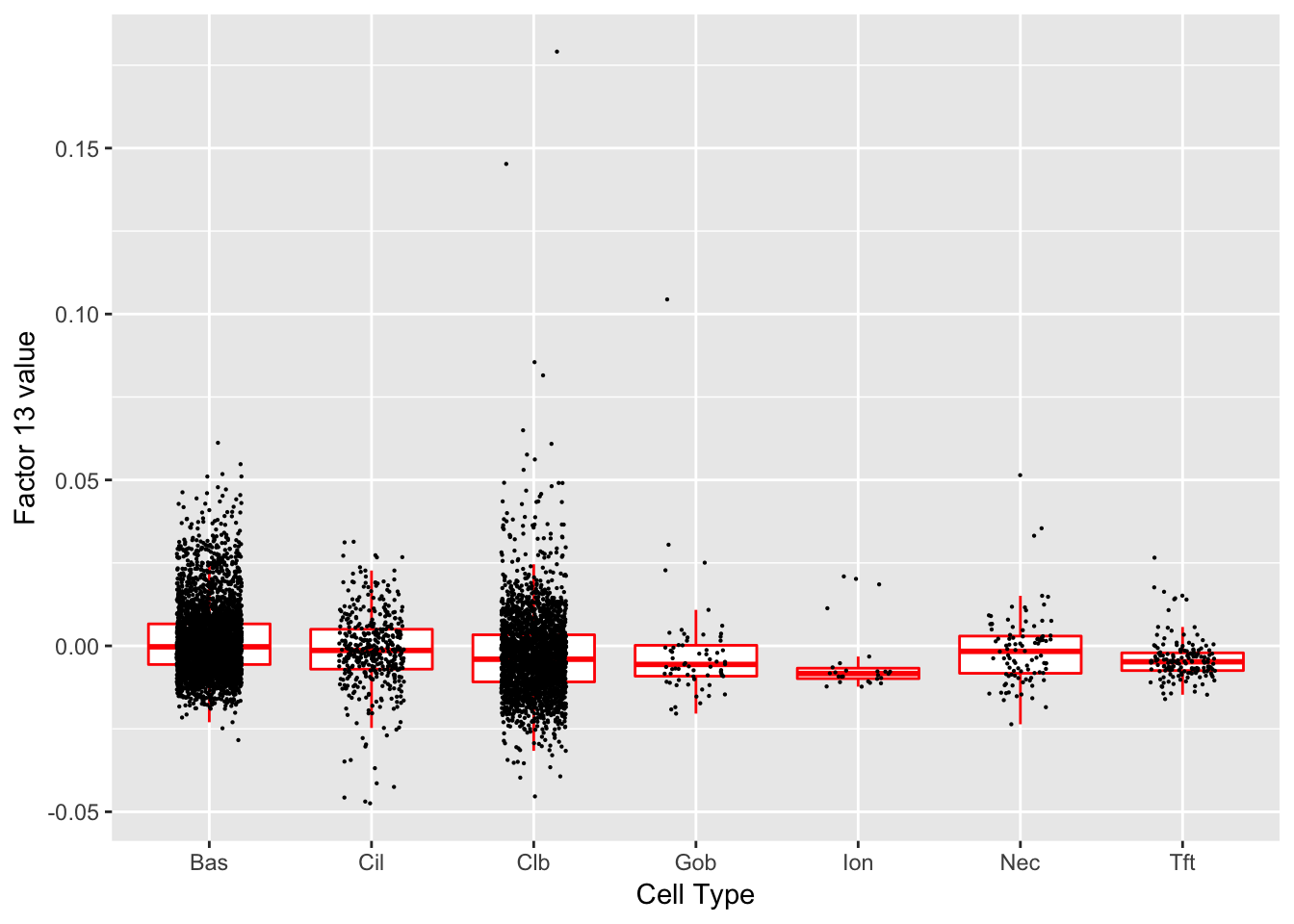
Factor 13 (PVE: 0.0027)
Expand here to see past versions of factors2-10.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 1 | Nfia | Club | 5.591073 |
| 39 | Aqp3 | Basal (Marker) | 7.844628 |
| 41 | Dapl1 | Basal (Marker) | 6.546861 |
| 46 | Cyp2f2 | Club (Marker) | 10.639600 |
| 48 | Cbr2 | Club (Marker) | 5.402642 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0019886 | antigen processing and presentation of e… | 14 | 4 | 0.08 | 7.9e-07 |
| GO:0002495 | antigen processing and presentation of p… | 18 | 4 | 0.10 | 2.4e-06 |
| GO:0002504 | antigen processing and presentation of p… | 19 | 4 | 0.10 | 3.0e-06 |
| GO:0002478 | antigen processing and presentation of e… | 21 | 4 | 0.11 | 4.6e-06 |
| GO:0019884 | antigen processing and presentation of e… | 26 | 4 | 0.14 | 1.1e-05 |
| GO:0006518 | peptide metabolic process | 682 | 14 | 3.72 | 1.8e-05 |
| GO:0043603 | cellular amide metabolic process | 802 | 14 | 4.37 | 0.00010 |
