Backfits
Jason Willwerscheid
8/17/2019
Last updated: 2019-09-04
Checks: 6 0
Knit directory: scFLASH/
This reproducible R Markdown analysis was created with workflowr (version 1.2.0). The Report tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20181103) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/initialization/
Ignored: data/Ensembl2Reactome.txt
Ignored: data/droplet.rds
Ignored: data/mus_pathways.rds
Ignored: output/backfit/
Ignored: output/prior_type/
Ignored: output/pseudocount/
Ignored: output/pseudocount_redux/
Ignored: output/size_factors/
Ignored: output/var_type/
Untracked files:
Untracked: analysis/NBapprox.Rmd
Untracked: analysis/deleted.Rmd
Untracked: analysis/trachea4.Rmd
Untracked: code/missing_data.R
Untracked: code/pseudocount_redux/
Untracked: code/trachea4.R
Unstaged changes:
Modified: code/backfit/backfit_fits.R
Modified: code/sc_comparisons.R
Modified: code/utils.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bcfa0cc | Jason Willwerscheid | 2019-09-04 | wflow_publish(“analysis/backfit.Rmd”) |
| html | 851baf8 | Jason Willwerscheid | 2019-09-01 | Build site. |
| Rmd | fc78232 | Jason Willwerscheid | 2019-09-01 | wflow_publish(c(“analysis/backfit.Rmd”, “analysis/var_type.Rmd”)) |
| html | dbfc274 | Jason Willwerscheid | 2019-08-31 | Build site. |
| Rmd | b2a2d7a | Jason Willwerscheid | 2019-08-31 | wflow_publish(“analysis/backfit.Rmd”) |
| html | 4b18e3e | Jason Willwerscheid | 2019-08-17 | Build site. |
| Rmd | 2c164cb | Jason Willwerscheid | 2019-08-17 | wflow_publish(“analysis/backfit.Rmd”) |
Introduction
Although I’ve done much to speed up backfits, they remain slow relative to greedy fits. The question, then, is whether they’re worth the wait. Here, I’ll argue that while the quantitative improvements are relatively small — one can obtain a similar increase in ELBO in a fraction of the time by greedily adding a handful of additional factors — the qualitative improvements can be dramatic.
In this analysis, I take my preferred fit from a previous analysis of variance structures (the fit that “pre-scales” cells) and compare it against a backfitted version of the same. The code used to produce the fits can be viewed here.
source("./code/utils.R")
droplet <- readRDS("./data/droplet.rds")
droplet <- preprocess.droplet(droplet)
res <- readRDS("./output/backfit/backfit_fits.rds")
progress.g <- data.table::fread("./output/backfit/greedy_output.txt")
progress.b <- data.table::fread("./output/backfit/backfit_output.txt")Results: ELBO
As a quantitative measure of how much backfitting improves the fit, I plot the ELBO attained after each of the last eight greedy factors have been added and after each of the first 60 backfitting iterations (backfitting continues for a total of 100 iterations, but the ELBO remains mostly level after the first 50 or so iterations).
From the perspective of the ELBO, backfitting offers about as much improvement as the greedy addition of 5-6 factors. However, a single backfitting iteration takes almost 30 seconds, whereas the entire greedy fit can be performed in less than two minutes. If the prospect of an hours-long backfit is too daunting, then a partial backfit might be sufficient: most of the improvement occurs within the first ten backfitting iterations.
g.iter <- nrow(progress.g)
b.iter <- nrow(progress.b)
