Exploratory analysis and Quality Control of P2 mice dataset: Scgn Cre tdTomato
2024-11-27
Last updated: 2024-11-27
Checks: 7 0
Knit directory: Hanics_2024/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20241007) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e1fb0b4. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .cache/
Ignored: .config/
Ignored: .ipython/
Ignored: .jupyter/
Ignored: .nv/
Ignored: .snakemake/
Ignored: analysis/figure/
Ignored: cellbender/BSF_1105_Mouse_Cortex_SCGN_P02_1_filtered.h5ad
Ignored: cellbender/BSF_1105_Mouse_Cortex_SCGN_P02_1_output.h5
Ignored: cellbender/BSF_1105_Mouse_Cortex_SCGN_P02_1_output_filtered.h5
Ignored: cellbender/BSF_1105_Mouse_Cortex_SCGN_P02_1_output_posterior.h5
Ignored: cellranger_BSF_1105_Mouse_Cortex_SCGN_P02_1/
Ignored: ckpt.tar.gz
Ignored: data/SCP1290
Ignored: fastq/
Ignored: mm10_tdTomato
Ignored: scrublet/BSF_1105_Mouse_Cortex_SCGN_P02_1_initial_annotation.h5ad
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/01A-eda.Rmd) and HTML (docs/01A-eda.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | e1fb0b4 | Evgenii O. Tretiakov | 2024-11-27 | analysis of Scgn-Cre mice with use of developmental reference |
| Rmd | ec23581 | Evgenii O. Tretiakov | 2024-11-15 | initial workflowr pipeline for notebooks with use of all cells filtered after qc and cortex development dataset from single-cell portal as a reference on which we will project our data |
We will load and reprocess reference dataset of cortical development from (Di Bella et al. 2021).
# Create a vector with the stage of development for each object
stage_info <- c("E11.5", "E12.5", "E13.5", "E14.5", "E15.5", "E16", "E18.5", "E18", "P1", "P1", "E10", "E17.5", "P4")merged_cortex_2 <- SeuratObject::LoadSeuratRds(here::here("data/azimuth_integrated.rds"))
merged_cortex_2$cell_name <- Cells(merged_cortex_2)
merged_cortex_2An object of class Seurat
28186 features across 82415 samples within 5 assays
Active assay: RNA (27998 features, 2000 variable features)
25 layers present: data.E11.5, data.E12.5, data.E13.5, data.E14.5, data.E15.5, data.E16, data.E18.5, data.E18, data.P1, data.E10, data.E17.5, data.P4, scale.data, counts.E11.5, counts.E12.5, counts.E13.5, counts.E14.5, counts.E15.5, counts.E16, counts.E18.5, counts.E18, counts.P1, counts.E10, counts.E17.5, counts.P4
4 other assays present: prediction.score.class, prediction.score.cluster, prediction.score.subclass, prediction.score.cross_species_cluster
7 dimensional reductions calculated: pca, integrated_dr, ref.umap, integrated.cca, umap.cca, harmony, umap.harmonyorig_umap <- readr::read_tsv(
here("data/SCP1290/cluster/cluster_scDevSC.merged.umap.txt"),
skip = 2,
col_names = c("cell_name", "UMAP_1", "UMAP_2"),
col_types = list(col_character(), col_double(), col_double())
)
glimpse(orig_umap)Rows: 98,047
Columns: 3
$ cell_name <chr> "E10_v1_AAACCTGAGGGTCTCC-1", "E10_v1_AAACCTGCACAACGCC-1", "E…
$ UMAP_1 <dbl> -3.0025911, -3.6729214, -3.8859395, -3.9020242, -2.9312939, …
$ UMAP_2 <dbl> -10.453364, -6.552985, -10.773631, -10.869657, -10.769403, -…orig_umap %<>% tibble::column_to_rownames("cell_name")
orig_umap %<>% as.matrix()
orig_tsne <- readr::read_tsv(
here("data/SCP1290/cluster/cluster_scDevSC.merged.tsne.txt"),
skip = 2,
col_names = c("cell_name", "tSNE_1", "tSNE_2"),
col_types = list(col_character(), col_double(), col_double())
)
glimpse(orig_tsne)Rows: 98,047
Columns: 3
$ cell_name <chr> "E10_v1_AAACCTGAGGGTCTCC-1", "E10_v1_AAACCTGCACAACGCC-1", "E…
$ tSNE_1 <dbl> 15.442958, 10.373660, 14.828413, 16.307658, 18.062250, 13.72…
$ tSNE_2 <dbl> -19.603245, -17.062466, -20.102599, -20.003542, -18.636268, …orig_tsne %<>% tibble::column_to_rownames("cell_name")
orig_tsne %<>% as.matrix()
orig_metadata <- readr::read_tsv(here(
"data/SCP1290/metadata/metaData_scDevSC.txt"
))
orig_metadata %<>% dplyr::rename("cell_name" = "NAME")
orig_metadata_types <- orig_metadata[1, ] |> purrr::simplify()
orig_metadata %<>% dplyr::filter(!cell_name == "TYPE")
glimpse(orig_metadata)Rows: 98,047
Columns: 28
$ cell_name <chr> "E10_v1_AAACCTGAGGGTCTCC-…
$ orig_ident <chr> "E10", "E10", "E10", "E10…
$ nCount_RNA <chr> "1544", "1157", "2081", "…
$ nFeature_RNA <chr> "1022", "783", "1200", "1…
$ percent_mito <chr> "0.02007772", "0.01469317…
$ n_hkgene <chr> "51", "39", "67", "71", "…
$ S_Score <chr> "0.356987282", "0.4538538…
$ G2M_Score <chr> "0.330795055", "0.2605599…
$ Phase <chr> "S", "S", "S", "G2M", "S"…
$ CC_Difference <chr> "0.026192226", "0.1932938…
$ seurat_clusters <chr> "34", "34", "34", "37", "…
$ RNA_snn_res_1 <chr> "20", "20", "20", "20", "…
$ scrublet_doublet <chr> "FALSE", "FALSE", "FALSE"…
$ RNA_snn_res_2 <chr> "34", "34", "34", "37", "…
$ Doublet_intersect <chr> NA, NA, NA, NA, NA, NA, N…
$ Gral_cellType <chr> NA, NA, NA, NA, NA, NA, N…
$ New_cellType <chr> "Apical progenitors", "In…
$ biosample_id <chr> "E10", "E10", "E10", "E10…
$ donor_id <chr> "mouse_E10", "mouse_E10",…
$ species <chr> "NCBITaxon_10090", "NCBIT…
$ disease <chr> "PATO_0000461", "PATO_000…
$ disease__ontology_label <chr> "normal", "normal", "norm…
$ organ <chr> "UBERON_0008930", "UBERON…
$ organ__ontology_label <chr> "somatosensory cortex", "…
$ library_preparation_protocol <chr> "EFO_0009899", "EFO_00098…
$ library_preparation_protocol__ontology_label <chr> "10X 3' v2 sequencing", "…
$ sex <chr> "mixed", "mixed", "mixed"…
$ species__ontology_label <chr> "Mus musculus", "Mus musc…change_column_types <- function(df, types) {
for (col_name in names(types)) {
col_type <- types[col_name]
if (col_type == "character") {
df[[col_name]] <- as.character(df[[col_name]])
} else if (col_type == "numeric") {
df[[col_name]] <- as.numeric(df[[col_name]])
} else if (col_type == "integer") {
df[[col_name]] <- as.integer(df[[col_name]])
} else if (col_type == "logical") {
df[[col_name]] <- as.logical(df[[col_name]])
} else if (col_type == "factor") {
df[[col_name]] <- as.factor(df[[col_name]])
} else if (col_type == "group") {
df[[col_name]] <- as.factor(df[[col_name]])
} else {
warning(paste("Unknown type:", col_type, "for column", col_name))
}
}
return(df)
}
# Apply the function to the metadata
orig_metadata <- change_column_types(orig_metadata, orig_metadata_types)
# Print the modified metadata
glimpse(orig_metadata)Rows: 98,047
Columns: 28
$ cell_name <chr> "E10_v1_AAACCTGAGGGTCTCC-…
$ orig_ident <fct> E10, E10, E10, E10, E10, …
$ nCount_RNA <dbl> 1544, 1157, 2081, 2490, 2…
$ nFeature_RNA <dbl> 1022, 783, 1200, 1430, 14…
$ percent_mito <dbl> 0.020077720, 0.014693172,…
$ n_hkgene <dbl> 51, 39, 67, 71, 70, 50, 4…
$ S_Score <dbl> 0.35698728, 0.45385381, 0…
$ G2M_Score <dbl> 0.33079506, 0.26055995, 0…
$ Phase <fct> S, S, S, G2M, S, S, S, S,…
$ CC_Difference <dbl> 0.026192226, 0.193293862,…
$ seurat_clusters <fct> 34, 34, 34, 37, 37, 34, 4…
$ RNA_snn_res_1 <fct> 20, 20, 20, 20, 20, 20, 3…
$ scrublet_doublet <fct> FALSE, FALSE, FALSE, FALS…
$ RNA_snn_res_2 <fct> 34, 34, 34, 37, 37, 34, 4…
$ Doublet_intersect <fct> NA, NA, NA, NA, NA, NA, N…
$ Gral_cellType <fct> NA, NA, NA, NA, NA, NA, N…
$ New_cellType <fct> Apical progenitors, Inter…
$ biosample_id <fct> E10, E10, E10, E10, E10, …
$ donor_id <fct> mouse_E10, mouse_E10, mou…
$ species <fct> NCBITaxon_10090, NCBITaxo…
$ disease <fct> PATO_0000461, PATO_000046…
$ disease__ontology_label <fct> normal, normal, normal, n…
$ organ <fct> UBERON_0008930, UBERON_00…
$ organ__ontology_label <fct> somatosensory cortex, som…
$ library_preparation_protocol <fct> EFO_0009899, EFO_0009899,…
$ library_preparation_protocol__ontology_label <fct> 10X 3' v2 sequencing, 10X…
$ sex <fct> mixed, mixed, mixed, mixe…
$ species__ontology_label <fct> Mus musculus, Mus musculu…orig_srt <- Read10X(data.dir = here("data/SCP1290/expression/601ae2f4771a5b0d72588bfb"))
# Convert the log1p normalized matrix to a standard matrix if it's not already
normalized_matrix <- as.matrix(orig_srt)
# Reverse the log1p transformation to get the scaled count matrix
count_matrix <- expm1(normalized_matrix)
# Extract scaling factors
scaling_factors <- orig_metadata[orig_metadata$cell_name == colnames(count_matrix), ]$nCount_RNA / 1e4
# Multiply each column by its scaling factor and round the results (it's not necessary but just to be sure)
scaled_count_matrix <- sweep(count_matrix, 2, scaling_factors, FUN = "*")
scaled_count_matrix <- round(scaled_count_matrix)
# Convert the count matrix to a sparse matrix format (dgCMatrix) as needed
count_matrix_sparse <- as(scaled_count_matrix, "dgCMatrix")
# Create a Seurat object using the recovered count matrix
merged_cortex <- CreateSeuratObject(counts = count_matrix_sparse, meta.data = orig_metadata)
merged_cortex[["umap"]] <- CreateDimReducObject(embeddings = orig_umap, key = "UMAP_", assay = DefaultAssay(merged_cortex))
merged_cortex[["tsne"]] <- CreateDimReducObject(embeddings = orig_tsne, key = "tSNE_", assay = DefaultAssay(merged_cortex))
merged_cortex$stage <- merged_cortex$orig.ident
table(merged_cortex$New_cellType)
Apical progenitors Astrocytes Cajal Retzius cells
18491 2976 532
CThPN Cycling glial cells DL CPN
4607 1004 3106
DL_CPN_1 DL_CPN_2 Doublet
422 146 1854
Endothelial cells Ependymocytes Immature neurons
291 35 3092
Intermediate progenitors Interneurons Layer 4
8490 10469 5317
Layer 6b Low quality cells Microglia
194 4545 263
Migrating neurons NP Oligodendrocytes
12332 424 1098
Pericytes Red blood cells SCPN
236 330 2987
UL CPN VLMC
14041 765 Idents(merged_cortex) <- "New_cellType"
merged_cortex <- subset(merged_cortex, idents = c("Doublet", "Low quality cells", "Red blood cells"), invert = TRUE)
merged_cortex <-
Store_Palette_Seurat(
seurat_object = merged_cortex,
palette = rev(brewer.pal(n = 11, name = "Spectral")),
palette_name = "expr_Colour_Pal"
)
merged_cortex <- Store_Palette_Seurat(
seurat_object = merged_cortex,
palette = ggsci::pal_ucscgb("default")(length(levels(merged_cortex$New_cellType))),
palette_name = "types_Colour_Pal",
overwrite = T
)
names(merged_cortex@misc$types_Colour_Pal) <- levels(merged_cortex$New_cellType)
merged_cortex <- Store_Palette_Seurat(
seurat_object = merged_cortex,
palette = ggsci::pal_gsea("default")(length(levels(merged_cortex$stage))),
palette_name = "stage_Colour_Pal",
overwrite = T
)
names(merged_cortex@misc$stage_Colour_Pal) <- levels(merged_cortex$stage)
genes.embed <- c(
"Abcd1",
"Abcd2",
"Abcd3",
"Acaa1",
"Acaa2",
"Acox1",
"Agrn",
"Agt",
"Alcam",
"Aldh1a1",
"Aldh1l1",
"Aldoc",
"Angpt1",
"Apoe",
"App",
"Aqp4",
"Arf1",
"Bmp7",
"Bsg",
"Cacybp",
"Caf4",
"Ccl25",
"Ckb",
"Cnr1",
"Cnr2",
"Col4a5",
"Cst3",
"Dagla",
"Daglb",
"Decr2",
"Dcc",
"Dnm1",
"Drp1",
"Ech1",
"Efna5",
"Egfr",
"Enho",
"Eno1",
"Faah",
"Fgf1",
"Fgfr3",
"Fis1",
"Fos",
"Fth1",
"Ftl1",
"Gfap",
"Gja1",
"Gli1",
"Glul",
"Gnai2",
"Gnas",
"H2-K1",
"Hacd2",
"Hadhb",
"Hbegf",
"Hepacam",
"Hif1",
"Htra1",
"Igsf1",
"Il18",
"Il1rapl1",
"Itgav",
"Jam2",
"Lama2",
"Lamb2",
"Lcat",
"Lgi1",
"Lgi4",
"Lpcat3",
"Lrpap1",
"Lrrc4b",
"Lxn",
"Mdk",
"Mdv1",
"Mfn1",
"Mfn2",
"Mgll",
"Mief1",
"Napepld",
"Ncam1",
"Ncan",
"Ndrg2",
"Nfasc",
"Nfia",
"Nlgn3",
"Nrxn1",
"Nrxn2",
"Ntn1",
"Ntrk3",
"Opa1",
"Otp",
"Pex1",
"Pex10",
"Pex12",
"Pex13",
"Pex14",
"Pex16",
"Pex2",
"Pex26",
"Pex3",
"Pex6",
"Pkm",
"Pla2g7",
"Plcb1",
"Psap",
"Ptn",
"Pygb",
"Ralyl",
"Rgma",
"Rtn4",
"S100a1",
"S100a6",
"S100b",
"Siah1a",
"Siah1b",
"Scd2",
"Sdc2",
"Sema6a",
"Sema6d",
"Sgcd",
"Sirpa",
"Slc1a2",
"Slc1a3",
"Slc38a1",
"Slc4a4",
"Slc6a11",
"Slc7a10",
"Slit1",
"Slit2",
"Slitrk2",
"Sorbs1",
"Sox9",
"Sparc",
"Spon1",
"Tafa1",
"Timp3",
"Tkt",
"Trpv1",
"Vcam1",
"Vegfa"
) %>% .[. %in% rownames(merged_cortex)]
merged_cortex <- FindVariableFeatures(merged_cortex, nfeatures = 5000, verbose = FALSE)
merged_cortex <- NormalizeData(
merged_cortex,
features = rownames(merged_cortex),
verbose = FALSE
)
# Scale data
merged_cortex <- ScaleData(
merged_cortex,
features = rownames(merged_cortex),
verbose = FALSE
)# Create DimPlot
p1 <- DimPlot(
merged_cortex,
reduction = "umap",
group.by = c("stage", "New_cellType"),
combine = FALSE, label.size = 2,
alpha = 0.7,
cols = c(merged_cortex@misc$types_Colour_Pal, merged_cortex@misc$stage_Colour_Pal)
)
p2 <- DimPlot(
merged_cortex,
reduction = "tsne",
group.by = c("stage", "New_cellType"),
combine = FALSE, label.size = 2,
alpha = 0.7,
cols = c(merged_cortex@misc$types_Colour_Pal, merged_cortex@misc$stage_Colour_Pal)
)wrap_plots(c(p1, p2), ncol = 2, byrow = F)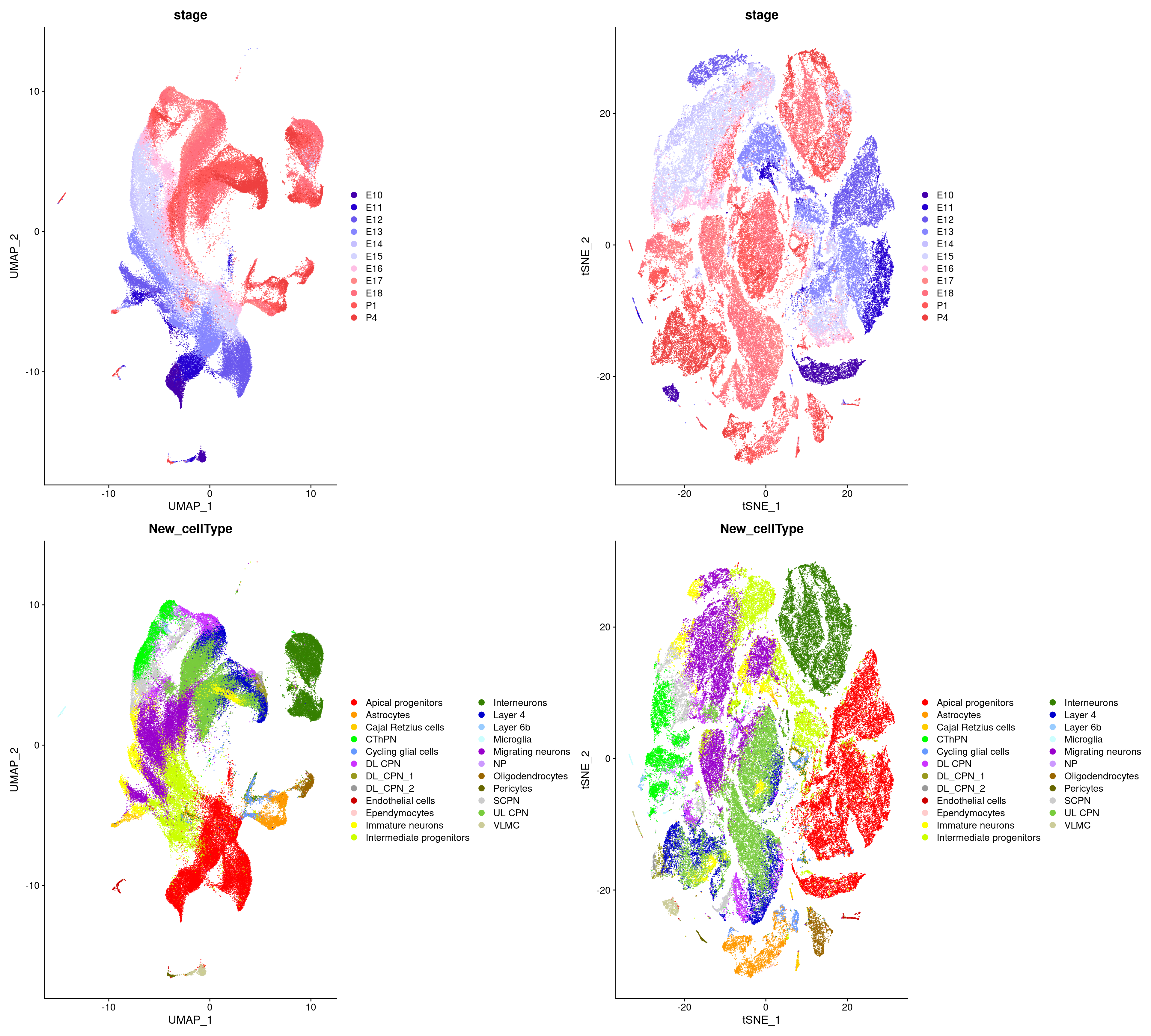
Introduction
In this document we are going to read in the RAW filtered counts matrix produced by Cell Ranger, the RNA filtered counts matrix, where we removed Ambient RNA using by CellBender at the false positive rate FPR=0.001 threshold and results of Cell Doublets call that was done using Scrublet then using summary statistics we determine which of those genes affected the most by our filtering procedure visualising results by scCustomize package and derive several categories of low quality cells using set of manually adjusted threshold parameters. Next, we use filtered high quality dataset to perform initial annotation using Seurat, leidenalg and clustree packages and deduce stable multi-resolution reconcile clustering tree with mrtree that we need to identify major cell groups for further analysis.
Set QC parameters
For the quality control we going to use set of well-known technical parameters reflecting sources of bias in data such as total mRNA content, percentage of mitochondrial mRNA content, fraction of molecules aligned to ribosomal genes, hemoglobine genes transcripts and overall cell complexity, which is determined as ratio between number of observed genes per molecule in logarithmic scale. As for doublets, we will use default Scrublet results.
Combined analysis of scRNA-seq dataset derived from the hanics2024-cortex-tdtomato
samples_table <- readr::read_tsv(here("samples.tsv")) %>% arrange(Run)
srr_set <- samples_table$Run
scrublet <-
purrr::reduce(
srr_set %>% map(~ read_scrublet(.x, fpr = cb_fpr)),
bind_rows
)
options(Seurat.object.assay.version = "v5")
cell_bender_merged <-
Read_CellBender_h5_Mat(
file_name = here(
"cellbender", glue::glue("{srr_set}_output_filtered.h5")
)
)
cell_ranger_merged <-
Read10X_h5(
filename = here(
"cellranger_BSF_1105_Mouse_Cortex_SCGN_P02_1/outs",
"filtered_feature_bc_matrix.h5"
)
)
cell_intersect <- intersect(
x = colnames(x = cell_bender_merged),
y = colnames(x = cell_ranger_merged)
)
cell_bender_merged <- cell_bender_merged[, cell_intersect]
combined_srt <- CreateSeuratObject(
counts = cell_bender_merged,
min.cells = 3,
min.features = 200
)
cell_names_seurat <- colnames(x = combined_srt)
gene_names_seurat <- rownames(x = combined_srt)
counts <- CreateAssay5Object(
counts = cell_ranger_merged, min.cells = 0,
min.features = 0
)
counts <- subset(
x = counts, cells = Cells(x = combined_srt),
features = rownames(x = combined_srt)
)
combined_srt[["RAW"]] <- counts
rm(cell_bender_merged, cell_ranger_merged)
combined_srtAn object of class Seurat
39038 features across 10078 samples within 2 assays
Active assay: RNA (19519 features, 0 variable features)
1 layer present: counts
1 other assay present: RAWcombined_srt@assays$RNA
Assay (v5) data with 19519 features for 10078 cells
First 10 features:
Xkr4, Gm1992, Gm19938, Gm37381, Rp1, Sox17, Gm37587, Mrpl15, Lypla1,
Tcea1
Layers:
counts
$RAW
Assay (v5) data with 19519 features for 10078 cells
First 10 features:
Xkr4, Gm1992, Gm19938, Gm37381, Rp1, Sox17, Gm37587, Mrpl15, Lypla1,
Tcea1
Layers:
counts Idents(object = combined_srt) <- "WT"
Idents(object = combined_srt, cells = WhichCells(combined_srt@assays$RAW, expression = tdTomato > 0)) <- "Scgn_Cre"
combined_srt$Scgn_tdTomato <- Idents(combined_srt)
plan(sequential)
invisible(gc())
options(future.globals.maxSize = 999999 * 1024^2)
set.seed(seed = reseed)
plan(multisession, workers = n_cores)Elimination of ambient RNA
Difference between assays
| orig.ident | nCount_RNA | nFeature_RNA | nCount_RAW | nFeature_RAW | Scgn_tdTomato | nFeature_Diff | nCount_Diff | |
|---|---|---|---|---|---|---|---|---|
| ACCCTCACAATAGGGC-1 | SeuratProject | 56962 | 8121 | 57010 | 8122 | WT | 1 | 48 |
| CGGGACTGTGTTACAC-1 | SeuratProject | 31199 | 6950 | 31246 | 6951 | WT | 1 | 47 |
| CTCATGCCAACGACAG-1 | SeuratProject | 27900 | 6033 | 27942 | 6033 | WT | 0 | 42 |
| CTTTCGGGTTCATCGA-1 | SeuratProject | 27802 | 6399 | 27846 | 6399 | Scgn_Cre | 0 | 44 |
| CCTCAGTAGATAGCTA-1 | SeuratProject | 23506 | 5371 | 23564 | 5379 | Scgn_Cre | 8 | 58 |
Median statistics of difference
| Scgn_tdTomato | Median_nCount_RNA | Median_nFeature_RNA | Median_nCount_Diff | Median_nFeature_Diff |
|---|---|---|---|---|
| Scgn_Cre | 2208.5 | 1185.5 | 77 | 25.5 |
| WT | 1066.5 | 628.0 | 85 | 35.0 |
| Totals (All Cells) | 1069.0 | 629.0 | 85 | 35.0 |
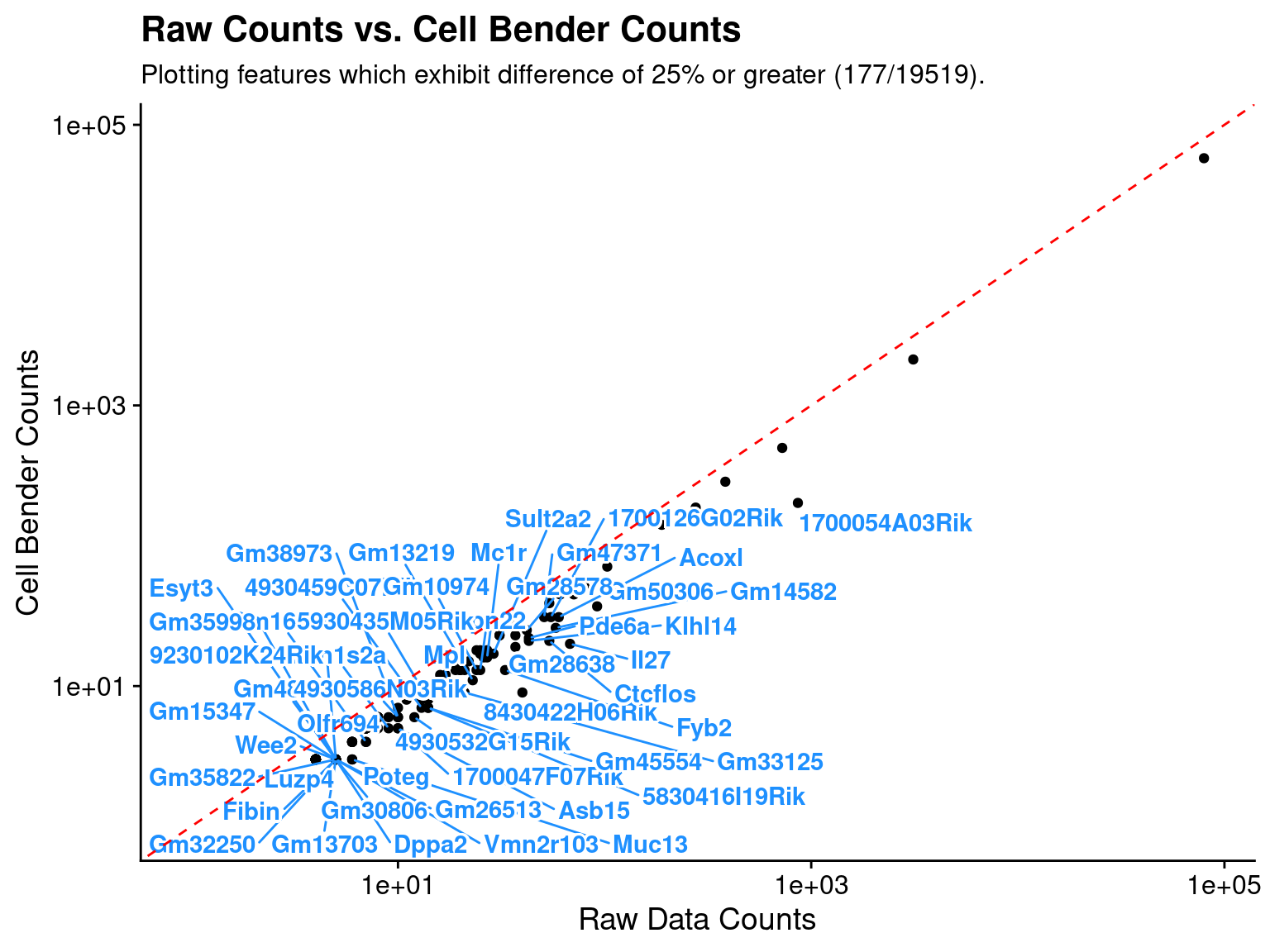
Top 20 leakage genes
| Raw_Counts | CellBender_Counts | Count_Diff | Pct_Diff | |
|---|---|---|---|---|
| 8430422H06Rik | 40 | 9 | 31 | 77.50000 |
| 1700054A03Rik | 862 | 202 | 660 | 76.56613 |
| Il27 | 68 | 20 | 48 | 70.58824 |
| Ctcflos | 54 | 21 | 33 | 61.11111 |
| Fyb2 | 33 | 13 | 20 | 60.60606 |
| Gm50306 | 92 | 37 | 55 | 59.78261 |
| Gm33125 | 21 | 9 | 12 | 57.14286 |
| Gm14582 | 58 | 26 | 32 | 55.17241 |
| Gm13219 | 23 | 11 | 12 | 52.17391 |
| Klhl14 | 43 | 21 | 22 | 51.16279 |
| 4930532G15Rik | 12 | 6 | 6 | 50.00000 |
| 1700047F07Rik | 10 | 5 | 5 | 50.00000 |
| Mpl | 18 | 9 | 9 | 50.00000 |
| 5830416I19Rik | 14 | 7 | 7 | 50.00000 |
| Asb15 | 10 | 5 | 5 | 50.00000 |
| Poteg | 6 | 3 | 3 | 50.00000 |
| Gm45554 | 14 | 7 | 7 | 50.00000 |
| Muc13 | 6 | 3 | 3 | 50.00000 |
| Pde6a | 43 | 22 | 21 | 48.83721 |
| Gm28638 | 37 | 19 | 18 | 48.64865 |
Plot feature differences
In addition to returning the data.frame it can be useful to visually examine the changes/trends after running CellBender.
Quality Control for different types of features
QC Violin plots
combined_srt <-
Store_Palette_Seurat(
seurat_object = combined_srt,
palette = rev(brewer.pal(n = 11, name = "RdYlGn")),
palette_name = "mdat_Colour_Pal"
)
combined_srt <-
Store_Palette_Seurat(
seurat_object = combined_srt,
palette = rev(brewer.pal(n = 11, name = "Spectral")),
palette_name = "expr_Colour_Pal"
)
combined_srt <-
Store_Palette_Seurat(
seurat_object = combined_srt,
palette = qc_palette,
palette_name = "qc_Colour_Pal"
)
combined_srt <-
Add_Mito_Ribo(combined_srt, species = "mouse")
combined_srt[["percent_hb"]] <-
PercentageFeatureSet(combined_srt, pattern = "^Hb[^(p)]")
combined_srt <-
Add_Cell_Complexity(combined_srt)
# Visualize QC metrics as a violin plot
p1 <-
QC_Plots_Complexity(
combined_srt,
high_cutoff = high_cutoff_complexity,
plot_median = TRUE,
color_seed = reseed,
ggplot_default_colors = TRUE
)
p2 <-
QC_Plots_Genes(
combined_srt,
low_cutoff = low_cutoff_gene,
high_cutoff = high_cutoff_gene,
plot_median = TRUE,
plot_title = "Genes per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p3 <-
QC_Plots_UMIs(
combined_srt,
low_cutoff = low_cutoff_umis,
high_cutoff = high_cutoff_umis,
plot_median = TRUE,
plot_title = "UMIs per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p4 <-
QC_Plots_Mito(
combined_srt,
high_cutoff = high_cutoff_pc_mt,
plot_median = TRUE,
plot_title = "Mito genes % per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p5 <-
QC_Plots_Feature(
combined_srt,
feature = "percent_ribo",
high_cutoff = high_cutoff_pc_ribo,
plot_median = TRUE,
y_axis_label = "% Ribosomal Genes Counts",
plot_title = "Ribo genes % per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p6 <-
QC_Plots_Feature(
combined_srt,
feature = "percent_hb",
high_cutoff = high_cutoff_pc_hb,
plot_median = TRUE,
y_axis_label = "% Hemoglobin Genes Counts",