| GO:0048002 | antigen processing and presentation of p… | 49 | 4 | 0.27 | 0.00014 |
| GO:0009636 | response to toxic substance | 94 | 5 | 0.51 | 0.00016 |
| GO:0002250 | adaptive immune response | 301 | 8 | 1.64 | 0.00023 |
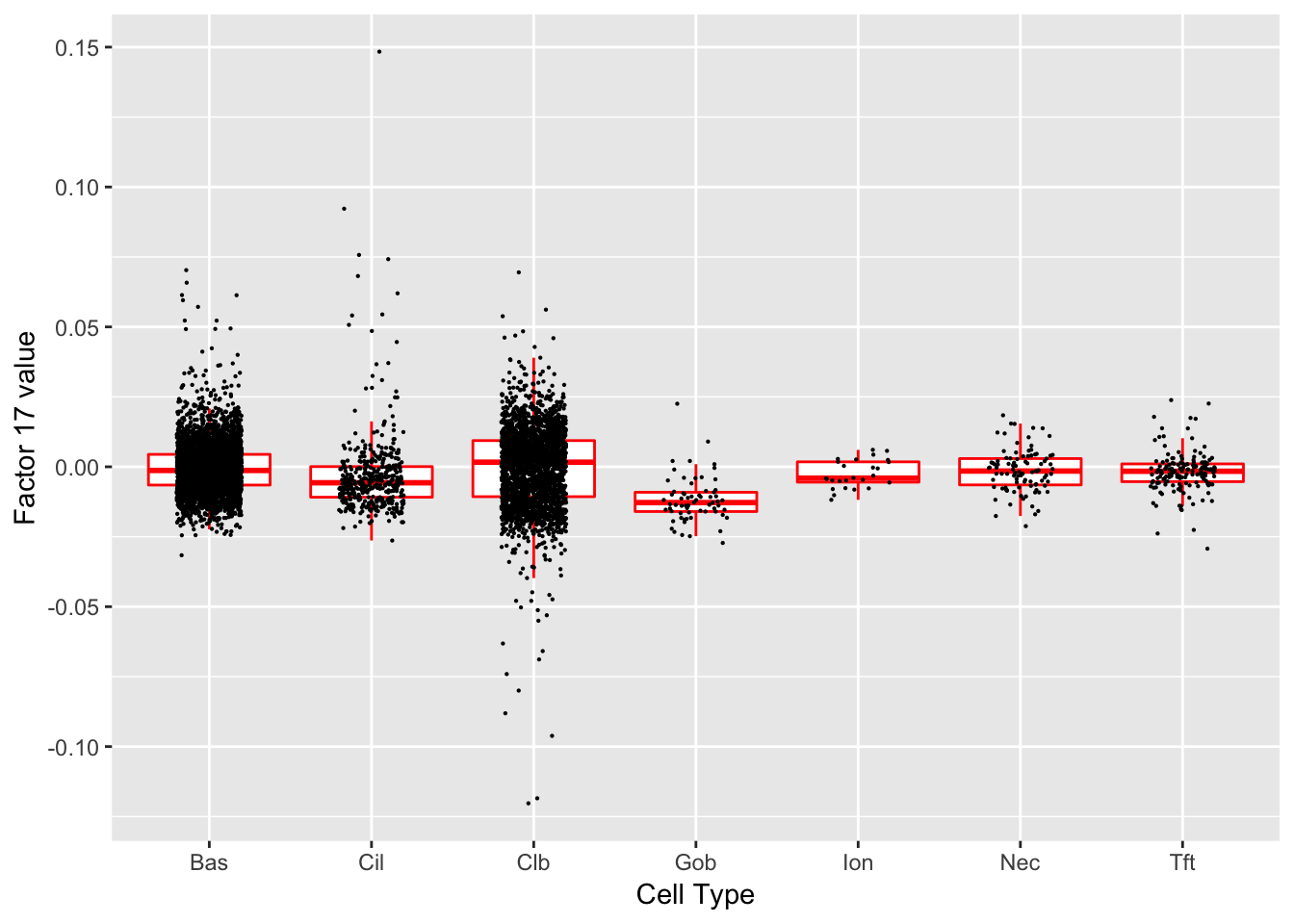
Factor 17 (PVE: 0.0003)
Expand here to see past versions of factors2-11.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0034213 | quinolinate catabolic process | 1 | 1 | 0.00 | 0.0012 |
| GO:0050976 | detection of mechanical stimulus involve… | 2 | 1 | 0.00 | 0.0023 |
| GO:1904106 | protein localization to microvillus | 2 | 1 | 0.00 | 0.0023 |
| GO:0023041 | neuronal signal transduction | 4 | 1 | 0.00 | 0.0046 |
| GO:0050975 | sensory perception of touch | 4 | 1 | 0.00 | 0.0046 |
| GO:0072526 | pyridine-containing compound catabolic p… | 4 | 1 | 0.00 | 0.0046 |
| GO:1904970 | brush border assembly | 4 | 1 | 0.00 | 0.0046 |
| GO:0002250 | adaptive immune response | 301 | 3 | 0.35 | 0.0047 |
| GO:0009582 | detection of abiotic stimulus | 91 | 2 | 0.11 | 0.0049 |
