PICRUSt Analyses
Bag-of-Genes
Last updated: 2021-04-29
Checks: 6 1
Knit directory: esoph-micro-cancer-workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200916) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2072df5. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/
Unstaged changes:
Modified: analysis/picrust-analyses.Rmd
Modified: analysis/picrust-campy.Rmd
Modified: analysis/picrust-fuso.Rmd
Modified: analysis/picrust-prevo.Rmd
Modified: analysis/picrust-strepto.Rmd
Modified: output/picrust_ec_data_results.csv
Modified: output/picrust_ec_stratefied_campy_data_results.csv
Modified: output/picrust_ec_stratefied_fuso_data_results.csv
Modified: output/picrust_ec_stratefied_prevo_data_results.csv
Modified: output/picrust_ec_stratefied_strepto_data_results.csv
Modified: output/picrust_ko_data_results.csv
Modified: output/picrust_ko_stratefied_campy_data_results.csv
Modified: output/picrust_ko_stratefied_fuso_data_results.csv
Modified: output/picrust_ko_stratefied_prevo_data_results.csv
Modified: output/picrust_ko_stratefied_strepto_data_results.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/picrust-analyses.Rmd) and HTML (docs/picrust-analyses.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2072df5 | noah-padgett | 2021-04-22 | fixed the issue picrust analysis code |
| html | 2072df5 | noah-padgett | 2021-04-22 | fixed the issue picrust analysis code |
| Rmd | ea63e54 | noah-padgett | 2021-04-15 | updated picrust results |
| html | ea63e54 | noah-padgett | 2021-04-15 | updated picrust results |
| Rmd | b11a4b5 | noah-padgett | 2021-03-25 | updated picrust analyses and results figures |
| html | b11a4b5 | noah-padgett | 2021-03-25 | updated picrust analyses and results figures |
| Rmd | 9315cce | noah-padgett | 2021-03-18 | updated picrust results |
| html | 9315cce | noah-padgett | 2021-03-18 | updated picrust results |
| Rmd | cb5f6e8 | noah-padgett | 2021-03-14 | picrust analyses updated |
| html | cb5f6e8 | noah-padgett | 2021-03-14 | picrust analyses updated |
| Rmd | cb97ca6 | noah-padgett | 2021-02-25 | updated figures violin |
For PICRUSt analysis:
Use file KO_metagenome_out > pred_metagenome_unstrat_descrip.tsv to conduct the following tests/measures:
Mean relative abundance in Tumors
Standard error of mean in Tumors
Mean relative abundance in Non-tumors
Standard error of mean in Non-tumors
GLM differential abundance analysis between Tumor vs Non-tumor (Patient=Fixed effect)
- P-value
- DR corrected q-value
Use file EC_metagenome_out > pred_metagenome_unstrat_descrip to conduct the same analyses as described for A.
Use file pathways_out > path_abun_unstrat_descrip to conduct the same analyses as in A. Part 1 EC Data
pi.dat <- readr::read_tsv("data/PICRUST/ec_pred_metagenome_unstrat_descrip.tsv")
-- Column specification --------------------------------------------------------
cols(
.default = col_double(),
`function` = col_character(),
description = col_character()
)
i Use `spec()` for the full column specifications.# aggregate descriptions to get 1 description per row - unique;y defined.
pi.dat<- pi.dat %>%
group_by(description) %>%
summarise(across(`1.S37.Jun172016`:`99.D01.S37.Jun232016`,.fns = sum))
# long format
pi.dat <- pi.dat %>%
pivot_longer(
cols=`1.S37.Jun172016`:`99.D01.S37.Jun232016`,
names_to = "ID",
values_to = "Abundance"
)
d <- as.data.frame(dat.16s)
mydata <- full_join(pi.dat, d)Joining, by = "ID"Warning in class(x) <- c(setdiff(subclass, tibble_class), tibble_class): Setting
class(x) to multiple strings ("tbl_df", "tbl", ...); result will no longer be an
S4 objectmydata <- mydata %>%
mutate(
ID.n = as.numeric(as.factor(ID)),
description.n = as.numeric(as.factor(description))
) %>%
group_by(ID) %>%
mutate(
dem = ifelse(sum(Abundance)==0, 1, sum(Abundance)),
RelAbundance = Abundance/dem*100) %>%
ungroup()%>%
group_by(description)%>%
mutate(avgRA = mean(RelAbundance))Relative Abundance
d <- mydata %>%
filter(description=="DNA-directed DNA polymerase")
t.test(d$RelAbundance ~ d$tumor.cat)
Welch Two Sample t-test
data: d$RelAbundance by d$tumor.cat
t = -1.5, df = 147, p-value = 0.13
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.137544 0.018669
sample estimates:
mean in group Non-Tumor mean in group Tumor
1.3784 1.4378 # Run on all descriptions
tb.ra1 <- mydata %>%
group_by(description) %>%
summarise(ng = n(),
Overall.M = mean(RelAbundance),
Overall.SE = sd(RelAbundance)/sqrt(ng))
tb.ra2m <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(M = mean(RelAbundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = M)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2se <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(),SE = sd(RelAbundance)/sqrt(ng)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = SE)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2var <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(), VAR = var(RelAbundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = VAR)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2ng <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n()) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = ng)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra <- left_join(tb.ra1, tb.ra2m)Joining, by = "description"tb.ra <- cbind(tb.ra, tb.ra2se[,-1])
tb.ra <- cbind(tb.ra, tb.ra2var[,-1])
tb.ra <- cbind(tb.ra, tb.ra2ng[,-1])
colnames(tb.ra) <- c("description", "ng", "Overall Mean", "Overall SE", "Non-Tumor Mean", "Tumor Mean", "Non-Tumor SE", "Tumor SE","Non-Tumor Var", "Tumor Var", "Non-Tumor Ng", "Tumor Ng")
tb.ra <- tb.ra %>%
arrange(desc(`Overall Mean`))
tb.ra <- tb.ra[, c("description", "Overall Mean", "Overall SE","Tumor Mean","Tumor Var", "Tumor SE","Tumor Ng", "Non-Tumor Mean","Non-Tumor Var", "Non-Tumor SE", "Non-Tumor Ng")]
# compute t-test
tb.ra <- tb.ra %>%
mutate(
SEpooled = sqrt(`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`),
t = (`Tumor Mean` - `Non-Tumor Mean`)/(SEpooled),
df = ((`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`)**2)/(((`Tumor Var`/`Tumor Ng`)**2)/(`Tumor Ng`-1) + ((`Non-Tumor Var`/`Non-Tumor Ng`)**2)/(`Non-Tumor Ng`-1)),
p = pt(q = abs(t), df=df, lower.tail = F)*2,
fdr_p = p.adjust(p, method="fdr")
)
kable(tb.ra, format="html", digits=5, caption="EC Data Average Relative Abundance of Each Description (sorted in descending order)") %>%
kable_styling(full_width = T) %>%
scroll_box(width = "100%", height="600px")| description | Overall Mean | Overall SE | Tumor Mean | Tumor Var | Tumor SE | Tumor Ng | Non-Tumor Mean | Non-Tumor Var | Non-Tumor SE | Non-Tumor Ng | SEpooled | t | df | p | fdr_p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA-directed DNA polymerase | 1.40285 | 0.01990 | 1.43784 | 0.05430 | 0.02890 | 65 | 1.37840 | 0.06757 | 0.02695 | 93 | 0.03952 | 1.50391 | 146.594 | 0.13476 | 0.70632 |
| DNA helicase | 1.35380 | 0.02046 | 1.39396 | 0.06314 | 0.03117 | 65 | 1.32573 | 0.06699 | 0.02684 | 93 | 0.04113 | 1.65890 | 140.402 | 0.09937 | 0.70632 |
| Histidine kinase | 0.85520 | 0.01720 | 0.83011 | 0.04863 | 0.02735 | 65 | 0.87273 | 0.04519 | 0.02204 | 93 | 0.03513 | -1.21317 | 134.628 | 0.22719 | 0.70632 |
| Peptidylprolyl isomerase | 0.69459 | 0.01303 | 0.69696 | 0.02777 | 0.02067 | 65 | 0.69294 | 0.02646 | 0.01687 | 93 | 0.02668 | 0.15065 | 135.749 | 0.88048 | 0.96590 |
| Protein-N(pi)-phosphohistidine–sugar phosphotransferase | 0.66839 | 0.02883 | 0.70639 | 0.12814 | 0.04440 | 65 | 0.64183 | 0.13320 | 0.03785 | 93 | 0.05834 | 1.10661 | 139.539 | 0.27037 | 0.70632 |
| DNA-directed RNA polymerase | 0.58448 | 0.00817 | 0.60065 | 0.00947 | 0.01207 | 65 | 0.57318 | 0.01108 | 0.01092 | 93 | 0.01627 | 1.68809 | 144.335 | 0.09355 | 0.70632 |
| NADH:ubiquinone reductase (H(+)-translocating) | 0.50907 | 0.02256 | 0.46077 | 0.07585 | 0.03416 | 65 | 0.54283 | 0.08164 | 0.02963 | 93 | 0.04522 | -1.81459 | 141.000 | 0.07171 | 0.70632 |
| Site-specific DNA-methyltransferase (adenine-specific) | 0.41749 | 0.01228 | 0.41812 | 0.01753 | 0.01642 | 65 | 0.41705 | 0.02849 | 0.01750 | 93 | 0.02400 | 0.04458 | 153.864 | 0.96450 | 0.99193 |
| 3-oxoacyl-[acyl-carrier-protein] reductase | 0.39404 | 0.01105 | 0.37447 | 0.01988 | 0.01749 | 65 | 0.40772 | 0.01864 | 0.01416 | 93 | 0.02250 | -1.47742 | 135.026 | 0.14189 | 0.70632 |
| H(+)-transporting two-sector ATPase | 0.38226 | 0.00446 | 0.38992 | 0.00282 | 0.00659 | 65 | 0.37691 | 0.00332 | 0.00597 | 93 | 0.00890 | 1.46290 | 144.511 | 0.14566 | 0.70632 |
| Iron-chelate-transporting ATPase | 0.36934 | 0.01075 | 0.39130 | 0.02565 | 0.01987 | 65 | 0.35400 | 0.01274 | 0.01170 | 93 | 0.02306 | 1.61789 | 107.154 | 0.10863 | 0.70632 |
| 8-oxo-dGTP diphosphatase | 0.36658 | 0.01443 | 0.36312 | 0.02895 | 0.02111 | 65 | 0.36899 | 0.03602 | 0.01968 | 93 | 0.02886 | -0.20358 | 146.583 | 0.83897 | 0.95674 |
| Glutaminyl-tRNA synthase (glutamine-hydrolyzing) | 0.36556 | 0.00849 | 0.36636 | 0.01166 | 0.01339 | 65 | 0.36500 | 0.01133 | 0.01104 | 93 | 0.01735 | 0.07823 | 136.614 | 0.93776 | 0.99049 |
| Asparaginyl-tRNA synthase (glutamine-hydrolyzing) | 0.36556 | 0.00849 | 0.36636 | 0.01166 | 0.01339 | 65 | 0.36499 | 0.01132 | 0.01104 | 93 | 0.01735 | 0.07896 | 136.606 | 0.93718 | 0.99049 |
| RNA helicase | 0.35964 | 0.00817 | 0.36207 | 0.01211 | 0.01365 | 65 | 0.35794 | 0.00957 | 0.01014 | 93 | 0.01701 | 0.24279 | 127.238 | 0.80856 | 0.94436 |
| Acetyl-CoA carboxylase | 0.34683 | 0.00744 | 0.34790 | 0.00748 | 0.01073 | 65 | 0.34608 | 0.00973 | 0.01023 | 93 | 0.01482 | 0.12246 | 148.118 | 0.90270 | 0.97462 |
| Serine-type D-Ala-D-Ala carboxypeptidase | 0.34488 | 0.00795 | 0.33720 | 0.00785 | 0.01099 | 65 | 0.35025 | 0.01152 | 0.01113 | 93 | 0.01564 | -0.83410 | 151.643 | 0.40554 | 0.75734 |
| 23S rRNA pseudouridine(1911/1915/1917) synthase | 0.34241 | 0.01063 | 0.35084 | 0.01849 | 0.01687 | 65 | 0.33652 | 0.01751 | 0.01372 | 93 | 0.02174 | 0.65867 | 135.472 | 0.51122 | 0.81548 |
| Phosphoglycerate mutase (2,3-diphosphoglycerate-independent) | 0.34040 | 0.01162 | 0.34666 | 0.02387 | 0.01916 | 65 | 0.33603 | 0.01973 | 0.01457 | 93 | 0.02407 | 0.44141 | 129.292 | 0.65966 | 0.88943 |
| DNA topoisomerase (ATP-hydrolyzing) | 0.33352 | 0.00617 | 0.34192 | 0.00596 | 0.00958 | 65 | 0.32764 | 0.00605 | 0.00806 | 93 | 0.01252 | 1.14057 | 138.486 | 0.25602 | 0.70632 |
| Polar-amino-acid-transporting ATPase | 0.33051 | 0.01151 | 0.33536 | 0.01691 | 0.01613 | 65 | 0.32712 | 0.02391 | 0.01604 | 93 | 0.02274 | 0.36236 | 150.652 | 0.71759 | 0.92120 |
| Type I site-specific deoxyribonuclease | 0.33035 | 0.01390 | 0.32919 | 0.02556 | 0.01983 | 65 | 0.33116 | 0.03433 | 0.01921 | 93 | 0.02761 | -0.07126 | 149.141 | 0.94328 | 0.99049 |
| Carbamoyl-phosphate synthase (glutamine-hydrolyzing) | 0.32347 | 0.00786 | 0.32687 | 0.01189 | 0.01352 | 65 | 0.32110 | 0.00838 | 0.00949 | 93 | 0.01652 | 0.34925 | 121.993 | 0.72750 | 0.92120 |
| Ribonuclease H | 0.32044 | 0.00573 | 0.32154 | 0.00560 | 0.00928 | 65 | 0.31967 | 0.00497 | 0.00731 | 93 | 0.01181 | 0.15869 | 132.533 | 0.87416 | 0.96378 |
| Cysteine desulfurase | 0.31992 | 0.00821 | 0.32021 | 0.01093 | 0.01297 | 65 | 0.31972 | 0.01057 | 0.01066 | 93 | 0.01679 | 0.02895 | 136.373 | 0.97695 | 0.99435 |
| Ribonucleoside-diphosphate reductase | 0.31803 | 0.00560 | 0.31832 | 0.00673 | 0.01018 | 65 | 0.31782 | 0.00376 | 0.00636 | 93 | 0.01200 | 0.04174 | 111.839 | 0.96678 | 0.99193 |
| Acetolactate synthase | 0.31196 | 0.00610 | 0.31167 | 0.00728 | 0.01058 | 65 | 0.31216 | 0.00498 | 0.00731 | 93 | 0.01287 | -0.03827 | 120.595 | 0.96953 | 0.99263 |
| Phenylalanine–tRNA ligase | 0.30482 | 0.00401 | 0.31142 | 0.00234 | 0.00600 | 65 | 0.30021 | 0.00267 | 0.00535 | 93 | 0.00804 | 1.39369 | 143.331 | 0.16557 | 0.70632 |
| Exodeoxyribonuclease VII | 0.30206 | 0.00480 | 0.31293 | 0.00385 | 0.00770 | 65 | 0.29447 | 0.00340 | 0.00605 | 93 | 0.00979 | 1.88548 | 132.344 | 0.06156 | 0.70632 |
| DNA-(apurinic or apyrimidinic site) lyase | 0.29656 | 0.00451 | 0.30520 | 0.00281 | 0.00657 | 65 | 0.29052 | 0.00345 | 0.00609 | 93 | 0.00896 | 1.63878 | 146.098 | 0.10341 | 0.70632 |
| Undecaprenyl-diphosphate phosphatase | 0.29175 | 0.00463 | 0.28580 | 0.00276 | 0.00651 | 65 | 0.29591 | 0.00382 | 0.00641 | 93 | 0.00914 | -1.10584 | 150.037 | 0.27056 | 0.70632 |
| DNA topoisomerase | 0.28866 | 0.00739 | 0.28711 | 0.00662 | 0.01009 | 65 | 0.28975 | 0.01013 | 0.01043 | 93 | 0.01452 | -0.18181 | 152.634 | 0.85597 | 0.96186 |
| Peroxiredoxin | 0.28834 | 0.00433 | 0.28918 | 0.00310 | 0.00691 | 65 | 0.28775 | 0.00288 | 0.00557 | 93 | 0.00887 | 0.16154 | 134.626 | 0.87191 | 0.96378 |
| Tryptophan synthase | 0.27600 | 0.00691 | 0.26814 | 0.00806 | 0.01113 | 65 | 0.28149 | 0.00719 | 0.00879 | 93 | 0.01419 | -0.94154 | 132.798 | 0.34814 | 0.72019 |
| Non-specific serine/threonine protein kinase | 0.27068 | 0.00844 | 0.26691 | 0.00988 | 0.01233 | 65 | 0.27332 | 0.01233 | 0.01151 | 93 | 0.01687 | -0.37974 | 146.680 | 0.70469 | 0.91393 |
| [Formate-C-acetyltransferase]-activating enzyme | 0.26762 | 0.01144 | 0.28195 | 0.02038 | 0.01771 | 65 | 0.25760 | 0.02083 | 0.01497 | 93 | 0.02319 | 1.05011 | 138.814 | 0.29549 | 0.70632 |
| Thioredoxin-disulfide reductase | 0.26523 | 0.00584 | 0.26674 | 0.00471 | 0.00851 | 65 | 0.26417 | 0.00593 | 0.00798 | 93 | 0.01167 | 0.21975 | 147.000 | 0.82637 | 0.95262 |
| Protein-tyrosine-phosphatase | 0.26095 | 0.00981 | 0.25955 | 0.01959 | 0.01736 | 65 | 0.26192 | 0.01232 | 0.01151 | 93 | 0.02083 | -0.11381 | 116.916 | 0.90959 | 0.97756 |
| Uracil phosphoribosyltransferase | 0.25837 | 0.00534 | 0.26836 | 0.00431 | 0.00814 | 65 | 0.25139 | 0.00456 | 0.00700 | 93 | 0.01074 | 1.57986 | 140.306 | 0.11639 | 0.70632 |
| Ribosomal-protein-alanine N-acetyltransferase | 0.25763 | 0.00951 | 0.26541 | 0.01161 | 0.01336 | 65 | 0.25220 | 0.01623 | 0.01321 | 93 | 0.01879 | 0.70268 | 150.329 | 0.48334 | 0.80031 |
| Alcohol dehydrogenase | 0.25583 | 0.00818 | 0.24408 | 0.01128 | 0.01317 | 65 | 0.26405 | 0.01002 | 0.01038 | 93 | 0.01677 | -1.19055 | 132.580 | 0.23596 | 0.70632 |
| Glycine–tRNA ligase | 0.25521 | 0.00414 | 0.26721 | 0.00224 | 0.00588 | 65 | 0.24683 | 0.00288 | 0.00557 | 93 | 0.00810 | 2.51725 | 147.724 | 0.01289 | 0.64530 |
| Glycerol-3-phosphate 1-O-acyltransferase | 0.25432 | 0.00500 | 0.26553 | 0.00336 | 0.00719 | 65 | 0.24649 | 0.00426 | 0.00677 | 93 | 0.00987 | 1.92802 | 147.311 | 0.05578 | 0.70632 |
| GTP diphosphokinase | 0.24922 | 0.00601 | 0.25425 | 0.00591 | 0.00954 | 65 | 0.24570 | 0.00558 | 0.00775 | 93 | 0.01229 | 0.69551 | 135.338 | 0.48793 | 0.80164 |
| UDP-glucose 4-epimerase | 0.24634 | 0.00526 | 0.24714 | 0.00446 | 0.00829 | 65 | 0.24578 | 0.00435 | 0.00684 | 93 | 0.01074 | 0.12677 | 136.686 | 0.89931 | 0.97344 |
| Coproporphyrinogen dehydrogenase | 0.24295 | 0.00598 | 0.25331 | 0.00779 | 0.01094 | 65 | 0.23571 | 0.00411 | 0.00665 | 93 | 0.01280 | 1.37475 | 109.534 | 0.17202 | 0.70632 |
| Glutamate–ammonia ligase | 0.24191 | 0.00815 | 0.23879 | 0.01141 | 0.01325 | 65 | 0.24410 | 0.00995 | 0.01035 | 93 | 0.01681 | -0.31543 | 131.769 | 0.75294 | 0.92944 |
| Pyruvate dehydrogenase (acetyl-transferring) | 0.23873 | 0.00867 | 0.22797 | 0.01175 | 0.01345 | 65 | 0.24626 | 0.01195 | 0.01133 | 93 | 0.01759 | -1.04032 | 138.593 | 0.30000 | 0.70632 |
| Transketolase | 0.22806 | 0.00546 | 0.22924 | 0.00508 | 0.00884 | 65 | 0.22723 | 0.00449 | 0.00695 | 93 | 0.01125 | 0.17937 | 132.367 | 0.85792 | 0.96186 |
| Peptide deformylase | 0.22635 | 0.00491 | 0.22801 | 0.00430 | 0.00813 | 65 | 0.22519 | 0.00351 | 0.00614 | 93 | 0.01019 | 0.27744 | 128.784 | 0.78189 | 0.93675 |
| 3-isopropylmalate dehydratase | 0.22481 | 0.00579 | 0.21562 | 0.00648 | 0.00998 | 65 | 0.23124 | 0.00443 | 0.00690 | 93 | 0.01214 | -1.28675 | 120.682 | 0.20064 | 0.70632 |
| (R)-2-methylmalate dehydratase | 0.22477 | 0.00579 | 0.21559 | 0.00647 | 0.00998 | 65 | 0.23118 | 0.00444 | 0.00691 | 93 | 0.01214 | -1.28460 | 120.708 | 0.20139 | 0.70632 |
| Anthranilate synthase | 0.22397 | 0.00556 | 0.21346 | 0.00595 | 0.00956 | 65 | 0.23131 | 0.00407 | 0.00662 | 93 | 0.01163 | -1.53476 | 120.721 | 0.12746 | 0.70632 |
| L-serine ammonia-lyase | 0.22167 | 0.00503 | 0.22499 | 0.00430 | 0.00813 | 65 | 0.21935 | 0.00381 | 0.00640 | 93 | 0.01035 | 0.54513 | 132.506 | 0.58658 | 0.85579 |
| Signal peptidase I | 0.22025 | 0.00447 | 0.22050 | 0.00373 | 0.00758 | 65 | 0.22008 | 0.00280 | 0.00549 | 93 | 0.00936 | 0.04495 | 124.803 | 0.96422 | 0.99193 |
| Peptide-methionine (S)-S-oxide reductase | 0.21753 | 0.00494 | 0.22395 | 0.00358 | 0.00743 | 65 | 0.21305 | 0.00403 | 0.00658 | 93 | 0.00992 | 1.09820 | 142.767 | 0.27397 | 0.70632 |
| Fumarate reductase (quinol) | 0.21549 | 0.00596 | 0.21577 | 0.00397 | 0.00781 | 65 | 0.21530 | 0.00682 | 0.00856 | 93 | 0.01159 | 0.04078 | 154.769 | 0.96753 | 0.99193 |
| Phosphoglycolate phosphatase | 0.21287 | 0.00605 | 0.20665 | 0.00703 | 0.01040 | 65 | 0.21721 | 0.00492 | 0.00727 | 93 | 0.01269 | -0.83245 | 121.668 | 0.40679 | 0.75900 |
| N-acetylmuramoyl-L-alanine amidase | 0.20735 | 0.00771 | 0.20448 | 0.00738 | 0.01066 | 65 | 0.20936 | 0.01089 | 0.01082 | 93 | 0.01519 | -0.32135 | 151.784 | 0.74839 | 0.92904 |
| Ribose-phosphate diphosphokinase | 0.20604 | 0.00515 | 0.21258 | 0.00430 | 0.00813 | 65 | 0.20148 | 0.00411 | 0.00665 | 93 | 0.01050 | 1.05720 | 135.884 | 0.29230 | 0.70632 |
| Monosaccharide-transporting ATPase | 0.20526 | 0.00992 | 0.20694 | 0.02296 | 0.01879 | 65 | 0.20409 | 0.01056 | 0.01066 | 93 | 0.02161 | 0.13188 | 104.278 | 0.89534 | 0.97163 |
| Phosphoribosylformylglycinamidine synthase | 0.20418 | 0.00500 | 0.20688 | 0.00508 | 0.00884 | 65 | 0.20229 | 0.00320 | 0.00587 | 93 | 0.01061 | 0.43195 | 117.009 | 0.66657 | 0.89299 |
| 4-hydroxy-tetrahydrodipicolinate synthase | 0.20303 | 0.00291 | 0.19802 | 0.00108 | 0.00408 | 65 | 0.20653 | 0.00150 | 0.00401 | 93 | 0.00572 | -1.48796 | 150.046 | 0.13886 | 0.70632 |
| 3-deoxy-7-phosphoheptulonate synthase | 0.20294 | 0.00454 | 0.20678 | 0.00333 | 0.00716 | 65 | 0.20026 | 0.00322 | 0.00589 | 93 | 0.00927 | 0.70336 | 136.376 | 0.48303 | 0.80031 |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase | 0.20277 | 0.00471 | 0.20662 | 0.00361 | 0.00745 | 65 | 0.20007 | 0.00344 | 0.00608 | 93 | 0.00962 | 0.68083 | 135.774 | 0.49714 | 0.80780 |
| Long-chain-fatty-acid–CoA ligase | 0.20123 | 0.00844 | 0.19108 | 0.00977 | 0.01226 | 65 | 0.20832 | 0.01229 | 0.01150 | 93 | 0.01681 | -1.02584 | 146.984 | 0.30665 | 0.70632 |
| Peptide-methionine (R)-S-oxide reductase | 0.19561 | 0.00471 | 0.20474 | 0.00310 | 0.00690 | 65 | 0.18923 | 0.00373 | 0.00633 | 93 | 0.00937 | 1.65521 | 145.388 | 0.10004 | 0.70632 |
| Methionyl aminopeptidase | 0.19385 | 0.00229 | 0.19255 | 0.00075 | 0.00340 | 65 | 0.19475 | 0.00089 | 0.00309 | 93 | 0.00459 | -0.47944 | 144.487 | 0.63235 | 0.88317 |
| Shikimate dehydrogenase | 0.19275 | 0.00560 | 0.20332 | 0.00786 | 0.01100 | 65 | 0.18537 | 0.00284 | 0.00553 | 93 | 0.01231 | 1.45800 | 96.164 | 0.14810 | 0.70632 |
| Dihydrofolate synthase | 0.19214 | 0.00475 | 0.19631 | 0.00323 | 0.00705 | 65 | 0.18923 | 0.00382 | 0.00641 | 93 | 0.00953 | 0.74303 | 144.667 | 0.45867 | 0.78637 |
| Tetrahydrofolate synthase | 0.19214 | 0.00475 | 0.19631 | 0.00323 | 0.00705 | 65 | 0.18923 | 0.00382 | 0.00641 | 93 | 0.00953 | 0.74303 | 144.667 | 0.45867 | 0.78637 |
| Exodeoxyribonuclease III | 0.19136 | 0.00300 | 0.19610 | 0.00165 | 0.00503 | 65 | 0.18805 | 0.00126 | 0.00368 | 93 | 0.00624 | 1.29037 | 125.864 | 0.19929 | 0.70632 |
| Glutathione transferase | 0.19043 | 0.01837 | 0.17271 | 0.05273 | 0.02848 | 65 | 0.20282 | 0.05389 | 0.02407 | 93 | 0.03729 | -0.80730 | 138.816 | 0.42087 | 0.76494 |
| Protein-serine/threonine phosphatase | 0.18941 | 0.00570 | 0.18533 | 0.00507 | 0.00883 | 65 | 0.19226 | 0.00520 | 0.00748 | 93 | 0.01157 | -0.59884 | 138.979 | 0.55026 | 0.84415 |
| Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | 0.18840 | 0.00257 | 0.18962 | 0.00088 | 0.00368 | 65 | 0.18755 | 0.00116 | 0.00354 | 93 | 0.00510 | 0.40555 | 148.697 | 0.68565 | 0.90745 |
| Dihydrolipoyl dehydrogenase | 0.18802 | 0.00507 | 0.17855 | 0.00444 | 0.00827 | 65 | 0.19464 | 0.00374 | 0.00634 | 93 | 0.01042 | -1.54424 | 130.148 | 0.12496 | 0.70632 |
| 1-acylglycerol-3-phosphate O-acyltransferase | 0.18713 | 0.00336 | 0.18527 | 0.00223 | 0.00585 | 65 | 0.18844 | 0.00149 | 0.00401 | 93 | 0.00709 | -0.44575 | 119.787 | 0.65658 | 0.88943 |
| Inorganic diphosphatase | 0.18661 | 0.00447 | 0.19450 | 0.00442 | 0.00825 | 65 | 0.18109 | 0.00224 | 0.00490 | 93 | 0.00959 | 1.39778 | 107.899 | 0.16505 | 0.70632 |
| Aspartate kinase | 0.18582 | 0.00354 | 0.18143 | 0.00190 | 0.00540 | 65 | 0.18888 | 0.00204 | 0.00469 | 93 | 0.00715 | -1.04270 | 141.099 | 0.29887 | 0.70632 |
| Fumarate hydratase | 0.18550 | 0.00626 | 0.18130 | 0.00612 | 0.00970 | 65 | 0.18843 | 0.00629 | 0.00822 | 93 | 0.01272 | -0.56058 | 139.063 | 0.57598 | 0.85190 |
| Arsenate reductase (glutaredoxin) | 0.18265 | 0.00373 | 0.18628 | 0.00199 | 0.00553 | 65 | 0.18011 | 0.00236 | 0.00504 | 93 | 0.00748 | 0.82610 | 144.875 | 0.41010 | 0.76070 |
| Chorismate mutase | 0.18095 | 0.00397 | 0.17999 | 0.00329 | 0.00712 | 65 | 0.18162 | 0.00196 | 0.00459 | 93 | 0.00847 | -0.19356 | 114.509 | 0.84686 | 0.95939 |
| Acetyl-CoA C-acetyltransferase | 0.18062 | 0.01010 | 0.16447 | 0.01234 | 0.01378 | 65 | 0.19190 | 0.01862 | 0.01415 | 93 | 0.01975 | -1.38856 | 152.309 | 0.16699 | 0.70632 |
| Cysteine synthase | 0.18016 | 0.00244 | 0.18247 | 0.00099 | 0.00391 | 65 | 0.17854 | 0.00091 | 0.00313 | 93 | 0.00501 | 0.78351 | 134.003 | 0.43471 | 0.77079 |
| GMP synthase (glutamine-hydrolyzing) | 0.18005 | 0.00293 | 0.18098 | 0.00110 | 0.00411 | 65 | 0.17941 | 0.00155 | 0.00408 | 93 | 0.00579 | 0.27072 | 150.497 | 0.78698 | 0.93675 |
| Exodeoxyribonuclease V | 0.17957 | 0.00547 | 0.18273 | 0.00484 | 0.00863 | 65 | 0.17736 | 0.00469 | 0.00710 | 93 | 0.01117 | 0.48013 | 136.485 | 0.63190 | 0.88317 |
| Phosphoribosylglycinamide formyltransferase | 0.17937 | 0.00254 | 0.18771 | 0.00113 | 0.00417 | 65 | 0.17353 | 0.00087 | 0.00306 | 93 | 0.00517 | 2.74146 | 126.239 | 0.00701 | 0.62334 |
| Lysine–tRNA ligase | 0.17902 | 0.00263 | 0.18190 | 0.00109 | 0.00409 | 65 | 0.17700 | 0.00110 | 0.00344 | 93 | 0.00535 | 0.91604 | 138.399 | 0.36124 | 0.72316 |
| Superoxide dismutase | 0.17870 | 0.00640 | 0.17039 | 0.00757 | 0.01079 | 65 | 0.18451 | 0.00570 | 0.00783 | 93 | 0.01333 | -1.05893 | 125.037 | 0.29167 | 0.70632 |
| Phosphate-transporting ATPase | 0.17713 | 0.00678 | 0.17322 | 0.00729 | 0.01059 | 65 | 0.17987 | 0.00731 | 0.00887 | 93 | 0.01381 | -0.48156 | 137.987 | 0.63088 | 0.88317 |
| N(6)-L-threonylcarbamoyladenine synthase | 0.17695 | 0.00376 | 0.18344 | 0.00230 | 0.00594 | 65 | 0.17240 | 0.00216 | 0.00482 | 93 | 0.00765 | 1.44297 | 135.187 | 0.15134 | 0.70632 |
| Phosphatidate cytidylyltransferase | 0.17694 | 0.00253 | 0.18372 | 0.00140 | 0.00464 | 65 | 0.17220 | 0.00069 | 0.00273 | 93 | 0.00538 | 2.13881 | 107.022 | 0.03473 | 0.67652 |
| Alanine racemase | 0.17522 | 0.00314 | 0.18274 | 0.00163 | 0.00501 | 65 | 0.16996 | 0.00145 | 0.00394 | 93 | 0.00638 | 2.00347 | 132.438 | 0.04717 | 0.69348 |
| Xaa-Pro aminopeptidase | 0.17501 | 0.00336 | 0.17959 | 0.00202 | 0.00557 | 65 | 0.17180 | 0.00162 | 0.00417 | 93 | 0.00696 | 1.11912 | 127.984 | 0.26519 | 0.70632 |
| D-alanine–D-alanine ligase | 0.17450 | 0.00271 | 0.17553 | 0.00086 | 0.00363 | 65 | 0.17378 | 0.00138 | 0.00386 | 93 | 0.00530 | 0.33038 | 153.729 | 0.74157 | 0.92652 |
| Fructose-bisphosphate aldolase | 0.17352 | 0.00358 | 0.17820 | 0.00246 | 0.00615 | 65 | 0.17024 | 0.00172 | 0.00430 | 93 | 0.00750 | 1.06177 | 121.622 | 0.29044 | 0.70632 |
| Methylated-DNA–[protein]-cysteine S-methyltransferase | 0.17195 | 0.00238 | 0.16932 | 0.00074 | 0.00337 | 65 | 0.17379 | 0.00100 | 0.00328 | 93 | 0.00470 | -0.95128 | 149.433 | 0.34300 | 0.72019 |
| 3-hydroxyacyl-[acyl-carrier-protein] dehydratase | 0.17172 | 0.00279 | 0.17559 | 0.00150 | 0.00480 | 65 | 0.16902 | 0.00104 | 0.00334 | 93 | 0.00585 | 1.12309 | 121.272 | 0.26362 | 0.70632 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase III | 0.17167 | 0.00340 | 0.17825 | 0.00224 | 0.00588 | 65 | 0.16707 | 0.00150 | 0.00402 | 93 | 0.00712 | 1.57067 | 119.726 | 0.11890 | 0.70632 |
| Endopeptidase Clp | 0.17114 | 0.00331 | 0.17314 | 0.00188 | 0.00538 | 65 | 0.16974 | 0.00163 | 0.00419 | 93 | 0.00682 | 0.49860 | 131.549 | 0.61889 | 0.87573 |
| Dihydroorotase | 0.17000 | 0.00225 | 0.16886 | 0.00065 | 0.00317 | 65 | 0.17080 | 0.00091 | 0.00313 | 93 | 0.00446 | -0.43570 | 150.322 | 0.66368 | 0.89024 |
| XTP/dITP diphosphatase | 0.16979 | 0.00417 | 0.16859 | 0.00289 | 0.00667 | 65 | 0.17062 | 0.00268 | 0.00536 | 93 | 0.00856 | -0.23780 | 134.482 | 0.81240 | 0.94519 |
| Cystathionine beta-lyase | 0.16881 | 0.00640 | 0.17926 | 0.00903 | 0.01178 | 65 | 0.16151 | 0.00463 | 0.00706 | 93 | 0.01374 | 1.29238 | 108.435 | 0.19897 | 0.70632 |
| Aspartate–tRNA ligase | 0.16843 | 0.00342 | 0.17302 | 0.00166 | 0.00506 | 65 | 0.16523 | 0.00197 | 0.00460 | 93 | 0.00684 | 1.13967 | 144.825 | 0.25631 | 0.70632 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase II | 0.16699 | 0.00276 | 0.16759 | 0.00082 | 0.00355 | 65 | 0.16657 | 0.00148 | 0.00399 | 93 | 0.00534 | 0.18979 | 155.353 | 0.84972 | 0.96036 |
| Beta-galactosidase | 0.16681 | 0.00870 | 0.16762 | 0.01043 | 0.01267 | 65 | 0.16625 | 0.01316 | 0.01190 | 93 | 0.01738 | 0.07874 | 147.074 | 0.93734 | 0.99049 |
| Ribose-5-phosphate isomerase | 0.16558 | 0.00332 | 0.16999 | 0.00180 | 0.00527 | 65 | 0.16249 | 0.00170 | 0.00427 | 93 | 0.00678 | 1.10445 | 135.178 | 0.27136 | 0.70632 |
| IMP dehydrogenase | 0.16497 | 0.00261 | 0.16761 | 0.00102 | 0.00396 | 65 | 0.16313 | 0.00112 | 0.00347 | 93 | 0.00527 | 0.85012 | 141.747 | 0.39669 | 0.74876 |
| Alanine–tRNA ligase | 0.16473 | 0.00295 | 0.17007 | 0.00172 | 0.00514 | 65 | 0.16099 | 0.00111 | 0.00345 | 93 | 0.00620 | 1.46579 | 118.039 | 0.14537 | 0.70632 |
| Triose-phosphate isomerase | 0.16404 | 0.00372 | 0.17236 | 0.00321 | 0.00703 | 65 | 0.15823 | 0.00141 | 0.00389 | 93 | 0.00803 | 1.75890 | 102.634 | 0.08157 | 0.70632 |
| tRNA dimethylallyltransferase | 0.16381 | 0.00329 | 0.16531 | 0.00141 | 0.00466 | 65 | 0.16276 | 0.00193 | 0.00456 | 93 | 0.00652 | 0.39100 | 149.756 | 0.69636 | 0.91007 |
| Glycine hydroxymethyltransferase | 0.16264 | 0.00279 | 0.16456 | 0.00151 | 0.00482 | 65 | 0.16130 | 0.00104 | 0.00334 | 93 | 0.00586 | 0.55690 | 120.925 | 0.57863 | 0.85190 |
| Glycerophosphodiester phosphodiesterase | 0.16261 | 0.00505 | 0.16522 | 0.00424 | 0.00807 | 65 | 0.16079 | 0.00393 | 0.00650 | 93 | 0.01036 | 0.42667 | 134.551 | 0.67030 | 0.89503 |
| Cysteine–tRNA ligase | 0.16260 | 0.00397 | 0.17383 | 0.00389 | 0.00774 | 65 | 0.15474 | 0.00139 | 0.00386 | 93 | 0.00865 | 2.20776 | 95.706 | 0.02965 | 0.67652 |
| Guanylate kinase | 0.16215 | 0.00377 | 0.16963 | 0.00292 | 0.00670 | 65 | 0.15692 | 0.00174 | 0.00432 | 93 | 0.00797 | 1.59391 | 114.592 | 0.11371 | 0.70632 |
| IMP cyclohydrolase | 0.16175 | 0.00336 | 0.16548 | 0.00153 | 0.00485 | 65 | 0.15914 | 0.00196 | 0.00459 | 93 | 0.00667 | 0.95028 | 147.612 | 0.34352 | 0.72019 |
| Phosphoribosylaminoimidazolecarboxamide formyltransferase | 0.16175 | 0.00336 | 0.16548 | 0.00153 | 0.00485 | 65 | 0.15914 | 0.00196 | 0.00459 | 93 | 0.00667 | 0.95028 | 147.612 | 0.34352 | 0.72019 |
| Phosphopyruvate hydratase | 0.16140 | 0.00428 | 0.16537 | 0.00301 | 0.00680 | 65 | 0.15863 | 0.00283 | 0.00551 | 93 | 0.00876 | 0.77055 | 135.117 | 0.44232 | 0.77520 |
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–2,6-diaminopimelate ligase | 0.16103 | 0.00426 | 0.16791 | 0.00328 | 0.00710 | 65 | 0.15621 | 0.00255 | 0.00524 | 93 | 0.00882 | 1.32573 | 126.575 | 0.18732 | 0.70632 |
| Glutamine–fructose-6-phosphate transaminase (isomerizing) | 0.16035 | 0.00197 | 0.16080 | 0.00058 | 0.00298 | 65 | 0.16004 | 0.00065 | 0.00264 | 93 | 0.00398 | 0.19144 | 142.632 | 0.84846 | 0.96012 |
| 16S rRNA (cytidine(1402)-2’-O)-methyltransferase | 0.16020 | 0.00253 | 0.16249 | 0.00088 | 0.00369 | 65 | 0.15859 | 0.00110 | 0.00344 | 93 | 0.00504 | 0.77149 | 146.729 | 0.44165 | 0.77520 |
| [Acyl-carrier-protein] S-malonyltransferase | 0.15983 | 0.00186 | 0.16248 | 0.00044 | 0.00261 | 65 | 0.15798 | 0.00061 | 0.00257 | 93 | 0.00366 | 1.22949 | 150.185 | 0.22081 | 0.70632 |
| Enoyl-[acyl-carrier-protein] reductase (NADH) | 0.15979 | 0.00330 | 0.16063 | 0.00213 | 0.00572 | 65 | 0.15921 | 0.00146 | 0.00396 | 93 | 0.00696 | 0.20426 | 120.774 | 0.83849 | 0.95674 |
| 3-dehydroquinate dehydratase | 0.15940 | 0.00181 | 0.16007 | 0.00057 | 0.00297 | 65 | 0.15893 | 0.00048 | 0.00228 | 93 | 0.00374 | 0.30373 | 129.968 | 0.76182 | 0.93350 |
| 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase | 0.15930 | 0.00271 | 0.15714 | 0.00135 | 0.00455 | 65 | 0.16081 | 0.00104 | 0.00334 | 93 | 0.00565 | -0.65021 | 126.046 | 0.51674 | 0.81889 |
| DNA ligase (NAD(+)) | 0.15835 | 0.00159 | 0.16217 | 0.00027 | 0.00203 | 65 | 0.15568 | 0.00048 | 0.00228 | 93 | 0.00305 | 2.13028 | 155.344 | 0.03472 | 0.67652 |
| Dephospho-CoA kinase | 0.15822 | 0.00302 | 0.16155 | 0.00122 | 0.00433 | 65 | 0.15589 | 0.00160 | 0.00415 | 93 | 0.00600 | 0.94405 | 148.394 | 0.34668 | 0.72019 |
| UDP-N-acetylmuramoyl-tripeptide–D-alanyl-D-alanine ligase | 0.15814 | 0.00310 | 0.16110 | 0.00124 | 0.00437 | 65 | 0.15608 | 0.00171 | 0.00429 | 93 | 0.00612 | 0.82118 | 149.978 | 0.41285 | 0.76224 |
| Pyruvate kinase | 0.15814 | 0.00207 | 0.16371 | 0.00047 | 0.00268 | 65 | 0.15424 | 0.00079 | 0.00292 | 93 | 0.00396 | 2.38761 | 154.505 | 0.01817 | 0.64530 |
| Leucine–tRNA ligase | 0.15805 | 0.00258 | 0.16076 | 0.00094 | 0.00380 | 65 | 0.15616 | 0.00113 | 0.00349 | 93 | 0.00516 | 0.89113 | 145.508 | 0.37433 | 0.73204 |
| Thiamine-phosphate diphosphorylase | 0.15785 | 0.00398 | 0.16127 | 0.00261 | 0.00633 | 65 | 0.15546 | 0.00244 | 0.00512 | 93 | 0.00814 | 0.71328 | 134.917 | 0.47690 | 0.79541 |
| Methenyltetrahydrofolate cyclohydrolase | 0.15722 | 0.00207 | 0.15830 | 0.00055 | 0.00290 | 65 | 0.15648 | 0.00078 | 0.00289 | 93 | 0.00410 | 0.44410 | 150.671 | 0.65761 | 0.88943 |
| Methylenetetrahydrofolate dehydrogenase (NADP(+)) | 0.15722 | 0.00207 | 0.15830 | 0.00055 | 0.00290 | 65 | 0.15648 | 0.00078 | 0.00289 | 93 | 0.00410 | 0.44410 | 150.671 | 0.65761 | 0.88943 |
| Purine-nucleoside phosphorylase | 0.15679 | 0.00693 | 0.15817 | 0.00703 | 0.01040 | 65 | 0.15583 | 0.00806 | 0.00931 | 93 | 0.01396 | 0.16824 | 143.563 | 0.86663 | 0.96221 |
| Methionyl-tRNA formyltransferase | 0.15603 | 0.00212 | 0.15928 | 0.00072 | 0.00333 | 65 | 0.15376 | 0.00070 | 0.00273 | 93 | 0.00431 | 1.27964 | 136.228 | 0.20285 | 0.70632 |
| 16S rRNA pseudouridine(516) synthase | 0.15544 | 0.00537 | 0.16446 | 0.00412 | 0.00796 | 65 | 0.14913 | 0.00481 | 0.00719 | 93 | 0.01073 | 1.42936 | 144.305 | 0.15506 | 0.70632 |
| Glucose-6-phosphate isomerase | 0.15529 | 0.00318 | 0.15867 | 0.00160 | 0.00497 | 65 | 0.15292 | 0.00160 | 0.00414 | 93 | 0.00647 | 0.88966 | 137.642 | 0.37520 | 0.73229 |
| (2E,6E)-farnesyl diphosphate synthase | 0.15507 | 0.00194 | 0.15791 | 0.00052 | 0.00283 | 65 | 0.15308 | 0.00064 | 0.00262 | 93 | 0.00386 | 1.25201 | 146.049 | 0.21257 | 0.70632 |
| Dimethylallyltranstransferase | 0.15507 | 0.00194 | 0.15791 | 0.00052 | 0.00283 | 65 | 0.15308 | 0.00064 | 0.00262 | 93 | 0.00386 | 1.25201 | 146.049 | 0.21257 | 0.70632 |
| Shikimate kinase | 0.15485 | 0.00342 | 0.15876 | 0.00226 | 0.00590 | 65 | 0.15212 | 0.00156 | 0.00410 | 93 | 0.00719 | 0.92349 | 121.123 | 0.35759 | 0.72131 |
| Phosphoglycerate dehydrogenase | 0.15480 | 0.00409 | 0.15233 | 0.00306 | 0.00686 | 65 | 0.15654 | 0.00238 | 0.00506 | 93 | 0.00852 | -0.49376 | 126.562 | 0.62233 | 0.87883 |
| Signal peptidase II | 0.15460 | 0.00255 | 0.15891 | 0.00111 | 0.00413 | 65 | 0.15160 | 0.00096 | 0.00321 | 93 | 0.00523 | 1.39735 | 131.326 | 0.16467 | 0.70632 |
| UDP-N-acetylmuramate–L-alanine ligase | 0.15403 | 0.00256 | 0.15807 | 0.00096 | 0.00384 | 65 | 0.15120 | 0.00108 | 0.00341 | 93 | 0.00513 | 1.33869 | 142.915 | 0.18280 | 0.70632 |
| Methionine–tRNA ligase | 0.15394 | 0.00247 | 0.15793 | 0.00093 | 0.00378 | 65 | 0.15114 | 0.00098 | 0.00325 | 93 | 0.00498 | 1.36242 | 140.461 | 0.17525 | 0.70632 |
| Tryptophan–tRNA ligase | 0.15375 | 0.00215 | 0.15768 | 0.00052 | 0.00283 | 65 | 0.15100 | 0.00086 | 0.00305 | 93 | 0.00416 | 1.60640 | 154.225 | 0.11023 | 0.70632 |
| Pantothenate kinase | 0.15352 | 0.00230 | 0.15517 | 0.00070 | 0.00327 | 65 | 0.15237 | 0.00094 | 0.00317 | 93 | 0.00456 | 0.61478 | 149.252 | 0.53964 | 0.83754 |
| Peptide chain release factor N(5)-glutamine methyltransferase | 0.15349 | 0.00208 | 0.15498 | 0.00067 | 0.00321 | 65 | 0.15244 | 0.00070 | 0.00275 | 93 | 0.00423 | 0.60023 | 139.803 | 0.54932 | 0.84333 |
| Succinate dehydrogenase (quinone) | 0.15329 | 0.00528 | 0.14332 | 0.00419 | 0.00803 | 65 | 0.16027 | 0.00449 | 0.00695 | 93 | 0.01062 | -1.59677 | 140.805 | 0.11256 | 0.70632 |
| Valine–tRNA ligase | 0.15317 | 0.00193 | 0.15578 | 0.00056 | 0.00294 | 65 | 0.15136 | 0.00060 | 0.00255 | 93 | 0.00389 | 1.13680 | 140.906 | 0.25755 | 0.70632 |
| Threonine–tRNA ligase | 0.15308 | 0.00203 | 0.15558 | 0.00062 | 0.00308 | 65 | 0.15134 | 0.00067 | 0.00269 | 93 | 0.00409 | 1.03626 | 141.454 | 0.30185 | 0.70632 |
| Isoleucine–tRNA ligase | 0.15308 | 0.00210 | 0.15597 | 0.00064 | 0.00315 | 65 | 0.15106 | 0.00073 | 0.00280 | 93 | 0.00421 | 1.16528 | 143.048 | 0.24585 | 0.70632 |
| Proline–tRNA ligase | 0.15283 | 0.00207 | 0.15555 | 0.00062 | 0.00309 | 65 | 0.15093 | 0.00072 | 0.00277 | 93 | 0.00415 | 1.11388 | 143.852 | 0.26719 | 0.70632 |
| Homoserine dehydrogenase | 0.15258 | 0.00258 | 0.15032 | 0.00138 | 0.00461 | 65 | 0.15417 | 0.00082 | 0.00297 | 93 | 0.00549 | -0.70004 | 114.439 | 0.48532 | 0.80111 |
| Threonine ammonia-lyase | 0.15255 | 0.00406 | 0.14833 | 0.00262 | 0.00635 | 65 | 0.15549 | 0.00260 | 0.00528 | 93 | 0.00826 | -0.86787 | 137.455 | 0.38698 | 0.73966 |
| Orotidine-5’-phosphate decarboxylase | 0.15241 | 0.00246 | 0.15680 | 0.00088 | 0.00367 | 65 | 0.14935 | 0.00100 | 0.00327 | 93 | 0.00492 | 1.51499 | 143.229 | 0.13198 | 0.70632 |
| Hypoxanthine phosphoribosyltransferase | 0.15229 | 0.00347 | 0.15987 | 0.00211 | 0.00570 | 65 | 0.14700 | 0.00171 | 0.00428 | 93 | 0.00713 | 1.80526 | 128.274 | 0.07338 | 0.70632 |
| tRNA pseudouridine(38-40) synthase | 0.15220 | 0.00243 | 0.15558 | 0.00086 | 0.00364 | 65 | 0.14984 | 0.00098 | 0.00325 | 93 | 0.00488 | 1.17704 | 143.284 | 0.24113 | 0.70632 |
| Phosphoribosylamine–glycine ligase | 0.15202 | 0.00278 | 0.15528 | 0.00115 | 0.00421 | 65 | 0.14974 | 0.00126 | 0.00369 | 93 | 0.00560 | 0.99000 | 141.865 | 0.32386 | 0.70874 |
| Orotate phosphoribosyltransferase | 0.15190 | 0.00241 | 0.15495 | 0.00084 | 0.00359 | 65 | 0.14978 | 0.00097 | 0.00323 | 93 | 0.00483 | 1.07082 | 144.039 | 0.28604 | 0.70632 |
| Arginine–tRNA ligase | 0.15189 | 0.00234 | 0.15434 | 0.00077 | 0.00343 | 65 | 0.15018 | 0.00094 | 0.00318 | 93 | 0.00468 | 0.88987 | 146.072 | 0.37500 | 0.73229 |
| Adenylate kinase | 0.15172 | 0.00210 | 0.15414 | 0.00069 | 0.00325 | 65 | 0.15002 | 0.00071 | 0.00276 | 93 | 0.00426 | 0.96717 | 139.273 | 0.33514 | 0.71823 |
| Tyrosine–tRNA ligase | 0.15167 | 0.00229 | 0.15420 | 0.00073 | 0.00335 | 65 | 0.14990 | 0.00090 | 0.00312 | 93 | 0.00458 | 0.93790 | 146.419 | 0.34984 | 0.72019 |
| acetylglucosaminyltransferase | 0.15164 | 0.00208 | 0.15471 | 0.00062 | 0.00309 | 65 | 0.14949 | 0.00073 | 0.00280 | 93 | 0.00417 | 1.25355 | 144.350 | 0.21203 | 0.70632 |
| 16S rRNA (cytosine(1402)-N(4))-methyltransferase | 0.15159 | 0.00241 | 0.15500 | 0.00084 | 0.00360 | 65 | 0.14921 | 0.00097 | 0.00323 | 93 | 0.00483 | 1.19922 | 143.590 | 0.23242 | 0.70632 |
| Dihydrofolate reductase | 0.15157 | 0.00201 | 0.15635 | 0.00059 | 0.00301 | 65 | 0.14824 | 0.00065 | 0.00265 | 93 | 0.00401 | 2.02326 | 142.233 | 0.04492 | 0.69348 |
| Adenylosuccinate synthase | 0.15133 | 0.00205 | 0.15439 | 0.00065 | 0.00315 | 65 | 0.14920 | 0.00068 | 0.00270 | 93 | 0.00415 | 1.25010 | 139.760 | 0.21335 | 0.70632 |
| Histidine–tRNA ligase | 0.15126 | 0.00233 | 0.15409 | 0.00073 | 0.00335 | 65 | 0.14928 | 0.00094 | 0.00319 | 93 | 0.00462 | 1.04162 | 148.028 | 0.29928 | 0.70632 |
| Aminoacyl-tRNA hydrolase | 0.15126 | 0.00234 | 0.15477 | 0.00074 | 0.00337 | 65 | 0.14880 | 0.00095 | 0.00319 | 93 | 0.00464 | 1.28753 | 147.801 | 0.19992 | 0.70632 |
| C-terminal processing peptidase | 0.15105 | 0.00483 | 0.14736 | 0.00292 | 0.00670 | 65 | 0.15362 | 0.00424 | 0.00676 | 93 | 0.00952 | -0.65765 | 151.380 | 0.51176 | 0.81548 |
| Ribulose-phosphate 3-epimerase | 0.15104 | 0.00236 | 0.15500 | 0.00080 | 0.00350 | 65 | 0.14827 | 0.00093 | 0.00317 | 93 | 0.00472 | 1.42390 | 144.321 | 0.15663 | 0.70632 |
| Ribonuclease III | 0.15099 | 0.00208 | 0.15464 | 0.00063 | 0.00311 | 65 | 0.14844 | 0.00072 | 0.00278 | 93 | 0.00417 | 1.48724 | 143.412 | 0.13915 | 0.70632 |
| UDP-N-acetylmuramate dehydrogenase | 0.15087 | 0.00237 | 0.15415 | 0.00076 | 0.00343 | 65 | 0.14857 | 0.00097 | 0.00324 | 93 | 0.00471 | 1.18418 | 147.497 | 0.23825 | 0.70632 |
| NAD(+) kinase | 0.15087 | 0.00232 | 0.15371 | 0.00071 | 0.00331 | 65 | 0.14888 | 0.00094 | 0.00319 | 93 | 0.00459 | 1.05006 | 148.745 | 0.29539 | 0.70632 |
| GTP cyclohydrolase I | 0.15073 | 0.00271 | 0.15265 | 0.00158 | 0.00493 | 65 | 0.14939 | 0.00088 | 0.00307 | 93 | 0.00581 | 0.56027 | 111.564 | 0.57642 | 0.85190 |
| Phosphoglycerate kinase | 0.15070 | 0.00237 | 0.15409 | 0.00077 | 0.00345 | 65 | 0.14834 | 0.00097 | 0.00323 | 93 | 0.00472 | 1.21623 | 146.760 | 0.22585 | 0.70632 |
| 16S rRNA (adenine(1518)-N(6)/adenine(1519)-N(6))-dimethyltransferase | 0.15069 | 0.00236 | 0.15438 | 0.00079 | 0.00348 | 65 | 0.14811 | 0.00094 | 0.00318 | 93 | 0.00471 | 1.33016 | 145.203 | 0.18555 | 0.70632 |
| tRNA (guanine(37)-N(1))-methyltransferase | 0.15039 | 0.00235 | 0.15437 | 0.00079 | 0.00350 | 65 | 0.14762 | 0.00092 | 0.00315 | 93 | 0.00470 | 1.43490 | 143.923 | 0.15348 | 0.70632 |
| 23S rRNA pseudouridine(2605) synthase | 0.15038 | 0.00244 | 0.15254 | 0.00099 | 0.00390 | 65 | 0.14887 | 0.00092 | 0.00314 | 93 | 0.00501 | 0.73258 | 134.644 | 0.46509 | 0.78908 |
| Methionine adenosyltransferase | 0.15033 | 0.00232 | 0.15354 | 0.00075 | 0.00339 | 65 | 0.14809 | 0.00092 | 0.00315 | 93 | 0.00463 | 1.17750 | 146.369 | 0.24091 | 0.70632 |
| Cu(+) exporting ATPase | 0.15029 | 0.00262 | 0.15307 | 0.00075 | 0.00339 | 65 | 0.14835 | 0.00132 | 0.00377 | 93 | 0.00507 | 0.93160 | 155.145 | 0.35299 | 0.72019 |
| 4-hydroxy-tetrahydrodipicolinate reductase | 0.15029 | 0.00325 | 0.14763 | 0.00175 | 0.00519 | 65 | 0.15214 | 0.00162 | 0.00418 | 93 | 0.00666 | -0.67701 | 134.421 | 0.49956 | 0.80830 |
| tRNA (guanine(46)-N(7))-methyltransferase | 0.15024 | 0.00247 | 0.15450 | 0.00090 | 0.00372 | 65 | 0.14726 | 0.00100 | 0.00327 | 93 | 0.00495 | 1.46220 | 142.174 | 0.14589 | 0.70632 |
| Phosphoglucomutase (alpha-D-glucose-1,6-bisphosphate-dependent) | 0.15020 | 0.00258 | 0.15199 | 0.00086 | 0.00363 | 65 | 0.14895 | 0.00120 | 0.00359 | 93 | 0.00511 | 0.59677 | 150.292 | 0.55156 | 0.84431 |
| UMP/CMP kinase | 0.15018 | 0.00296 | 0.15503 | 0.00130 | 0.00447 | 65 | 0.14678 | 0.00143 | 0.00392 | 93 | 0.00595 | 1.38720 | 141.993 | 0.16755 | 0.70632 |
| Pantetheine-phosphate adenylyltransferase | 0.15011 | 0.00230 | 0.15339 | 0.00073 | 0.00336 | 65 | 0.14782 | 0.00091 | 0.00312 | 93 | 0.00458 | 1.21405 | 146.420 | 0.22668 | 0.70632 |
| Glycerol-3-phosphate dehydrogenase (NAD(P)(+)) | 0.15009 | 0.00229 | 0.15343 | 0.00074 | 0.00337 | 65 | 0.14775 | 0.00089 | 0.00310 | 93 | 0.00457 | 1.24179 | 145.587 | 0.21631 | 0.70632 |
| Phospho-N-acetylmuramoyl-pentapeptide-transferase | 0.15006 | 0.00227 | 0.15327 | 0.00074 | 0.00337 | 65 | 0.14782 | 0.00087 | 0.00306 | 93 | 0.00455 | 1.19782 | 144.528 | 0.23295 | 0.70632 |
| 23S rRNA (uracil(1939)-C(5))-methyltransferase | 0.15002 | 0.00330 | 0.15161 | 0.00159 | 0.00494 | 65 | 0.14890 | 0.00184 | 0.00445 | 93 | 0.00665 | 0.40842 | 143.972 | 0.68357 | 0.90657 |
| tRNA pseudouridine(55) synthase | 0.15001 | 0.00210 | 0.15416 | 0.00061 | 0.00307 | 65 | 0.14710 | 0.00075 | 0.00284 | 93 | 0.00418 | 1.69015 | 145.963 | 0.09313 | 0.70632 |
| ADP-ribose diphosphatase | 0.14989 | 0.00371 | 0.16190 | 0.00206 | 0.00563 | 65 | 0.14149 | 0.00211 | 0.00477 | 93 | 0.00737 | 2.76853 | 139.001 | 0.00640 | 0.62334 |
| UMP kinase | 0.14984 | 0.00231 | 0.15315 | 0.00075 | 0.00339 | 65 | 0.14753 | 0.00091 | 0.00313 | 93 | 0.00462 | 1.21795 | 145.899 | 0.22521 | 0.70632 |
| Serine–tRNA ligase | 0.14982 | 0.00231 | 0.15324 | 0.00075 | 0.00339 | 65 | 0.14744 | 0.00091 | 0.00313 | 93 | 0.00461 | 1.25692 | 145.823 | 0.21079 | 0.70632 |
| UDP-N-acetylmuramoyl-L-alanine–D-glutamate ligase | 0.14975 | 0.00230 | 0.15328 | 0.00073 | 0.00336 | 65 | 0.14727 | 0.00090 | 0.00312 | 93 | 0.00458 | 1.31079 | 146.264 | 0.19198 | 0.70632 |
| diphosphate specific) | 0.14973 | 0.00234 | 0.15391 | 0.00079 | 0.00350 | 65 | 0.14681 | 0.00090 | 0.00312 | 93 | 0.00468 | 1.51692 | 143.308 | 0.13149 | 0.70632 |
| Phosphopantothenate–cysteine ligase | 0.14971 | 0.00203 | 0.15305 | 0.00059 | 0.00303 | 65 | 0.14737 | 0.00069 | 0.00272 | 93 | 0.00407 | 1.39549 | 143.779 | 0.16502 | 0.70632 |
| Adenylosuccinate lyase | 0.14967 | 0.00233 | 0.15319 | 0.00076 | 0.00342 | 65 | 0.14721 | 0.00092 | 0.00315 | 93 | 0.00465 | 1.28741 | 145.765 | 0.19999 | 0.70632 |
| Phosphopantothenoylcysteine decarboxylase | 0.14963 | 0.00203 | 0.15302 | 0.00060 | 0.00303 | 65 | 0.14726 | 0.00068 | 0.00271 | 93 | 0.00406 | 1.41935 | 143.567 | 0.15796 | 0.70632 |
| 16S rRNA (guanine(527)-N(7))-methyltransferase | 0.14954 | 0.00231 | 0.15333 | 0.00072 | 0.00334 | 65 | 0.14689 | 0.00092 | 0.00314 | 93 | 0.00458 | 1.40503 | 147.176 | 0.16212 | 0.70632 |
| FAD synthetase | 0.14952 | 0.00231 | 0.15310 | 0.00075 | 0.00339 | 65 | 0.14702 | 0.00090 | 0.00311 | 93 | 0.00460 | 1.32367 | 145.534 | 0.18769 | 0.70632 |
| Riboflavin kinase | 0.14952 | 0.00231 | 0.15310 | 0.00075 | 0.00339 | 65 | 0.14702 | 0.00090 | 0.00311 | 93 | 0.00460 | 1.32367 | 145.534 | 0.18769 | 0.70632 |
| tRNA(Ile)-lysidine synthetase | 0.14915 | 0.00231 | 0.15320 | 0.00071 | 0.00332 | 65 | 0.14631 | 0.00092 | 0.00314 | 93 | 0.00457 | 1.50784 | 147.664 | 0.13373 | 0.70632 |
| 2-oxoglutarate synthase | 0.14892 | 0.01178 | 0.13250 | 0.01687 | 0.01611 | 65 | 0.16039 | 0.02534 | 0.01651 | 93 | 0.02306 | -1.20896 | 152.213 | 0.22855 | 0.70632 |
| 6-phosphofructokinase | 0.14884 | 0.00566 | 0.15387 | 0.00562 | 0.00930 | 65 | 0.14533 | 0.00470 | 0.00711 | 93 | 0.01171 | 0.72928 | 129.801 | 0.46714 | 0.79096 |
| Glucokinase | 0.14841 | 0.00343 | 0.14815 | 0.00204 | 0.00561 | 65 | 0.14859 | 0.00176 | 0.00435 | 93 | 0.00709 | -0.06213 | 131.069 | 0.95055 | 0.99049 |
| Asparaginase | 0.14801 | 0.00294 | 0.15198 | 0.00108 | 0.00408 | 65 | 0.14524 | 0.00156 | 0.00410 | 93 | 0.00578 | 1.16475 | 151.297 | 0.24595 | 0.70632 |
| Phosphoribosylaminoimidazolesuccinocarboxamide synthase | 0.14788 | 0.00247 | 0.15139 | 0.00083 | 0.00357 | 65 | 0.14543 | 0.00106 | 0.00337 | 93 | 0.00491 | 1.21339 | 147.517 | 0.22692 | 0.70632 |
| Nicotinate phosphoribosyltransferase | 0.14775 | 0.00258 | 0.15088 | 0.00114 | 0.00418 | 65 | 0.14556 | 0.00099 | 0.00327 | 93 | 0.00531 | 1.00348 | 131.783 | 0.31747 | 0.70632 |
| CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase | 0.14735 | 0.00329 | 0.15567 | 0.00201 | 0.00556 | 65 | 0.14153 | 0.00144 | 0.00393 | 93 | 0.00681 | 2.07664 | 122.759 | 0.03992 | 0.67652 |
| Amidophosphoribosyltransferase | 0.14714 | 0.00241 | 0.14903 | 0.00089 | 0.00370 | 65 | 0.14582 | 0.00094 | 0.00318 | 93 | 0.00488 | 0.65885 | 140.351 | 0.51107 | 0.81548 |
| Biotin–[acetyl-CoA-carboxylase] ligase | 0.14679 | 0.00243 | 0.15071 | 0.00084 | 0.00360 | 65 | 0.14404 | 0.00098 | 0.00325 | 93 | 0.00485 | 1.37503 | 144.193 | 0.17126 | 0.70632 |
| S-adenosylmethionine:tRNA ribosyltransferase-isomerase | 0.14678 | 0.00379 | 0.14619 | 0.00226 | 0.00589 | 65 | 0.14719 | 0.00230 | 0.00497 | 93 | 0.00771 | -0.12947 | 138.579 | 0.89717 | 0.97163 |
| Ribonucleoside-triphosphate reductase | 0.14647 | 0.00647 | 0.15283 | 0.00642 | 0.00994 | 65 | 0.14203 | 0.00676 | 0.00852 | 93 | 0.01309 | 0.82427 | 140.053 | 0.41119 | 0.76116 |
| Serine O-acetyltransferase | 0.14621 | 0.00328 | 0.14438 | 0.00216 | 0.00576 | 65 | 0.14749 | 0.00139 | 0.00386 | 93 | 0.00694 | -0.44879 | 117.851 | 0.65441 | 0.88943 |
| Ribonuclease P | 0.14620 | 0.00264 | 0.15124 | 0.00083 | 0.00358 | 65 | 0.14268 | 0.00127 | 0.00369 | 93 | 0.00514 | 1.66496 | 152.473 | 0.09797 | 0.70632 |
| Aspartate-semialdehyde dehydrogenase | 0.14591 | 0.00247 | 0.14595 | 0.00114 | 0.00419 | 65 | 0.14589 | 0.00085 | 0.00302 | 93 | 0.00516 | 0.01220 | 124.416 | 0.99028 | 0.99736 |
| 5-formyltetrahydrofolate cyclo-ligase | 0.14534 | 0.00285 | 0.15174 | 0.00115 | 0.00421 | 65 | 0.14086 | 0.00134 | 0.00379 | 93 | 0.00566 | 1.92118 | 143.965 | 0.05669 | 0.70632 |
| Glutamate synthase (NADH) | 0.14512 | 0.00600 | 0.13052 | 0.00525 | 0.00898 | 65 | 0.15532 | 0.00581 | 0.00790 | 93 | 0.01197 | -2.07264 | 142.181 | 0.04001 | 0.67652 |
| Glutamate synthase (NADPH) | 0.14512 | 0.00600 | 0.13052 | 0.00525 | 0.00898 | 65 | 0.15532 | 0.00581 | 0.00790 | 93 | 0.01197 | -2.07264 | 142.181 | 0.04001 | 0.67652 |
| 6-phospho-beta-glucosidase | 0.14498 | 0.00842 | 0.15074 | 0.01105 | 0.01304 | 65 | 0.14096 | 0.01139 | 0.01107 | 93 | 0.01710 | 0.57189 | 139.171 | 0.56832 | 0.85024 |
| Glutamate racemase | 0.14452 | 0.00299 | 0.14780 | 0.00148 | 0.00477 | 65 | 0.14222 | 0.00137 | 0.00384 | 93 | 0.00613 | 0.90937 | 134.465 | 0.36478 | 0.72472 |
| tRNA-guanine(34) transglycosylase | 0.14449 | 0.00249 | 0.14606 | 0.00091 | 0.00373 | 65 | 0.14340 | 0.00104 | 0.00334 | 93 | 0.00501 | 0.53158 | 143.425 | 0.59584 | 0.86170 |
| 3-phosphoshikimate 1-carboxyvinyltransferase | 0.14403 | 0.00238 | 0.14511 | 0.00103 | 0.00398 | 65 | 0.14328 | 0.00081 | 0.00295 | 93 | 0.00496 | 0.36962 | 126.870 | 0.71228 | 0.92039 |
| 23S rRNA (adenine(2503)-C(2))-methyltransferase | 0.14402 | 0.00270 | 0.14593 | 0.00121 | 0.00432 | 65 | 0.14269 | 0.00112 | 0.00347 | 93 | 0.00554 | 0.58487 | 134.456 | 0.55961 | 0.85024 |
| 5-(carboxyamino)imidazole ribonucleotide mutase | 0.14393 | 0.00237 | 0.14805 | 0.00076 | 0.00341 | 65 | 0.14105 | 0.00097 | 0.00323 | 93 | 0.00470 | 1.48959 | 147.770 | 0.13846 | 0.70632 |
| Phosphoribosylformylglycinamidine cyclo-ligase | 0.14342 | 0.00250 | 0.14618 | 0.00095 | 0.00382 | 65 | 0.14150 | 0.00102 | 0.00331 | 93 | 0.00505 | 0.92603 | 140.867 | 0.35601 | 0.72019 |
| 16S rRNA (uracil(1498)-N(3))-methyltransferase | 0.14313 | 0.00237 | 0.14628 | 0.00080 | 0.00352 | 65 | 0.14093 | 0.00094 | 0.00318 | 93 | 0.00474 | 1.12941 | 144.209 | 0.26060 | 0.70632 |
| Succinyl-diaminopimelate desuccinylase | 0.14263 | 0.00575 | 0.15236 | 0.00725 | 0.01056 | 65 | 0.13584 | 0.00374 | 0.00635 | 93 | 0.01232 | 1.34062 | 108.700 | 0.18284 | 0.70632 |
| Aspartate carbamoyltransferase | 0.14255 | 0.00241 | 0.14547 | 0.00088 | 0.00367 | 65 | 0.14051 | 0.00095 | 0.00320 | 93 | 0.00487 | 1.01950 | 141.453 | 0.30971 | 0.70632 |
| Chorismate synthase | 0.14251 | 0.00256 | 0.14408 | 0.00111 | 0.00414 | 65 | 0.14140 | 0.00098 | 0.00325 | 93 | 0.00526 | 0.50907 | 132.401 | 0.61155 | 0.87289 |
| 16S rRNA (cytosine(967)-C(5))-methyltransferase | 0.14228 | 0.00220 | 0.14637 | 0.00065 | 0.00315 | 65 | 0.13942 | 0.00084 | 0.00301 | 93 | 0.00436 | 1.59687 | 148.114 | 0.11243 | 0.70632 |
| Uracil-DNA glycosylase | 0.14227 | 0.00299 | 0.14743 | 0.00113 | 0.00417 | 65 | 0.13867 | 0.00160 | 0.00415 | 93 | 0.00588 | 1.48910 | 150.677 | 0.13855 | 0.70632 |
| Adenine phosphoribosyltransferase | 0.14202 | 0.00344 | 0.15003 | 0.00229 | 0.00594 | 65 | 0.13642 | 0.00151 | 0.00403 | 93 | 0.00718 | 1.89621 | 119.053 | 0.06036 | 0.70632 |
| Polyribonucleotide nucleotidyltransferase | 0.14177 | 0.00282 | 0.14368 | 0.00136 | 0.00458 | 65 | 0.14044 | 0.00119 | 0.00358 | 93 | 0.00581 | 0.55669 | 132.011 | 0.57868 | 0.85190 |
| Glutathione peroxidase | 0.14176 | 0.00483 | 0.14202 | 0.00378 | 0.00762 | 65 | 0.14158 | 0.00367 | 0.00628 | 93 | 0.00988 | 0.04443 | 136.592 | 0.96463 | 0.99193 |
| Prepilin peptidase | 0.14124 | 0.00275 | 0.14709 | 0.00098 | 0.00389 | 65 | 0.13716 | 0.00131 | 0.00375 | 93 | 0.00540 | 1.83865 | 148.899 | 0.06796 | 0.70632 |
| Phosphoribosylanthranilate isomerase | 0.14092 | 0.00433 | 0.13383 | 0.00300 | 0.00680 | 65 | 0.14587 | 0.00290 | 0.00559 | 93 | 0.00880 | -1.36866 | 136.358 | 0.17336 | 0.70632 |
| Nicotinate-nucleotide adenylyltransferase | 0.14058 | 0.00274 | 0.14309 | 0.00098 | 0.00388 | 65 | 0.13882 | 0.00133 | 0.00378 | 93 | 0.00542 | 0.78906 | 149.609 | 0.43133 | 0.76880 |
| 23S rRNA (guanosine(2251)-2’-O)-methyltransferase | 0.14054 | 0.00204 | 0.14507 | 0.00055 | 0.00292 | 65 | 0.13737 | 0.00071 | 0.00276 | 93 | 0.00402 | 1.91788 | 147.438 | 0.05706 | 0.70632 |
| 5’-nucleotidase | 0.14028 | 0.00418 | 0.13302 | 0.00276 | 0.00651 | 65 | 0.14535 | 0.00272 | 0.00541 | 93 | 0.00846 | -1.45571 | 137.322 | 0.14776 | 0.70632 |
| 23S rRNA (pseudouridine(1915)-N(3))-methyltransferase | 0.13995 | 0.00236 | 0.14295 | 0.00084 | 0.00359 | 65 | 0.13785 | 0.00091 | 0.00312 | 93 | 0.00476 | 1.07104 | 141.151 | 0.28598 | 0.70632 |
| Diaminopimelate decarboxylase | 0.13991 | 0.00255 | 0.13838 | 0.00108 | 0.00408 | 65 | 0.14098 | 0.00099 | 0.00327 | 93 | 0.00523 | -0.49652 | 134.025 | 0.62034 | 0.87719 |
| Cyclic pyranopterin phosphate synthase | 0.13987 | 0.00474 | 0.14019 | 0.00374 | 0.00759 | 65 | 0.13965 | 0.00347 | 0.00611 | 93 | 0.00974 | 0.05606 | 134.500 | 0.95538 | 0.99147 |
| Acetate kinase | 0.13959 | 0.00439 | 0.14606 | 0.00316 | 0.00697 | 65 | 0.13507 | 0.00296 | 0.00564 | 93 | 0.00897 | 1.22583 | 134.918 | 0.22240 | 0.70632 |
| Thymidylate synthase | 0.13950 | 0.00261 | 0.14121 | 0.00119 | 0.00427 | 65 | 0.13830 | 0.00100 | 0.00328 | 93 | 0.00539 | 0.54028 | 130.296 | 0.58993 | 0.85771 |
| 3-dehydroquinate synthase | 0.13932 | 0.00260 | 0.13626 | 0.00140 | 0.00464 | 65 | 0.14146 | 0.00084 | 0.00301 | 93 | 0.00553 | -0.93977 | 114.890 | 0.34931 | 0.72019 |
| Phosphate acetyltransferase | 0.13928 | 0.00315 | 0.14145 | 0.00169 | 0.00510 | 65 | 0.13775 | 0.00150 | 0.00401 | 93 | 0.00649 | 0.57020 | 132.538 | 0.56951 | 0.85024 |
| Threonine synthase | 0.13909 | 0.00278 | 0.13821 | 0.00140 | 0.00465 | 65 | 0.13971 | 0.00110 | 0.00344 | 93 | 0.00578 | -0.25933 | 126.767 | 0.79580 | 0.94089 |
| GTP cyclohydrolase II | 0.13899 | 0.00441 | 0.14001 | 0.00298 | 0.00677 | 65 | 0.13828 | 0.00317 | 0.00584 | 93 | 0.00894 | 0.19365 | 140.551 | 0.84673 | 0.95939 |
| CTP synthase (glutamine hydrolyzing) | 0.13895 | 0.00235 | 0.13530 | 0.00119 | 0.00428 | 65 | 0.14150 | 0.00065 | 0.00264 | 93 | 0.00503 | -1.23299 | 110.841 | 0.22019 | 0.70632 |
| 3’(2’),5’-bisphosphate nucleotidase | 0.13874 | 0.00272 | 0.14112 | 0.00103 | 0.00398 | 65 | 0.13708 | 0.00127 | 0.00369 | 93 | 0.00543 | 0.74363 | 146.135 | 0.45830 | 0.78637 |
| dUTP diphosphatase | 0.13847 | 0.00228 | 0.14352 | 0.00077 | 0.00345 | 65 | 0.13495 | 0.00083 | 0.00299 | 93 | 0.00457 | 1.87873 | 140.980 | 0.06235 | 0.70632 |
| Glycogen phosphorylase | 0.13823 | 0.00523 | 0.14449 | 0.00462 | 0.00843 | 65 | 0.13386 | 0.00412 | 0.00666 | 93 | 0.01074 | 0.98924 | 132.736 | 0.32435 | 0.70906 |
| L-threonylcarbamoyladenylate synthase | 0.13815 | 0.00253 | 0.14149 | 0.00091 | 0.00374 | 65 | 0.13582 | 0.00108 | 0.00340 | 93 | 0.00506 | 1.12287 | 144.780 | 0.26335 | 0.70632 |
| Carbonate dehydratase | 0.13746 | 0.00393 | 0.13274 | 0.00248 | 0.00618 | 65 | 0.14076 | 0.00241 | 0.00510 | 93 | 0.00801 | -1.00149 | 136.703 | 0.31836 | 0.70632 |
| Sortase A | 0.13684 | 0.00748 | 0.14165 | 0.01001 | 0.01241 | 65 | 0.13347 | 0.00810 | 0.00933 | 93 | 0.01553 | 0.52656 | 128.314 | 0.59941 | 0.86318 |
| Homoserine kinase | 0.13676 | 0.00225 | 0.14085 | 0.00079 | 0.00348 | 65 | 0.13390 | 0.00079 | 0.00292 | 93 | 0.00454 | 1.53122 | 138.126 | 0.12800 | 0.70632 |
| Glucose-1-phosphate adenylyltransferase | 0.13663 | 0.00693 | 0.14279 | 0.00752 | 0.01076 | 65 | 0.13232 | 0.00768 | 0.00909 | 93 | 0.01408 | 0.74311 | 138.763 | 0.45867 | 0.78637 |
| Phosphoenolpyruvate–protein phosphotransferase | 0.13612 | 0.00284 | 0.14342 | 0.00142 | 0.00467 | 65 | 0.13102 | 0.00113 | 0.00349 | 93 | 0.00583 | 2.12606 | 127.677 | 0.03542 | 0.67652 |
| Mannose-6-phosphate isomerase | 0.13603 | 0.00336 | 0.13253 | 0.00169 | 0.00509 | 65 | 0.13847 | 0.00186 | 0.00447 | 93 | 0.00678 | -0.87631 | 141.968 | 0.38234 | 0.73815 |
| Branched-chain-amino-acid transaminase | 0.13603 | 0.00292 | 0.13296 | 0.00161 | 0.00497 | 65 | 0.13817 | 0.00117 | 0.00355 | 93 | 0.00611 | -0.85283 | 123.528 | 0.39541 | 0.74766 |
| UTP–glucose-1-phosphate uridylyltransferase | 0.13551 | 0.00258 | 0.13800 | 0.00101 | 0.00395 | 65 | 0.13378 | 0.00108 | 0.00341 | 93 | 0.00521 | 0.81081 | 140.554 | 0.41884 | 0.76408 |
| Aldose 1-epimerase | 0.13474 | 0.00413 | 0.13503 | 0.00242 | 0.00610 | 65 | 0.13454 | 0.00292 | 0.00560 | 93 | 0.00828 | 0.05903 | 145.464 | 0.95301 | 0.99049 |
| Nicotinamide-nucleotide amidase | 0.13452 | 0.00256 | 0.13710 | 0.00090 | 0.00371 | 65 | 0.13272 | 0.00113 | 0.00349 | 93 | 0.00509 | 0.85952 | 147.095 | 0.39145 | 0.74318 |
| Dihydroneopterin aldolase | 0.13444 | 0.00271 | 0.13464 | 0.00143 | 0.00469 | 65 | 0.13429 | 0.00099 | 0.00327 | 93 | 0.00571 | 0.06144 | 121.389 | 0.95111 | 0.99049 |
| Dihydropteroate synthase | 0.13435 | 0.00257 | 0.13352 | 0.00119 | 0.00428 | 65 | 0.13493 | 0.00095 | 0.00319 | 93 | 0.00534 | -0.26306 | 127.524 | 0.79293 | 0.93908 |
| Cytochrome-c oxidase | 0.13396 | 0.01164 | 0.11092 | 0.02086 | 0.01792 | 65 | 0.15007 | 0.02141 | 0.01517 | 93 | 0.02348 | -1.66757 | 138.984 | 0.09765 | 0.70632 |
| Nitrate reductase | 0.13394 | 0.00698 | 0.13706 | 0.00828 | 0.01129 | 65 | 0.13176 | 0.00736 | 0.00890 | 93 | 0.01437 | 0.36900 | 132.605 | 0.71272 | 0.92039 |
| Prephenate dehydratase | 0.13361 | 0.00276 | 0.13251 | 0.00164 | 0.00502 | 65 | 0.13438 | 0.00091 | 0.00314 | 93 | 0.00592 | -0.31670 | 111.875 | 0.75206 | 0.92944 |
| Phosphoglucosamine mutase | 0.13336 | 0.00230 | 0.13883 | 0.00064 | 0.00313 | 65 | 0.12953 | 0.00095 | 0.00320 | 93 | 0.00448 | 2.07694 | 151.997 | 0.03949 | 0.67652 |
| Lactoylglutathione lyase | 0.13322 | 0.00360 | 0.14087 | 0.00239 | 0.00607 | 65 | 0.12788 | 0.00176 | 0.00435 | 93 | 0.00747 | 1.73995 | 124.041 | 0.08435 | 0.70632 |
| UDP-N-acetylglucosamine diphosphorylase | 0.13275 | 0.00230 | 0.13630 | 0.00083 | 0.00357 | 65 | 0.13027 | 0.00084 | 0.00300 | 93 | 0.00466 | 1.29076 | 138.481 | 0.19894 | 0.70632 |
| tRNA (cytidine(34)-2’-O)-methyltransferase | 0.13190 | 0.00218 | 0.13786 | 0.00060 | 0.00304 | 65 | 0.12774 | 0.00082 | 0.00297 | 93 | 0.00425 | 2.38250 | 149.701 | 0.01845 | 0.64530 |
| Biotin carboxylase | 0.13190 | 0.00312 | 0.12840 | 0.00171 | 0.00513 | 65 | 0.13434 | 0.00142 | 0.00391 | 93 | 0.00645 | -0.92102 | 129.588 | 0.35875 | 0.72286 |
| dTMP kinase | 0.13182 | 0.00229 | 0.13594 | 0.00082 | 0.00354 | 65 | 0.12894 | 0.00082 | 0.00298 | 93 | 0.00463 | 1.51298 | 138.274 | 0.13257 | 0.70632 |
| Glucosamine-1-phosphate N-acetyltransferase | 0.13171 | 0.00231 | 0.13598 | 0.00082 | 0.00356 | 65 | 0.12873 | 0.00085 | 0.00302 | 93 | 0.00466 | 1.55589 | 139.090 | 0.12201 | 0.70632 |
| Pyridoxal kinase | 0.13115 | 0.00373 | 0.13051 | 0.00193 | 0.00545 | 65 | 0.13159 | 0.00241 | 0.00509 | 93 | 0.00746 | -0.14501 | 146.663 | 0.88490 | 0.96726 |
| Histidinol-phosphate transaminase | 0.13065 | 0.00372 | 0.12172 | 0.00247 | 0.00616 | 65 | 0.13688 | 0.00193 | 0.00455 | 93 | 0.00766 | -1.97971 | 126.703 | 0.04990 | 0.70331 |
| Fructose-bisphosphatase | 0.13045 | 0.00371 | 0.13531 | 0.00284 | 0.00661 | 65 | 0.12706 | 0.00170 | 0.00428 | 93 | 0.00787 | 1.04904 | 114.851 | 0.29636 | 0.70632 |
| Formate C-acetyltransferase | 0.12933 | 0.00609 | 0.13579 | 0.00547 | 0.00918 | 65 | 0.12482 | 0.00614 | 0.00812 | 93 | 0.01226 | 0.89499 | 142.684 | 0.37230 | 0.73139 |
| Dihydroxy-acid dehydratase | 0.12925 | 0.00376 | 0.11878 | 0.00208 | 0.00565 | 65 | 0.13658 | 0.00223 | 0.00490 | 93 | 0.00748 | -2.38079 | 140.920 | 0.01861 | 0.64530 |
| Cob(I)yrinic acid a,c-diamide adenosyltransferase | 0.12888 | 0.00629 | 0.13981 | 0.00816 | 0.01120 | 65 | 0.12124 | 0.00487 | 0.00723 | 93 | 0.01334 | 1.39233 | 114.616 | 0.16652 | 0.70632 |
| L-lactate dehydrogenase | 0.12846 | 0.00982 | 0.14448 | 0.02348 | 0.01900 | 65 | 0.11727 | 0.00936 | 0.01003 | 93 | 0.02149 | 1.26646 | 99.287 | 0.20831 | 0.70632 |
| Aconitate hydratase | 0.12843 | 0.00651 | 0.12046 | 0.00690 | 0.01030 | 65 | 0.13400 | 0.00656 | 0.00840 | 93 | 0.01329 | -1.01837 | 135.631 | 0.31031 | 0.70632 |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | 0.12817 | 0.00328 | 0.12565 | 0.00163 | 0.00501 | 65 | 0.12993 | 0.00176 | 0.00435 | 93 | 0.00664 | -0.64414 | 141.096 | 0.52053 | 0.82181 |
| Geranylgeranyl diphosphate synthase | 0.12810 | 0.00347 | 0.13314 | 0.00180 | 0.00526 | 65 | 0.12457 | 0.00196 | 0.00459 | 93 | 0.00698 | 1.22724 | 141.424 | 0.22177 | 0.70632 |
| Beta-lactamase | 0.12782 | 0.00429 | 0.12576 | 0.00289 | 0.00666 | 65 | 0.12926 | 0.00296 | 0.00564 | 93 | 0.00873 | -0.40064 | 138.963 | 0.68930 | 0.90847 |
| Hydroxymethylpyrimidine kinase | 0.12766 | 0.00300 | 0.13198 | 0.00165 | 0.00503 | 65 | 0.12464 | 0.00125 | 0.00367 | 93 | 0.00623 | 1.17772 | 125.511 | 0.24113 | 0.70632 |
| Phosphomethylpyrimidine kinase | 0.12766 | 0.00300 | 0.13198 | 0.00165 | 0.00503 | 65 | 0.12464 | 0.00125 | 0.00367 | 93 | 0.00623 | 1.17772 | 125.511 | 0.24113 | 0.70632 |
| DNA (cytosine-5-)-methyltransferase | 0.12755 | 0.00579 | 0.12324 | 0.00353 | 0.00737 | 65 | 0.13056 | 0.00656 | 0.00840 | 93 | 0.01117 | -0.65546 | 155.605 | 0.51314 | 0.81548 |
| Succinate–CoA ligase (ADP-forming) | 0.12754 | 0.00515 | 0.12044 | 0.00449 | 0.00831 | 65 | 0.13250 | 0.00396 | 0.00653 | 93 | 0.01057 | -1.14097 | 132.233 | 0.25595 | 0.70632 |
| Tripeptide aminopeptidase | 0.12744 | 0.00553 | 0.13155 | 0.00541 | 0.00912 | 65 | 0.12457 | 0.00447 | 0.00693 | 93 | 0.01146 | 0.60904 | 129.295 | 0.54356 | 0.84121 |
| Pyrroline-5-carboxylate reductase | 0.12728 | 0.00273 | 0.12669 | 0.00130 | 0.00447 | 65 | 0.12769 | 0.00110 | 0.00344 | 93 | 0.00564 | -0.17773 | 130.253 | 0.85921 | 0.96186 |
| Exopolyphosphatase | 0.12507 | 0.00481 | 0.13599 | 0.00446 | 0.00828 | 65 | 0.11744 | 0.00300 | 0.00568 | 93 | 0.01004 | 1.84676 | 119.922 | 0.06725 | 0.70632 |
| Guanosine-5’-triphosphate,3’-diphosphate diphosphatase | 0.12507 | 0.00481 | 0.13599 | 0.00446 | 0.00828 | 65 | 0.11744 | 0.00300 | 0.00568 | 93 | 0.01004 | 1.84676 | 119.922 | 0.06725 | 0.70632 |
| Dihydrolipoyllysine-residue acetyltransferase | 0.12504 | 0.00437 | 0.12168 | 0.00293 | 0.00671 | 65 | 0.12738 | 0.00309 | 0.00577 | 93 | 0.00885 | -0.64472 | 140.236 | 0.52016 | 0.82181 |
| Formate–tetrahydrofolate ligase | 0.12490 | 0.00504 | 0.12972 | 0.00404 | 0.00788 | 65 | 0.12153 | 0.00402 | 0.00657 | 93 | 0.01026 | 0.79805 | 137.621 | 0.42622 | 0.76494 |
| DNA-3-methyladenine glycosylase I | 0.12478 | 0.00270 | 0.12635 | 0.00120 | 0.00430 | 65 | 0.12369 | 0.00113 | 0.00349 | 93 | 0.00553 | 0.48055 | 135.233 | 0.63161 | 0.88317 |
| O-acetylhomoserine aminocarboxypropyltransferase | 0.12453 | 0.00454 | 0.11695 | 0.00314 | 0.00695 | 65 | 0.12983 | 0.00331 | 0.00597 | 93 | 0.00916 | -1.40685 | 140.126 | 0.16169 | 0.70632 |
| Beta-N-acetylhexosaminidase | 0.12398 | 0.00448 | 0.11761 | 0.00272 | 0.00647 | 65 | 0.12843 | 0.00347 | 0.00611 | 93 | 0.00890 | -1.21621 | 147.422 | 0.22585 | 0.70632 |
| DNA-formamidopyrimidine glycosylase | 0.12394 | 0.00349 | 0.12443 | 0.00220 | 0.00582 | 65 | 0.12360 | 0.00176 | 0.00435 | 93 | 0.00727 | 0.11406 | 127.643 | 0.90937 | 0.97756 |
| Glucose-6-phosphate dehydrogenase (NAD(P)(+)) | 0.12387 | 0.00374 | 0.12308 | 0.00247 | 0.00616 | 65 | 0.12442 | 0.00206 | 0.00471 | 93 | 0.00775 | -0.17368 | 129.849 | 0.86239 | 0.96186 |
| Glucose-6-phosphate dehydrogenase (NADP(+)) | 0.12387 | 0.00374 | 0.12308 | 0.00247 | 0.00616 | 65 | 0.12442 | 0.00206 | 0.00471 | 93 | 0.00775 | -0.17368 | 129.849 | 0.86239 | 0.96186 |
| Aminodeoxychorismate synthase | 0.12364 | 0.00440 | 0.12336 | 0.00380 | 0.00765 | 65 | 0.12384 | 0.00258 | 0.00527 | 93 | 0.00928 | -0.05193 | 120.331 | 0.95867 | 0.99187 |
| Adenosylhomocysteine nucleosidase | 0.12237 | 0.00367 | 0.12424 | 0.00210 | 0.00568 | 65 | 0.12106 | 0.00217 | 0.00483 | 93 | 0.00746 | 0.42654 | 139.287 | 0.67037 | 0.89503 |
| Phosphoglycerate mutase (2,3-diphosphoglycerate-dependent) | 0.12167 | 0.00529 | 0.12989 | 0.00553 | 0.00923 | 65 | 0.11593 | 0.00362 | 0.00624 | 93 | 0.01114 | 1.25324 | 118.649 | 0.21258 | 0.70632 |
| Enoyl-CoA hydratase | 0.12147 | 0.00871 | 0.11100 | 0.01157 | 0.01334 | 65 | 0.12879 | 0.01225 | 0.01148 | 93 | 0.01760 | -1.01071 | 140.322 | 0.31390 | 0.70632 |
| Peptidase Do | 0.12147 | 0.00323 | 0.11840 | 0.00153 | 0.00485 | 65 | 0.12361 | 0.00174 | 0.00433 | 93 | 0.00650 | -0.80152 | 143.277 | 0.42416 | 0.76494 |
| dTDP-glucose 4,6-dehydratase | 0.12139 | 0.00350 | 0.13328 | 0.00248 | 0.00618 | 65 | 0.11308 | 0.00141 | 0.00390 | 93 | 0.00731 | 2.76447 | 112.681 | 0.00666 | 0.62334 |
| Glycerol kinase | 0.12137 | 0.00206 | 0.12217 | 0.00053 | 0.00285 | 65 | 0.12081 | 0.00078 | 0.00289 | 93 | 0.00406 | 0.33580 | 151.824 | 0.73749 | 0.92470 |
| 2-isopropylmalate synthase | 0.12086 | 0.00322 | 0.11628 | 0.00192 | 0.00543 | 65 | 0.12406 | 0.00144 | 0.00394 | 93 | 0.00671 | -1.16081 | 124.966 | 0.24793 | 0.70632 |
| 1-phosphofructokinase | 0.12067 | 0.00397 | 0.12736 | 0.00205 | 0.00562 | 65 | 0.11599 | 0.00278 | 0.00546 | 93 | 0.00784 | 1.44989 | 149.392 | 0.14919 | 0.70632 |
| 5-methyltetrahydropteroyltriglutamate–homocysteine S-methyltransferase | 0.11885 | 0.00378 | 0.12240 | 0.00237 | 0.00604 | 65 | 0.11636 | 0.00219 | 0.00485 | 93 | 0.00775 | 0.77918 | 134.339 | 0.43724 | 0.77235 |
| Fructokinase | 0.11875 | 0.00319 | 0.11850 | 0.00172 | 0.00515 | 65 | 0.11893 | 0.00155 | 0.00408 | 93 | 0.00657 | -0.06615 | 133.051 | 0.94735 | 0.99049 |
| Uridine kinase | 0.11858 | 0.00517 | 0.11835 | 0.00429 | 0.00812 | 65 | 0.11875 | 0.00423 | 0.00675 | 93 | 0.01056 | -0.03744 | 137.273 | 0.97019 | 0.99273 |
| Endopeptidase La | 0.11857 | 0.00498 | 0.12601 | 0.00569 | 0.00935 | 65 | 0.11338 | 0.00267 | 0.00536 | 93 | 0.01078 | 1.17202 | 105.007 | 0.24384 | 0.70632 |
| 1-deoxy-D-xylulose-5-phosphate synthase | 0.11839 | 0.00396 | 0.12388 | 0.00310 | 0.00691 | 65 | 0.11456 | 0.00204 | 0.00468 | 93 | 0.00835 | 1.11744 | 118.893 | 0.26606 | 0.70632 |
| 3,4-dihydroxy-2-butanone-4-phosphate synthase | 0.11632 | 0.00342 | 0.11852 | 0.00188 | 0.00538 | 65 | 0.11479 | 0.00183 | 0.00444 | 93 | 0.00697 | 0.53500 | 136.668 | 0.59352 | 0.86115 |
| Selenocysteine lyase | 0.11596 | 0.00365 | 0.11448 | 0.00221 | 0.00583 | 65 | 0.11700 | 0.00205 | 0.00470 | 93 | 0.00749 | -0.33736 | 134.613 | 0.73637 | 0.92385 |
| Glutamate 5-kinase | 0.11481 | 0.00253 | 0.11586 | 0.00108 | 0.00408 | 65 | 0.11408 | 0.00097 | 0.00322 | 93 | 0.00520 | 0.34187 | 132.722 | 0.73299 | 0.92120 |
| Uroporphyrinogen-III C-methyltransferase | 0.11393 | 0.00474 | 0.11719 | 0.00362 | 0.00746 | 65 | 0.11164 | 0.00352 | 0.00616 | 93 | 0.00967 | 0.57388 | 136.749 | 0.56699 | 0.85024 |
| Glutamate dehydrogenase (NADP(+)) | 0.11391 | 0.00444 | 0.10974 | 0.00315 | 0.00696 | 65 | 0.11683 | 0.00310 | 0.00577 | 93 | 0.00904 | -0.78463 | 137.246 | 0.43403 | 0.77079 |
| 6-phosphogluconolactonase | 0.11382 | 0.00360 | 0.11582 | 0.00228 | 0.00593 | 65 | 0.11242 | 0.00191 | 0.00453 | 93 | 0.00746 | 0.45590 | 129.787 | 0.64922 | 0.88832 |
| Galactokinase | 0.11360 | 0.00439 | 0.12051 | 0.00290 | 0.00668 | 65 | 0.10877 | 0.00311 | 0.00579 | 93 | 0.00883 | 1.32930 | 140.913 | 0.18590 | 0.70632 |
| Glucosamine-6-phosphate deaminase | 0.11352 | 0.00479 | 0.11568 | 0.00329 | 0.00712 | 65 | 0.11201 | 0.00388 | 0.00646 | 93 | 0.00961 | 0.38195 | 144.648 | 0.70306 | 0.91393 |
| Phosphogluconate dehydrogenase (NAD(+)-dependent, decarboxylating) | 0.11325 | 0.00333 | 0.11216 | 0.00195 | 0.00547 | 65 | 0.11401 | 0.00163 | 0.00419 | 93 | 0.00689 | -0.26848 | 129.916 | 0.78875 | 0.93676 |
| Phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating) | 0.11325 | 0.00333 | 0.11216 | 0.00195 | 0.00547 | 65 | 0.11401 | 0.00163 | 0.00419 | 93 | 0.00689 | -0.26848 | 129.916 | 0.78875 | 0.93676 |
| Ketol-acid reductoisomerase (NADP(+)) | 0.11300 | 0.00279 | 0.10918 | 0.00154 | 0.00487 | 65 | 0.11567 | 0.00101 | 0.00330 | 93 | 0.00589 | -1.10168 | 118.838 | 0.27283 | 0.70632 |
| Kdo(2)-lipid IV(A) lauroyltransferase | 0.11280 | 0.00455 | 0.11806 | 0.00326 | 0.00709 | 65 | 0.10913 | 0.00328 | 0.00594 | 93 | 0.00925 | 0.96607 | 138.084 | 0.33570 | 0.71823 |
| 23S rRNA (cytosine(1962)-C(5))-methyltransferase | 0.11268 | 0.00314 | 0.11279 | 0.00179 | 0.00525 | 65 | 0.11260 | 0.00141 | 0.00390 | 93 | 0.00654 | 0.02957 | 127.179 | 0.97645 | 0.99435 |
| Glutamate-5-semialdehyde dehydrogenase | 0.11244 | 0.00266 | 0.11337 | 0.00118 | 0.00426 | 65 | 0.11179 | 0.00108 | 0.00341 | 93 | 0.00546 | 0.29076 | 133.987 | 0.77168 | 0.93540 |
| Deoxyribose-phosphate aldolase | 0.11205 | 0.00412 | 0.11317 | 0.00291 | 0.00669 | 65 | 0.11127 | 0.00255 | 0.00524 | 93 | 0.00850 | 0.22376 | 131.956 | 0.82329 | 0.95060 |
| Prephenate dehydrogenase | 0.11194 | 0.00342 | 0.10926 | 0.00212 | 0.00572 | 65 | 0.11382 | 0.00166 | 0.00422 | 93 | 0.00711 | -0.64036 | 126.705 | 0.52309 | 0.82182 |
| Glycerate 3-kinase | 0.11180 | 0.00305 | 0.11238 | 0.00151 | 0.00482 | 65 | 0.11139 | 0.00146 | 0.00396 | 93 | 0.00624 | 0.15910 | 136.367 | 0.87383 | 0.96378 |
| Xanthine phosphoribosyltransferase | 0.11082 | 0.00332 | 0.11080 | 0.00196 | 0.00549 | 65 | 0.11083 | 0.00161 | 0.00416 | 93 | 0.00688 | -0.00340 | 129.072 | 0.99729 | 0.99921 |
| Phosphoserine phosphatase | 0.11080 | 0.00350 | 0.10582 | 0.00198 | 0.00553 | 65 | 0.11429 | 0.00190 | 0.00451 | 93 | 0.00714 | -1.18667 | 135.865 | 0.23743 | 0.70632 |
| tRNA(adenine(34)) deaminase | 0.11036 | 0.00392 | 0.10965 | 0.00256 | 0.00628 | 65 | 0.11086 | 0.00237 | 0.00505 | 93 | 0.00806 | -0.15010 | 134.379 | 0.88091 | 0.96590 |
| D-alanine–poly(phosphoribitol) ligase | 0.10949 | 0.00745 | 0.11157 | 0.00943 | 0.01205 | 65 | 0.10804 | 0.00839 | 0.00950 | 93 | 0.01534 | 0.23017 | 132.654 | 0.81832 | 0.94790 |
| 2-oxoacid oxidoreductase (ferredoxin) | 0.10896 | 0.00785 | 0.09545 | 0.00767 | 0.01086 | 65 | 0.11841 | 0.01104 | 0.01090 | 93 | 0.01539 | -1.49229 | 151.152 | 0.13771 | 0.70632 |
| CCA tRNA nucleotidyltransferase | 0.10891 | 0.00276 | 0.11621 | 0.00107 | 0.00406 | 65 | 0.10381 | 0.00124 | 0.00366 | 93 | 0.00546 | 2.26947 | 143.991 | 0.02473 | 0.67652 |
| 1,4-alpha-glucan branching enzyme | 0.10886 | 0.00366 | 0.11093 | 0.00223 | 0.00585 | 65 | 0.10741 | 0.00206 | 0.00470 | 93 | 0.00751 | 0.46852 | 134.375 | 0.64017 | 0.88568 |
| 3-isopropylmalate dehydrogenase | 0.10874 | 0.00284 | 0.10578 | 0.00159 | 0.00495 | 65 | 0.11082 | 0.00106 | 0.00338 | 93 | 0.00599 | -0.84171 | 119.413 | 0.40163 | 0.75271 |
| Holo-[acyl-carrier-protein] synthase | 0.10858 | 0.00383 | 0.11342 | 0.00232 | 0.00597 | 65 | 0.10519 | 0.00231 | 0.00499 | 93 | 0.00778 | 1.05715 | 137.772 | 0.29229 | 0.70632 |
| N-acetylglucosamine-6-phosphate deacetylase | 0.10855 | 0.00355 | 0.10874 | 0.00173 | 0.00517 | 65 | 0.10841 | 0.00219 | 0.00486 | 93 | 0.00709 | 0.04625 | 147.174 | 0.96317 | 0.99193 |
| Inositol-phosphate phosphatase | 0.10845 | 0.00380 | 0.10242 | 0.00216 | 0.00576 | 65 | 0.11266 | 0.00235 | 0.00503 | 93 | 0.00765 | -1.33993 | 141.574 | 0.18241 | 0.70632 |
| Glutamate–tRNA ligase | 0.10832 | 0.00371 | 0.10893 | 0.00198 | 0.00552 | 65 | 0.10789 | 0.00233 | 0.00501 | 93 | 0.00746 | 0.13980 | 144.572 | 0.88901 | 0.96887 |
| Methylenetetrahydrofolate reductase (NAD(P)H) | 0.10800 | 0.00320 | 0.10608 | 0.00194 | 0.00546 | 65 | 0.10935 | 0.00141 | 0.00389 | 93 | 0.00671 | -0.48723 | 123.336 | 0.62696 | 0.88183 |
| Beta-glucosidase | 0.10752 | 0.00718 | 0.09650 | 0.00492 | 0.00870 | 65 | 0.11522 | 0.01032 | 0.01054 | 93 | 0.01366 | -1.36977 | 155.985 | 0.17273 | 0.70632 |
| Glycerol-3-phosphate dehydrogenase | 0.10666 | 0.00422 | 0.10623 | 0.00263 | 0.00636 | 65 | 0.10696 | 0.00296 | 0.00565 | 93 | 0.00851 | -0.08516 | 142.852 | 0.93225 | 0.98985 |
| 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | 0.10653 | 0.00333 | 0.10802 | 0.00217 | 0.00577 | 65 | 0.10549 | 0.00148 | 0.00399 | 93 | 0.00702 | 0.36069 | 120.549 | 0.71897 | 0.92120 |
| 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase | 0.10600 | 0.00242 | 0.10873 | 0.00088 | 0.00367 | 65 | 0.10410 | 0.00096 | 0.00321 | 93 | 0.00488 | 0.94922 | 141.677 | 0.34413 | 0.72019 |
| Cytidine deaminase | 0.10586 | 0.00416 | 0.10466 | 0.00288 | 0.00666 | 65 | 0.10670 | 0.00267 | 0.00536 | 93 | 0.00854 | -0.23940 | 134.488 | 0.81116 | 0.94519 |
| 5-(carboxyamino)imidazole ribonucleotide synthase | 0.10580 | 0.00347 | 0.10600 | 0.00237 | 0.00604 | 65 | 0.10567 | 0.00159 | 0.00414 | 93 | 0.00732 | 0.04422 | 119.795 | 0.96481 | 0.99193 |
| Indole-3-glycerol-phosphate synthase | 0.10555 | 0.00277 | 0.10056 | 0.00124 | 0.00437 | 65 | 0.10904 | 0.00117 | 0.00355 | 93 | 0.00563 | -1.50631 | 135.340 | 0.13432 | 0.70632 |
| Anthranilate phosphoribosyltransferase | 0.10537 | 0.00276 | 0.10116 | 0.00142 | 0.00468 | 65 | 0.10832 | 0.00104 | 0.00334 | 93 | 0.00575 | -1.24369 | 123.577 | 0.21597 | 0.70632 |
| Oligonucleotidase | 0.10480 | 0.00432 | 0.10888 | 0.00305 | 0.00685 | 65 | 0.10195 | 0.00288 | 0.00557 | 93 | 0.00883 | 0.78397 | 135.289 | 0.43443 | 0.77079 |
| Starch synthase | 0.10451 | 0.00351 | 0.10637 | 0.00203 | 0.00559 | 65 | 0.10321 | 0.00191 | 0.00453 | 93 | 0.00719 | 0.43906 | 134.996 | 0.66132 | 0.88968 |
| Galactose-6-phosphate isomerase | 0.10451 | 0.00795 | 0.10101 | 0.00767 | 0.01086 | 65 | 0.10695 | 0.01167 | 0.01120 | 93 | 0.01560 | -0.38013 | 152.523 | 0.70438 | 0.91393 |
| Ferrochelatase | 0.10372 | 0.00349 | 0.09842 | 0.00189 | 0.00539 | 65 | 0.10742 | 0.00194 | 0.00457 | 93 | 0.00707 | -1.27324 | 139.086 | 0.20506 | 0.70632 |
| dTDP-4-dehydrorhamnose 3,5-epimerase | 0.10306 | 0.00373 | 0.11256 | 0.00310 | 0.00691 | 65 | 0.09642 | 0.00148 | 0.00399 | 93 | 0.00798 | 2.02133 | 105.692 | 0.04577 | 0.69348 |
| Asparagine–tRNA ligase | 0.10295 | 0.00385 | 0.10736 | 0.00234 | 0.00600 | 65 | 0.09987 | 0.00235 | 0.00503 | 93 | 0.00783 | 0.95608 | 137.997 | 0.34070 | 0.72019 |
| NAD(P)(+) transhydrogenase (Re/Si-specific) | 0.10185 | 0.00719 | 0.09609 | 0.00850 | 0.01143 | 65 | 0.10588 | 0.00800 | 0.00928 | 93 | 0.01472 | -0.66458 | 135.213 | 0.50745 | 0.81454 |
| 16S rRNA (guanine(1207)-N(2))-methyltransferase | 0.10145 | 0.00354 | 0.10540 | 0.00173 | 0.00516 | 65 | 0.09869 | 0.00217 | 0.00483 | 93 | 0.00706 | 0.95021 | 146.853 | 0.34357 | 0.72019 |
| 4-alpha-glucanotransferase | 0.10145 | 0.00419 | 0.10633 | 0.00290 | 0.00667 | 65 | 0.09804 | 0.00270 | 0.00539 | 93 | 0.00858 | 0.96657 | 134.802 | 0.33549 | 0.71823 |
| NAD(+) synthase | 0.10141 | 0.00329 | 0.10563 | 0.00169 | 0.00510 | 65 | 0.09847 | 0.00172 | 0.00430 | 93 | 0.00667 | 1.07361 | 138.634 | 0.28486 | 0.70632 |
| dCMP deaminase | 0.10114 | 0.00435 | 0.10568 | 0.00309 | 0.00689 | 65 | 0.09797 | 0.00294 | 0.00562 | 93 | 0.00889 | 0.86716 | 135.658 | 0.38739 | 0.73978 |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | 0.10099 | 0.00306 | 0.10132 | 0.00167 | 0.00507 | 65 | 0.10077 | 0.00136 | 0.00383 | 93 | 0.00635 | 0.08617 | 128.745 | 0.93147 | 0.98985 |
| 6,7-dimethyl-8-ribityllumazine synthase | 0.10095 | 0.00283 | 0.09926 | 0.00120 | 0.00430 | 65 | 0.10212 | 0.00132 | 0.00377 | 93 | 0.00572 | -0.49986 | 141.886 | 0.61795 | 0.87497 |
| 16S rRNA (cytidine(1409)-2’-O)-methyltransferase | 0.10046 | 0.00381 | 0.10535 | 0.00239 | 0.00606 | 65 | 0.09704 | 0.00222 | 0.00488 | 93 | 0.00778 | 1.06683 | 134.674 | 0.28796 | 0.70632 |
| 23S rRNA (cytidine(1920)-2’-O)-methyltransferase | 0.10046 | 0.00381 | 0.10535 | 0.00239 | 0.00606 | 65 | 0.09704 | 0.00222 | 0.00488 | 93 | 0.00778 | 1.06683 | 134.674 | 0.28796 | 0.70632 |
| Argininosuccinate lyase | 0.10025 | 0.00249 | 0.09764 | 0.00097 | 0.00387 | 65 | 0.10208 | 0.00099 | 0.00327 | 93 | 0.00506 | -0.87709 | 138.706 | 0.38196 | 0.73808 |
| NADH dehydrogenase | 0.09999 | 0.00352 | 0.09221 | 0.00176 | 0.00521 | 65 | 0.10542 | 0.00204 | 0.00468 | 93 | 0.00700 | -1.88539 | 143.941 | 0.06139 | 0.70632 |
| Diacylglycerol kinase (ATP) | 0.09985 | 0.00303 | 0.10153 | 0.00149 | 0.00479 | 65 | 0.09868 | 0.00144 | 0.00393 | 93 | 0.00620 | 0.46004 | 136.335 | 0.64622 | 0.88749 |
| Pyridoxal 5’-phosphate synthase (glutamine hydrolyzing) | 0.09974 | 0.00528 | 0.09146 | 0.00361 | 0.00746 | 65 | 0.10552 | 0.00491 | 0.00727 | 93 | 0.01041 | -1.35092 | 149.493 | 0.17876 | 0.70632 |
| Beta-fructofuranosidase | 0.09904 | 0.00509 | 0.10731 | 0.00512 | 0.00888 | 65 | 0.09327 | 0.00334 | 0.00600 | 93 | 0.01071 | 1.31147 | 118.542 | 0.19223 | 0.70632 |
| Membrane alanyl aminopeptidase | 0.09870 | 0.00400 | 0.09859 | 0.00292 | 0.00670 | 65 | 0.09878 | 0.00228 | 0.00495 | 93 | 0.00833 | -0.02282 | 126.710 | 0.98183 | 0.99457 |
| Ornithine carbamoyltransferase | 0.09870 | 0.00318 | 0.10369 | 0.00164 | 0.00502 | 65 | 0.09521 | 0.00155 | 0.00408 | 93 | 0.00647 | 1.31099 | 135.465 | 0.19208 | 0.70632 |
| S-ribosylhomocysteine lyase | 0.09849 | 0.00417 | 0.10026 | 0.00283 | 0.00660 | 65 | 0.09725 | 0.00273 | 0.00542 | 93 | 0.00853 | 0.35279 | 136.299 | 0.72479 | 0.92120 |
| Maltose O-acetyltransferase | 0.09846 | 0.00594 | 0.10145 | 0.00752 | 0.01076 | 65 | 0.09637 | 0.00426 | 0.00677 | 93 | 0.01271 | 0.39952 | 112.438 | 0.69027 | 0.90855 |
| tRNA-2-methylthio-N(6)-dimethylallyladenosine synthase | 0.09845 | 0.00304 | 0.09804 | 0.00162 | 0.00500 | 65 | 0.09873 | 0.00137 | 0.00383 | 93 | 0.00630 | -0.10911 | 130.137 | 0.91329 | 0.97955 |
| (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ferredoxin) | 0.09805 | 0.00273 | 0.09785 | 0.00130 | 0.00447 | 65 | 0.09819 | 0.00110 | 0.00344 | 93 | 0.00564 | -0.06118 | 130.412 | 0.95131 | 0.99049 |
| (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (flavodoxin) | 0.09805 | 0.00273 | 0.09785 | 0.00130 | 0.00447 | 65 | 0.09819 | 0.00110 | 0.00344 | 93 | 0.00564 | -0.06118 | 130.412 | 0.95131 | 0.99049 |
| Repressor LexA | 0.09804 | 0.00288 | 0.10315 | 0.00136 | 0.00458 | 65 | 0.09447 | 0.00126 | 0.00368 | 93 | 0.00587 | 1.47676 | 134.285 | 0.14208 | 0.70632 |
| dTDP-4-dehydrorhamnose reductase | 0.09802 | 0.00299 | 0.10319 | 0.00150 | 0.00481 | 65 | 0.09440 | 0.00134 | 0.00379 | 93 | 0.00612 | 1.43565 | 132.602 | 0.15346 | 0.70632 |
| 2-iminobutanoate/2-iminopropanoate deaminase | 0.09794 | 0.00420 | 0.09835 | 0.00285 | 0.00662 | 65 | 0.09765 | 0.00277 | 0.00546 | 93 | 0.00858 | 0.08231 | 136.670 | 0.93452 | 0.98985 |
| Phosphinothricin acetyltransferase | 0.09791 | 0.00263 | 0.09582 | 0.00133 | 0.00452 | 65 | 0.09938 | 0.00093 | 0.00317 | 93 | 0.00552 | -0.64483 | 121.888 | 0.52025 | 0.82181 |
| CDP-diacylglycerol–serine O-phosphatidyltransferase | 0.09783 | 0.00307 | 0.09656 | 0.00159 | 0.00495 | 65 | 0.09871 | 0.00144 | 0.00393 | 93 | 0.00632 | -0.34076 | 133.396 | 0.73382 | 0.92120 |
| Glucose-1-phosphate thymidylyltransferase | 0.09714 | 0.00282 | 0.09921 | 0.00141 | 0.00465 | 65 | 0.09570 | 0.00117 | 0.00354 | 93 | 0.00585 | 0.60116 | 129.329 | 0.54879 | 0.84333 |
| Diadenylate cyclase | 0.09690 | 0.00412 | 0.10292 | 0.00282 | 0.00659 | 65 | 0.09269 | 0.00256 | 0.00525 | 93 | 0.00842 | 1.21375 | 133.547 | 0.22699 | 0.70632 |
| Cystathionine gamma-synthase | 0.09668 | 0.00359 | 0.10125 | 0.00257 | 0.00629 | 65 | 0.09349 | 0.00166 | 0.00423 | 93 | 0.00758 | 1.02389 | 118.055 | 0.30798 | 0.70632 |
| NAD(P)H dehydrogenase (quinone) | 0.09661 | 0.00505 | 0.09646 | 0.00415 | 0.00799 | 65 | 0.09672 | 0.00398 | 0.00654 | 93 | 0.01033 | -0.02498 | 135.975 | 0.98011 | 0.99457 |
| 1-deoxy-D-xylulose-5-phosphate reductoisomerase | 0.09646 | 0.00274 | 0.09656 | 0.00126 | 0.00441 | 65 | 0.09639 | 0.00115 | 0.00352 | 93 | 0.00564 | 0.03061 | 133.789 | 0.97563 | 0.99435 |
| Formate dehydrogenase | 0.09633 | 0.00609 | 0.09778 | 0.00621 | 0.00977 | 65 | 0.09532 | 0.00567 | 0.00781 | 93 | 0.01251 | 0.19663 | 133.799 | 0.84441 | 0.95939 |
| Uroporphyrinogen-III synthase | 0.09597 | 0.00319 | 0.09553 | 0.00134 | 0.00455 | 65 | 0.09627 | 0.00181 | 0.00441 | 93 | 0.00634 | -0.11675 | 149.262 | 0.90721 | 0.97700 |
| Thymidine kinase | 0.09564 | 0.00355 | 0.09415 | 0.00206 | 0.00563 | 65 | 0.09669 | 0.00196 | 0.00459 | 93 | 0.00726 | -0.34987 | 135.765 | 0.72698 | 0.92120 |
| 5-amino-6-(5-phosphoribosylamino)uracil reductase | 0.09517 | 0.00269 | 0.09497 | 0.00122 | 0.00433 | 65 | 0.09531 | 0.00110 | 0.00344 | 93 | 0.00553 | -0.06131 | 133.354 | 0.95120 | 0.99049 |
| 23S rRNA pseudouridine(746) synthase | 0.09515 | 0.00449 | 0.09221 | 0.00315 | 0.00696 | 65 | 0.09721 | 0.00323 | 0.00589 | 93 | 0.00912 | -0.54794 | 138.997 | 0.58461 | 0.85509 |
| tRNA pseudouridine(32) synthase | 0.09515 | 0.00449 | 0.09221 | 0.00315 | 0.00696 | 65 | 0.09721 | 0.00323 | 0.00589 | 93 | 0.00912 | -0.54794 | 138.997 | 0.58461 | 0.85509 |
| Diaminohydroxyphosphoribosylaminopyrimidine deaminase | 0.09458 | 0.00267 | 0.09463 | 0.00122 | 0.00433 | 65 | 0.09455 | 0.00108 | 0.00341 | 93 | 0.00551 | 0.01556 | 132.535 | 0.98761 | 0.99660 |
| Cadmium-exporting ATPase | 0.09445 | 0.00421 | 0.10084 | 0.00358 | 0.00742 | 65 | 0.08998 | 0.00224 | 0.00491 | 93 | 0.00889 | 1.22187 | 116.746 | 0.22422 | 0.70632 |
| Zinc-exporting ATPase | 0.09445 | 0.00421 | 0.10084 | 0.00358 | 0.00742 | 65 | 0.08998 | 0.00224 | 0.00491 | 93 | 0.00889 | 1.22187 | 116.746 | 0.22422 | 0.70632 |
| Nucleoside-diphosphate kinase | 0.09431 | 0.00227 | 0.09494 | 0.00107 | 0.00406 | 65 | 0.09387 | 0.00064 | 0.00262 | 93 | 0.00484 | 0.22006 | 114.639 | 0.82621 | 0.95262 |
| Dihydroorotate dehydrogenase (NAD(+)) | 0.09384 | 0.00432 | 0.09815 | 0.00290 | 0.00668 | 65 | 0.09083 | 0.00300 | 0.00568 | 93 | 0.00877 | 0.83439 | 139.349 | 0.40549 | 0.75734 |
| Argininosuccinate synthase | 0.09362 | 0.00236 | 0.09365 | 0.00101 | 0.00394 | 65 | 0.09360 | 0.00080 | 0.00294 | 93 | 0.00491 | 0.00946 | 127.507 | 0.99246 | 0.99860 |
| Thiamine diphosphokinase | 0.09307 | 0.00419 | 0.09599 | 0.00293 | 0.00671 | 65 | 0.09103 | 0.00268 | 0.00537 | 93 | 0.00859 | 0.57698 | 133.944 | 0.56492 | 0.85024 |
| Peptidoglycan glycosyltransferase | 0.09295 | 0.00308 | 0.09682 | 0.00164 | 0.00502 | 65 | 0.09024 | 0.00140 | 0.00388 | 93 | 0.00635 | 1.03604 | 130.846 | 0.30209 | 0.70632 |
| 2-dehydropantoate 2-reductase | 0.09261 | 0.00416 | 0.09531 | 0.00344 | 0.00727 | 65 | 0.09072 | 0.00226 | 0.00494 | 93 | 0.00879 | 0.52292 | 118.981 | 0.60201 | 0.86633 |
| 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | 0.09249 | 0.00328 | 0.09118 | 0.00172 | 0.00514 | 65 | 0.09340 | 0.00170 | 0.00427 | 93 | 0.00669 | -0.33194 | 137.353 | 0.74044 | 0.92600 |
| Histidine ammonia-lyase | 0.09233 | 0.00467 | 0.09129 | 0.00374 | 0.00759 | 65 | 0.09306 | 0.00328 | 0.00594 | 93 | 0.00964 | -0.18359 | 131.970 | 0.85462 | 0.96150 |
| Ribonuclease Z | 0.09233 | 0.00359 | 0.09168 | 0.00219 | 0.00580 | 65 | 0.09278 | 0.00195 | 0.00458 | 93 | 0.00739 | -0.14913 | 132.647 | 0.88167 | 0.96591 |
| Amino-acid N-acetyltransferase | 0.09193 | 0.00467 | 0.08831 | 0.00376 | 0.00760 | 65 | 0.09445 | 0.00325 | 0.00591 | 93 | 0.00963 | -0.63744 | 131.363 | 0.52495 | 0.82264 |
| Alpha-amylase | 0.09132 | 0.00394 | 0.08653 | 0.00233 | 0.00599 | 65 | 0.09467 | 0.00254 | 0.00523 | 93 | 0.00795 | -1.02396 | 141.487 | 0.30760 | 0.70632 |
| UDP-glucose–hexose-1-phosphate uridylyltransferase | 0.09028 | 0.00381 | 0.09961 | 0.00236 | 0.00603 | 65 | 0.08376 | 0.00216 | 0.00482 | 93 | 0.00772 | 2.05437 | 133.920 | 0.04188 | 0.68639 |
| Riboflavin synthase | 0.09022 | 0.00283 | 0.09103 | 0.00138 | 0.00461 | 65 | 0.08965 | 0.00119 | 0.00358 | 93 | 0.00584 | 0.23799 | 131.251 | 0.81226 | 0.94519 |
| D-lactate dehydrogenase | 0.09011 | 0.00635 | 0.10350 | 0.00866 | 0.01155 | 65 | 0.08075 | 0.00461 | 0.00704 | 93 | 0.01352 | 1.68247 | 109.916 | 0.09532 | 0.70632 |
| Calcium-transporting ATPase | 0.08979 | 0.00501 | 0.09196 | 0.00433 | 0.00816 | 65 | 0.08826 | 0.00375 | 0.00635 | 93 | 0.01034 | 0.35780 | 131.412 | 0.72107 | 0.92120 |
| Diaminopimelate epimerase | 0.08973 | 0.00254 | 0.09080 | 0.00111 | 0.00414 | 65 | 0.08898 | 0.00097 | 0.00322 | 93 | 0.00525 | 0.34692 | 131.585 | 0.72921 | 0.92120 |
| Pyruvate synthase | 0.08959 | 0.01013 | 0.11141 | 0.02661 | 0.02023 | 65 | 0.07434 | 0.00856 | 0.00959 | 93 | 0.02239 | 1.65540 | 92.747 | 0.10122 | 0.70632 |
| UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing) | 0.08958 | 0.00326 | 0.08798 | 0.00194 | 0.00546 | 65 | 0.09070 | 0.00151 | 0.00402 | 93 | 0.00678 | -0.40108 | 126.402 | 0.68904 | 0.90847 |
| Biotin synthase | 0.08946 | 0.00352 | 0.09241 | 0.00222 | 0.00585 | 65 | 0.08739 | 0.00178 | 0.00437 | 93 | 0.00730 | 0.68657 | 127.770 | 0.49360 | 0.80568 |
| 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase | 0.08942 | 0.00281 | 0.08857 | 0.00129 | 0.00446 | 65 | 0.09002 | 0.00123 | 0.00364 | 93 | 0.00576 | -0.25095 | 135.835 | 0.80223 | 0.94263 |
| Demethylmenaquinone methyltransferase | 0.08942 | 0.00281 | 0.08857 | 0.00129 | 0.00446 | 65 | 0.09002 | 0.00123 | 0.00364 | 93 | 0.00576 | -0.25095 | 135.835 | 0.80223 | 0.94263 |
| Uroporphyrinogen decarboxylase | 0.08911 | 0.00324 | 0.08319 | 0.00157 | 0.00492 | 65 | 0.09325 | 0.00169 | 0.00427 | 93 | 0.00651 | -1.54595 | 141.017 | 0.12436 | 0.70632 |
| Glutamate-1-semialdehyde 2,1-aminomutase | 0.08831 | 0.00325 | 0.09091 | 0.00141 | 0.00465 | 65 | 0.08649 | 0.00186 | 0.00447 | 93 | 0.00645 | 0.68516 | 148.573 | 0.49431 | 0.80568 |
| Phosphatidylserine decarboxylase | 0.08821 | 0.00258 | 0.08816 | 0.00123 | 0.00435 | 65 | 0.08824 | 0.00094 | 0.00319 | 93 | 0.00539 | -0.01527 | 125.919 | 0.98784 | 0.99660 |
| Aminodeoxychorismate lyase | 0.08778 | 0.00297 | 0.08651 | 0.00141 | 0.00465 | 65 | 0.08867 | 0.00140 | 0.00388 | 93 | 0.00606 | -0.35660 | 137.535 | 0.72194 | 0.92120 |
| Transaldolase | 0.08752 | 0.00275 | 0.08331 | 0.00123 | 0.00434 | 65 | 0.09046 | 0.00117 | 0.00354 | 93 | 0.00561 | -1.27625 | 135.719 | 0.20405 | 0.70632 |
| Crossover junction endodeoxyribonuclease | 0.08742 | 0.00298 | 0.08950 | 0.00151 | 0.00481 | 65 | 0.08596 | 0.00135 | 0.00381 | 93 | 0.00614 | 0.57597 | 132.991 | 0.56561 | 0.85024 |
| Potassium-transporting ATPase | 0.08672 | 0.00743 | 0.07590 | 0.00862 | 0.01151 | 65 | 0.09428 | 0.00876 | 0.00971 | 93 | 0.01506 | -1.22059 | 138.594 | 0.22431 | 0.70632 |
| Alpha-L-fucosidase | 0.08597 | 0.00594 | 0.07774 | 0.00373 | 0.00758 | 65 | 0.09172 | 0.00685 | 0.00858 | 93 | 0.01145 | -1.22111 | 155.507 | 0.22389 | 0.70632 |
| Acetylornithine transaminase | 0.08589 | 0.00284 | 0.08181 | 0.00135 | 0.00455 | 65 | 0.08874 | 0.00121 | 0.00361 | 93 | 0.00581 | -1.19188 | 133.242 | 0.23543 | 0.70632 |
| Phosphoribosyl-AMP cyclohydrolase | 0.08580 | 0.00241 | 0.08158 | 0.00106 | 0.00403 | 65 | 0.08876 | 0.00080 | 0.00294 | 93 | 0.00499 | -1.43913 | 125.391 | 0.15261 | 0.70632 |
| Xaa-Pro dipeptidase | 0.08559 | 0.00371 | 0.08566 | 0.00220 | 0.00581 | 65 | 0.08555 | 0.00218 | 0.00484 | 93 | 0.00757 | 0.01389 | 137.587 | 0.98893 | 0.99660 |
| isomerase | 0.08540 | 0.00233 | 0.08255 | 0.00095 | 0.00382 | 65 | 0.08739 | 0.00079 | 0.00292 | 93 | 0.00480 | -1.00681 | 129.848 | 0.31590 | 0.70632 |
| Fe(3+)-transporting ATPase | 0.08531 | 0.00416 | 0.08816 | 0.00332 | 0.00715 | 65 | 0.08333 | 0.00235 | 0.00503 | 93 | 0.00874 | 0.55220 | 122.182 | 0.58182 | 0.85342 |
| Phosphoribosyl-ATP diphosphatase | 0.08495 | 0.00236 | 0.08134 | 0.00106 | 0.00403 | 65 | 0.08746 | 0.00076 | 0.00285 | 93 | 0.00494 | -1.23944 | 122.666 | 0.21755 | 0.70632 |
| Methylmalonyl-CoA mutase | 0.08473 | 0.00652 | 0.07974 | 0.00775 | 0.01092 | 65 | 0.08822 | 0.00603 | 0.00805 | 93 | 0.01357 | -0.62551 | 126.520 | 0.53276 | 0.83089 |
| N-acetyl-gamma-glutamyl-phosphate reductase | 0.08470 | 0.00267 | 0.08140 | 0.00135 | 0.00455 | 65 | 0.08700 | 0.00098 | 0.00325 | 93 | 0.00559 | -1.00228 | 123.506 | 0.31817 | 0.70632 |
| 7-cyano-7-deazaguanine synthase | 0.08463 | 0.00256 | 0.07954 | 0.00112 | 0.00416 | 65 | 0.08819 | 0.00096 | 0.00321 | 93 | 0.00525 | -1.64618 | 130.696 | 0.10213 | 0.70632 |
| Acetate–CoA ligase | 0.08450 | 0.00563 | 0.07138 | 0.00450 | 0.00832 | 65 | 0.09367 | 0.00520 | 0.00748 | 93 | 0.01119 | -1.99209 | 143.821 | 0.04825 | 0.69870 |
| PreQ(1) synthase | 0.08449 | 0.00255 | 0.07884 | 0.00111 | 0.00413 | 65 | 0.08845 | 0.00095 | 0.00319 | 93 | 0.00522 | -1.84088 | 130.802 | 0.06791 | 0.70632 |
| Aldehyde dehydrogenase (NAD(+)) | 0.08428 | 0.00636 | 0.06689 | 0.00469 | 0.00849 | 65 | 0.09644 | 0.00729 | 0.00885 | 93 | 0.01226 | -2.40962 | 152.995 | 0.01716 | 0.64530 |
| tRNA (adenine(22)-N(1))-methyltransferase | 0.08413 | 0.00350 | 0.08652 | 0.00215 | 0.00575 | 65 | 0.08247 | 0.00179 | 0.00439 | 93 | 0.00723 | 0.55927 | 129.769 | 0.57694 | 0.85190 |
| Polyamine-transporting ATPase | 0.08399 | 0.00386 | 0.08833 | 0.00272 | 0.00646 | 65 | 0.08095 | 0.00211 | 0.00477 | 93 | 0.00803 | 0.91784 | 126.526 | 0.36045 | 0.72316 |
| Methionine synthase | 0.08384 | 0.00374 | 0.08256 | 0.00222 | 0.00584 | 65 | 0.08473 | 0.00223 | 0.00490 | 93 | 0.00762 | -0.28365 | 138.233 | 0.77711 | 0.93675 |
| Catalase | 0.08377 | 0.00664 | 0.07514 | 0.00630 | 0.00984 | 65 | 0.08979 | 0.00741 | 0.00893 | 93 | 0.01329 | -1.10223 | 144.540 | 0.27220 | 0.70632 |
| Phosphoserine transaminase | 0.08368 | 0.00244 | 0.08009 | 0.00112 | 0.00415 | 65 | 0.08620 | 0.00081 | 0.00296 | 93 | 0.00510 | -1.19707 | 123.300 | 0.23358 | 0.70632 |
| Succinate-semialdehyde dehydrogenase (NAD(P)(+)) | 0.08341 | 0.00553 | 0.08108 | 0.00494 | 0.00872 | 65 | 0.08505 | 0.00482 | 0.00720 | 93 | 0.01130 | -0.35073 | 136.808 | 0.72633 | 0.92120 |
| Acyl-[acyl-carrier-protein]–UDP-N-acetylglucosamine O-acyltransferase | 0.08239 | 0.00393 | 0.08339 | 0.00229 | 0.00593 | 65 | 0.08168 | 0.00257 | 0.00526 | 93 | 0.00792 | 0.21617 | 142.750 | 0.82917 | 0.95262 |
| Homoserine O-succinyltransferase | 0.08204 | 0.00409 | 0.08201 | 0.00291 | 0.00669 | 65 | 0.08206 | 0.00248 | 0.00516 | 93 | 0.00845 | -0.00616 | 130.591 | 0.99509 | 0.99921 |
| Phosphoenolpyruvate carboxykinase (ATP) | 0.08191 | 0.00308 | 0.08493 | 0.00176 | 0.00521 | 65 | 0.07980 | 0.00132 | 0.00376 | 93 | 0.00642 | 0.79916 | 124.612 | 0.42572 | 0.76494 |
| Molybdopterin molybdotransferase | 0.08175 | 0.00348 | 0.07945 | 0.00200 | 0.00555 | 65 | 0.08336 | 0.00187 | 0.00449 | 93 | 0.00714 | -0.54751 | 134.932 | 0.58493 | 0.85509 |
| Imidazoleglycerol-phosphate dehydratase | 0.08164 | 0.00237 | 0.07729 | 0.00108 | 0.00408 | 65 | 0.08467 | 0.00074 | 0.00283 | 93 | 0.00496 | -1.48864 | 120.936 | 0.13919 | 0.70632 |
| Mannose-1-phosphate guanylyltransferase | 0.08151 | 0.00420 | 0.07341 | 0.00209 | 0.00567 | 65 | 0.08718 | 0.00323 | 0.00589 | 93 | 0.00818 | -1.68290 | 152.806 | 0.09444 | 0.70632 |
| ATP phosphoribosyltransferase | 0.08141 | 0.00234 | 0.07736 | 0.00109 | 0.00410 | 65 | 0.08424 | 0.00070 | 0.00274 | 93 | 0.00493 | -1.39440 | 117.513 | 0.16583 | 0.70632 |
| Phosphomannomutase | 0.08136 | 0.00361 | 0.07631 | 0.00209 | 0.00567 | 65 | 0.08488 | 0.00203 | 0.00467 | 93 | 0.00735 | -1.16624 | 136.460 | 0.24555 | 0.70632 |
| Amidase | 0.08112 | 0.00644 | 0.07323 | 0.00468 | 0.00849 | 65 | 0.08663 | 0.00787 | 0.00920 | 93 | 0.01251 | -1.07021 | 154.420 | 0.28619 | 0.70632 |
| Acylphosphatase | 0.08101 | 0.00351 | 0.08426 | 0.00214 | 0.00574 | 65 | 0.07874 | 0.00182 | 0.00442 | 93 | 0.00724 | 0.76216 | 130.468 | 0.44734 | 0.77823 |
| Acetylglutamate kinase | 0.08089 | 0.00222 | 0.07760 | 0.00102 | 0.00395 | 65 | 0.08319 | 0.00061 | 0.00257 | 93 | 0.00472 | -1.18574 | 115.154 | 0.23817 | 0.70632 |
| Citrate (Si)-synthase | 0.08086 | 0.00331 | 0.07020 | 0.00163 | 0.00500 | 65 | 0.08831 | 0.00169 | 0.00427 | 93 | 0.00657 | -2.75465 | 139.652 | 0.00666 | 0.62334 |
| Phosphoenolpyruvate carboxylase | 0.08082 | 0.00284 | 0.08170 | 0.00135 | 0.00455 | 65 | 0.08021 | 0.00124 | 0.00365 | 93 | 0.00583 | 0.25588 | 134.142 | 0.79843 | 0.94138 |
| Lipoate–protein ligase | 0.08081 | 0.00493 | 0.08076 | 0.00515 | 0.00890 | 65 | 0.08084 | 0.00297 | 0.00565 | 93 | 0.01054 | -0.00707 | 113.251 | 0.99437 | 0.99921 |
| Histidinol-phosphatase | 0.08064 | 0.00412 | 0.08724 | 0.00371 | 0.00755 | 65 | 0.07604 | 0.00195 | 0.00458 | 93 | 0.00884 | 1.26776 | 109.443 | 0.20757 | 0.70632 |
| Hydroxymethylbilane synthase | 0.08046 | 0.00264 | 0.08387 | 0.00109 | 0.00410 | 65 | 0.07809 | 0.00111 | 0.00346 | 93 | 0.00536 | 1.07772 | 138.564 | 0.28303 | 0.70632 |
| Porphobilinogen synthase | 0.08046 | 0.00268 | 0.08352 | 0.00110 | 0.00412 | 65 | 0.07832 | 0.00116 | 0.00353 | 93 | 0.00543 | 0.95882 | 139.863 | 0.33931 | 0.72019 |
| Histidinol dehydrogenase | 0.08040 | 0.00249 | 0.07603 | 0.00115 | 0.00421 | 65 | 0.08346 | 0.00084 | 0.00300 | 93 | 0.00517 | -1.43680 | 123.452 | 0.15331 | 0.70632 |
| Oxaloacetate decarboxylase | 0.07994 | 0.00601 | 0.09135 | 0.00699 | 0.01037 | 65 | 0.07197 | 0.00473 | 0.00713 | 93 | 0.01259 | 1.54022 | 120.115 | 0.12614 | 0.70632 |
| Isocitrate dehydrogenase (NADP(+)) | 0.07946 | 0.00369 | 0.07323 | 0.00230 | 0.00595 | 65 | 0.08382 | 0.00203 | 0.00467 | 93 | 0.00756 | -1.40035 | 132.157 | 0.16375 | 0.70632 |
| Assimilatory sulfite reductase (NADPH) | 0.07940 | 0.00565 | 0.07626 | 0.00521 | 0.00895 | 65 | 0.08159 | 0.00497 | 0.00731 | 93 | 0.01156 | -0.46134 | 135.789 | 0.64529 | 0.88749 |
| 3-hydroxyisobutyrate dehydrogenase | 0.07909 | 0.00502 | 0.07151 | 0.00354 | 0.00738 | 65 | 0.08438 | 0.00426 | 0.00677 | 93 | 0.01001 | -1.28600 | 145.473 | 0.20049 | 0.70632 |
| 6-carboxytetrahydropterin synthase | 0.07908 | 0.00253 | 0.07393 | 0.00108 | 0.00408 | 65 | 0.08268 | 0.00094 | 0.00317 | 93 | 0.00517 | -1.69562 | 131.525 | 0.09232 | 0.70632 |
| 6-pyruvoyltetrahydropterin synthase | 0.07908 | 0.00253 | 0.07393 | 0.00108 | 0.00408 | 65 | 0.08268 | 0.00094 | 0.00317 | 93 | 0.00517 | -1.69562 | 131.525 | 0.09232 | 0.70632 |
| Pullulanase | 0.07906 | 0.00534 | 0.07409 | 0.00411 | 0.00795 | 65 | 0.08254 | 0.00479 | 0.00718 | 93 | 0.01071 | -0.78878 | 144.222 | 0.43154 | 0.76880 |
| Ribonuclease M5 | 0.07895 | 0.00383 | 0.08487 | 0.00250 | 0.00620 | 65 | 0.07482 | 0.00217 | 0.00483 | 93 | 0.00786 | 1.27832 | 131.553 | 0.20339 | 0.70632 |
| Bleomycin hydrolase | 0.07879 | 0.00436 | 0.07878 | 0.00292 | 0.00670 | 65 | 0.07879 | 0.00310 | 0.00577 | 93 | 0.00884 | -0.00117 | 140.496 | 0.99907 | 0.99947 |
| Acetaldehyde dehydrogenase (acetylating) | 0.07858 | 0.00374 | 0.08211 | 0.00213 | 0.00573 | 65 | 0.07612 | 0.00227 | 0.00494 | 93 | 0.00756 | 0.79233 | 140.562 | 0.42951 | 0.76781 |
| Leucyl aminopeptidase | 0.07853 | 0.00376 | 0.07505 | 0.00227 | 0.00591 | 65 | 0.08097 | 0.00222 | 0.00489 | 93 | 0.00767 | -0.77165 | 136.964 | 0.44165 | 0.77520 |
| Glutathione-disulfide reductase | 0.07847 | 0.00366 | 0.08365 | 0.00274 | 0.00649 | 65 | 0.07484 | 0.00167 | 0.00423 | 93 | 0.00775 | 1.13655 | 115.517 | 0.25808 | 0.70632 |
| Glycine dehydrogenase (aminomethyl-transferring) | 0.07796 | 0.00384 | 0.06962 | 0.00217 | 0.00578 | 65 | 0.08378 | 0.00238 | 0.00506 | 93 | 0.00768 | -1.84315 | 141.765 | 0.06740 | 0.70632 |
| Asparagine synthase (glutamine-hydrolyzing) | 0.07793 | 0.00596 | 0.07096 | 0.00620 | 0.00977 | 65 | 0.08279 | 0.00522 | 0.00749 | 93 | 0.01231 | -0.96178 | 130.108 | 0.33795 | 0.72019 |
| UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | 0.07783 | 0.00394 | 0.08428 | 0.00326 | 0.00708 | 65 | 0.07332 | 0.00186 | 0.00447 | 93 | 0.00837 | 1.30853 | 112.683 | 0.19336 | 0.70632 |
| Valine–pyruvate transaminase | 0.07728 | 0.00323 | 0.07746 | 0.00173 | 0.00516 | 65 | 0.07716 | 0.00161 | 0.00416 | 93 | 0.00663 | 0.04550 | 134.684 | 0.96377 | 0.99193 |
| 2-hydroxymuconate tautomerase | 0.07721 | 0.00359 | 0.07603 | 0.00197 | 0.00550 | 65 | 0.07804 | 0.00211 | 0.00476 | 93 | 0.00727 | -0.27680 | 140.786 | 0.78234 | 0.93675 |
| 16S rRNA (guanine(966)-N(2))-methyltransferase | 0.07719 | 0.00270 | 0.07596 | 0.00113 | 0.00416 | 65 | 0.07804 | 0.00119 | 0.00357 | 93 | 0.00548 | -0.37871 | 140.102 | 0.70548 | 0.91416 |
| Thiosulfate sulfurtransferase | 0.07712 | 0.00471 | 0.07259 | 0.00396 | 0.00780 | 65 | 0.08029 | 0.00321 | 0.00587 | 93 | 0.00977 | -0.78881 | 128.436 | 0.43168 | 0.76880 |
| (FADH(2)-oxidizing) | 0.07669 | 0.00355 | 0.07857 | 0.00200 | 0.00554 | 65 | 0.07538 | 0.00201 | 0.00465 | 93 | 0.00723 | 0.44201 | 138.118 | 0.65917 | 0.88943 |
| Ribokinase | 0.07642 | 0.00419 | 0.07785 | 0.00356 | 0.00740 | 65 | 0.07542 | 0.00224 | 0.00491 | 93 | 0.00888 | 0.27313 | 116.985 | 0.78524 | 0.93675 |
| Pantoate–beta-alanine ligase (AMP-forming) | 0.07627 | 0.00262 | 0.07123 | 0.00111 | 0.00412 | 65 | 0.07980 | 0.00105 | 0.00337 | 93 | 0.00532 | -1.60941 | 135.824 | 0.10985 | 0.70632 |
| Glutamyl-tRNA reductase | 0.07609 | 0.00267 | 0.08192 | 0.00108 | 0.00408 | 65 | 0.07202 | 0.00113 | 0.00349 | 93 | 0.00537 | 1.84356 | 139.670 | 0.06737 | 0.70632 |
| UDP-glucose 6-dehydrogenase | 0.07497 | 0.00368 | 0.06856 | 0.00180 | 0.00526 | 65 | 0.07946 | 0.00234 | 0.00502 | 93 | 0.00727 | -1.49875 | 148.144 | 0.13607 | 0.70632 |
| Aspartate transaminase | 0.07497 | 0.00411 | 0.07104 | 0.00254 | 0.00625 | 65 | 0.07771 | 0.00278 | 0.00546 | 93 | 0.00830 | -0.80350 | 141.660 | 0.42303 | 0.76494 |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase | 0.07474 | 0.00361 | 0.07868 | 0.00222 | 0.00585 | 65 | 0.07199 | 0.00195 | 0.00458 | 93 | 0.00743 | 0.89943 | 132.030 | 0.37006 | 0.73093 |
| Cyclopropane-fatty-acyl-phospholipid synthase | 0.07395 | 0.00504 | 0.07234 | 0.00420 | 0.00804 | 65 | 0.07507 | 0.00393 | 0.00650 | 93 | 0.01034 | -0.26327 | 134.962 | 0.79275 | 0.93908 |
| Heptaprenyl diphosphate synthase | 0.07354 | 0.00477 | 0.08613 | 0.00528 | 0.00901 | 65 | 0.06474 | 0.00226 | 0.00493 | 93 | 0.01027 | 2.08269 | 101.643 | 0.03979 | 0.67652 |
| Alanine transaminase | 0.07343 | 0.00308 | 0.07283 | 0.00151 | 0.00483 | 65 | 0.07385 | 0.00151 | 0.00403 | 93 | 0.00629 | -0.16215 | 137.720 | 0.87142 | 0.96378 |
| Dolichyl-phosphate beta-D-mannosyltransferase | 0.07328 | 0.00300 | 0.07390 | 0.00146 | 0.00474 | 65 | 0.07284 | 0.00141 | 0.00390 | 93 | 0.00614 | 0.17273 | 136.395 | 0.86312 | 0.96186 |
| Succinyldiaminopimelate transaminase | 0.07327 | 0.00429 | 0.07362 | 0.00280 | 0.00657 | 65 | 0.07302 | 0.00302 | 0.00570 | 93 | 0.00870 | 0.06850 | 141.114 | 0.94548 | 0.99049 |
| Phosphopentomutase | 0.07324 | 0.00374 | 0.07361 | 0.00221 | 0.00584 | 65 | 0.07299 | 0.00223 | 0.00490 | 93 | 0.00762 | 0.08162 | 138.243 | 0.93506 | 0.98985 |
| Xanthine dehydrogenase | 0.07302 | 0.00774 | 0.06700 | 0.00850 | 0.01144 | 65 | 0.07722 | 0.01022 | 0.01048 | 93 | 0.01551 | -0.65890 | 145.339 | 0.51100 | 0.81548 |
| Glutamate–cysteine ligase | 0.07298 | 0.00409 | 0.07878 | 0.00359 | 0.00744 | 65 | 0.06892 | 0.00196 | 0.00459 | 93 | 0.00874 | 1.12795 | 110.902 | 0.26178 | 0.70632 |
| Urocanate hydratase | 0.07292 | 0.00321 | 0.07522 | 0.00248 | 0.00618 | 65 | 0.07131 | 0.00105 | 0.00335 | 93 | 0.00703 | 0.55651 | 101.150 | 0.57909 | 0.85191 |
| Hydroxyethylthiazole kinase | 0.07248 | 0.00369 | 0.06985 | 0.00193 | 0.00544 | 65 | 0.07431 | 0.00233 | 0.00501 | 93 | 0.00740 | -0.60351 | 145.601 | 0.54711 | 0.84303 |
| N-acylglucosamine-6-phosphate 2-epimerase | 0.07218 | 0.00358 | 0.07806 | 0.00198 | 0.00552 | 65 | 0.06807 | 0.00204 | 0.00468 | 93 | 0.00724 | 1.38178 | 139.214 | 0.16925 | 0.70632 |
| Nicotinate-nucleotide–dimethylbenzimidazole phosphoribosyltransferase | 0.07192 | 0.00386 | 0.07530 | 0.00278 | 0.00654 | 65 | 0.06955 | 0.00206 | 0.00471 | 93 | 0.00806 | 0.71412 | 124.280 | 0.47649 | 0.79541 |
| dGTPase | 0.07176 | 0.00293 | 0.07115 | 0.00149 | 0.00480 | 65 | 0.07219 | 0.00128 | 0.00371 | 93 | 0.00606 | -0.17100 | 130.816 | 0.86449 | 0.96186 |
| Gamma-glutamyltransferase | 0.07175 | 0.00583 | 0.06954 | 0.00548 | 0.00918 | 65 | 0.07329 | 0.00534 | 0.00758 | 93 | 0.01190 | -0.31559 | 136.723 | 0.75279 | 0.92944 |
| Glutathione hydrolase | 0.07175 | 0.00583 | 0.06954 | 0.00548 | 0.00918 | 65 | 0.07329 | 0.00534 | 0.00758 | 93 | 0.01190 | -0.31559 | 136.723 | 0.75279 | 0.92944 |
| DNA-3-methyladenine glycosylase II | 0.07163 | 0.00456 | 0.07055 | 0.00363 | 0.00748 | 65 | 0.07238 | 0.00307 | 0.00575 | 93 | 0.00943 | -0.19409 | 130.303 | 0.84641 | 0.95939 |
| Imidazolonepropionase | 0.07122 | 0.00316 | 0.07350 | 0.00245 | 0.00614 | 65 | 0.06963 | 0.00098 | 0.00324 | 93 | 0.00694 | 0.55864 | 99.265 | 0.57767 | 0.85190 |
| Arabinose-5-phosphate isomerase | 0.07113 | 0.00304 | 0.07472 | 0.00153 | 0.00485 | 65 | 0.06862 | 0.00142 | 0.00391 | 93 | 0.00623 | 0.98056 | 134.691 | 0.32857 | 0.71151 |
| 1,4-dihydroxy-2-naphthoate polyprenyltransferase | 0.07094 | 0.00355 | 0.07454 | 0.00236 | 0.00602 | 65 | 0.06841 | 0.00175 | 0.00434 | 93 | 0.00742 | 0.82567 | 124.323 | 0.41058 | 0.76070 |
| Protein-glutamate methylesterase | 0.07068 | 0.00734 | 0.06662 | 0.00806 | 0.01113 | 65 | 0.07351 | 0.00889 | 0.00978 | 93 | 0.01482 | -0.46482 | 142.030 | 0.64277 | 0.88749 |
| 3-deoxy-8-phosphooctulonate synthase | 0.07061 | 0.00308 | 0.07411 | 0.00158 | 0.00493 | 65 | 0.06816 | 0.00144 | 0.00393 | 93 | 0.00630 | 0.94481 | 133.812 | 0.34646 | 0.72019 |
| 3-deoxy-manno-octulosonate cytidylyltransferase | 0.07054 | 0.00306 | 0.07359 | 0.00156 | 0.00489 | 65 | 0.06840 | 0.00144 | 0.00393 | 93 | 0.00627 | 0.82767 | 134.327 | 0.40932 | 0.76070 |
| Polyphosphate kinase | 0.07023 | 0.00265 | 0.06931 | 0.00107 | 0.00406 | 65 | 0.07087 | 0.00114 | 0.00351 | 93 | 0.00537 | -0.29125 | 140.617 | 0.77129 | 0.93540 |
| Lipid-A-disaccharide synthase | 0.07006 | 0.00304 | 0.07335 | 0.00154 | 0.00487 | 65 | 0.06776 | 0.00140 | 0.00388 | 93 | 0.00623 | 0.89897 | 133.711 | 0.37029 | 0.73093 |
| Glutarate-semialdehyde dehydrogenase | 0.07002 | 0.00475 | 0.06751 | 0.00357 | 0.00741 | 65 | 0.07177 | 0.00358 | 0.00621 | 93 | 0.00967 | -0.44127 | 137.979 | 0.65971 | 0.88943 |
| Succinate-semialdehyde dehydrogenase (NADP(+)) | 0.07002 | 0.00475 | 0.06751 | 0.00357 | 0.00741 | 65 | 0.07177 | 0.00358 | 0.00621 | 93 | 0.00967 | -0.44127 | 137.979 | 0.65971 | 0.88943 |
| Thiamine-phosphate kinase | 0.06990 | 0.00271 | 0.06904 | 0.00132 | 0.00450 | 65 | 0.07049 | 0.00107 | 0.00339 | 93 | 0.00563 | -0.25733 | 128.339 | 0.79734 | 0.94100 |
| Medium-chain acyl-CoA dehydrogenase | 0.06957 | 0.00823 | 0.05071 | 0.00615 | 0.00973 | 65 | 0.08275 | 0.01354 | 0.01207 | 93 | 0.01550 | -2.06703 | 155.831 | 0.04039 | 0.67652 |
| Pyrimidine-nucleoside phosphorylase | 0.06922 | 0.00370 | 0.07053 | 0.00225 | 0.00589 | 65 | 0.06830 | 0.00213 | 0.00478 | 93 | 0.00759 | 0.29463 | 135.361 | 0.76873 | 0.93519 |
| Trans-2-decenoyl-[acyl-carrier-protein] isomerase | 0.06881 | 0.00300 | 0.06860 | 0.00132 | 0.00451 | 65 | 0.06896 | 0.00150 | 0.00402 | 93 | 0.00604 | -0.05917 | 143.299 | 0.95290 | 0.99049 |
| Protein-disulfide reductase | 0.06865 | 0.00409 | 0.06785 | 0.00282 | 0.00659 | 65 | 0.06921 | 0.00254 | 0.00523 | 93 | 0.00841 | -0.16164 | 133.194 | 0.87184 | 0.96378 |
| Alpha-glucosidase | 0.06837 | 0.00438 | 0.06740 | 0.00300 | 0.00679 | 65 | 0.06906 | 0.00310 | 0.00577 | 93 | 0.00891 | -0.18610 | 139.247 | 0.85264 | 0.96065 |
| Glutamate–tRNA(Gln) ligase | 0.06820 | 0.00392 | 0.06883 | 0.00255 | 0.00626 | 65 | 0.06775 | 0.00236 | 0.00504 | 93 | 0.00804 | 0.13402 | 134.519 | 0.89358 | 0.97093 |
| Sarcosine oxidase | 0.06814 | 0.00962 | 0.06022 | 0.01219 | 0.01369 | 65 | 0.07367 | 0.01638 | 0.01327 | 93 | 0.01907 | -0.70506 | 149.139 | 0.48187 | 0.79962 |
| Isopentenyl-diphosphate Delta-isomerase | 0.06802 | 0.00361 | 0.06905 | 0.00216 | 0.00576 | 65 | 0.06730 | 0.00201 | 0.00465 | 93 | 0.00740 | 0.23556 | 134.737 | 0.81413 | 0.94567 |
| Nitronate monooxygenase | 0.06794 | 0.00539 | 0.05995 | 0.00347 | 0.00731 | 65 | 0.07352 | 0.00533 | 0.00757 | 93 | 0.01052 | -1.28903 | 152.725 | 0.19934 | 0.70632 |
| N-acetylglucosaminephosphotransferase | 0.06791 | 0.00316 | 0.06979 | 0.00173 | 0.00516 | 65 | 0.06660 | 0.00149 | 0.00400 | 93 | 0.00653 | 0.48826 | 131.215 | 0.62618 | 0.88183 |
| Ferredoxin–NADP(+) reductase | 0.06775 | 0.00384 | 0.06740 | 0.00231 | 0.00596 | 65 | 0.06799 | 0.00237 | 0.00504 | 93 | 0.00781 | -0.07458 | 138.913 | 0.94065 | 0.99049 |
| Dethiobiotin synthase | 0.06734 | 0.00318 | 0.07002 | 0.00176 | 0.00520 | 65 | 0.06546 | 0.00150 | 0.00402 | 93 | 0.00657 | 0.69329 | 130.724 | 0.48936 | 0.80257 |
| 3-deoxy-manno-octulosonate-8-phosphatase | 0.06733 | 0.00312 | 0.07187 | 0.00159 | 0.00494 | 65 | 0.06416 | 0.00149 | 0.00400 | 93 | 0.00636 | 1.21350 | 135.122 | 0.22705 | 0.70632 |
| Pyruvate, water dikinase | 0.06709 | 0.00292 | 0.06802 | 0.00157 | 0.00491 | 65 | 0.06643 | 0.00121 | 0.00360 | 93 | 0.00609 | 0.26178 | 126.018 | 0.79392 | 0.93972 |
| Protein-glutamate O-methyltransferase | 0.06697 | 0.00649 | 0.06327 | 0.00651 | 0.01001 | 65 | 0.06956 | 0.00682 | 0.00856 | 93 | 0.01317 | -0.47770 | 139.832 | 0.63361 | 0.88317 |
| Tetraacyldisaccharide 4’-kinase | 0.06696 | 0.00295 | 0.06924 | 0.00150 | 0.00480 | 65 | 0.06537 | 0.00130 | 0.00373 | 93 | 0.00608 | 0.63613 | 131.436 | 0.52580 | 0.82336 |
| Malate dehydrogenase (oxaloacetate-decarboxylating) | 0.06675 | 0.00397 | 0.07442 | 0.00385 | 0.00769 | 65 | 0.06140 | 0.00151 | 0.00403 | 93 | 0.00869 | 1.49907 | 98.822 | 0.13704 | 0.70632 |
| 23S rRNA (guanine(745)-N(1))-methyltransferase | 0.06658 | 0.00323 | 0.06794 | 0.00172 | 0.00514 | 65 | 0.06563 | 0.00161 | 0.00416 | 93 | 0.00661 | 0.34822 | 135.126 | 0.72821 | 0.92120 |
| [Ribosomal protein S12] (aspartate(89)-C(3))-methylthiotransferase | 0.06633 | 0.00284 | 0.06573 | 0.00124 | 0.00437 | 65 | 0.06674 | 0.00130 | 0.00375 | 93 | 0.00576 | -0.17562 | 140.024 | 0.86084 | 0.96186 |
| 2’,3’-cyclic-nucleotide 2’-phosphodiesterase | 0.06605 | 0.00417 | 0.06150 | 0.00219 | 0.00580 | 65 | 0.06924 | 0.00315 | 0.00582 | 93 | 0.00822 | -0.94223 | 151.177 | 0.34758 | 0.72019 |
| 3’-nucleotidase | 0.06605 | 0.00417 | 0.06150 | 0.00219 | 0.00580 | 65 | 0.06924 | 0.00315 | 0.00582 | 93 | 0.00822 | -0.94223 | 151.177 | 0.34758 | 0.72019 |
| NADPH:quinone reductase | 0.06595 | 0.00556 | 0.05570 | 0.00391 | 0.00776 | 65 | 0.07312 | 0.00547 | 0.00767 | 93 | 0.01091 | -1.59622 | 150.344 | 0.11254 | 0.70632 |
| Lipoyl synthase | 0.06590 | 0.00272 | 0.06147 | 0.00132 | 0.00450 | 65 | 0.06899 | 0.00106 | 0.00338 | 93 | 0.00563 | -1.33704 | 128.216 | 0.18358 | 0.70632 |
| Alpha-galactosidase | 0.06579 | 0.00435 | 0.06750 | 0.00353 | 0.00737 | 65 | 0.06459 | 0.00265 | 0.00534 | 93 | 0.00910 | 0.32028 | 124.953 | 0.74929 | 0.92904 |
| Sulfate adenylyltransferase | 0.06565 | 0.00478 | 0.05699 | 0.00263 | 0.00636 | 65 | 0.07170 | 0.00425 | 0.00676 | 93 | 0.00928 | -1.58568 | 153.758 | 0.11487 | 0.70632 |
| Phosphomethylpyrimidine synthase | 0.06514 | 0.00269 | 0.06673 | 0.00116 | 0.00423 | 65 | 0.06404 | 0.00114 | 0.00350 | 93 | 0.00549 | 0.48934 | 136.966 | 0.62538 | 0.88154 |
| Glutamine–tRNA ligase | 0.06508 | 0.00279 | 0.06608 | 0.00117 | 0.00423 | 65 | 0.06439 | 0.00129 | 0.00373 | 93 | 0.00564 | 0.29939 | 142.186 | 0.76508 | 0.93350 |
| Aminomethyltransferase | 0.06480 | 0.00360 | 0.05876 | 0.00195 | 0.00548 | 65 | 0.06902 | 0.00209 | 0.00474 | 93 | 0.00725 | -1.41570 | 140.735 | 0.15907 | 0.70632 |
| Thiazole synthase | 0.06374 | 0.00246 | 0.06532 | 0.00115 | 0.00421 | 65 | 0.06264 | 0.00082 | 0.00298 | 93 | 0.00515 | 0.51953 | 122.760 | 0.60432 | 0.86814 |
| Malate dehydrogenase | 0.06369 | 0.00288 | 0.05875 | 0.00112 | 0.00415 | 65 | 0.06714 | 0.00143 | 0.00392 | 93 | 0.00571 | -1.47090 | 147.486 | 0.14345 | 0.70632 |
| (Kdo)-lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase | 0.06360 | 0.00273 | 0.06263 | 0.00111 | 0.00413 | 65 | 0.06428 | 0.00124 | 0.00365 | 93 | 0.00552 | -0.29915 | 142.583 | 0.76526 | 0.93350 |
| (Kdo)(2)-lipid IV(A) (2-8) 3-deoxy-D-manno-octulosonic acid transferase | 0.06360 | 0.00273 | 0.06263 | 0.00111 | 0.00413 | 65 | 0.06428 | 0.00124 | 0.00365 | 93 | 0.00552 | -0.29915 | 142.583 | 0.76526 | 0.93350 |
| (Kdo)(3)-lipid IV(A) (2-4) 3-deoxy-D-manno-octulosonic acid transferase | 0.06360 | 0.00273 | 0.06263 | 0.00111 | 0.00413 | 65 | 0.06428 | 0.00124 | 0.00365 | 93 | 0.00552 | -0.29915 | 142.583 | 0.76526 | 0.93350 |
| Lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase | 0.06360 | 0.00273 | 0.06263 | 0.00111 | 0.00413 | 65 | 0.06428 | 0.00124 | 0.00365 | 93 | 0.00552 | -0.29915 | 142.583 | 0.76526 | 0.93350 |
| S-adenosylhomocysteine deaminase | 0.06312 | 0.00286 | 0.06215 | 0.00121 | 0.00431 | 65 | 0.06380 | 0.00137 | 0.00384 | 93 | 0.00577 | -0.28549 | 143.084 | 0.77568 | 0.93675 |
| S-methyl-5’-thioadenosine deaminase | 0.06312 | 0.00286 | 0.06215 | 0.00121 | 0.00431 | 65 | 0.06380 | 0.00137 | 0.00384 | 93 | 0.00577 | -0.28549 | 143.084 | 0.77568 | 0.93675 |
| Magnesium-importing ATPase | 0.06305 | 0.00333 | 0.06228 | 0.00168 | 0.00509 | 65 | 0.06359 | 0.00182 | 0.00443 | 93 | 0.00674 | -0.19413 | 141.234 | 0.84636 | 0.95939 |
| N-acetylneuraminate lyase | 0.06279 | 0.00355 | 0.06629 | 0.00216 | 0.00576 | 65 | 0.06035 | 0.00189 | 0.00451 | 93 | 0.00732 | 0.81108 | 131.839 | 0.41878 | 0.76408 |
| Ferroxidase | 0.06263 | 0.00591 | 0.06163 | 0.00617 | 0.00974 | 65 | 0.06332 | 0.00514 | 0.00743 | 93 | 0.01225 | -0.13820 | 129.615 | 0.89029 | 0.96887 |
| Adenylate cyclase | 0.06249 | 0.00381 | 0.05565 | 0.00137 | 0.00458 | 65 | 0.06728 | 0.00292 | 0.00560 | 93 | 0.00724 | -1.60815 | 155.949 | 0.10983 | 0.70632 |
| Cu(2+)-exporting ATPase | 0.06231 | 0.00289 | 0.06271 | 0.00123 | 0.00434 | 65 | 0.06202 | 0.00141 | 0.00389 | 93 | 0.00583 | 0.11858 | 143.532 | 0.90577 | 0.97700 |
| 3-methyl-2-oxobutanoate hydroxymethyltransferase | 0.06228 | 0.00251 | 0.05559 | 0.00093 | 0.00379 | 65 | 0.06696 | 0.00100 | 0.00327 | 93 | 0.00501 | -2.27022 | 140.614 | 0.02471 | 0.67652 |
| Adenosylmethionine–8-amino-7-oxononanoate transaminase | 0.06206 | 0.00273 | 0.06257 | 0.00126 | 0.00441 | 65 | 0.06170 | 0.00113 | 0.00349 | 93 | 0.00562 | 0.15499 | 132.920 | 0.87707 | 0.96548 |
| Alkaline phosphatase | 0.06183 | 0.00557 | 0.05157 | 0.00491 | 0.00869 | 65 | 0.06901 | 0.00481 | 0.00719 | 93 | 0.01128 | -1.54594 | 137.026 | 0.12442 | 0.70632 |
| Epoxyqueuosine reductase | 0.06161 | 0.00235 | 0.05676 | 0.00093 | 0.00378 | 65 | 0.06501 | 0.00081 | 0.00295 | 93 | 0.00480 | -1.71949 | 131.663 | 0.08788 | 0.70632 |
| Arginine decarboxylase | 0.06148 | 0.00317 | 0.05894 | 0.00180 | 0.00526 | 65 | 0.06325 | 0.00146 | 0.00396 | 93 | 0.00658 | -0.65532 | 128.462 | 0.51343 | 0.81548 |
| NAD(+) diphosphatase | 0.06135 | 0.00262 | 0.06215 | 0.00114 | 0.00419 | 65 | 0.06079 | 0.00105 | 0.00337 | 93 | 0.00537 | 0.25309 | 134.440 | 0.80058 | 0.94263 |
| Urease | 0.06124 | 0.00627 | 0.04704 | 0.00430 | 0.00813 | 65 | 0.07117 | 0.00738 | 0.00891 | 93 | 0.01206 | -2.00073 | 154.748 | 0.04717 | 0.69348 |
| GMP reductase | 0.06095 | 0.00358 | 0.06156 | 0.00224 | 0.00588 | 65 | 0.06053 | 0.00189 | 0.00451 | 93 | 0.00741 | 0.13987 | 130.156 | 0.88898 | 0.96887 |
| Pyruvate carboxylase | 0.06083 | 0.00380 | 0.05970 | 0.00233 | 0.00599 | 65 | 0.06162 | 0.00228 | 0.00495 | 93 | 0.00777 | -0.24775 | 136.867 | 0.80470 | 0.94350 |
| 3-mercaptopyruvate sulfurtransferase | 0.06073 | 0.00377 | 0.05427 | 0.00227 | 0.00591 | 65 | 0.06524 | 0.00220 | 0.00486 | 93 | 0.00765 | -1.43525 | 136.505 | 0.15350 | 0.70632 |
| Glutamate N-acetyltransferase | 0.06059 | 0.00321 | 0.05637 | 0.00167 | 0.00507 | 65 | 0.06355 | 0.00160 | 0.00415 | 93 | 0.00655 | -1.09702 | 135.957 | 0.27457 | 0.70632 |
| Prolyl aminopeptidase | 0.06057 | 0.00441 | 0.06036 | 0.00341 | 0.00725 | 65 | 0.06071 | 0.00288 | 0.00556 | 93 | 0.00914 | -0.03816 | 130.226 | 0.96962 | 0.99263 |
| All-trans-octaprenyl-diphosphate synthase | 0.06021 | 0.00263 | 0.05758 | 0.00113 | 0.00418 | 65 | 0.06205 | 0.00108 | 0.00340 | 93 | 0.00539 | -0.82975 | 135.537 | 0.40814 | 0.76070 |
| 3-hydroxyacyl-CoA dehydrogenase | 0.06003 | 0.00468 | 0.05307 | 0.00350 | 0.00733 | 65 | 0.06489 | 0.00341 | 0.00606 | 93 | 0.00951 | -1.24301 | 136.839 | 0.21599 | 0.70632 |
| L-glutamate gamma-semialdehyde dehydrogenase | 0.05974 | 0.00383 | 0.05511 | 0.00194 | 0.00546 | 65 | 0.06297 | 0.00259 | 0.00528 | 93 | 0.00759 | -1.03548 | 149.010 | 0.30212 | 0.70632 |
| Aspartate ammonia-lyase | 0.05956 | 0.00297 | 0.05813 | 0.00116 | 0.00422 | 65 | 0.06055 | 0.00156 | 0.00410 | 93 | 0.00589 | -0.41054 | 149.286 | 0.68200 | 0.90520 |
| Phosphatidylglycerol–membrane-oligosaccharide glycerophosphotransferase | 0.05955 | 0.00499 | 0.06539 | 0.00572 | 0.00938 | 65 | 0.05546 | 0.00270 | 0.00539 | 93 | 0.01082 | 0.91748 | 105.258 | 0.36099 | 0.72316 |
| Sirohydrochlorin ferrochelatase | 0.05931 | 0.00281 | 0.06233 | 0.00136 | 0.00458 | 65 | 0.05719 | 0.00118 | 0.00356 | 93 | 0.00580 | 0.88617 | 131.338 | 0.37715 | 0.73348 |
| Aspartate–ammonia ligase | 0.05876 | 0.00351 | 0.06387 | 0.00224 | 0.00587 | 65 | 0.05518 | 0.00174 | 0.00432 | 93 | 0.00729 | 1.19324 | 126.451 | 0.23501 | 0.70632 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase I | 0.05874 | 0.00404 | 0.05495 | 0.00260 | 0.00633 | 65 | 0.06138 | 0.00257 | 0.00526 | 93 | 0.00823 | -0.78095 | 137.345 | 0.43617 | 0.77161 |
| D-sedoheptulose 7-phosphate isomerase | 0.05860 | 0.00353 | 0.06904 | 0.00246 | 0.00616 | 65 | 0.05131 | 0.00152 | 0.00404 | 93 | 0.00737 | 2.40696 | 116.098 | 0.01766 | 0.64530 |
| Glutamate dehydrogenase | 0.05850 | 0.00295 | 0.05850 | 0.00153 | 0.00486 | 65 | 0.05850 | 0.00129 | 0.00372 | 93 | 0.00611 | 0.00066 | 129.959 | 0.99947 | 0.99947 |
| 3-oxoacid CoA-transferase | 0.05814 | 0.00526 | 0.05037 | 0.00394 | 0.00778 | 65 | 0.06358 | 0.00466 | 0.00708 | 93 | 0.01052 | -1.25536 | 144.753 | 0.21137 | 0.70632 |
| HslU–HslV peptidase | 0.05800 | 0.00261 | 0.05870 | 0.00127 | 0.00442 | 65 | 0.05751 | 0.00096 | 0.00320 | 93 | 0.00546 | 0.21760 | 124.979 | 0.82810 | 0.95262 |
| Homoserine O-acetyltransferase | 0.05795 | 0.00375 | 0.05502 | 0.00227 | 0.00591 | 65 | 0.06000 | 0.00220 | 0.00486 | 93 | 0.00765 | -0.65058 | 136.467 | 0.51641 | 0.81889 |
| Nicotinate-nucleotide diphosphorylase (carboxylating) | 0.05790 | 0.00275 | 0.05558 | 0.00125 | 0.00438 | 65 | 0.05952 | 0.00116 | 0.00354 | 93 | 0.00563 | -0.69951 | 134.782 | 0.48544 | 0.80111 |
| NAD(P)H-hydrate epimerase | 0.05748 | 0.00286 | 0.05791 | 0.00119 | 0.00428 | 65 | 0.05718 | 0.00139 | 0.00386 | 93 | 0.00576 | 0.12611 | 144.188 | 0.89982 | 0.97349 |
| Isochorismate synthase | 0.05727 | 0.00288 | 0.05571 | 0.00146 | 0.00474 | 65 | 0.05836 | 0.00121 | 0.00361 | 93 | 0.00596 | -0.44450 | 129.342 | 0.65742 | 0.88943 |
| Precorrin-2 dehydrogenase | 0.05691 | 0.00261 | 0.06157 | 0.00131 | 0.00449 | 65 | 0.05366 | 0.00090 | 0.00311 | 93 | 0.00546 | 1.44804 | 120.942 | 0.15019 | 0.70632 |
| Tagatose-bisphosphate aldolase | 0.05652 | 0.00370 | 0.05972 | 0.00232 | 0.00598 | 65 | 0.05428 | 0.00207 | 0.00472 | 93 | 0.00762 | 0.71365 | 132.836 | 0.47669 | 0.79541 |
| 2-dehydro-3-deoxy-phosphogluconate aldolase | 0.05645 | 0.00316 | 0.05694 | 0.00201 | 0.00557 | 65 | 0.05610 | 0.00128 | 0.00371 | 93 | 0.00669 | 0.12442 | 117.472 | 0.90119 | 0.97349 |
| Molybdate-transporting ATPase | 0.05631 | 0.00260 | 0.05575 | 0.00110 | 0.00412 | 65 | 0.05671 | 0.00106 | 0.00338 | 93 | 0.00533 | -0.17965 | 136.286 | 0.85770 | 0.96186 |
| tRNA nucleotidyltransferase | 0.05628 | 0.00256 | 0.05505 | 0.00121 | 0.00431 | 65 | 0.05714 | 0.00093 | 0.00315 | 93 | 0.00534 | -0.39136 | 125.703 | 0.69619 | 0.91007 |
| 4-hydroxybenzoate polyprenyltransferase | 0.05620 | 0.00253 | 0.05586 | 0.00112 | 0.00416 | 65 | 0.05644 | 0.00095 | 0.00320 | 93 | 0.00524 | -0.11026 | 130.445 | 0.91237 | 0.97907 |
| 4-hydroxy-3-polyprenylbenzoate decarboxylase | 0.05611 | 0.00412 | 0.05804 | 0.00281 | 0.00657 | 65 | 0.05475 | 0.00263 | 0.00531 | 93 | 0.00845 | 0.38907 | 134.944 | 0.69784 | 0.91007 |
| (4S)-4-hydroxy-2-oxoglutarate aldolase | 0.05609 | 0.00315 | 0.05656 | 0.00202 | 0.00557 | 65 | 0.05577 | 0.00127 | 0.00370 | 93 | 0.00669 | 0.11839 | 117.126 | 0.90596 | 0.97700 |
| DNA ligase (ATP) | 0.05544 | 0.00455 | 0.04964 | 0.00313 | 0.00694 | 65 | 0.05949 | 0.00337 | 0.00602 | 93 | 0.00919 | -1.07295 | 141.104 | 0.28512 | 0.70632 |
| tRNA (N(6)-L-threonylcarbamoyladenosine(37)-C(2))-methylthiotransferase | 0.05529 | 0.00347 | 0.05424 | 0.00214 | 0.00574 | 65 | 0.05602 | 0.00175 | 0.00434 | 93 | 0.00719 | -0.24643 | 128.686 | 0.80574 | 0.94351 |
| Catechol 2,3-dioxygenase | 0.05521 | 0.00387 | 0.04871 | 0.00197 | 0.00551 | 65 | 0.05976 | 0.00261 | 0.00530 | 93 | 0.00764 | -1.44571 | 148.717 | 0.15036 | 0.70632 |
| Pyroglutamyl-peptidase I | 0.05482 | 0.00283 | 0.05692 | 0.00150 | 0.00481 | 65 | 0.05336 | 0.00111 | 0.00346 | 93 | 0.00593 | 0.60141 | 124.318 | 0.54866 | 0.84333 |
| Enoyl-[acyl-carrier-protein] reductase (NADPH, Si-specific) | 0.05452 | 0.00277 | 0.05046 | 0.00116 | 0.00422 | 65 | 0.05736 | 0.00124 | 0.00366 | 93 | 0.00559 | -1.23608 | 140.817 | 0.21849 | 0.70632 |
| Adenosylcobalamin/alpha-ribazole phosphatase | 0.05439 | 0.00282 | 0.05620 | 0.00137 | 0.00460 | 65 | 0.05313 | 0.00119 | 0.00357 | 93 | 0.00582 | 0.52738 | 131.294 | 0.59882 | 0.86318 |
| Diamine N-acetyltransferase | 0.05425 | 0.00328 | 0.05197 | 0.00139 | 0.00463 | 65 | 0.05585 | 0.00193 | 0.00455 | 93 | 0.00649 | -0.59739 | 150.027 | 0.55115 | 0.84429 |
| 4-nitrophenylphosphatase | 0.05403 | 0.00351 | 0.05884 | 0.00219 | 0.00581 | 65 | 0.05068 | 0.00177 | 0.00437 | 93 | 0.00727 | 1.12355 | 128.297 | 0.26330 | 0.70632 |
| Xaa-Pro dipeptidyl-peptidase | 0.05388 | 0.00418 | 0.05352 | 0.00264 | 0.00637 | 65 | 0.05413 | 0.00288 | 0.00556 | 93 | 0.00846 | -0.07239 | 141.585 | 0.94239 | 0.99049 |
| Dihydroorotate oxidase (fumarate) | 0.05382 | 0.00336 | 0.05538 | 0.00180 | 0.00526 | 65 | 0.05274 | 0.00180 | 0.00439 | 93 | 0.00685 | 0.38492 | 137.823 | 0.70089 | 0.91306 |
| Acetolactate decarboxylase | 0.05362 | 0.00358 | 0.05597 | 0.00217 | 0.00578 | 65 | 0.05198 | 0.00193 | 0.00455 | 93 | 0.00736 | 0.54259 | 132.520 | 0.58833 | 0.85698 |
| Spermidine synthase | 0.05361 | 0.00251 | 0.05050 | 0.00096 | 0.00384 | 65 | 0.05578 | 0.00102 | 0.00331 | 93 | 0.00507 | -1.04131 | 140.479 | 0.29952 | 0.70632 |
| Nitrite reductase (NADH) | 0.05355 | 0.00443 | 0.04822 | 0.00310 | 0.00691 | 65 | 0.05728 | 0.00310 | 0.00577 | 93 | 0.00900 | -1.00584 | 137.826 | 0.31626 | 0.70632 |
| Polynucleotide adenylyltransferase | 0.05351 | 0.00287 | 0.05196 | 0.00145 | 0.00473 | 65 | 0.05459 | 0.00121 | 0.00360 | 93 | 0.00595 | -0.44190 | 129.541 | 0.65930 | 0.88943 |
| 3-hydroxybutyryl-CoA dehydrogenase | 0.05339 | 0.00418 | 0.05194 | 0.00259 | 0.00631 | 65 | 0.05439 | 0.00290 | 0.00559 | 93 | 0.00843 | -0.29087 | 142.738 | 0.77157 | 0.93540 |
| Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP(+)) | 0.05338 | 0.00276 | 0.04834 | 0.00108 | 0.00407 | 65 | 0.05690 | 0.00127 | 0.00370 | 93 | 0.00550 | -1.55558 | 144.626 | 0.12199 | 0.70632 |
| Dihydrolipoyllysine-residue succinyltransferase | 0.05336 | 0.00285 | 0.04970 | 0.00131 | 0.00449 | 65 | 0.05591 | 0.00127 | 0.00369 | 93 | 0.00581 | -1.06863 | 136.362 | 0.28713 | 0.70632 |
| Pyruvate, phosphate dikinase | 0.05325 | 0.00325 | 0.05265 | 0.00188 | 0.00537 | 65 | 0.05366 | 0.00154 | 0.00407 | 93 | 0.00674 | -0.15109 | 128.960 | 0.88014 | 0.96590 |
| Adenosylcobinamide-GDP ribazoletransferase | 0.05284 | 0.00271 | 0.05475 | 0.00134 | 0.00455 | 65 | 0.05151 | 0.00105 | 0.00336 | 93 | 0.00565 | 0.57213 | 126.683 | 0.56825 | 0.85024 |
| Hydroxymethylglutaryl-CoA reductase | 0.05282 | 0.00346 | 0.05601 | 0.00203 | 0.00559 | 65 | 0.05060 | 0.00180 | 0.00440 | 93 | 0.00712 | 0.76146 | 132.473 | 0.44773 | 0.77823 |
| 1,4-dihydroxy-2-naphthoyl-CoA synthase | 0.05281 | 0.00279 | 0.05358 | 0.00144 | 0.00471 | 65 | 0.05227 | 0.00110 | 0.00344 | 93 | 0.00583 | 0.22552 | 125.660 | 0.82194 | 0.95060 |
| Adenosylcobinamide-phosphate guanylyltransferase | 0.05259 | 0.00281 | 0.05422 | 0.00136 | 0.00457 | 65 | 0.05145 | 0.00118 | 0.00356 | 93 | 0.00579 | 0.47784 | 131.373 | 0.63356 | 0.88317 |
| Adenosylcobinamide kinase | 0.05258 | 0.00281 | 0.05422 | 0.00136 | 0.00457 | 65 | 0.05144 | 0.00118 | 0.00356 | 93 | 0.00579 | 0.47891 | 131.388 | 0.63280 | 0.88317 |
| 23S rRNA pseudouridine(955/2504/2580) synthase | 0.05249 | 0.00290 | 0.05592 | 0.00145 | 0.00472 | 65 | 0.05009 | 0.00125 | 0.00367 | 93 | 0.00598 | 0.97465 | 131.276 | 0.33153 | 0.71442 |
| Formyltetrahydrofolate deformylase | 0.05232 | 0.00450 | 0.04850 | 0.00292 | 0.00670 | 65 | 0.05499 | 0.00342 | 0.00606 | 93 | 0.00903 | -0.71844 | 144.377 | 0.47365 | 0.79541 |
| Adenosylcobyric acid synthase (glutamine-hydrolyzing) | 0.05215 | 0.00359 | 0.05973 | 0.00309 | 0.00690 | 65 | 0.04686 | 0.00125 | 0.00367 | 93 | 0.00781 | 1.64656 | 99.808 | 0.10279 | 0.70632 |
| Diphosphomevalonate decarboxylase | 0.05211 | 0.00348 | 0.05586 | 0.00205 | 0.00561 | 65 | 0.04949 | 0.00182 | 0.00442 | 93 | 0.00715 | 0.89198 | 132.649 | 0.37402 | 0.73204 |
| Mevalonate kinase | 0.05208 | 0.00348 | 0.05578 | 0.00205 | 0.00561 | 65 | 0.04949 | 0.00182 | 0.00443 | 93 | 0.00715 | 0.88004 | 132.638 | 0.38043 | 0.73587 |
| 4-carboxymuconolactone decarboxylase | 0.05200 | 0.00410 | 0.04521 | 0.00245 | 0.00614 | 65 | 0.05675 | 0.00276 | 0.00545 | 93 | 0.00821 | -1.40594 | 142.933 | 0.16191 | 0.70632 |
| Oxoglutarate dehydrogenase (succinyl-transferring) | 0.05196 | 0.00290 | 0.04929 | 0.00136 | 0.00458 | 65 | 0.05382 | 0.00131 | 0.00375 | 93 | 0.00592 | -0.76453 | 136.028 | 0.44588 | 0.77823 |
| 7-carboxy-7-deazaguanine synthase | 0.05156 | 0.00236 | 0.04825 | 0.00082 | 0.00354 | 65 | 0.05388 | 0.00092 | 0.00315 | 93 | 0.00474 | -1.18741 | 142.950 | 0.23704 | 0.70632 |
| Hydroxymethylglutaryl-CoA synthase | 0.05148 | 0.00346 | 0.05490 | 0.00202 | 0.00557 | 65 | 0.04910 | 0.00181 | 0.00441 | 93 | 0.00710 | 0.81705 | 133.050 | 0.41536 | 0.76374 |
| Methylcrotonoyl-CoA carboxylase | 0.05139 | 0.00554 | 0.04527 | 0.00451 | 0.00833 | 65 | 0.05566 | 0.00510 | 0.00741 | 93 | 0.01115 | -0.93150 | 142.985 | 0.35316 | 0.72019 |
| o-succinylbenzoate–CoA ligase | 0.05131 | 0.00275 | 0.05223 | 0.00143 | 0.00469 | 65 | 0.05067 | 0.00104 | 0.00335 | 93 | 0.00577 | 0.27102 | 123.556 | 0.78682 | 0.93675 |
| Glycerate dehydrogenase | 0.05115 | 0.00297 | 0.05680 | 0.00152 | 0.00484 | 65 | 0.04720 | 0.00127 | 0.00370 | 93 | 0.00609 | 1.57528 | 129.850 | 0.11763 | 0.70632 |
| 4-hydroxythreonine-4-phosphate dehydrogenase | 0.05097 | 0.00247 | 0.04791 | 0.00087 | 0.00365 | 65 | 0.05310 | 0.00104 | 0.00334 | 93 | 0.00495 | -1.04865 | 145.074 | 0.29608 | 0.70632 |
| Molybdenum cofactor guanylyltransferase | 0.05092 | 0.00245 | 0.04980 | 0.00094 | 0.00380 | 65 | 0.05170 | 0.00097 | 0.00323 | 93 | 0.00499 | -0.38042 | 139.307 | 0.70422 | 0.91393 |
| Phosphomevalonate kinase | 0.05068 | 0.00349 | 0.05407 | 0.00206 | 0.00563 | 65 | 0.04830 | 0.00184 | 0.00445 | 93 | 0.00718 | 0.80325 | 132.862 | 0.42327 | 0.76494 |
| Aromatic-amino-acid transaminase | 0.05067 | 0.00304 | 0.05619 | 0.00163 | 0.00501 | 65 | 0.04681 | 0.00132 | 0.00377 | 93 | 0.00627 | 1.49623 | 128.473 | 0.13705 | 0.70632 |
| 8-amino-7-oxononanoate synthase | 0.05052 | 0.00298 | 0.04959 | 0.00152 | 0.00483 | 65 | 0.05117 | 0.00134 | 0.00380 | 93 | 0.00614 | -0.25699 | 132.456 | 0.79758 | 0.94100 |
| L-threonine aldolase | 0.05030 | 0.00308 | 0.05127 | 0.00172 | 0.00515 | 65 | 0.04962 | 0.00136 | 0.00383 | 93 | 0.00641 | 0.25690 | 127.342 | 0.79767 | 0.94100 |
| Quinolinate synthase | 0.05010 | 0.00256 | 0.04853 | 0.00108 | 0.00408 | 65 | 0.05120 | 0.00101 | 0.00330 | 93 | 0.00525 | -0.50741 | 135.081 | 0.61269 | 0.87289 |
| Bacterial non-heme ferritin | 0.04971 | 0.00355 | 0.04954 | 0.00205 | 0.00562 | 65 | 0.04982 | 0.00198 | 0.00461 | 93 | 0.00727 | -0.03864 | 136.295 | 0.96923 | 0.99263 |
| Phosphatidylglycerophosphatase | 0.04952 | 0.00338 | 0.05458 | 0.00186 | 0.00534 | 65 | 0.04599 | 0.00177 | 0.00436 | 93 | 0.00689 | 1.24609 | 135.715 | 0.21488 | 0.70632 |
| L-aspartate oxidase | 0.04947 | 0.00258 | 0.04864 | 0.00110 | 0.00411 | 65 | 0.05006 | 0.00103 | 0.00333 | 93 | 0.00529 | -0.26918 | 135.069 | 0.78820 | 0.93676 |
| Malonate-semialdehyde dehydrogenase (acetylating) | 0.04932 | 0.00519 | 0.04196 | 0.00341 | 0.00725 | 65 | 0.05447 | 0.00484 | 0.00721 | 93 | 0.01022 | -1.22349 | 150.705 | 0.22305 | 0.70632 |
| Methylmalonate-semialdehyde dehydrogenase (CoA acylating) | 0.04932 | 0.00519 | 0.04196 | 0.00341 | 0.00725 | 65 | 0.05447 | 0.00484 | 0.00721 | 93 | 0.01022 | -1.22349 | 150.705 | 0.22305 | 0.70632 |
| synthase | 0.04927 | 0.00283 | 0.04897 | 0.00148 | 0.00477 | 65 | 0.04948 | 0.00112 | 0.00348 | 93 | 0.00590 | -0.08671 | 125.393 | 0.93104 | 0.98985 |
| Molybdopterin synthase | 0.04927 | 0.00272 | 0.04506 | 0.00110 | 0.00411 | 65 | 0.05221 | 0.00121 | 0.00360 | 93 | 0.00546 | -1.30735 | 141.838 | 0.19321 | 0.70632 |
| Gluconokinase | 0.04925 | 0.00308 | 0.05137 | 0.00211 | 0.00570 | 65 | 0.04777 | 0.00108 | 0.00341 | 93 | 0.00664 | 0.54187 | 108.423 | 0.58902 | 0.85698 |
| dCTP deaminase | 0.04922 | 0.00267 | 0.04779 | 0.00122 | 0.00434 | 65 | 0.05022 | 0.00106 | 0.00338 | 93 | 0.00550 | -0.44220 | 131.662 | 0.65907 | 0.88943 |
| Peptidyl-dipeptidase Dcp | 0.04914 | 0.00373 | 0.04277 | 0.00173 | 0.00516 | 65 | 0.05359 | 0.00250 | 0.00519 | 93 | 0.00732 | -1.47859 | 151.212 | 0.14133 | 0.70632 |
| Purine nucleosidase | 0.04896 | 0.00450 | 0.04820 | 0.00347 | 0.00731 | 65 | 0.04949 | 0.00305 | 0.00572 | 93 | 0.00929 | -0.13954 | 132.004 | 0.88924 | 0.96887 |
| Adenosylcobinamide-phosphate synthase | 0.04890 | 0.00271 | 0.05194 | 0.00130 | 0.00447 | 65 | 0.04678 | 0.00106 | 0.00337 | 93 | 0.00560 | 0.92050 | 128.686 | 0.35904 | 0.72286 |
| Carbon-monoxide dehydrogenase (acceptor) | 0.04887 | 0.00457 | 0.04378 | 0.00264 | 0.00637 | 65 | 0.05242 | 0.00376 | 0.00636 | 93 | 0.00900 | -0.95882 | 150.906 | 0.33918 | 0.72019 |
| Undecaprenol kinase | 0.04875 | 0.00330 | 0.05012 | 0.00183 | 0.00531 | 65 | 0.04780 | 0.00166 | 0.00423 | 93 | 0.00678 | 0.34251 | 133.554 | 0.73251 | 0.92120 |
| Alanine dehydrogenase | 0.04836 | 0.00256 | 0.04012 | 0.00084 | 0.00360 | 65 | 0.05412 | 0.00110 | 0.00344 | 93 | 0.00498 | -2.81112 | 148.375 | 0.00560 | 0.62334 |
| Dodecanoyl-[acyl-carrier-protein] hydrolase | 0.04826 | 0.00352 | 0.05206 | 0.00213 | 0.00572 | 65 | 0.04560 | 0.00185 | 0.00445 | 93 | 0.00725 | 0.89143 | 131.531 | 0.37433 | 0.73204 |
| Proline dehydrogenase | 0.04788 | 0.00345 | 0.04675 | 0.00183 | 0.00531 | 65 | 0.04867 | 0.00194 | 0.00456 | 93 | 0.00700 | -0.27349 | 140.265 | 0.78488 | 0.93675 |
| Carbamate kinase | 0.04777 | 0.00277 | 0.05124 | 0.00130 | 0.00447 | 65 | 0.04534 | 0.00115 | 0.00352 | 93 | 0.00569 | 1.03811 | 132.572 | 0.30111 | 0.70632 |
| Acetyl-CoA C-acyltransferase | 0.04750 | 0.00389 | 0.04241 | 0.00222 | 0.00585 | 65 | 0.05107 | 0.00251 | 0.00520 | 93 | 0.00782 | -1.10658 | 142.967 | 0.27033 | 0.70632 |
| Glutamyl aminopeptidase | 0.04733 | 0.00356 | 0.04763 | 0.00187 | 0.00537 | 65 | 0.04712 | 0.00212 | 0.00477 | 93 | 0.00718 | 0.07034 | 143.004 | 0.94402 | 0.99049 |
| UDP-2,3-diacylglucosamine diphosphatase | 0.04731 | 0.00250 | 0.04612 | 0.00095 | 0.00383 | 65 | 0.04814 | 0.00103 | 0.00332 | 93 | 0.00507 | -0.40000 | 141.037 | 0.68976 | 0.90851 |
| FMN reductase (NADPH) | 0.04714 | 0.00385 | 0.03849 | 0.00187 | 0.00537 | 65 | 0.05319 | 0.00260 | 0.00529 | 93 | 0.00754 | -1.95075 | 150.149 | 0.05295 | 0.70632 |
| Aspartate 1-decarboxylase | 0.04707 | 0.00251 | 0.04094 | 0.00076 | 0.00343 | 65 | 0.05135 | 0.00113 | 0.00348 | 93 | 0.00489 | -2.13100 | 151.812 | 0.03470 | 0.67652 |
| Dihydroorotate dehydrogenase (quinone) | 0.04683 | 0.00270 | 0.04534 | 0.00119 | 0.00427 | 65 | 0.04787 | 0.00114 | 0.00350 | 93 | 0.00552 | -0.45674 | 135.994 | 0.64859 | 0.88803 |
| Ribonuclease E | 0.04628 | 0.00275 | 0.04587 | 0.00130 | 0.00448 | 65 | 0.04658 | 0.00113 | 0.00349 | 93 | 0.00568 | -0.12481 | 131.487 | 0.90086 | 0.97349 |
| Lipoyl(octanoyl) transferase | 0.04616 | 0.00270 | 0.04496 | 0.00127 | 0.00442 | 65 | 0.04699 | 0.00108 | 0.00341 | 93 | 0.00559 | -0.36406 | 130.735 | 0.71640 | 0.92120 |
| NAD(+) synthase (glutamine-hydrolyzing) | 0.04600 | 0.00273 | 0.04362 | 0.00109 | 0.00410 | 65 | 0.04766 | 0.00124 | 0.00365 | 93 | 0.00549 | -0.73571 | 143.208 | 0.46311 | 0.78699 |
| [Glutamate–ammonia-ligase] adenylyltransferase | 0.04492 | 0.00261 | 0.04433 | 0.00119 | 0.00428 | 65 | 0.04533 | 0.00101 | 0.00330 | 93 | 0.00540 | -0.18610 | 130.468 | 0.85265 | 0.96065 |
| Cobyrinate a,c-diamide synthase (glutamine-hydrolyzing) | 0.04483 | 0.00257 | 0.04847 | 0.00133 | 0.00452 | 65 | 0.04229 | 0.00084 | 0.00300 | 93 | 0.00543 | 1.13977 | 116.893 | 0.25671 | 0.70632 |
| Hydrogenobyrinic acid a,c-diamide synthase (glutamine-hydrolyzing) | 0.04483 | 0.00257 | 0.04847 | 0.00133 | 0.00452 | 65 | 0.04229 | 0.00084 | 0.00300 | 93 | 0.00543 | 1.13977 | 116.893 | 0.25671 | 0.70632 |
| Acetylornithine deacetylase | 0.04473 | 0.00351 | 0.04362 | 0.00184 | 0.00532 | 65 | 0.04550 | 0.00204 | 0.00469 | 93 | 0.00709 | -0.26543 | 142.302 | 0.79106 | 0.93897 |
| Adenosylmethionine decarboxylase | 0.04472 | 0.00300 | 0.04613 | 0.00175 | 0.00519 | 65 | 0.04373 | 0.00121 | 0.00361 | 93 | 0.00632 | 0.37849 | 121.088 | 0.70573 | 0.91416 |
| N-acetyldiaminopimelate deacetylase | 0.04465 | 0.00320 | 0.04609 | 0.00167 | 0.00507 | 65 | 0.04365 | 0.00160 | 0.00414 | 93 | 0.00655 | 0.37265 | 135.832 | 0.70999 | 0.91799 |
| Alpha,alpha-phosphotrehalase | 0.04465 | 0.00274 | 0.04736 | 0.00154 | 0.00487 | 65 | 0.04276 | 0.00094 | 0.00318 | 93 | 0.00582 | 0.79128 | 115.618 | 0.43040 | 0.76847 |
| Aspartyl aminopeptidase | 0.04434 | 0.00271 | 0.05214 | 0.00146 | 0.00475 | 65 | 0.03889 | 0.00088 | 0.00308 | 93 | 0.00566 | 2.34225 | 115.167 | 0.02089 | 0.64928 |
| Cobaltochelatase | 0.04422 | 0.00340 | 0.03923 | 0.00135 | 0.00456 | 65 | 0.04771 | 0.00215 | 0.00481 | 93 | 0.00663 | -1.27932 | 153.420 | 0.20272 | 0.70632 |
| Deoxyribonuclease IV | 0.04414 | 0.00264 | 0.04510 | 0.00127 | 0.00442 | 65 | 0.04347 | 0.00100 | 0.00328 | 93 | 0.00550 | 0.29549 | 126.968 | 0.76810 | 0.93519 |
| Cobalt-precorrin 5A hydrolase | 0.04408 | 0.00299 | 0.05086 | 0.00172 | 0.00514 | 65 | 0.03934 | 0.00116 | 0.00353 | 93 | 0.00624 | 1.84719 | 119.883 | 0.06719 | 0.70632 |
| Maltose 6’-phosphate phosphatase | 0.04390 | 0.00330 | 0.04573 | 0.00161 | 0.00498 | 65 | 0.04263 | 0.00181 | 0.00441 | 93 | 0.00666 | 0.46556 | 142.741 | 0.64224 | 0.88744 |
| Formimidoylglutamase | 0.04384 | 0.00255 | 0.04322 | 0.00104 | 0.00399 | 65 | 0.04428 | 0.00103 | 0.00332 | 93 | 0.00520 | -0.20381 | 137.480 | 0.83880 | 0.95674 |
| Sirohydrochlorin cobaltochelatase | 0.04371 | 0.00365 | 0.04989 | 0.00276 | 0.00652 | 65 | 0.03939 | 0.00163 | 0.00419 | 93 | 0.00775 | 1.35548 | 114.310 | 0.17794 | 0.70632 |
| Protein disulfide-isomerase | 0.04362 | 0.00372 | 0.04449 | 0.00227 | 0.00591 | 65 | 0.04301 | 0.00216 | 0.00482 | 93 | 0.00763 | 0.19510 | 135.595 | 0.84561 | 0.95939 |
| Adenosylhomocysteinase | 0.04362 | 0.00263 | 0.04208 | 0.00113 | 0.00416 | 65 | 0.04469 | 0.00107 | 0.00340 | 93 | 0.00537 | -0.48414 | 135.718 | 0.62906 | 0.88303 |
| Malate dehydrogenase (quinone) | 0.04359 | 0.00367 | 0.04153 | 0.00213 | 0.00572 | 65 | 0.04503 | 0.00215 | 0.00481 | 93 | 0.00747 | -0.46876 | 138.447 | 0.63998 | 0.88568 |
| 2-dehydro-3-deoxygluconokinase | 0.04335 | 0.00290 | 0.04107 | 0.00189 | 0.00540 | 65 | 0.04495 | 0.00095 | 0.00320 | 93 | 0.00627 | -0.61813 | 107.559 | 0.53779 | 0.83586 |
| Ribonuclease D | 0.04334 | 0.00266 | 0.04115 | 0.00105 | 0.00403 | 65 | 0.04487 | 0.00116 | 0.00353 | 93 | 0.00536 | -0.69372 | 141.971 | 0.48899 | 0.80257 |
| L-ribulose-5-phosphate 4-epimerase | 0.04322 | 0.00267 | 0.04702 | 0.00146 | 0.00475 | 65 | 0.04055 | 0.00088 | 0.00308 | 93 | 0.00566 | 1.14328 | 115.053 | 0.25530 | 0.70632 |
| Carboxynorspermidine decarboxylase | 0.04318 | 0.00246 | 0.03771 | 0.00076 | 0.00341 | 65 | 0.04700 | 0.00107 | 0.00339 | 93 | 0.00481 | -1.93207 | 150.707 | 0.05523 | 0.70632 |
| Citryl-CoA lyase | 0.04312 | 0.00290 | 0.04372 | 0.00118 | 0.00426 | 65 | 0.04271 | 0.00144 | 0.00394 | 93 | 0.00580 | 0.17555 | 146.038 | 0.86089 | 0.96186 |
| Hydroxyacylglutathione hydrolase | 0.04304 | 0.00269 | 0.04529 | 0.00139 | 0.00463 | 65 | 0.04147 | 0.00098 | 0.00324 | 93 | 0.00565 | 0.67702 | 121.786 | 0.49968 | 0.80830 |
| 6-phospho-beta-galactosidase | 0.04282 | 0.00324 | 0.04223 | 0.00156 | 0.00490 | 65 | 0.04323 | 0.00175 | 0.00434 | 93 | 0.00655 | -0.15252 | 142.627 | 0.87899 | 0.96579 |
| Tagatose-6-phosphate kinase | 0.04266 | 0.00317 | 0.04435 | 0.00157 | 0.00492 | 65 | 0.04148 | 0.00162 | 0.00417 | 93 | 0.00645 | 0.44593 | 139.034 | 0.65634 | 0.88943 |
| Xylulokinase | 0.04248 | 0.00319 | 0.03820 | 0.00122 | 0.00432 | 65 | 0.04547 | 0.00188 | 0.00449 | 93 | 0.00624 | -1.16692 | 152.859 | 0.24506 | 0.70632 |
| D-glycero-alpha-D-manno-heptose-1,7-bisphosphate 7-phosphatase | 0.04243 | 0.00228 | 0.04455 | 0.00088 | 0.00367 | 65 | 0.04096 | 0.00079 | 0.00292 | 93 | 0.00469 | 0.76680 | 133.277 | 0.44456 | 0.77733 |
| D-glycero-beta-D-manno-heptose 1,7-bisphosphate 7-phosphatase | 0.04243 | 0.00228 | 0.04455 | 0.00088 | 0.00367 | 65 | 0.04096 | 0.00079 | 0.00292 | 93 | 0.00469 | 0.76680 | 133.277 | 0.44456 | 0.77733 |
| Hydroxylamine reductase | 0.04241 | 0.00352 | 0.04706 | 0.00238 | 0.00605 | 65 | 0.03916 | 0.00166 | 0.00423 | 93 | 0.00738 | 1.07053 | 121.716 | 0.28650 | 0.70632 |
| Propionyl-CoA carboxylase | 0.04237 | 0.00410 | 0.03348 | 0.00159 | 0.00495 | 65 | 0.04857 | 0.00334 | 0.00599 | 93 | 0.00777 | -1.94189 | 155.983 | 0.05395 | 0.70632 |
| Selenide, water dikinase | 0.04210 | 0.00235 | 0.04670 | 0.00113 | 0.00417 | 65 | 0.03889 | 0.00067 | 0.00269 | 93 | 0.00496 | 1.57479 | 114.348 | 0.11807 | 0.70632 |
| Glutamate formimidoyltransferase | 0.04204 | 0.00358 | 0.04844 | 0.00307 | 0.00687 | 65 | 0.03756 | 0.00127 | 0.00369 | 93 | 0.00780 | 1.39437 | 100.396 | 0.16628 | 0.70632 |
| Tellurite methyltransferase | 0.04173 | 0.00330 | 0.04102 | 0.00147 | 0.00475 | 65 | 0.04223 | 0.00192 | 0.00455 | 93 | 0.00657 | -0.18528 | 148.362 | 0.85326 | 0.96065 |
| Exo-alpha-sialidase | 0.04169 | 0.00317 | 0.03910 | 0.00134 | 0.00453 | 65 | 0.04350 | 0.00178 | 0.00437 | 93 | 0.00630 | -0.69755 | 148.856 | 0.48655 | 0.80150 |
| Protein-L-isoaspartate(D-aspartate) O-methyltransferase | 0.04136 | 0.00349 | 0.03794 | 0.00197 | 0.00550 | 65 | 0.04374 | 0.00191 | 0.00453 | 93 | 0.00713 | -0.81382 | 136.457 | 0.41717 | 0.76408 |
| Aminopyrimidine aminohydrolase | 0.04133 | 0.00251 | 0.03683 | 0.00071 | 0.00330 | 65 | 0.04448 | 0.00118 | 0.00357 | 93 | 0.00486 | -1.57384 | 154.373 | 0.11757 | 0.70632 |
| Nicotinamidase | 0.04133 | 0.00221 | 0.03916 | 0.00065 | 0.00315 | 65 | 0.04284 | 0.00086 | 0.00304 | 93 | 0.00438 | -0.84114 | 148.747 | 0.40162 | 0.75271 |
| Glycerol dehydrogenase | 0.04103 | 0.00254 | 0.04537 | 0.00129 | 0.00445 | 65 | 0.03800 | 0.00083 | 0.00298 | 93 | 0.00536 | 1.37690 | 117.874 | 0.17115 | 0.70632 |
| Precorrin-2 C(20)-methyltransferase | 0.04103 | 0.00273 | 0.04654 | 0.00150 | 0.00480 | 65 | 0.03717 | 0.00092 | 0.00315 | 93 | 0.00575 | 1.63082 | 116.050 | 0.10564 | 0.70632 |
| Cobalt-precorrin-4 methyltransferase | 0.04097 | 0.00272 | 0.04662 | 0.00148 | 0.00477 | 65 | 0.03702 | 0.00092 | 0.00315 | 93 | 0.00572 | 1.67937 | 116.380 | 0.09576 | 0.70632 |
| Precorrin-4 C(11)-methyltransferase | 0.04097 | 0.00272 | 0.04662 | 0.00148 | 0.00477 | 65 | 0.03702 | 0.00092 | 0.00315 | 93 | 0.00572 | 1.67937 | 116.380 | 0.09576 | 0.70632 |
| N-acetylmuramic acid 6-phosphate etherase | 0.04090 | 0.00246 | 0.04058 | 0.00117 | 0.00424 | 65 | 0.04113 | 0.00082 | 0.00296 | 93 | 0.00517 | -0.10696 | 121.427 | 0.91499 | 0.98088 |
| Cobalt-precorrin-8 methylmutase | 0.04082 | 0.00271 | 0.04640 | 0.00149 | 0.00478 | 65 | 0.03692 | 0.00091 | 0.00313 | 93 | 0.00571 | 1.65919 | 115.879 | 0.09978 | 0.70632 |
| Precorrin-8X methylmutase | 0.04082 | 0.00271 | 0.04640 | 0.00149 | 0.00478 | 65 | 0.03692 | 0.00091 | 0.00313 | 93 | 0.00571 | 1.65919 | 115.879 | 0.09978 | 0.70632 |
| 2-iminoacetate synthase | 0.04077 | 0.00345 | 0.04687 | 0.00208 | 0.00566 | 65 | 0.03650 | 0.00172 | 0.00430 | 93 | 0.00711 | 1.45885 | 129.176 | 0.14703 | 0.70632 |
| Choline dehydrogenase | 0.04075 | 0.00403 | 0.03716 | 0.00249 | 0.00618 | 65 | 0.04326 | 0.00263 | 0.00532 | 93 | 0.00816 | -0.74734 | 140.271 | 0.45611 | 0.78637 |
| Glycine C-acetyltransferase | 0.04053 | 0.00267 | 0.03658 | 0.00087 | 0.00366 | 65 | 0.04329 | 0.00130 | 0.00373 | 93 | 0.00523 | -1.28345 | 152.024 | 0.20129 | 0.70632 |
| Lysyltransferase | 0.04045 | 0.00277 | 0.04386 | 0.00148 | 0.00477 | 65 | 0.03807 | 0.00103 | 0.00332 | 93 | 0.00581 | 0.99728 | 121.350 | 0.32061 | 0.70632 |
| Malonyl-[acyl-carrier protein] O-methyltransferase | 0.04045 | 0.00278 | 0.04435 | 0.00143 | 0.00469 | 65 | 0.03772 | 0.00106 | 0.00338 | 93 | 0.00579 | 1.14542 | 124.416 | 0.25424 | 0.70632 |
| Membrane dipeptidase | 0.04037 | 0.00244 | 0.03887 | 0.00100 | 0.00392 | 65 | 0.04141 | 0.00091 | 0.00312 | 93 | 0.00501 | -0.50750 | 133.713 | 0.61264 | 0.87289 |
| Hydroxymethylglutaryl-CoA lyase | 0.03998 | 0.00369 | 0.03773 | 0.00199 | 0.00553 | 65 | 0.04155 | 0.00228 | 0.00496 | 93 | 0.00742 | -0.51427 | 143.694 | 0.60786 | 0.87001 |
| o-succinylbenzoate synthase | 0.03993 | 0.00289 | 0.03999 | 0.00144 | 0.00471 | 65 | 0.03989 | 0.00125 | 0.00367 | 93 | 0.00597 | 0.01749 | 131.619 | 0.98607 | 0.99660 |
| Dipeptidyl-peptidase IV | 0.03993 | 0.00325 | 0.03539 | 0.00113 | 0.00418 | 65 | 0.04310 | 0.00203 | 0.00467 | 93 | 0.00627 | -1.22982 | 155.265 | 0.22063 | 0.70632 |
| Aspartate racemase | 0.03993 | 0.00345 | 0.04355 | 0.00273 | 0.00648 | 65 | 0.03739 | 0.00129 | 0.00373 | 93 | 0.00748 | 0.82357 | 105.386 | 0.41205 | 0.76209 |
| Glutaconyl-CoA decarboxylase | 0.03990 | 0.00412 | 0.04827 | 0.00355 | 0.00739 | 65 | 0.03406 | 0.00203 | 0.00468 | 93 | 0.00874 | 1.62601 | 112.976 | 0.10673 | 0.70632 |
| Pyridoxal 5’-phosphate synthase | 0.03980 | 0.00270 | 0.03842 | 0.00126 | 0.00440 | 65 | 0.04076 | 0.00108 | 0.00341 | 93 | 0.00557 | -0.42052 | 131.154 | 0.67479 | 0.89887 |
| N-carbamoylputrescine amidase | 0.03977 | 0.00222 | 0.03524 | 0.00060 | 0.00305 | 65 | 0.04294 | 0.00089 | 0.00309 | 93 | 0.00434 | -1.77136 | 151.672 | 0.07851 | 0.70632 |
| Cobalt-factor II C(20)-methyltransferase | 0.03972 | 0.00273 | 0.04561 | 0.00150 | 0.00481 | 65 | 0.03560 | 0.00093 | 0.00316 | 93 | 0.00575 | 1.73932 | 116.241 | 0.08463 | 0.70632 |
| 2-polyprenyl-6-hydroxyphenol methylase | 0.03929 | 0.00332 | 0.03820 | 0.00188 | 0.00538 | 65 | 0.04005 | 0.00166 | 0.00423 | 93 | 0.00685 | -0.27085 | 132.312 | 0.78693 | 0.93675 |
| 3-demethylubiquinol 3-O-methyltransferase | 0.03929 | 0.00332 | 0.03820 | 0.00188 | 0.00538 | 65 | 0.04005 | 0.00166 | 0.00423 | 93 | 0.00685 | -0.27085 | 132.312 | 0.78693 | 0.93675 |
| 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) | 0.03900 | 0.00378 | 0.02960 | 0.00145 | 0.00472 | 65 | 0.04557 | 0.00275 | 0.00543 | 93 | 0.00720 | -2.21894 | 155.755 | 0.02794 | 0.67652 |
| Precorrin-3B C(17)-methyltransferase | 0.03889 | 0.00270 | 0.04434 | 0.00146 | 0.00474 | 65 | 0.03508 | 0.00092 | 0.00314 | 93 | 0.00568 | 1.62838 | 116.844 | 0.10614 | 0.70632 |
| Formimidoyltetrahydrofolate cyclodeaminase | 0.03872 | 0.00351 | 0.04528 | 0.00301 | 0.00680 | 65 | 0.03414 | 0.00117 | 0.00355 | 93 | 0.00767 | 1.45209 | 98.543 | 0.14965 | 0.70632 |
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–L-lysine ligase | 0.03862 | 0.00304 | 0.03764 | 0.00132 | 0.00450 | 65 | 0.03930 | 0.00157 | 0.00411 | 93 | 0.00610 | -0.27304 | 145.192 | 0.78521 | 0.93675 |
| S-methyl-5-thioribose-1-phosphate isomerase | 0.03859 | 0.00247 | 0.04413 | 0.00121 | 0.00432 | 65 | 0.03471 | 0.00077 | 0.00287 | 93 | 0.00518 | 1.81742 | 117.142 | 0.07171 | 0.70632 |
| [Protein-PII] uridylyltransferase | 0.03839 | 0.00265 | 0.03713 | 0.00114 | 0.00419 | 65 | 0.03927 | 0.00109 | 0.00343 | 93 | 0.00541 | -0.39570 | 135.994 | 0.69295 | 0.90946 |
| Glutaminase | 0.03812 | 0.00302 | 0.04301 | 0.00229 | 0.00594 | 65 | 0.03471 | 0.00083 | 0.00299 | 93 | 0.00665 | 1.24835 | 96.233 | 0.21493 | 0.70632 |
| Cobalt-precorrin-6A reductase | 0.03796 | 0.00255 | 0.04435 | 0.00140 | 0.00463 | 65 | 0.03349 | 0.00073 | 0.00280 | 93 | 0.00542 | 2.00683 | 109.230 | 0.04724 | 0.69348 |
| Precorrin-6A reductase | 0.03796 | 0.00255 | 0.04435 | 0.00140 | 0.00463 | 65 | 0.03349 | 0.00073 | 0.00280 | 93 | 0.00542 | 2.00683 | 109.230 | 0.04724 | 0.69348 |
| Arginine deiminase | 0.03770 | 0.00204 | 0.04082 | 0.00079 | 0.00350 | 65 | 0.03552 | 0.00056 | 0.00245 | 93 | 0.00427 | 1.24117 | 121.660 | 0.21693 | 0.70632 |
| L-lactate dehydrogenase (cytochrome) | 0.03767 | 0.00283 | 0.03481 | 0.00124 | 0.00437 | 65 | 0.03967 | 0.00129 | 0.00372 | 93 | 0.00574 | -0.84604 | 139.449 | 0.39898 | 0.74973 |
| Ornithine decarboxylase | 0.03706 | 0.00258 | 0.03958 | 0.00119 | 0.00427 | 65 | 0.03530 | 0.00096 | 0.00321 | 93 | 0.00535 | 0.80011 | 128.238 | 0.42513 | 0.76494 |
| 4-hydroxyphenylpyruvate dioxygenase | 0.03700 | 0.00385 | 0.03492 | 0.00235 | 0.00601 | 65 | 0.03845 | 0.00236 | 0.00504 | 93 | 0.00785 | -0.45036 | 138.159 | 0.65315 | 0.88943 |
| 3-hydroxybutyryl-CoA epimerase | 0.03697 | 0.00309 | 0.03496 | 0.00139 | 0.00462 | 65 | 0.03838 | 0.00160 | 0.00415 | 93 | 0.00621 | -0.55186 | 143.811 | 0.58190 | 0.85342 |
| Adenosine deaminase | 0.03694 | 0.00252 | 0.03788 | 0.00103 | 0.00397 | 65 | 0.03629 | 0.00100 | 0.00328 | 93 | 0.00515 | 0.30994 | 136.887 | 0.75707 | 0.93065 |
| Protoporphyrinogen oxidase | 0.03664 | 0.00266 | 0.03366 | 0.00095 | 0.00382 | 65 | 0.03873 | 0.00123 | 0.00364 | 93 | 0.00528 | -0.96122 | 148.052 | 0.33801 | 0.72019 |
| 2,5-didehydrogluconate reductase (2-dehydro-L-gulonate-forming) | 0.03660 | 0.00274 | 0.03602 | 0.00126 | 0.00441 | 65 | 0.03701 | 0.00114 | 0.00350 | 93 | 0.00563 | -0.17508 | 133.324 | 0.86128 | 0.96186 |
| Acid phosphatase | 0.03631 | 0.00302 | 0.03584 | 0.00137 | 0.00459 | 65 | 0.03663 | 0.00151 | 0.00402 | 93 | 0.00610 | -0.13100 | 141.915 | 0.89596 | 0.97163 |
| Pimeloyl-[acyl-carrier protein] methyl ester esterase | 0.03610 | 0.00258 | 0.04072 | 0.00126 | 0.00440 | 65 | 0.03287 | 0.00089 | 0.00309 | 93 | 0.00538 | 1.46129 | 122.235 | 0.14650 | 0.70632 |
| Lysophospholipase | 0.03593 | 0.00283 | 0.03105 | 0.00111 | 0.00413 | 65 | 0.03934 | 0.00136 | 0.00382 | 93 | 0.00563 | -1.47409 | 146.121 | 0.14261 | 0.70632 |
| Saccharopine dehydrogenase (NAD(+), L-lysine-forming) | 0.03582 | 0.00262 | 0.03126 | 0.00086 | 0.00363 | 65 | 0.03900 | 0.00124 | 0.00365 | 93 | 0.00514 | -1.50333 | 151.262 | 0.13484 | 0.70632 |
| Ethanolamine ammonia-lyase | 0.03581 | 0.00422 | 0.04305 | 0.00397 | 0.00781 | 65 | 0.03074 | 0.00198 | 0.00462 | 93 | 0.00908 | 1.35635 | 107.432 | 0.17783 | 0.70632 |
| Pyruvate oxidase | 0.03578 | 0.00281 | 0.03488 | 0.00120 | 0.00429 | 65 | 0.03641 | 0.00130 | 0.00373 | 93 | 0.00569 | -0.26872 | 141.178 | 0.78854 | 0.93676 |
| Succinyl-CoA:acetate CoA-transferase | 0.03555 | 0.00251 | 0.03515 | 0.00093 | 0.00378 | 65 | 0.03583 | 0.00106 | 0.00337 | 93 | 0.00506 | -0.13608 | 143.263 | 0.89195 | 0.96995 |
| UDP-N-acetylmuramoylpentapeptide-lysine N(6)-alanyltransferase | 0.03555 | 0.00299 | 0.03495 | 0.00122 | 0.00433 | 65 | 0.03596 | 0.00157 | 0.00410 | 93 | 0.00597 | -0.17007 | 147.642 | 0.86519 | 0.96213 |
| Molybdenum cofactor cytidylyltransferase | 0.03554 | 0.00226 | 0.03414 | 0.00092 | 0.00376 | 65 | 0.03652 | 0.00073 | 0.00281 | 93 | 0.00469 | -0.50764 | 127.707 | 0.61258 | 0.87289 |
| (R,R)-butanediol dehydrogenase | 0.03553 | 0.00270 | 0.03431 | 0.00088 | 0.00368 | 65 | 0.03638 | 0.00135 | 0.00381 | 93 | 0.00530 | -0.38965 | 152.755 | 0.69734 | 0.91007 |
| Diacetyl reductase ((R)-acetoin forming) | 0.03553 | 0.00270 | 0.03431 | 0.00088 | 0.00368 | 65 | 0.03638 | 0.00135 | 0.00381 | 93 | 0.00530 | -0.38965 | 152.755 | 0.69734 | 0.91007 |
| Ornithine cyclodeaminase | 0.03552 | 0.00284 | 0.03016 | 0.00096 | 0.00384 | 65 | 0.03926 | 0.00148 | 0.00399 | 93 | 0.00554 | -1.64272 | 152.893 | 0.10250 | 0.70632 |
| Short-chain acyl-CoA dehydrogenase | 0.03534 | 0.00460 | 0.04606 | 0.00605 | 0.00965 | 65 | 0.02784 | 0.00135 | 0.00380 | 93 | 0.01037 | 1.75607 | 84.018 | 0.08272 | 0.70632 |
| Magnesium chelatase | 0.03518 | 0.00291 | 0.03760 | 0.00166 | 0.00506 | 65 | 0.03349 | 0.00111 | 0.00346 | 93 | 0.00613 | 0.67176 | 119.621 | 0.50303 | 0.81115 |
| Diguanylate cyclase | 0.03516 | 0.00448 | 0.03258 | 0.00278 | 0.00654 | 65 | 0.03696 | 0.00347 | 0.00611 | 93 | 0.00895 | -0.48955 | 146.756 | 0.62519 | 0.88154 |
| UDP-galactopyranose mutase | 0.03497 | 0.00288 | 0.03696 | 0.00149 | 0.00479 | 65 | 0.03357 | 0.00119 | 0.00357 | 93 | 0.00598 | 0.56691 | 127.503 | 0.57177 | 0.85122 |
| Malate synthase | 0.03497 | 0.00300 | 0.03148 | 0.00164 | 0.00502 | 65 | 0.03740 | 0.00128 | 0.00370 | 93 | 0.00624 | -0.94947 | 126.597 | 0.34419 | 0.72019 |
| 23S rRNA pseudouridine(2604) synthase | 0.03465 | 0.00319 | 0.03586 | 0.00179 | 0.00525 | 65 | 0.03380 | 0.00149 | 0.00401 | 93 | 0.00660 | 0.31196 | 129.714 | 0.75558 | 0.93065 |
| S-(hydroxymethyl)glutathione dehydrogenase | 0.03450 | 0.00323 | 0.02828 | 0.00114 | 0.00419 | 65 | 0.03885 | 0.00198 | 0.00461 | 93 | 0.00623 | -1.69626 | 154.883 | 0.09185 | 0.70632 |
| mannosaminyltransferase | 0.03448 | 0.00222 | 0.03199 | 0.00077 | 0.00344 | 65 | 0.03622 | 0.00079 | 0.00292 | 93 | 0.00451 | -0.93954 | 139.323 | 0.34908 | 0.72019 |
| Cobalt-precorrin-5B (C(1))-methyltransferase | 0.03445 | 0.00254 | 0.04054 | 0.00140 | 0.00464 | 65 | 0.03020 | 0.00072 | 0.00277 | 93 | 0.00541 | 1.91172 | 108.322 | 0.05855 | 0.70632 |
| Guanosine-3’,5’-bis(diphosphate) 3’-diphosphatase | 0.03433 | 0.00244 | 0.03843 | 0.00116 | 0.00423 | 65 | 0.03147 | 0.00078 | 0.00289 | 93 | 0.00513 | 1.35711 | 119.727 | 0.17730 | 0.70632 |
| Pyridoxine 5’-phosphate synthase | 0.03401 | 0.00236 | 0.03097 | 0.00082 | 0.00355 | 65 | 0.03614 | 0.00092 | 0.00315 | 93 | 0.00475 | -1.09044 | 142.676 | 0.27736 | 0.70632 |
| 23S rRNA (uridine(2552)-2’-O)-methyltransferase | 0.03366 | 0.00243 | 0.03280 | 0.00100 | 0.00392 | 65 | 0.03426 | 0.00090 | 0.00310 | 93 | 0.00500 | -0.29254 | 133.088 | 0.77033 | 0.93537 |
| Anhydro-N-acetylmuramic acid kinase | 0.03361 | 0.00246 | 0.03277 | 0.00104 | 0.00401 | 65 | 0.03419 | 0.00091 | 0.00313 | 93 | 0.00508 | -0.27941 | 131.714 | 0.78037 | 0.93675 |
| Triphosphoribosyl-dephospho-CoA synthase | 0.03351 | 0.00301 | 0.04150 | 0.00189 | 0.00539 | 65 | 0.02792 | 0.00106 | 0.00337 | 93 | 0.00636 | 2.13434 | 112.045 | 0.03499 | 0.67652 |
| Oligopeptidase A | 0.03349 | 0.00268 | 0.03398 | 0.00115 | 0.00420 | 65 | 0.03315 | 0.00115 | 0.00351 | 93 | 0.00547 | 0.15171 | 137.816 | 0.87963 | 0.96590 |
| Choloylglycine hydrolase | 0.03348 | 0.00424 | 0.03493 | 0.00349 | 0.00733 | 65 | 0.03247 | 0.00243 | 0.00511 | 93 | 0.00893 | 0.27526 | 121.365 | 0.78359 | 0.93675 |
| D-glycero-beta-D-manno-heptose-7-phosphate kinase | 0.03339 | 0.00213 | 0.03554 | 0.00075 | 0.00339 | 65 | 0.03189 | 0.00070 | 0.00275 | 93 | 0.00436 | 0.83622 | 135.167 | 0.40450 | 0.75675 |
| D-glycero-beta-D-manno-heptose 1-phosphate adenylyltransferase | 0.03339 | 0.00213 | 0.03554 | 0.00075 | 0.00339 | 65 | 0.03189 | 0.00070 | 0.00275 | 93 | 0.00436 | 0.83622 | 135.167 | 0.40450 | 0.75675 |
| Betaine-aldehyde dehydrogenase | 0.03333 | 0.00307 | 0.02981 | 0.00134 | 0.00454 | 65 | 0.03578 | 0.00159 | 0.00413 | 93 | 0.00614 | -0.97214 | 144.754 | 0.33260 | 0.71544 |
| 4-aminobutyrate–2-oxoglutarate transaminase | 0.03332 | 0.00316 | 0.03083 | 0.00152 | 0.00483 | 65 | 0.03506 | 0.00163 | 0.00419 | 93 | 0.00639 | -0.66082 | 141.055 | 0.50980 | 0.81548 |
| (S)-2-hydroxy-acid oxidase | 0.03319 | 0.00300 | 0.03348 | 0.00116 | 0.00422 | 65 | 0.03299 | 0.00162 | 0.00417 | 93 | 0.00593 | 0.08264 | 150.262 | 0.93425 | 0.98985 |
| Glutaconate CoA-transferase | 0.03318 | 0.00418 | 0.04128 | 0.00364 | 0.00748 | 65 | 0.02752 | 0.00211 | 0.00476 | 93 | 0.00887 | 1.55071 | 113.399 | 0.12376 | 0.70632 |
| Hydroxypyruvate isomerase | 0.03312 | 0.00341 | 0.02846 | 0.00178 | 0.00523 | 65 | 0.03639 | 0.00187 | 0.00449 | 93 | 0.00689 | -1.15013 | 140.013 | 0.25205 | 0.70632 |
| Isoquinoline 1-oxidoreductase | 0.03293 | 0.00490 | 0.02718 | 0.00242 | 0.00610 | 65 | 0.03695 | 0.00475 | 0.00715 | 93 | 0.00940 | -1.04026 | 155.923 | 0.29983 | 0.70632 |
| Carboxylesterase | 0.03283 | 0.00329 | 0.03159 | 0.00161 | 0.00498 | 65 | 0.03370 | 0.00179 | 0.00439 | 93 | 0.00664 | -0.31741 | 142.274 | 0.75140 | 0.92944 |
| Deoxyribodipyrimidine photo-lyase | 0.03279 | 0.00282 | 0.03024 | 0.00124 | 0.00437 | 65 | 0.03458 | 0.00127 | 0.00370 | 93 | 0.00572 | -0.75758 | 139.122 | 0.44998 | 0.78036 |
| GDP-mannose 4,6-dehydratase | 0.03275 | 0.00229 | 0.02759 | 0.00050 | 0.00277 | 65 | 0.03635 | 0.00103 | 0.00333 | 93 | 0.00434 | -2.01867 | 155.998 | 0.04523 | 0.69348 |
| Triacylglycerol lipase | 0.03263 | 0.00323 | 0.03157 | 0.00152 | 0.00483 | 65 | 0.03338 | 0.00175 | 0.00434 | 93 | 0.00650 | -0.27746 | 143.749 | 0.78183 | 0.93675 |
| Muramoyltetrapeptide carboxypeptidase | 0.03262 | 0.00220 | 0.03356 | 0.00069 | 0.00327 | 65 | 0.03195 | 0.00082 | 0.00298 | 93 | 0.00442 | 0.36429 | 144.918 | 0.71617 | 0.92120 |
| Neopullulanase | 0.03242 | 0.00263 | 0.03163 | 0.00089 | 0.00371 | 65 | 0.03298 | 0.00125 | 0.00366 | 93 | 0.00521 | -0.25942 | 150.302 | 0.79567 | 0.94089 |
| Threonine-phosphate decarboxylase | 0.03220 | 0.00281 | 0.03777 | 0.00167 | 0.00507 | 65 | 0.02832 | 0.00093 | 0.00316 | 93 | 0.00597 | 1.58192 | 111.825 | 0.11649 | 0.70632 |
| D-ribose pyranase | 0.03200 | 0.00264 | 0.03530 | 0.00147 | 0.00476 | 65 | 0.02970 | 0.00084 | 0.00301 | 93 | 0.00563 | 0.99334 | 112.957 | 0.32267 | 0.70833 |
| Diaminopimelate dehydrogenase | 0.03199 | 0.00278 | 0.03224 | 0.00133 | 0.00453 | 65 | 0.03182 | 0.00116 | 0.00353 | 93 | 0.00574 | 0.07248 | 131.733 | 0.94233 | 0.99049 |
| ADP-glyceromanno-heptose 6-epimerase | 0.03196 | 0.00242 | 0.03741 | 0.00117 | 0.00425 | 65 | 0.02815 | 0.00073 | 0.00280 | 93 | 0.00509 | 1.81934 | 116.451 | 0.07143 | 0.70632 |
| Malonyl-S-ACP decarboxylase | 0.03192 | 0.00442 | 0.03293 | 0.00299 | 0.00678 | 65 | 0.03122 | 0.00320 | 0.00587 | 93 | 0.00896 | 0.19094 | 140.832 | 0.84885 | 0.96012 |
| Arginase | 0.03182 | 0.00273 | 0.02720 | 0.00120 | 0.00429 | 65 | 0.03504 | 0.00115 | 0.00352 | 93 | 0.00555 | -1.41391 | 136.116 | 0.15967 | 0.70632 |
| N-carbamoyl-L-amino-acid hydrolase | 0.03178 | 0.00269 | 0.02799 | 0.00083 | 0.00358 | 65 | 0.03442 | 0.00135 | 0.00381 | 93 | 0.00523 | -1.23063 | 153.812 | 0.22034 | 0.70632 |
| Dipeptidase E | 0.03173 | 0.00248 | 0.03563 | 0.00130 | 0.00447 | 65 | 0.02900 | 0.00073 | 0.00281 | 93 | 0.00528 | 1.25603 | 112.353 | 0.21171 | 0.70632 |
| Phosphonate-transporting ATPase | 0.03169 | 0.00265 | 0.03467 | 0.00134 | 0.00454 | 65 | 0.02962 | 0.00096 | 0.00321 | 93 | 0.00556 | 0.90841 | 122.450 | 0.36545 | 0.72472 |
| Isocitrate lyase | 0.03162 | 0.00269 | 0.02708 | 0.00109 | 0.00409 | 65 | 0.03479 | 0.00116 | 0.00353 | 93 | 0.00541 | -1.42602 | 140.625 | 0.15608 | 0.70632 |
| Homocysteine S-methyltransferase | 0.03155 | 0.00230 | 0.02976 | 0.00085 | 0.00362 | 65 | 0.03280 | 0.00083 | 0.00299 | 93 | 0.00470 | -0.64512 | 136.902 | 0.51993 | 0.82181 |
| 23S rRNA (adenine(2030)-N(6))-methyltransferase | 0.03136 | 0.00235 | 0.03103 | 0.00096 | 0.00383 | 65 | 0.03158 | 0.00082 | 0.00298 | 93 | 0.00485 | -0.11458 | 131.242 | 0.90895 | 0.97756 |
| Agmatine deiminase | 0.03117 | 0.00214 | 0.02916 | 0.00074 | 0.00338 | 65 | 0.03258 | 0.00071 | 0.00277 | 93 | 0.00437 | -0.78231 | 136.164 | 0.43539 | 0.77087 |
| 3-hydroxybutyrate dehydrogenase | 0.03113 | 0.00314 | 0.02586 | 0.00142 | 0.00467 | 65 | 0.03482 | 0.00164 | 0.00421 | 93 | 0.00628 | -1.42607 | 144.048 | 0.15601 | 0.70632 |
| Oligo-1,6-glucosidase | 0.03106 | 0.00343 | 0.03389 | 0.00248 | 0.00618 | 65 | 0.02908 | 0.00143 | 0.00393 | 93 | 0.00732 | 0.65638 | 113.272 | 0.51291 | 0.81548 |
| Glucose-6-phosphate 1-epimerase | 0.03105 | 0.00292 | 0.03309 | 0.00147 | 0.00476 | 65 | 0.02963 | 0.00128 | 0.00371 | 93 | 0.00603 | 0.57261 | 131.651 | 0.56788 | 0.85024 |
| Dextransucrase | 0.03087 | 0.00529 | 0.03102 | 0.00262 | 0.00635 | 65 | 0.03076 | 0.00571 | 0.00784 | 93 | 0.01009 | 0.02488 | 155.879 | 0.98018 | 0.99457 |
| Leucyltransferase | 0.03057 | 0.00232 | 0.02877 | 0.00091 | 0.00374 | 65 | 0.03183 | 0.00082 | 0.00297 | 93 | 0.00477 | -0.64179 | 133.220 | 0.52211 | 0.82182 |
| Oligopeptidase B | 0.03052 | 0.00225 | 0.03034 | 0.00094 | 0.00380 | 65 | 0.03065 | 0.00071 | 0.00277 | 93 | 0.00471 | -0.06539 | 125.392 | 0.94796 | 0.99049 |
| Ribosomal protein L3 N(5)-glutamine methyltransferase | 0.03047 | 0.00237 | 0.03148 | 0.00095 | 0.00382 | 65 | 0.02976 | 0.00085 | 0.00303 | 93 | 0.00487 | 0.35336 | 133.224 | 0.72438 | 0.92120 |
| L-fuculose-phosphate aldolase | 0.03046 | 0.00249 | 0.03589 | 0.00152 | 0.00484 | 65 | 0.02667 | 0.00058 | 0.00249 | 93 | 0.00544 | 1.69418 | 97.584 | 0.09342 | 0.70632 |
| Methylglyoxal synthase | 0.03046 | 0.00289 | 0.03325 | 0.00204 | 0.00560 | 65 | 0.02851 | 0.00083 | 0.00299 | 93 | 0.00635 | 0.74596 | 100.058 | 0.45744 | 0.78637 |
| 23S rRNA pseudouridine(2457) synthase | 0.03024 | 0.00238 | 0.03139 | 0.00095 | 0.00382 | 65 | 0.02944 | 0.00086 | 0.00305 | 93 | 0.00489 | 0.39883 | 133.599 | 0.69065 | 0.90855 |
| Bis(5’-nucleosyl)-tetraphosphatase (symmetrical) | 0.03022 | 0.00237 | 0.03142 | 0.00096 | 0.00383 | 65 | 0.02938 | 0.00085 | 0.00302 | 93 | 0.00488 | 0.41718 | 132.575 | 0.67722 | 0.90040 |
| Phosphoadenylyl-sulfate reductase (thioredoxin) | 0.03020 | 0.00223 | 0.02570 | 0.00069 | 0.00326 | 65 | 0.03335 | 0.00084 | 0.00300 | 93 | 0.00443 | -1.72664 | 145.741 | 0.08635 | 0.70632 |
| Glycine oxidase | 0.03011 | 0.00247 | 0.02720 | 0.00086 | 0.00364 | 65 | 0.03215 | 0.00103 | 0.00334 | 93 | 0.00494 | -1.00274 | 145.235 | 0.31765 | 0.70632 |
| Acetate CoA-transferase | 0.03004 | 0.00610 | 0.04477 | 0.01132 | 0.01319 | 65 | 0.01974 | 0.00189 | 0.00451 | 93 | 0.01394 | 1.79539 | 79.060 | 0.07641 | 0.70632 |
| Uridine phosphorylase | 0.02990 | 0.00247 | 0.02977 | 0.00091 | 0.00374 | 65 | 0.02998 | 0.00101 | 0.00330 | 93 | 0.00499 | -0.04168 | 142.413 | 0.96682 | 0.99193 |
| Taurine dioxygenase | 0.02978 | 0.00400 | 0.02721 | 0.00205 | 0.00562 | 65 | 0.03157 | 0.00288 | 0.00557 | 93 | 0.00791 | -0.55146 | 150.502 | 0.58213 | 0.85342 |
| (S,S)-butanediol dehydrogenase | 0.02976 | 0.00300 | 0.02924 | 0.00197 | 0.00550 | 65 | 0.03013 | 0.00106 | 0.00338 | 93 | 0.00645 | -0.13886 | 110.451 | 0.88981 | 0.96887 |
| Diacetyl reductase ((S)-acetoin forming) | 0.02976 | 0.00300 | 0.02924 | 0.00197 | 0.00550 | 65 | 0.03013 | 0.00106 | 0.00338 | 93 | 0.00645 | -0.13886 | 110.451 | 0.88981 | 0.96887 |
| Butyrate–acetoacetate CoA-transferase | 0.02964 | 0.00608 | 0.04451 | 0.01131 | 0.01319 | 65 | 0.01926 | 0.00184 | 0.00445 | 93 | 0.01392 | 1.81376 | 78.694 | 0.07353 | 0.70632 |
| 2-methylisocitrate dehydratase | 0.02950 | 0.00288 | 0.02932 | 0.00123 | 0.00435 | 65 | 0.02963 | 0.00138 | 0.00386 | 93 | 0.00581 | -0.05369 | 142.869 | 0.95726 | 0.99147 |
| Alkanesulfonate monooxygenase | 0.02932 | 0.00373 | 0.02409 | 0.00144 | 0.00470 | 65 | 0.03297 | 0.00271 | 0.00540 | 93 | 0.00716 | -1.23963 | 155.715 | 0.21698 | 0.70632 |
| Nitric oxide dioxygenase | 0.02929 | 0.00248 | 0.02675 | 0.00105 | 0.00401 | 65 | 0.03107 | 0.00092 | 0.00315 | 93 | 0.00510 | -0.84767 | 132.146 | 0.39815 | 0.74887 |
| Heme-transporting ATPase | 0.02923 | 0.00218 | 0.02914 | 0.00085 | 0.00362 | 65 | 0.02928 | 0.00069 | 0.00272 | 93 | 0.00453 | -0.03092 | 128.236 | 0.97539 | 0.99435 |
| S-formylglutathione hydrolase | 0.02913 | 0.00248 | 0.02732 | 0.00092 | 0.00376 | 65 | 0.03039 | 0.00101 | 0.00330 | 93 | 0.00500 | -0.61429 | 141.978 | 0.54000 | 0.83754 |
| Glyceraldehyde-3-phosphate dehydrogenase (NADP(+)) | 0.02895 | 0.00204 | 0.02970 | 0.00061 | 0.00305 | 65 | 0.02842 | 0.00070 | 0.00275 | 93 | 0.00411 | 0.30975 | 144.059 | 0.75720 | 0.93065 |
| Coproporphyrinogen oxidase | 0.02865 | 0.00257 | 0.02741 | 0.00102 | 0.00397 | 65 | 0.02952 | 0.00106 | 0.00338 | 93 | 0.00521 | -0.40603 | 139.463 | 0.68534 | 0.90745 |
| Glucan 1,6-alpha-glucosidase | 0.02826 | 0.00237 | 0.02897 | 0.00078 | 0.00346 | 65 | 0.02777 | 0.00097 | 0.00324 | 93 | 0.00474 | 0.25393 | 146.784 | 0.79991 | 0.94259 |
| L-seryl-tRNA(Sec) selenium transferase | 0.02817 | 0.00240 | 0.03095 | 0.00103 | 0.00397 | 65 | 0.02623 | 0.00082 | 0.00298 | 93 | 0.00497 | 0.95048 | 127.923 | 0.34366 | 0.72019 |
| Propanediol dehydratase | 0.02809 | 0.00350 | 0.03272 | 0.00248 | 0.00618 | 65 | 0.02485 | 0.00155 | 0.00408 | 93 | 0.00740 | 1.06357 | 116.551 | 0.28972 | 0.70632 |
| Hydrogenase (acceptor) | 0.02807 | 0.00306 | 0.03008 | 0.00177 | 0.00521 | 65 | 0.02667 | 0.00129 | 0.00373 | 93 | 0.00641 | 0.53287 | 123.706 | 0.59508 | 0.86170 |
| Sulfate-transporting ATPase | 0.02803 | 0.00237 | 0.02594 | 0.00079 | 0.00348 | 65 | 0.02950 | 0.00096 | 0.00322 | 93 | 0.00474 | -0.75069 | 145.883 | 0.45405 | 0.78548 |
| Carboxymethylenebutenolidase | 0.02802 | 0.00295 | 0.02239 | 0.00105 | 0.00402 | 65 | 0.03195 | 0.00158 | 0.00412 | 93 | 0.00576 | -1.65857 | 152.185 | 0.09926 | 0.70632 |
| Quaternary-amine-transporting ATPase | 0.02796 | 0.00340 | 0.02949 | 0.00206 | 0.00563 | 65 | 0.02689 | 0.00169 | 0.00426 | 93 | 0.00706 | 0.36771 | 128.753 | 0.71369 | 0.92057 |
| 50S ribosomal protein L16 3-hydroxylase | 0.02790 | 0.00230 | 0.02917 | 0.00087 | 0.00366 | 65 | 0.02701 | 0.00082 | 0.00297 | 93 | 0.00471 | 0.45833 | 135.258 | 0.64745 | 0.88762 |
| Glutaryl-CoA dehydrogenase (ETF) | 0.02776 | 0.00273 | 0.02347 | 0.00107 | 0.00406 | 65 | 0.03076 | 0.00124 | 0.00365 | 93 | 0.00546 | -1.33598 | 143.889 | 0.18366 | 0.70632 |
| Fumarylacetoacetase | 0.02770 | 0.00284 | 0.02544 | 0.00125 | 0.00439 | 65 | 0.02929 | 0.00130 | 0.00373 | 93 | 0.00576 | -0.66746 | 139.302 | 0.50558 | 0.81278 |
| Kanamycin kinase | 0.02767 | 0.00191 | 0.02807 | 0.00057 | 0.00296 | 65 | 0.02740 | 0.00058 | 0.00251 | 93 | 0.00388 | 0.17193 | 138.969 | 0.86374 | 0.96186 |
| DNA ligase (ATP or NAD(+)) | 0.02755 | 0.00225 | 0.02710 | 0.00086 | 0.00365 | 65 | 0.02786 | 0.00076 | 0.00285 | 93 | 0.00463 | -0.16506 | 131.936 | 0.86915 | 0.96353 |
| DNA ligase (ATP, ADP or GTP) | 0.02755 | 0.00225 | 0.02710 | 0.00086 | 0.00365 | 65 | 0.02786 | 0.00076 | 0.00285 | 93 | 0.00463 | -0.16506 | 131.936 | 0.86915 | 0.96353 |
| Hippurate hydrolase | 0.02738 | 0.00307 | 0.02190 | 0.00093 | 0.00378 | 65 | 0.03122 | 0.00186 | 0.00447 | 93 | 0.00585 | -1.59215 | 155.971 | 0.11337 | 0.70632 |
| Electron-transferring-flavoprotein dehydrogenase | 0.02738 | 0.00253 | 0.02492 | 0.00096 | 0.00384 | 65 | 0.02910 | 0.00106 | 0.00337 | 93 | 0.00511 | -0.81670 | 141.853 | 0.41547 | 0.76374 |
| ATP diphosphatase | 0.02737 | 0.00240 | 0.02705 | 0.00089 | 0.00371 | 65 | 0.02760 | 0.00093 | 0.00317 | 93 | 0.00488 | -0.11296 | 139.743 | 0.91022 | 0.97775 |
| Guanine deaminase | 0.02711 | 0.00258 | 0.02522 | 0.00096 | 0.00385 | 65 | 0.02844 | 0.00111 | 0.00346 | 93 | 0.00518 | -0.62052 | 143.756 | 0.53590 | 0.83395 |
| Protocatechuate 3,4-dioxygenase | 0.02706 | 0.00336 | 0.02597 | 0.00177 | 0.00522 | 65 | 0.02783 | 0.00181 | 0.00442 | 93 | 0.00684 | -0.27156 | 138.977 | 0.78637 | 0.93675 |
| Oleate hydratase | 0.02703 | 0.00327 | 0.02931 | 0.00220 | 0.00582 | 65 | 0.02544 | 0.00135 | 0.00381 | 93 | 0.00695 | 0.55727 | 115.716 | 0.57842 | 0.85190 |
| 23S rRNA (guanine(2069)-N(7))-methyltransferase | 0.02696 | 0.00240 | 0.02825 | 0.00088 | 0.00367 | 65 | 0.02607 | 0.00094 | 0.00319 | 93 | 0.00486 | 0.44823 | 140.992 | 0.65468 | 0.88943 |
| 23S rRNA (guanine(2445)-N(2))-methyltransferase | 0.02696 | 0.00240 | 0.02825 | 0.00088 | 0.00367 | 65 | 0.02607 | 0.00094 | 0.00319 | 93 | 0.00486 | 0.44823 | 140.992 | 0.65468 | 0.88943 |
| Phosphogluconate dehydratase | 0.02683 | 0.00287 | 0.02525 | 0.00120 | 0.00429 | 65 | 0.02794 | 0.00139 | 0.00386 | 93 | 0.00578 | -0.46549 | 143.942 | 0.64228 | 0.88744 |
| Glutathione synthase | 0.02683 | 0.00231 | 0.02570 | 0.00088 | 0.00368 | 65 | 0.02761 | 0.00082 | 0.00298 | 93 | 0.00473 | -0.40464 | 134.996 | 0.68638 | 0.90745 |
| Gamma-glutamyl hercynylcysteine S-oxide hydrolase | 0.02681 | 0.00228 | 0.02657 | 0.00077 | 0.00345 | 65 | 0.02699 | 0.00086 | 0.00305 | 93 | 0.00460 | -0.09100 | 142.446 | 0.92762 | 0.98985 |
| GDP-L-fucose synthase | 0.02679 | 0.00236 | 0.02146 | 0.00061 | 0.00306 | 65 | 0.03051 | 0.00105 | 0.00336 | 93 | 0.00454 | -1.99244 | 154.791 | 0.04808 | 0.69870 |
| tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase | 0.02679 | 0.00266 | 0.02758 | 0.00106 | 0.00404 | 65 | 0.02623 | 0.00118 | 0.00356 | 93 | 0.00538 | 0.25082 | 142.300 | 0.80231 | 0.94263 |
| Adenylyl-sulfate kinase | 0.02663 | 0.00280 | 0.02035 | 0.00077 | 0.00345 | 65 | 0.03103 | 0.00154 | 0.00406 | 93 | 0.00533 | -2.00392 | 155.956 | 0.04681 | 0.69348 |
| Superoxide reductase | 0.02662 | 0.00255 | 0.02852 | 0.00124 | 0.00436 | 65 | 0.02530 | 0.00088 | 0.00308 | 93 | 0.00534 | 0.60368 | 122.625 | 0.54717 | 0.84303 |
| Lactaldehyde reductase | 0.02657 | 0.00347 | 0.03467 | 0.00314 | 0.00695 | 65 | 0.02091 | 0.00098 | 0.00325 | 93 | 0.00767 | 1.79273 | 91.924 | 0.07631 | 0.70632 |
| Sialate O-acetylesterase | 0.02640 | 0.00278 | 0.02083 | 0.00061 | 0.00307 | 65 | 0.03030 | 0.00162 | 0.00418 | 93 | 0.00519 | -1.82655 | 153.733 | 0.06971 | 0.70632 |
| (S)-2-haloacid dehalogenase | 0.02639 | 0.00208 | 0.02101 | 0.00074 | 0.00337 | 65 | 0.03014 | 0.00062 | 0.00259 | 93 | 0.00425 | -2.14897 | 130.105 | 0.03349 | 0.67652 |
| 2,4-dienoyl-CoA reductase (NADPH) | 0.02626 | 0.00273 | 0.02512 | 0.00106 | 0.00404 | 65 | 0.02705 | 0.00127 | 0.00370 | 93 | 0.00548 | -0.35184 | 145.233 | 0.72547 | 0.92120 |
| Glycerol-3-phosphate oxidase | 0.02601 | 0.00250 | 0.02367 | 0.00075 | 0.00339 | 65 | 0.02764 | 0.00116 | 0.00352 | 93 | 0.00489 | -0.81248 | 152.832 | 0.41778 | 0.76408 |
| Fructan beta-fructosidase | 0.02576 | 0.00199 | 0.02288 | 0.00046 | 0.00267 | 65 | 0.02776 | 0.00073 | 0.00281 | 93 | 0.00388 | -1.25928 | 153.382 | 0.20984 | 0.70632 |
| 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase | 0.02564 | 0.00233 | 0.02681 | 0.00114 | 0.00418 | 65 | 0.02482 | 0.00067 | 0.00268 | 93 | 0.00497 | 0.40114 | 113.941 | 0.68907 | 0.90847 |
| D-Ala-D-Ala dipeptidase | 0.02546 | 0.00197 | 0.02149 | 0.00043 | 0.00258 | 65 | 0.02824 | 0.00072 | 0.00278 | 93 | 0.00380 | -1.77709 | 154.280 | 0.07752 | 0.70632 |
| Type III site-specific deoxyribonuclease | 0.02538 | 0.00254 | 0.02669 | 0.00091 | 0.00373 | 65 | 0.02446 | 0.00110 | 0.00344 | 93 | 0.00508 | 0.43812 | 145.784 | 0.66195 | 0.88968 |
| Methionine gamma-lyase | 0.02515 | 0.00190 | 0.02695 | 0.00080 | 0.00352 | 65 | 0.02389 | 0.00041 | 0.00210 | 93 | 0.00410 | 0.74670 | 108.357 | 0.45686 | 0.78637 |
| Prolyl oligopeptidase | 0.02514 | 0.00197 | 0.02332 | 0.00048 | 0.00271 | 65 | 0.02641 | 0.00071 | 0.00277 | 93 | 0.00387 | -0.79776 | 152.201 | 0.42625 | 0.76494 |
| 3-dehydro-L-gulonate-6-phosphate decarboxylase | 0.02511 | 0.00214 | 0.02756 | 0.00096 | 0.00383 | 65 | 0.02339 | 0.00057 | 0.00247 | 93 | 0.00456 | 0.91387 | 114.459 | 0.36271 | 0.72448 |
| tRNA(1)(Val) (adenine(37)-N(6))-methyltransferase | 0.02496 | 0.00217 | 0.02419 | 0.00066 | 0.00318 | 65 | 0.02549 | 0.00081 | 0.00296 | 93 | 0.00434 | -0.30050 | 146.429 | 0.76422 | 0.93350 |
| Cytosine deaminase | 0.02493 | 0.00240 | 0.02381 | 0.00089 | 0.00370 | 65 | 0.02572 | 0.00093 | 0.00316 | 93 | 0.00486 | -0.39258 | 139.549 | 0.69523 | 0.91004 |
| Gluconate 5-dehydrogenase | 0.02492 | 0.00248 | 0.02652 | 0.00140 | 0.00465 | 65 | 0.02380 | 0.00068 | 0.00271 | 93 | 0.00538 | 0.50494 | 106.482 | 0.61465 | 0.87323 |
| Quinol–cytochrome-c reductase | 0.02482 | 0.00223 | 0.02323 | 0.00083 | 0.00358 | 65 | 0.02594 | 0.00076 | 0.00287 | 93 | 0.00458 | -0.59168 | 134.054 | 0.55506 | 0.84600 |
| L(+)-tartrate dehydratase | 0.02477 | 0.00289 | 0.02726 | 0.00191 | 0.00542 | 65 | 0.02304 | 0.00092 | 0.00315 | 93 | 0.00627 | 0.67341 | 105.993 | 0.50215 | 0.81035 |
| tRNA (cytidine(32)/uridine(32)-2’-O)-methyltransferase | 0.02465 | 0.00216 | 0.02618 | 0.00074 | 0.00337 | 65 | 0.02358 | 0.00074 | 0.00282 | 93 | 0.00440 | 0.59164 | 138.028 | 0.55506 | 0.84600 |
| tRNA (uracil(54)-C(5))-methyltransferase | 0.02464 | 0.00197 | 0.02547 | 0.00061 | 0.00307 | 65 | 0.02407 | 0.00062 | 0.00257 | 93 | 0.00401 | 0.34951 | 138.087 | 0.72724 | 0.92120 |
| AMP nucleosidase | 0.02449 | 0.00233 | 0.02219 | 0.00087 | 0.00366 | 65 | 0.02609 | 0.00085 | 0.00302 | 93 | 0.00475 | -0.82145 | 136.661 | 0.41282 | 0.76224 |
| Chloride peroxidase | 0.02441 | 0.00313 | 0.02271 | 0.00157 | 0.00491 | 65 | 0.02560 | 0.00155 | 0.00409 | 93 | 0.00639 | -0.45310 | 137.550 | 0.65119 | 0.88943 |
| Exodeoxyribonuclease I | 0.02426 | 0.00217 | 0.02548 | 0.00078 | 0.00346 | 65 | 0.02342 | 0.00072 | 0.00279 | 93 | 0.00445 | 0.46306 | 134.650 | 0.64407 | 0.88749 |
| Methylisocitrate lyase | 0.02422 | 0.00225 | 0.02357 | 0.00084 | 0.00360 | 65 | 0.02467 | 0.00078 | 0.00289 | 93 | 0.00462 | -0.23879 | 134.295 | 0.81163 | 0.94519 |
| tRNA pseudouridine(13) synthase | 0.02416 | 0.00218 | 0.02550 | 0.00073 | 0.00335 | 65 | 0.02322 | 0.00077 | 0.00287 | 93 | 0.00441 | 0.51623 | 140.001 | 0.60651 | 0.86933 |
| Methylmalonyl-CoA epimerase | 0.02411 | 0.00181 | 0.02039 | 0.00049 | 0.00273 | 65 | 0.02671 | 0.00053 | 0.00238 | 93 | 0.00362 | -1.74549 | 141.344 | 0.08307 | 0.70632 |
| Diglucosyl diacylglycerol synthase (1,2-linking) | 0.02410 | 0.00204 | 0.02657 | 0.00068 | 0.00322 | 65 | 0.02237 | 0.00065 | 0.00264 | 93 | 0.00417 | 1.00913 | 136.066 | 0.31470 | 0.70632 |
| Arginine N-succinyltransferase | 0.02401 | 0.00324 | 0.02496 | 0.00173 | 0.00516 | 65 | 0.02334 | 0.00163 | 0.00419 | 93 | 0.00665 | 0.24341 | 135.182 | 0.80806 | 0.94429 |
| Carboxypeptidase Taq | 0.02391 | 0.00233 | 0.02724 | 0.00137 | 0.00460 | 65 | 0.02159 | 0.00049 | 0.00230 | 93 | 0.00514 | 1.09870 | 95.841 | 0.27465 | 0.70632 |
| Precorrin-6B C(5,15)-methyltransferase (decarboxylating) | 0.02389 | 0.00232 | 0.02331 | 0.00125 | 0.00439 | 65 | 0.02429 | 0.00058 | 0.00249 | 93 | 0.00504 | -0.19612 | 104.402 | 0.84489 | 0.95939 |
| Vanillate monooxygenase | 0.02383 | 0.00334 | 0.02225 | 0.00175 | 0.00519 | 65 | 0.02493 | 0.00178 | 0.00438 | 93 | 0.00679 | -0.39493 | 138.575 | 0.69350 | 0.90946 |
| 5’ to 3’ exodeoxyribonuclease (nucleoside 3’-phosphate-forming) | 0.02381 | 0.00214 | 0.02655 | 0.00099 | 0.00390 | 65 | 0.02191 | 0.00054 | 0.00240 | 93 | 0.00458 | 1.01361 | 110.803 | 0.31298 | 0.70632 |
| Arginyltransferase | 0.02375 | 0.00232 | 0.02218 | 0.00091 | 0.00375 | 65 | 0.02485 | 0.00081 | 0.00295 | 93 | 0.00477 | -0.55872 | 132.518 | 0.57729 | 0.85190 |
| Coniferyl-aldehyde dehydrogenase | 0.02369 | 0.00257 | 0.02198 | 0.00097 | 0.00386 | 65 | 0.02489 | 0.00110 | 0.00344 | 93 | 0.00517 | -0.56442 | 143.108 | 0.57335 | 0.85190 |
| 4a-hydroxytetrahydrobiopterin dehydratase | 0.02366 | 0.00226 | 0.02050 | 0.00085 | 0.00361 | 65 | 0.02588 | 0.00077 | 0.00288 | 93 | 0.00462 | -1.16340 | 133.778 | 0.24674 | 0.70632 |
| Quinoprotein glucose dehydrogenase (PQQ, quinone) | 0.02343 | 0.00277 | 0.02234 | 0.00114 | 0.00418 | 65 | 0.02419 | 0.00128 | 0.00371 | 93 | 0.00559 | -0.33155 | 142.960 | 0.74071 | 0.92600 |
| 3-oxoadipate enol-lactonase | 0.02338 | 0.00262 | 0.02050 | 0.00097 | 0.00385 | 65 | 0.02540 | 0.00117 | 0.00354 | 93 | 0.00524 | -0.93611 | 145.587 | 0.35077 | 0.72019 |
| Chlorite O(2)-lyase | 0.02331 | 0.00199 | 0.02087 | 0.00062 | 0.00309 | 65 | 0.02502 | 0.00063 | 0.00260 | 93 | 0.00403 | -1.03019 | 138.410 | 0.30472 | 0.70632 |
| Protein-glutamine glutaminase | 0.02323 | 0.00217 | 0.02058 | 0.00072 | 0.00332 | 65 | 0.02508 | 0.00077 | 0.00287 | 93 | 0.00439 | -1.02513 | 140.928 | 0.30706 | 0.70632 |
| Diaminobutyrate–2-oxoglutarate transaminase | 0.02313 | 0.00298 | 0.02194 | 0.00119 | 0.00428 | 65 | 0.02396 | 0.00157 | 0.00410 | 93 | 0.00593 | -0.34086 | 148.512 | 0.73369 | 0.92120 |
| Menaquinol oxidase (H(+)-transporting) | 0.02301 | 0.00615 | 0.01290 | 0.00243 | 0.00611 | 65 | 0.03009 | 0.00840 | 0.00950 | 93 | 0.01130 | -1.52151 | 147.537 | 0.13027 | 0.70632 |
| Poly(glycerol-phosphate) alpha-glucosyltransferase | 0.02294 | 0.00279 | 0.02410 | 0.00180 | 0.00527 | 65 | 0.02214 | 0.00085 | 0.00302 | 93 | 0.00607 | 0.32261 | 105.057 | 0.74763 | 0.92904 |
| Hydroxypyruvate reductase | 0.02280 | 0.00261 | 0.01784 | 0.00069 | 0.00327 | 65 | 0.02627 | 0.00132 | 0.00377 | 93 | 0.00499 | -1.68690 | 155.774 | 0.09362 | 0.70632 |
| 16S rRNA (guanine(1516)-N(2))-methyltransferase | 0.02262 | 0.00200 | 0.02435 | 0.00065 | 0.00317 | 65 | 0.02141 | 0.00062 | 0.00258 | 93 | 0.00409 | 0.71851 | 135.570 | 0.47368 | 0.79541 |
| Rhomboid protease | 0.02251 | 0.00302 | 0.02636 | 0.00229 | 0.00594 | 65 | 0.01982 | 0.00085 | 0.00303 | 93 | 0.00667 | 0.98141 | 97.072 | 0.32883 | 0.71151 |
| Methylglutaconyl-CoA hydratase | 0.02236 | 0.00221 | 0.02095 | 0.00081 | 0.00354 | 65 | 0.02335 | 0.00075 | 0.00283 | 93 | 0.00453 | -0.52848 | 133.974 | 0.59804 | 0.86297 |
| 2-methylcitrate synthase | 0.02234 | 0.00223 | 0.02265 | 0.00082 | 0.00356 | 65 | 0.02212 | 0.00076 | 0.00286 | 93 | 0.00457 | 0.11549 | 134.460 | 0.90823 | 0.97756 |
| Glycerol-3-phosphate cytidylyltransferase | 0.02233 | 0.00220 | 0.02610 | 0.00107 | 0.00405 | 65 | 0.01969 | 0.00055 | 0.00243 | 93 | 0.00472 | 1.35710 | 108.545 | 0.17756 | 0.70632 |
| Protein arginine kinase | 0.02227 | 0.00210 | 0.02320 | 0.00091 | 0.00375 | 65 | 0.02161 | 0.00055 | 0.00244 | 93 | 0.00447 | 0.35488 | 115.371 | 0.72333 | 0.92120 |
| D-amino-acid transaminase | 0.02226 | 0.00188 | 0.01995 | 0.00058 | 0.00299 | 65 | 0.02388 | 0.00054 | 0.00241 | 93 | 0.00384 | -1.02569 | 134.632 | 0.30688 | 0.70632 |
| 3-carboxy-cis,cis-muconate cycloisomerase | 0.02224 | 0.00387 | 0.01986 | 0.00188 | 0.00537 | 65 | 0.02389 | 0.00273 | 0.00542 | 93 | 0.00763 | -0.52839 | 151.463 | 0.59800 | 0.86297 |
| Indolepyruvate ferredoxin oxidoreductase | 0.02216 | 0.00227 | 0.01899 | 0.00071 | 0.00330 | 65 | 0.02437 | 0.00089 | 0.00309 | 93 | 0.00452 | -1.18946 | 146.878 | 0.23618 | 0.70632 |
| Nitrite reductase (cytochrome; ammonia-forming) | 0.02212 | 0.00221 | 0.02293 | 0.00071 | 0.00330 | 65 | 0.02155 | 0.00082 | 0.00297 | 93 | 0.00444 | 0.31122 | 144.124 | 0.75608 | 0.93065 |
| LL-diaminopimelate aminotransferase | 0.02205 | 0.00223 | 0.02204 | 0.00089 | 0.00370 | 65 | 0.02206 | 0.00072 | 0.00279 | 93 | 0.00463 | -0.00448 | 128.550 | 0.99643 | 0.99921 |
| Altronate dehydratase | 0.02203 | 0.00182 | 0.02266 | 0.00066 | 0.00319 | 65 | 0.02159 | 0.00044 | 0.00217 | 93 | 0.00385 | 0.27929 | 119.188 | 0.78051 | 0.93675 |
| 23S rRNA (cytidine(2498)-2’-O)-methyltransferase | 0.02197 | 0.00207 | 0.02383 | 0.00071 | 0.00330 | 65 | 0.02066 | 0.00065 | 0.00265 | 93 | 0.00423 | 0.74760 | 134.302 | 0.45601 | 0.78637 |
| Agmatinase | 0.02177 | 0.00191 | 0.02072 | 0.00055 | 0.00291 | 65 | 0.02250 | 0.00060 | 0.00254 | 93 | 0.00386 | -0.45846 | 141.330 | 0.64733 | 0.88762 |
| Citrate CoA-transferase | 0.02174 | 0.00215 | 0.02823 | 0.00112 | 0.00416 | 65 | 0.01720 | 0.00041 | 0.00210 | 93 | 0.00466 | 2.36543 | 96.575 | 0.02001 | 0.64530 |
| Isovaleryl-CoA dehydrogenase | 0.02174 | 0.00225 | 0.01953 | 0.00082 | 0.00355 | 65 | 0.02328 | 0.00079 | 0.00291 | 93 | 0.00459 | -0.81726 | 136.217 | 0.41521 | 0.76374 |
| Hydroxyisourate hydrolase | 0.02173 | 0.00251 | 0.01908 | 0.00082 | 0.00356 | 65 | 0.02358 | 0.00111 | 0.00346 | 93 | 0.00496 | -0.90786 | 149.394 | 0.36542 | 0.72472 |
| Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase | 0.02135 | 0.00236 | 0.02316 | 0.00145 | 0.00473 | 65 | 0.02008 | 0.00049 | 0.00229 | 93 | 0.00525 | 0.58515 | 94.046 | 0.55985 | 0.85024 |
| Leucine dehydrogenase | 0.02111 | 0.00216 | 0.01874 | 0.00072 | 0.00334 | 65 | 0.02276 | 0.00074 | 0.00283 | 93 | 0.00438 | -0.91927 | 139.085 | 0.35955 | 0.72286 |
| Penicillin amidase | 0.02094 | 0.00238 | 0.01913 | 0.00084 | 0.00361 | 65 | 0.02220 | 0.00094 | 0.00318 | 93 | 0.00481 | -0.64002 | 142.377 | 0.52319 | 0.82182 |
| Cytochrome-c peroxidase | 0.02091 | 0.00202 | 0.02142 | 0.00062 | 0.00308 | 65 | 0.02056 | 0.00067 | 0.00268 | 93 | 0.00408 | 0.21144 | 141.301 | 0.83285 | 0.95544 |
| Aldehyde dehydrogenase (NADP(+)) | 0.02089 | 0.00315 | 0.02032 | 0.00141 | 0.00465 | 65 | 0.02129 | 0.00170 | 0.00427 | 93 | 0.00632 | -0.15342 | 145.499 | 0.87828 | 0.96551 |
| Formyl-CoA transferase | 0.02089 | 0.00262 | 0.01791 | 0.00112 | 0.00415 | 65 | 0.02297 | 0.00106 | 0.00338 | 93 | 0.00536 | -0.94512 | 135.471 | 0.34628 | 0.72019 |
| Phenylacetate–CoA ligase | 0.02086 | 0.00217 | 0.01644 | 0.00045 | 0.00264 | 65 | 0.02394 | 0.00093 | 0.00316 | 93 | 0.00412 | -1.82042 | 155.999 | 0.07061 | 0.70632 |
| Phosphoribosyl 1,2-cyclic phosphate phosphodiesterase | 0.02057 | 0.00163 | 0.01634 | 0.00027 | 0.00205 | 65 | 0.02352 | 0.00050 | 0.00233 | 93 | 0.00310 | -2.31436 | 155.519 | 0.02196 | 0.65070 |
| Cephalosporin-C deacetylase | 0.02047 | 0.00195 | 0.02031 | 0.00053 | 0.00286 | 65 | 0.02058 | 0.00066 | 0.00266 | 93 | 0.00390 | -0.07001 | 146.600 | 0.94428 | 0.99049 |
| Stearoyl-CoA 9-desaturase | 0.02042 | 0.00292 | 0.01839 | 0.00130 | 0.00446 | 65 | 0.02183 | 0.00140 | 0.00388 | 93 | 0.00591 | -0.58081 | 141.121 | 0.56230 | 0.85024 |
| Cystathionine gamma-lyase | 0.02035 | 0.00217 | 0.01807 | 0.00064 | 0.00314 | 65 | 0.02195 | 0.00082 | 0.00296 | 93 | 0.00432 | -0.89865 | 147.297 | 0.37031 | 0.73093 |
| Lysine decarboxylase | 0.02029 | 0.00173 | 0.01623 | 0.00028 | 0.00208 | 65 | 0.02313 | 0.00059 | 0.00252 | 93 | 0.00327 | -2.11193 | 155.975 | 0.03629 | 0.67652 |
| Cobalt-precorrin-7 (C(5))-methyltransferase | 0.02027 | 0.00226 | 0.02699 | 0.00122 | 0.00433 | 65 | 0.01558 | 0.00047 | 0.00226 | 93 | 0.00489 | 2.33517 | 98.412 | 0.02157 | 0.65070 |
| Glutamate carboxypeptidase | 0.02012 | 0.00179 | 0.01699 | 0.00038 | 0.00240 | 65 | 0.02230 | 0.00059 | 0.00252 | 93 | 0.00348 | -1.52891 | 153.180 | 0.12835 | 0.70632 |
| Cobalt-precorrin-6B (C(15))-methyltransferase (decarboxylating) | 0.01999 | 0.00226 | 0.02690 | 0.00122 | 0.00433 | 65 | 0.01516 | 0.00047 | 0.00225 | 93 | 0.00488 | 2.40476 | 98.190 | 0.01806 | 0.64530 |
| Alpha-mannosidase | 0.01997 | 0.00180 | 0.02041 | 0.00062 | 0.00309 | 65 | 0.01966 | 0.00044 | 0.00217 | 93 | 0.00378 | 0.19846 | 121.967 | 0.84302 | 0.95939 |
| Maleylacetoacetate isomerase | 0.01994 | 0.00214 | 0.01884 | 0.00076 | 0.00342 | 65 | 0.02072 | 0.00070 | 0.00275 | 93 | 0.00439 | -0.42777 | 134.495 | 0.66950 | 0.89503 |
| 3-oxoadipate CoA-transferase | 0.01992 | 0.00345 | 0.01671 | 0.00083 | 0.00356 | 65 | 0.02217 | 0.00262 | 0.00530 | 93 | 0.00639 | -0.85519 | 149.907 | 0.39381 | 0.74598 |
| Pyruvate dehydrogenase (quinone) | 0.01987 | 0.00225 | 0.01887 | 0.00075 | 0.00340 | 65 | 0.02057 | 0.00084 | 0.00301 | 93 | 0.00454 | -0.37308 | 142.748 | 0.70964 | 0.91799 |
| Tetrahydrodipicolinate N-acetyltransferase | 0.01983 | 0.00197 | 0.01999 | 0.00088 | 0.00368 | 65 | 0.01973 | 0.00043 | 0.00215 | 93 | 0.00426 | 0.06103 | 106.516 | 0.95145 | 0.99049 |
| Homogentisate 1,2-dioxygenase | 0.01932 | 0.00206 | 0.01828 | 0.00074 | 0.00337 | 65 | 0.02005 | 0.00063 | 0.00260 | 93 | 0.00426 | -0.41560 | 130.764 | 0.67839 | 0.90139 |
| D-serine ammonia-lyase | 0.01931 | 0.00254 | 0.02336 | 0.00137 | 0.00459 | 65 | 0.01649 | 0.00077 | 0.00287 | 93 | 0.00542 | 1.26762 | 112.034 | 0.20756 | 0.70632 |
| Gluconate 2-dehydrogenase | 0.01920 | 0.00222 | 0.01886 | 0.00076 | 0.00342 | 65 | 0.01944 | 0.00081 | 0.00294 | 93 | 0.00451 | -0.13010 | 140.457 | 0.89667 | 0.97163 |
| Diaminopropionate ammonia-lyase | 0.01918 | 0.00193 | 0.02115 | 0.00075 | 0.00340 | 65 | 0.01781 | 0.00048 | 0.00227 | 93 | 0.00409 | 0.81827 | 117.342 | 0.41486 | 0.76374 |
| Mannonate dehydratase | 0.01896 | 0.00202 | 0.01902 | 0.00093 | 0.00378 | 65 | 0.01892 | 0.00046 | 0.00222 | 93 | 0.00438 | 0.02327 | 106.832 | 0.98148 | 0.99457 |
| Glutamate dehydrogenase (NAD(P)(+)) | 0.01882 | 0.00164 | 0.01818 | 0.00040 | 0.00247 | 65 | 0.01926 | 0.00045 | 0.00221 | 93 | 0.00331 | -0.32784 | 143.482 | 0.74351 | 0.92676 |
| S-methyl-5’-thioadenosine phosphorylase | 0.01881 | 0.00165 | 0.01878 | 0.00047 | 0.00269 | 65 | 0.01883 | 0.00041 | 0.00209 | 93 | 0.00340 | -0.01526 | 131.288 | 0.98784 | 0.99660 |
| Nitrite reductase (NO-forming) | 0.01870 | 0.00188 | 0.01902 | 0.00054 | 0.00289 | 65 | 0.01848 | 0.00057 | 0.00248 | 93 | 0.00381 | 0.14150 | 140.256 | 0.88768 | 0.96887 |
| Sucrose phosphorylase | 0.01863 | 0.00168 | 0.01951 | 0.00062 | 0.00309 | 65 | 0.01802 | 0.00033 | 0.00189 | 93 | 0.00362 | 0.41067 | 110.411 | 0.68211 | 0.90520 |
| Beta-alanine–pyruvate transaminase | 0.01856 | 0.00209 | 0.01737 | 0.00072 | 0.00332 | 65 | 0.01940 | 0.00067 | 0.00269 | 93 | 0.00428 | -0.47566 | 135.143 | 0.63509 | 0.88317 |
| 2-hydroxy-3-oxopropionate reductase | 0.01840 | 0.00186 | 0.01530 | 0.00044 | 0.00260 | 65 | 0.02057 | 0.00061 | 0.00257 | 93 | 0.00365 | -1.44052 | 150.191 | 0.15180 | 0.70632 |
| Gluconate 2-dehydrogenase (acceptor) | 0.01837 | 0.00387 | 0.01667 | 0.00178 | 0.00523 | 65 | 0.01956 | 0.00280 | 0.00549 | 93 | 0.00758 | -0.38084 | 153.231 | 0.70385 | 0.91393 |
| Mannitol-1-phosphate 5-dehydrogenase | 0.01830 | 0.00190 | 0.01908 | 0.00082 | 0.00356 | 65 | 0.01776 | 0.00040 | 0.00207 | 93 | 0.00412 | 0.32078 | 106.234 | 0.74901 | 0.92904 |
| 2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA isomerase | 0.01828 | 0.00213 | 0.01281 | 0.00034 | 0.00227 | 65 | 0.02210 | 0.00095 | 0.00320 | 93 | 0.00392 | -2.37032 | 152.500 | 0.01902 | 0.64530 |
| RNA ligase (ATP) | 0.01826 | 0.00175 | 0.01651 | 0.00044 | 0.00261 | 65 | 0.01949 | 0.00052 | 0.00236 | 93 | 0.00352 | -0.84488 | 144.222 | 0.39958 | 0.75019 |
| Poly(beta-D-mannuronate) lyase | 0.01821 | 0.00284 | 0.01896 | 0.00141 | 0.00465 | 65 | 0.01768 | 0.00120 | 0.00359 | 93 | 0.00588 | 0.21804 | 130.727 | 0.82774 | 0.95262 |
| Sulfur carrier protein ThiS adenylyltransferase | 0.01811 | 0.00198 | 0.02359 | 0.00093 | 0.00378 | 65 | 0.01427 | 0.00037 | 0.00200 | 93 | 0.00428 | 2.17766 | 99.218 | 0.03180 | 0.67652 |
| tRNA pseudouridine(65) synthase | 0.01802 | 0.00196 | 0.01917 | 0.00076 | 0.00343 | 65 | 0.01721 | 0.00050 | 0.00232 | 93 | 0.00414 | 0.47307 | 118.850 | 0.63703 | 0.88482 |
| Nickel-transporting ATPase | 0.01796 | 0.00349 | 0.02391 | 0.00309 | 0.00689 | 65 | 0.01380 | 0.00108 | 0.00341 | 93 | 0.00769 | 1.31407 | 95.231 | 0.19198 | 0.70632 |
| Maltose alpha-D-glucosyltransferase | 0.01795 | 0.00246 | 0.01735 | 0.00087 | 0.00365 | 65 | 0.01837 | 0.00103 | 0.00333 | 93 | 0.00494 | -0.20680 | 145.078 | 0.83645 | 0.95674 |
| Adenine deaminase | 0.01790 | 0.00227 | 0.01532 | 0.00084 | 0.00360 | 65 | 0.01971 | 0.00080 | 0.00292 | 93 | 0.00464 | -0.94504 | 135.332 | 0.34632 | 0.72019 |
| Diglucosyl diacylglycerol synthase (1,6-linking) | 0.01790 | 0.00170 | 0.01864 | 0.00061 | 0.00306 | 65 | 0.01739 | 0.00036 | 0.00197 | 93 | 0.00364 | 0.34495 | 114.293 | 0.73076 | 0.92120 |
| Arylsulfatase | 0.01780 | 0.00203 | 0.01321 | 0.00016 | 0.00155 | 65 | 0.02100 | 0.00097 | 0.00324 | 93 | 0.00359 | -2.17011 | 129.410 | 0.03182 | 0.67652 |
| Fructose-6-phosphate phosphoketolase | 0.01765 | 0.00250 | 0.02001 | 0.00144 | 0.00471 | 65 | 0.01600 | 0.00067 | 0.00269 | 93 | 0.00542 | 0.73972 | 104.693 | 0.46113 | 0.78637 |
| Phosphoketolase | 0.01765 | 0.00250 | 0.02001 | 0.00144 | 0.00471 | 65 | 0.01600 | 0.00067 | 0.00269 | 93 | 0.00542 | 0.73972 | 104.693 | 0.46113 | 0.78637 |
| Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase | 0.01730 | 0.00338 | 0.00886 | 0.00060 | 0.00304 | 65 | 0.02320 | 0.00257 | 0.00525 | 93 | 0.00607 | -2.36179 | 141.233 | 0.01955 | 0.64530 |
| Acyl-homoserine-lactone acylase | 0.01727 | 0.00298 | 0.01699 | 0.00113 | 0.00416 | 65 | 0.01747 | 0.00161 | 0.00416 | 93 | 0.00588 | -0.08192 | 150.933 | 0.93482 | 0.98985 |
| Beta-phosphoglucomutase | 0.01710 | 0.00256 | 0.02170 | 0.00161 | 0.00498 | 65 | 0.01388 | 0.00062 | 0.00258 | 93 | 0.00561 | 1.39321 | 97.906 | 0.16671 | 0.70632 |
| Acetoacetyl-CoA reductase | 0.01700 | 0.00273 | 0.01500 | 0.00128 | 0.00444 | 65 | 0.01840 | 0.00112 | 0.00347 | 93 | 0.00563 | -0.60302 | 131.864 | 0.54753 | 0.84303 |
| UDP-glucuronate 4-epimerase | 0.01693 | 0.00171 | 0.01515 | 0.00044 | 0.00262 | 65 | 0.01818 | 0.00047 | 0.00225 | 93 | 0.00345 | -0.87991 | 140.367 | 0.38041 | 0.73587 |
| Phenylalanine 4-monooxygenase | 0.01684 | 0.00198 | 0.01617 | 0.00069 | 0.00326 | 65 | 0.01730 | 0.00057 | 0.00249 | 93 | 0.00410 | -0.27568 | 129.727 | 0.78323 | 0.93675 |
| 4-phosphoerythronate dehydrogenase | 0.01673 | 0.00162 | 0.01678 | 0.00039 | 0.00246 | 65 | 0.01670 | 0.00043 | 0.00215 | 93 | 0.00327 | 0.02388 | 141.551 | 0.98098 | 0.99457 |
| Lactocepin | 0.01671 | 0.00148 | 0.01714 | 0.00039 | 0.00246 | 65 | 0.01640 | 0.00032 | 0.00184 | 93 | 0.00308 | 0.24125 | 127.855 | 0.80975 | 0.94519 |
| Glycerone kinase | 0.01663 | 0.00159 | 0.01428 | 0.00030 | 0.00215 | 65 | 0.01828 | 0.00047 | 0.00225 | 93 | 0.00311 | -1.28626 | 153.121 | 0.20029 | 0.70632 |
| Diphosphate–fructose-6-phosphate 1-phosphotransferase | 0.01642 | 0.00167 | 0.01440 | 0.00031 | 0.00219 | 65 | 0.01784 | 0.00053 | 0.00239 | 93 | 0.00324 | -1.06178 | 154.727 | 0.28999 | 0.70632 |
| Phospholipase A(1) | 0.01640 | 0.00178 | 0.01547 | 0.00043 | 0.00257 | 65 | 0.01705 | 0.00056 | 0.00245 | 93 | 0.00355 | -0.44518 | 147.959 | 0.65684 | 0.88943 |
| Phospholipase A(2) | 0.01640 | 0.00178 | 0.01547 | 0.00043 | 0.00257 | 65 | 0.01705 | 0.00056 | 0.00245 | 93 | 0.00355 | -0.44518 | 147.959 | 0.65684 | 0.88943 |
| 5-dehydro-4-deoxyglucarate dehydratase | 0.01631 | 0.00287 | 0.01461 | 0.00098 | 0.00388 | 65 | 0.01750 | 0.00153 | 0.00405 | 93 | 0.00562 | -0.51481 | 153.043 | 0.60743 | 0.86999 |
| [Isocitrate dehydrogenase (NADP(+))] kinase | 0.01625 | 0.00183 | 0.01735 | 0.00063 | 0.00311 | 65 | 0.01549 | 0.00046 | 0.00223 | 93 | 0.00383 | 0.48781 | 123.873 | 0.62654 | 0.88183 |
| Nitric-oxide reductase (cytochrome c) | 0.01621 | 0.00167 | 0.01594 | 0.00048 | 0.00272 | 65 | 0.01640 | 0.00042 | 0.00213 | 93 | 0.00345 | -0.13192 | 131.694 | 0.89525 | 0.97163 |
| L-iditol 2-dehydrogenase | 0.01615 | 0.00203 | 0.01418 | 0.00068 | 0.00323 | 65 | 0.01752 | 0.00063 | 0.00261 | 93 | 0.00416 | -0.80258 | 134.700 | 0.42363 | 0.76494 |
| Chorismate lyase | 0.01607 | 0.00174 | 0.01789 | 0.00052 | 0.00284 | 65 | 0.01479 | 0.00045 | 0.00220 | 93 | 0.00359 | 0.86345 | 131.085 | 0.38947 | 0.74308 |
| Rubredoxin–NAD(+) reductase | 0.01606 | 0.00212 | 0.01771 | 0.00078 | 0.00347 | 65 | 0.01491 | 0.00066 | 0.00267 | 93 | 0.00438 | 0.63907 | 130.508 | 0.52390 | 0.82182 |
| Cellulase | 0.01605 | 0.00165 | 0.01137 | 0.00031 | 0.00219 | 65 | 0.01932 | 0.00049 | 0.00230 | 93 | 0.00318 | -2.50220 | 153.184 | 0.01339 | 0.64530 |
| Ribitol-5-phosphate 2-dehydrogenase | 0.01603 | 0.00168 | 0.01351 | 0.00029 | 0.00213 | 65 | 0.01780 | 0.00055 | 0.00243 | 93 | 0.00323 | -1.32629 | 155.635 | 0.18669 | 0.70632 |
| Catalase peroxidase | 0.01599 | 0.00179 | 0.01444 | 0.00053 | 0.00286 | 65 | 0.01707 | 0.00049 | 0.00230 | 93 | 0.00367 | -0.71719 | 134.464 | 0.47450 | 0.79541 |
| Acireductone dioxygenase (Fe(2+)-requiring) | 0.01598 | 0.00195 | 0.01578 | 0.00064 | 0.00314 | 65 | 0.01612 | 0.00058 | 0.00250 | 93 | 0.00401 | -0.08516 | 133.462 | 0.93226 | 0.98985 |
| Acireductone dioxygenase (Ni(2+)-requiring) | 0.01598 | 0.00195 | 0.01578 | 0.00064 | 0.00314 | 65 | 0.01612 | 0.00058 | 0.00250 | 93 | 0.00401 | -0.08516 | 133.462 | 0.93226 | 0.98985 |
| Phosphoenolpyruvate carboxykinase (GTP) | 0.01587 | 0.00161 | 0.01454 | 0.00036 | 0.00237 | 65 | 0.01681 | 0.00044 | 0.00219 | 93 | 0.00322 | -0.70507 | 145.919 | 0.48189 | 0.79962 |
| 5’-deoxynucleotidase | 0.01581 | 0.00156 | 0.01819 | 0.00049 | 0.00275 | 65 | 0.01414 | 0.00030 | 0.00180 | 93 | 0.00329 | 1.22845 | 115.899 | 0.22177 | 0.70632 |
| Starch synthase (maltosyl-transferring) | 0.01578 | 0.00196 | 0.01613 | 0.00062 | 0.00309 | 65 | 0.01554 | 0.00061 | 0.00256 | 93 | 0.00401 | 0.14891 | 137.058 | 0.88184 | 0.96591 |
| D-aspartate ligase | 0.01560 | 0.00286 | 0.01663 | 0.00126 | 0.00440 | 65 | 0.01488 | 0.00133 | 0.00378 | 93 | 0.00580 | 0.30188 | 140.327 | 0.76319 | 0.93350 |
| Methylaspartate mutase | 0.01560 | 0.00341 | 0.02297 | 0.00306 | 0.00686 | 65 | 0.01045 | 0.00095 | 0.00320 | 93 | 0.00757 | 1.65493 | 91.772 | 0.10135 | 0.70632 |
| Succinylglutamate-semialdehyde dehydrogenase | 0.01559 | 0.00205 | 0.01570 | 0.00063 | 0.00312 | 65 | 0.01552 | 0.00069 | 0.00273 | 93 | 0.00415 | 0.04466 | 141.713 | 0.96444 | 0.99193 |
| Chitinase | 0.01549 | 0.00271 | 0.01376 | 0.00114 | 0.00418 | 65 | 0.01669 | 0.00118 | 0.00356 | 93 | 0.00549 | -0.53400 | 139.474 | 0.59419 | 0.86154 |
| CDP-4-dehydro-6-deoxyglucose reductase | 0.01538 | 0.00175 | 0.01608 | 0.00050 | 0.00279 | 65 | 0.01489 | 0.00048 | 0.00227 | 93 | 0.00359 | 0.33321 | 135.447 | 0.73949 | 0.92595 |
| Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase | 0.01530 | 0.00193 | 0.01144 | 0.00040 | 0.00247 | 65 | 0.01800 | 0.00071 | 0.00277 | 93 | 0.00371 | -1.76778 | 155.344 | 0.07906 | 0.70632 |
| Nicotinamide-nucleotide adenylyltransferase | 0.01530 | 0.00159 | 0.01698 | 0.00044 | 0.00261 | 65 | 0.01413 | 0.00037 | 0.00199 | 93 | 0.00328 | 0.86901 | 129.896 | 0.38644 | 0.73961 |
| 15-cis-phytoene synthase | 0.01528 | 0.00162 | 0.01156 | 0.00027 | 0.00202 | 65 | 0.01787 | 0.00051 | 0.00234 | 93 | 0.00309 | -2.04362 | 155.800 | 0.04268 | 0.69348 |
| Ribonuclease T(2) | 0.01525 | 0.00153 | 0.01277 | 0.00028 | 0.00207 | 65 | 0.01698 | 0.00043 | 0.00215 | 93 | 0.00298 | -1.41408 | 152.906 | 0.15937 | 0.70632 |
| Dihydropyrimidinase | 0.01515 | 0.00168 | 0.01233 | 0.00037 | 0.00238 | 65 | 0.01712 | 0.00050 | 0.00231 | 93 | 0.00331 | -1.44551 | 149.223 | 0.15041 | 0.70632 |
| Rhamnulokinase | 0.01505 | 0.00174 | 0.01688 | 0.00078 | 0.00347 | 65 | 0.01377 | 0.00027 | 0.00170 | 93 | 0.00386 | 0.80552 | 94.476 | 0.42254 | 0.76494 |
| Teichoic-acid-transporting ATPase | 0.01504 | 0.00237 | 0.01761 | 0.00121 | 0.00432 | 65 | 0.01325 | 0.00066 | 0.00266 | 93 | 0.00507 | 0.85940 | 110.697 | 0.39198 | 0.74318 |
| Alpha,alpha-trehalose-phosphate synthase (UDP-forming) | 0.01502 | 0.00173 | 0.01139 | 0.00037 | 0.00240 | 65 | 0.01756 | 0.00053 | 0.00239 | 93 | 0.00338 | -1.82225 | 150.702 | 0.07040 | 0.70632 |
| Propionate–CoA ligase | 0.01498 | 0.00195 | 0.01270 | 0.00052 | 0.00282 | 65 | 0.01657 | 0.00066 | 0.00266 | 93 | 0.00388 | -0.99944 | 147.412 | 0.31922 | 0.70632 |
| Monoamine oxidase | 0.01497 | 0.00205 | 0.01242 | 0.00054 | 0.00289 | 65 | 0.01676 | 0.00075 | 0.00284 | 93 | 0.00405 | -1.07178 | 149.834 | 0.28554 | 0.70632 |
| 23S rRNA (uracil(747)-C(5))-methyltransferase | 0.01488 | 0.00160 | 0.01644 | 0.00045 | 0.00262 | 65 | 0.01378 | 0.00037 | 0.00200 | 93 | 0.00330 | 0.80748 | 129.650 | 0.42087 | 0.76494 |
| Benzoate 1,2-dioxygenase | 0.01483 | 0.00236 | 0.01652 | 0.00117 | 0.00424 | 65 | 0.01364 | 0.00069 | 0.00272 | 93 | 0.00504 | 0.57127 | 113.856 | 0.56894 | 0.85024 |
| Lysozyme | 0.01482 | 0.00174 | 0.01463 | 0.00052 | 0.00282 | 65 | 0.01494 | 0.00045 | 0.00220 | 93 | 0.00358 | -0.08681 | 131.632 | 0.93096 | 0.98985 |
| Octanoyl-[GcvH]:protein N-octanoyltransferase | 0.01479 | 0.00183 | 0.01369 | 0.00070 | 0.00328 | 65 | 0.01555 | 0.00041 | 0.00211 | 93 | 0.00390 | -0.47573 | 114.239 | 0.63517 | 0.88317 |
| Arsenite-transporting ATPase | 0.01477 | 0.00322 | 0.02022 | 0.00264 | 0.00637 | 65 | 0.01095 | 0.00092 | 0.00315 | 93 | 0.00711 | 1.30485 | 95.089 | 0.19509 | 0.70632 |
| Thiosulfate dehydrogenase | 0.01461 | 0.00203 | 0.01376 | 0.00056 | 0.00294 | 65 | 0.01521 | 0.00072 | 0.00278 | 93 | 0.00405 | -0.35781 | 147.572 | 0.72100 | 0.92120 |
| Glyoxylate reductase | 0.01456 | 0.00143 | 0.01214 | 0.00031 | 0.00218 | 65 | 0.01625 | 0.00033 | 0.00187 | 93 | 0.00287 | -1.43087 | 140.473 | 0.15469 | 0.70632 |
| Exoribonuclease II | 0.01451 | 0.00170 | 0.01450 | 0.00041 | 0.00252 | 65 | 0.01452 | 0.00049 | 0.00230 | 93 | 0.00341 | -0.00457 | 144.817 | 0.99636 | 0.99921 |
| UDP-N-acetylglucosamine kinase | 0.01447 | 0.00155 | 0.01345 | 0.00026 | 0.00200 | 65 | 0.01518 | 0.00047 | 0.00224 | 93 | 0.00300 | -0.57680 | 155.289 | 0.56491 | 0.85024 |
| Trehalose-phosphatase | 0.01438 | 0.00165 | 0.01093 | 0.00037 | 0.00239 | 65 | 0.01679 | 0.00046 | 0.00222 | 93 | 0.00326 | -1.79471 | 146.410 | 0.07476 | 0.70632 |
| Tryptophanase | 0.01437 | 0.00208 | 0.01912 | 0.00105 | 0.00401 | 65 | 0.01105 | 0.00041 | 0.00211 | 93 | 0.00453 | 1.77973 | 98.858 | 0.07819 | 0.70632 |
| Levansucrase | 0.01436 | 0.00304 | 0.01601 | 0.00210 | 0.00569 | 65 | 0.01321 | 0.00103 | 0.00333 | 93 | 0.00659 | 0.42455 | 106.600 | 0.67202 | 0.89609 |
| 2-methylcitrate dehydratase | 0.01427 | 0.00181 | 0.01325 | 0.00053 | 0.00286 | 65 | 0.01498 | 0.00051 | 0.00234 | 93 | 0.00370 | -0.46872 | 135.767 | 0.64002 | 0.88568 |
| Maltose phosphorylase | 0.01419 | 0.00245 | 0.01767 | 0.00147 | 0.00476 | 65 | 0.01175 | 0.00058 | 0.00250 | 93 | 0.00538 | 1.09928 | 98.970 | 0.27431 | 0.70632 |
| Cystathionine beta-synthase | 0.01417 | 0.00158 | 0.01269 | 0.00034 | 0.00230 | 65 | 0.01520 | 0.00043 | 0.00215 | 93 | 0.00315 | -0.79793 | 146.761 | 0.42620 | 0.76494 |
| Nitrogenase | 0.01413 | 0.00252 | 0.01927 | 0.00164 | 0.00503 | 65 | 0.01055 | 0.00053 | 0.00239 | 93 | 0.00557 | 1.56544 | 92.927 | 0.12088 | 0.70632 |
| Citrate lyase holo-[acyl-carrier protein] synthase | 0.01406 | 0.00241 | 0.02008 | 0.00146 | 0.00475 | 65 | 0.00985 | 0.00050 | 0.00232 | 93 | 0.00528 | 1.93752 | 94.481 | 0.05567 | 0.70632 |
| Glucarate dehydratase | 0.01403 | 0.00200 | 0.01343 | 0.00061 | 0.00307 | 65 | 0.01444 | 0.00065 | 0.00265 | 93 | 0.00406 | -0.25044 | 140.530 | 0.80261 | 0.94263 |
| Mannan endo-1,4-beta-mannosidase | 0.01403 | 0.00179 | 0.01107 | 0.00022 | 0.00183 | 65 | 0.01609 | 0.00070 | 0.00274 | 93 | 0.00329 | -1.52183 | 149.741 | 0.13016 | 0.70632 |
| Acetyl-S-ACP:malonate ACP transferase | 0.01402 | 0.00184 | 0.01446 | 0.00055 | 0.00290 | 65 | 0.01371 | 0.00053 | 0.00240 | 93 | 0.00376 | 0.19833 | 137.032 | 0.84308 | 0.95939 |
| Trans-2-enoyl-CoA reductase (NAD(+)) | 0.01401 | 0.00167 | 0.01471 | 0.00042 | 0.00255 | 65 | 0.01352 | 0.00046 | 0.00221 | 93 | 0.00338 | 0.35292 | 141.033 | 0.72468 | 0.92120 |
| Micrococcal nuclease | 0.01400 | 0.00193 | 0.01134 | 0.00033 | 0.00224 | 65 | 0.01586 | 0.00077 | 0.00287 | 93 | 0.00364 | -1.24108 | 155.338 | 0.21645 | 0.70632 |
| (2,3-dihydroxybenzoyl)adenylate synthase | 0.01394 | 0.00175 | 0.01315 | 0.00053 | 0.00286 | 65 | 0.01448 | 0.00045 | 0.00221 | 93 | 0.00361 | -0.36765 | 130.673 | 0.71373 | 0.92057 |
| N-succinylarginine dihydrolase | 0.01392 | 0.00167 | 0.01431 | 0.00046 | 0.00266 | 65 | 0.01364 | 0.00043 | 0.00215 | 93 | 0.00343 | 0.19548 | 134.910 | 0.84531 | 0.95939 |
| Glycine reductase | 0.01390 | 0.00189 | 0.01791 | 0.00094 | 0.00380 | 65 | 0.01109 | 0.00030 | 0.00178 | 93 | 0.00420 | 1.62578 | 92.224 | 0.10741 | 0.70632 |
| 4-hydroxybenzoate 3-monooxygenase | 0.01382 | 0.00181 | 0.01210 | 0.00042 | 0.00255 | 65 | 0.01502 | 0.00059 | 0.00251 | 93 | 0.00357 | -0.81752 | 150.138 | 0.41492 | 0.76374 |
| Beta-glucuronidase | 0.01378 | 0.00153 | 0.01461 | 0.00042 | 0.00253 | 65 | 0.01321 | 0.00034 | 0.00192 | 93 | 0.00318 | 0.43892 | 129.111 | 0.66145 | 0.88968 |
| NAD(P)(+) transhydrogenase (Si-specific) | 0.01378 | 0.00164 | 0.01413 | 0.00044 | 0.00259 | 65 | 0.01354 | 0.00042 | 0.00212 | 93 | 0.00335 | 0.17623 | 136.016 | 0.86038 | 0.96186 |
| Dipeptidyl-peptidase III | 0.01375 | 0.00159 | 0.01183 | 0.00027 | 0.00203 | 65 | 0.01509 | 0.00049 | 0.00230 | 93 | 0.00307 | -1.06082 | 155.494 | 0.29042 | 0.70632 |
| Carbon-monoxide dehydrogenase (ferredoxin) | 0.01373 | 0.00168 | 0.01634 | 0.00066 | 0.00318 | 65 | 0.01190 | 0.00029 | 0.00178 | 93 | 0.00364 | 1.21908 | 103.257 | 0.22559 | 0.70632 |
| Butyrate kinase | 0.01372 | 0.00154 | 0.01188 | 0.00030 | 0.00214 | 65 | 0.01501 | 0.00043 | 0.00215 | 93 | 0.00303 | -1.03248 | 151.428 | 0.30349 | 0.70632 |
