cone_pmf1
zihao12
2021-06-08
Last updated: 2021-06-08
Checks: 7 0
Knit directory: ebpmf_data_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200511) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version fa23ffb. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/ebpmf_bg_tutorial_cache/
Ignored: analysis/ebpmf_wbg_model_intro_cache/
Ignored: analysis/ebpmf_wbg_simulate_big_data2_cache/
Ignored: analysis/ebpmf_wbg_simulate_big_data3_cache/
Ignored: analysis/ebpmf_wbg_simulate_big_data_cache/
Ignored: analysis/ebpmf_wbg_simulation_cache/
Ignored: analysis/investigate_np_ebpmf_wbg_cache/
Ignored: analysis/pmf_greedy_experiment_cache/
Ignored: analysis/sla_data_analysis_k10_cache/
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Ignored: output/News/.DS_Store
Ignored: topicView-app/.DS_Store
Untracked files:
Untracked: analysis/draft.Rmd
Untracked: analysis/draft2.Rmd
Untracked: analysis/ebpmf_wbg_simulate_correlated.Rmd
Untracked: analysis/ebpmf_wbg_simulation_big.Rmd
Untracked: analysis/ebpmf_wbg_simulation_big2_more.Rmd
Untracked: analysis/heatmap.Rmd
Untracked: analysis/investigate_largeK.Rmd
Untracked: analysis/investigate_news_topics.Rmd
Untracked: analysis/poissonmix_vs_pmf.Rmd
Untracked: analysis/simulate_data_bg.Rmd
Untracked: analysis/simulate_data_bg2.Rmd
Untracked: analysis/summary_sla_news_nips.Rmd
Untracked: analysis/test.R
Untracked: data/cell_data.csv
Untracked: data/sim/init_random.sim_bg_block_n1100_p2100_K50.Rds
Untracked: data/sim/simulated_data_small.RData
Untracked: data/sim/simulated_data_small2.RData
Untracked: output/poissonmix_vs_pmf.RDS
Untracked: output/poissonmix_vs_pmf.RData
Untracked: output/sim/v0.4.5/exper2/init_random.sim_bg_block_n1100_p2100_K50.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter100_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter10_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter10_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter10_init_random2.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter10_pmf_bg_K50_maxiter10_from_truth_scaled.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter10_pmf_bg_K50_maxiter10_from_truth_scaled0.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter10_pmf_bg_K50_maxiter10_from_truth_scaled1.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter1_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter20_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter2_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter30_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter3_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter40_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter4_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter50_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter5_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter5_pmf_bg_K50_maxiter10_from_truth_scaled.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter5_pmf_bg_K50_maxiter10_from_truth_scaled0.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter5_pmf_bg_K50_maxiter10_from_truth_scaled1.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter60_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter6_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter70_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter7_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter80_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter8_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter90_init_random.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter9_from_truth.Rds
Untracked: output/sim/v0.4.5/exper2/sim_bg_block_n1100_p2100_K50_fastTopics_K50_50em_950_scd.Rds
Untracked: output/smallsim2_1.RData
Untracked: script/Rplots.pdf
Untracked: script/init_ebpmf_wbg_from_pmf_bg.R
Untracked: script/init_ebpmf_wbg_random.R
Untracked: script/save_volcano_plot.R
Untracked: topicView-app/app_utils.R
Untracked: topicView-app/data/
Untracked: topicView-app/output/
Untracked: topicView-app/rsconnect/
Unstaged changes:
Modified: analysis/ebpmf_wbg_simulate_big_data2.Rmd
Deleted: analysis/sla_data_analysis_k10.Rmd
Deleted: analysis/sla_data_analysis_k5.Rmd
Deleted: analysis/sla_data_analysis_k50.Rmd
Deleted: data/SLA/SCC2016/Code/APL/compCM.m
Deleted: data/SLA/SCC2016/Code/APL/compMuI.m
Deleted: data/SLA/SCC2016/Code/APL/compParamErr2.m
Deleted: data/SLA/SCC2016/Code/APL/cpl4c.m
Deleted: data/SLA/SCC2016/Code/APL/cplEstimParam.m
Deleted: data/SLA/SCC2016/Code/APL/cpl_basic_demo_PJ.m
Deleted: data/SLA/SCC2016/Code/APL/cpl_demo.m
Deleted: data/SLA/SCC2016/Code/APL/cpl_demo2a.m
Deleted: data/SLA/SCC2016/Code/APL/dcBlkMod.m
Deleted: data/SLA/SCC2016/Code/APL/dcBlkMod2.m
Deleted: data/SLA/SCC2016/Code/APL/dcBlkMod3.m
Deleted: data/SLA/SCC2016/Code/APL/dcbm_nmi_beta_D.m
Deleted: data/SLA/SCC2016/Code/APL/dcbm_nmi_lambda_D.m
Deleted: data/SLA/SCC2016/Code/APL/dcbm_time_vs_n_D.m
Deleted: data/SLA/SCC2016/Code/APL/genDCBlkMod.c
Deleted: data/SLA/SCC2016/Code/APL/genDCBlkMod.mexa64
Deleted: data/SLA/SCC2016/Code/APL/genDCBlkMod2.m
Deleted: data/SLA/SCC2016/Code/APL/initLabel5b.m
Deleted: data/SLA/SCC2016/Code/BCPL/ProfileLike.m
Deleted: data/SLA/SCC2016/Code/BCPL/calCri1.m
Deleted: data/SLA/SCC2016/Code/BCPL/calCri2.m
Deleted: data/SLA/SCC2016/Code/BCPL/mutiExp.m
Deleted: data/SLA/SCC2016/Code/MatlabCode.m
Deleted: data/SLA/SCC2016/Code/NewmanSM/NewmanSM.m
Deleted: data/SLA/SCC2016/Code/coauthorThresh2GiantAdj.txt
Deleted: data/SLA/SCC2016/Code/coauthorThresh2GiantCommLabelK2Matlab.txt
Deleted: data/SLA/SCC2016/Code/functions.R
Deleted: data/SLA/SCC2016/Code/main.R
Deleted: data/SLA/SCC2016/Data/authorList.txt
Deleted: data/SLA/SCC2016/Data/authorPaperBiadj.txt
Deleted: data/SLA/SCC2016/Data/paperCitAdj.txt
Deleted: data/SLA/SCC2016/Data/paperList.txt
Deleted: data/SLA/SCC2016/ReadMe.txt
Modified: data/sim/docword.sim_bg_block_n1100_p2100_K50.txt
Deleted: data/sim/init.sim_bg_block_n1100_p2100_K50.Rds
Modified: data/sim/truth.sim_bg_block_n1100_p2100_K50.Rds
Deleted: data/uci_BoW.sh
Deleted: data/uci_BoW/docword.kos.txt
Deleted: data/uci_BoW/readme.txt
Deleted: data/uci_BoW/vocab.kos.txt
Deleted: output/sim/v0.4.5/fit_sim_bg_block_n1100_p2100_K50_ebpmf_wbg_maxiter_5000.Rout
Deleted: output/sim/v0.4.5/fit_sim_bg_block_n1100_p2100_K50_ebpmf_wbg_maxiter_5000_from_truth.Rout