| GO:0009581 | detection of external stimulus | 92 | 2 | 0.11 | 0.0050 |
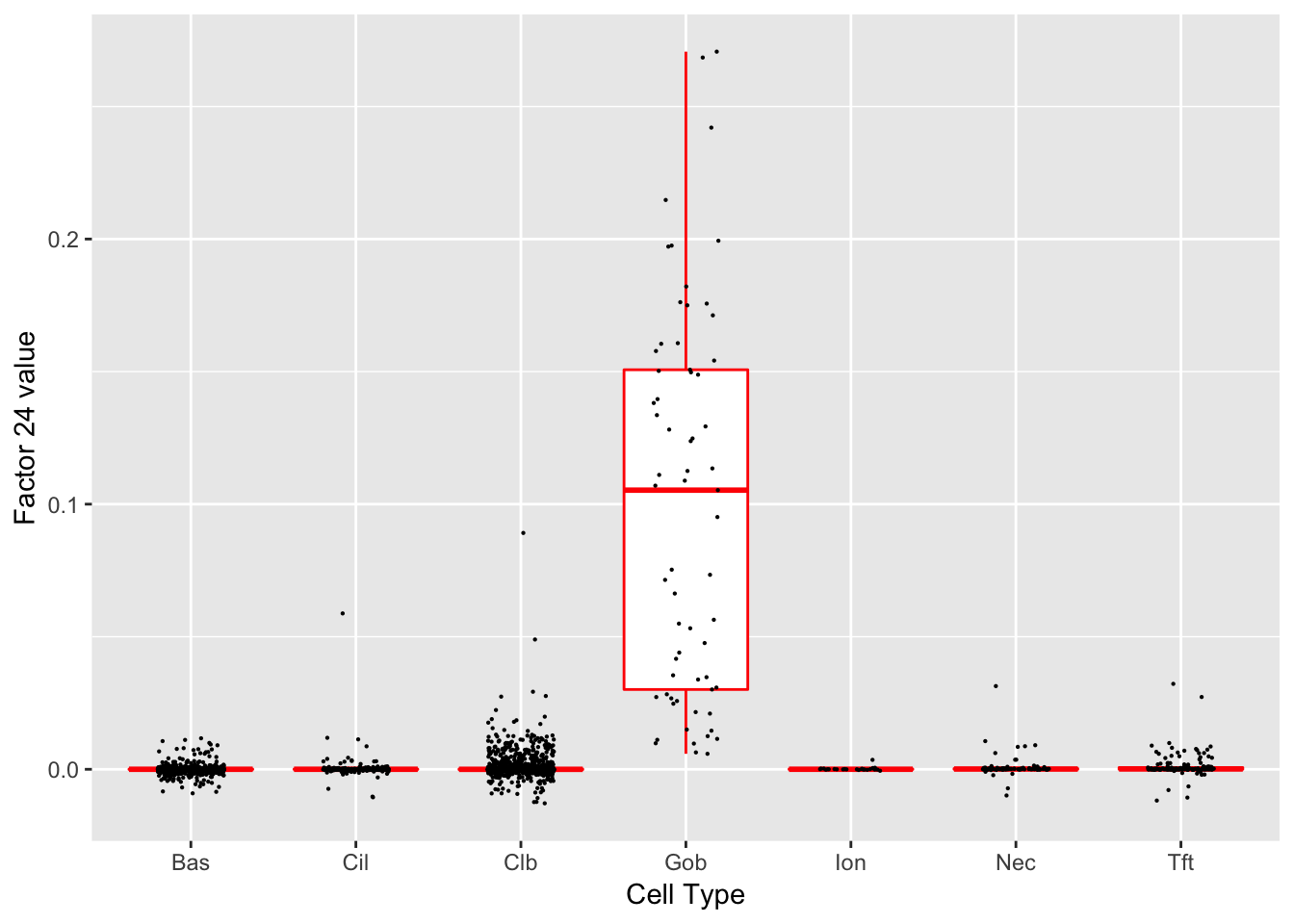
Factor 24 (PVE: 0.0006)
Expand here to see past versions of factors2-12.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 26 | Sox9 | Tuft-2 | 5.658985 |
| 27 | Gp2 | Goblet | 27.331516 |
| 28 | Tff1 | Goblet-1 | 13.907192 |
| 29 | Tff2 | Goblet-1 | 12.207553 |
| 30 | Lman1l | Goblet-1 | 22.949418 |
| 33 | Dcpp1 | Goblet-2 | 9.848094 |
| 34 | Dcpp2 | Goblet-2 | 10.711118 |
| 35 | Dcpp3 | Goblet-2 | 11.463935 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0019722 | calcium-mediated signaling | 125 | 6 | 0.67 | 5.4e-05 |
| GO:0006884 | cell volume homeostasis | 17 | 3 | 0.09 | 9.4e-05 |
| GO:0019932 | second-messenger-mediated signaling | 207 | 7 | 1.10 | 0.00012 |
| GO:0001580 | detection of chemical stimulus involved … | 19 | 3 | 0.10 | 0.00013 |
| GO:0050913 | sensory perception of bitter taste | 21 | 3 | 0.11 | 0.00018 |
| GO:0050912 | detection of chemical stimulus involved … | 23 | 3 | 0.12 | 0.00024 |
