model hic
Yunqi Yang
8/5/2020
Last updated: 2020-08-05
Checks: 6 1
Knit directory: gene_level_fine_mapping/
This reproducible R Markdown analysis was created with workflowr (version 1.6.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200622) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/nicholeyang/Desktop/gene_level_fine_mapping/data/train_add_hic.RData | data/train_add_hic.RData |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2bbd873. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Untracked files:
Untracked: data/hic_eqtl.RData
Untracked: data/train_add_hic.RData
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/model_hic.Rmd) and HTML (docs/model_hic.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2bbd873 | yunqiyang0215 | 2020-08-05 | wflow_publish(“analysis/model_hic.Rmd”) |
| html | addf17d | yunqiyang0215 | 2020-08-05 | Build site. |
| Rmd | 1622d52 | yunqiyang0215 | 2020-08-05 | wflow_publish(“analysis/model_hic.Rmd”) |
load("/Users/nicholeyang/Desktop/gene_level_fine_mapping/data/train_add_hic.RData")data summary
head(dat_add_hic) gene_name snp_loc38 variant_id UTR5 UTR3 exon intron
1 A1BG chr19:57866502 chr19_57866502_T_C_b38 0 0 0 0
2 A1BG chr19:58059544 chr19_58059544_T_G_b38 0 0 0 0
3 A1BG chr19:58170494 chr19_58170494_G_T_b38 0 0 0 0
4 A1BG chr19:58228973 chr19_58228973_T_G_b38 0 0 0 0
5 A1BG chr19:58330182 chr19_58330182_C_T_b38 0 0 0 0
6 A1BG chr19:58359927 chr19_58359927_G_A_b38 0 0 0 0
upstream tss_dist_to_snp y Mon Mac0 Mac1 Mac2 Neu MK EP Ery FoeT nCD4 tCD4
1 0 481132 0 0 0 0 0 0 0 0 0 0 0 0
2 0 288090 0 0 0 0 0 0 0 0 0 0 0 0
3 0 177140 0 0 0 0 0 0 0 0 0 0 0 0
4 0 118661 0 0 0 0 0 0 0 0 0 0 0 0
5 0 17452 1 0 0 0 0 0 0 0 0 0 0 0
6 0 6428 1 0 0 0 0 0 0 0 0 0 0 0
aCD4 naCD4 nCD8 tCD8 nB tB
1 0 0 0 0 0 0
2 0 0 0 0 0 0
3 0 0 0 0 0 0
4 0 0 0 0 0 0
5 0 0 0 0 0 0
6 0 0 0 0 0 0hist(dat_add_hic$tss_dist_to_snp)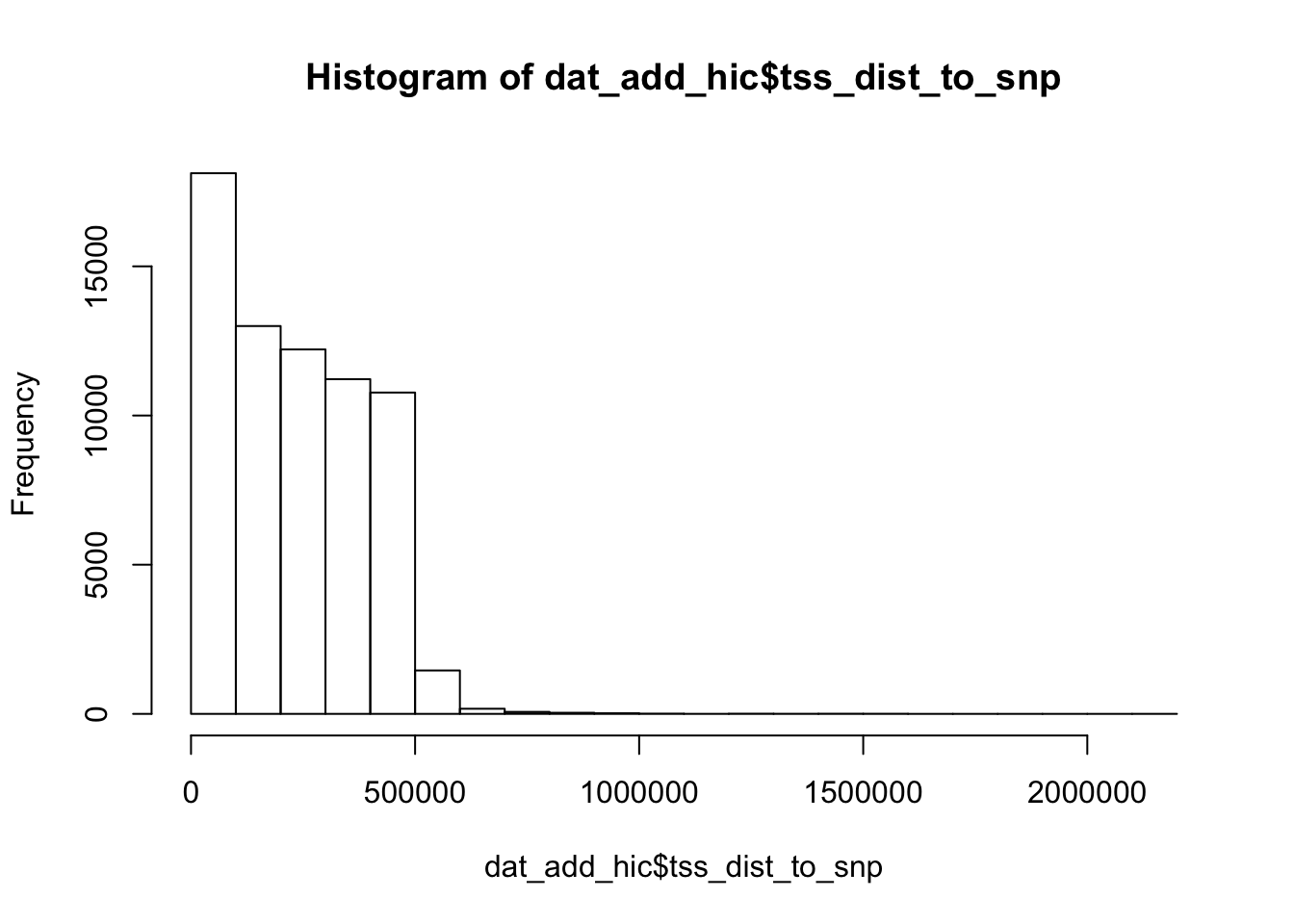
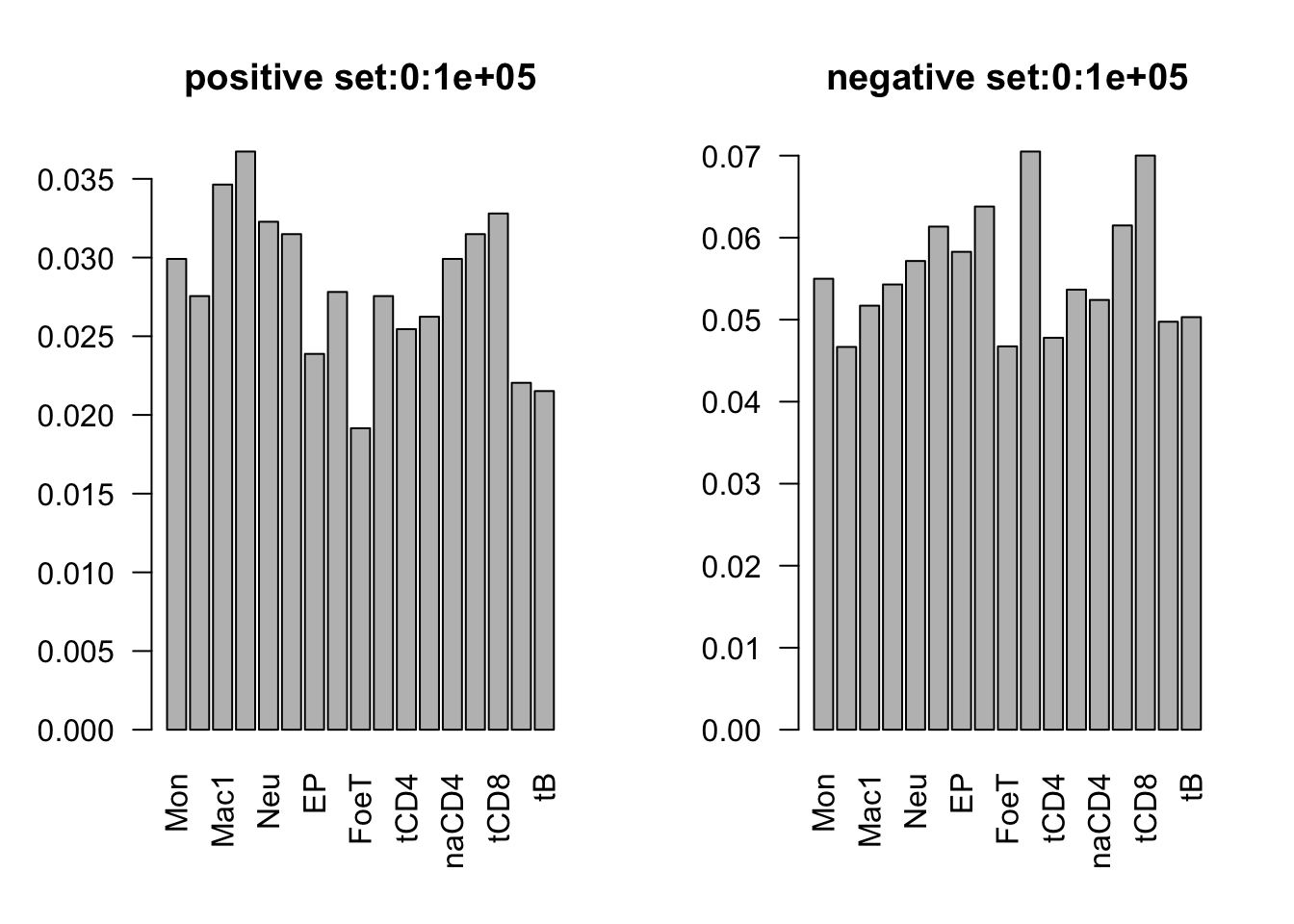
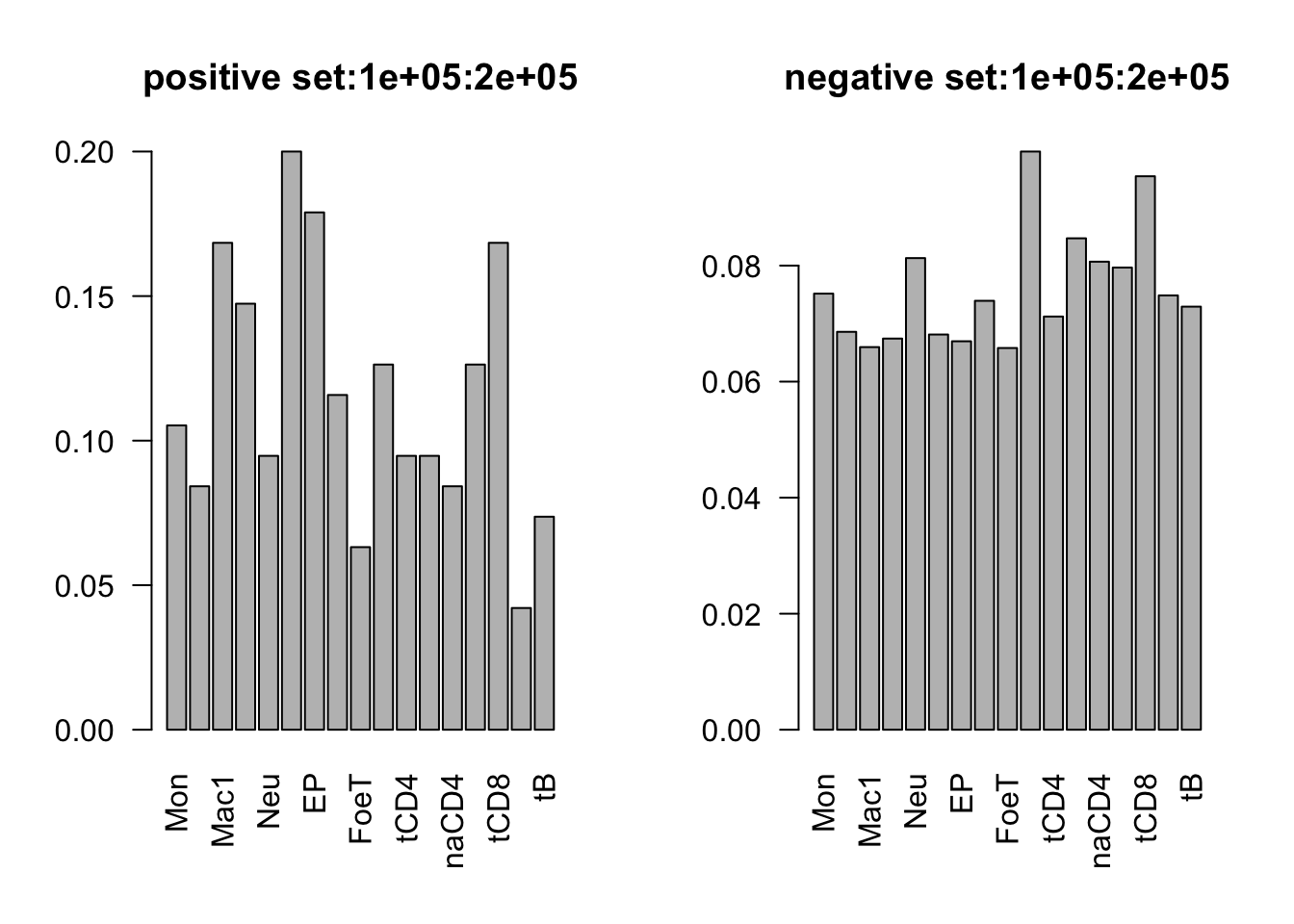
range(dat_add_hic$tss_dist_to_snp)[1] 0 2176373plot the proportion of features in the positive/negative set
dist_range = c(0, 1e5, 2e5, 3e5, 4e5, 5e5, 3e6)
for (i in 1:6){
par(mfrow = c(1,2))
sub_dat = dat_add_hic[dat_add_hic$tss_dist_to_snp %in% seq(dist_range[i], dist_range[i+1], by = 1), ]
# proportion of features in the positive set
tot_count = apply(sub_dat[sub_dat$y==1, c(11:27)], 2, function(x) sum(x == 1))