progress.g$elapsed.time <- (res$greedy$elapsed.time * 1:g.iter / g.iter) / 60
progress.b$elapsed.time <- (res$greedy$elapsed.time
+ res$backfit$elapsed.time * 1:b.iter / b.iter) / 60
# Only the final iteration for each greedy factor is needed.
progress.g <- subset(progress.g, c((progress.g$Factor[1:(nrow(progress.g) - 1)]
!= progress.g$Factor[2:nrow(progress.g)]), TRUE))
progress.df <- rbind(progress.g, progress.b)
ggplot(subset(progress.df, !(Factor %in% as.character(1:12)) & Iter < 61),
aes(x = elapsed.time, y = Obj, color = Type)) +
geom_point() +
labs(x = "elapsed time (min)", y = "ELBO (unadjusted)",
title = "Greedy factors 13-20 and first 60 backfitting iterations")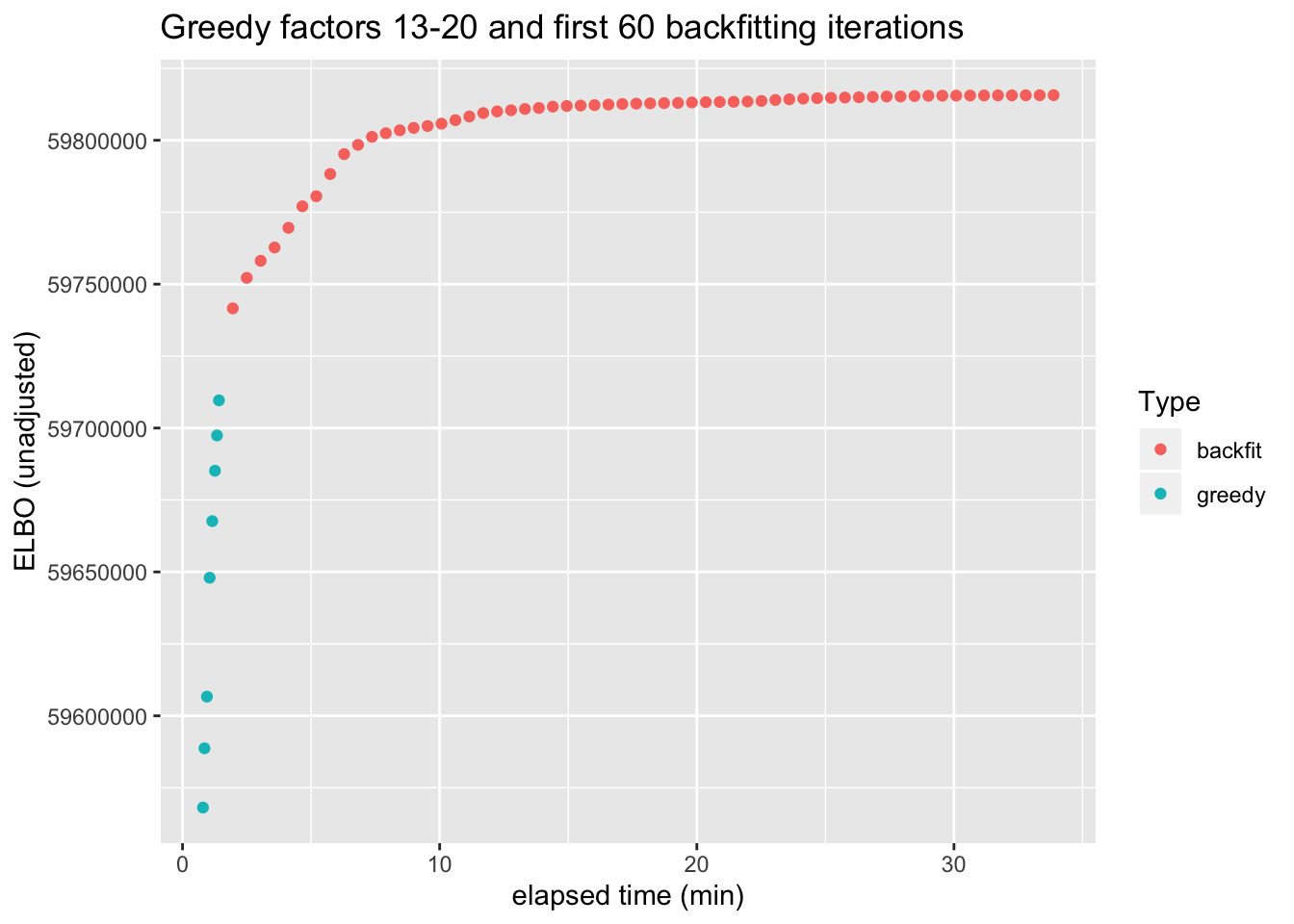
Results: Factor comparisons
The ELBO does not tell the whole story. Plotting the factors reveals clear improvements in the backfitted factors. Most obviously, factors 3, 6, 8, 11, 16, and 17 are much sparser and are more easily readable as cell-type specific factors. In addition, factor 2 more clearly indexes a basal-to-club gradation without ciliated cells mucking up the situation.