plot_title = "Hemoglobin genes % per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
wrap_plots(p1, p2, p3, p4, p5, p6, ncol = 3)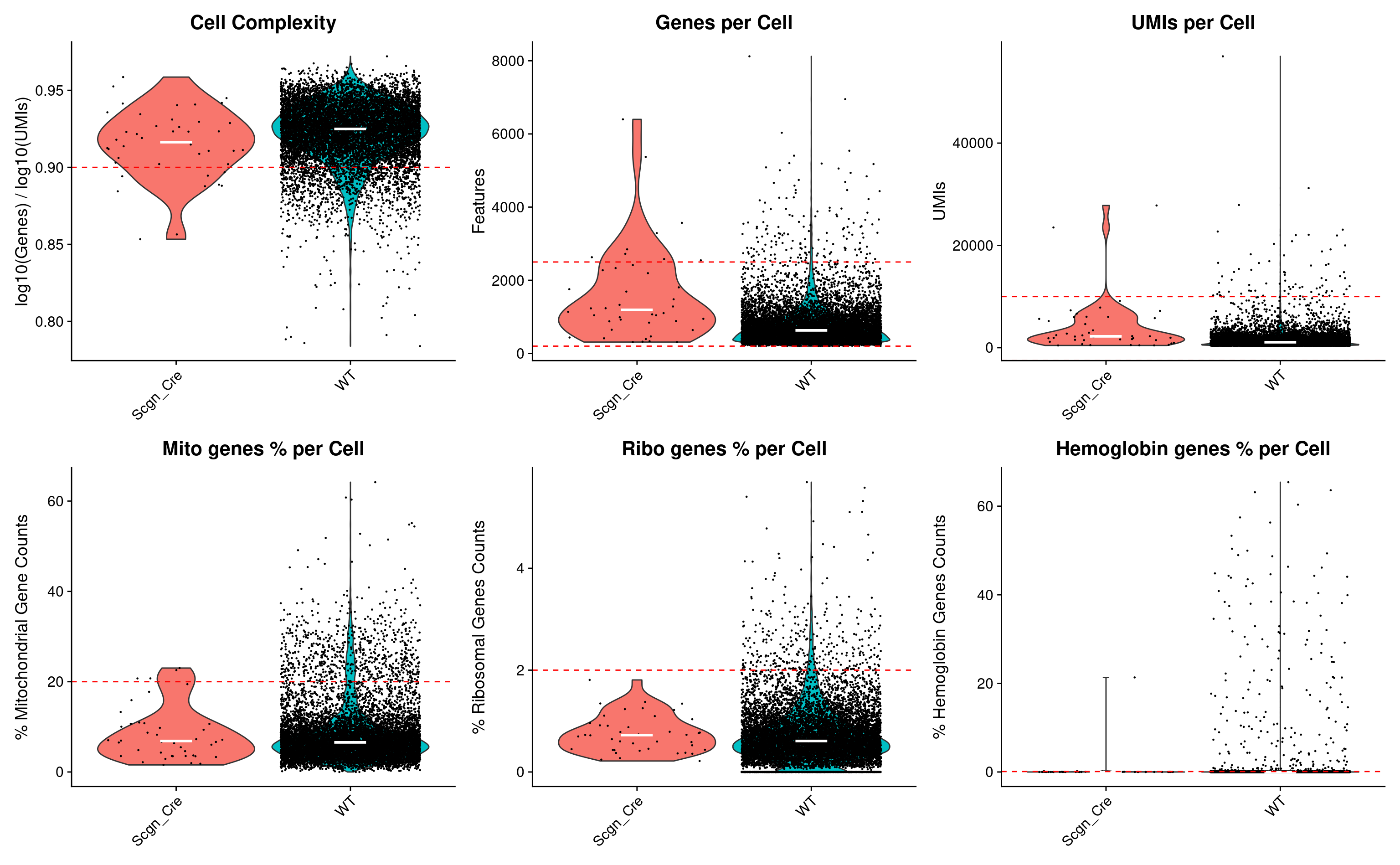
QC Scatter plots
plot1 <-
QC_Plot_GenevsFeature(
seurat_object = combined_srt,
feature1 = "percent_mito",
low_cutoff_gene = low_cutoff_gene,
high_cutoff_gene = high_cutoff_gene,
high_cutoff_feature = high_cutoff_pc_mt,
color_seed = reseed,
ggplot_default_colors = TRUE,
pt.size = 0.3,
shuffle_seed = reseed
) &
scale_y_log10()
plot2 <-
QC_Plot_UMIvsGene(
seurat_object = combined_srt,
low_cutoff_gene = low_cutoff_gene,
high_cutoff_gene = high_cutoff_gene,
low_cutoff_UMI = low_cutoff_umis,
high_cutoff_UMI = high_cutoff_umis,
color_seed = reseed,
ggplot_default_colors = TRUE,
pt.size = 0.3,
shuffle_seed = reseed
) &
scale_x_log10() & scale_y_log10()
plot3 <-
QC_Plot_GenevsFeature(
seurat_object = combined_srt,
feature1 = "percent_ribo",
low_cutoff_gene = low_cutoff_gene,
high_cutoff_gene = high_cutoff_gene,
high_cutoff_feature = high_cutoff_pc_ribo,
color_seed = reseed,
ggplot_default_colors = TRUE,
pt.size = 0.3,
shuffle_seed = reseed
) &
scale_y_log10()
plot4 <-
FeatureScatter(
combined_srt,
feature1 = "percent_ribo",
feature2 = "percent_mito",
shuffle = TRUE,
pt.size = 0.3,
seed = reseed
) +
geom_hline(yintercept = high_cutoff_pc_mt, color = "red", linetype = "dashed") +
geom_vline(xintercept = high_cutoff_pc_ribo, color = "blue", linetype = "dashed")
(plot1 + plot2) / (plot3 + plot4)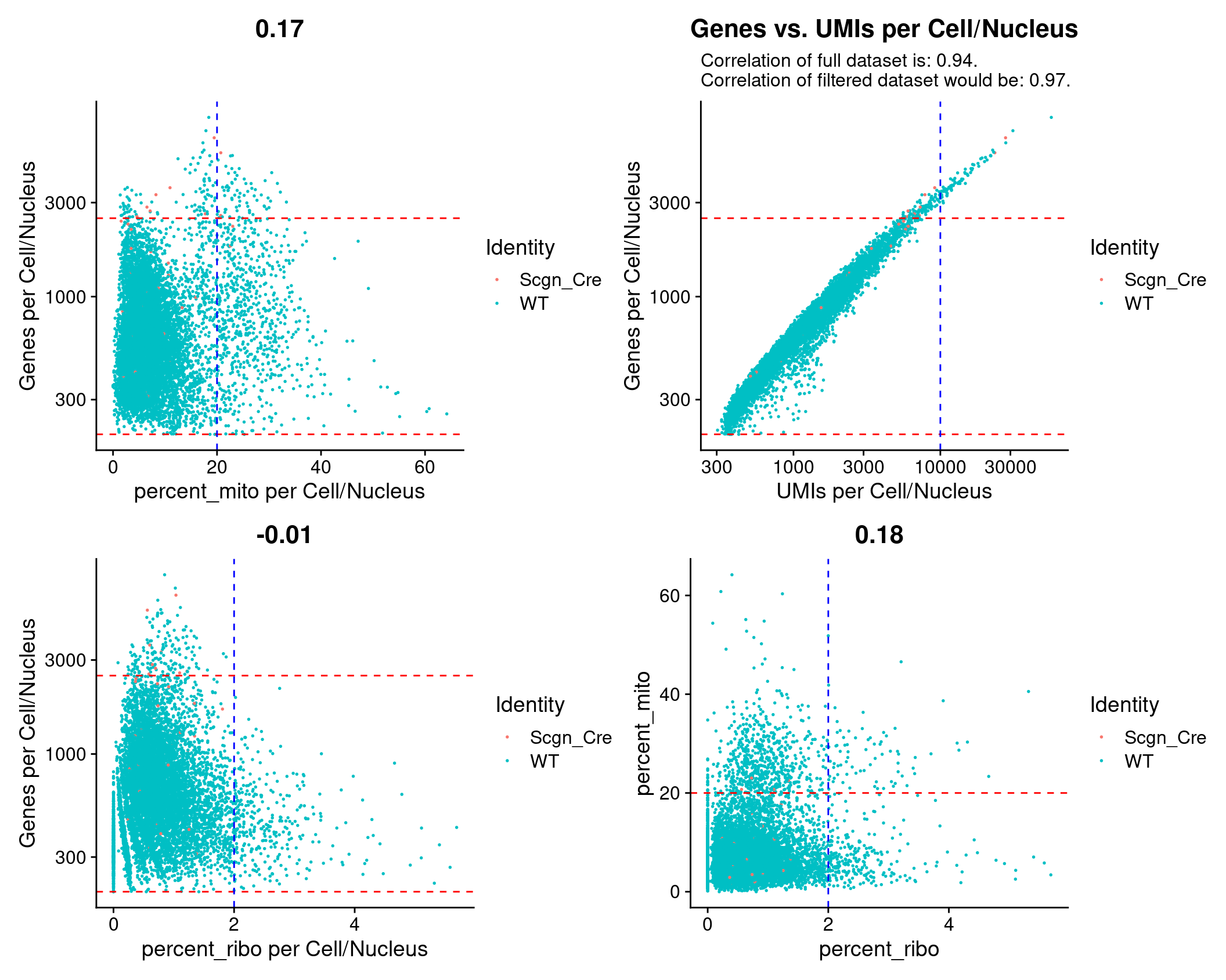
QC Scatter mito-threshold
QC_Plot_UMIvsGene(
seurat_object = combined_srt,
meta_gradient_name = "percent_mito",
low_cutoff_gene = low_cutoff_gene,
high_cutoff_gene = high_cutoff_gene,
low_cutoff_UMI = low_cutoff_umis,
high_cutoff_UMI = high_cutoff_umis,
meta_gradient_low_cutoff = high_cutoff_pc_mt,
meta_gradient_color = combined_srt@misc$mdat_Colour_Pal,
combination = TRUE,
color_seed = reseed,
ggplot_default_colors = TRUE,
pt.size = 1,
shuffle_seed = reseed
) &
scale_x_log10() & scale_y_log10()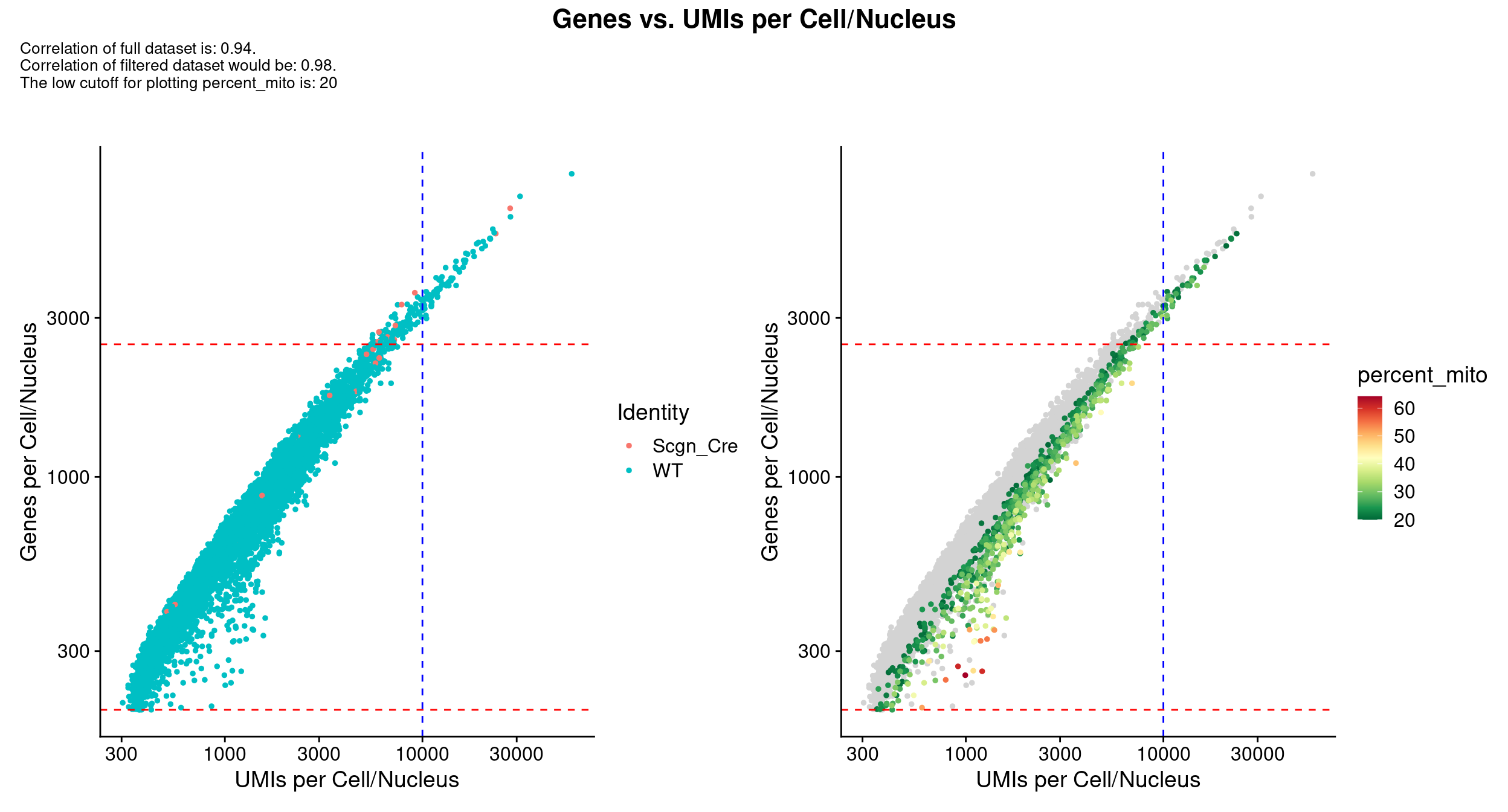
QC Scatter ribo-threshold
QC_Plot_UMIvsGene(
seurat_object = combined_srt,
meta_gradient_name = "percent_ribo",
low_cutoff_gene = low_cutoff_gene,
high_cutoff_gene = high_cutoff_gene,
low_cutoff_UMI = low_cutoff_umis,
high_cutoff_UMI = high_cutoff_umis,
meta_gradient_low_cutoff = high_cutoff_pc_ribo,
meta_gradient_color = combined_srt@misc$mdat_Colour_Pal,
combination = TRUE,
color_seed = reseed,
ggplot_default_colors = TRUE,
pt.size = 1,
shuffle_seed = reseed
) &
scale_x_log10() & scale_y_log10()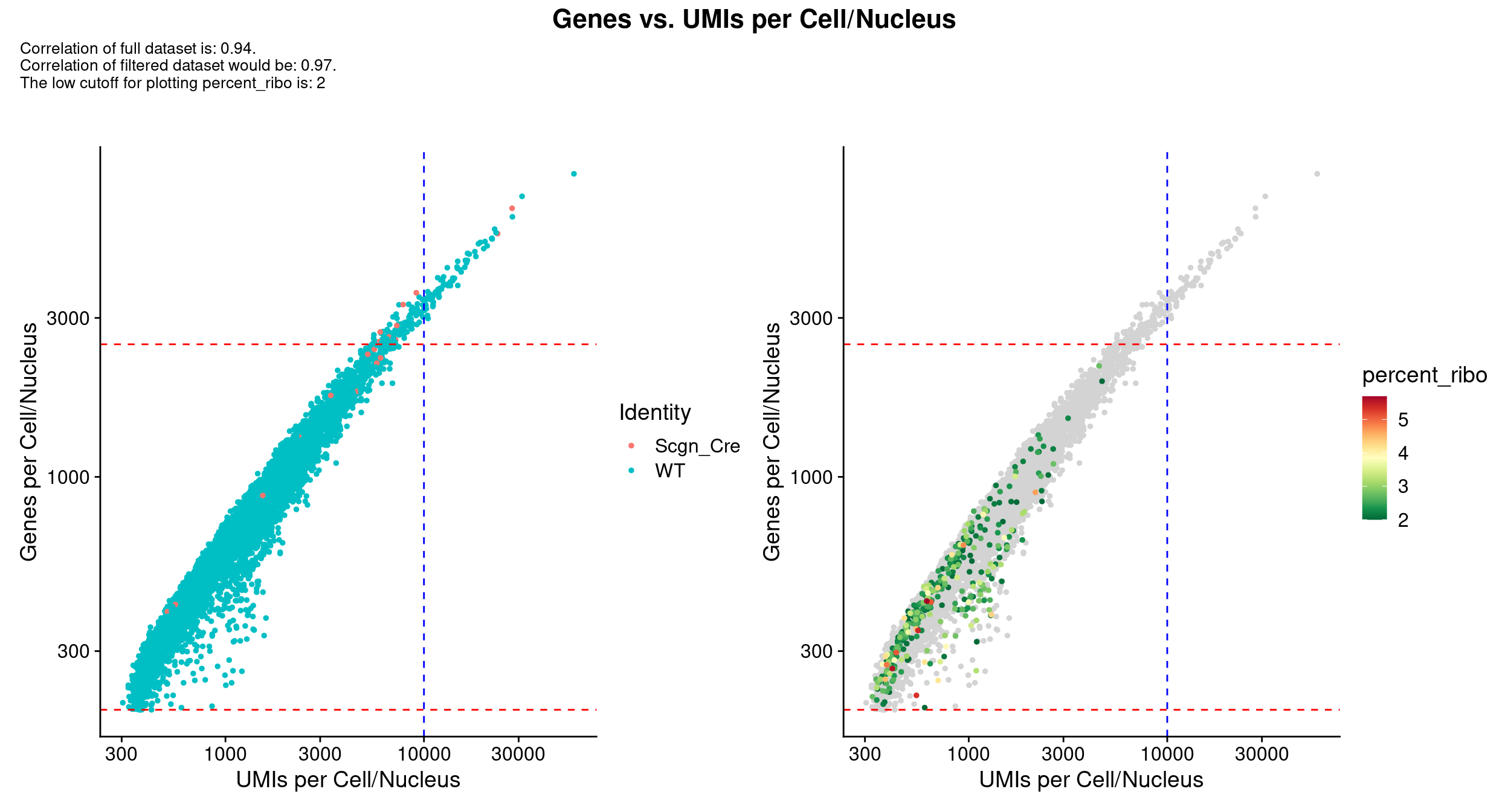
QC Scatter complexity-threshold
QC_Plot_UMIvsGene(
seurat_object = combined_srt,
meta_gradient_name = "log10GenesPerUMI",
low_cutoff_gene = low_cutoff_gene,
high_cutoff_gene = high_cutoff_gene,
low_cutoff_UMI = low_cutoff_umis,
high_cutoff_UMI = high_cutoff_umis,
meta_gradient_low_cutoff = high_cutoff_complexity,
meta_gradient_color = combined_srt@misc$mdat_Colour_Pal,
combination = TRUE,
color_seed = reseed,
ggplot_default_colors = TRUE,
pt.size = 1,
shuffle_seed = reseed
) &
scale_x_log10() & scale_y_log10()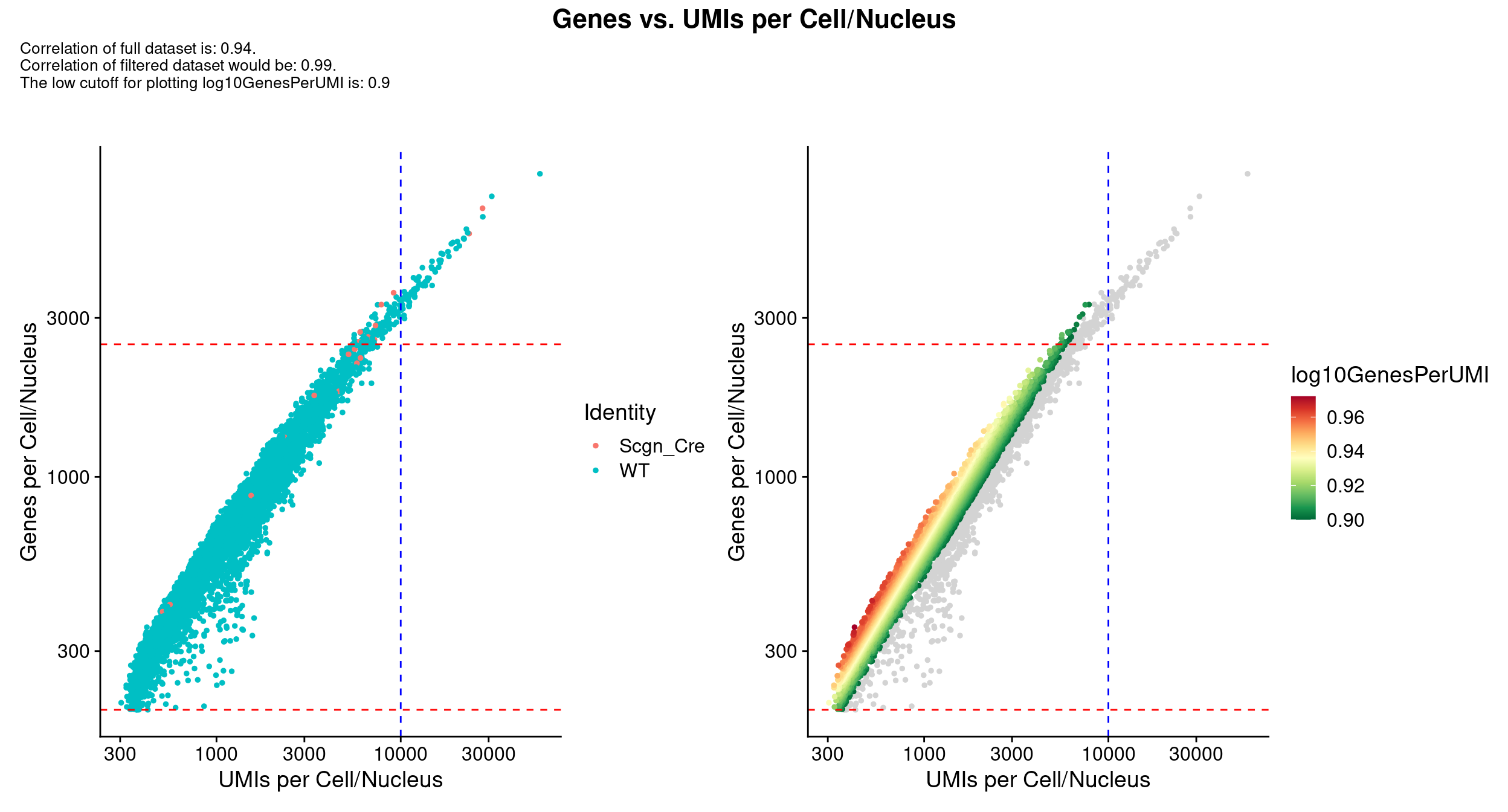
QC Scatter doublets by sample
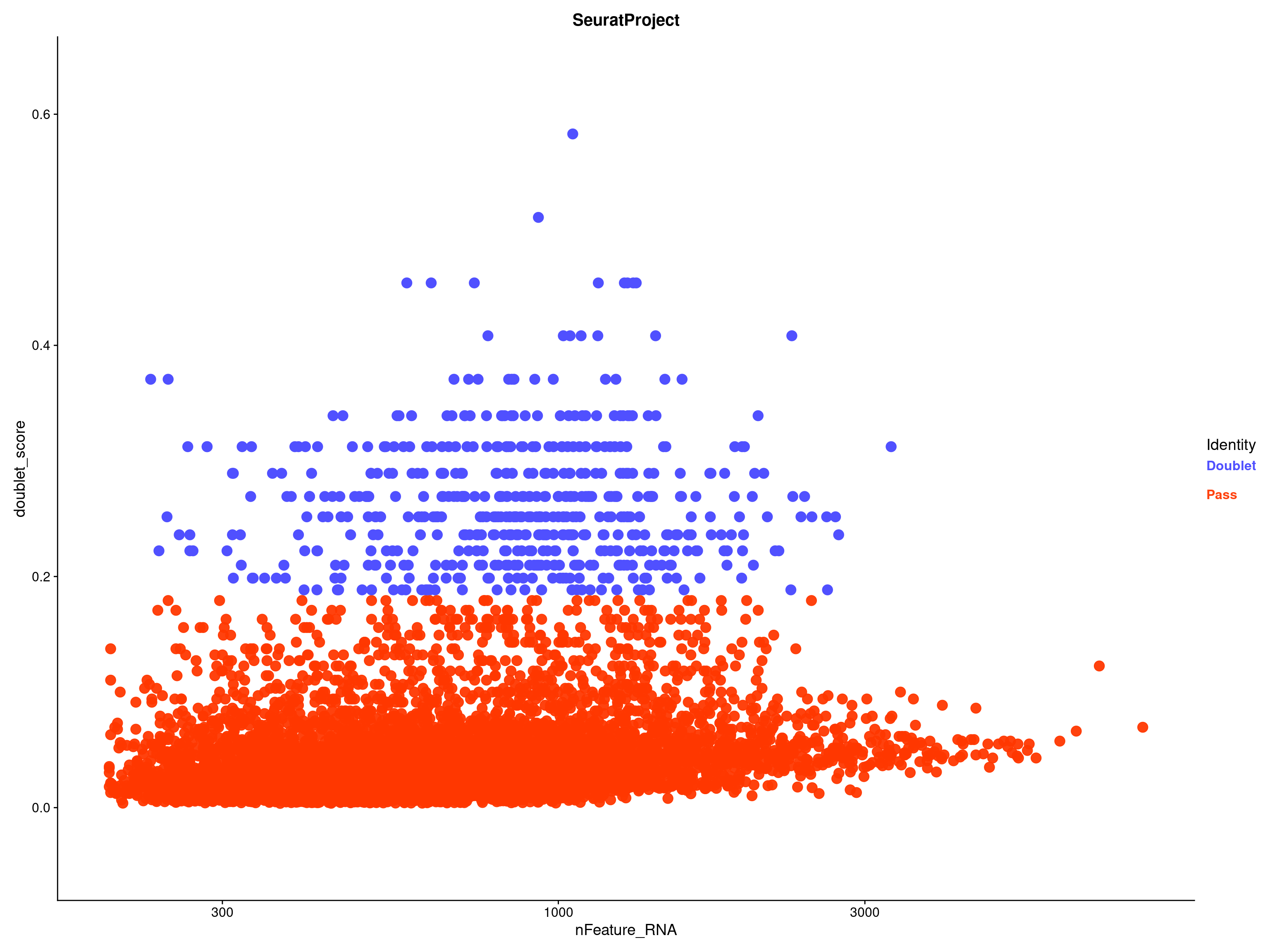
QC Scatter doublets-threshold
QC_Plot_UMIvsGene(
seurat_object = combined_srt,
meta_gradient_name = "doublet_score",
low_cutoff_gene = low_cutoff_gene,
high_cutoff_gene = high_cutoff_gene,
low_cutoff_UMI = low_cutoff_umis,
high_cutoff_UMI = high_cutoff_umis,
meta_gradient_low_cutoff = high_cutoff_doublet_score,
meta_gradient_color = combined_srt@misc$mdat_Colour_Pal,
combination = TRUE,
color_seed = reseed,
ggplot_default_colors = TRUE,
pt.size = 1,
shuffle_seed = reseed
) &
scale_x_log10() & scale_y_log10()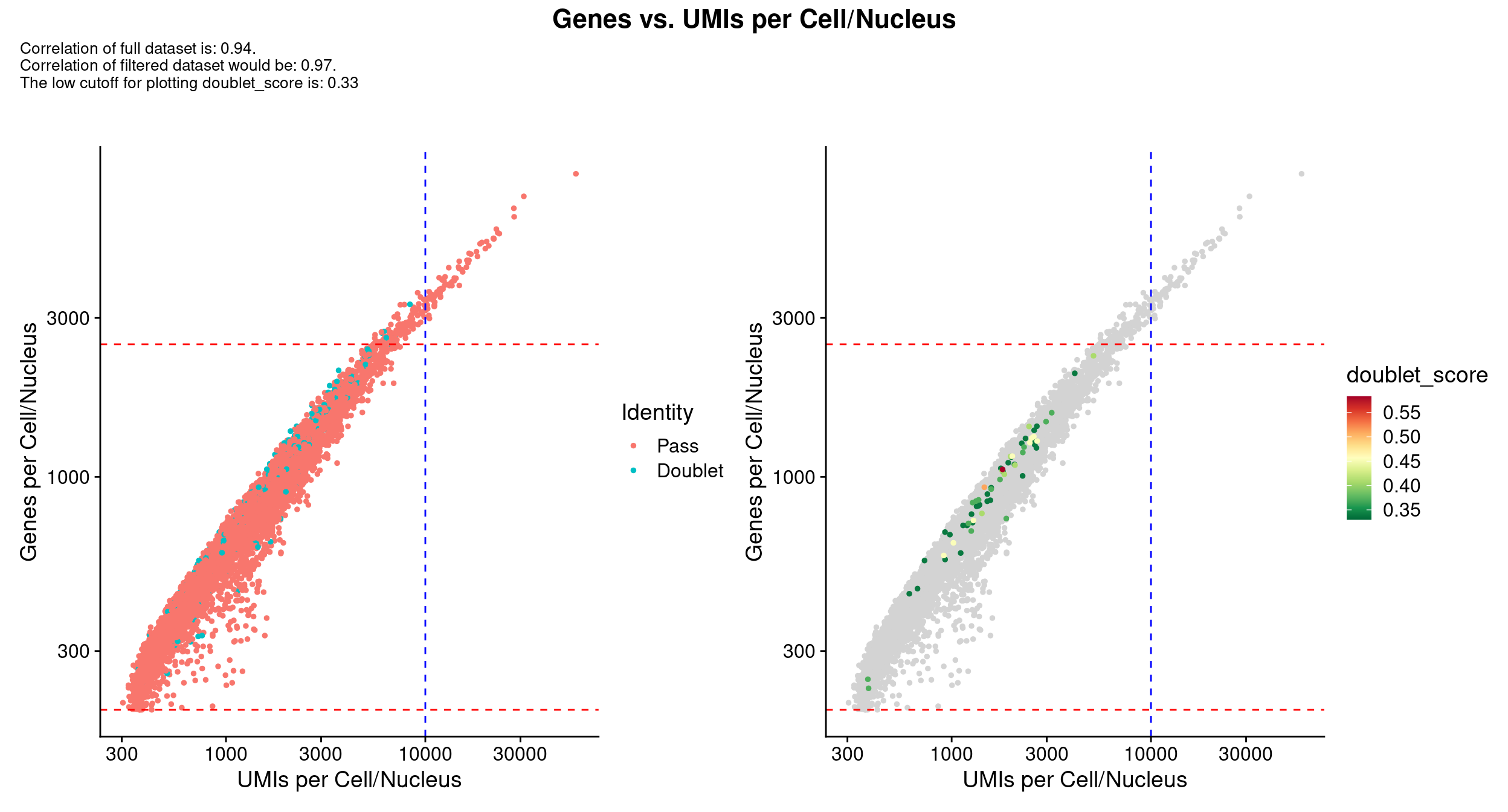
Apply QC thresholds to derive categories
combined_srt$QC <-
ifelse(
combined_srt@meta.data$log10GenesPerUMI < high_cutoff_complexity &
combined_srt@meta.data$QC == "Pass",
"Low_Complexity",
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$log10GenesPerUMI < high_cutoff_complexity &
combined_srt@meta.data$QC != "Pass" &
combined_srt@meta.data$QC != "Low_Complexity",
paste("Low_Complexity", combined_srt@meta.data$QC, sep = ","),
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$nFeature_RNA < low_cutoff_gene &
combined_srt@meta.data$QC == "Pass",
"Low_nFeature",
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$nFeature_RNA < low_cutoff_gene &
combined_srt@meta.data$QC != "Pass" &
combined_srt@meta.data$QC != "Low_nFeature",
paste("Low_nFeature", combined_srt@meta.data$QC, sep = ","),
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$percent_mito > high_cutoff_pc_mt &
combined_srt@meta.data$QC == "Pass",
"High_MT",
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$percent_mito > high_cutoff_pc_mt &
combined_srt@meta.data$QC != "Pass" &
combined_srt@meta.data$QC != "High_MT",
paste("High_MT", combined_srt@meta.data$QC, sep = ","),
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$nCount_RNA > high_cutoff_umis &
combined_srt@meta.data$QC == "Pass",
"High_UMIs",
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$nCount_RNA > high_cutoff_umis &
combined_srt@meta.data$QC != "Pass" &
combined_srt@meta.data$QC != "High_UMIs",
paste("High_UMIs", combined_srt@meta.data$QC, sep = ","),
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$percent_ribo > high_cutoff_pc_ribo &
combined_srt@meta.data$QC == "Pass",
"High_Ribo",
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$percent_ribo > high_cutoff_pc_ribo &
combined_srt@meta.data$QC != "Pass" &
combined_srt@meta.data$QC != "High_Ribo",
paste("High_Ribo", combined_srt@meta.data$QC, sep = ","),
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$percent_hb > high_cutoff_pc_hb &
combined_srt@meta.data$QC == "Pass",
"High_Hgb",
combined_srt@meta.data$QC
)
combined_srt$QC <-
ifelse(
combined_srt@meta.data$percent_hb > high_cutoff_pc_hb &
combined_srt@meta.data$QC != "Pass" &
combined_srt@meta.data$QC != "High_Hgb",
paste("High_Hgb", combined_srt@meta.data$QC, sep = ","),
combined_srt@meta.data$QC
)
table(combined_srt$QC)
Doublet
435
High_Hgb
480
High_Hgb,Doublet
56
High_Hgb,High_MT
4
High_Hgb,High_MT,Doublet
2
High_Hgb,High_MT,Low_Complexity
3
High_Hgb,High_Ribo
27
High_Hgb,High_Ribo,Doublet
1
High_Hgb,High_Ribo,High_MT
1
High_Hgb,High_Ribo,High_MT,Low_Complexity
1
High_Hgb,High_Ribo,High_MT,Low_Complexity,Doublet
1
High_Hgb,High_Ribo,Low_Complexity
2
High_Hgb,High_Ribo,Low_Complexity,Doublet
1
High_Hgb,High_UMIs,High_MT,Low_Complexity
2
High_Hgb,High_UMIs,Low_Complexity
1
High_Hgb,Low_Complexity
72
High_Hgb,Low_Complexity,Doublet
9
High_MT
144
High_MT,Doublet
11
High_MT,Low_Complexity
462
High_MT,Low_Complexity,Doublet
7
High_Ribo
216
High_Ribo,Doublet
4
High_Ribo,High_MT
7
High_Ribo,High_MT,Doublet
2
High_Ribo,High_MT,Low_Complexity
62
High_Ribo,Low_Complexity
7
High_UMIs,High_MT,Low_Complexity
38
High_UMIs,Low_Complexity
33
Low_Complexity
430
Low_Complexity,Doublet
15
Pass
7542 Let’s see how Scrublet score match distributed across our categories
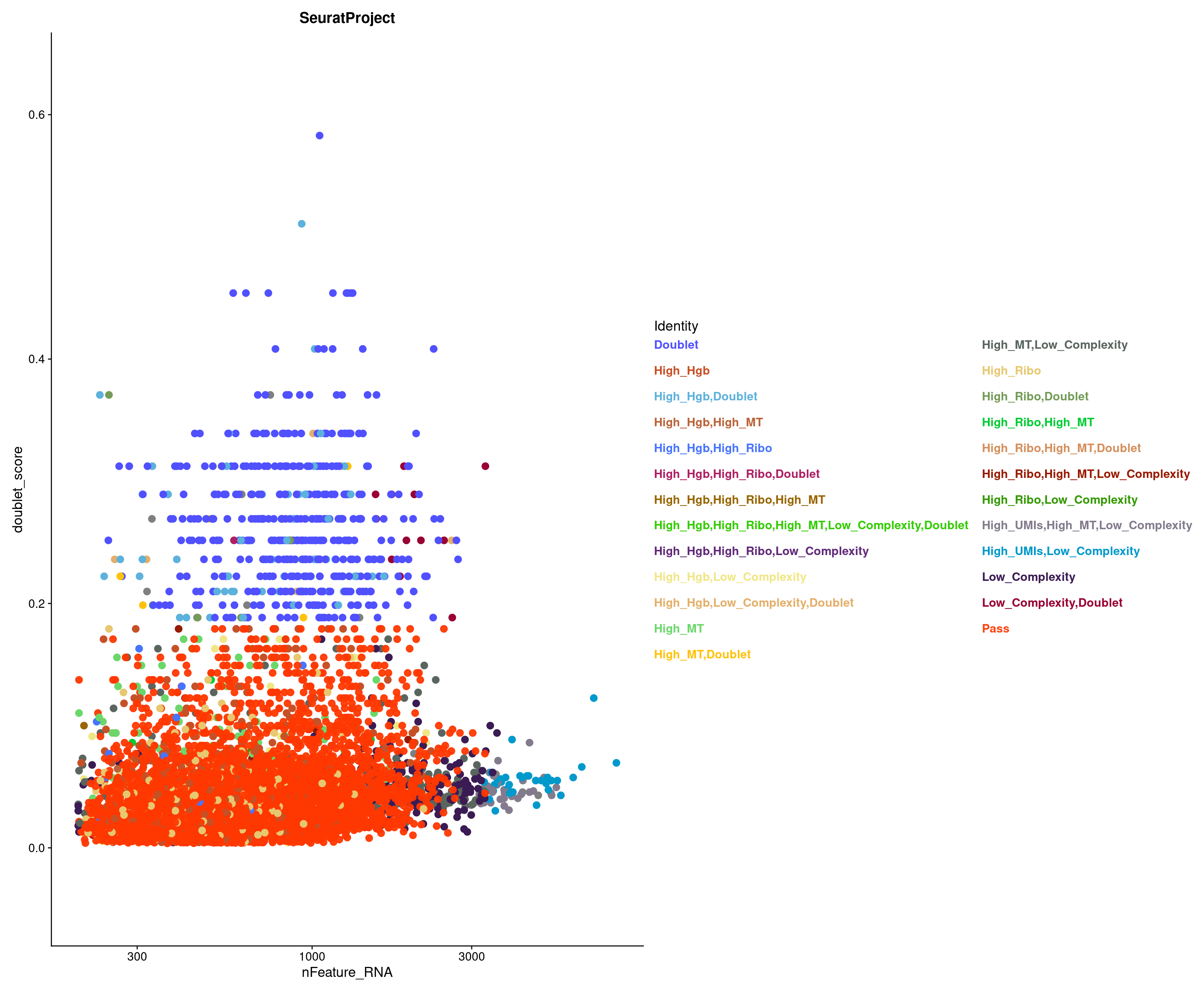
subset tdTomato
Idents(combined_srt) <- combined_srt$QC
DefaultAssay(combined_srt) <- "RNA"
cells <- WhichCells(combined_srt, idents = "Pass")
combined_srt <- subset(combined_srt, idents = "Pass")
Idents(combined_srt) <- combined_srt$Scgn_tdTomatoVisualize QC metrics as a violin plot again after subset
p1 <-
QC_Plots_Complexity(
seurat_object = combined_srt,
plot_median = TRUE,
color_seed = reseed,
ggplot_default_colors = TRUE
)
p2 <-
QC_Plots_Genes(
seurat_object = combined_srt,
low_cutoff = low_cutoff_gene,
high_cutoff = high_cutoff_gene,
plot_median = TRUE,
plot_title = "Genes per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p3 <-
QC_Plots_UMIs(
seurat_object = combined_srt,
low_cutoff = low_cutoff_umis,
high_cutoff = high_cutoff_umis,
plot_median = TRUE,
plot_title = "UMIs per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p4 <-
QC_Plots_Mito(
seurat_object = combined_srt,
high_cutoff = high_cutoff_pc_mt,
plot_median = TRUE,
plot_title = "Mito genes % per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p5 <-
QC_Plots_Feature(
seurat_object = combined_srt,
feature = "percent_ribo",
high_cutoff = high_cutoff_pc_ribo,
plot_median = TRUE,
y_axis_label = "% Ribosomal Genes Counts",
plot_title = "Ribo genes % per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p6 <-
QC_Plots_Feature(
seurat_object = combined_srt,
feature = "percent_hb",
high_cutoff = high_cutoff_pc_hb,
plot_median = TRUE,
y_axis_label = "% Hemoglobin Genes Counts",
plot_title = "Hemoglobin genes % per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
wrap_plots(p1, p2, p3, p4, p5, p6, ncol = 3)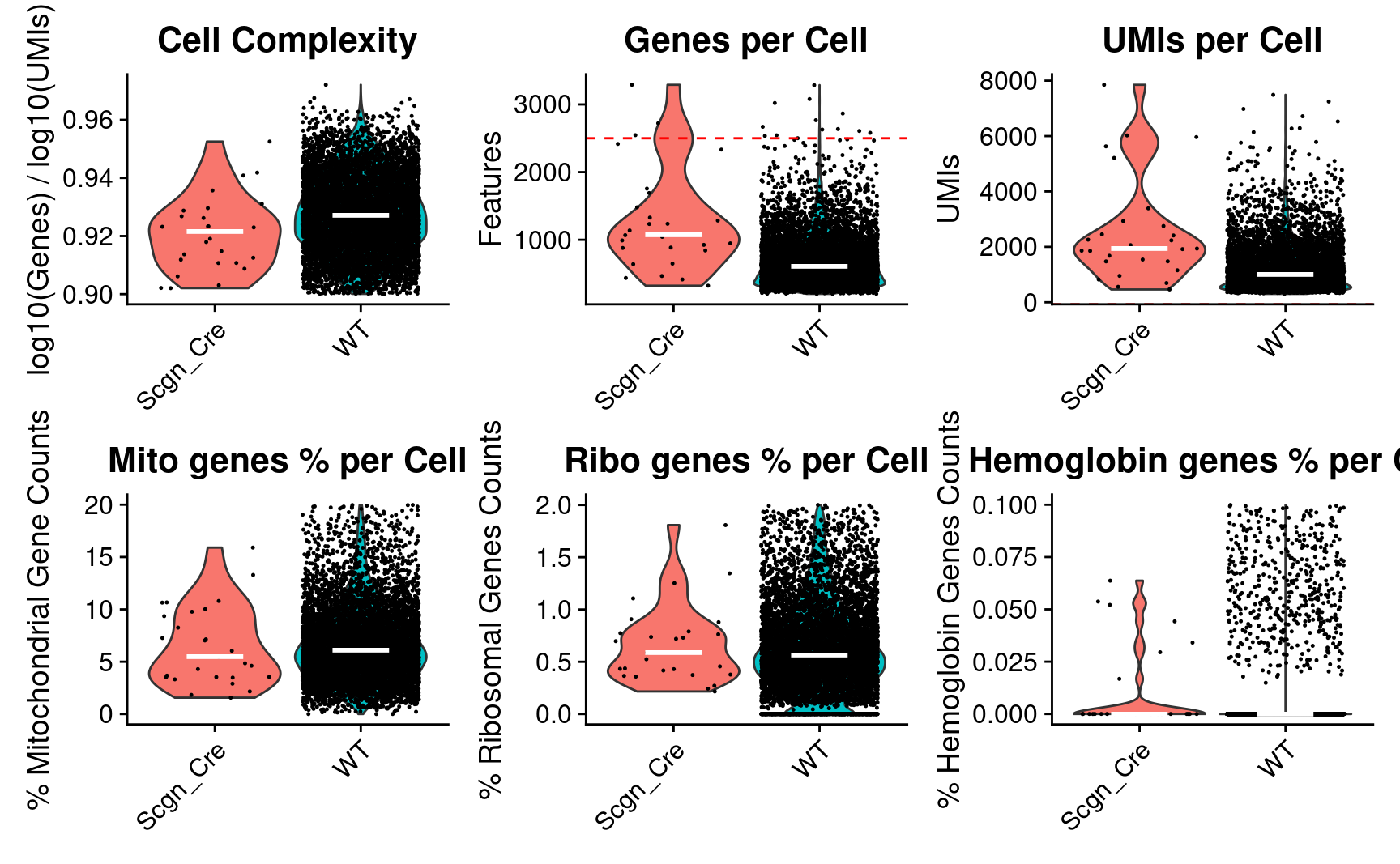
Reevaluate after subsetting low-quality cells
Apply SCTransform pipeline
plan("sequential")
invisible(gc())
set.seed(reseed)
plan(multisession, workers = n_cores)
# normalize and run dimensionality reduction on control dataset
npcs <- 100
metadata <- combined_srt@meta.data
rownames(metadata) <- colnames(combined_srt)
combined_srt <-
SCTransform(
combined_srt,
vst.flavor = "v2",
ncells = ncol(combined_srt),
variable.features.n = 5000,
vars.to.regress = c(
"log10GenesPerUMI",
"percent_mito_ribo"
),
return.only.var.genes = FALSE,
seed.use = reseed,
verbose = FALSE
)
hvg <- VariableFeatures(combined_srt)
var_regex <- "^Hla-|^Ig[hjkl]|^Rna|^mt-|^Rp[sl]|^Hb[^(p)]|^Gm"
hvg <- hvg[str_detect(pattern = var_regex, string = hvg, negate = TRUE)]
keep_genes <-
c("tdTomato", "Scgn", gene_int, hvg) %>%
unique() %>%
.[!. %in% housekeeping_mouse] %>%
.[!. %in% sex_genes] %>%
.[!. %in% stress_genes]
glimpse(keep_genes) chr [1:5981] "tdTomato" "Scgn" "Adcyap1r1" "Avpr1a" "Calcr" "Calcrl" ...out_of_hvg <- keep_genes[!keep_genes %in% hvg]
kable_material(
kable(out_of_hvg, "html"),
bootstrap_options = c(
"bordered",
"condensed",
"responsive",
"striped"
),
position = "left",
font_size = 14
)| x |
|---|
| tdTomato |
| Scgn |
| Avpr1a |
| Calcr |
| Cckar |
| Cckbr |
| Crhr1 |
| Crhr2 |
| Esr1 |
| Galr1 |
| Galr2 |
| Ghrhr |
| Ghsr |
| Glp1r |
| Gpr55 |
| Gpr83 |
| Gpr149 |
| Grpr |
| Hcrtr1 |
| Hcrtr2 |
| Insrr |
| Lepr |
| Mc1r |
| Mc3r |
| Mc4r |
| Mchr1 |
| Nmbr |
| Nmur1 |
| Nmur2 |
| Npffr2 |
| Npr1 |
| Npr2 |
| Npr3 |
| Npsr1 |
| Npsr2 |
| Npy1r |
| Npy2r |
| Npy5r |
| Ntsr1 |
| Oprd1 |
| Oprk1 |
| Oprl1 |
| Oprm1 |
| Oxtr |
| Prlhr |
| Prlr |
| Prokr2 |
| Qrfpr |
| Rxfp1 |
| Rxfp2 |
| Sstr1 |
| Sstr2 |
| Sstr3 |
| Tacr1 |
| Tacr3 |
| Trhr |
| Trhr2 |
| Tshr |
| Vipr1 |
| Vipr2 |
| Adcyap1 |
| Agrp |
| Avp |
| Bdnf |
| Cartpt |
| Cntf |
| Crh |
| Gal |
| Ghrh |
| Ghrl |
| Grp |
| Hcrt |
| Kiss1 |
| Lep |
| Nmb |
| Nms |
| Nmu |
| Npvf |
| Nts |
| Oxt |
| Pdyn |
| Pmch |
| Pnoc |
| Pomc |
| Qrfp |
| Rln1 |
| Rln3 |
| Sst |
| Tac2 |
| Trh |
| Adra1a |
| Adra1b |
| Adra1d |
| Adra2a |
| Adra2b |
| Adra2c |
| Adrb1 |
| Adrb2 |
| Adrb3 |
| Adrbk1 |
| Adrbk2 |
| Adora2b |
| Adora3 |
| Chrm1 |
| Chrm2 |
| Chrm4 |
| Chrm5 |
| Chrna1 |
| Chrna2 |
| Chrna3 |
| Chrna4 |
| Chrna5 |
| Chrna6 |
| Chrna7 |
| Chrna9 |
| Chrna10 |
| Chrnb2 |
| Chrnb3 |
| Chrnd |
| Chrng |
| Grin2d |
| Grin3b |
| Grm2 |
| Grm4 |
| Grm6 |
| Gabra1 |
| Gabra3 |
| Gabra5 |
| Gabra6 |
| Gabrg1 |
| Gabrg2 |
| Gabrd |
| Gabre |
| Gabrp |
| Gabrq |
| Gabrr1 |
| Gabrr2 |
| Gabrr3 |
| Drd1 |
| Drd3 |
| Drd4 |
| Drd5 |
| Htr1a |
| Htr1b |
| Htr1d |
| Htr1f |
| Htr2a |
| Htr2b |
| Htr3a |
| Htr3b |
| Htr4 |
| Htr5a |
| Htr5b |
| Htr6 |
| Htr7 |
| Gnao2 |
| Gnasxl |
| Gnb2 |
| Gng3 |
| Gng4 |
| Gng5 |
| Gng8 |
| Gng10 |
| Gng11 |
| Gng13 |
| Gngt1 |
| Gngt2 |
| P2rx1 |
| P2rx2 |
| P2rx3 |
| P2rx5 |
| P2rx6 |
| P2rx7 |
| P2ry1 |
| P2ry2 |
| P2ry4 |
| P2ry6 |
| P2ry12 |
| P2ry13 |
| P2ry14 |
| Ryr1 |
| Cnr1 |
| Cnr2 |
| Dagla |
| Daglb |
| Napepld |
| Trpv1 |
| Pparg |
| Ltk |
| Mdk |
| Fam150a |
| Fam150b |
| Sim1 |
| Slc2a3 |
| Drp1 |
| Mid49 |
| Mid51 |
| Ppargc1b |
| Nrf2 |
| Tfam |
| Prdx3 |
| Uqcrc2 |
| Cox4i2 |
| Bak1 |
| Bax |
| Bnip3 |
| Casp9 |
| Cybb |
| Gpx1 |
| Nfe2l2 |
| Nox4 |
| Polg |
| Polg2 |
| Pink1 |
| Park2 |
| Ogg1 |
| Mutyh |
| Sod3 |
| Ucp1 |
| Ucp2 |
| Ucp3 |
| Ucp4 |
| Ucp5 |
| Fgf21 |
| Klb |
| Bdh1 |
| Bdh2 |
| Hmgcs2 |
| Gck |
| Pkm |
| Pdha2 |
| Pdk1 |
| Pdk2 |
| Pdk4 |
| Cpt1a |
| Cpt1b |
| Cpt2 |
| Acsl4 |
| Acsl5 |
| Scd1 |
| Acaa1 |
| Acaa2 |
| Hadhb |
| Mc2r |
| Alx1 |
| Arid3a |
| Arid3b |
| Arid3c |
| Bcl3 |
| Barhl1 |
| Barx2 |
| Bmi1 |
| Ctbp2 |
| Cebpa |
| Cebpb |
| Cebpe |
| Cops2 |
| Cited1 |
| Cited2 |
| Cited4 |
| Dbp |
| Dpf2 |
| Ddx5 |
| Ddit3 |
| Ets2 |
| E2f1 |
| E2f2 |
| E2f4 |
| E2f5 |
| E2f6 |
| E2f7 |
| Elf3 |
| Elf4 |
| Elf5 |
| Elk1 |
| Eid2 |
| Ewsr1 |
| Fev |
| Fezf2 |
| Gabpa |
| Gata1 |
| Gata2 |
| Gata3 |
| Gata4 |
| Gata5 |
| Gata6 |
| Gli1 |
| Glis2 |
| Gsx1 |
| Gsx2 |
| Hmx2 |
| Hnf1b |
| Ikzf1 |
| Ikzf3 |
| Isl1 |
| Irx3 |
| Irx4 |
| Irx6 |
| Kat2a |
| Kat5 |
| Klf1 |
| Klf10 |
| Klf15 |
| Klf16 |
| Klf5 |
| Lhx1 |
| Lhx2 |
| Lhx3 |
| Lhx4 |
| Lmx1b |
| Mkl2 |
| Mybbp1a |
| Mycbp |
| Maz |
| Mdfi |
| Nkx3-1 |
| Nkx2-1 |
| Nkx2-3 |
| Nkx2-6 |
| Nkx2-9 |
| Nkx3-2 |
| Nkx6-1 |
| Nobox |
| Nanog |
| Nab2 |
| Paxip1 |
| Pou2af1 |
| Pou3f2 |
| Pou3f4 |
| Pou4f1 |
| Pou4f3 |
| Pou5f1 |
| Prdm1 |
| Rest |
| Rcor2 |
| Rrn3 |
| Spdef |
| Sertad1 |
| Smyd1 |
| Smad9 |
| Snw1 |
| Sox13 |
| Sox14 |
| Sox15 |
| Sox17 |
| Sox21 |
| Sox3 |
| Smarca1 |
| Smarcd2 |
| Smarce1 |
| Sp7 |
| Spib |
| Spic |
| Tal1 |
| Tal2 |
| Tlx1 |
| Tbx1 |
| Tbx19 |
| Tbx2 |
| Tbx20 |
| Tbx21 |
| Tbx22 |
| Tbx3 |
| Tbx5 |
| Tbx6 |
| Tbr1 |
| Tbpl1 |
| Taf10 |
| Taf11 |
| Taf12 |
| Taf4b |
| Taf6 |
| Taf7l |
| Taf8 |
| Taf1a |
| Taf1c |
| Tead2 |
| Tead3 |
| Tead4 |
| Tgif1 |
| Tgif2lx1 |
| Traf4 |
| Txk |
| Snrpc |
| Wt1 |
| Xbp1 |
| Ascl1 |
| Ascl3 |
| Atf5 |
| Aes |
| Ar |
| Arx |
| Ahr |
| Atoh1 |
| Atoh7 |
| Atoh8 |
| Aire |
| Relb |
| Bhlhe41 |
| Blzf1 |
| Batf |
| Bmp4 |
| Bmp7 |
| T |
| Abl1 |
| Creb3l3 |
| Creb3l4 |
| Camta2 |
| Cic |
| Cdx1 |
| Cdx2 |
| Cdc6 |
| Creg1 |
| Clock |
| Ciita |
| Crx |
| Crxos |
| Cbfa2t3 |
| Cdk9 |
| Cdkn1a |
| Cdkn2a |
| Ddb2 |
| Dmbx1 |
| Dlx2 |
| Dlx3 |
| Dlx4 |
| Dlx5 |
| Dlx6 |
| Egr2 |
| Egr3 |
| Egr4 |
| Emx1 |
| Emx2 |
| En1 |
| En2 |
| Ezh1 |
| Eomes |
| Esr2 |
| Esrra |
| Esrrb |
| Ehf |
| Etv3 |
| Etv4 |
| Evx2 |
| Esx1 |
| Figla |
| Foxa1 |
| Foxa2 |
| Foxc1 |
| Foxd1 |
| Foxd2 |
| Foxd4 |
| Foxe1 |
| Foxf1 |
| Foxf2 |
| Foxg1 |
| Foxh1 |
| Foxi1 |
| Foxk1 |
| Foxl1 |
| Foxl2 |
| Foxm1 |
| Foxo4 |
| Foxo6 |
| Foxp3 |
| Fosl2 |
| Fhl2 |
| Fus |
| Gbx2 |
| Gtf2h2 |
| Gtf2h4 |
| Gtf2a1l |
| Gtf3a |
| Gcm1 |
| Gcm2 |
| Gmeb1 |
| Gsc |
| Grhl1 |
| Grhl3 |
| Gadd45a |
| Gfi1 |
| Hes2 |
| Hes3 |
| Hes5 |
| Hes6 |
| Hes7 |
| Hey1 |
| Hand1 |
| Hand2 |
| Hsbp1 |
| Hsf4 |
| Helt |
| Hhex |
| Hnf4a |
| Hnrnph2 |
| Hnrnpu |
| Hmga1 |
| Hdac6 |
| Hoxa1 |
| Hoxa10 |
| Hoxa11 |
| Hoxa2 |
| Hoxa5 |
| Hoxa7 |
| Hoxa9 |
| Hoxb1 |
| Hoxb13 |
| Hoxb2 |
| Hoxb3 |
| Hoxb4 |
| Hoxb5 |
| Hoxb7 |
| Hoxb8 |
| Hoxb9 |
| Hoxc10 |
| Hoxc11 |
| Hoxc4 |
| Hoxc6 |
| Hoxd1 |
| Hoxd10 |
| Hoxd12 |
| Hoxd13 |
| Hoxd3 |
| Hoxd4 |
| Hoxd8 |
| Hoxd9 |
| Hesx1 |
| Hcfc1 |
| Hic1 |
| Id3 |
| Isl2 |
| Itgb3bp |
| Ifi204 |
| Irf3 |
| Irf5 |
| Irf7 |
| Irf8 |
| Irf9 |
| Jade1 |
| Lbx1 |
| Lztfl1 |
| Lcor |
| Maml1 |
| Mnt |
| Med12 |
| Med17 |
| Mn1 |
| Meox1 |
| Meox2 |
| Mesp2 |
| Mta1 |
| Mta2 |
| Mapk7 |
| Msx1 |
| Msx2 |
| Msc |
| Myb |
| Mybl1 |
| Mybl2 |
| Myc |
| Mzf1 |
| Mllt1 |
| Mllt11 |
| Mef2b |
| Myf5 |
| Myf6 |
| Myog |
| Nhlh1 |
| Nhlh2 |
| Neurod2 |
| Neurod4 |
| Neurod6 |
| Neurog1 |
| Neurog3 |
| Npas1 |
| Npas4 |
| Notch3 |
| Noto |
| Sp100 |
| Nfatc1 |
| Nfatc4 |
| Nfkb2 |
| Nfkbie |
| Nfe2 |
| Nfil3 |
| Nufip1 |
| Gm6740 |
| Ncor1 |
| Ncoa4 |
| Nrip3 |
| Nr0b1 |
| Nr1d1 |
| Nr1h3 |
| Nr1h4 |
| Nr1h5 |
| Nr1i2 |
| Nr1i3 |
| Nr2c1 |
| Nr2e1 |
| Nr2e3 |
| Nr2f6 |
| Nr5a1 |
| Nr5a2 |
| Nfya |
| Nfyb |
| Npm1 |
| Onecut1 |
| Onecut3 |
| Otx2 |
| Ovol1 |
| Ovol2 |
| Pax1 |
| Pax2 |
| Pax3 |
| Pax4 |
| Pax5 |
| Pax6 |
| Pax7 |
| Pax8 |
| Pax9 |
| Prop1 |
| Prrx1 |
| Prrx2 |
| Prrxl1 |
| Phox2b |
| Pitx1 |
| Pitx2 |
| Pitx3 |
| Ptf1a |
| Pdx1 |
| Per2 |
| Ppara |
| Pgs1 |
| Plag1 |
| Plagl1 |
| Plagl2 |
| Parp1 |
| Parp12 |
| Pabpn1 |
| Pcbp2 |
| Pcgf2 |
| Pqbp1 |
| Polr2k |
| Pbx4 |
| Pgr |
| Pdcd7 |
| Pml |
| Pin1 |
| Pias3 |
| Pcbd1 |
| Rbpjl |
| Rsl1 |
| Rfx1 |
| Rfx5 |
| Rfxank |
| Rfxap |
| Rhox11 |
| Rhox4e |
| Rhox5 |
| Rhox8 |
| Rel |
| Rax |
| Rbbp7 |
| Rbl1 |
| Rara |
| Rarg |
| Rxra |
| Rxrb |
| Rpl7 |
| Rpl7a |
| Ring1 |
| Rnf2 |
| Rnf25 |
| Rnf4 |
| Runx2 |
| Runx3 |
| Sall1 |
| Sall2 |
| Sall3 |
| Sall4 |
| Scx |
| Scrt1 |
| Srf |
| Scml2 |
| Sry |
| Stat4 |
| Stat5a |
| Stat6 |
| Six1 |
| Six2 |
| Six3 |
| Six4 |
| Sim2 |
| Sirt3 |
| Snapc4 |
| Snai1 |
| Snai2 |
| Snai3 |
| Spz1 |
| Spi1 |
| Sra1 |
| Srebf1 |
| Strn |
| Supt4a |
| Suv39h1 |
| Suv39h2 |
| Tdg |
| Thra |
| Tcea2 |
| Tceb2 |
| Tceb3 |
| Tcf12 |
| Tcf15 |
| Tcf7 |
| Tfap2b |
| Tfap2a |
| Tfap2e |
| Tfap4 |
| Tfdp1 |
| Tfe3 |
| Tfec |
| Tada3 |
| Tle2 |
| Tle6 |
| Trp53 |
| Trp63 |
| Mdm4 |
| Tgfb1i1 |
| Trib3 |
| Trim28 |
| Trim30a |
| Tsc2 |
| Twist1 |
| Twist2 |
| Utf1 |
| Mafb |
| Maff |
| Mafg |
| Mafk |
| Mycn |
| Rela |
| Vav1 |
| Vax1 |
| Vgll2 |
| Vsx2 |
| Vdr |
| Vhl |
| Zrsr1 |
| Zc3h8 |
| Zbtb14 |
| Zbtb17 |
| Zbtb7b |
| Zscan21 |
| Zfp110 |
| Zfp239 |
| Zfp263 |
| Zfp354a |
| Zfp422 |
| Zfp467 |
| Zfp472 |
| Zfp503 |
| Zfp60 |
| Zfp64 |
| Zfp68 |
| Zik1 |
| Zic2 |
| Zic3 |
| Zic5 |
| Zmynd10 |
| Ahrr |
| Aip |
| Alx3 |
| Alx4 |
| Atbf1 |
| Neurog2 |
| Hnrpd |
| Bapx1 |
| Bard1 |
| Bcl6b |
| Bnc1 |
| Brca1 |
| C2ta |
| Catnb |
| Cbfa2t1h |
| Cbx2 |
| Ccnk |
| Cdk2 |
| Cdk4 |
| Cdkn2b |
| Cdkn2c |
| Cdkn2d |
| Cdx4 |
| Chx10 |
| Cnbp1 |
| Crebl1 |
| Cutl1 |
| Cutl2 |
| Dbx1 |
| Pcbd |
| Ddef1 |
| Dlx1 |
| Dnmt1 |
| Dnmt3a |
| Drg1 |
| Ebf2 |
| Ercc2 |
| Ercc3 |
| Etsrp71 |
| Evi1 |
| Evx1 |
| Ewsh |
| Fusip1 |
| Fem1a |
| Fem1b |
| Fkhl18 |
| Foxb2 |
| Fxr1h |
| Gcn5l2 |
| Dsip1 |
| Gli |
| Glrp1 |
| Gsh1 |
| Gsh2 |
| Baz1a |
| Hcls1 |
| Hdgfrp2 |
| Hes1 |
| Hey2 |
| Foxj1 |
| Foxf1a |
| Hipk2 |
| Hlx |
| Hlxb9 |
| Hmgn2 |
| Hmg20b |
| Hmgb3 |
| Hmga2 |
| Hmx1 |
| Foxa3 |
| Hnf4 |
| Hnrpab |
| Hoxa11s |
| Hoxa13 |
| Hoxa4 |
| Hoxa6 |
| Hoxb6 |
| Hoxc12 |
| Hoxc13 |
| Hoxc5 |
| Hoxc8 |
| Hoxc9 |
| Hoxd11 |
| hr |
| Icsbp1 |
| Idb1 |
| Idb2 |
| Idb3 |
| Idb4 |
| Ifi16 |
| Ikbkg |
| Irf4 |
| Irx1 |
| Irx2 |
| Isgf3g |
| Jund1 |
| Bteb1 |
| Labx |
| Laf4 |
| Lbx1h |
| Lbx2h |
| Lhx5 |
| Lhx6 |
| Lhx8 |
| Lhx9 |
| Lmo2 |
| Lmyc1 |
| Zbtb7 |
| Lyl1 |
| Mad |
| Mad3 |
| Mad4 |
| Madh1 |
| Madh2 |
| Madh3 |
| Madh4 |
| Madh5 |
| Madh6 |
| Madh7 |
| Maf |
| Ascl2 |
| Mbd3 |
| Mcm3 |
| Mcm2 |
| Mcm4 |
| Mcm5 |
| Mcm6 |
| Mcm7 |
| Rab8a |
| Mist1 |
| Miz1 |
| Mllt10 |
| Mllt2h |
| Mpl |
| Mrg1 |
| Mrg2 |
| Msl31 |
| Msx3 |
| Mycs |
| Naca |
| Ndn |
| Nedd8 |
| Nfatc2ip |
| Nfkbib |
| Nfkbil1 |
| Nkx2-2 |
| Nmyc1 |
| Notch4 |
| Uhrf1 |
| Musk |
| Og2x |
| Og9x |
| Orc2l |
| Otp |
| Otx1 |
| Pcaf |
| Ipf1 |
| Pem |
| Pit1 |
| Papola |
| Pou2f3 |
| Pou3f-rs1 |
| Pou4f-rs1 |
| Pou4f2 |
| Pparbp |
| Ppp5c |
| Mapk8ip |
| Psmc3 |
| Psx1 |
| Ptma |
| Rbpsuh |
| Rbpsuhl |
| Recc1 |
| Rnf12 |
| Rorc |
| Rpo1-1 |
| Rpo1-2 |
| Rpo1-3 |
| Rpo1-4 |
| Polr2c |
| Polr2j |
| Rpo2tc1 |
| Trim30 |
| Ruvbl2 |
| Rxrg |
| Nkx1-1 |
| Sfpi1 |
| Sfrs5 |
| Shox2 |
| Siah2 |
| Six6 |
| Skd3 |
| Smarca3 |
| Ighmbp2 |
| Jarid1c |
| Sox12 |
| Sox19 |
| Srst |
| Bhlhb2 |
| Strm |
| Supt4h |
| Supt4h2 |
| Supt5h |
| Tbx13 |
| Tbx14 |
| Tbx15 |
| Tbx4 |
| Tcea3 |
| Tcf1 |
| Tcf2 |
| Tcf21 |
| Zfhx1a |
| Tcfap2a |
| Tcfap2b |
| Tcfap2c |
| Tcfcp2 |
| Tcfe2a |
| Tcfeb |