Deleted: output/sim/v0.4.5/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter3.Rds
Deleted: output/sim/v0.4.5/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter5000.Rds
Deleted: output/sim/v0.4.5/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter5000_from_truth.Rds
Deleted: output/sim/v0.4.5/sim_bg_block_n1100_p2100_K50_ebpmf_wbg_K50_maxiter50_from_truth2.Rds
Deleted: output/uci_BoW/v0.3.8/fit_kos_ebpmf_bg_K20_maxiter_1000.Rout
Deleted: output/uci_BoW/v0.3.8/fit_kos_ebpmf_bg_K20_maxiter_500.Rout
Deleted: output/uci_BoW/v0.3.8/fit_kos_ebpmf_bg_K20_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.8/kos_ebpmf_bg_K20_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.8/kos_ebpmf_bg_K20_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.8/kos_ebpmf_bg_K20_maxiter5000.Rds
Deleted: output/uci_BoW/v0.3.8/kos_ebpmf_bg_K2_maxiter10.Rds
Deleted: output/uci_BoW/v0.3.9/fit_kos_ebpmf_bg_K100_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_ebpmf_bg_K20_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_ebpmf_bg_K50_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_ebpmf_bg_initLF_K100_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_ebpmf_bg_initLF_K20_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_ebpmf_bg_initLF_K300_maxiter_1000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_ebpmf_bg_initLF_K500_maxiter_1000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_ebpmf_bg_initLF_K50_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_pmf_initLF_K100_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_pmf_initLF_K20_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_pmf_initLF_K300_maxiter_1000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_pmf_initLF_K500_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/fit_kos_pmf_initLF_K50_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K100_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K100_maxiter1500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K100_maxiter2000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K100_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter1500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter2000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter2500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter3000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter3500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter4000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter4500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K20_maxiter5000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K50_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K50_maxiter1500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K50_maxiter2000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K50_maxiter2500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K50_maxiter3000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K50_maxiter3500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K50_maxiter4000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K50_maxiter4500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_K50_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K100_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K100_maxiter1500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K100_maxiter2000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K100_maxiter2500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K100_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter10.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter1500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter2000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter2500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter3000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter3500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter4000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter4500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter5.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K20_maxiter5000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter100.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter200.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter300.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter400.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter600.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter700.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter800.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K300_maxiter900.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter1500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter2000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter2500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter3000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter3500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter4000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter4500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_ebpmf_bg_initLF50_K50_maxiter5000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_init_nmf_K100_iter50.Rds
Deleted: output/uci_BoW/v0.3.9/kos_init_nmf_K20_iter50.Rds
Deleted: output/uci_BoW/v0.3.9/kos_init_nmf_K300_iter50.Rds
Deleted: output/uci_BoW/v0.3.9/kos_init_nmf_K500_iter50.Rds
Deleted: output/uci_BoW/v0.3.9/kos_init_nmf_K50_iter50.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter1500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter2000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter2500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter3000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter3500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter4000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter4500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K100_maxiter5000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter10.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter1500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter2000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter2500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter3000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter3500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter4000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter4500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter5.