plot.factors(res$greedy, droplet$cell.type, kset = 1:20, title = "Greedy")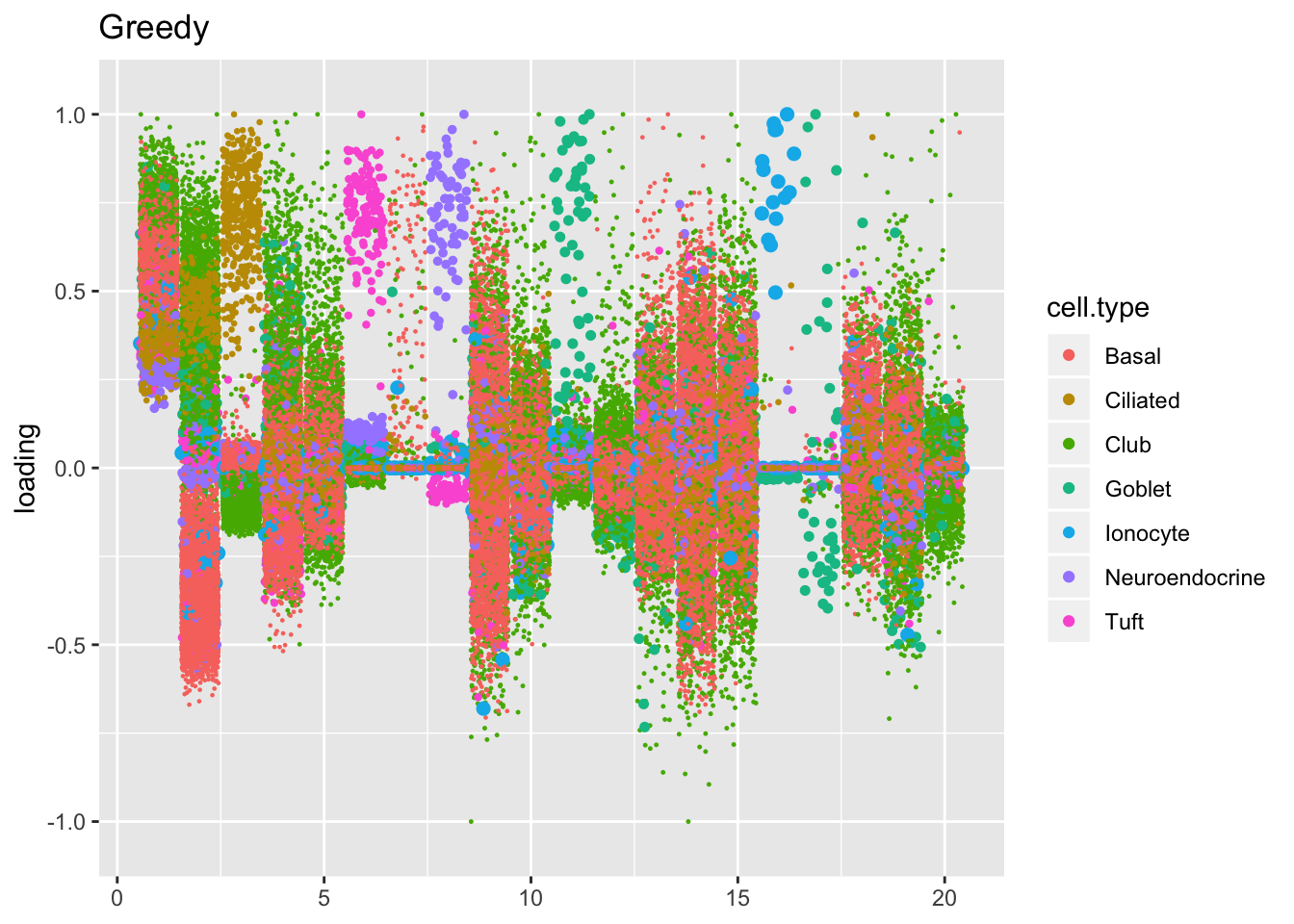
plot.factors(res$backfit, droplet$cell.type, kset = 1:20, title = "Backfit")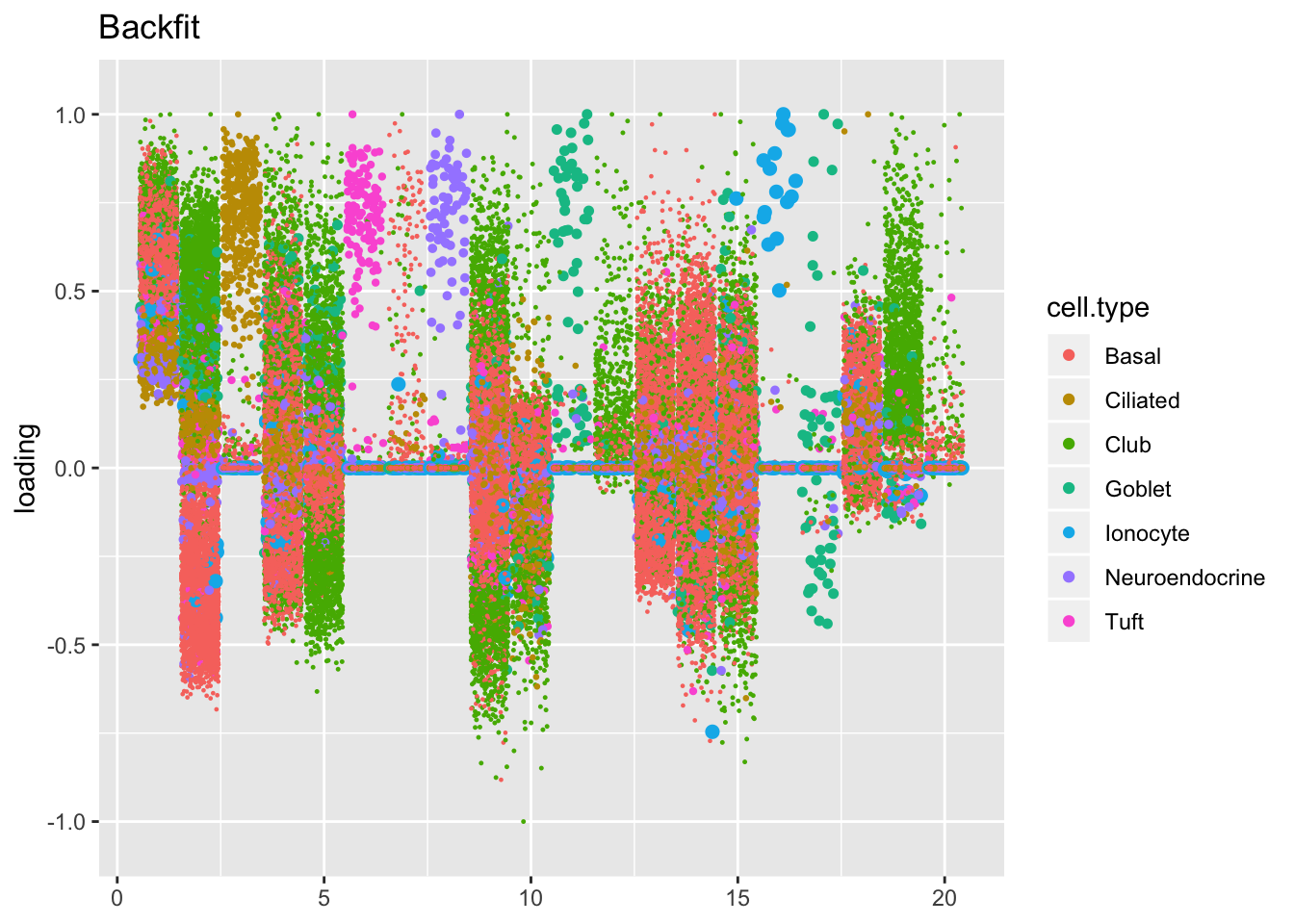
Factors above the dashed line in the plot below are factors that have increased in importance, while factors below the line have diminished in importance. Most interestingly, backfitting uncovers a large club-cell specific factor that is relatively unimportant in the greedy fit (factor 19).
pve.df <- data.frame(k = 1:res$greedy$fl$n.factors,
greedy = res$greedy$fl$pve,
backfit = res$backfit$fl$pve)