| Tcfec |
| Tcfl1 |
| Tcfl4 |
| Tgfb1i4 |
| Tgif |
| Thrsp |
| Tieg1 |
| Titf1 |
| Tnfaip3 |
| Trip6 |
| Trp73 |
| Ube1c |
| Uncx4.1 |
| Wbp7 |
| Nsep1 |
| Zfa |
| Zfp1 |
| Zfp100 |
| Zfp13 |
| Rnf110 |
| Zfp161 |
| Zfp162 |
| Zfp2 |
| Zfp27 |
| Zfp29 |
| Zfp35 |
| Zfp37 |
| Zipro1 |
| Zfp40 |
| Zfp42 |
| Zfp46 |
| Zfp51 |
| Zfp52 |
| Zfp57 |
| Zfp59 |
| Zfp9 |
| Zfp92 |
| Zfp93 |
| Zfp94 |
| Zfp95 |
| Zfp96 |
| Zfp97 |
| Zim1 |
| Zfpn1a1 |
| Zfpn1a2 |
| Zfpn1a3 |
| Ash2l |
| Bpnt1 |
| Copeb |
| Creg |
| Gcl |
| Nr0b2 |
| Thrap4 |
| Vax2 |
| Whsc2 |
| Zfhx1b |
| AW210570 |
| Map3k12 |
| Zfp146 |
| Irebf1 |
| Cops5 |
| Zfp275 |
| Tlx3 |
| Ing4 |
| Zfp385 |
| Neud4 |
| Pdlim4 |
| Fbxl10 |
| Foxe3 |
| Zfp238 |
| Hnf4g |
| Zfp354c |
| Abt1 |
| Cbx8 |
| Sirt6 |
| Nrbf2 |
| Lsm4 |
| Dmrt1 |
| Solh |
| Keap1 |
| Nsbp1 |
| Ppp2r1a |
| D19Ertd675e |
| D1Ertd161e |
| D15Ertd417e |
| D11Ertd530e |
| Hrb2 |
| D12Ertd748e |
| Zfp535 |
| D11Bwg0517e |
| Psmd10 |
| Htatip2 |
| Insm1 |
| Rab25 |
| Deaf1 |
| Pdlim1 |
| Irf6 |
| Myst4 |
| Irx5 |
| Hils1 |
| Lrrc6 |
| Mllt7 |
| Zfp108 |
| Madh9 |
| D1Bwg0491e |
| Ttrap |
| Heyl |
| Zfp278 |
| Zfp386 |
| Hdac7a |
| Nupr1 |
| Zfp113 |
| Mint |
| Csda |
| Zfp288 |
| Gbif |
| Ruvbl1 |
| Papolb |
| Pmfbp1 |
| Zfp235 |
| Rps6ka4 |
| Ankrd2 |
| Zfp111 |
| Garnl1 |
| Insm2 |
| Zfp109 |
| Hcngp |
| Th1l |
| Ehox |
| Zfp112 |
| Fhl5 |
| Hic2 |
| A730008L03Rik |
| Rog |
| Wbscr14 |
| Piasy |
| Rab2 |
| Tnrc11 |
| Ureb1 |
| Carm1 |
| Zfp191 |
| Bhlhb5 |
| Nrip2 |
| Sap30 |
| Gas41 |
| Sp5 |
| D10Jhu82e |
| Nmi |
| Asb1 |
| Asb4 |
| Asb2 |
| 2300009P13Rik |
| 1110005A23Rik |
| Znrd1 |
| Crsp9 |
| 3100002L24Rik |
| Polr2e |
| Polr2l |
| 5730410I19Rik |
| Mcm8 |
| 4733401N12Rik |
| 4921520G13Rik |
| 6130401J04Rik |
| 1200013F24Rik |
| 2610016F04Rik |
| Fank1 |
| 2310043K02Rik |
| 2400009B11Rik |
| Polr3d |
| Nrarp |
| Psmd9 |
| Pfdn1 |
| Zfp99 |
| Lass4 |
| 3110004H13Rik |
| 3110031B13Rik |
| 1810007M14Rik |
| Zfp606 |
| Dedd2 |
| Vdrip |
| 1110035L05Rik |
| 3632451O06Rik |
| Xab2 |
| 4930430A15Rik |
| Rabl3 |
| 1700020N01Rik |
| Polr2g |
| Sec14l2 |
| 2410018C20Rik |
| Mki67ip |
| Ssxb1 |
| Nudt12 |
| 3110024A21Rik |
| Gtf2e2 |
| Sirt5 |
| Phf5a |
| Ankra2 |
| 1110033I14Rik |
| 1110054N06Rik |
| Tulp4 |
| Asb11 |
| 1190004M21Rik |
| 1500031N24Rik |
| Cnot8 |
| 1810037G04Rik |
| Mll5 |
| 2810407K09Rik |
| 2610028L19Rik |
| Ing1l |
| Asb9 |
| 1700001F22Rik |
| 1700014N06Rik |
| 2300002D11Rik |
| 2310020P08Rik |
| 2310042L19Rik |
| Trip13 |
| Thrap6 |
| 1810060D16Rik |
| Zfp219 |
| Polr2i |
| 2810021J22Rik |
| 1700030J15Rik |
| 3010019O03Rik |
| 2610303A01Rik |
| 5730461K03Rik |
| Crsp7 |
| 5730521P14Rik |
| 4631416I11Rik |
| Hmgb2l1 |
| 4921509B22Rik |
| 4931423N10Rik |
| Rnf134 |
| Zfp597 |
| 4933416E05Rik |
| 4933426I21Rik |
| Fbxo24 |
| Dmrtc2 |
| 4933429H19Rik |
| Ches1 |
| Obox1 |
| Pogk |
| 9130012O13Rik |
| 1300004C11Rik |
| 1300019N10Rik |
| Ing3 |
| 1110001J12Rik |
| 1110020M19Rik |
| Gtf2a1lf |
| Zfp297b |
| Phf7 |
| 1700012B18Rik |
| 2310076O14Rik |
| Lass5 |
| Ddx54 |
| Ckn1 |
| Phf10 |
| Harp |
| 2600014C22Rik |
| 2810405K07Rik |
| 2610524B01Rik |
| Mms19l |
| Lsm11 |
| Nkd2 |
| Asb6 |
| 2410004N05Rik |
| Ankrd3 |
| 2600017A12Rik |
| Zfp131 |
| 2700043M03Rik |
| 2700067D09Rik |
| Zfp248 |
| B3Gat3 |
| Skz1 |
| 2810455B10Rik |
| 2900054J07Rik |
| 1700065O13Rik |
| Mbd3l1 |
| 1700123A16Rik |
| 1700123J19Rik |
| 4933409K03Rik |
| 4933417L02Rik |
| D8Ertd69e |
| Gasz |
| 1200006M05Rik |
| 1300003B13Rik |
| Zdhhc16 |
| Gtf2e1 |
| 1700086D15Rik |
| Hod |
| Cxxc1 |
| Hnrpr |
| Xrcc3 |
| Zfp84 |
| Hsf2bp |
| 4933430F08Rik |
| Zfp336 |
| 9130417I07Rik |
| 9130423L19Rik |
| Narg1 |
| 4930532L20Rik |
| 4930539I12Rik |
| Zbtb3 |
| 4930564N15Rik |
| 4930548G07Rik |
| Sirt4 |
| Hspb9 |
| 1700013G10Rik |
| 2010005A06Rik |
| Jarid1b |
| 4932409F11Rik |
| Mitc1 |
| Zfp198 |
| 5830417I10Rik |
| Asb5 |
| 1110011F09Rik |
| Tbx18 |
| 1700012M14Rik |
| Mrsb |
| 1700012H05Rik |
| 2210008I11Rik |
| Snip1 |
| 2410141K09Rik |
| Jmjd2c |
| 2810438M17Rik |
| Lass2 |
| Ssbp4 |
| 4921524J06Rik |
| 6030426L16Rik |
| 6720457D02Rik |
| Zfp142 |
| 9430034D17Rik |
| C030011J08Rik |
| C330002I19Rik |
| 9230102N17Rik |
| 9230110K08Rik |
| A030003K21Rik |
| 4930522L14Rik |
| 5730467H21Rik |
| 2410141M05Rik |
| 4921504N20Rik |
| 1700090G07Rik |
| 9530049C15Rik |
| C330022B21Rik |
| C330013J21Rik |
| Brpf1 |
| 0610009M14Rik |
| Asb15 |
| Sp2 |
| 9130019O22Rik |
| Polr3h |
| Zfp319 |
| Bhlhb3 |
| Abtb1 |
| 2210021A15Rik |
| AA408868 |
| Zfp202 |
| Htatip |
| Zfp297 |
| Jundm2 |
| Bat4 |
| Tcfcp2l1 |
| Sp6 |
| Lin28 |
| Cml3 |
| Zfp192 |
| Rbm9 |
| Sirt1 |
| Wdr9 |
| Cecr6 |
| Pcqap |
| Ptges2 |
| 2310058J06Rik |
| 2810405L04Rik |
| Taf13 |
| Hkr3 |
| AF013969 |
| Aprin |
| AW538212 |
| Tada3l |
| 2310058A11Rik |
| Hfh7 |
| Zfp119 |
| Psx2 |
| Rab15 |
| C730024G01Rik |
| C330003B14Rik |
| AI481750 |
| Tcf19 |
| A630042L21Rik |
| D330024H06Rik |
| Gsh5 |
| Myt2 |
| 8030445B08Rik |
| Zfp339 |
| Ankrd1 |
| Bmyc |
| Nkx2-7 |
| Surb7 |
| Zdhhc15 |
| Supt3h |
| Tbx10 |
| Lmo1 |
| Zfp98 |
| Hfh5 |
| Hfh6 |
| Hfh3 |
| Tcfap4 |
| Gsh3 |
| Dbx2 |
| Nol1 |
| Bat8 |
| Sprm1 |
| Mjd |
| Hoxb3s |
| Nztf2 |
| Sdccag33 |
| Pou5f1-rs10 |
| Mop3 |
| Zfp295 |
| Brdt |
| Pawr |
| Foxn4 |
| Sox16 |
| Otx3 |
| Centb5 |
| Rem2 |
| Zfp287 |
| Rnpc2 |
| Zfp617 |
| Clp1 |
| Bhc80 |
| MGC39058 |
| Jmjd2b |
| Gscl |
| A930021G21Rik |
| 9030612M13Rik |
| Kbtbd9 |
| Sirt7 |
| D5Ertd679e |
| DXImx41e |
| Tcfe3 |
| Mlr1 |
| 9430065N20Rik |
| 6720489N17Rik |
| Zswim4 |
| BC055310 |
| Mlr2 |
| A830025F02Rik |
| Centg3 |
| Sox30 |
| Mll |
| Myocd |
| Jarid1a |
| Thrap5 |
| Cart1 |
| Gls2 |
| 9630006B20Rik |
| Tada2l |
| Myst2 |
| 6030408C04Rik |
| D130006K24Rik |
| 6430502M16Rik |
| Btf3 |
| Glmr |
| 6720456H20Rik |
| F830020C16Rik |
| C730048E16Rik |
| Mkl1 |
| ORF63 |
| 6720480D16Rik |
| Zfp523 |
| Hszfp36 |
| AI255170 |
| AU018122 |
| Fbxl11 |
| Dmrt2 |
| Taf5 |
| Zfp281 |
| Elys |
| Tcfap2d |
| Pms1 |
| XPMC2H |
| 5830435C13Rik |
| Zdhhc5 |
| Ebf4 |
| Nkx2-4 |
| 6820402O20Rik |
| Tgif2 |
| Zfp334 |
| 3632413B07Rik |
| Taf4a |
| D3Jfr1 |
| Ddx58 |
| Zfp189 |
| Glis1 |
| Jmjd2a |
| C330039G02Rik |
| B930041F14Rik |
| Gbx1 |
| Mll3 |
| A730098D12Rik |
| AI591476 |
| LOC232337 |
| 5730403M16Rik |
| A230102I05Rik |
| Hkr2 |
| 6430596G11Rik |
| 6330581L23Rik |
| Zf |
| Zfp553 |
| BC026432 |
| LOC233987 |
| Zfp612 |
| Crsp6 |
| Zfp426 |
| Ankrd25 |
| BC005471 |
| Zfp145 |
| 6230410P16Rik |
| AW610627 |
| BC021921 |
| Tceal1 |
| BC024063 |
| G431002C21Rik |
| Zfp454 |
| 3526402J09Rik |
| Homez |
| 6030449J21 |
| 9930016F01Rik |
| 6030490I01Rik |
| BC031441 |
| Fem1c |
| BC024969 |
| 6430585N13Rik |
| Dmrt3 |
| Carf |
| Lass6 |
| D430039N05Rik |
| Dmrta1 |
| Dmrta2 |
| E130309B19Rik |
| 4732429I09Rik |
| Zfp537 |
| D030014N22Rik |
| Hip14l |
| Myst3 |
| D230022C05Rik |
| Bsx |
| 6030424L22Rik |
| Tgifx1 |
| D030011N01Rik |
| Obox3 |
| Obox5 |
| 4921515A04Rik |
| G431001E03Rik |
| Zbtb1 |
| 5730589K01Rik |
| Gtf3c4 |
| Cyln2 |
| A630035I11Rik |
| Gcdh |
| B430306D02Rik |
| BC024139 |
| A730032D07Rik |
| Zfp398 |
| Tcfl5 |
| Ankrd5 |
| 4921509E05Rik |
| 6330583I20Rik |
| A830014H24Rik |
| 5830403E09Rik |
| E430039K05Rik |
| 6330416L07Rik |
| 4932422E22Rik |
| D130026O16Rik |
| D830019J24Rik |
| B930011H20Rik |
| Lba1 |
| A630089N07Rik |
| A730019I05Rik |
| A830023I12Rik |
| Rfxdc1 |
| A730012O14Rik |
| AW146020 |
| Zfp82 |
| Tcfap2e |
| D030022P06Rik |
| Tieg2 |
| 2810021G02Rik |
| 9130211I03Rik |
| Mzf6d |
| Cri2 |
| Abcd1 |
| Abcd2 |
| Aldh1l1 |
| App |
| Caf4 |
| Ccl25 |
| Col4a5 |
| Decr2 |
| Ech1 |
| Enho |
| Hbegf |
| Hif1 |
| Igsf1 |
| Lamb2 |
| Lcat |
| Lgi4 |
| Lrpap1 |
| Lxn |
| Mdv1 |
| Mief1 |
| Ntn1 |
| Pex10 |
| Pex12 |
| Pex16 |
| Pex2 |
| Pex26 |
| Pex6 |
| Psap |
| Rtn4 |
| S100a1 |
| S100a6 |
| Slitrk2 |
| Chat |
| Slc18a3 |
| Ache |
| Slc5a7 |
| Gnaz |
| Th |
| Slc6a3 |
| Slc18a2 |
| Slc17a8 |
| Slc1a6 |
| Slc32a1 |
| Mbnl3 |
| Pgf |
| Irs4 |
| Gpr101 |
| Agtr1 |
hvg <- hvg[hvg %in% keep_genes]
combined_srt <- combined_srt %>%
RunPCA(features = keep_genes, npcs = npcs, seed.use = reseed, verbose = FALSE)source(here(src_dir, "genes.R"))
npr %<>% .[. %in% rownames(GetAssayData(combined_srt, slot = "scale.data"))]
np %<>% .[. %in% rownames(GetAssayData(combined_srt, slot = "scale.data"))]
neurotrans %<>% .[. %in% rownames(GetAssayData(combined_srt, slot = "scale.data"))]
glut %<>% .[. %in% rownames(GetAssayData(combined_srt, slot = "scale.data"))]
gaba %<>% .[. %in% rownames(GetAssayData(combined_srt, slot = "scale.data"))]
dopam %<>% .[. %in% rownames(GetAssayData(combined_srt, slot = "scale.data"))]
ach %<>% .[. %in% rownames(GetAssayData(combined_srt, slot = "scale.data"))]
genes.embed %<>% .[. %in% rownames(GetAssayData(combined_srt, slot = "scale.data"))]Derive dimensional reductions and clusters of filtered dataset
print(combined_srt[["pca"]], dims = 1:5, nfeatures = 5)PC_ 1
Positive: Plp1, Pcdh9, Tmeff2, Mbp, Pde4b
Negative: Rbfox1, Meg3, Cntnap2, Celf2, Nrg3
PC_ 2
Positive: Plp1, Rbfox1, Pcdh9, Meg3, Nkain2
Negative: Slc1a2, Flt1, Atp1a2, Gpc5, Ptprg
PC_ 3
Positive: Flt1, Ptprg, Slco1a4, Mecom, Cldn5
Negative: Slc1a2, Gpc5, Lsamp, Nrxn1, Atp1a2
PC_ 4
Positive: Cfap299, Dnah6, Dnah12, Cfap46, Syne1
Negative: Slc1a2, Flt1, Gpc5, Lsamp, Slco1a4
PC_ 5
Positive: Rbfox1, Pde10a, Phactr1, Celf2, Grm5
Negative: Kcnma1, Nwd2, Asic2, Trpm3, Scube1 PCA gene loadings
VizDimLoadings(combined_srt, dims = 1:4, reduction = "pca")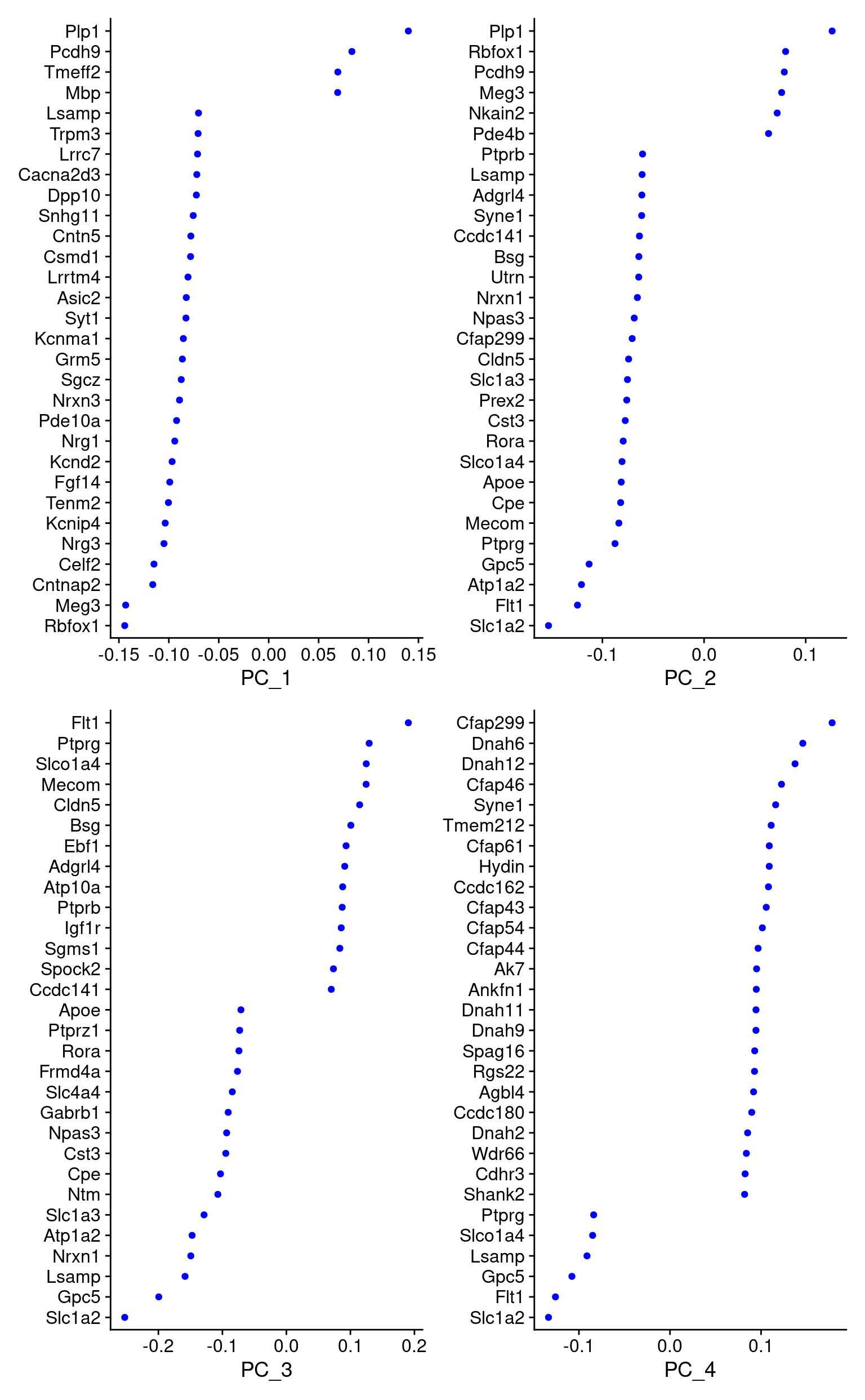
Heatmap
DimHeatmap(combined_srt, dims = 1:15, cells = 500, balanced = TRUE)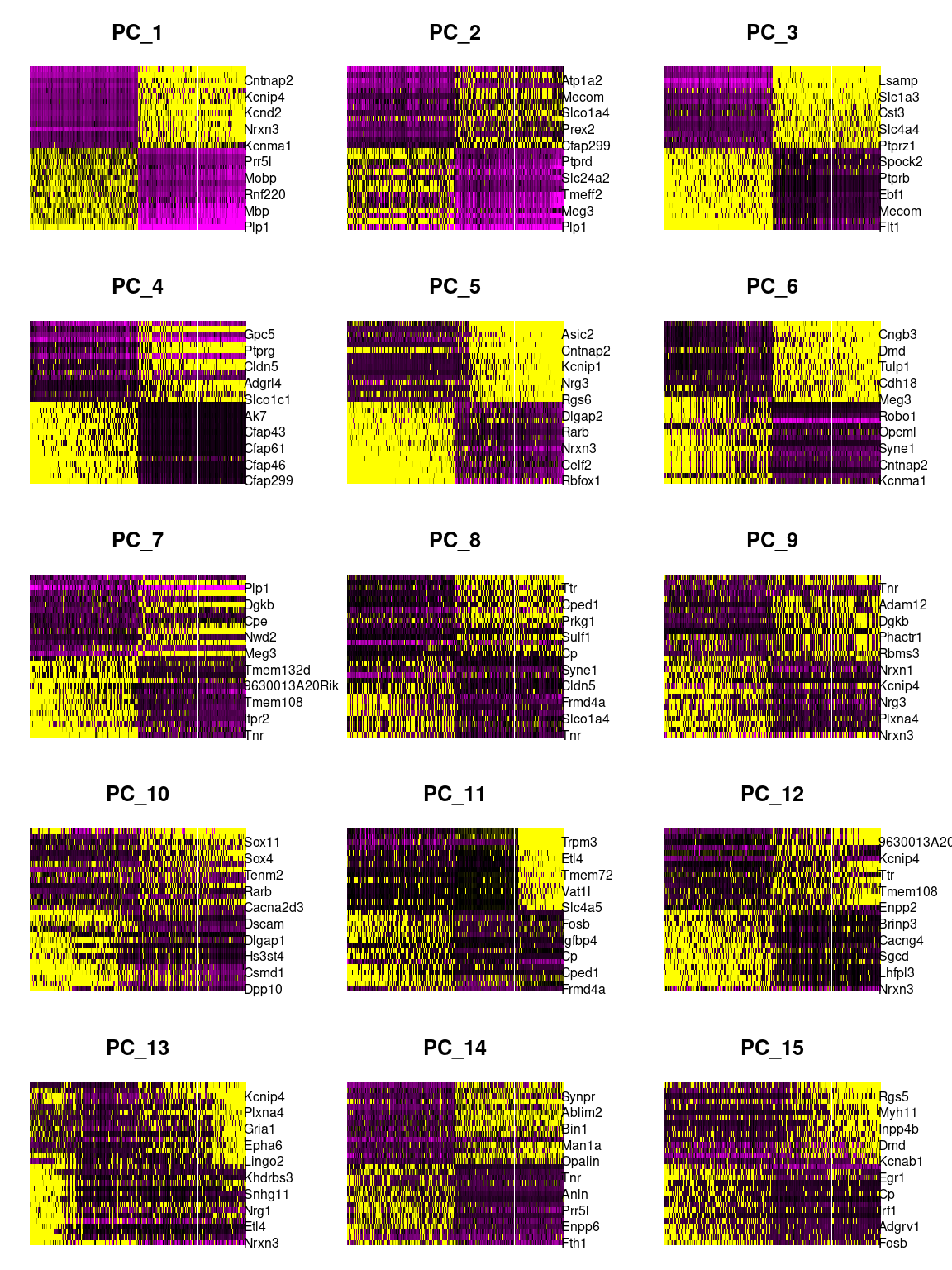
Elbow
ElbowPlot(combined_srt, ndims = npcs)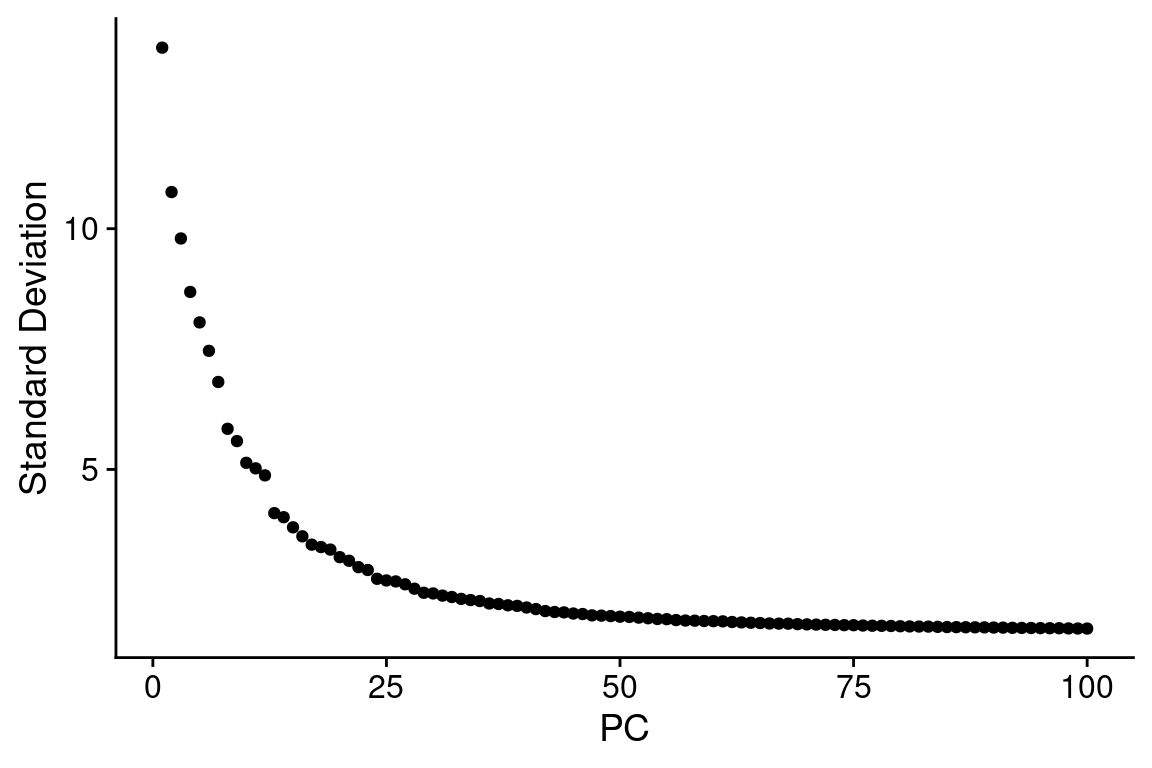
plan("sequential")
invisible(gc())
set.seed(reseed)
plan(multicore, workers = n_cores)
selected_pcs <-
seq_len(50)
if (!file.exists(here(data_dir, glue::glue("{project}-init/{project}-init-umap-search.Rds")))) {
umap_example <- scDEED(
input_data = combined_srt,
K = length(selected_pcs),
n_neighbors = seq(from = 15, to = 35, by = 10),
min.dist = c(0.01, 0.05, 0.1, 0.25, 0.5, 0.8),
reduction.method = "umap",
default_assay = "SCT"
)
dir.create(here(data_dir, sprintf("%s-init", project)))
readr::write_rds(
x = umap_example,
file = here(data_dir, glue::glue("{project}-init/{project}-init-umap-search.Rds"))
)
} else {
umap_example <-
read_rds(here(data_dir, glue::glue("{project}-init/{project}-init-umap-search.Rds")))
}plan(sequential)
invisible(gc())
set.seed(seed = reseed)
plan(multisession, workers = n_cores)
registerDoParallel(cores = n_cores)
if (!file.exists(here(data_dir, glue::glue("{project}-init/{project}-init-tsne-search.Rds")))) {
tsne_example <- scDEED(
combined_srt,
K = length(selected_pcs),
reduction.method = "tsne",
default_assay = "SCT"
)
dir.create(here(data_dir, sprintf("%s-init", project)))
readr::write_rds(
x = tsne_example,
file = here(data_dir, glue::glue("{project}-init/{project}-init-tsne-search.Rds"))
)
} else {
tsne_example <-
read_rds(here(data_dir, glue::glue("{project}-init/{project}-init-tsne-search.Rds")))
}plan(sequential)
invisible(gc())
set.seed(seed = reseed)
plan(multisession, workers = n_cores)
combined_srt <-
combined_srt |>
FindNeighbors(
dims = selected_pcs,
k.param = umap_example$num_dubious |>
dplyr::slice_min(order_by = number_dubious_cells, n = 1) |>
pull(n_neighbors),
annoy.metric = "euclidean",
n.trees = 100,
verbose = FALSE
) |>
RunUMAP(
dims = selected_pcs,
reduction.name = "umap",
reduction.key = "UMAP_",
return.model = FALSE,
umap.method = "uwot",
n.epochs = 1000L,
n.neighbors = umap_example$num_dubious |>
dplyr::slice_min(order_by = number_dubious_cells, n = 1) |>
pull(n_neighbors),
min.dist = umap_example$num_dubious |>
dplyr::slice_min(order_by = number_dubious_cells, n = 1) |>
pull(min.dist),
seed.use = reseed,
verbose = FALSE
)
combined_srt <-
RunTSNE(
combined_srt,
reduction = "pca",
dims = selected_pcs,
seed.use = reseed,
reduction.name = "tsne",
reduction.key = "tSNE_",
perplexity = tsne_example$num_dubious |>
dplyr::slice_min(order_by = number_dubious_cells, n = 1) |>
pull(perplexity) |> as.integer()
)
pacmap <- reticulate::import("pacmap")
# Initialize PaCMAP instance
reducer <- pacmap$PaCMAP(
n_components = 2L,
MN_ratio = 0.5,
FP_ratio = 2.0,
apply_pca = FALSE
)
# Perform dimensionality Reduction
pacmap_embedding <-
reducer$fit_transform(Embeddings(combined_srt[["pca"]])[, selected_pcs])
colnames(pacmap_embedding) <- paste0("PaCMAP_", 1:2)
rownames(pacmap_embedding) <- colnames(combined_srt)
# We will now store this as a custom dimensional reduction called 'pacmap'
combined_srt[["pacmap"]] <-
CreateDimReducObject(
embeddings = pacmap_embedding,
key = "PaCMAP_",
assay = DefaultAssay(combined_srt)
)Mitochondrial genes expression
UMAP
FeaturePlot_scCustom(
combined_srt,
features = "percent_mito",
label.size = 4,
repel = TRUE,
pt.size = 1,
label = TRUE,
colors_use = combined_srt@misc$mdat_Colour_Pal,
order = TRUE,
alpha_na_exp = 0.1,
alpha_exp = 0.45
) &
theme(plot.title = element_text(size = 16))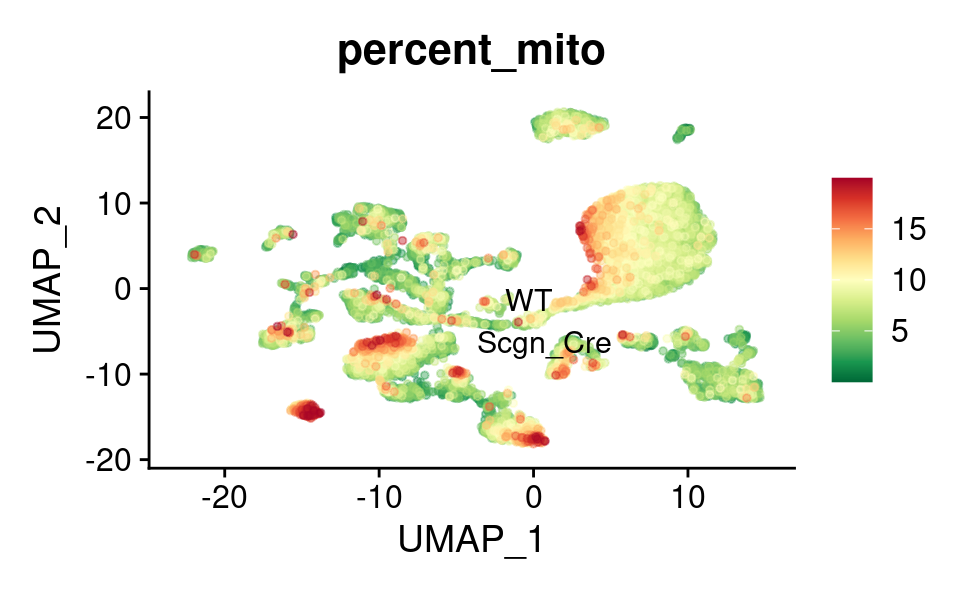
PaCMAP
FeaturePlot_scCustom(
combined_srt,
features = "percent_mito",
reduction = "pacmap",
label.size = 4,
repel = TRUE,
pt.size = 1,
label = TRUE,
colors_use = combined_srt@misc$mdat_Colour_Pal,
order = TRUE,
alpha_na_exp = 0.1,
alpha_exp = 0.45
) &
theme(plot.title = element_text(size = 16))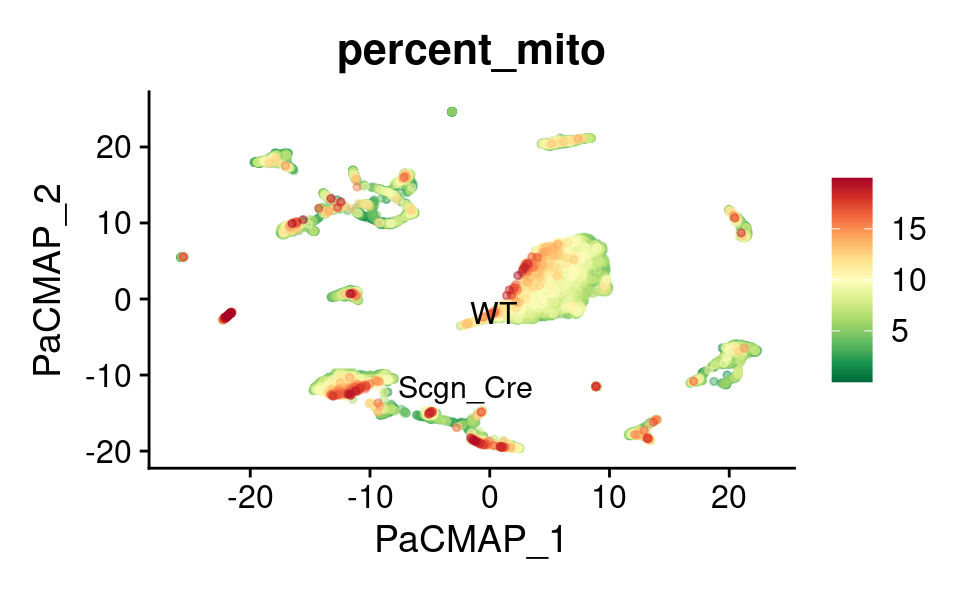
tSNE
FeaturePlot_scCustom(
combined_srt,
features = "percent_mito",
reduction = "tsne",
label.size = 4,
repel = TRUE,
pt.size = 1,
label = TRUE,
colors_use = combined_srt@misc$mdat_Colour_Pal,
order = TRUE,
alpha_na_exp = 0.1,
alpha_exp = 0.45
) &
theme(plot.title = element_text(size = 16))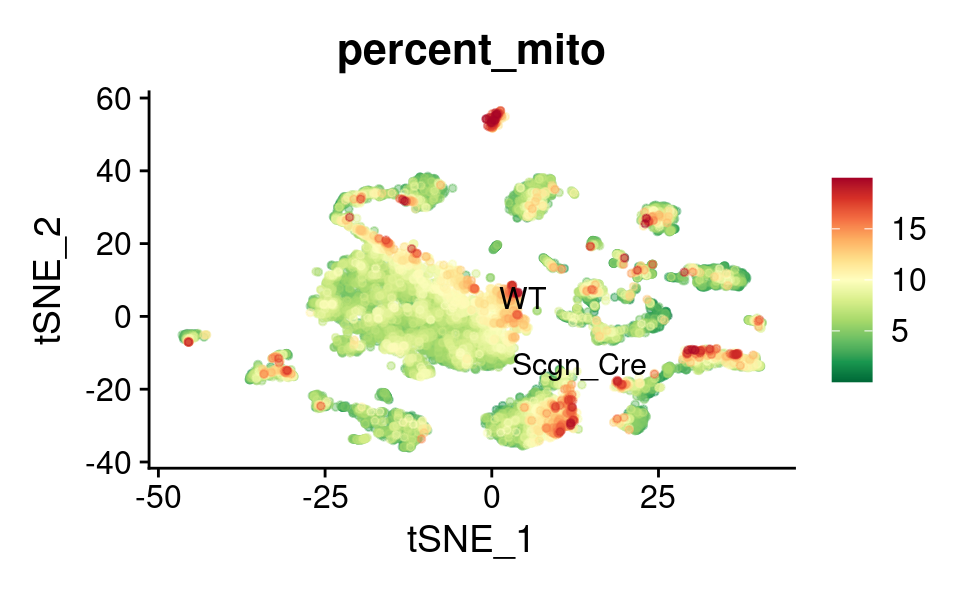
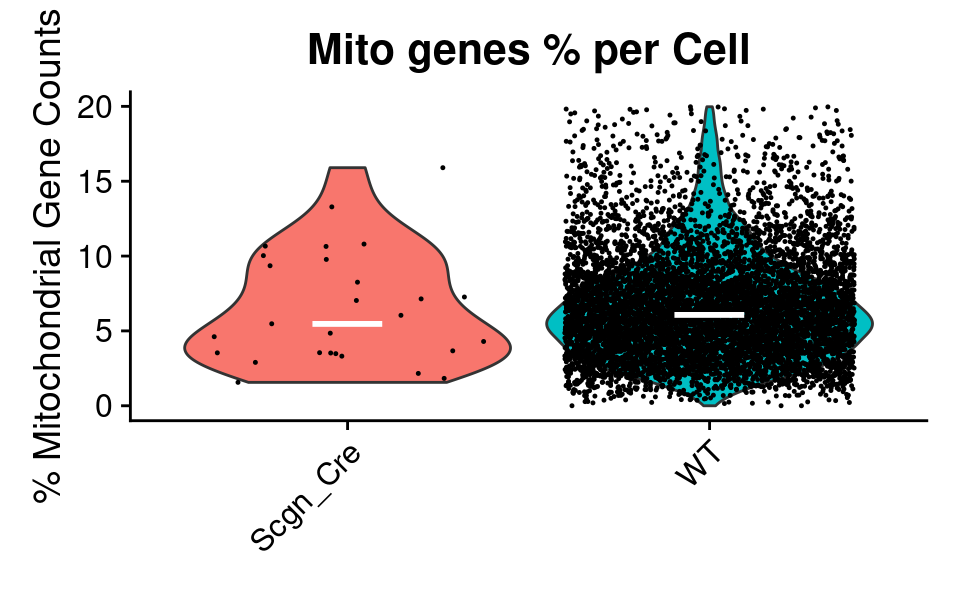
Violin
QC_Plots_Mito(
combined_srt,
high_cutoff = high_cutoff_pc_mt,
plot_median = TRUE,
plot_title = "Mito genes % per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
Ribosomal genes expression
UMAP
FeaturePlot_scCustom(
combined_srt,
features = "percent_ribo",
label.size = 4,
repel = TRUE,
pt.size = 1,
label = TRUE,
colors_use = combined_srt@misc$mdat_Colour_Pal,
order = TRUE,
alpha_na_exp = 0.1,
alpha_exp = 0.45
) &
theme(plot.title = element_text(size = 16))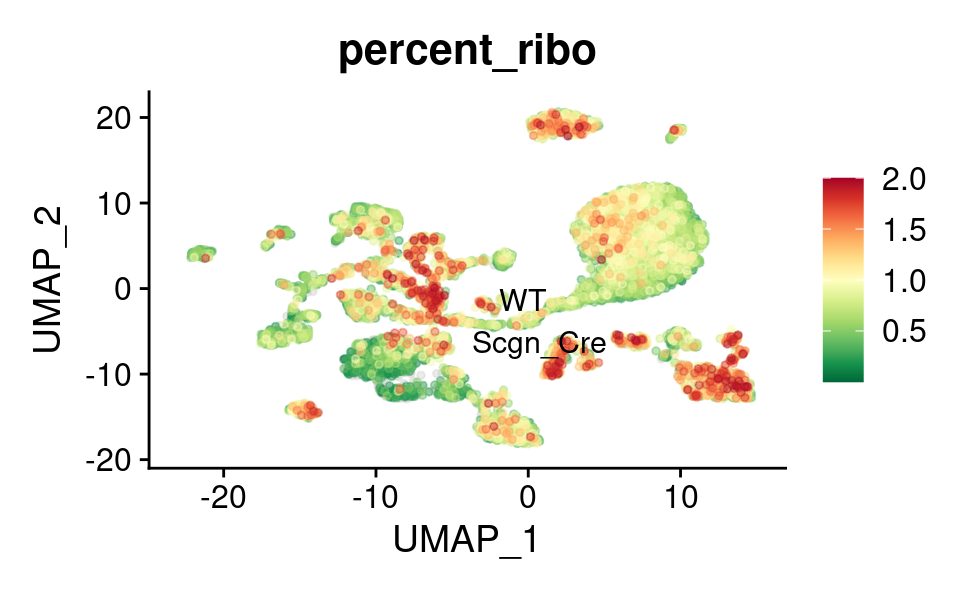
PaCMAP
FeaturePlot_scCustom(
combined_srt,
features = "percent_ribo",
reduction = "pacmap",
label.size = 4,
repel = TRUE,
pt.size = 1,
label = TRUE,
colors_use = combined_srt@misc$mdat_Colour_Pal,
order = TRUE,
alpha_na_exp = 0.1,
alpha_exp = 0.45
) &
theme(plot.title = element_text(size = 16))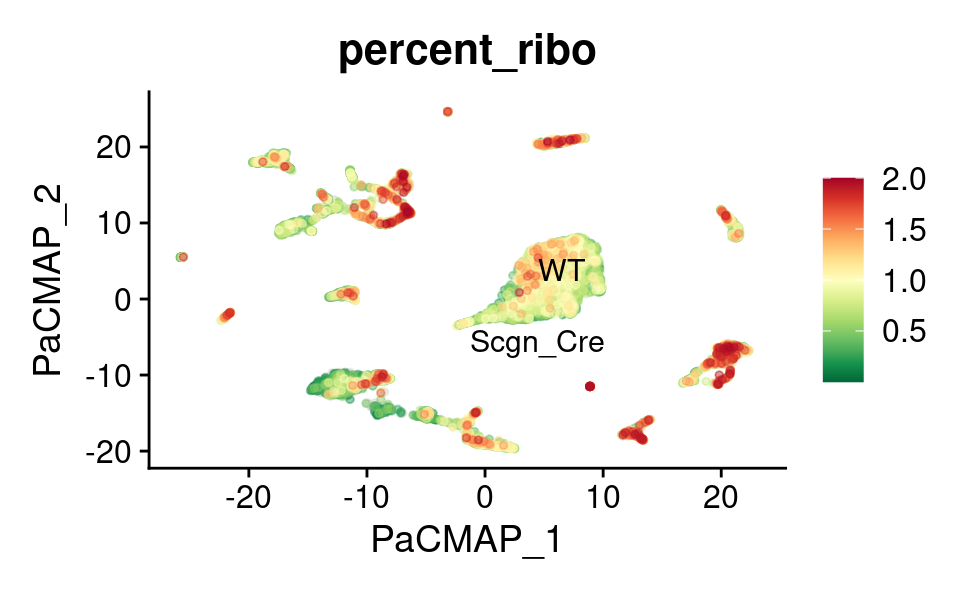
tSNE
FeaturePlot_scCustom(
combined_srt,
features = "percent_ribo",
reduction = "tsne",
label.size = 4,
repel = TRUE,
pt.size = 1,
label = TRUE,
colors_use = combined_srt@misc$mdat_Colour_Pal,
order = TRUE,
alpha_na_exp = 0.1,
alpha_exp = 0.45
) &
theme(plot.title = element_text(size = 16))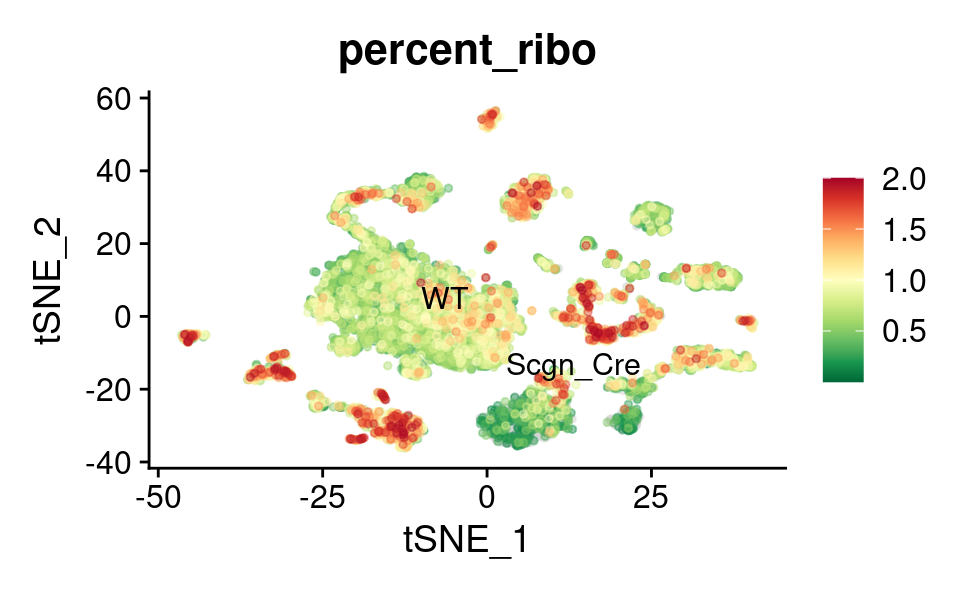
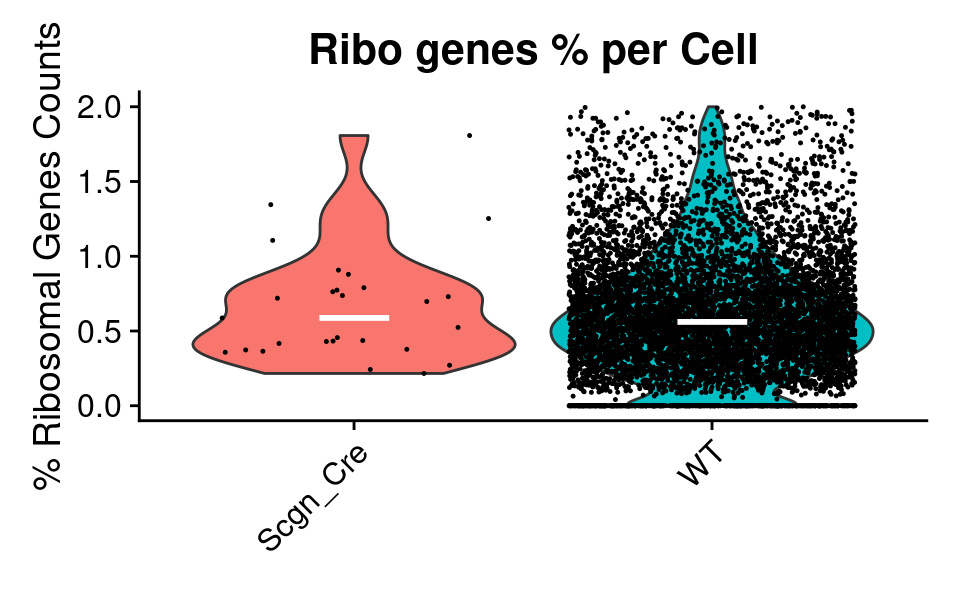
Violin
QC_Plots_Feature(
combined_srt,
feature = "percent_ribo",
plot_median = TRUE,
high_cutoff = high_cutoff_pc_ribo,
y_axis_label = "% Ribosomal Genes Counts",
plot_title = "Ribo genes % per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
Total UMIs and Genes
p1 <-
QC_Plots_Genes(
combined_srt,
low_cutoff = low_cutoff_gene,
high_cutoff = high_cutoff_gene,
plot_median = TRUE,
plot_title = "Genes per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p2 <-
QC_Plots_UMIs(
combined_srt,
low_cutoff = low_cutoff_umis,
high_cutoff = high_cutoff_umis,
plot_median = TRUE,
plot_title = "UMIs per Cell",
color_seed = reseed,
ggplot_default_colors = TRUE
)
p1 | p2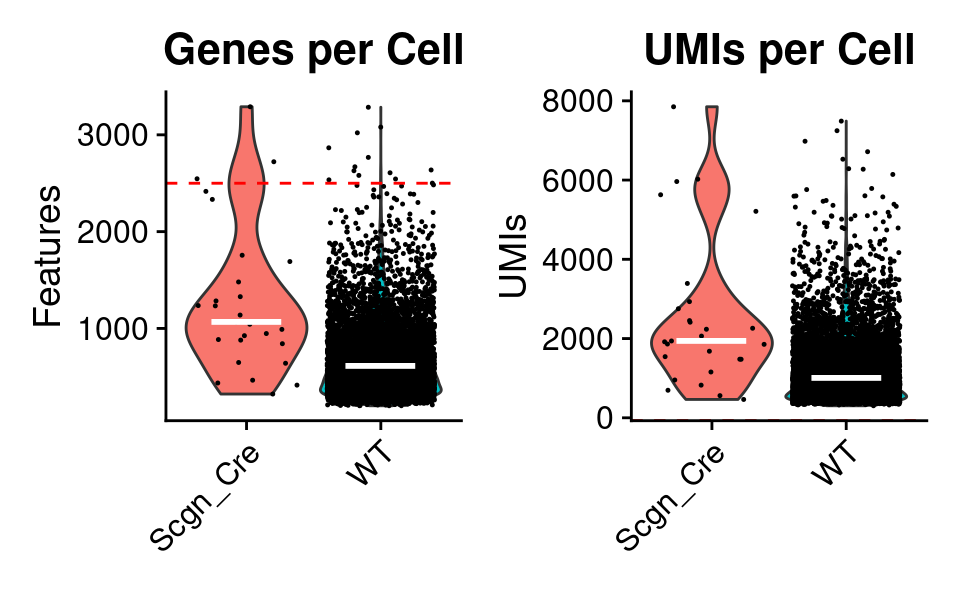
plan("sequential")
invisible(gc())
set.seed(reseed)
plan(multicore, workers = n_cores)
combined_srt <-
CellCycleScoring(
combined_srt,
s.features = str_to_sentence(
cc.genes.updated.2019$s.genes
) %>%
.[. %in% rownames(combined_srt)],
g2m.features = str_to_sentence(
cc.genes.updated.2019$g2m.genes
) %>%
.[. %in% rownames(combined_srt)],
assay = "SCT"
)
table(combined_srt[[]]$Phase)
G1 G2M S
3450 2320 1772 Cell Cycle genes expression
UMAP
FeaturePlot_scCustom(
combined_srt,
features = c("S.Score", "G2M.Score"),
reduction = "umap",
label.size = 4,
repel = TRUE,
pt.size = 1,
label = TRUE,
colors_use = combined_srt@misc$mdat_Colour_Pal,
na_cutoff = NA,
order = TRUE,
alpha_na_exp = 0.1,
alpha_exp = 0.45
) &
theme(plot.title = element_text(size = 16))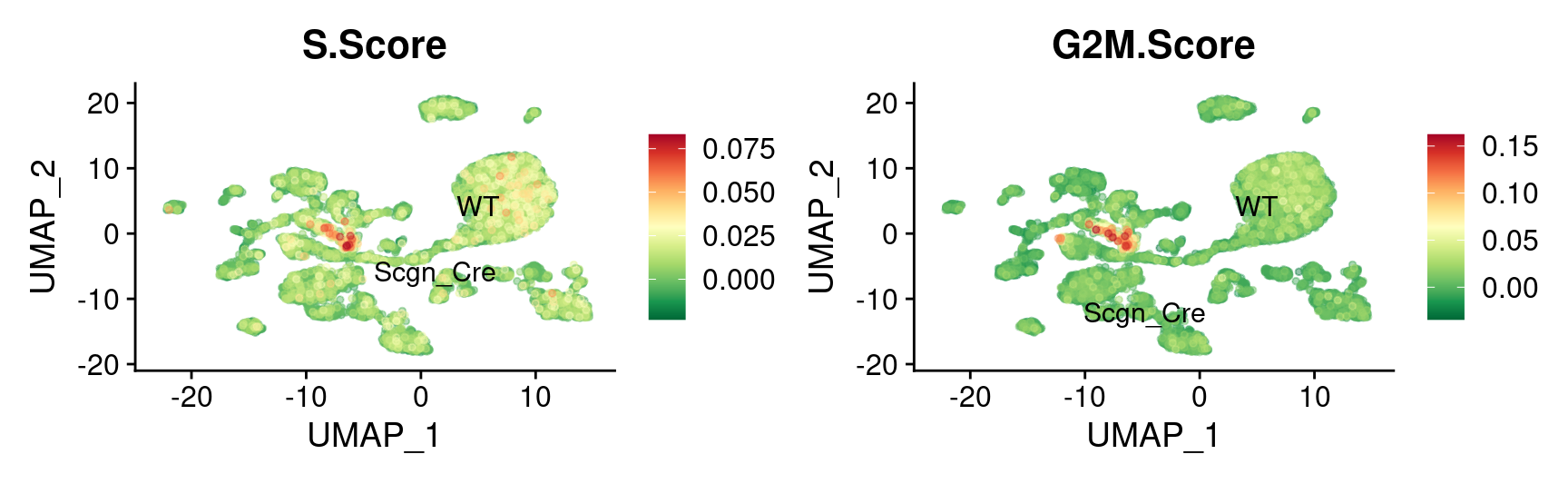
PaCMAP
FeaturePlot_scCustom(
combined_srt,
features = c("S.Score", "G2M.Score"),
reduction = "pacmap",
label.size = 4,
repel = TRUE,
pt.size = 1,
label = TRUE,
colors_use = combined_srt@misc$mdat_Colour_Pal,
na_cutoff = NA,
order = TRUE,
alpha_na_exp = 0.1,
alpha_exp = 0.45
) &
theme(plot.title = element_text(size = 16))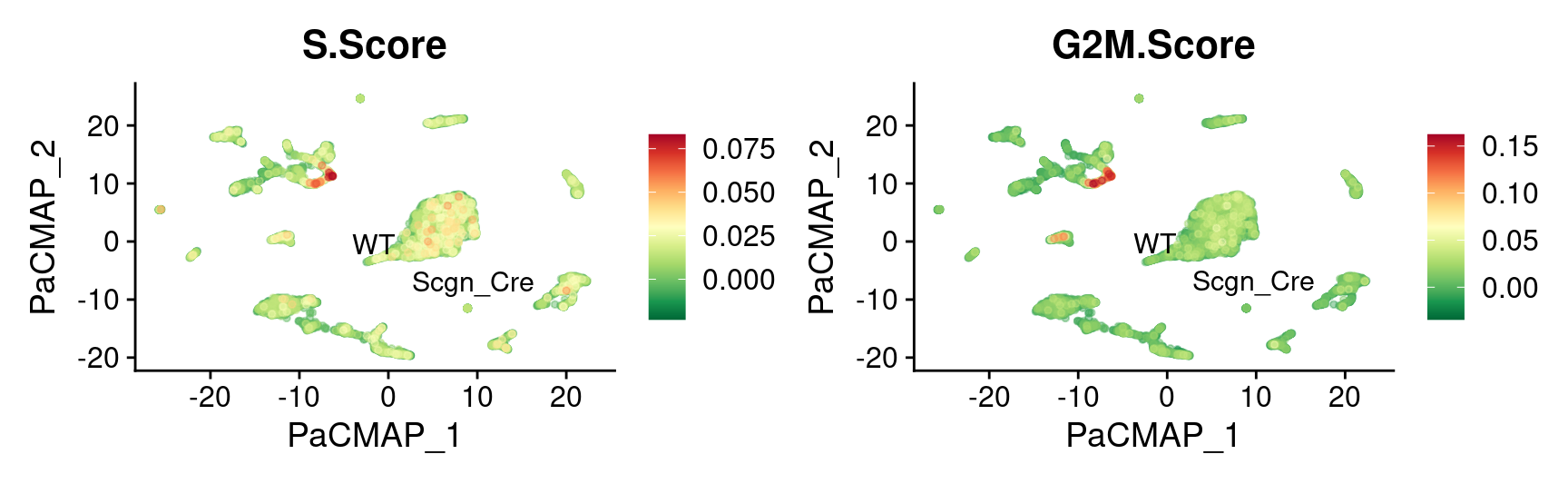
tSNE
FeaturePlot_scCustom(
combined_srt,
features = c("S.Score", "G2M.Score"),
reduction = "tsne",
label.size = 4,
repel = TRUE,
pt.size = 1,
label = TRUE,
colors_use = combined_srt@misc$mdat_Colour_Pal,
na_cutoff = NA,
order = TRUE,
alpha_na_exp = 0.1,
alpha_exp = 0.45
) &
theme(plot.title = element_text(size = 16))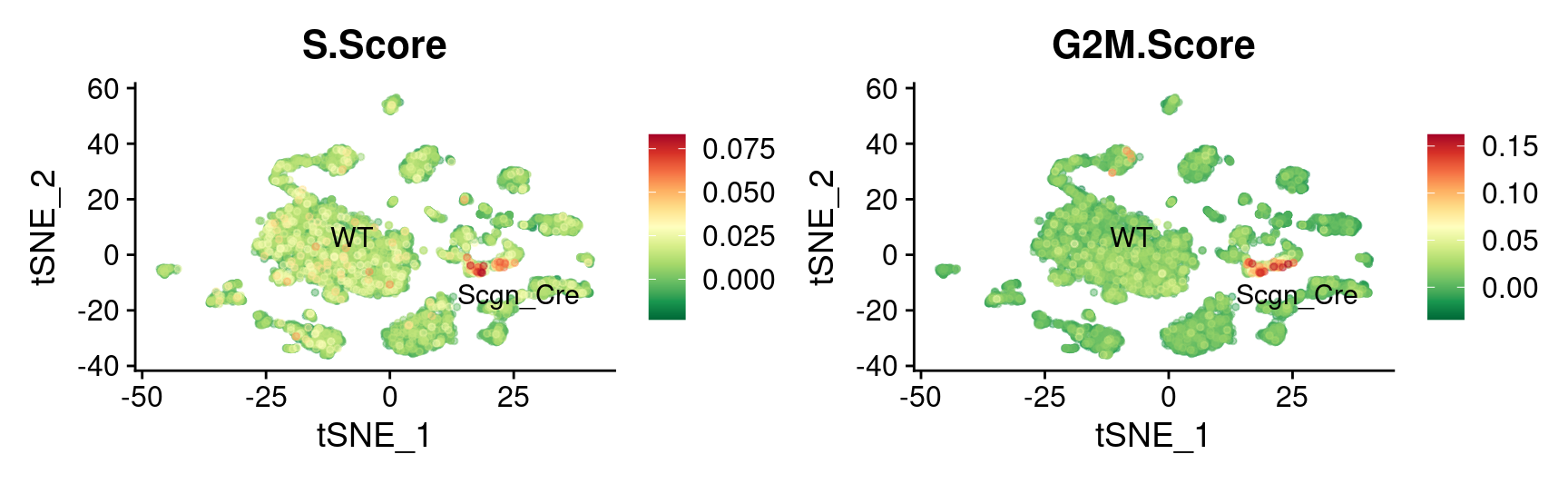
Violin
plt_s_phase <- VlnPlot_scCustom(seurat_object = combined_srt, features = c("S.Score"), plot_median = TRUE) & NoLegend()
plt_g2m_phase <- VlnPlot_scCustom(seurat_object = combined_srt, features = c("G2M.Score"), plot_median = TRUE) & NoLegend()
(plt_s_phase | plt_g2m_phase)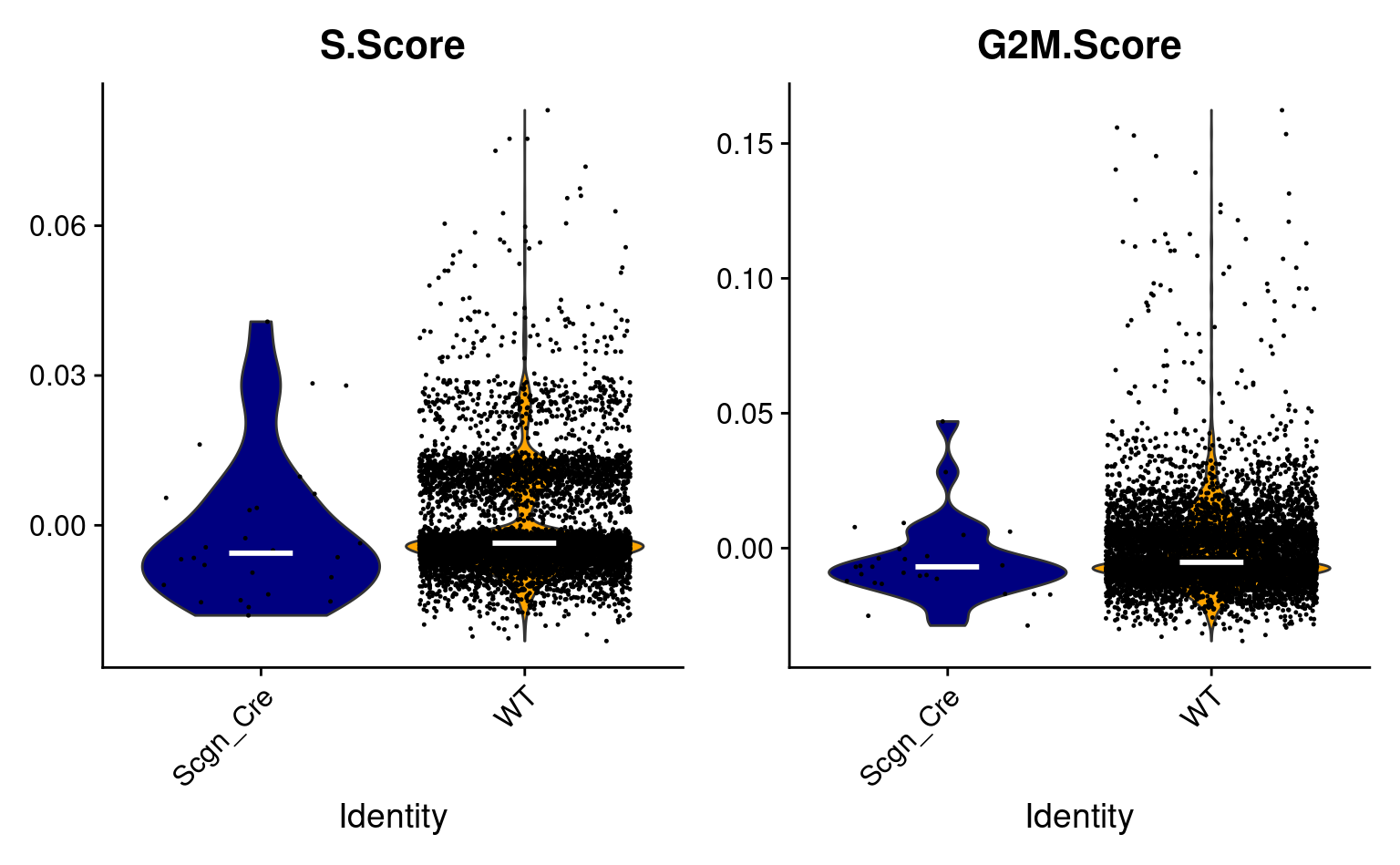
Plot by source after clean up
UMAP
pl_emb_comb_batch <- DimPlot_scCustom(
seurat_object = combined_srt,
reduction = "umap",
group.by = "Scgn_tdTomato",
pt.size = 1,
ggplot_default_colors = TRUE,
color_seed = reseed,
shuffle = TRUE,
seed = reseed,
repel = TRUE,
label = TRUE,
label.size = 5
) + NoLegend()
pl_emb_comb_batch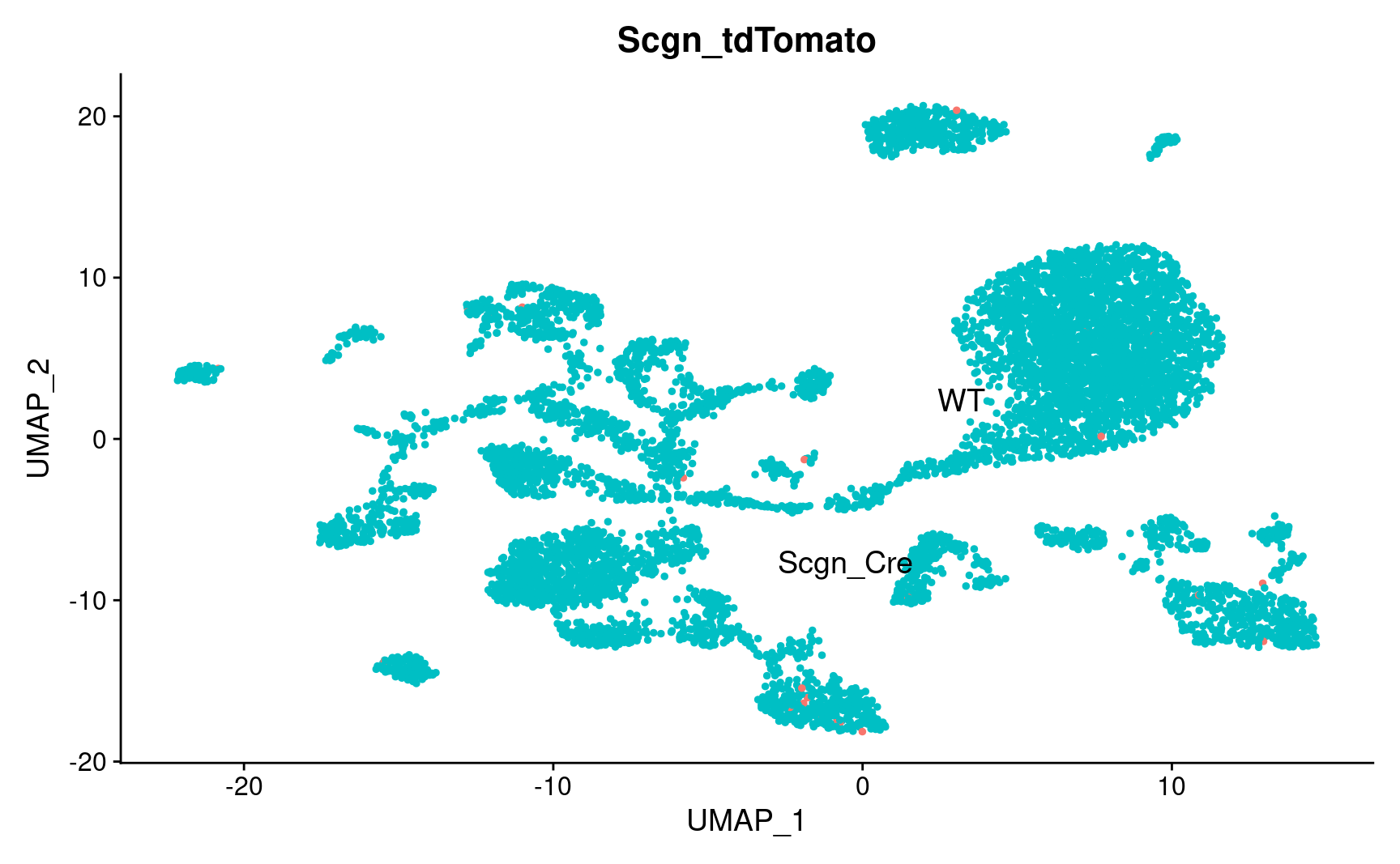
PaCMAP
pl_emb_comb_batch <- DimPlot_scCustom(
seurat_object = combined_srt,
reduction = "pacmap",
group.by = "Scgn_tdTomato",
pt.size = 1,
ggplot_default_colors = TRUE,
color_seed = reseed,
shuffle = TRUE,
seed = reseed,
repel = TRUE,
label = TRUE,
label.size = 5
) + NoLegend()
pl_emb_comb_batch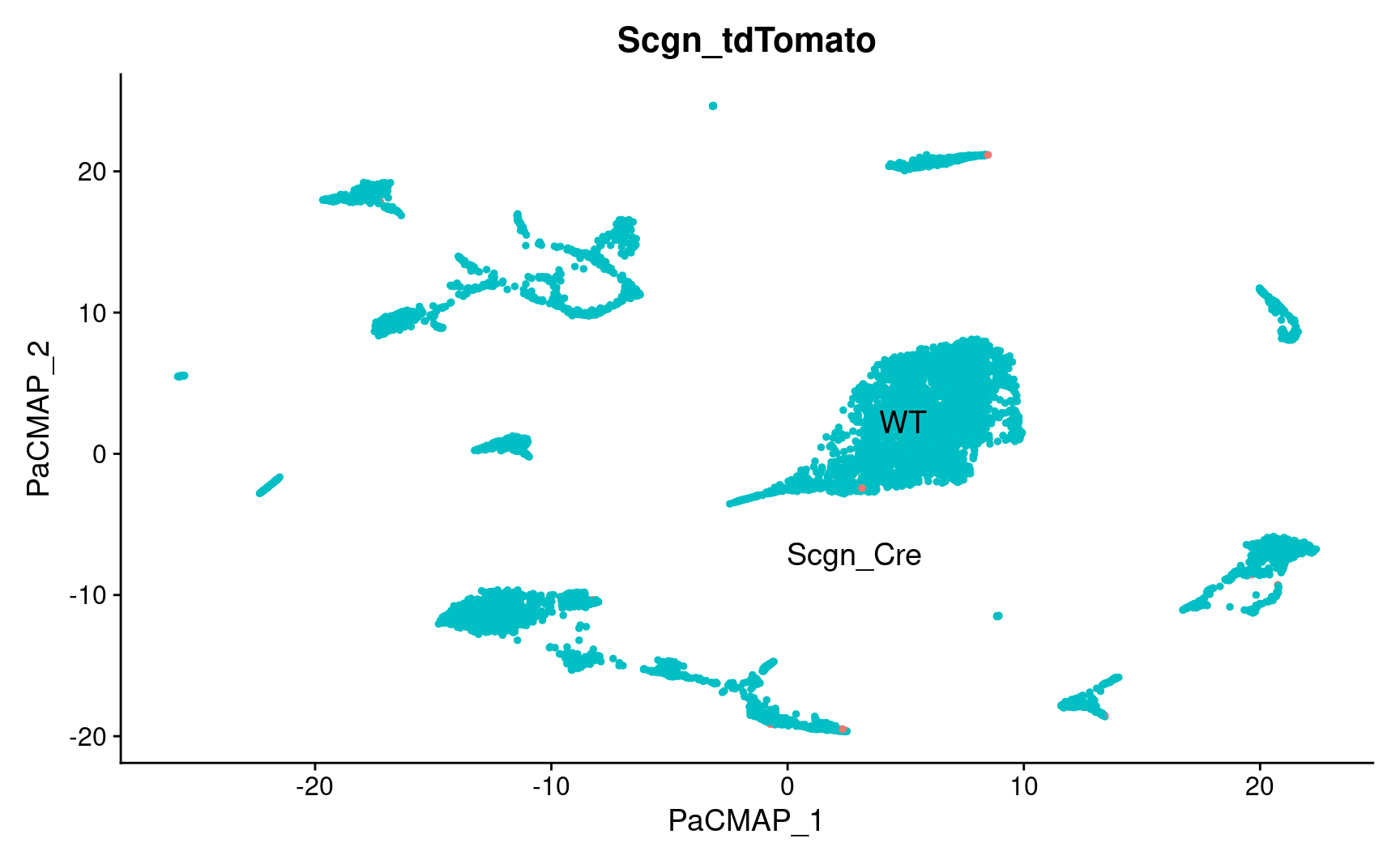
tSNE
pl_emb_comb_batch <- DimPlot_scCustom(
seurat_object = combined_srt,
reduction = "tsne",
group.by = "Scgn_tdTomato",
pt.size = 1,
ggplot_default_colors = TRUE,
color_seed = reseed,
shuffle = TRUE,
seed = reseed,
repel = TRUE,
label = TRUE,
label.size = 5
) + NoLegend()
pl_emb_comb_batch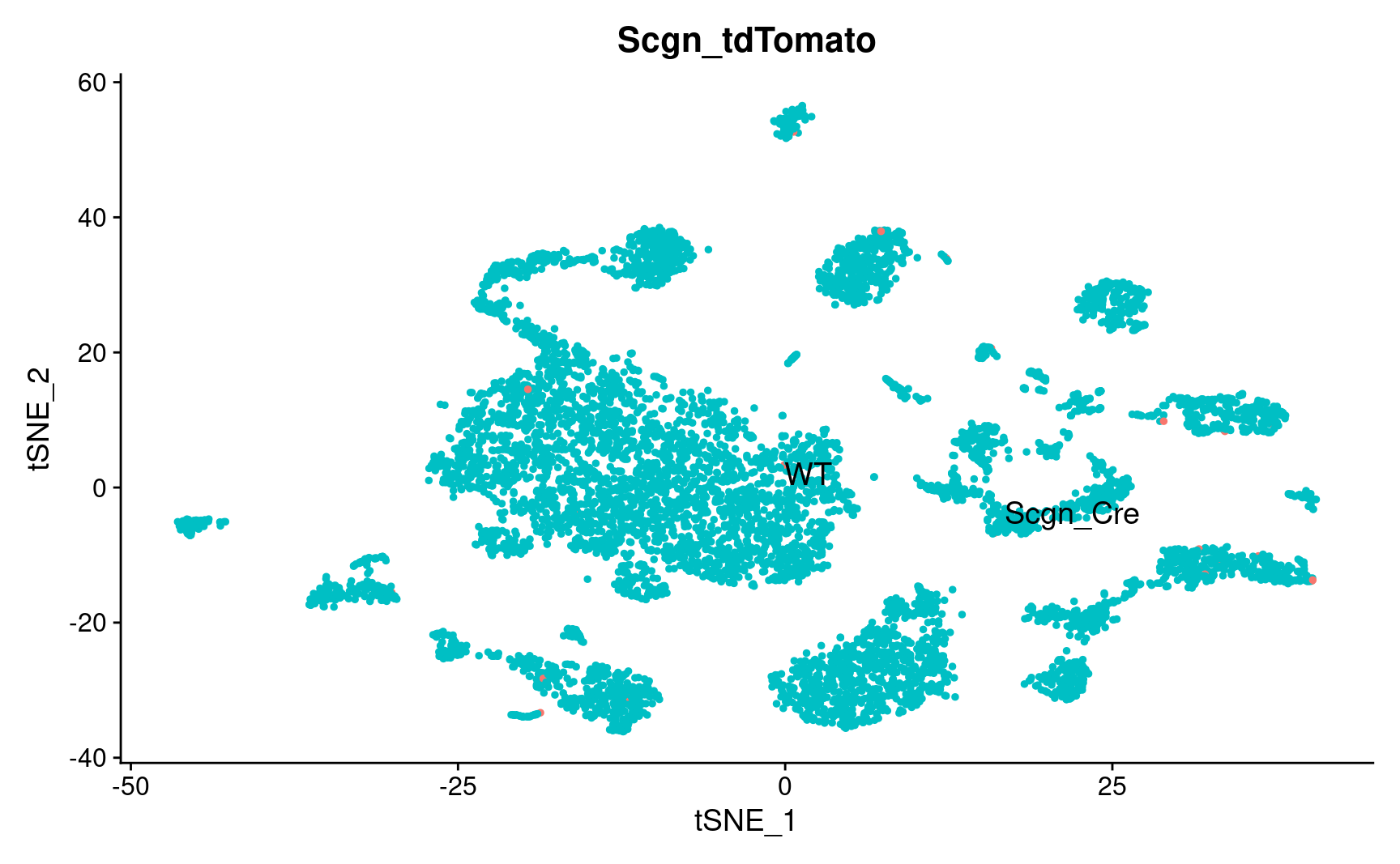
Clustering tree
Standard
Coloured by clustering resolution.
plan("sequential")
invisible(gc())
set.seed(reseed)
plan(multicore, workers = n_cores)
metadata <- combined_srt@meta.data
rownames(metadata) <- colnames(combined_srt)
resolutions <-
modularity_event_sampling(
A = combined_srt@graphs$SCT_snn,
n.res = 10,
gamma.min = 0.2,
gamma.max = 2.000001
) # sample based on the similarity matrix
plan("sequential")
invisible(gc())
set.seed(reseed)
plan(multicore, workers = n_cores)
# clustering using Suerat
combined_srt <- combined_srt %>%
FindClusters(
algorithm = "leiden",
partition.type = "ModularityVertexPartition",
method = "igraph",
n.iter = -1,
resolution = resolutions,
random.seed = reseed,
verbose = FALSE
)
ref_labels <- combined_srt$seurat_clusters
# initial cluster tree from Seurat flat clustering
plot_clustree(
labelmat = combined_srt@meta.data,
prefix = "SCT_snn_res.",
ref_labels = ref_labels,
plot.ref = FALSE,
layout = "sugiyama",
use_core_edges = FALSE
)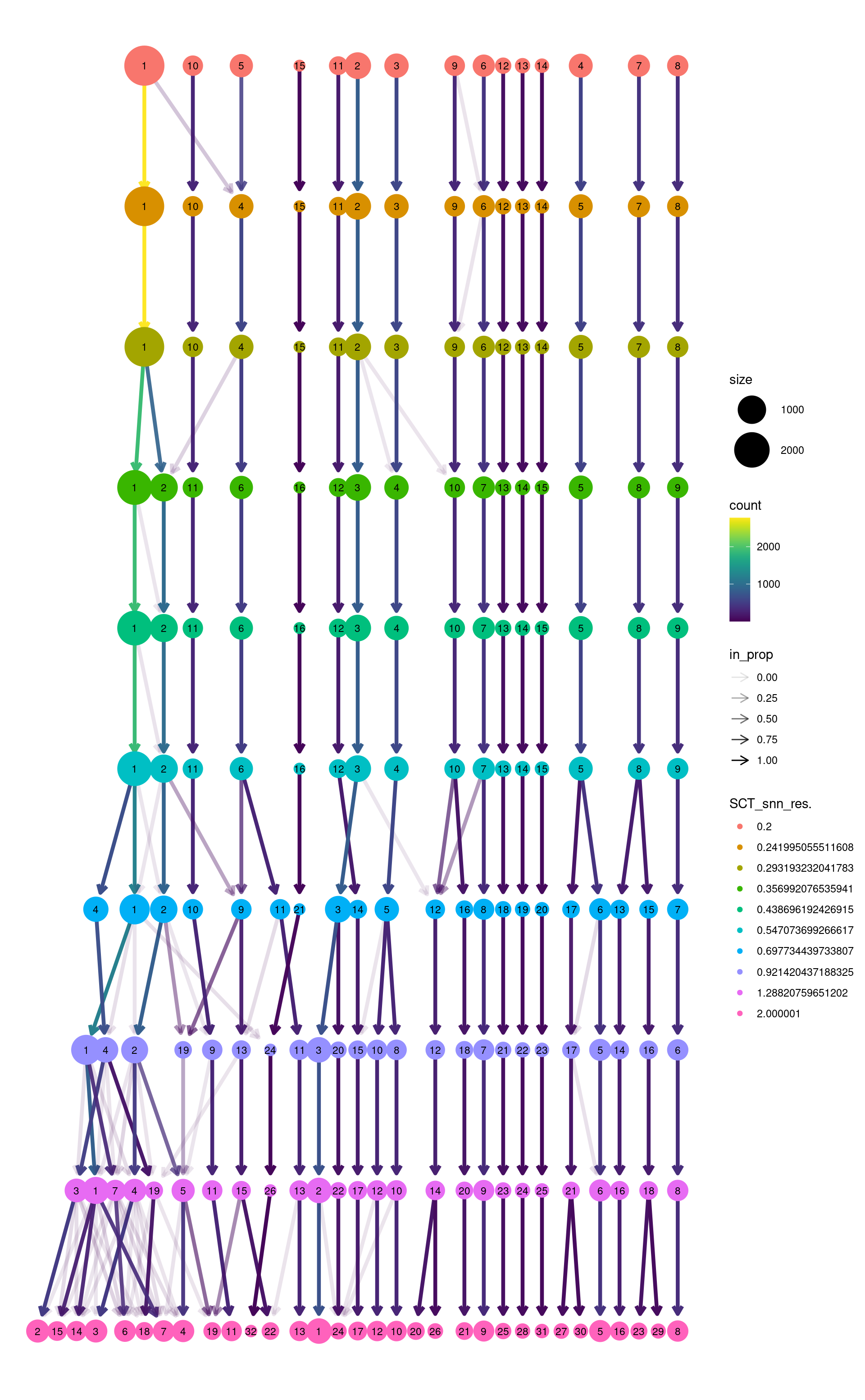
Stability
Coloured by the SC3 stability metric.