barplot(tot_count/dim(sub_dat[sub_dat$y==1, ])[1], las = 2, main = paste('positive set', dist_range[i], dist_range[i+1], sep = ':'))
# proportion of features in the negative set
tot_count = apply(sub_dat[sub_dat$y==0, c(11:27)], 2, function(x) sum(x == 1))
barplot(tot_count/dim(sub_dat[sub_dat$y==0, ])[1], las = 2,
main = paste('negative set', dist_range[i], dist_range[i+1], sep = ':'))
}
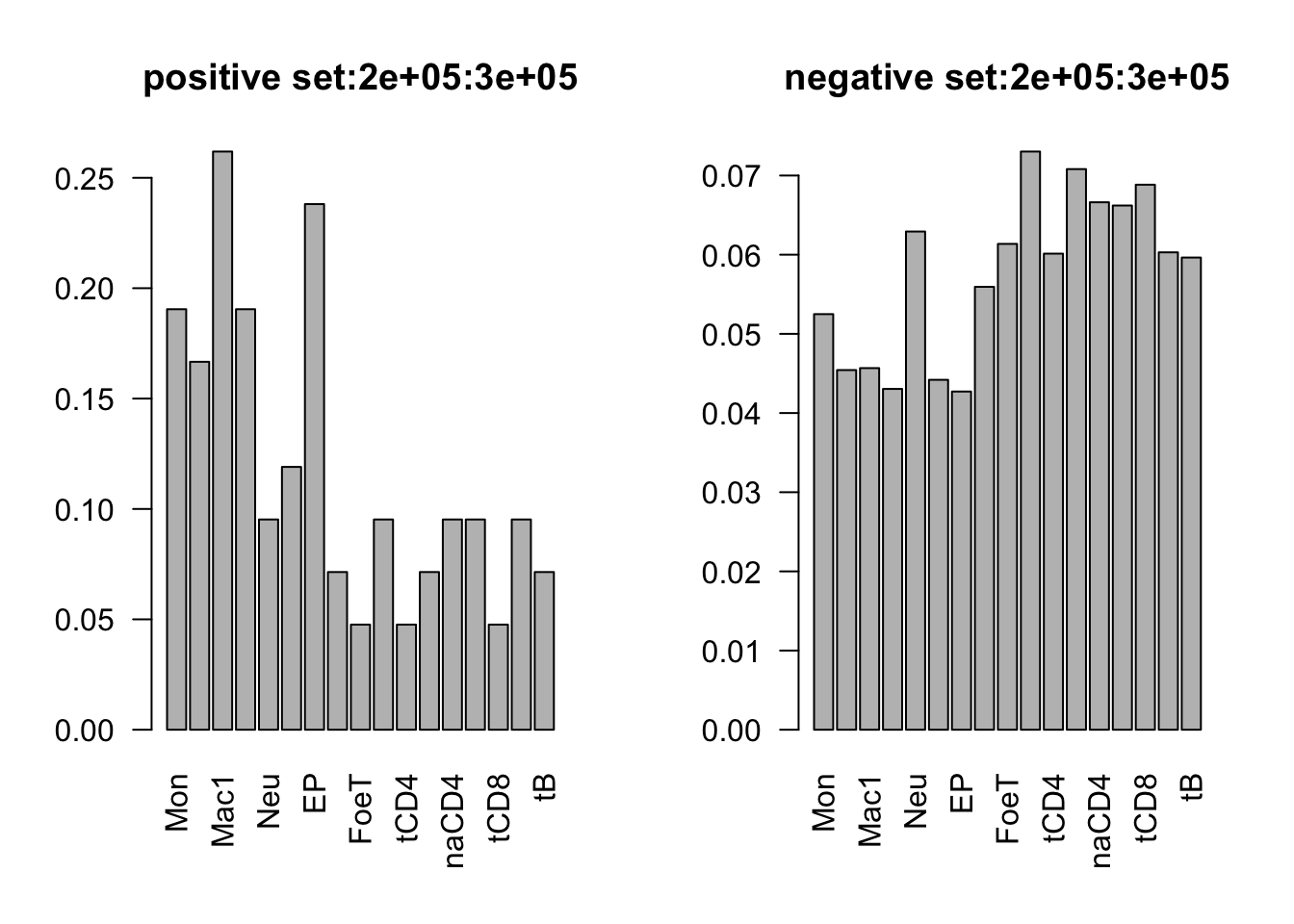
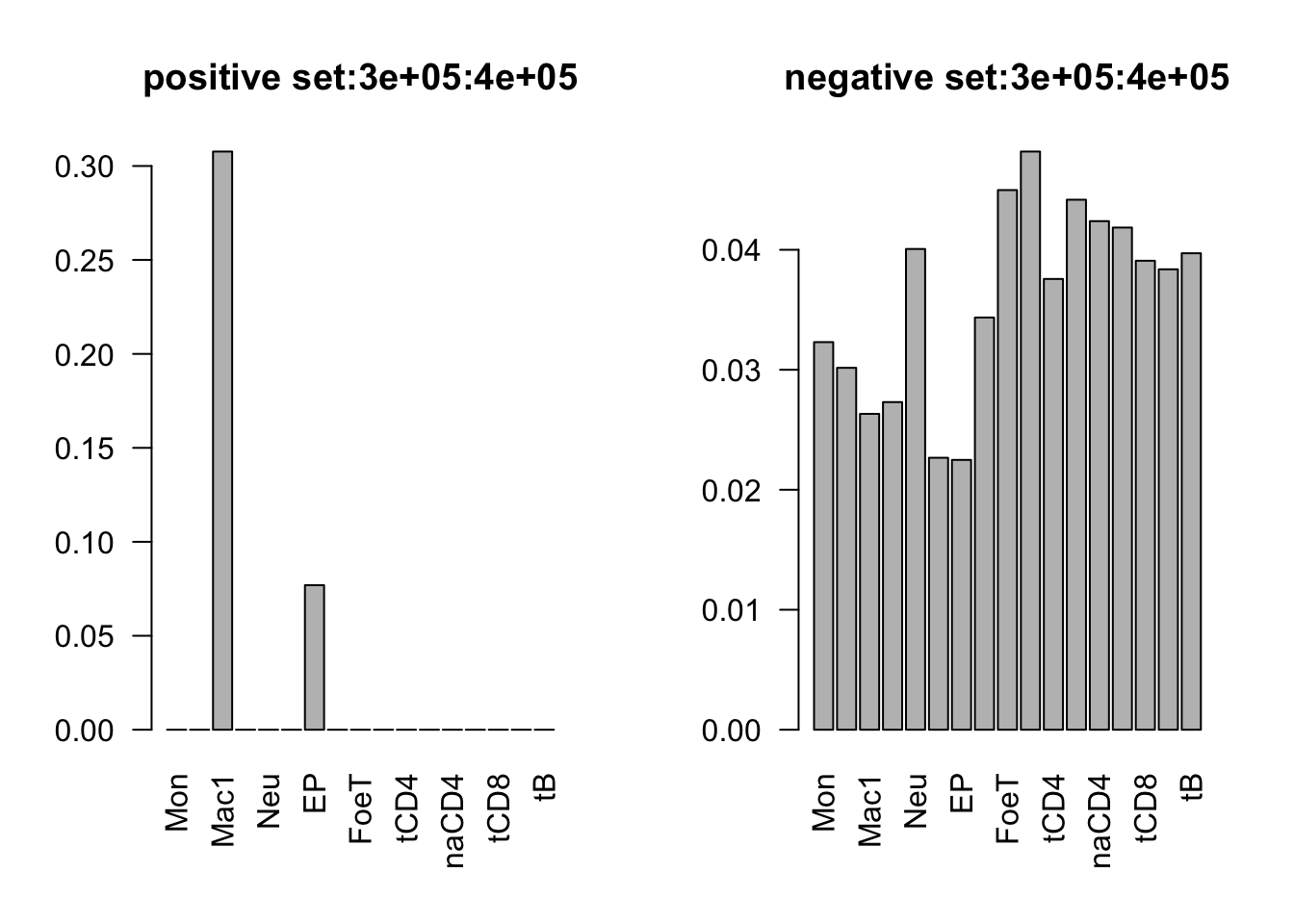
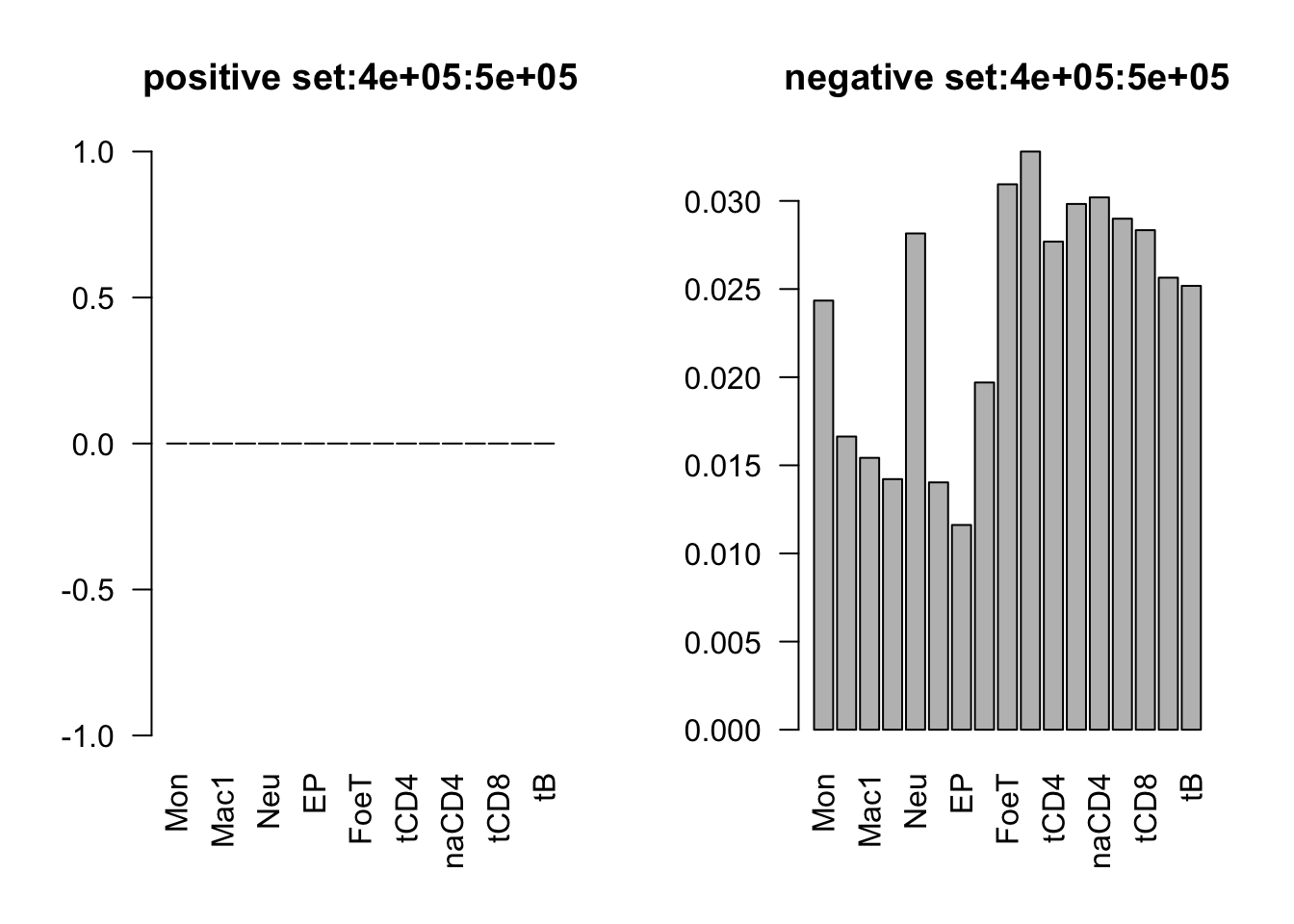
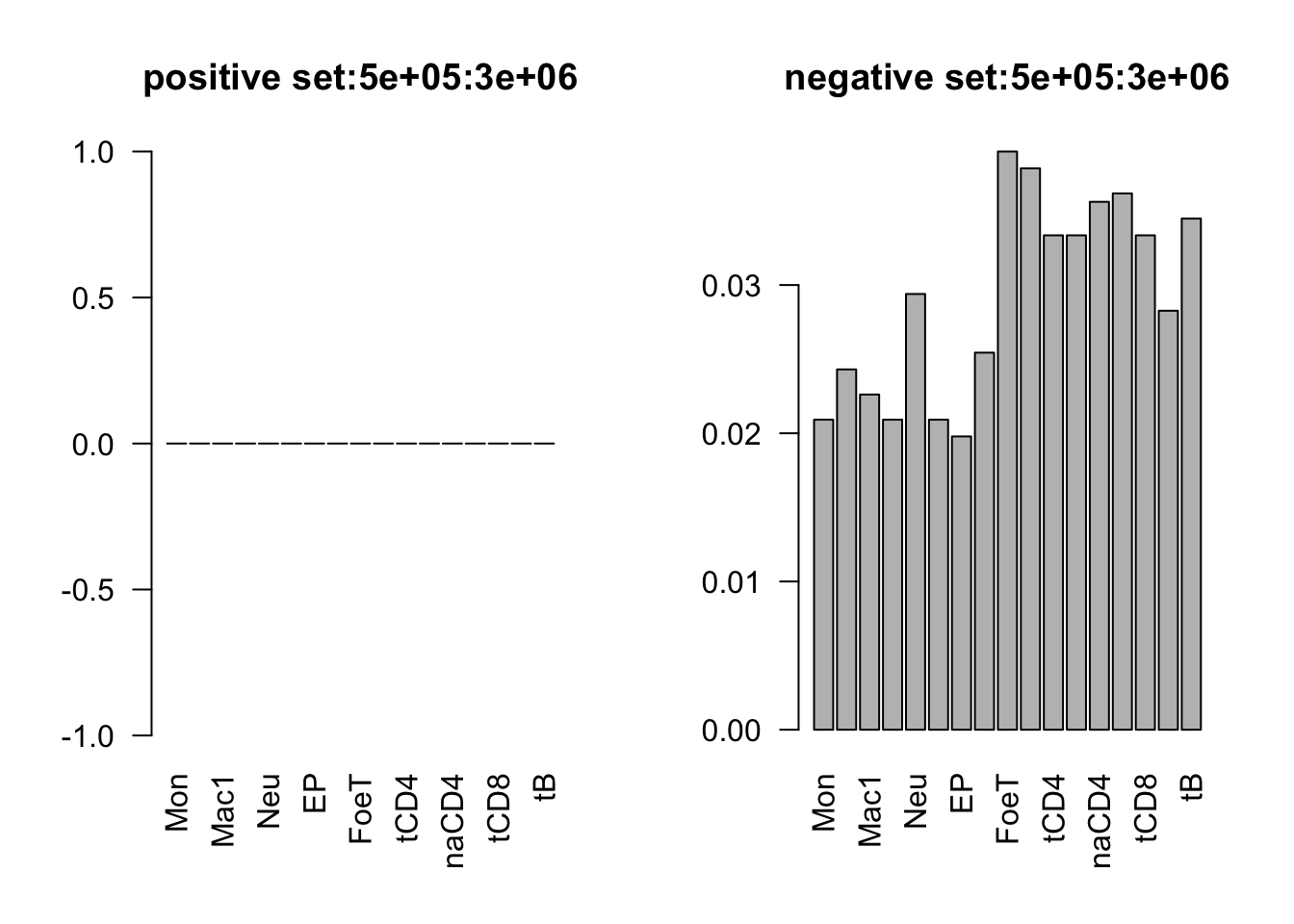
by varaible logistic regression
only Mac1 and Mac2 features are not significant
for (i in 11: 27){
fit = glm( dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
print(summary(fit))
}
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3524 -0.3524 -0.3524 -0.3524 2.5238
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.74794 0.01663 -165.252 < 2e-16 ***
dat_add_hic[, i] -0.39445 0.09042 -4.362 1.29e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30185 on 67104 degrees of freedom
AIC: 30189
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3518 -0.3518 -0.3518 -0.3518 2.5079