| Phosphate butyryltransferase | 0.01369 | 0.00153 | 0.01214 | 0.00029 | 0.00212 | 65 | 0.01477 | 0.00043 | 0.00214 | 93 | 0.00301 | -0.87365 | 151.384 | 0.38369 | 0.73873 |
| Isochorismatase | 0.01368 | 0.00182 | 0.01256 | 0.00052 | 0.00283 | 65 | 0.01446 | 0.00053 | 0.00238 | 93 | 0.00370 | -0.51345 | 138.221 | 0.60845 | 0.87028 |
| N-acetylglucosamine kinase | 0.01368 | 0.00168 | 0.01412 | 0.00045 | 0.00262 | 65 | 0.01337 | 0.00045 | 0.00220 | 93 | 0.00342 | 0.21824 | 138.176 | 0.82756 | 0.95262 |
| Glutamate synthase (ferredoxin) | 0.01363 | 0.00141 | 0.01183 | 0.00027 | 0.00206 | 65 | 0.01490 | 0.00034 | 0.00192 | 93 | 0.00281 | -1.09004 | 146.731 | 0.27748 | 0.70632 |
| N-acetylneuraminate synthase | 0.01356 | 0.00156 | 0.01376 | 0.00047 | 0.00268 | 65 | 0.01342 | 0.00033 | 0.00189 | 93 | 0.00328 | 0.10363 | 122.300 | 0.91763 | 0.98197 |
| Geranoyl-CoA carboxylase | 0.01352 | 0.00218 | 0.01139 | 0.00057 | 0.00297 | 65 | 0.01501 | 0.00087 | 0.00306 | 93 | 0.00427 | -0.84749 | 152.512 | 0.39805 | 0.74887 |
| 5-carboxymethyl-2-hydroxymuconate Delta-isomerase | 0.01346 | 0.00222 | 0.00940 | 0.00040 | 0.00249 | 65 | 0.01630 | 0.00102 | 0.00332 | 93 | 0.00415 | -1.66354 | 154.414 | 0.09823 | 0.70632 |
| Inositol-3-phosphate synthase | 0.01342 | 0.00134 | 0.01195 | 0.00024 | 0.00193 | 65 | 0.01445 | 0.00032 | 0.00185 | 93 | 0.00267 | -0.93602 | 148.391 | 0.35079 | 0.72019 |
| 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | 0.01332 | 0.00175 | 0.01250 | 0.00052 | 0.00284 | 65 | 0.01390 | 0.00046 | 0.00222 | 93 | 0.00360 | -0.38929 | 132.032 | 0.69769 | 0.91007 |
| L-arabinose isomerase | 0.01332 | 0.00188 | 0.01450 | 0.00072 | 0.00332 | 65 | 0.01250 | 0.00045 | 0.00221 | 93 | 0.00399 | 0.50159 | 117.091 | 0.61689 | 0.87407 |
| FMN reductase (NAD(P)H) | 0.01328 | 0.00157 | 0.01240 | 0.00030 | 0.00215 | 65 | 0.01390 | 0.00045 | 0.00221 | 93 | 0.00308 | -0.48477 | 152.191 | 0.62854 | 0.88303 |
| Xylose isomerase | 0.01328 | 0.00128 | 0.01153 | 0.00027 | 0.00203 | 65 | 0.01450 | 0.00025 | 0.00164 | 93 | 0.00261 | -1.13538 | 134.910 | 0.25823 | 0.70632 |
| Ureidoglycolate lyase | 0.01317 | 0.00174 | 0.01191 | 0.00047 | 0.00270 | 65 | 0.01405 | 0.00049 | 0.00228 | 93 | 0.00353 | -0.60630 | 139.009 | 0.54530 | 0.84290 |
| 5,6-dimethylbenzimidazole synthase | 0.01305 | 0.00167 | 0.01191 | 0.00041 | 0.00253 | 65 | 0.01385 | 0.00046 | 0.00222 | 93 | 0.00336 | -0.57518 | 142.116 | 0.56608 | 0.85024 |
| Beta-lysine 5,6-aminomutase | 0.01301 | 0.00308 | 0.02057 | 0.00281 | 0.00657 | 65 | 0.00773 | 0.00054 | 0.00240 | 93 | 0.00700 | 1.83488 | 81.277 | 0.07018 | 0.70632 |
| Succinate-semialdehyde dehydrogenase (NAD(+)) | 0.01301 | 0.00171 | 0.01302 | 0.00045 | 0.00263 | 65 | 0.01301 | 0.00047 | 0.00226 | 93 | 0.00347 | 0.00218 | 140.105 | 0.99827 | 0.99921 |
| D-chiro-inositol 1-dehydrogenase | 0.01300 | 0.00208 | 0.01003 | 0.00041 | 0.00250 | 65 | 0.01508 | 0.00088 | 0.00307 | 93 | 0.00396 | -1.27501 | 155.896 | 0.20420 | 0.70632 |
| Inositol 2-dehydrogenase | 0.01300 | 0.00208 | 0.01003 | 0.00041 | 0.00250 | 65 | 0.01508 | 0.00088 | 0.00307 | 93 | 0.00396 | -1.27501 | 155.896 | 0.20420 | 0.70632 |
| Polyphosphate–glucose phosphotransferase | 0.01298 | 0.00158 | 0.01328 | 0.00036 | 0.00237 | 65 | 0.01277 | 0.00042 | 0.00213 | 93 | 0.00318 | 0.15854 | 143.850 | 0.87425 | 0.96378 |
| Molybdopterin adenylyltransferase | 0.01297 | 0.00153 | 0.01234 | 0.00028 | 0.00206 | 65 | 0.01340 | 0.00044 | 0.00217 | 93 | 0.00299 | -0.35356 | 153.315 | 0.72416 | 0.92120 |
| Catechol 1,2-dioxygenase | 0.01285 | 0.00162 | 0.01225 | 0.00048 | 0.00273 | 65 | 0.01327 | 0.00037 | 0.00199 | 93 | 0.00338 | -0.30044 | 125.648 | 0.76434 | 0.93350 |
| ATP adenylyltransferase | 0.01277 | 0.00140 | 0.01310 | 0.00034 | 0.00229 | 65 | 0.01254 | 0.00029 | 0.00178 | 93 | 0.00290 | 0.19303 | 131.450 | 0.84724 | 0.95939 |
| Isocitrate dehydrogenase (NAD(+)) | 0.01277 | 0.00196 | 0.01676 | 0.00095 | 0.00383 | 65 | 0.00998 | 0.00035 | 0.00195 | 93 | 0.00430 | 1.57742 | 97.077 | 0.11795 | 0.70632 |
| Deoxyribonuclease I | 0.01274 | 0.00197 | 0.01528 | 0.00081 | 0.00352 | 65 | 0.01096 | 0.00048 | 0.00227 | 93 | 0.00419 | 1.02928 | 114.456 | 0.30552 | 0.70632 |
| Dihydrouracil dehydrogenase (NAD(+)) | 0.01271 | 0.00187 | 0.01188 | 0.00072 | 0.00332 | 65 | 0.01330 | 0.00045 | 0.00219 | 93 | 0.00397 | -0.35589 | 116.520 | 0.72256 | 0.92120 |
| 5-dehydro-4-deoxy-D-glucuronate isomerase | 0.01271 | 0.00170 | 0.01368 | 0.00075 | 0.00340 | 65 | 0.01203 | 0.00026 | 0.00166 | 93 | 0.00378 | 0.43748 | 94.375 | 0.66276 | 0.88968 |
| Beta-ketodecanoyl-[acyl-carrier-protein] synthase | 0.01260 | 0.00164 | 0.01292 | 0.00046 | 0.00265 | 65 | 0.01238 | 0.00040 | 0.00208 | 93 | 0.00337 | 0.15940 | 132.489 | 0.87360 | 0.96378 |
| Dodecenoyl-CoA isomerase | 0.01256 | 0.00160 | 0.01332 | 0.00042 | 0.00255 | 65 | 0.01203 | 0.00039 | 0.00206 | 93 | 0.00327 | 0.39490 | 134.774 | 0.69354 | 0.90946 |
| 3-deoxy-D-manno-octulosonic acid kinase | 0.01256 | 0.00165 | 0.01352 | 0.00046 | 0.00266 | 65 | 0.01188 | 0.00041 | 0.00211 | 93 | 0.00340 | 0.48232 | 133.291 | 0.63037 | 0.88317 |
| Phospholipase C | 0.01253 | 0.00174 | 0.01094 | 0.00047 | 0.00269 | 65 | 0.01364 | 0.00049 | 0.00229 | 93 | 0.00354 | -0.76376 | 139.476 | 0.44630 | 0.77823 |
| Type II site-specific deoxyribonuclease | 0.01252 | 0.00234 | 0.00944 | 0.00019 | 0.00169 | 65 | 0.01467 | 0.00133 | 0.00379 | 93 | 0.00415 | -1.26058 | 125.210 | 0.20980 | 0.70632 |
| Succinylglutamate desuccinylase | 0.01246 | 0.00161 | 0.01310 | 0.00043 | 0.00257 | 65 | 0.01202 | 0.00040 | 0.00207 | 93 | 0.00330 | 0.32639 | 134.395 | 0.74464 | 0.92762 |
| Precorrin-3B synthase | 0.01245 | 0.00183 | 0.01185 | 0.00052 | 0.00282 | 65 | 0.01288 | 0.00055 | 0.00242 | 93 | 0.00372 | -0.27657 | 140.314 | 0.78252 | 0.93675 |
| Tryptophan 2,3-dioxygenase | 0.01241 | 0.00165 | 0.00950 | 0.00038 | 0.00241 | 65 | 0.01445 | 0.00046 | 0.00222 | 93 | 0.00328 | -1.51222 | 145.462 | 0.13265 | 0.70632 |
| (1->4)-alpha-D-glucan 1-alpha-D-glucosylmutase | 0.01241 | 0.00159 | 0.01172 | 0.00042 | 0.00255 | 65 | 0.01290 | 0.00039 | 0.00204 | 93 | 0.00327 | -0.36153 | 133.688 | 0.71827 | 0.92120 |
| Kynureninase | 0.01236 | 0.00166 | 0.01020 | 0.00043 | 0.00257 | 65 | 0.01387 | 0.00044 | 0.00216 | 93 | 0.00336 | -1.08962 | 138.353 | 0.27777 | 0.70632 |
| [Citrate (pro-3S)-lyase] ligase | 0.01230 | 0.00193 | 0.01798 | 0.00102 | 0.00395 | 65 | 0.00833 | 0.00026 | 0.00167 | 93 | 0.00429 | 2.24846 | 86.859 | 0.02708 | 0.67652 |
| Malonate decarboxylase holo-[acyl-carrier protein] synthase | 0.01229 | 0.00166 | 0.01290 | 0.00047 | 0.00270 | 65 | 0.01186 | 0.00041 | 0.00210 | 93 | 0.00342 | 0.30475 | 131.553 | 0.76104 | 0.93350 |
| 4-alpha-D-((1->4)-alpha-D-glucano)trehalose trehalohydrolase | 0.01226 | 0.00157 | 0.01151 | 0.00043 | 0.00256 | 65 | 0.01279 | 0.00037 | 0.00200 | 93 | 0.00325 | -0.39440 | 131.524 | 0.69393 | 0.90946 |
| Glucose-1-phosphatase | 0.01225 | 0.00140 | 0.01278 | 0.00031 | 0.00220 | 65 | 0.01188 | 0.00031 | 0.00182 | 93 | 0.00285 | 0.31522 | 137.209 | 0.75307 | 0.92944 |
| Gluconolactonase | 0.01221 | 0.00172 | 0.00769 | 0.00023 | 0.00186 | 65 | 0.01537 | 0.00061 | 0.00257 | 93 | 0.00317 | -2.41875 | 153.264 | 0.01675 | 0.64530 |
| L-threonine kinase | 0.01221 | 0.00146 | 0.01369 | 0.00044 | 0.00261 | 65 | 0.01118 | 0.00027 | 0.00169 | 93 | 0.00311 | 0.80886 | 115.037 | 0.42026 | 0.76494 |
| Haloacetate dehalogenase | 0.01212 | 0.00132 | 0.01131 | 0.00028 | 0.00206 | 65 | 0.01269 | 0.00028 | 0.00172 | 93 | 0.00269 | -0.51190 | 137.851 | 0.60954 | 0.87124 |
| Alpha-D-xyloside xylohydrolase | 0.01203 | 0.00187 | 0.00862 | 0.00022 | 0.00184 | 65 | 0.01441 | 0.00077 | 0.00289 | 93 | 0.00342 | -1.69104 | 147.208 | 0.09295 | 0.70632 |
| Phenylacetyl-CoA 1,2-epoxidase | 0.01200 | 0.00160 | 0.01041 | 0.00031 | 0.00218 | 65 | 0.01311 | 0.00048 | 0.00226 | 93 | 0.00314 | -0.86171 | 152.861 | 0.39020 | 0.74318 |
| N-methylhydantoinase (ATP-hydrolyzing) | 0.01199 | 0.00224 | 0.00833 | 0.00035 | 0.00230 | 65 | 0.01454 | 0.00110 | 0.00344 | 93 | 0.00414 | -1.49834 | 149.754 | 0.13615 | 0.70632 |
| 4-hydroxy-2-oxoheptanedioate aldolase | 0.01198 | 0.00180 | 0.00732 | 0.00017 | 0.00160 | 65 | 0.01524 | 0.00073 | 0.00281 | 93 | 0.00323 | -2.45032 | 140.045 | 0.01551 | 0.64530 |
| 3-oxoadipyl-CoA thiolase | 0.01195 | 0.00192 | 0.01226 | 0.00055 | 0.00290 | 65 | 0.01173 | 0.00062 | 0.00258 | 93 | 0.00388 | 0.13792 | 142.982 | 0.89050 | 0.96887 |
| Ferredoxin–NAD(+) reductase | 0.01189 | 0.00156 | 0.00677 | 0.00014 | 0.00146 | 65 | 0.01547 | 0.00053 | 0.00238 | 93 | 0.00279 | -3.11780 | 144.788 | 0.00220 | 0.62334 |
| D-malate dehydrogenase (decarboxylating) | 0.01186 | 0.00207 | 0.00719 | 0.00020 | 0.00176 | 65 | 0.01513 | 0.00099 | 0.00327 | 93 | 0.00371 | -2.14118 | 136.786 | 0.03403 | 0.67652 |
| Tartrate decarboxylase | 0.01186 | 0.00207 | 0.00719 | 0.00020 | 0.00176 | 65 | 0.01513 | 0.00099 | 0.00327 | 93 | 0.00371 | -2.14118 | 136.786 | 0.03403 | 0.67652 |
| Tartrate dehydrogenase | 0.01186 | 0.00207 | 0.00719 | 0.00020 | 0.00176 | 65 | 0.01513 | 0.00099 | 0.00327 | 93 | 0.00371 | -2.14118 | 136.786 | 0.03403 | 0.67652 |
| 1-aminocyclopropane-1-carboxylate deaminase | 0.01178 | 0.00162 | 0.01268 | 0.00046 | 0.00266 | 65 | 0.01115 | 0.00039 | 0.00204 | 93 | 0.00335 | 0.45690 | 129.934 | 0.64851 | 0.88803 |
| N-acylglucosamine 2-epimerase | 0.01173 | 0.00128 | 0.00971 | 0.00013 | 0.00142 | 65 | 0.01314 | 0.00035 | 0.00194 | 93 | 0.00240 | -1.42812 | 153.740 | 0.15529 | 0.70632 |
| Galactarate dehydratase | 0.01173 | 0.00161 | 0.01097 | 0.00038 | 0.00243 | 65 | 0.01226 | 0.00043 | 0.00216 | 93 | 0.00325 | -0.39592 | 142.729 | 0.69276 | 0.90946 |
| Glucuronate isomerase | 0.01172 | 0.00146 | 0.01060 | 0.00039 | 0.00244 | 65 | 0.01250 | 0.00031 | 0.00181 | 93 | 0.00304 | -0.62519 | 127.005 | 0.53297 | 0.83089 |
| N-hydroxyarylamine O-acetyltransferase | 0.01160 | 0.00171 | 0.00907 | 0.00036 | 0.00236 | 65 | 0.01337 | 0.00053 | 0.00238 | 93 | 0.00336 | -1.28054 | 151.377 | 0.20231 | 0.70632 |
| Xenobiotic-transporting ATPase | 0.01158 | 0.00192 | 0.01022 | 0.00067 | 0.00322 | 65 | 0.01254 | 0.00053 | 0.00238 | 93 | 0.00400 | -0.58083 | 126.724 | 0.56238 | 0.85024 |
| Acyl-CoA 6-desaturase | 0.01154 | 0.00175 | 0.00957 | 0.00031 | 0.00218 | 65 | 0.01292 | 0.00061 | 0.00256 | 93 | 0.00336 | -0.99982 | 155.940 | 0.31894 | 0.70632 |
| Nitrilase | 0.01144 | 0.00139 | 0.00999 | 0.00022 | 0.00183 | 65 | 0.01245 | 0.00037 | 0.00199 | 93 | 0.00270 | -0.91048 | 154.492 | 0.36399 | 0.72472 |
| Pyrroloquinoline-quinone synthase | 0.01142 | 0.00160 | 0.01160 | 0.00044 | 0.00261 | 65 | 0.01129 | 0.00038 | 0.00203 | 93 | 0.00330 | 0.09273 | 131.366 | 0.92626 | 0.98985 |
| Erythrose-4-phosphate dehydrogenase | 0.01135 | 0.00154 | 0.01206 | 0.00040 | 0.00247 | 65 | 0.01085 | 0.00037 | 0.00198 | 93 | 0.00317 | 0.38363 | 134.328 | 0.70186 | 0.91306 |
| Ferredoxin–nitrite reductase | 0.01119 | 0.00127 | 0.01157 | 0.00029 | 0.00210 | 65 | 0.01092 | 0.00024 | 0.00160 | 93 | 0.00264 | 0.24878 | 129.448 | 0.80393 | 0.94350 |
| Formaldehyde dehydrogenase | 0.01101 | 0.00193 | 0.01194 | 0.00059 | 0.00302 | 65 | 0.01036 | 0.00059 | 0.00252 | 93 | 0.00393 | 0.40261 | 137.676 | 0.68786 | 0.90827 |
| Beta-mannosidase | 0.01099 | 0.00150 | 0.00860 | 0.00034 | 0.00228 | 65 | 0.01266 | 0.00037 | 0.00199 | 93 | 0.00303 | -1.34303 | 141.595 | 0.18141 | 0.70632 |
| Cellulose synthase (UDP-forming) | 0.01095 | 0.00146 | 0.01023 | 0.00024 | 0.00192 | 65 | 0.01144 | 0.00040 | 0.00208 | 93 | 0.00284 | -0.42608 | 154.431 | 0.67064 | 0.89503 |
| 1,4-beta-mannosyl-N-acetylglucosamine phosphorylase | 0.01094 | 0.00121 | 0.00893 | 0.00013 | 0.00140 | 65 | 0.01235 | 0.00030 | 0.00181 | 93 | 0.00228 | -1.49604 | 155.178 | 0.13667 | 0.70632 |
| Beta-1,4-mannooligosaccharide phosphorylase | 0.01094 | 0.00121 | 0.00893 | 0.00013 | 0.00140 | 65 | 0.01235 | 0.00030 | 0.00181 | 93 | 0.00228 | -1.49604 | 155.178 | 0.13667 | 0.70632 |
| tRNA (adenine(57)-N(1)/adenine(58)-N(1))-methyltransferase | 0.01094 | 0.00131 | 0.01144 | 0.00030 | 0.00216 | 65 | 0.01059 | 0.00025 | 0.00164 | 93 | 0.00271 | 0.31402 | 129.035 | 0.75402 | 0.92944 |
| tRNA (adenine(58)-N(1))-methyltransferase | 0.01094 | 0.00131 | 0.01144 | 0.00030 | 0.00216 | 65 | 0.01059 | 0.00025 | 0.00164 | 93 | 0.00271 | 0.31402 | 129.035 | 0.75402 | 0.92944 |
| Trimethylamine-N-oxide reductase (cytochrome c) | 0.01092 | 0.00205 | 0.00856 | 0.00040 | 0.00248 | 65 | 0.01256 | 0.00084 | 0.00301 | 93 | 0.00390 | -1.02480 | 155.984 | 0.30704 | 0.70632 |
| Propanal dehydrogenase (CoA-propanoylating) | 0.01090 | 0.00118 | 0.01235 | 0.00028 | 0.00208 | 65 | 0.00989 | 0.00017 | 0.00137 | 93 | 0.00249 | 0.98445 | 116.191 | 0.32694 | 0.71125 |
| Poly(3-hydroxybutyrate) depolymerase | 0.01088 | 0.00158 | 0.00903 | 0.00038 | 0.00243 | 65 | 0.01217 | 0.00040 | 0.00207 | 93 | 0.00319 | -0.98183 | 139.455 | 0.32788 | 0.71151 |
| Thymidylate synthase (FAD) | 0.01084 | 0.00152 | 0.01021 | 0.00032 | 0.00220 | 65 | 0.01127 | 0.00040 | 0.00208 | 93 | 0.00303 | -0.34867 | 147.403 | 0.72784 | 0.92120 |
| Alkane 1-monooxygenase | 0.01082 | 0.00224 | 0.00795 | 0.00022 | 0.00183 | 65 | 0.01282 | 0.00119 | 0.00357 | 93 | 0.00402 | -1.21284 | 133.431 | 0.22734 | 0.70632 |
| Hyaluronoglucosaminidase | 0.01079 | 0.00172 | 0.01092 | 0.00077 | 0.00344 | 65 | 0.01069 | 0.00026 | 0.00168 | 93 | 0.00382 | 0.05922 | 94.471 | 0.95290 | 0.99049 |
| Ribosylnicotinamide kinase | 0.01070 | 0.00135 | 0.01291 | 0.00034 | 0.00230 | 65 | 0.00917 | 0.00025 | 0.00164 | 93 | 0.00282 | 1.32434 | 123.216 | 0.18784 | 0.70632 |
| 6,7-dihydropteridine reductase | 0.01068 | 0.00124 | 0.01165 | 0.00026 | 0.00199 | 65 | 0.01000 | 0.00024 | 0.00160 | 93 | 0.00255 | 0.64467 | 134.309 | 0.52024 | 0.82181 |
| Ribulose-bisphosphate carboxylase | 0.01065 | 0.00123 | 0.01033 | 0.00025 | 0.00196 | 65 | 0.01088 | 0.00024 | 0.00160 | 93 | 0.00253 | -0.21891 | 135.669 | 0.82705 | 0.95262 |
| Adenosine kinase | 0.01065 | 0.00155 | 0.00902 | 0.00034 | 0.00228 | 65 | 0.01180 | 0.00041 | 0.00211 | 93 | 0.00310 | -0.89518 | 146.202 | 0.37216 | 0.73139 |
| Diacylglycerol diphosphate phosphatase | 0.01065 | 0.00134 | 0.01097 | 0.00026 | 0.00200 | 65 | 0.01042 | 0.00030 | 0.00181 | 93 | 0.00269 | 0.20345 | 144.349 | 0.83907 | 0.95674 |
| Phosphatidate phosphatase | 0.01065 | 0.00134 | 0.01097 | 0.00026 | 0.00200 | 65 | 0.01042 | 0.00030 | 0.00181 | 93 | 0.00269 | 0.20345 | 144.349 | 0.83907 | 0.95674 |
| Lysine 2,3-aminomutase | 0.01064 | 0.00174 | 0.01224 | 0.00073 | 0.00336 | 65 | 0.00952 | 0.00030 | 0.00179 | 93 | 0.00381 | 0.71546 | 99.889 | 0.47599 | 0.79541 |
| Tartronate-semialdehyde synthase | 0.01064 | 0.00174 | 0.01043 | 0.00046 | 0.00266 | 65 | 0.01079 | 0.00050 | 0.00231 | 93 | 0.00352 | -0.10349 | 141.239 | 0.91772 | 0.98197 |
| DNA oxidative demethylase | 0.01059 | 0.00137 | 0.00851 | 0.00020 | 0.00176 | 65 | 0.01204 | 0.00036 | 0.00197 | 93 | 0.00264 | -1.33781 | 155.294 | 0.18291 | 0.70632 |
| Quinate dehydrogenase (quinone) | 0.01055 | 0.00166 | 0.01001 | 0.00034 | 0.00228 | 65 | 0.01092 | 0.00051 | 0.00234 | 93 | 0.00327 | -0.27838 | 152.393 | 0.78110 | 0.93675 |
| D-lactate dehydratase | 0.01049 | 0.00153 | 0.00886 | 0.00032 | 0.00222 | 65 | 0.01163 | 0.00041 | 0.00209 | 93 | 0.00305 | -0.90972 | 147.178 | 0.36446 | 0.72472 |
| 4-hydroxybenzoyl-CoA thioesterase | 0.01040 | 0.00149 | 0.01032 | 0.00038 | 0.00241 | 65 | 0.01046 | 0.00034 | 0.00190 | 93 | 0.00307 | -0.04784 | 132.804 | 0.96191 | 0.99193 |
| Trans-aconitate 2-methyltransferase | 0.01037 | 0.00136 | 0.00960 | 0.00030 | 0.00215 | 65 | 0.01091 | 0.00029 | 0.00178 | 93 | 0.00278 | -0.46906 | 137.013 | 0.63977 | 0.88568 |
| Non-reducing end alpha-L-arabinofuranosidase | 0.01034 | 0.00170 | 0.00587 | 0.00011 | 0.00128 | 65 | 0.01346 | 0.00068 | 0.00271 | 93 | 0.00300 | -2.53146 | 128.565 | 0.01256 | 0.64530 |
| Cardiolipin synthase (CMP-forming) | 0.01025 | 0.00123 | 0.00687 | 0.00011 | 0.00128 | 65 | 0.01262 | 0.00032 | 0.00185 | 93 | 0.00225 | -2.55854 | 151.156 | 0.01149 | 0.64530 |
| Mannitol 2-dehydrogenase | 0.01024 | 0.00173 | 0.00905 | 0.00038 | 0.00241 | 65 | 0.01108 | 0.00055 | 0.00243 | 93 | 0.00342 | -0.59310 | 151.462 | 0.55400 | 0.84560 |
| 8-oxoguanine deaminase | 0.01024 | 0.00219 | 0.00969 | 0.00062 | 0.00310 | 65 | 0.01062 | 0.00086 | 0.00305 | 93 | 0.00435 | -0.21580 | 150.114 | 0.82943 | 0.95262 |
| Beta-aspartyl-peptidase | 0.01023 | 0.00149 | 0.00857 | 0.00035 | 0.00233 | 65 | 0.01140 | 0.00035 | 0.00195 | 93 | 0.00303 | -0.92997 | 137.914 | 0.35401 | 0.72019 |
| Ornithine aminotransferase | 0.01022 | 0.00143 | 0.00608 | 0.00012 | 0.00134 | 65 | 0.01311 | 0.00045 | 0.00220 | 93 | 0.00258 | -2.72802 | 144.354 | 0.00716 | 0.62334 |
| Muconolactone Delta-isomerase | 0.01020 | 0.00139 | 0.01055 | 0.00037 | 0.00239 | 65 | 0.00995 | 0.00026 | 0.00168 | 93 | 0.00292 | 0.20506 | 122.084 | 0.83786 | 0.95674 |
| Ribose 1,5-bisphosphate phosphokinase | 0.01020 | 0.00139 | 0.00942 | 0.00028 | 0.00209 | 65 | 0.01074 | 0.00032 | 0.00187 | 93 | 0.00281 | -0.47071 | 143.291 | 0.63857 | 0.88568 |
| Galactoside O-acetyltransferase | 0.01018 | 0.00126 | 0.00949 | 0.00027 | 0.00203 | 65 | 0.01066 | 0.00024 | 0.00162 | 93 | 0.00260 | -0.45160 | 133.533 | 0.65229 | 0.88943 |
| All-trans-zeta-carotene desaturase | 0.01016 | 0.00193 | 0.00912 | 0.00050 | 0.00278 | 65 | 0.01089 | 0.00066 | 0.00266 | 93 | 0.00385 | -0.45955 | 148.116 | 0.64652 | 0.88749 |
| Phytoene desaturase (lycopene-forming) | 0.01016 | 0.00193 | 0.00912 | 0.00050 | 0.00278 | 65 | 0.01089 | 0.00066 | 0.00266 | 93 | 0.00385 | -0.45955 | 148.116 | 0.64652 | 0.88749 |
| Phytoene desaturase (neurosporene-forming) | 0.01016 | 0.00193 | 0.00912 | 0.00050 | 0.00278 | 65 | 0.01089 | 0.00066 | 0.00266 | 93 | 0.00385 | -0.45955 | 148.116 | 0.64652 | 0.88749 |
| Phytoene desaturase (zeta-carotene-forming) | 0.01016 | 0.00193 | 0.00912 | 0.00050 | 0.00278 | 65 | 0.01089 | 0.00066 | 0.00266 | 93 | 0.00385 | -0.45955 | 148.116 | 0.64652 | 0.88749 |
| 2-oxopent-4-enoate hydratase | 0.01012 | 0.00131 | 0.00879 | 0.00025 | 0.00196 | 65 | 0.01105 | 0.00029 | 0.00175 | 93 | 0.00263 | -0.85942 | 143.633 | 0.39154 | 0.74318 |
| UDP-N-acetyl-D-mannosamine dehydrogenase | 0.01011 | 0.00119 | 0.00952 | 0.00021 | 0.00179 | 65 | 0.01052 | 0.00024 | 0.00159 | 93 | 0.00239 | -0.41894 | 143.196 | 0.67589 | 0.89920 |
| Taurine-transporting ATPase | 0.01004 | 0.00129 | 0.00881 | 0.00019 | 0.00173 | 65 | 0.01089 | 0.00031 | 0.00182 | 93 | 0.00251 | -0.82623 | 153.539 | 0.40995 | 0.76070 |
| 23S rRNA (adenine(1618)-N(6))-methyltransferase | 0.01000 | 0.00149 | 0.01155 | 0.00037 | 0.00238 | 65 | 0.00892 | 0.00034 | 0.00190 | 93 | 0.00305 | 0.86141 | 133.739 | 0.39055 | 0.74318 |
| Uronate dehydrogenase | 0.00996 | 0.00162 | 0.00896 | 0.00037 | 0.00237 | 65 | 0.01065 | 0.00046 | 0.00222 | 93 | 0.00324 | -0.52062 | 146.708 | 0.60342 | 0.86777 |
| Muconate cycloisomerase | 0.00995 | 0.00135 | 0.01017 | 0.00033 | 0.00226 | 65 | 0.00979 | 0.00026 | 0.00167 | 93 | 0.00280 | 0.13381 | 126.667 | 0.89377 | 0.97093 |
| Methylthioribulose 1-phosphate dehydratase | 0.00993 | 0.00148 | 0.00996 | 0.00041 | 0.00252 | 65 | 0.00992 | 0.00030 | 0.00181 | 93 | 0.00310 | 0.01376 | 123.911 | 0.98904 | 0.99660 |
| Acetoacetate–CoA ligase | 0.00993 | 0.00135 | 0.00836 | 0.00033 | 0.00225 | 65 | 0.01103 | 0.00026 | 0.00168 | 93 | 0.00280 | -0.95465 | 127.575 | 0.34156 | 0.72019 |
| (2Z,6E)-farnesyl diphosphate synthase | 0.00989 | 0.00138 | 0.00995 | 0.00029 | 0.00211 | 65 | 0.00986 | 0.00031 | 0.00184 | 93 | 0.00280 | 0.03195 | 141.320 | 0.97456 | 0.99435 |
| Hydrogen dehydrogenase (NADP(+)) | 0.00986 | 0.00205 | 0.00904 | 0.00045 | 0.00264 | 65 | 0.01043 | 0.00082 | 0.00297 | 93 | 0.00397 | -0.35056 | 155.379 | 0.72640 | 0.92120 |
| Carnitine 3-dehydrogenase | 0.00985 | 0.00175 | 0.01028 | 0.00046 | 0.00265 | 65 | 0.00955 | 0.00051 | 0.00234 | 93 | 0.00354 | 0.20562 | 142.372 | 0.83738 | 0.95674 |
| Protoporphyrinogen IX dehydrogenase (menaquinone) | 0.00985 | 0.00127 | 0.01074 | 0.00030 | 0.00214 | 65 | 0.00922 | 0.00023 | 0.00157 | 93 | 0.00265 | 0.57293 | 126.114 | 0.56771 | 0.85024 |
| Tryptophan 7-halogenase | 0.00973 | 0.00230 | 0.00606 | 0.00035 | 0.00232 | 65 | 0.01230 | 0.00117 | 0.00355 | 93 | 0.00424 | -1.47297 | 148.491 | 0.14288 | 0.70632 |
| Aryl-alcohol dehydrogenase | 0.00967 | 0.00160 | 0.01010 | 0.00048 | 0.00272 | 65 | 0.00937 | 0.00035 | 0.00195 | 93 | 0.00335 | 0.21567 | 123.666 | 0.82960 | 0.95262 |
| L-fucose mutarotase | 0.00966 | 0.00117 | 0.00973 | 0.00028 | 0.00206 | 65 | 0.00962 | 0.00018 | 0.00137 | 93 | 0.00248 | 0.04212 | 117.285 | 0.96647 | 0.99193 |
| Fructuronate reductase | 0.00962 | 0.00126 | 0.00959 | 0.00039 | 0.00244 | 65 | 0.00963 | 0.00016 | 0.00129 | 93 | 0.00276 | -0.01478 | 99.470 | 0.98824 | 0.99660 |
| Gamma-guanidinobutyraldehyde dehydrogenase | 0.00958 | 0.00155 | 0.01070 | 0.00041 | 0.00251 | 65 | 0.00879 | 0.00036 | 0.00196 | 93 | 0.00319 | 0.60055 | 131.665 | 0.54917 | 0.84333 |
| N-acylneuraminate cytidylyltransferase | 0.00956 | 0.00128 | 0.00884 | 0.00026 | 0.00201 | 65 | 0.01007 | 0.00026 | 0.00166 | 93 | 0.00260 | -0.47055 | 136.684 | 0.63871 | 0.88568 |
| Tryptophan 2-monooxygenase | 0.00945 | 0.00206 | 0.00852 | 0.00045 | 0.00262 | 65 | 0.01010 | 0.00083 | 0.00299 | 93 | 0.00398 | -0.39520 | 155.618 | 0.69324 | 0.90946 |
| D-lactate dehydrogenase (cytochrome) | 0.00944 | 0.00134 | 0.00399 | 0.00007 | 0.00106 | 65 | 0.01325 | 0.00040 | 0.00207 | 93 | 0.00233 | -3.97599 | 133.416 | 0.00011 | 0.13911 |
| Thiopurine S-methyltransferase | 0.00944 | 0.00147 | 0.01050 | 0.00038 | 0.00241 | 65 | 0.00869 | 0.00032 | 0.00187 | 93 | 0.00304 | 0.59501 | 131.126 | 0.55286 | 0.84479 |
| Deoxyadenosine kinase | 0.00943 | 0.00153 | 0.00753 | 0.00027 | 0.00205 | 65 | 0.01076 | 0.00044 | 0.00217 | 93 | 0.00299 | -1.07908 | 153.701 | 0.28224 | 0.70632 |
| 6-carboxyhexanoate–CoA ligase | 0.00936 | 0.00115 | 0.00971 | 0.00025 | 0.00196 | 65 | 0.00912 | 0.00018 | 0.00139 | 93 | 0.00241 | 0.24394 | 123.050 | 0.80768 | 0.94429 |
| S-methyl-5-thioribose kinase | 0.00934 | 0.00169 | 0.01292 | 0.00083 | 0.00357 | 65 | 0.00683 | 0.00018 | 0.00138 | 93 | 0.00383 | 1.59369 | 83.353 | 0.11479 | 0.70632 |
| 3-(3-hydroxy-phenyl)propanoic acid hydroxylase | 0.00932 | 0.00160 | 0.00557 | 0.00017 | 0.00161 | 65 | 0.01194 | 0.00056 | 0.00245 | 93 | 0.00293 | -2.17046 | 148.835 | 0.03156 | 0.67652 |
| Glyoxylate reductase (NADP(+)) | 0.00928 | 0.00123 | 0.00677 | 0.00017 | 0.00161 | 65 | 0.01103 | 0.00029 | 0.00175 | 93 | 0.00238 | -1.78864 | 154.566 | 0.07563 | 0.70632 |
| 3-hydroxy acid dehydrogenase | 0.00927 | 0.00128 | 0.01042 | 0.00028 | 0.00207 | 65 | 0.00847 | 0.00025 | 0.00163 | 93 | 0.00263 | 0.73861 | 132.921 | 0.46144 | 0.78637 |
| Benzaldehyde dehydrogenase (NAD(+)) | 0.00926 | 0.00153 | 0.00807 | 0.00034 | 0.00229 | 65 | 0.01010 | 0.00039 | 0.00206 | 93 | 0.00308 | -0.65700 | 143.987 | 0.51223 | 0.81548 |
| Myo-inosose-2 dehydratase | 0.00921 | 0.00124 | 0.00763 | 0.00016 | 0.00157 | 65 | 0.01032 | 0.00030 | 0.00179 | 93 | 0.00238 | -1.12922 | 155.567 | 0.26054 | 0.70632 |
| 4-hydroxyproline epimerase | 0.00920 | 0.00136 | 0.00782 | 0.00019 | 0.00172 | 65 | 0.01016 | 0.00036 | 0.00198 | 93 | 0.00262 | -0.89116 | 155.711 | 0.37422 | 0.73204 |
| Thymidine phosphorylase | 0.00915 | 0.00112 | 0.00706 | 0.00011 | 0.00130 | 65 | 0.01062 | 0.00025 | 0.00165 | 93 | 0.00210 | -1.69340 | 155.466 | 0.09238 | 0.70632 |
| 3-phytase | 0.00915 | 0.00145 | 0.00735 | 0.00032 | 0.00220 | 65 | 0.01040 | 0.00034 | 0.00192 | 93 | 0.00292 | -1.04156 | 141.330 | 0.29940 | 0.70632 |
| Spore photoproduct lyase | 0.00906 | 0.00170 | 0.01147 | 0.00073 | 0.00335 | 65 | 0.00738 | 0.00026 | 0.00168 | 93 | 0.00375 | 1.08863 | 95.996 | 0.27904 | 0.70632 |
| Phenylacetaldehyde dehydrogenase | 0.00901 | 0.00162 | 0.00905 | 0.00041 | 0.00251 | 65 | 0.00898 | 0.00042 | 0.00213 | 93 | 0.00329 | 0.02042 | 139.351 | 0.98373 | 0.99598 |
| Allophanate hydrolase | 0.00894 | 0.00127 | 0.00851 | 0.00024 | 0.00191 | 65 | 0.00924 | 0.00027 | 0.00170 | 93 | 0.00256 | -0.28637 | 143.159 | 0.77501 | 0.93675 |
| Maleylpyruvate isomerase | 0.00891 | 0.00148 | 0.00591 | 0.00010 | 0.00125 | 65 | 0.01100 | 0.00051 | 0.00233 | 93 | 0.00265 | -1.92177 | 136.309 | 0.05672 | 0.70632 |
| Chloramphenicol O-acetyltransferase | 0.00891 | 0.00137 | 0.00655 | 0.00011 | 0.00130 | 65 | 0.01055 | 0.00042 | 0.00213 | 93 | 0.00250 | -1.60043 | 144.347 | 0.11169 | 0.70632 |
| L-threonine 3-dehydrogenase | 0.00885 | 0.00140 | 0.00826 | 0.00029 | 0.00213 | 65 | 0.00927 | 0.00033 | 0.00187 | 93 | 0.00283 | -0.35359 | 142.222 | 0.72417 | 0.92120 |
| (S)-ureidoglycine aminohydrolase | 0.00880 | 0.00155 | 0.00850 | 0.00035 | 0.00231 | 65 | 0.00901 | 0.00040 | 0.00208 | 93 | 0.00311 | -0.16414 | 143.953 | 0.86985 | 0.96353 |
| Phosphate propanoyltransferase | 0.00877 | 0.00116 | 0.01014 | 0.00028 | 0.00207 | 65 | 0.00781 | 0.00017 | 0.00133 | 93 | 0.00246 | 0.94562 | 114.539 | 0.34633 | 0.72019 |
| Choline-sulfatase | 0.00876 | 0.00143 | 0.00925 | 0.00034 | 0.00227 | 65 | 0.00842 | 0.00032 | 0.00185 | 93 | 0.00293 | 0.28374 | 135.609 | 0.77704 | 0.93675 |
| Glycine dehydrogenase (cyanide-forming) | 0.00868 | 0.00256 | 0.00798 | 0.00089 | 0.00370 | 65 | 0.00918 | 0.00115 | 0.00351 | 93 | 0.00510 | -0.23550 | 147.876 | 0.81414 | 0.94567 |
| Aminodeoxyfutalosine synthase | 0.00868 | 0.00202 | 0.00979 | 0.00093 | 0.00379 | 65 | 0.00790 | 0.00045 | 0.00219 | 93 | 0.00438 | 0.43107 | 105.708 | 0.66730 | 0.89333 |
| Deoxyribonuclease V | 0.00866 | 0.00138 | 0.00831 | 0.00031 | 0.00217 | 65 | 0.00891 | 0.00030 | 0.00179 | 93 | 0.00282 | -0.21131 | 136.895 | 0.83296 | 0.95544 |
| Acireductone synthase | 0.00864 | 0.00142 | 0.00964 | 0.00039 | 0.00245 | 65 | 0.00794 | 0.00027 | 0.00171 | 93 | 0.00299 | 0.57039 | 121.224 | 0.56947 | 0.85024 |
| UDP-sugar diphosphatase | 0.00861 | 0.00135 | 0.00906 | 0.00021 | 0.00180 | 65 | 0.00829 | 0.00034 | 0.00192 | 93 | 0.00264 | 0.29501 | 153.975 | 0.76839 | 0.93519 |
| Glucose-1-phosphate cytidylyltransferase | 0.00851 | 0.00112 | 0.00840 | 0.00033 | 0.00224 | 65 | 0.00859 | 0.00011 | 0.00110 | 93 | 0.00249 | -0.07538 | 94.703 | 0.94007 | 0.99049 |
| Precorrin-6A synthase (deacetylating) | 0.00850 | 0.00122 | 0.00755 | 0.00019 | 0.00173 | 65 | 0.00917 | 0.00027 | 0.00169 | 93 | 0.00242 | -0.66594 | 149.869 | 0.50648 | 0.81359 |
| Putrescine aminotransferase | 0.00843 | 0.00246 | 0.00738 | 0.00079 | 0.00349 | 65 | 0.00916 | 0.00108 | 0.00340 | 93 | 0.00488 | -0.36451 | 149.424 | 0.71599 | 0.92120 |
| 2-aminoethylphosphonate–pyruvate transaminase | 0.00840 | 0.00126 | 0.00679 | 0.00015 | 0.00151 | 65 | 0.00953 | 0.00032 | 0.00186 | 93 | 0.00240 | -1.14403 | 155.859 | 0.25437 | 0.70632 |
| 3-fumarylpyruvate hydrolase | 0.00839 | 0.00137 | 0.00465 | 0.00008 | 0.00114 | 65 | 0.01100 | 0.00043 | 0.00215 | 93 | 0.00243 | -2.61252 | 135.419 | 0.01000 | 0.64530 |
| Arylformamidase | 0.00833 | 0.00135 | 0.00413 | 0.00007 | 0.00101 | 65 | 0.01126 | 0.00042 | 0.00214 | 93 | 0.00237 | -3.01829 | 128.756 | 0.00307 | 0.62334 |
| 5-aminovalerate transaminase | 0.00830 | 0.00143 | 0.00935 | 0.00035 | 0.00233 | 65 | 0.00756 | 0.00031 | 0.00182 | 93 | 0.00295 | 0.60595 | 132.057 | 0.54559 | 0.84290 |
| Beta-carotene 3-hydroxylase | 0.00824 | 0.00139 | 0.00850 | 0.00029 | 0.00211 | 65 | 0.00807 | 0.00032 | 0.00186 | 93 | 0.00281 | 0.15404 | 142.618 | 0.87779 | 0.96548 |
| Nitrile hydratase | 0.00823 | 0.00174 | 0.00517 | 0.00022 | 0.00186 | 65 | 0.01037 | 0.00065 | 0.00264 | 93 | 0.00323 | -1.60830 | 152.052 | 0.10984 | 0.70632 |
| Glutamin-(asparagin-)ase | 0.00816 | 0.00134 | 0.00661 | 0.00018 | 0.00166 | 65 | 0.00925 | 0.00036 | 0.00195 | 93 | 0.00256 | -1.03068 | 155.947 | 0.30429 | 0.70632 |
| Tagaturonate reductase | 0.00815 | 0.00122 | 0.00763 | 0.00034 | 0.00229 | 65 | 0.00851 | 0.00016 | 0.00132 | 93 | 0.00264 | -0.33389 | 105.633 | 0.73912 | 0.92595 |
| Creatininase | 0.00811 | 0.00133 | 0.00445 | 0.00009 | 0.00116 | 65 | 0.01067 | 0.00040 | 0.00207 | 93 | 0.00237 | -2.62118 | 139.327 | 0.00974 | 0.64530 |
| Aminoglycoside N(3’)-acetyltransferase | 0.00809 | 0.00144 | 0.00890 | 0.00042 | 0.00255 | 65 | 0.00752 | 0.00026 | 0.00168 | 93 | 0.00305 | 0.45243 | 116.437 | 0.65180 | 0.88943 |
| Phosphonoacetaldehyde hydrolase | 0.00806 | 0.00127 | 0.00694 | 0.00017 | 0.00160 | 65 | 0.00884 | 0.00032 | 0.00185 | 93 | 0.00245 | -0.77683 | 155.783 | 0.43844 | 0.77367 |
| PepB aminopeptidase | 0.00796 | 0.00121 | 0.00908 | 0.00026 | 0.00199 | 65 | 0.00718 | 0.00021 | 0.00152 | 93 | 0.00250 | 0.76104 | 129.893 | 0.44801 | 0.77823 |
| Formimidoylglutamate deiminase | 0.00796 | 0.00129 | 0.00715 | 0.00020 | 0.00175 | 65 | 0.00852 | 0.00031 | 0.00182 | 93 | 0.00252 | -0.54422 | 152.826 | 0.58709 | 0.85593 |
| CDP-diacylglycerol diphosphatase | 0.00796 | 0.00147 | 0.00753 | 0.00031 | 0.00218 | 65 | 0.00825 | 0.00036 | 0.00198 | 93 | 0.00294 | -0.24500 | 144.735 | 0.80681 | 0.94388 |
| tRNA(Met) cytidine acetyltransferase | 0.00794 | 0.00121 | 0.00899 | 0.00026 | 0.00198 | 65 | 0.00721 | 0.00022 | 0.00152 | 93 | 0.00250 | 0.71213 | 130.093 | 0.47766 | 0.79572 |
| Lauroyl-Kdo(2)-lipid IV(A) myristoyltransferase | 0.00792 | 0.00121 | 0.00899 | 0.00026 | 0.00199 | 65 | 0.00718 | 0.00021 | 0.00152 | 93 | 0.00250 | 0.72373 | 129.848 | 0.47054 | 0.79541 |
| Nicotinate dehydrogenase (cytochrome) | 0.00790 | 0.00171 | 0.00606 | 0.00024 | 0.00194 | 65 | 0.00919 | 0.00062 | 0.00258 | 93 | 0.00322 | -0.97179 | 154.449 | 0.33267 | 0.71544 |
| 3D-(3,5/4)-trihydroxycyclohexane-1,2-dione acylhydrolase (decyclizing) | 0.00787 | 0.00103 | 0.00646 | 0.00013 | 0.00142 | 65 | 0.00885 | 0.00019 | 0.00143 | 93 | 0.00202 | -1.18524 | 151.435 | 0.23778 | 0.70632 |
| decaprenol beta-1,5/1,6-galactofuranosyltransferase | 0.00785 | 0.00129 | 0.00809 | 0.00028 | 0.00206 | 65 | 0.00768 | 0.00026 | 0.00166 | 93 | 0.00265 | 0.15455 | 134.705 | 0.87741 | 0.96548 |
| S-methyl-5’-thioinosine phosphorylase | 0.00784 | 0.00139 | 0.00889 | 0.00037 | 0.00239 | 65 | 0.00711 | 0.00026 | 0.00168 | 93 | 0.00292 | 0.60915 | 122.069 | 0.54356 | 0.84121 |
| Guanidinobutyrase | 0.00784 | 0.00133 | 0.00877 | 0.00030 | 0.00215 | 65 | 0.00719 | 0.00027 | 0.00169 | 93 | 0.00274 | 0.57492 | 132.500 | 0.56632 | 0.85024 |
| L-fucose isomerase | 0.00779 | 0.00111 | 0.00861 | 0.00027 | 0.00205 | 65 | 0.00721 | 0.00014 | 0.00124 | 93 | 0.00240 | 0.58452 | 108.976 | 0.56008 | 0.85024 |
| Acylaminoacyl-peptidase | 0.00777 | 0.00159 | 0.01075 | 0.00070 | 0.00329 | 65 | 0.00568 | 0.00019 | 0.00141 | 93 | 0.00358 | 1.41652 | 87.806 | 0.16016 | 0.70632 |
| Unsaturated chondroitin disaccharide hydrolase | 0.00774 | 0.00150 | 0.00789 | 0.00042 | 0.00253 | 65 | 0.00764 | 0.00032 | 0.00185 | 93 | 0.00313 | 0.08217 | 125.923 | 0.93465 | 0.98985 |
| 5-deoxy-glucuronate isomerase | 0.00773 | 0.00100 | 0.00618 | 0.00012 | 0.00137 | 65 | 0.00882 | 0.00018 | 0.00141 | 93 | 0.00196 | -1.34313 | 152.484 | 0.18123 | 0.70632 |
| (S)-citramalyl-CoA lyase | 0.00773 | 0.00135 | 0.00766 | 0.00027 | 0.00203 | 65 | 0.00777 | 0.00031 | 0.00182 | 93 | 0.00272 | -0.04122 | 143.555 | 0.96718 | 0.99193 |
| Rhamnulose-1-phosphate aldolase | 0.00768 | 0.00136 | 0.00812 | 0.00053 | 0.00285 | 65 | 0.00737 | 0.00013 | 0.00120 | 93 | 0.00309 | 0.24069 | 87.032 | 0.81036 | 0.94519 |
| (S)-3-amino-2-methylpropionate transaminase | 0.00767 | 0.00122 | 0.00453 | 0.00008 | 0.00113 | 65 | 0.00986 | 0.00033 | 0.00188 | 93 | 0.00220 | -2.42403 | 143.719 | 0.01659 | 0.64530 |
| NADH peroxidase | 0.00767 | 0.00160 | 0.01003 | 0.00068 | 0.00323 | 65 | 0.00601 | 0.00022 | 0.00152 | 93 | 0.00357 | 1.12430 | 92.452 | 0.26380 | 0.70632 |
| 5-dehydro-2-deoxygluconokinase | 0.00766 | 0.00099 | 0.00619 | 0.00012 | 0.00136 | 65 | 0.00868 | 0.00018 | 0.00139 | 93 | 0.00195 | -1.27858 | 152.056 | 0.20299 | 0.70632 |
| Aminobutyraldehyde dehydrogenase | 0.00764 | 0.00149 | 0.00666 | 0.00021 | 0.00179 | 65 | 0.00832 | 0.00045 | 0.00220 | 93 | 0.00284 | -0.58746 | 155.923 | 0.55774 | 0.84948 |
| ligase | 0.00761 | 0.00115 | 0.00752 | 0.00024 | 0.00194 | 65 | 0.00767 | 0.00018 | 0.00141 | 93 | 0.00239 | -0.06105 | 125.282 | 0.95141 | 0.99049 |
| Trans-feruloyl-CoA synthase | 0.00760 | 0.00149 | 0.00381 | 0.00009 | 0.00120 | 65 | 0.01025 | 0.00052 | 0.00236 | 93 | 0.00265 | -2.43270 | 133.127 | 0.01631 | 0.64530 |
| N-acylmannosamine kinase | 0.00760 | 0.00121 | 0.00763 | 0.00023 | 0.00187 | 65 | 0.00757 | 0.00024 | 0.00159 | 93 | 0.00245 | 0.02428 | 139.598 | 0.98067 | 0.99457 |
| Allantoin racemase | 0.00759 | 0.00130 | 0.00613 | 0.00016 | 0.00157 | 65 | 0.00861 | 0.00034 | 0.00192 | 93 | 0.00248 | -1.00059 | 155.965 | 0.31858 | 0.70632 |
| Oxalyl-CoA decarboxylase | 0.00758 | 0.00276 | 0.00966 | 0.00245 | 0.00614 | 65 | 0.00612 | 0.00035 | 0.00193 | 93 | 0.00644 | 0.54971 | 76.791 | 0.58412 | 0.85509 |
| D-inositol-3-phosphate glycosyltransferase | 0.00758 | 0.00114 | 0.00750 | 0.00024 | 0.00193 | 65 | 0.00763 | 0.00018 | 0.00141 | 93 | 0.00239 | -0.05401 | 125.435 | 0.95701 | 0.99147 |
| Mycothiol S-conjugate amidase | 0.00757 | 0.00115 | 0.00752 | 0.00024 | 0.00194 | 65 | 0.00760 | 0.00019 | 0.00141 | 93 | 0.00240 | -0.03066 | 125.445 | 0.97559 | 0.99435 |
| Mycothiol synthase | 0.00757 | 0.00115 | 0.00752 | 0.00024 | 0.00194 | 65 | 0.00760 | 0.00019 | 0.00141 | 93 | 0.00240 | -0.03066 | 125.445 | 0.97559 | 0.99435 |
| Diaminobutyrate decarboxylase | 0.00747 | 0.00115 | 0.00635 | 0.00013 | 0.00143 | 65 | 0.00825 | 0.00026 | 0.00168 | 93 | 0.00220 | -0.85850 | 155.912 | 0.39193 | 0.74318 |
| 1,6-dihydroxycyclohexa-2,4-diene-1-carboxylate dehydrogenase | 0.00741 | 0.00116 | 0.00851 | 0.00028 | 0.00208 | 65 | 0.00664 | 0.00016 | 0.00132 | 93 | 0.00247 | 0.75676 | 113.277 | 0.45076 | 0.78108 |
| Sugar-phosphatase | 0.00741 | 0.00116 | 0.00651 | 0.00014 | 0.00149 | 65 | 0.00803 | 0.00026 | 0.00167 | 93 | 0.00224 | -0.67884 | 155.338 | 0.49825 | 0.80830 |
| Allantoate deiminase | 0.00733 | 0.00156 | 0.00494 | 0.00016 | 0.00157 | 65 | 0.00900 | 0.00054 | 0.00241 | 93 | 0.00288 | -1.41028 | 148.397 | 0.16055 | 0.70632 |
| 2-deoxy-D-gluconate 3-dehydrogenase | 0.00732 | 0.00112 | 0.00566 | 0.00020 | 0.00174 | 65 | 0.00847 | 0.00020 | 0.00146 | 93 | 0.00228 | -1.23479 | 138.350 | 0.21900 | 0.70632 |
| D-cysteine desulfhydrase | 0.00728 | 0.00112 | 0.00706 | 0.00019 | 0.00172 | 65 | 0.00744 | 0.00021 | 0.00149 | 93 | 0.00228 | -0.16384 | 141.061 | 0.87009 | 0.96353 |
| Levanase | 0.00727 | 0.00105 | 0.00647 | 0.00011 | 0.00129 | 65 | 0.00783 | 0.00022 | 0.00155 | 93 | 0.00202 | -0.67291 | 155.995 | 0.50200 | 0.81035 |
| deacetylase | 0.00725 | 0.00123 | 0.00725 | 0.00024 | 0.00193 | 65 | 0.00726 | 0.00024 | 0.00161 | 93 | 0.00252 | -0.00603 | 137.627 | 0.99520 | 0.99921 |
| IgA-specific serine endopeptidase | 0.00724 | 0.00130 | 0.00834 | 0.00034 | 0.00227 | 65 | 0.00646 | 0.00022 | 0.00153 | 93 | 0.00274 | 0.68726 | 118.541 | 0.49326 | 0.80568 |
| Mycothione reductase | 0.00723 | 0.00114 | 0.00701 | 0.00024 | 0.00193 | 65 | 0.00738 | 0.00018 | 0.00141 | 93 | 0.00239 | -0.15837 | 125.548 | 0.87442 | 0.96378 |
| S-(hydroxymethyl)mycothiol dehydrogenase | 0.00720 | 0.00116 | 0.00699 | 0.00025 | 0.00196 | 65 | 0.00735 | 0.00019 | 0.00142 | 93 | 0.00242 | -0.14618 | 125.333 | 0.88401 | 0.96726 |
| N-acetylglucosaminyl-diphospho-decaprenol L-rhamnosyltransferase | 0.00719 | 0.00118 | 0.00753 | 0.00024 | 0.00194 | 65 | 0.00696 | 0.00021 | 0.00149 | 93 | 0.00245 | 0.23217 | 130.515 | 0.81677 | 0.94663 |
| Glycerol-3-phosphate-transporting ATPase | 0.00719 | 0.00112 | 0.00368 | 0.00006 | 0.00097 | 65 | 0.00964 | 0.00028 | 0.00173 | 93 | 0.00198 | -3.00576 | 139.068 | 0.00314 | 0.62334 |
| 3-aminobutyryl-CoA ammonia-lyase | 0.00712 | 0.00161 | 0.01093 | 0.00073 | 0.00335 | 65 | 0.00447 | 0.00017 | 0.00135 | 93 | 0.00362 | 1.78739 | 84.749 | 0.07745 | 0.70632 |
| N(4)-(beta-N-acetylglucosaminyl)-L-asparaginase | 0.00712 | 0.00132 | 0.00667 | 0.00031 | 0.00218 | 65 | 0.00744 | 0.00025 | 0.00165 | 93 | 0.00274 | -0.27852 | 128.608 | 0.78106 | 0.93675 |
| Phosphatidylcholine synthase | 0.00705 | 0.00102 | 0.00682 | 0.00017 | 0.00164 | 65 | 0.00721 | 0.00016 | 0.00132 | 93 | 0.00210 | -0.18589 | 134.537 | 0.85281 | 0.96065 |
| Formamidase | 0.00704 | 0.00134 | 0.00495 | 0.00017 | 0.00163 | 65 | 0.00851 | 0.00036 | 0.00197 | 93 | 0.00256 | -1.39322 | 155.978 | 0.16553 | 0.70632 |
| CDP-glucose 4,6-dehydratase | 0.00704 | 0.00109 | 0.00777 | 0.00033 | 0.00225 | 65 | 0.00653 | 0.00009 | 0.00097 | 93 | 0.00245 | 0.50663 | 88.059 | 0.61368 | 0.87303 |
| Serralysin | 0.00703 | 0.00168 | 0.00480 | 0.00021 | 0.00178 | 65 | 0.00859 | 0.00061 | 0.00256 | 93 | 0.00312 | -1.21340 | 151.541 | 0.22686 | 0.70632 |
| Peptidyl-dipeptidase A | 0.00702 | 0.00136 | 0.00623 | 0.00031 | 0.00218 | 65 | 0.00757 | 0.00028 | 0.00175 | 93 | 0.00279 | -0.48186 | 134.265 | 0.63069 | 0.88317 |
| Haloalkane dehalogenase | 0.00701 | 0.00134 | 0.00659 | 0.00031 | 0.00218 | 65 | 0.00730 | 0.00027 | 0.00170 | 93 | 0.00276 | -0.25874 | 131.597 | 0.79624 | 0.94089 |
| Long-chain-acyl-CoA dehydrogenase | 0.00696 | 0.00138 | 0.00528 | 0.00013 | 0.00141 | 65 | 0.00813 | 0.00042 | 0.00213 | 93 | 0.00256 | -1.11205 | 149.113 | 0.26790 | 0.70632 |
| Cyclohexanone monooxygenase | 0.00695 | 0.00155 | 0.00400 | 0.00005 | 0.00087 | 65 | 0.00902 | 0.00060 | 0.00255 | 93 | 0.00269 | -1.86676 | 112.310 | 0.06454 | 0.70632 |
| Aminoglycoside 6’-N-acetyltransferase | 0.00694 | 0.00118 | 0.00618 | 0.00017 | 0.00162 | 65 | 0.00748 | 0.00026 | 0.00166 | 93 | 0.00232 | -0.55663 | 152.284 | 0.57860 | 0.85190 |
| Salicylate 1-monooxygenase | 0.00694 | 0.00166 | 0.00323 | 0.00004 | 0.00079 | 65 | 0.00953 | 0.00069 | 0.00273 | 93 | 0.00284 | -2.21455 | 106.860 | 0.02891 | 0.67652 |
| Isochorismate lyase | 0.00692 | 0.00096 | 0.00606 | 0.00011 | 0.00133 | 65 | 0.00751 | 0.00017 | 0.00134 | 93 | 0.00189 | -0.76982 | 151.488 | 0.44261 | 0.77520 |
| D-glycero-alpha-D-manno-heptose-7-phosphate kinase | 0.00690 | 0.00178 | 0.00950 | 0.00104 | 0.00399 | 65 | 0.00508 | 0.00012 | 0.00116 | 93 | 0.00416 | 1.06358 | 74.827 | 0.29094 | 0.70632 |
| Caffeoyl-CoA O-methyltransferase | 0.00678 | 0.00156 | 0.01065 | 0.00071 | 0.00329 | 65 | 0.00407 | 0.00015 | 0.00126 | 93 | 0.00353 | 1.86551 | 82.820 | 0.06565 | 0.70632 |
| L-rhamnose mutarotase | 0.00676 | 0.00095 | 0.00448 | 0.00007 | 0.00101 | 65 | 0.00836 | 0.00019 | 0.00143 | 93 | 0.00175 | -2.21593 | 152.188 | 0.02818 | 0.67652 |
| 6-aminohexanoate-oligomer exohydrolase | 0.00674 | 0.00134 | 0.00350 | 0.00010 | 0.00122 | 65 | 0.00901 | 0.00040 | 0.00208 | 93 | 0.00241 | -2.28415 | 142.279 | 0.02384 | 0.67652 |
| Long-chain-fatty-acid–[acyl-carrier-protein] ligase | 0.00672 | 0.00087 | 0.00849 | 0.00017 | 0.00163 | 65 | 0.00548 | 0.00008 | 0.00094 | 93 | 0.00188 | 1.60637 | 105.428 | 0.11118 | 0.70632 |
| Alpha-methylacyl-CoA racemase | 0.00667 | 0.00135 | 0.00355 | 0.00006 | 0.00096 | 65 | 0.00884 | 0.00044 | 0.00217 | 93 | 0.00237 | -2.23311 | 124.443 | 0.02733 | 0.67652 |
| Pitrilysin | 0.00664 | 0.00113 | 0.00747 | 0.00021 | 0.00178 | 65 | 0.00606 | 0.00020 | 0.00146 | 93 | 0.00230 | 0.61358 | 136.047 | 0.54052 | 0.83772 |
| Ceramidase | 0.00660 | 0.00114 | 0.00693 | 0.00025 | 0.00196 | 65 | 0.00637 | 0.00018 | 0.00138 | 93 | 0.00240 | 0.23413 | 122.392 | 0.81527 | 0.94645 |
| L-erythro-3,5-diaminohexanoate dehydrogenase | 0.00659 | 0.00154 | 0.01037 | 0.00070 | 0.00329 | 65 | 0.00395 | 0.00014 | 0.00121 | 93 | 0.00351 | 1.82942 | 81.516 | 0.07099 | 0.70632 |
| Glutamate decarboxylase | 0.00658 | 0.00141 | 0.00629 | 0.00039 | 0.00245 | 65 | 0.00678 | 0.00027 | 0.00170 | 93 | 0.00298 | -0.16439 | 120.891 | 0.86970 | 0.96353 |
| Ste24 endopeptidase | 0.00657 | 0.00111 | 0.00590 | 0.00012 | 0.00136 | 65 | 0.00704 | 0.00025 | 0.00163 | 93 | 0.00212 | -0.53883 | 155.997 | 0.59077 | 0.85791 |
| Arabinogalactan endo-beta-1,4-galactanase | 0.00650 | 0.00102 | 0.00451 | 0.00005 | 0.00089 | 65 | 0.00789 | 0.00024 | 0.00160 | 93 | 0.00184 | -1.84036 | 138.827 | 0.06785 | 0.70632 |
| 2-dehydro-3-deoxygalactonokinase | 0.00650 | 0.00102 | 0.00355 | 0.00006 | 0.00093 | 65 | 0.00856 | 0.00023 | 0.00158 | 93 | 0.00184 | -2.73244 | 142.452 | 0.00708 | 0.62334 |
| 5-oxopent-3-ene-1,2,5-tricarboxylate decarboxylase | 0.00649 | 0.00144 | 0.00487 | 0.00019 | 0.00171 | 65 | 0.00763 | 0.00042 | 0.00213 | 93 | 0.00273 | -1.01110 | 155.787 | 0.31354 | 0.70632 |
| Streptomycin 6-kinase | 0.00645 | 0.00130 | 0.00598 | 0.00029 | 0.00210 | 65 | 0.00678 | 0.00025 | 0.00165 | 93 | 0.00267 | -0.30219 | 132.556 | 0.76298 | 0.93350 |
| Gamma-D-glutamyl-meso-diaminopimelate peptidase | 0.00642 | 0.00128 | 0.00598 | 0.00033 | 0.00225 | 65 | 0.00674 | 0.00021 | 0.00151 | 93 | 0.00271 | -0.27990 | 118.123 | 0.78005 | 0.93675 |
| Serine 3-dehydrogenase (NADP(+)) | 0.00642 | 0.00132 | 0.00580 | 0.00030 | 0.00216 | 65 | 0.00685 | 0.00026 | 0.00166 | 93 | 0.00273 | -0.38399 | 130.238 | 0.70162 | 0.91306 |
| Acyl-[acyl-carrier-protein]–phospholipid O-acyltransferase | 0.00639 | 0.00087 | 0.00847 | 0.00017 | 0.00163 | 65 | 0.00494 | 0.00008 | 0.00092 | 93 | 0.00187 | 1.89007 | 103.708 | 0.06154 | 0.70632 |
| Allantoinase | 0.00636 | 0.00102 | 0.00505 | 0.00013 | 0.00142 | 65 | 0.00727 | 0.00019 | 0.00142 | 93 | 0.00201 | -1.10697 | 151.083 | 0.27007 | 0.70632 |
| Pteridine reductase | 0.00632 | 0.00130 | 0.00641 | 0.00030 | 0.00214 | 65 | 0.00626 | 0.00025 | 0.00163 | 93 | 0.00269 | 0.05650 | 129.606 | 0.95503 | 0.99147 |
| Alcohol dehydrogenase (cytochrome c) | 0.00630 | 0.00114 | 0.00805 | 0.00034 | 0.00227 | 65 | 0.00508 | 0.00011 | 0.00110 | 93 | 0.00253 | 1.17693 | 93.911 | 0.24220 | 0.70632 |
| Urea carboxylase | 0.00627 | 0.00095 | 0.00552 | 0.00011 | 0.00128 | 65 | 0.00680 | 0.00017 | 0.00135 | 93 | 0.00186 | -0.68562 | 153.638 | 0.49399 | 0.80568 |
| Lycopene beta-cyclase | 0.00626 | 0.00132 | 0.00485 | 0.00021 | 0.00182 | 65 | 0.00725 | 0.00032 | 0.00184 | 93 | 0.00259 | -0.92593 | 151.726 | 0.35595 | 0.72019 |
| Diacylglycerol O-acyltransferase | 0.00624 | 0.00131 | 0.00345 | 0.00004 | 0.00079 | 65 | 0.00819 | 0.00043 | 0.00214 | 93 | 0.00228 | -2.08053 | 115.501 | 0.03969 | 0.67652 |
| 2-dehydro-3-deoxy-6-phosphogalactonate aldolase | 0.00623 | 0.00099 | 0.00337 | 0.00005 | 0.00091 | 65 | 0.00823 | 0.00022 | 0.00153 | 93 | 0.00178 | -2.72063 | 142.862 | 0.00733 | 0.62334 |
| D-stereospecific aminopeptidase | 0.00623 | 0.00120 | 0.00315 | 0.00006 | 0.00100 | 65 | 0.00838 | 0.00033 | 0.00188 | 93 | 0.00213 | -2.45477 | 135.406 | 0.01537 | 0.64530 |
| Thiosulfate dehydrogenase (quinone) | 0.00622 | 0.00115 | 0.00331 | 0.00009 | 0.00117 | 65 | 0.00826 | 0.00028 | 0.00174 | 93 | 0.00210 | -2.36316 | 149.672 | 0.01941 | 0.64530 |
| GDP-mannose 6-dehydrogenase | 0.00621 | 0.00112 | 0.00724 | 0.00026 | 0.00199 | 65 | 0.00548 | 0.00016 | 0.00129 | 93 | 0.00238 | 0.74177 | 115.072 | 0.45974 | 0.78637 |
| Nucleoside deoxyribosyltransferase | 0.00618 | 0.00170 | 0.00764 | 0.00071 | 0.00330 | 65 | 0.00516 | 0.00028 | 0.00175 | 93 | 0.00374 | 0.66315 | 99.493 | 0.50877 | 0.81548 |
| Glucose 1-dehydrogenase (NAD(P)(+)) | 0.00611 | 0.00110 | 0.00438 | 0.00012 | 0.00133 | 65 | 0.00733 | 0.00024 | 0.00161 | 93 | 0.00209 | -1.41152 | 155.991 | 0.16008 | 0.70632 |
| Lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase | 0.00607 | 0.00156 | 0.00466 | 0.00020 | 0.00177 | 65 | 0.00706 | 0.00052 | 0.00235 | 93 | 0.00294 | -0.81552 | 154.403 | 0.41603 | 0.76408 |
| 3-hydroxyanthranilate 3,4-dioxygenase | 0.00606 | 0.00132 | 0.00560 | 0.00028 | 0.00208 | 65 | 0.00638 | 0.00028 | 0.00172 | 93 | 0.00270 | -0.28999 | 136.983 | 0.77227 | 0.93557 |
| Tyrosine phenol-lyase | 0.00598 | 0.00153 | 0.00955 | 0.00068 | 0.00324 | 65 | 0.00349 | 0.00014 | 0.00123 | 93 | 0.00347 | 1.74660 | 82.665 | 0.08442 | 0.70632 |
| Acylpyruvate hydrolase | 0.00587 | 0.00108 | 0.00662 | 0.00024 | 0.00192 | 65 | 0.00535 | 0.00015 | 0.00126 | 93 | 0.00230 | 0.55231 | 115.844 | 0.58180 | 0.85342 |
| Nitric-oxide synthase (NAD(P)H) | 0.00583 | 0.00116 | 0.00300 | 0.00009 | 0.00118 | 65 | 0.00781 | 0.00029 | 0.00177 | 93 | 0.00213 | -2.25449 | 149.397 | 0.02562 | 0.67652 |
| Itaconyl-CoA hydratase | 0.00582 | 0.00118 | 0.00605 | 0.00020 | 0.00176 | 65 | 0.00565 | 0.00023 | 0.00159 | 93 | 0.00237 | 0.16907 | 144.226 | 0.86598 | 0.96221 |
| Isohexenylglutaconyl-CoA hydratase | 0.00582 | 0.00105 | 0.00527 | 0.00013 | 0.00142 | 65 | 0.00620 | 0.00021 | 0.00149 | 93 | 0.00206 | -0.45264 | 153.297 | 0.65145 | 0.88943 |
| Chorismate dehydratase | 0.00581 | 0.00122 | 0.00592 | 0.00026 | 0.00201 | 65 | 0.00574 | 0.00022 | 0.00154 | 93 | 0.00253 | 0.07042 | 130.185 | 0.94396 | 0.99049 |
| 4-hydroxy-2-oxovalerate aldolase | 0.00580 | 0.00097 | 0.00493 | 0.00009 | 0.00120 | 65 | 0.00641 | 0.00019 | 0.00142 | 93 | 0.00186 | -0.80042 | 155.986 | 0.42469 | 0.76494 |
| Dihydrokaempferol 4-reductase | 0.00580 | 0.00094 | 0.00440 | 0.00007 | 0.00106 | 65 | 0.00678 | 0.00019 | 0.00141 | 93 | 0.00177 | -1.34695 | 154.263 | 0.17997 | 0.70632 |
| 2-phosphosulfolactate phosphatase | 0.00579 | 0.00091 | 0.00410 | 0.00008 | 0.00111 | 65 | 0.00697 | 0.00016 | 0.00133 | 93 | 0.00173 | -1.65462 | 156.000 | 0.10001 | 0.70632 |
| Kynurenine 3-monooxygenase | 0.00572 | 0.00128 | 0.00564 | 0.00028 | 0.00208 | 65 | 0.00577 | 0.00025 | 0.00163 | 93 | 0.00264 | -0.04624 | 131.728 | 0.96319 | 0.99193 |
| L-rhamnonate dehydratase | 0.00570 | 0.00146 | 0.00881 | 0.00065 | 0.00317 | 65 | 0.00352 | 0.00010 | 0.00106 | 93 | 0.00335 | 1.57868 | 78.291 | 0.11844 | 0.70632 |
| Pectinesterase | 0.00569 | 0.00105 | 0.00317 | 0.00003 | 0.00064 | 65 | 0.00744 | 0.00027 | 0.00171 | 93 | 0.00183 | -2.33400 | 116.168 | 0.02131 | 0.65070 |
| SpoIVB peptidase | 0.00561 | 0.00117 | 0.00282 | 0.00008 | 0.00110 | 65 | 0.00756 | 0.00031 | 0.00182 | 93 | 0.00213 | -2.23107 | 143.722 | 0.02723 | 0.67652 |
| Alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase | 0.00561 | 0.00098 | 0.00264 | 0.00004 | 0.00078 | 65 | 0.00768 | 0.00022 | 0.00154 | 93 | 0.00173 | -2.91451 | 132.290 | 0.00418 | 0.62334 |
| N-acetylornithine carbamoyltransferase | 0.00557 | 0.00128 | 0.00532 | 0.00028 | 0.00208 | 65 | 0.00574 | 0.00025 | 0.00162 | 93 | 0.00264 | -0.15917 | 131.571 | 0.87378 | 0.96378 |
| 5-carboxymethyl-2-hydroxymuconic-semialdehyde dehydrogenase | 0.00552 | 0.00095 | 0.00294 | 0.00005 | 0.00089 | 65 | 0.00733 | 0.00020 | 0.00147 | 93 | 0.00172 | -2.55918 | 144.073 | 0.01152 | 0.64530 |
| Cyanophycin synthase (L-arginine-adding) | 0.00552 | 0.00136 | 0.00479 | 0.00017 | 0.00161 | 65 | 0.00602 | 0.00038 | 0.00202 | 93 | 0.00258 | -0.47649 | 155.671 | 0.63439 | 0.88317 |
| Cyanophycin synthase (L-aspartate-adding) | 0.00552 | 0.00136 | 0.00479 | 0.00017 | 0.00161 | 65 | 0.00602 | 0.00038 | 0.00202 | 93 | 0.00258 | -0.47649 | 155.671 | 0.63439 | 0.88317 |
| Cyclic dehypoxanthinyl futalosine synthase | 0.00548 | 0.00120 | 0.00589 | 0.00026 | 0.00201 | 65 | 0.00520 | 0.00021 | 0.00149 | 93 | 0.00250 | 0.27741 | 127.217 | 0.78192 | 0.93675 |
| Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase | 0.00547 | 0.00106 | 0.00486 | 0.00012 | 0.00133 | 65 | 0.00589 | 0.00022 | 0.00154 | 93 | 0.00204 | -0.50349 | 155.837 | 0.61533 | 0.87356 |
| GPR endopeptidase | 0.00546 | 0.00117 | 0.00282 | 0.00008 | 0.00110 | 65 | 0.00730 | 0.00031 | 0.00182 | 93 | 0.00212 | -2.11186 | 143.869 | 0.03643 | 0.67652 |
| N-acyl-D-amino-acid deacylase | 0.00542 | 0.00083 | 0.00454 | 0.00007 | 0.00100 | 65 | 0.00604 | 0.00014 | 0.00122 | 93 | 0.00158 | -0.95234 | 155.980 | 0.34240 | 0.72019 |
| Tropinone reductase I | 0.00540 | 0.00128 | 0.00531 | 0.00028 | 0.00208 | 65 | 0.00547 | 0.00024 | 0.00162 | 93 | 0.00264 | -0.06135 | 131.463 | 0.95117 | 0.99049 |
| Sorbitol-6-phosphate 2-dehydrogenase | 0.00538 | 0.00097 | 0.00648 | 0.00023 | 0.00190 | 65 | 0.00462 | 0.00009 | 0.00098 | 93 | 0.00213 | 0.86970 | 97.500 | 0.38660 | 0.73961 |
| D-arginine dehydrogenase | 0.00537 | 0.00096 | 0.00590 | 0.00020 | 0.00175 | 65 | 0.00500 | 0.00011 | 0.00108 | 93 | 0.00206 | 0.43766 | 111.172 | 0.66248 | 0.88968 |
| 1-piperideine-2-carboxylate/1-pyrroline-2-carboxylate reductase (NADPH) | 0.00533 | 0.00109 | 0.00511 | 0.00014 | 0.00145 | 65 | 0.00548 | 0.00022 | 0.00155 | 93 | 0.00212 | -0.17519 | 153.938 | 0.86116 | 0.96186 |
| L-rhamnose isomerase | 0.00530 | 0.00076 | 0.00342 | 0.00003 | 0.00070 | 65 | 0.00661 | 0.00013 | 0.00119 | 93 | 0.00138 | -2.31287 | 142.450 | 0.02216 | 0.65070 |
| Heme oxygenase (staphylobilin-producing) | 0.00526 | 0.00154 | 0.00332 | 0.00016 | 0.00157 | 65 | 0.00661 | 0.00053 | 0.00238 | 93 | 0.00285 | -1.15404 | 148.935 | 0.25033 | 0.70632 |
| 2.7.11.30 and 2.7.12.1 | 0.00518 | 0.00149 | 0.00895 | 0.00068 | 0.00324 | 65 | 0.00254 | 0.00010 | 0.00106 | 93 | 0.00341 | 1.87832 | 77.671 | 0.06409 | 0.70632 |
| NADPH dehydrogenase | 0.00517 | 0.00101 | 0.00259 | 0.00006 | 0.00093 | 65 | 0.00698 | 0.00023 | 0.00157 | 93 | 0.00183 | -2.39768 | 142.663 | 0.01779 | 0.64530 |
| Ceramide glucosyltransferase | 0.00517 | 0.00146 | 0.00897 | 0.00066 | 0.00318 | 65 | 0.00251 | 0.00010 | 0.00104 | 93 | 0.00335 | 1.93305 | 77.674 | 0.05687 | 0.70632 |
| Anthranilate 1,2-dioxygenase (deaminating, decarboxylating) | 0.00516 | 0.00138 | 0.00324 | 0.00014 | 0.00146 | 65 | 0.00650 | 0.00041 | 0.00211 | 93 | 0.00256 | -1.27265 | 151.353 | 0.20510 | 0.70632 |
| Hyaluronate lyase | 0.00516 | 0.00128 | 0.00681 | 0.00055 | 0.00290 | 65 | 0.00401 | 0.00006 | 0.00081 | 93 | 0.00301 | 0.93095 | 74.126 | 0.35490 | 0.72019 |
| Endo-1,4-beta-xylanase | 0.00511 | 0.00085 | 0.00332 | 0.00006 | 0.00092 | 65 | 0.00636 | 0.00015 | 0.00128 | 93 | 0.00158 | -1.92464 | 153.037 | 0.05613 | 0.70632 |
| Galactonate dehydratase | 0.00510 | 0.00089 | 0.00317 | 0.00006 | 0.00094 | 65 | 0.00645 | 0.00017 | 0.00135 | 93 | 0.00165 | -1.98764 | 151.747 | 0.04865 | 0.69964 |
| Homocysteine desulfhydrase | 0.00509 | 0.00099 | 0.00333 | 0.00011 | 0.00130 | 65 | 0.00632 | 0.00018 | 0.00140 | 93 | 0.00191 | -1.56458 | 154.182 | 0.11973 | 0.70632 |
| UDP-N-acetylglucosamine 6-dehydrogenase | 0.00509 | 0.00094 | 0.00415 | 0.00009 | 0.00120 | 65 | 0.00574 | 0.00017 | 0.00136 | 93 | 0.00181 | -0.88180 | 155.510 | 0.37925 | 0.73587 |
| Arabinan endo-1,5-alpha-L-arabinosidase | 0.00506 | 0.00111 | 0.00325 | 0.00003 | 0.00073 | 65 | 0.00633 | 0.00030 | 0.00180 | 93 | 0.00195 | -1.58423 | 120.165 | 0.11577 | 0.70632 |
| Formylmethanofuran dehydrogenase | 0.00504 | 0.00145 | 0.00878 | 0.00066 | 0.00318 | 65 | 0.00242 | 0.00009 | 0.00101 | 93 | 0.00333 | 1.90754 | 76.915 | 0.06018 | 0.70632 |
| Dehydrogluconokinase | 0.00501 | 0.00108 | 0.00497 | 0.00014 | 0.00145 | 65 | 0.00503 | 0.00022 | 0.00154 | 93 | 0.00211 | -0.02599 | 153.861 | 0.97930 | 0.99457 |
| Allantoicase | 0.00497 | 0.00091 | 0.00309 | 0.00006 | 0.00094 | 65 | 0.00628 | 0.00018 | 0.00139 | 93 | 0.00168 | -1.89796 | 150.193 | 0.05962 | 0.70632 |
| Phosphogluconate 2-dehydrogenase | 0.00496 | 0.00108 | 0.00486 | 0.00013 | 0.00143 | 65 | 0.00502 | 0.00022 | 0.00154 | 93 | 0.00210 | -0.07569 | 154.314 | 0.93976 | 0.99049 |
| Deoxycytidine kinase | 0.00491 | 0.00109 | 0.00296 | 0.00009 | 0.00119 | 65 | 0.00628 | 0.00025 | 0.00164 | 93 | 0.00203 | -1.63470 | 153.407 | 0.10416 | 0.70632 |
| 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase | 0.00490 | 0.00076 | 0.00467 | 0.00007 | 0.00102 | 65 | 0.00506 | 0.00011 | 0.00109 | 93 | 0.00149 | -0.26305 | 153.896 | 0.79286 | 0.93908 |
| Oxepin-CoA hydrolase | 0.00490 | 0.00076 | 0.00467 | 0.00007 | 0.00102 | 65 | 0.00506 | 0.00011 | 0.00109 | 93 | 0.00149 | -0.26305 | 153.896 | 0.79286 | 0.93908 |
| D-threo-aldose 1-dehydrogenase | 0.00488 | 0.00084 | 0.00309 | 0.00007 | 0.00104 | 65 | 0.00614 | 0.00014 | 0.00122 | 93 | 0.00161 | -1.89633 | 155.934 | 0.05977 | 0.70632 |
| Nitrous-oxide reductase | 0.00481 | 0.00074 | 0.00374 | 0.00004 | 0.00075 | 65 | 0.00556 | 0.00012 | 0.00113 | 93 | 0.00136 | -1.34146 | 149.445 | 0.18181 | 0.70632 |
| 2’,3’-cyclic-nucleotide 3’-phosphodiesterase | 0.00480 | 0.00146 | 0.00843 | 0.00066 | 0.00319 | 65 | 0.00227 | 0.00010 | 0.00103 | 93 | 0.00335 | 1.83681 | 77.558 | 0.07007 | 0.70632 |
| 1,3-beta-galactosyl-N-acetylhexosamine phosphorylase | 0.00476 | 0.00104 | 0.00557 | 0.00027 | 0.00203 | 65 | 0.00420 | 0.00011 | 0.00107 | 93 | 0.00230 | 0.59826 | 99.396 | 0.55103 | 0.84429 |
| Deoxyguanosine kinase | 0.00475 | 0.00109 | 0.00292 | 0.00009 | 0.00120 | 65 | 0.00603 | 0.00025 | 0.00164 | 93 | 0.00203 | -1.53170 | 153.424 | 0.12766 | 0.70632 |
| Sulfoacetaldehyde dehydrogenase (acylating) | 0.00472 | 0.00145 | 0.00828 | 0.00066 | 0.00319 | 65 | 0.00223 | 0.00009 | 0.00101 | 93 | 0.00334 | 1.80882 | 76.903 | 0.07439 | 0.70632 |
| Cyanase | 0.00468 | 0.00083 | 0.00414 | 0.00013 | 0.00144 | 65 | 0.00506 | 0.00009 | 0.00099 | 93 | 0.00175 | -0.52893 | 120.311 | 0.59783 | 0.86297 |
| Propionate CoA-transferase | 0.00464 | 0.00074 | 0.00396 | 0.00008 | 0.00108 | 65 | 0.00513 | 0.00009 | 0.00100 | 93 | 0.00148 | -0.79336 | 145.772 | 0.42886 | 0.76781 |
| All-trans-retinol 13,14-reductase | 0.00460 | 0.00094 | 0.00318 | 0.00003 | 0.00068 | 65 | 0.00560 | 0.00021 | 0.00151 | 93 | 0.00166 | -1.46270 | 125.496 | 0.14605 | 0.70632 |
| UDP-4-amino-4-deoxy-L-arabinose formyltransferase | 0.00455 | 0.00104 | 0.00389 | 0.00011 | 0.00130 | 65 | 0.00500 | 0.00021 | 0.00151 | 93 | 0.00200 | -0.55841 | 155.859 | 0.57737 | 0.85190 |
| UDP-glucuronic acid oxidase (UDP-4-keto-hexauronic acid decarboxylating) | 0.00455 | 0.00104 | 0.00389 | 0.00011 | 0.00130 | 65 | 0.00500 | 0.00021 | 0.00151 | 93 | 0.00200 | -0.55841 | 155.859 | 0.57737 | 0.85190 |
| Indolepyruvate decarboxylase | 0.00451 | 0.00089 | 0.00344 | 0.00008 | 0.00114 | 65 | 0.00526 | 0.00015 | 0.00128 | 93 | 0.00171 | -1.06387 | 155.333 | 0.28904 | 0.70632 |
| Cytosol alanyl aminopeptidase | 0.00451 | 0.00116 | 0.00450 | 0.00020 | 0.00174 | 65 | 0.00452 | 0.00022 | 0.00155 | 93 | 0.00233 | -0.01013 | 143.299 | 0.99193 | 0.99855 |
| Maleate isomerase | 0.00451 | 0.00091 | 0.00488 | 0.00017 | 0.00163 | 65 | 0.00425 | 0.00010 | 0.00105 | 93 | 0.00194 | 0.32089 | 114.218 | 0.74888 | 0.92904 |
| 1,3-propanediol dehydrogenase | 0.00450 | 0.00123 | 0.00540 | 0.00037 | 0.00237 | 65 | 0.00388 | 0.00015 | 0.00129 | 93 | 0.00270 | 0.56410 | 101.048 | 0.57394 | 0.85190 |
| dTDP-4-amino-4,6-dideoxygalactose transaminase | 0.00445 | 0.00084 | 0.00357 | 0.00007 | 0.00102 | 65 | 0.00506 | 0.00014 | 0.00123 | 93 | 0.00160 | -0.93412 | 156.000 | 0.35169 | 0.72019 |
| 3-dehydroshikimate dehydratase | 0.00445 | 0.00092 | 0.00371 | 0.00008 | 0.00110 | 65 | 0.00496 | 0.00018 | 0.00137 | 93 | 0.00176 | -0.71270 | 155.720 | 0.47710 | 0.79541 |
| Aspartate 4-decarboxylase | 0.00445 | 0.00098 | 0.00525 | 0.00027 | 0.00204 | 65 | 0.00388 | 0.00007 | 0.00087 | 93 | 0.00222 | 0.61809 | 87.275 | 0.53813 | 0.83586 |
| UDP-glucuronate decarboxylase | 0.00443 | 0.00066 | 0.00184 | 0.00001 | 0.00029 | 65 | 0.00623 | 0.00011 | 0.00107 | 93 | 0.00111 | -3.97069 | 104.829 | 0.00013 | 0.13911 |
| 23S rRNA (adenine(2085)-N(6))-dimethyltransferase | 0.00442 | 0.00072 | 0.00504 | 0.00012 | 0.00136 | 65 | 0.00398 | 0.00005 | 0.00077 | 93 | 0.00156 | 0.67770 | 103.742 | 0.49947 | 0.80830 |
| Cholesterol oxidase | 0.00441 | 0.00169 | 0.00217 | 0.00003 | 0.00066 | 65 | 0.00597 | 0.00074 | 0.00282 | 93 | 0.00290 | -1.31129 | 101.969 | 0.19270 | 0.70632 |
| Maltose-6’-phosphate glucosidase | 0.00433 | 0.00066 | 0.00477 | 0.00006 | 0.00094 | 65 | 0.00402 | 0.00008 | 0.00091 | 93 | 0.00131 | 0.57532 | 149.466 | 0.56594 | 0.85024 |
| Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P-lyase | 0.00432 | 0.00084 | 0.00220 | 0.00004 | 0.00076 | 65 | 0.00580 | 0.00016 | 0.00131 | 93 | 0.00152 | -2.37113 | 141.232 | 0.01908 | 0.64530 |
| 3-ketosteroid 9-alpha-monooxygenase | 0.00431 | 0.00169 | 0.00184 | 0.00003 | 0.00070 | 65 | 0.00604 | 0.00074 | 0.00282 | 93 | 0.00291 | -1.44645 | 103.043 | 0.15109 | 0.70632 |
| CoA:oxalate CoA-transferase | 0.00430 | 0.00108 | 0.00446 | 0.00019 | 0.00172 | 65 | 0.00418 | 0.00018 | 0.00138 | 93 | 0.00221 | 0.12963 | 134.680 | 0.89705 | 0.97163 |
| Selenate reductase | 0.00427 | 0.00100 | 0.00471 | 0.00023 | 0.00189 | 65 | 0.00396 | 0.00011 | 0.00108 | 93 | 0.00217 | 0.34539 | 105.137 | 0.73049 | 0.92120 |
| Succinylornithine transaminase | 0.00424 | 0.00081 | 0.00342 | 0.00004 | 0.00082 | 65 | 0.00482 | 0.00015 | 0.00125 | 93 | 0.00150 | -0.93252 | 148.274 | 0.35258 | 0.72019 |
| Pseudouridine kinase | 0.00421 | 0.00071 | 0.00391 | 0.00006 | 0.00099 | 65 | 0.00442 | 0.00009 | 0.00099 | 93 | 0.00140 | -0.36456 | 150.953 | 0.71595 | 0.92120 |
| UDP-N-acetylglucosamine 4-epimerase | 0.00421 | 0.00076 | 0.00411 | 0.00007 | 0.00104 | 65 | 0.00428 | 0.00011 | 0.00107 | 93 | 0.00149 | -0.11780 | 152.594 | 0.90638 | 0.97700 |
| Mercury(II) reductase | 0.00417 | 0.00086 | 0.00437 | 0.00014 | 0.00149 | 65 | 0.00403 | 0.00010 | 0.00103 | 93 | 0.00181 | 0.18816 | 120.721 | 0.85106 | 0.96065 |
| Assimilatory sulfite reductase (ferredoxin) | 0.00417 | 0.00063 | 0.00337 | 0.00003 | 0.00071 | 65 | 0.00473 | 0.00008 | 0.00095 | 93 | 0.00118 | -1.15400 | 154.233 | 0.25029 | 0.70632 |
| Chitin disaccharide deacetylase | 0.00416 | 0.00074 | 0.00349 | 0.00007 | 0.00103 | 65 | 0.00463 | 0.00010 | 0.00103 | 93 | 0.00146 | -0.78622 | 150.940 | 0.43297 | 0.77045 |
| Succinyl-CoA–L-malate CoA-transferase | 0.00415 | 0.00091 | 0.00452 | 0.00014 | 0.00149 | 65 | 0.00389 | 0.00012 | 0.00114 | 93 | 0.00188 | 0.33284 | 130.106 | 0.73979 | 0.92595 |
| Unsaturated rhamnogalacturonyl hydrolase | 0.00415 | 0.00101 | 0.00189 | 0.00002 | 0.00049 | 65 | 0.00573 | 0.00026 | 0.00166 | 93 | 0.00173 | -2.21569 | 107.748 | 0.02882 | 0.67652 |
| Urocanate reductase | 0.00413 | 0.00190 | 0.00275 | 0.00011 | 0.00129 | 65 | 0.00509 | 0.00089 | 0.00310 | 93 | 0.00336 | -0.69726 | 121.529 | 0.48697 | 0.80150 |
| Dextranase | 0.00413 | 0.00098 | 0.00419 | 0.00006 | 0.00099 | 65 | 0.00408 | 0.00021 | 0.00151 | 93 | 0.00181 | 0.06269 | 148.886 | 0.95010 | 0.99049 |
| 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase | 0.00412 | 0.00072 | 0.00253 | 0.00003 | 0.00065 | 65 | 0.00523 | 0.00012 | 0.00113 | 93 | 0.00130 | -2.07167 | 140.765 | 0.04012 | 0.67652 |
| Nicotinamide phosphoribosyltransferase | 0.00409 | 0.00066 | 0.00401 | 0.00004 | 0.00080 | 65 | 0.00414 | 0.00009 | 0.00098 | 93 | 0.00127 | -0.10589 | 155.931 | 0.91581 | 0.98126 |
| glycyltransferase | 0.00407 | 0.00158 | 0.00355 | 0.00028 | 0.00206 | 65 | 0.00443 | 0.00048 | 0.00227 | 93 | 0.00306 | -0.28557 | 154.886 | 0.77559 | 0.93675 |
| 4-hydroxyphenylacetate 3-monooxygenase | 0.00406 | 0.00098 | 0.00238 | 0.00004 | 0.00079 | 65 | 0.00524 | 0.00023 | 0.00156 | 93 | 0.00175 | -1.63395 | 132.392 | 0.10465 | 0.70632 |
| Arogenate dehydratase | 0.00404 | 0.00086 | 0.00218 | 0.00004 | 0.00079 | 65 | 0.00534 | 0.00017 | 0.00134 | 93 | 0.00156 | -2.02706 | 142.325 | 0.04452 | 0.69348 |
| Phosphatidylinositol alpha-mannosyltransferase | 0.00401 | 0.00054 | 0.00397 | 0.00004 | 0.00075 | 65 | 0.00404 | 0.00005 | 0.00075 | 93 | 0.00107 | -0.06567 | 150.817 | 0.94773 | 0.99049 |
| L-fuculokinase | 0.00396 | 0.00094 | 0.00507 | 0.00026 | 0.00202 | 65 | 0.00319 | 0.00005 | 0.00076 | 93 | 0.00216 | 0.87480 | 82.445 | 0.38423 | 0.73901 |
| Rhamnogalacturonan endolyase | 0.00396 | 0.00100 | 0.00172 | 0.00003 | 0.00071 | 65 | 0.00552 | 0.00024 | 0.00161 | 93 | 0.00176 | -2.15883 | 124.756 | 0.03278 | 0.67652 |
| UDP-2-acetamido-2,6-beta-L-arabino-hexul-4-ose reductase | 0.00395 | 0.00077 | 0.00246 | 0.00005 | 0.00085 | 65 | 0.00499 | 0.00013 | 0.00116 | 93 | 0.00144 | -1.75178 | 153.528 | 0.08181 | 0.70632 |
| Cyanophycinase | 0.00395 | 0.00116 | 0.00307 | 0.00009 | 0.00119 | 65 | 0.00456 | 0.00030 | 0.00178 | 93 | 0.00214 | -0.69753 | 149.377 | 0.48656 | 0.80150 |
| tRNA (guanosine(18)-2’-O)-methyltransferase | 0.00395 | 0.00063 | 0.00328 | 0.00003 | 0.00070 | 65 | 0.00442 | 0.00009 | 0.00096 | 93 | 0.00119 | -0.95705 | 153.722 | 0.34004 | 0.72019 |
| 7-alpha-hydroxysteroid dehydrogenase | 0.00394 | 0.00085 | 0.00288 | 0.00006 | 0.00098 | 65 | 0.00469 | 0.00015 | 0.00128 | 93 | 0.00161 | -1.12368 | 154.854 | 0.26289 | 0.70632 |
| Aryl-sulfate sulfotransferase | 0.00394 | 0.00170 | 0.00207 | 0.00006 | 0.00100 | 65 | 0.00524 | 0.00073 | 0.00281 | 93 | 0.00298 | -1.06611 | 114.152 | 0.28863 | 0.70632 |
| Hyaluronan synthase | 0.00393 | 0.00111 | 0.00443 | 0.00024 | 0.00192 | 65 | 0.00358 | 0.00016 | 0.00133 | 93 | 0.00234 | 0.36509 | 120.798 | 0.71569 | 0.92120 |
| 5-aminolevulinate synthase | 0.00390 | 0.00074 | 0.00164 | 0.00001 | 0.00042 | 65 | 0.00548 | 0.00014 | 0.00121 | 93 | 0.00128 | -3.00745 | 113.121 | 0.00325 | 0.62334 |
| Chondroitin AC lyase | 0.00389 | 0.00123 | 0.00325 | 0.00020 | 0.00174 | 65 | 0.00433 | 0.00027 | 0.00171 | 93 | 0.00244 | -0.44311 | 149.985 | 0.65832 | 0.88943 |
| Hexokinase | 0.00385 | 0.00095 | 0.00458 | 0.00023 | 0.00187 | 65 | 0.00333 | 0.00008 | 0.00095 | 93 | 0.00209 | 0.59676 | 97.080 | 0.55206 | 0.84446 |
| Dimethylargininase | 0.00384 | 0.00107 | 0.00384 | 0.00019 | 0.00173 | 65 | 0.00384 | 0.00018 | 0.00138 | 93 | 0.00221 | -0.00253 | 133.655 | 0.99798 | 0.99921 |
| Undecaprenyl-phosphate galactose phosphotransferase | 0.00383 | 0.00097 | 0.00502 | 0.00024 | 0.00194 | 65 | 0.00300 | 0.00008 | 0.00094 | 93 | 0.00216 | 0.93369 | 94.076 | 0.35286 | 0.72019 |
| Procollagen-proline dioxygenase | 0.00383 | 0.00074 | 0.00227 | 0.00004 | 0.00078 | 65 | 0.00492 | 0.00012 | 0.00113 | 93 | 0.00137 | -1.93514 | 151.445 | 0.05484 | 0.70632 |
| Glucan 1,3-beta-glucosidase | 0.00379 | 0.00070 | 0.00315 | 0.00005 | 0.00092 | 65 | 0.00424 | 0.00009 | 0.00099 | 93 | 0.00135 | -0.80487 | 154.360 | 0.42213 | 0.76494 |
| Amylosucrase | 0.00376 | 0.00061 | 0.00318 | 0.00003 | 0.00072 | 65 | 0.00417 | 0.00008 | 0.00092 | 93 | 0.00116 | -0.84771 | 155.364 | 0.39790 | 0.74887 |
| Hydrogen:quinone oxidoreductase | 0.00372 | 0.00144 | 0.00296 | 0.00008 | 0.00112 | 65 | 0.00425 | 0.00050 | 0.00232 | 93 | 0.00257 | -0.50257 | 129.568 | 0.61612 | 0.87356 |
| Choline monooxygenase | 0.00371 | 0.00086 | 0.00413 | 0.00014 | 0.00149 | 65 | 0.00342 | 0.00010 | 0.00102 | 93 | 0.00181 | 0.39354 | 120.005 | 0.69462 | 0.90981 |
| Zeaxanthin glucosyltransferase | 0.00371 | 0.00122 | 0.00340 | 0.00020 | 0.00175 | 65 | 0.00392 | 0.00027 | 0.00169 | 93 | 0.00243 | -0.21672 | 148.989 | 0.82872 | 0.95262 |
| Glutathionylspermidine amidase | 0.00369 | 0.00073 | 0.00538 | 0.00013 | 0.00144 | 65 | 0.00251 | 0.00005 | 0.00072 | 93 | 0.00161 | 1.79028 | 96.252 | 0.07655 | 0.70632 |
| Glutathionylspermidine synthase | 0.00369 | 0.00073 | 0.00538 | 0.00013 | 0.00144 | 65 | 0.00251 | 0.00005 | 0.00072 | 93 | 0.00161 | 1.79028 | 96.252 | 0.07655 | 0.70632 |
| Acyl-CoA oxidase | 0.00369 | 0.00080 | 0.00224 | 0.00005 | 0.00086 | 65 | 0.00470 | 0.00013 | 0.00120 | 93 | 0.00148 | -1.66676 | 152.565 | 0.09761 | 0.70632 |
| Methylmalonyl-CoA carboxytransferase | 0.00366 | 0.00097 | 0.00450 | 0.00023 | 0.00189 | 65 | 0.00308 | 0.00009 | 0.00100 | 93 | 0.00213 | 0.66977 | 99.285 | 0.50456 | 0.81236 |
| L-fuconate dehydratase | 0.00366 | 0.00060 | 0.00342 | 0.00005 | 0.00087 | 65 | 0.00383 | 0.00006 | 0.00083 | 93 | 0.00120 | -0.34149 | 147.533 | 0.73322 | 0.92120 |
| Xylan 1,4-beta-xylosidase | 0.00365 | 0.00105 | 0.00185 | 0.00002 | 0.00058 | 65 | 0.00491 | 0.00028 | 0.00173 | 93 | 0.00182 | -1.67993 | 111.783 | 0.09576 | 0.70632 |
| Deoxyhypusine synthase | 0.00360 | 0.00056 | 0.00315 | 0.00003 | 0.00072 | 65 | 0.00392 | 0.00006 | 0.00081 | 93 | 0.00109 | -0.70978 | 155.379 | 0.47891 | 0.79654 |
| Ribulokinase | 0.00357 | 0.00059 | 0.00283 | 0.00005 | 0.00088 | 65 | 0.00409 | 0.00006 | 0.00080 | 93 | 0.00119 | -1.06249 | 144.587 | 0.28979 | 0.70632 |
| Sulfoquinovose isomerase | 0.00357 | 0.00057 | 0.00382 | 0.00005 | 0.00091 | 65 | 0.00339 | 0.00005 | 0.00074 | 93 | 0.00118 | 0.36213 | 135.683 | 0.71782 | 0.92120 |
| 3-hydroxybutyryl-CoA dehydratase | 0.00354 | 0.00055 | 0.00268 | 0.00004 | 0.00074 | 65 | 0.00415 | 0.00006 | 0.00077 | 93 | 0.00107 | -1.37902 | 153.218 | 0.16990 | 0.70632 |
| CDP-glycerol glycerophosphotransferase | 0.00352 | 0.00067 | 0.00252 | 0.00005 | 0.00088 | 65 | 0.00422 | 0.00009 | 0.00096 | 93 | 0.00131 | -1.30053 | 154.652 | 0.19535 | 0.70632 |
| Alpha-N-acetylglucosaminidase | 0.00346 | 0.00090 | 0.00181 | 0.00002 | 0.00058 | 65 | 0.00462 | 0.00020 | 0.00146 | 93 | 0.00157 | -1.79164 | 118.689 | 0.07574 | 0.70632 |
| Glycolaldehyde dehydrogenase | 0.00344 | 0.00066 | 0.00362 | 0.00008 | 0.00110 | 65 | 0.00331 | 0.00006 | 0.00083 | 93 | 0.00138 | 0.22370 | 127.930 | 0.82334 | 0.95060 |
| Lactaldehyde dehydrogenase | 0.00344 | 0.00066 | 0.00362 | 0.00008 | 0.00110 | 65 | 0.00331 | 0.00006 | 0.00083 | 93 | 0.00138 | 0.22370 | 127.930 | 0.82334 | 0.95060 |
| Protocatechuate 4,5-dioxygenase | 0.00343 | 0.00158 | 0.00082 | 0.00001 | 0.00038 | 65 | 0.00525 | 0.00066 | 0.00266 | 93 | 0.00269 | -1.64642 | 95.712 | 0.10296 | 0.70632 |
| 2,6-beta-fructan 6-levanbiohydrolase | 0.00342 | 0.00097 | 0.00366 | 0.00007 | 0.00102 | 65 | 0.00326 | 0.00021 | 0.00150 | 93 | 0.00182 | 0.22421 | 150.771 | 0.82290 | 0.95060 |
| Thiazole tautomerase | 0.00342 | 0.00086 | 0.00190 | 0.00007 | 0.00102 | 65 | 0.00449 | 0.00015 | 0.00128 | 93 | 0.00163 | -1.58811 | 155.683 | 0.11429 | 0.70632 |
| Feruloyl esterase | 0.00339 | 0.00119 | 0.00161 | 0.00001 | 0.00040 | 65 | 0.00464 | 0.00037 | 0.00200 | 93 | 0.00203 | -1.49095 | 99.202 | 0.13915 | 0.70632 |
| Glutamate–putrescine ligase | 0.00338 | 0.00064 | 0.00317 | 0.00004 | 0.00079 | 65 | 0.00353 | 0.00008 | 0.00094 | 93 | 0.00122 | -0.29367 | 155.989 | 0.76940 | 0.93519 |
| Flavin reductase (NADH) | 0.00337 | 0.00067 | 0.00430 | 0.00009 | 0.00119 | 65 | 0.00272 | 0.00005 | 0.00077 | 93 | 0.00142 | 1.11265 | 114.299 | 0.26820 | 0.70632 |
| N-formylglutamate deformylase | 0.00336 | 0.00101 | 0.00211 | 0.00003 | 0.00066 | 65 | 0.00424 | 0.00025 | 0.00165 | 93 | 0.00178 | -1.19729 | 119.239 | 0.23357 | 0.70632 |
| RNA-directed DNA polymerase | 0.00335 | 0.00091 | 0.00400 | 0.00023 | 0.00188 | 65 | 0.00290 | 0.00006 | 0.00081 | 93 | 0.00205 | 0.53954 | 87.849 | 0.59088 | 0.85791 |
| 5-oxoprolinase (ATP-hydrolyzing) | 0.00335 | 0.00099 | 0.00120 | 0.00001 | 0.00047 | 65 | 0.00486 | 0.00025 | 0.00164 | 93 | 0.00171 | -2.14278 | 106.846 | 0.03440 | 0.67652 |
| Anthraniloyl-CoA monooxygenase | 0.00333 | 0.00074 | 0.00211 | 0.00004 | 0.00081 | 65 | 0.00418 | 0.00012 | 0.00112 | 93 | 0.00138 | -1.49425 | 153.151 | 0.13717 | 0.70632 |
| Lipopolysaccharide glucosyltransferase I | 0.00332 | 0.00140 | 0.00208 | 0.00004 | 0.00074 | 65 | 0.00419 | 0.00050 | 0.00232 | 93 | 0.00243 | -0.86951 | 110.083 | 0.38646 | 0.73961 |
| Alpha-glucuronidase | 0.00332 | 0.00061 | 0.00216 | 0.00002 | 0.00057 | 65 | 0.00413 | 0.00008 | 0.00094 | 93 | 0.00110 | -1.79297 | 144.346 | 0.07507 | 0.70632 |
| Acyl-homoserine-lactone synthase | 0.00331 | 0.00068 | 0.00316 | 0.00006 | 0.00100 | 65 | 0.00342 | 0.00008 | 0.00093 | 93 | 0.00137 | -0.19291 | 146.508 | 0.84730 | 0.95939 |
| Sedoheptulose-bisphosphatase | 0.00325 | 0.00062 | 0.00129 | 0.00001 | 0.00035 | 65 | 0.00462 | 0.00009 | 0.00100 | 93 | 0.00106 | -3.15110 | 113.699 | 0.00208 | 0.62334 |
| 4-hydroxybutyrate dehydrogenase | 0.00325 | 0.00066 | 0.00317 | 0.00007 | 0.00105 | 65 | 0.00330 | 0.00007 | 0.00084 | 93 | 0.00135 | -0.09790 | 134.199 | 0.92215 | 0.98606 |
| Glutamyl endopeptidase | 0.00318 | 0.00131 | 0.00090 | 0.00002 | 0.00058 | 65 | 0.00477 | 0.00044 | 0.00218 | 93 | 0.00226 | -1.71704 | 104.653 | 0.08893 | 0.70632 |
| Cyclohexyl-isocyanide hydratase | 0.00317 | 0.00071 | 0.00151 | 0.00003 | 0.00070 | 65 | 0.00433 | 0.00011 | 0.00110 | 93 | 0.00130 | -2.16878 | 146.701 | 0.03171 | 0.67652 |
| Phosphatidyl-N-methylethanolamine N-methyltransferase | 0.00317 | 0.00086 | 0.00126 | 0.00001 | 0.00039 | 65 | 0.00450 | 0.00019 | 0.00142 | 93 | 0.00147 | -2.20976 | 105.864 | 0.02928 | 0.67652 |
| Phosphatidylethanolamine N-methyltransferase | 0.00317 | 0.00086 | 0.00126 | 0.00001 | 0.00039 | 65 | 0.00450 | 0.00019 | 0.00142 | 93 | 0.00147 | -2.20976 | 105.864 | 0.02928 | 0.67652 |
| Glucosamine kinase | 0.00316 | 0.00080 | 0.00121 | 0.00001 | 0.00045 | 65 | 0.00452 | 0.00016 | 0.00130 | 93 | 0.00138 | -2.40411 | 113.040 | 0.01784 | 0.64530 |
| Ribitol 2-dehydrogenase | 0.00313 | 0.00079 | 0.00187 | 0.00005 | 0.00087 | 65 | 0.00402 | 0.00013 | 0.00120 | 93 | 0.00148 | -1.44819 | 153.163 | 0.14961 | 0.70632 |
| Pectin lyase | 0.00313 | 0.00124 | 0.00177 | 0.00011 | 0.00130 | 65 | 0.00408 | 0.00034 | 0.00191 | 93 | 0.00231 | -1.00346 | 150.326 | 0.31725 | 0.70632 |
| 3-oxosteroid 1-dehydrogenase | 0.00313 | 0.00116 | 0.00166 | 0.00003 | 0.00068 | 65 | 0.00416 | 0.00034 | 0.00191 | 93 | 0.00203 | -1.22928 | 114.357 | 0.22149 | 0.70632 |
| 4-O-beta-D-mannosyl-D-glucose phosphorylase | 0.00311 | 0.00060 | 0.00161 | 0.00001 | 0.00046 | 65 | 0.00416 | 0.00008 | 0.00095 | 93 | 0.00106 | -2.41723 | 130.507 | 0.01702 | 0.64530 |
| Thimet oligopeptidase | 0.00310 | 0.00062 | 0.00248 | 0.00003 | 0.00068 | 65 | 0.00354 | 0.00008 | 0.00093 | 93 | 0.00115 | -0.91666 | 153.695 | 0.36076 | 0.72316 |
| D-galactose 1-dehydrogenase | 0.00306 | 0.00059 | 0.00150 | 0.00002 | 0.00051 | 65 | 0.00415 | 0.00008 | 0.00092 | 93 | 0.00105 | -2.51972 | 138.773 | 0.01288 | 0.64530 |
| L-lysine N(6)-monooxygenase (NADPH) | 0.00303 | 0.00060 | 0.00249 | 0.00003 | 0.00067 | 65 | 0.00341 | 0.00008 | 0.00090 | 93 | 0.00112 | -0.82666 | 154.189 | 0.40971 | 0.76070 |
| 4-hydroxybutanoyl-CoA dehydratase | 0.00301 | 0.00061 | 0.00213 | 0.00003 | 0.00069 | 65 | 0.00362 | 0.00008 | 0.00091 | 93 | 0.00114 | -1.30479 | 154.549 | 0.19390 | 0.70632 |
| Vinylacetyl-CoA Delta-isomerase | 0.00301 | 0.00061 | 0.00213 | 0.00003 | 0.00069 | 65 | 0.00362 | 0.00008 | 0.00091 | 93 | 0.00114 | -1.30479 | 154.549 | 0.19390 | 0.70632 |
| Lipid II:glycine glycyltransferase | 0.00301 | 0.00090 | 0.00275 | 0.00011 | 0.00129 | 65 | 0.00318 | 0.00014 | 0.00125 | 93 | 0.00179 | -0.23981 | 149.105 | 0.81081 | 0.94519 |
| UDP-4-amino-4-deoxy-L-arabinose aminotransferase | 0.00299 | 0.00070 | 0.00311 | 0.00007 | 0.00105 | 65 | 0.00290 | 0.00008 | 0.00093 | 93 | 0.00140 | 0.14535 | 142.847 | 0.88464 | 0.96726 |
| ADP-dependent NAD(P)H-hydrate dehydratase | 0.00297 | 0.00070 | 0.00362 | 0.00008 | 0.00111 | 65 | 0.00251 | 0.00007 | 0.00089 | 93 | 0.00143 | 0.77904 | 134.376 | 0.43732 | 0.77235 |
| Opine dehydrogenase | 0.00295 | 0.00085 | 0.00242 | 0.00007 | 0.00105 | 65 | 0.00333 | 0.00015 | 0.00125 | 93 | 0.00164 | -0.55769 | 155.996 | 0.57785 | 0.85190 |
| Pectate lyase | 0.00291 | 0.00087 | 0.00170 | 0.00003 | 0.00069 | 65 | 0.00376 | 0.00018 | 0.00140 | 93 | 0.00156 | -1.32396 | 130.918 | 0.18782 | 0.70632 |
| 3,4-dihydroxyphenylacetate 2,3-dioxygenase | 0.00290 | 0.00077 | 0.00177 | 0.00004 | 0.00076 | 65 | 0.00370 | 0.00013 | 0.00119 | 93 | 0.00141 | -1.37444 | 147.195 | 0.17140 | 0.70632 |
| Choline-phosphate cytidylyltransferase | 0.00290 | 0.00059 | 0.00245 | 0.00003 | 0.00069 | 65 | 0.00321 | 0.00007 | 0.00087 | 93 | 0.00111 | -0.68640 | 155.528 | 0.49348 | 0.80568 |
| Arogenate dehydrogenase | 0.00289 | 0.00058 | 0.00108 | 0.00001 | 0.00030 | 65 | 0.00416 | 0.00008 | 0.00095 | 93 | 0.00099 | -3.09015 | 109.316 | 0.00254 | 0.62334 |
| Streptomycin 3’’-adenylyltransferase | 0.00289 | 0.00059 | 0.00237 | 0.00003 | 0.00065 | 65 | 0.00325 | 0.00007 | 0.00089 | 93 | 0.00111 | -0.79483 | 153.754 | 0.42794 | 0.76731 |
| 6-phospho-3-hexuloisomerase | 0.00288 | 0.00055 | 0.00127 | 0.00001 | 0.00036 | 65 | 0.00401 | 0.00007 | 0.00089 | 93 | 0.00096 | -2.85402 | 119.211 | 0.00509 | 0.62334 |
| Primary-amine oxidase | 0.00286 | 0.00057 | 0.00246 | 0.00005 | 0.00092 | 65 | 0.00314 | 0.00005 | 0.00073 | 93 | 0.00117 | -0.57937 | 133.471 | 0.56332 | 0.85024 |
| Trans-feruloyl-CoA hydratase | 0.00284 | 0.00072 | 0.00213 | 0.00005 | 0.00084 | 65 | 0.00334 | 0.00011 | 0.00107 | 93 | 0.00136 | -0.88749 | 155.354 | 0.37619 | 0.73229 |
| Vanillin synthase | 0.00284 | 0.00072 | 0.00213 | 0.00005 | 0.00084 | 65 | 0.00334 | 0.00011 | 0.00107 | 93 | 0.00136 | -0.88749 | 155.354 | 0.37619 | 0.73229 |
| Glucosylceramidase | 0.00282 | 0.00044 | 0.00275 | 0.00002 | 0.00057 | 65 | 0.00287 | 0.00004 | 0.00063 | 93 | 0.00085 | -0.14298 | 154.637 | 0.88649 | 0.96849 |
| 3-dehydro-L-gulonate 2-dehydrogenase | 0.00280 | 0.00083 | 0.00365 | 0.00022 | 0.00183 | 65 | 0.00220 | 0.00003 | 0.00060 | 93 | 0.00192 | 0.75164 | 78.138 | 0.45452 | 0.78566 |
| Trehalose O-mycolyltransferase | 0.00277 | 0.00106 | 0.00074 | 0.00001 | 0.00031 | 65 | 0.00419 | 0.00029 | 0.00177 | 93 | 0.00180 | -1.91163 | 97.614 | 0.05886 | 0.70632 |
| Alpha-L-rhamnosidase | 0.00275 | 0.00053 | 0.00174 | 0.00001 | 0.00045 | 65 | 0.00346 | 0.00007 | 0.00084 | 93 | 0.00096 | -1.79612 | 135.939 | 0.07470 | 0.70632 |
| Anthranilate–CoA ligase | 0.00275 | 0.00073 | 0.00143 | 0.00002 | 0.00057 | 65 | 0.00367 | 0.00013 | 0.00117 | 93 | 0.00130 | -1.71551 | 130.513 | 0.08862 | 0.70632 |
| 4-hydroxy-4-methyl-2-oxoglutarate aldolase | 0.00262 | 0.00059 | 0.00146 | 0.00002 | 0.00052 | 65 | 0.00344 | 0.00008 | 0.00092 | 93 | 0.00106 | -1.87425 | 138.884 | 0.06300 | 0.70632 |
| dTDP-6-deoxy-L-talose 4-dehydrogenase (NAD(+)) | 0.00262 | 0.00037 | 0.00320 | 0.00002 | 0.00058 | 65 | 0.00221 | 0.00002 | 0.00047 | 93 | 0.00075 | 1.32070 | 134.950 | 0.18884 | 0.70632 |
| Methylaspartate ammonia-lyase | 0.00259 | 0.00060 | 0.00261 | 0.00004 | 0.00077 | 65 | 0.00258 | 0.00007 | 0.00086 | 93 | 0.00115 | 0.02992 | 155.437 | 0.97617 | 0.99435 |
| Dimethylglycine dehydrogenase | 0.00259 | 0.00080 | 0.00077 | 0.00003 | 0.00064 | 65 | 0.00386 | 0.00015 | 0.00127 | 93 | 0.00142 | -2.17028 | 132.595 | 0.03177 | 0.67652 |
| Adenylyl-sulfate reductase | 0.00259 | 0.00064 | 0.00261 | 0.00008 | 0.00111 | 65 | 0.00257 | 0.00005 | 0.00077 | 93 | 0.00135 | 0.02921 | 120.828 | 0.97675 | 0.99435 |
| Proline racemase | 0.00258 | 0.00070 | 0.00222 | 0.00004 | 0.00080 | 65 | 0.00283 | 0.00010 | 0.00106 | 93 | 0.00133 | -0.46325 | 154.685 | 0.64384 | 0.88749 |
| 4-oxalmesaconate hydratase | 0.00256 | 0.00058 | 0.00101 | 0.00001 | 0.00035 | 65 | 0.00365 | 0.00008 | 0.00093 | 93 | 0.00100 | -2.63703 | 116.641 | 0.00950 | 0.64530 |
| Enoyl-[acyl-carrier-protein] reductase | 0.00256 | 0.00081 | 0.00056 | 0.00000 | 0.00024 | 65 | 0.00396 | 0.00017 | 0.00135 | 93 | 0.00137 | -2.48770 | 97.985 | 0.01455 | 0.64530 |
| D-proline reductase (dithiol) | 0.00255 | 0.00042 | 0.00234 | 0.00002 | 0.00060 | 65 | 0.00270 | 0.00003 | 0.00059 | 93 | 0.00084 | -0.43020 | 149.814 | 0.66767 | 0.89333 |
| Aureolysin | 0.00254 | 0.00080 | 0.00187 | 0.00007 | 0.00103 | 65 | 0.00301 | 0.00012 | 0.00115 | 93 | 0.00154 | -0.73799 | 155.367 | 0.46163 | 0.78637 |
| 2-keto-myo-inositol isomerase | 0.00254 | 0.00069 | 0.00179 | 0.00004 | 0.00078 | 65 | 0.00306 | 0.00010 | 0.00103 | 93 | 0.00129 | -0.97727 | 154.624 | 0.32996 | 0.71250 |
| UDP-N-acetylglucosamine 4,6-dehydratase (inverting) | 0.00247 | 0.00073 | 0.00162 | 0.00001 | 0.00043 | 65 | 0.00306 | 0.00013 | 0.00120 | 93 | 0.00128 | -1.13377 | 114.921 | 0.25925 | 0.70632 |
| Phosphoenolpyruvate mutase | 0.00246 | 0.00062 | 0.00223 | 0.00005 | 0.00088 | 65 | 0.00262 | 0.00007 | 0.00086 | 93 | 0.00123 | -0.31400 | 149.867 | 0.75396 | 0.92944 |
| Microbial collagenase | 0.00245 | 0.00087 | 0.00174 | 0.00006 | 0.00097 | 65 | 0.00296 | 0.00016 | 0.00131 | 93 | 0.00163 | -0.74604 | 154.115 | 0.45678 | 0.78637 |
| Alpha,alpha-trehalase | 0.00243 | 0.00052 | 0.00193 | 0.00002 | 0.00057 | 65 | 0.00278 | 0.00006 | 0.00079 | 93 | 0.00098 | -0.87196 | 153.217 | 0.38459 | 0.73901 |
| Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | 0.00242 | 0.00051 | 0.00102 | 0.00001 | 0.00030 | 65 | 0.00340 | 0.00006 | 0.00082 | 93 | 0.00088 | -2.71514 | 115.731 | 0.00764 | 0.62334 |
| 4-methylaminobutanoate oxidase (formaldehyde-forming) | 0.00239 | 0.00069 | 0.00087 | 0.00003 | 0.00064 | 65 | 0.00346 | 0.00011 | 0.00107 | 93 | 0.00125 | -2.06585 | 143.420 | 0.04064 | 0.67652 |
| CoB–CoM heterodisulfide reductase | 0.00239 | 0.00056 | 0.00238 | 0.00004 | 0.00081 | 65 | 0.00240 | 0.00006 | 0.00077 | 93 | 0.00112 | -0.01373 | 147.697 | 0.98906 | 0.99660 |
| 5’-phosphate synthase | 0.00238 | 0.00054 | 0.00211 | 0.00005 | 0.00091 | 65 | 0.00257 | 0.00004 | 0.00068 | 93 | 0.00113 | -0.40543 | 127.856 | 0.68584 | 0.90745 |
| Arginine–pyruvate transaminase | 0.00238 | 0.00051 | 0.00235 | 0.00004 | 0.00082 | 65 | 0.00241 | 0.00004 | 0.00066 | 93 | 0.00106 | -0.05961 | 135.036 | 0.95255 | 0.99049 |
| 3-hexulose-6-phosphate synthase | 0.00235 | 0.00054 | 0.00075 | 0.00001 | 0.00029 | 65 | 0.00348 | 0.00007 | 0.00087 | 93 | 0.00092 | -2.97162 | 111.372 | 0.00363 | 0.62334 |
| Ferredoxin hydrogenase | 0.00233 | 0.00117 | 0.00261 | 0.00039 | 0.00244 | 65 | 0.00213 | 0.00010 | 0.00103 | 93 | 0.00265 | 0.18230 | 86.783 | 0.85577 | 0.96186 |
| RNA 3’-terminal-phosphate cyclase (ATP) | 0.00231 | 0.00048 | 0.00225 | 0.00003 | 0.00062 | 65 | 0.00234 | 0.00004 | 0.00069 | 93 | 0.00093 | -0.09163 | 155.238 | 0.92711 | 0.98985 |
| Alanine–glyoxylate transaminase | 0.00230 | 0.00064 | 0.00103 | 0.00001 | 0.00031 | 65 | 0.00319 | 0.00010 | 0.00105 | 93 | 0.00110 | -1.97382 | 107.369 | 0.05097 | 0.70632 |
| Serine–glyoxylate transaminase | 0.00230 | 0.00064 | 0.00103 | 0.00001 | 0.00031 | 65 | 0.00319 | 0.00010 | 0.00105 | 93 | 0.00110 | -1.96839 | 107.366 | 0.05160 | 0.70632 |
| Serine–pyruvate transaminase | 0.00230 | 0.00064 | 0.00103 | 0.00001 | 0.00031 | 65 | 0.00319 | 0.00010 | 0.00105 | 93 | 0.00110 | -1.96839 | 107.366 | 0.05160 | 0.70632 |
| L-proline amide hydrolase | 0.00225 | 0.00069 | 0.00157 | 0.00004 | 0.00078 | 65 | 0.00273 | 0.00010 | 0.00103 | 93 | 0.00129 | -0.89921 | 154.481 | 0.36994 | 0.73093 |
| Gentisate 1,2-dioxygenase | 0.00225 | 0.00064 | 0.00092 | 0.00000 | 0.00025 | 65 | 0.00318 | 0.00011 | 0.00107 | 93 | 0.00110 | -2.06099 | 101.688 | 0.04186 | 0.68639 |
| N-succinylornithine carbamoyltransferase | 0.00224 | 0.00043 | 0.00153 | 0.00001 | 0.00038 | 65 | 0.00274 | 0.00004 | 0.00068 | 93 | 0.00078 | -1.54759 | 139.396 | 0.12399 | 0.70632 |
| Trimethylamine monooxygenase | 0.00223 | 0.00065 | 0.00137 | 0.00003 | 0.00071 | 65 | 0.00283 | 0.00009 | 0.00099 | 93 | 0.00121 | -1.19841 | 152.862 | 0.23261 | 0.70632 |
| Alpha,alpha-trehalose phosphorylase | 0.00219 | 0.00057 | 0.00278 | 0.00007 | 0.00105 | 65 | 0.00178 | 0.00004 | 0.00062 | 93 | 0.00122 | 0.81442 | 107.321 | 0.41721 | 0.76408 |
| Aspartate dehydrogenase | 0.00219 | 0.00042 | 0.00150 | 0.00002 | 0.00051 | 65 | 0.00267 | 0.00004 | 0.00062 | 93 | 0.00080 | -1.46981 | 155.944 | 0.14363 | 0.70632 |
| 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase | 0.00219 | 0.00076 | 0.00076 | 0.00002 | 0.00060 | 65 | 0.00319 | 0.00014 | 0.00121 | 93 | 0.00135 | -1.79240 | 131.368 | 0.07537 | 0.70632 |
| Proteasome endopeptidase complex | 0.00219 | 0.00071 | 0.00169 | 0.00002 | 0.00061 | 65 | 0.00254 | 0.00012 | 0.00113 | 93 | 0.00128 | -0.66128 | 137.041 | 0.50954 | 0.81548 |
| Aminocarboxymuconate-semialdehyde decarboxylase | 0.00218 | 0.00056 | 0.00092 | 0.00001 | 0.00033 | 65 | 0.00305 | 0.00008 | 0.00091 | 93 | 0.00097 | -2.19478 | 114.898 | 0.03019 | 0.67652 |
| sn-glycerol-1-phosphate dehydrogenase | 0.00217 | 0.00036 | 0.00194 | 0.00001 | 0.00043 | 65 | 0.00233 | 0.00003 | 0.00053 | 93 | 0.00068 | -0.57108 | 155.968 | 0.56877 | 0.85024 |
| CMP-N,N’-diacetyllegionaminic acid synthase | 0.00216 | 0.00069 | 0.00161 | 0.00003 | 0.00070 | 65 | 0.00255 | 0.00011 | 0.00107 | 93 | 0.00128 | -0.73615 | 148.770 | 0.46280 | 0.78699 |
| FAD-dependent urate hydroxylase | 0.00216 | 0.00052 | 0.00142 | 0.00001 | 0.00045 | 65 | 0.00268 | 0.00006 | 0.00083 | 93 | 0.00095 | -1.32996 | 137.631 | 0.18573 | 0.70632 |
| Proton-exporting ATPase | 0.00215 | 0.00090 | 0.00213 | 0.00016 | 0.00158 | 65 | 0.00217 | 0.00010 | 0.00106 | 93 | 0.00190 | -0.02278 | 117.949 | 0.98187 | 0.99457 |
| Stearoyl-[acyl-carrier-protein] 9-desaturase | 0.00215 | 0.00054 | 0.00060 | 0.00000 | 0.00023 | 65 | 0.00323 | 0.00007 | 0.00089 | 93 | 0.00092 | -2.85515 | 103.480 | 0.00520 | 0.62334 |
| 3-methylfumaryl-CoA hydratase | 0.00212 | 0.00052 | 0.00101 | 0.00001 | 0.00030 | 65 | 0.00289 | 0.00007 | 0.00085 | 93 | 0.00091 | -2.07418 | 113.863 | 0.04032 | 0.67652 |
| Cinnamoyl-CoA:phenyllactate CoA-transferase | 0.00212 | 0.00081 | 0.00329 | 0.00021 | 0.00182 | 65 | 0.00130 | 0.00002 | 0.00051 | 93 | 0.00189 | 1.05148 | 74.078 | 0.29646 | 0.70632 |
| Kojibiose phosphorylase | 0.00212 | 0.00081 | 0.00328 | 0.00022 | 0.00184 | 65 | 0.00130 | 0.00002 | 0.00051 | 93 | 0.00190 | 1.03621 | 73.878 | 0.30349 | 0.70632 |
| 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione monooxygenase | 0.00208 | 0.00070 | 0.00108 | 0.00001 | 0.00037 | 65 | 0.00277 | 0.00013 | 0.00117 | 93 | 0.00122 | -1.38231 | 109.462 | 0.16969 | 0.70632 |
| Tungstate-importing ATPase | 0.00207 | 0.00054 | 0.00203 | 0.00003 | 0.00062 | 65 | 0.00209 | 0.00006 | 0.00081 | 93 | 0.00102 | -0.06073 | 155.128 | 0.95165 | 0.99049 |
| Phosphoribulokinase | 0.00204 | 0.00042 | 0.00180 | 0.00002 | 0.00058 | 65 | 0.00222 | 0.00003 | 0.00060 | 93 | 0.00083 | -0.50249 | 152.085 | 0.61605 | 0.87356 |
| Unspecific monooxygenase | 0.00204 | 0.00056 | 0.00082 | 0.00001 | 0.00029 | 65 | 0.00289 | 0.00008 | 0.00092 | 93 | 0.00096 | -2.14831 | 109.716 | 0.03389 | 0.67652 |
| Dimethylsulfoxide reductase | 0.00203 | 0.00070 | 0.00108 | 0.00001 | 0.00032 | 65 | 0.00270 | 0.00013 | 0.00116 | 93 | 0.00121 | -1.33978 | 105.178 | 0.18321 | 0.70632 |
| Malonyl-CoA decarboxylase | 0.00203 | 0.00057 | 0.00089 | 0.00000 | 0.00026 | 65 | 0.00283 | 0.00008 | 0.00094 | 93 | 0.00098 | -1.98675 | 105.611 | 0.04954 | 0.70331 |
| Cholest-4-en-3-one 26-monooxygenase | 0.00203 | 0.00099 | 0.00043 | 0.00000 | 0.00023 | 65 | 0.00315 | 0.00026 | 0.00167 | 93 | 0.00169 | -1.61749 | 95.422 | 0.10907 | 0.70632 |
| Betaine reductase | 0.00202 | 0.00045 | 0.00218 | 0.00003 | 0.00066 | 65 | 0.00191 | 0.00004 | 0.00062 | 93 | 0.00090 | 0.29332 | 147.073 | 0.76969 | 0.93519 |
| Ectoine synthase | 0.00202 | 0.00044 | 0.00133 | 0.00001 | 0.00042 | 65 | 0.00250 | 0.00004 | 0.00068 | 93 | 0.00080 | -1.46095 | 144.764 | 0.14620 | 0.70632 |
| Acetoacetate decarboxylase | 0.00199 | 0.00064 | 0.00080 | 0.00000 | 0.00022 | 65 | 0.00282 | 0.00011 | 0.00107 | 93 | 0.00109 | -1.84694 | 99.763 | 0.06772 | 0.70632 |
| 6-hydroxynicotinate 3-monooxygenase | 0.00198 | 0.00065 | 0.00097 | 0.00003 | 0.00065 | 65 | 0.00269 | 0.00009 | 0.00100 | 93 | 0.00119 | -1.45194 | 147.895 | 0.14864 | 0.70632 |
| CoA-disulfide reductase | 0.00197 | 0.00079 | 0.00175 | 0.00007 | 0.00103 | 65 | 0.00213 | 0.00012 | 0.00113 | 93 | 0.00153 | -0.25178 | 154.885 | 0.80155 | 0.94263 |
| Aliphatic aldoxime dehydratase | 0.00197 | 0.00070 | 0.00090 | 0.00003 | 0.00065 | 65 | 0.00272 | 0.00011 | 0.00109 | 93 | 0.00127 | -1.43174 | 143.190 | 0.15440 | 0.70632 |
| Benzoylformate decarboxylase | 0.00197 | 0.00064 | 0.00103 | 0.00001 | 0.00032 | 65 | 0.00262 | 0.00010 | 0.00106 | 93 | 0.00111 | -1.43744 | 108.569 | 0.15347 | 0.70632 |
| N-acetylneuraminate epimerase | 0.00196 | 0.00054 | 0.00161 | 0.00003 | 0.00066 | 65 | 0.00222 | 0.00006 | 0.00079 | 93 | 0.00103 | -0.59446 | 155.999 | 0.55307 | 0.84479 |
| Limonene-1,2-epoxide hydrolase | 0.00196 | 0.00128 | 0.00042 | 0.00001 | 0.00028 | 65 | 0.00304 | 0.00044 | 0.00217 | 93 | 0.00218 | -1.19991 | 95.069 | 0.23316 | 0.70632 |
| UDP-N-acetyl-2-amino-2-deoxyglucuronate dehydrogenase | 0.00196 | 0.00071 | 0.00149 | 0.00002 | 0.00053 | 65 | 0.00229 | 0.00012 | 0.00115 | 93 | 0.00127 | -0.62254 | 126.829 | 0.53470 | 0.83299 |
| Steroid Delta-isomerase | 0.00195 | 0.00098 | 0.00060 | 0.00000 | 0.00026 | 65 | 0.00289 | 0.00026 | 0.00166 | 93 | 0.00168 | -1.36312 | 96.638 | 0.17601 | 0.70632 |
| Dye decolorizing peroxidase | 0.00193 | 0.00047 | 0.00119 | 0.00001 | 0.00043 | 65 | 0.00245 | 0.00005 | 0.00073 | 93 | 0.00085 | -1.48456 | 142.706 | 0.13987 | 0.70632 |
| Lysine 6-dehydrogenase | 0.00192 | 0.00073 | 0.00071 | 0.00002 | 0.00060 | 65 | 0.00278 | 0.00013 | 0.00117 | 93 | 0.00131 | -1.57636 | 133.530 | 0.11731 | 0.70632 |
| Alkylglycerone-phosphate synthase | 0.00192 | 0.00054 | 0.00098 | 0.00001 | 0.00037 | 65 | 0.00257 | 0.00007 | 0.00088 | 93 | 0.00096 | -1.66394 | 122.067 | 0.09869 | 0.70632 |
| 3-oxo-5-alpha-steroid 4-dehydrogenase (acceptor) | 0.00190 | 0.00098 | 0.00091 | 0.00001 | 0.00036 | 65 | 0.00259 | 0.00025 | 0.00165 | 93 | 0.00169 | -0.99714 | 100.765 | 0.32109 | 0.70632 |
| Succinate-semialdehyde dehydrogenase (acetylating) | 0.00190 | 0.00054 | 0.00160 | 0.00003 | 0.00067 | 65 | 0.00210 | 0.00006 | 0.00080 | 93 | 0.00104 | -0.48142 | 155.995 | 0.63089 | 0.88317 |
| 1,4-dihydroxy-2-naphthoyl-CoA hydrolase | 0.00187 | 0.00038 | 0.00170 | 0.00002 | 0.00053 | 65 | 0.00199 | 0.00003 | 0.00054 | 93 | 0.00076 | -0.38378 | 152.545 | 0.70167 | 0.91306 |
| Peroxyureidoacrylate/ureidoacrylate amidohydrolase | 0.00187 | 0.00039 | 0.00175 | 0.00003 | 0.00068 | 65 | 0.00195 | 0.00002 | 0.00047 | 93 | 0.00083 | -0.24700 | 121.664 | 0.80532 | 0.94351 |
| Mycoredoxin | 0.00187 | 0.00044 | 0.00141 | 0.00002 | 0.00050 | 65 | 0.00219 | 0.00004 | 0.00067 | 93 | 0.00083 | -0.93276 | 154.558 | 0.35240 | 0.72019 |
| ADP-ribosyl-[dinitrogen reductase] hydrolase | 0.00186 | 0.00044 | 0.00225 | 0.00003 | 0.00070 | 65 | 0.00159 | 0.00003 | 0.00057 | 93 | 0.00090 | 0.73672 | 134.684 | 0.46257 | 0.78699 |
| CDP-ribitol ribitolphosphotransferase | 0.00185 | 0.00065 | 0.00086 | 0.00001 | 0.00038 | 65 | 0.00255 | 0.00011 | 0.00108 | 93 | 0.00114 | -1.48271 | 113.718 | 0.14092 | 0.70632 |
| Diaminobutyrate acetyltransferase | 0.00183 | 0.00042 | 0.00115 | 0.00001 | 0.00039 | 65 | 0.00230 | 0.00004 | 0.00066 | 93 | 0.00077 | -1.50017 | 142.420 | 0.13578 | 0.70632 |
| Aminodeoxyfutalosine nucleosidase | 0.00182 | 0.00072 | 0.00139 | 0.00002 | 0.00056 | 65 | 0.00213 | 0.00012 | 0.00116 | 93 | 0.00129 | -0.57498 | 129.395 | 0.56631 | 0.85024 |
| Spermidine dehydrogenase | 0.00182 | 0.00065 | 0.00111 | 0.00003 | 0.00070 | 65 | 0.00232 | 0.00009 | 0.00100 | 93 | 0.00122 | -0.98998 | 151.962 | 0.32376 | 0.70874 |
| Maleamate amidohydrolase | 0.00181 | 0.00063 | 0.00107 | 0.00003 | 0.00068 | 65 | 0.00234 | 0.00009 | 0.00096 | 93 | 0.00118 | -1.07683 | 152.402 | 0.28326 | 0.70632 |
| Chondroitin-sulfate-ABC endolyase | 0.00181 | 0.00043 | 0.00118 | 0.00001 | 0.00030 | 65 | 0.00225 | 0.00005 | 0.00070 | 93 | 0.00077 | -1.39943 | 122.470 | 0.16421 | 0.70632 |
| Chondroitin-sulfate-ABC exolyase | 0.00181 | 0.00043 | 0.00118 | 0.00001 | 0.00030 | 65 | 0.00225 | 0.00005 | 0.00070 | 93 | 0.00077 | -1.39943 | 122.470 | 0.16421 | 0.70632 |
| Xaa-Xaa-Pro tripeptidyl-peptidase | 0.00181 | 0.00054 | 0.00160 | 0.00003 | 0.00067 | 65 | 0.00196 | 0.00006 | 0.00080 | 93 | 0.00104 | -0.34330 | 155.987 | 0.73184 | 0.92120 |
| Tetraprenyl-beta-curcumene synthase | 0.00181 | 0.00049 | 0.00034 | 0.00000 | 0.00015 | 65 | 0.00283 | 0.00006 | 0.00081 | 93 | 0.00082 | -3.02902 | 98.528 | 0.00313 | 0.62334 |
| UDP-N,N’-diacetylbacillosamine 2-epimerase (hydrolyzing) | 0.00180 | 0.00067 | 0.00129 | 0.00003 | 0.00065 | 65 | 0.00215 | 0.00010 | 0.00105 | 93 | 0.00124 | -0.69520 | 145.150 | 0.48804 | 0.80164 |
| Pseudaminic acid synthase | 0.00178 | 0.00071 | 0.00098 | 0.00001 | 0.00040 | 65 | 0.00234 | 0.00013 | 0.00117 | 93 | 0.00124 | -1.10798 | 112.198 | 0.27024 | 0.70632 |
| Oxalate decarboxylase | 0.00177 | 0.00070 | 0.00107 | 0.00002 | 0.00058 | 65 | 0.00226 | 0.00012 | 0.00112 | 93 | 0.00126 | -0.94149 | 134.390 | 0.34814 | 0.72019 |
| Aromatic-L-amino-acid decarboxylase | 0.00175 | 0.00046 | 0.00282 | 0.00006 | 0.00099 | 65 | 0.00101 | 0.00001 | 0.00036 | 93 | 0.00105 | 1.72384 | 81.503 | 0.08853 | 0.70632 |
| GDP-perosamine synthase | 0.00174 | 0.00071 | 0.00088 | 0.00002 | 0.00056 | 65 | 0.00234 | 0.00012 | 0.00113 | 93 | 0.00127 | -1.15456 | 131.620 | 0.25036 | 0.70632 |
| Putrescine oxidase | 0.00174 | 0.00048 | 0.00107 | 0.00001 | 0.00042 | 65 | 0.00220 | 0.00005 | 0.00076 | 93 | 0.00087 | -1.30176 | 137.776 | 0.19517 | 0.70632 |
| Isonocardicin synthase | 0.00172 | 0.00061 | 0.00155 | 0.00005 | 0.00087 | 65 | 0.00184 | 0.00007 | 0.00084 | 93 | 0.00121 | -0.23549 | 149.284 | 0.81415 | 0.94567 |
| HycI peptidase | 0.00172 | 0.00038 | 0.00203 | 0.00003 | 0.00071 | 65 | 0.00150 | 0.00002 | 0.00041 | 93 | 0.00082 | 0.65058 | 104.851 | 0.51674 | 0.81889 |
| Coenzyme F420-0:L-glutamate ligase | 0.00171 | 0.00042 | 0.00135 | 0.00002 | 0.00050 | 65 | 0.00197 | 0.00004 | 0.00062 | 93 | 0.00080 | -0.77340 | 155.823 | 0.44045 | 0.77520 |
| Coenzyme F420-1:gamma-L-glutamate ligase | 0.00171 | 0.00042 | 0.00135 | 0.00002 | 0.00050 | 65 | 0.00197 | 0.00004 | 0.00062 | 93 | 0.00080 | -0.77340 | 155.823 | 0.44045 | 0.77520 |
| 2,5-dihydroxypyridine 5,6-dioxygenase | 0.00169 | 0.00062 | 0.00085 | 0.00003 | 0.00064 | 65 | 0.00227 | 0.00008 | 0.00095 | 93 | 0.00115 | -1.24056 | 150.212 | 0.21670 | 0.70632 |
| Ureidoglycolate dehydrogenase (NAD(+)) | 0.00168 | 0.00069 | 0.00079 | 0.00002 | 0.00056 | 65 | 0.00229 | 0.00011 | 0.00110 | 93 | 0.00123 | -1.21651 | 132.568 | 0.22595 | 0.70632 |
| Oligogalacturonide lyase | 0.00167 | 0.00058 | 0.00084 | 0.00001 | 0.00039 | 65 | 0.00226 | 0.00008 | 0.00094 | 93 | 0.00102 | -1.38997 | 120.699 | 0.16709 | 0.70632 |
| N-formylmaleamate deformylase | 0.00165 | 0.00062 | 0.00088 | 0.00003 | 0.00065 | 65 | 0.00220 | 0.00008 | 0.00095 | 93 | 0.00115 | -1.13952 | 150.528 | 0.25630 | 0.70632 |
| L-xylulokinase | 0.00165 | 0.00030 | 0.00139 | 0.00001 | 0.00043 | 65 | 0.00184 | 0.00002 | 0.00041 | 93 | 0.00060 | -0.76115 | 147.428 | 0.44778 | 0.77823 |
| Glucosyl-3-phosphoglycerate synthase | 0.00164 | 0.00044 | 0.00140 | 0.00002 | 0.00059 | 65 | 0.00181 | 0.00004 | 0.00062 | 93 | 0.00086 | -0.47505 | 153.072 | 0.63543 | 0.88317 |
| Mannosylglycerate hydrolase | 0.00163 | 0.00049 | 0.00168 | 0.00005 | 0.00084 | 65 | 0.00160 | 0.00003 | 0.00060 | 93 | 0.00103 | 0.08259 | 124.385 | 0.93431 | 0.98985 |
| 5-dehydro-2-deoxyphosphogluconate aldolase | 0.00162 | 0.00061 | 0.00169 | 0.00010 | 0.00126 | 65 | 0.00157 | 0.00003 | 0.00057 | 93 | 0.00138 | 0.08361 | 90.467 | 0.93355 | 0.98985 |
| UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine N-acetyltransferase | 0.00162 | 0.00070 | 0.00096 | 0.00001 | 0.00040 | 65 | 0.00208 | 0.00012 | 0.00116 | 93 | 0.00122 | -0.91286 | 112.592 | 0.36327 | 0.72448 |
| UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine transaminase | 0.00162 | 0.00070 | 0.00096 | 0.00001 | 0.00040 | 65 | 0.00208 | 0.00012 | 0.00116 | 93 | 0.00122 | -0.91286 | 112.592 | 0.36327 | 0.72448 |
| Homospermidine synthase | 0.00161 | 0.00046 | 0.00037 | 0.00000 | 0.00011 | 65 | 0.00249 | 0.00006 | 0.00077 | 93 | 0.00078 | -2.71274 | 95.692 | 0.00791 | 0.62334 |
| Glycogen(starch) synthase | 0.00161 | 0.00038 | 0.00096 | 0.00000 | 0.00020 | 65 | 0.00207 | 0.00004 | 0.00063 | 93 | 0.00066 | -1.67723 | 109.439 | 0.09635 | 0.70632 |
| Ribonuclease T(1) | 0.00161 | 0.00052 | 0.00096 | 0.00001 | 0.00033 | 65 | 0.00207 | 0.00007 | 0.00085 | 93 | 0.00091 | -1.21341 | 117.440 | 0.22741 | 0.70632 |
| L-ribulose-5-phosphate 3-epimerase | 0.00160 | 0.00032 | 0.00201 | 0.00003 | 0.00064 | 65 | 0.00132 | 0.00001 | 0.00031 | 93 | 0.00071 | 0.96914 | 94.064 | 0.33496 | 0.71823 |
| o-pyrocatechuate decarboxylase | 0.00159 | 0.00062 | 0.00078 | 0.00003 | 0.00064 | 65 | 0.00216 | 0.00008 | 0.00095 | 93 | 0.00115 | -1.19702 | 150.043 | 0.23319 | 0.70632 |
| (R)-citramalate synthase | 0.00159 | 0.00041 | 0.00107 | 0.00001 | 0.00047 | 65 | 0.00196 | 0.00004 | 0.00062 | 93 | 0.00078 | -1.14134 | 154.283 | 0.25550 | 0.70632 |
| Cutinase | 0.00157 | 0.00061 | 0.00017 | 0.00000 | 0.00013 | 65 | 0.00254 | 0.00010 | 0.00102 | 93 | 0.00103 | -2.30881 | 94.803 | 0.02313 | 0.66972 |
| 2-phospho-L-lactate guanylyltransferase | 0.00155 | 0.00039 | 0.00097 | 0.00001 | 0.00033 | 65 | 0.00196 | 0.00004 | 0.00062 | 93 | 0.00070 | -1.41411 | 134.956 | 0.15963 | 0.70632 |
| 3-carboxyethylcatechol 2,3-dioxygenase | 0.00155 | 0.00045 | 0.00093 | 0.00001 | 0.00036 | 65 | 0.00199 | 0.00005 | 0.00072 | 93 | 0.00080 | -1.31639 | 132.080 | 0.19032 | 0.70632 |
| Sarcosine/dimethylglycine N-methyltransferase | 0.00155 | 0.00062 | 0.00075 | 0.00003 | 0.00064 | 65 | 0.00211 | 0.00008 | 0.00095 | 93 | 0.00115 | -1.17974 | 149.995 | 0.23997 | 0.70632 |
| Cyclic-guanylate-specific phosphodiesterase | 0.00153 | 0.00051 | 0.00107 | 0.00001 | 0.00032 | 65 | 0.00186 | 0.00007 | 0.00084 | 93 | 0.00090 | -0.88281 | 116.197 | 0.37916 | 0.73587 |
| Glycine/sarcosine N-methyltransferase | 0.00151 | 0.00062 | 0.00078 | 0.00003 | 0.00064 | 65 | 0.00203 | 0.00008 | 0.00095 | 93 | 0.00115 | -1.08659 | 150.083 | 0.27896 | 0.70632 |
| Aldehyde oxygenase (deformylating) | 0.00150 | 0.00062 | 0.00075 | 0.00003 | 0.00064 | 65 | 0.00203 | 0.00008 | 0.00095 | 93 | 0.00115 | -1.11161 | 150.073 | 0.26808 | 0.70632 |
| Exo-poly-alpha-galacturonosidase | 0.00150 | 0.00062 | 0.00075 | 0.00003 | 0.00064 | 65 | 0.00203 | 0.00008 | 0.00095 | 93 | 0.00115 | -1.11161 | 150.073 | 0.26808 | 0.70632 |
| 2-hydroxyhexa-2,4-dienoate hydratase | 0.00149 | 0.00035 | 0.00211 | 0.00003 | 0.00065 | 65 | 0.00107 | 0.00001 | 0.00038 | 93 | 0.00075 | 1.38042 | 106.643 | 0.17034 | 0.70632 |
| Protein-histidine pros-kinase | 0.00149 | 0.00053 | 0.00070 | 0.00001 | 0.00036 | 65 | 0.00204 | 0.00007 | 0.00087 | 93 | 0.00094 | -1.43149 | 121.430 | 0.15486 | 0.70632 |
| Decaprenylphospho-beta-D-erythro-pentofuranosid-2-ulose 2-reductase | 0.00149 | 0.00039 | 0.00124 | 0.00001 | 0.00045 | 65 | 0.00166 | 0.00003 | 0.00059 | 93 | 0.00074 | -0.56686 | 154.421 | 0.57164 | 0.85122 |
| Decaprenyl-phosphate phosphoribosyltransferase | 0.00146 | 0.00039 | 0.00117 | 0.00001 | 0.00044 | 65 | 0.00166 | 0.00003 | 0.00059 | 93 | 0.00074 | -0.65633 | 154.279 | 0.51259 | 0.81548 |
| Decaprenylphospho-beta-D-ribofuranose 2-dehydrogenase | 0.00146 | 0.00039 | 0.00117 | 0.00001 | 0.00044 | 65 | 0.00166 | 0.00003 | 0.00059 | 93 | 0.00074 | -0.65633 | 154.279 | 0.51259 | 0.81548 |
| Factor independent urate hydroxylase | 0.00146 | 0.00044 | 0.00075 | 0.00001 | 0.00033 | 65 | 0.00196 | 0.00005 | 0.00072 | 93 | 0.00079 | -1.52895 | 126.664 | 0.12877 | 0.70632 |
| galactofuranosyltransferase | 0.00145 | 0.00040 | 0.00120 | 0.00001 | 0.00048 | 65 | 0.00163 | 0.00003 | 0.00060 | 93 | 0.00076 | -0.55774 | 155.788 | 0.57782 | 0.85190 |
| 4-hydroxy-2-oxohexanoate aldolase | 0.00143 | 0.00036 | 0.00141 | 0.00002 | 0.00050 | 65 | 0.00145 | 0.00002 | 0.00051 | 93 | 0.00071 | -0.05405 | 151.671 | 0.95697 | 0.99147 |
| IgA-specific metalloendopeptidase | 0.00143 | 0.00048 | 0.00140 | 0.00003 | 0.00069 | 65 | 0.00145 | 0.00004 | 0.00066 | 93 | 0.00096 | -0.04452 | 149.004 | 0.96455 | 0.99193 |
| Inosine kinase | 0.00143 | 0.00037 | 0.00176 | 0.00002 | 0.00062 | 65 | 0.00119 | 0.00002 | 0.00045 | 93 | 0.00077 | 0.74167 | 126.011 | 0.45967 | 0.78637 |
| Bacterial leucyl aminopeptidase | 0.00142 | 0.00042 | 0.00077 | 0.00001 | 0.00033 | 65 | 0.00188 | 0.00004 | 0.00067 | 93 | 0.00075 | -1.47569 | 130.362 | 0.14244 | 0.70632 |
| Crotonobetainyl-CoA hydratase | 0.00142 | 0.00031 | 0.00039 | 0.00000 | 0.00010 | 65 | 0.00214 | 0.00002 | 0.00050 | 93 | 0.00051 | -3.40450 | 99.348 | 0.00096 | 0.62334 |
| Pyrimidine monooxygenase | 0.00142 | 0.00034 | 0.00143 | 0.00002 | 0.00055 | 65 | 0.00141 | 0.00002 | 0.00044 | 93 | 0.00071 | 0.02728 | 132.970 | 0.97828 | 0.99457 |
| Aquacobalamin reductase | 0.00142 | 0.00037 | 0.00174 | 0.00002 | 0.00062 | 65 | 0.00119 | 0.00002 | 0.00045 | 93 | 0.00077 | 0.71402 | 126.037 | 0.47653 | 0.79541 |
| Riboflavin reductase (NAD(P)H) | 0.00142 | 0.00037 | 0.00174 | 0.00002 | 0.00062 | 65 | 0.00119 | 0.00002 | 0.00045 | 93 | 0.00077 | 0.71402 | 126.037 | 0.47653 | 0.79541 |
| Limonene 1,2-monooxygenase | 0.00142 | 0.00040 | 0.00081 | 0.00001 | 0.00031 | 65 | 0.00184 | 0.00004 | 0.00065 | 93 | 0.00072 | -1.44085 | 129.446 | 0.15204 | 0.70632 |
| Creatinase | 0.00141 | 0.00039 | 0.00190 | 0.00003 | 0.00069 | 65 | 0.00107 | 0.00002 | 0.00045 | 93 | 0.00083 | 0.99994 | 115.581 | 0.31943 | 0.70632 |
| 2-phospho-L-lactate transferase | 0.00141 | 0.00039 | 0.00089 | 0.00001 | 0.00032 | 65 | 0.00178 | 0.00004 | 0.00062 | 93 | 0.00070 | -1.26882 | 134.955 | 0.20669 | 0.70632 |
| 2-oxoglutarate decarboxylase | 0.00141 | 0.00039 | 0.00116 | 0.00001 | 0.00044 | 65 | 0.00159 | 0.00003 | 0.00059 | 93 | 0.00074 | -0.58143 | 154.520 | 0.56180 | 0.85024 |
| Aryl-alcohol dehydrogenase (NADP(+)) | 0.00141 | 0.00040 | 0.00061 | 0.00000 | 0.00015 | 65 | 0.00196 | 0.00004 | 0.00067 | 93 | 0.00069 | -1.96159 | 101.393 | 0.05255 | 0.70632 |
| Triphosphatase | 0.00138 | 0.00036 | 0.00165 | 0.00002 | 0.00061 | 65 | 0.00119 | 0.00002 | 0.00045 | 93 | 0.00076 | 0.60561 | 127.836 | 0.54585 | 0.84290 |
| 2-oxo-3-hexenedioate decarboxylase | 0.00134 | 0.00041 | 0.00070 | 0.00000 | 0.00024 | 65 | 0.00178 | 0.00004 | 0.00067 | 93 | 0.00071 | -1.51198 | 113.576 | 0.13332 | 0.70632 |
| [Acyl-carrier-protein] phosphodiesterase | 0.00133 | 0.00036 | 0.00152 | 0.00002 | 0.00060 | 65 | 0.00119 | 0.00002 | 0.00045 | 93 | 0.00075 | 0.43701 | 128.062 | 0.66284 | 0.88968 |
| 4-phytase | 0.00133 | 0.00034 | 0.00124 | 0.00002 | 0.00054 | 65 | 0.00139 | 0.00002 | 0.00044 | 93 | 0.00070 | -0.20636 | 136.196 | 0.83682 | 0.95674 |
| hydrolase | 0.00131 | 0.00041 | 0.00076 | 0.00001 | 0.00031 | 65 | 0.00170 | 0.00004 | 0.00065 | 93 | 0.00072 | -1.30342 | 128.019 | 0.19477 | 0.70632 |
| Formate dehydrogenase (NADP(+)) | 0.00131 | 0.00030 | 0.00114 | 0.00001 | 0.00041 | 65 | 0.00143 | 0.00002 | 0.00042 | 93 | 0.00059 | -0.48906 | 152.403 | 0.62550 | 0.88154 |
| 3-oxo-5-alpha-steroid 4-dehydrogenase (NADP(+)) | 0.00130 | 0.00037 | 0.00060 | 0.00000 | 0.00027 | 65 | 0.00179 | 0.00003 | 0.00059 | 93 | 0.00065 | -1.82488 | 127.577 | 0.07036 | 0.70632 |
| Galactan 5-O-arabinofuranosyltransferase | 0.00130 | 0.00039 | 0.00078 | 0.00001 | 0.00038 | 65 | 0.00166 | 0.00004 | 0.00061 | 93 | 0.00072 | -1.22547 | 145.097 | 0.22238 | 0.70632 |
| Maltokinase | 0.00129 | 0.00024 | 0.00114 | 0.00001 | 0.00031 | 65 | 0.00140 | 0.00001 | 0.00036 | 93 | 0.00047 | -0.54681 | 155.885 | 0.58529 | 0.85509 |
| 3-alpha(or 20-beta)-hydroxysteroid dehydrogenase | 0.00128 | 0.00039 | 0.00026 | 0.00000 | 0.00011 | 65 | 0.00198 | 0.00004 | 0.00066 | 93 | 0.00066 | -2.58928 | 96.751 | 0.01110 | 0.64530 |
| L-idonate 5-dehydrogenase (NAD(P)(+)) | 0.00127 | 0.00029 | 0.00088 | 0.00001 | 0.00030 | 65 | 0.00155 | 0.00002 | 0.00045 | 93 | 0.00054 | -1.24131 | 149.655 | 0.21643 | 0.70632 |
| Arabinofuranan 3-O-arabinosyltransferase | 0.00127 | 0.00038 | 0.00077 | 0.00001 | 0.00038 | 65 | 0.00161 | 0.00003 | 0.00059 | 93 | 0.00070 | -1.19818 | 147.157 | 0.23277 | 0.70632 |
| 4-hydroxymandelate oxidase | 0.00126 | 0.00049 | 0.00103 | 0.00001 | 0.00041 | 65 | 0.00142 | 0.00006 | 0.00079 | 93 | 0.00089 | -0.43698 | 133.879 | 0.66283 | 0.88968 |
| 3-phenylpropanoate dioxygenase | 0.00126 | 0.00054 | 0.00066 | 0.00000 | 0.00022 | 65 | 0.00167 | 0.00008 | 0.00091 | 93 | 0.00093 | -1.08569 | 102.254 | 0.28017 | 0.70632 |
| Salicylate 5-hydroxylase | 0.00125 | 0.00079 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00206 | 0.00017 | 0.00133 | 93 | 0.00134 | -1.46329 | 92.453 | 0.14678 | 0.70632 |
| Endothelin-converting enzyme 1 | 0.00125 | 0.00033 | 0.00096 | 0.00001 | 0.00043 | 65 | 0.00145 | 0.00002 | 0.00047 | 93 | 0.00064 | -0.77559 | 154.937 | 0.43917 | 0.77432 |
| dioxygenase | 0.00125 | 0.00040 | 0.00074 | 0.00001 | 0.00031 | 65 | 0.00160 | 0.00004 | 0.00064 | 93 | 0.00071 | -1.20653 | 129.503 | 0.22981 | 0.70632 |
| 4-coumaroyl-homoserine lactone synthase | 0.00124 | 0.00035 | 0.00087 | 0.00003 | 0.00067 | 65 | 0.00149 | 0.00001 | 0.00038 | 93 | 0.00077 | -0.80401 | 104.030 | 0.42322 | 0.76494 |
| Isovaleryl-homoserine lactone synthase | 0.00124 | 0.00035 | 0.00087 | 0.00003 | 0.00067 | 65 | 0.00149 | 0.00001 | 0.00038 | 93 | 0.00077 | -0.80401 | 104.030 | 0.42322 | 0.76494 |
| Formate dehydrogenase-N | 0.00124 | 0.00039 | 0.00126 | 0.00002 | 0.00050 | 65 | 0.00122 | 0.00003 | 0.00056 | 93 | 0.00075 | 0.05312 | 155.555 | 0.95770 | 0.99147 |
| Glycerol dehydratase | 0.00124 | 0.00102 | 0.00263 | 0.00040 | 0.00247 | 65 | 0.00026 | 0.00000 | 0.00020 | 93 | 0.00247 | 0.95669 | 64.812 | 0.34228 | 0.72019 |
| Benzoyl-CoA-dihydrodiol lyase | 0.00123 | 0.00044 | 0.00034 | 0.00000 | 0.00012 | 65 | 0.00186 | 0.00005 | 0.00074 | 93 | 0.00075 | -2.02242 | 96.987 | 0.04589 | 0.69348 |
| Hydroxybutyrate-dimer hydrolase | 0.00123 | 0.00055 | 0.00032 | 0.00000 | 0.00014 | 65 | 0.00187 | 0.00008 | 0.00093 | 93 | 0.00094 | -1.64242 | 96.337 | 0.10376 | 0.70632 |
| Aspartate–tRNA(Asn) ligase | 0.00122 | 0.00043 | 0.00095 | 0.00003 | 0.00070 | 65 | 0.00141 | 0.00003 | 0.00054 | 93 | 0.00088 | -0.51860 | 129.483 | 0.60493 | 0.86817 |
| Benzoate–CoA ligase | 0.00122 | 0.00044 | 0.00031 | 0.00000 | 0.00011 | 65 | 0.00186 | 0.00005 | 0.00074 | 93 | 0.00075 | -2.07641 | 96.141 | 0.04052 | 0.67652 |
| L-lysine 6-transaminase | 0.00119 | 0.00038 | 0.00069 | 0.00000 | 0.00024 | 65 | 0.00154 | 0.00004 | 0.00062 | 93 | 0.00067 | -1.27327 | 117.779 | 0.20543 | 0.70632 |
| 2-hydroxymuconate-6-semialdehyde dehydrogenase | 0.00117 | 0.00038 | 0.00090 | 0.00001 | 0.00033 | 65 | 0.00136 | 0.00003 | 0.00060 | 93 | 0.00068 | -0.68583 | 137.534 | 0.49398 | 0.80568 |
| Aminomuconate-semialdehyde dehydrogenase | 0.00117 | 0.00038 | 0.00090 | 0.00001 | 0.00033 | 65 | 0.00136 | 0.00003 | 0.00060 | 93 | 0.00068 | -0.68583 | 137.534 | 0.49398 | 0.80568 |
| 2’-hydroxybiphenyl-2-sulfinate desulfinase | 0.00117 | 0.00054 | 0.00057 | 0.00000 | 0.00019 | 65 | 0.00158 | 0.00008 | 0.00090 | 93 | 0.00092 | -1.09315 | 100.010 | 0.27695 | 0.70632 |
| L-histidine N(alpha)-methyltransferase | 0.00116 | 0.00038 | 0.00064 | 0.00001 | 0.00028 | 65 | 0.00153 | 0.00003 | 0.00061 | 93 | 0.00067 | -1.32914 | 126.611 | 0.18619 | 0.70632 |
| Vitamin B12-transporting ATPase | 0.00116 | 0.00033 | 0.00130 | 0.00002 | 0.00051 | 65 | 0.00106 | 0.00002 | 0.00044 | 93 | 0.00067 | 0.35501 | 139.795 | 0.72312 | 0.92120 |
| Sulfofructose kinase | 0.00116 | 0.00043 | 0.00035 | 0.00000 | 0.00012 | 65 | 0.00173 | 0.00005 | 0.00072 | 93 | 0.00073 | -1.89647 | 97.134 | 0.06087 | 0.70632 |
| 2,3-diketo-5-methylthiopentyl-1-phosphate enolase | 0.00115 | 0.00042 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00187 | 0.00005 | 0.00070 | 93 | 0.00070 | -2.50212 | 93.704 | 0.01408 | 0.64530 |
| Lipopolysaccharide N-acetylmannosaminouronosyltransferase | 0.00115 | 0.00033 | 0.00118 | 0.00002 | 0.00051 | 65 | 0.00113 | 0.00002 | 0.00045 | 93 | 0.00068 | 0.07287 | 141.835 | 0.94202 | 0.99049 |
| Protein-glutamine gamma-glutamyltransferase | 0.00115 | 0.00042 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00187 | 0.00005 | 0.00070 | 93 | 0.00070 | -2.50477 | 93.706 | 0.01398 | 0.64530 |
| L-arabinose 1-dehydrogenase (NAD(P)(+)) | 0.00115 | 0.00040 | 0.00021 | 0.00000 | 0.00007 | 65 | 0.00180 | 0.00004 | 0.00067 | 93 | 0.00067 | -2.35800 | 93.740 | 0.02045 | 0.64530 |
| UDP-2-acetamido-3-amino-2,3-dideoxy-glucuronate N-acetyltransferase | 0.00114 | 0.00025 | 0.00108 | 0.00001 | 0.00039 | 65 | 0.00118 | 0.00001 | 0.00032 | 93 | 0.00051 | -0.19377 | 137.392 | 0.84664 | 0.95939 |
| Taurine–2-oxoglutarate transaminase | 0.00113 | 0.00025 | 0.00105 | 0.00001 | 0.00044 | 65 | 0.00119 | 0.00001 | 0.00029 | 93 | 0.00053 | -0.27431 | 116.085 | 0.78434 | 0.93675 |
| N-acyl-D-glutamate deacylase | 0.00113 | 0.00042 | 0.00091 | 0.00002 | 0.00050 | 65 | 0.00128 | 0.00004 | 0.00063 | 93 | 0.00080 | -0.46242 | 155.543 | 0.64443 | 0.88749 |
| Squalene synthase | 0.00113 | 0.00064 | 0.00067 | 0.00002 | 0.00060 | 65 | 0.00144 | 0.00009 | 0.00100 | 93 | 0.00116 | -0.66083 | 143.720 | 0.50978 | 0.81548 |
| Thiamine kinase | 0.00112 | 0.00033 | 0.00119 | 0.00002 | 0.00051 | 65 | 0.00106 | 0.00002 | 0.00044 | 93 | 0.00067 | 0.18626 | 140.365 | 0.85251 | 0.96065 |
| 4-aminobutyrate–pyruvate transaminase | 0.00112 | 0.00037 | 0.00042 | 0.00000 | 0.00018 | 65 | 0.00160 | 0.00003 | 0.00061 | 93 | 0.00063 | -1.86087 | 107.607 | 0.06549 | 0.70632 |
| Methylamine dehydrogenase (amicyanin) | 0.00111 | 0.00040 | 0.00127 | 0.00002 | 0.00051 | 65 | 0.00100 | 0.00003 | 0.00058 | 93 | 0.00077 | 0.35436 | 155.347 | 0.72355 | 0.92120 |
| Arsenate-mycothiol transferase | 0.00111 | 0.00038 | 0.00081 | 0.00001 | 0.00041 | 65 | 0.00132 | 0.00003 | 0.00058 | 93 | 0.00071 | -0.72868 | 151.883 | 0.46732 | 0.79096 |
| dTDP-4-amino-4,6-dideoxy-D-galactose acyltransferase | 0.00111 | 0.00033 | 0.00118 | 0.00002 | 0.00051 | 65 | 0.00106 | 0.00002 | 0.00044 | 93 | 0.00067 | 0.17131 | 140.326 | 0.86423 | 0.96186 |
| Kdo(2)-lipid IV(A) palmitoleoyltransferase | 0.00111 | 0.00033 | 0.00118 | 0.00002 | 0.00051 | 65 | 0.00106 | 0.00002 | 0.00044 | 93 | 0.00067 | 0.17131 | 140.326 | 0.86423 | 0.96186 |
| TDP-N-acetylfucosamine:lipid II N-acetylfucosaminyltransferase | 0.00111 | 0.00033 | 0.00118 | 0.00002 | 0.00051 | 65 | 0.00106 | 0.00002 | 0.00044 | 93 | 0.00067 | 0.17131 | 140.326 | 0.86423 | 0.96186 |
| 2-hydroxy-6-oxonona-2,4-dienedioate hydrolase | 0.00110 | 0.00037 | 0.00051 | 0.00000 | 0.00020 | 65 | 0.00151 | 0.00003 | 0.00061 | 93 | 0.00064 | -1.56470 | 110.285 | 0.12052 | 0.70632 |
| [Trimethylamine–corrinoid protein] Co-methyltransferase | 0.00109 | 0.00052 | 0.00021 | 0.00000 | 0.00018 | 65 | 0.00171 | 0.00007 | 0.00087 | 93 | 0.00089 | -1.68339 | 99.319 | 0.09544 | 0.70632 |
| Scyllo-inositol 2-dehydrogenase (NADP(+)) | 0.00109 | 0.00033 | 0.00072 | 0.00001 | 0.00036 | 65 | 0.00135 | 0.00002 | 0.00050 | 93 | 0.00062 | -1.01572 | 152.429 | 0.31137 | 0.70632 |
| Phosphatidylinositol diacylglycerol-lyase | 0.00108 | 0.00034 | 0.00035 | 0.00000 | 0.00017 | 65 | 0.00159 | 0.00003 | 0.00057 | 93 | 0.00059 | -2.09705 | 107.493 | 0.03833 | 0.67652 |
| UDP-2,3-diacetamido-2,3-dideoxyglucuronic acid 2-epimerase | 0.00108 | 0.00035 | 0.00046 | 0.00000 | 0.00015 | 65 | 0.00150 | 0.00003 | 0.00059 | 93 | 0.00061 | -1.71248 | 104.361 | 0.08978 | 0.70632 |
| Gamma-glutamyl hercynylcysteine S-oxide synthase | 0.00107 | 0.00037 | 0.00058 | 0.00000 | 0.00027 | 65 | 0.00141 | 0.00003 | 0.00060 | 93 | 0.00065 | -1.27392 | 125.839 | 0.20504 | 0.70632 |
| N-acetylhexosamine 1-kinase | 0.00105 | 0.00023 | 0.00100 | 0.00001 | 0.00032 | 65 | 0.00109 | 0.00001 | 0.00032 | 93 | 0.00045 | -0.18509 | 150.717 | 0.85341 | 0.96065 |
| Acetone carboxylase | 0.00104 | 0.00067 | 0.00037 | 0.00000 | 0.00024 | 65 | 0.00151 | 0.00012 | 0.00113 | 93 | 0.00116 | -0.98355 | 100.049 | 0.32771 | 0.71151 |
| Squalene–hopanol cyclase | 0.00104 | 0.00029 | 0.00093 | 0.00001 | 0.00040 | 65 | 0.00112 | 0.00002 | 0.00041 | 93 | 0.00057 | -0.32445 | 151.609 | 0.74604 | 0.92773 |
| Squalene–hopene cyclase | 0.00104 | 0.00029 | 0.00093 | 0.00001 | 0.00040 | 65 | 0.00112 | 0.00002 | 0.00041 | 93 | 0.00057 | -0.32445 | 151.609 | 0.74604 | 0.92773 |
| GDP-4-dehydro-6-deoxy-D-mannose reductase | 0.00104 | 0.00028 | 0.00071 | 0.00000 | 0.00027 | 65 | 0.00127 | 0.00002 | 0.00043 | 93 | 0.00051 | -1.09727 | 147.652 | 0.27431 | 0.70632 |
| 16S rRNA (cytosine(1407)-C(5))-methyltransferase | 0.00102 | 0.00028 | 0.00127 | 0.00002 | 0.00051 | 65 | 0.00085 | 0.00001 | 0.00032 | 93 | 0.00060 | 0.69055 | 112.342 | 0.49128 | 0.80508 |
| 1,4-N-acetyl-D-galactosaminyltransferase | 0.00102 | 0.00027 | 0.00130 | 0.00002 | 0.00053 | 65 | 0.00083 | 0.00001 | 0.00029 | 93 | 0.00060 | 0.79380 | 101.344 | 0.42917 | 0.76781 |
| Cyanuric acid amidohydrolase | 0.00102 | 0.00039 | 0.00054 | 0.00000 | 0.00025 | 65 | 0.00135 | 0.00004 | 0.00063 | 93 | 0.00068 | -1.19840 | 119.656 | 0.23313 | 0.70632 |
| UDP-sulfoquinovose synthase | 0.00101 | 0.00028 | 0.00048 | 0.00000 | 0.00016 | 65 | 0.00139 | 0.00002 | 0.00047 | 93 | 0.00049 | -1.84060 | 113.976 | 0.06828 | 0.70632 |
| 5-guanidino-2-oxopentanoate decarboxylase | 0.00101 | 0.00030 | 0.00150 | 0.00002 | 0.00056 | 65 | 0.00067 | 0.00001 | 0.00033 | 93 | 0.00065 | 1.28316 | 108.364 | 0.20217 | 0.70632 |
| D-sorbitol dehydrogenase (acceptor) | 0.00101 | 0.00031 | 0.00083 | 0.00001 | 0.00044 | 65 | 0.00113 | 0.00002 | 0.00043 | 93 | 0.00061 | -0.48455 | 150.455 | 0.62870 | 0.88303 |
| UDP-N-acetylgalactosamine diphosphorylase | 0.00100 | 0.00035 | 0.00031 | 0.00000 | 0.00016 | 65 | 0.00148 | 0.00003 | 0.00058 | 93 | 0.00060 | -1.95330 | 105.118 | 0.05344 | 0.70632 |
| Enamidase | 0.00099 | 0.00043 | 0.00019 | 0.00000 | 0.00007 | 65 | 0.00154 | 0.00005 | 0.00072 | 93 | 0.00073 | -1.86405 | 93.497 | 0.06545 | 0.70632 |
| D-psicose 3-epimerase | 0.00099 | 0.00024 | 0.00030 | 0.00000 | 0.00017 | 65 | 0.00147 | 0.00001 | 0.00039 | 93 | 0.00043 | -2.73839 | 122.183 | 0.00710 | 0.62334 |
| D-tagatose 3-epimerase | 0.00099 | 0.00024 | 0.00030 | 0.00000 | 0.00017 | 65 | 0.00147 | 0.00001 | 0.00039 | 93 | 0.00043 | -2.73839 | 122.183 | 0.00710 | 0.62334 |
| Undecaprenyl-diphosphooligosaccharide–protein glycotransferase | 0.00098 | 0.00027 | 0.00128 | 0.00002 | 0.00052 | 65 | 0.00078 | 0.00001 | 0.00027 | 93 | 0.00059 | 0.84858 | 98.484 | 0.39817 | 0.74887 |
| L-gulonate 5-dehydrogenase | 0.00098 | 0.00028 | 0.00108 | 0.00001 | 0.00047 | 65 | 0.00091 | 0.00001 | 0.00035 | 93 | 0.00059 | 0.28143 | 126.553 | 0.77884 | 0.93675 |
| Sulfoacetaldehyde reductase | 0.00098 | 0.00037 | 0.00097 | 0.00002 | 0.00060 | 65 | 0.00098 | 0.00002 | 0.00047 | 93 | 0.00076 | -0.00261 | 131.315 | 0.99792 | 0.99921 |
| 2-pyrone-4,6-dicarboxylate lactonase | 0.00098 | 0.00047 | 0.00017 | 0.00000 | 0.00008 | 65 | 0.00153 | 0.00006 | 0.00079 | 93 | 0.00079 | -1.71238 | 93.886 | 0.09013 | 0.70632 |
| 6-aminohexanoate-cyclic-dimer hydrolase | 0.00095 | 0.00041 | 0.00030 | 0.00000 | 0.00011 | 65 | 0.00140 | 0.00004 | 0.00068 | 93 | 0.00069 | -1.59908 | 96.943 | 0.11306 | 0.70632 |
| L-arabinonate dehydratase | 0.00095 | 0.00050 | 0.00033 | 0.00000 | 0.00021 | 65 | 0.00138 | 0.00006 | 0.00083 | 93 | 0.00086 | -1.22195 | 103.409 | 0.22450 | 0.70632 |
| 2-dehydro-3-deoxyglucarate aldolase | 0.00092 | 0.00025 | 0.00113 | 0.00001 | 0.00044 | 65 | 0.00078 | 0.00001 | 0.00030 | 93 | 0.00054 | 0.64133 | 118.573 | 0.52255 | 0.82182 |
| Maleylacetate reductase | 0.00091 | 0.00040 | 0.00032 | 0.00000 | 0.00012 | 65 | 0.00132 | 0.00004 | 0.00067 | 93 | 0.00068 | -1.45026 | 98.281 | 0.15017 | 0.70632 |
| Cellobiose phosphorylase | 0.00090 | 0.00029 | 0.00068 | 0.00001 | 0.00034 | 65 | 0.00105 | 0.00002 | 0.00043 | 93 | 0.00055 | -0.66829 | 155.508 | 0.50494 | 0.81236 |
| N,N’-diacetyllegionaminate synthase | 0.00090 | 0.00029 | 0.00071 | 0.00001 | 0.00035 | 65 | 0.00103 | 0.00002 | 0.00042 | 93 | 0.00055 | -0.58298 | 155.940 | 0.56075 | 0.85024 |
| Alpha,alpha-trehalose synthase | 0.00090 | 0.00022 | 0.00064 | 0.00000 | 0.00019 | 65 | 0.00107 | 0.00001 | 0.00034 | 93 | 0.00039 | -1.09888 | 137.837 | 0.27374 | 0.70632 |
| Erythritol kinase | 0.00089 | 0.00038 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00146 | 0.00004 | 0.00065 | 93 | 0.00065 | -2.11877 | 92.725 | 0.03678 | 0.67652 |
| Glutaryl-CoA dehydrogenase (acceptor) | 0.00089 | 0.00030 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00144 | 0.00002 | 0.00050 | 93 | 0.00050 | -2.71230 | 92.940 | 0.00796 | 0.62334 |
| 4-hydroxyacetophenone monooxygenase | 0.00089 | 0.00026 | 0.00020 | 0.00000 | 0.00007 | 65 | 0.00136 | 0.00002 | 0.00043 | 93 | 0.00043 | -2.67094 | 96.208 | 0.00888 | 0.64530 |
| 2-methylfumaryl-CoA isomerase | 0.00087 | 0.00034 | 0.00038 | 0.00000 | 0.00013 | 65 | 0.00121 | 0.00003 | 0.00057 | 93 | 0.00058 | -1.42972 | 101.951 | 0.15585 | 0.70632 |
| Sulfopropanediol 3-dehydrogenase | 0.00086 | 0.00030 | 0.00047 | 0.00001 | 0.00041 | 65 | 0.00113 | 0.00002 | 0.00042 | 93 | 0.00059 | -1.12519 | 152.138 | 0.26228 | 0.70632 |
| Gamma-butyrobetaine dioxygenase | 0.00086 | 0.00027 | 0.00086 | 0.00001 | 0.00036 | 65 | 0.00086 | 0.00001 | 0.00038 | 93 | 0.00052 | 0.00798 | 152.871 | 0.99364 | 0.99921 |
| 3-oxo-5,6-didehydrosuberyl-CoA thiolase | 0.00085 | 0.00024 | 0.00081 | 0.00001 | 0.00038 | 65 | 0.00088 | 0.00001 | 0.00032 | 93 | 0.00050 | -0.14573 | 136.470 | 0.88435 | 0.96726 |
| Aldehyde ferredoxin oxidoreductase | 0.00085 | 0.00031 | 0.00096 | 0.00002 | 0.00057 | 65 | 0.00077 | 0.00001 | 0.00034 | 93 | 0.00066 | 0.29583 | 108.878 | 0.76793 | 0.93519 |
| Non-specific protein-tyrosine kinase | 0.00085 | 0.00042 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00134 | 0.00005 | 0.00071 | 93 | 0.00071 | -1.68004 | 93.112 | 0.09630 | 0.70632 |
| Phenol 2-monooxygenase | 0.00085 | 0.00031 | 0.00084 | 0.00001 | 0.00048 | 65 | 0.00085 | 0.00002 | 0.00041 | 93 | 0.00063 | -0.01858 | 138.945 | 0.98520 | 0.99660 |
| Ribosylpyrimidine nucleosidase | 0.00084 | 0.00019 | 0.00085 | 0.00000 | 0.00022 | 65 | 0.00083 | 0.00001 | 0.00028 | 93 | 0.00035 | 0.05500 | 155.900 | 0.95621 | 0.99147 |
| 4-alpha-N-acetylgalactosaminyltransferase | 0.00083 | 0.00025 | 0.00109 | 0.00002 | 0.00050 | 65 | 0.00065 | 0.00001 | 0.00025 | 93 | 0.00056 | 0.78402 | 94.855 | 0.43498 | 0.77079 |
| Alcohol dehydrogenase (azurin) | 0.00082 | 0.00024 | 0.00035 | 0.00000 | 0.00011 | 65 | 0.00116 | 0.00001 | 0.00039 | 93 | 0.00041 | -1.98537 | 107.191 | 0.04966 | 0.70331 |
| dTDP-L-rhamnose 4-epimerase | 0.00082 | 0.00021 | 0.00054 | 0.00000 | 0.00019 | 65 | 0.00102 | 0.00001 | 0.00033 | 93 | 0.00038 | -1.26263 | 141.738 | 0.20880 | 0.70632 |
| Endoglycosylceramidase | 0.00082 | 0.00064 | 0.00020 | 0.00000 | 0.00014 | 65 | 0.00125 | 0.00011 | 0.00108 | 93 | 0.00109 | -0.96217 | 94.991 | 0.33841 | 0.72019 |
| 3-hydroxyisobutyryl-CoA hydrolase | 0.00082 | 0.00036 | 0.00044 | 0.00001 | 0.00028 | 65 | 0.00108 | 0.00003 | 0.00058 | 93 | 0.00064 | -1.00737 | 129.182 | 0.31564 | 0.70632 |
| Sulfite dehydrogenase | 0.00081 | 0.00046 | 0.00014 | 0.00000 | 0.00008 | 65 | 0.00129 | 0.00006 | 0.00078 | 93 | 0.00078 | -1.46307 | 94.062 | 0.14678 | 0.70632 |
| Sporulenol synthase | 0.00081 | 0.00031 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00127 | 0.00002 | 0.00051 | 93 | 0.00052 | -2.16921 | 95.395 | 0.03255 | 0.67652 |
| UDP-arabinose 4-epimerase | 0.00081 | 0.00035 | 0.00033 | 0.00000 | 0.00013 | 65 | 0.00114 | 0.00003 | 0.00059 | 93 | 0.00061 | -1.34793 | 101.347 | 0.18069 | 0.70632 |
| Thermolysin | 0.00080 | 0.00035 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00134 | 0.00003 | 0.00060 | 93 | 0.00060 | -2.18611 | 92.343 | 0.03134 | 0.67652 |
| 1,5-anhydro-D-fructose reductase (1,5-anhydro-D-mannitol-forming) | 0.00079 | 0.00035 | 0.00032 | 0.00000 | 0.00023 | 65 | 0.00112 | 0.00003 | 0.00058 | 93 | 0.00062 | -1.29291 | 120.317 | 0.19852 | 0.70632 |
| Glucan 1,4-alpha-glucosidase | 0.00079 | 0.00028 | 0.00077 | 0.00001 | 0.00037 | 65 | 0.00080 | 0.00001 | 0.00039 | 93 | 0.00054 | -0.04567 | 153.299 | 0.96364 | 0.99193 |
| Histidine decarboxylase | 0.00079 | 0.00043 | 0.00108 | 0.00004 | 0.00080 | 65 | 0.00059 | 0.00002 | 0.00046 | 93 | 0.00092 | 0.53301 | 105.343 | 0.59515 | 0.86170 |
| Pseudaminic acid cytidylyltransferase | 0.00078 | 0.00020 | 0.00079 | 0.00001 | 0.00031 | 65 | 0.00078 | 0.00001 | 0.00027 | 93 | 0.00041 | 0.02582 | 139.055 | 0.97944 | 0.99457 |
| Glucose-6-phosphate dehydrogenase (coenzyme-F420) | 0.00078 | 0.00034 | 0.00041 | 0.00000 | 0.00021 | 65 | 0.00105 | 0.00003 | 0.00056 | 93 | 0.00059 | -1.07803 | 115.672 | 0.28326 | 0.70632 |
| (d)CTP diphosphatase | 0.00078 | 0.00024 | 0.00088 | 0.00001 | 0.00039 | 65 | 0.00071 | 0.00001 | 0.00030 | 93 | 0.00049 | 0.34398 | 129.545 | 0.73142 | 0.92120 |
| 2-hydroxy-4-carboxymuconate semialdehyde hemiacetal dehydrogenase | 0.00078 | 0.00040 | 0.00015 | 0.00000 | 0.00008 | 65 | 0.00122 | 0.00004 | 0.00067 | 93 | 0.00068 | -1.57829 | 94.416 | 0.11784 | 0.70632 |
| transferase | 0.00078 | 0.00025 | 0.00101 | 0.00002 | 0.00050 | 65 | 0.00062 | 0.00001 | 0.00025 | 93 | 0.00056 | 0.70809 | 95.306 | 0.48062 | 0.79876 |
| 6-pyruvoyltetrahydropterin 2’-reductase | 0.00078 | 0.00025 | 0.00101 | 0.00002 | 0.00050 | 65 | 0.00061 | 0.00001 | 0.00025 | 93 | 0.00056 | 0.71452 | 95.323 | 0.47665 | 0.79541 |
| acetylgalactosaminyltransferase | 0.00078 | 0.00025 | 0.00101 | 0.00002 | 0.00050 | 65 | 0.00061 | 0.00001 | 0.00025 | 93 | 0.00056 | 0.71452 | 95.323 | 0.47665 | 0.79541 |
| UDP-N-acetylbacillosamine N-acetyltransferase | 0.00078 | 0.00025 | 0.00101 | 0.00002 | 0.00050 | 65 | 0.00061 | 0.00001 | 0.00025 | 93 | 0.00056 | 0.71452 | 95.323 | 0.47665 | 0.79541 |
| UDP-N-acetylbacillosamine transaminase | 0.00078 | 0.00025 | 0.00101 | 0.00002 | 0.00050 | 65 | 0.00061 | 0.00001 | 0.00025 | 93 | 0.00056 | 0.71452 | 95.323 | 0.47665 | 0.79541 |
| UDP-N-acetylglucosamine 4,6-dehydratase (configuration-retaining) | 0.00078 | 0.00025 | 0.00101 | 0.00002 | 0.00050 | 65 | 0.00061 | 0.00001 | 0.00025 | 93 | 0.00056 | 0.71452 | 95.323 | 0.47665 | 0.79541 |
| 4,4’-diapophytoene desaturase (4,4’-diapolycopene-forming) | 0.00077 | 0.00033 | 0.00024 | 0.00000 | 0.00015 | 65 | 0.00114 | 0.00003 | 0.00056 | 93 | 0.00058 | -1.56248 | 104.650 | 0.12119 | 0.70632 |
| Diapolycopene oxygenase | 0.00077 | 0.00033 | 0.00024 | 0.00000 | 0.00015 | 65 | 0.00114 | 0.00003 | 0.00056 | 93 | 0.00058 | -1.56248 | 104.650 | 0.12119 | 0.70632 |
| Lysostaphin | 0.00077 | 0.00033 | 0.00024 | 0.00000 | 0.00015 | 65 | 0.00114 | 0.00003 | 0.00056 | 93 | 0.00058 | -1.56248 | 104.650 | 0.12119 | 0.70632 |
| Staphopain | 0.00077 | 0.00033 | 0.00024 | 0.00000 | 0.00015 | 65 | 0.00114 | 0.00003 | 0.00056 | 93 | 0.00058 | -1.56248 | 104.650 | 0.12119 | 0.70632 |
| L-gulonolactone oxidase | 0.00077 | 0.00035 | 0.00037 | 0.00000 | 0.00015 | 65 | 0.00105 | 0.00003 | 0.00059 | 93 | 0.00061 | -1.12148 | 102.940 | 0.26469 | 0.70632 |
| S-(hydroxymethyl)glutathione synthase | 0.00077 | 0.00024 | 0.00039 | 0.00000 | 0.00017 | 65 | 0.00104 | 0.00001 | 0.00039 | 93 | 0.00042 | -1.52977 | 123.885 | 0.12862 | 0.70632 |
| UDP-2-acetamido-2-deoxy-ribo-hexuluronate aminotransferase | 0.00077 | 0.00024 | 0.00084 | 0.00001 | 0.00044 | 65 | 0.00071 | 0.00001 | 0.00027 | 93 | 0.00052 | 0.24813 | 109.267 | 0.80450 | 0.94350 |
| Fatty-acid peroxygenase | 0.00077 | 0.00025 | 0.00105 | 0.00002 | 0.00051 | 65 | 0.00057 | 0.00000 | 0.00022 | 93 | 0.00056 | 0.87318 | 88.647 | 0.38493 | 0.73901 |
| NADPH–hemoprotein reductase | 0.00076 | 0.00024 | 0.00055 | 0.00000 | 0.00026 | 65 | 0.00091 | 0.00001 | 0.00036 | 93 | 0.00044 | -0.81068 | 152.422 | 0.41881 | 0.76408 |
| Hydrogen dehydrogenase | 0.00076 | 0.00029 | 0.00064 | 0.00001 | 0.00035 | 65 | 0.00084 | 0.00002 | 0.00043 | 93 | 0.00055 | -0.35457 | 155.986 | 0.72339 | 0.92120 |
| Nicotine blue oxidoreductase | 0.00074 | 0.00018 | 0.00072 | 0.00001 | 0.00030 | 65 | 0.00075 | 0.00000 | 0.00022 | 93 | 0.00037 | -0.06784 | 125.819 | 0.94602 | 0.99049 |
| Gamma-glutamyl-gamma-aminobutyrate hydrolase | 0.00073 | 0.00023 | 0.00077 | 0.00001 | 0.00038 | 65 | 0.00070 | 0.00001 | 0.00029 | 93 | 0.00048 | 0.15493 | 130.476 | 0.87711 | 0.96548 |
| Phosphonopyruvate decarboxylase | 0.00072 | 0.00017 | 0.00057 | 0.00000 | 0.00017 | 65 | 0.00083 | 0.00001 | 0.00027 | 93 | 0.00032 | -0.82615 | 144.535 | 0.41008 | 0.76070 |
| specific) | 0.00072 | 0.00040 | 0.00025 | 0.00000 | 0.00017 | 65 | 0.00104 | 0.00004 | 0.00067 | 93 | 0.00070 | -1.14389 | 103.960 | 0.25530 | 0.70632 |
| D-ornithine 4,5-aminomutase | 0.00071 | 0.00018 | 0.00114 | 0.00001 | 0.00037 | 65 | 0.00042 | 0.00000 | 0.00016 | 93 | 0.00040 | 1.80016 | 87.052 | 0.07530 | 0.70632 |
| Formaldehyde dismutase | 0.00071 | 0.00035 | 0.00035 | 0.00000 | 0.00024 | 65 | 0.00096 | 0.00003 | 0.00057 | 93 | 0.00062 | -0.98879 | 122.667 | 0.32471 | 0.70914 |
| Co-methyltransferase | 0.00071 | 0.00035 | 0.00042 | 0.00001 | 0.00035 | 65 | 0.00091 | 0.00003 | 0.00053 | 93 | 0.00064 | -0.75813 | 148.497 | 0.44958 | 0.78030 |
| Aminopeptidase B | 0.00070 | 0.00028 | 0.00027 | 0.00000 | 0.00026 | 65 | 0.00100 | 0.00002 | 0.00043 | 93 | 0.00051 | -1.43262 | 143.735 | 0.15414 | 0.70632 |
| Malyl-CoA lyase | 0.00070 | 0.00025 | 0.00026 | 0.00000 | 0.00011 | 65 | 0.00100 | 0.00002 | 0.00042 | 93 | 0.00044 | -1.69839 | 104.740 | 0.09240 | 0.70632 |
| L-rhamnose 1-dehydrogenase | 0.00068 | 0.00019 | 0.00036 | 0.00000 | 0.00016 | 65 | 0.00091 | 0.00001 | 0.00029 | 93 | 0.00033 | -1.64306 | 137.238 | 0.10266 | 0.70632 |
| L-rhamnose 1-dehydrogenase (NAD(P)(+)) | 0.00068 | 0.00019 | 0.00036 | 0.00000 | 0.00016 | 65 | 0.00091 | 0.00001 | 0.00029 | 93 | 0.00033 | -1.64306 | 137.238 | 0.10266 | 0.70632 |
| L-rhamnose 1-dehydrogenase (NADP(+)) | 0.00068 | 0.00019 | 0.00036 | 0.00000 | 0.00016 | 65 | 0.00091 | 0.00001 | 0.00029 | 93 | 0.00033 | -1.64306 | 137.238 | 0.10266 | 0.70632 |
| Hydroxyquinol 1,2-dioxygenase | 0.00068 | 0.00023 | 0.00055 | 0.00000 | 0.00025 | 65 | 0.00077 | 0.00001 | 0.00034 | 93 | 0.00043 | -0.52653 | 154.063 | 0.59928 | 0.86318 |
| Glucose-fructose oxidoreductase | 0.00067 | 0.00018 | 0.00024 | 0.00000 | 0.00009 | 65 | 0.00097 | 0.00001 | 0.00030 | 93 | 0.00031 | -2.35296 | 108.084 | 0.02043 | 0.64530 |
| (2S)-methylsuccinyl-CoA dehydrogenase | 0.00066 | 0.00022 | 0.00030 | 0.00000 | 0.00013 | 65 | 0.00092 | 0.00001 | 0.00036 | 93 | 0.00038 | -1.62229 | 113.950 | 0.10751 | 0.70632 |
| Sterol 14-alpha-demethylase | 0.00066 | 0.00033 | 0.00013 | 0.00000 | 0.00008 | 65 | 0.00102 | 0.00003 | 0.00056 | 93 | 0.00056 | -1.59038 | 95.464 | 0.11506 | 0.70632 |
| Acyl-CoA hydrolase | 0.00064 | 0.00018 | 0.00058 | 0.00000 | 0.00026 | 65 | 0.00068 | 0.00001 | 0.00025 | 93 | 0.00036 | -0.28083 | 148.176 | 0.77924 | 0.93675 |
| Endo-alpha-N-acetylgalactosaminidase | 0.00063 | 0.00034 | 0.00099 | 0.00004 | 0.00079 | 65 | 0.00038 | 0.00000 | 0.00014 | 93 | 0.00080 | 0.76394 | 67.821 | 0.44755 | 0.77823 |
| 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate dehydrogenase | 0.00063 | 0.00027 | 0.00033 | 0.00000 | 0.00011 | 65 | 0.00084 | 0.00002 | 0.00045 | 93 | 0.00047 | -1.08569 | 102.254 | 0.28017 | 0.70632 |
| D-arabinitol 4-dehydrogenase | 0.00063 | 0.00039 | 0.00014 | 0.00000 | 0.00006 | 65 | 0.00096 | 0.00004 | 0.00066 | 93 | 0.00066 | -1.24323 | 93.504 | 0.21689 | 0.70632 |
| Aliphatic nitrilase | 0.00062 | 0.00021 | 0.00042 | 0.00000 | 0.00016 | 65 | 0.00076 | 0.00001 | 0.00034 | 93 | 0.00038 | -0.89590 | 129.300 | 0.37197 | 0.73139 |
| Phytanoyl-CoA dioxygenase | 0.00061 | 0.00034 | 0.00051 | 0.00001 | 0.00032 | 65 | 0.00069 | 0.00003 | 0.00053 | 93 | 0.00062 | -0.28532 | 143.800 | 0.77581 | 0.93675 |
| D-glycero-alpha-D-manno-heptose 1-phosphate guanylyltransferase | 0.00060 | 0.00015 | 0.00040 | 0.00000 | 0.00015 | 65 | 0.00073 | 0.00001 | 0.00024 | 93 | 0.00028 | -1.16928 | 145.446 | 0.24420 | 0.70632 |
| Valine dehydrogenase (NAD(+)) | 0.00059 | 0.00034 | 0.00040 | 0.00000 | 0.00024 | 65 | 0.00073 | 0.00003 | 0.00054 | 93 | 0.00060 | -0.54627 | 125.569 | 0.58585 | 0.85531 |
| Propionate kinase | 0.00058 | 0.00020 | 0.00049 | 0.00000 | 0.00021 | 65 | 0.00065 | 0.00001 | 0.00031 | 93 | 0.00038 | -0.41960 | 151.472 | 0.67537 | 0.89908 |
| Receptor protein-tyrosine kinase | 0.00058 | 0.00020 | 0.00025 | 0.00000 | 0.00015 | 65 | 0.00080 | 0.00001 | 0.00032 | 93 | 0.00036 | -1.53891 | 125.303 | 0.12635 | 0.70632 |
| 3-phospho-methyltransferase | 0.00058 | 0.00027 | 0.00129 | 0.00003 | 0.00063 | 65 | 0.00008 | 0.00000 | 0.00005 | 93 | 0.00064 | 1.89621 | 64.867 | 0.06239 | 0.70632 |
| Polymannosyl GlcNAc-diphospho-ditrans,octacis-undecaprenol kinase | 0.00058 | 0.00027 | 0.00129 | 0.00003 | 0.00063 | 65 | 0.00008 | 0.00000 | 0.00005 | 93 | 0.00064 | 1.89621 | 64.867 | 0.06239 | 0.70632 |
| 3(or 17)-beta-hydroxysteroid dehydrogenase | 0.00057 | 0.00022 | 0.00059 | 0.00001 | 0.00030 | 65 | 0.00056 | 0.00001 | 0.00031 | 93 | 0.00043 | 0.06734 | 153.310 | 0.94640 | 0.99049 |
| Neprilysin | 0.00057 | 0.00033 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00090 | 0.00003 | 0.00055 | 93 | 0.00056 | -1.42312 | 94.887 | 0.15798 | 0.70632 |
| GalNAc(5)-diNAcBac-PP-undecaprenol beta-1,3-glucosyltransferase | 0.00057 | 0.00019 | 0.00058 | 0.00001 | 0.00030 | 65 | 0.00056 | 0.00001 | 0.00024 | 93 | 0.00039 | 0.05089 | 133.670 | 0.95949 | 0.99187 |
| UDP-2,4-diacetamido-2,4,6-trideoxy-beta-L-altropyranose hydrolase | 0.00057 | 0.00019 | 0.00058 | 0.00001 | 0.00030 | 65 | 0.00056 | 0.00001 | 0.00024 | 93 | 0.00039 | 0.05089 | 133.670 | 0.95949 | 0.99187 |
| Mannosyl-3-phosphoglycerate phosphatase | 0.00057 | 0.00020 | 0.00042 | 0.00000 | 0.00017 | 65 | 0.00067 | 0.00001 | 0.00031 | 93 | 0.00036 | -0.70129 | 136.364 | 0.48431 | 0.80111 |
| 23S rRNA (guanine(1835)-N(2))-methyltransferase | 0.00056 | 0.00018 | 0.00055 | 0.00000 | 0.00020 | 65 | 0.00057 | 0.00001 | 0.00028 | 93 | 0.00034 | -0.06569 | 152.316 | 0.94771 | 0.99049 |
| Lactate 2-monooxygenase | 0.00056 | 0.00021 | 0.00012 | 0.00000 | 0.00006 | 65 | 0.00087 | 0.00001 | 0.00035 | 93 | 0.00036 | -2.11626 | 97.090 | 0.03688 | 0.67652 |
| Phenylalanine dehydrogenase | 0.00056 | 0.00025 | 0.00010 | 0.00000 | 0.00006 | 65 | 0.00088 | 0.00002 | 0.00042 | 93 | 0.00042 | -1.84251 | 95.373 | 0.06851 | 0.70632 |
| Enterobacter ribonuclease | 0.00056 | 0.00019 | 0.00043 | 0.00000 | 0.00018 | 65 | 0.00065 | 0.00001 | 0.00029 | 93 | 0.00034 | -0.63867 | 145.621 | 0.52404 | 0.82182 |
| Gingipain R | 0.00054 | 0.00025 | 0.00047 | 0.00001 | 0.00041 | 65 | 0.00059 | 0.00001 | 0.00032 | 93 | 0.00052 | -0.22700 | 131.514 | 0.82077 | 0.95023 |
| Methanogen homoaconitase | 0.00054 | 0.00038 | 0.00024 | 0.00000 | 0.00019 | 65 | 0.00075 | 0.00004 | 0.00063 | 93 | 0.00066 | -0.77118 | 107.434 | 0.44230 | 0.77520 |
| Homocitrate synthase | 0.00054 | 0.00020 | 0.00027 | 0.00000 | 0.00009 | 65 | 0.00073 | 0.00001 | 0.00033 | 93 | 0.00035 | -1.32558 | 106.234 | 0.18782 | 0.70632 |
| D-ribulokinase | 0.00054 | 0.00032 | 0.00017 | 0.00000 | 0.00007 | 65 | 0.00080 | 0.00003 | 0.00054 | 93 | 0.00055 | -1.14353 | 94.971 | 0.25569 | 0.70632 |
| Putrescine carbamoyltransferase | 0.00054 | 0.00019 | 0.00059 | 0.00000 | 0.00027 | 65 | 0.00051 | 0.00001 | 0.00027 | 93 | 0.00038 | 0.21014 | 150.142 | 0.83384 | 0.95593 |
| Benzil reductase ((S)-benzoin forming) | 0.00053 | 0.00022 | 0.00028 | 0.00000 | 0.00018 | 65 | 0.00070 | 0.00001 | 0.00035 | 93 | 0.00039 | -1.09881 | 134.496 | 0.27381 | 0.70632 |
| 2’-hydroxyisoflavone reductase | 0.00052 | 0.00025 | 0.00007 | 0.00000 | 0.00005 | 65 | 0.00084 | 0.00002 | 0.00042 | 93 | 0.00042 | -1.83379 | 94.231 | 0.06984 | 0.70632 |
| Phosphosulfolactate synthase | 0.00052 | 0.00018 | 0.00024 | 0.00000 | 0.00013 | 65 | 0.00072 | 0.00001 | 0.00029 | 93 | 0.00031 | -1.51978 | 124.737 | 0.13110 | 0.70632 |
| 3,4-dehydroadipyl-CoA semialdehyde dehydrogenase (NADP(+)) | 0.00052 | 0.00038 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00086 | 0.00004 | 0.00064 | 93 | 0.00065 | -1.28204 | 92.292 | 0.20304 | 0.70632 |
| 5-(hydroxymethyl)furfural oxidase | 0.00052 | 0.00020 | 0.00037 | 0.00000 | 0.00014 | 65 | 0.00062 | 0.00001 | 0.00033 | 93 | 0.00036 | -0.67692 | 121.996 | 0.49974 | 0.80830 |
| L-cysteate sulfo-lyase | 0.00052 | 0.00020 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00060 | 0.00001 | 0.00031 | 93 | 0.00036 | -0.55206 | 143.114 | 0.58177 | 0.85342 |
| Ferric-chelate reductase (NADPH) | 0.00052 | 0.00018 | 0.00044 | 0.00000 | 0.00018 | 65 | 0.00057 | 0.00001 | 0.00028 | 93 | 0.00033 | -0.40477 | 148.107 | 0.68623 | 0.90745 |
| L-arabinonolactonase | 0.00052 | 0.00032 | 0.00031 | 0.00000 | 0.00021 | 65 | 0.00066 | 0.00003 | 0.00053 | 93 | 0.00057 | -0.63223 | 118.550 | 0.52845 | 0.82630 |
| N-carbamoyl-D-amino-acid hydrolase | 0.00051 | 0.00021 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00073 | 0.00001 | 0.00034 | 93 | 0.00035 | -1.54007 | 100.868 | 0.12667 | 0.70632 |
| 2-dehydro-3-deoxy-L-arabinonate dehydratase | 0.00051 | 0.00039 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00081 | 0.00004 | 0.00065 | 93 | 0.00066 | -1.13063 | 92.851 | 0.26112 | 0.70632 |
| 2-hydroxymuconate-6-semialdehyde hydrolase | 0.00051 | 0.00023 | 0.00033 | 0.00000 | 0.00014 | 65 | 0.00063 | 0.00001 | 0.00037 | 93 | 0.00040 | -0.75261 | 116.654 | 0.45320 | 0.78466 |
| Crotonyl-CoA carboxylase/reductase | 0.00051 | 0.00021 | 0.00023 | 0.00000 | 0.00011 | 65 | 0.00070 | 0.00001 | 0.00035 | 93 | 0.00037 | -1.27284 | 110.082 | 0.20576 | 0.70632 |
| 2-alkenal reductase (NAD(P)(+)) | 0.00050 | 0.00021 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00075 | 0.00001 | 0.00035 | 93 | 0.00035 | -1.68304 | 96.856 | 0.09559 | 0.70632 |
| CDP-paratose 2-epimerase | 0.00050 | 0.00016 | 0.00014 | 0.00000 | 0.00007 | 65 | 0.00075 | 0.00001 | 0.00027 | 93 | 0.00028 | -2.20575 | 105.555 | 0.02957 | 0.67652 |
| Cellobionic acid phosphorylase | 0.00050 | 0.00032 | 0.00030 | 0.00000 | 0.00021 | 65 | 0.00064 | 0.00003 | 0.00053 | 93 | 0.00057 | -0.60431 | 118.707 | 0.54679 | 0.84303 |
| 4,5-dihydroxyphthalate decarboxylase | 0.00049 | 0.00021 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00073 | 0.00001 | 0.00036 | 93 | 0.00036 | -1.61468 | 96.712 | 0.10964 | 0.70632 |
| Histone acetyltransferase | 0.00049 | 0.00016 | 0.00038 | 0.00000 | 0.00010 | 65 | 0.00057 | 0.00001 | 0.00026 | 93 | 0.00028 | -0.68404 | 116.164 | 0.49531 | 0.80655 |
| Trans-L-3-hydroxyproline dehydratase | 0.00048 | 0.00020 | 0.00048 | 0.00001 | 0.00029 | 65 | 0.00048 | 0.00001 | 0.00027 | 93 | 0.00039 | -0.01523 | 145.855 | 0.98787 | 0.99660 |
| 2-methylfumaryl-CoA hydratase | 0.00047 | 0.00021 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00070 | 0.00001 | 0.00035 | 93 | 0.00036 | -1.56132 | 100.069 | 0.12160 | 0.70632 |
| Mandelamide amidase | 0.00047 | 0.00022 | 0.00034 | 0.00000 | 0.00012 | 65 | 0.00056 | 0.00001 | 0.00037 | 93 | 0.00039 | -0.56835 | 110.890 | 0.57095 | 0.85122 |
| [Lysine-biosynthesis-protein LysW]–L-2-aminoadipate ligase | 0.00047 | 0.00023 | 0.00023 | 0.00000 | 0.00017 | 65 | 0.00064 | 0.00001 | 0.00038 | 93 | 0.00042 | -1.00607 | 127.125 | 0.31629 | 0.70632 |
| (+)-trans-carveol dehydrogenase | 0.00047 | 0.00021 | 0.00050 | 0.00001 | 0.00047 | 65 | 0.00045 | 0.00000 | 0.00017 | 93 | 0.00050 | 0.10344 | 80.934 | 0.91787 | 0.98197 |
| D-xylulose reductase | 0.00047 | 0.00016 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00074 | 0.00001 | 0.00026 | 93 | 0.00027 | -2.45999 | 97.546 | 0.01565 | 0.64530 |
| 3’,5’-cyclic-nucleotide phosphodiesterase | 0.00047 | 0.00015 | 0.00047 | 0.00000 | 0.00026 | 65 | 0.00046 | 0.00000 | 0.00017 | 93 | 0.00032 | 0.03164 | 116.243 | 0.97481 | 0.99435 |
| Oligosaccharide reducing-end xylanase | 0.00045 | 0.00021 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00077 | 0.00001 | 0.00036 | 93 | 0.00036 | -2.11287 | 92.052 | 0.03732 | 0.67652 |
| NAD(+) ADP-ribosyltransferase | 0.00044 | 0.00021 | 0.00038 | 0.00000 | 0.00025 | 65 | 0.00049 | 0.00001 | 0.00031 | 93 | 0.00040 | -0.28393 | 155.839 | 0.77684 | 0.93675 |
| Alditol oxidase | 0.00044 | 0.00018 | 0.00064 | 0.00001 | 0.00039 | 65 | 0.00029 | 0.00000 | 0.00012 | 93 | 0.00041 | 0.85247 | 75.651 | 0.39665 | 0.74876 |
| 2-aminomuconate deaminase | 0.00044 | 0.00032 | 0.00017 | 0.00000 | 0.00011 | 65 | 0.00062 | 0.00003 | 0.00054 | 93 | 0.00055 | -0.82700 | 99.397 | 0.41022 | 0.76070 |
| Futalosine hydrolase | 0.00042 | 0.00020 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00066 | 0.00001 | 0.00034 | 93 | 0.00035 | -1.67700 | 98.418 | 0.09672 | 0.70632 |
| Acetate–CoA ligase (ADP-forming) | 0.00042 | 0.00031 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00069 | 0.00003 | 0.00053 | 93 | 0.00053 | -1.24419 | 92.331 | 0.21658 | 0.70632 |
| N(6)-hydroxylysine O-acetyltransferase | 0.00042 | 0.00013 | 0.00061 | 0.00000 | 0.00026 | 65 | 0.00029 | 0.00000 | 0.00012 | 93 | 0.00029 | 1.09878 | 91.334 | 0.27475 | 0.70632 |
| (S)-3,5-dihydroxyphenylglycine transaminase | 0.00042 | 0.00020 | 0.00038 | 0.00000 | 0.00022 | 65 | 0.00044 | 0.00001 | 0.00030 | 93 | 0.00037 | -0.17439 | 153.476 | 0.86179 | 0.96186 |
| Pyridoxine 4-dehydrogenase | 0.00042 | 0.00020 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00060 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.33571 | 97.339 | 0.18476 | 0.70632 |
| Guanylate cyclase | 0.00042 | 0.00020 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00060 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.35454 | 96.965 | 0.17871 | 0.70632 |
| Heparin-sulfate lyase | 0.00041 | 0.00020 | 0.00026 | 0.00000 | 0.00013 | 65 | 0.00052 | 0.00001 | 0.00032 | 93 | 0.00035 | -0.74276 | 118.705 | 0.45909 | 0.78637 |
| 4-hydroxybenzoate–CoA ligase | 0.00041 | 0.00020 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00059 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.30855 | 97.338 | 0.19377 | 0.70632 |
| Hygromycin-B 7’’-O-kinase | 0.00041 | 0.00020 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00059 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.30855 | 97.338 | 0.19377 | 0.70632 |
| Mandelate racemase | 0.00041 | 0.00020 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00059 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.30855 | 97.338 | 0.19377 | 0.70632 |
| GDP-mannose:cellobiosyl-diphosphopolyprenol alpha-mannosyltransferase | 0.00041 | 0.00014 | 0.00039 | 0.00000 | 0.00024 | 65 | 0.00042 | 0.00000 | 0.00018 | 93 | 0.00030 | -0.08504 | 128.778 | 0.93236 | 0.98985 |
| Coenzyme gamma-F420-2:alpha-L-glutamate ligase | 0.00041 | 0.00022 | 0.00026 | 0.00000 | 0.00021 | 65 | 0.00051 | 0.00001 | 0.00035 | 93 | 0.00041 | -0.62579 | 142.161 | 0.53246 | 0.83089 |
| Acetyl-CoA hydrolase | 0.00041 | 0.00032 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00060 | 0.00003 | 0.00054 | 93 | 0.00054 | -0.85545 | 95.448 | 0.39445 | 0.74652 |
| Anthranilate 3-monooxygenase (FAD) | 0.00040 | 0.00020 | 0.00037 | 0.00001 | 0.00033 | 65 | 0.00043 | 0.00001 | 0.00024 | 93 | 0.00041 | -0.12542 | 124.334 | 0.90040 | 0.97349 |
| L-galactose 1-dehydrogenase | 0.00040 | 0.00020 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00058 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.29661 | 96.971 | 0.19784 | 0.70632 |
| Aspartate–prephenate aminotransferase | 0.00040 | 0.00022 | 0.00023 | 0.00000 | 0.00009 | 65 | 0.00051 | 0.00001 | 0.00037 | 93 | 0.00038 | -0.74041 | 102.789 | 0.46074 | 0.78637 |
| Glutamate–prephenate aminotransferase | 0.00040 | 0.00022 | 0.00023 | 0.00000 | 0.00009 | 65 | 0.00051 | 0.00001 | 0.00037 | 93 | 0.00038 | -0.74041 | 102.789 | 0.46074 | 0.78637 |
| 4-oxalomesaconate tautomerase | 0.00039 | 0.00011 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00059 | 0.00000 | 0.00019 | 93 | 0.00019 | -2.57931 | 101.179 | 0.01134 | 0.64530 |
| Oligosaccharide 4-alpha-D-glucosyltransferase | 0.00039 | 0.00031 | 0.00017 | 0.00000 | 0.00010 | 65 | 0.00053 | 0.00003 | 0.00052 | 93 | 0.00053 | -0.68016 | 99.312 | 0.49798 | 0.80830 |
| D-amino-acid oxidase | 0.00038 | 0.00015 | 0.00036 | 0.00000 | 0.00024 | 65 | 0.00039 | 0.00000 | 0.00018 | 93 | 0.00030 | -0.08859 | 126.098 | 0.92955 | 0.98985 |
| L-carnitine CoA-transferase | 0.00037 | 0.00016 | 0.00031 | 0.00000 | 0.00010 | 65 | 0.00042 | 0.00001 | 0.00026 | 93 | 0.00027 | -0.42408 | 118.246 | 0.67228 | 0.89609 |
| Peptide-aspartate beta-dioxygenase | 0.00036 | 0.00027 | 0.00011 | 0.00000 | 0.00008 | 65 | 0.00054 | 0.00002 | 0.00046 | 93 | 0.00047 | -0.93130 | 96.942 | 0.35401 | 0.72019 |
| Crotonyl-CoA reductase | 0.00035 | 0.00013 | 0.00058 | 0.00001 | 0.00029 | 65 | 0.00019 | 0.00000 | 0.00008 | 93 | 0.00030 | 1.28291 | 74.428 | 0.20350 | 0.70632 |
| Sulfolactaldehyde 3-reductase | 0.00035 | 0.00016 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00046 | 0.00001 | 0.00026 | 93 | 0.00028 | -0.96589 | 110.091 | 0.33622 | 0.71823 |
| Galactitol-1-phosphate 5-dehydrogenase | 0.00034 | 0.00017 | 0.00022 | 0.00000 | 0.00012 | 65 | 0.00042 | 0.00001 | 0.00027 | 93 | 0.00030 | -0.69681 | 126.892 | 0.48720 | 0.80150 |
| Tyrosinase | 0.00033 | 0.00012 | 0.00039 | 0.00000 | 0.00024 | 65 | 0.00030 | 0.00000 | 0.00012 | 93 | 0.00027 | 0.32952 | 97.032 | 0.74248 | 0.92676 |
| Clostripain | 0.00033 | 0.00033 | 0.00079 | 0.00004 | 0.00079 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00079 | 0.99891 | 64.000 | 0.32160 | 0.70672 |
| Gentamicin 2’-N-acetyltransferase | 0.00033 | 0.00012 | 0.00032 | 0.00000 | 0.00024 | 65 | 0.00033 | 0.00000 | 0.00012 | 93 | 0.00027 | -0.03125 | 96.367 | 0.97513 | 0.99435 |
| 2-keto-3-deoxy-L-rhamnonate aldolase | 0.00032 | 0.00016 | 0.00017 | 0.00000 | 0.00008 | 65 | 0.00042 | 0.00001 | 0.00027 | 93 | 0.00028 | -0.91603 | 107.179 | 0.36171 | 0.72341 |
| Monogalactosyldiacylglycerol synthase | 0.00031 | 0.00012 | 0.00011 | 0.00000 | 0.00005 | 65 | 0.00046 | 0.00000 | 0.00020 | 93 | 0.00021 | -1.66991 | 103.777 | 0.09795 | 0.70632 |
| Aldehyde dehydrogenase (FAD-independent) | 0.00031 | 0.00017 | 0.00035 | 0.00001 | 0.00035 | 65 | 0.00028 | 0.00000 | 0.00017 | 93 | 0.00038 | 0.17933 | 93.937 | 0.85806 | 0.96186 |
| Sulfofructosephosphate aldolase | 0.00030 | 0.00016 | 0.00012 | 0.00000 | 0.00006 | 65 | 0.00043 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.15604 | 101.484 | 0.25038 | 0.70632 |
| Glutaminyl-peptide cyclotransferase | 0.00030 | 0.00019 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00049 | 0.00001 | 0.00033 | 93 | 0.00033 | -1.44326 | 92.276 | 0.15233 | 0.70632 |
| Peptidyl-Lys metalloendopeptidase | 0.00029 | 0.00014 | 0.00046 | 0.00001 | 0.00031 | 65 | 0.00017 | 0.00000 | 0.00010 | 93 | 0.00032 | 0.88201 | 78.399 | 0.38047 | 0.73587 |
| Chitin deacetylase | 0.00029 | 0.00014 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00049 | 0.00001 | 0.00024 | 93 | 0.00024 | -2.02812 | 92.000 | 0.04544 | 0.69348 |
| Taurine–pyruvate aminotransferase | 0.00029 | 0.00014 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00049 | 0.00001 | 0.00024 | 93 | 0.00024 | -2.02812 | 92.000 | 0.04544 | 0.69348 |
| Cob(II)yrinic acid a,c-diamide reductase | 0.00029 | 0.00013 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00046 | 0.00000 | 0.00023 | 93 | 0.00023 | -1.87196 | 94.907 | 0.06429 | 0.70632 |
| D-alanine–D-serine ligase | 0.00028 | 0.00017 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00048 | 0.00001 | 0.00028 | 93 | 0.00028 | -1.71551 | 92.000 | 0.08962 | 0.70632 |
| N(5)-(carboxyethyl)ornithine synthase | 0.00028 | 0.00013 | 0.00033 | 0.00000 | 0.00025 | 65 | 0.00025 | 0.00000 | 0.00014 | 93 | 0.00028 | 0.29350 | 101.847 | 0.76974 | 0.93519 |
| Glycine amidinotransferase | 0.00028 | 0.00007 | 0.00032 | 0.00000 | 0.00009 | 65 | 0.00025 | 0.00000 | 0.00010 | 93 | 0.00013 | 0.49467 | 154.474 | 0.62154 | 0.87830 |
| Polygalacturonase | 0.00028 | 0.00014 | 0.00022 | 0.00000 | 0.00015 | 65 | 0.00032 | 0.00000 | 0.00022 | 93 | 0.00026 | -0.40336 | 150.992 | 0.68726 | 0.90804 |
| (S)-sulfolactate dehydrogenase | 0.00028 | 0.00009 | 0.00031 | 0.00000 | 0.00015 | 65 | 0.00025 | 0.00000 | 0.00011 | 93 | 0.00019 | 0.31624 | 126.709 | 0.75234 | 0.92944 |
| Ubiquinol oxidase (non-electrogenic) | 0.00027 | 0.00016 | 0.00034 | 0.00001 | 0.00034 | 65 | 0.00022 | 0.00000 | 0.00014 | 93 | 0.00037 | 0.32888 | 85.596 | 0.74305 | 0.92676 |
| (2R)-3-sulfolactate dehydrogenase (NADP(+)) | 0.00027 | 0.00012 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00041 | 0.00000 | 0.00020 | 93 | 0.00021 | -1.54908 | 105.966 | 0.12435 | 0.70632 |
| 2,6-dihydroxypyridine 3-monooxygenase | 0.00027 | 0.00012 | 0.00023 | 0.00000 | 0.00023 | 65 | 0.00029 | 0.00000 | 0.00013 | 93 | 0.00027 | -0.20576 | 102.362 | 0.83739 | 0.95674 |
| Ferredoxin:protochlorophyllide reductase (ATP-dependent) | 0.00026 | 0.00014 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00042 | 0.00001 | 0.00023 | 93 | 0.00023 | -1.68431 | 93.535 | 0.09546 | 0.70632 |
| Omptin | 0.00026 | 0.00015 | 0.00015 | 0.00000 | 0.00008 | 65 | 0.00033 | 0.00001 | 0.00025 | 93 | 0.00026 | -0.66967 | 109.447 | 0.50448 | 0.81236 |
| Glutamate carboxypeptidase II | 0.00026 | 0.00013 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00041 | 0.00000 | 0.00022 | 93 | 0.00023 | -1.61863 | 94.341 | 0.10887 | 0.70632 |
| Vancomycin aglycone glucosyltransferase | 0.00026 | 0.00012 | 0.00031 | 0.00000 | 0.00024 | 65 | 0.00022 | 0.00000 | 0.00011 | 93 | 0.00026 | 0.34602 | 90.849 | 0.73013 | 0.92120 |
| Hydroxymethylglutaryl-CoA reductase (NADPH) | 0.00026 | 0.00009 | 0.00016 | 0.00000 | 0.00009 | 65 | 0.00032 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.96456 | 147.561 | 0.33634 | 0.71823 |
| Methylmalonyl-CoA decarboxylase | 0.00025 | 0.00015 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00035 | 0.00001 | 0.00026 | 93 | 0.00026 | -0.92828 | 102.094 | 0.35545 | 0.72019 |
| Quinate/shikimate dehydrogenase | 0.00025 | 0.00015 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00035 | 0.00001 | 0.00026 | 93 | 0.00026 | -0.92828 | 102.094 | 0.35545 | 0.72019 |
| Pentachlorophenol monooxygenase | 0.00025 | 0.00012 | 0.00037 | 0.00000 | 0.00025 | 65 | 0.00017 | 0.00000 | 0.00011 | 93 | 0.00027 | 0.71658 | 87.042 | 0.47555 | 0.79541 |
| 4-methylphenol dehydrogenase (hydroxylating) | 0.00025 | 0.00017 | 0.00058 | 0.00001 | 0.00042 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00042 | 1.34500 | 64.082 | 0.18337 | 0.70632 |
| Quorum-quenching N-acyl-homoserine lactonase | 0.00025 | 0.00015 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00040 | 0.00001 | 0.00026 | 93 | 0.00026 | -1.39137 | 93.815 | 0.16741 | 0.70632 |
| Homoisocitrate dehydrogenase | 0.00025 | 0.00019 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00038 | 0.00001 | 0.00032 | 93 | 0.00032 | -0.98195 | 98.627 | 0.32853 | 0.71151 |
| Azobenzene reductase | 0.00024 | 0.00015 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00038 | 0.00001 | 0.00026 | 93 | 0.00026 | -1.24188 | 96.911 | 0.21728 | 0.70632 |
| Exo-1,4-beta-D-glucosaminidase | 0.00024 | 0.00009 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00000 | 0.00015 | 93 | 0.00015 | -2.25828 | 96.708 | 0.02618 | 0.67652 |
| 2-enoate reductase | 0.00024 | 0.00010 | 0.00010 | 0.00000 | 0.00008 | 65 | 0.00034 | 0.00000 | 0.00016 | 93 | 0.00018 | -1.30624 | 127.600 | 0.19382 | 0.70632 |
| 2,3-dihydroxybenzoate 3,4-dioxygenase | 0.00024 | 0.00012 | 0.00033 | 0.00000 | 0.00025 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00028 | 0.57990 | 93.504 | 0.56337 | 0.85024 |
| L-2-hydroxyglutarate dehydrogenase | 0.00023 | 0.00014 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00033 | 0.00000 | 0.00023 | 93 | 0.00025 | -0.98799 | 116.043 | 0.32521 | 0.70949 |
| Sulfite oxidase | 0.00023 | 0.00008 | 0.00023 | 0.00000 | 0.00013 | 65 | 0.00022 | 0.00000 | 0.00010 | 93 | 0.00017 | 0.04126 | 126.054 | 0.96716 | 0.99193 |
| Carboxynorspermidine synthase | 0.00023 | 0.00016 | 0.00037 | 0.00001 | 0.00035 | 65 | 0.00013 | 0.00000 | 0.00013 | 93 | 0.00037 | 0.64035 | 82.275 | 0.52373 | 0.82182 |
| CO-methylating acetyl-CoA synthase | 0.00023 | 0.00012 | 0.00014 | 0.00000 | 0.00012 | 65 | 0.00029 | 0.00000 | 0.00018 | 93 | 0.00021 | -0.69997 | 148.544 | 0.48504 | 0.80111 |
| Phosphonoacetate hydrolase | 0.00023 | 0.00010 | 0.00016 | 0.00000 | 0.00011 | 65 | 0.00027 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.57119 | 154.396 | 0.56870 | 0.85024 |
| Delta(7)-sterol 5(6)-desaturase | 0.00022 | 0.00013 | 0.00037 | 0.00001 | 0.00029 | 65 | 0.00011 | 0.00000 | 0.00008 | 93 | 0.00030 | 0.87733 | 73.476 | 0.38317 | 0.73839 |
| 5,10-methylenetetrahydromethanopterin reductase | 0.00022 | 0.00019 | 0.00047 | 0.00001 | 0.00047 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00047 | 0.91320 | 64.327 | 0.36455 | 0.72472 |
| Heparin lyase | 0.00021 | 0.00010 | 0.00014 | 0.00000 | 0.00007 | 65 | 0.00026 | 0.00000 | 0.00016 | 93 | 0.00017 | -0.70297 | 119.585 | 0.48344 | 0.80031 |
| Thermitase | 0.00021 | 0.00007 | 0.00020 | 0.00000 | 0.00010 | 65 | 0.00022 | 0.00000 | 0.00010 | 93 | 0.00014 | -0.18948 | 153.303 | 0.84997 | 0.96036 |
| Nucleoside-triphosphate phosphatase | 0.00021 | 0.00011 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00028 | 0.00000 | 0.00018 | 93 | 0.00021 | -0.78459 | 148.378 | 0.43395 | 0.77079 |
| Diadenosine hexaphosphate hydrolase (ATP-forming) | 0.00021 | 0.00019 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00031 | 0.00001 | 0.00031 | 93 | 0.00032 | -0.79302 | 98.745 | 0.42967 | 0.76781 |
| 2-furoyl-CoA dehydrogenase | 0.00021 | 0.00011 | 0.00034 | 0.00000 | 0.00023 | 65 | 0.00012 | 0.00000 | 0.00008 | 93 | 0.00024 | 0.89735 | 79.963 | 0.37223 | 0.73139 |
| 3-hydroxybenzoate synthase | 0.00020 | 0.00008 | 0.00022 | 0.00000 | 0.00013 | 65 | 0.00019 | 0.00000 | 0.00010 | 93 | 0.00016 | 0.17312 | 131.527 | 0.86282 | 0.96186 |
| Catechol O-methyltransferase | 0.00019 | 0.00007 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00031 | 0.00000 | 0.00012 | 93 | 0.00012 | -2.39889 | 96.168 | 0.01837 | 0.64530 |
| 3-beta-hydroxy-Delta(5)-steroid dehydrogenase | 0.00019 | 0.00007 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00031 | 0.00000 | 0.00012 | 93 | 0.00012 | -2.41830 | 96.327 | 0.01747 | 0.64530 |
| Trans,polycis-decaprenyl diphosphate synthase | 0.00019 | 0.00007 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00031 | 0.00000 | 0.00012 | 93 | 0.00012 | -2.41830 | 96.327 | 0.01747 | 0.64530 |
| Phospholipase D | 0.00019 | 0.00008 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00030 | 0.00000 | 0.00013 | 93 | 0.00014 | -2.04259 | 99.807 | 0.04373 | 0.69348 |
| Chlorophyllide a reductase | 0.00019 | 0.00012 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00031 | 0.00000 | 0.00021 | 93 | 0.00021 | -1.40637 | 92.596 | 0.16296 | 0.70632 |
| Saccharopine dehydrogenase (NADP(+), L-glutamate-forming) | 0.00019 | 0.00015 | 0.00035 | 0.00001 | 0.00035 | 65 | 0.00008 | 0.00000 | 0.00008 | 93 | 0.00035 | 0.76444 | 70.099 | 0.44717 | 0.77823 |
| Citrate–CoA ligase | 0.00019 | 0.00013 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00026 | 0.00000 | 0.00022 | 93 | 0.00024 | -0.71950 | 118.079 | 0.47325 | 0.79541 |
| GTP cyclohydrolase IV | 0.00019 | 0.00013 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00026 | 0.00000 | 0.00022 | 93 | 0.00024 | -0.71950 | 118.079 | 0.47325 | 0.79541 |
| 3-deoxy-alpha-D-manno-octulosonate 8-oxidase | 0.00018 | 0.00012 | 0.00017 | 0.00000 | 0.00017 | 65 | 0.00019 | 0.00000 | 0.00015 | 93 | 0.00023 | -0.06896 | 143.631 | 0.94511 | 0.99049 |
| Aerobactin synthase | 0.00018 | 0.00007 | 0.00026 | 0.00000 | 0.00014 | 65 | 0.00013 | 0.00000 | 0.00007 | 93 | 0.00016 | 0.81179 | 96.785 | 0.41890 | 0.76408 |
| N(2)-citryl-N(6)-acetyl-N(6)-hydroxylysine synthase | 0.00018 | 0.00007 | 0.00026 | 0.00000 | 0.00014 | 65 | 0.00013 | 0.00000 | 0.00007 | 93 | 0.00016 | 0.81179 | 96.785 | 0.41890 | 0.76408 |
| 4-coumarate–CoA ligase | 0.00018 | 0.00008 | 0.00011 | 0.00000 | 0.00009 | 65 | 0.00023 | 0.00000 | 0.00012 | 93 | 0.00015 | -0.80277 | 155.142 | 0.42333 | 0.76494 |
| Microsomal epoxide hydrolase | 0.00018 | 0.00010 | 0.00036 | 0.00000 | 0.00025 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00025 | 1.21632 | 66.329 | 0.22818 | 0.70632 |
| 3-dehydro-glucose-6-phosphate–glutamate transaminase | 0.00018 | 0.00013 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00028 | 0.00000 | 0.00022 | 93 | 0.00022 | -1.13792 | 93.906 | 0.25805 | 0.70632 |
| Kanosamine-6-phosphate phosphatase | 0.00018 | 0.00013 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00028 | 0.00000 | 0.00022 | 93 | 0.00022 | -1.13792 | 93.906 | 0.25805 | 0.70632 |
| Arylamine N-acetyltransferase | 0.00018 | 0.00007 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00028 | 0.00000 | 0.00012 | 93 | 0.00012 | -2.21187 | 96.413 | 0.02934 | 0.67652 |
| Guanidinopropionase | 0.00018 | 0.00009 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00029 | 0.00000 | 0.00016 | 93 | 0.00016 | -1.69355 | 93.585 | 0.09368 | 0.70632 |
| Sedoheptulokinase | 0.00017 | 0.00010 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00029 | 0.00000 | 0.00017 | 93 | 0.00017 | -1.70207 | 92.267 | 0.09211 | 0.70632 |
| 2-amino-4-deoxychorismate synthase | 0.00017 | 0.00007 | 0.00018 | 0.00000 | 0.00011 | 65 | 0.00017 | 0.00000 | 0.00010 | 93 | 0.00015 | 0.08743 | 142.652 | 0.93045 | 0.98985 |
| (R)-amidase | 0.00017 | 0.00009 | 0.00024 | 0.00000 | 0.00017 | 65 | 0.00012 | 0.00000 | 0.00008 | 93 | 0.00019 | 0.62099 | 94.059 | 0.53611 | 0.83395 |
| UDP-N-acetylglucosamine 2-epimerase (hydrolyzing) | 0.00017 | 0.00010 | 0.00034 | 0.00000 | 0.00024 | 65 | 0.00006 | 0.00000 | 0.00005 | 93 | 0.00024 | 1.13956 | 68.836 | 0.25842 | 0.70632 |
| Insulysin | 0.00017 | 0.00013 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00028 | 0.00000 | 0.00022 | 93 | 0.00022 | -1.22149 | 92.151 | 0.22502 | 0.70632 |
| 3-hydroxy-D-aspartate aldolase | 0.00017 | 0.00010 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00022 | 0.00000 | 0.00015 | 93 | 0.00017 | -0.73346 | 131.307 | 0.46459 | 0.78886 |
| Carnitine O-acetyltransferase | 0.00017 | 0.00008 | 0.00014 | 0.00000 | 0.00010 | 65 | 0.00019 | 0.00000 | 0.00011 | 93 | 0.00015 | -0.32783 | 155.976 | 0.74348 | 0.92676 |
| Cyclopentanol dehydrogenase | 0.00016 | 0.00008 | 0.00026 | 0.00000 | 0.00017 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00018 | 0.96771 | 73.816 | 0.33635 | 0.71823 |
| (3S)-malyl-CoA thioesterase | 0.00016 | 0.00011 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00027 | 0.00000 | 0.00019 | 93 | 0.00019 | -1.43183 | 92.000 | 0.15558 | 0.70632 |
| Non-reducing end beta-L-arabinofuranosidase | 0.00016 | 0.00010 | 0.00017 | 0.00000 | 0.00017 | 65 | 0.00015 | 0.00000 | 0.00011 | 93 | 0.00020 | 0.09191 | 113.565 | 0.92693 | 0.98985 |
| D-glucosaminate-6-phosphate ammonia lyase | 0.00015 | 0.00010 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00025 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.33021 | 93.636 | 0.18668 | 0.70632 |
| Fluoroacetyl-CoA thioesterase | 0.00015 | 0.00009 | 0.00031 | 0.00000 | 0.00021 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00022 | 1.25693 | 69.326 | 0.21300 | 0.70632 |
| Sphingomyelin phosphodiesterase | 0.00015 | 0.00008 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00024 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.62893 | 96.175 | 0.10660 | 0.70632 |
| Sulfoacetaldehyde acetyltransferase | 0.00015 | 0.00009 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00025 | 0.00000 | 0.00015 | 93 | 0.00016 | -1.59017 | 92.187 | 0.11522 | 0.70632 |
| Coenzyme F420 hydrogenase | 0.00015 | 0.00006 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00022 | 0.00000 | 0.00009 | 93 | 0.00010 | -1.66518 | 112.594 | 0.09866 | 0.70632 |
| (2R)-sulfolactate sulfo-lyase | 0.00015 | 0.00007 | 0.00015 | 0.00000 | 0.00011 | 65 | 0.00014 | 0.00000 | 0.00008 | 93 | 0.00014 | 0.07264 | 125.973 | 0.94221 | 0.99049 |
| Transferred entry: 3.4.22.32 and 3.4.22.33 | 0.00015 | 0.00008 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00025 | 0.00000 | 0.00014 | 93 | 0.00014 | -1.82495 | 92.000 | 0.07125 | 0.70632 |
| N,N-dimethylformamidase | 0.00015 | 0.00007 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00024 | 0.00000 | 0.00012 | 93 | 0.00012 | -2.04511 | 92.330 | 0.04369 | 0.69348 |
| 2-methylacyl-CoA dehydrogenase | 0.00014 | 0.00008 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00023 | 0.00000 | 0.00014 | 93 | 0.00014 | -1.57814 | 93.207 | 0.11792 | 0.70632 |
| 2-beta-glucuronyltransferase | 0.00014 | 0.00006 | 0.00018 | 0.00000 | 0.00012 | 65 | 0.00011 | 0.00000 | 0.00007 | 93 | 0.00014 | 0.53187 | 105.176 | 0.59594 | 0.86170 |
| Dolichyl-phosphate-mannose–protein mannosyltransferase | 0.00014 | 0.00005 | 0.00014 | 0.00000 | 0.00005 | 65 | 0.00014 | 0.00000 | 0.00007 | 93 | 0.00009 | -0.05759 | 154.441 | 0.95415 | 0.99119 |
| 2,2-dialkylglycine decarboxylase (pyruvate) | 0.00014 | 0.00007 | 0.00012 | 0.00000 | 0.00009 | 65 | 0.00015 | 0.00000 | 0.00009 | 93 | 0.00013 | -0.23253 | 150.596 | 0.81644 | 0.94663 |
| 8-amino-3,8-dideoxy-alpha-D-manno-octulosonate transaminase | 0.00014 | 0.00009 | 0.00017 | 0.00000 | 0.00017 | 65 | 0.00011 | 0.00000 | 0.00008 | 93 | 0.00019 | 0.30819 | 94.208 | 0.75862 | 0.93185 |
| Pseudolysin | 0.00014 | 0.00007 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00023 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.86942 | 92.000 | 0.06475 | 0.70632 |
| Aldehyde dehydrogenase (NAD(P)(+)) | 0.00014 | 0.00006 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00018 | 0.00000 | 0.00009 | 93 | 0.00010 | -1.06657 | 124.741 | 0.28823 | 0.70632 |
| 5,6,7,8-tetrahydromethanopterin hydro-lyase | 0.00013 | 0.00006 | 0.00020 | 0.00000 | 0.00011 | 65 | 0.00009 | 0.00000 | 0.00006 | 93 | 0.00012 | 0.88996 | 95.075 | 0.37573 | 0.73229 |
| Geranyl diphosphate 2-C-methyltransferase | 0.00013 | 0.00008 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00021 | 0.00000 | 0.00014 | 93 | 0.00014 | -1.30652 | 97.667 | 0.19444 | 0.70632 |
| Indoleamine 2,3-dioxygenase | 0.00013 | 0.00007 | 0.00018 | 0.00000 | 0.00017 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00017 | 0.52016 | 74.588 | 0.60449 | 0.86814 |
| CDP-diacylglycerol–inositol 3-phosphatidyltransferase | 0.00012 | 0.00006 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00020 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.62047 | 98.749 | 0.10832 | 0.70632 |
| Site-specific DNA-methyltransferase (cytosine-N(4)-specific) | 0.00012 | 0.00010 | 0.00027 | 0.00000 | 0.00025 | 65 | 0.00003 | 0.00000 | 0.00001 | 93 | 0.00025 | 0.95824 | 64.216 | 0.34153 | 0.72019 |
| 6-methylsalicylic acid synthase | 0.00012 | 0.00006 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00021 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.90072 | 92.000 | 0.06047 | 0.70632 |
| Abequosyltransferase | 0.00012 | 0.00007 | 0.00018 | 0.00000 | 0.00017 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00017 | 0.64211 | 73.735 | 0.52279 | 0.82182 |
| Cis-3,4-dihydrophenanthrene-3,4-diol dehydrogenase | 0.00012 | 0.00007 | 0.00018 | 0.00000 | 0.00017 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00017 | 0.64211 | 73.735 | 0.52279 | 0.82182 |
| 4-hydroxy-2-oxoglutarate aldolase | 0.00012 | 0.00007 | 0.00022 | 0.00000 | 0.00017 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00018 | 0.98900 | 67.274 | 0.32621 | 0.71093 |
| Serine racemase | 0.00011 | 0.00007 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00019 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.54952 | 92.000 | 0.12469 | 0.70632 |
| Sucrose-phosphate synthase | 0.00011 | 0.00006 | 0.00018 | 0.00000 | 0.00012 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00013 | 0.88230 | 85.446 | 0.38009 | 0.73587 |
| Prostaglandin-endoperoxide synthase | 0.00011 | 0.00010 | 0.00023 | 0.00000 | 0.00023 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00023 | 0.87417 | 65.163 | 0.38524 | 0.73901 |
| Fumarate reductase (CoM/CoB) | 0.00011 | 0.00008 | 0.00017 | 0.00000 | 0.00017 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00018 | 0.58787 | 81.909 | 0.55824 | 0.84961 |
| ADP-specific glucokinase | 0.00011 | 0.00004 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00009 | 0.00000 | 0.00006 | 93 | 0.00009 | 0.50674 | 134.179 | 0.61317 | 0.87289 |
| ADP-specific phosphofructokinase | 0.00011 | 0.00004 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00009 | 0.00000 | 0.00006 | 93 | 0.00009 | 0.50674 | 134.179 | 0.61317 | 0.87289 |
| Hydroxydechloroatrazine ethylaminohydrolase | 0.00011 | 0.00006 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00016 | 0.00000 | 0.00010 | 93 | 0.00011 | -1.27819 | 105.058 | 0.20400 | 0.70632 |
| Formylmethanofuran–tetrahydromethanopterin N-formyltransferase | 0.00010 | 0.00005 | 0.00019 | 0.00000 | 0.00011 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00012 | 1.23457 | 74.657 | 0.22086 | 0.70632 |
| Methenyltetrahydromethanopterin cyclohydrolase | 0.00010 | 0.00005 | 0.00019 | 0.00000 | 0.00011 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00012 | 1.23457 | 74.657 | 0.22086 | 0.70632 |
| NADH oxidase (H(2)O(2)-forming) | 0.00010 | 0.00005 | 0.00016 | 0.00000 | 0.00010 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00011 | 0.92701 | 79.309 | 0.35674 | 0.72028 |
| 2,4-dichlorophenol 6-monooxygenase | 0.00010 | 0.00010 | 0.00023 | 0.00000 | 0.00023 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00023 | 0.96004 | 64.197 | 0.34064 | 0.72019 |
| Glycerate 2-kinase | 0.00010 | 0.00007 | 0.00017 | 0.00000 | 0.00017 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00018 | 0.71275 | 71.039 | 0.47834 | 0.79622 |
| Sedolisin | 0.00010 | 0.00010 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00017 | 0.00000 | 0.00017 | 93 | 0.00017 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Xanthomonalisin | 0.00010 | 0.00010 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00017 | 0.00000 | 0.00017 | 93 | 0.00017 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Aclacinomycin methylesterase | 0.00010 | 0.00010 | 0.00023 | 0.00000 | 0.00023 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00023 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Carboxypeptidase T | 0.00010 | 0.00010 | 0.00023 | 0.00000 | 0.00023 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00023 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Monoterpene epsilon-lactone hydrolase | 0.00010 | 0.00010 | 0.00023 | 0.00000 | 0.00023 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00023 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Ethanolaminephosphotransferase | 0.00010 | 0.00008 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.16117 | 92.447 | 0.24856 | 0.70632 |
| Vibriolysin | 0.00009 | 0.00006 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.47511 | 94.070 | 0.14352 | 0.70632 |
| Glucuronoarabinoxylan endo-1,4-beta-xylanase | 0.00009 | 0.00004 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00009 | 0.00000 | 0.00006 | 93 | 0.00008 | -0.11085 | 155.766 | 0.91188 | 0.97903 |
| Beta-glucoside kinase | 0.00009 | 0.00004 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00008 | -0.24597 | 148.307 | 0.80604 | 0.94351 |
| Choline oxidase | 0.00009 | 0.00005 | 0.00015 | 0.00000 | 0.00011 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00011 | 0.92762 | 81.322 | 0.35635 | 0.72019 |
| (Ara-f)(3)-Hyp beta-L-arabinobiosidase | 0.00009 | 0.00006 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00015 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.34888 | 92.000 | 0.18069 | 0.70632 |
| Gallate dioxygenase | 0.00009 | 0.00004 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00008 | -0.35900 | 145.983 | 0.72011 | 0.92120 |
| Aminopeptidase S | 0.00009 | 0.00004 | 0.00012 | 0.00000 | 0.00009 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00010 | 0.58171 | 86.448 | 0.56227 | 0.85024 |
| L-iduronidase | 0.00008 | 0.00005 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00012 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.02744 | 110.120 | 0.30646 | 0.70632 |
| Succinyl-CoA–D-citramalate CoA-transferase | 0.00008 | 0.00005 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00013 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.39362 | 93.942 | 0.16672 | 0.70632 |
| 4,4’-diapophytoene synthase | 0.00008 | 0.00003 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00010 | 0.00000 | 0.00005 | 93 | 0.00006 | -0.89437 | 144.183 | 0.37262 | 0.73139 |
| Aspergillus nuclease S(1) | 0.00008 | 0.00005 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00012 | 0.00000 | 0.00008 | 93 | 0.00009 | -1.27338 | 96.519 | 0.20594 | 0.70632 |
| Bilirubin oxidase | 0.00008 | 0.00006 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00013 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.30612 | 92.509 | 0.19475 | 0.70632 |
| Plasminogen activator Pla | 0.00008 | 0.00004 | 0.00016 | 0.00000 | 0.00009 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00009 | 1.59875 | 67.113 | 0.11457 | 0.70632 |
| (NAD(P)H) | 0.00008 | 0.00005 | 0.00004 | 0.00000 | 0.00004 | 65 | 0.00010 | 0.00000 | 0.00009 | 93 | 0.00010 | -0.56776 | 133.600 | 0.57115 | 0.85122 |
| Alkanal monooxygenase (FMN-linked) | 0.00008 | 0.00006 | 0.00019 | 0.00000 | 0.00015 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00015 | 1.21723 | 64.000 | 0.22799 | 0.70632 |
| Ferredoxin–nitrate reductase | 0.00008 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00013 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.51819 | 92.000 | 0.13239 | 0.70632 |
| (S)-mandelate dehydrogenase | 0.00008 | 0.00004 | 0.00016 | 0.00000 | 0.00009 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00009 | 1.59099 | 65.390 | 0.11644 | 0.70632 |
| Terephthalate 1,2-dioxygenase | 0.00007 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00012 | 0.56503 | 83.895 | 0.57356 | 0.85190 |
| Chitosanase | 0.00007 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00012 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.40016 | 92.000 | 0.16483 | 0.70632 |
| (R)-citramalyl-CoA lyase | 0.00007 | 0.00005 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00006 | 0.00000 | 0.00005 | 93 | 0.00010 | 0.37384 | 107.832 | 0.70926 | 0.91799 |
| Sulfide-cytochrome-c reductase (flavocytochrome c) | 0.00007 | 0.00007 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00012 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Cysteate synthase | 0.00007 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00012 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.32140 | 92.000 | 0.18964 | 0.70632 |
| Cysteine dioxygenase | 0.00007 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00012 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.32847 | 92.000 | 0.18731 | 0.70632 |
| 4-hydroxybenzoyl-CoA reductase | 0.00007 | 0.00007 | 0.00017 | 0.00000 | 0.00017 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00017 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Phosphonate dehydrogenase | 0.00007 | 0.00004 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00008 | 0.00000 | 0.00007 | 93 | 0.00008 | -0.23253 | 145.233 | 0.81646 | 0.94663 |
| 2-furoate–CoA ligase | 0.00007 | 0.00004 | 0.00011 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00008 | 0.89735 | 79.963 | 0.37223 | 0.73139 |
| 2-dehydro-3-deoxy-D-arabinonate dehydratase | 0.00007 | 0.00004 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00008 | 0.32517 | 113.924 | 0.74565 | 0.92773 |
| Magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase | 0.00007 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00011 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.54722 | 92.592 | 0.12522 | 0.70632 |
| Lipopolysaccharide 3-alpha-galactosyltransferase | 0.00007 | 0.00005 | 0.00013 | 0.00000 | 0.00011 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00011 | 0.93838 | 64.668 | 0.35154 | 0.72019 |
| Alkylmercury lyase | 0.00007 | 0.00003 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00008 | 0.77257 | 85.142 | 0.44192 | 0.77520 |
| 5-exo-hydroxycamphor dehydrogenase | 0.00007 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.30652 | 97.667 | 0.19444 | 0.70632 |
| 6-hydroxypseudooxynicotine dehydrogenase | 0.00007 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.30652 | 97.667 | 0.19444 | 0.70632 |
| Carboxymethylproline synthase | 0.00007 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.30652 | 97.667 | 0.19444 | 0.70632 |
| Polyvinyl alcohol dehydrogenase (cytochrome) | 0.00007 | 0.00003 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00010 | 0.00000 | 0.00005 | 93 | 0.00006 | -1.29247 | 121.066 | 0.19865 | 0.70632 |
| Inulin fructotransferase (DFA-I-forming) | 0.00006 | 0.00005 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00009 | 0.00000 | 0.00008 | 93 | 0.00008 | -0.73056 | 112.972 | 0.46656 | 0.79095 |
| Demethylspheroidene O-methyltransferase | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.40637 | 92.596 | 0.16296 | 0.70632 |
| Malate–CoA ligase | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00009 | 0.51663 | 99.803 | 0.60656 | 0.86933 |
| Lipoyl amidotransferase | 0.00006 | 0.00006 | 0.00014 | 0.00000 | 0.00013 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00013 | 0.98766 | 64.140 | 0.32703 | 0.71125 |
| 3-dehydroquinate synthase II | 0.00006 | 0.00006 | 0.00014 | 0.00000 | 0.00013 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00013 | 0.99428 | 64.286 | 0.32381 | 0.70874 |
| 1-hydroxycarotenoid 3,4-desaturase | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.46558 | 92.000 | 0.14617 | 0.70632 |
| Chlorophyll synthase | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.46558 | 92.000 | 0.14617 | 0.70632 |
| Geranylgeranyl diphosphate reductase | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.46558 | 92.000 | 0.14617 | 0.70632 |
| Alcohol dehydrogenase (NADP(+)) | 0.00006 | 0.00003 | 0.00010 | 0.00000 | 0.00006 | 65 | 0.00003 | 0.00000 | 0.00001 | 93 | 0.00007 | 1.13039 | 69.440 | 0.26220 | 0.70632 |
| Plastoquinol–plastocyanin reductase | 0.00006 | 0.00003 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00006 | -1.30556 | 107.582 | 0.19449 | 0.70632 |
| 3-hydroxypropionyl-CoA dehydratase | 0.00006 | 0.00003 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00006 | 0.63428 | 91.871 | 0.52748 | 0.82538 |
| All-trans-retinol dehydrogenase (NAD(+)) | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00009 | 0.50573 | 89.552 | 0.61429 | 0.87323 |
| Allose kinase | 0.00006 | 0.00004 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00006 | 0.00000 | 0.00005 | 93 | 0.00007 | 0.00319 | 144.184 | 0.99746 | 0.99921 |
| Fucokinase | 0.00006 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00010 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.77623 | 92.000 | 0.07900 | 0.70632 |
| Isopenicillin-N epimerase | 0.00005 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00009 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.06113 | 92.000 | 0.29141 | 0.70632 |
| Inositol oxygenase | 0.00005 | 0.00005 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00008 | 0.00000 | 0.00008 | 93 | 0.00008 | -0.67470 | 107.670 | 0.50131 | 0.81022 |
| Glycoprotein endo-alpha-1,2-mannosidase | 0.00005 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00008 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.22359 | 101.684 | 0.22393 | 0.70632 |
| Aminodeoxyfutalosine deaminase | 0.00005 | 0.00002 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00006 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.30351 | 155.087 | 0.76191 | 0.93350 |
| (3S,4R)-3,4-dihydroxycyclohexa-1,5-diene-1,4-dicarboxylate dehydrogenase | 0.00005 | 0.00004 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00007 | 0.16840 | 132.342 | 0.86653 | 0.96221 |
| 1-alkyl-2-acetylglycerophosphocholine esterase | 0.00005 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| diphospho-ditrans,octacis-undecaprenol 4-beta-mannosyltransferase | 0.00005 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.55912 | 92.000 | 0.12240 | 0.70632 |
| Scyllo-inositol 2-dehydrogenase (NAD(+)) | 0.00005 | 0.00003 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 0.87705 | 86.424 | 0.38289 | 0.73839 |
| N,N’-diacetylchitobiose phosphorylase | 0.00005 | 0.00003 | 0.00011 | 0.00000 | 0.00008 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00008 | 1.44840 | 64.000 | 0.15239 | 0.70632 |
| tRNA (pseudouridine(54)-N(1))-methyltransferase | 0.00005 | 0.00003 | 0.00011 | 0.00000 | 0.00008 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00008 | 1.44840 | 64.000 | 0.15239 | 0.70632 |
| 1L-myo-inositol 1-phosphate cytidylyltransferase | 0.00005 | 0.00003 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00006 | 0.00000 | 0.00005 | 93 | 0.00006 | -0.43234 | 130.932 | 0.66620 | 0.89299 |
| Phenylalanine ammonia-lyase | 0.00004 | 0.00002 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00005 | 1.21816 | 74.469 | 0.22701 | 0.70632 |
| Aclacinomycin-A oxidase | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Aclacinomycin-N oxidase | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Methane monooxygenase (soluble) | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Protein-S-isoprenylcysteine O-methyltransferase | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.39740 | 92.000 | 0.16565 | 0.70632 |
| Undecaprenyl-phosphate glucose phosphotransferase | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.39740 | 92.000 | 0.16565 | 0.70632 |
| Hydroxyacid-oxoacid transhydrogenase | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.03489 | 92.000 | 0.30343 | 0.70632 |
| Cellulose 1,4-beta-cellobiosidase (non-reducing end) | 0.00004 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00005 | 0.54208 | 121.506 | 0.58876 | 0.85698 |
| Peptidylglycine monooxygenase | 0.00004 | 0.00002 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 1.03913 | 74.758 | 0.30210 | 0.70632 |
| NADH oxidase (H(2)O-forming) | 0.00004 | 0.00002 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00004 | 1.66315 | 67.747 | 0.10090 | 0.70632 |
| Arabinonate dehydratase | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00006 | -0.92044 | 93.085 | 0.35972 | 0.72286 |
| NAD(+)–dinitrogen-reductase ADP-D-ribosyltransferase | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Aspartoacylase | 0.00004 | 0.00003 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00008 | 1.21723 | 64.000 | 0.22799 | 0.70632 |
| Deoxynucleoside kinase | 0.00004 | 0.00003 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00008 | 1.21723 | 64.000 | 0.22799 | 0.70632 |
| Long-chain-fatty-acid–luciferin-component ligase | 0.00004 | 0.00003 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00008 | 1.21723 | 64.000 | 0.22799 | 0.70632 |
| Long-chain-fatty-acyl-CoA reductase | 0.00004 | 0.00003 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00008 | 1.21723 | 64.000 | 0.22799 | 0.70632 |
| N-acetylglucosamine-6-sulfatase | 0.00004 | 0.00003 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00008 | 1.21723 | 64.000 | 0.22799 | 0.70632 |
| Cerebroside-sulfatase | 0.00004 | 0.00003 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00006 | 0.63157 | 71.032 | 0.52969 | 0.82762 |
| Butyrate–CoA ligase | 0.00004 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00006 | 0.00000 | 0.00005 | 93 | 0.00005 | -0.93069 | 97.517 | 0.35431 | 0.72019 |
| D-alanine–(R)-lactate ligase | 0.00004 | 0.00004 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00009 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| 1,3,6,8-tetrahydroxynaphthalene synthase | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Carotenoid 1,2-hydratase | 0.00003 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00005 | 93 | 0.00005 | -0.97934 | 93.106 | 0.32995 | 0.71250 |
| Methanol dehydrogenase (cytochrome c) | 0.00003 | 0.00003 | 0.00008 | 0.00000 | 0.00008 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00008 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Magnesium protoporphyrin IX methyltransferase | 0.00003 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00005 | 93 | 0.00005 | -0.97768 | 93.259 | 0.33076 | 0.71350 |
| Fucosylgalactoside 3-alpha-galactosyltransferase | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.24384 | 93.644 | 0.21666 | 0.70632 |
| Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.24384 | 93.644 | 0.21666 | 0.70632 |
| N-acylneuraminate-9-phosphate synthase | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.24384 | 93.644 | 0.21666 | 0.70632 |
| Streptomycin 3’’-kinase | 0.00003 | 0.00002 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00006 | 0.80259 | 67.198 | 0.42504 | 0.76494 |
| Gentamicin 3’-N-acetyltransferase | 0.00003 | 0.00002 | 0.00004 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.45040 | 81.374 | 0.65362 | 0.88943 |
| Protein geranylgeranyltransferase type II | 0.00003 | 0.00003 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00006 | 0.92976 | 64.594 | 0.35596 | 0.72019 |
| N-acetylgalactosamine-4-sulfatase | 0.00003 | 0.00001 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 1.12739 | 70.336 | 0.26341 | 0.70632 |
| Subtilisin | 0.00003 | 0.00002 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.57181 | 64.000 | 0.12093 | 0.70632 |
| 4-hydroxyphenylacetate decarboxylase | 0.00003 | 0.00001 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 1.34735 | 72.680 | 0.18205 | 0.70632 |
| Dissimilatory sulfite reductase | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.08243 | 93.740 | 0.28184 | 0.70632 |
| Hydrogensulfite reductase | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.08243 | 93.740 | 0.28184 | 0.70632 |
| Pulcherriminic acid synthase | 0.00002 | 0.00002 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.35604 | 142.089 | 0.72234 | 0.92120 |
| Mannosyl-3-phosphoglycerate synthase | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00005 | 0.86246 | 66.177 | 0.39155 | 0.74318 |
| Bis(5’-nucleosyl)-tetraphosphatase (asymmetrical) | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00005 | 0.90701 | 65.024 | 0.36775 | 0.72861 |
| Tyrosine 3-monooxygenase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Arsenate reductase (azurin) | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00005 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Dimethyl sulfide:cytochrome c2 reductase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Dimethylamine dehydrogenase | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.68329 | 101.027 | 0.49599 | 0.80655 |
| Trimethylamine dehydrogenase | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.68329 | 101.027 | 0.49599 | 0.80655 |
| Tetracenomycin A2 monooxygenase-diooxygenase | 0.00002 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.94514 | 100.161 | 0.34686 | 0.72019 |
| Lipopolysaccharide N-acetylglucosaminyltransferase | 0.00002 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.49433 | 148.871 | 0.13721 | 0.70632 |
| CDP-L-myo-inositol myo-inositolphosphotransferase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| (-)-germacrene D synthase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Cyclo(L-tyrosyl-L-tyrosyl) synthase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Geosmin synthase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Germacradienol synthase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| L-asparagine oxygenase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Pentalenene oxygenase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Pentalenic acid synthase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Snapalysin | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Streptogrisin B | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Dihydromethanopterin reductase (NAD(P)(+)) | 0.00002 | 0.00002 | 0.00004 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Glucosylglycerol-phosphate synthase | 0.00002 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.14643 | 64.336 | 0.25586 | 0.70632 |
| C5a peptidase | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.06200 | 131.407 | 0.95066 | 0.99049 |
| N-acetylphosphatidylethanolamine-hydrolyzing phospholipase D | 0.00002 | 0.00002 | 0.00004 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Aklaviketone reductase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00002 | -1.12672 | 103.818 | 0.26246 | 0.70632 |
| Enoyl-CoA hydratase 2 | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.33963 | 70.098 | 0.18469 | 0.70632 |
| Pentalenolactone synthase | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Vesicle-fusing ATPase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.08243 | 93.740 | 0.28184 | 0.70632 |
| Phosphoethanolamine N-methyltransferase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Mannosyl-oligosaccharide 1,2-alpha-mannosidase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| [Histone H3]-lysine-36 demethylase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| All-trans-8’-apo-beta-carotenal 15,15’-oxygenase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Arachidonate 15-lipoxygenase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| 2-hydroxy-6-oxo-6-(2-aminophenyl)hexa-2,4-dienoate hydrolase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 0.93204 | 72.770 | 0.35440 | 0.72019 |
| 2,6-dioxo-6-phenylhexa-3-enoate hydrolase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 0.93204 | 72.770 | 0.35440 | 0.72019 |
| Carbazole 1,9a-dioxygenase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 0.93204 | 72.770 | 0.35440 | 0.72019 |
| hydratase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 0.93204 | 72.770 | 0.35440 | 0.72019 |
| L-serine 3-dehydrogenase (NAD(+)) | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 0.93204 | 72.770 | 0.35440 | 0.72019 |
| Glucuronokinase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 1-phosphatidylinositol-4-phosphate 5-kinase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| 9,9’-di-cis-zeta-carotene desaturase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| 3-dehydrosphinganine reductase | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -0.34155 | 155.569 | 0.73315 | 0.92120 |
| Prolycopene isomerase | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -0.34155 | 155.569 | 0.73315 | 0.92120 |
| Ammonia monooxygenase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.41779 | 92.000 | 0.15963 | 0.70632 |
| Methane monooxygenase (particulate) | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.41779 | 92.000 | 0.15963 | 0.70632 |
| Ureidoglycolate amidohydrolase | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | 0.31023 | 112.931 | 0.75696 | 0.93065 |
| Formate–phosphoribosylaminoimidazolecarboxamide ligase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Transferred entry: 4.6.1.16 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Cyclo(L-leucyl-L-leucyl) synthase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Glucose-6-phosphate 3-dehydrogenase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Lysine–8-amino-7-oxononanoate transaminase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Pimeloyl-[acyl-carrier protein] synthase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Rhamnogalacturonan exolyase | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Tyrosine ammonia-lyase | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.85153 | 92.000 | 0.06730 | 0.70632 |
| Bacillolysin | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.18801 | 92.000 | 0.23789 | 0.70632 |
| UDP-N-acetylglucosamine 3-dehydrogenase | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.47052 | 92.000 | 0.14483 | 0.70632 |
| Cyclomaltodextrinase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Quercetin 2,3-dioxygenase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Cyclomaltodextrin glucanotransferase | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| O-ureido-D-serine cyclo-ligase | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+)) (phosphorylating) | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.29111 | 92.000 | 0.19990 | 0.70632 |
| Succinyl-CoA:(R)-benzylsuccinate CoA-transferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | -0.11686 | 155.965 | 0.90712 | 0.97700 |
| L-ascorbate oxidase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 2-(3-amino-3-carboxypropyl)histidine synthase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate synthase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 2,3-bis-O-geranylgeranyl-sn-glycero-phospholipid reductase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 2,3-bis-O-geranylgeranyl-sn-glycerol 1-phosphate reductase (NAD(P)H) | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5’-phosphate reductase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 4-phosphopantoate–beta-alanine ligase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Archaetidylinositol phosphate synthase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| CTP-dependent riboflavin kinase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| D-aminoacyl-tRNA deacylase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Deoxyhypusine monooxygenase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Diphthine synthase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Dolichyl-diphosphooligosaccharide–protein glycotransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Geranylgeranylglycerol-phosphate geranylgeranyltransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Glucosamine-phosphate N-acetyltransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| GTP cyclohydrolase IIa | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Isopentenyl phosphate kinase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Pantoate kinase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Phosphoglycerol geranylgeranyltransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| rRNA small subunit pseudouridine methyltransferase Nep1 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Tetrahydromethanopterin S-methyltransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| tRNA-guanine(15) transglycosylase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| tRNA (cytidine(56)-2’-O)-methyltransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| tRNA (guanine(26)-N(2))-dimethyltransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| tRNA (guanine(26)-N(2)/guanine(27)-N(2))-dimethyltransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| tRNA 4-demethylwyosine synthase (AdoMet-dependent) | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| tRNA(Ile)(2)-agmatinylcytidine synthase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 2-amino-5-chlorophenol 1,6-dioxygenase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 2-aminophenol 1,6-dioxygenase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Betaine–homocysteine S-methyltransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| NAD(+) glycohydrolase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| Bontoxilysin | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.41680 | 92.000 | 0.15992 | 0.70632 |
| Iduronate-2-sulfatase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
| 2-(acetamidomethylene)succinate hydrolase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| 3-hydroxy-2-methylpyridine-4,5-dicarboxylate 4-decarboxylase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| 3-hydroxy-2-methylpyridinecarboxylate dioxygenase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| 4-pyridoxolactonase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Dihydroanticapsin 7-dehydrogenase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Hygromycin B 4-O-kinase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Pyridoxal 4-dehydrogenase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Pyridoxamine–pyruvate transaminase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Pyridoxine 4-oxidase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Succinate–hydroxymethylglutarate CoA-transferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Dimethylpropiothetin dethiomethylase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.38377 | 92.000 | 0.16978 | 0.70632 |
| Divinyl chlorophyllide a 8-vinyl-reductase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.38377 | 92.000 | 0.16978 | 0.70632 |
| Spheroidene monooxygenase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.38377 | 92.000 | 0.16978 | 0.70632 |
| Licheninase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Ribose 1,5-bisphosphate isomerase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.70632 |
| Pyruvate decarboxylase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.70632 |
plot.dat <- tb.ra %>%
arrange(desc(`Overall Mean`)) %>%
slice_head(n=50)%>%
mutate(
description = fct_reorder(description, `Overall Mean`),
description = factor(description, levels = levels(description), ordered=T)
) %>%
arrange(description) %>%
mutate(
id = 1:n(),
step = ifelse(id%%2 == 0, 1, 0),
Mean_diff = `Tumor Mean` - `Non-Tumor Mean`,
diff_se = SEpooled,
Mean_diff_ll = Mean_diff - qt(0.975, df)*diff_se,
Mean_diff_ul = Mean_diff + qt(0.975, df)*diff_se
) %>%
pivot_longer(
cols=contains("Mean"),
names_to = "group",
values_to = "mean"
)
p1.d <- plot.dat %>%
filter(group %in% c("Tumor Mean","Non-Tumor Mean")) %>%
mutate(
group = ifelse(group == "Tumor Mean", "Tumor", "Non-Tumor"),
col = ifelse(step == 1, "grey90", "white"),
h=1, w=Inf
)
p1 <- ggplot()+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_bar(data=p1.d,
aes(x=mean, y=description,
group=group, color=group,
fill=group),
stat="identity",position = "dodge",
alpha = 1)+
labs(x="Mean Proportion (%)")+
theme_classic()+
theme(
legend.position = "bottom",
plot.margin = unit(c(1,0,1,1), "lines")
)
p2.d <- plot.dat %>%
filter(group %in% c("Mean_diff", "Mean_diff_ll", "Mean_diff_ul")) %>%
pivot_wider(
names_from = group,
values_from = mean
) %>%
mutate(
group = ifelse(Mean_diff > 0, "Tumor", "Non-Tumor"),
p = sprintf("%.3f", round(p,3)),
ll = min(Mean_diff_ll)-0.01,
ul = max(Mean_diff_ul)+0.01
)
p2<-ggplot(p2.d, aes(x=Mean_diff, y=description))+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_vline(xintercept = 0, linetype="dashed", alpha=0.5)+
geom_segment(aes(x=Mean_diff_ll, y=description, xend=Mean_diff_ul, yend=description))+
geom_point(aes(fill=group, color=group))+
geom_text(aes(label=p, x=unique(ul)+0.1))+
coord_cartesian(xlim = c(unique(p2.d$ll), unique(p2.d$ul)),
clip = 'off') +
annotate("text", x=unique(p2.d$ul)+0.2,y = 25,
angle=90,
label="p-value (uncorrected)")+
labs(x="Mean Difference in Proportions")+
theme_classic()+
theme(
legend.position = "bottom",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
plot.margin = unit(c(1,4,1,0), "lines")
)
# plot
p1 + p2 +
plot_annotation(title="EC Data: 50 most abundant descriptions")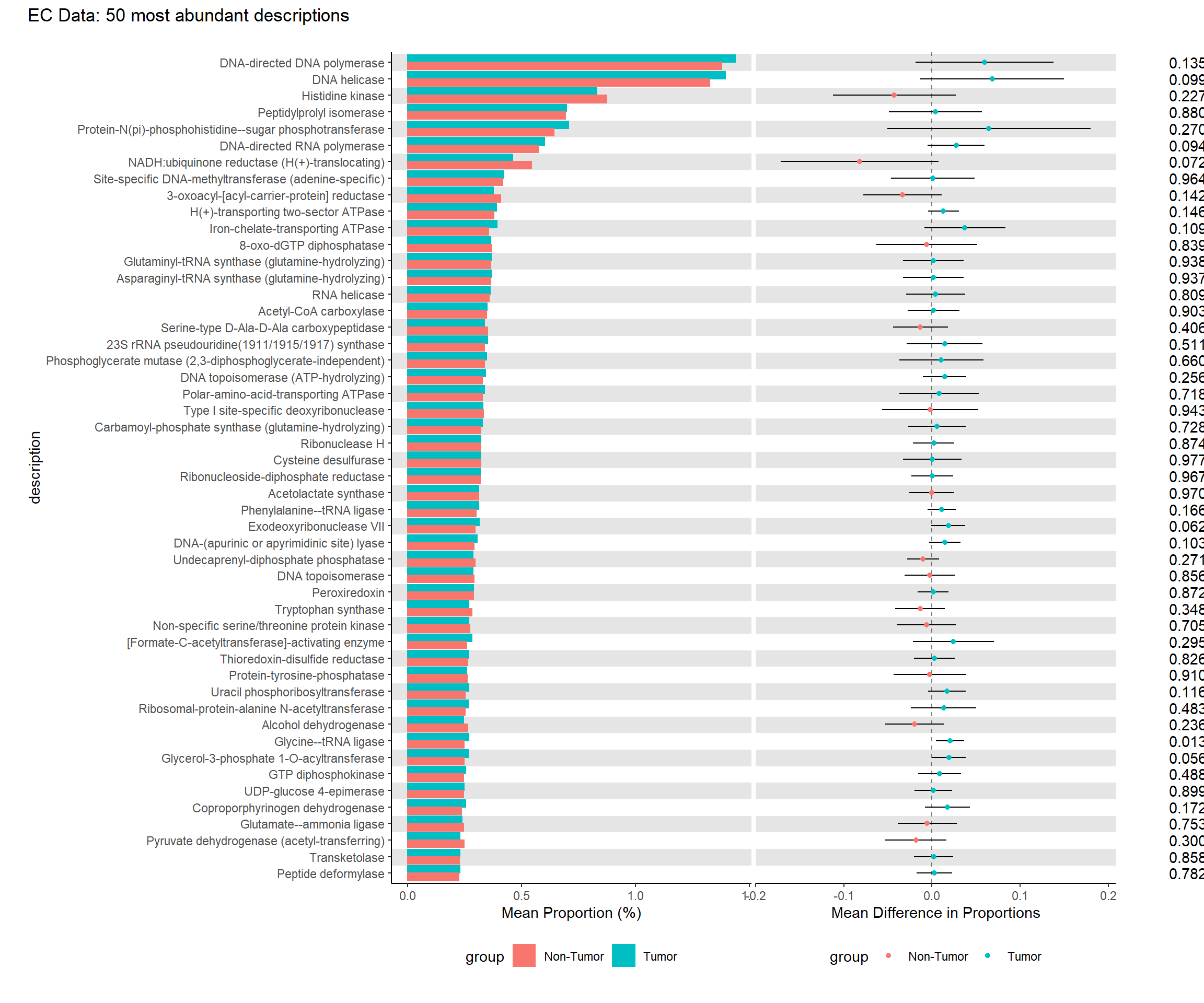
Modeling Difference between Tumor and Non-Tumor
For the modeling, we used a generalized linear mixed model (GLMM).
First, we looked at the biserial correlation between the abundance of each description and the tumor status.
tb <- mydata %>%
group_by(description)%>%
summarise(
r = cor(tumor, Abundance)
) %>%
mutate(
M=mean(r)
)
ggplot(tb, aes(x=r))+
geom_density()+
geom_vline(aes(xintercept = M))+
labs(x="Biserial Correlation",title="Relationship between description abundance and tumor (tumor vs. non-tumor)")+
theme(panel.grid = element_blank())
Next, we need the data to be on an interpretable scale. First, let’s use the raw abundance like scale.
p <- ggplot(mydata, aes(x=Abundance))+
geom_density()
p
mydata <- mydata %>%
mutate(Abundance.dich=ifelse(Abundance==0, 0, 1))
table(mydata$Abundance.dich)
0 1
126115 207897 Let’s first run models by description so that we can avoid the nesting issue initially. We will come back to this to conduct the final model (it will be more powerful).
DESCRIPTIONS <- unique(mydata$description)
i<-1
dat0 <- mydata %>% filter(description==DESCRIPTIONS[i])
# quasipossion (approximately negative binom) give an approx. answer.
fit0 <- glm(
Abundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
summary(fit0)
Call:
glm(formula = Abundance ~ 1 + tumor, family = quasipoisson(link = "log"),
data = dat0)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.415 -0.415 -0.415 0.000 7.530
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.453 0.768 -3.19 0.0017 **
tumor -17.849 4187.194 0.00 0.9966
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 4.7179)
Null deviance: 81.002 on 157 degrees of freedom
Residual deviance: 72.522 on 156 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 18NExt, let’s model the percent relative abundance. This will allow us to make inference about the difference average relative abundance. Which is a simpler interpretation than trying to model differences in the log relative abundance which will need to be interpreted as the multiplicative relative change.