plot_clustree(
labelmat = combined_srt@meta.data,
prefix = "SCT_snn_res.",
node_colour = "sc3_stability",
plot.ref = FALSE,
layout = "sugiyama",
use_core_edges = FALSE
)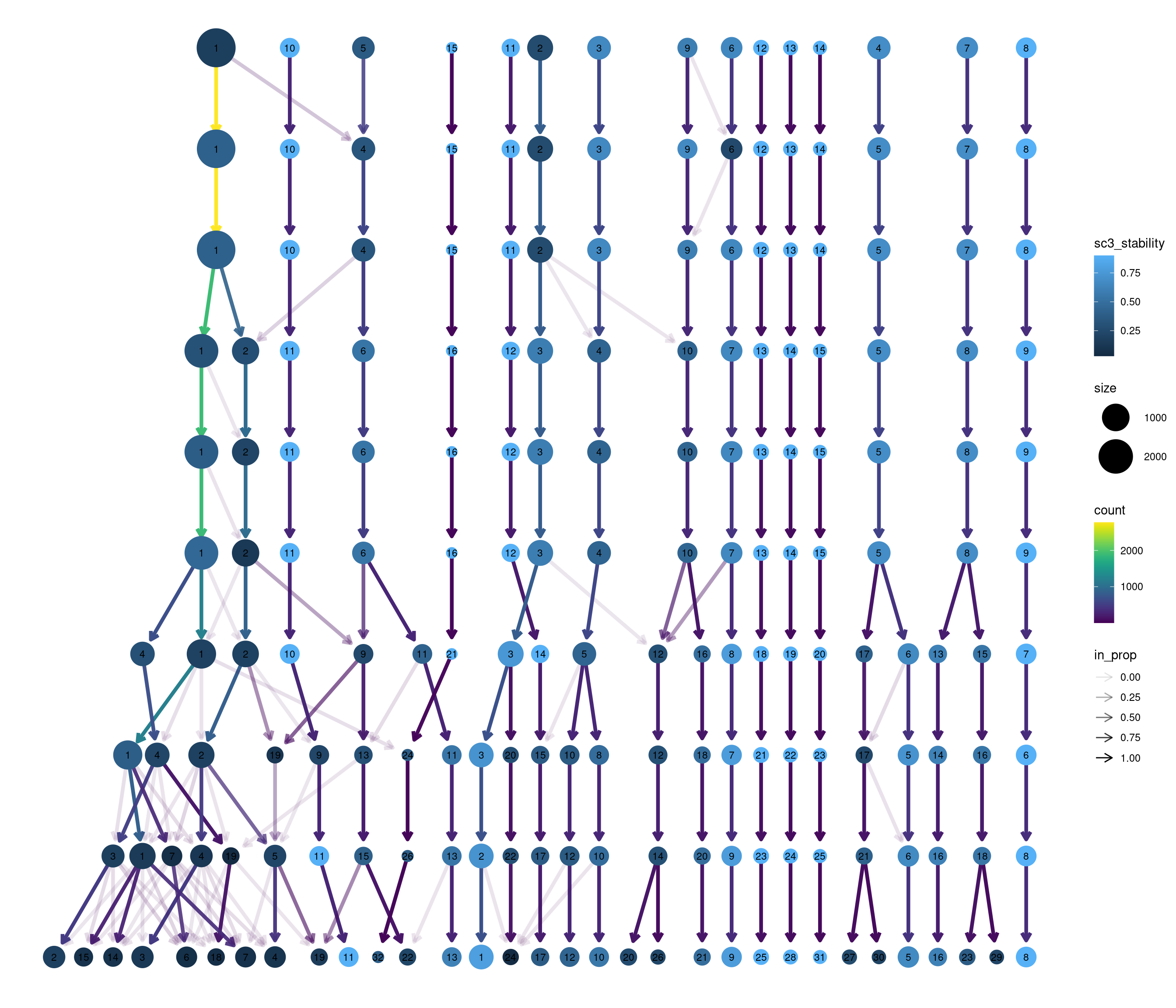
Reconcile clustering tree
MRTree
Adjusted Multiresolution Rand Index (AMRI)
# Adjusted Multiresolution Rand Index (AMRI)
ks_flat <- apply(
out$labelmat.flat,
2,
FUN = function(x) {
length(unique(x))
}
)
ks_mrtree <- apply(
out$labelmat.mrtree,
2,
FUN = function(x) {
length(unique(x))
}
)
amri_flat <- sapply(seq_len(ncol(out$labelmat.flat)), function(i) {
AMRI(out$labelmat.flat[, i], ref_labels)$amri
})
amri_flat <- aggregate(amri_flat, by = list(k = ks_flat), FUN = mean)
amri_recon <- sapply(seq_len(ncol(out$labelmat.mrtree)), function(i) {
AMRI(out$labelmat.mrtree[, i], ref_labels)$amri
})
df <- rbind(
data.frame(
k = amri_flat$k,
amri = amri_flat$x,
method = "Seurat flat"
),
data.frame(k = ks_mrtree, amri = amri_recon, method = "MRtree")
)
ggplot2::ggplot(data = df, aes(x = k, y = amri, color = method)) +
geom_line() +
theme_bw()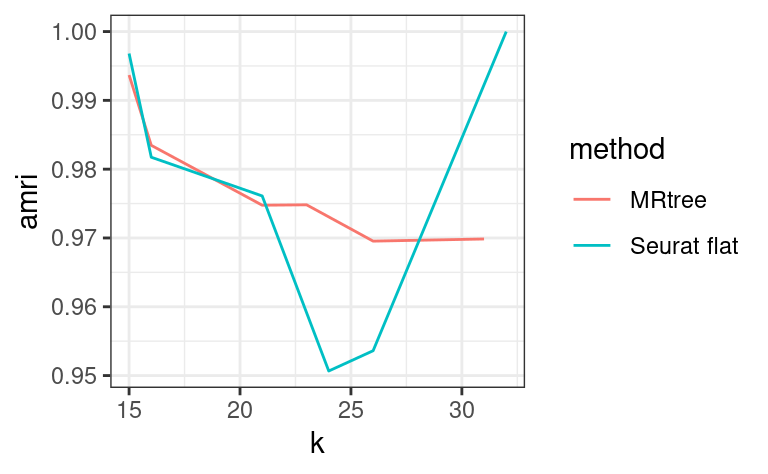
Stability
stab_out <- stability_plot(out)
stab_out$plot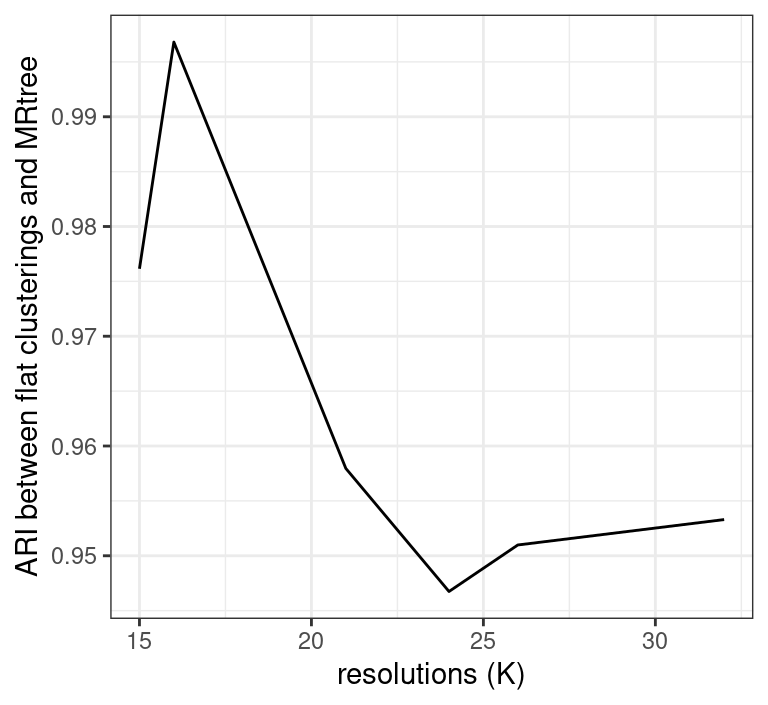
kable_material(
kable(
stab_out$df,
"html"
),
bootstrap_options = c(
"bordered",
"condensed",
"responsive",
"striped"
),
position = "left",
font_size = 14
)| resolution | ari |
|---|---|
| 15 | 0.9761475 |
| 16 | 0.9968033 |
| 21 | 0.9579564 |
| 24 | 0.9467431 |
| 26 | 0.9509819 |
| 32 | 0.9532974 |
res_k <- select_resolution(stab_out$df)
kable_material(
kable(
table(
out$labelmat.mrtree[, which.min(
abs(as.integer(
str_remove(dimnames(
out$labelmat.mrtree
)[[2]], "K")
) - res_k)
)]
),
"html"
),
bootstrap_options = c(
"bordered",
"condensed",
"responsive",
"striped"
),
position = "left",
font_size = 14
)| Var1 | Freq |
|---|---|
| 1 | 1891 |
| 2 | 971 |
| 3 | 808 |
| 4 | 587 |
| 5 | 547 |
| 6 | 492 |
| 7 | 414 |
| 8 | 410 |
| 9 | 348 |
| 10 | 306 |
| 11 | 290 |
| 12 | 202 |
| 13 | 104 |
| 14 | 77 |
| 15 | 59 |
| 16 | 36 |
Selected clustering resolution
UMAP
combined_srt$k_tree <- out$labelmat.mrtree[, which.min(
abs(as.integer(
str_remove(dimnames(
out$labelmat.mrtree
)[[2]], "K")
) - res_k)
)] %>%
as.numeric() %>%
as.factor()
p1 <-
DimPlot_scCustom(
combined_srt,
pt.size = 1,
ggplot_default_colors = TRUE,
color_seed = reseed,
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + ggtitle("Unsupervised overclustering") + NoLegend()
p2 <-
DimPlot_scCustom(
combined_srt,
group.by = "k_tree",
pt.size = 1,
ggplot_default_colors = TRUE,
color_seed = reseed,
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + ggtitle("MRTree") + NoLegend()
p1 | p2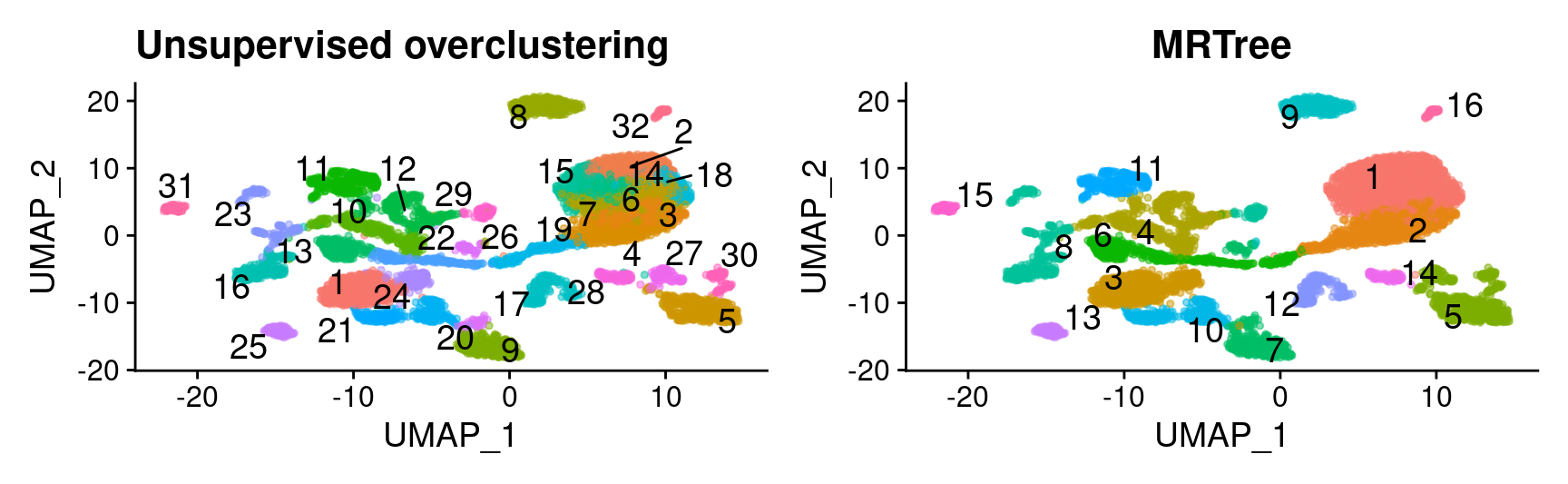
PaCMAP
p1 <-
DimPlot_scCustom(
combined_srt,
pt.size = 1,
ggplot_default_colors = TRUE,
color_seed = reseed,
reduction = "pacmap",
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + ggtitle("Unsupervised overclustering") + NoLegend()
p2 <-
DimPlot_scCustom(
combined_srt,
group.by = "k_tree",
pt.size = 1,
ggplot_default_colors = TRUE,
color_seed = reseed,
reduction = "pacmap",
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + ggtitle("MRTree") + NoLegend()
p1 | p2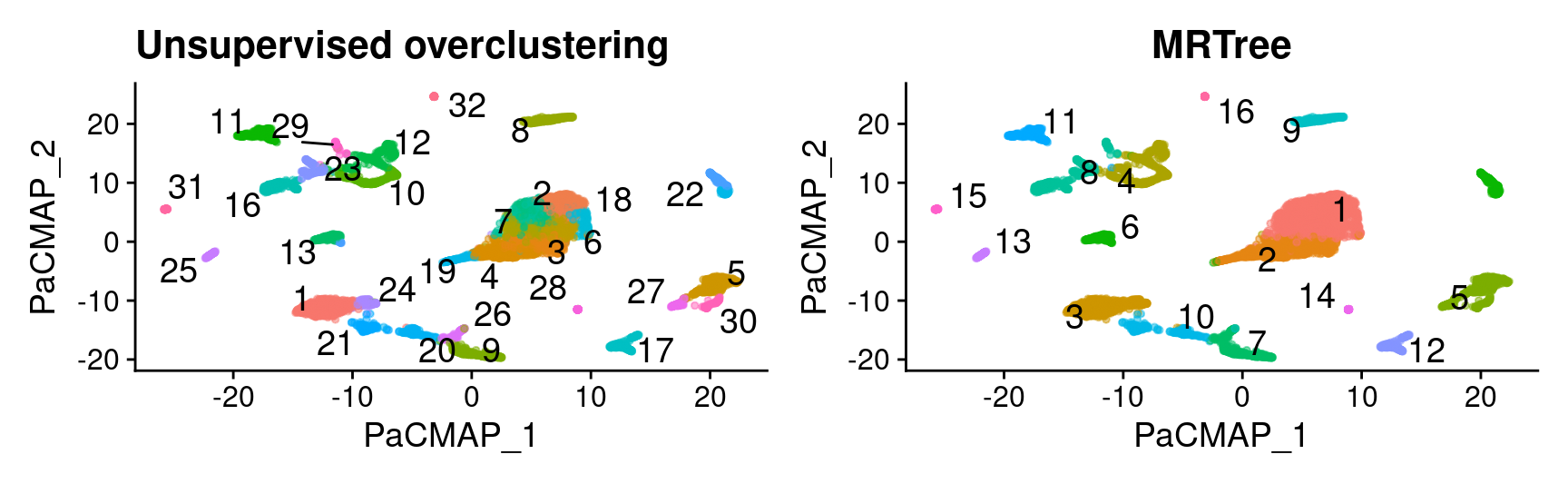
tSNE
p1 <-
DimPlot_scCustom(
combined_srt,
pt.size = 1,
ggplot_default_colors = TRUE,
color_seed = reseed,
reduction = "tsne",
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + ggtitle("Unsupervised overclustering") + NoLegend()
p2 <-
DimPlot_scCustom(
combined_srt,
group.by = "k_tree",
pt.size = 1,
ggplot_default_colors = TRUE,
color_seed = reseed,
reduction = "tsne",
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + ggtitle("MRTree") + NoLegend()
p1 | p2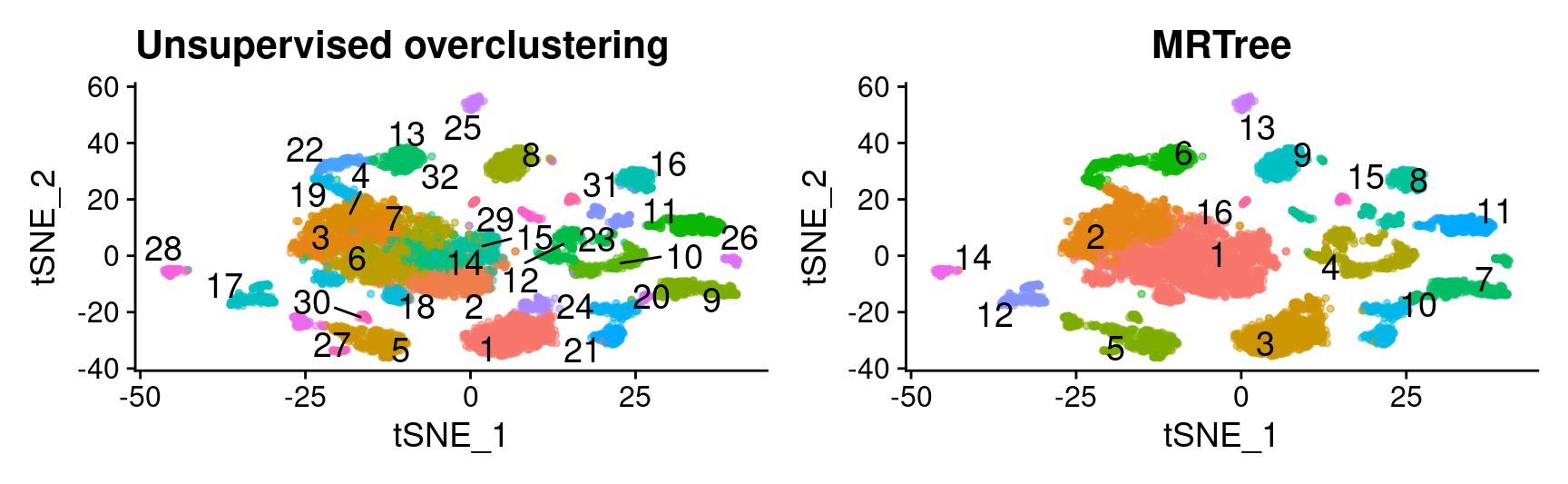
tdTomato across clusters
VlnPlot_scCustom(seurat_object = combined_srt, assay = "RNA", layer = "counts", features = c("tdTomato"), plot_median = TRUE) & NoLegend()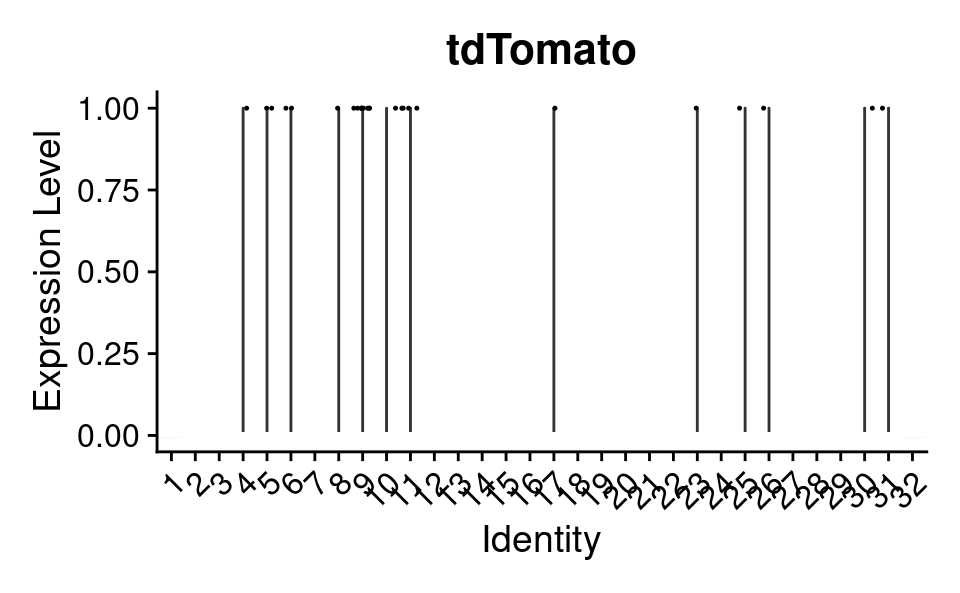
UMAP plot of gene expression
src_list <- map(genes.embed, function(gene) {
src <- c(
"### {{gene}} {.unnumbered}",
"```{r pl-umap-expression-{{gene}}}",
"FeaturePlot_scCustom(",
"seurat_object = combined_srt, features = '{{gene}}',",
"pt.size = 1, order = TRUE,",
"colors_use = combined_srt@misc$expr_Colour_Pal,",
"alpha_na_exp = 0.1, alpha_exp = 0.45) +",
"ggtitle(sprintf('%s: ', '{{gene}}')) +",
"theme(plot.title = element_text(size = 24))",
"```",
""
)
knitr::knit_expand(text = src)
})
out <- knitr::knit_child(text = unlist(src_list), options = list(cache = FALSE))Abcd1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Abcd1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Abcd1')) +
theme(plot.title = element_text(size = 24))
Abcd2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Abcd2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Abcd2')) +
theme(plot.title = element_text(size = 24))
Abcd3
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Abcd3',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Abcd3')) +
theme(plot.title = element_text(size = 24))
Acaa2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Acaa2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Acaa2')) +
theme(plot.title = element_text(size = 24))
Acox1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Acox1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Acox1')) +
theme(plot.title = element_text(size = 24))
Agrn
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Agrn',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Agrn')) +
theme(plot.title = element_text(size = 24))
Agt
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Agt',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Agt')) +
theme(plot.title = element_text(size = 24))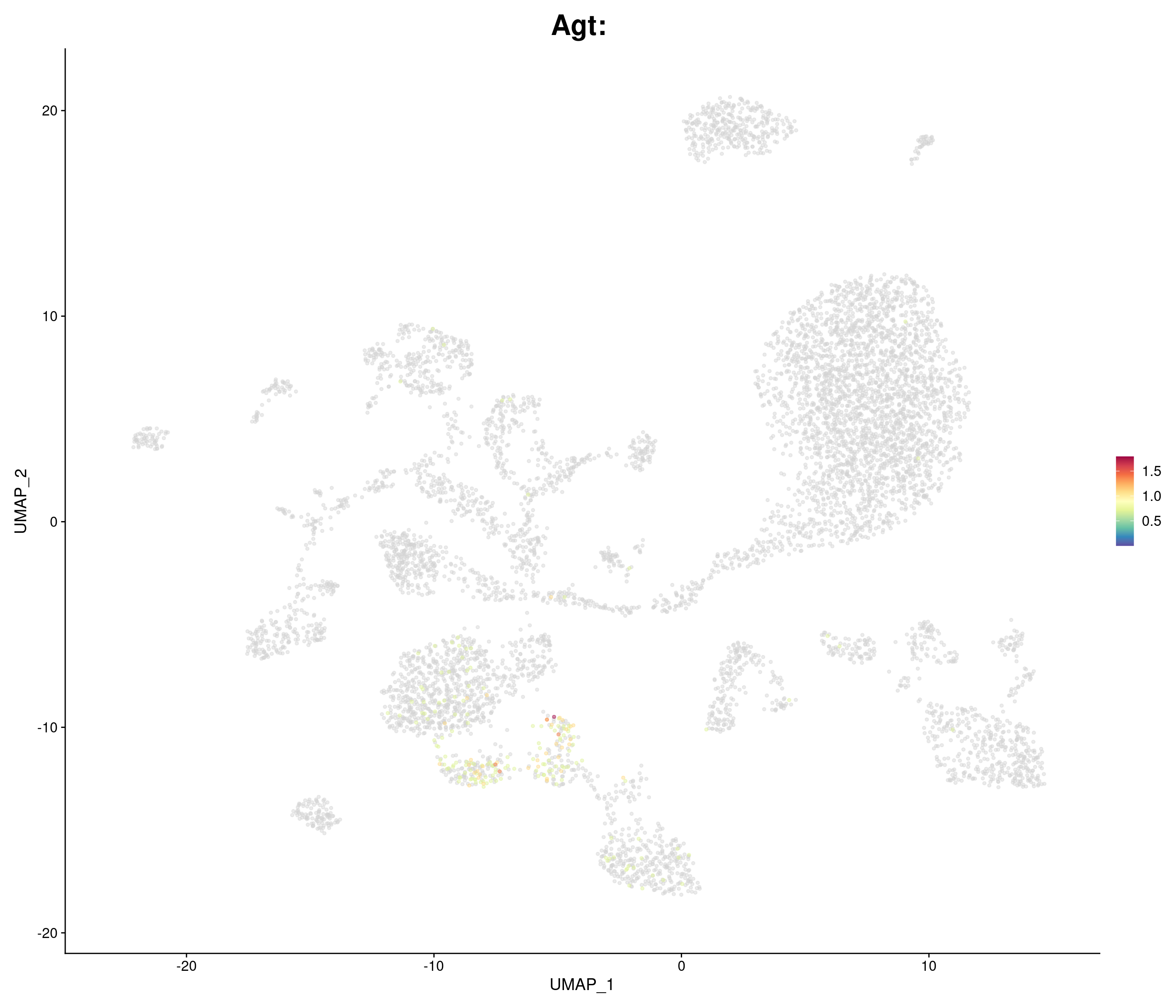
Alcam
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Alcam',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Alcam')) +
theme(plot.title = element_text(size = 24))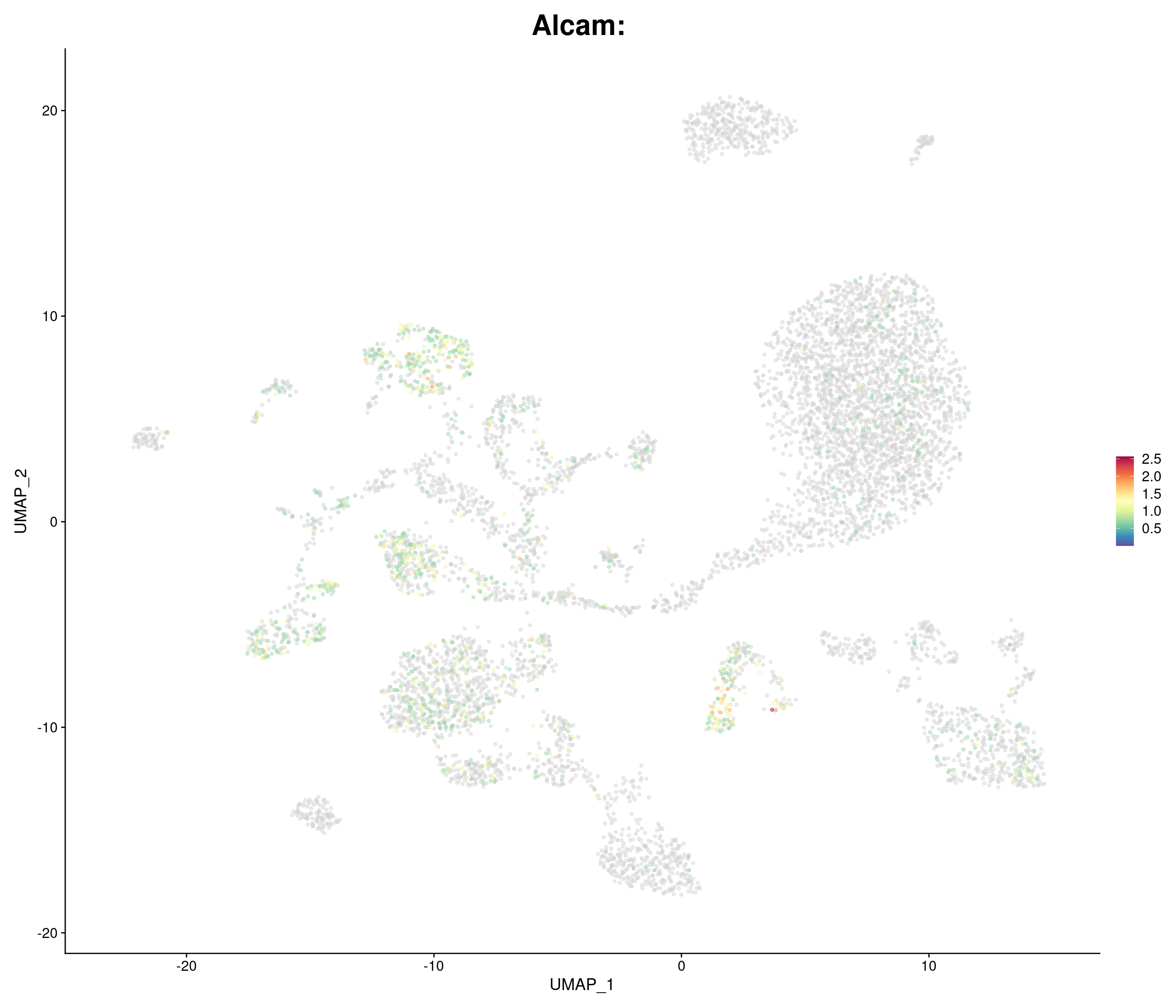
Aldh1a1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Aldh1a1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Aldh1a1')) +
theme(plot.title = element_text(size = 24))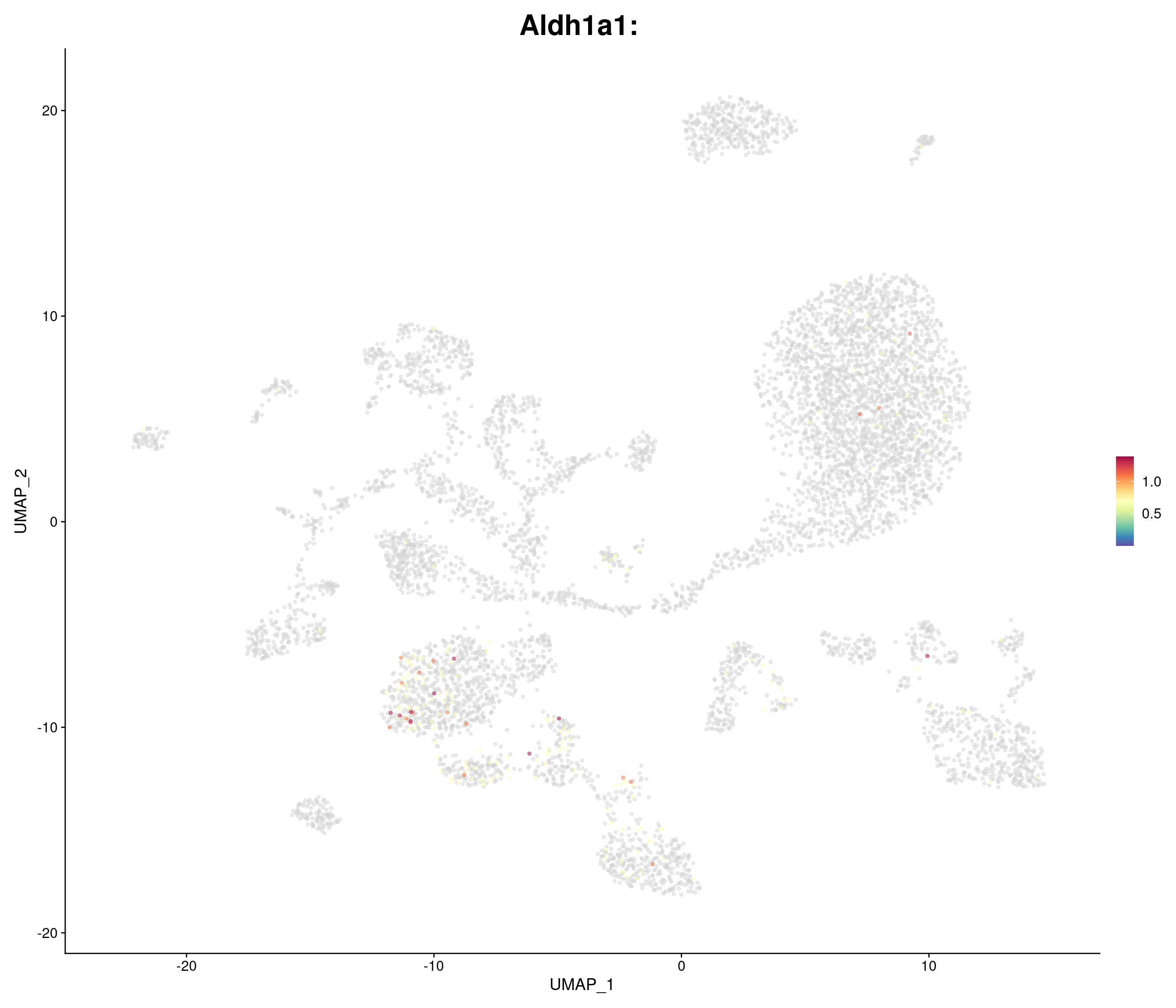
Aldh1l1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Aldh1l1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Aldh1l1')) +
theme(plot.title = element_text(size = 24))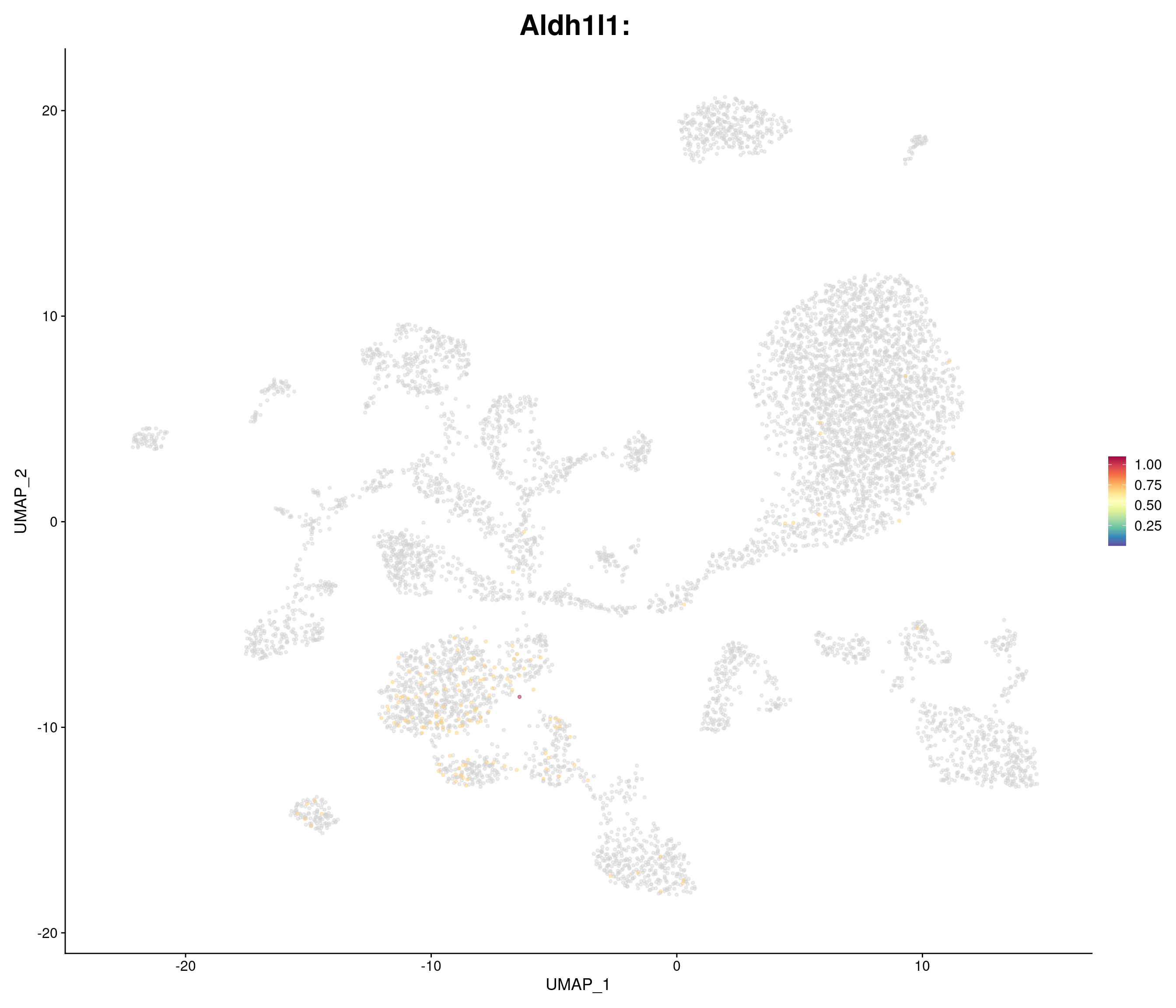
Aldoc
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Aldoc',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Aldoc')) +
theme(plot.title = element_text(size = 24))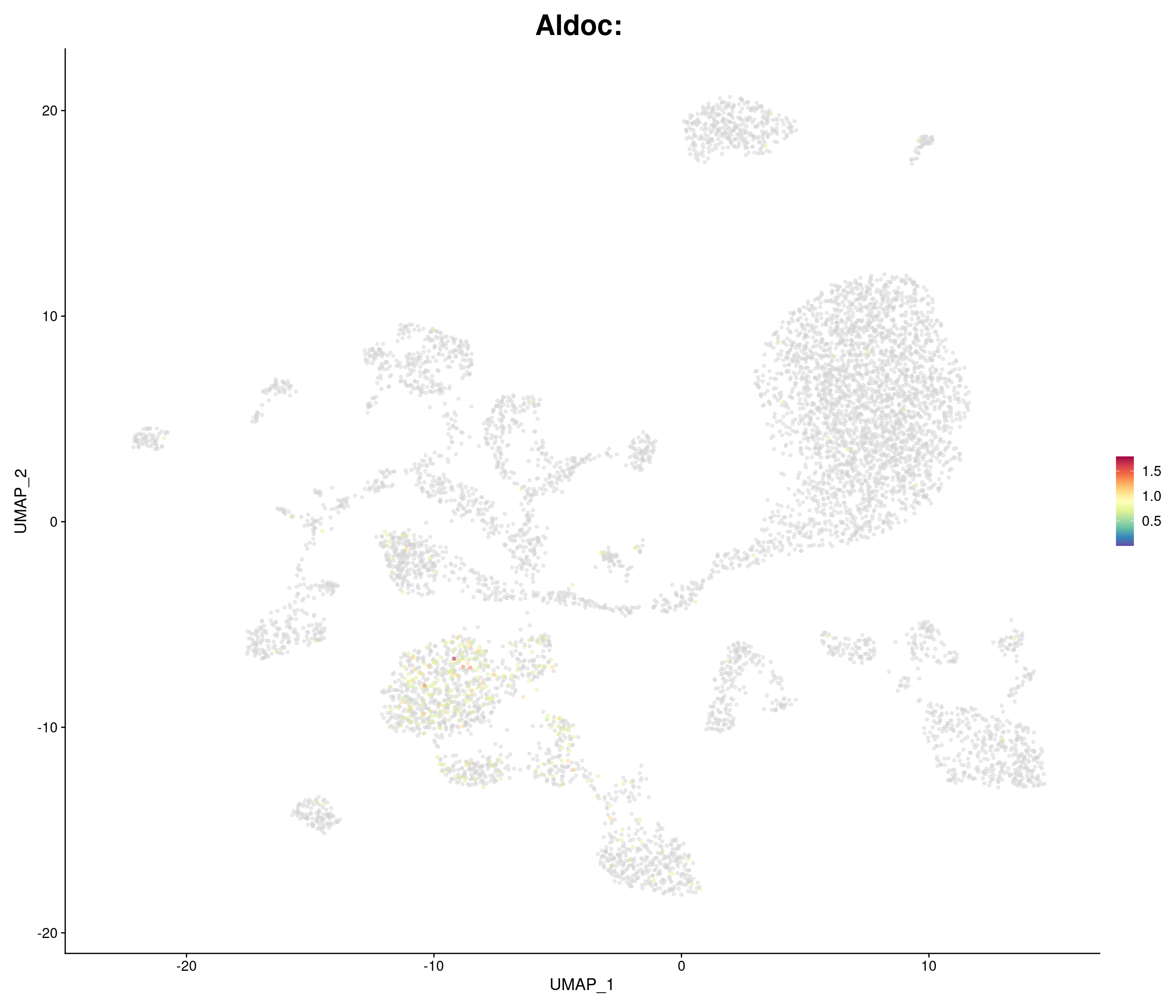
Angpt1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Angpt1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Angpt1')) +
theme(plot.title = element_text(size = 24))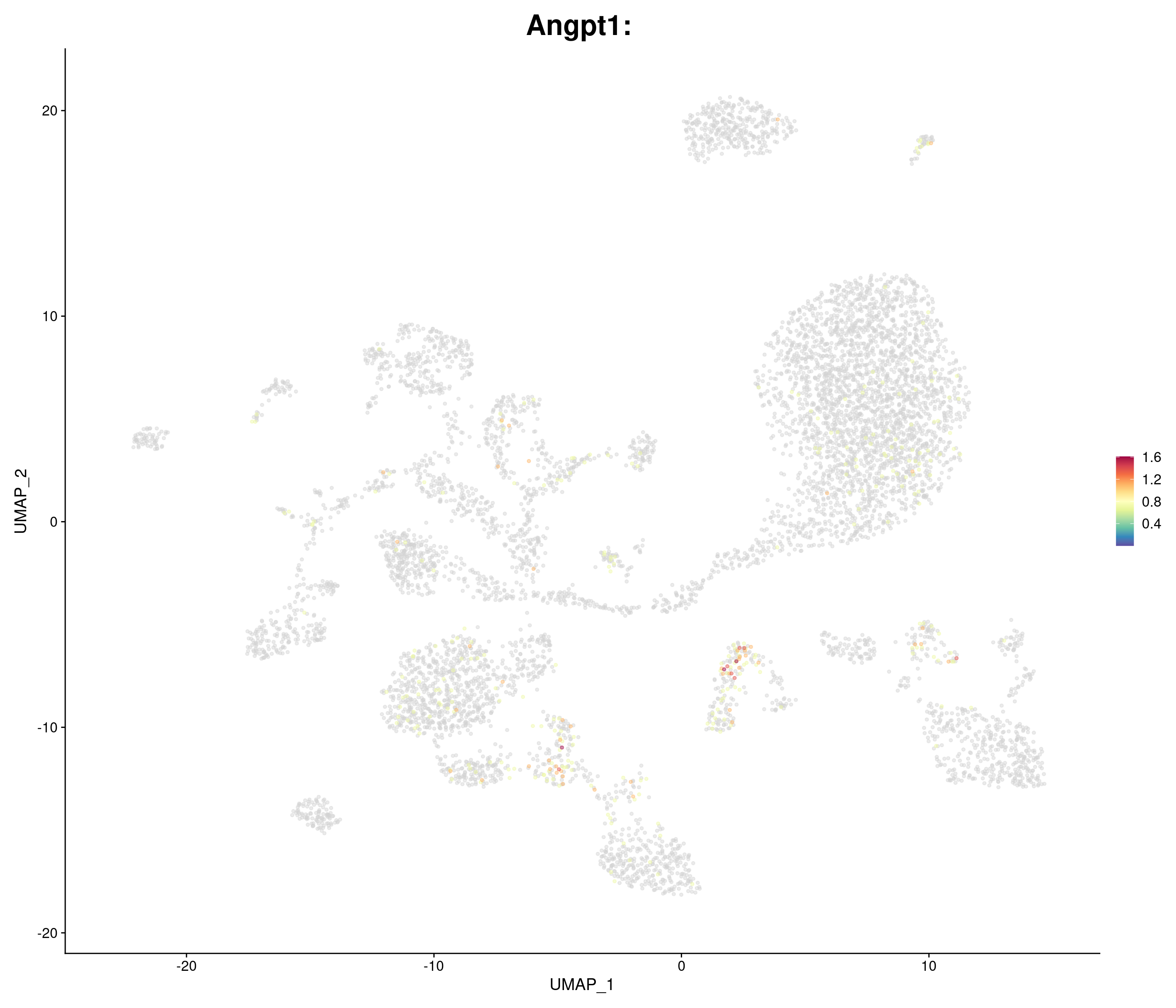
Apoe
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Apoe',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Apoe')) +
theme(plot.title = element_text(size = 24))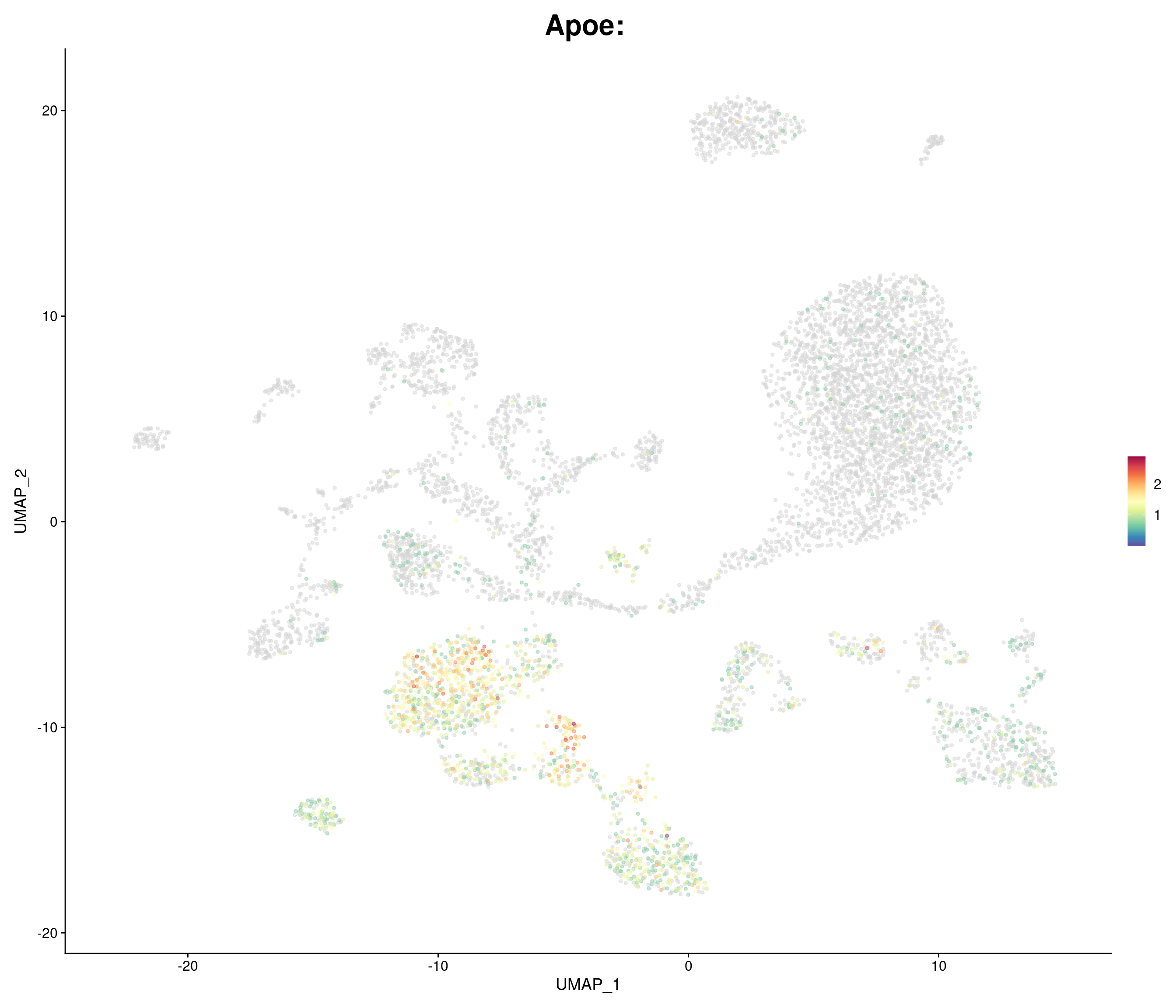
App
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'App',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'App')) +
theme(plot.title = element_text(size = 24))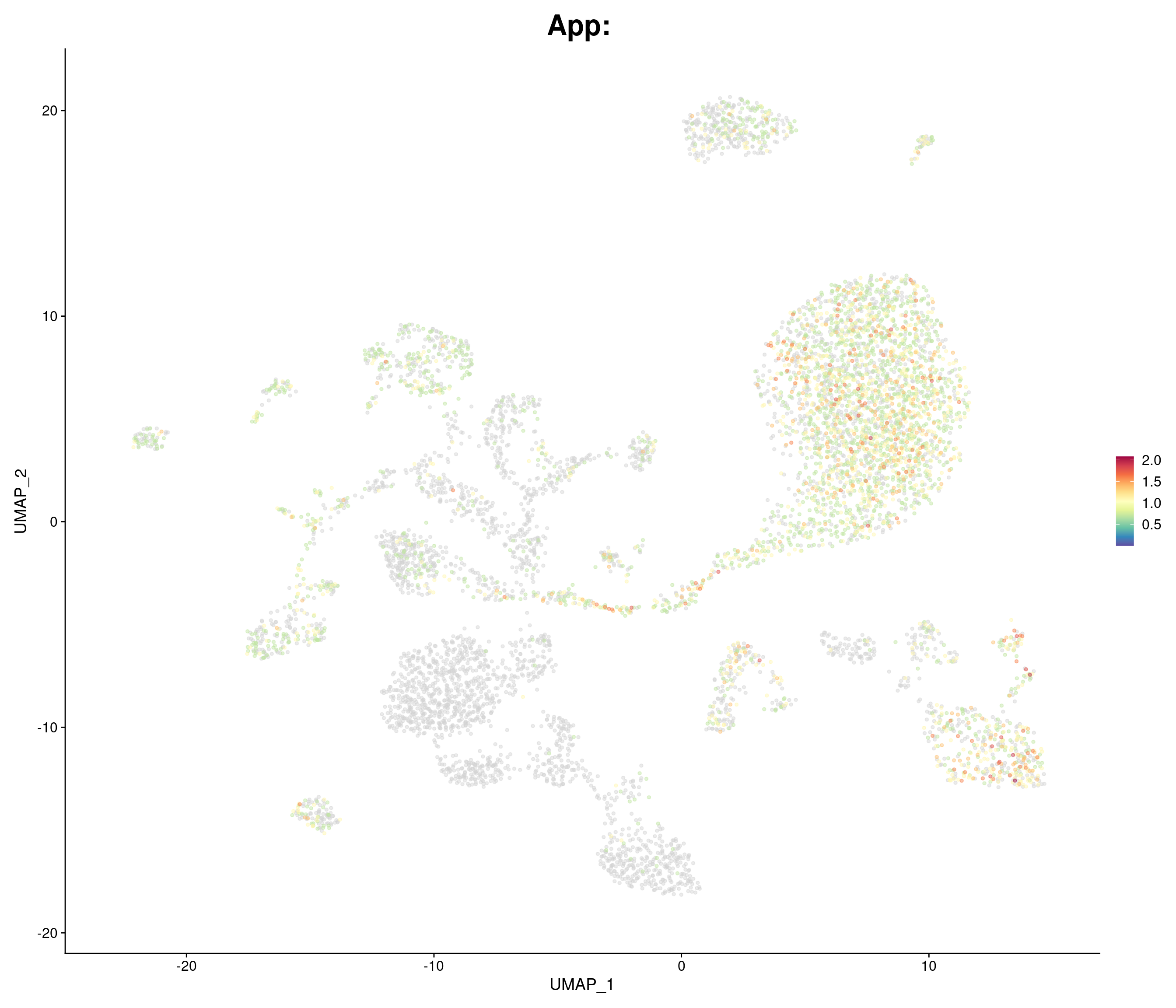
Aqp4
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Aqp4',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Aqp4')) +
theme(plot.title = element_text(size = 24))
Arf1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Arf1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Arf1')) +
theme(plot.title = element_text(size = 24))
Bmp7
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Bmp7',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Bmp7')) +
theme(plot.title = element_text(size = 24))
Bsg
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Bsg',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Bsg')) +
theme(plot.title = element_text(size = 24))
Ccl25
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ccl25',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ccl25')) +
theme(plot.title = element_text(size = 24))
Ckb
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ckb',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ckb')) +
theme(plot.title = element_text(size = 24))
Cnr1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Cnr1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Cnr1')) +
theme(plot.title = element_text(size = 24))
Col4a5
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Col4a5',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Col4a5')) +
theme(plot.title = element_text(size = 24))
Cst3
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Cst3',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Cst3')) +
theme(plot.title = element_text(size = 24))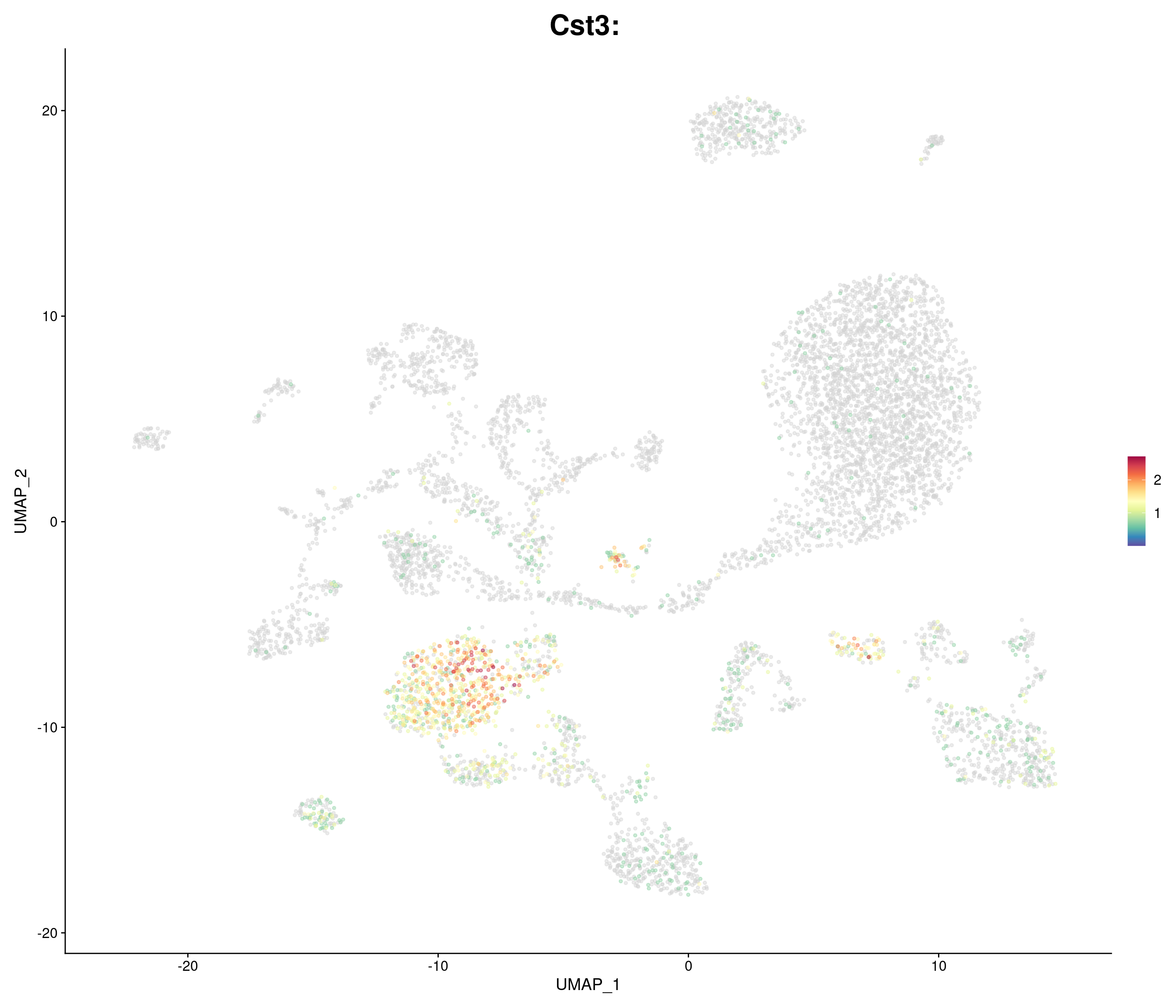
Dagla
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Dagla',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Dagla')) +
theme(plot.title = element_text(size = 24))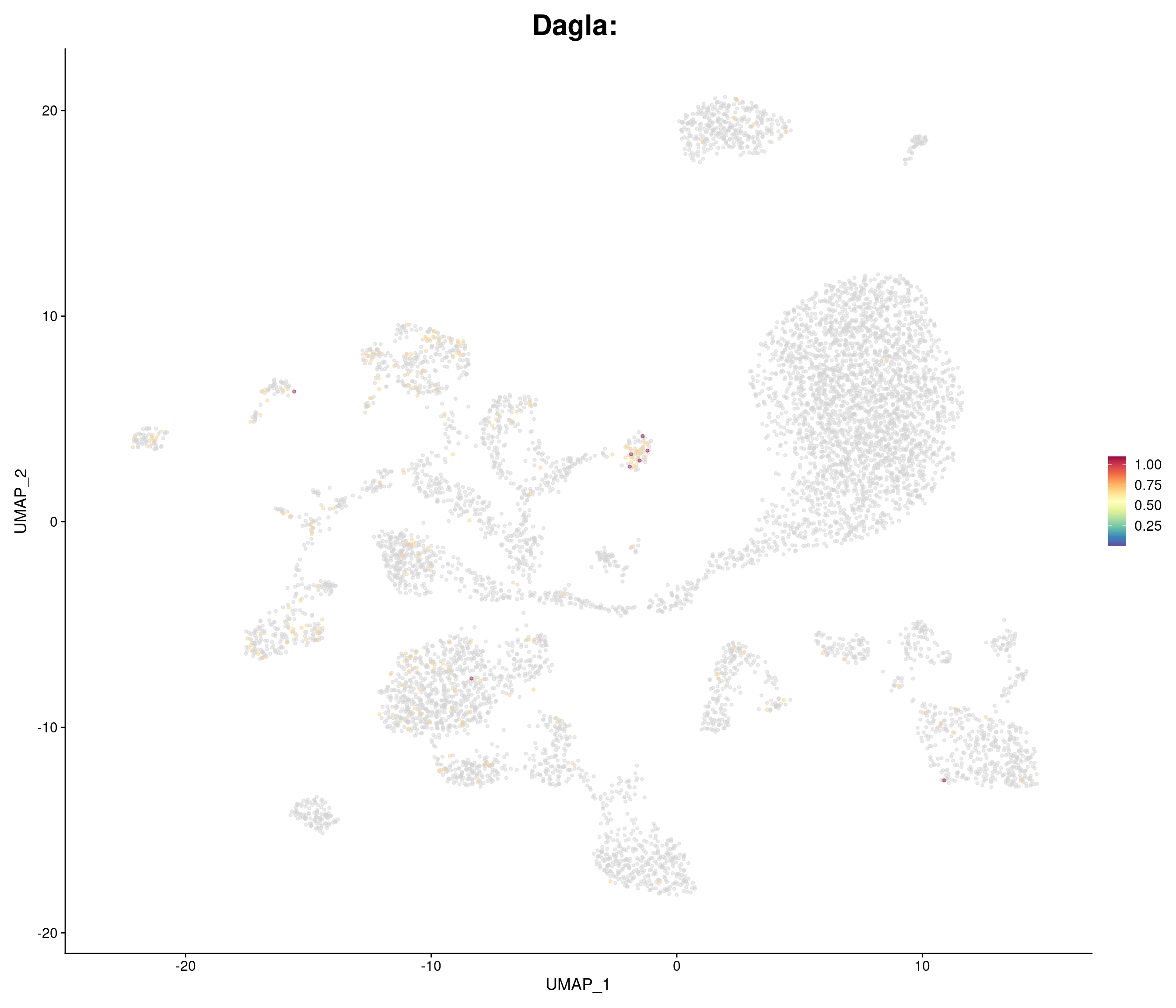
Daglb
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Daglb',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Daglb')) +
theme(plot.title = element_text(size = 24))
Decr2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Decr2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Decr2')) +
theme(plot.title = element_text(size = 24))
Dcc
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Dcc',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Dcc')) +
theme(plot.title = element_text(size = 24))
Dnm1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Dnm1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Dnm1')) +
theme(plot.title = element_text(size = 24))
Ech1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ech1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ech1')) +
theme(plot.title = element_text(size = 24))
Efna5
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Efna5',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Efna5')) +
theme(plot.title = element_text(size = 24))
Egfr
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Egfr',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Egfr')) +
theme(plot.title = element_text(size = 24))
Enho
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Enho',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Enho')) +