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K20_maxiter5000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter100.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter200.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter300.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter400.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter600.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter700.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter800.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K300_maxiter900.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter1000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter1500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter2000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter2500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter3000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter3500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter4000.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter4500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter500.Rds
Deleted: output/uci_BoW/v0.3.9/kos_pmf_initLF50_K50_maxiter5000.Rds
Deleted: output/uci_BoW/v0.4.2/fit_kos_ebpmf_wbg_initLF_K100_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.4.2/fit_kos_ebpmf_wbg_initLF_K20_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.4.2/fit_kos_ebpmf_wbg_initLF_K50_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.4.2/fit_kos_ebpmf_wbg_initL_K100_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.4.2/fit_kos_ebpmf_wbg_initL_K20_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.4.2/fit_kos_ebpmf_wbg_initL_K50_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter10.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter1000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter1500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter2000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter2500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter3000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter3500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter4000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter4500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K20_maxiter5000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter1000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter1500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter2000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter2500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter3000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter3500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter4000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter4500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initL50_K50_maxiter5000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K100_maxiter1000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K100_maxiter1500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K100_maxiter2000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K100_maxiter2500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K100_maxiter500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter10.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter1000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter1500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter2000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter2500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter3000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter3500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter4000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter4500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K20_maxiter5000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K3_maxiter10.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter1000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter1500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter2000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter2500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter3000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter3500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter4000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter4500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter500.Rds
Deleted: output/uci_BoW/v0.4.2/kos_ebpmf_wbg_initLF50_K50_maxiter5000.Rds
Deleted: output/uci_BoW/v0.4.2/kos_init_nmf_K100_iter50.Rds
Deleted: output/uci_BoW/v0.4.2/kos_init_nmf_K20_iter50.Rds
Deleted: output/uci_BoW/v0.4.2/kos_init_nmf_K300_iter50.Rds
Deleted: output/uci_BoW/v0.4.2/kos_init_nmf_K3_iter50.Rds
Deleted: output/uci_BoW/v0.4.2/kos_init_nmf_K500_iter50.Rds
Deleted: output/uci_BoW/v0.4.2/kos_init_nmf_K50_iter50.Rds
Deleted: output/uci_BoW/v0.4.4/fit_kos_np_ebpmf_wbg_initLF_K100_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.4.4/fit_kos_np_ebpmf_wbg_initLF_K20_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.4.4/fit_kos_np_ebpmf_wbg_initLF_K50_maxiter_5000.Rout
Deleted: output/uci_BoW/v0.4.4/kos_np_ebpmf_wbg_initLF50_K100_maxiter500.Rds
Deleted: output/uci_BoW/v0.4.4/kos_np_ebpmf_wbg_initLF50_K20_maxiter500.Rds
Deleted: output/uci_BoW/v0.4.4/kos_np_ebpmf_wbg_initLF50_K50_maxiter500.Rds
Modified: topicView-app/app.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/cone_pmf1.Rmd) and HTML (docs/cone_pmf1.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | fa23ffb | zihao12 | 2021-06-08 | cone_pmf1.Rmd |
Summary
- Consider the Poisson Matrix Factorization problem: \(X \sim \text{Pois}(\Lambda); \Lambda = L F^T; L, F \geq 0\) with \(\text{dim}(X) = (n, p), n \ll p\). I use the notation \(\text{cone}(A) := \{\sum_k w_k \mathbf{a}_k: w_k \geq 0\} = \{A \mathbf{w}: \mathbf{w} \geq 0\}\) where \(\text{dim}(A) = (n, K)\)
- In many such applications with \(n \ll p\), I think it’s probably reasonable to assume the factors lie in the convex cone of samples. In other words, each factor is a weighted (non-negative) average of rows of data. This is a generalization of the “anchor sample” assumption (symmetric to the more famous “anchor word” assumption).