| GO:0030968 | endoplasmic reticulum unfolded protein r… | 60 | 4 | 0.32 | 0.00029 |
| GO:0050907 | detection of chemical stimulus involved … | 26 | 3 | 0.14 | 0.00035 |
| GO:0034620 | cellular response to unfolded protein | 66 | 4 | 0.35 | 0.00042 |
| GO:0032808 | lacrimal gland development | 7 | 2 | 0.04 | 0.00058 |
Factor 29 (PVE: 0.0001)
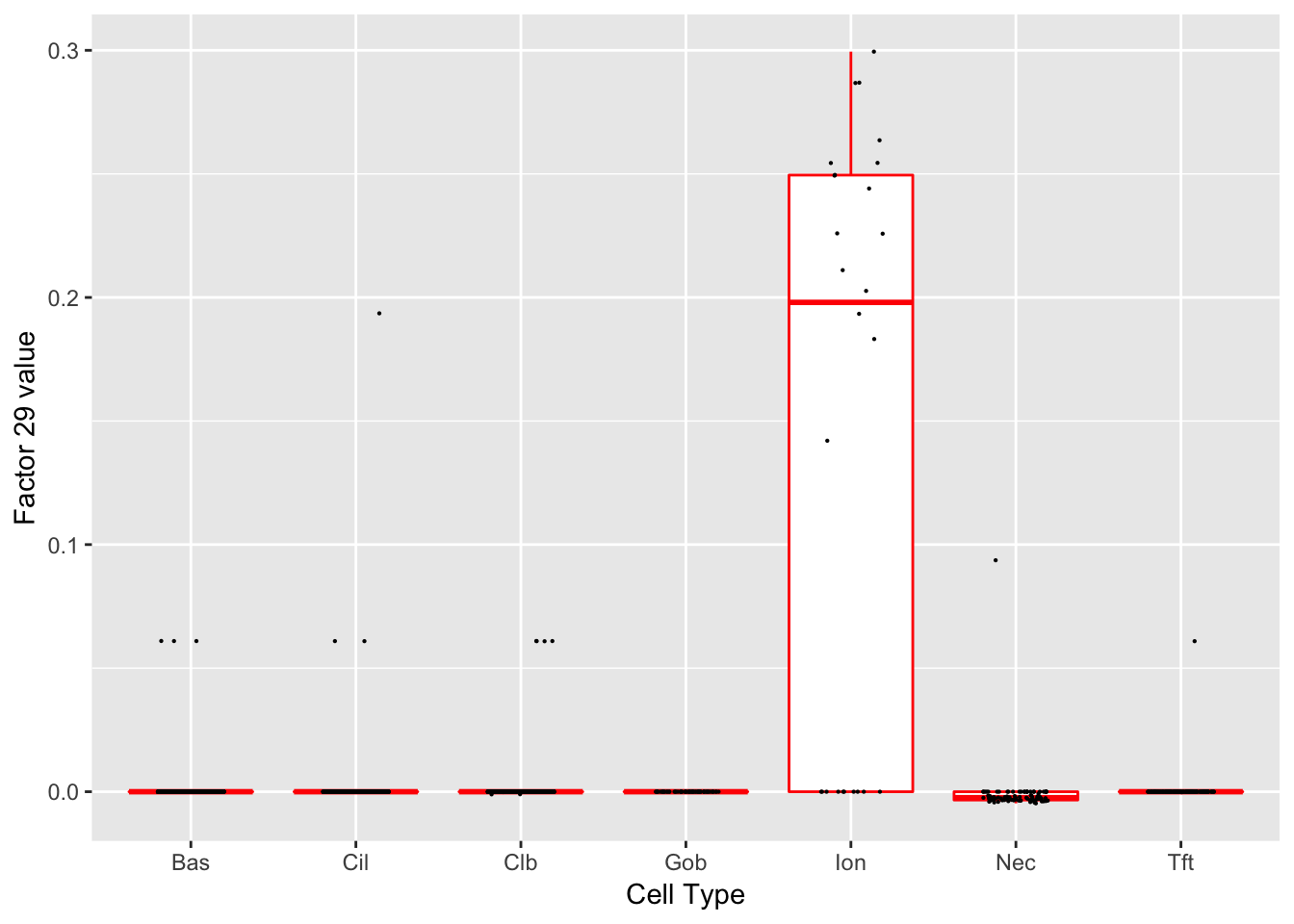
Expand here to see past versions of factors2-13.png:
| Version | Author | Date |
|---|---|---|
| f2b86c1 | Jason Willwerscheid | 2018-12-05 |
Significant genes that are also mentioned in the paper:
| gene | type | t.val | |
|---|---|---|---|
| 4 | Ascl3 | Ionocyte | 27.53504 |
| 37 | Atp6v0d2 | Ionocyte | 22.27301 |
| 38 | Cftr | Ionocyte | 17.94190 |
Gene Ontology terms:
| GO.ID | Term | Annotated | Significant | Expected | classic |
|---|---|---|---|---|---|
| GO:0007600 | sensory perception | 382 | 8 | 1.00 | 5.7e-06 |
| GO:0050877 | nervous system process | 698 | 10 | 1.84 | 9.6e-06 |
