survival_example
Yunqi Yang
2/1/2023
Last updated: 2023-02-01
Checks: 7 0
Knit directory: survival-susie/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230201) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 12b150a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rproj.user/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/survival_example.Rmd) and HTML (docs/survival_example.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 12b150a | yunqiyang0215 | 2023-02-01 | wflow_publish("analysis/survival_example.Rmd") |
Description: run a survival analysis example from:
https://rstudio-pubs-static.s3.amazonaws.com/5896_8f0fed2ccbbd42489276e554a05af87e.html
Data description: NCCTG Lung Cancer Data
Description: Survival in patients with advanced lung cancer from the North Central Cancer Treatment Group. Performance scores rate how well the patient can perform usual daily activities.
inst: Institution code time: Survival time in days status: censoring status 1=censored, 2=dead age: Age in years sex: Male=1 Female=2 ph.ecog: ECOG performance score (0=good 5=dead) ph.karno: Karnofsky performance score (bad=0-good=100) rated by physician pat.karno: Karnofsky performance score as rated by patient meal.cal: Calories consumed at meals wt.loss: Weight loss in last six months
## Load survival package
library(survival)
## List datasets in survival package
# data(package = "survival")
## Load lung data
data(lung)Warning in data(lung): data set 'lung' not foundhead(lung) inst time status age sex ph.ecog ph.karno pat.karno meal.cal wt.loss
1 3 306 2 74 1 1 90 100 1175 NA
2 3 455 2 68 1 0 90 90 1225 15
3 3 1010 1 56 1 0 90 90 NA 15
4 5 210 2 57 1 1 90 60 1150 11
5 1 883 2 60 1 0 100 90 NA 0
6 12 1022 1 74 1 1 50 80 513 0Create a survival object
## Add survival object. status == 2 is death
lung$SurvObj <- with(lung, Surv(time, status == 2))
## Check data
head(lung) inst time status age sex ph.ecog ph.karno pat.karno meal.cal wt.loss SurvObj
1 3 306 2 74 1 1 90 100 1175 NA 306
2 3 455 2 68 1 0 90 90 1225 15 455
3 3 1010 1 56 1 0 90 90 NA 15 1010+
4 5 210 2 57 1 1 90 60 1150 11 210
5 1 883 2 60 1 0 100 90 NA 0 883
6 12 1022 1 74 1 1 50 80 513 0 1022+Cox regression using coxph
## Fit Cox regression: age, sex, Karnofsky performance score, wt loss
res.cox1 <- coxph(SurvObj ~ age + sex + ph.karno + wt.loss, data = lung)
res.cox1Call:
coxph(formula = SurvObj ~ age + sex + ph.karno + wt.loss, data = lung)
coef exp(coef) se(coef) z p
age 0.015140 1.015255 0.009837 1.539 0.12379
sex -0.513955 0.598125 0.174410 -2.947 0.00321
ph.karno -0.012871 0.987211 0.006184 -2.081 0.03741
wt.loss -0.002246 0.997757 0.006357 -0.353 0.72389
Likelihood ratio test=18.84 on 4 df, p=0.0008436
n= 214, number of events= 152
(14 observations deleted due to missingness)## Check for violation of proportional hazard (constant HR over time)
(res.zph1 <- cox.zph(res.cox1)) chisq df p
age 0.90304 1 0.3420
sex 3.24299 1 0.0717
ph.karno 7.98433 1 0.0047
wt.loss 0.00382 1 0.9507
GLOBAL 10.73439 4 0.0297## Displays a graph of the scaled Schoenfeld residuals, along with a smooth curve.