theme(plot.title = element_text(size = 24))
Eno1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Eno1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Eno1')) +
theme(plot.title = element_text(size = 24))
Faah
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Faah',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Faah')) +
theme(plot.title = element_text(size = 24))
Fgf1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Fgf1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Fgf1')) +
theme(plot.title = element_text(size = 24))
Fgfr3
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Fgfr3',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Fgfr3')) +
theme(plot.title = element_text(size = 24))
Fis1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Fis1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Fis1')) +
theme(plot.title = element_text(size = 24))
Fos
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Fos',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Fos')) +
theme(plot.title = element_text(size = 24))
Fth1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Fth1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Fth1')) +
theme(plot.title = element_text(size = 24))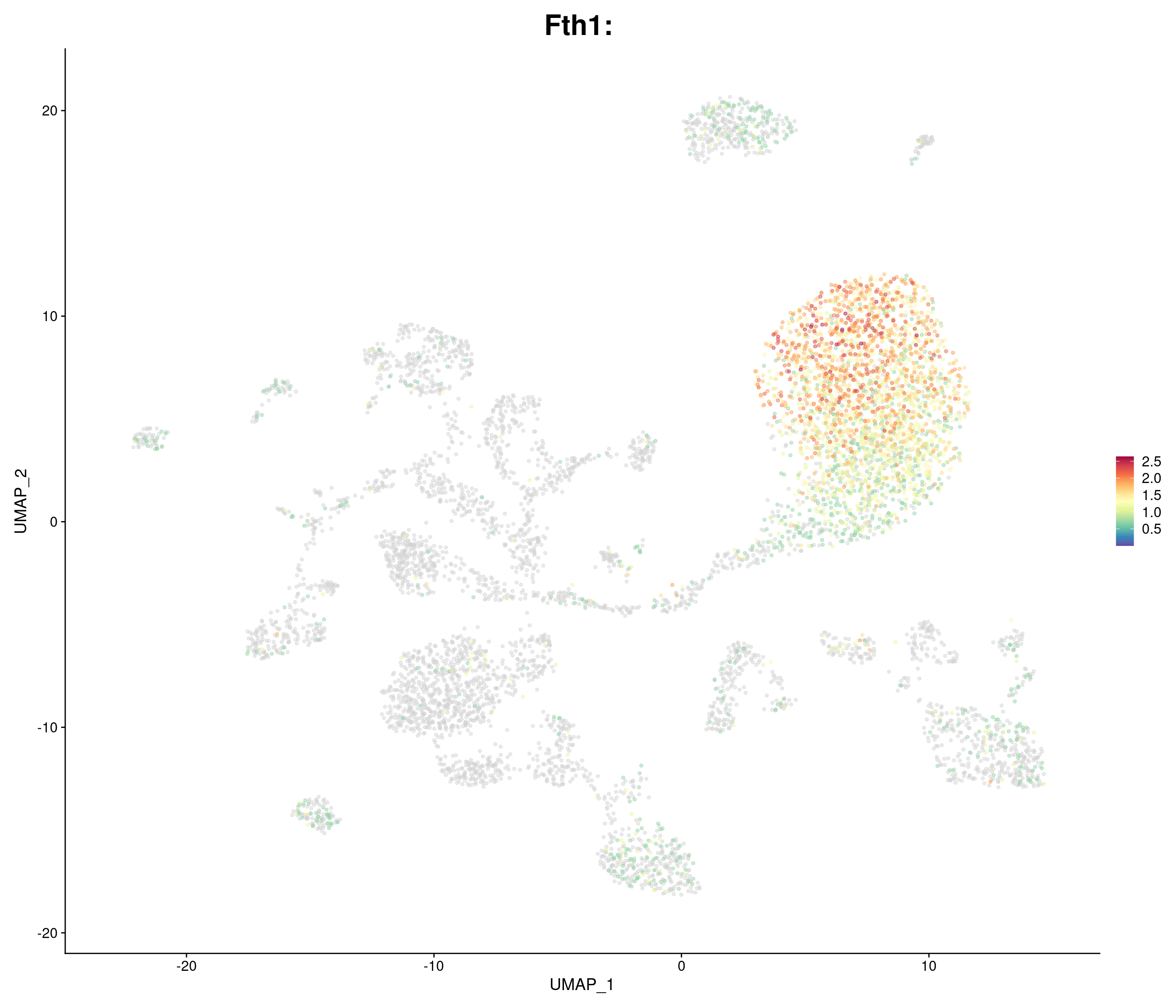
Ftl1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ftl1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ftl1')) +
theme(plot.title = element_text(size = 24))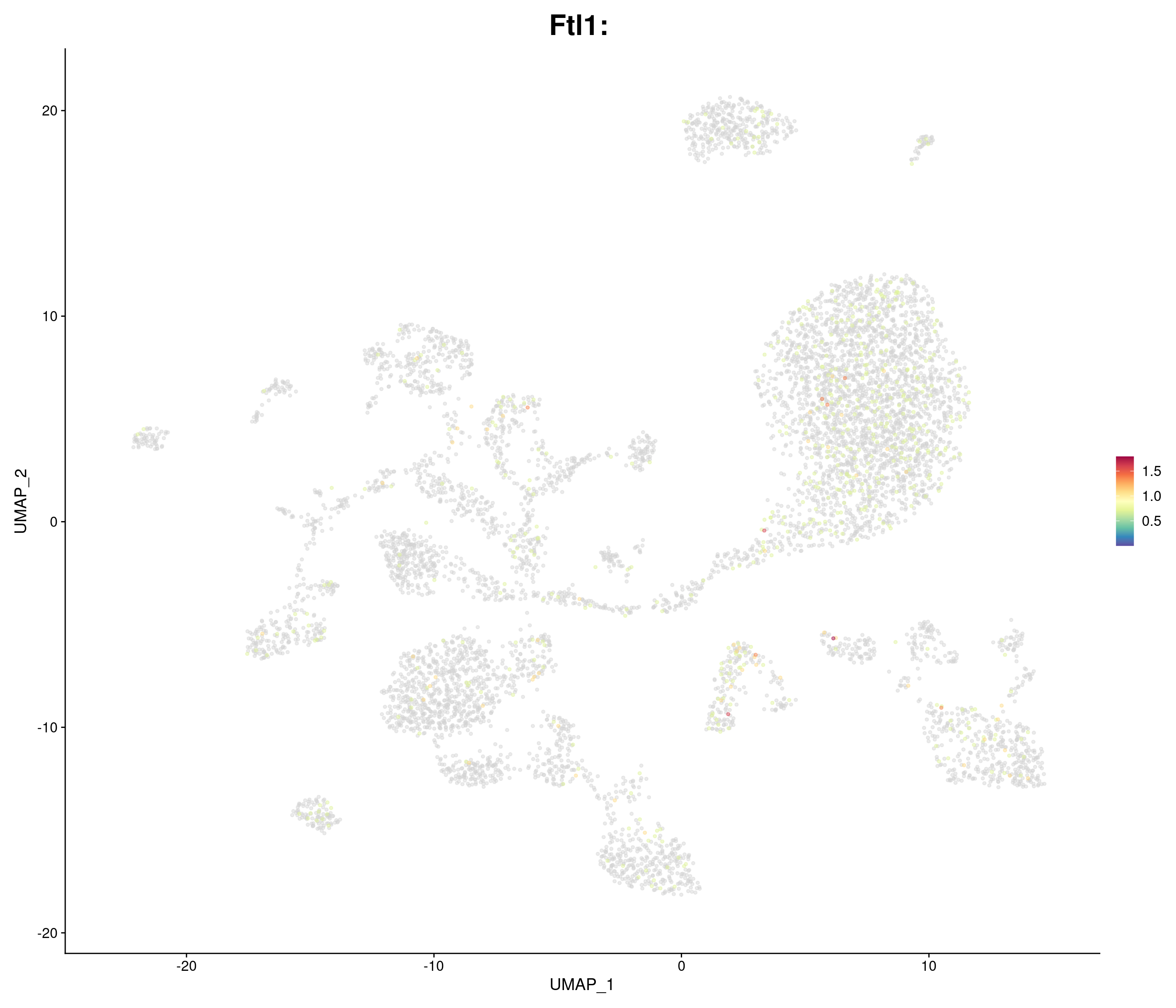
Gfap
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Gfap',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Gfap')) +
theme(plot.title = element_text(size = 24))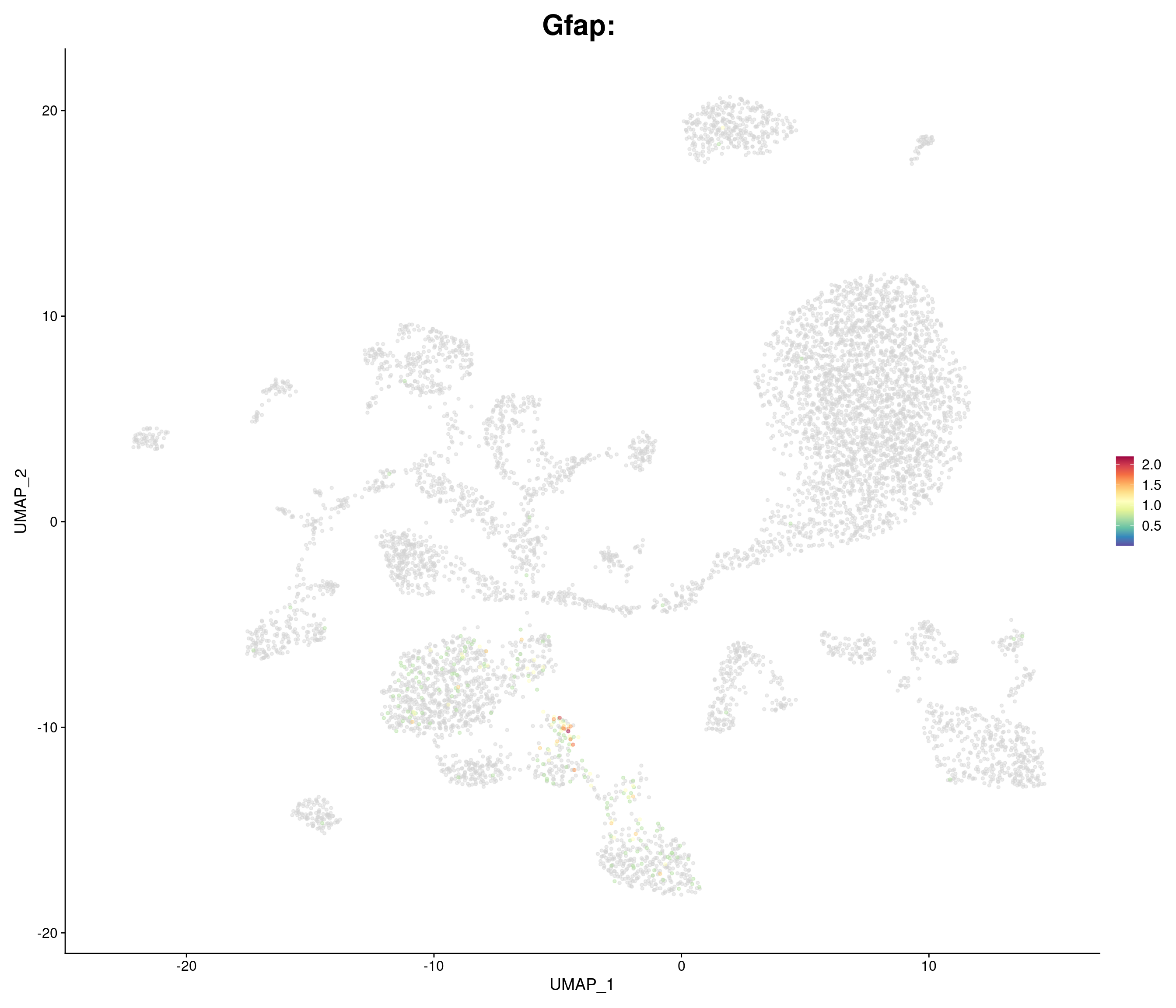
Gja1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Gja1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Gja1')) +
theme(plot.title = element_text(size = 24))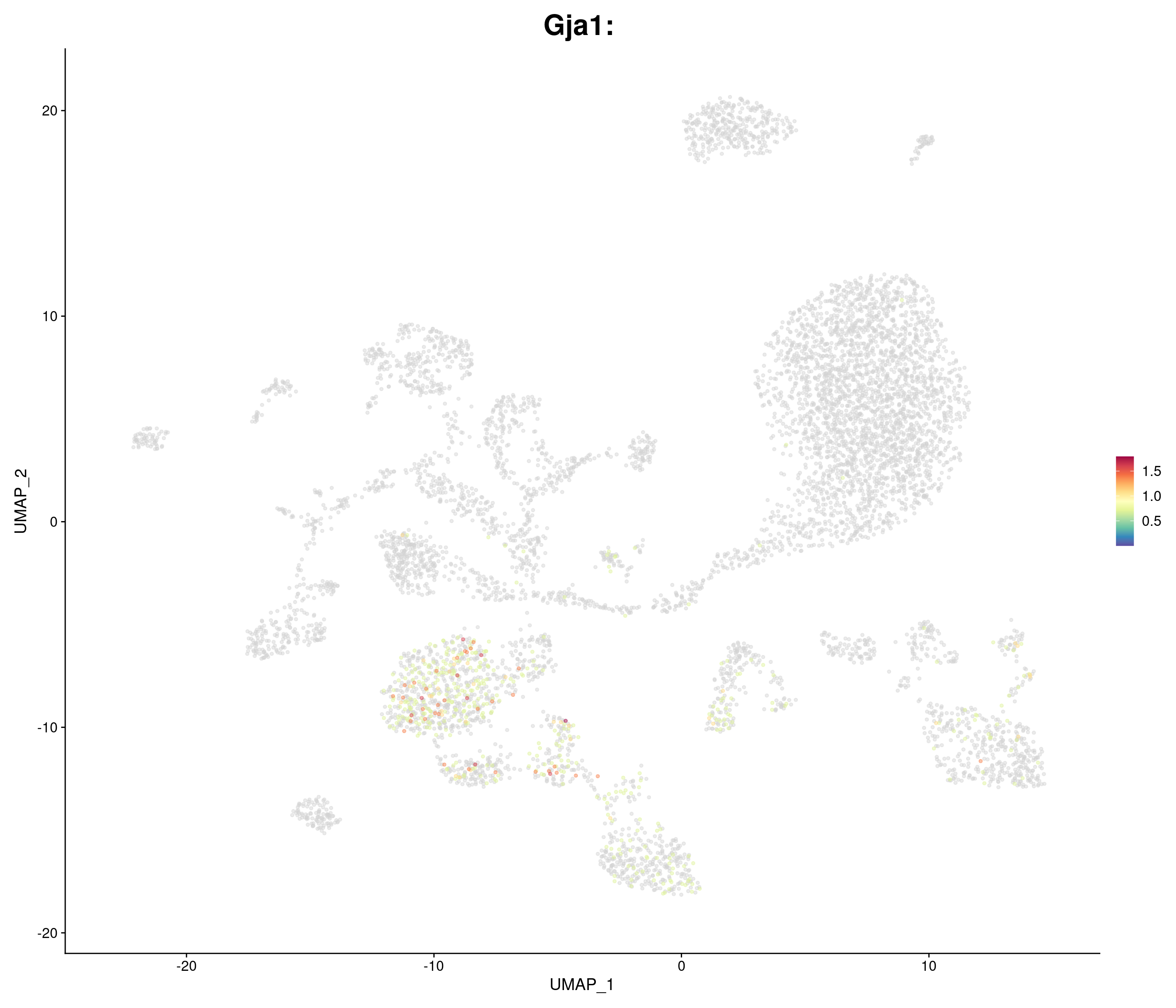
Gli1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Gli1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Gli1')) +
theme(plot.title = element_text(size = 24))
Glul
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Glul',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Glul')) +
theme(plot.title = element_text(size = 24))
Gnai2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Gnai2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Gnai2')) +
theme(plot.title = element_text(size = 24))
Gnas
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Gnas',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Gnas')) +
theme(plot.title = element_text(size = 24))
H2-K1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'H2-K1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'H2-K1')) +
theme(plot.title = element_text(size = 24))
Hacd2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Hacd2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Hacd2')) +
theme(plot.title = element_text(size = 24))
Hadhb
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Hadhb',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Hadhb')) +
theme(plot.title = element_text(size = 24))
Hbegf
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Hbegf',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Hbegf')) +
theme(plot.title = element_text(size = 24))
Hepacam
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Hepacam',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Hepacam')) +
theme(plot.title = element_text(size = 24))
Htra1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Htra1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Htra1')) +
theme(plot.title = element_text(size = 24))
Igsf1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Igsf1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Igsf1')) +
theme(plot.title = element_text(size = 24))
Il18
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Il18',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Il18')) +
theme(plot.title = element_text(size = 24))
Il1rapl1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Il1rapl1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Il1rapl1')) +
theme(plot.title = element_text(size = 24))
Itgav
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Itgav',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Itgav')) +
theme(plot.title = element_text(size = 24))
Jam2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Jam2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Jam2')) +
theme(plot.title = element_text(size = 24))
Lama2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Lama2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Lama2')) +
theme(plot.title = element_text(size = 24))
Lamb2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Lamb2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Lamb2')) +
theme(plot.title = element_text(size = 24))
Lcat
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Lcat',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Lcat')) +
theme(plot.title = element_text(size = 24))
Lgi1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Lgi1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Lgi1')) +
theme(plot.title = element_text(size = 24))
Lgi4
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Lgi4',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Lgi4')) +
theme(plot.title = element_text(size = 24))
Lpcat3
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Lpcat3',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Lpcat3')) +
theme(plot.title = element_text(size = 24))
Lrpap1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Lrpap1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Lrpap1')) +
theme(plot.title = element_text(size = 24))
Lrrc4b
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Lrrc4b',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Lrrc4b')) +
theme(plot.title = element_text(size = 24))
Lxn
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Lxn',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Lxn')) +
theme(plot.title = element_text(size = 24))
Mdk
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Mdk',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Mdk')) +
theme(plot.title = element_text(size = 24))
Mfn1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Mfn1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Mfn1')) +
theme(plot.title = element_text(size = 24))
Mfn2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Mfn2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Mfn2')) +
theme(plot.title = element_text(size = 24))
Mgll
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Mgll',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Mgll')) +
theme(plot.title = element_text(size = 24))
Mief1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Mief1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Mief1')) +
theme(plot.title = element_text(size = 24))
Napepld
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Napepld',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Napepld')) +
theme(plot.title = element_text(size = 24))
Ncam1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ncam1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ncam1')) +
theme(plot.title = element_text(size = 24))
Ncan
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ncan',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ncan')) +
theme(plot.title = element_text(size = 24))
Ndrg2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ndrg2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ndrg2')) +
theme(plot.title = element_text(size = 24))
Nfasc
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Nfasc',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Nfasc')) +
theme(plot.title = element_text(size = 24))
Nfia
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Nfia',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Nfia')) +
theme(plot.title = element_text(size = 24))
Nlgn3
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Nlgn3',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Nlgn3')) +
theme(plot.title = element_text(size = 24))
Nrxn1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Nrxn1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Nrxn1')) +
theme(plot.title = element_text(size = 24))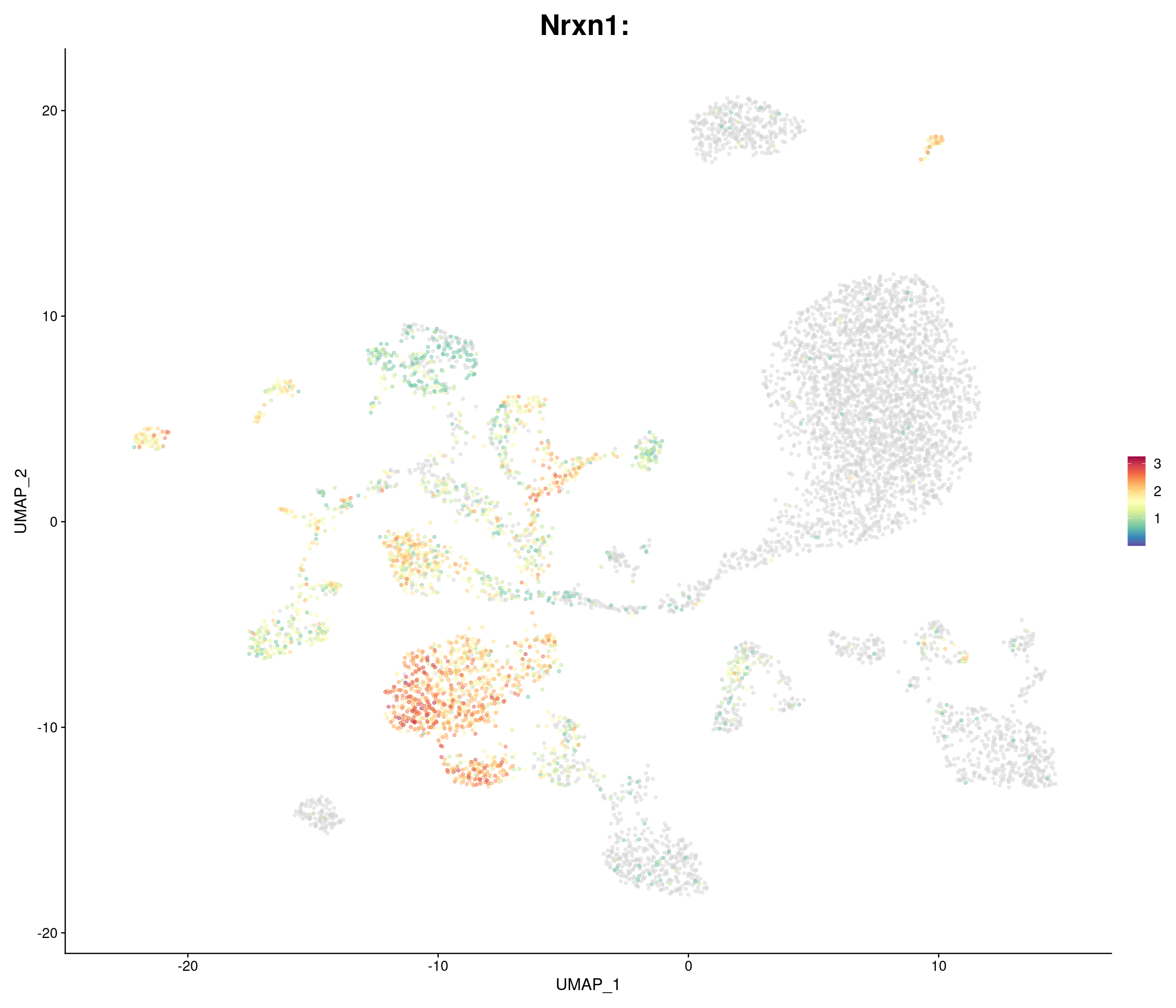
Nrxn2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Nrxn2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Nrxn2')) +
theme(plot.title = element_text(size = 24))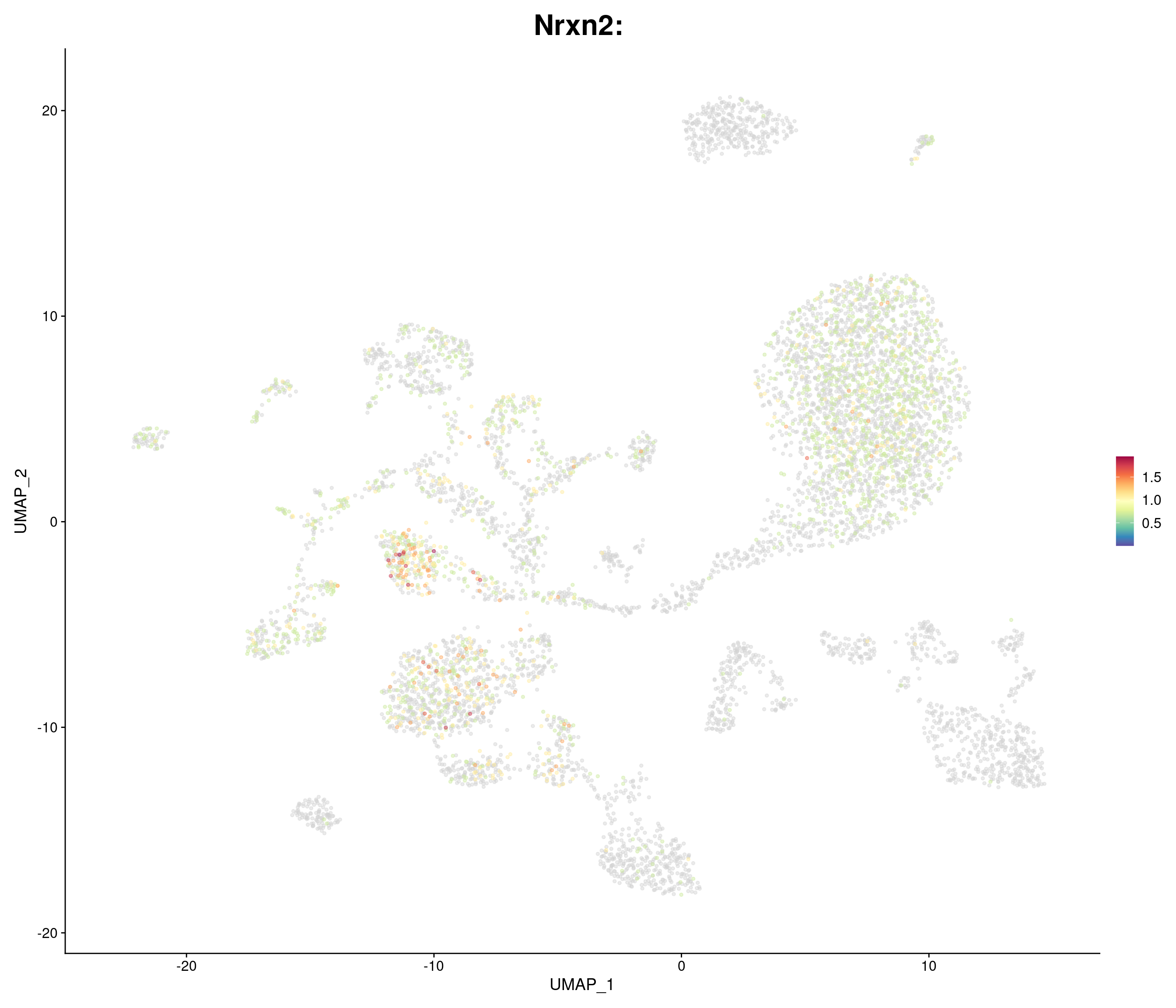
Ntn1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ntn1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ntn1')) +
theme(plot.title = element_text(size = 24))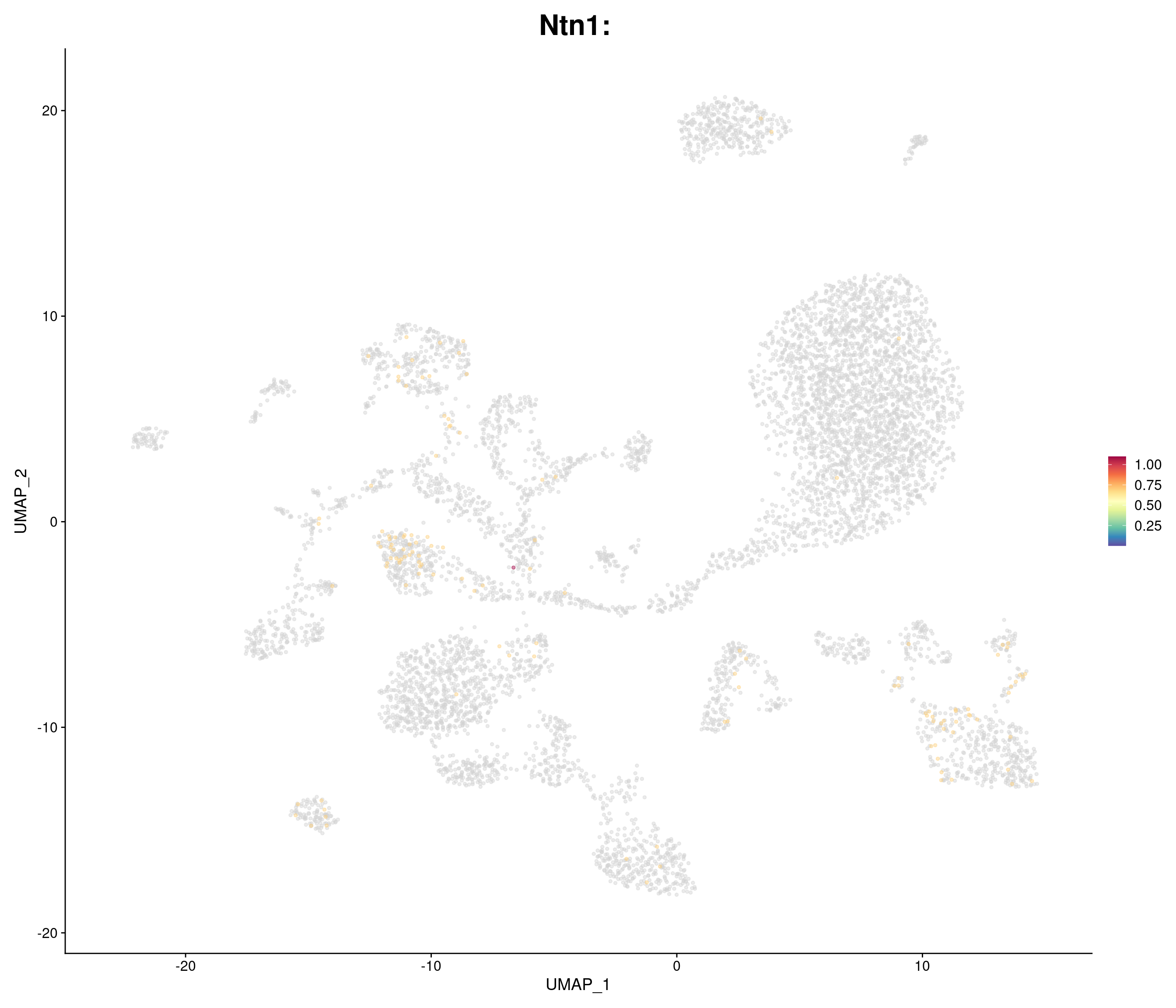
Ntrk3
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ntrk3',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ntrk3')) +
theme(plot.title = element_text(size = 24))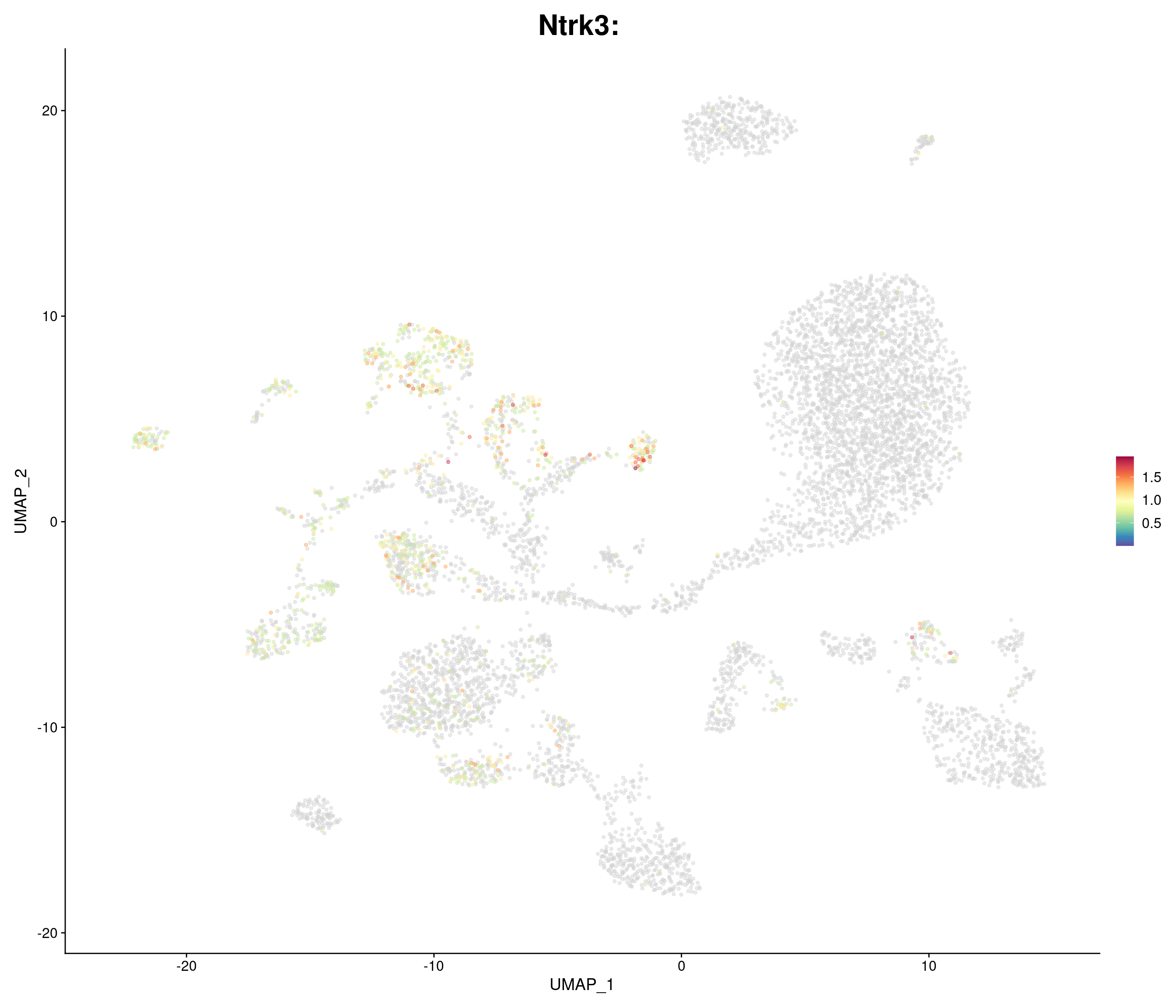
Opa1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Opa1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Opa1')) +
theme(plot.title = element_text(size = 24))
Pex1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex1')) +
theme(plot.title = element_text(size = 24))
Pex10
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex10',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex10')) +
theme(plot.title = element_text(size = 24))
Pex12
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex12',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex12')) +
theme(plot.title = element_text(size = 24))
Pex13
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex13',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex13')) +
theme(plot.title = element_text(size = 24))
Pex14
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex14',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex14')) +
theme(plot.title = element_text(size = 24))
Pex16
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex16',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex16')) +
theme(plot.title = element_text(size = 24))
Pex2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex2')) +
theme(plot.title = element_text(size = 24))
Pex26
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex26',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex26')) +
theme(plot.title = element_text(size = 24))
Pex3