results.out <- as.data.frame(matrix(ncol=4, nrow=length(DESCRIPTIONS)))
colnames(results.out) <- c("description", "Est", "SE", "p")
#results.out$description <-DESCRIPTIONS
i <- 1
for(i in 1:length(DESCRIPTIONS)){
#for(i in 1:5){
dat0 <- mydata %>%
filter(description == DESCRIPTIONS[i])%>%
mutate(RelAbundance= RelAbundance*100)
fit0 <- glm(
RelAbundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
fit.sum <- summary(fit0)
results.out[i, 1] <- DESCRIPTIONS[i]
results.out[i, 2:4] <- fit.sum$coefficients[2, c(1,2,4)]
}
results.out$fdr_p <- p.adjust(results.out$p, method="fdr")
kable(results.out, format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="600px")| description | Est | SE | p | fdr_p |
|---|---|---|---|---|
| (-)-germacrene D synthase | -17.486 | 3491.164 | 0.996 | 0.999 |
| (+)-trans-carveol dehydrogenase | 0.108 | 0.903 | 0.905 | 0.999 |
| (1->4)-alpha-D-glucan 1-alpha-D-glucosylmutase | -0.096 | 0.264 | 0.717 | 0.999 |
| (2,3-dihydroxybenzoyl)adenylate synthase | -0.096 | 0.259 | 0.711 | 0.999 |
| (2E,6E)-farnesyl diphosphate synthase | 0.031 | 0.025 | 0.221 | 0.806 |
| (2R)-3-sulfolactate dehydrogenase (NADP(+)) | -1.601 | 1.182 | 0.178 | 0.806 |
| (2R)-sulfolactate sulfo-lyase | 0.068 | 0.913 | 0.941 | 0.999 |
| (2S)-methylsuccinyl-CoA dehydrogenase | -1.104 | 0.748 | 0.142 | 0.806 |
| (2Z,6E)-farnesyl diphosphate synthase | 0.009 | 0.285 | 0.975 | 0.999 |
| (3S)-malyl-CoA thioesterase | -17.709 | 2725.574 | 0.995 | 0.999 |
| (3S,4R)-3,4-dihydroxycyclohexa-1,5-diene-1,4-dicarboxylate dehydrogenase | 0.241 | 1.424 | 0.866 | 0.999 |
| (4S)-4-hydroxy-2-oxoglutarate aldolase | 0.014 | 0.114 | 0.902 | 0.999 |
| (Ara-f)(3)-Hyp beta-L-arabinobiosidase | -18.092 | 3503.766 | 0.996 | 0.999 |
| (d)CTP diphosphatase | 0.212 | 0.607 | 0.728 | 0.999 |
| (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ferredoxin) | -0.004 | 0.057 | 0.951 | 0.999 |
| (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (flavodoxin) | -0.004 | 0.057 | 0.951 | 0.999 |
| (FADH(2)-oxidizing) | 0.042 | 0.094 | 0.659 | 0.975 |
| (Kdo)-lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase | -0.026 | 0.088 | 0.767 | 0.999 |
| (Kdo)(2)-lipid IV(A) (2-8) 3-deoxy-D-manno-octulosonic acid transferase | -0.026 | 0.088 | 0.767 | 0.999 |
| (Kdo)(3)-lipid IV(A) (2-4) 3-deoxy-D-manno-octulosonic acid transferase | -0.026 | 0.088 | 0.767 | 0.999 |
| (NAD(P)H) | -0.808 | 1.552 | 0.604 | 0.957 |
| (R)-2-methylmalate dehydratase | -0.070 | 0.053 | 0.188 | 0.806 |
| (R)-amidase | 0.678 | 0.985 | 0.492 | 0.905 |
| (R)-citramalate synthase | -0.607 | 0.571 | 0.290 | 0.820 |
| (R)-citramalyl-CoA lyase | 0.503 | 1.269 | 0.692 | 0.995 |
| (R,R)-butanediol dehydrogenase | -0.058 | 0.155 | 0.707 | 0.999 |
| (S)-2-haloacid dehalogenase | -0.361 | 0.170 | 0.036 | 0.780 |
| (S)-2-hydroxy-acid oxidase | 0.015 | 0.184 | 0.936 | 0.999 |
| (S)-3-amino-2-methylpropionate transaminase | -0.777 | 0.350 | 0.028 | 0.780 |
| (S)-3,5-dihydroxyphenylglycine transaminase | -0.159 | 0.975 | 0.871 | 0.999 |
| (S)-citramalyl-CoA lyase | -0.015 | 0.357 | 0.968 | 0.999 |
| (S)-mandelate dehydrogenase | 2.169 | 1.034 | 0.038 | 0.780 |
| (S)-sulfolactate dehydrogenase | 0.214 | 0.663 | 0.747 | 0.999 |
| (S)-ureidoglycine aminohydrolase | -0.058 | 0.360 | 0.871 | 0.999 |
| (S,S)-butanediol dehydrogenase | -0.030 | 0.207 | 0.884 | 0.999 |
| [Acyl-carrier-protein] phosphodiesterase | 0.244 | 0.549 | 0.658 | 0.974 |
| [Acyl-carrier-protein] S-malonyltransferase | 0.028 | 0.024 | 0.235 | 0.806 |
| [Citrate (pro-3S)-lyase] ligase | 0.769 | 0.298 | 0.011 | 0.780 |
| [Formate-C-acetyltransferase]-activating enzyme | 0.090 | 0.086 | 0.297 | 0.820 |
| [Glutamate–ammonia-ligase] adenylyltransferase | -0.022 | 0.119 | 0.851 | 0.999 |
| [Histone H3]-lysine-36 demethylase | -17.083 | 2854.434 | 0.995 | 0.999 |
| [Isocitrate dehydrogenase (NADP(+))] kinase | 0.114 | 0.227 | 0.616 | 0.963 |
| [Lysine-biosynthesis-protein LysW]–L-2-aminoadipate ligase | -1.048 | 1.155 | 0.365 | 0.845 |
| [Protein-PII] uridylyltransferase | -0.056 | 0.141 | 0.692 | 0.995 |
| [Ribosomal protein S12] (aspartate(89)-C(3))-methylthiotransferase | -0.015 | 0.087 | 0.861 | 0.999 |
| [Trimethylamine–corrinoid protein] Co-methyltransferase | -2.110 | 1.516 | 0.166 | 0.806 |
| 1-acylglycerol-3-phosphate O-acyltransferase | -0.017 | 0.037 | 0.645 | 0.973 |
| 1-alkyl-2-acetylglycerophosphocholine esterase | -17.513 | 3538.142 | 0.996 | 0.999 |
| 1-aminocyclopropane-1-carboxylate deaminase | 0.129 | 0.277 | 0.643 | 0.973 |
| 1-deoxy-D-xylulose-5-phosphate reductoisomerase | 0.002 | 0.058 | 0.975 | 0.999 |
| 1-deoxy-D-xylulose-5-phosphate synthase | 0.078 | 0.067 | 0.247 | 0.806 |
| 1-hydroxycarotenoid 3,4-desaturase | -17.728 | 2688.993 | 0.995 | 0.999 |
| 1-phosphatidylinositol-4-phosphate 5-kinase | 18.195 | 2901.073 | 0.995 | 0.999 |
| 1-phosphofructokinase | 0.093 | 0.067 | 0.162 | 0.806 |
| 1-piperideine-2-carboxylate/1-pyrroline-2-carboxylate reductase (NADPH) | -0.070 | 0.417 | 0.867 | 0.999 |
| 1,3-beta-galactosyl-N-acetylhexosamine phosphorylase | 0.283 | 0.430 | 0.511 | 0.913 |
| 1,3-propanediol dehydrogenase | 0.331 | 0.537 | 0.538 | 0.924 |
| 1,3,6,8-tetrahydroxynaphthalene synthase | -17.179 | 2994.595 | 0.995 | 0.999 |
| 1,4-alpha-glucan branching enzyme | 0.032 | 0.068 | 0.638 | 0.973 |
| 1,4-beta-mannosyl-N-acetylglucosamine phosphorylase | -0.324 | 0.229 | 0.160 | 0.806 |
| 1,4-dihydroxy-2-naphthoate polyprenyltransferase | 0.086 | 0.101 | 0.397 | 0.862 |
| 1,4-dihydroxy-2-naphthoyl-CoA hydrolase | -0.157 | 0.423 | 0.711 | 0.999 |
| 1,4-dihydroxy-2-naphthoyl-CoA synthase | 0.025 | 0.107 | 0.818 | 0.999 |
| 1,4-N-acetyl-D-galactosaminyltransferase | 0.454 | 0.524 | 0.388 | 0.853 |
| 1,5-anhydro-D-fructose reductase (1,5-anhydro-D-mannitol-forming) | -1.260 | 1.098 | 0.253 | 0.811 |
| 1,6-dihydroxycyclohexa-2,4-diene-1-carboxylate dehydrogenase | 0.248 | 0.311 | 0.425 | 0.865 |
| 15-cis-phytoene synthase | -0.435 | 0.225 | 0.054 | 0.780 |
| 16S rRNA (adenine(1518)-N(6)/adenine(1519)-N(6))-dimethyltransferase | 0.041 | 0.032 | 0.193 | 0.806 |
| 16S rRNA (cytidine(1402)-2’-O)-methyltransferase | 0.024 | 0.032 | 0.451 | 0.878 |
| 16S rRNA (cytidine(1409)-2’-O)-methyltransferase | 0.082 | 0.076 | 0.285 | 0.820 |
| 16S rRNA (cytosine(1402)-N(4))-methyltransferase | 0.038 | 0.032 | 0.238 | 0.806 |
| 16S rRNA (cytosine(1407)-C(5))-methyltransferase | 0.397 | 0.546 | 0.468 | 0.885 |
| 16S rRNA (cytosine(967)-C(5))-methyltransferase | 0.049 | 0.031 | 0.121 | 0.806 |
| 16S rRNA (guanine(1207)-N(2))-methyltransferase | 0.066 | 0.071 | 0.354 | 0.844 |
| 16S rRNA (guanine(1516)-N(2))-methyltransferase | 0.129 | 0.179 | 0.472 | 0.888 |
| 16S rRNA (guanine(527)-N(7))-methyltransferase | 0.043 | 0.031 | 0.171 | 0.806 |
| 16S rRNA (guanine(966)-N(2))-methyltransferase | -0.027 | 0.072 | 0.707 | 0.999 |
| 16S rRNA (uracil(1498)-N(3))-methyltransferase | 0.037 | 0.034 | 0.267 | 0.820 |
| 16S rRNA pseudouridine(516) synthase | 0.098 | 0.070 | 0.162 | 0.806 |
| 1L-myo-inositol 1-phosphate cytidylyltransferase | -0.620 | 1.555 | 0.691 | 0.995 |
| 2’-hydroxybiphenyl-2-sulfinate desulfinase | -1.012 | 1.000 | 0.313 | 0.820 |
| 2’-hydroxyisoflavone reductase | -2.529 | 1.728 | 0.145 | 0.806 |
| 2’,3’-cyclic-nucleotide 2’-phosphodiesterase | -0.119 | 0.130 | 0.362 | 0.844 |
| 2’,3’-cyclic-nucleotide 3’-phosphodiesterase | 1.312 | 0.617 | 0.035 | 0.780 |
| 2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA isomerase | -0.546 | 0.247 | 0.029 | 0.780 |
| 2-(3-amino-3-carboxypropyl)histidine synthase | -16.940 | 2657.012 | 0.995 | 0.999 |
| 2-(acetamidomethylene)succinate hydrolase | 17.690 | 2253.589 | 0.994 | 0.999 |
| 2-alkenal reductase (NAD(P)(+)) | -1.599 | 1.054 | 0.131 | 0.806 |
| 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate synthase | -16.940 | 2657.012 | 0.995 | 0.999 |
| 2-amino-4-deoxychorismate synthase | 0.074 | 0.852 | 0.931 | 0.999 |
| 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase | -0.023 | 0.035 | 0.507 | 0.913 |
| 2-amino-5-chlorophenol 1,6-dioxygenase | -17.840 | 4168.155 | 0.997 | 0.999 |
| 2-aminoethylphosphonate–pyruvate transaminase | -0.339 | 0.313 | 0.280 | 0.820 |
| 2-aminomuconate deaminase | -1.308 | 1.726 | 0.450 | 0.877 |
| 2-aminophenol 1,6-dioxygenase | -17.840 | 4168.155 | 0.997 | 0.999 |
| 2-beta-glucuronyltransferase | 0.512 | 0.899 | 0.570 | 0.938 |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | 0.005 | 0.062 | 0.930 | 0.999 |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | -0.033 | 0.052 | 0.523 | 0.915 |
| 2-dehydro-3-deoxy-6-phosphogalactonate aldolase | -0.891 | 0.361 | 0.015 | 0.780 |
| 2-dehydro-3-deoxy-D-arabinonate dehydratase | 0.356 | 1.044 | 0.733 | 0.999 |
| 2-dehydro-3-deoxy-L-arabinonate dehydratase | -2.435 | 2.607 | 0.352 | 0.842 |
| 2-dehydro-3-deoxy-phosphogluconate aldolase | 0.015 | 0.114 | 0.897 | 0.999 |
| 2-dehydro-3-deoxygalactonokinase | -0.882 | 0.355 | 0.014 | 0.780 |
| 2-dehydro-3-deoxyglucarate aldolase | 0.364 | 0.548 | 0.508 | 0.913 |
| 2-dehydro-3-deoxygluconokinase | -0.090 | 0.139 | 0.517 | 0.913 |
| 2-dehydropantoate 2-reductase | 0.049 | 0.091 | 0.587 | 0.950 |
| 2-deoxy-D-gluconate 3-dehydrogenase | -0.403 | 0.335 | 0.231 | 0.806 |
| 2-enoate reductase | -1.219 | 1.057 | 0.250 | 0.808 |
| 2-furoate–CoA ligase | 1.044 | 0.990 | 0.293 | 0.820 |
| 2-furoyl-CoA dehydrogenase | 1.044 | 0.990 | 0.293 | 0.820 |
| 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase | -1.431 | 0.945 | 0.132 | 0.806 |
| 2-hydroxy-3-oxopropionate reductase | -0.296 | 0.212 | 0.165 | 0.806 |
| 2-hydroxy-4-carboxymuconate semialdehyde hemiacetal dehydrogenase | -2.081 | 1.531 | 0.176 | 0.806 |
| 2-hydroxy-6-oxo-6-(2-aminophenyl)hexa-2,4-dienoate hydrolase | 1.546 | 1.393 | 0.269 | 0.820 |
| 2-hydroxy-6-oxonona-2,4-dienedioate hydrolase | -1.092 | 0.764 | 0.155 | 0.806 |
| 2-hydroxyhexa-2,4-dienoate hydratase | 0.679 | 0.471 | 0.151 | 0.806 |
| 2-hydroxymuconate-6-semialdehyde dehydrogenase | -0.420 | 0.663 | 0.528 | 0.918 |
| 2-hydroxymuconate-6-semialdehyde hydrolase | -0.647 | 0.934 | 0.490 | 0.902 |
| 2-hydroxymuconate tautomerase | -0.026 | 0.095 | 0.784 | 0.999 |
| 2-iminoacetate synthase | 0.250 | 0.169 | 0.142 | 0.806 |
| 2-iminobutanoate/2-iminopropanoate deaminase | 0.007 | 0.087 | 0.934 | 0.999 |
| 2-isopropylmalate synthase | -0.065 | 0.055 | 0.237 | 0.806 |
| 2-keto-3-deoxy-L-rhamnonate aldolase | -0.936 | 1.108 | 0.400 | 0.863 |
| 2-keto-myo-inositol isomerase | -0.533 | 0.583 | 0.362 | 0.844 |
| 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase | -0.016 | 0.064 | 0.801 | 0.999 |
| 2-methylacyl-CoA dehydrogenase | -3.022 | 2.625 | 0.251 | 0.809 |
| 2-methylcitrate dehydratase | -0.123 | 0.262 | 0.640 | 0.973 |
| 2-methylcitrate synthase | 0.024 | 0.203 | 0.908 | 0.999 |
| 2-methylfumaryl-CoA hydratase | -1.570 | 1.124 | 0.164 | 0.806 |
| 2-methylfumaryl-CoA isomerase | -1.156 | 0.879 | 0.190 | 0.806 |
| 2-methylisocitrate dehydratase | -0.011 | 0.199 | 0.958 | 0.999 |
| 2-oxo-3-hexenedioate decarboxylase | -0.930 | 0.670 | 0.167 | 0.806 |
| 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase | -0.726 | 0.381 | 0.058 | 0.780 |
| 2-oxoacid oxidoreductase (ferredoxin) | -0.216 | 0.149 | 0.150 | 0.806 |
| 2-oxoglutarate decarboxylase | -0.316 | 0.577 | 0.585 | 0.948 |
| 2-oxoglutarate synthase | -0.191 | 0.163 | 0.244 | 0.806 |
| 2-oxopent-4-enoate hydratase | -0.229 | 0.271 | 0.399 | 0.863 |
| 2-phospho-L-lactate guanylyltransferase | -0.703 | 0.540 | 0.195 | 0.806 |
| 2-phospho-L-lactate transferase | -0.691 | 0.591 | 0.244 | 0.806 |
| 2-phosphosulfolactate phosphatase | -0.530 | 0.341 | 0.122 | 0.806 |
| 2-polyprenyl-6-hydroxyphenol methylase | -0.047 | 0.173 | 0.785 | 0.999 |
| 2-pyrone-4,6-dicarboxylate lactonase | -2.177 | 1.492 | 0.146 | 0.806 |
| 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase | 0.077 | 0.183 | 0.674 | 0.984 |
| 2,2-dialkylglycine decarboxylase (pyruvate) | -0.228 | 1.011 | 0.822 | 0.999 |
| 2,3-bis-O-geranylgeranyl-sn-glycero-phospholipid reductase | -16.940 | 2657.012 | 0.995 | 0.999 |
| 2,3-bis-O-geranylgeranyl-sn-glycerol 1-phosphate reductase (NAD(P)H) | -16.940 | 2657.012 | 0.995 | 0.999 |
| 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | -0.106 | 0.271 | 0.695 | 0.995 |
| 2,3-dihydroxybenzoate 3,4-dioxygenase | 0.662 | 1.026 | 0.519 | 0.913 |
| 2,3-diketo-5-methylthiopentyl-1-phosphate enolase | -2.714 | 1.401 | 0.055 | 0.780 |
| 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | -0.024 | 0.072 | 0.740 | 0.999 |
| 2,4-dichlorophenol 6-monooxygenase | 3.239 | 2.889 | 0.264 | 0.820 |
| 2,4-dienoyl-CoA reductase (NADPH) | -0.074 | 0.213 | 0.729 | 0.999 |
| 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5’-phosphate reductase | -16.940 | 2657.012 | 0.995 | 0.999 |
| 2,5-didehydrogluconate reductase (2-dehydro-L-gulonate-forming) | -0.027 | 0.153 | 0.860 | 0.999 |
| 2,5-dihydroxypyridine 5,6-dioxygenase | -0.989 | 0.896 | 0.271 | 0.820 |
| 2,6-beta-fructan 6-levanbiohydrolase | 0.118 | 0.584 | 0.840 | 0.999 |
| 2,6-dihydroxypyridine 3-monooxygenase | -0.211 | 0.984 | 0.830 | 0.999 |
| 2,6-dioxo-6-phenylhexa-3-enoate hydrolase | 1.546 | 1.393 | 0.269 | 0.820 |
| 2.7.11.30 and 2.7.12.1 | 1.258 | 0.574 | 0.030 | 0.780 |
| 23S rRNA (adenine(1618)-N(6))-methyltransferase | 0.258 | 0.300 | 0.390 | 0.857 |
| 23S rRNA (adenine(2030)-N(6))-methyltransferase | -0.018 | 0.153 | 0.908 | 0.999 |
| 23S rRNA (adenine(2085)-N(6))-dimethyltransferase | 0.236 | 0.321 | 0.464 | 0.884 |
| 23S rRNA (adenine(2503)-C(2))-methyltransferase | 0.022 | 0.038 | 0.557 | 0.936 |
| 23S rRNA (cytidine(1920)-2’-O)-methyltransferase | 0.082 | 0.076 | 0.285 | 0.820 |
| 23S rRNA (cytidine(2498)-2’-O)-methyltransferase | 0.142 | 0.190 | 0.453 | 0.881 |
| 23S rRNA (cytosine(1962)-C(5))-methyltransferase | 0.002 | 0.057 | 0.976 | 0.999 |
| 23S rRNA (guanine(1835)-N(2))-methyltransferase | -0.040 | 0.663 | 0.952 | 0.999 |
| 23S rRNA (guanine(2069)-N(7))-methyltransferase | 0.080 | 0.181 | 0.657 | 0.974 |
| 23S rRNA (guanine(2445)-N(2))-methyltransferase | 0.080 | 0.181 | 0.657 | 0.974 |
| 23S rRNA (guanine(745)-N(1))-methyltransferase | 0.034 | 0.098 | 0.727 | 0.999 |
| 23S rRNA (guanosine(2251)-2’-O)-methyltransferase | 0.055 | 0.029 | 0.063 | 0.780 |
| 23S rRNA (pseudouridine(1915)-N(3))-methyltransferase | 0.036 | 0.034 | 0.289 | 0.820 |
| 23S rRNA (uracil(1939)-C(5))-methyltransferase | 0.018 | 0.045 | 0.688 | 0.995 |
| 23S rRNA (uracil(747)-C(5))-methyltransferase | 0.177 | 0.216 | 0.414 | 0.865 |
| 23S rRNA (uridine(2552)-2’-O)-methyltransferase | -0.044 | 0.148 | 0.768 | 0.999 |
| 23S rRNA pseudouridine(1911/1915/1917) synthase | 0.042 | 0.063 | 0.509 | 0.913 |
| 23S rRNA pseudouridine(2457) synthase | 0.064 | 0.159 | 0.688 | 0.995 |
| 23S rRNA pseudouridine(2604) synthase | 0.059 | 0.186 | 0.751 | 0.999 |
| 23S rRNA pseudouridine(2605) synthase | 0.024 | 0.033 | 0.462 | 0.884 |
| 23S rRNA pseudouridine(746) synthase | -0.053 | 0.097 | 0.585 | 0.948 |
| 23S rRNA pseudouridine(955/2504/2580) synthase | 0.110 | 0.111 | 0.325 | 0.827 |
| 3’-nucleotidase | -0.119 | 0.130 | 0.362 | 0.844 |
| 3’(2’),5’-bisphosphate nucleotidase | 0.029 | 0.040 | 0.467 | 0.884 |
| 3’,5’-cyclic-nucleotide phosphodiesterase | 0.021 | 0.650 | 0.974 | 0.999 |
| 3-(3-hydroxy-phenyl)propanoic acid hydroxylase | -0.763 | 0.383 | 0.048 | 0.780 |
| 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate dehydrogenase | -0.931 | 0.928 | 0.317 | 0.823 |
| 3-alpha(or 20-beta)-hydroxysteroid dehydrogenase | -2.016 | 0.910 | 0.028 | 0.780 |
| 3-aminobutyryl-CoA ammonia-lyase | 0.895 | 0.436 | 0.042 | 0.780 |
| 3-beta-hydroxy-Delta(5)-steroid dehydrogenase | -2.559 | 1.370 | 0.064 | 0.780 |
| 3-carboxy-cis,cis-muconate cycloisomerase | -0.185 | 0.361 | 0.609 | 0.960 |
| 3-carboxyethylcatechol 2,3-dioxygenase | -0.762 | 0.630 | 0.228 | 0.806 |
| 3-dehydro-glucose-6-phosphate–glutamate transaminase | -2.363 | 2.511 | 0.348 | 0.840 |
| 3-dehydro-L-gulonate-6-phosphate decarboxylase | 0.164 | 0.170 | 0.337 | 0.833 |
| 3-dehydro-L-gulonate 2-dehydrogenase | 0.505 | 0.552 | 0.362 | 0.844 |
| 3-dehydroquinate dehydratase | 0.007 | 0.023 | 0.758 | 0.999 |
| 3-dehydroquinate synthase | -0.037 | 0.038 | 0.329 | 0.829 |
| 3-dehydroquinate synthase II | 3.093 | 2.611 | 0.238 | 0.806 |
| 3-dehydroshikimate dehydratase | -0.291 | 0.432 | 0.501 | 0.911 |
| 3-dehydrosphinganine reductase | -0.417 | 1.284 | 0.746 | 0.999 |
| 3-demethylubiquinol 3-O-methyltransferase | -0.047 | 0.173 | 0.785 | 0.999 |
| 3-deoxy-7-phosphoheptulonate synthase | 0.032 | 0.045 | 0.482 | 0.897 |
| 3-deoxy-8-phosphooctulonate synthase | 0.084 | 0.088 | 0.342 | 0.837 |
| 3-deoxy-alpha-D-manno-octulosonate 8-oxidase | -0.089 | 1.300 | 0.946 | 0.999 |
| 3-deoxy-D-manno-octulosonic acid kinase | 0.129 | 0.266 | 0.628 | 0.971 |
| 3-deoxy-manno-octulosonate-8-phosphatase | 0.114 | 0.093 | 0.225 | 0.806 |
| 3-deoxy-manno-octulosonate cytidylyltransferase | 0.073 | 0.088 | 0.406 | 0.865 |
| 3-fumarylpyruvate hydrolase | -0.861 | 0.361 | 0.018 | 0.780 |
| 3-hexulose-6-phosphate synthase | -1.540 | 0.596 | 0.011 | 0.780 |
| 3-hydroxy-2-methylpyridine-4,5-dicarboxylate 4-decarboxylase | 17.690 | 2253.589 | 0.994 | 0.999 |
| 3-hydroxy-2-methylpyridinecarboxylate dioxygenase | 17.690 | 2253.589 | 0.994 | 0.999 |
| 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione monooxygenase | -0.940 | 0.739 | 0.205 | 0.806 |
| 3-hydroxy-D-aspartate aldolase | -0.858 | 1.279 | 0.503 | 0.913 |
| 3-hydroxy acid dehydrogenase | 0.207 | 0.278 | 0.459 | 0.884 |
| 3-hydroxyacyl-[acyl-carrier-protein] dehydratase | 0.038 | 0.033 | 0.247 | 0.806 |
| 3-hydroxyacyl-CoA dehydrogenase | -0.201 | 0.163 | 0.218 | 0.806 |
| 3-hydroxyanthranilate 3,4-dioxygenase | -0.131 | 0.452 | 0.772 | 0.999 |
| 3-hydroxybenzoate synthase | 0.141 | 0.803 | 0.861 | 0.999 |
| 3-hydroxybutyrate dehydrogenase | -0.297 | 0.213 | 0.165 | 0.806 |
| 3-hydroxybutyryl-CoA dehydratase | -0.438 | 0.333 | 0.190 | 0.806 |
| 3-hydroxybutyryl-CoA dehydrogenase | -0.046 | 0.160 | 0.774 | 0.999 |
| 3-hydroxybutyryl-CoA epimerase | -0.093 | 0.171 | 0.587 | 0.949 |
| 3-hydroxyisobutyrate dehydrogenase | -0.166 | 0.131 | 0.208 | 0.806 |
| 3-hydroxyisobutyryl-CoA hydrolase | -0.910 | 0.991 | 0.360 | 0.844 |
| 3-hydroxypropionyl-CoA dehydratase | 0.656 | 0.920 | 0.477 | 0.893 |
| 3-isopropylmalate dehydratase | -0.070 | 0.053 | 0.187 | 0.806 |
| 3-isopropylmalate dehydrogenase | -0.047 | 0.054 | 0.386 | 0.852 |
| 3-ketosteroid 9-alpha-monooxygenase | -1.190 | 0.897 | 0.187 | 0.806 |
| 3-mercaptopyruvate sulfurtransferase | -0.184 | 0.129 | 0.155 | 0.806 |
| 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) | -0.431 | 0.205 | 0.037 | 0.780 |
| 3-methyl-2-oxobutanoate hydroxymethyltransferase | -0.186 | 0.083 | 0.026 | 0.780 |
| 3-methylfumaryl-CoA hydratase | -1.049 | 0.554 | 0.060 | 0.780 |
| 3-oxo-5-alpha-steroid 4-dehydrogenase (acceptor) | -1.049 | 1.138 | 0.358 | 0.844 |
| 3-oxo-5-alpha-steroid 4-dehydrogenase (NADP(+)) | -1.091 | 0.666 | 0.103 | 0.806 |
| 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase | -0.081 | 0.318 | 0.801 | 0.999 |
| 3-oxo-5,6-didehydrosuberyl-CoA thiolase | -0.086 | 0.588 | 0.884 | 0.999 |
| 3-oxoacid CoA-transferase | -0.233 | 0.189 | 0.220 | 0.806 |
| 3-oxoacyl-[acyl-carrier-protein] reductase | -0.085 | 0.057 | 0.140 | 0.806 |
| 3-oxoadipate CoA-transferase | -0.283 | 0.355 | 0.427 | 0.865 |
| 3-oxoadipate enol-lactonase | -0.214 | 0.234 | 0.360 | 0.844 |
| 3-oxoadipyl-CoA thiolase | 0.045 | 0.327 | 0.892 | 0.999 |
| 3-oxosteroid 1-dehydrogenase | -0.919 | 0.814 | 0.261 | 0.820 |
| 3-phenylpropanoate dioxygenase | -0.931 | 0.928 | 0.317 | 0.823 |
| 3-phospho-methyltransferase | 2.793 | 1.233 | 0.025 | 0.780 |
| 3-phosphoshikimate 1-carboxyvinyltransferase | 0.013 | 0.034 | 0.706 | 0.999 |
| 3-phytase | -0.346 | 0.340 | 0.310 | 0.820 |
| 3(or 17)-beta-hydroxysteroid dehydrogenase | 0.051 | 0.785 | 0.949 | 0.999 |
| 3,4-dehydroadipyl-CoA semialdehyde dehydrogenase (NADP(+)) | -3.225 | 3.528 | 0.362 | 0.844 |
| 3,4-dihydroxy-2-butanone-4-phosphate synthase | 0.032 | 0.060 | 0.593 | 0.951 |
| 3,4-dihydroxyphenylacetate 2,3-dioxygenase | -0.740 | 0.585 | 0.208 | 0.806 |
| 3D-(3,5/4)-trihydroxycyclohexane-1,2-dione acylhydrolase (decyclizing) | -0.315 | 0.275 | 0.254 | 0.813 |
| 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase | 0.043 | 0.046 | 0.348 | 0.840 |
| 4-alpha-D-((1->4)-alpha-D-glucano)trehalose trehalohydrolase | -0.106 | 0.265 | 0.691 | 0.995 |
| 4-alpha-glucanotransferase | 0.081 | 0.084 | 0.332 | 0.831 |
| 4-alpha-N-acetylgalactosaminyltransferase | 0.519 | 0.593 | 0.383 | 0.852 |
| 4-aminobutyrate–2-oxoglutarate transaminase | -0.128 | 0.196 | 0.513 | 0.913 |
| 4-aminobutyrate–pyruvate transaminase | -1.332 | 0.795 | 0.096 | 0.806 |
| 4-carboxymuconolactone decarboxylase | -0.227 | 0.164 | 0.168 | 0.806 |
| 4-coumarate–CoA ligase | -0.715 | 0.966 | 0.461 | 0.884 |
| 4-coumaroyl-homoserine lactone synthase | -0.536 | 0.687 | 0.436 | 0.869 |
| 4-hydroxy-2-oxoglutarate aldolase | 1.593 | 1.203 | 0.188 | 0.806 |
| 4-hydroxy-2-oxoheptanedioate aldolase | -0.733 | 0.325 | 0.026 | 0.780 |
| 4-hydroxy-2-oxohexanoate aldolase | -0.027 | 0.516 | 0.958 | 0.999 |
| 4-hydroxy-2-oxovalerate aldolase | -0.264 | 0.346 | 0.448 | 0.875 |
| 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | 0.024 | 0.063 | 0.709 | 0.999 |
| 4-hydroxy-3-polyprenylbenzoate decarboxylase | 0.058 | 0.149 | 0.696 | 0.995 |
| 4-hydroxy-4-methyl-2-oxoglutarate aldolase | -0.859 | 0.503 | 0.090 | 0.806 |
| 4-hydroxy-tetrahydrodipicolinate reductase | -0.030 | 0.044 | 0.497 | 0.905 |
| 4-hydroxy-tetrahydrodipicolinate synthase | -0.042 | 0.029 | 0.150 | 0.806 |
| 4-hydroxyacetophenone monooxygenase | -1.901 | 0.815 | 0.021 | 0.780 |
| 4-hydroxybenzoate–CoA ligase | -1.369 | 1.141 | 0.232 | 0.806 |
| 4-hydroxybenzoate 3-monooxygenase | -0.216 | 0.272 | 0.428 | 0.866 |
| 4-hydroxybenzoate polyprenyltransferase | -0.010 | 0.092 | 0.911 | 0.999 |
| 4-hydroxybenzoyl-CoA reductase | 18.243 | 2971.595 | 0.995 | 0.999 |
| 4-hydroxybenzoyl-CoA thioesterase | -0.014 | 0.292 | 0.962 | 0.999 |
| 4-hydroxybutanoyl-CoA dehydratase | -0.529 | 0.433 | 0.224 | 0.806 |
| 4-hydroxybutyrate dehydrogenase | -0.041 | 0.415 | 0.922 | 0.999 |
| 4-hydroxymandelate oxidase | -0.320 | 0.799 | 0.690 | 0.995 |
| 4-hydroxyphenylacetate 3-monooxygenase | -0.791 | 0.527 | 0.136 | 0.806 |
| 4-hydroxyphenylacetate decarboxylase | 1.578 | 0.989 | 0.112 | 0.806 |
| 4-hydroxyphenylpyruvate dioxygenase | -0.096 | 0.214 | 0.654 | 0.973 |
| 4-hydroxyproline epimerase | -0.261 | 0.307 | 0.396 | 0.861 |
| 4-hydroxythreonine-4-phosphate dehydrogenase | -0.103 | 0.100 | 0.303 | 0.820 |
| 4-methylaminobutanoate oxidase (formaldehyde-forming) | -1.381 | 0.804 | 0.088 | 0.806 |
| 4-methylphenol dehydrogenase (hydroxylating) | 3.700 | 2.578 | 0.153 | 0.806 |
| 4-nitrophenylphosphatase | 0.149 | 0.130 | 0.254 | 0.813 |
| 4-O-beta-D-mannosyl-D-glucose phosphorylase | -0.952 | 0.434 | 0.030 | 0.780 |
| 4-oxalmesaconate hydratase | -1.282 | 0.546 | 0.020 | 0.780 |
| 4-oxalomesaconate tautomerase | -1.878 | 0.853 | 0.029 | 0.780 |
| 4-phosphoerythronate dehydrogenase | 0.005 | 0.197 | 0.981 | 0.999 |
| 4-phosphopantoate–beta-alanine ligase | -16.940 | 2657.012 | 0.995 | 0.999 |
| 4-phytase | -0.109 | 0.528 | 0.837 | 0.999 |
| 4-pyridoxolactonase | 17.690 | 2253.589 | 0.994 | 0.999 |
| 4,4’-diapophytoene desaturase (4,4’-diapolycopene-forming) | -1.552 | 1.123 | 0.169 | 0.806 |
| 4,4’-diapophytoene synthase | -0.812 | 0.993 | 0.415 | 0.865 |
| 4,5-dihydroxyphthalate decarboxylase | -1.578 | 1.081 | 0.146 | 0.806 |
| 4a-hydroxytetrahydrobiopterin dehydratase | -0.233 | 0.201 | 0.248 | 0.808 |
| 5’-deoxynucleotidase | 0.252 | 0.195 | 0.200 | 0.806 |
| 5’-nucleotidase | -0.089 | 0.061 | 0.148 | 0.806 |
| 5’-phosphate synthase | -0.196 | 0.480 | 0.683 | 0.992 |
| 5’ to 3’ exodeoxyribonuclease (nucleoside 3’-phosphate-forming) | 0.192 | 0.178 | 0.283 | 0.820 |
| 5-(carboxyamino)imidazole ribonucleotide mutase | 0.048 | 0.033 | 0.148 | 0.806 |
| 5-(carboxyamino)imidazole ribonucleotide synthase | 0.003 | 0.067 | 0.964 | 0.999 |
| 5-(hydroxymethyl)furfural oxidase | -0.505 | 0.813 | 0.536 | 0.924 |
| 5-amino-6-(5-phosphoribosylamino)uracil reductase | -0.004 | 0.058 | 0.951 | 0.999 |
| 5-aminolevulinate synthase | -1.208 | 0.445 | 0.007 | 0.780 |
| 5-aminovalerate transaminase | 0.212 | 0.348 | 0.542 | 0.927 |
| 5-carboxymethyl-2-hydroxymuconate Delta-isomerase | -0.551 | 0.354 | 0.122 | 0.806 |
| 5-carboxymethyl-2-hydroxymuconic-semialdehyde dehydrogenase | -0.914 | 0.395 | 0.022 | 0.780 |
| 5-dehydro-2-deoxygluconokinase | -0.338 | 0.274 | 0.220 | 0.806 |
| 5-dehydro-2-deoxyphosphogluconate aldolase | 0.071 | 0.761 | 0.926 | 0.999 |
| 5-dehydro-4-deoxy-D-glucuronate isomerase | 0.129 | 0.266 | 0.629 | 0.971 |
| 5-dehydro-4-deoxyglucarate dehydratase | -0.181 | 0.363 | 0.620 | 0.965 |
| 5-deoxy-glucuronate isomerase | -0.355 | 0.275 | 0.198 | 0.806 |
| 5-exo-hydroxycamphor dehydrogenase | -2.143 | 1.972 | 0.279 | 0.820 |
| 5-formyltetrahydrofolate cyclo-ligase | 0.074 | 0.039 | 0.060 | 0.780 |
| 5-guanidino-2-oxopentanoate decarboxylase | 0.806 | 0.618 | 0.194 | 0.806 |
| 5-methyltetrahydropteroyltriglutamate–homocysteine S-methyltransferase | 0.051 | 0.064 | 0.434 | 0.869 |
| 5-oxopent-3-ene-1,2,5-tricarboxylate decarboxylase | -0.449 | 0.471 | 0.342 | 0.837 |
| 5-oxoprolinase (ATP-hydrolyzing) | -1.400 | 0.730 | 0.057 | 0.780 |
| 5,10-methylenetetrahydromethanopterin reductase | 2.458 | 1.997 | 0.220 | 0.806 |
| 5,6-dimethylbenzimidazole synthase | -0.150 | 0.264 | 0.570 | 0.938 |
| 5,6,7,8-tetrahydromethanopterin hydro-lyase | 0.805 | 0.832 | 0.335 | 0.833 |
| 50S ribosomal protein L16 3-hydroxylase | 0.077 | 0.167 | 0.646 | 0.973 |
| 6-aminohexanoate-cyclic-dimer hydrolase | -1.553 | 1.072 | 0.150 | 0.806 |
| 6-aminohexanoate-oligomer exohydrolase | -0.946 | 0.459 | 0.041 | 0.780 |
| 6-carboxyhexanoate–CoA ligase | 0.062 | 0.248 | 0.802 | 0.999 |
| 6-carboxytetrahydropterin synthase | -0.112 | 0.065 | 0.089 | 0.806 |
| 6-hydroxynicotinate 3-monooxygenase | -1.023 | 0.794 | 0.200 | 0.806 |
| 6-hydroxypseudooxynicotine dehydrogenase | -2.143 | 1.972 | 0.279 | 0.820 |
| 6-methylsalicylic acid synthase | -18.427 | 2940.855 | 0.995 | 0.999 |
| 6-phospho-3-hexuloisomerase | -1.149 | 0.447 | 0.011 | 0.780 |
| 6-phospho-beta-galactosidase | -0.023 | 0.155 | 0.880 | 0.999 |
| 6-phospho-beta-glucosidase | 0.067 | 0.118 | 0.570 | 0.938 |
| 6-phosphofructokinase | 0.057 | 0.077 | 0.459 | 0.884 |
| 6-phosphogluconolactonase | 0.030 | 0.064 | 0.644 | 0.973 |
| 6-pyruvoyltetrahydropterin 2’-reductase | 0.500 | 0.628 | 0.427 | 0.865 |
| 6-pyruvoyltetrahydropterin synthase | -0.112 | 0.065 | 0.089 | 0.806 |
| 6,7-dihydropteridine reductase | 0.152 | 0.235 | 0.518 | 0.913 |
| 6,7-dimethyl-8-ribityllumazine synthase | -0.028 | 0.057 | 0.621 | 0.966 |
| 7-alpha-hydroxysteroid dehydrogenase | -0.487 | 0.462 | 0.293 | 0.820 |
| 7-carboxy-7-deazaguanine synthase | -0.110 | 0.094 | 0.242 | 0.806 |
| 7-cyano-7-deazaguanine synthase | -0.103 | 0.062 | 0.098 | 0.806 |
| 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase | -16.940 | 2657.012 | 0.995 | 0.999 |
| 8-amino-3,8-dideoxy-alpha-D-manno-octulosonate transaminase | 0.420 | 1.214 | 0.730 | 0.999 |
| 8-amino-7-oxononanoate synthase | -0.031 | 0.121 | 0.795 | 0.999 |
| 8-oxo-dGTP diphosphatase | -0.016 | 0.080 | 0.842 | 0.999 |
| 8-oxoguanine deaminase | -0.092 | 0.440 | 0.834 | 0.999 |
| 9,9’-di-cis-zeta-carotene desaturase | 18.195 | 2901.073 | 0.995 | 0.999 |
| Abequosyltransferase | 0.936 | 1.152 | 0.418 | 0.865 |
| Acetaldehyde dehydrogenase (acetylating) | 0.076 | 0.096 | 0.433 | 0.869 |
| Acetate–CoA ligase | -0.272 | 0.139 | 0.053 | 0.780 |
| Acetate–CoA ligase (ADP-forming) | -2.906 | 3.063 | 0.344 | 0.838 |
| Acetate CoA-transferase | 0.819 | 0.377 | 0.031 | 0.780 |
| Acetate kinase | 0.078 | 0.063 | 0.220 | 0.806 |
| Acetoacetate–CoA ligase | -0.278 | 0.291 | 0.341 | 0.837 |
| Acetoacetate decarboxylase | -1.256 | 0.740 | 0.092 | 0.806 |
| Acetoacetyl-CoA reductase | -0.204 | 0.338 | 0.547 | 0.930 |
| Acetolactate decarboxylase | 0.074 | 0.135 | 0.584 | 0.948 |
| Acetolactate synthase | -0.002 | 0.040 | 0.968 | 0.999 |
| Acetone carboxylase | -1.396 | 1.560 | 0.372 | 0.852 |
| Acetyl-CoA C-acetyltransferase | -0.154 | 0.115 | 0.181 | 0.806 |
| Acetyl-CoA C-acyltransferase | -0.186 | 0.170 | 0.277 | 0.820 |
| Acetyl-CoA carboxylase | 0.005 | 0.044 | 0.905 | 0.999 |
| Acetyl-CoA hydrolase | -1.517 | 1.943 | 0.436 | 0.869 |
| Acetyl-S-ACP:malonate ACP transferase | 0.053 | 0.267 | 0.843 | 0.999 |
| acetylgalactosaminyltransferase | 0.500 | 0.628 | 0.427 | 0.865 |
| acetylglucosaminyltransferase | 0.034 | 0.028 | 0.219 | 0.806 |
| Acetylglutamate kinase | -0.070 | 0.056 | 0.219 | 0.806 |
| Acetylornithine deacetylase | -0.042 | 0.161 | 0.793 | 0.999 |
| Acetylornithine transaminase | -0.081 | 0.068 | 0.232 | 0.806 |
| Acid phosphatase | -0.022 | 0.170 | 0.897 | 0.999 |
| Acireductone dioxygenase (Fe(2+)-requiring) | -0.021 | 0.249 | 0.932 | 0.999 |
| Acireductone dioxygenase (Ni(2+)-requiring) | -0.021 | 0.249 | 0.932 | 0.999 |
| Acireductone synthase | 0.195 | 0.330 | 0.556 | 0.936 |
| Aclacinomycin-A oxidase | -17.411 | 3363.343 | 0.996 | 0.999 |
| Aclacinomycin-N oxidase | -17.411 | 3363.343 | 0.996 | 0.999 |
| Aclacinomycin methylesterase | 18.543 | 3451.904 | 0.996 | 0.999 |
| Aconitate hydratase | -0.106 | 0.104 | 0.309 | 0.820 |
| Acyl-[acyl-carrier-protein]–phospholipid O-acyltransferase | 0.540 | 0.266 | 0.044 | 0.780 |
| Acyl-[acyl-carrier-protein]–UDP-N-acetylglucosamine O-acyltransferase | 0.021 | 0.097 | 0.831 | 0.999 |
| Acyl-CoA 6-desaturase | -0.301 | 0.316 | 0.343 | 0.837 |
| Acyl-CoA hydrolase | -0.160 | 0.581 | 0.784 | 0.999 |
| Acyl-CoA oxidase | -0.742 | 0.485 | 0.128 | 0.806 |
| Acyl-homoserine-lactone acylase | -0.028 | 0.352 | 0.937 | 0.999 |
| Acyl-homoserine-lactone synthase | -0.080 | 0.423 | 0.850 | 0.999 |
| Acylaminoacyl-peptidase | 0.638 | 0.391 | 0.105 | 0.806 |
| Acylphosphatase | 0.068 | 0.088 | 0.440 | 0.869 |
| Acylpyruvate hydrolase | 0.213 | 0.367 | 0.563 | 0.936 |
| Adenine deaminase | -0.252 | 0.268 | 0.349 | 0.840 |
| Adenine phosphoribosyltransferase | 0.095 | 0.048 | 0.050 | 0.780 |
| Adenosine deaminase | 0.043 | 0.139 | 0.757 | 0.999 |
| Adenosine kinase | -0.269 | 0.307 | 0.383 | 0.852 |
| Adenosylcobalamin/alpha-ribazole phosphatase | 0.056 | 0.105 | 0.594 | 0.951 |
| Adenosylcobinamide-GDP ribazoletransferase | 0.061 | 0.104 | 0.559 | 0.936 |
| Adenosylcobinamide-phosphate guanylyltransferase | 0.052 | 0.108 | 0.629 | 0.971 |
| Adenosylcobinamide-phosphate synthase | 0.105 | 0.111 | 0.350 | 0.840 |
| Adenosylcobinamide kinase | 0.053 | 0.108 | 0.628 | 0.971 |
| Adenosylcobyric acid synthase (glutamine-hydrolyzing) | 0.243 | 0.134 | 0.073 | 0.787 |
| Adenosylhomocysteinase | -0.060 | 0.123 | 0.628 | 0.971 |
| Adenosylhomocysteine nucleosidase | 0.026 | 0.061 | 0.671 | 0.982 |
| Adenosylmethionine–8-amino-7-oxononanoate transaminase | 0.014 | 0.090 | 0.876 | 0.999 |
| Adenosylmethionine decarboxylase | 0.053 | 0.136 | 0.696 | 0.995 |
| Adenylate cyclase | -0.190 | 0.125 | 0.130 | 0.806 |
| Adenylate kinase | 0.027 | 0.028 | 0.336 | 0.833 |
| Adenylosuccinate lyase | 0.040 | 0.032 | 0.208 | 0.806 |
| Adenylosuccinate synthase | 0.034 | 0.027 | 0.215 | 0.806 |
| Adenylyl-sulfate kinase | -0.422 | 0.222 | 0.059 | 0.780 |
| Adenylyl-sulfate reductase | 0.015 | 0.503 | 0.976 | 0.999 |
| ADP-dependent NAD(P)H-hydrate dehydratase | 0.366 | 0.476 | 0.442 | 0.870 |
| ADP-glyceromanno-heptose 6-epimerase | 0.284 | 0.149 | 0.059 | 0.780 |
| ADP-ribose diphosphatase | 0.135 | 0.049 | 0.007 | 0.780 |
| ADP-ribosyl-[dinitrogen reductase] hydrolase | 0.349 | 0.478 | 0.467 | 0.884 |
| ADP-specific glucokinase | 0.413 | 0.829 | 0.619 | 0.965 |
| ADP-specific phosphofructokinase | 0.413 | 0.829 | 0.619 | 0.965 |
| Aerobactin synthase | 0.697 | 0.786 | 0.377 | 0.852 |
| Agmatinase | -0.082 | 0.180 | 0.650 | 0.973 |
| Agmatine deiminase | -0.111 | 0.142 | 0.435 | 0.869 |
| Aklaviketone reductase | -1.712 | 1.754 | 0.330 | 0.829 |
| Alanine–glyoxylate transaminase | -1.131 | 0.626 | 0.073 | 0.787 |
| Alanine–tRNA ligase | 0.055 | 0.036 | 0.129 | 0.806 |
| Alanine dehydrogenase | -0.299 | 0.110 | 0.007 | 0.780 |
| Alanine racemase | 0.073 | 0.036 | 0.045 | 0.780 |
| Alanine transaminase | -0.014 | 0.086 | 0.871 | 0.999 |
| Alcohol dehydrogenase | -0.079 | 0.065 | 0.232 | 0.806 |
| Alcohol dehydrogenase (azurin) | -1.210 | 0.669 | 0.073 | 0.787 |
| Alcohol dehydrogenase (cytochrome c) | 0.461 | 0.349 | 0.188 | 0.806 |
| Alcohol dehydrogenase (NADP(+)) | 1.256 | 0.843 | 0.138 | 0.806 |
| Aldehyde dehydrogenase (FAD-independent) | 0.221 | 1.104 | 0.842 | 0.999 |
| Aldehyde dehydrogenase (NAD(+)) | -0.366 | 0.158 | 0.022 | 0.780 |
| Aldehyde dehydrogenase (NAD(P)(+)) | -0.933 | 0.959 | 0.332 | 0.831 |
| Aldehyde dehydrogenase (NADP(+)) | -0.047 | 0.309 | 0.880 | 0.999 |
| Aldehyde ferredoxin oxidoreductase | 0.227 | 0.717 | 0.752 | 0.999 |
| Aldehyde oxygenase (deformylating) | -0.995 | 1.007 | 0.325 | 0.827 |
| Alditol oxidase | 0.782 | 0.736 | 0.290 | 0.820 |
| Aldose 1-epimerase | 0.004 | 0.062 | 0.954 | 0.999 |
| Aliphatic aldoxime dehydratase | -1.102 | 0.874 | 0.209 | 0.806 |
| Aliphatic nitrilase | -0.592 | 0.716 | 0.410 | 0.865 |
| Alkaline phosphatase | -0.291 | 0.191 | 0.129 | 0.806 |
| Alkanal monooxygenase (FMN-linked) | 18.315 | 2531.149 | 0.994 | 0.999 |
| Alkane 1-monooxygenase | -0.478 | 0.428 | 0.265 | 0.820 |
| Alkanesulfonate monooxygenase | -0.314 | 0.265 | 0.239 | 0.806 |
| Alkylglycerone-phosphate synthase | -0.968 | 0.638 | 0.131 | 0.806 |
| Alkylmercury lyase | 0.896 | 1.012 | 0.377 | 0.852 |
| All-trans-8’-apo-beta-carotenal 15,15’-oxygenase | -17.083 | 2854.434 | 0.995 | 0.999 |
| All-trans-octaprenyl-diphosphate synthase | -0.075 | 0.090 | 0.406 | 0.865 |
| All-trans-retinol 13,14-reductase | -0.567 | 0.421 | 0.180 | 0.806 |
| All-trans-retinol dehydrogenase (NAD(+)) | 0.807 | 1.421 | 0.571 | 0.938 |
| All-trans-zeta-carotene desaturase | -0.177 | 0.395 | 0.654 | 0.973 |
| Allantoate deiminase | -0.599 | 0.458 | 0.193 | 0.806 |
| Allantoicase | -0.708 | 0.405 | 0.082 | 0.806 |
| Allantoin racemase | -0.340 | 0.358 | 0.344 | 0.838 |
| Allantoinase | -0.364 | 0.342 | 0.288 | 0.820 |
| Allophanate hydrolase | -0.083 | 0.292 | 0.777 | 0.999 |
| Allose kinase | 0.004 | 1.317 | 0.997 | 0.999 |
| Alpha-amylase | -0.090 | 0.088 | 0.311 | 0.820 |
| Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P-lyase | -0.967 | 0.453 | 0.034 | 0.780 |
| Alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase | -1.068 | 0.409 | 0.010 | 0.780 |
| Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase | -0.963 | 0.453 | 0.035 | 0.780 |
| Alpha-D-xyloside xylohydrolase | -0.514 | 0.327 | 0.118 | 0.806 |
| Alpha-galactosidase | 0.044 | 0.134 | 0.743 | 0.999 |
| Alpha-glucosidase | -0.024 | 0.131 | 0.853 | 0.999 |
| Alpha-glucuronidase | -0.650 | 0.392 | 0.100 | 0.806 |
| Alpha-L-fucosidase | -0.165 | 0.142 | 0.245 | 0.806 |
| Alpha-L-rhamnosidase | -0.688 | 0.416 | 0.100 | 0.806 |
| Alpha-mannosidase | 0.037 | 0.182 | 0.838 | 0.999 |
| Alpha-methylacyl-CoA racemase | -0.912 | 0.447 | 0.043 | 0.780 |
| Alpha-N-acetylglucosaminidase | -0.939 | 0.572 | 0.103 | 0.806 |
| Alpha,alpha-phosphotrehalase | 0.102 | 0.123 | 0.408 | 0.865 |
| Alpha,alpha-trehalase | -0.366 | 0.447 | 0.415 | 0.865 |
| Alpha,alpha-trehalose-phosphate synthase (UDP-forming) | -0.432 | 0.248 | 0.083 | 0.806 |
| Alpha,alpha-trehalose phosphorylase | 0.444 | 0.511 | 0.386 | 0.852 |
| Alpha,alpha-trehalose synthase | -0.511 | 0.503 | 0.311 | 0.820 |
| Altronate dehydratase | 0.049 | 0.168 | 0.772 | 0.999 |
| Amidase | -0.168 | 0.163 | 0.305 | 0.820 |
| Amidophosphoribosyltransferase | 0.022 | 0.033 | 0.513 | 0.913 |
| Amino-acid N-acetyltransferase | -0.067 | 0.104 | 0.520 | 0.913 |
| Aminoacyl-tRNA hydrolase | 0.039 | 0.031 | 0.211 | 0.806 |
| Aminobutyraldehyde dehydrogenase | -0.224 | 0.402 | 0.579 | 0.946 |
| Aminocarboxymuconate-semialdehyde decarboxylase | -1.199 | 0.606 | 0.050 | 0.780 |
| Aminodeoxychorismate lyase | -0.025 | 0.069 | 0.722 | 0.999 |
| Aminodeoxychorismate synthase | -0.004 | 0.073 | 0.957 | 0.999 |
| Aminodeoxyfutalosine deaminase | -0.276 | 0.951 | 0.772 | 0.999 |
| Aminodeoxyfutalosine nucleosidase | -0.427 | 0.810 | 0.599 | 0.953 |
| Aminodeoxyfutalosine synthase | 0.214 | 0.461 | 0.643 | 0.973 |
| Aminoglycoside 6’-N-acetyltransferase | -0.190 | 0.353 | 0.591 | 0.951 |
| Aminoglycoside N(3’)-acetyltransferase | 0.168 | 0.355 | 0.636 | 0.973 |
| Aminomethyltransferase | -0.161 | 0.115 | 0.163 | 0.806 |
| Aminomuconate-semialdehyde dehydrogenase | -0.420 | 0.663 | 0.528 | 0.918 |
| Aminopeptidase B | -1.300 | 1.072 | 0.227 | 0.806 |
| Aminopeptidase S | 0.679 | 1.010 | 0.502 | 0.912 |
| Aminopyrimidine aminohydrolase | -0.189 | 0.125 | 0.132 | 0.806 |
| Ammonia monooxygenase | -17.712 | 2757.136 | 0.995 | 0.999 |
| AMP nucleosidase | -0.162 | 0.198 | 0.414 | 0.865 |
| Amylosucrase | -0.270 | 0.337 | 0.425 | 0.865 |
| Anhydro-N-acetylmuramic acid kinase | -0.042 | 0.150 | 0.778 | 0.999 |
| Anthranilate–CoA ligase | -0.942 | 0.604 | 0.121 | 0.806 |
| Anthranilate 1,2-dioxygenase (deaminating, decarboxylating) | -0.697 | 0.594 | 0.242 | 0.806 |
| Anthranilate 3-monooxygenase (FAD) | -0.129 | 1.009 | 0.898 | 0.999 |
| Anthranilate phosphoribosyltransferase | -0.068 | 0.054 | 0.204 | 0.806 |
| Anthranilate synthase | -0.080 | 0.051 | 0.116 | 0.806 |
| Anthraniloyl-CoA monooxygenase | -0.683 | 0.495 | 0.169 | 0.806 |
| Aquacobalamin reductase | 0.377 | 0.522 | 0.471 | 0.888 |
| Arabinan endo-1,5-alpha-L-arabinosidase | -0.668 | 0.458 | 0.147 | 0.806 |
| Arabinofuranan 3-O-arabinosyltransferase | -0.735 | 0.667 | 0.272 | 0.820 |
| Arabinogalactan endo-beta-1,4-galactanase | -0.559 | 0.328 | 0.091 | 0.806 |
| Arabinonate dehydratase | -2.566 | 3.464 | 0.460 | 0.884 |
| Arabinose-5-phosphate isomerase | 0.085 | 0.086 | 0.325 | 0.827 |
| Arachidonate 15-lipoxygenase | 18.299 | 3055.837 | 0.995 | 0.999 |
| Archaetidylinositol phosphate synthase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Arginase | -0.253 | 0.181 | 0.163 | 0.806 |
| Arginine–pyruvate transaminase | -0.026 | 0.442 | 0.952 | 0.999 |
| Arginine–tRNA ligase | 0.027 | 0.031 | 0.384 | 0.852 |
| Arginine decarboxylase | -0.071 | 0.106 | 0.506 | 0.913 |
| Arginine deiminase | 0.139 | 0.108 | 0.201 | 0.806 |
| Arginine N-succinyltransferase | 0.067 | 0.274 | 0.807 | 0.999 |
| Argininosuccinate lyase | -0.044 | 0.051 | 0.383 | 0.852 |
| Argininosuccinate synthase | 0.000 | 0.051 | 0.992 | 0.999 |
| Arginyltransferase | -0.113 | 0.202 | 0.574 | 0.941 |
| Arogenate dehydratase | -0.894 | 0.486 | 0.068 | 0.780 |
| Arogenate dehydrogenase | -1.345 | 0.486 | 0.006 | 0.780 |
| Aromatic-amino-acid transaminase | 0.183 | 0.120 | 0.130 | 0.806 |
| Aromatic-L-amino-acid decarboxylase | 1.029 | 0.514 | 0.047 | 0.780 |
| Arsenate-mycothiol transferase | -0.494 | 0.726 | 0.497 | 0.905 |
| Arsenate reductase (azurin) | 17.978 | 2602.989 | 0.994 | 0.999 |
| Arsenate reductase (glutaredoxin) | 0.034 | 0.041 | 0.418 | 0.865 |
| Arsenite-transporting ATPase | 0.613 | 0.424 | 0.150 | 0.806 |
| Aryl-alcohol dehydrogenase | 0.074 | 0.334 | 0.824 | 0.999 |
| Aryl-alcohol dehydrogenase (NADP(+)) | -1.167 | 0.646 | 0.073 | 0.787 |
| Aryl-sulfate sulfotransferase | -0.931 | 0.951 | 0.329 | 0.829 |
| Arylamine N-acetyltransferase | -2.468 | 1.417 | 0.083 | 0.806 |
| Arylformamidase | -1.004 | 0.368 | 0.007 | 0.780 |
| Arylsulfatase | -0.463 | 0.232 | 0.048 | 0.780 |
| Asparaginase | 0.045 | 0.040 | 0.262 | 0.820 |
| Asparagine–tRNA ligase | 0.072 | 0.076 | 0.341 | 0.837 |
| Asparagine synthase (glutamine-hydrolyzing) | -0.154 | 0.159 | 0.334 | 0.832 |
| Asparaginyl-tRNA synthase (glutamine-hydrolyzing) | 0.004 | 0.047 | 0.937 | 0.999 |
| Aspartate–ammonia ligase | 0.146 | 0.120 | 0.224 | 0.806 |
| Aspartate–prephenate aminotransferase | -0.787 | 1.153 | 0.496 | 0.905 |
| Aspartate–tRNA ligase | 0.046 | 0.041 | 0.264 | 0.820 |
| Aspartate–tRNA(Asn) ligase | -0.393 | 0.772 | 0.611 | 0.960 |
| Aspartate-semialdehyde dehydrogenase | 0.000 | 0.034 | 0.990 | 0.999 |
| Aspartate 1-decarboxylase | -0.227 | 0.110 | 0.041 | 0.780 |
| Aspartate 4-decarboxylase | 0.302 | 0.425 | 0.478 | 0.895 |
| Aspartate ammonia-lyase | -0.041 | 0.102 | 0.690 | 0.995 |
| Aspartate carbamoyltransferase | 0.035 | 0.034 | 0.313 | 0.820 |
| Aspartate dehydrogenase | -0.578 | 0.420 | 0.171 | 0.806 |
| Aspartate kinase | -0.040 | 0.039 | 0.302 | 0.820 |
| Aspartate racemase | 0.152 | 0.172 | 0.377 | 0.852 |
| Aspartate transaminase | -0.090 | 0.113 | 0.427 | 0.865 |
| Aspartoacylase | 18.622 | 2950.870 | 0.995 | 0.999 |
| Aspartyl aminopeptidase | 0.293 | 0.119 | 0.015 | 0.780 |
| Aspergillus nuclease S(1) | -2.219 | 2.106 | 0.294 | 0.820 |
| Assimilatory sulfite reductase (ferredoxin) | -0.340 | 0.314 | 0.280 | 0.820 |
| Assimilatory sulfite reductase (NADPH) | -0.068 | 0.146 | 0.644 | 0.973 |
| ATP adenylyltransferase | 0.044 | 0.223 | 0.845 | 0.999 |
| ATP diphosphatase | -0.020 | 0.179 | 0.911 | 0.999 |
| ATP phosphoribosyltransferase | -0.085 | 0.059 | 0.151 | 0.806 |
| Aureolysin | -0.476 | 0.680 | 0.486 | 0.899 |
| Azobenzene reductase | -1.989 | 1.867 | 0.288 | 0.820 |
| Bacillolysin | -17.434 | 2863.190 | 0.995 | 0.999 |
| Bacterial leucyl aminopeptidase | -0.885 | 0.657 | 0.180 | 0.806 |
| Bacterial non-heme ferritin | -0.006 | 0.146 | 0.969 | 0.999 |
| Benzaldehyde dehydrogenase (NAD(+)) | -0.224 | 0.346 | 0.519 | 0.913 |
| Benzil reductase ((S)-benzoin forming) | -0.935 | 0.938 | 0.320 | 0.826 |
| Benzoate–CoA ligase | -1.805 | 0.983 | 0.068 | 0.780 |
| Benzoate 1,2-dioxygenase | 0.191 | 0.318 | 0.548 | 0.930 |
| Benzoyl-CoA-dihydrodiol lyase | -1.691 | 0.937 | 0.073 | 0.787 |
| Benzoylformate decarboxylase | -0.935 | 0.706 | 0.188 | 0.806 |
| Beta-1,4-mannooligosaccharide phosphorylase | -0.324 | 0.229 | 0.160 | 0.806 |
| Beta-alanine–pyruvate transaminase | -0.111 | 0.232 | 0.634 | 0.973 |
| Beta-aspartyl-peptidase | -0.285 | 0.310 | 0.360 | 0.844 |
| Beta-carotene 3-hydroxylase | 0.052 | 0.344 | 0.879 | 0.999 |
| Beta-fructofuranosidase | 0.140 | 0.103 | 0.174 | 0.806 |
| Beta-galactosidase | 0.008 | 0.106 | 0.939 | 0.999 |
| Beta-glucosidase | -0.177 | 0.137 | 0.196 | 0.806 |
| Beta-glucoside kinase | -0.225 | 0.990 | 0.820 | 0.999 |
| Beta-glucuronidase | 0.100 | 0.225 | 0.656 | 0.974 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase I | -0.111 | 0.142 | 0.436 | 0.869 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase II | 0.006 | 0.034 | 0.857 | 0.999 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase III | 0.065 | 0.040 | 0.105 | 0.806 |
| Beta-ketodecanoyl-[acyl-carrier-protein] synthase | 0.042 | 0.264 | 0.872 | 0.999 |
| Beta-lactamase | -0.027 | 0.069 | 0.690 | 0.995 |
| Beta-lysine 5,6-aminomutase | 0.979 | 0.456 | 0.033 | 0.780 |
| Beta-mannosidase | -0.387 | 0.296 | 0.193 | 0.806 |
| Beta-N-acetylhexosaminidase | -0.088 | 0.074 | 0.235 | 0.806 |
| Beta-phosphoglucomutase | 0.446 | 0.290 | 0.126 | 0.806 |
| Betaine–homocysteine S-methyltransferase | -17.840 | 4168.155 | 0.997 | 0.999 |
| Betaine-aldehyde dehydrogenase | -0.183 | 0.191 | 0.340 | 0.837 |
| Betaine reductase | 0.130 | 0.456 | 0.776 | 0.999 |
| Bilirubin oxidase | -3.253 | 3.540 | 0.360 | 0.844 |
| Biotin–[acetyl-CoA-carboxylase] ligase | 0.045 | 0.033 | 0.177 | 0.806 |
| Biotin carboxylase | -0.045 | 0.048 | 0.351 | 0.842 |
| Biotin synthase | 0.056 | 0.080 | 0.484 | 0.899 |
| Bis(5’-nucleosyl)-tetraphosphatase (asymmetrical) | 2.415 | 2.069 | 0.245 | 0.806 |
| Bis(5’-nucleosyl)-tetraphosphatase (symmetrical) | 0.067 | 0.159 | 0.674 | 0.984 |
| Bleomycin hydrolase | 0.000 | 0.113 | 0.999 | 0.999 |
| Bontoxilysin | -17.546 | 2539.415 | 0.994 | 0.999 |
| Branched-chain-amino-acid transaminase | -0.038 | 0.044 | 0.383 | 0.852 |
| Butyrate–acetoacetate CoA-transferase | 0.838 | 0.381 | 0.029 | 0.780 |
| Butyrate–CoA ligase | -1.862 | 2.295 | 0.418 | 0.865 |
| Butyrate kinase | -0.234 | 0.234 | 0.319 | 0.824 |
| C-terminal processing peptidase | -0.042 | 0.065 | 0.525 | 0.916 |
| C5a peptidase | 0.064 | 1.026 | 0.950 | 0.999 |
| Cadmium-exporting ATPase | 0.114 | 0.089 | 0.203 | 0.806 |
| Caffeoyl-CoA O-methyltransferase | 0.962 | 0.445 | 0.032 | 0.780 |
| Calcium-transporting ATPase | 0.041 | 0.113 | 0.717 | 0.999 |
| Carbamate kinase | 0.122 | 0.117 | 0.296 | 0.820 |
| Carbamoyl-phosphate synthase (glutamine-hydrolyzing) | 0.018 | 0.049 | 0.719 | 0.999 |
| Carbazole 1,9a-dioxygenase | 1.546 | 1.393 | 0.269 | 0.820 |
| Carbon-monoxide dehydrogenase (acceptor) | -0.180 | 0.193 | 0.353 | 0.843 |
| Carbon-monoxide dehydrogenase (ferredoxin) | 0.317 | 0.239 | 0.187 | 0.806 |
| Carbonate dehydratase | -0.059 | 0.058 | 0.317 | 0.823 |
| Carboxylesterase | -0.065 | 0.205 | 0.754 | 0.999 |
| Carboxymethylenebutenolidase | -0.355 | 0.223 | 0.113 | 0.806 |
| Carboxymethylproline synthase | -2.143 | 1.972 | 0.279 | 0.820 |
| Carboxynorspermidine decarboxylase | -0.220 | 0.117 | 0.063 | 0.780 |
| Carboxynorspermidine synthase | 1.036 | 1.404 | 0.462 | 0.884 |
| Carboxypeptidase T | 18.543 | 3451.904 | 0.996 | 0.999 |
| Carboxypeptidase Taq | 0.232 | 0.190 | 0.224 | 0.806 |
| Cardiolipin synthase (CMP-forming) | -0.608 | 0.256 | 0.019 | 0.780 |
| Carnitine 3-dehydrogenase | 0.073 | 0.361 | 0.839 | 0.999 |
| Carnitine O-acetyltransferase | -0.301 | 0.965 | 0.756 | 0.999 |
| Carotenoid 1,2-hydratase | -2.615 | 3.349 | 0.436 | 0.869 |
| Catalase | -0.178 | 0.164 | 0.280 | 0.820 |
| Catalase peroxidase | -0.167 | 0.233 | 0.474 | 0.889 |
| Catechol 1,2-dioxygenase | -0.080 | 0.260 | 0.760 | 0.999 |
| Catechol 2,3-dioxygenase | -0.204 | 0.145 | 0.160 | 0.806 |
| Catechol O-methyltransferase | -2.569 | 1.387 | 0.066 | 0.780 |
| CCA tRNA nucleotidyltransferase | 0.113 | 0.051 | 0.027 | 0.780 |
| CDP-4-dehydro-6-deoxyglucose reductase | 0.077 | 0.231 | 0.738 | 0.999 |
| CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase | 0.095 | 0.044 | 0.034 | 0.780 |
| CDP-diacylglycerol–inositol 3-phosphatidyltransferase | -2.259 | 1.730 | 0.194 | 0.806 |
| CDP-diacylglycerol–serine O-phosphatidyltransferase | -0.022 | 0.064 | 0.732 | 0.999 |
| CDP-diacylglycerol diphosphatase | -0.091 | 0.379 | 0.809 | 0.999 |
| CDP-glucose 4,6-dehydratase | 0.174 | 0.303 | 0.566 | 0.936 |
| CDP-glycerol glycerophosphotransferase | -0.514 | 0.418 | 0.221 | 0.806 |
| CDP-L-myo-inositol myo-inositolphosphotransferase | -17.501 | 3517.810 | 0.996 | 0.999 |
| CDP-paratose 2-epimerase | -1.712 | 0.902 | 0.059 | 0.780 |
| CDP-ribitol ribitolphosphotransferase | -1.091 | 0.808 | 0.179 | 0.806 |
| Cellobionic acid phosphorylase | -0.770 | 1.385 | 0.579 | 0.946 |
| Cellobiose phosphorylase | -0.428 | 0.679 | 0.530 | 0.920 |
| Cellulase | -0.530 | 0.224 | 0.019 | 0.780 |
| Cellulose 1,4-beta-cellobiosidase (non-reducing end) | 0.652 | 1.214 | 0.592 | 0.951 |
| Cellulose synthase (UDP-forming) | -0.112 | 0.273 | 0.683 | 0.992 |
| Cephalosporin-C deacetylase | -0.013 | 0.195 | 0.945 | 0.999 |
| Ceramidase | 0.084 | 0.349 | 0.809 | 0.999 |
| Ceramide glucosyltransferase | 1.275 | 0.567 | 0.026 | 0.780 |
| Cerebroside-sulfatase | 1.046 | 1.272 | 0.412 | 0.865 |
| Chitin deacetylase | -18.294 | 2578.334 | 0.994 | 0.999 |
| Chitin disaccharide deacetylase | -0.284 | 0.373 | 0.448 | 0.875 |
| Chitinase | -0.193 | 0.365 | 0.597 | 0.952 |
| Chitosanase | -17.909 | 3081.137 | 0.995 | 0.999 |
| Chloramphenicol O-acetyltransferase | -0.476 | 0.320 | 0.139 | 0.806 |
| Chloride peroxidase | -0.120 | 0.265 | 0.652 | 0.973 |
| Chlorite O(2)-lyase | -0.182 | 0.177 | 0.307 | 0.820 |
| Chlorophyll synthase | -17.728 | 2688.993 | 0.995 | 0.999 |
| Chlorophyllide a reductase | -3.248 | 3.287 | 0.325 | 0.827 |
| Cholest-4-en-3-one 26-monooxygenase | -2.002 | 1.431 | 0.164 | 0.806 |
| Cholesterol oxidase | -1.014 | 0.837 | 0.228 | 0.806 |
| Choline-phosphate cytidylyltransferase | -0.271 | 0.419 | 0.518 | 0.913 |
| Choline-sulfatase | 0.094 | 0.331 | 0.776 | 0.999 |
| Choline dehydrogenase | -0.152 | 0.205 | 0.459 | 0.884 |
| Choline monooxygenase | 0.189 | 0.462 | 0.683 | 0.992 |
| Choline oxidase | 1.209 | 1.153 | 0.296 | 0.820 |
| Choloylglycine hydrolase | 0.073 | 0.256 | 0.776 | 0.999 |
| Chondroitin-sulfate-ABC endolyase | -0.644 | 0.500 | 0.200 | 0.806 |
| Chondroitin-sulfate-ABC exolyase | -0.644 | 0.500 | 0.200 | 0.806 |
| Chondroitin AC lyase | -0.286 | 0.667 | 0.668 | 0.979 |
| Chorismate dehydratase | 0.031 | 0.427 | 0.943 | 0.999 |
| Chorismate lyase | 0.190 | 0.218 | 0.384 | 0.852 |
| Chorismate mutase | -0.009 | 0.045 | 0.840 | 0.999 |
| Chorismate synthase | 0.019 | 0.036 | 0.607 | 0.960 |
| Cinnamoyl-CoA:phenyllactate CoA-transferase | 0.927 | 0.699 | 0.187 | 0.806 |
| Cis-3,4-dihydrophenanthrene-3,4-diol dehydrogenase | 0.936 | 1.152 | 0.418 | 0.865 |
| Citrate–CoA ligase | -1.093 | 1.677 | 0.515 | 0.913 |
| Citrate (Si)-synthase | -0.229 | 0.084 | 0.007 | 0.780 |
| Citrate CoA-transferase | 0.495 | 0.189 | 0.010 | 0.780 |
| Citrate lyase holo-[acyl-carrier protein] synthase | 0.713 | 0.334 | 0.034 | 0.780 |
| Citryl-CoA lyase | 0.024 | 0.137 | 0.863 | 0.999 |
| Clostripain | 6.303 | 12.523 | 0.615 | 0.963 |
| CMP-N,N’-diacetyllegionaminic acid synthase | -0.462 | 0.673 | 0.494 | 0.905 |
| CO-methylating acetyl-CoA synthase | -0.728 | 1.131 | 0.521 | 0.913 |
| Co-methyltransferase | -0.766 | 1.103 | 0.488 | 0.901 |
| CoA-disulfide reductase | -0.200 | 0.826 | 0.809 | 0.999 |
| CoA:oxalate CoA-transferase | 0.066 | 0.507 | 0.896 | 0.999 |
| CoB–CoM heterodisulfide reductase | -0.006 | 0.480 | 0.989 | 0.999 |
| Cob(I)yrinic acid a,c-diamide adenosyltransferase | 0.142 | 0.097 | 0.145 | 0.806 |
| Cob(II)yrinic acid a,c-diamide reductase | -2.491 | 1.668 | 0.137 | 0.806 |
| Cobalt-factor II C(20)-methyltransferase | 0.248 | 0.136 | 0.070 | 0.787 |
| Cobalt-precorrin-4 methyltransferase | 0.231 | 0.131 | 0.081 | 0.806 |
| Cobalt-precorrin-5B (C(1))-methyltransferase | 0.294 | 0.144 | 0.042 | 0.780 |
| Cobalt-precorrin-6A reductase | 0.281 | 0.131 | 0.034 | 0.780 |
| Cobalt-precorrin-6B (C(15))-methyltransferase (decarboxylating) | 0.573 | 0.217 | 0.009 | 0.780 |
| Cobalt-precorrin-7 (C(5))-methyltransferase | 0.550 | 0.214 | 0.011 | 0.780 |
| Cobalt-precorrin-8 methylmutase | 0.229 | 0.131 | 0.084 | 0.806 |
| Cobalt-precorrin 5A hydrolase | 0.257 | 0.134 | 0.057 | 0.780 |
| Cobaltochelatase | -0.196 | 0.159 | 0.219 | 0.806 |
| Cobyrinate a,c-diamide synthase (glutamine-hydrolyzing) | 0.136 | 0.114 | 0.235 | 0.806 |
| Coenzyme F420-0:L-glutamate ligase | -0.378 | 0.517 | 0.466 | 0.884 |
| Coenzyme F420-1:gamma-L-glutamate ligase | -0.378 | 0.517 | 0.466 | 0.884 |
| Coenzyme F420 hydrogenase | -1.432 | 0.977 | 0.145 | 0.806 |
| Coenzyme gamma-F420-2:alpha-L-glutamate ligase | -0.691 | 1.197 | 0.565 | 0.936 |
| Coniferyl-aldehyde dehydrogenase | -0.125 | 0.223 | 0.578 | 0.945 |
| Coproporphyrinogen dehydrogenase | 0.072 | 0.049 | 0.146 | 0.806 |
| Coproporphyrinogen oxidase | -0.074 | 0.184 | 0.686 | 0.995 |
| Creatinase | 0.572 | 0.559 | 0.307 | 0.820 |
| Creatininase | -0.875 | 0.367 | 0.018 | 0.780 |
| Crossover junction endodeoxyribonuclease | 0.040 | 0.069 | 0.562 | 0.936 |
| Crotonobetainyl-CoA hydratase | -1.693 | 0.563 | 0.003 | 0.780 |
| Crotonyl-CoA carboxylase/reductase | -1.100 | 0.946 | 0.247 | 0.806 |
| Crotonyl-CoA reductase | 1.111 | 0.698 | 0.114 | 0.806 |
| CTP-dependent riboflavin kinase | -16.940 | 2657.012 | 0.995 | 0.999 |
| CTP synthase (glutamine hydrolyzing) | -0.045 | 0.035 | 0.197 | 0.806 |
| Cu(+) exporting ATPase | 0.031 | 0.035 | 0.378 | 0.852 |
| Cu(2+)-exporting ATPase | 0.011 | 0.095 | 0.907 | 0.999 |
| Cutinase | -2.715 | 1.546 | 0.081 | 0.806 |
| Cyanase | -0.202 | 0.375 | 0.591 | 0.951 |
| Cyanophycin synthase (L-arginine-adding) | -0.229 | 0.508 | 0.653 | 0.973 |
| Cyanophycin synthase (L-aspartate-adding) | -0.229 | 0.508 | 0.653 | 0.973 |
| Cyanophycinase | -0.397 | 0.610 | 0.516 | 0.913 |
| Cyanuric acid amidohydrolase | -0.921 | 0.840 | 0.274 | 0.820 |
| Cyclic-guanylate-specific phosphodiesterase | -0.558 | 0.690 | 0.420 | 0.865 |
| Cyclic dehypoxanthinyl futalosine synthase | 0.125 | 0.442 | 0.777 | 0.999 |
| Cyclic pyranopterin phosphate synthase | 0.004 | 0.069 | 0.955 | 0.999 |
| Cyclo(L-leucyl-L-leucyl) synthase | 17.953 | 2571.043 | 0.994 | 0.999 |
| Cyclo(L-tyrosyl-L-tyrosyl) synthase | -17.486 | 3491.164 | 0.996 | 0.999 |
| Cyclohexanone monooxygenase | -0.813 | 0.473 | 0.088 | 0.806 |
| Cyclohexyl-isocyanide hydratase | -1.053 | 0.549 | 0.057 | 0.780 |
| Cyclomaltodextrin glucanotransferase | 18.682 | 3701.820 | 0.996 | 0.999 |
| Cyclomaltodextrinase | -17.352 | 3264.645 | 0.996 | 0.999 |
| Cyclopentanol dehydrogenase | 1.065 | 0.877 | 0.227 | 0.806 |
| Cyclopropane-fatty-acyl-phospholipid synthase | -0.037 | 0.140 | 0.792 | 0.999 |
| Cystathionine beta-lyase | 0.104 | 0.076 | 0.170 | 0.806 |
| Cystathionine beta-synthase | -0.181 | 0.231 | 0.435 | 0.869 |
| Cystathionine gamma-lyase | -0.195 | 0.221 | 0.381 | 0.852 |
| Cystathionine gamma-synthase | 0.080 | 0.075 | 0.287 | 0.820 |
| Cysteate synthase | -17.886 | 3227.774 | 0.996 | 0.999 |
| Cysteine–tRNA ligase | 0.116 | 0.048 | 0.016 | 0.780 |
| Cysteine desulfurase | 0.002 | 0.052 | 0.977 | 0.999 |
| Cysteine dioxygenase | -17.885 | 3209.069 | 0.996 | 0.999 |
| Cysteine synthase | 0.022 | 0.028 | 0.431 | 0.869 |
| Cytidine deaminase | -0.019 | 0.080 | 0.810 | 0.999 |
| Cytochrome-c oxidase | -0.302 | 0.184 | 0.103 | 0.806 |
| Cytochrome-c peroxidase | 0.041 | 0.196 | 0.834 | 0.999 |
| Cytosine deaminase | -0.077 | 0.197 | 0.696 | 0.995 |
| Cytosol alanyl aminopeptidase | -0.005 | 0.522 | 0.992 | 0.999 |
| D-Ala-D-Ala dipeptidase | -0.273 | 0.160 | 0.090 | 0.806 |
| D-alanine–(R)-lactate ligase | 18.542 | 3451.607 | 0.996 | 0.999 |
| D-alanine–D-alanine ligase | 0.010 | 0.032 | 0.752 | 0.999 |
| D-alanine–D-serine ligase | -18.269 | 3009.878 | 0.995 | 0.999 |
| D-alanine–poly(phosphoribitol) ligase | 0.032 | 0.138 | 0.816 | 0.999 |
| D-amino-acid oxidase | -0.072 | 0.797 | 0.928 | 0.999 |
| D-amino-acid transaminase | -0.180 | 0.176 | 0.307 | 0.820 |
| D-aminoacyl-tRNA deacylase | -16.940 | 2657.012 | 0.995 | 0.999 |
| D-arabinitol 4-dehydrogenase | -1.922 | 1.748 | 0.273 | 0.820 |
| D-arginine dehydrogenase | 0.166 | 0.357 | 0.643 | 0.973 |
| D-aspartate ligase | 0.111 | 0.371 | 0.765 | 0.999 |
| D-chiro-inositol 1-dehydrogenase | -0.408 | 0.338 | 0.230 | 0.806 |
| D-cysteine desulfhydrase | -0.051 | 0.316 | 0.871 | 0.999 |
| D-galactose 1-dehydrogenase | -1.017 | 0.451 | 0.025 | 0.780 |
| D-glucosaminate-6-phosphate ammonia lyase | -2.675 | 2.574 | 0.300 | 0.820 |
| D-glycero-alpha-D-manno-heptose-1,7-bisphosphate 7-phosphatase | 0.084 | 0.109 | 0.440 | 0.869 |
| D-glycero-alpha-D-manno-heptose-7-phosphate kinase | 0.627 | 0.471 | 0.185 | 0.806 |
| D-glycero-alpha-D-manno-heptose 1-phosphate guanylyltransferase | -0.589 | 0.543 | 0.280 | 0.820 |
| D-glycero-beta-D-manno-heptose-7-phosphate kinase | 0.108 | 0.129 | 0.402 | 0.863 |
| D-glycero-beta-D-manno-heptose 1-phosphate adenylyltransferase | 0.108 | 0.129 | 0.402 | 0.863 |
| D-glycero-beta-D-manno-heptose 1,7-bisphosphate 7-phosphatase | 0.084 | 0.109 | 0.440 | 0.869 |
| D-inositol-3-phosphate glycosyltransferase | -0.017 | 0.308 | 0.956 | 0.999 |
| D-lactate dehydratase | -0.272 | 0.307 | 0.376 | 0.852 |
| D-lactate dehydrogenase | 0.248 | 0.138 | 0.075 | 0.791 |
| D-lactate dehydrogenase (cytochrome) | -1.202 | 0.344 | 0.001 | 0.663 |
| D-malate dehydrogenase (decarboxylating) | -0.745 | 0.378 | 0.051 | 0.780 |
| D-ornithine 4,5-aminomutase | 1.007 | 0.503 | 0.047 | 0.780 |
| D-proline reductase (dithiol) | -0.143 | 0.342 | 0.676 | 0.984 |
| D-psicose 3-epimerase | -1.585 | 0.692 | 0.023 | 0.780 |
| D-ribose pyranase | 0.173 | 0.164 | 0.295 | 0.820 |
| D-ribulokinase | -1.526 | 1.461 | 0.298 | 0.820 |
| D-sedoheptulose 7-phosphate isomerase | 0.297 | 0.118 | 0.013 | 0.780 |
| D-serine ammonia-lyase | 0.348 | 0.260 | 0.182 | 0.806 |
| D-sorbitol dehydrogenase (acceptor) | -0.304 | 0.649 | 0.640 | 0.973 |
| D-stereospecific aminopeptidase | -0.978 | 0.441 | 0.028 | 0.780 |
| D-tagatose 3-epimerase | -1.585 | 0.692 | 0.023 | 0.780 |
| D-threo-aldose 1-dehydrogenase | -0.686 | 0.391 | 0.081 | 0.806 |
| D-xylulose reductase | -2.180 | 1.072 | 0.044 | 0.780 |
| dCMP deaminase | 0.076 | 0.087 | 0.385 | 0.852 |
| dCTP deaminase | -0.050 | 0.111 | 0.655 | 0.974 |
| deacetylase | -0.002 | 0.347 | 0.995 | 0.999 |
| decaprenol beta-1,5/1,6-galactofuranosyltransferase | 0.052 | 0.334 | 0.877 | 0.999 |
| Decaprenyl-phosphate phosphoribosyltransferase | -0.347 | 0.562 | 0.538 | 0.924 |
| Decaprenylphospho-beta-D-erythro-pentofuranosid-2-ulose 2-reductase | -0.293 | 0.549 | 0.595 | 0.951 |
| Decaprenylphospho-beta-D-ribofuranose 2-dehydrogenase | -0.347 | 0.562 | 0.538 | 0.924 |
| Dehydrogluconokinase | -0.011 | 0.440 | 0.980 | 0.999 |
| Delta(7)-sterol 5(6)-desaturase | 1.212 | 1.112 | 0.277 | 0.820 |
| Demethylmenaquinone methyltransferase | -0.016 | 0.064 | 0.801 | 0.999 |
| Demethylspheroidene O-methyltransferase | -3.248 | 3.287 | 0.325 | 0.827 |
| Deoxyadenosine kinase | -0.356 | 0.344 | 0.302 | 0.820 |
| Deoxycytidine kinase | -0.752 | 0.501 | 0.136 | 0.806 |
| Deoxyguanosine kinase | -0.725 | 0.514 | 0.161 | 0.806 |
| Deoxyhypusine monooxygenase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Deoxyhypusine synthase | -0.220 | 0.323 | 0.498 | 0.906 |
| Deoxynucleoside kinase | 18.622 | 2950.870 | 0.995 | 0.999 |
| Deoxyribodipyrimidine photo-lyase | -0.134 | 0.178 | 0.452 | 0.879 |
| Deoxyribonuclease I | 0.332 | 0.307 | 0.282 | 0.820 |
| Deoxyribonuclease IV | 0.037 | 0.121 | 0.763 | 0.999 |
| Deoxyribonuclease V | -0.069 | 0.327 | 0.833 | 0.999 |
| Deoxyribose-phosphate aldolase | 0.017 | 0.075 | 0.821 | 0.999 |
| Dephospho-CoA kinase | 0.036 | 0.039 | 0.359 | 0.844 |
| Dethiobiotin synthase | 0.067 | 0.096 | 0.483 | 0.897 |
| Dextranase | 0.027 | 0.483 | 0.955 | 0.999 |
| Dextransucrase | 0.008 | 0.349 | 0.981 | 0.999 |
| dGTPase | -0.014 | 0.083 | 0.863 | 0.999 |
| Diacetyl reductase ((R)-acetoin forming) | -0.058 | 0.155 | 0.707 | 0.999 |
| Diacetyl reductase ((S)-acetoin forming) | -0.030 | 0.207 | 0.884 | 0.999 |
| Diacylglycerol diphosphate phosphatase | 0.051 | 0.256 | 0.841 | 0.999 |
| Diacylglycerol kinase (ATP) | 0.028 | 0.062 | 0.645 | 0.973 |
| Diacylglycerol O-acyltransferase | -0.864 | 0.452 | 0.058 | 0.780 |
| Diadenosine hexaphosphate hydrolase (ATP-forming) | -1.648 | 2.332 | 0.481 | 0.897 |
| Diadenylate cyclase | 0.105 | 0.086 | 0.223 | 0.806 |
| Diamine N-acetyltransferase | -0.072 | 0.124 | 0.562 | 0.936 |
| Diaminobutyrate–2-oxoglutarate transaminase | -0.088 | 0.264 | 0.739 | 0.999 |
| Diaminobutyrate acetyltransferase | -0.691 | 0.499 | 0.169 | 0.806 |
| Diaminobutyrate decarboxylase | -0.261 | 0.319 | 0.415 | 0.865 |
| Diaminohydroxyphosphoribosylaminopyrimidine deaminase | 0.001 | 0.058 | 0.987 | 0.999 |
| Diaminopimelate decarboxylase | -0.019 | 0.037 | 0.618 | 0.964 |
| Diaminopimelate dehydrogenase | 0.013 | 0.177 | 0.942 | 0.999 |
| Diaminopimelate epimerase | 0.020 | 0.058 | 0.726 | 0.999 |
| Diaminopropionate ammonia-lyase | 0.172 | 0.201 | 0.394 | 0.861 |
| Diapolycopene oxygenase | -1.552 | 1.123 | 0.169 | 0.806 |
| Diglucosyl diacylglycerol synthase (1,2-linking) | 0.172 | 0.171 | 0.315 | 0.822 |
| Diglucosyl diacylglycerol synthase (1,6-linking) | 0.070 | 0.192 | 0.718 | 0.999 |
| Diguanylate cyclase | -0.126 | 0.263 | 0.632 | 0.972 |
| Dihydroanticapsin 7-dehydrogenase | 17.690 | 2253.589 | 0.994 | 0.999 |
| Dihydrofolate reductase | 0.053 | 0.027 | 0.047 | 0.780 |
| Dihydrofolate synthase | 0.037 | 0.050 | 0.466 | 0.884 |
| Dihydrokaempferol 4-reductase | -0.432 | 0.342 | 0.208 | 0.806 |
| Dihydrolipoyl dehydrogenase | -0.086 | 0.055 | 0.120 | 0.806 |
| Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase | -0.453 | 0.270 | 0.095 | 0.806 |
| Dihydrolipoyllysine-residue acetyltransferase | -0.046 | 0.071 | 0.522 | 0.914 |
| Dihydrolipoyllysine-residue succinyltransferase | -0.118 | 0.110 | 0.287 | 0.820 |
| Dihydromethanopterin reductase (NAD(P)(+)) | 18.780 | 3887.052 | 0.996 | 0.999 |
| Dihydroneopterin aldolase | 0.003 | 0.041 | 0.949 | 0.999 |
| Dihydroorotase | -0.011 | 0.027 | 0.673 | 0.984 |
| Dihydroorotate dehydrogenase (NAD(+)) | 0.077 | 0.093 | 0.407 | 0.865 |
| Dihydroorotate dehydrogenase (quinone) | -0.054 | 0.118 | 0.647 | 0.973 |
| Dihydroorotate oxidase (fumarate) | 0.049 | 0.127 | 0.701 | 0.999 |
| Dihydropteroate synthase | -0.010 | 0.039 | 0.789 | 0.999 |
| Dihydropyrimidinase | -0.328 | 0.234 | 0.163 | 0.806 |
| Dihydrouracil dehydrogenase (NAD(+)) | -0.112 | 0.306 | 0.714 | 0.999 |
| Dihydroxy-acid dehydratase | -0.140 | 0.059 | 0.019 | 0.780 |
| Dimethyl sulfide:cytochrome c2 reductase | -17.618 | 3729.235 | 0.996 | 0.999 |
| Dimethylallyltranstransferase | 0.031 | 0.025 | 0.221 | 0.806 |
| Dimethylamine dehydrogenase | 0.672 | 0.917 | 0.465 | 0.884 |
| Dimethylargininase | -0.001 | 0.570 | 0.998 | 0.999 |
| Dimethylglycine dehydrogenase | -1.613 | 0.921 | 0.082 | 0.806 |
| Dimethylpropiothetin dethiomethylase | -17.226 | 2215.245 | 0.994 | 0.999 |
| Dimethylsulfoxide reductase | -0.914 | 0.739 | 0.218 | 0.806 |
| dioxygenase | -0.765 | 0.690 | 0.269 | 0.820 |
| Dipeptidase E | 0.206 | 0.155 | 0.186 | 0.806 |
| Dipeptidyl-peptidase III | -0.243 | 0.240 | 0.312 | 0.820 |
| Dipeptidyl-peptidase IV | -0.197 | 0.167 | 0.241 | 0.806 |
| Diphosphate–fructose-6-phosphate 1-phosphotransferase | -0.214 | 0.210 | 0.310 | 0.820 |
| diphosphate specific) | 0.047 | 0.032 | 0.136 | 0.806 |
| diphospho-ditrans,octacis-undecaprenol 4-beta-mannosyltransferase | -17.505 | 2261.205 | 0.994 | 0.999 |
| Diphosphomevalonate decarboxylase | 0.121 | 0.135 | 0.369 | 0.849 |
| Diphthine synthase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Dissimilatory sulfite reductase | -2.499 | 2.858 | 0.383 | 0.852 |
| Divinyl chlorophyllide a 8-vinyl-reductase | -17.226 | 2215.245 | 0.994 | 0.999 |
| DNA-(apurinic or apyrimidinic site) lyase | 0.049 | 0.031 | 0.110 | 0.806 |
| DNA-3-methyladenine glycosylase I | 0.021 | 0.044 | 0.630 | 0.971 |
| DNA-3-methyladenine glycosylase II | -0.026 | 0.130 | 0.844 | 0.999 |
| DNA-directed DNA polymerase | 0.042 | 0.029 | 0.143 | 0.806 |
| DNA-directed RNA polymerase | 0.047 | 0.028 | 0.098 | 0.806 |
| DNA-formamidopyrimidine glycosylase | 0.007 | 0.057 | 0.907 | 0.999 |
| DNA (cytosine-5-)-methyltransferase | -0.058 | 0.093 | 0.534 | 0.924 |
| DNA helicase | 0.050 | 0.030 | 0.101 | 0.806 |
| DNA ligase (ATP or NAD(+)) | -0.028 | 0.167 | 0.868 | 0.999 |
| DNA ligase (ATP) | -0.181 | 0.171 | 0.290 | 0.820 |
| DNA ligase (ATP, ADP or GTP) | -0.028 | 0.167 | 0.868 | 0.999 |
| DNA ligase (NAD(+)) | 0.041 | 0.020 | 0.045 | 0.780 |
| DNA oxidative demethylase | -0.347 | 0.272 | 0.203 | 0.806 |
| DNA topoisomerase | -0.009 | 0.052 | 0.861 | 0.999 |
| DNA topoisomerase (ATP-hydrolyzing) | 0.043 | 0.037 | 0.257 | 0.817 |
| Dodecanoyl-[acyl-carrier-protein] hydrolase | 0.133 | 0.147 | 0.368 | 0.848 |
| Dodecenoyl-CoA isomerase | 0.102 | 0.257 | 0.692 | 0.995 |
| Dolichyl-diphosphooligosaccharide–protein glycotransferase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Dolichyl-phosphate-mannose–protein mannosyltransferase | -0.036 | 0.668 | 0.957 | 0.999 |
| Dolichyl-phosphate beta-D-mannosyltransferase | 0.014 | 0.083 | 0.863 | 0.999 |
| dTDP-4-amino-4,6-dideoxy-D-galactose acyltransferase | 0.102 | 0.603 | 0.865 | 0.999 |
| dTDP-4-amino-4,6-dideoxygalactose transaminase | -0.350 | 0.395 | 0.377 | 0.852 |
| dTDP-4-dehydrorhamnose 3,5-epimerase | 0.155 | 0.071 | 0.031 | 0.780 |
| dTDP-4-dehydrorhamnose reductase | 0.089 | 0.061 | 0.149 | 0.806 |
| dTDP-6-deoxy-L-talose 4-dehydrogenase (NAD(+)) | 0.371 | 0.285 | 0.195 | 0.806 |
| dTDP-glucose 4,6-dehydratase | 0.164 | 0.056 | 0.004 | 0.780 |
| dTDP-L-rhamnose 4-epimerase | -0.634 | 0.543 | 0.245 | 0.806 |
| dTMP kinase | 0.053 | 0.035 | 0.133 | 0.806 |
| dUTP diphosphatase | 0.062 | 0.033 | 0.064 | 0.780 |
| Dye decolorizing peroxidase | -0.725 | 0.531 | 0.174 | 0.806 |
| Ectoine synthase | -0.628 | 0.465 | 0.178 | 0.806 |
| Electron-transferring-flavoprotein dehydrogenase | -0.155 | 0.192 | 0.420 | 0.865 |
| Enamidase | -2.093 | 1.297 | 0.109 | 0.806 |
| Endo-1,4-beta-xylanase | -0.650 | 0.364 | 0.076 | 0.796 |
| Endo-alpha-N-acetylgalactosaminidase | 0.963 | 0.926 | 0.300 | 0.820 |
| Endoglycosylceramidase | -1.824 | 2.136 | 0.394 | 0.861 |
| Endopeptidase Clp | 0.020 | 0.039 | 0.614 | 0.963 |
| Endopeptidase La | 0.106 | 0.084 | 0.209 | 0.806 |
| Endothelin-converting enzyme 1 | -0.416 | 0.563 | 0.461 | 0.884 |
| Enoyl-[acyl-carrier-protein] reductase | -1.963 | 0.921 | 0.035 | 0.780 |
| Enoyl-[acyl-carrier-protein] reductase (NADH) | 0.009 | 0.042 | 0.833 | 0.999 |
| Enoyl-[acyl-carrier-protein] reductase (NADPH, Si-specific) | -0.128 | 0.105 | 0.222 | 0.806 |
| Enoyl-CoA hydratase | -0.149 | 0.148 | 0.317 | 0.823 |
| Enoyl-CoA hydratase 2 | 1.988 | 1.303 | 0.129 | 0.806 |
| Enterobacter ribonuclease | -0.415 | 0.699 | 0.554 | 0.934 |
| Epoxyqueuosine reductase | -0.136 | 0.078 | 0.085 | 0.806 |
| Erythritol kinase | -2.828 | 1.735 | 0.105 | 0.806 |
| Erythrose-4-phosphate dehydrogenase | 0.106 | 0.275 | 0.700 | 0.999 |
| Ethanolamine ammonia-lyase | 0.337 | 0.232 | 0.148 | 0.806 |
| Ethanolaminephosphotransferase | -3.202 | 3.867 | 0.409 | 0.865 |
| Exo-1,4-beta-D-glucosaminidase | -2.289 | 1.243 | 0.067 | 0.780 |
| Exo-alpha-sialidase | -0.107 | 0.156 | 0.497 | 0.905 |
| Exo-poly-alpha-galacturonosidase | -0.995 | 1.007 | 0.325 | 0.827 |
| Exodeoxyribonuclease I | 0.084 | 0.181 | 0.642 | 0.973 |
| Exodeoxyribonuclease III | 0.042 | 0.032 | 0.188 | 0.806 |
| Exodeoxyribonuclease V | 0.030 | 0.062 | 0.631 | 0.971 |
| Exodeoxyribonuclease VII | 0.061 | 0.032 | 0.058 | 0.780 |
| Exopolyphosphatase | 0.147 | 0.076 | 0.057 | 0.780 |
| Exoribonuclease II | -0.001 | 0.239 | 0.996 | 0.999 |
| Factor independent urate hydroxylase | -0.958 | 0.688 | 0.166 | 0.806 |
| FAD-dependent urate hydroxylase | -0.635 | 0.517 | 0.221 | 0.806 |
| FAD synthetase | 0.041 | 0.031 | 0.196 | 0.806 |
| Fatty-acid peroxygenase | 0.620 | 0.620 | 0.319 | 0.824 |
| Fe(3+)-transporting ATPase | 0.056 | 0.099 | 0.569 | 0.938 |
| Ferredoxin–NAD(+) reductase | -0.827 | 0.291 | 0.005 | 0.780 |
| Ferredoxin–NADP(+) reductase | -0.009 | 0.116 | 0.941 | 0.999 |
| Ferredoxin–nitrate reductase | -17.955 | 2907.409 | 0.995 | 0.999 |
| Ferredoxin–nitrite reductase | 0.058 | 0.231 | 0.800 | 0.999 |
| Ferredoxin hydrogenase | 0.204 | 0.981 | 0.835 | 0.999 |
| Ferredoxin:protochlorophyllide reductase (ATP-dependent) | -2.655 | 2.006 | 0.188 | 0.806 |
| Ferric-chelate reductase (NADPH) | -0.269 | 0.717 | 0.708 | 0.999 |
| Ferrochelatase | -0.088 | 0.069 | 0.206 | 0.806 |
| Ferroxidase | -0.027 | 0.193 | 0.889 | 0.999 |
| Feruloyl esterase | -1.060 | 0.768 | 0.170 | 0.806 |
| Flavin reductase (NADH) | 0.457 | 0.394 | 0.248 | 0.808 |
| Fluoroacetyl-CoA thioesterase | 1.988 | 1.356 | 0.145 | 0.806 |
| FMN reductase (NAD(P)H) | -0.114 | 0.242 | 0.639 | 0.973 |
| FMN reductase (NADPH) | -0.323 | 0.171 | 0.061 | 0.780 |
| Formaldehyde dehydrogenase | 0.142 | 0.354 | 0.689 | 0.995 |
| Formaldehyde dismutase | -1.005 | 1.118 | 0.370 | 0.850 |
| Formamidase | -0.542 | 0.415 | 0.193 | 0.806 |
| Formate–phosphoribosylaminoimidazolecarboxamide ligase | -17.633 | 3757.583 | 0.996 | 0.999 |
| Formate–tetrahydrofolate ligase | 0.065 | 0.082 | 0.426 | 0.865 |
| Formate C-acetyltransferase | 0.084 | 0.095 | 0.378 | 0.852 |
| Formate dehydrogenase | 0.025 | 0.128 | 0.843 | 0.999 |
| Formate dehydrogenase-N | 0.032 | 0.641 | 0.960 | 0.999 |
| Formate dehydrogenase (NADP(+)) | -0.226 | 0.478 | 0.637 | 0.973 |
| Formimidoylglutamase | -0.024 | 0.119 | 0.839 | 0.999 |
| Formimidoylglutamate deiminase | -0.176 | 0.334 | 0.600 | 0.954 |
| Formimidoyltetrahydrofolate cyclodeaminase | 0.282 | 0.176 | 0.111 | 0.806 |
| Formyl-CoA transferase | -0.249 | 0.265 | 0.349 | 0.840 |
| Formylmethanofuran–tetrahydromethanopterin N-formyltransferase | 1.444 | 0.995 | 0.149 | 0.806 |
| Formylmethanofuran dehydrogenase | 1.287 | 0.576 | 0.027 | 0.780 |
| Formyltetrahydrofolate deformylase | -0.126 | 0.177 | 0.480 | 0.896 |
| Fructan beta-fructosidase | -0.193 | 0.159 | 0.226 | 0.806 |
| Fructokinase | -0.004 | 0.055 | 0.947 | 0.999 |
| Fructose-6-phosphate phosphoketolase | 0.224 | 0.280 | 0.425 | 0.865 |
| Fructose-bisphosphatase | 0.063 | 0.057 | 0.273 | 0.820 |
| Fructose-bisphosphate aldolase | 0.046 | 0.042 | 0.274 | 0.820 |
| Fructuronate reductase | -0.004 | 0.266 | 0.987 | 0.999 |
| Fucokinase | -17.672 | 2157.420 | 0.993 | 0.999 |
| Fucosylgalactoside 3-alpha-galactosyltransferase | -2.654 | 2.718 | 0.330 | 0.829 |
| Fumarate hydratase | -0.039 | 0.069 | 0.577 | 0.945 |
| Fumarate reductase (CoM/CoB) | 0.987 | 1.444 | 0.495 | 0.905 |
| Fumarate reductase (quinol) | 0.002 | 0.056 | 0.969 | 0.999 |
| Fumarylacetoacetase | -0.141 | 0.212 | 0.508 | 0.913 |
| Futalosine hydrolase | -2.087 | 1.489 | 0.163 | 0.806 |
| Galactan 5-O-arabinofuranosyltransferase | -0.759 | 0.675 | 0.262 | 0.820 |
| Galactarate dehydratase | -0.111 | 0.283 | 0.696 | 0.995 |
| Galactitol-1-phosphate 5-dehydrogenase | -0.667 | 1.039 | 0.522 | 0.914 |
| galactofuranosyltransferase | -0.304 | 0.575 | 0.598 | 0.953 |
| Galactokinase | 0.103 | 0.078 | 0.190 | 0.806 |
| Galactonate dehydratase | -0.711 | 0.388 | 0.069 | 0.781 |
| Galactose-6-phosphate isomerase | -0.057 | 0.155 | 0.714 | 0.999 |
| Galactoside O-acetyltransferase | -0.117 | 0.257 | 0.651 | 0.973 |
| Gallate dioxygenase | -0.333 | 0.999 | 0.740 | 0.999 |
| GalNAc(5)-diNAcBac-PP-undecaprenol beta-1,3-glucosyltransferase | 0.035 | 0.673 | 0.959 | 0.999 |
| Gamma-butyrobetaine dioxygenase | 0.005 | 0.633 | 0.994 | 0.999 |
| Gamma-D-glutamyl-meso-diaminopimelate peptidase | -0.120 | 0.415 | 0.773 | 0.999 |
| Gamma-glutamyl-gamma-aminobutyrate hydrolase | 0.102 | 0.647 | 0.875 | 0.999 |
| Gamma-glutamyl hercynylcysteine S-oxide hydrolase | -0.016 | 0.174 | 0.928 | 0.999 |
| Gamma-glutamyl hercynylcysteine S-oxide synthase | -0.895 | 0.768 | 0.246 | 0.806 |
| Gamma-glutamyltransferase | -0.053 | 0.166 | 0.752 | 0.999 |
| Gamma-guanidinobutyraldehyde dehydrogenase | 0.197 | 0.325 | 0.545 | 0.928 |
| GDP-4-dehydro-6-deoxy-D-mannose reductase | -0.579 | 0.569 | 0.310 | 0.820 |
| GDP-L-fucose synthase | -0.352 | 0.185 | 0.059 | 0.780 |
| GDP-mannose 4,6-dehydratase | -0.276 | 0.144 | 0.057 | 0.780 |
| GDP-mannose 6-dehydrogenase | 0.279 | 0.358 | 0.437 | 0.869 |
| GDP-mannose:cellobiosyl-diphosphopolyprenol alpha-mannosyltransferase | -0.063 | 0.724 | 0.931 | 0.999 |
| GDP-perosamine synthase | -0.976 | 0.933 | 0.297 | 0.820 |
| Gentamicin 2’-N-acetyltransferase | -0.026 | 0.763 | 0.973 | 0.999 |
| Gentamicin 3’-N-acetyltransferase | 0.653 | 1.214 | 0.591 | 0.951 |
| Gentisate 1,2-dioxygenase | -1.240 | 0.657 | 0.061 | 0.780 |
| Geosmin synthase | -17.486 | 3491.164 | 0.996 | 0.999 |
| Geranoyl-CoA carboxylase | -0.276 | 0.337 | 0.415 | 0.865 |
| Geranyl diphosphate 2-C-methyltransferase | -2.143 | 1.973 | 0.279 | 0.820 |
| Geranylgeranyl diphosphate reductase | -17.728 | 2688.993 | 0.995 | 0.999 |
| Geranylgeranyl diphosphate synthase | 0.067 | 0.055 | 0.226 | 0.806 |
| Geranylgeranylglycerol-phosphate geranylgeranyltransferase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Germacradienol synthase | -17.486 | 3491.164 | 0.996 | 0.999 |
| Gingipain R | -0.224 | 0.984 | 0.821 | 0.999 |
| Glucan 1,3-beta-glucosidase | -0.297 | 0.385 | 0.441 | 0.869 |
| Glucan 1,4-alpha-glucosidase | -0.032 | 0.717 | 0.965 | 0.999 |
| Glucan 1,6-alpha-glucosidase | 0.042 | 0.171 | 0.804 | 0.999 |
| Glucarate dehydratase | -0.073 | 0.293 | 0.804 | 0.999 |
| Glucokinase | -0.003 | 0.047 | 0.950 | 0.999 |
| Gluconate 2-dehydrogenase | -0.031 | 0.237 | 0.897 | 0.999 |
| Gluconate 2-dehydrogenase (acceptor) | -0.160 | 0.435 | 0.714 | 0.999 |
| Gluconate 5-dehydrogenase | 0.108 | 0.200 | 0.589 | 0.951 |
| Gluconokinase | 0.073 | 0.126 | 0.565 | 0.936 |
| Gluconolactonase | -0.692 | 0.310 | 0.027 | 0.780 |
| Glucosamine-1-phosphate N-acetyltransferase | 0.055 | 0.035 | 0.123 | 0.806 |
| Glucosamine-6-phosphate deaminase | 0.032 | 0.086 | 0.707 | 0.999 |
| Glucosamine-phosphate N-acetyltransferase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Glucosamine kinase | -1.319 | 0.615 | 0.034 | 0.780 |
| Glucose-1-phosphatase | 0.073 | 0.231 | 0.753 | 0.999 |
| Glucose-1-phosphate adenylyltransferase | 0.076 | 0.103 | 0.460 | 0.884 |
| Glucose-1-phosphate cytidylyltransferase | -0.022 | 0.270 | 0.935 | 0.999 |
| Glucose-1-phosphate thymidylyltransferase | 0.036 | 0.059 | 0.542 | 0.926 |
| Glucose-6-phosphate 1-epimerase | 0.110 | 0.190 | 0.563 | 0.936 |
| Glucose-6-phosphate 3-dehydrogenase | 17.953 | 2571.043 | 0.994 | 0.999 |
| Glucose-6-phosphate dehydrogenase (coenzyme-F420) | -0.944 | 0.955 | 0.325 | 0.827 |
| Glucose-6-phosphate dehydrogenase (NAD(P)(+)) | -0.011 | 0.062 | 0.860 | 0.999 |
| Glucose-6-phosphate dehydrogenase (NADP(+)) | -0.011 | 0.062 | 0.860 | 0.999 |
| Glucose-6-phosphate isomerase | 0.037 | 0.042 | 0.375 | 0.852 |
| Glucose-fructose oxidoreductase | -1.400 | 0.667 | 0.037 | 0.780 |
| Glucose 1-dehydrogenase (NAD(P)(+)) | -0.515 | 0.388 | 0.186 | 0.806 |
| Glucosyl-3-phosphoglycerate synthase | -0.255 | 0.557 | 0.647 | 0.973 |
| Glucosylceramidase | -0.043 | 0.316 | 0.891 | 0.999 |
| Glucosylglycerol-phosphate synthase | 3.153 | 2.362 | 0.184 | 0.806 |
| Glucuronate isomerase | -0.165 | 0.261 | 0.528 | 0.918 |
| Glucuronoarabinoxylan endo-1,4-beta-xylanase | -0.095 | 0.907 | 0.917 | 0.999 |
| Glucuronokinase | -17.862 | 4214.639 | 0.997 | 0.999 |
| Glutaconate CoA-transferase | 0.405 | 0.249 | 0.106 | 0.806 |
| Glutaconyl-CoA decarboxylase | 0.349 | 0.204 | 0.089 | 0.806 |
| Glutamate–ammonia ligase | -0.022 | 0.069 | 0.750 | 0.999 |
| Glutamate–cysteine ligase | 0.134 | 0.112 | 0.233 | 0.806 |
| Glutamate–prephenate aminotransferase | -0.787 | 1.153 | 0.496 | 0.905 |
| Glutamate–putrescine ligase | -0.107 | 0.385 | 0.781 | 0.999 |
| Glutamate–tRNA ligase | 0.010 | 0.070 | 0.891 | 0.999 |
| Glutamate–tRNA(Gln) ligase | 0.016 | 0.117 | 0.893 | 0.999 |
| Glutamate-1-semialdehyde 2,1-aminomutase | 0.050 | 0.075 | 0.506 | 0.913 |
| Glutamate-5-semialdehyde dehydrogenase | 0.014 | 0.048 | 0.770 | 0.999 |
| Glutamate 5-kinase | 0.015 | 0.045 | 0.730 | 0.999 |
| Glutamate carboxypeptidase | -0.272 | 0.185 | 0.143 | 0.806 |
| Glutamate carboxypeptidase II | -2.266 | 1.673 | 0.178 | 0.806 |
| Glutamate decarboxylase | -0.075 | 0.443 | 0.866 | 0.999 |
| Glutamate dehydrogenase | 0.000 | 0.103 | 0.999 | 0.999 |
| Glutamate dehydrogenase (NAD(P)(+)) | -0.058 | 0.179 | 0.746 | 0.999 |
| Glutamate dehydrogenase (NADP(+)) | -0.063 | 0.080 | 0.434 | 0.869 |
| Glutamate formimidoyltransferase | 0.254 | 0.166 | 0.129 | 0.806 |
| Glutamate N-acetyltransferase | -0.120 | 0.109 | 0.274 | 0.820 |
| Glutamate racemase | 0.038 | 0.042 | 0.361 | 0.844 |
| Glutamate synthase (ferredoxin) | -0.231 | 0.216 | 0.288 | 0.820 |
| Glutamate synthase (NADH) | -0.174 | 0.085 | 0.042 | 0.780 |
| Glutamate synthase (NADPH) | -0.174 | 0.085 | 0.042 | 0.780 |
| Glutamin-(asparagin-)ase | -0.337 | 0.343 | 0.329 | 0.829 |
| Glutaminase | 0.214 | 0.155 | 0.169 | 0.806 |
| Glutamine–fructose-6-phosphate transaminase (isomerizing) | 0.005 | 0.025 | 0.850 | 0.999 |
| Glutamine–tRNA ligase | 0.026 | 0.087 | 0.767 | 0.999 |
| Glutaminyl-peptide cyclotransferase | -3.240 | 3.159 | 0.307 | 0.820 |
| Glutaminyl-tRNA synthase (glutamine-hydrolyzing) | 0.004 | 0.047 | 0.938 | 0.999 |
| Glutamyl-tRNA reductase | 0.129 | 0.070 | 0.069 | 0.781 |
| Glutamyl aminopeptidase | 0.011 | 0.153 | 0.945 | 0.999 |
| Glutamyl endopeptidase | -1.669 | 1.118 | 0.137 | 0.806 |
| Glutarate-semialdehyde dehydrogenase | -0.061 | 0.139 | 0.660 | 0.975 |
| Glutaryl-CoA dehydrogenase (acceptor) | -2.764 | 1.313 | 0.037 | 0.780 |
| Glutaryl-CoA dehydrogenase (ETF) | -0.271 | 0.207 | 0.192 | 0.806 |
| Glutathione-disulfide reductase | 0.111 | 0.093 | 0.235 | 0.806 |
| Glutathione hydrolase | -0.053 | 0.166 | 0.752 | 0.999 |
| Glutathione peroxidase | 0.003 | 0.069 | 0.965 | 0.999 |
| Glutathione synthase | -0.072 | 0.177 | 0.685 | 0.994 |
| Glutathione transferase | -0.161 | 0.200 | 0.423 | 0.865 |
| Glutathionylspermidine amidase | 0.765 | 0.393 | 0.054 | 0.780 |
| Glutathionylspermidine synthase | 0.765 | 0.393 | 0.054 | 0.780 |
| Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+)) (phosphorylating) | -17.257 | 2411.517 | 0.994 | 0.999 |
| Glyceraldehyde-3-phosphate dehydrogenase (NADP(+)) | 0.044 | 0.144 | 0.761 | 0.999 |
| Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | 0.011 | 0.028 | 0.693 | 0.995 |
| Glycerate 2-kinase | 1.317 | 1.449 | 0.365 | 0.845 |
| Glycerate 3-kinase | 0.009 | 0.056 | 0.873 | 0.999 |
| Glycerate dehydrogenase | 0.185 | 0.116 | 0.112 | 0.806 |
| Glycerol-3-phosphate-transporting ATPase | -0.963 | 0.355 | 0.007 | 0.780 |
| Glycerol-3-phosphate 1-O-acyltransferase | 0.074 | 0.040 | 0.062 | 0.780 |
| Glycerol-3-phosphate cytidylyltransferase | 0.282 | 0.194 | 0.149 | 0.806 |
| Glycerol-3-phosphate dehydrogenase | -0.007 | 0.081 | 0.933 | 0.999 |
| Glycerol-3-phosphate dehydrogenase (NAD(P)(+)) | 0.038 | 0.031 | 0.224 | 0.806 |
| Glycerol-3-phosphate oxidase | -0.155 | 0.198 | 0.433 | 0.869 |
| Glycerol dehydratase | 2.303 | 1.806 | 0.204 | 0.806 |
| Glycerol dehydrogenase | 0.177 | 0.123 | 0.153 | 0.806 |
| Glycerol kinase | 0.011 | 0.035 | 0.746 | 0.999 |
| Glycerone kinase | -0.247 | 0.199 | 0.217 | 0.806 |
| Glycerophosphodiester phosphodiesterase | 0.027 | 0.063 | 0.668 | 0.979 |
| Glycine–tRNA ligase | 0.079 | 0.032 | 0.015 | 0.780 |
| Glycine amidinotransferase | 0.227 | 0.491 | 0.644 | 0.973 |
| Glycine C-acetyltransferase | -0.168 | 0.135 | 0.216 | 0.806 |
| Glycine dehydrogenase (aminomethyl-transferring) | -0.185 | 0.102 | 0.070 | 0.787 |
| Glycine dehydrogenase (cyanide-forming) | -0.140 | 0.609 | 0.818 | 0.999 |
| Glycine hydroxymethyltransferase | 0.020 | 0.035 | 0.565 | 0.936 |
| Glycine oxidase | -0.167 | 0.170 | 0.326 | 0.827 |
| Glycine reductase | 0.480 | 0.261 | 0.068 | 0.780 |
| Glycine/sarcosine N-methyltransferase | -0.957 | 0.985 | 0.333 | 0.831 |
| Glycogen phosphorylase | 0.076 | 0.076 | 0.319 | 0.824 |
| Glycogen(starch) synthase | -0.765 | 0.495 | 0.124 | 0.806 |
| Glycolaldehyde dehydrogenase | 0.089 | 0.390 | 0.820 | 0.999 |
| Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase | -2.654 | 2.718 | 0.330 | 0.829 |
| Glycoprotein endo-alpha-1,2-mannosidase | -1.861 | 1.782 | 0.298 | 0.820 |
| glycyltransferase | -0.220 | 0.803 | 0.785 | 0.999 |
| Glyoxylate reductase | -0.291 | 0.207 | 0.161 | 0.806 |
| Glyoxylate reductase (NADP(+)) | -0.487 | 0.287 | 0.092 | 0.806 |
| GMP reductase | 0.017 | 0.119 | 0.887 | 0.999 |
| GMP synthase (glutamine-hydrolyzing) | 0.009 | 0.033 | 0.793 | 0.999 |
| GPR endopeptidase | -0.952 | 0.501 | 0.059 | 0.780 |
| GTP cyclohydrolase I | 0.022 | 0.037 | 0.556 | 0.936 |
| GTP cyclohydrolase II | 0.012 | 0.065 | 0.848 | 0.999 |
| GTP cyclohydrolase IIa | -16.940 | 2657.012 | 0.995 | 0.999 |
| GTP cyclohydrolase IV | -1.093 | 1.677 | 0.515 | 0.913 |
| GTP diphosphokinase | 0.034 | 0.049 | 0.486 | 0.899 |
| Guanidinobutyrase | 0.198 | 0.342 | 0.563 | 0.936 |
| Guanidinopropionase | -2.770 | 2.134 | 0.196 | 0.806 |
| Guanine deaminase | -0.120 | 0.196 | 0.541 | 0.926 |
| Guanosine-3’,5’-bis(diphosphate) 3’-diphosphatase | 0.200 | 0.142 | 0.160 | 0.806 |
| Guanosine-5’-triphosphate,3’-diphosphate diphosphatase | 0.147 | 0.076 | 0.057 | 0.780 |
| Guanylate cyclase | -1.425 | 1.151 | 0.217 | 0.806 |
| Guanylate kinase | 0.078 | 0.047 | 0.096 | 0.806 |
| H(+)-transporting two-sector ATPase | 0.034 | 0.024 | 0.152 | 0.806 |
| Haloacetate dehalogenase | -0.115 | 0.225 | 0.610 | 0.960 |
| Haloalkane dehalogenase | -0.103 | 0.394 | 0.794 | 0.999 |
| Heme-transporting ATPase | -0.005 | 0.152 | 0.975 | 0.999 |
| Heme oxygenase (staphylobilin-producing) | -0.688 | 0.646 | 0.289 | 0.820 |
| Heparin-sulfate lyase | -0.688 | 1.006 | 0.495 | 0.905 |
| Heparin lyase | -0.628 | 0.972 | 0.519 | 0.913 |
| Heptaprenyl diphosphate synthase | 0.286 | 0.126 | 0.024 | 0.780 |
| Hexokinase | 0.318 | 0.481 | 0.509 | 0.913 |
| Hippurate hydrolase | -0.355 | 0.234 | 0.132 | 0.806 |
| Histidine–tRNA ligase | 0.032 | 0.031 | 0.311 | 0.820 |
| Histidine ammonia-lyase | -0.019 | 0.103 | 0.853 | 0.999 |
| Histidine decarboxylase | 0.610 | 1.080 | 0.573 | 0.941 |
| Histidine kinase | -0.050 | 0.041 | 0.224 | 0.806 |
| Histidinol-phosphatase | 0.137 | 0.102 | 0.179 | 0.806 |
| Histidinol-phosphate transaminase | -0.117 | 0.058 | 0.046 | 0.780 |
| Histidinol dehydrogenase | -0.093 | 0.063 | 0.143 | 0.806 |
| Histone acetyltransferase | -0.411 | 0.661 | 0.535 | 0.924 |
| Holo-[acyl-carrier-protein] synthase | 0.075 | 0.071 | 0.292 | 0.820 |
| Homocitrate synthase | -0.984 | 0.806 | 0.224 | 0.806 |
| Homocysteine desulfhydrase | -0.640 | 0.439 | 0.147 | 0.806 |
| Homocysteine S-methyltransferase | -0.097 | 0.150 | 0.520 | 0.913 |
| Homogentisate 1,2-dioxygenase | -0.092 | 0.220 | 0.675 | 0.984 |
| Homoisocitrate dehydrogenase | -1.831 | 2.142 | 0.394 | 0.861 |
| Homoserine dehydrogenase | -0.025 | 0.035 | 0.466 | 0.884 |
| Homoserine kinase | 0.051 | 0.033 | 0.128 | 0.806 |
| Homoserine O-acetyltransferase | -0.087 | 0.133 | 0.516 | 0.913 |
| Homoserine O-succinyltransferase | -0.001 | 0.102 | 0.995 | 0.999 |
| Homospermidine synthase | -1.912 | 0.806 | 0.019 | 0.780 |
| HslU–HslV peptidase | 0.020 | 0.092 | 0.824 | 0.999 |
| Hyaluronan synthase | 0.214 | 0.566 | 0.706 | 0.999 |
| Hyaluronate lyase | 0.530 | 0.455 | 0.246 | 0.806 |
| Hyaluronoglucosaminidase | 0.021 | 0.323 | 0.948 | 0.999 |
| HycI peptidase | 0.304 | 0.432 | 0.483 | 0.897 |
| hydratase | 1.546 | 1.393 | 0.269 | 0.820 |
| Hydrogen dehydrogenase | -0.267 | 0.792 | 0.737 | 0.999 |
| Hydrogen dehydrogenase (NADP(+)) | -0.143 | 0.428 | 0.738 | 0.999 |
| Hydrogen:quinone oxidoreductase | -0.362 | 0.789 | 0.646 | 0.973 |
| Hydrogenase (acceptor) | 0.121 | 0.220 | 0.584 | 0.948 |
| Hydrogenobyrinic acid a,c-diamide synthase (glutamine-hydrolyzing) | 0.136 | 0.114 | 0.235 | 0.806 |
| Hydrogensulfite reductase | -2.499 | 2.858 | 0.383 | 0.852 |
| hydrolase | -0.809 | 0.676 | 0.233 | 0.806 |
| Hydroxyacid-oxoacid transhydrogenase | -17.379 | 3197.270 | 0.996 | 0.999 |
| Hydroxyacylglutathione hydrolase | 0.088 | 0.126 | 0.485 | 0.899 |
| Hydroxybutyrate-dimer hydrolase | -1.764 | 1.210 | 0.147 | 0.806 |
| Hydroxydechloroatrazine ethylaminohydrolase | -1.777 | 1.630 | 0.277 | 0.820 |
| Hydroxyethylthiazole kinase | -0.062 | 0.104 | 0.553 | 0.934 |
| Hydroxyisourate hydrolase | -0.212 | 0.240 | 0.378 | 0.852 |
| Hydroxylamine reductase | 0.184 | 0.166 | 0.270 | 0.820 |
| Hydroxymethylbilane synthase | 0.071 | 0.066 | 0.284 | 0.820 |
| Hydroxymethylglutaryl-CoA lyase | -0.096 | 0.190 | 0.612 | 0.961 |
| Hydroxymethylglutaryl-CoA reductase | 0.102 | 0.132 | 0.443 | 0.870 |
| Hydroxymethylglutaryl-CoA reductase (NADPH) | -0.719 | 0.810 | 0.376 | 0.852 |
| Hydroxymethylglutaryl-CoA synthase | 0.112 | 0.136 | 0.411 | 0.865 |
| Hydroxymethylpyrimidine kinase | 0.057 | 0.047 | 0.229 | 0.806 |
| Hydroxypyruvate isomerase | -0.246 | 0.216 | 0.258 | 0.818 |
| Hydroxypyruvate reductase | -0.387 | 0.241 | 0.111 | 0.806 |
| Hydroxyquinol 1,2-dioxygenase | -0.345 | 0.697 | 0.621 | 0.966 |
| Hygromycin-B 7’’-O-kinase | -1.369 | 1.141 | 0.232 | 0.806 |
| Hygromycin B 4-O-kinase | 17.690 | 2253.589 | 0.994 | 0.999 |
| Hypoxanthine phosphoribosyltransferase | 0.084 | 0.046 | 0.067 | 0.780 |
| Iduronate-2-sulfatase | -17.519 | 3549.753 | 0.996 | 0.999 |
| IgA-specific metalloendopeptidase | -0.030 | 0.688 | 0.965 | 0.999 |
| IgA-specific serine endopeptidase | 0.256 | 0.358 | 0.475 | 0.891 |
| Imidazoleglycerol-phosphate dehydratase | -0.091 | 0.060 | 0.128 | 0.806 |
| Imidazolonepropionase | 0.054 | 0.089 | 0.545 | 0.928 |
| IMP cyclohydrolase | 0.039 | 0.042 | 0.355 | 0.844 |
| IMP dehydrogenase | 0.027 | 0.032 | 0.401 | 0.863 |
| Indole-3-glycerol-phosphate synthase | -0.081 | 0.054 | 0.133 | 0.806 |
| Indoleamine 2,3-dioxygenase | 0.681 | 1.042 | 0.514 | 0.913 |
| Indolepyruvate decarboxylase | -0.425 | 0.420 | 0.313 | 0.820 |
| Indolepyruvate ferredoxin oxidoreductase | -0.249 | 0.214 | 0.247 | 0.806 |
| Inorganic diphosphatase | 0.071 | 0.048 | 0.138 | 0.806 |
| Inosine kinase | 0.389 | 0.519 | 0.455 | 0.883 |
| Inositol-3-phosphate synthase | -0.190 | 0.208 | 0.362 | 0.844 |
| Inositol-phosphate phosphatase | -0.095 | 0.072 | 0.186 | 0.806 |
| Inositol 2-dehydrogenase | -0.408 | 0.338 | 0.230 | 0.806 |
| Inositol oxygenase | -1.216 | 1.983 | 0.541 | 0.926 |
| Insulysin | -3.435 | 4.121 | 0.406 | 0.865 |
| Inulin fructotransferase (DFA-I-forming) | -1.175 | 1.776 | 0.509 | 0.913 |
| Iron-chelate-transporting ATPase | 0.100 | 0.058 | 0.086 | 0.806 |
| Isochorismatase | -0.141 | 0.275 | 0.610 | 0.960 |
| Isochorismate lyase | -0.215 | 0.288 | 0.457 | 0.884 |
| Isochorismate synthase | -0.046 | 0.103 | 0.652 | 0.973 |
| Isocitrate dehydrogenase (NAD(+)) | 0.518 | 0.297 | 0.083 | 0.806 |
| Isocitrate dehydrogenase (NADP(+)) | -0.135 | 0.096 | 0.161 | 0.806 |
| Isocitrate lyase | -0.251 | 0.178 | 0.162 | 0.806 |
| Isohexenylglutaconyl-CoA hydratase | -0.163 | 0.374 | 0.663 | 0.977 |
| Isoleucine–tRNA ligase | 0.032 | 0.028 | 0.251 | 0.809 |
| isomerase | -0.057 | 0.056 | 0.309 | 0.820 |
| Isonocardicin synthase | -0.169 | 0.734 | 0.819 | 0.999 |
| Isopenicillin-N epimerase | -17.612 | 3503.932 | 0.996 | 0.999 |
| Isopentenyl-diphosphate Delta-isomerase | 0.026 | 0.108 | 0.813 | 0.999 |
| Isopentenyl phosphate kinase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Isoquinoline 1-oxidoreductase | -0.307 | 0.310 | 0.324 | 0.827 |
| Isovaleryl-CoA dehydrogenase | -0.176 | 0.215 | 0.416 | 0.865 |
| Isovaleryl-homoserine lactone synthase | -0.536 | 0.687 | 0.436 | 0.869 |
| Itaconyl-CoA hydratase | 0.068 | 0.411 | 0.868 | 0.999 |
| Kanamycin kinase | 0.024 | 0.140 | 0.864 | 0.999 |
| Kanosamine-6-phosphate phosphatase | -2.363 | 2.511 | 0.348 | 0.840 |
| Kdo(2)-lipid IV(A) lauroyltransferase | 0.079 | 0.082 | 0.336 | 0.833 |
| Kdo(2)-lipid IV(A) palmitoleoyltransferase | 0.102 | 0.603 | 0.865 | 0.999 |
| Ketol-acid reductoisomerase (NADP(+)) | -0.058 | 0.051 | 0.256 | 0.817 |
| Kojibiose phosphorylase | 0.921 | 0.703 | 0.192 | 0.806 |
| Kynureninase | -0.307 | 0.286 | 0.285 | 0.820 |
| Kynurenine 3-monooxygenase | -0.021 | 0.458 | 0.963 | 0.999 |
| L-2-hydroxyglutarate dehydrogenase | -1.344 | 1.538 | 0.384 | 0.852 |
| L-arabinonate dehydratase | -1.431 | 1.302 | 0.273 | 0.820 |
| L-arabinonolactonase | -0.778 | 1.338 | 0.562 | 0.936 |
| L-arabinose 1-dehydrogenase (NAD(P)(+)) | -2.133 | 1.053 | 0.045 | 0.780 |
| L-arabinose isomerase | 0.148 | 0.283 | 0.601 | 0.954 |
| L-ascorbate oxidase | -16.946 | 2665.572 | 0.995 | 0.999 |
| L-asparagine oxygenase | -17.486 | 3491.164 | 0.996 | 0.999 |
| L-aspartate oxidase | -0.029 | 0.107 | 0.787 | 0.999 |
| L-carnitine CoA-transferase | -0.324 | 0.845 | 0.702 | 0.999 |
| L-cysteate sulfo-lyase | -0.405 | 0.792 | 0.610 | 0.960 |
| L-erythro-3,5-diaminohexanoate dehydrogenase | 0.964 | 0.450 | 0.034 | 0.780 |
| L-fuconate dehydratase | -0.114 | 0.339 | 0.738 | 0.999 |
| L-fucose isomerase | 0.178 | 0.285 | 0.533 | 0.924 |
| L-fucose mutarotase | 0.011 | 0.246 | 0.965 | 0.999 |
| L-fuculokinase | 0.465 | 0.449 | 0.301 | 0.820 |
| L-fuculose-phosphate aldolase | 0.297 | 0.158 | 0.063 | 0.780 |
| L-galactose 1-dehydrogenase | -1.392 | 1.171 | 0.236 | 0.806 |
| L-glutamate gamma-semialdehyde dehydrogenase | -0.133 | 0.132 | 0.314 | 0.820 |
| L-gulonate 5-dehydrogenase | 0.167 | 0.579 | 0.774 | 0.999 |
| L-gulonolactone oxidase | -1.056 | 1.021 | 0.303 | 0.820 |
| L-histidine N(alpha)-methyltransferase | -0.869 | 0.714 | 0.226 | 0.806 |
| L-iditol 2-dehydrogenase | -0.211 | 0.264 | 0.425 | 0.865 |
| L-idonate 5-dehydrogenase (NAD(P)(+)) | -0.567 | 0.491 | 0.250 | 0.808 |
| L-iduronidase | -1.476 | 1.632 | 0.367 | 0.847 |
| L-lactate dehydrogenase | 0.209 | 0.150 | 0.166 | 0.806 |
| L-lactate dehydrogenase (cytochrome) | -0.131 | 0.155 | 0.401 | 0.863 |
| L-lysine 6-transaminase | -0.804 | 0.687 | 0.243 | 0.806 |
| L-lysine N(6)-monooxygenase (NADPH) | -0.317 | 0.408 | 0.438 | 0.869 |
| L-proline amide hydrolase | -0.553 | 0.659 | 0.402 | 0.863 |
| L-rhamnonate dehydratase | 0.916 | 0.479 | 0.058 | 0.780 |
| L-rhamnose 1-dehydrogenase | -0.931 | 0.626 | 0.139 | 0.806 |
| L-rhamnose 1-dehydrogenase (NAD(P)(+)) | -0.931 | 0.626 | 0.139 | 0.806 |
| L-rhamnose 1-dehydrogenase (NADP(+)) | -0.931 | 0.626 | 0.139 | 0.806 |
| L-rhamnose isomerase | -0.659 | 0.308 | 0.034 | 0.780 |
| L-rhamnose mutarotase | -0.623 | 0.303 | 0.041 | 0.780 |
| L-ribulose-5-phosphate 3-epimerase | 0.420 | 0.386 | 0.278 | 0.820 |
| L-ribulose-5-phosphate 4-epimerase | 0.148 | 0.123 | 0.231 | 0.806 |
| L-serine 3-dehydrogenase (NAD(+)) | 1.546 | 1.393 | 0.269 | 0.820 |
| L-serine ammonia-lyase | 0.025 | 0.046 | 0.582 | 0.948 |
| L-seryl-tRNA(Sec) selenium transferase | 0.165 | 0.171 | 0.334 | 0.832 |
| L-threonine 3-dehydrogenase | -0.114 | 0.327 | 0.727 | 0.999 |
| L-threonine aldolase | 0.033 | 0.124 | 0.793 | 0.999 |
| L-threonine kinase | 0.203 | 0.239 | 0.396 | 0.861 |
| L-threonylcarbamoyladenylate synthase | 0.041 | 0.037 | 0.271 | 0.820 |
| L-xylulokinase | -0.284 | 0.383 | 0.459 | 0.884 |
| L(+)-tartrate dehydratase | 0.168 | 0.232 | 0.470 | 0.887 |
| Lactaldehyde dehydrogenase | 0.089 | 0.390 | 0.820 | 0.999 |
| Lactaldehyde reductase | 0.505 | 0.249 | 0.044 | 0.780 |
| Lactate 2-monooxygenase | -2.010 | 1.112 | 0.072 | 0.787 |
| Lactocepin | 0.044 | 0.180 | 0.806 | 0.999 |
| Lactoylglutathione lyase | 0.097 | 0.054 | 0.075 | 0.791 |
| Lauroyl-Kdo(2)-lipid IV(A) myristoyltransferase | 0.225 | 0.307 | 0.465 | 0.884 |
| Leucine–tRNA ligase | 0.029 | 0.033 | 0.383 | 0.852 |
| Leucine dehydrogenase | -0.194 | 0.213 | 0.363 | 0.844 |
| Leucyl aminopeptidase | -0.076 | 0.098 | 0.441 | 0.869 |
| Leucyltransferase | -0.101 | 0.157 | 0.519 | 0.913 |
| Levanase | -0.190 | 0.298 | 0.524 | 0.916 |
| Levansucrase | 0.192 | 0.421 | 0.649 | 0.973 |
| Licheninase | 18.480 | 3345.951 | 0.996 | 0.999 |
| ligase | -0.019 | 0.308 | 0.950 | 0.999 |
| Limonene-1,2-epoxide hydrolase | -1.981 | 1.899 | 0.298 | 0.820 |
| Limonene 1,2-monooxygenase | -0.828 | 0.627 | 0.189 | 0.806 |
| Lipid-A-disaccharide synthase | 0.079 | 0.088 | 0.366 | 0.846 |
| Lipid II:glycine glycyltransferase | -0.145 | 0.620 | 0.815 | 0.999 |
| Lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase | -0.026 | 0.088 | 0.767 | 0.999 |
| Lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase | -0.416 | 0.542 | 0.445 | 0.871 |
| Lipoate–protein ligase | -0.001 | 0.124 | 0.994 | 0.999 |
| Lipopolysaccharide 3-alpha-galactosyltransferase | 1.652 | 1.219 | 0.177 | 0.806 |
| Lipopolysaccharide glucosyltransferase I | -0.703 | 0.879 | 0.425 | 0.865 |
| Lipopolysaccharide N-acetylglucosaminyltransferase | -0.866 | 0.639 | 0.177 | 0.806 |
| Lipopolysaccharide N-acetylmannosaminouronosyltransferase | 0.043 | 0.589 | 0.943 | 0.999 |
| Lipoyl amidotransferase | 2.952 | 2.379 | 0.216 | 0.806 |
| Lipoyl synthase | -0.115 | 0.085 | 0.177 | 0.806 |
| Lipoyl(octanoyl) transferase | -0.044 | 0.120 | 0.713 | 0.999 |
| LL-diaminopimelate aminotransferase | -0.001 | 0.206 | 0.996 | 0.999 |
| Long-chain-acyl-CoA dehydrogenase | -0.431 | 0.416 | 0.301 | 0.820 |
| Long-chain-fatty-acid–[acyl-carrier-protein] ligase | 0.439 | 0.254 | 0.086 | 0.806 |
| Long-chain-fatty-acid–CoA ligase | -0.086 | 0.086 | 0.316 | 0.823 |
| Long-chain-fatty-acid–luciferin-component ligase | 18.622 | 2950.870 | 0.995 | 0.999 |
| Long-chain-fatty-acyl-CoA reductase | 18.622 | 2950.870 | 0.995 | 0.999 |
| Lycopene beta-cyclase | -0.402 | 0.452 | 0.376 | 0.852 |
| Lysine–8-amino-7-oxononanoate transaminase | 17.953 | 2571.043 | 0.994 | 0.999 |
| Lysine–tRNA ligase | 0.027 | 0.030 | 0.362 | 0.844 |
| Lysine 2,3-aminomutase | 0.252 | 0.321 | 0.434 | 0.869 |
| Lysine 6-dehydrogenase | -1.370 | 1.021 | 0.182 | 0.806 |
| Lysine decarboxylase | -0.354 | 0.177 | 0.047 | 0.780 |
| Lysophospholipase | -0.237 | 0.164 | 0.151 | 0.806 |
| Lysostaphin | -1.552 | 1.123 | 0.169 | 0.806 |
| Lysozyme | -0.021 | 0.239 | 0.930 | 0.999 |
| Lysyltransferase | 0.142 | 0.137 | 0.303 | 0.820 |
| Magnesium-importing ATPase | -0.021 | 0.108 | 0.847 | 0.999 |
| Magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase | -3.343 | 3.146 | 0.290 | 0.820 |
| Magnesium chelatase | 0.116 | 0.166 | 0.486 | 0.899 |
| Magnesium protoporphyrin IX methyltransferase | -2.553 | 3.245 | 0.433 | 0.869 |
| Malate–CoA ligase | 0.742 | 1.343 | 0.581 | 0.948 |
| Malate dehydrogenase | -0.134 | 0.093 | 0.152 | 0.806 |
| Malate dehydrogenase (oxaloacetate-decarboxylating) | 0.192 | 0.117 | 0.102 | 0.806 |
| Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP(+)) | -0.163 | 0.106 | 0.128 | 0.806 |
| Malate dehydrogenase (quinone) | -0.081 | 0.173 | 0.641 | 0.973 |
| Malate synthase | -0.172 | 0.179 | 0.338 | 0.834 |
| Maleamate amidohydrolase | -0.782 | 0.795 | 0.326 | 0.827 |
| Maleate isomerase | 0.137 | 0.405 | 0.736 | 0.999 |
| Maleylacetate reductase | -1.402 | 1.059 | 0.187 | 0.806 |
| Maleylacetoacetate isomerase | -0.095 | 0.221 | 0.668 | 0.979 |
| Maleylpyruvate isomerase | -0.621 | 0.350 | 0.078 | 0.806 |
| Malonate-semialdehyde dehydrogenase (acetylating) | -0.261 | 0.220 | 0.237 | 0.806 |
| Malonate decarboxylase holo-[acyl-carrier protein] synthase | 0.084 | 0.273 | 0.758 | 0.999 |
| Malonyl-[acyl-carrier protein] O-methyltransferase | 0.162 | 0.137 | 0.241 | 0.806 |
| Malonyl-CoA decarboxylase | -1.159 | 0.638 | 0.071 | 0.787 |
| Malonyl-S-ACP decarboxylase | 0.053 | 0.282 | 0.850 | 0.999 |
| Maltokinase | -0.203 | 0.390 | 0.603 | 0.957 |
| Maltose-6’-phosphate glucosidase | 0.172 | 0.310 | 0.580 | 0.947 |
| Maltose 6’-phosphate phosphatase | 0.070 | 0.153 | 0.646 | 0.973 |
| Maltose alpha-D-glucosyltransferase | -0.057 | 0.281 | 0.839 | 0.999 |
| Maltose O-acetyltransferase | 0.051 | 0.122 | 0.674 | 0.984 |
| Maltose phosphorylase | 0.408 | 0.337 | 0.228 | 0.806 |
| Malyl-CoA lyase | -1.353 | 0.882 | 0.127 | 0.806 |
| Mandelamide amidase | -0.492 | 0.950 | 0.605 | 0.959 |
| Mandelate racemase | -1.369 | 1.141 | 0.232 | 0.806 |
| Mannan endo-1,4-beta-mannosidase | -0.373 | 0.263 | 0.158 | 0.806 |
| Mannitol-1-phosphate 5-dehydrogenase | 0.072 | 0.209 | 0.732 | 0.999 |
| Mannitol 2-dehydrogenase | -0.202 | 0.351 | 0.566 | 0.936 |
| Mannonate dehydratase | 0.005 | 0.217 | 0.980 | 0.999 |
| mannosaminyltransferase | -0.124 | 0.133 | 0.351 | 0.842 |
| Mannose-1-phosphate guanylyltransferase | -0.172 | 0.106 | 0.106 | 0.806 |
| Mannose-6-phosphate isomerase | -0.044 | 0.050 | 0.386 | 0.852 |
| Mannosyl-3-phosphoglycerate phosphatase | -0.466 | 0.720 | 0.518 | 0.913 |
| Mannosyl-3-phosphoglycerate synthase | 2.038 | 1.820 | 0.264 | 0.820 |
| Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase | 0.142 | 0.219 | 0.517 | 0.913 |
| Mannosyl-oligosaccharide 1,2-alpha-mannosidase | -17.136 | 2931.202 | 0.995 | 0.999 |
| Mannosylglycerate hydrolase | 0.052 | 0.614 | 0.932 | 0.999 |
| Medium-chain acyl-CoA dehydrogenase | -0.490 | 0.252 | 0.054 | 0.780 |
| Membrane alanyl aminopeptidase | -0.002 | 0.083 | 0.981 | 0.999 |
| Membrane dipeptidase | -0.063 | 0.124 | 0.610 | 0.960 |
| Menaquinol oxidase (H(+)-transporting) | -0.847 | 0.612 | 0.168 | 0.806 |
| Mercury(II) reductase | 0.081 | 0.415 | 0.846 | 0.999 |
| Methane monooxygenase (particulate) | -17.712 | 2757.136 | 0.995 | 0.999 |
| Methane monooxygenase (soluble) | -17.395 | 3335.511 | 0.996 | 0.999 |
| Methanogen homoaconitase | -1.125 | 1.594 | 0.482 | 0.897 |
| Methanol dehydrogenase (cytochrome c) | 18.473 | 3334.173 | 0.996 | 0.999 |
| Methenyltetrahydrofolate cyclohydrolase | 0.012 | 0.027 | 0.667 | 0.979 |
| Methenyltetrahydromethanopterin cyclohydrolase | 1.444 | 0.995 | 0.149 | 0.806 |
| Methionine–tRNA ligase | 0.044 | 0.032 | 0.178 | 0.806 |
| Methionine adenosyltransferase | 0.036 | 0.031 | 0.250 | 0.808 |
| Methionine gamma-lyase | 0.120 | 0.151 | 0.427 | 0.865 |
| Methionine synthase | -0.026 | 0.091 | 0.777 | 0.999 |
| Methionyl-tRNA formyltransferase | 0.035 | 0.027 | 0.201 | 0.806 |
| Methionyl aminopeptidase | -0.011 | 0.024 | 0.637 | 0.973 |
| Methylamine dehydrogenase (amicyanin) | 0.242 | 0.738 | 0.743 | 0.999 |
| Methylaspartate ammonia-lyase | 0.013 | 0.469 | 0.977 | 0.999 |
| Methylaspartate mutase | 0.788 | 0.429 | 0.068 | 0.780 |
| Methylated-DNA–[protein]-cysteine S-methyltransferase | -0.026 | 0.028 | 0.355 | 0.844 |
| Methylcrotonoyl-CoA carboxylase | -0.207 | 0.225 | 0.360 | 0.844 |
| Methylenetetrahydrofolate dehydrogenase (NADP(+)) | 0.012 | 0.027 | 0.667 | 0.979 |
| Methylenetetrahydrofolate reductase (NAD(P)H) | -0.030 | 0.061 | 0.617 | 0.964 |
| Methylglutaconyl-CoA hydratase | -0.108 | 0.204 | 0.596 | 0.952 |
| Methylglyoxal synthase | 0.154 | 0.189 | 0.416 | 0.865 |
| Methylisocitrate lyase | -0.046 | 0.190 | 0.810 | 0.999 |
| Methylmalonate-semialdehyde dehydrogenase (CoA acylating) | -0.261 | 0.220 | 0.237 | 0.806 |
| Methylmalonyl-CoA carboxytransferase | 0.382 | 0.518 | 0.462 | 0.884 |
| Methylmalonyl-CoA decarboxylase | -1.179 | 1.383 | 0.395 | 0.861 |
| Methylmalonyl-CoA epimerase | -0.270 | 0.157 | 0.088 | 0.806 |
| Methylmalonyl-CoA mutase | -0.101 | 0.159 | 0.525 | 0.916 |
| Methylthioribulose 1-phosphate dehydratase | 0.004 | 0.303 | 0.989 | 0.999 |
| Mevalonate kinase | 0.120 | 0.135 | 0.376 | 0.852 |
| Microbial collagenase | -0.531 | 0.761 | 0.486 | 0.899 |
| Micrococcal nuclease | -0.336 | 0.287 | 0.243 | 0.806 |
| Microsomal epoxide hydrolase | 1.900 | 1.182 | 0.110 | 0.806 |
| Molybdate-transporting ATPase | -0.017 | 0.094 | 0.857 | 0.999 |
| Molybdenum cofactor cytidylyltransferase | -0.067 | 0.131 | 0.606 | 0.960 |
| Molybdenum cofactor guanylyltransferase | -0.037 | 0.099 | 0.705 | 0.999 |
| Molybdopterin adenylyltransferase | -0.082 | 0.241 | 0.734 | 0.999 |
| Molybdopterin molybdotransferase | -0.048 | 0.087 | 0.583 | 0.948 |
| Molybdopterin synthase | -0.147 | 0.114 | 0.198 | 0.806 |
| Monoamine oxidase | -0.300 | 0.289 | 0.300 | 0.820 |
| Monogalactosyldiacylglycerol synthase | -1.464 | 0.979 | 0.137 | 0.806 |
| Monosaccharide-transporting ATPase | 0.014 | 0.098 | 0.888 | 0.999 |
| Monoterpene epsilon-lactone hydrolase | 18.543 | 3451.904 | 0.996 | 0.999 |
| Muconate cycloisomerase | 0.038 | 0.275 | 0.891 | 0.999 |
| Muconolactone Delta-isomerase | 0.058 | 0.275 | 0.832 | 0.999 |
| Muramoyltetrapeptide carboxypeptidase | 0.049 | 0.137 | 0.721 | 0.999 |
| Mycoredoxin | -0.439 | 0.501 | 0.382 | 0.852 |
| Mycothiol S-conjugate amidase | -0.010 | 0.309 | 0.975 | 0.999 |
| Mycothiol synthase | -0.010 | 0.309 | 0.975 | 0.999 |
| Mycothione reductase | -0.053 | 0.325 | 0.872 | 0.999 |
| Myo-inosose-2 dehydratase | -0.302 | 0.280 | 0.283 | 0.820 |
| N-acetyl-gamma-glutamyl-phosphate reductase | -0.067 | 0.065 | 0.306 | 0.820 |
| N-acetyldiaminopimelate deacetylase | 0.054 | 0.145 | 0.709 | 0.999 |
| N-acetylgalactosamine-4-sulfatase | 1.384 | 0.959 | 0.151 | 0.806 |
| N-acetylglucosamine-6-phosphate deacetylase | 0.003 | 0.067 | 0.964 | 0.999 |
| N-acetylglucosamine-6-sulfatase | 18.622 | 2950.870 | 0.995 | 0.999 |
| N-acetylglucosamine kinase | 0.054 | 0.249 | 0.828 | 0.999 |
| N-acetylglucosaminephosphotransferase | 0.047 | 0.094 | 0.621 | 0.966 |
| N-acetylglucosaminyl-diphospho-decaprenol L-rhamnosyltransferase | 0.078 | 0.333 | 0.814 | 0.999 |
| N-acetylhexosamine 1-kinase | -0.080 | 0.446 | 0.857 | 0.999 |
| N-acetylmuramic acid 6-phosphate etherase | -0.014 | 0.123 | 0.912 | 0.999 |
| N-acetylmuramoyl-L-alanine amidase | -0.024 | 0.076 | 0.756 | 0.999 |
| N-acetylneuraminate epimerase | -0.322 | 0.570 | 0.573 | 0.941 |
| N-acetylneuraminate lyase | 0.094 | 0.114 | 0.413 | 0.865 |
| N-acetylneuraminate synthase | 0.025 | 0.234 | 0.915 | 0.999 |
| N-acetylornithine carbamoyltransferase | -0.076 | 0.473 | 0.872 | 0.999 |
| N-acetylphosphatidylethanolamine-hydrolyzing phospholipase D | 18.701 | 3735.802 | 0.996 | 0.999 |
| N-acyl-D-amino-acid deacylase | -0.286 | 0.317 | 0.368 | 0.847 |
| N-acyl-D-glutamate deacylase | -0.343 | 0.785 | 0.663 | 0.977 |
| N-acylglucosamine-6-phosphate 2-epimerase | 0.137 | 0.100 | 0.172 | 0.806 |
| N-acylglucosamine 2-epimerase | -0.302 | 0.226 | 0.182 | 0.806 |
| N-acylmannosamine kinase | 0.008 | 0.324 | 0.981 | 0.999 |
| N-acylneuraminate-9-phosphate synthase | -2.654 | 2.718 | 0.330 | 0.829 |
| N-acylneuraminate cytidylyltransferase | -0.130 | 0.276 | 0.639 | 0.973 |
| N-carbamoyl-D-amino-acid hydrolase | -1.349 | 0.961 | 0.163 | 0.806 |
| N-carbamoyl-L-amino-acid hydrolase | -0.207 | 0.174 | 0.238 | 0.806 |
| N-carbamoylputrescine amidase | -0.197 | 0.115 | 0.088 | 0.806 |
| N-formylglutamate deformylase | -0.699 | 0.634 | 0.272 | 0.820 |
| N-formylmaleamate deformylase | -0.914 | 0.892 | 0.307 | 0.820 |
| N-hydroxyarylamine O-acetyltransferase | -0.388 | 0.315 | 0.220 | 0.806 |
| N-methylhydantoinase (ATP-hydrolyzing) | -0.556 | 0.399 | 0.165 | 0.806 |
| N-succinylarginine dihydrolase | 0.048 | 0.244 | 0.844 | 0.999 |
| N-succinylornithine carbamoyltransferase | -0.584 | 0.408 | 0.154 | 0.806 |
| N(2)-citryl-N(6)-acetyl-N(6)-hydroxylysine synthase | 0.697 | 0.786 | 0.377 | 0.852 |
| N(4)-(beta-N-acetylglucosaminyl)-L-asparaginase | -0.108 | 0.383 | 0.778 | 0.999 |
| N(5)-(carboxyethyl)ornithine synthase | 0.291 | 0.909 | 0.749 | 0.999 |
| N(6)-hydroxylysine O-acetyltransferase | 0.749 | 0.609 | 0.221 | 0.806 |
| N(6)-L-threonylcarbamoyladenine synthase | 0.062 | 0.043 | 0.149 | 0.806 |
| N,N’-diacetylchitobiose phosphorylase | 18.828 | 2749.550 | 0.995 | 0.999 |
| N,N’-diacetyllegionaminate synthase | -0.371 | 0.671 | 0.581 | 0.947 |
| N,N-dimethylformamidase | -3.899 | 3.176 | 0.221 | 0.806 |
| NAD(+)–dinitrogen-reductase ADP-D-ribosyltransferase | -17.305 | 3190.000 | 0.996 | 0.999 |
| NAD(+) ADP-ribosyltransferase | -0.261 | 0.973 | 0.788 | 0.999 |
| NAD(+) diphosphatase | 0.022 | 0.087 | 0.799 | 0.999 |
| NAD(+) glycohydrolase | -17.563 | 3628.687 | 0.996 | 0.999 |
| NAD(+) kinase | 0.032 | 0.031 | 0.308 | 0.820 |
| NAD(+) synthase | 0.070 | 0.066 | 0.286 | 0.820 |
| NAD(+) synthase (glutamine-hydrolyzing) | -0.089 | 0.122 | 0.468 | 0.885 |
| NAD(P)(+) transhydrogenase (Re/Si-specific) | -0.097 | 0.145 | 0.506 | 0.913 |
| NAD(P)(+) transhydrogenase (Si-specific) | 0.043 | 0.241 | 0.860 | 0.999 |
| NAD(P)H-hydrate epimerase | 0.013 | 0.101 | 0.901 | 0.999 |
| NAD(P)H dehydrogenase (quinone) | -0.003 | 0.107 | 0.980 | 0.999 |
| NADH dehydrogenase | -0.134 | 0.072 | 0.065 | 0.780 |
| NADH oxidase (H(2)O-forming) | 2.090 | 1.045 | 0.047 | 0.780 |
| NADH oxidase (H(2)O(2)-forming) | 0.993 | 0.900 | 0.272 | 0.820 |
| NADH peroxidase | 0.511 | 0.403 | 0.206 | 0.806 |
| NADH:ubiquinone reductase (H(+)-translocating) | -0.164 | 0.091 | 0.074 | 0.788 |
| NADPH–hemoprotein reductase | -0.501 | 0.661 | 0.450 | 0.877 |
| NADPH dehydrogenase | -0.989 | 0.460 | 0.033 | 0.780 |
| NADPH:quinone reductase | -0.272 | 0.176 | 0.124 | 0.806 |
| Neopullulanase | -0.042 | 0.166 | 0.801 | 0.999 |
| Neprilysin | -2.153 | 1.783 | 0.229 | 0.806 |
| Nickel-transporting ATPase | 0.549 | 0.376 | 0.146 | 0.806 |
| Nicotinamidase | -0.090 | 0.109 | 0.412 | 0.865 |
| Nicotinamide-nucleotide adenylyltransferase | 0.184 | 0.209 | 0.379 | 0.852 |
| Nicotinamide-nucleotide amidase | 0.032 | 0.039 | 0.402 | 0.863 |
| Nicotinamide phosphoribosyltransferase | -0.033 | 0.331 | 0.921 | 0.999 |
| Nicotinate-nucleotide–dimethylbenzimidazole phosphoribosyltransferase | 0.079 | 0.108 | 0.464 | 0.884 |
| Nicotinate-nucleotide adenylyltransferase | 0.030 | 0.040 | 0.444 | 0.871 |
| Nicotinate-nucleotide diphosphorylase (carboxylating) | -0.068 | 0.097 | 0.483 | 0.897 |
| Nicotinate dehydrogenase (cytochrome) | -0.417 | 0.456 | 0.362 | 0.844 |
| Nicotinate phosphoribosyltransferase | 0.036 | 0.035 | 0.311 | 0.820 |
| Nicotine blue oxidoreductase | -0.034 | 0.493 | 0.945 | 0.999 |
| Nitrate reductase | 0.039 | 0.106 | 0.710 | 0.999 |
| Nitric-oxide reductase (cytochrome c) | -0.028 | 0.211 | 0.894 | 0.999 |
| Nitric-oxide synthase (NAD(P)H) | -0.955 | 0.473 | 0.045 | 0.780 |
| Nitric oxide dioxygenase | -0.150 | 0.176 | 0.395 | 0.861 |
| Nitrilase | -0.220 | 0.251 | 0.383 | 0.852 |
| Nitrile hydratase | -0.696 | 0.469 | 0.140 | 0.806 |
| Nitrite reductase (cytochrome; ammonia-forming) | 0.062 | 0.203 | 0.760 | 0.999 |
| Nitrite reductase (NADH) | -0.172 | 0.172 | 0.318 | 0.824 |
| Nitrite reductase (NO-forming) | 0.029 | 0.204 | 0.888 | 0.999 |
| Nitrogenase | 0.602 | 0.343 | 0.081 | 0.806 |
| Nitronate monooxygenase | -0.204 | 0.164 | 0.215 | 0.806 |
| Nitrous-oxide reductase | -0.398 | 0.318 | 0.213 | 0.806 |
| Non-reducing end alpha-L-arabinofuranosidase | -0.830 | 0.358 | 0.022 | 0.780 |
| Non-reducing end beta-L-arabinofuranosidase | 0.115 | 1.191 | 0.923 | 0.999 |
| Non-specific protein-tyrosine kinase | -2.221 | 1.551 | 0.154 | 0.806 |
| Non-specific serine/threonine protein kinase | -0.024 | 0.064 | 0.710 | 0.999 |
| Nucleoside-diphosphate kinase | 0.011 | 0.049 | 0.818 | 0.999 |
| Nucleoside-triphosphate phosphatase | -0.890 | 1.255 | 0.479 | 0.896 |
| Nucleoside deoxyribosyltransferase | 0.392 | 0.538 | 0.467 | 0.884 |
| O-acetylhomoserine aminocarboxypropyltransferase | -0.105 | 0.075 | 0.164 | 0.806 |
| o-pyrocatechuate decarboxylase | -1.016 | 0.958 | 0.291 | 0.820 |
| o-succinylbenzoate–CoA ligase | 0.030 | 0.109 | 0.781 | 0.999 |
| o-succinylbenzoate synthase | 0.003 | 0.148 | 0.986 | 0.999 |
| O-ureido-D-serine cyclo-ligase | 18.682 | 3701.820 | 0.996 | 0.999 |
| Octanoyl-[GcvH]:protein N-octanoyltransferase | -0.127 | 0.258 | 0.623 | 0.967 |
| Oleate hydratase | 0.142 | 0.242 | 0.560 | 0.936 |
| Oligo-1,6-glucosidase | 0.153 | 0.221 | 0.489 | 0.902 |
| Oligogalacturonide lyase | -0.984 | 0.777 | 0.207 | 0.806 |
| Oligonucleotidase | 0.066 | 0.083 | 0.432 | 0.869 |
| Oligopeptidase A | 0.025 | 0.163 | 0.880 | 0.999 |
| Oligopeptidase B | -0.010 | 0.151 | 0.947 | 0.999 |
| Oligosaccharide 4-alpha-D-glucosyltransferase | -1.129 | 1.797 | 0.531 | 0.920 |
| Oligosaccharide reducing-end xylanase | -4.839 | 4.902 | 0.325 | 0.827 |
| Omptin | -0.768 | 1.245 | 0.538 | 0.924 |
| Opine dehydrogenase | -0.320 | 0.605 | 0.597 | 0.952 |
| Ornithine aminotransferase | -0.769 | 0.307 | 0.013 | 0.780 |
| Ornithine carbamoyltransferase | 0.085 | 0.065 | 0.190 | 0.806 |
| Ornithine cyclodeaminase | -0.264 | 0.166 | 0.115 | 0.806 |
| Ornithine decarboxylase | 0.114 | 0.140 | 0.416 | 0.865 |
| Orotate phosphoribosyltransferase | 0.034 | 0.032 | 0.293 | 0.820 |
| Orotidine-5’-phosphate decarboxylase | 0.049 | 0.033 | 0.137 | 0.806 |
| Oxalate decarboxylase | -0.744 | 0.859 | 0.388 | 0.853 |
| Oxaloacetate decarboxylase | 0.239 | 0.149 | 0.112 | 0.806 |
| Oxalyl-CoA decarboxylase | 0.456 | 0.679 | 0.502 | 0.912 |
| Oxepin-CoA hydrolase | -0.081 | 0.318 | 0.801 | 0.999 |
| Oxoglutarate dehydrogenase (succinyl-transferring) | -0.088 | 0.115 | 0.445 | 0.871 |
| Pantetheine-phosphate adenylyltransferase | 0.037 | 0.031 | 0.236 | 0.806 |
| Pantoate–beta-alanine ligase (AMP-forming) | -0.114 | 0.070 | 0.109 | 0.806 |
| Pantoate kinase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Pantothenate kinase | 0.018 | 0.030 | 0.550 | 0.932 |
| Pectate lyase | -0.797 | 0.656 | 0.226 | 0.806 |
| Pectin lyase | -0.838 | 0.918 | 0.363 | 0.844 |
| Pectinesterase | -0.852 | 0.397 | 0.033 | 0.780 |
| Penicillin amidase | -0.149 | 0.236 | 0.528 | 0.918 |
| Pentachlorophenol monooxygenase | 0.758 | 0.923 | 0.413 | 0.865 |
| Pentalenene oxygenase | -17.486 | 3491.164 | 0.996 | 0.999 |
| Pentalenic acid synthase | -17.486 | 3491.164 | 0.996 | 0.999 |
| Pentalenolactone synthase | 18.521 | 3415.516 | 0.996 | 0.999 |
| PepB aminopeptidase | 0.235 | 0.305 | 0.442 | 0.870 |
| Peptidase Do | -0.043 | 0.054 | 0.429 | 0.867 |
| Peptide-aspartate beta-dioxygenase | -1.625 | 1.941 | 0.404 | 0.864 |
| Peptide-methionine (R)-S-oxide reductase | 0.079 | 0.049 | 0.106 | 0.806 |
| Peptide-methionine (S)-S-oxide reductase | 0.050 | 0.046 | 0.279 | 0.820 |
| Peptide chain release factor N(5)-glutamine methyltransferase | 0.016 | 0.028 | 0.551 | 0.932 |
| Peptide deformylase | 0.012 | 0.044 | 0.778 | 0.999 |
| Peptidoglycan glycosyltransferase | 0.070 | 0.067 | 0.295 | 0.820 |
| Peptidyl-dipeptidase A | -0.196 | 0.406 | 0.631 | 0.971 |
| Peptidyl-dipeptidase Dcp | -0.225 | 0.157 | 0.154 | 0.806 |
| Peptidyl-Lys metalloendopeptidase | 0.968 | 0.912 | 0.290 | 0.820 |
| Peptidylglycine monooxygenase | 1.557 | 1.308 | 0.236 | 0.806 |
| Peptidylprolyl isomerase | 0.006 | 0.038 | 0.880 | 0.999 |
| Peroxiredoxin | 0.005 | 0.031 | 0.871 | 0.999 |
| Peroxyureidoacrylate/ureidoacrylate amidohydrolase | -0.110 | 0.436 | 0.800 | 0.999 |
| Phenol 2-monooxygenase | -0.014 | 0.744 | 0.985 | 0.999 |
| Phenylacetaldehyde dehydrogenase | 0.007 | 0.366 | 0.984 | 0.999 |
| Phenylacetate–CoA ligase | -0.376 | 0.218 | 0.086 | 0.806 |
| Phenylacetyl-CoA 1,2-epoxidase | -0.231 | 0.277 | 0.407 | 0.865 |
| Phenylalanine–tRNA ligase | 0.037 | 0.027 | 0.171 | 0.806 |
| Phenylalanine 4-monooxygenase | -0.067 | 0.241 | 0.780 | 0.999 |
| Phenylalanine ammonia-lyase | 1.433 | 0.995 | 0.152 | 0.806 |
| Phenylalanine dehydrogenase | -2.184 | 1.410 | 0.123 | 0.806 |
| Phosphate-transporting ATPase | -0.038 | 0.078 | 0.631 | 0.971 |
| Phosphate acetyltransferase | 0.026 | 0.046 | 0.565 | 0.936 |
| Phosphate butyryltransferase | -0.196 | 0.232 | 0.398 | 0.863 |
| Phosphate propanoyltransferase | 0.261 | 0.262 | 0.321 | 0.827 |
| Phosphatidate cytidylyltransferase | 0.065 | 0.028 | 0.024 | 0.780 |
| Phosphatidate phosphatase | 0.051 | 0.256 | 0.841 | 0.999 |
| Phosphatidyl-N-methylethanolamine N-methyltransferase | -1.278 | 0.637 | 0.047 | 0.780 |
| Phosphatidylcholine synthase | -0.056 | 0.298 | 0.852 | 0.999 |
| Phosphatidylethanolamine N-methyltransferase | -1.278 | 0.637 | 0.047 | 0.780 |
| Phosphatidylglycerol–membrane-oligosaccharide glycerophosphotransferase | 0.165 | 0.167 | 0.324 | 0.827 |
| Phosphatidylglycerophosphatase | 0.171 | 0.137 | 0.214 | 0.806 |
| Phosphatidylinositol alpha-mannosyltransferase | -0.017 | 0.274 | 0.949 | 0.999 |
| Phosphatidylinositol diacylglycerol-lyase | -1.512 | 0.818 | 0.066 | 0.780 |
| Phosphatidylserine decarboxylase | -0.001 | 0.060 | 0.988 | 0.999 |
| Phosphinothricin acetyltransferase | -0.036 | 0.055 | 0.508 | 0.913 |
| Phospho-N-acetylmuramoyl-pentapeptide-transferase | 0.036 | 0.031 | 0.240 | 0.806 |
| Phosphoadenylyl-sulfate reductase (thioredoxin) | -0.261 | 0.154 | 0.093 | 0.806 |
| Phosphoenolpyruvate–protein phosphotransferase | 0.090 | 0.042 | 0.031 | 0.780 |
| Phosphoenolpyruvate carboxykinase (ATP) | 0.062 | 0.076 | 0.413 | 0.865 |
| Phosphoenolpyruvate carboxykinase (GTP) | -0.145 | 0.210 | 0.489 | 0.902 |
| Phosphoenolpyruvate carboxylase | 0.018 | 0.071 | 0.797 | 0.999 |
| Phosphoenolpyruvate mutase | -0.159 | 0.520 | 0.760 | 0.999 |
| Phosphoethanolamine N-methyltransferase | -17.155 | 2958.490 | 0.995 | 0.999 |
| Phosphoglucomutase (alpha-D-glucose-1,6-bisphosphate-dependent) | 0.020 | 0.035 | 0.563 | 0.936 |
| Phosphogluconate 2-dehydrogenase | -0.032 | 0.443 | 0.942 | 0.999 |
| Phosphogluconate dehydratase | -0.101 | 0.220 | 0.646 | 0.973 |
| Phosphogluconate dehydrogenase (NAD(+)-dependent, decarboxylating) | -0.016 | 0.060 | 0.785 | 0.999 |
| Phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating) | -0.016 | 0.060 | 0.785 | 0.999 |
| Phosphoglucosamine mutase | 0.069 | 0.035 | 0.048 | 0.780 |
| Phosphoglycerate dehydrogenase | -0.027 | 0.054 | 0.615 | 0.963 |
| Phosphoglycerate kinase | 0.038 | 0.032 | 0.236 | 0.806 |
| Phosphoglycerate mutase (2,3-diphosphoglycerate-dependent) | 0.114 | 0.087 | 0.194 | 0.806 |
| Phosphoglycerate mutase (2,3-diphosphoglycerate-independent) | 0.031 | 0.069 | 0.654 | 0.973 |
| Phosphoglycerol geranylgeranyltransferase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Phosphoglycolate phosphatase | -0.050 | 0.058 | 0.393 | 0.861 |
| Phosphoketolase | 0.224 | 0.280 | 0.425 | 0.865 |
| Phospholipase A(1) | -0.097 | 0.223 | 0.664 | 0.977 |
| Phospholipase A(2) | -0.097 | 0.223 | 0.664 | 0.977 |
| Phospholipase C | -0.221 | 0.292 | 0.451 | 0.878 |
| Phospholipase D | -2.404 | 1.524 | 0.117 | 0.806 |
| Phosphomannomutase | -0.106 | 0.091 | 0.245 | 0.806 |
| Phosphomethylpyrimidine kinase | 0.057 | 0.047 | 0.229 | 0.806 |
| Phosphomethylpyrimidine synthase | 0.041 | 0.084 | 0.625 | 0.969 |
| Phosphomevalonate kinase | 0.113 | 0.139 | 0.419 | 0.865 |
| Phosphonate-transporting ATPase | 0.157 | 0.168 | 0.350 | 0.840 |
| Phosphonate dehydrogenase | -0.263 | 1.225 | 0.830 | 0.999 |
| Phosphonoacetaldehyde hydrolase | -0.242 | 0.326 | 0.460 | 0.884 |
| Phosphonoacetate hydrolase | -0.503 | 0.940 | 0.593 | 0.951 |
| Phosphonopyruvate decarboxylase | -0.379 | 0.494 | 0.445 | 0.871 |
| Phosphopantothenate–cysteine ligase | 0.038 | 0.027 | 0.171 | 0.806 |
| Phosphopantothenoylcysteine decarboxylase | 0.038 | 0.027 | 0.163 | 0.806 |
| Phosphopentomutase | 0.008 | 0.104 | 0.935 | 0.999 |
| Phosphopyruvate hydratase | 0.042 | 0.054 | 0.440 | 0.869 |
| Phosphoribosyl-AMP cyclohydrolase | -0.084 | 0.057 | 0.144 | 0.806 |
| Phosphoribosyl-ATP diphosphatase | -0.073 | 0.057 | 0.205 | 0.806 |
| Phosphoribosyl 1,2-cyclic phosphate phosphodiesterase | -0.364 | 0.165 | 0.029 | 0.780 |
| Phosphoribosylamine–glycine ligase | 0.036 | 0.037 | 0.328 | 0.829 |
| Phosphoribosylaminoimidazolecarboxamide formyltransferase | 0.039 | 0.042 | 0.355 | 0.844 |
| Phosphoribosylaminoimidazolesuccinocarboxamide synthase | 0.040 | 0.034 | 0.238 | 0.806 |
| Phosphoribosylanthranilate isomerase | -0.086 | 0.063 | 0.172 | 0.806 |
| Phosphoribosylformylglycinamidine cyclo-ligase | 0.033 | 0.035 | 0.359 | 0.844 |
| Phosphoribosylformylglycinamidine synthase | 0.022 | 0.050 | 0.653 | 0.973 |
| Phosphoribosylglycinamide formyltransferase | 0.079 | 0.028 | 0.006 | 0.780 |
| Phosphoribulokinase | -0.210 | 0.431 | 0.627 | 0.971 |
| Phosphoserine phosphatase | -0.077 | 0.065 | 0.236 | 0.806 |
| Phosphoserine transaminase | -0.073 | 0.060 | 0.222 | 0.806 |
| Phosphosulfolactate synthase | -1.091 | 0.797 | 0.173 | 0.806 |
| Phytanoyl-CoA dioxygenase | -0.297 | 1.127 | 0.792 | 0.999 |
| Phytoene desaturase (lycopene-forming) | -0.177 | 0.395 | 0.654 | 0.973 |
| Phytoene desaturase (neurosporene-forming) | -0.177 | 0.395 | 0.654 | 0.973 |
| Phytoene desaturase (zeta-carotene-forming) | -0.177 | 0.395 | 0.654 | 0.973 |
| Pimeloyl-[acyl-carrier protein] methyl ester esterase | 0.214 | 0.142 | 0.133 | 0.806 |
| Pimeloyl-[acyl-carrier protein] synthase | 17.953 | 2571.043 | 0.994 | 0.999 |
| Pitrilysin | 0.210 | 0.343 | 0.541 | 0.926 |
| Plasminogen activator Pla | 2.057 | 1.035 | 0.049 | 0.780 |
| Plastoquinol–plastocyanin reductase | -1.724 | 1.549 | 0.267 | 0.820 |
| Polar-amino-acid-transporting ATPase | 0.025 | 0.071 | 0.726 | 0.999 |
| Poly(3-hydroxybutyrate) depolymerase | -0.298 | 0.309 | 0.335 | 0.833 |
| Poly(beta-D-mannuronate) lyase | 0.070 | 0.316 | 0.825 | 0.999 |
| Poly(glycerol-phosphate) alpha-glucosyltransferase | 0.085 | 0.245 | 0.730 | 0.999 |
| Polyamine-transporting ATPase | 0.087 | 0.093 | 0.349 | 0.840 |
| Polygalacturonase | -0.395 | 1.048 | 0.707 | 0.999 |
| Polymannosyl GlcNAc-diphospho-ditrans,octacis-undecaprenol kinase | 2.793 | 1.233 | 0.025 | 0.780 |
| Polynucleotide adenylyltransferase | -0.049 | 0.110 | 0.654 | 0.973 |
| Polyphosphate–glucose phosphotransferase | 0.039 | 0.248 | 0.876 | 0.999 |
| Polyphosphate kinase | -0.022 | 0.077 | 0.772 | 0.999 |
| Polyribonucleotide nucleotidyltransferase | 0.023 | 0.040 | 0.574 | 0.941 |
| Polyvinyl alcohol dehydrogenase (cytochrome) | -1.478 | 1.335 | 0.270 | 0.820 |
| Porphobilinogen synthase | 0.064 | 0.067 | 0.342 | 0.837 |
| Potassium-transporting ATPase | -0.217 | 0.179 | 0.228 | 0.806 |
| Precorrin-2 C(20)-methyltransferase | 0.225 | 0.131 | 0.089 | 0.806 |
| Precorrin-2 dehydrogenase | 0.137 | 0.092 | 0.135 | 0.806 |
| Precorrin-3B C(17)-methyltransferase | 0.234 | 0.137 | 0.090 | 0.806 |
| Precorrin-3B synthase | -0.083 | 0.302 | 0.784 | 0.999 |
| Precorrin-4 C(11)-methyltransferase | 0.231 | 0.131 | 0.081 | 0.806 |
| Precorrin-6A reductase | 0.281 | 0.131 | 0.034 | 0.780 |
| Precorrin-6A synthase (deacetylating) | -0.193 | 0.298 | 0.518 | 0.913 |
| Precorrin-6B C(5,15)-methyltransferase (decarboxylating) | -0.042 | 0.199 | 0.835 | 0.999 |
| Precorrin-8X methylmutase | 0.229 | 0.131 | 0.084 | 0.806 |
| Prephenate dehydratase | -0.014 | 0.042 | 0.740 | 0.999 |
| Prephenate dehydrogenase | -0.041 | 0.062 | 0.514 | 0.913 |
| Prepilin peptidase | 0.070 | 0.039 | 0.076 | 0.795 |
| PreQ(1) synthase | -0.115 | 0.062 | 0.065 | 0.780 |
| Primary-amine oxidase | -0.244 | 0.423 | 0.565 | 0.936 |
| Procollagen-proline dioxygenase | -0.775 | 0.438 | 0.078 | 0.806 |
| Proline–tRNA ligase | 0.030 | 0.027 | 0.274 | 0.820 |
| Proline dehydrogenase | -0.040 | 0.147 | 0.786 | 0.999 |
| Proline racemase | -0.245 | 0.561 | 0.664 | 0.977 |
| Prolycopene isomerase | -0.417 | 1.284 | 0.746 | 0.999 |
| Prolyl aminopeptidase | -0.006 | 0.149 | 0.969 | 0.999 |
| Prolyl oligopeptidase | -0.124 | 0.161 | 0.441 | 0.869 |
| Propanal dehydrogenase (CoA-propanoylating) | 0.222 | 0.215 | 0.304 | 0.820 |
| Propanediol dehydratase | 0.275 | 0.248 | 0.268 | 0.820 |
| Propionate–CoA ligase | -0.266 | 0.273 | 0.331 | 0.831 |
| Propionate CoA-transferase | -0.259 | 0.334 | 0.439 | 0.869 |
| Propionate kinase | -0.280 | 0.716 | 0.696 | 0.995 |
| Propionyl-CoA carboxylase | -0.372 | 0.202 | 0.068 | 0.780 |
| Prostaglandin-endoperoxide synthase | 2.105 | 1.785 | 0.240 | 0.806 |
| Proteasome endopeptidase complex | -0.407 | 0.668 | 0.543 | 0.927 |
| Protein-disulfide reductase | -0.020 | 0.122 | 0.871 | 0.999 |
| Protein-glutamate methylesterase | -0.098 | 0.214 | 0.646 | 0.973 |
| Protein-glutamate O-methyltransferase | -0.095 | 0.199 | 0.635 | 0.973 |
| Protein-glutamine gamma-glutamyltransferase | -2.730 | 1.413 | 0.055 | 0.780 |
| Protein-glutamine glutaminase | -0.198 | 0.195 | 0.312 | 0.820 |
| Protein-histidine pros-kinase | -1.079 | 0.833 | 0.197 | 0.806 |
| Protein-L-isoaspartate(D-aspartate) O-methyltransferase | -0.142 | 0.175 | 0.417 | 0.865 |
| Protein-N(pi)-phosphohistidine–sugar phosphotransferase | 0.096 | 0.087 | 0.273 | 0.820 |
| Protein-S-isoprenylcysteine O-methyltransferase | -17.382 | 2371.984 | 0.994 | 0.999 |
| Protein-serine/threonine phosphatase | -0.037 | 0.061 | 0.551 | 0.932 |
| Protein-tyrosine-phosphatase | -0.009 | 0.077 | 0.906 | 0.999 |
| Protein arginine kinase | 0.071 | 0.191 | 0.710 | 0.999 |
| Protein disulfide-isomerase | 0.034 | 0.174 | 0.845 | 0.999 |
| Protein geranylgeranyltransferase type II | 2.687 | 2.295 | 0.243 | 0.806 |
| Protocatechuate 3,4-dioxygenase | -0.069 | 0.255 | 0.787 | 0.999 |
| Protocatechuate 4,5-dioxygenase | -1.853 | 1.277 | 0.149 | 0.806 |
| Proton-exporting ATPase | -0.020 | 0.853 | 0.981 | 0.999 |
| Protoporphyrinogen IX dehydrogenase (menaquinone) | 0.152 | 0.260 | 0.558 | 0.936 |
| Protoporphyrinogen oxidase | -0.140 | 0.149 | 0.348 | 0.840 |
| Pseudaminic acid cytidylyltransferase | 0.014 | 0.526 | 0.979 | 0.999 |
| Pseudaminic acid synthase | -0.876 | 0.860 | 0.310 | 0.820 |
| Pseudolysin | -17.542 | 1920.349 | 0.993 | 0.999 |
| Pseudouridine kinase | -0.123 | 0.346 | 0.724 | 0.999 |
| Pteridine reductase | 0.024 | 0.418 | 0.954 | 0.999 |
| Pulcherriminic acid synthase | -0.533 | 1.614 | 0.742 | 0.999 |
| Pullulanase | -0.108 | 0.139 | 0.438 | 0.869 |
| Purine-nucleoside phosphorylase | 0.015 | 0.090 | 0.868 | 0.999 |
| Purine nucleosidase | -0.027 | 0.188 | 0.888 | 0.999 |
| Putrescine aminotransferase | -0.216 | 0.608 | 0.723 | 0.999 |
| Putrescine carbamoyltransferase | 0.147 | 0.727 | 0.840 | 0.999 |
| Putrescine oxidase | -0.721 | 0.602 | 0.233 | 0.806 |
| Pyridoxal 4-dehydrogenase | 17.690 | 2253.589 | 0.994 | 0.999 |
| Pyridoxal 5’-phosphate synthase | -0.059 | 0.139 | 0.671 | 0.982 |
| Pyridoxal 5’-phosphate synthase (glutamine hydrolyzing) | -0.143 | 0.109 | 0.190 | 0.806 |
| Pyridoxal kinase | -0.008 | 0.058 | 0.887 | 0.999 |
| Pyridoxamine–pyruvate transaminase | 17.690 | 2253.589 | 0.994 | 0.999 |
| Pyridoxine 4-dehydrogenase | -1.384 | 1.131 | 0.223 | 0.806 |
| Pyridoxine 4-oxidase | 17.690 | 2253.589 | 0.994 | 0.999 |
| Pyridoxine 5’-phosphate synthase | -0.155 | 0.143 | 0.283 | 0.820 |
| Pyrimidine-nucleoside phosphorylase | 0.032 | 0.109 | 0.768 | 0.999 |
| Pyrimidine monooxygenase | 0.014 | 0.492 | 0.978 | 0.999 |
| Pyroglutamyl-peptidase I | 0.065 | 0.105 | 0.537 | 0.924 |
| Pyrroline-5-carboxylate reductase | -0.008 | 0.044 | 0.857 | 0.999 |
| Pyrroloquinoline-quinone synthase | 0.027 | 0.285 | 0.925 | 0.999 |
| Pyruvate carboxylase | -0.032 | 0.128 | 0.804 | 0.999 |
| Pyruvate decarboxylase | -16.954 | 2676.034 | 0.995 | 0.999 |
| Pyruvate dehydrogenase (acetyl-transferring) | -0.077 | 0.074 | 0.301 | 0.820 |
| Pyruvate dehydrogenase (quinone) | -0.086 | 0.233 | 0.712 | 0.999 |
| Pyruvate kinase | 0.060 | 0.026 | 0.024 | 0.780 |
| Pyruvate oxidase | -0.043 | 0.161 | 0.790 | 0.999 |
| Pyruvate synthase | 0.405 | 0.217 | 0.064 | 0.780 |
| Pyruvate, phosphate dikinase | -0.019 | 0.125 | 0.878 | 0.999 |
| Pyruvate, water dikinase | 0.024 | 0.088 | 0.789 | 0.999 |
| Quaternary-amine-transporting ATPase | 0.092 | 0.246 | 0.708 | 0.999 |
| Quercetin 2,3-dioxygenase | -17.352 | 3264.645 | 0.996 | 0.999 |
| Quinate dehydrogenase (quinone) | -0.087 | 0.323 | 0.788 | 0.999 |
| Quinate/shikimate dehydrogenase | -1.179 | 1.383 | 0.395 | 0.861 |
| Quinol–cytochrome-c reductase | -0.110 | 0.186 | 0.553 | 0.934 |
| Quinolinate synthase | -0.053 | 0.105 | 0.611 | 0.960 |
| Quinoprotein glucose dehydrogenase (PQQ, quinone) | -0.080 | 0.243 | 0.743 | 0.999 |
| Quorum-quenching N-acyl-homoserine lactonase | -2.454 | 2.167 | 0.259 | 0.820 |
| Receptor protein-tyrosine kinase | -1.146 | 0.833 | 0.171 | 0.806 |
| Repressor LexA | 0.088 | 0.059 | 0.139 | 0.806 |
| Rhamnogalacturonan endolyase | -1.164 | 0.604 | 0.056 | 0.780 |
| Rhamnogalacturonan exolyase | 17.953 | 2571.043 | 0.994 | 0.999 |
| Rhamnulokinase | 0.204 | 0.227 | 0.371 | 0.850 |
| Rhamnulose-1-phosphate aldolase | 0.096 | 0.354 | 0.786 | 0.999 |
| Rhomboid protease | 0.285 | 0.262 | 0.278 | 0.820 |
| Ribitol-5-phosphate 2-dehydrogenase | -0.275 | 0.217 | 0.207 | 0.806 |
| Ribitol 2-dehydrogenase | -0.765 | 0.576 | 0.187 | 0.806 |
| Riboflavin kinase | 0.041 | 0.031 | 0.196 | 0.806 |
| Riboflavin reductase (NAD(P)H) | 0.377 | 0.522 | 0.471 | 0.888 |
| Riboflavin synthase | 0.015 | 0.064 | 0.810 | 0.999 |
| Ribokinase | 0.032 | 0.111 | 0.776 | 0.999 |
| Ribonuclease D | -0.086 | 0.126 | 0.493 | 0.905 |
| Ribonuclease E | -0.015 | 0.121 | 0.900 | 0.999 |
| Ribonuclease H | 0.006 | 0.036 | 0.873 | 0.999 |
| Ribonuclease III | 0.041 | 0.028 | 0.144 | 0.806 |
| Ribonuclease M5 | 0.126 | 0.097 | 0.198 | 0.806 |
| Ribonuclease P | 0.058 | 0.036 | 0.112 | 0.806 |
| Ribonuclease T(1) | -0.766 | 0.686 | 0.266 | 0.820 |
| Ribonuclease T(2) | -0.285 | 0.209 | 0.175 | 0.806 |
| Ribonuclease Z | -0.012 | 0.079 | 0.880 | 0.999 |
| Ribonucleoside-diphosphate reductase | 0.002 | 0.036 | 0.965 | 0.999 |
| Ribonucleoside-triphosphate reductase | 0.073 | 0.089 | 0.414 | 0.865 |
| Ribose-5-phosphate isomerase | 0.045 | 0.041 | 0.269 | 0.820 |
| Ribose-phosphate diphosphokinase | 0.054 | 0.051 | 0.290 | 0.820 |
| Ribose 1,5-bisphosphate isomerase | 18.464 | 3319.693 | 0.996 | 0.999 |
| Ribose 1,5-bisphosphate phosphokinase | -0.131 | 0.282 | 0.643 | 0.973 |
| Ribosomal-protein-alanine N-acetyltransferase | 0.051 | 0.075 | 0.497 | 0.905 |
| Ribosomal protein L3 N(5)-glutamine methyltransferase | 0.056 | 0.158 | 0.722 | 0.999 |
| Ribosylnicotinamide kinase | 0.342 | 0.253 | 0.177 | 0.806 |
| Ribosylpyrimidine nucleosidase | 0.023 | 0.452 | 0.959 | 0.999 |
| Ribulokinase | -0.369 | 0.357 | 0.304 | 0.820 |
| Ribulose-bisphosphate carboxylase | -0.052 | 0.238 | 0.826 | 0.999 |
| Ribulose-phosphate 3-epimerase | 0.044 | 0.032 | 0.163 | 0.806 |
| RNA-directed DNA polymerase | 0.323 | 0.521 | 0.536 | 0.924 |
| RNA 3’-terminal-phosphate cyclase (ATP) | -0.037 | 0.425 | 0.931 | 0.999 |
| RNA helicase | 0.011 | 0.046 | 0.804 | 0.999 |
| RNA ligase (ATP) | -0.166 | 0.199 | 0.407 | 0.865 |
| rRNA small subunit pseudouridine methyltransferase Nep1 | -16.940 | 2657.012 | 0.995 | 0.999 |
| Rubredoxin–NAD(+) reductase | 0.172 | 0.266 | 0.518 | 0.913 |
| S-(hydroxymethyl)glutathione dehydrogenase | -0.317 | 0.195 | 0.106 | 0.806 |
| S-(hydroxymethyl)glutathione synthase | -0.984 | 0.707 | 0.166 | 0.806 |
| S-(hydroxymethyl)mycothiol dehydrogenase | -0.049 | 0.330 | 0.881 | 0.999 |
| S-adenosylhomocysteine deaminase | -0.026 | 0.093 | 0.778 | 0.999 |
| S-adenosylmethionine:tRNA ribosyltransferase-isomerase | -0.007 | 0.053 | 0.897 | 0.999 |
| S-formylglutathione hydrolase | -0.107 | 0.175 | 0.544 | 0.928 |
| S-methyl-5’-thioadenosine deaminase | -0.026 | 0.093 | 0.778 | 0.999 |
| S-methyl-5’-thioadenosine phosphorylase | -0.003 | 0.179 | 0.988 | 0.999 |
| S-methyl-5’-thioinosine phosphorylase | 0.224 | 0.355 | 0.530 | 0.920 |
| S-methyl-5-thioribose-1-phosphate isomerase | 0.240 | 0.126 | 0.059 | 0.780 |
| S-methyl-5-thioribose kinase | 0.638 | 0.339 | 0.062 | 0.780 |
| S-ribosylhomocysteine lyase | 0.030 | 0.086 | 0.724 | 0.999 |
| Saccharopine dehydrogenase (NAD(+), L-lysine-forming) | -0.221 | 0.152 | 0.147 | 0.806 |
| Saccharopine dehydrogenase (NADP(+), L-glutamate-forming) | 1.525 | 1.581 | 0.336 | 0.833 |
| Salicylate 1-monooxygenase | -1.081 | 0.532 | 0.044 | 0.780 |
| Salicylate 5-hydroxylase | -3.009 | 2.762 | 0.278 | 0.820 |
| Sarcosine oxidase | -0.202 | 0.293 | 0.493 | 0.905 |
| Sarcosine/dimethylglycine N-methyltransferase | -1.033 | 0.992 | 0.299 | 0.820 |
| Scyllo-inositol 2-dehydrogenase (NAD(+)) | 1.187 | 1.252 | 0.345 | 0.838 |
| Scyllo-inositol 2-dehydrogenase (NADP(+)) | -0.628 | 0.666 | 0.347 | 0.840 |
| Sedoheptulokinase | -3.822 | 3.651 | 0.297 | 0.820 |
| Sedoheptulose-bisphosphatase | -1.279 | 0.453 | 0.005 | 0.780 |
| Sedolisin | -17.206 | 3034.891 | 0.995 | 0.999 |
| Selenate reductase | 0.174 | 0.466 | 0.710 | 0.999 |
| Selenide, water dikinase | 0.183 | 0.110 | 0.099 | 0.806 |
| Selenocysteine lyase | -0.022 | 0.064 | 0.735 | 0.999 |
| Serine–glyoxylate transaminase | -1.129 | 0.627 | 0.074 | 0.787 |
| Serine–pyruvate transaminase | -1.129 | 0.627 | 0.074 | 0.787 |
| Serine–tRNA ligase | 0.039 | 0.031 | 0.219 | 0.806 |
| Serine-type D-Ala-D-Ala carboxypeptidase | -0.038 | 0.047 | 0.421 | 0.865 |
| Serine 3-dehydrogenase (NADP(+)) | -0.166 | 0.429 | 0.699 | 0.998 |
| Serine O-acetyltransferase | -0.021 | 0.046 | 0.642 | 0.973 |
| Serine racemase | -17.363 | 2119.006 | 0.993 | 0.999 |
| Serralysin | -0.581 | 0.515 | 0.261 | 0.820 |
| Shikimate dehydrogenase | 0.092 | 0.058 | 0.111 | 0.806 |
| Shikimate kinase | 0.043 | 0.045 | 0.341 | 0.837 |
| Short-chain acyl-CoA dehydrogenase | 0.503 | 0.244 | 0.040 | 0.780 |
| Sialate O-acetylesterase | -0.375 | 0.219 | 0.088 | 0.806 |
| Signal peptidase I | 0.002 | 0.041 | 0.963 | 0.999 |
| Signal peptidase II | 0.047 | 0.033 | 0.159 | 0.806 |
| Sirohydrochlorin cobaltochelatase | 0.236 | 0.166 | 0.155 | 0.806 |
| Sirohydrochlorin ferrochelatase | 0.086 | 0.096 | 0.371 | 0.850 |
| Site-specific DNA-methyltransferase (adenine-specific) | 0.003 | 0.060 | 0.966 | 0.999 |
| Site-specific DNA-methyltransferase (cytosine-N(4)-specific) | 2.347 | 1.772 | 0.187 | 0.806 |
| sn-glycerol-1-phosphate dehydrogenase | -0.184 | 0.339 | 0.589 | 0.951 |
| Snapalysin | -17.486 | 3491.164 | 0.996 | 0.999 |
| Sorbitol-6-phosphate 2-dehydrogenase | 0.338 | 0.351 | 0.337 | 0.833 |
| Sortase A | 0.059 | 0.111 | 0.592 | 0.951 |
| specific) | -1.442 | 1.406 | 0.306 | 0.820 |
| Spermidine dehydrogenase | -0.733 | 0.806 | 0.364 | 0.844 |
| Spermidine synthase | -0.099 | 0.096 | 0.302 | 0.820 |
| Spheroidene monooxygenase | -17.226 | 2215.245 | 0.994 | 0.999 |
| Sphingomyelin phosphodiesterase | -2.262 | 1.686 | 0.182 | 0.806 |
| SpoIVB peptidase | -0.987 | 0.494 | 0.047 | 0.780 |
| Spore photoproduct lyase | 0.440 | 0.363 | 0.227 | 0.806 |
| Sporulenol synthase | -2.135 | 1.162 | 0.068 | 0.780 |
| Squalene–hopanol cyclase | -0.181 | 0.576 | 0.753 | 0.999 |
| Squalene–hopene cyclase | -0.181 | 0.576 | 0.753 | 0.999 |
| Squalene synthase | -0.761 | 1.255 | 0.545 | 0.928 |
| Staphopain | -1.552 | 1.123 | 0.169 | 0.806 |
| Starch synthase | 0.030 | 0.068 | 0.659 | 0.975 |
| Starch synthase (maltosyl-transferring) | 0.038 | 0.253 | 0.882 | 0.999 |
| Ste24 endopeptidase | -0.177 | 0.346 | 0.610 | 0.960 |
| Stearoyl-[acyl-carrier-protein] 9-desaturase | -1.679 | 0.674 | 0.014 | 0.780 |
| Stearoyl-CoA 9-desaturase | -0.171 | 0.298 | 0.566 | 0.936 |
| Steroid Delta-isomerase | -1.567 | 1.270 | 0.219 | 0.806 |
| Sterol 14-alpha-demethylase | -2.061 | 1.512 | 0.175 | 0.806 |
| Streptogrisin B | -17.486 | 3491.164 | 0.996 | 0.999 |
| Streptomycin 3’’-adenylyltransferase | -0.315 | 0.422 | 0.457 | 0.884 |
| Streptomycin 3’’-kinase | 1.674 | 1.572 | 0.288 | 0.820 |
| Streptomycin 6-kinase | -0.127 | 0.416 | 0.761 | 0.999 |
| Subtilisin | 18.282 | 1927.540 | 0.992 | 0.999 |
| Succinate–CoA ligase (ADP-forming) | -0.095 | 0.083 | 0.252 | 0.809 |
| Succinate–hydroxymethylglutarate CoA-transferase | 17.690 | 2253.589 | 0.994 | 0.999 |
| Succinate-semialdehyde dehydrogenase (acetylating) | -0.272 | 0.595 | 0.648 | 0.973 |
| Succinate-semialdehyde dehydrogenase (NAD(+)) | 0.001 | 0.268 | 0.998 | 0.999 |
| Succinate-semialdehyde dehydrogenase (NAD(P)(+)) | -0.048 | 0.136 | 0.726 | 0.999 |
| Succinate-semialdehyde dehydrogenase (NADP(+)) | -0.061 | 0.139 | 0.660 | 0.975 |
| Succinate dehydrogenase (quinone) | -0.112 | 0.071 | 0.115 | 0.806 |
| Succinyl-CoA–D-citramalate CoA-transferase | -2.682 | 2.479 | 0.281 | 0.820 |
| Succinyl-CoA–L-malate CoA-transferase | 0.149 | 0.441 | 0.736 | 0.999 |
| Succinyl-CoA:(R)-benzylsuccinate CoA-transferase | -0.166 | 1.496 | 0.912 | 0.999 |
| Succinyl-CoA:acetate CoA-transferase | -0.019 | 0.144 | 0.893 | 0.999 |
| Succinyl-diaminopimelate desuccinylase | 0.115 | 0.080 | 0.155 | 0.806 |
| Succinyldiaminopimelate transaminase | 0.008 | 0.119 | 0.946 | 0.999 |
| Succinylglutamate-semialdehyde dehydrogenase | 0.012 | 0.268 | 0.965 | 0.999 |
| Succinylglutamate desuccinylase | 0.086 | 0.262 | 0.743 | 0.999 |
| Succinylornithine transaminase | -0.342 | 0.394 | 0.387 | 0.852 |
| Sucrose-phosphate synthase | 0.995 | 1.000 | 0.321 | 0.827 |
| Sucrose phosphorylase | 0.079 | 0.182 | 0.664 | 0.977 |
| Sugar-phosphatase | -0.210 | 0.323 | 0.517 | 0.913 |
| Sulfate-transporting ATPase | -0.129 | 0.174 | 0.462 | 0.884 |
| Sulfate adenylyltransferase | -0.230 | 0.150 | 0.129 | 0.806 |
| Sulfide-cytochrome-c reductase (flavocytochrome c) | -17.895 | 4282.949 | 0.997 | 0.999 |
| Sulfite dehydrogenase | -2.212 | 1.786 | 0.217 | 0.806 |
| Sulfite oxidase | 0.030 | 0.713 | 0.966 | 0.999 |
| Sulfoacetaldehyde acetyltransferase | -3.930 | 4.117 | 0.341 | 0.837 |
| Sulfoacetaldehyde dehydrogenase (acylating) | 1.310 | 0.620 | 0.036 | 0.780 |
| Sulfoacetaldehyde reductase | -0.002 | 0.770 | 0.998 | 0.999 |
| Sulfofructose kinase | -1.604 | 0.940 | 0.090 | 0.806 |
| Sulfofructosephosphate aldolase | -1.309 | 1.241 | 0.293 | 0.820 |
| Sulfolactaldehyde 3-reductase | -0.872 | 0.980 | 0.375 | 0.852 |
| Sulfopropanediol 3-dehydrogenase | -0.875 | 0.871 | 0.317 | 0.823 |
| Sulfoquinovose isomerase | 0.118 | 0.326 | 0.717 | 0.999 |
| Sulfur carrier protein ThiS adenylyltransferase | 0.502 | 0.210 | 0.018 | 0.780 |
| Superoxide dismutase | -0.080 | 0.074 | 0.281 | 0.820 |
| Superoxide reductase | 0.120 | 0.192 | 0.534 | 0.924 |
| synthase | -0.010 | 0.117 | 0.929 | 0.999 |
| Tagatose-6-phosphate kinase | 0.067 | 0.151 | 0.657 | 0.974 |
| Tagatose-bisphosphate aldolase | 0.095 | 0.132 | 0.472 | 0.888 |
| Tagaturonate reductase | -0.109 | 0.311 | 0.726 | 0.999 |
| Tartrate decarboxylase | -0.745 | 0.378 | 0.051 | 0.780 |
| Tartrate dehydrogenase | -0.745 | 0.378 | 0.051 | 0.780 |
| Tartronate-semialdehyde synthase | -0.034 | 0.334 | 0.918 | 0.999 |
| Taurine–2-oxoglutarate transaminase | -0.129 | 0.457 | 0.777 | 0.999 |
| Taurine–pyruvate aminotransferase | -18.294 | 2578.334 | 0.994 | 0.999 |
| Taurine-transporting ATPase | -0.211 | 0.265 | 0.427 | 0.865 |
| Taurine dioxygenase | -0.149 | 0.277 | 0.592 | 0.951 |
| TDP-N-acetylfucosamine:lipid II N-acetylfucosaminyltransferase | 0.102 | 0.603 | 0.865 | 0.999 |
| Teichoic-acid-transporting ATPase | 0.284 | 0.311 | 0.362 | 0.844 |
| Tellurite methyltransferase | -0.029 | 0.162 | 0.857 | 0.999 |
| Terephthalate 1,2-dioxygenase | 0.934 | 1.436 | 0.516 | 0.913 |
| Tetraacyldisaccharide 4’-kinase | 0.057 | 0.089 | 0.520 | 0.913 |
| Tetracenomycin A2 monooxygenase-diooxygenase | -1.715 | 2.065 | 0.408 | 0.865 |
| Tetrahydrodipicolinate N-acetyltransferase | 0.013 | 0.202 | 0.948 | 0.999 |
| Tetrahydrofolate synthase | 0.037 | 0.050 | 0.466 | 0.884 |
| Tetrahydromethanopterin S-methyltransferase | -16.940 | 2657.012 | 0.995 | 0.999 |
| Tetraprenyl-beta-curcumene synthase | -2.107 | 0.836 | 0.013 | 0.780 |
| Thermitase | -0.130 | 0.711 | 0.855 | 0.999 |
| Thermolysin | -3.672 | 2.634 | 0.165 | 0.806 |
| Thiamine-phosphate diphosphorylase | 0.037 | 0.051 | 0.474 | 0.889 |
| Thiamine-phosphate kinase | -0.021 | 0.079 | 0.794 | 0.999 |
| Thiamine diphosphokinase | 0.053 | 0.091 | 0.562 | 0.936 |
| Thiamine kinase | 0.111 | 0.600 | 0.854 | 0.999 |
| Thiazole synthase | 0.042 | 0.078 | 0.593 | 0.951 |
| Thiazole tautomerase | -0.861 | 0.601 | 0.154 | 0.806 |
| Thimet oligopeptidase | -0.355 | 0.413 | 0.391 | 0.857 |
| Thiopurine S-methyltransferase | 0.189 | 0.315 | 0.548 | 0.930 |
| Thioredoxin-disulfide reductase | 0.010 | 0.045 | 0.830 | 0.999 |
| Thiosulfate dehydrogenase | -0.100 | 0.286 | 0.726 | 0.999 |
| Thiosulfate dehydrogenase (quinone) | -0.915 | 0.430 | 0.035 | 0.780 |
| Thiosulfate sulfurtransferase | -0.101 | 0.126 | 0.425 | 0.865 |
| Threonine–tRNA ligase | 0.028 | 0.027 | 0.306 | 0.820 |
| Threonine-phosphate decarboxylase | 0.288 | 0.172 | 0.096 | 0.806 |
| Threonine ammonia-lyase | -0.047 | 0.054 | 0.387 | 0.852 |
| Threonine synthase | -0.011 | 0.041 | 0.791 | 0.999 |
| Thymidine kinase | -0.027 | 0.076 | 0.726 | 0.999 |
| Thymidine phosphorylase | -0.409 | 0.256 | 0.112 | 0.806 |
| Thymidylate synthase | 0.021 | 0.038 | 0.584 | 0.948 |
| Thymidylate synthase (FAD) | -0.098 | 0.288 | 0.733 | 0.999 |
| Trans-2-decenoyl-[acyl-carrier-protein] isomerase | -0.005 | 0.089 | 0.953 | 0.999 |
| Trans-2-enoyl-CoA reductase (NAD(+)) | 0.084 | 0.241 | 0.727 | 0.999 |
| Trans-2,3-dihydro-3-hydroxyanthranilate isomerase | -1.206 | 0.494 | 0.016 | 0.780 |
| Trans-aconitate 2-methyltransferase | -0.128 | 0.272 | 0.640 | 0.973 |
| Trans-feruloyl-CoA hydratase | -0.448 | 0.537 | 0.405 | 0.865 |
| Trans-feruloyl-CoA synthase | -0.990 | 0.451 | 0.029 | 0.780 |
| Trans-L-3-hydroxyproline dehydratase | -0.012 | 0.831 | 0.988 | 0.999 |
| Trans,polycis-decaprenyl diphosphate synthase | -2.559 | 1.370 | 0.064 | 0.780 |
| Transaldolase | -0.082 | 0.064 | 0.202 | 0.806 |
| transferase | 0.494 | 0.626 | 0.431 | 0.869 |
| Transferred entry: 3.4.22.32 and 3.4.22.33 | -17.603 | 2028.355 | 0.993 | 0.999 |
| Transferred entry: 4.6.1.16 | -17.633 | 3757.583 | 0.996 | 0.999 |
| Transketolase | 0.009 | 0.049 | 0.856 | 0.999 |
| Trehalose-phosphatase | -0.429 | 0.248 | 0.086 | 0.806 |
| Trehalose O-mycolyltransferase | -1.730 | 1.021 | 0.092 | 0.806 |
| Triacylglycerol lipase | -0.056 | 0.203 | 0.784 | 0.999 |
| Trimethylamine-N-oxide reductase (cytochrome c) | -0.383 | 0.395 | 0.333 | 0.832 |
| Trimethylamine dehydrogenase | 0.672 | 0.917 | 0.465 | 0.884 |
| Trimethylamine monooxygenase | -0.723 | 0.655 | 0.272 | 0.820 |
| Triose-phosphate isomerase | 0.086 | 0.045 | 0.059 | 0.780 |
| Tripeptide aminopeptidase | 0.055 | 0.088 | 0.536 | 0.924 |
| Triphosphatase | 0.324 | 0.530 | 0.541 | 0.926 |
| Triphosphoribosyl-dephospho-CoA synthase | 0.396 | 0.176 | 0.026 | 0.780 |
| tRNA-2-methylthio-N(6)-dimethylallyladenosine synthase | -0.007 | 0.063 | 0.912 | 0.999 |
| tRNA-guanine(15) transglycosylase | -16.940 | 2657.012 | 0.995 | 0.999 |
| tRNA-guanine(34) transglycosylase | 0.018 | 0.035 | 0.600 | 0.954 |
| tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase | 0.050 | 0.202 | 0.804 | 0.999 |
| tRNA (adenine(22)-N(1))-methyltransferase | 0.048 | 0.084 | 0.570 | 0.938 |
| tRNA (adenine(57)-N(1)/adenine(58)-N(1))-methyltransferase | 0.077 | 0.242 | 0.750 | 0.999 |
| tRNA (adenine(58)-N(1))-methyltransferase | 0.077 | 0.242 | 0.750 | 0.999 |
| tRNA (cytidine(32)/uridine(32)-2’-O)-methyltransferase | 0.105 | 0.177 | 0.556 | 0.936 |
| tRNA (cytidine(34)-2’-O)-methyltransferase | 0.076 | 0.033 | 0.022 | 0.780 |
| tRNA (cytidine(56)-2’-O)-methyltransferase | -16.940 | 2657.012 | 0.995 | 0.999 |
| tRNA (guanine(26)-N(2))-dimethyltransferase | -16.940 | 2657.012 | 0.995 | 0.999 |
| tRNA (guanine(26)-N(2)/guanine(27)-N(2))-dimethyltransferase | -16.940 | 2657.012 | 0.995 | 0.999 |
| tRNA (guanine(37)-N(1))-methyltransferase | 0.045 | 0.032 | 0.159 | 0.806 |
| tRNA (guanine(46)-N(7))-methyltransferase | 0.048 | 0.033 | 0.150 | 0.806 |
| tRNA (guanosine(18)-2’-O)-methyltransferase | -0.298 | 0.332 | 0.370 | 0.850 |
| tRNA (N(6)-L-threonylcarbamoyladenosine(37)-C(2))-methylthiotransferase | -0.032 | 0.128 | 0.802 | 0.999 |
| tRNA (pseudouridine(54)-N(1))-methyltransferase | 18.828 | 2749.550 | 0.995 | 0.999 |
| tRNA (uracil(54)-C(5))-methyltransferase | 0.057 | 0.162 | 0.727 | 0.999 |
| tRNA 4-demethylwyosine synthase (AdoMet-dependent) | -16.940 | 2657.012 | 0.995 | 0.999 |
| tRNA dimethylallyltransferase | 0.016 | 0.041 | 0.704 | 0.999 |
| tRNA nucleotidyltransferase | -0.037 | 0.093 | 0.690 | 0.995 |
| tRNA pseudouridine(13) synthase | 0.094 | 0.183 | 0.609 | 0.960 |
| tRNA pseudouridine(32) synthase | -0.053 | 0.097 | 0.585 | 0.948 |
| tRNA pseudouridine(38-40) synthase | 0.038 | 0.032 | 0.247 | 0.806 |
| tRNA pseudouridine(55) synthase | 0.047 | 0.028 | 0.099 | 0.806 |
| tRNA pseudouridine(65) synthase | 0.108 | 0.219 | 0.623 | 0.967 |
| tRNA(1)(Val) (adenine(37)-N(6))-methyltransferase | -0.053 | 0.178 | 0.768 | 0.999 |
| tRNA(adenine(34)) deaminase | -0.011 | 0.073 | 0.880 | 0.999 |
| tRNA(Ile)-lysidine synthetase | 0.046 | 0.031 | 0.143 | 0.806 |
| tRNA(Ile)(2)-agmatinylcytidine synthase | -16.940 | 2657.012 | 0.995 | 0.999 |
| tRNA(Met) cytidine acetyltransferase | 0.221 | 0.306 | 0.472 | 0.888 |
| Tropinone reductase I | -0.030 | 0.484 | 0.951 | 0.999 |
| Tryptophan–tRNA ligase | 0.043 | 0.028 | 0.127 | 0.806 |
| Tryptophan 2-monooxygenase | -0.169 | 0.449 | 0.706 | 0.999 |
| Tryptophan 2,3-dioxygenase | -0.420 | 0.288 | 0.146 | 0.806 |
| Tryptophan 7-halogenase | -0.708 | 0.522 | 0.177 | 0.806 |
| Tryptophan synthase | -0.049 | 0.051 | 0.344 | 0.838 |
| Tryptophanase | 0.548 | 0.281 | 0.053 | 0.780 |
| Tungstate-importing ATPase | -0.030 | 0.531 | 0.955 | 0.999 |
| Type I site-specific deoxyribonuclease | -0.006 | 0.086 | 0.945 | 0.999 |
| Type II site-specific deoxyribonuclease | -0.441 | 0.382 | 0.250 | 0.808 |
| Type III site-specific deoxyribonuclease | 0.087 | 0.203 | 0.668 | 0.979 |
| Tyrosinase | 0.264 | 0.723 | 0.716 | 0.999 |
| Tyrosine–tRNA ligase | 0.028 | 0.031 | 0.359 | 0.844 |
| Tyrosine 3-monooxygenase | -17.718 | 3921.059 | 0.996 | 0.999 |
| Tyrosine ammonia-lyase | -17.488 | 1887.792 | 0.993 | 0.999 |
| Tyrosine phenol-lyase | 1.007 | 0.500 | 0.046 | 0.780 |
| Ubiquinol oxidase (non-electrogenic) | 0.439 | 1.146 | 0.702 | 0.999 |
| UDP-2-acetamido-2-deoxy-ribo-hexuluronate aminotransferase | 0.165 | 0.625 | 0.792 | 0.999 |
| UDP-2-acetamido-2,6-beta-L-arabino-hexul-4-ose reductase | -0.706 | 0.437 | 0.108 | 0.806 |
| UDP-2-acetamido-3-amino-2,3-dideoxy-glucuronate N-acetyltransferase | -0.087 | 0.448 | 0.847 | 0.999 |
| UDP-2,3-diacetamido-2,3-dideoxyglucuronic acid 2-epimerase | -1.175 | 0.749 | 0.119 | 0.806 |
| UDP-2,3-diacylglucosamine diphosphatase | -0.043 | 0.108 | 0.692 | 0.995 |
| UDP-2,4-diacetamido-2,4,6-trideoxy-beta-L-altropyranose hydrolase | 0.035 | 0.673 | 0.959 | 0.999 |
| UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | 0.139 | 0.101 | 0.169 | 0.806 |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase | 0.089 | 0.098 | 0.364 | 0.844 |
| UDP-4-amino-4-deoxy-L-arabinose aminotransferase | 0.068 | 0.473 | 0.886 | 0.999 |
| UDP-4-amino-4-deoxy-L-arabinose formyltransferase | -0.252 | 0.473 | 0.595 | 0.951 |
| UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine N-acetyltransferase | -0.774 | 0.921 | 0.402 | 0.863 |
| UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine transaminase | -0.774 | 0.921 | 0.402 | 0.863 |
| UDP-arabinose 4-epimerase | -1.254 | 1.016 | 0.219 | 0.806 |
| UDP-galactopyranose mutase | 0.096 | 0.166 | 0.563 | 0.936 |
| UDP-glucose–hexose-1-phosphate uridylyltransferase | 0.173 | 0.084 | 0.041 | 0.780 |
| UDP-glucose 4-epimerase | 0.006 | 0.043 | 0.899 | 0.999 |
| UDP-glucose 6-dehydrogenase | -0.148 | 0.101 | 0.145 | 0.806 |
| UDP-glucuronate 4-epimerase | -0.183 | 0.210 | 0.385 | 0.852 |
| UDP-glucuronate decarboxylase | -1.219 | 0.336 | 0.000 | 0.663 |
| UDP-glucuronic acid oxidase (UDP-4-keto-hexauronic acid decarboxylating) | -0.252 | 0.473 | 0.595 | 0.951 |
| UDP-N-acetyl-2-amino-2-deoxyglucuronate dehydrogenase | -0.425 | 0.745 | 0.569 | 0.938 |
| UDP-N-acetyl-D-mannosamine dehydrogenase | -0.100 | 0.242 | 0.679 | 0.989 |
| UDP-N-acetylbacillosamine N-acetyltransferase | 0.500 | 0.628 | 0.427 | 0.865 |
| UDP-N-acetylbacillosamine transaminase | 0.500 | 0.628 | 0.427 | 0.865 |
| UDP-N-acetylgalactosamine diphosphorylase | -1.560 | 0.906 | 0.087 | 0.806 |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase | 0.032 | 0.047 | 0.495 | 0.905 |
| UDP-N-acetylglucosamine 2-epimerase (hydrolyzing) | 1.766 | 1.243 | 0.157 | 0.806 |
| UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing) | -0.030 | 0.074 | 0.683 | 0.992 |
| UDP-N-acetylglucosamine 3-dehydrogenase | -17.399 | 2273.679 | 0.994 | 0.999 |
| UDP-N-acetylglucosamine 4-epimerase | -0.042 | 0.368 | 0.910 | 0.999 |
| UDP-N-acetylglucosamine 4,6-dehydratase (configuration-retaining) | 0.500 | 0.628 | 0.427 | 0.865 |
| UDP-N-acetylglucosamine 4,6-dehydratase (inverting) | -0.639 | 0.614 | 0.299 | 0.820 |
| UDP-N-acetylglucosamine 6-dehydrogenase | -0.325 | 0.386 | 0.401 | 0.863 |
| UDP-N-acetylglucosamine diphosphorylase | 0.045 | 0.035 | 0.199 | 0.806 |
| UDP-N-acetylglucosamine kinase | -0.121 | 0.220 | 0.582 | 0.948 |
| UDP-N-acetylmuramate–L-alanine ligase | 0.044 | 0.034 | 0.188 | 0.806 |
| UDP-N-acetylmuramate dehydrogenase | 0.037 | 0.032 | 0.249 | 0.808 |
| UDP-N-acetylmuramoyl-L-alanine–D-glutamate ligase | 0.040 | 0.031 | 0.201 | 0.806 |
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–2,6-diaminopimelate ligase | 0.072 | 0.053 | 0.177 | 0.806 |
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–L-lysine ligase | -0.043 | 0.161 | 0.788 | 0.999 |
| UDP-N-acetylmuramoyl-tripeptide–D-alanyl-D-alanine ligase | 0.032 | 0.040 | 0.427 | 0.865 |
| UDP-N-acetylmuramoylpentapeptide-lysine N(6)-alanyltransferase | -0.029 | 0.172 | 0.868 | 0.999 |
| UDP-N,N’-diacetylbacillosamine 2-epimerase (hydrolyzing) | -0.511 | 0.792 | 0.520 | 0.913 |
| UDP-sugar diphosphatase | 0.090 | 0.319 | 0.779 | 0.999 |
| UDP-sulfoquinovose synthase | -1.065 | 0.635 | 0.095 | 0.806 |
| UMP kinase | 0.037 | 0.031 | 0.234 | 0.806 |
| UMP/CMP kinase | 0.055 | 0.040 | 0.172 | 0.806 |
| Undecaprenol kinase | 0.047 | 0.137 | 0.730 | 0.999 |
| Undecaprenyl-diphosphate phosphatase | -0.035 | 0.032 | 0.284 | 0.820 |
| Undecaprenyl-diphosphooligosaccharide–protein glycotransferase | 0.498 | 0.534 | 0.352 | 0.842 |
| Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase | -0.192 | 0.399 | 0.632 | 0.972 |
| Undecaprenyl-phosphate galactose phosphotransferase | 0.513 | 0.490 | 0.297 | 0.820 |
| Undecaprenyl-phosphate glucose phosphotransferase | -17.382 | 2371.984 | 0.994 | 0.999 |
| Unsaturated chondroitin disaccharide hydrolase | 0.033 | 0.394 | 0.933 | 0.999 |
| Unsaturated rhamnogalacturonyl hydrolase | -1.107 | 0.546 | 0.044 | 0.780 |
| Unspecific monooxygenase | -1.262 | 0.651 | 0.054 | 0.780 |
| Uracil-DNA glycosylase | 0.061 | 0.043 | 0.152 | 0.806 |
| Uracil phosphoribosyltransferase | 0.065 | 0.042 | 0.118 | 0.806 |
| Urea carboxylase | -0.208 | 0.314 | 0.510 | 0.913 |
| Urease | -0.414 | 0.217 | 0.058 | 0.780 |
| Ureidoglycolate amidohydrolase | 0.457 | 1.410 | 0.746 | 0.999 |
| Ureidoglycolate dehydrogenase (NAD(+)) | -1.059 | 0.971 | 0.277 | 0.820 |
| Ureidoglycolate lyase | -0.165 | 0.275 | 0.548 | 0.930 |
| Uridine kinase | -0.003 | 0.089 | 0.970 | 0.999 |
| Uridine phosphorylase | -0.007 | 0.169 | 0.967 | 0.999 |
| Urocanate hydratase | 0.053 | 0.089 | 0.548 | 0.930 |
| Urocanate reductase | -0.615 | 0.960 | 0.522 | 0.914 |
| Uronate dehydrogenase | -0.173 | 0.338 | 0.610 | 0.960 |
| Uroporphyrinogen-III C-methyltransferase | 0.049 | 0.084 | 0.566 | 0.936 |
| Uroporphyrinogen-III synthase | -0.008 | 0.068 | 0.910 | 0.999 |
| Uroporphyrinogen decarboxylase | -0.114 | 0.074 | 0.127 | 0.806 |
| UTP–glucose-1-phosphate uridylyltransferase | 0.031 | 0.039 | 0.421 | 0.865 |
| Valine–pyruvate transaminase | 0.004 | 0.085 | 0.964 | 0.999 |
| Valine–tRNA ligase | 0.029 | 0.025 | 0.261 | 0.820 |
| Valine dehydrogenase (NAD(+)) | -0.596 | 1.185 | 0.616 | 0.963 |
| Vancomycin aglycone glucosyltransferase | 0.347 | 0.882 | 0.695 | 0.995 |
| Vanillate monooxygenase | -0.114 | 0.289 | 0.694 | 0.995 |
| Vanillin synthase | -0.448 | 0.537 | 0.405 | 0.865 |
| Vesicle-fusing ATPase | -2.499 | 2.858 | 0.383 | 0.852 |
| Vibriolysin | -2.706 | 2.378 | 0.257 | 0.817 |
| Vinylacetyl-CoA Delta-isomerase | -0.529 | 0.433 | 0.224 | 0.806 |
| Vitamin B12-transporting ATPase | 0.202 | 0.577 | 0.726 | 0.999 |
| Xaa-Pro aminopeptidase | 0.044 | 0.039 | 0.255 | 0.815 |
| Xaa-Pro dipeptidase | 0.001 | 0.088 | 0.989 | 0.999 |
| Xaa-Pro dipeptidyl-peptidase | -0.011 | 0.158 | 0.943 | 0.999 |
| Xaa-Xaa-Pro tripeptidyl-peptidase | -0.201 | 0.617 | 0.745 | 0.999 |
| Xanthine dehydrogenase | -0.142 | 0.219 | 0.518 | 0.913 |
| Xanthine phosphoribosyltransferase | 0.000 | 0.061 | 0.997 | 0.999 |
| Xanthomonalisin | -17.206 | 3034.891 | 0.995 | 0.999 |
| Xenobiotic-transporting ATPase | -0.205 | 0.350 | 0.559 | 0.936 |
| XTP/dITP diphosphatase | -0.012 | 0.050 | 0.811 | 0.999 |
| Xylan 1,4-beta-xylosidase | -0.978 | 0.635 | 0.125 | 0.806 |
| Xylose isomerase | -0.229 | 0.203 | 0.260 | 0.820 |
| Xylulokinase | -0.174 | 0.155 | 0.261 | 0.820 |
| Zeaxanthin glucosyltransferase | -0.144 | 0.682 | 0.833 | 0.999 |
| Zinc-exporting ATPase | 0.114 | 0.089 | 0.203 | 0.806 |
# merge with tb.ra for full results table
full.res <- left_join(tb.ra, results.out, by = "description")
write.csv(full.res, "output/picrust_ec_data_results.csv", row.names = F)Part 2 KO Data
pi.dat <- readr::read_tsv("data/PICRUST/ko_pred_metagenome_unstrat_descrip.tsv")
-- Column specification --------------------------------------------------------
cols(
.default = col_double(),
`function` = col_character(),
description = col_character()
)
i Use `spec()` for the full column specifications.# aggregate descriptions to get 1 description per row - unique;y defined.
pi.dat <- pi.dat %>%
group_by(description) %>%
summarise(across(`1.S37.Jun172016`:`99.D01.S37.Jun232016`,.fns = sum))
# transform to long format
pi.dat <- pi.dat %>%
pivot_longer(
cols=`1.S37.Jun172016`:`99.D01.S37.Jun232016`,
names_to = "ID",
values_to = "Abundance"
)
d <- as.data.frame(dat.16s)
mydata <- full_join(pi.dat, d)Joining, by = "ID"Warning in class(x) <- c(setdiff(subclass, tibble_class), tibble_class): Setting
class(x) to multiple strings ("tbl_df", "tbl", ...); result will no longer be an
S4 objectmydata <- mydata %>%
mutate(
ID.n = as.numeric(as.factor(ID)),
description.n = as.numeric(as.factor(description))
) %>%
group_by(ID) %>%
mutate(
dem = ifelse(sum(Abundance)==0, 1, sum(Abundance)),
RelAbundance = Abundance/dem*100) %>%
ungroup()%>%
group_by(description)%>%
mutate(avgRA = mean(RelAbundance))Relative Abundance
tb.ra1 <- mydata %>%
group_by(description) %>%
summarise(ng = n(),
Overall.M = mean(RelAbundance),
Overall.SE = sd(RelAbundance)/sqrt(ng))
tb.ra2m <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(M = mean(RelAbundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = M)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2se <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(),SE = sd(RelAbundance)/sqrt(ng)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = SE)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2var <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(), VAR = var(RelAbundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = VAR)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2ng <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n()) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = ng)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra <- left_join(tb.ra1, tb.ra2m)Joining, by = "description"tb.ra <- cbind(tb.ra, tb.ra2se[,-1])
tb.ra <- cbind(tb.ra, tb.ra2var[,-1])
tb.ra <- cbind(tb.ra, tb.ra2ng[,-1])
colnames(tb.ra) <- c("description", "ng", "Overall Mean", "Overall SE", "Non-Tumor Mean", "Tumor Mean", "Non-Tumor SE", "Tumor SE","Non-Tumor Var", "Tumor Var", "Non-Tumor Ng", "Tumor Ng")
tb.ra <- tb.ra %>%
arrange(desc(`Overall Mean`))
tb.ra <- tb.ra[, c("description", "Overall Mean", "Overall SE","Tumor Mean","Tumor Var", "Tumor SE","Tumor Ng", "Non-Tumor Mean","Non-Tumor Var", "Non-Tumor SE", "Non-Tumor Ng")]
# compute t-test
tb.ra <- tb.ra %>%
mutate(
SEpooled = sqrt(`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`),
t = (`Tumor Mean` - `Non-Tumor Mean`)/(SEpooled),
df = ((`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`)**2)/(((`Tumor Var`/`Tumor Ng`)**2)/(`Tumor Ng`-1) + ((`Non-Tumor Var`/`Non-Tumor Ng`)**2)/(`Non-Tumor Ng`-1)),
p = pt(q = abs(t), df=df, lower.tail = F)*2,
fdr_p = p.adjust(p, method="fdr")
)
kable(tb.ra, format="html", digits=5, caption="KO Data Average Relative Abundance of Each Description (sorted in descending order)") %>%
kable_styling(full_width = T) %>%
scroll_box(width = "100%", height="600px")| description | Overall Mean | Overall SE | Tumor Mean | Tumor Var | Tumor SE | Tumor Ng | Non-Tumor Mean | Non-Tumor Var | Non-Tumor SE | Non-Tumor Ng | SEpooled | t | df | p | fdr_p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABC-2.A; ABC-2 type transport system ATP-binding protein | 0.43629 | 0.01518 | 0.42850 | 0.03308 | 0.02256 | 65 | 0.44173 | 0.03904 | 0.02049 | 93 | 0.03048 | -0.43414 | 144.660 | 0.66484 | 0.91100 |
| ABC-2.P; ABC-2 type transport system permease protein | 0.41765 | 0.01276 | 0.40461 | 0.02271 | 0.01869 | 65 | 0.42676 | 0.02789 | 0.01732 | 93 | 0.02548 | -0.86903 | 146.116 | 0.38626 | 0.80262 |
| ABCB-BAC; ATP-binding cassette, subfamily B, bacterial | 0.35364 | 0.01235 | 0.36484 | 0.02514 | 0.01967 | 65 | 0.34581 | 0.02347 | 0.01589 | 93 | 0.02528 | 0.75282 | 134.836 | 0.45287 | 0.83438 |
| SPP; sucrose-6-phosphatase [EC:3.1.3.24] | 0.34403 | 0.01528 | 0.35668 | 0.03599 | 0.02353 | 65 | 0.33520 | 0.03769 | 0.02013 | 93 | 0.03097 | 0.69378 | 139.852 | 0.48897 | 0.84788 |
| ABC.CD.P; putative ABC transport system permease protein | 0.31124 | 0.00935 | 0.31596 | 0.01112 | 0.01308 | 65 | 0.30795 | 0.01581 | 0.01304 | 93 | 0.01847 | 0.43374 | 150.800 | 0.66510 | 0.91117 |
| ABC.CD.A; putative ABC transport system ATP-binding protein | 0.30766 | 0.01039 | 0.30801 | 0.01322 | 0.01426 | 65 | 0.30742 | 0.01993 | 0.01464 | 93 | 0.02044 | 0.02857 | 152.293 | 0.97725 | 0.99626 |
| ABC.PA.S; polar amino acid transport system substrate-binding protein | 0.23066 | 0.00638 | 0.23917 | 0.00495 | 0.00873 | 65 | 0.22471 | 0.00744 | 0.00894 | 93 | 0.01250 | 1.15697 | 152.200 | 0.24910 | 0.77376 |
| rpoE; RNA polymerase sigma-70 factor, ECF subfamily | 0.22266 | 0.00926 | 0.19849 | 0.01000 | 0.01240 | 65 | 0.23954 | 0.01545 | 0.01289 | 93 | 0.01789 | -2.29492 | 152.852 | 0.02310 | 0.59764 |
| ABC.FEV.P; iron complex transport system permease protein | 0.21291 | 0.00616 | 0.22260 | 0.00770 | 0.01088 | 65 | 0.20613 | 0.00478 | 0.00717 | 93 | 0.01303 | 1.26319 | 116.313 | 0.20905 | 0.77376 |
| TC.FEV.OM; iron complex outermembrane recepter protein | 0.19827 | 0.00962 | 0.19974 | 0.01458 | 0.01498 | 65 | 0.19723 | 0.01482 | 0.01262 | 93 | 0.01959 | 0.12807 | 138.567 | 0.89828 | 0.97377 |
| fabG; 3-oxoacyl-[acyl-carrier protein] reductase [EC:1.1.1.100] | 0.19694 | 0.00490 | 0.18640 | 0.00373 | 0.00758 | 65 | 0.20431 | 0.00375 | 0.00635 | 93 | 0.00988 | -1.81163 | 138.031 | 0.07222 | 0.74128 |
| lacI, galR; LacI family transcriptional regulator | 0.19070 | 0.00643 | 0.19100 | 0.00942 | 0.01204 | 65 | 0.19049 | 0.00459 | 0.00702 | 93 | 0.01394 | 0.03627 | 106.384 | 0.97113 | 0.99386 |
| ABC.PE.S; peptide/nickel transport system substrate-binding protein | 0.18855 | 0.00758 | 0.19652 | 0.01187 | 0.01351 | 65 | 0.18297 | 0.00718 | 0.00879 | 93 | 0.01612 | 0.84039 | 115.222 | 0.40243 | 0.80926 |
| ABC.FEV.A; iron complex transport system ATP-binding protein [EC:3.6.3.34] | 0.18807 | 0.00562 | 0.19896 | 0.00695 | 0.01034 | 65 | 0.18046 | 0.00354 | 0.00617 | 93 | 0.01204 | 1.53614 | 108.138 | 0.12742 | 0.77376 |
| mutT, NUDT15, MTH2; 8-oxo-dGTP diphosphatase [EC:3.6.1.55] | 0.18701 | 0.00750 | 0.18505 | 0.00795 | 0.01106 | 65 | 0.18838 | 0.00964 | 0.01018 | 93 | 0.01503 | -0.22161 | 145.691 | 0.82493 | 0.95506 |
| K07052; uncharacterized protein | 0.18483 | 0.00957 | 0.18356 | 0.01225 | 0.01373 | 65 | 0.18572 | 0.01619 | 0.01320 | 93 | 0.01904 | -0.11375 | 148.632 | 0.90959 | 0.97664 |
| ABC.FEV.S; iron complex transport system substrate-binding protein | 0.17980 | 0.00582 | 0.18878 | 0.00717 | 0.01051 | 65 | 0.17353 | 0.00405 | 0.00660 | 93 | 0.01241 | 1.22945 | 112.308 | 0.22147 | 0.77376 |
| ABC.PA.P; polar amino acid transport system permease protein | 0.17814 | 0.00565 | 0.18314 | 0.00410 | 0.00794 | 65 | 0.17465 | 0.00573 | 0.00785 | 93 | 0.01117 | 0.76090 | 150.348 | 0.44791 | 0.83372 |
| rluD; 23S rRNA pseudouridine1911/1915/1917 synthase [EC:5.4.99.23] | 0.17554 | 0.00570 | 0.17956 | 0.00535 | 0.00907 | 65 | 0.17273 | 0.00502 | 0.00734 | 93 | 0.01167 | 0.58556 | 134.990 | 0.55915 | 0.86813 |
| ABC.PA.A; polar amino acid transport system ATP-binding protein [EC:3.6.3.21] | 0.16598 | 0.00572 | 0.16859 | 0.00433 | 0.00816 | 65 | 0.16416 | 0.00581 | 0.00791 | 93 | 0.01137 | 0.38929 | 149.102 | 0.69761 | 0.91825 |
| gpmB; probable phosphoglycerate mutase [EC:5.4.2.12] | 0.13908 | 0.00645 | 0.14409 | 0.00748 | 0.01073 | 65 | 0.13559 | 0.00599 | 0.00802 | 93 | 0.01340 | 0.63467 | 127.791 | 0.52678 | 0.85844 |
| trxB, TRR; thioredoxin reductase (NADPH) [EC:1.8.1.9] | 0.13487 | 0.00304 | 0.13537 | 0.00135 | 0.00456 | 65 | 0.13453 | 0.00154 | 0.00408 | 93 | 0.00611 | 0.13813 | 143.449 | 0.89033 | 0.97064 |
| K07090; uncharacterized protein | 0.13458 | 0.00548 | 0.12804 | 0.00454 | 0.00836 | 65 | 0.13914 | 0.00487 | 0.00724 | 93 | 0.01106 | -1.00356 | 140.857 | 0.31731 | 0.77376 |
| E3.1.3.48; protein-tyrosine phosphatase [EC:3.1.3.48] | 0.13266 | 0.00518 | 0.13169 | 0.00543 | 0.00914 | 65 | 0.13333 | 0.00346 | 0.00610 | 93 | 0.01099 | -0.14855 | 117.491 | 0.88217 | 0.96944 |
| uvrD, pcrA; DNA helicase II / ATP-dependent DNA helicase PcrA [EC:3.6.4.12] | 0.12897 | 0.00326 | 0.13175 | 0.00181 | 0.00527 | 65 | 0.12703 | 0.00160 | 0.00415 | 93 | 0.00671 | 0.70353 | 132.432 | 0.48296 | 0.84354 |
| acpP; acyl carrier protein | 0.12629 | 0.00217 | 0.12649 | 0.00080 | 0.00351 | 65 | 0.12615 | 0.00071 | 0.00277 | 93 | 0.00447 | 0.07572 | 132.688 | 0.93976 | 0.98527 |
| ssb; single-strand DNA-binding protein | 0.12524 | 0.00284 | 0.12792 | 0.00119 | 0.00429 | 65 | 0.12337 | 0.00133 | 0.00378 | 93 | 0.00572 | 0.79630 | 142.442 | 0.42718 | 0.82661 |
| hemN, hemZ; oxygen-independent coproporphyrinogen III oxidase [EC:1.3.98.3] | 0.12357 | 0.00325 | 0.12856 | 0.00223 | 0.00586 | 65 | 0.12009 | 0.00127 | 0.00369 | 93 | 0.00692 | 1.22261 | 112.634 | 0.22403 | 0.77376 |
| ecfT; energy-coupling factor transport system permease protein | 0.12326 | 0.00592 | 0.12841 | 0.00523 | 0.00897 | 65 | 0.11967 | 0.00576 | 0.00787 | 93 | 0.01194 | 0.73237 | 141.931 | 0.46515 | 0.83786 |
| ecfA2; energy-coupling factor transport system ATP-binding protein [EC:3.6.3.-] | 0.12218 | 0.00552 | 0.13211 | 0.00527 | 0.00900 | 65 | 0.11523 | 0.00445 | 0.00692 | 93 | 0.01135 | 1.48719 | 130.276 | 0.13938 | 0.77376 |
| mcp; methyl-accepting chemotaxis protein | 0.12206 | 0.00983 | 0.11505 | 0.01801 | 0.01665 | 65 | 0.12696 | 0.01344 | 0.01202 | 93 | 0.02053 | -0.58018 | 124.593 | 0.56284 | 0.86813 |
| galE, GALE; UDP-glucose 4-epimerase [EC:5.1.3.2] | 0.12148 | 0.00294 | 0.12262 | 0.00133 | 0.00452 | 65 | 0.12069 | 0.00140 | 0.00389 | 93 | 0.00596 | 0.32442 | 140.255 | 0.74611 | 0.93202 |
| glnA, GLUL; glutamine synthetase [EC:6.3.1.2] | 0.12122 | 0.00364 | 0.11922 | 0.00228 | 0.00593 | 65 | 0.12262 | 0.00198 | 0.00462 | 93 | 0.00751 | -0.45125 | 131.531 | 0.65255 | 0.90537 |
| E2.2.1.1, tktA, tktB; transketolase [EC:2.2.1.1] | 0.11633 | 0.00296 | 0.11672 | 0.00148 | 0.00478 | 65 | 0.11605 | 0.00133 | 0.00379 | 93 | 0.00610 | 0.10918 | 133.158 | 0.91322 | 0.97664 |
| PDF, def; peptide deformylase [EC:3.5.1.88] | 0.11528 | 0.00264 | 0.11596 | 0.00125 | 0.00439 | 65 | 0.11480 | 0.00100 | 0.00328 | 93 | 0.00548 | 0.21051 | 127.669 | 0.83361 | 0.95506 |
| metQ; D-methionine transport system substrate-binding protein | 0.11454 | 0.00319 | 0.12333 | 0.00212 | 0.00570 | 65 | 0.10839 | 0.00118 | 0.00357 | 93 | 0.00673 | 2.22068 | 111.898 | 0.02839 | 0.61334 |
| K07025; putative hydrolase of the HAD superfamily | 0.11290 | 0.00258 | 0.10653 | 0.00109 | 0.00409 | 65 | 0.11735 | 0.00099 | 0.00326 | 93 | 0.00523 | -2.07033 | 133.558 | 0.04035 | 0.63822 |
| recG; ATP-dependent DNA helicase RecG [EC:3.6.4.12] | 0.11275 | 0.00356 | 0.11627 | 0.00212 | 0.00571 | 65 | 0.11029 | 0.00193 | 0.00456 | 93 | 0.00731 | 0.81729 | 133.701 | 0.41522 | 0.81730 |
| lepB; signal peptidase I [EC:3.4.21.89] | 0.11207 | 0.00241 | 0.11199 | 0.00107 | 0.00407 | 65 | 0.11212 | 0.00082 | 0.00297 | 93 | 0.00503 | -0.02671 | 125.599 | 0.97874 | 0.99657 |
| trkA, ktrA; trk system potassium uptake protein | 0.11140 | 0.00362 | 0.11576 | 0.00215 | 0.00575 | 65 | 0.10835 | 0.00202 | 0.00466 | 93 | 0.00740 | 1.00118 | 135.083 | 0.31853 | 0.77376 |
| E4.3.1.17, sdaA, sdaB, tdcG; L-serine dehydratase [EC:4.3.1.17] | 0.11084 | 0.00272 | 0.11217 | 0.00129 | 0.00446 | 65 | 0.10990 | 0.00109 | 0.00343 | 93 | 0.00562 | 0.40449 | 130.436 | 0.68652 | 0.91567 |
| ABC.PE.A1; peptide/nickel transport system ATP-binding protein | 0.10821 | 0.00686 | 0.11364 | 0.01064 | 0.01279 | 65 | 0.10441 | 0.00525 | 0.00752 | 93 | 0.01484 | 0.62184 | 106.957 | 0.53537 | 0.86207 |
| trxA; thioredoxin 1 | 0.10802 | 0.00212 | 0.10924 | 0.00084 | 0.00360 | 65 | 0.10716 | 0.00062 | 0.00258 | 93 | 0.00443 | 0.47006 | 123.955 | 0.63914 | 0.89853 |
| gph; phosphoglycolate phosphatase [EC:3.1.3.18] | 0.10726 | 0.00297 | 0.10378 | 0.00167 | 0.00506 | 65 | 0.10970 | 0.00120 | 0.00359 | 93 | 0.00620 | -0.95474 | 122.920 | 0.34158 | 0.78559 |
| cspA; cold shock protein (beta-ribbon, CspA family) | 0.10717 | 0.00437 | 0.10781 | 0.00296 | 0.00674 | 65 | 0.10672 | 0.00308 | 0.00576 | 93 | 0.00887 | 0.12332 | 139.685 | 0.90203 | 0.97641 |
| oppA, mppA; oligopeptide transport system substrate-binding protein | 0.10669 | 0.00677 | 0.10225 | 0.00569 | 0.00935 | 65 | 0.10979 | 0.00839 | 0.00950 | 93 | 0.01333 | -0.56567 | 151.761 | 0.57245 | 0.87292 |
| PRPS, prsA; ribose-phosphate pyrophosphokinase [EC:2.7.6.1] | 0.10510 | 0.00274 | 0.10824 | 0.00124 | 0.00437 | 65 | 0.10291 | 0.00115 | 0.00352 | 93 | 0.00561 | 0.95058 | 134.540 | 0.34352 | 0.78597 |
| parA, soj; chromosome partitioning protein | 0.10491 | 0.00360 | 0.10055 | 0.00189 | 0.00539 | 65 | 0.10796 | 0.00217 | 0.00483 | 93 | 0.00724 | -1.02410 | 143.612 | 0.30751 | 0.77376 |
| yidC, spoIIIJ, OXA1, ccfA; YidC/Oxa1 family membrane protein insertase | 0.10458 | 0.00277 | 0.10686 | 0.00125 | 0.00438 | 65 | 0.10298 | 0.00120 | 0.00359 | 93 | 0.00566 | 0.68596 | 136.039 | 0.49391 | 0.84797 |
| purL, PFAS; phosphoribosylformylglycinamidine synthase [EC:6.3.5.3] | 0.10408 | 0.00271 | 0.10511 | 0.00143 | 0.00469 | 65 | 0.10335 | 0.00099 | 0.00326 | 93 | 0.00571 | 0.30803 | 121.197 | 0.75859 | 0.93719 |
| pbuG; putative MFS transporter, AGZA family, xanthine/uracil permease | 0.10358 | 0.00311 | 0.11359 | 0.00214 | 0.00573 | 65 | 0.09658 | 0.00100 | 0.00327 | 93 | 0.00660 | 2.57661 | 104.768 | 0.01137 | 0.59095 |
| murA; UDP-N-acetylglucosamine 1-carboxyvinyltransferase [EC:2.5.1.7] | 0.10333 | 0.00252 | 0.10513 | 0.00105 | 0.00402 | 65 | 0.10207 | 0.00098 | 0.00325 | 93 | 0.00517 | 0.59196 | 134.826 | 0.55487 | 0.86806 |
| fusA, GFM, EFG; elongation factor G | 0.10325 | 0.00260 | 0.10664 | 0.00106 | 0.00405 | 65 | 0.10088 | 0.00107 | 0.00339 | 93 | 0.00528 | 1.09066 | 138.118 | 0.27732 | 0.77376 |
| iscS, NFS1; cysteine desulfurase [EC:2.8.1.7] | 0.10314 | 0.00309 | 0.10430 | 0.00153 | 0.00486 | 65 | 0.10232 | 0.00151 | 0.00403 | 93 | 0.00631 | 0.31384 | 137.233 | 0.75412 | 0.93665 |
| ftsK, spoIIIE; DNA segregation ATPase FtsK/SpoIIIE, S-DNA-T family | 0.10312 | 0.00267 | 0.10118 | 0.00134 | 0.00455 | 65 | 0.10449 | 0.00099 | 0.00326 | 93 | 0.00559 | -0.59193 | 123.793 | 0.55498 | 0.86806 |
| dapA; 4-hydroxy-tetrahydrodipicolinate synthase [EC:4.3.3.7] | 0.10272 | 0.00149 | 0.09994 | 0.00029 | 0.00210 | 65 | 0.10467 | 0.00039 | 0.00206 | 93 | 0.00294 | -1.61125 | 149.883 | 0.10923 | 0.77376 |
| ACSL, fadD; long-chain acyl-CoA synthetase [EC:6.2.1.3] | 0.10247 | 0.00446 | 0.09663 | 0.00258 | 0.00629 | 65 | 0.10655 | 0.00354 | 0.00617 | 93 | 0.00881 | -1.12517 | 149.846 | 0.26232 | 0.77376 |
| tatD; TatD DNase family protein [EC:3.1.21.-] | 0.10226 | 0.00108 | 0.10444 | 0.00014 | 0.00146 | 65 | 0.10074 | 0.00021 | 0.00150 | 93 | 0.00210 | 1.76551 | 152.245 | 0.07948 | 0.75920 |
| ABC.PE.P1; peptide/nickel transport system permease protein | 0.09981 | 0.00603 | 0.10268 | 0.00787 | 0.01100 | 65 | 0.09781 | 0.00431 | 0.00681 | 93 | 0.01294 | 0.37672 | 111.109 | 0.70710 | 0.92004 |
| ABC.PE.P; peptide/nickel transport system permease protein | 0.09877 | 0.00606 | 0.10174 | 0.00785 | 0.01099 | 65 | 0.09670 | 0.00442 | 0.00689 | 93 | 0.01298 | 0.38805 | 112.187 | 0.69871 | 0.91844 |
| tuf, TUFM; elongation factor Tu | 0.09863 | 0.00167 | 0.10049 | 0.00035 | 0.00231 | 65 | 0.09733 | 0.00050 | 0.00233 | 93 | 0.00328 | 0.96448 | 151.361 | 0.33634 | 0.78254 |
| map; methionyl aminopeptidase [EC:3.4.11.18] | 0.09856 | 0.00139 | 0.09753 | 0.00027 | 0.00202 | 65 | 0.09927 | 0.00034 | 0.00191 | 93 | 0.00278 | -0.62720 | 147.403 | 0.53150 | 0.86136 |
| folC; dihydrofolate synthase / folylpolyglutamate synthase [EC:6.3.2.12 6.3.2.17] | 0.09800 | 0.00254 | 0.10000 | 0.00096 | 0.00384 | 65 | 0.09660 | 0.00106 | 0.00338 | 93 | 0.00512 | 0.66331 | 142.288 | 0.50821 | 0.84956 |
| ABC.SS.P; simple sugar transport system permease protein | 0.09748 | 0.00414 | 0.09805 | 0.00298 | 0.00678 | 65 | 0.09709 | 0.00254 | 0.00523 | 93 | 0.00856 | 0.11272 | 130.719 | 0.91043 | 0.97664 |
| aroE; shikimate dehydrogenase [EC:1.1.1.25] | 0.09737 | 0.00290 | 0.10227 | 0.00206 | 0.00563 | 65 | 0.09395 | 0.00080 | 0.00294 | 93 | 0.00635 | 1.31041 | 98.633 | 0.19310 | 0.77376 |
| uvrA; excinuclease ABC subunit A | 0.09718 | 0.00163 | 0.09722 | 0.00041 | 0.00250 | 65 | 0.09716 | 0.00044 | 0.00216 | 93 | 0.00331 | 0.01688 | 140.812 | 0.98656 | 0.99745 |
| E3.1.11.2, xthA; exodeoxyribonuclease III [EC:3.1.11.2] | 0.09699 | 0.00159 | 0.09901 | 0.00043 | 0.00259 | 65 | 0.09558 | 0.00038 | 0.00201 | 93 | 0.00328 | 1.04748 | 131.488 | 0.29680 | 0.77376 |
| E2.2.1.6L, ilvB, ilvG, ilvI; acetolactate synthase I/II/III large subunit [EC:2.2.1.6] | 0.09641 | 0.00191 | 0.09639 | 0.00056 | 0.00292 | 65 | 0.09643 | 0.00060 | 0.00254 | 93 | 0.00388 | -0.01029 | 141.169 | 0.99180 | 0.99884 |
| TC.AGCS; alanine or glycine:cation symporter, AGCS family | 0.09640 | 0.00470 | 0.10770 | 0.00560 | 0.00928 | 65 | 0.08850 | 0.00191 | 0.00454 | 93 | 0.01033 | 1.85765 | 94.465 | 0.06633 | 0.73334 |
| K08303; putative protease [EC:3.4.-.-] | 0.09635 | 0.00333 | 0.09901 | 0.00196 | 0.00549 | 65 | 0.09450 | 0.00162 | 0.00417 | 93 | 0.00689 | 0.65351 | 129.250 | 0.51459 | 0.85216 |
| GAPDH, gapA; glyceraldehyde 3-phosphate dehydrogenase [EC:1.2.1.12] | 0.09565 | 0.00144 | 0.09596 | 0.00027 | 0.00204 | 65 | 0.09543 | 0.00037 | 0.00199 | 93 | 0.00285 | 0.18446 | 149.404 | 0.85390 | 0.95848 |
| rpoD; RNA polymerase primary sigma factor | 0.09544 | 0.00267 | 0.09677 | 0.00103 | 0.00398 | 65 | 0.09450 | 0.00121 | 0.00361 | 93 | 0.00537 | 0.42350 | 144.581 | 0.67256 | 0.91184 |
| hupB; DNA-binding protein HU-beta | 0.09531 | 0.00244 | 0.09398 | 0.00065 | 0.00316 | 65 | 0.09624 | 0.00116 | 0.00352 | 93 | 0.00473 | -0.47607 | 155.218 | 0.63469 | 0.89696 |
| hsdS; type I restriction enzyme, S subunit [EC:3.1.21.3] | 0.09523 | 0.00423 | 0.09428 | 0.00239 | 0.00607 | 65 | 0.09589 | 0.00315 | 0.00582 | 93 | 0.00841 | -0.19078 | 148.560 | 0.84896 | 0.95712 |
| DLD, lpd, pdhD; dihydrolipoamide dehydrogenase [EC:1.8.1.4] | 0.09510 | 0.00259 | 0.08993 | 0.00112 | 0.00416 | 65 | 0.09871 | 0.00099 | 0.00327 | 93 | 0.00529 | -1.65939 | 132.377 | 0.09940 | 0.76942 |
| secA; preprotein translocase subunit SecA | 0.09505 | 0.00208 | 0.09441 | 0.00059 | 0.00301 | 65 | 0.09549 | 0.00076 | 0.00285 | 93 | 0.00415 | -0.26129 | 147.552 | 0.79423 | 0.94783 |
| tonB; periplasmic protein TonB | 0.09487 | 0.00569 | 0.09430 | 0.00543 | 0.00914 | 65 | 0.09528 | 0.00495 | 0.00730 | 93 | 0.01169 | -0.08382 | 133.791 | 0.93333 | 0.98428 |
| plsC; 1-acyl-sn-glycerol-3-phosphate acyltransferase [EC:2.3.1.51] | 0.09425 | 0.00162 | 0.09332 | 0.00053 | 0.00285 | 65 | 0.09490 | 0.00034 | 0.00191 | 93 | 0.00343 | -0.46174 | 117.983 | 0.64512 | 0.90290 |
| parB, spo0J; chromosome partitioning protein, ParB family | 0.09357 | 0.00294 | 0.08917 | 0.00139 | 0.00462 | 65 | 0.09664 | 0.00135 | 0.00381 | 93 | 0.00598 | -1.24781 | 136.652 | 0.21423 | 0.77376 |
| K07729; putative transcriptional regulator | 0.09344 | 0.00592 | 0.08893 | 0.00457 | 0.00839 | 65 | 0.09659 | 0.00625 | 0.00820 | 93 | 0.01173 | -0.65273 | 149.671 | 0.51493 | 0.85216 |
| GST, gst; glutathione S-transferase [EC:2.5.1.18] | 0.09234 | 0.00861 | 0.08280 | 0.01133 | 0.01320 | 65 | 0.09901 | 0.01199 | 0.01135 | 93 | 0.01741 | -0.93089 | 140.293 | 0.35351 | 0.79231 |
| clsA_B; cardiolipin synthase A/B [EC:2.7.8.-] | 0.09212 | 0.00229 | 0.09430 | 0.00092 | 0.00375 | 65 | 0.09059 | 0.00078 | 0.00289 | 93 | 0.00473 | 0.78498 | 130.427 | 0.43389 | 0.82874 |
| secY; preprotein translocase subunit SecY | 0.09167 | 0.00227 | 0.09189 | 0.00066 | 0.00320 | 65 | 0.09152 | 0.00092 | 0.00315 | 93 | 0.00449 | 0.08351 | 150.110 | 0.93356 | 0.98428 |
| trkH, trkG, ktrB; trk system potassium uptake protein | 0.09152 | 0.00340 | 0.09717 | 0.00203 | 0.00559 | 65 | 0.08757 | 0.00166 | 0.00423 | 93 | 0.00701 | 1.36911 | 128.830 | 0.17335 | 0.77376 |
| guaA, GMPS; GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2] | 0.09129 | 0.00148 | 0.09152 | 0.00028 | 0.00208 | 65 | 0.09113 | 0.00039 | 0.00206 | 93 | 0.00292 | 0.13479 | 150.418 | 0.89296 | 0.97159 |
| RP-L33, MRPL33, rpmG; large subunit ribosomal protein L33 | 0.09114 | 0.00224 | 0.09336 | 0.00096 | 0.00385 | 65 | 0.08959 | 0.00067 | 0.00269 | 93 | 0.00469 | 0.80446 | 121.705 | 0.42270 | 0.82322 |
| ybgC; acyl-CoA thioester hydrolase [EC:3.1.2.-] | 0.09072 | 0.00344 | 0.08406 | 0.00184 | 0.00532 | 65 | 0.09537 | 0.00187 | 0.00448 | 93 | 0.00695 | -1.62656 | 138.542 | 0.10610 | 0.77376 |
| crcB, FEX; fluoride exporter | 0.09042 | 0.00292 | 0.08770 | 0.00152 | 0.00483 | 65 | 0.09232 | 0.00123 | 0.00364 | 93 | 0.00605 | -0.76313 | 128.559 | 0.44679 | 0.83371 |
| KAE1, tsaD, QRI7; N6-L-threonylcarbamoyladenine synthase [EC:2.3.1.234] | 0.09037 | 0.00209 | 0.09348 | 0.00072 | 0.00332 | 65 | 0.08820 | 0.00067 | 0.00268 | 93 | 0.00427 | 1.23675 | 134.707 | 0.21833 | 0.77376 |
| E2.3.1.9, atoB; acetyl-CoA C-acetyltransferase [EC:2.3.1.9] | 0.09001 | 0.00492 | 0.08122 | 0.00265 | 0.00639 | 65 | 0.09615 | 0.00459 | 0.00702 | 93 | 0.00949 | -1.57263 | 154.832 | 0.11784 | 0.77376 |
| E2.7.7.41, CDS1, CDS2, cdsA; phosphatidate cytidylyltransferase [EC:2.7.7.41] | 0.08966 | 0.00133 | 0.09292 | 0.00038 | 0.00242 | 65 | 0.08737 | 0.00020 | 0.00145 | 93 | 0.00282 | 1.96669 | 108.750 | 0.05177 | 0.68802 |
| pstB; phosphate transport system ATP-binding protein [EC:3.6.3.27] | 0.08965 | 0.00347 | 0.08772 | 0.00194 | 0.00546 | 65 | 0.09100 | 0.00190 | 0.00452 | 93 | 0.00709 | -0.46344 | 137.025 | 0.64379 | 0.90196 |
| trpB; tryptophan synthase beta chain [EC:4.2.1.20] | 0.08936 | 0.00260 | 0.08720 | 0.00112 | 0.00416 | 65 | 0.09087 | 0.00104 | 0.00335 | 93 | 0.00534 | -0.68889 | 134.542 | 0.49208 | 0.84797 |
| relA; GTP pyrophosphokinase [EC:2.7.6.5] | 0.08920 | 0.00213 | 0.09205 | 0.00071 | 0.00331 | 65 | 0.08721 | 0.00072 | 0.00278 | 93 | 0.00432 | 1.11935 | 138.212 | 0.26493 | 0.77376 |
| pepP; Xaa-Pro aminopeptidase [EC:3.4.11.9] | 0.08916 | 0.00190 | 0.09131 | 0.00062 | 0.00309 | 65 | 0.08766 | 0.00054 | 0.00241 | 93 | 0.00392 | 0.93224 | 131.843 | 0.35291 | 0.79194 |
| argR, ahrC; transcriptional regulator of arginine metabolism | 0.08911 | 0.00454 | 0.09118 | 0.00323 | 0.00705 | 65 | 0.08766 | 0.00330 | 0.00596 | 93 | 0.00923 | 0.38058 | 138.894 | 0.70409 | 0.92004 |
| ddl; D-alanine-D-alanine ligase [EC:6.3.2.4] | 0.08867 | 0.00148 | 0.08904 | 0.00028 | 0.00207 | 65 | 0.08840 | 0.00040 | 0.00207 | 93 | 0.00293 | 0.21880 | 151.019 | 0.82710 | 0.95506 |
| clpP, CLPP; ATP-dependent Clp protease, protease subunit [EC:3.4.21.92] | 0.08736 | 0.00187 | 0.08811 | 0.00059 | 0.00301 | 65 | 0.08684 | 0.00053 | 0.00239 | 93 | 0.00385 | 0.33067 | 133.455 | 0.74141 | 0.93032 |
| dnaQ; DNA polymerase III subunit epsilon [EC:2.7.7.7] | 0.08721 | 0.00193 | 0.08589 | 0.00061 | 0.00307 | 65 | 0.08813 | 0.00057 | 0.00249 | 93 | 0.00395 | -0.56665 | 134.955 | 0.57189 | 0.87284 |
| rdgB; XTP/dITP diphosphohydrolase [EC:3.6.1.66] | 0.08697 | 0.00233 | 0.08607 | 0.00089 | 0.00370 | 65 | 0.08760 | 0.00085 | 0.00302 | 93 | 0.00478 | -0.31870 | 135.782 | 0.75044 | 0.93465 |
| alr; alanine racemase [EC:5.1.1.1] | 0.08662 | 0.00163 | 0.09079 | 0.00046 | 0.00267 | 65 | 0.08371 | 0.00037 | 0.00200 | 93 | 0.00333 | 2.12246 | 128.141 | 0.03572 | 0.62411 |
| metI; D-methionine transport system permease protein | 0.08660 | 0.00194 | 0.09019 | 0.00056 | 0.00294 | 65 | 0.08409 | 0.00061 | 0.00255 | 93 | 0.00389 | 1.56697 | 141.092 | 0.11936 | 0.77376 |
| dnaJ; molecular chaperone DnaJ | 0.08627 | 0.00156 | 0.08751 | 0.00039 | 0.00245 | 65 | 0.08540 | 0.00038 | 0.00203 | 93 | 0.00318 | 0.66472 | 137.208 | 0.50734 | 0.84956 |
| fabH; 3-oxoacyl-[acyl-carrier-protein] synthase III [EC:2.3.1.180] | 0.08625 | 0.00193 | 0.08876 | 0.00074 | 0.00337 | 65 | 0.08449 | 0.00048 | 0.00228 | 93 | 0.00407 | 1.05215 | 118.893 | 0.29487 | 0.77376 |
| URA4, pyrC; dihydroorotase [EC:3.5.2.3] | 0.08606 | 0.00114 | 0.08528 | 0.00017 | 0.00162 | 65 | 0.08661 | 0.00023 | 0.00157 | 93 | 0.00225 | -0.59259 | 149.143 | 0.55435 | 0.86806 |
| aspS; aspartyl-tRNA synthetase [EC:6.1.1.12] | 0.08595 | 0.00190 | 0.08811 | 0.00054 | 0.00287 | 65 | 0.08444 | 0.00060 | 0.00253 | 93 | 0.00383 | 0.95893 | 142.214 | 0.33922 | 0.78433 |
| gyrA; DNA gyrase subunit A [EC:5.99.1.3] | 0.08512 | 0.00171 | 0.08709 | 0.00047 | 0.00268 | 65 | 0.08375 | 0.00046 | 0.00222 | 93 | 0.00349 | 0.95630 | 137.100 | 0.34060 | 0.78491 |
| gyrB; DNA gyrase subunit B [EC:5.99.1.3] | 0.08502 | 0.00185 | 0.08697 | 0.00053 | 0.00285 | 65 | 0.08366 | 0.00055 | 0.00243 | 93 | 0.00375 | 0.88311 | 139.694 | 0.37870 | 0.79852 |
| carB, CPA2; carbamoyl-phosphate synthase large subunit [EC:6.3.5.5] | 0.08483 | 0.00221 | 0.08573 | 0.00093 | 0.00378 | 65 | 0.08419 | 0.00067 | 0.00268 | 93 | 0.00463 | 0.33250 | 123.032 | 0.74008 | 0.92969 |
| RP-L31, rpmE; large subunit ribosomal protein L31 | 0.08474 | 0.00136 | 0.08637 | 0.00031 | 0.00219 | 65 | 0.08361 | 0.00028 | 0.00173 | 93 | 0.00279 | 0.98640 | 132.476 | 0.32573 | 0.77376 |
| rnj; ribonuclease J [EC:3.1.-.-] | 0.08472 | 0.00373 | 0.08691 | 0.00224 | 0.00587 | 65 | 0.08319 | 0.00219 | 0.00486 | 93 | 0.00761 | 0.48860 | 137.014 | 0.62591 | 0.89255 |
| mrcA; penicillin-binding protein 1A [EC:2.4.1.129 3.4.16.4] | 0.08448 | 0.00210 | 0.08335 | 0.00059 | 0.00301 | 65 | 0.08526 | 0.00078 | 0.00289 | 93 | 0.00417 | -0.45984 | 148.542 | 0.64630 | 0.90314 |
| mnmA, trmU; tRNA-uridine 2-sulfurtransferase [EC:2.8.1.13] | 0.08437 | 0.00240 | 0.08924 | 0.00123 | 0.00436 | 65 | 0.08096 | 0.00067 | 0.00268 | 93 | 0.00511 | 1.61796 | 110.681 | 0.10852 | 0.77376 |
| ftsH, hflB; cell division protease FtsH [EC:3.4.24.-] | 0.08431 | 0.00180 | 0.08582 | 0.00058 | 0.00298 | 65 | 0.08325 | 0.00047 | 0.00225 | 93 | 0.00373 | 0.68741 | 128.453 | 0.49306 | 0.84797 |
| fabF; 3-oxoacyl-[acyl-carrier-protein] synthase II [EC:2.3.1.179] | 0.08420 | 0.00126 | 0.08441 | 0.00017 | 0.00164 | 65 | 0.08406 | 0.00031 | 0.00182 | 93 | 0.00245 | 0.14374 | 155.050 | 0.88589 | 0.97012 |
| IMPDH, guaB; IMP dehydrogenase [EC:1.1.1.205] | 0.08415 | 0.00155 | 0.08524 | 0.00036 | 0.00236 | 65 | 0.08338 | 0.00039 | 0.00206 | 93 | 0.00313 | 0.59588 | 141.292 | 0.55221 | 0.86788 |
| E1.17.4.1A, nrdA, nrdE; ribonucleoside-diphosphate reductase alpha chain [EC:1.17.4.1] | 0.08401 | 0.00168 | 0.08330 | 0.00054 | 0.00289 | 65 | 0.08451 | 0.00039 | 0.00204 | 93 | 0.00354 | -0.34135 | 122.305 | 0.73343 | 0.92674 |
| AARS, alaS; alanyl-tRNA synthetase [EC:6.1.1.7] | 0.08393 | 0.00165 | 0.08643 | 0.00051 | 0.00281 | 65 | 0.08218 | 0.00037 | 0.00199 | 93 | 0.00344 | 1.23434 | 122.540 | 0.21944 | 0.77376 |
| miaA, TRIT1; tRNA dimethylallyltransferase [EC:2.5.1.75] | 0.08375 | 0.00190 | 0.08425 | 0.00048 | 0.00272 | 65 | 0.08340 | 0.00064 | 0.00262 | 93 | 0.00378 | 0.22585 | 148.737 | 0.82163 | 0.95506 |
| TPI, tpiA; triosephosphate isomerase (TIM) [EC:5.3.1.1] | 0.08369 | 0.00203 | 0.08774 | 0.00093 | 0.00377 | 65 | 0.08086 | 0.00045 | 0.00219 | 93 | 0.00436 | 1.57666 | 105.913 | 0.11786 | 0.77376 |
| K06889; uncharacterized protein | 0.08365 | 0.00361 | 0.08527 | 0.00272 | 0.00647 | 65 | 0.08252 | 0.00161 | 0.00416 | 93 | 0.00769 | 0.35759 | 114.414 | 0.72131 | 0.92156 |
| topA; DNA topoisomerase I [EC:5.99.1.2] | 0.08325 | 0.00114 | 0.08475 | 0.00011 | 0.00130 | 65 | 0.08221 | 0.00027 | 0.00170 | 93 | 0.00214 | 1.18789 | 155.001 | 0.23669 | 0.77376 |
| RP-S14, MRPS14, rpsN; small subunit ribosomal protein S14 | 0.08279 | 0.00195 | 0.08540 | 0.00071 | 0.00331 | 65 | 0.08097 | 0.00052 | 0.00237 | 93 | 0.00407 | 1.08603 | 123.777 | 0.27958 | 0.77376 |
| cysK; cysteine synthase A [EC:2.5.1.47] | 0.08277 | 0.00132 | 0.08391 | 0.00037 | 0.00237 | 65 | 0.08198 | 0.00021 | 0.00151 | 93 | 0.00281 | 0.68615 | 113.195 | 0.49402 | 0.84797 |
| purH; phosphoribosylaminoimidazolecarboxamide formyltransferase / IMP cyclohydrolase [EC:2.1.2.3 3.5.4.10] | 0.08268 | 0.00196 | 0.08433 | 0.00053 | 0.00284 | 65 | 0.08152 | 0.00066 | 0.00267 | 93 | 0.00390 | 0.72026 | 147.037 | 0.47251 | 0.84050 |
| E2.7.4.8, gmk; guanylate kinase [EC:2.7.4.8] | 0.08265 | 0.00203 | 0.08627 | 0.00083 | 0.00358 | 65 | 0.08013 | 0.00052 | 0.00236 | 93 | 0.00429 | 1.42927 | 116.397 | 0.15560 | 0.77376 |
| comEC; competence protein ComEC | 0.08251 | 0.00185 | 0.08612 | 0.00057 | 0.00295 | 65 | 0.07998 | 0.00051 | 0.00235 | 93 | 0.00377 | 1.62846 | 133.461 | 0.10579 | 0.77376 |
| glyA, SHMT; glycine hydroxymethyltransferase [EC:2.1.2.1] | 0.08235 | 0.00138 | 0.08311 | 0.00037 | 0.00240 | 65 | 0.08182 | 0.00026 | 0.00166 | 93 | 0.00292 | 0.43920 | 120.482 | 0.66130 | 0.90968 |
| ENO, eno; enolase [EC:4.2.1.11] | 0.08227 | 0.00228 | 0.08412 | 0.00086 | 0.00364 | 65 | 0.08097 | 0.00080 | 0.00292 | 93 | 0.00467 | 0.67428 | 134.335 | 0.50129 | 0.84942 |
| E3.1.4.46, glpQ, ugpQ; glycerophosphoryl diester phosphodiesterase [EC:3.1.4.46] | 0.08213 | 0.00255 | 0.08325 | 0.00110 | 0.00411 | 65 | 0.08135 | 0.00099 | 0.00326 | 93 | 0.00524 | 0.36125 | 133.112 | 0.71848 | 0.92092 |
| hsdM; type I restriction enzyme M protein [EC:2.1.1.72] | 0.08199 | 0.00287 | 0.08152 | 0.00110 | 0.00412 | 65 | 0.08232 | 0.00145 | 0.00395 | 93 | 0.00571 | -0.13867 | 148.383 | 0.88990 | 0.97048 |
| efp; elongation factor P | 0.08195 | 0.00154 | 0.08360 | 0.00036 | 0.00234 | 65 | 0.08080 | 0.00039 | 0.00204 | 93 | 0.00310 | 0.90144 | 141.523 | 0.36888 | 0.79587 |
| dacC, dacA, dacD; serine-type D-Ala-D-Ala carboxypeptidase (penicillin-binding protein 5/6) [EC:3.4.16.4] | 0.08194 | 0.00272 | 0.08296 | 0.00132 | 0.00451 | 65 | 0.08123 | 0.00107 | 0.00340 | 93 | 0.00565 | 0.30533 | 128.527 | 0.76061 | 0.93791 |
| metN; D-methionine transport system ATP-binding protein | 0.08169 | 0.00162 | 0.08605 | 0.00049 | 0.00275 | 65 | 0.07865 | 0.00034 | 0.00193 | 93 | 0.00336 | 2.20524 | 121.886 | 0.02931 | 0.61334 |
| rnhB; ribonuclease HII [EC:3.1.26.4] | 0.08169 | 0.00169 | 0.08255 | 0.00035 | 0.00231 | 65 | 0.08109 | 0.00052 | 0.00237 | 93 | 0.00331 | 0.44048 | 152.317 | 0.66022 | 0.90939 |
| rsmI; 16S rRNA (cytidine1402-2’-O)-methyltransferase [EC:2.1.1.198] | 0.08168 | 0.00146 | 0.08261 | 0.00031 | 0.00217 | 65 | 0.08103 | 0.00036 | 0.00198 | 93 | 0.00293 | 0.53833 | 145.060 | 0.59117 | 0.87896 |
| dnaB; replicative DNA helicase [EC:3.6.4.12] | 0.08163 | 0.00116 | 0.08264 | 0.00018 | 0.00167 | 65 | 0.08094 | 0.00023 | 0.00159 | 93 | 0.00230 | 0.73859 | 147.867 | 0.46133 | 0.83584 |
| polA; DNA polymerase I [EC:2.7.7.7] | 0.08148 | 0.00147 | 0.08268 | 0.00033 | 0.00226 | 65 | 0.08065 | 0.00035 | 0.00193 | 93 | 0.00297 | 0.68158 | 139.945 | 0.49663 | 0.84797 |
| glmS, GFPT; glucosamine—fructose-6-phosphate aminotransferase (isomerizing) [EC:2.6.1.16] | 0.08141 | 0.00110 | 0.08149 | 0.00019 | 0.00170 | 65 | 0.08136 | 0.00020 | 0.00146 | 93 | 0.00224 | 0.06150 | 139.847 | 0.95105 | 0.98792 |
| groEL, HSPD1; chaperonin GroEL | 0.08103 | 0.00152 | 0.08225 | 0.00037 | 0.00237 | 65 | 0.08017 | 0.00037 | 0.00199 | 93 | 0.00310 | 0.67273 | 138.194 | 0.50224 | 0.84944 |
| coaE; dephospho-CoA kinase [EC:2.7.1.24] | 0.08083 | 0.00175 | 0.08229 | 0.00042 | 0.00254 | 65 | 0.07980 | 0.00053 | 0.00239 | 93 | 0.00349 | 0.71322 | 147.226 | 0.47684 | 0.84209 |
| ABC.MS.S; multiple sugar transport system substrate-binding protein | 0.08072 | 0.00442 | 0.07831 | 0.00378 | 0.00763 | 65 | 0.08241 | 0.00263 | 0.00532 | 93 | 0.00930 | -0.44122 | 121.368 | 0.65984 | 0.90939 |
| LARS, leuS; leucyl-tRNA synthetase [EC:6.1.1.4] | 0.08064 | 0.00153 | 0.08181 | 0.00034 | 0.00229 | 65 | 0.07981 | 0.00039 | 0.00206 | 93 | 0.00308 | 0.64976 | 143.857 | 0.51688 | 0.85216 |
| murE; UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–2,6-diaminopimelate ligase [EC:6.3.2.13] | 0.08059 | 0.00221 | 0.08349 | 0.00090 | 0.00372 | 65 | 0.07857 | 0.00068 | 0.00271 | 93 | 0.00460 | 1.07000 | 125.555 | 0.28667 | 0.77376 |
| E6.5.1.2, ligA, ligB; DNA ligase (NAD+) [EC:6.5.1.2] | 0.08043 | 0.00097 | 0.08219 | 0.00012 | 0.00136 | 65 | 0.07919 | 0.00017 | 0.00134 | 93 | 0.00191 | 1.57040 | 150.095 | 0.11843 | 0.77376 |
| PK, pyk; pyruvate kinase [EC:2.7.1.40] | 0.08031 | 0.00118 | 0.08294 | 0.00017 | 0.00162 | 65 | 0.07847 | 0.00025 | 0.00164 | 93 | 0.00230 | 1.94083 | 151.812 | 0.05413 | 0.70115 |
| ftsZ; cell division protein FtsZ | 0.08029 | 0.00171 | 0.08298 | 0.00052 | 0.00282 | 65 | 0.07841 | 0.00042 | 0.00213 | 93 | 0.00353 | 1.29508 | 128.986 | 0.19761 | 0.77376 |
| KARS, lysS; lysyl-tRNA synthetase, class II [EC:6.1.1.6] | 0.08019 | 0.00158 | 0.08277 | 0.00041 | 0.00252 | 65 | 0.07839 | 0.00038 | 0.00202 | 93 | 0.00323 | 1.35610 | 134.270 | 0.17734 | 0.77376 |
| carA, CPA1; carbamoyl-phosphate synthase small subunit [EC:6.3.5.5] | 0.07996 | 0.00211 | 0.08043 | 0.00089 | 0.00370 | 65 | 0.07964 | 0.00058 | 0.00251 | 93 | 0.00447 | 0.17808 | 118.724 | 0.85896 | 0.96086 |
| folD; methylenetetrahydrofolate dehydrogenase (NADP+) / methenyltetrahydrofolate cyclohydrolase [EC:1.5.1.5 3.5.4.9] | 0.07990 | 0.00116 | 0.08033 | 0.00020 | 0.00174 | 65 | 0.07959 | 0.00023 | 0.00156 | 93 | 0.00234 | 0.31443 | 143.684 | 0.75365 | 0.93643 |
| thiE; thiamine-phosphate pyrophosphorylase [EC:2.5.1.3] | 0.07989 | 0.00224 | 0.08137 | 0.00081 | 0.00353 | 65 | 0.07886 | 0.00078 | 0.00289 | 93 | 0.00457 | 0.54839 | 136.068 | 0.58433 | 0.87720 |
| smf; DNA processing protein | 0.07983 | 0.00167 | 0.08330 | 0.00050 | 0.00278 | 65 | 0.07741 | 0.00039 | 0.00204 | 93 | 0.00345 | 1.70839 | 126.136 | 0.09002 | 0.76567 |
| ycaJ; putative ATPase | 0.07978 | 0.00177 | 0.08151 | 0.00049 | 0.00276 | 65 | 0.07857 | 0.00050 | 0.00231 | 93 | 0.00360 | 0.81731 | 137.988 | 0.41516 | 0.81730 |
| exbB; biopolymer transport protein ExbB | 0.07953 | 0.00369 | 0.08064 | 0.00220 | 0.00582 | 65 | 0.07875 | 0.00213 | 0.00479 | 93 | 0.00753 | 0.25026 | 136.528 | 0.80276 | 0.94869 |
| greA; transcription elongation factor GreA | 0.07951 | 0.00215 | 0.08011 | 0.00095 | 0.00383 | 65 | 0.07909 | 0.00058 | 0.00251 | 93 | 0.00458 | 0.22407 | 115.788 | 0.82310 | 0.95506 |
| dnaK, HSPA9; molecular chaperone DnaK | 0.07948 | 0.00139 | 0.08142 | 0.00028 | 0.00206 | 65 | 0.07813 | 0.00032 | 0.00186 | 93 | 0.00278 | 1.18375 | 144.203 | 0.23846 | 0.77376 |
| MTFMT, fmt; methionyl-tRNA formyltransferase [EC:2.1.2.9] | 0.07947 | 0.00126 | 0.08092 | 0.00026 | 0.00198 | 65 | 0.07846 | 0.00025 | 0.00163 | 93 | 0.00257 | 0.95827 | 136.536 | 0.33962 | 0.78448 |
| dnaN; DNA polymerase III subunit beta [EC:2.7.7.7] | 0.07944 | 0.00167 | 0.08296 | 0.00051 | 0.00280 | 65 | 0.07697 | 0.00038 | 0.00203 | 93 | 0.00346 | 1.73337 | 124.962 | 0.08550 | 0.76567 |
| clpX, CLPX; ATP-dependent Clp protease ATP-binding subunit ClpX | 0.07943 | 0.00162 | 0.08178 | 0.00044 | 0.00261 | 65 | 0.07778 | 0.00039 | 0.00206 | 93 | 0.00333 | 1.20149 | 132.569 | 0.23170 | 0.77376 |
| xseA; exodeoxyribonuclease VII large subunit [EC:3.1.11.6] | 0.07919 | 0.00154 | 0.08261 | 0.00046 | 0.00266 | 65 | 0.07681 | 0.00031 | 0.00182 | 93 | 0.00322 | 1.80167 | 119.534 | 0.07412 | 0.74128 |
| K08884; serine/threonine protein kinase, bacterial [EC:2.7.11.1] | 0.07910 | 0.00349 | 0.08033 | 0.00186 | 0.00535 | 65 | 0.07824 | 0.00199 | 0.00463 | 93 | 0.00708 | 0.29508 | 140.702 | 0.76836 | 0.93958 |
| tig; trigger factor | 0.07905 | 0.00154 | 0.08104 | 0.00039 | 0.00246 | 65 | 0.07767 | 0.00036 | 0.00198 | 93 | 0.00316 | 1.06693 | 134.411 | 0.28792 | 0.77376 |
| K07007; uncharacterized protein | 0.07901 | 0.00191 | 0.07766 | 0.00071 | 0.00330 | 65 | 0.07995 | 0.00049 | 0.00230 | 93 | 0.00402 | -0.57060 | 121.318 | 0.56933 | 0.87135 |
| rsuA; 16S rRNA pseudouridine516 synthase [EC:5.4.99.19] | 0.07897 | 0.00277 | 0.08349 | 0.00111 | 0.00414 | 65 | 0.07582 | 0.00128 | 0.00371 | 93 | 0.00555 | 1.38030 | 143.604 | 0.16964 | 0.77376 |
| lspA; signal peptidase II [EC:3.4.23.36] | 0.07887 | 0.00147 | 0.08083 | 0.00037 | 0.00237 | 65 | 0.07750 | 0.00033 | 0.00187 | 93 | 0.00302 | 1.10228 | 132.867 | 0.27233 | 0.77376 |
| lysC; aspartate kinase [EC:2.7.2.4] | 0.07881 | 0.00195 | 0.07700 | 0.00062 | 0.00308 | 65 | 0.08008 | 0.00059 | 0.00252 | 93 | 0.00398 | -0.77288 | 135.964 | 0.44093 | 0.83186 |
| FARSB, pheT; phenylalanyl-tRNA synthetase beta chain [EC:6.1.1.20] | 0.07874 | 0.00120 | 0.08018 | 0.00022 | 0.00185 | 65 | 0.07774 | 0.00023 | 0.00157 | 93 | 0.00243 | 1.00496 | 139.058 | 0.31666 | 0.77376 |
| ftsW, spoVE; cell division protein FtsW | 0.07873 | 0.00134 | 0.07880 | 0.00026 | 0.00200 | 65 | 0.07868 | 0.00031 | 0.00181 | 93 | 0.00270 | 0.04706 | 144.662 | 0.96253 | 0.99023 |
| murF; UDP-N-acetylmuramoyl-tripeptide–D-alanyl-D-alanine ligase [EC:6.3.2.10] | 0.07870 | 0.00158 | 0.07974 | 0.00033 | 0.00227 | 65 | 0.07797 | 0.00044 | 0.00218 | 93 | 0.00315 | 0.56361 | 148.721 | 0.57387 | 0.87294 |
| PTS-Man-EIIB, manX; PTS system, mannose-specific IIB component [EC:2.7.1.191] | 0.07868 | 0.00465 | 0.08104 | 0.00304 | 0.00684 | 65 | 0.07703 | 0.00372 | 0.00632 | 93 | 0.00931 | 0.43111 | 145.994 | 0.66702 | 0.91145 |
| murC; UDP-N-acetylmuramate–alanine ligase [EC:6.3.2.8] | 0.07859 | 0.00149 | 0.08044 | 0.00034 | 0.00227 | 65 | 0.07730 | 0.00036 | 0.00198 | 93 | 0.00301 | 1.04245 | 141.221 | 0.29898 | 0.77376 |
| MARS, metG; methionyl-tRNA synthetase [EC:6.1.1.10] | 0.07849 | 0.00143 | 0.08034 | 0.00032 | 0.00222 | 65 | 0.07719 | 0.00032 | 0.00187 | 93 | 0.00290 | 1.08611 | 138.267 | 0.27932 | 0.77376 |
| serA, PHGDH; D-3-phosphoglycerate dehydrogenase / 2-oxoglutarate reductase [EC:1.1.1.95 1.1.1.399] | 0.07836 | 0.00210 | 0.07689 | 0.00081 | 0.00353 | 65 | 0.07938 | 0.00062 | 0.00259 | 93 | 0.00437 | -0.56941 | 125.975 | 0.57009 | 0.87135 |
| WARS, trpS; tryptophanyl-tRNA synthetase [EC:6.1.1.2] | 0.07831 | 0.00126 | 0.08007 | 0.00020 | 0.00175 | 65 | 0.07708 | 0.00029 | 0.00176 | 93 | 0.00248 | 1.20534 | 151.330 | 0.22995 | 0.77376 |
| hemK, prmC, HEMK; release factor glutamine methyltransferase [EC:2.1.1.297] | 0.07821 | 0.00126 | 0.07878 | 0.00025 | 0.00195 | 65 | 0.07781 | 0.00025 | 0.00165 | 93 | 0.00255 | 0.37897 | 139.277 | 0.70529 | 0.92004 |
| GPI, pgi; glucose-6-phosphate isomerase [EC:5.3.1.9] | 0.07821 | 0.00178 | 0.08013 | 0.00052 | 0.00284 | 65 | 0.07687 | 0.00049 | 0.00229 | 93 | 0.00364 | 0.89460 | 134.629 | 0.37260 | 0.79809 |
| ATPF0B, atpF; F-type H+-transporting ATPase subunit b | 0.07806 | 0.00141 | 0.07825 | 0.00026 | 0.00200 | 65 | 0.07792 | 0.00036 | 0.00196 | 93 | 0.00280 | 0.11500 | 149.895 | 0.90860 | 0.97664 |
| TARS, thrS; threonyl-tRNA synthetase [EC:6.1.1.3] | 0.07796 | 0.00121 | 0.07906 | 0.00023 | 0.00188 | 65 | 0.07720 | 0.00024 | 0.00159 | 93 | 0.00246 | 0.75559 | 139.128 | 0.45117 | 0.83438 |
| IARS, ileS; isoleucyl-tRNA synthetase [EC:6.1.1.5] | 0.07795 | 0.00124 | 0.07927 | 0.00024 | 0.00192 | 65 | 0.07703 | 0.00025 | 0.00164 | 93 | 0.00252 | 0.88910 | 139.740 | 0.37548 | 0.79810 |
| VARS, valS; valyl-tRNA synthetase [EC:6.1.1.9] | 0.07794 | 0.00116 | 0.07913 | 0.00021 | 0.00181 | 65 | 0.07711 | 0.00021 | 0.00151 | 93 | 0.00236 | 0.85783 | 137.762 | 0.39247 | 0.80459 |
| dnaE; DNA polymerase III subunit alpha [EC:2.7.7.7] | 0.07792 | 0.00124 | 0.07948 | 0.00024 | 0.00193 | 65 | 0.07682 | 0.00024 | 0.00161 | 93 | 0.00251 | 1.05744 | 137.925 | 0.29216 | 0.77376 |
| uvrC; excinuclease ABC subunit C | 0.07790 | 0.00119 | 0.07936 | 0.00022 | 0.00184 | 65 | 0.07688 | 0.00023 | 0.00156 | 93 | 0.00241 | 1.02906 | 139.102 | 0.30524 | 0.77376 |
| E2.5.1.54, aroF, aroG, aroH; 3-deoxy-7-phosphoheptulonate synthase [EC:2.5.1.54] | 0.07789 | 0.00290 | 0.07694 | 0.00139 | 0.00462 | 65 | 0.07855 | 0.00129 | 0.00373 | 93 | 0.00594 | -0.27075 | 134.709 | 0.78700 | 0.94470 |
| priA; primosomal protein N’ (replication factor Y) (superfamily II helicase) [EC:3.6.4.-] | 0.07787 | 0.00119 | 0.07934 | 0.00022 | 0.00184 | 65 | 0.07684 | 0.00023 | 0.00156 | 93 | 0.00241 | 1.03693 | 139.143 | 0.30157 | 0.77376 |
| PARS, proS; prolyl-tRNA synthetase [EC:6.1.1.15] | 0.07782 | 0.00123 | 0.07905 | 0.00023 | 0.00188 | 65 | 0.07696 | 0.00025 | 0.00163 | 93 | 0.00249 | 0.83797 | 140.693 | 0.40347 | 0.81012 |
| recA; recombination protein RecA | 0.07779 | 0.00123 | 0.07896 | 0.00022 | 0.00185 | 65 | 0.07697 | 0.00025 | 0.00165 | 93 | 0.00248 | 0.80547 | 143.074 | 0.42189 | 0.82295 |
| dnaX; DNA polymerase III subunit gamma/tau [EC:2.7.7.7] | 0.07769 | 0.00123 | 0.07923 | 0.00022 | 0.00184 | 65 | 0.07661 | 0.00025 | 0.00164 | 93 | 0.00247 | 1.06512 | 143.592 | 0.28861 | 0.77376 |
| E2.7.1.71, aroK, aroL; shikimate kinase [EC:2.7.1.71] | 0.07764 | 0.00189 | 0.07940 | 0.00067 | 0.00322 | 65 | 0.07640 | 0.00049 | 0.00230 | 93 | 0.00395 | 0.75882 | 123.569 | 0.44941 | 0.83438 |
| truA, PUS1; tRNA pseudouridine38-40 synthase [EC:5.4.99.12] | 0.07762 | 0.00141 | 0.07914 | 0.00030 | 0.00215 | 65 | 0.07655 | 0.00033 | 0.00187 | 93 | 0.00285 | 0.90852 | 141.340 | 0.36515 | 0.79587 |
| pyrF; orotidine-5’-phosphate decarboxylase [EC:4.1.1.23] | 0.07748 | 0.00145 | 0.07961 | 0.00032 | 0.00221 | 65 | 0.07598 | 0.00034 | 0.00191 | 93 | 0.00292 | 1.24077 | 140.462 | 0.21676 | 0.77376 |
| RARS, argS; arginyl-tRNA synthetase [EC:6.1.1.19] | 0.07743 | 0.00136 | 0.07850 | 0.00027 | 0.00205 | 65 | 0.07667 | 0.00031 | 0.00182 | 93 | 0.00274 | 0.66771 | 142.779 | 0.50539 | 0.84956 |
| RP-L28, MRPL28, rpmB; large subunit ribosomal protein L28 | 0.07737 | 0.00135 | 0.07855 | 0.00027 | 0.00204 | 65 | 0.07655 | 0.00030 | 0.00179 | 93 | 0.00271 | 0.73820 | 142.167 | 0.46161 | 0.83596 |
| uvrB; excinuclease ABC subunit B | 0.07732 | 0.00125 | 0.07884 | 0.00023 | 0.00187 | 65 | 0.07626 | 0.00026 | 0.00166 | 93 | 0.00251 | 1.03127 | 142.837 | 0.30416 | 0.77376 |
| mraW, rsmH; 16S rRNA (cytosine1402-N4)-methyltransferase [EC:2.1.1.199] | 0.07731 | 0.00140 | 0.07886 | 0.00030 | 0.00214 | 65 | 0.07623 | 0.00032 | 0.00186 | 93 | 0.00283 | 0.92524 | 141.415 | 0.35642 | 0.79399 |
| YARS, tyrS; tyrosyl-tRNA synthetase [EC:6.1.1.1] | 0.07730 | 0.00133 | 0.07841 | 0.00026 | 0.00200 | 65 | 0.07652 | 0.00030 | 0.00178 | 93 | 0.00268 | 0.70414 | 143.097 | 0.48249 | 0.84354 |
| adk, AK; adenylate kinase [EC:2.7.4.3] | 0.07727 | 0.00125 | 0.07837 | 0.00025 | 0.00196 | 65 | 0.07651 | 0.00025 | 0.00162 | 93 | 0.00255 | 0.72885 | 137.155 | 0.46733 | 0.83837 |
| CARS, cysS; cysteinyl-tRNA synthetase [EC:6.1.1.16] | 0.07725 | 0.00123 | 0.07885 | 0.00022 | 0.00186 | 65 | 0.07613 | 0.00025 | 0.00164 | 93 | 0.00248 | 1.09707 | 142.623 | 0.27446 | 0.77376 |
| murG; UDP-N-acetylglucosamine–N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase [EC:2.4.1.227] | 0.07724 | 0.00124 | 0.07863 | 0.00023 | 0.00189 | 65 | 0.07627 | 0.00025 | 0.00165 | 93 | 0.00251 | 0.93926 | 141.855 | 0.34920 | 0.79120 |
| pyrE; orotate phosphoribosyltransferase [EC:2.4.2.10] | 0.07723 | 0.00142 | 0.07864 | 0.00030 | 0.00215 | 65 | 0.07624 | 0.00033 | 0.00188 | 93 | 0.00286 | 0.83829 | 141.471 | 0.40328 | 0.81012 |
| rpoB; DNA-directed RNA polymerase subunit beta [EC:2.7.7.6] | 0.07719 | 0.00120 | 0.07900 | 0.00021 | 0.00181 | 65 | 0.07592 | 0.00024 | 0.00159 | 93 | 0.00241 | 1.27851 | 141.816 | 0.20316 | 0.77376 |
| obgE, cgtA; GTPase [EC:3.6.5.-] | 0.07715 | 0.00124 | 0.07872 | 0.00023 | 0.00188 | 65 | 0.07605 | 0.00025 | 0.00164 | 93 | 0.00250 | 1.07021 | 141.770 | 0.28634 | 0.77376 |
| groES, HSPE1; chaperonin GroES | 0.07712 | 0.00133 | 0.07850 | 0.00025 | 0.00197 | 65 | 0.07616 | 0.00030 | 0.00179 | 93 | 0.00267 | 0.87919 | 144.642 | 0.38076 | 0.79887 |
| mfd; transcription-repair coupling factor (superfamily II helicase) [EC:3.6.4.-] | 0.07711 | 0.00132 | 0.07850 | 0.00025 | 0.00197 | 65 | 0.07614 | 0.00030 | 0.00178 | 93 | 0.00265 | 0.88743 | 144.608 | 0.37632 | 0.79810 |
| purA, ADSS; adenylosuccinate synthase [EC:6.3.4.4] | 0.07711 | 0.00123 | 0.07846 | 0.00024 | 0.00191 | 65 | 0.07616 | 0.00024 | 0.00162 | 93 | 0.00251 | 0.92175 | 138.814 | 0.35826 | 0.79496 |
| infA; translation initiation factor IF-1 | 0.07709 | 0.00131 | 0.07878 | 0.00024 | 0.00191 | 65 | 0.07591 | 0.00030 | 0.00178 | 93 | 0.00261 | 1.09623 | 146.601 | 0.27477 | 0.77376 |
| HARS, hisS; histidyl-tRNA synthetase [EC:6.1.1.21] | 0.07709 | 0.00135 | 0.07836 | 0.00026 | 0.00200 | 65 | 0.07621 | 0.00031 | 0.00181 | 93 | 0.00270 | 0.79598 | 144.542 | 0.42735 | 0.82664 |
| infB, MTIF2; translation initiation factor IF-2 | 0.07703 | 0.00139 | 0.07874 | 0.00027 | 0.00205 | 65 | 0.07584 | 0.00033 | 0.00187 | 93 | 0.00278 | 1.04592 | 145.043 | 0.29734 | 0.77376 |
| rpe, RPE; ribulose-phosphate 3-epimerase [EC:5.1.3.1] | 0.07702 | 0.00138 | 0.07884 | 0.00028 | 0.00209 | 65 | 0.07576 | 0.00031 | 0.00183 | 93 | 0.00278 | 1.10908 | 142.018 | 0.26927 | 0.77376 |
| fabD; [acyl-carrier-protein] S-malonyltransferase [EC:2.3.1.39] | 0.07695 | 0.00122 | 0.07767 | 0.00022 | 0.00184 | 65 | 0.07644 | 0.00025 | 0.00163 | 93 | 0.00246 | 0.49811 | 142.902 | 0.61917 | 0.88981 |
| murB; UDP-N-acetylmuramate dehydrogenase [EC:1.3.1.98] | 0.07694 | 0.00138 | 0.07841 | 0.00027 | 0.00205 | 65 | 0.07591 | 0.00032 | 0.00187 | 93 | 0.00277 | 0.90233 | 144.817 | 0.36838 | 0.79587 |
| ppnK, NADK; NAD+ kinase [EC:2.7.1.23] | 0.07693 | 0.00136 | 0.07816 | 0.00026 | 0.00199 | 65 | 0.07606 | 0.00032 | 0.00184 | 93 | 0.00271 | 0.77661 | 146.106 | 0.43865 | 0.83152 |
| rnc, DROSHA, RNT1; ribonuclease III [EC:3.1.26.3] | 0.07690 | 0.00124 | 0.07859 | 0.00023 | 0.00190 | 65 | 0.07572 | 0.00025 | 0.00164 | 93 | 0.00250 | 1.14761 | 140.611 | 0.25308 | 0.77376 |
| PGK, pgk; phosphoglycerate kinase [EC:2.7.2.3] | 0.07687 | 0.00139 | 0.07838 | 0.00028 | 0.00206 | 65 | 0.07581 | 0.00033 | 0.00187 | 93 | 0.00278 | 0.92520 | 144.549 | 0.35641 | 0.79399 |
| ksgA; 16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase [EC:2.1.1.182] | 0.07685 | 0.00138 | 0.07854 | 0.00028 | 0.00208 | 65 | 0.07567 | 0.00032 | 0.00184 | 93 | 0.00278 | 1.03329 | 142.557 | 0.30322 | 0.77376 |
| hprT, hpt, HPRT1; hypoxanthine phosphoribosyltransferase [EC:2.4.2.8] | 0.07682 | 0.00187 | 0.08039 | 0.00060 | 0.00303 | 65 | 0.07433 | 0.00051 | 0.00235 | 93 | 0.00383 | 1.58276 | 131.204 | 0.11588 | 0.77376 |
| TC.LIVCS; branched-chain amino acid:cation transporter, LIVCS family | 0.07680 | 0.00308 | 0.08472 | 0.00171 | 0.00513 | 65 | 0.07127 | 0.00129 | 0.00372 | 93 | 0.00634 | 2.11969 | 124.969 | 0.03601 | 0.62411 |
| rpoA; DNA-directed RNA polymerase subunit alpha [EC:2.7.7.6] | 0.07676 | 0.00138 | 0.07835 | 0.00028 | 0.00208 | 65 | 0.07565 | 0.00031 | 0.00184 | 93 | 0.00277 | 0.97530 | 142.569 | 0.33106 | 0.77780 |
| E1.17.4.1B, nrdB, nrdF; ribonucleoside-diphosphate reductase beta chain [EC:1.17.4.1] | 0.07673 | 0.00172 | 0.07732 | 0.00055 | 0.00290 | 65 | 0.07632 | 0.00041 | 0.00211 | 93 | 0.00359 | 0.27692 | 125.079 | 0.78230 | 0.94310 |
| purD; phosphoribosylamine—glycine ligase [EC:6.3.4.13] | 0.07672 | 0.00160 | 0.07855 | 0.00040 | 0.00248 | 65 | 0.07544 | 0.00041 | 0.00209 | 93 | 0.00324 | 0.95859 | 138.484 | 0.33944 | 0.78433 |
| trmD; tRNA (guanine37-N1)-methyltransferase [EC:2.1.1.228] | 0.07669 | 0.00137 | 0.07853 | 0.00028 | 0.00208 | 65 | 0.07541 | 0.00031 | 0.00182 | 93 | 0.00277 | 1.12468 | 141.548 | 0.26263 | 0.77376 |
| smpB; SsrA-binding protein | 0.07669 | 0.00136 | 0.07823 | 0.00027 | 0.00204 | 65 | 0.07561 | 0.00031 | 0.00181 | 93 | 0.00273 | 0.96007 | 143.069 | 0.33864 | 0.78407 |
| RP-S18, MRPS18, rpsR; small subunit ribosomal protein S18 | 0.07667 | 0.00134 | 0.07802 | 0.00026 | 0.00201 | 65 | 0.07572 | 0.00030 | 0.00179 | 93 | 0.00269 | 0.85425 | 142.772 | 0.39440 | 0.80459 |
| metK; S-adenosylmethionine synthetase [EC:2.5.1.6] | 0.07666 | 0.00136 | 0.07809 | 0.00027 | 0.00203 | 65 | 0.07566 | 0.00031 | 0.00183 | 93 | 0.00273 | 0.89146 | 143.932 | 0.37417 | 0.79810 |
| radA, sms; DNA repair protein RadA/Sms | 0.07666 | 0.00142 | 0.07816 | 0.00028 | 0.00207 | 65 | 0.07561 | 0.00034 | 0.00192 | 93 | 0.00283 | 0.90300 | 146.336 | 0.36801 | 0.79587 |
| dapB; 4-hydroxy-tetrahydrodipicolinate reductase [EC:1.17.1.8] | 0.07666 | 0.00179 | 0.07516 | 0.00053 | 0.00287 | 65 | 0.07771 | 0.00049 | 0.00229 | 93 | 0.00367 | -0.69528 | 133.994 | 0.48809 | 0.84678 |
| RP-L17, MRPL17, rplQ; large subunit ribosomal protein L17 | 0.07664 | 0.00136 | 0.07838 | 0.00027 | 0.00203 | 65 | 0.07543 | 0.00031 | 0.00183 | 93 | 0.00273 | 1.08066 | 143.967 | 0.28165 | 0.77376 |
| trmB, METTL1; tRNA (guanine-N7-)-methyltransferase [EC:2.1.1.33] | 0.07663 | 0.00143 | 0.07862 | 0.00031 | 0.00220 | 65 | 0.07524 | 0.00033 | 0.00188 | 93 | 0.00289 | 1.16908 | 140.070 | 0.24436 | 0.77376 |
| nusG; transcriptional antiterminator NusG | 0.07663 | 0.00135 | 0.07795 | 0.00027 | 0.00202 | 65 | 0.07570 | 0.00031 | 0.00181 | 93 | 0.00272 | 0.82637 | 143.497 | 0.40996 | 0.81416 |
| rsgA, engC; ribosome biogenesis GTPase / thiamine phosphate phosphatase [EC:3.6.1.- 3.1.3.100] | 0.07661 | 0.00160 | 0.07930 | 0.00039 | 0.00244 | 65 | 0.07472 | 0.00041 | 0.00211 | 93 | 0.00323 | 1.41851 | 140.651 | 0.15825 | 0.77376 |
| cmk; CMP/dCMP kinase [EC:2.7.4.25] | 0.07660 | 0.00165 | 0.07887 | 0.00041 | 0.00251 | 65 | 0.07501 | 0.00044 | 0.00219 | 93 | 0.00333 | 1.16197 | 141.487 | 0.24720 | 0.77376 |
| ruvB; holliday junction DNA helicase RuvB [EC:3.6.4.12] | 0.07659 | 0.00133 | 0.07828 | 0.00026 | 0.00198 | 65 | 0.07541 | 0.00030 | 0.00179 | 93 | 0.00267 | 1.07228 | 144.174 | 0.28539 | 0.77376 |
| RP-S4, rpsD; small subunit ribosomal protein S4 | 0.07659 | 0.00136 | 0.07838 | 0.00028 | 0.00206 | 65 | 0.07534 | 0.00030 | 0.00180 | 93 | 0.00273 | 1.11439 | 141.723 | 0.26700 | 0.77376 |
| E3.4.21.102, prc, ctpA; carboxyl-terminal processing protease [EC:3.4.21.102] | 0.07657 | 0.00255 | 0.07437 | 0.00078 | 0.00347 | 65 | 0.07811 | 0.00121 | 0.00360 | 93 | 0.00500 | -0.74723 | 152.862 | 0.45607 | 0.83438 |
| gpsA; glycerol-3-phosphate dehydrogenase (NAD(P)+) [EC:1.1.1.94] | 0.07654 | 0.00135 | 0.07803 | 0.00026 | 0.00202 | 65 | 0.07550 | 0.00030 | 0.00180 | 93 | 0.00270 | 0.93698 | 143.422 | 0.35035 | 0.79120 |
| PTH1, pth, spoVC; peptidyl-tRNA hydrolase, PTH1 family [EC:3.1.1.29] | 0.07654 | 0.00135 | 0.07824 | 0.00026 | 0.00201 | 65 | 0.07535 | 0.00030 | 0.00181 | 93 | 0.00271 | 1.06785 | 143.896 | 0.28737 | 0.77376 |
| mraY; phospho-N-acetylmuramoyl-pentapeptide-transferase [EC:2.7.8.13] | 0.07653 | 0.00134 | 0.07795 | 0.00026 | 0.00202 | 65 | 0.07554 | 0.00030 | 0.00178 | 93 | 0.00269 | 0.89554 | 142.570 | 0.37201 | 0.79738 |
| pflA, pflC, pflE; pyruvate formate lyase activating enzyme [EC:1.97.1.4] | 0.07653 | 0.00364 | 0.08030 | 0.00215 | 0.00575 | 65 | 0.07390 | 0.00207 | 0.00472 | 93 | 0.00744 | 0.86060 | 136.116 | 0.39097 | 0.80384 |
| E2.7.7.3A, coaD, kdtB; pantetheine-phosphate adenylyltransferase [EC:2.7.7.3] | 0.07652 | 0.00134 | 0.07801 | 0.00026 | 0.00201 | 65 | 0.07549 | 0.00030 | 0.00180 | 93 | 0.00270 | 0.93581 | 143.432 | 0.35094 | 0.79120 |
| RP-L24, MRPL24, rplX; large subunit ribosomal protein L24 | 0.07652 | 0.00134 | 0.07794 | 0.00027 | 0.00202 | 65 | 0.07553 | 0.00030 | 0.00179 | 93 | 0.00270 | 0.89259 | 142.845 | 0.37358 | 0.79810 |
| dnaG; DNA primase [EC:2.7.7.-] | 0.07650 | 0.00134 | 0.07808 | 0.00026 | 0.00201 | 65 | 0.07540 | 0.00030 | 0.00179 | 93 | 0.00269 | 0.99370 | 143.237 | 0.32204 | 0.77376 |
| rumA; 23S rRNA (uracil1939-C5)-methyltransferase [EC:2.1.1.190] | 0.07647 | 0.00181 | 0.07709 | 0.00048 | 0.00272 | 65 | 0.07603 | 0.00055 | 0.00243 | 93 | 0.00365 | 0.29187 | 143.516 | 0.77081 | 0.94023 |
| GRPE; molecular chaperone GrpE | 0.07646 | 0.00134 | 0.07809 | 0.00026 | 0.00201 | 65 | 0.07532 | 0.00030 | 0.00180 | 93 | 0.00270 | 1.02490 | 143.725 | 0.30713 | 0.77376 |
| rluB; 23S rRNA pseudouridine2605 synthase [EC:5.4.99.22] | 0.07644 | 0.00133 | 0.07734 | 0.00029 | 0.00212 | 65 | 0.07582 | 0.00027 | 0.00171 | 93 | 0.00272 | 0.55685 | 134.873 | 0.57855 | 0.87532 |
| era, ERAL1; GTPase | 0.07641 | 0.00124 | 0.07811 | 0.00023 | 0.00187 | 65 | 0.07522 | 0.00025 | 0.00165 | 93 | 0.00250 | 1.15822 | 142.324 | 0.24871 | 0.77376 |
| truB, PUS4, TRUB1; tRNA pseudouridine55 synthase [EC:5.4.99.25] | 0.07641 | 0.00125 | 0.07835 | 0.00023 | 0.00187 | 65 | 0.07505 | 0.00026 | 0.00167 | 93 | 0.00251 | 1.31370 | 143.023 | 0.19105 | 0.77376 |
| pyrH; uridylate kinase [EC:2.7.4.22] | 0.07640 | 0.00135 | 0.07789 | 0.00027 | 0.00203 | 65 | 0.07536 | 0.00030 | 0.00181 | 93 | 0.00272 | 0.93140 | 143.395 | 0.35321 | 0.79206 |
| SARS, serS; seryl-tRNA synthetase [EC:6.1.1.11] | 0.07640 | 0.00135 | 0.07794 | 0.00027 | 0.00203 | 65 | 0.07532 | 0.00030 | 0.00181 | 93 | 0.00272 | 0.96553 | 143.237 | 0.33591 | 0.78225 |
| FARSA, pheS; phenylalanyl-tRNA synthetase alpha chain [EC:6.1.1.20] | 0.07637 | 0.00134 | 0.07802 | 0.00026 | 0.00200 | 65 | 0.07522 | 0.00030 | 0.00180 | 93 | 0.00269 | 1.04053 | 143.659 | 0.29984 | 0.77376 |
| ATPF1B, atpD; F-type H+-transporting ATPase subunit beta [EC:3.6.3.14] | 0.07636 | 0.00125 | 0.07768 | 0.00023 | 0.00187 | 65 | 0.07544 | 0.00026 | 0.00168 | 93 | 0.00251 | 0.89185 | 143.684 | 0.37397 | 0.79810 |
| murD; UDP-N-acetylmuramoylalanine–D-glutamate ligase [EC:6.3.2.9] | 0.07635 | 0.00135 | 0.07796 | 0.00026 | 0.00201 | 65 | 0.07523 | 0.00030 | 0.00180 | 93 | 0.00270 | 1.00805 | 143.681 | 0.31512 | 0.77376 |
| ychF; ribosome-binding ATPase | 0.07635 | 0.00135 | 0.07799 | 0.00026 | 0.00201 | 65 | 0.07521 | 0.00030 | 0.00181 | 93 | 0.00270 | 1.02761 | 143.732 | 0.30586 | 0.77376 |
| SRP54, ffh; signal recognition particle subunit SRP54 [EC:3.6.5.4] | 0.07635 | 0.00135 | 0.07794 | 0.00027 | 0.00202 | 65 | 0.07524 | 0.00030 | 0.00181 | 93 | 0.00271 | 0.99772 | 143.543 | 0.32010 | 0.77376 |
| rbfA; ribosome-binding factor A | 0.07635 | 0.00135 | 0.07799 | 0.00026 | 0.00201 | 65 | 0.07520 | 0.00030 | 0.00181 | 93 | 0.00270 | 1.02971 | 143.733 | 0.30487 | 0.77376 |
| uppS; undecaprenyl diphosphate synthase [EC:2.5.1.31] | 0.07634 | 0.00136 | 0.07829 | 0.00028 | 0.00207 | 65 | 0.07497 | 0.00030 | 0.00180 | 93 | 0.00274 | 1.20816 | 140.846 | 0.22901 | 0.77376 |
| purB, ADSL; adenylosuccinate lyase [EC:4.3.2.2] | 0.07631 | 0.00136 | 0.07790 | 0.00027 | 0.00203 | 65 | 0.07520 | 0.00031 | 0.00182 | 93 | 0.00272 | 0.98962 | 143.617 | 0.32403 | 0.77376 |
| RP-L1, MRPL1, rplA; large subunit ribosomal protein L1 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00669 | 143.227 | 0.31578 | 0.77376 |
| RP-L10, MRPL10, rplJ; large subunit ribosomal protein L10 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00669 | 143.227 | 0.31578 | 0.77376 |
| RP-L13, MRPL13, rplM; large subunit ribosomal protein L13 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00669 | 143.227 | 0.31578 | 0.77376 |
| RP-L3, MRPL3, rplC; large subunit ribosomal protein L3 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00669 | 143.227 | 0.31578 | 0.77376 |
| RP-L6, MRPL6, rplF; large subunit ribosomal protein L6 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00669 | 143.227 | 0.31578 | 0.77376 |
| RP-S12, MRPS12, rpsL; small subunit ribosomal protein S12 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00669 | 143.227 | 0.31578 | 0.77376 |
| RP-S3, rpsC; small subunit ribosomal protein S3 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00669 | 143.227 | 0.31578 | 0.77376 |
| RP-S5, MRPS5, rpsE; small subunit ribosomal protein S5 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00669 | 143.227 | 0.31578 | 0.77376 |
| RP-S8, rpsH; small subunit ribosomal protein S8 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00669 | 143.227 | 0.31578 | 0.77376 |
| RP-L7, MRPL12, rplL; large subunit ribosomal protein L7/L12 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00898 | 143.223 | 0.31468 | 0.77376 |
| nusA; N utilization substance protein A | 0.07629 | 0.00135 | 0.07789 | 0.00027 | 0.00203 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00326 | 143.177 | 0.31743 | 0.77376 |
| frr, MRRF, RRF; ribosome recycling factor | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00786 | 143.225 | 0.31522 | 0.77376 |
| prfB; peptide chain release factor 2 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00786 | 143.225 | 0.31522 | 0.77376 |
| RP-L20, MRPL20, rplT; large subunit ribosomal protein L20 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00786 | 143.225 | 0.31522 | 0.77376 |
| RP-L27, MRPL27, rpmA; large subunit ribosomal protein L27 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00786 | 143.225 | 0.31522 | 0.77376 |
| RP-L4, MRPL4, rplD; large subunit ribosomal protein L4 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00786 | 143.225 | 0.31522 | 0.77376 |
| tsf, TSFM; elongation factor Ts | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00786 | 143.225 | 0.31522 | 0.77376 |
| RP-L21, MRPL21, rplU; large subunit ribosomal protein L21 | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07516 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00842 | 143.222 | 0.31495 | 0.77376 |
| holA; DNA polymerase III subunit delta [EC:2.7.7.7] | 0.07629 | 0.00136 | 0.07787 | 0.00027 | 0.00204 | 65 | 0.07518 | 0.00031 | 0.00181 | 93 | 0.00273 | 0.98539 | 142.844 | 0.32610 | 0.77376 |
| ftsY; fused signal recognition particle receptor | 0.07629 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07516 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.01140 | 143.218 | 0.31353 | 0.77376 |
| RP-L2, MRPL2, rplB; large subunit ribosomal protein L2 | 0.07628 | 0.00135 | 0.07788 | 0.00027 | 0.00203 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 0.99705 | 143.084 | 0.32042 | 0.77376 |
| RP-S2, MRPS2, rpsB; small subunit ribosomal protein S2 | 0.07628 | 0.00135 | 0.07788 | 0.00027 | 0.00203 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 0.99705 | 143.084 | 0.32042 | 0.77376 |
| engA, der; GTPase | 0.07628 | 0.00135 | 0.07788 | 0.00027 | 0.00203 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 0.99821 | 143.082 | 0.31986 | 0.77376 |
| prfA, MTRF1, MRF1; peptide chain release factor 1 | 0.07628 | 0.00135 | 0.07788 | 0.00027 | 0.00203 | 65 | 0.07517 | 0.00030 | 0.00180 | 93 | 0.00271 | 0.99821 | 143.082 | 0.31986 | 0.77376 |
| RP-S16, MRPS16, rpsP; small subunit ribosomal protein S16 | 0.07628 | 0.00135 | 0.07790 | 0.00027 | 0.00203 | 65 | 0.07515 | 0.00030 | 0.00181 | 93 | 0.00271 | 1.01080 | 143.245 | 0.31381 | 0.77376 |
| recR; recombination protein RecR | 0.07627 | 0.00135 | 0.07790 | 0.00027 | 0.00202 | 65 | 0.07513 | 0.00030 | 0.00181 | 93 | 0.00271 | 1.02112 | 143.390 | 0.30892 | 0.77376 |
| RP-L29, rpmC; large subunit ribosomal protein L29 | 0.07627 | 0.00135 | 0.07787 | 0.00027 | 0.00203 | 65 | 0.07516 | 0.00030 | 0.00180 | 93 | 0.00271 | 0.99917 | 143.068 | 0.31940 | 0.77376 |
| RP-L5, MRPL5, rplE; large subunit ribosomal protein L5 | 0.07627 | 0.00135 | 0.07787 | 0.00027 | 0.00203 | 65 | 0.07516 | 0.00030 | 0.00180 | 93 | 0.00271 | 0.99917 | 143.068 | 0.31940 | 0.77376 |
| RP-S9, MRPS9, rpsI; small subunit ribosomal protein S9 | 0.07627 | 0.00135 | 0.07787 | 0.00027 | 0.00203 | 65 | 0.07516 | 0.00030 | 0.00180 | 93 | 0.00271 | 0.99917 | 143.068 | 0.31940 | 0.77376 |
| infC, MTIF3; translation initiation factor IF-3 | 0.07627 | 0.00135 | 0.07787 | 0.00027 | 0.00202 | 65 | 0.07515 | 0.00030 | 0.00181 | 93 | 0.00271 | 1.00290 | 143.345 | 0.31760 | 0.77376 |
| ruvA; holliday junction DNA helicase RuvA [EC:3.6.4.12] | 0.07626 | 0.00135 | 0.07785 | 0.00027 | 0.00203 | 65 | 0.07515 | 0.00030 | 0.00180 | 93 | 0.00272 | 0.99062 | 142.910 | 0.32354 | 0.77376 |
| recN; DNA repair protein RecN (Recombination protein N) | 0.07626 | 0.00135 | 0.07789 | 0.00027 | 0.00202 | 65 | 0.07512 | 0.00030 | 0.00181 | 93 | 0.00271 | 1.02224 | 143.368 | 0.30839 | 0.77376 |
| nudF; ADP-ribose pyrophosphatase [EC:3.6.1.13] | 0.07626 | 0.00196 | 0.08224 | 0.00058 | 0.00299 | 65 | 0.07207 | 0.00058 | 0.00251 | 93 | 0.00390 | 2.60540 | 138.036 | 0.01018 | 0.58284 |
| RP-L23, MRPL23, rplW; large subunit ribosomal protein L23 | 0.07625 | 0.00135 | 0.07782 | 0.00027 | 0.00204 | 65 | 0.07516 | 0.00030 | 0.00181 | 93 | 0.00272 | 0.97377 | 142.639 | 0.33182 | 0.77825 |
| RP-S10, MRPS10, rpsJ; small subunit ribosomal protein S10 | 0.07625 | 0.00135 | 0.07782 | 0.00027 | 0.00204 | 65 | 0.07516 | 0.00030 | 0.00181 | 93 | 0.00272 | 0.97377 | 142.639 | 0.33182 | 0.77825 |
| RP-L35, MRPL35, rpmI; large subunit ribosomal protein L35 | 0.07625 | 0.00135 | 0.07786 | 0.00027 | 0.00202 | 65 | 0.07513 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.00640 | 143.162 | 0.31592 | 0.77376 |
| gidB, rsmG; 16S rRNA (guanine527-N7)-methyltransferase [EC:2.1.1.170] | 0.07625 | 0.00135 | 0.07798 | 0.00026 | 0.00200 | 65 | 0.07504 | 0.00031 | 0.00181 | 93 | 0.00270 | 1.08967 | 144.415 | 0.27767 | 0.77376 |
| RP-L19, MRPL19, rplS; large subunit ribosomal protein L19 | 0.07624 | 0.00135 | 0.07788 | 0.00027 | 0.00203 | 65 | 0.07510 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.02606 | 143.035 | 0.30659 | 0.77376 |
| RP-S20, rpsT; small subunit ribosomal protein S20 | 0.07624 | 0.00135 | 0.07784 | 0.00027 | 0.00203 | 65 | 0.07512 | 0.00030 | 0.00181 | 93 | 0.00272 | 1.00218 | 143.234 | 0.31795 | 0.77376 |
| ribF; riboflavin kinase / FMN adenylyltransferase [EC:2.7.1.26 2.7.7.2] | 0.07624 | 0.00135 | 0.07787 | 0.00027 | 0.00202 | 65 | 0.07509 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.02522 | 142.945 | 0.30699 | 0.77376 |
| RP-S7, MRPS7, rpsG; small subunit ribosomal protein S7 | 0.07623 | 0.00135 | 0.07779 | 0.00027 | 0.00204 | 65 | 0.07515 | 0.00030 | 0.00181 | 93 | 0.00273 | 0.96632 | 142.480 | 0.33552 | 0.78186 |
| RP-L11, MRPL11, rplK; large subunit ribosomal protein L11 | 0.07623 | 0.00135 | 0.07789 | 0.00027 | 0.00202 | 65 | 0.07507 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.03928 | 143.095 | 0.30043 | 0.77376 |
| RP-L14, MRPL14, rplN; large subunit ribosomal protein L14 | 0.07623 | 0.00135 | 0.07789 | 0.00027 | 0.00202 | 65 | 0.07507 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.03928 | 143.095 | 0.30043 | 0.77376 |