- From numerical experiments with both simulated and real data, I find:
- first, this assumption probably holds true in many datasets;
- second, even when we do MLE without using this assumption, the fitted \(F_{\text{mle}}\) can be projected to the \(\text{cone}(X^{t})\) without much loss of information.
- third, the projection coefficients (\(\mathbf{w}_k\) in \(\text{Proj}_{\text{cone}(X^t)}(\mathbf{f}_k) = X^t \mathbf{w}_k\)) is like applying soft-thresholding to loading \(\mathbf{l}_k\). It’s very sparse and interpretable: for each factor (topic) we can say it’s weighted average of a few “important” documents.
- Thus this assumption should be useful for better interpretability as well as develop more efficient and stable algorithm. I haven’t figured out a way to implement this, but parameterizing \(f_k\) by \(w_k\) alone is reducing \(p\) parameters to \(n\), so possibly making an easier optimization problem. Also we might impose further assumptions on \(w_k\)… (many possible things to do)
rm(list = ls())
library(NNLM)
library(nnls)
source("code/smallsim2_functions.R")
source("code/misc.R")
source("code/util.R")
set.seed(123)Simulated Data
- I find in many of my simulated data, we can easily estimate very high dimensional \(F\) with very few samples.
- I think it’s because the true Factors lie in convex cone of samples; and the construction of \(L\) makes many “anchor documents”. Therefore, even very few samples contain most of useful information of factors.
- Below the full data is \(\text{dim}(X) = (300, 1000)\). But in fact we can estimate \(F\) well with only 60 samples. I use functions here
n <- 300
m <- 1000
k <- 3
n_sample = 60
doc_len = m ## average number of words in each document
S <- 15*diag(k) - 2
F <- simulate_factors(m = m, k = k)
L <- simulate_loadings(n,k,S)
s <- simulate_sizes(n = n, doc_len = doc_len)
X <- simulate_multinom_counts(L,F,s)I initialize with 50 EM runs on small data; then fit on small and large data (NNLM::nnmf supports initializing with only factors).
fit0 = nnmf(A = X[1:n_sample, ], k = k, loss = "mkl", method = "lee", max.iter = 50)
fit_small = nnmf(A = X[1:n_sample, ], k = k, loss = "mkl", method = "scd", init = list(H = fit0$H), max.iter = 200)
fit_big = nnmf(A = X, k = k, loss = "mkl", method = "scd", init = list(H = fit0$H), max.iter = 200)
fit_small.m = get_multinom_from_pnmf(F = t(fit_small$H), L = fit_small$W)
fit_big.m = get_multinom_from_pnmf(F = t(fit_big$H), L = fit_big$W)
idx_small = match_topics(F1 = F, F2 = fit_small.m$F)