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex3',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex3')) +
theme(plot.title = element_text(size = 24))
Pex6
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pex6',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pex6')) +
theme(plot.title = element_text(size = 24))
Pkm
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pkm',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pkm')) +
theme(plot.title = element_text(size = 24))
Pla2g7
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pla2g7',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pla2g7')) +
theme(plot.title = element_text(size = 24))
Plcb1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Plcb1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Plcb1')) +
theme(plot.title = element_text(size = 24))
Psap
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Psap',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Psap')) +
theme(plot.title = element_text(size = 24))
Ptn
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ptn',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ptn')) +
theme(plot.title = element_text(size = 24))
Pygb
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Pygb',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Pygb')) +
theme(plot.title = element_text(size = 24))
Ralyl
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Ralyl',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Ralyl')) +
theme(plot.title = element_text(size = 24))
Rgma
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Rgma',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Rgma')) +
theme(plot.title = element_text(size = 24))
Rtn4
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Rtn4',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Rtn4')) +
theme(plot.title = element_text(size = 24))
S100a1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'S100a1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'S100a1')) +
theme(plot.title = element_text(size = 24))
S100a6
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'S100a6',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'S100a6')) +
theme(plot.title = element_text(size = 24))
S100b
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'S100b',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'S100b')) +
theme(plot.title = element_text(size = 24))
Scd2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Scd2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Scd2')) +
theme(plot.title = element_text(size = 24))
Sdc2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Sdc2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Sdc2')) +
theme(plot.title = element_text(size = 24))
Sema6a
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Sema6a',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Sema6a')) +
theme(plot.title = element_text(size = 24))
Sema6d
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Sema6d',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Sema6d')) +
theme(plot.title = element_text(size = 24))
Sgcd
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Sgcd',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Sgcd')) +
theme(plot.title = element_text(size = 24))
Sirpa
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Sirpa',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Sirpa')) +
theme(plot.title = element_text(size = 24))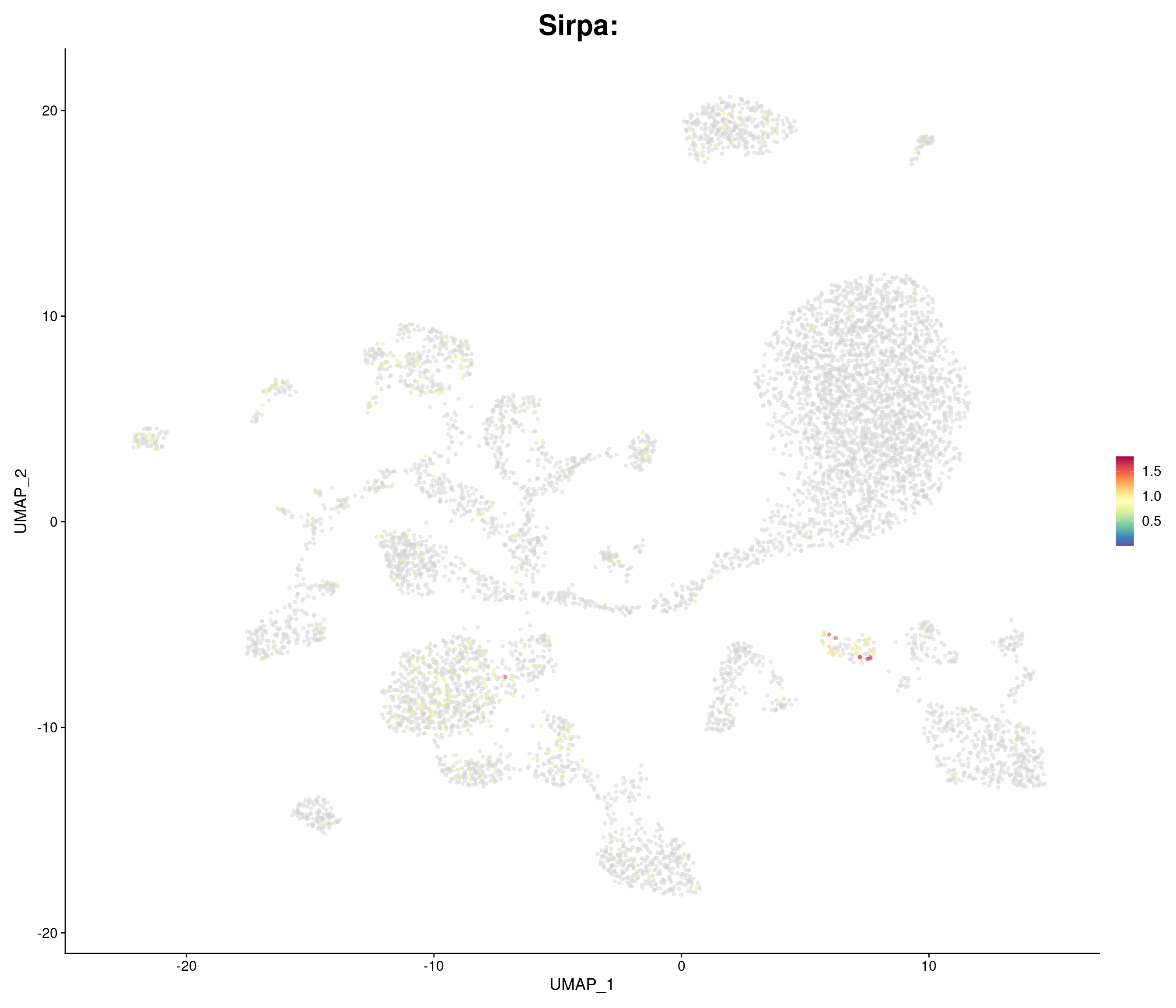
Slc1a2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Slc1a2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Slc1a2')) +
theme(plot.title = element_text(size = 24))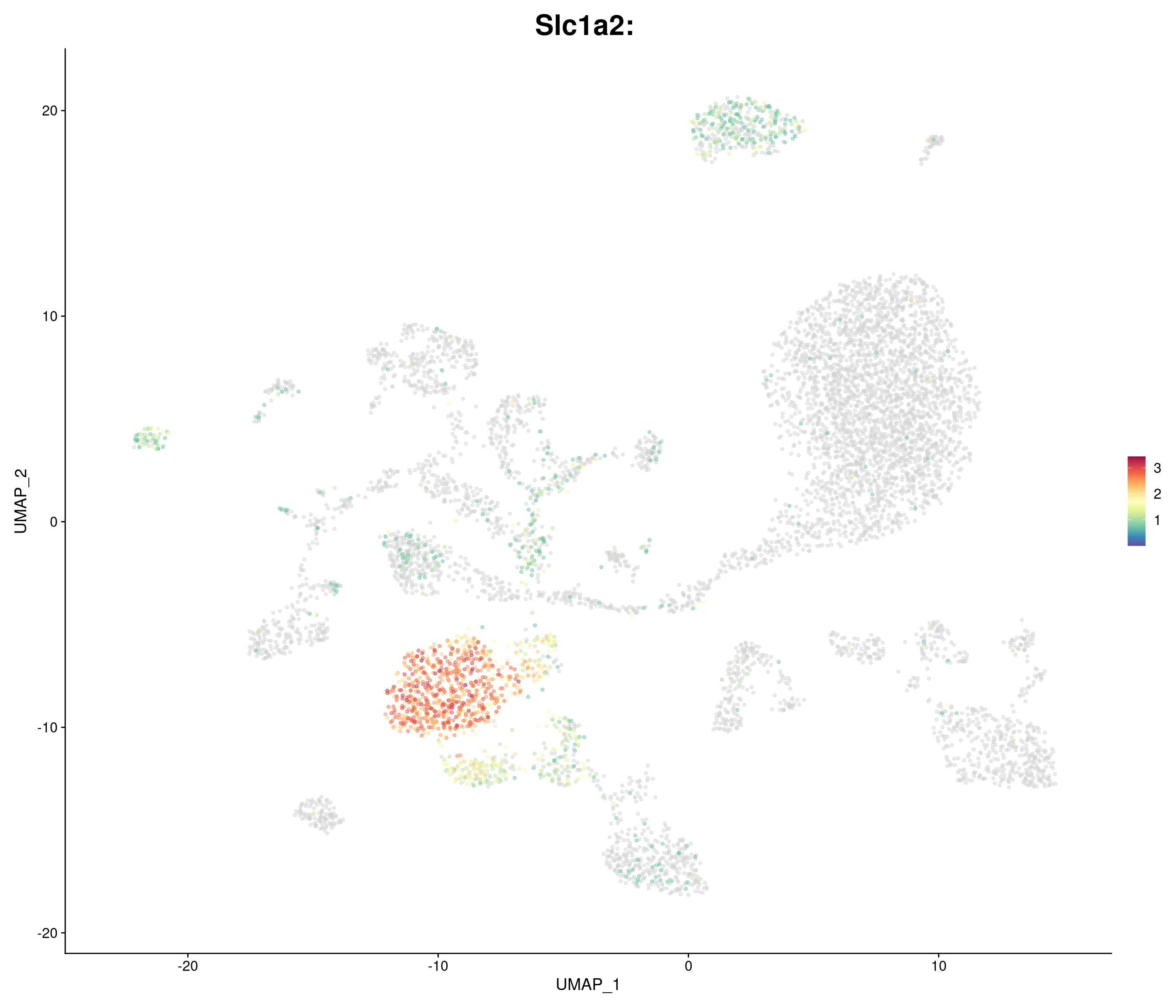
Slc1a3
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Slc1a3',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Slc1a3')) +
theme(plot.title = element_text(size = 24))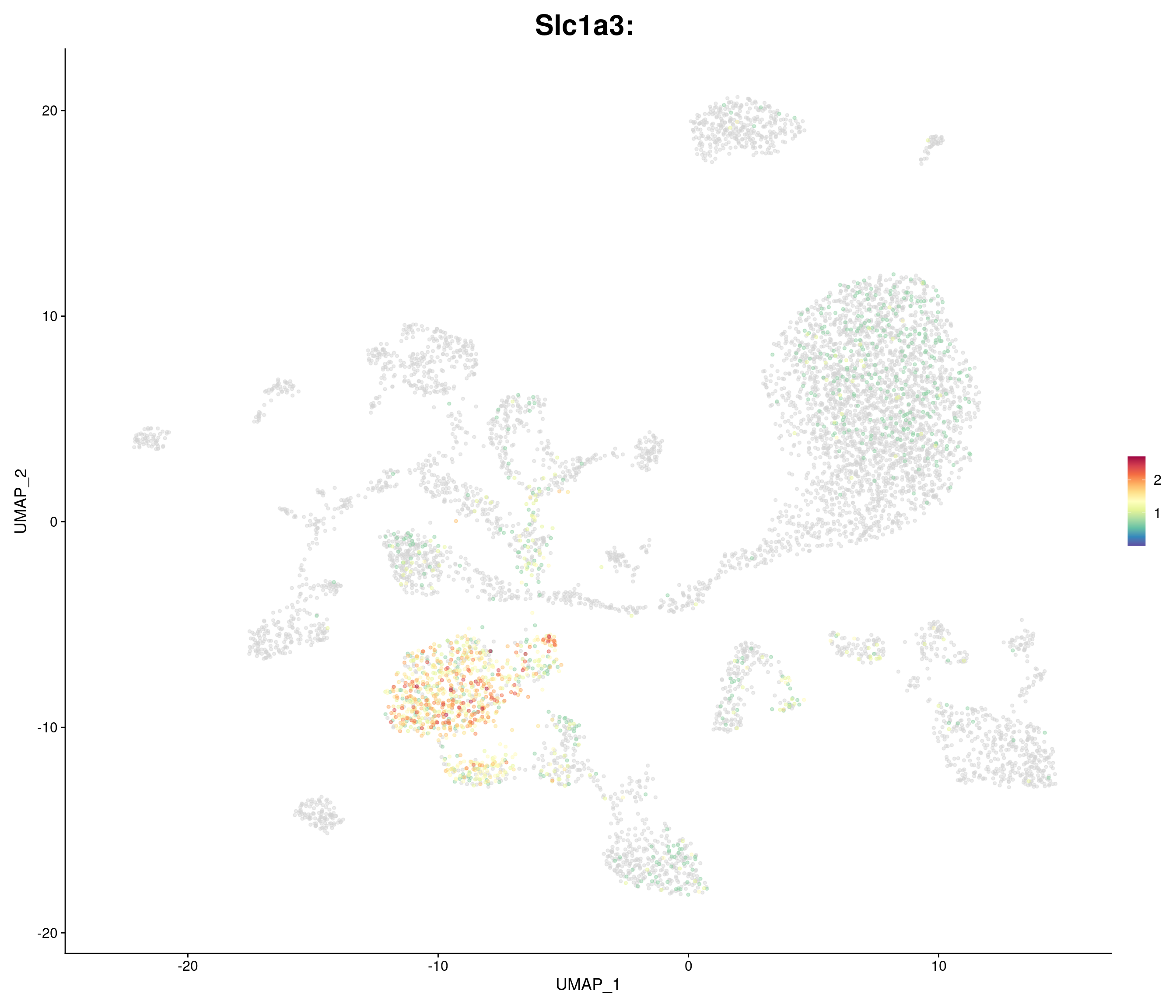
Slc38a1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Slc38a1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Slc38a1')) +
theme(plot.title = element_text(size = 24))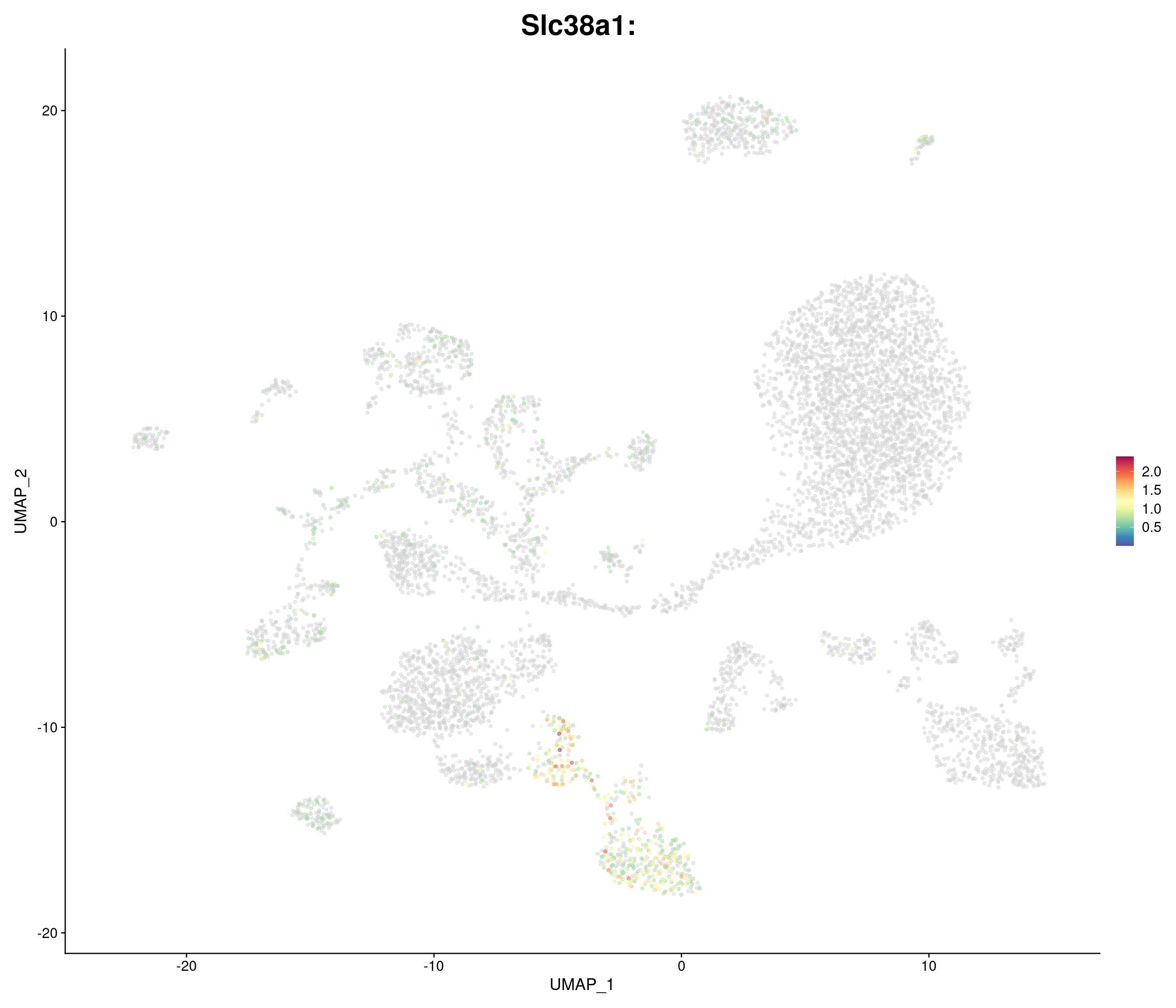
Slc4a4
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Slc4a4',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Slc4a4')) +
theme(plot.title = element_text(size = 24))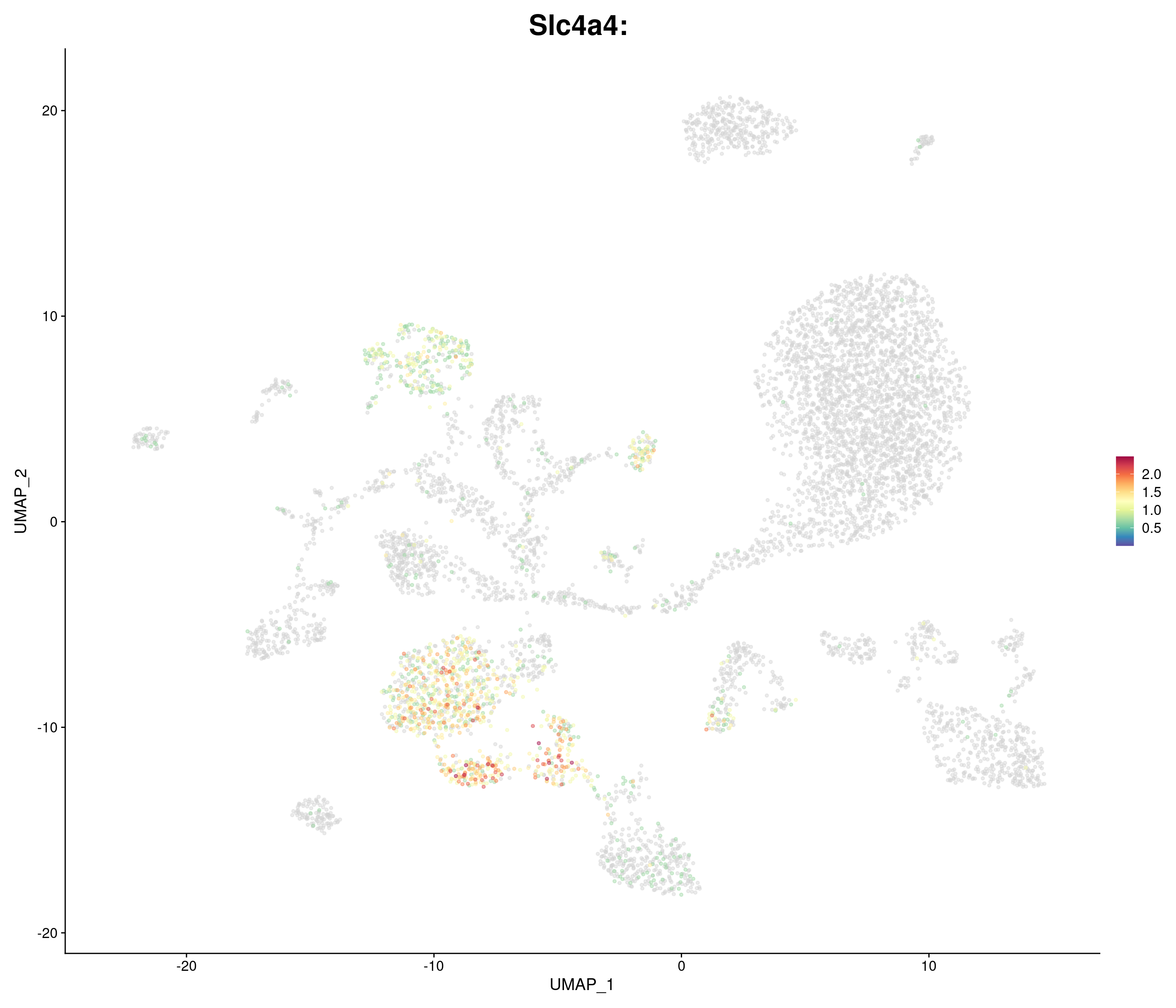
Slc6a11
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Slc6a11',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Slc6a11')) +
theme(plot.title = element_text(size = 24))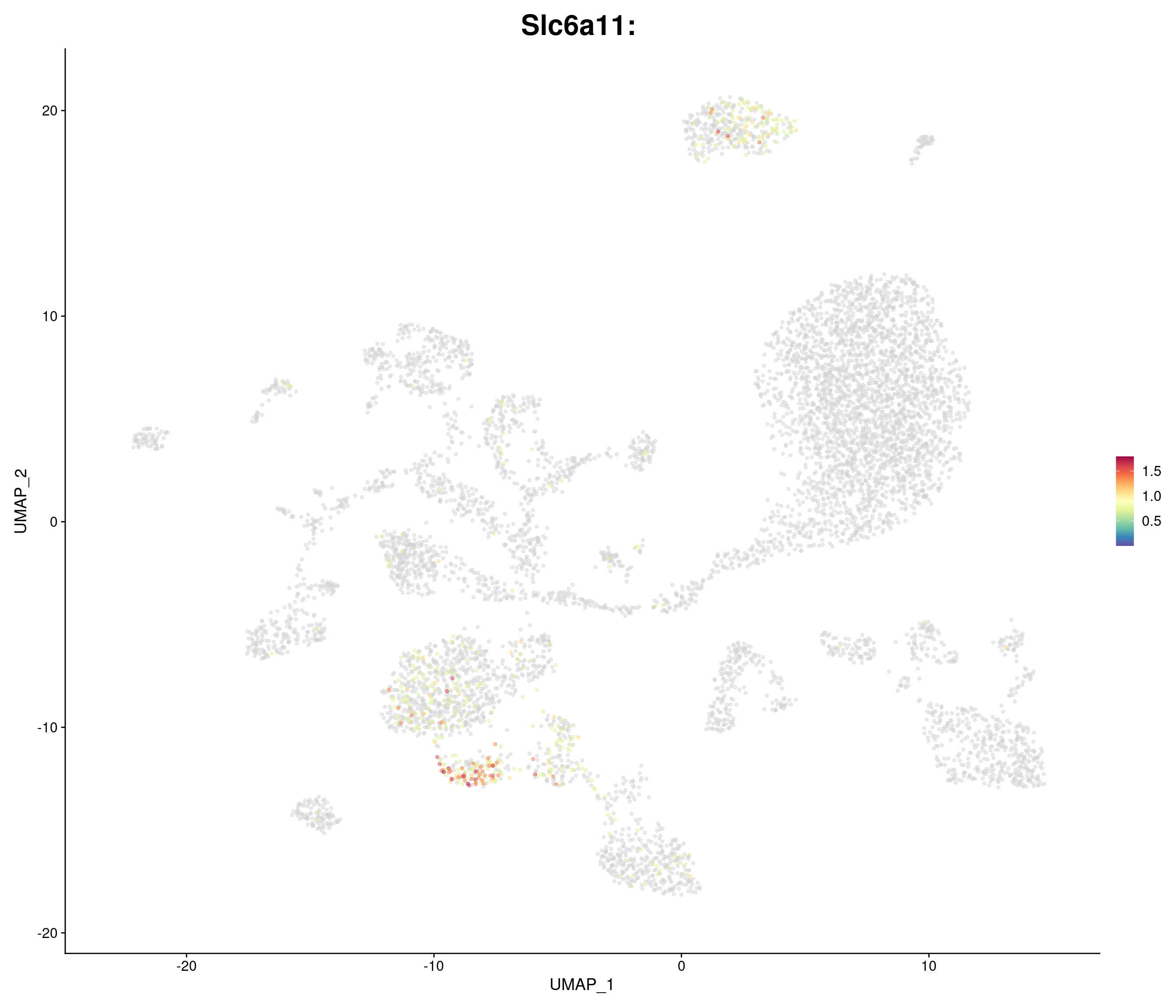
Slc7a10
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Slc7a10',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Slc7a10')) +
theme(plot.title = element_text(size = 24))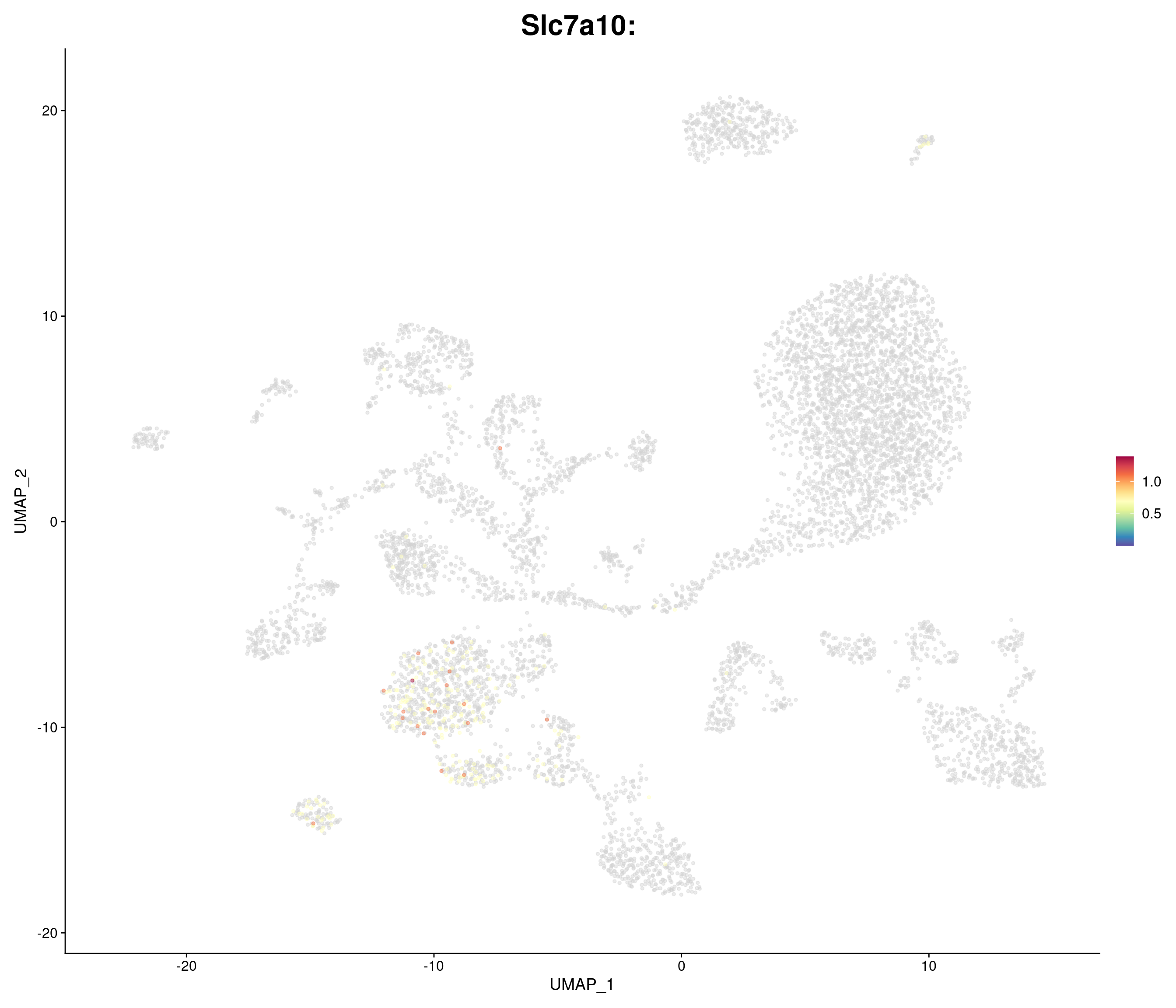
Slit1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Slit1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Slit1')) +
theme(plot.title = element_text(size = 24))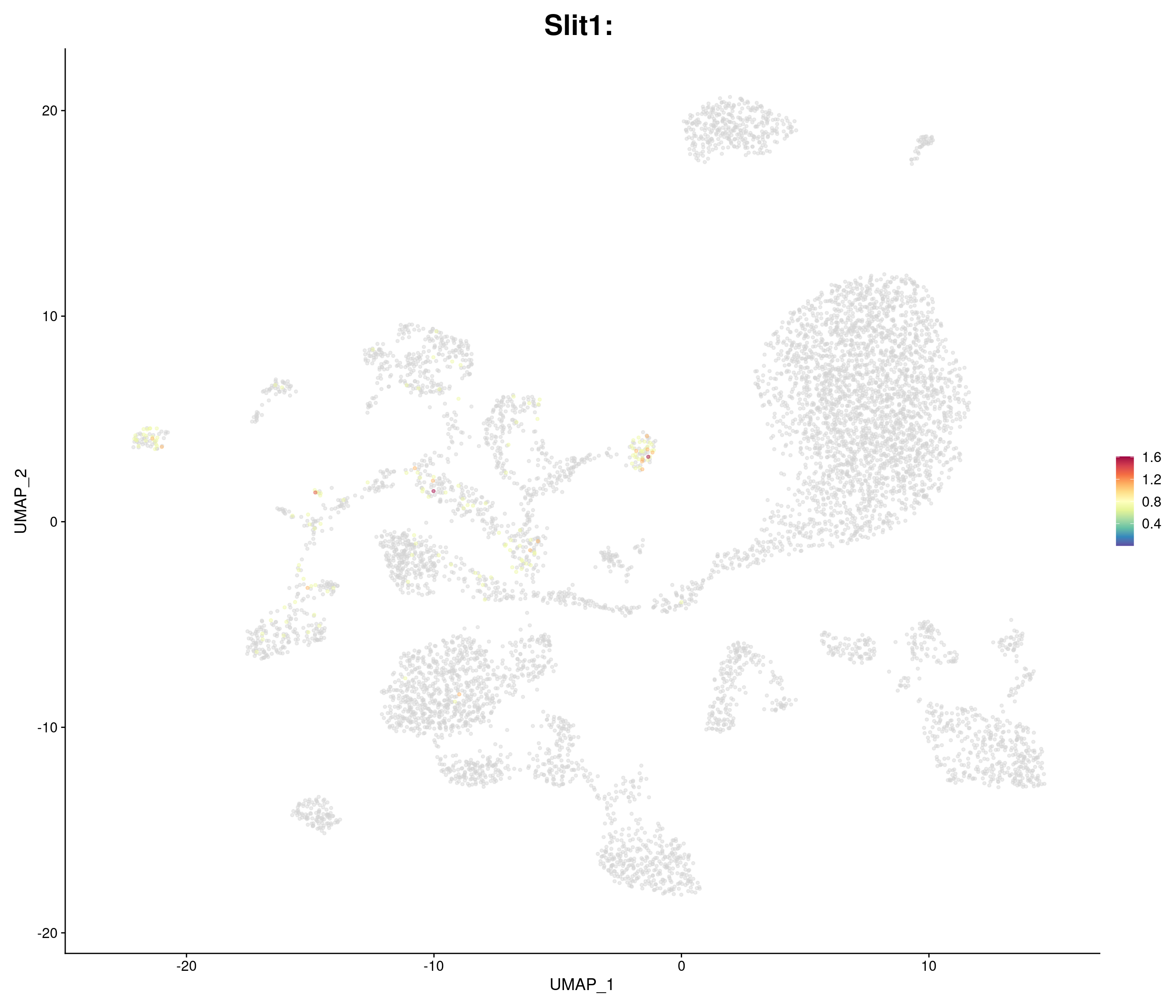
Slit2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Slit2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Slit2')) +
theme(plot.title = element_text(size = 24))
Slitrk2
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Slitrk2',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Slitrk2')) +
theme(plot.title = element_text(size = 24))
Sorbs1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Sorbs1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Sorbs1')) +
theme(plot.title = element_text(size = 24))
Sox9
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Sox9',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Sox9')) +
theme(plot.title = element_text(size = 24))
Sparc
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Sparc',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Sparc')) +
theme(plot.title = element_text(size = 24))
Spon1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Spon1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Spon1')) +
theme(plot.title = element_text(size = 24))
Tafa1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Tafa1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Tafa1')) +
theme(plot.title = element_text(size = 24))
Timp3
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Timp3',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Timp3')) +
theme(plot.title = element_text(size = 24))
Tkt
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Tkt',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Tkt')) +
theme(plot.title = element_text(size = 24))
Vcam1
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Vcam1',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Vcam1')) +
theme(plot.title = element_text(size = 24))
Vegfa
FeaturePlot_scCustom(
seurat_object = combined_srt, features = 'Vegfa',
pt.size = 1, order = TRUE,
colors_use = combined_srt@misc$expr_Colour_Pal,
alpha_na_exp = 0.1, alpha_exp = 0.45) +
ggtitle(sprintf('%s: ', 'Vegfa')) +
theme(plot.title = element_text(size = 24))
Dot-plot of gene expression
Transcription Factors
plt_g_tf <- transcription_factors %>% .[. %in% hvg]SCT
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "SCT",
features = plt_g_tf,
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "G")
)
RNA
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "RNA",
features = plt_g_tf,
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "E")
)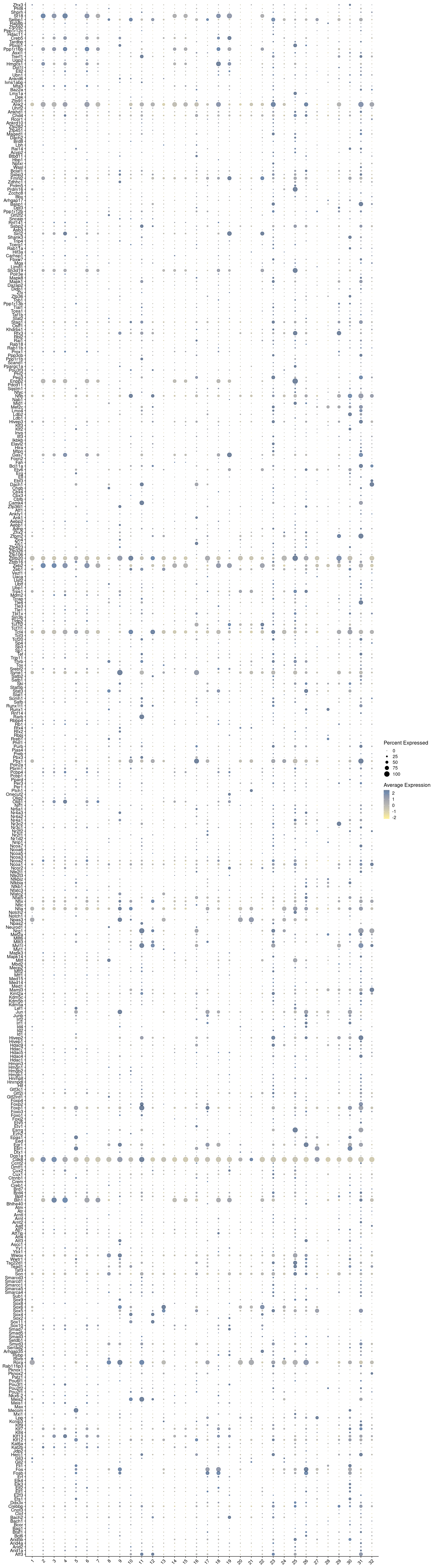
Glutamate
plt_g_glut <- c(glut, glutr) %>% .[. %in% hvg]SCT
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "SCT",
features = plt_g_glut,
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "G")
)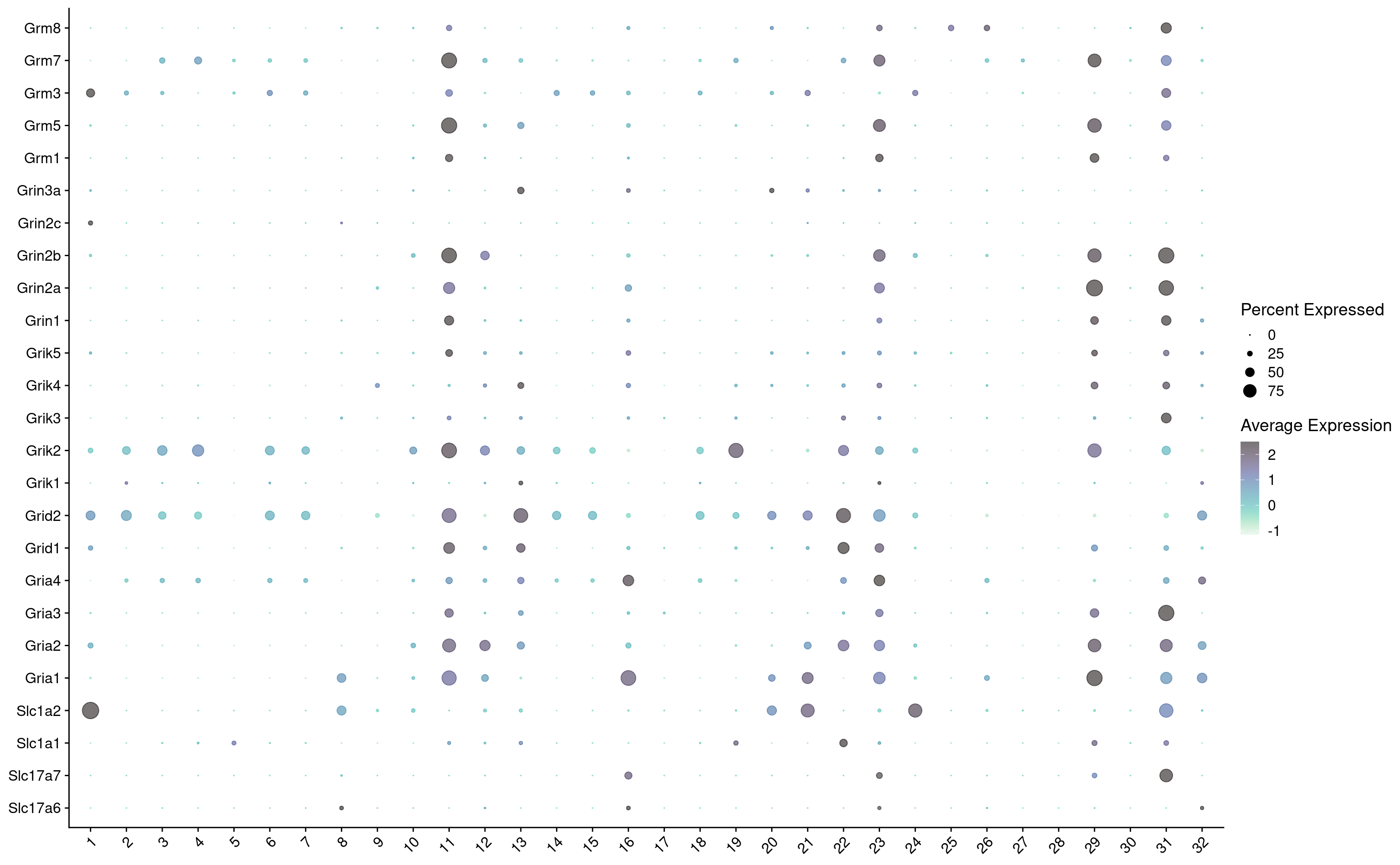
RNA
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "RNA",
features = plt_g_glut,
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "E")
)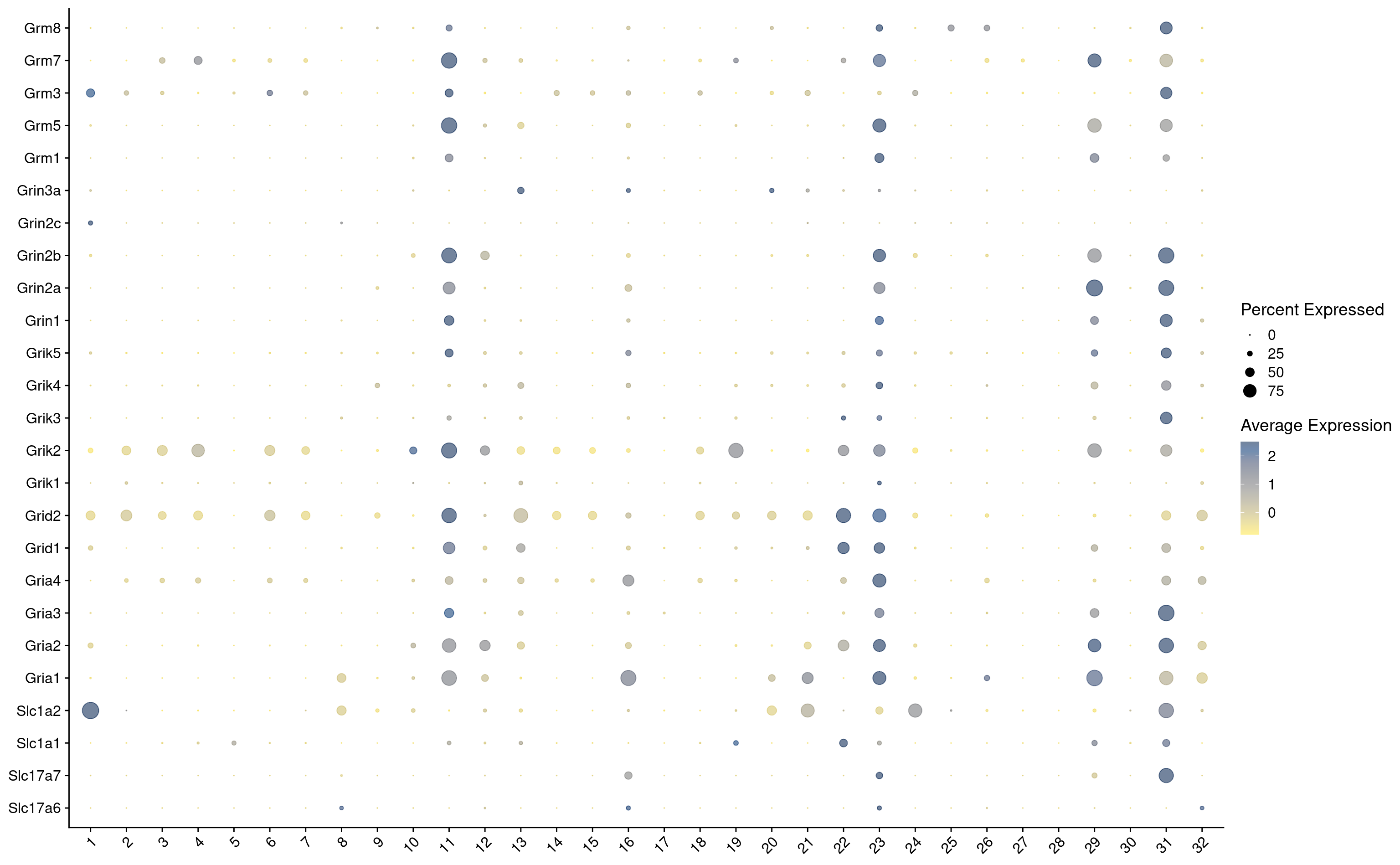
GABA
plt_g_gaba <- c(gaba, gabar) %>% .[. %in% hvg]SCT
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "SCT",
features = unique(plt_g_gaba),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "G")
)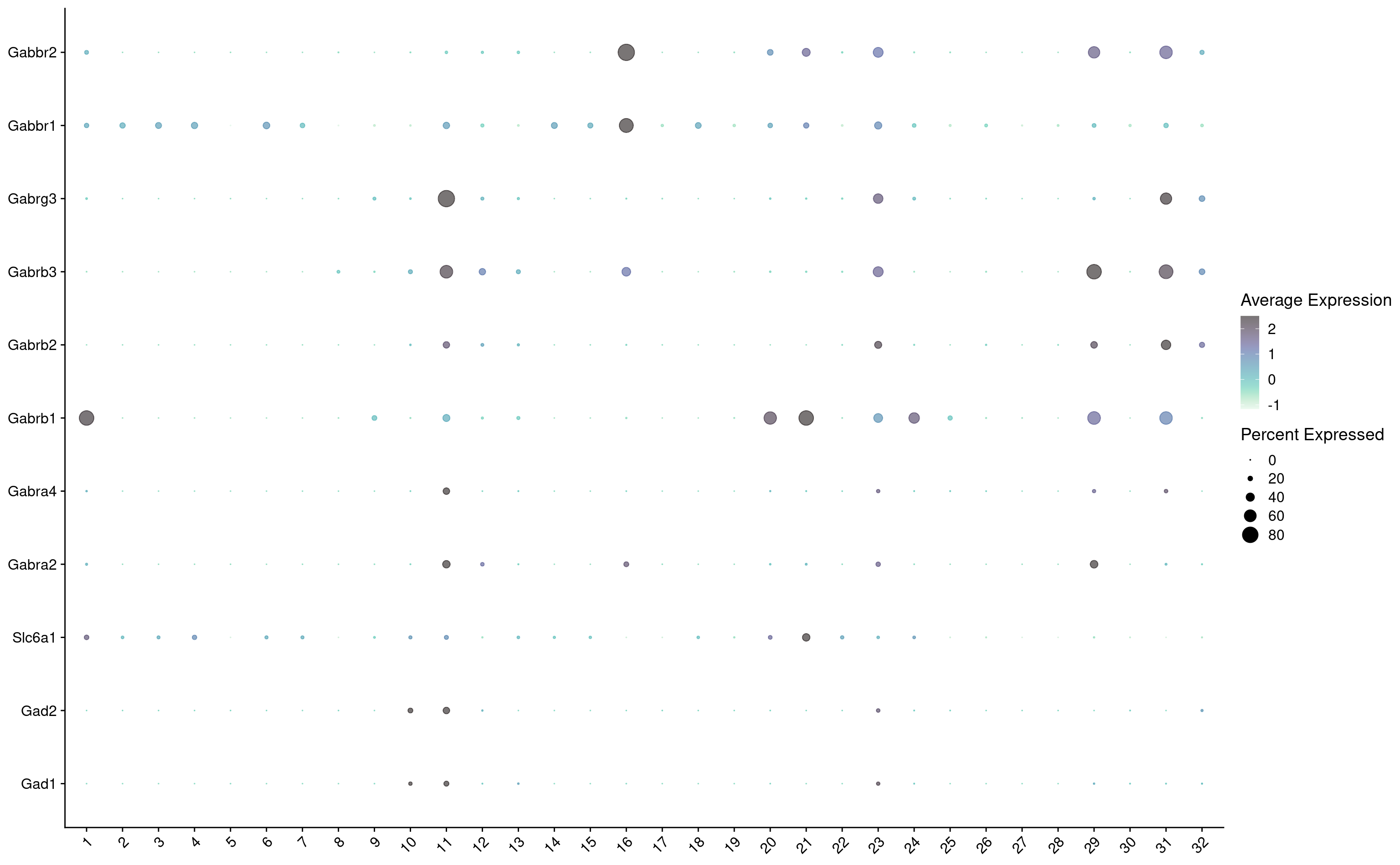
RNA
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "RNA",
features = unique(plt_g_gaba),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "E")
)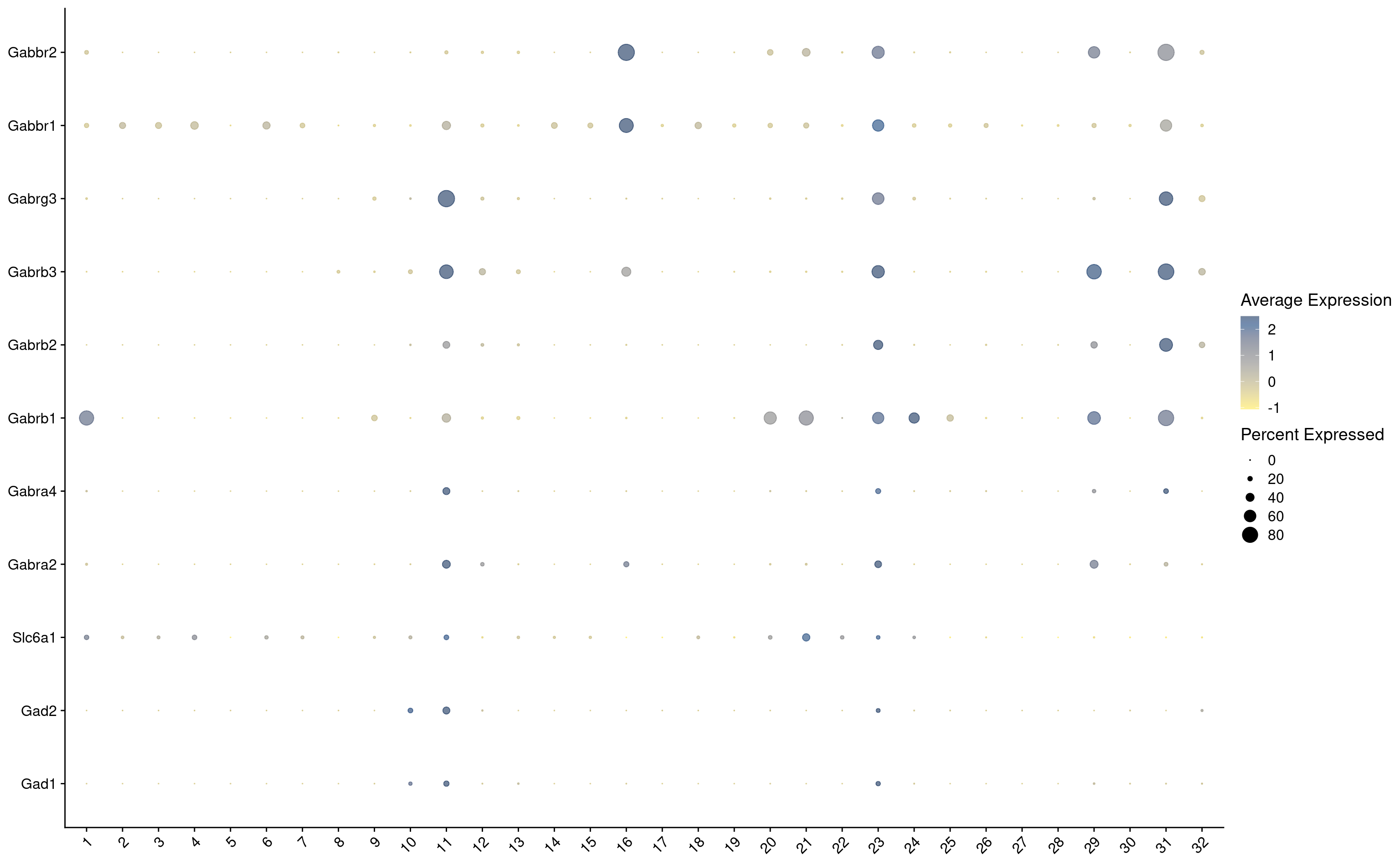
Neuropeptides
plt_g_np <- c(np) %>% .[. %in% hvg]SCT
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "SCT",
features = unique(plt_g_np),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "G")
)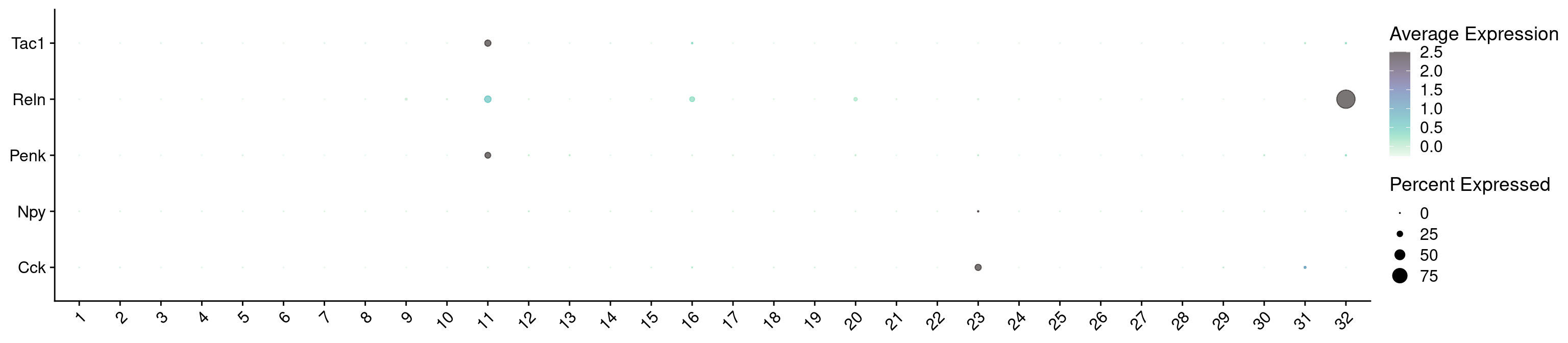
RNA
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "RNA",
features = unique(plt_g_np),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "E")
)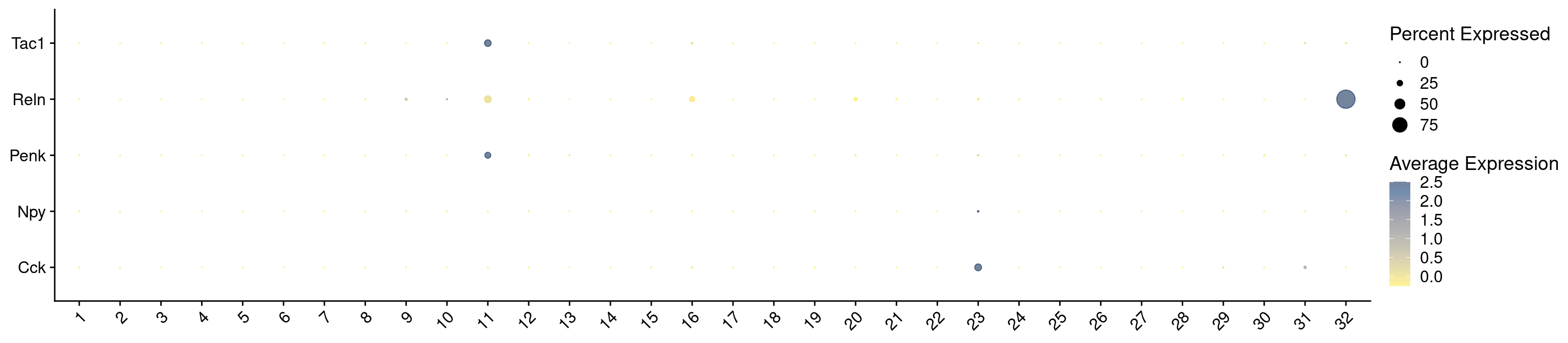
Neuropeptide receptors
plt_g_npr <- c(npr) %>% .[. %in% hvg]SCT
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "SCT",
features = unique(plt_g_npr),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "G")
)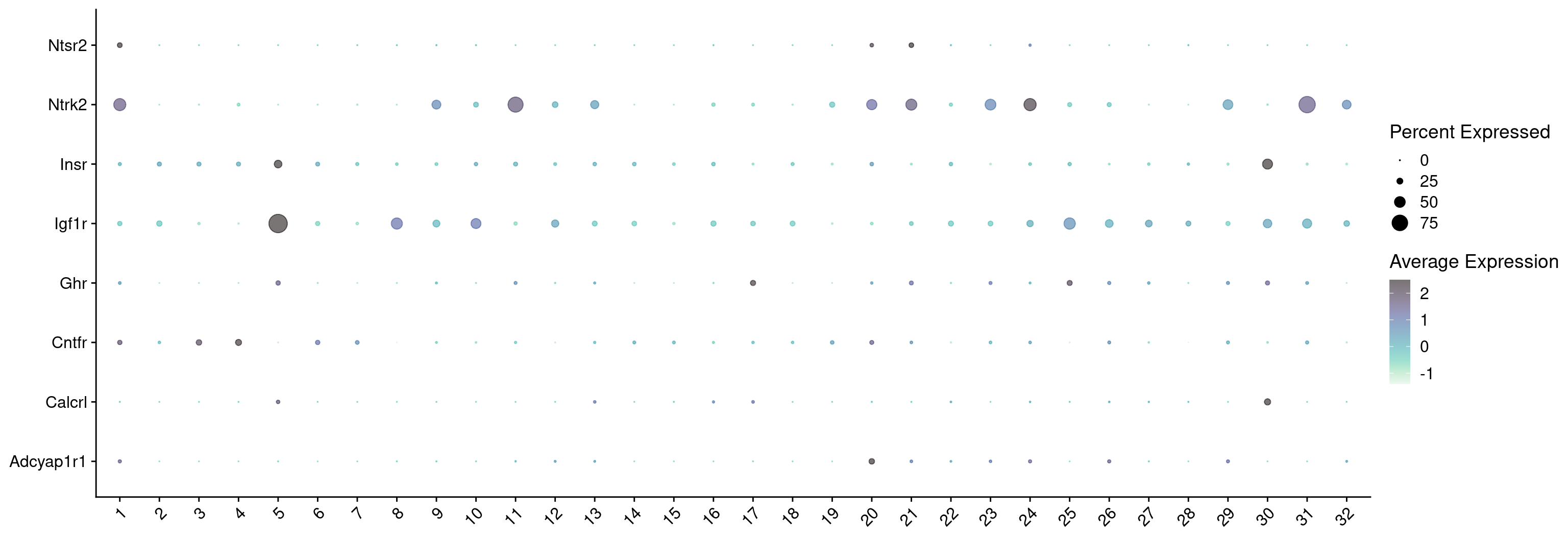
RNA
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "RNA",
features = unique(plt_g_npr),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "E")
)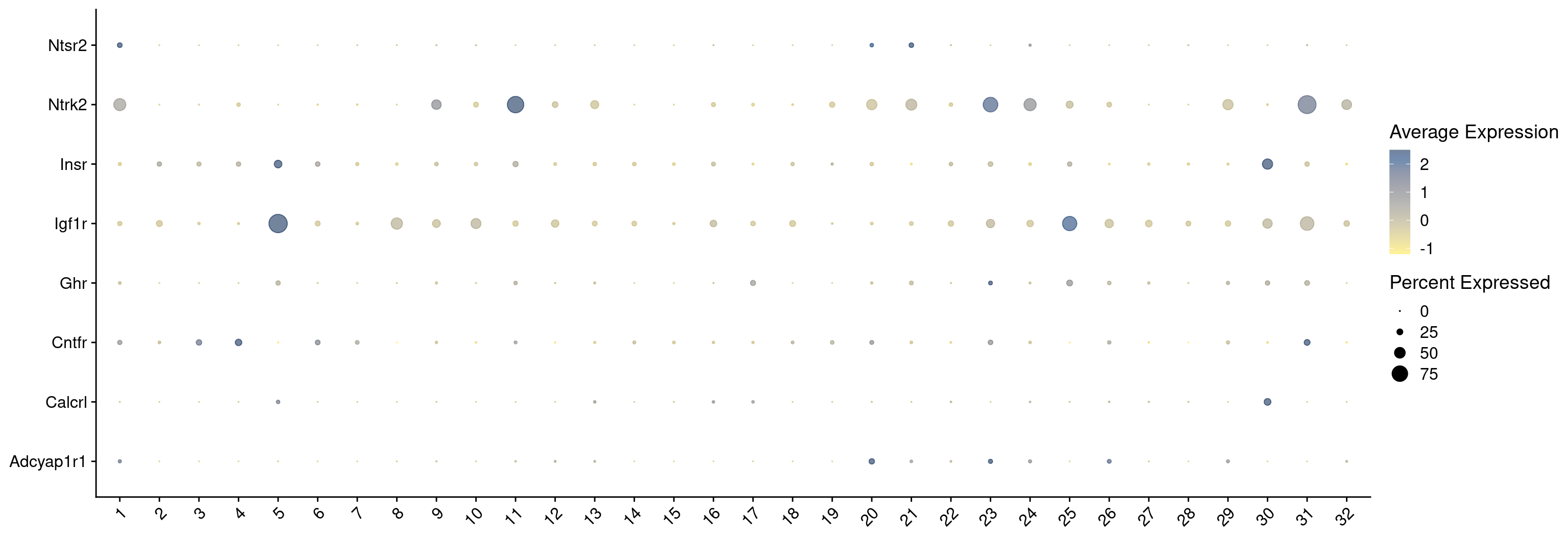
Neuromediators receptors
plt_g_nmr <- c(nmr) %>%
.[. %in% hvg] %>%
.[!. %in% c(plt_g_glut, plt_g_gaba)]SCT
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "SCT",
features = unique(plt_g_nmr),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "G")
)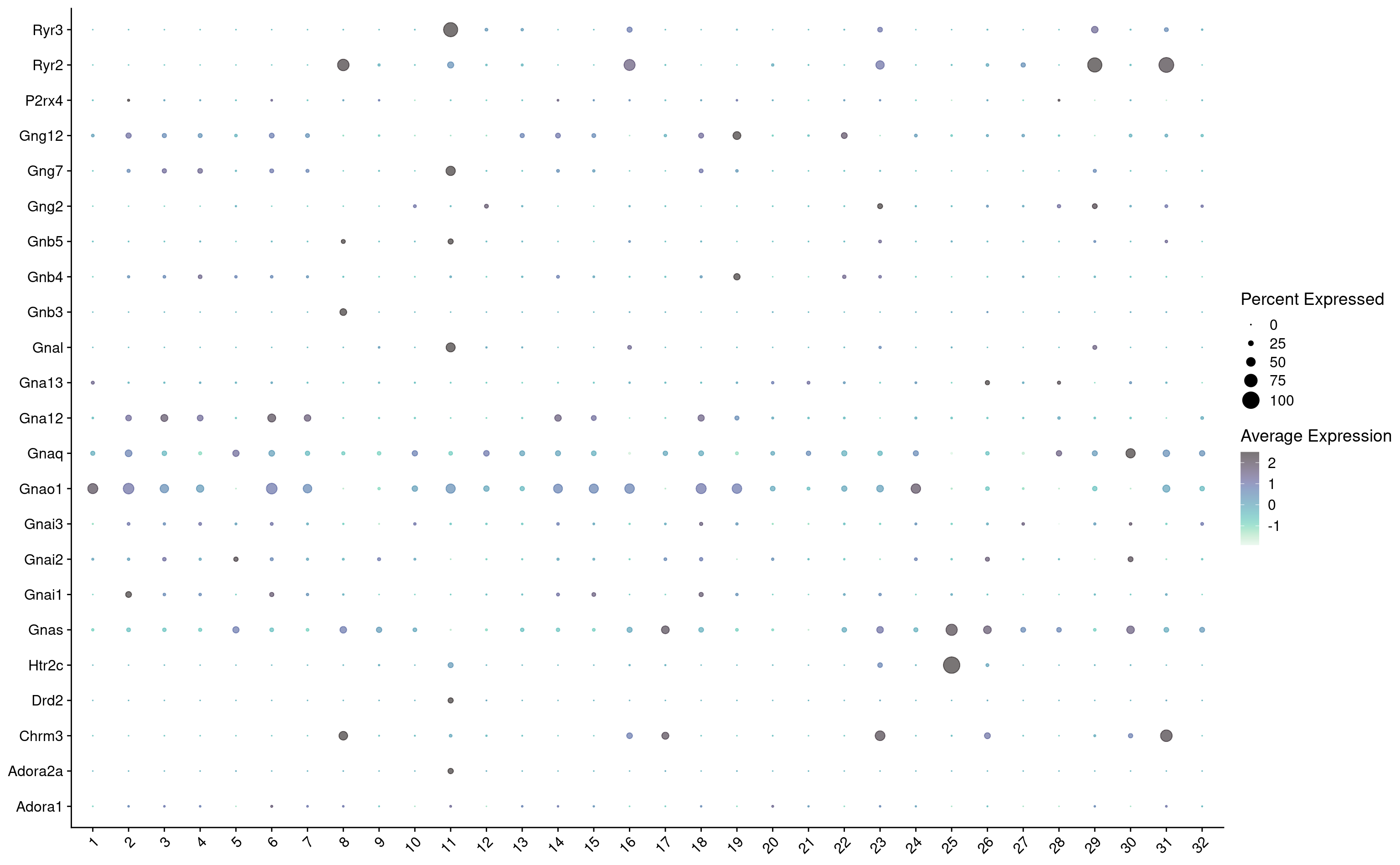
RNA
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "RNA",
features = unique(plt_g_nmr),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "E")
)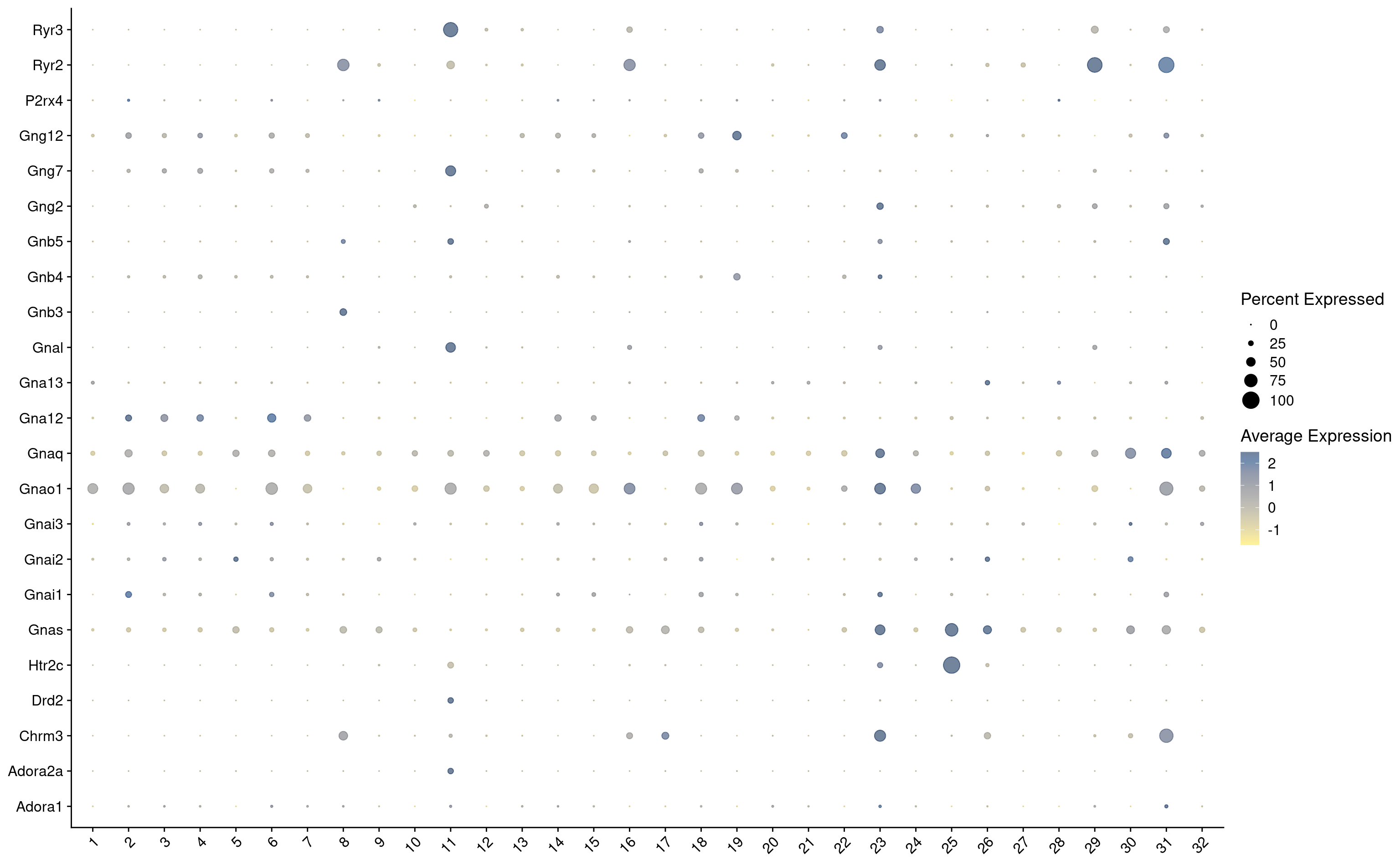
Cannabinoids
plt_g_ecb <- c(cnbn) %>% .[. %in% hvg]SCT
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "SCT",
features = unique(plt_g_ecb),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "G")
)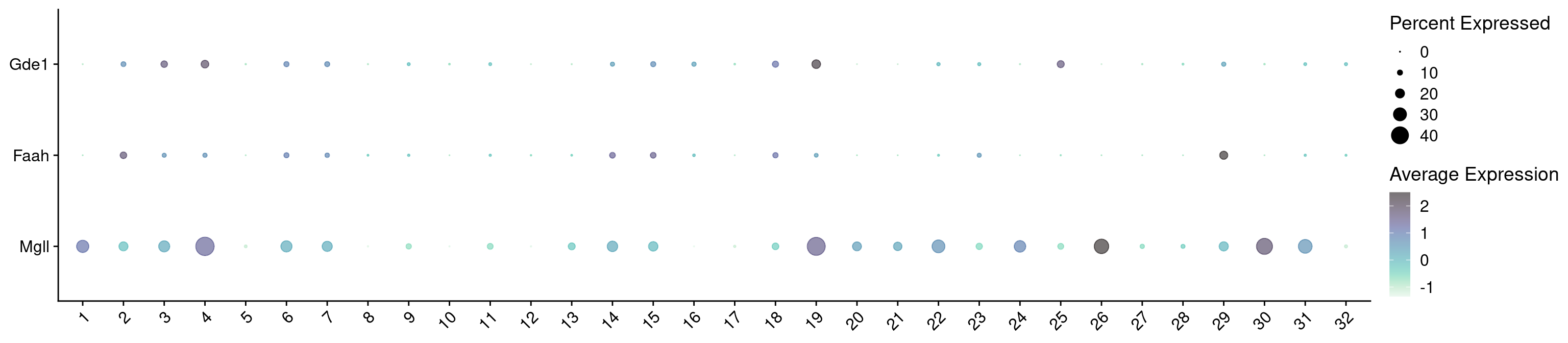
RNA
DotPlot_scCustom(
seurat_object = combined_srt,
assay = "RNA",
features = unique(plt_g_ecb),
flip_axes = TRUE,
x_lab_rotate = TRUE,
colors_use = viridis(n = 30, alpha = .55, direction = -1, option = "E")
)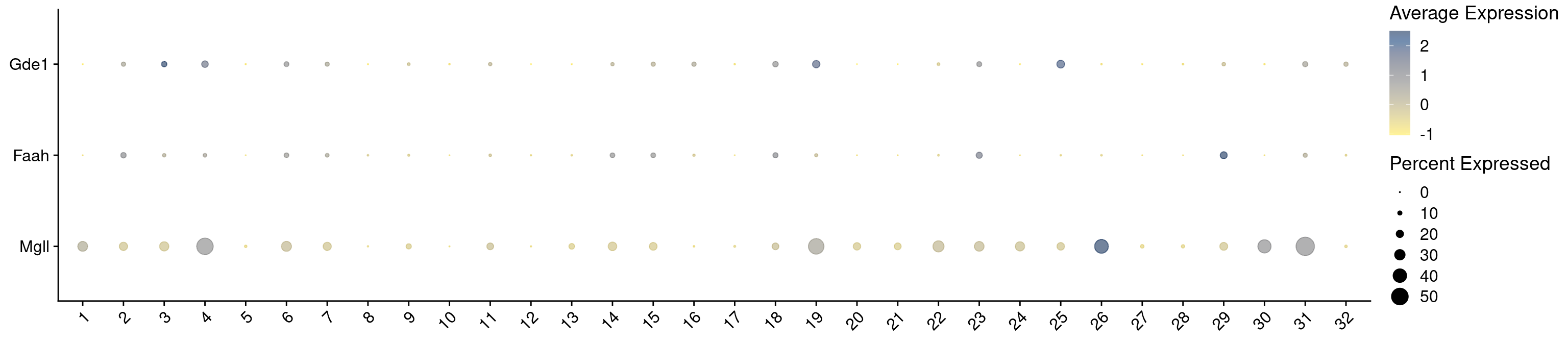
Differential gene expression (DGE)
We see the spread of our targets across derived clusters, which isn’t optimal. Lets see if we will see some significant hits with proper statistical testing.