ggplot(pve.df, aes(x = greedy, y = backfit)) +
geom_text(label = pve.df$k, size = 3) +
geom_abline(slope = 1, linetype = "dashed") +
scale_x_log10() + scale_y_log10() +
labs(title = "Proportion of variance explained")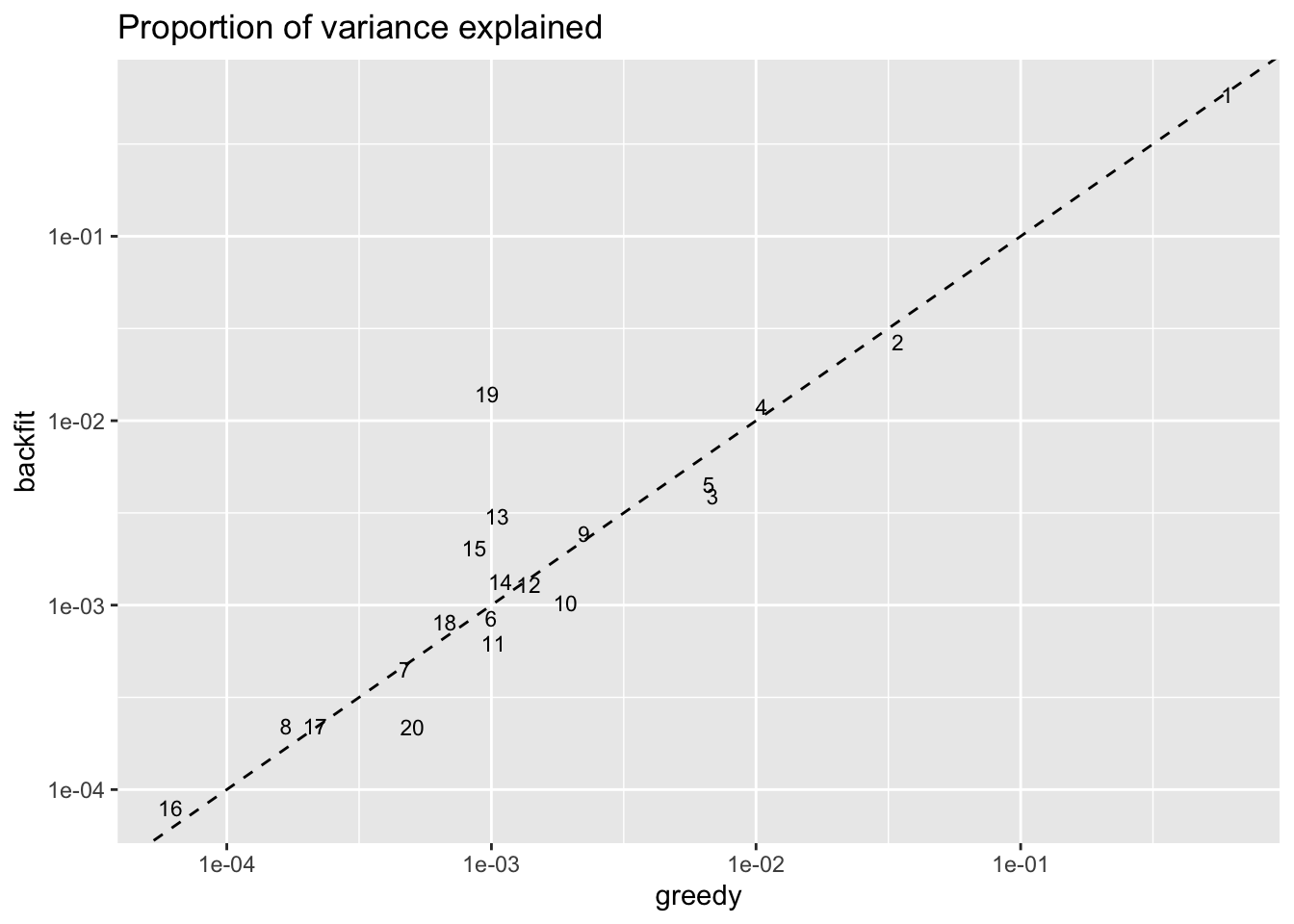
sessionInfo()R version 3.5.3 (2019-03-11)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] flashier_0.1.15 ggplot2_3.2.0 Matrix_1.2-15
loaded via a namespace (and not attached):
[1] Rcpp_1.0.1 plyr_1.8.4 compiler_3.5.3
[4] pillar_1.3.1 git2r_0.25.2 workflowr_1.2.0
[7] iterators_1.0.10 tools_3.5.3 digest_0.6.18
[10] evaluate_0.13 tibble_2.1.1 gtable_0.3.0
[13] lattice_0.20-38 pkgconfig_2.0.2 rlang_0.3.1
[16] foreach_1.4.4 parallel_3.5.3 yaml_2.2.0
[19] ebnm_0.1-24 xfun_0.6 withr_2.1.2
[22] stringr_1.4.0 dplyr_0.8.0.1 knitr_1.22
[25] fs_1.2.7 rprojroot_1.3-2 grid_3.5.3
[28] tidyselect_0.2.5 data.table_1.12.2 glue_1.3.1
[31] R6_2.4.0 rmarkdown_1.12 mixsqp_0.1-119
[34] reshape2_1.4.3 ashr_2.2-38 purrr_0.3.2
[37] magrittr_1.5 whisker_0.3-2 MASS_7.3-51.1
[40] codetools_0.2-16 backports_1.1.3 scales_1.0.0
[43] htmltools_0.3.6 assertthat_0.2.1 colorspace_1.4-1
[46] labeling_0.3 stringi_1.4.3 pscl_1.5.2
[49] doParallel_1.0.14 lazyeval_0.2.2 munsell_0.5.0
[52] truncnorm_1.0-8 SQUAREM_2017.10-1 crayon_1.3.4