idx_big = match_topics(F1 = F, F2 = fit_big.m$F)Can see the small data recovers \(L, F\) well
model = fit_small.m
idx = idx_small
par(mfrow = c(3,2))
compare_truth_fitted(model, idx, L, F)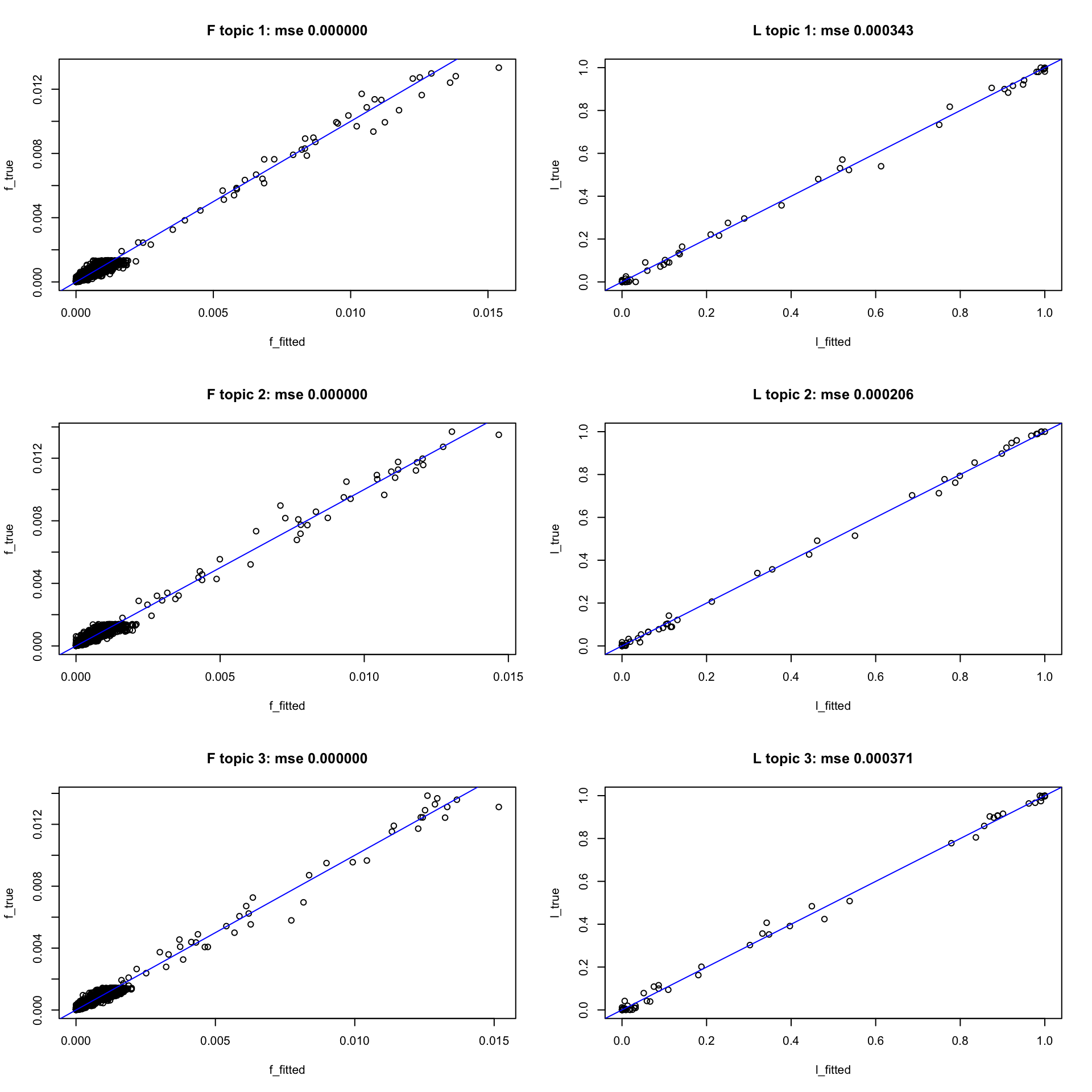
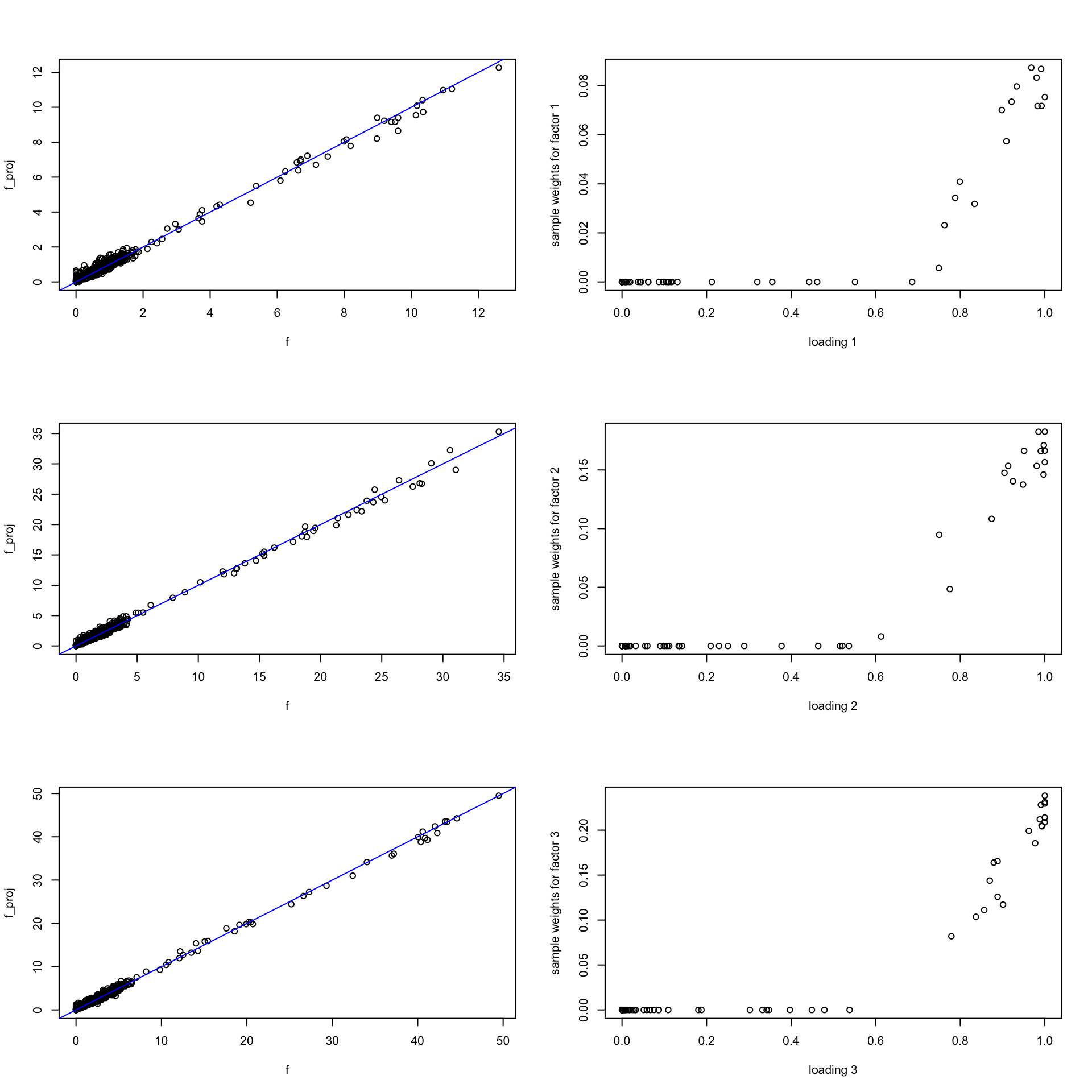
- Below I computed \(\text{Proj}_{\text{cone}(X^t)}(\hat{\mathbf{f}}_k) = X^t \mathbf{w}_k\) by
nnlsfunction (solving \(\text{min} \ |\mathbf{f}_k - X^t \mathbf{w}_k|_2^2 \ s.t. \mathbf{w}_k \geq 0\)). It returns both the projected \(f_k\) and weights \(w_k\). Here \(\hat{\mathbf{f}}_k\) is the MLE fit using the small data. - We can see:
- \(\mathbf{f}_k, \text{Proj}_{\text{cone}(X^t)}(\hat{\mathbf{f}}_k)\) are quite similar (at least on important words/genes/features)
- \(\mathbf{w}_k\) looks like applying soft-thresholding to \(l_k\)!
par(mfrow = c(3, 2))
for(i in 1:k){
f = t(fit_small$H)[,i]
l = fit_small.m$L[,i]
f_proj = nnls(A = t(X[1:n_sample,]), b = f)
plot(f, f_proj$fitted, ylab = "f_proj", cex.lab = 1)
abline(a = 0, b = 1, col = "blue")
plot(l, f_proj$x, cex = 1, cex.lab = 1,
xlab = sprintf("loading %d", i), ylab = sprintf("sample weights for factor %d", i))
}
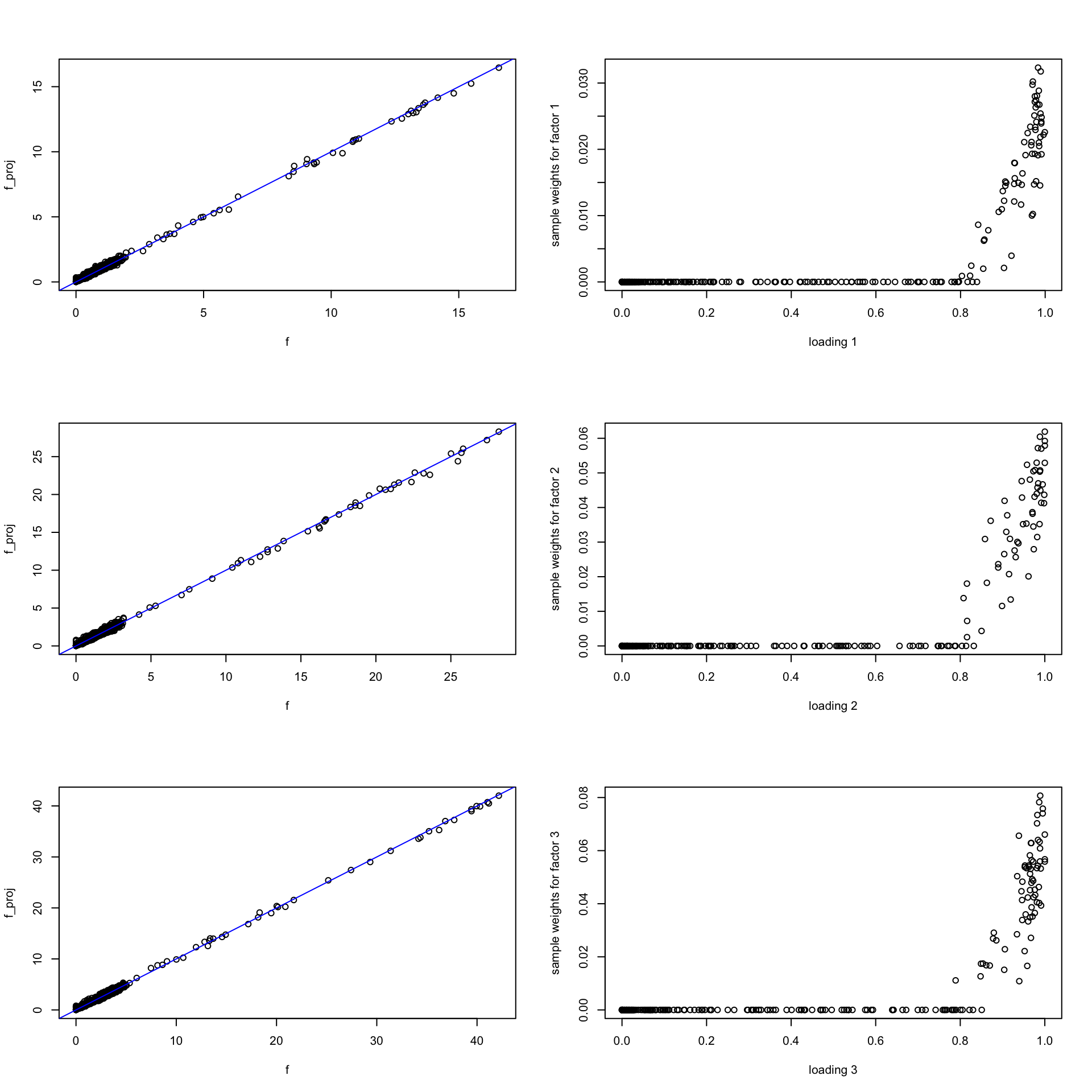
Do the same thing for fit on bigger data. Result is similar.