| GO:0055067 | monovalent inorganic cation homeostasis | 123 | 5 | 0.32 | 1.7e-05 |
| GO:0003008 | system process | 1209 | 12 | 3.18 | 4.3e-05 |
| GO:0006821 | chloride transport | 78 | 4 | 0.21 | 5.1e-05 |
| GO:0007191 | adenylate cyclase-activating dopamine re… | 5 | 2 | 0.01 | 6.7e-05 |
| GO:0030321 | transepithelial chloride transport | 5 | 2 | 0.01 | 6.7e-05 |
| GO:1901617 | organic hydroxy compound biosynthetic pr… | 166 | 5 | 0.44 | 7.1e-05 |
| GO:0030004 | cellular monovalent inorganic cation hom… | 91 | 4 | 0.24 | 9.3e-05 |
| GO:0042421 | norepinephrine biosynthetic process | 6 | 2 | 0.02 | 1e-04 |
Session information
sessionInfo()
#> R version 3.4.3 (2017-11-30)
#> Platform: x86_64-apple-darwin15.6.0 (64-bit)
#> Running under: macOS High Sierra 10.13.6
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] stats4 parallel stats graphics grDevices utils datasets
#> [8] methods base
#>
#> other attached packages:
#> [1] org.Mm.eg.db_3.5.0 topGO_2.30.1 SparseM_1.77
#> [4] GO.db_3.5.0 AnnotationDbi_1.40.0 IRanges_2.12.0