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.75131 0.01660 -165.729 < 2e-16 ***
dat_add_hic[, i] -0.34953 0.09477 -3.688 0.000226 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30191 on 67104 degrees of freedom
AIC: 30195
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3499 -0.3499 -0.3499 -0.3499 2.3864
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.76281 0.01669 -165.549 <2e-16 ***
dat_add_hic[, i] -0.02499 0.08241 -0.303 0.762
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30206 on 67104 degrees of freedom
AIC: 30210
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.350 -0.350 -0.350 -0.350 2.392
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.76224 0.01669 -165.534 <2e-16 ***
dat_add_hic[, i] -0.03887 0.08262 -0.471 0.638
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30206 on 67104 degrees of freedom
AIC: 30210
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3533 -0.3533 -0.3533 -0.3533 2.5568
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.74263 0.01664 -164.820 < 2e-16 ***
dat_add_hic[, i] -0.48706 0.08897 -5.475 4.38e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30172 on 67104 degrees of freedom
AIC: 30176
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.351 -0.351 -0.351 -0.351 2.448
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.75638 0.01665 -165.542 <2e-16 ***
dat_add_hic[, i] -0.18843 0.08710 -2.163 0.0305 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30201 on 67104 degrees of freedom
AIC: 30205
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3518 -0.3518 -0.3518 -0.3518 2.5059
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.75165 0.01660 -165.773 < 2e-16 ***
dat_add_hic[, i] -0.34396 0.09517 -3.614 0.000301 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30192 on 67104 degrees of freedom
AIC: 30196
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3533 -0.3533 -0.3533 -0.3533 2.5757
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.74261 0.01661 -165.162 < 2e-16 ***
dat_add_hic[, i] -0.53767 0.09441 -5.695 1.24e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30168 on 67104 degrees of freedom
AIC: 30172
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.355 -0.355 -0.355 -0.355 2.718
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.73287 0.01653 -165.354 <2e-16 ***