| RP-L15, MRPL15, rplO; large subunit ribosomal protein L15 | 0.07623 | 0.00135 | 0.07789 | 0.00027 | 0.00202 | 65 | 0.07507 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.04008 | 143.097 | 0.30006 | 0.77376 |
| RP-L18, MRPL18, rplR; large subunit ribosomal protein L18 | 0.07623 | 0.00135 | 0.07788 | 0.00027 | 0.00202 | 65 | 0.07507 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.03740 | 143.084 | 0.30130 | 0.77376 |
| RP-S13, rpsM; small subunit ribosomal protein S13 | 0.07623 | 0.00135 | 0.07789 | 0.00027 | 0.00202 | 65 | 0.07507 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.04082 | 143.118 | 0.29972 | 0.77376 |
| RP-S19, rpsS; small subunit ribosomal protein S19 | 0.07621 | 0.00135 | 0.07785 | 0.00027 | 0.00203 | 65 | 0.07507 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.02336 | 142.898 | 0.30787 | 0.77376 |
| rimM; 16S rRNA processing protein RimM | 0.07621 | 0.00135 | 0.07788 | 0.00027 | 0.00202 | 65 | 0.07504 | 0.00031 | 0.00181 | 93 | 0.00271 | 1.04489 | 143.663 | 0.29783 | 0.77376 |
| RP-L22, MRPL22, rplV; large subunit ribosomal protein L22 | 0.07620 | 0.00135 | 0.07784 | 0.00027 | 0.00202 | 65 | 0.07506 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.02863 | 143.221 | 0.30539 | 0.77376 |
| RP-S17, MRPS17, rpsQ; small subunit ribosomal protein S17 | 0.07620 | 0.00135 | 0.07786 | 0.00027 | 0.00203 | 65 | 0.07504 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.03830 | 143.047 | 0.30088 | 0.77376 |
| RP-S6, MRPS6, rpsF; small subunit ribosomal protein S6 | 0.07619 | 0.00136 | 0.07782 | 0.00027 | 0.00204 | 65 | 0.07506 | 0.00031 | 0.00181 | 93 | 0.00273 | 1.00915 | 142.884 | 0.31461 | 0.77376 |
| typA, bipA; GTP-binding protein | 0.07619 | 0.00135 | 0.07783 | 0.00027 | 0.00203 | 65 | 0.07504 | 0.00030 | 0.00180 | 93 | 0.00271 | 1.03029 | 142.909 | 0.30461 | 0.77376 |
| ruvX; putative holliday junction resolvase [EC:3.1.-.-] | 0.07614 | 0.00135 | 0.07785 | 0.00027 | 0.00202 | 65 | 0.07494 | 0.00030 | 0.00181 | 93 | 0.00271 | 1.07441 | 143.340 | 0.28444 | 0.77376 |
| rimI; [ribosomal protein S18]-alanine N-acetyltransferase [EC:2.3.1.266] | 0.07611 | 0.00188 | 0.07963 | 0.00048 | 0.00273 | 65 | 0.07365 | 0.00061 | 0.00255 | 93 | 0.00374 | 1.60258 | 146.840 | 0.11118 | 0.77376 |
| lepA; GTP-binding protein LepA | 0.07610 | 0.00134 | 0.07773 | 0.00026 | 0.00201 | 65 | 0.07496 | 0.00030 | 0.00179 | 93 | 0.00269 | 1.02913 | 143.226 | 0.30516 | 0.77376 |
| RP-S11, MRPS11, rpsK; small subunit ribosomal protein S11 | 0.07610 | 0.00135 | 0.07773 | 0.00027 | 0.00203 | 65 | 0.07496 | 0.00030 | 0.00181 | 93 | 0.00272 | 1.02015 | 143.342 | 0.30938 | 0.77376 |
| RP-L32, MRPL32, rpmF; large subunit ribosomal protein L32 | 0.07606 | 0.00136 | 0.07786 | 0.00026 | 0.00202 | 65 | 0.07480 | 0.00031 | 0.00182 | 93 | 0.00272 | 1.12632 | 144.148 | 0.26190 | 0.77376 |
| RP-S15, MRPS15, rpsO; small subunit ribosomal protein S15 | 0.07606 | 0.00136 | 0.07747 | 0.00027 | 0.00203 | 65 | 0.07507 | 0.00031 | 0.00181 | 93 | 0.00272 | 0.88064 | 143.392 | 0.37999 | 0.79852 |
| RP-L16, MRPL16, rplP; large subunit ribosomal protein L16 | 0.07604 | 0.00134 | 0.07772 | 0.00026 | 0.00201 | 65 | 0.07486 | 0.00030 | 0.00179 | 93 | 0.00269 | 1.06299 | 143.088 | 0.28958 | 0.77376 |
| tilS, mesJ; tRNA(Ile)-lysidine synthase [EC:6.3.4.19] | 0.07599 | 0.00134 | 0.07786 | 0.00026 | 0.00199 | 65 | 0.07468 | 0.00030 | 0.00180 | 93 | 0.00268 | 1.18603 | 144.726 | 0.23756 | 0.77376 |
| copA, ATP7; Cu+-exporting ATPase [EC:3.6.3.54] | 0.07597 | 0.00129 | 0.07729 | 0.00019 | 0.00169 | 65 | 0.07504 | 0.00031 | 0.00184 | 93 | 0.00250 | 0.90382 | 154.496 | 0.36750 | 0.79587 |
| holB; DNA polymerase III subunit delta’ [EC:2.7.7.7] | 0.07594 | 0.00135 | 0.07775 | 0.00027 | 0.00202 | 65 | 0.07467 | 0.00030 | 0.00179 | 93 | 0.00270 | 1.13800 | 142.671 | 0.25703 | 0.77376 |
| rpoC; DNA-directed RNA polymerase subunit beta’ [EC:2.7.7.6] | 0.07588 | 0.00136 | 0.07789 | 0.00026 | 0.00200 | 65 | 0.07448 | 0.00031 | 0.00184 | 93 | 0.00271 | 1.25592 | 145.772 | 0.21115 | 0.77376 |
| yggS, PROSC; PLP dependent protein | 0.07588 | 0.00139 | 0.07665 | 0.00031 | 0.00219 | 65 | 0.07534 | 0.00031 | 0.00181 | 93 | 0.00284 | 0.45965 | 137.135 | 0.64650 | 0.90318 |
| nusB; N utilization substance protein B | 0.07586 | 0.00137 | 0.07715 | 0.00028 | 0.00208 | 65 | 0.07495 | 0.00031 | 0.00182 | 93 | 0.00277 | 0.79627 | 141.522 | 0.42721 | 0.82661 |
| FBA, fbaA; fructose-bisphosphate aldolase, class II [EC:4.1.2.13] | 0.07585 | 0.00203 | 0.07718 | 0.00084 | 0.00359 | 65 | 0.07492 | 0.00053 | 0.00238 | 93 | 0.00431 | 0.52435 | 116.719 | 0.60103 | 0.88272 |
| yajC; preprotein translocase subunit YajC | 0.07584 | 0.00139 | 0.07808 | 0.00029 | 0.00212 | 65 | 0.07428 | 0.00031 | 0.00184 | 93 | 0.00281 | 1.35348 | 140.731 | 0.17807 | 0.77376 |
| K07088; uncharacterized protein | 0.07578 | 0.00278 | 0.08186 | 0.00120 | 0.00429 | 65 | 0.07154 | 0.00121 | 0.00360 | 93 | 0.00560 | 1.84196 | 138.201 | 0.06762 | 0.73437 |
| RP-L9, MRPL9, rplI; large subunit ribosomal protein L9 | 0.07563 | 0.00134 | 0.07725 | 0.00026 | 0.00201 | 65 | 0.07450 | 0.00030 | 0.00178 | 93 | 0.00268 | 1.02777 | 142.907 | 0.30579 | 0.77376 |
| ATPF1A, atpA; F-type H+-transporting ATPase subunit alpha [EC:3.6.3.14] | 0.07562 | 0.00136 | 0.07701 | 0.00026 | 0.00199 | 65 | 0.07465 | 0.00031 | 0.00183 | 93 | 0.00271 | 0.87083 | 145.639 | 0.38528 | 0.80258 |
| ATPF0C, atpE; F-type H+-transporting ATPase subunit c | 0.07559 | 0.00135 | 0.07696 | 0.00026 | 0.00200 | 65 | 0.07463 | 0.00031 | 0.00183 | 93 | 0.00271 | 0.86146 | 145.445 | 0.39040 | 0.80384 |
| ATPF0A, atpB; F-type H+-transporting ATPase subunit a | 0.07558 | 0.00135 | 0.07699 | 0.00026 | 0.00199 | 65 | 0.07460 | 0.00031 | 0.00183 | 93 | 0.00271 | 0.88153 | 145.400 | 0.37949 | 0.79852 |
| ATPF1G, atpG; F-type H+-transporting ATPase subunit gamma | 0.07556 | 0.00135 | 0.07695 | 0.00026 | 0.00200 | 65 | 0.07459 | 0.00031 | 0.00183 | 93 | 0.00271 | 0.86864 | 145.283 | 0.38647 | 0.80262 |
| ATPF1E, atpC; F-type H+-transporting ATPase subunit epsilon | 0.07554 | 0.00135 | 0.07695 | 0.00026 | 0.00200 | 65 | 0.07456 | 0.00031 | 0.00183 | 93 | 0.00271 | 0.88127 | 145.276 | 0.37962 | 0.79852 |
| tsaE; tRNA threonylcarbamoyladenosine biosynthesis protein TsaE | 0.07547 | 0.00142 | 0.07783 | 0.00027 | 0.00206 | 65 | 0.07382 | 0.00035 | 0.00193 | 93 | 0.00282 | 1.42426 | 146.931 | 0.15649 | 0.77376 |
| E3.5.1.1, ansA, ansB; L-asparaginase [EC:3.5.1.1] | 0.07543 | 0.00164 | 0.07721 | 0.00034 | 0.00230 | 65 | 0.07418 | 0.00048 | 0.00228 | 93 | 0.00324 | 0.93651 | 150.357 | 0.35051 | 0.79120 |
| ABCC-BAC; ATP-binding cassette, subfamily C, bacterial | 0.07539 | 0.00371 | 0.07649 | 0.00179 | 0.00525 | 65 | 0.07462 | 0.00246 | 0.00514 | 93 | 0.00735 | 0.25540 | 149.698 | 0.79877 | 0.94822 |
| pstS; phosphate transport system substrate-binding protein | 0.07531 | 0.00177 | 0.07301 | 0.00061 | 0.00307 | 65 | 0.07691 | 0.00041 | 0.00211 | 93 | 0.00373 | -1.04717 | 119.989 | 0.29713 | 0.77376 |
| NTH; endonuclease III [EC:4.2.99.18] | 0.07526 | 0.00140 | 0.07631 | 0.00030 | 0.00216 | 65 | 0.07452 | 0.00032 | 0.00185 | 93 | 0.00284 | 0.63327 | 139.876 | 0.52759 | 0.85909 |
| rtpR; ribonucleoside-triphosphate reductase (thioredoxin) [EC:1.17.4.2] | 0.07521 | 0.00335 | 0.07838 | 0.00173 | 0.00516 | 65 | 0.07299 | 0.00181 | 0.00441 | 93 | 0.00679 | 0.79271 | 139.667 | 0.42929 | 0.82749 |
| pncB, NAPRT1; nicotinate phosphoribosyltransferase [EC:6.3.4.21] | 0.07516 | 0.00144 | 0.07664 | 0.00036 | 0.00236 | 65 | 0.07412 | 0.00031 | 0.00182 | 93 | 0.00298 | 0.84756 | 130.521 | 0.39823 | 0.80598 |
| radC; DNA repair protein RadC | 0.07514 | 0.00123 | 0.07686 | 0.00016 | 0.00159 | 65 | 0.07393 | 0.00029 | 0.00176 | 93 | 0.00237 | 1.23411 | 154.988 | 0.21903 | 0.77376 |
| znuA; zinc transport system substrate-binding protein | 0.07512 | 0.00267 | 0.08066 | 0.00106 | 0.00404 | 65 | 0.07125 | 0.00115 | 0.00351 | 93 | 0.00535 | 1.75945 | 141.229 | 0.08067 | 0.75920 |
| pfkA, PFK; 6-phosphofructokinase 1 [EC:2.7.1.11] | 0.07500 | 0.00305 | 0.07795 | 0.00161 | 0.00497 | 65 | 0.07294 | 0.00137 | 0.00384 | 93 | 0.00628 | 0.79689 | 130.746 | 0.42696 | 0.82661 |
| purF, PPAT; amidophosphoribosyltransferase [EC:2.4.2.14] | 0.07498 | 0.00139 | 0.07574 | 0.00030 | 0.00215 | 65 | 0.07444 | 0.00031 | 0.00184 | 93 | 0.00283 | 0.46043 | 139.907 | 0.64592 | 0.90314 |
| E4.3.1.19, ilvA, tdcB; threonine dehydratase [EC:4.3.1.19] | 0.07497 | 0.00181 | 0.07236 | 0.00048 | 0.00271 | 65 | 0.07680 | 0.00054 | 0.00242 | 93 | 0.00363 | -1.22523 | 143.391 | 0.22250 | 0.77376 |
| glk; glucokinase [EC:2.7.1.2] | 0.07497 | 0.00166 | 0.07456 | 0.00046 | 0.00267 | 65 | 0.07526 | 0.00042 | 0.00213 | 93 | 0.00342 | -0.20459 | 133.667 | 0.83820 | 0.95506 |
| queA; S-adenosylmethionine:tRNA ribosyltransferase-isomerase [EC:2.4.99.17] | 0.07491 | 0.00211 | 0.07438 | 0.00068 | 0.00324 | 65 | 0.07528 | 0.00072 | 0.00278 | 93 | 0.00427 | -0.21098 | 140.114 | 0.83321 | 0.95506 |
| purC; phosphoribosylaminoimidazole-succinocarboxamide synthase [EC:6.3.2.6] | 0.07485 | 0.00139 | 0.07653 | 0.00028 | 0.00207 | 65 | 0.07367 | 0.00032 | 0.00187 | 93 | 0.00279 | 1.02331 | 144.117 | 0.30788 | 0.77376 |
| recJ; single-stranded-DNA-specific exonuclease [EC:3.1.-.-] | 0.07484 | 0.00184 | 0.07844 | 0.00063 | 0.00312 | 65 | 0.07232 | 0.00046 | 0.00222 | 93 | 0.00383 | 1.59832 | 123.173 | 0.11253 | 0.77376 |
| rimP; ribosome maturation factor RimP | 0.07481 | 0.00135 | 0.07645 | 0.00027 | 0.00203 | 65 | 0.07367 | 0.00030 | 0.00179 | 93 | 0.00271 | 1.02837 | 142.620 | 0.30552 | 0.77376 |
| secG; preprotein translocase subunit SecG | 0.07475 | 0.00157 | 0.07635 | 0.00035 | 0.00233 | 65 | 0.07363 | 0.00042 | 0.00212 | 93 | 0.00315 | 0.86215 | 144.817 | 0.39003 | 0.80359 |
| xseB; exodeoxyribonuclease VII small subunit [EC:3.1.11.6] | 0.07475 | 0.00134 | 0.07649 | 0.00027 | 0.00203 | 65 | 0.07353 | 0.00030 | 0.00178 | 93 | 0.00270 | 1.09252 | 142.112 | 0.27645 | 0.77376 |
| RP-L30, MRPL30, rpmD; large subunit ribosomal protein L30 | 0.07464 | 0.00136 | 0.07664 | 0.00027 | 0.00204 | 65 | 0.07324 | 0.00031 | 0.00181 | 93 | 0.00273 | 1.24677 | 143.071 | 0.21452 | 0.77376 |
| rnpA; ribonuclease P protein component [EC:3.1.26.5] | 0.07458 | 0.00148 | 0.07690 | 0.00028 | 0.00209 | 65 | 0.07296 | 0.00039 | 0.00204 | 93 | 0.00292 | 1.34594 | 149.513 | 0.18036 | 0.77376 |
| PTS-Man-EIIC, manY; PTS system, mannose-specific IIC component | 0.07455 | 0.00465 | 0.07610 | 0.00300 | 0.00679 | 65 | 0.07347 | 0.00375 | 0.00635 | 93 | 0.00930 | 0.28260 | 146.783 | 0.77788 | 0.94269 |
| upp, UPRT; uracil phosphoribosyltransferase [EC:2.4.2.9] | 0.07449 | 0.00139 | 0.07642 | 0.00028 | 0.00206 | 65 | 0.07314 | 0.00032 | 0.00186 | 93 | 0.00277 | 1.18022 | 144.240 | 0.23985 | 0.77376 |
| asd; aspartate-semialdehyde dehydrogenase [EC:1.2.1.11] | 0.07439 | 0.00141 | 0.07424 | 0.00037 | 0.00237 | 65 | 0.07450 | 0.00029 | 0.00175 | 93 | 0.00295 | -0.08884 | 126.634 | 0.92935 | 0.98183 |
| pgsA, PGS1; CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase [EC:2.7.8.5] | 0.07432 | 0.00162 | 0.07839 | 0.00049 | 0.00274 | 65 | 0.07148 | 0.00035 | 0.00194 | 93 | 0.00336 | 2.05732 | 122.436 | 0.04178 | 0.64661 |
| gidA, mnmG, MTO1; tRNA uridine 5-carboxymethylaminomethyl modification enzyme | 0.07425 | 0.00177 | 0.07806 | 0.00061 | 0.00307 | 65 | 0.07159 | 0.00041 | 0.00209 | 93 | 0.00371 | 1.74278 | 119.324 | 0.08395 | 0.76567 |
| cysE; serine O-acetyltransferase [EC:2.3.1.30] | 0.07421 | 0.00169 | 0.07315 | 0.00058 | 0.00298 | 65 | 0.07495 | 0.00037 | 0.00200 | 93 | 0.00359 | -0.50335 | 117.680 | 0.61566 | 0.88878 |
| dnaA; chromosomal replication initiator protein | 0.07416 | 0.00133 | 0.07385 | 0.00034 | 0.00229 | 65 | 0.07437 | 0.00024 | 0.00161 | 93 | 0.00280 | -0.18448 | 122.510 | 0.85394 | 0.95848 |
| MTHFS; 5-formyltetrahydrofolate cyclo-ligase [EC:6.3.3.2] | 0.07413 | 0.00159 | 0.07721 | 0.00037 | 0.00239 | 65 | 0.07198 | 0.00042 | 0.00212 | 93 | 0.00319 | 1.63790 | 142.663 | 0.10365 | 0.77376 |
| corA; magnesium transporter | 0.07398 | 0.00140 | 0.07265 | 0.00031 | 0.00220 | 65 | 0.07491 | 0.00031 | 0.00182 | 93 | 0.00286 | -0.79188 | 137.136 | 0.42980 | 0.82749 |
| murI; glutamate racemase [EC:5.1.1.3] | 0.07391 | 0.00167 | 0.07541 | 0.00046 | 0.00266 | 65 | 0.07286 | 0.00043 | 0.00214 | 93 | 0.00342 | 0.74594 | 134.348 | 0.45701 | 0.83438 |
| ATPF1D, atpH; F-type H+-transporting ATPase subunit delta | 0.07387 | 0.00136 | 0.07552 | 0.00027 | 0.00205 | 65 | 0.07272 | 0.00031 | 0.00182 | 93 | 0.00274 | 1.02053 | 142.965 | 0.30920 | 0.77376 |
| K07058; membrane protein | 0.07384 | 0.00147 | 0.07290 | 0.00037 | 0.00240 | 65 | 0.07451 | 0.00032 | 0.00185 | 93 | 0.00303 | -0.53066 | 130.640 | 0.59656 | 0.88059 |
| tgt, QTRT1; queuine tRNA-ribosyltransferase [EC:2.4.2.29] | 0.07369 | 0.00143 | 0.07431 | 0.00031 | 0.00218 | 65 | 0.07326 | 0.00033 | 0.00190 | 93 | 0.00289 | 0.36503 | 141.311 | 0.71563 | 0.92004 |
| birA; BirA family transcriptional regulator, biotin operon repressor / biotin—[acetyl-CoA-carboxylase] ligase [EC:6.3.4.15] | 0.07364 | 0.00142 | 0.07523 | 0.00030 | 0.00216 | 65 | 0.07253 | 0.00033 | 0.00188 | 93 | 0.00286 | 0.94733 | 141.248 | 0.34509 | 0.78739 |
| deaD, cshA; ATP-dependent RNA helicase DeaD [EC:3.6.4.13] | 0.07364 | 0.00181 | 0.07453 | 0.00056 | 0.00292 | 65 | 0.07302 | 0.00050 | 0.00231 | 93 | 0.00373 | 0.40649 | 132.745 | 0.68504 | 0.91533 |
| PPIB, ppiB; peptidyl-prolyl cis-trans isomerase B (cyclophilin B) [EC:5.2.1.8] | 0.07360 | 0.00165 | 0.07563 | 0.00038 | 0.00241 | 65 | 0.07218 | 0.00046 | 0.00223 | 93 | 0.00328 | 1.05151 | 145.938 | 0.29476 | 0.77376 |
| ARSC1, arsC; arsenate reductase [EC:1.20.4.1] | 0.07348 | 0.00146 | 0.07554 | 0.00021 | 0.00180 | 65 | 0.07204 | 0.00042 | 0.00213 | 93 | 0.00279 | 1.25803 | 155.985 | 0.21026 | 0.77376 |
| purE; 5-(carboxyamino)imidazole ribonucleotide mutase [EC:5.4.99.18] | 0.07341 | 0.00138 | 0.07530 | 0.00027 | 0.00203 | 65 | 0.07209 | 0.00032 | 0.00186 | 93 | 0.00275 | 1.16799 | 145.563 | 0.24472 | 0.77376 |
| rlmN; 23S rRNA (adenine2503-C2)-methyltransferase [EC:2.1.1.192] | 0.07337 | 0.00152 | 0.07406 | 0.00038 | 0.00240 | 65 | 0.07289 | 0.00036 | 0.00197 | 93 | 0.00311 | 0.37742 | 136.012 | 0.70645 | 0.92004 |
| aroA; 3-phosphoshikimate 1-carboxyvinyltransferase [EC:2.5.1.19] | 0.07336 | 0.00136 | 0.07375 | 0.00033 | 0.00226 | 65 | 0.07309 | 0.00027 | 0.00170 | 93 | 0.00283 | 0.23308 | 128.270 | 0.81607 | 0.95487 |
| mscL; large conductance mechanosensitive channel | 0.07321 | 0.00130 | 0.07512 | 0.00027 | 0.00205 | 65 | 0.07188 | 0.00026 | 0.00168 | 93 | 0.00265 | 1.22200 | 136.179 | 0.22382 | 0.77376 |
| dtd, DTD1; D-tyrosyl-tRNA(Tyr) deacylase [EC:3.1.-.-] | 0.07315 | 0.00150 | 0.07581 | 0.00031 | 0.00217 | 65 | 0.07129 | 0.00038 | 0.00202 | 93 | 0.00297 | 1.52419 | 146.381 | 0.12962 | 0.77376 |
| K07010; putative glutamine amidotransferase | 0.07306 | 0.00276 | 0.07907 | 0.00107 | 0.00406 | 65 | 0.06886 | 0.00127 | 0.00369 | 93 | 0.00549 | 1.86197 | 144.631 | 0.06464 | 0.73334 |
| RP-L34, MRPL34, rpmH; large subunit ribosomal protein L34 | 0.07302 | 0.00160 | 0.07627 | 0.00031 | 0.00219 | 65 | 0.07075 | 0.00046 | 0.00222 | 93 | 0.00312 | 1.76903 | 151.564 | 0.07890 | 0.75800 |
| rsmE; 16S rRNA (uracil1498-N3)-methyltransferase [EC:2.1.1.193] | 0.07299 | 0.00137 | 0.07440 | 0.00028 | 0.00207 | 65 | 0.07200 | 0.00031 | 0.00183 | 93 | 0.00276 | 0.86839 | 142.394 | 0.38664 | 0.80262 |
| recO; DNA repair protein RecO (recombination protein O) | 0.07285 | 0.00160 | 0.07604 | 0.00050 | 0.00277 | 65 | 0.07062 | 0.00033 | 0.00188 | 93 | 0.00335 | 1.61857 | 119.234 | 0.10818 | 0.77376 |
| pncC; nicotinamide-nucleotide amidase [EC:3.5.1.42] | 0.07282 | 0.00182 | 0.07460 | 0.00053 | 0.00287 | 65 | 0.07158 | 0.00052 | 0.00236 | 93 | 0.00371 | 0.81407 | 136.571 | 0.41702 | 0.81866 |
| aroC; chorismate synthase [EC:4.2.3.5] | 0.07268 | 0.00146 | 0.07328 | 0.00036 | 0.00235 | 65 | 0.07226 | 0.00032 | 0.00186 | 93 | 0.00300 | 0.34046 | 133.274 | 0.73405 | 0.92701 |
| HINT1, hinT, hit; histidine triad (HIT) family protein | 0.07266 | 0.00162 | 0.07405 | 0.00041 | 0.00251 | 65 | 0.07169 | 0.00042 | 0.00212 | 93 | 0.00328 | 0.71843 | 138.964 | 0.47370 | 0.84050 |
| pyrB, PYR2; aspartate carbamoyltransferase catalytic subunit [EC:2.1.3.2] | 0.07264 | 0.00139 | 0.07398 | 0.00029 | 0.00213 | 65 | 0.07171 | 0.00032 | 0.00184 | 93 | 0.00282 | 0.80429 | 140.756 | 0.42258 | 0.82322 |
| recF; DNA replication and repair protein RecF | 0.07261 | 0.00149 | 0.07463 | 0.00031 | 0.00217 | 65 | 0.07120 | 0.00038 | 0.00201 | 93 | 0.00296 | 1.15611 | 146.067 | 0.24952 | 0.77376 |
| DHFR, folA; dihydrofolate reductase [EC:1.5.1.3] | 0.07260 | 0.00141 | 0.07429 | 0.00032 | 0.00222 | 65 | 0.07142 | 0.00031 | 0.00183 | 93 | 0.00288 | 0.99702 | 136.560 | 0.32052 | 0.77376 |
| purM; phosphoribosylformylglycinamidine cyclo-ligase [EC:6.3.3.1] | 0.07260 | 0.00144 | 0.07409 | 0.00032 | 0.00223 | 65 | 0.07155 | 0.00033 | 0.00189 | 93 | 0.00292 | 0.86996 | 139.196 | 0.38582 | 0.80258 |
| UNG, UDG; uracil-DNA glycosylase [EC:3.2.2.27] | 0.07250 | 0.00166 | 0.07494 | 0.00036 | 0.00235 | 65 | 0.07079 | 0.00049 | 0.00229 | 93 | 0.00328 | 1.26497 | 149.458 | 0.20785 | 0.77376 |
| bcrC; undecaprenyl-diphosphatase [EC:3.6.1.27] | 0.07246 | 0.00170 | 0.06854 | 0.00044 | 0.00261 | 65 | 0.07520 | 0.00046 | 0.00221 | 93 | 0.00342 | -1.94431 | 139.157 | 0.05388 | 0.70048 |
| exbD; biopolymer transport protein ExbD | 0.07237 | 0.00421 | 0.07324 | 0.00297 | 0.00676 | 65 | 0.07176 | 0.00272 | 0.00541 | 93 | 0.00865 | 0.17068 | 134.026 | 0.86474 | 0.96263 |
| APRT, apt; adenine phosphoribosyltransferase [EC:2.4.2.7] | 0.07231 | 0.00184 | 0.07617 | 0.00064 | 0.00313 | 65 | 0.06962 | 0.00045 | 0.00219 | 93 | 0.00382 | 1.71176 | 121.699 | 0.08949 | 0.76567 |
| pnp, PNPT1; polyribonucleotide nucleotidyltransferase [EC:2.7.7.8] | 0.07230 | 0.00158 | 0.07305 | 0.00042 | 0.00255 | 65 | 0.07177 | 0.00038 | 0.00202 | 93 | 0.00325 | 0.39129 | 132.899 | 0.69621 | 0.91772 |
| dapE; succinyl-diaminopimelate desuccinylase [EC:3.5.1.18] | 0.07224 | 0.00294 | 0.07713 | 0.00194 | 0.00546 | 65 | 0.06883 | 0.00096 | 0.00321 | 93 | 0.00634 | 1.31008 | 106.825 | 0.19298 | 0.77376 |
| amiABC; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | 0.07220 | 0.00260 | 0.07184 | 0.00114 | 0.00418 | 65 | 0.07245 | 0.00103 | 0.00333 | 93 | 0.00535 | -0.11508 | 133.572 | 0.90856 | 0.97664 |
| rsmB, sun; 16S rRNA (cytosine967-C5)-methyltransferase [EC:2.1.1.176] | 0.07216 | 0.00118 | 0.07411 | 0.00019 | 0.00173 | 65 | 0.07079 | 0.00023 | 0.00159 | 93 | 0.00235 | 1.41372 | 145.361 | 0.15958 | 0.77376 |
| K07082; UPF0755 protein | 0.07209 | 0.00140 | 0.07326 | 0.00029 | 0.00210 | 65 | 0.07127 | 0.00033 | 0.00189 | 93 | 0.00282 | 0.70397 | 143.913 | 0.48259 | 0.84354 |
| E3.2.1.86B, bglA; 6-phospho-beta-glucosidase [EC:3.2.1.86] | 0.07198 | 0.00429 | 0.07547 | 0.00288 | 0.00665 | 65 | 0.06954 | 0.00294 | 0.00562 | 93 | 0.00871 | 0.68094 | 138.755 | 0.49704 | 0.84797 |
| ABCF3; ATP-binding cassette, subfamily F, member 3 | 0.07188 | 0.00147 | 0.07282 | 0.00033 | 0.00225 | 65 | 0.07122 | 0.00036 | 0.00196 | 93 | 0.00298 | 0.53608 | 141.243 | 0.59274 | 0.87944 |
| ybeB; ribosome-associated protein | 0.07180 | 0.00142 | 0.07138 | 0.00037 | 0.00238 | 65 | 0.07210 | 0.00028 | 0.00175 | 93 | 0.00295 | -0.24423 | 125.961 | 0.80745 | 0.94987 |
| nadD; nicotinate-nucleotide adenylyltransferase [EC:2.7.7.18] | 0.07166 | 0.00152 | 0.07277 | 0.00032 | 0.00222 | 65 | 0.07088 | 0.00040 | 0.00208 | 93 | 0.00304 | 0.62348 | 147.103 | 0.53393 | 0.86207 |
| ogt, MGMT; methylated-DNA-[protein]-cysteine S-methyltransferase [EC:2.1.1.63] | 0.07155 | 0.00149 | 0.07227 | 0.00034 | 0.00228 | 65 | 0.07105 | 0.00036 | 0.00198 | 93 | 0.00301 | 0.40581 | 141.177 | 0.68549 | 0.91564 |
| ackA; acetate kinase [EC:2.7.2.1] | 0.07155 | 0.00231 | 0.07465 | 0.00089 | 0.00370 | 65 | 0.06938 | 0.00081 | 0.00295 | 93 | 0.00474 | 1.11360 | 133.717 | 0.26745 | 0.77376 |
| recX; regulatory protein | 0.07149 | 0.00140 | 0.07369 | 0.00028 | 0.00206 | 65 | 0.06995 | 0.00033 | 0.00189 | 93 | 0.00280 | 1.33644 | 145.271 | 0.18349 | 0.77376 |
| bacA; undecaprenyl-diphosphatase [EC:3.6.1.27] | 0.07144 | 0.00155 | 0.07146 | 0.00039 | 0.00244 | 65 | 0.07142 | 0.00038 | 0.00203 | 93 | 0.00317 | 0.01400 | 137.182 | 0.98885 | 0.99801 |
| rlmB; 23S rRNA (guanosine2251-2’-O)-methyltransferase [EC:2.1.1.185] | 0.07138 | 0.00113 | 0.07357 | 0.00019 | 0.00169 | 65 | 0.06986 | 0.00021 | 0.00151 | 93 | 0.00227 | 1.63657 | 143.537 | 0.10391 | 0.77376 |
| livK; branched-chain amino acid transport system substrate-binding protein | 0.07126 | 0.00420 | 0.06336 | 0.00149 | 0.00479 | 65 | 0.07678 | 0.00363 | 0.00625 | 93 | 0.00788 | -1.70372 | 154.991 | 0.09044 | 0.76567 |
| rodA, mrdB; rod shape determining protein RodA | 0.07120 | 0.00146 | 0.07397 | 0.00027 | 0.00206 | 65 | 0.06925 | 0.00037 | 0.00200 | 93 | 0.00287 | 1.64341 | 149.437 | 0.10240 | 0.77376 |
| rlmH; 23S rRNA (pseudouridine1915-N3)-methyltransferase [EC:2.1.1.177] | 0.07116 | 0.00132 | 0.07257 | 0.00027 | 0.00204 | 65 | 0.07017 | 0.00028 | 0.00174 | 93 | 0.00268 | 0.89589 | 139.522 | 0.37185 | 0.79738 |
| pdtaS; two-component system, sensor histidine kinase PdtaS [EC:2.7.13.3] | 0.07111 | 0.00472 | 0.07892 | 0.00582 | 0.00946 | 65 | 0.06565 | 0.00189 | 0.00451 | 93 | 0.01048 | 1.26708 | 92.993 | 0.20829 | 0.77376 |
| phoH, phoL; phosphate starvation-inducible protein PhoH and related proteins | 0.07111 | 0.00146 | 0.07076 | 0.00039 | 0.00244 | 65 | 0.07135 | 0.00031 | 0.00182 | 93 | 0.00305 | -0.19617 | 127.525 | 0.84479 | 0.95575 |
| gpx; glutathione peroxidase [EC:1.11.1.9] | 0.07111 | 0.00231 | 0.07089 | 0.00083 | 0.00357 | 65 | 0.07126 | 0.00086 | 0.00304 | 93 | 0.00469 | -0.07827 | 139.498 | 0.93772 | 0.98459 |
| purN; phosphoribosylglycinamide formyltransferase 1 [EC:2.1.2.2] | 0.07097 | 0.00154 | 0.07510 | 0.00043 | 0.00259 | 65 | 0.06808 | 0.00032 | 0.00185 | 93 | 0.00318 | 2.20485 | 123.802 | 0.02931 | 0.61334 |
| RP-S1, rpsA; small subunit ribosomal protein S1 | 0.07090 | 0.00152 | 0.07089 | 0.00047 | 0.00268 | 65 | 0.07091 | 0.00029 | 0.00178 | 93 | 0.00322 | -0.00788 | 117.086 | 0.99372 | 0.99919 |
| mreB; rod shape-determining protein MreB and related proteins | 0.07088 | 0.00302 | 0.07632 | 0.00169 | 0.00509 | 65 | 0.06708 | 0.00125 | 0.00367 | 93 | 0.00627 | 1.47171 | 124.291 | 0.14363 | 0.77376 |
| thrC; threonine synthase [EC:4.2.3.1] | 0.07080 | 0.00153 | 0.07019 | 0.00042 | 0.00255 | 65 | 0.07122 | 0.00034 | 0.00191 | 93 | 0.00318 | -0.32255 | 127.919 | 0.74756 | 0.93207 |
| rseP; regulator of sigma E protease [EC:3.4.24.-] | 0.07069 | 0.00142 | 0.07217 | 0.00030 | 0.00215 | 65 | 0.06966 | 0.00033 | 0.00190 | 93 | 0.00287 | 0.87549 | 142.419 | 0.38278 | 0.80064 |
| pyrG, CTPS; CTP synthase [EC:6.3.4.2] | 0.07064 | 0.00130 | 0.06867 | 0.00035 | 0.00233 | 65 | 0.07201 | 0.00021 | 0.00148 | 93 | 0.00276 | -1.20988 | 113.563 | 0.22884 | 0.77376 |
| dut, DUT; dUTP pyrophosphatase [EC:3.6.1.23] | 0.07053 | 0.00130 | 0.07293 | 0.00026 | 0.00198 | 65 | 0.06885 | 0.00027 | 0.00170 | 93 | 0.00261 | 1.56353 | 139.890 | 0.12019 | 0.77376 |
| dinB; DNA polymerase IV [EC:2.7.7.7] | 0.07049 | 0.00140 | 0.06995 | 0.00032 | 0.00221 | 65 | 0.07086 | 0.00031 | 0.00181 | 93 | 0.00286 | -0.31829 | 136.276 | 0.75075 | 0.93483 |
| mnmE, trmE, MSS1; tRNA modification GTPase [EC:3.6.-.-] | 0.07045 | 0.00144 | 0.07214 | 0.00032 | 0.00220 | 65 | 0.06927 | 0.00034 | 0.00191 | 93 | 0.00291 | 0.98314 | 140.896 | 0.32722 | 0.77376 |
| K07133; uncharacterized protein | 0.07043 | 0.00554 | 0.07735 | 0.00609 | 0.00968 | 65 | 0.06559 | 0.00398 | 0.00655 | 93 | 0.01169 | 1.00655 | 118.619 | 0.31620 | 0.77376 |
| mutS; DNA mismatch repair protein MutS | 0.07041 | 0.00132 | 0.07214 | 0.00026 | 0.00202 | 65 | 0.06919 | 0.00028 | 0.00174 | 93 | 0.00267 | 1.10584 | 140.595 | 0.27069 | 0.77376 |
| thyA, TYMS; thymidylate synthase [EC:2.1.1.45] | 0.07036 | 0.00140 | 0.07119 | 0.00035 | 0.00232 | 65 | 0.06977 | 0.00028 | 0.00174 | 93 | 0.00290 | 0.48918 | 128.406 | 0.62555 | 0.89228 |
| tsaC, rimN, SUA5; L-threonylcarbamoyladenylate synthase [EC:2.7.7.87] | 0.07034 | 0.00141 | 0.07189 | 0.00029 | 0.00211 | 65 | 0.06926 | 0.00033 | 0.00189 | 93 | 0.00283 | 0.92581 | 143.453 | 0.35610 | 0.79399 |
| PYG, glgP; glycogen phosphorylase [EC:2.4.1.1] | 0.07030 | 0.00272 | 0.07344 | 0.00125 | 0.00439 | 65 | 0.06811 | 0.00112 | 0.00346 | 93 | 0.00559 | 0.95209 | 132.731 | 0.34278 | 0.78567 |
| SOD2; superoxide dismutase, Fe-Mn family [EC:1.15.1.1] | 0.07026 | 0.00207 | 0.06699 | 0.00077 | 0.00345 | 65 | 0.07254 | 0.00060 | 0.00254 | 93 | 0.00428 | -1.29675 | 126.240 | 0.19708 | 0.77376 |
| glgC; glucose-1-phosphate adenylyltransferase [EC:2.7.7.27] | 0.07010 | 0.00356 | 0.07311 | 0.00199 | 0.00553 | 65 | 0.06800 | 0.00202 | 0.00466 | 93 | 0.00723 | 0.70652 | 138.610 | 0.48105 | 0.84354 |
| hsdR; type I restriction enzyme, R subunit [EC:3.1.21.3] | 0.07004 | 0.00290 | 0.07138 | 0.00111 | 0.00414 | 65 | 0.06911 | 0.00149 | 0.00401 | 93 | 0.00576 | 0.39507 | 149.057 | 0.69335 | 0.91677 |
| rnr, vacB; ribonuclease R [EC:3.1.-.-] | 0.06979 | 0.00140 | 0.07155 | 0.00029 | 0.00212 | 65 | 0.06856 | 0.00032 | 0.00186 | 93 | 0.00282 | 1.06264 | 142.149 | 0.28975 | 0.77376 |
| msrAB; peptide methionine sulfoxide reductase msrA/msrB [EC:1.8.4.11 1.8.4.12] | 0.06972 | 0.00372 | 0.07396 | 0.00238 | 0.00605 | 65 | 0.06675 | 0.00206 | 0.00471 | 93 | 0.00767 | 0.94021 | 131.425 | 0.34884 | 0.79120 |
| engB; GTP-binding protein | 0.06970 | 0.00144 | 0.07154 | 0.00030 | 0.00214 | 65 | 0.06841 | 0.00035 | 0.00194 | 93 | 0.00289 | 1.08055 | 144.437 | 0.28170 | 0.77376 |
| prmA; ribosomal protein L11 methyltransferase [EC:2.1.1.-] | 0.06967 | 0.00140 | 0.07149 | 0.00030 | 0.00213 | 65 | 0.06840 | 0.00032 | 0.00185 | 93 | 0.00283 | 1.09285 | 141.074 | 0.27632 | 0.77376 |
| hflX; GTPase | 0.06940 | 0.00126 | 0.07059 | 0.00025 | 0.00195 | 65 | 0.06858 | 0.00026 | 0.00166 | 93 | 0.00256 | 0.78306 | 139.485 | 0.43492 | 0.82874 |
| GLPF; glycerol uptake facilitator protein | 0.06937 | 0.00309 | 0.07281 | 0.00148 | 0.00477 | 65 | 0.06696 | 0.00153 | 0.00405 | 93 | 0.00626 | 0.93430 | 139.238 | 0.35177 | 0.79120 |
| mutL; DNA mismatch repair protein MutL | 0.06936 | 0.00141 | 0.07108 | 0.00030 | 0.00216 | 65 | 0.06816 | 0.00032 | 0.00187 | 93 | 0.00285 | 1.02598 | 141.000 | 0.30666 | 0.77376 |
| E2.6.1.42, ilvE; branched-chain amino acid aminotransferase [EC:2.6.1.42] | 0.06929 | 0.00159 | 0.06750 | 0.00046 | 0.00266 | 65 | 0.07055 | 0.00036 | 0.00196 | 93 | 0.00330 | -0.92407 | 126.502 | 0.35721 | 0.79437 |
| mreC; rod shape-determining protein MreC | 0.06916 | 0.00151 | 0.07161 | 0.00030 | 0.00215 | 65 | 0.06744 | 0.00040 | 0.00206 | 93 | 0.00298 | 1.39943 | 148.504 | 0.16377 | 0.77376 |
| galM, GALM; aldose 1-epimerase [EC:5.1.3.3] | 0.06901 | 0.00224 | 0.06901 | 0.00071 | 0.00331 | 65 | 0.06901 | 0.00085 | 0.00303 | 93 | 0.00449 | -0.00102 | 145.121 | 0.99919 | 0.99957 |
| aroB; 3-dehydroquinate synthase [EC:4.2.3.4] | 0.06896 | 0.00155 | 0.06736 | 0.00047 | 0.00268 | 65 | 0.07008 | 0.00032 | 0.00186 | 93 | 0.00326 | -0.83356 | 121.242 | 0.40617 | 0.81189 |
| TC.APA; basic amino acid/polyamine antiporter, APA family | 0.06896 | 0.00435 | 0.07162 | 0.00440 | 0.00823 | 65 | 0.06710 | 0.00203 | 0.00467 | 93 | 0.00946 | 0.47828 | 104.363 | 0.63345 | 0.89696 |
| UGP2, galU, galF; UTP–glucose-1-phosphate uridylyltransferase [EC:2.7.7.9] | 0.06873 | 0.00135 | 0.06990 | 0.00029 | 0.00211 | 65 | 0.06791 | 0.00029 | 0.00176 | 93 | 0.00275 | 0.72618 | 137.380 | 0.46897 | 0.83862 |
| opuBD; osmoprotectant transport system permease protein | 0.06869 | 0.00260 | 0.07085 | 0.00120 | 0.00430 | 65 | 0.06717 | 0.00099 | 0.00326 | 93 | 0.00540 | 0.68216 | 128.841 | 0.49636 | 0.84797 |
| lysA; diaminopimelate decarboxylase [EC:4.1.1.20] | 0.06868 | 0.00165 | 0.06777 | 0.00045 | 0.00262 | 65 | 0.06932 | 0.00042 | 0.00213 | 93 | 0.00338 | -0.45755 | 135.537 | 0.64801 | 0.90350 |
| gatA, QRSL1; aspartyl-tRNA(Asn)/glutamyl-tRNA(Gln) amidotransferase subunit A [EC:6.3.5.6 6.3.5.7] | 0.06853 | 0.00179 | 0.06608 | 0.00037 | 0.00238 | 65 | 0.07025 | 0.00060 | 0.00255 | 93 | 0.00349 | -1.19253 | 153.958 | 0.23489 | 0.77376 |
| folP; dihydropteroate synthase [EC:2.5.1.15] | 0.06838 | 0.00143 | 0.06775 | 0.00036 | 0.00235 | 65 | 0.06883 | 0.00030 | 0.00180 | 93 | 0.00296 | -0.36320 | 129.931 | 0.71704 | 0.92035 |
| comEA; competence protein ComEA | 0.06800 | 0.00151 | 0.07284 | 0.00028 | 0.00208 | 65 | 0.06461 | 0.00039 | 0.00205 | 93 | 0.00292 | 2.81395 | 150.062 | 0.00555 | 0.54022 |
| tex; protein Tex | 0.06780 | 0.00126 | 0.07025 | 0.00025 | 0.00195 | 65 | 0.06610 | 0.00025 | 0.00165 | 93 | 0.00255 | 1.62564 | 138.602 | 0.10630 | 0.77376 |
| glmM; phosphoglucosamine mutase [EC:5.4.2.10] | 0.06775 | 0.00125 | 0.07043 | 0.00020 | 0.00177 | 65 | 0.06588 | 0.00027 | 0.00170 | 93 | 0.00246 | 1.85261 | 148.495 | 0.06592 | 0.73334 |
| GLO1, gloA; lactoylglutathione lyase [EC:4.4.1.5] | 0.06761 | 0.00187 | 0.07129 | 0.00064 | 0.00313 | 65 | 0.06504 | 0.00048 | 0.00228 | 93 | 0.00387 | 1.61411 | 125.224 | 0.10902 | 0.77376 |
| arsR; ArsR family transcriptional regulator, arsenate/arsenite/antimonite-responsive transcriptional repressor | 0.06745 | 0.00254 | 0.06778 | 0.00095 | 0.00383 | 65 | 0.06722 | 0.00108 | 0.00341 | 93 | 0.00513 | 0.10914 | 143.218 | 0.91324 | 0.97664 |
| ybeY, yqfG; probable rRNA maturation factor | 0.06739 | 0.00138 | 0.07008 | 0.00026 | 0.00200 | 65 | 0.06551 | 0.00033 | 0.00188 | 93 | 0.00274 | 1.66897 | 147.073 | 0.09725 | 0.76673 |
| PTS-HPR; phosphocarrier protein | 0.06735 | 0.00155 | 0.06882 | 0.00038 | 0.00243 | 65 | 0.06633 | 0.00038 | 0.00203 | 93 | 0.00316 | 0.78580 | 137.777 | 0.43333 | 0.82874 |
| fur, zur, furB; Fur family transcriptional regulator, ferric uptake regulator | 0.06714 | 0.00221 | 0.06640 | 0.00072 | 0.00334 | 65 | 0.06766 | 0.00081 | 0.00295 | 93 | 0.00445 | -0.28364 | 142.401 | 0.77710 | 0.94269 |
| trmL, cspR; tRNA (cytidine/uridine-2’-O-)-methyltransferase [EC:2.1.1.207] | 0.06694 | 0.00118 | 0.06989 | 0.00019 | 0.00172 | 65 | 0.06487 | 0.00023 | 0.00158 | 93 | 0.00233 | 2.15420 | 145.442 | 0.03287 | 0.62130 |
| xerD; integrase/recombinase XerD | 0.06694 | 0.00242 | 0.06824 | 0.00140 | 0.00465 | 65 | 0.06603 | 0.00059 | 0.00253 | 93 | 0.00529 | 0.41683 | 101.303 | 0.67768 | 0.91311 |
| tmk, DTYMK; dTMP kinase [EC:2.7.4.9] | 0.06687 | 0.00123 | 0.06888 | 0.00024 | 0.00193 | 65 | 0.06547 | 0.00023 | 0.00158 | 93 | 0.00249 | 1.36451 | 135.779 | 0.17466 | 0.77376 |
| ABC.MS.P1; multiple sugar transport system permease protein | 0.06685 | 0.00404 | 0.06338 | 0.00331 | 0.00713 | 65 | 0.06927 | 0.00208 | 0.00473 | 93 | 0.00856 | -0.68787 | 117.005 | 0.49290 | 0.84797 |
| pdxK, pdxY; pyridoxine kinase [EC:2.7.1.35] | 0.06682 | 0.00197 | 0.06645 | 0.00056 | 0.00293 | 65 | 0.06708 | 0.00066 | 0.00266 | 93 | 0.00396 | -0.15900 | 144.553 | 0.87389 | 0.96576 |
| glmU; bifunctional UDP-N-acetylglucosamine pyrophosphorylase / Glucosamine-1-phosphate N-acetyltransferase [EC:2.7.7.23 2.3.1.157] | 0.06682 | 0.00124 | 0.06891 | 0.00025 | 0.00194 | 65 | 0.06536 | 0.00024 | 0.00160 | 93 | 0.00251 | 1.41087 | 136.305 | 0.16056 | 0.77376 |
| E2.3.1.54, pflD; formate C-acetyltransferase [EC:2.3.1.54] | 0.06656 | 0.00317 | 0.06979 | 0.00150 | 0.00480 | 65 | 0.06430 | 0.00166 | 0.00422 | 93 | 0.00640 | 0.85821 | 142.111 | 0.39222 | 0.80459 |
| recQ; ATP-dependent DNA helicase RecQ [EC:3.6.4.12] | 0.06640 | 0.00226 | 0.06750 | 0.00066 | 0.00319 | 65 | 0.06563 | 0.00091 | 0.00313 | 93 | 0.00447 | 0.41860 | 149.986 | 0.67611 | 0.91266 |
| livM; branched-chain amino acid transport system permease protein | 0.06628 | 0.00380 | 0.05921 | 0.00124 | 0.00437 | 65 | 0.07121 | 0.00296 | 0.00564 | 93 | 0.00714 | -1.68156 | 155.196 | 0.09467 | 0.76609 |
| fabZ; 3-hydroxyacyl-[acyl-carrier-protein] dehydratase [EC:4.2.1.59] | 0.06627 | 0.00154 | 0.06915 | 0.00051 | 0.00279 | 65 | 0.06426 | 0.00027 | 0.00172 | 93 | 0.00328 | 1.49046 | 110.612 | 0.13895 | 0.77376 |
| rpoZ; DNA-directed RNA polymerase subunit omega [EC:2.7.7.6] | 0.06621 | 0.00125 | 0.06922 | 0.00021 | 0.00178 | 65 | 0.06411 | 0.00026 | 0.00168 | 93 | 0.00245 | 2.08539 | 147.614 | 0.03876 | 0.63196 |
| secE; preprotein translocase subunit SecE | 0.06620 | 0.00143 | 0.06944 | 0.00028 | 0.00209 | 65 | 0.06394 | 0.00035 | 0.00193 | 93 | 0.00284 | 1.93513 | 146.000 | 0.05491 | 0.70519 |
| ftsQ; cell division protein FtsQ | 0.06612 | 0.00122 | 0.06835 | 0.00024 | 0.00191 | 65 | 0.06456 | 0.00023 | 0.00157 | 93 | 0.00247 | 1.53360 | 136.767 | 0.12744 | 0.77376 |
| ABC.MS.P; multiple sugar transport system permease protein | 0.06611 | 0.00368 | 0.06050 | 0.00210 | 0.00569 | 65 | 0.07002 | 0.00215 | 0.00481 | 93 | 0.00745 | -1.27748 | 138.828 | 0.20357 | 0.77376 |
| livF; branched-chain amino acid transport system ATP-binding protein | 0.06608 | 0.00380 | 0.05904 | 0.00123 | 0.00435 | 65 | 0.07101 | 0.00299 | 0.00567 | 93 | 0.00714 | -1.67640 | 154.986 | 0.09568 | 0.76609 |
| RP-S21, MRPS21, rpsU; small subunit ribosomal protein S21 | 0.06604 | 0.00147 | 0.06556 | 0.00038 | 0.00240 | 65 | 0.06637 | 0.00032 | 0.00186 | 93 | 0.00304 | -0.26714 | 130.913 | 0.78978 | 0.94606 |
| K08998; uncharacterized protein | 0.06603 | 0.00192 | 0.06756 | 0.00057 | 0.00295 | 65 | 0.06496 | 0.00060 | 0.00254 | 93 | 0.00389 | 0.66843 | 140.479 | 0.50496 | 0.84956 |
| PTS-Fru-EIIC, fruA; PTS system, fructose-specific IIC component | 0.06594 | 0.00239 | 0.07078 | 0.00122 | 0.00433 | 65 | 0.06256 | 0.00066 | 0.00267 | 93 | 0.00509 | 1.61575 | 110.891 | 0.10899 | 0.77376 |
| hisC; histidinol-phosphate aminotransferase [EC:2.6.1.9] | 0.06593 | 0.00185 | 0.06117 | 0.00059 | 0.00300 | 65 | 0.06925 | 0.00049 | 0.00228 | 93 | 0.00377 | -2.14093 | 129.298 | 0.03416 | 0.62130 |
| LDH, ldh; L-lactate dehydrogenase [EC:1.1.1.27] | 0.06589 | 0.00504 | 0.07407 | 0.00621 | 0.00977 | 65 | 0.06018 | 0.00244 | 0.00512 | 93 | 0.01103 | 1.25940 | 98.781 | 0.21085 | 0.77376 |
| mutY; A/G-specific adenine glycosylase [EC:3.2.2.31] | 0.06585 | 0.00161 | 0.06373 | 0.00052 | 0.00283 | 65 | 0.06733 | 0.00033 | 0.00189 | 93 | 0.00340 | -1.05844 | 117.784 | 0.29202 | 0.77376 |
| lemA; LemA protein | 0.06573 | 0.00175 | 0.06761 | 0.00044 | 0.00260 | 65 | 0.06441 | 0.00052 | 0.00236 | 93 | 0.00351 | 0.91144 | 144.461 | 0.36358 | 0.79515 |
| TC.MATE, SLC47A, norM, mdtK, dinF; multidrug resistance protein, MATE family | 0.06560 | 0.00199 | 0.06290 | 0.00070 | 0.00328 | 65 | 0.06749 | 0.00057 | 0.00248 | 93 | 0.00411 | -1.11578 | 128.750 | 0.26659 | 0.77376 |
| DNMT1, dcm; DNA (cytosine-5)-methyltransferase 1 [EC:2.1.1.37] | 0.06554 | 0.00307 | 0.06306 | 0.00103 | 0.00397 | 65 | 0.06727 | 0.00182 | 0.00443 | 93 | 0.00595 | -0.70805 | 155.179 | 0.47998 | 0.84324 |
| pepT; tripeptide aminopeptidase [EC:3.4.11.4] | 0.06552 | 0.00289 | 0.06755 | 0.00148 | 0.00477 | 65 | 0.06411 | 0.00122 | 0.00362 | 93 | 0.00599 | 0.57427 | 129.271 | 0.56679 | 0.87051 |
| ilvD; dihydroxy-acid dehydratase [EC:4.2.1.9] | 0.06546 | 0.00192 | 0.05999 | 0.00054 | 0.00288 | 65 | 0.06928 | 0.00058 | 0.00251 | 93 | 0.00382 | -2.43302 | 141.184 | 0.01622 | 0.59095 |
| rapZ; RNase adapter protein RapZ | 0.06538 | 0.00130 | 0.06884 | 0.00022 | 0.00183 | 65 | 0.06297 | 0.00029 | 0.00176 | 93 | 0.00254 | 2.31477 | 148.566 | 0.02199 | 0.59447 |
| dps; starvation-inducible DNA-binding protein | 0.06520 | 0.00142 | 0.06596 | 0.00030 | 0.00215 | 65 | 0.06466 | 0.00034 | 0.00190 | 93 | 0.00287 | 0.44978 | 142.734 | 0.65355 | 0.90621 |
| smc; chromosome segregation protein | 0.06491 | 0.00131 | 0.06697 | 0.00025 | 0.00196 | 65 | 0.06347 | 0.00028 | 0.00175 | 93 | 0.00263 | 1.33264 | 143.097 | 0.18477 | 0.77376 |
| spoU; RNA methyltransferase, TrmH family | 0.06491 | 0.00211 | 0.06639 | 0.00071 | 0.00331 | 65 | 0.06386 | 0.00070 | 0.00275 | 93 | 0.00430 | 0.58796 | 137.369 | 0.55752 | 0.86813 |
| prfC; peptide chain release factor 3 | 0.06476 | 0.00139 | 0.06464 | 0.00036 | 0.00234 | 65 | 0.06484 | 0.00027 | 0.00170 | 93 | 0.00289 | -0.06840 | 125.485 | 0.94558 | 0.98674 |
| proC; pyrroline-5-carboxylate reductase [EC:1.5.1.2] | 0.06466 | 0.00145 | 0.06426 | 0.00037 | 0.00239 | 65 | 0.06494 | 0.00031 | 0.00183 | 93 | 0.00301 | -0.22524 | 129.764 | 0.82215 | 0.95506 |
| GCH1, folE; GTP cyclohydrolase IA [EC:3.5.4.16] | 0.06441 | 0.00133 | 0.06495 | 0.00031 | 0.00218 | 65 | 0.06404 | 0.00026 | 0.00168 | 93 | 0.00276 | 0.33213 | 130.511 | 0.74032 | 0.92969 |
| E1.1.1.3; homoserine dehydrogenase [EC:1.1.1.3] | 0.06431 | 0.00158 | 0.06331 | 0.00049 | 0.00273 | 65 | 0.06501 | 0.00033 | 0.00190 | 93 | 0.00333 | -0.50913 | 120.982 | 0.61159 | 0.88722 |
| fhs; formate–tetrahydrofolate ligase [EC:6.3.4.3] | 0.06430 | 0.00265 | 0.06669 | 0.00112 | 0.00415 | 65 | 0.06263 | 0.00111 | 0.00345 | 93 | 0.00540 | 0.75237 | 137.243 | 0.45312 | 0.83438 |
| thiD; hydroxymethylpyrimidine/phosphomethylpyrimidine kinase [EC:2.7.1.49 2.7.4.7] | 0.06426 | 0.00164 | 0.06607 | 0.00048 | 0.00273 | 65 | 0.06299 | 0.00038 | 0.00203 | 93 | 0.00340 | 0.90446 | 127.438 | 0.36746 | 0.79587 |
| nrdR; transcriptional repressor NrdR | 0.06409 | 0.00117 | 0.06729 | 0.00020 | 0.00174 | 65 | 0.06186 | 0.00022 | 0.00153 | 93 | 0.00232 | 2.34286 | 142.189 | 0.02052 | 0.59095 |
| scpB; segregation and condensation protein B | 0.06403 | 0.00130 | 0.06618 | 0.00025 | 0.00198 | 65 | 0.06252 | 0.00027 | 0.00171 | 93 | 0.00261 | 1.39719 | 140.892 | 0.16455 | 0.77376 |
| livH; branched-chain amino acid transport system permease protein | 0.06396 | 0.00364 | 0.05690 | 0.00108 | 0.00408 | 65 | 0.06890 | 0.00276 | 0.00545 | 93 | 0.00681 | -1.76298 | 154.336 | 0.07988 | 0.75920 |
| ppx-gppA; exopolyphosphatase / guanosine-5’-triphosphate,3’-diphosphate pyrophosphatase [EC:3.6.1.11 3.6.1.40] | 0.06386 | 0.00258 | 0.06933 | 0.00129 | 0.00446 | 65 | 0.06004 | 0.00086 | 0.00304 | 93 | 0.00540 | 1.71983 | 119.451 | 0.08805 | 0.76567 |
| K09747; uncharacterized protein | 0.06373 | 0.00123 | 0.06609 | 0.00021 | 0.00181 | 65 | 0.06209 | 0.00025 | 0.00164 | 93 | 0.00244 | 1.64015 | 144.346 | 0.10315 | 0.77376 |
| metY; O-acetylhomoserine (thiol)-lyase [EC:2.5.1.49] | 0.06373 | 0.00242 | 0.05967 | 0.00089 | 0.00371 | 65 | 0.06657 | 0.00094 | 0.00318 | 93 | 0.00488 | -1.41184 | 139.944 | 0.16022 | 0.77376 |
| tag; DNA-3-methyladenine glycosylase I [EC:3.2.2.20] | 0.06344 | 0.00147 | 0.06420 | 0.00036 | 0.00235 | 65 | 0.06291 | 0.00033 | 0.00188 | 93 | 0.00301 | 0.42927 | 133.739 | 0.66842 | 0.91165 |
| livG; branched-chain amino acid transport system ATP-binding protein | 0.06331 | 0.00360 | 0.05664 | 0.00107 | 0.00406 | 65 | 0.06797 | 0.00270 | 0.00539 | 93 | 0.00675 | -1.67929 | 154.523 | 0.09512 | 0.76609 |
| scpA; segregation and condensation protein A | 0.06325 | 0.00136 | 0.06545 | 0.00027 | 0.00205 | 65 | 0.06172 | 0.00030 | 0.00180 | 93 | 0.00273 | 1.36355 | 141.987 | 0.17487 | 0.77376 |
| ftsX; cell division transport system permease protein | 0.06309 | 0.00168 | 0.06367 | 0.00044 | 0.00260 | 65 | 0.06268 | 0.00046 | 0.00222 | 93 | 0.00342 | 0.29054 | 139.628 | 0.77183 | 0.94083 |
| RP-L36, MRPL36, rpmJ; large subunit ribosomal protein L36 | 0.06300 | 0.00179 | 0.06652 | 0.00048 | 0.00272 | 65 | 0.06054 | 0.00051 | 0.00235 | 93 | 0.00360 | 1.66186 | 140.544 | 0.09877 | 0.76942 |
| sufB; Fe-S cluster assembly protein SufB | 0.06296 | 0.00232 | 0.06185 | 0.00082 | 0.00355 | 65 | 0.06373 | 0.00088 | 0.00307 | 93 | 0.00469 | -0.40168 | 140.652 | 0.68853 | 0.91567 |
| mutM, fpg; formamidopyrimidine-DNA glycosylase [EC:3.2.2.23 4.2.99.18] | 0.06295 | 0.00185 | 0.06306 | 0.00061 | 0.00306 | 65 | 0.06287 | 0.00049 | 0.00231 | 93 | 0.00384 | 0.04976 | 128.420 | 0.96039 | 0.98903 |
| DLAT, aceF, pdhC; pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12] | 0.06285 | 0.00216 | 0.06092 | 0.00070 | 0.00328 | 65 | 0.06419 | 0.00076 | 0.00287 | 93 | 0.00435 | -0.75191 | 141.764 | 0.45335 | 0.83438 |
| ybaK, ebsC; Cys-tRNA(Pro)/Cys-tRNA(Cys) deacylase [EC:3.1.1.-] | 0.06269 | 0.00150 | 0.06678 | 0.00031 | 0.00220 | 65 | 0.05983 | 0.00037 | 0.00200 | 93 | 0.00297 | 2.34117 | 144.670 | 0.02059 | 0.59095 |
| topB; DNA topoisomerase III [EC:5.99.1.2] | 0.06266 | 0.00369 | 0.06056 | 0.00170 | 0.00511 | 65 | 0.06412 | 0.00249 | 0.00517 | 93 | 0.00727 | -0.49040 | 151.618 | 0.62456 | 0.89208 |
| cynT, can; carbonic anhydrase [EC:4.2.1.1] | 0.06245 | 0.00181 | 0.05902 | 0.00059 | 0.00302 | 65 | 0.06485 | 0.00045 | 0.00221 | 93 | 0.00374 | -1.55563 | 125.536 | 0.12231 | 0.77376 |
| G6PD, zwf; glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49 1.1.1.363] | 0.06238 | 0.00183 | 0.06186 | 0.00060 | 0.00303 | 65 | 0.06274 | 0.00049 | 0.00228 | 93 | 0.00379 | -0.23201 | 128.544 | 0.81690 | 0.95487 |
| ABC.X4.S; putative ABC transport system substrate-binding protein | 0.06237 | 0.00288 | 0.06630 | 0.00145 | 0.00472 | 65 | 0.05963 | 0.00121 | 0.00361 | 93 | 0.00594 | 1.12307 | 129.925 | 0.26348 | 0.77376 |
| iscU, nifU; nitrogen fixation protein NifU and related proteins | 0.06219 | 0.00150 | 0.06567 | 0.00032 | 0.00223 | 65 | 0.05976 | 0.00037 | 0.00198 | 93 | 0.00298 | 1.98156 | 143.062 | 0.04944 | 0.67483 |
| trpF; phosphoribosylanthranilate isomerase [EC:5.3.1.24] | 0.06218 | 0.00223 | 0.05812 | 0.00074 | 0.00338 | 65 | 0.06503 | 0.00080 | 0.00293 | 93 | 0.00448 | -1.54304 | 141.074 | 0.12506 | 0.77376 |
| apbE; FAD:protein FMN transferase [EC:2.7.1.180] | 0.06216 | 0.00187 | 0.06636 | 0.00049 | 0.00274 | 65 | 0.05923 | 0.00058 | 0.00249 | 93 | 0.00370 | 1.92395 | 144.833 | 0.05632 | 0.70577 |
| K07095; uncharacterized protein | 0.06201 | 0.00283 | 0.06460 | 0.00123 | 0.00435 | 65 | 0.06020 | 0.00130 | 0.00374 | 93 | 0.00573 | 0.76765 | 140.294 | 0.44398 | 0.83186 |
| PGAM, gpmA; 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase [EC:5.4.2.11] | 0.06197 | 0.00271 | 0.06608 | 0.00145 | 0.00473 | 65 | 0.05911 | 0.00094 | 0.00318 | 93 | 0.00570 | 1.22327 | 118.340 | 0.22366 | 0.77376 |
| lgt, umpA; phosphatidylglycerol:prolipoprotein diacylglycerol transferase [EC:2.-.-.-] | 0.06192 | 0.00123 | 0.06464 | 0.00022 | 0.00184 | 65 | 0.06002 | 0.00025 | 0.00163 | 93 | 0.00246 | 1.87696 | 142.498 | 0.06257 | 0.73012 |
| patB, malY; cystathione beta-lyase [EC:4.4.1.8] | 0.06188 | 0.00288 | 0.06542 | 0.00149 | 0.00480 | 65 | 0.05940 | 0.00118 | 0.00356 | 93 | 0.00597 | 1.00837 | 127.190 | 0.31519 | 0.77376 |
| E4.2.1.46, rfbB, rffG; dTDP-glucose 4,6-dehydratase [EC:4.2.1.46] | 0.06176 | 0.00185 | 0.06756 | 0.00067 | 0.00320 | 65 | 0.05771 | 0.00042 | 0.00212 | 93 | 0.00384 | 2.56421 | 116.854 | 0.01161 | 0.59095 |
| mtnN, mtn, pfs; adenosylhomocysteine nucleosidase [EC:3.2.2.9] | 0.06160 | 0.00190 | 0.06265 | 0.00059 | 0.00300 | 65 | 0.06086 | 0.00057 | 0.00247 | 93 | 0.00389 | 0.45931 | 136.528 | 0.64674 | 0.90333 |
| pgm; phosphoglucomutase [EC:5.4.2.2] | 0.06156 | 0.00180 | 0.06285 | 0.00051 | 0.00280 | 65 | 0.06066 | 0.00051 | 0.00235 | 93 | 0.00366 | 0.59840 | 138.117 | 0.55055 | 0.86727 |
| msmX, msmK, malK, sugC, ggtA, msiK; multiple sugar transport system ATP-binding protein | 0.06150 | 0.00226 | 0.05737 | 0.00080 | 0.00350 | 65 | 0.06438 | 0.00080 | 0.00293 | 93 | 0.00456 | -1.53627 | 138.070 | 0.12676 | 0.77376 |
| acrA, mexA, adeI, smeD, mtrC, cmeA; membrane fusion protein, multidrug efflux system | 0.06148 | 0.00358 | 0.05775 | 0.00202 | 0.00557 | 65 | 0.06408 | 0.00204 | 0.00468 | 93 | 0.00728 | -0.87021 | 138.309 | 0.38569 | 0.80258 |
| glpK, GK; glycerol kinase [EC:2.7.1.30] | 0.06143 | 0.00106 | 0.06174 | 0.00015 | 0.00150 | 65 | 0.06121 | 0.00020 | 0.00147 | 93 | 0.00210 | 0.25266 | 149.937 | 0.80087 | 0.94855 |
| htpX; heat shock protein HtpX [EC:3.4.24.-] | 0.06142 | 0.00164 | 0.06507 | 0.00041 | 0.00251 | 65 | 0.05887 | 0.00042 | 0.00214 | 93 | 0.00330 | 1.88230 | 139.436 | 0.06188 | 0.73001 |
| fruK; 1-phosphofructokinase [EC:2.7.1.56] | 0.06142 | 0.00205 | 0.06481 | 0.00056 | 0.00294 | 65 | 0.05905 | 0.00073 | 0.00280 | 93 | 0.00406 | 1.41769 | 147.914 | 0.15838 | 0.77376 |
| degP, htrA; serine protease Do [EC:3.4.21.107] | 0.06141 | 0.00162 | 0.05978 | 0.00039 | 0.00243 | 65 | 0.06256 | 0.00044 | 0.00218 | 93 | 0.00327 | -0.85153 | 143.435 | 0.39590 | 0.80459 |
| PTS-EI.PTSI, ptsI; phosphotransferase system, enzyme I, PtsI [EC:2.7.3.9] | 0.06141 | 0.00208 | 0.06550 | 0.00075 | 0.00339 | 65 | 0.05854 | 0.00062 | 0.00259 | 93 | 0.00427 | 1.63172 | 129.684 | 0.10516 | 0.77376 |
| K09861; uncharacterized protein | 0.06140 | 0.00191 | 0.06501 | 0.00057 | 0.00295 | 65 | 0.05888 | 0.00057 | 0.00248 | 93 | 0.00385 | 1.58918 | 138.150 | 0.11430 | 0.77376 |
| leuA, IMS; 2-isopropylmalate synthase [EC:2.3.3.13] | 0.06126 | 0.00167 | 0.05876 | 0.00051 | 0.00279 | 65 | 0.06301 | 0.00040 | 0.00206 | 93 | 0.00347 | -1.22389 | 126.574 | 0.22327 | 0.77376 |
| ftsA; cell division protein FtsA | 0.06126 | 0.00142 | 0.06130 | 0.00036 | 0.00235 | 65 | 0.06123 | 0.00029 | 0.00177 | 93 | 0.00294 | 0.02425 | 128.498 | 0.98069 | 0.99661 |
| udk, UCK; uridine kinase [EC:2.7.1.48] | 0.06104 | 0.00272 | 0.06083 | 0.00118 | 0.00426 | 65 | 0.06118 | 0.00118 | 0.00356 | 93 | 0.00555 | -0.06342 | 137.731 | 0.94952 | 0.98774 |
| pepO; putative endopeptidase [EC:3.4.24.-] | 0.06085 | 0.00306 | 0.05970 | 0.00158 | 0.00494 | 65 | 0.06166 | 0.00142 | 0.00391 | 93 | 0.00629 | -0.31244 | 132.978 | 0.75520 | 0.93665 |
| uup; ABC transport system ATP-binding/permease protein | 0.06082 | 0.00146 | 0.05966 | 0.00040 | 0.00249 | 65 | 0.06163 | 0.00029 | 0.00177 | 93 | 0.00306 | -0.64453 | 123.379 | 0.52043 | 0.85574 |
| PTS-Man-EIID, manZ; PTS system, mannose-specific IID component | 0.06078 | 0.00375 | 0.06322 | 0.00215 | 0.00575 | 65 | 0.05907 | 0.00229 | 0.00496 | 93 | 0.00760 | 0.54668 | 140.439 | 0.58547 | 0.87720 |
| hslO; molecular chaperone Hsp33 | 0.06048 | 0.00127 | 0.06324 | 0.00023 | 0.00189 | 65 | 0.05854 | 0.00026 | 0.00168 | 93 | 0.00253 | 1.85554 | 142.661 | 0.06558 | 0.73334 |
| E2.7.1.4, scrK; fructokinase [EC:2.7.1.4] | 0.06047 | 0.00171 | 0.06024 | 0.00049 | 0.00275 | 65 | 0.06062 | 0.00045 | 0.00219 | 93 | 0.00352 | -0.10859 | 133.699 | 0.91369 | 0.97664 |
| dxs; 1-deoxy-D-xylulose-5-phosphate synthase [EC:2.2.1.7] | 0.06030 | 0.00210 | 0.06290 | 0.00084 | 0.00360 | 65 | 0.05848 | 0.00059 | 0.00252 | 93 | 0.00439 | 1.00488 | 121.888 | 0.31695 | 0.77376 |
| ftsE; cell division transport system ATP-binding protein | 0.06020 | 0.00174 | 0.05899 | 0.00052 | 0.00283 | 65 | 0.06104 | 0.00045 | 0.00220 | 93 | 0.00358 | -0.57420 | 131.487 | 0.56681 | 0.87051 |
| ppnN; pyrimidine/purine-5’-nucleotide nucleosidase [EC:3.2.2.10 3.2.2.-] | 0.06014 | 0.00202 | 0.06107 | 0.00074 | 0.00337 | 65 | 0.05949 | 0.00058 | 0.00250 | 93 | 0.00420 | 0.37636 | 126.842 | 0.70728 | 0.92004 |
| metE; 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase [EC:2.1.1.14] | 0.06013 | 0.00193 | 0.06190 | 0.00063 | 0.00311 | 65 | 0.05889 | 0.00057 | 0.00247 | 93 | 0.00397 | 0.75735 | 133.339 | 0.45018 | 0.83438 |
| lon; ATP-dependent Lon protease [EC:3.4.21.53] | 0.06009 | 0.00258 | 0.06360 | 0.00150 | 0.00481 | 65 | 0.05763 | 0.00074 | 0.00281 | 93 | 0.00557 | 1.07218 | 106.598 | 0.28606 | 0.77376 |
| folK; 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase [EC:2.7.6.3] | 0.05998 | 0.00171 | 0.05780 | 0.00044 | 0.00262 | 65 | 0.06150 | 0.00047 | 0.00225 | 93 | 0.00345 | -1.07046 | 140.286 | 0.28625 | 0.77376 |
| pstA; phosphate transport system permease protein | 0.05990 | 0.00172 | 0.05765 | 0.00050 | 0.00278 | 65 | 0.06146 | 0.00044 | 0.00219 | 93 | 0.00353 | -1.07707 | 132.483 | 0.28341 | 0.77376 |
| mbtN, fadE14; acyl-ACP dehydrogenase [EC:1.3.99.-] | 0.05963 | 0.00536 | 0.05023 | 0.00353 | 0.00737 | 65 | 0.06620 | 0.00520 | 0.00747 | 93 | 0.01050 | -1.52184 | 151.714 | 0.13013 | 0.77376 |
| K07040; uncharacterized protein | 0.05961 | 0.00159 | 0.06051 | 0.00046 | 0.00267 | 65 | 0.05899 | 0.00036 | 0.00195 | 93 | 0.00331 | 0.45876 | 125.735 | 0.64720 | 0.90344 |
| sufS; cysteine desulfurase / selenocysteine lyase [EC:2.8.1.7 4.4.1.16] | 0.05949 | 0.00195 | 0.05863 | 0.00064 | 0.00314 | 65 | 0.06008 | 0.00058 | 0.00251 | 93 | 0.00402 | -0.36000 | 133.814 | 0.71941 | 0.92108 |
| mscS; small conductance mechanosensitive channel | 0.05924 | 0.00165 | 0.06262 | 0.00041 | 0.00250 | 65 | 0.05687 | 0.00044 | 0.00217 | 93 | 0.00331 | 1.73741 | 140.914 | 0.08450 | 0.76567 |
| TC.AAT; amino acid transporter, AAT family | 0.05912 | 0.00465 | 0.06220 | 0.00458 | 0.00840 | 65 | 0.05697 | 0.00263 | 0.00532 | 93 | 0.00994 | 0.52582 | 113.012 | 0.60005 | 0.88240 |
| gatC, GATC; aspartyl-tRNA(Asn)/glutamyl-tRNA(Gln) amidotransferase subunit C [EC:6.3.5.6 6.3.5.7] | 0.05904 | 0.00156 | 0.06028 | 0.00040 | 0.00248 | 65 | 0.05818 | 0.00037 | 0.00201 | 93 | 0.00319 | 0.65655 | 134.908 | 0.51259 | 0.85122 |
| plsY; glycerol-3-phosphate acyltransferase PlsY [EC:2.3.1.15] | 0.05882 | 0.00165 | 0.06118 | 0.00040 | 0.00247 | 65 | 0.05718 | 0.00045 | 0.00220 | 93 | 0.00331 | 1.20861 | 142.855 | 0.22881 | 0.77376 |
| sufC; Fe-S cluster assembly ATP-binding protein | 0.05873 | 0.00200 | 0.05774 | 0.00064 | 0.00314 | 65 | 0.05942 | 0.00064 | 0.00261 | 93 | 0.00409 | -0.41248 | 137.454 | 0.68063 | 0.91355 |
| GGPS; geranylgeranyl diphosphate synthase, type II [EC:2.5.1.1 2.5.1.10 2.5.1.29] | 0.05864 | 0.00188 | 0.06135 | 0.00054 | 0.00289 | 65 | 0.05675 | 0.00057 | 0.00247 | 93 | 0.00380 | 1.20894 | 139.957 | 0.22872 | 0.77376 |
| galK; galactokinase [EC:2.7.1.6] | 0.05850 | 0.00231 | 0.06190 | 0.00081 | 0.00353 | 65 | 0.05611 | 0.00086 | 0.00304 | 93 | 0.00465 | 1.24372 | 140.299 | 0.21568 | 0.77376 |
| gatB, PET112; aspartyl-tRNA(Asn)/glutamyl-tRNA(Gln) amidotransferase subunit B [EC:6.3.5.6 6.3.5.7] | 0.05850 | 0.00153 | 0.05976 | 0.00039 | 0.00246 | 65 | 0.05761 | 0.00036 | 0.00196 | 93 | 0.00315 | 0.68472 | 133.387 | 0.49471 | 0.84797 |
| nagB, GNPDA; glucosamine-6-phosphate deaminase [EC:3.5.99.6] | 0.05842 | 0.00251 | 0.05936 | 0.00090 | 0.00372 | 65 | 0.05777 | 0.00108 | 0.00340 | 93 | 0.00504 | 0.31630 | 145.295 | 0.75223 | 0.93565 |
| pyrP, uraA; uracil permease | 0.05827 | 0.00197 | 0.06164 | 0.00055 | 0.00292 | 65 | 0.05591 | 0.00065 | 0.00264 | 93 | 0.00393 | 1.45790 | 144.315 | 0.14704 | 0.77376 |
| pstC; phosphate transport system permease protein | 0.05826 | 0.00172 | 0.05629 | 0.00056 | 0.00295 | 65 | 0.05964 | 0.00040 | 0.00208 | 93 | 0.00361 | -0.92938 | 122.478 | 0.35452 | 0.79354 |
| E1.4.1.4, gdhA; glutamate dehydrogenase (NADP+) [EC:1.4.1.4] | 0.05823 | 0.00233 | 0.05602 | 0.00086 | 0.00365 | 65 | 0.05978 | 0.00086 | 0.00303 | 93 | 0.00474 | -0.79291 | 137.401 | 0.42920 | 0.82749 |
| proB; glutamate 5-kinase [EC:2.7.2.11] | 0.05820 | 0.00132 | 0.05868 | 0.00030 | 0.00215 | 65 | 0.05786 | 0.00026 | 0.00168 | 93 | 0.00273 | 0.29816 | 131.457 | 0.76605 | 0.93900 |
| coaBC, dfp; phosphopantothenoylcysteine decarboxylase / phosphopantothenate—cysteine ligase [EC:4.1.1.36 6.3.2.5] | 0.05815 | 0.00141 | 0.05990 | 0.00031 | 0.00217 | 65 | 0.05693 | 0.00032 | 0.00187 | 93 | 0.00286 | 1.03620 | 140.410 | 0.30189 | 0.77376 |
| deoC, DERA; deoxyribose-phosphate aldolase [EC:4.1.2.4] | 0.05760 | 0.00216 | 0.05814 | 0.00081 | 0.00353 | 65 | 0.05722 | 0.00070 | 0.00274 | 93 | 0.00447 | 0.20604 | 131.263 | 0.83708 | 0.95506 |
| PGD, gnd, gntZ; 6-phosphogluconate dehydrogenase [EC:1.1.1.44 1.1.1.343] | 0.05752 | 0.00173 | 0.05694 | 0.00053 | 0.00286 | 65 | 0.05792 | 0.00044 | 0.00217 | 93 | 0.00359 | -0.27303 | 129.361 | 0.78527 | 0.94445 |
| ilvC; ketol-acid reductoisomerase [EC:1.1.1.86] | 0.05732 | 0.00147 | 0.05528 | 0.00042 | 0.00255 | 65 | 0.05874 | 0.00028 | 0.00174 | 93 | 0.00309 | -1.12168 | 119.490 | 0.26425 | 0.77376 |
| pyrR; pyrimidine operon attenuation protein / uracil phosphoribosyltransferase [EC:2.4.2.9] | 0.05725 | 0.00190 | 0.06022 | 0.00057 | 0.00296 | 65 | 0.05518 | 0.00056 | 0.00246 | 93 | 0.00385 | 1.30777 | 137.601 | 0.19313 | 0.77376 |
| leuD, IPMI-S; 3-isopropylmalate/(R)-2-methylmalate dehydratase small subunit [EC:4.2.1.33 4.2.1.35] | 0.05713 | 0.00149 | 0.05483 | 0.00044 | 0.00260 | 65 | 0.05874 | 0.00029 | 0.00176 | 93 | 0.00314 | -1.24410 | 118.871 | 0.21591 | 0.77376 |
| afuA, fbpA; iron(III) transport system substrate-binding protein | 0.05706 | 0.00232 | 0.05945 | 0.00108 | 0.00407 | 65 | 0.05540 | 0.00070 | 0.00274 | 93 | 0.00491 | 0.82589 | 118.058 | 0.41053 | 0.81416 |
| proA; glutamate-5-semialdehyde dehydrogenase [EC:1.2.1.41] | 0.05706 | 0.00140 | 0.05746 | 0.00033 | 0.00225 | 65 | 0.05678 | 0.00030 | 0.00179 | 93 | 0.00288 | 0.23661 | 133.150 | 0.81333 | 0.95343 |
| ispD; 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [EC:2.7.7.60] | 0.05703 | 0.00175 | 0.05659 | 0.00043 | 0.00256 | 65 | 0.05734 | 0.00053 | 0.00239 | 93 | 0.00351 | -0.21456 | 146.705 | 0.83040 | 0.95506 |
| ABC.X4.P; putative ABC transport system permease protein | 0.05703 | 0.00275 | 0.05982 | 0.00116 | 0.00422 | 65 | 0.05508 | 0.00123 | 0.00363 | 93 | 0.00557 | 0.85124 | 140.467 | 0.39608 | 0.80459 |
| bioY; biotin transport system substrate-specific component | 0.05700 | 0.00245 | 0.05921 | 0.00105 | 0.00402 | 65 | 0.05545 | 0.00088 | 0.00307 | 93 | 0.00506 | 0.74391 | 129.915 | 0.45828 | 0.83438 |
| K03710; GntR family transcriptional regulator | 0.05695 | 0.00304 | 0.05448 | 0.00165 | 0.00504 | 65 | 0.05868 | 0.00134 | 0.00379 | 93 | 0.00631 | -0.66650 | 128.236 | 0.50629 | 0.84956 |
| lpxL, htrB; Kdo2-lipid IVA lauroyltransferase [EC:2.3.1.241] | 0.05694 | 0.00229 | 0.05937 | 0.00082 | 0.00356 | 65 | 0.05524 | 0.00084 | 0.00300 | 93 | 0.00466 | 0.88777 | 138.607 | 0.37620 | 0.79810 |
| accC; acetyl-CoA carboxylase, biotin carboxylase subunit [EC:6.4.1.2 6.3.4.14] | 0.05691 | 0.00158 | 0.05575 | 0.00044 | 0.00259 | 65 | 0.05772 | 0.00036 | 0.00198 | 93 | 0.00326 | -0.60625 | 129.570 | 0.54541 | 0.86444 |
| TC.NSS; neurotransmitter:Na+ symporter, NSS family | 0.05673 | 0.00293 | 0.05981 | 0.00153 | 0.00485 | 65 | 0.05458 | 0.00123 | 0.00364 | 93 | 0.00607 | 0.86053 | 128.050 | 0.39111 | 0.80388 |
| rlmI; 23S rRNA (cytosine1962-C5)-methyltransferase [EC:2.1.1.191] | 0.05672 | 0.00153 | 0.05659 | 0.00042 | 0.00256 | 65 | 0.05682 | 0.00033 | 0.00189 | 93 | 0.00318 | -0.07367 | 126.842 | 0.94139 | 0.98528 |
| tadA; tRNA(adenine34) deaminase [EC:3.5.4.33] | 0.05670 | 0.00206 | 0.05619 | 0.00070 | 0.00329 | 65 | 0.05706 | 0.00066 | 0.00266 | 93 | 0.00423 | -0.20564 | 134.832 | 0.83739 | 0.95506 |
| znuB; zinc transport system permease protein | 0.05665 | 0.00164 | 0.05940 | 0.00042 | 0.00254 | 65 | 0.05473 | 0.00042 | 0.00213 | 93 | 0.00331 | 1.41157 | 138.285 | 0.16032 | 0.77376 |
| leuC, IPMI-L; 3-isopropylmalate/(R)-2-methylmalate dehydratase large subunit [EC:4.2.1.33 4.2.1.35] | 0.05661 | 0.00154 | 0.05406 | 0.00044 | 0.00260 | 65 | 0.05839 | 0.00032 | 0.00187 | 93 | 0.00320 | -1.35502 | 123.914 | 0.17788 | 0.77376 |
| plsX; glycerol-3-phosphate acyltransferase PlsX [EC:2.3.1.15] | 0.05659 | 0.00152 | 0.05951 | 0.00035 | 0.00232 | 65 | 0.05455 | 0.00037 | 0.00200 | 93 | 0.00306 | 1.62062 | 140.765 | 0.10734 | 0.77376 |
| ABC.X4.A; putative ABC transport system ATP-binding protein | 0.05640 | 0.00269 | 0.05922 | 0.00112 | 0.00415 | 65 | 0.05442 | 0.00117 | 0.00355 | 93 | 0.00546 | 0.88066 | 139.860 | 0.38001 | 0.79852 |
| parC; topoisomerase IV subunit A [EC:5.99.1.-] | 0.05639 | 0.00149 | 0.05487 | 0.00042 | 0.00255 | 65 | 0.05745 | 0.00030 | 0.00180 | 93 | 0.00312 | -0.82666 | 122.208 | 0.41004 | 0.81416 |
| dinG; ATP-dependent DNA helicase DinG [EC:3.6.4.12] | 0.05625 | 0.00156 | 0.05853 | 0.00043 | 0.00256 | 65 | 0.05465 | 0.00035 | 0.00194 | 93 | 0.00321 | 1.20837 | 128.976 | 0.22912 | 0.77376 |
| trpE; anthranilate synthase component I [EC:4.1.3.27] | 0.05619 | 0.00152 | 0.05372 | 0.00043 | 0.00258 | 65 | 0.05792 | 0.00032 | 0.00185 | 93 | 0.00317 | -1.32148 | 123.871 | 0.18878 | 0.77376 |
| npdA; NAD-dependent deacetylase [EC:3.5.1.-] | 0.05616 | 0.00201 | 0.05883 | 0.00064 | 0.00313 | 65 | 0.05430 | 0.00064 | 0.00262 | 93 | 0.00408 | 1.10906 | 137.735 | 0.26934 | 0.77376 |
| clpC; ATP-dependent Clp protease ATP-binding subunit ClpC | 0.05615 | 0.00227 | 0.05630 | 0.00086 | 0.00364 | 65 | 0.05605 | 0.00079 | 0.00292 | 93 | 0.00466 | 0.05537 | 134.239 | 0.95593 | 0.98792 |
| PTS-Cel-EIIC, celB, chbC; PTS system, cellobiose-specific IIC component | 0.05611 | 0.00498 | 0.06339 | 0.00475 | 0.00855 | 65 | 0.05102 | 0.00332 | 0.00598 | 93 | 0.01043 | 1.18598 | 121.594 | 0.23794 | 0.77376 |
| rnhA, RNASEH1; ribonuclease HI [EC:3.1.26.4] | 0.05608 | 0.00170 | 0.05609 | 0.00056 | 0.00293 | 65 | 0.05608 | 0.00039 | 0.00206 | 93 | 0.00358 | 0.00415 | 121.911 | 0.99670 | 0.99951 |
| nrdG; anaerobic ribonucleoside-triphosphate reductase activating protein [EC:1.97.1.4] | 0.05598 | 0.00235 | 0.05937 | 0.00085 | 0.00361 | 65 | 0.05361 | 0.00088 | 0.00308 | 93 | 0.00474 | 1.21384 | 139.645 | 0.22686 | 0.77376 |
| manA, MPI; mannose-6-phosphate isomerase [EC:5.3.1.8] | 0.05591 | 0.00197 | 0.05520 | 0.00063 | 0.00311 | 65 | 0.05640 | 0.00061 | 0.00255 | 93 | 0.00403 | -0.29727 | 136.250 | 0.76672 | 0.93900 |
| hlyIII; hemolysin III | 0.05577 | 0.00142 | 0.05716 | 0.00033 | 0.00226 | 65 | 0.05480 | 0.00031 | 0.00182 | 93 | 0.00290 | 0.81218 | 134.740 | 0.41812 | 0.81941 |
| accD; acetyl-CoA carboxylase carboxyl transferase subunit beta [EC:6.4.1.2 2.1.3.15] | 0.05577 | 0.00139 | 0.05730 | 0.00030 | 0.00213 | 65 | 0.05470 | 0.00031 | 0.00183 | 93 | 0.00281 | 0.92692 | 140.019 | 0.35556 | 0.79399 |
| acpS; holo-[acyl-carrier protein] synthase [EC:2.7.8.7] | 0.05572 | 0.00201 | 0.05809 | 0.00064 | 0.00314 | 65 | 0.05406 | 0.00063 | 0.00261 | 93 | 0.00408 | 0.98720 | 137.373 | 0.32528 | 0.77376 |
| parE; topoisomerase IV subunit B [EC:5.99.1.-] | 0.05566 | 0.00147 | 0.05424 | 0.00041 | 0.00251 | 65 | 0.05665 | 0.00029 | 0.00178 | 93 | 0.00308 | -0.78150 | 122.929 | 0.43601 | 0.83006 |
| E2.2.1.6S, ilvH, ilvN; acetolactate synthase I/III small subunit [EC:2.2.1.6] | 0.05558 | 0.00183 | 0.05474 | 0.00066 | 0.00319 | 65 | 0.05616 | 0.00044 | 0.00217 | 93 | 0.00386 | -0.36756 | 119.233 | 0.71385 | 0.92004 |
| accB, bccP; acetyl-CoA carboxylase biotin carboxyl carrier protein | 0.05549 | 0.00162 | 0.05444 | 0.00042 | 0.00254 | 65 | 0.05622 | 0.00041 | 0.00210 | 93 | 0.00330 | -0.53826 | 136.961 | 0.59127 | 0.87896 |
| yhbY; RNA-binding protein | 0.05540 | 0.00142 | 0.05992 | 0.00024 | 0.00193 | 65 | 0.05225 | 0.00035 | 0.00194 | 93 | 0.00273 | 2.80488 | 151.245 | 0.00569 | 0.54022 |
| opuA; osmoprotectant transport system ATP-binding protein | 0.05540 | 0.00211 | 0.05796 | 0.00082 | 0.00355 | 65 | 0.05361 | 0.00062 | 0.00259 | 93 | 0.00439 | 0.99103 | 125.519 | 0.32358 | 0.77376 |
| znuC; zinc transport system ATP-binding protein [EC:3.6.3.-] | 0.05538 | 0.00161 | 0.05839 | 0.00042 | 0.00253 | 65 | 0.05328 | 0.00040 | 0.00207 | 93 | 0.00327 | 1.56403 | 136.134 | 0.12013 | 0.77376 |
| hlpA, ompH; outer membrane protein | 0.05535 | 0.00385 | 0.05533 | 0.00189 | 0.00539 | 65 | 0.05536 | 0.00268 | 0.00537 | 93 | 0.00761 | -0.00465 | 150.725 | 0.99630 | 0.99944 |
| recD; exodeoxyribonuclease V alpha subunit [EC:3.1.11.5] | 0.05535 | 0.00151 | 0.05698 | 0.00038 | 0.00243 | 65 | 0.05421 | 0.00034 | 0.00192 | 93 | 0.00310 | 0.89430 | 132.931 | 0.37278 | 0.79810 |
| E3.1.3.16; protein phosphatase [EC:3.1.3.16] | 0.05532 | 0.00226 | 0.05397 | 0.00086 | 0.00363 | 65 | 0.05627 | 0.00078 | 0.00290 | 93 | 0.00464 | -0.49379 | 133.914 | 0.62227 | 0.89150 |
| ABC.SN.S; NitT/TauT family transport system substrate-binding protein | 0.05532 | 0.00354 | 0.04804 | 0.00171 | 0.00513 | 65 | 0.06040 | 0.00213 | 0.00479 | 93 | 0.00702 | -1.76246 | 146.659 | 0.08008 | 0.75920 |
| chrA; chromate transporter | 0.05522 | 0.00315 | 0.05386 | 0.00154 | 0.00487 | 65 | 0.05618 | 0.00160 | 0.00414 | 93 | 0.00639 | -0.36274 | 139.420 | 0.71735 | 0.92035 |
| leuB, IMDH; 3-isopropylmalate dehydrogenase [EC:1.1.1.85] | 0.05521 | 0.00150 | 0.05357 | 0.00043 | 0.00258 | 65 | 0.05635 | 0.00030 | 0.00179 | 93 | 0.00315 | -0.88621 | 121.010 | 0.37726 | 0.79815 |
| EARS, gltX; glutamyl-tRNA synthetase [EC:6.1.1.17] | 0.05521 | 0.00199 | 0.05526 | 0.00056 | 0.00293 | 65 | 0.05517 | 0.00068 | 0.00270 | 93 | 0.00398 | 0.02101 | 145.503 | 0.98326 | 0.99706 |
| lacZ; beta-galactosidase [EC:3.2.1.23] | 0.05510 | 0.00365 | 0.05436 | 0.00165 | 0.00504 | 65 | 0.05562 | 0.00245 | 0.00514 | 93 | 0.00719 | -0.17484 | 152.003 | 0.86144 | 0.96206 |
| cca; tRNA nucleotidyltransferase (CCA-adding enzyme) [EC:2.7.7.72 3.1.3.- 3.1.4.-] | 0.05507 | 0.00141 | 0.05871 | 0.00029 | 0.00210 | 65 | 0.05253 | 0.00032 | 0.00185 | 93 | 0.00280 | 2.20782 | 142.382 | 0.02886 | 0.61334 |
| nagA, AMDHD2; N-acetylglucosamine-6-phosphate deacetylase [EC:3.5.1.25] | 0.05476 | 0.00174 | 0.05483 | 0.00043 | 0.00258 | 65 | 0.05471 | 0.00051 | 0.00235 | 93 | 0.00349 | 0.03688 | 144.815 | 0.97063 | 0.99349 |
| E3.1.3.25, IMPA, suhB; myo-inositol-1(or 4)-monophosphatase [EC:3.1.3.25] | 0.05459 | 0.00190 | 0.05134 | 0.00052 | 0.00284 | 65 | 0.05687 | 0.00059 | 0.00253 | 93 | 0.00380 | -1.45397 | 143.049 | 0.14814 | 0.77376 |
| K07461; putative endonuclease | 0.05456 | 0.00155 | 0.05800 | 0.00029 | 0.00210 | 65 | 0.05215 | 0.00043 | 0.00215 | 93 | 0.00301 | 1.94367 | 152.146 | 0.05378 | 0.70048 |
| metF, MTHFR; methylenetetrahydrofolate reductase (NADPH) [EC:1.5.1.20] | 0.05450 | 0.00158 | 0.05334 | 0.00047 | 0.00269 | 65 | 0.05532 | 0.00034 | 0.00192 | 93 | 0.00330 | -0.59826 | 123.789 | 0.55076 | 0.86739 |
| cdd, CDA; cytidine deaminase [EC:3.5.4.5] | 0.05445 | 0.00219 | 0.05377 | 0.00080 | 0.00352 | 65 | 0.05492 | 0.00073 | 0.00281 | 93 | 0.00450 | -0.25621 | 133.703 | 0.79818 | 0.94822 |
| hrcA; heat-inducible transcriptional repressor | 0.05437 | 0.00180 | 0.05597 | 0.00051 | 0.00280 | 65 | 0.05326 | 0.00051 | 0.00235 | 93 | 0.00365 | 0.74352 | 138.369 | 0.45843 | 0.83438 |
| ispH, lytB; 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase [EC:1.17.7.4] | 0.05427 | 0.00179 | 0.05480 | 0.00060 | 0.00305 | 65 | 0.05389 | 0.00045 | 0.00219 | 93 | 0.00376 | 0.24284 | 124.239 | 0.80853 | 0.95002 |
| K15977; putative oxidoreductase | 0.05423 | 0.00379 | 0.05122 | 0.00243 | 0.00612 | 65 | 0.05634 | 0.00217 | 0.00483 | 93 | 0.00779 | -0.65684 | 132.677 | 0.51242 | 0.85122 |
| sepF; cell division inhibitor SepF | 0.05409 | 0.00248 | 0.06021 | 0.00119 | 0.00428 | 65 | 0.04982 | 0.00079 | 0.00291 | 93 | 0.00517 | 2.00967 | 119.015 | 0.04673 | 0.66255 |
| rny; ribonucrease Y [EC:3.1.-.-] | 0.05408 | 0.00234 | 0.05613 | 0.00087 | 0.00366 | 65 | 0.05265 | 0.00086 | 0.00304 | 93 | 0.00476 | 0.73167 | 137.383 | 0.46562 | 0.83786 |
| nrnA; bifunctional oligoribonuclease and PAP phosphatase NrnA [EC:3.1.3.7 3.1.13.3] | 0.05400 | 0.00228 | 0.05596 | 0.00084 | 0.00360 | 65 | 0.05263 | 0.00080 | 0.00294 | 93 | 0.00465 | 0.71748 | 135.841 | 0.47431 | 0.84073 |
| ispE; 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase [EC:2.7.1.148] | 0.05400 | 0.00133 | 0.05516 | 0.00026 | 0.00199 | 65 | 0.05319 | 0.00030 | 0.00180 | 93 | 0.00268 | 0.73632 | 144.296 | 0.46273 | 0.83654 |
| PRDX2_4, ahpC; peroxiredoxin (alkyl hydroperoxide reductase subunit C) [EC:1.11.1.15] | 0.05392 | 0.00147 | 0.05621 | 0.00032 | 0.00223 | 65 | 0.05231 | 0.00036 | 0.00196 | 93 | 0.00296 | 1.31532 | 141.979 | 0.19052 | 0.77376 |
| K02483; two-component system, OmpR family, response regulator | 0.05389 | 0.00445 | 0.04750 | 0.00349 | 0.00732 | 65 | 0.05836 | 0.00286 | 0.00555 | 93 | 0.00919 | -1.18293 | 128.970 | 0.23901 | 0.77376 |
| glxK, garK; glycerate 2-kinase [EC:2.7.1.165] | 0.05386 | 0.00178 | 0.05418 | 0.00054 | 0.00287 | 65 | 0.05365 | 0.00048 | 0.00228 | 93 | 0.00367 | 0.14536 | 133.388 | 0.88464 | 0.96984 |
| ribBA; 3,4-dihydroxy 2-butanone 4-phosphate synthase / GTP cyclohydrolase II [EC:4.1.99.12 3.5.4.25] | 0.05386 | 0.00181 | 0.05392 | 0.00054 | 0.00289 | 65 | 0.05382 | 0.00051 | 0.00234 | 93 | 0.00372 | 0.02769 | 134.952 | 0.97795 | 0.99657 |
| K07001; NTE family protein | 0.05382 | 0.00298 | 0.04959 | 0.00127 | 0.00441 | 65 | 0.05678 | 0.00149 | 0.00400 | 93 | 0.00596 | -1.20641 | 144.556 | 0.22963 | 0.77376 |
| ABC.SS.A; simple sugar transport system ATP-binding protein [EC:3.6.3.17] | 0.05382 | 0.00224 | 0.05315 | 0.00090 | 0.00372 | 65 | 0.05429 | 0.00072 | 0.00279 | 93 | 0.00465 | -0.24450 | 128.000 | 0.80724 | 0.94987 |
| yggT; YggT family protein | 0.05352 | 0.00167 | 0.05536 | 0.00043 | 0.00258 | 65 | 0.05224 | 0.00045 | 0.00220 | 93 | 0.00339 | 0.92010 | 139.724 | 0.35911 | 0.79496 |
| oppF; oligopeptide transport system ATP-binding protein | 0.05349 | 0.00340 | 0.05352 | 0.00150 | 0.00480 | 65 | 0.05347 | 0.00207 | 0.00472 | 93 | 0.00673 | 0.00712 | 149.996 | 0.99433 | 0.99919 |
| glyS; glycyl-tRNA synthetase beta chain [EC:6.1.1.14] | 0.05349 | 0.00137 | 0.05770 | 0.00025 | 0.00194 | 65 | 0.05054 | 0.00031 | 0.00183 | 93 | 0.00267 | 2.68205 | 147.466 | 0.00815 | 0.56121 |
| E5.3.1.26, lacA, lacB; galactose-6-phosphate isomerase [EC:5.3.1.26] | 0.05342 | 0.00405 | 0.05172 | 0.00200 | 0.00554 | 65 | 0.05461 | 0.00304 | 0.00572 | 93 | 0.00796 | -0.36388 | 152.544 | 0.71645 | 0.92004 |
| purK; 5-(carboxyamino)imidazole ribonucleotide synthase [EC:6.3.4.18] | 0.05341 | 0.00177 | 0.05343 | 0.00062 | 0.00309 | 65 | 0.05339 | 0.00041 | 0.00211 | 93 | 0.00374 | 0.01023 | 119.621 | 0.99185 | 0.99884 |
| mutS2; DNA mismatch repair protein MutS2 | 0.05340 | 0.00231 | 0.05553 | 0.00087 | 0.00367 | 65 | 0.05190 | 0.00083 | 0.00299 | 93 | 0.00473 | 0.76617 | 135.512 | 0.44491 | 0.83274 |
| K00243; uncharacterized protein | 0.05328 | 0.00193 | 0.05670 | 0.00054 | 0.00287 | 65 | 0.05089 | 0.00062 | 0.00258 | 93 | 0.00386 | 1.50664 | 143.674 | 0.13410 | 0.77376 |
| GBE1, glgB; 1,4-alpha-glucan branching enzyme [EC:2.4.1.18] | 0.05324 | 0.00186 | 0.05425 | 0.00055 | 0.00291 | 65 | 0.05253 | 0.00055 | 0.00243 | 93 | 0.00380 | 0.45211 | 137.713 | 0.65190 | 0.90504 |
| gltD; glutamate synthase (NADPH/NADH) small chain [EC:1.4.1.13 1.4.1.14] | 0.05298 | 0.00225 | 0.04848 | 0.00080 | 0.00351 | 65 | 0.05613 | 0.00078 | 0.00290 | 93 | 0.00455 | -1.68173 | 136.968 | 0.09490 | 0.76609 |
| trpG; anthranilate synthase component II [EC:4.1.3.27] | 0.05288 | 0.00151 | 0.05089 | 0.00045 | 0.00264 | 65 | 0.05426 | 0.00029 | 0.00178 | 93 | 0.00318 | -1.06108 | 118.323 | 0.29082 | 0.77376 |
| ABC.PE.A; peptide/nickel transport system ATP-binding protein | 0.05287 | 0.00402 | 0.05493 | 0.00316 | 0.00697 | 65 | 0.05144 | 0.00216 | 0.00482 | 93 | 0.00847 | 0.41238 | 120.658 | 0.68079 | 0.91355 |
| glyQ; glycyl-tRNA synthetase alpha chain [EC:6.1.1.14] | 0.05279 | 0.00140 | 0.05705 | 0.00026 | 0.00199 | 65 | 0.04981 | 0.00033 | 0.00188 | 93 | 0.00274 | 2.64666 | 147.410 | 0.00901 | 0.58112 |
| cydB; cytochrome bd ubiquinol oxidase subunit II [EC:1.10.3.14] | 0.05268 | 0.00169 | 0.05163 | 0.00045 | 0.00264 | 65 | 0.05342 | 0.00046 | 0.00222 | 93 | 0.00345 | -0.51907 | 138.168 | 0.60454 | 0.88434 |
| K06950; uncharacterized protein | 0.05266 | 0.00232 | 0.05401 | 0.00109 | 0.00410 | 65 | 0.05172 | 0.00069 | 0.00273 | 93 | 0.00492 | 0.46546 | 117.092 | 0.64247 | 0.90098 |
| yfbK; Ca-activated chloride channel homolog | 0.05259 | 0.00290 | 0.05053 | 0.00121 | 0.00432 | 65 | 0.05404 | 0.00142 | 0.00390 | 93 | 0.00582 | -0.60340 | 144.160 | 0.54719 | 0.86509 |
| NARS, asnS; asparaginyl-tRNA synthetase [EC:6.1.1.22] | 0.05259 | 0.00199 | 0.05473 | 0.00063 | 0.00310 | 65 | 0.05110 | 0.00063 | 0.00260 | 93 | 0.00405 | 0.89827 | 137.975 | 0.37060 | 0.79606 |
| hemH, FECH; protoporphyrin/coproporphyrin ferrochelatase [EC:4.99.1.1 4.99.1.9] | 0.05252 | 0.00180 | 0.04977 | 0.00051 | 0.00280 | 65 | 0.05444 | 0.00051 | 0.00235 | 93 | 0.00365 | -1.27901 | 138.205 | 0.20304 | 0.77376 |
| rfbC, rmlC; dTDP-4-dehydrorhamnose 3,5-epimerase [EC:5.1.3.13] | 0.05245 | 0.00195 | 0.05703 | 0.00082 | 0.00355 | 65 | 0.04925 | 0.00043 | 0.00215 | 93 | 0.00415 | 1.87532 | 109.439 | 0.06341 | 0.73289 |
| yhbH; putative sigma-54 modulation protein | 0.05239 | 0.00169 | 0.05451 | 0.00046 | 0.00266 | 65 | 0.05090 | 0.00044 | 0.00218 | 93 | 0.00344 | 1.04990 | 136.043 | 0.29563 | 0.77376 |
| yesM; two-component system, sensor histidine kinase YesM [EC:2.7.13.3] | 0.05233 | 0.00395 | 0.05595 | 0.00315 | 0.00697 | 65 | 0.04979 | 0.00199 | 0.00463 | 93 | 0.00836 | 0.73672 | 117.167 | 0.46277 | 0.83654 |
| accA; acetyl-CoA carboxylase carboxyl transferase subunit alpha [EC:6.4.1.2 2.1.3.15] | 0.05218 | 0.00142 | 0.05326 | 0.00030 | 0.00216 | 65 | 0.05143 | 0.00034 | 0.00190 | 93 | 0.00287 | 0.63810 | 142.240 | 0.52444 | 0.85728 |
| rbsB; ribose transport system substrate-binding protein | 0.05213 | 0.00363 | 0.05043 | 0.00323 | 0.00705 | 65 | 0.05332 | 0.00130 | 0.00374 | 93 | 0.00798 | -0.36114 | 99.575 | 0.71876 | 0.92092 |
| K07483; transposase | 0.05210 | 0.00229 | 0.05702 | 0.00102 | 0.00395 | 65 | 0.04866 | 0.00067 | 0.00269 | 93 | 0.00478 | 1.74722 | 119.222 | 0.08317 | 0.76567 |
| comEB; dCMP deaminase [EC:3.5.4.12] | 0.05207 | 0.00229 | 0.05433 | 0.00085 | 0.00361 | 65 | 0.05050 | 0.00081 | 0.00296 | 93 | 0.00467 | 0.82043 | 136.048 | 0.41341 | 0.81655 |
| thrB1; homoserine kinase [EC:2.7.1.39] | 0.05203 | 0.00199 | 0.05252 | 0.00068 | 0.00323 | 65 | 0.05169 | 0.00060 | 0.00254 | 93 | 0.00411 | 0.20279 | 132.288 | 0.83961 | 0.95506 |
| trpA; tryptophan synthase alpha chain [EC:4.2.1.20] | 0.05203 | 0.00141 | 0.04984 | 0.00036 | 0.00234 | 65 | 0.05357 | 0.00028 | 0.00173 | 93 | 0.00291 | -1.28116 | 126.743 | 0.20248 | 0.77376 |
| fecR; transmembrane sensor | 0.05201 | 0.00682 | 0.05064 | 0.00718 | 0.01051 | 65 | 0.05298 | 0.00754 | 0.00900 | 93 | 0.01384 | -0.16883 | 139.972 | 0.86617 | 0.96296 |
| mreD; rod shape-determining protein MreD | 0.05201 | 0.00139 | 0.05437 | 0.00033 | 0.00227 | 65 | 0.05036 | 0.00028 | 0.00175 | 93 | 0.00286 | 1.40156 | 130.549 | 0.16342 | 0.77376 |
| malQ; 4-alpha-glucanotransferase [EC:2.4.1.25] | 0.05189 | 0.00220 | 0.05424 | 0.00079 | 0.00349 | 65 | 0.05026 | 0.00075 | 0.00284 | 93 | 0.00450 | 0.88480 | 135.721 | 0.37783 | 0.79852 |
| cydA; cytochrome bd ubiquinol oxidase subunit I [EC:1.10.3.14] | 0.05187 | 0.00171 | 0.05168 | 0.00048 | 0.00271 | 65 | 0.05200 | 0.00046 | 0.00222 | 93 | 0.00350 | -0.09118 | 135.841 | 0.92749 | 0.98137 |
| clpB; ATP-dependent Clp protease ATP-binding subunit ClpB | 0.05167 | 0.00147 | 0.05195 | 0.00035 | 0.00233 | 65 | 0.05149 | 0.00034 | 0.00191 | 93 | 0.00302 | 0.15280 | 136.250 | 0.87878 | 0.96759 |
| tlyA; 23S rRNA (cytidine1920-2’-O)/16S rRNA (cytidine1409-2’-O)-methyltransferase [EC:2.1.1.226 2.1.1.227] | 0.05160 | 0.00199 | 0.05402 | 0.00066 | 0.00319 | 65 | 0.04991 | 0.00060 | 0.00255 | 93 | 0.00408 | 1.00659 | 133.950 | 0.31594 | 0.77376 |
| E2.1.1.72; site-specific DNA-methyltransferase (adenine-specific) [EC:2.1.1.72] | 0.05157 | 0.00319 | 0.04807 | 0.00115 | 0.00421 | 65 | 0.05402 | 0.00193 | 0.00456 | 93 | 0.00620 | -0.95956 | 154.407 | 0.33878 | 0.78412 |
| nadE; NAD+ synthase [EC:6.3.1.5] | 0.05150 | 0.00170 | 0.05364 | 0.00046 | 0.00266 | 65 | 0.05001 | 0.00045 | 0.00220 | 93 | 0.00345 | 1.05258 | 136.864 | 0.29439 | 0.77376 |
| trpD; anthranilate phosphoribosyltransferase [EC:2.4.2.18] | 0.05143 | 0.00142 | 0.04894 | 0.00038 | 0.00242 | 65 | 0.05318 | 0.00027 | 0.00171 | 93 | 0.00297 | -1.42809 | 122.590 | 0.15581 | 0.77376 |
| fixA, etfB; electron transfer flavoprotein beta subunit | 0.05135 | 0.00237 | 0.05407 | 0.00118 | 0.00426 | 65 | 0.04945 | 0.00069 | 0.00272 | 93 | 0.00505 | 0.91527 | 113.662 | 0.36199 | 0.79496 |
| K06911; uncharacterized protein | 0.05133 | 0.00370 | 0.04808 | 0.00211 | 0.00569 | 65 | 0.05361 | 0.00221 | 0.00488 | 93 | 0.00749 | -0.73819 | 139.964 | 0.46164 | 0.83596 |
| K07124; uncharacterized protein | 0.05132 | 0.00165 | 0.05320 | 0.00045 | 0.00262 | 65 | 0.05000 | 0.00042 | 0.00212 | 93 | 0.00337 | 0.94834 | 134.718 | 0.34466 | 0.78711 |
| ribH, RIB4; 6,7-dimethyl-8-ribityllumazine synthase [EC:2.5.1.78] | 0.05126 | 0.00149 | 0.05022 | 0.00032 | 0.00223 | 65 | 0.05198 | 0.00037 | 0.00200 | 93 | 0.00300 | -0.58560 | 143.677 | 0.55907 | 0.86813 |
| rimJ; [ribosomal protein S5]-alanine N-acetyltransferase [EC:2.3.1.267] | 0.05102 | 0.00344 | 0.05078 | 0.00143 | 0.00469 | 65 | 0.05119 | 0.00219 | 0.00485 | 93 | 0.00675 | -0.06004 | 152.662 | 0.95220 | 0.98792 |
| ABC.CD.TX; HlyD family secretion protein | 0.05101 | 0.00229 | 0.04780 | 0.00077 | 0.00344 | 65 | 0.05326 | 0.00087 | 0.00305 | 93 | 0.00460 | -1.18609 | 142.852 | 0.23756 | 0.77376 |
| INV, sacA; beta-fructofuranosidase [EC:3.2.1.26] | 0.05085 | 0.00263 | 0.05505 | 0.00137 | 0.00459 | 65 | 0.04792 | 0.00090 | 0.00311 | 93 | 0.00554 | 1.28777 | 118.861 | 0.20033 | 0.77376 |
| tyrA2; prephenate dehydrogenase [EC:1.3.1.12] | 0.05083 | 0.00188 | 0.05000 | 0.00062 | 0.00309 | 65 | 0.05141 | 0.00052 | 0.00236 | 93 | 0.00389 | -0.36109 | 129.720 | 0.71862 | 0.92092 |
| fixB, etfA; electron transfer flavoprotein alpha subunit | 0.05083 | 0.00235 | 0.05367 | 0.00117 | 0.00425 | 65 | 0.04884 | 0.00066 | 0.00266 | 93 | 0.00501 | 0.96348 | 112.088 | 0.33738 | 0.78354 |
| czcD, zitB; cobalt-zinc-cadmium efflux system protein | 0.05077 | 0.00191 | 0.04822 | 0.00058 | 0.00300 | 65 | 0.05256 | 0.00057 | 0.00248 | 93 | 0.00389 | -1.11249 | 137.075 | 0.26787 | 0.77376 |
| rsmC; 16S rRNA (guanine1207-N2)-methyltransferase [EC:2.1.1.172] | 0.05071 | 0.00166 | 0.05267 | 0.00038 | 0.00240 | 65 | 0.04934 | 0.00047 | 0.00226 | 93 | 0.00330 | 1.00826 | 147.167 | 0.31499 | 0.77376 |
| K07491; putative transposase | 0.05064 | 0.00343 | 0.05273 | 0.00231 | 0.00596 | 65 | 0.04917 | 0.00156 | 0.00409 | 93 | 0.00723 | 0.49183 | 120.055 | 0.62374 | 0.89159 |
| thiJ; protein deglycase [EC:3.5.1.124] | 0.05058 | 0.00271 | 0.05366 | 0.00127 | 0.00442 | 65 | 0.04842 | 0.00108 | 0.00341 | 93 | 0.00559 | 0.93712 | 130.646 | 0.35043 | 0.79120 |
| luxS; S-ribosylhomocysteine lyase [EC:4.4.1.21] | 0.05056 | 0.00217 | 0.05143 | 0.00077 | 0.00344 | 65 | 0.04995 | 0.00073 | 0.00281 | 93 | 0.00444 | 0.33263 | 135.775 | 0.73992 | 0.92969 |
| argH, ASL; argininosuccinate lyase [EC:4.3.2.1] | 0.05055 | 0.00126 | 0.04908 | 0.00025 | 0.00195 | 65 | 0.05157 | 0.00025 | 0.00164 | 93 | 0.00255 | -0.97828 | 138.177 | 0.32965 | 0.77554 |
| xpt; xanthine phosphoribosyltransferase [EC:2.4.2.22] | 0.05043 | 0.00180 | 0.05090 | 0.00058 | 0.00299 | 65 | 0.05011 | 0.00046 | 0.00224 | 93 | 0.00373 | 0.21368 | 127.703 | 0.83114 | 0.95506 |
| maf; septum formation protein | 0.05043 | 0.00177 | 0.04875 | 0.00049 | 0.00273 | 65 | 0.05160 | 0.00050 | 0.00233 | 93 | 0.00359 | -0.79416 | 139.472 | 0.42845 | 0.82737 |
| E2.3.1.8, pta; phosphate acetyltransferase [EC:2.3.1.8] | 0.05041 | 0.00219 | 0.05183 | 0.00081 | 0.00352 | 65 | 0.04942 | 0.00073 | 0.00280 | 93 | 0.00450 | 0.53527 | 133.470 | 0.59335 | 0.87985 |
| ridA, tdcF, RIDA; 2-iminobutanoate/2-iminopropanoate deaminase [EC:3.5.99.10] | 0.05041 | 0.00220 | 0.05051 | 0.00078 | 0.00347 | 65 | 0.05034 | 0.00076 | 0.00287 | 93 | 0.00450 | 0.03832 | 136.778 | 0.96949 | 0.99292 |
| K06890; uncharacterized protein | 0.05039 | 0.00251 | 0.05171 | 0.00120 | 0.00429 | 65 | 0.04947 | 0.00086 | 0.00304 | 93 | 0.00526 | 0.42472 | 122.726 | 0.67179 | 0.91184 |
| cidA; holin-like protein | 0.05033 | 0.00197 | 0.05238 | 0.00060 | 0.00305 | 65 | 0.04889 | 0.00062 | 0.00259 | 93 | 0.00400 | 0.87253 | 139.236 | 0.38442 | 0.80197 |
| comM; magnesium chelatase family protein | 0.05032 | 0.00202 | 0.05295 | 0.00082 | 0.00354 | 65 | 0.04848 | 0.00053 | 0.00238 | 93 | 0.00427 | 1.04878 | 117.986 | 0.29642 | 0.77376 |
| miaB; tRNA-2-methylthio-N6-dimethylallyladenosine synthase [EC:2.8.4.3] | 0.05029 | 0.00164 | 0.04986 | 0.00046 | 0.00266 | 65 | 0.05059 | 0.00041 | 0.00210 | 93 | 0.00338 | -0.21510 | 132.782 | 0.83002 | 0.95506 |
| maa; maltose O-acetyltransferase [EC:2.3.1.79] | 0.05029 | 0.00307 | 0.05181 | 0.00203 | 0.00559 | 65 | 0.04923 | 0.00113 | 0.00348 | 93 | 0.00658 | 0.39221 | 111.589 | 0.69565 | 0.91769 |
| polC; DNA polymerase III subunit alpha, Gram-positive type [EC:2.7.7.7] | 0.05029 | 0.00251 | 0.05561 | 0.00117 | 0.00424 | 65 | 0.04656 | 0.00086 | 0.00304 | 93 | 0.00522 | 1.73419 | 123.855 | 0.08537 | 0.76567 |
| tatA; sec-independent protein translocase protein TatA | 0.05003 | 0.00153 | 0.04686 | 0.00043 | 0.00257 | 65 | 0.05225 | 0.00032 | 0.00186 | 93 | 0.00318 | -1.69561 | 124.651 | 0.09246 | 0.76567 |
| glgA; starch synthase [EC:2.4.1.21] | 0.05000 | 0.00186 | 0.05097 | 0.00055 | 0.00292 | 65 | 0.04932 | 0.00055 | 0.00243 | 93 | 0.00380 | 0.43535 | 137.349 | 0.66399 | 0.91038 |
| gcpE, ispG; (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase [EC:1.17.7.1 1.17.7.3] | 0.04995 | 0.00148 | 0.04963 | 0.00037 | 0.00238 | 65 | 0.05018 | 0.00034 | 0.00190 | 93 | 0.00304 | -0.18172 | 133.905 | 0.85608 | 0.95933 |
| pepN; aminopeptidase N [EC:3.4.11.2] | 0.04994 | 0.00204 | 0.04983 | 0.00077 | 0.00344 | 65 | 0.05002 | 0.00059 | 0.00251 | 93 | 0.00426 | -0.04302 | 125.633 | 0.96575 | 0.99101 |
| OTC, argF, argI; ornithine carbamoyltransferase [EC:2.1.3.3] | 0.04981 | 0.00152 | 0.05232 | 0.00039 | 0.00244 | 65 | 0.04805 | 0.00034 | 0.00192 | 93 | 0.00311 | 1.37217 | 132.477 | 0.17233 | 0.77376 |
| K07497; putative transposase | 0.04979 | 0.00321 | 0.05125 | 0.00171 | 0.00512 | 65 | 0.04876 | 0.00158 | 0.00412 | 93 | 0.00658 | 0.37926 | 134.521 | 0.70510 | 0.92004 |
| phoU; phosphate transport system protein | 0.04972 | 0.00168 | 0.04744 | 0.00049 | 0.00274 | 65 | 0.05132 | 0.00041 | 0.00210 | 93 | 0.00346 | -1.12101 | 130.105 | 0.26435 | 0.77376 |
| PTS-Scr-EIIC, scrA, sacP, sacX, ptsS; PTS system, sucrose-specific IIC component | 0.04963 | 0.00269 | 0.05376 | 0.00131 | 0.00448 | 65 | 0.04674 | 0.00102 | 0.00332 | 93 | 0.00558 | 1.26026 | 126.829 | 0.20989 | 0.77376 |
| K09762; uncharacterized protein | 0.04958 | 0.00211 | 0.05324 | 0.00073 | 0.00336 | 65 | 0.04703 | 0.00067 | 0.00269 | 93 | 0.00430 | 1.44251 | 133.806 | 0.15150 | 0.77376 |
| lexA; repressor LexA [EC:3.4.21.88] | 0.04954 | 0.00145 | 0.05200 | 0.00035 | 0.00232 | 65 | 0.04781 | 0.00032 | 0.00185 | 93 | 0.00297 | 1.41237 | 133.916 | 0.16016 | 0.77376 |
| rfbD, rmlD; dTDP-4-dehydrorhamnose reductase [EC:1.1.1.133] | 0.04946 | 0.00150 | 0.05187 | 0.00036 | 0.00237 | 65 | 0.04777 | 0.00034 | 0.00192 | 93 | 0.00305 | 1.34456 | 135.218 | 0.18102 | 0.77376 |
| E2.7.7.24, rfbA, rffH; glucose-1-phosphate thymidylyltransferase [EC:2.7.7.24] | 0.04938 | 0.00150 | 0.05023 | 0.00038 | 0.00242 | 65 | 0.04879 | 0.00034 | 0.00191 | 93 | 0.00309 | 0.46827 | 132.557 | 0.64036 | 0.89936 |
| ACO, acnA; aconitate hydratase [EC:4.2.1.3] | 0.04935 | 0.00191 | 0.04536 | 0.00061 | 0.00307 | 65 | 0.05214 | 0.00054 | 0.00242 | 93 | 0.00390 | -1.73706 | 132.685 | 0.08470 | 0.76567 |
| pat; phosphinothricin acetyltransferase [EC:2.3.1.183] | 0.04932 | 0.00129 | 0.04827 | 0.00033 | 0.00227 | 65 | 0.05006 | 0.00021 | 0.00152 | 93 | 0.00273 | -0.65691 | 117.566 | 0.51252 | 0.85122 |
| yfiH; polyphenol oxidase [EC:1.10.3.-] | 0.04924 | 0.00144 | 0.04959 | 0.00038 | 0.00242 | 65 | 0.04900 | 0.00029 | 0.00178 | 93 | 0.00301 | 0.19850 | 126.008 | 0.84297 | 0.95519 |
| dxr; 1-deoxy-D-xylulose-5-phosphate reductoisomerase [EC:1.1.1.267] | 0.04915 | 0.00149 | 0.04897 | 0.00036 | 0.00234 | 65 | 0.04927 | 0.00035 | 0.00193 | 93 | 0.00303 | -0.09916 | 136.871 | 0.92116 | 0.97905 |
| ylxR; uncharacterized protein | 0.04912 | 0.00205 | 0.05163 | 0.00072 | 0.00332 | 65 | 0.04737 | 0.00063 | 0.00260 | 93 | 0.00422 | 1.01126 | 131.892 | 0.31375 | 0.77376 |
| metB; cystathionine gamma-synthase [EC:2.5.1.48] | 0.04904 | 0.00183 | 0.05135 | 0.00067 | 0.00320 | 65 | 0.04742 | 0.00044 | 0.00217 | 93 | 0.00386 | 1.01608 | 118.738 | 0.31166 | 0.77376 |
| spxA; regulatory protein spx | 0.04904 | 0.00330 | 0.04868 | 0.00159 | 0.00494 | 65 | 0.04929 | 0.00183 | 0.00444 | 93 | 0.00664 | -0.09134 | 143.742 | 0.92735 | 0.98137 |
| lrp; Lrp/AsnC family transcriptional regulator, leucine-responsive regulatory protein | 0.04885 | 0.00318 | 0.04562 | 0.00146 | 0.00474 | 65 | 0.05111 | 0.00170 | 0.00427 | 93 | 0.00638 | -0.86164 | 144.055 | 0.39032 | 0.80384 |
| tdk, TK; thymidine kinase [EC:2.7.1.21] | 0.04878 | 0.00183 | 0.04796 | 0.00055 | 0.00291 | 65 | 0.04936 | 0.00052 | 0.00236 | 93 | 0.00374 | -0.37478 | 135.031 | 0.70841 | 0.92004 |
| umuC; DNA polymerase V | 0.04876 | 0.00201 | 0.04861 | 0.00069 | 0.00325 | 65 | 0.04886 | 0.00061 | 0.00255 | 93 | 0.00414 | -0.06110 | 132.157 | 0.95137 | 0.98792 |
| tpx; thiol peroxidase, atypical 2-Cys peroxiredoxin [EC:1.11.1.15] | 0.04873 | 0.00195 | 0.04959 | 0.00063 | 0.00310 | 65 | 0.04813 | 0.00059 | 0.00252 | 93 | 0.00400 | 0.36720 | 135.100 | 0.71405 | 0.92004 |
| rpiA; ribose 5-phosphate isomerase A [EC:5.3.1.6] | 0.04852 | 0.00212 | 0.04907 | 0.00081 | 0.00352 | 65 | 0.04813 | 0.00065 | 0.00264 | 93 | 0.00440 | 0.21320 | 127.929 | 0.83151 | 0.95506 |
| pnuC; nicotinamide mononucleotide transporter | 0.04844 | 0.00186 | 0.04817 | 0.00067 | 0.00321 | 65 | 0.04862 | 0.00047 | 0.00224 | 93 | 0.00392 | -0.11557 | 121.623 | 0.90819 | 0.97664 |
| pyrDI; dihydroorotate dehydrogenase (NAD+) catalytic subunit [EC:1.3.1.14] | 0.04840 | 0.00228 | 0.05049 | 0.00080 | 0.00351 | 65 | 0.04693 | 0.00083 | 0.00299 | 93 | 0.00461 | 0.77206 | 139.557 | 0.44138 | 0.83186 |
| ccdA; cytochrome c-type biogenesis protein | 0.04828 | 0.00292 | 0.04754 | 0.00128 | 0.00444 | 65 | 0.04879 | 0.00141 | 0.00390 | 93 | 0.00591 | -0.21070 | 142.010 | 0.83342 | 0.95506 |
| moxR; MoxR-like ATPase [EC:3.6.3.-] | 0.04815 | 0.00276 | 0.04424 | 0.00138 | 0.00462 | 65 | 0.05089 | 0.00107 | 0.00340 | 93 | 0.00573 | -1.16046 | 126.307 | 0.24805 | 0.77376 |
| ABC.SN.A; NitT/TauT family transport system ATP-binding protein | 0.04811 | 0.00295 | 0.04130 | 0.00105 | 0.00402 | 65 | 0.05287 | 0.00156 | 0.00409 | 93 | 0.00574 | -2.01721 | 151.947 | 0.04543 | 0.65981 |
| zntA; Cd2+/Zn2+-exporting ATPase [EC:3.6.3.3 3.6.3.5] | 0.04811 | 0.00217 | 0.05116 | 0.00094 | 0.00380 | 65 | 0.04598 | 0.00061 | 0.00256 | 93 | 0.00458 | 1.12943 | 118.357 | 0.26100 | 0.77376 |
| TC.CPA1; monovalent cation:H+ antiporter, CPA1 family | 0.04804 | 0.00226 | 0.04889 | 0.00115 | 0.00420 | 65 | 0.04745 | 0.00058 | 0.00250 | 93 | 0.00489 | 0.29391 | 108.005 | 0.76939 | 0.93993 |
| ribD; diaminohydroxyphosphoribosylaminopyrimidine deaminase / 5-amino-6-(5-phosphoribosylamino)uracil reductase [EC:3.5.4.26 1.1.1.193] | 0.04803 | 0.00141 | 0.04787 | 0.00033 | 0.00224 | 65 | 0.04815 | 0.00031 | 0.00183 | 93 | 0.00289 | -0.09676 | 135.819 | 0.92306 | 0.97947 |
| mraZ; MraZ protein | 0.04801 | 0.00145 | 0.04839 | 0.00035 | 0.00232 | 65 | 0.04775 | 0.00033 | 0.00188 | 93 | 0.00298 | 0.21381 | 135.181 | 0.83102 | 0.95506 |
| sufD; Fe-S cluster assembly protein SufD | 0.04800 | 0.00190 | 0.04595 | 0.00062 | 0.00309 | 65 | 0.04944 | 0.00054 | 0.00240 | 93 | 0.00392 | -0.89040 | 131.231 | 0.37488 | 0.79810 |
| K06960; uncharacterized protein | 0.04791 | 0.00206 | 0.05120 | 0.00066 | 0.00317 | 65 | 0.04560 | 0.00068 | 0.00270 | 93 | 0.00417 | 1.34325 | 139.492 | 0.18137 | 0.77376 |
| jag; spoIIIJ-associated protein | 0.04785 | 0.00206 | 0.05124 | 0.00065 | 0.00316 | 65 | 0.04548 | 0.00068 | 0.00271 | 93 | 0.00416 | 1.38416 | 140.004 | 0.16851 | 0.77376 |
| thiN, TPK1, THI80; thiamine pyrophosphokinase [EC:2.7.6.2] | 0.04783 | 0.00218 | 0.04924 | 0.00079 | 0.00348 | 65 | 0.04684 | 0.00073 | 0.00280 | 93 | 0.00447 | 0.53656 | 134.541 | 0.59246 | 0.87921 |
| ndk, NME; nucleoside-diphosphate kinase [EC:2.7.4.6] | 0.04772 | 0.00116 | 0.04796 | 0.00028 | 0.00208 | 65 | 0.04756 | 0.00017 | 0.00135 | 93 | 0.00248 | 0.16228 | 114.769 | 0.87137 | 0.96576 |
| yjbB; phosphate:Na+ symporter | 0.04771 | 0.00188 | 0.04673 | 0.00054 | 0.00288 | 65 | 0.04839 | 0.00058 | 0.00250 | 93 | 0.00381 | -0.43628 | 141.020 | 0.66330 | 0.91006 |
| aroQ, qutE; 3-dehydroquinate dehydratase II [EC:4.2.1.10] | 0.04769 | 0.00163 | 0.04817 | 0.00045 | 0.00264 | 65 | 0.04735 | 0.00040 | 0.00207 | 93 | 0.00336 | 0.24608 | 132.190 | 0.80600 | 0.94958 |
| srtA; sortase A [EC:3.4.22.70] | 0.04764 | 0.00261 | 0.04798 | 0.00125 | 0.00438 | 65 | 0.04740 | 0.00096 | 0.00322 | 93 | 0.00544 | 0.10720 | 126.146 | 0.91480 | 0.97664 |
| bdhAB; butanol dehydrogenase [EC:1.1.1.-] | 0.04756 | 0.00286 | 0.04706 | 0.00111 | 0.00414 | 65 | 0.04791 | 0.00143 | 0.00392 | 93 | 0.00570 | -0.14809 | 147.620 | 0.88248 | 0.96962 |
| fabK; enoyl-[acyl-carrier protein] reductase II [EC:1.3.1.9] | 0.04753 | 0.00232 | 0.05120 | 0.00099 | 0.00391 | 65 | 0.04496 | 0.00075 | 0.00283 | 93 | 0.00483 | 1.29126 | 124.828 | 0.19900 | 0.77376 |
| rluA; tRNA pseudouridine32 synthase / 23S rRNA pseudouridine746 synthase [EC:5.4.99.28 5.4.99.29] | 0.04750 | 0.00215 | 0.04579 | 0.00070 | 0.00329 | 65 | 0.04869 | 0.00075 | 0.00285 | 93 | 0.00435 | -0.66705 | 140.742 | 0.50583 | 0.84956 |
| argG, ASS1; argininosuccinate synthase [EC:6.3.4.5] | 0.04750 | 0.00122 | 0.04742 | 0.00027 | 0.00204 | 65 | 0.04756 | 0.00022 | 0.00152 | 93 | 0.00255 | -0.05545 | 127.523 | 0.95587 | 0.98792 |
| rho; transcription termination factor Rho | 0.04744 | 0.00140 | 0.04800 | 0.00036 | 0.00235 | 65 | 0.04705 | 0.00028 | 0.00172 | 93 | 0.00292 | 0.32562 | 125.985 | 0.74525 | 0.93189 |
| folB; 7,8-dihydroneopterin aldolase/epimerase/oxygenase [EC:4.1.2.25 5.1.99.8 1.13.11.81] | 0.04741 | 0.00138 | 0.04626 | 0.00035 | 0.00231 | 65 | 0.04821 | 0.00027 | 0.00170 | 93 | 0.00287 | -0.68172 | 126.490 | 0.49666 | 0.84797 |
| K07126; uncharacterized protein | 0.04730 | 0.00380 | 0.04539 | 0.00105 | 0.00402 | 65 | 0.04864 | 0.00316 | 0.00583 | 93 | 0.00708 | -0.45977 | 151.203 | 0.64634 | 0.90314 |
| acm; lysozyme | 0.04727 | 0.00323 | 0.04428 | 0.00131 | 0.00448 | 65 | 0.04936 | 0.00190 | 0.00452 | 93 | 0.00636 | -0.79810 | 151.384 | 0.42606 | 0.82652 |
| pyrDII; dihydroorotate dehydrogenase electron transfer subunit | 0.04723 | 0.00231 | 0.04844 | 0.00085 | 0.00362 | 65 | 0.04638 | 0.00084 | 0.00301 | 93 | 0.00471 | 0.43698 | 137.541 | 0.66281 | 0.91006 |
| rnz; ribonuclease Z [EC:3.1.26.11] | 0.04723 | 0.00187 | 0.04683 | 0.00059 | 0.00302 | 65 | 0.04751 | 0.00053 | 0.00239 | 93 | 0.00385 | -0.17663 | 132.957 | 0.86007 | 0.96137 |
| ytmI; uncharacterized N-acetyltransferase [EC:2.3.1.-] | 0.04708 | 0.00650 | 0.06207 | 0.01278 | 0.01402 | 65 | 0.03660 | 0.00225 | 0.00491 | 93 | 0.01486 | 1.71411 | 79.847 | 0.09039 | 0.76567 |
| BCP, PRXQ, DOT5; peroxiredoxin Q/BCP [EC:1.11.1.15] | 0.04693 | 0.00151 | 0.04351 | 0.00037 | 0.00239 | 65 | 0.04932 | 0.00035 | 0.00193 | 93 | 0.00307 | -1.89249 | 134.569 | 0.06057 | 0.73001 |
| K07098; uncharacterized protein | 0.04689 | 0.00240 | 0.04391 | 0.00081 | 0.00352 | 65 | 0.04897 | 0.00097 | 0.00324 | 93 | 0.00479 | -1.05650 | 145.507 | 0.29249 | 0.77376 |
| TC.FEV.OM3, tbpA, hemR, lbpA, hpuB, bhuR, hugA, hmbR; hemoglobin/transferrin/lactoferrin receptor protein | 0.04677 | 0.00427 | 0.04820 | 0.00344 | 0.00728 | 65 | 0.04577 | 0.00252 | 0.00521 | 93 | 0.00895 | 0.27107 | 123.734 | 0.78679 | 0.94461 |
| dapD; 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase [EC:2.3.1.117] | 0.04669 | 0.00167 | 0.04595 | 0.00044 | 0.00262 | 65 | 0.04721 | 0.00044 | 0.00217 | 93 | 0.00340 | -0.37117 | 137.132 | 0.71108 | 0.92004 |
| mrdA; penicillin-binding protein 2 [EC:3.4.16.4] | 0.04658 | 0.00203 | 0.04832 | 0.00059 | 0.00302 | 65 | 0.04536 | 0.00070 | 0.00274 | 93 | 0.00408 | 0.72663 | 144.452 | 0.46863 | 0.83837 |
| panE, apbA; 2-dehydropantoate 2-reductase [EC:1.1.1.169] | 0.04654 | 0.00206 | 0.04776 | 0.00085 | 0.00361 | 65 | 0.04568 | 0.00055 | 0.00244 | 93 | 0.00436 | 0.47623 | 118.413 | 0.63479 | 0.89696 |
| tatC; sec-independent protein translocase protein TatC | 0.04647 | 0.00138 | 0.04332 | 0.00033 | 0.00225 | 65 | 0.04866 | 0.00027 | 0.00172 | 93 | 0.00283 | -1.88333 | 129.665 | 0.06190 | 0.73001 |
| bglX; beta-glucosidase [EC:3.2.1.21] | 0.04641 | 0.00327 | 0.04118 | 0.00103 | 0.00398 | 65 | 0.05006 | 0.00214 | 0.00479 | 93 | 0.00623 | -1.42580 | 155.997 | 0.15592 | 0.77376 |
| galT, GALT; UDPglucose–hexose-1-phosphate uridylyltransferase [EC:2.7.7.12] | 0.04631 | 0.00196 | 0.05102 | 0.00063 | 0.00312 | 65 | 0.04302 | 0.00057 | 0.00248 | 93 | 0.00398 | 2.00890 | 133.291 | 0.04657 | 0.66255 |
| ecfA1; energy-coupling factor transport system ATP-binding protein [EC:3.6.3.-] | 0.04626 | 0.00222 | 0.04966 | 0.00078 | 0.00346 | 65 | 0.04389 | 0.00078 | 0.00289 | 93 | 0.00451 | 1.28088 | 137.815 | 0.20239 | 0.77376 |
| yraN; putative endonuclease | 0.04624 | 0.00162 | 0.04795 | 0.00043 | 0.00257 | 65 | 0.04503 | 0.00041 | 0.00209 | 93 | 0.00331 | 0.88096 | 135.483 | 0.37990 | 0.79852 |
| potD; spermidine/putrescine transport system substrate-binding protein | 0.04622 | 0.00211 | 0.04932 | 0.00079 | 0.00348 | 65 | 0.04405 | 0.00064 | 0.00263 | 93 | 0.00436 | 1.20691 | 128.873 | 0.22968 | 0.77376 |
| bcr, tcaB; MFS transporter, DHA1 family, multidrug resistance protein | 0.04621 | 0.00220 | 0.04393 | 0.00072 | 0.00334 | 65 | 0.04780 | 0.00079 | 0.00292 | 93 | 0.00443 | -0.87237 | 141.727 | 0.38448 | 0.80197 |
| padR; PadR family transcriptional regulator, regulatory protein PadR | 0.04616 | 0.00266 | 0.04371 | 0.00083 | 0.00356 | 65 | 0.04787 | 0.00132 | 0.00377 | 93 | 0.00519 | -0.80060 | 153.586 | 0.42460 | 0.82505 |
| rbgA; ribosome biogenesis GTPase A | 0.04603 | 0.00199 | 0.04968 | 0.00065 | 0.00317 | 65 | 0.04348 | 0.00060 | 0.00254 | 93 | 0.00406 | 1.52755 | 134.042 | 0.12898 | 0.77376 |
| PDHB, pdhB; pyruvate dehydrogenase E1 component beta subunit [EC:1.2.4.1] | 0.04603 | 0.00206 | 0.04411 | 0.00059 | 0.00301 | 65 | 0.04737 | 0.00073 | 0.00280 | 93 | 0.00412 | -0.79121 | 146.380 | 0.43010 | 0.82749 |
| hutH, HAL; histidine ammonia-lyase [EC:4.3.1.3] | 0.04603 | 0.00219 | 0.04551 | 0.00086 | 0.00364 | 65 | 0.04639 | 0.00069 | 0.00272 | 93 | 0.00455 | -0.19324 | 127.835 | 0.84708 | 0.95682 |
| E3.6.3.8; Ca2+-transporting ATPase [EC:3.6.3.8] | 0.04603 | 0.00257 | 0.04705 | 0.00113 | 0.00418 | 65 | 0.04531 | 0.00099 | 0.00326 | 93 | 0.00530 | 0.32845 | 131.588 | 0.74309 | 0.93090 |
| PDHA, pdhA; pyruvate dehydrogenase E1 component alpha subunit [EC:1.2.4.1] | 0.04602 | 0.00207 | 0.04400 | 0.00059 | 0.00302 | 65 | 0.04743 | 0.00074 | 0.00281 | 93 | 0.00413 | -0.83312 | 146.548 | 0.40614 | 0.81189 |
| ribE, RIB5; riboflavin synthase [EC:2.5.1.9] | 0.04571 | 0.00147 | 0.04593 | 0.00036 | 0.00236 | 65 | 0.04555 | 0.00033 | 0.00188 | 93 | 0.00302 | 0.12683 | 133.745 | 0.89927 | 0.97435 |
| lolC_E; lipoprotein-releasing system permease protein | 0.04569 | 0.00231 | 0.04588 | 0.00082 | 0.00354 | 65 | 0.04556 | 0.00087 | 0.00307 | 93 | 0.00469 | 0.06823 | 140.768 | 0.94570 | 0.98674 |
| wecB; UDP-N-acetylglucosamine 2-epimerase (non-hydrolysing) [EC:5.1.3.14] | 0.04554 | 0.00168 | 0.04477 | 0.00053 | 0.00285 | 65 | 0.04608 | 0.00039 | 0.00205 | 93 | 0.00351 | -0.37363 | 124.457 | 0.70932 | 0.92004 |
| bioB; biotin synthase [EC:2.8.1.6] | 0.04545 | 0.00185 | 0.04684 | 0.00061 | 0.00307 | 65 | 0.04449 | 0.00050 | 0.00231 | 93 | 0.00385 | 0.61165 | 128.353 | 0.54185 | 0.86207 |
| dapF; diaminopimelate epimerase [EC:5.1.1.7] | 0.04544 | 0.00130 | 0.04587 | 0.00029 | 0.00212 | 65 | 0.04514 | 0.00026 | 0.00166 | 93 | 0.00269 | 0.27159 | 131.765 | 0.78636 | 0.94458 |
| mlaE, linK; phospholipid/cholesterol/gamma-HCH transport system permease protein | 0.04535 | 0.00256 | 0.04063 | 0.00056 | 0.00293 | 65 | 0.04865 | 0.00135 | 0.00381 | 93 | 0.00480 | -1.66978 | 155.086 | 0.09698 | 0.76673 |
| oppD; oligopeptide transport system ATP-binding protein | 0.04532 | 0.00218 | 0.04556 | 0.00072 | 0.00332 | 65 | 0.04515 | 0.00079 | 0.00291 | 93 | 0.00441 | 0.09359 | 141.805 | 0.92557 | 0.98041 |
| ubiE; demethylmenaquinone methyltransferase / 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase [EC:2.1.1.163 2.1.1.201] | 0.04532 | 0.00146 | 0.04471 | 0.00034 | 0.00229 | 65 | 0.04574 | 0.00034 | 0.00190 | 93 | 0.00297 | -0.34488 | 137.286 | 0.73071 | 0.92479 |
| fklB; FKBP-type peptidyl-prolyl cis-trans isomerase FklB [EC:5.2.1.8] | 0.04528 | 0.00321 | 0.04353 | 0.00120 | 0.00429 | 65 | 0.04651 | 0.00194 | 0.00457 | 93 | 0.00627 | -0.47466 | 153.849 | 0.63570 | 0.89696 |
| phnA; protein PhnA | 0.04528 | 0.00181 | 0.04525 | 0.00054 | 0.00288 | 65 | 0.04530 | 0.00051 | 0.00234 | 93 | 0.00371 | -0.01341 | 135.086 | 0.98932 | 0.99801 |
| K09790; uncharacterized protein | 0.04526 | 0.00160 | 0.04704 | 0.00039 | 0.00244 | 65 | 0.04402 | 0.00041 | 0.00211 | 93 | 0.00323 | 0.93526 | 140.409 | 0.35126 | 0.79120 |
| rmuC; DNA recombination protein RmuC | 0.04516 | 0.00145 | 0.04378 | 0.00038 | 0.00241 | 65 | 0.04613 | 0.00030 | 0.00180 | 93 | 0.00301 | -0.77697 | 127.645 | 0.43861 | 0.83152 |
| hemE, UROD; uroporphyrinogen decarboxylase [EC:4.1.1.37] | 0.04512 | 0.00166 | 0.04196 | 0.00041 | 0.00250 | 65 | 0.04734 | 0.00045 | 0.00219 | 93 | 0.00332 | -1.61775 | 141.941 | 0.10794 | 0.77376 |
| hisH; glutamine amidotransferase [EC:2.4.2.-] | 0.04503 | 0.00135 | 0.04291 | 0.00036 | 0.00236 | 65 | 0.04651 | 0.00024 | 0.00159 | 93 | 0.00284 | -1.26489 | 118.437 | 0.20839 | 0.77376 |
| TC.HAE1; hydrophobic/amphiphilic exporter-1 (mainly G- bacteria), HAE1 family | 0.04501 | 0.00221 | 0.04013 | 0.00066 | 0.00318 | 65 | 0.04843 | 0.00083 | 0.00299 | 93 | 0.00437 | -1.90030 | 147.097 | 0.05935 | 0.72498 |
| afuB, fbpB; iron(III) transport system permease protein | 0.04498 | 0.00235 | 0.04725 | 0.00105 | 0.00402 | 65 | 0.04339 | 0.00075 | 0.00283 | 93 | 0.00492 | 0.78526 | 122.335 | 0.43382 | 0.82874 |
| fnr; CRP/FNR family transcriptional regulator, anaerobic regulatory protein | 0.04497 | 0.00248 | 0.04674 | 0.00113 | 0.00417 | 65 | 0.04373 | 0.00087 | 0.00306 | 93 | 0.00518 | 0.58068 | 126.061 | 0.56250 | 0.86813 |
| yesN; two-component system, response regulator YesN | 0.04491 | 0.00328 | 0.04834 | 0.00235 | 0.00602 | 65 | 0.04252 | 0.00124 | 0.00366 | 93 | 0.00704 | 0.82739 | 109.622 | 0.40981 | 0.81416 |
| ypsC; putative N6-adenine-specific DNA methylase [EC:2.1.1.-] | 0.04490 | 0.00195 | 0.04424 | 0.00065 | 0.00316 | 65 | 0.04536 | 0.00058 | 0.00249 | 93 | 0.00402 | -0.28027 | 132.668 | 0.77971 | 0.94269 |
| K07112; uncharacterized protein | 0.04487 | 0.00262 | 0.04297 | 0.00109 | 0.00409 | 65 | 0.04619 | 0.00109 | 0.00343 | 93 | 0.00533 | -0.60396 | 138.157 | 0.54686 | 0.86503 |
| potC; spermidine/putrescine transport system permease protein | 0.04486 | 0.00220 | 0.04813 | 0.00101 | 0.00394 | 65 | 0.04257 | 0.00059 | 0.00253 | 93 | 0.00468 | 1.18902 | 114.161 | 0.23690 | 0.77376 |
| psd, PISD; phosphatidylserine decarboxylase [EC:4.1.1.65] | 0.04478 | 0.00137 | 0.04459 | 0.00033 | 0.00227 | 65 | 0.04491 | 0.00027 | 0.00171 | 93 | 0.00284 | -0.11486 | 128.509 | 0.90874 | 0.97664 |
| mlaD, linM; phospholipid/cholesterol/gamma-HCH transport system substrate-binding protein | 0.04476 | 0.00261 | 0.04055 | 0.00058 | 0.00299 | 65 | 0.04770 | 0.00141 | 0.00389 | 93 | 0.00490 | -1.45891 | 155.026 | 0.14661 | 0.77376 |
| xerC; integrase/recombinase XerC | 0.04475 | 0.00152 | 0.04261 | 0.00036 | 0.00234 | 65 | 0.04625 | 0.00037 | 0.00200 | 93 | 0.00308 | -1.18106 | 139.656 | 0.23959 | 0.77376 |
| K00666; fatty-acyl-CoA synthase [EC:6.2.1.-] | 0.04460 | 0.00417 | 0.03465 | 0.00175 | 0.00519 | 65 | 0.05156 | 0.00335 | 0.00601 | 93 | 0.00794 | -2.12987 | 155.812 | 0.03475 | 0.62130 |
| tlyC; putative hemolysin | 0.04456 | 0.00177 | 0.04783 | 0.00047 | 0.00269 | 65 | 0.04227 | 0.00050 | 0.00232 | 93 | 0.00355 | 1.56476 | 140.599 | 0.11989 | 0.77376 |
| fruR2, fruR; DeoR family transcriptional regulator, fructose operon transcriptional repressor | 0.04454 | 0.00216 | 0.04595 | 0.00070 | 0.00328 | 65 | 0.04356 | 0.00077 | 0.00288 | 93 | 0.00436 | 0.54810 | 142.015 | 0.58448 | 0.87720 |
| hemL; glutamate-1-semialdehyde 2,1-aminomutase [EC:5.4.3.8] | 0.04452 | 0.00162 | 0.04570 | 0.00034 | 0.00230 | 65 | 0.04369 | 0.00047 | 0.00225 | 93 | 0.00322 | 0.62512 | 149.661 | 0.53285 | 0.86207 |
| K07139; uncharacterized protein | 0.04437 | 0.00219 | 0.04555 | 0.00081 | 0.00353 | 65 | 0.04355 | 0.00072 | 0.00279 | 93 | 0.00450 | 0.44647 | 132.622 | 0.65599 | 0.90679 |
| agrA, blpR, fsrA; two-component system, LytTR family, response regulator AgrA | 0.04434 | 0.00422 | 0.04117 | 0.00181 | 0.00528 | 65 | 0.04656 | 0.00352 | 0.00615 | 93 | 0.00811 | -0.66552 | 155.875 | 0.50670 | 0.84956 |
| pqqL; zinc protease [EC:3.4.24.-] | 0.04426 | 0.00295 | 0.04150 | 0.00113 | 0.00417 | 65 | 0.04619 | 0.00155 | 0.00409 | 93 | 0.00584 | -0.80254 | 149.878 | 0.42351 | 0.82432 |
| K07030; uncharacterized protein | 0.04420 | 0.00204 | 0.04712 | 0.00069 | 0.00327 | 65 | 0.04216 | 0.00063 | 0.00260 | 93 | 0.00418 | 1.18723 | 133.476 | 0.23724 | 0.77376 |
| feoB; ferrous iron transport protein B | 0.04413 | 0.00236 | 0.04422 | 0.00069 | 0.00327 | 65 | 0.04407 | 0.00102 | 0.00331 | 93 | 0.00465 | 0.03173 | 151.588 | 0.97473 | 0.99546 |
| ABC.GLN1.P; putative glutamine transport system permease protein | 0.04413 | 0.00353 | 0.04526 | 0.00199 | 0.00554 | 65 | 0.04333 | 0.00197 | 0.00460 | 93 | 0.00720 | 0.26824 | 137.300 | 0.78892 | 0.94606 |
| thiI; tRNA uracil 4-sulfurtransferase [EC:2.8.1.4] | 0.04409 | 0.00177 | 0.04569 | 0.00050 | 0.00276 | 65 | 0.04297 | 0.00049 | 0.00230 | 93 | 0.00360 | 0.75787 | 137.534 | 0.44983 | 0.83438 |
| ppaC; manganese-dependent inorganic pyrophosphatase [EC:3.6.1.1] | 0.04408 | 0.00204 | 0.04765 | 0.00070 | 0.00329 | 65 | 0.04158 | 0.00062 | 0.00258 | 93 | 0.00418 | 1.45087 | 132.184 | 0.14919 | 0.77376 |
| trpC; indole-3-glycerol phosphate synthase [EC:4.1.1.48] | 0.04400 | 0.00136 | 0.04098 | 0.00031 | 0.00219 | 65 | 0.04611 | 0.00027 | 0.00172 | 93 | 0.00278 | -1.84411 | 132.195 | 0.06741 | 0.73334 |
| ruvC; crossover junction endodeoxyribonuclease RuvC [EC:3.1.22.4] | 0.04393 | 0.00158 | 0.04488 | 0.00041 | 0.00252 | 65 | 0.04326 | 0.00038 | 0.00202 | 93 | 0.00323 | 0.49950 | 134.159 | 0.61824 | 0.88903 |
| ndh; NADH dehydrogenase [EC:1.6.99.3] | 0.04382 | 0.00158 | 0.04156 | 0.00037 | 0.00240 | 65 | 0.04539 | 0.00040 | 0.00209 | 93 | 0.00318 | -1.20651 | 141.310 | 0.22964 | 0.77376 |
| ABC.SN.P; NitT/TauT family transport system permease protein | 0.04371 | 0.00297 | 0.03857 | 0.00135 | 0.00456 | 65 | 0.04731 | 0.00140 | 0.00388 | 93 | 0.00599 | -1.46015 | 139.236 | 0.14650 | 0.77376 |
| K09787; uncharacterized protein | 0.04367 | 0.00206 | 0.04746 | 0.00072 | 0.00333 | 65 | 0.04102 | 0.00063 | 0.00260 | 93 | 0.00422 | 1.52522 | 131.625 | 0.12960 | 0.77376 |
| CHO1, pssA; CDP-diacylglycerol—serine O-phosphatidyltransferase [EC:2.7.8.8] | 0.04366 | 0.00151 | 0.04237 | 0.00039 | 0.00246 | 65 | 0.04456 | 0.00034 | 0.00190 | 93 | 0.00311 | -0.70360 | 130.897 | 0.48293 | 0.84354 |
| pepQ; Xaa-Pro dipeptidase [EC:3.4.13.9] | 0.04356 | 0.00188 | 0.04357 | 0.00058 | 0.00298 | 65 | 0.04355 | 0.00056 | 0.00245 | 93 | 0.00385 | 0.00586 | 136.310 | 0.99533 | 0.99944 |
| vanY; zinc D-Ala-D-Ala carboxypeptidase [EC:3.4.17.14] | 0.04356 | 0.00270 | 0.04245 | 0.00105 | 0.00401 | 65 | 0.04433 | 0.00123 | 0.00364 | 93 | 0.00542 | -0.34613 | 144.605 | 0.72975 | 0.92426 |
| E2.2.1.2, talA, talB; transaldolase [EC:2.2.1.2] | 0.04351 | 0.00134 | 0.04150 | 0.00030 | 0.00215 | 65 | 0.04492 | 0.00027 | 0.00171 | 93 | 0.00275 | -1.24236 | 133.477 | 0.21628 | 0.77376 |
| nrdI; protein involved in ribonucleotide reduction | 0.04322 | 0.00257 | 0.04399 | 0.00116 | 0.00423 | 65 | 0.04267 | 0.00097 | 0.00322 | 93 | 0.00532 | 0.24789 | 129.489 | 0.80461 | 0.94931 |
| hisA; phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase [EC:5.3.1.16] | 0.04319 | 0.00119 | 0.04167 | 0.00025 | 0.00195 | 65 | 0.04426 | 0.00021 | 0.00151 | 93 | 0.00246 | -1.05137 | 130.978 | 0.29503 | 0.77376 |
| potB; spermidine/putrescine transport system permease protein | 0.04313 | 0.00197 | 0.04587 | 0.00071 | 0.00331 | 65 | 0.04121 | 0.00055 | 0.00242 | 93 | 0.00410 | 1.13711 | 125.963 | 0.25765 | 0.77376 |
| TC.POT; proton-dependent oligopeptide transporter, POT family | 0.04308 | 0.00221 | 0.04123 | 0.00091 | 0.00373 | 65 | 0.04438 | 0.00068 | 0.00271 | 93 | 0.00461 | -0.68330 | 124.927 | 0.49568 | 0.84797 |
| cutC; copper homeostasis protein | 0.04293 | 0.00182 | 0.04418 | 0.00049 | 0.00275 | 65 | 0.04206 | 0.00055 | 0.00244 | 93 | 0.00368 | 0.57794 | 142.775 | 0.56422 | 0.86944 |
| trmK; tRNA (adenine22-N1)-methyltransferase [EC:2.1.1.217] | 0.04293 | 0.00181 | 0.04422 | 0.00058 | 0.00300 | 65 | 0.04202 | 0.00048 | 0.00227 | 93 | 0.00376 | 0.58335 | 128.964 | 0.56068 | 0.86813 |
| afuC, fbpC; iron(III) transport system ATP-binding protein [EC:3.6.3.30] | 0.04290 | 0.00205 | 0.04435 | 0.00083 | 0.00358 | 65 | 0.04189 | 0.00055 | 0.00244 | 93 | 0.00433 | 0.56895 | 119.461 | 0.57046 | 0.87143 |
| queC; 7-cyano-7-deazaguanine synthase [EC:6.3.4.20] | 0.04289 | 0.00134 | 0.04021 | 0.00030 | 0.00215 | 65 | 0.04477 | 0.00026 | 0.00169 | 93 | 0.00274 | -1.66983 | 132.159 | 0.09732 | 0.76673 |
| potA; spermidine/putrescine transport system ATP-binding protein [EC:3.6.3.31] | 0.04288 | 0.00196 | 0.04506 | 0.00070 | 0.00328 | 65 | 0.04137 | 0.00054 | 0.00242 | 93 | 0.00407 | 0.90566 | 126.570 | 0.36683 | 0.79587 |
| cas2; CRISPR-associated protein Cas2 | 0.04279 | 0.00214 | 0.04701 | 0.00083 | 0.00357 | 65 | 0.03983 | 0.00064 | 0.00263 | 93 | 0.00443 | 1.61939 | 126.383 | 0.10786 | 0.77376 |
| sigH; RNA polymerase sporulation-specific sigma factor | 0.04278 | 0.00348 | 0.04264 | 0.00167 | 0.00507 | 65 | 0.04288 | 0.00211 | 0.00476 | 93 | 0.00696 | -0.03370 | 147.041 | 0.97316 | 0.99509 |
| queF; 7-cyano-7-deazaguanine reductase [EC:1.7.1.13] | 0.04271 | 0.00133 | 0.03969 | 0.00029 | 0.00213 | 65 | 0.04482 | 0.00026 | 0.00167 | 93 | 0.00271 | -1.89508 | 132.394 | 0.06026 | 0.73001 |
| K11145; ribonuclease III family protein [EC:3.1.26.-] | 0.04271 | 0.00203 | 0.04585 | 0.00069 | 0.00325 | 65 | 0.04051 | 0.00062 | 0.00257 | 93 | 0.00415 | 1.28981 | 133.016 | 0.19936 | 0.77376 |
| argC; N-acetyl-gamma-glutamyl-phosphate reductase [EC:1.2.1.38] | 0.04257 | 0.00129 | 0.04081 | 0.00032 | 0.00220 | 65 | 0.04381 | 0.00022 | 0.00155 | 93 | 0.00269 | -1.11505 | 122.187 | 0.26702 | 0.77376 |
| ftsI; cell division protein FtsI (penicillin-binding protein 3) [EC:3.4.16.4] | 0.04256 | 0.00164 | 0.04317 | 0.00044 | 0.00259 | 65 | 0.04214 | 0.00042 | 0.00213 | 93 | 0.00335 | 0.30778 | 136.388 | 0.75872 | 0.93719 |
| hisF; cyclase [EC:4.1.3.-] | 0.04243 | 0.00135 | 0.04011 | 0.00036 | 0.00236 | 65 | 0.04405 | 0.00023 | 0.00158 | 93 | 0.00284 | -1.38888 | 117.895 | 0.16749 | 0.77376 |
| serC, PSAT1; phosphoserine aminotransferase [EC:2.6.1.52] | 0.04237 | 0.00126 | 0.04037 | 0.00029 | 0.00210 | 65 | 0.04376 | 0.00022 | 0.00155 | 93 | 0.00261 | -1.29712 | 126.485 | 0.19695 | 0.77376 |
| metH, MTR; 5-methyltetrahydrofolate–homocysteine methyltransferase [EC:2.1.1.13] | 0.04232 | 0.00188 | 0.04144 | 0.00054 | 0.00289 | 65 | 0.04293 | 0.00057 | 0.00248 | 93 | 0.00381 | -0.39038 | 140.114 | 0.69685 | 0.91823 |
| metA; homoserine O-succinyltransferase/O-acetyltransferase [EC:2.3.1.46 2.3.1.31] | 0.04229 | 0.00214 | 0.04233 | 0.00081 | 0.00352 | 65 | 0.04226 | 0.00068 | 0.00270 | 93 | 0.00444 | 0.01540 | 130.113 | 0.98773 | 0.99801 |
| addA; ATP-dependent helicase/nuclease subunit A [EC:3.1.-.- 3.6.4.12] | 0.04220 | 0.00196 | 0.04196 | 0.00067 | 0.00321 | 65 | 0.04236 | 0.00056 | 0.00246 | 93 | 0.00405 | -0.09910 | 130.003 | 0.92121 | 0.97905 |
| hprK, ptsK; HPr kinase/phosphorylase [EC:2.7.11.- 2.7.4.-] | 0.04215 | 0.00181 | 0.04422 | 0.00055 | 0.00290 | 65 | 0.04070 | 0.00050 | 0.00231 | 93 | 0.00371 | 0.94874 | 133.585 | 0.34447 | 0.78711 |
| fdoG, fdhF, fdwA; formate dehydrogenase major subunit [EC:1.17.1.9] | 0.04213 | 0.00250 | 0.04319 | 0.00106 | 0.00404 | 65 | 0.04139 | 0.00094 | 0.00319 | 93 | 0.00515 | 0.34818 | 132.596 | 0.72826 | 0.92350 |
| serB, PSPH; phosphoserine phosphatase [EC:3.1.3.3] | 0.04210 | 0.00159 | 0.03984 | 0.00045 | 0.00264 | 65 | 0.04368 | 0.00036 | 0.00197 | 93 | 0.00329 | -1.16695 | 127.695 | 0.24540 | 0.77376 |
| K09155; uncharacterized protein | 0.04209 | 0.00237 | 0.04626 | 0.00099 | 0.00391 | 65 | 0.03917 | 0.00081 | 0.00295 | 93 | 0.00490 | 1.44933 | 128.591 | 0.14968 | 0.77376 |
| lpxA; UDP-N-acetylglucosamine acyltransferase [EC:2.3.1.129] | 0.04203 | 0.00210 | 0.04232 | 0.00063 | 0.00312 | 65 | 0.04183 | 0.00075 | 0.00284 | 93 | 0.00422 | 0.11591 | 144.799 | 0.90789 | 0.97664 |
| pepD; dipeptidase D [EC:3.4.13.-] | 0.04199 | 0.00350 | 0.04800 | 0.00246 | 0.00616 | 65 | 0.03780 | 0.00154 | 0.00407 | 93 | 0.00738 | 1.38324 | 116.567 | 0.16924 | 0.77376 |
| ACSS, acs; acetyl-CoA synthetase [EC:6.2.1.1] | 0.04195 | 0.00271 | 0.03502 | 0.00100 | 0.00392 | 65 | 0.04678 | 0.00123 | 0.00363 | 93 | 0.00534 | -2.20154 | 146.128 | 0.02927 | 0.61334 |
| fhaB; filamentous hemagglutinin | 0.04181 | 0.00620 | 0.05479 | 0.00898 | 0.01176 | 65 | 0.03273 | 0.00391 | 0.00648 | 93 | 0.01342 | 1.64331 | 102.272 | 0.10339 | 0.77376 |
| oppC; oligopeptide transport system permease protein | 0.04180 | 0.00206 | 0.04114 | 0.00065 | 0.00317 | 65 | 0.04227 | 0.00069 | 0.00272 | 93 | 0.00417 | -0.27144 | 140.203 | 0.78645 | 0.94458 |
| E4.1.1.49, pckA; phosphoenolpyruvate carboxykinase (ATP) [EC:4.1.1.49] | 0.04176 | 0.00162 | 0.04326 | 0.00049 | 0.00273 | 65 | 0.04071 | 0.00037 | 0.00199 | 93 | 0.00338 | 0.75368 | 125.300 | 0.45246 | 0.83438 |
| ALDH; aldehyde dehydrogenase (NAD+) [EC:1.2.1.3] | 0.04175 | 0.00310 | 0.03274 | 0.00107 | 0.00406 | 65 | 0.04805 | 0.00175 | 0.00434 | 93 | 0.00594 | -2.57581 | 154.032 | 0.01094 | 0.58284 |
| deoD; purine-nucleoside phosphorylase [EC:2.4.2.1] | 0.04156 | 0.00196 | 0.04363 | 0.00056 | 0.00295 | 65 | 0.04011 | 0.00064 | 0.00262 | 93 | 0.00394 | 0.89305 | 143.027 | 0.37333 | 0.79810 |
| ispF; 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [EC:4.6.1.12] | 0.04152 | 0.00141 | 0.04240 | 0.00032 | 0.00222 | 65 | 0.04091 | 0.00031 | 0.00183 | 93 | 0.00288 | 0.51603 | 136.616 | 0.60666 | 0.88555 |
| oppB; oligopeptide transport system permease protein | 0.04150 | 0.00205 | 0.04096 | 0.00065 | 0.00316 | 65 | 0.04188 | 0.00069 | 0.00272 | 93 | 0.00417 | -0.21966 | 140.217 | 0.82646 | 0.95506 |
| por, nifJ; pyruvate-ferredoxin/flavodoxin oxidoreductase [EC:1.2.7.1 1.2.7.-] | 0.04145 | 0.00507 | 0.05369 | 0.00676 | 0.01020 | 65 | 0.03289 | 0.00203 | 0.00467 | 93 | 0.01122 | 1.85388 | 90.913 | 0.06700 | 0.73334 |
| mgtC; putative Mg2+ transporter-C (MgtC) family protein | 0.04144 | 0.00211 | 0.04319 | 0.00079 | 0.00349 | 65 | 0.04022 | 0.00064 | 0.00263 | 93 | 0.00437 | 0.68057 | 128.497 | 0.49737 | 0.84814 |
| moeA; molybdopterin molybdotransferase [EC:2.10.1.1] | 0.04138 | 0.00180 | 0.04007 | 0.00052 | 0.00284 | 65 | 0.04230 | 0.00051 | 0.00234 | 93 | 0.00368 | -0.60482 | 136.601 | 0.54630 | 0.86444 |
| acyP; acylphosphatase [EC:3.6.1.7] | 0.04137 | 0.00181 | 0.04293 | 0.00057 | 0.00297 | 65 | 0.04028 | 0.00048 | 0.00227 | 93 | 0.00374 | 0.70974 | 129.933 | 0.47914 | 0.84314 |
| dinJ; DNA-damage-inducible protein J | 0.04136 | 0.00380 | 0.04908 | 0.00398 | 0.00783 | 65 | 0.03596 | 0.00104 | 0.00335 | 93 | 0.00851 | 1.54146 | 87.546 | 0.12681 | 0.77376 |
| lplA, lplJ; lipoate—protein ligase [EC:6.3.1.20] | 0.04131 | 0.00252 | 0.04129 | 0.00135 | 0.00455 | 65 | 0.04133 | 0.00077 | 0.00289 | 93 | 0.00539 | -0.00613 | 113.079 | 0.99512 | 0.99944 |
| MFS.CP; MFS transporter, CP family, cyanate transporter | 0.04118 | 0.00206 | 0.03960 | 0.00058 | 0.00299 | 65 | 0.04228 | 0.00073 | 0.00281 | 93 | 0.00410 | -0.65279 | 147.154 | 0.51491 | 0.85216 |
| hisG; ATP phosphoribosyltransferase [EC:2.4.2.17] | 0.04112 | 0.00118 | 0.03899 | 0.00028 | 0.00207 | 65 | 0.04262 | 0.00018 | 0.00138 | 93 | 0.00249 | -1.45730 | 117.330 | 0.14770 | 0.77376 |
| argB; acetylglutamate kinase [EC:2.7.2.8] | 0.04086 | 0.00112 | 0.03908 | 0.00026 | 0.00198 | 65 | 0.04210 | 0.00016 | 0.00130 | 93 | 0.00237 | -1.27189 | 115.717 | 0.20596 | 0.77376 |
| katE, CAT, catB, srpA; catalase [EC:1.11.1.6] | 0.04079 | 0.00308 | 0.03637 | 0.00135 | 0.00456 | 65 | 0.04388 | 0.00159 | 0.00414 | 93 | 0.00616 | -1.22052 | 144.637 | 0.22425 | 0.77376 |
| msrA; peptide-methionine (S)-S-oxide reductase [EC:1.8.4.11] | 0.04078 | 0.00224 | 0.03843 | 0.00079 | 0.00348 | 65 | 0.04243 | 0.00079 | 0.00292 | 93 | 0.00455 | -0.88027 | 138.245 | 0.38024 | 0.79852 |
| PPIA; peptidyl-prolyl cis-trans isomerase A (cyclophilin A) [EC:5.2.1.8] | 0.04077 | 0.00164 | 0.04126 | 0.00048 | 0.00270 | 65 | 0.04042 | 0.00040 | 0.00207 | 93 | 0.00340 | 0.24529 | 129.787 | 0.80662 | 0.94982 |
| mlaF, linL, mkl; phospholipid/cholesterol/gamma-HCH transport system ATP-binding protein | 0.04076 | 0.00181 | 0.03915 | 0.00054 | 0.00287 | 65 | 0.04189 | 0.00050 | 0.00233 | 93 | 0.00370 | -0.74388 | 135.121 | 0.45824 | 0.83438 |
| amt, AMT, MEP; ammonium transporter, Amt family | 0.04076 | 0.00212 | 0.03794 | 0.00063 | 0.00311 | 65 | 0.04272 | 0.00076 | 0.00286 | 93 | 0.00423 | -1.13046 | 145.485 | 0.26014 | 0.77376 |
| pepF, pepB; oligoendopeptidase F [EC:3.4.24.-] | 0.04075 | 0.00184 | 0.04045 | 0.00050 | 0.00278 | 65 | 0.04097 | 0.00056 | 0.00245 | 93 | 0.00371 | -0.14074 | 142.578 | 0.88827 | 0.97039 |
| TC.SULP; sulfate permease, SulP family | 0.04070 | 0.00220 | 0.03912 | 0.00080 | 0.00351 | 65 | 0.04181 | 0.00075 | 0.00284 | 93 | 0.00452 | -0.59532 | 134.860 | 0.55263 | 0.86788 |
| hisD; histidinol dehydrogenase [EC:1.1.1.23] | 0.04069 | 0.00126 | 0.03837 | 0.00030 | 0.00214 | 65 | 0.04232 | 0.00022 | 0.00153 | 93 | 0.00263 | -1.50537 | 123.793 | 0.13478 | 0.77376 |
| CS, gltA; citrate synthase [EC:2.3.3.1] | 0.04068 | 0.00166 | 0.03509 | 0.00039 | 0.00244 | 65 | 0.04459 | 0.00043 | 0.00216 | 93 | 0.00326 | -2.91629 | 142.524 | 0.00412 | 0.52172 |
| BLMH, pepC; bleomycin hydrolase [EC:3.4.22.40] | 0.04065 | 0.00229 | 0.04056 | 0.00079 | 0.00350 | 65 | 0.04072 | 0.00087 | 0.00305 | 93 | 0.00464 | -0.03448 | 141.472 | 0.97254 | 0.99471 |
| hemC, HMBS; hydroxymethylbilane synthase [EC:2.5.1.61] | 0.04065 | 0.00134 | 0.04224 | 0.00028 | 0.00206 | 65 | 0.03953 | 0.00029 | 0.00176 | 93 | 0.00271 | 0.99957 | 139.825 | 0.31924 | 0.77376 |
| hemB, ALAD; porphobilinogen synthase [EC:4.2.1.24] | 0.04063 | 0.00135 | 0.04207 | 0.00028 | 0.00207 | 65 | 0.03961 | 0.00030 | 0.00178 | 93 | 0.00273 | 0.89892 | 140.395 | 0.37023 | 0.79587 |
| perR; Fur family transcriptional regulator, peroxide stress response regulator | 0.04062 | 0.00199 | 0.04427 | 0.00070 | 0.00329 | 65 | 0.03807 | 0.00056 | 0.00246 | 93 | 0.00410 | 1.51225 | 127.935 | 0.13294 | 0.77376 |
| ppc; phosphoenolpyruvate carboxylase [EC:4.1.1.31] | 0.04061 | 0.00140 | 0.04098 | 0.00033 | 0.00224 | 65 | 0.04036 | 0.00030 | 0.00180 | 93 | 0.00287 | 0.21578 | 134.006 | 0.82949 | 0.95506 |
| pulA; pullulanase [EC:3.2.1.41] | 0.04061 | 0.00273 | 0.03809 | 0.00108 | 0.00408 | 65 | 0.04237 | 0.00125 | 0.00367 | 93 | 0.00549 | -0.78125 | 143.850 | 0.43594 | 0.83006 |
| lctP; lactate permease | 0.04061 | 0.00244 | 0.04082 | 0.00111 | 0.00413 | 65 | 0.04046 | 0.00084 | 0.00300 | 93 | 0.00510 | 0.07132 | 125.093 | 0.94326 | 0.98634 |
| rnmV; ribonuclease M5 [EC:3.1.26.8] | 0.04052 | 0.00197 | 0.04356 | 0.00067 | 0.00321 | 65 | 0.03839 | 0.00057 | 0.00249 | 93 | 0.00406 | 1.27389 | 131.121 | 0.20495 | 0.77376 |
| yhgE; putative membrane protein | 0.04045 | 0.00223 | 0.03896 | 0.00075 | 0.00341 | 65 | 0.04150 | 0.00082 | 0.00297 | 93 | 0.00452 | -0.56143 | 141.399 | 0.57540 | 0.87389 |
| K07043; uncharacterized protein | 0.04035 | 0.00167 | 0.04063 | 0.00047 | 0.00270 | 65 | 0.04015 | 0.00042 | 0.00213 | 93 | 0.00344 | 0.14076 | 132.485 | 0.88828 | 0.97039 |
| cas1; CRISP-associated protein Cas1 | 0.04032 | 0.00192 | 0.04292 | 0.00062 | 0.00308 | 65 | 0.03850 | 0.00055 | 0.00244 | 93 | 0.00393 | 1.12486 | 133.108 | 0.26267 | 0.77376 |
| K09157; uncharacterized protein | 0.04019 | 0.00201 | 0.04277 | 0.00068 | 0.00323 | 65 | 0.03839 | 0.00061 | 0.00256 | 93 | 0.00412 | 1.06488 | 133.057 | 0.28886 | 0.77376 |
| E3.5.1.4, amiE; amidase [EC:3.5.1.4] | 0.04011 | 0.00318 | 0.03590 | 0.00105 | 0.00402 | 65 | 0.04305 | 0.00197 | 0.00460 | 93 | 0.00611 | -1.17051 | 155.675 | 0.24359 | 0.77376 |
| E4.2.1.2B, fumC, FH; fumarate hydratase, class II [EC:4.2.1.2] | 0.04002 | 0.00203 | 0.03957 | 0.00061 | 0.00305 | 65 | 0.04033 | 0.00069 | 0.00272 | 93 | 0.00409 | -0.18673 | 143.087 | 0.85214 | 0.95838 |
| K06940; uncharacterized protein | 0.03999 | 0.00238 | 0.03884 | 0.00092 | 0.00376 | 65 | 0.04080 | 0.00088 | 0.00308 | 93 | 0.00486 | -0.40343 | 136.178 | 0.68726 | 0.91567 |
| queD, ptpS, PTS; 6-pyruvoyltetrahydropterin/6-carboxytetrahydropterin synthase [EC:4.2.3.12 4.1.2.50] | 0.03999 | 0.00129 | 0.03728 | 0.00028 | 0.00207 | 65 | 0.04188 | 0.00025 | 0.00163 | 93 | 0.00264 | -1.74740 | 132.433 | 0.08289 | 0.76567 |
| dacA; diadenylate cyclase [EC:2.7.7.85] | 0.03996 | 0.00201 | 0.04112 | 0.00071 | 0.00330 | 65 | 0.03915 | 0.00060 | 0.00254 | 93 | 0.00416 | 0.47405 | 130.413 | 0.63626 | 0.89696 |
| pheA2; prephenate dehydratase [EC:4.2.1.51] | 0.03992 | 0.00196 | 0.03828 | 0.00063 | 0.00312 | 65 | 0.04106 | 0.00059 | 0.00253 | 93 | 0.00401 | -0.69141 | 135.029 | 0.49049 | 0.84797 |
| agrC, blpH, fsrC; two-component system, LytTR family, sensor histidine kinase AgrC [EC:2.7.13.3] | 0.03979 | 0.00409 | 0.03716 | 0.00168 | 0.00508 | 65 | 0.04163 | 0.00332 | 0.00598 | 93 | 0.00785 | -0.56903 | 155.943 | 0.57015 | 0.87135 |
| sdhA, frdA; succinate dehydrogenase / fumarate reductase, flavoprotein subunit [EC:1.3.5.1 1.3.5.4] | 0.03977 | 0.00142 | 0.03689 | 0.00029 | 0.00213 | 65 | 0.04178 | 0.00033 | 0.00188 | 93 | 0.00284 | -1.72316 | 142.499 | 0.08703 | 0.76567 |
| sppA; protease IV [EC:3.4.21.-] | 0.03975 | 0.00199 | 0.04145 | 0.00090 | 0.00373 | 65 | 0.03856 | 0.00044 | 0.00217 | 93 | 0.00432 | 0.66955 | 106.333 | 0.50459 | 0.84956 |
| IDH1, IDH2, icd; isocitrate dehydrogenase [EC:1.1.1.42] | 0.03952 | 0.00175 | 0.03622 | 0.00051 | 0.00279 | 65 | 0.04182 | 0.00046 | 0.00222 | 93 | 0.00357 | -1.57037 | 133.312 | 0.11870 | 0.77376 |
| lpxD; UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase [EC:2.3.1.191] | 0.03941 | 0.00202 | 0.04251 | 0.00084 | 0.00359 | 65 | 0.03724 | 0.00050 | 0.00233 | 93 | 0.00428 | 1.23260 | 114.879 | 0.22024 | 0.77376 |
| gcvH, GCSH; glycine cleavage system H protein | 0.03941 | 0.00151 | 0.03853 | 0.00046 | 0.00267 | 65 | 0.04001 | 0.00029 | 0.00176 | 93 | 0.00320 | -0.46254 | 116.305 | 0.64456 | 0.90243 |
| GSR, gor; glutathione reductase (NADPH) [EC:1.8.1.7] | 0.03939 | 0.00183 | 0.04197 | 0.00069 | 0.00325 | 65 | 0.03758 | 0.00041 | 0.00211 | 93 | 0.00387 | 1.13343 | 114.902 | 0.25939 | 0.77376 |
| mmsB, HIBADH; 3-hydroxyisobutyrate dehydrogenase [EC:1.1.1.31] | 0.03936 | 0.00246 | 0.03531 | 0.00083 | 0.00358 | 65 | 0.04218 | 0.00103 | 0.00333 | 93 | 0.00488 | -1.40696 | 146.431 | 0.16156 | 0.77376 |
| trmFO, gid; methylenetetrahydrofolate–tRNA-(uracil-5-)-methyltransferase [EC:2.1.1.74] | 0.03935 | 0.00183 | 0.04029 | 0.00053 | 0.00287 | 65 | 0.03868 | 0.00053 | 0.00239 | 93 | 0.00373 | 0.43154 | 137.696 | 0.66675 | 0.91126 |
| punA, PNP; purine-nucleoside phosphorylase [EC:2.4.2.1] | 0.03927 | 0.00199 | 0.03777 | 0.00060 | 0.00303 | 65 | 0.04033 | 0.00065 | 0.00264 | 93 | 0.00402 | -0.63581 | 141.377 | 0.52593 | 0.85821 |
| praC, xylH; 4-oxalocrotonate tautomerase [EC:5.3.2.6] | 0.03927 | 0.00183 | 0.03866 | 0.00052 | 0.00284 | 65 | 0.03970 | 0.00054 | 0.00240 | 93 | 0.00372 | -0.28139 | 139.060 | 0.77883 | 0.94269 |
| addB; ATP-dependent helicase/nuclease subunit B [EC:3.1.-.- 3.6.4.12] | 0.03922 | 0.00185 | 0.03997 | 0.00062 | 0.00310 | 65 | 0.03869 | 0.00049 | 0.00230 | 93 | 0.00386 | 0.33257 | 127.109 | 0.74001 | 0.92969 |
| msbA; ATP-binding cassette, subfamily B, bacterial MsbA [EC:3.6.3.-] | 0.03918 | 0.00157 | 0.03944 | 0.00040 | 0.00250 | 65 | 0.03900 | 0.00039 | 0.00204 | 93 | 0.00322 | 0.13575 | 135.861 | 0.89222 | 0.97147 |
| rex; redox-sensing transcriptional repressor | 0.03918 | 0.00194 | 0.03956 | 0.00063 | 0.00312 | 65 | 0.03891 | 0.00057 | 0.00249 | 93 | 0.00399 | 0.16299 | 133.585 | 0.87077 | 0.96546 |
| CARP, pepA; leucyl aminopeptidase [EC:3.4.11.1] | 0.03907 | 0.00178 | 0.03717 | 0.00050 | 0.00278 | 65 | 0.04040 | 0.00050 | 0.00232 | 93 | 0.00362 | -0.89276 | 137.637 | 0.37354 | 0.79810 |
| K06915; uncharacterized protein | 0.03905 | 0.00216 | 0.04200 | 0.00126 | 0.00441 | 65 | 0.03699 | 0.00037 | 0.00201 | 93 | 0.00484 | 1.03614 | 90.576 | 0.30290 | 0.77376 |
| kch, trkA, mthK, pch; voltage-gated potassium channel | 0.03905 | 0.00158 | 0.03716 | 0.00044 | 0.00261 | 65 | 0.04037 | 0.00036 | 0.00198 | 93 | 0.00327 | -0.98119 | 128.890 | 0.32834 | 0.77376 |
| ysxB; uncharacterized protein | 0.03903 | 0.00190 | 0.04251 | 0.00060 | 0.00304 | 65 | 0.03660 | 0.00054 | 0.00242 | 93 | 0.00389 | 1.52320 | 133.371 | 0.13008 | 0.77376 |
| rsmD; 16S rRNA (guanine966-N2)-methyltransferase [EC:2.1.1.171] | 0.03892 | 0.00137 | 0.03823 | 0.00029 | 0.00210 | 65 | 0.03941 | 0.00030 | 0.00181 | 93 | 0.00277 | -0.42683 | 140.490 | 0.67015 | 0.91184 |
| E5.2.1.8; peptidylprolyl isomerase [EC:5.2.1.8] | 0.03876 | 0.00242 | 0.03969 | 0.00099 | 0.00390 | 65 | 0.03811 | 0.00090 | 0.00310 | 93 | 0.00498 | 0.31906 | 133.489 | 0.75018 | 0.93449 |
| panC; pantoate–beta-alanine ligase [EC:6.3.2.1] | 0.03866 | 0.00136 | 0.03594 | 0.00029 | 0.00210 | 65 | 0.04057 | 0.00029 | 0.00177 | 93 | 0.00274 | -1.69076 | 138.572 | 0.09313 | 0.76567 |
| rbsK, RBKS; ribokinase [EC:2.7.1.15] | 0.03866 | 0.00214 | 0.03942 | 0.00095 | 0.00383 | 65 | 0.03813 | 0.00057 | 0.00247 | 93 | 0.00456 | 0.28231 | 114.534 | 0.77822 | 0.94269 |
| RP-L25, rplY; large subunit ribosomal protein L25 | 0.03865 | 0.00143 | 0.03676 | 0.00037 | 0.00237 | 65 | 0.03998 | 0.00029 | 0.00177 | 93 | 0.00296 | -1.08599 | 127.506 | 0.27953 | 0.77376 |
| glpA, glpD; glycerol-3-phosphate dehydrogenase [EC:1.1.5.3] | 0.03850 | 0.00155 | 0.03782 | 0.00039 | 0.00244 | 65 | 0.03897 | 0.00037 | 0.00201 | 93 | 0.00316 | -0.36467 | 136.194 | 0.71593 | 0.92004 |
| hemA; glutamyl-tRNA reductase [EC:1.2.1.70] | 0.03842 | 0.00135 | 0.04126 | 0.00027 | 0.00205 | 65 | 0.03643 | 0.00029 | 0.00177 | 93 | 0.00271 | 1.78341 | 140.392 | 0.07668 | 0.75063 |
| asnB, ASNS; asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4] | 0.03836 | 0.00280 | 0.03471 | 0.00138 | 0.00461 | 65 | 0.04092 | 0.00114 | 0.00350 | 93 | 0.00579 | -1.07137 | 129.123 | 0.28600 | 0.77376 |
| phoR; two-component system, OmpR family, phosphate regulon sensor histidine kinase PhoR [EC:2.7.13.3] | 0.03833 | 0.00160 | 0.03412 | 0.00027 | 0.00206 | 65 | 0.04127 | 0.00048 | 0.00227 | 93 | 0.00306 | -2.33536 | 154.994 | 0.02081 | 0.59147 |
| K08987; putative membrane protein | 0.03833 | 0.00178 | 0.03964 | 0.00057 | 0.00295 | 65 | 0.03742 | 0.00046 | 0.00222 | 93 | 0.00369 | 0.60085 | 128.364 | 0.54900 | 0.86621 |
| rpiB; ribose 5-phosphate isomerase B [EC:5.3.1.6] | 0.03823 | 0.00213 | 0.03920 | 0.00085 | 0.00361 | 65 | 0.03756 | 0.00064 | 0.00262 | 93 | 0.00446 | 0.36714 | 125.184 | 0.71413 | 0.92004 |
| moaA, CNX2; GTP 3’,8-cyclase [EC:4.1.99.22] | 0.03822 | 0.00139 | 0.03815 | 0.00035 | 0.00232 | 65 | 0.03826 | 0.00028 | 0.00174 | 93 | 0.00290 | -0.03689 | 127.932 | 0.97063 | 0.99349 |
| modA; molybdate transport system substrate-binding protein | 0.03819 | 0.00161 | 0.03591 | 0.00034 | 0.00230 | 65 | 0.03979 | 0.00045 | 0.00220 | 93 | 0.00319 | -1.21753 | 148.433 | 0.22534 | 0.77376 |
| hepST; heptaprenyl diphosphate synthase [EC:2.5.1.30] | 0.03784 | 0.00245 | 0.04412 | 0.00138 | 0.00460 | 65 | 0.03345 | 0.00061 | 0.00256 | 93 | 0.00527 | 2.02561 | 102.984 | 0.04539 | 0.65981 |
| scrR; LacI family transcriptional regulator, sucrose operon repressor | 0.03784 | 0.00252 | 0.03929 | 0.00130 | 0.00446 | 65 | 0.03683 | 0.00081 | 0.00295 | 93 | 0.00535 | 0.46020 | 116.669 | 0.64623 | 0.90314 |
| UGDH, ugd; UDPglucose 6-dehydrogenase [EC:1.1.1.22] | 0.03773 | 0.00180 | 0.03429 | 0.00042 | 0.00254 | 65 | 0.04013 | 0.00057 | 0.00247 | 93 | 0.00354 | -1.65039 | 149.407 | 0.10096 | 0.77376 |
| sdhB, frdB; succinate dehydrogenase / fumarate reductase, iron-sulfur subunit [EC:1.3.5.1 1.3.5.4] | 0.03763 | 0.00139 | 0.03485 | 0.00027 | 0.00206 | 65 | 0.03957 | 0.00032 | 0.00185 | 93 | 0.00277 | -1.70417 | 144.022 | 0.09051 | 0.76567 |
| DPM1; dolichol-phosphate mannosyltransferase [EC:2.4.1.83] | 0.03763 | 0.00160 | 0.03771 | 0.00041 | 0.00251 | 65 | 0.03757 | 0.00041 | 0.00210 | 93 | 0.00327 | 0.04365 | 137.841 | 0.96525 | 0.99101 |
| K06885; uncharacterized protein | 0.03760 | 0.00200 | 0.03856 | 0.00067 | 0.00321 | 65 | 0.03693 | 0.00061 | 0.00256 | 93 | 0.00411 | 0.39791 | 133.797 | 0.69133 | 0.91676 |
| deoB; phosphopentomutase [EC:5.4.2.7] | 0.03759 | 0.00193 | 0.03781 | 0.00060 | 0.00303 | 65 | 0.03743 | 0.00059 | 0.00252 | 93 | 0.00394 | 0.09517 | 137.297 | 0.92432 | 0.97984 |
| rarD; chloramphenicol-sensitive protein RarD | 0.03758 | 0.00170 | 0.03710 | 0.00052 | 0.00284 | 65 | 0.03792 | 0.00041 | 0.00210 | 93 | 0.00353 | -0.23420 | 126.899 | 0.81521 | 0.95409 |
| troR; DtxR family transcriptional regulator, Mn-dependent transcriptional regulator | 0.03738 | 0.00219 | 0.03890 | 0.00079 | 0.00348 | 65 | 0.03633 | 0.00074 | 0.00282 | 93 | 0.00448 | 0.57303 | 135.014 | 0.56758 | 0.87063 |
| SAM50, TOB55, bamA; outer membrane protein insertion porin family | 0.03732 | 0.00160 | 0.03821 | 0.00042 | 0.00255 | 65 | 0.03671 | 0.00040 | 0.00207 | 93 | 0.00329 | 0.45700 | 135.463 | 0.64841 | 0.90350 |
| HEXA_B; hexosaminidase [EC:3.2.1.52] | 0.03729 | 0.00233 | 0.03505 | 0.00060 | 0.00304 | 65 | 0.03885 | 0.00105 | 0.00335 | 93 | 0.00452 | -0.83792 | 154.953 | 0.40337 | 0.81012 |
| K07146; UPF0176 protein | 0.03728 | 0.00177 | 0.03586 | 0.00052 | 0.00283 | 65 | 0.03827 | 0.00048 | 0.00227 | 93 | 0.00363 | -0.66399 | 134.493 | 0.50784 | 0.84956 |
| ampS, pepS, ampT; aminopeptidase [EC:3.4.11.-] | 0.03716 | 0.00197 | 0.03595 | 0.00062 | 0.00310 | 65 | 0.03801 | 0.00061 | 0.00256 | 93 | 0.00402 | -0.51149 | 137.006 | 0.60983 | 0.88693 |
| thiM; hydroxyethylthiazole kinase [EC:2.7.1.50] | 0.03713 | 0.00190 | 0.03577 | 0.00051 | 0.00281 | 65 | 0.03809 | 0.00061 | 0.00257 | 93 | 0.00381 | -0.60908 | 145.057 | 0.54342 | 0.86313 |
| E2.7.6.5X; putative GTP pyrophosphokinase [EC:2.7.6.5] | 0.03710 | 0.00213 | 0.03685 | 0.00077 | 0.00345 | 65 | 0.03728 | 0.00068 | 0.00271 | 93 | 0.00439 | -0.09670 | 132.344 | 0.92311 | 0.97947 |
| gltS; glutamate:Na+ symporter, ESS family | 0.03701 | 0.00299 | 0.04484 | 0.00212 | 0.00571 | 65 | 0.03154 | 0.00086 | 0.00305 | 93 | 0.00647 | 2.05426 | 100.039 | 0.04256 | 0.65361 |
| alaA; alanine-synthesizing transaminase [EC:2.6.1.66 2.6.1.2] | 0.03697 | 0.00155 | 0.03660 | 0.00038 | 0.00243 | 65 | 0.03723 | 0.00038 | 0.00203 | 93 | 0.00317 | -0.20005 | 137.841 | 0.84174 | 0.95506 |
| lptB; lipopolysaccharide export system ATP-binding protein [EC:3.6.3.-] | 0.03697 | 0.00158 | 0.03807 | 0.00041 | 0.00252 | 65 | 0.03621 | 0.00038 | 0.00203 | 93 | 0.00323 | 0.57503 | 134.503 | 0.56623 | 0.87051 |
| hutU, UROC1; urocanate hydratase [EC:4.2.1.49] | 0.03682 | 0.00162 | 0.03788 | 0.00063 | 0.00312 | 65 | 0.03608 | 0.00027 | 0.00171 | 93 | 0.00355 | 0.50731 | 101.744 | 0.61304 | 0.88788 |
| E2.4.2.21, cobU, cobT; nicotinate-nucleotide–dimethylbenzimidazole phosphoribosyltransferase [EC:2.4.2.21] | 0.03677 | 0.00204 | 0.03843 | 0.00078 | 0.00346 | 65 | 0.03560 | 0.00057 | 0.00248 | 93 | 0.00426 | 0.66455 | 123.826 | 0.50757 | 0.84956 |
| bmpA, bmpB, tmpC; basic membrane protein A and related proteins | 0.03674 | 0.00179 | 0.03581 | 0.00045 | 0.00264 | 65 | 0.03738 | 0.00055 | 0.00244 | 93 | 0.00359 | -0.43841 | 145.849 | 0.66174 | 0.90968 |
| nanE; N-acylglucosamine-6-phosphate 2-epimerase [EC:5.1.3.9] | 0.03672 | 0.00184 | 0.03986 | 0.00051 | 0.00281 | 65 | 0.03452 | 0.00055 | 0.00243 | 93 | 0.00371 | 1.43835 | 140.576 | 0.15256 | 0.77376 |
| dgt; dGTPase [EC:3.1.5.1] | 0.03666 | 0.00156 | 0.03620 | 0.00041 | 0.00252 | 65 | 0.03699 | 0.00036 | 0.00198 | 93 | 0.00320 | -0.24688 | 132.418 | 0.80538 | 0.94958 |
| cfa; cyclopropane-fatty-acyl-phospholipid synthase [EC:2.1.1.79] | 0.03665 | 0.00241 | 0.03561 | 0.00094 | 0.00379 | 65 | 0.03738 | 0.00091 | 0.00313 | 93 | 0.00492 | -0.35942 | 136.795 | 0.71983 | 0.92108 |
| gshA; glutamate–cysteine ligase [EC:6.3.2.2] | 0.03663 | 0.00207 | 0.03956 | 0.00095 | 0.00382 | 65 | 0.03459 | 0.00048 | 0.00228 | 93 | 0.00445 | 1.11794 | 108.283 | 0.26607 | 0.77376 |
| xdhC; xanthine dehydrogenase accessory factor | 0.03659 | 0.00225 | 0.03660 | 0.00101 | 0.00394 | 65 | 0.03658 | 0.00067 | 0.00267 | 93 | 0.00476 | 0.00312 | 118.977 | 0.99752 | 0.99955 |
| menA; 1,4-dihydroxy-2-naphthoate octaprenyltransferase [EC:2.5.1.74 2.5.1.-] | 0.03657 | 0.00187 | 0.03831 | 0.00064 | 0.00314 | 65 | 0.03536 | 0.00049 | 0.00229 | 93 | 0.00389 | 0.76083 | 125.581 | 0.44818 | 0.83372 |
| adhE; acetaldehyde dehydrogenase / alcohol dehydrogenase [EC:1.2.1.10 1.1.1.1] | 0.03651 | 0.00198 | 0.03885 | 0.00064 | 0.00315 | 65 | 0.03488 | 0.00060 | 0.00255 | 93 | 0.00405 | 0.98112 | 134.983 | 0.32829 | 0.77376 |
| ABC-2.TX; HlyD family secretion protein | 0.03649 | 0.00210 | 0.03334 | 0.00052 | 0.00284 | 65 | 0.03870 | 0.00081 | 0.00295 | 93 | 0.00410 | -1.30964 | 152.949 | 0.19228 | 0.77376 |
| mrp, NUBPL; ATP-binding protein involved in chromosome partitioning | 0.03637 | 0.00135 | 0.03342 | 0.00026 | 0.00202 | 65 | 0.03843 | 0.00030 | 0.00178 | 93 | 0.00269 | -1.86220 | 142.594 | 0.06463 | 0.73334 |
| TC.PIT; inorganic phosphate transporter, PiT family | 0.03626 | 0.00172 | 0.03419 | 0.00049 | 0.00275 | 65 | 0.03771 | 0.00045 | 0.00220 | 93 | 0.00352 | -0.99958 | 133.956 | 0.31932 | 0.77376 |
| lptG; lipopolysaccharide export system permease protein | 0.03624 | 0.00156 | 0.03718 | 0.00041 | 0.00253 | 65 | 0.03559 | 0.00037 | 0.00199 | 93 | 0.00321 | 0.49177 | 132.554 | 0.62369 | 0.89159 |
| K07166; ACT domain-containing protein | 0.03623 | 0.00194 | 0.03810 | 0.00065 | 0.00317 | 65 | 0.03493 | 0.00056 | 0.00246 | 93 | 0.00401 | 0.78972 | 131.208 | 0.43112 | 0.82763 |
| gspE; general secretion pathway protein E | 0.03622 | 0.00253 | 0.03756 | 0.00098 | 0.00389 | 65 | 0.03527 | 0.00105 | 0.00335 | 93 | 0.00513 | 0.44563 | 140.524 | 0.65655 | 0.90679 |
| sdhC, frdC; succinate dehydrogenase / fumarate reductase, cytochrome b subunit | 0.03615 | 0.00135 | 0.03363 | 0.00025 | 0.00197 | 65 | 0.03791 | 0.00031 | 0.00183 | 93 | 0.00268 | -1.59406 | 146.278 | 0.11308 | 0.77376 |
| TC.FEV.OM1, fhuE, fpvA, fptA; outer-membrane receptor for ferric coprogen and ferric-rhodotorulic acid | 0.03614 | 0.00443 | 0.03460 | 0.00291 | 0.00669 | 65 | 0.03721 | 0.00327 | 0.00593 | 93 | 0.00893 | -0.29212 | 142.777 | 0.77062 | 0.94017 |
| hutI, AMDHD1; imidazolonepropionase [EC:3.5.2.7] | 0.03604 | 0.00161 | 0.03707 | 0.00063 | 0.00311 | 65 | 0.03532 | 0.00026 | 0.00168 | 93 | 0.00353 | 0.49457 | 100.823 | 0.62198 | 0.89150 |
| ppsR; [pyruvate, water dikinase]-phosphate phosphotransferase / [pyruvate, water dikinase] kinase [EC:2.7.4.28 2.7.11.33] | 0.03597 | 0.00116 | 0.03480 | 0.00026 | 0.00198 | 65 | 0.03680 | 0.00018 | 0.00140 | 93 | 0.00243 | -0.82601 | 122.514 | 0.41040 | 0.81416 |
| K06975; uncharacterized protein | 0.03596 | 0.00144 | 0.03794 | 0.00034 | 0.00228 | 65 | 0.03458 | 0.00032 | 0.00186 | 93 | 0.00294 | 1.14224 | 135.547 | 0.25537 | 0.77376 |
| kdsA; 2-dehydro-3-deoxyphosphooctonate aldolase (KDO 8-P synthase) [EC:2.5.1.55] | 0.03578 | 0.00159 | 0.03739 | 0.00041 | 0.00252 | 65 | 0.03465 | 0.00039 | 0.00206 | 93 | 0.00325 | 0.84366 | 135.916 | 0.40034 | 0.80762 |
| kdsB; 3-deoxy-manno-octulosonate cytidylyltransferase (CMP-KDO synthetase) [EC:2.7.7.38] | 0.03577 | 0.00159 | 0.03717 | 0.00041 | 0.00252 | 65 | 0.03479 | 0.00040 | 0.00206 | 93 | 0.00325 | 0.73093 | 136.160 | 0.46608 | 0.83830 |
| kdsD, kpsF; arabinose-5-phosphate isomerase [EC:5.3.1.13] | 0.03568 | 0.00158 | 0.03732 | 0.00040 | 0.00249 | 65 | 0.03453 | 0.00039 | 0.00205 | 93 | 0.00322 | 0.86708 | 136.346 | 0.38742 | 0.80262 |
| acrB, mexB, adeJ, smeE, mtrD, cmeB; multidrug efflux pump | 0.03563 | 0.00320 | 0.03404 | 0.00172 | 0.00514 | 65 | 0.03673 | 0.00157 | 0.00411 | 93 | 0.00658 | -0.40938 | 133.844 | 0.68291 | 0.91495 |
| pdp; pyrimidine-nucleoside phosphorylase [EC:2.4.2.2] | 0.03556 | 0.00191 | 0.03627 | 0.00061 | 0.00305 | 65 | 0.03507 | 0.00056 | 0.00246 | 93 | 0.00392 | 0.30595 | 134.583 | 0.76012 | 0.93791 |
| lpxB; lipid-A-disaccharide synthase [EC:2.4.1.182] | 0.03553 | 0.00158 | 0.03705 | 0.00041 | 0.00250 | 65 | 0.03447 | 0.00039 | 0.00204 | 93 | 0.00323 | 0.79936 | 135.675 | 0.42548 | 0.82605 |
| ppk; polyphosphate kinase [EC:2.7.4.1] | 0.03549 | 0.00135 | 0.03490 | 0.00027 | 0.00204 | 65 | 0.03590 | 0.00030 | 0.00180 | 93 | 0.00272 | -0.36662 | 142.213 | 0.71445 | 0.92004 |
| thiL; thiamine-monophosphate kinase [EC:2.7.4.16] | 0.03547 | 0.00141 | 0.03484 | 0.00035 | 0.00231 | 65 | 0.03591 | 0.00029 | 0.00178 | 93 | 0.00291 | -0.36709 | 130.419 | 0.71414 | 0.92004 |
| msrB; peptide-methionine (R)-S-oxide reductase [EC:1.8.4.12] | 0.03543 | 0.00182 | 0.03540 | 0.00064 | 0.00313 | 65 | 0.03546 | 0.00045 | 0.00219 | 93 | 0.00382 | -0.01491 | 121.915 | 0.98813 | 0.99801 |
| TC.FEV.OM2, cirA, cfrA, hmuR; outer membrane receptor for ferrienterochelin and colicins | 0.03542 | 0.00290 | 0.03275 | 0.00117 | 0.00425 | 65 | 0.03729 | 0.00144 | 0.00393 | 93 | 0.00579 | -0.78304 | 146.051 | 0.43487 | 0.82874 |
| TC.GNTP; gluconate:H+ symporter, GntP family | 0.03535 | 0.00223 | 0.04097 | 0.00091 | 0.00374 | 65 | 0.03141 | 0.00067 | 0.00268 | 93 | 0.00460 | 2.07721 | 123.848 | 0.03985 | 0.63577 |
| sstT; serine/threonine transporter | 0.03533 | 0.00159 | 0.03615 | 0.00042 | 0.00254 | 65 | 0.03475 | 0.00039 | 0.00204 | 93 | 0.00326 | 0.42759 | 134.289 | 0.66963 | 0.91175 |
| rng, cafA; ribonuclease G [EC:3.1.26.-] | 0.03527 | 0.00153 | 0.03695 | 0.00037 | 0.00239 | 65 | 0.03410 | 0.00037 | 0.00199 | 93 | 0.00311 | 0.91513 | 137.772 | 0.36172 | 0.79496 |
| mgtE; magnesium transporter | 0.03525 | 0.00182 | 0.03821 | 0.00060 | 0.00304 | 65 | 0.03319 | 0.00046 | 0.00223 | 93 | 0.00377 | 1.33377 | 126.078 | 0.18468 | 0.77376 |
| ggt; gamma-glutamyltranspeptidase / glutathione hydrolase [EC:2.3.2.2 3.4.19.13] | 0.03514 | 0.00277 | 0.03385 | 0.00125 | 0.00438 | 65 | 0.03604 | 0.00120 | 0.00359 | 93 | 0.00567 | -0.38730 | 136.226 | 0.69914 | 0.91867 |
| coaX; type III pantothenate kinase [EC:2.7.1.33] | 0.03512 | 0.00142 | 0.03532 | 0.00035 | 0.00233 | 65 | 0.03498 | 0.00030 | 0.00179 | 93 | 0.00294 | 0.11796 | 130.168 | 0.90628 | 0.97664 |
| gltX; nondiscriminating glutamyl-tRNA synthetase [EC:6.1.1.24] | 0.03494 | 0.00202 | 0.03533 | 0.00068 | 0.00324 | 65 | 0.03466 | 0.00062 | 0.00259 | 93 | 0.00414 | 0.15949 | 133.821 | 0.87352 | 0.96576 |
| K06871; uncharacterized protein | 0.03491 | 0.00265 | 0.03454 | 0.00124 | 0.00437 | 65 | 0.03517 | 0.00103 | 0.00332 | 93 | 0.00549 | -0.11448 | 129.225 | 0.90904 | 0.97664 |
| sbcC, rad50; DNA repair protein SbcC/Rad50 | 0.03491 | 0.00147 | 0.03922 | 0.00036 | 0.00236 | 65 | 0.03190 | 0.00031 | 0.00181 | 93 | 0.00298 | 2.45775 | 130.279 | 0.01529 | 0.59095 |
| wecA, tagO, rfe; UDP-GlcNAc:undecaprenyl-phosphate/decaprenyl-phosphate GlcNAc-1-phosphate transferase [EC:2.7.8.33 2.7.8.35] | 0.03483 | 0.00163 | 0.03581 | 0.00047 | 0.00268 | 65 | 0.03414 | 0.00039 | 0.00204 | 93 | 0.00337 | 0.49611 | 129.640 | 0.62066 | 0.89082 |
| gabD; succinate-semialdehyde dehydrogenase / glutarate-semialdehyde dehydrogenase [EC:1.2.1.16 1.2.1.79 1.2.1.20] | 0.03482 | 0.00234 | 0.03325 | 0.00084 | 0.00359 | 65 | 0.03591 | 0.00090 | 0.00311 | 93 | 0.00475 | -0.56193 | 140.950 | 0.57506 | 0.87389 |
| patA; aminotransferase [EC:2.6.1.-] | 0.03477 | 0.00288 | 0.03741 | 0.00197 | 0.00551 | 65 | 0.03292 | 0.00086 | 0.00304 | 93 | 0.00629 | 0.71443 | 102.278 | 0.47659 | 0.84209 |
| motB; chemotaxis protein MotB | 0.03473 | 0.00200 | 0.03425 | 0.00077 | 0.00344 | 65 | 0.03506 | 0.00054 | 0.00241 | 93 | 0.00420 | -0.19279 | 121.724 | 0.84745 | 0.95682 |
| K07105; uncharacterized protein | 0.03472 | 0.00190 | 0.03588 | 0.00058 | 0.00297 | 65 | 0.03391 | 0.00057 | 0.00248 | 93 | 0.00387 | 0.51013 | 137.465 | 0.61078 | 0.88722 |
| idi, IDI; isopentenyl-diphosphate Delta-isomerase [EC:5.3.3.2] | 0.03457 | 0.00184 | 0.03513 | 0.00056 | 0.00294 | 65 | 0.03418 | 0.00052 | 0.00237 | 93 | 0.00377 | 0.25047 | 134.412 | 0.80261 | 0.94869 |
| nhaC; Na+:H+ antiporter, NhaC family | 0.03455 | 0.00400 | 0.04658 | 0.00438 | 0.00821 | 65 | 0.02615 | 0.00109 | 0.00342 | 93 | 0.00889 | 2.29904 | 86.332 | 0.02392 | 0.59764 |
| purR; purine operon repressor | 0.03452 | 0.00201 | 0.03519 | 0.00069 | 0.00325 | 65 | 0.03406 | 0.00061 | 0.00255 | 93 | 0.00413 | 0.27369 | 132.447 | 0.78475 | 0.94445 |
| divIVA; cell division initiation protein | 0.03451 | 0.00189 | 0.03568 | 0.00065 | 0.00316 | 65 | 0.03369 | 0.00051 | 0.00235 | 93 | 0.00394 | 0.50732 | 127.426 | 0.61281 | 0.88788 |
| ACADM, acd; acyl-CoA dehydrogenase [EC:1.3.8.7] | 0.03441 | 0.00412 | 0.02441 | 0.00136 | 0.00458 | 65 | 0.04140 | 0.00352 | 0.00615 | 93 | 0.00767 | -2.21527 | 154.144 | 0.02821 | 0.61334 |
| ctsR; transcriptional regulator of stress and heat shock response | 0.03440 | 0.00190 | 0.03542 | 0.00059 | 0.00302 | 65 | 0.03369 | 0.00056 | 0.00245 | 93 | 0.00389 | 0.44576 | 135.408 | 0.65648 | 0.90679 |
| licD; lipopolysaccharide cholinephosphotransferase [EC:2.7.8.-] | 0.03437 | 0.00223 | 0.03539 | 0.00078 | 0.00347 | 65 | 0.03366 | 0.00079 | 0.00292 | 93 | 0.00454 | 0.38277 | 138.301 | 0.70248 | 0.92004 |
| ME2, sfcA, maeA; malate dehydrogenase (oxaloacetate-decarboxylating) [EC:1.1.1.38] | 0.03433 | 0.00205 | 0.03817 | 0.00101 | 0.00395 | 65 | 0.03165 | 0.00041 | 0.00211 | 93 | 0.00447 | 1.45671 | 100.095 | 0.14833 | 0.77376 |
| coaA; type I pantothenate kinase [EC:2.7.1.33] | 0.03424 | 0.00192 | 0.03612 | 0.00064 | 0.00313 | 65 | 0.03292 | 0.00055 | 0.00243 | 93 | 0.00396 | 0.81013 | 131.144 | 0.41933 | 0.81991 |
| TC.ZIP, zupT, ZRT3, ZIP2; zinc transporter, ZIP family | 0.03417 | 0.00139 | 0.03257 | 0.00030 | 0.00215 | 65 | 0.03528 | 0.00031 | 0.00182 | 93 | 0.00281 | -0.96421 | 138.882 | 0.33662 | 0.78282 |
| PTS-Bgl-EIIC, bglF, bglP; PTS system, beta-glucoside-specific IIC component | 0.03416 | 0.00297 | 0.04048 | 0.00192 | 0.00543 | 65 | 0.02974 | 0.00099 | 0.00326 | 93 | 0.00634 | 1.69375 | 108.630 | 0.09318 | 0.76567 |
| K07009; uncharacterized protein | 0.03415 | 0.00208 | 0.03481 | 0.00075 | 0.00340 | 65 | 0.03369 | 0.00064 | 0.00263 | 93 | 0.00430 | 0.26128 | 131.019 | 0.79428 | 0.94783 |
| kdsC; 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (KDO 8-P phosphatase) [EC:3.1.3.45] | 0.03415 | 0.00162 | 0.03632 | 0.00042 | 0.00255 | 65 | 0.03263 | 0.00041 | 0.00210 | 93 | 0.00330 | 1.11819 | 136.723 | 0.26545 | 0.77376 |
| ctpE; cation-transporting P-type ATPase E [EC:3.6.3.-] | 0.03415 | 0.00217 | 0.03498 | 0.00077 | 0.00345 | 65 | 0.03357 | 0.00073 | 0.00280 | 93 | 0.00444 | 0.31709 | 135.255 | 0.75166 | 0.93532 |
| modB; molybdate transport system permease protein | 0.03412 | 0.00132 | 0.03342 | 0.00028 | 0.00209 | 65 | 0.03461 | 0.00027 | 0.00171 | 93 | 0.00271 | -0.43949 | 136.099 | 0.66100 | 0.90968 |
| malZ; alpha-glucosidase [EC:3.2.1.20] | 0.03412 | 0.00214 | 0.03364 | 0.00073 | 0.00335 | 65 | 0.03445 | 0.00073 | 0.00280 | 93 | 0.00436 | -0.18434 | 138.055 | 0.85402 | 0.95848 |
| secD; preprotein translocase subunit SecD | 0.03403 | 0.00132 | 0.03687 | 0.00032 | 0.00224 | 65 | 0.03204 | 0.00023 | 0.00158 | 93 | 0.00274 | 1.76210 | 122.725 | 0.08054 | 0.75920 |
| secF; preprotein translocase subunit SecF | 0.03399 | 0.00131 | 0.03685 | 0.00032 | 0.00223 | 65 | 0.03199 | 0.00023 | 0.00158 | 93 | 0.00273 | 1.77897 | 122.600 | 0.07772 | 0.75469 |
| lpxK; tetraacyldisaccharide 4’-kinase [EC:2.7.1.130] | 0.03389 | 0.00152 | 0.03489 | 0.00039 | 0.00245 | 65 | 0.03318 | 0.00035 | 0.00194 | 93 | 0.00312 | 0.54797 | 133.070 | 0.58463 | 0.87720 |
| bioD; dethiobiotin synthetase [EC:6.3.3.3] | 0.03386 | 0.00160 | 0.03515 | 0.00044 | 0.00261 | 65 | 0.03296 | 0.00038 | 0.00202 | 93 | 0.00330 | 0.66286 | 130.923 | 0.50859 | 0.84956 |
| fpr; ferredoxin/flavodoxin—NADP+ reductase [EC:1.18.1.2 1.19.1.1] | 0.03385 | 0.00189 | 0.03342 | 0.00053 | 0.00286 | 65 | 0.03415 | 0.00059 | 0.00252 | 93 | 0.00381 | -0.19317 | 142.379 | 0.84710 | 0.95682 |
| murJ, mviN; putative peptidoglycan lipid II flippase | 0.03379 | 0.00182 | 0.03664 | 0.00073 | 0.00335 | 65 | 0.03180 | 0.00038 | 0.00202 | 93 | 0.00391 | 1.23674 | 109.079 | 0.21884 | 0.77376 |
| queH; epoxyqueuosine reductase [EC:1.17.99.6] | 0.03376 | 0.00168 | 0.03620 | 0.00046 | 0.00267 | 65 | 0.03206 | 0.00043 | 0.00215 | 93 | 0.00343 | 1.20889 | 134.781 | 0.22882 | 0.77376 |
| pps, ppsA; pyruvate, water dikinase [EC:2.7.9.2] | 0.03371 | 0.00144 | 0.03416 | 0.00039 | 0.00245 | 65 | 0.03340 | 0.00029 | 0.00177 | 93 | 0.00302 | 0.25119 | 124.694 | 0.80208 | 0.94855 |
| rimO; ribosomal protein S12 methylthiotransferase [EC:2.8.4.4] | 0.03367 | 0.00148 | 0.03322 | 0.00033 | 0.00226 | 65 | 0.03398 | 0.00036 | 0.00197 | 93 | 0.00300 | -0.25196 | 141.490 | 0.80144 | 0.94855 |
| gcvA; LysR family transcriptional regulator, glycine cleavage system transcriptional activator | 0.03362 | 0.00316 | 0.03178 | 0.00153 | 0.00486 | 65 | 0.03490 | 0.00162 | 0.00418 | 93 | 0.00641 | -0.48702 | 140.242 | 0.62700 | 0.89265 |
| pilD, pppA; leader peptidase (prepilin peptidase) / N-methyltransferase [EC:3.4.23.43 2.1.1.-] | 0.03362 | 0.00141 | 0.03689 | 0.00037 | 0.00240 | 65 | 0.03133 | 0.00027 | 0.00169 | 93 | 0.00293 | 1.89626 | 122.354 | 0.06028 | 0.73001 |
| yqeH; 30S ribosome assembly GTPase | 0.03361 | 0.00196 | 0.03581 | 0.00064 | 0.00314 | 65 | 0.03208 | 0.00059 | 0.00251 | 93 | 0.00402 | 0.92881 | 133.915 | 0.35466 | 0.79359 |
| K06878; tRNA-binding protein | 0.03361 | 0.00191 | 0.03313 | 0.00056 | 0.00294 | 65 | 0.03394 | 0.00059 | 0.00252 | 93 | 0.00387 | -0.21157 | 140.198 | 0.83275 | 0.95506 |
| rlmA1; 23S rRNA (guanine745-N1)-methyltransferase [EC:2.1.1.187] | 0.03357 | 0.00162 | 0.03427 | 0.00043 | 0.00258 | 65 | 0.03308 | 0.00040 | 0.00208 | 93 | 0.00331 | 0.35928 | 134.958 | 0.71994 | 0.92108 |
| ncd2, npd; nitronate monooxygenase [EC:1.13.12.16] | 0.03354 | 0.00262 | 0.02925 | 0.00077 | 0.00343 | 65 | 0.03654 | 0.00130 | 0.00373 | 93 | 0.00507 | -1.43658 | 154.547 | 0.15286 | 0.77376 |
| feoA; ferrous iron transport protein A | 0.03350 | 0.00222 | 0.03545 | 0.00075 | 0.00340 | 65 | 0.03214 | 0.00080 | 0.00293 | 93 | 0.00449 | 0.73732 | 140.530 | 0.46216 | 0.83654 |
| FUCA; alpha-L-fucosidase [EC:3.2.1.51] | 0.03346 | 0.00242 | 0.02967 | 0.00056 | 0.00294 | 65 | 0.03612 | 0.00117 | 0.00354 | 93 | 0.00460 | -1.40217 | 155.994 | 0.16285 | 0.77376 |
| TC.BASS; bile acid:Na+ symporter, BASS family | 0.03345 | 0.00148 | 0.03127 | 0.00030 | 0.00216 | 65 | 0.03497 | 0.00037 | 0.00200 | 93 | 0.00294 | -1.25968 | 146.182 | 0.20979 | 0.77376 |
| divIC, divA; cell division protein DivIC | 0.03327 | 0.00195 | 0.03440 | 0.00066 | 0.00318 | 65 | 0.03248 | 0.00057 | 0.00247 | 93 | 0.00403 | 0.47540 | 131.314 | 0.63529 | 0.89696 |
| lipA; lipoyl synthase [EC:2.8.1.8] | 0.03322 | 0.00137 | 0.03078 | 0.00032 | 0.00222 | 65 | 0.03493 | 0.00028 | 0.00173 | 93 | 0.00282 | -1.47280 | 131.485 | 0.14319 | 0.77376 |
| K00375; GntR family transcriptional regulator / MocR family aminotransferase | 0.03322 | 0.00209 | 0.03224 | 0.00066 | 0.00319 | 65 | 0.03390 | 0.00071 | 0.00277 | 93 | 0.00423 | -0.39268 | 140.830 | 0.69515 | 0.91738 |
| adhP; alcohol dehydrogenase, propanol-preferring [EC:1.1.1.1] | 0.03318 | 0.00174 | 0.03281 | 0.00052 | 0.00282 | 65 | 0.03343 | 0.00046 | 0.00221 | 93 | 0.00358 | -0.17185 | 132.187 | 0.86382 | 0.96263 |
| sbcD, mre11; DNA repair protein SbcD/Mre11 | 0.03317 | 0.00131 | 0.03508 | 0.00033 | 0.00226 | 65 | 0.03184 | 0.00023 | 0.00157 | 93 | 0.00275 | 1.17471 | 121.349 | 0.24241 | 0.77376 |
| hisB; imidazoleglycerol-phosphate dehydratase [EC:4.2.1.19] | 0.03308 | 0.00119 | 0.03083 | 0.00026 | 0.00201 | 65 | 0.03466 | 0.00019 | 0.00144 | 93 | 0.00247 | -1.55427 | 124.055 | 0.12267 | 0.77376 |
| ampG; MFS transporter, PAT family, beta-lactamase induction signal transducer AmpG | 0.03305 | 0.00200 | 0.03079 | 0.00046 | 0.00267 | 65 | 0.03463 | 0.00075 | 0.00284 | 93 | 0.00390 | -0.98439 | 153.811 | 0.32647 | 0.77376 |
| ganP; arabinogalactan oligomer / maltooligosaccharide transport system permease protein | 0.03304 | 0.00216 | 0.03176 | 0.00061 | 0.00307 | 65 | 0.03393 | 0.00083 | 0.00299 | 93 | 0.00428 | -0.50670 | 149.455 | 0.61311 | 0.88788 |
| rpoN; RNA polymerase sigma-54 factor | 0.03302 | 0.00155 | 0.03132 | 0.00041 | 0.00250 | 65 | 0.03422 | 0.00036 | 0.00197 | 93 | 0.00319 | -0.90965 | 132.480 | 0.36466 | 0.79587 |
| QARS, glnS; glutaminyl-tRNA synthetase [EC:6.1.1.18] | 0.03302 | 0.00146 | 0.03339 | 0.00031 | 0.00219 | 65 | 0.03276 | 0.00036 | 0.00196 | 93 | 0.00293 | 0.21612 | 143.432 | 0.82920 | 0.95506 |
| K07137; uncharacterized protein | 0.03297 | 0.00235 | 0.03574 | 0.00101 | 0.00393 | 65 | 0.03104 | 0.00078 | 0.00290 | 93 | 0.00488 | 0.96244 | 126.306 | 0.33767 | 0.78354 |
| K07015; uncharacterized protein | 0.03296 | 0.00194 | 0.03303 | 0.00065 | 0.00316 | 65 | 0.03291 | 0.00057 | 0.00247 | 93 | 0.00401 | 0.03032 | 131.657 | 0.97586 | 0.99571 |
| MMAB, pduO; cob(I)alamin adenosyltransferase [EC:2.5.1.17] | 0.03292 | 0.00147 | 0.03372 | 0.00038 | 0.00240 | 65 | 0.03236 | 0.00032 | 0.00187 | 93 | 0.00304 | 0.44566 | 131.202 | 0.65657 | 0.90679 |
| thiC; phosphomethylpyrimidine synthase [EC:4.1.99.17] | 0.03285 | 0.00135 | 0.03353 | 0.00029 | 0.00212 | 65 | 0.03238 | 0.00029 | 0.00177 | 93 | 0.00276 | 0.41461 | 137.872 | 0.67907 | 0.91311 |
| hemD, UROS; uroporphyrinogen-III synthase [EC:4.2.1.75] | 0.03275 | 0.00140 | 0.02906 | 0.00026 | 0.00201 | 65 | 0.03533 | 0.00033 | 0.00189 | 93 | 0.00276 | -2.27588 | 147.104 | 0.02430 | 0.59764 |
| qor, CRYZ; NADPH2:quinone reductase [EC:1.6.5.5] | 0.03275 | 0.00280 | 0.02723 | 0.00091 | 0.00375 | 65 | 0.03661 | 0.00144 | 0.00394 | 93 | 0.00544 | -1.72629 | 153.281 | 0.08631 | 0.76567 |
| moaC, CNX3; cyclic pyranopterin monophosphate synthase [EC:4.6.1.17] | 0.03274 | 0.00123 | 0.03296 | 0.00024 | 0.00191 | 65 | 0.03259 | 0.00024 | 0.00162 | 93 | 0.00250 | 0.15070 | 138.827 | 0.88043 | 0.96815 |
| HSP90A, htpG; molecular chaperone HtpG | 0.03271 | 0.00173 | 0.03315 | 0.00052 | 0.00282 | 65 | 0.03240 | 0.00044 | 0.00218 | 93 | 0.00357 | 0.21199 | 131.030 | 0.83244 | 0.95506 |
| cpdB; 2’,3’-cyclic-nucleotide 2’-phosphodiesterase / 3’-nucleotidase [EC:3.1.4.16 3.1.3.6] | 0.03268 | 0.00189 | 0.03082 | 0.00052 | 0.00283 | 65 | 0.03397 | 0.00060 | 0.00253 | 93 | 0.00380 | -0.83117 | 143.587 | 0.40726 | 0.81200 |
| K07089; uncharacterized protein | 0.03265 | 0.00178 | 0.03228 | 0.00051 | 0.00280 | 65 | 0.03290 | 0.00050 | 0.00232 | 93 | 0.00363 | -0.17064 | 137.145 | 0.86476 | 0.96263 |
| tsaB; tRNA threonylcarbamoyladenosine biosynthesis protein TsaB | 0.03264 | 0.00137 | 0.03155 | 0.00032 | 0.00221 | 65 | 0.03340 | 0.00028 | 0.00174 | 93 | 0.00281 | -0.65777 | 132.234 | 0.51183 | 0.85122 |
| recU; recombination protein U | 0.03262 | 0.00197 | 0.03293 | 0.00065 | 0.00315 | 65 | 0.03239 | 0.00060 | 0.00254 | 93 | 0.00405 | 0.13237 | 134.579 | 0.89489 | 0.97195 |
| PTS-Cel-EIIB, celA, chbB; PTS system, cellobiose-specific IIB component [EC:2.7.1.196 2.7.1.205] | 0.03261 | 0.00290 | 0.03786 | 0.00240 | 0.00608 | 65 | 0.02894 | 0.00056 | 0.00246 | 93 | 0.00656 | 1.36030 | 85.050 | 0.17733 | 0.77376 |
| rpoE; DNA-directed RNA polymerase subunit delta | 0.03256 | 0.00193 | 0.03351 | 0.00062 | 0.00309 | 65 | 0.03190 | 0.00057 | 0.00248 | 93 | 0.00396 | 0.40766 | 134.303 | 0.68417 | 0.91533 |
| ldhA; D-lactate dehydrogenase [EC:1.1.1.28] | 0.03254 | 0.00298 | 0.03928 | 0.00213 | 0.00572 | 65 | 0.02783 | 0.00085 | 0.00303 | 93 | 0.00647 | 1.76812 | 99.385 | 0.08011 | 0.75920 |
| sucC; succinyl-CoA synthetase beta subunit [EC:6.2.1.5] | 0.03252 | 0.00131 | 0.03053 | 0.00028 | 0.00208 | 65 | 0.03391 | 0.00026 | 0.00169 | 93 | 0.00268 | -1.26000 | 135.217 | 0.20984 | 0.77376 |
| mdh; malate dehydrogenase [EC:1.1.1.37] | 0.03249 | 0.00148 | 0.02979 | 0.00029 | 0.00212 | 65 | 0.03438 | 0.00038 | 0.00203 | 93 | 0.00293 | -1.56621 | 148.393 | 0.11943 | 0.77376 |
| surE; 5’-nucleotidase [EC:3.1.3.5] | 0.03247 | 0.00139 | 0.03145 | 0.00030 | 0.00215 | 65 | 0.03319 | 0.00031 | 0.00182 | 93 | 0.00282 | -0.61932 | 138.886 | 0.53672 | 0.86207 |
| proX; glycine betaine/proline transport system substrate-binding protein | 0.03246 | 0.00442 | 0.03417 | 0.00309 | 0.00689 | 65 | 0.03127 | 0.00312 | 0.00580 | 93 | 0.00901 | 0.32278 | 138.357 | 0.74735 | 0.93207 |
| prsA; foldase protein PrsA [EC:5.2.1.8] | 0.03244 | 0.00197 | 0.03180 | 0.00062 | 0.00309 | 65 | 0.03288 | 0.00062 | 0.00258 | 93 | 0.00402 | -0.26770 | 138.015 | 0.78933 | 0.94606 |
| gcvT, AMT; aminomethyltransferase [EC:2.1.2.10] | 0.03229 | 0.00170 | 0.02909 | 0.00043 | 0.00258 | 65 | 0.03454 | 0.00047 | 0.00224 | 93 | 0.00342 | -1.59470 | 141.258 | 0.11301 | 0.77376 |
| E4.1.3.3, nanA, NPL; N-acetylneuraminate lyase [EC:4.1.3.3] | 0.03225 | 0.00183 | 0.03396 | 0.00056 | 0.00295 | 65 | 0.03105 | 0.00050 | 0.00232 | 93 | 0.00375 | 0.77373 | 132.652 | 0.44047 | 0.83186 |
| kdtA, waaA; 3-deoxy-D-manno-octulosonic-acid transferase [EC:2.4.99.12 2.4.99.13 2.4.99.14 2.4.99.15] | 0.03222 | 0.00142 | 0.03161 | 0.00029 | 0.00212 | 65 | 0.03265 | 0.00034 | 0.00190 | 93 | 0.00285 | -0.36630 | 143.812 | 0.71468 | 0.92004 |
| thiG; thiazole synthase [EC:2.8.1.10] | 0.03219 | 0.00124 | 0.03282 | 0.00029 | 0.00210 | 65 | 0.03174 | 0.00021 | 0.00151 | 93 | 0.00258 | 0.41928 | 124.371 | 0.67573 | 0.91266 |
| ppa; inorganic pyrophosphatase [EC:3.6.1.1] | 0.03213 | 0.00161 | 0.03344 | 0.00048 | 0.00270 | 65 | 0.03121 | 0.00037 | 0.00199 | 93 | 0.00336 | 0.66239 | 126.306 | 0.50893 | 0.84956 |
| E3.1.3.5; 5’-nucleotidase [EC:3.1.3.5] | 0.03204 | 0.00138 | 0.02896 | 0.00029 | 0.00212 | 65 | 0.03419 | 0.00030 | 0.00179 | 93 | 0.00277 | -1.88420 | 138.642 | 0.06163 | 0.73001 |
| mtaD; 5-methylthioadenosine/S-adenosylhomocysteine deaminase [EC:3.5.4.31 3.5.4.28] | 0.03191 | 0.00145 | 0.03143 | 0.00031 | 0.00220 | 65 | 0.03225 | 0.00035 | 0.00193 | 93 | 0.00293 | -0.28000 | 142.068 | 0.77989 | 0.94269 |
| paaI; acyl-CoA thioesterase [EC:3.1.2.-] | 0.03188 | 0.00152 | 0.03222 | 0.00040 | 0.00248 | 65 | 0.03163 | 0.00035 | 0.00194 | 93 | 0.00315 | 0.18839 | 131.813 | 0.85086 | 0.95754 |
| K17318, lplA; putative aldouronate transport system substrate-binding protein | 0.03181 | 0.00244 | 0.03383 | 0.00112 | 0.00415 | 65 | 0.03039 | 0.00082 | 0.00297 | 93 | 0.00510 | 0.67511 | 123.617 | 0.50087 | 0.84900 |
| sucD; succinyl-CoA synthetase alpha subunit [EC:6.2.1.5] | 0.03174 | 0.00128 | 0.02980 | 0.00027 | 0.00203 | 65 | 0.03310 | 0.00025 | 0.00165 | 93 | 0.00261 | -1.26414 | 135.376 | 0.20835 | 0.77376 |
| mgtA, mgtB; Mg2+-importing ATPase [EC:3.6.3.2] | 0.03172 | 0.00167 | 0.03141 | 0.00043 | 0.00256 | 65 | 0.03194 | 0.00045 | 0.00221 | 93 | 0.00338 | -0.15724 | 140.442 | 0.87528 | 0.96693 |
| zapA; cell division protein ZapA | 0.03160 | 0.00132 | 0.03072 | 0.00031 | 0.00217 | 65 | 0.03222 | 0.00026 | 0.00166 | 93 | 0.00273 | -0.54673 | 129.657 | 0.58550 | 0.87720 |
| copB; Cu2+-exporting ATPase [EC:3.6.3.4] | 0.03155 | 0.00145 | 0.03156 | 0.00030 | 0.00214 | 65 | 0.03155 | 0.00036 | 0.00196 | 93 | 0.00290 | 0.00459 | 145.078 | 0.99634 | 0.99944 |
| cbf, cbf1; 3’-5’ exoribonuclease [EC:3.1.-.-] | 0.03152 | 0.00186 | 0.03156 | 0.00054 | 0.00288 | 65 | 0.03149 | 0.00055 | 0.00244 | 93 | 0.00378 | 0.01859 | 139.117 | 0.98519 | 0.99718 |
| thiS; sulfur carrier protein | 0.03152 | 0.00126 | 0.03199 | 0.00026 | 0.00202 | 65 | 0.03119 | 0.00024 | 0.00162 | 93 | 0.00259 | 0.31159 | 134.101 | 0.75583 | 0.93669 |
| ABC.SP.S; putative spermidine/putrescine transport system substrate-binding protein | 0.03146 | 0.00409 | 0.02913 | 0.00211 | 0.00570 | 65 | 0.03308 | 0.00303 | 0.00571 | 93 | 0.00806 | -0.48943 | 151.103 | 0.62525 | 0.89228 |
| panB; 3-methyl-2-oxobutanoate hydroxymethyltransferase [EC:2.1.2.11] | 0.03143 | 0.00127 | 0.02782 | 0.00022 | 0.00185 | 65 | 0.03395 | 0.00026 | 0.00169 | 93 | 0.00250 | -2.44997 | 145.017 | 0.01548 | 0.59095 |
| lplC; putative aldouronate transport system permease protein | 0.03142 | 0.00231 | 0.03254 | 0.00092 | 0.00377 | 65 | 0.03063 | 0.00079 | 0.00292 | 93 | 0.00477 | 0.39985 | 131.137 | 0.68991 | 0.91567 |
| desK; two-component system, NarL family, sensor histidine kinase DesK [EC:2.7.13.3] | 0.03140 | 0.00194 | 0.02909 | 0.00052 | 0.00283 | 65 | 0.03302 | 0.00064 | 0.00263 | 93 | 0.00386 | -1.01963 | 146.383 | 0.30959 | 0.77376 |
| surA; peptidyl-prolyl cis-trans isomerase SurA [EC:5.2.1.8] | 0.03138 | 0.00164 | 0.02915 | 0.00034 | 0.00228 | 65 | 0.03294 | 0.00048 | 0.00228 | 93 | 0.00322 | -1.17876 | 151.029 | 0.24035 | 0.77376 |
| K09125; uncharacterized protein | 0.03136 | 0.00158 | 0.03192 | 0.00047 | 0.00268 | 65 | 0.03097 | 0.00035 | 0.00194 | 93 | 0.00331 | 0.28594 | 124.896 | 0.77539 | 0.94269 |
| pgl; 6-phosphogluconolactonase [EC:3.1.1.31] | 0.03136 | 0.00205 | 0.03290 | 0.00080 | 0.00352 | 65 | 0.03028 | 0.00058 | 0.00249 | 93 | 0.00431 | 0.60752 | 122.695 | 0.54463 | 0.86388 |
| comFC; competence protein ComFC | 0.03124 | 0.00185 | 0.03379 | 0.00058 | 0.00300 | 65 | 0.02945 | 0.00051 | 0.00235 | 93 | 0.00381 | 1.13806 | 132.081 | 0.25716 | 0.77376 |
| endA; DNA-entry nuclease | 0.03119 | 0.00250 | 0.03326 | 0.00106 | 0.00403 | 65 | 0.02974 | 0.00095 | 0.00319 | 93 | 0.00514 | 0.68437 | 133.003 | 0.49493 | 0.84797 |
| K01436; amidohydrolase [EC:3.5.1.-] | 0.03118 | 0.00179 | 0.03224 | 0.00077 | 0.00345 | 65 | 0.03044 | 0.00032 | 0.00185 | 93 | 0.00392 | 0.45865 | 100.494 | 0.64748 | 0.90344 |
| E1.7.1.7, guaC; GMP reductase [EC:1.7.1.7] | 0.03117 | 0.00184 | 0.03153 | 0.00060 | 0.00303 | 65 | 0.03092 | 0.00050 | 0.00231 | 93 | 0.00381 | 0.16063 | 129.378 | 0.87263 | 0.96576 |
| K07177; Lon-like protease | 0.03117 | 0.00190 | 0.03176 | 0.00063 | 0.00312 | 65 | 0.03075 | 0.00053 | 0.00239 | 93 | 0.00393 | 0.25597 | 130.199 | 0.79837 | 0.94822 |
| bioA; adenosylmethionine—8-amino-7-oxononanoate aminotransferase [EC:2.6.1.62] | 0.03113 | 0.00134 | 0.03131 | 0.00031 | 0.00217 | 65 | 0.03099 | 0.00027 | 0.00172 | 93 | 0.00277 | 0.11458 | 132.899 | 0.90895 | 0.97664 |
| pphA; serine/threonine protein phosphatase 1 [EC:3.1.3.16] | 0.03105 | 0.00168 | 0.03192 | 0.00047 | 0.00269 | 65 | 0.03044 | 0.00044 | 0.00217 | 93 | 0.00346 | 0.42985 | 134.643 | 0.66799 | 0.91150 |
| E3.6.1.22, NUDT12, nudC; NAD+ diphosphatase [EC:3.6.1.22] | 0.03105 | 0.00135 | 0.03130 | 0.00029 | 0.00212 | 65 | 0.03088 | 0.00029 | 0.00175 | 93 | 0.00275 | 0.15287 | 137.209 | 0.87873 | 0.96759 |
| gpmI; 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [EC:5.4.2.12] | 0.03102 | 0.00149 | 0.02932 | 0.00034 | 0.00229 | 65 | 0.03221 | 0.00036 | 0.00197 | 93 | 0.00302 | -0.95780 | 140.484 | 0.33981 | 0.78466 |
| lplB; putative aldouronate transport system permease protein | 0.03101 | 0.00231 | 0.03217 | 0.00093 | 0.00378 | 65 | 0.03021 | 0.00079 | 0.00292 | 93 | 0.00477 | 0.41121 | 130.736 | 0.68159 | 0.91425 |
| queG; epoxyqueuosine reductase [EC:1.17.99.6] | 0.03099 | 0.00116 | 0.02838 | 0.00022 | 0.00184 | 65 | 0.03282 | 0.00020 | 0.00148 | 93 | 0.00236 | -1.87733 | 134.279 | 0.06264 | 0.73012 |
| E3.2.1.22B, galA, rafA; alpha-galactosidase [EC:3.2.1.22] | 0.03097 | 0.00223 | 0.03137 | 0.00093 | 0.00379 | 65 | 0.03068 | 0.00069 | 0.00272 | 93 | 0.00466 | 0.14712 | 123.994 | 0.88328 | 0.96974 |
| dadA; D-amino-acid dehydrogenase [EC:1.4.5.1] | 0.03093 | 0.00282 | 0.02658 | 0.00107 | 0.00406 | 65 | 0.03396 | 0.00137 | 0.00384 | 93 | 0.00559 | -1.31894 | 147.544 | 0.18923 | 0.77376 |
| cheY; two-component system, chemotaxis family, chemotaxis protein CheY | 0.03090 | 0.00223 | 0.02765 | 0.00080 | 0.00350 | 65 | 0.03317 | 0.00078 | 0.00289 | 93 | 0.00454 | -1.21487 | 136.768 | 0.22651 | 0.77376 |
| waaF, rfaF; heptosyltransferase II [EC:2.4.-.-] | 0.03082 | 0.00204 | 0.03645 | 0.00088 | 0.00367 | 65 | 0.02689 | 0.00047 | 0.00226 | 93 | 0.00431 | 2.21603 | 110.558 | 0.02874 | 0.61334 |
| rbsC; ribose transport system permease protein | 0.03057 | 0.00323 | 0.02940 | 0.00246 | 0.00615 | 65 | 0.03139 | 0.00110 | 0.00343 | 93 | 0.00704 | -0.28227 | 103.167 | 0.77830 | 0.94269 |
| E3.1.3.97; 3’,5’-nucleoside bisphosphate phosphatase [EC:3.1.3.97] | 0.03048 | 0.00148 | 0.03192 | 0.00036 | 0.00235 | 65 | 0.02948 | 0.00034 | 0.00190 | 93 | 0.00303 | 0.80462 | 134.995 | 0.42245 | 0.82321 |
| K07027; glycosyltransferase 2 family protein | 0.03046 | 0.00164 | 0.02969 | 0.00040 | 0.00249 | 65 | 0.03100 | 0.00045 | 0.00219 | 93 | 0.00332 | -0.39493 | 142.151 | 0.69349 | 0.91677 |
| mecA1_2; adapter protein MecA 1/2 | 0.03043 | 0.00184 | 0.03061 | 0.00060 | 0.00304 | 65 | 0.03030 | 0.00050 | 0.00232 | 93 | 0.00382 | 0.08070 | 129.439 | 0.93581 | 0.98459 |
| lysE, argO; L-lysine exporter family protein LysE/ArgO | 0.03042 | 0.00154 | 0.03197 | 0.00040 | 0.00247 | 65 | 0.02933 | 0.00036 | 0.00198 | 93 | 0.00316 | 0.83426 | 134.134 | 0.40562 | 0.81189 |
| ispB; octaprenyl-diphosphate synthase [EC:2.5.1.90] | 0.03041 | 0.00134 | 0.02894 | 0.00029 | 0.00210 | 65 | 0.03144 | 0.00028 | 0.00174 | 93 | 0.00273 | -0.91622 | 137.122 | 0.36116 | 0.79496 |
| aspA; aspartate ammonia-lyase [EC:4.3.1.1] | 0.03040 | 0.00157 | 0.02959 | 0.00032 | 0.00222 | 65 | 0.03097 | 0.00044 | 0.00218 | 93 | 0.00311 | -0.44520 | 149.803 | 0.65682 | 0.90689 |
| vicR; two-component system, OmpR family, response regulator VicR | 0.03037 | 0.00194 | 0.03005 | 0.00062 | 0.00310 | 65 | 0.03058 | 0.00059 | 0.00251 | 93 | 0.00399 | -0.13319 | 135.087 | 0.89424 | 0.97180 |
| AMY, amyA, malS; alpha-amylase [EC:3.2.1.1] | 0.03036 | 0.00179 | 0.02913 | 0.00045 | 0.00264 | 65 | 0.03121 | 0.00055 | 0.00243 | 93 | 0.00358 | -0.58048 | 145.546 | 0.56249 | 0.86813 |
| argJ; glutamate N-acetyltransferase / amino-acid N-acetyltransferase [EC:2.3.1.35 2.3.1.1] | 0.03035 | 0.00153 | 0.02815 | 0.00039 | 0.00244 | 65 | 0.03188 | 0.00036 | 0.00196 | 93 | 0.00313 | -1.19263 | 134.121 | 0.23512 | 0.77376 |
| bfr; bacterioferritin [EC:1.16.3.1] | 0.03025 | 0.00274 | 0.02954 | 0.00132 | 0.00451 | 65 | 0.03074 | 0.00110 | 0.00344 | 93 | 0.00567 | -0.21194 | 129.688 | 0.83249 | 0.95506 |
| TST, MPST, sseA; thiosulfate/3-mercaptopyruvate sulfurtransferase [EC:2.8.1.1 2.8.1.2] | 0.03019 | 0.00183 | 0.02672 | 0.00051 | 0.00279 | 65 | 0.03263 | 0.00054 | 0.00241 | 93 | 0.00369 | -1.60349 | 140.749 | 0.11107 | 0.77376 |
| asnA; aspartate–ammonia ligase [EC:6.3.1.1] | 0.03014 | 0.00180 | 0.03271 | 0.00058 | 0.00300 | 65 | 0.02834 | 0.00046 | 0.00222 | 93 | 0.00373 | 1.16903 | 127.012 | 0.24458 | 0.77376 |
| pip; proline iminopeptidase [EC:3.4.11.5] | 0.03009 | 0.00217 | 0.02988 | 0.00083 | 0.00357 | 65 | 0.03023 | 0.00069 | 0.00272 | 93 | 0.00449 | -0.07967 | 129.741 | 0.93662 | 0.98459 |
| nfdA; N-substituted formamide deformylase [EC:3.5.1.91] | 0.03007 | 0.00204 | 0.03071 | 0.00074 | 0.00338 | 65 | 0.02963 | 0.00060 | 0.00255 | 93 | 0.00423 | 0.25492 | 128.428 | 0.79920 | 0.94840 |
| potF; putrescine transport system substrate-binding protein | 0.03005 | 0.00387 | 0.03330 | 0.00259 | 0.00631 | 65 | 0.02778 | 0.00221 | 0.00488 | 93 | 0.00798 | 0.69208 | 130.823 | 0.49011 | 0.84797 |
| K13653; AraC family transcriptional regulator | 0.03001 | 0.00160 | 0.03199 | 0.00044 | 0.00261 | 65 | 0.02863 | 0.00038 | 0.00202 | 93 | 0.00330 | 1.02047 | 131.012 | 0.30939 | 0.77376 |
| vicK; two-component system, OmpR family, sensor histidine kinase VicK [EC:2.7.13.3] | 0.03001 | 0.00194 | 0.02991 | 0.00062 | 0.00309 | 65 | 0.03008 | 0.00058 | 0.00249 | 93 | 0.00397 | -0.04101 | 134.799 | 0.96735 | 0.99206 |
| TC.KEF; monovalent cation:H+ antiporter-2, CPA2 family | 0.02995 | 0.00162 | 0.02704 | 0.00038 | 0.00242 | 65 | 0.03198 | 0.00043 | 0.00215 | 93 | 0.00324 | -1.52763 | 142.935 | 0.12881 | 0.77376 |
| putP; sodium/proline symporter | 0.02990 | 0.00126 | 0.03129 | 0.00027 | 0.00205 | 65 | 0.02894 | 0.00024 | 0.00159 | 93 | 0.00259 | 0.90471 | 131.351 | 0.36728 | 0.79587 |
| pntA; NAD(P) transhydrogenase subunit alpha [EC:1.6.1.2] | 0.02987 | 0.00208 | 0.02775 | 0.00071 | 0.00331 | 65 | 0.03136 | 0.00066 | 0.00267 | 93 | 0.00425 | -0.84843 | 134.649 | 0.39771 | 0.80562 |
| ABC-2.OM, wza; polysaccharide biosynthesis/export protein | 0.02983 | 0.00162 | 0.02782 | 0.00030 | 0.00216 | 65 | 0.03123 | 0.00049 | 0.00230 | 93 | 0.00316 | -1.08158 | 153.809 | 0.28113 | 0.77376 |
| dnaI; primosomal protein DnaI | 0.02982 | 0.00194 | 0.02984 | 0.00062 | 0.00309 | 65 | 0.02982 | 0.00058 | 0.00250 | 93 | 0.00398 | 0.00477 | 134.742 | 0.99620 | 0.99944 |
| K09976; uncharacterized protein | 0.02981 | 0.00194 | 0.02981 | 0.00062 | 0.00310 | 65 | 0.02980 | 0.00058 | 0.00250 | 93 | 0.00398 | 0.00274 | 134.697 | 0.99782 | 0.99955 |
| comFA; competence protein ComFA | 0.02980 | 0.00194 | 0.02980 | 0.00062 | 0.00310 | 65 | 0.02980 | 0.00058 | 0.00250 | 93 | 0.00398 | 0.00083 | 134.687 | 0.99934 | 0.99957 |
| comGA; competence protein ComGA | 0.02980 | 0.00194 | 0.02980 | 0.00062 | 0.00310 | 65 | 0.02980 | 0.00058 | 0.00250 | 93 | 0.00398 | 0.00083 | 134.687 | 0.99934 | 0.99957 |
| comGC; competence protein ComGC | 0.02980 | 0.00194 | 0.02980 | 0.00062 | 0.00310 | 65 | 0.02980 | 0.00058 | 0.00250 | 93 | 0.00398 | 0.00083 | 134.687 | 0.99934 | 0.99957 |
| cshB; ATP-dependent RNA helicase CshB [EC:3.6.4.13] | 0.02980 | 0.00194 | 0.02980 | 0.00062 | 0.00310 | 65 | 0.02980 | 0.00058 | 0.00250 | 93 | 0.00398 | 0.00083 | 134.687 | 0.99934 | 0.99957 |
| gmhA, lpcA; D-sedoheptulose 7-phosphate isomerase [EC:5.3.1.28] | 0.02975 | 0.00181 | 0.03499 | 0.00064 | 0.00314 | 65 | 0.02609 | 0.00040 | 0.00207 | 93 | 0.00377 | 2.36308 | 116.452 | 0.01978 | 0.59095 |
| rlpA; rare lipoprotein A | 0.02973 | 0.00153 | 0.02823 | 0.00037 | 0.00237 | 65 | 0.03078 | 0.00037 | 0.00200 | 93 | 0.00310 | -0.82212 | 138.443 | 0.41242 | 0.81593 |
| TC.OOP; OmpA-OmpF porin, OOP family | 0.02973 | 0.00182 | 0.03304 | 0.00057 | 0.00295 | 65 | 0.02742 | 0.00049 | 0.00229 | 93 | 0.00373 | 1.50533 | 131.060 | 0.13465 | 0.77376 |
| lptF; lipopolysaccharide export system permease protein | 0.02973 | 0.00145 | 0.02990 | 0.00037 | 0.00239 | 65 | 0.02961 | 0.00031 | 0.00182 | 93 | 0.00300 | 0.09421 | 129.360 | 0.92509 | 0.98041 |
| codY; transcriptional pleiotropic repressor | 0.02969 | 0.00182 | 0.03016 | 0.00054 | 0.00287 | 65 | 0.02936 | 0.00052 | 0.00237 | 93 | 0.00373 | 0.21684 | 136.774 | 0.82866 | 0.95506 |
| lolD; lipoprotein-releasing system ATP-binding protein [EC:3.6.3.-] | 0.02966 | 0.00145 | 0.02988 | 0.00036 | 0.00237 | 65 | 0.02951 | 0.00032 | 0.00184 | 93 | 0.00300 | 0.12299 | 131.433 | 0.90230 | 0.97641 |
| dnaD; DNA replication protein | 0.02962 | 0.00194 | 0.02964 | 0.00062 | 0.00310 | 65 | 0.02960 | 0.00058 | 0.00250 | 93 | 0.00398 | 0.01060 | 134.715 | 0.99156 | 0.99884 |
| comGB; competence protein ComGB | 0.02958 | 0.00194 | 0.02963 | 0.00062 | 0.00310 | 65 | 0.02954 | 0.00058 | 0.00250 | 93 | 0.00398 | 0.02077 | 134.796 | 0.98346 | 0.99706 |
| fucP; MFS transporter, FHS family, L-fucose permease | 0.02956 | 0.00176 | 0.02836 | 0.00044 | 0.00260 | 65 | 0.03041 | 0.00052 | 0.00237 | 93 | 0.00352 | -0.58170 | 145.070 | 0.56167 | 0.86813 |
| ppiD; peptidyl-prolyl cis-trans isomerase D [EC:5.2.1.8] | 0.02955 | 0.00145 | 0.03001 | 0.00037 | 0.00238 | 65 | 0.02923 | 0.00031 | 0.00183 | 93 | 0.00300 | 0.25970 | 130.174 | 0.79550 | 0.94791 |
| dnaB; replication initiation and membrane attachment protein | 0.02952 | 0.00194 | 0.02956 | 0.00062 | 0.00309 | 65 | 0.02949 | 0.00058 | 0.00250 | 93 | 0.00397 | 0.01858 | 135.061 | 0.98520 | 0.99718 |
| cobA, btuR; cob(I)alamin adenosyltransferase [EC:2.5.1.17] | 0.02949 | 0.00176 | 0.03175 | 0.00063 | 0.00311 | 65 | 0.02792 | 0.00040 | 0.00206 | 93 | 0.00373 | 1.02697 | 116.967 | 0.30655 | 0.77376 |
| emrA; membrane fusion protein, multidrug efflux system | 0.02948 | 0.00253 | 0.02852 | 0.00091 | 0.00375 | 65 | 0.03016 | 0.00109 | 0.00342 | 93 | 0.00507 | -0.32396 | 145.003 | 0.74643 | 0.93202 |
| K09117; uncharacterized protein | 0.02945 | 0.00152 | 0.02894 | 0.00040 | 0.00248 | 65 | 0.02980 | 0.00034 | 0.00192 | 93 | 0.00313 | -0.27362 | 130.794 | 0.78481 | 0.94445 |
| dltA; D-alanine–poly(phosphoribitol) ligase subunit 1 [EC:6.1.1.13] | 0.02942 | 0.00197 | 0.02983 | 0.00064 | 0.00314 | 65 | 0.02914 | 0.00060 | 0.00255 | 93 | 0.00404 | 0.17135 | 135.134 | 0.86420 | 0.96263 |
| fliC; flagellin | 0.02939 | 0.00251 | 0.02700 | 0.00122 | 0.00433 | 65 | 0.03106 | 0.00085 | 0.00302 | 93 | 0.00528 | -0.76869 | 121.344 | 0.44357 | 0.83186 |
| FTR, FTH1, efeU; high-affinity iron transporter | 0.02939 | 0.00128 | 0.03137 | 0.00027 | 0.00203 | 65 | 0.02800 | 0.00025 | 0.00165 | 93 | 0.00261 | 1.28966 | 135.659 | 0.19936 | 0.77376 |
| AIBP, nnrE; NAD(P)H-hydrate epimerase [EC:5.1.99.6] | 0.02939 | 0.00152 | 0.02949 | 0.00033 | 0.00226 | 65 | 0.02931 | 0.00039 | 0.00206 | 93 | 0.00305 | 0.05883 | 144.970 | 0.95317 | 0.98792 |
| hslR; ribosome-associated heat shock protein Hsp15 | 0.02936 | 0.00125 | 0.02796 | 0.00025 | 0.00197 | 65 | 0.03033 | 0.00025 | 0.00163 | 93 | 0.00255 | -0.93061 | 137.164 | 0.35369 | 0.79236 |
| nrdH; glutaredoxin-like protein NrdH | 0.02932 | 0.00177 | 0.03051 | 0.00052 | 0.00283 | 65 | 0.02848 | 0.00048 | 0.00227 | 93 | 0.00363 | 0.55946 | 134.359 | 0.57678 | 0.87389 |
| cheW; purine-binding chemotaxis protein CheW | 0.02921 | 0.00245 | 0.02655 | 0.00095 | 0.00383 | 65 | 0.03106 | 0.00095 | 0.00319 | 93 | 0.00498 | -0.90449 | 137.663 | 0.36732 | 0.79587 |
| K06867; uncharacterized protein | 0.02920 | 0.00336 | 0.03346 | 0.00275 | 0.00651 | 65 | 0.02623 | 0.00110 | 0.00344 | 93 | 0.00736 | 0.98275 | 99.361 | 0.32812 | 0.77376 |
| hslU; ATP-dependent HslUV protease ATP-binding subunit HslU | 0.02917 | 0.00130 | 0.02952 | 0.00032 | 0.00220 | 65 | 0.02894 | 0.00023 | 0.00159 | 93 | 0.00271 | 0.21427 | 124.371 | 0.83068 | 0.95506 |
| penP; beta-lactamase class A [EC:3.5.2.6] | 0.02916 | 0.00212 | 0.02950 | 0.00074 | 0.00338 | 65 | 0.02892 | 0.00070 | 0.00274 | 93 | 0.00434 | 0.13247 | 135.129 | 0.89481 | 0.97195 |
| hslV, clpQ; ATP-dependent HslUV protease, peptidase subunit HslV [EC:3.4.25.2] | 0.02913 | 0.00129 | 0.02943 | 0.00031 | 0.00219 | 65 | 0.02892 | 0.00023 | 0.00159 | 93 | 0.00270 | 0.18875 | 124.995 | 0.85059 | 0.95754 |
| K06929; uncharacterized protein | 0.02909 | 0.00178 | 0.02850 | 0.00050 | 0.00277 | 65 | 0.02951 | 0.00051 | 0.00233 | 93 | 0.00362 | -0.27898 | 138.625 | 0.78067 | 0.94288 |
| capA, pgsA; gamma-polyglutamate biosynthesis protein CapA | 0.02900 | 0.00168 | 0.02774 | 0.00046 | 0.00265 | 65 | 0.02988 | 0.00044 | 0.00219 | 93 | 0.00343 | -0.62369 | 136.732 | 0.53387 | 0.86207 |
| nadC, QPRT; nicotinate-nucleotide pyrophosphorylase (carboxylating) [EC:2.4.2.19] | 0.02899 | 0.00135 | 0.02765 | 0.00029 | 0.00212 | 65 | 0.02992 | 0.00028 | 0.00175 | 93 | 0.00275 | -0.82395 | 136.537 | 0.41141 | 0.81492 |
| dhaL; phosphoenolpyruvate—glycerone phosphotransferase subunit DhaL [EC:2.7.1.121] | 0.02899 | 0.00166 | 0.03139 | 0.00044 | 0.00259 | 65 | 0.02731 | 0.00043 | 0.00216 | 93 | 0.00337 | 1.21227 | 137.394 | 0.22749 | 0.77376 |
| paaF, echA; enoyl-CoA hydratase [EC:4.2.1.17] | 0.02892 | 0.00332 | 0.02267 | 0.00120 | 0.00429 | 65 | 0.03329 | 0.00210 | 0.00475 | 93 | 0.00640 | -1.65855 | 155.029 | 0.09923 | 0.76942 |
| fabB; 3-oxoacyl-[acyl-carrier-protein] synthase I [EC:2.3.1.41] | 0.02891 | 0.00189 | 0.02689 | 0.00057 | 0.00297 | 65 | 0.03032 | 0.00056 | 0.00246 | 93 | 0.00386 | -0.88778 | 137.254 | 0.37621 | 0.79810 |
| dltB; membrane protein involved in D-alanine export | 0.02873 | 0.00196 | 0.02911 | 0.00064 | 0.00313 | 65 | 0.02847 | 0.00059 | 0.00252 | 93 | 0.00402 | 0.16145 | 134.700 | 0.87198 | 0.96576 |
| NRT, narK, nrtP, nasA; MFS transporter, NNP family, nitrate/nitrite transporter | 0.02872 | 0.00175 | 0.02842 | 0.00049 | 0.00275 | 65 | 0.02892 | 0.00048 | 0.00227 | 93 | 0.00357 | -0.13986 | 136.883 | 0.88898 | 0.97039 |
| metX; homoserine O-acetyltransferase/O-succinyltransferase [EC:2.3.1.31 2.3.1.46] | 0.02870 | 0.00179 | 0.02707 | 0.00051 | 0.00279 | 65 | 0.02984 | 0.00051 | 0.00234 | 93 | 0.00364 | -0.76050 | 138.241 | 0.44825 | 0.83372 |
| cybB; cytochrome b561 | 0.02868 | 0.00291 | 0.02768 | 0.00132 | 0.00451 | 65 | 0.02937 | 0.00135 | 0.00382 | 93 | 0.00591 | -0.28475 | 138.912 | 0.77626 | 0.94269 |
| mtaB; threonylcarbamoyladenosine tRNA methylthiotransferase MtaB [EC:2.8.4.5] | 0.02865 | 0.00184 | 0.02802 | 0.00059 | 0.00301 | 65 | 0.02908 | 0.00050 | 0.00232 | 93 | 0.00380 | -0.27979 | 130.492 | 0.78008 | 0.94269 |
| aroD; 3-dehydroquinate dehydratase I [EC:4.2.1.10] | 0.02855 | 0.00153 | 0.02760 | 0.00035 | 0.00232 | 65 | 0.02922 | 0.00039 | 0.00204 | 93 | 0.00309 | -0.52381 | 142.434 | 0.60123 | 0.88272 |
| cheR; chemotaxis protein methyltransferase CheR [EC:2.1.1.80] | 0.02853 | 0.00252 | 0.02693 | 0.00103 | 0.00399 | 65 | 0.02965 | 0.00099 | 0.00326 | 93 | 0.00515 | -0.52723 | 135.834 | 0.59889 | 0.88240 |
| korB, oorB, oforB; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit beta [EC:1.2.7.3 1.2.7.11] | 0.02850 | 0.00211 | 0.02488 | 0.00055 | 0.00292 | 65 | 0.03103 | 0.00080 | 0.00294 | 93 | 0.00414 | -1.48558 | 151.347 | 0.13947 | 0.77376 |
| ubiB, aarF; ubiquinone biosynthesis protein | 0.02844 | 0.00160 | 0.02762 | 0.00041 | 0.00252 | 65 | 0.02901 | 0.00040 | 0.00208 | 93 | 0.00327 | -0.42503 | 136.708 | 0.67148 | 0.91184 |
| pepDA, pepDB; dipeptidase [EC:3.4.-.-] | 0.02835 | 0.00437 | 0.03268 | 0.00448 | 0.00830 | 65 | 0.02532 | 0.00200 | 0.00464 | 93 | 0.00951 | 0.77296 | 103.241 | 0.44131 | 0.83186 |
| korA, oorA, oforA; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit alpha [EC:1.2.7.3 1.2.7.11] | 0.02833 | 0.00212 | 0.02483 | 0.00055 | 0.00291 | 65 | 0.03078 | 0.00081 | 0.00295 | 93 | 0.00415 | -1.43336 | 151.676 | 0.15381 | 0.77376 |
| modC; molybdate transport system ATP-binding protein [EC:3.6.3.29] | 0.02832 | 0.00130 | 0.02795 | 0.00027 | 0.00204 | 65 | 0.02858 | 0.00027 | 0.00169 | 93 | 0.00265 | -0.23597 | 136.872 | 0.81381 | 0.95371 |
| rph; ribonuclease PH [EC:2.7.7.56] | 0.02825 | 0.00127 | 0.02750 | 0.00030 | 0.00213 | 65 | 0.02877 | 0.00023 | 0.00158 | 93 | 0.00265 | -0.47881 | 126.709 | 0.63290 | 0.89696 |
| pbuX; xanthine permease | 0.02820 | 0.00179 | 0.02898 | 0.00059 | 0.00300 | 65 | 0.02765 | 0.00046 | 0.00222 | 93 | 0.00373 | 0.35674 | 126.649 | 0.72188 | 0.92156 |
| betT, betS; choline/glycine/proline betaine transport protein | 0.02814 | 0.00244 | 0.02892 | 0.00093 | 0.00379 | 65 | 0.02760 | 0.00095 | 0.00320 | 93 | 0.00496 | 0.26701 | 138.715 | 0.78986 | 0.94606 |
| emrB; MFS transporter, DHA2 family, multidrug resistance protein | 0.02813 | 0.00275 | 0.02617 | 0.00105 | 0.00402 | 65 | 0.02950 | 0.00131 | 0.00375 | 93 | 0.00550 | -0.60502 | 146.658 | 0.54610 | 0.86444 |
| cycB, ganO; arabinogalactan oligomer / maltooligosaccharide transport system substrate-binding protein | 0.02813 | 0.00162 | 0.02867 | 0.00040 | 0.00249 | 65 | 0.02775 | 0.00043 | 0.00214 | 93 | 0.00329 | 0.27977 | 140.100 | 0.78007 | 0.94269 |
| manC, cpsB; mannose-1-phosphate guanylyltransferase [EC:2.7.7.13] | 0.02808 | 0.00175 | 0.02480 | 0.00035 | 0.00233 | 65 | 0.03036 | 0.00057 | 0.00247 | 93 | 0.00339 | -1.63891 | 153.667 | 0.10328 | 0.77376 |
| ubiA; 4-hydroxybenzoate polyprenyltransferase [EC:2.5.1.39] | 0.02806 | 0.00122 | 0.02781 | 0.00026 | 0.00201 | 65 | 0.02824 | 0.00022 | 0.00153 | 93 | 0.00253 | -0.16800 | 129.413 | 0.86684 | 0.96327 |
| dpo; DNA polymerase bacteriophage-type [EC:2.7.7.7] | 0.02804 | 0.00179 | 0.02819 | 0.00061 | 0.00307 | 65 | 0.02794 | 0.00044 | 0.00217 | 93 | 0.00376 | 0.06627 | 122.933 | 0.94727 | 0.98674 |
| rbsA; ribose transport system ATP-binding protein [EC:3.6.3.17] | 0.02802 | 0.00277 | 0.02741 | 0.00180 | 0.00526 | 65 | 0.02845 | 0.00081 | 0.00296 | 93 | 0.00603 | -0.17248 | 103.644 | 0.86340 | 0.96263 |
| AROA1, aroA; chorismate mutase [EC:5.4.99.5] | 0.02801 | 0.00172 | 0.02743 | 0.00046 | 0.00265 | 65 | 0.02842 | 0.00048 | 0.00228 | 93 | 0.00349 | -0.28578 | 140.316 | 0.77547 | 0.94269 |
| dacB; serine-type D-Ala-D-Ala carboxypeptidase/endopeptidase (penicillin-binding protein 4) [EC:3.4.16.4 3.4.21.-] | 0.02799 | 0.00140 | 0.02638 | 0.00026 | 0.00200 | 65 | 0.02911 | 0.00035 | 0.00193 | 93 | 0.00278 | -0.98299 | 148.698 | 0.32721 | 0.77376 |
| pcp; pyroglutamyl-peptidase [EC:3.4.19.3] | 0.02795 | 0.00145 | 0.02901 | 0.00039 | 0.00244 | 65 | 0.02721 | 0.00029 | 0.00178 | 93 | 0.00302 | 0.59620 | 125.337 | 0.55212 | 0.86788 |
| PTS-Man-EIIA, manX; PTS system, mannose-specific IIA component [EC:2.7.1.191] | 0.02793 | 0.00181 | 0.02795 | 0.00053 | 0.00286 | 65 | 0.02791 | 0.00052 | 0.00236 | 93 | 0.00371 | 0.01142 | 136.904 | 0.99091 | 0.99847 |
| ygaC; uncharacterized protein | 0.02792 | 0.00178 | 0.02733 | 0.00047 | 0.00269 | 65 | 0.02834 | 0.00053 | 0.00238 | 93 | 0.00359 | -0.28012 | 142.794 | 0.77979 | 0.94269 |
| mntH; manganese transport protein | 0.02792 | 0.00127 | 0.02639 | 0.00031 | 0.00217 | 65 | 0.02899 | 0.00022 | 0.00154 | 93 | 0.00266 | -0.97751 | 122.880 | 0.33024 | 0.77640 |
| eda; 2-dehydro-3-deoxyphosphogluconate aldolase / (4S)-4-hydroxy-2-oxoglutarate aldolase [EC:4.1.2.14 4.1.3.42] | 0.02790 | 0.00149 | 0.02799 | 0.00046 | 0.00265 | 65 | 0.02785 | 0.00028 | 0.00174 | 93 | 0.00317 | 0.04395 | 115.880 | 0.96502 | 0.99101 |
| hisZ; ATP phosphoribosyltransferase regulatory subunit | 0.02789 | 0.00119 | 0.02601 | 0.00026 | 0.00199 | 65 | 0.02920 | 0.00020 | 0.00146 | 93 | 0.00246 | -1.29285 | 126.111 | 0.19843 | 0.77376 |
| nuoG; NADH-quinone oxidoreductase subunit G [EC:1.6.5.3] | 0.02784 | 0.00135 | 0.02771 | 0.00032 | 0.00221 | 65 | 0.02793 | 0.00027 | 0.00171 | 93 | 0.00280 | -0.07790 | 130.804 | 0.93802 | 0.98459 |
| fabI; enoyl-[acyl-carrier protein] reductase I [EC:1.3.1.9 1.3.1.10] | 0.02775 | 0.00142 | 0.02551 | 0.00030 | 0.00214 | 65 | 0.02931 | 0.00033 | 0.00188 | 93 | 0.00284 | -1.33657 | 142.020 | 0.18350 | 0.77376 |
| pilT; twitching motility protein PilT | 0.02774 | 0.00133 | 0.02890 | 0.00029 | 0.00211 | 65 | 0.02693 | 0.00027 | 0.00171 | 93 | 0.00271 | 0.72688 | 135.045 | 0.46856 | 0.83837 |
| speG, SAT; diamine N-acetyltransferase [EC:2.3.1.57] | 0.02760 | 0.00165 | 0.02637 | 0.00035 | 0.00234 | 65 | 0.02846 | 0.00048 | 0.00228 | 93 | 0.00327 | -0.63865 | 149.604 | 0.52403 | 0.85711 |
| aceE; pyruvate dehydrogenase E1 component [EC:1.2.4.1] | 0.02759 | 0.00209 | 0.02642 | 0.00060 | 0.00303 | 65 | 0.02841 | 0.00076 | 0.00286 | 93 | 0.00417 | -0.47588 | 147.506 | 0.63486 | 0.89696 |
| E3.1.3.41; 4-nitrophenyl phosphatase [EC:3.1.3.41] | 0.02755 | 0.00178 | 0.02998 | 0.00057 | 0.00295 | 65 | 0.02586 | 0.00046 | 0.00222 | 93 | 0.00369 | 1.11634 | 128.112 | 0.26636 | 0.77376 |
| E3.1.3.15B; histidinol-phosphatase (PHP family) [EC:3.1.3.15] | 0.02754 | 0.00215 | 0.03078 | 0.00104 | 0.00400 | 65 | 0.02527 | 0.00051 | 0.00234 | 93 | 0.00463 | 1.18785 | 106.574 | 0.23753 | 0.77376 |
| pepX; X-Pro dipeptidyl-peptidase [EC:3.4.14.11] | 0.02753 | 0.00213 | 0.02742 | 0.00069 | 0.00326 | 65 | 0.02761 | 0.00074 | 0.00283 | 93 | 0.00431 | -0.04477 | 141.088 | 0.96435 | 0.99101 |
| aspB; aspartate aminotransferase [EC:2.6.1.1] | 0.02753 | 0.00188 | 0.02417 | 0.00048 | 0.00272 | 65 | 0.02988 | 0.00060 | 0.00255 | 93 | 0.00372 | -1.53427 | 147.028 | 0.12711 | 0.77376 |
| fer; ferredoxin | 0.02751 | 0.00164 | 0.02614 | 0.00039 | 0.00246 | 65 | 0.02847 | 0.00045 | 0.00219 | 93 | 0.00330 | -0.70723 | 143.136 | 0.48057 | 0.84354 |
| pyrD; dihydroorotate dehydrogenase (fumarate) [EC:1.3.98.1] | 0.02751 | 0.00172 | 0.02832 | 0.00047 | 0.00269 | 65 | 0.02694 | 0.00047 | 0.00224 | 93 | 0.00350 | 0.39417 | 137.411 | 0.69407 | 0.91701 |
| ppdK; pyruvate, orthophosphate dikinase [EC:2.7.9.1] | 0.02750 | 0.00170 | 0.02707 | 0.00050 | 0.00278 | 65 | 0.02779 | 0.00043 | 0.00215 | 93 | 0.00352 | -0.20590 | 130.717 | 0.83719 | 0.95506 |
| fadL; long-chain fatty acid transport protein | 0.02746 | 0.00191 | 0.02836 | 0.00052 | 0.00284 | 65 | 0.02682 | 0.00062 | 0.00258 | 93 | 0.00384 | 0.39969 | 144.757 | 0.68997 | 0.91567 |
| PTS-Cel-EIIA, celC, chbA; PTS system, cellobiose-specific IIA component [EC:2.7.1.196 2.7.1.205] | 0.02745 | 0.00230 | 0.03143 | 0.00147 | 0.00475 | 65 | 0.02466 | 0.00039 | 0.00204 | 93 | 0.00517 | 1.31042 | 87.670 | 0.19348 | 0.77376 |
| cobC, phpB; alpha-ribazole phosphatase [EC:3.1.3.73] | 0.02744 | 0.00143 | 0.02826 | 0.00035 | 0.00231 | 65 | 0.02686 | 0.00031 | 0.00182 | 93 | 0.00294 | 0.47310 | 132.539 | 0.63692 | 0.89696 |
| alsD, budA, aldC; acetolactate decarboxylase [EC:4.1.1.5] | 0.02744 | 0.00183 | 0.02864 | 0.00057 | 0.00297 | 65 | 0.02659 | 0.00051 | 0.00233 | 93 | 0.00378 | 0.54363 | 132.276 | 0.58761 | 0.87732 |
| lpxC; UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase [EC:3.5.1.108] | 0.02736 | 0.00127 | 0.02991 | 0.00030 | 0.00214 | 65 | 0.02557 | 0.00022 | 0.00155 | 93 | 0.00264 | 1.64510 | 124.855 | 0.10247 | 0.77376 |
| pdxS, pdx1; pyridoxal 5’-phosphate synthase pdxS subunit [EC:4.3.3.6] | 0.02731 | 0.00151 | 0.02608 | 0.00035 | 0.00233 | 65 | 0.02817 | 0.00037 | 0.00199 | 93 | 0.00306 | -0.68166 | 139.441 | 0.49658 | 0.84797 |
| menB; naphthoate synthase [EC:4.1.3.36] | 0.02727 | 0.00148 | 0.02761 | 0.00040 | 0.00247 | 65 | 0.02704 | 0.00031 | 0.00183 | 93 | 0.00308 | 0.18329 | 126.752 | 0.85486 | 0.95861 |
| K06872; uncharacterized protein | 0.02723 | 0.00153 | 0.02522 | 0.00037 | 0.00240 | 65 | 0.02863 | 0.00036 | 0.00198 | 93 | 0.00311 | -1.09722 | 136.763 | 0.27447 | 0.77376 |
| clpE; ATP-dependent Clp protease ATP-binding subunit ClpE | 0.02718 | 0.00208 | 0.02854 | 0.00076 | 0.00343 | 65 | 0.02622 | 0.00063 | 0.00261 | 93 | 0.00431 | 0.53913 | 129.430 | 0.59072 | 0.87896 |
| speE, SRM; spermidine synthase [EC:2.5.1.16] | 0.02713 | 0.00127 | 0.02544 | 0.00024 | 0.00194 | 65 | 0.02831 | 0.00026 | 0.00167 | 93 | 0.00256 | -1.12217 | 140.742 | 0.26370 | 0.77376 |
| slyD; FKBP-type peptidyl-prolyl cis-trans isomerase SlyD [EC:5.2.1.8] | 0.02709 | 0.00145 | 0.02785 | 0.00035 | 0.00231 | 65 | 0.02656 | 0.00032 | 0.00186 | 93 | 0.00297 | 0.43589 | 134.415 | 0.66362 | 0.91006 |
| cstA; carbon starvation protein | 0.02701 | 0.00151 | 0.02585 | 0.00037 | 0.00239 | 65 | 0.02783 | 0.00036 | 0.00196 | 93 | 0.00309 | -0.63916 | 135.962 | 0.52379 | 0.85711 |
| gspG; general secretion pathway protein G | 0.02700 | 0.00212 | 0.02882 | 0.00081 | 0.00353 | 65 | 0.02573 | 0.00064 | 0.00263 | 93 | 0.00441 | 0.70011 | 127.388 | 0.48514 | 0.84422 |
| paaH, hbd, fadB, mmgB; 3-hydroxybutyryl-CoA dehydrogenase [EC:1.1.1.157] | 0.02699 | 0.00212 | 0.02603 | 0.00065 | 0.00317 | 65 | 0.02766 | 0.00076 | 0.00285 | 93 | 0.00426 | -0.38346 | 143.969 | 0.70195 | 0.92004 |
| mvaA; hydroxymethylglutaryl-CoA reductase [EC:1.1.1.88] | 0.02697 | 0.00176 | 0.02862 | 0.00053 | 0.00286 | 65 | 0.02581 | 0.00047 | 0.00224 | 93 | 0.00363 | 0.77412 | 131.989 | 0.44024 | 0.83186 |
| dltD; D-alanine transfer protein | 0.02695 | 0.00191 | 0.02783 | 0.00063 | 0.00312 | 65 | 0.02633 | 0.00054 | 0.00242 | 93 | 0.00395 | 0.37931 | 130.886 | 0.70507 | 0.92004 |
| lacD; tagatose 1,6-diphosphate aldolase [EC:4.1.2.40] | 0.02689 | 0.00190 | 0.02866 | 0.00063 | 0.00310 | 65 | 0.02565 | 0.00053 | 0.00239 | 93 | 0.00392 | 0.77055 | 130.522 | 0.44237 | 0.83186 |
| E2.7.8.26, cobS, cobV; adenosylcobinamide-GDP ribazoletransferase [EC:2.7.8.26] | 0.02688 | 0.00140 | 0.02772 | 0.00035 | 0.00232 | 65 | 0.02629 | 0.00029 | 0.00176 | 93 | 0.00291 | 0.49167 | 128.853 | 0.62379 | 0.89159 |
| yghU, yfcG; GSH-dependent disulfide-bond oxidoreductase [EC:1.8.4.-] | 0.02683 | 0.00182 | 0.02361 | 0.00043 | 0.00256 | 65 | 0.02909 | 0.00059 | 0.00251 | 93 | 0.00359 | -1.52901 | 149.918 | 0.12837 | 0.77376 |
| fbp3; fructose-1,6-bisphosphatase III [EC:3.1.3.11] | 0.02678 | 0.00181 | 0.02771 | 0.00058 | 0.00300 | 65 | 0.02613 | 0.00047 | 0.00226 | 93 | 0.00375 | 0.42222 | 128.562 | 0.67357 | 0.91184 |
| gspD; general secretion pathway protein D | 0.02676 | 0.00190 | 0.02775 | 0.00055 | 0.00292 | 65 | 0.02606 | 0.00059 | 0.00251 | 93 | 0.00385 | 0.43898 | 140.319 | 0.66135 | 0.90968 |
| E1.1.1.40, maeB; malate dehydrogenase (oxaloacetate-decarboxylating)(NADP+) [EC:1.1.1.40] | 0.02675 | 0.00136 | 0.02403 | 0.00025 | 0.00196 | 65 | 0.02866 | 0.00031 | 0.00184 | 93 | 0.00269 | -1.72237 | 146.965 | 0.08711 | 0.76567 |
| cobP, cobU; adenosylcobinamide kinase / adenosylcobinamide-phosphate guanylyltransferase [EC:2.7.1.156 2.7.7.62] | 0.02674 | 0.00145 | 0.02744 | 0.00035 | 0.00233 | 65 | 0.02625 | 0.00032 | 0.00186 | 93 | 0.00298 | 0.39977 | 133.701 | 0.68996 | 0.91567 |
| PTS-Glc-EIIC, ptsG; PTS system, glucose-specific IIC component | 0.02674 | 0.00171 | 0.02883 | 0.00047 | 0.00270 | 65 | 0.02528 | 0.00046 | 0.00222 | 93 | 0.00349 | 1.01620 | 136.309 | 0.31133 | 0.77376 |
| pcnB; poly(A) polymerase [EC:2.7.7.19] | 0.02671 | 0.00141 | 0.02581 | 0.00035 | 0.00230 | 65 | 0.02735 | 0.00029 | 0.00177 | 93 | 0.00291 | -0.52982 | 130.348 | 0.59714 | 0.88102 |
| ampC; beta-lactamase class C [EC:3.5.2.6] | 0.02666 | 0.00145 | 0.02658 | 0.00030 | 0.00213 | 65 | 0.02672 | 0.00036 | 0.00197 | 93 | 0.00290 | -0.04678 | 145.874 | 0.96275 | 0.99031 |
| DLST, sucB; 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide succinyltransferase) [EC:2.3.1.61] | 0.02665 | 0.00142 | 0.02463 | 0.00031 | 0.00218 | 65 | 0.02806 | 0.00032 | 0.00186 | 93 | 0.00286 | -1.19521 | 139.352 | 0.23404 | 0.77376 |
| frdA; fumarate reductase flavoprotein subunit [EC:1.3.5.4] | 0.02664 | 0.00301 | 0.03146 | 0.00160 | 0.00497 | 65 | 0.02327 | 0.00130 | 0.00374 | 93 | 0.00622 | 1.31746 | 128.441 | 0.19003 | 0.77376 |
| nhaA; Na+:H+ antiporter, NhaA family | 0.02662 | 0.00134 | 0.02674 | 0.00031 | 0.00220 | 65 | 0.02654 | 0.00027 | 0.00170 | 93 | 0.00278 | 0.07355 | 130.978 | 0.94148 | 0.98528 |
| MVD, mvaD; diphosphomevalonate decarboxylase [EC:4.1.1.33] | 0.02661 | 0.00178 | 0.02855 | 0.00054 | 0.00287 | 65 | 0.02526 | 0.00047 | 0.00225 | 93 | 0.00365 | 0.90095 | 132.291 | 0.36925 | 0.79587 |
| ezrA; septation ring formation regulator | 0.02661 | 0.00184 | 0.02708 | 0.00060 | 0.00303 | 65 | 0.02628 | 0.00049 | 0.00231 | 93 | 0.00380 | 0.21094 | 129.505 | 0.83327 | 0.95506 |
| E2.7.1.36, MVK, mvaK1; mevalonate kinase [EC:2.7.1.36] | 0.02660 | 0.00178 | 0.02851 | 0.00054 | 0.00287 | 65 | 0.02526 | 0.00047 | 0.00226 | 93 | 0.00365 | 0.88854 | 132.289 | 0.37587 | 0.79810 |
| PTS-Ula-EIIC, ulaA, sgaT; PTS system, ascorbate-specific IIC component | 0.02657 | 0.00181 | 0.02824 | 0.00072 | 0.00333 | 65 | 0.02541 | 0.00038 | 0.00201 | 93 | 0.00389 | 0.72816 | 109.111 | 0.46807 | 0.83837 |
| cobQ, cbiP; adenosylcobyric acid synthase [EC:6.3.5.10] | 0.02650 | 0.00183 | 0.03024 | 0.00079 | 0.00349 | 65 | 0.02389 | 0.00034 | 0.00190 | 93 | 0.00398 | 1.59672 | 101.382 | 0.11344 | 0.77376 |
| ltaS; lipoteichoic acid synthase [EC:2.7.8.20] | 0.02650 | 0.00257 | 0.02968 | 0.00155 | 0.00488 | 65 | 0.02427 | 0.00069 | 0.00272 | 93 | 0.00559 | 0.96634 | 103.017 | 0.33614 | 0.78250 |
| menE; O-succinylbenzoic acid—CoA ligase [EC:6.2.1.26] | 0.02649 | 0.00145 | 0.02690 | 0.00039 | 0.00246 | 65 | 0.02621 | 0.00029 | 0.00178 | 93 | 0.00304 | 0.22905 | 124.621 | 0.81921 | 0.95506 |
| slt; soluble lytic murein transglycosylase [EC:4.2.2.-] | 0.02649 | 0.00123 | 0.02613 | 0.00025 | 0.00195 | 65 | 0.02674 | 0.00024 | 0.00160 | 93 | 0.00253 | -0.23860 | 136.208 | 0.81177 | 0.95229 |
| macA; membrane fusion protein, macrolide-specific efflux system | 0.02649 | 0.00192 | 0.02746 | 0.00062 | 0.00309 | 65 | 0.02580 | 0.00056 | 0.00245 | 93 | 0.00394 | 0.42120 | 132.996 | 0.67429 | 0.91194 |
| dltC; D-alanine–poly(phosphoribitol) ligase subunit 2 [EC:6.1.1.13] | 0.02645 | 0.00185 | 0.02721 | 0.00060 | 0.00304 | 65 | 0.02592 | 0.00050 | 0.00232 | 93 | 0.00382 | 0.33936 | 129.929 | 0.73489 | 0.92721 |
| dsbD; thiol:disulfide interchange protein DsbD [EC:1.8.1.8] | 0.02644 | 0.00141 | 0.02576 | 0.00031 | 0.00218 | 65 | 0.02692 | 0.00032 | 0.00185 | 93 | 0.00286 | -0.40843 | 139.046 | 0.68359 | 0.91514 |
| rnhC; ribonuclease HIII [EC:3.1.26.4] | 0.02643 | 0.00183 | 0.02551 | 0.00051 | 0.00279 | 65 | 0.02708 | 0.00055 | 0.00243 | 93 | 0.00371 | -0.42222 | 141.338 | 0.67350 | 0.91184 |
| GSP13; general stress protein 13 | 0.02642 | 0.00173 | 0.02629 | 0.00043 | 0.00258 | 65 | 0.02651 | 0.00051 | 0.00234 | 93 | 0.00348 | -0.06115 | 144.323 | 0.95133 | 0.98792 |
| virD4, lvhD4; type IV secretion system protein VirD4 | 0.02635 | 0.00293 | 0.02815 | 0.00156 | 0.00489 | 65 | 0.02509 | 0.00123 | 0.00364 | 93 | 0.00610 | 0.50141 | 127.244 | 0.61695 | 0.88903 |
| cheB; two-component system, chemotaxis family, protein-glutamate methylesterase/glutaminase [EC:3.1.1.61 3.5.1.44] | 0.02632 | 0.00235 | 0.02468 | 0.00090 | 0.00372 | 65 | 0.02747 | 0.00086 | 0.00304 | 93 | 0.00481 | -0.58023 | 135.964 | 0.56272 | 0.86813 |
| lldG; L-lactate dehydrogenase complex protein LldG | 0.02631 | 0.00154 | 0.02787 | 0.00041 | 0.00250 | 65 | 0.02522 | 0.00035 | 0.00195 | 93 | 0.00317 | 0.83485 | 131.861 | 0.40531 | 0.81189 |
| E2.3.3.10; hydroxymethylglutaryl-CoA synthase [EC:2.3.3.10] | 0.02628 | 0.00177 | 0.02804 | 0.00053 | 0.00285 | 65 | 0.02505 | 0.00047 | 0.00225 | 93 | 0.00363 | 0.82583 | 132.653 | 0.41038 | 0.81416 |
| comGD; competence protein ComGD | 0.02624 | 0.00181 | 0.02662 | 0.00057 | 0.00297 | 65 | 0.02597 | 0.00049 | 0.00229 | 93 | 0.00375 | 0.17269 | 130.433 | 0.86317 | 0.96263 |
| tamB; translocation and assembly module TamB | 0.02623 | 0.00170 | 0.02669 | 0.00044 | 0.00260 | 65 | 0.02591 | 0.00048 | 0.00226 | 93 | 0.00344 | 0.22720 | 141.323 | 0.82059 | 0.95506 |
| coiA; competence protein CoiA | 0.02621 | 0.00184 | 0.02747 | 0.00059 | 0.00301 | 65 | 0.02533 | 0.00050 | 0.00231 | 93 | 0.00379 | 0.56431 | 130.204 | 0.57351 | 0.87292 |
| ganQ; arabinogalactan oligomer / maltooligosaccharide transport system permease protein | 0.02619 | 0.00157 | 0.02581 | 0.00036 | 0.00235 | 65 | 0.02645 | 0.00041 | 0.00210 | 93 | 0.00315 | -0.20521 | 143.431 | 0.83770 | 0.95506 |
| vgrG; type VI secretion system secreted protein VgrG | 0.02616 | 0.00284 | 0.02845 | 0.00137 | 0.00459 | 65 | 0.02456 | 0.00121 | 0.00361 | 93 | 0.00584 | 0.66585 | 132.380 | 0.50666 | 0.84956 |
| pabC; 4-amino-4-deoxychorismate lyase [EC:4.1.3.38] | 0.02612 | 0.00134 | 0.02708 | 0.00030 | 0.00215 | 65 | 0.02545 | 0.00028 | 0.00173 | 93 | 0.00276 | 0.59030 | 134.245 | 0.55598 | 0.86813 |
| rluC; 23S rRNA pseudouridine955/2504/2580 synthase [EC:5.4.99.24] | 0.02610 | 0.00141 | 0.02775 | 0.00034 | 0.00230 | 65 | 0.02495 | 0.00030 | 0.00178 | 93 | 0.00291 | 0.96373 | 131.066 | 0.33696 | 0.78334 |
| hprA; glycerate dehydrogenase [EC:1.1.1.29] | 0.02609 | 0.00155 | 0.02887 | 0.00041 | 0.00251 | 65 | 0.02414 | 0.00035 | 0.00194 | 93 | 0.00318 | 1.48868 | 130.972 | 0.13898 | 0.77376 |
| comC; leader peptidase (prepilin peptidase) / N-methyltransferase [EC:3.4.23.43 2.1.1.-] | 0.02607 | 0.00173 | 0.02662 | 0.00052 | 0.00282 | 65 | 0.02569 | 0.00044 | 0.00218 | 93 | 0.00357 | 0.25951 | 130.701 | 0.79565 | 0.94791 |
| TC.DASS; divalent anion:Na+ symporter, DASS family | 0.02600 | 0.00221 | 0.02862 | 0.00096 | 0.00384 | 65 | 0.02417 | 0.00064 | 0.00263 | 93 | 0.00466 | 0.95399 | 119.668 | 0.34201 | 0.78567 |
| pbp1b; penicillin-binding protein 1B | 0.02599 | 0.00173 | 0.02589 | 0.00043 | 0.00258 | 65 | 0.02606 | 0.00051 | 0.00234 | 93 | 0.00348 | -0.04985 | 144.636 | 0.96031 | 0.98903 |
| PGLS, pgl, devB; 6-phosphogluconolactonase [EC:3.1.1.31] | 0.02596 | 0.00134 | 0.02530 | 0.00030 | 0.00213 | 65 | 0.02642 | 0.00028 | 0.00173 | 93 | 0.00275 | -0.40811 | 135.250 | 0.68384 | 0.91529 |
| oxyR; LysR family transcriptional regulator, hydrogen peroxide-inducible genes activator | 0.02591 | 0.00124 | 0.02403 | 0.00023 | 0.00188 | 65 | 0.02722 | 0.00025 | 0.00165 | 93 | 0.00250 | -1.27610 | 141.949 | 0.20400 | 0.77376 |
| E2.7.4.2, mvaK2; phosphomevalonate kinase [EC:2.7.4.2] | 0.02589 | 0.00178 | 0.02764 | 0.00054 | 0.00288 | 65 | 0.02466 | 0.00048 | 0.00227 | 93 | 0.00367 | 0.81476 | 132.490 | 0.41667 | 0.81842 |
| licT, bglG; beta-glucoside operon transcriptional antiterminator | 0.02585 | 0.00170 | 0.02687 | 0.00053 | 0.00286 | 65 | 0.02514 | 0.00041 | 0.00211 | 93 | 0.00355 | 0.48720 | 126.492 | 0.62696 | 0.89265 |
| queE; 7-carboxy-7-deazaguanine synthase [EC:4.3.99.3] | 0.02582 | 0.00116 | 0.02414 | 0.00020 | 0.00174 | 65 | 0.02700 | 0.00022 | 0.00154 | 93 | 0.00233 | -1.22733 | 142.792 | 0.22172 | 0.77376 |
| OGDH, sucA; 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2] | 0.02581 | 0.00141 | 0.02438 | 0.00032 | 0.00221 | 65 | 0.02682 | 0.00031 | 0.00183 | 93 | 0.00287 | -0.85089 | 137.233 | 0.39631 | 0.80459 |
| lacR; DeoR family transcriptional regulator, lactose phosphotransferase system repressor | 0.02578 | 0.00192 | 0.02503 | 0.00049 | 0.00275 | 65 | 0.02631 | 0.00065 | 0.00265 | 93 | 0.00382 | -0.33712 | 148.805 | 0.73650 | 0.92721 |
| TC.SSS; solute:Na+ symporter, SSS family | 0.02576 | 0.00166 | 0.02546 | 0.00046 | 0.00267 | 65 | 0.02598 | 0.00042 | 0.00212 | 93 | 0.00341 | -0.15233 | 133.400 | 0.87916 | 0.96779 |
| ybcJ; ribosome-associated protein | 0.02574 | 0.00150 | 0.02892 | 0.00045 | 0.00262 | 65 | 0.02352 | 0.00028 | 0.00175 | 93 | 0.00315 | 1.71132 | 117.389 | 0.08966 | 0.76567 |
| pilC; type IV pilus assembly protein PilC | 0.02570 | 0.00125 | 0.02742 | 0.00029 | 0.00211 | 65 | 0.02449 | 0.00022 | 0.00153 | 93 | 0.00260 | 1.12501 | 125.247 | 0.26274 | 0.77376 |
| pdxA; 4-hydroxythreonine-4-phosphate dehydrogenase [EC:1.1.1.262] | 0.02565 | 0.00124 | 0.02392 | 0.00021 | 0.00178 | 65 | 0.02686 | 0.00027 | 0.00170 | 93 | 0.00246 | -1.19750 | 148.303 | 0.23302 | 0.77376 |
| ATOX1, ATX1, copZ, golB; copper chaperone | 0.02563 | 0.00139 | 0.02361 | 0.00030 | 0.00215 | 65 | 0.02703 | 0.00030 | 0.00181 | 93 | 0.00281 | -1.21719 | 138.322 | 0.22560 | 0.77376 |
| bamD; outer membrane protein assembly factor BamD | 0.02562 | 0.00125 | 0.02401 | 0.00024 | 0.00192 | 65 | 0.02674 | 0.00025 | 0.00165 | 93 | 0.00253 | -1.07665 | 140.410 | 0.28348 | 0.77376 |
| ftnA, ftn; ferritin [EC:1.16.3.2] | 0.02559 | 0.00184 | 0.02549 | 0.00055 | 0.00291 | 65 | 0.02566 | 0.00053 | 0.00239 | 93 | 0.00377 | -0.04367 | 136.306 | 0.96523 | 0.99101 |
| purU; formyltetrahydrofolate deformylase [EC:3.5.1.10] | 0.02554 | 0.00209 | 0.02356 | 0.00064 | 0.00314 | 65 | 0.02692 | 0.00072 | 0.00279 | 93 | 0.00420 | -0.80001 | 142.800 | 0.42503 | 0.82542 |
| argD; acetylornithine/N-succinyldiaminopimelate aminotransferase [EC:2.6.1.11 2.6.1.17] | 0.02552 | 0.00135 | 0.02625 | 0.00029 | 0.00211 | 65 | 0.02501 | 0.00029 | 0.00176 | 93 | 0.00275 | 0.44986 | 137.927 | 0.65351 | 0.90621 |
| ssuA; sulfonate transport system substrate-binding protein | 0.02551 | 0.00282 | 0.02349 | 0.00099 | 0.00391 | 65 | 0.02692 | 0.00145 | 0.00395 | 93 | 0.00556 | -0.61803 | 151.532 | 0.53748 | 0.86207 |
| mgs, bgsB; 1,2-diacylglycerol 3-alpha-glucosyltransferase [EC:2.4.1.337] | 0.02549 | 0.00178 | 0.02760 | 0.00053 | 0.00287 | 65 | 0.02401 | 0.00047 | 0.00226 | 93 | 0.00365 | 0.98470 | 132.499 | 0.32657 | 0.77376 |
| ENDOG; endonuclease G, mitochondrial | 0.02548 | 0.00206 | 0.02377 | 0.00058 | 0.00299 | 65 | 0.02668 | 0.00073 | 0.00281 | 93 | 0.00410 | -0.70979 | 147.166 | 0.47896 | 0.84314 |
| menD; 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase [EC:2.2.1.9] | 0.02547 | 0.00149 | 0.02524 | 0.00041 | 0.00250 | 65 | 0.02563 | 0.00032 | 0.00184 | 93 | 0.00311 | -0.12799 | 126.404 | 0.89836 | 0.97377 |
| mobA; molybdenum cofactor guanylyltransferase [EC:2.7.7.77] | 0.02545 | 0.00120 | 0.02486 | 0.00022 | 0.00186 | 65 | 0.02587 | 0.00023 | 0.00158 | 93 | 0.00244 | -0.41380 | 139.282 | 0.67966 | 0.91311 |
| fdhD; FdhD protein | 0.02544 | 0.00118 | 0.02398 | 0.00024 | 0.00191 | 65 | 0.02646 | 0.00021 | 0.00150 | 93 | 0.00242 | -1.02627 | 132.227 | 0.30664 | 0.77376 |
| K07220; uncharacterized protein | 0.02542 | 0.00135 | 0.02443 | 0.00030 | 0.00214 | 65 | 0.02612 | 0.00029 | 0.00176 | 93 | 0.00277 | -0.61185 | 136.422 | 0.54166 | 0.86207 |
| comGF; competence protein ComGF | 0.02539 | 0.00177 | 0.02636 | 0.00053 | 0.00286 | 65 | 0.02471 | 0.00047 | 0.00226 | 93 | 0.00364 | 0.45484 | 132.710 | 0.64997 | 0.90409 |
| acpD, azoR; FMN-dependent NADH-azoreductase [EC:1.7.1.17] | 0.02536 | 0.00175 | 0.02409 | 0.00054 | 0.00288 | 65 | 0.02625 | 0.00045 | 0.00220 | 93 | 0.00362 | -0.59412 | 129.749 | 0.55347 | 0.86788 |
| glnR; MerR family transcriptional regulator, glutamine synthetase repressor | 0.02536 | 0.00181 | 0.02440 | 0.00048 | 0.00272 | 65 | 0.02603 | 0.00055 | 0.00243 | 93 | 0.00365 | -0.44577 | 143.304 | 0.65644 | 0.90679 |
| nadA; quinolinate synthase [EC:2.5.1.72] | 0.02530 | 0.00130 | 0.02434 | 0.00027 | 0.00203 | 65 | 0.02597 | 0.00027 | 0.00169 | 93 | 0.00264 | -0.61724 | 137.550 | 0.53810 | 0.86207 |
| E3.4.16.4; D-alanyl-D-alanine carboxypeptidase [EC:3.4.16.4] | 0.02527 | 0.00210 | 0.02339 | 0.00059 | 0.00302 | 65 | 0.02659 | 0.00077 | 0.00287 | 93 | 0.00417 | -0.76906 | 147.824 | 0.44308 | 0.83186 |
| catE; catechol 2,3-dioxygenase [EC:1.13.11.2] | 0.02527 | 0.00181 | 0.02330 | 0.00046 | 0.00266 | 65 | 0.02665 | 0.00056 | 0.00245 | 93 | 0.00361 | -0.92820 | 145.612 | 0.35484 | 0.79373 |
| ltaE; threonine aldolase [EC:4.1.2.48] | 0.02527 | 0.00152 | 0.02562 | 0.00042 | 0.00253 | 65 | 0.02502 | 0.00034 | 0.00190 | 93 | 0.00316 | 0.19022 | 128.125 | 0.84943 | 0.95712 |
| bioF; 8-amino-7-oxononanoate synthase [EC:2.3.1.47] | 0.02517 | 0.00147 | 0.02455 | 0.00036 | 0.00235 | 65 | 0.02559 | 0.00033 | 0.00188 | 93 | 0.00302 | -0.34394 | 134.012 | 0.73143 | 0.92524 |
| cheA; two-component system, chemotaxis family, sensor kinase CheA [EC:2.7.13.3] | 0.02517 | 0.00214 | 0.02278 | 0.00070 | 0.00328 | 65 | 0.02683 | 0.00074 | 0.00281 | 93 | 0.00432 | -0.93715 | 140.025 | 0.35030 | 0.79120 |
| dedA; membrane-associated protein | 0.02515 | 0.00190 | 0.02654 | 0.00073 | 0.00335 | 65 | 0.02418 | 0.00046 | 0.00223 | 93 | 0.00403 | 0.58438 | 117.251 | 0.56009 | 0.86813 |
| tyrB; aromatic-amino-acid transaminase [EC:2.6.1.57] | 0.02515 | 0.00143 | 0.02786 | 0.00037 | 0.00237 | 65 | 0.02326 | 0.00029 | 0.00177 | 93 | 0.00296 | 1.55382 | 127.457 | 0.12271 | 0.77376 |
| NQO1; NAD(P)H dehydrogenase (quinone) [EC:1.6.5.2] | 0.02512 | 0.00145 | 0.02538 | 0.00032 | 0.00223 | 65 | 0.02495 | 0.00034 | 0.00191 | 93 | 0.00294 | 0.14726 | 139.687 | 0.88314 | 0.96974 |
| htsT; energy-coupling factor transport system substrate-specific component | 0.02512 | 0.00179 | 0.02568 | 0.00057 | 0.00297 | 65 | 0.02473 | 0.00046 | 0.00223 | 93 | 0.00371 | 0.25724 | 127.953 | 0.79741 | 0.94810 |
| K07005; uncharacterized protein | 0.02506 | 0.00246 | 0.02868 | 0.00123 | 0.00436 | 65 | 0.02253 | 0.00076 | 0.00285 | 93 | 0.00521 | 1.18118 | 115.723 | 0.23995 | 0.77376 |
| pbp2A; penicillin-binding protein 2A [EC:2.4.1.129 3.4.16.4] | 0.02506 | 0.00179 | 0.02637 | 0.00055 | 0.00292 | 65 | 0.02415 | 0.00048 | 0.00227 | 93 | 0.00369 | 0.60268 | 131.417 | 0.54776 | 0.86549 |
| minD; septum site-determining protein MinD | 0.02505 | 0.00117 | 0.02580 | 0.00025 | 0.00197 | 65 | 0.02453 | 0.00020 | 0.00145 | 93 | 0.00244 | 0.51868 | 126.512 | 0.60489 | 0.88465 |
| czcA; cobalt-zinc-cadmium resistance protein CzcA | 0.02505 | 0.00317 | 0.02358 | 0.00179 | 0.00525 | 65 | 0.02607 | 0.00146 | 0.00397 | 93 | 0.00658 | -0.37867 | 128.790 | 0.70556 | 0.92004 |
| dcp; peptidyl-dipeptidase Dcp [EC:3.4.15.5] | 0.02504 | 0.00191 | 0.02160 | 0.00043 | 0.00257 | 65 | 0.02744 | 0.00067 | 0.00268 | 93 | 0.00372 | -1.57175 | 152.940 | 0.11808 | 0.77376 |
| ccmH; cytochrome c-type biogenesis protein CcmH | 0.02503 | 0.00210 | 0.02450 | 0.00073 | 0.00335 | 65 | 0.02541 | 0.00068 | 0.00271 | 93 | 0.00431 | -0.21158 | 134.751 | 0.83275 | 0.95506 |
| K06962; uncharacterized protein | 0.02502 | 0.00149 | 0.02605 | 0.00041 | 0.00252 | 65 | 0.02430 | 0.00031 | 0.00184 | 93 | 0.00312 | 0.56019 | 125.368 | 0.57635 | 0.87389 |
| MOCS3, UBA4; adenylyltransferase and sulfurtransferase [EC:2.7.7.80 2.8.1.11] | 0.02501 | 0.00151 | 0.02296 | 0.00031 | 0.00219 | 65 | 0.02644 | 0.00039 | 0.00205 | 93 | 0.00300 | -1.15930 | 147.024 | 0.24821 | 0.77376 |
| pcaC; 4-carboxymuconolactone decarboxylase [EC:4.1.1.44] | 0.02500 | 0.00195 | 0.02172 | 0.00054 | 0.00289 | 65 | 0.02729 | 0.00064 | 0.00261 | 93 | 0.00390 | -1.43155 | 144.393 | 0.15443 | 0.77376 |
| nadB; L-aspartate oxidase [EC:1.4.3.16] | 0.02497 | 0.00130 | 0.02438 | 0.00027 | 0.00205 | 65 | 0.02538 | 0.00027 | 0.00170 | 93 | 0.00266 | -0.37672 | 137.242 | 0.70697 | 0.92004 |
| cysJ; sulfite reductase (NADPH) flavoprotein alpha-component [EC:1.8.1.2] | 0.02495 | 0.00202 | 0.02428 | 0.00066 | 0.00318 | 65 | 0.02543 | 0.00064 | 0.00262 | 93 | 0.00412 | -0.27828 | 136.784 | 0.78122 | 0.94304 |
| dgkA; undecaprenol kinase [EC:2.7.1.66] | 0.02490 | 0.00168 | 0.02561 | 0.00048 | 0.00271 | 65 | 0.02440 | 0.00043 | 0.00215 | 93 | 0.00346 | 0.35143 | 133.318 | 0.72582 | 0.92177 |
| E2.7.1.12, gntK, idnK; gluconokinase [EC:2.7.1.12] | 0.02488 | 0.00156 | 0.02590 | 0.00055 | 0.00290 | 65 | 0.02417 | 0.00027 | 0.00172 | 93 | 0.00337 | 0.51188 | 107.746 | 0.60978 | 0.88693 |
| dgkA, DGK; diacylglycerol kinase (ATP) [EC:2.7.1.107] | 0.02487 | 0.00149 | 0.02486 | 0.00036 | 0.00234 | 65 | 0.02487 | 0.00035 | 0.00194 | 93 | 0.00304 | -0.00555 | 137.101 | 0.99558 | 0.99944 |
| cbiB, cobD; adenosylcobinamide-phosphate synthase [EC:6.3.1.10] | 0.02481 | 0.00138 | 0.02626 | 0.00034 | 0.00227 | 65 | 0.02379 | 0.00028 | 0.00173 | 93 | 0.00285 | 0.86757 | 129.240 | 0.38724 | 0.80262 |
| manB; phosphomannomutase [EC:5.4.2.8] | 0.02478 | 0.00141 | 0.02282 | 0.00027 | 0.00204 | 65 | 0.02615 | 0.00035 | 0.00193 | 93 | 0.00281 | -1.18644 | 147.637 | 0.23735 | 0.77376 |
| ABC.SP.A; putative spermidine/putrescine transport system ATP-binding protein | 0.02475 | 0.00321 | 0.02203 | 0.00126 | 0.00441 | 65 | 0.02666 | 0.00189 | 0.00451 | 93 | 0.00631 | -0.73440 | 152.196 | 0.46383 | 0.83699 |
| glnB; nitrogen regulatory protein P-II 1 | 0.02472 | 0.00153 | 0.02331 | 0.00039 | 0.00246 | 65 | 0.02570 | 0.00035 | 0.00194 | 93 | 0.00314 | -0.76238 | 132.725 | 0.44719 | 0.83372 |
| yajQ; cyclic-di-GMP-binding protein | 0.02471 | 0.00125 | 0.02460 | 0.00027 | 0.00205 | 65 | 0.02479 | 0.00023 | 0.00158 | 93 | 0.00259 | -0.07349 | 130.343 | 0.94153 | 0.98528 |
| ABC.SP.P; putative spermidine/putrescine transport system permease protein | 0.02468 | 0.00334 | 0.02148 | 0.00133 | 0.00452 | 65 | 0.02691 | 0.00207 | 0.00471 | 93 | 0.00653 | -0.83189 | 152.959 | 0.40677 | 0.81189 |
| MCH; medium-chain acyl-[acyl-carrier-protein] hydrolase [EC:3.1.2.21] | 0.02465 | 0.00180 | 0.02662 | 0.00056 | 0.00292 | 65 | 0.02328 | 0.00048 | 0.00227 | 93 | 0.00370 | 0.90240 | 131.194 | 0.36850 | 0.79587 |
| ABC.GLN1.S; putative glutamine transport system substrate-binding protein | 0.02463 | 0.00208 | 0.02555 | 0.00071 | 0.00330 | 65 | 0.02399 | 0.00068 | 0.00270 | 93 | 0.00426 | 0.36705 | 136.114 | 0.71415 | 0.92004 |
| tldD; TldD protein | 0.02461 | 0.00206 | 0.02332 | 0.00073 | 0.00336 | 65 | 0.02550 | 0.00063 | 0.00261 | 93 | 0.00425 | -0.51294 | 131.385 | 0.60885 | 0.88667 |
| dcd; dCTP deaminase [EC:3.5.4.13] | 0.02458 | 0.00131 | 0.02371 | 0.00029 | 0.00210 | 65 | 0.02519 | 0.00026 | 0.00168 | 93 | 0.00269 | -0.54895 | 133.921 | 0.58395 | 0.87720 |
| fliY; cystine transport system substrate-binding protein | 0.02458 | 0.00155 | 0.02789 | 0.00048 | 0.00272 | 65 | 0.02227 | 0.00030 | 0.00179 | 93 | 0.00326 | 1.72284 | 116.306 | 0.08758 | 0.76567 |
| MOCS2B, moaE; molybdopterin synthase catalytic subunit [EC:2.8.1.12] | 0.02456 | 0.00134 | 0.02234 | 0.00026 | 0.00200 | 65 | 0.02612 | 0.00030 | 0.00179 | 93 | 0.00268 | -1.40783 | 143.649 | 0.16134 | 0.77376 |
| phnE; phosphonate transport system permease protein | 0.02456 | 0.00197 | 0.02619 | 0.00064 | 0.00314 | 65 | 0.02342 | 0.00059 | 0.00253 | 93 | 0.00403 | 0.68860 | 134.433 | 0.49226 | 0.84797 |
| yafQ; mRNA interferase YafQ [EC:3.1.-.-] | 0.02454 | 0.00209 | 0.02504 | 0.00085 | 0.00361 | 65 | 0.02419 | 0.00059 | 0.00252 | 93 | 0.00440 | 0.19304 | 121.343 | 0.84725 | 0.95682 |
| nagZ; beta-N-acetylhexosaminidase [EC:3.2.1.52] | 0.02453 | 0.00125 | 0.02332 | 0.00028 | 0.00209 | 65 | 0.02537 | 0.00022 | 0.00154 | 93 | 0.00260 | -0.78607 | 126.390 | 0.43330 | 0.82874 |
| mlaA, vacJ; phospholipid-binding lipoprotein MlaA | 0.02446 | 0.00175 | 0.02712 | 0.00051 | 0.00280 | 65 | 0.02260 | 0.00046 | 0.00222 | 93 | 0.00357 | 1.26351 | 133.069 | 0.20862 | 0.77376 |
| PTS-Ntr-EIIA, ptsN; PTS system, nitrogen regulatory IIA component [EC:2.7.1.-] | 0.02441 | 0.00185 | 0.02936 | 0.00085 | 0.00362 | 65 | 0.02094 | 0.00030 | 0.00179 | 93 | 0.00404 | 2.08267 | 95.156 | 0.03996 | 0.63577 |
| zapE; cell division protein ZapE | 0.02440 | 0.00214 | 0.02396 | 0.00077 | 0.00343 | 65 | 0.02471 | 0.00070 | 0.00274 | 93 | 0.00439 | -0.17009 | 133.748 | 0.86520 | 0.96270 |
| lysX2; putative lysine transport system permease protein | 0.02440 | 0.00160 | 0.02488 | 0.00037 | 0.00240 | 65 | 0.02406 | 0.00043 | 0.00215 | 93 | 0.00322 | 0.25332 | 143.642 | 0.80038 | 0.94855 |
| nuoA; NADH-quinone oxidoreductase subunit A [EC:1.6.5.3] | 0.02430 | 0.00119 | 0.02149 | 0.00020 | 0.00174 | 65 | 0.02627 | 0.00023 | 0.00159 | 93 | 0.00235 | -2.03211 | 145.000 | 0.04397 | 0.65981 |
| motA; chemotaxis protein MotA | 0.02429 | 0.00199 | 0.02317 | 0.00077 | 0.00344 | 65 | 0.02507 | 0.00053 | 0.00238 | 93 | 0.00419 | -0.45382 | 120.716 | 0.65077 | 0.90423 |
| iunH; purine nucleosidase [EC:3.2.2.1] | 0.02423 | 0.00221 | 0.02384 | 0.00085 | 0.00362 | 65 | 0.02451 | 0.00072 | 0.00278 | 93 | 0.00457 | -0.14612 | 130.266 | 0.88405 | 0.96974 |
| nuoL; NADH-quinone oxidoreductase subunit L [EC:1.6.5.3] | 0.02423 | 0.00119 | 0.02121 | 0.00019 | 0.00170 | 65 | 0.02635 | 0.00024 | 0.00161 | 93 | 0.00234 | -2.19446 | 147.947 | 0.02976 | 0.61356 |
| pepA; glutamyl aminopeptidase [EC:3.4.11.7] | 0.02420 | 0.00182 | 0.02436 | 0.00049 | 0.00274 | 65 | 0.02409 | 0.00055 | 0.00244 | 93 | 0.00367 | 0.07312 | 143.118 | 0.94181 | 0.98528 |
| nuoM; NADH-quinone oxidoreductase subunit M [EC:1.6.5.3] | 0.02418 | 0.00118 | 0.02124 | 0.00019 | 0.00170 | 65 | 0.02623 | 0.00023 | 0.00158 | 93 | 0.00233 | -2.14197 | 146.472 | 0.03385 | 0.62130 |
| nuoH; NADH-quinone oxidoreductase subunit H [EC:1.6.5.3] | 0.02417 | 0.00118 | 0.02121 | 0.00019 | 0.00170 | 65 | 0.02624 | 0.00023 | 0.00159 | 93 | 0.00233 | -2.16257 | 146.669 | 0.03220 | 0.62130 |
| nasT; two-component system, response regulator / RNA-binding antiterminator | 0.02413 | 0.00132 | 0.02620 | 0.00034 | 0.00229 | 65 | 0.02268 | 0.00022 | 0.00155 | 93 | 0.00277 | 1.27179 | 118.811 | 0.20593 | 0.77376 |
| pdxT, pdx2; 5’-phosphate synthase pdxT subunit [EC:4.3.3.6] | 0.02411 | 0.00137 | 0.02088 | 0.00023 | 0.00188 | 65 | 0.02637 | 0.00034 | 0.00191 | 93 | 0.00268 | -2.05270 | 151.745 | 0.04182 | 0.64661 |
| nuoN; NADH-quinone oxidoreductase subunit N [EC:1.6.5.3] | 0.02411 | 0.00117 | 0.02121 | 0.00019 | 0.00170 | 65 | 0.02613 | 0.00023 | 0.00156 | 93 | 0.00231 | -2.13162 | 145.544 | 0.03472 | 0.62130 |
| arcC; carbamate kinase [EC:2.7.2.2] | 0.02409 | 0.00139 | 0.02595 | 0.00034 | 0.00229 | 65 | 0.02279 | 0.00028 | 0.00173 | 93 | 0.00287 | 1.10339 | 129.042 | 0.27191 | 0.77376 |
| nuoB; NADH-quinone oxidoreductase subunit B [EC:1.6.5.3] | 0.02409 | 0.00116 | 0.02110 | 0.00019 | 0.00169 | 65 | 0.02617 | 0.00022 | 0.00154 | 93 | 0.00229 | -2.21398 | 145.139 | 0.02839 | 0.61334 |
| panD; aspartate 1-decarboxylase [EC:4.1.1.11] | 0.02403 | 0.00130 | 0.02076 | 0.00020 | 0.00175 | 65 | 0.02631 | 0.00031 | 0.00181 | 93 | 0.00252 | -2.20512 | 152.867 | 0.02894 | 0.61334 |
| nuoI; NADH-quinone oxidoreductase subunit I [EC:1.6.5.3] | 0.02402 | 0.00116 | 0.02137 | 0.00019 | 0.00172 | 65 | 0.02587 | 0.00022 | 0.00153 | 93 | 0.00231 | -1.95008 | 143.054 | 0.05312 | 0.69853 |
| nuoJ; NADH-quinone oxidoreductase subunit J [EC:1.6.5.3] | 0.02394 | 0.00115 | 0.02108 | 0.00018 | 0.00169 | 65 | 0.02595 | 0.00022 | 0.00153 | 93 | 0.00228 | -2.13643 | 144.720 | 0.03433 | 0.62130 |
| ahpF; alkyl hydroperoxide reductase subunit F [EC:1.6.4.-] | 0.02392 | 0.00121 | 0.02335 | 0.00021 | 0.00178 | 65 | 0.02431 | 0.00025 | 0.00165 | 93 | 0.00243 | -0.39564 | 146.110 | 0.69295 | 0.91677 |
| lptD, imp, ostA; LPS-assembly protein | 0.02391 | 0.00119 | 0.02395 | 0.00023 | 0.00188 | 65 | 0.02389 | 0.00022 | 0.00153 | 93 | 0.00243 | 0.02454 | 135.661 | 0.98046 | 0.99657 |
| mmsA, iolA, ALDH6A1; malonate-semialdehyde dehydrogenase (acetylating) / methylmalonate-semialdehyde dehydrogenase [EC:1.2.1.18 1.2.1.27] | 0.02387 | 0.00242 | 0.02013 | 0.00074 | 0.00337 | 65 | 0.02649 | 0.00105 | 0.00336 | 93 | 0.00475 | -1.33714 | 150.749 | 0.18319 | 0.77376 |
| mtsC; iron/zinc/manganese/copper transport system permease protein | 0.02387 | 0.00182 | 0.02370 | 0.00049 | 0.00275 | 65 | 0.02398 | 0.00055 | 0.00244 | 93 | 0.00368 | -0.07800 | 142.806 | 0.93794 | 0.98459 |
| mtsA; iron/zinc/manganese/copper transport system substrate-binding protein | 0.02386 | 0.00182 | 0.02369 | 0.00049 | 0.00275 | 65 | 0.02398 | 0.00055 | 0.00244 | 93 | 0.00368 | -0.07881 | 142.781 | 0.93729 | 0.98459 |
| agaS; tagatose-6-phosphate ketose/aldose isomerase [EC:5.-.-.-] | 0.02386 | 0.00149 | 0.02476 | 0.00034 | 0.00227 | 65 | 0.02322 | 0.00036 | 0.00198 | 93 | 0.00301 | 0.51062 | 141.215 | 0.61041 | 0.88722 |
| bgaB, lacA; beta-galactosidase [EC:3.2.1.23] | 0.02385 | 0.00162 | 0.02161 | 0.00033 | 0.00225 | 65 | 0.02542 | 0.00047 | 0.00226 | 93 | 0.00319 | -1.19405 | 151.173 | 0.23433 | 0.77376 |
| SCO1_2; protein SCO1/2 | 0.02384 | 0.00203 | 0.02095 | 0.00066 | 0.00319 | 65 | 0.02586 | 0.00064 | 0.00263 | 93 | 0.00413 | -1.18784 | 136.820 | 0.23695 | 0.77376 |
| lptA; lipopolysaccharide export system protein LptA | 0.02379 | 0.00118 | 0.02384 | 0.00023 | 0.00188 | 65 | 0.02376 | 0.00022 | 0.00153 | 93 | 0.00243 | 0.03062 | 135.093 | 0.97562 | 0.99571 |
| mtsB; iron/zinc/manganese/copper transport system ATP-binding protein | 0.02379 | 0.00182 | 0.02362 | 0.00049 | 0.00275 | 65 | 0.02391 | 0.00055 | 0.00244 | 93 | 0.00367 | -0.07961 | 142.701 | 0.93666 | 0.98459 |
| lpxH; UDP-2,3-diacylglucosamine hydrolase [EC:3.6.1.54] | 0.02372 | 0.00124 | 0.02298 | 0.00023 | 0.00186 | 65 | 0.02424 | 0.00026 | 0.00167 | 93 | 0.00250 | -0.50409 | 143.517 | 0.61497 | 0.88842 |
| K08974; putative membrane protein | 0.02369 | 0.00152 | 0.02512 | 0.00036 | 0.00237 | 65 | 0.02269 | 0.00037 | 0.00199 | 93 | 0.00309 | 0.78328 | 138.288 | 0.43480 | 0.82874 |
| msmE; raffinose/stachyose/melibiose transport system substrate-binding protein | 0.02369 | 0.00235 | 0.02286 | 0.00120 | 0.00429 | 65 | 0.02426 | 0.00065 | 0.00264 | 93 | 0.00504 | -0.27767 | 110.663 | 0.78178 | 0.94310 |
| yuiF; putative amino acid transporter | 0.02368 | 0.00167 | 0.02824 | 0.00065 | 0.00317 | 65 | 0.02050 | 0.00027 | 0.00171 | 93 | 0.00360 | 2.15093 | 100.600 | 0.03388 | 0.62130 |
| K07160; UPF0271 protein | 0.02368 | 0.00150 | 0.02375 | 0.00036 | 0.00236 | 65 | 0.02363 | 0.00036 | 0.00196 | 93 | 0.00307 | 0.03937 | 137.206 | 0.96865 | 0.99265 |
| dhaK; phosphoenolpyruvate—glycerone phosphotransferase subunit DhaK [EC:2.7.1.121] | 0.02367 | 0.00148 | 0.02699 | 0.00042 | 0.00255 | 65 | 0.02134 | 0.00028 | 0.00175 | 93 | 0.00309 | 1.83231 | 119.938 | 0.06939 | 0.73522 |
| putA; RHH-type transcriptional regulator, proline utilization regulon repressor / proline dehydrogenase / delta 1-pyrroline-5-carboxylate dehydrogenase [EC:1.5.5.2 1.2.1.88] | 0.02360 | 0.00163 | 0.02300 | 0.00041 | 0.00252 | 65 | 0.02403 | 0.00043 | 0.00214 | 93 | 0.00331 | -0.31112 | 139.432 | 0.75618 | 0.93669 |
| pabB; para-aminobenzoate synthetase component I [EC:2.6.1.85] | 0.02359 | 0.00137 | 0.02391 | 0.00031 | 0.00218 | 65 | 0.02336 | 0.00029 | 0.00177 | 93 | 0.00281 | 0.19691 | 135.047 | 0.84419 | 0.95539 |
| E6.3.5.1, NADSYN1, QNS1, nadE; NAD+ synthase (glutamine-hydrolysing) [EC:6.3.5.1] | 0.02355 | 0.00143 | 0.02216 | 0.00029 | 0.00211 | 65 | 0.02453 | 0.00035 | 0.00194 | 93 | 0.00286 | -0.82726 | 145.508 | 0.40945 | 0.81416 |
| dksA; DnaK suppressor protein | 0.02355 | 0.00188 | 0.02300 | 0.00055 | 0.00291 | 65 | 0.02394 | 0.00057 | 0.00248 | 93 | 0.00382 | -0.24710 | 139.629 | 0.80519 | 0.94958 |
| K17076, lysY; putative lysine transport system ATP-binding protein [EC:3.6.3.-] | 0.02352 | 0.00155 | 0.02408 | 0.00035 | 0.00233 | 65 | 0.02313 | 0.00040 | 0.00207 | 93 | 0.00312 | 0.30373 | 142.935 | 0.76177 | 0.93813 |
| PTS-Ula-EIIB, ulaB, sgaB; PTS system, ascorbate-specific IIB component [EC:2.7.1.194] | 0.02351 | 0.00173 | 0.02470 | 0.00067 | 0.00320 | 65 | 0.02267 | 0.00034 | 0.00190 | 93 | 0.00373 | 0.54407 | 107.822 | 0.58752 | 0.87732 |
| metC; cystathionine beta-lyase [EC:4.4.1.8] | 0.02350 | 0.00148 | 0.02540 | 0.00046 | 0.00267 | 65 | 0.02218 | 0.00027 | 0.00169 | 93 | 0.00316 | 1.02019 | 113.168 | 0.30981 | 0.77376 |
| pheA; chorismate mutase / prephenate dehydratase [EC:5.4.99.5 4.2.1.51] | 0.02350 | 0.00131 | 0.02560 | 0.00037 | 0.00238 | 65 | 0.02203 | 0.00020 | 0.00148 | 93 | 0.00281 | 1.27153 | 111.230 | 0.20619 | 0.77376 |
| cmoA; tRNA (cmo5U34)-methyltransferase [EC:2.1.1.-] | 0.02347 | 0.00143 | 0.02471 | 0.00028 | 0.00206 | 65 | 0.02260 | 0.00035 | 0.00195 | 93 | 0.00284 | 0.74206 | 147.535 | 0.45923 | 0.83438 |
| treR2, treR; GntR family transcriptional regulator, trehalose operon transcriptional repressor | 0.02343 | 0.00137 | 0.02495 | 0.00036 | 0.00236 | 65 | 0.02236 | 0.00025 | 0.00164 | 93 | 0.00287 | 0.90066 | 121.066 | 0.36956 | 0.79587 |
| fadA, fadI; acetyl-CoA acyltransferase [EC:2.3.1.16] | 0.02342 | 0.00195 | 0.02061 | 0.00050 | 0.00278 | 65 | 0.02539 | 0.00066 | 0.00267 | 93 | 0.00386 | -1.23833 | 148.422 | 0.21755 | 0.77376 |
| GARS, glyS1; glycyl-tRNA synthetase [EC:6.1.1.14] | 0.02339 | 0.00141 | 0.02078 | 0.00027 | 0.00203 | 65 | 0.02521 | 0.00034 | 0.00191 | 93 | 0.00279 | -1.58742 | 147.124 | 0.11456 | 0.77376 |
| flA; adenosyl-fluoride synthase [EC:2.5.1.63] | 0.02338 | 0.00167 | 0.02284 | 0.00038 | 0.00243 | 65 | 0.02376 | 0.00049 | 0.00229 | 93 | 0.00334 | -0.27342 | 147.406 | 0.78492 | 0.94445 |
| ABC.CYST.P; cystine transport system permease protein | 0.02338 | 0.00158 | 0.02577 | 0.00049 | 0.00275 | 65 | 0.02171 | 0.00032 | 0.00187 | 93 | 0.00332 | 1.22395 | 119.197 | 0.22338 | 0.77376 |
| ald; alanine dehydrogenase [EC:1.4.1.1] | 0.02333 | 0.00130 | 0.01904 | 0.00020 | 0.00174 | 65 | 0.02633 | 0.00030 | 0.00178 | 93 | 0.00249 | -2.92719 | 152.298 | 0.00395 | 0.52084 |
| DHODH, pyrD; dihydroorotate dehydrogenase [EC:1.3.5.2] | 0.02333 | 0.00133 | 0.02244 | 0.00028 | 0.00207 | 65 | 0.02395 | 0.00028 | 0.00174 | 93 | 0.00270 | -0.55635 | 138.271 | 0.57887 | 0.87532 |
| nuoK; NADH-quinone oxidoreductase subunit K [EC:1.6.5.3] | 0.02327 | 0.00116 | 0.02066 | 0.00019 | 0.00172 | 65 | 0.02510 | 0.00022 | 0.00154 | 93 | 0.00231 | -1.92052 | 143.496 | 0.05678 | 0.70711 |
| MUT; methylmalonyl-CoA mutase [EC:5.4.99.2] | 0.02323 | 0.00185 | 0.02031 | 0.00054 | 0.00289 | 65 | 0.02527 | 0.00054 | 0.00241 | 93 | 0.00376 | -1.32131 | 137.803 | 0.18859 | 0.77376 |
| lldF; L-lactate dehydrogenase complex protein LldF | 0.02322 | 0.00135 | 0.02283 | 0.00031 | 0.00219 | 65 | 0.02350 | 0.00028 | 0.00172 | 93 | 0.00279 | -0.23912 | 132.243 | 0.81138 | 0.95214 |
| mtgA; monofunctional glycosyltransferase [EC:2.4.1.129] | 0.02322 | 0.00115 | 0.02208 | 0.00021 | 0.00180 | 65 | 0.02401 | 0.00021 | 0.00149 | 93 | 0.00234 | -0.82597 | 136.923 | 0.41026 | 0.81416 |
| hipA; serine/threonine-protein kinase HipA [EC:2.7.11.1] | 0.02320 | 0.00166 | 0.02218 | 0.00037 | 0.00239 | 65 | 0.02391 | 0.00048 | 0.00228 | 93 | 0.00330 | -0.52499 | 148.035 | 0.60038 | 0.88240 |
| E1.1.1.1, adh; alcohol dehydrogenase [EC:1.1.1.1] | 0.02319 | 0.00154 | 0.02023 | 0.00034 | 0.00230 | 65 | 0.02526 | 0.00039 | 0.00205 | 93 | 0.00308 | -1.63354 | 143.409 | 0.10455 | 0.77376 |
| lldE; L-lactate dehydrogenase complex protein LldE | 0.02311 | 0.00135 | 0.02278 | 0.00031 | 0.00219 | 65 | 0.02334 | 0.00028 | 0.00173 | 93 | 0.00279 | -0.20059 | 132.623 | 0.84133 | 0.95506 |
| comX1_2; competence protein ComX | 0.02310 | 0.00207 | 0.02197 | 0.00051 | 0.00280 | 65 | 0.02390 | 0.00080 | 0.00293 | 93 | 0.00405 | -0.47714 | 153.175 | 0.63394 | 0.89696 |
| K06999; phospholipase/carboxylesterase | 0.02309 | 0.00172 | 0.02254 | 0.00056 | 0.00293 | 65 | 0.02348 | 0.00041 | 0.00210 | 93 | 0.00360 | -0.26026 | 124.041 | 0.79509 | 0.94791 |
| lipB; lipoyl(octanoyl) transferase [EC:2.3.1.181] | 0.02309 | 0.00134 | 0.02232 | 0.00030 | 0.00216 | 65 | 0.02363 | 0.00027 | 0.00171 | 93 | 0.00276 | -0.47486 | 133.116 | 0.63566 | 0.89696 |
| pilB; type IV pilus assembly protein PilB | 0.02308 | 0.00127 | 0.02543 | 0.00032 | 0.00221 | 65 | 0.02143 | 0.00021 | 0.00151 | 93 | 0.00267 | 1.49871 | 119.636 | 0.13658 | 0.77376 |
| phnB; PhnB protein | 0.02307 | 0.00143 | 0.02213 | 0.00032 | 0.00223 | 65 | 0.02373 | 0.00032 | 0.00186 | 93 | 0.00290 | -0.55119 | 137.806 | 0.58240 | 0.87718 |
| rne; ribonuclease E [EC:3.1.26.12] | 0.02297 | 0.00133 | 0.02266 | 0.00030 | 0.00216 | 65 | 0.02319 | 0.00026 | 0.00169 | 93 | 0.00274 | -0.19170 | 131.835 | 0.84827 | 0.95712 |