plan("sequential")
invisible(gc())
set.seed(reseed)
plan(multicore, workers = n_cores)
Idents(combined_srt) <- "k_tree"
combined_srt <-
PrepSCTFindMarkers(combined_srt, assay = "SCT")logistic regression
markers_logreg <-
FindAllMarkers(
combined_srt,
assay = "SCT",
verbose = FALSE,
random.seed = reseed,
only.pos = TRUE,
min.pct = 0.2,
base = 10,
logfc.threshold = 0.2,
densify = TRUE,
test.use = "LR"
)
# markers_logreg %>%
# pull(gene) %>%
# gconvert(
# .,
# organism = "mmusculus",
# target = "MGI",
# numeric_ns = "",
# mthreshold = Inf,
# filter_na = TRUE
# ) %>%
# select(name, description) %>%
# right_join(markers_logreg, by = c("gene" = "name"))
write_csv(
markers_logreg,
here(
tables_dir,
docname,
sprintf(
"%s_all_mrk-logreg_sct-combined-whole_dataset-fpr_%s.csv",
project, cb_fpr
)
)
)
markers_logreg %>%
group_by(cluster) %>%
slice_max(n = 20, order_by = avg_log10FC) %>%
kable("html") %>%
kable_material(
bootstrap_options = c(
"bordered",
"condensed",
"responsive",
"striped"
),
position = "left",
font_size = 14
)| p_val | avg_log10FC | pct.1 | pct.2 | p_val_adj | cluster | gene |
|---|---|---|---|---|---|---|
| 0 | 1.6209648 | 0.297 | 0.009 | 0.0e+00 | 1 | Hapln2 |
| 0 | 1.3190035 | 0.353 | 0.019 | 0.0e+00 | 1 | Anln |
| 0 | 1.1383225 | 0.737 | 0.080 | 0.0e+00 | 1 | Trf |
| 0 | 1.0522038 | 0.234 | 0.022 | 0.0e+00 | 1 | Hcn2 |
| 0 | 1.0374340 | 0.206 | 0.019 | 0.0e+00 | 1 | Kcnk13 |
| 0 | 1.0350104 | 0.863 | 0.125 | 0.0e+00 | 1 | Prr5l |
| 0 | 1.0181583 | 0.464 | 0.047 | 0.0e+00 | 1 | Apod |
| 0 | 0.9779225 | 0.200 | 0.022 | 0.0e+00 | 1 | Fgd3 |
| 0 | 0.9768928 | 0.959 | 0.215 | 0.0e+00 | 1 | Fth1 |
| 0 | 0.9106105 | 0.363 | 0.048 | 0.0e+00 | 1 | Rftn1 |
| 0 | 0.8949042 | 0.346 | 0.047 | 0.0e+00 | 1 | Carns1 |
| 0 | 0.8908094 | 0.208 | 0.028 | 0.0e+00 | 1 | Adam19 |
| 0 | 0.8851835 | 0.363 | 0.042 | 0.0e+00 | 1 | Enpp6 |
| 0 | 0.8805379 | 0.489 | 0.076 | 0.0e+00 | 1 | Gatm |
| 0 | 0.8596478 | 0.209 | 0.029 | 0.0e+00 | 1 | Dock5 |
| 0 | 0.8399110 | 0.392 | 0.069 | 0.0e+00 | 1 | Tmem117 |
| 0 | 0.8303882 | 0.282 | 0.048 | 0.0e+00 | 1 | Kctd3 |
| 0 | 0.8241909 | 0.293 | 0.043 | 0.0e+00 | 1 | Ndrg1 |
| 0 | 0.8104694 | 0.386 | 0.064 | 0.0e+00 | 1 | Slco3a1 |
| 0 | 0.8062539 | 0.736 | 0.134 | 0.0e+00 | 1 | Gsn |
| 0 | 1.2036073 | 0.264 | 0.019 | 0.0e+00 | 2 | Gm32633 |
| 0 | 1.0164049 | 0.218 | 0.022 | 0.0e+00 | 2 | Tnni1 |
| 0 | 0.9838140 | 0.260 | 0.028 | 0.0e+00 | 2 | C030029H02Rik |
| 0 | 0.9631501 | 0.591 | 0.082 | 0.0e+00 | 2 | Synpr |
| 0 | 0.9062614 | 0.575 | 0.083 | 0.0e+00 | 2 | Opalin |
| 0 | 0.8853478 | 0.679 | 0.116 | 0.0e+00 | 2 | Ablim2 |
| 0 | 0.8579505 | 0.454 | 0.098 | 0.0e+00 | 2 | Ptgds |
| 0 | 0.8219893 | 0.600 | 0.108 | 0.0e+00 | 2 | Sh3gl3 |
| 0 | 0.8192472 | 0.455 | 0.077 | 0.0e+00 | 2 | Tmod1 |
| 0 | 0.8179436 | 0.485 | 0.106 | 0.0e+00 | 2 | Fam214a |
| 0 | 0.8046050 | 0.530 | 0.109 | 0.0e+00 | 2 | Man1a |
| 0 | 0.7922918 | 0.366 | 0.063 | 0.0e+00 | 2 | Mcam |
| 0 | 0.7804220 | 0.458 | 0.086 | 0.0e+00 | 2 | 1700047M11Rik |
| 0 | 0.7648205 | 0.661 | 0.134 | 0.0e+00 | 2 | Lgi3 |
| 0 | 0.7601628 | 0.688 | 0.164 | 0.0e+00 | 2 | Prickle1 |
| 0 | 0.7584761 | 0.400 | 0.078 | 0.0e+00 | 2 | Ninj2 |
| 0 | 0.7552239 | 0.961 | 0.316 | 0.0e+00 | 2 | Bin1 |
| 0 | 0.7483199 | 0.586 | 0.118 | 0.0e+00 | 2 | Olig1 |
| 0 | 0.7456168 | 0.252 | 0.047 | 0.0e+00 | 2 | Tyro3 |
| 0 | 0.7392018 | 0.844 | 0.200 | 0.0e+00 | 2 | Ano4 |
| 0 | 1.6853299 | 0.970 | 0.106 | 0.0e+00 | 3 | Slc1a2 |
| 0 | 1.3345483 | 0.302 | 0.019 | 0.0e+00 | 3 | Htra1 |
| 0 | 1.3145373 | 0.816 | 0.081 | 0.0e+00 | 3 | Gpc5 |
| 0 | 1.3055228 | 0.873 | 0.108 | 0.0e+00 | 3 | Slc1a3 |
| 0 | 1.2553896 | 0.798 | 0.093 | 0.0e+00 | 3 | Cst3 |
| 0 | 1.2132916 | 0.204 | 0.014 | 0.0e+00 | 3 | F3 |
| 0 | 1.1829051 | 0.442 | 0.040 | 0.0e+00 | 3 | Mertk |
| 0 | 1.1761343 | 0.210 | 0.016 | 0.0e+00 | 3 | Aldoc |
| 0 | 1.1416954 | 0.635 | 0.068 | 0.0e+00 | 3 | Prex2 |
| 0 | 1.1252629 | 0.632 | 0.095 | 0.0e+00 | 3 | Tspan7 |
| 0 | 1.1044352 | 0.304 | 0.027 | 0.0e+00 | 3 | Nwd1 |
| 0 | 1.0728424 | 0.927 | 0.137 | 0.0e+00 | 3 | Atp1a2 |
| 0 | 1.0599244 | 0.722 | 0.094 | 0.0e+00 | 3 | Gabrb1 |
| 0 | 1.0358371 | 0.434 | 0.055 | 0.0e+00 | 3 | Rgs20 |
| 0 | 1.0306564 | 0.376 | 0.048 | 0.0e+00 | 3 | Rorb |
| 0 | 1.0154493 | 0.337 | 0.039 | 0.0e+00 | 3 | Cspg5 |
| 0 | 1.0092837 | 0.262 | 0.030 | 0.0e+00 | 3 | Gli3 |
| 0 | 1.0084536 | 0.859 | 0.174 | 0.0e+00 | 3 | Cpe |
| 0 | 1.0061784 | 0.351 | 0.046 | 0.0e+00 | 3 | Gpr37l1 |
| 0 | 1.0054572 | 0.234 | 0.025 | 0.0e+00 | 3 | Arhgef26 |
| 0 | 1.7002947 | 0.555 | 0.019 | 0.0e+00 | 4 | Sox11 |
| 0 | 1.5624399 | 0.245 | 0.012 | 0.0e+00 | 4 | Dlx6os1 |
| 0 | 1.3690852 | 0.313 | 0.016 | 0.0e+00 | 4 | Dcx |
| 0 | 1.3006627 | 0.235 | 0.028 | 0.0e+00 | 4 | Unc5d |
| 0 | 1.1928454 | 0.208 | 0.017 | 0.0e+00 | 4 | Epha3 |
| 0 | 1.1640244 | 0.559 | 0.064 | 0.0e+00 | 4 | Sox4 |
| 0 | 1.1107389 | 0.305 | 0.046 | 0.0e+00 | 4 | Gm26871 |
| 0 | 1.0614437 | 0.414 | 0.068 | 0.0e+00 | 4 | Plxna4 |
| 0 | 1.0569533 | 0.279 | 0.033 | 0.0e+00 | 4 | Bcl11a |
| 0 | 1.0367877 | 0.247 | 0.034 | 0.0e+00 | 4 | Tiam2 |
| 0 | 1.0314614 | 0.257 | 0.028 | 0.0e+00 | 4 | Dpysl3 |
| 0 | 1.0283565 | 0.288 | 0.050 | 0.0e+00 | 4 | Ccnd2 |
| 0 | 1.0169078 | 0.511 | 0.173 | 0.0e+00 | 4 | Nrxn3 |
| 0 | 0.9979383 | 0.336 | 0.051 | 0.0e+00 | 4 | 2610307P16Rik |
| 0 | 0.9975689 | 0.506 | 0.095 | 0.0e+00 | 4 | Meis2 |
| 0 | 0.9975689 | 0.334 | 0.049 | 0.0e+00 | 4 | 5730522E02Rik |
| 0 | 0.9935625 | 0.215 | 0.024 | 0.0e+00 | 4 | Srrm4 |
| 0 | 0.9734674 | 0.428 | 0.071 | 0.0e+00 | 4 | Nol4 |
| 0 | 0.9535298 | 0.216 | 0.061 | 0.0e+00 | 4 | Adarb2 |
| 0 | 0.9268525 | 0.390 | 0.090 | 0.0e+00 | 4 | Robo2 |
| 0 | 2.6740745 | 0.792 | 0.007 | 0.0e+00 | 5 | Flt1 |
| 0 | 2.5981621 | 0.550 | 0.003 | 0.0e+00 | 5 | Adgrl4 |
| 0 | 2.5421669 | 0.283 | 0.001 | 0.0e+00 | 5 | Zfp366 |
| 0 | 2.5027505 | 0.601 | 0.004 | 0.0e+00 | 5 | Cldn5 |
| 0 | 2.3406675 | 0.623 | 0.008 | 0.0e+00 | 5 | Slco1a4 |
| 0 | 2.3036211 | 0.303 | 0.002 | 0.0e+00 | 5 | Apold1 |
| 0 | 2.2665012 | 0.336 | 0.002 | 0.0e+00 | 5 | Tek |
| 0 | 2.1792421 | 0.654 | 0.011 | 0.0e+00 | 5 | Mecom |
| 0 | 2.1583102 | 0.541 | 0.007 | 0.0e+00 | 5 | Ptprb |
| 0 | 2.1238337 | 0.256 | 0.002 | 0.0e+00 | 5 | Fgd5 |
| 0 | 2.1210408 | 0.236 | 0.003 | 0.0e+00 | 5 | Rgs5 |
| 0 | 2.1163457 | 0.272 | 0.002 | 0.0e+00 | 5 | Egfl7 |
| 0 | 2.1001698 | 0.399 | 0.004 | 0.0e+00 | 5 | Abcb1a |
| 0 | 2.0947915 | 0.267 | 0.002 | 0.0e+00 | 5 | Vwf |
| 0 | 2.0438165 | 0.252 | 0.003 | 0.0e+00 | 5 | Erg |
| 0 | 2.0390486 | 0.393 | 0.005 | 0.0e+00 | 5 | Eng |
| 0 | 2.0362193 | 0.433 | 0.006 | 0.0e+00 | 5 | Cyyr1 |
| 0 | 1.9902948 | 0.229 | 0.002 | 0.0e+00 | 5 | Ly6c1 |
| 0 | 1.9713962 | 0.525 | 0.011 | 0.0e+00 | 5 | Atp10a |
| 0 | 1.9712036 | 0.238 | 0.003 | 0.0e+00 | 5 | Hspa12b |
| 0 | 1.6258295 | 0.917 | 0.087 | 0.0e+00 | 6 | Tnr |
| 0 | 1.5686199 | 0.201 | 0.009 | 0.0e+00 | 6 | Gm36988 |
| 0 | 1.5101472 | 0.459 | 0.017 | 0.0e+00 | 6 | Tmem132d |
| 0 | 1.5091262 | 0.224 | 0.009 | 0.0e+00 | 6 | 4930588A03Rik |
| 0 | 1.4581971 | 0.431 | 0.030 | 0.0e+00 | 6 | Lhfpl3 |
| 0 | 1.4491361 | 0.201 | 0.007 | 0.0e+00 | 6 | Pdgfra |
| 0 | 1.4216137 | 0.488 | 0.038 | 0.0e+00 | 6 | Pcdh15 |
| 0 | 1.3720107 | 0.329 | 0.025 | 0.0e+00 | 6 | Arhgap24 |
| 0 | 1.3492160 | 0.346 | 0.020 | 0.0e+00 | 6 | Vcan |
| 0 | 1.3362456 | 0.278 | 0.017 | 0.0e+00 | 6 | 9630013A20Rik |
| 0 | 1.3298121 | 0.242 | 0.014 | 0.0e+00 | 6 | Megf11 |
| 0 | 1.3174237 | 0.209 | 0.012 | 0.0e+00 | 6 | Sema3d |
| 0 | 1.3079245 | 0.305 | 0.024 | 0.0e+00 | 6 | Gm13052 |
| 0 | 1.2794788 | 0.783 | 0.102 | 0.0e+00 | 6 | Dscam |
| 0 | 1.2409114 | 0.559 | 0.064 | 0.0e+00 | 6 | Xylt1 |
| 0 | 1.2369910 | 0.616 | 0.075 | 0.0e+00 | 6 | Itpr2 |
| 0 | 1.2045885 | 0.425 | 0.026 | 0.0e+00 | 6 | Nxph1 |
| 0 | 1.1973263 | 0.372 | 0.032 | 0.0e+00 | 6 | Ppfibp1 |
| 0 | 1.1626581 | 0.350 | 0.032 | 0.0e+00 | 6 | Sez6l |
| 0 | 1.1529330 | 0.927 | 0.240 | 0.0e+00 | 6 | Opcml |
| 0 | 3.1934150 | 0.432 | 0.000 | 0.0e+00 | 7 | Ccdc180 |
| 0 | 2.8891799 | 0.498 | 0.001 | 0.0e+00 | 7 | Tmem212 |
| 0 | 2.8343931 | 0.408 | 0.001 | 0.0e+00 | 7 | Cdhr3 |
| 0 | 2.7500722 | 0.225 | 0.000 | 0.0e+00 | 7 | Lrrc43 |
| 0 | 2.7186822 | 0.633 | 0.002 | 0.0e+00 | 7 | Dnah12 |
| 0 | 2.7148944 | 0.425 | 0.001 | 0.0e+00 | 7 | Lrrc36 |
| 0 | 2.6171556 | 0.710 | 0.004 | 0.0e+00 | 7 | Cfap299 |
| 0 | 2.4744696 | 0.546 | 0.003 | 0.0e+00 | 7 | Hydin |
| 0 | 2.4519866 | 0.316 | 0.001 | 0.0e+00 | 7 | Gm10714 |
| 0 | 2.4262990 | 0.336 | 0.002 | 0.0e+00 | 7 | Cfap77 |
| 0 | 2.3891957 | 0.367 | 0.002 | 0.0e+00 | 7 | 3300002A11Rik |
| 0 | 2.3870027 | 0.635 | 0.005 | 0.0e+00 | 7 | Dnah6 |
| 0 | 2.3751465 | 0.273 | 0.001 | 0.0e+00 | 7 | Ttc29 |
| 0 | 2.3538233 | 0.367 | 0.002 | 0.0e+00 | 7 | Spag17 |
| 0 | 2.3209470 | 0.541 | 0.004 | 0.0e+00 | 7 | Cfap43 |
| 0 | 2.3013603 | 0.348 | 0.002 | 0.0e+00 | 7 | Armc4 |
| 0 | 2.3004253 | 0.261 | 0.001 | 0.0e+00 | 7 | Gm28729 |
| 0 | 2.2681520 | 0.307 | 0.002 | 0.0e+00 | 7 | Wdr49 |
| 0 | 2.2680486 | 0.517 | 0.004 | 0.0e+00 | 7 | Ak7 |
| 0 | 2.2655898 | 0.348 | 0.002 | 0.0e+00 | 7 | Dnah3 |
| 0 | 2.9773434 | 0.580 | 0.003 | 0.0e+00 | 8 | Nwd2 |
| 0 | 2.2290479 | 0.563 | 0.007 | 0.0e+00 | 8 | Cpne4 |
| 0 | 2.1624853 | 0.254 | 0.002 | 0.0e+00 | 8 | Robo3 |
| 0 | 2.1485549 | 0.498 | 0.009 | 0.0e+00 | 8 | Scube1 |
| 0 | 1.7294480 | 0.229 | 0.005 | 0.0e+00 | 8 | Lrrc3b |
| 0 | 1.6740830 | 0.293 | 0.008 | 0.0e+00 | 8 | Necab2 |
| 0 | 1.6500159 | 0.512 | 0.025 | 0.0e+00 | 8 | Vav3 |
| 0 | 1.6184173 | 0.229 | 0.007 | 0.0e+00 | 8 | Necab3 |
| 0 | 1.5683338 | 0.878 | 0.192 | 0.0e+00 | 8 | Kcnma1 |
| 0 | 1.5597677 | 0.456 | 0.027 | 0.0e+00 | 8 | Syt9 |
| 0 | 1.5235827 | 0.222 | 0.007 | 0.0e+00 | 8 | Kcnj6 |
| 0 | 1.5024617 | 0.466 | 0.028 | 0.0e+00 | 8 | Kcnip1 |
| 0 | 1.4905451 | 0.356 | 0.011 | 0.0e+00 | 8 | Slc17a7 |
| 0 | 1.4894285 | 0.229 | 0.008 | 0.0e+00 | 8 | Cntnap4 |
| 0 | 1.4752967 | 0.266 | 0.012 | 0.0e+00 | 8 | Cplx1 |
| 0 | 1.4721310 | 0.578 | 0.027 | 0.0e+00 | 8 | St6galnac5 |
| 0 | 1.4557125 | 0.327 | 0.015 | 0.0e+00 | 8 | Pld5 |
| 0 | 1.4517236 | 0.710 | 0.108 | 0.0e+00 | 8 | Asic2 |
| 0 | 1.4512808 | 0.261 | 0.011 | 0.0e+00 | 8 | Cygb |
| 0 | 1.4279518 | 0.800 | 0.066 | 0.0e+00 | 8 | Cntnap2 |
| 0 | 2.8327592 | 0.828 | 0.002 | 0.0e+00 | 9 | Ush2a |
| 0 | 2.6812950 | 0.457 | 0.001 | 0.0e+00 | 9 | Gm32884 |
| 0 | 2.6238452 | 0.759 | 0.003 | 0.0e+00 | 9 | Gm44196 |
| 0 | 2.4972348 | 0.210 | 0.001 | 0.0e+00 | 9 | Gm47480 |
| 0 | 2.4737537 | 0.477 | 0.002 | 0.0e+00 | 9 | Rbp3 |
| 0 | 2.4659283 | 0.664 | 0.003 | 0.0e+00 | 9 | Tulp1 |
| 0 | 2.4321096 | 0.580 | 0.003 | 0.0e+00 | 9 | Gm31615 |
| 0 | 2.3798492 | 0.761 | 0.005 | 0.0e+00 | 9 | Cngb3 |
| 0 | 2.3603583 | 0.322 | 0.001 | 0.0e+00 | 9 | Pde6c |
| 0 | 2.3453544 | 0.213 | 0.001 | 0.0e+00 | 9 | Crx |
| 0 | 2.3345075 | 0.457 | 0.002 | 0.0e+00 | 9 | Cplx3 |
| 0 | 2.3163142 | 0.707 | 0.005 | 0.0e+00 | 9 | Rpgrip1 |
| 0 | 2.2194671 | 0.661 | 0.007 | 0.0e+00 | 9 | Sag |
| 0 | 2.2034174 | 0.385 | 0.003 | 0.0e+00 | 9 | Gnb3 |
| 0 | 2.1796524 | 0.362 | 0.003 | 0.0e+00 | 9 | Mpp4 |
| 0 | 2.1183660 | 0.261 | 0.002 | 0.0e+00 | 9 | Tph1 |
| 0 | 2.0650539 | 0.313 | 0.003 | 0.0e+00 | 9 | Samsn1 |
| 0 | 1.9600035 | 0.210 | 0.002 | 0.0e+00 | 9 | Fabp12 |
| 0 | 1.8924073 | 0.460 | 0.008 | 0.0e+00 | 9 | Col7a1 |
| 0 | 1.8808223 | 0.284 | 0.005 | 0.0e+00 | 9 | Tc2n |
| 0 | 1.6802022 | 0.389 | 0.010 | 0.0e+00 | 10 | Agt |
| 0 | 1.2595827 | 0.471 | 0.039 | 0.0e+00 | 10 | Slc6a11 |
| 0 | 1.2024848 | 0.235 | 0.017 | 0.0e+00 | 10 | Aqp4 |
| 0 | 1.0237024 | 0.824 | 0.135 | 0.0e+00 | 10 | Slc4a4 |
| 0 | 0.9736382 | 0.356 | 0.041 | 0.0e+00 | 10 | Slc7a11 |
| 0 | 0.9453237 | 0.392 | 0.075 | 0.0e+00 | 10 | Slc38a1 |
| 0 | 0.9427607 | 0.330 | 0.042 | 0.0e+00 | 10 | Bmpr1b |
| 0 | 0.9219528 | 0.536 | 0.089 | 0.0e+00 | 10 | Sfxn5 |
| 0 | 0.9118849 | 0.657 | 0.106 | 0.0e+00 | 10 | Sparcl1 |
| 0 | 0.9112603 | 0.418 | 0.080 | 0.0e+00 | 10 | Camk2g |
| 0 | 0.8984468 | 0.225 | 0.031 | 0.0e+00 | 10 | Pla2g7 |
| 0 | 0.8813931 | 0.892 | 0.238 | 0.0e+00 | 10 | Npas3 |
| 0 | 0.8805419 | 0.297 | 0.072 | 0.0e+00 | 10 | Ptch1 |
| 0 | 0.8776880 | 0.261 | 0.043 | 0.0e+00 | 10 | Lgi1 |
| 0 | 0.8709505 | 0.225 | 0.034 | 0.0e+00 | 10 | Lrig1 |
| 0 | 0.8610014 | 0.356 | 0.060 | 0.0e+00 | 10 | Lhfp |
| 0 | 0.8311888 | 0.252 | 0.035 | 0.0e+00 | 10 | Slc39a12 |
| 0 | 0.8311279 | 0.356 | 0.074 | 0.0e+00 | 10 | Nhsl1 |
| 0 | 0.8182101 | 0.693 | 0.139 | 0.0e+00 | 10 | Gabrb1 |
| 0 | 0.8147879 | 0.288 | 0.065 | 0.0e+00 | 10 | Pdgfd |
| 0 | 2.4672568 | 0.359 | 0.001 | 0.0e+00 | 11 | Gm10754 |
| 0 | 2.3669018 | 0.355 | 0.002 | 0.0e+00 | 11 | Scn4b |
| 0 | 2.3274787 | 0.283 | 0.002 | 0.0e+00 | 11 | Drd2 |
| 0 | 2.2583978 | 0.300 | 0.002 | 0.0e+00 | 11 | Tac1 |
| 0 | 2.2475670 | 0.293 | 0.002 | 0.0e+00 | 11 | Adora2a |
| 0 | 2.2197916 | 0.548 | 0.004 | 0.0e+00 | 11 | Cpne5 |
| 0 | 2.1556245 | 0.276 | 0.002 | 0.0e+00 | 11 | Penk |
| 0 | 2.1513875 | 0.276 | 0.002 | 0.0e+00 | 11 | Gpr88 |
| 0 | 2.0850209 | 0.348 | 0.003 | 0.0e+00 | 11 | Actn2 |
| 0 | 1.9799164 | 0.483 | 0.006 | 0.0e+00 | 11 | Pde1b |
| 0 | 1.9679351 | 0.390 | 0.004 | 0.0e+00 | 11 | Gprin3 |
| 0 | 1.9518151 | 0.221 | 0.002 | 0.0e+00 | 11 | Sh3rf2 |
| 0 | 1.8953844 | 0.217 | 0.003 | 0.0e+00 | 11 | Gabrd |
| 0 | 1.8572430 | 0.841 | 0.021 | 0.0e+00 | 11 | Rgs9 |
| 0 | 1.8527769 | 0.821 | 0.021 | 0.0e+00 | 11 | Camk4 |
| 0 | 1.8136088 | 0.410 | 0.007 | 0.0e+00 | 11 | Ppp1r1b |
| 0 | 1.7104495 | 0.855 | 0.031 | 0.0e+00 | 11 | Ryr3 |
| 0 | 1.6961049 | 0.369 | 0.009 | 0.0e+00 | 11 | 6530403H02Rik |
| 0 | 1.6662470 | 0.810 | 0.027 | 0.0e+00 | 11 | Caln1 |
| 0 | 1.6620185 | 0.293 | 0.007 | 0.0e+00 | 11 | Gm15155 |
| 0 | 2.6603095 | 0.391 | 0.002 | 0.0e+00 | 12 | Bnc2 |
| 0 | 2.4869158 | 0.322 | 0.001 | 0.0e+00 | 12 | Aox3 |
| 0 | 2.3702104 | 0.302 | 0.001 | 0.0e+00 | 12 | Lum |
| 0 | 2.0484613 | 0.723 | 0.012 | 0.0e+00 | 12 | Cped1 |
| 0 | 1.9761963 | 0.351 | 0.005 | 0.0e+00 | 12 | Alpl |
| 0 | 1.9678300 | 0.342 | 0.005 | 0.0e+00 | 12 | Olfr1033 |
| 0 | 1.8736091 | 0.332 | 0.004 | 0.0e+00 | 12 | Col1a1 |
| 0 | 1.8227453 | 0.381 | 0.006 | 0.0e+00 | 12 | Eya2 |
| 0 | 1.7833348 | 0.465 | 0.009 | 0.0e+00 | 12 | Col1a2 |
| 0 | 1.7696043 | 0.297 | 0.005 | 0.0e+00 | 12 | Adamts12 |
| 0 | 1.7389683 | 0.267 | 0.007 | 0.0e+00 | 12 | Cemip |
| 0 | 1.6999254 | 0.728 | 0.042 | 0.0e+00 | 12 | Adam12 |
| 0 | 1.6638853 | 0.535 | 0.015 | 0.0e+00 | 12 | Igfbp4 |
| 0 | 1.6091933 | 0.218 | 0.006 | 0.0e+00 | 12 | Tbx15 |
| 0 | 1.4962272 | 0.213 | 0.007 | 0.0e+00 | 12 | Col26a1 |
| 0 | 1.4354060 | 0.292 | 0.011 | 0.0e+00 | 12 | Mrc2 |
| 0 | 1.3227017 | 0.248 | 0.016 | 0.0e+00 | 12 | Bmper |
| 0 | 1.3181814 | 0.252 | 0.013 | 0.0e+00 | 12 | Igf2 |
| 0 | 1.2630748 | 0.500 | 0.042 | 0.0e+00 | 12 | Slc7a11 |
| 0 | 1.2049570 | 0.203 | 0.013 | 0.0e+00 | 12 | Cfh |
| 0 | 2.5921917 | 1.000 | 0.023 | 0.0e+00 | 13 | Htr2c |
| 0 | 2.5050882 | 0.606 | 0.002 | 0.0e+00 | 13 | Gmnc |
| 0 | 2.2771864 | 0.683 | 0.004 | 0.0e+00 | 13 | Tmem72 |
| 0 | 2.1958815 | 0.654 | 0.005 | 0.0e+00 | 13 | Kl |
| 0 | 2.0885060 | 0.394 | 0.004 | 0.0e+00 | 13 | Prlr |
| 0 | 2.0843895 | 0.625 | 0.007 | 0.0e+00 | 13 | Rbm47 |
| 0 | 2.0652762 | 0.625 | 0.006 | 0.0e+00 | 13 | Slc4a5 |
| 0 | 2.0247612 | 1.000 | 0.250 | 0.0e+00 | 13 | Ttr |
| 0 | 2.0157908 | 0.625 | 0.008 | 0.0e+00 | 13 | Clic6 |
| 0 | 1.9793616 | 0.433 | 0.005 | 0.0e+00 | 13 | Car12 |
| 0 | 1.9513328 | 0.471 | 0.006 | 0.0e+00 | 13 | Folr1 |
| 0 | 1.9336041 | 0.337 | 0.004 | 0.0e+00 | 13 | Baiap2l1 |
| 0 | 1.8399450 | 0.510 | 0.007 | 0.0e+00 | 13 | Lmx1a |
| 0 | 1.8272706 | 0.260 | 0.004 | 0.0e+00 | 13 | Slc13a4 |
| 0 | 1.7517605 | 0.404 | 0.008 | 0.0e+00 | 13 | Steap2 |
| 0 | 1.7263930 | 0.288 | 0.006 | 0.0e+00 | 13 | Trpv4 |
| 0 | 1.7242829 | 0.567 | 0.012 | 0.0e+00 | 13 | Igf2 |
| 0 | 1.7178401 | 0.413 | 0.008 | 0.0e+00 | 13 | Fap |
| 0 | 1.6968150 | 0.567 | 0.014 | 0.0e+00 | 13 | Ecrg4 |
| 0 | 1.6642038 | 0.923 | 0.062 | 0.0e+00 | 13 | Esrrg |
| 0 | 2.0834491 | 0.506 | 0.006 | 0.0e+00 | 14 | Cd83 |
| 0 | 1.9139884 | 0.403 | 0.006 | 0.0e+00 | 14 | Plek |
| 0 | 1.7016959 | 0.494 | 0.010 | 0.0e+00 | 14 | Inpp5d |
| 0 | 1.6046859 | 0.260 | 0.007 | 0.0e+00 | 14 | Apbb1ip |
| 0 | 1.5448902 | 0.208 | 0.006 | 0.0e+00 | 14 | Tnfaip3 |
| 0 | 1.4079602 | 0.234 | 0.010 | 0.0e+00 | 14 | Dock8 |
| 0 | 1.3844791 | 0.416 | 0.021 | 0.0e+00 | 14 | Pde3b |
| 0 | 1.3816313 | 0.377 | 0.019 | 0.0e+00 | 14 | Lyn |
| 0 | 1.3412955 | 0.494 | 0.025 | 0.0e+00 | 14 | Runx1 |
| 0 | 1.3044370 | 0.247 | 0.013 | 0.0e+00 | 14 | Maf |
| 0 | 1.2949720 | 0.481 | 0.043 | 0.0e+00 | 14 | Sirpa |
| 0 | 1.2803905 | 0.338 | 0.019 | 0.0e+00 | 14 | Adap2 |
| 0 | 1.2586054 | 0.312 | 0.020 | 0.0e+00 | 14 | Itgb5 |
| 0 | 1.2154211 | 0.532 | 0.053 | 0.0e+00 | 14 | Lrmda |
| 0 | 1.2008347 | 0.312 | 0.027 | 0.0e+00 | 14 | Entpd1 |
| 0 | 1.1120943 | 0.532 | 0.063 | 0.0e+00 | 14 | Mef2c |
| 0 | 1.0997167 | 0.208 | 0.017 | 0.0e+00 | 14 | Rel |
| 0 | 1.0925925 | 0.377 | 0.048 | 0.0e+00 | 14 | Mapkapk2 |
| 0 | 1.0627096 | 0.234 | 0.025 | 0.0e+00 | 14 | Ctsz |
| 0 | 1.0502364 | 0.649 | 0.115 | 0.0e+00 | 14 | Ptprj |
| 0 | 3.4456464 | 0.288 | 0.000 | 0.0e+00 | 15 | Rprm |
| 0 | 2.1820846 | 0.966 | 0.023 | 0.0e+00 | 15 | Hs3st4 |
| 0 | 2.1800638 | 0.542 | 0.004 | 0.0e+00 | 15 | Ipcef1 |
| 0 | 2.0306731 | 1.000 | 0.079 | 0.0e+00 | 15 | Dpp10 |
| 0 | 1.9094037 | 0.441 | 0.007 | 0.0e+00 | 15 | Vxn |
| 0 | 1.8621917 | 0.458 | 0.007 | 0.0e+00 | 15 | Ttc9b |
| 0 | 1.8521677 | 0.339 | 0.005 | 0.0e+00 | 15 | Tbr1 |
| 0 | 1.8230439 | 0.407 | 0.008 | 0.0e+00 | 15 | Htr1f |
| 0 | 1.8223971 | 0.288 | 0.005 | 0.0e+00 | 15 | Hs3st2 |
| 0 | 1.7874705 | 0.407 | 0.008 | 0.0e+00 | 15 | Hpcal4 |
| 0 | 1.7178729 | 0.390 | 0.009 | 0.0e+00 | 15 | Npas4 |
| 0 | 1.6370979 | 0.763 | 0.024 | 0.0e+00 | 15 | Slc17a7 |
| 0 | 1.6307976 | 0.458 | 0.012 | 0.0e+00 | 15 | Nptx1 |
| 0 | 1.6196684 | 0.763 | 0.024 | 0.0e+00 | 15 | Tmem178 |
| 0 | 1.5863354 | 0.780 | 0.028 | 0.0e+00 | 15 | A830018L16Rik |
| 0 | 1.5781789 | 0.559 | 0.015 | 0.0e+00 | 15 | Vsnl1 |
| 0 | 1.5757974 | 0.288 | 0.008 | 0.0e+00 | 15 | Trabd2b |
| 0 | 1.5630161 | 0.881 | 0.039 | 0.0e+00 | 15 | Pde1a |
| 0 | 1.5591557 | 0.254 | 0.007 | 0.0e+00 | 15 | 1110008P14Rik |
| 0 | 1.5588618 | 0.949 | 0.059 | 0.0e+00 | 15 | Etl4 |
| 0 | 2.4362784 | 0.972 | 0.029 | 0.0e+00 | 16 | Reln |
| 0 | 2.0333160 | 0.611 | 0.007 | 0.0e+00 | 16 | Trp73 |
| 0 | 1.9122723 | 0.556 | 0.009 | 0.0e+00 | 16 | Ndnf |
| 0 | 1.8833775 | 0.472 | 0.007 | 0.0e+00 | 16 | Ebf3 |
| 0 | 1.7624574 | 0.972 | 0.056 | 0.0e+00 | 16 | Clstn2 |
| 0 | 1.7272174 | 0.861 | 0.040 | 0.0e+00 | 16 | Thsd7b |
| 0 | 1.6811718 | 0.972 | 0.060 | 0.0e+00 | 16 | Kcnh7 |
| 0 | 1.6201361 | 0.833 | 0.032 | 0.0e+00 | 16 | Cdh4 |
| 0 | 1.4951973 | 0.750 | 0.043 | 0.0e+00 | 16 | Dync1i1 |
| 0 | 1.4740080 | 0.583 | 0.030 | 0.0e+00 | 16 | Epha3 |
| 0 | 1.4344995 | 0.361 | 0.014 | 0.0e+00 | 16 | Col11a1 |
| 0 | 1.3958127 | 0.389 | 0.021 | 0.0e+00 | 16 | Hs3st5 |
| 0 | 1.3791710 | 0.444 | 0.026 | 0.0e+00 | 16 | Cacna2d2 |
| 0 | 1.3780536 | 0.472 | 0.022 | 0.0e+00 | 16 | Kcnc2 |
| 0 | 1.3647438 | 1.000 | 0.102 | 0.0e+00 | 16 | Cntnap2 |
| 0 | 1.2944836 | 0.278 | 0.017 | 1.3e-06 | 16 | Plxnd1 |
| 0 | 1.2918764 | 0.528 | 0.043 | 0.0e+00 | 16 | Plekha7 |
| 0 | 1.2752528 | 0.306 | 0.022 | 2.0e-07 | 16 | 5330417C22Rik |
| 0 | 1.2703730 | 0.389 | 0.017 | 1.9e-06 | 16 | Eln |
| 0 | 1.2590954 | 0.833 | 0.101 | 0.0e+00 | 16 | Kcnb2 |
Dot-plot
Heatmap
top10 <-
markers_logreg %>%
group_by(cluster) %>%
top_n(n = 10, wt = avg_log10FC)
DoHeatmap(combined_srt, features = top10$gene) + NoLegend()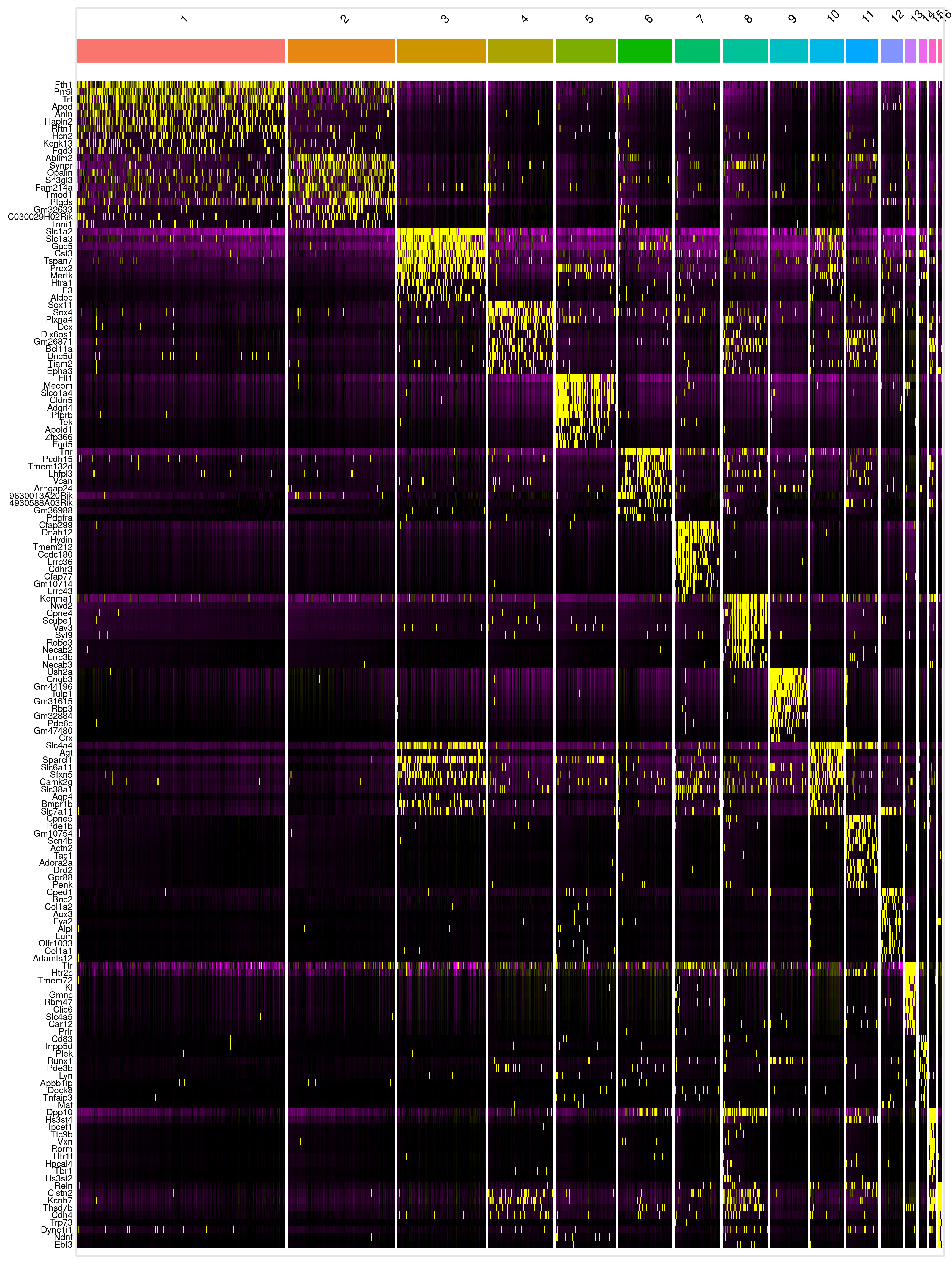
MAST
plan("sequential")
invisible(gc())
set.seed(reseed)
plan(multicore, workers = n_cores)
markers_MAST <-
FindAllMarkers(
combined_srt,
assay = "SCT",
verbose = FALSE,
random.seed = reseed,
only.pos = TRUE,
min.pct = 0.1,
base = 10,
logfc.threshold = 0.2,
test.use = "MAST"
)
# markers_MAST %>%
# pull(gene) %>%
# gconvert(
# .,
# organism = "mmusculus",
# target = "MGI",
# numeric_ns = "",
# mthreshold = Inf,
# filter_na = TRUE
# ) %>%
# select(name, description) %>%
# right_join(markers_MAST, by = c("gene" = "name"))
write_csv(
markers_MAST,
here(
tables_dir,
docname,
sprintf(
"%s_all_mrk-MAST_sct-combined-whole_dataset-fpr_%s.csv",
project, cb_fpr
)
)
)
markers_MAST %>%
group_by(cluster) %>%
slice_max(n = 20, order_by = avg_log10FC) %>%
kable("html") %>%
kable_material(
bootstrap_options = c(
"bordered",
"condensed",
"responsive",
"striped"
),
position = "left",
font_size = 14
)| p_val | avg_log10FC | pct.1 | pct.2 | p_val_adj | cluster | gene |
|---|---|---|---|---|---|---|
| 0.0e+00 | 1.6209648 | 0.297 | 0.009 | 0.0000000 | 1 | Hapln2 |
| 0.0e+00 | 1.3190035 | 0.353 | 0.019 | 0.0000000 | 1 | Anln |
| 0.0e+00 | 1.3135830 | 0.177 | 0.009 | 0.0000000 | 1 | A330049N07Rik |
| 0.0e+00 | 1.1383225 | 0.737 | 0.080 | 0.0000000 | 1 | Trf |
| 0.0e+00 | 1.0892322 | 0.147 | 0.012 | 0.0000000 | 1 | Cyp2j12 |
| 0.0e+00 | 1.0522038 | 0.234 | 0.022 | 0.0000000 | 1 | Hcn2 |
| 0.0e+00 | 1.0374340 | 0.206 | 0.019 | 0.0000000 | 1 | Kcnk13 |
| 0.0e+00 | 1.0350104 | 0.863 | 0.125 | 0.0000000 | 1 | Prr5l |
| 0.0e+00 | 1.0181583 | 0.464 | 0.047 | 0.0000000 | 1 | Apod |
| 0.0e+00 | 0.9779225 | 0.200 | 0.022 | 0.0000000 | 1 | Fgd3 |
| 0.0e+00 | 0.9768928 | 0.959 | 0.215 | 0.0000000 | 1 | Fth1 |
| 0.0e+00 | 0.9718602 | 0.137 | 0.015 | 0.0000000 | 1 | Gjb1 |
| 0.0e+00 | 0.9106105 | 0.363 | 0.048 | 0.0000000 | 1 | Rftn1 |
| 0.0e+00 | 0.8949042 | 0.346 | 0.047 | 0.0000000 | 1 | Carns1 |
| 0.0e+00 | 0.8908094 | 0.208 | 0.028 | 0.0000000 | 1 | Adam19 |
| 0.0e+00 | 0.8853827 | 0.131 | 0.018 | 0.0000000 | 1 | Mboat1 |
| 0.0e+00 | 0.8851835 | 0.363 | 0.042 | 0.0000000 | 1 | Enpp6 |
| 0.0e+00 | 0.8850288 | 0.158 | 0.022 | 0.0000000 | 1 | B3galt5 |
| 0.0e+00 | 0.8805379 | 0.489 | 0.076 | 0.0000000 | 1 | Gatm |
| 0.0e+00 | 0.8596478 | 0.209 | 0.029 | 0.0000000 | 1 | Dock5 |
| 0.0e+00 | 1.3209802 | 0.147 | 0.008 | 0.0000000 | 2 | Rasal1 |
| 0.0e+00 | 1.2036073 | 0.264 | 0.019 | 0.0000000 | 2 | Gm32633 |
| 0.0e+00 | 1.1419077 | 0.156 | 0.012 | 0.0000000 | 2 | Tmem141 |
| 0.0e+00 | 1.0566926 | 0.163 | 0.016 | 0.0000000 | 2 | Onecut2 |
| 0.0e+00 | 1.0200060 | 0.131 | 0.014 | 0.0000000 | 2 | Tssc4 |
| 0.0e+00 | 1.0164049 | 0.218 | 0.022 | 0.0000000 | 2 | Tnni1 |
| 0.0e+00 | 0.9838140 | 0.260 | 0.028 | 0.0000000 | 2 | C030029H02Rik |
| 0.0e+00 | 0.9753300 | 0.189 | 0.023 | 0.0000000 | 2 | Ctps |
| 0.0e+00 | 0.9631501 | 0.591 | 0.082 | 0.0000000 | 2 | Synpr |
| 0.0e+00 | 0.9388717 | 0.174 | 0.021 | 0.0000000 | 2 | Hebp1 |
| 0.0e+00 | 0.9062614 | 0.575 | 0.083 | 0.0000000 | 2 | Opalin |
| 0.0e+00 | 0.8853478 | 0.679 | 0.116 | 0.0000000 | 2 | Ablim2 |
| 0.0e+00 | 0.8693303 | 0.103 | 0.014 | 0.0000000 | 2 | Arhgef19 |
| 0.0e+00 | 0.8579505 | 0.454 | 0.098 | 0.0000000 | 2 | Ptgds |
| 0.0e+00 | 0.8280960 | 0.180 | 0.028 | 0.0000000 | 2 | Erbb3 |
| 0.0e+00 | 0.8219893 | 0.600 | 0.108 | 0.0000000 | 2 | Sh3gl3 |
| 0.0e+00 | 0.8192472 | 0.455 | 0.077 | 0.0000000 | 2 | Tmod1 |
| 0.0e+00 | 0.8179436 | 0.485 | 0.106 | 0.0000000 | 2 | Fam214a |
| 0.0e+00 | 0.8046050 | 0.530 | 0.109 | 0.0000000 | 2 | Man1a |
| 0.0e+00 | 0.7922918 | 0.366 | 0.063 | 0.0000000 | 2 | Mcam |
| 0.0e+00 | 1.6853299 | 0.970 | 0.106 | 0.0000000 | 3 | Slc1a2 |
| 0.0e+00 | 1.5067922 | 0.189 | 0.007 | 0.0000000 | 3 | Grin2c |
| 0.0e+00 | 1.4327451 | 0.194 | 0.010 | 0.0000000 | 3 | Gm12239 |
| 0.0e+00 | 1.4249665 | 0.101 | 0.004 | 0.0000000 | 3 | Gm26512 |
| 0.0e+00 | 1.3345483 | 0.302 | 0.019 | 0.0000000 | 3 | Htra1 |
| 0.0e+00 | 1.3145373 | 0.816 | 0.081 | 0.0000000 | 3 | Gpc5 |
| 0.0e+00 | 1.3055228 | 0.873 | 0.108 | 0.0000000 | 3 | Slc1a3 |
| 0.0e+00 | 1.2643648 | 0.181 | 0.010 | 0.0000000 | 3 | Ntsr2 |
| 0.0e+00 | 1.2615391 | 0.167 | 0.009 | 0.0000000 | 3 | Acsbg1 |
| 0.0e+00 | 1.2553896 | 0.798 | 0.093 | 0.0000000 | 3 | Cst3 |
| 0.0e+00 | 1.2414453 | 0.168 | 0.010 | 0.0000000 | 3 | Fgfr3 |
| 0.0e+00 | 1.2132916 | 0.204 | 0.014 | 0.0000000 | 3 | F3 |
| 0.0e+00 | 1.1987239 | 0.160 | 0.011 | 0.0000000 | 3 | Gldc |
| 0.0e+00 | 1.1858164 | 0.145 | 0.010 | 0.0000000 | 3 | Acot11 |
| 0.0e+00 | 1.1829051 | 0.442 | 0.040 | 0.0000000 | 3 | Mertk |
| 0.0e+00 | 1.1786231 | 0.127 | 0.008 | 0.0000000 | 3 | Cbs |
| 0.0e+00 | 1.1767296 | 0.161 | 0.011 | 0.0000000 | 3 | Gm6145 |
| 0.0e+00 | 1.1761343 | 0.210 | 0.016 | 0.0000000 | 3 | Aldoc |
| 0.0e+00 | 1.1416954 | 0.635 | 0.068 | 0.0000000 | 3 | Prex2 |
| 0.0e+00 | 1.1252629 | 0.632 | 0.095 | 0.0000000 | 3 | Tspan7 |
| 0.0e+00 | 1.8790844 | 0.194 | 0.002 | 0.0000000 | 4 | Igfbpl1 |
| 0.0e+00 | 1.7387708 | 0.147 | 0.003 | 0.0000000 | 4 | Xist |
| 0.0e+00 | 1.7033905 | 0.119 | 0.002 | 0.0000000 | 4 | Mki67 |
| 0.0e+00 | 1.7002947 | 0.555 | 0.019 | 0.0000000 | 4 | Sox11 |
| 0.0e+00 | 1.6476903 | 0.101 | 0.002 | 0.0000000 | 4 | Dlx1 |
| 0.0e+00 | 1.6200189 | 0.143 | 0.003 | 0.0000000 | 4 | Top2a |
| 0.0e+00 | 1.5624399 | 0.245 | 0.012 | 0.0000000 | 4 | Dlx6os1 |
| 0.0e+00 | 1.3819476 | 0.170 | 0.011 | 0.0000000 | 4 | Gm29260 |
| 0.0e+00 | 1.3746890 | 0.153 | 0.013 | 0.0000000 | 4 | Satb2 |
| 0.0e+00 | 1.3690852 | 0.313 | 0.016 | 0.0000000 | 4 | Dcx |
| 0.0e+00 | 1.3483601 | 0.107 | 0.005 | 0.0000000 | 4 | Mex3a |
| 0.0e+00 | 1.3077422 | 0.136 | 0.007 | 0.0000000 | 4 | Tubb2b |
| 0.0e+00 | 1.3006627 | 0.235 | 0.028 | 0.0000000 | 4 | Unc5d |
| 0.0e+00 | 1.1928454 | 0.208 | 0.017 | 0.0000000 | 4 | Epha3 |
| 0.0e+00 | 1.1680802 | 0.124 | 0.009 | 0.0000000 | 4 | Mpped1 |
| 0.0e+00 | 1.1640244 | 0.559 | 0.064 | 0.0000000 | 4 | Sox4 |
| 0.0e+00 | 1.1452113 | 0.123 | 0.009 | 0.0000000 | 4 | Sox1ot |
| 0.0e+00 | 1.1243517 | 0.167 | 0.015 | 0.0000000 | 4 | Lmnb1 |
| 0.0e+00 | 1.1107389 | 0.305 | 0.046 | 0.0000000 | 4 | Gm26871 |
| 0.0e+00 | 1.1054509 | 0.126 | 0.011 | 0.0000000 | 4 | Hmgb2 |
| 0.0e+00 | 2.7402688 | 0.188 | 0.000 | 0.0000000 | 5 | Edn1 |
| 0.0e+00 | 2.6740745 | 0.792 | 0.007 | 0.0000000 | 5 | Flt1 |
| 0.0e+00 | 2.5981621 | 0.550 | 0.003 | 0.0000000 | 5 | Adgrl4 |
| 0.0e+00 | 2.5421669 | 0.283 | 0.001 | 0.0000000 | 5 | Zfp366 |
| 0.0e+00 | 2.5027505 | 0.601 | 0.004 | 0.0000000 | 5 | Cldn5 |
| 0.0e+00 | 2.3406675 | 0.623 | 0.008 | 0.0000000 | 5 | Slco1a4 |
| 0.0e+00 | 2.3036211 | 0.303 | 0.002 | 0.0000000 | 5 | Apold1 |
| 0.0e+00 | 2.2665012 | 0.336 | 0.002 | 0.0000000 | 5 | Tek |
| 0.0e+00 | 2.1792421 | 0.654 | 0.011 | 0.0000000 | 5 | Mecom |
| 0.0e+00 | 2.1583102 | 0.541 | 0.007 | 0.0000000 | 5 | Ptprb |
| 0.0e+00 | 2.1238337 | 0.256 | 0.002 | 0.0000000 | 5 | Fgd5 |
| 0.0e+00 | 2.1210408 | 0.236 | 0.003 | 0.0000000 | 5 | Rgs5 |
| 0.0e+00 | 2.1163457 | 0.272 | 0.002 | 0.0000000 | 5 | Egfl7 |
| 0.0e+00 | 2.1001698 | 0.399 | 0.004 | 0.0000000 | 5 | Abcb1a |
| 0.0e+00 | 2.0947915 | 0.267 | 0.002 | 0.0000000 | 5 | Vwf |
| 0.0e+00 | 2.0438165 | 0.252 | 0.003 | 0.0000000 | 5 | Erg |
| 0.0e+00 | 2.0390486 | 0.393 | 0.005 | 0.0000000 | 5 | Eng |
| 0.0e+00 | 2.0362193 | 0.433 | 0.006 | 0.0000000 | 5 | Cyyr1 |
| 0.0e+00 | 1.9999062 | 0.148 | 0.001 | 0.0000000 | 5 | Tbx3os1 |
| 0.0e+00 | 1.9902948 | 0.229 | 0.002 | 0.0000000 | 5 | Ly6c1 |
| 0.0e+00 | 1.6919400 | 0.199 | 0.004 | 0.0000000 | 6 | Gpr17 |
| 0.0e+00 | 1.6258295 | 0.917 | 0.087 | 0.0000000 | 6 | Tnr |
| 0.0e+00 | 1.5686199 | 0.201 | 0.009 | 0.0000000 | 6 | Gm36988 |
| 0.0e+00 | 1.5101472 | 0.459 | 0.017 | 0.0000000 | 6 | Tmem132d |
| 0.0e+00 | 1.5091262 | 0.224 | 0.009 | 0.0000000 | 6 | 4930588A03Rik |
| 0.0e+00 | 1.4581971 | 0.431 | 0.030 | 0.0000000 | 6 | Lhfpl3 |
| 0.0e+00 | 1.4491361 | 0.201 | 0.007 | 0.0000000 | 6 | Pdgfra |
| 0.0e+00 | 1.4216137 | 0.488 | 0.038 | 0.0000000 | 6 | Pcdh15 |
| 0.0e+00 | 1.4069489 | 0.114 | 0.004 | 0.0000000 | 6 | Shc4 |
| 0.0e+00 | 1.3766695 | 0.199 | 0.009 | 0.0000000 | 6 | Cspg4 |
| 0.0e+00 | 1.3720107 | 0.329 | 0.025 | 0.0000000 | 6 | Arhgap24 |
| 0.0e+00 | 1.3492160 | 0.346 | 0.020 | 0.0000000 | 6 | Vcan |
| 0.0e+00 | 1.3362456 | 0.278 | 0.017 | 0.0000000 | 6 | 9630013A20Rik |
| 0.0e+00 | 1.3298121 | 0.242 | 0.014 | 0.0000000 | 6 | Megf11 |
| 0.0e+00 | 1.3174237 | 0.209 | 0.012 | 0.0000000 | 6 | Sema3d |
| 0.0e+00 | 1.3079245 | 0.305 | 0.024 | 0.0000000 | 6 | Gm13052 |
| 0.0e+00 | 1.2794788 | 0.783 | 0.102 | 0.0000000 | 6 | Dscam |
| 0.0e+00 | 1.2409114 | 0.559 | 0.064 | 0.0000000 | 6 | Xylt1 |
| 0.0e+00 | 1.2369910 | 0.616 | 0.075 | 0.0000000 | 6 | Itpr2 |
| 0.0e+00 | 1.2045885 | 0.425 | 0.026 | 0.0000000 | 6 | Nxph1 |
| 0.0e+00 | 3.1934150 | 0.432 | 0.000 | 0.0000000 | 7 | Ccdc180 |
| 0.0e+00 | 2.8891799 | 0.498 | 0.001 | 0.0000000 | 7 | Tmem212 |
| 0.0e+00 | 2.8343931 | 0.408 | 0.001 | 0.0000000 | 7 | Cdhr3 |
| 0.0e+00 | 2.7500722 | 0.225 | 0.000 | 0.0000000 | 7 | Lrrc43 |
| 0.0e+00 | 2.7186822 | 0.633 | 0.002 | 0.0000000 | 7 | Dnah12 |
| 0.0e+00 | 2.7148944 | 0.425 | 0.001 | 0.0000000 | 7 | Lrrc36 |
| 0.0e+00 | 2.6171556 | 0.710 | 0.004 | 0.0000000 | 7 | Cfap299 |
| 0.0e+00 | 2.4744696 | 0.546 | 0.003 | 0.0000000 | 7 | Hydin |
| 0.0e+00 | 2.4519866 | 0.316 | 0.001 | 0.0000000 | 7 | Gm10714 |
| 0.0e+00 | 2.4262990 | 0.336 | 0.002 | 0.0000000 | 7 | Cfap77 |
| 0.0e+00 | 2.3891957 | 0.367 | 0.002 | 0.0000000 | 7 | 3300002A11Rik |
| 0.0e+00 | 2.3870027 | 0.635 | 0.005 | 0.0000000 | 7 | Dnah6 |
| 0.0e+00 | 2.3751465 | 0.273 | 0.001 | 0.0000000 | 7 | Ttc29 |
| 0.0e+00 | 2.3538233 | 0.367 | 0.002 | 0.0000000 | 7 | Spag17 |
| 0.0e+00 | 2.3499107 | 0.186 | 0.001 | 0.0000000 | 7 | Ttll6 |
| 0.0e+00 | 2.3209470 | 0.541 | 0.004 | 0.0000000 | 7 | Cfap43 |
| 0.0e+00 | 2.3151486 | 0.162 | 0.001 | 0.0000000 | 7 | Zfp474 |
| 0.0e+00 | 2.3013603 | 0.348 | 0.002 | 0.0000000 | 7 | Armc4 |
| 0.0e+00 | 2.3004253 | 0.261 | 0.001 | 0.0000000 | 7 | Gm28729 |
| 0.0e+00 | 2.2681520 | 0.307 | 0.002 | 0.0000000 | 7 | Wdr49 |
| 0.0e+00 | 2.9773434 | 0.580 | 0.003 | 0.0000000 | 8 | Nwd2 |
| 0.0e+00 | 2.8936400 | 0.185 | 0.000 | 0.0000000 | 8 | D130079A08Rik |
| 0.0e+00 | 2.2290479 | 0.563 | 0.007 | 0.0000000 | 8 | Cpne4 |
| 0.0e+00 | 2.1698464 | 0.149 | 0.001 | 0.0000000 | 8 | Tac2 |
| 0.0e+00 | 2.1624853 | 0.254 | 0.002 | 0.0000000 | 8 | Robo3 |
| 0.0e+00 | 2.1485549 | 0.498 | 0.009 | 0.0000000 | 8 | Scube1 |
| 0.0e+00 | 2.1335332 | 0.183 | 0.001 | 0.0000000 | 8 | Lrrc55 |
| 0.0e+00 | 2.0420598 | 0.193 | 0.002 | 0.0000000 | 8 | D130009I18Rik |
| 0.0e+00 | 1.9393975 | 0.139 | 0.002 | 0.0000000 | 8 | Fibcd1 |
| 0.0e+00 | 1.7450925 | 0.178 | 0.006 | 0.0000000 | 8 | Il1rapl2 |
| 0.0e+00 | 1.7294480 | 0.229 | 0.005 | 0.0000000 | 8 | Lrrc3b |
| 0.0e+00 | 1.6740830 | 0.293 | 0.008 | 0.0000000 | 8 | Necab2 |
| 0.0e+00 | 1.6500159 | 0.512 | 0.025 | 0.0000000 | 8 | Vav3 |
| 0.0e+00 | 1.6461928 | 0.102 | 0.003 | 0.0000000 | 8 | Cck |
| 0.0e+00 | 1.6316341 | 0.146 | 0.004 | 0.0000000 | 8 | Syt13 |
| 0.0e+00 | 1.6184173 | 0.229 | 0.007 | 0.0000000 | 8 | Necab3 |
| 0.0e+00 | 1.6003241 | 0.154 | 0.004 | 0.0000000 | 8 | Cpne9 |
| 0.0e+00 | 1.5736422 | 0.129 | 0.004 | 0.0000000 | 8 | Htr4 |
| 0.0e+00 | 1.5683338 | 0.878 | 0.192 | 0.0000000 | 8 | Kcnma1 |
| 0.0e+00 | 1.5667633 | 0.105 | 0.003 | 0.0000000 | 8 | Calb2 |
| 0.0e+00 | 2.8327592 | 0.828 | 0.002 | 0.0000000 | 9 | Ush2a |
| 0.0e+00 | 2.6812950 | 0.457 | 0.001 | 0.0000000 | 9 | Gm32884 |
| 0.0e+00 | 2.6238452 | 0.759 | 0.003 | 0.0000000 | 9 | Gm44196 |
| 0.0e+00 | 2.4972348 | 0.210 | 0.001 | 0.0000000 | 9 | Gm47480 |
| 0.0e+00 | 2.4737537 | 0.477 | 0.002 | 0.0000000 | 9 | Rbp3 |
| 0.0e+00 | 2.4659283 | 0.664 | 0.003 | 0.0000000 | 9 | Tulp1 |
| 0.0e+00 | 2.4321096 | 0.580 | 0.003 | 0.0000000 | 9 | Gm31615 |
| 0.0e+00 | 2.3798492 | 0.761 | 0.005 | 0.0000000 | 9 | Cngb3 |
| 0.0e+00 | 2.3603583 | 0.322 | 0.001 | 0.0000000 | 9 | Pde6c |
| 0.0e+00 | 2.3453544 | 0.213 | 0.001 | 0.0000000 | 9 | Crx |
| 0.0e+00 | 2.3395249 | 0.198 | 0.001 | 0.0000000 | 9 | Lhx4 |
| 0.0e+00 | 2.3345075 | 0.457 | 0.002 | 0.0000000 | 9 | Cplx3 |
| 0.0e+00 | 2.3163142 | 0.707 | 0.005 | 0.0000000 | 9 | Rpgrip1 |
| 0.0e+00 | 2.2194671 | 0.661 | 0.007 | 0.0000000 | 9 | Sag |
| 0.0e+00 | 2.2034174 | 0.385 | 0.003 | 0.0000000 | 9 | Gnb3 |
| 0.0e+00 | 2.1796524 | 0.362 | 0.003 | 0.0000000 | 9 | Mpp4 |
| 0.0e+00 | 2.1183660 | 0.261 | 0.002 | 0.0000000 | 9 | Tph1 |
| 0.0e+00 | 2.0650539 | 0.313 | 0.003 | 0.0000000 | 9 | Samsn1 |
| 0.0e+00 | 1.9600035 | 0.210 | 0.002 | 0.0000000 | 9 | Fabp12 |
| 0.0e+00 | 1.8924073 | 0.460 | 0.008 | 0.0000000 | 9 | Col7a1 |
| 0.0e+00 | 1.6802022 | 0.389 | 0.010 | 0.0000000 | 10 | Agt |
| 0.0e+00 | 1.2595827 | 0.471 | 0.039 | 0.0000000 | 10 | Slc6a11 |
| 0.0e+00 | 1.2566637 | 0.124 | 0.007 | 0.0000000 | 10 | Gm33680 |
| 0.0e+00 | 1.2024848 | 0.235 | 0.017 | 0.0000000 | 10 | Aqp4 |
| 0.0e+00 | 1.1942527 | 0.199 | 0.015 | 0.0000000 | 10 | Atp13a4 |
| 0.0e+00 | 1.1585337 | 0.170 | 0.012 | 0.0000000 | 10 | Etnppl |
| 0.0e+00 | 1.1258256 | 0.180 | 0.022 | 0.0000000 | 10 | Gfap |
| 0.0e+00 | 1.0237024 | 0.824 | 0.135 | 0.0000000 | 10 | Slc4a4 |
| 0.0e+00 | 1.0125599 | 0.118 | 0.011 | 0.0000000 | 10 | Itih3 |
| 0.0e+00 | 0.9736382 | 0.356 | 0.041 | 0.0000000 | 10 | Slc7a11 |
| 0.0e+00 | 0.9478084 | 0.196 | 0.024 | 0.0000000 | 10 | Adcy8 |
| 0.0e+00 | 0.9453237 | 0.392 | 0.075 | 0.0000000 | 10 | Slc38a1 |
| 0.0e+00 | 0.9427607 | 0.330 | 0.042 | 0.0000000 | 10 | Bmpr1b |
| 0.0e+00 | 0.9219528 | 0.536 | 0.089 | 0.0000000 | 10 | Sfxn5 |
| 0.0e+00 | 0.9118849 | 0.657 | 0.106 | 0.0000000 | 10 | Sparcl1 |
| 0.0e+00 | 0.9112603 | 0.418 | 0.080 | 0.0000000 | 10 | Camk2g |
| 0.0e+00 | 0.9076513 | 0.121 | 0.015 | 0.0000000 | 10 | Cdh22 |
| 0.0e+00 | 0.8984468 | 0.225 | 0.031 | 0.0000000 | 10 | Pla2g7 |
| 0.0e+00 | 0.8966559 | 0.157 | 0.023 | 0.0000000 | 10 | Psd2 |
| 0.0e+00 | 0.8813931 | 0.892 | 0.238 | 0.0000000 | 10 | Npas3 |
| 0.0e+00 | 2.4672568 | 0.359 | 0.001 | 0.0000000 | 11 | Gm10754 |
| 0.0e+00 | 2.3669018 | 0.355 | 0.002 | 0.0000000 | 11 | Scn4b |
| 0.0e+00 | 2.3274787 | 0.283 | 0.002 | 0.0000000 | 11 | Drd2 |
| 0.0e+00 | 2.2583978 | 0.300 | 0.002 | 0.0000000 | 11 | Tac1 |
| 0.0e+00 | 2.2475670 | 0.293 | 0.002 | 0.0000000 | 11 | Adora2a |
| 0.0e+00 | 2.2197916 | 0.548 | 0.004 | 0.0000000 | 11 | Cpne5 |
| 0.0e+00 | 2.1556245 | 0.276 | 0.002 | 0.0000000 | 11 | Penk |
| 0.0e+00 | 2.1513875 | 0.276 | 0.002 | 0.0000000 | 11 | Gpr88 |
| 0.0e+00 | 2.0850209 | 0.348 | 0.003 | 0.0000000 | 11 | Actn2 |
| 0.0e+00 | 1.9799164 | 0.483 | 0.006 | 0.0000000 | 11 | Pde1b |