par(mfrow = c(2,2))
plot(res.zph1)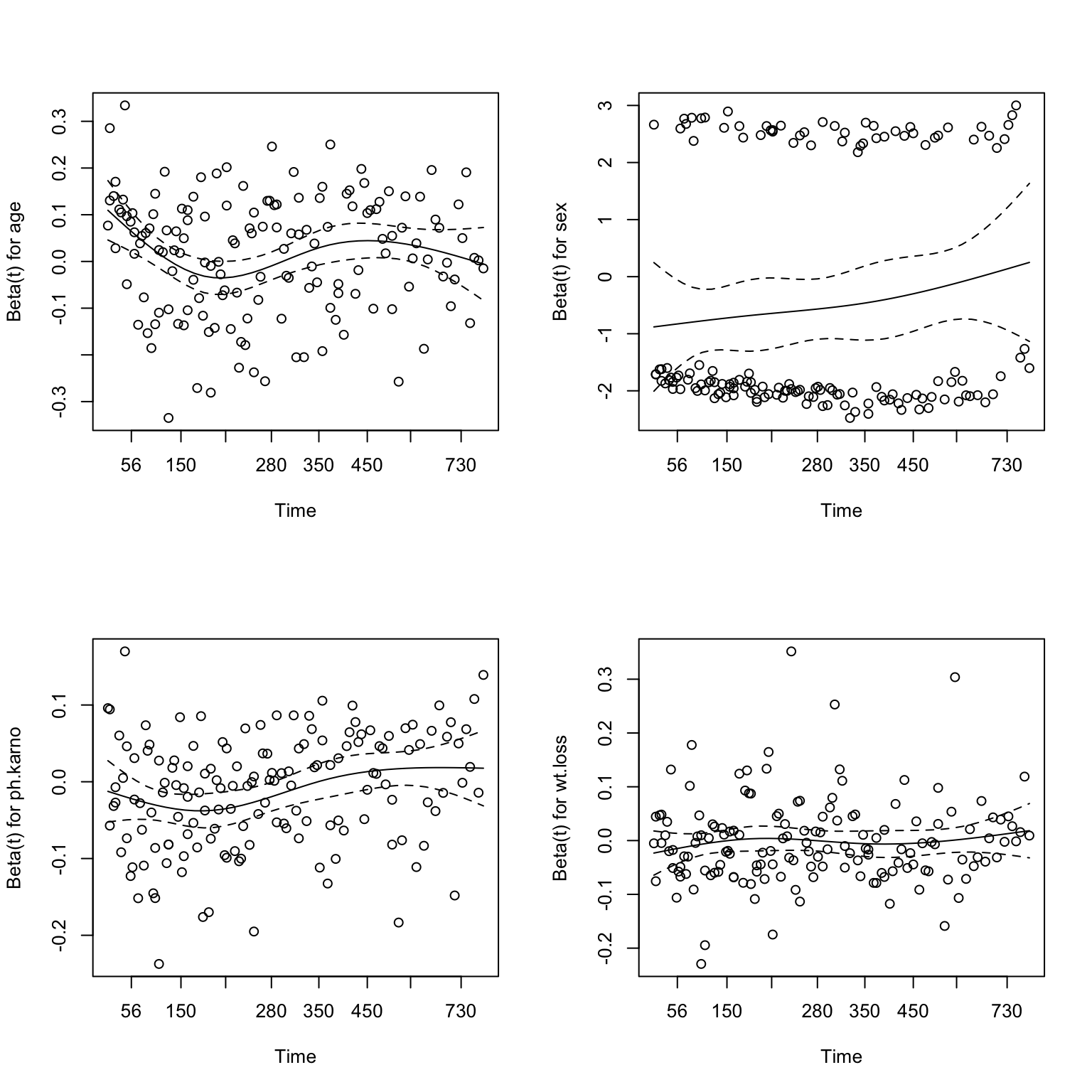
sessionInfo()R version 4.1.1 (2021-08-10)
Platform: x86_64-apple-darwin20.6.0 (64-bit)
Running under: macOS Monterey 12.0.1
Matrix products: default
BLAS: /usr/local/Cellar/openblas/0.3.18/lib/libopenblasp-r0.3.18.dylib
LAPACK: /usr/local/Cellar/r/4.1.1_1/lib/R/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] survival_3.2-11 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.8.3 highr_0.9 pillar_1.6.4 compiler_4.1.1
[5] bslib_0.4.1 later_1.3.0 jquerylib_0.1.4 git2r_0.28.0
[9] tools_4.1.1 digest_0.6.28 lattice_0.20-44 jsonlite_1.7.2
[13] evaluate_0.14 lifecycle_1.0.1 tibble_3.1.5 pkgconfig_2.0.3
[17] rlang_1.0.6 Matrix_1.5-3 cli_3.1.0 rstudioapi_0.13
[21] yaml_2.2.1 xfun_0.27 fastmap_1.1.0 stringr_1.4.0
[25] knitr_1.36 fs_1.5.0 vctrs_0.3.8 sass_0.4.4
[29] grid_4.1.1 rprojroot_2.0.2 glue_1.4.2 R6_2.5.1
[33] fansi_0.5.0 rmarkdown_2.11 magrittr_2.0.1 whisker_0.4
[37] splines_4.1.1 promises_1.2.0.1 ellipsis_0.3.2 htmltools_0.5.2
[41] httpuv_1.6.3 utf8_1.2.2 stringi_1.7.5 cachem_1.0.6
[45] crayon_1.4.1