| dagK; diacylglycerol kinase (ATP) [EC:2.7.1.107] | 0.02292 | 0.00158 | 0.02416 | 0.00048 | 0.00271 | 65 | 0.02206 | 0.00034 | 0.00190 | 93 | 0.00331 | 0.63449 | 121.724 | 0.52695 | 0.85845 |
| dapL; N-acetyldiaminopimelate deacetylase [EC:3.5.1.47] | 0.02282 | 0.00163 | 0.02356 | 0.00043 | 0.00259 | 65 | 0.02231 | 0.00041 | 0.00211 | 93 | 0.00334 | 0.37490 | 135.748 | 0.70832 | 0.92004 |
| tadB; tight adherence protein B | 0.02282 | 0.00173 | 0.02270 | 0.00057 | 0.00297 | 65 | 0.02291 | 0.00041 | 0.00209 | 93 | 0.00363 | -0.05750 | 122.339 | 0.95424 | 0.98792 |
| speD, AMD1; S-adenosylmethionine decarboxylase [EC:4.1.1.50] | 0.02280 | 0.00157 | 0.02349 | 0.00049 | 0.00273 | 65 | 0.02232 | 0.00033 | 0.00189 | 93 | 0.00332 | 0.35091 | 120.464 | 0.72627 | 0.92177 |
| ABC.GLN1.A; putative glutamine transport system ATP-binding protein [EC:3.6.3.-] | 0.02280 | 0.00189 | 0.02350 | 0.00054 | 0.00289 | 65 | 0.02232 | 0.00059 | 0.00251 | 93 | 0.00383 | 0.30800 | 141.291 | 0.75853 | 0.93719 |
| cobB-cbiA; cobyrinic acid a,c-diamide synthase [EC:6.3.5.9 6.3.5.11] | 0.02277 | 0.00132 | 0.02454 | 0.00035 | 0.00231 | 65 | 0.02153 | 0.00022 | 0.00155 | 93 | 0.00278 | 1.08320 | 117.981 | 0.28093 | 0.77376 |
| treC; trehalose-6-phosphate hydrolase [EC:3.2.1.93] | 0.02276 | 0.00141 | 0.02419 | 0.00041 | 0.00252 | 65 | 0.02176 | 0.00025 | 0.00163 | 93 | 0.00300 | 0.81080 | 114.977 | 0.41915 | 0.81980 |
| msmF; raffinose/stachyose/melibiose transport system permease protein | 0.02275 | 0.00218 | 0.02211 | 0.00112 | 0.00415 | 65 | 0.02320 | 0.00051 | 0.00233 | 93 | 0.00476 | -0.22798 | 103.526 | 0.82011 | 0.95506 |
| nfo; deoxyribonuclease IV [EC:3.1.21.2] | 0.02272 | 0.00137 | 0.02320 | 0.00034 | 0.00229 | 65 | 0.02239 | 0.00027 | 0.00171 | 93 | 0.00286 | 0.28221 | 127.230 | 0.77824 | 0.94269 |
| cpaF, tadA; pilus assembly protein CpaF | 0.02269 | 0.00171 | 0.02239 | 0.00059 | 0.00300 | 65 | 0.02291 | 0.00038 | 0.00203 | 93 | 0.00362 | -0.14287 | 118.702 | 0.88663 | 0.97020 |
| dsbA; thiol:disulfide interchange protein DsbA | 0.02266 | 0.00183 | 0.02331 | 0.00056 | 0.00293 | 65 | 0.02221 | 0.00051 | 0.00235 | 93 | 0.00376 | 0.29331 | 134.131 | 0.76974 | 0.93993 |
| DNPEP; aspartyl aminopeptidase [EC:3.4.11.21] | 0.02262 | 0.00140 | 0.02651 | 0.00039 | 0.00244 | 65 | 0.01990 | 0.00024 | 0.00160 | 93 | 0.00292 | 2.26181 | 116.143 | 0.02557 | 0.59941 |
| PTS-Ula-EIIA, ulaC, sgaA; PTS system, ascorbate-specific IIA component [EC:2.7.1.194] | 0.02261 | 0.00123 | 0.02376 | 0.00029 | 0.00212 | 65 | 0.02181 | 0.00020 | 0.00148 | 93 | 0.00259 | 0.75111 | 121.396 | 0.45404 | 0.83438 |
| iscA; iron-sulfur cluster assembly protein | 0.02258 | 0.00124 | 0.01999 | 0.00022 | 0.00186 | 65 | 0.02440 | 0.00025 | 0.00165 | 93 | 0.00248 | -1.77755 | 142.865 | 0.07760 | 0.75461 |
| liaS; two-component system, NarL family, sensor histidine kinase LiaS [EC:2.7.13.3] | 0.02258 | 0.00171 | 0.02263 | 0.00044 | 0.00261 | 65 | 0.02255 | 0.00048 | 0.00227 | 93 | 0.00345 | 0.02245 | 141.207 | 0.98212 | 0.99680 |
| mltD, dniR; membrane-bound lytic murein transglycosylase D [EC:4.2.2.-] | 0.02253 | 0.00145 | 0.02157 | 0.00033 | 0.00227 | 65 | 0.02320 | 0.00034 | 0.00190 | 93 | 0.00296 | -0.55132 | 138.078 | 0.58231 | 0.87718 |
| cycA; D-serine/D-alanine/glycine transporter | 0.02252 | 0.00159 | 0.02139 | 0.00041 | 0.00252 | 65 | 0.02331 | 0.00039 | 0.00205 | 93 | 0.00325 | -0.58936 | 135.606 | 0.55660 | 0.86813 |
| E4.2.1.2A, fumA, fumB; fumarate hydratase, class I [EC:4.2.1.2] | 0.02252 | 0.00118 | 0.02014 | 0.00019 | 0.00170 | 65 | 0.02418 | 0.00024 | 0.00160 | 93 | 0.00234 | -1.72749 | 147.260 | 0.08618 | 0.76567 |
| mapP; maltose 6’-phosphate phosphatase [EC:3.1.3.90] | 0.02250 | 0.00169 | 0.02346 | 0.00043 | 0.00257 | 65 | 0.02183 | 0.00047 | 0.00226 | 93 | 0.00342 | 0.47572 | 142.064 | 0.63500 | 0.89696 |
| csrA; carbon storage regulator | 0.02250 | 0.00185 | 0.02208 | 0.00049 | 0.00274 | 65 | 0.02280 | 0.00058 | 0.00250 | 93 | 0.00371 | -0.19301 | 145.203 | 0.84722 | 0.95682 |
| cvpA; membrane protein required for colicin V production | 0.02248 | 0.00131 | 0.02368 | 0.00032 | 0.00221 | 65 | 0.02164 | 0.00024 | 0.00161 | 93 | 0.00274 | 0.74564 | 125.087 | 0.45728 | 0.83438 |
| tatB; sec-independent protein translocase protein TatB | 0.02243 | 0.00131 | 0.02154 | 0.00029 | 0.00210 | 65 | 0.02305 | 0.00026 | 0.00167 | 93 | 0.00268 | -0.56528 | 133.752 | 0.57283 | 0.87292 |
| mtsT; energy-coupling factor transport system substrate-specific component | 0.02241 | 0.00154 | 0.02240 | 0.00033 | 0.00224 | 65 | 0.02242 | 0.00042 | 0.00211 | 93 | 0.00308 | -0.00704 | 147.357 | 0.99439 | 0.99919 |
| K02484; two-component system, OmpR family, sensor kinase [EC:2.7.13.3] | 0.02241 | 0.00171 | 0.02101 | 0.00044 | 0.00261 | 65 | 0.02339 | 0.00048 | 0.00227 | 93 | 0.00346 | -0.68617 | 141.281 | 0.49373 | 0.84797 |
| msmG; raffinose/stachyose/melibiose transport system permease protein | 0.02236 | 0.00223 | 0.02187 | 0.00115 | 0.00421 | 65 | 0.02270 | 0.00053 | 0.00240 | 93 | 0.00484 | -0.17091 | 104.665 | 0.86463 | 0.96263 |
| glnE; [glutamine synthetase] adenylyltransferase / [glutamine synthetase]-adenylyl-L-tyrosine phosphorylase [EC:2.7.7.42 2.7.7.89] | 0.02236 | 0.00128 | 0.02194 | 0.00028 | 0.00207 | 65 | 0.02265 | 0.00025 | 0.00163 | 93 | 0.00264 | -0.27172 | 132.080 | 0.78626 | 0.94458 |
| czcB; membrane fusion protein, cobalt-zinc-cadmium efflux system | 0.02233 | 0.00312 | 0.02120 | 0.00173 | 0.00516 | 65 | 0.02311 | 0.00143 | 0.00392 | 93 | 0.00648 | -0.29470 | 129.312 | 0.76870 | 0.93982 |
| liaR; two-component system, NarL family, response regulator LiaR | 0.02232 | 0.00164 | 0.02226 | 0.00040 | 0.00249 | 65 | 0.02236 | 0.00044 | 0.00218 | 93 | 0.00331 | -0.03192 | 141.704 | 0.97458 | 0.99546 |
| K07727; putative transcriptional regulator | 0.02226 | 0.00145 | 0.02032 | 0.00026 | 0.00202 | 65 | 0.02362 | 0.00037 | 0.00201 | 93 | 0.00285 | -1.15898 | 150.668 | 0.24830 | 0.77376 |
| fabM; trans-2-decenoyl-[acyl-carrier protein] isomerase [EC:5.3.3.14] | 0.02225 | 0.00166 | 0.02236 | 0.00041 | 0.00252 | 65 | 0.02218 | 0.00045 | 0.00221 | 93 | 0.00335 | 0.05277 | 141.779 | 0.95799 | 0.98896 |
| hrpA; ATP-dependent helicase HrpA [EC:3.6.4.13] | 0.02224 | 0.00136 | 0.02311 | 0.00033 | 0.00225 | 65 | 0.02163 | 0.00027 | 0.00171 | 93 | 0.00283 | 0.52389 | 129.030 | 0.60126 | 0.88272 |
| phnD; phosphonate transport system substrate-binding protein | 0.02223 | 0.00172 | 0.02347 | 0.00049 | 0.00273 | 65 | 0.02136 | 0.00045 | 0.00221 | 93 | 0.00352 | 0.60021 | 134.970 | 0.54937 | 0.86645 |
| comE; two-component system, LytTR family, response regulator ComE | 0.02220 | 0.00201 | 0.02106 | 0.00047 | 0.00270 | 65 | 0.02299 | 0.00076 | 0.00285 | 93 | 0.00393 | -0.49196 | 153.500 | 0.62345 | 0.89159 |
| relE, stbE; mRNA interferase RelE/StbE | 0.02218 | 0.00254 | 0.02713 | 0.00165 | 0.00504 | 65 | 0.01873 | 0.00056 | 0.00245 | 93 | 0.00560 | 1.49912 | 94.276 | 0.13718 | 0.77376 |
| nuoF; NADH-quinone oxidoreductase subunit F [EC:1.6.5.3] | 0.02217 | 0.00139 | 0.02092 | 0.00031 | 0.00219 | 65 | 0.02304 | 0.00030 | 0.00179 | 93 | 0.00283 | -0.75040 | 136.003 | 0.45431 | 0.83438 |
| K07270; glycosyl transferase, family 25 | 0.02217 | 0.00279 | 0.02197 | 0.00073 | 0.00336 | 65 | 0.02230 | 0.00158 | 0.00413 | 93 | 0.00532 | -0.06257 | 155.914 | 0.95019 | 0.98774 |
| citE; citrate lyase subunit beta / citryl-CoA lyase [EC:4.1.3.34] | 0.02214 | 0.00152 | 0.02231 | 0.00031 | 0.00218 | 65 | 0.02203 | 0.00041 | 0.00210 | 93 | 0.00303 | 0.09182 | 148.524 | 0.92696 | 0.98128 |
| K07481; transposase, IS5 family | 0.02214 | 0.00233 | 0.02092 | 0.00051 | 0.00279 | 65 | 0.02298 | 0.00111 | 0.00346 | 93 | 0.00444 | -0.46399 | 155.836 | 0.64330 | 0.90146 |
| cas3; CRISPR-associated endonuclease/helicase Cas3 [EC:3.1.-.- 3.6.4.-] | 0.02207 | 0.00147 | 0.02537 | 0.00040 | 0.00248 | 65 | 0.01977 | 0.00029 | 0.00177 | 93 | 0.00305 | 1.83540 | 123.300 | 0.06886 | 0.73514 |
| macB; macrolide transport system ATP-binding/permease protein [EC:3.6.3.-] | 0.02205 | 0.00199 | 0.02193 | 0.00068 | 0.00324 | 65 | 0.02214 | 0.00060 | 0.00254 | 93 | 0.00411 | -0.04976 | 132.083 | 0.96039 | 0.98903 |
| nspC; carboxynorspermidine decarboxylase [EC:4.1.1.96] | 0.02205 | 0.00129 | 0.01925 | 0.00021 | 0.00179 | 65 | 0.02400 | 0.00029 | 0.00178 | 93 | 0.00252 | -1.88533 | 150.659 | 0.06131 | 0.73001 |
| ppiC; peptidyl-prolyl cis-trans isomerase C [EC:5.2.1.8] | 0.02204 | 0.00192 | 0.02237 | 0.00065 | 0.00317 | 65 | 0.02181 | 0.00054 | 0.00240 | 93 | 0.00398 | 0.14017 | 128.874 | 0.88874 | 0.97039 |
| ABC.SP.P1; putative spermidine/putrescine transport system permease protein | 0.02201 | 0.00297 | 0.01940 | 0.00106 | 0.00404 | 65 | 0.02383 | 0.00164 | 0.00420 | 93 | 0.00583 | -0.76002 | 152.821 | 0.44841 | 0.83379 |
| lhr; ATP-dependent helicase Lhr and Lhr-like helicase [EC:3.6.4.-] | 0.02201 | 0.00190 | 0.02113 | 0.00052 | 0.00282 | 65 | 0.02262 | 0.00061 | 0.00256 | 93 | 0.00381 | -0.38922 | 144.652 | 0.69769 | 0.91825 |
| argE; acetylornithine deacetylase [EC:3.5.1.16] | 0.02195 | 0.00165 | 0.02130 | 0.00040 | 0.00249 | 65 | 0.02240 | 0.00045 | 0.00220 | 93 | 0.00332 | -0.33015 | 142.517 | 0.74177 | 0.93043 |
| dctA; aerobic C4-dicarboxylate transport protein | 0.02195 | 0.00214 | 0.01864 | 0.00063 | 0.00312 | 65 | 0.02425 | 0.00078 | 0.00289 | 93 | 0.00425 | -1.31903 | 146.025 | 0.18922 | 0.77376 |
| HSP20; HSP20 family protein | 0.02193 | 0.00188 | 0.02062 | 0.00055 | 0.00290 | 65 | 0.02284 | 0.00057 | 0.00248 | 93 | 0.00382 | -0.58397 | 139.712 | 0.56018 | 0.86813 |
| kdgK; 2-dehydro-3-deoxygluconokinase [EC:2.7.1.45] | 0.02192 | 0.00147 | 0.02066 | 0.00048 | 0.00272 | 65 | 0.02280 | 0.00025 | 0.00164 | 93 | 0.00317 | -0.67377 | 108.912 | 0.50188 | 0.84942 |
| E3.2.1.85, lacG; 6-phospho-beta-galactosidase [EC:3.2.1.85] | 0.02190 | 0.00166 | 0.02164 | 0.00041 | 0.00251 | 65 | 0.02207 | 0.00046 | 0.00222 | 93 | 0.00335 | -0.12920 | 142.428 | 0.89738 | 0.97338 |
| tolR; biopolymer transport protein TolR | 0.02189 | 0.00180 | 0.02018 | 0.00050 | 0.00278 | 65 | 0.02308 | 0.00052 | 0.00236 | 93 | 0.00365 | -0.79494 | 139.347 | 0.42800 | 0.82733 |
| ygfZ; tRNA-modifying protein YgfZ | 0.02186 | 0.00130 | 0.02129 | 0.00028 | 0.00209 | 65 | 0.02227 | 0.00026 | 0.00166 | 93 | 0.00267 | -0.36576 | 133.643 | 0.71512 | 0.92004 |
| PTS-Lac-EIIC, lacE; PTS system, lactose-specific IIC component | 0.02186 | 0.00166 | 0.02161 | 0.00041 | 0.00251 | 65 | 0.02204 | 0.00046 | 0.00221 | 93 | 0.00335 | -0.12807 | 142.434 | 0.89827 | 0.97377 |
| acuB; acetoin utilization protein AcuB | 0.02183 | 0.00155 | 0.02116 | 0.00036 | 0.00236 | 65 | 0.02231 | 0.00040 | 0.00206 | 93 | 0.00313 | -0.36654 | 141.776 | 0.71451 | 0.92004 |
| lacC; tagatose 6-phosphate kinase [EC:2.7.1.144] | 0.02181 | 0.00162 | 0.02268 | 0.00041 | 0.00251 | 65 | 0.02119 | 0.00042 | 0.00213 | 93 | 0.00329 | 0.45375 | 139.207 | 0.65072 | 0.90423 |
| PTS-Lac-EIIA, lacF; PTS system, lactose-specific IIA component [EC:2.7.1.207] | 0.02180 | 0.00165 | 0.02154 | 0.00041 | 0.00250 | 65 | 0.02197 | 0.00045 | 0.00221 | 93 | 0.00334 | -0.12968 | 142.369 | 0.89701 | 0.97318 |
| E3.3.1.1, ahcY; adenosylhomocysteinase [EC:3.3.1.1] | 0.02178 | 0.00127 | 0.02091 | 0.00027 | 0.00202 | 65 | 0.02238 | 0.00025 | 0.00164 | 93 | 0.00260 | -0.56197 | 135.532 | 0.57506 | 0.87389 |
| hutG; formiminoglutamase [EC:3.5.3.8] | 0.02176 | 0.00122 | 0.02139 | 0.00024 | 0.00193 | 65 | 0.02201 | 0.00023 | 0.00158 | 93 | 0.00249 | -0.25118 | 136.376 | 0.80206 | 0.94855 |
| glgX; glycogen debranching enzyme [EC:3.2.1.196] | 0.02164 | 0.00132 | 0.02366 | 0.00032 | 0.00223 | 65 | 0.02022 | 0.00024 | 0.00159 | 93 | 0.00274 | 1.25327 | 123.564 | 0.21247 | 0.77376 |
| hfq; host factor-I protein | 0.02163 | 0.00122 | 0.02044 | 0.00025 | 0.00195 | 65 | 0.02247 | 0.00023 | 0.00156 | 93 | 0.00249 | -0.81258 | 133.991 | 0.41790 | 0.81941 |
| sufE; cysteine desulfuration protein SufE | 0.02161 | 0.00114 | 0.02002 | 0.00018 | 0.00167 | 65 | 0.02273 | 0.00022 | 0.00155 | 93 | 0.00228 | -1.19107 | 146.077 | 0.23556 | 0.77376 |
| rnd; ribonuclease D [EC:3.1.13.5] | 0.02159 | 0.00130 | 0.02037 | 0.00025 | 0.00195 | 65 | 0.02245 | 0.00029 | 0.00175 | 93 | 0.00262 | -0.79187 | 143.755 | 0.42974 | 0.82749 |
| K07150; uncharacterized protein | 0.02157 | 0.00159 | 0.02301 | 0.00050 | 0.00279 | 65 | 0.02056 | 0.00033 | 0.00188 | 93 | 0.00336 | 0.72978 | 118.628 | 0.46696 | 0.83837 |
| gspF; general secretion pathway protein F | 0.02154 | 0.00184 | 0.02248 | 0.00055 | 0.00290 | 65 | 0.02087 | 0.00053 | 0.00240 | 93 | 0.00376 | 0.42786 | 136.886 | 0.66943 | 0.91165 |
| NEU1; sialidase-1 [EC:3.2.1.18] | 0.02146 | 0.00163 | 0.02008 | 0.00035 | 0.00232 | 65 | 0.02243 | 0.00047 | 0.00225 | 93 | 0.00324 | -0.72431 | 149.113 | 0.47001 | 0.83870 |
| gmhB; D-glycero-D-manno-heptose 1,7-bisphosphate phosphatase [EC:3.1.3.82 3.1.3.83] | 0.02145 | 0.00116 | 0.02252 | 0.00023 | 0.00188 | 65 | 0.02071 | 0.00020 | 0.00147 | 93 | 0.00238 | 0.75720 | 131.979 | 0.45028 | 0.83438 |
| APOD; apolipoprotein D and lipocalin family protein | 0.02139 | 0.00261 | 0.02068 | 0.00113 | 0.00418 | 65 | 0.02188 | 0.00104 | 0.00335 | 93 | 0.00535 | -0.22526 | 134.182 | 0.82212 | 0.95506 |
| mqo; malate dehydrogenase (quinone) [EC:1.1.5.4] | 0.02136 | 0.00175 | 0.02024 | 0.00048 | 0.00272 | 65 | 0.02215 | 0.00049 | 0.00230 | 93 | 0.00356 | -0.53652 | 139.047 | 0.59245 | 0.87921 |
| xylB, XYLB; xylulokinase [EC:2.7.1.17] | 0.02134 | 0.00156 | 0.01915 | 0.00030 | 0.00213 | 65 | 0.02287 | 0.00044 | 0.00218 | 93 | 0.00305 | -1.21972 | 152.087 | 0.22446 | 0.77376 |
| hcp; hydroxylamine reductase [EC:1.7.99.1] | 0.02134 | 0.00188 | 0.02402 | 0.00065 | 0.00317 | 65 | 0.01946 | 0.00049 | 0.00229 | 93 | 0.00391 | 1.16603 | 124.642 | 0.24583 | 0.77376 |
| rhtA; inner membrane transporter RhtA | 0.02132 | 0.00199 | 0.02154 | 0.00072 | 0.00333 | 65 | 0.02116 | 0.00057 | 0.00247 | 93 | 0.00414 | 0.09102 | 127.128 | 0.92762 | 0.98137 |
| gloB, HAGH; hydroxyacylglutathione hydrolase [EC:3.1.2.6] | 0.02130 | 0.00130 | 0.02233 | 0.00033 | 0.00225 | 65 | 0.02058 | 0.00023 | 0.00156 | 93 | 0.00274 | 0.63803 | 121.199 | 0.52466 | 0.85728 |
| ybaZ; methylated-DNA-protein-cysteine methyltransferase related protein | 0.02129 | 0.00099 | 0.02094 | 0.00016 | 0.00156 | 65 | 0.02153 | 0.00015 | 0.00128 | 93 | 0.00202 | -0.29054 | 136.610 | 0.77185 | 0.94083 |
| tehB; tellurite methyltransferase [EC:2.1.1.265] | 0.02127 | 0.00168 | 0.02093 | 0.00038 | 0.00242 | 65 | 0.02151 | 0.00049 | 0.00231 | 93 | 0.00334 | -0.17245 | 148.020 | 0.86332 | 0.96263 |
| kup; KUP system potassium uptake protein | 0.02121 | 0.00138 | 0.02006 | 0.00029 | 0.00212 | 65 | 0.02201 | 0.00031 | 0.00183 | 93 | 0.00280 | -0.69730 | 140.633 | 0.48676 | 0.84553 |
| PTS-Gat-EIIC, gatC, sgcC; PTS system, galactitol-specific IIC component | 0.02121 | 0.00221 | 0.02240 | 0.00099 | 0.00391 | 65 | 0.02037 | 0.00062 | 0.00259 | 93 | 0.00469 | 0.43329 | 116.894 | 0.66561 | 0.91126 |
| clpS; ATP-dependent Clp protease adaptor protein ClpS | 0.02117 | 0.00131 | 0.01941 | 0.00026 | 0.00200 | 65 | 0.02240 | 0.00028 | 0.00172 | 93 | 0.00264 | -1.13281 | 140.297 | 0.25923 | 0.77376 |
| cydC; ATP-binding cassette, subfamily C, bacterial CydC | 0.02117 | 0.00143 | 0.02218 | 0.00043 | 0.00258 | 65 | 0.02046 | 0.00025 | 0.00165 | 93 | 0.00306 | 0.56028 | 114.083 | 0.57639 | 0.87389 |
| gldA; glycerol dehydrogenase [EC:1.1.1.6] | 0.02117 | 0.00132 | 0.02343 | 0.00035 | 0.00232 | 65 | 0.01959 | 0.00022 | 0.00155 | 93 | 0.00279 | 1.37729 | 117.327 | 0.17105 | 0.77376 |
| K09780; uncharacterized protein | 0.02117 | 0.00123 | 0.02335 | 0.00027 | 0.00206 | 65 | 0.01964 | 0.00021 | 0.00149 | 93 | 0.00254 | 1.46196 | 125.172 | 0.14626 | 0.77376 |
| selD, SEPHS; selenide, water dikinase [EC:2.7.9.3] | 0.02115 | 0.00116 | 0.02338 | 0.00028 | 0.00207 | 65 | 0.01959 | 0.00016 | 0.00132 | 93 | 0.00246 | 1.54157 | 113.917 | 0.12595 | 0.77376 |
| thiH; 2-iminoacetate synthase [EC:4.1.99.19] | 0.02115 | 0.00181 | 0.02423 | 0.00057 | 0.00296 | 65 | 0.01899 | 0.00048 | 0.00226 | 93 | 0.00372 | 1.40934 | 129.845 | 0.16113 | 0.77376 |
| cydD; ATP-binding cassette, subfamily C, bacterial CydD | 0.02113 | 0.00143 | 0.02216 | 0.00043 | 0.00258 | 65 | 0.02040 | 0.00025 | 0.00165 | 93 | 0.00306 | 0.57488 | 113.988 | 0.56650 | 0.87051 |
| tenA; thiaminase (transcriptional activator TenA) [EC:3.5.99.2] | 0.02112 | 0.00130 | 0.01874 | 0.00019 | 0.00170 | 65 | 0.02278 | 0.00031 | 0.00184 | 93 | 0.00251 | -1.61308 | 154.282 | 0.10877 | 0.77376 |
| ykoE; energy-coupling factor transport system substrate-specific component | 0.02112 | 0.00169 | 0.02226 | 0.00059 | 0.00302 | 65 | 0.02033 | 0.00036 | 0.00197 | 93 | 0.00360 | 0.53540 | 115.558 | 0.59341 | 0.87985 |
| murQ; N-acetylmuramic acid 6-phosphate etherase [EC:4.2.1.126] | 0.02095 | 0.00126 | 0.02070 | 0.00030 | 0.00216 | 65 | 0.02113 | 0.00022 | 0.00154 | 93 | 0.00265 | -0.15931 | 123.225 | 0.87369 | 0.96576 |
| flgG; flagellar basal-body rod protein FlgG | 0.02095 | 0.00163 | 0.01765 | 0.00041 | 0.00252 | 65 | 0.02326 | 0.00042 | 0.00211 | 93 | 0.00329 | -1.70596 | 138.150 | 0.09026 | 0.76567 |
| ytfE, scdA; regulator of cell morphogenesis and NO signaling | 0.02093 | 0.00155 | 0.01913 | 0.00031 | 0.00219 | 65 | 0.02220 | 0.00042 | 0.00213 | 93 | 0.00306 | -1.00178 | 149.339 | 0.31807 | 0.77376 |
| gcdA; glutaconyl-CoA decarboxylase [EC:4.1.1.70] | 0.02088 | 0.00218 | 0.02519 | 0.00100 | 0.00392 | 65 | 0.01786 | 0.00057 | 0.00248 | 93 | 0.00464 | 1.58055 | 112.773 | 0.11678 | 0.77376 |
| cheV; two-component system, chemotaxis family, chemotaxis protein CheV | 0.02086 | 0.00234 | 0.02059 | 0.00074 | 0.00338 | 65 | 0.02105 | 0.00096 | 0.00321 | 93 | 0.00466 | -0.09962 | 147.686 | 0.92078 | 0.97905 |
| pncA; nicotinamidase/pyrazinamidase [EC:3.5.1.19 3.5.1.-] | 0.02085 | 0.00111 | 0.01967 | 0.00016 | 0.00157 | 65 | 0.02167 | 0.00022 | 0.00154 | 93 | 0.00220 | -0.91278 | 150.066 | 0.36282 | 0.79496 |
| sulD; dihydroneopterin aldolase / 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase [EC:4.1.2.25 2.7.6.3] | 0.02084 | 0.00145 | 0.02185 | 0.00032 | 0.00222 | 65 | 0.02012 | 0.00035 | 0.00193 | 93 | 0.00294 | 0.58871 | 140.988 | 0.55700 | 0.86813 |
| fimD, fimC, mrkC, htrE, cssD; outer membrane usher protein | 0.02083 | 0.00239 | 0.01950 | 0.00081 | 0.00353 | 65 | 0.02176 | 0.00098 | 0.00325 | 93 | 0.00479 | -0.47085 | 145.667 | 0.63845 | 0.89822 |
| sprL; SprT-like protein | 0.02074 | 0.00158 | 0.02226 | 0.00050 | 0.00278 | 65 | 0.01968 | 0.00032 | 0.00186 | 93 | 0.00334 | 0.77155 | 117.713 | 0.44193 | 0.83186 |
| niaX; niacin transporter | 0.02074 | 0.00161 | 0.02146 | 0.00040 | 0.00249 | 65 | 0.02023 | 0.00042 | 0.00212 | 93 | 0.00327 | 0.37466 | 139.484 | 0.70848 | 0.92004 |
| cobM, cbiF; precorrin-4/cobalt-precorrin-4 C11-methyltransferase [EC:2.1.1.133 2.1.1.271] | 0.02072 | 0.00136 | 0.02356 | 0.00038 | 0.00241 | 65 | 0.01873 | 0.00023 | 0.00156 | 93 | 0.00287 | 1.68167 | 114.859 | 0.09535 | 0.76609 |
| kbl, GCAT; glycine C-acetyltransferase [EC:2.3.1.29] | 0.02071 | 0.00139 | 0.01866 | 0.00023 | 0.00190 | 65 | 0.02214 | 0.00035 | 0.00194 | 93 | 0.00272 | -1.28472 | 152.016 | 0.20084 | 0.77376 |
| prdX, proX; Ala-tRNA(Pro) deacylase [EC:3.1.1.-] | 0.02069 | 0.00120 | 0.02303 | 0.00032 | 0.00221 | 65 | 0.01906 | 0.00016 | 0.00132 | 93 | 0.00257 | 1.54197 | 108.485 | 0.12599 | 0.77376 |
| pilA; type IV pilus assembly protein PilA | 0.02066 | 0.00201 | 0.02048 | 0.00069 | 0.00327 | 65 | 0.02079 | 0.00061 | 0.00256 | 93 | 0.00415 | -0.07478 | 132.124 | 0.94051 | 0.98528 |
| cobH-cbiC; precorrin-8X/cobalt-precorrin-8 methylmutase [EC:5.4.99.61 5.4.99.60] | 0.02064 | 0.00136 | 0.02345 | 0.00038 | 0.00242 | 65 | 0.01867 | 0.00022 | 0.00155 | 93 | 0.00287 | 1.66197 | 114.324 | 0.09926 | 0.76942 |
| lptC; lipopolysaccharide export system protein LptC | 0.02061 | 0.00117 | 0.02083 | 0.00023 | 0.00188 | 65 | 0.02046 | 0.00021 | 0.00151 | 93 | 0.00241 | 0.15204 | 134.033 | 0.87939 | 0.96779 |
| DPP4, CD26; dipeptidyl-peptidase 4 [EC:3.4.14.5] | 0.02057 | 0.00173 | 0.01808 | 0.00031 | 0.00218 | 65 | 0.02231 | 0.00058 | 0.00250 | 93 | 0.00332 | -1.27412 | 155.711 | 0.20452 | 0.77376 |
| menC; O-succinylbenzoate synthase [EC:4.2.1.113] | 0.02052 | 0.00150 | 0.02053 | 0.00038 | 0.00243 | 65 | 0.02052 | 0.00034 | 0.00190 | 93 | 0.00309 | 0.00315 | 131.986 | 0.99749 | 0.99955 |
| pntB; NAD(P) transhydrogenase subunit beta [EC:1.6.1.2] | 0.02048 | 0.00151 | 0.01935 | 0.00035 | 0.00231 | 65 | 0.02127 | 0.00037 | 0.00199 | 93 | 0.00305 | -0.62833 | 140.252 | 0.53081 | 0.86136 |
| K07003; uncharacterized protein | 0.02046 | 0.00182 | 0.02069 | 0.00051 | 0.00280 | 65 | 0.02030 | 0.00054 | 0.00241 | 93 | 0.00370 | 0.10560 | 140.493 | 0.91605 | 0.97681 |
| dsbB; disulfide bond formation protein DsbB | 0.02041 | 0.00144 | 0.01952 | 0.00031 | 0.00219 | 65 | 0.02104 | 0.00034 | 0.00192 | 93 | 0.00291 | -0.52501 | 142.092 | 0.60039 | 0.88240 |
| ydhP; MFS transporter, DHA1 family, inner membrane transport protein | 0.02038 | 0.00216 | 0.01897 | 0.00067 | 0.00321 | 65 | 0.02136 | 0.00079 | 0.00291 | 93 | 0.00434 | -0.55057 | 144.536 | 0.58278 | 0.87720 |
| czcC; outer membrane protein, cobalt-zinc-cadmium efflux system | 0.02038 | 0.00310 | 0.01946 | 0.00170 | 0.00511 | 65 | 0.02102 | 0.00141 | 0.00389 | 93 | 0.00643 | -0.24285 | 129.416 | 0.80851 | 0.95002 |
| minC; septum site-determining protein MinC | 0.02034 | 0.00124 | 0.02150 | 0.00030 | 0.00215 | 65 | 0.01954 | 0.00020 | 0.00147 | 93 | 0.00261 | 0.75419 | 119.409 | 0.45222 | 0.83438 |
| gltB; glutamate synthase (NADPH/NADH) large chain [EC:1.4.1.13 1.4.1.14] | 0.02034 | 0.00145 | 0.01737 | 0.00031 | 0.00219 | 65 | 0.02241 | 0.00034 | 0.00190 | 93 | 0.00290 | -1.73542 | 141.217 | 0.08485 | 0.76567 |
| purT; phosphoribosylglycinamide formyltransferase 2 [EC:2.1.2.2] | 0.02033 | 0.00104 | 0.02007 | 0.00016 | 0.00158 | 65 | 0.02051 | 0.00018 | 0.00139 | 93 | 0.00210 | -0.20843 | 141.949 | 0.83519 | 0.95506 |
| orn, REX2, REXO2; oligoribonuclease [EC:3.1.-.-] | 0.02032 | 0.00120 | 0.02102 | 0.00026 | 0.00200 | 65 | 0.01984 | 0.00021 | 0.00150 | 93 | 0.00250 | 0.47092 | 127.951 | 0.63850 | 0.89822 |
| lnt; apolipoprotein N-acyltransferase [EC:2.3.1.-] | 0.02029 | 0.00135 | 0.01910 | 0.00028 | 0.00209 | 65 | 0.02112 | 0.00029 | 0.00177 | 93 | 0.00273 | -0.73879 | 138.982 | 0.46128 | 0.83584 |
| E2.1.1.77, pcm; protein-L-isoaspartate(D-aspartate) O-methyltransferase [EC:2.1.1.77] | 0.02028 | 0.00166 | 0.01840 | 0.00043 | 0.00257 | 65 | 0.02160 | 0.00044 | 0.00217 | 93 | 0.00336 | -0.95094 | 138.564 | 0.34329 | 0.78583 |
| fldA, nifF, isiB; flavodoxin I | 0.02027 | 0.00217 | 0.02338 | 0.00103 | 0.00398 | 65 | 0.01811 | 0.00054 | 0.00241 | 93 | 0.00465 | 1.13270 | 109.359 | 0.25982 | 0.77376 |
| aguB; N-carbamoylputrescine amidase [EC:3.5.1.53] | 0.02024 | 0.00114 | 0.01774 | 0.00015 | 0.00152 | 65 | 0.02199 | 0.00024 | 0.00160 | 93 | 0.00221 | -1.92375 | 153.425 | 0.05624 | 0.70577 |
| tolA; colicin import membrane protein | 0.02023 | 0.00115 | 0.02103 | 0.00025 | 0.00195 | 65 | 0.01967 | 0.00018 | 0.00140 | 93 | 0.00240 | 0.56517 | 124.027 | 0.57298 | 0.87292 |
| nuoE; NADH-quinone oxidoreductase subunit E [EC:1.6.5.3] | 0.02021 | 0.00116 | 0.01873 | 0.00023 | 0.00188 | 65 | 0.02125 | 0.00020 | 0.00146 | 93 | 0.00238 | -1.05985 | 131.322 | 0.29116 | 0.77376 |
| PTS-Glc-EIIA, crr; PTS system, sugar-specific IIA component [EC:2.7.1.-] | 0.02018 | 0.00144 | 0.02266 | 0.00050 | 0.00276 | 65 | 0.01845 | 0.00021 | 0.00149 | 93 | 0.00314 | 1.34110 | 100.904 | 0.18290 | 0.77376 |
| K09962; uncharacterized protein | 0.02014 | 0.00135 | 0.01874 | 0.00024 | 0.00192 | 65 | 0.02111 | 0.00032 | 0.00186 | 93 | 0.00267 | -0.88944 | 149.090 | 0.37520 | 0.79810 |
| clpL; ATP-dependent Clp protease ATP-binding subunit ClpL | 0.02013 | 0.00158 | 0.02036 | 0.00038 | 0.00243 | 65 | 0.01997 | 0.00041 | 0.00209 | 93 | 0.00320 | 0.11887 | 140.557 | 0.90555 | 0.97664 |
| tcyM; L-cystine transport system permease protein | 0.02012 | 0.00195 | 0.01792 | 0.00041 | 0.00250 | 65 | 0.02166 | 0.00074 | 0.00282 | 93 | 0.00377 | -0.99354 | 155.458 | 0.32199 | 0.77376 |
| DPEP; membrane dipeptidase [EC:3.4.13.19] | 0.02011 | 0.00118 | 0.01927 | 0.00023 | 0.00188 | 65 | 0.02070 | 0.00021 | 0.00151 | 93 | 0.00241 | -0.59116 | 134.467 | 0.55540 | 0.86813 |
| racD; aspartate racemase [EC:5.1.1.13] | 0.02008 | 0.00173 | 0.02195 | 0.00070 | 0.00329 | 65 | 0.01877 | 0.00031 | 0.00184 | 93 | 0.00377 | 0.84410 | 103.252 | 0.40056 | 0.80783 |
| cobI-cbiL; precorrin-2/cobalt-factor-2 C20-methyltransferase [EC:2.1.1.130 2.1.1.151] | 0.02006 | 0.00137 | 0.02306 | 0.00038 | 0.00243 | 65 | 0.01797 | 0.00023 | 0.00156 | 93 | 0.00289 | 1.76330 | 114.347 | 0.08052 | 0.75920 |
| mprF, fmtC; phosphatidylglycerol lysyltransferase [EC:2.3.2.3] | 0.02003 | 0.00134 | 0.02176 | 0.00036 | 0.00235 | 65 | 0.01882 | 0.00023 | 0.00158 | 93 | 0.00283 | 1.03718 | 118.368 | 0.30176 | 0.77376 |
| bioC; malonyl-CoA O-methyltransferase [EC:2.1.1.197] | 0.02002 | 0.00136 | 0.02191 | 0.00034 | 0.00230 | 65 | 0.01871 | 0.00025 | 0.00165 | 93 | 0.00283 | 1.13166 | 124.264 | 0.25996 | 0.77376 |
| K09858; SEC-C motif domain protein | 0.02001 | 0.00124 | 0.02064 | 0.00026 | 0.00201 | 65 | 0.01956 | 0.00023 | 0.00158 | 93 | 0.00256 | 0.42189 | 132.331 | 0.67379 | 0.91184 |
| cysP, sbp; sulfate transport system substrate-binding protein | 0.01992 | 0.00199 | 0.01862 | 0.00056 | 0.00295 | 65 | 0.02083 | 0.00067 | 0.00269 | 93 | 0.00399 | -0.55466 | 145.031 | 0.57998 | 0.87573 |
| hisI; phosphoribosyl-AMP cyclohydrolase [EC:3.5.4.19] | 0.01991 | 0.00116 | 0.01807 | 0.00021 | 0.00179 | 65 | 0.02120 | 0.00021 | 0.00151 | 93 | 0.00234 | -1.33388 | 138.713 | 0.18443 | 0.77376 |
| cbiK; sirohydrochlorin cobaltochelatase [EC:4.99.1.3] | 0.01984 | 0.00190 | 0.02422 | 0.00075 | 0.00340 | 65 | 0.01679 | 0.00043 | 0.00214 | 93 | 0.00402 | 1.84779 | 112.408 | 0.06726 | 0.73334 |
| zmpB; zinc metalloprotease ZmpB [EC:3.4.24.-] | 0.01981 | 0.00196 | 0.01769 | 0.00040 | 0.00249 | 65 | 0.02128 | 0.00075 | 0.00283 | 93 | 0.00377 | -0.95243 | 155.582 | 0.34236 | 0.78567 |
| SLC13A2_3_5; solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2/3/5 | 0.01977 | 0.00165 | 0.02051 | 0.00043 | 0.00256 | 65 | 0.01926 | 0.00044 | 0.00217 | 93 | 0.00336 | 0.37414 | 138.916 | 0.70887 | 0.92004 |
| lysP; lysine-specific permease | 0.01975 | 0.00143 | 0.01966 | 0.00043 | 0.00257 | 65 | 0.01982 | 0.00025 | 0.00164 | 93 | 0.00305 | -0.05096 | 113.659 | 0.95944 | 0.98903 |
| betA, CHDH; choline dehydrogenase [EC:1.1.99.1] | 0.01972 | 0.00189 | 0.01782 | 0.00053 | 0.00287 | 65 | 0.02105 | 0.00058 | 0.00251 | 93 | 0.00381 | -0.84788 | 141.614 | 0.39794 | 0.80577 |
| fsr; MFS transporter, FSR family, fosmidomycin resistance protein | 0.01971 | 0.00124 | 0.01906 | 0.00029 | 0.00212 | 65 | 0.02017 | 0.00021 | 0.00151 | 93 | 0.00261 | -0.42833 | 123.498 | 0.66916 | 0.91165 |
| murE; UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-L-lysine ligase [EC:6.3.2.7] | 0.01971 | 0.00155 | 0.01926 | 0.00034 | 0.00230 | 65 | 0.02003 | 0.00041 | 0.00209 | 93 | 0.00311 | -0.24486 | 144.939 | 0.80691 | 0.94987 |
| pdxH, PNPO; pyridoxamine 5’-phosphate oxidase [EC:1.4.3.5] | 0.01969 | 0.00131 | 0.01884 | 0.00029 | 0.00211 | 65 | 0.02029 | 0.00026 | 0.00168 | 93 | 0.00269 | -0.53544 | 133.353 | 0.59324 | 0.87985 |
| K07038; inner membrane protein | 0.01969 | 0.00171 | 0.01954 | 0.00042 | 0.00255 | 65 | 0.01980 | 0.00049 | 0.00230 | 93 | 0.00343 | -0.07581 | 144.221 | 0.93967 | 0.98527 |
| arcD, lysl, lysP; arginine:ornithine antiporter / lysine permease | 0.01968 | 0.00182 | 0.02164 | 0.00072 | 0.00333 | 65 | 0.01831 | 0.00039 | 0.00204 | 93 | 0.00391 | 0.85198 | 110.229 | 0.39607 | 0.80459 |
| lacT; transcriptional antiterminator | 0.01963 | 0.00160 | 0.01969 | 0.00038 | 0.00241 | 65 | 0.01959 | 0.00043 | 0.00216 | 93 | 0.00323 | 0.03019 | 143.492 | 0.97596 | 0.99571 |
| K07085; putative transport protein | 0.01959 | 0.00156 | 0.02074 | 0.00038 | 0.00243 | 65 | 0.01879 | 0.00039 | 0.00205 | 93 | 0.00318 | 0.61337 | 138.806 | 0.54064 | 0.86207 |
| E4.1.3.4, HMGCL, hmgL; hydroxymethylglutaryl-CoA lyase [EC:4.1.3.4] | 0.01958 | 0.00178 | 0.01828 | 0.00044 | 0.00262 | 65 | 0.02049 | 0.00054 | 0.00241 | 93 | 0.00356 | -0.62312 | 145.718 | 0.53418 | 0.86207 |
| K06910; uncharacterized protein | 0.01958 | 0.00119 | 0.02016 | 0.00024 | 0.00194 | 65 | 0.01918 | 0.00021 | 0.00152 | 93 | 0.00246 | 0.40022 | 132.021 | 0.68964 | 0.91567 |
| TC.DCUC, dcuC, dcuD; C4-dicarboxylate transporter, DcuC family | 0.01951 | 0.00231 | 0.02075 | 0.00082 | 0.00356 | 65 | 0.01865 | 0.00086 | 0.00305 | 93 | 0.00469 | 0.44785 | 139.855 | 0.65495 | 0.90679 |
| vcaM; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.01951 | 0.00206 | 0.01925 | 0.00060 | 0.00305 | 65 | 0.01969 | 0.00073 | 0.00280 | 93 | 0.00414 | -0.10688 | 145.506 | 0.91503 | 0.97664 |
| K07496; putative transposase | 0.01944 | 0.00397 | 0.01956 | 0.00371 | 0.00755 | 65 | 0.01935 | 0.00168 | 0.00425 | 93 | 0.00866 | 0.02407 | 103.665 | 0.98084 | 0.99661 |
| rnfG; electron transport complex protein RnfG | 0.01943 | 0.00148 | 0.02216 | 0.00039 | 0.00245 | 65 | 0.01753 | 0.00031 | 0.00182 | 93 | 0.00305 | 1.51799 | 127.177 | 0.13150 | 0.77376 |
| K07054; uncharacterized protein | 0.01941 | 0.00111 | 0.01916 | 0.00022 | 0.00185 | 65 | 0.01959 | 0.00018 | 0.00138 | 93 | 0.00231 | -0.18444 | 127.720 | 0.85396 | 0.95848 |
| hisE; phosphoribosyl-ATP pyrophosphohydrolase [EC:3.6.1.31] | 0.01937 | 0.00108 | 0.01788 | 0.00019 | 0.00173 | 65 | 0.02041 | 0.00018 | 0.00138 | 93 | 0.00221 | -1.14171 | 133.844 | 0.25561 | 0.77376 |
| TC.CITMHS; citrate-Mg2+:H+ or citrate-Ca2+:H+ symporter, CitMHS family | 0.01937 | 0.00142 | 0.02083 | 0.00033 | 0.00224 | 65 | 0.01835 | 0.00031 | 0.00183 | 93 | 0.00289 | 0.85665 | 136.134 | 0.39314 | 0.80459 |
| mtnA; methylthioribose-1-phosphate isomerase [EC:5.3.1.23] | 0.01935 | 0.00122 | 0.02207 | 0.00030 | 0.00214 | 65 | 0.01745 | 0.00019 | 0.00141 | 93 | 0.00257 | 1.79836 | 116.476 | 0.07471 | 0.74200 |
| sixA; phosphohistidine phosphatase [EC:3.1.3.-] | 0.01931 | 0.00115 | 0.01895 | 0.00022 | 0.00182 | 65 | 0.01956 | 0.00021 | 0.00149 | 93 | 0.00236 | -0.25961 | 135.787 | 0.79556 | 0.94791 |
| mazF, ndoA, chpA; mRNA interferase MazF [EC:3.1.-.-] | 0.01930 | 0.00179 | 0.01852 | 0.00059 | 0.00301 | 65 | 0.01984 | 0.00046 | 0.00222 | 93 | 0.00374 | -0.35263 | 126.369 | 0.72495 | 0.92177 |
| cobK-cbiJ; precorrin-6A/cobalt-precorrin-6A reductase [EC:1.3.1.54 1.3.1.106] | 0.01926 | 0.00130 | 0.02247 | 0.00036 | 0.00236 | 65 | 0.01702 | 0.00019 | 0.00143 | 93 | 0.00276 | 1.97680 | 109.095 | 0.05059 | 0.68163 |
| pqiA; paraquat-inducible protein A | 0.01923 | 0.00235 | 0.01966 | 0.00071 | 0.00331 | 65 | 0.01893 | 0.00099 | 0.00327 | 93 | 0.00465 | 0.15690 | 150.284 | 0.87553 | 0.96694 |
| atpI; ATP synthase protein I | 0.01922 | 0.00121 | 0.01966 | 0.00026 | 0.00199 | 65 | 0.01891 | 0.00021 | 0.00151 | 93 | 0.00250 | 0.29889 | 129.025 | 0.76551 | 0.93900 |
| ubiG; 2-polyprenyl-6-hydroxyphenyl methylase / 3-demethylubiquinone-9 3-methyltransferase [EC:2.1.1.222 2.1.1.64] | 0.01919 | 0.00156 | 0.01851 | 0.00041 | 0.00251 | 65 | 0.01967 | 0.00037 | 0.00199 | 93 | 0.00321 | -0.36036 | 132.978 | 0.71915 | 0.92108 |
| sugE; quaternary ammonium compound-resistance protein SugE | 0.01919 | 0.00160 | 0.01856 | 0.00047 | 0.00269 | 65 | 0.01962 | 0.00036 | 0.00197 | 93 | 0.00334 | -0.31820 | 125.673 | 0.75086 | 0.93483 |
| glsA, GLS; glutaminase [EC:3.5.1.2] | 0.01913 | 0.00151 | 0.02157 | 0.00059 | 0.00300 | 65 | 0.01743 | 0.00020 | 0.00148 | 93 | 0.00335 | 1.23660 | 95.081 | 0.21928 | 0.77376 |
| eptC; heptose-I-phosphate ethanolaminephosphotransferase [EC:2.7.8.-] | 0.01912 | 0.00166 | 0.01941 | 0.00045 | 0.00264 | 65 | 0.01893 | 0.00042 | 0.00214 | 93 | 0.00339 | 0.14163 | 135.147 | 0.88759 | 0.97039 |
| aqpZ; aquaporin Z | 0.01910 | 0.00099 | 0.01652 | 0.00013 | 0.00140 | 65 | 0.02091 | 0.00017 | 0.00135 | 93 | 0.00195 | -2.25427 | 148.778 | 0.02564 | 0.59941 |
| TC.NCS2; nucleobase:cation symporter-2, NCS2 family | 0.01904 | 0.00141 | 0.01983 | 0.00044 | 0.00260 | 65 | 0.01849 | 0.00023 | 0.00157 | 93 | 0.00304 | 0.43905 | 108.932 | 0.66149 | 0.90968 |
| yaeR; glyoxylase I family protein | 0.01903 | 0.00116 | 0.01794 | 0.00020 | 0.00175 | 65 | 0.01979 | 0.00022 | 0.00154 | 93 | 0.00233 | -0.79422 | 142.451 | 0.42839 | 0.82737 |
| PPOX, hemY; protoporphyrinogen/coproporphyrinogen III oxidase [EC:1.3.3.4 1.3.3.15] | 0.01902 | 0.00140 | 0.01743 | 0.00027 | 0.00203 | 65 | 0.02013 | 0.00034 | 0.00190 | 93 | 0.00278 | -0.96955 | 146.769 | 0.33387 | 0.77992 |
| arcA; arginine deiminase [EC:3.5.3.6] | 0.01900 | 0.00102 | 0.02061 | 0.00020 | 0.00176 | 65 | 0.01788 | 0.00014 | 0.00121 | 93 | 0.00214 | 1.27225 | 120.379 | 0.20573 | 0.77376 |
| cusR, copR, silR; two-component system, OmpR family, copper resistance phosphate regulon response regulator CusR | 0.01898 | 0.00239 | 0.01912 | 0.00087 | 0.00366 | 65 | 0.01888 | 0.00093 | 0.00316 | 93 | 0.00484 | 0.04990 | 140.656 | 0.96027 | 0.98903 |
| malY, malT; maltose/moltooligosaccharide transporter | 0.01893 | 0.00201 | 0.01974 | 0.00095 | 0.00383 | 65 | 0.01837 | 0.00043 | 0.00214 | 93 | 0.00438 | 0.31138 | 103.218 | 0.75614 | 0.93669 |
| tolC; outer membrane protein | 0.01893 | 0.00130 | 0.01646 | 0.00026 | 0.00198 | 65 | 0.02066 | 0.00027 | 0.00170 | 93 | 0.00261 | -1.60777 | 139.969 | 0.11014 | 0.77376 |
| hisIE; phosphoribosyl-ATP pyrophosphohydrolase / phosphoribosyl-AMP cyclohydrolase [EC:3.6.1.31 3.5.4.19] | 0.01892 | 0.00126 | 0.01800 | 0.00023 | 0.00190 | 65 | 0.01957 | 0.00026 | 0.00168 | 93 | 0.00254 | -0.62144 | 142.719 | 0.53530 | 0.86207 |
| patA, rscA, lmrC, satA; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.01891 | 0.00150 | 0.01932 | 0.00034 | 0.00228 | 65 | 0.01862 | 0.00037 | 0.00199 | 93 | 0.00303 | 0.23427 | 141.753 | 0.81511 | 0.95409 |
| patB, rscB, lmrC, satB; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.01891 | 0.00150 | 0.01932 | 0.00034 | 0.00228 | 65 | 0.01861 | 0.00037 | 0.00199 | 93 | 0.00303 | 0.23488 | 141.760 | 0.81464 | 0.95409 |
| moaD, cysO; sulfur-carrier protein | 0.01889 | 0.00117 | 0.01710 | 0.00020 | 0.00174 | 65 | 0.02013 | 0.00023 | 0.00157 | 93 | 0.00234 | -1.29345 | 144.243 | 0.19792 | 0.77376 |
| glnD; [protein-PII] uridylyltransferase [EC:2.7.7.59] | 0.01888 | 0.00126 | 0.01816 | 0.00025 | 0.00198 | 65 | 0.01938 | 0.00025 | 0.00163 | 93 | 0.00256 | -0.47864 | 136.723 | 0.63296 | 0.89696 |
| opuC; osmoprotectant transport system substrate-binding protein | 0.01885 | 0.00159 | 0.01947 | 0.00045 | 0.00263 | 65 | 0.01842 | 0.00037 | 0.00198 | 93 | 0.00330 | 0.31762 | 128.508 | 0.75129 | 0.93519 |
| grxD, GLRX5; monothiol glutaredoxin | 0.01884 | 0.00150 | 0.01840 | 0.00039 | 0.00244 | 65 | 0.01915 | 0.00034 | 0.00191 | 93 | 0.00310 | -0.24242 | 132.067 | 0.80883 | 0.95002 |
| eta; exfoliative toxin A/B | 0.01880 | 0.00155 | 0.01862 | 0.00035 | 0.00230 | 65 | 0.01892 | 0.00041 | 0.00209 | 93 | 0.00311 | -0.09699 | 144.694 | 0.92287 | 0.97947 |
| mltB; membrane-bound lytic murein transglycosylase B [EC:4.2.2.-] | 0.01877 | 0.00157 | 0.01947 | 0.00042 | 0.00254 | 65 | 0.01828 | 0.00037 | 0.00199 | 93 | 0.00323 | 0.36912 | 131.731 | 0.71263 | 0.92004 |
| LYSN; 2-aminoadipate transaminase [EC:2.6.1.-] | 0.01875 | 0.00190 | 0.01952 | 0.00070 | 0.00328 | 65 | 0.01822 | 0.00048 | 0.00228 | 93 | 0.00400 | 0.32751 | 121.103 | 0.74385 | 0.93099 |
| ytrA; GntR family transcriptional regulator | 0.01875 | 0.00196 | 0.01517 | 0.00030 | 0.00214 | 65 | 0.02125 | 0.00081 | 0.00295 | 93 | 0.00365 | -1.66503 | 153.267 | 0.09795 | 0.76722 |
| K07011; uncharacterized protein | 0.01875 | 0.00131 | 0.01676 | 0.00018 | 0.00168 | 65 | 0.02013 | 0.00033 | 0.00189 | 93 | 0.00253 | -1.33399 | 155.400 | 0.18416 | 0.77376 |
| murN; alanine adding enzyme [EC:2.3.2.-] | 0.01874 | 0.00156 | 0.01797 | 0.00033 | 0.00224 | 65 | 0.01928 | 0.00043 | 0.00215 | 93 | 0.00310 | -0.42270 | 148.547 | 0.67313 | 0.91184 |
| dsbC; thiol:disulfide interchange protein DsbC [EC:5.3.4.1] | 0.01873 | 0.00148 | 0.01919 | 0.00033 | 0.00226 | 65 | 0.01842 | 0.00036 | 0.00197 | 93 | 0.00300 | 0.25606 | 141.338 | 0.79828 | 0.94822 |
| ARSC2, arsC; arsenate reductase [EC:1.20.4.1] | 0.01870 | 0.00113 | 0.01814 | 0.00022 | 0.00185 | 65 | 0.01908 | 0.00019 | 0.00142 | 93 | 0.00233 | -0.40421 | 130.597 | 0.68672 | 0.91567 |
| fliL; flagellar FliL protein | 0.01868 | 0.00157 | 0.01792 | 0.00044 | 0.00260 | 65 | 0.01920 | 0.00036 | 0.00197 | 93 | 0.00326 | -0.39379 | 129.191 | 0.69439 | 0.91715 |
| ybbN; putative thioredoxin | 0.01867 | 0.00113 | 0.01852 | 0.00023 | 0.00187 | 65 | 0.01878 | 0.00019 | 0.00141 | 93 | 0.00235 | -0.10823 | 128.575 | 0.91398 | 0.97664 |
| aadK; aminoglycoside 6-adenylyltransferase [EC:2.7.7.-] | 0.01864 | 0.00153 | 0.01815 | 0.00033 | 0.00224 | 65 | 0.01899 | 0.00040 | 0.00208 | 93 | 0.00305 | -0.27310 | 146.185 | 0.78516 | 0.94445 |
| ribA, RIB1; GTP cyclohydrolase II [EC:3.5.4.25] | 0.01864 | 0.00125 | 0.01923 | 0.00028 | 0.00206 | 65 | 0.01822 | 0.00023 | 0.00157 | 93 | 0.00260 | 0.38809 | 129.647 | 0.69859 | 0.91844 |
| PTS-Tre-EIIC, treB; PTS system, trehalose-specific IIC component | 0.01863 | 0.00135 | 0.02061 | 0.00040 | 0.00247 | 65 | 0.01724 | 0.00021 | 0.00149 | 93 | 0.00289 | 1.16789 | 109.296 | 0.24539 | 0.77376 |
| asnC; Lrp/AsnC family transcriptional regulator, regulator for asnA, asnC and gidA | 0.01859 | 0.00168 | 0.01728 | 0.00032 | 0.00223 | 65 | 0.01952 | 0.00054 | 0.00240 | 93 | 0.00328 | -0.68256 | 154.196 | 0.49591 | 0.84797 |
| fdx; ferredoxin, 2Fe-2S | 0.01859 | 0.00120 | 0.01796 | 0.00023 | 0.00190 | 65 | 0.01903 | 0.00023 | 0.00156 | 93 | 0.00245 | -0.43619 | 136.553 | 0.66339 | 0.91006 |
| E4.1.1.17, ODC1, speC, speF; ornithine decarboxylase [EC:4.1.1.17] | 0.01858 | 0.00126 | 0.01982 | 0.00029 | 0.00210 | 65 | 0.01772 | 0.00023 | 0.00156 | 93 | 0.00262 | 0.80271 | 127.238 | 0.42364 | 0.82432 |
| narH, narY, nxrB; nitrate reductase / nitrite oxidoreductase, beta subunit [EC:1.7.5.1 1.7.99.-] | 0.01856 | 0.00115 | 0.01944 | 0.00024 | 0.00193 | 65 | 0.01794 | 0.00018 | 0.00141 | 93 | 0.00239 | 0.62755 | 125.324 | 0.53144 | 0.86136 |
| ATPVD, ntpD, atpD; V/A-type H+/Na+-transporting ATPase subunit D | 0.01855 | 0.00143 | 0.02210 | 0.00043 | 0.00258 | 65 | 0.01608 | 0.00023 | 0.00158 | 93 | 0.00303 | 1.98829 | 110.344 | 0.04926 | 0.67429 |
| LYS1; saccharopine dehydrogenase (NAD+, L-lysine forming) [EC:1.5.1.7] | 0.01855 | 0.00138 | 0.01617 | 0.00024 | 0.00191 | 65 | 0.02021 | 0.00034 | 0.00192 | 93 | 0.00271 | -1.48978 | 151.163 | 0.13836 | 0.77376 |
| K01163; uncharacterized protein | 0.01855 | 0.00159 | 0.02116 | 0.00048 | 0.00272 | 65 | 0.01672 | 0.00034 | 0.00190 | 93 | 0.00332 | 1.33892 | 121.616 | 0.18310 | 0.77376 |
| lldD; L-lactate dehydrogenase (cytochrome) [EC:1.1.2.3] | 0.01852 | 0.00135 | 0.01697 | 0.00028 | 0.00208 | 65 | 0.01960 | 0.00030 | 0.00179 | 93 | 0.00274 | -0.96125 | 140.331 | 0.33808 | 0.78383 |
| btuB; vitamin B12 transporter | 0.01852 | 0.00115 | 0.01864 | 0.00022 | 0.00183 | 65 | 0.01843 | 0.00021 | 0.00149 | 93 | 0.00236 | 0.08744 | 135.149 | 0.93045 | 0.98255 |
| ycgQ; putative membrane protein | 0.01851 | 0.00148 | 0.01783 | 0.00030 | 0.00215 | 65 | 0.01899 | 0.00038 | 0.00203 | 93 | 0.00296 | -0.39306 | 147.682 | 0.69484 | 0.91715 |
| secDF; SecD/SecF fusion protein | 0.01847 | 0.00131 | 0.01520 | 0.00022 | 0.00184 | 65 | 0.02076 | 0.00029 | 0.00178 | 93 | 0.00256 | -2.17033 | 148.943 | 0.03157 | 0.61842 |
| algH; putative transcriptional regulator | 0.01847 | 0.00121 | 0.01749 | 0.00025 | 0.00198 | 65 | 0.01915 | 0.00022 | 0.00153 | 93 | 0.00250 | -0.66399 | 130.558 | 0.50787 | 0.84956 |
| GLDC, gcvP; glycine dehydrogenase [EC:1.4.4.2] | 0.01847 | 0.00131 | 0.01795 | 0.00028 | 0.00208 | 65 | 0.01882 | 0.00027 | 0.00170 | 93 | 0.00269 | -0.32301 | 135.847 | 0.74719 | 0.93207 |
| cbiG; cobalt-precorrin 5A hydrolase [EC:3.7.1.12] | 0.01845 | 0.00128 | 0.02173 | 0.00034 | 0.00229 | 65 | 0.01616 | 0.00019 | 0.00143 | 93 | 0.00270 | 2.06985 | 111.717 | 0.04077 | 0.64345 |
| narI, narV; nitrate reductase gamma subunit [EC:1.7.5.1 1.7.99.-] | 0.01844 | 0.00115 | 0.01931 | 0.00024 | 0.00194 | 65 | 0.01783 | 0.00019 | 0.00141 | 93 | 0.00240 | 0.61939 | 125.546 | 0.53678 | 0.86207 |
| pbp2B, penA; penicillin-binding protein 2B | 0.01842 | 0.00153 | 0.01798 | 0.00032 | 0.00222 | 65 | 0.01873 | 0.00041 | 0.00210 | 93 | 0.00306 | -0.24436 | 147.659 | 0.80729 | 0.94987 |
| ATPVB, ntpB, atpB; V/A-type H+/Na+-transporting ATPase subunit B | 0.01841 | 0.00141 | 0.02185 | 0.00042 | 0.00255 | 65 | 0.01601 | 0.00023 | 0.00157 | 93 | 0.00300 | 1.94929 | 110.759 | 0.05379 | 0.70048 |
| add, ADA; adenosine deaminase [EC:3.5.4.4] | 0.01841 | 0.00124 | 0.01881 | 0.00024 | 0.00193 | 65 | 0.01813 | 0.00025 | 0.00163 | 93 | 0.00253 | 0.26711 | 138.334 | 0.78978 | 0.94606 |
| ATPVK, ntpK, atpK; V/A-type H+/Na+-transporting ATPase subunit K | 0.01841 | 0.00141 | 0.02185 | 0.00042 | 0.00255 | 65 | 0.01600 | 0.00023 | 0.00157 | 93 | 0.00299 | 1.95408 | 110.580 | 0.05322 | 0.69853 |
| dusB; tRNA-dihydrouridine synthase B [EC:1.-.-.-] | 0.01840 | 0.00125 | 0.01830 | 0.00025 | 0.00194 | 65 | 0.01847 | 0.00025 | 0.00163 | 93 | 0.00254 | -0.06982 | 138.168 | 0.94444 | 0.98674 |
| glpX; fructose-1,6-bisphosphatase II [EC:3.1.3.11] | 0.01840 | 0.00127 | 0.01998 | 0.00032 | 0.00221 | 65 | 0.01730 | 0.00021 | 0.00150 | 93 | 0.00267 | 1.00130 | 118.751 | 0.31872 | 0.77376 |
| yhfA; putative redox protein | 0.01840 | 0.00164 | 0.01729 | 0.00035 | 0.00232 | 65 | 0.01918 | 0.00048 | 0.00226 | 93 | 0.00324 | -0.58300 | 149.560 | 0.56077 | 0.86813 |
| ATPVI, ntpI, atpI; V/A-type H+/Na+-transporting ATPase subunit I | 0.01840 | 0.00141 | 0.02183 | 0.00042 | 0.00255 | 65 | 0.01600 | 0.00023 | 0.00157 | 93 | 0.00299 | 1.94535 | 110.527 | 0.05427 | 0.70167 |
| argK; LAO/AO transport system kinase [EC:2.7.-.-] | 0.01838 | 0.00160 | 0.01750 | 0.00050 | 0.00277 | 65 | 0.01900 | 0.00035 | 0.00193 | 93 | 0.00338 | -0.44345 | 121.133 | 0.65823 | 0.90829 |
| narG, narZ, nxrA; nitrate reductase / nitrite oxidoreductase, alpha subunit [EC:1.7.5.1 1.7.99.-] | 0.01838 | 0.00115 | 0.01925 | 0.00024 | 0.00194 | 65 | 0.01777 | 0.00018 | 0.00141 | 93 | 0.00240 | 0.61541 | 125.355 | 0.53940 | 0.86207 |
| ATPVA, ntpA, atpA; V/A-type H+/Na+-transporting ATPase subunit A [EC:3.6.3.14 3.6.3.15] | 0.01836 | 0.00141 | 0.02182 | 0.00042 | 0.00255 | 65 | 0.01594 | 0.00023 | 0.00156 | 93 | 0.00299 | 1.96594 | 109.921 | 0.05183 | 0.68802 |
| oadB; oxaloacetate decarboxylase, beta subunit [EC:4.1.1.3] | 0.01830 | 0.00172 | 0.02111 | 0.00059 | 0.00302 | 65 | 0.01634 | 0.00038 | 0.00202 | 93 | 0.00363 | 1.31553 | 117.515 | 0.19089 | 0.77376 |
| yrbG; cation:H+ antiporter | 0.01827 | 0.00115 | 0.01780 | 0.00018 | 0.00168 | 65 | 0.01860 | 0.00023 | 0.00156 | 93 | 0.00230 | -0.34667 | 146.462 | 0.72933 | 0.92412 |
| ABC.ZM.S; zinc/manganese transport system substrate-binding protein | 0.01826 | 0.00186 | 0.02057 | 0.00070 | 0.00329 | 65 | 0.01665 | 0.00043 | 0.00215 | 93 | 0.00393 | 0.99697 | 115.785 | 0.32086 | 0.77376 |
| E1.2.3.3, poxL; pyruvate oxidase [EC:1.2.3.3] | 0.01825 | 0.00143 | 0.01778 | 0.00031 | 0.00218 | 65 | 0.01858 | 0.00034 | 0.00190 | 93 | 0.00289 | -0.27861 | 141.598 | 0.78095 | 0.94296 |
| pabBC; para-aminobenzoate synthetase / 4-amino-4-deoxychorismate lyase [EC:2.6.1.85 4.1.3.38] | 0.01822 | 0.00149 | 0.01638 | 0.00025 | 0.00198 | 65 | 0.01950 | 0.00042 | 0.00212 | 93 | 0.00290 | -1.07661 | 153.979 | 0.28334 | 0.77376 |
| ciaR; two-component system, OmpR family, response regulator CiaR | 0.01819 | 0.00153 | 0.01800 | 0.00032 | 0.00221 | 65 | 0.01832 | 0.00041 | 0.00210 | 93 | 0.00304 | -0.10827 | 147.901 | 0.91393 | 0.97664 |
| dam; DNA adenine methylase [EC:2.1.1.72] | 0.01818 | 0.00140 | 0.01887 | 0.00033 | 0.00226 | 65 | 0.01770 | 0.00030 | 0.00178 | 93 | 0.00288 | 0.40330 | 132.571 | 0.68738 | 0.91567 |
| murM; serine/alanine adding enzyme [EC:2.3.2.10] | 0.01818 | 0.00152 | 0.01790 | 0.00032 | 0.00221 | 65 | 0.01837 | 0.00041 | 0.00209 | 93 | 0.00304 | -0.15445 | 147.560 | 0.87746 | 0.96737 |
| ABC-2.CYL.P, cylB; multidrug/hemolysin transport system permease protein | 0.01818 | 0.00149 | 0.01803 | 0.00031 | 0.00217 | 65 | 0.01828 | 0.00039 | 0.00204 | 93 | 0.00298 | -0.08379 | 147.066 | 0.93334 | 0.98428 |
| pta; phosphate acetyltransferase [EC:2.3.1.8] | 0.01816 | 0.00122 | 0.01871 | 0.00021 | 0.00181 | 65 | 0.01778 | 0.00025 | 0.00164 | 93 | 0.00245 | 0.37757 | 144.580 | 0.70630 | 0.92004 |
| K09807; uncharacterized protein | 0.01816 | 0.00142 | 0.01655 | 0.00034 | 0.00228 | 65 | 0.01928 | 0.00031 | 0.00182 | 93 | 0.00292 | -0.93539 | 133.861 | 0.35127 | 0.79120 |
| marC; multiple antibiotic resistance protein | 0.01814 | 0.00122 | 0.01728 | 0.00022 | 0.00182 | 65 | 0.01874 | 0.00025 | 0.00165 | 93 | 0.00246 | -0.59463 | 144.378 | 0.55302 | 0.86788 |
| secB; preprotein translocase subunit SecB | 0.01814 | 0.00125 | 0.01774 | 0.00026 | 0.00199 | 65 | 0.01843 | 0.00024 | 0.00162 | 93 | 0.00257 | -0.26900 | 135.554 | 0.78834 | 0.94581 |
| ABC-2.CYL.A, cylA; multidrug/hemolysin transport system ATP-binding protein | 0.01812 | 0.00149 | 0.01798 | 0.00031 | 0.00217 | 65 | 0.01822 | 0.00039 | 0.00204 | 93 | 0.00298 | -0.07945 | 147.029 | 0.93678 | 0.98459 |
| ciaH; two-component system, OmpR family, sensor histidine kinase CiaH [EC:2.7.13.3] | 0.01812 | 0.00153 | 0.01796 | 0.00032 | 0.00220 | 65 | 0.01823 | 0.00041 | 0.00210 | 93 | 0.00304 | -0.09056 | 147.966 | 0.92796 | 0.98143 |
| MPG; DNA-3-methyladenine glycosylase [EC:3.2.2.21] | 0.01811 | 0.00139 | 0.01934 | 0.00036 | 0.00234 | 65 | 0.01725 | 0.00027 | 0.00170 | 93 | 0.00289 | 0.72451 | 124.773 | 0.47011 | 0.83870 |
| PPCS, COAB; phosphopantothenate—cysteine ligase (ATP) [EC:6.3.2.51] | 0.01810 | 0.00152 | 0.01789 | 0.00031 | 0.00220 | 65 | 0.01824 | 0.00041 | 0.00209 | 93 | 0.00303 | -0.11639 | 147.877 | 0.90750 | 0.97664 |
| fic; cell filamentation protein | 0.01808 | 0.00138 | 0.01720 | 0.00029 | 0.00210 | 65 | 0.01870 | 0.00031 | 0.00183 | 93 | 0.00279 | -0.53788 | 141.640 | 0.59150 | 0.87896 |
| PPCDC, coaC; phosphopantothenoylcysteine decarboxylase [EC:4.1.1.36] | 0.01808 | 0.00152 | 0.01788 | 0.00031 | 0.00220 | 65 | 0.01822 | 0.00041 | 0.00209 | 93 | 0.00304 | -0.11326 | 147.866 | 0.90998 | 0.97664 |
| cobN; cobaltochelatase CobN [EC:6.6.1.2] | 0.01805 | 0.00146 | 0.01740 | 0.00030 | 0.00215 | 65 | 0.01850 | 0.00037 | 0.00199 | 93 | 0.00293 | -0.37608 | 145.985 | 0.70740 | 0.92004 |
| estA; putative tributyrin esterase [EC:3.1.1.-] | 0.01804 | 0.00153 | 0.01785 | 0.00032 | 0.00220 | 65 | 0.01818 | 0.00041 | 0.00209 | 93 | 0.00304 | -0.10990 | 147.873 | 0.91263 | 0.97664 |
| pbp2X; penicillin-binding protein 2X | 0.01804 | 0.00153 | 0.01785 | 0.00032 | 0.00220 | 65 | 0.01818 | 0.00041 | 0.00209 | 93 | 0.00304 | -0.10990 | 147.873 | 0.91263 | 0.97664 |
| BDH, butB; (R,R)-butanediol dehydrogenase / meso-butanediol dehydrogenase / diacetyl reductase [EC:1.1.1.4 1.1.1.- 1.1.1.303] | 0.01804 | 0.00134 | 0.01747 | 0.00023 | 0.00189 | 65 | 0.01844 | 0.00032 | 0.00185 | 93 | 0.00265 | -0.36477 | 149.685 | 0.71580 | 0.92004 |
| ssuE; FMN reductase [EC:1.5.1.38] | 0.01801 | 0.00178 | 0.01537 | 0.00043 | 0.00257 | 65 | 0.01985 | 0.00054 | 0.00242 | 93 | 0.00353 | -1.26982 | 147.085 | 0.20615 | 0.77376 |
| glf; UDP-galactopyranose mutase [EC:5.4.99.9] | 0.01800 | 0.00149 | 0.01894 | 0.00040 | 0.00247 | 65 | 0.01735 | 0.00032 | 0.00185 | 93 | 0.00309 | 0.51420 | 127.899 | 0.60800 | 0.88637 |
| spoVB; stage V sporulation protein B | 0.01791 | 0.00180 | 0.01917 | 0.00069 | 0.00327 | 65 | 0.01703 | 0.00039 | 0.00204 | 93 | 0.00385 | 0.55402 | 111.731 | 0.58067 | 0.87573 |
| HPD, hppD; 4-hydroxyphenylpyruvate dioxygenase [EC:1.13.11.27] | 0.01788 | 0.00180 | 0.01670 | 0.00051 | 0.00279 | 65 | 0.01870 | 0.00052 | 0.00237 | 93 | 0.00366 | -0.54634 | 139.364 | 0.58571 | 0.87720 |
| phbC, phaC; polyhydroxyalkanoate synthase [EC:2.3.1.-] | 0.01784 | 0.00178 | 0.01656 | 0.00055 | 0.00290 | 65 | 0.01874 | 0.00047 | 0.00226 | 93 | 0.00368 | -0.59298 | 131.440 | 0.55422 | 0.86806 |
| flhA; flagellar biosynthesis protein FlhA | 0.01779 | 0.00129 | 0.01550 | 0.00026 | 0.00200 | 65 | 0.01939 | 0.00026 | 0.00168 | 93 | 0.00261 | -1.48899 | 138.118 | 0.13877 | 0.77376 |
| tagA, tarA; N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase [EC:2.4.1.187] | 0.01778 | 0.00116 | 0.01652 | 0.00021 | 0.00180 | 65 | 0.01867 | 0.00021 | 0.00151 | 93 | 0.00235 | -0.91526 | 138.239 | 0.36165 | 0.79496 |
| ihfA, himA; integration host factor subunit alpha | 0.01778 | 0.00114 | 0.01705 | 0.00021 | 0.00181 | 65 | 0.01830 | 0.00020 | 0.00148 | 93 | 0.00234 | -0.53319 | 136.189 | 0.59477 | 0.88005 |
| aroP; aromatic amino acid transport protein AroP | 0.01777 | 0.00156 | 0.01805 | 0.00033 | 0.00224 | 65 | 0.01758 | 0.00043 | 0.00215 | 93 | 0.00311 | 0.15179 | 148.368 | 0.87956 | 0.96779 |
| E4.3.1.12, ocd; ornithine cyclodeaminase [EC:4.3.1.12] | 0.01774 | 0.00139 | 0.01501 | 0.00023 | 0.00188 | 65 | 0.01964 | 0.00035 | 0.00195 | 93 | 0.00271 | -1.70958 | 152.811 | 0.08937 | 0.76567 |
| mocA; molybdenum cofactor cytidylyltransferase [EC:2.7.7.76] | 0.01774 | 0.00110 | 0.01694 | 0.00022 | 0.00183 | 65 | 0.01829 | 0.00017 | 0.00137 | 93 | 0.00228 | -0.59072 | 127.852 | 0.55575 | 0.86813 |
| ACADS, bcd; butyryl-CoA dehydrogenase [EC:1.3.8.1] | 0.01773 | 0.00231 | 0.02300 | 0.00152 | 0.00484 | 65 | 0.01404 | 0.00035 | 0.00194 | 93 | 0.00521 | 1.71889 | 84.640 | 0.08929 | 0.76567 |
| K07035; uncharacterized protein | 0.01771 | 0.00157 | 0.02126 | 0.00052 | 0.00284 | 65 | 0.01524 | 0.00029 | 0.00176 | 93 | 0.00334 | 1.80265 | 111.437 | 0.07415 | 0.74128 |
| minE; cell division topological specificity factor | 0.01770 | 0.00120 | 0.01953 | 0.00030 | 0.00216 | 65 | 0.01642 | 0.00018 | 0.00137 | 93 | 0.00256 | 1.21371 | 113.400 | 0.22738 | 0.77376 |
| qseC; two-component system, OmpR family, sensor histidine kinase QseC [EC:2.7.13.3] | 0.01770 | 0.00185 | 0.01721 | 0.00041 | 0.00252 | 65 | 0.01804 | 0.00063 | 0.00260 | 93 | 0.00362 | -0.22653 | 152.569 | 0.82109 | 0.95506 |
| waaC, rfaC; heptosyltransferase I [EC:2.4.-.-] | 0.01767 | 0.00118 | 0.01912 | 0.00020 | 0.00176 | 65 | 0.01666 | 0.00023 | 0.00157 | 93 | 0.00236 | 1.04025 | 143.227 | 0.29998 | 0.77376 |
| cbiD; cobalt-precorrin-5B (C1)-methyltransferase [EC:2.1.1.195] | 0.01766 | 0.00132 | 0.02074 | 0.00037 | 0.00239 | 65 | 0.01551 | 0.00020 | 0.00145 | 93 | 0.00280 | 1.86500 | 109.499 | 0.06486 | 0.73334 |
| aarC, cat1; succinyl-CoA:acetate CoA-transferase [EC:2.8.3.18] | 0.01765 | 0.00120 | 0.01747 | 0.00022 | 0.00183 | 65 | 0.01778 | 0.00024 | 0.00160 | 93 | 0.00243 | -0.13065 | 141.315 | 0.89624 | 0.97276 |
| nudH; putative (di)nucleoside polyphosphate hydrolase [EC:3.6.1.-] | 0.01765 | 0.00121 | 0.01700 | 0.00023 | 0.00190 | 65 | 0.01809 | 0.00023 | 0.00158 | 93 | 0.00247 | -0.43994 | 137.456 | 0.66067 | 0.90968 |
| dppA; dipeptide transport system substrate-binding protein | 0.01762 | 0.00211 | 0.01726 | 0.00058 | 0.00298 | 65 | 0.01787 | 0.00080 | 0.00293 | 93 | 0.00417 | -0.14609 | 150.026 | 0.88405 | 0.96974 |
| phoB1, phoP; two-component system, OmpR family, alkaline phosphatase synthesis response regulator PhoP | 0.01761 | 0.00125 | 0.01672 | 0.00028 | 0.00207 | 65 | 0.01824 | 0.00023 | 0.00156 | 93 | 0.00260 | -0.58744 | 128.578 | 0.55794 | 0.86813 |
| pheB; chorismate mutase [EC:5.4.99.5] | 0.01761 | 0.00134 | 0.01911 | 0.00039 | 0.00245 | 65 | 0.01655 | 0.00021 | 0.00150 | 93 | 0.00287 | 0.89052 | 110.487 | 0.37512 | 0.79810 |
| rnfD; electron transport complex protein RnfD | 0.01761 | 0.00152 | 0.02183 | 0.00048 | 0.00271 | 65 | 0.01465 | 0.00027 | 0.00171 | 93 | 0.00320 | 2.24257 | 112.551 | 0.02688 | 0.61112 |
| fliNY, fliN; flagellar motor switch protein FliN/FliY | 0.01760 | 0.00131 | 0.01500 | 0.00024 | 0.00193 | 65 | 0.01943 | 0.00029 | 0.00176 | 93 | 0.00261 | -1.69414 | 145.102 | 0.09238 | 0.76567 |
| cusS, copS, silS; two-component system, OmpR family, heavy metal sensor histidine kinase CusS [EC:2.7.13.3] | 0.01758 | 0.00221 | 0.01758 | 0.00075 | 0.00341 | 65 | 0.01758 | 0.00079 | 0.00291 | 93 | 0.00448 | -0.00059 | 139.711 | 0.99953 | 0.99957 |
| yejA; microcin C transport system substrate-binding protein | 0.01756 | 0.00224 | 0.01643 | 0.00066 | 0.00320 | 65 | 0.01835 | 0.00089 | 0.00310 | 93 | 0.00445 | -0.43046 | 149.127 | 0.66749 | 0.91150 |
| K07071; uncharacterized protein | 0.01749 | 0.00131 | 0.01582 | 0.00025 | 0.00196 | 65 | 0.01866 | 0.00028 | 0.00175 | 93 | 0.00262 | -1.08338 | 143.216 | 0.28046 | 0.77376 |
| narJ, narW; nitrate reductase molybdenum cofactor assembly chaperone NarJ/NarW | 0.01743 | 0.00111 | 0.01826 | 0.00023 | 0.00188 | 65 | 0.01685 | 0.00017 | 0.00135 | 93 | 0.00231 | 0.61098 | 124.267 | 0.54233 | 0.86262 |
| fliR; flagellar biosynthetic protein FliR | 0.01743 | 0.00126 | 0.01555 | 0.00026 | 0.00199 | 65 | 0.01874 | 0.00025 | 0.00163 | 93 | 0.00257 | -1.23856 | 136.380 | 0.21763 | 0.77376 |
| flhB; flagellar biosynthetic protein FlhB | 0.01741 | 0.00127 | 0.01549 | 0.00026 | 0.00199 | 65 | 0.01875 | 0.00025 | 0.00164 | 93 | 0.00258 | -1.26679 | 136.363 | 0.20739 | 0.77376 |
| fliP; flagellar biosynthetic protein FliP | 0.01740 | 0.00127 | 0.01542 | 0.00026 | 0.00201 | 65 | 0.01879 | 0.00025 | 0.00164 | 93 | 0.00259 | -1.30029 | 136.009 | 0.19570 | 0.77376 |
| pfpI; protease I [EC:3.5.1.124] | 0.01739 | 0.00151 | 0.01555 | 0.00037 | 0.00240 | 65 | 0.01868 | 0.00035 | 0.00194 | 93 | 0.00308 | -1.01751 | 134.795 | 0.31073 | 0.77376 |
| iscR; Rrf2 family transcriptional regulator, iron-sulfur cluster assembly transcription factor | 0.01738 | 0.00115 | 0.01783 | 0.00020 | 0.00177 | 65 | 0.01707 | 0.00021 | 0.00152 | 93 | 0.00233 | 0.32679 | 139.945 | 0.74432 | 0.93142 |
| pal; peptidoglycan-associated lipoprotein | 0.01738 | 0.00123 | 0.01647 | 0.00024 | 0.00193 | 65 | 0.01801 | 0.00024 | 0.00160 | 93 | 0.00251 | -0.61397 | 137.458 | 0.54025 | 0.86207 |
| tesB; acyl-CoA thioesterase II [EC:3.1.2.-] | 0.01736 | 0.00150 | 0.01627 | 0.00023 | 0.00187 | 65 | 0.01812 | 0.00045 | 0.00220 | 93 | 0.00289 | -0.64133 | 155.936 | 0.52225 | 0.85628 |
| TC.NCS1; nucleobase:cation symporter-1, NCS1 family | 0.01735 | 0.00259 | 0.01593 | 0.00086 | 0.00363 | 65 | 0.01834 | 0.00120 | 0.00360 | 93 | 0.00511 | -0.47037 | 150.516 | 0.63878 | 0.89824 |
| mdtG; MFS transporter, DHA1 family, multidrug resistance protein | 0.01733 | 0.00250 | 0.02001 | 0.00127 | 0.00441 | 65 | 0.01546 | 0.00080 | 0.00293 | 93 | 0.00529 | 0.85957 | 116.953 | 0.39179 | 0.80459 |
| E2.6.1.11, argD; acetylornithine aminotransferase [EC:2.6.1.11] | 0.01732 | 0.00103 | 0.01459 | 0.00014 | 0.00146 | 65 | 0.01924 | 0.00018 | 0.00139 | 93 | 0.00202 | -2.30391 | 147.864 | 0.02262 | 0.59764 |
| PTS-Aga-EIIA, agaF; PTS system, N-acetylgalactosamine-specific IIA component [EC:2.7.1.-] | 0.01732 | 0.00136 | 0.01697 | 0.00025 | 0.00195 | 65 | 0.01757 | 0.00033 | 0.00188 | 93 | 0.00271 | -0.22112 | 148.888 | 0.82530 | 0.95506 |
| SOD1; superoxide dismutase, Cu-Zn family [EC:1.15.1.1] | 0.01731 | 0.00149 | 0.01593 | 0.00036 | 0.00235 | 65 | 0.01828 | 0.00034 | 0.00192 | 93 | 0.00304 | -0.77330 | 135.897 | 0.44069 | 0.83186 |
| rpoH; RNA polymerase sigma-32 factor | 0.01731 | 0.00123 | 0.01621 | 0.00023 | 0.00190 | 65 | 0.01807 | 0.00025 | 0.00163 | 93 | 0.00250 | -0.74301 | 140.137 | 0.45872 | 0.83438 |
| ACR3, arsB; arsenite transporter | 0.01730 | 0.00144 | 0.01680 | 0.00033 | 0.00224 | 65 | 0.01765 | 0.00033 | 0.00188 | 93 | 0.00293 | -0.29260 | 138.253 | 0.77027 | 0.94009 |
| flgK; flagellar hook-associated protein 1 FlgK | 0.01729 | 0.00138 | 0.01547 | 0.00031 | 0.00218 | 65 | 0.01856 | 0.00029 | 0.00177 | 93 | 0.00281 | -1.10107 | 135.554 | 0.27282 | 0.77376 |
| tamA; translocation and assembly module TamA | 0.01729 | 0.00127 | 0.01696 | 0.00027 | 0.00203 | 65 | 0.01752 | 0.00025 | 0.00164 | 93 | 0.00261 | -0.21661 | 134.932 | 0.82884 | 0.95506 |
| kdpD; two-component system, OmpR family, sensor histidine kinase KdpD [EC:2.7.13.3] | 0.01727 | 0.00160 | 0.01411 | 0.00033 | 0.00226 | 65 | 0.01948 | 0.00045 | 0.00220 | 93 | 0.00315 | -1.70357 | 149.335 | 0.09054 | 0.76567 |
| FBP, fbp; fructose-1,6-bisphosphatase I [EC:3.1.3.11] | 0.01722 | 0.00119 | 0.01709 | 0.00023 | 0.00187 | 65 | 0.01731 | 0.00023 | 0.00156 | 93 | 0.00243 | -0.09147 | 137.517 | 0.92725 | 0.98137 |
| nemR; TetR/AcrR family transcriptional regulator, transcriptional repressor for nem operon | 0.01721 | 0.00195 | 0.01352 | 0.00035 | 0.00233 | 65 | 0.01979 | 0.00076 | 0.00285 | 93 | 0.00369 | -1.70096 | 155.939 | 0.09094 | 0.76567 |
| bshA; L-malate glycosyltransferase [EC:2.4.1.-] | 0.01721 | 0.00195 | 0.02118 | 0.00108 | 0.00407 | 65 | 0.01444 | 0.00026 | 0.00167 | 93 | 0.00440 | 1.53076 | 85.661 | 0.12951 | 0.77376 |
| K06996; uncharacterized protein | 0.01719 | 0.00203 | 0.01546 | 0.00066 | 0.00319 | 65 | 0.01841 | 0.00065 | 0.00264 | 93 | 0.00414 | -0.71291 | 137.136 | 0.47711 | 0.84209 |
| csn1, cas9; CRISPR-associated endonuclease Csn1 [EC:3.1.-.-] | 0.01716 | 0.00113 | 0.01717 | 0.00022 | 0.00183 | 65 | 0.01716 | 0.00019 | 0.00145 | 93 | 0.00233 | 0.00361 | 132.704 | 0.99712 | 0.99951 |
| K09781; uncharacterized protein | 0.01713 | 0.00117 | 0.01690 | 0.00024 | 0.00191 | 65 | 0.01729 | 0.00021 | 0.00149 | 93 | 0.00242 | -0.16122 | 131.472 | 0.87217 | 0.96576 |
| AROA2, aroA; 3-deoxy-7-phosphoheptulonate synthase [EC:2.5.1.54] | 0.01713 | 0.00167 | 0.02098 | 0.00062 | 0.00309 | 65 | 0.01443 | 0.00031 | 0.00181 | 93 | 0.00359 | 1.82681 | 106.759 | 0.07052 | 0.73788 |
| frmA, ADH5, adhC; S-(hydroxymethyl)glutathione dehydrogenase / alcohol dehydrogenase [EC:1.1.1.284 1.1.1.1] | 0.01706 | 0.00165 | 0.01366 | 0.00025 | 0.00197 | 65 | 0.01943 | 0.00054 | 0.00242 | 93 | 0.00312 | -1.84666 | 155.936 | 0.06669 | 0.73334 |
| flgE; flagellar hook protein FlgE | 0.01705 | 0.00129 | 0.01497 | 0.00024 | 0.00191 | 65 | 0.01850 | 0.00028 | 0.00173 | 93 | 0.00258 | -1.36435 | 144.460 | 0.17458 | 0.77376 |
| aceB, glcB; malate synthase [EC:2.3.3.9] | 0.01704 | 0.00142 | 0.01511 | 0.00036 | 0.00234 | 65 | 0.01839 | 0.00029 | 0.00177 | 93 | 0.00293 | -1.11655 | 128.825 | 0.26627 | 0.77376 |
| ydjE; MFS transporter, putative metabolite:H+ symporter | 0.01698 | 0.00117 | 0.01576 | 0.00020 | 0.00177 | 65 | 0.01784 | 0.00022 | 0.00155 | 93 | 0.00235 | -0.88412 | 141.956 | 0.37813 | 0.79852 |
| elaA; ElaA protein | 0.01697 | 0.00116 | 0.01839 | 0.00034 | 0.00227 | 65 | 0.01597 | 0.00013 | 0.00117 | 93 | 0.00256 | 0.94846 | 97.722 | 0.34523 | 0.78739 |
| spoT; GTP diphosphokinase / guanosine-3’,5’-bis(diphosphate) 3’-diphosphatase [EC:2.7.6.5 3.1.7.2] | 0.01694 | 0.00118 | 0.01895 | 0.00027 | 0.00206 | 65 | 0.01553 | 0.00018 | 0.00139 | 93 | 0.00248 | 1.37856 | 118.646 | 0.17063 | 0.77376 |
| glcD; glycolate oxidase [EC:1.1.3.15] | 0.01694 | 0.00159 | 0.01674 | 0.00029 | 0.00210 | 65 | 0.01708 | 0.00049 | 0.00229 | 93 | 0.00310 | -0.10664 | 154.529 | 0.91521 | 0.97664 |
| dusA; tRNA-dihydrouridine synthase A [EC:1.-.-.-] | 0.01692 | 0.00113 | 0.01688 | 0.00021 | 0.00179 | 65 | 0.01694 | 0.00020 | 0.00146 | 93 | 0.00231 | -0.02345 | 135.692 | 0.98133 | 0.99661 |
| oprM, emhC, ttgC, cusC, adeK, smeF, mtrE, cmeC, gesC; outer membrane protein, multidrug efflux system | 0.01691 | 0.00171 | 0.01600 | 0.00042 | 0.00254 | 65 | 0.01754 | 0.00050 | 0.00232 | 93 | 0.00344 | -0.44842 | 145.093 | 0.65452 | 0.90679 |
| DOPA; 4,5-DOPA dioxygenase extradiol [EC:1.13.11.-] | 0.01690 | 0.00109 | 0.01691 | 0.00017 | 0.00164 | 65 | 0.01689 | 0.00020 | 0.00147 | 93 | 0.00220 | 0.01254 | 143.973 | 0.99001 | 0.99801 |
| gmhC, hldE, waaE, rfaE; D-beta-D-heptose 7-phosphate kinase / D-beta-D-heptose 1-phosphate adenosyltransferase [EC:2.7.1.167 2.7.7.70] | 0.01689 | 0.00108 | 0.01797 | 0.00019 | 0.00173 | 65 | 0.01613 | 0.00018 | 0.00138 | 93 | 0.00221 | 0.82763 | 134.132 | 0.40935 | 0.81416 |
| K09153; uncharacterized protein | 0.01687 | 0.00123 | 0.01802 | 0.00026 | 0.00202 | 65 | 0.01607 | 0.00022 | 0.00155 | 93 | 0.00254 | 0.76956 | 130.204 | 0.44296 | 0.83186 |
| E3.5.1.24; choloylglycine hydrolase [EC:3.5.1.24] | 0.01687 | 0.00216 | 0.01757 | 0.00091 | 0.00375 | 65 | 0.01638 | 0.00062 | 0.00258 | 93 | 0.00455 | 0.26269 | 120.191 | 0.79324 | 0.94783 |
| ihfB, himD; integration host factor subunit beta | 0.01687 | 0.00119 | 0.01638 | 0.00024 | 0.00191 | 65 | 0.01721 | 0.00021 | 0.00152 | 93 | 0.00244 | -0.33737 | 133.516 | 0.73636 | 0.92721 |
| cbpA; curved DNA-binding protein | 0.01687 | 0.00107 | 0.01554 | 0.00015 | 0.00152 | 65 | 0.01779 | 0.00020 | 0.00148 | 93 | 0.00212 | -1.06268 | 149.317 | 0.28964 | 0.77376 |
| rluF; 23S rRNA pseudouridine2604 synthase [EC:5.4.99.21] | 0.01686 | 0.00148 | 0.01741 | 0.00039 | 0.00246 | 65 | 0.01648 | 0.00032 | 0.00185 | 93 | 0.00308 | 0.30262 | 128.265 | 0.76267 | 0.93813 |
| fliQ; flagellar biosynthetic protein FliQ | 0.01685 | 0.00118 | 0.01489 | 0.00023 | 0.00188 | 65 | 0.01821 | 0.00021 | 0.00152 | 93 | 0.00242 | -1.37375 | 134.738 | 0.17180 | 0.77376 |
| K09768; uncharacterized protein | 0.01681 | 0.00126 | 0.01592 | 0.00030 | 0.00216 | 65 | 0.01742 | 0.00022 | 0.00153 | 93 | 0.00265 | -0.56636 | 123.035 | 0.57218 | 0.87289 |
| K07571; S1 RNA binding domain protein | 0.01680 | 0.00122 | 0.01771 | 0.00033 | 0.00224 | 65 | 0.01617 | 0.00017 | 0.00137 | 93 | 0.00262 | 0.58883 | 110.098 | 0.55718 | 0.86813 |
| K07017; uncharacterized protein | 0.01680 | 0.00193 | 0.01719 | 0.00062 | 0.00308 | 65 | 0.01654 | 0.00057 | 0.00248 | 93 | 0.00396 | 0.16401 | 134.512 | 0.86997 | 0.96495 |
| sdhD, frdD; succinate dehydrogenase / fumarate reductase, membrane anchor subunit | 0.01679 | 0.00124 | 0.01577 | 0.00024 | 0.00194 | 65 | 0.01749 | 0.00024 | 0.00161 | 93 | 0.00252 | -0.68315 | 137.267 | 0.49567 | 0.84797 |
| fkpA; FKBP-type peptidyl-prolyl cis-trans isomerase FkpA [EC:5.2.1.8] | 0.01676 | 0.00163 | 0.01619 | 0.00040 | 0.00248 | 65 | 0.01716 | 0.00044 | 0.00216 | 93 | 0.00329 | -0.29380 | 141.651 | 0.76934 | 0.93993 |
| pdxJ; pyridoxine 5-phosphate synthase [EC:2.6.99.2] | 0.01675 | 0.00113 | 0.01510 | 0.00018 | 0.00166 | 65 | 0.01790 | 0.00021 | 0.00152 | 93 | 0.00225 | -1.24635 | 145.415 | 0.21464 | 0.77376 |
| tolQ; biopolymer transport protein TolQ | 0.01675 | 0.00129 | 0.01572 | 0.00026 | 0.00198 | 65 | 0.01747 | 0.00027 | 0.00171 | 93 | 0.00262 | -0.66833 | 140.431 | 0.50502 | 0.84956 |
| K09930; uncharacterized protein | 0.01674 | 0.00158 | 0.01676 | 0.00045 | 0.00264 | 65 | 0.01673 | 0.00036 | 0.00196 | 93 | 0.00329 | 0.00938 | 126.927 | 0.99253 | 0.99908 |
| kefB; glutathione-regulated potassium-efflux system protein KefB | 0.01673 | 0.00152 | 0.01624 | 0.00035 | 0.00232 | 65 | 0.01708 | 0.00038 | 0.00201 | 93 | 0.00307 | -0.27146 | 141.173 | 0.78644 | 0.94458 |
| dapdh; diaminopimelate dehydrogenase [EC:1.4.1.16] | 0.01673 | 0.00149 | 0.01682 | 0.00037 | 0.00240 | 65 | 0.01666 | 0.00033 | 0.00190 | 93 | 0.00306 | 0.05133 | 132.948 | 0.95914 | 0.98903 |
| K06994; putative drug exporter of the RND superfamily | 0.01673 | 0.00225 | 0.01208 | 0.00024 | 0.00191 | 65 | 0.01997 | 0.00118 | 0.00355 | 93 | 0.00403 | -1.95519 | 136.409 | 0.05261 | 0.69316 |
| flgC; flagellar basal-body rod protein FlgC | 0.01671 | 0.00120 | 0.01466 | 0.00023 | 0.00187 | 65 | 0.01815 | 0.00023 | 0.00156 | 93 | 0.00243 | -1.43481 | 137.970 | 0.15361 | 0.77376 |
| mglB; methyl-galactoside transport system substrate-binding protein | 0.01670 | 0.00158 | 0.02068 | 0.00059 | 0.00302 | 65 | 0.01392 | 0.00024 | 0.00162 | 93 | 0.00343 | 1.97358 | 100.424 | 0.05118 | 0.68675 |
| PTS-Gat-EIIA, gatA, sgcA; PTS system, galactitol-specific IIA component [EC:2.7.1.200] | 0.01669 | 0.00139 | 0.01721 | 0.00034 | 0.00229 | 65 | 0.01633 | 0.00028 | 0.00174 | 93 | 0.00288 | 0.30362 | 129.219 | 0.76191 | 0.93813 |
| K06926; uncharacterized protein | 0.01669 | 0.00267 | 0.02199 | 0.00199 | 0.00553 | 65 | 0.01299 | 0.00051 | 0.00233 | 93 | 0.00600 | 1.50043 | 86.846 | 0.13713 | 0.77376 |
| cysQ, MET22, BPNT1; 3’(2’), 5’-bisphosphate nucleotidase [EC:3.1.3.7] | 0.01668 | 0.00112 | 0.01566 | 0.00020 | 0.00174 | 65 | 0.01739 | 0.00020 | 0.00147 | 93 | 0.00228 | -0.75813 | 139.119 | 0.44965 | 0.83438 |
| hppA; K(+)-stimulated pyrophosphate-energized sodium pump [EC:3.6.1.1] | 0.01667 | 0.00124 | 0.01636 | 0.00028 | 0.00209 | 65 | 0.01689 | 0.00021 | 0.00152 | 93 | 0.00258 | -0.20722 | 125.077 | 0.83617 | 0.95506 |
| cobD; threonine-phosphate decarboxylase [EC:4.1.1.81] | 0.01667 | 0.00146 | 0.01949 | 0.00044 | 0.00261 | 65 | 0.01469 | 0.00025 | 0.00165 | 93 | 0.00309 | 1.55205 | 113.108 | 0.12344 | 0.77376 |
| gmd, GMDS; GDPmannose 4,6-dehydratase [EC:4.2.1.47] | 0.01666 | 0.00117 | 0.01395 | 0.00013 | 0.00140 | 65 | 0.01855 | 0.00027 | 0.00172 | 93 | 0.00221 | -2.07754 | 155.905 | 0.03939 | 0.63577 |
| pmbA; PmbA protein | 0.01665 | 0.00119 | 0.01615 | 0.00023 | 0.00189 | 65 | 0.01700 | 0.00022 | 0.00153 | 93 | 0.00243 | -0.34891 | 135.293 | 0.72770 | 0.92341 |
| pdhR; GntR family transcriptional regulator, transcriptional repressor for pyruvate dehydrogenase complex | 0.01664 | 0.00165 | 0.01583 | 0.00039 | 0.00243 | 65 | 0.01720 | 0.00046 | 0.00223 | 93 | 0.00330 | -0.41514 | 145.202 | 0.67865 | 0.91311 |
| lolA; outer membrane lipoprotein carrier protein | 0.01664 | 0.00116 | 0.01670 | 0.00021 | 0.00179 | 65 | 0.01659 | 0.00022 | 0.00153 | 93 | 0.00236 | 0.04961 | 139.651 | 0.96050 | 0.98903 |
| ybdG, mscM; miniconductance mechanosensitive channel | 0.01661 | 0.00116 | 0.01539 | 0.00017 | 0.00162 | 65 | 0.01746 | 0.00024 | 0.00162 | 93 | 0.00229 | -0.90475 | 151.226 | 0.36704 | 0.79587 |
| fliI; flagellum-specific ATP synthase [EC:3.6.3.14] | 0.01655 | 0.00118 | 0.01457 | 0.00023 | 0.00187 | 65 | 0.01792 | 0.00021 | 0.00151 | 93 | 0.00240 | -1.39436 | 135.063 | 0.16550 | 0.77376 |
| terC; tellurite resistance protein TerC | 0.01653 | 0.00118 | 0.01630 | 0.00024 | 0.00190 | 65 | 0.01669 | 0.00021 | 0.00151 | 93 | 0.00243 | -0.16127 | 133.418 | 0.87212 | 0.96576 |
| anmK; anhydro-N-acetylmuramic acid kinase [EC:2.7.1.170] | 0.01652 | 0.00118 | 0.01600 | 0.00024 | 0.00191 | 65 | 0.01689 | 0.00021 | 0.00151 | 93 | 0.00243 | -0.36962 | 132.939 | 0.71226 | 0.92004 |
| rlmE, rrmJ, ftsJ; 23S rRNA (uridine2552-2’-O)-methyltransferase [EC:2.1.1.166] | 0.01652 | 0.00116 | 0.01601 | 0.00023 | 0.00186 | 65 | 0.01688 | 0.00021 | 0.00149 | 93 | 0.00238 | -0.36277 | 133.861 | 0.71735 | 0.92035 |
| tolB; TolB protein | 0.01652 | 0.00119 | 0.01576 | 0.00023 | 0.00187 | 65 | 0.01704 | 0.00022 | 0.00154 | 93 | 0.00242 | -0.52763 | 136.959 | 0.59861 | 0.88240 |
| yvaK; carboxylesterase [EC:3.1.1.1] | 0.01649 | 0.00163 | 0.01589 | 0.00041 | 0.00251 | 65 | 0.01691 | 0.00043 | 0.00215 | 93 | 0.00331 | -0.30740 | 139.944 | 0.75900 | 0.93719 |
| gmhD, rfaD; ADP-L-glycero-D-manno-heptose 6-epimerase [EC:5.1.3.20] | 0.01645 | 0.00125 | 0.01919 | 0.00031 | 0.00218 | 65 | 0.01453 | 0.00020 | 0.00145 | 93 | 0.00262 | 1.77661 | 117.082 | 0.07823 | 0.75550 |
| lptE, rlpB; LPS-assembly lipoprotein | 0.01640 | 0.00117 | 0.01599 | 0.00023 | 0.00187 | 65 | 0.01669 | 0.00021 | 0.00150 | 93 | 0.00240 | -0.29219 | 134.305 | 0.77059 | 0.94017 |
| ssuB; sulfonate transport system ATP-binding protein [EC:3.6.3.-] | 0.01638 | 0.00138 | 0.01548 | 0.00026 | 0.00202 | 65 | 0.01701 | 0.00033 | 0.00188 | 93 | 0.00276 | -0.55581 | 146.527 | 0.57919 | 0.87546 |
| mlaC; phospholipid transport system substrate-binding protein | 0.01637 | 0.00123 | 0.01700 | 0.00027 | 0.00202 | 65 | 0.01594 | 0.00022 | 0.00154 | 93 | 0.00254 | 0.41797 | 129.256 | 0.67666 | 0.91266 |
| emrE, qac, mmr, smr; small multidrug resistance pump | 0.01636 | 0.00121 | 0.01564 | 0.00023 | 0.00186 | 65 | 0.01687 | 0.00024 | 0.00159 | 93 | 0.00245 | -0.49795 | 139.983 | 0.61930 | 0.88981 |
| ldcA; muramoyltetrapeptide carboxypeptidase [EC:3.4.17.13] | 0.01636 | 0.00108 | 0.01686 | 0.00017 | 0.00163 | 65 | 0.01601 | 0.00019 | 0.00144 | 93 | 0.00218 | 0.38952 | 142.556 | 0.69747 | 0.91825 |
| prlC; oligopeptidase A [EC:3.4.24.70] | 0.01634 | 0.00127 | 0.01655 | 0.00026 | 0.00199 | 65 | 0.01619 | 0.00025 | 0.00165 | 93 | 0.00259 | 0.14004 | 137.256 | 0.88884 | 0.97039 |
| ssuC; sulfonate transport system permease protein | 0.01632 | 0.00151 | 0.01538 | 0.00034 | 0.00228 | 65 | 0.01697 | 0.00038 | 0.00201 | 93 | 0.00304 | -0.52570 | 142.215 | 0.59991 | 0.88240 |
| pepE; dipeptidase E [EC:3.4.13.21] | 0.01632 | 0.00127 | 0.01824 | 0.00034 | 0.00228 | 65 | 0.01497 | 0.00020 | 0.00146 | 93 | 0.00270 | 1.20758 | 113.851 | 0.22971 | 0.77376 |
| holC; DNA polymerase III subunit chi [EC:2.7.7.7] | 0.01631 | 0.00116 | 0.01588 | 0.00023 | 0.00186 | 65 | 0.01661 | 0.00021 | 0.00149 | 93 | 0.00239 | -0.30565 | 133.828 | 0.76035 | 0.93791 |
| cptB; antitoxin CptB | 0.01629 | 0.00116 | 0.01598 | 0.00023 | 0.00186 | 65 | 0.01651 | 0.00021 | 0.00149 | 93 | 0.00239 | -0.22480 | 133.895 | 0.82248 | 0.95506 |
| betB, gbsA; betaine-aldehyde dehydrogenase [EC:1.2.1.8] | 0.01629 | 0.00148 | 0.01442 | 0.00030 | 0.00215 | 65 | 0.01760 | 0.00037 | 0.00201 | 93 | 0.00294 | -1.08128 | 146.663 | 0.28135 | 0.77376 |
| ATPVF, ntpF, atpF; V/A-type H+/Na+-transporting ATPase subunit F | 0.01629 | 0.00134 | 0.01994 | 0.00041 | 0.00250 | 65 | 0.01374 | 0.00019 | 0.00141 | 93 | 0.00287 | 2.16174 | 104.014 | 0.03293 | 0.62130 |
| dnaC; DNA replication protein DnaC | 0.01629 | 0.00173 | 0.01746 | 0.00057 | 0.00297 | 65 | 0.01547 | 0.00041 | 0.00210 | 93 | 0.00364 | 0.54762 | 122.446 | 0.58495 | 0.87720 |
| tctE; two-component system, OmpR family, sensor histidine kinase TctE [EC:2.7.13.3] | 0.01628 | 0.00223 | 0.01505 | 0.00072 | 0.00334 | 65 | 0.01714 | 0.00083 | 0.00299 | 93 | 0.00448 | -0.46689 | 143.673 | 0.64129 | 0.90011 |
| bolA; BolA family transcriptional regulator, general stress-responsive regulator | 0.01628 | 0.00116 | 0.01600 | 0.00023 | 0.00187 | 65 | 0.01648 | 0.00021 | 0.00149 | 93 | 0.00239 | -0.20019 | 133.881 | 0.84163 | 0.95506 |
| fliE; flagellar hook-basal body complex protein FliE | 0.01628 | 0.00119 | 0.01411 | 0.00023 | 0.00187 | 65 | 0.01779 | 0.00022 | 0.00154 | 93 | 0.00242 | -1.51807 | 136.170 | 0.13131 | 0.77376 |
| moaB; molybdopterin adenylyltransferase [EC:2.7.7.75] | 0.01627 | 0.00111 | 0.01596 | 0.00022 | 0.00182 | 65 | 0.01649 | 0.00018 | 0.00139 | 93 | 0.00229 | -0.22766 | 129.933 | 0.82027 | 0.95506 |
| pgpA; phosphatidylglycerophosphatase A [EC:3.1.3.27] | 0.01626 | 0.00120 | 0.01864 | 0.00027 | 0.00202 | 65 | 0.01460 | 0.00020 | 0.00146 | 93 | 0.00249 | 1.62508 | 124.336 | 0.10668 | 0.77376 |
| cobA-hemD; uroporphyrinogen III methyltransferase / synthase [EC:2.1.1.107 4.2.1.75] | 0.01625 | 0.00133 | 0.01906 | 0.00037 | 0.00238 | 65 | 0.01429 | 0.00021 | 0.00150 | 93 | 0.00282 | 1.69230 | 112.695 | 0.09335 | 0.76567 |
| gltP, gltT; proton glutamate symport protein | 0.01624 | 0.00131 | 0.01462 | 0.00021 | 0.00181 | 65 | 0.01737 | 0.00031 | 0.00183 | 93 | 0.00258 | -1.06885 | 151.758 | 0.28683 | 0.77376 |
| ttcA; tRNA 2-thiocytidine biosynthesis protein TtcA | 0.01622 | 0.00122 | 0.01554 | 0.00021 | 0.00181 | 65 | 0.01670 | 0.00026 | 0.00166 | 93 | 0.00245 | -0.47449 | 145.355 | 0.63586 | 0.89696 |
| fdoI, fdsG; formate dehydrogenase subunit gamma | 0.01622 | 0.00126 | 0.01757 | 0.00031 | 0.00220 | 65 | 0.01528 | 0.00020 | 0.00148 | 93 | 0.00265 | 0.86390 | 118.513 | 0.38939 | 0.80347 |
| saeS; two-component system, OmpR family, sensor histidine kinase SaeS [EC:2.7.13.3] | 0.01622 | 0.00140 | 0.01488 | 0.00024 | 0.00193 | 65 | 0.01715 | 0.00036 | 0.00197 | 93 | 0.00276 | -0.82529 | 152.104 | 0.41050 | 0.81416 |
| K07118; uncharacterized protein | 0.01621 | 0.00137 | 0.01746 | 0.00040 | 0.00249 | 65 | 0.01533 | 0.00022 | 0.00155 | 93 | 0.00293 | 0.72503 | 111.548 | 0.46995 | 0.83870 |
| K10907; aminotransferase [EC:2.6.1.-] | 0.01621 | 0.00159 | 0.01719 | 0.00047 | 0.00269 | 65 | 0.01552 | 0.00035 | 0.00194 | 93 | 0.00332 | 0.50067 | 124.558 | 0.61748 | 0.88903 |
| lip, TGL2; triacylglycerol lipase [EC:3.1.1.3] | 0.01620 | 0.00161 | 0.01557 | 0.00037 | 0.00238 | 65 | 0.01663 | 0.00044 | 0.00218 | 93 | 0.00322 | -0.32921 | 145.333 | 0.74247 | 0.93050 |
| pccA; periplasmic copper chaperone A | 0.01618 | 0.00154 | 0.01497 | 0.00034 | 0.00230 | 65 | 0.01702 | 0.00040 | 0.00207 | 93 | 0.00309 | -0.66475 | 143.737 | 0.50727 | 0.84956 |
| rbsD; D-ribose pyranase [EC:5.4.99.62] | 0.01615 | 0.00133 | 0.01788 | 0.00038 | 0.00243 | 65 | 0.01494 | 0.00021 | 0.00150 | 93 | 0.00286 | 1.03049 | 110.975 | 0.30502 | 0.77376 |
| ccmG, dsbE; cytochrome c biogenesis protein CcmG, thiol:disulfide interchange protein DsbE | 0.01614 | 0.00136 | 0.01603 | 0.00032 | 0.00222 | 65 | 0.01622 | 0.00028 | 0.00173 | 93 | 0.00281 | -0.06616 | 131.356 | 0.94735 | 0.98674 |
| fctD; glutamate formiminotransferase [EC:2.1.2.5] | 0.01611 | 0.00159 | 0.01954 | 0.00068 | 0.00324 | 65 | 0.01372 | 0.00019 | 0.00143 | 93 | 0.00354 | 1.64352 | 89.027 | 0.10380 | 0.77376 |
| mmuM, BHMT2; homocysteine S-methyltransferase [EC:2.1.1.10] | 0.01611 | 0.00117 | 0.01524 | 0.00022 | 0.00185 | 65 | 0.01672 | 0.00021 | 0.00151 | 93 | 0.00239 | -0.61688 | 135.707 | 0.53835 | 0.86207 |
| recB; exodeoxyribonuclease V beta subunit [EC:3.1.11.5] | 0.01609 | 0.00123 | 0.01668 | 0.00024 | 0.00194 | 65 | 0.01568 | 0.00024 | 0.00159 | 93 | 0.00251 | 0.39617 | 136.334 | 0.69260 | 0.91677 |
| K06956; uncharacterized protein | 0.01609 | 0.00102 | 0.01679 | 0.00019 | 0.00170 | 65 | 0.01560 | 0.00015 | 0.00126 | 93 | 0.00212 | 0.55969 | 126.629 | 0.57668 | 0.87389 |
| LYS5, acpT; 4’-phosphopantetheinyl transferase [EC:2.7.8.-] | 0.01604 | 0.00122 | 0.01604 | 0.00023 | 0.00189 | 65 | 0.01604 | 0.00024 | 0.00161 | 93 | 0.00249 | 0.00201 | 139.706 | 0.99840 | 0.99957 |
| hyi, gip; hydroxypyruvate isomerase [EC:5.3.1.22] | 0.01604 | 0.00160 | 0.01358 | 0.00039 | 0.00243 | 65 | 0.01775 | 0.00042 | 0.00211 | 93 | 0.00322 | -1.29396 | 141.121 | 0.19779 | 0.77376 |
| rnfC; electron transport complex protein RnfC | 0.01603 | 0.00150 | 0.01946 | 0.00044 | 0.00261 | 65 | 0.01364 | 0.00028 | 0.00175 | 93 | 0.00314 | 1.85463 | 117.848 | 0.06615 | 0.73334 |
| hflK; membrane protease subunit HflK [EC:3.4.-.-] | 0.01602 | 0.00132 | 0.01508 | 0.00027 | 0.00204 | 65 | 0.01668 | 0.00028 | 0.00173 | 93 | 0.00268 | -0.59863 | 139.280 | 0.55039 | 0.86721 |
| flgB; flagellar basal-body rod protein FlgB | 0.01601 | 0.00117 | 0.01426 | 0.00024 | 0.00190 | 65 | 0.01723 | 0.00020 | 0.00147 | 93 | 0.00241 | -1.23560 | 130.854 | 0.21882 | 0.77376 |
| phrB; deoxyribodipyrimidine photo-lyase [EC:4.1.99.3] | 0.01601 | 0.00134 | 0.01458 | 0.00027 | 0.00204 | 65 | 0.01700 | 0.00029 | 0.00177 | 93 | 0.00270 | -0.89333 | 140.875 | 0.37321 | 0.79810 |
| coxB, ctaC; cytochrome c oxidase subunit II [EC:1.9.3.1] | 0.01600 | 0.00151 | 0.01173 | 0.00026 | 0.00198 | 65 | 0.01899 | 0.00041 | 0.00211 | 93 | 0.00290 | -2.50829 | 153.763 | 0.01317 | 0.59095 |
| sfsA; sugar fermentation stimulation protein A | 0.01600 | 0.00100 | 0.01736 | 0.00017 | 0.00162 | 65 | 0.01505 | 0.00015 | 0.00127 | 93 | 0.00206 | 1.11810 | 132.001 | 0.26555 | 0.77376 |
| E3.5.3.1, rocF, arg; arginase [EC:3.5.3.1] | 0.01600 | 0.00136 | 0.01373 | 0.00031 | 0.00218 | 65 | 0.01759 | 0.00028 | 0.00173 | 93 | 0.00279 | -1.38317 | 132.973 | 0.16893 | 0.77376 |
| IMA, malL; oligo-1,6-glucosidase [EC:3.2.1.10] | 0.01600 | 0.00178 | 0.01741 | 0.00066 | 0.00319 | 65 | 0.01501 | 0.00039 | 0.00205 | 93 | 0.00379 | 0.63487 | 114.316 | 0.52678 | 0.85844 |
| oadA; oxaloacetate decarboxylase, alpha subunit [EC:4.1.1.3] | 0.01598 | 0.00140 | 0.01887 | 0.00042 | 0.00255 | 65 | 0.01395 | 0.00022 | 0.00155 | 93 | 0.00298 | 1.64830 | 109.743 | 0.10215 | 0.77376 |
| speA; arginine decarboxylase [EC:4.1.1.19] | 0.01592 | 0.00103 | 0.01533 | 0.00015 | 0.00150 | 65 | 0.01634 | 0.00019 | 0.00141 | 93 | 0.00206 | -0.49133 | 147.409 | 0.62393 | 0.89160 |
| whiB1_2_3_4; WhiB family transcriptional regulator, redox-sensing transcriptional regulator | 0.01592 | 0.00214 | 0.01516 | 0.00046 | 0.00267 | 65 | 0.01645 | 0.00091 | 0.00313 | 93 | 0.00411 | -0.31253 | 155.934 | 0.75506 | 0.93665 |
| ATPVC, ntpC, atpC; V/A-type H+/Na+-transporting ATPase subunit C | 0.01591 | 0.00132 | 0.01971 | 0.00040 | 0.00248 | 65 | 0.01325 | 0.00017 | 0.00136 | 93 | 0.00283 | 2.28566 | 101.588 | 0.02435 | 0.59764 |
| pydC; beta-ureidopropionase / N-carbamoyl-L-amino-acid hydrolase [EC:3.5.1.6 3.5.1.87] | 0.01590 | 0.00131 | 0.01404 | 0.00020 | 0.00177 | 65 | 0.01720 | 0.00032 | 0.00184 | 93 | 0.00256 | -1.23451 | 152.892 | 0.21891 | 0.77376 |
| proP; MFS transporter, MHS family, proline/betaine transporter | 0.01590 | 0.00182 | 0.01455 | 0.00040 | 0.00249 | 65 | 0.01684 | 0.00061 | 0.00256 | 93 | 0.00357 | -0.63961 | 152.310 | 0.52338 | 0.85708 |
| MET8; precorrin-2 dehydrogenase / sirohydrochlorin ferrochelatase [EC:1.3.1.76 4.99.1.4] | 0.01586 | 0.00131 | 0.01794 | 0.00039 | 0.00245 | 65 | 0.01440 | 0.00018 | 0.00140 | 93 | 0.00283 | 1.25230 | 104.904 | 0.21325 | 0.77376 |
| K07485; transposase | 0.01576 | 0.00232 | 0.02118 | 0.00153 | 0.00485 | 65 | 0.01198 | 0.00036 | 0.00196 | 93 | 0.00523 | 1.75929 | 84.935 | 0.08213 | 0.76567 |
| hns; DNA-binding protein H-NS | 0.01576 | 0.00189 | 0.01478 | 0.00057 | 0.00296 | 65 | 0.01644 | 0.00057 | 0.00247 | 93 | 0.00386 | -0.43069 | 137.735 | 0.66737 | 0.91150 |
| K07037; uncharacterized protein | 0.01575 | 0.00145 | 0.01693 | 0.00042 | 0.00254 | 65 | 0.01493 | 0.00027 | 0.00171 | 93 | 0.00307 | 0.64988 | 118.356 | 0.51703 | 0.85220 |
| ompW; outer membrane protein | 0.01574 | 0.00161 | 0.01471 | 0.00041 | 0.00251 | 65 | 0.01645 | 0.00042 | 0.00211 | 93 | 0.00328 | -0.53053 | 138.657 | 0.59659 | 0.88059 |
| fliG; flagellar motor switch protein FliG | 0.01574 | 0.00114 | 0.01397 | 0.00022 | 0.00185 | 65 | 0.01697 | 0.00019 | 0.00143 | 93 | 0.00234 | -1.28229 | 130.915 | 0.20201 | 0.77376 |
| yabN; tetrapyrrole methylase family protein / MazG family protein | 0.01573 | 0.00111 | 0.01468 | 0.00022 | 0.00182 | 65 | 0.01647 | 0.00018 | 0.00140 | 93 | 0.00230 | -0.77891 | 130.053 | 0.43745 | 0.83071 |
| K07023; putative hydrolases of HD superfamily | 0.01571 | 0.00132 | 0.01425 | 0.00032 | 0.00222 | 65 | 0.01673 | 0.00025 | 0.00162 | 93 | 0.00275 | -0.90583 | 125.833 | 0.36676 | 0.79587 |
| fliA; RNA polymerase sigma factor for flagellar operon FliA | 0.01570 | 0.00120 | 0.01477 | 0.00023 | 0.00188 | 65 | 0.01636 | 0.00023 | 0.00157 | 93 | 0.00244 | -0.64991 | 137.622 | 0.51683 | 0.85216 |
| E2.4.1.5; dextransucrase [EC:2.4.1.5] | 0.01570 | 0.00265 | 0.01586 | 0.00068 | 0.00324 | 65 | 0.01559 | 0.00142 | 0.00391 | 93 | 0.00507 | 0.05281 | 155.995 | 0.95795 | 0.98896 |
| coxA, ctaD; cytochrome c oxidase subunit I [EC:1.9.3.1] | 0.01570 | 0.00156 | 0.01156 | 0.00025 | 0.00196 | 65 | 0.01858 | 0.00046 | 0.00223 | 93 | 0.00296 | -2.36781 | 155.599 | 0.01912 | 0.59095 |
| phnC; phosphonate transport system ATP-binding protein [EC:3.6.3.28] | 0.01569 | 0.00129 | 0.01716 | 0.00032 | 0.00222 | 65 | 0.01466 | 0.00022 | 0.00154 | 93 | 0.00270 | 0.92582 | 120.959 | 0.35638 | 0.79399 |
| K09963; uncharacterized protein | 0.01567 | 0.00180 | 0.01775 | 0.00080 | 0.00350 | 65 | 0.01423 | 0.00031 | 0.00183 | 93 | 0.00395 | 0.89093 | 98.692 | 0.37514 | 0.79810 |
| fliF; flagellar M-ring protein FliF | 0.01567 | 0.00115 | 0.01414 | 0.00023 | 0.00189 | 65 | 0.01674 | 0.00019 | 0.00144 | 93 | 0.00238 | -1.08968 | 129.619 | 0.27787 | 0.77376 |
| clpA; ATP-dependent Clp protease ATP-binding subunit ClpA | 0.01567 | 0.00121 | 0.01412 | 0.00022 | 0.00186 | 65 | 0.01675 | 0.00024 | 0.00160 | 93 | 0.00245 | -1.07109 | 140.149 | 0.28597 | 0.77376 |
| flgD; flagellar basal-body rod modification protein FlgD | 0.01564 | 0.00116 | 0.01397 | 0.00023 | 0.00189 | 65 | 0.01680 | 0.00020 | 0.00146 | 93 | 0.00239 | -1.18458 | 130.812 | 0.23833 | 0.77376 |
| ftsB; cell division protein FtsB | 0.01563 | 0.00111 | 0.01678 | 0.00020 | 0.00176 | 65 | 0.01483 | 0.00019 | 0.00142 | 93 | 0.00226 | 0.86530 | 134.986 | 0.38841 | 0.80262 |
| glnG, ntrC; two-component system, NtrC family, nitrogen regulation response regulator GlnG | 0.01560 | 0.00130 | 0.01652 | 0.00032 | 0.00221 | 65 | 0.01496 | 0.00023 | 0.00158 | 93 | 0.00272 | 0.57130 | 123.588 | 0.56884 | 0.87135 |
| saeR; two-component system, OmpR family, response regulator SaeR | 0.01558 | 0.00137 | 0.01475 | 0.00024 | 0.00191 | 65 | 0.01616 | 0.00034 | 0.00192 | 93 | 0.00271 | -0.52061 | 151.083 | 0.60340 | 0.88434 |
| fliM; flagellar motor switch protein FliM | 0.01557 | 0.00113 | 0.01381 | 0.00022 | 0.00185 | 65 | 0.01680 | 0.00019 | 0.00142 | 93 | 0.00233 | -1.27925 | 130.395 | 0.20308 | 0.77376 |
| pilU; twitching motility protein PilU | 0.01555 | 0.00178 | 0.01538 | 0.00044 | 0.00260 | 65 | 0.01567 | 0.00055 | 0.00243 | 93 | 0.00356 | -0.08260 | 146.719 | 0.93428 | 0.98428 |
| mgsA; methylglyoxal synthase [EC:4.2.3.3] | 0.01554 | 0.00147 | 0.01693 | 0.00052 | 0.00284 | 65 | 0.01457 | 0.00022 | 0.00153 | 93 | 0.00323 | 0.73110 | 100.490 | 0.46642 | 0.83837 |
| thiT; thiamine transporter | 0.01553 | 0.00118 | 0.01668 | 0.00027 | 0.00205 | 65 | 0.01473 | 0.00018 | 0.00140 | 93 | 0.00248 | 0.78137 | 119.391 | 0.43613 | 0.83006 |
| acrR, smeT; TetR/AcrR family transcriptional regulator, acrAB operon repressor | 0.01552 | 0.00093 | 0.01550 | 0.00016 | 0.00155 | 65 | 0.01554 | 0.00013 | 0.00116 | 93 | 0.00194 | -0.02276 | 127.943 | 0.98188 | 0.99680 |
| K06938; uncharacterized protein | 0.01552 | 0.00168 | 0.01490 | 0.00043 | 0.00257 | 65 | 0.01595 | 0.00046 | 0.00223 | 93 | 0.00340 | -0.30976 | 141.043 | 0.75720 | 0.93719 |
| mod; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | 0.01551 | 0.00230 | 0.01393 | 0.00021 | 0.00179 | 65 | 0.01661 | 0.00128 | 0.00370 | 93 | 0.00411 | -0.65041 | 129.976 | 0.51657 | 0.85216 |
| fucA; L-fuculose-phosphate aldolase [EC:4.1.2.17] | 0.01549 | 0.00127 | 0.01814 | 0.00039 | 0.00246 | 65 | 0.01363 | 0.00015 | 0.00128 | 93 | 0.00277 | 1.63066 | 98.261 | 0.10616 | 0.77376 |
| udp, UPP; uridine phosphorylase [EC:2.4.2.3] | 0.01546 | 0.00129 | 0.01532 | 0.00024 | 0.00193 | 65 | 0.01556 | 0.00028 | 0.00174 | 93 | 0.00260 | -0.09244 | 143.966 | 0.92648 | 0.98122 |
| rnfE; electron transport complex protein RnfE | 0.01545 | 0.00131 | 0.01840 | 0.00031 | 0.00219 | 65 | 0.01339 | 0.00024 | 0.00160 | 93 | 0.00271 | 1.84744 | 125.733 | 0.06703 | 0.73334 |
| rnfA; electron transport complex protein RnfA | 0.01544 | 0.00131 | 0.01838 | 0.00031 | 0.00219 | 65 | 0.01338 | 0.00024 | 0.00160 | 93 | 0.00271 | 1.84511 | 125.785 | 0.06737 | 0.73334 |
| rlmJ; 23S rRNA (adenine2030-N6)-methyltransferase [EC:2.1.1.266] | 0.01543 | 0.00113 | 0.01518 | 0.00022 | 0.00183 | 65 | 0.01561 | 0.00019 | 0.00144 | 93 | 0.00233 | -0.18394 | 132.220 | 0.85434 | 0.95858 |
| mrr; restriction system protein | 0.01543 | 0.00118 | 0.01521 | 0.00028 | 0.00207 | 65 | 0.01557 | 0.00018 | 0.00139 | 93 | 0.00249 | -0.14410 | 117.777 | 0.88567 | 0.97002 |
| E4.1.3.1, aceA; isocitrate lyase [EC:4.1.3.1] | 0.01542 | 0.00127 | 0.01303 | 0.00024 | 0.00192 | 65 | 0.01708 | 0.00026 | 0.00168 | 93 | 0.00255 | -1.59083 | 141.800 | 0.11388 | 0.77376 |
| K06894; uncharacterized protein | 0.01541 | 0.00146 | 0.01652 | 0.00035 | 0.00231 | 65 | 0.01464 | 0.00033 | 0.00188 | 93 | 0.00298 | 0.63031 | 135.585 | 0.52955 | 0.86094 |
| iciA; LysR family transcriptional regulator, chromosome initiation inhibitor | 0.01541 | 0.00150 | 0.01480 | 0.00037 | 0.00237 | 65 | 0.01583 | 0.00035 | 0.00195 | 93 | 0.00307 | -0.33438 | 136.164 | 0.73860 | 0.92914 |
| lolB; outer membrane lipoprotein LolB | 0.01540 | 0.00122 | 0.01600 | 0.00024 | 0.00192 | 65 | 0.01498 | 0.00023 | 0.00158 | 93 | 0.00249 | 0.41045 | 136.481 | 0.68212 | 0.91478 |
| K09769; uncharacterized protein | 0.01540 | 0.00133 | 0.01632 | 0.00041 | 0.00252 | 65 | 0.01475 | 0.00019 | 0.00141 | 93 | 0.00289 | 0.54514 | 103.432 | 0.58683 | 0.87731 |
| aguA; agmatine deiminase [EC:3.5.3.12] | 0.01537 | 0.00101 | 0.01424 | 0.00016 | 0.00158 | 65 | 0.01616 | 0.00016 | 0.00132 | 93 | 0.00206 | -0.93138 | 138.046 | 0.35328 | 0.79206 |
| hflC; membrane protease subunit HflC [EC:3.4.-.-] | 0.01536 | 0.00118 | 0.01421 | 0.00021 | 0.00181 | 65 | 0.01617 | 0.00023 | 0.00156 | 93 | 0.00239 | -0.81958 | 140.283 | 0.41385 | 0.81663 |
| E2.1.1.131, cobJ, cbiH; precorrin-3B C17-methyltransferase [EC:2.1.1.131] | 0.01534 | 0.00142 | 0.01810 | 0.00044 | 0.00259 | 65 | 0.01341 | 0.00023 | 0.00158 | 93 | 0.00303 | 1.54743 | 110.021 | 0.12463 | 0.77376 |
| hcaT; MFS transporter, PPP family, 3-phenylpropionic acid transporter | 0.01534 | 0.00107 | 0.01552 | 0.00020 | 0.00174 | 65 | 0.01520 | 0.00017 | 0.00136 | 93 | 0.00220 | 0.14589 | 131.854 | 0.88423 | 0.96974 |
| E4.3.1.4; formiminotetrahydrofolate cyclodeaminase [EC:4.3.1.4] | 0.01532 | 0.00157 | 0.01876 | 0.00067 | 0.00321 | 65 | 0.01291 | 0.00019 | 0.00141 | 93 | 0.00350 | 1.67004 | 88.876 | 0.09843 | 0.76942 |
| K06995; uncharacterized protein | 0.01530 | 0.00221 | 0.01517 | 0.00071 | 0.00331 | 65 | 0.01539 | 0.00082 | 0.00297 | 93 | 0.00445 | -0.05008 | 143.690 | 0.96012 | 0.98903 |
| K09158; uncharacterized protein | 0.01530 | 0.00115 | 0.01596 | 0.00022 | 0.00184 | 65 | 0.01484 | 0.00020 | 0.00147 | 93 | 0.00235 | 0.47721 | 133.604 | 0.63399 | 0.89696 |
| glbN; hemoglobin | 0.01529 | 0.00136 | 0.01129 | 0.00020 | 0.00176 | 65 | 0.01809 | 0.00034 | 0.00191 | 93 | 0.00259 | -2.62425 | 154.473 | 0.00956 | 0.58112 |
| asp2; accessory secretory protein Asp2 | 0.01526 | 0.00138 | 0.01397 | 0.00023 | 0.00187 | 65 | 0.01616 | 0.00035 | 0.00195 | 93 | 0.00271 | -0.80787 | 152.959 | 0.42042 | 0.82181 |
| ccoP; cytochrome c oxidase cbb3-type subunit III | 0.01525 | 0.00176 | 0.01488 | 0.00045 | 0.00263 | 65 | 0.01551 | 0.00052 | 0.00238 | 93 | 0.00355 | -0.17817 | 144.156 | 0.85884 | 0.96086 |
| asp1; accessory secretory protein Asp1 | 0.01525 | 0.00138 | 0.01395 | 0.00023 | 0.00188 | 65 | 0.01615 | 0.00035 | 0.00195 | 93 | 0.00271 | -0.81233 | 152.946 | 0.41786 | 0.81941 |
| pilE; type IV pilus assembly protein PilE | 0.01523 | 0.00142 | 0.01533 | 0.00031 | 0.00218 | 65 | 0.01516 | 0.00033 | 0.00188 | 93 | 0.00288 | 0.05908 | 140.464 | 0.95297 | 0.98792 |
| K07044; uncharacterized protein | 0.01521 | 0.00288 | 0.01149 | 0.00056 | 0.00293 | 65 | 0.01782 | 0.00184 | 0.00445 | 93 | 0.00532 | -1.18853 | 148.805 | 0.23652 | 0.77376 |
| E4.6.1.1; adenylate cyclase [EC:4.6.1.1] | 0.01520 | 0.00189 | 0.01104 | 0.00018 | 0.00165 | 65 | 0.01810 | 0.00082 | 0.00296 | 93 | 0.00339 | -2.08092 | 138.806 | 0.03928 | 0.63577 |
| fdoH, fdsB; formate dehydrogenase iron-sulfur subunit | 0.01519 | 0.00121 | 0.01621 | 0.00030 | 0.00213 | 65 | 0.01448 | 0.00019 | 0.00142 | 93 | 0.00256 | 0.67391 | 117.267 | 0.50170 | 0.84942 |
| gspL; general secretion pathway protein L | 0.01518 | 0.00176 | 0.01387 | 0.00043 | 0.00256 | 65 | 0.01610 | 0.00054 | 0.00241 | 93 | 0.00351 | -0.63491 | 147.314 | 0.52647 | 0.85844 |
| cggR; central glycolytic genes regulator | 0.01518 | 0.00118 | 0.01525 | 0.00028 | 0.00206 | 65 | 0.01513 | 0.00018 | 0.00140 | 93 | 0.00249 | 0.04896 | 119.335 | 0.96103 | 0.98943 |
| K07006; uncharacterized protein | 0.01518 | 0.00152 | 0.01440 | 0.00028 | 0.00209 | 65 | 0.01572 | 0.00043 | 0.00214 | 93 | 0.00299 | -0.44091 | 152.317 | 0.65990 | 0.90939 |
| slyX; SlyX protein | 0.01518 | 0.00113 | 0.01516 | 0.00021 | 0.00179 | 65 | 0.01519 | 0.00020 | 0.00147 | 93 | 0.00231 | -0.01506 | 136.111 | 0.98800 | 0.99801 |
| rutR; TetR/AcrR family transcriptional regulator | 0.01515 | 0.00206 | 0.01534 | 0.00071 | 0.00330 | 65 | 0.01502 | 0.00066 | 0.00266 | 93 | 0.00424 | 0.07402 | 134.520 | 0.94110 | 0.98528 |
| K07018; uncharacterized protein | 0.01515 | 0.00117 | 0.01462 | 0.00025 | 0.00194 | 65 | 0.01552 | 0.00020 | 0.00145 | 93 | 0.00242 | -0.37132 | 127.683 | 0.71101 | 0.92004 |
| ABC.CYST.A; cystine transport system ATP-binding protein [EC:3.6.3.-] | 0.01514 | 0.00113 | 0.01645 | 0.00023 | 0.00189 | 65 | 0.01422 | 0.00018 | 0.00140 | 93 | 0.00235 | 0.94990 | 127.169 | 0.34397 | 0.78632 |
| greB; transcription elongation factor GreB | 0.01512 | 0.00114 | 0.01551 | 0.00022 | 0.00184 | 65 | 0.01484 | 0.00020 | 0.00145 | 93 | 0.00234 | 0.28449 | 132.656 | 0.77648 | 0.94269 |
| E1.1.1.30, bdh; 3-hydroxybutyrate dehydrogenase [EC:1.1.1.30] | 0.01509 | 0.00148 | 0.01233 | 0.00030 | 0.00216 | 65 | 0.01702 | 0.00037 | 0.00199 | 93 | 0.00294 | -1.59411 | 145.624 | 0.11308 | 0.77376 |
| E5.1.3.15; glucose-6-phosphate 1-epimerase [EC:5.1.3.15] | 0.01509 | 0.00135 | 0.01608 | 0.00032 | 0.00221 | 65 | 0.01440 | 0.00027 | 0.00170 | 93 | 0.00279 | 0.60173 | 130.616 | 0.54839 | 0.86565 |
| ccoN; cytochrome c oxidase cbb3-type subunit I [EC:1.9.3.1] | 0.01509 | 0.00196 | 0.01455 | 0.00053 | 0.00285 | 65 | 0.01546 | 0.00066 | 0.00267 | 93 | 0.00390 | -0.23175 | 146.995 | 0.81705 | 0.95487 |
| ptrB; oligopeptidase B [EC:3.4.21.83] | 0.01506 | 0.00108 | 0.01489 | 0.00022 | 0.00183 | 65 | 0.01518 | 0.00016 | 0.00132 | 93 | 0.00226 | -0.13127 | 124.794 | 0.89577 | 0.97255 |
| kdpB; K+-transporting ATPase ATPase B chain [EC:3.6.3.12] | 0.01506 | 0.00134 | 0.01312 | 0.00027 | 0.00203 | 65 | 0.01641 | 0.00029 | 0.00177 | 93 | 0.00269 | -1.22298 | 141.750 | 0.22337 | 0.77376 |
| higB-1; toxin HigB-1 | 0.01506 | 0.00134 | 0.01494 | 0.00034 | 0.00229 | 65 | 0.01514 | 0.00025 | 0.00164 | 93 | 0.00281 | -0.07015 | 124.102 | 0.94418 | 0.98673 |
| asp3; accessory secretory protein Asp3 | 0.01504 | 0.00137 | 0.01345 | 0.00021 | 0.00180 | 65 | 0.01615 | 0.00035 | 0.00195 | 93 | 0.00266 | -1.01468 | 154.387 | 0.31185 | 0.77376 |
| fimT; type IV fimbrial biogenesis protein FimT | 0.01502 | 0.00153 | 0.01526 | 0.00040 | 0.00248 | 65 | 0.01486 | 0.00035 | 0.00195 | 93 | 0.00315 | 0.12733 | 132.359 | 0.89888 | 0.97408 |
| erpA; iron-sulfur cluster insertion protein | 0.01501 | 0.00112 | 0.01531 | 0.00021 | 0.00181 | 65 | 0.01481 | 0.00019 | 0.00144 | 93 | 0.00231 | 0.21875 | 133.512 | 0.82718 | 0.95506 |
| K09940; uncharacterized protein | 0.01500 | 0.00149 | 0.01481 | 0.00039 | 0.00244 | 65 | 0.01513 | 0.00033 | 0.00188 | 93 | 0.00308 | -0.10436 | 130.681 | 0.91704 | 0.97692 |
| nirB; nitrite reductase (NADH) large subunit [EC:1.7.1.15] | 0.01499 | 0.00126 | 0.01339 | 0.00023 | 0.00189 | 65 | 0.01612 | 0.00026 | 0.00168 | 93 | 0.00253 | -1.08155 | 142.760 | 0.28128 | 0.77376 |
| pabA; para-aminobenzoate synthetase component II [EC:2.6.1.85] | 0.01499 | 0.00119 | 0.01661 | 0.00029 | 0.00211 | 65 | 0.01385 | 0.00018 | 0.00139 | 93 | 0.00253 | 1.09068 | 116.143 | 0.27767 | 0.77376 |
| recC; exodeoxyribonuclease V gamma subunit [EC:3.1.11.5] | 0.01498 | 0.00119 | 0.01556 | 0.00022 | 0.00186 | 65 | 0.01457 | 0.00023 | 0.00156 | 93 | 0.00243 | 0.40936 | 138.111 | 0.68291 | 0.91495 |
| benE; benzoate membrane transport protein | 0.01498 | 0.00147 | 0.01390 | 0.00030 | 0.00216 | 65 | 0.01572 | 0.00037 | 0.00200 | 93 | 0.00294 | -0.61823 | 146.086 | 0.53738 | 0.86207 |
| cysH; phosphoadenosine phosphosulfate reductase [EC:1.8.4.8 1.8.4.10] | 0.01497 | 0.00109 | 0.01261 | 0.00016 | 0.00156 | 65 | 0.01663 | 0.00020 | 0.00148 | 93 | 0.00215 | -1.87281 | 147.655 | 0.06307 | 0.73016 |
| thiO; glycine oxidase [EC:1.4.3.19] | 0.01497 | 0.00121 | 0.01340 | 0.00020 | 0.00177 | 65 | 0.01607 | 0.00025 | 0.00164 | 93 | 0.00241 | -1.10788 | 146.223 | 0.26973 | 0.77376 |
| aat; leucyl/phenylalanyl-tRNA—protein transferase [EC:2.3.2.6] | 0.01496 | 0.00110 | 0.01395 | 0.00020 | 0.00175 | 65 | 0.01567 | 0.00018 | 0.00141 | 93 | 0.00224 | -0.76811 | 134.771 | 0.44377 | 0.83186 |
| E3.1.3.1, phoA, phoB; alkaline phosphatase [EC:3.1.3.1] | 0.01496 | 0.00138 | 0.01151 | 0.00024 | 0.00191 | 65 | 0.01737 | 0.00033 | 0.00189 | 93 | 0.00269 | -2.18083 | 150.152 | 0.03075 | 0.61611 |
| prpF; 2-methylaconitate isomerase [EC:5.3.3.-] | 0.01495 | 0.00157 | 0.01462 | 0.00041 | 0.00250 | 65 | 0.01519 | 0.00038 | 0.00202 | 93 | 0.00321 | -0.17542 | 135.139 | 0.86101 | 0.96180 |
| K06973; uncharacterized protein | 0.01495 | 0.00117 | 0.01446 | 0.00026 | 0.00200 | 65 | 0.01529 | 0.00019 | 0.00143 | 93 | 0.00246 | -0.33765 | 123.648 | 0.73620 | 0.92721 |
| fliD; flagellar hook-associated protein 2 | 0.01493 | 0.00124 | 0.01456 | 0.00028 | 0.00208 | 65 | 0.01518 | 0.00022 | 0.00153 | 93 | 0.00259 | -0.24223 | 126.323 | 0.80900 | 0.95002 |
| TC.CIC; chloride channel protein, CIC family | 0.01493 | 0.00101 | 0.01364 | 0.00013 | 0.00143 | 65 | 0.01583 | 0.00018 | 0.00140 | 93 | 0.00200 | -1.09158 | 149.773 | 0.27677 | 0.77376 |
| ubiH; 2-octaprenyl-6-methoxyphenol hydroxylase [EC:1.14.13.-] | 0.01492 | 0.00118 | 0.01389 | 0.00020 | 0.00175 | 65 | 0.01564 | 0.00023 | 0.00159 | 93 | 0.00236 | -0.74077 | 144.650 | 0.46004 | 0.83438 |
| prmB; ribosomal protein L3 glutamine methyltransferase [EC:2.1.1.298] | 0.01491 | 0.00113 | 0.01537 | 0.00021 | 0.00182 | 65 | 0.01459 | 0.00019 | 0.00145 | 93 | 0.00232 | 0.33316 | 133.615 | 0.73954 | 0.92969 |
| K07032; uncharacterized protein | 0.01490 | 0.00142 | 0.01460 | 0.00034 | 0.00228 | 65 | 0.01510 | 0.00031 | 0.00182 | 93 | 0.00292 | -0.16907 | 133.889 | 0.86600 | 0.96292 |
| phoD; alkaline phosphatase D [EC:3.1.3.1] | 0.01488 | 0.00152 | 0.01324 | 0.00036 | 0.00236 | 65 | 0.01603 | 0.00037 | 0.00198 | 93 | 0.00309 | -0.90119 | 138.230 | 0.36905 | 0.79587 |
| puuB, ordL; gamma-glutamylputrescine oxidase [EC:1.4.3.-] | 0.01487 | 0.00150 | 0.01461 | 0.00030 | 0.00217 | 65 | 0.01506 | 0.00039 | 0.00206 | 93 | 0.00299 | -0.14947 | 147.781 | 0.88139 | 0.96901 |
| fliS; flagellar protein FliS | 0.01487 | 0.00116 | 0.01341 | 0.00021 | 0.00180 | 65 | 0.01589 | 0.00022 | 0.00152 | 93 | 0.00235 | -1.05494 | 139.026 | 0.29328 | 0.77376 |
| sspB; stringent starvation protein B | 0.01486 | 0.00114 | 0.01537 | 0.00021 | 0.00181 | 65 | 0.01450 | 0.00020 | 0.00147 | 93 | 0.00234 | 0.37460 | 135.347 | 0.70855 | 0.92004 |
| K07223; putative iron-dependent peroxidase | 0.01485 | 0.00108 | 0.01541 | 0.00019 | 0.00172 | 65 | 0.01447 | 0.00018 | 0.00139 | 93 | 0.00221 | 0.42817 | 134.569 | 0.66921 | 0.91165 |
| pmm-pgm; phosphomannomutase / phosphoglucomutase [EC:5.4.2.8 5.4.2.2] | 0.01484 | 0.00145 | 0.01433 | 0.00036 | 0.00236 | 65 | 0.01519 | 0.00032 | 0.00185 | 93 | 0.00300 | -0.28491 | 132.100 | 0.77616 | 0.94269 |
| hemY; HemY protein | 0.01483 | 0.00118 | 0.01491 | 0.00023 | 0.00188 | 65 | 0.01478 | 0.00021 | 0.00151 | 93 | 0.00242 | 0.05420 | 134.418 | 0.95686 | 0.98854 |
| slo; thiol-activated cytolysin | 0.01483 | 0.00144 | 0.01328 | 0.00024 | 0.00194 | 65 | 0.01592 | 0.00039 | 0.00204 | 93 | 0.00282 | -0.93662 | 153.465 | 0.35043 | 0.79120 |
| mltA; membrane-bound lytic murein transglycosylase A [EC:4.2.2.-] | 0.01483 | 0.00124 | 0.01396 | 0.00024 | 0.00192 | 65 | 0.01544 | 0.00025 | 0.00164 | 93 | 0.00252 | -0.58356 | 139.558 | 0.56046 | 0.86813 |
| flgL; flagellar hook-associated protein 3 FlgL | 0.01480 | 0.00111 | 0.01355 | 0.00022 | 0.00183 | 65 | 0.01566 | 0.00018 | 0.00139 | 93 | 0.00230 | -0.91725 | 129.358 | 0.36072 | 0.79496 |
| rluE; 23S rRNA pseudouridine2457 synthase [EC:5.4.99.20] | 0.01479 | 0.00113 | 0.01532 | 0.00021 | 0.00182 | 65 | 0.01442 | 0.00020 | 0.00145 | 93 | 0.00233 | 0.38500 | 133.738 | 0.70085 | 0.91978 |
| corC; magnesium and cobalt transporter | 0.01479 | 0.00112 | 0.01519 | 0.00021 | 0.00180 | 65 | 0.01451 | 0.00019 | 0.00144 | 93 | 0.00230 | 0.29356 | 134.020 | 0.76955 | 0.93993 |
| apaH; bis(5’-nucleosyl)-tetraphosphatase (symmetrical) [EC:3.6.1.41] | 0.01478 | 0.00113 | 0.01533 | 0.00022 | 0.00182 | 65 | 0.01440 | 0.00019 | 0.00144 | 93 | 0.00232 | 0.40057 | 132.976 | 0.68938 | 0.91567 |
| nlpD; lipoprotein NlpD | 0.01478 | 0.00112 | 0.01534 | 0.00021 | 0.00181 | 65 | 0.01439 | 0.00019 | 0.00144 | 93 | 0.00231 | 0.40954 | 133.792 | 0.68280 | 0.91495 |
| gudB, rocG; glutamate dehydrogenase [EC:1.4.1.2] | 0.01478 | 0.00121 | 0.01545 | 0.00029 | 0.00212 | 65 | 0.01431 | 0.00019 | 0.00143 | 93 | 0.00255 | 0.44260 | 118.381 | 0.65886 | 0.90880 |
| menF; menaquinone-specific isochorismate synthase [EC:5.4.4.2] | 0.01476 | 0.00122 | 0.01553 | 0.00031 | 0.00218 | 65 | 0.01422 | 0.00018 | 0.00140 | 93 | 0.00259 | 0.50907 | 114.430 | 0.61169 | 0.88722 |
| efeO; iron uptake system component EfeO | 0.01475 | 0.00101 | 0.01357 | 0.00013 | 0.00142 | 65 | 0.01558 | 0.00018 | 0.00139 | 93 | 0.00199 | -1.00825 | 149.880 | 0.31496 | 0.77376 |
| mpl; UDP-N-acetylmuramate: L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase [EC:6.3.2.45] | 0.01475 | 0.00113 | 0.01542 | 0.00022 | 0.00184 | 65 | 0.01428 | 0.00019 | 0.00143 | 93 | 0.00234 | 0.48924 | 131.332 | 0.62549 | 0.89228 |
| metR; LysR family transcriptional regulator, regulator for metE and metH | 0.01473 | 0.00112 | 0.01526 | 0.00021 | 0.00181 | 65 | 0.01435 | 0.00019 | 0.00143 | 93 | 0.00231 | 0.39694 | 133.217 | 0.69204 | 0.91676 |
| K07002; uncharacterized protein | 0.01471 | 0.00145 | 0.01271 | 0.00021 | 0.00178 | 65 | 0.01612 | 0.00042 | 0.00211 | 93 | 0.00276 | -1.23352 | 155.988 | 0.21924 | 0.77376 |
| TC.BAT2; bacterial/archaeal transporter family-2 protein | 0.01468 | 0.00133 | 0.01308 | 0.00020 | 0.00176 | 65 | 0.01580 | 0.00033 | 0.00189 | 93 | 0.00259 | -1.05130 | 154.175 | 0.29476 | 0.77376 |
| sspA, mglA; stringent starvation protein A | 0.01468 | 0.00113 | 0.01521 | 0.00021 | 0.00180 | 65 | 0.01431 | 0.00019 | 0.00145 | 93 | 0.00231 | 0.38839 | 134.301 | 0.69835 | 0.91844 |
| cysD; sulfate adenylyltransferase subunit 2 [EC:2.7.7.4] | 0.01468 | 0.00120 | 0.01276 | 0.00014 | 0.00147 | 65 | 0.01602 | 0.00029 | 0.00175 | 93 | 0.00229 | -1.42316 | 155.996 | 0.15669 | 0.77376 |
| yjgA; ribosome-associated protein | 0.01467 | 0.00113 | 0.01525 | 0.00021 | 0.00181 | 65 | 0.01427 | 0.00019 | 0.00144 | 93 | 0.00231 | 0.42386 | 133.852 | 0.67235 | 0.91184 |
| PTS-Gat-EIIB, gatB, sgcB; PTS system, galactitol-specific IIB component [EC:2.7.1.200] | 0.01467 | 0.00110 | 0.01500 | 0.00023 | 0.00188 | 65 | 0.01444 | 0.00017 | 0.00133 | 93 | 0.00231 | 0.24306 | 122.828 | 0.80837 | 0.95002 |
| hda; DnaA-homolog protein | 0.01466 | 0.00113 | 0.01521 | 0.00021 | 0.00181 | 65 | 0.01427 | 0.00019 | 0.00145 | 93 | 0.00231 | 0.40545 | 133.980 | 0.68580 | 0.91567 |
| bamE, smpA; outer membrane protein assembly factor BamE | 0.01465 | 0.00113 | 0.01527 | 0.00021 | 0.00181 | 65 | 0.01422 | 0.00019 | 0.00145 | 93 | 0.00232 | 0.45142 | 133.895 | 0.65242 | 0.90536 |
| djlA; DnaJ like chaperone protein | 0.01463 | 0.00107 | 0.01460 | 0.00017 | 0.00160 | 65 | 0.01466 | 0.00019 | 0.00144 | 93 | 0.00216 | -0.02567 | 143.747 | 0.97956 | 0.99657 |
| ccmF; cytochrome c-type biogenesis protein CcmF | 0.01460 | 0.00105 | 0.01431 | 0.00020 | 0.00175 | 65 | 0.01480 | 0.00016 | 0.00131 | 93 | 0.00218 | -0.22255 | 127.616 | 0.82424 | 0.95506 |
| CYC; cytochrome c | 0.01458 | 0.00161 | 0.01097 | 0.00024 | 0.00191 | 65 | 0.01711 | 0.00052 | 0.00236 | 93 | 0.00303 | -2.02530 | 155.833 | 0.04454 | 0.65981 |
| fis; Fis family transcriptional regulator, factor for inversion stimulation protein | 0.01456 | 0.00112 | 0.01527 | 0.00021 | 0.00181 | 65 | 0.01407 | 0.00019 | 0.00144 | 93 | 0.00231 | 0.51939 | 133.221 | 0.60435 | 0.88434 |
| rep; ATP-dependent DNA helicase Rep [EC:3.6.4.12] | 0.01456 | 0.00112 | 0.01527 | 0.00021 | 0.00181 | 65 | 0.01407 | 0.00019 | 0.00144 | 93 | 0.00231 | 0.51939 | 133.221 | 0.60435 | 0.88434 |
| ubiD; 4-hydroxy-3-polyprenylbenzoate decarboxylase [EC:4.1.1.98] | 0.01454 | 0.00111 | 0.01492 | 0.00019 | 0.00173 | 65 | 0.01428 | 0.00020 | 0.00146 | 93 | 0.00226 | 0.27954 | 138.781 | 0.78024 | 0.94269 |
| gspJ; general secretion pathway protein J | 0.01453 | 0.00167 | 0.01331 | 0.00040 | 0.00247 | 65 | 0.01538 | 0.00048 | 0.00226 | 93 | 0.00335 | -0.61722 | 145.408 | 0.53805 | 0.86207 |
| gapN; glyceraldehyde-3-phosphate dehydrogenase (NADP+) [EC:1.2.1.9] | 0.01453 | 0.00102 | 0.01496 | 0.00015 | 0.00153 | 65 | 0.01423 | 0.00017 | 0.00136 | 93 | 0.00205 | 0.35782 | 143.068 | 0.72101 | 0.92156 |
| TC.SMR3; small multidrug resistance family-3 protein | 0.01453 | 0.00103 | 0.01469 | 0.00017 | 0.00164 | 65 | 0.01441 | 0.00016 | 0.00132 | 93 | 0.00210 | 0.13232 | 134.786 | 0.89493 | 0.97195 |
| ccmB; heme exporter protein B | 0.01451 | 0.00104 | 0.01443 | 0.00020 | 0.00177 | 65 | 0.01457 | 0.00015 | 0.00128 | 93 | 0.00218 | -0.06411 | 124.894 | 0.94898 | 0.98754 |
| hmp, YHB1; nitric oxide dioxygenase [EC:1.14.12.17] | 0.01451 | 0.00121 | 0.01320 | 0.00025 | 0.00197 | 65 | 0.01543 | 0.00021 | 0.00152 | 93 | 0.00249 | -0.89647 | 130.537 | 0.37165 | 0.79738 |
| virK; uncharacterized protein | 0.01451 | 0.00142 | 0.01609 | 0.00035 | 0.00234 | 65 | 0.01340 | 0.00030 | 0.00178 | 93 | 0.00294 | 0.91216 | 129.654 | 0.36338 | 0.79496 |
| PTS-Nag-EIIC, nagE; PTS system, N-acetylglucosamine-specific IIC component | 0.01450 | 0.00107 | 0.01569 | 0.00025 | 0.00196 | 65 | 0.01367 | 0.00013 | 0.00120 | 93 | 0.00230 | 0.87809 | 110.168 | 0.38181 | 0.80009 |
| fliH; flagellar assembly protein FliH | 0.01449 | 0.00112 | 0.01348 | 0.00022 | 0.00185 | 65 | 0.01520 | 0.00018 | 0.00140 | 93 | 0.00232 | -0.74207 | 129.056 | 0.45939 | 0.83438 |
| gspK; general secretion pathway protein K | 0.01448 | 0.00167 | 0.01326 | 0.00039 | 0.00246 | 65 | 0.01533 | 0.00047 | 0.00226 | 93 | 0.00334 | -0.62018 | 145.387 | 0.53611 | 0.86207 |
| K06923; uncharacterized protein | 0.01448 | 0.00112 | 0.01459 | 0.00024 | 0.00193 | 65 | 0.01439 | 0.00017 | 0.00136 | 93 | 0.00236 | 0.08500 | 122.523 | 0.93240 | 0.98400 |
| ccmE; cytochrome c-type biogenesis protein CcmE | 0.01447 | 0.00104 | 0.01432 | 0.00020 | 0.00175 | 65 | 0.01457 | 0.00015 | 0.00128 | 93 | 0.00217 | -0.11407 | 126.096 | 0.90936 | 0.97664 |
| ccmC; heme exporter protein C | 0.01446 | 0.00104 | 0.01433 | 0.00020 | 0.00175 | 65 | 0.01456 | 0.00015 | 0.00128 | 93 | 0.00217 | -0.10372 | 125.929 | 0.91756 | 0.97705 |
| xlyAB; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | 0.01446 | 0.00155 | 0.01396 | 0.00039 | 0.00244 | 65 | 0.01480 | 0.00038 | 0.00203 | 93 | 0.00317 | -0.26799 | 137.465 | 0.78911 | 0.94606 |
| tadC; tight adherence protein C | 0.01445 | 0.00110 | 0.01373 | 0.00020 | 0.00176 | 65 | 0.01495 | 0.00018 | 0.00141 | 93 | 0.00226 | -0.54227 | 133.768 | 0.58853 | 0.87736 |
| tauD; taurine dioxygenase [EC:1.14.11.17] | 0.01443 | 0.00195 | 0.01300 | 0.00044 | 0.00261 | 65 | 0.01542 | 0.00071 | 0.00276 | 93 | 0.00380 | -0.63862 | 153.607 | 0.52402 | 0.85711 |
| dexB; glucan 1,6-alpha-glucosidase [EC:3.2.1.70] | 0.01442 | 0.00121 | 0.01479 | 0.00020 | 0.00176 | 65 | 0.01416 | 0.00025 | 0.00164 | 93 | 0.00241 | 0.26296 | 146.630 | 0.79295 | 0.94783 |
| mglC; methyl-galactoside transport system permease protein | 0.01442 | 0.00126 | 0.01781 | 0.00034 | 0.00230 | 65 | 0.01205 | 0.00018 | 0.00138 | 93 | 0.00268 | 2.15030 | 108.645 | 0.03375 | 0.62130 |
| oprO_P; phosphate-selective porin OprO and OprP | 0.01442 | 0.00179 | 0.01443 | 0.00055 | 0.00291 | 65 | 0.01441 | 0.00049 | 0.00229 | 93 | 0.00370 | 0.00509 | 132.410 | 0.99594 | 0.99944 |
| fdxA; ferredoxin | 0.01441 | 0.00119 | 0.01321 | 0.00023 | 0.00189 | 65 | 0.01526 | 0.00022 | 0.00154 | 93 | 0.00243 | -0.84228 | 135.427 | 0.40111 | 0.80823 |
| mglA; methyl-galactoside transport system ATP-binding protein [EC:3.6.3.17] | 0.01441 | 0.00126 | 0.01781 | 0.00034 | 0.00230 | 65 | 0.01204 | 0.00018 | 0.00137 | 93 | 0.00268 | 2.15710 | 108.295 | 0.03321 | 0.62130 |
| gspI; general secretion pathway protein I | 0.01441 | 0.00166 | 0.01306 | 0.00039 | 0.00244 | 65 | 0.01536 | 0.00048 | 0.00226 | 93 | 0.00333 | -0.68946 | 146.171 | 0.49163 | 0.84797 |
| ccmA; heme exporter protein A [EC:3.6.3.41] | 0.01441 | 0.00105 | 0.01431 | 0.00020 | 0.00175 | 65 | 0.01448 | 0.00016 | 0.00130 | 93 | 0.00218 | -0.07783 | 127.278 | 0.93809 | 0.98459 |
| glpR; DeoR family transcriptional regulator, glycerol-3-phosphate regulon repressor | 0.01440 | 0.00098 | 0.01507 | 0.00018 | 0.00166 | 65 | 0.01393 | 0.00013 | 0.00120 | 93 | 0.00205 | 0.55487 | 124.187 | 0.57998 | 0.87573 |
| srtB; sortase B [EC:3.4.22.70] | 0.01436 | 0.00136 | 0.01405 | 0.00037 | 0.00239 | 65 | 0.01457 | 0.00024 | 0.00161 | 93 | 0.00288 | -0.18186 | 118.131 | 0.85600 | 0.95933 |
| K09704; uncharacterized protein | 0.01434 | 0.00101 | 0.01354 | 0.00014 | 0.00145 | 65 | 0.01490 | 0.00018 | 0.00138 | 93 | 0.00200 | -0.68028 | 147.935 | 0.49739 | 0.84814 |
| cyoC; cytochrome o ubiquinol oxidase subunit III | 0.01433 | 0.00147 | 0.01318 | 0.00034 | 0.00229 | 65 | 0.01514 | 0.00034 | 0.00191 | 93 | 0.00299 | -0.65766 | 137.592 | 0.51185 | 0.85122 |
| yagU; putative membrane protein | 0.01431 | 0.00170 | 0.01603 | 0.00057 | 0.00297 | 65 | 0.01310 | 0.00038 | 0.00201 | 93 | 0.00359 | 0.81486 | 118.855 | 0.41678 | 0.81842 |
| ssuD; alkanesulfonate monooxygenase [EC:1.14.14.5] | 0.01428 | 0.00181 | 0.01155 | 0.00031 | 0.00219 | 65 | 0.01619 | 0.00066 | 0.00266 | 93 | 0.00345 | -1.34496 | 155.969 | 0.18059 | 0.77376 |
| acnB; aconitate hydratase 2 / 2-methylisocitrate dehydratase [EC:4.2.1.3 4.2.1.99] | 0.01428 | 0.00134 | 0.01415 | 0.00026 | 0.00202 | 65 | 0.01437 | 0.00030 | 0.00179 | 93 | 0.00270 | -0.08105 | 142.916 | 0.93552 | 0.98459 |
| entC; isochorismate synthase [EC:5.4.4.2] | 0.01427 | 0.00106 | 0.01274 | 0.00014 | 0.00149 | 65 | 0.01535 | 0.00020 | 0.00146 | 93 | 0.00208 | -1.25079 | 149.896 | 0.21296 | 0.77376 |
| impL, vasK, icmF; type VI secretion system protein ImpL | 0.01427 | 0.00214 | 0.01566 | 0.00065 | 0.00317 | 65 | 0.01329 | 0.00077 | 0.00288 | 93 | 0.00429 | 0.55169 | 144.709 | 0.58201 | 0.87698 |
| kdpE; two-component system, OmpR family, KDP operon response regulator KdpE | 0.01421 | 0.00119 | 0.01218 | 0.00020 | 0.00176 | 65 | 0.01562 | 0.00024 | 0.00159 | 93 | 0.00238 | -1.44947 | 144.283 | 0.14938 | 0.77376 |
| frmB, ESD, fghA; S-formylglutathione hydrolase [EC:3.1.2.12] | 0.01419 | 0.00118 | 0.01321 | 0.00020 | 0.00176 | 65 | 0.01488 | 0.00023 | 0.00158 | 93 | 0.00236 | -0.70542 | 143.718 | 0.48169 | 0.84354 |
| E2.8.3.5B, scoB; 3-oxoacid CoA-transferase subunit B [EC:2.8.3.5] | 0.01413 | 0.00128 | 0.01206 | 0.00021 | 0.00182 | 65 | 0.01559 | 0.00029 | 0.00175 | 93 | 0.00253 | -1.39870 | 148.877 | 0.16398 | 0.77376 |
| pilF; type IV pilus assembly protein PilF | 0.01413 | 0.00112 | 0.01485 | 0.00021 | 0.00180 | 65 | 0.01364 | 0.00019 | 0.00144 | 93 | 0.00231 | 0.52432 | 134.340 | 0.60092 | 0.88272 |
| E2.8.3.5A, scoA; 3-oxoacid CoA-transferase subunit A [EC:2.8.3.5] | 0.01413 | 0.00128 | 0.01204 | 0.00022 | 0.00183 | 65 | 0.01559 | 0.00029 | 0.00176 | 93 | 0.00253 | -1.40112 | 148.648 | 0.16326 | 0.77376 |
| wrbA; NAD(P)H dehydrogenase (quinone) [EC:1.6.5.2] | 0.01412 | 0.00124 | 0.01299 | 0.00023 | 0.00187 | 65 | 0.01491 | 0.00026 | 0.00166 | 93 | 0.00250 | -0.77086 | 142.670 | 0.44207 | 0.83186 |
| cobA; uroporphyrin-III C-methyltransferase [EC:2.1.1.107] | 0.01411 | 0.00128 | 0.01149 | 0.00020 | 0.00173 | 65 | 0.01594 | 0.00030 | 0.00178 | 93 | 0.00249 | -1.79029 | 152.396 | 0.07539 | 0.74536 |
| efeB; deferrochelatase/peroxidase EfeB [EC:1.11.1.-] | 0.01410 | 0.00098 | 0.01297 | 0.00012 | 0.00135 | 65 | 0.01489 | 0.00017 | 0.00136 | 93 | 0.00192 | -1.00154 | 151.531 | 0.31816 | 0.77376 |
| ABC.X2.P; putative ABC transport system permease protein | 0.01408 | 0.00118 | 0.01312 | 0.00023 | 0.00188 | 65 | 0.01475 | 0.00022 | 0.00153 | 93 | 0.00242 | -0.67119 | 135.676 | 0.50324 | 0.84956 |
| SLC10A7, P7; solute carrier family 10 (sodium/bile acid cotransporter), member 7 | 0.01406 | 0.00110 | 0.01417 | 0.00023 | 0.00187 | 65 | 0.01399 | 0.00017 | 0.00134 | 93 | 0.00230 | 0.08071 | 123.877 | 0.93580 | 0.98459 |
| GDH2; glutamate dehydrogenase [EC:1.4.1.2] | 0.01406 | 0.00142 | 0.01337 | 0.00031 | 0.00217 | 65 | 0.01454 | 0.00033 | 0.00190 | 93 | 0.00288 | -0.40675 | 141.762 | 0.68480 | 0.91533 |
| ABC.SS.S; simple sugar transport system substrate-binding protein | 0.01405 | 0.00140 | 0.01203 | 0.00023 | 0.00189 | 65 | 0.01546 | 0.00036 | 0.00198 | 93 | 0.00274 | -1.25266 | 153.168 | 0.21224 | 0.77376 |
| bacA; vitamin B12/bleomycin/antimicrobial peptide transport system ATP-binding/permease protein | 0.01402 | 0.00128 | 0.01302 | 0.00023 | 0.00189 | 65 | 0.01472 | 0.00028 | 0.00172 | 93 | 0.00256 | -0.66417 | 144.751 | 0.50764 | 0.84956 |
| yciA; acyl-CoA thioesterase YciA [EC:3.1.2.-] | 0.01400 | 0.00103 | 0.01388 | 0.00017 | 0.00163 | 65 | 0.01409 | 0.00017 | 0.00134 | 93 | 0.00211 | -0.09803 | 136.683 | 0.92205 | 0.97910 |
| cysG; uroporphyrin-III C-methyltransferase / precorrin-2 dehydrogenase / sirohydrochlorin ferrochelatase [EC:2.1.1.107 1.3.1.76 4.99.1.4] | 0.01399 | 0.00116 | 0.01451 | 0.00022 | 0.00185 | 65 | 0.01362 | 0.00021 | 0.00149 | 93 | 0.00238 | 0.37203 | 134.736 | 0.71046 | 0.92004 |
| K07119; uncharacterized protein | 0.01397 | 0.00139 | 0.01158 | 0.00023 | 0.00187 | 65 | 0.01564 | 0.00036 | 0.00195 | 93 | 0.00271 | -1.49704 | 153.009 | 0.13644 | 0.77376 |
| dfx; superoxide reductase [EC:1.15.1.2] | 0.01395 | 0.00136 | 0.01490 | 0.00035 | 0.00231 | 65 | 0.01328 | 0.00025 | 0.00165 | 93 | 0.00284 | 0.56926 | 123.847 | 0.57021 | 0.87135 |
| ubiJ; ubiquinone biosynthesis protein UbiJ | 0.01390 | 0.00111 | 0.01457 | 0.00020 | 0.00176 | 65 | 0.01344 | 0.00019 | 0.00143 | 93 | 0.00227 | 0.50101 | 135.115 | 0.61718 | 0.88903 |
| CPOX, hemF; coproporphyrinogen III oxidase [EC:1.3.3.3] | 0.01390 | 0.00120 | 0.01321 | 0.00022 | 0.00185 | 65 | 0.01438 | 0.00023 | 0.00159 | 93 | 0.00244 | -0.48209 | 140.156 | 0.63049 | 0.89558 |
| K08973; putative membrane protein | 0.01388 | 0.00111 | 0.01273 | 0.00019 | 0.00173 | 65 | 0.01469 | 0.00020 | 0.00145 | 93 | 0.00226 | -0.86483 | 138.415 | 0.38863 | 0.80262 |
| selA; L-seryl-tRNA(Ser) seleniumtransferase [EC:2.9.1.1] | 0.01385 | 0.00114 | 0.01520 | 0.00024 | 0.00190 | 65 | 0.01291 | 0.00019 | 0.00141 | 93 | 0.00237 | 0.96727 | 126.996 | 0.33525 | 0.78186 |
| E3.1.1.45; carboxymethylenebutenolidase [EC:3.1.1.45] | 0.01385 | 0.00147 | 0.01082 | 0.00024 | 0.00191 | 65 | 0.01597 | 0.00040 | 0.00208 | 93 | 0.00283 | -1.82357 | 154.576 | 0.07015 | 0.73522 |
| rnfB; electron transport complex protein RnfB | 0.01384 | 0.00105 | 0.01436 | 0.00019 | 0.00170 | 65 | 0.01348 | 0.00017 | 0.00134 | 93 | 0.00217 | 0.40319 | 132.705 | 0.68746 | 0.91567 |
| pilQ; type IV pilus assembly protein PilQ | 0.01383 | 0.00114 | 0.01408 | 0.00022 | 0.00185 | 65 | 0.01365 | 0.00020 | 0.00145 | 93 | 0.00235 | 0.17932 | 132.148 | 0.85796 | 0.96050 |
| SIAE; sialate O-acetylesterase [EC:3.1.1.53] | 0.01382 | 0.00149 | 0.01084 | 0.00017 | 0.00164 | 65 | 0.01590 | 0.00047 | 0.00224 | 93 | 0.00278 | -1.81988 | 153.521 | 0.07072 | 0.73788 |
| K07120; uncharacterized protein | 0.01382 | 0.00134 | 0.01391 | 0.00031 | 0.00220 | 65 | 0.01375 | 0.00027 | 0.00169 | 93 | 0.00278 | 0.05974 | 130.606 | 0.95245 | 0.98792 |
| K07000; uncharacterized protein | 0.01382 | 0.00098 | 0.01361 | 0.00013 | 0.00140 | 65 | 0.01396 | 0.00017 | 0.00135 | 93 | 0.00195 | -0.17861 | 148.583 | 0.85849 | 0.96064 |
| TSTA3, fcl; GDP-L-fucose synthase [EC:1.1.1.271] | 0.01380 | 0.00123 | 0.01107 | 0.00017 | 0.00159 | 65 | 0.01571 | 0.00028 | 0.00175 | 93 | 0.00236 | -1.96330 | 154.738 | 0.05140 | 0.68789 |
| tcyK; L-cystine transport system substrate-binding protein | 0.01379 | 0.00131 | 0.01269 | 0.00019 | 0.00172 | 65 | 0.01456 | 0.00033 | 0.00187 | 93 | 0.00254 | -0.73406 | 154.666 | 0.46402 | 0.83699 |
| cyoA; cytochrome o ubiquinol oxidase subunit II [EC:1.10.3.10] | 0.01379 | 0.00149 | 0.01294 | 0.00036 | 0.00235 | 65 | 0.01438 | 0.00034 | 0.00192 | 93 | 0.00304 | -0.47360 | 136.070 | 0.63654 | 0.89696 |
| doc; death on curing protein | 0.01378 | 0.00099 | 0.01344 | 0.00016 | 0.00157 | 65 | 0.01401 | 0.00015 | 0.00127 | 93 | 0.00202 | -0.28326 | 135.008 | 0.77741 | 0.94269 |
| ada; AraC family transcriptional regulator, regulatory protein of adaptative response / methylated-DNA-[protein]-cysteine methyltransferase [EC:2.1.1.63] | 0.01378 | 0.00109 | 0.01206 | 0.00016 | 0.00159 | 65 | 0.01498 | 0.00020 | 0.00147 | 93 | 0.00217 | -1.34782 | 146.111 | 0.17980 | 0.77376 |
| ispZ; intracellular septation protein | 0.01376 | 0.00105 | 0.01351 | 0.00018 | 0.00168 | 65 | 0.01394 | 0.00017 | 0.00136 | 93 | 0.00216 | -0.19750 | 135.373 | 0.84374 | 0.95519 |
| tusA, sirA; tRNA 2-thiouridine synthesizing protein A [EC:2.8.1.-] | 0.01375 | 0.00117 | 0.01313 | 0.00019 | 0.00172 | 65 | 0.01418 | 0.00023 | 0.00159 | 93 | 0.00234 | -0.44715 | 145.867 | 0.65543 | 0.90679 |
| pbpC; penicillin-binding protein 1C [EC:2.4.1.129] | 0.01374 | 0.00121 | 0.01479 | 0.00027 | 0.00204 | 65 | 0.01301 | 0.00020 | 0.00148 | 93 | 0.00252 | 0.70625 | 125.301 | 0.48134 | 0.84354 |
| ftsL; cell division protein FtsL | 0.01373 | 0.00109 | 0.01450 | 0.00021 | 0.00178 | 65 | 0.01319 | 0.00018 | 0.00138 | 93 | 0.00226 | 0.58315 | 131.272 | 0.56079 | 0.86813 |
| rodZ; cytoskeleton protein RodZ | 0.01372 | 0.00117 | 0.01454 | 0.00023 | 0.00188 | 65 | 0.01314 | 0.00021 | 0.00150 | 93 | 0.00240 | 0.58024 | 134.147 | 0.56272 | 0.86813 |
| fucO; lactaldehyde reductase [EC:1.1.1.77] | 0.01372 | 0.00177 | 0.01780 | 0.00080 | 0.00351 | 65 | 0.01086 | 0.00026 | 0.00169 | 93 | 0.00390 | 1.78069 | 93.460 | 0.07821 | 0.75550 |
| E4.2.1.2AB, fumB; fumarate hydratase subunit beta [EC:4.2.1.2] | 0.01372 | 0.00138 | 0.01443 | 0.00035 | 0.00233 | 65 | 0.01321 | 0.00027 | 0.00170 | 93 | 0.00288 | 0.42208 | 125.536 | 0.67369 | 0.91184 |
| nqrE; Na+-transporting NADH:ubiquinone oxidoreductase subunit E [EC:1.6.5.8] | 0.01371 | 0.00111 | 0.01369 | 0.00017 | 0.00160 | 65 | 0.01373 | 0.00021 | 0.00152 | 93 | 0.00221 | -0.01764 | 147.639 | 0.98595 | 0.99729 |
| nqrC; Na+-transporting NADH:ubiquinone oxidoreductase subunit C [EC:1.6.5.8] | 0.01371 | 0.00111 | 0.01369 | 0.00017 | 0.00160 | 65 | 0.01373 | 0.00021 | 0.00152 | 93 | 0.00221 | -0.02056 | 147.663 | 0.98363 | 0.99706 |
| nqrB; Na+-transporting NADH:ubiquinone oxidoreductase subunit B [EC:1.6.5.8] | 0.01371 | 0.00111 | 0.01369 | 0.00017 | 0.00160 | 65 | 0.01373 | 0.00021 | 0.00152 | 93 | 0.00221 | -0.01985 | 147.659 | 0.98419 | 0.99706 |
| nqrD; Na+-transporting NADH:ubiquinone oxidoreductase subunit D [EC:1.6.5.8] | 0.01371 | 0.00111 | 0.01369 | 0.00017 | 0.00160 | 65 | 0.01373 | 0.00021 | 0.00152 | 93 | 0.00221 | -0.01985 | 147.659 | 0.98419 | 0.99706 |
| nqrF; Na+-transporting NADH:ubiquinone oxidoreductase subunit F [EC:1.6.5.8] | 0.01371 | 0.00111 | 0.01369 | 0.00017 | 0.00160 | 65 | 0.01373 | 0.00021 | 0.00152 | 93 | 0.00221 | -0.01912 | 147.646 | 0.98477 | 0.99718 |
| nqrA; Na+-transporting NADH:ubiquinone oxidoreductase subunit A [EC:1.6.5.8] | 0.01371 | 0.00111 | 0.01368 | 0.00017 | 0.00161 | 65 | 0.01373 | 0.00021 | 0.00152 | 93 | 0.00221 | -0.02145 | 147.636 | 0.98291 | 0.99706 |
| E4.2.1.2AA, fumA; fumarate hydratase subunit alpha [EC:4.2.1.2] | 0.01370 | 0.00138 | 0.01441 | 0.00035 | 0.00233 | 65 | 0.01320 | 0.00027 | 0.00170 | 93 | 0.00288 | 0.41977 | 125.517 | 0.67537 | 0.91264 |
| grxC, GLRX, GLRX2; glutaredoxin 3 | 0.01370 | 0.00116 | 0.01244 | 0.00019 | 0.00172 | 65 | 0.01457 | 0.00023 | 0.00156 | 93 | 0.00232 | -0.92137 | 144.459 | 0.35839 | 0.79496 |
| hipO; hippurate hydrolase [EC:3.5.1.32] | 0.01365 | 0.00154 | 0.01064 | 0.00021 | 0.00180 | 65 | 0.01575 | 0.00048 | 0.00227 | 93 | 0.00290 | -1.76239 | 155.613 | 0.07997 | 0.75920 |
| rhlE; ATP-dependent RNA helicase RhlE [EC:3.6.4.13] | 0.01364 | 0.00120 | 0.01218 | 0.00021 | 0.00178 | 65 | 0.01467 | 0.00024 | 0.00162 | 93 | 0.00240 | -1.03390 | 144.663 | 0.30291 | 0.77376 |
| flgA; flagella basal body P-ring formation protein FlgA | 0.01364 | 0.00120 | 0.01307 | 0.00024 | 0.00191 | 65 | 0.01404 | 0.00022 | 0.00154 | 93 | 0.00246 | -0.39499 | 134.799 | 0.69348 | 0.91677 |
| kdpA; K+-transporting ATPase ATPase A chain [EC:3.6.3.12] | 0.01364 | 0.00113 | 0.01175 | 0.00020 | 0.00176 | 65 | 0.01496 | 0.00020 | 0.00147 | 93 | 0.00229 | -1.39918 | 138.118 | 0.16400 | 0.77376 |
| coxC, ctaE; cytochrome c oxidase subunit III [EC:1.9.3.1] | 0.01364 | 0.00123 | 0.01086 | 0.00022 | 0.00183 | 65 | 0.01558 | 0.00025 | 0.00163 | 93 | 0.00245 | -1.92706 | 143.136 | 0.05595 | 0.70577 |
| K09862; uncharacterized protein | 0.01363 | 0.00106 | 0.01324 | 0.00017 | 0.00164 | 65 | 0.01390 | 0.00018 | 0.00139 | 93 | 0.00215 | -0.30844 | 139.146 | 0.75821 | 0.93719 |
| ycfD; 50S ribosomal protein L16 3-hydroxylase [EC:1.14.11.47] | 0.01360 | 0.00109 | 0.01420 | 0.00019 | 0.00173 | 65 | 0.01318 | 0.00018 | 0.00141 | 93 | 0.00223 | 0.45532 | 135.592 | 0.64961 | 0.90402 |
| cysI; sulfite reductase (NADPH) hemoprotein beta-component [EC:1.8.1.2] | 0.01357 | 0.00106 | 0.01249 | 0.00017 | 0.00160 | 65 | 0.01433 | 0.00019 | 0.00142 | 93 | 0.00214 | -0.86287 | 143.104 | 0.38965 | 0.80353 |
| tctA; putative tricarboxylic transport membrane protein | 0.01356 | 0.00128 | 0.01331 | 0.00027 | 0.00206 | 65 | 0.01373 | 0.00025 | 0.00163 | 93 | 0.00262 | -0.15967 | 133.201 | 0.87338 | 0.96576 |
| ispA; farnesyl diphosphate synthase [EC:2.5.1.1 2.5.1.10] | 0.01356 | 0.00110 | 0.01232 | 0.00018 | 0.00165 | 65 | 0.01442 | 0.00020 | 0.00147 | 93 | 0.00221 | -0.94992 | 143.093 | 0.34375 | 0.78610 |
| pyrI; aspartate carbamoyltransferase regulatory subunit | 0.01356 | 0.00117 | 0.01364 | 0.00021 | 0.00179 | 65 | 0.01350 | 0.00022 | 0.00154 | 93 | 0.00236 | 0.05992 | 140.740 | 0.95231 | 0.98792 |
| GCDH, gcdH; glutaryl-CoA dehydrogenase [EC:1.3.8.6] | 0.01355 | 0.00131 | 0.01126 | 0.00024 | 0.00191 | 65 | 0.01516 | 0.00029 | 0.00177 | 93 | 0.00261 | -1.49502 | 145.982 | 0.13707 | 0.77376 |
| COX15, ctaA; cytochrome c oxidase assembly protein subunit 15 | 0.01354 | 0.00116 | 0.01066 | 0.00020 | 0.00175 | 65 | 0.01556 | 0.00021 | 0.00152 | 93 | 0.00231 | -2.11538 | 141.012 | 0.03616 | 0.62411 |
| ligD; bifunctional non-homologous end joining protein LigD [EC:6.5.1.1] | 0.01353 | 0.00143 | 0.01073 | 0.00021 | 0.00181 | 65 | 0.01549 | 0.00039 | 0.00206 | 93 | 0.00274 | -1.73792 | 155.557 | 0.08420 | 0.76567 |
| ohyA, sph; oleate hydratase [EC:4.2.1.53] | 0.01353 | 0.00164 | 0.01465 | 0.00056 | 0.00293 | 65 | 0.01274 | 0.00034 | 0.00190 | 93 | 0.00349 | 0.54694 | 115.106 | 0.58548 | 0.87720 |
| LIG1; DNA ligase 1 [EC:6.5.1.1 6.5.1.6 6.5.1.7] | 0.01353 | 0.00107 | 0.01321 | 0.00020 | 0.00173 | 65 | 0.01375 | 0.00017 | 0.00137 | 93 | 0.00221 | -0.24415 | 132.871 | 0.80749 | 0.94987 |
| proV; glycine betaine/proline transport system ATP-binding protein [EC:3.6.3.32] | 0.01348 | 0.00159 | 0.01422 | 0.00046 | 0.00266 | 65 | 0.01297 | 0.00036 | 0.00196 | 93 | 0.00331 | 0.37866 | 126.422 | 0.70557 | 0.92004 |
| fxsA; UPF0716 protein FxsA | 0.01347 | 0.00101 | 0.01345 | 0.00016 | 0.00158 | 65 | 0.01348 | 0.00016 | 0.00131 | 93 | 0.00206 | -0.01540 | 137.250 | 0.98774 | 0.99801 |
| spoIID; stage II sporulation protein D | 0.01346 | 0.00123 | 0.01420 | 0.00030 | 0.00214 | 65 | 0.01294 | 0.00020 | 0.00146 | 93 | 0.00259 | 0.48815 | 119.190 | 0.62634 | 0.89255 |
| K09806; uncharacterized protein | 0.01345 | 0.00107 | 0.01415 | 0.00019 | 0.00171 | 65 | 0.01295 | 0.00017 | 0.00137 | 93 | 0.00219 | 0.54699 | 134.321 | 0.58529 | 0.87720 |
| abgT; aminobenzoyl-glutamate transport protein | 0.01344 | 0.00180 | 0.01671 | 0.00081 | 0.00353 | 65 | 0.01116 | 0.00029 | 0.00178 | 93 | 0.00395 | 1.40509 | 96.259 | 0.16321 | 0.77376 |
| kdpC; K+-transporting ATPase ATPase C chain [EC:3.6.3.12] | 0.01344 | 0.00112 | 0.01157 | 0.00019 | 0.00171 | 65 | 0.01475 | 0.00020 | 0.00146 | 93 | 0.00225 | -1.41286 | 139.800 | 0.15992 | 0.77376 |
| mlaB; phospholipid transport system transporter-binding protein | 0.01341 | 0.00108 | 0.01418 | 0.00020 | 0.00175 | 65 | 0.01286 | 0.00018 | 0.00138 | 93 | 0.00223 | 0.59408 | 132.837 | 0.55347 | 0.86788 |
| modE; molybdate transport system regulatory protein | 0.01340 | 0.00093 | 0.01408 | 0.00017 | 0.00164 | 65 | 0.01292 | 0.00011 | 0.00111 | 93 | 0.00197 | 0.58624 | 118.650 | 0.55883 | 0.86813 |
| ubiX, bsdB, PAD1; flavin prenyltransferase [EC:2.5.1.129] | 0.01340 | 0.00099 | 0.01390 | 0.00017 | 0.00162 | 65 | 0.01305 | 0.00015 | 0.00125 | 93 | 0.00205 | 0.41477 | 131.036 | 0.67899 | 0.91311 |
| K14645; serine protease [EC:3.4.21.-] | 0.01339 | 0.00204 | 0.01411 | 0.00083 | 0.00357 | 65 | 0.01290 | 0.00055 | 0.00243 | 93 | 0.00432 | 0.28095 | 119.444 | 0.77923 | 0.94269 |
| rgpA; rhamnosyltransferase [EC:2.4.1.-] | 0.01338 | 0.00137 | 0.01425 | 0.00025 | 0.00196 | 65 | 0.01277 | 0.00034 | 0.00190 | 93 | 0.00273 | 0.54310 | 149.216 | 0.58787 | 0.87732 |
| K09801; uncharacterized protein | 0.01338 | 0.00107 | 0.01365 | 0.00019 | 0.00172 | 65 | 0.01318 | 0.00017 | 0.00136 | 93 | 0.00220 | 0.21423 | 133.122 | 0.83069 | 0.95506 |
| qseB; two-component system, OmpR family, response regulator QseB | 0.01337 | 0.00145 | 0.01296 | 0.00026 | 0.00201 | 65 | 0.01367 | 0.00038 | 0.00203 | 93 | 0.00286 | -0.24839 | 151.492 | 0.80417 | 0.94931 |
| cyoD; cytochrome o ubiquinol oxidase subunit IV | 0.01337 | 0.00144 | 0.01232 | 0.00034 | 0.00227 | 65 | 0.01411 | 0.00033 | 0.00187 | 93 | 0.00295 | -0.60661 | 136.558 | 0.54512 | 0.86426 |
| E3.5.4.3, guaD; guanine deaminase [EC:3.5.4.3] | 0.01334 | 0.00123 | 0.01238 | 0.00022 | 0.00186 | 65 | 0.01401 | 0.00025 | 0.00164 | 93 | 0.00248 | -0.66002 | 142.304 | 0.51031 | 0.85013 |
| phoB; two-component system, OmpR family, phosphate regulon response regulator PhoB | 0.01334 | 0.00115 | 0.01194 | 0.00020 | 0.00175 | 65 | 0.01431 | 0.00022 | 0.00152 | 93 | 0.00232 | -1.02089 | 141.387 | 0.30905 | 0.77376 |
| cyoB; cytochrome o ubiquinol oxidase subunit I [EC:1.10.3.10] | 0.01333 | 0.00144 | 0.01230 | 0.00034 | 0.00227 | 65 | 0.01405 | 0.00033 | 0.00188 | 93 | 0.00295 | -0.59666 | 136.751 | 0.55172 | 0.86788 |
| fruA; fructan beta-fructosidase [EC:3.2.1.80] | 0.01333 | 0.00104 | 0.01183 | 0.00013 | 0.00140 | 65 | 0.01437 | 0.00020 | 0.00147 | 93 | 0.00203 | -1.25595 | 153.364 | 0.21104 | 0.77376 |
| plsB; glycerol-3-phosphate O-acyltransferase [EC:2.3.1.15] | 0.01332 | 0.00113 | 0.01365 | 0.00020 | 0.00174 | 65 | 0.01309 | 0.00021 | 0.00150 | 93 | 0.00230 | 0.24654 | 140.484 | 0.80562 | 0.94958 |
| mazG; nucleoside triphosphate diphosphatase [EC:3.6.1.9] | 0.01331 | 0.00112 | 0.01312 | 0.00020 | 0.00173 | 65 | 0.01344 | 0.00020 | 0.00148 | 93 | 0.00228 | -0.14326 | 139.777 | 0.88629 | 0.97020 |
| E3.8.1.2; 2-haloacid dehalogenase [EC:3.8.1.2] | 0.01331 | 0.00105 | 0.01055 | 0.00019 | 0.00170 | 65 | 0.01523 | 0.00016 | 0.00130 | 93 | 0.00213 | -2.18981 | 129.710 | 0.03033 | 0.61413 |
| ETFDH; electron-transferring-flavoprotein dehydrogenase [EC:1.5.5.1] | 0.01328 | 0.00119 | 0.01198 | 0.00021 | 0.00179 | 65 | 0.01418 | 0.00023 | 0.00158 | 93 | 0.00239 | -0.91985 | 142.216 | 0.35921 | 0.79496 |
| E1.1.3.21; alpha-glycerophosphate oxidase [EC:1.1.3.21] | 0.01327 | 0.00127 | 0.01209 | 0.00019 | 0.00172 | 65 | 0.01409 | 0.00030 | 0.00179 | 93 | 0.00249 | -0.80513 | 152.875 | 0.42199 | 0.82295 |
| actP; cation/acetate symporter | 0.01326 | 0.00136 | 0.01146 | 0.00023 | 0.00189 | 65 | 0.01452 | 0.00033 | 0.00189 | 93 | 0.00267 | -1.14245 | 150.736 | 0.25508 | 0.77376 |
| egtC; gamma-glutamyl hercynylcysteine S-oxide hydrolase [EC:3.5.1.118] | 0.01325 | 0.00112 | 0.01307 | 0.00018 | 0.00167 | 65 | 0.01338 | 0.00021 | 0.00151 | 93 | 0.00225 | -0.13781 | 144.137 | 0.89058 | 0.97076 |
| dppD; dipeptide transport system ATP-binding protein | 0.01323 | 0.00100 | 0.01345 | 0.00015 | 0.00152 | 65 | 0.01307 | 0.00016 | 0.00133 | 93 | 0.00202 | 0.18348 | 141.688 | 0.85469 | 0.95858 |
| gluQ; glutamyl-Q tRNA(Asp) synthetase [EC:6.1.1.-] | 0.01323 | 0.00108 | 0.01242 | 0.00019 | 0.00170 | 65 | 0.01379 | 0.00018 | 0.00140 | 93 | 0.00221 | -0.61977 | 136.788 | 0.53644 | 0.86207 |
| flgH; flagellar L-ring protein precursor FlgH | 0.01321 | 0.00111 | 0.01250 | 0.00021 | 0.00180 | 65 | 0.01371 | 0.00019 | 0.00142 | 93 | 0.00229 | -0.52605 | 132.498 | 0.59973 | 0.88240 |
| flgI; flagellar P-ring protein precursor FlgI | 0.01321 | 0.00111 | 0.01255 | 0.00021 | 0.00181 | 65 | 0.01368 | 0.00019 | 0.00141 | 93 | 0.00229 | -0.49321 | 131.931 | 0.62268 | 0.89150 |
| dnaE2; error-prone DNA polymerase [EC:2.7.7.7] | 0.01320 | 0.00121 | 0.01188 | 0.00028 | 0.00206 | 65 | 0.01412 | 0.00020 | 0.00146 | 93 | 0.00253 | -0.88758 | 123.025 | 0.37650 | 0.79810 |
| licR; lichenan operon transcriptional antiterminator | 0.01319 | 0.00104 | 0.01224 | 0.00014 | 0.00147 | 65 | 0.01385 | 0.00019 | 0.00144 | 93 | 0.00206 | -0.78561 | 149.986 | 0.43333 | 0.82874 |
| mrcB; penicillin-binding protein 1B [EC:2.4.1.129 3.4.16.4] | 0.01318 | 0.00116 | 0.01405 | 0.00021 | 0.00180 | 65 | 0.01258 | 0.00022 | 0.00153 | 93 | 0.00236 | 0.62274 | 138.975 | 0.53448 | 0.86207 |
| menH; 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase [EC:4.2.99.20] | 0.01314 | 0.00119 | 0.01376 | 0.00030 | 0.00214 | 65 | 0.01270 | 0.00017 | 0.00137 | 93 | 0.00254 | 0.41715 | 113.931 | 0.67735 | 0.91302 |
| ybbJ; inner membrane protein | 0.01311 | 0.00105 | 0.01302 | 0.00018 | 0.00164 | 65 | 0.01317 | 0.00018 | 0.00137 | 93 | 0.00214 | -0.07107 | 137.901 | 0.94344 | 0.98634 |
| rlmL, rlmK; 23S rRNA (guanine2445-N2)-methyltransferase / 23S rRNA (guanine2069-N7)-methyltransferase [EC:2.1.1.173 2.1.1.264] | 0.01309 | 0.00112 | 0.01373 | 0.00019 | 0.00172 | 65 | 0.01265 | 0.00021 | 0.00149 | 93 | 0.00227 | 0.47360 | 141.032 | 0.63652 | 0.89696 |
| mnmC; tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein [EC:2.1.1.61 1.5.-.-] | 0.01309 | 0.00125 | 0.01349 | 0.00024 | 0.00190 | 65 | 0.01281 | 0.00026 | 0.00166 | 93 | 0.00252 | 0.26954 | 141.549 | 0.78791 | 0.94546 |
| ATPVE, ntpE, atpE; V/A-type H+/Na+-transporting ATPase subunit E | 0.01308 | 0.00113 | 0.01474 | 0.00027 | 0.00203 | 65 | 0.01191 | 0.00015 | 0.00129 | 93 | 0.00241 | 1.17546 | 113.036 | 0.24228 | 0.77376 |
| coxS; aerobic carbon-monoxide dehydrogenase small subunit [EC:1.2.5.3] | 0.01306 | 0.00133 | 0.01150 | 0.00020 | 0.00177 | 65 | 0.01415 | 0.00033 | 0.00190 | 93 | 0.00259 | -1.02126 | 154.120 | 0.30873 | 0.77376 |
| gshB; glutathione synthase [EC:6.3.2.3] | 0.01305 | 0.00109 | 0.01240 | 0.00019 | 0.00172 | 65 | 0.01350 | 0.00019 | 0.00141 | 93 | 0.00223 | -0.49701 | 136.081 | 0.61998 | 0.89023 |
| K09950; uncharacterized protein | 0.01304 | 0.00173 | 0.01373 | 0.00051 | 0.00281 | 65 | 0.01255 | 0.00045 | 0.00219 | 93 | 0.00357 | 0.32877 | 131.546 | 0.74286 | 0.93077 |
| res; type III restriction enzyme [EC:3.1.21.5] | 0.01301 | 0.00130 | 0.01365 | 0.00024 | 0.00193 | 65 | 0.01256 | 0.00029 | 0.00175 | 93 | 0.00260 | 0.41644 | 144.852 | 0.67770 | 0.91311 |
| tcyN; L-cystine transport system ATP-binding protein [EC:3.6.3.-] | 0.01300 | 0.00120 | 0.01172 | 0.00016 | 0.00157 | 65 | 0.01390 | 0.00028 | 0.00172 | 93 | 0.00233 | -0.93727 | 154.689 | 0.35008 | 0.79120 |
| wbpO; UDP-N-acetyl-D-galactosamine dehydrogenase [EC:1.1.1.-] | 0.01300 | 0.00111 | 0.01448 | 0.00027 | 0.00204 | 65 | 0.01197 | 0.00014 | 0.00124 | 93 | 0.00238 | 1.05546 | 109.566 | 0.29354 | 0.77376 |
| E6.4.1.4B; 3-methylcrotonyl-CoA carboxylase beta subunit [EC:6.4.1.4] | 0.01299 | 0.00138 | 0.01137 | 0.00028 | 0.00207 | 65 | 0.01412 | 0.00031 | 0.00184 | 93 | 0.00277 | -0.99305 | 142.763 | 0.32237 | 0.77376 |
| rdgC; recombination associated protein RdgC | 0.01299 | 0.00106 | 0.01391 | 0.00020 | 0.00175 | 65 | 0.01235 | 0.00017 | 0.00133 | 93 | 0.00220 | 0.70925 | 129.519 | 0.47944 | 0.84314 |
| K08972; putative membrane protein | 0.01299 | 0.00111 | 0.01312 | 0.00024 | 0.00192 | 65 | 0.01290 | 0.00017 | 0.00134 | 93 | 0.00234 | 0.09197 | 121.465 | 0.92688 | 0.98128 |
| E3.4.13.-; D-alanyl-D-alanine dipeptidase [EC:3.4.13.-] | 0.01299 | 0.00126 | 0.01140 | 0.00019 | 0.00170 | 65 | 0.01410 | 0.00029 | 0.00178 | 93 | 0.00246 | -1.10022 | 153.230 | 0.27296 | 0.77376 |
| nudB, ntpA; dihydroneopterin triphosphate diphosphatase [EC:3.6.1.67] | 0.01296 | 0.00100 | 0.01312 | 0.00015 | 0.00153 | 65 | 0.01285 | 0.00016 | 0.00133 | 93 | 0.00202 | 0.13381 | 141.282 | 0.89374 | 0.97159 |
| pbpG; serine-type D-Ala-D-Ala endopeptidase (penicillin-binding protein 7) [EC:3.4.21.-] | 0.01295 | 0.00118 | 0.01346 | 0.00022 | 0.00183 | 65 | 0.01260 | 0.00022 | 0.00154 | 93 | 0.00239 | 0.35922 | 138.792 | 0.71998 | 0.92108 |
| vanX; zinc D-Ala-D-Ala dipeptidase [EC:3.4.13.22] | 0.01294 | 0.00100 | 0.01086 | 0.00011 | 0.00128 | 65 | 0.01439 | 0.00019 | 0.00142 | 93 | 0.00191 | -1.84607 | 155.118 | 0.06679 | 0.73334 |
| gspM; general secretion pathway protein M | 0.01294 | 0.00159 | 0.01138 | 0.00035 | 0.00234 | 65 | 0.01402 | 0.00043 | 0.00215 | 93 | 0.00318 | -0.83053 | 145.718 | 0.40760 | 0.81200 |
| yfiC, trmX; tRNA1Val (adenine37-N6)-methyltransferase [EC:2.1.1.223] | 0.01293 | 0.00114 | 0.01247 | 0.00018 | 0.00165 | 65 | 0.01326 | 0.00023 | 0.00156 | 93 | 0.00227 | -0.34562 | 147.636 | 0.73012 | 0.92426 |
| edd; phosphogluconate dehydratase [EC:4.2.1.12] | 0.01291 | 0.00133 | 0.01209 | 0.00026 | 0.00199 | 65 | 0.01348 | 0.00030 | 0.00179 | 93 | 0.00267 | -0.51918 | 144.081 | 0.60443 | 0.88434 |
| butA, budC; meso-butanediol dehydrogenase / (S,S)-butanediol dehydrogenase / diacetyl reductase [EC:1.1.1.- 1.1.1.76 1.1.1.304] | 0.01290 | 0.00120 | 0.01202 | 0.00024 | 0.00190 | 65 | 0.01351 | 0.00022 | 0.00154 | 93 | 0.00245 | -0.60485 | 135.133 | 0.54629 | 0.86444 |
| rhlB; ATP-dependent RNA helicase RhlB [EC:3.6.4.13] | 0.01288 | 0.00114 | 0.01359 | 0.00020 | 0.00173 | 65 | 0.01238 | 0.00021 | 0.00151 | 93 | 0.00230 | 0.52524 | 141.168 | 0.60024 | 0.88240 |
| E2.7.13.3; histidine kinase [EC:2.7.13.3] | 0.01288 | 0.00107 | 0.01141 | 0.00014 | 0.00148 | 65 | 0.01391 | 0.00021 | 0.00150 | 93 | 0.00211 | -1.18582 | 151.947 | 0.23754 | 0.77376 |
| thrB2; homoserine kinase type II [EC:2.7.1.39] | 0.01283 | 0.00108 | 0.01450 | 0.00025 | 0.00196 | 65 | 0.01166 | 0.00014 | 0.00123 | 93 | 0.00231 | 1.22935 | 111.985 | 0.22152 | 0.77376 |
| aer; aerotaxis receptor | 0.01283 | 0.00170 | 0.01299 | 0.00045 | 0.00263 | 65 | 0.01272 | 0.00047 | 0.00224 | 93 | 0.00346 | 0.07861 | 139.229 | 0.93746 | 0.98459 |
| ABC.ZM.P; zinc/manganese transport system permease protein | 0.01282 | 0.00128 | 0.01317 | 0.00034 | 0.00227 | 65 | 0.01258 | 0.00021 | 0.00150 | 93 | 0.00272 | 0.21851 | 116.523 | 0.82742 | 0.95506 |
| nuoCD; NADH-quinone oxidoreductase subunit C/D [EC:1.6.5.3] | 0.01282 | 0.00095 | 0.01193 | 0.00011 | 0.00128 | 65 | 0.01343 | 0.00017 | 0.00134 | 93 | 0.00185 | -0.81081 | 153.154 | 0.41873 | 0.81980 |
| ycaO; ribosomal protein S12 methylthiotransferase accessory factor | 0.01278 | 0.00127 | 0.01279 | 0.00024 | 0.00193 | 65 | 0.01278 | 0.00027 | 0.00169 | 93 | 0.00257 | 0.00354 | 141.747 | 0.99718 | 0.99951 |
| ulaG; L-ascorbate 6-phosphate lactonase [EC:3.1.1.-] | 0.01277 | 0.00107 | 0.01410 | 0.00024 | 0.00192 | 65 | 0.01184 | 0.00014 | 0.00122 | 93 | 0.00227 | 0.99207 | 112.961 | 0.32328 | 0.77376 |
| xanP; xanthine permease XanP | 0.01274 | 0.00101 | 0.01268 | 0.00013 | 0.00141 | 65 | 0.01278 | 0.00018 | 0.00140 | 93 | 0.00199 | -0.05110 | 150.736 | 0.95932 | 0.98903 |
| E4.4.1.11; methionine-gamma-lyase [EC:4.4.1.11] | 0.01274 | 0.00096 | 0.01365 | 0.00021 | 0.00179 | 65 | 0.01210 | 0.00010 | 0.00105 | 93 | 0.00207 | 0.75026 | 107.040 | 0.45474 | 0.83438 |
| PREP; prolyl oligopeptidase [EC:3.4.21.26] | 0.01274 | 0.00099 | 0.01170 | 0.00011 | 0.00132 | 65 | 0.01347 | 0.00018 | 0.00141 | 93 | 0.00193 | -0.91601 | 153.811 | 0.36109 | 0.79496 |
| pycB; pyruvate carboxylase subunit B [EC:6.4.1.1] | 0.01274 | 0.00109 | 0.01200 | 0.00014 | 0.00149 | 65 | 0.01325 | 0.00022 | 0.00154 | 93 | 0.00214 | -0.57980 | 152.858 | 0.56290 | 0.86813 |
| idnO; gluconate 5-dehydrogenase [EC:1.1.1.69] | 0.01273 | 0.00126 | 0.01339 | 0.00036 | 0.00234 | 65 | 0.01227 | 0.00018 | 0.00140 | 93 | 0.00273 | 0.41382 | 108.145 | 0.67983 | 0.91311 |
| hflD; high frequency lysogenization protein | 0.01272 | 0.00113 | 0.01344 | 0.00019 | 0.00173 | 65 | 0.01222 | 0.00021 | 0.00150 | 93 | 0.00229 | 0.53357 | 140.952 | 0.59448 | 0.88005 |
| K09794; uncharacterized protein | 0.01270 | 0.00124 | 0.01398 | 0.00027 | 0.00205 | 65 | 0.01182 | 0.00022 | 0.00155 | 93 | 0.00257 | 0.84152 | 128.531 | 0.40162 | 0.80850 |
| cdaR; carbohydrate diacid regulator | 0.01270 | 0.00145 | 0.01417 | 0.00046 | 0.00267 | 65 | 0.01168 | 0.00024 | 0.00161 | 93 | 0.00312 | 0.79974 | 109.179 | 0.42560 | 0.82605 |
| fadH; 2,4-dienoyl-CoA reductase (NADPH2) [EC:1.3.1.34] | 0.01270 | 0.00130 | 0.01206 | 0.00023 | 0.00189 | 65 | 0.01315 | 0.00029 | 0.00178 | 93 | 0.00260 | -0.41949 | 147.186 | 0.67547 | 0.91264 |
| K02477; two-component system, LytTR family, response regulator | 0.01269 | 0.00136 | 0.01295 | 0.00027 | 0.00204 | 65 | 0.01251 | 0.00031 | 0.00182 | 93 | 0.00273 | 0.16095 | 143.340 | 0.87236 | 0.96576 |
| tcyL; L-cystine transport system permease protein | 0.01269 | 0.00128 | 0.01108 | 0.00017 | 0.00160 | 65 | 0.01381 | 0.00032 | 0.00186 | 93 | 0.00246 | -1.10786 | 155.850 | 0.26963 | 0.77376 |
| amgK; N-acetylmuramate 1-kinase [EC:2.7.1.221] | 0.01268 | 0.00110 | 0.01173 | 0.00019 | 0.00173 | 65 | 0.01334 | 0.00019 | 0.00142 | 93 | 0.00224 | -0.71863 | 136.505 | 0.47360 | 0.84050 |
| gspH; general secretion pathway protein H | 0.01266 | 0.00161 | 0.01157 | 0.00037 | 0.00240 | 65 | 0.01342 | 0.00044 | 0.00218 | 93 | 0.00324 | -0.57089 | 144.631 | 0.56896 | 0.87135 |
| cpoA; 1,2-diacylglycerol-3-alpha-glucose alpha-1,2-galactosyltransferase [EC:2.4.1.-] | 0.01265 | 0.00137 | 0.01358 | 0.00040 | 0.00248 | 65 | 0.01200 | 0.00022 | 0.00155 | 93 | 0.00293 | 0.53963 | 111.786 | 0.59052 | 0.87896 |
| zur; Fur family transcriptional regulator, zinc uptake regulator | 0.01264 | 0.00106 | 0.01217 | 0.00018 | 0.00165 | 65 | 0.01296 | 0.00018 | 0.00138 | 93 | 0.00215 | -0.37036 | 138.382 | 0.71168 | 0.92004 |
| K09791; uncharacterized protein | 0.01262 | 0.00101 | 0.01287 | 0.00018 | 0.00169 | 65 | 0.01245 | 0.00014 | 0.00125 | 93 | 0.00210 | 0.19973 | 126.844 | 0.84201 | 0.95506 |
| spoVD; stage V sporulation protein D (sporulation-specific penicillin-binding protein) | 0.01262 | 0.00130 | 0.01295 | 0.00032 | 0.00224 | 65 | 0.01238 | 0.00023 | 0.00158 | 93 | 0.00274 | 0.20507 | 122.623 | 0.83786 | 0.95506 |
| K07004; uncharacterized protein | 0.01260 | 0.00089 | 0.01181 | 0.00012 | 0.00136 | 65 | 0.01316 | 0.00013 | 0.00119 | 93 | 0.00181 | -0.74947 | 141.356 | 0.45482 | 0.83438 |
| ccoO; cytochrome c oxidase cbb3-type subunit II | 0.01260 | 0.00138 | 0.01255 | 0.00031 | 0.00219 | 65 | 0.01264 | 0.00030 | 0.00180 | 93 | 0.00283 | -0.02984 | 136.275 | 0.97624 | 0.99582 |
| hcp; type VI secretion system secreted protein Hcp | 0.01255 | 0.00155 | 0.01429 | 0.00038 | 0.00243 | 65 | 0.01134 | 0.00038 | 0.00201 | 93 | 0.00316 | 0.93353 | 137.100 | 0.35219 | 0.79159 |
| comD; two-component system, LytTR family, sensor histidine kinase ComD [EC:2.7.13.3] | 0.01254 | 0.00124 | 0.01115 | 0.00018 | 0.00167 | 65 | 0.01351 | 0.00029 | 0.00176 | 93 | 0.00243 | -0.97037 | 153.592 | 0.33339 | 0.77957 |
| flgJ; flagellar protein FlgJ | 0.01252 | 0.00119 | 0.01273 | 0.00024 | 0.00191 | 65 | 0.01238 | 0.00022 | 0.00154 | 93 | 0.00245 | 0.14450 | 134.412 | 0.88532 | 0.97001 |
| yejB; microcin C transport system permease protein | 0.01252 | 0.00144 | 0.01174 | 0.00026 | 0.00201 | 65 | 0.01307 | 0.00038 | 0.00202 | 93 | 0.00285 | -0.46714 | 151.093 | 0.64107 | 0.89999 |
| flgM; negative regulator of flagellin synthesis FlgM | 0.01249 | 0.00107 | 0.01215 | 0.00020 | 0.00176 | 65 | 0.01273 | 0.00017 | 0.00134 | 93 | 0.00222 | -0.26195 | 129.204 | 0.79378 | 0.94783 |
| pcoB, copB; copper resistance protein B | 0.01248 | 0.00152 | 0.01260 | 0.00039 | 0.00246 | 65 | 0.01240 | 0.00035 | 0.00193 | 93 | 0.00313 | 0.06168 | 132.394 | 0.95091 | 0.98792 |
| MCEE, epi; methylmalonyl-CoA/ethylmalonyl-CoA epimerase [EC:5.1.99.1] | 0.01248 | 0.00096 | 0.01055 | 0.00014 | 0.00146 | 65 | 0.01383 | 0.00015 | 0.00125 | 93 | 0.00192 | -1.70308 | 140.372 | 0.09077 | 0.76567 |
| fabA; 3-hydroxyacyl-[acyl-carrier protein] dehydratase / trans-2-decenoyl-[acyl-carrier protein] isomerase [EC:4.2.1.59 5.3.3.14] | 0.01245 | 0.00100 | 0.01225 | 0.00017 | 0.00161 | 65 | 0.01259 | 0.00016 | 0.00129 | 93 | 0.00206 | -0.16413 | 134.478 | 0.86987 | 0.96495 |
| rraA, menG; regulator of ribonuclease activity A | 0.01244 | 0.00105 | 0.01208 | 0.00017 | 0.00163 | 65 | 0.01269 | 0.00018 | 0.00137 | 93 | 0.00213 | -0.28591 | 138.786 | 0.77537 | 0.94269 |
| thrA; bifunctional aspartokinase / homoserine dehydrogenase 1 [EC:2.7.2.4 1.1.1.3] | 0.01243 | 0.00093 | 0.01209 | 0.00012 | 0.00137 | 65 | 0.01267 | 0.00015 | 0.00126 | 93 | 0.00186 | -0.30908 | 145.097 | 0.75771 | 0.93719 |
| murU; N-acetyl-alpha-D-muramate 1-phosphate uridylyltransferase [EC:2.7.7.99] | 0.01243 | 0.00109 | 0.01145 | 0.00019 | 0.00172 | 65 | 0.01312 | 0.00018 | 0.00141 | 93 | 0.00222 | -0.75270 | 136.053 | 0.45293 | 0.83438 |
| larC; pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickel chelatase [EC:4.99.1.12] | 0.01243 | 0.00131 | 0.01351 | 0.00033 | 0.00225 | 65 | 0.01167 | 0.00023 | 0.00158 | 93 | 0.00275 | 0.66879 | 122.159 | 0.50489 | 0.84956 |
| fliOZ, fliO; flagellar protein FliO/FliZ | 0.01242 | 0.00105 | 0.01206 | 0.00020 | 0.00177 | 65 | 0.01268 | 0.00015 | 0.00129 | 93 | 0.00219 | -0.28681 | 125.366 | 0.77473 | 0.94269 |
| PC, pyc; pyruvate carboxylase [EC:6.4.1.1] | 0.01242 | 0.00105 | 0.01198 | 0.00024 | 0.00193 | 65 | 0.01273 | 0.00013 | 0.00117 | 93 | 0.00226 | -0.33222 | 109.482 | 0.74036 | 0.92969 |
| evgS, bvgS; two-component system, NarL family, sensor histidine kinase EvgS [EC:2.7.13.3] | 0.01240 | 0.00186 | 0.01216 | 0.00050 | 0.00278 | 65 | 0.01258 | 0.00058 | 0.00250 | 93 | 0.00374 | -0.11246 | 144.044 | 0.91062 | 0.97664 |
| pilM; type IV pilus assembly protein PilM | 0.01240 | 0.00104 | 0.01191 | 0.00018 | 0.00167 | 65 | 0.01274 | 0.00017 | 0.00135 | 93 | 0.00214 | -0.38476 | 134.774 | 0.70103 | 0.91983 |
| K07045; uncharacterized protein | 0.01239 | 0.00156 | 0.01011 | 0.00027 | 0.00203 | 65 | 0.01398 | 0.00046 | 0.00222 | 93 | 0.00301 | -1.28218 | 154.693 | 0.20170 | 0.77376 |
| glnL, ntrB; two-component system, NtrC family, nitrogen regulation sensor histidine kinase GlnL [EC:2.7.13.3] | 0.01237 | 0.00119 | 0.01131 | 0.00021 | 0.00182 | 65 | 0.01312 | 0.00023 | 0.00158 | 93 | 0.00241 | -0.75229 | 140.904 | 0.45313 | 0.83438 |
| dgs, bgsA; 1,2-diacylglycerol-3-alpha-glucose alpha-1,2-glucosyltransferase [EC:2.4.1.208] | 0.01234 | 0.00104 | 0.01363 | 0.00018 | 0.00165 | 65 | 0.01144 | 0.00017 | 0.00134 | 93 | 0.00213 | 1.02969 | 135.477 | 0.30499 | 0.77376 |
| cas4; CRISPR-associated exonuclease Cas4 [EC:3.1.12.1] | 0.01233 | 0.00111 | 0.01365 | 0.00026 | 0.00200 | 65 | 0.01140 | 0.00015 | 0.00127 | 93 | 0.00237 | 0.95229 | 113.375 | 0.34297 | 0.78567 |
| mltF; membrane-bound lytic murein transglycosylase F [EC:4.2.2.-] | 0.01233 | 0.00106 | 0.01245 | 0.00017 | 0.00160 | 65 | 0.01224 | 0.00019 | 0.00143 | 93 | 0.00214 | 0.09553 | 143.264 | 0.92403 | 0.97981 |
| yejE; microcin C transport system permease protein | 0.01231 | 0.00143 | 0.01131 | 0.00025 | 0.00195 | 65 | 0.01301 | 0.00038 | 0.00202 | 93 | 0.00280 | -0.60885 | 152.751 | 0.54353 | 0.86313 |
| E3.4.17.19; carboxypeptidase Taq [EC:3.4.17.19] | 0.01225 | 0.00118 | 0.01386 | 0.00035 | 0.00232 | 65 | 0.01113 | 0.00013 | 0.00118 | 93 | 0.00260 | 1.04938 | 96.870 | 0.29661 | 0.77376 |
| tesA; acyl-CoA thioesterase I [EC:3.1.2.- 3.1.1.5] | 0.01225 | 0.00118 | 0.01121 | 0.00021 | 0.00179 | 65 | 0.01298 | 0.00023 | 0.00157 | 93 | 0.00238 | -0.74222 | 141.570 | 0.45918 | 0.83438 |
| crp; CRP/FNR family transcriptional regulator, cyclic AMP receptor protein | 0.01224 | 0.00101 | 0.01301 | 0.00016 | 0.00158 | 65 | 0.01170 | 0.00016 | 0.00132 | 93 | 0.00206 | 0.63618 | 137.478 | 0.52572 | 0.85808 |
| lpxO; beta-hydroxylase [EC:1.14.11.-] | 0.01223 | 0.00145 | 0.01138 | 0.00026 | 0.00200 | 65 | 0.01282 | 0.00038 | 0.00203 | 93 | 0.00285 | -0.50412 | 151.663 | 0.61491 | 0.88842 |
| dld; D-lactate dehydrogenase (quinone) [EC:1.1.5.12] | 0.01222 | 0.00137 | 0.01221 | 0.00031 | 0.00217 | 65 | 0.01223 | 0.00029 | 0.00178 | 93 | 0.00281 | -0.00915 | 135.900 | 0.99271 | 0.99912 |
| eptA, pmrC; lipid A ethanolaminephosphotransferase [EC:2.7.8.43] | 0.01219 | 0.00140 | 0.01124 | 0.00029 | 0.00213 | 65 | 0.01285 | 0.00032 | 0.00186 | 93 | 0.00282 | -0.57312 | 141.513 | 0.56748 | 0.87063 |
| trmA; tRNA (uracil-5-)-methyltransferase [EC:2.1.1.35] | 0.01218 | 0.00096 | 0.01256 | 0.00014 | 0.00149 | 65 | 0.01192 | 0.00015 | 0.00126 | 93 | 0.00195 | 0.33035 | 138.885 | 0.74163 | 0.93043 |
| K09975; uncharacterized protein | 0.01218 | 0.00098 | 0.01149 | 0.00014 | 0.00146 | 65 | 0.01266 | 0.00016 | 0.00131 | 93 | 0.00196 | -0.59922 | 144.011 | 0.54997 | 0.86680 |
| rnt; ribonuclease T [EC:3.1.13.-] | 0.01215 | 0.00102 | 0.01282 | 0.00017 | 0.00160 | 65 | 0.01168 | 0.00017 | 0.00134 | 93 | 0.00208 | 0.54645 | 137.804 | 0.58564 | 0.87720 |
| codA; cytosine deaminase [EC:3.5.4.1] | 0.01213 | 0.00114 | 0.01154 | 0.00020 | 0.00177 | 65 | 0.01253 | 0.00021 | 0.00150 | 93 | 0.00232 | -0.42804 | 138.705 | 0.66928 | 0.91165 |
| gabP; GABA permease | 0.01212 | 0.00192 | 0.01210 | 0.00053 | 0.00287 | 65 | 0.01214 | 0.00062 | 0.00259 | 93 | 0.00386 | -0.01094 | 144.275 | 0.99129 | 0.99871 |
| hemX; uroporphyrin-III C-methyltransferase [EC:2.1.1.107] | 0.01211 | 0.00102 | 0.01303 | 0.00018 | 0.00167 | 65 | 0.01146 | 0.00016 | 0.00130 | 93 | 0.00211 | 0.74147 | 131.619 | 0.45973 | 0.83438 |
| flhF; flagellar biosynthesis protein FlhF | 0.01210 | 0.00106 | 0.01140 | 0.00019 | 0.00171 | 65 | 0.01259 | 0.00017 | 0.00135 | 93 | 0.00218 | -0.54457 | 133.041 | 0.58696 | 0.87731 |
| ccoQ; cytochrome c oxidase cbb3-type subunit IV | 0.01210 | 0.00135 | 0.01217 | 0.00030 | 0.00213 | 65 | 0.01206 | 0.00029 | 0.00176 | 93 | 0.00276 | 0.03957 | 136.530 | 0.96849 | 0.99264 |
| hscB, HSCB, HSC20; molecular chaperone HscB | 0.01208 | 0.00102 | 0.01272 | 0.00017 | 0.00162 | 65 | 0.01163 | 0.00016 | 0.00131 | 93 | 0.00209 | 0.52091 | 135.146 | 0.60328 | 0.88434 |
| rgpB; rhamnosyltransferase [EC:2.4.1.-] | 0.01208 | 0.00107 | 0.01327 | 0.00020 | 0.00176 | 65 | 0.01124 | 0.00017 | 0.00134 | 93 | 0.00221 | 0.91763 | 129.867 | 0.36051 | 0.79496 |
| UQCRFS1, RIP1, petA; ubiquinol-cytochrome c reductase iron-sulfur subunit [EC:1.10.2.2] | 0.01207 | 0.00105 | 0.01119 | 0.00018 | 0.00167 | 65 | 0.01269 | 0.00017 | 0.00136 | 93 | 0.00215 | -0.70105 | 135.333 | 0.48448 | 0.84354 |
| hscA; molecular chaperone HscA | 0.01206 | 0.00102 | 0.01270 | 0.00017 | 0.00162 | 65 | 0.01162 | 0.00016 | 0.00131 | 93 | 0.00208 | 0.52106 | 135.273 | 0.60318 | 0.88434 |
| cld; chlorite dismutase [EC:1.13.11.49] | 0.01206 | 0.00104 | 0.01080 | 0.00017 | 0.00163 | 65 | 0.01294 | 0.00017 | 0.00136 | 93 | 0.00212 | -1.00426 | 137.609 | 0.31702 | 0.77376 |
| K07487; transposase | 0.01206 | 0.00364 | 0.02022 | 0.00411 | 0.00796 | 65 | 0.00635 | 0.00063 | 0.00260 | 93 | 0.00837 | 1.65701 | 77.781 | 0.10155 | 0.77376 |
| fadJ; 3-hydroxyacyl-CoA dehydrogenase / enoyl-CoA hydratase / 3-hydroxybutyryl-CoA epimerase [EC:1.1.1.35 4.2.1.17 5.1.2.3] | 0.01205 | 0.00102 | 0.01057 | 0.00013 | 0.00140 | 65 | 0.01309 | 0.00019 | 0.00143 | 93 | 0.00200 | -1.25709 | 152.000 | 0.21065 | 0.77376 |
| K09895; uncharacterized protein | 0.01205 | 0.00102 | 0.01282 | 0.00017 | 0.00160 | 65 | 0.01151 | 0.00017 | 0.00134 | 93 | 0.00209 | 0.62740 | 138.010 | 0.53143 | 0.86136 |
| pcoD; copper resistance protein D | 0.01204 | 0.00144 | 0.00982 | 0.00028 | 0.00208 | 65 | 0.01360 | 0.00036 | 0.00197 | 93 | 0.00286 | -1.32388 | 147.596 | 0.18759 | 0.77376 |
| nfuA; Fe/S biogenesis protein NfuA | 0.01204 | 0.00103 | 0.01280 | 0.00017 | 0.00160 | 65 | 0.01151 | 0.00017 | 0.00134 | 93 | 0.00209 | 0.61951 | 138.183 | 0.53660 | 0.86207 |
| cvrA, nhaP2; cell volume regulation protein A | 0.01203 | 0.00107 | 0.01439 | 0.00026 | 0.00199 | 65 | 0.01039 | 0.00013 | 0.00116 | 93 | 0.00230 | 1.73729 | 106.616 | 0.08522 | 0.76567 |
| zipA; cell division protein ZipA | 0.01203 | 0.00102 | 0.01280 | 0.00017 | 0.00160 | 65 | 0.01149 | 0.00017 | 0.00134 | 93 | 0.00208 | 0.62764 | 138.079 | 0.53128 | 0.86136 |
| K09902; uncharacterized protein | 0.01202 | 0.00102 | 0.01280 | 0.00017 | 0.00160 | 65 | 0.01148 | 0.00017 | 0.00134 | 93 | 0.00208 | 0.63358 | 138.095 | 0.52740 | 0.85898 |
| K07121; uncharacterized protein | 0.01202 | 0.00103 | 0.01281 | 0.00017 | 0.00159 | 65 | 0.01146 | 0.00017 | 0.00135 | 93 | 0.00209 | 0.64709 | 138.831 | 0.51864 | 0.85403 |
| PTS-Dgl-EIIC, gamP; PTS system, D-glucosamine-specific IIC component | 0.01200 | 0.00104 | 0.01125 | 0.00019 | 0.00169 | 65 | 0.01253 | 0.00016 | 0.00132 | 93 | 0.00214 | -0.59662 | 131.325 | 0.55179 | 0.86788 |
| K07039; uncharacterized protein | 0.01200 | 0.00137 | 0.01215 | 0.00031 | 0.00218 | 65 | 0.01188 | 0.00029 | 0.00176 | 93 | 0.00281 | 0.09622 | 134.706 | 0.92349 | 0.97972 |
| chrR, NQR; chromate reductase, NAD(P)H dehydrogenase (quinone) | 0.01199 | 0.00136 | 0.01157 | 0.00032 | 0.00221 | 65 | 0.01229 | 0.00028 | 0.00174 | 93 | 0.00281 | -0.25719 | 132.435 | 0.79743 | 0.94810 |
| trmJ; tRNA (cytidine32/uridine32-2’-O)-methyltransferase [EC:2.1.1.200] | 0.01199 | 0.00102 | 0.01274 | 0.00016 | 0.00159 | 65 | 0.01146 | 0.00017 | 0.00134 | 93 | 0.00208 | 0.61238 | 138.568 | 0.54129 | 0.86207 |
| ampD; N-acetyl-anhydromuramoyl-L-alanine amidase [EC:3.5.1.28] | 0.01198 | 0.00101 | 0.01256 | 0.00017 | 0.00160 | 65 | 0.01158 | 0.00016 | 0.00132 | 93 | 0.00207 | 0.47382 | 136.296 | 0.63639 | 0.89696 |
| K07164; uncharacterized protein | 0.01197 | 0.00100 | 0.01025 | 0.00010 | 0.00125 | 65 | 0.01318 | 0.00019 | 0.00144 | 93 | 0.00191 | -1.53385 | 155.787 | 0.12710 | 0.77376 |
| ABC.ZM.A; zinc/manganese transport system ATP-binding protein | 0.01196 | 0.00128 | 0.01230 | 0.00034 | 0.00228 | 65 | 0.01172 | 0.00021 | 0.00149 | 93 | 0.00272 | 0.21278 | 115.375 | 0.83188 | 0.95506 |
| ABC.X2.A; putative ABC transport system ATP-binding protein | 0.01195 | 0.00117 | 0.01152 | 0.00022 | 0.00186 | 65 | 0.01225 | 0.00021 | 0.00150 | 93 | 0.00239 | -0.30559 | 135.131 | 0.76039 | 0.93791 |
| K07506; AraC family transcriptional regulator | 0.01193 | 0.00143 | 0.01182 | 0.00034 | 0.00230 | 65 | 0.01200 | 0.00031 | 0.00183 | 93 | 0.00294 | -0.06161 | 133.431 | 0.95097 | 0.98792 |
| cobL; precorrin-6Y C5,15-methyltransferase (decarboxylating) [EC:2.1.1.132] | 0.01192 | 0.00115 | 0.01154 | 0.00031 | 0.00217 | 65 | 0.01219 | 0.00014 | 0.00123 | 93 | 0.00250 | -0.26086 | 104.288 | 0.79471 | 0.94783 |
| CYTB, petB; ubiquinol-cytochrome c reductase cytochrome b subunit | 0.01188 | 0.00104 | 0.01110 | 0.00018 | 0.00166 | 65 | 0.01242 | 0.00016 | 0.00133 | 93 | 0.00213 | -0.61936 | 134.055 | 0.53673 | 0.86207 |
| adh2; alcohol dehydrogenase [EC:1.1.1.-] | 0.01187 | 0.00105 | 0.01085 | 0.00016 | 0.00157 | 65 | 0.01257 | 0.00018 | 0.00140 | 93 | 0.00210 | -0.82062 | 143.344 | 0.41323 | 0.81655 |
| chpA; chemosensory pili system protein ChpA (sensor histidine kinase/response regulator) | 0.01186 | 0.00172 | 0.01139 | 0.00042 | 0.00254 | 65 | 0.01219 | 0.00051 | 0.00234 | 93 | 0.00345 | -0.23274 | 145.560 | 0.81629 | 0.95487 |
| amn; AMP nucleosidase [EC:3.2.2.4] | 0.01185 | 0.00108 | 0.01064 | 0.00019 | 0.00169 | 65 | 0.01270 | 0.00018 | 0.00139 | 93 | 0.00219 | -0.93911 | 136.605 | 0.34933 | 0.79120 |
| fliJ; flagellar FliJ protein | 0.01184 | 0.00101 | 0.01156 | 0.00019 | 0.00170 | 65 | 0.01203 | 0.00015 | 0.00125 | 93 | 0.00211 | -0.22175 | 126.235 | 0.82486 | 0.95506 |
| TC.CNT; concentrative nucleoside transporter, CNT family | 0.01181 | 0.00107 | 0.01217 | 0.00018 | 0.00165 | 65 | 0.01155 | 0.00018 | 0.00141 | 93 | 0.00217 | 0.28475 | 139.393 | 0.77626 | 0.94269 |
| cpo; non-heme chloroperoxidase [EC:1.11.1.10] | 0.01181 | 0.00145 | 0.01078 | 0.00033 | 0.00227 | 65 | 0.01252 | 0.00034 | 0.00190 | 93 | 0.00296 | -0.58829 | 138.343 | 0.55730 | 0.86813 |
| nirD; nitrite reductase (NADH) small subunit [EC:1.7.1.15] | 0.01181 | 0.00102 | 0.01054 | 0.00016 | 0.00157 | 65 | 0.01269 | 0.00016 | 0.00133 | 93 | 0.00206 | -1.04425 | 138.624 | 0.29819 | 0.77376 |
| E6.4.1.4A; 3-methylcrotonyl-CoA carboxylase alpha subunit [EC:6.4.1.4] | 0.01181 | 0.00124 | 0.01018 | 0.00021 | 0.00181 | 65 | 0.01294 | 0.00026 | 0.00169 | 93 | 0.00247 | -1.11616 | 146.406 | 0.26618 | 0.77376 |
| sbcB, exoI; exodeoxyribonuclease I [EC:3.1.11.1] | 0.01178 | 0.00102 | 0.01236 | 0.00017 | 0.00163 | 65 | 0.01137 | 0.00016 | 0.00131 | 93 | 0.00209 | 0.47306 | 134.500 | 0.63694 | 0.89696 |
| truD, PUS7; tRNA pseudouridine13 synthase [EC:5.4.99.27] | 0.01176 | 0.00103 | 0.01241 | 0.00016 | 0.00158 | 65 | 0.01131 | 0.00017 | 0.00136 | 93 | 0.00208 | 0.52612 | 140.883 | 0.59963 | 0.88240 |
| yoeB; toxin YoeB [EC:3.1.-.-] | 0.01174 | 0.00122 | 0.01450 | 0.00035 | 0.00233 | 65 | 0.00981 | 0.00015 | 0.00125 | 93 | 0.00265 | 1.77246 | 100.502 | 0.07935 | 0.75920 |
| pilH; twitching motility two-component system response regulator PilH | 0.01174 | 0.00141 | 0.01155 | 0.00034 | 0.00228 | 65 | 0.01187 | 0.00030 | 0.00179 | 93 | 0.00290 | -0.10854 | 132.070 | 0.91373 | 0.97664 |
| PITRM1, PreP, CYM1; presequence protease [EC:3.4.24.-] | 0.01174 | 0.00105 | 0.01272 | 0.00024 | 0.00194 | 65 | 0.01104 | 0.00012 | 0.00115 | 93 | 0.00226 | 0.74444 | 108.012 | 0.45823 | 0.83438 |
| prpB; methylisocitrate lyase [EC:4.1.3.30] | 0.01173 | 0.00106 | 0.01135 | 0.00019 | 0.00169 | 65 | 0.01200 | 0.00017 | 0.00136 | 93 | 0.00217 | -0.30159 | 134.564 | 0.76343 | 0.93836 |
| tagE; poly(glycerol-phosphate) alpha-glucosyltransferase [EC:2.4.1.52] | 0.01170 | 0.00143 | 0.01236 | 0.00048 | 0.00271 | 65 | 0.01125 | 0.00022 | 0.00152 | 93 | 0.00311 | 0.35657 | 103.449 | 0.72214 | 0.92156 |
| nuoD; NADH-quinone oxidoreductase subunit D [EC:1.6.5.3] | 0.01167 | 0.00101 | 0.00934 | 0.00013 | 0.00141 | 65 | 0.01330 | 0.00018 | 0.00138 | 93 | 0.00198 | -2.00085 | 149.732 | 0.04722 | 0.66255 |
| selB, EEFSEC; selenocysteine-specific elongation factor | 0.01167 | 0.00098 | 0.01366 | 0.00020 | 0.00177 | 65 | 0.01028 | 0.00011 | 0.00111 | 93 | 0.00209 | 1.61849 | 111.870 | 0.10837 | 0.77376 |
| yccA; modulator of FtsH protease | 0.01165 | 0.00103 | 0.01236 | 0.00016 | 0.00158 | 65 | 0.01116 | 0.00017 | 0.00136 | 93 | 0.00209 | 0.57694 | 140.336 | 0.56490 | 0.86988 |
| K09167; uncharacterized protein | 0.01165 | 0.00114 | 0.01099 | 0.00021 | 0.00180 | 65 | 0.01211 | 0.00020 | 0.00147 | 93 | 0.00233 | -0.48121 | 136.056 | 0.63114 | 0.89595 |
| K07019; uncharacterized protein | 0.01164 | 0.00100 | 0.01164 | 0.00017 | 0.00160 | 65 | 0.01165 | 0.00016 | 0.00130 | 93 | 0.00206 | -0.00454 | 135.054 | 0.99638 | 0.99944 |
| yneE, BEST; ion channel-forming bestrophin family protein | 0.01161 | 0.00130 | 0.01096 | 0.00022 | 0.00186 | 65 | 0.01207 | 0.00030 | 0.00180 | 93 | 0.00259 | -0.43183 | 149.011 | 0.66649 | 0.91126 |
| bfd; bacterioferritin-associated ferredoxin | 0.01161 | 0.00101 | 0.01171 | 0.00017 | 0.00163 | 65 | 0.01154 | 0.00015 | 0.00128 | 93 | 0.00207 | 0.08004 | 132.260 | 0.93633 | 0.98459 |
| ku; DNA end-binding protein Ku | 0.01159 | 0.00114 | 0.00986 | 0.00019 | 0.00171 | 65 | 0.01280 | 0.00021 | 0.00152 | 93 | 0.00229 | -1.28273 | 142.911 | 0.20166 | 0.77376 |
| arsH; arsenical resistance protein ArsH | 0.01157 | 0.00139 | 0.01079 | 0.00033 | 0.00226 | 65 | 0.01212 | 0.00029 | 0.00177 | 93 | 0.00287 | -0.46562 | 132.042 | 0.64226 | 0.90098 |
| K09949; uncharacterized protein | 0.01157 | 0.00116 | 0.01365 | 0.00032 | 0.00221 | 65 | 0.01012 | 0.00014 | 0.00122 | 93 | 0.00252 | 1.39869 | 102.723 | 0.16492 | 0.77376 |
| kgtP; MFS transporter, MHS family, alpha-ketoglutarate permease | 0.01157 | 0.00124 | 0.01004 | 0.00020 | 0.00176 | 65 | 0.01264 | 0.00027 | 0.00171 | 93 | 0.00245 | -1.06148 | 149.391 | 0.29018 | 0.77376 |
| algI; alginate O-acetyltransferase complex protein AlgI | 0.01157 | 0.00137 | 0.01270 | 0.00030 | 0.00214 | 65 | 0.01078 | 0.00030 | 0.00179 | 93 | 0.00279 | 0.68840 | 137.760 | 0.49236 | 0.84797 |
| mcsB; protein arginine kinase [EC:2.7.14.1] | 0.01155 | 0.00109 | 0.01205 | 0.00025 | 0.00196 | 65 | 0.01120 | 0.00015 | 0.00127 | 93 | 0.00233 | 0.36526 | 114.868 | 0.71559 | 0.92004 |
| CYC1, CYT1, petC; ubiquinol-cytochrome c reductase cytochrome c1 subunit | 0.01154 | 0.00103 | 0.01094 | 0.00018 | 0.00166 | 65 | 0.01196 | 0.00016 | 0.00132 | 93 | 0.00212 | -0.48097 | 133.207 | 0.63132 | 0.89602 |
| nrfA; nitrite reductase (cytochrome c-552) [EC:1.7.2.2] | 0.01153 | 0.00117 | 0.01189 | 0.00020 | 0.00173 | 65 | 0.01127 | 0.00024 | 0.00159 | 93 | 0.00235 | 0.26611 | 145.451 | 0.79053 | 0.94658 |
| E2.6.1.83; LL-diaminopimelate aminotransferase [EC:2.6.1.83] | 0.01152 | 0.00118 | 0.01150 | 0.00025 | 0.00195 | 65 | 0.01154 | 0.00020 | 0.00148 | 93 | 0.00244 | -0.01297 | 129.019 | 0.98967 | 0.99801 |
| PCBD, phhB; 4a-hydroxytetrahydrobiopterin dehydratase [EC:4.2.1.96] | 0.01152 | 0.00107 | 0.00980 | 0.00019 | 0.00169 | 65 | 0.01272 | 0.00018 | 0.00138 | 93 | 0.00218 | -1.34033 | 135.813 | 0.18237 | 0.77376 |
| dat; D-alanine transaminase [EC:2.6.1.21] | 0.01152 | 0.00098 | 0.01035 | 0.00016 | 0.00157 | 65 | 0.01234 | 0.00014 | 0.00124 | 93 | 0.00201 | -0.99283 | 132.883 | 0.32260 | 0.77376 |
| ATE1; arginyl-tRNA—protein transferase [EC:2.3.2.8] | 0.01151 | 0.00109 | 0.01064 | 0.00020 | 0.00175 | 65 | 0.01212 | 0.00018 | 0.00140 | 93 | 0.00224 | -0.66458 | 133.605 | 0.50747 | 0.84956 |
| EIF1, SUI1; translation initiation factor 1 | 0.01151 | 0.00098 | 0.01175 | 0.00016 | 0.00155 | 65 | 0.01134 | 0.00015 | 0.00127 | 93 | 0.00200 | 0.20547 | 136.254 | 0.83751 | 0.95506 |
| dld; D-lactate dehydrogenase | 0.01151 | 0.00096 | 0.01227 | 0.00016 | 0.00159 | 65 | 0.01097 | 0.00013 | 0.00120 | 93 | 0.00199 | 0.65229 | 129.193 | 0.51537 | 0.85216 |
| fadE; acyl-CoA dehydrogenase [EC:1.3.99.-] | 0.01151 | 0.00138 | 0.01095 | 0.00026 | 0.00200 | 65 | 0.01190 | 0.00033 | 0.00189 | 93 | 0.00275 | -0.34580 | 147.552 | 0.72998 | 0.92426 |
| pilN; type IV pilus assembly protein PilN | 0.01151 | 0.00106 | 0.01163 | 0.00020 | 0.00174 | 65 | 0.01142 | 0.00017 | 0.00134 | 93 | 0.00219 | 0.09395 | 130.663 | 0.92529 | 0.98041 |
| gluP; rhomboid protease GluP [EC:3.4.21.105] | 0.01148 | 0.00154 | 0.01342 | 0.00060 | 0.00303 | 65 | 0.01013 | 0.00022 | 0.00154 | 93 | 0.00340 | 0.96828 | 96.943 | 0.33532 | 0.78186 |
| E1.2.1.68; coniferyl-aldehyde dehydrogenase [EC:1.2.1.68] | 0.01148 | 0.00123 | 0.01052 | 0.00021 | 0.00181 | 65 | 0.01215 | 0.00026 | 0.00166 | 93 | 0.00245 | -0.66800 | 145.514 | 0.50519 | 0.84956 |
| PTS-Fru1-EIID, levG; PTS system, fructose-specific IID component | 0.01148 | 0.00116 | 0.01005 | 0.00015 | 0.00150 | 65 | 0.01248 | 0.00026 | 0.00168 | 93 | 0.00225 | -1.07971 | 155.209 | 0.28195 | 0.77376 |
| copC, pcoC; copper resistance protein C | 0.01147 | 0.00138 | 0.00967 | 0.00028 | 0.00208 | 65 | 0.01273 | 0.00031 | 0.00184 | 93 | 0.00278 | -1.10369 | 142.235 | 0.27159 | 0.77376 |
| astA; arginine N-succinyltransferase [EC:2.3.1.109] | 0.01146 | 0.00150 | 0.01192 | 0.00037 | 0.00240 | 65 | 0.01114 | 0.00035 | 0.00194 | 93 | 0.00308 | 0.25224 | 134.812 | 0.80124 | 0.94855 |
| tagD; glycerol-3-phosphate cytidylyltransferase [EC:2.7.7.39] | 0.01145 | 0.00113 | 0.01332 | 0.00028 | 0.00206 | 65 | 0.01014 | 0.00015 | 0.00126 | 93 | 0.00241 | 1.31998 | 110.117 | 0.18958 | 0.77376 |
| TC.BAT1; bacterial/archaeal transporter family protein | 0.01144 | 0.00096 | 0.01133 | 0.00019 | 0.00169 | 65 | 0.01152 | 0.00012 | 0.00114 | 93 | 0.00204 | -0.09707 | 118.180 | 0.92283 | 0.97947 |
| pilO; type IV pilus assembly protein PilO | 0.01143 | 0.00099 | 0.01143 | 0.00018 | 0.00164 | 65 | 0.01143 | 0.00014 | 0.00123 | 93 | 0.00205 | 0.00258 | 128.057 | 0.99794 | 0.99955 |
| K07138; uncharacterized protein | 0.01142 | 0.00131 | 0.01206 | 0.00032 | 0.00220 | 65 | 0.01098 | 0.00024 | 0.00161 | 93 | 0.00273 | 0.39588 | 125.405 | 0.69286 | 0.91677 |
| efrA; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.01142 | 0.00104 | 0.01212 | 0.00018 | 0.00165 | 65 | 0.01093 | 0.00017 | 0.00135 | 93 | 0.00213 | 0.56060 | 135.737 | 0.57600 | 0.87389 |
| efrB; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.01142 | 0.00104 | 0.01212 | 0.00018 | 0.00165 | 65 | 0.01093 | 0.00017 | 0.00135 | 93 | 0.00213 | 0.56060 | 135.737 | 0.57600 | 0.87389 |
| cysU; sulfate transport system permease protein | 0.01141 | 0.00096 | 0.01025 | 0.00012 | 0.00136 | 65 | 0.01222 | 0.00016 | 0.00133 | 93 | 0.00190 | -1.03734 | 149.750 | 0.30125 | 0.77376 |
| cysW; sulfate transport system permease protein | 0.01141 | 0.00096 | 0.01025 | 0.00012 | 0.00136 | 65 | 0.01222 | 0.00016 | 0.00133 | 93 | 0.00190 | -1.03734 | 149.750 | 0.30125 | 0.77376 |
| hrpB; ATP-dependent helicase HrpB [EC:3.6.4.13] | 0.01141 | 0.00105 | 0.01080 | 0.00018 | 0.00164 | 65 | 0.01183 | 0.00017 | 0.00136 | 93 | 0.00213 | -0.48052 | 137.302 | 0.63162 | 0.89607 |
| dusC; tRNA-dihydrouridine synthase C [EC:1.-.-.-] | 0.01140 | 0.00100 | 0.01205 | 0.00016 | 0.00158 | 65 | 0.01095 | 0.00015 | 0.00129 | 93 | 0.00204 | 0.53814 | 135.957 | 0.59136 | 0.87896 |
| arsB; arsenical pump membrane protein | 0.01140 | 0.00109 | 0.00996 | 0.00015 | 0.00154 | 65 | 0.01240 | 0.00021 | 0.00151 | 93 | 0.00216 | -1.13234 | 149.899 | 0.25930 | 0.77376 |
| cheD; chemotaxis protein CheD [EC:3.5.1.44] | 0.01138 | 0.00103 | 0.01003 | 0.00017 | 0.00160 | 65 | 0.01233 | 0.00017 | 0.00135 | 93 | 0.00209 | -1.09748 | 139.062 | 0.27433 | 0.77376 |
| argAB; amino-acid N-acetyltransferase [EC:2.3.1.1] | 0.01136 | 0.00099 | 0.01210 | 0.00016 | 0.00156 | 65 | 0.01085 | 0.00015 | 0.00129 | 93 | 0.00202 | 0.61765 | 136.758 | 0.53784 | 0.86207 |
| chlD, bchD; magnesium chelatase subunit D [EC:6.6.1.1] | 0.01136 | 0.00126 | 0.01221 | 0.00032 | 0.00222 | 65 | 0.01076 | 0.00020 | 0.00148 | 93 | 0.00267 | 0.54287 | 117.402 | 0.58825 | 0.87732 |
| yefM; antitoxin YefM | 0.01135 | 0.00091 | 0.01032 | 0.00008 | 0.00111 | 65 | 0.01207 | 0.00017 | 0.00133 | 93 | 0.00174 | -1.00865 | 155.999 | 0.31471 | 0.77376 |
| aldB; aldehyde dehydrogenase [EC:1.2.1.-] | 0.01135 | 0.00118 | 0.00985 | 0.00020 | 0.00175 | 65 | 0.01240 | 0.00024 | 0.00159 | 93 | 0.00236 | -1.07454 | 144.773 | 0.28437 | 0.77376 |
| flgF; flagellar basal-body rod protein FlgF | 0.01135 | 0.00116 | 0.01065 | 0.00022 | 0.00183 | 65 | 0.01183 | 0.00021 | 0.00150 | 93 | 0.00237 | -0.49965 | 136.649 | 0.61813 | 0.88903 |
| imuB; protein ImuB | 0.01134 | 0.00104 | 0.01039 | 0.00020 | 0.00177 | 65 | 0.01201 | 0.00015 | 0.00125 | 93 | 0.00217 | -0.74363 | 122.633 | 0.45852 | 0.83438 |
| mcsA; protein arginine kinase activator | 0.01132 | 0.00104 | 0.01161 | 0.00022 | 0.00182 | 65 | 0.01112 | 0.00014 | 0.00124 | 93 | 0.00220 | 0.21924 | 118.883 | 0.82684 | 0.95506 |
| glpT; MFS transporter, OPA family, glycerol-3-phosphate transporter | 0.01132 | 0.00092 | 0.01146 | 0.00016 | 0.00156 | 65 | 0.01122 | 0.00012 | 0.00112 | 93 | 0.00192 | 0.12598 | 124.266 | 0.89995 | 0.97494 |
| fimA; major type 1 subunit fimbrin (pilin) | 0.01131 | 0.00160 | 0.00975 | 0.00017 | 0.00160 | 65 | 0.01241 | 0.00057 | 0.00248 | 93 | 0.00295 | -0.90355 | 147.536 | 0.36771 | 0.79587 |
| algA, xanB, rfbA, wbpW, pslB; mannose-1-phosphate guanylyltransferase / mannose-6-phosphate isomerase [EC:2.7.7.13 5.3.1.8] | 0.01128 | 0.00141 | 0.01034 | 0.00028 | 0.00207 | 65 | 0.01193 | 0.00034 | 0.00191 | 93 | 0.00282 | -0.56462 | 145.798 | 0.57320 | 0.87292 |
| fliK; flagellar hook-length control protein FliK | 0.01128 | 0.00104 | 0.01139 | 0.00019 | 0.00171 | 65 | 0.01120 | 0.00016 | 0.00131 | 93 | 0.00216 | 0.08867 | 130.207 | 0.92948 | 0.98183 |
| flhG, fleN; flagellar biosynthesis protein FlhG | 0.01127 | 0.00101 | 0.01050 | 0.00018 | 0.00165 | 65 | 0.01181 | 0.00015 | 0.00128 | 93 | 0.00209 | -0.62808 | 131.319 | 0.53104 | 0.86136 |
| ccmD; heme exporter protein D | 0.01127 | 0.00099 | 0.01165 | 0.00016 | 0.00159 | 65 | 0.01100 | 0.00015 | 0.00127 | 93 | 0.00204 | 0.31616 | 133.835 | 0.75237 | 0.93565 |
| rgpF; rhamnosyltransferase [EC:2.4.1.-] | 0.01126 | 0.00103 | 0.01198 | 0.00017 | 0.00160 | 65 | 0.01076 | 0.00017 | 0.00134 | 93 | 0.00209 | 0.58650 | 138.342 | 0.55850 | 0.86813 |
| impG, vasA; type VI secretion system protein ImpG | 0.01126 | 0.00142 | 0.01223 | 0.00028 | 0.00208 | 65 | 0.01057 | 0.00035 | 0.00193 | 93 | 0.00284 | 0.58437 | 146.248 | 0.55987 | 0.86813 |
| K07078; uncharacterized protein | 0.01124 | 0.00119 | 0.01247 | 0.00031 | 0.00218 | 65 | 0.01037 | 0.00016 | 0.00133 | 93 | 0.00255 | 0.82349 | 109.975 | 0.41201 | 0.81559 |
| tctB; putative tricarboxylic transport membrane protein | 0.01122 | 0.00115 | 0.01138 | 0.00024 | 0.00193 | 65 | 0.01111 | 0.00019 | 0.00143 | 93 | 0.00240 | 0.11294 | 126.906 | 0.91026 | 0.97664 |
| nuoC; NADH-quinone oxidoreductase subunit C [EC:1.6.5.3] | 0.01122 | 0.00099 | 0.00915 | 0.00013 | 0.00141 | 65 | 0.01266 | 0.00017 | 0.00134 | 93 | 0.00195 | -1.79813 | 147.660 | 0.07420 | 0.74128 |
| K09160; uncharacterized protein | 0.01122 | 0.00095 | 0.01104 | 0.00014 | 0.00146 | 65 | 0.01134 | 0.00015 | 0.00125 | 93 | 0.00193 | -0.15739 | 139.677 | 0.87517 | 0.96693 |
| alkA; DNA-3-methyladenine glycosylase II [EC:3.2.2.21] | 0.01122 | 0.00111 | 0.00877 | 0.00015 | 0.00149 | 65 | 0.01292 | 0.00023 | 0.00156 | 93 | 0.00216 | -1.92138 | 152.969 | 0.05654 | 0.70577 |
| gcd; quinoprotein glucose dehydrogenase [EC:1.1.5.2] | 0.01121 | 0.00128 | 0.01064 | 0.00024 | 0.00193 | 65 | 0.01161 | 0.00027 | 0.00171 | 93 | 0.00257 | -0.38001 | 142.702 | 0.70451 | 0.92004 |
| citF; citrate lyase subunit alpha / citrate CoA-transferase [EC:2.8.3.10] | 0.01121 | 0.00110 | 0.01454 | 0.00030 | 0.00213 | 65 | 0.00888 | 0.00011 | 0.00109 | 93 | 0.00239 | 2.36468 | 97.019 | 0.02004 | 0.59095 |
| yahK; uncharacterized zinc-type alcohol dehydrogenase-like protein [EC:1.-.-.-] | 0.01121 | 0.00125 | 0.00977 | 0.00018 | 0.00167 | 65 | 0.01221 | 0.00029 | 0.00177 | 93 | 0.00243 | -0.99845 | 153.470 | 0.31963 | 0.77376 |
| ectB, dat; diaminobutyrate-2-oxoglutarate transaminase [EC:2.6.1.76] | 0.01119 | 0.00139 | 0.01061 | 0.00026 | 0.00201 | 65 | 0.01160 | 0.00034 | 0.00190 | 93 | 0.00276 | -0.35628 | 147.714 | 0.72214 | 0.92156 |
| apaG; ApaG protein | 0.01119 | 0.00100 | 0.01044 | 0.00018 | 0.00165 | 65 | 0.01171 | 0.00015 | 0.00127 | 93 | 0.00208 | -0.60955 | 130.447 | 0.54322 | 0.86313 |
| cysA; sulfate transport system ATP-binding protein [EC:3.6.3.25] | 0.01119 | 0.00092 | 0.01027 | 0.00012 | 0.00136 | 65 | 0.01183 | 0.00014 | 0.00124 | 93 | 0.00184 | -0.84551 | 145.013 | 0.39922 | 0.80678 |
| hutC; GntR family transcriptional regulator, histidine utilization repressor | 0.01118 | 0.00115 | 0.01021 | 0.00019 | 0.00172 | 65 | 0.01186 | 0.00022 | 0.00154 | 93 | 0.00231 | -0.71749 | 143.351 | 0.47424 | 0.84073 |
| phoH2; PhoH-like ATPase | 0.01118 | 0.00101 | 0.01023 | 0.00017 | 0.00164 | 65 | 0.01184 | 0.00015 | 0.00128 | 93 | 0.00208 | -0.77528 | 131.857 | 0.43956 | 0.83156 |
| rseC; sigma-E factor negative regulatory protein RseC | 0.01118 | 0.00107 | 0.01342 | 0.00025 | 0.00195 | 65 | 0.00960 | 0.00013 | 0.00119 | 93 | 0.00228 | 1.67747 | 109.800 | 0.09630 | 0.76667 |
| crt; enoyl-CoA hydratase [EC:4.2.1.17] | 0.01116 | 0.00111 | 0.01302 | 0.00030 | 0.00215 | 65 | 0.00985 | 0.00012 | 0.00112 | 93 | 0.00243 | 1.30735 | 98.468 | 0.19414 | 0.77376 |
| algR; two-component system, LytTR family, response regulator AlgR | 0.01115 | 0.00113 | 0.01105 | 0.00020 | 0.00174 | 65 | 0.01122 | 0.00021 | 0.00149 | 93 | 0.00229 | -0.07452 | 140.154 | 0.94070 | 0.98528 |
| rsmJ; 16S rRNA (guanine1516-N2)-methyltransferase [EC:2.1.1.242] | 0.01114 | 0.00097 | 0.01200 | 0.00015 | 0.00154 | 65 | 0.01054 | 0.00015 | 0.00126 | 93 | 0.00199 | 0.73611 | 135.904 | 0.46293 | 0.83654 |
| alpA; prophage regulatory protein | 0.01112 | 0.00118 | 0.01194 | 0.00027 | 0.00205 | 65 | 0.01055 | 0.00018 | 0.00141 | 93 | 0.00249 | 0.56050 | 119.869 | 0.57619 | 0.87389 |
| mdaB; modulator of drug activity B | 0.01110 | 0.00091 | 0.01194 | 0.00015 | 0.00153 | 65 | 0.01052 | 0.00012 | 0.00113 | 93 | 0.00190 | 0.74356 | 126.438 | 0.45852 | 0.83438 |
| aes; acetyl esterase [EC:3.1.1.-] | 0.01110 | 0.00116 | 0.00902 | 0.00015 | 0.00151 | 65 | 0.01255 | 0.00026 | 0.00166 | 93 | 0.00225 | -1.57037 | 154.849 | 0.11837 | 0.77376 |
| atoE; short-chain fatty acids transporter | 0.01110 | 0.00111 | 0.01263 | 0.00025 | 0.00197 | 65 | 0.01003 | 0.00015 | 0.00128 | 93 | 0.00235 | 1.10669 | 115.575 | 0.27073 | 0.77376 |
| citD; citrate lyase subunit gamma (acyl carrier protein) | 0.01108 | 0.00110 | 0.01441 | 0.00029 | 0.00211 | 65 | 0.00876 | 0.00011 | 0.00109 | 93 | 0.00237 | 2.37968 | 98.025 | 0.01926 | 0.59095 |
| yaeJ; ribosome-associated protein | 0.01108 | 0.00105 | 0.01033 | 0.00019 | 0.00169 | 65 | 0.01161 | 0.00016 | 0.00133 | 93 | 0.00215 | -0.59485 | 132.478 | 0.55296 | 0.86788 |
| aphA; kanamycin kinase [EC:2.7.1.95] | 0.01108 | 0.00088 | 0.01162 | 0.00013 | 0.00140 | 65 | 0.01070 | 0.00012 | 0.00115 | 93 | 0.00181 | 0.51041 | 136.489 | 0.61059 | 0.88722 |
| slyB; outer membrane lipoprotein SlyB | 0.01108 | 0.00097 | 0.01202 | 0.00016 | 0.00158 | 65 | 0.01042 | 0.00014 | 0.00122 | 93 | 0.00200 | 0.79748 | 130.914 | 0.42662 | 0.82661 |
| PTS-HPR.FRUB, fruB, fpr; phosphocarrier protein FPr | 0.01106 | 0.00134 | 0.01128 | 0.00024 | 0.00191 | 65 | 0.01091 | 0.00032 | 0.00186 | 93 | 0.00267 | 0.13678 | 149.260 | 0.89139 | 0.97141 |
| speB; agmatinase [EC:3.5.3.11] | 0.01104 | 0.00097 | 0.01040 | 0.00014 | 0.00147 | 65 | 0.01150 | 0.00016 | 0.00129 | 93 | 0.00196 | -0.55952 | 142.109 | 0.57669 | 0.87389 |
| oprB; porin | 0.01099 | 0.00145 | 0.01021 | 0.00026 | 0.00202 | 65 | 0.01154 | 0.00038 | 0.00203 | 93 | 0.00286 | -0.46283 | 151.457 | 0.64415 | 0.90228 |
| dctD; two-component system, NtrC family, C4-dicarboxylate transport response regulator DctD | 0.01096 | 0.00177 | 0.01115 | 0.00048 | 0.00271 | 65 | 0.01082 | 0.00051 | 0.00235 | 93 | 0.00359 | 0.09197 | 140.690 | 0.92685 | 0.98128 |
| chlI, bchI; magnesium chelatase subunit I [EC:6.6.1.1] | 0.01093 | 0.00090 | 0.01168 | 0.00015 | 0.00152 | 65 | 0.01041 | 0.00011 | 0.00109 | 93 | 0.00187 | 0.67774 | 124.200 | 0.49920 | 0.84856 |
| fimV; pilus assembly protein FimV | 0.01092 | 0.00130 | 0.01172 | 0.00030 | 0.00214 | 65 | 0.01037 | 0.00025 | 0.00163 | 93 | 0.00269 | 0.50206 | 129.229 | 0.61648 | 0.88903 |
| K09929; uncharacterized protein | 0.01088 | 0.00136 | 0.01182 | 0.00032 | 0.00222 | 65 | 0.01023 | 0.00028 | 0.00173 | 93 | 0.00281 | 0.56405 | 131.584 | 0.57368 | 0.87292 |
| ompR; two-component system, OmpR family, phosphate regulon response regulator OmpR | 0.01088 | 0.00130 | 0.00974 | 0.00023 | 0.00188 | 65 | 0.01168 | 0.00030 | 0.00178 | 93 | 0.00259 | -0.74795 | 147.950 | 0.45567 | 0.83438 |
| ybdL; methionine transaminase [EC:2.6.1.88] | 0.01082 | 0.00113 | 0.01068 | 0.00020 | 0.00177 | 65 | 0.01091 | 0.00020 | 0.00148 | 93 | 0.00231 | -0.09880 | 137.618 | 0.92144 | 0.97905 |
| mexK; multidrug efflux pump | 0.01081 | 0.00123 | 0.01122 | 0.00024 | 0.00193 | 65 | 0.01053 | 0.00024 | 0.00159 | 93 | 0.00250 | 0.27702 | 136.418 | 0.78219 | 0.94310 |
| oxlT; MFS transporter, OFA family, oxalate/formate antiporter | 0.01081 | 0.00099 | 0.01046 | 0.00016 | 0.00158 | 65 | 0.01105 | 0.00015 | 0.00128 | 93 | 0.00203 | -0.29132 | 134.712 | 0.77126 | 0.94061 |
| pafA; proteasome accessory factor A [EC:6.3.1.19] | 0.01081 | 0.00137 | 0.01080 | 0.00032 | 0.00221 | 65 | 0.01082 | 0.00029 | 0.00176 | 93 | 0.00283 | -0.00723 | 133.521 | 0.99424 | 0.99919 |
| rcsC; two-component system, NarL family, capsular synthesis sensor histidine kinase RcsC [EC:2.7.13.3] | 0.01080 | 0.00222 | 0.00991 | 0.00062 | 0.00308 | 65 | 0.01142 | 0.00090 | 0.00312 | 93 | 0.00438 | -0.34449 | 151.642 | 0.73095 | 0.92481 |
| liuC; methylglutaconyl-CoA hydratase [EC:4.2.1.18] | 0.01079 | 0.00103 | 0.01002 | 0.00018 | 0.00165 | 65 | 0.01133 | 0.00017 | 0.00133 | 93 | 0.00212 | -0.61992 | 135.165 | 0.53636 | 0.86207 |
| K07093; uncharacterized protein | 0.01079 | 0.00130 | 0.00987 | 0.00024 | 0.00194 | 65 | 0.01143 | 0.00028 | 0.00175 | 93 | 0.00261 | -0.59525 | 144.147 | 0.55261 | 0.86788 |
| proW; glycine betaine/proline transport system permease protein | 0.01079 | 0.00139 | 0.01068 | 0.00032 | 0.00223 | 65 | 0.01086 | 0.00029 | 0.00177 | 93 | 0.00285 | -0.06319 | 133.528 | 0.94971 | 0.98774 |
| prpC; 2-methylcitrate synthase [EC:2.3.3.5] | 0.01077 | 0.00104 | 0.01088 | 0.00018 | 0.00166 | 65 | 0.01070 | 0.00017 | 0.00134 | 93 | 0.00214 | 0.08280 | 134.871 | 0.93413 | 0.98428 |
| clsC; cardiolipin synthase C [EC:2.7.8.-] | 0.01077 | 0.00105 | 0.01122 | 0.00018 | 0.00167 | 65 | 0.01046 | 0.00017 | 0.00136 | 93 | 0.00216 | 0.35493 | 135.914 | 0.72319 | 0.92156 |
| hexR; RpiR family transcriptional regulator, carbohydrate utilization regulator | 0.01075 | 0.00137 | 0.01190 | 0.00031 | 0.00219 | 65 | 0.00996 | 0.00029 | 0.00176 | 93 | 0.00281 | 0.69026 | 134.118 | 0.49123 | 0.84797 |
| paiB; transcriptional regulator | 0.01075 | 0.00106 | 0.00933 | 0.00019 | 0.00169 | 65 | 0.01174 | 0.00017 | 0.00135 | 93 | 0.00216 | -1.11382 | 133.575 | 0.26736 | 0.77376 |
| rimK; ribosomal protein S6–L-glutamate ligase [EC:6.3.2.-] | 0.01073 | 0.00098 | 0.01167 | 0.00017 | 0.00160 | 65 | 0.01008 | 0.00014 | 0.00124 | 93 | 0.00202 | 0.78719 | 131.006 | 0.43260 | 0.82862 |
| K06978; uncharacterized protein | 0.01073 | 0.00136 | 0.00819 | 0.00028 | 0.00207 | 65 | 0.01251 | 0.00030 | 0.00178 | 93 | 0.00273 | -1.57953 | 140.293 | 0.11647 | 0.77376 |
| slyA; MarR family transcriptional regulator, transcriptional regulator for hemolysin | 0.01072 | 0.00128 | 0.01041 | 0.00024 | 0.00190 | 65 | 0.01093 | 0.00028 | 0.00173 | 93 | 0.00257 | -0.20161 | 144.713 | 0.84050 | 0.95506 |
| exuT; MFS transporter, ACS family, hexuronate transporter | 0.01072 | 0.00112 | 0.00964 | 0.00020 | 0.00173 | 65 | 0.01147 | 0.00020 | 0.00147 | 93 | 0.00227 | -0.80546 | 138.885 | 0.42193 | 0.82295 |
| soxB; sarcosine oxidase, subunit beta [EC:1.5.3.1] | 0.01071 | 0.00172 | 0.00906 | 0.00033 | 0.00224 | 65 | 0.01186 | 0.00057 | 0.00247 | 93 | 0.00333 | -0.84086 | 154.968 | 0.40172 | 0.80850 |
| impK, ompA, vasF, dotU; type VI secretion system protein ImpK | 0.01070 | 0.00127 | 0.01158 | 0.00023 | 0.00187 | 65 | 0.01009 | 0.00027 | 0.00172 | 93 | 0.00254 | 0.58736 | 145.653 | 0.55787 | 0.86813 |
| rcsB; two-component system, NarL family, captular synthesis response regulator RcsB | 0.01070 | 0.00183 | 0.00973 | 0.00055 | 0.00291 | 65 | 0.01137 | 0.00052 | 0.00237 | 93 | 0.00375 | -0.43617 | 135.493 | 0.66341 | 0.91006 |
| pbpB; penicillin-binding protein 2B | 0.01068 | 0.00131 | 0.01103 | 0.00040 | 0.00247 | 65 | 0.01043 | 0.00019 | 0.00143 | 93 | 0.00285 | 0.20896 | 105.634 | 0.83488 | 0.95506 |
| hypC; hydrogenase expression/formation protein HypC | 0.01067 | 0.00099 | 0.01029 | 0.00014 | 0.00147 | 65 | 0.01094 | 0.00016 | 0.00133 | 93 | 0.00199 | -0.32809 | 144.199 | 0.74332 | 0.93099 |
| lpxC-fabZ; UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase / 3-hydroxyacyl-[acyl-carrier-protein] dehydratase [EC:3.5.1.108 4.2.1.59] | 0.01067 | 0.00115 | 0.01001 | 0.00017 | 0.00160 | 65 | 0.01113 | 0.00024 | 0.00161 | 93 | 0.00227 | -0.49655 | 151.107 | 0.62023 | 0.89039 |
| kptA; putative RNA 2’-phosphotransferase [EC:2.7.1.-] | 0.01066 | 0.00181 | 0.01564 | 0.00101 | 0.00395 | 65 | 0.00718 | 0.00015 | 0.00128 | 93 | 0.00415 | 2.04053 | 77.510 | 0.04470 | 0.65981 |
| lapC; membrane fusion protein, adhesin transport system | 0.01066 | 0.00157 | 0.01129 | 0.00036 | 0.00237 | 65 | 0.01021 | 0.00041 | 0.00209 | 93 | 0.00316 | 0.34168 | 142.639 | 0.73310 | 0.92649 |
| rlmM; 23S rRNA (cytidine2498-2’-O)-methyltransferase [EC:2.1.1.186] | 0.01064 | 0.00097 | 0.01156 | 0.00016 | 0.00155 | 65 | 0.01000 | 0.00014 | 0.00124 | 93 | 0.00199 | 0.78549 | 133.932 | 0.43356 | 0.82874 |
| pcaB; 3-carboxy-cis,cis-muconate cycloisomerase [EC:5.5.1.2] | 0.01060 | 0.00178 | 0.00946 | 0.00040 | 0.00248 | 65 | 0.01140 | 0.00057 | 0.00248 | 93 | 0.00351 | -0.55369 | 150.965 | 0.58061 | 0.87573 |
| dcuB; anaerobic C4-dicarboxylate transporter DcuB | 0.01060 | 0.00121 | 0.01091 | 0.00025 | 0.00195 | 65 | 0.01038 | 0.00022 | 0.00155 | 93 | 0.00249 | 0.21136 | 133.270 | 0.83293 | 0.95506 |
| uraH, pucM, hiuH; 5-hydroxyisourate hydrolase [EC:3.5.2.17] | 0.01059 | 0.00120 | 0.00920 | 0.00018 | 0.00168 | 65 | 0.01157 | 0.00026 | 0.00167 | 93 | 0.00237 | -0.99886 | 150.474 | 0.31947 | 0.77376 |
| impJ, vasE; type VI secretion system protein ImpJ | 0.01059 | 0.00137 | 0.01142 | 0.00025 | 0.00197 | 65 | 0.01001 | 0.00033 | 0.00189 | 93 | 0.00273 | 0.51425 | 148.251 | 0.60784 | 0.88637 |
| fitB; toxin FitB [EC:3.1.-.-] | 0.01058 | 0.00108 | 0.00898 | 0.00016 | 0.00158 | 65 | 0.01170 | 0.00020 | 0.00146 | 93 | 0.00215 | -1.26157 | 146.042 | 0.20912 | 0.77376 |
| phnP; phosphoribosyl 1,2-cyclic phosphate phosphodiesterase [EC:3.1.4.55] | 0.01058 | 0.00086 | 0.00839 | 0.00008 | 0.00108 | 65 | 0.01211 | 0.00014 | 0.00123 | 93 | 0.00163 | -2.27500 | 155.637 | 0.02427 | 0.59764 |
| cbiE; cobalt-precorrin-7 (C5)-methyltransferase [EC:2.1.1.289] | 0.01055 | 0.00117 | 0.01398 | 0.00032 | 0.00223 | 65 | 0.00815 | 0.00013 | 0.00119 | 93 | 0.00253 | 2.30159 | 99.939 | 0.02343 | 0.59764 |
| zapB; cell division protein ZapB | 0.01053 | 0.00097 | 0.01153 | 0.00016 | 0.00155 | 65 | 0.00983 | 0.00015 | 0.00125 | 93 | 0.00199 | 0.85632 | 134.850 | 0.39334 | 0.80459 |
| cyaY; CyaY protein | 0.01053 | 0.00095 | 0.01138 | 0.00015 | 0.00153 | 65 | 0.00993 | 0.00013 | 0.00120 | 93 | 0.00195 | 0.74819 | 132.369 | 0.45567 | 0.83438 |
| cas6; CRISPR-associated endoribonuclease Cas6 [EC:3.1.-.-] | 0.01051 | 0.00103 | 0.01186 | 0.00025 | 0.00195 | 65 | 0.00957 | 0.00011 | 0.00109 | 93 | 0.00223 | 1.02558 | 103.224 | 0.30749 | 0.77376 |
| IVD, ivd; isovaleryl-CoA dehydrogenase [EC:1.3.8.4] | 0.01049 | 0.00106 | 0.00929 | 0.00018 | 0.00165 | 65 | 0.01133 | 0.00018 | 0.00138 | 93 | 0.00215 | -0.94719 | 137.676 | 0.34520 | 0.78739 |
| paaK; phenylacetate-CoA ligase [EC:6.2.1.30] | 0.01048 | 0.00108 | 0.00823 | 0.00011 | 0.00133 | 65 | 0.01205 | 0.00023 | 0.00156 | 93 | 0.00205 | -1.86155 | 155.941 | 0.06455 | 0.73334 |
| cah; cephalosporin-C deacetylase [EC:3.1.1.41] | 0.01048 | 0.00100 | 0.01038 | 0.00014 | 0.00146 | 65 | 0.01054 | 0.00017 | 0.00136 | 93 | 0.00199 | -0.07819 | 146.925 | 0.93779 | 0.98459 |
| dcuA; anaerobic C4-dicarboxylate transporter DcuA | 0.01047 | 0.00102 | 0.00943 | 0.00012 | 0.00138 | 65 | 0.01120 | 0.00019 | 0.00144 | 93 | 0.00200 | -0.88715 | 152.899 | 0.37639 | 0.79810 |
| cysB; LysR family transcriptional regulator, cys regulon transcriptional activator | 0.01047 | 0.00095 | 0.01147 | 0.00015 | 0.00154 | 65 | 0.00976 | 0.00013 | 0.00119 | 93 | 0.00195 | 0.87717 | 130.829 | 0.38200 | 0.80026 |
| fadN; 3-hydroxyacyl-CoA dehydrogenase [EC:1.1.1.35] | 0.01046 | 0.00129 | 0.00774 | 0.00020 | 0.00176 | 65 | 0.01237 | 0.00030 | 0.00179 | 93 | 0.00251 | -1.84545 | 151.723 | 0.06692 | 0.73334 |
| nfeD; membrane-bound serine protease (ClpP class) | 0.01046 | 0.00093 | 0.01123 | 0.00020 | 0.00174 | 65 | 0.00992 | 0.00009 | 0.00101 | 93 | 0.00201 | 0.64926 | 106.153 | 0.51757 | 0.85268 |
| K08981; putative membrane protein | 0.01046 | 0.00103 | 0.00965 | 0.00017 | 0.00162 | 65 | 0.01102 | 0.00017 | 0.00134 | 93 | 0.00211 | -0.64630 | 137.061 | 0.51917 | 0.85449 |
| nemA; N-ethylmaleimide reductase [EC:1.-.-.-] | 0.01045 | 0.00127 | 0.00838 | 0.00018 | 0.00168 | 65 | 0.01190 | 0.00030 | 0.00179 | 93 | 0.00246 | -1.43054 | 153.917 | 0.15459 | 0.77376 |
| korD, oorD; 2-oxoglutarate ferredoxin oxidoreductase subunit delta [EC:1.2.7.3] | 0.01045 | 0.00116 | 0.00957 | 0.00016 | 0.00158 | 65 | 0.01106 | 0.00025 | 0.00165 | 93 | 0.00228 | -0.65113 | 152.958 | 0.51594 | 0.85216 |
| gcvPA; glycine dehydrogenase subunit 1 [EC:1.4.4.2] | 0.01044 | 0.00100 | 0.00848 | 0.00012 | 0.00138 | 65 | 0.01181 | 0.00018 | 0.00138 | 93 | 0.00195 | -1.70285 | 151.046 | 0.09065 | 0.76567 |
| fdhE; FdhE protein | 0.01044 | 0.00099 | 0.01140 | 0.00016 | 0.00159 | 65 | 0.00977 | 0.00015 | 0.00127 | 93 | 0.00203 | 0.80325 | 133.757 | 0.42326 | 0.82407 |
| impB; type VI secretion system protein ImpB | 0.01044 | 0.00136 | 0.01102 | 0.00022 | 0.00186 | 65 | 0.01003 | 0.00034 | 0.00192 | 93 | 0.00267 | 0.37043 | 152.738 | 0.71158 | 0.92004 |
| fhaC; hemolysin activation/secretion protein | 0.01043 | 0.00305 | 0.00746 | 0.00072 | 0.00332 | 65 | 0.01250 | 0.00200 | 0.00464 | 93 | 0.00571 | -0.88351 | 152.839 | 0.37835 | 0.79852 |
| E1.11.1.5; cytochrome c peroxidase [EC:1.11.1.5] | 0.01042 | 0.00100 | 0.01060 | 0.00015 | 0.00150 | 65 | 0.01030 | 0.00017 | 0.00135 | 93 | 0.00202 | 0.14849 | 143.558 | 0.88216 | 0.96944 |
| cbiN; cobalt/nickel transport protein | 0.01042 | 0.00170 | 0.01461 | 0.00078 | 0.00346 | 65 | 0.00749 | 0.00021 | 0.00152 | 93 | 0.00378 | 1.88465 | 88.723 | 0.06275 | 0.73012 |
| ynfM; MFS transporter, YNFM family, putative membrane transport protein | 0.01042 | 0.00100 | 0.00993 | 0.00016 | 0.00157 | 65 | 0.01076 | 0.00016 | 0.00131 | 93 | 0.00205 | -0.40736 | 137.865 | 0.68438 | 0.91533 |
| K09921; uncharacterized protein | 0.01041 | 0.00095 | 0.01126 | 0.00016 | 0.00158 | 65 | 0.00982 | 0.00013 | 0.00118 | 93 | 0.00198 | 0.72941 | 127.653 | 0.46709 | 0.83837 |
| nsrR; Rrf2 family transcriptional regulator, nitric oxide-sensitive transcriptional repressor | 0.01041 | 0.00098 | 0.00837 | 0.00011 | 0.00129 | 65 | 0.01184 | 0.00018 | 0.00138 | 93 | 0.00189 | -1.83124 | 153.865 | 0.06900 | 0.73522 |
| cbiT; cobalt-precorrin-6B (C15)-methyltransferase [EC:2.1.1.196] | 0.01041 | 0.00117 | 0.01393 | 0.00032 | 0.00223 | 65 | 0.00795 | 0.00013 | 0.00119 | 93 | 0.00253 | 2.36524 | 99.772 | 0.01995 | 0.59095 |
| gcvPB; glycine dehydrogenase subunit 2 [EC:1.4.4.2] | 0.01040 | 0.00100 | 0.00841 | 0.00012 | 0.00138 | 65 | 0.01179 | 0.00018 | 0.00138 | 93 | 0.00195 | -1.73124 | 150.984 | 0.08545 | 0.76567 |
| frc; formyl-CoA transferase [EC:2.8.3.16] | 0.01039 | 0.00132 | 0.00879 | 0.00028 | 0.00207 | 65 | 0.01150 | 0.00027 | 0.00171 | 93 | 0.00269 | -1.00638 | 136.929 | 0.31601 | 0.77376 |
| osmC; osmotically inducible protein OsmC | 0.01038 | 0.00110 | 0.00978 | 0.00019 | 0.00171 | 65 | 0.01080 | 0.00020 | 0.00145 | 93 | 0.00224 | -0.45212 | 138.963 | 0.65189 | 0.90504 |
| dctB; two-component system, NtrC family, C4-dicarboxylate transport sensor histidine kinase DctB [EC:2.7.13.3] | 0.01036 | 0.00157 | 0.01063 | 0.00039 | 0.00245 | 65 | 0.01018 | 0.00040 | 0.00207 | 93 | 0.00320 | 0.13915 | 138.569 | 0.88954 | 0.97048 |
| korC, oorC; 2-oxoglutarate ferredoxin oxidoreductase subunit gamma [EC:1.2.7.3] | 0.01036 | 0.00117 | 0.00948 | 0.00017 | 0.00160 | 65 | 0.01097 | 0.00025 | 0.00165 | 93 | 0.00229 | -0.65111 | 152.623 | 0.51596 | 0.85216 |
| tctD; two-component system, OmpR family, response regulator TctD | 0.01034 | 0.00137 | 0.00969 | 0.00025 | 0.00195 | 65 | 0.01080 | 0.00033 | 0.00189 | 93 | 0.00271 | -0.40955 | 149.061 | 0.68272 | 0.91495 |
| ybjD; putative ATP-dependent endonuclease of the OLD family | 0.01033 | 0.00105 | 0.01270 | 0.00025 | 0.00195 | 65 | 0.00867 | 0.00012 | 0.00114 | 93 | 0.00226 | 1.78240 | 106.510 | 0.07753 | 0.75461 |
| UMF1; MFS transporter, UMF1 family | 0.01032 | 0.00101 | 0.00915 | 0.00019 | 0.00173 | 65 | 0.01115 | 0.00014 | 0.00123 | 93 | 0.00212 | -0.94058 | 123.027 | 0.34876 | 0.79120 |
| E4.1.1.18, ldcC, cadA; lysine decarboxylase [EC:4.1.1.18] | 0.01031 | 0.00087 | 0.00826 | 0.00007 | 0.00105 | 65 | 0.01174 | 0.00015 | 0.00127 | 93 | 0.00165 | -2.11206 | 155.994 | 0.03627 | 0.62411 |
| K09967; uncharacterized protein | 0.01029 | 0.00134 | 0.00961 | 0.00029 | 0.00210 | 65 | 0.01077 | 0.00029 | 0.00175 | 93 | 0.00274 | -0.42127 | 137.812 | 0.67421 | 0.91194 |
| malT; LuxR family transcriptional regulator, maltose regulon positive regulatory protein | 0.01028 | 0.00143 | 0.01145 | 0.00035 | 0.00233 | 65 | 0.00946 | 0.00030 | 0.00181 | 93 | 0.00295 | 0.67624 | 131.191 | 0.50008 | 0.84899 |
| hepA; ATP-dependent helicase HepA [EC:3.6.4.-] | 0.01028 | 0.00104 | 0.01104 | 0.00017 | 0.00159 | 65 | 0.00974 | 0.00018 | 0.00137 | 93 | 0.00210 | 0.61859 | 140.575 | 0.53719 | 0.86207 |
| bamB; outer membrane protein assembly factor BamB | 0.01027 | 0.00109 | 0.01042 | 0.00019 | 0.00169 | 65 | 0.01017 | 0.00019 | 0.00143 | 93 | 0.00221 | 0.11175 | 138.419 | 0.91119 | 0.97664 |
| K07576; metallo-beta-lactamase family protein | 0.01025 | 0.00099 | 0.01163 | 0.00022 | 0.00182 | 65 | 0.00929 | 0.00011 | 0.00110 | 93 | 0.00213 | 1.09759 | 109.148 | 0.27480 | 0.77376 |
| E1.4.1.9; leucine dehydrogenase [EC:1.4.1.9] | 0.01024 | 0.00102 | 0.00897 | 0.00016 | 0.00156 | 65 | 0.01112 | 0.00017 | 0.00134 | 93 | 0.00205 | -1.04753 | 140.217 | 0.29666 | 0.77376 |
| MAN2C1; alpha-mannosidase [EC:3.2.1.24] | 0.01023 | 0.00092 | 0.01046 | 0.00016 | 0.00158 | 65 | 0.01007 | 0.00012 | 0.00111 | 93 | 0.00194 | 0.20196 | 122.021 | 0.84028 | 0.95506 |
| regB, regS, actS; two-component system, sensor histidine kinase RegB [EC:2.7.13.3] | 0.01023 | 0.00113 | 0.00935 | 0.00020 | 0.00176 | 65 | 0.01084 | 0.00020 | 0.00147 | 93 | 0.00230 | -0.65069 | 137.531 | 0.51633 | 0.85216 |
| TC.GPH; glycoside/pentoside/hexuronide:cation symporter, GPH family | 0.01022 | 0.00132 | 0.01183 | 0.00037 | 0.00239 | 65 | 0.00910 | 0.00021 | 0.00149 | 93 | 0.00282 | 0.96549 | 111.813 | 0.33638 | 0.78254 |
| dctM; C4-dicarboxylate transporter, DctM subunit | 0.01022 | 0.00112 | 0.01245 | 0.00031 | 0.00218 | 65 | 0.00866 | 0.00012 | 0.00112 | 93 | 0.00245 | 1.54831 | 97.362 | 0.12479 | 0.77376 |
| cpg; glutamate carboxypeptidase [EC:3.4.17.11] | 0.01021 | 0.00091 | 0.00863 | 0.00010 | 0.00123 | 65 | 0.01131 | 0.00015 | 0.00128 | 93 | 0.00178 | -1.51196 | 153.004 | 0.13261 | 0.77376 |
| COQ7; ubiquinone biosynthesis monooxygenase Coq7 [EC:1.14.13.-] | 0.01020 | 0.00104 | 0.00990 | 0.00019 | 0.00171 | 65 | 0.01042 | 0.00016 | 0.00131 | 93 | 0.00215 | -0.23942 | 130.207 | 0.81115 | 0.95214 |
| waaL, rfaL; O-antigen ligase [EC:2.4.1.-] | 0.01020 | 0.00109 | 0.01197 | 0.00023 | 0.00190 | 65 | 0.00897 | 0.00016 | 0.00130 | 93 | 0.00230 | 1.30535 | 119.411 | 0.19428 | 0.77376 |
| pilW; type IV pilus assembly protein PilW | 0.01017 | 0.00100 | 0.01037 | 0.00017 | 0.00164 | 65 | 0.01003 | 0.00015 | 0.00127 | 93 | 0.00207 | 0.16294 | 131.266 | 0.87082 | 0.96546 |
| dapH, dapD; tetrahydrodipicolinate N-acetyltransferase [EC:2.3.1.89] | 0.01017 | 0.00101 | 0.01024 | 0.00023 | 0.00188 | 65 | 0.01013 | 0.00011 | 0.00111 | 93 | 0.00218 | 0.05113 | 107.131 | 0.95932 | 0.98903 |
| nudE; ADP-ribose diphosphatase [EC:3.6.1.-] | 0.01017 | 0.00097 | 0.01129 | 0.00016 | 0.00156 | 65 | 0.00938 | 0.00014 | 0.00125 | 93 | 0.00199 | 0.96213 | 133.985 | 0.33772 | 0.78354 |
| K07318; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | 0.01016 | 0.00169 | 0.01266 | 0.00069 | 0.00326 | 65 | 0.00842 | 0.00028 | 0.00174 | 93 | 0.00370 | 1.14499 | 100.082 | 0.25494 | 0.77376 |
| impC; type VI secretion system protein ImpC | 0.01016 | 0.00119 | 0.01073 | 0.00020 | 0.00175 | 65 | 0.00976 | 0.00025 | 0.00163 | 93 | 0.00239 | 0.40746 | 146.579 | 0.68426 | 0.91533 |
| yejF; microcin C transport system ATP-binding protein | 0.01015 | 0.00107 | 0.00919 | 0.00013 | 0.00142 | 65 | 0.01082 | 0.00022 | 0.00153 | 93 | 0.00209 | -0.77814 | 154.331 | 0.43768 | 0.83071 |
| pcaR; IclR family transcriptional regulator, pca regulon regulatory protein | 0.01015 | 0.00127 | 0.00897 | 0.00020 | 0.00177 | 65 | 0.01098 | 0.00029 | 0.00177 | 93 | 0.00250 | -0.80090 | 150.925 | 0.42445 | 0.82505 |
| pcaD; 3-oxoadipate enol-lactonase [EC:3.1.1.24] | 0.01010 | 0.00114 | 0.00914 | 0.00019 | 0.00173 | 65 | 0.01078 | 0.00022 | 0.00152 | 93 | 0.00230 | -0.70868 | 142.473 | 0.47968 | 0.84324 |
| dapC; N-succinyldiaminopimelate aminotransferase [EC:2.6.1.17] | 0.01010 | 0.00107 | 0.00984 | 0.00017 | 0.00162 | 65 | 0.01028 | 0.00019 | 0.00142 | 93 | 0.00216 | -0.20186 | 141.605 | 0.84032 | 0.95506 |
| creD; inner membrane protein | 0.01010 | 0.00106 | 0.01186 | 0.00024 | 0.00194 | 65 | 0.00887 | 0.00013 | 0.00118 | 93 | 0.00227 | 1.31956 | 109.722 | 0.18973 | 0.77376 |
| ispDF; 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase / 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [EC:2.7.7.60 4.6.1.12] | 0.01008 | 0.00086 | 0.00939 | 0.00012 | 0.00137 | 65 | 0.01056 | 0.00011 | 0.00110 | 93 | 0.00176 | -0.66550 | 134.766 | 0.50687 | 0.84956 |
| spoVG; stage V sporulation protein G | 0.01006 | 0.00119 | 0.01141 | 0.00031 | 0.00217 | 65 | 0.00911 | 0.00017 | 0.00134 | 93 | 0.00255 | 0.90308 | 110.862 | 0.36844 | 0.79587 |
| K07157; uncharacterized protein | 0.01004 | 0.00102 | 0.00896 | 0.00017 | 0.00164 | 65 | 0.01080 | 0.00016 | 0.00130 | 93 | 0.00209 | -0.88193 | 133.053 | 0.37940 | 0.79852 |
| aidB; putative acyl-CoA dehydrogenase | 0.01004 | 0.00102 | 0.00920 | 0.00016 | 0.00158 | 65 | 0.01062 | 0.00017 | 0.00133 | 93 | 0.00207 | -0.68823 | 138.541 | 0.49246 | 0.84797 |
| rimL; ribosomal-protein-serine acetyltransferase [EC:2.3.1.-] | 0.01003 | 0.00108 | 0.00851 | 0.00011 | 0.00131 | 65 | 0.01109 | 0.00023 | 0.00159 | 93 | 0.00206 | -1.25405 | 155.967 | 0.21170 | 0.77376 |
| E3.5.1.11; penicillin amidase [EC:3.5.1.11] | 0.01002 | 0.00110 | 0.00909 | 0.00018 | 0.00167 | 65 | 0.01067 | 0.00020 | 0.00147 | 93 | 0.00222 | -0.70901 | 142.391 | 0.47948 | 0.84314 |
| phaF; multicomponent K+:H+ antiporter subunit F | 0.01000 | 0.00103 | 0.00899 | 0.00016 | 0.00158 | 65 | 0.01072 | 0.00017 | 0.00136 | 93 | 0.00208 | -0.83246 | 140.349 | 0.40656 | 0.81189 |
| ureC; urease subunit alpha [EC:3.5.1.5] | 0.01000 | 0.00108 | 0.00719 | 0.00011 | 0.00131 | 65 | 0.01196 | 0.00023 | 0.00156 | 93 | 0.00204 | -2.33795 | 156.000 | 0.02066 | 0.59095 |
| bioH; pimeloyl-[acyl-carrier protein] methyl ester esterase [EC:3.1.1.85] | 0.00999 | 0.00095 | 0.01045 | 0.00015 | 0.00151 | 65 | 0.00967 | 0.00014 | 0.00124 | 93 | 0.00195 | 0.40004 | 135.975 | 0.68975 | 0.91567 |
| panF; sodium/pantothenate symporter | 0.00999 | 0.00109 | 0.01195 | 0.00027 | 0.00203 | 65 | 0.00863 | 0.00013 | 0.00119 | 93 | 0.00235 | 1.41075 | 106.570 | 0.16123 | 0.77376 |
| desR; two-component system, NarL family, response regulator DesR | 0.00996 | 0.00114 | 0.00874 | 0.00016 | 0.00157 | 65 | 0.01082 | 0.00023 | 0.00159 | 93 | 0.00224 | -0.93165 | 151.423 | 0.35300 | 0.79194 |
| flp, pilA; pilus assembly protein Flp/PilA | 0.00995 | 0.00110 | 0.00763 | 0.00013 | 0.00140 | 65 | 0.01157 | 0.00023 | 0.00159 | 93 | 0.00212 | -1.86066 | 155.501 | 0.06468 | 0.73334 |
| fiu; catecholate siderophore receptor | 0.00994 | 0.00113 | 0.00936 | 0.00019 | 0.00171 | 65 | 0.01035 | 0.00021 | 0.00151 | 93 | 0.00228 | -0.43241 | 142.495 | 0.66610 | 0.91126 |
| aldH; NADP-dependent aldehyde dehydrogenase [EC:1.2.1.4] | 0.00993 | 0.00146 | 0.00967 | 0.00031 | 0.00217 | 65 | 0.01012 | 0.00036 | 0.00197 | 93 | 0.00293 | -0.15379 | 144.464 | 0.87799 | 0.96759 |
| asmA; AsmA protein | 0.00993 | 0.00189 | 0.00944 | 0.00048 | 0.00272 | 65 | 0.01027 | 0.00063 | 0.00260 | 93 | 0.00376 | -0.22041 | 148.501 | 0.82585 | 0.95506 |
| SCD, desC; stearoyl-CoA desaturase (Delta-9 desaturase) [EC:1.14.19.1] | 0.00993 | 0.00137 | 0.00884 | 0.00028 | 0.00208 | 65 | 0.01069 | 0.00031 | 0.00182 | 93 | 0.00276 | -0.67069 | 142.005 | 0.50351 | 0.84956 |
| gspC; general secretion pathway protein C | 0.00992 | 0.00108 | 0.00926 | 0.00017 | 0.00162 | 65 | 0.01038 | 0.00020 | 0.00145 | 93 | 0.00218 | -0.51621 | 143.611 | 0.60650 | 0.88555 |
| ureF; urease accessory protein | 0.00990 | 0.00107 | 0.00700 | 0.00011 | 0.00127 | 65 | 0.01193 | 0.00023 | 0.00156 | 93 | 0.00202 | -2.44283 | 155.922 | 0.01569 | 0.59095 |
| ureG; urease accessory protein | 0.00990 | 0.00107 | 0.00699 | 0.00011 | 0.00127 | 65 | 0.01194 | 0.00023 | 0.00156 | 93 | 0.00202 | -2.45187 | 155.911 | 0.01532 | 0.59095 |
| E4.3.1.15; diaminopropionate ammonia-lyase [EC:4.3.1.15] | 0.00989 | 0.00099 | 0.01096 | 0.00020 | 0.00176 | 65 | 0.00915 | 0.00012 | 0.00115 | 93 | 0.00211 | 0.85586 | 115.617 | 0.39385 | 0.80459 |
| virB11, lvhB11; type IV secretion system protein VirB11 | 0.00989 | 0.00146 | 0.00932 | 0.00034 | 0.00227 | 65 | 0.01029 | 0.00034 | 0.00192 | 93 | 0.00298 | -0.32606 | 138.770 | 0.74487 | 0.93176 |
| K09924; uncharacterized protein | 0.00989 | 0.00095 | 0.00937 | 0.00014 | 0.00147 | 65 | 0.01025 | 0.00014 | 0.00125 | 93 | 0.00192 | -0.45504 | 139.180 | 0.64979 | 0.90409 |
| yjaB; putative acetyltransferase [EC:2.3.1.-] | 0.00988 | 0.00100 | 0.00975 | 0.00015 | 0.00151 | 65 | 0.00997 | 0.00017 | 0.00134 | 93 | 0.00203 | -0.10909 | 142.918 | 0.91329 | 0.97664 |
| cmoB; tRNA (mo5U34)-methyltransferase [EC:2.1.1.-] | 0.00988 | 0.00104 | 0.01060 | 0.00015 | 0.00151 | 65 | 0.00937 | 0.00019 | 0.00142 | 93 | 0.00208 | 0.59225 | 147.076 | 0.55459 | 0.86806 |
| ALDO; fructose-bisphosphate aldolase, class I [EC:4.1.2.13] | 0.00986 | 0.00114 | 0.01162 | 0.00028 | 0.00208 | 65 | 0.00863 | 0.00015 | 0.00127 | 93 | 0.00243 | 1.22887 | 109.799 | 0.22175 | 0.77376 |
| flhB2; flagellar biosynthesis protein | 0.00983 | 0.00097 | 0.00879 | 0.00014 | 0.00146 | 65 | 0.01056 | 0.00015 | 0.00129 | 93 | 0.00194 | -0.91198 | 142.552 | 0.36332 | 0.79496 |
| carD; CarD family transcriptional regulator | 0.00982 | 0.00092 | 0.00820 | 0.00011 | 0.00133 | 65 | 0.01095 | 0.00015 | 0.00126 | 93 | 0.00183 | -1.50364 | 147.536 | 0.13481 | 0.77376 |
| cheZ; chemotaxis protein CheZ | 0.00982 | 0.00109 | 0.00984 | 0.00019 | 0.00173 | 65 | 0.00980 | 0.00019 | 0.00142 | 93 | 0.00223 | 0.02035 | 136.259 | 0.98380 | 0.99706 |
| mtaP, MTAP; 5’-methylthioadenosine phosphorylase [EC:2.4.2.28] | 0.00979 | 0.00087 | 0.00976 | 0.00013 | 0.00143 | 65 | 0.00981 | 0.00011 | 0.00110 | 93 | 0.00181 | -0.02631 | 130.612 | 0.97905 | 0.99657 |
| K07148; uncharacterized protein | 0.00978 | 0.00145 | 0.00690 | 0.00016 | 0.00157 | 65 | 0.01179 | 0.00045 | 0.00220 | 93 | 0.00270 | -1.81212 | 152.819 | 0.07193 | 0.74066 |
| PTS-Mtl-EIIC, mtlA, cmtA; PTS system, mannitol-specific IIC component | 0.00974 | 0.00099 | 0.00998 | 0.00021 | 0.00182 | 65 | 0.00957 | 0.00012 | 0.00112 | 93 | 0.00213 | 0.19402 | 110.535 | 0.84652 | 0.95682 |
| GLUD1_2, gdhA; glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3] | 0.00970 | 0.00085 | 0.00935 | 0.00011 | 0.00129 | 65 | 0.00995 | 0.00012 | 0.00114 | 93 | 0.00172 | -0.34773 | 142.691 | 0.72855 | 0.92350 |
| mdoH; membrane glycosyltransferase [EC:2.4.1.-] | 0.00969 | 0.00125 | 0.00910 | 0.00022 | 0.00186 | 65 | 0.01011 | 0.00027 | 0.00169 | 93 | 0.00251 | -0.40178 | 145.076 | 0.68844 | 0.91567 |
| fimC; fimbrial chaperone protein | 0.00969 | 0.00131 | 0.00950 | 0.00029 | 0.00210 | 65 | 0.00982 | 0.00026 | 0.00168 | 93 | 0.00269 | -0.11713 | 133.708 | 0.90693 | 0.97664 |
| sohB; serine protease SohB [EC:3.4.21.-] | 0.00969 | 0.00090 | 0.01034 | 0.00013 | 0.00143 | 65 | 0.00923 | 0.00013 | 0.00116 | 93 | 0.00184 | 0.60365 | 135.435 | 0.54709 | 0.86509 |
| sqr; sulfide:quinone oxidoreductase [EC:1.8.5.4] | 0.00968 | 0.00096 | 0.00894 | 0.00014 | 0.00149 | 65 | 0.01020 | 0.00015 | 0.00127 | 93 | 0.00196 | -0.64282 | 139.180 | 0.52140 | 0.85613 |
| K06907; uncharacterized protein | 0.00968 | 0.00146 | 0.00856 | 0.00023 | 0.00189 | 65 | 0.01046 | 0.00041 | 0.00210 | 93 | 0.00283 | -0.67006 | 155.087 | 0.50382 | 0.84956 |
| K07486; transposase | 0.00966 | 0.00220 | 0.01378 | 0.00146 | 0.00474 | 65 | 0.00677 | 0.00027 | 0.00172 | 93 | 0.00504 | 1.39119 | 80.908 | 0.16798 | 0.77376 |
| potH; putrescine transport system permease protein | 0.00965 | 0.00096 | 0.00948 | 0.00015 | 0.00150 | 65 | 0.00977 | 0.00015 | 0.00126 | 93 | 0.00196 | -0.14654 | 138.066 | 0.88371 | 0.96974 |
| phaA; multicomponent K+:H+ antiporter subunit A | 0.00965 | 0.00101 | 0.00889 | 0.00016 | 0.00157 | 65 | 0.01018 | 0.00016 | 0.00132 | 93 | 0.00205 | -0.62957 | 138.318 | 0.53001 | 0.86094 |
| impH, vasB; type VI secretion system protein ImpH | 0.00965 | 0.00119 | 0.01052 | 0.00019 | 0.00173 | 65 | 0.00903 | 0.00024 | 0.00162 | 93 | 0.00237 | 0.62770 | 146.954 | 0.53118 | 0.86136 |
| K09919; uncharacterized protein | 0.00964 | 0.00099 | 0.00881 | 0.00017 | 0.00161 | 65 | 0.01022 | 0.00015 | 0.00127 | 93 | 0.00205 | -0.68993 | 132.571 | 0.49144 | 0.84797 |
| comA; ATP-binding cassette, subfamily C, bacterial, competence factor transporting protein [EC:3.4.22.-] | 0.00962 | 0.00099 | 0.00941 | 0.00013 | 0.00142 | 65 | 0.00977 | 0.00017 | 0.00137 | 93 | 0.00197 | -0.18346 | 148.947 | 0.85469 | 0.95858 |
| dsdA; D-serine dehydratase [EC:4.3.1.18] | 0.00961 | 0.00125 | 0.01160 | 0.00034 | 0.00229 | 65 | 0.00822 | 0.00018 | 0.00140 | 93 | 0.00268 | 1.26018 | 109.881 | 0.21028 | 0.77376 |
| pilV; type IV pilus assembly protein PilV | 0.00961 | 0.00101 | 0.00974 | 0.00018 | 0.00165 | 65 | 0.00952 | 0.00015 | 0.00127 | 93 | 0.00209 | 0.10503 | 130.621 | 0.91651 | 0.97681 |
| maiA, GSTZ1; maleylacetoacetate isomerase [EC:5.2.1.2] | 0.00960 | 0.00100 | 0.00897 | 0.00016 | 0.00159 | 65 | 0.01004 | 0.00015 | 0.00129 | 93 | 0.00204 | -0.52540 | 135.388 | 0.60016 | 0.88240 |
| phaG; multicomponent K+:H+ antiporter subunit G | 0.00959 | 0.00100 | 0.00893 | 0.00016 | 0.00156 | 65 | 0.01006 | 0.00016 | 0.00131 | 93 | 0.00204 | -0.55677 | 138.163 | 0.57859 | 0.87532 |
| trpCF; indole-3-glycerol phosphate synthase / phosphoribosylanthranilate isomerase [EC:4.1.1.48 5.3.1.24] | 0.00958 | 0.00096 | 0.00991 | 0.00015 | 0.00152 | 65 | 0.00935 | 0.00015 | 0.00125 | 93 | 0.00197 | 0.28426 | 136.852 | 0.77664 | 0.94269 |
| tsr; methyl-accepting chemotaxis protein I, serine sensor receptor | 0.00958 | 0.00162 | 0.00765 | 0.00027 | 0.00203 | 65 | 0.01093 | 0.00051 | 0.00235 | 93 | 0.00311 | -1.05587 | 155.795 | 0.29266 | 0.77376 |
| comB; competence factor transport accessory protein ComB | 0.00958 | 0.00099 | 0.00935 | 0.00013 | 0.00142 | 65 | 0.00974 | 0.00017 | 0.00137 | 93 | 0.00197 | -0.19825 | 148.864 | 0.84312 | 0.95519 |
| yegD; hypothetical chaperone protein | 0.00957 | 0.00097 | 0.00899 | 0.00016 | 0.00155 | 65 | 0.00998 | 0.00014 | 0.00124 | 93 | 0.00199 | -0.49802 | 133.950 | 0.61929 | 0.88981 |
| poxB; pyruvate dehydrogenase (quinone) [EC:1.2.5.1] | 0.00956 | 0.00104 | 0.00901 | 0.00016 | 0.00156 | 65 | 0.00995 | 0.00018 | 0.00139 | 93 | 0.00210 | -0.44813 | 143.213 | 0.65473 | 0.90679 |
| phaE; multicomponent K+:H+ antiporter subunit E | 0.00955 | 0.00100 | 0.00889 | 0.00016 | 0.00157 | 65 | 0.01000 | 0.00016 | 0.00131 | 93 | 0.00204 | -0.54338 | 138.104 | 0.58774 | 0.87732 |
| folE2; GTP cyclohydrolase IB [EC:3.5.4.16] | 0.00954 | 0.00091 | 0.01014 | 0.00014 | 0.00146 | 65 | 0.00912 | 0.00013 | 0.00116 | 93 | 0.00187 | 0.54469 | 133.150 | 0.58688 | 0.87731 |
| hhoB, degS; serine protease DegS [EC:3.4.21.-] | 0.00954 | 0.00090 | 0.01036 | 0.00013 | 0.00143 | 65 | 0.00897 | 0.00013 | 0.00116 | 93 | 0.00184 | 0.75436 | 135.282 | 0.45194 | 0.83438 |
| hypA, hybF; hydrogenase nickel incorporation protein HypA/HybF | 0.00953 | 0.00089 | 0.00962 | 0.00013 | 0.00141 | 65 | 0.00947 | 0.00013 | 0.00116 | 93 | 0.00183 | 0.08287 | 136.223 | 0.93408 | 0.98428 |
| cpdA; 3’,5’-cyclic-AMP phosphodiesterase [EC:3.1.4.53] | 0.00953 | 0.00091 | 0.01032 | 0.00013 | 0.00143 | 65 | 0.00897 | 0.00013 | 0.00117 | 93 | 0.00185 | 0.73203 | 136.211 | 0.46541 | 0.83786 |
| phaC; multicomponent K+:H+ antiporter subunit C | 0.00953 | 0.00100 | 0.00889 | 0.00016 | 0.00157 | 65 | 0.00997 | 0.00016 | 0.00131 | 93 | 0.00204 | -0.53183 | 138.012 | 0.59570 | 0.88005 |
| phaD; multicomponent K+:H+ antiporter subunit D | 0.00953 | 0.00100 | 0.00889 | 0.00016 | 0.00157 | 65 | 0.00997 | 0.00016 | 0.00131 | 93 | 0.00204 | -0.53183 | 138.012 | 0.59570 | 0.88005 |
| E2.4.1.7; sucrose phosphorylase [EC:2.4.1.7] | 0.00953 | 0.00086 | 0.00999 | 0.00016 | 0.00156 | 65 | 0.00920 | 0.00009 | 0.00096 | 93 | 0.00184 | 0.42532 | 110.776 | 0.67143 | 0.91184 |
| RP-L7A, rplGB; large subunit ribosomal protein L7A | 0.00952 | 0.00092 | 0.00912 | 0.00015 | 0.00154 | 65 | 0.00981 | 0.00012 | 0.00114 | 93 | 0.00192 | -0.35933 | 126.817 | 0.71994 | 0.92108 |
| metZ; O-succinylhomoserine sulfhydrylase [EC:2.5.1.-] | 0.00949 | 0.00089 | 0.00872 | 0.00012 | 0.00134 | 65 | 0.01003 | 0.00013 | 0.00120 | 93 | 0.00180 | -0.72704 | 143.714 | 0.46838 | 0.83837 |
| mnhD, mrpD; multicomponent Na+:H+ antiporter subunit D | 0.00949 | 0.00115 | 0.00700 | 0.00013 | 0.00140 | 65 | 0.01123 | 0.00026 | 0.00168 | 93 | 0.00218 | -1.94127 | 156.000 | 0.05403 | 0.70113 |
| gltI, aatJ; glutamate/aspartate transport system substrate-binding protein | 0.00949 | 0.00128 | 0.00791 | 0.00014 | 0.00149 | 65 | 0.01059 | 0.00034 | 0.00191 | 93 | 0.00242 | -1.10462 | 155.282 | 0.27103 | 0.77376 |
| tusE, dsrC; tRNA 2-thiouridine synthesizing protein E [EC:2.8.1.-] | 0.00947 | 0.00090 | 0.01031 | 0.00013 | 0.00143 | 65 | 0.00888 | 0.00013 | 0.00117 | 93 | 0.00184 | 0.77446 | 135.886 | 0.44000 | 0.83186 |
| tusD, dsrE; tRNA 2-thiouridine synthesizing protein D [EC:2.8.1.-] | 0.00946 | 0.00090 | 0.01031 | 0.00013 | 0.00143 | 65 | 0.00887 | 0.00013 | 0.00117 | 93 | 0.00184 | 0.77985 | 135.937 | 0.43684 | 0.83048 |
| ureD, ureH; urease accessory protein | 0.00946 | 0.00103 | 0.00683 | 0.00010 | 0.00124 | 65 | 0.01129 | 0.00021 | 0.00149 | 93 | 0.00194 | -2.29838 | 155.999 | 0.02287 | 0.59764 |
| K09798; uncharacterized protein | 0.00946 | 0.00078 | 0.00990 | 0.00011 | 0.00128 | 65 | 0.00914 | 0.00009 | 0.00098 | 93 | 0.00161 | 0.47346 | 130.418 | 0.63668 | 0.89696 |
| regA, regR, actR; two-component system, response regulator RegA | 0.00943 | 0.00100 | 0.00862 | 0.00017 | 0.00160 | 65 | 0.01000 | 0.00015 | 0.00128 | 93 | 0.00205 | -0.67347 | 133.415 | 0.50181 | 0.84942 |
| potG; putrescine transport system ATP-binding protein | 0.00941 | 0.00094 | 0.00928 | 0.00014 | 0.00149 | 65 | 0.00951 | 0.00014 | 0.00122 | 93 | 0.00192 | -0.11784 | 135.848 | 0.90637 | 0.97664 |
| lacY; MFS transporter, OHS family, lactose permease | 0.00940 | 0.00081 | 0.00937 | 0.00013 | 0.00142 | 65 | 0.00942 | 0.00008 | 0.00095 | 93 | 0.00171 | -0.02746 | 117.716 | 0.97814 | 0.99657 |
| mtlD; mannitol-1-phosphate 5-dehydrogenase [EC:1.1.1.17] | 0.00939 | 0.00098 | 0.00974 | 0.00021 | 0.00182 | 65 | 0.00915 | 0.00011 | 0.00108 | 93 | 0.00211 | 0.28350 | 107.510 | 0.77734 | 0.94269 |
| pilP; type IV pilus assembly protein PilP | 0.00937 | 0.00093 | 0.01018 | 0.00016 | 0.00157 | 65 | 0.00880 | 0.00012 | 0.00114 | 93 | 0.00194 | 0.71150 | 124.701 | 0.47810 | 0.84266 |
| potI; putrescine transport system permease protein | 0.00936 | 0.00093 | 0.00929 | 0.00014 | 0.00149 | 65 | 0.00941 | 0.00013 | 0.00120 | 93 | 0.00191 | -0.05818 | 134.975 | 0.95369 | 0.98792 |
| thiF; sulfur carrier protein ThiS adenylyltransferase [EC:2.7.7.73] | 0.00936 | 0.00102 | 0.01214 | 0.00024 | 0.00193 | 65 | 0.00741 | 0.00010 | 0.00104 | 93 | 0.00219 | 2.15746 | 100.252 | 0.03336 | 0.62130 |
| FLOT; flotillin | 0.00936 | 0.00071 | 0.00801 | 0.00005 | 0.00091 | 65 | 0.01030 | 0.00009 | 0.00101 | 93 | 0.00136 | -1.68218 | 154.986 | 0.09455 | 0.76609 |
| dkgB; 2,5-diketo-D-gluconate reductase B [EC:1.1.1.346] | 0.00935 | 0.00105 | 0.00930 | 0.00017 | 0.00162 | 65 | 0.00938 | 0.00018 | 0.00139 | 93 | 0.00213 | -0.03589 | 140.092 | 0.97142 | 0.99401 |
| HGD, hmgA; homogentisate 1,2-dioxygenase [EC:1.13.11.5] | 0.00933 | 0.00096 | 0.00870 | 0.00016 | 0.00156 | 65 | 0.00978 | 0.00014 | 0.00122 | 93 | 0.00198 | -0.54359 | 131.548 | 0.58765 | 0.87732 |
| ugtP; processive 1,2-diacylglycerol beta-glucosyltransferase [EC:2.4.1.315] | 0.00933 | 0.00090 | 0.00973 | 0.00017 | 0.00162 | 65 | 0.00906 | 0.00010 | 0.00104 | 93 | 0.00192 | 0.35117 | 114.226 | 0.72611 | 0.92177 |
| hypB; hydrogenase nickel incorporation protein HypB | 0.00932 | 0.00089 | 0.00947 | 0.00013 | 0.00141 | 65 | 0.00922 | 0.00012 | 0.00115 | 93 | 0.00182 | 0.13586 | 135.544 | 0.89213 | 0.97147 |
| rnk; regulator of nucleoside diphosphate kinase | 0.00932 | 0.00095 | 0.00903 | 0.00016 | 0.00155 | 65 | 0.00953 | 0.00013 | 0.00120 | 93 | 0.00197 | -0.25620 | 131.074 | 0.79820 | 0.94822 |
| hypF; hydrogenase maturation protein HypF | 0.00931 | 0.00088 | 0.00929 | 0.00013 | 0.00144 | 65 | 0.00932 | 0.00012 | 0.00112 | 93 | 0.00182 | -0.01737 | 131.654 | 0.98616 | 0.99729 |
| zurR, zur; Fur family transcriptional regulator, zinc uptake regulator | 0.00930 | 0.00104 | 0.00950 | 0.00025 | 0.00196 | 65 | 0.00916 | 0.00012 | 0.00113 | 93 | 0.00226 | 0.15372 | 105.248 | 0.87812 | 0.96759 |
| vasG, clpV; type VI secretion system protein VasG | 0.00929 | 0.00118 | 0.00996 | 0.00018 | 0.00166 | 65 | 0.00883 | 0.00025 | 0.00164 | 93 | 0.00233 | 0.48524 | 150.551 | 0.62821 | 0.89381 |
| smvA, qacA, lfrA; MFS transporter, DHA2 family, multidrug resistance protein | 0.00929 | 0.00099 | 0.00850 | 0.00015 | 0.00152 | 65 | 0.00984 | 0.00016 | 0.00131 | 93 | 0.00200 | -0.66917 | 140.293 | 0.50449 | 0.84956 |
| mnhA, mrpA; multicomponent Na+:H+ antiporter subunit A | 0.00928 | 0.00124 | 0.00630 | 0.00011 | 0.00131 | 65 | 0.01136 | 0.00033 | 0.00187 | 93 | 0.00229 | -2.21433 | 151.803 | 0.02830 | 0.61334 |
| mnhC, mrpC; multicomponent Na+:H+ antiporter subunit C | 0.00928 | 0.00115 | 0.00695 | 0.00013 | 0.00139 | 65 | 0.01091 | 0.00026 | 0.00167 | 93 | 0.00218 | -1.81815 | 155.998 | 0.07096 | 0.73909 |
| disA; diadenylate cyclase [EC:2.7.7.85] | 0.00927 | 0.00109 | 0.01122 | 0.00027 | 0.00205 | 65 | 0.00792 | 0.00012 | 0.00115 | 93 | 0.00235 | 1.40364 | 103.730 | 0.16341 | 0.77376 |
| paaG; 2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA isomerase [EC:5.3.3.18] | 0.00926 | 0.00111 | 0.00629 | 0.00008 | 0.00110 | 65 | 0.01133 | 0.00026 | 0.00169 | 93 | 0.00201 | -2.50504 | 148.341 | 0.01332 | 0.59095 |
| ureE; urease accessory protein | 0.00926 | 0.00102 | 0.00671 | 0.00010 | 0.00124 | 65 | 0.01103 | 0.00020 | 0.00148 | 93 | 0.00193 | -2.23803 | 156.000 | 0.02664 | 0.61112 |
| K08989; putative membrane protein | 0.00925 | 0.00077 | 0.00865 | 0.00010 | 0.00123 | 65 | 0.00967 | 0.00009 | 0.00099 | 93 | 0.00158 | -0.64048 | 134.834 | 0.52295 | 0.85702 |
| bamC; outer membrane protein assembly factor BamC | 0.00924 | 0.00088 | 0.01047 | 0.00014 | 0.00147 | 65 | 0.00838 | 0.00011 | 0.00109 | 93 | 0.00183 | 1.14326 | 127.119 | 0.25508 | 0.77376 |
| mnhE, mrpE; multicomponent Na+:H+ antiporter subunit E | 0.00923 | 0.00115 | 0.00689 | 0.00013 | 0.00139 | 65 | 0.01086 | 0.00026 | 0.00167 | 93 | 0.00218 | -1.82495 | 156.000 | 0.06992 | 0.73522 |
| mnhG, mrpG; multicomponent Na+:H+ antiporter subunit G | 0.00922 | 0.00115 | 0.00690 | 0.00013 | 0.00139 | 65 | 0.01084 | 0.00026 | 0.00168 | 93 | 0.00218 | -1.81166 | 155.997 | 0.07196 | 0.74066 |
| K06906; uncharacterized protein | 0.00921 | 0.00187 | 0.00715 | 0.00029 | 0.00211 | 65 | 0.01065 | 0.00073 | 0.00281 | 93 | 0.00351 | -0.99770 | 154.498 | 0.31998 | 0.77376 |
| envZ; two-component system, OmpR family, osmolarity sensor histidine kinase EnvZ [EC:2.7.13.3] | 0.00921 | 0.00099 | 0.00828 | 0.00014 | 0.00146 | 65 | 0.00985 | 0.00017 | 0.00134 | 93 | 0.00198 | -0.79364 | 145.080 | 0.42870 | 0.82749 |
| rseA; sigma-E factor negative regulatory protein RseA | 0.00920 | 0.00088 | 0.01003 | 0.00013 | 0.00143 | 65 | 0.00862 | 0.00012 | 0.00112 | 93 | 0.00181 | 0.77892 | 132.154 | 0.43742 | 0.83071 |
| ghrB; glyoxylate/hydroxypyruvate/2-ketogluconate reductase [EC:1.1.1.79 1.1.1.81 1.1.1.215] | 0.00919 | 0.00103 | 0.00897 | 0.00016 | 0.00158 | 65 | 0.00934 | 0.00017 | 0.00136 | 93 | 0.00208 | -0.17417 | 140.723 | 0.86198 | 0.96251 |
| pilJ; twitching motility protein PilJ | 0.00916 | 0.00099 | 0.00902 | 0.00016 | 0.00158 | 65 | 0.00926 | 0.00015 | 0.00127 | 93 | 0.00203 | -0.12127 | 134.221 | 0.90366 | 0.97641 |
| mdtB; multidrug efflux pump | 0.00916 | 0.00106 | 0.00908 | 0.00018 | 0.00165 | 65 | 0.00922 | 0.00018 | 0.00138 | 93 | 0.00215 | -0.06734 | 138.056 | 0.94641 | 0.98674 |
| yagR; xanthine dehydrogenase YagR molybdenum-binding subunit [EC:1.17.1.4] | 0.00915 | 0.00111 | 0.00848 | 0.00018 | 0.00166 | 65 | 0.00961 | 0.00021 | 0.00149 | 93 | 0.00223 | -0.50582 | 144.028 | 0.61376 | 0.88788 |
| pilG; twitching motility two-component system response regulator PilG | 0.00915 | 0.00099 | 0.00907 | 0.00016 | 0.00159 | 65 | 0.00920 | 0.00015 | 0.00127 | 93 | 0.00203 | -0.06028 | 134.181 | 0.95202 | 0.98792 |
| yjdF; putative membrane protein | 0.00914 | 0.00085 | 0.00864 | 0.00010 | 0.00122 | 65 | 0.00949 | 0.00013 | 0.00117 | 93 | 0.00168 | -0.49956 | 148.425 | 0.61813 | 0.88903 |
| tyrA1; chorismate mutase [EC:5.4.99.5] | 0.00914 | 0.00090 | 0.00839 | 0.00012 | 0.00133 | 65 | 0.00966 | 0.00014 | 0.00121 | 93 | 0.00180 | -0.70343 | 144.752 | 0.48292 | 0.84354 |
| cydCD; ATP-binding cassette, subfamily C, bacterial CydCD | 0.00913 | 0.00094 | 0.01029 | 0.00018 | 0.00167 | 65 | 0.00832 | 0.00011 | 0.00108 | 93 | 0.00199 | 0.99023 | 115.090 | 0.32414 | 0.77376 |
| hypE; hydrogenase expression/formation protein HypE | 0.00912 | 0.00088 | 0.00921 | 0.00013 | 0.00142 | 65 | 0.00905 | 0.00012 | 0.00111 | 93 | 0.00181 | 0.08812 | 131.985 | 0.92991 | 0.98213 |
| ade; adenine deaminase [EC:3.5.4.2] | 0.00911 | 0.00113 | 0.00776 | 0.00021 | 0.00181 | 65 | 0.01005 | 0.00020 | 0.00145 | 93 | 0.00232 | -0.98809 | 134.019 | 0.32489 | 0.77376 |
| pilI; twitching motility protein PilI | 0.00910 | 0.00099 | 0.00905 | 0.00016 | 0.00158 | 65 | 0.00914 | 0.00015 | 0.00127 | 93 | 0.00203 | -0.04442 | 134.029 | 0.96464 | 0.99101 |
| hypD; hydrogenase expression/formation protein HypD | 0.00909 | 0.00087 | 0.00933 | 0.00013 | 0.00142 | 65 | 0.00892 | 0.00011 | 0.00111 | 93 | 0.00180 | 0.22993 | 132.009 | 0.81850 | 0.95506 |
| E3.1.6.1, aslA; arylsulfatase [EC:3.1.6.1] | 0.00909 | 0.00107 | 0.00664 | 0.00004 | 0.00077 | 65 | 0.01080 | 0.00028 | 0.00172 | 93 | 0.00189 | -2.20051 | 125.482 | 0.02960 | 0.61356 |
| sotB; MFS transporter, DHA1 family, L-arabinose/isopropyl-beta-D-thiogalactopyranoside export protein | 0.00908 | 0.00092 | 0.00960 | 0.00012 | 0.00137 | 65 | 0.00871 | 0.00014 | 0.00125 | 93 | 0.00185 | 0.48120 | 144.865 | 0.63110 | 0.89595 |
| gntR; LacI family transcriptional regulator, gluconate utilization system Gnt-I transcriptional repressor | 0.00907 | 0.00124 | 0.00807 | 0.00019 | 0.00169 | 65 | 0.00978 | 0.00028 | 0.00174 | 93 | 0.00243 | -0.70272 | 152.550 | 0.48330 | 0.84354 |
| ABC-2.LPSE.A; lipopolysaccharide transport system ATP-binding protein | 0.00905 | 0.00111 | 0.00819 | 0.00015 | 0.00154 | 65 | 0.00965 | 0.00022 | 0.00154 | 93 | 0.00218 | -0.66886 | 151.219 | 0.50460 | 0.84956 |
| bccA, pccA; acetyl-CoA/propionyl-CoA carboxylase, biotin carboxylase, biotin carboxyl carrier protein [EC:6.4.1.2 6.4.1.3 6.3.4.14] | 0.00904 | 0.00100 | 0.00842 | 0.00012 | 0.00135 | 65 | 0.00948 | 0.00019 | 0.00142 | 93 | 0.00196 | -0.54249 | 153.354 | 0.58827 | 0.87732 |
| glnK; nitrogen regulatory protein P-II 2 | 0.00904 | 0.00092 | 0.00830 | 0.00013 | 0.00142 | 65 | 0.00956 | 0.00014 | 0.00121 | 93 | 0.00186 | -0.67892 | 139.334 | 0.49832 | 0.84856 |
| xfp, xpk; xylulose-5-phosphate/fructose-6-phosphate phosphoketolase [EC:4.1.2.9 4.1.2.22] | 0.00902 | 0.00128 | 0.01018 | 0.00038 | 0.00241 | 65 | 0.00821 | 0.00018 | 0.00138 | 93 | 0.00277 | 0.71011 | 104.863 | 0.47921 | 0.84314 |
| K06905; uncharacterized protein | 0.00901 | 0.00137 | 0.00775 | 0.00018 | 0.00167 | 65 | 0.00989 | 0.00038 | 0.00202 | 93 | 0.00262 | -0.81546 | 155.974 | 0.41605 | 0.81772 |
| PCCB, pccB; propionyl-CoA carboxylase beta chain [EC:6.4.1.3 2.1.3.15] | 0.00900 | 0.00089 | 0.00676 | 0.00009 | 0.00118 | 65 | 0.01056 | 0.00014 | 0.00125 | 93 | 0.00171 | -2.22045 | 153.655 | 0.02785 | 0.61334 |
| garR, glxR; 2-hydroxy-3-oxopropionate reductase [EC:1.1.1.60] | 0.00900 | 0.00089 | 0.00742 | 0.00010 | 0.00123 | 65 | 0.01010 | 0.00014 | 0.00123 | 93 | 0.00174 | -1.53940 | 150.830 | 0.12580 | 0.77376 |
| RTCB, rtcB; tRNA-splicing ligase RtcB (3’-phosphate/5’-hydroxy nucleic acid ligase) [EC:6.5.1.8] | 0.00898 | 0.00084 | 0.00801 | 0.00010 | 0.00123 | 65 | 0.00966 | 0.00012 | 0.00114 | 93 | 0.00168 | -0.98649 | 146.207 | 0.32552 | 0.77376 |
| K08999; uncharacterized protein | 0.00897 | 0.00081 | 0.00758 | 0.00006 | 0.00098 | 65 | 0.00994 | 0.00013 | 0.00118 | 93 | 0.00154 | -1.53494 | 155.994 | 0.12682 | 0.77376 |
| eamA; O-acetylserine/cysteine efflux transporter | 0.00893 | 0.00134 | 0.00766 | 0.00021 | 0.00180 | 65 | 0.00982 | 0.00034 | 0.00190 | 93 | 0.00262 | -0.82376 | 153.524 | 0.41135 | 0.81492 |
| dppC; dipeptide transport system permease protein | 0.00893 | 0.00089 | 0.00874 | 0.00010 | 0.00126 | 65 | 0.00906 | 0.00014 | 0.00124 | 93 | 0.00177 | -0.18569 | 149.714 | 0.85294 | 0.95848 |
| glcU; glucose uptake protein | 0.00893 | 0.00158 | 0.01030 | 0.00056 | 0.00293 | 65 | 0.00797 | 0.00028 | 0.00174 | 93 | 0.00341 | 0.68373 | 107.806 | 0.49561 | 0.84797 |
| ihk; two-component system, OmpR family, sensor kinase Ihk [EC:2.7.13.3] | 0.00893 | 0.00080 | 0.00933 | 0.00010 | 0.00127 | 65 | 0.00864 | 0.00010 | 0.00104 | 93 | 0.00164 | 0.42225 | 135.677 | 0.67351 | 0.91184 |
| truC; tRNA pseudouridine65 synthase [EC:5.4.99.26] | 0.00893 | 0.00095 | 0.00949 | 0.00018 | 0.00168 | 65 | 0.00853 | 0.00012 | 0.00112 | 93 | 0.00202 | 0.47357 | 117.840 | 0.63668 | 0.89696 |
| irr; two-component system, OmpR family, response regulator Irr | 0.00892 | 0.00080 | 0.00934 | 0.00010 | 0.00127 | 65 | 0.00863 | 0.00010 | 0.00104 | 93 | 0.00164 | 0.43616 | 135.833 | 0.66341 | 0.91006 |