| 0.0e+00 | 1.9679351 | 0.390 | 0.004 | 0.0000000 | 11 | Gprin3 |
| 0.0e+00 | 1.9518151 | 0.221 | 0.002 | 0.0000000 | 11 | Sh3rf2 |
| 0.0e+00 | 1.8953844 | 0.217 | 0.003 | 0.0000000 | 11 | Gabrd |
| 0.0e+00 | 1.8572430 | 0.841 | 0.021 | 0.0000000 | 11 | Rgs9 |
| 0.0e+00 | 1.8527769 | 0.821 | 0.021 | 0.0000000 | 11 | Camk4 |
| 0.0e+00 | 1.8264891 | 0.197 | 0.003 | 0.0000000 | 11 | Syndig1l |
| 0.0e+00 | 1.8136088 | 0.410 | 0.007 | 0.0000000 | 11 | Ppp1r1b |
| 0.0e+00 | 1.7104495 | 0.855 | 0.031 | 0.0000000 | 11 | Ryr3 |
| 0.0e+00 | 1.6990898 | 0.148 | 0.003 | 0.0000000 | 11 | Akap5 |
| 0.0e+00 | 1.6961049 | 0.369 | 0.009 | 0.0000000 | 11 | 6530403H02Rik |
| 0.0e+00 | 2.6603095 | 0.391 | 0.002 | 0.0000000 | 12 | Bnc2 |
| 0.0e+00 | 2.4869158 | 0.322 | 0.001 | 0.0000000 | 12 | Aox3 |
| 0.0e+00 | 2.3702104 | 0.302 | 0.001 | 0.0000000 | 12 | Lum |
| 0.0e+00 | 2.0484613 | 0.723 | 0.012 | 0.0000000 | 12 | Cped1 |
| 0.0e+00 | 1.9761963 | 0.351 | 0.005 | 0.0000000 | 12 | Alpl |
| 0.0e+00 | 1.9678300 | 0.342 | 0.005 | 0.0000000 | 12 | Olfr1033 |
| 0.0e+00 | 1.8736091 | 0.332 | 0.004 | 0.0000000 | 12 | Col1a1 |
| 0.0e+00 | 1.8227453 | 0.381 | 0.006 | 0.0000000 | 12 | Eya2 |
| 0.0e+00 | 1.7833348 | 0.465 | 0.009 | 0.0000000 | 12 | Col1a2 |
| 0.0e+00 | 1.7696043 | 0.297 | 0.005 | 0.0000000 | 12 | Adamts12 |
| 0.0e+00 | 1.7389683 | 0.267 | 0.007 | 0.0000000 | 12 | Cemip |
| 0.0e+00 | 1.7000067 | 0.183 | 0.004 | 0.0000000 | 12 | Efemp1 |
| 0.0e+00 | 1.6999254 | 0.728 | 0.042 | 0.0000000 | 12 | Adam12 |
| 0.0e+00 | 1.6638853 | 0.535 | 0.015 | 0.0000000 | 12 | Igfbp4 |
| 0.0e+00 | 1.6091933 | 0.218 | 0.006 | 0.0000000 | 12 | Tbx15 |
| 0.0e+00 | 1.5710686 | 0.183 | 0.005 | 0.0000000 | 12 | Col12a1 |
| 0.0e+00 | 1.4962272 | 0.213 | 0.007 | 0.0000000 | 12 | Col26a1 |
| 0.0e+00 | 1.4407348 | 0.193 | 0.007 | 0.0000000 | 12 | Cpxm1 |
| 0.0e+00 | 1.4354060 | 0.292 | 0.011 | 0.0000000 | 12 | Mrc2 |
| 0.0e+00 | 1.3961527 | 0.173 | 0.007 | 0.0000000 | 12 | Fxyd5 |
| 0.0e+00 | 3.0585428 | 0.135 | 0.000 | 0.0000000 | 13 | Sostdc1 |
| 0.0e+00 | 2.5921917 | 1.000 | 0.023 | 0.0000000 | 13 | Htr2c |
| 0.0e+00 | 2.5050882 | 0.606 | 0.002 | 0.0000000 | 13 | Gmnc |
| 0.0e+00 | 2.4564828 | 0.106 | 0.000 | 0.0000000 | 13 | Aqp1 |
| 0.0e+00 | 2.2771864 | 0.683 | 0.004 | 0.0000000 | 13 | Tmem72 |
| 0.0e+00 | 2.1958815 | 0.654 | 0.005 | 0.0000000 | 13 | Kl |
| 0.0e+00 | 2.0885060 | 0.394 | 0.004 | 0.0000000 | 13 | Prlr |
| 0.0e+00 | 2.0843895 | 0.625 | 0.007 | 0.0000000 | 13 | Rbm47 |
| 0.0e+00 | 2.0652762 | 0.625 | 0.006 | 0.0000000 | 13 | Slc4a5 |
| 0.0e+00 | 2.0247612 | 1.000 | 0.250 | 0.0000000 | 13 | Ttr |
| 0.0e+00 | 2.0157908 | 0.625 | 0.008 | 0.0000000 | 13 | Clic6 |
| 0.0e+00 | 1.9793616 | 0.433 | 0.005 | 0.0000000 | 13 | Car12 |
| 0.0e+00 | 1.9513328 | 0.471 | 0.006 | 0.0000000 | 13 | Folr1 |
| 0.0e+00 | 1.9336041 | 0.337 | 0.004 | 0.0000000 | 13 | Baiap2l1 |
| 0.0e+00 | 1.8399450 | 0.510 | 0.007 | 0.0000000 | 13 | Lmx1a |
| 0.0e+00 | 1.8272706 | 0.260 | 0.004 | 0.0000000 | 13 | Slc13a4 |
| 0.0e+00 | 1.7517605 | 0.404 | 0.008 | 0.0000000 | 13 | Steap2 |
| 0.0e+00 | 1.7263930 | 0.288 | 0.006 | 0.0000000 | 13 | Trpv4 |
| 0.0e+00 | 1.7242829 | 0.567 | 0.012 | 0.0000000 | 13 | Igf2 |
| 0.0e+00 | 1.7178401 | 0.413 | 0.008 | 0.0000000 | 13 | Fap |
| 0.0e+00 | 2.7994524 | 0.156 | 0.000 | 0.0000000 | 14 | Cx3cr1 |
| 0.0e+00 | 2.5885991 | 0.143 | 0.000 | 0.0000000 | 14 | Fyb |
| 0.0e+00 | 2.4258718 | 0.130 | 0.000 | 0.0000000 | 14 | Ly86 |
| 0.0e+00 | 2.3844791 | 0.117 | 0.000 | 0.0000000 | 14 | Nlrp3 |
| 0.0e+00 | 2.3289618 | 0.130 | 0.001 | 0.0000000 | 14 | Laptm5 |
| 0.0e+00 | 2.1828337 | 0.117 | 0.001 | 0.0000000 | 14 | Dock2 |
| 0.0e+00 | 2.0834491 | 0.506 | 0.006 | 0.0000000 | 14 | Cd83 |
| 0.0e+00 | 1.9139884 | 0.403 | 0.006 | 0.0000000 | 14 | Plek |
| 0.0e+00 | 1.7016959 | 0.494 | 0.010 | 0.0000000 | 14 | Inpp5d |
| 0.0e+00 | 1.6185623 | 0.104 | 0.003 | 0.0000128 | 14 | Rcsd1 |
| 0.0e+00 | 1.6046859 | 0.260 | 0.007 | 0.0000000 | 14 | Apbb1ip |
| 0.0e+00 | 1.5448902 | 0.208 | 0.006 | 0.0000000 | 14 | Tnfaip3 |
| 0.0e+00 | 1.4079602 | 0.234 | 0.010 | 0.0000000 | 14 | Dock8 |
| 0.0e+00 | 1.3844791 | 0.416 | 0.021 | 0.0000000 | 14 | Pde3b |
| 0.0e+00 | 1.3816313 | 0.377 | 0.019 | 0.0000000 | 14 | Lyn |
| 0.0e+00 | 1.3412955 | 0.494 | 0.025 | 0.0000000 | 14 | Runx1 |
| 0.0e+00 | 1.3044370 | 0.247 | 0.013 | 0.0000000 | 14 | Maf |
| 0.0e+00 | 1.2949720 | 0.481 | 0.043 | 0.0000000 | 14 | Sirpa |
| 0.0e+00 | 1.2803905 | 0.338 | 0.019 | 0.0000000 | 14 | Adap2 |
| 0.0e+00 | 1.2705357 | 0.182 | 0.010 | 0.0000000 | 14 | Lair1 |
| 0.0e+00 | 3.4456464 | 0.288 | 0.000 | 0.0000000 | 15 | Rprm |
| 0.0e+00 | 2.9161371 | 0.169 | 0.000 | 0.0000000 | 15 | Nxph3 |
| 0.0e+00 | 2.4042537 | 0.153 | 0.001 | 0.0000000 | 15 | Galnt9 |
| 0.0e+00 | 2.1820846 | 0.966 | 0.023 | 0.0000000 | 15 | Hs3st4 |
| 0.0e+00 | 2.1800638 | 0.542 | 0.004 | 0.0000000 | 15 | Ipcef1 |
| 0.0e+00 | 2.1612157 | 0.102 | 0.001 | 0.0000075 | 15 | Col5a1 |
| 0.0e+00 | 2.0306731 | 1.000 | 0.079 | 0.0000000 | 15 | Dpp10 |
| 0.0e+00 | 1.9094037 | 0.441 | 0.007 | 0.0000000 | 15 | Vxn |
| 0.0e+00 | 1.8621917 | 0.458 | 0.007 | 0.0000000 | 15 | Ttc9b |
| 0.0e+00 | 1.8521677 | 0.339 | 0.005 | 0.0000000 | 15 | Tbr1 |
| 0.0e+00 | 1.8230439 | 0.407 | 0.008 | 0.0000000 | 15 | Htr1f |
| 0.0e+00 | 1.8223971 | 0.288 | 0.005 | 0.0000000 | 15 | Hs3st2 |
| 0.0e+00 | 1.7874705 | 0.407 | 0.008 | 0.0000000 | 15 | Hpcal4 |
| 0.0e+00 | 1.7178729 | 0.390 | 0.009 | 0.0000000 | 15 | Npas4 |
| 0.0e+00 | 1.6370979 | 0.763 | 0.024 | 0.0000000 | 15 | Slc17a7 |
| 0.0e+00 | 1.6307976 | 0.458 | 0.012 | 0.0000000 | 15 | Nptx1 |
| 0.0e+00 | 1.6196684 | 0.763 | 0.024 | 0.0000000 | 15 | Tmem178 |
| 0.0e+00 | 1.5863354 | 0.780 | 0.028 | 0.0000000 | 15 | A830018L16Rik |
| 0.0e+00 | 1.5781789 | 0.559 | 0.015 | 0.0000000 | 15 | Vsnl1 |
| 0.0e+00 | 1.5757974 | 0.288 | 0.008 | 0.0000000 | 15 | Trabd2b |
| 0.0e+00 | 2.4362784 | 0.972 | 0.029 | 0.0000000 | 16 | Reln |
| 0.0e+00 | 2.0333160 | 0.611 | 0.007 | 0.0000000 | 16 | Trp73 |
| 0.0e+00 | 1.9122723 | 0.556 | 0.009 | 0.0000000 | 16 | Ndnf |
| 0.0e+00 | 1.8833775 | 0.472 | 0.007 | 0.0000000 | 16 | Ebf3 |
| 0.0e+00 | 1.7624574 | 0.972 | 0.056 | 0.0000000 | 16 | Clstn2 |
| 0.0e+00 | 1.7272174 | 0.861 | 0.040 | 0.0000000 | 16 | Thsd7b |
| 0.0e+00 | 1.6811718 | 0.972 | 0.060 | 0.0000000 | 16 | Kcnh7 |
| 0.0e+00 | 1.6201361 | 0.833 | 0.032 | 0.0000000 | 16 | Cdh4 |
| 0.0e+00 | 1.4951973 | 0.750 | 0.043 | 0.0000000 | 16 | Dync1i1 |
| 0.0e+00 | 1.4740080 | 0.194 | 0.007 | 0.0006911 | 16 | Tbr1 |
| 0.0e+00 | 1.4740080 | 0.583 | 0.030 | 0.0000000 | 16 | Epha3 |
| 0.0e+00 | 1.4344995 | 0.361 | 0.014 | 0.0000000 | 16 | Col11a1 |
| 0.0e+00 | 1.3958127 | 0.389 | 0.021 | 0.0000000 | 16 | Hs3st5 |
| 0.0e+00 | 1.3791710 | 0.444 | 0.026 | 0.0000000 | 16 | Cacna2d2 |
| 0.0e+00 | 1.3780536 | 0.472 | 0.022 | 0.0000000 | 16 | Kcnc2 |
| 0.0e+00 | 1.3647438 | 1.000 | 0.102 | 0.0000000 | 16 | Cntnap2 |
| 4.0e-07 | 1.3413825 | 0.194 | 0.010 | 0.0076880 | 16 | Ajap1 |
| 5.8e-06 | 1.3253550 | 0.139 | 0.009 | 0.0988710 | 16 | Tmem200a |
| 7.9e-06 | 1.2949724 | 0.167 | 0.010 | 0.1357362 | 16 | Syndig1l |
| 0.0e+00 | 1.2944836 | 0.278 | 0.017 | 0.0000001 | 16 | Plxnd1 |
Dot-plot
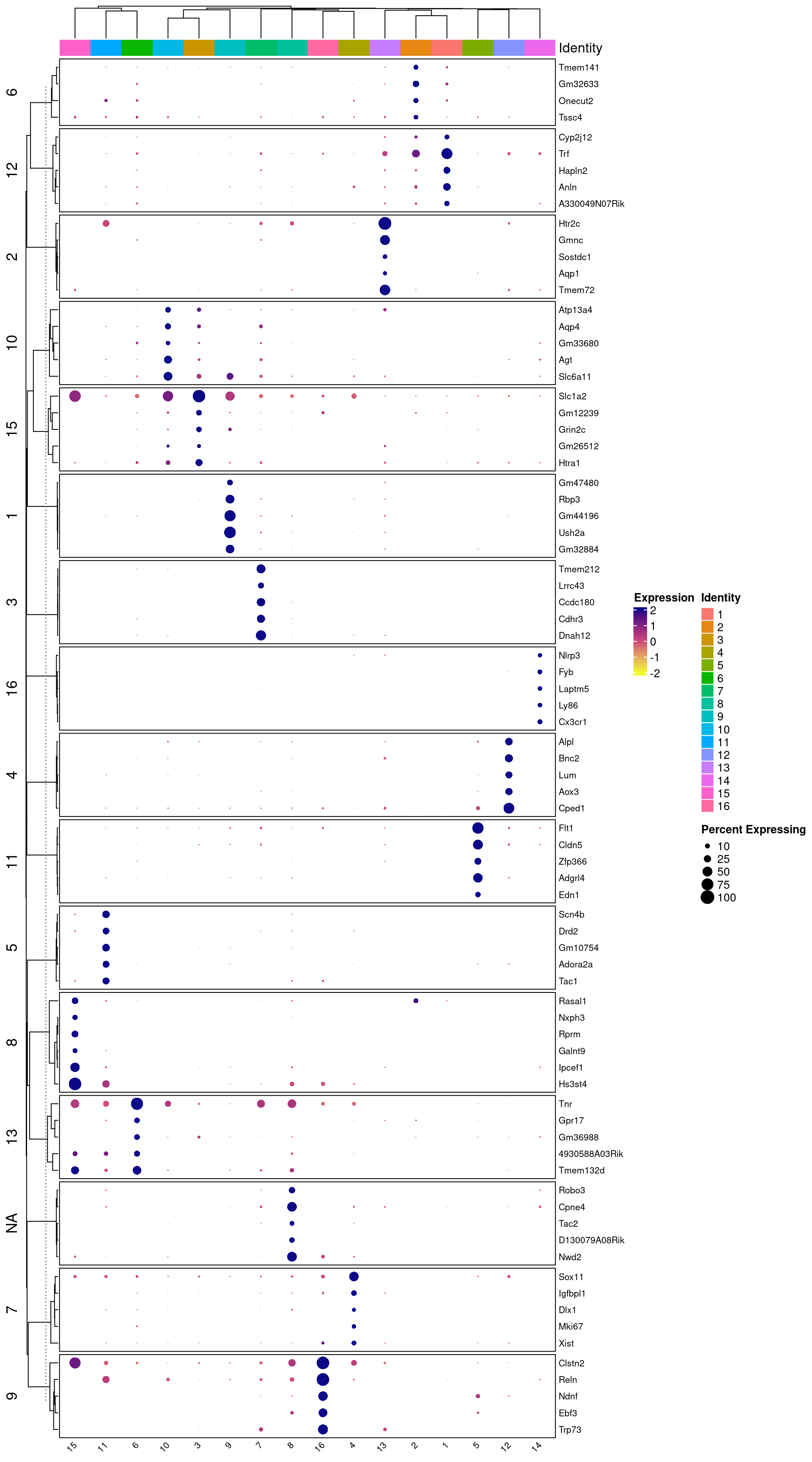
Heatmap
top10 <-
markers_MAST %>%
group_by(cluster) %>%
top_n(n = 10, wt = avg_log10FC)
DoHeatmap(combined_srt, features = top10$gene) + NoLegend()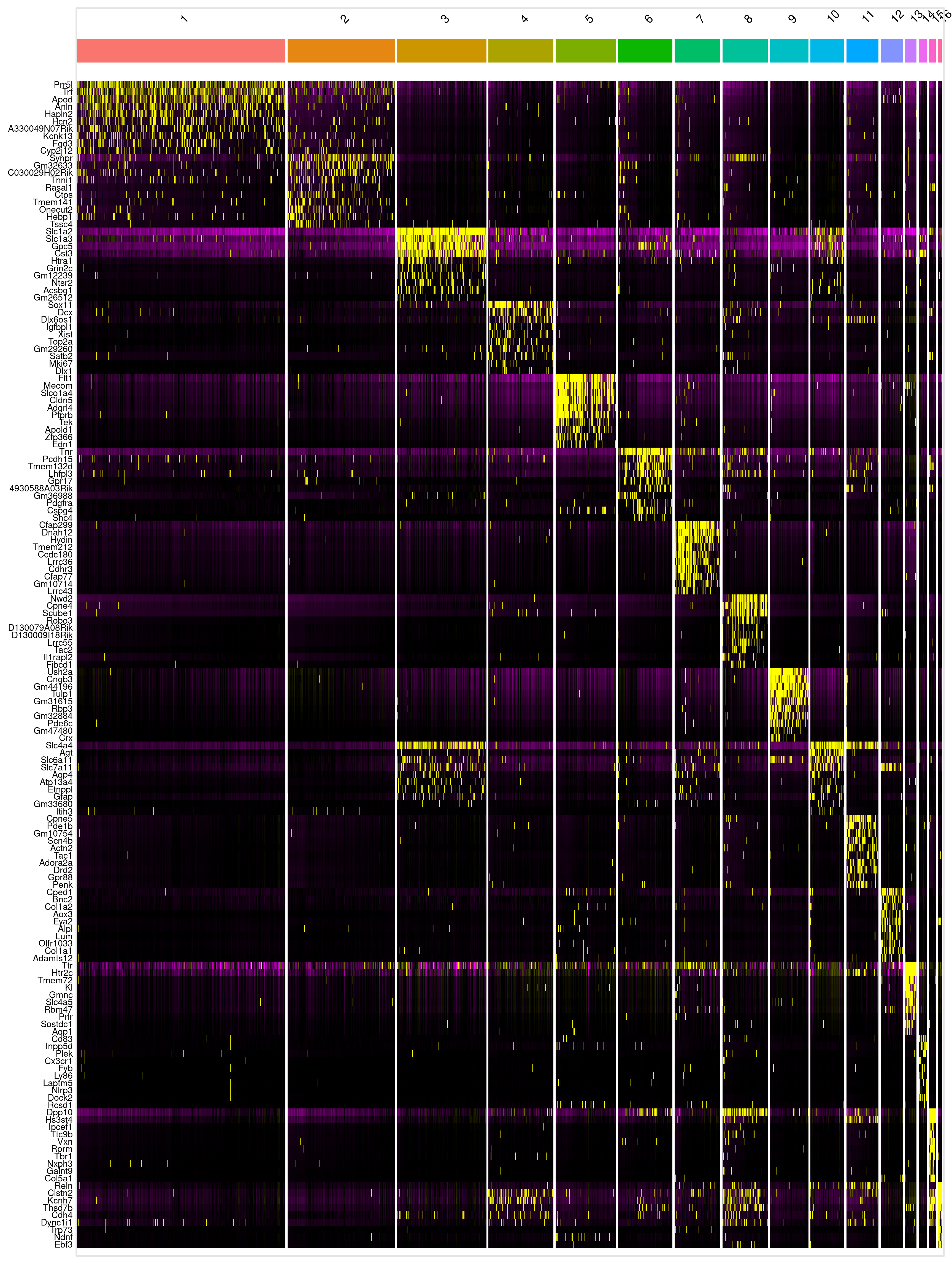
# Run PCA
merged_cortex <- RunPCA(merged_cortex, npcs = 20, verbose = FALSE)
# Set cell type annotations as identities if available
Idents(merged_cortex) <- "New_cellType"
ElbowPlot(merged_cortex, ndims = 20)
invisible(gc())
set.seed(reseed)
# registerDoParallel(cores = availableCores())
selected_pcs <-
seq_len(20)
if (!file.exists(here(data_dir, glue::glue("{project}-init/{project}-init-umap-search-ref.Rds")))) {
umap_example <- scDEED(
input_data = merged_cortex,
K = 20,
n_neighbors = seq(from = 15, to = 55, by = 20),
min.dist = c(0.01, 0.05, 0.1, 0.25, 0.5, 0.8),
reduction.method = "umap",
default_assay = "RNA"
)
dir.create(here(data_dir, sprintf("%s-init", project)))
readr::write_rds(
x = umap_example,
file = here(data_dir, glue::glue("{project}-init/{project}-init-umap-search-ref.Rds"))
)
} else {
umap_example <-
read_rds(here(data_dir, glue::glue("{project}-init/{project}-init-umap-search-ref.Rds")))
}invisible(gc())
set.seed(seed = reseed)
merged_cortex <-
merged_cortex |>
FindNeighbors(
dims = selected_pcs,
k.param = umap_example$num_dubious |>
dplyr::slice_min(
order_by = c(number_dubious_cells),
n = 1
) |>
dplyr::slice_min(
order_by = c(min.dist),
n = 1
) |>
pull(n_neighbors),
annoy.metric = "euclidean",
n.trees = 100,
verbose = FALSE
)
merged_cortex <-
merged_cortex |>
RunUMAP(
dims = selected_pcs,
reduction.name = "umap",
reduction.key = "UMAP_",
return.model = TRUE,
n.epochs = 1000L,
n.neighbors = umap_example$num_dubious |>
dplyr::slice_min(
order_by = c(number_dubious_cells),
n = 1
) |>
dplyr::slice_min(
order_by = c(min.dist),
n = 1
) |>
pull(n_neighbors),
min.dist = umap_example$num_dubious |>
dplyr::slice_min(
order_by = c(number_dubious_cells),
n = 1
) |>
dplyr::slice_min(
order_by = c(min.dist),
n = 1
) |>
pull(min.dist),
seed.use = reseed,
verbose = FALSE
)DefaultAssay(combined_srt) <- "RNA"
# Normalize the data
combined_srt <- NormalizeData(combined_srt, verbose = FALSE)
# Identify variable features
combined_srt <- FindVariableFeatures(combined_srt, selection.method = "vst", nfeatures = 5000, verbose = FALSE)
# Scale the data
combined_srt <- ScaleData(combined_srt, features = keep_genes, verbose = FALSE)# Find transfer anchors
anchors <- FindTransferAnchors(
reference = merged_cortex,
query = combined_srt,
dims = selected_pcs,
reference.reduction = "pca"
)
# Map the query data onto the reference UMAP and transfer cell type annotations
combined_srt <- MapQuery(
anchorset = anchors,
reference = merged_cortex,
query = combined_srt,
refdata = list(New_cellType = "New_cellType"), # Transfer cell type labels
reference.reduction = "pca",
reduction.model = "umap"
)
# The predicted cell types are stored in combined_srt$predicted.New_cellType
# The projected UMAP coordinates are in combined_srt[["ref.umap"]]# Plot the reference UMAP colored by cell types
p1 <- DimPlot_scCustom(
seurat_object = merged_cortex,
reduction = "umap",
group.by = "New_cellType",
pt.size = 1,
colors_use = merged_cortex@misc$types_Colour_Pal,
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + ggtitle("Reference: Cell Type Annotations")
# Plot the query cells projected onto the reference UMAP, colored by the predicted cell types
p2 <- DimPlot_scCustom(
seurat_object = combined_srt,
reduction = "ref.umap",
group.by = "predicted.New_cellType",
pt.size = 1,
colors_use = merged_cortex@misc$types_Colour_Pal,
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + NoLegend() + ggtitle("Query: Transferred Cell Type Labels")
# Combine the plots
p1 + p2 + plot_layout(guides = "collect")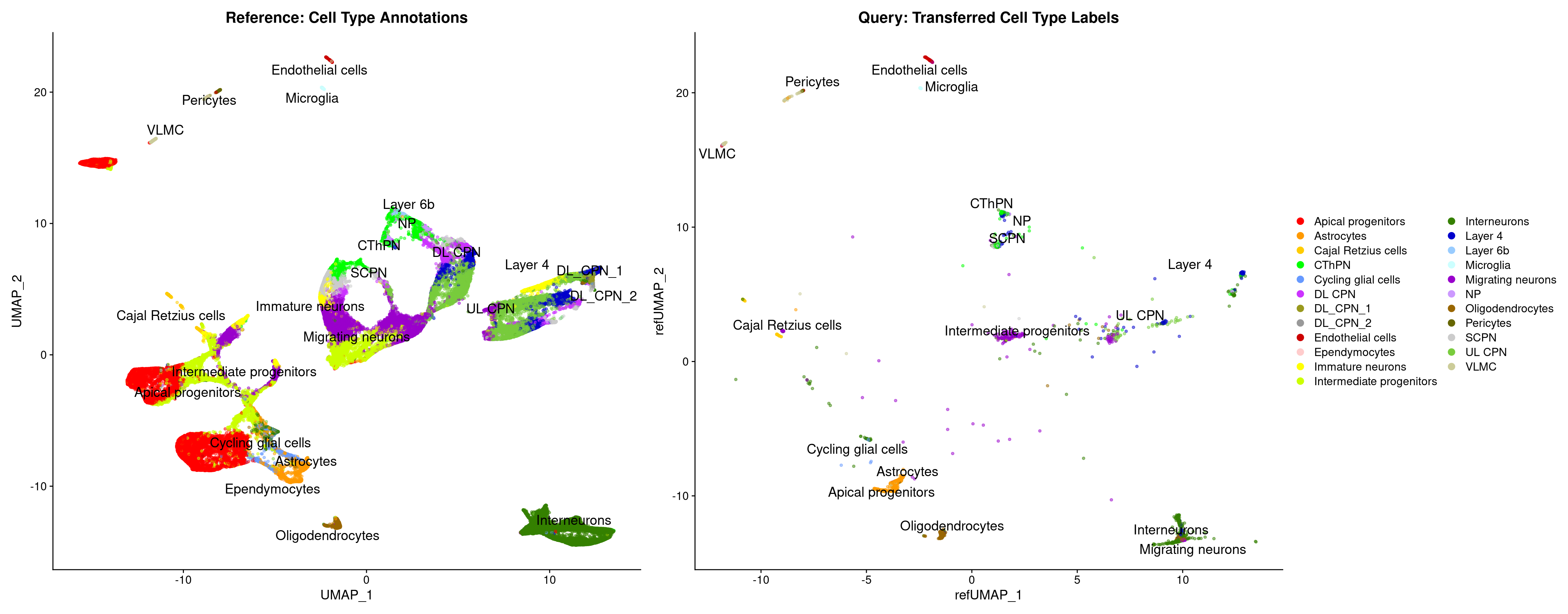
# Plot the projected UMAP, coloring by Scgn expression
FeaturePlot_scCustom(
seurat_object = merged_cortex,
features = c("Scgn"),
reduction = "umap",
split.by = "stage",
pt.size = 2,
alpha_exp = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5,
num_columns = 6
)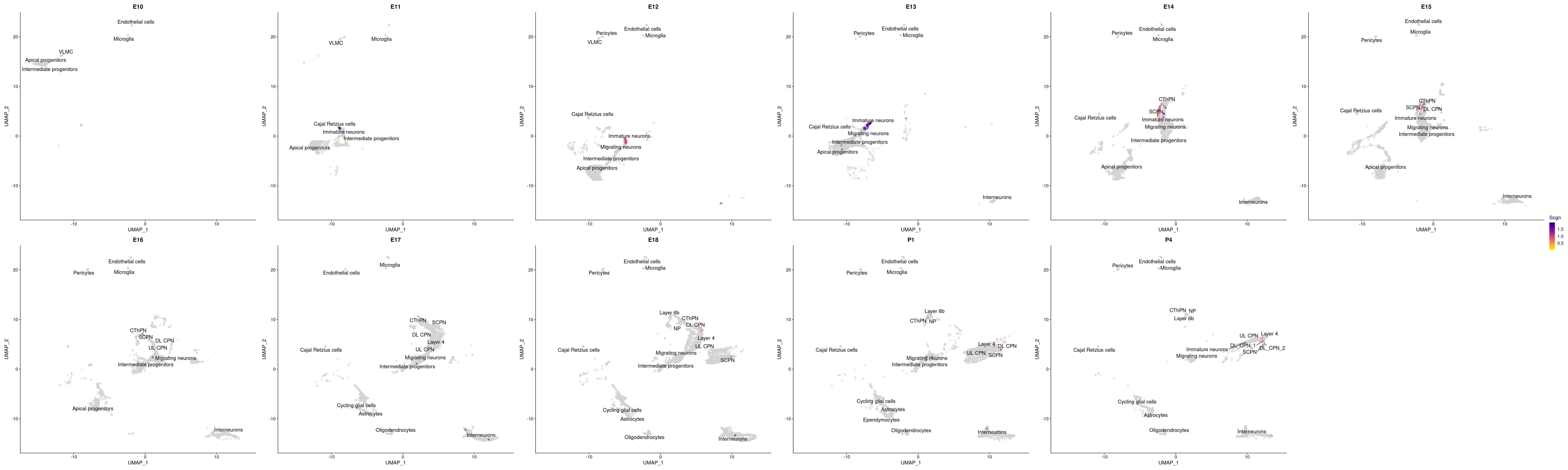
# Plot the projected UMAP, coloring by tdTomato status
DimPlot(
combined_srt,
reduction = "ref.umap",
group.by = "predicted.New_cellType",
split.by = "Scgn_tdTomato",
pt.size = 1,
cols = merged_cortex@misc$types_Colour_Pal,
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + ggtitle("tdTomato Status in Query Cells")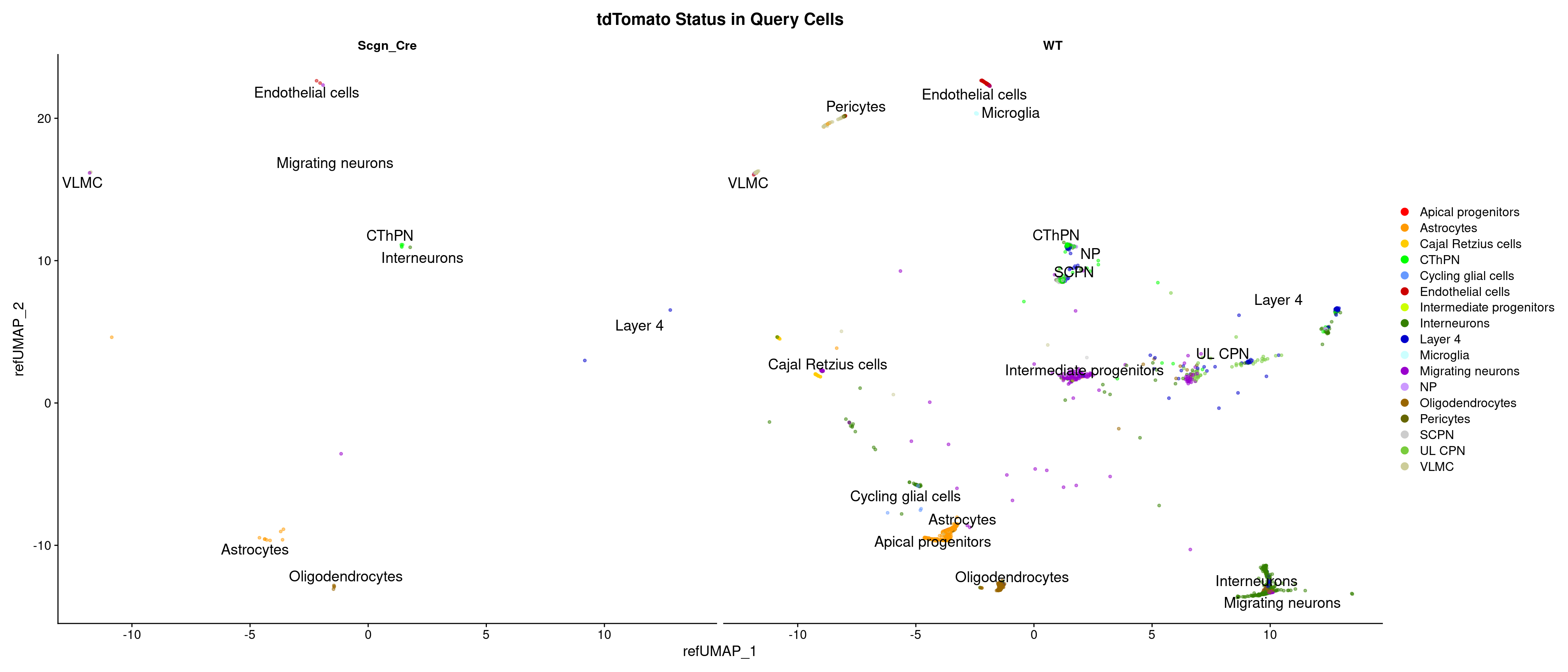
# Plot the projected UMAP, coloring by tdTomato status
DimPlot_scCustom(
seurat_object = combined_srt,
reduction = "ref.umap",
group.by = "k_tree",
split.by = "Scgn_tdTomato",
pt.size = 1,
colors_use = merged_cortex@misc$types_Colour_Pal,
shuffle = TRUE,
seed = reseed,
alpha = 0.5,
repel = TRUE,
label = TRUE,
label.size = 5
) + ggtitle("tdTomato Status in Query Cells")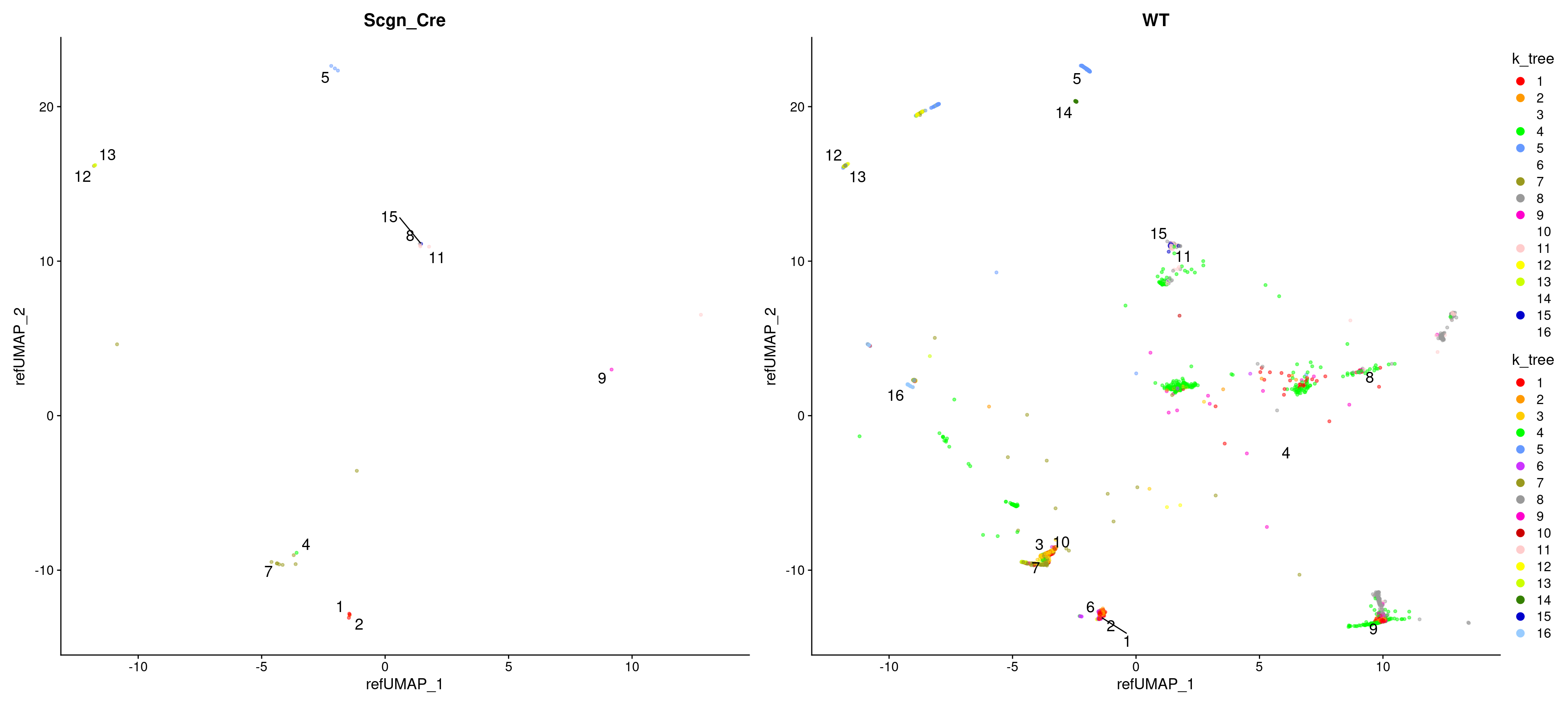
Idents(subset(combined_srt, subset = Scgn_tdTomato == "Scgn_Cre")) |> table()
1 2 4 5 7 8 9 11 12 13 15
3 1 1 3 10 1 1 4 1 1 1 subset(combined_srt, subset = Scgn_tdTomato == "Scgn_Cre")$k_tree |> table()
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
3 1 0 1 3 0 10 1 1 0 4 1 1 0 1 0 The most represented cluster’s markers are Mki67, Top2a, Dlx1, and Dlx6
Summary
Parameters
This table describes parameters used and set in this document.
params <- list(
list(
Parameter = "high_cutoff_umis",
Value = high_cutoff_umis,
Description = "Maximum threshold for total counts"
),
list(
Parameter = "low_cutoff_gene",
Value = low_cutoff_gene,
Description = "Minimum threshold for total features"
),
list(
Parameter = "high_cutoff_gene",
Value = high_cutoff_gene,
Description = "Maximum threshold for total features"
),
list(
Parameter = "high_cutoff_pc_mt",
Value = high_cutoff_pc_mt,
Description = "Maximum threshold for percentage counts mitochondrial"
),
list(
Parameter = "high_cutoff_pc_ribo",
Value = high_cutoff_pc_ribo,
Description = "Maximum threshold for percentage counts ribosomal"
),
list(
Parameter = "high_cutoff_pc_hb",
Value = high_cutoff_pc_hb,
Description = "Maximum threshold for percentage counts hemoglobin"
),
list(
Parameter = "high_cutoff_complexity",
Value = high_cutoff_complexity,
Description = "Maximum threshold for cells complexity"
),
list(
Parameter = "n_cells",
Value = ncol(combined_srt),
Description = "Number of cells in the filtered dataset"
),
list(
Parameter = "n_genes",
Value = nrow(combined_srt),
Description = "Number of genes in the filtered dataset"
),
list(
Parameter = "median_genes",
Value = median(Matrix::colSums(GetAssayData(
combined_srt,
slot = "counts", assay = "RNA"
) != 0)),
Description = paste(
"Median number of expressed genes per cell in the",
"filtered dataset"
)
),
list(
Parameter = "median_counts",
Value = median(Matrix::colSums(GetAssayData(
combined_srt,
slot = "counts", assay = "RNA"
))),
Description = paste(
"Median number of counts per cell in the filtered",
"dataset"
)
),
unlist(purrr::map(srr_set, n_cells_per_file))
)
params <- jsonlite::toJSON(params, pretty = TRUE)
knitr::kable(jsonlite::fromJSON(params))
|
|
|
|
|
|
|
|
|
|
|
|
Output files
This table describes the output files produced by this document. Right click and Save Link As… to download the results.
saveRDS(
object = combined_srt,
file = here(data_dir, sprintf("%s-whole_dataset-fpr_%s-clusters.Rds", project, cb_fpr))
)dir.create(here(tables_dir, docname), showWarnings = FALSE)
for (sample in unique(combined_srt@meta.data$Scgn_tdTomato)) {
cells <- combined_srt@meta.data %>%
as.data.frame() %>%
filter(Scgn_tdTomato == sample) %>%
select(cell_name)
readr::write_tsv(cells,
print(here(
tables_dir, docname,
glue::glue("cell_names-{sample}.tsv")
)),
col_names = FALSE
)
}readr::write_lines(params, here(tables_dir, docname, "parameters.json"))
knitr::kable(data.frame(
File = c(
get_download_link("parameters.json", here(tables_dir, docname)),
purrr::map_chr(
srr_set,
~ get_download_link(
file = sprintf("cell_names-%s.tsv", .x),
folder = here(tables_dir, docname)
)
),
get_download_link(sprintf(
"%s_all_mrk-logreg_sct-combined-whole_dataset-fpr_%s.csv",
project, cb_fpr
), here(tables_dir, docname)),
get_download_link(sprintf(
"%s_all_mrk-MAST_sct-combined-whole_dataset-fpr_%s.csv",
project, cb_fpr
), here(tables_dir, docname)),
get_download_link(sprintf(
"combined-top5_logreg-umap-whole_dataset-fpr_%s.pdf",
cb_fpr
), plots_dir),
get_download_link(sprintf(
"combined-top5_MAST-umap-whole_dataset-fpr_%s.pdf",
cb_fpr
), plots_dir)
),
Description = c(
"Parameters set and used in this analysis",
purrr::map_chr(srr_set, ~ sprintf("cell_names-%s.tsv", .x)),
"DGE with logreg test",
"DGE with MAST test",
"UMAP embeddings of top5 genes per cluster from logreg test",
"UMAP embeddings of top5 genes per cluster from MAST test"
)
))| File | Description |
|---|---|
| parameters.json | Parameters set and used in this analysis |
| cell_names-BSF_1105_Mouse_Cortex_SCGN_P02_1.tsv | cell_names-BSF_1105_Mouse_Cortex_SCGN_P02_1.tsv |
| hanics2024-cortex-tdtomato_all_mrk-logreg_sct-combined-whole_dataset-fpr_0.001.csv | DGE with logreg test |
| hanics2024-cortex-tdtomato_all_mrk-MAST_sct-combined-whole_dataset-fpr_0.001.csv | DGE with MAST test |
| combined-top5_logreg-umap-whole_dataset-fpr_0.001.pdf | UMAP embeddings of top5 genes per cluster from logreg test |
| combined-top5_MAST-umap-whole_dataset-fpr_0.001.pdf | UMAP embeddings of top5 genes per cluster from MAST test |
Session information
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.4.1 (2024-06-14)
os Ubuntu 22.04.5 LTS
system x86_64, linux-gnu
ui X11
language en_US:en
collate en_US.UTF-8
ctype en_US.UTF-8
tz Etc/UTC
date 2024-11-28
pandoc 2.9.2.1 @ /usr/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
abind 1.4-8 2024-09-12 [1] RSPM
ape 5.8 2024-04-11 [1] RSPM (R 4.4.1)
aplot 0.2.3 2024-06-17 [1] RSPM (R 4.4.0)
backports 1.5.0 2024-05-23 [1] RSPM
base64enc 0.1-3 2015-07-28 [1] RSPM (R 4.4.1)
beeswarm 0.4.0 2021-06-01 [1] RSPM (R 4.4.0)
Biobase 2.64.0 2024-04-30 [1] RSPM (R 4.4.1)
BiocGenerics 0.50.0 2024-04-30 [1] RSPM (R 4.4.1)
BiocManager 1.30.25 2024-08-28 [1] RSPM (R 4.4.0)
bit 4.5.0 2024-09-20 [1] RSPM
bit64 4.5.2 2024-09-22 [1] RSPM
bitops 1.0-9 2024-10-03 [1] RSPM
bslib 0.8.0 2024-07-29 [1] RSPM (R 4.4.1)
cachem 1.1.0 2024-05-16 [1] RSPM (R 4.4.1)
Cairo 1.6-2 2023-11-28 [1] RSPM
callr 3.7.6 2024-03-25 [1] RSPM (R 4.4.1)
checkmate 2.3.2 2024-07-29 [1] RSPM (R 4.4.0)
circlize 0.4.16 2024-10-12 [1] Github (jokergoo/circlize@9b21578)
cli 3.6.3 2024-06-21 [1] RSPM (R 4.4.1)
clue 0.3-65 2023-09-23 [1] RSPM (R 4.4.0)
cluster 2.1.6 2023-12-01 [1] CRAN (R 4.4.1)
clusterGeneration 1.3.8 2023-08-16 [1] RSPM (R 4.4.0)
clustree * 0.5.1 2023-11-05 [1] RSPM (R 4.4.0)
coda 0.19-4.1 2024-01-31 [1] RSPM
codetools 0.2-20 2024-03-31 [1] CRAN (R 4.4.1)
colorspace 2.1-1 2024-07-26 [1] RSPM (R 4.4.1)
combinat 0.0-8 2012-10-29 [1] RSPM (R 4.4.0)
ComplexHeatmap 2.18.0 2023-10-24 [1] RSPM (R 4.4.1)
cowplot 1.1.3 2024-01-22 [1] RSPM
crayon 1.5.3 2024-06-20 [1] RSPM (R 4.4.1)
data.table 1.16.2 2024-10-10 [1] RSPM (R 4.4.1)
data.tree 1.1.0 2023-11-12 [1] RSPM (R 4.4.0)
DelayedArray 0.28.0 2023-10-24 [1] RSPM (R 4.4.1)
deldir 2.0-4 2024-02-28 [1] RSPM
dendextend 1.18.0 2024-10-05 [1] RSPM (R 4.4.0)
DEoptim 2.2-8 2022-11-11 [1] RSPM (R 4.4.0)
devtools 2.4.5 2022-10-11 [1] RSPM
digest 0.6.37 2024-08-19 [1] RSPM (R 4.4.1)
doParallel * 1.0.17 2022-02-07 [1] RSPM (R 4.4.1)
dotCall64 1.2 2024-10-04 [1] RSPM
dplyr * 1.1.4 2023-11-17 [1] RSPM (R 4.4.1)
ellipsis 0.3.2 2021-04-29 [1] RSPM
evaluate 1.0.1 2024-10-10 [1] RSPM (R 4.4.1)
expm 1.0-0 2024-08-19 [1] RSPM (R 4.4.0)
fansi 1.0.6 2023-12-08 [1] RSPM (R 4.4.1)
farver 2.1.2 2024-05-13 [1] RSPM (R 4.4.1)
fastDummies 1.7.4 2024-08-16 [1] RSPM
fastmap 1.2.0 2024-05-15 [1] RSPM (R 4.4.1)
fastmatch 1.1-4 2023-08-18 [1] RSPM (R 4.4.0)
fitdistrplus 1.2-1 2024-07-12 [1] RSPM
forcats * 1.0.0 2023-01-29 [1] RSPM
foreach * 1.5.2 2022-02-02 [1] RSPM (R 4.4.1)
fs 1.6.4 2024-04-25 [1] RSPM (R 4.4.1)
future * 1.34.0 2024-07-29 [1] RSPM
future.apply 1.11.2 2024-03-28 [1] RSPM
generics 0.1.3 2022-07-05 [1] RSPM (R 4.4.1)
GenomeInfoDb 1.40.1 2024-05-24 [1] RSPM (R 4.4.1)
GenomeInfoDbData 1.2.11 2024-10-12 [1] RSPM (R 4.4.1)
GenomicRanges 1.56.1 2024-06-12 [1] RSPM (R 4.4.1)
GetoptLong 1.0.5 2020-12-15 [1] RSPM (R 4.4.0)
getPass 0.2-4 2023-12-10 [1] RSPM
ggbeeswarm 0.7.2 2024-10-12 [1] Github (eclarke/ggbeeswarm@ce2da8a)
ggforce 0.5.0 2024-10-12 [1] Github (thomasp85/ggforce@9292822)
ggfun 0.1.6 2024-08-28 [1] RSPM (R 4.4.0)
ggh4x 0.2.8.9000 2024-10-12 [1] Github (teunbrand/ggh4x@e797c7c)
ggimage 0.3.3 2023-06-19 [1] RSPM (R 4.4.0)
ggmin 0.0.0.9000 2024-10-12 [1] Github (sjessa/ggmin@8ada274)
ggplot2 * 3.5.1 2024-04-23 [1] RSPM (R 4.4.1)
ggplotify 0.1.2 2023-08-09 [1] RSPM (R 4.4.0)
ggprism 1.0.5 2024-10-12 [1] Github (csdaw/ggprism@b6e6c0e)
ggraph * 2.2.1.9000 2024-10-12 [1] Github (thomasp85/ggraph@9a0bfb1)
ggrastr 1.0.2 2024-10-12 [1] Github (VPetukhov/ggrastr@50ca3e0)
ggrepel 0.9.6 2024-10-12 [1] Github (slowkow/ggrepel@e94776b)
ggridges 0.5.6 2024-01-23 [1] RSPM
ggsci 3.2.0 2024-10-12 [1] Github (nanxstats/ggsci@b5bf1fd)
ggtree 3.13.1 2024-10-12 [1] Github (YuLab-SMU/ggtree@01b29ef)
git2r 0.33.0 2023-11-26 [1] RSPM
glmGamPoi * 1.16.0 2024-04-30 [1] RSPM (R 4.4.1)
GlobalOptions 0.1.2 2020-06-10 [1] RSPM (R 4.4.0)
globals 0.16.3 2024-03-08 [1] RSPM
glue 1.8.0 2024-09-30 [1] RSPM (R 4.4.1)
goftest 1.2-3 2021-10-07 [1] RSPM
gprofiler2 * 0.2.3 2024-02-23 [1] RSPM (R 4.4.0)
graphlayouts 1.2.0 2024-09-24 [1] RSPM (R 4.4.0)
gridExtra 2.3 2017-09-09 [1] RSPM
gridGraphics 0.5-1 2020-12-13 [1] RSPM (R 4.4.0)
gtable 0.3.5 2024-04-22 [1] RSPM (R 4.4.1)
hdf5r 1.3.11 2024-07-07 [1] RSPM (R 4.4.1)
here * 1.0.1 2020-12-13 [1] RSPM
highr 0.11 2024-05-26 [1] RSPM (R 4.4.1)
hms 1.1.3 2023-03-21 [1] RSPM
htmltools 0.5.8.1 2024-04-04 [1] RSPM (R 4.4.1)
htmlwidgets 1.6.4 2023-12-06 [1] RSPM (R 4.4.1)
httpuv 1.6.15 2024-03-26 [1] RSPM (R 4.4.1)
httr 1.4.7 2023-08-15 [1] RSPM (R 4.4.1)
ica 1.0-3 2022-07-08 [1] RSPM
igraph 2.0.3 2024-03-13 [1] RSPM
IRanges 2.38.1 2024-07-03 [1] RSPM (R 4.4.1)
irlba 2.3.5.1 2022-10-03 [1] RSPM
iterators * 1.0.14 2022-02-05 [1] RSPM (R 4.4.1)
janitor 2.2.0.9000 2024-10-12 [1] Github (sfirke/janitor@709b2ab)
jquerylib 0.1.4 2021-04-26 [1] RSPM (R 4.4.1)
jsonlite 1.8.9 2024-09-20 [1] RSPM (R 4.4.1)
kableExtra * 1.4.0 2024-01-24 [1] RSPM (R 4.4.0)
KernSmooth 2.23-24 2024-05-17 [1] CRAN (R 4.4.1)
knitr * 1.48 2024-07-07 [1] RSPM
ks 1.14.2 2024-01-15 [1] RSPM (R 4.4.0)
labeling 0.4.3 2023-08-29 [1] RSPM (R 4.4.1)
later 1.3.2 2023-12-06 [1] RSPM (R 4.4.1)
lattice 0.22-6 2024-03-20 [1] CRAN (R 4.4.1)
lazyeval 0.2.2 2019-03-15 [1] RSPM (R 4.4.1)
leiden 0.4.3.1 2023-11-17 [1] RSPM
lifecycle 1.0.4 2023-11-07 [1] RSPM (R 4.4.1)
listenv 0.9.1 2024-01-29 [1] RSPM
lmtest 0.9-40 2022-03-21 [1] RSPM (R 4.4.1)
lubridate * 1.9.3 2023-09-27 [1] RSPM
magick 2.8.5 2024-09-20 [1] RSPM
magrittr * 2.0.3 2022-03-30 [1] RSPM (R 4.4.1)
maps 3.4.2 2023-12-15 [1] RSPM (R 4.4.0)
MASS 7.3-60.2 2024-04-26 [1] CRAN (R 4.4.1)
MAST 1.30.0 2024-04-30 [1] RSPM (R 4.4.1)
Matrix 1.7-0 2024-04-26 [1] CRAN (R 4.4.1)
MatrixGenerics 1.14.0 2023-10-24 [1] RSPM (R 4.4.1)
matrixStats 1.4.1 2024-09-08 [1] RSPM
mclust 6.1.1 2024-04-29 [1] RSPM
memoise 2.0.1 2021-11-26 [1] RSPM (R 4.4.1)
mime 0.12 2021-09-28 [1] RSPM (R 4.4.1)
miniUI 0.1.1.1 2018-05-18 [1] RSPM
mnormt 2.1.1 2022-09-26 [1] RSPM (R 4.4.1)
mrtree * 0.0.0.9000 2024-10-12 [1] Github (pengminshi/mrtree@720f469)
munsell 0.5.1 2024-04-01 [1] RSPM (R 4.4.1)
mvtnorm 1.3-1 2024-09-03 [1] RSPM
Nebulosa * 1.14.0 2024-04-30 [1] RSPM (R 4.4.1)
nlme 3.1-166 2024-08-14 [1] RSPM
numDeriv 2016.8-1.1 2019-06-06 [1] RSPM (R 4.4.1)
optimParallel 1.0-2 2021-02-11 [1] RSPM (R 4.4.0)
paletteer 1.6.0 2024-01-21 [1] RSPM
parallelly 1.38.0 2024-07-27 [1] RSPM
patchwork * 1.3.0.9000 2024-10-12 [1] Github (thomasp85/patchwork@2695a9f)
pbapply 1.7-2 2023-06-27 [1] RSPM
phangorn 2.12.1 2024-09-17 [1] RSPM (R 4.4.0)
phytools 2.3-0 2024-06-13 [1] RSPM (R 4.4.0)
pillar 1.9.0 2023-03-22 [1] RSPM (R 4.4.1)
pkgbuild 1.4.4 2024-03-17 [1] RSPM (R 4.4.1)
pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.4.1)
pkgload 1.4.0 2024-06-28 [1] RSPM (R 4.4.1)
plotly 4.10.4 2024-01-13 [1] RSPM
plyr 1.8.9 2023-10-02 [1] RSPM
png 0.1-8 2022-11-29 [1] RSPM
polyclip 1.10-7 2024-07-23 [1] RSPM
pracma 2.4.4 2023-11-10 [1] RSPM (R 4.4.0)
prettyunits 1.2.0 2023-09-24 [1] RSPM
prismatic 1.1.2 2024-04-10 [1] RSPM
processx 3.8.4 2024-03-16 [1] RSPM
profvis 0.4.0 2024-09-20 [1] RSPM
progress 1.2.3 2023-12-06 [1] RSPM (R 4.4.0)
progressr 0.14.0 2023-08-10 [1] RSPM
promises 1.3.0 2024-04-05 [1] RSPM (R 4.4.1)
proxy 0.4-27 2022-06-09 [1] RSPM (R 4.4.0)
ps 1.8.0 2024-09-12 [1] RSPM (R 4.4.1)
purrr * 1.0.2 2023-08-10 [1] RSPM (R 4.4.1)
qs * 0.27.2 2024-10-01 [1] RSPM (R 4.4.0)
quadprog 1.5-8 2019-11-20 [1] RSPM
R.methodsS3 1.8.2 2022-06-13 [1] RSPM (R 4.4.0)
R.oo 1.26.0 2024-01-24 [1] RSPM (R 4.4.0)
R.utils 2.12.3 2023-11-18 [1] RSPM (R 4.4.0)
R6 2.5.1 2021-08-19 [1] RSPM (R 4.4.1)
RANN 2.6.2 2024-08-25 [1] RSPM
RApiSerialize 0.1.4 2024-09-28 [1] RSPM (R 4.4.0)
RColorBrewer * 1.1-3 2022-04-03 [1] RSPM
Rcpp 1.0.13 2024-07-17 [1] RSPM (R 4.4.1)
RcppAnnoy 0.0.22 2024-01-23 [1] RSPM
RcppHNSW 0.6.0 2024-02-04 [1] RSPM
RcppParallel 5.1.9 2024-08-19 [1] RSPM (R 4.4.0)
RCurl 1.98-1.16 2024-07-11 [1] RSPM
readr * 2.1.5 2024-01-10 [1] RSPM
rematch2 2.1.2 2020-05-01 [1] RSPM (R 4.4.1)
remotes 2.5.0 2024-03-17 [1] RSPM
repr 1.1.7 2024-03-22 [1] RSPM
reshape2 1.4.4 2020-04-09 [1] RSPM
reticulate * 1.39.0 2024-09-05 [1] RSPM
rjson 0.2.23 2024-09-16 [1] RSPM (R 4.4.0)
rlang 1.1.4 2024-06-04 [1] RSPM (R 4.4.1)
rmarkdown 2.28 2024-08-17 [1] RSPM
ROCR 1.0-11 2020-05-02 [1] RSPM
rprojroot 2.0.4 2023-11-05 [1] RSPM (R 4.4.1)
RSpectra 0.16-2 2024-07-18 [1] RSPM
rstudioapi 0.16.0 2024-03-24 [1] RSPM
rsvd 1.0.5 2021-04-16 [1] RSPM (R 4.4.0)
Rtsne 0.17 2023-12-07 [1] RSPM
S4Arrays 1.2.1 2024-03-04 [1] RSPM (R 4.4.1)
S4Vectors 0.40.2 2023-11-23 [1] RSPM (R 4.4.1)
sass 0.4.9 2024-03-15 [1] RSPM (R 4.4.1)
scales 1.3.0 2023-11-28 [1] RSPM (R 4.4.1)
scattermore 1.2 2023-06-12 [1] RSPM
scatterplot3d 0.3-44 2023-05-05 [1] RSPM (R 4.4.0)
scBubbletree * 1.6.0 2024-04-30 [1] RSPM (R 4.4.1)
scCustomize * 2.1.2 2024-10-12 [1] Github (samuel-marsh/scCustomize@fc7a282)
scDEED * 0.1.0 2024-10-12 [1] Github (JSB-UCLA/scDEED@25282e8)
sctransform * 0.4.1 2023-10-19 [1] RSPM
sessioninfo 1.2.2 2021-12-06 [1] RSPM
Seurat * 5.1.0.9006 2024-10-14 [1] Github (satijalab/seurat@63a7b1a)
SeuratDisk * 0.0.0.9021 2024-10-12 [1] Github (mojaveazure/seurat-disk@877d4e1)
SeuratObject * 5.0.99.9001 2024-10-14 [1] Github (satijalab/seurat-object@1a140c7)
SeuratWrappers * 0.3.5 2024-10-12 [1] Github (satijalab/seurat-wrappers@8d46d6c)
shape 1.4.6.1 2024-02-23 [1] RSPM
shiny 1.9.1 2024-08-01 [1] RSPM (R 4.4.1)
SingleCellExperiment 1.24.0 2023-10-24 [1] RSPM (R 4.4.1)
skimr * 2.1.5 2024-10-12 [1] Github (ropensci/skimr@d5126aa)
snakecase 0.11.1 2023-08-27 [1] RSPM (R 4.4.0)
sp * 2.1-4 2024-04-30 [1] RSPM
spam 2.11-0 2024-10-03 [1] RSPM
SparseArray 1.2.4 2024-02-11 [1] RSPM (R 4.4.1)
spatstat.data 3.1-2 2024-06-21 [1] RSPM
spatstat.explore 3.3-2 2024-08-21 [1] RSPM
spatstat.geom 3.3-3 2024-09-18 [1] RSPM
spatstat.random 3.3-2 2024-09-18 [1] RSPM
spatstat.sparse 3.1-0 2024-06-21 [1] RSPM
spatstat.univar 3.0-1 2024-09-05 [1] RSPM
spatstat.utils 3.1-0 2024-08-17 [1] RSPM
stringfish 0.16.0 2023-11-28 [1] RSPM (R 4.4.0)
stringi 1.8.4 2024-05-06 [1] RSPM (R 4.4.1)
stringr * 1.5.1 2023-11-14 [1] RSPM (R 4.4.1)
SummarizedExperiment 1.32.0 2023-10-24 [1] RSPM (R 4.4.1)
survival 3.7-0 2024-06-05 [1] RSPM
svglite 2.1.3 2023-12-08 [1] RSPM
SymSim 0.0.0.9000 2024-10-12 [1] Github (YosefLab/SymSim@76a674b)
systemfonts 1.1.0 2024-05-15 [1] RSPM
tensor 1.5 2012-05-05 [1] RSPM
tibble * 3.2.1 2023-03-20 [1] RSPM (R 4.4.1)
tidygraph 1.3.1 2024-01-30 [1] RSPM
tidyr * 1.3.1 2024-01-24 [1] RSPM (R 4.4.1)
tidyselect 1.2.1 2024-03-11 [1] RSPM (R 4.4.1)
tidytree 0.4.6 2023-12-12 [1] RSPM (R 4.4.0)
tidyverse * 2.0.0.9000 2024-10-12 [1] Github (tidyverse/tidyverse@62f32d4)
timechange 0.3.0 2024-01-18 [1] RSPM
treeio 1.26.0 2023-10-24 [1] RSPM (R 4.4.1)
tweenr 2.0.3 2024-02-26 [1] RSPM (R 4.4.0)
tzdb 0.4.0 2023-05-12 [1] RSPM
UCSC.utils 1.0.0 2024-04-30 [1] RSPM (R 4.4.1)
urlchecker 1.0.1 2021-11-30 [1] RSPM
usethis 3.0.0 2024-07-29 [1] RSPM
utf8 1.2.4 2023-10-22 [1] RSPM (R 4.4.1)
uwot 0.2.2 2024-04-21 [1] RSPM
vctrs 0.6.5 2023-12-01 [1] RSPM (R 4.4.1)
vipor 0.4.7 2023-12-18 [1] RSPM (R 4.4.0)
viridis * 0.6.5 2024-01-29 [1] RSPM
viridisLite * 0.4.2 2023-05-02 [1] RSPM (R 4.4.1)
vroom 1.6.5 2023-12-05 [1] RSPM
whisker 0.4.1 2022-12-05 [1] RSPM
withr 3.0.1 2024-07-31 [1] RSPM (R 4.4.1)
workflowr * 1.7.1 2023-08-23 [1] RSPM
xfun 0.48 2024-10-03 [1] RSPM (R 4.4.1)
xml2 1.3.6 2023-12-04 [1] RSPM (R 4.4.1)
xtable 1.8-4 2019-04-21 [1] RSPM (R 4.4.1)
XVector 0.42.0 2023-10-24 [1] RSPM (R 4.4.1)
yaml 2.3.10 2024-07-26 [1] RSPM
yulab.utils 0.1.7 2024-08-26 [1] RSPM (R 4.4.0)
zeallot * 0.1.0 2018-01-28 [1] RSPM
zlibbioc 1.48.2 2024-03-13 [1] RSPM (R 4.4.1)
zoo 1.8-12 2023-04-13 [1] RSPM (R 4.4.1)
[1] /opt/R/4.4.1/lib/R/library
─ Python configuration ───────────────────────────────────────────────────────
python: /opt/python/3.12/bin/python
libpython: /opt/python/3.12/lib/libpython3.12.so
pythonhome: /opt/python/3.12:/opt/python/3.12
version: 3.12.7 | packaged by conda-forge | (main, Oct 4 2024, 16:05:46) [GCC 13.3.0]
numpy: /opt/python/3.12/lib/python3.12/site-packages/numpy
numpy_version: 1.26.4
pacmap: /opt/python/3.12/lib/python3.12/site-packages/pacmap
NOTE: Python version was forced by RETICULATE_PYTHON
──────────────────────────────────────────────────────────────────────────────Di Bella, Daniela J., Ehsan Habibi, Robert R. Stickels, Gabriele Scalia, Juliana Brown, Payman Yadollahpour, Sung Min Yang, et al. 2021. “Molecular Logic of Cellular Diversification in the Mouse Cerebral Cortex.” Nature 595 (7868): 554–59. https://doi.org/10.1038/s41586-021-03670-5.
sessionInfo()R version 4.4.1 (2024-06-14)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.5 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] scBubbletree_1.6.0 gprofiler2_0.2.3 Nebulosa_1.14.0
[4] scDEED_0.1.0 mrtree_0.0.0.9000 scCustomize_2.1.2
[7] qs_0.27.2 patchwork_1.3.0.9000 clustree_0.5.1
[10] ggraph_2.2.1.9000 glmGamPoi_1.16.0 sctransform_0.4.1
[13] SeuratDisk_0.0.0.9021 SeuratWrappers_0.3.5 Seurat_5.1.0.9006
[16] SeuratObject_5.0.99.9001 sp_2.1-4 doParallel_1.0.17
[19] iterators_1.0.14 foreach_1.5.2 reticulate_1.39.0
[22] kableExtra_1.4.0 zeallot_0.1.0 future_1.34.0
[25] skimr_2.1.5 magrittr_2.0.3 lubridate_1.9.3
[28] forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4
[31] purrr_1.0.2 readr_2.1.5 tidyr_1.3.1
[34] tibble_3.2.1 ggplot2_3.5.1 tidyverse_2.0.0.9000
[37] viridis_0.6.5 viridisLite_0.4.2 RColorBrewer_1.1-3
[40] knitr_1.48 here_1.0.1 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] IRanges_2.38.1 R.methodsS3_1.8.2
[3] vroom_1.6.5 progress_1.2.3
[5] urlchecker_1.0.1 goftest_1.2-3
[7] phytools_2.3-0 vctrs_0.6.5
[9] spatstat.random_3.3-2 RApiSerialize_0.1.4
[11] proxy_0.4-27 digest_0.6.37
[13] png_0.1-8 shape_1.4.6.1
[15] git2r_0.33.0 ggrepel_0.9.6
[17] deldir_2.0-4 parallelly_1.38.0
[19] combinat_0.0-8 magick_2.8.5
[21] MASS_7.3-60.2 MAST_1.30.0
[23] reshape2_1.4.4 httpuv_1.6.15
[25] BiocGenerics_0.50.0 withr_3.0.1
[27] ggrastr_1.0.2 xfun_0.48
[29] ggfun_0.1.6 ellipsis_0.3.2
[31] survival_3.7-0 memoise_2.0.1
[33] ggbeeswarm_0.7.2 janitor_2.2.0.9000
[35] profvis_0.4.0 ggsci_3.2.0
[37] systemfonts_1.1.0 tidytree_0.4.6
[39] zoo_1.8-12 GlobalOptions_0.1.2
[41] DEoptim_2.2-8 pbapply_1.7-2
[43] R.oo_1.26.0 prettyunits_1.2.0
[45] rematch2_2.1.2 promises_1.3.0
[47] scatterplot3d_0.3-44 httr_1.4.7
[49] globals_0.16.3 fitdistrplus_1.2-1
[51] ps_1.8.0 stringfish_0.16.0
[53] rstudioapi_0.16.0 UCSC.utils_1.0.0
[55] miniUI_0.1.1.1 generics_0.1.3
[57] base64enc_0.1-3 processx_3.8.4
[59] S4Vectors_0.40.2 repr_1.1.7
[61] zlibbioc_1.48.2 polyclip_1.10-7
[63] GenomeInfoDbData_1.2.11 quadprog_1.5-8
[65] SparseArray_1.2.4 pracma_2.4.4
[67] xtable_1.8-4 evaluate_1.0.1
[69] S4Arrays_1.2.1 hms_1.1.3
[71] GenomicRanges_1.56.1 irlba_2.3.5.1
[73] colorspace_2.1-1 hdf5r_1.3.11
[75] ROCR_1.0-11 spatstat.data_3.1-2
[77] lmtest_0.9-40 snakecase_0.11.1
[79] later_1.3.2 ggtree_3.13.1
[81] lattice_0.22-6 spatstat.geom_3.3-3
[83] future.apply_1.11.2 getPass_0.2-4
[85] scattermore_1.2 cowplot_1.1.3
[87] matrixStats_1.4.1 RcppAnnoy_0.0.22
[89] pillar_1.9.0 nlme_3.1-166
[91] compiler_4.4.1 RSpectra_0.16-2
[93] stringi_1.8.4 devtools_2.4.5
[95] tensor_1.5 SummarizedExperiment_1.32.0
[97] dendextend_1.18.0 plyr_1.8.9
[99] crayon_1.5.3 abind_1.4-8
[101] gridGraphics_0.5-1 graphlayouts_1.2.0
[103] bit_4.5.0 fastmatch_1.1-4
[105] whisker_0.4.1 codetools_0.2-20
[107] bslib_0.8.0 paletteer_1.6.0
[109] GetoptLong_1.0.5 plotly_4.10.4
[111] mime_0.12 splines_4.4.1
[113] circlize_0.4.16 Rcpp_1.0.13
[115] fastDummies_1.7.4 prismatic_1.1.2
[117] utf8_1.2.4 clue_0.3-65
[119] fs_1.6.4 listenv_0.9.1
[121] checkmate_2.3.2 pkgbuild_1.4.4
[123] expm_1.0-0 ggplotify_0.1.2
[125] Matrix_1.7-0 callr_3.7.6
[127] tzdb_0.4.0 svglite_2.1.3
[129] tweenr_2.0.3 pkgconfig_2.0.3
[131] tools_4.4.1 cachem_1.1.0
[133] numDeriv_2016.8-1.1 fastmap_1.2.0
[135] rmarkdown_2.28 scales_1.3.0
[137] grid_4.4.1 usethis_3.0.0
[139] ica_1.0-3 sass_0.4.9
[141] coda_0.19-4.1 ggprism_1.0.5
[143] BiocManager_1.30.25 dotCall64_1.2
[145] RANN_2.6.2 ggimage_0.3.3
[147] farver_2.1.2 tidygraph_1.3.1
[149] yaml_2.3.10 MatrixGenerics_1.14.0
[151] cli_3.6.3 stats4_4.4.1
[153] leiden_0.4.3.1 lifecycle_1.0.4
[155] uwot_0.2.2 Biobase_2.64.0
[157] mvtnorm_1.3-1 sessioninfo_1.2.2
[159] backports_1.5.0 rjson_0.2.23
[161] timechange_0.3.0 gtable_0.3.5
[163] ggridges_0.5.6 progressr_0.14.0
[165] ape_5.8 jsonlite_1.8.9
[167] RcppHNSW_0.6.0 bitops_1.0-9
[169] bit64_4.5.2 Rtsne_0.17
[171] yulab.utils_0.1.7 spatstat.utils_3.1-0
[173] RcppParallel_5.1.9 highr_0.11
[175] jquerylib_0.1.4 spatstat.univar_3.0-1
[177] R.utils_2.12.3 lazyeval_0.2.2
[179] shiny_1.9.1 htmltools_0.5.8.1
[181] data.tree_1.1.0 glue_1.8.0
[183] spam_2.11-0 SymSim_0.0.0.9000
[185] XVector_0.42.0 RCurl_1.98-1.16
[187] treeio_1.26.0 rprojroot_2.0.4
[189] mclust_6.1.1 ks_1.14.2
[191] mnormt_2.1.1 gridExtra_2.3
[193] igraph_2.0.3 R6_2.5.1
[195] SingleCellExperiment_1.24.0 labeling_0.4.3
[197] ggh4x_0.2.8.9000 cluster_2.1.6
[199] pkgload_1.4.0 aplot_0.2.3
[201] GenomeInfoDb_1.40.1 DelayedArray_0.28.0
[203] tidyselect_1.2.1 vipor_0.4.7
[205] maps_3.4.2 ggforce_0.5.0
[207] xml2_1.3.6 rsvd_1.0.5
[209] munsell_0.5.1 KernSmooth_2.23-24
[211] optimParallel_1.0-2 data.table_1.16.2
[213] ComplexHeatmap_2.18.0 htmlwidgets_1.6.4
[215] ggmin_0.0.0.9000 rlang_1.1.4
[217] clusterGeneration_1.3.8 spatstat.sparse_3.1-0
[219] spatstat.explore_3.3-2 remotes_2.5.0
[221] Cairo_1.6-2 phangorn_2.12.1
[223] fansi_1.0.6 beeswarm_0.4.0