#> [7] S4Vectors_0.16.0 Biobase_2.38.0 graph_1.56.0
#> [10] BiocGenerics_0.24.0 ggplot2_3.1.0
#>
#> loaded via a namespace (and not attached):
#> [1] xfun_0.4 lattice_0.20-35 colorspace_1.3-2
#> [4] htmltools_0.3.6 yaml_2.2.0 blob_1.1.0
#> [7] rlang_0.3.0.1 R.oo_1.21.0 pillar_1.2.1
#> [10] glue_1.3.0 withr_2.1.2.9000 DBI_0.7
#> [13] R.utils_2.6.0 bit64_0.9-7 bindrcpp_0.2
#> [16] matrixStats_0.54.0 bindr_0.1 plyr_1.8.4
#> [19] stringr_1.3.1 munsell_0.5.0 gtable_0.2.0
#> [22] workflowr_1.0.1 flashier_0.1.0 R.methodsS3_1.7.1
#> [25] evaluate_0.12 memoise_1.1.0 labeling_0.3
#> [28] knitr_1.21.6 highr_0.7 Rcpp_1.0.0
#> [31] scales_1.0.0 backports_1.1.2 bit_1.1-12
#> [34] digest_0.6.18 stringi_1.2.4 dplyr_0.7.4
#> [37] grid_3.4.3 rprojroot_1.3-2 tools_3.4.3
#> [40] magrittr_1.5 lazyeval_0.2.1 tibble_1.4.2
#> [43] RSQLite_2.0 whisker_0.3-2 pkgconfig_2.0.1
#> [46] assertthat_0.2.0 rmarkdown_1.11 R6_2.3.0
#> [49] git2r_0.21.0 compiler_3.4.3This reproducible R Markdown analysis was created with workflowr 1.0.1