| K06877; DEAD/DEAH box helicase domain-containing protein | 0.00891 | 0.00096 | 0.00775 | 0.00013 | 0.00144 | 65 | 0.00972 | 0.00016 | 0.00129 | 93 | 0.00193 | -1.01987 | 143.907 | 0.30950 | 0.77376 |
| E2.6.1.18; beta-alanine–pyruvate transaminase [EC:2.6.1.18] | 0.00890 | 0.00097 | 0.00824 | 0.00015 | 0.00154 | 65 | 0.00937 | 0.00015 | 0.00126 | 93 | 0.00199 | -0.56972 | 135.829 | 0.56981 | 0.87135 |
| nikC; nickel transport system permease protein | 0.00888 | 0.00100 | 0.01063 | 0.00024 | 0.00193 | 65 | 0.00765 | 0.00010 | 0.00103 | 93 | 0.00219 | 1.36042 | 100.261 | 0.17675 | 0.77376 |
| rseB; sigma-E factor negative regulatory protein RseB | 0.00887 | 0.00086 | 0.00963 | 0.00012 | 0.00138 | 65 | 0.00833 | 0.00011 | 0.00109 | 93 | 0.00176 | 0.73573 | 132.669 | 0.46319 | 0.83677 |
| thpR; RNA 2’,3’-cyclic 3’-phosphodiesterase [EC:3.1.4.58] | 0.00887 | 0.00084 | 0.00694 | 0.00008 | 0.00112 | 65 | 0.01021 | 0.00013 | 0.00118 | 93 | 0.00163 | -2.00992 | 153.723 | 0.04619 | 0.66255 |
| K09729; uncharacterized protein | 0.00886 | 0.00104 | 0.01145 | 0.00027 | 0.00203 | 65 | 0.00705 | 0.00010 | 0.00102 | 93 | 0.00228 | 1.93132 | 95.948 | 0.05640 | 0.70577 |
| flaG; flagellar protein FlaG | 0.00886 | 0.00092 | 0.00846 | 0.00013 | 0.00144 | 65 | 0.00913 | 0.00013 | 0.00120 | 93 | 0.00188 | -0.35605 | 137.622 | 0.72235 | 0.92156 |
| ABC-2.LPSE.P; lipopolysaccharide transport system permease protein | 0.00885 | 0.00089 | 0.00862 | 0.00012 | 0.00136 | 65 | 0.00901 | 0.00013 | 0.00119 | 93 | 0.00181 | -0.21798 | 141.410 | 0.82776 | 0.95506 |
| epmA, poxA; elongation factor P–(R)-beta-lysine ligase [EC:6.3.1.-] | 0.00885 | 0.00087 | 0.00857 | 0.00011 | 0.00131 | 65 | 0.00905 | 0.00013 | 0.00118 | 93 | 0.00176 | -0.27387 | 144.063 | 0.78458 | 0.94445 |
| K09775; uncharacterized protein | 0.00885 | 0.00083 | 0.00832 | 0.00012 | 0.00138 | 65 | 0.00922 | 0.00010 | 0.00103 | 93 | 0.00172 | -0.52006 | 127.787 | 0.60392 | 0.88434 |
| mdoG; periplasmic glucans biosynthesis protein | 0.00884 | 0.00107 | 0.00837 | 0.00017 | 0.00160 | 65 | 0.00916 | 0.00019 | 0.00143 | 93 | 0.00215 | -0.36734 | 143.333 | 0.71391 | 0.92004 |
| omp31; outer membrane immunogenic protein | 0.00884 | 0.00164 | 0.00444 | 0.00008 | 0.00113 | 65 | 0.01191 | 0.00065 | 0.00263 | 93 | 0.00287 | -2.60487 | 123.120 | 0.01032 | 0.58284 |
| mexT; LysR family transcriptional regulator, mexEF-oprN operon transcriptional activator | 0.00884 | 0.00159 | 0.00951 | 0.00037 | 0.00240 | 65 | 0.00837 | 0.00042 | 0.00213 | 93 | 0.00321 | 0.35447 | 142.989 | 0.72351 | 0.92162 |
| betI; TetR/AcrR family transcriptional regulator, transcriptional repressor of bet genes | 0.00883 | 0.00092 | 0.00859 | 0.00013 | 0.00141 | 65 | 0.00900 | 0.00014 | 0.00122 | 93 | 0.00186 | -0.22015 | 141.338 | 0.82607 | 0.95506 |
| cysZ; CysZ protein | 0.00882 | 0.00085 | 0.00948 | 0.00012 | 0.00136 | 65 | 0.00836 | 0.00011 | 0.00108 | 93 | 0.00174 | 0.64248 | 133.536 | 0.52166 | 0.85613 |
| phoN; acid phosphatase (class A) [EC:3.1.3.2] | 0.00882 | 0.00098 | 0.00854 | 0.00011 | 0.00127 | 65 | 0.00902 | 0.00019 | 0.00142 | 93 | 0.00191 | -0.25161 | 155.183 | 0.80167 | 0.94855 |
| K09792; uncharacterized protein | 0.00881 | 0.00099 | 0.00894 | 0.00014 | 0.00144 | 65 | 0.00872 | 0.00017 | 0.00135 | 93 | 0.00197 | 0.10999 | 146.602 | 0.91257 | 0.97664 |
| ybaO; Lrp/AsnC family transcriptional regulator | 0.00880 | 0.00108 | 0.00757 | 0.00014 | 0.00148 | 65 | 0.00967 | 0.00021 | 0.00151 | 93 | 0.00211 | -0.99259 | 151.889 | 0.32249 | 0.77376 |
| nirK; nitrite reductase (NO-forming) [EC:1.7.2.1] | 0.00880 | 0.00094 | 0.00937 | 0.00014 | 0.00149 | 65 | 0.00841 | 0.00014 | 0.00122 | 93 | 0.00192 | 0.50038 | 136.076 | 0.61762 | 0.88903 |
| ureA; urease subunit gamma [EC:3.5.1.5] | 0.00880 | 0.00099 | 0.00666 | 0.00010 | 0.00126 | 65 | 0.01030 | 0.00019 | 0.00141 | 93 | 0.00189 | -1.92456 | 155.349 | 0.05611 | 0.70577 |
| rsbW; serine/threonine-protein kinase RsbW [EC:2.7.11.1] | 0.00877 | 0.00108 | 0.00984 | 0.00024 | 0.00192 | 65 | 0.00803 | 0.00015 | 0.00126 | 93 | 0.00229 | 0.79211 | 115.830 | 0.42991 | 0.82749 |
| urtA; urea transport system substrate-binding protein | 0.00877 | 0.00103 | 0.00714 | 0.00013 | 0.00139 | 65 | 0.00991 | 0.00020 | 0.00145 | 93 | 0.00201 | -1.37610 | 152.977 | 0.17080 | 0.77376 |
| eutS; ethanolamine utilization protein EutS | 0.00876 | 0.00102 | 0.01134 | 0.00026 | 0.00198 | 65 | 0.00695 | 0.00010 | 0.00102 | 93 | 0.00223 | 1.96792 | 97.465 | 0.05192 | 0.68802 |
| K07336; PKHD-type hydroxylase [EC:1.14.11.-] | 0.00875 | 0.00095 | 0.00863 | 0.00016 | 0.00159 | 65 | 0.00884 | 0.00013 | 0.00118 | 93 | 0.00198 | -0.10862 | 126.996 | 0.91368 | 0.97664 |
| lapB; ATP-binding cassette, subfamily C, bacterial LapB | 0.00875 | 0.00110 | 0.00960 | 0.00019 | 0.00173 | 65 | 0.00815 | 0.00019 | 0.00142 | 93 | 0.00224 | 0.64633 | 136.319 | 0.51915 | 0.85449 |
| K09701; uncharacterized protein | 0.00874 | 0.00090 | 0.00825 | 0.00014 | 0.00146 | 65 | 0.00908 | 0.00012 | 0.00114 | 93 | 0.00185 | -0.44658 | 131.908 | 0.65591 | 0.90679 |
| malK, mtlK, thuK; multiple sugar transport system ATP-binding protein [EC:3.6.3.-] | 0.00874 | 0.00103 | 0.00631 | 0.00009 | 0.00120 | 65 | 0.01043 | 0.00022 | 0.00152 | 93 | 0.00194 | -2.12354 | 155.598 | 0.03529 | 0.62411 |
| eutB; ethanolamine ammonia-lyase large subunit [EC:4.3.1.7] | 0.00874 | 0.00103 | 0.01046 | 0.00024 | 0.00192 | 65 | 0.00753 | 0.00011 | 0.00110 | 93 | 0.00222 | 1.32287 | 105.186 | 0.18875 | 0.77376 |
| K09857; uncharacterized protein | 0.00874 | 0.00120 | 0.00887 | 0.00019 | 0.00170 | 65 | 0.00864 | 0.00026 | 0.00166 | 93 | 0.00238 | 0.09537 | 149.787 | 0.92415 | 0.97981 |
| mdtA; membrane fusion protein, multidrug efflux system | 0.00872 | 0.00094 | 0.00855 | 0.00015 | 0.00150 | 65 | 0.00883 | 0.00014 | 0.00121 | 93 | 0.00193 | -0.14523 | 134.463 | 0.88475 | 0.96984 |
| PTS-Fru-EIIA, fruB; PTS system, fructose-specific IIA component [EC:2.7.1.202] | 0.00871 | 0.00147 | 0.01234 | 0.00070 | 0.00327 | 65 | 0.00617 | 0.00008 | 0.00095 | 93 | 0.00341 | 1.81169 | 74.923 | 0.07404 | 0.74128 |
| treS; maltose alpha-D-glucosyltransferase / alpha-amylase [EC:5.4.99.16 3.2.1.1] | 0.00870 | 0.00114 | 0.00837 | 0.00019 | 0.00169 | 65 | 0.00894 | 0.00022 | 0.00154 | 93 | 0.00228 | -0.24925 | 144.915 | 0.80352 | 0.94911 |
| mobB; molybdopterin-guanine dinucleotide biosynthesis adapter protein | 0.00869 | 0.00080 | 0.00805 | 0.00009 | 0.00119 | 65 | 0.00914 | 0.00011 | 0.00109 | 93 | 0.00161 | -0.67378 | 144.839 | 0.50153 | 0.84942 |
| dctP; C4-dicarboxylate-binding protein DctP | 0.00869 | 0.00130 | 0.00887 | 0.00034 | 0.00227 | 65 | 0.00856 | 0.00022 | 0.00155 | 93 | 0.00275 | 0.11180 | 119.230 | 0.91117 | 0.97664 |
| K09912; uncharacterized protein | 0.00868 | 0.00094 | 0.00901 | 0.00015 | 0.00150 | 65 | 0.00844 | 0.00014 | 0.00122 | 93 | 0.00193 | 0.29723 | 135.240 | 0.76674 | 0.93900 |
| pilX; type IV pilus assembly protein PilX | 0.00867 | 0.00095 | 0.00911 | 0.00016 | 0.00156 | 65 | 0.00837 | 0.00013 | 0.00120 | 93 | 0.00197 | 0.37396 | 130.573 | 0.70904 | 0.92004 |
| pgmB; beta-phosphoglucomutase [EC:5.4.2.6] | 0.00867 | 0.00130 | 0.01101 | 0.00042 | 0.00254 | 65 | 0.00704 | 0.00016 | 0.00130 | 93 | 0.00285 | 1.38997 | 97.601 | 0.16770 | 0.77376 |
| ofaC, arfC; arthrofactin-type cyclic lipopeptide synthetase C | 0.00867 | 0.00289 | 0.00766 | 0.00108 | 0.00407 | 65 | 0.00938 | 0.00149 | 0.00401 | 93 | 0.00572 | -0.30029 | 150.068 | 0.76437 | 0.93888 |
| eutC; ethanolamine ammonia-lyase small subunit [EC:4.3.1.7] | 0.00867 | 0.00103 | 0.01044 | 0.00024 | 0.00192 | 65 | 0.00744 | 0.00011 | 0.00110 | 93 | 0.00222 | 1.35357 | 105.297 | 0.17877 | 0.77376 |
| eutP; ethanolamine utilization protein EutP | 0.00866 | 0.00102 | 0.01134 | 0.00026 | 0.00198 | 65 | 0.00679 | 0.00010 | 0.00101 | 93 | 0.00223 | 2.04285 | 97.195 | 0.04377 | 0.65981 |
| cyaA; adenylate cyclase, class 1 [EC:4.6.1.1] | 0.00865 | 0.00099 | 0.00958 | 0.00015 | 0.00153 | 65 | 0.00800 | 0.00016 | 0.00130 | 93 | 0.00200 | 0.78965 | 139.365 | 0.43107 | 0.82763 |
| tctC; putative tricarboxylic transport membrane protein | 0.00864 | 0.00117 | 0.00693 | 0.00015 | 0.00149 | 65 | 0.00984 | 0.00027 | 0.00169 | 93 | 0.00225 | -1.28829 | 155.469 | 0.19956 | 0.77376 |
| fes; enterochelin esterase and related enzymes | 0.00864 | 0.00087 | 0.00776 | 0.00009 | 0.00118 | 65 | 0.00926 | 0.00014 | 0.00123 | 93 | 0.00170 | -0.87864 | 153.011 | 0.38097 | 0.79908 |
| pilR, pehR; two-component system, NtrC family, response regulator PilR | 0.00863 | 0.00090 | 0.00871 | 0.00013 | 0.00144 | 65 | 0.00857 | 0.00012 | 0.00116 | 93 | 0.00185 | 0.07831 | 134.544 | 0.93770 | 0.98459 |
| gshA, ybdK; glutamate—cysteine ligase / carboxylate-amine ligase [EC:6.3.2.2 6.3.-.-] | 0.00862 | 0.00097 | 0.00832 | 0.00012 | 0.00137 | 65 | 0.00883 | 0.00017 | 0.00135 | 93 | 0.00192 | -0.26509 | 149.979 | 0.79130 | 0.94705 |
| pfp, PFP; diphosphate-dependent phosphofructokinase [EC:2.7.1.90] | 0.00862 | 0.00090 | 0.00752 | 0.00009 | 0.00116 | 65 | 0.00939 | 0.00016 | 0.00129 | 93 | 0.00174 | -1.07757 | 155.109 | 0.28290 | 0.77376 |
| pbpA; penicillin-binding protein A | 0.00862 | 0.00100 | 0.00830 | 0.00014 | 0.00149 | 65 | 0.00884 | 0.00017 | 0.00135 | 93 | 0.00201 | -0.26482 | 144.061 | 0.79153 | 0.94716 |
| lacS, galP, rafP; lactose/raffinose/galactose permease | 0.00861 | 0.00148 | 0.01103 | 0.00058 | 0.00300 | 65 | 0.00692 | 0.00017 | 0.00137 | 93 | 0.00329 | 1.24676 | 90.665 | 0.21570 | 0.77376 |
| pcaK; MFS transporter, AAHS family, 4-hydroxybenzoate transporter | 0.00860 | 0.00123 | 0.00841 | 0.00020 | 0.00176 | 65 | 0.00874 | 0.00027 | 0.00170 | 93 | 0.00244 | -0.13335 | 148.853 | 0.89410 | 0.97180 |
| dppF; dipeptide transport system ATP-binding protein | 0.00860 | 0.00089 | 0.00863 | 0.00010 | 0.00127 | 65 | 0.00858 | 0.00014 | 0.00123 | 93 | 0.00176 | 0.03131 | 149.158 | 0.97507 | 0.99566 |
| algL; poly(beta-D-mannuronate) lyase [EC:4.2.2.3] | 0.00859 | 0.00130 | 0.00892 | 0.00030 | 0.00214 | 65 | 0.00836 | 0.00025 | 0.00164 | 93 | 0.00269 | 0.20723 | 129.907 | 0.83615 | 0.95506 |
| psiE; protein PsiE | 0.00858 | 0.00120 | 0.00939 | 0.00029 | 0.00210 | 65 | 0.00802 | 0.00019 | 0.00143 | 93 | 0.00254 | 0.53902 | 119.229 | 0.59088 | 0.87896 |
| urtD; urea transport system ATP-binding protein | 0.00858 | 0.00102 | 0.00711 | 0.00013 | 0.00139 | 65 | 0.00961 | 0.00019 | 0.00142 | 93 | 0.00199 | -1.25293 | 152.013 | 0.21216 | 0.77376 |
| urtB; urea transport system permease protein | 0.00858 | 0.00102 | 0.00711 | 0.00013 | 0.00139 | 65 | 0.00961 | 0.00019 | 0.00142 | 93 | 0.00199 | -1.25473 | 152.016 | 0.21150 | 0.77376 |
| urtC; urea transport system permease protein | 0.00858 | 0.00102 | 0.00711 | 0.00013 | 0.00139 | 65 | 0.00961 | 0.00019 | 0.00142 | 93 | 0.00199 | -1.25598 | 152.008 | 0.21105 | 0.77376 |
| dppB; dipeptide transport system permease protein | 0.00858 | 0.00089 | 0.00859 | 0.00010 | 0.00127 | 65 | 0.00857 | 0.00014 | 0.00123 | 93 | 0.00176 | 0.01459 | 149.167 | 0.98837 | 0.99801 |
| cusA, silA; Cu(I)/Ag(I) efflux system membrane protein CusA/SilA | 0.00858 | 0.00091 | 0.00820 | 0.00012 | 0.00134 | 65 | 0.00884 | 0.00014 | 0.00123 | 93 | 0.00182 | -0.35145 | 145.522 | 0.72576 | 0.92177 |
| urtE; urea transport system ATP-binding protein | 0.00858 | 0.00102 | 0.00708 | 0.00012 | 0.00139 | 65 | 0.00962 | 0.00019 | 0.00142 | 93 | 0.00199 | -1.27933 | 152.271 | 0.20273 | 0.77376 |
| K03791; putative chitinase | 0.00857 | 0.00149 | 0.00717 | 0.00027 | 0.00204 | 65 | 0.00956 | 0.00041 | 0.00210 | 93 | 0.00292 | -0.81546 | 152.506 | 0.41608 | 0.81772 |
| merR; MerR family transcriptional regulator, mercuric resistance operon regulatory protein | 0.00857 | 0.00128 | 0.00828 | 0.00028 | 0.00208 | 65 | 0.00878 | 0.00025 | 0.00163 | 93 | 0.00264 | -0.18729 | 131.984 | 0.85172 | 0.95806 |
| linN; cholesterol transport system auxiliary component | 0.00857 | 0.00096 | 0.00790 | 0.00016 | 0.00155 | 65 | 0.00904 | 0.00014 | 0.00121 | 93 | 0.00197 | -0.57896 | 131.849 | 0.56360 | 0.86877 |
| E3.4.21.96; lactocepin [EC:3.4.21.96] | 0.00856 | 0.00076 | 0.00880 | 0.00010 | 0.00126 | 65 | 0.00839 | 0.00008 | 0.00094 | 93 | 0.00157 | 0.26145 | 128.032 | 0.79417 | 0.94783 |
| K07080; uncharacterized protein | 0.00856 | 0.00088 | 0.00785 | 0.00014 | 0.00145 | 65 | 0.00905 | 0.00011 | 0.00110 | 93 | 0.00182 | -0.66380 | 129.254 | 0.50800 | 0.84956 |
| fecA; Fe(3+) dicitrate transport protein | 0.00853 | 0.00124 | 0.00693 | 0.00009 | 0.00114 | 65 | 0.00965 | 0.00035 | 0.00195 | 93 | 0.00226 | -1.20737 | 142.129 | 0.22930 | 0.77376 |
| p19, ftrA; periplasmic iron binding protein | 0.00853 | 0.00098 | 0.01221 | 0.00024 | 0.00194 | 65 | 0.00595 | 0.00007 | 0.00087 | 93 | 0.00213 | 2.94009 | 90.054 | 0.00417 | 0.52172 |
| hofQ; protein transport protein HofQ | 0.00852 | 0.00090 | 0.00936 | 0.00015 | 0.00150 | 65 | 0.00794 | 0.00012 | 0.00112 | 93 | 0.00187 | 0.75623 | 127.880 | 0.45090 | 0.83438 |
| virB4, lvhB4; type IV secretion system protein VirB4 | 0.00852 | 0.00106 | 0.00885 | 0.00029 | 0.00211 | 65 | 0.00828 | 0.00010 | 0.00103 | 93 | 0.00235 | 0.24331 | 94.369 | 0.80830 | 0.95002 |
| TC.DAACS; dicarboxylate/amino acid:cation (Na+ or H+) symporter, DAACS family | 0.00852 | 0.00076 | 0.00781 | 0.00008 | 0.00113 | 65 | 0.00901 | 0.00010 | 0.00103 | 93 | 0.00152 | -0.79096 | 145.045 | 0.43026 | 0.82749 |
| dbpA; ATP-independent RNA helicase DbpA [EC:3.6.4.13] | 0.00851 | 0.00092 | 0.00882 | 0.00015 | 0.00151 | 65 | 0.00829 | 0.00013 | 0.00117 | 93 | 0.00191 | 0.27730 | 131.156 | 0.78198 | 0.94310 |
| E3.5.1.28D, amiD; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | 0.00849 | 0.00102 | 0.01122 | 0.00025 | 0.00195 | 65 | 0.00659 | 0.00010 | 0.00103 | 93 | 0.00221 | 2.10016 | 99.021 | 0.03826 | 0.63091 |
| DAK, TKFC; triose/dihydroxyacetone kinase / FAD-AMP lyase (cyclizing) [EC:2.7.1.28 2.7.1.29 4.6.1.15] | 0.00849 | 0.00081 | 0.00727 | 0.00008 | 0.00109 | 65 | 0.00934 | 0.00012 | 0.00115 | 93 | 0.00158 | -1.31045 | 153.451 | 0.19200 | 0.77376 |
| gctB; glutaconate CoA-transferase, subunit B [EC:2.8.3.12] | 0.00849 | 0.00109 | 0.01053 | 0.00024 | 0.00191 | 65 | 0.00706 | 0.00015 | 0.00127 | 93 | 0.00229 | 1.51575 | 116.929 | 0.13228 | 0.77376 |
| COX11, ctaG; cytochrome c oxidase assembly protein subunit 11 | 0.00849 | 0.00092 | 0.00764 | 0.00015 | 0.00152 | 65 | 0.00907 | 0.00012 | 0.00115 | 93 | 0.00190 | -0.75028 | 129.453 | 0.45445 | 0.83438 |
| ureB; urease subunit beta [EC:3.5.1.5] | 0.00848 | 0.00098 | 0.00660 | 0.00010 | 0.00125 | 65 | 0.00980 | 0.00018 | 0.00141 | 93 | 0.00188 | -1.70173 | 155.394 | 0.09081 | 0.76567 |
| dgcB; diguanylate cyclase [EC:2.7.7.65] | 0.00848 | 0.00120 | 0.00806 | 0.00021 | 0.00180 | 65 | 0.00877 | 0.00024 | 0.00162 | 93 | 0.00242 | -0.29431 | 143.897 | 0.76894 | 0.93993 |
| larE; pyridinium-3,5-biscarboxylic acid mononucleotide sulfurtransferase | 0.00847 | 0.00083 | 0.00885 | 0.00013 | 0.00140 | 65 | 0.00820 | 0.00010 | 0.00103 | 93 | 0.00174 | 0.37575 | 126.738 | 0.70773 | 0.92004 |
| K07075; uncharacterized protein | 0.00847 | 0.00096 | 0.00880 | 0.00016 | 0.00155 | 65 | 0.00823 | 0.00014 | 0.00123 | 93 | 0.00197 | 0.29107 | 133.224 | 0.77145 | 0.94067 |
| AXY8, FUC95A, afcA; alpha-L-fucosidase 2 [EC:3.2.1.51] | 0.00845 | 0.00079 | 0.00712 | 0.00006 | 0.00097 | 65 | 0.00937 | 0.00013 | 0.00116 | 93 | 0.00151 | -1.48759 | 156.000 | 0.13888 | 0.77376 |
| cpaA, tadV; prepilin peptidase CpaA [EC:3.4.23.43] | 0.00845 | 0.00094 | 0.00781 | 0.00014 | 0.00147 | 65 | 0.00889 | 0.00014 | 0.00122 | 93 | 0.00191 | -0.56913 | 137.166 | 0.57020 | 0.87135 |
| K06908; uncharacterized protein | 0.00843 | 0.00128 | 0.00749 | 0.00017 | 0.00163 | 65 | 0.00909 | 0.00032 | 0.00185 | 93 | 0.00247 | -0.64754 | 155.514 | 0.51824 | 0.85358 |
| iorB; isoquinoline 1-oxidoreductase subunit beta [EC:1.3.99.16] | 0.00842 | 0.00127 | 0.00688 | 0.00016 | 0.00155 | 65 | 0.00949 | 0.00033 | 0.00187 | 93 | 0.00243 | -1.07349 | 155.993 | 0.28471 | 0.77376 |
| sdaC; serine transporter | 0.00838 | 0.00097 | 0.00817 | 0.00011 | 0.00131 | 65 | 0.00853 | 0.00018 | 0.00138 | 93 | 0.00190 | -0.18758 | 153.537 | 0.85146 | 0.95792 |
| rsbU_P; phosphoserine phosphatase RsbU/P [EC:3.1.3.3] | 0.00838 | 0.00110 | 0.00867 | 0.00021 | 0.00180 | 65 | 0.00818 | 0.00018 | 0.00140 | 93 | 0.00228 | 0.21597 | 131.540 | 0.82934 | 0.95506 |
| pnbA; para-nitrobenzyl esterase [EC:3.1.1.-] | 0.00837 | 0.00130 | 0.00521 | 0.00008 | 0.00114 | 65 | 0.01059 | 0.00039 | 0.00204 | 93 | 0.00233 | -2.30381 | 139.251 | 0.02271 | 0.59764 |
| pilS, pehS; two-component system, NtrC family, sensor histidine kinase PilS [EC:2.7.13.3] | 0.00837 | 0.00089 | 0.00843 | 0.00013 | 0.00143 | 65 | 0.00832 | 0.00012 | 0.00113 | 93 | 0.00183 | 0.06269 | 132.673 | 0.95011 | 0.98774 |
| dkgA; 2,5-diketo-D-gluconate reductase A [EC:1.1.1.346] | 0.00837 | 0.00090 | 0.00818 | 0.00014 | 0.00148 | 65 | 0.00850 | 0.00012 | 0.00113 | 93 | 0.00186 | -0.17094 | 129.740 | 0.86453 | 0.96263 |
| hyaD, hybD; hydrogenase maturation protease [EC:3.4.23.-] | 0.00836 | 0.00086 | 0.00859 | 0.00013 | 0.00141 | 65 | 0.00819 | 0.00011 | 0.00109 | 93 | 0.00178 | 0.22526 | 131.197 | 0.82213 | 0.95506 |
| pspA; phage shock protein A | 0.00835 | 0.00081 | 0.00778 | 0.00011 | 0.00129 | 65 | 0.00875 | 0.00010 | 0.00105 | 93 | 0.00166 | -0.58803 | 135.748 | 0.55749 | 0.86813 |
| E5.1.3.6; UDP-glucuronate 4-epimerase [EC:5.1.3.6] | 0.00831 | 0.00081 | 0.00732 | 0.00010 | 0.00122 | 65 | 0.00900 | 0.00011 | 0.00108 | 93 | 0.00163 | -1.02730 | 142.636 | 0.30602 | 0.77376 |
| norB; nitric oxide reductase subunit B [EC:1.7.2.5] | 0.00829 | 0.00087 | 0.00812 | 0.00013 | 0.00141 | 65 | 0.00841 | 0.00011 | 0.00110 | 93 | 0.00179 | -0.15995 | 131.649 | 0.87317 | 0.96576 |
| int; integrase | 0.00828 | 0.00092 | 0.00772 | 0.00013 | 0.00143 | 65 | 0.00867 | 0.00014 | 0.00121 | 93 | 0.00187 | -0.50303 | 139.347 | 0.61574 | 0.88878 |
| mscK, kefA, aefA; potassium-dependent mechanosensitive channel | 0.00827 | 0.00085 | 0.00917 | 0.00012 | 0.00135 | 65 | 0.00764 | 0.00011 | 0.00110 | 93 | 0.00174 | 0.88185 | 135.800 | 0.37942 | 0.79852 |
| pilZ; type IV pilus assembly protein PilZ | 0.00827 | 0.00088 | 0.00843 | 0.00013 | 0.00143 | 65 | 0.00815 | 0.00012 | 0.00112 | 93 | 0.00182 | 0.15279 | 132.090 | 0.87880 | 0.96759 |
| SORD, gutB; L-iditol 2-dehydrogenase [EC:1.1.1.14] | 0.00824 | 0.00104 | 0.00724 | 0.00018 | 0.00167 | 65 | 0.00894 | 0.00017 | 0.00134 | 93 | 0.00214 | -0.79317 | 134.194 | 0.42908 | 0.82749 |
| phbB; acetoacetyl-CoA reductase [EC:1.1.1.36] | 0.00823 | 0.00127 | 0.00712 | 0.00027 | 0.00206 | 65 | 0.00900 | 0.00024 | 0.00161 | 93 | 0.00261 | -0.71830 | 131.962 | 0.47384 | 0.84050 |
| mdcD; malonate decarboxylase beta subunit [EC:4.1.1.87] | 0.00822 | 0.00120 | 0.00830 | 0.00021 | 0.00179 | 65 | 0.00817 | 0.00024 | 0.00162 | 93 | 0.00242 | 0.05530 | 144.299 | 0.95597 | 0.98792 |
| yajG; uncharacterized lipoprotein | 0.00822 | 0.00083 | 0.00929 | 0.00012 | 0.00133 | 65 | 0.00747 | 0.00010 | 0.00106 | 93 | 0.00170 | 1.06886 | 133.002 | 0.28707 | 0.77376 |
| prkA; serine protein kinase | 0.00821 | 0.00098 | 0.00749 | 0.00014 | 0.00146 | 65 | 0.00872 | 0.00016 | 0.00131 | 93 | 0.00196 | -0.62301 | 143.998 | 0.53426 | 0.86207 |
| vasD, lip; type VI secretion system protein VasD | 0.00821 | 0.00109 | 0.00942 | 0.00017 | 0.00160 | 65 | 0.00735 | 0.00020 | 0.00147 | 93 | 0.00218 | 0.95021 | 145.440 | 0.34358 | 0.78597 |
| K09777; uncharacterized protein | 0.00820 | 0.00084 | 0.00813 | 0.00013 | 0.00142 | 65 | 0.00825 | 0.00010 | 0.00102 | 93 | 0.00175 | -0.06629 | 124.430 | 0.94725 | 0.98674 |
| yagS; xanthine dehydrogenase YagS FAD-binding subunit [EC:1.17.1.4] | 0.00820 | 0.00098 | 0.00757 | 0.00014 | 0.00148 | 65 | 0.00864 | 0.00016 | 0.00132 | 93 | 0.00198 | -0.54242 | 143.171 | 0.58837 | 0.87732 |
| pdxB; erythronate-4-phosphate dehydrogenase [EC:1.1.1.290] | 0.00820 | 0.00077 | 0.00819 | 0.00009 | 0.00115 | 65 | 0.00821 | 0.00010 | 0.00103 | 93 | 0.00154 | -0.00870 | 143.250 | 0.99307 | 0.99919 |
| mmuP; S-methylmethionine transporter | 0.00820 | 0.00101 | 0.00825 | 0.00013 | 0.00143 | 65 | 0.00816 | 0.00018 | 0.00140 | 93 | 0.00201 | 0.04500 | 149.768 | 0.96416 | 0.99101 |
| mtfA; MtfA peptidase | 0.00820 | 0.00092 | 0.00821 | 0.00015 | 0.00151 | 65 | 0.00819 | 0.00013 | 0.00116 | 93 | 0.00190 | 0.01157 | 130.339 | 0.99079 | 0.99847 |
| ubiI; 2-octaprenylphenol hydroxylase [EC:1.14.13.-] | 0.00818 | 0.00098 | 0.00912 | 0.00014 | 0.00149 | 65 | 0.00753 | 0.00016 | 0.00130 | 93 | 0.00198 | 0.80495 | 141.235 | 0.42220 | 0.82295 |
| yfbR; 5’-deoxynucleotidase [EC:3.1.3.89] | 0.00818 | 0.00081 | 0.00941 | 0.00014 | 0.00144 | 65 | 0.00732 | 0.00008 | 0.00094 | 93 | 0.00172 | 1.21694 | 115.065 | 0.22612 | 0.77376 |
| tarJ; ribitol-5-phosphate 2-dehydrogenase (NADP+) [EC:1.1.1.405] | 0.00818 | 0.00085 | 0.00690 | 0.00008 | 0.00108 | 65 | 0.00907 | 0.00014 | 0.00123 | 93 | 0.00164 | -1.32734 | 155.656 | 0.18634 | 0.77376 |
| virB9, lvhB9; type IV secretion system protein VirB9 | 0.00817 | 0.00104 | 0.00776 | 0.00025 | 0.00196 | 65 | 0.00846 | 0.00012 | 0.00113 | 93 | 0.00226 | -0.31084 | 105.671 | 0.75653 | 0.93680 |
| rstA; two-component system, OmpR family, response regulator RstA | 0.00817 | 0.00090 | 0.00832 | 0.00013 | 0.00141 | 65 | 0.00806 | 0.00013 | 0.00117 | 93 | 0.00183 | 0.13968 | 137.696 | 0.88911 | 0.97039 |
| pvdQ, quiP; acyl-homoserine-lactone acylase [EC:3.5.1.97] | 0.00814 | 0.00137 | 0.00805 | 0.00024 | 0.00193 | 65 | 0.00820 | 0.00034 | 0.00190 | 93 | 0.00271 | -0.05464 | 150.105 | 0.95650 | 0.98831 |
| yhjG; AsmA family protein | 0.00814 | 0.00093 | 0.00803 | 0.00015 | 0.00151 | 65 | 0.00821 | 0.00013 | 0.00117 | 93 | 0.00191 | -0.09362 | 131.218 | 0.92556 | 0.98041 |
| K07577; putative mRNA 3-end processing factor | 0.00814 | 0.00089 | 0.00720 | 0.00013 | 0.00140 | 65 | 0.00879 | 0.00012 | 0.00115 | 93 | 0.00181 | -0.87632 | 136.704 | 0.38240 | 0.80037 |
| ndpA; nucleoid-associated protein | 0.00813 | 0.00085 | 0.00932 | 0.00013 | 0.00140 | 65 | 0.00730 | 0.00011 | 0.00107 | 93 | 0.00176 | 1.14474 | 129.671 | 0.25443 | 0.77376 |
| uspA; universal stress protein A | 0.00813 | 0.00085 | 0.00910 | 0.00012 | 0.00136 | 65 | 0.00746 | 0.00011 | 0.00108 | 93 | 0.00174 | 0.94564 | 132.904 | 0.34605 | 0.78846 |
| cpaB, rcpC; pilus assembly protein CpaB | 0.00813 | 0.00088 | 0.00661 | 0.00010 | 0.00125 | 65 | 0.00919 | 0.00013 | 0.00120 | 93 | 0.00173 | -1.48892 | 148.856 | 0.13863 | 0.77376 |
| hupA; DNA-binding protein HU-alpha | 0.00813 | 0.00086 | 0.00917 | 0.00012 | 0.00137 | 65 | 0.00741 | 0.00011 | 0.00110 | 93 | 0.00175 | 1.00341 | 134.149 | 0.31747 | 0.77376 |
| E4.1.1.32, pckA, PCK; phosphoenolpyruvate carboxykinase (GTP) [EC:4.1.1.32] | 0.00813 | 0.00084 | 0.00738 | 0.00010 | 0.00122 | 65 | 0.00865 | 0.00012 | 0.00114 | 93 | 0.00167 | -0.76523 | 146.313 | 0.44536 | 0.83300 |
| K09989; uncharacterized protein | 0.00812 | 0.00088 | 0.00742 | 0.00013 | 0.00139 | 65 | 0.00861 | 0.00012 | 0.00115 | 93 | 0.00180 | -0.65913 | 137.314 | 0.51092 | 0.85070 |
| rsbV; anti-sigma B factor antagonist | 0.00811 | 0.00100 | 0.00887 | 0.00022 | 0.00183 | 65 | 0.00759 | 0.00012 | 0.00112 | 93 | 0.00215 | 0.59440 | 110.194 | 0.55346 | 0.86788 |
| hisB; imidazoleglycerol-phosphate dehydratase / histidinol-phosphatase [EC:4.2.1.19 3.1.3.15] | 0.00810 | 0.00083 | 0.00808 | 0.00011 | 0.00130 | 65 | 0.00812 | 0.00011 | 0.00108 | 93 | 0.00169 | -0.02370 | 136.722 | 0.98113 | 0.99661 |
| virB10, lvhB10; type IV secretion system protein VirB10 | 0.00808 | 0.00112 | 0.00735 | 0.00030 | 0.00216 | 65 | 0.00860 | 0.00013 | 0.00117 | 93 | 0.00245 | -0.50775 | 100.885 | 0.61273 | 0.88788 |
| pldA; phospholipase A1/A2 [EC:3.1.1.32 3.1.1.4] | 0.00808 | 0.00085 | 0.00758 | 0.00010 | 0.00121 | 65 | 0.00843 | 0.00013 | 0.00118 | 93 | 0.00169 | -0.50303 | 149.363 | 0.61569 | 0.88878 |
| yagT; xanthine dehydrogenase YagT iron-sulfur-binding subunit | 0.00807 | 0.00097 | 0.00763 | 0.00015 | 0.00151 | 65 | 0.00838 | 0.00015 | 0.00126 | 93 | 0.00197 | -0.38258 | 138.236 | 0.70262 | 0.92004 |
| phhA, PAH; phenylalanine-4-hydroxylase [EC:1.14.16.1] | 0.00806 | 0.00092 | 0.00766 | 0.00015 | 0.00151 | 65 | 0.00835 | 0.00012 | 0.00115 | 93 | 0.00190 | -0.36491 | 129.879 | 0.71578 | 0.92004 |
| larB; pyridinium-3,5-biscarboxylic acid mononucleotide synthase [EC:2.5.1.143] | 0.00806 | 0.00082 | 0.00852 | 0.00013 | 0.00141 | 65 | 0.00774 | 0.00009 | 0.00099 | 93 | 0.00173 | 0.45619 | 122.357 | 0.64906 | 0.90350 |
| nrfH; cytochrome c nitrite reductase small subunit | 0.00806 | 0.00106 | 0.00809 | 0.00016 | 0.00155 | 65 | 0.00804 | 0.00019 | 0.00144 | 93 | 0.00212 | 0.02061 | 146.343 | 0.98358 | 0.99706 |
| rstB; two-component system, OmpR family, sensor histidine kinase RstB [EC:2.7.13.3] | 0.00806 | 0.00089 | 0.00814 | 0.00013 | 0.00141 | 65 | 0.00800 | 0.00013 | 0.00116 | 93 | 0.00182 | 0.07500 | 136.570 | 0.94033 | 0.98528 |
| E3.2.1.4; endoglucanase [EC:3.2.1.4] | 0.00804 | 0.00082 | 0.00561 | 0.00007 | 0.00106 | 65 | 0.00973 | 0.00012 | 0.00116 | 93 | 0.00157 | -2.62069 | 154.526 | 0.00965 | 0.58112 |
| glpE; thiosulfate sulfurtransferase [EC:2.8.1.1] | 0.00800 | 0.00084 | 0.00898 | 0.00012 | 0.00135 | 65 | 0.00732 | 0.00010 | 0.00106 | 93 | 0.00172 | 0.96215 | 132.315 | 0.33773 | 0.78354 |
| E1.2.7.8; indolepyruvate ferredoxin oxidoreductase [EC:1.2.7.8] | 0.00799 | 0.00100 | 0.00716 | 0.00015 | 0.00151 | 65 | 0.00858 | 0.00016 | 0.00132 | 93 | 0.00201 | -0.70357 | 141.697 | 0.48286 | 0.84354 |
| PTS-HPR.PTSH, ptsH; phosphocarrier protein HPr | 0.00797 | 0.00104 | 0.01077 | 0.00026 | 0.00199 | 65 | 0.00602 | 0.00011 | 0.00106 | 93 | 0.00225 | 2.11194 | 100.172 | 0.03718 | 0.63026 |
| evgA, bvgA; two-component system, NarL family, response regulator EvgA | 0.00797 | 0.00153 | 0.00786 | 0.00030 | 0.00215 | 65 | 0.00805 | 0.00042 | 0.00214 | 93 | 0.00303 | -0.06035 | 150.692 | 0.95195 | 0.98792 |
| mioC; MioC protein | 0.00796 | 0.00084 | 0.00899 | 0.00012 | 0.00135 | 65 | 0.00725 | 0.00011 | 0.00107 | 93 | 0.00172 | 1.01002 | 132.601 | 0.31433 | 0.77376 |
| lytS; two-component system, LytTR family, sensor histidine kinase LytS [EC:2.7.13.3] | 0.00796 | 0.00085 | 0.00821 | 0.00015 | 0.00150 | 65 | 0.00778 | 0.00009 | 0.00100 | 93 | 0.00180 | 0.23902 | 117.863 | 0.81151 | 0.95214 |
| csdA; cysteine sulfinate desulfinase [EC:4.4.1.-] | 0.00795 | 0.00085 | 0.00884 | 0.00012 | 0.00134 | 65 | 0.00733 | 0.00011 | 0.00111 | 93 | 0.00174 | 0.86725 | 137.420 | 0.38732 | 0.80262 |
| K09898; uncharacterized protein | 0.00794 | 0.00084 | 0.00902 | 0.00012 | 0.00135 | 65 | 0.00719 | 0.00011 | 0.00107 | 93 | 0.00172 | 1.05823 | 132.771 | 0.29187 | 0.77376 |
| tusC, dsrF; tRNA 2-thiouridine synthesizing protein C | 0.00794 | 0.00084 | 0.00901 | 0.00012 | 0.00135 | 65 | 0.00718 | 0.00011 | 0.00107 | 93 | 0.00172 | 1.06203 | 132.676 | 0.29015 | 0.77376 |
| cysM; cysteine synthase B [EC:2.5.1.47] | 0.00790 | 0.00086 | 0.00776 | 0.00012 | 0.00137 | 65 | 0.00800 | 0.00011 | 0.00111 | 93 | 0.00176 | -0.13618 | 134.718 | 0.89189 | 0.97141 |
| perM; putative permease | 0.00790 | 0.00084 | 0.00890 | 0.00012 | 0.00134 | 65 | 0.00720 | 0.00011 | 0.00106 | 93 | 0.00171 | 0.99152 | 132.930 | 0.32323 | 0.77376 |
| cusB, silB; membrane fusion protein, Cu(I)/Ag(I) efflux system | 0.00790 | 0.00086 | 0.00776 | 0.00011 | 0.00132 | 65 | 0.00800 | 0.00012 | 0.00114 | 93 | 0.00174 | -0.13630 | 140.609 | 0.89178 | 0.97141 |
| asl; D-aspartate ligase [EC:6.3.1.12] | 0.00789 | 0.00142 | 0.00850 | 0.00033 | 0.00225 | 65 | 0.00746 | 0.00031 | 0.00184 | 93 | 0.00290 | 0.35646 | 135.654 | 0.72205 | 0.92156 |
| mexL; TetR/AcrR family transcriptional regulator, mexJK operon transcriptional repressor | 0.00789 | 0.00091 | 0.00793 | 0.00014 | 0.00144 | 65 | 0.00785 | 0.00013 | 0.00117 | 93 | 0.00186 | 0.04308 | 135.377 | 0.96570 | 0.99101 |
| lapE; outer membrane protein, adhesin transport system | 0.00788 | 0.00089 | 0.00880 | 0.00014 | 0.00146 | 65 | 0.00724 | 0.00012 | 0.00112 | 93 | 0.00184 | 0.84649 | 129.963 | 0.39883 | 0.80624 |
| K06903; uncharacterized protein | 0.00788 | 0.00116 | 0.00654 | 0.00014 | 0.00147 | 65 | 0.00882 | 0.00026 | 0.00168 | 93 | 0.00224 | -1.02059 | 155.621 | 0.30903 | 0.77376 |
| katG; catalase-peroxidase [EC:1.11.1.21] | 0.00788 | 0.00087 | 0.00694 | 0.00012 | 0.00134 | 65 | 0.00853 | 0.00012 | 0.00114 | 93 | 0.00176 | -0.90267 | 139.406 | 0.36826 | 0.79587 |
| cpaE, tadZ; pilus assembly protein CpaE | 0.00788 | 0.00089 | 0.00629 | 0.00011 | 0.00127 | 65 | 0.00898 | 0.00014 | 0.00121 | 93 | 0.00176 | -1.53351 | 147.968 | 0.12728 | 0.77376 |
| hugZ, hutZ; heme iron utilization protein | 0.00787 | 0.00090 | 0.00712 | 0.00009 | 0.00120 | 65 | 0.00839 | 0.00015 | 0.00127 | 93 | 0.00175 | -0.72494 | 153.766 | 0.46959 | 0.83870 |
| ribB, RIB3; 3,4-dihydroxy 2-butanone 4-phosphate synthase [EC:4.1.99.12] | 0.00786 | 0.00076 | 0.00888 | 0.00011 | 0.00127 | 65 | 0.00715 | 0.00008 | 0.00094 | 93 | 0.00158 | 1.08910 | 126.512 | 0.27818 | 0.77376 |
| K09915; uncharacterized protein | 0.00785 | 0.00091 | 0.00802 | 0.00014 | 0.00145 | 65 | 0.00773 | 0.00013 | 0.00117 | 93 | 0.00186 | 0.15712 | 134.840 | 0.87538 | 0.96693 |
| cysN; sulfate adenylyltransferase subunit 1 [EC:2.7.7.4] | 0.00784 | 0.00067 | 0.00708 | 0.00005 | 0.00087 | 65 | 0.00837 | 0.00009 | 0.00096 | 93 | 0.00130 | -0.98944 | 154.980 | 0.32399 | 0.77376 |
| fixC; electron transfer flavoprotein-quinone oxidoreductase [EC:1.5.5.-] | 0.00784 | 0.00093 | 0.00989 | 0.00021 | 0.00181 | 65 | 0.00641 | 0.00008 | 0.00094 | 93 | 0.00203 | 1.71161 | 98.067 | 0.09013 | 0.76567 |
| osmY; hyperosmotically inducible periplasmic protein | 0.00783 | 0.00088 | 0.00825 | 0.00013 | 0.00142 | 65 | 0.00753 | 0.00012 | 0.00113 | 93 | 0.00181 | 0.39686 | 133.203 | 0.69211 | 0.91676 |
| bioG; pimeloyl-[acyl-carrier protein] methyl ester esterase [EC:3.1.1.85] | 0.00783 | 0.00103 | 0.00965 | 0.00024 | 0.00194 | 65 | 0.00656 | 0.00012 | 0.00111 | 93 | 0.00223 | 1.38357 | 105.207 | 0.16942 | 0.77376 |
| K09923; uncharacterized protein | 0.00783 | 0.00083 | 0.00883 | 0.00012 | 0.00134 | 65 | 0.00712 | 0.00011 | 0.00106 | 93 | 0.00171 | 1.00016 | 133.491 | 0.31904 | 0.77376 |
| K09908; uncharacterized protein | 0.00783 | 0.00083 | 0.00883 | 0.00012 | 0.00133 | 65 | 0.00712 | 0.00010 | 0.00105 | 93 | 0.00170 | 1.00535 | 132.775 | 0.31656 | 0.77376 |
| K07222; putative flavoprotein involved in K+ transport | 0.00782 | 0.00093 | 0.00708 | 0.00013 | 0.00143 | 65 | 0.00834 | 0.00014 | 0.00123 | 93 | 0.00189 | -0.66981 | 140.567 | 0.50408 | 0.84956 |
| xdhB; xanthine dehydrogenase large subunit [EC:1.17.1.4] | 0.00781 | 0.00118 | 0.00720 | 0.00019 | 0.00173 | 65 | 0.00824 | 0.00024 | 0.00162 | 93 | 0.00236 | -0.44095 | 146.826 | 0.65990 | 0.90939 |
| gpr; L-glyceraldehyde 3-phosphate reductase [EC:1.1.1.-] | 0.00781 | 0.00080 | 0.00714 | 0.00011 | 0.00131 | 65 | 0.00828 | 0.00009 | 0.00100 | 93 | 0.00165 | -0.69230 | 129.507 | 0.48999 | 0.84797 |
| aceK; isocitrate dehydrogenase kinase/phosphatase [EC:2.7.11.5 3.1.3.-] | 0.00780 | 0.00085 | 0.00829 | 0.00014 | 0.00145 | 65 | 0.00746 | 0.00010 | 0.00103 | 93 | 0.00178 | 0.46560 | 123.241 | 0.64232 | 0.90098 |
| gctA; glutaconate CoA-transferase, subunit A [EC:2.8.3.12] | 0.00780 | 0.00095 | 0.00981 | 0.00021 | 0.00178 | 65 | 0.00640 | 0.00010 | 0.00102 | 93 | 0.00205 | 1.66426 | 104.977 | 0.09904 | 0.76942 |
| PTS-EI.PTSP, ptsP; phosphotransferase system, enzyme I, PtsP [EC:2.7.3.9] | 0.00779 | 0.00083 | 0.00729 | 0.00010 | 0.00124 | 65 | 0.00814 | 0.00012 | 0.00111 | 93 | 0.00167 | -0.51130 | 143.656 | 0.60992 | 0.88693 |
| TC.PST; polysaccharide transporter, PST family | 0.00778 | 0.00093 | 0.00726 | 0.00019 | 0.00169 | 65 | 0.00815 | 0.00010 | 0.00106 | 93 | 0.00200 | -0.44910 | 111.668 | 0.65423 | 0.90679 |
| K09793; uncharacterized protein | 0.00778 | 0.00089 | 0.00620 | 0.00010 | 0.00122 | 65 | 0.00888 | 0.00014 | 0.00123 | 93 | 0.00173 | -1.54624 | 151.505 | 0.12413 | 0.77376 |
| tusB, dsrH; tRNA 2-thiouridine synthesizing protein B | 0.00777 | 0.00084 | 0.00886 | 0.00012 | 0.00136 | 65 | 0.00701 | 0.00011 | 0.00107 | 93 | 0.00173 | 1.06813 | 132.405 | 0.28741 | 0.77376 |
| tyrR; transcriptional regulator of aroF, aroG, tyrA and aromatic amino acid transport | 0.00776 | 0.00082 | 0.00862 | 0.00011 | 0.00132 | 65 | 0.00716 | 0.00010 | 0.00105 | 93 | 0.00169 | 0.86694 | 133.347 | 0.38753 | 0.80262 |
| lsa; lincosamide and streptogramin A transport system ATP-binding/permease protein | 0.00776 | 0.00119 | 0.00999 | 0.00039 | 0.00246 | 65 | 0.00620 | 0.00010 | 0.00106 | 93 | 0.00268 | 1.41614 | 87.768 | 0.16027 | 0.77376 |
| rubB, alkT; rubredoxin—NAD+ reductase [EC:1.18.1.1] | 0.00776 | 0.00099 | 0.00857 | 0.00018 | 0.00164 | 65 | 0.00719 | 0.00014 | 0.00124 | 93 | 0.00206 | 0.67401 | 128.750 | 0.50151 | 0.84942 |
| K06992; uncharacterized protein | 0.00775 | 0.00089 | 0.00749 | 0.00012 | 0.00139 | 65 | 0.00794 | 0.00013 | 0.00117 | 93 | 0.00181 | -0.24607 | 138.559 | 0.80599 | 0.94958 |
| coaW; type II pantothenate kinase [EC:2.7.1.33] | 0.00774 | 0.00089 | 0.00610 | 0.00008 | 0.00108 | 65 | 0.00888 | 0.00016 | 0.00130 | 93 | 0.00169 | -1.64581 | 155.994 | 0.10182 | 0.77376 |
| creA; CreA protein | 0.00773 | 0.00079 | 0.00759 | 0.00009 | 0.00119 | 65 | 0.00783 | 0.00010 | 0.00106 | 93 | 0.00159 | -0.15086 | 143.109 | 0.88030 | 0.96815 |
| aapJ, bztA; general L-amino acid transport system substrate-binding protein | 0.00773 | 0.00105 | 0.00628 | 0.00011 | 0.00131 | 65 | 0.00875 | 0.00022 | 0.00152 | 93 | 0.00200 | -1.22902 | 155.876 | 0.22092 | 0.77376 |
| kdgD; 5-dehydro-4-deoxyglucarate dehydratase [EC:4.2.1.41] | 0.00772 | 0.00132 | 0.00691 | 0.00021 | 0.00179 | 65 | 0.00829 | 0.00032 | 0.00186 | 93 | 0.00259 | -0.53329 | 152.784 | 0.59460 | 0.88005 |
| rhaB; rhamnulokinase [EC:2.7.1.5] | 0.00772 | 0.00088 | 0.00859 | 0.00020 | 0.00175 | 65 | 0.00711 | 0.00007 | 0.00088 | 93 | 0.00195 | 0.75566 | 96.082 | 0.45170 | 0.83438 |
| glpC; glycerol-3-phosphate dehydrogenase subunit C [EC:1.1.5.3] | 0.00772 | 0.00076 | 0.00771 | 0.00007 | 0.00105 | 65 | 0.00772 | 0.00011 | 0.00107 | 93 | 0.00150 | -0.00387 | 151.957 | 0.99692 | 0.99951 |
| E4.6.1.1B, cyaB; adenylate cyclase, class 2 [EC:4.6.1.1] | 0.00772 | 0.00082 | 0.00749 | 0.00010 | 0.00121 | 65 | 0.00787 | 0.00012 | 0.00112 | 93 | 0.00165 | -0.23192 | 145.913 | 0.81692 | 0.95487 |
| glpB; glycerol-3-phosphate dehydrogenase subunit B [EC:1.1.5.3] | 0.00771 | 0.00076 | 0.00772 | 0.00007 | 0.00105 | 65 | 0.00770 | 0.00011 | 0.00107 | 93 | 0.00150 | 0.01392 | 151.911 | 0.98891 | 0.99801 |
| K09165; uncharacterized protein | 0.00771 | 0.00091 | 0.00751 | 0.00014 | 0.00146 | 65 | 0.00784 | 0.00013 | 0.00117 | 93 | 0.00187 | -0.17656 | 134.077 | 0.86012 | 0.96137 |
| BCKDHB, bkdA2; 2-oxoisovalerate dehydrogenase E1 component beta subunit [EC:1.2.4.4] | 0.00769 | 0.00092 | 0.00552 | 0.00009 | 0.00117 | 65 | 0.00921 | 0.00016 | 0.00132 | 93 | 0.00176 | -2.09418 | 155.434 | 0.03787 | 0.63091 |
| tagH; teichoic acid transport system ATP-binding protein [EC:3.6.3.40] | 0.00769 | 0.00121 | 0.00897 | 0.00031 | 0.00220 | 65 | 0.00680 | 0.00018 | 0.00137 | 93 | 0.00259 | 0.83452 | 111.822 | 0.40577 | 0.81189 |
| glgE; starch synthase (maltosyl-transferring) [EC:2.4.99.16] | 0.00768 | 0.00091 | 0.00782 | 0.00014 | 0.00144 | 65 | 0.00758 | 0.00013 | 0.00119 | 93 | 0.00187 | 0.12963 | 136.334 | 0.89705 | 0.97318 |
| adrA; diguanylate cyclase [EC:2.7.7.65] | 0.00767 | 0.00127 | 0.00691 | 0.00019 | 0.00173 | 65 | 0.00819 | 0.00030 | 0.00179 | 93 | 0.00249 | -0.51315 | 152.874 | 0.60859 | 0.88654 |
| ubiC; chorismate–pyruvate lyase [EC:4.1.3.40] | 0.00766 | 0.00081 | 0.00857 | 0.00011 | 0.00133 | 65 | 0.00703 | 0.00010 | 0.00103 | 93 | 0.00168 | 0.91587 | 131.250 | 0.36141 | 0.79496 |
| K09958; uncharacterized protein | 0.00766 | 0.00088 | 0.00746 | 0.00013 | 0.00139 | 65 | 0.00780 | 0.00012 | 0.00113 | 93 | 0.00180 | -0.18438 | 135.534 | 0.85399 | 0.95848 |
| algZ; two-component system, LytTR family, sensor histidine kinase AlgZ [EC:2.7.13.3] | 0.00766 | 0.00086 | 0.00780 | 0.00012 | 0.00137 | 65 | 0.00756 | 0.00011 | 0.00111 | 93 | 0.00176 | 0.13979 | 135.131 | 0.88903 | 0.97039 |
| arfA; alternative ribosome-rescue factor | 0.00765 | 0.00082 | 0.00855 | 0.00012 | 0.00136 | 65 | 0.00703 | 0.00010 | 0.00102 | 93 | 0.00170 | 0.89328 | 128.454 | 0.37338 | 0.79810 |
| phoP; two-component system, OmpR family, response regulator PhoP | 0.00765 | 0.00097 | 0.00810 | 0.00015 | 0.00153 | 65 | 0.00734 | 0.00015 | 0.00127 | 93 | 0.00199 | 0.38208 | 137.666 | 0.70299 | 0.92004 |
| crtB; 15-cis-phytoene synthase [EC:2.5.1.32] | 0.00764 | 0.00082 | 0.00567 | 0.00006 | 0.00098 | 65 | 0.00902 | 0.00013 | 0.00119 | 93 | 0.00154 | -2.17990 | 155.962 | 0.03076 | 0.61611 |
| rumB; 23S rRNA (uracil747-C5)-methyltransferase [EC:2.1.1.189] | 0.00764 | 0.00083 | 0.00843 | 0.00012 | 0.00136 | 65 | 0.00709 | 0.00010 | 0.00105 | 93 | 0.00172 | 0.78415 | 130.140 | 0.43437 | 0.82874 |
| ribT; riboflavin biosynthesis RibT protein | 0.00763 | 0.00129 | 0.00877 | 0.00040 | 0.00249 | 65 | 0.00684 | 0.00017 | 0.00135 | 93 | 0.00283 | 0.67868 | 100.887 | 0.49890 | 0.84856 |
| mtnD, mtnZ, ADI1; 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase [EC:1.13.11.53 1.13.11.54] | 0.00762 | 0.00090 | 0.00746 | 0.00014 | 0.00144 | 65 | 0.00773 | 0.00012 | 0.00115 | 93 | 0.00184 | -0.14289 | 133.260 | 0.88660 | 0.97020 |
| E2.4.-.-; glycosyltransferase [EC:2.4.-.-] | 0.00762 | 0.00069 | 0.00746 | 0.00008 | 0.00108 | 65 | 0.00773 | 0.00008 | 0.00090 | 93 | 0.00140 | -0.19328 | 138.234 | 0.84702 | 0.95682 |
| trxC; thioredoxin 2 [EC:1.8.1.8] | 0.00761 | 0.00085 | 0.00757 | 0.00012 | 0.00136 | 65 | 0.00763 | 0.00011 | 0.00110 | 93 | 0.00175 | -0.03208 | 134.488 | 0.97445 | 0.99546 |
| E3.1.27.1; ribonuclease T2 [EC:3.1.27.1] | 0.00761 | 0.00076 | 0.00638 | 0.00007 | 0.00103 | 65 | 0.00846 | 0.00011 | 0.00106 | 93 | 0.00148 | -1.40244 | 152.411 | 0.16282 | 0.77376 |
| soxR; MerR family transcriptional regulator, redox-sensitive transcriptional activator SoxR | 0.00760 | 0.00087 | 0.00736 | 0.00012 | 0.00137 | 65 | 0.00777 | 0.00012 | 0.00113 | 93 | 0.00178 | -0.22610 | 136.631 | 0.82146 | 0.95506 |
| opuD, betL; glycine betaine transporter | 0.00759 | 0.00115 | 0.00666 | 0.00014 | 0.00147 | 65 | 0.00825 | 0.00026 | 0.00166 | 93 | 0.00222 | -0.71642 | 155.530 | 0.47480 | 0.84095 |
| cpaC, rcpA; pilus assembly protein CpaC | 0.00759 | 0.00088 | 0.00620 | 0.00010 | 0.00124 | 65 | 0.00856 | 0.00014 | 0.00121 | 93 | 0.00173 | -1.36241 | 149.761 | 0.17511 | 0.77376 |
| K09927; uncharacterized protein | 0.00758 | 0.00084 | 0.00681 | 0.00012 | 0.00133 | 65 | 0.00812 | 0.00011 | 0.00109 | 93 | 0.00172 | -0.75961 | 136.105 | 0.44880 | 0.83406 |
| K06985; aspartyl protease family protein | 0.00758 | 0.00097 | 0.00755 | 0.00019 | 0.00169 | 65 | 0.00760 | 0.00013 | 0.00116 | 93 | 0.00205 | -0.02271 | 120.270 | 0.98192 | 0.99680 |
| atoA; acetate CoA/acetoacetate CoA-transferase beta subunit [EC:2.8.3.8 2.8.3.9] | 0.00758 | 0.00154 | 0.01132 | 0.00072 | 0.00332 | 65 | 0.00496 | 0.00012 | 0.00115 | 93 | 0.00351 | 1.81207 | 79.598 | 0.07375 | 0.74128 |
| mdtC; multidrug efflux pump | 0.00757 | 0.00085 | 0.00762 | 0.00013 | 0.00140 | 65 | 0.00754 | 0.00010 | 0.00106 | 93 | 0.00176 | 0.04430 | 128.684 | 0.96473 | 0.99101 |
| atoD; acetate CoA/acetoacetate CoA-transferase alpha subunit [EC:2.8.3.8 2.8.3.9] | 0.00757 | 0.00153 | 0.01125 | 0.00071 | 0.00331 | 65 | 0.00500 | 0.00012 | 0.00115 | 93 | 0.00350 | 1.78321 | 79.674 | 0.07836 | 0.75554 |
| tagG; teichoic acid transport system permease protein | 0.00757 | 0.00118 | 0.00836 | 0.00028 | 0.00209 | 65 | 0.00702 | 0.00018 | 0.00138 | 93 | 0.00251 | 0.53343 | 116.734 | 0.59475 | 0.88005 |
| arsA, ASNA1, GET3; arsenite/tail-anchored protein-transporting ATPase [EC:3.6.3.16 3.6.3.-] | 0.00756 | 0.00166 | 0.01020 | 0.00066 | 0.00320 | 65 | 0.00572 | 0.00027 | 0.00170 | 93 | 0.00362 | 1.23661 | 99.765 | 0.21914 | 0.77376 |
| soxA; sarcosine oxidase, subunit alpha [EC:1.5.3.1] | 0.00755 | 0.00108 | 0.00658 | 0.00014 | 0.00146 | 65 | 0.00822 | 0.00022 | 0.00153 | 93 | 0.00211 | -0.77540 | 153.220 | 0.43930 | 0.83156 |
| pilY1; type IV pilus assembly protein PilY1 | 0.00754 | 0.00091 | 0.00747 | 0.00013 | 0.00144 | 65 | 0.00760 | 0.00013 | 0.00118 | 93 | 0.00186 | -0.06945 | 136.363 | 0.94473 | 0.98674 |
| zntB; zinc transporter | 0.00754 | 0.00111 | 0.00711 | 0.00016 | 0.00157 | 65 | 0.00784 | 0.00022 | 0.00155 | 93 | 0.00220 | -0.32951 | 150.030 | 0.74223 | 0.93050 |
| lipL; octanoyl-[GcvH]:protein N-octanoyltransferase [EC:2.3.1.204] | 0.00753 | 0.00092 | 0.00696 | 0.00018 | 0.00166 | 65 | 0.00793 | 0.00011 | 0.00107 | 93 | 0.00197 | -0.49370 | 114.367 | 0.62246 | 0.89150 |
| K09786; uncharacterized protein | 0.00753 | 0.00082 | 0.00683 | 0.00010 | 0.00126 | 65 | 0.00802 | 0.00011 | 0.00108 | 93 | 0.00166 | -0.71576 | 140.405 | 0.47533 | 0.84101 |
| gyaR, GOR1; glyoxylate reductase [EC:1.1.1.26] | 0.00751 | 0.00074 | 0.00630 | 0.00009 | 0.00115 | 65 | 0.00836 | 0.00008 | 0.00095 | 93 | 0.00149 | -1.38024 | 137.166 | 0.16976 | 0.77376 |
| E3.2.1.1A; alpha-amylase [EC:3.2.1.1] | 0.00750 | 0.00087 | 0.00650 | 0.00008 | 0.00109 | 65 | 0.00820 | 0.00015 | 0.00126 | 93 | 0.00167 | -1.02163 | 155.803 | 0.30854 | 0.77376 |
| ibpA; molecular chaperone IbpA | 0.00750 | 0.00084 | 0.00657 | 0.00010 | 0.00123 | 65 | 0.00815 | 0.00012 | 0.00115 | 93 | 0.00168 | -0.93497 | 146.526 | 0.35134 | 0.79120 |
| astD; succinylglutamic semialdehyde dehydrogenase [EC:1.2.1.71] | 0.00748 | 0.00095 | 0.00753 | 0.00014 | 0.00145 | 65 | 0.00744 | 0.00015 | 0.00127 | 93 | 0.00193 | 0.04498 | 141.483 | 0.96418 | 0.99101 |
| pigA, hemO; heme oxygenase (biliverdin-IX-beta and delta-forming) [EC:1.14.99.58] | 0.00747 | 0.00084 | 0.00731 | 0.00011 | 0.00132 | 65 | 0.00759 | 0.00011 | 0.00108 | 93 | 0.00171 | -0.16512 | 135.960 | 0.86910 | 0.96465 |
| DPYS, dht, hydA; dihydropyrimidinase [EC:3.5.2.2] | 0.00746 | 0.00081 | 0.00608 | 0.00009 | 0.00116 | 65 | 0.00843 | 0.00011 | 0.00111 | 93 | 0.00161 | -1.45885 | 148.005 | 0.14673 | 0.77376 |
| ascD, ddhD, rfbI; CDP-4-dehydro-6-deoxyglucose reductase, E3 [EC:1.17.1.1] | 0.00746 | 0.00083 | 0.00776 | 0.00011 | 0.00131 | 65 | 0.00725 | 0.00011 | 0.00108 | 93 | 0.00170 | 0.29733 | 136.050 | 0.76667 | 0.93900 |
| degQ, hhoA; serine protease DegQ [EC:3.4.21.-] | 0.00746 | 0.00079 | 0.00820 | 0.00012 | 0.00136 | 65 | 0.00695 | 0.00008 | 0.00096 | 93 | 0.00166 | 0.75567 | 122.396 | 0.45130 | 0.83438 |
| K13671; alpha-1,2-mannosyltransferase [EC:2.4.1.-] | 0.00746 | 0.00130 | 0.00722 | 0.00027 | 0.00204 | 65 | 0.00762 | 0.00026 | 0.00169 | 93 | 0.00265 | -0.15122 | 136.674 | 0.88002 | 0.96801 |
| DBT, bkdB; 2-oxoisovalerate dehydrogenase E2 component (dihydrolipoyl transacylase) [EC:2.3.1.168] | 0.00745 | 0.00092 | 0.00553 | 0.00009 | 0.00117 | 65 | 0.00879 | 0.00016 | 0.00131 | 93 | 0.00176 | -1.85407 | 155.325 | 0.06563 | 0.73334 |
| K15975; glyoxalase family protein | 0.00745 | 0.00130 | 0.00589 | 0.00021 | 0.00179 | 65 | 0.00853 | 0.00031 | 0.00182 | 93 | 0.00255 | -1.03573 | 151.869 | 0.30197 | 0.77376 |
| mnhB, mrpB; multicomponent Na+:H+ antiporter subunit B | 0.00744 | 0.00111 | 0.00543 | 0.00012 | 0.00134 | 65 | 0.00885 | 0.00025 | 0.00163 | 93 | 0.00211 | -1.62094 | 155.960 | 0.10705 | 0.77376 |
| mdcB; triphosphoribosyl-dephospho-CoA synthase [EC:2.4.2.52] | 0.00743 | 0.00100 | 0.00757 | 0.00015 | 0.00153 | 65 | 0.00734 | 0.00016 | 0.00133 | 93 | 0.00203 | 0.11354 | 141.142 | 0.90977 | 0.97664 |
| mdcC; malonate decarboxylase delta subunit | 0.00742 | 0.00100 | 0.00757 | 0.00015 | 0.00153 | 65 | 0.00732 | 0.00016 | 0.00133 | 93 | 0.00203 | 0.12457 | 141.094 | 0.90104 | 0.97565 |
| CYP81F; indol-3-yl-methylglucosinolate hydroxylase [EC:1.14.-.-] | 0.00742 | 0.00119 | 0.00533 | 0.00009 | 0.00120 | 65 | 0.00889 | 0.00031 | 0.00183 | 93 | 0.00219 | -1.62705 | 148.644 | 0.10584 | 0.77376 |
| slpA; FKBP-type peptidyl-prolyl cis-trans isomerase SlpA [EC:5.2.1.8] | 0.00740 | 0.00084 | 0.00757 | 0.00011 | 0.00133 | 65 | 0.00728 | 0.00011 | 0.00108 | 93 | 0.00171 | 0.17078 | 135.775 | 0.86465 | 0.96263 |
| creC; two-component system, OmpR family, sensor histidine kinase CreC [EC:2.7.13.3] | 0.00740 | 0.00087 | 0.00721 | 0.00012 | 0.00137 | 65 | 0.00752 | 0.00012 | 0.00113 | 93 | 0.00178 | -0.17738 | 136.141 | 0.85947 | 0.96122 |
| tnaA; tryptophanase [EC:4.1.99.1] | 0.00739 | 0.00107 | 0.00975 | 0.00027 | 0.00203 | 65 | 0.00575 | 0.00011 | 0.00111 | 93 | 0.00231 | 1.73473 | 101.638 | 0.08582 | 0.76567 |
| pezT; UDP-N-acetylglucosamine kinase [EC:2.7.1.176] | 0.00739 | 0.00079 | 0.00688 | 0.00007 | 0.00101 | 65 | 0.00775 | 0.00012 | 0.00114 | 93 | 0.00153 | -0.56972 | 155.386 | 0.56969 | 0.87135 |
| terD; tellurium resistance protein TerD | 0.00738 | 0.00143 | 0.00790 | 0.00052 | 0.00282 | 65 | 0.00701 | 0.00019 | 0.00143 | 93 | 0.00316 | 0.28087 | 96.688 | 0.77941 | 0.94269 |
| FAH, fahA; fumarylacetoacetase [EC:3.7.1.2] | 0.00738 | 0.00089 | 0.00711 | 0.00012 | 0.00136 | 65 | 0.00756 | 0.00013 | 0.00118 | 93 | 0.00180 | -0.25215 | 140.723 | 0.80129 | 0.94855 |
| hyaB, hybC; hydrogenase large subunit [EC:1.12.99.6] | 0.00737 | 0.00081 | 0.00787 | 0.00013 | 0.00139 | 65 | 0.00703 | 0.00009 | 0.00099 | 93 | 0.00170 | 0.49174 | 123.033 | 0.62378 | 0.89159 |
| CTH; cystathionine gamma-lyase [EC:4.4.1.1] | 0.00737 | 0.00093 | 0.00693 | 0.00012 | 0.00136 | 65 | 0.00768 | 0.00015 | 0.00127 | 93 | 0.00186 | -0.40182 | 146.502 | 0.68840 | 0.91567 |
| K09946; uncharacterized protein | 0.00736 | 0.00079 | 0.00748 | 0.00009 | 0.00118 | 65 | 0.00728 | 0.00010 | 0.00106 | 93 | 0.00158 | 0.12785 | 143.762 | 0.89845 | 0.97377 |
| msrP; methionine sulfoxide reductase catalytic subunit [EC:1.8.-.-] | 0.00736 | 0.00096 | 0.00694 | 0.00013 | 0.00143 | 65 | 0.00765 | 0.00016 | 0.00130 | 93 | 0.00193 | -0.36547 | 144.778 | 0.71529 | 0.92004 |
| umuD; DNA polymerase V [EC:3.4.21.-] | 0.00734 | 0.00094 | 0.00741 | 0.00013 | 0.00143 | 65 | 0.00729 | 0.00015 | 0.00125 | 93 | 0.00190 | 0.06416 | 141.689 | 0.94893 | 0.98754 |
| K06919; putative DNA primase/helicase | 0.00734 | 0.00085 | 0.00840 | 0.00013 | 0.00144 | 65 | 0.00660 | 0.00010 | 0.00105 | 93 | 0.00178 | 1.01112 | 125.438 | 0.31390 | 0.77376 |
| merT; mercuric ion transport protein | 0.00734 | 0.00087 | 0.00793 | 0.00013 | 0.00140 | 65 | 0.00692 | 0.00011 | 0.00110 | 93 | 0.00178 | 0.56644 | 132.279 | 0.57205 | 0.87289 |
| creB; two-component system, OmpR family, catabolic regulation response regulator CreB | 0.00733 | 0.00087 | 0.00721 | 0.00012 | 0.00137 | 65 | 0.00741 | 0.00012 | 0.00113 | 93 | 0.00178 | -0.11474 | 136.369 | 0.90882 | 0.97664 |
| bglG1; transcriptional antiterminator | 0.00733 | 0.00071 | 0.00721 | 0.00008 | 0.00112 | 65 | 0.00741 | 0.00008 | 0.00091 | 93 | 0.00145 | -0.13889 | 135.327 | 0.88974 | 0.97048 |
| iorA; isoquinoline 1-oxidoreductase subunit alpha [EC:1.3.99.16] | 0.00732 | 0.00105 | 0.00603 | 0.00011 | 0.00131 | 65 | 0.00823 | 0.00022 | 0.00152 | 93 | 0.00201 | -1.09765 | 155.858 | 0.27405 | 0.77376 |
| tfoX; DNA transformation protein and related proteins | 0.00732 | 0.00072 | 0.00681 | 0.00007 | 0.00107 | 65 | 0.00768 | 0.00009 | 0.00097 | 93 | 0.00144 | -0.60568 | 144.509 | 0.54568 | 0.86444 |
| K09928; uncharacterized protein | 0.00732 | 0.00084 | 0.00690 | 0.00010 | 0.00123 | 65 | 0.00761 | 0.00012 | 0.00115 | 93 | 0.00168 | -0.42194 | 146.918 | 0.67369 | 0.91184 |
| hspR; MerR family transcriptional regulator, heat shock protein HspR | 0.00732 | 0.00081 | 0.00710 | 0.00009 | 0.00117 | 65 | 0.00747 | 0.00012 | 0.00112 | 93 | 0.00162 | -0.22869 | 148.330 | 0.81942 | 0.95506 |
| rnb; exoribonuclease II [EC:3.1.13.1] | 0.00732 | 0.00085 | 0.00730 | 0.00011 | 0.00127 | 65 | 0.00733 | 0.00012 | 0.00115 | 93 | 0.00171 | -0.01727 | 144.107 | 0.98625 | 0.99729 |
| hyaA, hybO; hydrogenase small subunit [EC:1.12.99.6] | 0.00732 | 0.00081 | 0.00787 | 0.00013 | 0.00139 | 65 | 0.00693 | 0.00009 | 0.00098 | 93 | 0.00170 | 0.54847 | 122.623 | 0.58437 | 0.87720 |
| E3.2.1.14; chitinase [EC:3.2.1.14] | 0.00730 | 0.00124 | 0.00647 | 0.00024 | 0.00193 | 65 | 0.00788 | 0.00025 | 0.00163 | 93 | 0.00253 | -0.55945 | 138.648 | 0.57676 | 0.87389 |
| ppnP; purine/pyrimidine-nucleoside phosphorylase [EC:2.4.2.1 2.4.2.2] | 0.00730 | 0.00084 | 0.00739 | 0.00011 | 0.00129 | 65 | 0.00724 | 0.00012 | 0.00111 | 93 | 0.00170 | 0.08712 | 140.831 | 0.93070 | 0.98266 |
| K09932; uncharacterized protein | 0.00730 | 0.00112 | 0.00807 | 0.00022 | 0.00186 | 65 | 0.00676 | 0.00018 | 0.00140 | 93 | 0.00233 | 0.56174 | 128.778 | 0.57527 | 0.87389 |
| rutE; 3-hydroxypropanoate dehydrogenase [EC:1.1.1.-] | 0.00729 | 0.00087 | 0.00679 | 0.00014 | 0.00147 | 65 | 0.00764 | 0.00011 | 0.00107 | 93 | 0.00182 | -0.47121 | 124.994 | 0.63831 | 0.89822 |
| hyaC; Ni/Fe-hydrogenase 1 B-type cytochrome subunit | 0.00728 | 0.00084 | 0.00766 | 0.00013 | 0.00139 | 65 | 0.00702 | 0.00010 | 0.00104 | 93 | 0.00174 | 0.37054 | 127.543 | 0.71159 | 0.92004 |
| thiB, tbpA; thiamine transport system substrate-binding protein | 0.00728 | 0.00078 | 0.00757 | 0.00011 | 0.00130 | 65 | 0.00707 | 0.00009 | 0.00096 | 93 | 0.00162 | 0.30635 | 126.812 | 0.75984 | 0.93791 |
| PTS-Gut-EIIA, srlB; PTS system, glucitol/sorbitol-specific IIA component [EC:2.7.1.198] | 0.00727 | 0.00101 | 0.00893 | 0.00025 | 0.00196 | 65 | 0.00610 | 0.00010 | 0.00104 | 93 | 0.00222 | 1.27715 | 99.203 | 0.20453 | 0.77376 |
| E3.2.1.17; lysozyme [EC:3.2.1.17] | 0.00726 | 0.00084 | 0.00715 | 0.00012 | 0.00138 | 65 | 0.00734 | 0.00010 | 0.00105 | 93 | 0.00173 | -0.10894 | 129.681 | 0.91342 | 0.97664 |
| tyrP; tyrosine-specific transport protein | 0.00725 | 0.00118 | 0.00820 | 0.00024 | 0.00190 | 65 | 0.00659 | 0.00021 | 0.00151 | 93 | 0.00243 | 0.66010 | 133.146 | 0.51033 | 0.85013 |
| lctO; L-lactate oxidase [EC:1.1.3.2] | 0.00725 | 0.00081 | 0.00676 | 0.00008 | 0.00113 | 65 | 0.00760 | 0.00012 | 0.00113 | 93 | 0.00160 | -0.52651 | 150.571 | 0.59931 | 0.88240 |
| K07395; putative proteasome-type protease | 0.00725 | 0.00086 | 0.00696 | 0.00012 | 0.00134 | 65 | 0.00745 | 0.00012 | 0.00114 | 93 | 0.00176 | -0.27971 | 139.474 | 0.78011 | 0.94269 |
| MAO, aofH; monoamine oxidase [EC:1.4.3.4] | 0.00724 | 0.00096 | 0.00591 | 0.00012 | 0.00134 | 65 | 0.00817 | 0.00017 | 0.00133 | 93 | 0.00189 | -1.19584 | 150.798 | 0.23364 | 0.77376 |
| DPP3; dipeptidyl-peptidase III [EC:3.4.14.4] | 0.00723 | 0.00086 | 0.00619 | 0.00008 | 0.00108 | 65 | 0.00796 | 0.00014 | 0.00124 | 93 | 0.00165 | -1.07898 | 155.762 | 0.28226 | 0.77376 |
| sanA; SanA protein | 0.00723 | 0.00075 | 0.00757 | 0.00009 | 0.00115 | 65 | 0.00699 | 0.00009 | 0.00098 | 93 | 0.00152 | 0.37959 | 139.464 | 0.70482 | 0.92004 |
| ramB; XRE family transcriptional regulator, fatty acid utilization regulator | 0.00723 | 0.00094 | 0.00441 | 0.00008 | 0.00110 | 65 | 0.00920 | 0.00018 | 0.00137 | 93 | 0.00176 | -2.72889 | 155.717 | 0.00709 | 0.54612 |
| prpE; propionyl-CoA synthetase [EC:6.2.1.17] | 0.00723 | 0.00091 | 0.00602 | 0.00011 | 0.00130 | 65 | 0.00807 | 0.00015 | 0.00126 | 93 | 0.00181 | -1.13188 | 148.938 | 0.25950 | 0.77376 |
| ECM4, yqjG; glutathionyl-hydroquinone reductase [EC:1.8.5.7] | 0.00722 | 0.00084 | 0.00721 | 0.00012 | 0.00138 | 65 | 0.00723 | 0.00010 | 0.00106 | 93 | 0.00174 | -0.01217 | 130.425 | 0.99031 | 0.99816 |
| merP; periplasmic mercuric ion binding protein | 0.00722 | 0.00084 | 0.00782 | 0.00012 | 0.00137 | 65 | 0.00680 | 0.00011 | 0.00107 | 93 | 0.00173 | 0.59009 | 131.775 | 0.55614 | 0.86813 |
| mapA; maltose phosphorylase [EC:2.4.1.8] | 0.00721 | 0.00125 | 0.00897 | 0.00038 | 0.00243 | 65 | 0.00597 | 0.00015 | 0.00127 | 93 | 0.00274 | 1.09311 | 98.564 | 0.27701 | 0.77376 |
| sacB; levansucrase [EC:2.4.1.10] | 0.00720 | 0.00154 | 0.00810 | 0.00055 | 0.00290 | 65 | 0.00657 | 0.00026 | 0.00167 | 93 | 0.00334 | 0.45730 | 105.483 | 0.64839 | 0.90350 |
| cooS, acsA; anaerobic carbon-monoxide dehydrogenase catalytic subunit [EC:1.2.7.4] | 0.00719 | 0.00088 | 0.00854 | 0.00018 | 0.00167 | 65 | 0.00625 | 0.00008 | 0.00095 | 93 | 0.00192 | 1.19868 | 104.292 | 0.23337 | 0.77376 |
| E3.2.1.96; mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase [EC:3.2.1.96] | 0.00719 | 0.00079 | 0.00640 | 0.00007 | 0.00102 | 65 | 0.00774 | 0.00012 | 0.00113 | 93 | 0.00152 | -0.87597 | 154.923 | 0.38240 | 0.80037 |
| gspN; general secretion pathway protein N | 0.00719 | 0.00086 | 0.00659 | 0.00010 | 0.00124 | 65 | 0.00761 | 0.00013 | 0.00118 | 93 | 0.00171 | -0.59634 | 147.828 | 0.55186 | 0.86788 |
| K09973; uncharacterized protein | 0.00718 | 0.00083 | 0.00591 | 0.00010 | 0.00122 | 65 | 0.00806 | 0.00012 | 0.00113 | 93 | 0.00166 | -1.29837 | 145.573 | 0.19621 | 0.77376 |
| tetM, tetO; ribosomal protection tetracycline resistance protein | 0.00717 | 0.00078 | 0.00735 | 0.00011 | 0.00129 | 65 | 0.00705 | 0.00009 | 0.00098 | 93 | 0.00162 | 0.18127 | 128.621 | 0.85644 | 0.95945 |
| K09986; uncharacterized protein | 0.00717 | 0.00083 | 0.00647 | 0.00010 | 0.00124 | 65 | 0.00766 | 0.00012 | 0.00112 | 93 | 0.00167 | -0.70744 | 144.039 | 0.48044 | 0.84354 |
| buk; butyrate kinase [EC:2.7.2.7] | 0.00717 | 0.00082 | 0.00619 | 0.00008 | 0.00113 | 65 | 0.00785 | 0.00012 | 0.00114 | 93 | 0.00161 | -1.03293 | 151.866 | 0.30328 | 0.77376 |
| K06987; uncharacterized protein | 0.00717 | 0.00106 | 0.00673 | 0.00015 | 0.00150 | 65 | 0.00748 | 0.00020 | 0.00147 | 93 | 0.00210 | -0.35786 | 150.062 | 0.72095 | 0.92156 |
| glpG; GlpG protein | 0.00716 | 0.00075 | 0.00953 | 0.00013 | 0.00141 | 65 | 0.00551 | 0.00005 | 0.00077 | 93 | 0.00160 | 2.50519 | 101.475 | 0.01383 | 0.59095 |
| K07140; uncharacterized protein | 0.00716 | 0.00084 | 0.00695 | 0.00012 | 0.00136 | 65 | 0.00731 | 0.00011 | 0.00107 | 93 | 0.00174 | -0.20787 | 132.449 | 0.83565 | 0.95506 |
| nuc; micrococcal nuclease [EC:3.1.31.1] | 0.00715 | 0.00098 | 0.00584 | 0.00009 | 0.00116 | 65 | 0.00807 | 0.00020 | 0.00146 | 93 | 0.00186 | -1.19530 | 155.627 | 0.23379 | 0.77376 |
| sulA; cell division inhibitor SulA | 0.00715 | 0.00088 | 0.00775 | 0.00013 | 0.00141 | 65 | 0.00673 | 0.00012 | 0.00112 | 93 | 0.00181 | 0.56397 | 133.394 | 0.57372 | 0.87292 |
| grdA; glycine/sarcosine/betaine reductase complex component A [EC:1.21.4.2 1.21.4.3 1.21.4.4] | 0.00715 | 0.00097 | 0.00917 | 0.00024 | 0.00194 | 65 | 0.00574 | 0.00008 | 0.00092 | 93 | 0.00214 | 1.60055 | 92.699 | 0.11288 | 0.77376 |
| ptb; phosphate butyryltransferase [EC:2.3.1.19] | 0.00714 | 0.00081 | 0.00632 | 0.00008 | 0.00112 | 65 | 0.00772 | 0.00012 | 0.00114 | 93 | 0.00159 | -0.87605 | 151.823 | 0.38239 | 0.80037 |
| qorB; NAD(P)H dehydrogenase (quinone) [EC:1.6.5.2] | 0.00714 | 0.00095 | 0.00815 | 0.00026 | 0.00199 | 65 | 0.00643 | 0.00006 | 0.00083 | 93 | 0.00216 | 0.79867 | 86.365 | 0.42668 | 0.82661 |
| adiA; arginine decarboxylase [EC:4.1.1.19] | 0.00712 | 0.00084 | 0.00705 | 0.00013 | 0.00141 | 65 | 0.00717 | 0.00010 | 0.00104 | 93 | 0.00175 | -0.06682 | 126.466 | 0.94683 | 0.98674 |
| uidA, GUSB; beta-glucuronidase [EC:3.2.1.31] | 0.00712 | 0.00079 | 0.00758 | 0.00012 | 0.00133 | 65 | 0.00679 | 0.00009 | 0.00098 | 93 | 0.00165 | 0.47676 | 126.703 | 0.63435 | 0.89696 |
| E1.4.7.1; glutamate synthase (ferredoxin) [EC:1.4.7.1] | 0.00712 | 0.00075 | 0.00619 | 0.00008 | 0.00111 | 65 | 0.00777 | 0.00009 | 0.00101 | 93 | 0.00149 | -1.05758 | 144.802 | 0.29201 | 0.77376 |
| aapP, bztD; general L-amino acid transport system ATP-binding protein [EC:3.6.3.-] | 0.00711 | 0.00100 | 0.00659 | 0.00014 | 0.00145 | 65 | 0.00747 | 0.00018 | 0.00138 | 93 | 0.00200 | -0.43906 | 147.801 | 0.66126 | 0.90968 |
| nei; endonuclease VIII [EC:3.2.2.- 4.2.99.18] | 0.00711 | 0.00082 | 0.00684 | 0.00010 | 0.00125 | 65 | 0.00729 | 0.00011 | 0.00110 | 93 | 0.00166 | -0.27649 | 141.953 | 0.78257 | 0.94310 |
| sprT; SprT protein | 0.00709 | 0.00076 | 0.00820 | 0.00010 | 0.00127 | 65 | 0.00631 | 0.00008 | 0.00094 | 93 | 0.00158 | 1.19281 | 127.013 | 0.23517 | 0.77376 |
| gcvR; glycine cleavage system transcriptional repressor | 0.00708 | 0.00087 | 0.00758 | 0.00013 | 0.00141 | 65 | 0.00673 | 0.00011 | 0.00111 | 93 | 0.00180 | 0.46965 | 132.529 | 0.63938 | 0.89853 |
| puuE; 4-aminobutyrate aminotransferase [EC:2.6.1.19] | 0.00707 | 0.00095 | 0.00708 | 0.00022 | 0.00185 | 65 | 0.00707 | 0.00009 | 0.00097 | 93 | 0.00209 | 0.00687 | 98.617 | 0.99453 | 0.99919 |
| hutT; histidine transporter | 0.00707 | 0.00077 | 0.00679 | 0.00009 | 0.00117 | 65 | 0.00726 | 0.00010 | 0.00104 | 93 | 0.00156 | -0.29757 | 142.821 | 0.76646 | 0.93900 |
| K07168; CBS domain-containing membrane protein | 0.00706 | 0.00083 | 0.00671 | 0.00011 | 0.00129 | 65 | 0.00730 | 0.00011 | 0.00110 | 93 | 0.00169 | -0.34771 | 139.332 | 0.72858 | 0.92350 |
| ygiM; SH3 domain protein | 0.00703 | 0.00077 | 0.00847 | 0.00011 | 0.00129 | 65 | 0.00602 | 0.00008 | 0.00094 | 93 | 0.00159 | 1.53841 | 125.136 | 0.12647 | 0.77376 |
| K09802; uncharacterized protein | 0.00703 | 0.00095 | 0.00948 | 0.00022 | 0.00183 | 65 | 0.00531 | 0.00008 | 0.00094 | 93 | 0.00206 | 2.02432 | 97.647 | 0.04567 | 0.65981 |
| sdiA; LuxR family transcriptional regulator, quorum-sensing system regulator SdiA | 0.00702 | 0.00124 | 0.00570 | 0.00017 | 0.00160 | 65 | 0.00794 | 0.00030 | 0.00179 | 93 | 0.00240 | -0.93413 | 155.271 | 0.35169 | 0.79120 |
| INO1, ISYNA1; myo-inositol-1-phosphate synthase [EC:5.5.1.4] | 0.00700 | 0.00072 | 0.00619 | 0.00007 | 0.00101 | 65 | 0.00757 | 0.00009 | 0.00100 | 93 | 0.00142 | -0.96664 | 150.002 | 0.33528 | 0.78186 |
| nfrA2; FMN reductase [NAD(P)H] [EC:1.5.1.39] | 0.00699 | 0.00084 | 0.00649 | 0.00009 | 0.00114 | 65 | 0.00734 | 0.00013 | 0.00119 | 93 | 0.00165 | -0.51399 | 153.005 | 0.60800 | 0.88637 |
| troB, mntB, znuC; manganese/zinc/iron transport system ATP- binding protein | 0.00698 | 0.00104 | 0.00963 | 0.00028 | 0.00206 | 65 | 0.00513 | 0.00009 | 0.00099 | 93 | 0.00228 | 1.97137 | 93.875 | 0.05163 | 0.68802 |
| E5.4.99.2A, mcmA1; methylmalonyl-CoA mutase, N-terminal domain [EC:5.4.99.2] | 0.00696 | 0.00077 | 0.00751 | 0.00013 | 0.00139 | 65 | 0.00658 | 0.00007 | 0.00089 | 93 | 0.00165 | 0.55772 | 113.571 | 0.57813 | 0.87519 |
| mviM; virulence factor | 0.00696 | 0.00082 | 0.00728 | 0.00016 | 0.00155 | 65 | 0.00674 | 0.00007 | 0.00088 | 93 | 0.00178 | 0.30162 | 104.761 | 0.76354 | 0.93836 |
| entE, dhbE, vibE, mxcE; 2,3-dihydroxybenzoate-AMP ligase [EC:6.3.2.14 2.7.7.58] | 0.00695 | 0.00086 | 0.00651 | 0.00013 | 0.00140 | 65 | 0.00726 | 0.00011 | 0.00108 | 93 | 0.00177 | -0.42372 | 130.625 | 0.67246 | 0.91184 |
| tsdA; thiosulfate dehydrogenase [EC:1.8.2.2] | 0.00694 | 0.00093 | 0.00655 | 0.00012 | 0.00137 | 65 | 0.00721 | 0.00015 | 0.00127 | 93 | 0.00187 | -0.35697 | 146.425 | 0.72163 | 0.92156 |
| mipA, ompV; MipA family protein | 0.00693 | 0.00096 | 0.00872 | 0.00023 | 0.00186 | 65 | 0.00568 | 0.00009 | 0.00098 | 93 | 0.00210 | 1.44454 | 99.024 | 0.15174 | 0.77376 |
| E2.5.1.56, neuB; N-acetylneuraminate synthase [EC:2.5.1.56] | 0.00693 | 0.00081 | 0.00705 | 0.00013 | 0.00141 | 65 | 0.00685 | 0.00009 | 0.00097 | 93 | 0.00171 | 0.11764 | 120.378 | 0.90655 | 0.97664 |
| barA, gacS, varS; two-component system, NarL family, sensor histidine kinase BarA [EC:2.7.13.3] | 0.00693 | 0.00092 | 0.00738 | 0.00013 | 0.00140 | 65 | 0.00662 | 0.00014 | 0.00124 | 93 | 0.00187 | 0.40646 | 142.691 | 0.68502 | 0.91533 |
| pezA; HTH-type transcriptional regulator / antitoxin PezA | 0.00692 | 0.00078 | 0.00617 | 0.00007 | 0.00101 | 65 | 0.00745 | 0.00012 | 0.00113 | 93 | 0.00151 | -0.84385 | 155.366 | 0.40005 | 0.80762 |
| CBS; cystathionine beta-synthase [EC:4.2.1.22] | 0.00692 | 0.00074 | 0.00614 | 0.00007 | 0.00107 | 65 | 0.00746 | 0.00010 | 0.00102 | 93 | 0.00147 | -0.89160 | 147.836 | 0.37405 | 0.79810 |
| K09920; uncharacterized protein | 0.00691 | 0.00087 | 0.00742 | 0.00013 | 0.00142 | 65 | 0.00655 | 0.00011 | 0.00111 | 93 | 0.00180 | 0.48520 | 131.819 | 0.62834 | 0.89381 |
| araA; L-arabinose isomerase [EC:5.3.1.4] | 0.00689 | 0.00098 | 0.00748 | 0.00019 | 0.00172 | 65 | 0.00648 | 0.00013 | 0.00117 | 93 | 0.00208 | 0.47751 | 118.688 | 0.63388 | 0.89696 |
| mnhF, mrpF; multicomponent Na+:H+ antiporter subunit F | 0.00689 | 0.00113 | 0.00498 | 0.00013 | 0.00143 | 65 | 0.00822 | 0.00025 | 0.00163 | 93 | 0.00217 | -1.49066 | 155.616 | 0.13808 | 0.77376 |
| prpD; 2-methylcitrate dehydratase [EC:4.2.1.79] | 0.00687 | 0.00084 | 0.00630 | 0.00011 | 0.00133 | 65 | 0.00727 | 0.00011 | 0.00109 | 93 | 0.00172 | -0.56505 | 136.467 | 0.57297 | 0.87292 |
| BCKDHA, bkdA1; 2-oxoisovalerate dehydrogenase E1 component alpha subunit [EC:1.2.4.4] | 0.00687 | 0.00078 | 0.00475 | 0.00006 | 0.00093 | 65 | 0.00835 | 0.00012 | 0.00113 | 93 | 0.00146 | -2.46371 | 155.987 | 0.01484 | 0.59095 |
| E5.4.99.2B, mcmA2; methylmalonyl-CoA mutase, C-terminal domain [EC:5.4.99.2] | 0.00685 | 0.00076 | 0.00758 | 0.00012 | 0.00138 | 65 | 0.00634 | 0.00007 | 0.00087 | 93 | 0.00163 | 0.76302 | 112.255 | 0.44705 | 0.83372 |
| K07149; uncharacterized protein | 0.00684 | 0.00108 | 0.00576 | 0.00011 | 0.00128 | 65 | 0.00760 | 0.00024 | 0.00160 | 93 | 0.00205 | -0.90013 | 155.717 | 0.36944 | 0.79587 |
| entB, dhbB, vibB, mxcF; bifunctional isochorismate lyase / aryl carrier protein [EC:3.3.2.1 6.3.2.14] | 0.00683 | 0.00090 | 0.00621 | 0.00013 | 0.00139 | 65 | 0.00727 | 0.00013 | 0.00119 | 93 | 0.00183 | -0.57679 | 140.068 | 0.56501 | 0.86988 |
| glcF; glycolate oxidase iron-sulfur subunit | 0.00682 | 0.00081 | 0.00561 | 0.00009 | 0.00117 | 65 | 0.00767 | 0.00011 | 0.00110 | 93 | 0.00160 | -1.28192 | 147.378 | 0.20188 | 0.77376 |
| cysNC; bifunctional enzyme CysN/CysC [EC:2.7.7.4 2.7.1.25] | 0.00682 | 0.00092 | 0.00578 | 0.00009 | 0.00120 | 65 | 0.00754 | 0.00016 | 0.00131 | 93 | 0.00177 | -0.99137 | 154.794 | 0.32305 | 0.77376 |
| troA, mntA, znuA; manganese/zinc/iron transport system substrate-binding protein | 0.00681 | 0.00104 | 0.00979 | 0.00028 | 0.00206 | 65 | 0.00473 | 0.00009 | 0.00098 | 93 | 0.00229 | 2.20834 | 93.065 | 0.02968 | 0.61356 |
| ttuD; hydroxypyruvate reductase [EC:1.1.1.81] | 0.00680 | 0.00078 | 0.00572 | 0.00008 | 0.00111 | 65 | 0.00756 | 0.00011 | 0.00107 | 93 | 0.00154 | -1.19506 | 148.304 | 0.23397 | 0.77376 |
| mexJ; membrane fusion protein, multidrug efflux system | 0.00680 | 0.00084 | 0.00701 | 0.00012 | 0.00138 | 65 | 0.00665 | 0.00010 | 0.00105 | 93 | 0.00173 | 0.20842 | 129.418 | 0.83523 | 0.95506 |
| phoQ; two-component system, OmpR family, sensor histidine kinase PhoQ [EC:2.7.13.3] | 0.00680 | 0.00084 | 0.00729 | 0.00012 | 0.00137 | 65 | 0.00646 | 0.00010 | 0.00106 | 93 | 0.00173 | 0.47822 | 131.192 | 0.63329 | 0.89696 |
| cpxR; two-component system, OmpR family, response regulator CpxR | 0.00680 | 0.00088 | 0.00687 | 0.00011 | 0.00128 | 65 | 0.00675 | 0.00013 | 0.00120 | 93 | 0.00176 | 0.06841 | 147.170 | 0.94555 | 0.98674 |
| alaC; alanine-synthesizing transaminase [EC:2.6.1.-] | 0.00679 | 0.00076 | 0.00648 | 0.00009 | 0.00118 | 65 | 0.00701 | 0.00009 | 0.00100 | 93 | 0.00154 | -0.33830 | 138.603 | 0.73565 | 0.92721 |
| ampE; AmpE protein | 0.00677 | 0.00087 | 0.00718 | 0.00013 | 0.00140 | 65 | 0.00649 | 0.00011 | 0.00111 | 93 | 0.00179 | 0.38652 | 132.745 | 0.69973 | 0.91918 |
| apeE, estA, lip-1; outer membrane lipase/esterase | 0.00677 | 0.00095 | 0.00736 | 0.00016 | 0.00155 | 65 | 0.00636 | 0.00013 | 0.00120 | 93 | 0.00196 | 0.50955 | 131.145 | 0.61122 | 0.88722 |
| rhtB; homoserine/homoserine lactone efflux protein | 0.00675 | 0.00083 | 0.00647 | 0.00010 | 0.00126 | 65 | 0.00694 | 0.00011 | 0.00110 | 93 | 0.00167 | -0.28084 | 141.962 | 0.77924 | 0.94269 |
| bigA; putative surface-exposed virulence protein | 0.00674 | 0.00162 | 0.01147 | 0.00079 | 0.00349 | 65 | 0.00344 | 0.00013 | 0.00119 | 93 | 0.00369 | 2.17546 | 78.960 | 0.03259 | 0.62130 |
| ppgK; polyphosphate glucokinase [EC:2.7.1.63] | 0.00674 | 0.00084 | 0.00684 | 0.00010 | 0.00124 | 65 | 0.00667 | 0.00012 | 0.00114 | 93 | 0.00169 | 0.10497 | 145.485 | 0.91655 | 0.97681 |
| NAGK, nagK; N-acetylglucosamine kinase [EC:2.7.1.59] | 0.00673 | 0.00081 | 0.00695 | 0.00010 | 0.00126 | 65 | 0.00658 | 0.00010 | 0.00105 | 93 | 0.00164 | 0.22769 | 137.887 | 0.82023 | 0.95506 |
| gudD; glucarate dehydratase [EC:4.2.1.40] | 0.00673 | 0.00094 | 0.00644 | 0.00014 | 0.00146 | 65 | 0.00693 | 0.00014 | 0.00123 | 93 | 0.00191 | -0.26072 | 139.013 | 0.79469 | 0.94783 |
| fabV, ter; enoyl-[acyl-carrier protein] reductase / trans-2-enoyl-CoA reductase (NAD+) [EC:1.3.1.9 1.3.1.44] | 0.00673 | 0.00077 | 0.00710 | 0.00009 | 0.00119 | 65 | 0.00647 | 0.00009 | 0.00101 | 93 | 0.00156 | 0.40449 | 138.888 | 0.68647 | 0.91567 |
| PTS-Glv-EIIC, glvC, malP, aglA; PTS system, alpha-glucoside-specific IIC component | 0.00672 | 0.00082 | 0.00770 | 0.00015 | 0.00154 | 65 | 0.00604 | 0.00007 | 0.00090 | 93 | 0.00178 | 0.92859 | 106.157 | 0.35521 | 0.79399 |
| otsA; trehalose 6-phosphate synthase [EC:2.4.1.15 2.4.1.347] | 0.00672 | 0.00080 | 0.00530 | 0.00008 | 0.00111 | 65 | 0.00771 | 0.00012 | 0.00112 | 93 | 0.00157 | -1.52662 | 151.204 | 0.12895 | 0.77376 |
| mepS, spr; murein DD-endopeptidase / murein LD-carboxypeptidase [EC:3.4.-.- 3.4.17.13] | 0.00672 | 0.00074 | 0.00740 | 0.00010 | 0.00123 | 65 | 0.00624 | 0.00008 | 0.00093 | 93 | 0.00154 | 0.74944 | 129.172 | 0.45496 | 0.83438 |
| eat, eutP; ethanolamine permease | 0.00670 | 0.00096 | 0.00606 | 0.00012 | 0.00134 | 65 | 0.00716 | 0.00016 | 0.00133 | 93 | 0.00189 | -0.58130 | 150.383 | 0.56191 | 0.86813 |
| astB; succinylarginine dihydrolase [EC:3.5.3.23] | 0.00670 | 0.00078 | 0.00688 | 0.00010 | 0.00125 | 65 | 0.00658 | 0.00010 | 0.00101 | 93 | 0.00161 | 0.18495 | 135.337 | 0.85354 | 0.95848 |
| pobA; p-hydroxybenzoate 3-monooxygenase [EC:1.14.13.2] | 0.00670 | 0.00087 | 0.00577 | 0.00009 | 0.00119 | 65 | 0.00734 | 0.00014 | 0.00121 | 93 | 0.00170 | -0.92326 | 152.147 | 0.35733 | 0.79439 |
| xylA; xylose isomerase [EC:5.3.1.5] | 0.00669 | 0.00064 | 0.00580 | 0.00007 | 0.00101 | 65 | 0.00731 | 0.00006 | 0.00082 | 93 | 0.00130 | -1.15747 | 134.969 | 0.24912 | 0.77376 |
| sthA, udhA; NAD(P) transhydrogenase [EC:1.6.1.1] | 0.00667 | 0.00077 | 0.00681 | 0.00010 | 0.00122 | 65 | 0.00658 | 0.00009 | 0.00101 | 93 | 0.00158 | 0.14134 | 137.274 | 0.88781 | 0.97039 |
| adeR; two-component system, OmpR family, response regulator AdeR | 0.00667 | 0.00119 | 0.00607 | 0.00024 | 0.00191 | 65 | 0.00709 | 0.00022 | 0.00153 | 93 | 0.00245 | -0.41506 | 133.661 | 0.67877 | 0.91311 |
| gtsA, glcE; glucose/mannose transport system substrate-binding protein | 0.00667 | 0.00085 | 0.00566 | 0.00009 | 0.00119 | 65 | 0.00737 | 0.00013 | 0.00117 | 93 | 0.00167 | -1.02260 | 150.134 | 0.30814 | 0.77376 |
| K09926; uncharacterized protein | 0.00666 | 0.00077 | 0.00710 | 0.00010 | 0.00122 | 65 | 0.00635 | 0.00009 | 0.00099 | 93 | 0.00157 | 0.47613 | 135.267 | 0.63475 | 0.89696 |
| cwlK; peptidoglycan LD-endopeptidase CwlK [EC:3.4.-.-] | 0.00665 | 0.00109 | 0.00559 | 0.00011 | 0.00128 | 65 | 0.00739 | 0.00025 | 0.00163 | 93 | 0.00207 | -0.87015 | 155.498 | 0.38556 | 0.80258 |
| hasF, prtF; outer membrane protein, protease secretion system | 0.00665 | 0.00167 | 0.00567 | 0.00032 | 0.00223 | 65 | 0.00733 | 0.00053 | 0.00238 | 93 | 0.00326 | -0.50727 | 153.918 | 0.61269 | 0.88788 |
| mmr; MFS transporter, DHA2 family, methylenomycin A resistance protein | 0.00664 | 0.00071 | 0.00550 | 0.00004 | 0.00080 | 65 | 0.00744 | 0.00010 | 0.00106 | 93 | 0.00133 | -1.45753 | 154.634 | 0.14700 | 0.77376 |
| thiP; thiamine transport system permease protein | 0.00664 | 0.00076 | 0.00681 | 0.00010 | 0.00126 | 65 | 0.00652 | 0.00008 | 0.00095 | 93 | 0.00157 | 0.18496 | 128.633 | 0.85355 | 0.95848 |
| fleQ, flrA; sigma-54 dependent transcriptional regulator, flagellar regulatory protein | 0.00664 | 0.00088 | 0.00721 | 0.00013 | 0.00143 | 65 | 0.00625 | 0.00012 | 0.00111 | 93 | 0.00181 | 0.53197 | 131.627 | 0.59564 | 0.88005 |
| entA; 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase [EC:1.3.1.28] | 0.00664 | 0.00086 | 0.00618 | 0.00013 | 0.00139 | 65 | 0.00696 | 0.00011 | 0.00109 | 93 | 0.00177 | -0.43849 | 132.055 | 0.66175 | 0.90968 |
| mdcA; malonate decarboxylase alpha subunit [EC:2.3.1.187] | 0.00663 | 0.00084 | 0.00684 | 0.00012 | 0.00134 | 65 | 0.00648 | 0.00011 | 0.00110 | 93 | 0.00173 | 0.20886 | 136.068 | 0.83487 | 0.95506 |
| mdcE; malonate decarboxylase gamma subunit [EC:4.1.1.87] | 0.00663 | 0.00084 | 0.00684 | 0.00012 | 0.00134 | 65 | 0.00648 | 0.00011 | 0.00110 | 93 | 0.00173 | 0.20886 | 136.068 | 0.83487 | 0.95506 |
| K09938; uncharacterized protein | 0.00663 | 0.00088 | 0.00718 | 0.00013 | 0.00143 | 65 | 0.00624 | 0.00012 | 0.00112 | 93 | 0.00181 | 0.52215 | 131.947 | 0.60244 | 0.88425 |
| msrQ; methionine sulfoxide reductase heme-binding subunit | 0.00662 | 0.00079 | 0.00621 | 0.00010 | 0.00121 | 65 | 0.00690 | 0.00010 | 0.00105 | 93 | 0.00160 | -0.42805 | 141.131 | 0.66927 | 0.91165 |
| K07014; uncharacterized protein | 0.00661 | 0.00080 | 0.00704 | 0.00011 | 0.00128 | 65 | 0.00631 | 0.00010 | 0.00103 | 93 | 0.00164 | 0.44630 | 134.470 | 0.65610 | 0.90679 |
| tehA; tellurite resistance protein | 0.00660 | 0.00080 | 0.00698 | 0.00011 | 0.00130 | 65 | 0.00633 | 0.00009 | 0.00101 | 93 | 0.00165 | 0.39551 | 131.000 | 0.69311 | 0.91677 |
| ycdX; putative hydrolase | 0.00659 | 0.00093 | 0.00702 | 0.00018 | 0.00166 | 65 | 0.00630 | 0.00011 | 0.00109 | 93 | 0.00198 | 0.36411 | 116.129 | 0.71644 | 0.92004 |
| pcaH; protocatechuate 3,4-dioxygenase, beta subunit [EC:1.13.11.3] | 0.00659 | 0.00080 | 0.00618 | 0.00010 | 0.00123 | 65 | 0.00687 | 0.00011 | 0.00106 | 93 | 0.00163 | -0.41987 | 140.443 | 0.67522 | 0.91264 |
| cwlJ, sleB; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | 0.00658 | 0.00127 | 0.00391 | 0.00011 | 0.00131 | 65 | 0.00845 | 0.00035 | 0.00193 | 93 | 0.00233 | -1.94494 | 150.308 | 0.05365 | 0.70048 |
| K08995; putative membrane protein | 0.00657 | 0.00084 | 0.00580 | 0.00010 | 0.00123 | 65 | 0.00711 | 0.00012 | 0.00114 | 93 | 0.00167 | -0.77990 | 146.009 | 0.43671 | 0.83048 |
| uxaA; altronate hydrolase [EC:4.2.1.7] | 0.00655 | 0.00073 | 0.00631 | 0.00011 | 0.00129 | 65 | 0.00672 | 0.00007 | 0.00085 | 93 | 0.00154 | -0.26592 | 117.098 | 0.79077 | 0.94658 |
| kdgR; LacI family transcriptional regulator, kdg operon repressor | 0.00655 | 0.00144 | 0.00712 | 0.00045 | 0.00263 | 65 | 0.00615 | 0.00025 | 0.00163 | 93 | 0.00309 | 0.31215 | 111.114 | 0.75551 | 0.93665 |
| mogA; molybdopterin adenylyltransferase [EC:2.7.7.75] | 0.00654 | 0.00077 | 0.00622 | 0.00007 | 0.00104 | 65 | 0.00676 | 0.00011 | 0.00109 | 93 | 0.00150 | -0.36169 | 153.224 | 0.71808 | 0.92090 |
| troC, mntC, znuB; manganese/zinc/iron transport system permease protein | 0.00654 | 0.00104 | 0.00948 | 0.00028 | 0.00206 | 65 | 0.00448 | 0.00009 | 0.00098 | 93 | 0.00228 | 2.18705 | 92.688 | 0.03126 | 0.61842 |
| yfiF, trmG; RNA methyltransferase, TrmH family [EC:2.1.1.-] | 0.00652 | 0.00079 | 0.00685 | 0.00010 | 0.00126 | 65 | 0.00628 | 0.00010 | 0.00103 | 93 | 0.00162 | 0.35189 | 135.624 | 0.72547 | 0.92177 |
| iolG; myo-inositol 2-dehydrogenase / D-chiro-inositol 1-dehydrogenase [EC:1.1.1.18 1.1.1.369] | 0.00652 | 0.00107 | 0.00500 | 0.00010 | 0.00124 | 65 | 0.00758 | 0.00023 | 0.00159 | 93 | 0.00201 | -1.28328 | 155.316 | 0.20130 | 0.77376 |
| kduI; 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase [EC:5.3.1.17] | 0.00650 | 0.00086 | 0.00693 | 0.00019 | 0.00171 | 65 | 0.00621 | 0.00007 | 0.00085 | 93 | 0.00191 | 0.38048 | 95.649 | 0.70443 | 0.92004 |
| hpaF, hpcD; 5-carboxymethyl-2-hydroxymuconate isomerase [EC:5.3.3.10] | 0.00650 | 0.00103 | 0.00456 | 0.00009 | 0.00117 | 65 | 0.00785 | 0.00022 | 0.00154 | 93 | 0.00194 | -1.70307 | 154.820 | 0.09056 | 0.76567 |
| ttdB; L(+)-tartrate dehydratase beta subunit [EC:4.2.1.32] | 0.00649 | 0.00076 | 0.00715 | 0.00013 | 0.00142 | 65 | 0.00603 | 0.00006 | 0.00082 | 93 | 0.00164 | 0.68019 | 105.797 | 0.49787 | 0.84853 |
| K09778; uncharacterized protein | 0.00649 | 0.00100 | 0.00786 | 0.00022 | 0.00183 | 65 | 0.00553 | 0.00012 | 0.00112 | 93 | 0.00215 | 1.08058 | 110.148 | 0.28224 | 0.77376 |
| chpC; chemosensory pili system protein ChpC | 0.00649 | 0.00087 | 0.00694 | 0.00013 | 0.00142 | 65 | 0.00617 | 0.00011 | 0.00110 | 93 | 0.00180 | 0.42960 | 130.915 | 0.66820 | 0.91160 |
| K09131; uncharacterized protein | 0.00648 | 0.00073 | 0.00618 | 0.00008 | 0.00114 | 65 | 0.00669 | 0.00009 | 0.00097 | 93 | 0.00149 | -0.33787 | 139.444 | 0.73597 | 0.92721 |
| nasA; assimilatory nitrate reductase catalytic subunit [EC:1.7.99.-] | 0.00647 | 0.00078 | 0.00569 | 0.00008 | 0.00110 | 65 | 0.00702 | 0.00011 | 0.00107 | 93 | 0.00154 | -0.86417 | 149.623 | 0.38888 | 0.80291 |
| K09005; uncharacterized protein | 0.00647 | 0.00077 | 0.00526 | 0.00009 | 0.00115 | 65 | 0.00732 | 0.00010 | 0.00102 | 93 | 0.00154 | -1.34161 | 143.065 | 0.18185 | 0.77376 |
| adeS; two-component system, OmpR family, sensor histidine kinase AdeS [EC:2.7.13.3] | 0.00644 | 0.00119 | 0.00596 | 0.00024 | 0.00191 | 65 | 0.00678 | 0.00022 | 0.00153 | 93 | 0.00245 | -0.33426 | 133.634 | 0.73871 | 0.92914 |
| K06945; uncharacterized protein | 0.00644 | 0.00080 | 0.00592 | 0.00009 | 0.00117 | 65 | 0.00680 | 0.00011 | 0.00110 | 93 | 0.00160 | -0.54808 | 147.055 | 0.58447 | 0.87720 |
| E3.1.21.4; type II restriction enzyme [EC:3.1.21.4] | 0.00644 | 0.00119 | 0.00485 | 0.00005 | 0.00087 | 65 | 0.00755 | 0.00034 | 0.00192 | 93 | 0.00211 | -1.28013 | 126.336 | 0.20284 | 0.77376 |
| cah; carbonic anhydrase [EC:4.2.1.1] | 0.00643 | 0.00096 | 0.00724 | 0.00019 | 0.00171 | 65 | 0.00586 | 0.00011 | 0.00111 | 93 | 0.00203 | 0.67756 | 115.119 | 0.49941 | 0.84862 |
| E2.1.3.-; carbamoyltransferase [EC:2.1.3.-] | 0.00643 | 0.00113 | 0.00655 | 0.00018 | 0.00168 | 65 | 0.00635 | 0.00022 | 0.00154 | 93 | 0.00227 | 0.09056 | 145.207 | 0.92797 | 0.98143 |
| pcaG; protocatechuate 3,4-dioxygenase, alpha subunit [EC:1.13.11.3] | 0.00643 | 0.00079 | 0.00605 | 0.00010 | 0.00121 | 65 | 0.00669 | 0.00010 | 0.00105 | 93 | 0.00160 | -0.39792 | 140.659 | 0.69129 | 0.91676 |
| pduX; L-threonine kinase [EC:2.7.1.177] | 0.00642 | 0.00078 | 0.00720 | 0.00013 | 0.00140 | 65 | 0.00588 | 0.00008 | 0.00090 | 93 | 0.00166 | 0.79820 | 114.564 | 0.42640 | 0.82661 |
| yfiQ; acetyltransferase | 0.00642 | 0.00077 | 0.00560 | 0.00008 | 0.00114 | 65 | 0.00700 | 0.00010 | 0.00103 | 93 | 0.00154 | -0.90593 | 144.400 | 0.36648 | 0.79587 |
| otsB; trehalose 6-phosphate phosphatase [EC:3.1.3.12] | 0.00642 | 0.00076 | 0.00504 | 0.00008 | 0.00110 | 65 | 0.00738 | 0.00010 | 0.00102 | 93 | 0.00151 | -1.55578 | 146.141 | 0.12192 | 0.77376 |
| ttdA; L(+)-tartrate dehydratase alpha subunit [EC:4.2.1.32] | 0.00641 | 0.00075 | 0.00708 | 0.00013 | 0.00141 | 65 | 0.00595 | 0.00006 | 0.00082 | 93 | 0.00163 | 0.69281 | 106.148 | 0.48994 | 0.84797 |
| K02481; two-component system, NtrC family, response regulator | 0.00641 | 0.00094 | 0.00687 | 0.00019 | 0.00169 | 65 | 0.00609 | 0.00011 | 0.00108 | 93 | 0.00201 | 0.38808 | 113.463 | 0.69869 | 0.91844 |
| fepA, pfeA, iroN, pirA; ferric enterobactin receptor | 0.00640 | 0.00085 | 0.00651 | 0.00012 | 0.00135 | 65 | 0.00631 | 0.00011 | 0.00110 | 93 | 0.00174 | 0.11476 | 135.533 | 0.90880 | 0.97664 |
| K07726; putative transcriptional regulator | 0.00638 | 0.00075 | 0.00541 | 0.00009 | 0.00114 | 65 | 0.00705 | 0.00009 | 0.00099 | 93 | 0.00152 | -1.08632 | 141.107 | 0.27919 | 0.77376 |
| dsbG; thiol:disulfide interchange protein DsbG | 0.00637 | 0.00083 | 0.00664 | 0.00012 | 0.00136 | 65 | 0.00619 | 0.00010 | 0.00106 | 93 | 0.00172 | 0.26144 | 131.611 | 0.79416 | 0.94783 |
| uxuA; mannonate dehydratase [EC:4.2.1.8] | 0.00637 | 0.00077 | 0.00550 | 0.00010 | 0.00127 | 65 | 0.00698 | 0.00009 | 0.00097 | 93 | 0.00159 | -0.93065 | 129.561 | 0.35376 | 0.79236 |
| glpP; glycerol uptake operon antiterminator | 0.00635 | 0.00094 | 0.00702 | 0.00020 | 0.00174 | 65 | 0.00589 | 0.00010 | 0.00104 | 93 | 0.00202 | 0.55778 | 108.169 | 0.57815 | 0.87519 |
| K07070; uncharacterized protein | 0.00635 | 0.00075 | 0.00665 | 0.00009 | 0.00120 | 65 | 0.00614 | 0.00009 | 0.00098 | 93 | 0.00154 | 0.33228 | 135.870 | 0.74019 | 0.92969 |
| resD; two-component system, OmpR family, response regulator ResD | 0.00634 | 0.00076 | 0.00381 | 0.00003 | 0.00072 | 65 | 0.00811 | 0.00013 | 0.00117 | 93 | 0.00137 | -3.14010 | 144.778 | 0.00205 | 0.46257 |
| mtlR; mannitol operon transcriptional antiterminator | 0.00634 | 0.00093 | 0.00720 | 0.00022 | 0.00184 | 65 | 0.00574 | 0.00008 | 0.00091 | 93 | 0.00206 | 0.70832 | 95.480 | 0.48047 | 0.84354 |
| IDH3; isocitrate dehydrogenase (NAD+) [EC:1.1.1.41] | 0.00634 | 0.00096 | 0.00831 | 0.00023 | 0.00188 | 65 | 0.00496 | 0.00008 | 0.00094 | 93 | 0.00210 | 1.59323 | 95.499 | 0.11441 | 0.77376 |
| dedD; DedD protein | 0.00633 | 0.00074 | 0.00683 | 0.00009 | 0.00121 | 65 | 0.00598 | 0.00008 | 0.00094 | 93 | 0.00153 | 0.55320 | 131.983 | 0.58106 | 0.87593 |
| soxD; sarcosine oxidase, subunit delta [EC:1.5.3.1] | 0.00633 | 0.00096 | 0.00616 | 0.00014 | 0.00149 | 65 | 0.00644 | 0.00015 | 0.00127 | 93 | 0.00196 | -0.14245 | 139.527 | 0.88693 | 0.97032 |
| yprB; uncharacterized protein | 0.00632 | 0.00081 | 0.00606 | 0.00014 | 0.00144 | 65 | 0.00651 | 0.00008 | 0.00095 | 93 | 0.00173 | -0.25633 | 116.477 | 0.79815 | 0.94822 |
| codB; cytosine permease | 0.00632 | 0.00096 | 0.00590 | 0.00011 | 0.00132 | 65 | 0.00662 | 0.00017 | 0.00135 | 93 | 0.00189 | -0.38037 | 152.001 | 0.70420 | 0.92004 |
| miaE; tRNA-(ms[2]io[6]A)-hydroxylase [EC:1.-.-.-] | 0.00629 | 0.00078 | 0.00675 | 0.00010 | 0.00124 | 65 | 0.00597 | 0.00010 | 0.00101 | 93 | 0.00160 | 0.48822 | 136.241 | 0.62618 | 0.89255 |
| bluB; 5,6-dimethylbenzimidazole synthase [EC:1.13.11.79] | 0.00629 | 0.00078 | 0.00569 | 0.00009 | 0.00118 | 65 | 0.00670 | 0.00010 | 0.00105 | 93 | 0.00158 | -0.63996 | 142.734 | 0.52323 | 0.85706 |
| mobAB; molybdopterin-guanine dinucleotide biosynthesis protein [EC:2.7.7.77] | 0.00629 | 0.00078 | 0.00705 | 0.00013 | 0.00139 | 65 | 0.00575 | 0.00008 | 0.00091 | 93 | 0.00166 | 0.78596 | 115.404 | 0.43350 | 0.82874 |
| catA; catechol 1,2-dioxygenase [EC:1.13.11.1] | 0.00628 | 0.00078 | 0.00589 | 0.00011 | 0.00128 | 65 | 0.00656 | 0.00009 | 0.00098 | 93 | 0.00161 | -0.41229 | 129.982 | 0.68080 | 0.91355 |
| gltL, aatP; glutamate/aspartate transport system ATP-binding protein [EC:3.6.3.-] | 0.00628 | 0.00072 | 0.00626 | 0.00009 | 0.00114 | 65 | 0.00630 | 0.00008 | 0.00093 | 93 | 0.00148 | -0.03344 | 135.650 | 0.97337 | 0.99509 |
| citC; [citrate (pro-3S)-lyase] ligase [EC:6.2.1.22] | 0.00628 | 0.00098 | 0.00917 | 0.00026 | 0.00201 | 65 | 0.00426 | 0.00007 | 0.00085 | 93 | 0.00218 | 2.25263 | 87.090 | 0.02680 | 0.61112 |
| ofaB, arfB; arthrofactin-type cyclic lipopeptide synthetase B | 0.00628 | 0.00205 | 0.00547 | 0.00054 | 0.00288 | 65 | 0.00685 | 0.00076 | 0.00286 | 93 | 0.00406 | -0.33879 | 150.752 | 0.73524 | 0.92721 |
| cbiQ; cobalt/nickel transport system permease protein | 0.00628 | 0.00073 | 0.00704 | 0.00011 | 0.00131 | 65 | 0.00574 | 0.00006 | 0.00084 | 93 | 0.00156 | 0.83797 | 113.379 | 0.40381 | 0.81034 |
| K07099; uncharacterized protein | 0.00628 | 0.00068 | 0.00641 | 0.00009 | 0.00118 | 65 | 0.00618 | 0.00006 | 0.00083 | 93 | 0.00144 | 0.15910 | 122.203 | 0.87385 | 0.96576 |
| xdhA; xanthine dehydrogenase small subunit [EC:1.17.1.4] | 0.00624 | 0.00081 | 0.00581 | 0.00010 | 0.00123 | 65 | 0.00655 | 0.00011 | 0.00108 | 93 | 0.00163 | -0.45360 | 142.058 | 0.65081 | 0.90423 |
| ydhQ; GntR family transcriptional regulator | 0.00624 | 0.00094 | 0.00783 | 0.00021 | 0.00180 | 65 | 0.00513 | 0.00009 | 0.00099 | 93 | 0.00205 | 1.31437 | 101.710 | 0.19168 | 0.77376 |
| K07484; transposase | 0.00623 | 0.00121 | 0.00657 | 0.00030 | 0.00214 | 65 | 0.00600 | 0.00018 | 0.00141 | 93 | 0.00256 | 0.22200 | 115.949 | 0.82471 | 0.95506 |
| ilvM; acetolactate synthase II small subunit [EC:2.2.1.6] | 0.00622 | 0.00079 | 0.00682 | 0.00011 | 0.00129 | 65 | 0.00581 | 0.00010 | 0.00101 | 93 | 0.00164 | 0.62013 | 132.302 | 0.53624 | 0.86207 |
| faaH; fumarylacetoacetate (FAA) hydrolase [EC:3.7.1.2] | 0.00622 | 0.00083 | 0.00589 | 0.00012 | 0.00137 | 65 | 0.00645 | 0.00010 | 0.00103 | 93 | 0.00172 | -0.32659 | 128.032 | 0.74451 | 0.93149 |
| K09797; uncharacterized protein | 0.00622 | 0.00061 | 0.00550 | 0.00004 | 0.00079 | 65 | 0.00672 | 0.00007 | 0.00087 | 93 | 0.00117 | -1.04246 | 154.972 | 0.29882 | 0.77376 |
| dehH; haloacetate dehalogenase [EC:3.8.1.3] | 0.00622 | 0.00068 | 0.00585 | 0.00008 | 0.00109 | 65 | 0.00647 | 0.00007 | 0.00087 | 93 | 0.00139 | -0.44883 | 134.059 | 0.65428 | 0.90679 |
| gltJ, aatQ; glutamate/aspartate transport system permease protein | 0.00622 | 0.00071 | 0.00610 | 0.00008 | 0.00112 | 65 | 0.00630 | 0.00008 | 0.00093 | 93 | 0.00146 | -0.13414 | 137.578 | 0.89349 | 0.97159 |
| gltK, aatM; glutamate/aspartate transport system permease protein | 0.00622 | 0.00071 | 0.00610 | 0.00008 | 0.00112 | 65 | 0.00630 | 0.00008 | 0.00093 | 93 | 0.00146 | -0.13414 | 137.578 | 0.89349 | 0.97159 |
| sad; succinate-semialdehyde dehydrogenase [EC:1.2.1.16 1.2.1.24] | 0.00621 | 0.00079 | 0.00623 | 0.00010 | 0.00123 | 65 | 0.00620 | 0.00010 | 0.00104 | 93 | 0.00161 | 0.01754 | 138.843 | 0.98603 | 0.99729 |
| yjgM; putative acetyltransferase [EC:2.3.1.-] | 0.00621 | 0.00059 | 0.00559 | 0.00005 | 0.00086 | 65 | 0.00664 | 0.00006 | 0.00081 | 93 | 0.00118 | -0.88533 | 147.235 | 0.37742 | 0.79821 |
| cpbD; chitin-binding protein | 0.00620 | 0.00120 | 0.00578 | 0.00024 | 0.00192 | 65 | 0.00649 | 0.00022 | 0.00154 | 93 | 0.00246 | -0.28869 | 134.247 | 0.77327 | 0.94205 |
| cptA; toxin CptA | 0.00620 | 0.00079 | 0.00701 | 0.00011 | 0.00127 | 65 | 0.00564 | 0.00010 | 0.00102 | 93 | 0.00163 | 0.84160 | 133.911 | 0.40151 | 0.80850 |
| E4.1.1.19; arginine decarboxylase [EC:4.1.1.19] | 0.00620 | 0.00070 | 0.00538 | 0.00004 | 0.00082 | 65 | 0.00677 | 0.00010 | 0.00104 | 93 | 0.00132 | -1.05324 | 155.444 | 0.29387 | 0.77376 |
| kdkA; 3-deoxy-D-manno-octulosonic acid kinase [EC:2.7.1.166] | 0.00619 | 0.00079 | 0.00667 | 0.00011 | 0.00128 | 65 | 0.00586 | 0.00010 | 0.00101 | 93 | 0.00163 | 0.49287 | 132.993 | 0.62292 | 0.89159 |
| RENBP; N-acylglucosamine 2-epimerase [EC:5.1.3.8] | 0.00614 | 0.00068 | 0.00505 | 0.00004 | 0.00075 | 65 | 0.00690 | 0.00010 | 0.00103 | 93 | 0.00127 | -1.45628 | 153.400 | 0.14736 | 0.77376 |
| K09916; uncharacterized protein | 0.00614 | 0.00076 | 0.00658 | 0.00009 | 0.00119 | 65 | 0.00583 | 0.00009 | 0.00098 | 93 | 0.00154 | 0.48227 | 136.596 | 0.63039 | 0.89558 |
| selU; tRNA 2-selenouridine synthase [EC:2.9.1.-] | 0.00614 | 0.00067 | 0.00680 | 0.00009 | 0.00117 | 65 | 0.00568 | 0.00006 | 0.00079 | 93 | 0.00141 | 0.79521 | 119.310 | 0.42807 | 0.82733 |
| cysC; adenylylsulfate kinase [EC:2.7.1.25] | 0.00614 | 0.00080 | 0.00408 | 0.00005 | 0.00087 | 65 | 0.00757 | 0.00013 | 0.00119 | 93 | 0.00148 | -2.35775 | 153.588 | 0.01965 | 0.59095 |
| K09954; uncharacterized protein | 0.00614 | 0.00076 | 0.00652 | 0.00009 | 0.00119 | 65 | 0.00586 | 0.00009 | 0.00098 | 93 | 0.00155 | 0.42680 | 136.769 | 0.67020 | 0.91184 |
| agp; glucose-1-phosphatase [EC:3.1.3.10] | 0.00613 | 0.00067 | 0.00638 | 0.00007 | 0.00104 | 65 | 0.00596 | 0.00007 | 0.00087 | 93 | 0.00136 | 0.31351 | 138.021 | 0.75436 | 0.93665 |
| pgaC, icaA; poly-beta-1,6-N-acetyl-D-glucosamine synthase [EC:2.4.1.-] | 0.00613 | 0.00084 | 0.00490 | 0.00008 | 0.00108 | 65 | 0.00699 | 0.00013 | 0.00120 | 93 | 0.00161 | -1.29191 | 155.134 | 0.19831 | 0.77376 |
| nupG; MFS transporter, NHS family, nucleoside permease | 0.00613 | 0.00078 | 0.00550 | 0.00007 | 0.00102 | 65 | 0.00657 | 0.00012 | 0.00112 | 93 | 0.00152 | -0.70548 | 154.802 | 0.48157 | 0.84354 |
| abgA; aminobenzoyl-glutamate utilization protein A | 0.00612 | 0.00079 | 0.00640 | 0.00009 | 0.00117 | 65 | 0.00593 | 0.00011 | 0.00106 | 93 | 0.00158 | 0.29415 | 144.607 | 0.76907 | 0.93993 |
| pepD; putative serine protease PepD [EC:3.4.21.-] | 0.00612 | 0.00073 | 0.00635 | 0.00010 | 0.00122 | 65 | 0.00595 | 0.00008 | 0.00090 | 93 | 0.00152 | 0.26102 | 126.092 | 0.79450 | 0.94783 |
| xylS, yicI; alpha-D-xyloside xylohydrolase [EC:3.2.1.177] | 0.00611 | 0.00095 | 0.00437 | 0.00006 | 0.00094 | 65 | 0.00733 | 0.00020 | 0.00146 | 93 | 0.00174 | -1.70568 | 147.403 | 0.09017 | 0.76567 |
| osmB; osmotically inducible lipoprotein OsmB | 0.00611 | 0.00082 | 0.00682 | 0.00013 | 0.00142 | 65 | 0.00561 | 0.00009 | 0.00098 | 93 | 0.00173 | 0.69770 | 120.708 | 0.48671 | 0.84553 |
| K05952; uncharacterized protein | 0.00610 | 0.00071 | 0.00718 | 0.00009 | 0.00118 | 65 | 0.00535 | 0.00007 | 0.00088 | 93 | 0.00147 | 1.24707 | 126.771 | 0.21467 | 0.77376 |
| K09977; uncharacterized protein | 0.00610 | 0.00070 | 0.00665 | 0.00009 | 0.00116 | 65 | 0.00571 | 0.00007 | 0.00086 | 93 | 0.00144 | 0.65622 | 127.750 | 0.51287 | 0.85147 |
| cobC1, cobC; cobalamin biosynthetic protein CobC | 0.00609 | 0.00080 | 0.00530 | 0.00008 | 0.00113 | 65 | 0.00665 | 0.00011 | 0.00110 | 93 | 0.00157 | -0.85473 | 149.775 | 0.39406 | 0.80459 |
| gtsC, glcG; glucose/mannose transport system permease protein | 0.00608 | 0.00077 | 0.00494 | 0.00008 | 0.00111 | 65 | 0.00688 | 0.00010 | 0.00105 | 93 | 0.00153 | -1.26886 | 147.678 | 0.20649 | 0.77376 |
| entF; enterobactin synthetase component F [EC:6.3.2.14] | 0.00608 | 0.00082 | 0.00594 | 0.00013 | 0.00139 | 65 | 0.00617 | 0.00010 | 0.00101 | 93 | 0.00172 | -0.13448 | 125.176 | 0.89324 | 0.97159 |
| baeS, smeS; two-component system, OmpR family, sensor histidine kinase BaeS [EC:2.7.13.3] | 0.00606 | 0.00077 | 0.00502 | 0.00007 | 0.00106 | 65 | 0.00679 | 0.00011 | 0.00108 | 93 | 0.00152 | -1.16612 | 151.771 | 0.24540 | 0.77376 |
| idsA; geranylgeranyl diphosphate synthase, type I [EC:2.5.1.1 2.5.1.10 2.5.1.29] | 0.00606 | 0.00071 | 0.00603 | 0.00009 | 0.00115 | 65 | 0.00609 | 0.00008 | 0.00090 | 93 | 0.00147 | -0.04395 | 131.897 | 0.96501 | 0.99101 |
| E2.3.1.207; beta-ketodecanoyl-[acyl-carrier-protein] synthase [EC:2.3.1.207] | 0.00605 | 0.00077 | 0.00619 | 0.00010 | 0.00124 | 65 | 0.00596 | 0.00009 | 0.00098 | 93 | 0.00158 | 0.14434 | 132.819 | 0.88545 | 0.97001 |
| soxG; sarcosine oxidase, subunit gamma [EC:1.5.3.1] | 0.00605 | 0.00094 | 0.00565 | 0.00013 | 0.00139 | 65 | 0.00633 | 0.00015 | 0.00127 | 93 | 0.00188 | -0.36156 | 144.713 | 0.71821 | 0.92090 |
| endA; deoxyribonuclease I [EC:3.1.21.1] | 0.00605 | 0.00091 | 0.00727 | 0.00017 | 0.00162 | 65 | 0.00519 | 0.00010 | 0.00105 | 93 | 0.00193 | 1.07244 | 115.264 | 0.28576 | 0.77376 |
| K07234; uncharacterized protein involved in response to NO | 0.00604 | 0.00067 | 0.00612 | 0.00008 | 0.00108 | 65 | 0.00597 | 0.00007 | 0.00085 | 93 | 0.00137 | 0.10896 | 132.148 | 0.91340 | 0.97664 |
| fadB; 3-hydroxyacyl-CoA dehydrogenase / enoyl-CoA hydratase / 3-hydroxybutyryl-CoA epimerase / enoyl-CoA isomerase [EC:1.1.1.35 4.2.1.17 5.1.2.3 5.3.3.8] | 0.00603 | 0.00075 | 0.00640 | 0.00009 | 0.00119 | 65 | 0.00578 | 0.00009 | 0.00097 | 93 | 0.00153 | 0.40252 | 135.263 | 0.68794 | 0.91567 |
| TDO2, kynA; tryptophan 2,3-dioxygenase [EC:1.13.11.11] | 0.00603 | 0.00078 | 0.00452 | 0.00008 | 0.00112 | 65 | 0.00709 | 0.00010 | 0.00106 | 93 | 0.00155 | -1.66711 | 147.496 | 0.09761 | 0.76673 |
| treY, glgY; (1->4)-alpha-D-glucan 1-alpha-D-glucosylmutase [EC:5.4.99.15] | 0.00603 | 0.00075 | 0.00561 | 0.00009 | 0.00119 | 65 | 0.00633 | 0.00009 | 0.00097 | 93 | 0.00154 | -0.47063 | 135.784 | 0.63866 | 0.89824 |
| gpt; xanthine phosphoribosyltransferase [EC:2.4.2.22] | 0.00603 | 0.00074 | 0.00554 | 0.00006 | 0.00098 | 65 | 0.00637 | 0.00010 | 0.00105 | 93 | 0.00144 | -0.58200 | 154.155 | 0.56142 | 0.86813 |
| opsX; heptosyltransferase I [EC:2.4.-.-] | 0.00601 | 0.00080 | 0.00646 | 0.00011 | 0.00129 | 65 | 0.00570 | 0.00010 | 0.00102 | 93 | 0.00164 | 0.46096 | 133.047 | 0.64558 | 0.90314 |
| pqiB; paraquat-inducible protein B | 0.00601 | 0.00080 | 0.00597 | 0.00009 | 0.00119 | 65 | 0.00604 | 0.00011 | 0.00107 | 93 | 0.00160 | -0.04339 | 144.337 | 0.96545 | 0.99101 |
| waaY, rfaY; heptose II phosphotransferase [EC:2.7.1.-] | 0.00601 | 0.00153 | 0.00990 | 0.00070 | 0.00329 | 65 | 0.00329 | 0.00012 | 0.00115 | 93 | 0.00348 | 1.89886 | 79.856 | 0.06119 | 0.73001 |
| K07131; uncharacterized protein | 0.00601 | 0.00075 | 0.00526 | 0.00008 | 0.00112 | 65 | 0.00654 | 0.00009 | 0.00101 | 93 | 0.00151 | -0.84840 | 144.028 | 0.39763 | 0.80562 |
| gtsB, glcF; glucose/mannose transport system permease protein | 0.00600 | 0.00077 | 0.00493 | 0.00008 | 0.00111 | 65 | 0.00676 | 0.00010 | 0.00105 | 93 | 0.00153 | -1.19426 | 147.757 | 0.23429 | 0.77376 |
| KYNU, kynU; kynureninase [EC:3.7.1.3] | 0.00600 | 0.00078 | 0.00487 | 0.00009 | 0.00121 | 65 | 0.00679 | 0.00010 | 0.00103 | 93 | 0.00159 | -1.20960 | 139.802 | 0.22847 | 0.77376 |
| pspC; phage shock protein C | 0.00600 | 0.00091 | 0.00734 | 0.00022 | 0.00186 | 65 | 0.00507 | 0.00006 | 0.00083 | 93 | 0.00204 | 1.11721 | 89.869 | 0.26688 | 0.77376 |
| ureJ; urease accessory protein | 0.00600 | 0.00079 | 0.00555 | 0.00009 | 0.00120 | 65 | 0.00631 | 0.00010 | 0.00105 | 93 | 0.00160 | -0.47242 | 142.157 | 0.63735 | 0.89715 |
| uxaC; glucuronate isomerase [EC:5.3.1.12] | 0.00600 | 0.00075 | 0.00538 | 0.00010 | 0.00124 | 65 | 0.00642 | 0.00008 | 0.00093 | 93 | 0.00155 | -0.67067 | 127.614 | 0.50364 | 0.84956 |
| cbiO; cobalt/nickel transport system ATP-binding protein | 0.00599 | 0.00071 | 0.00632 | 0.00010 | 0.00127 | 65 | 0.00576 | 0.00006 | 0.00084 | 93 | 0.00152 | 0.36821 | 116.472 | 0.71339 | 0.92004 |
| E3.1.1.17, gnl, RGN; gluconolactonase [EC:3.1.1.17] | 0.00599 | 0.00085 | 0.00370 | 0.00005 | 0.00088 | 65 | 0.00760 | 0.00015 | 0.00128 | 93 | 0.00155 | -2.51243 | 151.115 | 0.01304 | 0.59095 |
| priB; primosomal replication protein N | 0.00598 | 0.00072 | 0.00655 | 0.00009 | 0.00120 | 65 | 0.00558 | 0.00008 | 0.00090 | 93 | 0.00150 | 0.64932 | 127.787 | 0.51730 | 0.85244 |
| cobG; precorrin-3B synthase [EC:1.14.13.83] | 0.00598 | 0.00085 | 0.00563 | 0.00011 | 0.00131 | 65 | 0.00622 | 0.00012 | 0.00113 | 93 | 0.00173 | -0.33854 | 140.573 | 0.73546 | 0.92721 |
| astE; succinylglutamate desuccinylase [EC:3.5.1.96] | 0.00597 | 0.00075 | 0.00628 | 0.00009 | 0.00120 | 65 | 0.00575 | 0.00009 | 0.00097 | 93 | 0.00154 | 0.34307 | 134.526 | 0.73208 | 0.92572 |
| ubiF; 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [EC:1.14.13.-] | 0.00597 | 0.00078 | 0.00634 | 0.00010 | 0.00122 | 65 | 0.00570 | 0.00010 | 0.00101 | 93 | 0.00159 | 0.40029 | 137.004 | 0.68957 | 0.91567 |
| cobW; cobalamin biosynthesis protein CobW | 0.00597 | 0.00091 | 0.00547 | 0.00011 | 0.00133 | 65 | 0.00631 | 0.00014 | 0.00125 | 93 | 0.00182 | -0.46052 | 147.041 | 0.64582 | 0.90314 |
| hpaI, hpcH; 4-hydroxy-2-oxoheptanedioate aldolase [EC:4.1.2.52] | 0.00596 | 0.00091 | 0.00356 | 0.00004 | 0.00077 | 65 | 0.00763 | 0.00019 | 0.00143 | 93 | 0.00162 | -2.50706 | 136.662 | 0.01335 | 0.59095 |
| E1.2.1.88; 1-pyrroline-5-carboxylate dehydrogenase [EC:1.2.1.88] | 0.00596 | 0.00106 | 0.00416 | 0.00007 | 0.00104 | 65 | 0.00721 | 0.00025 | 0.00163 | 93 | 0.00194 | -1.57705 | 147.118 | 0.11693 | 0.77376 |
| treZ, glgZ; maltooligosyltrehalose trehalohydrolase [EC:3.2.1.141] | 0.00596 | 0.00074 | 0.00550 | 0.00009 | 0.00120 | 65 | 0.00627 | 0.00008 | 0.00095 | 93 | 0.00153 | -0.50605 | 133.160 | 0.61366 | 0.88788 |
| wcaJ; putative colanic acid biosysnthesis UDP-glucose lipid carrier transferase | 0.00596 | 0.00060 | 0.00534 | 0.00004 | 0.00078 | 65 | 0.00638 | 0.00007 | 0.00086 | 93 | 0.00117 | -0.89360 | 154.944 | 0.37292 | 0.79810 |
| plc; phospholipase C [EC:3.1.4.3] | 0.00596 | 0.00082 | 0.00487 | 0.00010 | 0.00122 | 65 | 0.00671 | 0.00011 | 0.00110 | 93 | 0.00164 | -1.12082 | 143.508 | 0.26424 | 0.77376 |
| rpoS; RNA polymerase nonessential primary-like sigma factor | 0.00596 | 0.00075 | 0.00637 | 0.00010 | 0.00124 | 65 | 0.00566 | 0.00008 | 0.00094 | 93 | 0.00156 | 0.45689 | 128.869 | 0.64852 | 0.90350 |
| trbL; type IV secretion system protein TrbL | 0.00595 | 0.00099 | 0.00583 | 0.00013 | 0.00143 | 65 | 0.00604 | 0.00017 | 0.00135 | 93 | 0.00197 | -0.10650 | 147.564 | 0.91533 | 0.97664 |
| yaaU; MFS transporter, putative metabolite transport protein | 0.00594 | 0.00130 | 0.00660 | 0.00040 | 0.00247 | 65 | 0.00549 | 0.00018 | 0.00139 | 93 | 0.00283 | 0.39233 | 104.022 | 0.69562 | 0.91769 |
| glvR; RpiR family transcriptional regulator, glv operon transcriptional regulator | 0.00594 | 0.00078 | 0.00619 | 0.00012 | 0.00137 | 65 | 0.00577 | 0.00008 | 0.00092 | 93 | 0.00166 | 0.25790 | 118.292 | 0.79693 | 0.94800 |
| K06884; uncharacterized protein | 0.00592 | 0.00087 | 0.00689 | 0.00015 | 0.00150 | 65 | 0.00525 | 0.00010 | 0.00104 | 93 | 0.00183 | 0.89909 | 120.166 | 0.37040 | 0.79587 |
| K09118; uncharacterized protein | 0.00591 | 0.00074 | 0.00631 | 0.00011 | 0.00130 | 65 | 0.00564 | 0.00007 | 0.00087 | 93 | 0.00157 | 0.42800 | 117.839 | 0.66943 | 0.91165 |
| vanB; vanillate monooxygenase ferredoxin subunit | 0.00591 | 0.00087 | 0.00539 | 0.00012 | 0.00134 | 65 | 0.00627 | 0.00012 | 0.00115 | 93 | 0.00176 | -0.50100 | 139.915 | 0.61716 | 0.88903 |
| proQ; ProP effector | 0.00590 | 0.00072 | 0.00718 | 0.00009 | 0.00120 | 65 | 0.00501 | 0.00007 | 0.00088 | 93 | 0.00149 | 1.46350 | 126.766 | 0.14581 | 0.77376 |
| hasE, prtE; membrane fusion protein, protease secretion system | 0.00589 | 0.00142 | 0.00495 | 0.00022 | 0.00185 | 65 | 0.00654 | 0.00038 | 0.00203 | 93 | 0.00275 | -0.58146 | 154.792 | 0.56177 | 0.86813 |
| E2.7.7.53; ATP adenylyltransferase [EC:2.7.7.53] | 0.00588 | 0.00070 | 0.00603 | 0.00009 | 0.00115 | 65 | 0.00578 | 0.00007 | 0.00088 | 93 | 0.00145 | 0.17042 | 129.804 | 0.86495 | 0.96269 |
| baeR, smeR; two-component system, OmpR family, response regulator BaeR | 0.00588 | 0.00075 | 0.00500 | 0.00007 | 0.00105 | 65 | 0.00650 | 0.00010 | 0.00105 | 93 | 0.00148 | -1.01279 | 151.050 | 0.31278 | 0.77376 |
| abcA, bmrA; ATP-binding cassette, subfamily B, bacterial AbcA/BmrA [EC:3.6.3.44] | 0.00587 | 0.00097 | 0.00520 | 0.00018 | 0.00164 | 65 | 0.00634 | 0.00013 | 0.00120 | 93 | 0.00203 | -0.56422 | 125.584 | 0.57361 | 0.87292 |
| hcaD; 3-phenylpropionate/trans-cinnamate dioxygenase ferredoxin reductase component [EC:1.18.1.3] | 0.00586 | 0.00079 | 0.00318 | 0.00003 | 0.00068 | 65 | 0.00774 | 0.00014 | 0.00122 | 93 | 0.00140 | -3.26494 | 138.682 | 0.00138 | 0.46257 |
| resB, ccs1; cytochrome c biogenesis protein | 0.00586 | 0.00073 | 0.00415 | 0.00004 | 0.00075 | 65 | 0.00705 | 0.00012 | 0.00112 | 93 | 0.00135 | -2.15350 | 149.754 | 0.03288 | 0.62130 |
| mcrA; 5-methylcytosine-specific restriction enzyme A [EC:3.1.21.-] | 0.00586 | 0.00075 | 0.00539 | 0.00009 | 0.00118 | 65 | 0.00618 | 0.00009 | 0.00098 | 93 | 0.00153 | -0.51506 | 137.349 | 0.60734 | 0.88598 |
| K09956; uncharacterized protein | 0.00585 | 0.00076 | 0.00528 | 0.00008 | 0.00112 | 65 | 0.00625 | 0.00010 | 0.00103 | 93 | 0.00153 | -0.62999 | 145.734 | 0.52969 | 0.86094 |
| nirA; ferredoxin-nitrite reductase [EC:1.7.7.1] | 0.00584 | 0.00067 | 0.00605 | 0.00008 | 0.00112 | 65 | 0.00569 | 0.00007 | 0.00084 | 93 | 0.00140 | 0.25923 | 128.105 | 0.79588 | 0.94791 |
| mdcG; phosphoribosyl-dephospho-CoA transferase [EC:2.7.7.66] | 0.00583 | 0.00076 | 0.00611 | 0.00010 | 0.00125 | 65 | 0.00563 | 0.00009 | 0.00096 | 93 | 0.00158 | 0.30461 | 130.707 | 0.76115 | 0.93806 |
| ttuC, dmlA; tartrate dehydrogenase/decarboxylase / D-malate dehydrogenase [EC:1.1.1.93 4.1.1.73 1.1.1.83] | 0.00583 | 0.00100 | 0.00351 | 0.00005 | 0.00086 | 65 | 0.00744 | 0.00023 | 0.00158 | 93 | 0.00179 | -2.19045 | 137.186 | 0.03018 | 0.61413 |
| nucS; endonuclease [EC:3.1.-.-] | 0.00583 | 0.00075 | 0.00580 | 0.00008 | 0.00114 | 65 | 0.00585 | 0.00009 | 0.00100 | 93 | 0.00152 | -0.03328 | 142.095 | 0.97350 | 0.99509 |
| vasJ; type VI secretion system protein VasJ | 0.00582 | 0.00070 | 0.00704 | 0.00009 | 0.00116 | 65 | 0.00497 | 0.00007 | 0.00087 | 93 | 0.00145 | 1.42025 | 128.161 | 0.15796 | 0.77376 |
| lmrB; MFS transporter, DHA2 family, lincomycin resistance protein | 0.00582 | 0.00092 | 0.00641 | 0.00019 | 0.00172 | 65 | 0.00540 | 0.00009 | 0.00099 | 93 | 0.00199 | 0.50594 | 105.458 | 0.61396 | 0.88788 |
| nagD; NagD protein | 0.00579 | 0.00079 | 0.00727 | 0.00017 | 0.00160 | 65 | 0.00476 | 0.00005 | 0.00074 | 93 | 0.00176 | 1.42585 | 91.544 | 0.15731 | 0.77376 |
| K06921; uncharacterized protein | 0.00579 | 0.00095 | 0.00676 | 0.00019 | 0.00170 | 65 | 0.00511 | 0.00011 | 0.00109 | 93 | 0.00202 | 0.81762 | 114.649 | 0.41527 | 0.81730 |
| bglB; beta-glucosidase [EC:3.2.1.21] | 0.00578 | 0.00086 | 0.00515 | 0.00015 | 0.00150 | 65 | 0.00622 | 0.00010 | 0.00102 | 93 | 0.00181 | -0.59288 | 118.786 | 0.55439 | 0.86806 |
| aapM, bztC; general L-amino acid transport system permease protein | 0.00578 | 0.00075 | 0.00548 | 0.00008 | 0.00114 | 65 | 0.00599 | 0.00009 | 0.00099 | 93 | 0.00151 | -0.33518 | 141.158 | 0.73799 | 0.92891 |
| K07457; endonuclease III related protein | 0.00577 | 0.00082 | 0.00547 | 0.00007 | 0.00102 | 65 | 0.00598 | 0.00013 | 0.00119 | 93 | 0.00157 | -0.32348 | 155.914 | 0.74677 | 0.93207 |
| K07814; putative two-component system response regulator | 0.00577 | 0.00120 | 0.00411 | 0.00005 | 0.00090 | 65 | 0.00692 | 0.00035 | 0.00194 | 93 | 0.00214 | -1.31659 | 127.540 | 0.19034 | 0.77376 |
| oprD; imipenem/basic amino acid-specific outer membrane pore [EC:3.4.21.-] | 0.00576 | 0.00096 | 0.00736 | 0.00021 | 0.00179 | 65 | 0.00463 | 0.00010 | 0.00105 | 93 | 0.00208 | 1.31155 | 106.582 | 0.19249 | 0.77376 |
| potE; putrescine:ornithine antiporter | 0.00575 | 0.00067 | 0.00592 | 0.00007 | 0.00106 | 65 | 0.00563 | 0.00007 | 0.00086 | 93 | 0.00137 | 0.20965 | 135.162 | 0.83425 | 0.95506 |
| aapQ, bztB; general L-amino acid transport system permease protein | 0.00575 | 0.00075 | 0.00545 | 0.00008 | 0.00114 | 65 | 0.00596 | 0.00009 | 0.00099 | 93 | 0.00151 | -0.33810 | 141.558 | 0.73579 | 0.92721 |
| mp2; beta-1,4-mannooligosaccharide/beta-1,4-mannosyl-N-acetylglucosamine phosphorylase [EC:2.4.1.319 2.4.1.320] | 0.00574 | 0.00065 | 0.00466 | 0.00004 | 0.00074 | 65 | 0.00650 | 0.00009 | 0.00097 | 93 | 0.00122 | -1.51505 | 154.826 | 0.13180 | 0.77376 |
| cutA; periplasmic divalent cation tolerance protein | 0.00573 | 0.00072 | 0.00551 | 0.00009 | 0.00120 | 65 | 0.00587 | 0.00007 | 0.00090 | 93 | 0.00150 | -0.23921 | 127.800 | 0.81133 | 0.95214 |
| pucR; purine catabolism regulatory protein | 0.00572 | 0.00187 | 0.00331 | 0.00011 | 0.00130 | 65 | 0.00741 | 0.00086 | 0.00304 | 93 | 0.00330 | -1.24104 | 122.799 | 0.21696 | 0.77376 |
| yiaY; alcohol dehydrogenase [EC:1.1.1.1] | 0.00571 | 0.00072 | 0.00479 | 0.00006 | 0.00098 | 65 | 0.00636 | 0.00009 | 0.00100 | 93 | 0.00140 | -1.12093 | 152.077 | 0.26408 | 0.77376 |
| E1.14.19.3; linoleoyl-CoA desaturase [EC:1.14.19.3] | 0.00571 | 0.00087 | 0.00461 | 0.00007 | 0.00101 | 65 | 0.00647 | 0.00016 | 0.00131 | 93 | 0.00165 | -1.12834 | 155.229 | 0.26092 | 0.77376 |
| pqqE; pyrroloquinoline quinone biosynthesis protein E | 0.00570 | 0.00088 | 0.00571 | 0.00012 | 0.00135 | 65 | 0.00570 | 0.00013 | 0.00117 | 93 | 0.00178 | 0.00771 | 141.096 | 0.99386 | 0.99919 |
| ina; immune inhibitor A [EC:3.4.24.-] | 0.00569 | 0.00080 | 0.00438 | 0.00006 | 0.00094 | 65 | 0.00661 | 0.00013 | 0.00118 | 93 | 0.00151 | -1.47686 | 155.610 | 0.14173 | 0.77376 |
| smoE, mtlE; sorbitol/mannitol transport system substrate-binding protein | 0.00569 | 0.00100 | 0.00460 | 0.00011 | 0.00130 | 65 | 0.00645 | 0.00019 | 0.00144 | 93 | 0.00194 | -0.95448 | 155.084 | 0.34133 | 0.78526 |
| tauA; taurine transport system substrate-binding protein | 0.00568 | 0.00080 | 0.00499 | 0.00008 | 0.00108 | 65 | 0.00616 | 0.00012 | 0.00113 | 93 | 0.00156 | -0.75180 | 152.859 | 0.45332 | 0.83438 |
| pssA; CDP-diacylglycerol—serine O-phosphatidyltransferase [EC:2.7.8.8] | 0.00567 | 0.00068 | 0.00615 | 0.00007 | 0.00107 | 65 | 0.00534 | 0.00007 | 0.00087 | 93 | 0.00138 | 0.58803 | 135.490 | 0.55749 | 0.86813 |
| TRM61, GCD14; tRNA (adenine57-N1/adenine58-N1)-methyltransferase catalytic subunit [EC:2.1.1.219 2.1.1.220] | 0.00567 | 0.00069 | 0.00588 | 0.00008 | 0.00113 | 65 | 0.00552 | 0.00007 | 0.00088 | 93 | 0.00143 | 0.25569 | 130.679 | 0.79859 | 0.94822 |
| msrA, vmlR; macrolide transport system ATP-binding/permease protein | 0.00567 | 0.00090 | 0.00559 | 0.00015 | 0.00154 | 65 | 0.00572 | 0.00011 | 0.00109 | 93 | 0.00189 | -0.07052 | 122.910 | 0.94390 | 0.98658 |
| helY; ATP-dependent RNA helicase HelY [EC:3.6.4.-] | 0.00566 | 0.00069 | 0.00579 | 0.00008 | 0.00114 | 65 | 0.00557 | 0.00007 | 0.00087 | 93 | 0.00144 | 0.15167 | 130.253 | 0.87968 | 0.96779 |
| allA; ureidoglycolate lyase [EC:4.3.2.3] | 0.00566 | 0.00072 | 0.00501 | 0.00008 | 0.00110 | 65 | 0.00611 | 0.00008 | 0.00095 | 93 | 0.00145 | -0.75843 | 141.123 | 0.44946 | 0.83438 |
| ctpC; manganese/zinc-transporting P-type ATPase C [EC:3.6.3.-] | 0.00566 | 0.00104 | 0.00846 | 0.00029 | 0.00210 | 65 | 0.00370 | 0.00008 | 0.00096 | 93 | 0.00230 | 2.06551 | 90.635 | 0.04173 | 0.64661 |
| K07394; SM-20-related protein | 0.00566 | 0.00075 | 0.00591 | 0.00010 | 0.00121 | 65 | 0.00548 | 0.00009 | 0.00096 | 93 | 0.00155 | 0.27856 | 133.197 | 0.78101 | 0.94296 |
| E3.5.99.7; 1-aminocyclopropane-1-carboxylate deaminase [EC:3.5.99.7] | 0.00566 | 0.00075 | 0.00607 | 0.00010 | 0.00124 | 65 | 0.00537 | 0.00008 | 0.00095 | 93 | 0.00156 | 0.44821 | 129.700 | 0.65475 | 0.90679 |
| hya; hyaluronoglucosaminidase [EC:3.2.1.35] | 0.00565 | 0.00089 | 0.00566 | 0.00020 | 0.00176 | 65 | 0.00564 | 0.00008 | 0.00090 | 93 | 0.00198 | 0.01268 | 97.284 | 0.98991 | 0.99801 |
| K07182; CBS domain-containing protein | 0.00564 | 0.00077 | 0.00575 | 0.00010 | 0.00123 | 65 | 0.00557 | 0.00009 | 0.00098 | 93 | 0.00158 | 0.11357 | 133.773 | 0.90975 | 0.97664 |
| thyX, thy1; thymidylate synthase (FAD) [EC:2.1.1.148] | 0.00563 | 0.00080 | 0.00525 | 0.00009 | 0.00115 | 65 | 0.00590 | 0.00011 | 0.00110 | 93 | 0.00160 | -0.40636 | 148.335 | 0.68506 | 0.91533 |
| K07216; hemerythrin | 0.00563 | 0.00070 | 0.00467 | 0.00007 | 0.00100 | 65 | 0.00631 | 0.00008 | 0.00095 | 93 | 0.00138 | -1.18801 | 147.983 | 0.23673 | 0.77376 |
| spuC; putrescine—pyruvate transaminase [EC:2.6.1.113] | 0.00563 | 0.00091 | 0.00601 | 0.00013 | 0.00142 | 65 | 0.00535 | 0.00013 | 0.00120 | 93 | 0.00186 | 0.35530 | 138.456 | 0.72291 | 0.92156 |
| yhbS; putative acetyltransferase [EC:2.3.1.-] | 0.00562 | 0.00076 | 0.00458 | 0.00007 | 0.00102 | 65 | 0.00635 | 0.00011 | 0.00108 | 93 | 0.00148 | -1.20214 | 153.628 | 0.23116 | 0.77376 |
| E3.5.5.1; nitrilase [EC:3.5.5.1] | 0.00562 | 0.00067 | 0.00489 | 0.00005 | 0.00087 | 65 | 0.00613 | 0.00009 | 0.00096 | 93 | 0.00130 | -0.95681 | 154.738 | 0.34016 | 0.78467 |
| garD; galactarate dehydratase [EC:4.2.1.42] | 0.00561 | 0.00075 | 0.00522 | 0.00008 | 0.00113 | 65 | 0.00589 | 0.00010 | 0.00101 | 93 | 0.00152 | -0.44056 | 143.691 | 0.66019 | 0.90939 |
| cbiM; cobalt/nickel transport system permease protein | 0.00561 | 0.00068 | 0.00594 | 0.00009 | 0.00118 | 65 | 0.00539 | 0.00006 | 0.00081 | 93 | 0.00143 | 0.38562 | 119.680 | 0.70046 | 0.91953 |
| cysS1; cysteinyl-tRNA synthetase, unknown class [EC:6.1.1.16] | 0.00559 | 0.00152 | 0.00947 | 0.00071 | 0.00330 | 65 | 0.00288 | 0.00012 | 0.00111 | 93 | 0.00348 | 1.89329 | 78.701 | 0.06200 | 0.73001 |
| K09009; uncharacterized protein | 0.00559 | 0.00070 | 0.00574 | 0.00008 | 0.00114 | 65 | 0.00548 | 0.00007 | 0.00088 | 93 | 0.00144 | 0.17675 | 130.497 | 0.85998 | 0.96137 |
| K11905; type VI secretion system protein | 0.00559 | 0.00070 | 0.00682 | 0.00009 | 0.00116 | 65 | 0.00472 | 0.00007 | 0.00086 | 93 | 0.00144 | 1.46142 | 126.956 | 0.14637 | 0.77376 |
| nhoA; N-hydroxyarylamine O-acetyltransferase [EC:2.3.1.118] | 0.00556 | 0.00080 | 0.00432 | 0.00008 | 0.00110 | 65 | 0.00643 | 0.00012 | 0.00111 | 93 | 0.00157 | -1.35039 | 151.716 | 0.17890 | 0.77376 |
| K10253; DOPA 4,5-dioxygenase [EC:1.14.99.-] | 0.00554 | 0.00078 | 0.00590 | 0.00011 | 0.00130 | 65 | 0.00529 | 0.00009 | 0.00096 | 93 | 0.00162 | 0.37322 | 126.935 | 0.70961 | 0.92004 |
| mtr; tryptophan-specific transport protein | 0.00551 | 0.00075 | 0.00635 | 0.00010 | 0.00125 | 65 | 0.00493 | 0.00008 | 0.00092 | 93 | 0.00155 | 0.91434 | 125.828 | 0.36229 | 0.79496 |
| hpaH; 2-oxo-hept-3-ene-1,7-dioate hydratase [EC:4.2.1.-] | 0.00551 | 0.00116 | 0.00591 | 0.00034 | 0.00227 | 65 | 0.00522 | 0.00013 | 0.00118 | 93 | 0.00256 | 0.26998 | 98.166 | 0.78774 | 0.94543 |
| icmF; isobutyryl-CoA mutase [EC:5.4.99.13] | 0.00549 | 0.00062 | 0.00421 | 0.00003 | 0.00063 | 65 | 0.00639 | 0.00008 | 0.00095 | 93 | 0.00114 | -1.91928 | 149.215 | 0.05686 | 0.70711 |
| uspE; universal stress protein E | 0.00549 | 0.00091 | 0.00641 | 0.00011 | 0.00131 | 65 | 0.00484 | 0.00014 | 0.00124 | 93 | 0.00180 | 0.86967 | 147.357 | 0.38590 | 0.80258 |
| napA; periplasmic nitrate reductase NapA [EC:1.7.99.-] | 0.00547 | 0.00064 | 0.00619 | 0.00007 | 0.00107 | 65 | 0.00496 | 0.00006 | 0.00079 | 93 | 0.00133 | 0.92321 | 127.078 | 0.35765 | 0.79461 |
| troD, mntD, znuB; manganese/zinc/iron transport system permease protein | 0.00547 | 0.00094 | 0.00770 | 0.00023 | 0.00187 | 65 | 0.00390 | 0.00008 | 0.00091 | 93 | 0.00207 | 1.83177 | 94.142 | 0.07015 | 0.73522 |
| ATPVG, ahaH, atpH; V/A-type H+/Na+-transporting ATPase subunit G/H | 0.00547 | 0.00100 | 0.00843 | 0.00026 | 0.00200 | 65 | 0.00340 | 0.00008 | 0.00092 | 93 | 0.00220 | 2.28541 | 91.245 | 0.02461 | 0.59764 |
| napD; periplasmic nitrate reductase NapD | 0.00547 | 0.00064 | 0.00619 | 0.00007 | 0.00107 | 65 | 0.00496 | 0.00006 | 0.00079 | 93 | 0.00133 | 0.92793 | 127.063 | 0.35520 | 0.79399 |
| nadR; HTH-type transcriptional regulator, transcriptional repressor of NAD biosynthesis genes [EC:2.7.7.1 2.7.1.22] | 0.00546 | 0.00069 | 0.00660 | 0.00009 | 0.00118 | 65 | 0.00466 | 0.00006 | 0.00083 | 93 | 0.00144 | 1.34520 | 122.355 | 0.18105 | 0.77376 |
| mpa; proteasome-associated ATPase | 0.00546 | 0.00069 | 0.00549 | 0.00008 | 0.00111 | 65 | 0.00544 | 0.00007 | 0.00088 | 93 | 0.00142 | 0.02930 | 133.538 | 0.97667 | 0.99597 |
| pqqC; pyrroloquinoline-quinone synthase [EC:1.3.3.11] | 0.00546 | 0.00075 | 0.00552 | 0.00010 | 0.00121 | 65 | 0.00542 | 0.00008 | 0.00095 | 93 | 0.00154 | 0.06085 | 131.683 | 0.95157 | 0.98792 |
| alkB1_2, alkM; alkane 1-monooxygenase [EC:1.14.15.3] | 0.00546 | 0.00121 | 0.00387 | 0.00005 | 0.00089 | 65 | 0.00657 | 0.00036 | 0.00196 | 93 | 0.00215 | -1.25267 | 125.807 | 0.21265 | 0.77376 |
| epmC; elongation factor P hydroxylase [EC:1.14.-.-] | 0.00546 | 0.00069 | 0.00579 | 0.00007 | 0.00104 | 65 | 0.00523 | 0.00008 | 0.00092 | 93 | 0.00139 | 0.40287 | 142.784 | 0.68765 | 0.91567 |
| spo0A; two-component system, response regulator, stage 0 sporulation protein A | 0.00544 | 0.00073 | 0.00340 | 0.00002 | 0.00061 | 65 | 0.00687 | 0.00012 | 0.00115 | 93 | 0.00130 | -2.67045 | 136.090 | 0.00850 | 0.57196 |
| pqqD; pyrroloquinoline quinone biosynthesis protein D | 0.00544 | 0.00076 | 0.00586 | 0.00011 | 0.00131 | 65 | 0.00515 | 0.00008 | 0.00092 | 93 | 0.00160 | 0.44569 | 122.033 | 0.65661 | 0.90679 |
| wspE; two-component system, chemotaxis family, sensor histidine kinase and response regulator WspE | 0.00544 | 0.00104 | 0.00584 | 0.00015 | 0.00154 | 65 | 0.00515 | 0.00018 | 0.00141 | 93 | 0.00209 | 0.33172 | 145.136 | 0.74058 | 0.92979 |
| E3.2.1.25, MANBA, manB; beta-mannosidase [EC:3.2.1.25] | 0.00543 | 0.00072 | 0.00414 | 0.00007 | 0.00106 | 65 | 0.00633 | 0.00009 | 0.00097 | 93 | 0.00144 | -1.52218 | 145.042 | 0.13014 | 0.77376 |
| K06893; uncharacterized protein | 0.00543 | 0.00070 | 0.00552 | 0.00008 | 0.00108 | 65 | 0.00536 | 0.00008 | 0.00093 | 93 | 0.00142 | 0.10725 | 140.614 | 0.91474 | 0.97664 |
| epd; D-erythrose 4-phosphate dehydrogenase [EC:1.2.1.72] | 0.00542 | 0.00072 | 0.00577 | 0.00009 | 0.00115 | 65 | 0.00518 | 0.00008 | 0.00092 | 93 | 0.00148 | 0.39690 | 133.995 | 0.69207 | 0.91676 |
| kamA; lysine 2,3-aminomutase [EC:5.4.3.2] | 0.00542 | 0.00087 | 0.00620 | 0.00019 | 0.00169 | 65 | 0.00487 | 0.00008 | 0.00090 | 93 | 0.00191 | 0.69143 | 99.970 | 0.49090 | 0.84797 |
| nifH; nitrogenase iron protein NifH [EC:1.18.6.1] | 0.00541 | 0.00094 | 0.00763 | 0.00024 | 0.00194 | 65 | 0.00386 | 0.00006 | 0.00081 | 93 | 0.00210 | 1.79230 | 86.673 | 0.07657 | 0.75063 |
| flrC, fleR; two-component system, response regulator FlrC | 0.00541 | 0.00089 | 0.00578 | 0.00012 | 0.00135 | 65 | 0.00514 | 0.00013 | 0.00119 | 93 | 0.00180 | 0.35476 | 142.441 | 0.72329 | 0.92156 |
| pup; prokaryotic ubiquitin-like protein Pup | 0.00540 | 0.00069 | 0.00540 | 0.00008 | 0.00111 | 65 | 0.00541 | 0.00007 | 0.00088 | 93 | 0.00141 | -0.00723 | 133.521 | 0.99424 | 0.99919 |
| smp; membrane protein | 0.00539 | 0.00078 | 0.00609 | 0.00010 | 0.00123 | 65 | 0.00490 | 0.00010 | 0.00101 | 93 | 0.00160 | 0.74563 | 136.363 | 0.45717 | 0.83438 |
| bcsA; cellulose synthase (UDP-forming) [EC:2.4.1.12] | 0.00538 | 0.00071 | 0.00499 | 0.00005 | 0.00091 | 65 | 0.00565 | 0.00010 | 0.00102 | 93 | 0.00137 | -0.48019 | 155.287 | 0.63177 | 0.89609 |
| pgpB; phosphatidylglycerophosphatase B [EC:3.1.3.27 3.1.3.81 3.1.3.4 3.6.1.27] | 0.00537 | 0.00068 | 0.00554 | 0.00007 | 0.00101 | 65 | 0.00526 | 0.00008 | 0.00092 | 93 | 0.00136 | 0.21024 | 144.513 | 0.83378 | 0.95506 |
| CHAC, chaC; glutathione-specific gamma-glutamylcyclotransferase [EC:4.3.2.7] | 0.00537 | 0.00073 | 0.00401 | 0.00004 | 0.00079 | 65 | 0.00632 | 0.00011 | 0.00110 | 93 | 0.00136 | -1.70633 | 153.171 | 0.08997 | 0.76567 |
| puuC, aldH; 4-(gamma-glutamylamino)butanal dehydrogenase [EC:1.2.1.99] | 0.00537 | 0.00072 | 0.00502 | 0.00008 | 0.00110 | 65 | 0.00561 | 0.00008 | 0.00095 | 93 | 0.00146 | -0.40358 | 140.611 | 0.68713 | 0.91567 |
| K09909; uncharacterized protein | 0.00536 | 0.00069 | 0.00574 | 0.00007 | 0.00106 | 65 | 0.00510 | 0.00008 | 0.00092 | 93 | 0.00140 | 0.46008 | 141.146 | 0.64617 | 0.90314 |
| K09955; uncharacterized protein | 0.00536 | 0.00114 | 0.00250 | 0.00002 | 0.00051 | 65 | 0.00737 | 0.00033 | 0.00188 | 93 | 0.00195 | -2.50013 | 105.254 | 0.01396 | 0.59095 |
| nrtA, nasF, cynA; nitrate/nitrite transport system substrate-binding protein | 0.00536 | 0.00067 | 0.00514 | 0.00008 | 0.00109 | 65 | 0.00552 | 0.00007 | 0.00085 | 93 | 0.00138 | -0.27150 | 130.971 | 0.78643 | 0.94458 |
| higA; HTH-type transcriptional regulator / antitoxin HigA | 0.00536 | 0.00070 | 0.00454 | 0.00004 | 0.00082 | 65 | 0.00592 | 0.00010 | 0.00104 | 93 | 0.00132 | -1.04500 | 155.590 | 0.29764 | 0.77376 |
| K06860; putative heme uptake system protein | 0.00535 | 0.00076 | 0.00540 | 0.00009 | 0.00116 | 65 | 0.00531 | 0.00009 | 0.00101 | 93 | 0.00153 | 0.05765 | 141.288 | 0.95411 | 0.98792 |
| napB; cytochrome c-type protein NapB | 0.00534 | 0.00064 | 0.00619 | 0.00007 | 0.00107 | 65 | 0.00475 | 0.00006 | 0.00078 | 93 | 0.00133 | 1.09309 | 125.599 | 0.27645 | 0.77376 |
| PRODH; proline dehydrogenase [EC:1.5.-.-] | 0.00533 | 0.00089 | 0.00331 | 0.00005 | 0.00086 | 65 | 0.00675 | 0.00017 | 0.00137 | 93 | 0.00162 | -2.13114 | 145.971 | 0.03475 | 0.62130 |
| cat2, abfT; 4-hydroxybutyrate CoA-transferase [EC:2.8.3.-] | 0.00533 | 0.00095 | 0.00691 | 0.00020 | 0.00175 | 65 | 0.00423 | 0.00010 | 0.00104 | 93 | 0.00203 | 1.31767 | 107.787 | 0.19041 | 0.77376 |
| preA; dihydropyrimidine dehydrogenase (NAD+) subunit PreA [EC:1.3.1.1] | 0.00533 | 0.00072 | 0.00447 | 0.00007 | 0.00103 | 65 | 0.00593 | 0.00009 | 0.00099 | 93 | 0.00143 | -1.01920 | 148.250 | 0.30977 | 0.77376 |
| ABC.MN.P; manganese/iron transport system permease protein | 0.00531 | 0.00076 | 0.00569 | 0.00009 | 0.00120 | 65 | 0.00505 | 0.00009 | 0.00098 | 93 | 0.00155 | 0.41436 | 135.702 | 0.67927 | 0.91311 |
| mbtH, nocI; MbtH protein | 0.00531 | 0.00082 | 0.00526 | 0.00010 | 0.00125 | 65 | 0.00535 | 0.00011 | 0.00110 | 93 | 0.00166 | -0.05059 | 141.964 | 0.95972 | 0.98903 |
| phaZ; poly(3-hydroxybutyrate) depolymerase [EC:3.1.1.75] | 0.00531 | 0.00075 | 0.00431 | 0.00008 | 0.00113 | 65 | 0.00600 | 0.00009 | 0.00099 | 93 | 0.00150 | -1.12631 | 141.270 | 0.26194 | 0.77376 |
| K18333; L-fucose dehydrogenase | 0.00530 | 0.00068 | 0.00566 | 0.00010 | 0.00123 | 65 | 0.00505 | 0.00006 | 0.00078 | 93 | 0.00145 | 0.41814 | 112.908 | 0.67664 | 0.91266 |
| benK; MFS transporter, AAHS family, benzoate transport protein | 0.00530 | 0.00085 | 0.00525 | 0.00010 | 0.00124 | 65 | 0.00534 | 0.00012 | 0.00115 | 93 | 0.00170 | -0.05166 | 146.148 | 0.95887 | 0.98903 |
| mtrB; two-component system, OmpR family, sensor histidine kinase MtrB [EC:2.7.13.3] | 0.00530 | 0.00074 | 0.00519 | 0.00008 | 0.00111 | 65 | 0.00538 | 0.00009 | 0.00099 | 93 | 0.00149 | -0.12808 | 143.303 | 0.89827 | 0.97377 |
| napC; cytochrome c-type protein NapC | 0.00530 | 0.00065 | 0.00587 | 0.00008 | 0.00109 | 65 | 0.00490 | 0.00006 | 0.00081 | 93 | 0.00135 | 0.71269 | 127.309 | 0.47734 | 0.84209 |
| virB8, lvhB8; type IV secretion system protein VirB8 | 0.00528 | 0.00085 | 0.00527 | 0.00015 | 0.00152 | 65 | 0.00529 | 0.00009 | 0.00099 | 93 | 0.00181 | -0.01423 | 115.857 | 0.98867 | 0.99801 |
| aidA-I, misL; autotransporter family porin | 0.00528 | 0.00118 | 0.00503 | 0.00024 | 0.00191 | 65 | 0.00546 | 0.00021 | 0.00149 | 93 | 0.00242 | -0.17601 | 131.988 | 0.86055 | 0.96169 |
| cbpM; chaperone modulatory protein CbpM | 0.00528 | 0.00060 | 0.00538 | 0.00005 | 0.00091 | 65 | 0.00521 | 0.00006 | 0.00080 | 93 | 0.00121 | 0.13849 | 142.691 | 0.89005 | 0.97049 |
| yfiP; DTW domain-containing protein | 0.00527 | 0.00072 | 0.00555 | 0.00008 | 0.00113 | 65 | 0.00508 | 0.00008 | 0.00093 | 93 | 0.00147 | 0.32204 | 136.468 | 0.74792 | 0.93235 |
| glcE; glycolate oxidase FAD binding subunit | 0.00527 | 0.00069 | 0.00486 | 0.00008 | 0.00110 | 65 | 0.00556 | 0.00007 | 0.00089 | 93 | 0.00141 | -0.49419 | 135.327 | 0.62197 | 0.89150 |
| fruR1, fruR; LacI family transcriptional regulator, fructose operon transcriptional repressor | 0.00526 | 0.00071 | 0.00575 | 0.00008 | 0.00111 | 65 | 0.00492 | 0.00008 | 0.00092 | 93 | 0.00144 | 0.58065 | 137.426 | 0.56243 | 0.86813 |
| nfnB, nfsB; nitroreductase / dihydropteridine reductase [EC:1.-.-.- 1.5.1.34] | 0.00526 | 0.00061 | 0.00568 | 0.00006 | 0.00094 | 65 | 0.00496 | 0.00006 | 0.00080 | 93 | 0.00123 | 0.57979 | 139.422 | 0.56299 | 0.86813 |
| flgN; flagella synthesis protein FlgN | 0.00526 | 0.00075 | 0.00581 | 0.00009 | 0.00116 | 65 | 0.00487 | 0.00009 | 0.00099 | 93 | 0.00153 | 0.61286 | 139.421 | 0.54097 | 0.86207 |
| hcaC; 3-phenylpropionate/trans-cinnamate dioxygenase ferredoxin component | 0.00526 | 0.00073 | 0.00418 | 0.00007 | 0.00105 | 65 | 0.00601 | 0.00009 | 0.00100 | 93 | 0.00145 | -1.26483 | 147.652 | 0.20792 | 0.77376 |
| E2.7.1.20, ADK; adenosine kinase [EC:2.7.1.20] | 0.00525 | 0.00075 | 0.00434 | 0.00007 | 0.00106 | 65 | 0.00588 | 0.00010 | 0.00103 | 93 | 0.00148 | -1.04603 | 149.111 | 0.29724 | 0.77376 |
| pcaF; 3-oxoadipyl-CoA thiolase [EC:2.3.1.174] | 0.00524 | 0.00088 | 0.00542 | 0.00012 | 0.00134 | 65 | 0.00512 | 0.00013 | 0.00118 | 93 | 0.00178 | 0.16948 | 141.991 | 0.86566 | 0.96270 |
| lapA; lipopolysaccharide assembly protein A | 0.00524 | 0.00068 | 0.00564 | 0.00007 | 0.00103 | 65 | 0.00496 | 0.00008 | 0.00090 | 93 | 0.00137 | 0.49367 | 141.211 | 0.62231 | 0.89150 |
| narL; two-component system, NarL family, nitrate/nitrite response regulator NarL | 0.00524 | 0.00067 | 0.00408 | 0.00003 | 0.00073 | 65 | 0.00605 | 0.00010 | 0.00101 | 93 | 0.00125 | -1.57914 | 153.235 | 0.11636 | 0.77376 |
| cueR; MerR family transcriptional regulator, copper efflux regulator | 0.00524 | 0.00070 | 0.00552 | 0.00008 | 0.00111 | 65 | 0.00504 | 0.00008 | 0.00091 | 93 | 0.00144 | 0.33811 | 135.516 | 0.73580 | 0.92721 |
| gluB; glutamate transport system substrate-binding protein | 0.00522 | 0.00069 | 0.00519 | 0.00008 | 0.00112 | 65 | 0.00525 | 0.00007 | 0.00087 | 93 | 0.00142 | -0.04217 | 130.832 | 0.96643 | 0.99141 |
| citT; citrate:succinate antiporter | 0.00522 | 0.00068 | 0.00510 | 0.00007 | 0.00106 | 65 | 0.00531 | 0.00008 | 0.00090 | 93 | 0.00139 | -0.15659 | 139.181 | 0.87580 | 0.96703 |
| CRLS; cardiolipin synthase (CMP-forming) [EC:2.7.8.41] | 0.00522 | 0.00063 | 0.00345 | 0.00003 | 0.00063 | 65 | 0.00645 | 0.00008 | 0.00095 | 93 | 0.00114 | -2.63065 | 149.376 | 0.00942 | 0.58112 |
| mhqR; MarR family transcriptional regulator, 2-MHQ and catechol-resistance regulon repressor | 0.00521 | 0.00077 | 0.00311 | 0.00003 | 0.00069 | 65 | 0.00667 | 0.00013 | 0.00119 | 93 | 0.00138 | -2.58936 | 141.127 | 0.01062 | 0.58284 |
| acuI; acrylyl-CoA reductase (NADPH) [EC:1.3.1.-] | 0.00521 | 0.00071 | 0.00486 | 0.00008 | 0.00108 | 65 | 0.00545 | 0.00008 | 0.00094 | 93 | 0.00143 | -0.41520 | 141.643 | 0.67862 | 0.91311 |
| rbcL; ribulose-bisphosphate carboxylase large chain [EC:4.1.1.39] | 0.00520 | 0.00061 | 0.00523 | 0.00007 | 0.00105 | 65 | 0.00518 | 0.00005 | 0.00075 | 93 | 0.00129 | 0.04035 | 123.311 | 0.96788 | 0.99231 |
| fdhA; glutathione-independent formaldehyde dehydrogenase [EC:1.2.1.46] | 0.00520 | 0.00089 | 0.00567 | 0.00013 | 0.00140 | 65 | 0.00488 | 0.00012 | 0.00115 | 93 | 0.00181 | 0.43684 | 136.141 | 0.66292 | 0.91006 |
| lacA; galactoside O-acetyltransferase [EC:2.3.1.18] | 0.00520 | 0.00065 | 0.00487 | 0.00007 | 0.00105 | 65 | 0.00543 | 0.00006 | 0.00082 | 93 | 0.00133 | -0.42100 | 132.387 | 0.67444 | 0.91196 |
| K07020; uncharacterized protein | 0.00520 | 0.00072 | 0.00548 | 0.00009 | 0.00119 | 65 | 0.00500 | 0.00008 | 0.00090 | 93 | 0.00149 | 0.32471 | 129.097 | 0.74593 | 0.93202 |
| qrtT; energy-coupling factor transport system substrate-specific component | 0.00520 | 0.00063 | 0.00566 | 0.00005 | 0.00085 | 65 | 0.00487 | 0.00008 | 0.00090 | 93 | 0.00124 | 0.64203 | 153.562 | 0.52181 | 0.85617 |
| slmA, ttk; TetR/AcrR family transcriptional regulator | 0.00520 | 0.00067 | 0.00537 | 0.00007 | 0.00103 | 65 | 0.00508 | 0.00007 | 0.00089 | 93 | 0.00136 | 0.21184 | 139.902 | 0.83254 | 0.95506 |
| gudP; MFS transporter, ACS family, glucarate transporter | 0.00518 | 0.00067 | 0.00539 | 0.00008 | 0.00111 | 65 | 0.00503 | 0.00006 | 0.00083 | 93 | 0.00139 | 0.25948 | 127.408 | 0.79569 | 0.94791 |
| mtnE, mtnV; aminotransferase [EC:2.6.1.-] | 0.00518 | 0.00082 | 0.00498 | 0.00011 | 0.00129 | 65 | 0.00532 | 0.00011 | 0.00107 | 93 | 0.00168 | -0.20341 | 137.751 | 0.83911 | 0.95506 |
| rocD, OAT; ornithine–oxo-acid transaminase [EC:2.6.1.13] | 0.00518 | 0.00072 | 0.00306 | 0.00003 | 0.00068 | 65 | 0.00666 | 0.00011 | 0.00110 | 93 | 0.00130 | -2.77063 | 145.297 | 0.00633 | 0.54194 |
| gluD; glutamate transport system permease protein | 0.00518 | 0.00068 | 0.00509 | 0.00008 | 0.00111 | 65 | 0.00524 | 0.00007 | 0.00087 | 93 | 0.00141 | -0.10959 | 131.913 | 0.91290 | 0.97664 |
| pycA; pyruvate carboxylase subunit A [EC:6.4.1.1] | 0.00517 | 0.00086 | 0.00568 | 0.00011 | 0.00132 | 65 | 0.00482 | 0.00012 | 0.00114 | 93 | 0.00174 | 0.49722 | 140.592 | 0.61981 | 0.89016 |
| tauC; taurine transport system permease protein | 0.00517 | 0.00062 | 0.00433 | 0.00004 | 0.00082 | 65 | 0.00575 | 0.00007 | 0.00088 | 93 | 0.00120 | -1.19072 | 154.168 | 0.23559 | 0.77376 |
| alkB; DNA oxidative demethylase [EC:1.14.11.33] | 0.00516 | 0.00066 | 0.00410 | 0.00004 | 0.00083 | 65 | 0.00591 | 0.00008 | 0.00095 | 93 | 0.00126 | -1.43417 | 155.705 | 0.15353 | 0.77376 |
| abfA; alpha-N-arabinofuranosidase [EC:3.2.1.55] | 0.00516 | 0.00084 | 0.00291 | 0.00003 | 0.00063 | 65 | 0.00673 | 0.00016 | 0.00133 | 93 | 0.00147 | -2.59266 | 128.890 | 0.01062 | 0.58284 |
| lasT; tRNA/rRNA methyltransferase [EC:2.1.1.-] | 0.00515 | 0.00056 | 0.00384 | 0.00003 | 0.00071 | 65 | 0.00607 | 0.00006 | 0.00080 | 93 | 0.00107 | -2.08573 | 155.437 | 0.03864 | 0.63154 |
| fliW; flagellar assembly factor FliW | 0.00515 | 0.00075 | 0.00408 | 0.00008 | 0.00112 | 65 | 0.00590 | 0.00010 | 0.00101 | 93 | 0.00151 | -1.20158 | 144.101 | 0.23150 | 0.77376 |
| mtrA; two-component system, OmpR family, response regulator MtrA | 0.00515 | 0.00068 | 0.00517 | 0.00008 | 0.00111 | 65 | 0.00513 | 0.00007 | 0.00086 | 93 | 0.00140 | 0.03017 | 130.758 | 0.97598 | 0.99571 |
| pldB; lysophospholipase [EC:3.1.1.5] | 0.00514 | 0.00058 | 0.00376 | 0.00003 | 0.00071 | 65 | 0.00611 | 0.00007 | 0.00084 | 93 | 0.00111 | -2.12215 | 155.976 | 0.03540 | 0.62411 |
| E2.5.1.68; short-chain Z-isoprenyl diphosphate synthase [EC:2.5.1.68] | 0.00513 | 0.00074 | 0.00511 | 0.00008 | 0.00111 | 65 | 0.00515 | 0.00009 | 0.00099 | 93 | 0.00148 | -0.02604 | 143.317 | 0.97927 | 0.99657 |
| gntT; Gnt-I system high-affinity gluconate transporter | 0.00512 | 0.00064 | 0.00546 | 0.00008 | 0.00113 | 65 | 0.00489 | 0.00005 | 0.00076 | 93 | 0.00136 | 0.41479 | 118.251 | 0.67905 | 0.91311 |
| virB6, lvhB6; type IV secretion system protein VirB6 | 0.00512 | 0.00074 | 0.00486 | 0.00011 | 0.00130 | 65 | 0.00530 | 0.00007 | 0.00086 | 93 | 0.00156 | -0.27675 | 117.009 | 0.78246 | 0.94310 |
| wecC; UDP-N-acetyl-D-mannosaminuronic acid dehydrogenase [EC:1.1.1.336] | 0.00512 | 0.00060 | 0.00482 | 0.00005 | 0.00091 | 65 | 0.00532 | 0.00006 | 0.00081 | 93 | 0.00121 | -0.41486 | 142.969 | 0.67887 | 0.91311 |
| aspC; aspartate aminotransferase [EC:2.6.1.1] | 0.00511 | 0.00072 | 0.00580 | 0.00010 | 0.00122 | 65 | 0.00463 | 0.00007 | 0.00089 | 93 | 0.00151 | 0.77779 | 125.424 | 0.43816 | 0.83116 |
| K09974; uncharacterized protein | 0.00511 | 0.00072 | 0.00559 | 0.00011 | 0.00133 | 65 | 0.00478 | 0.00006 | 0.00080 | 93 | 0.00155 | 0.52764 | 108.643 | 0.59883 | 0.88240 |
| K07028; uncharacterized protein | 0.00511 | 0.00072 | 0.00495 | 0.00008 | 0.00113 | 65 | 0.00522 | 0.00008 | 0.00095 | 93 | 0.00147 | -0.18155 | 138.551 | 0.85620 | 0.95933 |
| frdB; fumarate reductase iron-sulfur subunit [EC:1.3.5.4] | 0.00510 | 0.00072 | 0.00527 | 0.00007 | 0.00101 | 65 | 0.00499 | 0.00009 | 0.00100 | 93 | 0.00143 | 0.19285 | 150.585 | 0.84734 | 0.95682 |
| frdC; fumarate reductase subunit C | 0.00510 | 0.00072 | 0.00527 | 0.00007 | 0.00101 | 65 | 0.00499 | 0.00009 | 0.00100 | 93 | 0.00143 | 0.19285 | 150.585 | 0.84734 | 0.95682 |
| thiQ; thiamine transport system ATP-binding protein | 0.00510 | 0.00064 | 0.00510 | 0.00007 | 0.00103 | 65 | 0.00511 | 0.00006 | 0.00081 | 93 | 0.00131 | -0.00707 | 132.080 | 0.99437 | 0.99919 |
| waaG, rfaG; UDP-glucose:(heptosyl)LPS alpha-1,3-glucosyltransferase [EC:2.4.1.-] | 0.00510 | 0.00067 | 0.00580 | 0.00008 | 0.00110 | 65 | 0.00461 | 0.00007 | 0.00085 | 93 | 0.00139 | 0.86073 | 131.473 | 0.39095 | 0.80384 |
| gluA; glutamate transport system ATP-binding protein [EC:3.6.3.-] | 0.00510 | 0.00068 | 0.00498 | 0.00008 | 0.00110 | 65 | 0.00517 | 0.00007 | 0.00086 | 93 | 0.00140 | -0.13558 | 132.184 | 0.89236 | 0.97147 |
| K09948; uncharacterized protein | 0.00509 | 0.00071 | 0.00505 | 0.00008 | 0.00111 | 65 | 0.00512 | 0.00008 | 0.00093 | 93 | 0.00145 | -0.04712 | 138.125 | 0.96249 | 0.99023 |
| K09937; uncharacterized protein | 0.00509 | 0.00070 | 0.00487 | 0.00008 | 0.00114 | 65 | 0.00523 | 0.00007 | 0.00089 | 93 | 0.00145 | -0.24866 | 131.910 | 0.80401 | 0.94931 |
| aotJ; arginine/ornithine transport system substrate-binding protein | 0.00508 | 0.00083 | 0.00535 | 0.00011 | 0.00128 | 65 | 0.00490 | 0.00011 | 0.00109 | 93 | 0.00168 | 0.26879 | 139.767 | 0.78849 | 0.94583 |
| smg; Smg protein | 0.00507 | 0.00069 | 0.00532 | 0.00009 | 0.00118 | 65 | 0.00490 | 0.00006 | 0.00083 | 93 | 0.00145 | 0.29031 | 122.644 | 0.77207 | 0.94093 |
| regX3; two-component system, OmpR family, response regulator RegX3 | 0.00507 | 0.00067 | 0.00498 | 0.00008 | 0.00108 | 65 | 0.00514 | 0.00007 | 0.00085 | 93 | 0.00137 | -0.11517 | 131.850 | 0.90848 | 0.97664 |
| rihC; non-specific riboncleoside hydrolase [EC:3.2.-.-] | 0.00506 | 0.00113 | 0.00571 | 0.00026 | 0.00202 | 65 | 0.00461 | 0.00016 | 0.00131 | 93 | 0.00240 | 0.45621 | 114.926 | 0.64910 | 0.90350 |
| norR; anaerobic nitric oxide reductase transcription regulator | 0.00505 | 0.00074 | 0.00551 | 0.00009 | 0.00120 | 65 | 0.00474 | 0.00008 | 0.00094 | 93 | 0.00153 | 0.50641 | 132.457 | 0.61341 | 0.88788 |
| pgaB; poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase [EC:3.5.1.-] | 0.00505 | 0.00070 | 0.00444 | 0.00007 | 0.00105 | 65 | 0.00548 | 0.00008 | 0.00094 | 93 | 0.00141 | -0.73608 | 144.048 | 0.46288 | 0.83654 |
| lapA; surface adhesion protein | 0.00505 | 0.00131 | 0.00461 | 0.00019 | 0.00173 | 65 | 0.00536 | 0.00033 | 0.00188 | 93 | 0.00255 | -0.29325 | 154.520 | 0.76972 | 0.93993 |
| yhdR; aspartate aminotransferase [EC:2.6.1.1] | 0.00504 | 0.00084 | 0.00576 | 0.00017 | 0.00164 | 65 | 0.00454 | 0.00007 | 0.00086 | 93 | 0.00185 | 0.65774 | 98.686 | 0.51224 | 0.85122 |
| hemG; menaquinone-dependent protoporphyrinogen oxidase [EC:1.3.5.3] | 0.00504 | 0.00065 | 0.00548 | 0.00008 | 0.00109 | 65 | 0.00473 | 0.00006 | 0.00081 | 93 | 0.00136 | 0.55769 | 126.954 | 0.57804 | 0.87519 |
| hchA; D-lactate dehydratase / protein deglycase [EC:4.2.1.130 3.5.1.124] | 0.00504 | 0.00071 | 0.00421 | 0.00007 | 0.00102 | 65 | 0.00561 | 0.00009 | 0.00098 | 93 | 0.00142 | -0.98731 | 148.297 | 0.32510 | 0.77376 |
| gcl; tartronate-semialdehyde synthase [EC:4.1.1.47] | 0.00504 | 0.00080 | 0.00493 | 0.00010 | 0.00123 | 65 | 0.00511 | 0.00010 | 0.00106 | 93 | 0.00162 | -0.10744 | 140.459 | 0.91460 | 0.97664 |
| quiA; quinate dehydrogenase (quinone) [EC:1.1.5.8] | 0.00503 | 0.00077 | 0.00480 | 0.00007 | 0.00106 | 65 | 0.00520 | 0.00011 | 0.00108 | 93 | 0.00152 | -0.25908 | 151.901 | 0.79592 | 0.94791 |
| gluC; glutamate transport system permease protein | 0.00502 | 0.00068 | 0.00494 | 0.00008 | 0.00110 | 65 | 0.00508 | 0.00007 | 0.00087 | 93 | 0.00140 | -0.09827 | 132.467 | 0.92186 | 0.97905 |
| K09384; uncharacterized protein | 0.00501 | 0.00110 | 0.00573 | 0.00026 | 0.00199 | 65 | 0.00451 | 0.00015 | 0.00125 | 93 | 0.00235 | 0.52020 | 112.207 | 0.60395 | 0.88434 |
| tam; trans-aconitate 2-methyltransferase [EC:2.1.1.144] | 0.00501 | 0.00064 | 0.00457 | 0.00006 | 0.00099 | 65 | 0.00532 | 0.00007 | 0.00085 | 93 | 0.00130 | -0.57362 | 139.533 | 0.56715 | 0.87063 |
| hscC; molecular chaperone HscC | 0.00501 | 0.00083 | 0.00483 | 0.00007 | 0.00103 | 65 | 0.00513 | 0.00014 | 0.00121 | 93 | 0.00159 | -0.19303 | 155.959 | 0.84719 | 0.95682 |
| hisN; histidinol-phosphatase [EC:3.1.3.15] | 0.00500 | 0.00067 | 0.00519 | 0.00008 | 0.00110 | 65 | 0.00487 | 0.00007 | 0.00085 | 93 | 0.00139 | 0.23441 | 131.297 | 0.81504 | 0.95409 |
| K07387; putative metalloprotease [EC:3.4.24.-] | 0.00500 | 0.00057 | 0.00415 | 0.00002 | 0.00062 | 65 | 0.00559 | 0.00007 | 0.00085 | 93 | 0.00106 | -1.36845 | 153.279 | 0.17317 | 0.77376 |
| utp; urea transporter | 0.00499 | 0.00067 | 0.00438 | 0.00007 | 0.00101 | 65 | 0.00542 | 0.00008 | 0.00091 | 93 | 0.00136 | -0.76694 | 144.072 | 0.44437 | 0.83236 |
| K06976; uncharacterized protein | 0.00499 | 0.00069 | 0.00393 | 0.00007 | 0.00102 | 65 | 0.00574 | 0.00008 | 0.00093 | 93 | 0.00138 | -1.30635 | 144.755 | 0.19350 | 0.77376 |
| E3.1.2.23; 4-hydroxybenzoyl-CoA thioesterase [EC:3.1.2.23] | 0.00498 | 0.00069 | 0.00490 | 0.00008 | 0.00112 | 65 | 0.00504 | 0.00007 | 0.00089 | 93 | 0.00143 | -0.10105 | 133.116 | 0.91966 | 0.97868 |
| iaaA, ASRGL1; beta-aspartyl-peptidase (threonine type) [EC:3.4.19.5] | 0.00498 | 0.00070 | 0.00409 | 0.00008 | 0.00108 | 65 | 0.00560 | 0.00008 | 0.00093 | 93 | 0.00143 | -1.05434 | 139.705 | 0.29355 | 0.77376 |
| nfrA1; FMN reductase (NADPH) [EC:1.5.1.38] | 0.00497 | 0.00064 | 0.00325 | 0.00003 | 0.00064 | 65 | 0.00618 | 0.00009 | 0.00097 | 93 | 0.00116 | -2.52298 | 148.650 | 0.01269 | 0.59095 |
| catC; muconolactone D-isomerase [EC:5.3.3.4] | 0.00497 | 0.00067 | 0.00508 | 0.00008 | 0.00113 | 65 | 0.00489 | 0.00006 | 0.00082 | 93 | 0.00139 | 0.13033 | 125.500 | 0.89652 | 0.97290 |
| pcaJ; 3-oxoadipate CoA-transferase, beta subunit [EC:2.8.3.6] | 0.00497 | 0.00086 | 0.00410 | 0.00005 | 0.00087 | 65 | 0.00558 | 0.00016 | 0.00132 | 93 | 0.00158 | -0.93494 | 149.124 | 0.35133 | 0.79120 |
| wzzB; chain length determinant protein (polysaccharide antigen chain regulator) | 0.00497 | 0.00075 | 0.00510 | 0.00008 | 0.00112 | 65 | 0.00487 | 0.00010 | 0.00102 | 93 | 0.00151 | 0.15446 | 144.792 | 0.87746 | 0.96737 |
| K07396; putative protein-disulfide isomerase | 0.00496 | 0.00075 | 0.00511 | 0.00008 | 0.00114 | 65 | 0.00487 | 0.00009 | 0.00099 | 93 | 0.00151 | 0.15962 | 141.658 | 0.87340 | 0.96576 |
| pobR; AraC family transcriptional regulator, transcriptional activator of pobA | 0.00496 | 0.00071 | 0.00469 | 0.00008 | 0.00109 | 65 | 0.00515 | 0.00008 | 0.00094 | 93 | 0.00144 | -0.32250 | 140.618 | 0.74755 | 0.93207 |
| pduP; propionaldehyde dehydrogenase [EC:1.2.1.87] | 0.00496 | 0.00061 | 0.00574 | 0.00008 | 0.00110 | 65 | 0.00441 | 0.00004 | 0.00069 | 93 | 0.00130 | 1.01999 | 112.665 | 0.30992 | 0.77376 |
| K09941; uncharacterized protein | 0.00495 | 0.00071 | 0.00492 | 0.00009 | 0.00117 | 65 | 0.00497 | 0.00007 | 0.00089 | 93 | 0.00147 | -0.03893 | 129.382 | 0.96901 | 0.99272 |
| pheA1; chorismate mutase [EC:5.4.99.5] | 0.00495 | 0.00072 | 0.00468 | 0.00008 | 0.00112 | 65 | 0.00514 | 0.00008 | 0.00095 | 93 | 0.00147 | -0.31225 | 139.612 | 0.75531 | 0.93665 |
| fucU, FUOM; L-fucose mutarotase [EC:5.1.3.29] | 0.00495 | 0.00059 | 0.00498 | 0.00007 | 0.00104 | 65 | 0.00492 | 0.00004 | 0.00070 | 93 | 0.00125 | 0.04568 | 117.559 | 0.96365 | 0.99101 |
| TC.AAA; ATP:ADP antiporter, AAA family | 0.00495 | 0.00075 | 0.00574 | 0.00011 | 0.00131 | 65 | 0.00439 | 0.00007 | 0.00088 | 93 | 0.00157 | 0.86019 | 118.210 | 0.39143 | 0.80429 |
| kaiC; circadian clock protein KaiC | 0.00494 | 0.00083 | 0.00439 | 0.00008 | 0.00112 | 65 | 0.00533 | 0.00013 | 0.00118 | 93 | 0.00163 | -0.57624 | 153.706 | 0.56529 | 0.87012 |
| crtI; phytoene desaturase [EC:1.3.99.26 1.3.99.28 1.3.99.29 1.3.99.31] | 0.00494 | 0.00090 | 0.00440 | 0.00011 | 0.00130 | 65 | 0.00531 | 0.00014 | 0.00124 | 93 | 0.00179 | -0.51160 | 148.060 | 0.60969 | 0.88693 |
| eutN; ethanolamine utilization protein EutN | 0.00493 | 0.00092 | 0.00664 | 0.00021 | 0.00182 | 65 | 0.00374 | 0.00008 | 0.00090 | 93 | 0.00203 | 1.43104 | 95.394 | 0.15569 | 0.77376 |
| pduE; propanediol dehydratase small subunit [EC:4.2.1.28] | 0.00493 | 0.00062 | 0.00572 | 0.00008 | 0.00110 | 65 | 0.00437 | 0.00005 | 0.00072 | 93 | 0.00131 | 1.02133 | 115.600 | 0.30923 | 0.77376 |
| pduC; propanediol dehydratase large subunit [EC:4.2.1.28] | 0.00493 | 0.00062 | 0.00572 | 0.00008 | 0.00110 | 65 | 0.00437 | 0.00005 | 0.00072 | 93 | 0.00131 | 1.02176 | 115.584 | 0.30903 | 0.77376 |
| pcaI; 3-oxoadipate CoA-transferase, alpha subunit [EC:2.8.3.6] | 0.00492 | 0.00085 | 0.00408 | 0.00005 | 0.00087 | 65 | 0.00551 | 0.00016 | 0.00131 | 93 | 0.00158 | -0.90973 | 149.092 | 0.36443 | 0.79587 |
| uxuB; fructuronate reductase [EC:1.1.1.57] | 0.00492 | 0.00064 | 0.00487 | 0.00010 | 0.00124 | 65 | 0.00496 | 0.00004 | 0.00067 | 93 | 0.00141 | -0.06621 | 101.281 | 0.94734 | 0.98674 |
| PTS-Mtl-EIIA, mtlA, cmtB; PTS system, mannitol-specific IIA component [EC:2.7.1.197] | 0.00491 | 0.00081 | 0.00561 | 0.00018 | 0.00164 | 65 | 0.00443 | 0.00006 | 0.00078 | 93 | 0.00182 | 0.65129 | 92.460 | 0.51648 | 0.85216 |
| E1.1.1.67, mtlK; mannitol 2-dehydrogenase [EC:1.1.1.67] | 0.00491 | 0.00081 | 0.00430 | 0.00008 | 0.00111 | 65 | 0.00534 | 0.00012 | 0.00114 | 93 | 0.00159 | -0.65655 | 152.038 | 0.51246 | 0.85122 |
| vsr; DNA mismatch endonuclease, patch repair protein [EC:3.1.-.-] | 0.00490 | 0.00061 | 0.00415 | 0.00004 | 0.00076 | 65 | 0.00543 | 0.00007 | 0.00089 | 93 | 0.00116 | -1.10313 | 155.925 | 0.27167 | 0.77376 |
| bioM; biotin transport system ATP-binding protein [EC:3.6.3.-] | 0.00490 | 0.00067 | 0.00470 | 0.00008 | 0.00112 | 65 | 0.00504 | 0.00006 | 0.00083 | 93 | 0.00140 | -0.24582 | 127.252 | 0.80622 | 0.94967 |
| senX3; two-component system, OmpR family, sensor histidine kinase SenX3 [EC:2.7.13.3] | 0.00490 | 0.00067 | 0.00485 | 0.00008 | 0.00109 | 65 | 0.00493 | 0.00007 | 0.00085 | 93 | 0.00138 | -0.05763 | 131.404 | 0.95413 | 0.98792 |
| K09705; uncharacterized protein | 0.00489 | 0.00070 | 0.00415 | 0.00008 | 0.00108 | 65 | 0.00541 | 0.00008 | 0.00093 | 93 | 0.00142 | -0.88714 | 140.510 | 0.37652 | 0.79810 |
| phnN; ribose 1,5-bisphosphokinase [EC:2.7.4.23] | 0.00489 | 0.00065 | 0.00449 | 0.00006 | 0.00097 | 65 | 0.00517 | 0.00007 | 0.00088 | 93 | 0.00131 | -0.52603 | 144.455 | 0.59967 | 0.88240 |
| bioW; 6-carboxyhexanoate–CoA ligase [EC:6.2.1.14] | 0.00489 | 0.00061 | 0.00509 | 0.00007 | 0.00105 | 65 | 0.00474 | 0.00005 | 0.00073 | 93 | 0.00128 | 0.27148 | 120.396 | 0.78649 | 0.94458 |
| neuA; N-acylneuraminate cytidylyltransferase [EC:2.7.7.43] | 0.00489 | 0.00066 | 0.00454 | 0.00007 | 0.00106 | 65 | 0.00513 | 0.00007 | 0.00084 | 93 | 0.00135 | -0.44061 | 133.706 | 0.66021 | 0.90939 |
| rstA1; phage replication initiation protein | 0.00488 | 0.00100 | 0.00427 | 0.00006 | 0.00098 | 65 | 0.00531 | 0.00023 | 0.00156 | 93 | 0.00184 | -0.56527 | 146.251 | 0.57276 | 0.87292 |
| polB; DNA polymerase II [EC:2.7.7.7] | 0.00487 | 0.00069 | 0.00549 | 0.00008 | 0.00111 | 65 | 0.00444 | 0.00007 | 0.00089 | 93 | 0.00142 | 0.73651 | 134.563 | 0.46270 | 0.83654 |
| yxjA, nupG; purine nucleoside transport protein | 0.00487 | 0.00086 | 0.00561 | 0.00016 | 0.00159 | 65 | 0.00435 | 0.00008 | 0.00094 | 93 | 0.00185 | 0.68032 | 107.452 | 0.49776 | 0.84853 |
| ttdT; L-tartrate/succinate antiporter | 0.00486 | 0.00061 | 0.00518 | 0.00007 | 0.00105 | 65 | 0.00463 | 0.00005 | 0.00075 | 93 | 0.00129 | 0.42719 | 123.461 | 0.66998 | 0.91184 |
| pduD; propanediol dehydratase medium subunit [EC:4.2.1.28] | 0.00486 | 0.00062 | 0.00572 | 0.00008 | 0.00110 | 65 | 0.00426 | 0.00005 | 0.00071 | 93 | 0.00131 | 1.11708 | 114.183 | 0.26631 | 0.77376 |
| stp1, pppA; serine/threonine protein phosphatase Stp1 [EC:3.1.3.16] | 0.00486 | 0.00102 | 0.00456 | 0.00012 | 0.00136 | 65 | 0.00506 | 0.00020 | 0.00145 | 93 | 0.00199 | -0.25221 | 153.727 | 0.80121 | 0.94855 |
| tauB; taurine transport system ATP-binding protein [EC:3.6.3.36] | 0.00486 | 0.00061 | 0.00424 | 0.00004 | 0.00081 | 65 | 0.00528 | 0.00007 | 0.00087 | 93 | 0.00119 | -0.87541 | 153.882 | 0.38271 | 0.80064 |
| pqqB; pyrroloquinoline quinone biosynthesis protein B | 0.00485 | 0.00069 | 0.00497 | 0.00008 | 0.00110 | 65 | 0.00477 | 0.00007 | 0.00088 | 93 | 0.00141 | 0.14058 | 133.556 | 0.88842 | 0.97039 |
| AACS, acsA; acetoacetyl-CoA synthetase [EC:6.2.1.16] | 0.00484 | 0.00065 | 0.00398 | 0.00007 | 0.00105 | 65 | 0.00545 | 0.00006 | 0.00082 | 93 | 0.00133 | -1.09948 | 131.056 | 0.27357 | 0.77376 |
| catB; muconate cycloisomerase [EC:5.5.1.1] | 0.00484 | 0.00065 | 0.00489 | 0.00007 | 0.00106 | 65 | 0.00481 | 0.00006 | 0.00082 | 93 | 0.00134 | 0.05642 | 130.470 | 0.95509 | 0.98792 |
| aotM; arginine/ornithine transport system permease protein | 0.00483 | 0.00087 | 0.00528 | 0.00011 | 0.00131 | 65 | 0.00452 | 0.00013 | 0.00117 | 93 | 0.00176 | 0.43031 | 142.934 | 0.66762 | 0.91150 |
| ycjF; putative membrane protein | 0.00483 | 0.00063 | 0.00459 | 0.00006 | 0.00098 | 65 | 0.00500 | 0.00006 | 0.00083 | 93 | 0.00129 | -0.32400 | 139.447 | 0.74643 | 0.93202 |
| prnA, rebH, ktzQ; tryptophan 7-halogenase [EC:1.14.19.9] | 0.00483 | 0.00114 | 0.00294 | 0.00008 | 0.00112 | 65 | 0.00615 | 0.00029 | 0.00176 | 93 | 0.00208 | -1.54074 | 147.262 | 0.12553 | 0.77376 |
| K06918; uncharacterized protein | 0.00483 | 0.00063 | 0.00458 | 0.00006 | 0.00098 | 65 | 0.00500 | 0.00006 | 0.00083 | 93 | 0.00128 | -0.33222 | 139.718 | 0.74022 | 0.92969 |
| mepA; penicillin-insensitive murein DD-endopeptidase [EC:3.4.24.-] | 0.00480 | 0.00065 | 0.00438 | 0.00006 | 0.00097 | 65 | 0.00510 | 0.00007 | 0.00088 | 93 | 0.00131 | -0.54553 | 144.253 | 0.58623 | 0.87731 |
| K09891; uncharacterized protein | 0.00480 | 0.00065 | 0.00529 | 0.00007 | 0.00105 | 65 | 0.00446 | 0.00006 | 0.00082 | 93 | 0.00133 | 0.61987 | 132.150 | 0.53641 | 0.86207 |
| priC; primosomal replication protein N’’ | 0.00480 | 0.00065 | 0.00528 | 0.00007 | 0.00105 | 65 | 0.00446 | 0.00006 | 0.00082 | 93 | 0.00133 | 0.61556 | 132.258 | 0.53924 | 0.86207 |
| E3.5.4.32; 8-oxoguanine deaminase [EC:3.5.4.32] | 0.00480 | 0.00100 | 0.00456 | 0.00013 | 0.00143 | 65 | 0.00497 | 0.00018 | 0.00139 | 93 | 0.00199 | -0.20159 | 149.364 | 0.84051 | 0.95506 |
| mtnK; 5-methylthioribose kinase [EC:2.7.1.100] | 0.00478 | 0.00085 | 0.00655 | 0.00021 | 0.00179 | 65 | 0.00354 | 0.00005 | 0.00072 | 93 | 0.00193 | 1.55809 | 84.737 | 0.12294 | 0.77376 |
| purR; LacI family transcriptional regulator, purine nucleotide synthesis repressor | 0.00478 | 0.00061 | 0.00483 | 0.00006 | 0.00096 | 65 | 0.00474 | 0.00006 | 0.00080 | 93 | 0.00125 | 0.06473 | 137.344 | 0.94849 | 0.98748 |
| mtnB; methylthioribulose-1-phosphate dehydratase [EC:4.2.1.109] | 0.00478 | 0.00069 | 0.00471 | 0.00009 | 0.00116 | 65 | 0.00483 | 0.00007 | 0.00084 | 93 | 0.00144 | -0.08430 | 124.571 | 0.93295 | 0.98428 |
| K07074; uncharacterized protein | 0.00477 | 0.00098 | 0.00547 | 0.00025 | 0.00198 | 65 | 0.00428 | 0.00008 | 0.00092 | 93 | 0.00218 | 0.54325 | 91.982 | 0.58827 | 0.87732 |
| udh; uronate dehydrogenase [EC:1.1.1.203] | 0.00477 | 0.00076 | 0.00426 | 0.00008 | 0.00110 | 65 | 0.00513 | 0.00010 | 0.00103 | 93 | 0.00151 | -0.57710 | 146.556 | 0.56475 | 0.86988 |
| nhaB; Na+:H+ antiporter, NhaB family | 0.00476 | 0.00063 | 0.00539 | 0.00007 | 0.00102 | 65 | 0.00433 | 0.00006 | 0.00080 | 93 | 0.00129 | 0.82398 | 131.790 | 0.41144 | 0.81492 |
| hydN; electron transport protein HydN | 0.00476 | 0.00061 | 0.00524 | 0.00008 | 0.00110 | 65 | 0.00443 | 0.00005 | 0.00070 | 93 | 0.00131 | 0.61512 | 113.541 | 0.53971 | 0.86207 |
| mexE; membrane fusion protein, multidrug efflux system | 0.00476 | 0.00086 | 0.00519 | 0.00011 | 0.00132 | 65 | 0.00445 | 0.00012 | 0.00114 | 93 | 0.00175 | 0.42006 | 140.731 | 0.67508 | 0.91264 |
| FTCD; glutamate formiminotransferase / formiminotetrahydrofolate cyclodeaminase [EC:2.1.2.5 4.3.1.4] | 0.00475 | 0.00067 | 0.00466 | 0.00006 | 0.00093 | 65 | 0.00482 | 0.00008 | 0.00093 | 93 | 0.00132 | -0.12526 | 150.649 | 0.90049 | 0.97521 |
| rlmF; 23S rRNA (adenine1618-N6)-methyltransferase [EC:2.1.1.181] | 0.00475 | 0.00069 | 0.00553 | 0.00008 | 0.00111 | 65 | 0.00421 | 0.00007 | 0.00087 | 93 | 0.00141 | 0.93009 | 132.924 | 0.35401 | 0.79265 |
| K07501; 3’-5’ exonuclease | 0.00475 | 0.00057 | 0.00456 | 0.00003 | 0.00071 | 65 | 0.00489 | 0.00007 | 0.00084 | 93 | 0.00110 | -0.29597 | 155.977 | 0.76765 | 0.93900 |
| K13652; AraC family transcriptional regulator | 0.00475 | 0.00094 | 0.00288 | 0.00006 | 0.00094 | 65 | 0.00605 | 0.00019 | 0.00144 | 93 | 0.00172 | -1.83742 | 148.663 | 0.06814 | 0.73473 |
| E1.1.1.90; aryl-alcohol dehydrogenase [EC:1.1.1.90] | 0.00474 | 0.00077 | 0.00492 | 0.00011 | 0.00133 | 65 | 0.00462 | 0.00008 | 0.00093 | 93 | 0.00162 | 0.18452 | 121.740 | 0.85391 | 0.95848 |
| K09922; uncharacterized protein | 0.00474 | 0.00062 | 0.00273 | 0.00002 | 0.00051 | 65 | 0.00614 | 0.00009 | 0.00097 | 93 | 0.00110 | -3.10566 | 135.193 | 0.00231 | 0.46257 |
| nikE; nickel transport system ATP-binding protein [EC:3.6.3.24] | 0.00472 | 0.00090 | 0.00626 | 0.00020 | 0.00175 | 65 | 0.00365 | 0.00008 | 0.00091 | 93 | 0.00197 | 1.31950 | 98.094 | 0.19007 | 0.77376 |
| LDHD, dld; D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4] | 0.00472 | 0.00067 | 0.00194 | 0.00002 | 0.00051 | 65 | 0.00666 | 0.00010 | 0.00104 | 93 | 0.00116 | -4.07445 | 131.438 | 0.00008 | 0.43014 |
| nhaR; LysR family transcriptional regulator, transcriptional activator of nhaA | 0.00471 | 0.00071 | 0.00430 | 0.00005 | 0.00088 | 65 | 0.00500 | 0.00010 | 0.00103 | 93 | 0.00136 | -0.51713 | 155.878 | 0.60580 | 0.88527 |
| virB3, lvhB3; type IV secretion system protein VirB3 | 0.00471 | 0.00077 | 0.00395 | 0.00012 | 0.00138 | 65 | 0.00524 | 0.00007 | 0.00088 | 93 | 0.00164 | -0.78611 | 113.483 | 0.43344 | 0.82874 |
| smtA; S-adenosylmethionine-dependent methyltransferase | 0.00471 | 0.00068 | 0.00535 | 0.00008 | 0.00109 | 65 | 0.00427 | 0.00007 | 0.00087 | 93 | 0.00140 | 0.77527 | 134.242 | 0.43954 | 0.83156 |
| E1.1.99.3A; gluconate 2-dehydrogenase alpha chain [EC:1.1.99.3] | 0.00470 | 0.00101 | 0.00427 | 0.00012 | 0.00138 | 65 | 0.00499 | 0.00019 | 0.00142 | 93 | 0.00198 | -0.36225 | 152.337 | 0.71767 | 0.92056 |
| K02478; two-component system, LytTR family, sensor kinase [EC:2.7.13.3] | 0.00469 | 0.00084 | 0.00640 | 0.00019 | 0.00171 | 65 | 0.00350 | 0.00005 | 0.00076 | 93 | 0.00188 | 1.54697 | 89.524 | 0.12540 | 0.77376 |
| fixX; ferredoxin like protein | 0.00469 | 0.00060 | 0.00497 | 0.00007 | 0.00105 | 65 | 0.00449 | 0.00005 | 0.00071 | 93 | 0.00127 | 0.37669 | 118.663 | 0.70707 | 0.92004 |
| cytR; LacI family transcriptional regulator, repressor for deo operon, udp, cdd, tsx, nupC, and nupG | 0.00468 | 0.00066 | 0.00382 | 0.00006 | 0.00099 | 65 | 0.00528 | 0.00007 | 0.00087 | 93 | 0.00132 | -1.10609 | 141.941 | 0.27056 | 0.77376 |
| K07401; selenoprotein W-related protein | 0.00467 | 0.00069 | 0.00483 | 0.00008 | 0.00109 | 65 | 0.00456 | 0.00007 | 0.00089 | 93 | 0.00141 | 0.19166 | 135.389 | 0.84830 | 0.95712 |
| virB2, lvhB2; type IV secretion system protein VirB2 | 0.00466 | 0.00081 | 0.00423 | 0.00016 | 0.00155 | 65 | 0.00497 | 0.00007 | 0.00086 | 93 | 0.00177 | -0.41919 | 102.507 | 0.67595 | 0.91266 |
| deoA, TYMP; thymidine phosphorylase [EC:2.4.2.4] | 0.00465 | 0.00057 | 0.00354 | 0.00003 | 0.00065 | 65 | 0.00543 | 0.00007 | 0.00085 | 93 | 0.00107 | -1.75818 | 154.911 | 0.08069 | 0.75920 |
| pduQ; 1-propanol dehydrogenase | 0.00465 | 0.00061 | 0.00539 | 0.00008 | 0.00110 | 65 | 0.00413 | 0.00005 | 0.00070 | 93 | 0.00130 | 0.96336 | 113.049 | 0.33742 | 0.78354 |
| K09939; uncharacterized protein | 0.00465 | 0.00067 | 0.00410 | 0.00007 | 0.00102 | 65 | 0.00503 | 0.00007 | 0.00090 | 93 | 0.00136 | -0.68244 | 142.127 | 0.49607 | 0.84797 |
| ydfG; 3-hydroxy acid dehydrogenase / malonic semialdehyde reductase [EC:1.1.1.381 1.1.1.-] | 0.00465 | 0.00064 | 0.00522 | 0.00007 | 0.00104 | 65 | 0.00425 | 0.00006 | 0.00082 | 93 | 0.00132 | 0.73956 | 132.647 | 0.46087 | 0.83546 |
| lcdH, cdhA; carnitine 3-dehydrogenase [EC:1.1.1.108] | 0.00465 | 0.00081 | 0.00486 | 0.00010 | 0.00122 | 65 | 0.00450 | 0.00011 | 0.00107 | 93 | 0.00163 | 0.22152 | 141.820 | 0.82501 | 0.95506 |
| iolE; inosose dehydratase [EC:4.2.1.44] | 0.00464 | 0.00063 | 0.00383 | 0.00004 | 0.00079 | 65 | 0.00521 | 0.00008 | 0.00092 | 93 | 0.00121 | -1.13818 | 155.901 | 0.25679 | 0.77376 |
| K07101; uncharacterized protein | 0.00463 | 0.00050 | 0.00468 | 0.00004 | 0.00079 | 65 | 0.00460 | 0.00004 | 0.00065 | 93 | 0.00102 | 0.08230 | 136.477 | 0.93453 | 0.98428 |
| PTS-Gut-EIIC, srlA; PTS system, glucitol/sorbitol-specific IIC component | 0.00463 | 0.00104 | 0.00519 | 0.00026 | 0.00200 | 65 | 0.00424 | 0.00011 | 0.00109 | 93 | 0.00228 | 0.41805 | 101.138 | 0.67680 | 0.91266 |
| bkdA; 2-oxoisovalerate dehydrogenase E1 component [EC:1.2.4.4] | 0.00461 | 0.00067 | 0.00421 | 0.00006 | 0.00099 | 65 | 0.00490 | 0.00008 | 0.00091 | 93 | 0.00134 | -0.50882 | 145.687 | 0.61165 | 0.88722 |
| xtmA; phage terminase small subunit | 0.00461 | 0.00076 | 0.00473 | 0.00011 | 0.00133 | 65 | 0.00453 | 0.00008 | 0.00091 | 93 | 0.00161 | 0.12027 | 119.858 | 0.90447 | 0.97658 |
| pagL; lipid A 3-O-deacylase [EC:3.1.-.-] | 0.00461 | 0.00075 | 0.00540 | 0.00011 | 0.00129 | 65 | 0.00405 | 0.00007 | 0.00090 | 93 | 0.00157 | 0.86292 | 121.149 | 0.38988 | 0.80353 |
| sigB; RNA polymerase sigma-B factor | 0.00460 | 0.00071 | 0.00318 | 0.00003 | 0.00072 | 65 | 0.00559 | 0.00011 | 0.00109 | 93 | 0.00130 | -1.84254 | 148.849 | 0.06739 | 0.73334 |
| pduL; phosphate propanoyltransferase [EC:2.3.1.222] | 0.00460 | 0.00061 | 0.00532 | 0.00008 | 0.00110 | 65 | 0.00409 | 0.00005 | 0.00070 | 93 | 0.00131 | 0.94627 | 112.934 | 0.34603 | 0.78846 |
| splB; spore photoproduct lyase [EC:4.1.99.14] | 0.00459 | 0.00085 | 0.00580 | 0.00018 | 0.00169 | 65 | 0.00374 | 0.00007 | 0.00084 | 93 | 0.00188 | 1.09352 | 95.442 | 0.27692 | 0.77376 |
| tccC; insecticidal toxin complex protein TccC | 0.00458 | 0.00142 | 0.00440 | 0.00027 | 0.00204 | 65 | 0.00471 | 0.00035 | 0.00195 | 93 | 0.00282 | -0.10756 | 148.333 | 0.91449 | 0.97664 |
| ABC.VB12.S1, btuF; vitamin B12 transport system substrate-binding protein | 0.00458 | 0.00068 | 0.00505 | 0.00008 | 0.00109 | 65 | 0.00425 | 0.00007 | 0.00086 | 93 | 0.00139 | 0.57308 | 132.698 | 0.56756 | 0.87063 |
| aphA; acid phosphatase (class B) [EC:3.1.3.2] | 0.00457 | 0.00063 | 0.00526 | 0.00006 | 0.00100 | 65 | 0.00409 | 0.00006 | 0.00081 | 93 | 0.00128 | 0.91361 | 135.824 | 0.36254 | 0.79496 |
| dhcR; LysR family transcriptional regulator, carnitine catabolism transcriptional activator | 0.00457 | 0.00065 | 0.00479 | 0.00007 | 0.00104 | 65 | 0.00442 | 0.00007 | 0.00084 | 93 | 0.00134 | 0.28197 | 135.208 | 0.77840 | 0.94269 |
| mqnE; aminodeoxyfutalosine synthase [EC:2.5.1.120] | 0.00456 | 0.00107 | 0.00511 | 0.00026 | 0.00201 | 65 | 0.00417 | 0.00013 | 0.00117 | 93 | 0.00232 | 0.40262 | 106.185 | 0.68803 | 0.91567 |
| mhpA; 3-(3-hydroxy-phenyl)propionate hydroxylase [EC:1.14.13.127] | 0.00456 | 0.00077 | 0.00268 | 0.00004 | 0.00076 | 65 | 0.00587 | 0.00013 | 0.00118 | 93 | 0.00140 | -2.27496 | 147.572 | 0.02435 | 0.59764 |
| etk-wzc; tyrosine-protein kinase Etk/Wzc [EC:2.7.10.-] | 0.00455 | 0.00060 | 0.00448 | 0.00005 | 0.00091 | 65 | 0.00460 | 0.00006 | 0.00079 | 93 | 0.00120 | -0.10115 | 141.511 | 0.91958 | 0.97868 |
| K07498; putative transposase | 0.00455 | 0.00112 | 0.00227 | 0.00007 | 0.00106 | 65 | 0.00614 | 0.00028 | 0.00173 | 93 | 0.00203 | -1.90567 | 145.033 | 0.05867 | 0.72183 |
| ghrA; glyoxylate/hydroxypyruvatereductase [EC:1.1.1.79 1.1.1.81] | 0.00454 | 0.00060 | 0.00328 | 0.00004 | 0.00077 | 65 | 0.00542 | 0.00007 | 0.00085 | 93 | 0.00115 | -1.86206 | 154.915 | 0.06449 | 0.73334 |
| algJ; alginate O-acetyltransferase complex protein AlgJ | 0.00454 | 0.00082 | 0.00476 | 0.00009 | 0.00118 | 65 | 0.00439 | 0.00012 | 0.00113 | 93 | 0.00163 | 0.22689 | 147.922 | 0.82082 | 0.95506 |
| napG; ferredoxin-type protein NapG | 0.00453 | 0.00064 | 0.00475 | 0.00006 | 0.00099 | 65 | 0.00438 | 0.00006 | 0.00083 | 93 | 0.00130 | 0.27928 | 138.456 | 0.78044 | 0.94277 |
| kauB; 4-guanidinobutyraldehyde dehydrogenase / NAD-dependent aldehyde dehydrogenase [EC:1.2.1.54 1.2.1.-] | 0.00452 | 0.00072 | 0.00507 | 0.00009 | 0.00117 | 65 | 0.00414 | 0.00008 | 0.00090 | 93 | 0.00148 | 0.63526 | 130.890 | 0.52636 | 0.85844 |
| nikA; nickel transport system substrate-binding protein | 0.00450 | 0.00085 | 0.00582 | 0.00019 | 0.00172 | 65 | 0.00357 | 0.00006 | 0.00081 | 93 | 0.00190 | 1.18214 | 92.332 | 0.24019 | 0.77376 |
| ada-alkA; AraC family transcriptional regulator, regulatory protein of adaptative response / DNA-3-methyladenine glycosylase II [EC:3.2.2.21] | 0.00449 | 0.00070 | 0.00384 | 0.00007 | 0.00102 | 65 | 0.00495 | 0.00008 | 0.00095 | 93 | 0.00140 | -0.79414 | 146.537 | 0.42840 | 0.82737 |
| abrB; transcriptional pleiotropic regulator of transition state genes | 0.00449 | 0.00087 | 0.00290 | 0.00006 | 0.00092 | 65 | 0.00561 | 0.00016 | 0.00133 | 93 | 0.00162 | -1.67387 | 151.682 | 0.09622 | 0.76667 |
| K09979; uncharacterized protein | 0.00449 | 0.00059 | 0.00501 | 0.00006 | 0.00092 | 65 | 0.00412 | 0.00006 | 0.00077 | 93 | 0.00120 | 0.73538 | 137.912 | 0.46336 | 0.83677 |
| K06955; uncharacterized protein | 0.00449 | 0.00070 | 0.00436 | 0.00007 | 0.00106 | 65 | 0.00458 | 0.00008 | 0.00094 | 93 | 0.00142 | -0.15638 | 142.269 | 0.87595 | 0.96703 |
| TPMT, tpmT; thiopurine S-methyltransferase [EC:2.1.1.67] | 0.00448 | 0.00068 | 0.00500 | 0.00008 | 0.00112 | 65 | 0.00412 | 0.00007 | 0.00086 | 93 | 0.00141 | 0.62945 | 130.446 | 0.53016 | 0.86094 |
| damX; DamX protein | 0.00447 | 0.00067 | 0.00506 | 0.00008 | 0.00108 | 65 | 0.00406 | 0.00007 | 0.00085 | 93 | 0.00138 | 0.72787 | 132.792 | 0.46797 | 0.83837 |
| napH; ferredoxin-type protein NapH | 0.00446 | 0.00062 | 0.00475 | 0.00006 | 0.00099 | 65 | 0.00425 | 0.00006 | 0.00081 | 93 | 0.00128 | 0.39020 | 135.354 | 0.69700 | 0.91823 |
| ABC.MN.A; manganese/iron transport system ATP-binding protein | 0.00445 | 0.00075 | 0.00480 | 0.00008 | 0.00108 | 65 | 0.00421 | 0.00010 | 0.00103 | 93 | 0.00149 | 0.38971 | 148.369 | 0.69731 | 0.91825 |
| iaaM; tryptophan 2-monooxygenase [EC:1.13.12.3] | 0.00444 | 0.00095 | 0.00403 | 0.00010 | 0.00122 | 65 | 0.00473 | 0.00017 | 0.00137 | 93 | 0.00183 | -0.37958 | 155.345 | 0.70478 | 0.92004 |
| nikB; nickel transport system permease protein | 0.00444 | 0.00085 | 0.00582 | 0.00019 | 0.00172 | 65 | 0.00348 | 0.00006 | 0.00081 | 93 | 0.00190 | 1.22849 | 92.412 | 0.22238 | 0.77376 |
| aotP; arginine/ornithine transport system ATP-binding protein [EC:3.6.3.-] | 0.00444 | 0.00072 | 0.00518 | 0.00009 | 0.00121 | 65 | 0.00393 | 0.00007 | 0.00090 | 93 | 0.00150 | 0.83059 | 127.077 | 0.40776 | 0.81209 |
| thrH; phosphoserine / homoserine phosphotransferase [EC:3.1.3.3 2.7.1.39] | 0.00444 | 0.00055 | 0.00432 | 0.00005 | 0.00090 | 65 | 0.00452 | 0.00005 | 0.00071 | 93 | 0.00114 | -0.17916 | 132.505 | 0.85808 | 0.96050 |
| atzF; allophanate hydrolase [EC:3.5.1.54] | 0.00444 | 0.00064 | 0.00411 | 0.00005 | 0.00091 | 65 | 0.00467 | 0.00007 | 0.00089 | 93 | 0.00127 | -0.43714 | 149.468 | 0.66264 | 0.91006 |
| rsd; regulator of sigma D | 0.00444 | 0.00068 | 0.00510 | 0.00008 | 0.00109 | 65 | 0.00398 | 0.00007 | 0.00086 | 93 | 0.00139 | 0.80620 | 133.406 | 0.42156 | 0.82295 |
| K07034; uncharacterized protein | 0.00443 | 0.00061 | 0.00482 | 0.00006 | 0.00096 | 65 | 0.00416 | 0.00006 | 0.00078 | 93 | 0.00124 | 0.53305 | 136.232 | 0.59487 | 0.88005 |
| xylC; benzaldehyde dehydrogenase (NAD) [EC:1.2.1.28] | 0.00442 | 0.00072 | 0.00381 | 0.00007 | 0.00106 | 65 | 0.00484 | 0.00009 | 0.00097 | 93 | 0.00144 | -0.71665 | 144.837 | 0.47475 | 0.84095 |
| entD; enterobactin synthetase component D [EC:6.3.2.14 2.7.8.-] | 0.00441 | 0.00068 | 0.00468 | 0.00008 | 0.00108 | 65 | 0.00423 | 0.00007 | 0.00088 | 93 | 0.00139 | 0.31910 | 136.370 | 0.75014 | 0.93449 |
| uxaA2; altronate dehydratase large subunit [EC:4.2.1.7] | 0.00441 | 0.00060 | 0.00486 | 0.00007 | 0.00105 | 65 | 0.00410 | 0.00005 | 0.00070 | 93 | 0.00126 | 0.59761 | 117.363 | 0.55125 | 0.86788 |
| K09918; uncharacterized protein | 0.00441 | 0.00068 | 0.00506 | 0.00008 | 0.00109 | 65 | 0.00396 | 0.00007 | 0.00087 | 93 | 0.00139 | 0.79336 | 133.762 | 0.42897 | 0.82749 |
| ABC.GGU.P, gguB; putative multiple sugar transport system permease protein | 0.00441 | 0.00056 | 0.00428 | 0.00007 | 0.00103 | 65 | 0.00450 | 0.00004 | 0.00062 | 93 | 0.00121 | -0.18628 | 109.091 | 0.85257 | 0.95848 |
| gbcA; glycine betaine catabolism A | 0.00440 | 0.00066 | 0.00445 | 0.00007 | 0.00104 | 65 | 0.00437 | 0.00007 | 0.00085 | 93 | 0.00135 | 0.05363 | 135.864 | 0.95731 | 0.98885 |
| lhpA; 4-hydroxyproline epimerase [EC:5.1.1.8] | 0.00440 | 0.00064 | 0.00374 | 0.00004 | 0.00081 | 65 | 0.00487 | 0.00008 | 0.00093 | 93 | 0.00123 | -0.91854 | 155.736 | 0.35975 | 0.79496 |
| pabAB; para-aminobenzoate synthetase [EC:2.6.1.85] | 0.00439 | 0.00066 | 0.00406 | 0.00007 | 0.00107 | 65 | 0.00463 | 0.00006 | 0.00083 | 93 | 0.00135 | -0.42358 | 131.183 | 0.67256 | 0.91184 |
| nikD; nickel transport system ATP-binding protein [EC:3.6.3.24] | 0.00437 | 0.00085 | 0.00589 | 0.00019 | 0.00172 | 65 | 0.00331 | 0.00006 | 0.00080 | 93 | 0.00190 | 1.35738 | 91.694 | 0.17799 | 0.77376 |
| ugpB; sn-glycerol 3-phosphate transport system substrate-binding protein | 0.00436 | 0.00063 | 0.00221 | 0.00002 | 0.00056 | 65 | 0.00586 | 0.00009 | 0.00098 | 93 | 0.00112 | -3.25520 | 140.160 | 0.00142 | 0.46257 |
| cdhR; AraC family transcriptional regulator, carnitine catabolism transcriptional activator | 0.00436 | 0.00080 | 0.00484 | 0.00010 | 0.00122 | 65 | 0.00402 | 0.00010 | 0.00106 | 93 | 0.00162 | 0.50881 | 140.612 | 0.61168 | 0.88722 |
| K07117; uncharacterized protein | 0.00436 | 0.00069 | 0.00358 | 0.00007 | 0.00104 | 65 | 0.00490 | 0.00008 | 0.00092 | 93 | 0.00139 | -0.95261 | 142.149 | 0.34241 | 0.78567 |
| E3.1.3.8; 3-phytase [EC:3.1.3.8] | 0.00435 | 0.00067 | 0.00346 | 0.00007 | 0.00102 | 65 | 0.00498 | 0.00007 | 0.00088 | 93 | 0.00134 | -1.12482 | 140.876 | 0.26258 | 0.77376 |
| rfbF; glucose-1-phosphate cytidylyltransferase [EC:2.7.7.33] | 0.00435 | 0.00057 | 0.00427 | 0.00008 | 0.00114 | 65 | 0.00441 | 0.00003 | 0.00056 | 93 | 0.00127 | -0.11056 | 95.053 | 0.91220 | 0.97664 |
| K09945; uncharacterized protein | 0.00435 | 0.00064 | 0.00475 | 0.00007 | 0.00103 | 65 | 0.00406 | 0.00006 | 0.00081 | 93 | 0.00131 | 0.52709 | 131.954 | 0.59902 | 0.88240 |
| lamB; maltoporin | 0.00434 | 0.00063 | 0.00359 | 0.00005 | 0.00083 | 65 | 0.00486 | 0.00008 | 0.00090 | 93 | 0.00123 | -1.03984 | 154.297 | 0.30004 | 0.77376 |
| antB; anti-repressor protein | 0.00433 | 0.00108 | 0.00423 | 0.00020 | 0.00173 | 65 | 0.00440 | 0.00018 | 0.00138 | 93 | 0.00222 | -0.07395 | 133.751 | 0.94116 | 0.98528 |
| ybfF; esterase [EC:3.1.-.-] | 0.00432 | 0.00062 | 0.00491 | 0.00007 | 0.00101 | 65 | 0.00390 | 0.00006 | 0.00079 | 93 | 0.00128 | 0.78802 | 131.244 | 0.43210 | 0.82814 |
| napF; ferredoxin-type protein NapF | 0.00432 | 0.00061 | 0.00470 | 0.00006 | 0.00099 | 65 | 0.00405 | 0.00006 | 0.00077 | 93 | 0.00126 | 0.52175 | 131.181 | 0.60272 | 0.88431 |
| TC.BCT; betaine/carnitine transporter, BCCT family | 0.00431 | 0.00068 | 0.00457 | 0.00010 | 0.00125 | 65 | 0.00413 | 0.00005 | 0.00077 | 93 | 0.00146 | 0.30172 | 110.737 | 0.76343 | 0.93836 |
| tdh; threonine 3-dehydrogenase [EC:1.1.1.103] | 0.00430 | 0.00066 | 0.00398 | 0.00006 | 0.00099 | 65 | 0.00453 | 0.00007 | 0.00088 | 93 | 0.00133 | -0.42195 | 143.584 | 0.67369 | 0.91184 |
| ampR; LysR family transcriptional regulator, regulator of gene expression of beta-lactamase | 0.00430 | 0.00068 | 0.00335 | 0.00007 | 0.00104 | 65 | 0.00497 | 0.00008 | 0.00090 | 93 | 0.00137 | -1.17823 | 141.003 | 0.24069 | 0.77376 |
| hasD, prtD, aprD; ATP-binding cassette, subfamily C, bacterial exporter for protease/lipase | 0.00429 | 0.00094 | 0.00349 | 0.00009 | 0.00115 | 65 | 0.00485 | 0.00018 | 0.00139 | 93 | 0.00180 | -0.75375 | 155.996 | 0.45214 | 0.83438 |
| K07231; putative iron-regulated protein | 0.00429 | 0.00068 | 0.00461 | 0.00008 | 0.00108 | 65 | 0.00407 | 0.00007 | 0.00087 | 93 | 0.00139 | 0.39337 | 134.521 | 0.69467 | 0.91715 |
| ABC.GGU.S, chvE; putative multiple sugar transport system substrate-binding protein | 0.00429 | 0.00061 | 0.00368 | 0.00006 | 0.00098 | 65 | 0.00471 | 0.00006 | 0.00079 | 93 | 0.00126 | -0.81902 | 135.134 | 0.41422 | 0.81712 |
| K09947; uncharacterized protein | 0.00428 | 0.00067 | 0.00446 | 0.00007 | 0.00107 | 65 | 0.00415 | 0.00007 | 0.00085 | 93 | 0.00137 | 0.22226 | 133.461 | 0.82445 | 0.95506 |
| pit; low-affinity inorganic phosphate transporter | 0.00428 | 0.00063 | 0.00479 | 0.00007 | 0.00100 | 65 | 0.00392 | 0.00006 | 0.00082 | 93 | 0.00130 | 0.66919 | 135.627 | 0.50451 | 0.84956 |
| vanA; vanillate monooxygenase [EC:1.14.13.82] | 0.00428 | 0.00063 | 0.00345 | 0.00004 | 0.00078 | 65 | 0.00485 | 0.00008 | 0.00092 | 93 | 0.00121 | -1.15828 | 155.977 | 0.24852 | 0.77376 |
| feaB; phenylacetaldehyde dehydrogenase [EC:1.2.1.39] | 0.00427 | 0.00074 | 0.00429 | 0.00009 | 0.00115 | 65 | 0.00426 | 0.00009 | 0.00098 | 93 | 0.00151 | 0.02014 | 139.340 | 0.98396 | 0.99706 |
| MAN; mannan endo-1,4-beta-mannosidase [EC:3.2.1.78] | 0.00427 | 0.00052 | 0.00377 | 0.00003 | 0.00066 | 65 | 0.00461 | 0.00005 | 0.00076 | 93 | 0.00100 | -0.83718 | 155.788 | 0.40377 | 0.81034 |
| ompX; outer membrane protein X | 0.00426 | 0.00057 | 0.00391 | 0.00004 | 0.00075 | 65 | 0.00451 | 0.00006 | 0.00082 | 93 | 0.00111 | -0.53327 | 154.910 | 0.59461 | 0.88005 |
| folM; dihydromonapterin reductase / dihydrofolate reductase [EC:1.5.1.50 1.5.1.3] | 0.00426 | 0.00067 | 0.00481 | 0.00008 | 0.00108 | 65 | 0.00387 | 0.00007 | 0.00085 | 93 | 0.00137 | 0.68659 | 132.457 | 0.49354 | 0.84797 |
| sitB; manganese/iron transport system ATP-binding protein | 0.00424 | 0.00061 | 0.00448 | 0.00006 | 0.00098 | 65 | 0.00407 | 0.00006 | 0.00078 | 93 | 0.00125 | 0.32514 | 132.501 | 0.74559 | 0.93199 |
| aefR; TetR/AcrR family transcriptional regulator, regulator of autoinduction and epiphytic fitness | 0.00423 | 0.00069 | 0.00337 | 0.00007 | 0.00101 | 65 | 0.00484 | 0.00008 | 0.00093 | 93 | 0.00137 | -1.07130 | 145.906 | 0.28580 | 0.77376 |
| K07068; uncharacterized protein | 0.00422 | 0.00094 | 0.00235 | 0.00002 | 0.00058 | 65 | 0.00553 | 0.00022 | 0.00153 | 93 | 0.00164 | -1.94229 | 117.197 | 0.05450 | 0.70330 |
| pflX; putative pyruvate formate lyase activating enzyme [EC:1.97.1.4] | 0.00422 | 0.00073 | 0.00446 | 0.00010 | 0.00126 | 65 | 0.00405 | 0.00007 | 0.00088 | 93 | 0.00153 | 0.26277 | 121.631 | 0.79317 | 0.94783 |
| K06953; uncharacterized protein | 0.00422 | 0.00068 | 0.00322 | 0.00007 | 0.00105 | 65 | 0.00492 | 0.00007 | 0.00088 | 93 | 0.00137 | -1.24094 | 138.446 | 0.21673 | 0.77376 |
| ylbA, UGHY; (S)-ureidoglycine aminohydrolase [EC:3.5.3.26] | 0.00421 | 0.00072 | 0.00403 | 0.00007 | 0.00107 | 65 | 0.00433 | 0.00009 | 0.00097 | 93 | 0.00144 | -0.20827 | 144.751 | 0.83531 | 0.95506 |
| narX; two-component system, NarL family, nitrate/nitrite sensor histidine kinase NarX [EC:2.7.13.3] | 0.00420 | 0.00056 | 0.00363 | 0.00003 | 0.00071 | 65 | 0.00460 | 0.00006 | 0.00081 | 93 | 0.00107 | -0.90027 | 155.587 | 0.36937 | 0.79587 |
| uxaB; tagaturonate reductase [EC:1.1.1.58] | 0.00419 | 0.00063 | 0.00391 | 0.00009 | 0.00116 | 65 | 0.00439 | 0.00004 | 0.00069 | 93 | 0.00135 | -0.35881 | 107.389 | 0.72044 | 0.92136 |
| glmS, mutS, mamA; methylaspartate mutase sigma subunit [EC:5.4.99.1] | 0.00419 | 0.00089 | 0.00604 | 0.00020 | 0.00175 | 65 | 0.00289 | 0.00007 | 0.00088 | 93 | 0.00195 | 1.61121 | 96.158 | 0.11041 | 0.77376 |
| sfnG; dimethylsulfone monooxygenase [EC:1.14.14.35] | 0.00419 | 0.00058 | 0.00356 | 0.00003 | 0.00073 | 65 | 0.00463 | 0.00007 | 0.00085 | 93 | 0.00112 | -0.95868 | 155.821 | 0.33920 | 0.78433 |
| erfK; L,D-transpeptidase ErfK/SrfK | 0.00418 | 0.00066 | 0.00469 | 0.00008 | 0.00108 | 65 | 0.00383 | 0.00007 | 0.00084 | 93 | 0.00137 | 0.62992 | 131.351 | 0.52984 | 0.86094 |
| arcB; two-component system, OmpR family, aerobic respiration control sensor histidine kinase ArcB [EC:2.7.13.3] | 0.00417 | 0.00062 | 0.00472 | 0.00007 | 0.00101 | 65 | 0.00379 | 0.00006 | 0.00078 | 93 | 0.00128 | 0.72127 | 131.174 | 0.47203 | 0.84037 |
| impA; type VI secretion system protein ImpA | 0.00417 | 0.00074 | 0.00378 | 0.00006 | 0.00094 | 65 | 0.00444 | 0.00011 | 0.00108 | 93 | 0.00143 | -0.45882 | 155.621 | 0.64701 | 0.90344 |
| flrB, fleS; two-component system, sensor histidine kinase FlrB [EC:2.7.13.3] | 0.00416 | 0.00068 | 0.00474 | 0.00008 | 0.00109 | 65 | 0.00376 | 0.00007 | 0.00086 | 93 | 0.00139 | 0.71012 | 133.330 | 0.47887 | 0.84314 |
| modF; molybdate transport system ATP-binding protein | 0.00416 | 0.00061 | 0.00429 | 0.00006 | 0.00099 | 65 | 0.00407 | 0.00006 | 0.00079 | 93 | 0.00126 | 0.17326 | 134.023 | 0.86271 | 0.96263 |
| argA; amino-acid N-acetyltransferase [EC:2.3.1.1] | 0.00415 | 0.00062 | 0.00381 | 0.00007 | 0.00105 | 65 | 0.00440 | 0.00005 | 0.00076 | 93 | 0.00130 | -0.45213 | 124.267 | 0.65197 | 0.90504 |
| atuG; citronellol/citronellal dehydrogenase | 0.00415 | 0.00069 | 0.00436 | 0.00008 | 0.00112 | 65 | 0.00400 | 0.00007 | 0.00087 | 93 | 0.00141 | 0.25274 | 131.263 | 0.80086 | 0.94855 |
| nagK; fumarylpyruvate hydrolase [EC:3.7.1.20] | 0.00415 | 0.00067 | 0.00226 | 0.00002 | 0.00054 | 65 | 0.00547 | 0.00010 | 0.00105 | 93 | 0.00119 | -2.70975 | 133.720 | 0.00762 | 0.54764 |
| aacC; aminoglycoside 3-N-acetyltransferase [EC:2.3.1.81] | 0.00415 | 0.00073 | 0.00455 | 0.00011 | 0.00129 | 65 | 0.00387 | 0.00007 | 0.00086 | 93 | 0.00155 | 0.43771 | 117.430 | 0.66240 | 0.91006 |
| K07338; uncharacterized protein | 0.00415 | 0.00067 | 0.00439 | 0.00007 | 0.00107 | 65 | 0.00398 | 0.00007 | 0.00085 | 93 | 0.00137 | 0.30331 | 133.282 | 0.76212 | 0.93813 |
| E3.1.21.7, nfi; deoxyribonuclease V [EC:3.1.21.7] | 0.00415 | 0.00064 | 0.00395 | 0.00007 | 0.00101 | 65 | 0.00428 | 0.00006 | 0.00083 | 93 | 0.00130 | -0.25806 | 136.641 | 0.79675 | 0.94795 |
| betC; choline-sulfatase [EC:3.1.6.6] | 0.00414 | 0.00066 | 0.00438 | 0.00007 | 0.00105 | 65 | 0.00398 | 0.00007 | 0.00085 | 93 | 0.00135 | 0.29259 | 135.433 | 0.77029 | 0.94009 |
| sitA; manganese/iron transport system substrate-binding protein | 0.00414 | 0.00061 | 0.00443 | 0.00006 | 0.00098 | 65 | 0.00393 | 0.00006 | 0.00077 | 93 | 0.00125 | 0.40031 | 132.265 | 0.68957 | 0.91567 |
| sitC; manganese/iron transport system permease protein | 0.00414 | 0.00061 | 0.00443 | 0.00006 | 0.00098 | 65 | 0.00393 | 0.00006 | 0.00077 | 93 | 0.00125 | 0.40031 | 132.265 | 0.68957 | 0.91567 |
| sitD; manganese/iron transport system permease protein | 0.00414 | 0.00061 | 0.00443 | 0.00006 | 0.00098 | 65 | 0.00393 | 0.00006 | 0.00077 | 93 | 0.00125 | 0.40031 | 132.265 | 0.68957 | 0.91567 |
| E3.5.2.10; creatinine amidohydrolase [EC:3.5.2.10] | 0.00413 | 0.00068 | 0.00222 | 0.00002 | 0.00059 | 65 | 0.00547 | 0.00010 | 0.00106 | 93 | 0.00121 | -2.68622 | 138.244 | 0.00811 | 0.56121 |
| sufI; suppressor of ftsI | 0.00413 | 0.00062 | 0.00436 | 0.00006 | 0.00097 | 65 | 0.00397 | 0.00006 | 0.00080 | 93 | 0.00126 | 0.31491 | 136.934 | 0.75331 | 0.93639 |
| dctQ; C4-dicarboxylate transporter, DctQ subunit | 0.00413 | 0.00063 | 0.00398 | 0.00006 | 0.00093 | 65 | 0.00423 | 0.00007 | 0.00085 | 93 | 0.00126 | -0.20421 | 145.491 | 0.83847 | 0.95506 |
| bofA; inhibitor of the pro-sigma K processing machinery | 0.00412 | 0.00065 | 0.00350 | 0.00008 | 0.00109 | 65 | 0.00455 | 0.00006 | 0.00081 | 93 | 0.00136 | -0.76817 | 128.075 | 0.44380 | 0.83186 |
| mtnC, ENOPH1; enolase-phosphatase E1 [EC:3.1.3.77] | 0.00411 | 0.00065 | 0.00455 | 0.00008 | 0.00113 | 65 | 0.00380 | 0.00006 | 0.00078 | 93 | 0.00137 | 0.54604 | 120.565 | 0.58604 | 0.87728 |
| ppkA; serine/threonine-protein kinase PpkA [EC:2.7.11.1] | 0.00410 | 0.00124 | 0.00348 | 0.00019 | 0.00170 | 65 | 0.00454 | 0.00028 | 0.00174 | 93 | 0.00243 | -0.43482 | 152.175 | 0.66431 | 0.91064 |
| cobF; precorrin-6A synthase [EC:2.1.1.152] | 0.00410 | 0.00058 | 0.00361 | 0.00004 | 0.00081 | 65 | 0.00444 | 0.00006 | 0.00080 | 93 | 0.00114 | -0.73466 | 150.053 | 0.46369 | 0.83699 |
| K06991; uncharacterized protein | 0.00409 | 0.00061 | 0.00383 | 0.00006 | 0.00097 | 65 | 0.00428 | 0.00006 | 0.00078 | 93 | 0.00125 | -0.36664 | 134.463 | 0.71446 | 0.92004 |
| bioN; biotin transport system permease protein | 0.00409 | 0.00063 | 0.00397 | 0.00007 | 0.00105 | 65 | 0.00417 | 0.00006 | 0.00079 | 93 | 0.00131 | -0.14937 | 128.520 | 0.88150 | 0.96901 |
| rraB; regulator of ribonuclease activity B | 0.00409 | 0.00061 | 0.00465 | 0.00007 | 0.00100 | 65 | 0.00369 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.75541 | 130.462 | 0.45137 | 0.83438 |
| spoVAE; stage V sporulation protein AE | 0.00409 | 0.00085 | 0.00151 | 0.00002 | 0.00056 | 65 | 0.00589 | 0.00017 | 0.00137 | 93 | 0.00148 | -2.95972 | 120.381 | 0.00371 | 0.49926 |
| glft2; galactofuranosylgalactofuranosylrhamnosyl-N-acetylglucosaminyl-diphospho-decaprenol beta-1,5/1,6-galactofuranosyltransferase [EC:2.4.1.288] | 0.00408 | 0.00069 | 0.00415 | 0.00008 | 0.00108 | 65 | 0.00404 | 0.00008 | 0.00091 | 93 | 0.00141 | 0.07315 | 138.310 | 0.94180 | 0.98528 |
| mef; MFS transporter, DHA3 family, macrolide efflux protein | 0.00408 | 0.00067 | 0.00366 | 0.00006 | 0.00093 | 65 | 0.00438 | 0.00008 | 0.00094 | 93 | 0.00132 | -0.54917 | 151.245 | 0.58370 | 0.87720 |
| bcpA; oxaloacetate decarboxylase [EC:4.1.1.3] | 0.00408 | 0.00066 | 0.00443 | 0.00007 | 0.00107 | 65 | 0.00384 | 0.00007 | 0.00084 | 93 | 0.00136 | 0.43859 | 132.184 | 0.66167 | 0.90968 |
| grcA; autonomous glycyl radical cofactor | 0.00408 | 0.00062 | 0.00459 | 0.00007 | 0.00101 | 65 | 0.00372 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.68374 | 129.964 | 0.49536 | 0.84797 |
| waaP, rfaP; heptose I phosphotransferase [EC:2.7.1.-] | 0.00407 | 0.00066 | 0.00458 | 0.00007 | 0.00107 | 65 | 0.00370 | 0.00007 | 0.00084 | 93 | 0.00136 | 0.64445 | 131.593 | 0.52041 | 0.85574 |
| eutR; AraC family transcriptional regulator, ethanolamine operon transcriptional activator | 0.00406 | 0.00067 | 0.00348 | 0.00007 | 0.00102 | 65 | 0.00447 | 0.00007 | 0.00088 | 93 | 0.00135 | -0.72826 | 140.118 | 0.46767 | 0.83837 |
| virB5, lvhB5; type IV secretion system protein VirB5 | 0.00406 | 0.00054 | 0.00377 | 0.00005 | 0.00091 | 65 | 0.00426 | 0.00004 | 0.00066 | 93 | 0.00113 | -0.42844 | 124.993 | 0.66907 | 0.91165 |
| paaX; phenylacetic acid degradation operon negative regulatory protein | 0.00405 | 0.00060 | 0.00317 | 0.00003 | 0.00066 | 65 | 0.00467 | 0.00008 | 0.00090 | 93 | 0.00111 | -1.34512 | 153.764 | 0.18057 | 0.77376 |
| cheC; chemotaxis protein CheC | 0.00405 | 0.00066 | 0.00332 | 0.00007 | 0.00107 | 65 | 0.00456 | 0.00007 | 0.00084 | 93 | 0.00136 | -0.90873 | 132.513 | 0.36514 | 0.79587 |
| phnW; 2-aminoethylphosphonate-pyruvate transaminase [EC:2.6.1.37] | 0.00405 | 0.00059 | 0.00327 | 0.00003 | 0.00071 | 65 | 0.00459 | 0.00007 | 0.00087 | 93 | 0.00112 | -1.17105 | 155.925 | 0.24336 | 0.77376 |
| frdD; fumarate reductase subunit D | 0.00405 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00369 | 0.00005 | 0.00077 | 93 | 0.00127 | 0.68533 | 129.799 | 0.49436 | 0.84797 |
| nrfC; protein NrfC | 0.00404 | 0.00062 | 0.00410 | 0.00006 | 0.00095 | 65 | 0.00400 | 0.00006 | 0.00083 | 93 | 0.00126 | 0.07772 | 141.278 | 0.93816 | 0.98459 |
| K09943; uncharacterized protein | 0.00404 | 0.00061 | 0.00384 | 0.00005 | 0.00084 | 65 | 0.00417 | 0.00007 | 0.00086 | 93 | 0.00120 | -0.27558 | 152.128 | 0.78324 | 0.94361 |
| artI; arginine transport system substrate-binding protein | 0.00404 | 0.00061 | 0.00458 | 0.00007 | 0.00101 | 65 | 0.00366 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.72671 | 130.179 | 0.46871 | 0.83837 |
| chpB; chemosensory pili system protein ChpB (putative protein-glutamate methylesterase) | 0.00403 | 0.00068 | 0.00455 | 0.00009 | 0.00115 | 65 | 0.00368 | 0.00006 | 0.00083 | 93 | 0.00142 | 0.61391 | 124.019 | 0.54040 | 0.86207 |
| fieF; ferrous-iron efflux pump FieF | 0.00403 | 0.00060 | 0.00454 | 0.00007 | 0.00101 | 65 | 0.00367 | 0.00005 | 0.00074 | 93 | 0.00125 | 0.69715 | 125.638 | 0.48699 | 0.84553 |
| trpR; TrpR family transcriptional regulator, trp operon repressor | 0.00403 | 0.00060 | 0.00452 | 0.00006 | 0.00098 | 65 | 0.00369 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.67507 | 132.730 | 0.50081 | 0.84900 |
| K07109; uncharacterized protein | 0.00403 | 0.00061 | 0.00444 | 0.00006 | 0.00097 | 65 | 0.00374 | 0.00006 | 0.00079 | 93 | 0.00125 | 0.55441 | 135.926 | 0.58021 | 0.87573 |
| pepB; PepB aminopeptidase [EC:3.4.11.23] | 0.00403 | 0.00061 | 0.00461 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.78015 | 129.959 | 0.43672 | 0.83048 |
| ahpD; alkyl hydroperoxide reductase subunit D | 0.00403 | 0.00065 | 0.00376 | 0.00007 | 0.00101 | 65 | 0.00421 | 0.00007 | 0.00085 | 93 | 0.00133 | -0.33895 | 138.603 | 0.73516 | 0.92721 |
| nlpI; lipoprotein NlpI | 0.00403 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00365 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.72249 | 129.913 | 0.47129 | 0.83971 |
| tyrA; chorismate mutase / prephenate dehydrogenase [EC:5.4.99.5 1.3.1.12] | 0.00402 | 0.00062 | 0.00459 | 0.00007 | 0.00102 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.76318 | 129.127 | 0.44675 | 0.83371 |
| nagL; maleylpyruvate isomerase [EC:5.2.1.4] | 0.00402 | 0.00068 | 0.00259 | 0.00002 | 0.00057 | 65 | 0.00502 | 0.00011 | 0.00108 | 93 | 0.00122 | -1.98106 | 135.840 | 0.04960 | 0.67522 |
| holD; DNA polymerase III subunit psi [EC:2.7.7.7] | 0.00402 | 0.00061 | 0.00459 | 0.00007 | 0.00100 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.76359 | 130.105 | 0.44649 | 0.83371 |
| tmcA; tRNA(Met) cytidine acetyltransferase [EC:2.3.1.193] | 0.00402 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00364 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.73200 | 130.140 | 0.46549 | 0.83786 |
| arcA; two-component system, OmpR family, aerobic respiration control protein ArcA | 0.00402 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00364 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.72721 | 129.966 | 0.46840 | 0.83837 |
| ilvY; LysR family transcriptional regulator, positive regulator for ilvC | 0.00401 | 0.00061 | 0.00457 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74762 | 130.033 | 0.45604 | 0.83438 |
| K08985; putative lipoprotein | 0.00401 | 0.00067 | 0.00460 | 0.00008 | 0.00108 | 65 | 0.00360 | 0.00007 | 0.00085 | 93 | 0.00138 | 0.73188 | 132.956 | 0.46553 | 0.83786 |
| sapA; cationic peptide transport system substrate-binding protein | 0.00401 | 0.00061 | 0.00457 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74733 | 130.035 | 0.45621 | 0.83438 |
| sapB; cationic peptide transport system permease protein | 0.00401 | 0.00061 | 0.00457 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74733 | 130.035 | 0.45621 | 0.83438 |
| sapC; cationic peptide transport system permease protein | 0.00401 | 0.00061 | 0.00457 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74733 | 130.035 | 0.45621 | 0.83438 |
| lpxM, msbB; lauroyl-Kdo2-lipid IVA myristoyltransferase [EC:2.3.1.243] | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74426 | 129.906 | 0.45806 | 0.83438 |
| fadR; GntR family transcriptional regulator, negative regulator for fad regulon and positive regulator of fabA | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| K09894; uncharacterized protein | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| K09896; uncharacterized protein | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| K09899; uncharacterized protein | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| K09904; uncharacterized protein | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| K09910; uncharacterized protein | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| lonH; Lon-like ATP-dependent protease [EC:3.4.21.-] | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| metJ; MetJ family transcriptional regulator, methionine regulon repressor | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| mltC; membrane-bound lytic murein transglycosylase C [EC:4.2.2.-] | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| mutH; DNA mismatch repair protein MutH | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| raiA; ribosome-associated inhibitor A | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| sapD; cationic peptide transport system ATP-binding protein | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| sapF; cationic peptide transport system ATP-binding protein | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| seqA; negative modulator of initiation of replication | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| srmB; ATP-dependent RNA helicase SrmB [EC:3.6.4.13] | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| yrdD; putative DNA topoisomerase | 0.00401 | 0.00061 | 0.00456 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00006 | 0.00077 | 93 | 0.00127 | 0.74104 | 130.032 | 0.46000 | 0.83438 |
| K13580; magnesium chelatase subunit ChlD-like protein | 0.00400 | 0.00067 | 0.00459 | 0.00008 | 0.00110 | 65 | 0.00358 | 0.00007 | 0.00084 | 93 | 0.00139 | 0.72468 | 129.415 | 0.46996 | 0.83870 |
| aotQ; arginine/ornithine transport system permease protein | 0.00399 | 0.00067 | 0.00450 | 0.00007 | 0.00107 | 65 | 0.00364 | 0.00007 | 0.00087 | 93 | 0.00138 | 0.62070 | 135.847 | 0.53584 | 0.86207 |
| nthA; nitrile hydratase subunit alpha [EC:4.2.1.84] | 0.00399 | 0.00083 | 0.00247 | 0.00005 | 0.00087 | 65 | 0.00505 | 0.00015 | 0.00127 | 93 | 0.00154 | -1.68151 | 150.988 | 0.09473 | 0.76609 |
| ABC.GGU.A, gguA; putative multiple sugar transport system ATP-binding protein [EC:3.6.3.17] | 0.00399 | 0.00055 | 0.00352 | 0.00006 | 0.00098 | 65 | 0.00432 | 0.00004 | 0.00064 | 93 | 0.00117 | -0.68399 | 115.453 | 0.49535 | 0.84797 |
| fucI; L-fucose/D-arabinose isomerase [EC:5.3.1.25 5.3.1.3] | 0.00399 | 0.00056 | 0.00441 | 0.00007 | 0.00103 | 65 | 0.00369 | 0.00004 | 0.00063 | 93 | 0.00121 | 0.59381 | 109.565 | 0.55387 | 0.86806 |
| nlpC; probable lipoprotein NlpC | 0.00399 | 0.00060 | 0.00436 | 0.00006 | 0.00097 | 65 | 0.00372 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.51391 | 132.865 | 0.60817 | 0.88643 |
| ycbB, glnL; two-component system, response regulator YcbB | 0.00398 | 0.00080 | 0.00497 | 0.00015 | 0.00152 | 65 | 0.00328 | 0.00007 | 0.00085 | 93 | 0.00174 | 0.97292 | 103.427 | 0.33286 | 0.77937 |
| APEH; acylaminoacyl-peptidase [EC:3.4.19.1] | 0.00398 | 0.00081 | 0.00544 | 0.00018 | 0.00165 | 65 | 0.00296 | 0.00005 | 0.00074 | 93 | 0.00181 | 1.37276 | 89.420 | 0.17326 | 0.77376 |
| wspA; methyl-accepting chemotaxis protein WspA | 0.00398 | 0.00067 | 0.00436 | 0.00007 | 0.00103 | 65 | 0.00371 | 0.00007 | 0.00089 | 93 | 0.00136 | 0.48284 | 140.591 | 0.62996 | 0.89536 |
| rbbA; ribosome-dependent ATPase | 0.00397 | 0.00057 | 0.00349 | 0.00005 | 0.00088 | 65 | 0.00431 | 0.00005 | 0.00076 | 93 | 0.00116 | -0.71091 | 140.703 | 0.47832 | 0.84283 |
| K06876; deoxyribodipyrimidine photolyase-related protein | 0.00397 | 0.00056 | 0.00455 | 0.00007 | 0.00101 | 65 | 0.00357 | 0.00004 | 0.00064 | 93 | 0.00120 | 0.81604 | 113.759 | 0.41618 | 0.81772 |
| smoF, mtlF; sorbitol/mannitol transport system permease protein | 0.00397 | 0.00064 | 0.00329 | 0.00005 | 0.00085 | 65 | 0.00445 | 0.00008 | 0.00090 | 93 | 0.00124 | -0.93475 | 153.582 | 0.35139 | 0.79120 |
| smoG, mtlG; sorbitol/mannitol transport system permease protein | 0.00397 | 0.00064 | 0.00329 | 0.00005 | 0.00085 | 65 | 0.00445 | 0.00008 | 0.00090 | 93 | 0.00124 | -0.93475 | 153.582 | 0.35139 | 0.79120 |
| mexF; multidrug efflux pump | 0.00397 | 0.00066 | 0.00455 | 0.00008 | 0.00109 | 65 | 0.00356 | 0.00006 | 0.00084 | 93 | 0.00137 | 0.72219 | 130.439 | 0.47147 | 0.83981 |
| ugpA; sn-glycerol 3-phosphate transport system permease protein | 0.00396 | 0.00057 | 0.00203 | 0.00002 | 0.00049 | 65 | 0.00532 | 0.00007 | 0.00089 | 93 | 0.00101 | -3.24803 | 138.280 | 0.00146 | 0.46257 |
| ugpE; sn-glycerol 3-phosphate transport system permease protein | 0.00396 | 0.00058 | 0.00203 | 0.00002 | 0.00049 | 65 | 0.00532 | 0.00007 | 0.00089 | 93 | 0.00102 | -3.23034 | 137.735 | 0.00155 | 0.46257 |
| K09965; uncharacterized protein | 0.00396 | 0.00064 | 0.00340 | 0.00006 | 0.00097 | 65 | 0.00435 | 0.00007 | 0.00085 | 93 | 0.00129 | -0.72963 | 141.805 | 0.46682 | 0.83837 |
| patA; putrescine aminotransferase [EC:2.6.1.82] | 0.00395 | 0.00113 | 0.00350 | 0.00017 | 0.00161 | 65 | 0.00427 | 0.00022 | 0.00155 | 93 | 0.00224 | -0.34091 | 148.663 | 0.73365 | 0.92686 |
| crtZ; beta-carotene 3-hydroxylase [EC:1.14.15.24] | 0.00395 | 0.00065 | 0.00406 | 0.00006 | 0.00098 | 65 | 0.00387 | 0.00007 | 0.00086 | 93 | 0.00131 | 0.14516 | 142.011 | 0.88479 | 0.96984 |
| E1.1.99.3G; gluconate 2-dehydrogenase gamma chain [EC:1.1.99.3] | 0.00395 | 0.00078 | 0.00366 | 0.00007 | 0.00107 | 65 | 0.00415 | 0.00011 | 0.00109 | 93 | 0.00153 | -0.31716 | 152.270 | 0.75155 | 0.93532 |
| oprN; outer membrane protein, multidrug efflux system | 0.00395 | 0.00066 | 0.00443 | 0.00008 | 0.00108 | 65 | 0.00361 | 0.00006 | 0.00084 | 93 | 0.00136 | 0.60083 | 131.278 | 0.54899 | 0.86621 |
| cotH; spore coat protein H | 0.00394 | 0.00063 | 0.00408 | 0.00005 | 0.00089 | 65 | 0.00385 | 0.00007 | 0.00086 | 93 | 0.00124 | 0.18820 | 149.132 | 0.85098 | 0.95754 |
| chaA, CAX; Ca2+:H+ antiporter | 0.00394 | 0.00057 | 0.00242 | 0.00003 | 0.00065 | 65 | 0.00501 | 0.00006 | 0.00083 | 93 | 0.00106 | -2.44261 | 155.507 | 0.01570 | 0.59095 |
| mshC; L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase [EC:6.3.1.13] | 0.00394 | 0.00061 | 0.00385 | 0.00007 | 0.00102 | 65 | 0.00400 | 0.00005 | 0.00075 | 93 | 0.00127 | -0.11362 | 126.703 | 0.90972 | 0.97664 |
| E2.7.1.-; kinase [EC:2.7.1.-] | 0.00394 | 0.00061 | 0.00313 | 0.00004 | 0.00076 | 65 | 0.00450 | 0.00007 | 0.00088 | 93 | 0.00116 | -1.17534 | 155.790 | 0.24165 | 0.77376 |
| iolD; 3D-(3,5/4)-trihydroxycyclohexane-1,2-dione acylhydrolase (decyclizing) [EC:3.7.1.22] | 0.00394 | 0.00051 | 0.00323 | 0.00003 | 0.00070 | 65 | 0.00443 | 0.00005 | 0.00071 | 93 | 0.00100 | -1.19592 | 151.120 | 0.23360 | 0.77376 |
| rhaD; rhamnulose-1-phosphate aldolase [EC:4.1.2.19] | 0.00392 | 0.00069 | 0.00410 | 0.00013 | 0.00143 | 65 | 0.00380 | 0.00004 | 0.00062 | 93 | 0.00156 | 0.19780 | 88.185 | 0.84366 | 0.95519 |
| fno; 8-hydroxy-5-deazaflavin:NADPH oxidoreductase [EC:1.5.1.40] | 0.00392 | 0.00064 | 0.00353 | 0.00008 | 0.00108 | 65 | 0.00420 | 0.00006 | 0.00078 | 93 | 0.00133 | -0.49918 | 124.588 | 0.61853 | 0.88926 |
| mshA; D-inositol-3-phosphate glycosyltransferase [EC:2.4.1.250] | 0.00392 | 0.00061 | 0.00384 | 0.00007 | 0.00101 | 65 | 0.00398 | 0.00005 | 0.00075 | 93 | 0.00126 | -0.10736 | 126.875 | 0.91468 | 0.97664 |
| ICP; inhibitor of cysteine peptidase | 0.00392 | 0.00066 | 0.00446 | 0.00008 | 0.00108 | 65 | 0.00355 | 0.00006 | 0.00083 | 93 | 0.00136 | 0.66804 | 130.903 | 0.50528 | 0.84956 |
| mca; mycothiol S-conjugate amidase [EC:3.5.1.115] | 0.00392 | 0.00061 | 0.00385 | 0.00007 | 0.00102 | 65 | 0.00396 | 0.00005 | 0.00075 | 93 | 0.00127 | -0.08275 | 126.859 | 0.93418 | 0.98428 |
| mshD; mycothiol synthase [EC:2.3.1.189] | 0.00392 | 0.00061 | 0.00385 | 0.00007 | 0.00102 | 65 | 0.00396 | 0.00005 | 0.00075 | 93 | 0.00127 | -0.08275 | 126.859 | 0.93418 | 0.98428 |
| aspQ, ansB, ansA; glutamin-(asparagin-)ase [EC:3.5.1.38] | 0.00391 | 0.00063 | 0.00314 | 0.00004 | 0.00078 | 65 | 0.00445 | 0.00008 | 0.00093 | 93 | 0.00121 | -1.07740 | 155.985 | 0.28296 | 0.77376 |
| algB; two-component system, NtrC family, response regulator AlgB | 0.00391 | 0.00066 | 0.00444 | 0.00008 | 0.00108 | 65 | 0.00354 | 0.00006 | 0.00084 | 93 | 0.00136 | 0.66676 | 131.272 | 0.50610 | 0.84956 |
| davT, gabT; 5-aminovalerate/4-aminobutyrate aminotransferase [EC:2.6.1.48 2.6.1.19] | 0.00391 | 0.00066 | 0.00442 | 0.00008 | 0.00108 | 65 | 0.00355 | 0.00006 | 0.00083 | 93 | 0.00136 | 0.64349 | 131.217 | 0.52103 | 0.85612 |
| ppdD; prepilin peptidase dependent protein D | 0.00391 | 0.00060 | 0.00437 | 0.00006 | 0.00097 | 65 | 0.00358 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.63757 | 133.507 | 0.52484 | 0.85728 |
| mvpT, vapB; antitoxin VapB | 0.00390 | 0.00061 | 0.00392 | 0.00008 | 0.00108 | 65 | 0.00389 | 0.00005 | 0.00071 | 93 | 0.00129 | 0.02593 | 116.154 | 0.97935 | 0.99657 |
| K09901; uncharacterized protein | 0.00390 | 0.00060 | 0.00438 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.66441 | 133.136 | 0.50758 | 0.84956 |
| focA; formate transporter | 0.00390 | 0.00060 | 0.00438 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.66107 | 133.271 | 0.50971 | 0.84956 |
| grxA; glutaredoxin 1 | 0.00390 | 0.00060 | 0.00438 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.66107 | 133.271 | 0.50971 | 0.84956 |
| K09897; uncharacterized protein | 0.00390 | 0.00060 | 0.00438 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.66107 | 133.271 | 0.50971 | 0.84956 |
| K09907; uncharacterized protein | 0.00390 | 0.00060 | 0.00438 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.66107 | 133.271 | 0.50971 | 0.84956 |
| mukB; chromosome partition protein MukB | 0.00390 | 0.00060 | 0.00438 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.66107 | 133.271 | 0.50971 | 0.84956 |
| mukE; chromosome partition protein MukE | 0.00390 | 0.00060 | 0.00438 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.66107 | 133.271 | 0.50971 | 0.84956 |
| mukF; chromosome partition protein MukF | 0.00390 | 0.00060 | 0.00438 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.66107 | 133.271 | 0.50971 | 0.84956 |
| npr; NADH peroxidase [EC:1.11.1.1] | 0.00390 | 0.00081 | 0.00507 | 0.00017 | 0.00163 | 65 | 0.00307 | 0.00006 | 0.00078 | 93 | 0.00181 | 1.10664 | 93.043 | 0.27130 | 0.77376 |
| K03653; N-glycosylase/DNA lyase [EC:3.2.2.- 4.2.99.18] | 0.00390 | 0.00088 | 0.00602 | 0.00021 | 0.00181 | 65 | 0.00241 | 0.00006 | 0.00078 | 93 | 0.00197 | 1.83354 | 87.740 | 0.07011 | 0.73522 |
| K09911; uncharacterized protein | 0.00390 | 0.00060 | 0.00437 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.65772 | 133.404 | 0.51185 | 0.85122 |
| K09917; uncharacterized protein | 0.00390 | 0.00060 | 0.00437 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.65772 | 133.404 | 0.51185 | 0.85122 |
| ppdB; prepilin peptidase dependent protein B | 0.00390 | 0.00060 | 0.00432 | 0.00006 | 0.00097 | 65 | 0.00360 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.58055 | 133.160 | 0.56253 | 0.86813 |
| cdgJ; c-di-GMP phosphodiesterase [EC:3.1.4.52] | 0.00389 | 0.00095 | 0.00296 | 0.00005 | 0.00086 | 65 | 0.00455 | 0.00021 | 0.00150 | 93 | 0.00173 | -0.91922 | 140.310 | 0.35956 | 0.79496 |
| impI, vasC; type VI secretion system protein ImpI | 0.00389 | 0.00064 | 0.00402 | 0.00006 | 0.00098 | 65 | 0.00380 | 0.00007 | 0.00084 | 93 | 0.00129 | 0.17115 | 140.252 | 0.86435 | 0.96263 |
| artM; arginine transport system permease protein | 0.00389 | 0.00060 | 0.00437 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.65241 | 133.302 | 0.51526 | 0.85216 |
| artP; arginine transport system ATP-binding protein [EC:3.6.3.-] | 0.00389 | 0.00060 | 0.00437 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.65241 | 133.302 | 0.51526 | 0.85216 |
| artQ; arginine transport system permease protein | 0.00389 | 0.00060 | 0.00437 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.65241 | 133.302 | 0.51526 | 0.85216 |
| glmE, mutE, mamB; methylaspartate mutase epsilon subunit [EC:5.4.99.1] | 0.00389 | 0.00087 | 0.00577 | 0.00020 | 0.00173 | 65 | 0.00257 | 0.00006 | 0.00082 | 93 | 0.00192 | 1.66726 | 92.541 | 0.09884 | 0.76942 |
| ofaA, arfA; arthrofactin-type cyclic lipopeptide synthetase A | 0.00389 | 0.00123 | 0.00328 | 0.00018 | 0.00169 | 65 | 0.00431 | 0.00028 | 0.00173 | 93 | 0.00242 | -0.42806 | 152.353 | 0.66921 | 0.91165 |
| narP; two-component system, NarL family, nitrate/nitrite response regulator NarP | 0.00389 | 0.00061 | 0.00442 | 0.00007 | 0.00102 | 65 | 0.00352 | 0.00005 | 0.00077 | 93 | 0.00127 | 0.71027 | 128.486 | 0.47882 | 0.84314 |
| hslJ; heat shock protein HslJ | 0.00388 | 0.00057 | 0.00422 | 0.00006 | 0.00095 | 65 | 0.00365 | 0.00005 | 0.00072 | 93 | 0.00119 | 0.47322 | 128.784 | 0.63686 | 0.89696 |
| agaR; DeoR family transcriptional regulator, aga operon transcriptional repressor | 0.00388 | 0.00061 | 0.00354 | 0.00006 | 0.00097 | 65 | 0.00412 | 0.00006 | 0.00078 | 93 | 0.00124 | -0.46502 | 135.286 | 0.64267 | 0.90098 |
| coxL, cutL; aerobic carbon-monoxide dehydrogenase large subunit [EC:1.2.5.3] | 0.00388 | 0.00100 | 0.00182 | 0.00002 | 0.00055 | 65 | 0.00532 | 0.00025 | 0.00164 | 93 | 0.00173 | -2.02143 | 112.049 | 0.04562 | 0.65981 |
| torZ; trimethylamine-N-oxide reductase (cytochrome c) [EC:1.7.2.3] | 0.00388 | 0.00071 | 0.00298 | 0.00004 | 0.00078 | 65 | 0.00451 | 0.00011 | 0.00107 | 93 | 0.00132 | -1.15850 | 153.317 | 0.24846 | 0.77376 |
| ansP; L-asparagine permease | 0.00387 | 0.00064 | 0.00362 | 0.00007 | 0.00102 | 65 | 0.00405 | 0.00006 | 0.00082 | 93 | 0.00131 | -0.33205 | 135.240 | 0.74036 | 0.92969 |
| ftsN; cell division protein FtsN | 0.00387 | 0.00060 | 0.00432 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.61527 | 133.126 | 0.53943 | 0.86207 |
| polX, dpx; DNA polymerase (family X) | 0.00387 | 0.00067 | 0.00211 | 0.00003 | 0.00063 | 65 | 0.00511 | 0.00010 | 0.00104 | 93 | 0.00122 | -2.46665 | 144.055 | 0.01481 | 0.59095 |
| hofB; protein transport protein HofB | 0.00387 | 0.00060 | 0.00432 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.61188 | 133.259 | 0.54166 | 0.86207 |
| hofC; protein transport protein HofC | 0.00387 | 0.00060 | 0.00432 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.61188 | 133.259 | 0.54166 | 0.86207 |
| ppdA; prepilin peptidase dependent protein A | 0.00387 | 0.00060 | 0.00432 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.61188 | 133.259 | 0.54166 | 0.86207 |
| ppdC; prepilin peptidase dependent protein C | 0.00387 | 0.00060 | 0.00432 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00077 | 93 | 0.00124 | 0.61188 | 133.259 | 0.54166 | 0.86207 |
| waaE, kdtX; (heptosyl)LPS beta-1,4-glucosyltransferase [EC:2.4.1.-] | 0.00387 | 0.00061 | 0.00446 | 0.00007 | 0.00101 | 65 | 0.00346 | 0.00005 | 0.00077 | 93 | 0.00127 | 0.79012 | 129.244 | 0.43091 | 0.82763 |
| iolB; 5-deoxy-glucuronate isomerase [EC:5.3.1.30] | 0.00387 | 0.00050 | 0.00310 | 0.00003 | 0.00068 | 65 | 0.00441 | 0.00004 | 0.00069 | 93 | 0.00097 | -1.35413 | 152.071 | 0.17771 | 0.77376 |
| cdh; CDP-diacylglycerol pyrophosphatase [EC:3.6.1.26] | 0.00385 | 0.00069 | 0.00363 | 0.00007 | 0.00101 | 65 | 0.00401 | 0.00008 | 0.00094 | 93 | 0.00138 | -0.27729 | 145.962 | 0.78195 | 0.94310 |
| wspC; chemotaxis protein methyltransferase WspC | 0.00385 | 0.00063 | 0.00439 | 0.00007 | 0.00103 | 65 | 0.00347 | 0.00006 | 0.00079 | 93 | 0.00130 | 0.70439 | 130.534 | 0.48245 | 0.84354 |
| phnX; phosphonoacetaldehyde hydrolase [EC:3.11.1.1] | 0.00385 | 0.00059 | 0.00332 | 0.00004 | 0.00075 | 65 | 0.00421 | 0.00007 | 0.00086 | 93 | 0.00114 | -0.77978 | 155.514 | 0.43670 | 0.83048 |
| sacC, levB; levanase [EC:3.2.1.65] | 0.00385 | 0.00057 | 0.00340 | 0.00003 | 0.00069 | 65 | 0.00416 | 0.00007 | 0.00084 | 93 | 0.00108 | -0.70349 | 155.942 | 0.48280 | 0.84354 |
| slp; outer membrane lipoprotein | 0.00385 | 0.00063 | 0.00389 | 0.00006 | 0.00100 | 65 | 0.00382 | 0.00006 | 0.00081 | 93 | 0.00129 | 0.05833 | 135.917 | 0.95358 | 0.98792 |
| K06954; uncharacterized protein | 0.00385 | 0.00064 | 0.00359 | 0.00006 | 0.00099 | 65 | 0.00403 | 0.00007 | 0.00084 | 93 | 0.00129 | -0.34329 | 139.224 | 0.73190 | 0.92566 |
| cbiL; nickel transport protein | 0.00385 | 0.00061 | 0.00394 | 0.00006 | 0.00095 | 65 | 0.00378 | 0.00006 | 0.00080 | 93 | 0.00124 | 0.13173 | 138.330 | 0.89539 | 0.97230 |
| wspD; chemotaxis-related protein WspD | 0.00384 | 0.00063 | 0.00438 | 0.00007 | 0.00103 | 65 | 0.00346 | 0.00006 | 0.00079 | 93 | 0.00130 | 0.70927 | 130.336 | 0.47943 | 0.84314 |
| oxc; oxalyl-CoA decarboxylase [EC:4.1.1.8] | 0.00384 | 0.00140 | 0.00492 | 0.00063 | 0.00312 | 65 | 0.00308 | 0.00009 | 0.00097 | 93 | 0.00327 | 0.56291 | 76.409 | 0.57514 | 0.87389 |
| iolC; 5-dehydro-2-deoxygluconokinase [EC:2.7.1.92] | 0.00383 | 0.00049 | 0.00310 | 0.00003 | 0.00068 | 65 | 0.00434 | 0.00004 | 0.00068 | 93 | 0.00096 | -1.29116 | 151.591 | 0.19861 | 0.77376 |
| SURF1, SHY1; surfeit locus 1 family protein | 0.00383 | 0.00072 | 0.00204 | 0.00002 | 0.00055 | 65 | 0.00508 | 0.00012 | 0.00115 | 93 | 0.00127 | -2.39205 | 129.421 | 0.01819 | 0.59095 |
| nupC; nucleoside transport protein | 0.00383 | 0.00082 | 0.00279 | 0.00005 | 0.00090 | 65 | 0.00456 | 0.00014 | 0.00124 | 93 | 0.00153 | -1.15610 | 153.184 | 0.24944 | 0.77376 |
| K09164; uncharacterized protein | 0.00383 | 0.00064 | 0.00383 | 0.00007 | 0.00106 | 65 | 0.00383 | 0.00006 | 0.00081 | 93 | 0.00133 | 0.00172 | 128.937 | 0.99863 | 0.99957 |
| impF; type VI secretion system protein ImpF | 0.00383 | 0.00068 | 0.00344 | 0.00005 | 0.00087 | 65 | 0.00410 | 0.00009 | 0.00097 | 93 | 0.00131 | -0.50547 | 155.184 | 0.61394 | 0.88788 |
| wspB; chemotaxis-related protein WspB | 0.00383 | 0.00063 | 0.00435 | 0.00007 | 0.00103 | 65 | 0.00346 | 0.00006 | 0.00079 | 93 | 0.00130 | 0.68605 | 130.446 | 0.49390 | 0.84797 |
| wspR; two-component system, chemotaxis family, response regulator WspR [EC:2.7.7.65] | 0.00383 | 0.00063 | 0.00435 | 0.00007 | 0.00103 | 65 | 0.00346 | 0.00006 | 0.00079 | 93 | 0.00130 | 0.68605 | 130.446 | 0.49390 | 0.84797 |
| SEC11, sipW; signal peptidase I [EC:3.4.21.89] | 0.00383 | 0.00094 | 0.00448 | 0.00013 | 0.00142 | 65 | 0.00337 | 0.00015 | 0.00126 | 93 | 0.00190 | 0.58471 | 143.138 | 0.55966 | 0.86813 |
| wspF; two-component system, chemotaxis family, response regulator WspF [EC:3.1.1.61] | 0.00382 | 0.00063 | 0.00434 | 0.00007 | 0.00102 | 65 | 0.00346 | 0.00006 | 0.00079 | 93 | 0.00129 | 0.67560 | 130.814 | 0.50049 | 0.84899 |
| aroH; chorismate mutase [EC:5.4.99.5] | 0.00382 | 0.00068 | 0.00332 | 0.00007 | 0.00103 | 65 | 0.00417 | 0.00008 | 0.00092 | 93 | 0.00138 | -0.61477 | 143.282 | 0.53968 | 0.86207 |
| gabT; 4-aminobutyrate aminotransferase / (S)-3-amino-2-methylpropionate transaminase [EC:2.6.1.19 2.6.1.22] | 0.00381 | 0.00062 | 0.00222 | 0.00002 | 0.00055 | 65 | 0.00493 | 0.00009 | 0.00097 | 93 | 0.00111 | -2.44442 | 140.221 | 0.01575 | 0.59095 |
| pimC; alpha-1,6-mannosyltransferase [EC:2.4.1.-] | 0.00381 | 0.00053 | 0.00421 | 0.00006 | 0.00092 | 65 | 0.00353 | 0.00004 | 0.00062 | 93 | 0.00111 | 0.61012 | 118.383 | 0.54295 | 0.86313 |
| eutH; ethanolamine transporter | 0.00381 | 0.00091 | 0.00582 | 0.00023 | 0.00189 | 65 | 0.00240 | 0.00006 | 0.00078 | 93 | 0.00205 | 1.66544 | 85.961 | 0.09947 | 0.76942 |
| ugl; unsaturated chondroitin disaccharide hydrolase [EC:3.2.1.180] | 0.00381 | 0.00072 | 0.00391 | 0.00010 | 0.00123 | 65 | 0.00373 | 0.00007 | 0.00087 | 93 | 0.00150 | 0.12071 | 122.841 | 0.90412 | 0.97658 |
| pgaA; biofilm PGA synthesis protein PgaA | 0.00380 | 0.00064 | 0.00334 | 0.00006 | 0.00100 | 65 | 0.00413 | 0.00007 | 0.00085 | 93 | 0.00131 | -0.60182 | 139.528 | 0.54827 | 0.86565 |
| narQ; two-component system, NarL family, nitrate/nitrite sensor histidine kinase NarQ [EC:2.7.13.3] | 0.00380 | 0.00061 | 0.00435 | 0.00007 | 0.00100 | 65 | 0.00342 | 0.00005 | 0.00076 | 93 | 0.00126 | 0.74455 | 129.116 | 0.45790 | 0.83438 |
| chr, crh; catabolite repression HPr-like protein | 0.00380 | 0.00065 | 0.00345 | 0.00009 | 0.00118 | 65 | 0.00404 | 0.00005 | 0.00074 | 93 | 0.00139 | -0.42622 | 111.440 | 0.67077 | 0.91184 |
| spoVS; stage V sporulation protein S | 0.00379 | 0.00067 | 0.00316 | 0.00008 | 0.00109 | 65 | 0.00424 | 0.00007 | 0.00084 | 93 | 0.00138 | -0.78385 | 130.078 | 0.43455 | 0.82874 |
| lldR; GntR family transcriptional regulator, L-lactate dehydrogenase operon regulator | 0.00379 | 0.00064 | 0.00339 | 0.00006 | 0.00098 | 65 | 0.00407 | 0.00007 | 0.00085 | 93 | 0.00130 | -0.51956 | 140.832 | 0.60419 | 0.88434 |
| yqhD; NADP-dependent alcohol dehydrogenase [EC:1.1.-.-] | 0.00379 | 0.00053 | 0.00354 | 0.00004 | 0.00077 | 65 | 0.00396 | 0.00005 | 0.00071 | 93 | 0.00105 | -0.40072 | 145.584 | 0.68921 | 0.91567 |
| comK; competence protein ComK | 0.00379 | 0.00084 | 0.00201 | 0.00004 | 0.00082 | 65 | 0.00503 | 0.00016 | 0.00129 | 93 | 0.00153 | -1.97635 | 146.535 | 0.04999 | 0.67826 |
| hutF; formimidoylglutamate deiminase [EC:3.5.3.13] | 0.00379 | 0.00060 | 0.00340 | 0.00004 | 0.00082 | 65 | 0.00406 | 0.00007 | 0.00085 | 93 | 0.00118 | -0.55572 | 152.443 | 0.57922 | 0.87546 |
| lldP, lctP; L-lactate permease | 0.00378 | 0.00064 | 0.00337 | 0.00006 | 0.00098 | 65 | 0.00407 | 0.00007 | 0.00085 | 93 | 0.00130 | -0.54363 | 140.630 | 0.58756 | 0.87732 |
| K08988; putative membrane protein | 0.00378 | 0.00055 | 0.00312 | 0.00003 | 0.00071 | 65 | 0.00424 | 0.00006 | 0.00079 | 93 | 0.00106 | -1.06599 | 155.198 | 0.28808 | 0.77376 |
| ddc; L-2,4-diaminobutyrate decarboxylase [EC:4.1.1.86] | 0.00377 | 0.00058 | 0.00321 | 0.00003 | 0.00073 | 65 | 0.00417 | 0.00007 | 0.00085 | 93 | 0.00112 | -0.85101 | 155.897 | 0.39607 | 0.80459 |
| mshB; N-acetyl-1-D-myo-inositol-2-amino-2-deoxy-alpha-D-glucopyranoside deacetylase [EC:3.5.1.103] | 0.00377 | 0.00066 | 0.00372 | 0.00007 | 0.00102 | 65 | 0.00380 | 0.00007 | 0.00087 | 93 | 0.00134 | -0.06509 | 139.595 | 0.94820 | 0.98747 |
| mdcR; LysR family transcriptional regulator, malonate utilization transcriptional regulator | 0.00377 | 0.00063 | 0.00399 | 0.00006 | 0.00097 | 65 | 0.00361 | 0.00006 | 0.00083 | 93 | 0.00128 | 0.29897 | 140.153 | 0.76541 | 0.93900 |
| bla2, blm, ccrA, blaB; metallo-beta-lactamase class B [EC:3.5.2.6] | 0.00375 | 0.00065 | 0.00334 | 0.00007 | 0.00105 | 65 | 0.00404 | 0.00006 | 0.00082 | 93 | 0.00133 | -0.52573 | 131.633 | 0.59996 | 0.88240 |
| mtr; mycothione reductase [EC:1.8.1.15] | 0.00375 | 0.00061 | 0.00360 | 0.00007 | 0.00102 | 65 | 0.00385 | 0.00005 | 0.00075 | 93 | 0.00126 | -0.19818 | 126.855 | 0.84322 | 0.95519 |
| fcs; feruloyl-CoA synthase [EC:6.2.1.34] | 0.00375 | 0.00074 | 0.00184 | 0.00002 | 0.00058 | 65 | 0.00509 | 0.00013 | 0.00118 | 93 | 0.00131 | -2.47729 | 130.989 | 0.01451 | 0.59095 |
| wbbL; N-acetylglucosaminyl-diphospho-decaprenol L-rhamnosyltransferase [EC:2.4.1.289] | 0.00374 | 0.00063 | 0.00387 | 0.00007 | 0.00102 | 65 | 0.00365 | 0.00006 | 0.00080 | 93 | 0.00130 | 0.16388 | 132.700 | 0.87007 | 0.96495 |
| lytT, lytR; two-component system, LytTR family, response regulator LytT | 0.00374 | 0.00063 | 0.00323 | 0.00006 | 0.00095 | 65 | 0.00409 | 0.00006 | 0.00083 | 93 | 0.00126 | -0.68104 | 142.393 | 0.49695 | 0.84797 |
| K09925; uncharacterized protein | 0.00374 | 0.00060 | 0.00353 | 0.00006 | 0.00096 | 65 | 0.00388 | 0.00006 | 0.00078 | 93 | 0.00124 | -0.28096 | 134.794 | 0.77918 | 0.94269 |
| E1.1.1.306; S-(hydroxymethyl)mycothiol dehydrogenase [EC:1.1.1.306] | 0.00374 | 0.00061 | 0.00359 | 0.00007 | 0.00103 | 65 | 0.00384 | 0.00005 | 0.00076 | 93 | 0.00128 | -0.18883 | 126.740 | 0.85053 | 0.95754 |
| coxM, cutM; aerobic carbon-monoxide dehydrogenase medium subunit [EC:1.2.5.3] | 0.00374 | 0.00088 | 0.00241 | 0.00004 | 0.00077 | 65 | 0.00467 | 0.00018 | 0.00138 | 93 | 0.00158 | -1.42664 | 138.900 | 0.15593 | 0.77376 |
| PTS-Gfr-EIIC, gfrC; PTS system, fructoselysine/glucoselysine-specific IIC component | 0.00373 | 0.00079 | 0.00513 | 0.00017 | 0.00163 | 65 | 0.00276 | 0.00004 | 0.00069 | 93 | 0.00177 | 1.34450 | 87.063 | 0.18228 | 0.77376 |
| mdcH; malonate decarboxylase epsilon subunit [EC:2.3.1.39] | 0.00373 | 0.00063 | 0.00397 | 0.00006 | 0.00097 | 65 | 0.00356 | 0.00006 | 0.00083 | 93 | 0.00128 | 0.32432 | 140.176 | 0.74618 | 0.93202 |
| acuC; acetoin utilization protein AcuC | 0.00373 | 0.00063 | 0.00190 | 0.00003 | 0.00064 | 65 | 0.00501 | 0.00008 | 0.00095 | 93 | 0.00115 | -2.70397 | 149.439 | 0.00765 | 0.54764 |
| fnbA; fibronectin-binding protein A | 0.00372 | 0.00097 | 0.00363 | 0.00006 | 0.00098 | 65 | 0.00379 | 0.00021 | 0.00151 | 93 | 0.00180 | -0.08676 | 148.116 | 0.93098 | 0.98280 |
| nrfD; protein NrfD | 0.00372 | 0.00060 | 0.00393 | 0.00006 | 0.00095 | 65 | 0.00357 | 0.00006 | 0.00077 | 93 | 0.00123 | 0.28966 | 135.675 | 0.77252 | 0.94131 |
| kduD; 2-dehydro-3-deoxy-D-gluconate 5-dehydrogenase [EC:1.1.1.127] | 0.00372 | 0.00058 | 0.00287 | 0.00005 | 0.00090 | 65 | 0.00431 | 0.00005 | 0.00075 | 93 | 0.00117 | -1.22630 | 137.020 | 0.22219 | 0.77376 |
| mtiP; 5’-methylthioinosine phosphorylase [EC:2.4.2.44] | 0.00372 | 0.00064 | 0.00418 | 0.00008 | 0.00110 | 65 | 0.00340 | 0.00006 | 0.00077 | 93 | 0.00134 | 0.58238 | 121.663 | 0.56139 | 0.86813 |
| E3.2.1.86A, celF; 6-phospho-beta-glucosidase [EC:3.2.1.86] | 0.00371 | 0.00069 | 0.00382 | 0.00014 | 0.00147 | 65 | 0.00363 | 0.00003 | 0.00056 | 93 | 0.00157 | 0.11872 | 83.023 | 0.90578 | 0.97664 |
| cpxA; two-component system, OmpR family, sensor histidine kinase CpxA [EC:2.7.13.3] | 0.00371 | 0.00060 | 0.00439 | 0.00007 | 0.00101 | 65 | 0.00324 | 0.00005 | 0.00074 | 93 | 0.00125 | 0.91747 | 126.029 | 0.36065 | 0.79496 |
| lhgO; L-2-hydroxyglutarate oxidase [EC:1.1.3.-] | 0.00370 | 0.00086 | 0.00336 | 0.00010 | 0.00123 | 65 | 0.00394 | 0.00013 | 0.00119 | 93 | 0.00171 | -0.33773 | 149.103 | 0.73604 | 0.92721 |
| gbuA; guanidinobutyrase [EC:3.5.3.7] | 0.00370 | 0.00061 | 0.00415 | 0.00006 | 0.00099 | 65 | 0.00338 | 0.00006 | 0.00078 | 93 | 0.00126 | 0.60478 | 132.035 | 0.54637 | 0.86444 |
| blt; MFS transporter, DHA1 family, multidrug resistance protein | 0.00370 | 0.00083 | 0.00435 | 0.00012 | 0.00135 | 65 | 0.00324 | 0.00010 | 0.00106 | 93 | 0.00172 | 0.64397 | 132.417 | 0.52071 | 0.85579 |
| rtcR; transcriptional regulatory protein RtcR | 0.00369 | 0.00062 | 0.00337 | 0.00005 | 0.00087 | 65 | 0.00391 | 0.00007 | 0.00087 | 93 | 0.00123 | -0.43958 | 151.034 | 0.66087 | 0.90968 |
| PHO; acid phosphatase [EC:3.1.3.2] | 0.00369 | 0.00066 | 0.00325 | 0.00007 | 0.00101 | 65 | 0.00399 | 0.00007 | 0.00087 | 93 | 0.00133 | -0.54978 | 140.745 | 0.58334 | 0.87720 |
| ychN; uncharacterized protein involved in oxidation of intracellular sulfur | 0.00369 | 0.00059 | 0.00434 | 0.00006 | 0.00098 | 65 | 0.00323 | 0.00005 | 0.00074 | 93 | 0.00123 | 0.90808 | 128.745 | 0.36553 | 0.79587 |
| hpxA; allantoin racemase [EC:5.1.99.3] | 0.00368 | 0.00063 | 0.00294 | 0.00004 | 0.00074 | 65 | 0.00419 | 0.00008 | 0.00094 | 93 | 0.00120 | -1.04067 | 155.537 | 0.29965 | 0.77376 |
| iga; IgA-specific serine endopeptidase [EC:3.4.21.72] | 0.00368 | 0.00066 | 0.00425 | 0.00009 | 0.00115 | 65 | 0.00327 | 0.00006 | 0.00078 | 93 | 0.00139 | 0.70396 | 118.751 | 0.48284 | 0.84354 |
| K07069; uncharacterized protein | 0.00367 | 0.00085 | 0.00582 | 0.00021 | 0.00179 | 65 | 0.00217 | 0.00004 | 0.00068 | 93 | 0.00192 | 1.90645 | 82.342 | 0.06008 | 0.73001 |
| PTS-MalGlc-EIIC, malX; PTS system, maltose/glucose-specific IIC component | 0.00367 | 0.00049 | 0.00435 | 0.00006 | 0.00093 | 65 | 0.00319 | 0.00003 | 0.00053 | 93 | 0.00107 | 1.08639 | 104.638 | 0.27980 | 0.77376 |
| allC; allantoate deiminase [EC:3.5.3.9] | 0.00366 | 0.00076 | 0.00248 | 0.00004 | 0.00080 | 65 | 0.00449 | 0.00013 | 0.00116 | 93 | 0.00141 | -1.42313 | 150.769 | 0.15676 | 0.77376 |
| adaB; methylated-DNA-[protein]-cysteine S-methyltransferase [EC:2.1.1.63] | 0.00366 | 0.00086 | 0.00369 | 0.00009 | 0.00120 | 65 | 0.00364 | 0.00013 | 0.00120 | 93 | 0.00169 | 0.03228 | 151.116 | 0.97429 | 0.99546 |
| RIOK1; RIO kinase 1 [EC:2.7.11.1] | 0.00366 | 0.00057 | 0.00421 | 0.00006 | 0.00099 | 65 | 0.00327 | 0.00004 | 0.00067 | 93 | 0.00119 | 0.78183 | 118.974 | 0.43587 | 0.83006 |
| ugpC; sn-glycerol 3-phosphate transport system ATP-binding protein [EC:3.6.3.20] | 0.00366 | 0.00058 | 0.00182 | 0.00001 | 0.00048 | 65 | 0.00494 | 0.00007 | 0.00090 | 93 | 0.00102 | -3.07422 | 135.754 | 0.00255 | 0.46257 |
| K09900; uncharacterized protein | 0.00365 | 0.00061 | 0.00420 | 0.00007 | 0.00101 | 65 | 0.00326 | 0.00005 | 0.00076 | 93 | 0.00126 | 0.74626 | 128.503 | 0.45688 | 0.83438 |
| rmf; ribosome modulation factor | 0.00365 | 0.00057 | 0.00452 | 0.00007 | 0.00101 | 65 | 0.00303 | 0.00004 | 0.00066 | 93 | 0.00121 | 1.23546 | 114.818 | 0.21918 | 0.77376 |
| kal; 3-aminobutyryl-CoA ammonia-lyase [EC:4.3.1.14] | 0.00364 | 0.00081 | 0.00554 | 0.00018 | 0.00169 | 65 | 0.00232 | 0.00004 | 0.00069 | 93 | 0.00182 | 1.76665 | 85.846 | 0.08084 | 0.75920 |
| mdoB; phosphoglycerol transferase [EC:2.7.8.20] | 0.00364 | 0.00063 | 0.00346 | 0.00007 | 0.00105 | 65 | 0.00377 | 0.00006 | 0.00078 | 93 | 0.00131 | -0.23649 | 127.423 | 0.81344 | 0.95343 |
| flbD; flagellar protein FlbD | 0.00364 | 0.00063 | 0.00339 | 0.00008 | 0.00108 | 65 | 0.00382 | 0.00006 | 0.00077 | 93 | 0.00133 | -0.32442 | 123.760 | 0.74617 | 0.93202 |
| cbiGH-cobJ; cobalt-precorrin 5A hydrolase / precorrin-3B C17-methyltransferase [EC:3.7.1.12 2.1.1.131] | 0.00364 | 0.00062 | 0.00390 | 0.00006 | 0.00097 | 65 | 0.00346 | 0.00006 | 0.00081 | 93 | 0.00127 | 0.35097 | 137.564 | 0.72615 | 0.92177 |
| prr; aminobutyraldehyde dehydrogenase [EC:1.2.1.19] | 0.00363 | 0.00069 | 0.00318 | 0.00005 | 0.00084 | 65 | 0.00394 | 0.00010 | 0.00101 | 93 | 0.00132 | -0.57733 | 155.996 | 0.56455 | 0.86975 |
| xylG; D-xylose transport system ATP-binding protein [EC:3.6.3.17] | 0.00362 | 0.00051 | 0.00268 | 0.00002 | 0.00052 | 65 | 0.00428 | 0.00006 | 0.00078 | 93 | 0.00094 | -1.70786 | 149.245 | 0.08974 | 0.76567 |
| HMOX1; heme oxygenase 1 [EC:1.14.14.18] | 0.00362 | 0.00065 | 0.00400 | 0.00009 | 0.00120 | 65 | 0.00335 | 0.00005 | 0.00073 | 93 | 0.00140 | 0.46892 | 109.549 | 0.64006 | 0.89931 |
| citXG; holo-ACP synthase / triphosphoribosyl-dephospho-CoA synthase [EC:2.7.7.61 2.4.2.52] | 0.00362 | 0.00096 | 0.00454 | 0.00022 | 0.00185 | 65 | 0.00297 | 0.00009 | 0.00100 | 93 | 0.00210 | 0.74457 | 101.221 | 0.45826 | 0.83438 |
| benD-xylL; dihydroxycyclohexadiene carboxylate dehydrogenase [EC:1.3.1.25 1.3.1.-] | 0.00361 | 0.00056 | 0.00409 | 0.00006 | 0.00098 | 65 | 0.00328 | 0.00004 | 0.00065 | 93 | 0.00118 | 0.68783 | 117.432 | 0.49292 | 0.84797 |
| agrB; accessory gene regulator B | 0.00361 | 0.00076 | 0.00363 | 0.00011 | 0.00130 | 65 | 0.00360 | 0.00008 | 0.00093 | 93 | 0.00160 | 0.01899 | 123.745 | 0.98488 | 0.99718 |
| algF; alginate O-acetyltransferase complex protein AlgF | 0.00361 | 0.00061 | 0.00418 | 0.00007 | 0.00102 | 65 | 0.00321 | 0.00005 | 0.00076 | 93 | 0.00127 | 0.76097 | 127.558 | 0.44808 | 0.83372 |
| nanK; N-acylmannosamine kinase [EC:2.7.1.60] | 0.00361 | 0.00060 | 0.00384 | 0.00006 | 0.00095 | 65 | 0.00345 | 0.00006 | 0.00077 | 93 | 0.00122 | 0.31962 | 135.937 | 0.74975 | 0.93430 |
| mlc; transcriptional regulator of PTS gene | 0.00361 | 0.00059 | 0.00425 | 0.00006 | 0.00097 | 65 | 0.00316 | 0.00005 | 0.00074 | 93 | 0.00122 | 0.89577 | 129.529 | 0.37204 | 0.79738 |
| E1.6.99.3; NADH dehydrogenase [EC:1.6.99.3] | 0.00360 | 0.00053 | 0.00201 | 0.00002 | 0.00050 | 65 | 0.00472 | 0.00006 | 0.00082 | 93 | 0.00096 | -2.83707 | 144.572 | 0.00521 | 0.54022 |
| rfbG; CDP-glucose 4,6-dehydratase [EC:4.2.1.45] | 0.00360 | 0.00055 | 0.00394 | 0.00009 | 0.00114 | 65 | 0.00336 | 0.00002 | 0.00050 | 93 | 0.00125 | 0.46564 | 88.749 | 0.64261 | 0.90098 |
| alg8; mannuronan synthase [EC:2.4.1.33] | 0.00359 | 0.00061 | 0.00414 | 0.00007 | 0.00102 | 65 | 0.00321 | 0.00005 | 0.00076 | 93 | 0.00127 | 0.72889 | 127.817 | 0.46740 | 0.83837 |
| algX; alginate biosynthesis protein AlgX | 0.00359 | 0.00061 | 0.00414 | 0.00007 | 0.00102 | 65 | 0.00321 | 0.00005 | 0.00076 | 93 | 0.00127 | 0.72889 | 127.817 | 0.46740 | 0.83837 |
| alg44; mannuronan synthase [EC:2.4.1.33] | 0.00359 | 0.00061 | 0.00414 | 0.00007 | 0.00102 | 65 | 0.00321 | 0.00005 | 0.00076 | 93 | 0.00127 | 0.73237 | 127.841 | 0.46528 | 0.83786 |
| algE; alginate production protein | 0.00359 | 0.00061 | 0.00414 | 0.00007 | 0.00102 | 65 | 0.00321 | 0.00005 | 0.00076 | 93 | 0.00127 | 0.73237 | 127.841 | 0.46528 | 0.83786 |
| algK; alginate biosynthesis protein AlgK | 0.00359 | 0.00061 | 0.00414 | 0.00007 | 0.00102 | 65 | 0.00321 | 0.00005 | 0.00076 | 93 | 0.00127 | 0.73237 | 127.841 | 0.46528 | 0.83786 |
| chnB; cyclohexanone monooxygenase [EC:1.14.13.22] | 0.00359 | 0.00084 | 0.00199 | 0.00001 | 0.00043 | 65 | 0.00471 | 0.00018 | 0.00139 | 93 | 0.00146 | -1.86560 | 109.010 | 0.06479 | 0.73334 |
| hddA; D-glycero-alpha-D-manno-heptose-7-phosphate kinase [EC:2.7.1.168] | 0.00356 | 0.00090 | 0.00487 | 0.00026 | 0.00202 | 65 | 0.00265 | 0.00003 | 0.00061 | 93 | 0.00211 | 1.05037 | 75.666 | 0.29689 | 0.77376 |
| pcaQ; LysR family transcriptional regulator, pca operon transcriptional activator | 0.00356 | 0.00058 | 0.00408 | 0.00007 | 0.00100 | 65 | 0.00319 | 0.00004 | 0.00069 | 93 | 0.00122 | 0.73654 | 120.627 | 0.46283 | 0.83654 |
| aauR; two-component system, response regulator AauR | 0.00355 | 0.00061 | 0.00404 | 0.00006 | 0.00099 | 65 | 0.00320 | 0.00005 | 0.00076 | 93 | 0.00125 | 0.67795 | 130.791 | 0.49900 | 0.84856 |
| aauS; two-component system, sensor histidine kinase AauS [EC:2.7.13.3] | 0.00355 | 0.00061 | 0.00404 | 0.00006 | 0.00099 | 65 | 0.00320 | 0.00005 | 0.00076 | 93 | 0.00125 | 0.67795 | 130.791 | 0.49900 | 0.84856 |
| wbpL; Fuc2NAc and GlcNAc transferase [EC:2.4.1.-] | 0.00354 | 0.00064 | 0.00409 | 0.00007 | 0.00104 | 65 | 0.00316 | 0.00006 | 0.00082 | 93 | 0.00132 | 0.70827 | 132.035 | 0.48003 | 0.84324 |
| benC-xylZ; benzoate/toluate 1,2-dioxygenase reductase component [EC:1.18.1.-] | 0.00354 | 0.00056 | 0.00398 | 0.00006 | 0.00099 | 65 | 0.00323 | 0.00004 | 0.00065 | 93 | 0.00119 | 0.63137 | 115.945 | 0.52904 | 0.86094 |
| ACADL; long-chain-acyl-CoA dehydrogenase [EC:1.3.8.8] | 0.00353 | 0.00071 | 0.00263 | 0.00003 | 0.00071 | 65 | 0.00416 | 0.00011 | 0.00111 | 93 | 0.00131 | -1.17014 | 147.193 | 0.24384 | 0.77376 |
| E3.5.1.49; formamidase [EC:3.5.1.49] | 0.00352 | 0.00068 | 0.00246 | 0.00004 | 0.00081 | 65 | 0.00427 | 0.00009 | 0.00099 | 93 | 0.00128 | -1.41439 | 155.923 | 0.15924 | 0.77376 |
| OGG1; N-glycosylase/DNA lyase [EC:3.2.2.- 4.2.99.18] | 0.00352 | 0.00062 | 0.00421 | 0.00010 | 0.00124 | 65 | 0.00304 | 0.00003 | 0.00060 | 93 | 0.00138 | 0.85475 | 94.454 | 0.39485 | 0.80459 |
| benB-xylY; benzoate/toluate 1,2-dioxygenase subunit beta [EC:1.14.12.10 1.14.12.-] | 0.00352 | 0.00057 | 0.00396 | 0.00006 | 0.00100 | 65 | 0.00322 | 0.00004 | 0.00066 | 93 | 0.00120 | 0.61459 | 117.032 | 0.54002 | 0.86207 |
| dcyD; D-cysteine desulfhydrase [EC:4.4.1.15] | 0.00352 | 0.00053 | 0.00338 | 0.00004 | 0.00080 | 65 | 0.00361 | 0.00005 | 0.00071 | 93 | 0.00108 | -0.21906 | 142.771 | 0.82691 | 0.95506 |
| pgaD; biofilm PGA synthesis protein PgaD | 0.00351 | 0.00064 | 0.00308 | 0.00006 | 0.00099 | 65 | 0.00381 | 0.00007 | 0.00085 | 93 | 0.00131 | -0.55656 | 139.535 | 0.57872 | 0.87532 |
| gatZ-kbaZ; D-tagatose-1,6-bisphosphate aldolase subunit GatZ/KbaZ | 0.00351 | 0.00061 | 0.00315 | 0.00006 | 0.00096 | 65 | 0.00376 | 0.00006 | 0.00080 | 93 | 0.00125 | -0.49235 | 137.500 | 0.62326 | 0.89159 |
| psuG; pseudouridylate synthase [EC:4.2.1.70] | 0.00351 | 0.00049 | 0.00218 | 0.00001 | 0.00047 | 65 | 0.00444 | 0.00005 | 0.00075 | 93 | 0.00089 | -2.53713 | 146.405 | 0.01222 | 0.59095 |
| benA-xylX; benzoate/toluate 1,2-dioxygenase subunit alpha [EC:1.14.12.10 1.14.12.-] | 0.00351 | 0.00056 | 0.00393 | 0.00006 | 0.00100 | 65 | 0.00321 | 0.00004 | 0.00066 | 93 | 0.00120 | 0.60559 | 117.359 | 0.54596 | 0.86444 |
| mvpA, vapC; tRNA(fMet)-specific endonuclease VapC [EC:3.1.-.-] | 0.00351 | 0.00038 | 0.00313 | 0.00002 | 0.00052 | 65 | 0.00377 | 0.00003 | 0.00054 | 93 | 0.00075 | -0.85528 | 153.274 | 0.39373 | 0.80459 |
| E1.14.13.1; salicylate hydroxylase [EC:1.14.13.1] | 0.00349 | 0.00084 | 0.00156 | 0.00001 | 0.00037 | 65 | 0.00484 | 0.00018 | 0.00139 | 93 | 0.00144 | -2.28446 | 104.965 | 0.02436 | 0.59764 |
| cheBR; two-component system, chemotaxis family, CheB/CheR fusion protein [EC:2.1.1.80 3.1.1.61] | 0.00348 | 0.00064 | 0.00306 | 0.00006 | 0.00093 | 65 | 0.00378 | 0.00007 | 0.00089 | 93 | 0.00128 | -0.55641 | 148.234 | 0.57877 | 0.87532 |
| eutA; ethanolamine utilization protein EutA | 0.00348 | 0.00086 | 0.00546 | 0.00020 | 0.00177 | 65 | 0.00209 | 0.00005 | 0.00074 | 93 | 0.00192 | 1.75537 | 86.626 | 0.08273 | 0.76567 |
| ascC, ddhC, rfbH; CDP-4-dehydro-6-deoxyglucose reductase, E1 [EC:1.17.1.1] | 0.00348 | 0.00060 | 0.00386 | 0.00009 | 0.00117 | 65 | 0.00321 | 0.00003 | 0.00061 | 93 | 0.00132 | 0.49601 | 98.735 | 0.62099 | 0.89091 |
| imuA; protein ImuA | 0.00347 | 0.00054 | 0.00211 | 0.00002 | 0.00059 | 65 | 0.00443 | 0.00006 | 0.00080 | 93 | 0.00100 | -2.33264 | 154.182 | 0.02096 | 0.59147 |
| eutJ; ethanolamine utilization protein EutJ | 0.00347 | 0.00081 | 0.00555 | 0.00019 | 0.00169 | 65 | 0.00202 | 0.00004 | 0.00066 | 93 | 0.00182 | 1.94572 | 83.317 | 0.05506 | 0.70519 |
| mntA; manganese transport system ATP-binding protein | 0.00347 | 0.00060 | 0.00329 | 0.00007 | 0.00101 | 65 | 0.00360 | 0.00005 | 0.00075 | 93 | 0.00126 | -0.24931 | 127.359 | 0.80352 | 0.94911 |
| bax; Bax protein | 0.00347 | 0.00080 | 0.00538 | 0.00019 | 0.00170 | 65 | 0.00213 | 0.00004 | 0.00066 | 93 | 0.00182 | 1.78787 | 83.221 | 0.07744 | 0.75461 |
| PTS-Gfr-EIIA, gfrA; PTS system, fructoselysine/glucoselysine-specific IIA component [EC:2.7.1.-] | 0.00347 | 0.00077 | 0.00495 | 0.00017 | 0.00163 | 65 | 0.00243 | 0.00004 | 0.00065 | 93 | 0.00175 | 1.43987 | 84.264 | 0.15361 | 0.77376 |
| E2.1.1.104; caffeoyl-CoA O-methyltransferase [EC:2.1.1.104] | 0.00346 | 0.00079 | 0.00539 | 0.00018 | 0.00166 | 65 | 0.00211 | 0.00004 | 0.00065 | 93 | 0.00178 | 1.84752 | 83.809 | 0.06820 | 0.73473 |
| acrR; TetR/AcrR family transcriptional regulator, multidrug resistance operon repressor | 0.00346 | 0.00060 | 0.00397 | 0.00006 | 0.00099 | 65 | 0.00310 | 0.00005 | 0.00075 | 93 | 0.00124 | 0.70295 | 129.642 | 0.48335 | 0.84354 |
| citG; triphosphoribosyl-dephospho-CoA synthase [EC:2.4.2.52] | 0.00346 | 0.00076 | 0.00551 | 0.00017 | 0.00163 | 65 | 0.00202 | 0.00003 | 0.00058 | 93 | 0.00173 | 2.01837 | 80.067 | 0.04690 | 0.66255 |
| hofD, hopD; leader peptidase HopD [EC:3.4.23.43] | 0.00345 | 0.00059 | 0.00389 | 0.00006 | 0.00095 | 65 | 0.00315 | 0.00005 | 0.00076 | 93 | 0.00121 | 0.60970 | 134.249 | 0.54309 | 0.86313 |
| nrfB; cytochrome c-type protein NrfB | 0.00345 | 0.00059 | 0.00379 | 0.00006 | 0.00095 | 65 | 0.00321 | 0.00005 | 0.00076 | 93 | 0.00121 | 0.48059 | 133.598 | 0.63160 | 0.89607 |
| nrfE; cytochrome c-type biogenesis protein NrfE | 0.00345 | 0.00059 | 0.00379 | 0.00006 | 0.00095 | 65 | 0.00321 | 0.00005 | 0.00076 | 93 | 0.00121 | 0.47702 | 133.741 | 0.63413 | 0.89696 |
| nfrB; bacteriophage N4 adsorption protein B | 0.00344 | 0.00063 | 0.00327 | 0.00007 | 0.00106 | 65 | 0.00357 | 0.00006 | 0.00078 | 93 | 0.00132 | -0.22626 | 125.735 | 0.82137 | 0.95506 |
| pcs; phosphatidylcholine synthase [EC:2.7.8.24] | 0.00344 | 0.00049 | 0.00332 | 0.00004 | 0.00078 | 65 | 0.00353 | 0.00004 | 0.00062 | 93 | 0.00100 | -0.21547 | 134.467 | 0.82973 | 0.95506 |
| eutL; ethanolamine utilization protein EutL | 0.00344 | 0.00085 | 0.00543 | 0.00020 | 0.00177 | 65 | 0.00204 | 0.00005 | 0.00074 | 93 | 0.00191 | 1.77138 | 86.689 | 0.08001 | 0.75920 |
| ASAH2; neutral ceramidase [EC:3.5.1.23] | 0.00343 | 0.00061 | 0.00357 | 0.00007 | 0.00103 | 65 | 0.00334 | 0.00005 | 0.00074 | 93 | 0.00127 | 0.18356 | 124.143 | 0.85466 | 0.95858 |
| ushA; 5’-nucleotidase / UDP-sugar diphosphatase [EC:3.1.3.5 3.6.1.45] | 0.00343 | 0.00057 | 0.00389 | 0.00005 | 0.00090 | 65 | 0.00311 | 0.00005 | 0.00074 | 93 | 0.00117 | 0.67145 | 136.317 | 0.50307 | 0.84956 |
| ydiY; putative salt-induced outer membrane protein | 0.00343 | 0.00061 | 0.00317 | 0.00007 | 0.00101 | 65 | 0.00362 | 0.00005 | 0.00077 | 93 | 0.00127 | -0.35442 | 129.341 | 0.72360 | 0.92162 |