dat_add_hic[, i] -0.93448 0.11373 -8.217 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30115 on 67104 degrees of freedom
AIC: 30119
Number of Fisher Scoring iterations: 6
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.356 -0.356 -0.356 -0.356 2.669
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.72682 0.01662 -164.109 <2e-16 ***
dat_add_hic[, i] -0.80680 0.09371 -8.609 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30111 on 67104 degrees of freedom
AIC: 30115
Number of Fisher Scoring iterations: 6
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3536 -0.3536 -0.3536 -0.3536 2.6059
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.74079 0.01658 -165.299 <2e-16 ***
dat_add_hic[, i] -0.62042 0.09920 -6.254 4e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30159 on 67104 degrees of freedom
AIC: 30163
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3548 -0.3548 -0.3548 -0.3548 2.6459
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.73371 0.01659 -164.750 < 2e-16 ***
dat_add_hic[, i] -0.73593 0.09737 -7.558 4.1e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30135 on 67104 degrees of freedom
AIC: 30139
Number of Fisher Scoring iterations: 6
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3538 -0.3538 -0.3538 -0.3538 2.5884
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.73957 0.01662 -164.835 < 2e-16 ***
dat_add_hic[, i] -0.57462 0.09215 -6.236 4.5e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30160 on 67104 degrees of freedom
AIC: 30164
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3537 -0.3537 -0.3537 -0.3537 2.5699
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.74059 0.01664 -164.687 < 2e-16 ***
dat_add_hic[, i] -0.52399 0.08890 -5.894 3.77e-09 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30166 on 67104 degrees of freedom
AIC: 30170
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3543 -0.3543 -0.3543 -0.3543 2.5827
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.73713 0.01666 -164.312 < 2e-16 ***
dat_add_hic[, i] -0.56181 0.08672 -6.478 9.28e-11 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30157 on 67104 degrees of freedom
AIC: 30161
Number of Fisher Scoring iterations: 5
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3545 -0.3545 -0.3545 -0.3545 2.6712
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.73575 0.01655 -165.309 < 2e-16 ***