par(mfrow = c(3, 2))
for(i in 1:k){
f = t(fit_big$H)[,i]
l = fit_big.m$L[,i]
f_proj = nnls(A = t(X), b = f)
plot(f, f_proj$fitted, ylab = "f_proj", cex.lab = 1)
abline(a = 0, b = 1, col = "blue")
plot(l, f_proj$x, cex = 1, cex.lab = 1,
xlab = sprintf("loading %d", i), ylab = sprintf("sample weights for factor %d", i))
}
Do the true factors lie in the convex cone of rows of samll subset of \(X\) ? Basically yes
par(mfrow = c(2, 2))
for(i in 1:k){
f = F[,i]
f_proj = nnls(A = t(X[1:n_sample,]), b = f)
plot(f, f_proj$fitted, ylab = "f_proj", cex.lab = 1)
abline(a = 0, b = 1, col = "blue")
}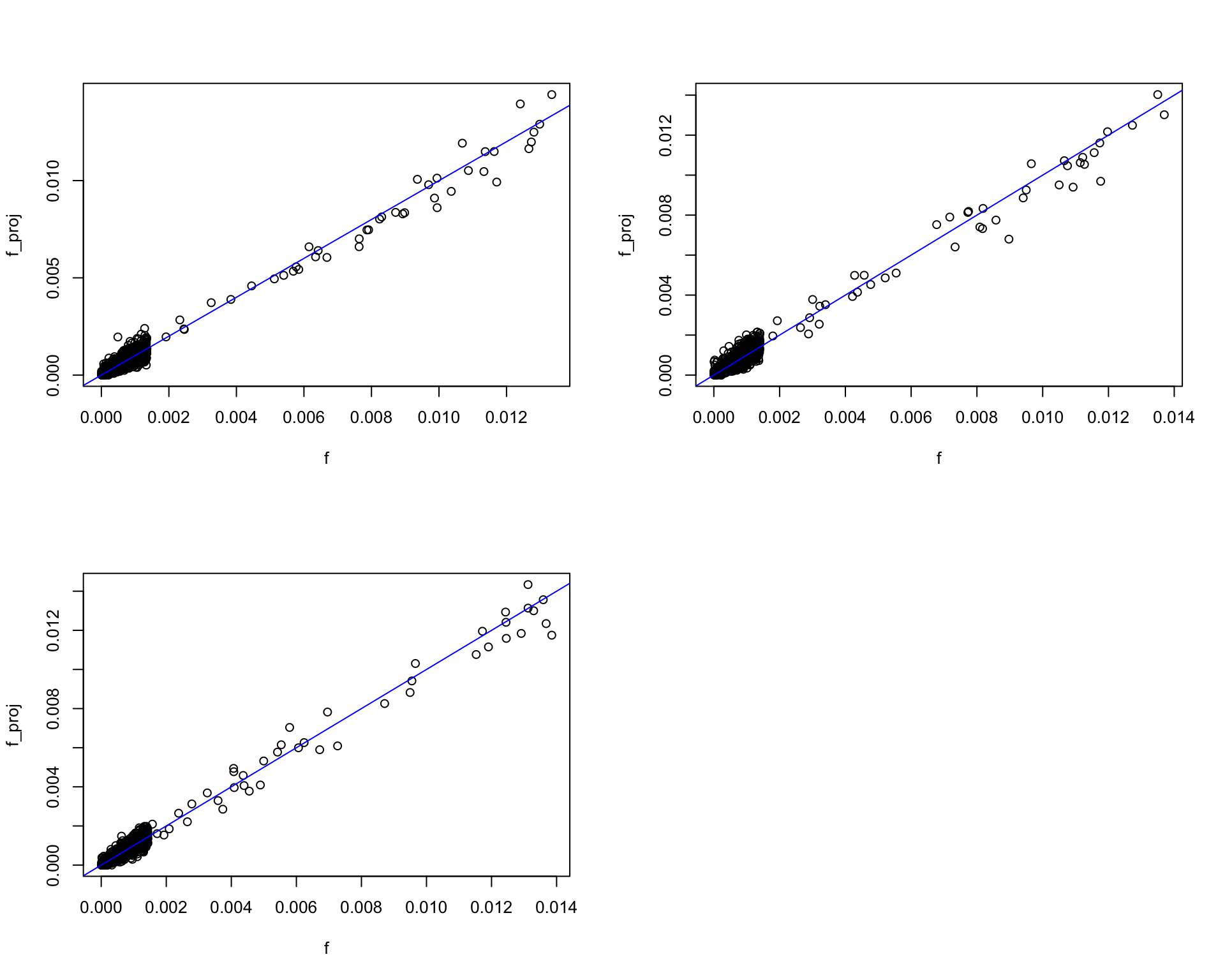
Comment
- we can see each factor (truth) is basically a weighted sum of a small subset of samples.