| impM; type VI secretion system protein ImpM | 0.00343 | 0.00061 | 0.00297 | 0.00003 | 0.00073 | 65 | 0.00374 | 0.00008 | 0.00090 | 93 | 0.00116 | -0.66579 | 155.868 | 0.50653 | 0.84956 |
| yhhL; putative membrane protein | 0.00342 | 0.00052 | 0.00434 | 0.00006 | 0.00094 | 65 | 0.00278 | 0.00003 | 0.00059 | 93 | 0.00111 | 1.40172 | 113.046 | 0.16374 | 0.77376 |
| AGA, aspG; N4-(beta-N-acetylglucosaminyl)-L-asparaginase [EC:3.5.1.26] | 0.00342 | 0.00061 | 0.00316 | 0.00007 | 0.00101 | 65 | 0.00360 | 0.00005 | 0.00076 | 93 | 0.00126 | -0.35270 | 127.926 | 0.72489 | 0.92177 |
| rhaM; L-rhamnose mutarotase [EC:5.1.3.32] | 0.00342 | 0.00048 | 0.00226 | 0.00002 | 0.00050 | 65 | 0.00423 | 0.00005 | 0.00072 | 93 | 0.00088 | -2.24415 | 151.729 | 0.02627 | 0.60721 |
| spoVAD; stage V sporulation protein AD | 0.00341 | 0.00070 | 0.00144 | 0.00002 | 0.00055 | 65 | 0.00479 | 0.00011 | 0.00110 | 93 | 0.00124 | -2.71465 | 132.088 | 0.00752 | 0.54764 |
| spoVAC; stage V sporulation protein AC | 0.00341 | 0.00069 | 0.00142 | 0.00002 | 0.00055 | 65 | 0.00480 | 0.00011 | 0.00109 | 93 | 0.00123 | -2.75891 | 132.757 | 0.00662 | 0.54612 |
| K09959; uncharacterized protein | 0.00341 | 0.00058 | 0.00149 | 0.00001 | 0.00041 | 65 | 0.00475 | 0.00008 | 0.00091 | 93 | 0.00100 | -3.25760 | 126.063 | 0.00144 | 0.46257 |
| K09799; uncharacterized protein | 0.00341 | 0.00058 | 0.00252 | 0.00002 | 0.00059 | 65 | 0.00403 | 0.00007 | 0.00088 | 93 | 0.00106 | -1.42118 | 150.035 | 0.15734 | 0.77376 |
| ulaR; DeoR family transcriptional regulator, ulaG and ulaABCDEF operon transcriptional repressor | 0.00340 | 0.00057 | 0.00354 | 0.00005 | 0.00086 | 65 | 0.00330 | 0.00005 | 0.00076 | 93 | 0.00115 | 0.21273 | 142.456 | 0.83184 | 0.95506 |
| osmE; osmotically inducible lipoprotein OsmE | 0.00339 | 0.00053 | 0.00428 | 0.00006 | 0.00095 | 65 | 0.00277 | 0.00003 | 0.00061 | 93 | 0.00112 | 1.34017 | 113.969 | 0.18286 | 0.77376 |
| dhaA; haloalkane dehalogenase [EC:3.8.1.5] | 0.00339 | 0.00062 | 0.00313 | 0.00007 | 0.00101 | 65 | 0.00357 | 0.00006 | 0.00079 | 93 | 0.00128 | -0.34263 | 132.345 | 0.73242 | 0.92598 |
| murEF; murE/murF fusion protein [EC:6.3.2.13 6.3.2.10] | 0.00338 | 0.00097 | 0.00408 | 0.00024 | 0.00192 | 65 | 0.00289 | 0.00009 | 0.00096 | 93 | 0.00215 | 0.55522 | 95.718 | 0.58004 | 0.87573 |
| eutQ; ethanolamine utilization protein EutQ | 0.00338 | 0.00085 | 0.00557 | 0.00021 | 0.00179 | 65 | 0.00184 | 0.00004 | 0.00068 | 93 | 0.00191 | 1.94957 | 82.553 | 0.05462 | 0.70353 |
| kdd; L-erythro-3,5-diaminohexanoate dehydrogenase [EC:1.4.1.11] | 0.00337 | 0.00078 | 0.00526 | 0.00018 | 0.00165 | 65 | 0.00205 | 0.00004 | 0.00063 | 93 | 0.00177 | 1.81288 | 82.510 | 0.07349 | 0.74128 |
| E3.2.1.89; arabinogalactan endo-1,4-beta-galactosidase [EC:3.2.1.89] | 0.00337 | 0.00053 | 0.00232 | 0.00001 | 0.00046 | 65 | 0.00410 | 0.00007 | 0.00084 | 93 | 0.00096 | -1.84569 | 137.635 | 0.06709 | 0.73334 |
| ulaD, sgaH, sgbH; 3-dehydro-L-gulonate-6-phosphate decarboxylase [EC:4.1.1.85] | 0.00336 | 0.00056 | 0.00380 | 0.00005 | 0.00087 | 65 | 0.00306 | 0.00005 | 0.00073 | 93 | 0.00113 | 0.64562 | 138.609 | 0.51959 | 0.85478 |
| paaE; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaE | 0.00336 | 0.00041 | 0.00324 | 0.00002 | 0.00056 | 65 | 0.00345 | 0.00003 | 0.00059 | 93 | 0.00081 | -0.25496 | 153.372 | 0.79909 | 0.94840 |
| atuC; geranyl-CoA carboxylase beta subunit [EC:6.4.1.5] | 0.00336 | 0.00053 | 0.00277 | 0.00003 | 0.00070 | 65 | 0.00378 | 0.00005 | 0.00075 | 93 | 0.00103 | -0.97908 | 153.770 | 0.32908 | 0.77446 |
| ptrA; protease III [EC:3.4.24.55] | 0.00336 | 0.00057 | 0.00379 | 0.00005 | 0.00090 | 65 | 0.00306 | 0.00005 | 0.00074 | 93 | 0.00117 | 0.62458 | 136.199 | 0.53329 | 0.86207 |
| parR; two-component system, OmpR family, response regulator ParR | 0.00336 | 0.00064 | 0.00292 | 0.00006 | 0.00100 | 65 | 0.00367 | 0.00007 | 0.00084 | 93 | 0.00131 | -0.57443 | 138.954 | 0.56660 | 0.87051 |
| parS; two-component system, OmpR family, sensor kinase ParS [EC:2.7.13.3] | 0.00336 | 0.00064 | 0.00292 | 0.00006 | 0.00100 | 65 | 0.00367 | 0.00007 | 0.00084 | 93 | 0.00131 | -0.57443 | 138.954 | 0.56660 | 0.87051 |
| ACE, CD143; peptidyl-dipeptidase A [EC:3.4.15.1] | 0.00336 | 0.00063 | 0.00294 | 0.00007 | 0.00101 | 65 | 0.00365 | 0.00006 | 0.00081 | 93 | 0.00129 | -0.54838 | 134.266 | 0.58434 | 0.87720 |
| E5.1.99.4, AMACR, mcr; alpha-methylacyl-CoA racemase [EC:5.1.99.4] | 0.00335 | 0.00071 | 0.00172 | 0.00001 | 0.00046 | 65 | 0.00449 | 0.00012 | 0.00116 | 93 | 0.00124 | -2.22657 | 118.648 | 0.02786 | 0.61334 |
| K09779; uncharacterized protein | 0.00335 | 0.00064 | 0.00228 | 0.00004 | 0.00083 | 65 | 0.00410 | 0.00008 | 0.00091 | 93 | 0.00123 | -1.47137 | 154.997 | 0.14322 | 0.77376 |
| citX; holo-ACP synthase [EC:2.7.7.61] | 0.00334 | 0.00076 | 0.00543 | 0.00017 | 0.00163 | 65 | 0.00188 | 0.00003 | 0.00057 | 93 | 0.00173 | 2.05500 | 79.456 | 0.04316 | 0.65559 |
| K07276; uncharacterized protein | 0.00334 | 0.00061 | 0.00305 | 0.00006 | 0.00096 | 65 | 0.00354 | 0.00006 | 0.00079 | 93 | 0.00124 | -0.39464 | 135.709 | 0.69373 | 0.91691 |
| kce; 3-keto-5-aminohexanoate cleavage enzyme [EC:2.3.1.247] | 0.00334 | 0.00078 | 0.00521 | 0.00018 | 0.00165 | 65 | 0.00203 | 0.00004 | 0.00063 | 93 | 0.00177 | 1.79903 | 82.475 | 0.07567 | 0.74536 |
| apgM; 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [EC:5.4.2.12] | 0.00334 | 0.00056 | 0.00320 | 0.00006 | 0.00098 | 65 | 0.00343 | 0.00004 | 0.00065 | 93 | 0.00118 | -0.19077 | 116.869 | 0.84904 | 0.95712 |
| K09984; uncharacterized protein | 0.00333 | 0.00049 | 0.00347 | 0.00005 | 0.00087 | 65 | 0.00323 | 0.00003 | 0.00058 | 93 | 0.00105 | 0.22657 | 118.065 | 0.82115 | 0.95506 |
| virB1; type IV secretion system protein VirB1 | 0.00333 | 0.00066 | 0.00331 | 0.00009 | 0.00117 | 65 | 0.00334 | 0.00006 | 0.00078 | 93 | 0.00140 | -0.02460 | 117.478 | 0.98041 | 0.99657 |
| kamD; beta-lysine 5,6-aminomutase alpha subunit [EC:5.4.3.3] | 0.00333 | 0.00078 | 0.00522 | 0.00018 | 0.00165 | 65 | 0.00201 | 0.00004 | 0.00062 | 93 | 0.00177 | 1.81790 | 82.275 | 0.07272 | 0.74128 |
| kamE; beta-lysine 5,6-aminomutase beta subunit [EC:5.4.3.3] | 0.00333 | 0.00078 | 0.00522 | 0.00018 | 0.00165 | 65 | 0.00201 | 0.00004 | 0.00062 | 93 | 0.00177 | 1.81790 | 82.275 | 0.07272 | 0.74128 |
| E4.1.1.15, gadB, gadA, GAD; glutamate decarboxylase [EC:4.1.1.15] | 0.00333 | 0.00071 | 0.00319 | 0.00010 | 0.00124 | 65 | 0.00343 | 0.00007 | 0.00084 | 93 | 0.00150 | -0.16112 | 118.885 | 0.87227 | 0.96576 |
| mdfA, cmr; MFS transporter, DHA1 family, multidrug/chloramphenicol efflux transport protein | 0.00333 | 0.00046 | 0.00347 | 0.00003 | 0.00066 | 65 | 0.00323 | 0.00004 | 0.00063 | 93 | 0.00092 | 0.26410 | 148.263 | 0.79207 | 0.94731 |
| prtC; serralysin [EC:3.4.24.40] | 0.00332 | 0.00078 | 0.00227 | 0.00004 | 0.00083 | 65 | 0.00406 | 0.00013 | 0.00119 | 93 | 0.00145 | -1.22940 | 151.482 | 0.22083 | 0.77376 |
| PTS-Fru-EIIB, fruA; PTS system, fructose-specific IIB component [EC:2.7.1.202] | 0.00332 | 0.00074 | 0.00450 | 0.00017 | 0.00164 | 65 | 0.00250 | 0.00003 | 0.00053 | 93 | 0.00172 | 1.15817 | 77.677 | 0.25034 | 0.77376 |
| oapA; opacity associated protein | 0.00332 | 0.00059 | 0.00373 | 0.00006 | 0.00095 | 65 | 0.00304 | 0.00005 | 0.00075 | 93 | 0.00121 | 0.57144 | 133.149 | 0.56867 | 0.87135 |
| STE24; STE24 endopeptidase [EC:3.4.24.84] | 0.00332 | 0.00056 | 0.00296 | 0.00003 | 0.00068 | 65 | 0.00358 | 0.00006 | 0.00082 | 93 | 0.00106 | -0.58116 | 155.998 | 0.56197 | 0.86813 |
| nrtB, nasE, cynB; nitrate/nitrite transport system permease protein | 0.00332 | 0.00049 | 0.00301 | 0.00003 | 0.00069 | 65 | 0.00353 | 0.00004 | 0.00069 | 93 | 0.00097 | -0.53759 | 150.881 | 0.59165 | 0.87896 |
| nrtC, nasD; nitrate/nitrite transport system ATP-binding protein [EC:3.6.3.-] | 0.00332 | 0.00049 | 0.00301 | 0.00003 | 0.00069 | 65 | 0.00353 | 0.00004 | 0.00069 | 93 | 0.00097 | -0.53759 | 150.881 | 0.59165 | 0.87896 |
| mhpD; 2-keto-4-pentenoate hydratase [EC:4.2.1.80] | 0.00332 | 0.00052 | 0.00214 | 0.00002 | 0.00052 | 65 | 0.00414 | 0.00006 | 0.00081 | 93 | 0.00096 | -2.09173 | 147.390 | 0.03818 | 0.63091 |
| E4.1.3.25; (S)-citramalyl-CoA lyase [EC:4.1.3.25] | 0.00331 | 0.00061 | 0.00350 | 0.00006 | 0.00095 | 65 | 0.00318 | 0.00006 | 0.00081 | 93 | 0.00124 | 0.25820 | 139.386 | 0.79663 | 0.94795 |
| ftrA; AraC family transcriptional regulator, transcriptional activator FtrA | 0.00331 | 0.00064 | 0.00305 | 0.00006 | 0.00099 | 65 | 0.00349 | 0.00007 | 0.00084 | 93 | 0.00130 | -0.34071 | 139.193 | 0.73384 | 0.92692 |
| E2.7.7.65; diguanylate cyclase [EC:2.7.7.65] | 0.00331 | 0.00061 | 0.00315 | 0.00006 | 0.00097 | 65 | 0.00341 | 0.00006 | 0.00079 | 93 | 0.00125 | -0.20695 | 135.654 | 0.83636 | 0.95506 |
| K06888; uncharacterized protein | 0.00331 | 0.00046 | 0.00213 | 0.00001 | 0.00047 | 65 | 0.00413 | 0.00005 | 0.00070 | 93 | 0.00085 | -2.35544 | 149.547 | 0.01980 | 0.59095 |
| K06986; uncharacterized protein | 0.00330 | 0.00050 | 0.00365 | 0.00005 | 0.00087 | 65 | 0.00306 | 0.00003 | 0.00059 | 93 | 0.00105 | 0.55395 | 118.368 | 0.58066 | 0.87573 |
| mntB; manganese transport system permease protein | 0.00329 | 0.00059 | 0.00327 | 0.00007 | 0.00101 | 65 | 0.00331 | 0.00005 | 0.00073 | 93 | 0.00124 | -0.02951 | 125.128 | 0.97651 | 0.99595 |
| E3.4.11.-; aminopeptidase [EC:3.4.11.-] | 0.00329 | 0.00079 | 0.00499 | 0.00018 | 0.00165 | 65 | 0.00210 | 0.00004 | 0.00068 | 93 | 0.00178 | 1.62198 | 85.811 | 0.10848 | 0.77376 |
| K09983; uncharacterized protein | 0.00328 | 0.00056 | 0.00190 | 0.00002 | 0.00054 | 65 | 0.00425 | 0.00007 | 0.00085 | 93 | 0.00101 | -2.32804 | 146.236 | 0.02128 | 0.59147 |
| soxY; sulfur-oxidizing protein SoxY | 0.00328 | 0.00054 | 0.00327 | 0.00004 | 0.00080 | 65 | 0.00329 | 0.00005 | 0.00073 | 93 | 0.00108 | -0.01932 | 144.760 | 0.98461 | 0.99718 |
| K07337; penicillin-binding protein activator | 0.00328 | 0.00062 | 0.00237 | 0.00003 | 0.00063 | 65 | 0.00392 | 0.00008 | 0.00095 | 93 | 0.00114 | -1.36043 | 149.464 | 0.17574 | 0.77376 |
| pchB; isochorismate pyruvate lyase [EC:4.2.99.21] | 0.00328 | 0.00046 | 0.00293 | 0.00003 | 0.00064 | 65 | 0.00352 | 0.00004 | 0.00064 | 93 | 0.00091 | -0.65032 | 151.589 | 0.51647 | 0.85216 |
| adeB; multidrug efflux pump | 0.00328 | 0.00061 | 0.00322 | 0.00006 | 0.00096 | 65 | 0.00332 | 0.00006 | 0.00078 | 93 | 0.00124 | -0.08055 | 135.511 | 0.93592 | 0.98459 |
| yebQ; MFS transporter, DHA2 family, multidrug resistance protein | 0.00328 | 0.00043 | 0.00213 | 0.00002 | 0.00048 | 65 | 0.00407 | 0.00004 | 0.00065 | 93 | 0.00081 | -2.41295 | 154.240 | 0.01700 | 0.59095 |
| atuB; citronellol/citronellal dehydrogenase | 0.00327 | 0.00052 | 0.00264 | 0.00003 | 0.00067 | 65 | 0.00372 | 0.00005 | 0.00074 | 93 | 0.00100 | -1.08387 | 155.028 | 0.28010 | 0.77376 |
| K09770; uncharacterized protein | 0.00327 | 0.00062 | 0.00271 | 0.00007 | 0.00103 | 65 | 0.00366 | 0.00006 | 0.00077 | 93 | 0.00128 | -0.74395 | 128.123 | 0.45827 | 0.83438 |
| cas5d; CRISPR-associated protein Cas5d | 0.00326 | 0.00050 | 0.00285 | 0.00003 | 0.00069 | 65 | 0.00355 | 0.00005 | 0.00070 | 93 | 0.00098 | -0.71788 | 151.636 | 0.47394 | 0.84050 |
| csd2, cas7; CRISPR-associated protein Csd2 | 0.00326 | 0.00050 | 0.00285 | 0.00003 | 0.00069 | 65 | 0.00355 | 0.00005 | 0.00070 | 93 | 0.00098 | -0.71788 | 151.636 | 0.47394 | 0.84050 |
| atuD; citronellyl-CoA dehydrogenase [EC:1.3.99.-] | 0.00326 | 0.00052 | 0.00272 | 0.00003 | 0.00069 | 65 | 0.00363 | 0.00005 | 0.00074 | 93 | 0.00101 | -0.89769 | 154.336 | 0.37075 | 0.79607 |
| wzzE; lipopolysaccharide biosynthesis protein WzzE | 0.00325 | 0.00056 | 0.00361 | 0.00005 | 0.00086 | 65 | 0.00300 | 0.00005 | 0.00074 | 93 | 0.00114 | 0.53722 | 140.203 | 0.59197 | 0.87898 |
| K09764; uncharacterized protein | 0.00325 | 0.00062 | 0.00260 | 0.00007 | 0.00105 | 65 | 0.00370 | 0.00005 | 0.00075 | 93 | 0.00129 | -0.85096 | 123.423 | 0.39644 | 0.80459 |
| fliT; flagellar protein FliT | 0.00324 | 0.00060 | 0.00222 | 0.00002 | 0.00057 | 65 | 0.00396 | 0.00008 | 0.00093 | 93 | 0.00109 | -1.59644 | 144.372 | 0.11258 | 0.77376 |
| csd1, cas8c; CRISPR-associated protein Csd1 | 0.00324 | 0.00050 | 0.00285 | 0.00003 | 0.00069 | 65 | 0.00352 | 0.00004 | 0.00069 | 93 | 0.00098 | -0.68790 | 151.159 | 0.49257 | 0.84797 |
| nylB; 6-aminohexanoate-oligomer exohydrolase [EC:3.5.1.46] | 0.00324 | 0.00063 | 0.00166 | 0.00002 | 0.00057 | 65 | 0.00435 | 0.00009 | 0.00099 | 93 | 0.00114 | -2.36214 | 140.451 | 0.01954 | 0.59095 |
| cgeB; spore maturation protein CgeB | 0.00324 | 0.00058 | 0.00309 | 0.00007 | 0.00100 | 65 | 0.00335 | 0.00005 | 0.00071 | 93 | 0.00123 | -0.21122 | 122.135 | 0.83307 | 0.95506 |
| pvdE; putative pyoverdin transport system ATP-binding/permease protein | 0.00323 | 0.00073 | 0.00286 | 0.00005 | 0.00088 | 65 | 0.00350 | 0.00011 | 0.00108 | 93 | 0.00139 | -0.45743 | 155.926 | 0.64800 | 0.90350 |
| aas; acyl-[acyl-carrier-protein]-phospholipid O-acyltransferase / long-chain-fatty-acid–[acyl-carrier-protein] ligase [EC:2.3.1.40 6.2.1.20] | 0.00322 | 0.00044 | 0.00426 | 0.00004 | 0.00082 | 65 | 0.00250 | 0.00002 | 0.00046 | 93 | 0.00094 | 1.87958 | 104.114 | 0.06296 | 0.73012 |
| yqgT; g-D-glutamyl-meso-diaminopimelate peptidase [EC:3.4.19.11] | 0.00322 | 0.00063 | 0.00304 | 0.00009 | 0.00114 | 65 | 0.00335 | 0.00005 | 0.00071 | 93 | 0.00135 | -0.22520 | 111.620 | 0.82223 | 0.95506 |
| chrR; putative transcriptional regulator | 0.00322 | 0.00061 | 0.00277 | 0.00006 | 0.00096 | 65 | 0.00353 | 0.00006 | 0.00079 | 93 | 0.00124 | -0.60939 | 136.549 | 0.54328 | 0.86313 |
| E3.1.11.5; exodeoxyribonuclease V [EC:3.1.11.5] | 0.00321 | 0.00044 | 0.00214 | 0.00001 | 0.00042 | 65 | 0.00396 | 0.00004 | 0.00068 | 93 | 0.00080 | -2.26331 | 145.429 | 0.02510 | 0.59941 |
| K09749; uncharacterized protein | 0.00321 | 0.00065 | 0.00305 | 0.00007 | 0.00103 | 65 | 0.00332 | 0.00007 | 0.00084 | 93 | 0.00133 | -0.20060 | 135.763 | 0.84131 | 0.95506 |
| rssB, hnr; two-component system, response regulator | 0.00321 | 0.00060 | 0.00315 | 0.00006 | 0.00098 | 65 | 0.00325 | 0.00005 | 0.00076 | 93 | 0.00124 | -0.07635 | 131.664 | 0.93926 | 0.98518 |
| pgpC; phosphatidylglycerophosphatase C [EC:3.1.3.27] | 0.00321 | 0.00060 | 0.00309 | 0.00006 | 0.00096 | 65 | 0.00328 | 0.00005 | 0.00076 | 93 | 0.00123 | -0.15454 | 133.315 | 0.87742 | 0.96737 |
| lpp; murein lipoprotein | 0.00320 | 0.00061 | 0.00316 | 0.00006 | 0.00097 | 65 | 0.00323 | 0.00006 | 0.00079 | 93 | 0.00125 | -0.05571 | 135.555 | 0.95566 | 0.98792 |
| arnE; undecaprenyl phosphate-alpha-L-ara4N flippase subunit ArnE | 0.00320 | 0.00051 | 0.00250 | 0.00003 | 0.00062 | 65 | 0.00369 | 0.00005 | 0.00076 | 93 | 0.00098 | -1.21225 | 155.974 | 0.22725 | 0.77376 |
| deoR; deoxyribonucleoside regulator | 0.00320 | 0.00073 | 0.00388 | 0.00015 | 0.00153 | 65 | 0.00272 | 0.00004 | 0.00063 | 93 | 0.00165 | 0.70057 | 85.711 | 0.48547 | 0.84459 |
| K09712; uncharacterized protein | 0.00320 | 0.00038 | 0.00334 | 0.00002 | 0.00051 | 65 | 0.00309 | 0.00003 | 0.00055 | 93 | 0.00075 | 0.33077 | 153.744 | 0.74127 | 0.93032 |
| K09766; uncharacterized protein | 0.00319 | 0.00078 | 0.00291 | 0.00008 | 0.00114 | 65 | 0.00339 | 0.00010 | 0.00105 | 93 | 0.00155 | -0.31279 | 146.063 | 0.75488 | 0.93665 |
| yscN, sctN, hrcN, ssaN; ATP synthase in type III secretion protein N [EC:3.6.3.14] | 0.00319 | 0.00072 | 0.00246 | 0.00006 | 0.00093 | 65 | 0.00370 | 0.00010 | 0.00104 | 93 | 0.00140 | -0.88631 | 155.073 | 0.37682 | 0.79810 |
| helD; DNA helicase IV [EC:3.6.4.12] | 0.00319 | 0.00060 | 0.00321 | 0.00006 | 0.00098 | 65 | 0.00318 | 0.00005 | 0.00077 | 93 | 0.00124 | 0.02021 | 132.063 | 0.98391 | 0.99706 |
| atuF; geranyl-CoA carboxylase alpha subunit [EC:6.4.1.5] | 0.00319 | 0.00052 | 0.00268 | 0.00003 | 0.00069 | 65 | 0.00355 | 0.00005 | 0.00074 | 93 | 0.00101 | -0.85827 | 153.909 | 0.39208 | 0.80459 |
| ssp; subtilase-type serine protease [EC:3.4.21.-] | 0.00319 | 0.00062 | 0.00281 | 0.00006 | 0.00096 | 65 | 0.00346 | 0.00006 | 0.00082 | 93 | 0.00126 | -0.51607 | 139.223 | 0.60662 | 0.88555 |
| rsbT; serine/threonine-protein kinase RsbT [EC:2.7.11.1] | 0.00319 | 0.00067 | 0.00278 | 0.00005 | 0.00091 | 65 | 0.00348 | 0.00008 | 0.00094 | 93 | 0.00131 | -0.53405 | 152.913 | 0.59408 | 0.88005 |
| adeA; membrane fusion protein, multidrug efflux system | 0.00318 | 0.00061 | 0.00307 | 0.00006 | 0.00097 | 65 | 0.00325 | 0.00006 | 0.00079 | 93 | 0.00125 | -0.14658 | 135.435 | 0.88368 | 0.96974 |
| mntC; manganese transport system substrate-binding protein | 0.00317 | 0.00059 | 0.00323 | 0.00007 | 0.00101 | 65 | 0.00313 | 0.00005 | 0.00072 | 93 | 0.00124 | 0.07912 | 123.610 | 0.93707 | 0.98459 |
| dgoK; 2-dehydro-3-deoxygalactonokinase [EC:2.7.1.58] | 0.00316 | 0.00049 | 0.00171 | 0.00001 | 0.00044 | 65 | 0.00418 | 0.00005 | 0.00076 | 93 | 0.00088 | -2.80574 | 141.333 | 0.00573 | 0.54022 |
| doxD; thiosulfate dehydrogenase [quinone] large subunit [EC:1.8.5.2] | 0.00316 | 0.00058 | 0.00168 | 0.00002 | 0.00060 | 65 | 0.00419 | 0.00007 | 0.00087 | 93 | 0.00106 | -2.37842 | 150.868 | 0.01864 | 0.59095 |
| K09981; uncharacterized protein | 0.00316 | 0.00061 | 0.00314 | 0.00007 | 0.00104 | 65 | 0.00317 | 0.00005 | 0.00075 | 93 | 0.00128 | -0.02244 | 124.639 | 0.98213 | 0.99680 |
| qoxB; cytochrome aa3-600 menaquinol oxidase subunit I [EC:1.10.3.12] | 0.00315 | 0.00076 | 0.00172 | 0.00004 | 0.00078 | 65 | 0.00415 | 0.00013 | 0.00117 | 93 | 0.00140 | -1.73980 | 149.464 | 0.08395 | 0.76567 |
| K08976; putative membrane protein | 0.00315 | 0.00060 | 0.00159 | 0.00002 | 0.00061 | 65 | 0.00423 | 0.00008 | 0.00091 | 93 | 0.00110 | -2.39849 | 149.536 | 0.01770 | 0.59095 |
| gerM; germination protein M | 0.00315 | 0.00060 | 0.00281 | 0.00009 | 0.00115 | 65 | 0.00338 | 0.00004 | 0.00063 | 93 | 0.00131 | -0.43266 | 101.518 | 0.66618 | 0.91126 |
| degS; two-component system, NarL family, sensor histidine kinase DegS [EC:2.7.13.3] | 0.00314 | 0.00063 | 0.00262 | 0.00007 | 0.00102 | 65 | 0.00350 | 0.00006 | 0.00079 | 93 | 0.00129 | -0.68281 | 130.317 | 0.49594 | 0.84797 |
| E2.4.2.6; nucleoside deoxyribosyltransferase [EC:2.4.2.6] | 0.00314 | 0.00087 | 0.00389 | 0.00018 | 0.00168 | 65 | 0.00261 | 0.00007 | 0.00089 | 93 | 0.00190 | 0.67169 | 99.255 | 0.50334 | 0.84956 |
| spo0M; sporulation-control protein | 0.00313 | 0.00064 | 0.00196 | 0.00003 | 0.00067 | 65 | 0.00395 | 0.00009 | 0.00098 | 93 | 0.00119 | -1.67723 | 150.718 | 0.09557 | 0.76609 |
| xylF; D-xylose transport system substrate-binding protein | 0.00313 | 0.00052 | 0.00185 | 0.00001 | 0.00047 | 65 | 0.00403 | 0.00006 | 0.00081 | 93 | 0.00093 | -2.33138 | 141.271 | 0.02115 | 0.59147 |
| PCCA, pccA; propionyl-CoA carboxylase alpha chain [EC:6.4.1.3] | 0.00313 | 0.00052 | 0.00146 | 0.00001 | 0.00035 | 65 | 0.00430 | 0.00006 | 0.00083 | 93 | 0.00090 | -3.14707 | 122.528 | 0.00207 | 0.46257 |
| mdtI; spermidine export protein MdtI | 0.00313 | 0.00060 | 0.00312 | 0.00006 | 0.00097 | 65 | 0.00313 | 0.00005 | 0.00076 | 93 | 0.00123 | -0.01379 | 131.781 | 0.98902 | 0.99801 |
| mdtJ; spermidine export protein MdtJ | 0.00313 | 0.00060 | 0.00312 | 0.00006 | 0.00097 | 65 | 0.00313 | 0.00005 | 0.00076 | 93 | 0.00123 | -0.01379 | 131.781 | 0.98902 | 0.99801 |
| cybC; soluble cytochrome b562 | 0.00312 | 0.00055 | 0.00257 | 0.00003 | 0.00067 | 65 | 0.00351 | 0.00006 | 0.00081 | 93 | 0.00105 | -0.88691 | 155.969 | 0.37649 | 0.79810 |
| fliB; lysine-N-methylase [EC:2.1.1.-] | 0.00312 | 0.00088 | 0.00399 | 0.00023 | 0.00186 | 65 | 0.00252 | 0.00005 | 0.00075 | 93 | 0.00201 | 0.73019 | 84.694 | 0.46729 | 0.83837 |
| hyuA; N-methylhydantoinase A [EC:3.5.2.14] | 0.00312 | 0.00060 | 0.00217 | 0.00002 | 0.00061 | 65 | 0.00379 | 0.00008 | 0.00091 | 93 | 0.00110 | -1.46740 | 149.798 | 0.14436 | 0.77376 |
| nac; LysR family transcriptional regulator, nitrogen assimilation regulatory protein | 0.00312 | 0.00049 | 0.00225 | 0.00002 | 0.00059 | 65 | 0.00372 | 0.00005 | 0.00072 | 93 | 0.00093 | -1.57407 | 155.904 | 0.11750 | 0.77376 |
| rfbN; rhamnosyltransferase [EC:2.4.1.-] | 0.00312 | 0.00060 | 0.00386 | 0.00010 | 0.00124 | 65 | 0.00260 | 0.00003 | 0.00054 | 93 | 0.00135 | 0.92729 | 88.399 | 0.35630 | 0.79399 |
| E6.3.4.6; urea carboxylase [EC:6.3.4.6] | 0.00312 | 0.00048 | 0.00270 | 0.00003 | 0.00062 | 65 | 0.00341 | 0.00004 | 0.00068 | 93 | 0.00092 | -0.76499 | 154.916 | 0.44544 | 0.83300 |
| golT; Au+-exporting ATPase [EC:3.6.1.-] | 0.00312 | 0.00062 | 0.00275 | 0.00006 | 0.00096 | 65 | 0.00338 | 0.00006 | 0.00081 | 93 | 0.00126 | -0.50114 | 138.778 | 0.61707 | 0.88903 |
| epsO; pyruvyl transferase EpsO [EC:2.-.-.-] | 0.00311 | 0.00058 | 0.00361 | 0.00006 | 0.00096 | 65 | 0.00276 | 0.00005 | 0.00072 | 93 | 0.00119 | 0.71237 | 128.125 | 0.47753 | 0.84209 |
| pbp5, pbp4, pbp3; penicillin-binding protein | 0.00311 | 0.00067 | 0.00308 | 0.00014 | 0.00144 | 65 | 0.00314 | 0.00003 | 0.00053 | 93 | 0.00154 | -0.03738 | 81.543 | 0.97028 | 0.99343 |
| yfbT, yniC; sugar-phosphatase [EC:3.1.3.23] | 0.00311 | 0.00050 | 0.00285 | 0.00003 | 0.00070 | 65 | 0.00330 | 0.00004 | 0.00070 | 93 | 0.00098 | -0.45645 | 150.958 | 0.64872 | 0.90350 |
| K07488; transposase | 0.00310 | 0.00056 | 0.00319 | 0.00006 | 0.00094 | 65 | 0.00303 | 0.00004 | 0.00069 | 93 | 0.00117 | 0.14136 | 125.215 | 0.88781 | 0.97039 |
| citA, tcuC; MFS transporter, MHS family, citrate/tricarballylate:H+ symporter | 0.00310 | 0.00071 | 0.00249 | 0.00006 | 0.00093 | 65 | 0.00352 | 0.00010 | 0.00102 | 93 | 0.00138 | -0.74613 | 154.962 | 0.45672 | 0.83438 |
| nosD; nitrous oxidase accessory protein | 0.00309 | 0.00048 | 0.00220 | 0.00002 | 0.00049 | 65 | 0.00372 | 0.00005 | 0.00073 | 93 | 0.00088 | -1.72145 | 150.224 | 0.08723 | 0.76567 |
| strB; streptomycin 6-kinase [EC:2.7.1.72] | 0.00309 | 0.00060 | 0.00281 | 0.00006 | 0.00097 | 65 | 0.00329 | 0.00005 | 0.00076 | 93 | 0.00123 | -0.38723 | 132.538 | 0.69920 | 0.91867 |
| hpaG; 5-oxopent-3-ene-1,2,5-tricarboxylate decarboxylase / 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase [EC:4.1.1.68 5.3.3.-] | 0.00309 | 0.00066 | 0.00232 | 0.00004 | 0.00080 | 65 | 0.00363 | 0.00009 | 0.00098 | 93 | 0.00127 | -1.03764 | 155.936 | 0.30104 | 0.77376 |
| paaA; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaA [EC:1.14.13.149] | 0.00307 | 0.00041 | 0.00257 | 0.00002 | 0.00053 | 65 | 0.00342 | 0.00003 | 0.00059 | 93 | 0.00080 | -1.05914 | 155.150 | 0.29118 | 0.77376 |
| sdh; serine 3-dehydrogenase (NADP+) [EC:1.1.1.276] | 0.00307 | 0.00061 | 0.00273 | 0.00006 | 0.00100 | 65 | 0.00331 | 0.00005 | 0.00076 | 93 | 0.00126 | -0.45762 | 129.872 | 0.64799 | 0.90350 |
| ykkC; paired small multidrug resistance pump | 0.00306 | 0.00077 | 0.00218 | 0.00004 | 0.00080 | 65 | 0.00367 | 0.00013 | 0.00119 | 93 | 0.00144 | -1.03633 | 150.091 | 0.30171 | 0.77376 |
| dmpA, dap; D-aminopeptidase [EC:3.4.11.19] | 0.00306 | 0.00058 | 0.00153 | 0.00001 | 0.00048 | 65 | 0.00412 | 0.00008 | 0.00091 | 93 | 0.00103 | -2.51046 | 135.004 | 0.01324 | 0.59095 |
| ykkD; paired small multidrug resistance pump | 0.00306 | 0.00077 | 0.00218 | 0.00004 | 0.00080 | 65 | 0.00367 | 0.00013 | 0.00119 | 93 | 0.00144 | -1.04231 | 150.089 | 0.29895 | 0.77376 |
| E4.1.99.2; tyrosine phenol-lyase [EC:4.1.99.2] | 0.00306 | 0.00077 | 0.00484 | 0.00017 | 0.00163 | 65 | 0.00181 | 0.00004 | 0.00064 | 93 | 0.00175 | 1.73211 | 83.648 | 0.08694 | 0.76567 |
| exaA; alcohol dehydrogenase (cytochrome c) [EC:1.1.2.8] | 0.00305 | 0.00054 | 0.00383 | 0.00007 | 0.00105 | 65 | 0.00251 | 0.00003 | 0.00053 | 93 | 0.00118 | 1.11433 | 96.801 | 0.26790 | 0.77376 |
| PTS-Gfr-EIID, gfrD; PTS system, fructoselysine/glucoselysine-specific IID component | 0.00305 | 0.00075 | 0.00448 | 0.00017 | 0.00162 | 65 | 0.00205 | 0.00003 | 0.00057 | 93 | 0.00172 | 1.41561 | 79.700 | 0.16079 | 0.77376 |
| sat, met3; sulfate adenylyltransferase [EC:2.7.7.4] | 0.00304 | 0.00058 | 0.00204 | 0.00003 | 0.00065 | 65 | 0.00374 | 0.00007 | 0.00086 | 93 | 0.00108 | -1.57310 | 154.428 | 0.11774 | 0.77376 |
| K16164; acylpyruvate hydrolase [EC:3.7.1.5] | 0.00304 | 0.00057 | 0.00341 | 0.00007 | 0.00101 | 65 | 0.00278 | 0.00004 | 0.00067 | 93 | 0.00121 | 0.51531 | 116.651 | 0.60731 | 0.88598 |
| nreC; two-component system, NarL family, response regulator NreC | 0.00304 | 0.00074 | 0.00370 | 0.00013 | 0.00144 | 65 | 0.00258 | 0.00005 | 0.00076 | 93 | 0.00163 | 0.68939 | 99.266 | 0.49218 | 0.84797 |
| mqnA; chorismate dehydratase [EC:4.2.1.151] | 0.00303 | 0.00064 | 0.00307 | 0.00007 | 0.00106 | 65 | 0.00301 | 0.00006 | 0.00080 | 93 | 0.00133 | 0.04856 | 129.087 | 0.96135 | 0.98960 |
| lcyB, crtL1, crtY; lycopene beta-cyclase [EC:5.5.1.19] | 0.00303 | 0.00062 | 0.00231 | 0.00005 | 0.00084 | 65 | 0.00354 | 0.00007 | 0.00087 | 93 | 0.00121 | -1.01461 | 152.641 | 0.31190 | 0.77376 |
| araR; GntR family transcriptional regulator, arabinose operon transcriptional repressor | 0.00303 | 0.00064 | 0.00354 | 0.00010 | 0.00126 | 65 | 0.00268 | 0.00004 | 0.00065 | 93 | 0.00142 | 0.60939 | 97.600 | 0.54368 | 0.86313 |
| aroG, aroA; 3-deoxy-7-phosphoheptulonate synthase / chorismate mutase [EC:2.5.1.54 5.4.99.5] | 0.00303 | 0.00059 | 0.00156 | 0.00002 | 0.00061 | 65 | 0.00406 | 0.00007 | 0.00089 | 93 | 0.00108 | -2.32246 | 150.584 | 0.02155 | 0.59153 |
| sbmA, bacA; peptide/bleomycin uptake transporter | 0.00303 | 0.00048 | 0.00219 | 0.00001 | 0.00043 | 65 | 0.00362 | 0.00005 | 0.00075 | 93 | 0.00087 | -1.64517 | 140.137 | 0.10218 | 0.77376 |
| dgoA; 2-dehydro-3-deoxyphosphogalactonate aldolase [EC:4.1.2.21] | 0.00303 | 0.00048 | 0.00163 | 0.00001 | 0.00043 | 65 | 0.00401 | 0.00005 | 0.00073 | 93 | 0.00085 | -2.79889 | 141.954 | 0.00584 | 0.54022 |
| gdh; glucose 1-dehydrogenase [EC:1.1.1.47] | 0.00302 | 0.00055 | 0.00215 | 0.00003 | 0.00066 | 65 | 0.00363 | 0.00006 | 0.00080 | 93 | 0.00104 | -1.42610 | 155.994 | 0.15584 | 0.77376 |
| norB, norC; MFS transporter, DHA2 family, multidrug resistance protein | 0.00302 | 0.00089 | 0.00208 | 0.00007 | 0.00107 | 65 | 0.00368 | 0.00016 | 0.00132 | 93 | 0.00170 | -0.94293 | 155.916 | 0.34718 | 0.78983 |
| cheX; chemotaxis protein CheX | 0.00302 | 0.00060 | 0.00307 | 0.00007 | 0.00103 | 65 | 0.00299 | 0.00005 | 0.00072 | 93 | 0.00126 | 0.06681 | 121.859 | 0.94684 | 0.98674 |
| PTR1; pteridine reductase [EC:1.5.1.33] | 0.00302 | 0.00060 | 0.00304 | 0.00006 | 0.00099 | 65 | 0.00301 | 0.00005 | 0.00075 | 93 | 0.00124 | 0.02568 | 128.595 | 0.97955 | 0.99657 |
| eutM; ethanolamine utilization protein EutM | 0.00302 | 0.00081 | 0.00517 | 0.00020 | 0.00175 | 65 | 0.00151 | 0.00003 | 0.00059 | 93 | 0.00184 | 1.98495 | 78.607 | 0.05064 | 0.68163 |
| TC.OMF; outer membrane factor, OMF family | 0.00302 | 0.00078 | 0.00492 | 0.00018 | 0.00165 | 65 | 0.00169 | 0.00003 | 0.00061 | 93 | 0.00176 | 1.84025 | 81.535 | 0.06937 | 0.73522 |
| nikR; CopG family transcriptional regulator, nickel-responsive regulator | 0.00302 | 0.00053 | 0.00201 | 0.00001 | 0.00042 | 65 | 0.00372 | 0.00007 | 0.00085 | 93 | 0.00095 | -1.79996 | 130.855 | 0.07417 | 0.74128 |
| comB; 2-phosphosulfolactate phosphatase [EC:3.1.3.71] | 0.00301 | 0.00048 | 0.00210 | 0.00002 | 0.00057 | 65 | 0.00365 | 0.00005 | 0.00071 | 93 | 0.00091 | -1.69733 | 155.741 | 0.09163 | 0.76567 |
| mas; Mce-associated membrane protein | 0.00301 | 0.00148 | 0.00065 | 0.00001 | 0.00032 | 65 | 0.00466 | 0.00058 | 0.00250 | 93 | 0.00252 | -1.59135 | 95.088 | 0.11485 | 0.77376 |
| golS; MerR family transcriptional regulator, gold-responsive activator of gol and ges genes | 0.00301 | 0.00060 | 0.00277 | 0.00006 | 0.00096 | 65 | 0.00318 | 0.00006 | 0.00078 | 93 | 0.00123 | -0.33846 | 134.840 | 0.73554 | 0.92721 |
| aroKB; shikimate kinase / 3-dehydroquinate synthase [EC:2.7.1.71 4.2.3.4] | 0.00301 | 0.00052 | 0.00295 | 0.00005 | 0.00090 | 65 | 0.00306 | 0.00004 | 0.00063 | 93 | 0.00110 | -0.09853 | 121.263 | 0.92168 | 0.97905 |
| natA; sodium transport system ATP-binding protein [EC:3.6.3.7] | 0.00300 | 0.00061 | 0.00281 | 0.00006 | 0.00097 | 65 | 0.00314 | 0.00006 | 0.00078 | 93 | 0.00125 | -0.27114 | 135.160 | 0.78670 | 0.94461 |
| hyuB; N-methylhydantoinase B [EC:3.5.2.14] | 0.00300 | 0.00057 | 0.00212 | 0.00002 | 0.00061 | 65 | 0.00362 | 0.00007 | 0.00086 | 93 | 0.00105 | -1.43196 | 152.406 | 0.15420 | 0.77376 |
| yxdL; putative ABC transport system ATP-binding protein | 0.00300 | 0.00077 | 0.00314 | 0.00010 | 0.00122 | 65 | 0.00290 | 0.00009 | 0.00100 | 93 | 0.00158 | 0.15289 | 136.474 | 0.87871 | 0.96759 |
| natB; sodium transport system permease protein | 0.00300 | 0.00061 | 0.00281 | 0.00006 | 0.00097 | 65 | 0.00313 | 0.00006 | 0.00079 | 93 | 0.00125 | -0.26096 | 135.204 | 0.79452 | 0.94783 |
| mntR; DtxR family transcriptional regulator, manganese transport regulator | 0.00299 | 0.00059 | 0.00295 | 0.00006 | 0.00097 | 65 | 0.00302 | 0.00005 | 0.00075 | 93 | 0.00123 | -0.06044 | 131.592 | 0.95190 | 0.98792 |
| pcrB; putative glycerol-1-phosphate prenyltransferase [EC:2.5.1.-] | 0.00299 | 0.00059 | 0.00167 | 0.00002 | 0.00061 | 65 | 0.00391 | 0.00007 | 0.00089 | 93 | 0.00108 | -2.07206 | 151.024 | 0.03996 | 0.63577 |
| cst2, cas7; CRISPR-associated protein Cst2 | 0.00299 | 0.00081 | 0.00497 | 0.00018 | 0.00167 | 65 | 0.00160 | 0.00004 | 0.00069 | 93 | 0.00181 | 1.86380 | 85.910 | 0.06577 | 0.73334 |
| wbqP; O-antigen biosynthesis protein WbqP | 0.00298 | 0.00063 | 0.00285 | 0.00008 | 0.00111 | 65 | 0.00308 | 0.00005 | 0.00074 | 93 | 0.00134 | -0.16782 | 117.742 | 0.86701 | 0.96327 |
| nos; nitric-oxide synthase, bacterial [EC:1.14.14.47] | 0.00297 | 0.00059 | 0.00152 | 0.00002 | 0.00061 | 65 | 0.00399 | 0.00007 | 0.00090 | 93 | 0.00108 | -2.27708 | 150.149 | 0.02419 | 0.59764 |
| K07097; uncharacterized protein | 0.00297 | 0.00076 | 0.00492 | 0.00017 | 0.00162 | 65 | 0.00161 | 0.00003 | 0.00057 | 93 | 0.00172 | 1.92205 | 79.899 | 0.05816 | 0.71816 |
| K07079; uncharacterized protein | 0.00296 | 0.00077 | 0.00213 | 0.00004 | 0.00081 | 65 | 0.00354 | 0.00013 | 0.00118 | 93 | 0.00143 | -0.98458 | 150.640 | 0.32641 | 0.77376 |
| eutT; ethanolamine utilization cobalamin adenosyltransferase [EC:2.5.1.17] | 0.00295 | 0.00081 | 0.00505 | 0.00020 | 0.00175 | 65 | 0.00148 | 0.00003 | 0.00059 | 93 | 0.00184 | 1.93609 | 78.642 | 0.05645 | 0.70577 |
| yscC, sctC, ssaC; type III secretion protein C | 0.00295 | 0.00073 | 0.00203 | 0.00004 | 0.00081 | 65 | 0.00358 | 0.00011 | 0.00110 | 93 | 0.00136 | -1.13618 | 153.753 | 0.25765 | 0.77376 |
| paaC; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaC [EC:1.14.13.149] | 0.00294 | 0.00040 | 0.00254 | 0.00002 | 0.00053 | 65 | 0.00322 | 0.00003 | 0.00057 | 93 | 0.00077 | -0.88117 | 154.213 | 0.37960 | 0.79852 |
| nifE; nitrogenase molybdenum-cofactor synthesis protein NifE | 0.00294 | 0.00060 | 0.00320 | 0.00007 | 0.00104 | 65 | 0.00276 | 0.00005 | 0.00071 | 93 | 0.00126 | 0.35116 | 119.641 | 0.72609 | 0.92177 |
| prpR; transcriptional regulator, propionate catabolism operon regulatory protein | 0.00294 | 0.00060 | 0.00271 | 0.00006 | 0.00096 | 65 | 0.00310 | 0.00006 | 0.00078 | 93 | 0.00124 | -0.31138 | 134.797 | 0.75599 | 0.93669 |
| algD; GDP-mannose 6-dehydrogenase [EC:1.1.1.132] | 0.00294 | 0.00052 | 0.00343 | 0.00006 | 0.00092 | 65 | 0.00260 | 0.00003 | 0.00060 | 93 | 0.00110 | 0.75699 | 115.331 | 0.45060 | 0.83438 |
| xylH; D-xylose transport system permease protein | 0.00294 | 0.00051 | 0.00180 | 0.00001 | 0.00046 | 65 | 0.00373 | 0.00006 | 0.00079 | 93 | 0.00091 | -2.11559 | 141.200 | 0.03613 | 0.62411 |
| xapB; MFS transporter, NHS family, xanthosine permease | 0.00293 | 0.00040 | 0.00252 | 0.00001 | 0.00045 | 65 | 0.00322 | 0.00003 | 0.00059 | 93 | 0.00075 | -0.93725 | 154.656 | 0.35009 | 0.79120 |
| gerKA; spore germination protein KA | 0.00293 | 0.00076 | 0.00122 | 0.00002 | 0.00062 | 65 | 0.00411 | 0.00013 | 0.00120 | 93 | 0.00135 | -2.13745 | 133.334 | 0.03439 | 0.62130 |
| paaB; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaB | 0.00292 | 0.00040 | 0.00249 | 0.00002 | 0.00052 | 65 | 0.00322 | 0.00003 | 0.00057 | 93 | 0.00077 | -0.94286 | 154.346 | 0.34723 | 0.78983 |
| paaD; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaD | 0.00292 | 0.00040 | 0.00249 | 0.00002 | 0.00052 | 65 | 0.00322 | 0.00003 | 0.00057 | 93 | 0.00077 | -0.94286 | 154.346 | 0.34723 | 0.78983 |
| K02479; two-component system, NarL family, response regulator | 0.00292 | 0.00062 | 0.00240 | 0.00006 | 0.00096 | 65 | 0.00329 | 0.00006 | 0.00081 | 93 | 0.00125 | -0.70817 | 138.971 | 0.48003 | 0.84324 |
| K17213; inositol transport system substrate-binding protein | 0.00292 | 0.00064 | 0.00172 | 0.00003 | 0.00067 | 65 | 0.00375 | 0.00009 | 0.00098 | 93 | 0.00118 | -1.71170 | 150.863 | 0.08901 | 0.76567 |
| spoIIP; stage II sporulation protein P | 0.00291 | 0.00060 | 0.00143 | 0.00002 | 0.00055 | 65 | 0.00395 | 0.00008 | 0.00093 | 93 | 0.00108 | -2.33569 | 143.277 | 0.02089 | 0.59147 |
| sdrC_D_E; serine-aspartate repeat-containing protein C/D/E | 0.00291 | 0.00089 | 0.00201 | 0.00007 | 0.00107 | 65 | 0.00353 | 0.00016 | 0.00131 | 93 | 0.00169 | -0.89749 | 155.928 | 0.37084 | 0.79607 |
| spoIVCA; site-specific DNA recombinase | 0.00291 | 0.00054 | 0.00266 | 0.00005 | 0.00090 | 65 | 0.00307 | 0.00004 | 0.00067 | 93 | 0.00112 | -0.36830 | 127.551 | 0.71326 | 0.92004 |
| HAAO; 3-hydroxyanthranilate 3,4-dioxygenase [EC:1.13.11.6] | 0.00290 | 0.00062 | 0.00263 | 0.00006 | 0.00096 | 65 | 0.00309 | 0.00006 | 0.00081 | 93 | 0.00125 | -0.36925 | 138.369 | 0.71250 | 0.92004 |
| K09795; uncharacterized protein | 0.00290 | 0.00041 | 0.00241 | 0.00002 | 0.00055 | 65 | 0.00324 | 0.00003 | 0.00059 | 93 | 0.00080 | -1.03023 | 154.270 | 0.30451 | 0.77376 |
| E3.1.1.11; pectinesterase [EC:3.1.1.11] | 0.00290 | 0.00054 | 0.00160 | 0.00001 | 0.00032 | 65 | 0.00380 | 0.00007 | 0.00087 | 93 | 0.00093 | -2.35821 | 115.232 | 0.02005 | 0.59095 |
| nosY; Cu-processing system permease protein | 0.00289 | 0.00053 | 0.00189 | 0.00001 | 0.00043 | 65 | 0.00359 | 0.00007 | 0.00084 | 93 | 0.00094 | -1.80364 | 133.315 | 0.07355 | 0.74128 |
| adiC; arginine:agmatine antiporter | 0.00289 | 0.00060 | 0.00260 | 0.00006 | 0.00095 | 65 | 0.00309 | 0.00006 | 0.00078 | 93 | 0.00123 | -0.39033 | 136.202 | 0.69690 | 0.91823 |
| pfeR, pirR; two-component system, OmpR family, response regulator PfeR | 0.00289 | 0.00054 | 0.00393 | 0.00007 | 0.00105 | 65 | 0.00216 | 0.00003 | 0.00053 | 93 | 0.00117 | 1.51172 | 96.941 | 0.13386 | 0.77376 |
| pfeS, pirS; two-component system, OmpR family, sensor histidine kinase PfeS [EC:2.7.13.3] | 0.00289 | 0.00054 | 0.00393 | 0.00007 | 0.00105 | 65 | 0.00216 | 0.00003 | 0.00053 | 93 | 0.00117 | 1.51172 | 96.941 | 0.13386 | 0.77376 |
| LRA3, yfaW; L-rhamnonate dehydratase [EC:4.2.1.90] | 0.00288 | 0.00073 | 0.00443 | 0.00016 | 0.00159 | 65 | 0.00180 | 0.00003 | 0.00054 | 93 | 0.00168 | 1.56841 | 79.026 | 0.12078 | 0.77376 |
| arr; rifampin ADP-ribosylating transferase | 0.00288 | 0.00059 | 0.00282 | 0.00006 | 0.00097 | 65 | 0.00291 | 0.00005 | 0.00074 | 93 | 0.00122 | -0.07406 | 128.956 | 0.94108 | 0.98528 |
| GLYK; D-glycerate 3-kinase [EC:2.7.1.31] | 0.00288 | 0.00060 | 0.00274 | 0.00006 | 0.00099 | 65 | 0.00297 | 0.00005 | 0.00075 | 93 | 0.00124 | -0.18096 | 129.165 | 0.85668 | 0.95956 |
| yxdM; putative ABC transport system permease protein | 0.00287 | 0.00078 | 0.00251 | 0.00007 | 0.00107 | 65 | 0.00312 | 0.00011 | 0.00110 | 93 | 0.00153 | -0.40009 | 152.411 | 0.68965 | 0.91567 |
| spoIIAA; stage II sporulation protein AA (anti-sigma F factor antagonist) | 0.00286 | 0.00059 | 0.00160 | 0.00002 | 0.00056 | 65 | 0.00375 | 0.00008 | 0.00092 | 93 | 0.00108 | -2.00129 | 144.565 | 0.04723 | 0.66255 |
| mqnC; cyclic dehypoxanthinyl futalosine synthase [EC:1.21.98.1] | 0.00286 | 0.00063 | 0.00306 | 0.00007 | 0.00106 | 65 | 0.00272 | 0.00006 | 0.00078 | 93 | 0.00131 | 0.25869 | 125.976 | 0.79630 | 0.94791 |
| ccrM; modification methylase [EC:2.1.1.72] | 0.00286 | 0.00042 | 0.00155 | 0.00001 | 0.00029 | 65 | 0.00378 | 0.00004 | 0.00068 | 93 | 0.00074 | -3.03264 | 123.522 | 0.00296 | 0.46257 |
| cynR; LysR family transcriptional regulator, cyn operon transcriptional activator | 0.00286 | 0.00042 | 0.00203 | 0.00001 | 0.00041 | 65 | 0.00344 | 0.00004 | 0.00065 | 93 | 0.00077 | -1.82542 | 146.622 | 0.06997 | 0.73522 |
| kinB; two-component system, NtrC family, sensor histidine kinase KinB [EC:2.7.13.3] | 0.00286 | 0.00051 | 0.00340 | 0.00005 | 0.00091 | 65 | 0.00248 | 0.00003 | 0.00059 | 93 | 0.00108 | 0.85822 | 114.906 | 0.39256 | 0.80459 |
| K09978; uncharacterized protein | 0.00285 | 0.00045 | 0.00322 | 0.00003 | 0.00072 | 65 | 0.00259 | 0.00003 | 0.00057 | 93 | 0.00092 | 0.68961 | 132.142 | 0.49165 | 0.84797 |
| resE; two-component system, OmpR family, sensor histidine kinase ResE [EC:2.7.13.3] | 0.00285 | 0.00059 | 0.00155 | 0.00002 | 0.00061 | 65 | 0.00375 | 0.00007 | 0.00089 | 93 | 0.00108 | -2.03991 | 150.978 | 0.04310 | 0.65559 |
| K06934; uncharacterized protein | 0.00284 | 0.00051 | 0.00217 | 0.00004 | 0.00076 | 65 | 0.00332 | 0.00004 | 0.00069 | 93 | 0.00103 | -1.12021 | 145.538 | 0.26447 | 0.77376 |
| E1.1.1.219; dihydroflavonol-4-reductase [EC:1.1.1.219] | 0.00284 | 0.00045 | 0.00215 | 0.00002 | 0.00051 | 65 | 0.00332 | 0.00004 | 0.00068 | 93 | 0.00085 | -1.37282 | 154.303 | 0.17180 | 0.77376 |
| cas5t; CRISPR-associated protein Cas5t | 0.00284 | 0.00080 | 0.00461 | 0.00018 | 0.00165 | 65 | 0.00160 | 0.00004 | 0.00069 | 93 | 0.00179 | 1.68724 | 86.503 | 0.09516 | 0.76609 |
| spoIVB; stage IV sporulation protein B [EC:3.4.21.116] | 0.00284 | 0.00059 | 0.00142 | 0.00002 | 0.00055 | 65 | 0.00383 | 0.00008 | 0.00092 | 93 | 0.00107 | -2.25449 | 143.763 | 0.02568 | 0.59941 |
| K09763; uncharacterized protein | 0.00284 | 0.00061 | 0.00160 | 0.00002 | 0.00061 | 65 | 0.00370 | 0.00008 | 0.00093 | 93 | 0.00111 | -1.88697 | 149.043 | 0.06111 | 0.73001 |
| arnT, pmrK; 4-amino-4-deoxy-L-arabinose transferase [EC:2.4.2.43] | 0.00283 | 0.00071 | 0.00222 | 0.00004 | 0.00082 | 65 | 0.00326 | 0.00011 | 0.00107 | 93 | 0.00135 | -0.77143 | 155.015 | 0.44163 | 0.83186 |
| PTS-Gfr-EIIB, gfrB; PTS system, fructoselysine/glucoselysine-specific IIB component [EC:2.7.1.-] | 0.00283 | 0.00074 | 0.00431 | 0.00017 | 0.00162 | 65 | 0.00180 | 0.00003 | 0.00056 | 93 | 0.00171 | 1.46586 | 79.298 | 0.14664 | 0.77376 |
| nosL; copper chaperone NosL | 0.00283 | 0.00047 | 0.00197 | 0.00001 | 0.00045 | 65 | 0.00343 | 0.00005 | 0.00072 | 93 | 0.00085 | -1.71575 | 145.215 | 0.08834 | 0.76567 |
| SERPINB; serpin B | 0.00283 | 0.00040 | 0.00259 | 0.00002 | 0.00052 | 65 | 0.00299 | 0.00003 | 0.00058 | 93 | 0.00078 | -0.52077 | 154.829 | 0.60327 | 0.88434 |
| cst1, cas8a; CRISPR-associated protein Cst1 | 0.00282 | 0.00080 | 0.00461 | 0.00018 | 0.00165 | 65 | 0.00157 | 0.00004 | 0.00069 | 93 | 0.00179 | 1.70120 | 86.527 | 0.09250 | 0.76567 |
| K09914; putative lipoprotein | 0.00282 | 0.00044 | 0.00226 | 0.00002 | 0.00052 | 65 | 0.00321 | 0.00004 | 0.00066 | 93 | 0.00084 | -1.13235 | 155.247 | 0.25923 | 0.77376 |
| cbl; LysR family transcriptional regulator, cys regulon transcriptional activator | 0.00281 | 0.00047 | 0.00231 | 0.00002 | 0.00051 | 65 | 0.00316 | 0.00005 | 0.00071 | 93 | 0.00087 | -0.96799 | 153.402 | 0.33457 | 0.78124 |
| srlD; sorbitol-6-phosphate 2-dehydrogenase [EC:1.1.1.140] | 0.00281 | 0.00052 | 0.00337 | 0.00007 | 0.00101 | 65 | 0.00241 | 0.00003 | 0.00052 | 93 | 0.00113 | 0.84784 | 98.381 | 0.39858 | 0.80621 |
| oqxA; membrane fusion protein, multidrug efflux system | 0.00281 | 0.00059 | 0.00289 | 0.00006 | 0.00096 | 65 | 0.00275 | 0.00005 | 0.00075 | 93 | 0.00122 | 0.11201 | 130.838 | 0.91099 | 0.97664 |
| atuE; isohexenylglutaconyl-CoA hydratase [EC:4.2.1.57] | 0.00281 | 0.00050 | 0.00252 | 0.00003 | 0.00067 | 65 | 0.00301 | 0.00005 | 0.00072 | 93 | 0.00098 | -0.49528 | 154.151 | 0.62111 | 0.89091 |
| DCAA; acyl-CoA dehydrogenase [EC:1.3.99.-] | 0.00280 | 0.00070 | 0.00109 | 0.00001 | 0.00037 | 65 | 0.00400 | 0.00012 | 0.00115 | 93 | 0.00121 | -2.39791 | 110.244 | 0.01817 | 0.59095 |
| folX; D-erythro-7,8-dihydroneopterin triphosphate epimerase [EC:5.1.99.7] | 0.00280 | 0.00047 | 0.00293 | 0.00004 | 0.00075 | 65 | 0.00271 | 0.00003 | 0.00061 | 93 | 0.00097 | 0.22544 | 135.462 | 0.82198 | 0.95506 |
| cbiX; sirohydrochlorin cobaltochelatase [EC:4.99.1.3] | 0.00280 | 0.00071 | 0.00159 | 0.00003 | 0.00064 | 65 | 0.00365 | 0.00011 | 0.00111 | 93 | 0.00128 | -1.60866 | 141.164 | 0.10993 | 0.77376 |
| qoxA; cytochrome aa3-600 menaquinol oxidase subunit II [EC:1.10.3.12] | 0.00280 | 0.00075 | 0.00161 | 0.00004 | 0.00077 | 65 | 0.00364 | 0.00012 | 0.00115 | 93 | 0.00139 | -1.46358 | 150.080 | 0.14540 | 0.77376 |
| qoxC; cytochrome aa3-600 menaquinol oxidase subunit III [EC:1.10.3.12] | 0.00280 | 0.00075 | 0.00161 | 0.00004 | 0.00077 | 65 | 0.00363 | 0.00012 | 0.00115 | 93 | 0.00139 | -1.46251 | 150.080 | 0.14569 | 0.77376 |
| qoxD; cytochrome aa3-600 menaquinol oxidase subunit IV [EC:1.10.3.12] | 0.00280 | 0.00075 | 0.00161 | 0.00004 | 0.00077 | 65 | 0.00363 | 0.00012 | 0.00115 | 93 | 0.00139 | -1.46251 | 150.080 | 0.14569 | 0.77376 |
| algG; mannuronan 5-epimerase [EC:5.1.3.37] | 0.00280 | 0.00051 | 0.00341 | 0.00006 | 0.00092 | 65 | 0.00237 | 0.00003 | 0.00058 | 93 | 0.00109 | 0.95268 | 112.647 | 0.34279 | 0.78567 |
| zraR, hydG; two-component system, NtrC family, response regulator HydG | 0.00279 | 0.00055 | 0.00180 | 0.00001 | 0.00039 | 65 | 0.00349 | 0.00007 | 0.00088 | 93 | 0.00097 | -1.74359 | 124.750 | 0.08369 | 0.76567 |
| acuA; acetoin utilization protein AcuA [EC:2.3.1.-] | 0.00279 | 0.00059 | 0.00145 | 0.00002 | 0.00061 | 65 | 0.00372 | 0.00007 | 0.00089 | 93 | 0.00108 | -2.10623 | 150.678 | 0.03684 | 0.62759 |
| oqxB; multidrug efflux pump | 0.00278 | 0.00059 | 0.00283 | 0.00006 | 0.00097 | 65 | 0.00275 | 0.00005 | 0.00075 | 93 | 0.00122 | 0.06323 | 130.766 | 0.94968 | 0.98774 |
| rihA; pyrimidine-specific ribonucleoside hydrolase [EC:3.2.-.-] | 0.00278 | 0.00073 | 0.00344 | 0.00016 | 0.00155 | 65 | 0.00231 | 0.00004 | 0.00063 | 93 | 0.00167 | 0.67873 | 85.442 | 0.49914 | 0.84856 |
| yaaH; spore germination protein | 0.00277 | 0.00075 | 0.00090 | 0.00002 | 0.00058 | 65 | 0.00408 | 0.00013 | 0.00120 | 93 | 0.00133 | -2.38872 | 130.355 | 0.01834 | 0.59095 |
| tilS-hprT; bifunctional protein TilS/HprT [EC:6.3.4.19 2.4.2.8] | 0.00276 | 0.00071 | 0.00380 | 0.00016 | 0.00157 | 65 | 0.00204 | 0.00002 | 0.00052 | 93 | 0.00165 | 1.06454 | 78.094 | 0.29036 | 0.77376 |
| gpr; spore protease [EC:3.4.24.78] | 0.00276 | 0.00059 | 0.00142 | 0.00002 | 0.00055 | 65 | 0.00370 | 0.00008 | 0.00092 | 93 | 0.00107 | -2.13241 | 143.956 | 0.03467 | 0.62130 |
| spoIIAB; stage II sporulation protein AB (anti-sigma F factor) [EC:2.7.11.1] | 0.00276 | 0.00059 | 0.00142 | 0.00002 | 0.00055 | 65 | 0.00370 | 0.00008 | 0.00092 | 93 | 0.00107 | -2.13241 | 143.956 | 0.03467 | 0.62130 |
| spoIVA; stage IV sporulation protein A | 0.00276 | 0.00059 | 0.00142 | 0.00002 | 0.00055 | 65 | 0.00370 | 0.00008 | 0.00092 | 93 | 0.00107 | -2.13241 | 143.956 | 0.03467 | 0.62130 |
| glgM; alpha-maltose-1-phosphate synthase [EC:2.4.1.342] | 0.00276 | 0.00035 | 0.00263 | 0.00002 | 0.00049 | 65 | 0.00285 | 0.00002 | 0.00048 | 93 | 0.00068 | -0.32362 | 150.768 | 0.74668 | 0.93207 |
| phnM; alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase [EC:3.6.1.63] | 0.00276 | 0.00048 | 0.00129 | 0.00001 | 0.00037 | 65 | 0.00378 | 0.00005 | 0.00075 | 93 | 0.00084 | -2.97182 | 130.868 | 0.00352 | 0.48387 |
| spoIIIAC; stage III sporulation protein AC | 0.00275 | 0.00059 | 0.00142 | 0.00002 | 0.00055 | 65 | 0.00369 | 0.00008 | 0.00091 | 93 | 0.00107 | -2.12459 | 143.991 | 0.03533 | 0.62411 |
| spoIIIAH; stage III sporulation protein AH | 0.00275 | 0.00059 | 0.00142 | 0.00002 | 0.00055 | 65 | 0.00369 | 0.00008 | 0.00091 | 93 | 0.00107 | -2.12459 | 143.991 | 0.03533 | 0.62411 |
| THI4, THI1; cysteine-dependent adenosine diphosphate thiazole synthase [EC:2.4.2.60] | 0.00275 | 0.00051 | 0.00284 | 0.00003 | 0.00071 | 65 | 0.00268 | 0.00005 | 0.00071 | 93 | 0.00100 | 0.15552 | 151.613 | 0.87661 | 0.96705 |
| kynB; arylformamidase [EC:3.5.1.9] | 0.00275 | 0.00052 | 0.00114 | 0.00001 | 0.00035 | 65 | 0.00387 | 0.00006 | 0.00083 | 93 | 0.00090 | -3.04675 | 122.006 | 0.00284 | 0.46257 |
| spoIIIAG; stage III sporulation protein AG | 0.00274 | 0.00059 | 0.00142 | 0.00002 | 0.00055 | 65 | 0.00367 | 0.00008 | 0.00091 | 93 | 0.00106 | -2.12525 | 144.540 | 0.03527 | 0.62411 |
| spoVT; AbrB family transcriptional regulator, stage V sporulation protein T | 0.00274 | 0.00059 | 0.00140 | 0.00002 | 0.00055 | 65 | 0.00368 | 0.00008 | 0.00091 | 93 | 0.00107 | -2.13794 | 144.264 | 0.03421 | 0.62130 |
| cphA; cyanophycin synthetase [EC:6.3.2.29 6.3.2.30] | 0.00274 | 0.00066 | 0.00236 | 0.00004 | 0.00079 | 65 | 0.00301 | 0.00009 | 0.00099 | 93 | 0.00126 | -0.51312 | 155.772 | 0.60860 | 0.88654 |
| glcT; transcriptional antiterminator | 0.00274 | 0.00051 | 0.00166 | 0.00003 | 0.00066 | 65 | 0.00349 | 0.00005 | 0.00073 | 93 | 0.00098 | -1.86126 | 154.962 | 0.06460 | 0.73334 |
| yscW, sctW; type III secretion protein W | 0.00274 | 0.00071 | 0.00225 | 0.00006 | 0.00093 | 65 | 0.00308 | 0.00010 | 0.00103 | 93 | 0.00139 | -0.59907 | 155.026 | 0.55000 | 0.86680 |
| rpfF; DSF synthase | 0.00274 | 0.00059 | 0.00266 | 0.00006 | 0.00096 | 65 | 0.00279 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.10750 | 131.068 | 0.91456 | 0.97664 |
| yscL, sctL; type III secretion protein L | 0.00273 | 0.00071 | 0.00223 | 0.00006 | 0.00092 | 65 | 0.00309 | 0.00010 | 0.00101 | 93 | 0.00137 | -0.62959 | 154.845 | 0.52989 | 0.86094 |
| fdsD; formate dehydrogenase subunit delta [EC:1.17.1.9] | 0.00272 | 0.00042 | 0.00232 | 0.00002 | 0.00059 | 65 | 0.00301 | 0.00003 | 0.00058 | 93 | 0.00083 | -0.83465 | 150.645 | 0.40524 | 0.81189 |
| K09859; uncharacterized protein | 0.00272 | 0.00061 | 0.00174 | 0.00002 | 0.00060 | 65 | 0.00340 | 0.00008 | 0.00094 | 93 | 0.00111 | -1.49765 | 147.004 | 0.13637 | 0.77376 |
| spoIIE; stage II sporulation protein E [EC:3.1.3.16] | 0.00272 | 0.00059 | 0.00136 | 0.00002 | 0.00055 | 65 | 0.00367 | 0.00008 | 0.00091 | 93 | 0.00106 | -2.18133 | 143.880 | 0.03078 | 0.61611 |
| rutF; flavin reductase [EC:1.5.1.-] | 0.00272 | 0.00049 | 0.00219 | 0.00002 | 0.00052 | 65 | 0.00309 | 0.00005 | 0.00074 | 93 | 0.00091 | -0.99470 | 152.258 | 0.32146 | 0.77376 |
| hpaE, hpcC; 5-carboxymethyl-2-hydroxymuconic-semialdehyde dehydrogenase [EC:1.2.1.60] | 0.00271 | 0.00046 | 0.00143 | 0.00001 | 0.00042 | 65 | 0.00361 | 0.00005 | 0.00071 | 93 | 0.00083 | -2.63355 | 142.752 | 0.00938 | 0.58112 |
| ich-P; itaconyl-CoA hydratase / mesaconyl-C4 CoA hydratase [EC:4.2.1.56 4.2.1.-] | 0.00271 | 0.00054 | 0.00284 | 0.00004 | 0.00081 | 65 | 0.00262 | 0.00005 | 0.00072 | 93 | 0.00109 | 0.20591 | 142.827 | 0.83715 | 0.95506 |
| KMO; kynurenine 3-monooxygenase [EC:1.14.13.9] | 0.00271 | 0.00059 | 0.00265 | 0.00006 | 0.00096 | 65 | 0.00275 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.08348 | 131.078 | 0.93360 | 0.98428 |
| lytE, cwlF; peptidoglycan DL-endopeptidase LytE [EC:3.4.-.-] | 0.00270 | 0.00077 | 0.00160 | 0.00004 | 0.00077 | 65 | 0.00347 | 0.00013 | 0.00119 | 93 | 0.00142 | -1.31962 | 148.263 | 0.18899 | 0.77376 |
| K10120, msmE; fructooligosaccharide transport system substrate-binding protein | 0.00270 | 0.00059 | 0.00397 | 0.00010 | 0.00125 | 65 | 0.00180 | 0.00002 | 0.00046 | 93 | 0.00133 | 1.62868 | 81.653 | 0.10723 | 0.77376 |
| ydiU; uncharacterized protein | 0.00270 | 0.00043 | 0.00243 | 0.00002 | 0.00055 | 65 | 0.00288 | 0.00004 | 0.00062 | 93 | 0.00083 | -0.54282 | 155.282 | 0.58803 | 0.87732 |
| PSRP4, RPS31; 30S ribosomal protein S31 | 0.00269 | 0.00059 | 0.00257 | 0.00006 | 0.00096 | 65 | 0.00278 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.17727 | 130.897 | 0.85957 | 0.96122 |
| mexI; multidrug efflux pump | 0.00269 | 0.00084 | 0.00238 | 0.00009 | 0.00120 | 65 | 0.00291 | 0.00012 | 0.00116 | 93 | 0.00167 | -0.32137 | 148.874 | 0.74838 | 0.93276 |
| capD; UDP-glucose 4-epimerase [EC:5.1.3.2] | 0.00269 | 0.00046 | 0.00183 | 0.00002 | 0.00050 | 65 | 0.00329 | 0.00005 | 0.00070 | 93 | 0.00086 | -1.69043 | 152.279 | 0.09299 | 0.76567 |
| SUPV3L1, SUV3; ATP-dependent RNA helicase SUPV3L1/SUV3 [EC:3.6.4.13] | 0.00269 | 0.00044 | 0.00165 | 0.00002 | 0.00052 | 65 | 0.00342 | 0.00004 | 0.00065 | 93 | 0.00084 | -2.11617 | 155.751 | 0.03592 | 0.62411 |
| braR, bceR; two-component system, OmpR family, response regulator protein BraR/BceR | 0.00269 | 0.00058 | 0.00282 | 0.00007 | 0.00105 | 65 | 0.00259 | 0.00004 | 0.00067 | 93 | 0.00124 | 0.18913 | 113.593 | 0.85033 | 0.95754 |
| braS, bceS; two-component system, OmpR family, sensor histidine kinase BraS/BceS [EC:2.7.13.3] | 0.00269 | 0.00058 | 0.00282 | 0.00007 | 0.00105 | 65 | 0.00259 | 0.00004 | 0.00067 | 93 | 0.00124 | 0.18913 | 113.593 | 0.85033 | 0.95754 |
| cmlA, cmlB, floR; MFS transporter, DHA1 family, florfenicol/chloramphenicol resistance protein | 0.00269 | 0.00059 | 0.00259 | 0.00006 | 0.00096 | 65 | 0.00275 | 0.00005 | 0.00075 | 93 | 0.00122 | -0.13448 | 131.328 | 0.89323 | 0.97159 |
| pcaT; MFS transporter, MHS family, dicarboxylic acid transporter PcaT | 0.00269 | 0.00059 | 0.00266 | 0.00005 | 0.00086 | 65 | 0.00270 | 0.00006 | 0.00081 | 93 | 0.00119 | -0.03971 | 147.454 | 0.96838 | 0.99264 |
| zapD; cell division protein ZapD | 0.00268 | 0.00042 | 0.00260 | 0.00002 | 0.00062 | 65 | 0.00274 | 0.00003 | 0.00058 | 93 | 0.00085 | -0.16529 | 146.679 | 0.86894 | 0.96463 |
| ynaI, mscMJ; MscS family membrane protein | 0.00268 | 0.00053 | 0.00222 | 0.00002 | 0.00059 | 65 | 0.00299 | 0.00006 | 0.00079 | 93 | 0.00099 | -0.78582 | 153.861 | 0.43318 | 0.82874 |
| K10121, msmF; fructooligosaccharide transport system permease protein | 0.00267 | 0.00059 | 0.00397 | 0.00010 | 0.00125 | 65 | 0.00177 | 0.00002 | 0.00046 | 93 | 0.00133 | 1.65787 | 81.652 | 0.10118 | 0.77376 |
| K10122, msmG; fructooligosaccharide transport system permease protein | 0.00267 | 0.00059 | 0.00397 | 0.00010 | 0.00125 | 65 | 0.00177 | 0.00002 | 0.00046 | 93 | 0.00133 | 1.65787 | 81.652 | 0.10118 | 0.77376 |
| qnr, mcbG; fluoroquinolone resistance protein | 0.00267 | 0.00059 | 0.00272 | 0.00006 | 0.00097 | 65 | 0.00264 | 0.00005 | 0.00075 | 93 | 0.00122 | 0.06704 | 130.447 | 0.94666 | 0.98674 |
| E3.5.1.81; N-acyl-D-amino-acid deacylase [EC:3.5.1.81] | 0.00267 | 0.00040 | 0.00224 | 0.00002 | 0.00048 | 65 | 0.00296 | 0.00003 | 0.00058 | 93 | 0.00076 | -0.95206 | 155.999 | 0.34254 | 0.78567 |
| spoIIR; stage II sporulation protein R | 0.00266 | 0.00057 | 0.00140 | 0.00002 | 0.00055 | 65 | 0.00354 | 0.00007 | 0.00087 | 93 | 0.00103 | -2.07128 | 146.757 | 0.04008 | 0.63577 |
| spoIIID; putative DeoR family transcriptional regulator, stage III sporulation protein D | 0.00266 | 0.00057 | 0.00138 | 0.00002 | 0.00055 | 65 | 0.00354 | 0.00007 | 0.00088 | 93 | 0.00104 | -2.08416 | 146.636 | 0.03888 | 0.63249 |
| yfaE; ferredoxin | 0.00265 | 0.00042 | 0.00308 | 0.00004 | 0.00074 | 65 | 0.00235 | 0.00002 | 0.00049 | 93 | 0.00089 | 0.82036 | 116.514 | 0.41369 | 0.81655 |
| aph3-II; aminoglycoside 3’-phosphotransferase II [EC:2.7.1.95] | 0.00265 | 0.00059 | 0.00240 | 0.00006 | 0.00096 | 65 | 0.00283 | 0.00005 | 0.00075 | 93 | 0.00122 | -0.35420 | 132.108 | 0.72376 | 0.92165 |
| E3.6.3.25; sulfate-transporting ATPase [EC:3.6.3.25] | 0.00265 | 0.00042 | 0.00243 | 0.00002 | 0.00057 | 65 | 0.00280 | 0.00003 | 0.00059 | 93 | 0.00082 | -0.45711 | 152.137 | 0.64824 | 0.90350 |
| E2.7.1.37; protein kinase [EC:2.7.1.37] | 0.00265 | 0.00075 | 0.00454 | 0.00017 | 0.00163 | 65 | 0.00133 | 0.00003 | 0.00055 | 93 | 0.00172 | 1.86879 | 78.557 | 0.06538 | 0.73334 |
| abgB; aminobenzoyl-glutamate utilization protein B | 0.00264 | 0.00043 | 0.00185 | 0.00002 | 0.00056 | 65 | 0.00320 | 0.00004 | 0.00062 | 93 | 0.00084 | -1.60323 | 155.114 | 0.11092 | 0.77376 |
| argF; N-acetylornithine carbamoyltransferase [EC:2.1.3.9] | 0.00264 | 0.00059 | 0.00249 | 0.00006 | 0.00096 | 65 | 0.00274 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.20478 | 130.923 | 0.83806 | 0.95506 |
| leuE; leucine efflux protein | 0.00264 | 0.00043 | 0.00250 | 0.00003 | 0.00063 | 65 | 0.00273 | 0.00003 | 0.00059 | 93 | 0.00086 | -0.27286 | 146.493 | 0.78535 | 0.94445 |
| exoX; exodeoxyribonuclease X [EC:3.1.11.-] | 0.00263 | 0.00043 | 0.00220 | 0.00002 | 0.00048 | 65 | 0.00293 | 0.00004 | 0.00065 | 93 | 0.00081 | -0.90748 | 154.316 | 0.36557 | 0.79587 |
| UGCG; ceramide glucosyltransferase [EC:2.4.1.80] | 0.00263 | 0.00074 | 0.00453 | 0.00017 | 0.00160 | 65 | 0.00130 | 0.00003 | 0.00054 | 93 | 0.00168 | 1.91966 | 78.566 | 0.05853 | 0.72139 |
| isdG, isdI; heme oxygenase (staphylobilin-producing) [EC:1.14.99.48] | 0.00263 | 0.00075 | 0.00168 | 0.00004 | 0.00080 | 65 | 0.00329 | 0.00012 | 0.00115 | 93 | 0.00140 | -1.15312 | 151.430 | 0.25068 | 0.77376 |
| yscD, sctD, ssaD; type III secretion protein D | 0.00263 | 0.00070 | 0.00209 | 0.00005 | 0.00092 | 65 | 0.00300 | 0.00010 | 0.00101 | 93 | 0.00137 | -0.66633 | 154.935 | 0.50619 | 0.84956 |
| K09964; uncharacterized protein | 0.00263 | 0.00039 | 0.00262 | 0.00002 | 0.00061 | 65 | 0.00263 | 0.00002 | 0.00051 | 93 | 0.00080 | -0.01825 | 137.457 | 0.98547 | 0.99723 |
| hysA, hylA, hylB; hyaluronate lyase [EC:4.2.2.1] | 0.00263 | 0.00065 | 0.00344 | 0.00014 | 0.00146 | 65 | 0.00206 | 0.00002 | 0.00041 | 93 | 0.00152 | 0.91543 | 74.294 | 0.36293 | 0.79496 |
| prfH; peptide chain release factor | 0.00262 | 0.00047 | 0.00307 | 0.00006 | 0.00096 | 65 | 0.00231 | 0.00002 | 0.00044 | 93 | 0.00105 | 0.72844 | 90.653 | 0.46822 | 0.83837 |
| K06977; uncharacterized protein | 0.00262 | 0.00059 | 0.00257 | 0.00006 | 0.00097 | 65 | 0.00265 | 0.00005 | 0.00075 | 93 | 0.00122 | -0.06951 | 130.743 | 0.94469 | 0.98674 |
| dauA; D-arginine dehydrogenase [EC:1.4.99.6] | 0.00261 | 0.00046 | 0.00280 | 0.00004 | 0.00081 | 65 | 0.00248 | 0.00003 | 0.00054 | 93 | 0.00097 | 0.32327 | 117.090 | 0.74707 | 0.93207 |
| vanR; GntR family transcriptional regulator, vanillate catabolism transcriptional regulator | 0.00261 | 0.00044 | 0.00200 | 0.00002 | 0.00057 | 65 | 0.00304 | 0.00004 | 0.00062 | 93 | 0.00085 | -1.23553 | 154.790 | 0.21850 | 0.77376 |
| fha1; type VI secretion system protein | 0.00261 | 0.00071 | 0.00203 | 0.00006 | 0.00092 | 65 | 0.00301 | 0.00010 | 0.00102 | 93 | 0.00137 | -0.71526 | 154.981 | 0.47552 | 0.84114 |
| yscU, sctU, hrcU, ssaU; type III secretion protein U | 0.00260 | 0.00053 | 0.00181 | 0.00002 | 0.00058 | 65 | 0.00316 | 0.00006 | 0.00080 | 93 | 0.00099 | -1.36880 | 153.519 | 0.17306 | 0.77376 |
| spoVAF; stage V sporulation protein AF | 0.00260 | 0.00058 | 0.00099 | 0.00002 | 0.00051 | 65 | 0.00373 | 0.00008 | 0.00091 | 93 | 0.00105 | -2.61769 | 139.618 | 0.00983 | 0.58169 |
| gcrA; GcrA cell cycle regulator | 0.00259 | 0.00054 | 0.00118 | 0.00001 | 0.00030 | 65 | 0.00358 | 0.00007 | 0.00088 | 93 | 0.00093 | -2.58786 | 113.031 | 0.01092 | 0.58284 |
| arnC, pmrF; undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase [EC:2.4.2.53] | 0.00259 | 0.00049 | 0.00234 | 0.00003 | 0.00062 | 65 | 0.00276 | 0.00005 | 0.00071 | 93 | 0.00094 | -0.44694 | 155.474 | 0.65554 | 0.90679 |
| mccB; cystathionine gamma-lyase / homocysteine desulfhydrase [EC:4.4.1.1 4.4.1.2] | 0.00259 | 0.00051 | 0.00169 | 0.00003 | 0.00067 | 65 | 0.00322 | 0.00005 | 0.00072 | 93 | 0.00098 | -1.56426 | 154.148 | 0.11981 | 0.77376 |
| nicA; nicotinate dehydrogenase subunit A [EC:1.17.2.1] | 0.00258 | 0.00052 | 0.00238 | 0.00003 | 0.00072 | 65 | 0.00272 | 0.00005 | 0.00072 | 93 | 0.00102 | -0.33796 | 151.148 | 0.73586 | 0.92721 |
| amhX; amidohydrolase [EC:3.5.1.-] | 0.00257 | 0.00093 | 0.00109 | 0.00005 | 0.00087 | 65 | 0.00361 | 0.00019 | 0.00145 | 93 | 0.00169 | -1.49657 | 143.895 | 0.13670 | 0.77376 |
| K06983; uncharacterized protein | 0.00257 | 0.00059 | 0.00253 | 0.00006 | 0.00096 | 65 | 0.00260 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.05864 | 130.834 | 0.95333 | 0.98792 |
| yusF; toprim domain protein | 0.00257 | 0.00058 | 0.00142 | 0.00002 | 0.00061 | 65 | 0.00338 | 0.00007 | 0.00088 | 93 | 0.00107 | -1.83011 | 151.453 | 0.06920 | 0.73522 |
| abnA; arabinan endo-1,5-alpha-L-arabinosidase [EC:3.2.1.99] | 0.00257 | 0.00054 | 0.00164 | 0.00001 | 0.00036 | 65 | 0.00323 | 0.00007 | 0.00088 | 93 | 0.00095 | -1.66980 | 121.320 | 0.09754 | 0.76673 |
| rpfC; two-component system, sensor histidine kinase RpfC [EC:2.7.13.3] | 0.00257 | 0.00059 | 0.00247 | 0.00006 | 0.00096 | 65 | 0.00264 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.14209 | 130.608 | 0.88723 | 0.97039 |
| K18133, porB; major outer membrane protein P.IB | 0.00257 | 0.00058 | 0.00284 | 0.00006 | 0.00094 | 65 | 0.00238 | 0.00005 | 0.00074 | 93 | 0.00119 | 0.38543 | 133.333 | 0.70053 | 0.91953 |
| gfrF; fructoselysine-6-phosphate deglycase | 0.00257 | 0.00073 | 0.00412 | 0.00017 | 0.00160 | 65 | 0.00149 | 0.00003 | 0.00053 | 93 | 0.00168 | 1.56493 | 78.040 | 0.12164 | 0.77376 |
| E5.3.4.1; protein disulfide-isomerase [EC:5.3.4.1] | 0.00257 | 0.00059 | 0.00242 | 0.00006 | 0.00096 | 65 | 0.00267 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.20860 | 131.003 | 0.83508 | 0.95506 |
| csb1; CRISPR-associated protein Csb1 | 0.00256 | 0.00056 | 0.00301 | 0.00006 | 0.00099 | 65 | 0.00224 | 0.00004 | 0.00064 | 93 | 0.00118 | 0.65596 | 114.869 | 0.51316 | 0.85155 |
| csb2; CRISPR-associated protein Csb2 | 0.00256 | 0.00056 | 0.00301 | 0.00006 | 0.00099 | 65 | 0.00224 | 0.00004 | 0.00064 | 93 | 0.00118 | 0.65596 | 114.869 | 0.51316 | 0.85155 |
| lysAC; bifunctional diaminopimelate decarboxylase / aspartate kinase [EC:4.1.1.20 2.7.2.4] | 0.00256 | 0.00059 | 0.00249 | 0.00006 | 0.00096 | 65 | 0.00260 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.09585 | 130.750 | 0.92379 | 0.97973 |
| TR1; tropinone reductase I [EC:1.1.1.206] | 0.00256 | 0.00059 | 0.00249 | 0.00006 | 0.00096 | 65 | 0.00260 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.09585 | 130.750 | 0.92379 | 0.97973 |
| parE1_3_4; toxin ParE1/3/4 | 0.00255 | 0.00041 | 0.00170 | 0.00002 | 0.00056 | 65 | 0.00315 | 0.00003 | 0.00058 | 93 | 0.00080 | -1.79361 | 152.460 | 0.07486 | 0.74200 |
| tetA; MFS transporter, DHA1 family, tetracycline resistance protein | 0.00255 | 0.00040 | 0.00140 | 0.00001 | 0.00032 | 65 | 0.00336 | 0.00004 | 0.00063 | 93 | 0.00071 | -2.76604 | 133.162 | 0.00648 | 0.54257 |
| gmuG; mannan endo-1,4-beta-mannosidase [EC:3.2.1.78] | 0.00255 | 0.00050 | 0.00161 | 0.00001 | 0.00038 | 65 | 0.00321 | 0.00006 | 0.00081 | 93 | 0.00089 | -1.80187 | 128.056 | 0.07392 | 0.74128 |
| wbpA; UDP-N-acetyl-D-glucosamine dehydrogenase [EC:1.1.1.136] | 0.00255 | 0.00046 | 0.00207 | 0.00002 | 0.00060 | 65 | 0.00288 | 0.00004 | 0.00066 | 93 | 0.00089 | -0.90250 | 155.021 | 0.36819 | 0.79587 |
| rpfG; two-component system, response regulator RpfG | 0.00254 | 0.00059 | 0.00247 | 0.00006 | 0.00096 | 65 | 0.00260 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.10771 | 130.659 | 0.91439 | 0.97664 |
| E1.6.99.1; NADPH2 dehydrogenase [EC:1.6.99.1] | 0.00254 | 0.00049 | 0.00128 | 0.00001 | 0.00045 | 65 | 0.00343 | 0.00005 | 0.00075 | 93 | 0.00088 | -2.44127 | 143.644 | 0.01585 | 0.59095 |
| csb3; CRISPR-associated protein Csb3 | 0.00254 | 0.00056 | 0.00298 | 0.00006 | 0.00100 | 65 | 0.00224 | 0.00004 | 0.00064 | 93 | 0.00119 | 0.62956 | 114.785 | 0.53024 | 0.86094 |
| divK; two-component system, cell cycle response regulator DivK | 0.00254 | 0.00043 | 0.00120 | 0.00001 | 0.00035 | 65 | 0.00347 | 0.00004 | 0.00067 | 93 | 0.00076 | -2.99823 | 134.821 | 0.00323 | 0.46257 |
| ABC.NGC.P1; N-acetylglucosamine transport system permease protein | 0.00253 | 0.00053 | 0.00303 | 0.00008 | 0.00109 | 65 | 0.00219 | 0.00002 | 0.00050 | 93 | 0.00119 | 0.70318 | 90.742 | 0.48375 | 0.84354 |
| rocR; arginine utilization regulatory protein | 0.00252 | 0.00092 | 0.00133 | 0.00005 | 0.00087 | 65 | 0.00335 | 0.00019 | 0.00144 | 93 | 0.00168 | -1.19875 | 144.206 | 0.23259 | 0.77376 |
| ndoAI; CopG family transcriptional regulator / antitoxin EndoAI | 0.00252 | 0.00055 | 0.00146 | 0.00002 | 0.00061 | 65 | 0.00326 | 0.00006 | 0.00084 | 93 | 0.00103 | -1.73951 | 153.457 | 0.08395 | 0.76567 |
| cotJC; spore coat protein JC | 0.00251 | 0.00057 | 0.00112 | 0.00002 | 0.00050 | 65 | 0.00349 | 0.00007 | 0.00089 | 93 | 0.00102 | -2.32453 | 138.641 | 0.02155 | 0.59153 |
| yojI; multidrug/microcin transport system ATP-binding/permease protein | 0.00250 | 0.00057 | 0.00250 | 0.00005 | 0.00084 | 65 | 0.00251 | 0.00006 | 0.00078 | 93 | 0.00114 | -0.00639 | 146.499 | 0.99491 | 0.99942 |
| sspH; small acid-soluble spore protein H (minor) | 0.00250 | 0.00071 | 0.00048 | 0.00001 | 0.00030 | 65 | 0.00391 | 0.00013 | 0.00116 | 93 | 0.00120 | -2.86926 | 104.313 | 0.00498 | 0.54022 |
| E3.2.1.8, xynA; endo-1,4-beta-xylanase [EC:3.2.1.8] | 0.00250 | 0.00041 | 0.00160 | 0.00001 | 0.00044 | 65 | 0.00312 | 0.00004 | 0.00061 | 93 | 0.00075 | -2.02047 | 152.501 | 0.04508 | 0.65981 |
| nfxB; TetR/AcrR family transcriptional regulator, mexCD-oprJ operon repressor | 0.00249 | 0.00058 | 0.00246 | 0.00006 | 0.00096 | 65 | 0.00252 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.05131 | 130.609 | 0.95915 | 0.98903 |
| dpkA, lhpD; delta1-piperideine-2-carboxylate reductase [EC:1.5.1.21] | 0.00249 | 0.00050 | 0.00241 | 0.00003 | 0.00067 | 65 | 0.00255 | 0.00005 | 0.00071 | 93 | 0.00098 | -0.14086 | 153.279 | 0.88816 | 0.97039 |
| ecpD; chaperone protein EcpD | 0.00249 | 0.00074 | 0.00193 | 0.00002 | 0.00056 | 65 | 0.00288 | 0.00013 | 0.00120 | 93 | 0.00132 | -0.71657 | 128.170 | 0.47495 | 0.84095 |
| mexG; transmembrane protein | 0.00249 | 0.00058 | 0.00238 | 0.00006 | 0.00096 | 65 | 0.00256 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.14441 | 130.459 | 0.88540 | 0.97001 |
| ABC.MR.TX; HlyD family secretion protein | 0.00248 | 0.00051 | 0.00129 | 0.00002 | 0.00049 | 65 | 0.00332 | 0.00006 | 0.00078 | 93 | 0.00092 | -2.21106 | 146.096 | 0.02859 | 0.61334 |
| dck; deoxyadenosine/deoxycytidine kinase [EC:2.7.1.76 2.7.1.74] | 0.00247 | 0.00054 | 0.00150 | 0.00002 | 0.00061 | 65 | 0.00316 | 0.00006 | 0.00081 | 93 | 0.00102 | -1.62905 | 154.427 | 0.10534 | 0.77376 |
| dgoD; galactonate dehydratase [EC:4.2.1.6] | 0.00247 | 0.00042 | 0.00153 | 0.00001 | 0.00045 | 65 | 0.00313 | 0.00004 | 0.00064 | 93 | 0.00078 | -2.04028 | 151.844 | 0.04306 | 0.65559 |
| cymR; Rrf2 family transcriptional regulator, cysteine metabolism repressor | 0.00247 | 0.00057 | 0.00141 | 0.00002 | 0.00061 | 65 | 0.00321 | 0.00007 | 0.00087 | 93 | 0.00106 | -1.69173 | 152.070 | 0.09275 | 0.76567 |
| K07161; uncharacterized protein | 0.00247 | 0.00057 | 0.00125 | 0.00001 | 0.00045 | 65 | 0.00332 | 0.00008 | 0.00091 | 93 | 0.00101 | -2.03879 | 130.746 | 0.04349 | 0.65903 |
| ycjT; hypothetical glycosyl hydrolase [EC:3.2.1.-] | 0.00246 | 0.00082 | 0.00356 | 0.00022 | 0.00183 | 65 | 0.00170 | 0.00003 | 0.00055 | 93 | 0.00191 | 0.97356 | 75.771 | 0.33337 | 0.77957 |
| kpsC, lipA; capsular polysaccharide export protein | 0.00246 | 0.00044 | 0.00263 | 0.00003 | 0.00063 | 65 | 0.00235 | 0.00003 | 0.00061 | 93 | 0.00087 | 0.32584 | 149.032 | 0.74500 | 0.93176 |
| aph, spcN; spectinomycin phosphotransferase | 0.00246 | 0.00058 | 0.00238 | 0.00006 | 0.00096 | 65 | 0.00252 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.10929 | 130.483 | 0.91314 | 0.97664 |
| K09190; uncharacterized protein | 0.00246 | 0.00045 | 0.00224 | 0.00002 | 0.00060 | 65 | 0.00261 | 0.00004 | 0.00064 | 93 | 0.00088 | -0.42392 | 154.180 | 0.67222 | 0.91184 |
| malE; maltose/maltodextrin transport system substrate-binding protein | 0.00246 | 0.00048 | 0.00186 | 0.00003 | 0.00064 | 65 | 0.00288 | 0.00004 | 0.00068 | 93 | 0.00093 | -1.09913 | 153.280 | 0.27343 | 0.77376 |
| smeA; membrane fusion protein, multidrug efflux system | 0.00246 | 0.00058 | 0.00238 | 0.00006 | 0.00096 | 65 | 0.00251 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.10489 | 130.486 | 0.91662 | 0.97681 |
| smeB; multidrug efflux pump | 0.00246 | 0.00058 | 0.00238 | 0.00006 | 0.00096 | 65 | 0.00251 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.10489 | 130.486 | 0.91662 | 0.97681 |
| smeC; outer membrane protein, multidrug efflux system | 0.00246 | 0.00058 | 0.00238 | 0.00006 | 0.00096 | 65 | 0.00251 | 0.00005 | 0.00074 | 93 | 0.00121 | -0.10489 | 130.486 | 0.91662 | 0.97681 |
| nreB; two-component system, NarL family, sensor histidine kinase NreB [EC:2.7.13.3] | 0.00246 | 0.00061 | 0.00257 | 0.00007 | 0.00103 | 65 | 0.00238 | 0.00005 | 0.00074 | 93 | 0.00127 | 0.14585 | 123.682 | 0.88427 | 0.96974 |
| ycbA, glnK; two-component system, sensor histidine kinase YcbA [EC:2.7.13.3] | 0.00245 | 0.00060 | 0.00272 | 0.00008 | 0.00108 | 65 | 0.00226 | 0.00005 | 0.00070 | 93 | 0.00129 | 0.35970 | 115.730 | 0.71973 | 0.92108 |
| CNP; 2’,3’-cyclic-nucleotide 3’-phosphodiesterase [EC:3.1.4.37] | 0.00245 | 0.00074 | 0.00427 | 0.00017 | 0.00160 | 65 | 0.00118 | 0.00003 | 0.00054 | 93 | 0.00169 | 1.82917 | 78.439 | 0.07117 | 0.73920 |
| mug; double-stranded uracil-DNA glycosylase [EC:3.2.2.28] | 0.00245 | 0.00041 | 0.00147 | 0.00001 | 0.00040 | 65 | 0.00313 | 0.00004 | 0.00063 | 93 | 0.00074 | -2.23068 | 147.800 | 0.02721 | 0.61334 |
| K15533; 1,3-beta-galactosyl-N-acetylhexosamine phosphorylase [EC:2.4.1.211] | 0.00245 | 0.00053 | 0.00285 | 0.00007 | 0.00102 | 65 | 0.00217 | 0.00003 | 0.00056 | 93 | 0.00116 | 0.58835 | 101.248 | 0.55761 | 0.86813 |
| glpS; glycerol transport system ATP-binding protein | 0.00244 | 0.00044 | 0.00161 | 0.00001 | 0.00048 | 65 | 0.00302 | 0.00004 | 0.00066 | 93 | 0.00081 | -1.73285 | 153.135 | 0.08513 | 0.76567 |
| hemX; HemX protein | 0.00244 | 0.00057 | 0.00135 | 0.00002 | 0.00061 | 65 | 0.00320 | 0.00007 | 0.00087 | 93 | 0.00106 | -1.74987 | 151.796 | 0.08216 | 0.76567 |
| K17325, glpT; glycerol transport system ATP-binding protein | 0.00244 | 0.00044 | 0.00161 | 0.00001 | 0.00048 | 65 | 0.00302 | 0.00004 | 0.00066 | 93 | 0.00081 | -1.73667 | 153.136 | 0.08446 | 0.76567 |
| paaZ; oxepin-CoA hydrolase / 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase [EC:3.3.2.12 1.2.1.91] | 0.00244 | 0.00038 | 0.00228 | 0.00002 | 0.00049 | 65 | 0.00255 | 0.00003 | 0.00055 | 93 | 0.00074 | -0.35192 | 155.212 | 0.72538 | 0.92177 |
| RETSAT; all-trans-retinol 13,14-reductase [EC:1.3.99.23] | 0.00244 | 0.00051 | 0.00165 | 0.00001 | 0.00035 | 65 | 0.00298 | 0.00006 | 0.00082 | 93 | 0.00090 | -1.48301 | 123.154 | 0.14063 | 0.77376 |
| glcG; glc operon protein GlcG | 0.00243 | 0.00044 | 0.00327 | 0.00005 | 0.00085 | 65 | 0.00185 | 0.00002 | 0.00044 | 93 | 0.00096 | 1.47576 | 97.151 | 0.14324 | 0.77376 |
| alc, ALLC; allantoicase [EC:3.5.3.4] | 0.00243 | 0.00044 | 0.00148 | 0.00001 | 0.00044 | 65 | 0.00309 | 0.00004 | 0.00068 | 93 | 0.00081 | -1.97896 | 148.452 | 0.04967 | 0.67522 |
| glpQ; glycerol transport system permease protein | 0.00243 | 0.00043 | 0.00161 | 0.00001 | 0.00048 | 65 | 0.00300 | 0.00004 | 0.00066 | 93 | 0.00081 | -1.71352 | 153.258 | 0.08864 | 0.76567 |
| glpV; glycerol transport system substrate-binding protein | 0.00243 | 0.00043 | 0.00161 | 0.00001 | 0.00048 | 65 | 0.00300 | 0.00004 | 0.00066 | 93 | 0.00081 | -1.71352 | 153.258 | 0.08864 | 0.76567 |
| K17322, glpP; glycerol transport system permease protein | 0.00243 | 0.00043 | 0.00161 | 0.00001 | 0.00048 | 65 | 0.00300 | 0.00004 | 0.00066 | 93 | 0.00081 | -1.71352 | 153.258 | 0.08864 | 0.76567 |
| nosZ; nitrous-oxide reductase [EC:1.7.2.4] | 0.00242 | 0.00036 | 0.00186 | 0.00001 | 0.00037 | 65 | 0.00281 | 0.00003 | 0.00056 | 93 | 0.00067 | -1.41377 | 148.828 | 0.15952 | 0.77376 |
| K15383; MtN3 and saliva related transmembrane protein | 0.00242 | 0.00044 | 0.00251 | 0.00004 | 0.00078 | 65 | 0.00235 | 0.00003 | 0.00052 | 93 | 0.00093 | 0.16411 | 117.753 | 0.86993 | 0.96495 |
| waaQ, rfaQ; heptosyltransferase III [EC:2.4.-.-] | 0.00242 | 0.00036 | 0.00255 | 0.00002 | 0.00060 | 65 | 0.00233 | 0.00002 | 0.00044 | 93 | 0.00074 | 0.29668 | 125.067 | 0.76721 | 0.93900 |
| hndD; NADP-reducing hydrogenase subunit HndD [EC:1.12.1.3] | 0.00241 | 0.00055 | 0.00266 | 0.00007 | 0.00102 | 65 | 0.00224 | 0.00003 | 0.00060 | 93 | 0.00119 | 0.35616 | 106.508 | 0.72243 | 0.92156 |
| pafC; proteasome accessory factor C | 0.00241 | 0.00033 | 0.00228 | 0.00001 | 0.00046 | 65 | 0.00251 | 0.00002 | 0.00045 | 93 | 0.00065 | -0.36087 | 149.337 | 0.71870 | 0.92092 |
| nfsA; nitroreductase [EC:1.-.-.-] | 0.00241 | 0.00042 | 0.00271 | 0.00004 | 0.00074 | 65 | 0.00221 | 0.00002 | 0.00049 | 93 | 0.00089 | 0.56815 | 115.754 | 0.57103 | 0.87172 |
| fwdE, fmdE; formylmethanofuran dehydrogenase subunit E [EC:1.2.7.12] | 0.00241 | 0.00073 | 0.00415 | 0.00017 | 0.00160 | 65 | 0.00119 | 0.00003 | 0.00052 | 93 | 0.00168 | 1.76483 | 77.701 | 0.08152 | 0.76350 |
| yscT, sctT, hrcT, ssaT; type III secretion protein T | 0.00241 | 0.00049 | 0.00177 | 0.00002 | 0.00058 | 65 | 0.00286 | 0.00005 | 0.00073 | 93 | 0.00094 | -1.16694 | 155.734 | 0.24502 | 0.77376 |
| mhpT; MFS transporter, AAHS family, 3-hydroxyphenylpropionic acid transporter | 0.00241 | 0.00041 | 0.00225 | 0.00002 | 0.00060 | 65 | 0.00252 | 0.00003 | 0.00057 | 93 | 0.00082 | -0.33741 | 147.841 | 0.73628 | 0.92721 |
| sauS; sulfoacetaldehyde dehydrogenase [EC:1.2.1.81] | 0.00241 | 0.00073 | 0.00419 | 0.00017 | 0.00160 | 65 | 0.00116 | 0.00003 | 0.00052 | 93 | 0.00168 | 1.79801 | 77.773 | 0.07606 | 0.74696 |
| spoIIIAE; stage III sporulation protein AE | 0.00240 | 0.00051 | 0.00121 | 0.00002 | 0.00051 | 65 | 0.00323 | 0.00006 | 0.00078 | 93 | 0.00094 | -2.15580 | 148.703 | 0.03271 | 0.62130 |
| TC.DME; drug/metabolite transporter, DME family | 0.00240 | 0.00047 | 0.00191 | 0.00002 | 0.00052 | 65 | 0.00274 | 0.00005 | 0.00071 | 93 | 0.00088 | -0.94191 | 153.723 | 0.34772 | 0.79069 |
| yqfD; similar to stage IV sporulation protein | 0.00240 | 0.00051 | 0.00121 | 0.00002 | 0.00051 | 65 | 0.00323 | 0.00006 | 0.00079 | 93 | 0.00094 | -2.14211 | 148.318 | 0.03382 | 0.62130 |
| yscR, sctR, hrcR, ssaR; type III secretion protein R | 0.00240 | 0.00049 | 0.00175 | 0.00002 | 0.00058 | 65 | 0.00285 | 0.00005 | 0.00073 | 93 | 0.00094 | -1.18287 | 155.745 | 0.23866 | 0.77376 |
| dgk; deoxyguanosine kinase [EC:2.7.1.113] | 0.00239 | 0.00054 | 0.00148 | 0.00002 | 0.00061 | 65 | 0.00303 | 0.00006 | 0.00081 | 93 | 0.00102 | -1.52364 | 154.441 | 0.12964 | 0.77376 |
| yscV, sctV, hrcV, ssaV, invA; type III secretion protein V | 0.00239 | 0.00049 | 0.00173 | 0.00002 | 0.00059 | 65 | 0.00285 | 0.00005 | 0.00073 | 93 | 0.00094 | -1.18928 | 155.759 | 0.23614 | 0.77376 |
| pct; propionate CoA-transferase [EC:2.8.3.1] | 0.00238 | 0.00038 | 0.00201 | 0.00002 | 0.00055 | 65 | 0.00264 | 0.00002 | 0.00051 | 93 | 0.00075 | -0.84216 | 146.316 | 0.40107 | 0.80823 |
| yscS, sctS, hrcS, ssaS; type III secretion protein S | 0.00238 | 0.00049 | 0.00175 | 0.00002 | 0.00058 | 65 | 0.00281 | 0.00005 | 0.00073 | 93 | 0.00094 | -1.13284 | 155.692 | 0.25902 | 0.77376 |
| yhhY; putative acetyltransferase [EC:2.3.1.-] | 0.00237 | 0.00043 | 0.00208 | 0.00003 | 0.00064 | 65 | 0.00257 | 0.00003 | 0.00058 | 93 | 0.00086 | -0.57283 | 144.884 | 0.56765 | 0.87063 |
| E1.1.1.122; D-threo-aldose 1-dehydrogenase [EC:1.1.1.122] | 0.00237 | 0.00040 | 0.00149 | 0.00002 | 0.00050 | 65 | 0.00298 | 0.00003 | 0.00057 | 93 | 0.00076 | -1.95802 | 155.726 | 0.05201 | 0.68802 |
| K07077; uncharacterized protein | 0.00236 | 0.00044 | 0.00124 | 0.00001 | 0.00039 | 65 | 0.00314 | 0.00004 | 0.00069 | 93 | 0.00079 | -2.38501 | 140.074 | 0.01842 | 0.59095 |
| gfrE; glucoselysine-6-phosphate deglycase | 0.00236 | 0.00073 | 0.00407 | 0.00017 | 0.00160 | 65 | 0.00116 | 0.00003 | 0.00052 | 93 | 0.00168 | 1.73734 | 77.742 | 0.08629 | 0.76567 |
| K15024; putative phosphotransacetylase [EC:2.3.1.8] | 0.00236 | 0.00067 | 0.00138 | 0.00002 | 0.00061 | 65 | 0.00304 | 0.00010 | 0.00106 | 93 | 0.00122 | -1.36411 | 140.768 | 0.17471 | 0.77376 |
| trpGD; anthranilate synthase/phosphoribosyltransferase [EC:4.1.3.27 2.4.2.18] | 0.00235 | 0.00039 | 0.00237 | 0.00002 | 0.00049 | 65 | 0.00234 | 0.00003 | 0.00056 | 93 | 0.00075 | 0.03747 | 155.709 | 0.97016 | 0.99343 |
| catA; chloramphenicol O-acetyltransferase type A [EC:2.3.1.28] | 0.00235 | 0.00057 | 0.00129 | 0.00001 | 0.00037 | 65 | 0.00309 | 0.00008 | 0.00093 | 93 | 0.00100 | -1.80059 | 119.553 | 0.07429 | 0.74128 |
| atuH; citronellyl-CoA synthetase [EC:6.2.1.-] | 0.00234 | 0.00042 | 0.00177 | 0.00001 | 0.00048 | 65 | 0.00273 | 0.00004 | 0.00063 | 93 | 0.00079 | -1.21248 | 154.692 | 0.22718 | 0.77376 |
| kguK; dehydrogluconokinase [EC:2.7.1.13] | 0.00234 | 0.00050 | 0.00235 | 0.00003 | 0.00067 | 65 | 0.00232 | 0.00005 | 0.00070 | 93 | 0.00097 | 0.02721 | 153.102 | 0.97833 | 0.99657 |
| MuB; ATP-dependent target DNA activator [EC:3.6.1.3] | 0.00234 | 0.00052 | 0.00197 | 0.00001 | 0.00044 | 65 | 0.00259 | 0.00006 | 0.00083 | 93 | 0.00094 | -0.66330 | 135.836 | 0.50826 | 0.84956 |
| oadG; oxaloacetate decarboxylase, gamma subunit [EC:4.1.1.3] | 0.00233 | 0.00048 | 0.00195 | 0.00003 | 0.00070 | 65 | 0.00260 | 0.00004 | 0.00066 | 93 | 0.00096 | -0.67287 | 147.528 | 0.50208 | 0.84942 |
| cotJB; spore coat protein JB | 0.00233 | 0.00052 | 0.00109 | 0.00002 | 0.00050 | 65 | 0.00320 | 0.00006 | 0.00079 | 93 | 0.00094 | -2.25289 | 145.908 | 0.02576 | 0.59941 |
| hemAT; heam-based aerotactic trancducer | 0.00233 | 0.00120 | 0.00134 | 0.00009 | 0.00117 | 65 | 0.00303 | 0.00032 | 0.00187 | 93 | 0.00220 | -0.76639 | 146.247 | 0.44468 | 0.83271 |
| sfa2; sigma-54 dependent transcriptional regulator | 0.00232 | 0.00046 | 0.00253 | 0.00004 | 0.00077 | 65 | 0.00217 | 0.00003 | 0.00056 | 93 | 0.00095 | 0.37592 | 126.113 | 0.70761 | 0.92004 |
| yidH; putative membrane protein | 0.00232 | 0.00046 | 0.00175 | 0.00001 | 0.00041 | 65 | 0.00271 | 0.00005 | 0.00072 | 93 | 0.00083 | -1.16846 | 140.270 | 0.24460 | 0.77376 |
| araF; L-arabinose transport system substrate-binding protein | 0.00231 | 0.00060 | 0.00167 | 0.00003 | 0.00067 | 65 | 0.00277 | 0.00008 | 0.00091 | 93 | 0.00113 | -0.96989 | 153.591 | 0.33363 | 0.77983 |
| araG; L-arabinose transport system ATP-binding protein [EC:3.6.3.17] | 0.00231 | 0.00060 | 0.00166 | 0.00003 | 0.00067 | 65 | 0.00277 | 0.00008 | 0.00091 | 93 | 0.00113 | -0.98217 | 153.574 | 0.32756 | 0.77376 |
| ermC, ermA; 23S rRNA (adenine-N6)-dimethyltransferase [EC:2.1.1.184] | 0.00231 | 0.00038 | 0.00263 | 0.00003 | 0.00071 | 65 | 0.00209 | 0.00002 | 0.00041 | 93 | 0.00081 | 0.65965 | 105.810 | 0.51091 | 0.85070 |
| E1.1.1.43; phosphogluconate 2-dehydrogenase [EC:1.1.1.43] | 0.00231 | 0.00049 | 0.00230 | 0.00003 | 0.00066 | 65 | 0.00232 | 0.00005 | 0.00070 | 93 | 0.00096 | -0.02461 | 153.659 | 0.98040 | 0.99657 |
| araH; L-arabinose transport system permease protein | 0.00231 | 0.00060 | 0.00167 | 0.00003 | 0.00067 | 65 | 0.00276 | 0.00008 | 0.00091 | 93 | 0.00113 | -0.96529 | 153.593 | 0.33592 | 0.78225 |
| dhaT; 1,3-propanediol dehydrogenase [EC:1.1.1.202] | 0.00231 | 0.00063 | 0.00277 | 0.00010 | 0.00122 | 65 | 0.00198 | 0.00004 | 0.00065 | 93 | 0.00139 | 0.57437 | 99.925 | 0.56701 | 0.87061 |
| csgG; curli production assembly/transport component CsgG | 0.00230 | 0.00039 | 0.00286 | 0.00004 | 0.00078 | 65 | 0.00192 | 0.00001 | 0.00038 | 93 | 0.00087 | 1.08313 | 93.594 | 0.28153 | 0.77376 |
| choD; cholesterol oxidase [EC:1.1.3.6] | 0.00230 | 0.00094 | 0.00107 | 0.00001 | 0.00033 | 65 | 0.00316 | 0.00023 | 0.00157 | 93 | 0.00160 | -1.30808 | 99.990 | 0.19385 | 0.77376 |
| K09822; uncharacterized protein | 0.00230 | 0.00047 | 0.00145 | 0.00002 | 0.00057 | 65 | 0.00289 | 0.00004 | 0.00069 | 93 | 0.00090 | -1.60559 | 155.938 | 0.11039 | 0.77376 |
| hasR; heme acquisition protein HasR | 0.00230 | 0.00069 | 0.00182 | 0.00005 | 0.00090 | 65 | 0.00263 | 0.00009 | 0.00100 | 93 | 0.00134 | -0.60855 | 154.970 | 0.54371 | 0.86313 |
| cynS; cyanate lyase [EC:4.2.1.104] | 0.00229 | 0.00039 | 0.00195 | 0.00003 | 0.00066 | 65 | 0.00253 | 0.00002 | 0.00049 | 93 | 0.00082 | -0.70091 | 127.010 | 0.48464 | 0.84358 |
| E2.7.1.76, dak; deoxyadenosine kinase [EC:2.7.1.76] | 0.00229 | 0.00059 | 0.00233 | 0.00005 | 0.00089 | 65 | 0.00226 | 0.00006 | 0.00079 | 93 | 0.00119 | 0.05284 | 142.438 | 0.95794 | 0.98896 |
| E3.4.11.14; cytosol alanyl aminopeptidase [EC:3.4.11.14] | 0.00229 | 0.00059 | 0.00229 | 0.00005 | 0.00089 | 65 | 0.00229 | 0.00006 | 0.00079 | 93 | 0.00119 | 0.00388 | 142.855 | 0.99691 | 0.99951 |
| ipdC; indolepyruvate decarboxylase [EC:4.1.1.74] | 0.00228 | 0.00045 | 0.00174 | 0.00002 | 0.00058 | 65 | 0.00266 | 0.00004 | 0.00065 | 93 | 0.00088 | -1.05424 | 155.168 | 0.29341 | 0.77376 |
| recT; recombination protein RecT | 0.00228 | 0.00054 | 0.00135 | 0.00001 | 0.00046 | 65 | 0.00293 | 0.00007 | 0.00085 | 93 | 0.00097 | -1.62891 | 136.582 | 0.10564 | 0.77376 |
| uhpC; MFS transporter, OPA family, sugar phosphate sensor protein UhpC | 0.00227 | 0.00054 | 0.00300 | 0.00007 | 0.00106 | 65 | 0.00177 | 0.00003 | 0.00053 | 93 | 0.00119 | 1.03813 | 96.314 | 0.30181 | 0.77376 |
| rbsU; putative ribose uptake protein | 0.00227 | 0.00056 | 0.00215 | 0.00005 | 0.00088 | 65 | 0.00235 | 0.00005 | 0.00074 | 93 | 0.00115 | -0.17581 | 137.588 | 0.86070 | 0.96170 |
| K11159; carotenoid cleavage dioxygenase | 0.00227 | 0.00040 | 0.00118 | 0.00001 | 0.00037 | 65 | 0.00303 | 0.00004 | 0.00061 | 93 | 0.00072 | -2.57452 | 143.599 | 0.01105 | 0.58351 |
| phnF; GntR family transcriptional regulator, phosphonate transport system regulatory protein | 0.00227 | 0.00043 | 0.00108 | 0.00001 | 0.00036 | 65 | 0.00310 | 0.00004 | 0.00067 | 93 | 0.00076 | -2.65385 | 136.321 | 0.00890 | 0.58112 |
| yydK; GntR family transcriptional regulator, transcriptional regulator of bglA | 0.00226 | 0.00056 | 0.00217 | 0.00005 | 0.00088 | 65 | 0.00233 | 0.00005 | 0.00073 | 93 | 0.00114 | -0.14144 | 137.700 | 0.88773 | 0.97039 |
| csn2; CRISPR-associated protein Csn2 | 0.00226 | 0.00042 | 0.00280 | 0.00003 | 0.00072 | 65 | 0.00188 | 0.00002 | 0.00050 | 93 | 0.00088 | 1.05079 | 121.288 | 0.29544 | 0.77376 |
| comFB; competence protein ComFB | 0.00226 | 0.00058 | 0.00240 | 0.00007 | 0.00102 | 65 | 0.00216 | 0.00004 | 0.00069 | 93 | 0.00124 | 0.18964 | 118.470 | 0.84991 | 0.95744 |
| UXS1, uxs; UDP-glucuronate decarboxylase [EC:4.1.1.35] | 0.00226 | 0.00034 | 0.00094 | 0.00000 | 0.00014 | 65 | 0.00318 | 0.00003 | 0.00054 | 93 | 0.00056 | -3.98427 | 104.726 | 0.00013 | 0.43014 |
| PTS-Nag-EIIB, nagE; PTS system, N-acetylglucosamine-specific IIB component [EC:2.7.1.193] | 0.00225 | 0.00038 | 0.00222 | 0.00002 | 0.00052 | 65 | 0.00227 | 0.00003 | 0.00054 | 93 | 0.00075 | -0.06262 | 152.484 | 0.95015 | 0.98774 |
| mccA; cystathionine beta-synthase (O-acetyl-L-serine) [EC:2.5.1.134] | 0.00225 | 0.00048 | 0.00115 | 0.00002 | 0.00053 | 65 | 0.00301 | 0.00005 | 0.00071 | 93 | 0.00089 | -2.09491 | 154.227 | 0.03781 | 0.63091 |
| asdA; aspartate 4-decarboxylase [EC:4.1.1.12] | 0.00224 | 0.00050 | 0.00264 | 0.00007 | 0.00103 | 65 | 0.00196 | 0.00002 | 0.00044 | 93 | 0.00112 | 0.60420 | 87.151 | 0.54728 | 0.86509 |
| spoIIIAA; stage III sporulation protein AA | 0.00224 | 0.00052 | 0.00108 | 0.00001 | 0.00047 | 65 | 0.00306 | 0.00006 | 0.00080 | 93 | 0.00093 | -2.12070 | 142.351 | 0.03568 | 0.62411 |
| spoIIIAD; stage III sporulation protein AD | 0.00224 | 0.00052 | 0.00108 | 0.00001 | 0.00047 | 65 | 0.00306 | 0.00006 | 0.00080 | 93 | 0.00093 | -2.12070 | 142.351 | 0.03568 | 0.62411 |
| nreA; nitrogen regulatory protein A | 0.00224 | 0.00059 | 0.00206 | 0.00006 | 0.00097 | 65 | 0.00237 | 0.00005 | 0.00074 | 93 | 0.00122 | -0.25967 | 129.841 | 0.79553 | 0.94791 |
| fas; fatty acid synthase, bacteria type [EC:2.3.1.-] | 0.00224 | 0.00031 | 0.00209 | 0.00001 | 0.00038 | 65 | 0.00234 | 0.00002 | 0.00046 | 93 | 0.00060 | -0.41827 | 155.983 | 0.67633 | 0.91266 |
| ndhF; NAD(P)H-quinone oxidoreductase subunit 5 [EC:1.6.5.3] | 0.00223 | 0.00047 | 0.00146 | 0.00002 | 0.00057 | 65 | 0.00277 | 0.00004 | 0.00069 | 93 | 0.00089 | -1.47431 | 155.965 | 0.14241 | 0.77376 |
| spoVR; stage V sporulation protein R | 0.00221 | 0.00048 | 0.00128 | 0.00002 | 0.00056 | 65 | 0.00286 | 0.00005 | 0.00072 | 93 | 0.00091 | -1.73599 | 155.229 | 0.08455 | 0.76567 |
| glcC; GntR family transcriptional regulator, glc operon transcriptional activator | 0.00221 | 0.00041 | 0.00300 | 0.00004 | 0.00081 | 65 | 0.00166 | 0.00002 | 0.00040 | 93 | 0.00090 | 1.47532 | 95.639 | 0.14341 | 0.77376 |
| glvA; maltose-6’-phosphate glucosidase [EC:3.2.1.122] | 0.00220 | 0.00033 | 0.00244 | 0.00002 | 0.00048 | 65 | 0.00204 | 0.00002 | 0.00045 | 93 | 0.00066 | 0.60769 | 147.010 | 0.54433 | 0.86381 |
| parM; plasmid segregation protein ParM | 0.00220 | 0.00048 | 0.00110 | 0.00001 | 0.00034 | 65 | 0.00297 | 0.00006 | 0.00078 | 93 | 0.00085 | -2.19693 | 123.076 | 0.02990 | 0.61413 |
| ABC-2.CPSE.P1; capsular polysaccharide transport system permease protein | 0.00219 | 0.00040 | 0.00201 | 0.00002 | 0.00056 | 65 | 0.00232 | 0.00003 | 0.00056 | 93 | 0.00079 | -0.40334 | 151.159 | 0.68727 | 0.91567 |
| gerKC; spore germination protein KC | 0.00219 | 0.00067 | 0.00084 | 0.00002 | 0.00058 | 65 | 0.00313 | 0.00011 | 0.00106 | 93 | 0.00121 | -1.89010 | 137.710 | 0.06085 | 0.73001 |
| uctC; CoA:oxalate CoA-transferase [EC:2.8.3.19] | 0.00218 | 0.00055 | 0.00228 | 0.00005 | 0.00088 | 65 | 0.00212 | 0.00005 | 0.00070 | 93 | 0.00112 | 0.14029 | 134.043 | 0.88864 | 0.97039 |
| yscQ, sctQ, hrcQ, ssaQ, spaO; type III secretion protein Q | 0.00217 | 0.00049 | 0.00173 | 0.00002 | 0.00059 | 65 | 0.00248 | 0.00005 | 0.00072 | 93 | 0.00093 | -0.81017 | 155.959 | 0.41908 | 0.81980 |
| K09386; uncharacterized protein | 0.00217 | 0.00051 | 0.00055 | 0.00000 | 0.00016 | 65 | 0.00330 | 0.00006 | 0.00083 | 93 | 0.00085 | -3.23767 | 98.953 | 0.00164 | 0.46257 |
| allB; allantoinase [EC:3.5.2.5] | 0.00217 | 0.00046 | 0.00181 | 0.00003 | 0.00068 | 65 | 0.00242 | 0.00004 | 0.00062 | 93 | 0.00092 | -0.66607 | 144.724 | 0.50643 | 0.84956 |
| ABC-2.CPSE.A; capsular polysaccharide transport system ATP-binding protein [EC:3.6.3.38] | 0.00216 | 0.00040 | 0.00199 | 0.00002 | 0.00056 | 65 | 0.00228 | 0.00003 | 0.00055 | 93 | 0.00079 | -0.37545 | 150.465 | 0.70786 | 0.92004 |
| wecE, rffA; dTDP-4-amino-4,6-dideoxygalactose transaminase [EC:2.6.1.59] | 0.00216 | 0.00040 | 0.00173 | 0.00002 | 0.00048 | 65 | 0.00246 | 0.00003 | 0.00060 | 93 | 0.00077 | -0.95882 | 155.843 | 0.33914 | 0.78433 |
| spoVFA; dipicolinate synthase subunit A | 0.00216 | 0.00048 | 0.00081 | 0.00001 | 0.00035 | 65 | 0.00310 | 0.00005 | 0.00077 | 93 | 0.00084 | -2.70655 | 126.461 | 0.00774 | 0.54764 |
| spoVFB; dipicolinate synthase subunit B | 0.00216 | 0.00048 | 0.00081 | 0.00001 | 0.00035 | 65 | 0.00310 | 0.00005 | 0.00077 | 93 | 0.00084 | -2.70655 | 126.461 | 0.00774 | 0.54764 |
| sir; sulfite reductase (ferredoxin) [EC:1.8.7.1] | 0.00216 | 0.00033 | 0.00172 | 0.00001 | 0.00036 | 65 | 0.00247 | 0.00002 | 0.00051 | 93 | 0.00062 | -1.19866 | 153.019 | 0.23251 | 0.77376 |
| parD1_3_4; antitoxin ParD1/3/4 | 0.00216 | 0.00039 | 0.00147 | 0.00001 | 0.00048 | 65 | 0.00264 | 0.00003 | 0.00057 | 93 | 0.00075 | -1.56595 | 155.992 | 0.11939 | 0.77376 |
| uhpB; two-component system, NarL family, sensor histidine kinase UhpB [EC:2.7.13.3] | 0.00215 | 0.00036 | 0.00231 | 0.00002 | 0.00058 | 65 | 0.00204 | 0.00002 | 0.00046 | 93 | 0.00074 | 0.36844 | 133.177 | 0.71313 | 0.92004 |
| rsbR; rsbT co-antagonist protein RsbR | 0.00215 | 0.00043 | 0.00185 | 0.00002 | 0.00054 | 65 | 0.00236 | 0.00004 | 0.00063 | 93 | 0.00083 | -0.61214 | 155.779 | 0.54134 | 0.86207 |
| psuK; pseudouridine kinase [EC:2.7.1.83] | 0.00215 | 0.00036 | 0.00200 | 0.00002 | 0.00050 | 65 | 0.00225 | 0.00002 | 0.00049 | 93 | 0.00071 | -0.35560 | 149.978 | 0.72264 | 0.92156 |
| nicE, maiA; maleate isomerase [EC:5.2.1.1] | 0.00215 | 0.00042 | 0.00230 | 0.00004 | 0.00075 | 65 | 0.00204 | 0.00002 | 0.00049 | 93 | 0.00090 | 0.29280 | 114.800 | 0.77020 | 0.94009 |
| K09966; uncharacterized protein | 0.00214 | 0.00036 | 0.00166 | 0.00001 | 0.00040 | 65 | 0.00248 | 0.00003 | 0.00054 | 93 | 0.00067 | -1.22704 | 154.424 | 0.22168 | 0.77376 |
| nosF; Cu-processing system ATP-binding protein | 0.00214 | 0.00036 | 0.00157 | 0.00001 | 0.00035 | 65 | 0.00254 | 0.00003 | 0.00055 | 93 | 0.00065 | -1.47993 | 146.410 | 0.14104 | 0.77376 |
| arnF; undecaprenyl phosphate-alpha-L-ara4N flippase subunit ArnF | 0.00214 | 0.00048 | 0.00187 | 0.00002 | 0.00061 | 65 | 0.00233 | 0.00004 | 0.00069 | 93 | 0.00092 | -0.50339 | 155.548 | 0.61541 | 0.88878 |
| arnA, pmrI; UDP-4-amino-4-deoxy-L-arabinose formyltransferase / UDP-glucuronic acid dehydrogenase (UDP-4-keto-hexauronic acid decarboxylating) [EC:2.1.2.13 1.1.1.305] | 0.00213 | 0.00048 | 0.00186 | 0.00002 | 0.00061 | 65 | 0.00232 | 0.00004 | 0.00069 | 93 | 0.00092 | -0.50071 | 155.540 | 0.61728 | 0.88903 |
| arnD; undecaprenyl phosphate-alpha-L-ara4FN deformylase [EC:3.5.1.-] | 0.00213 | 0.00048 | 0.00186 | 0.00002 | 0.00061 | 65 | 0.00232 | 0.00004 | 0.00069 | 93 | 0.00092 | -0.50071 | 155.540 | 0.61728 | 0.88903 |
| urdA; urocanate reductase [EC:1.3.99.33] | 0.00213 | 0.00098 | 0.00140 | 0.00003 | 0.00065 | 65 | 0.00264 | 0.00024 | 0.00160 | 93 | 0.00172 | -0.71839 | 120.411 | 0.47391 | 0.84050 |
| pvdA, SIDA; L-ornithine N5-monooxygenase [EC:1.14.13.195 1.14.13.196] | 0.00212 | 0.00042 | 0.00206 | 0.00002 | 0.00059 | 65 | 0.00216 | 0.00003 | 0.00058 | 93 | 0.00082 | -0.12329 | 149.981 | 0.90204 | 0.97641 |
| yedL; putative acetyltransferase [EC:2.3.1.-] | 0.00211 | 0.00040 | 0.00123 | 0.00001 | 0.00032 | 65 | 0.00273 | 0.00004 | 0.00064 | 93 | 0.00072 | -2.09715 | 131.562 | 0.03789 | 0.63091 |
| ygeS, xdhA; xanthine dehydrogenase molybdenum-binding subunit [EC:1.17.1.4] | 0.00211 | 0.00050 | 0.00236 | 0.00006 | 0.00099 | 65 | 0.00194 | 0.00002 | 0.00051 | 93 | 0.00111 | 0.37670 | 98.143 | 0.70721 | 0.92004 |
| yteR, yesR; unsaturated rhamnogalacturonyl hydrolase [EC:3.2.1.172] | 0.00211 | 0.00051 | 0.00094 | 0.00000 | 0.00025 | 65 | 0.00292 | 0.00007 | 0.00085 | 93 | 0.00088 | -2.24459 | 106.886 | 0.02685 | 0.61112 |
| nirC; nitrite transporter | 0.00211 | 0.00043 | 0.00221 | 0.00005 | 0.00088 | 65 | 0.00203 | 0.00001 | 0.00039 | 93 | 0.00097 | 0.19045 | 89.555 | 0.84939 | 0.95712 |
| astC; succinylornithine aminotransferase [EC:2.6.1.81] | 0.00211 | 0.00040 | 0.00170 | 0.00001 | 0.00041 | 65 | 0.00239 | 0.00004 | 0.00062 | 93 | 0.00074 | -0.92363 | 148.873 | 0.35718 | 0.79437 |
| hspQ; heat shock protein HspQ | 0.00210 | 0.00034 | 0.00124 | 0.00001 | 0.00030 | 65 | 0.00271 | 0.00003 | 0.00054 | 93 | 0.00061 | -2.38618 | 139.288 | 0.01837 | 0.59095 |
| malF; maltose/maltodextrin transport system permease protein | 0.00210 | 0.00046 | 0.00176 | 0.00003 | 0.00064 | 65 | 0.00234 | 0.00004 | 0.00063 | 93 | 0.00090 | -0.64996 | 150.092 | 0.51671 | 0.85216 |
| malG; maltose/maltodextrin transport system permease protein | 0.00210 | 0.00046 | 0.00176 | 0.00003 | 0.00064 | 65 | 0.00234 | 0.00004 | 0.00063 | 93 | 0.00090 | -0.64996 | 150.092 | 0.51671 | 0.85216 |
| dexA; dextranase [EC:3.2.1.11] | 0.00210 | 0.00049 | 0.00214 | 0.00002 | 0.00050 | 65 | 0.00207 | 0.00005 | 0.00075 | 93 | 0.00091 | 0.07764 | 149.846 | 0.93822 | 0.98459 |
| phnG; alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG [EC:2.7.8.37] | 0.00210 | 0.00040 | 0.00107 | 0.00001 | 0.00036 | 65 | 0.00282 | 0.00004 | 0.00062 | 93 | 0.00072 | -2.42953 | 140.864 | 0.01638 | 0.59095 |
| phnI; alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI [EC:2.7.8.37] | 0.00210 | 0.00040 | 0.00107 | 0.00001 | 0.00036 | 65 | 0.00282 | 0.00004 | 0.00062 | 93 | 0.00072 | -2.42953 | 140.864 | 0.01638 | 0.59095 |
| phnJ; alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase [EC:4.7.1.1] | 0.00210 | 0.00040 | 0.00107 | 0.00001 | 0.00036 | 65 | 0.00282 | 0.00004 | 0.00062 | 93 | 0.00072 | -2.42953 | 140.864 | 0.01638 | 0.59095 |
| phnK; putative phosphonate transport system ATP-binding protein | 0.00210 | 0.00040 | 0.00107 | 0.00001 | 0.00036 | 65 | 0.00282 | 0.00004 | 0.00062 | 93 | 0.00072 | -2.42953 | 140.864 | 0.01638 | 0.59095 |
| merA; mercuric reductase [EC:1.16.1.1] | 0.00210 | 0.00043 | 0.00218 | 0.00004 | 0.00074 | 65 | 0.00204 | 0.00002 | 0.00052 | 93 | 0.00091 | 0.15592 | 121.108 | 0.87636 | 0.96705 |
| pafB; proteasome accessory factor B | 0.00210 | 0.00030 | 0.00219 | 0.00001 | 0.00044 | 65 | 0.00203 | 0.00002 | 0.00041 | 93 | 0.00060 | 0.27276 | 147.012 | 0.78542 | 0.94445 |
| phnH; alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH [EC:2.7.8.37] | 0.00210 | 0.00040 | 0.00107 | 0.00001 | 0.00036 | 65 | 0.00281 | 0.00004 | 0.00063 | 93 | 0.00072 | -2.41327 | 140.820 | 0.01710 | 0.59095 |
| phnL; alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnL [EC:2.7.8.37] | 0.00209 | 0.00040 | 0.00107 | 0.00001 | 0.00036 | 65 | 0.00281 | 0.00004 | 0.00063 | 93 | 0.00072 | -2.40547 | 140.851 | 0.01745 | 0.59095 |
| PTS-HPR.PTSO, ptsO, npr; phosphocarrier protein NPr | 0.00209 | 0.00035 | 0.00202 | 0.00001 | 0.00045 | 65 | 0.00214 | 0.00002 | 0.00051 | 93 | 0.00068 | -0.17259 | 155.524 | 0.86320 | 0.96263 |
| pdaA; peptidoglycan-N-acetylmuramic acid deacetylase [EC:3.5.1.-] | 0.00209 | 0.00045 | 0.00104 | 0.00002 | 0.00051 | 65 | 0.00282 | 0.00004 | 0.00067 | 93 | 0.00084 | -2.10974 | 154.673 | 0.03649 | 0.62546 |
| pimA; phosphatidyl-myo-inositol alpha-mannosyltransferase [EC:2.4.1.345] | 0.00208 | 0.00029 | 0.00203 | 0.00001 | 0.00039 | 65 | 0.00212 | 0.00002 | 0.00041 | 93 | 0.00056 | -0.15971 | 153.510 | 0.87332 | 0.96576 |
| chuX; heme iron utilization protein | 0.00208 | 0.00046 | 0.00156 | 0.00002 | 0.00062 | 65 | 0.00245 | 0.00004 | 0.00066 | 93 | 0.00090 | -0.98037 | 153.899 | 0.32844 | 0.77376 |
| NAMPT; nicotinamide phosphoribosyltransferase [EC:2.4.2.12] | 0.00208 | 0.00034 | 0.00204 | 0.00001 | 0.00041 | 65 | 0.00211 | 0.00002 | 0.00050 | 93 | 0.00065 | -0.10696 | 155.981 | 0.91496 | 0.97664 |
| ntrY; two-component system, NtrC family, nitrogen regulation sensor histidine kinase NtrY [EC:2.7.13.3] | 0.00208 | 0.00039 | 0.00110 | 0.00001 | 0.00034 | 65 | 0.00276 | 0.00003 | 0.00060 | 93 | 0.00069 | -2.37831 | 139.652 | 0.01875 | 0.59095 |
| csgF; curli production assembly/transport component CsgF | 0.00207 | 0.00038 | 0.00276 | 0.00004 | 0.00078 | 65 | 0.00160 | 0.00001 | 0.00034 | 93 | 0.00085 | 1.36437 | 88.073 | 0.17593 | 0.77376 |
| spoIIGA; stage II sporulation protein GA (sporulation sigma-E factor processing peptidase) [EC:3.4.23.-] | 0.00207 | 0.00044 | 0.00112 | 0.00002 | 0.00051 | 65 | 0.00273 | 0.00004 | 0.00066 | 93 | 0.00083 | -1.93362 | 155.345 | 0.05498 | 0.70519 |
| rfbF, rhlC; rhamnosyltransferase [EC:2.4.1.-] | 0.00206 | 0.00054 | 0.00233 | 0.00005 | 0.00089 | 65 | 0.00187 | 0.00004 | 0.00068 | 93 | 0.00112 | 0.41399 | 130.175 | 0.67956 | 0.91311 |
| ntrX; two-component system, NtrC family, nitrogen regulation response regulator NtrX | 0.00205 | 0.00036 | 0.00115 | 0.00001 | 0.00035 | 65 | 0.00269 | 0.00003 | 0.00056 | 93 | 0.00065 | -2.34893 | 145.510 | 0.02017 | 0.59095 |
| nodI; lipooligosaccharide transport system ATP-binding protein | 0.00204 | 0.00040 | 0.00143 | 0.00001 | 0.00043 | 65 | 0.00247 | 0.00004 | 0.00062 | 93 | 0.00075 | -1.38595 | 151.676 | 0.16780 | 0.77376 |
| thiY; putative hydroxymethylpyrimidine transport system substrate-binding protein | 0.00204 | 0.00033 | 0.00152 | 0.00001 | 0.00039 | 65 | 0.00241 | 0.00002 | 0.00049 | 93 | 0.00062 | -1.43083 | 155.692 | 0.15448 | 0.77376 |
| GMPP; mannose-1-phosphate guanylyltransferase [EC:2.7.7.13] | 0.00204 | 0.00038 | 0.00116 | 0.00001 | 0.00037 | 65 | 0.00266 | 0.00003 | 0.00058 | 93 | 0.00069 | -2.17722 | 147.193 | 0.03106 | 0.61842 |
| wbpP; UDP-N-acetylglucosamine 4-epimerase [EC:5.1.3.7] | 0.00204 | 0.00036 | 0.00200 | 0.00002 | 0.00049 | 65 | 0.00207 | 0.00002 | 0.00050 | 93 | 0.00070 | -0.08967 | 152.170 | 0.92867 | 0.98176 |
| iclR; IclR family transcriptional regulator, acetate operon repressor | 0.00204 | 0.00034 | 0.00124 | 0.00001 | 0.00038 | 65 | 0.00260 | 0.00002 | 0.00052 | 93 | 0.00064 | -2.13601 | 153.471 | 0.03426 | 0.62130 |
| hasA; hyaluronan synthase [EC:2.4.1.212] | 0.00204 | 0.00057 | 0.00232 | 0.00007 | 0.00102 | 65 | 0.00184 | 0.00004 | 0.00067 | 93 | 0.00122 | 0.38961 | 116.699 | 0.69753 | 0.91825 |
| HK; hexokinase [EC:2.7.1.1] | 0.00204 | 0.00051 | 0.00240 | 0.00006 | 0.00099 | 65 | 0.00178 | 0.00002 | 0.00052 | 93 | 0.00112 | 0.56148 | 98.510 | 0.57574 | 0.87389 |
| EPS15; epidermal growth factor receptor substrate 15 | 0.00203 | 0.00050 | 0.00186 | 0.00002 | 0.00049 | 65 | 0.00215 | 0.00006 | 0.00078 | 93 | 0.00092 | -0.31275 | 146.048 | 0.75492 | 0.93665 |
| ner, nlp, sfsB; Ner family transcriptional regulator | 0.00203 | 0.00039 | 0.00213 | 0.00002 | 0.00057 | 65 | 0.00196 | 0.00003 | 0.00054 | 93 | 0.00078 | 0.22189 | 147.386 | 0.82471 | 0.95506 |
| dmlR; LysR family transcriptional regulator, transcriptional activator for dmlA | 0.00203 | 0.00043 | 0.00128 | 0.00001 | 0.00041 | 65 | 0.00255 | 0.00004 | 0.00066 | 93 | 0.00078 | -1.63389 | 146.016 | 0.10444 | 0.77376 |
| assT; arylsulfate sulfotransferase [EC:2.8.2.22] | 0.00203 | 0.00088 | 0.00105 | 0.00002 | 0.00050 | 65 | 0.00271 | 0.00019 | 0.00144 | 93 | 0.00153 | -1.08726 | 112.854 | 0.27924 | 0.77376 |
| fucK; L-fuculokinase [EC:2.7.1.51] | 0.00202 | 0.00048 | 0.00259 | 0.00007 | 0.00102 | 65 | 0.00163 | 0.00001 | 0.00039 | 93 | 0.00109 | 0.88706 | 82.864 | 0.37761 | 0.79837 |
| fixJ; two-component system, LuxR family, response regulator FixJ | 0.00202 | 0.00041 | 0.00105 | 0.00001 | 0.00038 | 65 | 0.00270 | 0.00004 | 0.00064 | 93 | 0.00074 | -2.21293 | 142.415 | 0.02849 | 0.61334 |
| wbdC, wbpZ; N-acetyl-alpha-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol 3-alpha-mannosyltransferase / rhamnosyltransferase [EC:2.4.1.348 2.4.1.-] | 0.00202 | 0.00040 | 0.00185 | 0.00002 | 0.00049 | 65 | 0.00214 | 0.00003 | 0.00059 | 93 | 0.00076 | -0.37025 | 155.974 | 0.71170 | 0.92004 |
| aac6-I, aacA7; aminoglycoside 6’-N-acetyltransferase I [EC:2.3.1.82] | 0.00202 | 0.00047 | 0.00226 | 0.00004 | 0.00080 | 65 | 0.00185 | 0.00003 | 0.00057 | 93 | 0.00099 | 0.41351 | 123.363 | 0.67995 | 0.91311 |
| rfbP; undecaprenyl-phosphate galactose phosphotransferase [EC:2.7.8.6] | 0.00201 | 0.00052 | 0.00262 | 0.00007 | 0.00102 | 65 | 0.00159 | 0.00002 | 0.00051 | 93 | 0.00114 | 0.90100 | 95.482 | 0.36985 | 0.79587 |
| vraR; two-component system, NarL family, vancomycin resistance associated response regulator VraR | 0.00201 | 0.00055 | 0.00252 | 0.00007 | 0.00103 | 65 | 0.00166 | 0.00003 | 0.00059 | 93 | 0.00119 | 0.72382 | 105.024 | 0.47079 | 0.83912 |
| spmA; spore maturation protein A | 0.00201 | 0.00040 | 0.00114 | 0.00001 | 0.00043 | 65 | 0.00261 | 0.00003 | 0.00060 | 93 | 0.00074 | -1.99242 | 153.044 | 0.04810 | 0.66712 |
| chbG; chitin disaccharide deacetylase [EC:3.5.1.105] | 0.00201 | 0.00035 | 0.00168 | 0.00002 | 0.00048 | 65 | 0.00224 | 0.00002 | 0.00048 | 93 | 0.00068 | -0.81737 | 151.168 | 0.41500 | 0.81730 |
| trmH; tRNA (guanosine-2’-O-)-methyltransferase [EC:2.1.1.34] | 0.00201 | 0.00033 | 0.00165 | 0.00001 | 0.00035 | 65 | 0.00226 | 0.00002 | 0.00049 | 93 | 0.00061 | -1.00164 | 152.507 | 0.31810 | 0.77376 |
| hisM; histidine transport system permease protein | 0.00200 | 0.00042 | 0.00169 | 0.00002 | 0.00049 | 65 | 0.00222 | 0.00004 | 0.00062 | 93 | 0.00079 | -0.67801 | 155.767 | 0.49877 | 0.84856 |
| rhtC; threonine efflux protein | 0.00200 | 0.00035 | 0.00191 | 0.00001 | 0.00045 | 65 | 0.00206 | 0.00002 | 0.00050 | 93 | 0.00067 | -0.22114 | 155.067 | 0.82527 | 0.95506 |
| fhuF; ferric iron reductase protein FhuF | 0.00200 | 0.00046 | 0.00165 | 0.00002 | 0.00056 | 65 | 0.00225 | 0.00004 | 0.00068 | 93 | 0.00088 | -0.68229 | 156.000 | 0.49606 | 0.84797 |
| ABC-2.CPSE.P; capsular polysaccharide transport system permease protein | 0.00199 | 0.00037 | 0.00195 | 0.00002 | 0.00056 | 65 | 0.00203 | 0.00002 | 0.00050 | 93 | 0.00075 | -0.10924 | 144.203 | 0.91316 | 0.97664 |
| hdhA; 7-alpha-hydroxysteroid dehydrogenase [EC:1.1.1.159] | 0.00199 | 0.00043 | 0.00145 | 0.00002 | 0.00049 | 65 | 0.00236 | 0.00004 | 0.00063 | 93 | 0.00080 | -1.14181 | 155.247 | 0.25529 | 0.77376 |
| fimG; minor fimbrial subunit | 0.00198 | 0.00047 | 0.00152 | 0.00002 | 0.00057 | 65 | 0.00231 | 0.00005 | 0.00070 | 93 | 0.00090 | -0.87957 | 155.860 | 0.38045 | 0.79870 |
| kpsS, lipB; capsular polysaccharide export protein | 0.00198 | 0.00037 | 0.00191 | 0.00002 | 0.00055 | 65 | 0.00203 | 0.00002 | 0.00050 | 93 | 0.00074 | -0.15923 | 145.031 | 0.87371 | 0.96576 |
| hisQ; histidine transport system permease protein | 0.00198 | 0.00041 | 0.00168 | 0.00002 | 0.00049 | 65 | 0.00219 | 0.00003 | 0.00060 | 93 | 0.00078 | -0.65309 | 155.967 | 0.51466 | 0.85216 |
| E2.3.1.37, ALAS; 5-aminolevulinate synthase [EC:2.3.1.37] | 0.00198 | 0.00038 | 0.00081 | 0.00000 | 0.00021 | 65 | 0.00279 | 0.00004 | 0.00062 | 93 | 0.00065 | -3.04690 | 111.521 | 0.00289 | 0.46257 |
| eco; ecotin | 0.00197 | 0.00029 | 0.00216 | 0.00001 | 0.00043 | 65 | 0.00184 | 0.00001 | 0.00039 | 93 | 0.00058 | 0.54744 | 143.994 | 0.58492 | 0.87720 |
| K06904; uncharacterized protein | 0.00197 | 0.00030 | 0.00112 | 0.00000 | 0.00021 | 65 | 0.00257 | 0.00002 | 0.00047 | 93 | 0.00052 | -2.80056 | 125.621 | 0.00591 | 0.54022 |
| hisP; histidine transport system ATP-binding protein [EC:3.6.3.21] | 0.00197 | 0.00040 | 0.00164 | 0.00001 | 0.00048 | 65 | 0.00219 | 0.00003 | 0.00060 | 93 | 0.00077 | -0.72130 | 155.623 | 0.47181 | 0.84020 |
| norQ; nitric oxide reductase NorQ protein | 0.00196 | 0.00037 | 0.00059 | 0.00000 | 0.00015 | 65 | 0.00293 | 0.00003 | 0.00060 | 93 | 0.00062 | -3.76422 | 103.871 | 0.00028 | 0.46257 |
| dgoT; MFS transporter, ACS family, D-galactonate transporter | 0.00196 | 0.00038 | 0.00198 | 0.00002 | 0.00059 | 65 | 0.00195 | 0.00002 | 0.00050 | 93 | 0.00077 | 0.04980 | 140.298 | 0.96036 | 0.98903 |
| rsbQ; sigma-B regulation protein RsbQ | 0.00196 | 0.00047 | 0.00156 | 0.00002 | 0.00054 | 65 | 0.00223 | 0.00005 | 0.00070 | 93 | 0.00089 | -0.75639 | 155.135 | 0.45056 | 0.83438 |
| ict-P; itaconate CoA-transferase [EC:2.8.3.- 2.8.3.22] | 0.00196 | 0.00042 | 0.00213 | 0.00003 | 0.00070 | 65 | 0.00183 | 0.00003 | 0.00053 | 93 | 0.00087 | 0.34480 | 128.800 | 0.73081 | 0.92479 |
| lysK; lysyl-tRNA synthetase, class I [EC:6.1.1.6] | 0.00195 | 0.00034 | 0.00089 | 0.00000 | 0.00024 | 65 | 0.00270 | 0.00003 | 0.00053 | 93 | 0.00058 | -3.09840 | 125.469 | 0.00240 | 0.46257 |
| pheC; cyclohexadienyl dehydratase [EC:4.2.1.51 4.2.1.91] | 0.00195 | 0.00041 | 0.00105 | 0.00001 | 0.00037 | 65 | 0.00258 | 0.00004 | 0.00064 | 93 | 0.00074 | -2.07174 | 141.884 | 0.04010 | 0.63577 |
| K09990; uncharacterized protein | 0.00194 | 0.00046 | 0.00097 | 0.00001 | 0.00034 | 65 | 0.00263 | 0.00005 | 0.00074 | 93 | 0.00082 | -2.03158 | 127.652 | 0.04427 | 0.65981 |
| blaI; BlaI family transcriptional regulator, penicillinase repressor | 0.00194 | 0.00047 | 0.00115 | 0.00002 | 0.00055 | 65 | 0.00249 | 0.00005 | 0.00070 | 93 | 0.00089 | -1.50445 | 155.504 | 0.13449 | 0.77376 |
| cphB; cyanophycinase [EC:3.4.15.6] | 0.00193 | 0.00055 | 0.00152 | 0.00002 | 0.00059 | 65 | 0.00222 | 0.00007 | 0.00084 | 93 | 0.00103 | -0.68101 | 152.008 | 0.49690 | 0.84797 |
| thiX; putative hydroxymethylpyrimidine transport system permease protein | 0.00193 | 0.00031 | 0.00144 | 0.00001 | 0.00037 | 65 | 0.00228 | 0.00002 | 0.00045 | 93 | 0.00059 | -1.42626 | 155.971 | 0.15579 | 0.77376 |
| K11312; cupin 2 domain-containing protein | 0.00193 | 0.00030 | 0.00273 | 0.00002 | 0.00053 | 65 | 0.00138 | 0.00001 | 0.00034 | 93 | 0.00063 | 2.15998 | 112.552 | 0.03290 | 0.62130 |
| K13522, nadM; bifunctional NMN adenylyltransferase/nudix hydrolase [EC:2.7.7.1 3.6.1.-] | 0.00193 | 0.00039 | 0.00145 | 0.00001 | 0.00037 | 65 | 0.00227 | 0.00004 | 0.00062 | 93 | 0.00072 | -1.13513 | 143.329 | 0.25822 | 0.77376 |
| hpaB; 4-hydroxyphenylacetate 3-monooxygenase [EC:1.14.14.9] | 0.00193 | 0.00050 | 0.00109 | 0.00001 | 0.00037 | 65 | 0.00251 | 0.00006 | 0.00081 | 93 | 0.00089 | -1.59233 | 126.063 | 0.11382 | 0.77376 |
| DDAH, ddaH; dimethylargininase [EC:3.5.3.18] | 0.00193 | 0.00054 | 0.00196 | 0.00005 | 0.00090 | 65 | 0.00191 | 0.00004 | 0.00067 | 93 | 0.00112 | 0.04411 | 128.381 | 0.96489 | 0.99101 |
| wbjC; UDP-2-acetamido-2,6-beta-L-arabino-hexul-4-ose reductase [EC:1.1.1.367] | 0.00192 | 0.00036 | 0.00121 | 0.00001 | 0.00041 | 65 | 0.00241 | 0.00003 | 0.00055 | 93 | 0.00068 | -1.75998 | 154.290 | 0.08039 | 0.75920 |
| bpeE; membrane fusion protein, multidrug efflux system | 0.00192 | 0.00036 | 0.00115 | 0.00001 | 0.00033 | 65 | 0.00245 | 0.00003 | 0.00057 | 93 | 0.00066 | -1.98575 | 140.564 | 0.04901 | 0.67403 |
| K06952; uncharacterized protein | 0.00192 | 0.00036 | 0.00092 | 0.00001 | 0.00033 | 65 | 0.00261 | 0.00003 | 0.00056 | 93 | 0.00065 | -2.60385 | 143.358 | 0.01019 | 0.58284 |
| P4HA; prolyl 4-hydroxylase [EC:1.14.11.2] | 0.00191 | 0.00037 | 0.00110 | 0.00001 | 0.00038 | 65 | 0.00248 | 0.00003 | 0.00056 | 93 | 0.00068 | -2.04391 | 149.593 | 0.04272 | 0.65457 |
| hcaR; LysR family transcriptional regulator, hca operon transcriptional activator | 0.00191 | 0.00045 | 0.00146 | 0.00001 | 0.00038 | 65 | 0.00222 | 0.00005 | 0.00071 | 93 | 0.00081 | -0.93906 | 136.389 | 0.34936 | 0.79120 |
| avtA; valine–pyruvate aminotransferase [EC:2.6.1.66] | 0.00190 | 0.00034 | 0.00229 | 0.00002 | 0.00059 | 65 | 0.00164 | 0.00002 | 0.00040 | 93 | 0.00072 | 0.90889 | 119.706 | 0.36523 | 0.79587 |
| fsaB, talC; fructose-6-phosphate aldolase 2 [EC:4.1.2.-] | 0.00190 | 0.00027 | 0.00223 | 0.00001 | 0.00044 | 65 | 0.00167 | 0.00001 | 0.00034 | 93 | 0.00056 | 0.99659 | 130.984 | 0.32080 | 0.77376 |
| E2.4.1.4; amylosucrase [EC:2.4.1.4] | 0.00190 | 0.00031 | 0.00159 | 0.00001 | 0.00035 | 65 | 0.00212 | 0.00002 | 0.00046 | 93 | 0.00058 | -0.91338 | 154.741 | 0.36246 | 0.79496 |
| adeC; outer membrane protein, multidrug efflux system | 0.00190 | 0.00040 | 0.00176 | 0.00002 | 0.00053 | 65 | 0.00200 | 0.00003 | 0.00057 | 93 | 0.00078 | -0.30268 | 154.254 | 0.76254 | 0.93813 |
| rocE, rocC; arginine/ornithine permease | 0.00190 | 0.00063 | 0.00134 | 0.00003 | 0.00062 | 65 | 0.00229 | 0.00009 | 0.00098 | 93 | 0.00116 | -0.81310 | 146.802 | 0.41748 | 0.81932 |
| K18335; 2-keto-3-deoxy-L-fuconate dehydrogenase [EC:1.1.1.-] | 0.00189 | 0.00035 | 0.00102 | 0.00001 | 0.00030 | 65 | 0.00250 | 0.00003 | 0.00055 | 93 | 0.00063 | -2.35747 | 137.441 | 0.01981 | 0.59095 |
| casC, cse4; CRISPR system Cascade subunit CasC | 0.00189 | 0.00028 | 0.00212 | 0.00001 | 0.00041 | 65 | 0.00174 | 0.00001 | 0.00038 | 93 | 0.00056 | 0.68272 | 146.624 | 0.49586 | 0.84797 |
| emrR, mprA; MarR family transcriptional regulator, negative regulator of the multidrug operon emrRAB | 0.00189 | 0.00036 | 0.00203 | 0.00002 | 0.00057 | 65 | 0.00179 | 0.00002 | 0.00046 | 93 | 0.00073 | 0.32755 | 133.839 | 0.74377 | 0.93099 |
| atl; bifunctional autolysin [EC:3.5.1.28 3.2.1.96] | 0.00189 | 0.00081 | 0.00264 | 0.00022 | 0.00185 | 65 | 0.00136 | 0.00002 | 0.00047 | 93 | 0.00191 | 0.66707 | 72.483 | 0.50685 | 0.84956 |
| xtmB; phage terminase large subunit | 0.00188 | 0.00045 | 0.00126 | 0.00001 | 0.00037 | 65 | 0.00232 | 0.00005 | 0.00072 | 93 | 0.00081 | -1.31629 | 134.196 | 0.19032 | 0.77376 |
| AARSD1, ALAX; misacylated tRNA(Ala) deacylase [EC:3.1.1.-] | 0.00188 | 0.00042 | 0.00079 | 0.00001 | 0.00034 | 65 | 0.00265 | 0.00004 | 0.00066 | 93 | 0.00075 | -2.48236 | 134.145 | 0.01429 | 0.59095 |
| rsbS; rsbT antagonist protein RsbS | 0.00188 | 0.00037 | 0.00172 | 0.00002 | 0.00053 | 65 | 0.00199 | 0.00002 | 0.00052 | 93 | 0.00074 | -0.36757 | 149.310 | 0.71371 | 0.92004 |
| dhaM; phosphoenolpyruvate—glycerone phosphotransferase subunit DhaM [EC:2.7.1.121] | 0.00188 | 0.00027 | 0.00178 | 0.00001 | 0.00039 | 65 | 0.00195 | 0.00001 | 0.00038 | 93 | 0.00054 | -0.30335 | 150.437 | 0.76204 | 0.93813 |
| hcnC; hydrogen cyanide synthase HcnC [EC:1.4.99.5] | 0.00188 | 0.00057 | 0.00174 | 0.00004 | 0.00083 | 65 | 0.00198 | 0.00006 | 0.00078 | 93 | 0.00114 | -0.21329 | 147.638 | 0.83140 | 0.95506 |
| hisJ; histidine transport system substrate-binding protein | 0.00188 | 0.00041 | 0.00152 | 0.00001 | 0.00047 | 65 | 0.00213 | 0.00004 | 0.00062 | 93 | 0.00078 | -0.77637 | 154.796 | 0.43871 | 0.83152 |
| K05967; uncharacterized protein | 0.00188 | 0.00064 | 0.00088 | 0.00002 | 0.00060 | 65 | 0.00257 | 0.00009 | 0.00101 | 93 | 0.00117 | -1.44445 | 142.910 | 0.15080 | 0.77376 |
| dltE; uncharacterized oxidoreductase [EC:1.-.-.-] | 0.00188 | 0.00037 | 0.00121 | 0.00001 | 0.00042 | 65 | 0.00235 | 0.00003 | 0.00056 | 93 | 0.00070 | -1.62873 | 154.583 | 0.10541 | 0.77376 |
| sam; S-adenosylmethionine uptake transporter | 0.00187 | 0.00035 | 0.00109 | 0.00001 | 0.00037 | 65 | 0.00242 | 0.00003 | 0.00053 | 93 | 0.00065 | -2.04804 | 152.001 | 0.04228 | 0.65074 |
| nifD; nitrogenase molybdenum-iron protein alpha chain [EC:1.18.6.1] | 0.00187 | 0.00053 | 0.00228 | 0.00007 | 0.00102 | 65 | 0.00159 | 0.00003 | 0.00057 | 93 | 0.00116 | 0.59187 | 103.571 | 0.55523 | 0.86813 |
| rhaA; L-rhamnose isomerase [EC:5.3.1.14] | 0.00187 | 0.00035 | 0.00102 | 0.00000 | 0.00026 | 65 | 0.00246 | 0.00003 | 0.00056 | 93 | 0.00062 | -2.32941 | 126.555 | 0.02142 | 0.59153 |
| fadB; enoyl-CoA hydratase [EC:4.2.1.17] | 0.00187 | 0.00064 | 0.00072 | 0.00002 | 0.00057 | 65 | 0.00266 | 0.00009 | 0.00100 | 93 | 0.00115 | -1.68046 | 140.757 | 0.09509 | 0.76609 |
| mtrA; AraC family transcriptional regulator, activator of mtrCDE | 0.00186 | 0.00048 | 0.00202 | 0.00005 | 0.00085 | 65 | 0.00175 | 0.00003 | 0.00056 | 93 | 0.00102 | 0.25826 | 115.971 | 0.79666 | 0.94795 |
| E3.2.1.58; glucan 1,3-beta-glucosidase [EC:3.2.1.58] | 0.00186 | 0.00033 | 0.00155 | 0.00001 | 0.00043 | 65 | 0.00209 | 0.00002 | 0.00047 | 93 | 0.00064 | -0.84026 | 154.798 | 0.40206 | 0.80895 |
| gsp; glutathionylspermidine amidase/synthetase [EC:3.5.1.78 6.3.1.8] | 0.00186 | 0.00037 | 0.00272 | 0.00003 | 0.00072 | 65 | 0.00125 | 0.00001 | 0.00036 | 93 | 0.00081 | 1.82554 | 95.151 | 0.07106 | 0.73909 |
| yihS; sulfoquinovose isomerase [EC:5.3.1.31] | 0.00185 | 0.00030 | 0.00199 | 0.00002 | 0.00048 | 65 | 0.00176 | 0.00001 | 0.00039 | 93 | 0.00062 | 0.37799 | 134.687 | 0.70603 | 0.92004 |
| rgpE; glucosyltransferase [EC:2.4.1.-] | 0.00185 | 0.00048 | 0.00189 | 0.00002 | 0.00049 | 65 | 0.00182 | 0.00005 | 0.00075 | 93 | 0.00090 | 0.07815 | 148.767 | 0.93781 | 0.98459 |
| cobS; cobaltochelatase CobS [EC:6.6.1.2] | 0.00184 | 0.00034 | 0.00087 | 0.00000 | 0.00022 | 65 | 0.00252 | 0.00003 | 0.00054 | 93 | 0.00058 | -2.83088 | 121.349 | 0.00544 | 0.54022 |
| fucD; L-fuconate dehydratase [EC:4.2.1.68] | 0.00184 | 0.00030 | 0.00171 | 0.00001 | 0.00044 | 65 | 0.00193 | 0.00002 | 0.00042 | 93 | 0.00060 | -0.36356 | 147.774 | 0.71671 | 0.92018 |
| coxD, ctaF; cytochrome c oxidase subunit IV [EC:1.9.3.1] | 0.00183 | 0.00042 | 0.00087 | 0.00001 | 0.00042 | 65 | 0.00250 | 0.00004 | 0.00065 | 93 | 0.00078 | -2.10065 | 148.251 | 0.03736 | 0.63026 |
| malM; maltose operon periplasmic protein | 0.00183 | 0.00044 | 0.00148 | 0.00002 | 0.00059 | 65 | 0.00208 | 0.00004 | 0.00063 | 93 | 0.00087 | -0.68890 | 153.750 | 0.49193 | 0.84797 |
| K07465; putative RecB family exonuclease | 0.00183 | 0.00028 | 0.00179 | 0.00001 | 0.00037 | 65 | 0.00186 | 0.00001 | 0.00040 | 93 | 0.00054 | -0.12259 | 154.691 | 0.90259 | 0.97641 |
| oprC, opcM; outer membrane protein, multidrug efflux system | 0.00182 | 0.00043 | 0.00134 | 0.00001 | 0.00043 | 65 | 0.00216 | 0.00004 | 0.00067 | 93 | 0.00080 | -1.03202 | 146.910 | 0.30376 | 0.77376 |
| DHPS, dys; deoxyhypusine synthase [EC:2.5.1.46] | 0.00182 | 0.00029 | 0.00157 | 0.00001 | 0.00036 | 65 | 0.00200 | 0.00002 | 0.00042 | 93 | 0.00055 | -0.78277 | 155.834 | 0.43495 | 0.82874 |
| yjgB; uncharacterized zinc-type alcohol dehydrogenase-like protein [EC:1.-.-.-] | 0.00182 | 0.00033 | 0.00162 | 0.00001 | 0.00040 | 65 | 0.00196 | 0.00002 | 0.00050 | 93 | 0.00063 | -0.53698 | 155.761 | 0.59205 | 0.87898 |
| cslA; chondroitin AC lyase [EC:4.2.2.5] | 0.00182 | 0.00056 | 0.00153 | 0.00004 | 0.00080 | 65 | 0.00202 | 0.00006 | 0.00078 | 93 | 0.00112 | -0.43310 | 149.414 | 0.66557 | 0.91126 |
| NAGLU; alpha-N-acetylglucosaminidase [EC:3.2.1.50] | 0.00182 | 0.00049 | 0.00093 | 0.00001 | 0.00030 | 65 | 0.00244 | 0.00006 | 0.00079 | 93 | 0.00085 | -1.77295 | 116.346 | 0.07885 | 0.75800 |
| csgE; curli production assembly/transport component CsgE | 0.00182 | 0.00035 | 0.00248 | 0.00004 | 0.00074 | 65 | 0.00135 | 0.00001 | 0.00029 | 93 | 0.00080 | 1.41358 | 83.850 | 0.16119 | 0.77376 |
| croR; 3-hydroxybutyryl-CoA dehydratase [EC:4.2.1.55] | 0.00181 | 0.00028 | 0.00136 | 0.00001 | 0.00037 | 65 | 0.00213 | 0.00001 | 0.00039 | 93 | 0.00054 | -1.43477 | 153.876 | 0.15338 | 0.77376 |
| torY; trimethylamine-N-oxide reductase (cytochrome c), cytochrome c-type subunit TorY | 0.00181 | 0.00045 | 0.00127 | 0.00002 | 0.00058 | 65 | 0.00219 | 0.00004 | 0.00065 | 93 | 0.00087 | -1.06288 | 155.239 | 0.28949 | 0.77376 |
| E1.3.3.6, ACOX1, ACOX3; acyl-CoA oxidase [EC:1.3.3.6] | 0.00181 | 0.00039 | 0.00109 | 0.00001 | 0.00041 | 65 | 0.00232 | 0.00003 | 0.00058 | 93 | 0.00071 | -1.71822 | 152.003 | 0.08779 | 0.76567 |
| flhC; flagellar transcriptional activator FlhC | 0.00181 | 0.00035 | 0.00167 | 0.00001 | 0.00043 | 65 | 0.00191 | 0.00002 | 0.00052 | 93 | 0.00067 | -0.35472 | 156.000 | 0.72328 | 0.92156 |
| thuG, sugB; trehalose/maltose transport system permease protein | 0.00181 | 0.00047 | 0.00194 | 0.00005 | 0.00092 | 65 | 0.00172 | 0.00002 | 0.00048 | 93 | 0.00104 | 0.20820 | 99.495 | 0.83549 | 0.95506 |
| spmB; spore maturation protein B | 0.00181 | 0.00040 | 0.00086 | 0.00001 | 0.00042 | 65 | 0.00247 | 0.00003 | 0.00060 | 93 | 0.00074 | -2.17354 | 151.981 | 0.03129 | 0.61842 |
| araB; L-ribulokinase [EC:2.7.1.16] | 0.00181 | 0.00030 | 0.00144 | 0.00001 | 0.00045 | 65 | 0.00206 | 0.00001 | 0.00040 | 93 | 0.00060 | -1.03246 | 141.963 | 0.30361 | 0.77376 |
| cobT; cobaltochelatase CobT [EC:6.6.1.2] | 0.00181 | 0.00034 | 0.00082 | 0.00000 | 0.00024 | 65 | 0.00249 | 0.00003 | 0.00054 | 93 | 0.00059 | -2.83878 | 124.779 | 0.00529 | 0.54022 |
| grxB; glutaredoxin 2 | 0.00181 | 0.00037 | 0.00226 | 0.00003 | 0.00072 | 65 | 0.00149 | 0.00001 | 0.00037 | 93 | 0.00081 | 0.93975 | 97.518 | 0.34967 | 0.79120 |
| xynB; xylan 1,4-beta-xylosidase [EC:3.2.1.37] | 0.00180 | 0.00050 | 0.00091 | 0.00001 | 0.00028 | 65 | 0.00243 | 0.00006 | 0.00082 | 93 | 0.00087 | -1.75120 | 113.061 | 0.08262 | 0.76567 |
| spoIVFB; stage IV sporulation protein FB [EC:3.4.24.-] | 0.00180 | 0.00043 | 0.00102 | 0.00002 | 0.00051 | 65 | 0.00235 | 0.00004 | 0.00063 | 93 | 0.00082 | -1.63623 | 155.894 | 0.10381 | 0.77376 |
| K11692, dctR; two-component system, CitB family, response regulator DctR | 0.00180 | 0.00063 | 0.00130 | 0.00003 | 0.00065 | 65 | 0.00216 | 0.00009 | 0.00097 | 93 | 0.00117 | -0.73410 | 149.745 | 0.46403 | 0.83699 |
| csgB; minor curlin subunit | 0.00180 | 0.00034 | 0.00229 | 0.00003 | 0.00070 | 65 | 0.00146 | 0.00001 | 0.00032 | 93 | 0.00077 | 1.08420 | 90.046 | 0.28117 | 0.77376 |
| fbaB; fructose-bisphosphate aldolase, class I [EC:4.1.2.13] | 0.00180 | 0.00032 | 0.00124 | 0.00001 | 0.00036 | 65 | 0.00219 | 0.00002 | 0.00047 | 93 | 0.00059 | -1.60235 | 155.172 | 0.11111 | 0.77376 |
| E2.3.1.20; diacylglycerol O-acyltransferase [EC:2.3.1.20] | 0.00180 | 0.00034 | 0.00136 | 0.00001 | 0.00037 | 65 | 0.00210 | 0.00002 | 0.00051 | 93 | 0.00063 | -1.16948 | 152.633 | 0.24403 | 0.77376 |
| tesC; thioesterase III [EC:3.1.2.-] | 0.00179 | 0.00034 | 0.00202 | 0.00002 | 0.00058 | 65 | 0.00163 | 0.00002 | 0.00040 | 93 | 0.00070 | 0.55340 | 121.714 | 0.58101 | 0.87593 |
| CMO; choline monooxygenase [EC:1.14.15.7] | 0.00179 | 0.00040 | 0.00194 | 0.00003 | 0.00068 | 65 | 0.00168 | 0.00002 | 0.00050 | 93 | 0.00084 | 0.30094 | 126.101 | 0.76396 | 0.93854 |
| cckA; two-component system, cell cycle sensor histidine kinase and response regulator CckA [EC:2.7.13.3] | 0.00179 | 0.00033 | 0.00081 | 0.00000 | 0.00024 | 65 | 0.00247 | 0.00003 | 0.00053 | 93 | 0.00058 | -2.84960 | 125.315 | 0.00512 | 0.54022 |
| rsfA; prespore-specific regulator | 0.00179 | 0.00051 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00268 | 0.00006 | 0.00083 | 93 | 0.00089 | -2.46043 | 116.360 | 0.01535 | 0.59095 |
| budC; meso-butanediol dehydrogenase / (S,S)-butanediol dehydrogenase / diacetyl reductase [EC:1.1.1.- 1.1.1.76 1.1.1.304] | 0.00178 | 0.00056 | 0.00244 | 0.00008 | 0.00114 | 65 | 0.00133 | 0.00002 | 0.00052 | 93 | 0.00125 | 0.88376 | 90.407 | 0.37917 | 0.79852 |
| K08983; putative membrane protein | 0.00178 | 0.00035 | 0.00198 | 0.00002 | 0.00055 | 65 | 0.00165 | 0.00002 | 0.00045 | 93 | 0.00071 | 0.46488 | 135.459 | 0.64277 | 0.90098 |
| tagF; CDP-glycerol glycerophosphotransferase [EC:2.7.8.12] | 0.00178 | 0.00034 | 0.00125 | 0.00001 | 0.00044 | 65 | 0.00214 | 0.00002 | 0.00048 | 93 | 0.00065 | -1.36739 | 155.018 | 0.17348 | 0.77376 |
| E2.7.11.1; non-specific serine/threonine protein kinase [EC:2.7.11.1] | 0.00177 | 0.00045 | 0.00137 | 0.00001 | 0.00043 | 65 | 0.00204 | 0.00005 | 0.00070 | 93 | 0.00082 | -0.81770 | 144.506 | 0.41487 | 0.81730 |
| flhD; flagellar transcriptional activator FlhD | 0.00176 | 0.00034 | 0.00155 | 0.00001 | 0.00039 | 65 | 0.00191 | 0.00002 | 0.00052 | 93 | 0.00065 | -0.55200 | 154.881 | 0.58174 | 0.87677 |
| tenI; thiazole tautomerase (transcriptional regulator TenI) [EC:5.3.99.10] | 0.00176 | 0.00045 | 0.00097 | 0.00002 | 0.00053 | 65 | 0.00230 | 0.00004 | 0.00066 | 93 | 0.00084 | -1.57713 | 155.688 | 0.11679 | 0.77376 |
| PTER, php; phosphotriesterase-related protein | 0.00175 | 0.00044 | 0.00077 | 0.00000 | 0.00023 | 65 | 0.00244 | 0.00005 | 0.00071 | 93 | 0.00075 | -2.23083 | 110.192 | 0.02772 | 0.61334 |
| K02480; two-component system, NarL family, sensor kinase [EC:2.7.13.3] | 0.00175 | 0.00042 | 0.00103 | 0.00001 | 0.00037 | 65 | 0.00226 | 0.00004 | 0.00066 | 93 | 0.00076 | -1.61479 | 138.647 | 0.10863 | 0.77376 |
| COQ10; coenzyme Q-binding protein COQ10 | 0.00175 | 0.00033 | 0.00073 | 0.00000 | 0.00019 | 65 | 0.00245 | 0.00003 | 0.00054 | 93 | 0.00057 | -3.00860 | 113.749 | 0.00323 | 0.46257 |
| K09985; uncharacterized protein | 0.00175 | 0.00033 | 0.00073 | 0.00000 | 0.00019 | 65 | 0.00245 | 0.00003 | 0.00054 | 93 | 0.00057 | -3.00860 | 113.749 | 0.00323 | 0.46257 |
| K09987; uncharacterized protein | 0.00175 | 0.00033 | 0.00073 | 0.00000 | 0.00019 | 65 | 0.00245 | 0.00003 | 0.00054 | 93 | 0.00057 | -3.00860 | 113.749 | 0.00323 | 0.46257 |
| ctrA; two-component system, cell cycle response regulator CtrA | 0.00174 | 0.00033 | 0.00072 | 0.00000 | 0.00019 | 65 | 0.00245 | 0.00003 | 0.00054 | 93 | 0.00057 | -3.02922 | 113.308 | 0.00304 | 0.46257 |
| hlg, luk; leukocidin/hemolysin toxin family protein | 0.00173 | 0.00079 | 0.00049 | 0.00001 | 0.00036 | 65 | 0.00260 | 0.00016 | 0.00132 | 93 | 0.00137 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| se; staphylococcal enterotoxin | 0.00173 | 0.00079 | 0.00049 | 0.00001 | 0.00036 | 65 | 0.00260 | 0.00016 | 0.00132 | 93 | 0.00137 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| lf2; levanbiose-producing levanase [EC:3.2.1.64] | 0.00173 | 0.00049 | 0.00186 | 0.00002 | 0.00052 | 65 | 0.00164 | 0.00005 | 0.00075 | 93 | 0.00091 | 0.24273 | 151.553 | 0.80854 | 0.95002 |
| araD, ulaF, sgaE, sgbE; L-ribulose-5-phosphate 4-epimerase [EC:5.1.3.4] | 0.00173 | 0.00030 | 0.00119 | 0.00000 | 0.00025 | 65 | 0.00211 | 0.00002 | 0.00048 | 93 | 0.00054 | -1.69238 | 135.616 | 0.09287 | 0.76567 |
| flbT; flagellar protein FlbT | 0.00173 | 0.00036 | 0.00059 | 0.00000 | 0.00014 | 65 | 0.00252 | 0.00003 | 0.00059 | 93 | 0.00061 | -3.17032 | 102.544 | 0.00201 | 0.46257 |
| chpT; histidine phosphotransferase ChpT | 0.00173 | 0.00033 | 0.00072 | 0.00000 | 0.00019 | 65 | 0.00243 | 0.00003 | 0.00053 | 93 | 0.00056 | -3.03551 | 113.982 | 0.00298 | 0.46257 |
| kinB; two-component system, sporulation sensor kinase B [EC:2.7.13.3] | 0.00173 | 0.00052 | 0.00017 | 0.00000 | 0.00008 | 65 | 0.00281 | 0.00007 | 0.00087 | 93 | 0.00088 | -3.01271 | 93.471 | 0.00333 | 0.46673 |
| crtX; zeaxanthin glucosyltransferase [EC:2.4.1.276] | 0.00172 | 0.00056 | 0.00160 | 0.00004 | 0.00081 | 65 | 0.00181 | 0.00006 | 0.00077 | 93 | 0.00111 | -0.19011 | 148.170 | 0.84948 | 0.95712 |
| chvI; two-component system, OmpR family, response regulator ChvI | 0.00172 | 0.00032 | 0.00081 | 0.00000 | 0.00024 | 65 | 0.00236 | 0.00002 | 0.00051 | 93 | 0.00056 | -2.73638 | 127.333 | 0.00710 | 0.54612 |
| gatY-kbaY; tagatose 1,6-diphosphate aldolase GatY/KbaY [EC:4.1.2.40] | 0.00172 | 0.00040 | 0.00152 | 0.00002 | 0.00054 | 65 | 0.00186 | 0.00003 | 0.00057 | 93 | 0.00079 | -0.43609 | 153.586 | 0.66338 | 0.91006 |
| chvG; two-component system, OmpR family, sensor histidine kinase ChvG [EC:2.7.13.3] | 0.00172 | 0.00032 | 0.00081 | 0.00000 | 0.00024 | 65 | 0.00235 | 0.00002 | 0.00051 | 93 | 0.00056 | -2.73467 | 127.041 | 0.00714 | 0.54612 |
| rgpI; glucosyltransferase [EC:2.4.1.-] | 0.00172 | 0.00048 | 0.00183 | 0.00002 | 0.00049 | 65 | 0.00164 | 0.00005 | 0.00075 | 93 | 0.00089 | 0.20772 | 148.982 | 0.83573 | 0.95506 |
| paaY; phenylacetic acid degradation protein | 0.00171 | 0.00031 | 0.00183 | 0.00001 | 0.00047 | 65 | 0.00163 | 0.00002 | 0.00041 | 93 | 0.00062 | 0.32778 | 141.862 | 0.74356 | 0.93099 |
| lrgA; holin-like protein | 0.00171 | 0.00045 | 0.00160 | 0.00003 | 0.00066 | 65 | 0.00179 | 0.00003 | 0.00061 | 93 | 0.00090 | -0.21459 | 145.614 | 0.83039 | 0.95506 |
| lrgB; holin-like protein LrgB | 0.00171 | 0.00045 | 0.00160 | 0.00003 | 0.00066 | 65 | 0.00179 | 0.00003 | 0.00061 | 93 | 0.00090 | -0.21459 | 145.614 | 0.83039 | 0.95506 |
| aldA; lactaldehyde dehydrogenase / glycolaldehyde dehydrogenase [EC:1.2.1.22 1.2.1.21] | 0.00171 | 0.00033 | 0.00180 | 0.00002 | 0.00055 | 65 | 0.00165 | 0.00002 | 0.00041 | 93 | 0.00069 | 0.22783 | 126.586 | 0.82014 | 0.95506 |
| ppaX; pyrophosphatase PpaX [EC:3.6.1.1] | 0.00171 | 0.00043 | 0.00102 | 0.00002 | 0.00051 | 65 | 0.00219 | 0.00004 | 0.00063 | 93 | 0.00082 | -1.43069 | 155.880 | 0.15452 | 0.77376 |
| bceA, vraD; bacitracin transport system ATP-binding protein | 0.00171 | 0.00046 | 0.00070 | 0.00001 | 0.00035 | 65 | 0.00241 | 0.00005 | 0.00074 | 93 | 0.00082 | -2.09102 | 127.759 | 0.03851 | 0.63091 |
| blaR1; bla regulator protein blaR1 | 0.00171 | 0.00044 | 0.00103 | 0.00002 | 0.00054 | 65 | 0.00217 | 0.00004 | 0.00065 | 93 | 0.00084 | -1.35549 | 155.996 | 0.17722 | 0.77376 |
| K02351; putative membrane protein | 0.00170 | 0.00038 | 0.00129 | 0.00001 | 0.00047 | 65 | 0.00200 | 0.00003 | 0.00055 | 93 | 0.00072 | -0.98161 | 155.950 | 0.32781 | 0.77376 |
| FAEB; feruloyl esterase [EC:3.1.1.73] | 0.00170 | 0.00059 | 0.00079 | 0.00000 | 0.00019 | 65 | 0.00234 | 0.00009 | 0.00099 | 93 | 0.00101 | -1.53770 | 98.898 | 0.12732 | 0.77376 |
| APEX1; AP endonuclease 1 [EC:4.2.99.18] | 0.00170 | 0.00030 | 0.00190 | 0.00001 | 0.00046 | 65 | 0.00157 | 0.00001 | 0.00039 | 93 | 0.00061 | 0.54013 | 139.719 | 0.58997 | 0.87894 |
| hmuS; putative hemin transport protein | 0.00170 | 0.00036 | 0.00151 | 0.00002 | 0.00051 | 65 | 0.00183 | 0.00002 | 0.00050 | 93 | 0.00072 | -0.45043 | 150.277 | 0.65305 | 0.90588 |
| yiaC; putative acetyltransferase [EC:2.3.1.-] | 0.00170 | 0.00033 | 0.00153 | 0.00002 | 0.00051 | 65 | 0.00182 | 0.00002 | 0.00043 | 93 | 0.00066 | -0.43345 | 138.580 | 0.66536 | 0.91126 |
| bpeF; multidrug efflux pump | 0.00169 | 0.00035 | 0.00107 | 0.00001 | 0.00032 | 65 | 0.00213 | 0.00003 | 0.00055 | 93 | 0.00063 | -1.67310 | 142.732 | 0.09650 | 0.76667 |
| ygfK; putative selenate reductase [EC:1.97.1.9] | 0.00169 | 0.00044 | 0.00209 | 0.00006 | 0.00093 | 65 | 0.00140 | 0.00001 | 0.00035 | 93 | 0.00100 | 0.69355 | 82.538 | 0.48991 | 0.84797 |
| hpaC; flavin reductase (NADH) [EC:1.5.1.36] | 0.00169 | 0.00033 | 0.00215 | 0.00002 | 0.00060 | 65 | 0.00137 | 0.00001 | 0.00038 | 93 | 0.00071 | 1.10724 | 114.075 | 0.27052 | 0.77376 |
| aguA; alpha-glucuronidase [EC:3.2.1.139] | 0.00169 | 0.00031 | 0.00107 | 0.00001 | 0.00028 | 65 | 0.00212 | 0.00002 | 0.00049 | 93 | 0.00056 | -1.86247 | 140.880 | 0.06462 | 0.73334 |
| puuA; gamma-glutamylputrescine synthase [EC:6.3.1.11] | 0.00168 | 0.00032 | 0.00158 | 0.00001 | 0.00039 | 65 | 0.00176 | 0.00002 | 0.00047 | 93 | 0.00061 | -0.28305 | 155.965 | 0.77752 | 0.94269 |
| uvrY, gacA, varA; two-component system, NarL family, invasion response regulator UvrY | 0.00168 | 0.00033 | 0.00183 | 0.00002 | 0.00051 | 65 | 0.00158 | 0.00002 | 0.00044 | 93 | 0.00067 | 0.37005 | 140.125 | 0.71190 | 0.92004 |
| hutG; N-formylglutamate deformylase [EC:3.5.1.68] | 0.00168 | 0.00050 | 0.00102 | 0.00001 | 0.00032 | 65 | 0.00214 | 0.00006 | 0.00082 | 93 | 0.00088 | -1.27374 | 117.830 | 0.20526 | 0.77376 |
| K09703; uncharacterized protein | 0.00168 | 0.00069 | 0.00095 | 0.00002 | 0.00056 | 65 | 0.00219 | 0.00011 | 0.00110 | 93 | 0.00123 | -1.00804 | 133.191 | 0.31526 | 0.77376 |
| nixA; high-affinity nickel-transport protein | 0.00168 | 0.00043 | 0.00082 | 0.00000 | 0.00022 | 65 | 0.00228 | 0.00005 | 0.00071 | 93 | 0.00074 | -1.97942 | 109.636 | 0.05028 | 0.67941 |
| OPLAH, OXP1, oplAH; 5-oxoprolinase (ATP-hydrolysing) [EC:3.5.2.9] | 0.00168 | 0.00049 | 0.00059 | 0.00000 | 0.00023 | 65 | 0.00244 | 0.00006 | 0.00081 | 93 | 0.00084 | -2.19593 | 106.128 | 0.03027 | 0.61413 |
| waaJ, rfaJ; UDP-glucose:(galactosyl)LPS alpha-1,2-glucosyltransferase [EC:2.4.1.58] | 0.00167 | 0.00070 | 0.00104 | 0.00001 | 0.00037 | 65 | 0.00211 | 0.00013 | 0.00117 | 93 | 0.00122 | -0.87117 | 109.951 | 0.38556 | 0.80258 |
| E1.14.13.40; anthraniloyl-CoA monooxygenase [EC:1.14.13.40] | 0.00167 | 0.00037 | 0.00104 | 0.00001 | 0.00039 | 65 | 0.00211 | 0.00003 | 0.00056 | 93 | 0.00069 | -1.55860 | 151.886 | 0.12117 | 0.77376 |
| porG; pyruvate ferredoxin oxidoreductase gamma subunit [EC:1.2.7.1] | 0.00167 | 0.00042 | 0.00137 | 0.00001 | 0.00036 | 65 | 0.00188 | 0.00004 | 0.00067 | 93 | 0.00076 | -0.67777 | 136.491 | 0.49906 | 0.84856 |
| quiC; 3-dehydroshikimate dehydratase [EC:4.2.1.118] | 0.00166 | 0.00033 | 0.00150 | 0.00001 | 0.00046 | 65 | 0.00178 | 0.00002 | 0.00047 | 93 | 0.00066 | -0.42188 | 152.185 | 0.67371 | 0.91184 |
| wapR; alpha-1,3-rhamnosyltransferase [EC:2.4.1.-] | 0.00166 | 0.00045 | 0.00119 | 0.00002 | 0.00055 | 65 | 0.00200 | 0.00004 | 0.00067 | 93 | 0.00087 | -0.93466 | 155.885 | 0.35141 | 0.79120 |
| ltrA; RNA-directed DNA polymerase [EC:2.7.7.49] | 0.00165 | 0.00044 | 0.00192 | 0.00005 | 0.00090 | 65 | 0.00147 | 0.00002 | 0.00041 | 93 | 0.00099 | 0.45519 | 91.157 | 0.65005 | 0.90409 |
| araJ; MFS transporter, DHA1 family, arabinose polymer utilization protein | 0.00165 | 0.00028 | 0.00124 | 0.00000 | 0.00027 | 65 | 0.00193 | 0.00002 | 0.00043 | 93 | 0.00051 | -1.37642 | 147.487 | 0.17078 | 0.77376 |
| E2.1.3.1-12S; methylmalonyl-CoA carboxyltransferase 12S subunit [EC:2.1.3.1] | 0.00164 | 0.00049 | 0.00207 | 0.00006 | 0.00097 | 65 | 0.00135 | 0.00002 | 0.00049 | 93 | 0.00109 | 0.66290 | 96.418 | 0.50898 | 0.84956 |
| kdpF; K+-transporting ATPase ATPase F chain | 0.00164 | 0.00035 | 0.00159 | 0.00002 | 0.00048 | 65 | 0.00167 | 0.00002 | 0.00050 | 93 | 0.00069 | -0.11006 | 152.482 | 0.91251 | 0.97664 |
| stk1; serine/threonine-protein kinase Stk1 [EC:2.7.11.-] | 0.00164 | 0.00036 | 0.00135 | 0.00002 | 0.00050 | 65 | 0.00184 | 0.00002 | 0.00051 | 93 | 0.00072 | -0.67920 | 151.705 | 0.49805 | 0.84856 |
| E1.5.3.1; sarcosine oxidase [EC:1.5.3.1] | 0.00164 | 0.00039 | 0.00101 | 0.00001 | 0.00036 | 65 | 0.00207 | 0.00003 | 0.00061 | 93 | 0.00070 | -1.50804 | 143.087 | 0.13375 | 0.77376 |
| K09805; uncharacterized protein | 0.00163 | 0.00042 | 0.00093 | 0.00000 | 0.00027 | 65 | 0.00213 | 0.00004 | 0.00069 | 93 | 0.00074 | -1.61148 | 118.848 | 0.10973 | 0.77376 |
| K02476; two-component system, CitB family, sensor kinase [EC:2.7.13.3] | 0.00163 | 0.00042 | 0.00107 | 0.00002 | 0.00053 | 65 | 0.00202 | 0.00003 | 0.00061 | 93 | 0.00081 | -1.18157 | 155.774 | 0.23918 | 0.77376 |
| dctR; two-component system, LuxR family, response regulator DctR | 0.00163 | 0.00047 | 0.00074 | 0.00000 | 0.00026 | 65 | 0.00224 | 0.00006 | 0.00078 | 93 | 0.00082 | -1.83262 | 112.293 | 0.06951 | 0.73522 |
| mgp; 4-O-beta-D-mannosyl-D-glucose phosphorylase [EC:2.4.1.281] | 0.00163 | 0.00032 | 0.00083 | 0.00000 | 0.00024 | 65 | 0.00218 | 0.00002 | 0.00051 | 93 | 0.00056 | -2.39730 | 129.519 | 0.01794 | 0.59095 |
| gerKB; spore germination protein KB | 0.00162 | 0.00044 | 0.00053 | 0.00001 | 0.00031 | 65 | 0.00239 | 0.00005 | 0.00071 | 93 | 0.00077 | -2.40410 | 124.260 | 0.01769 | 0.59095 |
| ygeT, xdhB; xanthine dehydrogenase FAD-binding subunit [EC:1.17.1.4] | 0.00162 | 0.00043 | 0.00200 | 0.00006 | 0.00093 | 65 | 0.00136 | 0.00001 | 0.00035 | 93 | 0.00099 | 0.64133 | 82.181 | 0.52309 | 0.85704 |
| nosR; NosR/NirI family transcriptional regulator, nitrous oxide reductase regulator | 0.00162 | 0.00034 | 0.00106 | 0.00001 | 0.00030 | 65 | 0.00201 | 0.00003 | 0.00054 | 93 | 0.00061 | -1.55462 | 137.850 | 0.12233 | 0.77376 |
| K17215; inositol transport system ATP-binding protein | 0.00161 | 0.00036 | 0.00084 | 0.00001 | 0.00037 | 65 | 0.00216 | 0.00003 | 0.00055 | 93 | 0.00066 | -1.98717 | 149.422 | 0.04873 | 0.67326 |
| yscJ, sctJ, hrcJ, ssaJ; type III secretion protein J | 0.00161 | 0.00035 | 0.00102 | 0.00001 | 0.00036 | 65 | 0.00203 | 0.00003 | 0.00054 | 93 | 0.00065 | -1.55454 | 150.379 | 0.12216 | 0.77376 |
| K07219; putative molybdopterin biosynthesis protein | 0.00161 | 0.00040 | 0.00059 | 0.00001 | 0.00031 | 65 | 0.00233 | 0.00004 | 0.00063 | 93 | 0.00070 | -2.47264 | 130.151 | 0.01470 | 0.59095 |
| kstD; 3-oxosteroid 1-dehydrogenase [EC:1.3.99.4] | 0.00161 | 0.00062 | 0.00082 | 0.00001 | 0.00034 | 65 | 0.00216 | 0.00010 | 0.00103 | 93 | 0.00108 | -1.23587 | 111.319 | 0.21911 | 0.77376 |
| pmtA; phosphatidylethanolamine/phosphatidyl-N-methylethanolamine N-methyltransferase [EC:2.1.1.17 2.1.1.71] | 0.00161 | 0.00044 | 0.00061 | 0.00000 | 0.00019 | 65 | 0.00230 | 0.00005 | 0.00073 | 93 | 0.00075 | -2.23955 | 104.292 | 0.02724 | 0.61334 |
| tar; methyl-accepting chemotaxis protein II, aspartate sensor receptor | 0.00161 | 0.00046 | 0.00103 | 0.00001 | 0.00031 | 65 | 0.00201 | 0.00005 | 0.00075 | 93 | 0.00081 | -1.21450 | 120.625 | 0.22693 | 0.77376 |
| catB; chloramphenicol O-acetyltransferase type B [EC:2.3.1.28] | 0.00160 | 0.00032 | 0.00131 | 0.00001 | 0.00037 | 65 | 0.00179 | 0.00002 | 0.00048 | 93 | 0.00060 | -0.79470 | 155.011 | 0.42801 | 0.82733 |
| K09991; uncharacterized protein | 0.00160 | 0.00030 | 0.00073 | 0.00000 | 0.00019 | 65 | 0.00220 | 0.00002 | 0.00049 | 93 | 0.00052 | -2.80136 | 118.172 | 0.00595 | 0.54022 |
| K09702; uncharacterized protein | 0.00159 | 0.00032 | 0.00112 | 0.00001 | 0.00042 | 65 | 0.00192 | 0.00002 | 0.00046 | 93 | 0.00063 | -1.27165 | 154.559 | 0.20541 | 0.77376 |
| ttr; acetyltransferase [EC:2.3.1.-] | 0.00159 | 0.00036 | 0.00127 | 0.00002 | 0.00048 | 65 | 0.00182 | 0.00002 | 0.00051 | 93 | 0.00070 | -0.78363 | 153.933 | 0.43446 | 0.82874 |
| ERCC3, XPB; DNA excision repair protein ERCC-3 [EC:3.6.4.12] | 0.00159 | 0.00027 | 0.00138 | 0.00001 | 0.00033 | 65 | 0.00174 | 0.00001 | 0.00040 | 93 | 0.00052 | -0.69163 | 156.000 | 0.49020 | 0.84797 |
| CBP3, UQCC; cytochrome b pre-mRNA-processing protein 3 | 0.00159 | 0.00030 | 0.00072 | 0.00000 | 0.00019 | 65 | 0.00220 | 0.00002 | 0.00049 | 93 | 0.00052 | -2.82397 | 117.656 | 0.00557 | 0.54022 |
| sspA; glutamyl endopeptidase [EC:3.4.21.19] | 0.00159 | 0.00065 | 0.00045 | 0.00001 | 0.00029 | 65 | 0.00239 | 0.00011 | 0.00107 | 93 | 0.00111 | -1.74934 | 105.133 | 0.08315 | 0.76567 |
| relB; RHH-type transcriptional regulator, rel operon repressor / antitoxin RelB | 0.00159 | 0.00033 | 0.00126 | 0.00001 | 0.00041 | 65 | 0.00182 | 0.00002 | 0.00048 | 93 | 0.00063 | -0.88042 | 155.767 | 0.37999 | 0.79852 |
| mcrC; 5-methylcytosine-specific restriction enzyme subunit McrC | 0.00159 | 0.00025 | 0.00195 | 0.00002 | 0.00048 | 65 | 0.00133 | 0.00001 | 0.00025 | 93 | 0.00054 | 1.13466 | 98.065 | 0.25928 | 0.77376 |
| K09935; uncharacterized protein | 0.00159 | 0.00026 | 0.00167 | 0.00001 | 0.00044 | 65 | 0.00153 | 0.00001 | 0.00032 | 93 | 0.00054 | 0.24533 | 124.377 | 0.80661 | 0.94982 |
| yydH; putative peptide zinc metalloprotease protein | 0.00159 | 0.00032 | 0.00135 | 0.00001 | 0.00045 | 65 | 0.00175 | 0.00002 | 0.00045 | 93 | 0.00063 | -0.63434 | 151.223 | 0.52682 | 0.85844 |
| lyxA; D-lyxose ketol-isomerase [EC:5.3.1.15] | 0.00159 | 0.00044 | 0.00185 | 0.00005 | 0.00092 | 65 | 0.00140 | 0.00001 | 0.00039 | 93 | 0.00100 | 0.44615 | 87.115 | 0.65660 | 0.90679 |
| cag7; cag pathogenicity island protein 7 | 0.00158 | 0.00102 | 0.00057 | 0.00001 | 0.00037 | 65 | 0.00229 | 0.00027 | 0.00171 | 93 | 0.00175 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| hpaA; neuraminyllactose-binding hemagglutinin | 0.00158 | 0.00102 | 0.00057 | 0.00001 | 0.00037 | 65 | 0.00229 | 0.00027 | 0.00171 | 93 | 0.00175 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| wcaF; putative colanic acid biosynthesis acetyltransferase WcaF [EC:2.3.1.-] | 0.00158 | 0.00036 | 0.00071 | 0.00001 | 0.00031 | 65 | 0.00219 | 0.00003 | 0.00057 | 93 | 0.00065 | -2.29587 | 137.873 | 0.02319 | 0.59764 |
| spoIIM; stage II sporulation protein M | 0.00158 | 0.00034 | 0.00077 | 0.00001 | 0.00042 | 65 | 0.00215 | 0.00002 | 0.00050 | 93 | 0.00065 | -2.11280 | 155.952 | 0.03621 | 0.62411 |
| MQCRB, qcrB, bfcB, petB; menaquinol-cytochrome c reductase cytochrome b subunit | 0.00158 | 0.00040 | 0.00051 | 0.00001 | 0.00030 | 65 | 0.00233 | 0.00004 | 0.00063 | 93 | 0.00070 | -2.59406 | 129.562 | 0.01058 | 0.58284 |
| MQCRC, qcrC, bfcC, petD; menaquinol-cytochrome c reductase cytochrome b/c subunit | 0.00158 | 0.00040 | 0.00051 | 0.00001 | 0.00030 | 65 | 0.00233 | 0.00004 | 0.00063 | 93 | 0.00070 | -2.59406 | 129.562 | 0.01058 | 0.58284 |
| gspK; glucosamine kinase [EC:2.7.1.8] | 0.00157 | 0.00039 | 0.00059 | 0.00000 | 0.00022 | 65 | 0.00226 | 0.00004 | 0.00064 | 93 | 0.00068 | -2.45891 | 112.157 | 0.01546 | 0.59095 |
| coxAC; cytochrome c oxidase subunit I+III [EC:1.9.3.1] | 0.00157 | 0.00032 | 0.00090 | 0.00001 | 0.00035 | 65 | 0.00204 | 0.00002 | 0.00049 | 93 | 0.00060 | -1.88564 | 153.368 | 0.06123 | 0.73001 |
| nrfG; formate-dependent nitrite reductase complex subunit NrfG | 0.00157 | 0.00044 | 0.00112 | 0.00002 | 0.00057 | 65 | 0.00188 | 0.00004 | 0.00062 | 93 | 0.00085 | -0.89567 | 154.356 | 0.37182 | 0.79738 |
| ictB; putative inorganic carbon (hco3(-)) transporter | 0.00157 | 0.00050 | 0.00202 | 0.00006 | 0.00099 | 65 | 0.00125 | 0.00002 | 0.00050 | 93 | 0.00111 | 0.69294 | 97.230 | 0.49000 | 0.84797 |
| abfD; 4-hydroxybutyryl-CoA dehydratase / vinylacetyl-CoA-Delta-isomerase [EC:4.2.1.120 5.3.3.3] | 0.00157 | 0.00032 | 0.00110 | 0.00001 | 0.00035 | 65 | 0.00189 | 0.00002 | 0.00048 | 93 | 0.00060 | -1.32407 | 153.637 | 0.18745 | 0.77376 |
| ascG; LacI family transcriptional regulator, asc operon repressor | 0.00156 | 0.00043 | 0.00211 | 0.00006 | 0.00093 | 65 | 0.00118 | 0.00001 | 0.00032 | 93 | 0.00098 | 0.93856 | 79.205 | 0.35081 | 0.79120 |
| lacF, araP; lactose/L-arabinose transport system permease protein | 0.00156 | 0.00044 | 0.00187 | 0.00006 | 0.00093 | 65 | 0.00135 | 0.00001 | 0.00037 | 93 | 0.00100 | 0.51797 | 84.639 | 0.60583 | 0.88527 |
| THOP1; thimet oligopeptidase [EC:3.4.24.15] | 0.00156 | 0.00031 | 0.00123 | 0.00001 | 0.00034 | 65 | 0.00179 | 0.00002 | 0.00047 | 93 | 0.00058 | -0.95658 | 153.409 | 0.34028 | 0.78470 |
| K07125; uncharacterized protein | 0.00156 | 0.00032 | 0.00186 | 0.00002 | 0.00052 | 65 | 0.00134 | 0.00002 | 0.00041 | 93 | 0.00067 | 0.77851 | 133.504 | 0.43765 | 0.83071 |
| iadA; beta-aspartyl-dipeptidase (metallo-type) [EC:3.4.19.-] | 0.00155 | 0.00034 | 0.00136 | 0.00002 | 0.00050 | 65 | 0.00169 | 0.00002 | 0.00045 | 93 | 0.00068 | -0.49760 | 144.945 | 0.61952 | 0.88993 |
| lacG, araQ; lactose/L-arabinose transport system permease protein | 0.00155 | 0.00044 | 0.00187 | 0.00006 | 0.00093 | 65 | 0.00133 | 0.00001 | 0.00037 | 93 | 0.00100 | 0.54017 | 84.010 | 0.59051 | 0.87896 |
| rcdA; regulator of CtrA degradation | 0.00155 | 0.00029 | 0.00072 | 0.00000 | 0.00019 | 65 | 0.00212 | 0.00002 | 0.00047 | 93 | 0.00050 | -2.78612 | 119.742 | 0.00620 | 0.54022 |
| qcrB; ubiquinol-cytochrome c reductase cytochrome b subunit | 0.00155 | 0.00029 | 0.00119 | 0.00001 | 0.00035 | 65 | 0.00180 | 0.00002 | 0.00043 | 93 | 0.00055 | -1.09298 | 155.827 | 0.27609 | 0.77376 |
| porA; pyruvate ferredoxin oxidoreductase alpha subunit [EC:1.2.7.1] | 0.00154 | 0.00041 | 0.00109 | 0.00000 | 0.00027 | 65 | 0.00186 | 0.00004 | 0.00067 | 93 | 0.00072 | -1.06743 | 119.953 | 0.28792 | 0.77376 |
| arlR; two-component system, OmpR family, response regulator ArlR | 0.00154 | 0.00051 | 0.00126 | 0.00002 | 0.00060 | 65 | 0.00174 | 0.00005 | 0.00075 | 93 | 0.00097 | -0.49181 | 155.739 | 0.62354 | 0.89159 |
| kefC; glutathione-regulated potassium-efflux system ancillary protein KefC | 0.00154 | 0.00033 | 0.00119 | 0.00001 | 0.00035 | 65 | 0.00178 | 0.00002 | 0.00051 | 93 | 0.00061 | -0.96020 | 150.923 | 0.33849 | 0.78407 |
| ctaG; putative membrane protein | 0.00153 | 0.00040 | 0.00047 | 0.00001 | 0.00030 | 65 | 0.00228 | 0.00004 | 0.00063 | 93 | 0.00070 | -2.59106 | 129.117 | 0.01067 | 0.58284 |
| qcrA; ubiquinol-cytochrome c reductase iron-sulfur subunit | 0.00153 | 0.00029 | 0.00119 | 0.00001 | 0.00035 | 65 | 0.00177 | 0.00002 | 0.00043 | 93 | 0.00055 | -1.04815 | 155.837 | 0.29619 | 0.77376 |
| qcrC; ubiquinol-cytochrome c reductase cytochrome c subunit | 0.00153 | 0.00029 | 0.00119 | 0.00001 | 0.00035 | 65 | 0.00177 | 0.00002 | 0.00043 | 93 | 0.00055 | -1.04815 | 155.837 | 0.29619 | 0.77376 |
| cpdR; two-component system, cell cycle response regulator CpdR | 0.00153 | 0.00029 | 0.00072 | 0.00000 | 0.00019 | 65 | 0.00210 | 0.00002 | 0.00047 | 93 | 0.00050 | -2.75188 | 119.837 | 0.00685 | 0.54612 |
| femX, fmhB; peptidoglycan pentaglycine glycine transferase (the first glycine) [EC:2.3.2.16] | 0.00153 | 0.00046 | 0.00141 | 0.00003 | 0.00066 | 65 | 0.00162 | 0.00004 | 0.00064 | 93 | 0.00092 | -0.22907 | 148.905 | 0.81913 | 0.95506 |
| gal; D-galactose 1-dehydrogenase [EC:1.1.1.48] | 0.00153 | 0.00029 | 0.00073 | 0.00000 | 0.00025 | 65 | 0.00209 | 0.00002 | 0.00045 | 93 | 0.00052 | -2.62179 | 137.159 | 0.00973 | 0.58112 |
| gltC; LysR family transcriptional regulator, transcription activator of glutamate synthase operon | 0.00153 | 0.00044 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00225 | 0.00005 | 0.00070 | 93 | 0.00077 | -2.28477 | 124.387 | 0.02402 | 0.59764 |
| rnhA-dnaQ; ribonuclease HI / DNA polymerase III subunit epsilon [EC:3.1.26.4 2.7.7.7] | 0.00153 | 0.00032 | 0.00130 | 0.00001 | 0.00037 | 65 | 0.00169 | 0.00002 | 0.00049 | 93 | 0.00061 | -0.64177 | 154.392 | 0.52197 | 0.85623 |
| fixL; two-component system, LuxR family, sensor kinase FixL [EC:2.7.13.3] | 0.00153 | 0.00032 | 0.00069 | 0.00000 | 0.00022 | 65 | 0.00211 | 0.00003 | 0.00052 | 93 | 0.00057 | -2.49716 | 121.402 | 0.01386 | 0.59095 |
| PCYT1; choline-phosphate cytidylyltransferase [EC:2.7.7.15] | 0.00152 | 0.00032 | 0.00128 | 0.00001 | 0.00036 | 65 | 0.00170 | 0.00002 | 0.00047 | 93 | 0.00060 | -0.70236 | 155.089 | 0.48351 | 0.84354 |
| shlB, hhdB, hpmB; hemolysin activation/secretion protein | 0.00152 | 0.00029 | 0.00192 | 0.00001 | 0.00044 | 65 | 0.00125 | 0.00001 | 0.00039 | 93 | 0.00059 | 1.13775 | 142.205 | 0.25714 | 0.77376 |
| asrA; anaerobic sulfite reductase subunit A | 0.00152 | 0.00039 | 0.00236 | 0.00005 | 0.00090 | 65 | 0.00094 | 0.00000 | 0.00022 | 93 | 0.00092 | 1.52843 | 71.971 | 0.13079 | 0.77376 |
| asrB; anaerobic sulfite reductase subunit B | 0.00152 | 0.00039 | 0.00236 | 0.00005 | 0.00090 | 65 | 0.00094 | 0.00000 | 0.00022 | 93 | 0.00092 | 1.52843 | 71.971 | 0.13079 | 0.77376 |
| asrC; anaerobic sulfite reductase subunit C | 0.00152 | 0.00039 | 0.00236 | 0.00005 | 0.00090 | 65 | 0.00094 | 0.00000 | 0.00022 | 93 | 0.00092 | 1.52843 | 71.971 | 0.13079 | 0.77376 |
| spoIIQ; stage II sporulation protein Q | 0.00152 | 0.00041 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00223 | 0.00004 | 0.00065 | 93 | 0.00072 | -2.40579 | 129.490 | 0.01755 | 0.59095 |
| cbe, mbe; cellobiose epimerase [EC:5.1.3.11] | 0.00152 | 0.00031 | 0.00078 | 0.00000 | 0.00024 | 65 | 0.00204 | 0.00002 | 0.00049 | 93 | 0.00055 | -2.28556 | 130.474 | 0.02389 | 0.59764 |
| inhA; cyclohexyl-isocyanide hydratase [EC:4.2.1.103] | 0.00152 | 0.00033 | 0.00071 | 0.00001 | 0.00032 | 65 | 0.00208 | 0.00002 | 0.00051 | 93 | 0.00061 | -2.26199 | 146.119 | 0.02517 | 0.59941 |
| iucD; lysine N6-hydroxylase [EC:1.14.13.59] | 0.00152 | 0.00030 | 0.00124 | 0.00001 | 0.00034 | 65 | 0.00171 | 0.00002 | 0.00045 | 93 | 0.00056 | -0.84239 | 154.255 | 0.40088 | 0.80823 |
| glpX-SEBP; fructose-1,6-bisphosphatase II / sedoheptulose-1,7-bisphosphatase [EC:3.1.3.11 3.1.3.37] | 0.00152 | 0.00030 | 0.00055 | 0.00000 | 0.00014 | 65 | 0.00219 | 0.00002 | 0.00049 | 93 | 0.00051 | -3.23037 | 107.675 | 0.00164 | 0.46257 |
| K16149; 1,4-alpha-glucan branching enzyme [EC:2.4.1.18] | 0.00152 | 0.00029 | 0.00134 | 0.00001 | 0.00036 | 65 | 0.00164 | 0.00002 | 0.00043 | 93 | 0.00056 | -0.54191 | 155.997 | 0.58866 | 0.87736 |
| E2.6.1.-E, patB; aminotransferase [EC:2.6.1.-] | 0.00152 | 0.00038 | 0.00172 | 0.00002 | 0.00052 | 65 | 0.00137 | 0.00003 | 0.00054 | 93 | 0.00075 | 0.46732 | 152.922 | 0.64094 | 0.89999 |
| arlS; two-component system, OmpR family, sensor histidine kinase ArlS [EC:2.7.13.3] | 0.00152 | 0.00051 | 0.00122 | 0.00002 | 0.00060 | 65 | 0.00173 | 0.00005 | 0.00076 | 93 | 0.00097 | -0.52608 | 155.751 | 0.59958 | 0.88240 |
| safA; morphogenetic protein associated with SpoVID | 0.00152 | 0.00041 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00222 | 0.00004 | 0.00065 | 93 | 0.00072 | -2.39557 | 129.575 | 0.01802 | 0.59095 |
| iorA; indolepyruvate ferredoxin oxidoreductase, alpha subunit [EC:1.2.7.8] | 0.00152 | 0.00027 | 0.00107 | 0.00000 | 0.00025 | 65 | 0.00183 | 0.00002 | 0.00041 | 93 | 0.00048 | -1.57902 | 144.008 | 0.11653 | 0.77376 |
| lmrS; MFS transporter, DHA2 family, multidrug resistance protein | 0.00152 | 0.00041 | 0.00128 | 0.00002 | 0.00055 | 65 | 0.00168 | 0.00003 | 0.00059 | 93 | 0.00081 | -0.48895 | 154.341 | 0.62557 | 0.89228 |
| bpeT; LysR family transcriptional regulator, regulator for bpeEF and oprC | 0.00152 | 0.00048 | 0.00126 | 0.00001 | 0.00041 | 65 | 0.00170 | 0.00006 | 0.00077 | 93 | 0.00087 | -0.49971 | 135.492 | 0.61809 | 0.88903 |
| cccA; cytochrome c550 | 0.00152 | 0.00040 | 0.00049 | 0.00001 | 0.00030 | 65 | 0.00223 | 0.00004 | 0.00064 | 93 | 0.00070 | -2.47432 | 128.983 | 0.01465 | 0.59095 |
| iorB; indolepyruvate ferredoxin oxidoreductase, beta subunit [EC:1.2.7.8] | 0.00151 | 0.00027 | 0.00107 | 0.00000 | 0.00025 | 65 | 0.00183 | 0.00002 | 0.00041 | 93 | 0.00048 | -1.56916 | 143.980 | 0.11880 | 0.77376 |
| lacE, araN; lactose/L-arabinose transport system substrate-binding protein | 0.00151 | 0.00043 | 0.00187 | 0.00006 | 0.00093 | 65 | 0.00127 | 0.00001 | 0.00034 | 93 | 0.00099 | 0.60805 | 81.362 | 0.54485 | 0.86403 |
| ebgA; evolved beta-galactosidase subunit alpha [EC:3.2.1.23] | 0.00151 | 0.00043 | 0.00216 | 0.00006 | 0.00094 | 65 | 0.00106 | 0.00001 | 0.00031 | 93 | 0.00099 | 1.11720 | 78.457 | 0.26732 | 0.77376 |
| nalD; TetR/AcrR family transcriptional regulator, repressor of the mexAB-oprM multidrug resistance operon | 0.00151 | 0.00034 | 0.00209 | 0.00003 | 0.00066 | 65 | 0.00111 | 0.00001 | 0.00036 | 93 | 0.00075 | 1.30869 | 100.488 | 0.19363 | 0.77376 |
| spoIIIAF; stage III sporulation protein AF | 0.00151 | 0.00034 | 0.00072 | 0.00001 | 0.00042 | 65 | 0.00206 | 0.00002 | 0.00048 | 93 | 0.00064 | -2.09941 | 155.674 | 0.03739 | 0.63026 |
| FAD6, desA; acyl-lipid omega-6 desaturase (Delta-12 desaturase) [EC:1.14.19.23 1.14.19.45] | 0.00151 | 0.00037 | 0.00062 | 0.00001 | 0.00032 | 65 | 0.00213 | 0.00003 | 0.00058 | 93 | 0.00067 | -2.25735 | 138.541 | 0.02555 | 0.59941 |
| rbtD; ribitol 2-dehydrogenase [EC:1.1.1.56] | 0.00151 | 0.00038 | 0.00089 | 0.00001 | 0.00040 | 65 | 0.00194 | 0.00003 | 0.00057 | 93 | 0.00070 | -1.50295 | 151.731 | 0.13493 | 0.77376 |
| kapB; kinase-associated protein B | 0.00150 | 0.00042 | 0.00096 | 0.00002 | 0.00053 | 65 | 0.00188 | 0.00003 | 0.00061 | 93 | 0.00080 | -1.14259 | 155.750 | 0.25496 | 0.77376 |
| odh; opine dehydrogenase [EC:1.5.1.28] | 0.00150 | 0.00044 | 0.00123 | 0.00002 | 0.00054 | 65 | 0.00170 | 0.00004 | 0.00064 | 93 | 0.00084 | -0.55454 | 155.980 | 0.58000 | 0.87573 |
| hasA; heme acquisition protein HasA | 0.00150 | 0.00045 | 0.00109 | 0.00002 | 0.00054 | 65 | 0.00179 | 0.00004 | 0.00066 | 93 | 0.00086 | -0.82275 | 155.878 | 0.41190 | 0.81559 |
| yrrT; putative AdoMet-dependent methyltransferase [EC:2.1.1.-] | 0.00150 | 0.00038 | 0.00068 | 0.00001 | 0.00029 | 65 | 0.00207 | 0.00003 | 0.00060 | 93 | 0.00067 | -2.06262 | 130.051 | 0.04114 | 0.64661 |
| K09803; uncharacterized protein | 0.00150 | 0.00030 | 0.00107 | 0.00001 | 0.00029 | 65 | 0.00179 | 0.00002 | 0.00046 | 93 | 0.00055 | -1.32244 | 146.221 | 0.18809 | 0.77376 |
| fimU; type IV fimbrial biogenesis protein FimU | 0.00150 | 0.00035 | 0.00196 | 0.00003 | 0.00066 | 65 | 0.00117 | 0.00001 | 0.00038 | 93 | 0.00076 | 1.03696 | 105.352 | 0.30213 | 0.77376 |
| ysiA, fadR; TetR/AcrR family transcriptional regulator, fatty acid metabolism regulator protein | 0.00150 | 0.00039 | 0.00044 | 0.00001 | 0.00030 | 65 | 0.00223 | 0.00004 | 0.00062 | 93 | 0.00069 | -2.58292 | 129.471 | 0.01091 | 0.58284 |
| PTS-Aga-EIIC, agaW; PTS system, N-acetylgalactosamine-specific IIC component | 0.00150 | 0.00025 | 0.00153 | 0.00001 | 0.00040 | 65 | 0.00147 | 0.00001 | 0.00033 | 93 | 0.00051 | 0.10211 | 136.537 | 0.91882 | 0.97808 |
| pleC; two-component system, cell cycle sensor histidine kinase PleC [EC:2.7.13.3] | 0.00149 | 0.00035 | 0.00053 | 0.00000 | 0.00014 | 65 | 0.00217 | 0.00003 | 0.00058 | 93 | 0.00059 | -2.76238 | 102.474 | 0.00680 | 0.54612 |
| SDS, SDH, CHA1; L-serine/L-threonine ammonia-lyase [EC:4.3.1.17 4.3.1.19] | 0.00149 | 0.00033 | 0.00183 | 0.00002 | 0.00053 | 65 | 0.00125 | 0.00002 | 0.00041 | 93 | 0.00068 | 0.85328 | 131.028 | 0.39506 | 0.80459 |
| steA, tetA46; ATP-binding cassette, subfamily B, tetracycline resistant protein | 0.00149 | 0.00038 | 0.00193 | 0.00004 | 0.00077 | 65 | 0.00118 | 0.00001 | 0.00035 | 93 | 0.00084 | 0.88226 | 91.162 | 0.37996 | 0.79852 |
| steB, tetB46; ATP-binding cassette, subfamily B, tetracycline resistant protein | 0.00149 | 0.00038 | 0.00193 | 0.00004 | 0.00077 | 65 | 0.00118 | 0.00001 | 0.00035 | 93 | 0.00084 | 0.88226 | 91.162 | 0.37996 | 0.79852 |
| dppA1; D-amino peptidase [EC:3.4.11.-] | 0.00149 | 0.00043 | 0.00077 | 0.00001 | 0.00035 | 65 | 0.00199 | 0.00004 | 0.00068 | 93 | 0.00077 | -1.59044 | 134.523 | 0.11408 | 0.77376 |
| K09004; uncharacterized protein | 0.00149 | 0.00037 | 0.00122 | 0.00001 | 0.00046 | 65 | 0.00167 | 0.00003 | 0.00054 | 93 | 0.00071 | -0.62220 | 155.965 | 0.53472 | 0.86207 |
| flaF; flagellar protein FlaF | 0.00148 | 0.00030 | 0.00052 | 0.00000 | 0.00013 | 65 | 0.00215 | 0.00002 | 0.00049 | 93 | 0.00050 | -3.23102 | 105.283 | 0.00165 | 0.46257 |
| hpxQ; 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase [EC:4.1.1.97] | 0.00148 | 0.00033 | 0.00084 | 0.00000 | 0.00024 | 65 | 0.00193 | 0.00003 | 0.00054 | 93 | 0.00059 | -1.84778 | 125.976 | 0.06698 | 0.73334 |
| tsx; nucleoside-specific channel-forming protein | 0.00148 | 0.00034 | 0.00134 | 0.00001 | 0.00044 | 65 | 0.00157 | 0.00002 | 0.00048 | 93 | 0.00066 | -0.35531 | 154.491 | 0.72284 | 0.92156 |
| fbp; diacylglycerol O-acyltransferase / trehalose O-mycolyltransferase [EC:2.3.1.20 2.3.1.122] | 0.00148 | 0.00059 | 0.00037 | 0.00000 | 0.00016 | 65 | 0.00225 | 0.00009 | 0.00099 | 93 | 0.00100 | -1.87547 | 96.632 | 0.06375 | 0.73334 |
| nrfF; formate-dependent nitrite reductase complex subunit NrfF | 0.00147 | 0.00043 | 0.00105 | 0.00002 | 0.00057 | 65 | 0.00177 | 0.00004 | 0.00062 | 93 | 0.00084 | -0.85436 | 154.468 | 0.39423 | 0.80459 |
| caiC; carnitine-CoA ligase [EC:6.2.1.48] | 0.00147 | 0.00035 | 0.00095 | 0.00000 | 0.00025 | 65 | 0.00183 | 0.00003 | 0.00056 | 93 | 0.00061 | -1.44391 | 126.268 | 0.15124 | 0.77376 |
| mhpE; 4-hydroxy 2-oxovalerate aldolase [EC:4.1.3.39] | 0.00147 | 0.00041 | 0.00064 | 0.00000 | 0.00017 | 65 | 0.00204 | 0.00004 | 0.00068 | 93 | 0.00070 | -2.00412 | 102.894 | 0.04769 | 0.66537 |
| kbaA; KinB signaling pathway activation protein | 0.00147 | 0.00039 | 0.00046 | 0.00001 | 0.00030 | 65 | 0.00217 | 0.00004 | 0.00063 | 93 | 0.00069 | -2.45769 | 129.463 | 0.01531 | 0.59095 |
| MQCRA, qcrA, bfcA, petC; menaquinol-cytochrome c reductase iron-sulfur subunit [EC:1.10.2.-] | 0.00147 | 0.00039 | 0.00046 | 0.00001 | 0.00030 | 65 | 0.00217 | 0.00004 | 0.00063 | 93 | 0.00069 | -2.45769 | 129.463 | 0.01531 | 0.59095 |
| 4hbD, abfH; 4-hydroxybutyrate dehydrogenase [EC:1.1.1.61] | 0.00147 | 0.00034 | 0.00154 | 0.00002 | 0.00053 | 65 | 0.00142 | 0.00002 | 0.00044 | 93 | 0.00069 | 0.16958 | 136.107 | 0.86559 | 0.96270 |
| spo0F; two-component system, response regulator, stage 0 sporulation protein F | 0.00147 | 0.00040 | 0.00051 | 0.00001 | 0.00032 | 65 | 0.00213 | 0.00004 | 0.00064 | 93 | 0.00071 | -2.27182 | 132.517 | 0.02471 | 0.59764 |
| tyrC; cyclohexadieny/prephenate dehydrogenase [EC:1.3.1.43 1.3.1.12] | 0.00146 | 0.00030 | 0.00053 | 0.00000 | 0.00014 | 65 | 0.00211 | 0.00002 | 0.00048 | 93 | 0.00050 | -3.14935 | 107.805 | 0.00212 | 0.46257 |
| vraS; two-component system, NarL family, vancomycin resistance sensor histidine kinase VraS [EC:2.7.13.3] | 0.00146 | 0.00049 | 0.00120 | 0.00002 | 0.00060 | 65 | 0.00164 | 0.00005 | 0.00071 | 93 | 0.00093 | -0.47623 | 155.990 | 0.63458 | 0.89696 |
| nagC; N-acetylglucosamine repressor | 0.00146 | 0.00026 | 0.00187 | 0.00001 | 0.00045 | 65 | 0.00117 | 0.00001 | 0.00030 | 93 | 0.00054 | 1.29279 | 118.108 | 0.19861 | 0.77376 |
| aadA; streptomycin 3"-adenylyltransferase [EC:2.7.7.47] | 0.00145 | 0.00030 | 0.00118 | 0.00001 | 0.00033 | 65 | 0.00164 | 0.00002 | 0.00045 | 93 | 0.00056 | -0.81990 | 153.363 | 0.41355 | 0.81655 |
| cotD; spore coat protein D | 0.00145 | 0.00040 | 0.00046 | 0.00001 | 0.00029 | 65 | 0.00215 | 0.00004 | 0.00063 | 93 | 0.00069 | -2.44180 | 125.757 | 0.01600 | 0.59095 |
| yncG; GST-like protein | 0.00145 | 0.00032 | 0.00107 | 0.00001 | 0.00037 | 65 | 0.00172 | 0.00002 | 0.00049 | 93 | 0.00061 | -1.05566 | 154.579 | 0.29277 | 0.77376 |
| vanW; vancomycin resistance protein VanW | 0.00145 | 0.00039 | 0.00092 | 0.00001 | 0.00037 | 65 | 0.00183 | 0.00004 | 0.00062 | 93 | 0.00072 | -1.26136 | 143.208 | 0.20923 | 0.77376 |
| K07807; uncharacterized protein | 0.00145 | 0.00032 | 0.00215 | 0.00003 | 0.00070 | 65 | 0.00097 | 0.00001 | 0.00025 | 93 | 0.00074 | 1.59697 | 80.872 | 0.11417 | 0.77376 |
| K07136; uncharacterized protein | 0.00145 | 0.00025 | 0.00107 | 0.00001 | 0.00028 | 65 | 0.00172 | 0.00001 | 0.00037 | 93 | 0.00047 | -1.39474 | 154.693 | 0.16510 | 0.77376 |
| PTS-Aga-EIIB, agaV; PTS system, N-acetylgalactosamine-specific IIB component [EC:2.7.1.-] | 0.00145 | 0.00024 | 0.00145 | 0.00001 | 0.00037 | 65 | 0.00144 | 0.00001 | 0.00032 | 93 | 0.00049 | 0.02340 | 140.172 | 0.98136 | 0.99661 |
| cpaD; pilus assembly protein CpaD | 0.00144 | 0.00029 | 0.00075 | 0.00000 | 0.00019 | 65 | 0.00192 | 0.00002 | 0.00048 | 93 | 0.00051 | -2.27433 | 119.926 | 0.02472 | 0.59764 |
| K07217; Mn-containing catalase | 0.00144 | 0.00026 | 0.00106 | 0.00001 | 0.00033 | 65 | 0.00171 | 0.00001 | 0.00037 | 93 | 0.00050 | -1.30879 | 155.568 | 0.19254 | 0.77376 |
| cotE; spore coat protein E | 0.00144 | 0.00039 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00209 | 0.00004 | 0.00062 | 93 | 0.00070 | -2.29041 | 131.985 | 0.02358 | 0.59764 |
| gerE; LuxR family transcriptional regulator, transcriptional regulator of spore coat protein | 0.00144 | 0.00039 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00209 | 0.00004 | 0.00062 | 93 | 0.00070 | -2.29041 | 131.985 | 0.02358 | 0.59764 |
| gerQ; spore germination protein Q | 0.00144 | 0.00039 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00209 | 0.00004 | 0.00062 | 93 | 0.00070 | -2.29041 | 131.985 | 0.02358 | 0.59764 |
| GBA, srfJ; glucosylceramidase [EC:3.2.1.45] | 0.00144 | 0.00022 | 0.00139 | 0.00001 | 0.00029 | 65 | 0.00147 | 0.00001 | 0.00032 | 93 | 0.00044 | -0.19043 | 155.048 | 0.84922 | 0.95712 |
| cccB; cytochrome c551 | 0.00144 | 0.00040 | 0.00048 | 0.00001 | 0.00030 | 65 | 0.00210 | 0.00004 | 0.00063 | 93 | 0.00070 | -2.31089 | 129.250 | 0.02242 | 0.59764 |
| porB; pyruvate ferredoxin oxidoreductase beta subunit [EC:1.2.7.1] | 0.00144 | 0.00041 | 0.00105 | 0.00001 | 0.00035 | 65 | 0.00170 | 0.00004 | 0.00066 | 93 | 0.00075 | -0.86968 | 134.723 | 0.38602 | 0.80258 |
| AOC3, AOC2, tynA; primary-amine oxidase [EC:1.4.3.21] | 0.00144 | 0.00028 | 0.00122 | 0.00001 | 0.00045 | 65 | 0.00159 | 0.00001 | 0.00037 | 93 | 0.00058 | -0.63209 | 134.971 | 0.52840 | 0.86020 |
| gerD; spore germination protein D | 0.00143 | 0.00039 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00208 | 0.00004 | 0.00062 | 93 | 0.00069 | -2.27987 | 132.082 | 0.02422 | 0.59764 |
| sspI; small acid-soluble spore protein I (minor) | 0.00143 | 0.00039 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00208 | 0.00004 | 0.00062 | 93 | 0.00069 | -2.27987 | 132.082 | 0.02422 | 0.59764 |
| ypeB; spore germination protein | 0.00143 | 0.00039 | 0.00050 | 0.00001 | 0.00031 | 65 | 0.00208 | 0.00004 | 0.00062 | 93 | 0.00069 | -2.27987 | 132.082 | 0.02422 | 0.59764 |
| higB; mRNA interferase HigB [EC:3.1.-.-] | 0.00143 | 0.00035 | 0.00099 | 0.00001 | 0.00037 | 65 | 0.00174 | 0.00003 | 0.00054 | 93 | 0.00065 | -1.15102 | 150.245 | 0.25155 | 0.77376 |
| E4.2.2.10; pectin lyase [EC:4.2.2.10] | 0.00143 | 0.00057 | 0.00081 | 0.00002 | 0.00060 | 65 | 0.00186 | 0.00007 | 0.00087 | 93 | 0.00105 | -1.00090 | 151.385 | 0.31847 | 0.77376 |
| dlgD; 3-dehydro-L-gulonate 2-dehydrogenase [EC:1.1.1.130] | 0.00143 | 0.00042 | 0.00186 | 0.00005 | 0.00092 | 65 | 0.00112 | 0.00001 | 0.00031 | 93 | 0.00097 | 0.76558 | 78.740 | 0.44621 | 0.83371 |
| K17214; inositol transport system permease protein | 0.00143 | 0.00033 | 0.00082 | 0.00001 | 0.00036 | 65 | 0.00185 | 0.00002 | 0.00050 | 93 | 0.00062 | -1.66471 | 153.484 | 0.09801 | 0.76722 |
| arnB, pmrH; UDP-4-amino-4-deoxy-L-arabinose-oxoglutarate aminotransferase [EC:2.6.1.87] | 0.00142 | 0.00032 | 0.00150 | 0.00002 | 0.00049 | 65 | 0.00137 | 0.00002 | 0.00043 | 93 | 0.00066 | 0.20302 | 141.281 | 0.83941 | 0.95506 |
| ramA; alpha-L-rhamnosidase [EC:3.2.1.40] | 0.00141 | 0.00027 | 0.00088 | 0.00000 | 0.00023 | 65 | 0.00178 | 0.00002 | 0.00043 | 93 | 0.00049 | -1.83735 | 135.545 | 0.06835 | 0.73473 |
| fosB; metallothiol transferase [EC:2.5.1.-] | 0.00141 | 0.00038 | 0.00053 | 0.00001 | 0.00032 | 65 | 0.00202 | 0.00003 | 0.00060 | 93 | 0.00068 | -2.17660 | 135.084 | 0.03125 | 0.61842 |
| uhpA; two-component system, NarL family, uhpT operon response regulator UhpA | 0.00141 | 0.00032 | 0.00140 | 0.00001 | 0.00047 | 65 | 0.00141 | 0.00002 | 0.00043 | 93 | 0.00064 | -0.00949 | 145.980 | 0.99244 | 0.99908 |
| comB4; ComB4 competence protein | 0.00141 | 0.00070 | 0.00094 | 0.00001 | 0.00048 | 65 | 0.00173 | 0.00012 | 0.00114 | 93 | 0.00124 | -0.64183 | 121.364 | 0.52220 | 0.85628 |
| adeN; TetR/AcrR family transcriptional regulator, repressor of the adeIJK operon | 0.00141 | 0.00031 | 0.00119 | 0.00001 | 0.00035 | 65 | 0.00156 | 0.00002 | 0.00046 | 93 | 0.00058 | -0.62494 | 154.635 | 0.53293 | 0.86207 |
| inlA; internalin A | 0.00141 | 0.00032 | 0.00130 | 0.00001 | 0.00035 | 65 | 0.00148 | 0.00002 | 0.00048 | 93 | 0.00060 | -0.29845 | 153.288 | 0.76576 | 0.93900 |
| braE, bceB; bacitracin transport system permease protein | 0.00140 | 0.00041 | 0.00109 | 0.00002 | 0.00053 | 65 | 0.00162 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.66554 | 155.155 | 0.50669 | 0.84956 |
| mqnD; 1,4-dihydroxy-6-naphthoate synthase [EC:1.14.-.-] | 0.00140 | 0.00040 | 0.00107 | 0.00001 | 0.00039 | 65 | 0.00164 | 0.00004 | 0.00062 | 93 | 0.00073 | -0.77604 | 146.579 | 0.43898 | 0.83156 |
| K07100; putative phosphoribosyl transferase | 0.00140 | 0.00042 | 0.00095 | 0.00001 | 0.00030 | 65 | 0.00172 | 0.00004 | 0.00068 | 93 | 0.00074 | -1.04503 | 124.289 | 0.29804 | 0.77376 |
| K06946; uncharacterized protein | 0.00140 | 0.00071 | 0.00055 | 0.00000 | 0.00025 | 65 | 0.00200 | 0.00013 | 0.00119 | 93 | 0.00122 | -1.19148 | 99.936 | 0.23629 | 0.77376 |
| ureAB; urease subunit gamma/beta [EC:3.5.1.5] | 0.00140 | 0.00046 | 0.00086 | 0.00001 | 0.00037 | 65 | 0.00178 | 0.00005 | 0.00073 | 93 | 0.00082 | -1.11398 | 132.599 | 0.26730 | 0.77376 |
| ttrS; two-component system, LuxR family, sensor histidine kinase TtrS [EC:2.7.13.3] | 0.00140 | 0.00094 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00234 | 0.00024 | 0.00159 | 93 | 0.00159 | -1.43120 | 92.112 | 0.15576 | 0.77376 |
| hpr; MarR family transcriptional regulator, protease production regulatory protein HPr | 0.00140 | 0.00039 | 0.00043 | 0.00001 | 0.00030 | 65 | 0.00208 | 0.00004 | 0.00062 | 93 | 0.00069 | -2.40235 | 129.877 | 0.01771 | 0.59095 |
| spo0B; stage 0 sporulation protein B (sporulation initiation phosphotransferase) [EC:2.7.-.-] | 0.00140 | 0.00039 | 0.00043 | 0.00001 | 0.00030 | 65 | 0.00208 | 0.00004 | 0.00062 | 93 | 0.00069 | -2.40235 | 129.877 | 0.01771 | 0.59095 |
| nnr; ADP-dependent NAD(P)H-hydrate dehydratase [EC:4.2.1.136] | 0.00139 | 0.00032 | 0.00172 | 0.00002 | 0.00052 | 65 | 0.00117 | 0.00002 | 0.00041 | 93 | 0.00066 | 0.82548 | 133.069 | 0.41058 | 0.81416 |
| cobZ, tcuA; tricarballylate dehydrogenase | 0.00139 | 0.00031 | 0.00068 | 0.00000 | 0.00020 | 65 | 0.00189 | 0.00002 | 0.00049 | 93 | 0.00053 | -2.26384 | 120.761 | 0.02537 | 0.59941 |
| spoIIIAB; stage III sporulation protein AB | 0.00139 | 0.00032 | 0.00070 | 0.00001 | 0.00042 | 65 | 0.00187 | 0.00002 | 0.00046 | 93 | 0.00062 | -1.88838 | 154.803 | 0.06085 | 0.73001 |
| K11691, dctS; two-component system, CitB family, sensor histidine kinase DctS [EC:2.7.13.3] | 0.00139 | 0.00062 | 0.00062 | 0.00002 | 0.00058 | 65 | 0.00193 | 0.00009 | 0.00097 | 93 | 0.00113 | -1.15855 | 142.593 | 0.24858 | 0.77376 |
| pilK; type IV pilus assembly protein PilK | 0.00138 | 0.00032 | 0.00195 | 0.00003 | 0.00062 | 65 | 0.00099 | 0.00001 | 0.00033 | 93 | 0.00071 | 1.36990 | 99.982 | 0.17379 | 0.77376 |
| lst; beta-galactosamide-alpha-2,3-sialyltransferase [EC:2.4.99.-] | 0.00138 | 0.00032 | 0.00113 | 0.00001 | 0.00035 | 65 | 0.00156 | 0.00002 | 0.00049 | 93 | 0.00060 | -0.71491 | 152.960 | 0.47576 | 0.84133 |
| K00184; prokaryotic molybdopterin-containing oxidoreductase family, iron-sulfur binding subunit | 0.00138 | 0.00031 | 0.00114 | 0.00001 | 0.00033 | 65 | 0.00154 | 0.00002 | 0.00048 | 93 | 0.00058 | -0.69015 | 150.317 | 0.49116 | 0.84797 |
| mtrB; transcription attenuation protein (tryptophan RNA-binding attenuator protein) | 0.00138 | 0.00039 | 0.00057 | 0.00001 | 0.00032 | 65 | 0.00194 | 0.00003 | 0.00061 | 93 | 0.00069 | -1.97534 | 134.795 | 0.05027 | 0.67941 |
| bceB, vraE; bacitracin transport system permease protein | 0.00138 | 0.00037 | 0.00062 | 0.00001 | 0.00032 | 65 | 0.00191 | 0.00003 | 0.00059 | 93 | 0.00067 | -1.92702 | 137.437 | 0.05604 | 0.70577 |
| kshA; 3-ketosteroid 9alpha-monooxygenase subunit A [EC:1.14.13.142] | 0.00138 | 0.00073 | 0.00051 | 0.00000 | 0.00020 | 65 | 0.00199 | 0.00014 | 0.00122 | 93 | 0.00124 | -1.19193 | 97.058 | 0.23619 | 0.77376 |
| hpaD, hpcB; 3,4-dihydroxyphenylacetate 2,3-dioxygenase [EC:1.13.11.15] | 0.00138 | 0.00036 | 0.00084 | 0.00001 | 0.00035 | 65 | 0.00175 | 0.00003 | 0.00055 | 93 | 0.00066 | -1.39090 | 147.143 | 0.16636 | 0.77376 |
| pel; pectate lyase [EC:4.2.2.2] | 0.00137 | 0.00040 | 0.00081 | 0.00001 | 0.00032 | 65 | 0.00177 | 0.00004 | 0.00065 | 93 | 0.00072 | -1.32669 | 131.705 | 0.18691 | 0.77376 |
| abmG; 2-aminobenzoate-CoA ligase [EC:6.2.1.32] | 0.00137 | 0.00036 | 0.00070 | 0.00001 | 0.00028 | 65 | 0.00184 | 0.00003 | 0.00057 | 93 | 0.00063 | -1.79060 | 130.495 | 0.07568 | 0.74536 |
| VCP, CDC48; transitional endoplasmic reticulum ATPase | 0.00137 | 0.00026 | 0.00089 | 0.00000 | 0.00021 | 65 | 0.00171 | 0.00002 | 0.00042 | 93 | 0.00047 | -1.74754 | 132.705 | 0.08286 | 0.76567 |
| fabY; acetoacetyl-[acyl-carrier protein] synthase [EC:2.3.1.180] | 0.00137 | 0.00032 | 0.00202 | 0.00002 | 0.00061 | 65 | 0.00092 | 0.00001 | 0.00033 | 93 | 0.00069 | 1.60135 | 101.217 | 0.11242 | 0.77376 |
| glpM; membrane protein GlpM | 0.00137 | 0.00033 | 0.00094 | 0.00001 | 0.00037 | 65 | 0.00166 | 0.00002 | 0.00050 | 93 | 0.00062 | -1.16628 | 153.691 | 0.24531 | 0.77376 |
| phd; antitoxin Phd | 0.00136 | 0.00042 | 0.00190 | 0.00005 | 0.00092 | 65 | 0.00099 | 0.00001 | 0.00032 | 93 | 0.00097 | 0.93468 | 79.754 | 0.35277 | 0.79194 |
| mal; methylaspartate ammonia-lyase [EC:4.3.1.2] | 0.00136 | 0.00032 | 0.00136 | 0.00001 | 0.00040 | 65 | 0.00136 | 0.00002 | 0.00047 | 93 | 0.00062 | 0.00567 | 155.781 | 0.99549 | 0.99944 |
| nodJ; lipooligosaccharide transport system permease protein | 0.00136 | 0.00031 | 0.00085 | 0.00000 | 0.00024 | 65 | 0.00172 | 0.00002 | 0.00050 | 93 | 0.00055 | -1.56254 | 128.262 | 0.12062 | 0.77376 |
| comP; two-component system, NarL family, sensor histidine kinase ComP [EC:2.7.13.3] | 0.00136 | 0.00088 | 0.00090 | 0.00004 | 0.00083 | 65 | 0.00168 | 0.00018 | 0.00138 | 93 | 0.00161 | -0.48770 | 143.595 | 0.62650 | 0.89255 |
| mgtA; phosphatidylinositol alpha 1,6-mannosyltransferase [EC:2.4.1.-] | 0.00135 | 0.00028 | 0.00105 | 0.00001 | 0.00034 | 65 | 0.00157 | 0.00002 | 0.00041 | 93 | 0.00053 | -0.97029 | 155.958 | 0.33340 | 0.77957 |
| thiZ; putative hydroxymethylpyrimidine transport system ATP-binding protein | 0.00135 | 0.00025 | 0.00123 | 0.00001 | 0.00036 | 65 | 0.00144 | 0.00001 | 0.00035 | 93 | 0.00050 | -0.42157 | 149.356 | 0.67395 | 0.91184 |
| pspE; phage shock protein E | 0.00135 | 0.00029 | 0.00148 | 0.00002 | 0.00055 | 65 | 0.00126 | 0.00001 | 0.00031 | 93 | 0.00063 | 0.35246 | 102.539 | 0.72521 | 0.92177 |
| wzxE; enterobacterial common antigen flippase | 0.00135 | 0.00032 | 0.00132 | 0.00001 | 0.00047 | 65 | 0.00137 | 0.00002 | 0.00043 | 93 | 0.00064 | -0.07477 | 145.493 | 0.94050 | 0.98528 |
| tupA, vupA; tungstate transport system substrate-binding protein | 0.00134 | 0.00029 | 0.00111 | 0.00001 | 0.00032 | 65 | 0.00151 | 0.00002 | 0.00045 | 93 | 0.00055 | -0.72292 | 152.761 | 0.47083 | 0.83912 |
| gerPF; spore germination protein PF | 0.00134 | 0.00045 | 0.00009 | 0.00000 | 0.00003 | 65 | 0.00222 | 0.00005 | 0.00075 | 93 | 0.00076 | -2.81655 | 92.362 | 0.00594 | 0.54022 |
| degU; two-component system, NarL family, response regulator DegU | 0.00134 | 0.00039 | 0.00055 | 0.00001 | 0.00033 | 65 | 0.00190 | 0.00003 | 0.00061 | 93 | 0.00070 | -1.93054 | 137.143 | 0.05560 | 0.70577 |
| K18383; trans-feruloyl-CoA hydratase / vanillin synthase [EC:4.2.1.101 4.1.2.41] | 0.00134 | 0.00033 | 0.00101 | 0.00001 | 0.00039 | 65 | 0.00157 | 0.00002 | 0.00049 | 93 | 0.00063 | -0.89046 | 155.658 | 0.37459 | 0.79810 |
| spoVAB; stage V sporulation protein AB | 0.00134 | 0.00033 | 0.00023 | 0.00000 | 0.00007 | 65 | 0.00211 | 0.00003 | 0.00055 | 93 | 0.00055 | -3.42672 | 95.340 | 0.00090 | 0.46257 |
| tll; dTDP-6-deoxy-L-talose 4-dehydrogenase (NAD+) [EC:1.1.1.339] | 0.00134 | 0.00019 | 0.00164 | 0.00001 | 0.00030 | 65 | 0.00112 | 0.00001 | 0.00024 | 93 | 0.00038 | 1.34314 | 132.016 | 0.18153 | 0.77376 |
| dmpB, xylE; catechol 2,3-dioxygenase [EC:1.13.11.2] | 0.00134 | 0.00031 | 0.00060 | 0.00000 | 0.00018 | 65 | 0.00185 | 0.00002 | 0.00051 | 93 | 0.00054 | -2.32837 | 113.875 | 0.02166 | 0.59153 |
| tetB; MFS transporter, DHA2 family, metal-tetracycline-proton antiporter | 0.00134 | 0.00027 | 0.00124 | 0.00001 | 0.00047 | 65 | 0.00140 | 0.00001 | 0.00033 | 93 | 0.00057 | -0.28696 | 122.121 | 0.77463 | 0.94269 |
| norG; GntR family transcriptional regulator, regulator for abcA and norABC | 0.00133 | 0.00038 | 0.00053 | 0.00001 | 0.00031 | 65 | 0.00190 | 0.00003 | 0.00060 | 93 | 0.00068 | -2.02476 | 133.625 | 0.04488 | 0.65981 |
| bpsB, srsB; methyltransferase | 0.00133 | 0.00038 | 0.00046 | 0.00001 | 0.00030 | 65 | 0.00194 | 0.00003 | 0.00061 | 93 | 0.00068 | -2.19250 | 131.491 | 0.03010 | 0.61413 |
| puuP; putrescine importer | 0.00133 | 0.00025 | 0.00168 | 0.00001 | 0.00043 | 65 | 0.00108 | 0.00001 | 0.00031 | 93 | 0.00053 | 1.12550 | 123.837 | 0.26255 | 0.77376 |
| kipA; antagonist of KipI | 0.00133 | 0.00030 | 0.00023 | 0.00000 | 0.00008 | 65 | 0.00209 | 0.00002 | 0.00050 | 93 | 0.00050 | -3.69710 | 97.173 | 0.00036 | 0.46257 |
| pucG; (S)-ureidoglycine—glyoxylate transaminase [EC:2.6.1.112] | 0.00133 | 0.00045 | 0.00157 | 0.00005 | 0.00086 | 65 | 0.00115 | 0.00002 | 0.00048 | 93 | 0.00098 | 0.42359 | 104.018 | 0.67274 | 0.91184 |
| mqsA; HTH-type transcriptional regulator / antitoxin MqsA | 0.00132 | 0.00032 | 0.00087 | 0.00001 | 0.00032 | 65 | 0.00164 | 0.00002 | 0.00049 | 93 | 0.00059 | -1.29544 | 148.485 | 0.19718 | 0.77376 |
| bceR; two-component system, OmpR family, bacitracin resistance response regulator BceR | 0.00132 | 0.00037 | 0.00058 | 0.00001 | 0.00032 | 65 | 0.00184 | 0.00003 | 0.00058 | 93 | 0.00067 | -1.88278 | 137.670 | 0.06184 | 0.73001 |
| spoVAA; stage V sporulation protein AA | 0.00132 | 0.00033 | 0.00020 | 0.00000 | 0.00007 | 65 | 0.00210 | 0.00003 | 0.00054 | 93 | 0.00054 | -3.50635 | 95.023 | 0.00070 | 0.46257 |
| rhaT; L-rhamnose-H+ transport protein | 0.00131 | 0.00029 | 0.00069 | 0.00000 | 0.00017 | 65 | 0.00175 | 0.00002 | 0.00048 | 93 | 0.00051 | -2.10795 | 115.235 | 0.03720 | 0.63026 |
| narT; MFS transporter, NNP family, putative nitrate transporter | 0.00131 | 0.00041 | 0.00097 | 0.00002 | 0.00053 | 65 | 0.00155 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.73064 | 155.165 | 0.46610 | 0.83830 |
| mdeA; MFS transporter, DHA2 family, multidrug resistance protein | 0.00131 | 0.00041 | 0.00099 | 0.00002 | 0.00053 | 65 | 0.00153 | 0.00003 | 0.00059 | 93 | 0.00080 | -0.67726 | 155.161 | 0.49925 | 0.84856 |
| fabL; enoyl-[acyl-carrier protein] reductase III [EC:1.3.1.104] | 0.00131 | 0.00041 | 0.00028 | 0.00000 | 0.00012 | 65 | 0.00202 | 0.00004 | 0.00068 | 93 | 0.00070 | -2.50493 | 98.079 | 0.01390 | 0.59095 |
| ligK, galC; 4-hydroxy-4-methyl-2-oxoglutarate aldolase [EC:4.1.3.17] | 0.00131 | 0.00029 | 0.00069 | 0.00000 | 0.00024 | 65 | 0.00174 | 0.00002 | 0.00046 | 93 | 0.00052 | -2.01653 | 133.591 | 0.04575 | 0.65981 |
| AFMID; arylformamidase [EC:3.5.1.9] | 0.00130 | 0.00029 | 0.00076 | 0.00000 | 0.00024 | 65 | 0.00168 | 0.00002 | 0.00046 | 93 | 0.00052 | -1.77468 | 135.069 | 0.07820 | 0.75550 |
| liaI; lia operon protein LiaI | 0.00130 | 0.00039 | 0.00047 | 0.00001 | 0.00031 | 65 | 0.00188 | 0.00004 | 0.00062 | 93 | 0.00070 | -2.01930 | 131.745 | 0.04548 | 0.65981 |
| cotJA; spore coat protein JA | 0.00130 | 0.00038 | 0.00039 | 0.00001 | 0.00028 | 65 | 0.00193 | 0.00003 | 0.00060 | 93 | 0.00066 | -2.31903 | 126.752 | 0.02199 | 0.59447 |
| agrD; AgrD protein | 0.00130 | 0.00041 | 0.00096 | 0.00002 | 0.00053 | 65 | 0.00153 | 0.00003 | 0.00059 | 93 | 0.00080 | -0.71921 | 155.165 | 0.47309 | 0.84050 |
| aur; aureolysin [EC:3.4.24.29] | 0.00130 | 0.00041 | 0.00096 | 0.00002 | 0.00053 | 65 | 0.00153 | 0.00003 | 0.00059 | 93 | 0.00080 | -0.71921 | 155.165 | 0.47309 | 0.84050 |
| braD, bceA; bacitracin transport system ATP-binding protein | 0.00130 | 0.00041 | 0.00096 | 0.00002 | 0.00053 | 65 | 0.00153 | 0.00003 | 0.00059 | 93 | 0.00080 | -0.71921 | 155.165 | 0.47309 | 0.84050 |
| hrtA; hemin transport system ATP-binding protein [EC:3.6.3.-] | 0.00130 | 0.00041 | 0.00096 | 0.00002 | 0.00053 | 65 | 0.00153 | 0.00003 | 0.00059 | 93 | 0.00080 | -0.71921 | 155.165 | 0.47309 | 0.84050 |
| pleD; two-component system, cell cycle response regulator [EC:2.7.7.65] | 0.00129 | 0.00029 | 0.00060 | 0.00000 | 0.00018 | 65 | 0.00178 | 0.00002 | 0.00047 | 93 | 0.00050 | -2.33830 | 115.926 | 0.02109 | 0.59147 |
| prdF; proline racemase [EC:5.1.1.4] | 0.00129 | 0.00034 | 0.00111 | 0.00001 | 0.00040 | 65 | 0.00142 | 0.00002 | 0.00050 | 93 | 0.00064 | -0.48544 | 155.545 | 0.62805 | 0.89377 |
| pdc; phenolic acid decarboxylase [EC:4.1.1.-] | 0.00129 | 0.00045 | 0.00141 | 0.00005 | 0.00085 | 65 | 0.00120 | 0.00002 | 0.00049 | 93 | 0.00098 | 0.21553 | 105.352 | 0.82977 | 0.95506 |
| dmsB; anaerobic dimethyl sulfoxide reductase subunit B (DMSO reductase iron- sulfur subunit) | 0.00129 | 0.00038 | 0.00069 | 0.00000 | 0.00018 | 65 | 0.00171 | 0.00004 | 0.00064 | 93 | 0.00066 | -1.54430 | 106.115 | 0.12549 | 0.77376 |
| K07046; L-fuconolactonase [EC:3.1.1.-] | 0.00128 | 0.00022 | 0.00109 | 0.00001 | 0.00031 | 65 | 0.00142 | 0.00001 | 0.00029 | 93 | 0.00043 | -0.75294 | 146.691 | 0.45269 | 0.83438 |
| ABC.MR; putative ABC transport system ATP-binding protein | 0.00128 | 0.00040 | 0.00102 | 0.00000 | 0.00027 | 65 | 0.00146 | 0.00004 | 0.00065 | 93 | 0.00070 | -0.62991 | 120.969 | 0.52994 | 0.86094 |
| kefG; glutathione-regulated potassium-efflux system ancillary protein KefG | 0.00128 | 0.00023 | 0.00118 | 0.00001 | 0.00032 | 65 | 0.00135 | 0.00001 | 0.00031 | 93 | 0.00045 | -0.36632 | 148.934 | 0.71464 | 0.92004 |
| K11527; two-component system, sensor histidine kinase and response regulator [EC:2.7.13.3] | 0.00128 | 0.00034 | 0.00059 | 0.00000 | 0.00017 | 65 | 0.00177 | 0.00003 | 0.00056 | 93 | 0.00058 | -2.03335 | 108.699 | 0.04445 | 0.65981 |
| casD, cse5; CRISPR system Cascade subunit CasD | 0.00127 | 0.00024 | 0.00138 | 0.00001 | 0.00036 | 65 | 0.00119 | 0.00001 | 0.00033 | 93 | 0.00049 | 0.40446 | 145.958 | 0.68647 | 0.91567 |
| pseB; UDP-N-acetylglucosamine 4,6-dehydratase [EC:4.2.1.115] | 0.00127 | 0.00037 | 0.00084 | 0.00000 | 0.00023 | 65 | 0.00157 | 0.00003 | 0.00061 | 93 | 0.00065 | -1.12864 | 116.582 | 0.26137 | 0.77376 |
| yitG, ymfD, yfmO; MFS transporter, ACDE family, multidrug resistance protein | 0.00126 | 0.00034 | 0.00021 | 0.00000 | 0.00008 | 65 | 0.00200 | 0.00003 | 0.00056 | 93 | 0.00056 | -3.17463 | 95.704 | 0.00202 | 0.46257 |
| casE, cse3; CRISPR system Cascade subunit CasE | 0.00126 | 0.00024 | 0.00136 | 0.00001 | 0.00036 | 65 | 0.00119 | 0.00001 | 0.00033 | 93 | 0.00049 | 0.36563 | 146.008 | 0.71517 | 0.92004 |
| aroP; aromatic amino acid permease | 0.00126 | 0.00025 | 0.00135 | 0.00001 | 0.00039 | 65 | 0.00120 | 0.00001 | 0.00034 | 93 | 0.00052 | 0.28473 | 141.030 | 0.77627 | 0.94269 |
| blaZ; beta-lactamase class A BlaZ [EC:3.5.2.6] | 0.00126 | 0.00041 | 0.00094 | 0.00002 | 0.00053 | 65 | 0.00148 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.68878 | 155.073 | 0.49199 | 0.84797 |
| sasG; surface protein G | 0.00126 | 0.00041 | 0.00094 | 0.00002 | 0.00053 | 65 | 0.00148 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.68878 | 155.073 | 0.49199 | 0.84797 |
| tupB, vupB; tungstate transport system permease protein | 0.00125 | 0.00028 | 0.00107 | 0.00001 | 0.00031 | 65 | 0.00139 | 0.00002 | 0.00043 | 93 | 0.00053 | -0.60242 | 152.792 | 0.54779 | 0.86549 |
| DMGDH; dimethylglycine dehydrogenase [EC:1.5.8.4] | 0.00125 | 0.00039 | 0.00036 | 0.00001 | 0.00030 | 65 | 0.00188 | 0.00004 | 0.00062 | 93 | 0.00069 | -2.20713 | 129.298 | 0.02907 | 0.61334 |
| K09145; uncharacterized protein | 0.00125 | 0.00043 | 0.00093 | 0.00002 | 0.00053 | 65 | 0.00148 | 0.00004 | 0.00062 | 93 | 0.00082 | -0.67560 | 155.889 | 0.50030 | 0.84899 |
| casA, cse1; CRISPR system Cascade subunit CasA | 0.00125 | 0.00024 | 0.00136 | 0.00001 | 0.00036 | 65 | 0.00118 | 0.00001 | 0.00033 | 93 | 0.00049 | 0.36389 | 145.942 | 0.71647 | 0.92004 |
| lpqC; polyhydroxybutyrate depolymerase | 0.00125 | 0.00025 | 0.00103 | 0.00001 | 0.00030 | 65 | 0.00141 | 0.00001 | 0.00036 | 93 | 0.00047 | -0.80533 | 155.989 | 0.42185 | 0.82295 |
| K07482; transposase, IS30 family | 0.00125 | 0.00028 | 0.00118 | 0.00001 | 0.00038 | 65 | 0.00130 | 0.00001 | 0.00040 | 93 | 0.00055 | -0.21096 | 153.000 | 0.83320 | 0.95506 |
| hxlB; 6-phospho-3-hexuloisomerase [EC:5.3.1.27] | 0.00125 | 0.00026 | 0.00060 | 0.00000 | 0.00018 | 65 | 0.00171 | 0.00002 | 0.00042 | 93 | 0.00046 | -2.41061 | 123.221 | 0.01740 | 0.59095 |
| K07317; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | 0.00125 | 0.00038 | 0.00091 | 0.00000 | 0.00024 | 65 | 0.00148 | 0.00004 | 0.00063 | 93 | 0.00067 | -0.85640 | 117.571 | 0.39352 | 0.80459 |
| matB; malonyl-CoA/methylmalonyl-CoA synthetase [EC:6.2.1.-] | 0.00125 | 0.00030 | 0.00076 | 0.00000 | 0.00024 | 65 | 0.00159 | 0.00002 | 0.00048 | 93 | 0.00053 | -1.54874 | 131.117 | 0.12385 | 0.77376 |
| nepI; MFS transporter, DHA1 family, purine ribonucleoside efflux pump | 0.00124 | 0.00024 | 0.00162 | 0.00001 | 0.00043 | 65 | 0.00097 | 0.00001 | 0.00027 | 93 | 0.00051 | 1.27571 | 112.794 | 0.20468 | 0.77376 |
| K09992; uncharacterized protein | 0.00124 | 0.00024 | 0.00076 | 0.00001 | 0.00028 | 65 | 0.00157 | 0.00001 | 0.00036 | 93 | 0.00046 | -1.77266 | 155.166 | 0.07825 | 0.75550 |
| FDH; formate dehydrogenase [EC:1.17.1.9] | 0.00124 | 0.00034 | 0.00044 | 0.00000 | 0.00018 | 65 | 0.00179 | 0.00003 | 0.00056 | 93 | 0.00058 | -2.30613 | 110.313 | 0.02297 | 0.59764 |
| mcrB; 5-methylcytosine-specific restriction enzyme B [EC:3.1.21.-] | 0.00124 | 0.00025 | 0.00170 | 0.00002 | 0.00054 | 65 | 0.00091 | 0.00000 | 0.00020 | 93 | 0.00057 | 1.38270 | 81.436 | 0.17054 | 0.77376 |
| insB; insertion element IS1 protein InsB | 0.00123 | 0.00073 | 0.00061 | 0.00001 | 0.00033 | 65 | 0.00167 | 0.00014 | 0.00122 | 93 | 0.00127 | -0.83991 | 105.075 | 0.40287 | 0.80963 |
| PTS-Aga-EIID, agaE; PTS system, N-acetylgalactosamine-specific IID component | 0.00123 | 0.00023 | 0.00113 | 0.00001 | 0.00034 | 65 | 0.00131 | 0.00001 | 0.00031 | 93 | 0.00046 | -0.38554 | 144.643 | 0.70040 | 0.91953 |
| rob; AraC family transcriptional regulator, mar-sox-rob regulon activator | 0.00123 | 0.00034 | 0.00134 | 0.00002 | 0.00056 | 65 | 0.00115 | 0.00002 | 0.00044 | 93 | 0.00071 | 0.26446 | 133.138 | 0.79183 | 0.94731 |
| K02475; two-component system, CitB family, response regulator | 0.00123 | 0.00040 | 0.00089 | 0.00002 | 0.00050 | 65 | 0.00147 | 0.00003 | 0.00059 | 93 | 0.00077 | -0.75369 | 155.955 | 0.45217 | 0.83438 |
| mqsR; motility quorum-sensing regulator / GCU-specific mRNA interferase toxin | 0.00123 | 0.00031 | 0.00086 | 0.00001 | 0.00032 | 65 | 0.00148 | 0.00002 | 0.00048 | 93 | 0.00058 | -1.07400 | 150.423 | 0.28454 | 0.77376 |
| rfaH; transcriptional antiterminator RfaH | 0.00122 | 0.00022 | 0.00144 | 0.00001 | 0.00039 | 65 | 0.00108 | 0.00001 | 0.00027 | 93 | 0.00047 | 0.76535 | 120.488 | 0.44556 | 0.83300 |
| adaA; AraC family transcriptional regulator, regulatory protein of adaptative response / methylphosphotriester-DNA alkyltransferase methyltransferase [EC:2.1.1.-] | 0.00122 | 0.00062 | 0.00072 | 0.00002 | 0.00058 | 65 | 0.00158 | 0.00009 | 0.00096 | 93 | 0.00113 | -0.76058 | 144.120 | 0.44815 | 0.83372 |
| cutF, nlpE; copper homeostasis protein (lipoprotein) | 0.00122 | 0.00025 | 0.00110 | 0.00001 | 0.00033 | 65 | 0.00131 | 0.00001 | 0.00036 | 93 | 0.00049 | -0.44435 | 155.194 | 0.65740 | 0.90734 |
| birA-coaX; biotin—[acetyl-CoA-carboxylase] ligase / type III pantothenate kinase [EC:6.3.4.15 2.7.1.33] | 0.00122 | 0.00029 | 0.00144 | 0.00002 | 0.00052 | 65 | 0.00107 | 0.00001 | 0.00034 | 93 | 0.00062 | 0.58168 | 115.883 | 0.56191 | 0.86813 |
| K06897; 7,8-dihydropterin-6-yl-methyl-4-(beta-D-ribofuranosyl)aminobenzene 5’-phosphate synthase [EC:2.5.1.105] | 0.00122 | 0.00028 | 0.00108 | 0.00001 | 0.00046 | 65 | 0.00132 | 0.00001 | 0.00035 | 93 | 0.00058 | -0.41163 | 128.599 | 0.68130 | 0.91403 |
| mhpF; acetaldehyde dehydrogenase [EC:1.2.1.10] | 0.00122 | 0.00038 | 0.00056 | 0.00000 | 0.00015 | 65 | 0.00168 | 0.00004 | 0.00064 | 93 | 0.00066 | -1.70103 | 102.119 | 0.09198 | 0.76567 |
| mprA; two-component system, OmpR family, response regulator MprA | 0.00122 | 0.00028 | 0.00096 | 0.00001 | 0.00034 | 65 | 0.00140 | 0.00002 | 0.00040 | 93 | 0.00053 | -0.82423 | 155.974 | 0.41106 | 0.81489 |
| ala; alanine dehydrogenase [EC:1.4.1.1] | 0.00122 | 0.00044 | 0.00137 | 0.00005 | 0.00084 | 65 | 0.00111 | 0.00002 | 0.00048 | 93 | 0.00097 | 0.26278 | 104.644 | 0.79323 | 0.94783 |
| yfkR; spore germination protein | 0.00122 | 0.00060 | 0.00065 | 0.00002 | 0.00055 | 65 | 0.00161 | 0.00008 | 0.00094 | 93 | 0.00109 | -0.88033 | 142.395 | 0.38016 | 0.79852 |
| phzF; trans-2,3-dihydro-3-hydroxyanthranilate isomerase [EC:5.3.3.17] | 0.00122 | 0.00026 | 0.00050 | 0.00000 | 0.00015 | 65 | 0.00172 | 0.00002 | 0.00041 | 93 | 0.00044 | -2.77801 | 114.361 | 0.00639 | 0.54194 |
| sspE; small acid-soluble spore protein E (minor gamma-type SASP) | 0.00122 | 0.00036 | 0.00049 | 0.00001 | 0.00031 | 65 | 0.00172 | 0.00003 | 0.00057 | 93 | 0.00065 | -1.87646 | 136.972 | 0.06272 | 0.73012 |
| hpaA; AraC family transcriptional regulator, 4-hydroxyphenylacetate 3-monooxygenase operon regulatory protein | 0.00121 | 0.00031 | 0.00100 | 0.00001 | 0.00039 | 65 | 0.00136 | 0.00002 | 0.00046 | 93 | 0.00060 | -0.59412 | 156.000 | 0.55329 | 0.86788 |
| malR; two-component system, CitB family, response regulator MalR | 0.00121 | 0.00028 | 0.00134 | 0.00002 | 0.00060 | 65 | 0.00112 | 0.00001 | 0.00024 | 93 | 0.00065 | 0.33868 | 84.064 | 0.73570 | 0.92721 |
| colA; microbial collagenase [EC:3.4.24.3] | 0.00121 | 0.00042 | 0.00087 | 0.00002 | 0.00049 | 65 | 0.00145 | 0.00004 | 0.00063 | 93 | 0.00080 | -0.71681 | 155.298 | 0.47457 | 0.84095 |
| yesW; rhamnogalacturonan endolyase [EC:4.2.2.23] | 0.00121 | 0.00040 | 0.00048 | 0.00000 | 0.00017 | 65 | 0.00172 | 0.00004 | 0.00066 | 93 | 0.00068 | -1.80871 | 103.654 | 0.07340 | 0.74128 |
| pecM; probable blue pigment (indigoidine) exporter | 0.00121 | 0.00026 | 0.00071 | 0.00000 | 0.00022 | 65 | 0.00156 | 0.00002 | 0.00041 | 93 | 0.00046 | -1.82778 | 136.678 | 0.06976 | 0.73522 |
| torC; trimethylamine-N-oxide reductase (cytochrome c), cytochrome c-type subunit TorC | 0.00120 | 0.00026 | 0.00149 | 0.00001 | 0.00047 | 65 | 0.00101 | 0.00001 | 0.00029 | 93 | 0.00055 | 0.87290 | 112.622 | 0.38457 | 0.80197 |
| IS15, IS26; transposase, IS6 family | 0.00120 | 0.00041 | 0.00072 | 0.00000 | 0.00025 | 65 | 0.00154 | 0.00004 | 0.00067 | 93 | 0.00071 | -1.13947 | 116.025 | 0.25685 | 0.77376 |
| casB, cse2; CRISPR system Cascade subunit CasB | 0.00120 | 0.00024 | 0.00126 | 0.00001 | 0.00036 | 65 | 0.00115 | 0.00001 | 0.00033 | 93 | 0.00049 | 0.21129 | 145.877 | 0.83296 | 0.95506 |
| vasI; type VI secretion system protein VasI | 0.00119 | 0.00032 | 0.00122 | 0.00002 | 0.00049 | 65 | 0.00118 | 0.00002 | 0.00043 | 93 | 0.00065 | 0.05673 | 142.322 | 0.95484 | 0.98792 |
| TREH, treA, treF; alpha,alpha-trehalase [EC:3.2.1.28] | 0.00119 | 0.00025 | 0.00094 | 0.00001 | 0.00028 | 65 | 0.00137 | 0.00001 | 0.00038 | 93 | 0.00047 | -0.90515 | 153.444 | 0.36681 | 0.79587 |
| kefF; glutathione-regulated potassium-efflux system ancillary protein KefF | 0.00119 | 0.00023 | 0.00111 | 0.00001 | 0.00032 | 65 | 0.00125 | 0.00001 | 0.00033 | 93 | 0.00046 | -0.31398 | 152.046 | 0.75397 | 0.93665 |
| iolI; 2-keto-myo-inositol isomerase [EC:5.3.99.11] | 0.00119 | 0.00031 | 0.00086 | 0.00001 | 0.00037 | 65 | 0.00143 | 0.00002 | 0.00047 | 93 | 0.00060 | -0.95845 | 155.381 | 0.33932 | 0.78433 |
| soxZ; sulfur-oxidizing protein SoxZ | 0.00119 | 0.00031 | 0.00067 | 0.00000 | 0.00023 | 65 | 0.00155 | 0.00002 | 0.00050 | 93 | 0.00055 | -1.60963 | 126.544 | 0.10997 | 0.77376 |
| pfkB; 6-phosphofructokinase 2 [EC:2.7.1.11] | 0.00119 | 0.00028 | 0.00047 | 0.00000 | 0.00012 | 65 | 0.00169 | 0.00002 | 0.00046 | 93 | 0.00048 | -2.53602 | 103.891 | 0.01270 | 0.59095 |
| nicB; nicotinate dehydrogenase subunit B [EC:1.17.2.1] | 0.00119 | 0.00035 | 0.00048 | 0.00001 | 0.00030 | 65 | 0.00169 | 0.00003 | 0.00055 | 93 | 0.00063 | -1.92611 | 137.046 | 0.05616 | 0.70577 |
| pspB; phage shock protein B | 0.00119 | 0.00023 | 0.00118 | 0.00001 | 0.00033 | 65 | 0.00119 | 0.00001 | 0.00031 | 93 | 0.00045 | -0.01320 | 145.782 | 0.98949 | 0.99801 |
| sirB; sirohydrochlorin ferrochelatase [EC:4.99.1.4] | 0.00118 | 0.00037 | 0.00037 | 0.00000 | 0.00028 | 65 | 0.00175 | 0.00003 | 0.00060 | 93 | 0.00066 | -2.09167 | 127.153 | 0.03846 | 0.63091 |
| AGXT; alanine-glyoxylate transaminase / serine-glyoxylate transaminase / serine-pyruvate transaminase [EC:2.6.1.44 2.6.1.45 2.6.1.51] | 0.00118 | 0.00033 | 0.00051 | 0.00000 | 0.00015 | 65 | 0.00165 | 0.00003 | 0.00055 | 93 | 0.00057 | -2.01171 | 105.329 | 0.04680 | 0.66255 |
| yabG; spore coat assemly protein | 0.00118 | 0.00031 | 0.00059 | 0.00001 | 0.00041 | 65 | 0.00159 | 0.00002 | 0.00043 | 93 | 0.00060 | -1.68240 | 152.895 | 0.09453 | 0.76609 |
| pepM; phosphoenolpyruvate phosphomutase [EC:5.4.2.9] | 0.00118 | 0.00028 | 0.00108 | 0.00001 | 0.00040 | 65 | 0.00125 | 0.00001 | 0.00039 | 93 | 0.00056 | -0.29624 | 149.345 | 0.76746 | 0.93900 |
| zipB; zinc and cadmium transporter | 0.00117 | 0.00031 | 0.00077 | 0.00001 | 0.00029 | 65 | 0.00146 | 0.00002 | 0.00048 | 93 | 0.00056 | -1.22577 | 142.427 | 0.22231 | 0.77376 |
| mazE, chpAI; antitoxin MazE | 0.00117 | 0.00026 | 0.00090 | 0.00001 | 0.00032 | 65 | 0.00136 | 0.00001 | 0.00038 | 93 | 0.00050 | -0.91414 | 155.998 | 0.36205 | 0.79496 |
| btaA; S-adenosylmethionine-diacylglycerol 3-amino-3-carboxypropyl transferase | 0.00117 | 0.00019 | 0.00124 | 0.00001 | 0.00035 | 65 | 0.00111 | 0.00000 | 0.00021 | 93 | 0.00041 | 0.31689 | 107.348 | 0.75195 | 0.93550 |
| lonB; ATP-dependent Lon protease [EC:3.4.21.53] | 0.00117 | 0.00031 | 0.00051 | 0.00001 | 0.00041 | 65 | 0.00162 | 0.00002 | 0.00043 | 93 | 0.00059 | -1.87407 | 153.019 | 0.06283 | 0.73012 |
| kinE; two-component system, sporulation sensor kinase E [EC:2.7.13.3] | 0.00116 | 0.00038 | 0.00040 | 0.00001 | 0.00030 | 65 | 0.00170 | 0.00003 | 0.00061 | 93 | 0.00068 | -1.90286 | 130.822 | 0.05926 | 0.72498 |
| aruH; arginine:pyruvate transaminase [EC:2.6.1.84] | 0.00116 | 0.00025 | 0.00111 | 0.00001 | 0.00038 | 65 | 0.00120 | 0.00001 | 0.00033 | 93 | 0.00051 | -0.17534 | 140.845 | 0.86107 | 0.96180 |
| glrK, qseE; two-component system, NtrC family, sensor histidine kinase GlrK [EC:2.7.13.3] | 0.00116 | 0.00031 | 0.00064 | 0.00000 | 0.00025 | 65 | 0.00152 | 0.00002 | 0.00049 | 93 | 0.00056 | -1.58865 | 133.355 | 0.11451 | 0.77376 |
| ereA_B; erythromycin esterase [EC:3.1.1.-] | 0.00116 | 0.00059 | 0.00070 | 0.00002 | 0.00055 | 65 | 0.00148 | 0.00008 | 0.00093 | 93 | 0.00108 | -0.72433 | 142.820 | 0.47005 | 0.83870 |
| pilL; type IV pili sensor histidine kinase and response regulator | 0.00116 | 0.00032 | 0.00107 | 0.00001 | 0.00042 | 65 | 0.00122 | 0.00002 | 0.00045 | 93 | 0.00062 | -0.24045 | 154.424 | 0.81030 | 0.95138 |
| fsaA, mipB; fructose-6-phosphate aldolase 1 [EC:4.1.2.-] | 0.00116 | 0.00017 | 0.00127 | 0.00001 | 0.00028 | 65 | 0.00108 | 0.00000 | 0.00020 | 93 | 0.00035 | 0.54633 | 123.188 | 0.58582 | 0.87720 |
| citB, tcuB; citrate/tricarballylate utilization protein | 0.00116 | 0.00029 | 0.00065 | 0.00000 | 0.00020 | 65 | 0.00152 | 0.00002 | 0.00047 | 93 | 0.00051 | -1.70357 | 122.654 | 0.09099 | 0.76567 |
| pcaL; 3-oxoadipate enol-lactonase / 4-carboxymuconolactone decarboxylase [EC:3.1.1.24 4.1.1.44] | 0.00116 | 0.00030 | 0.00052 | 0.00000 | 0.00024 | 65 | 0.00160 | 0.00002 | 0.00048 | 93 | 0.00054 | -2.00755 | 132.123 | 0.04673 | 0.66255 |
| pimB; phosphatidyl-myo-inositol dimannoside synthase [EC:2.4.1.346] | 0.00116 | 0.00023 | 0.00100 | 0.00000 | 0.00024 | 65 | 0.00126 | 0.00001 | 0.00036 | 93 | 0.00043 | -0.61835 | 149.735 | 0.53728 | 0.86207 |
| antA; anthranilate 1,2-dioxygenase (deaminating, decarboxylating) large subunit [EC:1.14.12.1] | 0.00115 | 0.00032 | 0.00074 | 0.00001 | 0.00033 | 65 | 0.00144 | 0.00002 | 0.00048 | 93 | 0.00059 | -1.19336 | 150.866 | 0.23460 | 0.77376 |
| argF; N-succinyl-L-ornithine transcarbamylase [EC:2.1.3.11] | 0.00115 | 0.00022 | 0.00077 | 0.00000 | 0.00020 | 65 | 0.00141 | 0.00001 | 0.00035 | 93 | 0.00040 | -1.56076 | 139.655 | 0.12084 | 0.77376 |
| antB; anthranilate 1,2-dioxygenase (deaminating, decarboxylating) small subunit [EC:1.14.12.1] | 0.00115 | 0.00032 | 0.00073 | 0.00001 | 0.00033 | 65 | 0.00144 | 0.00002 | 0.00048 | 93 | 0.00058 | -1.21774 | 150.682 | 0.22523 | 0.77376 |
| antC; anthranilate 1,2-dioxygenase reductase component [EC:1.18.1.-] | 0.00115 | 0.00032 | 0.00073 | 0.00001 | 0.00033 | 65 | 0.00144 | 0.00002 | 0.00048 | 93 | 0.00058 | -1.21774 | 150.682 | 0.22523 | 0.77376 |
| mabO; 4-methylaminobutanoate oxidase (formaldehyde-forming) [EC:1.5.3.19] | 0.00114 | 0.00032 | 0.00041 | 0.00001 | 0.00030 | 65 | 0.00166 | 0.00002 | 0.00050 | 93 | 0.00058 | -2.14017 | 142.786 | 0.03404 | 0.62130 |
| RTCA, rtcA; RNA 3’-terminal phosphate cyclase (ATP) [EC:6.5.1.4] | 0.00114 | 0.00024 | 0.00112 | 0.00001 | 0.00031 | 65 | 0.00115 | 0.00001 | 0.00034 | 93 | 0.00046 | -0.07494 | 155.029 | 0.94036 | 0.98528 |
| E1.13.11.4; gentisate 1,2-dioxygenase [EC:1.13.11.4] | 0.00114 | 0.00032 | 0.00045 | 0.00000 | 0.00012 | 65 | 0.00161 | 0.00003 | 0.00054 | 93 | 0.00055 | -2.09736 | 101.207 | 0.03845 | 0.63091 |
| K00185; prokaryotic molybdopterin-containing oxidoreductase family, membrane subunit | 0.00113 | 0.00024 | 0.00110 | 0.00001 | 0.00032 | 65 | 0.00115 | 0.00001 | 0.00035 | 93 | 0.00048 | -0.11125 | 154.388 | 0.91156 | 0.97664 |
| lysX1; putative lysine transport system substrate-binding protein | 0.00113 | 0.00036 | 0.00066 | 0.00001 | 0.00036 | 65 | 0.00146 | 0.00003 | 0.00055 | 93 | 0.00066 | -1.20411 | 149.093 | 0.23046 | 0.77376 |
| rzpD; prophage endopeptidase [EC:3.4.-.-] | 0.00113 | 0.00052 | 0.00100 | 0.00003 | 0.00067 | 65 | 0.00122 | 0.00005 | 0.00075 | 93 | 0.00100 | -0.21453 | 155.225 | 0.83042 | 0.95506 |
| ACMSD; aminocarboxymuconate-semialdehyde decarboxylase [EC:4.1.1.45] | 0.00113 | 0.00030 | 0.00046 | 0.00000 | 0.00016 | 65 | 0.00160 | 0.00002 | 0.00049 | 93 | 0.00051 | -2.21962 | 112.071 | 0.02846 | 0.61334 |
| sspF; small acid-soluble spore protein F (minor alpha/beta-type SASP) | 0.00113 | 0.00034 | 0.00044 | 0.00001 | 0.00030 | 65 | 0.00161 | 0.00003 | 0.00054 | 93 | 0.00062 | -1.88443 | 138.229 | 0.06161 | 0.73001 |
| yndD; spore germination protein | 0.00112 | 0.00035 | 0.00045 | 0.00001 | 0.00028 | 65 | 0.00160 | 0.00003 | 0.00056 | 93 | 0.00063 | -1.83752 | 132.742 | 0.06837 | 0.73473 |
| aacA; aminoglycoside 6’-N-acetyltransferase [EC:2.3.1.82] | 0.00112 | 0.00040 | 0.00044 | 0.00000 | 0.00013 | 65 | 0.00160 | 0.00004 | 0.00066 | 93 | 0.00068 | -1.70956 | 98.515 | 0.09049 | 0.76567 |
| alsE; D-allulose-6-phosphate 3-epimerase [EC:5.1.3.-] | 0.00112 | 0.00045 | 0.00127 | 0.00005 | 0.00086 | 65 | 0.00102 | 0.00002 | 0.00049 | 93 | 0.00099 | 0.25560 | 104.514 | 0.79876 | 0.94822 |
| PMA1, PMA2; H+-transporting ATPase [EC:3.6.3.6] | 0.00112 | 0.00047 | 0.00111 | 0.00004 | 0.00083 | 65 | 0.00113 | 0.00003 | 0.00056 | 93 | 0.00100 | -0.02498 | 118.086 | 0.98012 | 0.99657 |
| gspA; general secretion pathway protein A | 0.00112 | 0.00030 | 0.00087 | 0.00000 | 0.00027 | 65 | 0.00129 | 0.00002 | 0.00047 | 93 | 0.00055 | -0.77756 | 141.635 | 0.43813 | 0.83116 |
| araM, egsA; glycerol-1-phosphate dehydrogenase [NAD(P)+] [EC:1.1.1.261] | 0.00112 | 0.00018 | 0.00100 | 0.00000 | 0.00022 | 65 | 0.00120 | 0.00001 | 0.00027 | 93 | 0.00035 | -0.57442 | 155.964 | 0.56651 | 0.87051 |
| yfkN; 2’,3’-cyclic-nucleotide 2’-phosphodiesterase / 3’-nucleotidase / 5’-nucleotidase [EC:3.1.4.16 3.1.3.6 3.1.3.5] | 0.00111 | 0.00060 | 0.00065 | 0.00002 | 0.00058 | 65 | 0.00143 | 0.00008 | 0.00093 | 93 | 0.00110 | -0.70638 | 144.966 | 0.48109 | 0.84354 |
| rspA, manD; mannonate dehydratase [EC:4.2.1.8] | 0.00111 | 0.00019 | 0.00112 | 0.00001 | 0.00035 | 65 | 0.00110 | 0.00000 | 0.00021 | 93 | 0.00041 | 0.05078 | 107.637 | 0.95959 | 0.98903 |
| nadX, ASPDH; aspartate dehydrogenase [EC:1.4.1.21] | 0.00111 | 0.00021 | 0.00074 | 0.00000 | 0.00025 | 65 | 0.00136 | 0.00001 | 0.00031 | 93 | 0.00040 | -1.53030 | 155.773 | 0.12797 | 0.77376 |
| ygjK; putative isomerase | 0.00111 | 0.00035 | 0.00083 | 0.00001 | 0.00038 | 65 | 0.00130 | 0.00003 | 0.00053 | 93 | 0.00065 | -0.72818 | 153.286 | 0.46762 | 0.83837 |
| yxeO; putative amino-acid transport system ATP-binding protein [EC:3.6.3.-] | 0.00110 | 0.00025 | 0.00163 | 0.00002 | 0.00056 | 65 | 0.00074 | 0.00000 | 0.00016 | 93 | 0.00059 | 1.52214 | 74.184 | 0.13223 | 0.77376 |
| mtnX; 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase [EC:3.1.3.87] | 0.00110 | 0.00037 | 0.00038 | 0.00001 | 0.00030 | 65 | 0.00160 | 0.00003 | 0.00059 | 93 | 0.00066 | -1.84194 | 132.823 | 0.06771 | 0.73437 |
| tcuR; LysR family transcriptional regulator, regulatory protein for tcuABC | 0.00110 | 0.00033 | 0.00066 | 0.00000 | 0.00021 | 65 | 0.00140 | 0.00003 | 0.00054 | 93 | 0.00058 | -1.28290 | 117.002 | 0.20206 | 0.77376 |
| bisC; biotin/methionine sulfoxide reductase [EC:1.-.-.-] | 0.00110 | 0.00024 | 0.00068 | 0.00000 | 0.00022 | 65 | 0.00139 | 0.00001 | 0.00038 | 93 | 0.00044 | -1.59846 | 140.316 | 0.11219 | 0.77376 |
| glrR, qseF; two-component system, NtrC family, response regulator GlrR | 0.00110 | 0.00031 | 0.00064 | 0.00000 | 0.00025 | 65 | 0.00141 | 0.00002 | 0.00049 | 93 | 0.00055 | -1.39669 | 133.811 | 0.16482 | 0.77376 |
| mexH; membrane fusion protein, multidrug efflux system | 0.00109 | 0.00030 | 0.00091 | 0.00001 | 0.00041 | 65 | 0.00122 | 0.00002 | 0.00042 | 93 | 0.00058 | -0.52734 | 152.109 | 0.59873 | 0.88240 |
| napE; periplasmic nitrate reductase NapE | 0.00109 | 0.00024 | 0.00163 | 0.00002 | 0.00052 | 65 | 0.00072 | 0.00000 | 0.00019 | 93 | 0.00055 | 1.66627 | 81.894 | 0.09948 | 0.76942 |
| CYP125A; cholest-4-en-3-one 26-monooxygenase [EC:1.14.13.141] | 0.00109 | 0.00055 | 0.00022 | 0.00000 | 0.00012 | 65 | 0.00171 | 0.00008 | 0.00094 | 93 | 0.00094 | -1.58341 | 94.861 | 0.11666 | 0.77376 |
| MFS.SET; MFS transporter, SET family, sugar efflux transporter | 0.00109 | 0.00034 | 0.00087 | 0.00001 | 0.00039 | 65 | 0.00125 | 0.00002 | 0.00051 | 93 | 0.00065 | -0.58506 | 154.597 | 0.55936 | 0.86813 |
| E2.4.1.64; alpha,alpha-trehalose phosphorylase [EC:2.4.1.64] | 0.00109 | 0.00028 | 0.00139 | 0.00002 | 0.00053 | 65 | 0.00089 | 0.00001 | 0.00030 | 93 | 0.00061 | 0.81778 | 104.287 | 0.41535 | 0.81730 |
| fldA; cinnamoyl-CoA:phenyllactate CoA-transferase [EC:2.8.3.17] | 0.00109 | 0.00041 | 0.00168 | 0.00005 | 0.00091 | 65 | 0.00068 | 0.00001 | 0.00027 | 93 | 0.00095 | 1.05024 | 75.016 | 0.29698 | 0.77376 |
| kojP; kojibiose phosphorylase [EC:2.4.1.230] | 0.00109 | 0.00041 | 0.00168 | 0.00006 | 0.00092 | 65 | 0.00068 | 0.00001 | 0.00027 | 93 | 0.00096 | 1.03875 | 74.755 | 0.30227 | 0.77376 |
| meh; 3-methylfumaryl-CoA hydratase [EC:4.2.1.153] | 0.00109 | 0.00027 | 0.00050 | 0.00000 | 0.00015 | 65 | 0.00150 | 0.00002 | 0.00045 | 93 | 0.00047 | -2.11653 | 111.001 | 0.03653 | 0.62546 |
| dctS; two-component system, LuxR family, sensor histidine kinase DctS [EC:2.7.13.3] | 0.00109 | 0.00030 | 0.00074 | 0.00000 | 0.00026 | 65 | 0.00133 | 0.00002 | 0.00047 | 93 | 0.00054 | -1.09573 | 138.817 | 0.27510 | 0.77376 |
| pbpA; penicillin-binding protein 1 [EC:3.4.-.-] | 0.00108 | 0.00041 | 0.00094 | 0.00002 | 0.00054 | 65 | 0.00119 | 0.00003 | 0.00059 | 93 | 0.00080 | -0.31448 | 154.593 | 0.75358 | 0.93643 |
| hsaA; 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione monooxygenase [EC:1.14.14.12] | 0.00108 | 0.00039 | 0.00053 | 0.00000 | 0.00018 | 65 | 0.00147 | 0.00004 | 0.00065 | 93 | 0.00067 | -1.39854 | 105.718 | 0.16488 | 0.77376 |
| hcnA; hydrogen cyanide synthase HcnA [EC:1.4.99.5] | 0.00108 | 0.00030 | 0.00101 | 0.00001 | 0.00044 | 65 | 0.00114 | 0.00002 | 0.00041 | 93 | 0.00061 | -0.21309 | 146.241 | 0.83155 | 0.95506 |
| hcnB; hydrogen cyanide synthase HcnB [EC:1.4.99.5] | 0.00108 | 0.00030 | 0.00101 | 0.00001 | 0.00044 | 65 | 0.00114 | 0.00002 | 0.00041 | 93 | 0.00061 | -0.21309 | 146.241 | 0.83155 | 0.95506 |
| legF, ptmB; CMP-N,N’-diacetyllegionaminic acid synthase [EC:2.7.7.82] | 0.00108 | 0.00033 | 0.00082 | 0.00001 | 0.00036 | 65 | 0.00126 | 0.00002 | 0.00051 | 93 | 0.00062 | -0.71299 | 151.771 | 0.47695 | 0.84209 |
| dinD; DNA-damage-inducible protein D | 0.00108 | 0.00020 | 0.00078 | 0.00000 | 0.00020 | 65 | 0.00129 | 0.00001 | 0.00031 | 93 | 0.00037 | -1.37890 | 148.508 | 0.17000 | 0.77376 |
| pspF; psp operon transcriptional activator | 0.00108 | 0.00020 | 0.00101 | 0.00001 | 0.00029 | 65 | 0.00113 | 0.00001 | 0.00028 | 93 | 0.00041 | -0.31596 | 150.179 | 0.75247 | 0.93565 |
| hpxO; FAD-dependent urate hydroxylase [EC:1.14.13.113] | 0.00108 | 0.00026 | 0.00069 | 0.00000 | 0.00022 | 65 | 0.00135 | 0.00002 | 0.00042 | 93 | 0.00047 | -1.39579 | 135.149 | 0.16507 | 0.77376 |
| terZ; tellurium resistance protein TerZ | 0.00108 | 0.00024 | 0.00098 | 0.00001 | 0.00030 | 65 | 0.00114 | 0.00001 | 0.00035 | 93 | 0.00046 | -0.34231 | 155.971 | 0.73258 | 0.92601 |
| dmsC; anaerobic dimethyl sulfoxide reductase subunit C (DMSO reductase anchor subunit) | 0.00108 | 0.00022 | 0.00098 | 0.00000 | 0.00025 | 65 | 0.00115 | 0.00001 | 0.00034 | 93 | 0.00042 | -0.40842 | 153.818 | 0.68353 | 0.91514 |
| lasR; LuxR family transcriptional regulator, quorum-sensing system regulator LasR | 0.00108 | 0.00031 | 0.00086 | 0.00001 | 0.00036 | 65 | 0.00122 | 0.00002 | 0.00046 | 93 | 0.00058 | -0.61762 | 155.284 | 0.53773 | 0.86207 |
| iolH; myo-inositol catabolism protein IolH | 0.00108 | 0.00025 | 0.00100 | 0.00001 | 0.00030 | 65 | 0.00113 | 0.00001 | 0.00037 | 93 | 0.00047 | -0.27635 | 155.943 | 0.78265 | 0.94310 |
| rtcB; release factor H-coupled RctB family protein | 0.00108 | 0.00023 | 0.00099 | 0.00001 | 0.00032 | 65 | 0.00113 | 0.00001 | 0.00031 | 93 | 0.00045 | -0.31185 | 148.911 | 0.75559 | 0.93665 |
| dbh; DNA polymerase IV (archaeal DinB-like DNA polymerase) [EC:2.7.7.7] | 0.00107 | 0.00041 | 0.00167 | 0.00005 | 0.00092 | 65 | 0.00066 | 0.00001 | 0.00027 | 93 | 0.00096 | 1.06007 | 74.746 | 0.29253 | 0.77376 |
| E3.3.2.8; limonene-1,2-epoxide hydrolase [EC:3.3.2.8] | 0.00107 | 0.00072 | 0.00021 | 0.00000 | 0.00014 | 65 | 0.00167 | 0.00014 | 0.00122 | 93 | 0.00123 | -1.19181 | 94.591 | 0.23632 | 0.77376 |
| bpsA, srsA, pks11, pks10; alkylresorcinol/alkylpyrone synthase | 0.00107 | 0.00034 | 0.00047 | 0.00001 | 0.00030 | 65 | 0.00149 | 0.00003 | 0.00053 | 93 | 0.00061 | -1.67276 | 140.413 | 0.09660 | 0.76667 |
| iolT; MFS transporter, SP family, major inositol transporter | 0.00106 | 0.00018 | 0.00124 | 0.00001 | 0.00028 | 65 | 0.00094 | 0.00000 | 0.00023 | 93 | 0.00036 | 0.81658 | 134.029 | 0.41562 | 0.81731 |
| lmrA, yxaF; TetR/AcrR family transcriptional regulator, lmrAB and yxaGH operons repressor | 0.00106 | 0.00047 | 0.00056 | 0.00001 | 0.00030 | 65 | 0.00141 | 0.00005 | 0.00077 | 93 | 0.00082 | -1.03373 | 117.810 | 0.30338 | 0.77376 |
| tmm; trimethylamine monooxygenase [EC:1.14.13.148] | 0.00106 | 0.00030 | 0.00065 | 0.00001 | 0.00033 | 65 | 0.00134 | 0.00002 | 0.00045 | 93 | 0.00056 | -1.23257 | 153.358 | 0.21962 | 0.77376 |
| babA; outer membrane protein BabA | 0.00106 | 0.00068 | 0.00038 | 0.00000 | 0.00024 | 65 | 0.00153 | 0.00012 | 0.00114 | 93 | 0.00117 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cgtA; beta-1,4-N-acetylgalactosaminyltransferase [EC:2.4.1.-] | 0.00106 | 0.00068 | 0.00038 | 0.00000 | 0.00024 | 65 | 0.00153 | 0.00012 | 0.00114 | 93 | 0.00117 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| comB10; ComB10 competence protein | 0.00106 | 0.00068 | 0.00038 | 0.00000 | 0.00024 | 65 | 0.00153 | 0.00012 | 0.00114 | 93 | 0.00117 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| sabA; outer membrane protein SabA | 0.00106 | 0.00068 | 0.00038 | 0.00000 | 0.00024 | 65 | 0.00153 | 0.00012 | 0.00114 | 93 | 0.00117 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| TROVE2, SSA2; 60 kDa SS-A/Ro ribonucleoprotein | 0.00106 | 0.00025 | 0.00108 | 0.00001 | 0.00033 | 65 | 0.00103 | 0.00001 | 0.00036 | 93 | 0.00049 | 0.10303 | 154.330 | 0.91807 | 0.97744 |
| hndC; NADP-reducing hydrogenase subunit HndC [EC:1.12.1.3] | 0.00105 | 0.00025 | 0.00085 | 0.00000 | 0.00027 | 65 | 0.00119 | 0.00001 | 0.00038 | 93 | 0.00046 | -0.73508 | 152.201 | 0.46342 | 0.83677 |
| K07051; uncharacterized protein | 0.00105 | 0.00030 | 0.00079 | 0.00001 | 0.00040 | 65 | 0.00124 | 0.00002 | 0.00043 | 93 | 0.00059 | -0.77568 | 153.892 | 0.43913 | 0.83156 |
| laaA; L-proline amide hydrolase [EC:3.5.1.101] | 0.00105 | 0.00031 | 0.00074 | 0.00001 | 0.00036 | 65 | 0.00126 | 0.00002 | 0.00047 | 93 | 0.00059 | -0.88547 | 155.115 | 0.37727 | 0.79815 |
| femA; peptidoglycan pentaglycine glycine transferase (the second and third glycine) [EC:2.3.2.17] | 0.00105 | 0.00041 | 0.00091 | 0.00002 | 0.00053 | 65 | 0.00114 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.28320 | 154.893 | 0.77740 | 0.94269 |
| femB; peptidoglycan pentaglycine glycine transferase (the fourth and fifth glycine) [EC:2.3.2.18] | 0.00105 | 0.00041 | 0.00091 | 0.00002 | 0.00053 | 65 | 0.00114 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.28320 | 154.893 | 0.77740 | 0.94269 |
| soxA; sulfur-oxidizing protein SoxA | 0.00105 | 0.00029 | 0.00040 | 0.00000 | 0.00015 | 65 | 0.00150 | 0.00002 | 0.00047 | 93 | 0.00050 | -2.23095 | 109.019 | 0.02773 | 0.61334 |
| gsiD; glutathione transport system permease protein | 0.00105 | 0.00033 | 0.00074 | 0.00001 | 0.00033 | 65 | 0.00126 | 0.00002 | 0.00051 | 93 | 0.00061 | -0.84952 | 148.441 | 0.39696 | 0.80459 |
| norE; nitric oxide reductase NorE protein | 0.00104 | 0.00028 | 0.00039 | 0.00000 | 0.00012 | 65 | 0.00150 | 0.00002 | 0.00046 | 93 | 0.00048 | -2.32001 | 105.023 | 0.02227 | 0.59764 |
| ectC; L-ectoine synthase [EC:4.2.1.108] | 0.00104 | 0.00023 | 0.00066 | 0.00000 | 0.00021 | 65 | 0.00131 | 0.00001 | 0.00037 | 93 | 0.00042 | -1.54136 | 138.924 | 0.12550 | 0.77376 |
| grdB; glycine reductase complex component B subunit gamma [EC:1.21.4.2] | 0.00104 | 0.00023 | 0.00113 | 0.00001 | 0.00034 | 65 | 0.00098 | 0.00001 | 0.00031 | 93 | 0.00046 | 0.31073 | 145.217 | 0.75646 | 0.93680 |
| pbp3; penicillin-binding protein 3 [EC:3.4.-.-] | 0.00104 | 0.00041 | 0.00090 | 0.00002 | 0.00053 | 65 | 0.00114 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.29887 | 154.886 | 0.76544 | 0.93900 |
| norC; nitric oxide reductase subunit C | 0.00103 | 0.00028 | 0.00041 | 0.00000 | 0.00013 | 65 | 0.00147 | 0.00002 | 0.00047 | 93 | 0.00049 | -2.16006 | 106.504 | 0.03301 | 0.62130 |
| K06882; uncharacterized protein | 0.00103 | 0.00037 | 0.00084 | 0.00001 | 0.00030 | 65 | 0.00117 | 0.00003 | 0.00059 | 93 | 0.00066 | -0.48808 | 132.177 | 0.62630 | 0.89255 |
| hndA; NADP-reducing hydrogenase subunit HndA [EC:1.12.1.3] | 0.00103 | 0.00027 | 0.00075 | 0.00000 | 0.00023 | 65 | 0.00123 | 0.00002 | 0.00043 | 93 | 0.00049 | -0.98844 | 135.832 | 0.32469 | 0.77376 |
| tupC, vupC; tungstate transport system ATP-binding protein [EC:3.6.3.55] | 0.00103 | 0.00027 | 0.00101 | 0.00001 | 0.00031 | 65 | 0.00104 | 0.00002 | 0.00040 | 93 | 0.00051 | -0.05225 | 154.936 | 0.95839 | 0.98903 |
| norD; nitric oxide reductase NorD protein | 0.00103 | 0.00028 | 0.00040 | 0.00000 | 0.00013 | 65 | 0.00147 | 0.00002 | 0.00047 | 93 | 0.00049 | -2.19409 | 105.210 | 0.03043 | 0.61440 |
| MLYCD; malonyl-CoA decarboxylase [EC:4.1.1.9] | 0.00103 | 0.00029 | 0.00043 | 0.00000 | 0.00012 | 65 | 0.00145 | 0.00002 | 0.00048 | 93 | 0.00049 | -2.06511 | 104.350 | 0.04139 | 0.64661 |
| dmsA; anaerobic dimethyl sulfoxide reductase subunit A [EC:1.8.5.3] | 0.00103 | 0.00036 | 0.00054 | 0.00000 | 0.00016 | 65 | 0.00137 | 0.00003 | 0.00060 | 93 | 0.00062 | -1.34383 | 104.451 | 0.18191 | 0.77376 |
| PRK, prkB; phosphoribulokinase [EC:2.7.1.19] | 0.00103 | 0.00021 | 0.00089 | 0.00001 | 0.00029 | 65 | 0.00112 | 0.00001 | 0.00030 | 93 | 0.00042 | -0.53953 | 152.984 | 0.59030 | 0.87896 |
| gsiC; glutathione transport system permease protein | 0.00103 | 0.00032 | 0.00079 | 0.00001 | 0.00034 | 65 | 0.00119 | 0.00002 | 0.00049 | 93 | 0.00060 | -0.66936 | 150.391 | 0.50429 | 0.84956 |
| vanSC, vanSE, vanSG; two-component system, OmpR family, sensor histidine kinase VanS [EC:2.7.13.3] | 0.00102 | 0.00040 | 0.00052 | 0.00001 | 0.00029 | 65 | 0.00138 | 0.00004 | 0.00065 | 93 | 0.00071 | -1.20498 | 124.801 | 0.23049 | 0.77376 |
| podJ; localization factor PodJL | 0.00102 | 0.00025 | 0.00038 | 0.00000 | 0.00010 | 65 | 0.00147 | 0.00002 | 0.00041 | 93 | 0.00042 | -2.60005 | 103.801 | 0.01068 | 0.58284 |
| cdr; CoA-disulfide reductase [EC:1.8.1.14] | 0.00101 | 0.00041 | 0.00090 | 0.00002 | 0.00053 | 65 | 0.00110 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.24774 | 154.891 | 0.80467 | 0.94931 |
| K11442; putative uridylyltransferase [EC:2.7.7.-] | 0.00101 | 0.00041 | 0.00090 | 0.00002 | 0.00053 | 65 | 0.00110 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.24774 | 154.891 | 0.80467 | 0.94931 |
| K12055, parA; chromosome partitioning related protein ParA | 0.00101 | 0.00030 | 0.00099 | 0.00001 | 0.00040 | 65 | 0.00103 | 0.00002 | 0.00044 | 93 | 0.00059 | -0.07685 | 154.394 | 0.93884 | 0.98500 |
| hpxB; allantoinase [EC:3.5.2.5] | 0.00101 | 0.00027 | 0.00070 | 0.00000 | 0.00024 | 65 | 0.00123 | 0.00002 | 0.00043 | 93 | 0.00049 | -1.06643 | 137.225 | 0.28810 | 0.77376 |
| modD; molybdenum transport protein [EC:2.4.2.-] | 0.00101 | 0.00025 | 0.00112 | 0.00001 | 0.00040 | 65 | 0.00094 | 0.00001 | 0.00031 | 93 | 0.00051 | 0.34785 | 130.886 | 0.72851 | 0.92350 |
| K05937; uncharacterized protein | 0.00101 | 0.00041 | 0.00096 | 0.00002 | 0.00053 | 65 | 0.00105 | 0.00003 | 0.00059 | 93 | 0.00079 | -0.11520 | 154.939 | 0.90843 | 0.97664 |
| K01138; uncharacterized sulfatase [EC:3.1.6.-] | 0.00101 | 0.00026 | 0.00122 | 0.00001 | 0.00048 | 65 | 0.00087 | 0.00001 | 0.00030 | 93 | 0.00056 | 0.61247 | 114.017 | 0.54145 | 0.86207 |
| wcaI; colanic acid biosynthesis glycosyl transferase WcaI | 0.00101 | 0.00030 | 0.00053 | 0.00001 | 0.00030 | 65 | 0.00135 | 0.00002 | 0.00046 | 93 | 0.00055 | -1.48016 | 148.809 | 0.14094 | 0.77376 |
| kipI; inhibitor of KinA | 0.00101 | 0.00026 | 0.00015 | 0.00000 | 0.00005 | 65 | 0.00161 | 0.00002 | 0.00044 | 93 | 0.00044 | -3.32681 | 93.950 | 0.00125 | 0.46257 |
| adc; acetoacetate decarboxylase [EC:4.1.1.4] | 0.00101 | 0.00033 | 0.00039 | 0.00000 | 0.00011 | 65 | 0.00144 | 0.00003 | 0.00055 | 93 | 0.00056 | -1.86041 | 99.025 | 0.06580 | 0.73334 |
| PTS-Nag-EIIA, nagE; PTS system, N-acetylglucosamine-specific IIA component [EC:2.7.1.193] | 0.00101 | 0.00017 | 0.00108 | 0.00000 | 0.00025 | 65 | 0.00096 | 0.00000 | 0.00023 | 93 | 0.00034 | 0.35760 | 144.164 | 0.72117 | 0.92156 |
| tesI; 3-oxo-5alpha-steroid 4-dehydrogenase [EC:1.3.99.5] | 0.00101 | 0.00055 | 0.00045 | 0.00000 | 0.00018 | 65 | 0.00140 | 0.00008 | 0.00092 | 93 | 0.00094 | -1.01181 | 98.852 | 0.31410 | 0.77376 |
| K07454; putative restriction endonuclease | 0.00100 | 0.00027 | 0.00109 | 0.00001 | 0.00040 | 65 | 0.00095 | 0.00001 | 0.00036 | 93 | 0.00054 | 0.26745 | 143.613 | 0.78951 | 0.94606 |
| mdlC; benzoylformate decarboxylase [EC:4.1.1.7] | 0.00100 | 0.00033 | 0.00051 | 0.00000 | 0.00016 | 65 | 0.00135 | 0.00003 | 0.00055 | 93 | 0.00057 | -1.46931 | 106.656 | 0.14469 | 0.77376 |
| zot; zona occludens toxin | 0.00100 | 0.00026 | 0.00077 | 0.00000 | 0.00027 | 65 | 0.00117 | 0.00001 | 0.00039 | 93 | 0.00048 | -0.84693 | 151.107 | 0.39837 | 0.80602 |
| soxX; sulfur-oxidizing protein SoxX | 0.00100 | 0.00028 | 0.00039 | 0.00000 | 0.00015 | 65 | 0.00143 | 0.00002 | 0.00046 | 93 | 0.00048 | -2.13827 | 109.769 | 0.03471 | 0.62130 |
| FAB2, SSI2, desA1; acyl-[acyl-carrier-protein] desaturase [EC:1.14.19.2 1.14.19.11 1.14.19.26] | 0.00100 | 0.00027 | 0.00028 | 0.00000 | 0.00011 | 65 | 0.00150 | 0.00002 | 0.00045 | 93 | 0.00046 | -2.64797 | 102.380 | 0.00938 | 0.58112 |
| E1.11.1.19; dye decolorizing peroxidase [EC:1.11.1.19] | 0.00100 | 0.00025 | 0.00060 | 0.00000 | 0.00022 | 65 | 0.00128 | 0.00001 | 0.00039 | 93 | 0.00045 | -1.52257 | 137.489 | 0.13016 | 0.77376 |
| pdaD; arginine decarboxylase [EC:4.1.1.19] | 0.00100 | 0.00029 | 0.00085 | 0.00001 | 0.00034 | 65 | 0.00111 | 0.00002 | 0.00043 | 93 | 0.00055 | -0.47264 | 155.801 | 0.63713 | 0.89703 |
| ylnA, cysP; sulfate permease | 0.00100 | 0.00034 | 0.00052 | 0.00001 | 0.00028 | 65 | 0.00133 | 0.00003 | 0.00054 | 93 | 0.00061 | -1.34999 | 134.214 | 0.17929 | 0.77376 |
| K09161; uncharacterized protein | 0.00100 | 0.00023 | 0.00122 | 0.00001 | 0.00040 | 65 | 0.00085 | 0.00001 | 0.00027 | 93 | 0.00048 | 0.76950 | 117.497 | 0.44314 | 0.83186 |
| sucD; succinate-semialdehyde dehydrogenase [EC:1.2.1.76] | 0.00099 | 0.00029 | 0.00083 | 0.00001 | 0.00034 | 65 | 0.00111 | 0.00002 | 0.00043 | 93 | 0.00055 | -0.50909 | 155.808 | 0.61141 | 0.88722 |
| yhcO; ribonuclease inhibitor | 0.00099 | 0.00019 | 0.00100 | 0.00000 | 0.00027 | 65 | 0.00099 | 0.00001 | 0.00026 | 93 | 0.00038 | 0.03278 | 150.469 | 0.97389 | 0.99535 |
| COQ9; ubiquinone biosynthesis protein COQ9 | 0.00098 | 0.00022 | 0.00054 | 0.00000 | 0.00018 | 65 | 0.00129 | 0.00001 | 0.00035 | 93 | 0.00040 | -1.87528 | 133.633 | 0.06294 | 0.73012 |
| rhaT; rhamnose transport system ATP-binding protein [EC:3.6.3.17] | 0.00098 | 0.00024 | 0.00046 | 0.00000 | 0.00021 | 65 | 0.00135 | 0.00001 | 0.00038 | 93 | 0.00044 | -2.02285 | 137.877 | 0.04502 | 0.65981 |
| uspG; universal stress protein G | 0.00098 | 0.00029 | 0.00107 | 0.00001 | 0.00046 | 65 | 0.00092 | 0.00001 | 0.00038 | 93 | 0.00059 | 0.26353 | 137.842 | 0.79254 | 0.94770 |
| nanM; N-acetylneuraminate epimerase [EC:5.1.3.24] | 0.00098 | 0.00027 | 0.00082 | 0.00001 | 0.00033 | 65 | 0.00110 | 0.00001 | 0.00039 | 93 | 0.00051 | -0.54948 | 155.929 | 0.58346 | 0.87720 |
| tri; tricorn protease [EC:3.4.21.-] | 0.00098 | 0.00025 | 0.00059 | 0.00001 | 0.00030 | 65 | 0.00125 | 0.00001 | 0.00037 | 93 | 0.00048 | -1.39541 | 155.548 | 0.16488 | 0.77376 |
| AGPS, agpS; alkyldihydroxyacetonephosphate synthase [EC:2.5.1.26] | 0.00098 | 0.00028 | 0.00049 | 0.00000 | 0.00018 | 65 | 0.00132 | 0.00002 | 0.00046 | 93 | 0.00049 | -1.69199 | 119.769 | 0.09325 | 0.76567 |
| mprB; two-component system, OmpR family, sensor histidine kinase MprB [EC:2.7.13.3] | 0.00098 | 0.00038 | 0.00062 | 0.00000 | 0.00022 | 65 | 0.00123 | 0.00004 | 0.00062 | 93 | 0.00066 | -0.91827 | 113.761 | 0.36042 | 0.79496 |
| splA; transcriptional regulator of the spore photoproduct lyase operon | 0.00098 | 0.00033 | 0.00038 | 0.00000 | 0.00028 | 65 | 0.00140 | 0.00003 | 0.00052 | 93 | 0.00059 | -1.71679 | 135.346 | 0.08831 | 0.76567 |
| yafP; putative acetyltransferase [EC:2.3.1.-] | 0.00098 | 0.00018 | 0.00059 | 0.00000 | 0.00018 | 65 | 0.00125 | 0.00001 | 0.00028 | 93 | 0.00033 | -1.96227 | 145.877 | 0.05163 | 0.68802 |
| bceS; two-component system, OmpR family, bacitracin resistance sensor histidine kinase BceS [EC:2.7.13.3] | 0.00097 | 0.00034 | 0.00048 | 0.00001 | 0.00031 | 65 | 0.00132 | 0.00003 | 0.00053 | 93 | 0.00062 | -1.34465 | 141.611 | 0.18089 | 0.77376 |
| K13819; NifU-like protein | 0.00097 | 0.00036 | 0.00073 | 0.00001 | 0.00028 | 65 | 0.00114 | 0.00003 | 0.00058 | 93 | 0.00065 | -0.63520 | 129.146 | 0.52642 | 0.85844 |
| hxlA; 3-hexulose-6-phosphate synthase [EC:4.1.2.43] | 0.00097 | 0.00025 | 0.00032 | 0.00000 | 0.00014 | 65 | 0.00143 | 0.00002 | 0.00041 | 93 | 0.00043 | -2.56391 | 112.844 | 0.01166 | 0.59095 |
| ligB; protocatechuate 4,5-dioxygenase, beta chain [EC:1.13.11.8] | 0.00097 | 0.00040 | 0.00023 | 0.00000 | 0.00009 | 65 | 0.00149 | 0.00004 | 0.00067 | 93 | 0.00068 | -1.85611 | 95.569 | 0.06652 | 0.73334 |
| yqgE; MFS transporter, YQGE family, putative transporter | 0.00097 | 0.00025 | 0.00013 | 0.00000 | 0.00004 | 65 | 0.00155 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.37981 | 94.023 | 0.00106 | 0.46257 |
| mrx1; mycoredoxin [EC:1.20.4.3] | 0.00097 | 0.00024 | 0.00070 | 0.00000 | 0.00025 | 65 | 0.00116 | 0.00001 | 0.00037 | 93 | 0.00044 | -1.04293 | 149.664 | 0.29866 | 0.77376 |
| vat; virginiamycin A acetyltransferase [EC:2.3.1.-] | 0.00096 | 0.00027 | 0.00030 | 0.00000 | 0.00008 | 65 | 0.00143 | 0.00002 | 0.00046 | 93 | 0.00046 | -2.44789 | 97.066 | 0.01617 | 0.59095 |
| irr; Fur family transcriptional regulator, iron response regulator | 0.00096 | 0.00024 | 0.00032 | 0.00000 | 0.00009 | 65 | 0.00142 | 0.00002 | 0.00041 | 93 | 0.00042 | -2.64682 | 100.114 | 0.00944 | 0.58112 |
| mgrA; MarR family transcriptional regulator, multiple gene regulator MgrA | 0.00096 | 0.00038 | 0.00087 | 0.00002 | 0.00053 | 65 | 0.00103 | 0.00003 | 0.00053 | 93 | 0.00075 | -0.21562 | 151.581 | 0.82957 | 0.95506 |
| lysDH; lysine 6-dehydrogenase [EC:1.4.1.18] | 0.00096 | 0.00036 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00139 | 0.00003 | 0.00057 | 93 | 0.00064 | -1.61605 | 135.344 | 0.10841 | 0.77376 |
| tlp; small acid-soluble spore protein (thioredoxin-like protein) | 0.00096 | 0.00026 | 0.00012 | 0.00000 | 0.00004 | 65 | 0.00154 | 0.00002 | 0.00044 | 93 | 0.00044 | -3.22471 | 93.885 | 0.00174 | 0.46257 |
| gspB; general secretion pathway protein B | 0.00096 | 0.00029 | 0.00072 | 0.00000 | 0.00027 | 65 | 0.00113 | 0.00002 | 0.00046 | 93 | 0.00053 | -0.75731 | 141.145 | 0.45013 | 0.83438 |
| mdtN; membrane fusion protein, multidrug efflux system | 0.00096 | 0.00029 | 0.00083 | 0.00001 | 0.00040 | 65 | 0.00104 | 0.00001 | 0.00040 | 93 | 0.00057 | -0.37190 | 150.993 | 0.71049 | 0.92004 |
| mdtO; multidrug resistance protein MdtO | 0.00096 | 0.00029 | 0.00083 | 0.00001 | 0.00040 | 65 | 0.00104 | 0.00001 | 0.00040 | 93 | 0.00057 | -0.37190 | 150.993 | 0.71049 | 0.92004 |
| wcaH; colanic acid biosynthesis protein WcaH [EC:3.6.1.-] | 0.00096 | 0.00030 | 0.00056 | 0.00001 | 0.00031 | 65 | 0.00123 | 0.00002 | 0.00046 | 93 | 0.00056 | -1.21737 | 148.803 | 0.22539 | 0.77376 |
| K08961; chondroitin-sulfate-ABC endolyase/exolyase [EC:4.2.2.20 4.2.2.21] | 0.00095 | 0.00024 | 0.00061 | 0.00000 | 0.00016 | 65 | 0.00119 | 0.00001 | 0.00038 | 93 | 0.00041 | -1.40285 | 119.871 | 0.16325 | 0.77376 |
| mucK; MFS transporter, AAHS family, cis,cis-muconate transporter | 0.00095 | 0.00048 | 0.00061 | 0.00000 | 0.00025 | 65 | 0.00119 | 0.00006 | 0.00080 | 93 | 0.00084 | -0.70238 | 109.213 | 0.48394 | 0.84354 |
| fadK; acyl-CoA synthetase [EC:6.2.1.-] | 0.00095 | 0.00029 | 0.00080 | 0.00001 | 0.00040 | 65 | 0.00105 | 0.00001 | 0.00040 | 93 | 0.00056 | -0.44498 | 150.825 | 0.65697 | 0.90692 |
| E5.3.3.1; steroid Delta-isomerase [EC:5.3.3.1] | 0.00095 | 0.00054 | 0.00029 | 0.00000 | 0.00013 | 65 | 0.00141 | 0.00008 | 0.00092 | 93 | 0.00093 | -1.21000 | 95.641 | 0.22926 | 0.77376 |
| spsF; spore coat polysaccharide biosynthesis protein SpsF | 0.00095 | 0.00031 | 0.00077 | 0.00001 | 0.00030 | 65 | 0.00107 | 0.00002 | 0.00048 | 93 | 0.00056 | -0.53236 | 147.077 | 0.59528 | 0.88005 |
| hrtB; hemin transport system permease protein | 0.00095 | 0.00038 | 0.00086 | 0.00002 | 0.00053 | 65 | 0.00101 | 0.00003 | 0.00053 | 93 | 0.00075 | -0.19902 | 151.590 | 0.84252 | 0.95506 |
| sdrM; MFS transporter, DHA2 family, multidrug resistance protein | 0.00095 | 0.00038 | 0.00086 | 0.00002 | 0.00053 | 65 | 0.00101 | 0.00003 | 0.00053 | 93 | 0.00075 | -0.19902 | 151.590 | 0.84252 | 0.95506 |
| sgtA; monofunctional glycosyltransferase [EC:2.4.1.129] | 0.00095 | 0.00038 | 0.00086 | 0.00002 | 0.00053 | 65 | 0.00101 | 0.00003 | 0.00053 | 93 | 0.00075 | -0.19902 | 151.590 | 0.84252 | 0.95506 |
| sgtB; monofunctional glycosyltransferase [EC:2.4.1.129] | 0.00095 | 0.00038 | 0.00086 | 0.00002 | 0.00053 | 65 | 0.00101 | 0.00003 | 0.00053 | 93 | 0.00075 | -0.19902 | 151.590 | 0.84252 | 0.95506 |
| ptpA; Xaa-Xaa-Pro tripeptidyl-peptidase [EC:3.4.14.12] | 0.00095 | 0.00029 | 0.00083 | 0.00001 | 0.00034 | 65 | 0.00103 | 0.00002 | 0.00043 | 93 | 0.00055 | -0.37545 | 155.836 | 0.70783 | 0.92004 |
| K09137; uncharacterized protein | 0.00095 | 0.00025 | 0.00106 | 0.00001 | 0.00044 | 65 | 0.00087 | 0.00001 | 0.00029 | 93 | 0.00053 | 0.35396 | 117.343 | 0.72400 | 0.92170 |
| ectA; L-2,4-diaminobutyric acid acetyltransferase [EC:2.3.1.178] | 0.00095 | 0.00023 | 0.00057 | 0.00000 | 0.00019 | 65 | 0.00121 | 0.00001 | 0.00036 | 93 | 0.00041 | -1.57231 | 136.270 | 0.11820 | 0.77376 |
| hydA; quinone-reactive Ni/Fe-hydrogenase small subunit [EC:1.12.5.1] | 0.00095 | 0.00036 | 0.00075 | 0.00001 | 0.00028 | 65 | 0.00108 | 0.00003 | 0.00058 | 93 | 0.00065 | -0.51141 | 129.303 | 0.60994 | 0.88693 |
| hydB; quinone-reactive Ni/Fe-hydrogenase large subunit [EC:1.12.5.1] | 0.00095 | 0.00036 | 0.00075 | 0.00001 | 0.00028 | 65 | 0.00108 | 0.00003 | 0.00058 | 93 | 0.00065 | -0.51141 | 129.303 | 0.60994 | 0.88693 |
| bcrB; bacitracin transport system permease protein | 0.00094 | 0.00033 | 0.00027 | 0.00000 | 0.00018 | 65 | 0.00141 | 0.00003 | 0.00054 | 93 | 0.00057 | -2.01119 | 111.351 | 0.04672 | 0.66255 |
| vapC; ribonuclease VapC [EC:3.1.-.-] | 0.00094 | 0.00024 | 0.00067 | 0.00001 | 0.00035 | 65 | 0.00113 | 0.00001 | 0.00032 | 93 | 0.00047 | -0.98141 | 143.867 | 0.32804 | 0.77376 |
| gsiB; glutathione transport system substrate-binding protein | 0.00094 | 0.00032 | 0.00075 | 0.00001 | 0.00033 | 65 | 0.00108 | 0.00002 | 0.00049 | 93 | 0.00059 | -0.54470 | 150.190 | 0.58677 | 0.87731 |
| oxdA; aldoxime dehydratase [EC:4.99.1.5] | 0.00094 | 0.00033 | 0.00042 | 0.00001 | 0.00030 | 65 | 0.00130 | 0.00003 | 0.00052 | 93 | 0.00060 | -1.45585 | 140.887 | 0.14766 | 0.77376 |
| comA; two-component system, NarL family, competent response regulator ComA | 0.00094 | 0.00059 | 0.00067 | 0.00002 | 0.00056 | 65 | 0.00112 | 0.00008 | 0.00092 | 93 | 0.00107 | -0.42651 | 144.210 | 0.67037 | 0.91184 |