dat_add_hic[, i] -0.80332 0.10705 -7.504 6.17e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30134 on 67104 degrees of freedom
AIC: 30138
Number of Fisher Scoring iterations: 6
Call:
glm(formula = dat_add_hic$y ~ dat_add_hic[, i], family = "binomial")
Deviance Residuals:
Min 1Q Median 3Q Max
-0.3545 -0.3545 -0.3545 -0.3545 2.6708
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.73582 0.01655 -165.313 < 2e-16 ***
dat_add_hic[, i] -0.80199 0.10705 -7.492 6.79e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30206 on 67105 degrees of freedom
Residual deviance: 30134 on 67104 degrees of freedom
AIC: 30138
Number of Fisher Scoring iterations: 6
sessionInfo()R version 3.6.3 (2020-02-29)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.5
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_1.6.1
loaded via a namespace (and not attached):
[1] Rcpp_1.0.4 rprojroot_1.3-2 digest_0.6.25 later_1.0.0
[5] R6_2.4.1 backports_1.1.5 git2r_0.26.1 magrittr_1.5
[9] evaluate_0.14 highr_0.8 stringi_1.4.6 rlang_0.4.5
[13] fs_1.3.2 promises_1.1.0 whisker_0.4 rmarkdown_2.1
[17] tools_3.6.3 stringr_1.4.0 glue_1.3.2 httpuv_1.5.2
[21] xfun_0.12 yaml_2.2.1 compiler_3.6.3 htmltools_0.4.0
[25] knitr_1.28