- The small subset contains enough “useful” documents/cells to reconstruct factor. That’s why such a high dimensional problem can be estimated with so few samples.
Real Data
This is a document dataset I fitted before, with \((n, p) = (3430, 6906)\). (I did MLE fit using EM)
data = readRDS("output/kos_coneFactor.Rds")
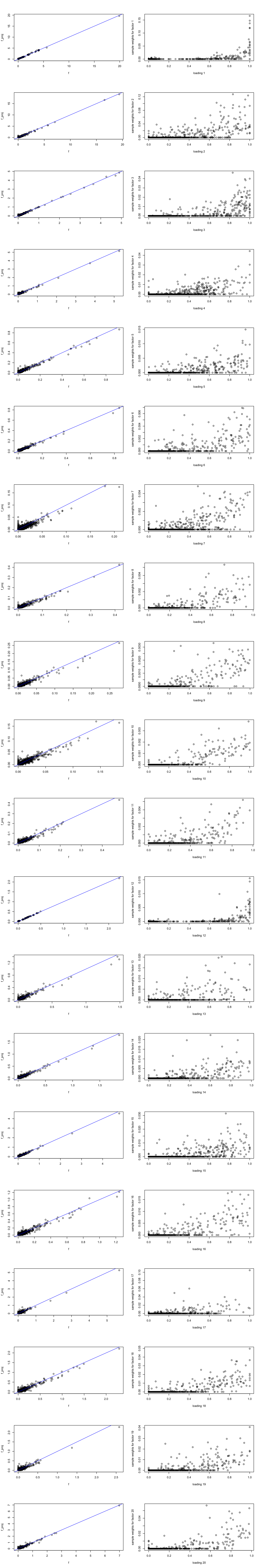
fit_kos.m = get_multinom_from_pnmf(F = data$pmf$F, L = data$pmf$L)- Below I look at the projection operator like in the section above
- The MLE fits basically lie in \(\text{cone}(X^t)\)
- The weights vs loading plot is a bit messier than in simulation data, but we can still see similar picture: \(w_k\)’s capture similar information as loadings, and are sparse and interpretable.
par(mfrow = c(20, 2))
for(i in 1:20){
f = data$pmf$F[,i]
l = fit_kos.m$L[,i]
f_proj = data$f_proj[[i]]
plot(f, f_proj$fitted, ylab = "f_proj", cex.lab = 1)
abline(a = 0, b = 1, col = "blue")
plot(l, f_proj$x, cex = 1, cex.lab = 1,
xlab = sprintf("loading %d", i), ylab = sprintf("sample weights for factor %d", i))
}
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] pheatmap_1.0.12 mvtnorm_1.1-0 nnls_1.4 NNLM_0.4.2
[5] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.5 knitr_1.28 whisker_0.3-2 magrittr_1.5
[5] munsell_0.5.0 colorspace_1.4-1 R6_2.4.1 rlang_0.4.5
[9] stringr_1.4.0 tools_3.5.1 grid_3.5.1 gtable_0.3.0
[13] xfun_0.8 git2r_0.26.1 htmltools_0.5.0 yaml_2.2.0
[17] digest_0.6.25 rprojroot_1.3-2 lifecycle_0.2.0 RColorBrewer_1.1-2
[21] later_1.1.0.1 promises_1.1.1 fs_1.3.1 glue_1.4.1
[25] evaluate_0.14 rmarkdown_2.1 stringi_1.6.2 compiler_3.5.1
[29] scales_1.1.1 backports_1.1.7 httpuv_1.5.4