| bcrA; bacitracin transport system ATP-binding protein | 0.00093 | 0.00032 | 0.00038 | 0.00000 | 0.00018 | 65 | 0.00132 | 0.00003 | 0.00052 | 93 | 0.00055 | -1.70539 | 113.272 | 0.09086 | 0.76567 |
| cbrT; energy-coupling factor transport system substrate-specific component | 0.00093 | 0.00020 | 0.00051 | 0.00000 | 0.00012 | 65 | 0.00123 | 0.00001 | 0.00034 | 93 | 0.00036 | -2.00828 | 114.213 | 0.04697 | 0.66255 |
| K09804; uncharacterized protein | 0.00093 | 0.00036 | 0.00072 | 0.00001 | 0.00028 | 65 | 0.00108 | 0.00003 | 0.00058 | 93 | 0.00065 | -0.56467 | 129.107 | 0.57328 | 0.87292 |
| spoVK; stage V sporulation protein K | 0.00093 | 0.00025 | 0.00017 | 0.00000 | 0.00007 | 65 | 0.00147 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.06821 | 97.252 | 0.00279 | 0.46257 |
| nicC; 6-hydroxynicotinate 3-monooxygenase [EC:1.14.13.114] | 0.00093 | 0.00030 | 0.00046 | 0.00001 | 0.00030 | 65 | 0.00126 | 0.00002 | 0.00046 | 93 | 0.00055 | -1.47499 | 148.296 | 0.14234 | 0.77376 |
| rutB; ureidoacrylate peracid hydrolase [EC:3.5.1.110] | 0.00093 | 0.00019 | 0.00086 | 0.00001 | 0.00033 | 65 | 0.00098 | 0.00001 | 0.00024 | 93 | 0.00040 | -0.30336 | 124.731 | 0.76212 | 0.93813 |
| basR; two-component system, OmpR family, response regulator BasR | 0.00093 | 0.00025 | 0.00082 | 0.00001 | 0.00029 | 65 | 0.00100 | 0.00001 | 0.00038 | 93 | 0.00048 | -0.37591 | 155.051 | 0.70750 | 0.92004 |
| tarL; CDP-ribitol ribitolphosphotransferase / teichoic acid ribitol-phosphate polymerase [EC:2.7.8.14 2.7.8.47] | 0.00093 | 0.00032 | 0.00043 | 0.00000 | 0.00019 | 65 | 0.00128 | 0.00003 | 0.00053 | 93 | 0.00056 | -1.51566 | 114.489 | 0.13236 | 0.77376 |
| draG; ADP-ribosyl-[dinitrogen reductase] hydrolase [EC:3.2.2.24] | 0.00093 | 0.00022 | 0.00109 | 0.00001 | 0.00033 | 65 | 0.00082 | 0.00001 | 0.00030 | 93 | 0.00045 | 0.60597 | 143.522 | 0.54549 | 0.86444 |
| stbD; antitoxin StbD | 0.00093 | 0.00021 | 0.00083 | 0.00000 | 0.00023 | 65 | 0.00100 | 0.00001 | 0.00031 | 93 | 0.00039 | -0.44615 | 153.375 | 0.65612 | 0.90679 |
| K09944; uncharacterized protein | 0.00093 | 0.00036 | 0.00071 | 0.00001 | 0.00028 | 65 | 0.00108 | 0.00003 | 0.00058 | 93 | 0.00065 | -0.58138 | 129.154 | 0.56200 | 0.86813 |
| K18284; adenosylhomocysteine/aminodeoxyfutalosine nucleosidase [EC:3.2.2.9 3.2.2.30] | 0.00093 | 0.00036 | 0.00071 | 0.00001 | 0.00028 | 65 | 0.00108 | 0.00003 | 0.00058 | 93 | 0.00065 | -0.58138 | 129.154 | 0.56200 | 0.86813 |
| NUDT14; UDP-sugar diphosphatase [EC:3.6.1.45] | 0.00093 | 0.00036 | 0.00071 | 0.00001 | 0.00028 | 65 | 0.00108 | 0.00003 | 0.00058 | 93 | 0.00065 | -0.58138 | 129.154 | 0.56200 | 0.86813 |
| spoIVFA; stage IV sporulation protein FA | 0.00092 | 0.00025 | 0.00017 | 0.00000 | 0.00007 | 65 | 0.00145 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.04721 | 97.268 | 0.00297 | 0.46257 |
| ytpB; tetraprenyl-beta-curcumene synthase [EC:4.2.3.130] | 0.00092 | 0.00025 | 0.00017 | 0.00000 | 0.00007 | 65 | 0.00145 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.04721 | 97.268 | 0.00297 | 0.46257 |
| dhbF; nonribosomal peptide synthetase DhbF | 0.00092 | 0.00029 | 0.00076 | 0.00001 | 0.00040 | 65 | 0.00104 | 0.00001 | 0.00040 | 93 | 0.00057 | -0.50455 | 151.055 | 0.61461 | 0.88827 |
| ybiC; uncharacterized oxidoreductase [EC:1.1.1.-] | 0.00092 | 0.00025 | 0.00049 | 0.00000 | 0.00019 | 65 | 0.00122 | 0.00001 | 0.00040 | 93 | 0.00044 | -1.66876 | 128.215 | 0.09760 | 0.76673 |
| menI; 1,4-dihydroxy-2-naphthoyl-CoA hydrolase [EC:3.1.2.28] | 0.00092 | 0.00019 | 0.00084 | 0.00000 | 0.00026 | 65 | 0.00098 | 0.00001 | 0.00027 | 93 | 0.00037 | -0.35900 | 152.471 | 0.72009 | 0.92108 |
| pinR; putative DNA-invertase from lambdoid prophage Rac | 0.00092 | 0.00029 | 0.00079 | 0.00001 | 0.00040 | 65 | 0.00101 | 0.00001 | 0.00040 | 93 | 0.00057 | -0.38278 | 150.821 | 0.70243 | 0.92004 |
| embC; arabinosyltransferase C [EC:2.4.2.-] | 0.00092 | 0.00038 | 0.00049 | 0.00000 | 0.00020 | 65 | 0.00122 | 0.00004 | 0.00062 | 93 | 0.00066 | -1.10224 | 110.339 | 0.27276 | 0.77376 |
| K06971; uncharacterized protein | 0.00092 | 0.00024 | 0.00032 | 0.00000 | 0.00018 | 65 | 0.00134 | 0.00001 | 0.00039 | 93 | 0.00043 | -2.37357 | 127.851 | 0.01911 | 0.59095 |
| mycP; membrane-anchored mycosin MYCP [EC:3.4.21.-] | 0.00092 | 0.00024 | 0.00046 | 0.00000 | 0.00019 | 65 | 0.00123 | 0.00001 | 0.00037 | 93 | 0.00042 | -1.83766 | 132.519 | 0.06835 | 0.73473 |
| lctB; potassium channel LctB | 0.00091 | 0.00025 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00145 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.09446 | 96.797 | 0.00258 | 0.46257 |
| K09166; uncharacterized protein | 0.00091 | 0.00031 | 0.00061 | 0.00001 | 0.00033 | 65 | 0.00112 | 0.00002 | 0.00048 | 93 | 0.00058 | -0.86769 | 150.993 | 0.38694 | 0.80262 |
| comX; competence protein ComX | 0.00091 | 0.00059 | 0.00060 | 0.00002 | 0.00055 | 65 | 0.00112 | 0.00008 | 0.00092 | 93 | 0.00107 | -0.48433 | 143.594 | 0.62889 | 0.89441 |
| pseI, neuB3; pseudaminic acid synthase [EC:2.5.1.97] | 0.00091 | 0.00036 | 0.00051 | 0.00000 | 0.00021 | 65 | 0.00119 | 0.00003 | 0.00059 | 93 | 0.00062 | -1.08736 | 113.692 | 0.27918 | 0.77376 |
| SASP-B, sspB; small acid-soluble spore protein B (major beta-type SASP) | 0.00090 | 0.00026 | 0.00012 | 0.00000 | 0.00006 | 65 | 0.00145 | 0.00002 | 0.00043 | 93 | 0.00043 | -3.06220 | 95.748 | 0.00285 | 0.46257 |
| mdlA, smdA; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.00090 | 0.00023 | 0.00094 | 0.00001 | 0.00032 | 65 | 0.00088 | 0.00001 | 0.00032 | 93 | 0.00045 | 0.13880 | 152.447 | 0.88979 | 0.97048 |
| mdlB, smdB; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.00090 | 0.00023 | 0.00094 | 0.00001 | 0.00032 | 65 | 0.00088 | 0.00001 | 0.00032 | 93 | 0.00045 | 0.13880 | 152.447 | 0.88979 | 0.97048 |
| hlyD, cyaD; hemolysin D | 0.00090 | 0.00019 | 0.00065 | 0.00001 | 0.00030 | 65 | 0.00108 | 0.00001 | 0.00025 | 93 | 0.00039 | -1.11471 | 139.487 | 0.26689 | 0.77376 |
| hlyB, cyaB; ATP-binding cassette, subfamily B, bacterial HlyB/CyaB | 0.00090 | 0.00018 | 0.00068 | 0.00000 | 0.00027 | 65 | 0.00105 | 0.00001 | 0.00025 | 93 | 0.00037 | -1.02012 | 146.833 | 0.30935 | 0.77376 |
| comER; competence protein ComER | 0.00090 | 0.00025 | 0.00010 | 0.00000 | 0.00004 | 65 | 0.00145 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.22973 | 93.595 | 0.00171 | 0.46257 |
| K09961; uncharacterized protein | 0.00090 | 0.00031 | 0.00072 | 0.00000 | 0.00026 | 65 | 0.00102 | 0.00002 | 0.00050 | 93 | 0.00056 | -0.54514 | 134.209 | 0.58656 | 0.87731 |
| PM20D1; carboxypeptidase PM20D1 [EC:3.4.17.-] | 0.00089 | 0.00020 | 0.00061 | 0.00000 | 0.00025 | 65 | 0.00109 | 0.00001 | 0.00029 | 93 | 0.00038 | -1.25019 | 155.952 | 0.21310 | 0.77376 |
| sspP, cotL; small acid-soluble spore protein P (minor) | 0.00089 | 0.00025 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00145 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.25248 | 93.331 | 0.00159 | 0.46257 |
| puo; putrescine oxidase [EC:1.4.3.10] | 0.00089 | 0.00025 | 0.00054 | 0.00000 | 0.00021 | 65 | 0.00114 | 0.00002 | 0.00041 | 93 | 0.00046 | -1.33346 | 133.645 | 0.18465 | 0.77376 |
| oxdD; oxalate decarboxylase [EC:4.1.1.2] | 0.00089 | 0.00034 | 0.00053 | 0.00001 | 0.00029 | 65 | 0.00114 | 0.00003 | 0.00055 | 93 | 0.00062 | -0.98190 | 135.296 | 0.32790 | 0.77376 |
| legG, neuC2; GDP/UDP-N,N’-diacetylbacillosamine 2-epimerase (hydrolysing) [EC:3.2.1.184] | 0.00089 | 0.00032 | 0.00065 | 0.00001 | 0.00033 | 65 | 0.00106 | 0.00002 | 0.00050 | 93 | 0.00060 | -0.67829 | 148.302 | 0.49864 | 0.84856 |
| repC; replication initiation protein RepC | 0.00089 | 0.00029 | 0.00024 | 0.00000 | 0.00012 | 65 | 0.00134 | 0.00002 | 0.00048 | 93 | 0.00050 | -2.21689 | 103.472 | 0.02882 | 0.61334 |
| cofE, fbiB; coenzyme F420-0:L-glutamate ligase / coenzyme F420-1:gamma-L-glutamate ligase [EC:6.3.2.31 6.3.2.34] | 0.00089 | 0.00023 | 0.00067 | 0.00000 | 0.00025 | 65 | 0.00104 | 0.00001 | 0.00034 | 93 | 0.00042 | -0.87208 | 153.235 | 0.38453 | 0.80197 |
| frsA; esterase FrsA [EC:3.1.-.-] | 0.00088 | 0.00026 | 0.00088 | 0.00001 | 0.00033 | 65 | 0.00089 | 0.00001 | 0.00037 | 93 | 0.00049 | -0.02697 | 155.610 | 0.97852 | 0.99657 |
| comB8; ComB8 competence protein | 0.00088 | 0.00038 | 0.00075 | 0.00001 | 0.00043 | 65 | 0.00097 | 0.00003 | 0.00058 | 93 | 0.00072 | -0.30795 | 154.009 | 0.75854 | 0.93719 |
| comB9; ComB9 competence protein | 0.00088 | 0.00038 | 0.00075 | 0.00001 | 0.00043 | 65 | 0.00097 | 0.00003 | 0.00058 | 93 | 0.00072 | -0.30795 | 154.009 | 0.75854 | 0.93719 |
| raxB, cvaB; ATP-binding cassette, subfamily B, bacterial RaxB | 0.00088 | 0.00024 | 0.00077 | 0.00001 | 0.00037 | 65 | 0.00096 | 0.00001 | 0.00032 | 93 | 0.00049 | -0.38019 | 139.879 | 0.70438 | 0.92004 |
| pksE; trans-AT polyketide synthase, acyltransferase and oxidoreductase domains | 0.00088 | 0.00028 | 0.00081 | 0.00001 | 0.00040 | 65 | 0.00092 | 0.00001 | 0.00039 | 93 | 0.00056 | -0.20476 | 149.333 | 0.83804 | 0.95506 |
| xanQ; xanthine permease XanQ | 0.00087 | 0.00029 | 0.00119 | 0.00002 | 0.00057 | 65 | 0.00065 | 0.00001 | 0.00029 | 93 | 0.00064 | 0.83352 | 96.394 | 0.40661 | 0.81189 |
| seaA; uncharacterized protein | 0.00087 | 0.00037 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00144 | 0.00004 | 0.00063 | 93 | 0.00063 | -2.17347 | 92.527 | 0.03230 | 0.62130 |
| solA; N-methyl-L-tryptophan oxidase [EC:1.5.3.-] | 0.00087 | 0.00032 | 0.00072 | 0.00001 | 0.00033 | 65 | 0.00097 | 0.00002 | 0.00049 | 93 | 0.00059 | -0.42280 | 150.119 | 0.67305 | 0.91184 |
| divJ; two-component system, cell cycle sensor histidine kinase DivJ [EC:2.7.13.3] | 0.00087 | 0.00023 | 0.00029 | 0.00000 | 0.00009 | 65 | 0.00127 | 0.00001 | 0.00038 | 93 | 0.00039 | -2.53234 | 101.888 | 0.01286 | 0.59095 |
| preT; dihydropyrimidine dehydrogenase (NAD+) subunit PreT [EC:1.3.1.1] | 0.00087 | 0.00034 | 0.00138 | 0.00004 | 0.00078 | 65 | 0.00051 | 0.00000 | 0.00018 | 93 | 0.00080 | 1.09425 | 70.509 | 0.27757 | 0.77376 |
| melA; alpha-galactosidase [EC:3.2.1.22] | 0.00087 | 0.00018 | 0.00023 | 0.00000 | 0.00008 | 65 | 0.00131 | 0.00001 | 0.00029 | 93 | 0.00030 | -3.59350 | 103.826 | 0.00050 | 0.46257 |
| hycI; hydrogenase 3 maturation protease [EC:3.4.23.51] | 0.00087 | 0.00019 | 0.00104 | 0.00001 | 0.00036 | 65 | 0.00074 | 0.00000 | 0.00020 | 93 | 0.00041 | 0.71353 | 101.799 | 0.47715 | 0.84209 |
| entS; MFS transporter, ENTS family, enterobactin (siderophore) exporter | 0.00086 | 0.00025 | 0.00073 | 0.00000 | 0.00026 | 65 | 0.00096 | 0.00001 | 0.00038 | 93 | 0.00046 | -0.50000 | 150.164 | 0.61781 | 0.88903 |
| K07492; putative transposase | 0.00086 | 0.00035 | 0.00037 | 0.00000 | 0.00022 | 65 | 0.00120 | 0.00003 | 0.00057 | 93 | 0.00061 | -1.35862 | 116.959 | 0.17688 | 0.77376 |
| K02482; two-component system, NtrC family, sensor kinase [EC:2.7.13.3] | 0.00086 | 0.00022 | 0.00049 | 0.00000 | 0.00018 | 65 | 0.00112 | 0.00001 | 0.00035 | 93 | 0.00040 | -1.55838 | 134.033 | 0.12150 | 0.77376 |
| per, rfbE; perosamine synthetase [EC:2.6.1.102] | 0.00086 | 0.00034 | 0.00045 | 0.00001 | 0.00028 | 65 | 0.00115 | 0.00003 | 0.00054 | 93 | 0.00061 | -1.15303 | 134.831 | 0.25094 | 0.77376 |
| gerPA; spore germination protein PA | 0.00086 | 0.00025 | 0.00010 | 0.00000 | 0.00004 | 65 | 0.00138 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.06517 | 93.751 | 0.00284 | 0.46257 |
| DDC, TDC; aromatic-L-amino-acid/L-tryptophan decarboxylase [EC:4.1.1.28 4.1.1.105] | 0.00085 | 0.00023 | 0.00136 | 0.00002 | 0.00048 | 65 | 0.00050 | 0.00000 | 0.00018 | 93 | 0.00051 | 1.69021 | 81.562 | 0.09481 | 0.76609 |
| kdgT; 2-keto-3-deoxygluconate permease | 0.00085 | 0.00015 | 0.00078 | 0.00000 | 0.00019 | 65 | 0.00090 | 0.00000 | 0.00021 | 93 | 0.00028 | -0.41269 | 155.699 | 0.68040 | 0.91354 |
| gerPB; spore germination protein PB | 0.00085 | 0.00025 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00138 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.08912 | 93.330 | 0.00264 | 0.46257 |
| gerPC; spore germination protein PC | 0.00085 | 0.00025 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00138 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.08912 | 93.330 | 0.00264 | 0.46257 |
| gerPE; spore germination protein PE | 0.00085 | 0.00025 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00138 | 0.00002 | 0.00042 | 93 | 0.00042 | -3.08912 | 93.330 | 0.00264 | 0.46257 |
| shlA, hhdA, hpmA; hemolysin | 0.00085 | 0.00021 | 0.00122 | 0.00001 | 0.00038 | 65 | 0.00059 | 0.00001 | 0.00023 | 93 | 0.00044 | 1.40311 | 111.321 | 0.16337 | 0.77376 |
| vanRC, vanRE, vanRG; two-component system, OmpR family, response regulator VanR | 0.00085 | 0.00034 | 0.00055 | 0.00001 | 0.00029 | 65 | 0.00106 | 0.00003 | 0.00053 | 93 | 0.00061 | -0.82544 | 137.631 | 0.41055 | 0.81416 |
| ligJ; 4-oxalmesaconate hydratase [EC:4.2.1.83] | 0.00085 | 0.00026 | 0.00032 | 0.00000 | 0.00016 | 65 | 0.00121 | 0.00002 | 0.00043 | 93 | 0.00045 | -1.96023 | 116.512 | 0.05236 | 0.69120 |
| cyaC, hlyC, rtxC; cytolysin-activating lysine-acyltransferase [EC:2.3.1.-] | 0.00085 | 0.00028 | 0.00075 | 0.00001 | 0.00040 | 65 | 0.00091 | 0.00001 | 0.00038 | 93 | 0.00056 | -0.29689 | 148.398 | 0.76697 | 0.93900 |
| K09706; uncharacterized protein | 0.00085 | 0.00026 | 0.00020 | 0.00000 | 0.00014 | 65 | 0.00130 | 0.00002 | 0.00043 | 93 | 0.00045 | -2.43869 | 109.387 | 0.01635 | 0.59095 |
| nicF; maleamate amidohydrolase [EC:3.5.1.107] | 0.00084 | 0.00029 | 0.00050 | 0.00001 | 0.00032 | 65 | 0.00108 | 0.00002 | 0.00044 | 93 | 0.00054 | -1.06934 | 153.398 | 0.28659 | 0.77376 |
| gpgS; glucosyl-3-phosphoglycerate synthase [EC:2.4.1.266] | 0.00084 | 0.00023 | 0.00069 | 0.00001 | 0.00029 | 65 | 0.00095 | 0.00001 | 0.00034 | 93 | 0.00045 | -0.58244 | 155.940 | 0.56111 | 0.86813 |
| PRSS15, PIM1; ATP-dependent Lon protease [EC:3.4.21.53] | 0.00084 | 0.00028 | 0.00083 | 0.00001 | 0.00041 | 65 | 0.00085 | 0.00001 | 0.00038 | 93 | 0.00056 | -0.03035 | 147.146 | 0.97583 | 0.99571 |
| spdH; spermidine dehydrogenase [EC:1.5.99.6] | 0.00084 | 0.00030 | 0.00052 | 0.00001 | 0.00032 | 65 | 0.00106 | 0.00002 | 0.00045 | 93 | 0.00056 | -0.98068 | 152.781 | 0.32830 | 0.77376 |
| spoVID; stage VI sporulation protein D | 0.00084 | 0.00025 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00132 | 0.00002 | 0.00041 | 93 | 0.00041 | -2.81980 | 97.287 | 0.00582 | 0.54022 |
| fixK; CRP/FNR family transcriptional regulator, nitrogen fixation regulation protein | 0.00084 | 0.00032 | 0.00033 | 0.00000 | 0.00010 | 65 | 0.00119 | 0.00003 | 0.00053 | 93 | 0.00054 | -1.57828 | 98.907 | 0.11769 | 0.77376 |
| K09982; uncharacterized protein | 0.00084 | 0.00017 | 0.00085 | 0.00000 | 0.00024 | 65 | 0.00083 | 0.00000 | 0.00023 | 93 | 0.00034 | 0.06887 | 147.554 | 0.94519 | 0.98674 |
| mngB; mannosylglycerate hydrolase [EC:3.2.1.170] | 0.00084 | 0.00025 | 0.00086 | 0.00001 | 0.00043 | 65 | 0.00082 | 0.00001 | 0.00031 | 93 | 0.00053 | 0.08175 | 125.781 | 0.93498 | 0.98459 |
| kshB; 3-ketosteroid 9alpha-monooxygenase subunit B [EC:1.14.13.142] | 0.00084 | 0.00023 | 0.00040 | 0.00000 | 0.00015 | 65 | 0.00114 | 0.00001 | 0.00038 | 93 | 0.00041 | -1.81976 | 119.157 | 0.07131 | 0.73944 |
| abiQ; protein AbiQ | 0.00084 | 0.00017 | 0.00112 | 0.00001 | 0.00035 | 65 | 0.00064 | 0.00000 | 0.00015 | 93 | 0.00038 | 1.24839 | 88.397 | 0.21519 | 0.77376 |
| lyxK; L-xylulokinase [EC:2.7.1.53] | 0.00083 | 0.00015 | 0.00070 | 0.00000 | 0.00022 | 65 | 0.00093 | 0.00000 | 0.00020 | 93 | 0.00030 | -0.75386 | 146.470 | 0.45214 | 0.83438 |
| citR; LysR family transcriptional regulator, repressor for citA | 0.00083 | 0.00032 | 0.00055 | 0.00001 | 0.00030 | 65 | 0.00103 | 0.00002 | 0.00050 | 93 | 0.00059 | -0.82542 | 143.995 | 0.41050 | 0.81416 |
| ycnJ; copper transport protein | 0.00083 | 0.00018 | 0.00038 | 0.00000 | 0.00014 | 65 | 0.00114 | 0.00001 | 0.00029 | 93 | 0.00032 | -2.36135 | 128.394 | 0.01971 | 0.59095 |
| pksJ; polyketide synthase PksJ | 0.00083 | 0.00028 | 0.00075 | 0.00001 | 0.00040 | 65 | 0.00088 | 0.00001 | 0.00039 | 93 | 0.00056 | -0.24727 | 148.685 | 0.80504 | 0.94958 |
| pseC; UDP-4-amino-4,6-dideoxy-L-N-acetyl-beta-L-altrosamine transaminase [EC:2.6.1.92] | 0.00083 | 0.00035 | 0.00050 | 0.00000 | 0.00021 | 65 | 0.00106 | 0.00003 | 0.00058 | 93 | 0.00062 | -0.89928 | 114.074 | 0.37040 | 0.79587 |
| pseH; UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine N-acetyltransferase [EC:2.3.1.202] | 0.00083 | 0.00035 | 0.00050 | 0.00000 | 0.00021 | 65 | 0.00106 | 0.00003 | 0.00058 | 93 | 0.00062 | -0.89928 | 114.074 | 0.37040 | 0.79587 |
| ogl; oligogalacturonide lyase [EC:4.2.2.6] | 0.00083 | 0.00028 | 0.00042 | 0.00000 | 0.00019 | 65 | 0.00111 | 0.00002 | 0.00045 | 93 | 0.00049 | -1.41989 | 122.860 | 0.15817 | 0.77376 |
| aglE, ggtB; alpha-glucoside transport system substrate-binding protein | 0.00082 | 0.00017 | 0.00065 | 0.00000 | 0.00021 | 65 | 0.00094 | 0.00001 | 0.00026 | 93 | 0.00033 | -0.87912 | 155.865 | 0.38069 | 0.79887 |
| E3.1.1.74; cutinase [EC:3.1.1.74] | 0.00082 | 0.00032 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00134 | 0.00003 | 0.00054 | 93 | 0.00055 | -2.30192 | 94.363 | 0.02354 | 0.59764 |
| rhaA; L-rhamnose isomerase / sugar isomerase [EC:5.3.1.14 5.3.1.-] | 0.00082 | 0.00020 | 0.00069 | 0.00000 | 0.00026 | 65 | 0.00091 | 0.00001 | 0.00028 | 93 | 0.00038 | -0.58229 | 154.319 | 0.56122 | 0.86813 |
| allD; ureidoglycolate dehydrogenase (NAD+) [EC:1.1.1.350] | 0.00082 | 0.00033 | 0.00040 | 0.00000 | 0.00028 | 65 | 0.00111 | 0.00003 | 0.00052 | 93 | 0.00059 | -1.21600 | 136.009 | 0.22609 | 0.77376 |
| hss; homospermidine synthase [EC:2.5.1.44] | 0.00082 | 0.00023 | 0.00018 | 0.00000 | 0.00005 | 65 | 0.00126 | 0.00001 | 0.00039 | 93 | 0.00039 | -2.74272 | 95.484 | 0.00728 | 0.54764 |
| E3.1.27.3; ribonuclease T1 [EC:3.1.27.3] | 0.00082 | 0.00026 | 0.00046 | 0.00000 | 0.00016 | 65 | 0.00106 | 0.00002 | 0.00043 | 93 | 0.00046 | -1.31316 | 115.428 | 0.19173 | 0.77376 |
| K09960; uncharacterized protein | 0.00082 | 0.00029 | 0.00056 | 0.00000 | 0.00019 | 65 | 0.00100 | 0.00002 | 0.00048 | 93 | 0.00052 | -0.84478 | 120.310 | 0.39991 | 0.80762 |
| thuF, sugA; trehalose/maltose transport system permease protein | 0.00082 | 0.00026 | 0.00036 | 0.00000 | 0.00014 | 65 | 0.00114 | 0.00002 | 0.00042 | 93 | 0.00044 | -1.76934 | 112.286 | 0.07955 | 0.75920 |
| sspK; small acid-soluble spore protein K (minor) | 0.00082 | 0.00025 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00133 | 0.00002 | 0.00041 | 93 | 0.00041 | -3.02955 | 93.057 | 0.00317 | 0.46257 |
| PTS-Fru2-EIIB; PTS system, fructose-specific IIB-like component [EC:2.7.1.-] | 0.00082 | 0.00025 | 0.00058 | 0.00000 | 0.00016 | 65 | 0.00098 | 0.00002 | 0.00041 | 93 | 0.00044 | -0.90201 | 119.105 | 0.36887 | 0.79587 |
| cimA; (R)-citramalate synthase [EC:2.3.1.182] | 0.00081 | 0.00021 | 0.00055 | 0.00000 | 0.00024 | 65 | 0.00100 | 0.00001 | 0.00032 | 93 | 0.00040 | -1.10906 | 154.758 | 0.26913 | 0.77376 |
| ulaE, sgaU, sgbU; L-ribulose-5-phosphate 3-epimerase [EC:5.1.3.22] | 0.00081 | 0.00016 | 0.00102 | 0.00001 | 0.00032 | 65 | 0.00067 | 0.00000 | 0.00015 | 93 | 0.00036 | 0.99882 | 91.947 | 0.32050 | 0.77376 |
| iolJ; 6-phospho-5-dehydro-2-deoxy-D-gluconate aldolase [EC:4.1.2.29] | 0.00081 | 0.00031 | 0.00085 | 0.00003 | 0.00063 | 65 | 0.00079 | 0.00001 | 0.00028 | 93 | 0.00069 | 0.08250 | 89.363 | 0.93444 | 0.98428 |
| mepM; murein DD-endopeptidase [EC:3.4.24.-] | 0.00081 | 0.00019 | 0.00091 | 0.00001 | 0.00031 | 65 | 0.00075 | 0.00001 | 0.00024 | 93 | 0.00040 | 0.40650 | 132.723 | 0.68503 | 0.91533 |
| atoC; two-component system, NtrC family, response regulator AtoC | 0.00081 | 0.00030 | 0.00067 | 0.00002 | 0.00054 | 65 | 0.00091 | 0.00001 | 0.00035 | 93 | 0.00065 | -0.37412 | 113.856 | 0.70901 | 0.92004 |
| hicB; antitoxin HicB | 0.00081 | 0.00023 | 0.00047 | 0.00000 | 0.00024 | 65 | 0.00105 | 0.00001 | 0.00036 | 93 | 0.00043 | -1.33076 | 149.413 | 0.18530 | 0.77376 |
| cofC; 2-phospho-L-lactate guanylyltransferase [EC:2.7.7.68] | 0.00081 | 0.00021 | 0.00049 | 0.00000 | 0.00016 | 65 | 0.00104 | 0.00001 | 0.00034 | 93 | 0.00038 | -1.45820 | 128.650 | 0.14722 | 0.77376 |
| dmpL, poxC; Phenol hydroxylase P1 protein | 0.00081 | 0.00020 | 0.00088 | 0.00000 | 0.00027 | 65 | 0.00076 | 0.00001 | 0.00027 | 93 | 0.00039 | 0.29617 | 150.826 | 0.76751 | 0.93900 |
| dmpM, poxB; phenol hydroxylase P2 protein | 0.00081 | 0.00020 | 0.00088 | 0.00000 | 0.00027 | 65 | 0.00076 | 0.00001 | 0.00027 | 93 | 0.00039 | 0.29617 | 150.826 | 0.76751 | 0.93900 |
| dmpN, poxD; phenol hydroxylase P3 protein [EC:1.14.13.-] | 0.00081 | 0.00020 | 0.00088 | 0.00000 | 0.00027 | 65 | 0.00076 | 0.00001 | 0.00027 | 93 | 0.00039 | 0.29617 | 150.826 | 0.76751 | 0.93900 |
| dmpO, poxE; phenol hydroxylase P4 protein | 0.00081 | 0.00020 | 0.00088 | 0.00000 | 0.00027 | 65 | 0.00076 | 0.00001 | 0.00027 | 93 | 0.00039 | 0.29617 | 150.826 | 0.76751 | 0.93900 |
| dmpP, poxF; phenol hydroxylase P5 protein | 0.00081 | 0.00020 | 0.00088 | 0.00000 | 0.00027 | 65 | 0.00076 | 0.00001 | 0.00027 | 93 | 0.00039 | 0.29617 | 150.826 | 0.76751 | 0.93900 |
| thuE; trehalose/maltose transport system substrate-binding protein | 0.00081 | 0.00026 | 0.00034 | 0.00000 | 0.00014 | 65 | 0.00114 | 0.00002 | 0.00042 | 93 | 0.00044 | -1.81296 | 112.177 | 0.07251 | 0.74128 |
| hyfE; hydrogenase-4 component E [EC:1.-.-.-] | 0.00081 | 0.00020 | 0.00092 | 0.00001 | 0.00036 | 65 | 0.00073 | 0.00000 | 0.00022 | 93 | 0.00042 | 0.45423 | 108.274 | 0.65057 | 0.90423 |
| hyfF; hydrogenase-4 component F [EC:1.-.-.-] | 0.00081 | 0.00020 | 0.00092 | 0.00001 | 0.00036 | 65 | 0.00073 | 0.00000 | 0.00022 | 93 | 0.00042 | 0.45423 | 108.274 | 0.65057 | 0.90423 |
| cotA; spore coat protein A, manganese oxidase [EC:1.16.3.3] | 0.00081 | 0.00032 | 0.00034 | 0.00000 | 0.00028 | 65 | 0.00113 | 0.00002 | 0.00051 | 93 | 0.00058 | -1.36848 | 136.616 | 0.17341 | 0.77376 |
| K09388; uncharacterized protein | 0.00080 | 0.00024 | 0.00015 | 0.00000 | 0.00008 | 65 | 0.00126 | 0.00002 | 0.00040 | 93 | 0.00041 | -2.69259 | 99.514 | 0.00832 | 0.56552 |
| csy3; CRISPR-associated protein Csy3 | 0.00080 | 0.00019 | 0.00092 | 0.00001 | 0.00030 | 65 | 0.00072 | 0.00001 | 0.00024 | 93 | 0.00038 | 0.53034 | 135.889 | 0.59674 | 0.88062 |
| PTS-Fru2-EIIC; PTS system, fructose-specific IIC-like component | 0.00080 | 0.00024 | 0.00072 | 0.00000 | 0.00022 | 65 | 0.00086 | 0.00001 | 0.00037 | 93 | 0.00043 | -0.30512 | 143.467 | 0.76071 | 0.93791 |
| rhlI, phzI, solI, cepI, tofI; acyl homoserine lactone synthase [EC:2.3.1.184] | 0.00080 | 0.00028 | 0.00074 | 0.00001 | 0.00040 | 65 | 0.00085 | 0.00001 | 0.00038 | 93 | 0.00056 | -0.20405 | 148.297 | 0.83860 | 0.95506 |
| GART; phosphoribosylamine–glycine ligase / phosphoribosylglycinamide formyltransferase / phosphoribosylformylglycinamidine cyclo-ligase [EC:6.3.4.13 2.1.2.2 6.3.3.1] | 0.00080 | 0.00024 | 0.00044 | 0.00000 | 0.00015 | 65 | 0.00105 | 0.00002 | 0.00040 | 93 | 0.00043 | -1.43798 | 115.104 | 0.15315 | 0.77376 |
| csm3; CRISPR-associated protein Csm3 | 0.00080 | 0.00023 | 0.00070 | 0.00000 | 0.00018 | 65 | 0.00087 | 0.00001 | 0.00037 | 93 | 0.00041 | -0.40920 | 129.746 | 0.68307 | 0.91498 |
| K09957; uncharacterized protein | 0.00080 | 0.00023 | 0.00053 | 0.00000 | 0.00022 | 65 | 0.00099 | 0.00001 | 0.00035 | 93 | 0.00041 | -1.10901 | 145.205 | 0.26926 | 0.77376 |
| nat; isonocardicin synthase [EC:2.5.1.38] | 0.00080 | 0.00028 | 0.00073 | 0.00001 | 0.00040 | 65 | 0.00084 | 0.00001 | 0.00038 | 93 | 0.00056 | -0.20476 | 148.477 | 0.83804 | 0.95506 |
| pksM; polyketide synthase PksM | 0.00080 | 0.00028 | 0.00073 | 0.00001 | 0.00040 | 65 | 0.00084 | 0.00001 | 0.00038 | 93 | 0.00056 | -0.20476 | 148.477 | 0.83804 | 0.95506 |
| rhlR, phzR; LuxR family transcriptional regulator, quorum-sensing system regulator RhlR | 0.00080 | 0.00028 | 0.00073 | 0.00001 | 0.00040 | 65 | 0.00084 | 0.00001 | 0.00038 | 93 | 0.00056 | -0.20476 | 148.477 | 0.83804 | 0.95506 |
| yeeJ; adhesin/invasin | 0.00079 | 0.00032 | 0.00063 | 0.00001 | 0.00034 | 65 | 0.00090 | 0.00002 | 0.00049 | 93 | 0.00060 | -0.45181 | 152.239 | 0.65205 | 0.90504 |
| mhpB; 2,3-dihydroxyphenylpropionate 1,2-dioxygenase [EC:1.13.11.16] | 0.00079 | 0.00024 | 0.00045 | 0.00000 | 0.00017 | 65 | 0.00103 | 0.00001 | 0.00038 | 93 | 0.00042 | -1.36079 | 126.407 | 0.17600 | 0.77376 |
| gatR; DeoR family transcriptional regulator, galactitol utilization operon repressor | 0.00079 | 0.00014 | 0.00084 | 0.00000 | 0.00021 | 65 | 0.00075 | 0.00000 | 0.00019 | 93 | 0.00028 | 0.30182 | 143.776 | 0.76322 | 0.93836 |
| rcnA; nickel/cobalt exporter | 0.00078 | 0.00018 | 0.00081 | 0.00001 | 0.00029 | 65 | 0.00077 | 0.00000 | 0.00022 | 93 | 0.00036 | 0.10490 | 130.191 | 0.91662 | 0.97681 |
| xylE; MFS transporter, SP family, xylose:H+ symportor | 0.00078 | 0.00021 | 0.00047 | 0.00000 | 0.00017 | 65 | 0.00100 | 0.00001 | 0.00034 | 93 | 0.00038 | -1.39402 | 133.380 | 0.16563 | 0.77376 |
| raxA; membrane fusion protein | 0.00078 | 0.00021 | 0.00060 | 0.00000 | 0.00025 | 65 | 0.00091 | 0.00001 | 0.00032 | 93 | 0.00040 | -0.75833 | 155.548 | 0.44940 | 0.83438 |
| ctpG; cation-transporting P-type ATPase G [EC:3.6.3.-] | 0.00078 | 0.00038 | 0.00053 | 0.00000 | 0.00024 | 65 | 0.00096 | 0.00004 | 0.00063 | 93 | 0.00067 | -0.63990 | 116.905 | 0.52349 | 0.85708 |
| nicX; 2,5-dihydroxypyridine 5,6-dioxygenase [EC:1.13.11.9] | 0.00078 | 0.00028 | 0.00040 | 0.00001 | 0.00030 | 65 | 0.00105 | 0.00002 | 0.00043 | 93 | 0.00053 | -1.24327 | 151.235 | 0.21569 | 0.77376 |
| yjeH; amino acid efflux transporter | 0.00078 | 0.00020 | 0.00088 | 0.00001 | 0.00034 | 65 | 0.00071 | 0.00001 | 0.00025 | 93 | 0.00042 | 0.40267 | 127.099 | 0.68787 | 0.91567 |
| ampH; serine-type D-Ala-D-Ala carboxypeptidase/endopeptidase [EC:3.4.16.4 3.4.21.-] | 0.00078 | 0.00024 | 0.00065 | 0.00000 | 0.00025 | 65 | 0.00087 | 0.00001 | 0.00037 | 93 | 0.00045 | -0.49449 | 151.039 | 0.62168 | 0.89150 |
| ectD; ectoine hydroxylase [EC:1.14.11.55] | 0.00078 | 0.00016 | 0.00048 | 0.00000 | 0.00018 | 65 | 0.00098 | 0.00001 | 0.00024 | 93 | 0.00030 | -1.67935 | 153.986 | 0.09511 | 0.76609 |
| lasI, luxI; acyl homoserine lactone synthase [EC:2.3.1.184] | 0.00077 | 0.00017 | 0.00073 | 0.00000 | 0.00026 | 65 | 0.00081 | 0.00000 | 0.00023 | 93 | 0.00035 | -0.23145 | 139.751 | 0.81730 | 0.95487 |
| dprE2; decaprenylphospho-beta-D-erythro-pentofuranosid-2-ulose 2-reductase [EC:1.1.1.333] | 0.00077 | 0.00021 | 0.00062 | 0.00000 | 0.00022 | 65 | 0.00088 | 0.00001 | 0.00033 | 93 | 0.00040 | -0.66683 | 149.282 | 0.50591 | 0.84956 |
| terA; tellurite resistance protein TerA | 0.00077 | 0.00023 | 0.00059 | 0.00000 | 0.00021 | 65 | 0.00090 | 0.00001 | 0.00037 | 93 | 0.00042 | -0.72448 | 141.411 | 0.46997 | 0.83870 |
| ABC.ARG.S, argT; lysine/arginine/ornithine transport system substrate-binding protein | 0.00077 | 0.00018 | 0.00073 | 0.00000 | 0.00026 | 65 | 0.00080 | 0.00001 | 0.00025 | 93 | 0.00036 | -0.19123 | 150.282 | 0.84861 | 0.95712 |
| sspO, cotK; small acid-soluble spore protein O (minor) | 0.00077 | 0.00024 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00125 | 0.00002 | 0.00041 | 93 | 0.00041 | -2.86130 | 93.333 | 0.00521 | 0.54022 |
| uaZ; urate oxidase [EC:1.7.3.3] | 0.00076 | 0.00024 | 0.00037 | 0.00000 | 0.00016 | 65 | 0.00104 | 0.00001 | 0.00039 | 93 | 0.00042 | -1.57756 | 120.886 | 0.11728 | 0.77376 |
| nicD; N-formylmaleamate deformylase [EC:3.5.1.106] | 0.00076 | 0.00028 | 0.00041 | 0.00001 | 0.00030 | 65 | 0.00101 | 0.00002 | 0.00043 | 93 | 0.00053 | -1.13173 | 151.574 | 0.25954 | 0.77376 |
| aroM; protein AroM | 0.00076 | 0.00031 | 0.00055 | 0.00001 | 0.00029 | 65 | 0.00091 | 0.00002 | 0.00049 | 93 | 0.00056 | -0.62895 | 142.814 | 0.53039 | 0.86098 |
| dprE1; decaprenylphospho-beta-D-ribofuranose 2-oxidase [EC:1.1.98.3] | 0.00076 | 0.00021 | 0.00058 | 0.00000 | 0.00022 | 65 | 0.00088 | 0.00001 | 0.00033 | 93 | 0.00039 | -0.75237 | 149.002 | 0.45302 | 0.83438 |
| K14136; decaprenyl-phosphate phosphoribosyltransferase [EC:2.4.2.45] | 0.00076 | 0.00021 | 0.00058 | 0.00000 | 0.00022 | 65 | 0.00088 | 0.00001 | 0.00033 | 93 | 0.00039 | -0.75237 | 149.002 | 0.45302 | 0.83438 |
| K09146; uncharacterized protein | 0.00075 | 0.00023 | 0.00043 | 0.00000 | 0.00019 | 65 | 0.00098 | 0.00001 | 0.00036 | 93 | 0.00041 | -1.34882 | 134.546 | 0.17966 | 0.77376 |
| glft1; rhamnopyranosyl-N-acetylglucosaminyl-diphospho-decaprenol beta-1,3/1,4-galactofuranosyltransferase [EC:2.4.1.287] | 0.00075 | 0.00022 | 0.00060 | 0.00000 | 0.00023 | 65 | 0.00086 | 0.00001 | 0.00033 | 93 | 0.00040 | -0.65824 | 152.434 | 0.51138 | 0.85122 |
| ligA; protocatechuate 4,5-dioxygenase, alpha chain [EC:1.13.11.8] | 0.00075 | 0.00039 | 0.00017 | 0.00000 | 0.00009 | 65 | 0.00116 | 0.00004 | 0.00066 | 93 | 0.00066 | -1.49489 | 95.516 | 0.13824 | 0.77376 |
| wbpB; UDP-N-acetyl-2-amino-2-deoxyglucuronate dehydrogenase [EC:1.1.1.335] | 0.00075 | 0.00035 | 0.00039 | 0.00000 | 0.00020 | 65 | 0.00100 | 0.00003 | 0.00058 | 93 | 0.00061 | -0.99949 | 112.752 | 0.31970 | 0.77376 |
| uvsE, UVE1; UV DNA damage endonuclease [EC:3.-.-.-] | 0.00074 | 0.00024 | 0.00060 | 0.00001 | 0.00041 | 65 | 0.00084 | 0.00001 | 0.00029 | 93 | 0.00050 | -0.48964 | 123.594 | 0.62525 | 0.89228 |
| mptA; alpha-1,6-mannosyltransferase [EC:2.4.1.-] | 0.00074 | 0.00021 | 0.00052 | 0.00000 | 0.00020 | 65 | 0.00090 | 0.00001 | 0.00033 | 93 | 0.00038 | -0.96954 | 144.787 | 0.33389 | 0.77992 |
| E2.7.13.1; protein-histidine pros-kinase [EC:2.7.13.1] | 0.00074 | 0.00026 | 0.00034 | 0.00000 | 0.00018 | 65 | 0.00102 | 0.00002 | 0.00043 | 93 | 0.00046 | -1.48158 | 121.207 | 0.14105 | 0.77376 |
| cotZ; spore coat protein Z | 0.00074 | 0.00033 | 0.00043 | 0.00001 | 0.00031 | 65 | 0.00096 | 0.00002 | 0.00052 | 93 | 0.00060 | -0.88636 | 143.793 | 0.37690 | 0.79810 |
| yndE; spore germination protein | 0.00074 | 0.00033 | 0.00035 | 0.00001 | 0.00028 | 65 | 0.00102 | 0.00003 | 0.00052 | 93 | 0.00059 | -1.12772 | 135.517 | 0.26143 | 0.77376 |
| E1.1.1.91; aryl-alcohol dehydrogenase (NADP+) [EC:1.1.1.91] | 0.00074 | 0.00022 | 0.00031 | 0.00000 | 0.00008 | 65 | 0.00104 | 0.00001 | 0.00037 | 93 | 0.00038 | -1.95486 | 99.902 | 0.05339 | 0.69953 |
| yndF; spore germination protein | 0.00074 | 0.00033 | 0.00035 | 0.00001 | 0.00028 | 65 | 0.00101 | 0.00003 | 0.00052 | 93 | 0.00059 | -1.12256 | 135.512 | 0.26361 | 0.77376 |
| sda; developmental checkpoint coupling sporulation initiation to replication initiation | 0.00074 | 0.00023 | 0.00014 | 0.00000 | 0.00007 | 65 | 0.00116 | 0.00001 | 0.00039 | 93 | 0.00040 | -2.56282 | 97.633 | 0.01191 | 0.59095 |
| spoIIB; stage II sporulation protein B | 0.00074 | 0.00024 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00121 | 0.00001 | 0.00040 | 93 | 0.00040 | -2.88984 | 92.874 | 0.00480 | 0.54022 |
| deoR; DeoR family transcriptional regulator, deoxyribose operon repressor | 0.00074 | 0.00022 | 0.00080 | 0.00001 | 0.00031 | 65 | 0.00069 | 0.00001 | 0.00030 | 93 | 0.00043 | 0.25674 | 149.687 | 0.79773 | 0.94822 |
| cofD; LPPG:FO 2-phospho-L-lactate transferase [EC:2.7.8.28] | 0.00074 | 0.00021 | 0.00044 | 0.00000 | 0.00016 | 65 | 0.00094 | 0.00001 | 0.00034 | 93 | 0.00038 | -1.31888 | 128.708 | 0.18955 | 0.77376 |
| lpcC; mannosyltransferase [EC:2.4.1.-] | 0.00074 | 0.00022 | 0.00013 | 0.00000 | 0.00004 | 65 | 0.00116 | 0.00001 | 0.00037 | 93 | 0.00037 | -2.74915 | 94.448 | 0.00716 | 0.54612 |
| limB; limonene 1,2-monooxygenase [EC:1.14.13.107] | 0.00074 | 0.00022 | 0.00040 | 0.00000 | 0.00015 | 65 | 0.00097 | 0.00001 | 0.00036 | 93 | 0.00039 | -1.48682 | 123.373 | 0.13961 | 0.77376 |
| DHBD; 2,3-dihydroxybenzoate decarboxylase [EC:4.1.1.46] | 0.00073 | 0.00028 | 0.00036 | 0.00001 | 0.00030 | 65 | 0.00099 | 0.00002 | 0.00043 | 93 | 0.00053 | -1.19759 | 151.058 | 0.23295 | 0.77376 |
| fbiC; FO synthase [EC:2.5.1.77] | 0.00073 | 0.00021 | 0.00045 | 0.00000 | 0.00016 | 65 | 0.00093 | 0.00001 | 0.00034 | 93 | 0.00038 | -1.28776 | 129.173 | 0.20013 | 0.77376 |
| kgd; 2-oxoglutarate decarboxylase [EC:4.1.1.71] | 0.00073 | 0.00021 | 0.00057 | 0.00000 | 0.00022 | 65 | 0.00084 | 0.00001 | 0.00033 | 93 | 0.00039 | -0.68368 | 149.380 | 0.49524 | 0.84797 |
| gerPD; spore germination protein PD | 0.00073 | 0.00024 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00120 | 0.00002 | 0.00040 | 93 | 0.00040 | -2.79987 | 93.042 | 0.00622 | 0.54022 |
| sspL; small acid-soluble spore protein L (minor) | 0.00073 | 0.00024 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00120 | 0.00002 | 0.00040 | 93 | 0.00040 | -2.79987 | 93.042 | 0.00622 | 0.54022 |
| cobIJ; precorrin-2 C20-methyltransferase / precorrin-3B C17-methyltransferase [EC:2.1.1.130 2.1.1.131] | 0.00073 | 0.00021 | 0.00047 | 0.00000 | 0.00017 | 65 | 0.00091 | 0.00001 | 0.00033 | 93 | 0.00038 | -1.16214 | 133.218 | 0.24726 | 0.77376 |
| hlyII; hemolysin II | 0.00073 | 0.00032 | 0.00021 | 0.00000 | 0.00014 | 65 | 0.00109 | 0.00003 | 0.00053 | 93 | 0.00055 | -1.61658 | 105.511 | 0.10895 | 0.77376 |
| iga; IgA-specific metalloendopeptidase [EC:3.4.24.13] | 0.00073 | 0.00024 | 0.00071 | 0.00001 | 0.00035 | 65 | 0.00074 | 0.00001 | 0.00034 | 93 | 0.00049 | -0.04784 | 149.645 | 0.96191 | 0.98988 |
| pfbA; plasmin and fibronectin-binding protein A | 0.00073 | 0.00024 | 0.00071 | 0.00001 | 0.00035 | 65 | 0.00074 | 0.00001 | 0.00034 | 93 | 0.00049 | -0.04784 | 149.645 | 0.96191 | 0.98988 |
| radD; DNA repair protein RadD | 0.00073 | 0.00018 | 0.00086 | 0.00001 | 0.00030 | 65 | 0.00063 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.59951 | 125.553 | 0.54991 | 0.86680 |
| dndC; DNA sulfur modification protein DndC | 0.00073 | 0.00018 | 0.00082 | 0.00001 | 0.00028 | 65 | 0.00066 | 0.00001 | 0.00023 | 93 | 0.00037 | 0.42996 | 136.300 | 0.66790 | 0.91150 |
| bphH, xylJ, tesE; 2-oxopent-4-enoate/cis-2-oxohex-4-enoate hydratase [EC:4.2.1.80 4.2.1.132] | 0.00072 | 0.00017 | 0.00100 | 0.00001 | 0.00030 | 65 | 0.00053 | 0.00000 | 0.00019 | 93 | 0.00036 | 1.32745 | 110.660 | 0.18709 | 0.77376 |
| caiD; crotonobetainyl-CoA hydratase [EC:4.2.1.149] | 0.00072 | 0.00016 | 0.00019 | 0.00000 | 0.00005 | 65 | 0.00109 | 0.00001 | 0.00026 | 93 | 0.00026 | -3.44639 | 98.810 | 0.00084 | 0.46257 |
| hyfB; hydrogenase-4 component B [EC:1.-.-.-] | 0.00072 | 0.00018 | 0.00085 | 0.00001 | 0.00035 | 65 | 0.00064 | 0.00000 | 0.00019 | 93 | 0.00040 | 0.53315 | 101.258 | 0.59510 | 0.88005 |
| mexC; membrane fusion protein, multidrug efflux system | 0.00072 | 0.00028 | 0.00042 | 0.00001 | 0.00030 | 65 | 0.00093 | 0.00002 | 0.00043 | 93 | 0.00053 | -0.97194 | 151.600 | 0.33263 | 0.77922 |
| mexD; multidrug efflux pump | 0.00072 | 0.00028 | 0.00042 | 0.00001 | 0.00030 | 65 | 0.00093 | 0.00002 | 0.00043 | 93 | 0.00053 | -0.97194 | 151.600 | 0.33263 | 0.77922 |
| wbdB, wbpY; mannosyl-N-acetyl-alpha-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol 3-alpha-mannosyltransferase / alpha-1,3-rhamnosyltransferase [EC:2.4.1.349 2.4.1.-] | 0.00072 | 0.00022 | 0.00096 | 0.00001 | 0.00035 | 65 | 0.00056 | 0.00001 | 0.00029 | 93 | 0.00045 | 0.90199 | 136.315 | 0.36865 | 0.79587 |
| E1.3.1.12; prephenate dehydrogenase [EC:1.3.1.12] | 0.00072 | 0.00020 | 0.00037 | 0.00000 | 0.00010 | 65 | 0.00097 | 0.00001 | 0.00033 | 93 | 0.00035 | -1.74215 | 108.048 | 0.08433 | 0.76567 |
| K09163; uncharacterized protein | 0.00072 | 0.00022 | 0.00038 | 0.00000 | 0.00018 | 65 | 0.00096 | 0.00001 | 0.00035 | 93 | 0.00040 | -1.48060 | 134.611 | 0.14105 | 0.77376 |
| trpEG; anthranilate synthase [EC:4.1.3.27] | 0.00072 | 0.00022 | 0.00013 | 0.00000 | 0.00004 | 65 | 0.00113 | 0.00001 | 0.00037 | 93 | 0.00037 | -2.65655 | 94.434 | 0.00927 | 0.58112 |
| K06887; uncharacterized protein | 0.00072 | 0.00022 | 0.00065 | 0.00001 | 0.00029 | 65 | 0.00076 | 0.00001 | 0.00032 | 93 | 0.00043 | -0.26415 | 154.518 | 0.79202 | 0.94731 |
| K09980; uncharacterized protein | 0.00072 | 0.00018 | 0.00080 | 0.00001 | 0.00029 | 65 | 0.00065 | 0.00000 | 0.00023 | 93 | 0.00037 | 0.40108 | 131.524 | 0.68901 | 0.91567 |
| sdmt; sarcosine/dimethylglycine N-methyltransferase [EC:2.1.1.157] | 0.00071 | 0.00028 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00097 | 0.00002 | 0.00043 | 93 | 0.00053 | -1.17613 | 151.005 | 0.24139 | 0.77376 |
| bofC; forespore regulator of the sigma-K checkpoint | 0.00071 | 0.00023 | 0.00014 | 0.00000 | 0.00007 | 65 | 0.00111 | 0.00001 | 0.00037 | 93 | 0.00038 | -2.54514 | 98.337 | 0.01248 | 0.59095 |
| mptB; alpha-1,6-mannosyltransferase [EC:2.4.1.-] | 0.00071 | 0.00021 | 0.00052 | 0.00000 | 0.00020 | 65 | 0.00085 | 0.00001 | 0.00033 | 93 | 0.00038 | -0.85544 | 145.250 | 0.39372 | 0.80459 |
| boxR, bzdR; XRE family transcriptional regulator, aerobic/anaerobic benzoate catabolism transcriptional regulator | 0.00071 | 0.00023 | 0.00018 | 0.00000 | 0.00006 | 65 | 0.00108 | 0.00001 | 0.00039 | 93 | 0.00040 | -2.27935 | 96.272 | 0.02485 | 0.59869 |
| dndD; DNA sulfur modification protein DndD | 0.00071 | 0.00018 | 0.00077 | 0.00001 | 0.00028 | 65 | 0.00066 | 0.00001 | 0.00023 | 93 | 0.00037 | 0.30116 | 137.417 | 0.76375 | 0.93846 |
| holE; DNA polymerase III subunit theta [EC:2.7.7.7] | 0.00071 | 0.00021 | 0.00082 | 0.00001 | 0.00039 | 65 | 0.00063 | 0.00001 | 0.00024 | 93 | 0.00046 | 0.40683 | 109.068 | 0.68493 | 0.91533 |
| bphI, xylK, nahM, tesG; 4-hydroxy-2-oxovalerate/4-hydroxy-2-oxohexanoate aldolase [EC:4.1.3.39 4.1.3.43] | 0.00071 | 0.00018 | 0.00068 | 0.00000 | 0.00024 | 65 | 0.00072 | 0.00001 | 0.00025 | 93 | 0.00035 | -0.11706 | 152.822 | 0.90697 | 0.97664 |
| bphJ, xylQ, nahO, tesF; acetaldehyde/propanal dehydrogenase [EC:1.2.1.10 1.2.1.87] | 0.00071 | 0.00018 | 0.00068 | 0.00000 | 0.00024 | 65 | 0.00072 | 0.00001 | 0.00025 | 93 | 0.00035 | -0.11706 | 152.822 | 0.90697 | 0.97664 |
| tqsA; AI-2 transport protein TqsA | 0.00071 | 0.00017 | 0.00076 | 0.00001 | 0.00030 | 65 | 0.00067 | 0.00000 | 0.00020 | 93 | 0.00036 | 0.24329 | 114.578 | 0.80822 | 0.95002 |
| pqqA; pyrroloquinoline quinone biosynthesis protein A | 0.00071 | 0.00028 | 0.00038 | 0.00001 | 0.00030 | 65 | 0.00094 | 0.00002 | 0.00043 | 93 | 0.00053 | -1.06277 | 151.131 | 0.28958 | 0.77376 |
| gsk; inosine kinase [EC:2.7.1.73] | 0.00071 | 0.00018 | 0.00088 | 0.00001 | 0.00031 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.79111 | 123.245 | 0.43040 | 0.82749 |
| vanRB, vanR, vanRD; two-component system, OmpR family, response regulator VanR | 0.00071 | 0.00030 | 0.00042 | 0.00001 | 0.00028 | 65 | 0.00091 | 0.00002 | 0.00047 | 93 | 0.00055 | -0.88414 | 142.202 | 0.37811 | 0.79852 |
| rutA; pyrimidine oxygenase [EC:1.14.99.46] | 0.00070 | 0.00017 | 0.00070 | 0.00000 | 0.00027 | 65 | 0.00071 | 0.00000 | 0.00022 | 93 | 0.00035 | -0.00963 | 135.472 | 0.99233 | 0.99908 |
| chpE; chemosensory pili system protein ChpE | 0.00070 | 0.00028 | 0.00037 | 0.00001 | 0.00030 | 65 | 0.00094 | 0.00002 | 0.00043 | 93 | 0.00053 | -1.08751 | 151.085 | 0.27854 | 0.77376 |
| fre, ubiB; aquacobalamin reductase / NAD(P)H-flavin reductase [EC:1.16.1.3 1.5.1.41] | 0.00070 | 0.00018 | 0.00087 | 0.00001 | 0.00031 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.76268 | 123.275 | 0.44711 | 0.83372 |
| hsaB; 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione monooxygenase reductase component [EC:1.5.1.-] | 0.00070 | 0.00022 | 0.00045 | 0.00000 | 0.00020 | 65 | 0.00088 | 0.00001 | 0.00035 | 93 | 0.00041 | -1.06579 | 140.040 | 0.28835 | 0.77376 |
| RGL4, rhiE; rhamnogalacturonan endolyase [EC:4.2.2.23] | 0.00070 | 0.00028 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00095 | 0.00002 | 0.00043 | 93 | 0.00052 | -1.14017 | 151.113 | 0.25602 | 0.77376 |
| atoS; two-component system, NtrC family, sensor histidine kinase AtoS [EC:2.7.13.3] | 0.00070 | 0.00035 | 0.00022 | 0.00000 | 0.00012 | 65 | 0.00103 | 0.00003 | 0.00058 | 93 | 0.00060 | -1.36511 | 99.994 | 0.17528 | 0.77376 |
| kinA; two-component system, sporulation sensor kinase A [EC:2.7.13.3] | 0.00070 | 0.00026 | 0.00017 | 0.00000 | 0.00007 | 65 | 0.00107 | 0.00002 | 0.00044 | 93 | 0.00044 | -2.02679 | 97.185 | 0.04542 | 0.65981 |
| rhlA; rhamnosyltransferase subunit A [EC:2.4.1.-] | 0.00070 | 0.00028 | 0.00036 | 0.00001 | 0.00030 | 65 | 0.00094 | 0.00002 | 0.00043 | 93 | 0.00053 | -1.10021 | 151.088 | 0.27299 | 0.77376 |
| ltnD; L-threonate 2-dehydrogenase [EC:1.1.1.411] | 0.00070 | 0.00020 | 0.00049 | 0.00000 | 0.00019 | 65 | 0.00084 | 0.00001 | 0.00032 | 93 | 0.00037 | -0.97314 | 144.204 | 0.33211 | 0.77868 |
| rutD; aminoacrylate hydrolase [EC:3.5.1.-] | 0.00070 | 0.00016 | 0.00063 | 0.00000 | 0.00025 | 65 | 0.00074 | 0.00000 | 0.00022 | 93 | 0.00033 | -0.34624 | 140.613 | 0.72968 | 0.92426 |
| dmpK, poxA; phenol hydroxylase P0 protein | 0.00070 | 0.00018 | 0.00081 | 0.00000 | 0.00027 | 65 | 0.00062 | 0.00001 | 0.00024 | 93 | 0.00036 | 0.53171 | 142.565 | 0.59575 | 0.88005 |
| dmpH, xylI, nahK; 2-oxo-3-hexenedioate decarboxylase [EC:4.1.1.77] | 0.00070 | 0.00022 | 0.00034 | 0.00000 | 0.00012 | 65 | 0.00094 | 0.00001 | 0.00037 | 93 | 0.00038 | -1.54903 | 109.474 | 0.12426 | 0.77376 |
| fldB; flavodoxin II | 0.00069 | 0.00018 | 0.00083 | 0.00001 | 0.00031 | 65 | 0.00060 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.61825 | 124.953 | 0.53754 | 0.86207 |
| scn, scin; staphylococcal complement inhibitor | 0.00069 | 0.00032 | 0.00020 | 0.00000 | 0.00014 | 65 | 0.00104 | 0.00003 | 0.00053 | 93 | 0.00055 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| gsmt; glycine/sarcosine N-methyltransferase [EC:2.1.1.156] | 0.00069 | 0.00028 | 0.00036 | 0.00001 | 0.00030 | 65 | 0.00092 | 0.00002 | 0.00043 | 93 | 0.00052 | -1.07232 | 151.128 | 0.28528 | 0.77376 |
| uhpT; MFS transporter, OPA family, hexose phosphate transport protein UhpT | 0.00069 | 0.00019 | 0.00042 | 0.00000 | 0.00015 | 65 | 0.00088 | 0.00001 | 0.00030 | 93 | 0.00033 | -1.36217 | 130.306 | 0.17549 | 0.77376 |
| uspF; universal stress protein F | 0.00069 | 0.00026 | 0.00075 | 0.00001 | 0.00038 | 65 | 0.00065 | 0.00001 | 0.00035 | 93 | 0.00052 | 0.20027 | 146.223 | 0.84155 | 0.95506 |
| tal-pgi; transaldolase / glucose-6-phosphate isomerase [EC:2.2.1.2 5.3.1.9] | 0.00069 | 0.00017 | 0.00048 | 0.00000 | 0.00021 | 65 | 0.00084 | 0.00001 | 0.00024 | 93 | 0.00032 | -1.14734 | 155.398 | 0.25300 | 0.77376 |
| csiD; protein CsiD | 0.00069 | 0.00018 | 0.00071 | 0.00000 | 0.00025 | 65 | 0.00068 | 0.00001 | 0.00025 | 93 | 0.00035 | 0.09838 | 150.466 | 0.92176 | 0.97905 |
| thiDE; hydroxymethylpyrimidine kinase / phosphomethylpyrimidine kinase / thiamine-phosphate diphosphorylase [EC:2.7.1.49 2.7.4.7 2.5.1.3] | 0.00069 | 0.00018 | 0.00083 | 0.00001 | 0.00037 | 65 | 0.00059 | 0.00000 | 0.00017 | 93 | 0.00041 | 0.57956 | 90.333 | 0.56365 | 0.86877 |
| pknG; serine/threonine-protein kinase PknG [EC:2.7.11.1] | 0.00069 | 0.00021 | 0.00044 | 0.00000 | 0.00019 | 65 | 0.00086 | 0.00001 | 0.00033 | 93 | 0.00038 | -1.11969 | 141.039 | 0.26475 | 0.77376 |
| AUP1; ancient ubiquitous protein 1 | 0.00069 | 0.00028 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00092 | 0.00002 | 0.00043 | 93 | 0.00052 | -1.09865 | 151.118 | 0.27367 | 0.77376 |
| chpD; AraC family transcriptional regulator, chemosensory pili system protein ChpD | 0.00069 | 0.00028 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00092 | 0.00002 | 0.00043 | 93 | 0.00052 | -1.09865 | 151.118 | 0.27367 | 0.77376 |
| K14331; fatty aldehyde decarbonylase [EC:4.1.99.5] | 0.00069 | 0.00028 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00092 | 0.00002 | 0.00043 | 93 | 0.00052 | -1.09865 | 151.118 | 0.27367 | 0.77376 |
| K16637, exoY; adenylate cyclase ExoY | 0.00069 | 0.00028 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00092 | 0.00002 | 0.00043 | 93 | 0.00052 | -1.09865 | 151.118 | 0.27367 | 0.77376 |
| K16638, exoU; exoenzyme U | 0.00069 | 0.00028 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00092 | 0.00002 | 0.00043 | 93 | 0.00052 | -1.09865 | 151.118 | 0.27367 | 0.77376 |
| pehX; exo-poly-alpha-galacturonosidase [EC:3.2.1.82] | 0.00069 | 0.00028 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00092 | 0.00002 | 0.00043 | 93 | 0.00052 | -1.09865 | 151.118 | 0.27367 | 0.77376 |
| rhaS; rhamnose transport system substrate-binding protein | 0.00069 | 0.00019 | 0.00027 | 0.00000 | 0.00016 | 65 | 0.00098 | 0.00001 | 0.00030 | 93 | 0.00034 | -2.04234 | 137.359 | 0.04303 | 0.65559 |
| E3.5.3.3; creatinase [EC:3.5.3.3] | 0.00069 | 0.00019 | 0.00091 | 0.00001 | 0.00033 | 65 | 0.00053 | 0.00000 | 0.00022 | 93 | 0.00040 | 0.95538 | 118.541 | 0.34133 | 0.78526 |
| ygiF; triphosphatase [EC:3.6.1.25] | 0.00068 | 0.00018 | 0.00083 | 0.00001 | 0.00031 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.65687 | 124.928 | 0.51247 | 0.85122 |
| cusF; Cu(I)/Ag(I) efflux system periplasmic protein CusF | 0.00068 | 0.00016 | 0.00087 | 0.00001 | 0.00029 | 65 | 0.00056 | 0.00000 | 0.00018 | 93 | 0.00034 | 0.92372 | 109.448 | 0.35766 | 0.79461 |
| crl; sigma factor-binding protein Crl | 0.00068 | 0.00018 | 0.00083 | 0.00001 | 0.00031 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.65205 | 124.911 | 0.51557 | 0.85216 |
| diaA; DnaA initiator-associating protein | 0.00068 | 0.00018 | 0.00083 | 0.00001 | 0.00031 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.65205 | 124.911 | 0.51557 | 0.85216 |
| syd; SecY interacting protein Syd | 0.00068 | 0.00018 | 0.00083 | 0.00001 | 0.00031 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.65205 | 124.911 | 0.51557 | 0.85216 |
| xni; protein Xni | 0.00068 | 0.00018 | 0.00083 | 0.00001 | 0.00031 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.65205 | 124.911 | 0.51557 | 0.85216 |
| zapC; cell division protein ZapC | 0.00068 | 0.00018 | 0.00083 | 0.00001 | 0.00031 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.65205 | 124.911 | 0.51557 | 0.85216 |
| hha; haemolysin expression modulating protein | 0.00068 | 0.00021 | 0.00065 | 0.00000 | 0.00026 | 65 | 0.00071 | 0.00001 | 0.00031 | 93 | 0.00040 | -0.16050 | 155.944 | 0.87269 | 0.96576 |
| K07504; predicted type IV restriction endonuclease | 0.00068 | 0.00011 | 0.00069 | 0.00000 | 0.00016 | 65 | 0.00067 | 0.00000 | 0.00015 | 93 | 0.00022 | 0.09702 | 150.281 | 0.92284 | 0.97947 |
| dinI; DNA-damage-inducible protein I | 0.00068 | 0.00021 | 0.00069 | 0.00000 | 0.00026 | 65 | 0.00068 | 0.00001 | 0.00030 | 93 | 0.00040 | 0.02592 | 155.807 | 0.97936 | 0.99657 |
| aftA; galactan 5-O-arabinofuranosyltransferase [EC:2.4.2.46] | 0.00068 | 0.00021 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00088 | 0.00001 | 0.00034 | 93 | 0.00039 | -1.27803 | 137.541 | 0.20339 | 0.77376 |
| hsaD; 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase [EC:3.7.1.17] | 0.00068 | 0.00022 | 0.00037 | 0.00000 | 0.00015 | 65 | 0.00089 | 0.00001 | 0.00035 | 93 | 0.00038 | -1.35799 | 121.766 | 0.17698 | 0.77376 |
| mdtH; MFS transporter, DHA1 family, multidrug resistance protein | 0.00068 | 0.00019 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00073 | 0.00001 | 0.00027 | 93 | 0.00037 | -0.38806 | 154.030 | 0.69850 | 0.91844 |
| K07280; outer membrane protein | 0.00068 | 0.00018 | 0.00070 | 0.00001 | 0.00028 | 65 | 0.00066 | 0.00001 | 0.00023 | 93 | 0.00036 | 0.12183 | 138.232 | 0.90321 | 0.97641 |
| K07033; uncharacterized protein | 0.00068 | 0.00017 | 0.00048 | 0.00000 | 0.00015 | 65 | 0.00081 | 0.00001 | 0.00027 | 93 | 0.00031 | -1.07064 | 136.335 | 0.28622 | 0.77376 |
| pagP, crcA; lipid IVA palmitoyltransferase [EC:2.3.1.251] | 0.00067 | 0.00018 | 0.00073 | 0.00001 | 0.00028 | 65 | 0.00064 | 0.00000 | 0.00023 | 93 | 0.00036 | 0.25114 | 133.152 | 0.80209 | 0.94855 |
| frcA; fructose transport system ATP-binding protein | 0.00067 | 0.00023 | 0.00017 | 0.00000 | 0.00012 | 65 | 0.00103 | 0.00001 | 0.00037 | 93 | 0.00039 | -2.17607 | 110.838 | 0.03167 | 0.61842 |
| MDH1; malate dehydrogenase [EC:1.1.1.37] | 0.00067 | 0.00018 | 0.00070 | 0.00001 | 0.00028 | 65 | 0.00065 | 0.00001 | 0.00023 | 93 | 0.00036 | 0.13388 | 138.274 | 0.89369 | 0.97159 |
| rcsF; RcsF protein | 0.00067 | 0.00018 | 0.00080 | 0.00001 | 0.00031 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.57541 | 124.316 | 0.56605 | 0.87051 |
| ttrB; tetrathionate reductase subunit B | 0.00067 | 0.00020 | 0.00069 | 0.00001 | 0.00032 | 65 | 0.00066 | 0.00001 | 0.00026 | 93 | 0.00041 | 0.05913 | 134.125 | 0.95294 | 0.98792 |
| fdrA; FdrA protein | 0.00067 | 0.00031 | 0.00055 | 0.00001 | 0.00030 | 65 | 0.00075 | 0.00002 | 0.00048 | 93 | 0.00057 | -0.35239 | 145.937 | 0.72506 | 0.92177 |
| pbuE; MFS transporter, DHA1 family, purine base/nucleoside efflux pump | 0.00067 | 0.00030 | 0.00037 | 0.00001 | 0.00028 | 65 | 0.00088 | 0.00002 | 0.00048 | 93 | 0.00056 | -0.91259 | 140.985 | 0.36302 | 0.79496 |
| aprX; serine protease AprX [EC:3.4.21.-] | 0.00067 | 0.00023 | 0.00016 | 0.00000 | 0.00007 | 65 | 0.00103 | 0.00001 | 0.00039 | 93 | 0.00039 | -2.21043 | 97.616 | 0.02941 | 0.61334 |
| aprA; adenylylsulfate reductase, subunit A [EC:1.8.99.2] | 0.00067 | 0.00017 | 0.00068 | 0.00001 | 0.00029 | 65 | 0.00066 | 0.00000 | 0.00020 | 93 | 0.00035 | 0.03492 | 119.829 | 0.97220 | 0.99451 |
| aprB; adenylylsulfate reductase, subunit B [EC:1.8.99.2] | 0.00067 | 0.00017 | 0.00068 | 0.00001 | 0.00029 | 65 | 0.00066 | 0.00000 | 0.00020 | 93 | 0.00035 | 0.03492 | 119.829 | 0.97220 | 0.99451 |
| E1.1.1.53; 3alpha(or 20beta)-hydroxysteroid dehydrogenase [EC:1.1.1.53] | 0.00067 | 0.00021 | 0.00013 | 0.00000 | 0.00005 | 65 | 0.00104 | 0.00001 | 0.00036 | 93 | 0.00036 | -2.51321 | 96.145 | 0.01363 | 0.59095 |
| pep2; maltokinase [EC:2.7.1.175] | 0.00067 | 0.00013 | 0.00058 | 0.00000 | 0.00015 | 65 | 0.00073 | 0.00000 | 0.00019 | 93 | 0.00024 | -0.61899 | 155.953 | 0.53682 | 0.86207 |
| hyfC; hydrogenase-4 component C [EC:1.-.-.-] | 0.00067 | 0.00018 | 0.00084 | 0.00001 | 0.00035 | 65 | 0.00055 | 0.00000 | 0.00019 | 93 | 0.00040 | 0.72769 | 99.504 | 0.46851 | 0.83837 |
| liaF; lia operon protein LiaF | 0.00067 | 0.00017 | 0.00023 | 0.00000 | 0.00013 | 65 | 0.00097 | 0.00001 | 0.00028 | 93 | 0.00031 | -2.39685 | 126.673 | 0.01800 | 0.59095 |
| csm1, cas10; CRISPR-associated protein Csm1 | 0.00067 | 0.00022 | 0.00057 | 0.00000 | 0.00015 | 65 | 0.00074 | 0.00001 | 0.00037 | 93 | 0.00039 | -0.43094 | 119.470 | 0.66729 | 0.91150 |
| appA; 4-phytase / acid phosphatase [EC:3.1.3.26 3.1.3.2] | 0.00066 | 0.00017 | 0.00061 | 0.00000 | 0.00026 | 65 | 0.00070 | 0.00000 | 0.00022 | 93 | 0.00034 | -0.25036 | 138.410 | 0.80268 | 0.94869 |
| aftC; arabinofuranan 3-O-arabinosyltransferase [EC:2.4.2.47] | 0.00066 | 0.00021 | 0.00038 | 0.00000 | 0.00018 | 65 | 0.00085 | 0.00001 | 0.00033 | 93 | 0.00038 | -1.24822 | 139.497 | 0.21404 | 0.77376 |
| glnQ; glutamine transport system ATP-binding protein [EC:3.6.3.-] | 0.00066 | 0.00014 | 0.00073 | 0.00000 | 0.00025 | 65 | 0.00061 | 0.00000 | 0.00017 | 93 | 0.00030 | 0.41403 | 119.691 | 0.67960 | 0.91311 |
| SRD5A1; 3-oxo-5-alpha-steroid 4-dehydrogenase 1 [EC:1.3.1.22] | 0.00066 | 0.00019 | 0.00030 | 0.00000 | 0.00014 | 65 | 0.00091 | 0.00001 | 0.00030 | 93 | 0.00033 | -1.83811 | 126.611 | 0.06839 | 0.73473 |
| acpH; acyl carrier protein phosphodiesterase [EC:3.1.4.14] | 0.00066 | 0.00018 | 0.00077 | 0.00001 | 0.00030 | 65 | 0.00058 | 0.00000 | 0.00022 | 93 | 0.00038 | 0.49115 | 125.111 | 0.62418 | 0.89178 |
| bmr; MFS transporter, DHA1 family, multidrug resistance protein | 0.00066 | 0.00032 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00087 | 0.00002 | 0.00050 | 93 | 0.00058 | -0.90079 | 143.877 | 0.36921 | 0.79587 |
| araC; AraC family transcriptional regulator, arabinose operon regulatory protein | 0.00066 | 0.00014 | 0.00062 | 0.00000 | 0.00022 | 65 | 0.00068 | 0.00000 | 0.00019 | 93 | 0.00029 | -0.23805 | 140.803 | 0.81219 | 0.95246 |
| E1.14.14.1; unspecific monooxygenase [EC:1.14.14.1] | 0.00066 | 0.00023 | 0.00013 | 0.00000 | 0.00005 | 65 | 0.00102 | 0.00001 | 0.00039 | 93 | 0.00040 | -2.23577 | 95.399 | 0.02770 | 0.61334 |
| tipF; cyclic-di-GMP phosphodiesterase, flagellum assembly factor TipF | 0.00065 | 0.00017 | 0.00032 | 0.00000 | 0.00010 | 65 | 0.00089 | 0.00001 | 0.00028 | 93 | 0.00029 | -1.91890 | 113.063 | 0.05752 | 0.71337 |
| zntR; MerR family transcriptional regulator, Zn(II)-responsive regulator of zntA | 0.00065 | 0.00017 | 0.00078 | 0.00001 | 0.00029 | 65 | 0.00057 | 0.00000 | 0.00022 | 93 | 0.00036 | 0.59443 | 128.633 | 0.55327 | 0.86788 |
| purNH; phosphoribosylglycinamide/phosphoribosylaminoimidazolecarboxamide formyltransferase [EC:2.1.2.2 2.1.2.3] | 0.00065 | 0.00016 | 0.00035 | 0.00000 | 0.00010 | 65 | 0.00086 | 0.00001 | 0.00027 | 93 | 0.00029 | -1.79302 | 114.852 | 0.07560 | 0.74536 |
| rutC; aminoacrylate peracid reductase | 0.00065 | 0.00016 | 0.00067 | 0.00000 | 0.00025 | 65 | 0.00064 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.08907 | 135.989 | 0.92915 | 0.98183 |
| ABC.NGC.P; N-acetylglucosamine transport system permease protein | 0.00065 | 0.00024 | 0.00062 | 0.00001 | 0.00044 | 65 | 0.00067 | 0.00001 | 0.00027 | 93 | 0.00051 | -0.08955 | 111.237 | 0.92881 | 0.98176 |
| ABC.NGC.S; N-acetylglucosamine transport system substrate-binding protein | 0.00065 | 0.00024 | 0.00062 | 0.00001 | 0.00044 | 65 | 0.00067 | 0.00001 | 0.00027 | 93 | 0.00051 | -0.08955 | 111.237 | 0.92881 | 0.98176 |
| rfbG; rhamnosyltransferase [EC:2.4.1.-] | 0.00065 | 0.00011 | 0.00077 | 0.00000 | 0.00021 | 65 | 0.00056 | 0.00000 | 0.00013 | 93 | 0.00024 | 0.84189 | 111.938 | 0.40164 | 0.80850 |
| idnD; L-idonate 5-dehydrogenase [EC:1.1.1.264] | 0.00064 | 0.00015 | 0.00043 | 0.00000 | 0.00015 | 65 | 0.00079 | 0.00000 | 0.00023 | 93 | 0.00027 | -1.32270 | 146.477 | 0.18800 | 0.77376 |
| hsaC; 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione 4,5-dioxygenase [EC:1.13.11.25] | 0.00064 | 0.00021 | 0.00036 | 0.00000 | 0.00015 | 65 | 0.00084 | 0.00001 | 0.00035 | 93 | 0.00038 | -1.26251 | 122.990 | 0.20916 | 0.77376 |
| gsiA; glutathione transport system ATP-binding protein | 0.00064 | 0.00015 | 0.00048 | 0.00000 | 0.00020 | 65 | 0.00076 | 0.00000 | 0.00021 | 93 | 0.00029 | -0.98578 | 153.091 | 0.32580 | 0.77376 |
| hmo, nocN; 4-hydroxymandelate oxidase [EC:1.1.3.46] | 0.00064 | 0.00025 | 0.00051 | 0.00000 | 0.00020 | 65 | 0.00073 | 0.00001 | 0.00039 | 93 | 0.00044 | -0.49589 | 133.266 | 0.62079 | 0.89082 |
| yraD; similar to spore coat protein | 0.00064 | 0.00021 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00105 | 0.00001 | 0.00036 | 93 | 0.00036 | -2.83296 | 92.531 | 0.00566 | 0.54022 |
| soxB; sulfur-oxidizing protein SoxB | 0.00063 | 0.00016 | 0.00029 | 0.00000 | 0.00011 | 65 | 0.00087 | 0.00001 | 0.00027 | 93 | 0.00029 | -1.99632 | 123.515 | 0.04810 | 0.66712 |
| cbiK; nickel transport protein | 0.00063 | 0.00020 | 0.00073 | 0.00001 | 0.00033 | 65 | 0.00056 | 0.00001 | 0.00026 | 93 | 0.00042 | 0.41553 | 132.173 | 0.67842 | 0.91311 |
| aftD; arabinofuranan 3-O-arabinosyltransferase [EC:2.4.2.-] | 0.00063 | 0.00021 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00079 | 0.00001 | 0.00032 | 93 | 0.00037 | -1.03932 | 140.403 | 0.30044 | 0.77376 |
| boxB; benzoyl-CoA 2,3-epoxidase subunit B [EC:1.14.13.208] | 0.00063 | 0.00022 | 0.00017 | 0.00000 | 0.00006 | 65 | 0.00095 | 0.00001 | 0.00037 | 93 | 0.00038 | -2.08155 | 96.745 | 0.04002 | 0.63577 |
| boxC; benzoyl-CoA-dihydrodiol lyase [EC:4.1.2.44] | 0.00063 | 0.00022 | 0.00017 | 0.00000 | 0.00006 | 65 | 0.00095 | 0.00001 | 0.00037 | 93 | 0.00038 | -2.08155 | 96.745 | 0.04002 | 0.63577 |
| nirS; nitrite reductase (NO-forming) / hydroxylamine reductase [EC:1.7.2.1 1.7.99.1] | 0.00063 | 0.00022 | 0.00029 | 0.00000 | 0.00012 | 65 | 0.00086 | 0.00001 | 0.00036 | 93 | 0.00038 | -1.51316 | 112.721 | 0.13304 | 0.77376 |
| aftB; arabinofuranosyltransferase [EC:2.4.2.-] | 0.00063 | 0.00021 | 0.00036 | 0.00000 | 0.00018 | 65 | 0.00081 | 0.00001 | 0.00033 | 93 | 0.00037 | -1.19669 | 138.370 | 0.23347 | 0.77376 |
| aspC, aspS; nondiscriminating aspartyl-tRNA synthetase [EC:6.1.1.23] | 0.00063 | 0.00022 | 0.00048 | 0.00001 | 0.00036 | 65 | 0.00073 | 0.00001 | 0.00028 | 93 | 0.00045 | -0.53918 | 131.217 | 0.59068 | 0.87896 |
| K07076; uncharacterized protein | 0.00062 | 0.00023 | 0.00071 | 0.00001 | 0.00042 | 65 | 0.00057 | 0.00001 | 0.00026 | 93 | 0.00049 | 0.27815 | 112.671 | 0.78141 | 0.94310 |
| K09776; uncharacterized protein | 0.00062 | 0.00017 | 0.00011 | 0.00000 | 0.00008 | 65 | 0.00098 | 0.00001 | 0.00029 | 93 | 0.00030 | -2.92865 | 104.478 | 0.00418 | 0.52172 |
| rhaP; rhamnose transport system permease protein | 0.00062 | 0.00018 | 0.00028 | 0.00000 | 0.00016 | 65 | 0.00086 | 0.00001 | 0.00029 | 93 | 0.00033 | -1.76156 | 141.115 | 0.08031 | 0.75920 |
| rof; Rho-binding antiterminator | 0.00062 | 0.00017 | 0.00066 | 0.00000 | 0.00026 | 65 | 0.00060 | 0.00000 | 0.00022 | 93 | 0.00034 | 0.19647 | 141.707 | 0.84452 | 0.95560 |
| lat; L-lysine 6-transaminase [EC:2.6.1.36] | 0.00062 | 0.00021 | 0.00034 | 0.00000 | 0.00012 | 65 | 0.00082 | 0.00001 | 0.00034 | 93 | 0.00036 | -1.31801 | 113.292 | 0.19016 | 0.77376 |
| MEMO1; MEMO1 family protein | 0.00062 | 0.00016 | 0.00075 | 0.00001 | 0.00030 | 65 | 0.00053 | 0.00000 | 0.00016 | 93 | 0.00035 | 0.64112 | 101.072 | 0.52290 | 0.85702 |
| traG; conjugal transfer mating pair stabilization protein TraG | 0.00062 | 0.00019 | 0.00063 | 0.00001 | 0.00034 | 65 | 0.00061 | 0.00000 | 0.00022 | 93 | 0.00041 | 0.05612 | 113.939 | 0.95535 | 0.98792 |
| sgrR; SgrR family transcriptional regulator | 0.00062 | 0.00017 | 0.00065 | 0.00000 | 0.00025 | 65 | 0.00060 | 0.00000 | 0.00023 | 93 | 0.00034 | 0.14288 | 143.427 | 0.88659 | 0.97020 |
| ureI; acid-activated urea channel | 0.00062 | 0.00034 | 0.00020 | 0.00000 | 0.00012 | 65 | 0.00091 | 0.00003 | 0.00057 | 93 | 0.00059 | -1.22420 | 100.260 | 0.22375 | 0.77376 |
| E1.12.7.2; ferredoxin hydrogenase [EC:1.12.7.2] | 0.00062 | 0.00050 | 0.00123 | 0.00010 | 0.00121 | 65 | 0.00019 | 0.00000 | 0.00011 | 93 | 0.00122 | 0.84950 | 65.102 | 0.39872 | 0.80624 |
| ECE; endothelin-converting enzyme [EC:3.4.24.71] | 0.00062 | 0.00016 | 0.00047 | 0.00000 | 0.00021 | 65 | 0.00072 | 0.00000 | 0.00023 | 93 | 0.00031 | -0.81716 | 155.074 | 0.41509 | 0.81730 |
| PRHOXNB, URAD; 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase [EC:4.1.1.97] | 0.00062 | 0.00018 | 0.00039 | 0.00000 | 0.00018 | 65 | 0.00077 | 0.00001 | 0.00028 | 93 | 0.00033 | -1.13634 | 147.334 | 0.25766 | 0.77376 |
| bjaI, rpaI, braI, rhiI; acyl-homoserine lactone synthase [EC:2.3.1.228 2.3.1.229 2.3.1.-] | 0.00061 | 0.00017 | 0.00042 | 0.00001 | 0.00032 | 65 | 0.00075 | 0.00000 | 0.00019 | 93 | 0.00037 | -0.87460 | 107.013 | 0.38375 | 0.80172 |
| cueO; blue copper oxidase | 0.00061 | 0.00020 | 0.00044 | 0.00000 | 0.00019 | 65 | 0.00073 | 0.00001 | 0.00031 | 93 | 0.00036 | -0.79064 | 145.712 | 0.43044 | 0.82749 |
| egtD; L-histidine Nalpha-methyltransferase [EC:2.1.1.44] | 0.00061 | 0.00021 | 0.00032 | 0.00000 | 0.00014 | 65 | 0.00082 | 0.00001 | 0.00034 | 93 | 0.00036 | -1.35612 | 120.996 | 0.17759 | 0.77376 |
| K07228; TrkA domain protein | 0.00061 | 0.00022 | 0.00039 | 0.00000 | 0.00018 | 65 | 0.00076 | 0.00001 | 0.00036 | 93 | 0.00040 | -0.91617 | 132.300 | 0.36125 | 0.79496 |
| E3.1.1.22; hydroxybutyrate-dimer hydrolase [EC:3.1.1.22] | 0.00061 | 0.00027 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00093 | 0.00002 | 0.00045 | 93 | 0.00046 | -1.68473 | 96.124 | 0.09528 | 0.76609 |
| fdnG; formate dehydrogenase-N, alpha subunit [EC:1.17.5.3] | 0.00061 | 0.00019 | 0.00063 | 0.00000 | 0.00025 | 65 | 0.00059 | 0.00001 | 0.00027 | 93 | 0.00037 | 0.10408 | 154.318 | 0.91724 | 0.97692 |
| cag12; cag pathogenicity island protein 12 | 0.00061 | 0.00035 | 0.00039 | 0.00000 | 0.00023 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00061 | -0.61634 | 119.253 | 0.53884 | 0.86207 |
| aphD; aminoglycoside 2’’-phosphotransferase [EC:2.7.1.190] | 0.00061 | 0.00030 | 0.00032 | 0.00000 | 0.00028 | 65 | 0.00081 | 0.00002 | 0.00048 | 93 | 0.00055 | -0.88375 | 141.425 | 0.37833 | 0.79852 |
| frcB; fructose transport system substrate-binding protein | 0.00061 | 0.00021 | 0.00017 | 0.00000 | 0.00012 | 65 | 0.00091 | 0.00001 | 0.00035 | 93 | 0.00037 | -2.00459 | 113.526 | 0.04739 | 0.66255 |
| frcC; fructose transport system permease protein | 0.00061 | 0.00021 | 0.00017 | 0.00000 | 0.00012 | 65 | 0.00091 | 0.00001 | 0.00035 | 93 | 0.00037 | -2.00459 | 113.526 | 0.04739 | 0.66255 |
| csiR; GntR family transcriptional regulator, carbon starvation induced regulator | 0.00061 | 0.00017 | 0.00071 | 0.00000 | 0.00025 | 65 | 0.00054 | 0.00000 | 0.00022 | 93 | 0.00033 | 0.51315 | 144.236 | 0.60863 | 0.88654 |
| dmpC, xylG, praB; aminomuconate-semialdehyde/2-hydroxymuconate-6-semialdehyde dehydrogenase [EC:1.2.1.32 1.2.1.85] | 0.00060 | 0.00020 | 0.00044 | 0.00000 | 0.00016 | 65 | 0.00072 | 0.00001 | 0.00033 | 93 | 0.00036 | -0.77951 | 129.181 | 0.43711 | 0.83054 |
| fucR; DeoR family transcriptional regulator, L-fucose operon activator | 0.00060 | 0.00017 | 0.00039 | 0.00000 | 0.00011 | 65 | 0.00075 | 0.00001 | 0.00028 | 93 | 0.00030 | -1.18274 | 117.213 | 0.23931 | 0.77376 |
| yihV; sulfofructose kinase [EC:2.7.1.184] | 0.00060 | 0.00023 | 0.00017 | 0.00000 | 0.00006 | 65 | 0.00090 | 0.00001 | 0.00039 | 93 | 0.00039 | -1.85429 | 96.502 | 0.06675 | 0.73334 |
| lmrP; MFS transporter, DHA1 family, multidrug resistance protein B | 0.00060 | 0.00020 | 0.00042 | 0.00000 | 0.00025 | 65 | 0.00073 | 0.00001 | 0.00028 | 93 | 0.00038 | -0.79989 | 155.205 | 0.42500 | 0.82542 |
| K09968; uncharacterized protein | 0.00060 | 0.00025 | 0.00014 | 0.00000 | 0.00005 | 65 | 0.00092 | 0.00002 | 0.00042 | 93 | 0.00042 | -1.85141 | 94.895 | 0.06722 | 0.73334 |
| nirC; cytochrome c55X | 0.00060 | 0.00022 | 0.00026 | 0.00000 | 0.00011 | 65 | 0.00083 | 0.00001 | 0.00036 | 93 | 0.00037 | -1.52253 | 109.697 | 0.13076 | 0.77376 |
| nirF; protein NirF | 0.00060 | 0.00022 | 0.00026 | 0.00000 | 0.00011 | 65 | 0.00083 | 0.00001 | 0.00036 | 93 | 0.00037 | -1.52253 | 109.697 | 0.13076 | 0.77376 |
| ccoNO; cytochrome c oxidase cbb3-type subunit I/II [EC:1.9.3.1] | 0.00060 | 0.00015 | 0.00053 | 0.00000 | 0.00015 | 65 | 0.00064 | 0.00001 | 0.00023 | 93 | 0.00028 | -0.39317 | 149.086 | 0.69475 | 0.91715 |
| mtnW; 2,3-diketo-5-methylthiopentyl-1-phosphate enolase [EC:5.3.2.5] | 0.00060 | 0.00022 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00097 | 0.00001 | 0.00036 | 93 | 0.00036 | -2.49769 | 93.532 | 0.01425 | 0.59095 |
| fadD36; fatty acid CoA ligase FadD36 | 0.00060 | 0.00036 | 0.00017 | 0.00000 | 0.00009 | 65 | 0.00089 | 0.00003 | 0.00061 | 93 | 0.00062 | -1.17652 | 95.638 | 0.24231 | 0.77376 |
| dszB; 2’-hydroxybiphenyl-2-sulfinate desulfinase [EC:3.13.1.3] | 0.00060 | 0.00028 | 0.00028 | 0.00000 | 0.00009 | 65 | 0.00082 | 0.00002 | 0.00047 | 93 | 0.00048 | -1.13416 | 98.940 | 0.25947 | 0.77376 |
| tgl; protein-glutamine gamma-glutamyltransferase [EC:2.3.2.13] | 0.00060 | 0.00022 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00097 | 0.00001 | 0.00036 | 93 | 0.00036 | -2.50011 | 93.534 | 0.01416 | 0.59095 |
| glnH; glutamine transport system substrate-binding protein | 0.00060 | 0.00014 | 0.00066 | 0.00000 | 0.00025 | 65 | 0.00055 | 0.00000 | 0.00017 | 93 | 0.00030 | 0.35677 | 118.195 | 0.72190 | 0.92156 |
| racA; chromosome-anchoring protein RacA | 0.00059 | 0.00022 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00097 | 0.00001 | 0.00036 | 93 | 0.00036 | -2.52764 | 93.400 | 0.01316 | 0.59095 |
| emrD; MFS transporter, DHA1 family, 2-module integral membrane pump EmrD | 0.00059 | 0.00017 | 0.00069 | 0.00000 | 0.00026 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00034 | 0.50944 | 136.242 | 0.61127 | 0.88722 |
| cruF; bisanhydrobacterioruberin hydratase [EC:4.2.1.161] | 0.00059 | 0.00023 | 0.00027 | 0.00000 | 0.00014 | 65 | 0.00081 | 0.00001 | 0.00037 | 93 | 0.00039 | -1.37223 | 115.520 | 0.17265 | 0.77376 |
| fadD32; fatty acid CoA ligase FadD32 | 0.00059 | 0.00020 | 0.00037 | 0.00000 | 0.00018 | 65 | 0.00075 | 0.00001 | 0.00032 | 93 | 0.00037 | -1.00694 | 140.140 | 0.31570 | 0.77376 |
| pks13; polyketide synthase 13 | 0.00059 | 0.00020 | 0.00037 | 0.00000 | 0.00018 | 65 | 0.00075 | 0.00001 | 0.00032 | 93 | 0.00037 | -1.00694 | 140.140 | 0.31570 | 0.77376 |
| hycH; formate hydrogenlyase maturation protein HycH | 0.00059 | 0.00013 | 0.00057 | 0.00000 | 0.00018 | 65 | 0.00060 | 0.00000 | 0.00018 | 93 | 0.00026 | -0.11706 | 150.561 | 0.90697 | 0.97664 |
| PTH2; peptidyl-tRNA hydrolase, PTH2 family [EC:3.1.1.29] | 0.00059 | 0.00015 | 0.00047 | 0.00000 | 0.00021 | 65 | 0.00067 | 0.00000 | 0.00021 | 93 | 0.00030 | -0.66080 | 152.590 | 0.50974 | 0.84956 |
| csy2; CRISPR-associated protein Csy2 | 0.00059 | 0.00015 | 0.00074 | 0.00000 | 0.00024 | 65 | 0.00048 | 0.00000 | 0.00019 | 93 | 0.00030 | 0.84985 | 131.203 | 0.39696 | 0.80459 |
| csy4, cas6f; CRISPR-associated endonuclease Csy4 [EC:3.1.-.-] | 0.00059 | 0.00015 | 0.00074 | 0.00000 | 0.00024 | 65 | 0.00048 | 0.00000 | 0.00019 | 93 | 0.00030 | 0.84985 | 131.203 | 0.39696 | 0.80459 |
| hyfD; hydrogenase-4 component D [EC:1.-.-.-] | 0.00058 | 0.00029 | 0.00091 | 0.00003 | 0.00064 | 65 | 0.00036 | 0.00000 | 0.00020 | 93 | 0.00067 | 0.83287 | 77.404 | 0.40748 | 0.81200 |
| E3.5.1.82; N-acyl-D-glutamate deacylase [EC:3.5.1.82] | 0.00058 | 0.00023 | 0.00044 | 0.00000 | 0.00024 | 65 | 0.00068 | 0.00001 | 0.00035 | 93 | 0.00042 | -0.57478 | 151.154 | 0.56629 | 0.87051 |
| lhpI; cis-L-3-hydroxyproline dehydratase [EC:4.2.1.171] | 0.00058 | 0.00016 | 0.00063 | 0.00000 | 0.00026 | 65 | 0.00055 | 0.00000 | 0.00019 | 93 | 0.00033 | 0.22141 | 125.827 | 0.82513 | 0.95506 |
| hndB; NADP-reducing hydrogenase subunit HndB [EC:1.12.1.3] | 0.00058 | 0.00020 | 0.00041 | 0.00000 | 0.00021 | 65 | 0.00071 | 0.00001 | 0.00030 | 93 | 0.00037 | -0.82103 | 153.235 | 0.41290 | 0.81655 |
| dpiA, citB; two-component system, CitB family, response regulator CitB | 0.00058 | 0.00019 | 0.00073 | 0.00001 | 0.00038 | 65 | 0.00048 | 0.00000 | 0.00018 | 93 | 0.00042 | 0.58177 | 91.351 | 0.56216 | 0.86813 |
| arsC; arsenate-mycothiol transferase [EC:2.8.4.2] | 0.00058 | 0.00021 | 0.00040 | 0.00000 | 0.00020 | 65 | 0.00071 | 0.00001 | 0.00032 | 93 | 0.00038 | -0.80075 | 145.533 | 0.42458 | 0.82505 |
| kdgR; IclR family transcriptional regulator, KDG regulon repressor | 0.00058 | 0.00015 | 0.00070 | 0.00001 | 0.00029 | 65 | 0.00049 | 0.00000 | 0.00016 | 93 | 0.00033 | 0.62522 | 102.485 | 0.53322 | 0.86207 |
| K17203, eryF; erythritol transport system permease protein | 0.00058 | 0.00021 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00092 | 0.00001 | 0.00035 | 93 | 0.00036 | -2.37075 | 93.179 | 0.01981 | 0.59095 |
| wbpD, wlbB; UDP-2-acetamido-3-amino-2,3-dideoxy-glucuronate N-acetyltransferase [EC:2.3.1.201] | 0.00058 | 0.00012 | 0.00054 | 0.00000 | 0.00019 | 65 | 0.00060 | 0.00000 | 0.00017 | 93 | 0.00025 | -0.23456 | 140.954 | 0.81489 | 0.95409 |
| ycfS; L,D-transpeptidase YcfS | 0.00058 | 0.00016 | 0.00065 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.39677 | 138.661 | 0.69214 | 0.91676 |
| eryE; erythritol transport system ATP-binding protein | 0.00058 | 0.00021 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00092 | 0.00001 | 0.00035 | 93 | 0.00036 | -2.35597 | 93.178 | 0.02057 | 0.59095 |
| K17202, eryG; erythritol transport system substrate-binding protein | 0.00058 | 0.00021 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00092 | 0.00001 | 0.00035 | 93 | 0.00036 | -2.35597 | 93.178 | 0.02057 | 0.59095 |
| vanSB, vanS, vanSD; two-component system, OmpR family, sensor histidine kinase VanS [EC:2.7.13.3] | 0.00057 | 0.00030 | 0.00031 | 0.00000 | 0.00028 | 65 | 0.00076 | 0.00002 | 0.00047 | 93 | 0.00054 | -0.83592 | 142.601 | 0.40460 | 0.81144 |
| ABC.VB12.A, btuD; vitamin B12 transport system ATP-binding protein [EC:3.6.3.33] | 0.00057 | 0.00016 | 0.00065 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.38203 | 138.647 | 0.70302 | 0.92004 |
| ABC.VB12.P, btuC; vitamin B12 transport system permease protein | 0.00057 | 0.00016 | 0.00065 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.38203 | 138.647 | 0.70302 | 0.92004 |
| metL; bifunctional aspartokinase / homoserine dehydrogenase 2 [EC:2.7.2.4 1.1.1.3] | 0.00057 | 0.00016 | 0.00065 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.38203 | 138.647 | 0.70302 | 0.92004 |
| ytfB; uncharacterized protein | 0.00057 | 0.00016 | 0.00065 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.38203 | 138.647 | 0.70302 | 0.92004 |
| chpS, chpBI; antitoxin ChpS | 0.00057 | 0.00022 | 0.00056 | 0.00000 | 0.00027 | 65 | 0.00058 | 0.00001 | 0.00033 | 93 | 0.00043 | -0.04421 | 155.949 | 0.96480 | 0.99101 |
| toa; taurine—2-oxoglutarate transaminase [EC:2.6.1.55] | 0.00057 | 0.00012 | 0.00052 | 0.00000 | 0.00022 | 65 | 0.00061 | 0.00000 | 0.00015 | 93 | 0.00026 | -0.30924 | 117.390 | 0.75768 | 0.93719 |
| psmB, prcB; proteasome beta subunit [EC:3.4.25.1] | 0.00057 | 0.00020 | 0.00042 | 0.00000 | 0.00015 | 65 | 0.00068 | 0.00001 | 0.00031 | 93 | 0.00035 | -0.75258 | 129.041 | 0.45308 | 0.83438 |
| cpxP; periplasmic protein CpxP | 0.00057 | 0.00016 | 0.00064 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.36721 | 138.602 | 0.71402 | 0.92004 |
| uspB; universal stress protein B | 0.00057 | 0.00016 | 0.00064 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.36721 | 138.602 | 0.71402 | 0.92004 |
| bmaC; fibronectin-binding autotransporter adhesin | 0.00057 | 0.00032 | 0.00013 | 0.00000 | 0.00005 | 65 | 0.00088 | 0.00003 | 0.00054 | 93 | 0.00054 | -1.39095 | 93.473 | 0.16754 | 0.77376 |
| mltE, emtA; membrane-bound lytic murein transglycosylase E [EC:4.2.2.-] | 0.00057 | 0.00017 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00056 | 0.00000 | 0.00022 | 93 | 0.00034 | 0.08534 | 142.433 | 0.93211 | 0.98385 |
| psmA, prcA; proteasome alpha subunit [EC:3.4.25.1] | 0.00057 | 0.00020 | 0.00042 | 0.00000 | 0.00015 | 65 | 0.00068 | 0.00001 | 0.00031 | 93 | 0.00035 | -0.74361 | 129.042 | 0.45846 | 0.83438 |
| mhpC; 2-hydroxy-6-oxonona-2,4-dienedioate hydrolase [EC:3.7.1.14] | 0.00057 | 0.00020 | 0.00025 | 0.00000 | 0.00010 | 65 | 0.00079 | 0.00001 | 0.00033 | 93 | 0.00035 | -1.55793 | 106.829 | 0.12221 | 0.77376 |
| wecG, rffM; UDP-N-acetyl-D-mannosaminouronate:lipid I N-acetyl-D-mannosaminouronosyltransferase [EC:2.4.1.180] | 0.00057 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00056 | 0.00000 | 0.00022 | 93 | 0.00033 | 0.09915 | 140.639 | 0.92116 | 0.97905 |
| mttB; trimethylamine—corrinoid protein Co-methyltransferase [EC:2.1.1.250] | 0.00057 | 0.00027 | 0.00011 | 0.00000 | 0.00009 | 65 | 0.00089 | 0.00002 | 0.00046 | 93 | 0.00047 | -1.68315 | 98.838 | 0.09550 | 0.76609 |
| symE; toxic protein SymE | 0.00057 | 0.00016 | 0.00057 | 0.00000 | 0.00024 | 65 | 0.00057 | 0.00000 | 0.00021 | 93 | 0.00032 | 0.00712 | 143.433 | 0.99433 | 0.99919 |
| sufA; Fe-S cluster assembly protein SufA | 0.00057 | 0.00016 | 0.00063 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.33426 | 138.537 | 0.73869 | 0.92914 |
| leuO; LysR family transcriptional regulator, transcriptional activator for leuABCD operon | 0.00057 | 0.00016 | 0.00064 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.36629 | 138.612 | 0.71471 | 0.92004 |
| terB; tellurite resistance protein TerB | 0.00056 | 0.00022 | 0.00048 | 0.00000 | 0.00020 | 65 | 0.00062 | 0.00001 | 0.00035 | 93 | 0.00040 | -0.35329 | 140.955 | 0.72440 | 0.92177 |
| araA; L-arabinose 1-dehydrogenase [EC:1.1.1.376] | 0.00056 | 0.00019 | 0.00010 | 0.00000 | 0.00003 | 65 | 0.00089 | 0.00001 | 0.00033 | 93 | 0.00033 | -2.38008 | 93.723 | 0.01933 | 0.59095 |
| K16881; mannose-1-phosphate guanylyltransferase / phosphomannomutase [EC:2.7.7.13 5.4.2.8] | 0.00056 | 0.00017 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00089 | 0.00001 | 0.00028 | 93 | 0.00028 | -2.82388 | 97.879 | 0.00575 | 0.54022 |
| iolF; MFS transporter, SP family, inositol transporter | 0.00056 | 0.00012 | 0.00035 | 0.00000 | 0.00011 | 65 | 0.00071 | 0.00000 | 0.00018 | 93 | 0.00021 | -1.69949 | 143.334 | 0.09140 | 0.76567 |
| egtB; gamma-glutamyl hercynylcysteine S-oxide synthase [EC:1.14.99.50] | 0.00056 | 0.00020 | 0.00029 | 0.00000 | 0.00013 | 65 | 0.00075 | 0.00001 | 0.00033 | 93 | 0.00036 | -1.30049 | 120.120 | 0.19592 | 0.77376 |
| allP; allantoin permease | 0.00056 | 0.00030 | 0.00034 | 0.00000 | 0.00028 | 65 | 0.00071 | 0.00002 | 0.00048 | 93 | 0.00055 | -0.66412 | 140.911 | 0.50770 | 0.84956 |
| kapD; sporulation inhibitor KapD | 0.00056 | 0.00022 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00091 | 0.00001 | 0.00036 | 93 | 0.00036 | -2.37330 | 93.419 | 0.01968 | 0.59095 |
| POP2; 4-aminobutyrate—pyruvate transaminase [EC:2.6.1.96] | 0.00056 | 0.00018 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00081 | 0.00001 | 0.00030 | 93 | 0.00031 | -1.90801 | 106.731 | 0.05908 | 0.72498 |
| prrA; two-component system, OmpR family, response regulator PrrA | 0.00056 | 0.00036 | 0.00017 | 0.00000 | 0.00009 | 65 | 0.00083 | 0.00003 | 0.00061 | 93 | 0.00061 | -1.07616 | 95.711 | 0.28456 | 0.77376 |
| glnP; glutamine transport system permease protein | 0.00056 | 0.00014 | 0.00063 | 0.00000 | 0.00025 | 65 | 0.00051 | 0.00000 | 0.00017 | 93 | 0.00030 | 0.38333 | 119.411 | 0.70216 | 0.92004 |
| wbpI, wlbD; UDP-GlcNAc3NAcA epimerase [EC:5.1.3.23] | 0.00056 | 0.00019 | 0.00024 | 0.00000 | 0.00008 | 65 | 0.00078 | 0.00001 | 0.00031 | 93 | 0.00032 | -1.70202 | 103.914 | 0.09174 | 0.76567 |
| mshA; MSHA pilin protein MshA | 0.00056 | 0.00023 | 0.00074 | 0.00001 | 0.00041 | 65 | 0.00043 | 0.00001 | 0.00027 | 93 | 0.00049 | 0.61746 | 117.516 | 0.53813 | 0.86207 |
| fdhA; formate dehydrogenase (NADP+) alpha subunit [EC:1.17.1.10] | 0.00056 | 0.00013 | 0.00059 | 0.00000 | 0.00022 | 65 | 0.00053 | 0.00000 | 0.00016 | 93 | 0.00027 | 0.24660 | 125.758 | 0.80562 | 0.94958 |
| rhaQ; rhamnose transport system permease protein | 0.00056 | 0.00018 | 0.00028 | 0.00000 | 0.00016 | 65 | 0.00075 | 0.00001 | 0.00028 | 93 | 0.00032 | -1.44438 | 143.232 | 0.15082 | 0.77376 |
| tus, tau; DNA replication terminus site-binding protein | 0.00055 | 0.00016 | 0.00060 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.23812 | 139.128 | 0.81214 | 0.95246 |
| ompF; outer membrane pore protein F | 0.00055 | 0.00016 | 0.00060 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.21721 | 139.078 | 0.82837 | 0.95506 |
| pbpD; penicillin-binding protein 4 [EC:2.4.1.129 3.4.16.4] | 0.00055 | 0.00030 | 0.00034 | 0.00001 | 0.00030 | 65 | 0.00070 | 0.00002 | 0.00046 | 93 | 0.00055 | -0.63931 | 147.627 | 0.52361 | 0.85709 |
| rusA; crossover junction endodeoxyribonuclease RusA [EC:3.1.22.4] | 0.00055 | 0.00022 | 0.00046 | 0.00000 | 0.00012 | 65 | 0.00061 | 0.00001 | 0.00036 | 93 | 0.00038 | -0.39166 | 110.903 | 0.69606 | 0.91772 |
| thiK; thiamine kinase [EC:2.7.1.89] | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.21424 | 139.187 | 0.83067 | 0.95506 |
| hcr; NADH oxidoreductase Hcr [EC:1.-.-.-] | 0.00055 | 0.00016 | 0.00060 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.24639 | 139.256 | 0.80574 | 0.94958 |
| traH; conjugative transfer pilus assembly protein TraH | 0.00055 | 0.00017 | 0.00054 | 0.00001 | 0.00029 | 65 | 0.00056 | 0.00000 | 0.00021 | 93 | 0.00036 | -0.06218 | 124.997 | 0.95052 | 0.98792 |
| yxeN; putative amino-acid transport system permease protein | 0.00055 | 0.00020 | 0.00079 | 0.00001 | 0.00044 | 65 | 0.00038 | 0.00000 | 0.00012 | 93 | 0.00046 | 0.89152 | 73.993 | 0.37554 | 0.79810 |
| K15976; putative NAD(P)H nitroreductase [EC:1.-.-.-] | 0.00055 | 0.00030 | 0.00035 | 0.00001 | 0.00030 | 65 | 0.00069 | 0.00002 | 0.00047 | 93 | 0.00055 | -0.61147 | 147.605 | 0.54183 | 0.86207 |
| gntU; Gnt-I system low-affinity gluconate transporter | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19795 | 139.142 | 0.84337 | 0.95519 |
| bssS; biofilm regulator BssS | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| hofM; pilus assembly protein HofM | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| hofN; pilus assembly protein HofN | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| lplT; MFS transporter, LPLT family, lysophospholipid transporter | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| lpxP; KDO2-lipid IV(A) palmitoleoyltransferase [EC:2.3.1.242] | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| nudJ; phosphatase NudJ [EC:3.6.1.-] | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| rcsD; two-component system, NarL family, sensor histidine kinase RcsD [EC:2.7.13.3] | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| rffC, wecD; dTDP-4-amino-4,6-dideoxy-D-galactose acyltransferase [EC:2.3.1.210] | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| secM; secretion monitor | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| tomB; hha toxicity modulator TomB | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| wecF, rffT; dTDP-N-acetylfucosamine:lipid II N-acetylfucosaminyltransferase [EC:2.4.1.325] | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| wzyE, rffT; enterobacterial common antigen polymerase [EC:2.4.1.-] | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| yfiM; putative lipoprotein | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.19942 | 139.146 | 0.84222 | 0.95506 |
| lsrK; autoinducer-2 kinase [EC:2.7.1.189] | 0.00055 | 0.00014 | 0.00049 | 0.00000 | 0.00020 | 65 | 0.00059 | 0.00000 | 0.00019 | 93 | 0.00027 | -0.33721 | 148.697 | 0.73643 | 0.92721 |
| csy1; CRISPR-associated protein Csy1 | 0.00055 | 0.00014 | 0.00065 | 0.00000 | 0.00023 | 65 | 0.00048 | 0.00000 | 0.00019 | 93 | 0.00029 | 0.57205 | 136.258 | 0.56823 | 0.87132 |
| feoC; ferrous iron transport protein C | 0.00055 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00052 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.20018 | 139.187 | 0.84163 | 0.95506 |
| FDFT1; farnesyl-diphosphate farnesyltransferase [EC:2.5.1.21] | 0.00055 | 0.00030 | 0.00034 | 0.00001 | 0.00030 | 65 | 0.00070 | 0.00002 | 0.00047 | 93 | 0.00055 | -0.65045 | 147.434 | 0.51641 | 0.85216 |
| plc; 1-phosphatidylinositol phosphodiesterase [EC:4.6.1.13] | 0.00054 | 0.00017 | 0.00017 | 0.00000 | 0.00008 | 65 | 0.00080 | 0.00001 | 0.00028 | 93 | 0.00029 | -2.14242 | 107.754 | 0.03441 | 0.62130 |
| nahK, lnpB; N-acetylhexosamine 1-kinase [EC:2.7.1.162] | 0.00054 | 0.00012 | 0.00052 | 0.00000 | 0.00017 | 65 | 0.00056 | 0.00000 | 0.00016 | 93 | 0.00024 | -0.15565 | 148.648 | 0.87652 | 0.96705 |
| K09740; uncharacterized protein | 0.00054 | 0.00022 | 0.00018 | 0.00000 | 0.00009 | 65 | 0.00079 | 0.00001 | 0.00036 | 93 | 0.00037 | -1.63984 | 103.591 | 0.10407 | 0.77376 |
| iolW; scyllo-inositol 2-dehydrogenase (NADP+) [EC:1.1.1.371] | 0.00054 | 0.00016 | 0.00035 | 0.00000 | 0.00018 | 65 | 0.00067 | 0.00001 | 0.00025 | 93 | 0.00031 | -1.05873 | 151.754 | 0.29141 | 0.77376 |
| vasL; type VI secretion system protein VasL | 0.00054 | 0.00016 | 0.00059 | 0.00000 | 0.00025 | 65 | 0.00050 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.26784 | 138.212 | 0.78922 | 0.94606 |
| hutP; hut operon positive regulatory protein | 0.00054 | 0.00021 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00087 | 0.00001 | 0.00035 | 93 | 0.00035 | -2.28266 | 93.494 | 0.02472 | 0.59764 |
| scrY; sucrose porin | 0.00054 | 0.00014 | 0.00068 | 0.00000 | 0.00024 | 65 | 0.00044 | 0.00000 | 0.00018 | 93 | 0.00030 | 0.78997 | 127.599 | 0.43101 | 0.82763 |
| ytrE; acetoin utilization transport system ATP-binding protein | 0.00054 | 0.00030 | 0.00041 | 0.00001 | 0.00031 | 65 | 0.00063 | 0.00002 | 0.00046 | 93 | 0.00056 | -0.39735 | 149.819 | 0.69167 | 0.91676 |
| ytrF; acetoin utilization transport system permease protein | 0.00054 | 0.00030 | 0.00041 | 0.00001 | 0.00031 | 65 | 0.00063 | 0.00002 | 0.00046 | 93 | 0.00056 | -0.39735 | 149.819 | 0.69167 | 0.91676 |
| togT, rhiT; oligogalacturonide transporter | 0.00054 | 0.00016 | 0.00059 | 0.00001 | 0.00035 | 65 | 0.00050 | 0.00000 | 0.00014 | 93 | 0.00038 | 0.26107 | 83.655 | 0.79468 | 0.94783 |
| whiB7; WhiB family transcriptional regulator, redox-sensing transcriptional regulator | 0.00054 | 0.00019 | 0.00038 | 0.00000 | 0.00016 | 65 | 0.00064 | 0.00001 | 0.00031 | 93 | 0.00035 | -0.74684 | 131.801 | 0.45649 | 0.83438 |
| traU; conjugal transfer pilus assembly protein TraU | 0.00054 | 0.00016 | 0.00041 | 0.00000 | 0.00020 | 65 | 0.00062 | 0.00001 | 0.00024 | 93 | 0.00031 | -0.68484 | 155.888 | 0.49446 | 0.84797 |
| atzD; cyanuric acid amidohydrolase [EC:3.5.2.15] | 0.00054 | 0.00021 | 0.00027 | 0.00000 | 0.00013 | 65 | 0.00072 | 0.00001 | 0.00035 | 93 | 0.00037 | -1.23705 | 114.758 | 0.21859 | 0.77376 |
| K07086; uncharacterized protein | 0.00053 | 0.00023 | 0.00012 | 0.00000 | 0.00006 | 65 | 0.00082 | 0.00001 | 0.00038 | 93 | 0.00039 | -1.80676 | 96.681 | 0.07391 | 0.74128 |
| ebr, qacEdelta1; small multidrug resistance pump | 0.00053 | 0.00014 | 0.00061 | 0.00000 | 0.00022 | 65 | 0.00048 | 0.00000 | 0.00019 | 93 | 0.00029 | 0.44250 | 139.135 | 0.65882 | 0.90880 |
| COQ4; ubiquinone biosynthesis protein COQ4 | 0.00053 | 0.00013 | 0.00035 | 0.00000 | 0.00013 | 65 | 0.00065 | 0.00000 | 0.00020 | 93 | 0.00024 | -1.25502 | 148.728 | 0.21144 | 0.77376 |
| spo0E; stage 0 sporulation regulatory protein | 0.00053 | 0.00030 | 0.00033 | 0.00000 | 0.00028 | 65 | 0.00067 | 0.00002 | 0.00047 | 93 | 0.00055 | -0.62147 | 142.298 | 0.53528 | 0.86207 |
| acxC; acetone carboxylase, gamma subunit [EC:6.4.1.6] | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag1; cag pathogenicity island protein 1 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag10; cag pathogenicity island protein 10 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag11; cag pathogenicity island protein 11 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag13; cag pathogenicity island protein 13 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag14; cag pathogenicity island protein 14 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag15; cag pathogenicity island protein 15 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag16; cag pathogenicity island protein 16 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag17; cag pathogenicity island protein 17 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag18; cag pathogenicity island protein 18 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag19; cag pathogenicity island protein 19 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag2; cag pathogenicity island protein 2 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag20; cag pathogenicity island protein 20 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag21; cag pathogenicity island protein 21 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag22; cag pathogenicity island protein 22 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag23; cag pathogenicity island protein 23 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag24; cag pathogenicity island protein 24 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag25; cag pathogenicity island protein 25 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag3; cag pathogenicity island protein 3 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag4; cag pathogenicity island protein 4 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag5; cag pathogenicity island protein 5 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag6; cag pathogenicity island protein 6 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag8; cag pathogenicity island protein 8 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cag9; cag pathogenicity island protein 9 | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| cagA; cytotoxicity-associated immunodominant antigen | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| comB6; ComB6 competence protein | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| hopB, alpB; outer membrane protein HopB/AlpB | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| hopC, alpA; outer membrane protein HopC/AlpA | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| hopZ; outer membrane protein HopZ | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| K07449; similar to archaeal holliday junction resolvase and Mrr protein | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| lpxE; lipid A 1-phosphatase [EC:3.1.3.-] | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| rpoBC; DNA-directed RNA polymerase subunit beta-beta’ [EC:2.7.7.6] | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| vacA; vacuolating cytotoxin | 0.00053 | 0.00034 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00076 | 0.00003 | 0.00057 | 93 | 0.00058 | -0.98237 | 100.382 | 0.32828 | 0.77376 |
| shc; squalene-hopene/tetraprenyl-beta-curcumene cyclase [EC:5.4.99.17 4.2.1.129] | 0.00053 | 0.00015 | 0.00046 | 0.00000 | 0.00020 | 65 | 0.00058 | 0.00000 | 0.00021 | 93 | 0.00029 | -0.40565 | 154.314 | 0.68556 | 0.91564 |
| pglH; GalNAc-alpha-(1->4)-GalNAc-alpha-(1->3)-diNAcBac-PP-undecaprenol alpha-1,4-N-acetyl-D-galactosaminyltransferase [EC:2.4.1.292] | 0.00053 | 0.00014 | 0.00067 | 0.00000 | 0.00026 | 65 | 0.00043 | 0.00000 | 0.00015 | 93 | 0.00030 | 0.78384 | 103.206 | 0.43493 | 0.82874 |
| mhpR; IclR family transcriptional regulator, mhp operon transcriptional activator | 0.00053 | 0.00014 | 0.00029 | 0.00000 | 0.00010 | 65 | 0.00069 | 0.00000 | 0.00023 | 93 | 0.00025 | -1.62789 | 121.728 | 0.10613 | 0.77376 |
| K07064; uncharacterized protein | 0.00053 | 0.00014 | 0.00067 | 0.00000 | 0.00026 | 65 | 0.00043 | 0.00000 | 0.00016 | 93 | 0.00030 | 0.80565 | 109.123 | 0.42220 | 0.82295 |
| csm2; CRISPR-associated protein Csm2 | 0.00053 | 0.00022 | 0.00032 | 0.00000 | 0.00011 | 65 | 0.00067 | 0.00001 | 0.00037 | 93 | 0.00038 | -0.91340 | 107.409 | 0.36308 | 0.79496 |
| maeN; malate:Na+ symporter | 0.00052 | 0.00015 | 0.00052 | 0.00000 | 0.00022 | 65 | 0.00053 | 0.00000 | 0.00020 | 93 | 0.00030 | -0.03339 | 144.373 | 0.97341 | 0.99509 |
| tylC, oleB, carA, srmB; macrolide transport system ATP-binding/permease protein | 0.00052 | 0.00012 | 0.00046 | 0.00000 | 0.00012 | 65 | 0.00057 | 0.00000 | 0.00019 | 93 | 0.00023 | -0.51084 | 147.027 | 0.61023 | 0.88717 |
| K09931; uncharacterized protein | 0.00052 | 0.00011 | 0.00041 | 0.00000 | 0.00015 | 65 | 0.00060 | 0.00000 | 0.00015 | 93 | 0.00021 | -0.87491 | 152.158 | 0.38300 | 0.80064 |
| frlD; fructoselysine 6-kinase [EC:2.7.1.218] | 0.00052 | 0.00019 | 0.00025 | 0.00000 | 0.00017 | 65 | 0.00071 | 0.00001 | 0.00029 | 93 | 0.00034 | -1.35041 | 142.561 | 0.17902 | 0.77376 |
| K11476, gntR; GntR family transcriptional regulator, gluconate operon transcriptional repressor | 0.00052 | 0.00029 | 0.00034 | 0.00000 | 0.00028 | 65 | 0.00065 | 0.00002 | 0.00046 | 93 | 0.00054 | -0.58502 | 143.471 | 0.55945 | 0.86813 |
| soxC; sulfane dehydrogenase subunit SoxC | 0.00052 | 0.00014 | 0.00031 | 0.00000 | 0.00012 | 65 | 0.00067 | 0.00000 | 0.00023 | 93 | 0.00025 | -1.43357 | 133.528 | 0.15403 | 0.77376 |
| tap; methyl-accepting chemotaxis protein IV, peptide sensor receptor | 0.00052 | 0.00016 | 0.00058 | 0.00000 | 0.00025 | 65 | 0.00048 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.28110 | 137.951 | 0.77906 | 0.94269 |
| hemDX; uroporphyrinogen III methyltransferase / synthase [EC:2.1.1.107 4.2.1.75] | 0.00052 | 0.00018 | 0.00048 | 0.00000 | 0.00018 | 65 | 0.00055 | 0.00001 | 0.00028 | 93 | 0.00034 | -0.22840 | 147.784 | 0.81965 | 0.95506 |
| epsD; glycosyltransferase EpsD [EC:2.4.-.-] | 0.00052 | 0.00011 | 0.00047 | 0.00000 | 0.00013 | 65 | 0.00055 | 0.00000 | 0.00016 | 93 | 0.00020 | -0.41429 | 155.928 | 0.67923 | 0.91311 |
| SQD1, sqdB; UDP-sulfoquinovose synthase [EC:3.13.1.1] | 0.00052 | 0.00015 | 0.00024 | 0.00000 | 0.00008 | 65 | 0.00072 | 0.00001 | 0.00024 | 93 | 0.00025 | -1.88113 | 112.518 | 0.06254 | 0.73012 |
| btaB; S-adenosylmethionine-diacylgycerolhomoserine-N-methlytransferase | 0.00052 | 0.00012 | 0.00023 | 0.00000 | 0.00009 | 65 | 0.00072 | 0.00000 | 0.00018 | 93 | 0.00021 | -2.37867 | 132.991 | 0.01880 | 0.59095 |
| vga; pleuromutilin/lincosamide/streptogramin A transport system ATP-binding/permease protein | 0.00052 | 0.00027 | 0.00070 | 0.00003 | 0.00063 | 65 | 0.00039 | 0.00000 | 0.00015 | 93 | 0.00065 | 0.48378 | 71.347 | 0.63002 | 0.89536 |
| fliZ; regulator of sigma S factor FliZ | 0.00051 | 0.00016 | 0.00056 | 0.00000 | 0.00025 | 65 | 0.00048 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.22518 | 139.319 | 0.82217 | 0.95506 |
| sinR; XRE family transcriptional regulator, master regulator for biofilm formation | 0.00051 | 0.00019 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00082 | 0.00001 | 0.00032 | 93 | 0.00032 | -2.33414 | 94.203 | 0.02171 | 0.59153 |
| cotF; spore coat protein F | 0.00051 | 0.00021 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00085 | 0.00001 | 0.00035 | 93 | 0.00035 | -2.34351 | 92.473 | 0.02125 | 0.59147 |
| rmd; GDP-4-dehydro-6-deoxy-D-mannose reductase [EC:1.1.1.281] | 0.00051 | 0.00013 | 0.00034 | 0.00000 | 0.00013 | 65 | 0.00063 | 0.00000 | 0.00021 | 93 | 0.00025 | -1.17274 | 146.473 | 0.24281 | 0.77376 |
| PGRP; peptidoglycan recognition protein | 0.00051 | 0.00017 | 0.00030 | 0.00000 | 0.00014 | 65 | 0.00065 | 0.00001 | 0.00027 | 93 | 0.00031 | -1.14420 | 135.862 | 0.25455 | 0.77376 |
| vpr; minor extracellular serine protease Vpr [EC:3.4.21.-] | 0.00051 | 0.00015 | 0.00014 | 0.00000 | 0.00007 | 65 | 0.00077 | 0.00001 | 0.00025 | 93 | 0.00026 | -2.36372 | 105.037 | 0.01993 | 0.59095 |
| csrD; RNase E specificity factor CsrD | 0.00051 | 0.00014 | 0.00064 | 0.00000 | 0.00026 | 65 | 0.00041 | 0.00000 | 0.00015 | 93 | 0.00030 | 0.76360 | 106.850 | 0.44679 | 0.83371 |
| rsmF; 16S rRNA (cytosine1407-C5)-methyltransferase [EC:2.1.1.178] | 0.00051 | 0.00014 | 0.00064 | 0.00000 | 0.00026 | 65 | 0.00041 | 0.00000 | 0.00015 | 93 | 0.00030 | 0.76360 | 106.850 | 0.44679 | 0.83371 |
| traV; conjugal transfer pilus assembly protein TraV | 0.00051 | 0.00015 | 0.00042 | 0.00000 | 0.00021 | 65 | 0.00057 | 0.00000 | 0.00021 | 93 | 0.00030 | -0.51717 | 152.955 | 0.60578 | 0.88527 |
| pglB; undecaprenyl-diphosphooligosaccharide—protein glycotransferase [EC:2.4.99.19] | 0.00051 | 0.00014 | 0.00065 | 0.00000 | 0.00026 | 65 | 0.00040 | 0.00000 | 0.00014 | 93 | 0.00030 | 0.84061 | 100.259 | 0.40257 | 0.80926 |
| ydgD; protease YdgD [EC:3.4.21.-] | 0.00051 | 0.00013 | 0.00043 | 0.00000 | 0.00019 | 65 | 0.00056 | 0.00000 | 0.00018 | 93 | 0.00026 | -0.51974 | 146.364 | 0.60403 | 0.88434 |
| UAP1; UDP-N-acetylglucosamine/UDP-N-acetylgalactosamine diphosphorylase [EC:2.7.7.23 2.7.7.83] | 0.00051 | 0.00017 | 0.00016 | 0.00000 | 0.00008 | 65 | 0.00075 | 0.00001 | 0.00029 | 93 | 0.00030 | -1.99078 | 105.467 | 0.04909 | 0.67403 |
| ebrA; multidrug resistance protein EbrA | 0.00051 | 0.00030 | 0.00033 | 0.00001 | 0.00030 | 65 | 0.00063 | 0.00002 | 0.00046 | 93 | 0.00055 | -0.54681 | 147.769 | 0.58533 | 0.87720 |
| ebrB; multidrug resistance protein EbrB | 0.00051 | 0.00030 | 0.00033 | 0.00001 | 0.00030 | 65 | 0.00063 | 0.00002 | 0.00046 | 93 | 0.00055 | -0.54681 | 147.769 | 0.58533 | 0.87720 |
| kas; beta-ketoacyl ACP synthase [EC:2.3.1.-] | 0.00051 | 0.00020 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00080 | 0.00001 | 0.00034 | 93 | 0.00034 | -2.09108 | 95.154 | 0.03919 | 0.63577 |
| dpe, lre; D-psicose/D-tagatose/L-ribulose 3-epimerase [EC:5.1.3.30 5.1.3.31] | 0.00050 | 0.00013 | 0.00015 | 0.00000 | 0.00008 | 65 | 0.00075 | 0.00000 | 0.00020 | 93 | 0.00022 | -2.79267 | 119.496 | 0.00609 | 0.54022 |
| sspN; small acid-soluble spore protein N (minor) | 0.00050 | 0.00016 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00080 | 0.00001 | 0.00027 | 93 | 0.00027 | -2.70179 | 94.547 | 0.00817 | 0.56121 |
| ena; enamidase [EC:3.5.2.18] | 0.00050 | 0.00021 | 0.00009 | 0.00000 | 0.00003 | 65 | 0.00079 | 0.00001 | 0.00036 | 93 | 0.00036 | -1.90565 | 93.457 | 0.05977 | 0.72880 |
| K09138; uncharacterized protein | 0.00050 | 0.00011 | 0.00048 | 0.00000 | 0.00011 | 65 | 0.00052 | 0.00000 | 0.00016 | 93 | 0.00020 | -0.21388 | 150.167 | 0.83093 | 0.95506 |
| TPS; trehalose 6-phosphate synthase/phosphatase [EC:2.4.1.15 3.1.3.12] | 0.00050 | 0.00015 | 0.00018 | 0.00000 | 0.00010 | 65 | 0.00072 | 0.00001 | 0.00025 | 93 | 0.00026 | -2.06318 | 117.753 | 0.04129 | 0.64661 |
| traC; conjugal transfer ATP-binding protein TraC | 0.00050 | 0.00015 | 0.00041 | 0.00000 | 0.00020 | 65 | 0.00056 | 0.00000 | 0.00021 | 93 | 0.00029 | -0.51144 | 152.884 | 0.60978 | 0.88693 |
| trbI; conjugal transfer pilin signal peptidase TrbI | 0.00050 | 0.00017 | 0.00051 | 0.00001 | 0.00030 | 65 | 0.00049 | 0.00000 | 0.00021 | 93 | 0.00037 | 0.07761 | 120.191 | 0.93827 | 0.98459 |
| E1.1.99.21; D-sorbitol dehydrogenase (acceptor) [EC:1.1.99.21] | 0.00050 | 0.00015 | 0.00040 | 0.00000 | 0.00021 | 65 | 0.00056 | 0.00000 | 0.00021 | 93 | 0.00030 | -0.55948 | 152.117 | 0.57666 | 0.87389 |
| hipB; HTH-type transcriptional regulator / antitoxin HipB | 0.00049 | 0.00016 | 0.00061 | 0.00001 | 0.00032 | 65 | 0.00041 | 0.00000 | 0.00016 | 93 | 0.00036 | 0.53774 | 95.726 | 0.59201 | 0.87898 |
| fimH; minor fimbrial subunit | 0.00049 | 0.00022 | 0.00033 | 0.00000 | 0.00013 | 65 | 0.00061 | 0.00001 | 0.00037 | 93 | 0.00039 | -0.72884 | 112.509 | 0.46761 | 0.83837 |
| ligI; 2-pyrone-4,6-dicarboxylate lactonase [EC:3.1.1.57] | 0.00049 | 0.00023 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00078 | 0.00001 | 0.00039 | 93 | 0.00039 | -1.75310 | 93.806 | 0.08285 | 0.76567 |
| cof; HMP-PP phosphatase [EC:3.6.1.-] | 0.00049 | 0.00015 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00055 | 0.00000 | 0.00022 | 93 | 0.00029 | -0.51906 | 155.631 | 0.60446 | 0.88434 |
| hdeA; acid stress chaperone HdeA | 0.00049 | 0.00018 | 0.00011 | 0.00000 | 0.00004 | 65 | 0.00076 | 0.00001 | 0.00031 | 93 | 0.00031 | -2.11039 | 94.796 | 0.03746 | 0.63026 |
| norW; nitric oxide reductase FlRd-NAD(+) reductase [EC:1.18.1.-] | 0.00049 | 0.00014 | 0.00063 | 0.00000 | 0.00026 | 65 | 0.00039 | 0.00000 | 0.00015 | 93 | 0.00030 | 0.77582 | 106.003 | 0.43958 | 0.83156 |
| hyfA; hydrogenase-4 component A [EC:1.-.-.-] | 0.00049 | 0.00016 | 0.00068 | 0.00001 | 0.00032 | 65 | 0.00035 | 0.00000 | 0.00015 | 93 | 0.00036 | 0.92720 | 90.779 | 0.35628 | 0.79399 |
| ecm; ethylmalonyl-CoA mutase [EC:5.4.99.63] | 0.00049 | 0.00014 | 0.00040 | 0.00000 | 0.00015 | 65 | 0.00055 | 0.00000 | 0.00022 | 93 | 0.00027 | -0.55408 | 152.648 | 0.58034 | 0.87573 |
| accD6; acetyl-CoA/propionyl-CoA carboxylase carboxyl transferase subunit [EC:6.4.1.2 6.4.1.3 2.1.3.15] | 0.00049 | 0.00020 | 0.00025 | 0.00000 | 0.00013 | 65 | 0.00065 | 0.00001 | 0.00032 | 93 | 0.00035 | -1.16171 | 121.921 | 0.24762 | 0.77376 |
| nylA; 6-aminohexanoate-cyclic-dimer hydrolase [EC:3.5.2.12] | 0.00049 | 0.00021 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00072 | 0.00001 | 0.00035 | 93 | 0.00036 | -1.61480 | 96.497 | 0.10962 | 0.77376 |
| asbF; 3-dehydroshikimate dehydratase [EC:4.2.1.118] | 0.00049 | 0.00029 | 0.00031 | 0.00000 | 0.00028 | 65 | 0.00061 | 0.00002 | 0.00046 | 93 | 0.00054 | -0.56151 | 143.400 | 0.57532 | 0.87389 |
| ybiV; sugar-phosphatase [EC:3.1.3.23] | 0.00049 | 0.00017 | 0.00032 | 0.00000 | 0.00010 | 65 | 0.00060 | 0.00001 | 0.00028 | 93 | 0.00030 | -0.94120 | 115.052 | 0.34857 | 0.79120 |
| norV; anaerobic nitric oxide reductase flavorubredoxin | 0.00048 | 0.00014 | 0.00062 | 0.00000 | 0.00026 | 65 | 0.00039 | 0.00000 | 0.00015 | 93 | 0.00030 | 0.74540 | 105.951 | 0.45768 | 0.83438 |
| K16150; glycogen synthase [EC:2.4.1.11] | 0.00048 | 0.00020 | 0.00017 | 0.00000 | 0.00007 | 65 | 0.00070 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.57864 | 101.177 | 0.11754 | 0.77376 |
| aruI; 5-guanidino-2-oxopentanoate decarboxylase [EC:4.1.1.75] | 0.00048 | 0.00014 | 0.00071 | 0.00000 | 0.00026 | 65 | 0.00033 | 0.00000 | 0.00016 | 93 | 0.00030 | 1.26563 | 111.704 | 0.20828 | 0.77376 |
| cas5h; CRISPR-associated protein Cas5h | 0.00048 | 0.00015 | 0.00052 | 0.00000 | 0.00025 | 65 | 0.00045 | 0.00000 | 0.00019 | 93 | 0.00031 | 0.22479 | 129.035 | 0.82250 | 0.95506 |
| csh2; CRISPR-associated protein Csh2 | 0.00048 | 0.00015 | 0.00052 | 0.00000 | 0.00025 | 65 | 0.00045 | 0.00000 | 0.00019 | 93 | 0.00031 | 0.22479 | 129.035 | 0.82250 | 0.95506 |
| treR; LacI family transcriptional regulator, trehalose operon repressor | 0.00048 | 0.00014 | 0.00059 | 0.00000 | 0.00026 | 65 | 0.00041 | 0.00000 | 0.00015 | 93 | 0.00030 | 0.61582 | 107.398 | 0.53932 | 0.86207 |
| PTS-Asc-EIIC, ascF; PTS system, beta-glucoside (arbutin/salicin/cellobiose)-specific IIC component | 0.00048 | 0.00013 | 0.00061 | 0.00000 | 0.00023 | 65 | 0.00039 | 0.00000 | 0.00014 | 93 | 0.00027 | 0.82315 | 109.458 | 0.41222 | 0.81576 |
| rspB; L-gulonate 5-dehydrogenase [EC:1.1.1.380] | 0.00048 | 0.00014 | 0.00053 | 0.00000 | 0.00023 | 65 | 0.00045 | 0.00000 | 0.00017 | 93 | 0.00029 | 0.30439 | 125.135 | 0.76134 | 0.93806 |
| melB; melibiose permease | 0.00048 | 0.00012 | 0.00028 | 0.00000 | 0.00007 | 65 | 0.00062 | 0.00000 | 0.00020 | 93 | 0.00022 | -1.55141 | 113.581 | 0.12358 | 0.77376 |
| prrB; two-component system, OmpR family, sensor histidine kinase PrrB [EC:2.7.13.3] | 0.00048 | 0.00036 | 0.00011 | 0.00000 | 0.00007 | 65 | 0.00074 | 0.00003 | 0.00061 | 93 | 0.00061 | -1.02629 | 94.714 | 0.30737 | 0.77376 |
| aglF, ggtC; alpha-glucoside transport system permease protein | 0.00048 | 0.00014 | 0.00037 | 0.00000 | 0.00020 | 65 | 0.00056 | 0.00000 | 0.00019 | 93 | 0.00027 | -0.71209 | 147.565 | 0.47753 | 0.84209 |
| aglG, ggtD; alpha-glucoside transport system permease protein | 0.00048 | 0.00014 | 0.00037 | 0.00000 | 0.00020 | 65 | 0.00056 | 0.00000 | 0.00019 | 93 | 0.00027 | -0.71209 | 147.565 | 0.47753 | 0.84209 |
| K15640, phoE; uncharacterized phosphatase | 0.00048 | 0.00015 | 0.00009 | 0.00000 | 0.00003 | 65 | 0.00075 | 0.00001 | 0.00024 | 93 | 0.00025 | -2.66171 | 95.197 | 0.00913 | 0.58112 |
| comGG; competence protein ComGG | 0.00048 | 0.00021 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00079 | 0.00001 | 0.00035 | 93 | 0.00035 | -2.20305 | 92.649 | 0.03007 | 0.61413 |
| epsF; glycosyltransferase EpsF [EC:2.4.-.-] | 0.00048 | 0.00029 | 0.00036 | 0.00001 | 0.00028 | 65 | 0.00056 | 0.00002 | 0.00046 | 93 | 0.00054 | -0.38121 | 144.415 | 0.70361 | 0.92004 |
| PTS-Mng-EIIC, mngA, hrsA; PTS system, 2-O-A-mannosyl-D-glycerate-specific IIC component | 0.00048 | 0.00019 | 0.00017 | 0.00000 | 0.00008 | 65 | 0.00069 | 0.00001 | 0.00031 | 93 | 0.00032 | -1.62852 | 103.029 | 0.10647 | 0.77376 |
| K08982; putative membrane protein | 0.00048 | 0.00014 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00068 | 0.00001 | 0.00023 | 93 | 0.00025 | -1.97058 | 112.964 | 0.05122 | 0.68675 |
| lsrF; 3-hydroxy-5-phosphonooxypentane-2,4-dione thiolase [EC:2.3.1.245] | 0.00047 | 0.00012 | 0.00032 | 0.00000 | 0.00010 | 65 | 0.00058 | 0.00000 | 0.00019 | 93 | 0.00022 | -1.20828 | 136.499 | 0.22903 | 0.77376 |
| dpiB, citA; two-component system, CitB family, cit operon sensor histidine kinase CitA [EC:2.7.13.3] | 0.00047 | 0.00014 | 0.00055 | 0.00000 | 0.00024 | 65 | 0.00042 | 0.00000 | 0.00016 | 93 | 0.00029 | 0.46843 | 115.684 | 0.64036 | 0.89936 |
| K16163; maleylpyruvate isomerase [EC:5.2.1.4] | 0.00047 | 0.00022 | 0.00033 | 0.00000 | 0.00016 | 65 | 0.00057 | 0.00001 | 0.00035 | 93 | 0.00039 | -0.64418 | 125.914 | 0.52063 | 0.85579 |
| traK; conjugal transfer pilus assembly protein TraK | 0.00047 | 0.00015 | 0.00042 | 0.00000 | 0.00021 | 65 | 0.00051 | 0.00000 | 0.00021 | 93 | 0.00029 | -0.31481 | 151.074 | 0.75334 | 0.93639 |
| isfD; sulfoacetaldehyde reductase [EC:1.1.1.313] | 0.00047 | 0.00018 | 0.00047 | 0.00001 | 0.00029 | 65 | 0.00047 | 0.00000 | 0.00022 | 93 | 0.00037 | 0.00054 | 129.083 | 0.99957 | 0.99957 |
| treT; trehalose synthase [EC:2.4.1.245] | 0.00047 | 0.00011 | 0.00034 | 0.00000 | 0.00010 | 65 | 0.00056 | 0.00000 | 0.00018 | 93 | 0.00021 | -1.08979 | 138.418 | 0.27770 | 0.77376 |
| eexE; membrane fusion protein, epimerase transport system | 0.00047 | 0.00016 | 0.00070 | 0.00001 | 0.00030 | 65 | 0.00031 | 0.00000 | 0.00016 | 93 | 0.00034 | 1.15840 | 99.780 | 0.24946 | 0.77376 |
| lpqH; ipoprotein LpqH | 0.00047 | 0.00021 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00075 | 0.00001 | 0.00035 | 93 | 0.00035 | -1.93066 | 94.857 | 0.05651 | 0.70577 |
| dsdC; LysR family transcriptional regulator, D-serine deaminase activator | 0.00047 | 0.00012 | 0.00065 | 0.00000 | 0.00024 | 65 | 0.00034 | 0.00000 | 0.00013 | 93 | 0.00027 | 1.16686 | 99.464 | 0.24606 | 0.77376 |
| fimB; type 1 fimbriae regulatory protein FimB | 0.00047 | 0.00016 | 0.00042 | 0.00000 | 0.00024 | 65 | 0.00050 | 0.00000 | 0.00022 | 93 | 0.00033 | -0.21832 | 147.574 | 0.82748 | 0.95506 |
| eexD; ATP-binding cassette, subfamily C, bacterial EexD | 0.00047 | 0.00015 | 0.00066 | 0.00001 | 0.00029 | 65 | 0.00033 | 0.00000 | 0.00016 | 93 | 0.00033 | 0.96808 | 103.457 | 0.33526 | 0.78186 |
| E1.3.1.32; maleylacetate reductase [EC:1.3.1.32] | 0.00046 | 0.00021 | 0.00016 | 0.00000 | 0.00006 | 65 | 0.00068 | 0.00001 | 0.00035 | 93 | 0.00035 | -1.46375 | 97.610 | 0.14648 | 0.77376 |
| E2.4.1.20; cellobiose phosphorylase [EC:2.4.1.20] | 0.00046 | 0.00015 | 0.00036 | 0.00000 | 0.00018 | 65 | 0.00054 | 0.00000 | 0.00022 | 93 | 0.00028 | -0.64544 | 155.685 | 0.51959 | 0.85478 |
| K13875, araC; L-arabonate dehydrase [EC:4.2.1.25] | 0.00046 | 0.00024 | 0.00016 | 0.00000 | 0.00010 | 65 | 0.00068 | 0.00002 | 0.00040 | 93 | 0.00042 | -1.24027 | 103.195 | 0.21769 | 0.77376 |
| malI; LacI family transcriptional regulator, maltose regulon regulatory protein | 0.00046 | 0.00013 | 0.00057 | 0.00000 | 0.00023 | 65 | 0.00038 | 0.00000 | 0.00015 | 93 | 0.00028 | 0.68179 | 111.593 | 0.49678 | 0.84797 |
| ugl; ureidoglycolate lyase [EC:4.3.2.3] | 0.00046 | 0.00016 | 0.00042 | 0.00000 | 0.00023 | 65 | 0.00049 | 0.00000 | 0.00023 | 93 | 0.00032 | -0.20417 | 149.290 | 0.83850 | 0.95506 |
| xapA; xanthosine phosphorylase [EC:2.4.2.-] | 0.00046 | 0.00017 | 0.00041 | 0.00000 | 0.00016 | 65 | 0.00050 | 0.00001 | 0.00027 | 93 | 0.00032 | -0.30287 | 142.754 | 0.76243 | 0.93813 |
| mct; 2-methylfumaryl-CoA isomerase [EC:5.4.1.3] | 0.00046 | 0.00019 | 0.00019 | 0.00000 | 0.00007 | 65 | 0.00065 | 0.00001 | 0.00032 | 93 | 0.00032 | -1.42888 | 99.880 | 0.15616 | 0.77376 |
| legI, neuB2; N,N’-diacetyllegionaminate synthase [EC:2.5.1.101] | 0.00046 | 0.00015 | 0.00036 | 0.00000 | 0.00018 | 65 | 0.00053 | 0.00000 | 0.00021 | 93 | 0.00028 | -0.59295 | 156.000 | 0.55407 | 0.86806 |
| ybcL; MFS transporter, DHA1 family, putative efflux transporter | 0.00046 | 0.00029 | 0.00032 | 0.00000 | 0.00028 | 65 | 0.00056 | 0.00002 | 0.00046 | 93 | 0.00054 | -0.46006 | 143.608 | 0.64617 | 0.90314 |
| traW; conjugal transfer pilus assembly protein TraW | 0.00046 | 0.00015 | 0.00041 | 0.00000 | 0.00020 | 65 | 0.00050 | 0.00000 | 0.00020 | 93 | 0.00029 | -0.29936 | 150.944 | 0.76508 | 0.93900 |
| hbhA; heparin binding hemagglutinin HbhA | 0.00046 | 0.00019 | 0.00026 | 0.00000 | 0.00013 | 65 | 0.00060 | 0.00001 | 0.00031 | 93 | 0.00034 | -1.01085 | 120.621 | 0.31411 | 0.77376 |
| traF; conjugal transfer pilus assembly protein TraF | 0.00046 | 0.00016 | 0.00052 | 0.00001 | 0.00029 | 65 | 0.00042 | 0.00000 | 0.00018 | 93 | 0.00034 | 0.28850 | 109.182 | 0.77351 | 0.94219 |
| comQ; competence protein ComQ | 0.00046 | 0.00029 | 0.00031 | 0.00000 | 0.00028 | 65 | 0.00057 | 0.00002 | 0.00046 | 93 | 0.00054 | -0.48276 | 143.599 | 0.63000 | 0.89536 |
| K09860; uncharacterized protein | 0.00046 | 0.00013 | 0.00055 | 0.00000 | 0.00024 | 65 | 0.00039 | 0.00000 | 0.00014 | 93 | 0.00028 | 0.56154 | 107.936 | 0.57560 | 0.87389 |
| prdB; D-proline reductase (dithiol) PrdB [EC:1.21.4.1] | 0.00046 | 0.00008 | 0.00040 | 0.00000 | 0.00010 | 65 | 0.00049 | 0.00000 | 0.00011 | 93 | 0.00015 | -0.58023 | 154.763 | 0.56260 | 0.86813 |
| garL; 2-dehydro-3-deoxyglucarate aldolase [EC:4.1.2.20] | 0.00046 | 0.00012 | 0.00056 | 0.00000 | 0.00022 | 65 | 0.00038 | 0.00000 | 0.00014 | 93 | 0.00026 | 0.67702 | 115.303 | 0.49975 | 0.84896 |
| hapE; 4-hydroxyacetophenone monooxygenase [EC:1.14.13.84] | 0.00046 | 0.00013 | 0.00010 | 0.00000 | 0.00003 | 65 | 0.00071 | 0.00000 | 0.00022 | 93 | 0.00023 | -2.67410 | 95.762 | 0.00881 | 0.58112 |
| E2.7.7.6; DNA-directed RNA polymerase [EC:2.7.7.6] | 0.00046 | 0.00016 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00072 | 0.00001 | 0.00026 | 93 | 0.00026 | -2.39685 | 95.143 | 0.01849 | 0.59095 |
| srfATE, srfAD, lchAD; external thioesterase TEII | 0.00045 | 0.00029 | 0.00030 | 0.00000 | 0.00028 | 65 | 0.00056 | 0.00002 | 0.00046 | 93 | 0.00054 | -0.47417 | 143.600 | 0.63610 | 0.89696 |
| sspD; small acid-soluble spore protein D (minor alpha/beta-type SASP) | 0.00045 | 0.00016 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00072 | 0.00001 | 0.00026 | 93 | 0.00026 | -2.45140 | 94.579 | 0.01607 | 0.59095 |
| vanRAc; two-component system, OmpR family, response regulator VanR | 0.00045 | 0.00012 | 0.00045 | 0.00000 | 0.00019 | 65 | 0.00045 | 0.00000 | 0.00015 | 93 | 0.00024 | -0.00808 | 134.831 | 0.99357 | 0.99919 |
| bmrR; MerR family transcriptional regulator, activator of bmr gene | 0.00045 | 0.00029 | 0.00030 | 0.00000 | 0.00028 | 65 | 0.00056 | 0.00002 | 0.00046 | 93 | 0.00054 | -0.49014 | 143.602 | 0.62478 | 0.89208 |
| lytF, cwlE; peptidoglycan DL-endopeptidase LytF [EC:3.4.-.-] | 0.00045 | 0.00029 | 0.00030 | 0.00000 | 0.00028 | 65 | 0.00056 | 0.00002 | 0.00046 | 93 | 0.00054 | -0.49014 | 143.602 | 0.62478 | 0.89208 |
| eryA; erythritol kinase (D-erythritol 1-phosphate-forming) [EC:2.7.1.215] | 0.00045 | 0.00019 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00073 | 0.00001 | 0.00032 | 93 | 0.00032 | -2.14673 | 92.759 | 0.03442 | 0.62130 |
| E3.2.1.123; endoglycosylceramidase [EC:3.2.1.123] | 0.00045 | 0.00036 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00069 | 0.00003 | 0.00061 | 93 | 0.00061 | -0.96175 | 94.544 | 0.33863 | 0.78407 |
| acd; glutaryl-CoA dehydrogenase (non-decarboxylating) [EC:1.3.99.32] | 0.00045 | 0.00015 | 0.00005 | 0.00000 | 0.00002 | 65 | 0.00073 | 0.00001 | 0.00025 | 93 | 0.00025 | -2.72464 | 92.941 | 0.00769 | 0.54764 |
| HXT; MFS transporter, SP family, sugar:H+ symporter | 0.00045 | 0.00010 | 0.00029 | 0.00000 | 0.00013 | 65 | 0.00056 | 0.00000 | 0.00015 | 93 | 0.00020 | -1.34908 | 155.991 | 0.17927 | 0.77376 |
| wbqV; O-antigen biosynthesis protein WbqV | 0.00045 | 0.00016 | 0.00013 | 0.00000 | 0.00005 | 65 | 0.00067 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.99583 | 97.535 | 0.04874 | 0.67326 |
| vanSAc; two-component system, OmpR family, sensor histidine kinase VanS [EC:2.7.13.3] | 0.00044 | 0.00012 | 0.00045 | 0.00000 | 0.00019 | 65 | 0.00044 | 0.00000 | 0.00015 | 93 | 0.00024 | 0.04070 | 133.745 | 0.96759 | 0.99216 |
| E2.7.10.2; non-specific protein-tyrosine kinase [EC:2.7.10.2] | 0.00044 | 0.00022 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00070 | 0.00001 | 0.00038 | 93 | 0.00038 | -1.66053 | 92.940 | 0.10018 | 0.77098 |
| hblAB; hemolysin BL binding component | 0.00044 | 0.00029 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00072 | 0.00002 | 0.00049 | 93 | 0.00050 | -1.33634 | 93.239 | 0.18469 | 0.77376 |
| OXCT; 3-oxoacid CoA-transferase [EC:2.8.3.5] | 0.00044 | 0.00015 | 0.00030 | 0.00000 | 0.00019 | 65 | 0.00054 | 0.00000 | 0.00021 | 93 | 0.00028 | -0.84751 | 155.458 | 0.39801 | 0.80577 |
| hpsN; sulfopropanediol 3-dehydrogenase [EC:1.1.1.308] | 0.00044 | 0.00015 | 0.00023 | 0.00000 | 0.00020 | 65 | 0.00059 | 0.00000 | 0.00022 | 93 | 0.00030 | -1.20216 | 154.641 | 0.23114 | 0.77376 |
| embB; arabinosyltransferase B [EC:2.4.2.-] | 0.00044 | 0.00019 | 0.00013 | 0.00000 | 0.00006 | 65 | 0.00066 | 0.00001 | 0.00032 | 93 | 0.00033 | -1.61928 | 98.274 | 0.10859 | 0.77376 |
| chvB, cgs, ndvB; cyclic beta-1,2-glucan synthetase [EC:2.4.1.-] | 0.00044 | 0.00017 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00070 | 0.00001 | 0.00029 | 93 | 0.00029 | -2.10069 | 93.828 | 0.03835 | 0.63091 |
| chpB, chpBK; mRNA interferase ChpB [EC:3.1.-.-] | 0.00044 | 0.00020 | 0.00024 | 0.00000 | 0.00013 | 65 | 0.00058 | 0.00001 | 0.00033 | 93 | 0.00035 | -0.95160 | 118.160 | 0.34324 | 0.78583 |
| doeX; Lrp/AsnC family transcriptional regulator, regulator of ectoine-degradation genes | 0.00044 | 0.00015 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00060 | 0.00001 | 0.00024 | 93 | 0.00027 | -1.54166 | 129.665 | 0.12559 | 0.77376 |
| cho; excinuclease Cho [EC:3.1.25.-] | 0.00043 | 0.00012 | 0.00044 | 0.00000 | 0.00019 | 65 | 0.00043 | 0.00000 | 0.00015 | 93 | 0.00025 | 0.07254 | 133.486 | 0.94228 | 0.98549 |
| aor; aldehyde:ferredoxin oxidoreductase [EC:1.2.7.5] | 0.00043 | 0.00016 | 0.00048 | 0.00001 | 0.00028 | 65 | 0.00040 | 0.00000 | 0.00018 | 93 | 0.00033 | 0.24938 | 111.044 | 0.80353 | 0.94911 |
| doeD; L-2,4-diaminobutyrate transaminase [EC:2.6.1.76] | 0.00043 | 0.00015 | 0.00021 | 0.00000 | 0.00012 | 65 | 0.00059 | 0.00001 | 0.00024 | 93 | 0.00027 | -1.42421 | 129.929 | 0.15678 | 0.77376 |
| ABC.VB1X.A; putative thiamine transport system ATP-binding protein | 0.00043 | 0.00012 | 0.00045 | 0.00000 | 0.00020 | 65 | 0.00042 | 0.00000 | 0.00015 | 93 | 0.00025 | 0.11565 | 128.570 | 0.90811 | 0.97664 |
| ABC.VB1X.P; putative thiamine transport system permease protein | 0.00043 | 0.00012 | 0.00045 | 0.00000 | 0.00020 | 65 | 0.00042 | 0.00000 | 0.00015 | 93 | 0.00025 | 0.11565 | 128.570 | 0.90811 | 0.97664 |
| ABC.VB1X.S; putative thiamine transport system substrate-binding protein | 0.00043 | 0.00012 | 0.00045 | 0.00000 | 0.00020 | 65 | 0.00042 | 0.00000 | 0.00015 | 93 | 0.00025 | 0.11565 | 128.570 | 0.90811 | 0.97664 |
| PTS-Mur-EIIC, murP; PTS system, N-acetylmuramic acid-specific IIC component | 0.00043 | 0.00016 | 0.00045 | 0.00000 | 0.00024 | 65 | 0.00042 | 0.00000 | 0.00021 | 93 | 0.00033 | 0.06628 | 142.028 | 0.94725 | 0.98674 |
| doeA; ectoine hydrolase [EC:3.5.4.44] | 0.00043 | 0.00015 | 0.00021 | 0.00000 | 0.00012 | 65 | 0.00059 | 0.00001 | 0.00024 | 93 | 0.00027 | -1.43431 | 129.919 | 0.15389 | 0.77376 |
| tsgA; MFS transporter, TsgA protein | 0.00043 | 0.00012 | 0.00049 | 0.00000 | 0.00020 | 65 | 0.00039 | 0.00000 | 0.00016 | 93 | 0.00026 | 0.39613 | 129.009 | 0.69266 | 0.91677 |
| ttrA; tetrathionate reductase subunit A | 0.00043 | 0.00011 | 0.00042 | 0.00000 | 0.00017 | 65 | 0.00044 | 0.00000 | 0.00014 | 93 | 0.00022 | -0.10410 | 139.127 | 0.91724 | 0.97692 |
| cbeA; cytoskeleton bundling-enhancing protein CbeA and related proteins | 0.00043 | 0.00029 | 0.00011 | 0.00000 | 0.00005 | 65 | 0.00066 | 0.00002 | 0.00048 | 93 | 0.00049 | -1.13385 | 93.975 | 0.25974 | 0.77376 |
| scsB; suppressor for copper-sensitivity B | 0.00043 | 0.00012 | 0.00063 | 0.00000 | 0.00026 | 65 | 0.00029 | 0.00000 | 0.00009 | 93 | 0.00028 | 1.22681 | 80.935 | 0.22345 | 0.77376 |
| rihB; ribosylpyrimidine nucleosidase [EC:3.2.2.8] | 0.00043 | 0.00009 | 0.00044 | 0.00000 | 0.00012 | 65 | 0.00042 | 0.00000 | 0.00013 | 93 | 0.00018 | 0.11085 | 155.480 | 0.91188 | 0.97664 |
| uxuR; GntR family transcriptional regulator, uxu operon transcriptional repressor | 0.00043 | 0.00010 | 0.00036 | 0.00000 | 0.00010 | 65 | 0.00047 | 0.00000 | 0.00016 | 93 | 0.00019 | -0.57430 | 147.236 | 0.56664 | 0.87051 |
| E1.14.13.7; phenol 2-monooxygenase [EC:1.14.13.7] | 0.00043 | 0.00016 | 0.00043 | 0.00000 | 0.00025 | 65 | 0.00043 | 0.00000 | 0.00020 | 93 | 0.00032 | -0.00463 | 136.109 | 0.99631 | 0.99944 |
| nifA; Nif-specific regulatory protein | 0.00043 | 0.00015 | 0.00044 | 0.00001 | 0.00032 | 65 | 0.00042 | 0.00000 | 0.00013 | 93 | 0.00034 | 0.07305 | 84.699 | 0.94194 | 0.98528 |
| E1.2.1.10; acetaldehyde dehydrogenase (acetylating) [EC:1.2.1.10] | 0.00042 | 0.00019 | 0.00051 | 0.00001 | 0.00033 | 65 | 0.00037 | 0.00000 | 0.00023 | 93 | 0.00040 | 0.34812 | 120.056 | 0.72836 | 0.92350 |
| cbtA; cytoskeleton-binding toxin CbtA and related proteins | 0.00042 | 0.00023 | 0.00026 | 0.00000 | 0.00017 | 65 | 0.00054 | 0.00001 | 0.00036 | 93 | 0.00040 | -0.69559 | 129.481 | 0.48793 | 0.84672 |
| vapB; antitoxin VapB | 0.00042 | 0.00012 | 0.00037 | 0.00000 | 0.00020 | 65 | 0.00046 | 0.00000 | 0.00015 | 93 | 0.00025 | -0.33490 | 126.794 | 0.73826 | 0.92908 |
| pglJ; N-acetylgalactosamine-N,N’-diacetylbacillosaminyl-diphospho-undecaprenol 4-alpha-N-acetylgalactosaminyltransferase [EC:2.4.1.291] | 0.00042 | 0.00013 | 0.00055 | 0.00000 | 0.00025 | 65 | 0.00033 | 0.00000 | 0.00013 | 93 | 0.00028 | 0.76703 | 96.631 | 0.44494 | 0.83274 |
| mdoC; glucans biosynthesis protein C [EC:2.1.-.-] | 0.00042 | 0.00012 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00044 | 0.00000 | 0.00015 | 93 | 0.00024 | -0.14689 | 131.412 | 0.88344 | 0.96974 |
| ERCC2, XPD; DNA excision repair protein ERCC-2 [EC:3.6.4.12] | 0.00042 | 0.00020 | 0.00081 | 0.00001 | 0.00046 | 65 | 0.00015 | 0.00000 | 0.00008 | 93 | 0.00047 | 1.42211 | 67.994 | 0.15957 | 0.77376 |
| manR; activator of the mannose operon, transcriptional antiterminator | 0.00042 | 0.00017 | 0.00020 | 0.00000 | 0.00010 | 65 | 0.00058 | 0.00001 | 0.00027 | 93 | 0.00029 | -1.30163 | 114.024 | 0.19567 | 0.77376 |
| PTS-Fru2-EIIA; PTS system, fructose-specific IIA-like component [EC:2.7.1.-] | 0.00042 | 0.00022 | 0.00021 | 0.00000 | 0.00010 | 65 | 0.00057 | 0.00001 | 0.00037 | 93 | 0.00038 | -0.96349 | 104.732 | 0.33752 | 0.78354 |
| fgd1; coenzyme F420-dependent glucose-6-phosphate dehydrogenase [EC:1.1.98.2] | 0.00042 | 0.00019 | 0.00021 | 0.00000 | 0.00011 | 65 | 0.00057 | 0.00001 | 0.00031 | 93 | 0.00033 | -1.08548 | 113.244 | 0.28001 | 0.77376 |
| paaJ; 3-oxo-5,6-didehydrosuberyl-CoA/3-oxoadipyl-CoA thiolase [EC:2.3.1.223 2.3.1.174] | 0.00042 | 0.00012 | 0.00041 | 0.00000 | 0.00019 | 65 | 0.00043 | 0.00000 | 0.00015 | 93 | 0.00025 | -0.10665 | 133.002 | 0.91523 | 0.97664 |
| drp35; lactonase [EC:3.1.1.-] | 0.00042 | 0.00017 | 0.00015 | 0.00000 | 0.00008 | 65 | 0.00061 | 0.00001 | 0.00028 | 93 | 0.00029 | -1.60312 | 105.737 | 0.11189 | 0.77376 |
| doeB; N2-acetyl-L-2,4-diaminobutanoate deacetylase [EC:3.5.1.125] | 0.00042 | 0.00014 | 0.00028 | 0.00000 | 0.00013 | 65 | 0.00052 | 0.00000 | 0.00023 | 93 | 0.00026 | -0.89047 | 140.599 | 0.37474 | 0.79810 |
| hdrB2; heterodisulfide reductase subunit B2 [EC:1.8.7.3 1.8.98.4 1.8.98.5 1.8.98.6] | 0.00042 | 0.00010 | 0.00043 | 0.00000 | 0.00015 | 65 | 0.00041 | 0.00000 | 0.00013 | 93 | 0.00020 | 0.06829 | 142.283 | 0.94565 | 0.98674 |
| wbiB; dTDP-L-rhamnose 4-epimerase [EC:5.1.3.25] | 0.00042 | 0.00011 | 0.00027 | 0.00000 | 0.00009 | 65 | 0.00053 | 0.00000 | 0.00017 | 93 | 0.00019 | -1.33357 | 137.178 | 0.18455 | 0.77376 |
| traE; conjugal transfer pilus assembly protein TraE | 0.00042 | 0.00012 | 0.00041 | 0.00000 | 0.00020 | 65 | 0.00043 | 0.00000 | 0.00016 | 93 | 0.00026 | -0.07097 | 130.336 | 0.94353 | 0.98634 |
| K09003; uncharacterized protein | 0.00042 | 0.00013 | 0.00018 | 0.00000 | 0.00007 | 65 | 0.00058 | 0.00000 | 0.00021 | 93 | 0.00022 | -1.79206 | 111.868 | 0.07583 | 0.74577 |
| phsA, psrA; thiosulfate reductase / polysulfide reductase chain A [EC:1.8.5.5] | 0.00042 | 0.00013 | 0.00054 | 0.00000 | 0.00025 | 65 | 0.00033 | 0.00000 | 0.00013 | 93 | 0.00028 | 0.75867 | 97.362 | 0.44988 | 0.83438 |
| mtlR; mannitol operon repressor | 0.00042 | 0.00012 | 0.00046 | 0.00000 | 0.00020 | 65 | 0.00039 | 0.00000 | 0.00015 | 93 | 0.00024 | 0.27444 | 127.596 | 0.78419 | 0.94430 |
| merC; mercuric ion transport protein | 0.00042 | 0.00014 | 0.00050 | 0.00000 | 0.00023 | 65 | 0.00036 | 0.00000 | 0.00018 | 93 | 0.00030 | 0.47613 | 131.751 | 0.63477 | 0.89696 |
| mshC; MSHA pilin protein MshC | 0.00042 | 0.00018 | 0.00047 | 0.00000 | 0.00024 | 65 | 0.00038 | 0.00001 | 0.00025 | 93 | 0.00035 | 0.25133 | 153.714 | 0.80190 | 0.94855 |
| mshD; MSHA pilin protein MshD | 0.00042 | 0.00018 | 0.00047 | 0.00000 | 0.00024 | 65 | 0.00038 | 0.00001 | 0.00025 | 93 | 0.00035 | 0.25133 | 153.714 | 0.80190 | 0.94855 |
| mshE; MSHA biogenesis protein MshE | 0.00042 | 0.00018 | 0.00047 | 0.00000 | 0.00024 | 65 | 0.00038 | 0.00001 | 0.00025 | 93 | 0.00035 | 0.25133 | 153.714 | 0.80190 | 0.94855 |
| mshG; MSHA biogenesis protein MshG | 0.00042 | 0.00018 | 0.00047 | 0.00000 | 0.00024 | 65 | 0.00038 | 0.00001 | 0.00025 | 93 | 0.00035 | 0.25133 | 153.714 | 0.80190 | 0.94855 |
| mshI; MSHA biogenesis protein MshI | 0.00042 | 0.00018 | 0.00047 | 0.00000 | 0.00024 | 65 | 0.00038 | 0.00001 | 0.00025 | 93 | 0.00035 | 0.25133 | 153.714 | 0.80190 | 0.94855 |
| mshL; MSHA biogenesis protein MshL | 0.00042 | 0.00018 | 0.00047 | 0.00000 | 0.00024 | 65 | 0.00038 | 0.00001 | 0.00025 | 93 | 0.00035 | 0.25133 | 153.714 | 0.80190 | 0.94855 |
| mshO; MSHA biogenesis protein MshO | 0.00042 | 0.00018 | 0.00047 | 0.00000 | 0.00024 | 65 | 0.00038 | 0.00001 | 0.00025 | 93 | 0.00035 | 0.25133 | 153.714 | 0.80190 | 0.94855 |
| ygeR; lipoprotein YgeR | 0.00042 | 0.00012 | 0.00049 | 0.00000 | 0.00021 | 65 | 0.00037 | 0.00000 | 0.00014 | 93 | 0.00025 | 0.48285 | 120.760 | 0.63008 | 0.89536 |
| qheDH, qbdA; quinohemoprotein ethanol dehydrogenase [EC:1.1.9.1] | 0.00042 | 0.00012 | 0.00017 | 0.00000 | 0.00006 | 65 | 0.00059 | 0.00000 | 0.00020 | 93 | 0.00021 | -2.01096 | 105.363 | 0.04688 | 0.66255 |
| traL; conjugal transfer pilus assembly protein TraL | 0.00042 | 0.00014 | 0.00032 | 0.00000 | 0.00015 | 65 | 0.00048 | 0.00000 | 0.00021 | 93 | 0.00026 | -0.63845 | 153.751 | 0.52413 | 0.85711 |
| npr; thermolysin [EC:3.4.24.27] | 0.00042 | 0.00019 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00069 | 0.00001 | 0.00031 | 93 | 0.00031 | -2.16818 | 92.297 | 0.03272 | 0.62130 |
| E1.14.11.1; gamma-butyrobetaine dioxygenase [EC:1.14.11.1] | 0.00041 | 0.00013 | 0.00041 | 0.00000 | 0.00017 | 65 | 0.00042 | 0.00000 | 0.00018 | 93 | 0.00025 | -0.02651 | 153.029 | 0.97888 | 0.99657 |
| galB; 4-oxalomesaconate hydratase [EC:4.2.1.83] | 0.00041 | 0.00012 | 0.00012 | 0.00000 | 0.00004 | 65 | 0.00062 | 0.00000 | 0.00020 | 93 | 0.00020 | -2.53880 | 98.133 | 0.01270 | 0.59095 |
| oxa; beta-lactamase class D [EC:3.5.2.6] | 0.00041 | 0.00012 | 0.00009 | 0.00000 | 0.00003 | 65 | 0.00064 | 0.00000 | 0.00021 | 93 | 0.00021 | -2.64125 | 96.423 | 0.00964 | 0.58112 |
| namH; UDP-MurNAc hydroxylase | 0.00041 | 0.00019 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00063 | 0.00001 | 0.00031 | 93 | 0.00032 | -1.60601 | 98.101 | 0.11149 | 0.77376 |
| hdrC2; heterodisulfide reductase subunit C2 [EC:1.8.7.3 1.8.98.4 1.8.98.5 1.8.98.6] | 0.00041 | 0.00010 | 0.00043 | 0.00000 | 0.00015 | 65 | 0.00040 | 0.00000 | 0.00013 | 93 | 0.00020 | 0.12009 | 141.883 | 0.90458 | 0.97658 |
| sqhC; sporulenol synthase [EC:4.2.1.137] | 0.00041 | 0.00016 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00065 | 0.00001 | 0.00026 | 93 | 0.00026 | -2.18148 | 95.264 | 0.03161 | 0.61842 |
| lsrC; AI-2 transport system permease protein | 0.00041 | 0.00012 | 0.00027 | 0.00000 | 0.00010 | 65 | 0.00051 | 0.00000 | 0.00019 | 93 | 0.00021 | -1.11803 | 134.162 | 0.26555 | 0.77376 |
| PTS-Dgl-EIIA, gamP; PTS system, D-glucosamine-specific IIA component [EC:2.7.1.-] | 0.00041 | 0.00020 | 0.00050 | 0.00001 | 0.00046 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00048 | 0.32974 | 76.189 | 0.74250 | 0.93050 |
| rirA; Rrf2 family transcriptional regulator, iron-responsive regulator | 0.00041 | 0.00017 | 0.00006 | 0.00000 | 0.00002 | 65 | 0.00066 | 0.00001 | 0.00029 | 93 | 0.00029 | -2.03083 | 93.227 | 0.04512 | 0.65981 |
| lsrA, ego; AI-2 transport system ATP-binding protein | 0.00041 | 0.00012 | 0.00027 | 0.00000 | 0.00010 | 65 | 0.00051 | 0.00000 | 0.00019 | 93 | 0.00021 | -1.13086 | 134.160 | 0.26013 | 0.77376 |
| lsrB; AI-2 transport system substrate-binding protein | 0.00041 | 0.00012 | 0.00027 | 0.00000 | 0.00010 | 65 | 0.00051 | 0.00000 | 0.00019 | 93 | 0.00021 | -1.13086 | 134.160 | 0.26013 | 0.77376 |
| lsrD; AI-2 transport system permease protein | 0.00041 | 0.00012 | 0.00027 | 0.00000 | 0.00010 | 65 | 0.00051 | 0.00000 | 0.00019 | 93 | 0.00021 | -1.13086 | 134.160 | 0.26013 | 0.77376 |
| crtQ; 4,4’-diaponeurosporenoate glycosyltransferase [EC:2.4.1.-] | 0.00041 | 0.00017 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00061 | 0.00001 | 0.00028 | 93 | 0.00029 | -1.65889 | 104.778 | 0.10013 | 0.77098 |
| K10212, crtO; glycosyl-4,4’-diaponeurosporenoate acyltransferase [EC:2.3.1.-] | 0.00041 | 0.00017 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00061 | 0.00001 | 0.00028 | 93 | 0.00029 | -1.65889 | 104.778 | 0.10013 | 0.77098 |
| GULO; L-gulonolactone oxidase [EC:1.1.3.8] | 0.00041 | 0.00020 | 0.00019 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.13538 | 101.355 | 0.25889 | 0.77376 |
| E1.8.2.1; sulfite dehydrogenase [EC:1.8.2.1] | 0.00041 | 0.00023 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00065 | 0.00001 | 0.00039 | 93 | 0.00039 | -1.49300 | 94.079 | 0.13878 | 0.77376 |
| hdrA2; heterodisulfide reductase subunit A2 [EC:1.8.7.3 1.8.98.4 1.8.98.5 1.8.98.6] | 0.00041 | 0.00010 | 0.00039 | 0.00000 | 0.00013 | 65 | 0.00043 | 0.00000 | 0.00013 | 93 | 0.00019 | -0.21730 | 151.934 | 0.82827 | 0.95506 |
| traB; conjugal transfer pilus assembly protein TraB | 0.00041 | 0.00013 | 0.00040 | 0.00000 | 0.00020 | 65 | 0.00042 | 0.00000 | 0.00018 | 93 | 0.00027 | -0.08033 | 140.909 | 0.93609 | 0.98459 |
| HIBCH; 3-hydroxyisobutyryl-CoA hydrolase [EC:3.1.2.4] | 0.00041 | 0.00018 | 0.00022 | 0.00000 | 0.00014 | 65 | 0.00054 | 0.00001 | 0.00029 | 93 | 0.00032 | -1.00053 | 130.102 | 0.31891 | 0.77376 |
| nadM; nicotinamide-nucleotide adenylyltransferase [EC:2.7.7.1] | 0.00041 | 0.00013 | 0.00060 | 0.00001 | 0.00029 | 65 | 0.00027 | 0.00000 | 0.00008 | 93 | 0.00030 | 1.09768 | 75.049 | 0.27585 | 0.77376 |
| artJ; arginine transport system substrate-binding protein | 0.00041 | 0.00012 | 0.00043 | 0.00000 | 0.00020 | 65 | 0.00039 | 0.00000 | 0.00015 | 93 | 0.00025 | 0.17126 | 130.581 | 0.86428 | 0.96263 |
| pseF; pseudaminic acid cytidylyltransferase [EC:2.7.7.81] | 0.00041 | 0.00010 | 0.00041 | 0.00000 | 0.00017 | 65 | 0.00040 | 0.00000 | 0.00014 | 93 | 0.00021 | 0.04319 | 136.561 | 0.96561 | 0.99101 |
| UXE, uxe; UDP-arabinose 4-epimerase [EC:5.1.3.5] | 0.00040 | 0.00018 | 0.00016 | 0.00000 | 0.00007 | 65 | 0.00058 | 0.00001 | 0.00029 | 93 | 0.00030 | -1.37581 | 101.156 | 0.17192 | 0.77376 |
| mepA; multidrug efflux pump | 0.00040 | 0.00016 | 0.00018 | 0.00000 | 0.00010 | 65 | 0.00056 | 0.00001 | 0.00027 | 93 | 0.00028 | -1.35963 | 115.727 | 0.17659 | 0.77376 |
| chbR, celD; AraC family transcriptional regulator, dual regulator of chb operon | 0.00040 | 0.00012 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00041 | 0.00000 | 0.00015 | 93 | 0.00024 | -0.11117 | 130.652 | 0.91166 | 0.97664 |
| ttuB; MFS transporter, ACS family, tartrate transporter | 0.00040 | 0.00014 | 0.00049 | 0.00000 | 0.00023 | 65 | 0.00034 | 0.00000 | 0.00019 | 93 | 0.00030 | 0.48769 | 134.556 | 0.62657 | 0.89255 |
| exoZ; exopolysaccharide production protein ExoZ | 0.00040 | 0.00013 | 0.00010 | 0.00000 | 0.00004 | 65 | 0.00061 | 0.00000 | 0.00022 | 93 | 0.00022 | -2.28158 | 99.223 | 0.02465 | 0.59764 |
| SGA1; glucoamylase [EC:3.2.1.3] | 0.00040 | 0.00014 | 0.00038 | 0.00000 | 0.00018 | 65 | 0.00042 | 0.00000 | 0.00021 | 93 | 0.00028 | -0.14734 | 155.717 | 0.88305 | 0.96974 |
| mshP; MSHA biogenesis protein MshP | 0.00040 | 0.00017 | 0.00044 | 0.00000 | 0.00022 | 65 | 0.00037 | 0.00001 | 0.00025 | 93 | 0.00034 | 0.18426 | 155.557 | 0.85405 | 0.95848 |
| pglC; undecaprenyl phosphate N,N’-diacetylbacillosamine 1-phosphate transferase [EC:2.7.8.36] | 0.00040 | 0.00013 | 0.00051 | 0.00000 | 0.00025 | 65 | 0.00032 | 0.00000 | 0.00013 | 93 | 0.00028 | 0.69521 | 97.071 | 0.48859 | 0.84744 |
| mcyA; microcystin synthetase protein McyA | 0.00040 | 0.00019 | 0.00051 | 0.00001 | 0.00033 | 65 | 0.00032 | 0.00000 | 0.00022 | 93 | 0.00040 | 0.45671 | 119.354 | 0.64871 | 0.90350 |
| pglA; N,N’-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase [EC:2.4.1.290] | 0.00040 | 0.00013 | 0.00051 | 0.00000 | 0.00025 | 65 | 0.00032 | 0.00000 | 0.00013 | 93 | 0.00028 | 0.70177 | 97.089 | 0.48450 | 0.84354 |
| pglD; UDP-N-acetylbacillosamine N-acetyltransferase [EC:2.3.1.203] | 0.00040 | 0.00013 | 0.00051 | 0.00000 | 0.00025 | 65 | 0.00032 | 0.00000 | 0.00013 | 93 | 0.00028 | 0.70177 | 97.089 | 0.48450 | 0.84354 |
| pglE; UDP-N-acetylbacillosamine transaminase [EC:2.6.1.34] | 0.00040 | 0.00013 | 0.00051 | 0.00000 | 0.00025 | 65 | 0.00032 | 0.00000 | 0.00013 | 93 | 0.00028 | 0.70177 | 97.089 | 0.48450 | 0.84354 |
| pglF; UDP-N-acetyl-D-glucosamine 4,6-dehydratase [EC:4.2.1.135] | 0.00040 | 0.00013 | 0.00051 | 0.00000 | 0.00025 | 65 | 0.00032 | 0.00000 | 0.00013 | 93 | 0.00028 | 0.70177 | 97.089 | 0.48450 | 0.84354 |
| psrB; polysulfide reductase chain B | 0.00040 | 0.00013 | 0.00051 | 0.00000 | 0.00025 | 65 | 0.00032 | 0.00000 | 0.00013 | 93 | 0.00028 | 0.70177 | 97.089 | 0.48450 | 0.84354 |
| psrC; polysulfide reductase chain C | 0.00040 | 0.00013 | 0.00051 | 0.00000 | 0.00025 | 65 | 0.00032 | 0.00000 | 0.00013 | 93 | 0.00028 | 0.70177 | 97.089 | 0.48450 | 0.84354 |
| hyfH; hydrogenase-4 component H | 0.00040 | 0.00016 | 0.00054 | 0.00001 | 0.00032 | 65 | 0.00030 | 0.00000 | 0.00015 | 93 | 0.00035 | 0.70491 | 91.404 | 0.48266 | 0.84354 |
| hdc, HDC; histidine decarboxylase [EC:4.1.1.22] | 0.00040 | 0.00022 | 0.00054 | 0.00001 | 0.00041 | 65 | 0.00029 | 0.00000 | 0.00023 | 93 | 0.00047 | 0.53401 | 103.897 | 0.59448 | 0.88005 |
| mshM; MSHA biogenesis protein MshM | 0.00040 | 0.00018 | 0.00047 | 0.00000 | 0.00024 | 65 | 0.00035 | 0.00001 | 0.00025 | 93 | 0.00035 | 0.35490 | 153.412 | 0.72315 | 0.92156 |
| srlR, gutR; DeoR family transcriptional regulator, glucitol operon repressor | 0.00040 | 0.00012 | 0.00043 | 0.00000 | 0.00020 | 65 | 0.00037 | 0.00000 | 0.00015 | 93 | 0.00025 | 0.20579 | 129.624 | 0.83728 | 0.95506 |
| bjaR1, rpaR, rhiR; LuxR family transcriptional regulator, quorum-sensing system regulator BjaR1 | 0.00040 | 0.00017 | 0.00005 | 0.00000 | 0.00002 | 65 | 0.00064 | 0.00001 | 0.00029 | 93 | 0.00029 | -2.06571 | 93.088 | 0.04164 | 0.64661 |
| afr; 1,5-anhydro-D-fructose reductase (1,5-anhydro-D-mannitol-forming) [EC:1.1.1.292] | 0.00039 | 0.00017 | 0.00016 | 0.00000 | 0.00012 | 65 | 0.00056 | 0.00001 | 0.00029 | 93 | 0.00031 | -1.30802 | 120.063 | 0.19336 | 0.77376 |
| bhsA; multiple stress resistance protein BhsA | 0.00039 | 0.00012 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00039 | 0.00000 | 0.00015 | 93 | 0.00025 | 0.01736 | 134.672 | 0.98617 | 0.99729 |
| lipV; lipase [EC:3.1.1.-] | 0.00039 | 0.00019 | 0.00014 | 0.00000 | 0.00006 | 65 | 0.00057 | 0.00001 | 0.00031 | 93 | 0.00032 | -1.37024 | 98.907 | 0.17371 | 0.77376 |
| lacK; lactose/L-arabinose transport system ATP-binding protein | 0.00039 | 0.00014 | 0.00012 | 0.00000 | 0.00006 | 65 | 0.00059 | 0.00001 | 0.00023 | 93 | 0.00024 | -1.93665 | 104.795 | 0.05548 | 0.70577 |
| ydfJ; MFS transporter, MHS family, metabolite:H+ symporter | 0.00039 | 0.00011 | 0.00044 | 0.00000 | 0.00019 | 65 | 0.00036 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.32752 | 123.471 | 0.74383 | 0.93099 |
| comGE; competence protein ComGE | 0.00039 | 0.00019 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00065 | 0.00001 | 0.00033 | 93 | 0.00033 | -1.91837 | 92.547 | 0.05815 | 0.71816 |
| ligC; 2-hydroxy-4-carboxymuconate semialdehyde hemiacetal dehydrogenase [EC:1.1.1.312] | 0.00039 | 0.00020 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00062 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.61556 | 94.317 | 0.10953 | 0.77376 |
| exoM; succinoglycan biosynthesis protein ExoM [EC:2.4.-.-] | 0.00039 | 0.00020 | 0.00014 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00033 | 93 | 0.00034 | -1.29993 | 100.053 | 0.19661 | 0.77376 |
| cypD_E, CYP102A2_3; cytochrome P450 / NADPH-cytochrome P450 reductase [EC:1.14.14.1 1.6.2.4] | 0.00039 | 0.00012 | 0.00027 | 0.00000 | 0.00013 | 65 | 0.00047 | 0.00000 | 0.00019 | 93 | 0.00023 | -0.89206 | 149.730 | 0.37379 | 0.79810 |
| yibL; ribosome-associated protein | 0.00039 | 0.00012 | 0.00045 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.39657 | 125.129 | 0.69236 | 0.91677 |
| yidP; GntR family transcriptional regulator, glv operon transcriptional regulator | 0.00039 | 0.00016 | 0.00030 | 0.00000 | 0.00012 | 65 | 0.00045 | 0.00001 | 0.00026 | 93 | 0.00028 | -0.51530 | 126.927 | 0.60724 | 0.88598 |
| K06937; 7,8-dihydro-6-hydroxymethylpterin dimethyltransferase [EC:2.1.1.-] | 0.00039 | 0.00011 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00060 | 0.00000 | 0.00017 | 93 | 0.00018 | -2.76002 | 113.140 | 0.00674 | 0.54612 |
| gfa; S-(hydroxymethyl)glutathione synthase [EC:4.4.1.22] | 0.00039 | 0.00012 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00053 | 0.00000 | 0.00020 | 93 | 0.00022 | -1.56885 | 121.009 | 0.11929 | 0.77376 |
| inhA; enoyl ACP reductase [EC:1.3.1.9] | 0.00039 | 0.00019 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00061 | 0.00001 | 0.00032 | 93 | 0.00032 | -1.68200 | 94.833 | 0.09586 | 0.76609 |
| K03822; putative long chain acyl-CoA synthase [EC:6.2.1.-] | 0.00039 | 0.00019 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00061 | 0.00001 | 0.00032 | 93 | 0.00032 | -1.68200 | 94.833 | 0.09586 | 0.76609 |
| mabA; beta-ketoacyl ACP reductase [EC:1.1.1.100] | 0.00039 | 0.00019 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00061 | 0.00001 | 0.00032 | 93 | 0.00032 | -1.68200 | 94.833 | 0.09586 | 0.76609 |
| eptB; KDO II ethanolaminephosphotransferase [EC:2.7.8.42] | 0.00039 | 0.00012 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00038 | 0.00000 | 0.00015 | 93 | 0.00024 | 0.08947 | 132.473 | 0.92885 | 0.98176 |
| motD; chemotaxis protein MotD | 0.00039 | 0.00017 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00062 | 0.00001 | 0.00029 | 93 | 0.00029 | -1.89528 | 93.372 | 0.06115 | 0.73001 |
| wbpE, wlbC; UDP-2-acetamido-2-deoxy-ribo-hexuluronate aminotransferase [EC:2.6.1.98] | 0.00039 | 0.00012 | 0.00042 | 0.00000 | 0.00022 | 65 | 0.00036 | 0.00000 | 0.00014 | 93 | 0.00026 | 0.23421 | 110.132 | 0.81525 | 0.95409 |
| dcuR; two-component system, CitB family, response regulator DcuR | 0.00039 | 0.00012 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00038 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.10405 | 127.605 | 0.91729 | 0.97692 |
| crtN; 4,4’-diapophytoene desaturase [EC:1.3.8.2] | 0.00039 | 0.00017 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00027 | 93 | 0.00028 | -1.58622 | 105.122 | 0.11569 | 0.77376 |
| crtP; diapolycopene oxygenase [EC:1.14.99.44] | 0.00039 | 0.00017 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00027 | 93 | 0.00028 | -1.58622 | 105.122 | 0.11569 | 0.77376 |
| graS; two-component system, OmpR family, sensor histidine kinase GraS [EC:2.7.13.3] | 0.00039 | 0.00017 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00027 | 93 | 0.00028 | -1.58622 | 105.122 | 0.11569 | 0.77376 |
| lytM; lysostaphin [EC:3.4.24.75] | 0.00039 | 0.00017 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00027 | 93 | 0.00028 | -1.58622 | 105.122 | 0.11569 | 0.77376 |
| norA; MFS transporter, DHA1 family, quinolone resistance protein | 0.00039 | 0.00017 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00027 | 93 | 0.00028 | -1.58622 | 105.122 | 0.11569 | 0.77376 |
| sspB2, sspP, scpA; staphopain A [EC:3.4.22.48] | 0.00039 | 0.00017 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00027 | 93 | 0.00028 | -1.58622 | 105.122 | 0.11569 | 0.77376 |
| vraG; cationic antimicrobial peptide transport system permease protein | 0.00039 | 0.00017 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00027 | 93 | 0.00028 | -1.58622 | 105.122 | 0.11569 | 0.77376 |
| ybdH; uncharacterized oxidoreductase [EC:1.1.-.-] | 0.00039 | 0.00009 | 0.00033 | 0.00000 | 0.00010 | 65 | 0.00043 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.59998 | 152.959 | 0.54941 | 0.86645 |
| nudG; (d)CTP diphosphatase [EC:3.6.1.65] | 0.00039 | 0.00012 | 0.00044 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.39535 | 125.170 | 0.69326 | 0.91677 |
| gutM; glucitol operon activator protein | 0.00038 | 0.00011 | 0.00041 | 0.00000 | 0.00019 | 65 | 0.00037 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.19203 | 125.398 | 0.84803 | 0.95712 |
| chiF; putative chitobiose transport system permease protein | 0.00038 | 0.00018 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00057 | 0.00001 | 0.00031 | 93 | 0.00032 | -1.40212 | 101.991 | 0.16391 | 0.77376 |
| K09124; uncharacterized protein | 0.00038 | 0.00012 | 0.00012 | 0.00000 | 0.00011 | 65 | 0.00057 | 0.00000 | 0.00020 | 93 | 0.00022 | -1.99474 | 137.018 | 0.04806 | 0.66712 |
| yxeM; putative amino-acid transport system substrate-binding protein | 0.00038 | 0.00011 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00038 | 0.00000 | 0.00012 | 93 | 0.00022 | 0.02464 | 115.306 | 0.98039 | 0.99657 |
| mpaA; protein MpaA | 0.00038 | 0.00012 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00037 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.13632 | 127.494 | 0.89178 | 0.97141 |
| CYP152A; fatty-acid peroxygenase [EC:1.11.2.4] | 0.00038 | 0.00012 | 0.00051 | 0.00000 | 0.00025 | 65 | 0.00029 | 0.00000 | 0.00011 | 93 | 0.00027 | 0.81156 | 91.253 | 0.41915 | 0.81980 |
| chiE; putative chitobiose transport system substrate-binding protein | 0.00038 | 0.00018 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00056 | 0.00001 | 0.00031 | 93 | 0.00032 | -1.39237 | 101.988 | 0.16684 | 0.77376 |
| mdo; formaldehyde dismutase / methanol dehydrogenase [EC:1.2.98.1 1.1.99.37] | 0.00038 | 0.00019 | 0.00017 | 0.00000 | 0.00012 | 65 | 0.00052 | 0.00001 | 0.00032 | 93 | 0.00034 | -1.03273 | 116.582 | 0.30387 | 0.77376 |
| ompC; outer membrane pore protein C | 0.00038 | 0.00012 | 0.00041 | 0.00000 | 0.00019 | 65 | 0.00036 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.20633 | 127.924 | 0.83686 | 0.95506 |
| doeC; aspartate-semialdehyde dehydrogenase [EC:1.2.1.-] | 0.00038 | 0.00012 | 0.00021 | 0.00000 | 0.00012 | 65 | 0.00050 | 0.00000 | 0.00019 | 93 | 0.00023 | -1.29351 | 143.241 | 0.19792 | 0.77376 |
| sigI; RNA polymerase sigma factor | 0.00038 | 0.00012 | 0.00012 | 0.00000 | 0.00005 | 65 | 0.00056 | 0.00000 | 0.00020 | 93 | 0.00021 | -2.18022 | 102.146 | 0.03154 | 0.61842 |
| puuR; HTH-type transcriptional regulator, repressor for puuD | 0.00038 | 0.00012 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00037 | 0.00000 | 0.00015 | 93 | 0.00024 | 0.10564 | 131.996 | 0.91603 | 0.97681 |
| lnuA_C_D_E, lin; lincosamide nucleotidyltransferase A/C/D/E | 0.00038 | 0.00024 | 0.00058 | 0.00002 | 0.00056 | 65 | 0.00024 | 0.00000 | 0.00011 | 93 | 0.00058 | 0.59522 | 69.194 | 0.55364 | 0.86795 |
| nboR; nicotine blue oxidoreductase [EC:1.1.1.328] | 0.00038 | 0.00009 | 0.00036 | 0.00000 | 0.00015 | 65 | 0.00039 | 0.00000 | 0.00011 | 93 | 0.00019 | -0.15630 | 130.953 | 0.87604 | 0.96703 |
| ynfE; Tat-targeted selenate reductase subunit YnfE [EC:1.97.1.9] | 0.00038 | 0.00012 | 0.00026 | 0.00000 | 0.00009 | 65 | 0.00046 | 0.00000 | 0.00018 | 93 | 0.00021 | -0.96072 | 131.956 | 0.33845 | 0.78407 |
| ybiS; L,D-transpeptidase YbiS | 0.00038 | 0.00011 | 0.00041 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.25236 | 126.591 | 0.80117 | 0.94855 |
| pspD; phage shock protein D | 0.00037 | 0.00011 | 0.00041 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.24433 | 126.406 | 0.80737 | 0.94987 |
| tabA; biofilm protein TabA | 0.00037 | 0.00011 | 0.00043 | 0.00000 | 0.00019 | 65 | 0.00034 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.39152 | 123.510 | 0.69608 | 0.91772 |
| E4.1.1.82; phosphonopyruvate decarboxylase [EC:4.1.1.82] | 0.00037 | 0.00009 | 0.00030 | 0.00000 | 0.00009 | 65 | 0.00043 | 0.00000 | 0.00014 | 93 | 0.00016 | -0.81659 | 145.805 | 0.41549 | 0.81730 |
| UMF2; MFS transporter, UMF2 family, putative MFS family transporter protein | 0.00037 | 0.00011 | 0.00041 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.23189 | 126.612 | 0.81700 | 0.95487 |
| ccdA; antitoxin CcdA | 0.00037 | 0.00011 | 0.00039 | 0.00000 | 0.00022 | 65 | 0.00036 | 0.00000 | 0.00011 | 93 | 0.00025 | 0.11602 | 94.779 | 0.90788 | 0.97664 |
| tatE; sec-independent protein translocase protein TatE | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.20495 | 126.646 | 0.83794 | 0.95506 |
| MKS1; Meckel syndrome type 1 protein | 0.00037 | 0.00014 | 0.00041 | 0.00000 | 0.00024 | 65 | 0.00035 | 0.00000 | 0.00018 | 93 | 0.00030 | 0.19783 | 125.937 | 0.84350 | 0.95519 |
| aaeA; p-hydroxybenzoic acid efflux pump subunit AaeA | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| aaeB; p-hydroxybenzoic acid efflux pump subunit AaeB | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| acrD; multidrug efflux pump | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| basS; two-component system, OmpR family, sensor histidine kinase BasS [EC:2.7.13.3] | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| dcuS; two-component system, CitB family, sensor histidine kinase DcuS [EC:2.7.13.3] | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| iap; alkaline phosphatase isozyme conversion protein [EC:3.4.11.-] | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| mdtD; MFS transporter, DHA2 family, multidrug resistance protein | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| mepH; murein DD-endopeptidase [EC:3.4.-.-] | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| msyB; acidic protein MsyB | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| proY; proline-specific permease ProY | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21131 | 126.573 | 0.83298 | 0.95506 |
| phnV; 2-aminoethylphosphonate transport system permease protein | 0.00037 | 0.00020 | 0.00028 | 0.00000 | 0.00018 | 65 | 0.00043 | 0.00001 | 0.00031 | 93 | 0.00036 | -0.43153 | 140.783 | 0.66675 | 0.91126 |
| cooC; CO dehydrogenase maturation factor | 0.00037 | 0.00016 | 0.00024 | 0.00000 | 0.00013 | 65 | 0.00047 | 0.00001 | 0.00026 | 93 | 0.00029 | -0.78773 | 130.446 | 0.43228 | 0.82825 |
| badA; benzoate-CoA ligase [EC:6.2.1.25] | 0.00037 | 0.00020 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00058 | 0.00001 | 0.00033 | 93 | 0.00033 | -1.50711 | 95.799 | 0.13507 | 0.77376 |
| ecnB; entericidin B | 0.00037 | 0.00011 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.17345 | 126.567 | 0.86258 | 0.96263 |
| lsrG; (4S)-4-hydroxy-5-phosphonooxypentane-2,3-dione isomerase [EC:5.3.1.32] | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.18843 | 126.600 | 0.85084 | 0.95754 |
| phoE; outer membrane pore protein E | 0.00037 | 0.00011 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.16665 | 126.793 | 0.86791 | 0.96364 |
| chaB; cation transport regulator | 0.00037 | 0.00011 | 0.00041 | 0.00000 | 0.00019 | 65 | 0.00034 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.26173 | 124.740 | 0.79396 | 0.94783 |
| shiA; MFS transporter, MHS family, shikimate and dehydroshikimate transport protein | 0.00037 | 0.00011 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.16747 | 125.639 | 0.86727 | 0.96329 |
| flhE; flagellar protein FlhE | 0.00037 | 0.00011 | 0.00043 | 0.00000 | 0.00019 | 65 | 0.00032 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.47314 | 122.460 | 0.63696 | 0.89696 |
| hdeB; acid stress chaperone HdeB | 0.00037 | 0.00013 | 0.00016 | 0.00000 | 0.00005 | 65 | 0.00051 | 0.00000 | 0.00021 | 93 | 0.00022 | -1.64184 | 100.665 | 0.10374 | 0.77376 |
| exuR; GntR family transcriptional regulator, hexuronate regulon transcriptional repressor | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21016 | 126.622 | 0.83388 | 0.95506 |
| ibpB; molecular chaperone IbpB | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21016 | 126.622 | 0.83388 | 0.95506 |
| uspC; universal stress protein C | 0.00037 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.21016 | 126.622 | 0.83388 | 0.95506 |
| chiG; putative chitobiose transport system permease protein | 0.00037 | 0.00018 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00054 | 0.00001 | 0.00031 | 93 | 0.00031 | -1.32389 | 102.126 | 0.18849 | 0.77376 |
| hyfG; hydrogenase-4 component G [EC:1.-.-.-] | 0.00037 | 0.00016 | 0.00053 | 0.00001 | 0.00032 | 65 | 0.00026 | 0.00000 | 0.00015 | 93 | 0.00035 | 0.77002 | 90.586 | 0.44329 | 0.83186 |
| rsbX; phosphoserine phosphatase RsbX [EC:3.1.3.3] | 0.00037 | 0.00014 | 0.00014 | 0.00000 | 0.00008 | 65 | 0.00053 | 0.00000 | 0.00023 | 93 | 0.00024 | -1.57977 | 113.890 | 0.11693 | 0.77376 |
| IAL; isopenicillin-N N-acyltransferase like protein | 0.00037 | 0.00012 | 0.00032 | 0.00000 | 0.00014 | 65 | 0.00040 | 0.00000 | 0.00017 | 93 | 0.00022 | -0.36726 | 155.804 | 0.71393 | 0.92004 |
| sbmC; DNA gyrase inhibitor | 0.00036 | 0.00011 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.17222 | 126.587 | 0.86354 | 0.96263 |
| gutQ; arabinose 5-phosphate isomerase [EC:5.3.1.13] | 0.00036 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00034 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.24812 | 125.417 | 0.80445 | 0.94931 |
| SELENBP1; selenium-binding protein 1 | 0.00036 | 0.00012 | 0.00018 | 0.00000 | 0.00008 | 65 | 0.00049 | 0.00000 | 0.00019 | 93 | 0.00021 | -1.48512 | 124.519 | 0.14004 | 0.77376 |
| xltB; putative xylitol transport system permease protein | 0.00036 | 0.00017 | 0.00010 | 0.00000 | 0.00004 | 65 | 0.00054 | 0.00001 | 0.00028 | 93 | 0.00028 | -1.55507 | 96.706 | 0.12320 | 0.77376 |
| feaR; AraC family transcriptional regulator, positive regulator of tynA and feaB | 0.00036 | 0.00011 | 0.00040 | 0.00000 | 0.00019 | 65 | 0.00033 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.27464 | 125.584 | 0.78404 | 0.94429 |
| salR; two-component system, NarL family, secretion system response regulator SalR | 0.00036 | 0.00020 | 0.00035 | 0.00000 | 0.00027 | 65 | 0.00037 | 0.00001 | 0.00028 | 93 | 0.00039 | -0.05147 | 152.976 | 0.95902 | 0.98903 |
| merE; mercuric ion transport protein | 0.00036 | 0.00020 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00055 | 0.00001 | 0.00034 | 93 | 0.00034 | -1.36524 | 94.499 | 0.17542 | 0.77376 |
| rcsA; LuxR family transcriptional regulator, capsular biosynthesis positive transcription factor | 0.00036 | 0.00011 | 0.00037 | 0.00000 | 0.00019 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.09391 | 127.153 | 0.92533 | 0.98041 |
| MFS.SP; MFS transporter, SP family, sugar porter, other | 0.00036 | 0.00014 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00054 | 0.00000 | 0.00023 | 93 | 0.00023 | -1.84023 | 103.376 | 0.06860 | 0.73473 |
| puuD; gamma-glutamyl-gamma-aminobutyrate hydrolase [EC:3.5.1.94] | 0.00036 | 0.00011 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00034 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.20753 | 125.973 | 0.83593 | 0.95506 |
| cebE; cellobiose transport system substrate-binding protein | 0.00036 | 0.00011 | 0.00038 | 0.00000 | 0.00020 | 65 | 0.00034 | 0.00000 | 0.00011 | 93 | 0.00023 | 0.16749 | 105.283 | 0.86731 | 0.96329 |
| nahAa, nagAa, ndoR, nbzAa, dntAa; naphthalene 1,2-dioxygenase ferredoxin reductase component [EC:1.18.1.7] | 0.00036 | 0.00020 | 0.00012 | 0.00000 | 0.00008 | 65 | 0.00053 | 0.00001 | 0.00034 | 93 | 0.00034 | -1.19163 | 102.704 | 0.23615 | 0.77376 |
| fdnH; formate dehydrogenase-N, beta subunit | 0.00036 | 0.00011 | 0.00042 | 0.00000 | 0.00020 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.47713 | 122.661 | 0.63412 | 0.89696 |
| fdnI; formate dehydrogenase-N, gamma subunit | 0.00036 | 0.00011 | 0.00042 | 0.00000 | 0.00020 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.47713 | 122.661 | 0.63412 | 0.89696 |
| rbcS; ribulose-bisphosphate carboxylase small chain [EC:4.1.1.39] | 0.00036 | 0.00012 | 0.00017 | 0.00000 | 0.00006 | 65 | 0.00049 | 0.00000 | 0.00019 | 93 | 0.00020 | -1.58759 | 109.160 | 0.11527 | 0.77376 |
| PEO1; twinkle protein [EC:3.6.4.12] | 0.00036 | 0.00015 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00054 | 0.00001 | 0.00025 | 93 | 0.00025 | -1.76429 | 98.334 | 0.08079 | 0.75920 |
| CYP51; sterol 14-demethylase [EC:1.14.13.70] | 0.00035 | 0.00018 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00056 | 0.00001 | 0.00031 | 93 | 0.00031 | -1.55853 | 94.899 | 0.12244 | 0.77376 |
| ywaD; aminopeptidase YwaD [EC:3.4.11.6 3.4.11.10] | 0.00035 | 0.00014 | 0.00013 | 0.00000 | 0.00013 | 65 | 0.00051 | 0.00000 | 0.00022 | 93 | 0.00026 | -1.47107 | 141.087 | 0.14350 | 0.77376 |
| mcl; malyl-CoA/(S)-citramalyl-CoA lyase [EC:4.1.3.24 4.1.3.25] | 0.00035 | 0.00013 | 0.00013 | 0.00000 | 0.00006 | 65 | 0.00051 | 0.00000 | 0.00021 | 93 | 0.00022 | -1.69765 | 105.281 | 0.09253 | 0.76567 |
| ldrA_B_C_D; small toxic polypeptide LdrA/B/C/D | 0.00035 | 0.00022 | 0.00018 | 0.00000 | 0.00012 | 65 | 0.00047 | 0.00001 | 0.00036 | 93 | 0.00038 | -0.77676 | 110.584 | 0.43896 | 0.83156 |
| galP; MFS transporter, SP family, galactose:H+ symporter | 0.00035 | 0.00011 | 0.00035 | 0.00000 | 0.00016 | 65 | 0.00035 | 0.00000 | 0.00015 | 93 | 0.00022 | -0.01849 | 145.956 | 0.98527 | 0.99718 |
| serB-plsC; putative phosphoserine phosphatase / 1-acylglycerol-3-phosphate O-acyltransferase [EC:3.1.3.3 2.3.1.51] | 0.00035 | 0.00018 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00054 | 0.00001 | 0.00031 | 93 | 0.00031 | -1.48769 | 95.120 | 0.14014 | 0.77376 |
| nudK; GDP-mannose pyrophosphatase NudK [EC:3.6.1.-] | 0.00035 | 0.00011 | 0.00039 | 0.00000 | 0.00019 | 65 | 0.00032 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.29596 | 123.898 | 0.76775 | 0.93900 |
| nudI; nucleoside triphosphatase [EC:3.6.1.-] | 0.00035 | 0.00011 | 0.00041 | 0.00000 | 0.00020 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.40169 | 121.896 | 0.68862 | 0.91567 |
| torA; trimethylamine-N-oxide reductase (cytochrome c) [EC:1.7.2.3] | 0.00035 | 0.00012 | 0.00043 | 0.00000 | 0.00021 | 65 | 0.00030 | 0.00000 | 0.00014 | 93 | 0.00026 | 0.52216 | 116.205 | 0.60255 | 0.88425 |
| dasB; N,N’-diacetylchitobiose transport system permease protein | 0.00035 | 0.00011 | 0.00052 | 0.00000 | 0.00023 | 65 | 0.00023 | 0.00000 | 0.00011 | 93 | 0.00025 | 1.10822 | 90.674 | 0.27070 | 0.77376 |
| impE; type VI secretion system protein ImpE | 0.00035 | 0.00012 | 0.00017 | 0.00000 | 0.00008 | 65 | 0.00047 | 0.00000 | 0.00020 | 93 | 0.00021 | -1.43910 | 117.192 | 0.15279 | 0.77376 |
| frvX; putative aminopeptidase FrvX [EC:3.4.11.-] | 0.00035 | 0.00011 | 0.00047 | 0.00000 | 0.00018 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00023 | 0.87420 | 124.801 | 0.38369 | 0.80172 |
| ttrR; two-component system, LuxR family, response regulator TtrR | 0.00035 | 0.00021 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00057 | 0.00001 | 0.00035 | 93 | 0.00035 | -1.52241 | 92.728 | 0.13131 | 0.77376 |
| hyfI; hydrogenase-4 component I [EC:1.-.-.-] | 0.00035 | 0.00016 | 0.00048 | 0.00001 | 0.00032 | 65 | 0.00026 | 0.00000 | 0.00015 | 93 | 0.00035 | 0.64334 | 90.765 | 0.52163 | 0.85613 |
| hyfJ; hydrogenase-4 component J [EC:1.-.-.-] | 0.00035 | 0.00016 | 0.00048 | 0.00001 | 0.00032 | 65 | 0.00026 | 0.00000 | 0.00015 | 93 | 0.00035 | 0.64334 | 90.765 | 0.52163 | 0.85613 |
| clfA; clumping factor A | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| clfB; clumping factor B | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| eap, map; protein Map | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| flr, flipr; FPRL1 inhibitory protein | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| fnbB; fibronectin-binding protein B | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| hld; delta-hemolysin | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| mecA; penicillin-binding protein 2 prime [EC:3.4.16.4] | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| mecR1; methicillin resistance protein | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| PTS-Bgl-EIIA, bglF, bglP; PTS system, beta-glucoside-specific IIA component [EC:2.7.1.-] | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| sak; staphylokinase | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| sbi; immunoglobulin G-binding protein Sbi | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| set; superantigen-like protein | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| spa; immunoglobulin G-binding protein A | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| sspB; staphopain B [EC:3.4.22.-] | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| yydI; putative peptide transport system ATP-binding protein | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| yydJ; putative peptide transport system permease protein | 0.00035 | 0.00016 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00052 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.54767 | 105.540 | 0.12470 | 0.77376 |
| fimF; minor fimbrial subunit | 0.00035 | 0.00015 | 0.00023 | 0.00000 | 0.00010 | 65 | 0.00042 | 0.00001 | 0.00025 | 93 | 0.00027 | -0.71357 | 117.134 | 0.47691 | 0.84209 |
| boxA; benzoyl-CoA 2,3-epoxidase subunit A [EC:1.14.13.208] | 0.00034 | 0.00019 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00056 | 0.00001 | 0.00033 | 93 | 0.00033 | -1.59726 | 92.785 | 0.11361 | 0.77376 |
| chqB; hydroxyquinol 1,2-dioxygenase [EC:1.13.11.37] | 0.00034 | 0.00012 | 0.00027 | 0.00000 | 0.00012 | 65 | 0.00040 | 0.00000 | 0.00018 | 93 | 0.00022 | -0.59402 | 152.022 | 0.55338 | 0.86788 |
| yhaV; toxin YhaV [EC:3.1.-.-] | 0.00034 | 0.00012 | 0.00023 | 0.00000 | 0.00012 | 65 | 0.00042 | 0.00000 | 0.00019 | 93 | 0.00023 | -0.80566 | 148.367 | 0.42173 | 0.82295 |
| adiY; AraC family transcriptional regulator, transcriptional activator of adiA | 0.00034 | 0.00022 | 0.00015 | 0.00000 | 0.00009 | 65 | 0.00047 | 0.00001 | 0.00036 | 93 | 0.00037 | -0.86710 | 103.422 | 0.38789 | 0.80262 |
| K09122; uncharacterized protein | 0.00034 | 0.00012 | 0.00027 | 0.00000 | 0.00015 | 65 | 0.00038 | 0.00000 | 0.00018 | 93 | 0.00024 | -0.47369 | 155.797 | 0.63638 | 0.89696 |
| LRA1; L-rhamnose 1-dehydrogenase [EC:1.1.1.378 1.1.1.377 1.1.1.173] | 0.00034 | 0.00009 | 0.00017 | 0.00000 | 0.00008 | 65 | 0.00045 | 0.00000 | 0.00015 | 93 | 0.00016 | -1.71570 | 135.750 | 0.08850 | 0.76567 |
| trg; methyl-accepting chemotaxis protein III, ribose and galactose sensor receptor | 0.00034 | 0.00011 | 0.00032 | 0.00000 | 0.00015 | 65 | 0.00035 | 0.00000 | 0.00015 | 93 | 0.00021 | -0.18027 | 153.261 | 0.85718 | 0.95994 |
| ftrB; CRP/FNR family transcriptional regulator, transcriptional activator FtrB | 0.00034 | 0.00018 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00054 | 0.00001 | 0.00030 | 93 | 0.00030 | -1.65128 | 92.860 | 0.10206 | 0.77376 |
| elaB; ElaB protein | 0.00034 | 0.00011 | 0.00037 | 0.00000 | 0.00019 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00023 | 0.25036 | 125.446 | 0.80272 | 0.94869 |
| bglH; carbohydrate-specific outer membrane porin | 0.00034 | 0.00016 | 0.00027 | 0.00000 | 0.00018 | 65 | 0.00038 | 0.00001 | 0.00025 | 93 | 0.00030 | -0.35920 | 152.657 | 0.71994 | 0.92108 |
| citS; two-component system, CitB family, sensor histidine kinase CitS [EC:2.7.13.3] | 0.00034 | 0.00013 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00053 | 0.00000 | 0.00022 | 93 | 0.00022 | -2.06337 | 95.555 | 0.04179 | 0.64661 |
| K11638, citT; two-component system, CitB family, response regulator CitT | 0.00034 | 0.00013 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00053 | 0.00000 | 0.00022 | 93 | 0.00022 | -2.06337 | 95.555 | 0.04179 | 0.64661 |
| phnS; 2-aminoethylphosphonate transport system substrate-binding protein | 0.00034 | 0.00019 | 0.00025 | 0.00000 | 0.00018 | 65 | 0.00040 | 0.00001 | 0.00031 | 93 | 0.00035 | -0.41768 | 140.609 | 0.67682 | 0.91266 |
| phnT; 2-aminoethylphosphonate transport system ATP-binding protein | 0.00034 | 0.00019 | 0.00025 | 0.00000 | 0.00018 | 65 | 0.00040 | 0.00001 | 0.00031 | 93 | 0.00035 | -0.41768 | 140.609 | 0.67682 | 0.91266 |
| phnU; 2-aminoethylphosphonate transport system permease protein | 0.00034 | 0.00019 | 0.00025 | 0.00000 | 0.00018 | 65 | 0.00040 | 0.00001 | 0.00031 | 93 | 0.00035 | -0.41768 | 140.609 | 0.67682 | 0.91266 |
| RON2; rhoptry neck protein 2 | 0.00034 | 0.00024 | 0.00058 | 0.00002 | 0.00056 | 65 | 0.00017 | 0.00000 | 0.00010 | 93 | 0.00057 | 0.71346 | 68.398 | 0.47799 | 0.84266 |
| phnO; aminoalkylphosphonate N-acetyltransferase [EC:2.3.1.-] | 0.00034 | 0.00011 | 0.00021 | 0.00000 | 0.00008 | 65 | 0.00042 | 0.00000 | 0.00018 | 93 | 0.00020 | -1.02909 | 126.159 | 0.30541 | 0.77376 |
| rpaB; two-component system, OmpR family, response regulator RpaB | 0.00033 | 0.00019 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00054 | 0.00001 | 0.00033 | 93 | 0.00033 | -1.51461 | 92.960 | 0.13326 | 0.77376 |
| baiB; bile acid-coenzyme A ligase [EC:6.2.1.7] | 0.00033 | 0.00011 | 0.00018 | 0.00000 | 0.00006 | 65 | 0.00044 | 0.00000 | 0.00018 | 93 | 0.00019 | -1.36266 | 113.584 | 0.17568 | 0.77376 |
| gudX; glucarate dehydratase-related protein | 0.00033 | 0.00011 | 0.00037 | 0.00000 | 0.00019 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.25250 | 124.517 | 0.80107 | 0.94855 |
| mcd; (2S)-methylsuccinyl-CoA dehydrogenase [EC:1.3.8.12] | 0.00033 | 0.00011 | 0.00015 | 0.00000 | 0.00006 | 65 | 0.00046 | 0.00000 | 0.00018 | 93 | 0.00019 | -1.65481 | 112.358 | 0.10075 | 0.77369 |
| cebF; cellobiose transport system permease protein | 0.00033 | 0.00010 | 0.00032 | 0.00000 | 0.00019 | 65 | 0.00034 | 0.00000 | 0.00011 | 93 | 0.00022 | -0.11491 | 108.420 | 0.90873 | 0.97664 |
| cebG; cellobiose transport system permease protein | 0.00033 | 0.00010 | 0.00032 | 0.00000 | 0.00019 | 65 | 0.00034 | 0.00000 | 0.00011 | 93 | 0.00022 | -0.11491 | 108.420 | 0.90873 | 0.97664 |
| gfo; glucose-fructose oxidoreductase [EC:1.1.99.28] | 0.00033 | 0.00009 | 0.00012 | 0.00000 | 0.00004 | 65 | 0.00048 | 0.00000 | 0.00015 | 93 | 0.00015 | -2.41354 | 107.648 | 0.01749 | 0.59095 |
| cooF; anaerobic carbon-monoxide dehydrogenase iron sulfur subunit | 0.00033 | 0.00018 | 0.00048 | 0.00001 | 0.00041 | 65 | 0.00023 | 0.00000 | 0.00010 | 93 | 0.00042 | 0.60371 | 72.113 | 0.54793 | 0.86551 |
| tuaG; teichuronic acid biosynthesis glycosyltransferase TuaG [EC:2.4.-.-] | 0.00033 | 0.00007 | 0.00020 | 0.00000 | 0.00006 | 65 | 0.00042 | 0.00000 | 0.00011 | 93 | 0.00013 | -1.69493 | 133.576 | 0.09242 | 0.76567 |
| ompN; outer membrane protein N | 0.00033 | 0.00010 | 0.00035 | 0.00000 | 0.00016 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00021 | 0.16996 | 140.782 | 0.86529 | 0.96270 |
| mnoP; high affinity Mn2+ porin | 0.00033 | 0.00012 | 0.00025 | 0.00000 | 0.00016 | 65 | 0.00038 | 0.00000 | 0.00018 | 93 | 0.00024 | -0.53089 | 154.952 | 0.59626 | 0.88047 |
| xltA; putative xylitol transport system ATP-binding protein | 0.00033 | 0.00016 | 0.00010 | 0.00000 | 0.00004 | 65 | 0.00048 | 0.00001 | 0.00028 | 93 | 0.00028 | -1.37738 | 96.838 | 0.17157 | 0.77376 |
| xltC; putative xylitol transport system substrate-binding protein | 0.00033 | 0.00016 | 0.00010 | 0.00000 | 0.00004 | 65 | 0.00048 | 0.00001 | 0.00028 | 93 | 0.00028 | -1.37738 | 96.838 | 0.17157 | 0.77376 |
| CYP142; cholest-4-en-3-one 26-monooxygenase [EC:1.14.13.221] | 0.00033 | 0.00018 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00051 | 0.00001 | 0.00031 | 93 | 0.00031 | -1.40384 | 94.940 | 0.16363 | 0.77376 |
| csm4; CRISPR-associated protein Csm4 | 0.00032 | 0.00008 | 0.00044 | 0.00000 | 0.00013 | 65 | 0.00024 | 0.00000 | 0.00009 | 93 | 0.00016 | 1.26731 | 124.418 | 0.20741 | 0.77376 |
| E3.1.2.20; acyl-CoA hydrolase [EC:3.1.2.20] | 0.00032 | 0.00009 | 0.00029 | 0.00000 | 0.00013 | 65 | 0.00035 | 0.00000 | 0.00012 | 93 | 0.00018 | -0.28182 | 146.224 | 0.77848 | 0.94269 |
| abgR; LysR family transcriptional regulator, regulator of abg operon | 0.00032 | 0.00011 | 0.00037 | 0.00000 | 0.00019 | 65 | 0.00029 | 0.00000 | 0.00014 | 93 | 0.00024 | 0.32547 | 123.784 | 0.74537 | 0.93189 |
| engCP, engBF, endoEF; endo-alpha-N-acetylgalactosaminidase [EC:3.2.1.97] | 0.00032 | 0.00017 | 0.00051 | 0.00001 | 0.00040 | 65 | 0.00019 | 0.00000 | 0.00007 | 93 | 0.00041 | 0.76732 | 67.840 | 0.44556 | 0.83300 |
| yggC; putative kinase | 0.00032 | 0.00009 | 0.00030 | 0.00000 | 0.00008 | 65 | 0.00034 | 0.00000 | 0.00015 | 93 | 0.00017 | -0.18014 | 141.728 | 0.85730 | 0.95994 |
| hofP; pilus assembly protein HofP | 0.00032 | 0.00011 | 0.00036 | 0.00000 | 0.00019 | 65 | 0.00029 | 0.00000 | 0.00013 | 93 | 0.00023 | 0.29605 | 123.877 | 0.76769 | 0.93900 |
| ABC.MN.S; manganese/iron transport system substrate-binding protein | 0.00032 | 0.00011 | 0.00016 | 0.00000 | 0.00005 | 65 | 0.00044 | 0.00000 | 0.00018 | 93 | 0.00019 | -1.50052 | 104.824 | 0.13649 | 0.77376 |
| badH; 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase [EC:1.1.1.-] | 0.00032 | 0.00008 | 0.00017 | 0.00000 | 0.00007 | 65 | 0.00042 | 0.00000 | 0.00013 | 93 | 0.00014 | -1.72572 | 136.115 | 0.08667 | 0.76567 |
| nodE; nodulation protein E [EC:2.3.1.-] | 0.00032 | 0.00009 | 0.00023 | 0.00000 | 0.00009 | 65 | 0.00038 | 0.00000 | 0.00015 | 93 | 0.00017 | -0.88223 | 146.777 | 0.37910 | 0.79852 |
| K05558; pyridoxamine 5’-phosphate oxidase family protein | 0.00032 | 0.00018 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00047 | 0.00001 | 0.00031 | 93 | 0.00031 | -1.15355 | 98.331 | 0.25148 | 0.77376 |
| torD; TorA specific chaperone | 0.00032 | 0.00011 | 0.00035 | 0.00000 | 0.00019 | 65 | 0.00030 | 0.00000 | 0.00014 | 93 | 0.00023 | 0.22554 | 128.220 | 0.82192 | 0.95506 |
| ribD2; 5-amino-6-(5-phosphoribosylamino)uracil reductase [EC:1.1.1.193] | 0.00032 | 0.00011 | 0.00017 | 0.00000 | 0.00006 | 65 | 0.00042 | 0.00000 | 0.00018 | 93 | 0.00019 | -1.33318 | 111.371 | 0.18519 | 0.77376 |
| K09934; uncharacterized protein | 0.00032 | 0.00012 | 0.00023 | 0.00000 | 0.00011 | 65 | 0.00038 | 0.00000 | 0.00018 | 93 | 0.00021 | -0.67583 | 140.797 | 0.50026 | 0.84899 |
| nagH; salicylate 5-hydroxylase small subunit [EC:1.14.13.172] | 0.00032 | 0.00020 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00052 | 0.00001 | 0.00033 | 93 | 0.00033 | -1.49832 | 92.436 | 0.13746 | 0.77376 |
| vdh; valine dehydrogenase (NAD+) [EC:1.4.1.23] | 0.00032 | 0.00019 | 0.00020 | 0.00000 | 0.00012 | 65 | 0.00040 | 0.00001 | 0.00031 | 93 | 0.00033 | -0.61463 | 118.751 | 0.53998 | 0.86207 |
| rhaS; AraC family transcriptional regulator, L-rhamnose operon regulatory protein RhaS | 0.00032 | 0.00011 | 0.00030 | 0.00000 | 0.00018 | 65 | 0.00033 | 0.00000 | 0.00015 | 93 | 0.00023 | -0.12259 | 137.783 | 0.90261 | 0.97641 |
| cpt; chloramphenicol 3-O phosphotransferase [EC:2.7.1.-] | 0.00032 | 0.00013 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00048 | 0.00000 | 0.00022 | 93 | 0.00022 | -1.82266 | 96.196 | 0.07146 | 0.73962 |
| dalD; D-arabinitol 4-dehydrogenase [EC:1.1.1.11] | 0.00031 | 0.00019 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00049 | 0.00001 | 0.00033 | 93 | 0.00033 | -1.26962 | 93.492 | 0.20737 | 0.77376 |
| nagG; salicylate 5-hydroxylase large subunit [EC:1.14.13.172] | 0.00031 | 0.00020 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00052 | 0.00001 | 0.00033 | 93 | 0.00033 | -1.48567 | 92.436 | 0.14077 | 0.77376 |
| nahAb, nagAb, ndoA, nbzAb, dntAb; naphthalene 1,2-dioxygenase ferredoxin component | 0.00031 | 0.00020 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00052 | 0.00001 | 0.00033 | 93 | 0.00033 | -1.48567 | 92.436 | 0.14077 | 0.77376 |
| nmpC, ompD; outer membrane porin protein LC | 0.00031 | 0.00015 | 0.00020 | 0.00000 | 0.00008 | 65 | 0.00039 | 0.00001 | 0.00024 | 93 | 0.00026 | -0.71961 | 110.652 | 0.47329 | 0.84050 |
| E3.5.5.7; aliphatic nitrilase [EC:3.5.5.7] | 0.00031 | 0.00011 | 0.00020 | 0.00000 | 0.00008 | 65 | 0.00039 | 0.00000 | 0.00018 | 93 | 0.00019 | -0.95754 | 125.127 | 0.34014 | 0.78467 |
| mdtP; outer membrane protein, multidrug efflux system | 0.00031 | 0.00016 | 0.00012 | 0.00000 | 0.00005 | 65 | 0.00044 | 0.00001 | 0.00027 | 93 | 0.00028 | -1.15977 | 97.885 | 0.24897 | 0.77376 |
| MME, CD10; neprilysin [EC:3.4.24.11] | 0.00031 | 0.00018 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00049 | 0.00001 | 0.00031 | 93 | 0.00031 | -1.40462 | 94.454 | 0.16341 | 0.77376 |
| aroDE, DHQ-SDH; 3-dehydroquinate dehydratase / shikimate dehydrogenase [EC:4.2.1.10 1.1.1.25] | 0.00031 | 0.00013 | 0.00037 | 0.00001 | 0.00029 | 65 | 0.00027 | 0.00000 | 0.00010 | 93 | 0.00031 | 0.34776 | 81.127 | 0.72892 | 0.92377 |
| dgoR; GntR family transcriptional regulator, galactonate operon transcriptional repressor | 0.00031 | 0.00009 | 0.00027 | 0.00000 | 0.00010 | 65 | 0.00033 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.35243 | 151.405 | 0.72500 | 0.92177 |
| K16153; glycogen phosphorylase/synthase [EC:2.4.1.1 2.4.1.11] | 0.00031 | 0.00007 | 0.00032 | 0.00000 | 0.00008 | 65 | 0.00030 | 0.00000 | 0.00010 | 93 | 0.00013 | 0.18209 | 155.942 | 0.85575 | 0.95929 |
| hcaB; 2,3-dihydroxy-2,3-dihydrophenylpropionate dehydrogenase [EC:1.3.1.87] | 0.00031 | 0.00013 | 0.00016 | 0.00000 | 0.00005 | 65 | 0.00041 | 0.00000 | 0.00022 | 93 | 0.00023 | -1.08947 | 102.117 | 0.27851 | 0.77376 |
| hcaE, hcaA1; 3-phenylpropionate/trans-cinnamate dioxygenase subunit alpha [EC:1.14.12.19] | 0.00031 | 0.00013 | 0.00016 | 0.00000 | 0.00005 | 65 | 0.00041 | 0.00000 | 0.00022 | 93 | 0.00023 | -1.08947 | 102.117 | 0.27851 | 0.77376 |
| hcaF, hcaA2; 3-phenylpropionate/trans-cinnamate dioxygenase subunit beta [EC:1.14.12.19] | 0.00031 | 0.00013 | 0.00016 | 0.00000 | 0.00005 | 65 | 0.00041 | 0.00000 | 0.00022 | 93 | 0.00023 | -1.08947 | 102.117 | 0.27851 | 0.77376 |
| hddC; D-glycero-alpha-D-manno-heptose 1-phosphate guanylyltransferase [EC:2.7.7.71] | 0.00031 | 0.00008 | 0.00020 | 0.00000 | 0.00007 | 65 | 0.00038 | 0.00000 | 0.00012 | 93 | 0.00014 | -1.22097 | 143.245 | 0.22410 | 0.77376 |
| bchE; anaerobic magnesium-protoporphyrin IX monomethyl ester cyclase [EC:1.21.98.3] | 0.00031 | 0.00018 | 0.00042 | 0.00001 | 0.00041 | 65 | 0.00023 | 0.00000 | 0.00011 | 93 | 0.00042 | 0.45920 | 72.753 | 0.64746 | 0.90344 |
| badI; 2-ketocyclohexanecarboxyl-CoA hydrolase [EC:3.1.2.-] | 0.00030 | 0.00019 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00047 | 0.00001 | 0.00032 | 93 | 0.00032 | -1.25828 | 94.892 | 0.21138 | 0.77376 |
| traN; conjugal transfer mating pair stabilization protein TraN | 0.00030 | 0.00010 | 0.00041 | 0.00000 | 0.00020 | 65 | 0.00023 | 0.00000 | 0.00009 | 93 | 0.00022 | 0.82151 | 87.136 | 0.41360 | 0.81655 |
| yfkQ; spore germination protein | 0.00030 | 0.00018 | 0.00042 | 0.00001 | 0.00040 | 65 | 0.00022 | 0.00000 | 0.00013 | 93 | 0.00043 | 0.48237 | 77.508 | 0.63090 | 0.89595 |
| occT, nocT; octopine/nopaline transport system substrate-binding protein | 0.00030 | 0.00017 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00046 | 0.00001 | 0.00029 | 93 | 0.00029 | -1.36060 | 96.295 | 0.17682 | 0.77376 |
| eamB; cysteine/O-acetylserine efflux protein | 0.00030 | 0.00011 | 0.00026 | 0.00000 | 0.00012 | 65 | 0.00033 | 0.00000 | 0.00016 | 93 | 0.00020 | -0.34581 | 152.092 | 0.72996 | 0.92426 |
| tspA; uncharacterized membrane protein | 0.00030 | 0.00010 | 0.00031 | 0.00000 | 0.00016 | 65 | 0.00029 | 0.00000 | 0.00012 | 93 | 0.00020 | 0.06472 | 130.868 | 0.94850 | 0.98748 |
| salK; two-component system, NarL family, secretion system sensor histidine kinase SalK | 0.00030 | 0.00020 | 0.00027 | 0.00000 | 0.00026 | 65 | 0.00032 | 0.00001 | 0.00028 | 93 | 0.00038 | -0.11296 | 153.361 | 0.91021 | 0.97664 |
| murR; RpiR family transcriptional regulator, murPQ operon repressor | 0.00030 | 0.00013 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00040 | 0.00000 | 0.00022 | 93 | 0.00023 | -1.10875 | 111.192 | 0.26993 | 0.77376 |
| pglI; GalNAc5-diNAcBac-PP-undecaprenol beta-1,3-glucosyltransferase [EC:2.4.1.293] | 0.00030 | 0.00010 | 0.00031 | 0.00000 | 0.00016 | 65 | 0.00029 | 0.00000 | 0.00012 | 93 | 0.00020 | 0.07552 | 130.903 | 0.93992 | 0.98527 |
| pseG; UDP-2,4-diacetamido-2,4,6-trideoxy-beta-L-altropyranose hydrolase [EC:3.6.1.57] | 0.00030 | 0.00010 | 0.00031 | 0.00000 | 0.00016 | 65 | 0.00029 | 0.00000 | 0.00012 | 93 | 0.00020 | 0.07552 | 130.903 | 0.93992 | 0.98527 |
| zraS, hydH; two-component system, NtrC family, sensor histidine kinase HydH [EC:2.7.13.3] | 0.00030 | 0.00010 | 0.00019 | 0.00000 | 0.00009 | 65 | 0.00037 | 0.00000 | 0.00016 | 93 | 0.00018 | -0.97208 | 139.734 | 0.33269 | 0.77922 |
| dppB1; dipeptide transport system permease protein | 0.00030 | 0.00014 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00045 | 0.00000 | 0.00023 | 93 | 0.00023 | -1.63912 | 95.889 | 0.10447 | 0.77376 |
| PNC1; nicotinamidase [EC:3.5.1.19] | 0.00030 | 0.00008 | 0.00035 | 0.00000 | 0.00011 | 65 | 0.00026 | 0.00000 | 0.00011 | 93 | 0.00016 | 0.54854 | 147.539 | 0.58415 | 0.87720 |
| ccdB; toxin CcdB | 0.00030 | 0.00010 | 0.00033 | 0.00000 | 0.00018 | 65 | 0.00028 | 0.00000 | 0.00010 | 93 | 0.00021 | 0.23031 | 99.700 | 0.81832 | 0.95506 |
| E2.7.10.1; receptor protein-tyrosine kinase [EC:2.7.10.1] | 0.00030 | 0.00010 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00042 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.58760 | 121.033 | 0.11498 | 0.77376 |
| expR; LuxR family transcriptional regulator, quorum-sensing system regulator ExpR | 0.00030 | 0.00015 | 0.00052 | 0.00001 | 0.00035 | 65 | 0.00014 | 0.00000 | 0.00008 | 93 | 0.00036 | 1.06434 | 70.770 | 0.29079 | 0.77376 |
| araE; MFS transporter, SP family, arabinose:H+ symporter | 0.00030 | 0.00009 | 0.00022 | 0.00000 | 0.00009 | 65 | 0.00035 | 0.00000 | 0.00015 | 93 | 0.00017 | -0.77133 | 146.452 | 0.44176 | 0.83186 |
| SASP-A, sspA; small acid-soluble spore protein A (major alpha-type SASP) | 0.00029 | 0.00016 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00045 | 0.00001 | 0.00026 | 93 | 0.00027 | -1.46698 | 96.433 | 0.14564 | 0.77376 |
| PHYH; phytanoyl-CoA hydroxylase [EC:1.14.11.18] | 0.00029 | 0.00016 | 0.00025 | 0.00000 | 0.00015 | 65 | 0.00033 | 0.00001 | 0.00025 | 93 | 0.00029 | -0.27548 | 145.308 | 0.78334 | 0.94361 |
| pksS; cytochrome P450 PksS | 0.00029 | 0.00012 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00045 | 0.00000 | 0.00020 | 93 | 0.00020 | -1.81322 | 95.588 | 0.07294 | 0.74128 |
| kinC; two-component system, sporulation sensor kinase C [EC:2.7.13.3] | 0.00029 | 0.00013 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00047 | 0.00000 | 0.00022 | 93 | 0.00022 | -1.93124 | 93.377 | 0.05649 | 0.70577 |
| cciR; LuxR family transcriptional regulator, quorum-sensing system regulator CciR | 0.00029 | 0.00009 | 0.00014 | 0.00000 | 0.00009 | 65 | 0.00040 | 0.00000 | 0.00013 | 93 | 0.00016 | -1.61306 | 147.262 | 0.10887 | 0.77376 |
| pagC; putatice virulence related protein PagC | 0.00029 | 0.00014 | 0.00021 | 0.00000 | 0.00017 | 65 | 0.00035 | 0.00000 | 0.00020 | 93 | 0.00026 | -0.55437 | 155.889 | 0.58012 | 0.87573 |
| hycE; formate hydrogenlyase subunit 5 | 0.00029 | 0.00009 | 0.00027 | 0.00000 | 0.00010 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.26688 | 152.912 | 0.78992 | 0.94606 |
| soxS; AraC family transcriptional regulator, mar-sox-rob regulon activator | 0.00029 | 0.00010 | 0.00025 | 0.00000 | 0.00011 | 65 | 0.00032 | 0.00000 | 0.00015 | 93 | 0.00018 | -0.34833 | 153.897 | 0.72807 | 0.92350 |
| E1.13.12.4; lactate 2-monooxygenase [EC:1.13.12.4] | 0.00029 | 0.00011 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00046 | 0.00000 | 0.00019 | 93 | 0.00019 | -2.10297 | 96.048 | 0.03808 | 0.63091 |
| fbcH; ubiquinol-cytochrome c reductase cytochrome b/c1 subunit | 0.00029 | 0.00011 | 0.00011 | 0.00000 | 0.00004 | 65 | 0.00042 | 0.00000 | 0.00019 | 93 | 0.00019 | -1.59137 | 102.148 | 0.11462 | 0.77376 |
| pgi1; glucose-6-phosphate isomerase, archaeal [EC:5.3.1.9] | 0.00029 | 0.00009 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00043 | 0.00000 | 0.00015 | 93 | 0.00016 | -2.25332 | 114.467 | 0.02614 | 0.60634 |
| prdA; D-proline reductase (dithiol) PrdA [EC:1.21.4.1] | 0.00029 | 0.00005 | 0.00027 | 0.00000 | 0.00007 | 65 | 0.00030 | 0.00000 | 0.00007 | 93 | 0.00010 | -0.36780 | 148.046 | 0.71355 | 0.92004 |
| prdD; D-proline reductase (dithiol)-stabilizing protein PrdD | 0.00029 | 0.00005 | 0.00027 | 0.00000 | 0.00007 | 65 | 0.00030 | 0.00000 | 0.00007 | 93 | 0.00010 | -0.36780 | 148.046 | 0.71355 | 0.92004 |
| prdE; D-proline reductase (dithiol)-stabilizing protein PrdE | 0.00029 | 0.00005 | 0.00027 | 0.00000 | 0.00007 | 65 | 0.00030 | 0.00000 | 0.00007 | 93 | 0.00010 | -0.36780 | 148.046 | 0.71355 | 0.92004 |
| fhlA; formate hydrogenlyase transcriptional activator | 0.00029 | 0.00009 | 0.00026 | 0.00000 | 0.00010 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.30150 | 152.938 | 0.76344 | 0.93836 |
| VGSC; voltage-gated sodium channel | 0.00029 | 0.00009 | 0.00010 | 0.00000 | 0.00006 | 65 | 0.00042 | 0.00000 | 0.00015 | 93 | 0.00016 | -2.03307 | 116.227 | 0.04432 | 0.65981 |
| wbyL; glycosyltransferase [EC:2.4.1.-] | 0.00029 | 0.00012 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00044 | 0.00000 | 0.00021 | 93 | 0.00021 | -1.67559 | 98.786 | 0.09698 | 0.76673 |
| recE; exodeoxyribonuclease VIII [EC:3.1.11.-] | 0.00029 | 0.00015 | 0.00016 | 0.00000 | 0.00008 | 65 | 0.00037 | 0.00001 | 0.00024 | 93 | 0.00025 | -0.83218 | 108.949 | 0.40713 | 0.81200 |
| mtfabH; beta-ketoacyl-[acyl-carrier-protein] synthase III [EC:2.3.1.180] | 0.00029 | 0.00018 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00045 | 0.00001 | 0.00031 | 93 | 0.00031 | -1.23376 | 94.525 | 0.22035 | 0.77376 |
| dhaR; transcriptional activator for dhaKLM operon | 0.00029 | 0.00011 | 0.00027 | 0.00000 | 0.00018 | 65 | 0.00030 | 0.00000 | 0.00014 | 93 | 0.00022 | -0.13382 | 133.598 | 0.89375 | 0.97159 |
| cadB; cadaverine:lysine antiporter | 0.00029 | 0.00011 | 0.00030 | 0.00000 | 0.00018 | 65 | 0.00028 | 0.00000 | 0.00014 | 93 | 0.00022 | 0.11266 | 130.579 | 0.91047 | 0.97664 |
| ecnA; entericidin A | 0.00029 | 0.00009 | 0.00024 | 0.00000 | 0.00009 | 65 | 0.00032 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.50595 | 149.580 | 0.61364 | 0.88788 |
| dasA; N,N’-diacetylchitobiose transport system substrate-binding protein | 0.00029 | 0.00010 | 0.00047 | 0.00000 | 0.00022 | 65 | 0.00016 | 0.00000 | 0.00006 | 93 | 0.00023 | 1.35180 | 74.530 | 0.18053 | 0.77376 |
| mshJ; MSHA biogenesis protein MshJ | 0.00029 | 0.00016 | 0.00029 | 0.00000 | 0.00016 | 65 | 0.00028 | 0.00001 | 0.00025 | 93 | 0.00029 | 0.01505 | 149.335 | 0.98801 | 0.99801 |
| mshN; MSHA biogenesis protein MshN | 0.00029 | 0.00016 | 0.00029 | 0.00000 | 0.00016 | 65 | 0.00028 | 0.00001 | 0.00025 | 93 | 0.00029 | 0.01505 | 149.335 | 0.98801 | 0.99801 |
| K09707; uncharacterized protein | 0.00029 | 0.00012 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00047 | 0.00000 | 0.00020 | 93 | 0.00020 | -2.27827 | 92.420 | 0.02501 | 0.59941 |
| virB7, lvhB7; type IV secretion system protein VirB7 | 0.00028 | 0.00016 | 0.00006 | 0.00000 | 0.00002 | 65 | 0.00044 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.38937 | 93.492 | 0.16802 | 0.77376 |
| epr; minor extracellular protease Epr [EC:3.4.21.-] | 0.00028 | 0.00013 | 0.00013 | 0.00000 | 0.00008 | 65 | 0.00039 | 0.00000 | 0.00021 | 93 | 0.00022 | -1.20079 | 118.978 | 0.23222 | 0.77376 |
| rgpA_B; gingipain R [EC:3.4.22.37] | 0.00028 | 0.00013 | 0.00025 | 0.00000 | 0.00022 | 65 | 0.00030 | 0.00000 | 0.00017 | 93 | 0.00028 | -0.19317 | 130.286 | 0.84713 | 0.95682 |
| pdh; phenylalanine dehydrogenase [EC:1.4.1.20] | 0.00028 | 0.00013 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00044 | 0.00000 | 0.00021 | 93 | 0.00021 | -1.84016 | 95.333 | 0.06886 | 0.73514 |
| rhaR; AraC family transcriptional regulator, L-rhamnose operon transcriptional activator RhaR | 0.00028 | 0.00011 | 0.00027 | 0.00000 | 0.00018 | 65 | 0.00029 | 0.00000 | 0.00014 | 93 | 0.00022 | -0.12217 | 131.435 | 0.90295 | 0.97641 |
| E3.1.3.70; mannosyl-3-phosphoglycerate phosphatase [EC:3.1.3.70] | 0.00028 | 0.00010 | 0.00021 | 0.00000 | 0.00008 | 65 | 0.00033 | 0.00000 | 0.00015 | 93 | 0.00018 | -0.69101 | 137.685 | 0.49072 | 0.84797 |
| cusC, silC; outer membrane protein, Cu(I)/Ag(I) efflux system | 0.00028 | 0.00010 | 0.00025 | 0.00000 | 0.00011 | 65 | 0.00030 | 0.00000 | 0.00015 | 93 | 0.00018 | -0.31214 | 154.146 | 0.75535 | 0.93665 |
| E1.1.1.51; 3(or 17)beta-hydroxysteroid dehydrogenase [EC:1.1.1.51] | 0.00028 | 0.00011 | 0.00029 | 0.00000 | 0.00014 | 65 | 0.00028 | 0.00000 | 0.00015 | 93 | 0.00021 | 0.03902 | 153.597 | 0.96893 | 0.99272 |
| yufL, malK; two-component system, CitB family, sensor histidine kinase MalK [EC:2.7.13.3] | 0.00028 | 0.00010 | 0.00008 | 0.00000 | 0.00003 | 65 | 0.00042 | 0.00000 | 0.00016 | 93 | 0.00017 | -2.04400 | 98.879 | 0.04361 | 0.65947 |
| K06883; uncharacterized protein | 0.00028 | 0.00011 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00045 | 0.00000 | 0.00019 | 93 | 0.00019 | -2.08468 | 96.488 | 0.03974 | 0.63577 |
| K07169; FHA domain-containing protein | 0.00028 | 0.00011 | 0.00016 | 0.00000 | 0.00007 | 65 | 0.00036 | 0.00000 | 0.00018 | 93 | 0.00019 | -1.07424 | 121.822 | 0.28484 | 0.77376 |
| wbdD, wbbD; O-antigen chain-terminating methyltransferase [EC:2.1.1.- 2.1.1.294 2.7.1.181] | 0.00028 | 0.00013 | 0.00062 | 0.00001 | 0.00031 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00031 | 1.89951 | 64.919 | 0.06194 | 0.73001 |
| E1.12.7.2L; ferredoxin hydrogenase large subunit [EC:1.12.7.2] | 0.00028 | 0.00014 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00045 | 0.00001 | 0.00023 | 93 | 0.00024 | -1.74244 | 94.562 | 0.08468 | 0.76567 |
| bdcA; cyclic-di-GMP-binding biofilm dispersal mediator protein | 0.00028 | 0.00010 | 0.00023 | 0.00000 | 0.00013 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00019 | -0.47433 | 154.330 | 0.63594 | 0.89696 |
| fimI; fimbrial protein | 0.00028 | 0.00009 | 0.00025 | 0.00000 | 0.00010 | 65 | 0.00029 | 0.00000 | 0.00013 | 93 | 0.00017 | -0.24213 | 155.115 | 0.80900 | 0.95002 |
| rlmG; 23S rRNA (guanine1835-N2)-methyltransferase [EC:2.1.1.174] | 0.00028 | 0.00009 | 0.00027 | 0.00000 | 0.00010 | 65 | 0.00028 | 0.00000 | 0.00013 | 93 | 0.00017 | -0.02350 | 154.139 | 0.98128 | 0.99661 |
| csx16; CRISPR-associated protein Csx16 | 0.00027 | 0.00008 | 0.00026 | 0.00000 | 0.00010 | 65 | 0.00028 | 0.00000 | 0.00012 | 93 | 0.00015 | -0.15491 | 155.797 | 0.87709 | 0.96737 |
| ail; attachment invasion locus protein | 0.00027 | 0.00014 | 0.00021 | 0.00000 | 0.00017 | 65 | 0.00032 | 0.00000 | 0.00020 | 93 | 0.00026 | -0.44188 | 155.940 | 0.65919 | 0.90906 |
| hycG; formate hydrogenlyase subunit 7 | 0.00027 | 0.00009 | 0.00022 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.54602 | 149.295 | 0.58587 | 0.87720 |
| ptcA; putrescine carbamoyltransferase [EC:2.1.3.6] | 0.00027 | 0.00010 | 0.00029 | 0.00000 | 0.00014 | 65 | 0.00026 | 0.00000 | 0.00014 | 93 | 0.00020 | 0.15558 | 152.872 | 0.87657 | 0.96705 |
| aglK; alpha-glucoside transport system ATP-binding protein | 0.00027 | 0.00011 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00045 | 0.00000 | 0.00018 | 93 | 0.00018 | -2.41470 | 92.966 | 0.01771 | 0.59095 |
| UMPS; uridine monophosphate synthetase [EC:2.4.2.10 4.1.1.23] | 0.00027 | 0.00010 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00033 | 0.00000 | 0.00016 | 93 | 0.00018 | -0.81774 | 128.792 | 0.41501 | 0.81730 |
| rna; ribonuclease I (enterobacter ribonuclease) [EC:3.1.27.6] | 0.00027 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.58913 | 148.858 | 0.55666 | 0.86813 |
| lsrR; lsr operon transcriptional repressor | 0.00027 | 0.00009 | 0.00026 | 0.00000 | 0.00010 | 65 | 0.00028 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.16797 | 152.890 | 0.86683 | 0.96327 |
| E1.3.1.45; 2’-hydroxyisoflavone reductase [EC:1.3.1.45] | 0.00027 | 0.00013 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00044 | 0.00000 | 0.00022 | 93 | 0.00022 | -1.83270 | 93.929 | 0.07002 | 0.73522 |
| dsdX; D-serine transporter | 0.00027 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.58156 | 149.310 | 0.56174 | 0.86813 |
| hycA; formate hydrogenlyase regulatory protein HycA | 0.00027 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.58156 | 149.310 | 0.56174 | 0.86813 |
| hycB; formate hydrogenlyase subunit 2 | 0.00027 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.58156 | 149.310 | 0.56174 | 0.86813 |
| hycC; formate hydrogenlyase subunit 3 | 0.00027 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.58156 | 149.310 | 0.56174 | 0.86813 |
| hycD; formate hydrogenlyase subunit 4 | 0.00027 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.58156 | 149.310 | 0.56174 | 0.86813 |
| hycF; formate hydrogenlyase subunit 6 | 0.00027 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00014 | 93 | 0.00017 | -0.58156 | 149.310 | 0.56174 | 0.86813 |
| wcaL, amsK; colanic acid/amylovoran biosynthesis glycosyltransferase [EC:2.4.-.-] | 0.00027 | 0.00009 | 0.00026 | 0.00000 | 0.00009 | 65 | 0.00028 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.16788 | 151.597 | 0.86690 | 0.96327 |
| hyaE; hydrogenase-1 operon protein HyaE | 0.00027 | 0.00012 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00042 | 0.00000 | 0.00020 | 93 | 0.00020 | -1.82144 | 96.274 | 0.07165 | 0.73962 |
| popA; two-component system, cell cycle response regulator PopA | 0.00027 | 0.00011 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00033 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.72388 | 126.489 | 0.47048 | 0.83905 |
| DIS3, RRP44; exosome complex exonuclease DIS3/RRP44 [EC:3.1.13.-] | 0.00027 | 0.00006 | 0.00032 | 0.00000 | 0.00008 | 65 | 0.00024 | 0.00000 | 0.00008 | 93 | 0.00011 | 0.71896 | 149.984 | 0.47328 | 0.84050 |
| ompU; outer membrane protein OmpU | 0.00027 | 0.00011 | 0.00028 | 0.00000 | 0.00019 | 65 | 0.00026 | 0.00000 | 0.00013 | 93 | 0.00023 | 0.07677 | 117.558 | 0.93894 | 0.98500 |
| E2.7.1.47; D-ribulokinase [EC:2.7.1.47] | 0.00027 | 0.00016 | 0.00009 | 0.00000 | 0.00003 | 65 | 0.00040 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.14925 | 94.979 | 0.25334 | 0.77376 |
| cadC; transcriptional activator of cad operon | 0.00027 | 0.00011 | 0.00026 | 0.00000 | 0.00018 | 65 | 0.00028 | 0.00000 | 0.00014 | 93 | 0.00022 | -0.08891 | 132.072 | 0.92929 | 0.98183 |
| hpaX; MFS transporter, ACS family, 4-hydroxyphenylacetate permease | 0.00027 | 0.00009 | 0.00038 | 0.00000 | 0.00019 | 65 | 0.00019 | 0.00000 | 0.00007 | 93 | 0.00020 | 0.90075 | 80.746 | 0.37040 | 0.79587 |
| comA; phosphosulfolactate synthase [EC:4.4.1.19] | 0.00027 | 0.00009 | 0.00012 | 0.00000 | 0.00007 | 65 | 0.00037 | 0.00000 | 0.00015 | 93 | 0.00016 | -1.53232 | 124.655 | 0.12798 | 0.77376 |
| CECR1, ADA2; adenosine deaminase CECR1 [EC:3.5.4.4] | 0.00027 | 0.00016 | 0.00023 | 0.00000 | 0.00015 | 65 | 0.00030 | 0.00001 | 0.00025 | 93 | 0.00029 | -0.23687 | 143.059 | 0.81310 | 0.95336 |
| hokA; protein HokA | 0.00027 | 0.00015 | 0.00011 | 0.00000 | 0.00005 | 65 | 0.00038 | 0.00001 | 0.00025 | 93 | 0.00025 | -1.08285 | 98.971 | 0.28151 | 0.77376 |
| yueD; benzil reductase ((S)-benzoin forming) [EC:1.1.1.320] | 0.00027 | 0.00011 | 0.00013 | 0.00000 | 0.00008 | 65 | 0.00036 | 0.00000 | 0.00018 | 93 | 0.00020 | -1.18927 | 128.029 | 0.23653 | 0.77376 |
| K07493; putative transposase | 0.00027 | 0.00010 | 0.00015 | 0.00000 | 0.00011 | 65 | 0.00035 | 0.00000 | 0.00015 | 93 | 0.00019 | -1.06033 | 154.815 | 0.29064 | 0.77376 |
| dauB; L-arginine dehydrogenase [EC:1.4.1.25] | 0.00027 | 0.00011 | 0.00028 | 0.00000 | 0.00018 | 65 | 0.00025 | 0.00000 | 0.00015 | 93 | 0.00023 | 0.13069 | 134.335 | 0.89621 | 0.97276 |
| togB; oligogalacturonide transport system substrate-binding protein | 0.00027 | 0.00009 | 0.00012 | 0.00000 | 0.00003 | 65 | 0.00037 | 0.00000 | 0.00014 | 93 | 0.00015 | -1.73799 | 102.499 | 0.08522 | 0.76567 |
| yjjG; 5’-nucleotidase [EC:3.1.3.5] | 0.00027 | 0.00009 | 0.00025 | 0.00000 | 0.00010 | 65 | 0.00028 | 0.00000 | 0.00013 | 93 | 0.00017 | -0.18211 | 155.105 | 0.85573 | 0.95929 |
| hfaA; holdfast attachment protein HfaA | 0.00027 | 0.00011 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00032 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.68214 | 126.499 | 0.49640 | 0.84797 |
| hfaB; holdfast attachment protein HfaB | 0.00027 | 0.00011 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00032 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.68214 | 126.499 | 0.49640 | 0.84797 |
| mauA; methylamine dehydrogenase light chain [EC:1.4.9.1] | 0.00027 | 0.00018 | 0.00011 | 0.00000 | 0.00005 | 65 | 0.00037 | 0.00001 | 0.00030 | 93 | 0.00031 | -0.84487 | 97.063 | 0.40026 | 0.80762 |
| rutG; putative pyrimidine permease RutG | 0.00027 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00030 | 0.00000 | 0.00014 | 93 | 0.00016 | -0.53212 | 149.912 | 0.59543 | 0.88005 |
| cuyA; L-cysteate sulfo-lyase [EC:4.4.1.25] | 0.00027 | 0.00010 | 0.00020 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00016 | 93 | 0.00019 | -0.63093 | 138.897 | 0.52912 | 0.86094 |
| boxD; 3,4-dehydroadipyl-CoA semialdehyde dehydrogenase [EC:1.2.1.77] | 0.00026 | 0.00019 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00044 | 0.00001 | 0.00032 | 93 | 0.00032 | -1.31022 | 92.284 | 0.19337 | 0.77376 |
| torT; periplasmic protein TorT | 0.00026 | 0.00010 | 0.00017 | 0.00000 | 0.00006 | 65 | 0.00033 | 0.00000 | 0.00016 | 93 | 0.00017 | -0.90961 | 120.320 | 0.36485 | 0.79587 |
| hmfH; 5-(hydroxymethyl)furfural/furfural oxidase [EC:1.1.3.47 1.1.3.-] | 0.00026 | 0.00011 | 0.00018 | 0.00000 | 0.00007 | 65 | 0.00032 | 0.00000 | 0.00017 | 93 | 0.00019 | -0.72580 | 118.474 | 0.46940 | 0.83870 |
| occM, nocM; octopine/nopaline transport system permease protein | 0.00026 | 0.00010 | 0.00016 | 0.00000 | 0.00009 | 65 | 0.00033 | 0.00000 | 0.00016 | 93 | 0.00018 | -0.93580 | 140.551 | 0.35098 | 0.79120 |
| E3.5.1.77; N-carbamoyl-D-amino-acid hydrolase [EC:3.5.1.77] | 0.00026 | 0.00011 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00038 | 0.00000 | 0.00018 | 93 | 0.00018 | -1.55603 | 99.941 | 0.12286 | 0.77376 |
| isp; major intracellular serine protease [EC:3.4.21.-] | 0.00026 | 0.00013 | 0.00010 | 0.00000 | 0.00004 | 65 | 0.00037 | 0.00000 | 0.00022 | 93 | 0.00022 | -1.22714 | 99.222 | 0.22268 | 0.77376 |
| E1.3.1.74; 2-alkenal reductase [EC:1.3.1.74] | 0.00026 | 0.00011 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00039 | 0.00000 | 0.00018 | 93 | 0.00018 | -1.69894 | 96.430 | 0.09256 | 0.76567 |
| hokE; protein HokE | 0.00026 | 0.00015 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00035 | 0.00001 | 0.00025 | 93 | 0.00026 | -0.86356 | 105.644 | 0.38978 | 0.80353 |
| gutA, gutP; probable glucitol transport protein GutA | 0.00026 | 0.00012 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00040 | 0.00000 | 0.00020 | 93 | 0.00021 | -1.65151 | 103.007 | 0.10168 | 0.77376 |
| ELP3, KAT9; elongator complex protein 3 [EC:2.3.1.48] | 0.00026 | 0.00008 | 0.00020 | 0.00000 | 0.00005 | 65 | 0.00030 | 0.00000 | 0.00014 | 93 | 0.00015 | -0.69321 | 116.428 | 0.48956 | 0.84797 |
| ccr; crotonyl-CoA carboxylase/reductase [EC:1.3.1.85] | 0.00026 | 0.00011 | 0.00012 | 0.00000 | 0.00006 | 65 | 0.00035 | 0.00000 | 0.00018 | 93 | 0.00019 | -1.27696 | 110.987 | 0.20428 | 0.77376 |
| gmr; c-di-GMP phosphodiesterase Gmr [EC:3.1.4.52] | 0.00026 | 0.00008 | 0.00020 | 0.00000 | 0.00008 | 65 | 0.00030 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.64998 | 142.031 | 0.51676 | 0.85216 |
| mdtL; MFS transporter, DHA1 family, multidrug resistance protein | 0.00026 | 0.00009 | 0.00022 | 0.00000 | 0.00009 | 65 | 0.00028 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.38812 | 150.851 | 0.69847 | 0.91844 |
| pdeA; c-di-GMP-specific phosphodiesterase [EC:3.1.4.52] | 0.00026 | 0.00011 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00031 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.60572 | 126.865 | 0.54578 | 0.86444 |
| csgD; LuxR family transcriptional regulator, csgAB operon transcriptional regulatory protein | 0.00026 | 0.00009 | 0.00022 | 0.00000 | 0.00009 | 65 | 0.00028 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.39964 | 150.963 | 0.68999 | 0.91567 |
| araD; 2-keto-3-deoxy-L-arabinonate dehydratase [EC:4.2.1.43] | 0.00026 | 0.00019 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00041 | 0.00001 | 0.00032 | 93 | 0.00033 | -1.15676 | 92.807 | 0.25034 | 0.77376 |
| pheP; phenylalanine-specific permease | 0.00026 | 0.00009 | 0.00022 | 0.00000 | 0.00009 | 65 | 0.00028 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36889 | 150.910 | 0.71273 | 0.92004 |
| ebgC; evolved beta-galactosidase subunit beta | 0.00025 | 0.00009 | 0.00030 | 0.00000 | 0.00011 | 65 | 0.00023 | 0.00000 | 0.00013 | 93 | 0.00017 | 0.40268 | 155.184 | 0.68774 | 0.91567 |
| yqjH; ferric-chelate reductase (NADPH) [EC:1.16.1.9] | 0.00025 | 0.00009 | 0.00022 | 0.00000 | 0.00009 | 65 | 0.00028 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36265 | 150.976 | 0.71737 | 0.92035 |
| cotX; spore coat protein X | 0.00025 | 0.00010 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00040 | 0.00000 | 0.00017 | 93 | 0.00017 | -2.15361 | 97.390 | 0.03374 | 0.62130 |
| pht5; 4,5-dihydroxyphthalate decarboxylase [EC:4.1.1.55] | 0.00025 | 0.00011 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00038 | 0.00000 | 0.00018 | 93 | 0.00019 | -1.62312 | 96.307 | 0.10783 | 0.77376 |
| rfbE; CDP-paratose 2-epimerase [EC:5.1.3.10] | 0.00025 | 0.00008 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00038 | 0.00000 | 0.00014 | 93 | 0.00014 | -2.23320 | 103.529 | 0.02769 | 0.61334 |
| dasC; N,N’-diacetylchitobiose transport system permease protein | 0.00025 | 0.00009 | 0.00039 | 0.00000 | 0.00021 | 65 | 0.00015 | 0.00000 | 0.00006 | 93 | 0.00022 | 1.09569 | 75.451 | 0.27670 | 0.77376 |
| togM; oligogalacturonide transport system permease protein | 0.00025 | 0.00008 | 0.00010 | 0.00000 | 0.00003 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00015 | -1.70517 | 101.829 | 0.09121 | 0.76567 |
| togN; oligogalacturonide transport system permease protein | 0.00025 | 0.00008 | 0.00010 | 0.00000 | 0.00003 | 65 | 0.00035 | 0.00000 | 0.00014 | 93 | 0.00015 | -1.70517 | 101.829 | 0.09121 | 0.76567 |
| dmpD, xylF; 2-hydroxymuconate-semialdehyde hydrolase [EC:3.7.1.9] | 0.00025 | 0.00011 | 0.00016 | 0.00000 | 0.00007 | 65 | 0.00031 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.77993 | 116.366 | 0.43701 | 0.83054 |
| lysX; [lysine-biosynthesis-protein LysW]—L-2-aminoadipate ligase [EC:6.3.2.43] | 0.00025 | 0.00013 | 0.00011 | 0.00000 | 0.00009 | 65 | 0.00034 | 0.00000 | 0.00020 | 93 | 0.00022 | -1.03488 | 123.482 | 0.30275 | 0.77376 |
| acrE; membrane fusion protein, multidrug efflux system | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| acrF; multidrug efflux pump | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| bssR; biofilm regulator BssR | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| dnaT; DNA replication protein DnaT | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| envR, acrS; TetR/AcrR family transcriptional regulator, acrEF/envCD operon repressor | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| ftnB; ferritin-like protein 2 | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| marA; AraC family transcriptional regulator, mar-sox-rob regulon activator | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| marB; multiple antibiotic resistance protein MarB | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| marR; MarR family transcriptional regulator, multiple antibiotic resistance protein MarR | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| stpA; DNA-binding protein StpA | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| tdcA; LysR family transcriptional regulator, tdc operon transcriptional activator | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| tdcC; threonine transporter | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| tdcD; propionate kinase [EC:2.7.2.15] | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| ynhG; L,D-transpeptidase YnhG | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.36429 | 150.892 | 0.71615 | 0.92004 |
| cssR; two-component system, OmpR family, response regulator CssR | 0.00025 | 0.00006 | 0.00021 | 0.00000 | 0.00008 | 65 | 0.00028 | 0.00000 | 0.00009 | 93 | 0.00012 | -0.61974 | 153.169 | 0.53635 | 0.86207 |
| oqxR; Rrf2 family transcriptional regulator, repressor of oqxAB | 0.00025 | 0.00009 | 0.00034 | 0.00000 | 0.00019 | 65 | 0.00019 | 0.00000 | 0.00007 | 93 | 0.00020 | 0.74739 | 81.005 | 0.45699 | 0.83438 |
| wzy; O-antigen polymerase [EC:2.4.1.-] | 0.00025 | 0.00011 | 0.00041 | 0.00000 | 0.00025 | 65 | 0.00013 | 0.00000 | 0.00008 | 93 | 0.00026 | 1.07429 | 76.959 | 0.28605 | 0.77376 |
| occQ, nocQ; octopine/nopaline transport system permease protein | 0.00025 | 0.00010 | 0.00016 | 0.00000 | 0.00009 | 65 | 0.00031 | 0.00000 | 0.00016 | 93 | 0.00018 | -0.83550 | 140.880 | 0.40485 | 0.81147 |
| K13874, araB; L-arabinonolactonase [EC:3.1.1.15] | 0.00025 | 0.00015 | 0.00015 | 0.00000 | 0.00010 | 65 | 0.00032 | 0.00001 | 0.00025 | 93 | 0.00027 | -0.62441 | 119.726 | 0.53355 | 0.86207 |
| ict-Y; itaconate CoA-transferase [EC:2.8.3.-] | 0.00025 | 0.00011 | 0.00017 | 0.00000 | 0.00007 | 65 | 0.00030 | 0.00000 | 0.00017 | 93 | 0.00019 | -0.66531 | 122.123 | 0.50711 | 0.84956 |
| sra; stationary-phase-induced ribosome-associated protein | 0.00025 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.41334 | 150.859 | 0.67995 | 0.91311 |
| nanEK; N-acetylmannosamine-6-phosphate 2-epimerase / N-acetylmannosamine kinase [EC:5.1.3.9 2.7.1.60] | 0.00025 | 0.00016 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00039 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.25920 | 93.059 | 0.21111 | 0.77376 |
| hypX, hoxX; putative two-component system protein, hydrogenase maturation factor HypX/HoxX | 0.00025 | 0.00018 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00038 | 0.00001 | 0.00031 | 93 | 0.00031 | -1.09297 | 94.478 | 0.27718 | 0.77376 |
| solR, cepR, tofR; LuxR family transcriptional regulator, quorum-sensing system regulator SolR | 0.00024 | 0.00016 | 0.00005 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.24478 | 93.139 | 0.21634 | 0.77376 |
| K05303; O-methyltransferase [EC:2.1.1.-] | 0.00024 | 0.00008 | 0.00012 | 0.00000 | 0.00006 | 65 | 0.00033 | 0.00000 | 0.00013 | 93 | 0.00014 | -1.46548 | 129.310 | 0.14522 | 0.77376 |
| bdcR; TetR/AcrR family transcriptional regulator, repressor for divergent bdcA | 0.00024 | 0.00009 | 0.00022 | 0.00000 | 0.00013 | 65 | 0.00026 | 0.00000 | 0.00013 | 93 | 0.00018 | -0.20644 | 153.526 | 0.83672 | 0.95506 |
| dge1; diguanylate cyclase [EC:2.7.7.65] | 0.00024 | 0.00011 | 0.00009 | 0.00000 | 0.00003 | 65 | 0.00035 | 0.00000 | 0.00018 | 93 | 0.00018 | -1.42129 | 96.711 | 0.15845 | 0.77376 |
| hokC_D; protein HokC/D | 0.00024 | 0.00015 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00037 | 0.00001 | 0.00025 | 93 | 0.00025 | -1.19119 | 96.072 | 0.23651 | 0.77376 |
| L3HYPDH; trans-L-3-hydroxyproline dehydratase [EC:4.2.1.77] | 0.00024 | 0.00010 | 0.00024 | 0.00000 | 0.00014 | 65 | 0.00025 | 0.00000 | 0.00014 | 93 | 0.00020 | -0.06713 | 147.216 | 0.94657 | 0.98674 |
| fimZ; two-component system, NarL family, response regulator, fimbrial Z protein, FimZ | 0.00024 | 0.00008 | 0.00021 | 0.00000 | 0.00008 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.41745 | 144.093 | 0.67697 | 0.91269 |
| attA1; mannopine transport system ATP-binding protein | 0.00024 | 0.00016 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.26315 | 93.095 | 0.20969 | 0.77376 |
| attA2; mannopine transport system permease protein | 0.00024 | 0.00016 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.26315 | 93.095 | 0.20969 | 0.77376 |
| attC; mannopine transport system substrate-binding protein | 0.00024 | 0.00016 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.26315 | 93.095 | 0.20969 | 0.77376 |
| btaE; hyaluronate-binding autotransporter adhesin | 0.00024 | 0.00016 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.26315 | 93.095 | 0.20969 | 0.77376 |
| btaF; ECM component-binding autotransporter adhesin | 0.00024 | 0.00016 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.26315 | 93.095 | 0.20969 | 0.77376 |
| hmfG; 2,5-furandicarboxylate decarboxylase 2 | 0.00024 | 0.00016 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.26315 | 93.095 | 0.20969 | 0.77376 |
| ptlF; type IV secretion system protein PtlF | 0.00024 | 0.00016 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00001 | 0.00027 | 93 | 0.00027 | -1.26315 | 93.095 | 0.20969 | 0.77376 |
| cotI; spore coat protein I | 0.00024 | 0.00017 | 0.00041 | 0.00001 | 0.00040 | 65 | 0.00013 | 0.00000 | 0.00009 | 93 | 0.00042 | 0.68294 | 70.548 | 0.49688 | 0.84797 |
| epsM; acetyltransferase EpsM [EC:2.3.1.-] | 0.00024 | 0.00008 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00026 | 0.00000 | 0.00012 | 93 | 0.00015 | -0.35475 | 153.328 | 0.72326 | 0.92156 |
| E1.1.1.275; (+)-trans-carveol dehydrogenase [EC:1.1.1.275] | 0.00024 | 0.00011 | 0.00025 | 0.00000 | 0.00023 | 65 | 0.00024 | 0.00000 | 0.00009 | 93 | 0.00025 | 0.04106 | 83.431 | 0.96735 | 0.99206 |
| E1.1.1.9; D-xylulose reductase [EC:1.1.1.9] | 0.00024 | 0.00008 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00038 | 0.00000 | 0.00013 | 93 | 0.00014 | -2.47325 | 97.614 | 0.01512 | 0.59095 |
| cotB; spore coat protein B | 0.00024 | 0.00015 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00038 | 0.00001 | 0.00026 | 93 | 0.00026 | -1.34486 | 93.711 | 0.18192 | 0.77376 |
| mch, mcd; 2-methylfumaryl-CoA hydratase [EC:4.2.1.148] | 0.00024 | 0.00011 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00036 | 0.00000 | 0.00018 | 93 | 0.00018 | -1.58532 | 99.479 | 0.11607 | 0.77376 |
| epsG; transmembrane protein EpsG | 0.00024 | 0.00009 | 0.00007 | 0.00000 | 0.00006 | 65 | 0.00036 | 0.00000 | 0.00014 | 93 | 0.00016 | -1.87174 | 126.222 | 0.06356 | 0.73331 |
| wlbA, bplA; UDP-N-acetyl-2-amino-2-deoxyglucuronate dehydrogenase [EC:1.1.1.335] | 0.00024 | 0.00009 | 0.00036 | 0.00000 | 0.00019 | 65 | 0.00015 | 0.00000 | 0.00007 | 93 | 0.00020 | 1.06997 | 80.645 | 0.28782 | 0.77376 |
| wcaK, amsJ; colanic acid/amylovoran biosynthesis protein | 0.00024 | 0.00008 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00026 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.37562 | 149.428 | 0.70773 | 0.92004 |
| amnD; 2-aminomuconate deaminase [EC:3.5.99.5] | 0.00024 | 0.00018 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00035 | 0.00001 | 0.00030 | 93 | 0.00031 | -0.84076 | 98.294 | 0.40252 | 0.80926 |
| fitD, mcf; insecticidal toxin | 0.00024 | 0.00010 | 0.00020 | 0.00000 | 0.00010 | 65 | 0.00026 | 0.00000 | 0.00015 | 93 | 0.00018 | -0.30730 | 152.213 | 0.75903 | 0.93719 |
| JEN; MFS transporter, SHS family, lactate transporter | 0.00024 | 0.00009 | 0.00024 | 0.00000 | 0.00017 | 65 | 0.00023 | 0.00000 | 0.00008 | 93 | 0.00019 | 0.04286 | 94.511 | 0.96590 | 0.99102 |
| merD; MerR family transcriptional regulator, mercuric resistance operon regulatory protein | 0.00024 | 0.00011 | 0.00034 | 0.00000 | 0.00022 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00025 | 0.72529 | 101.444 | 0.46994 | 0.83870 |
| hofO; pilus assembly protein HofO | 0.00024 | 0.00008 | 0.00019 | 0.00000 | 0.00008 | 65 | 0.00027 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.53201 | 143.011 | 0.59554 | 0.88005 |
| E2.4.1.321; cellobionic acid phosphorylase [EC:2.4.1.321] | 0.00024 | 0.00015 | 0.00014 | 0.00000 | 0.00010 | 65 | 0.00030 | 0.00001 | 0.00025 | 93 | 0.00027 | -0.59428 | 119.916 | 0.55344 | 0.86788 |
| mshQ; MSHA biogenesis protein MshQ | 0.00024 | 0.00015 | 0.00019 | 0.00000 | 0.00011 | 65 | 0.00027 | 0.00001 | 0.00024 | 93 | 0.00027 | -0.30433 | 124.868 | 0.76138 | 0.93806 |
| CTDSP; carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase [EC:3.1.3.16] | 0.00023 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00027 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.42311 | 155.842 | 0.67280 | 0.91184 |
| mdlY; mandelamide amidase [EC:3.5.1.86] | 0.00023 | 0.00011 | 0.00017 | 0.00000 | 0.00006 | 65 | 0.00028 | 0.00000 | 0.00018 | 93 | 0.00019 | -0.59220 | 109.871 | 0.55493 | 0.86806 |
| wbpX; alpha-1,2-rhamnosyltransferase [EC:2.4.1.-] | 0.00023 | 0.00010 | 0.00023 | 0.00000 | 0.00011 | 65 | 0.00024 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.06443 | 154.139 | 0.94871 | 0.98754 |
| rexA; oligosaccharide reducing-end xylanase [EC:3.2.1.156] | 0.00023 | 0.00011 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00039 | 0.00000 | 0.00018 | 93 | 0.00018 | -2.10795 | 92.051 | 0.03775 | 0.63091 |
| cpdP; 3’,5’-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17] | 0.00023 | 0.00007 | 0.00023 | 0.00000 | 0.00013 | 65 | 0.00023 | 0.00000 | 0.00009 | 93 | 0.00016 | -0.02883 | 120.033 | 0.97705 | 0.99621 |
| icmB, dotO; intracellular multiplication protein IcmB | 0.00023 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00026 | 0.00000 | 0.00015 | 93 | 0.00020 | -0.38770 | 155.774 | 0.69877 | 0.91844 |
| agaA; N-acetylgalactosamine-6-phosphate deacetylase [EC:3.5.1.25] | 0.00023 | 0.00008 | 0.00023 | 0.00000 | 0.00009 | 65 | 0.00023 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.00739 | 150.373 | 0.99411 | 0.99919 |
| cedA; cell division activator | 0.00023 | 0.00009 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00024 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.18624 | 151.966 | 0.85251 | 0.95848 |
| FANCM; fanconi anemia group M protein | 0.00023 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00025 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.32247 | 155.937 | 0.74753 | 0.93207 |
| lysW; alpha-aminoadipate/glutamate carrier protein LysW | 0.00023 | 0.00011 | 0.00015 | 0.00000 | 0.00009 | 65 | 0.00028 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.66883 | 135.285 | 0.50475 | 0.84956 |
| comZ; competence protein ComZ | 0.00023 | 0.00010 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00037 | 0.00000 | 0.00017 | 93 | 0.00017 | -1.99150 | 94.704 | 0.04931 | 0.67429 |
| occP, nocP; octopine/nopaline transport system ATP-binding protein [EC:3.6.3.-] | 0.00023 | 0.00008 | 0.00014 | 0.00000 | 0.00009 | 65 | 0.00029 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.94363 | 149.922 | 0.34687 | 0.78982 |
| traD; conjugal transfer pilus assembly protein TraD | 0.00022 | 0.00013 | 0.00014 | 0.00000 | 0.00012 | 65 | 0.00028 | 0.00000 | 0.00020 | 93 | 0.00023 | -0.63717 | 145.980 | 0.52501 | 0.85728 |
| uidB, gusB; glucuronide carrier protein | 0.00022 | 0.00010 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00035 | 0.00000 | 0.00017 | 93 | 0.00017 | -1.74427 | 98.006 | 0.08425 | 0.76567 |
| tfdA; alpha-ketoglutarate-dependent 2,4-dichlorophenoxyacetate dioxygenase [EC:1.14.11.-] | 0.00022 | 0.00010 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00033 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.42930 | 96.841 | 0.15614 | 0.77376 |
| mqnB; futalosine hydrolase [EC:3.2.2.26] | 0.00022 | 0.00011 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00035 | 0.00000 | 0.00018 | 93 | 0.00019 | -1.67166 | 97.085 | 0.09781 | 0.76722 |
| E3.1.2.1, ACH1; acetyl-CoA hydrolase [EC:3.1.2.1] | 0.00022 | 0.00018 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00033 | 0.00001 | 0.00030 | 93 | 0.00031 | -0.87424 | 94.862 | 0.38420 | 0.80197 |
| cmeR; TetR/AcrR family transcriptional regulator, cmeABC operon repressor | 0.00022 | 0.00009 | 0.00035 | 0.00000 | 0.00020 | 65 | 0.00014 | 0.00000 | 0.00005 | 93 | 0.00021 | 1.02380 | 72.132 | 0.30935 | 0.77376 |
| wcaB; putative colanic acid biosynthesis acetyltransferase WcaB [EC:2.3.1.-] | 0.00022 | 0.00008 | 0.00020 | 0.00000 | 0.00008 | 65 | 0.00024 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.23036 | 148.094 | 0.81813 | 0.95506 |
| K09700; uncharacterized protein | 0.00022 | 0.00009 | 0.00028 | 0.00000 | 0.00016 | 65 | 0.00018 | 0.00000 | 0.00011 | 93 | 0.00020 | 0.54118 | 117.854 | 0.58940 | 0.87828 |
| K14340; mannosyltransferase [EC:2.4.1.-] | 0.00022 | 0.00009 | 0.00036 | 0.00000 | 0.00021 | 65 | 0.00012 | 0.00000 | 0.00004 | 93 | 0.00021 | 1.13917 | 69.536 | 0.25854 | 0.77376 |
| cqsS; two-component system, CAI-1 autoinducer sensor kinase/phosphatase CqsS [EC:2.7.13.3 3.1.3.-] | 0.00022 | 0.00015 | 0.00017 | 0.00000 | 0.00008 | 65 | 0.00026 | 0.00001 | 0.00024 | 93 | 0.00026 | -0.35391 | 112.326 | 0.72407 | 0.92170 |
| acmB; methyl acetate hydrolase [EC:3.1.1.-] | 0.00022 | 0.00018 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00034 | 0.00001 | 0.00030 | 93 | 0.00031 | -0.94261 | 94.539 | 0.34828 | 0.79120 |
| cssS; two-component system, OmpR family, sensor histidine kinase CssS [EC:2.7.13.3] | 0.00022 | 0.00006 | 0.00014 | 0.00000 | 0.00006 | 65 | 0.00028 | 0.00000 | 0.00009 | 93 | 0.00010 | -1.30821 | 149.195 | 0.19281 | 0.77376 |
| ompG; outer membrane protein G | 0.00022 | 0.00008 | 0.00018 | 0.00000 | 0.00008 | 65 | 0.00025 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.40855 | 144.034 | 0.68348 | 0.91514 |
| aepZ; 2-aminoethylphosphonate-pyruvate transaminase | 0.00022 | 0.00006 | 0.00024 | 0.00000 | 0.00008 | 65 | 0.00021 | 0.00000 | 0.00009 | 93 | 0.00011 | 0.25886 | 155.194 | 0.79608 | 0.94791 |
| ERCC4, XPF; DNA excision repair protein ERCC-4 [EC:3.1.-.-] | 0.00022 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00024 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.29994 | 155.914 | 0.76462 | 0.93900 |
| xyoA, aldO; alditol oxidase [EC:1.1.3.41] | 0.00022 | 0.00009 | 0.00032 | 0.00000 | 0.00020 | 65 | 0.00015 | 0.00000 | 0.00006 | 93 | 0.00020 | 0.82888 | 76.573 | 0.40975 | 0.81416 |
| nifB; nitrogen fixation protein NifB | 0.00022 | 0.00006 | 0.00017 | 0.00000 | 0.00006 | 65 | 0.00025 | 0.00000 | 0.00010 | 93 | 0.00011 | -0.72760 | 142.817 | 0.46805 | 0.83837 |
| ebgR; LacI family transcriptional regulator, ebg operon repressor | 0.00022 | 0.00008 | 0.00022 | 0.00000 | 0.00009 | 65 | 0.00022 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.01973 | 150.076 | 0.98428 | 0.99706 |
| CTDSPL2; CTD small phosphatase-like protein 2 [EC:3.1.3.-] | 0.00022 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00024 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.28376 | 155.913 | 0.77697 | 0.94269 |
| dotB, traJ; defect in organelle trafficking protein DotB | 0.00022 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00024 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.28376 | 155.913 | 0.77697 | 0.94269 |
| dotC, traI; defect in organelle trafficking protein DotC | 0.00022 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00024 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.28376 | 155.913 | 0.77697 | 0.94269 |
| icmJ, dotN; intracellular multiplication protein IcmJ | 0.00022 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00024 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.28376 | 155.913 | 0.77697 | 0.94269 |
| icmL, traM, dotI; intracellular multiplication protein IcmL | 0.00022 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00024 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.28376 | 155.913 | 0.77697 | 0.94269 |
| icmO, trbC, dotL; intracellular multiplication protein IcmO | 0.00022 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00024 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.28376 | 155.913 | 0.77697 | 0.94269 |
| PARP; poly [ADP-ribose] polymerase [EC:2.4.2.30] | 0.00022 | 0.00010 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00024 | 0.00000 | 0.00015 | 93 | 0.00019 | -0.28376 | 155.913 | 0.77697 | 0.94269 |
| E3.4.11.10; bacterial leucyl aminopeptidase [EC:3.4.11.10] | 0.00022 | 0.00010 | 0.00011 | 0.00000 | 0.00007 | 65 | 0.00029 | 0.00000 | 0.00016 | 93 | 0.00018 | -1.00752 | 123.744 | 0.31565 | 0.77376 |
| dhbE; glycerol dehydratase small subunit [EC:4.2.1.30] | 0.00022 | 0.00017 | 0.00047 | 0.00001 | 0.00042 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00042 | 1.01180 | 64.720 | 0.31540 | 0.77376 |
| csgA; major curlin subunit | 0.00022 | 0.00008 | 0.00018 | 0.00000 | 0.00008 | 65 | 0.00025 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.46218 | 143.935 | 0.64465 | 0.90243 |
| csgC; curli production protein | 0.00022 | 0.00008 | 0.00018 | 0.00000 | 0.00008 | 65 | 0.00025 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.46218 | 143.935 | 0.64465 | 0.90243 |
| gspO; general secretion pathway protein O [EC:3.4.23.43 2.1.1.-] | 0.00022 | 0.00008 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00022 | 0.00000 | 0.00013 | 93 | 0.00016 | -0.12055 | 152.305 | 0.90421 | 0.97658 |
| torR; two-component system, OmpR family, torCAD operon response regulator TorR | 0.00022 | 0.00008 | 0.00016 | 0.00000 | 0.00006 | 65 | 0.00025 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.61326 | 129.853 | 0.54078 | 0.86207 |
| iolR; DeoR family transcriptional regulator, myo-inositol catabolism operon repressor | 0.00021 | 0.00017 | 0.00045 | 0.00001 | 0.00041 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00041 | 0.96537 | 64.957 | 0.33794 | 0.78377 |
| TRP1; anthranilate synthase / indole-3-glycerol phosphate synthase / phosphoribosylanthranilate isomerase [EC:4.1.3.27 4.1.1.48 5.3.1.24] | 0.00021 | 0.00012 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00034 | 0.00000 | 0.00020 | 93 | 0.00020 | -1.54627 | 93.081 | 0.12543 | 0.77376 |
| E1.1.1.65; pyridoxine 4-dehydrogenase [EC:1.1.1.65] | 0.00021 | 0.00010 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00031 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.34328 | 96.890 | 0.18232 | 0.77376 |
| raxST; sulfotransferase | 0.00021 | 0.00009 | 0.00018 | 0.00000 | 0.00010 | 65 | 0.00023 | 0.00000 | 0.00014 | 93 | 0.00018 | -0.28446 | 153.099 | 0.77644 | 0.94269 |
| araL; arabinose operon protein AraL | 0.00021 | 0.00012 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00034 | 0.00000 | 0.00020 | 93 | 0.00020 | -1.55504 | 93.073 | 0.12333 | 0.77376 |
| PHKA_B; phosphorylase kinase alpha/beta subunit | 0.00021 | 0.00012 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00034 | 0.00000 | 0.00020 | 93 | 0.00020 | -1.55504 | 93.073 | 0.12333 | 0.77376 |
| E4.6.1.2; guanylate cyclase, other [EC:4.6.1.2] | 0.00021 | 0.00010 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00031 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.36405 | 96.509 | 0.17572 | 0.77376 |
| hbaA; 4-hydroxybenzoate-CoA ligase [EC:6.2.1.27 6.2.1.25] | 0.00021 | 0.00010 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00031 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.31737 | 96.889 | 0.19082 | 0.77376 |
| hyg; hygromycin-B 7’’-O-kinase [EC:2.7.1.119] | 0.00021 | 0.00010 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00031 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.31737 | 96.889 | 0.19082 | 0.77376 |
| mdlA; mandelate racemase [EC:5.1.2.2] | 0.00021 | 0.00010 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00031 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.31737 | 96.889 | 0.19082 | 0.77376 |
| waaO, rfaI; UDP-glucose:(glucosyl)LPS alpha-1,3-glucosyltransferase [EC:2.4.1.-] | 0.00021 | 0.00013 | 0.00028 | 0.00000 | 0.00025 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00028 | 0.39382 | 93.067 | 0.69461 | 0.91715 |
| lysJ, argD; LysW-gamma-L-lysine/LysW-L-ornithine aminotransferase [EC:2.6.1.-] | 0.00021 | 0.00011 | 0.00014 | 0.00000 | 0.00009 | 65 | 0.00026 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.61196 | 135.314 | 0.54159 | 0.86207 |
| lysY, argC; LysW-gamma-L-alpha-aminoadipyl-6-phosphate/LysW-L-glutamyl-5-phosphate reductase [EC:1.2.1.-] | 0.00021 | 0.00011 | 0.00014 | 0.00000 | 0.00009 | 65 | 0.00026 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.61196 | 135.314 | 0.54159 | 0.86207 |
| lysZ, argB; LysW-gamma-L-alpha-aminoadipate/LysW-L-glutamate kinase [EC:2.7.2.-] | 0.00021 | 0.00011 | 0.00014 | 0.00000 | 0.00009 | 65 | 0.00026 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.61196 | 135.314 | 0.54159 | 0.86207 |
| tetR; TetR/AcrR family transcriptional regulator, tetracycline repressor protein | 0.00021 | 0.00008 | 0.00017 | 0.00000 | 0.00008 | 65 | 0.00023 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.40968 | 148.055 | 0.68264 | 0.91495 |
| ynfH; Tat-targeted selenate reductase subunit YnfH | 0.00021 | 0.00008 | 0.00011 | 0.00000 | 0.00004 | 65 | 0.00028 | 0.00000 | 0.00013 | 93 | 0.00014 | -1.23387 | 107.805 | 0.21993 | 0.77376 |
| fadD9; fatty acid CoA ligase FadD9 | 0.00021 | 0.00008 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00034 | 0.00000 | 0.00013 | 93 | 0.00013 | -2.51891 | 93.242 | 0.01347 | 0.59095 |
| GalDH; L-galactose dehydrogenase [EC:1.1.1.316] | 0.00021 | 0.00010 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00030 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.30661 | 96.515 | 0.19445 | 0.77376 |
| HDHD1; pseudouridine 5’-phosphatase [EC:3.1.3.96] | 0.00021 | 0.00009 | 0.00018 | 0.00000 | 0.00010 | 65 | 0.00022 | 0.00000 | 0.00014 | 93 | 0.00018 | -0.21664 | 153.156 | 0.82878 | 0.95506 |
| hsiF3; type VI secretion system lysozyme-related protein | 0.00021 | 0.00009 | 0.00018 | 0.00000 | 0.00010 | 65 | 0.00022 | 0.00000 | 0.00014 | 93 | 0.00018 | -0.21664 | 153.156 | 0.82878 | 0.95506 |
| lip3; type VI secretion system protein | 0.00021 | 0.00009 | 0.00018 | 0.00000 | 0.00010 | 65 | 0.00022 | 0.00000 | 0.00014 | 93 | 0.00018 | -0.21664 | 153.156 | 0.82878 | 0.95506 |
| iucB; acetyl CoA:N6-hydroxylysine acetyl transferase [EC:2.3.1.102] | 0.00021 | 0.00006 | 0.00029 | 0.00000 | 0.00013 | 65 | 0.00014 | 0.00000 | 0.00006 | 93 | 0.00014 | 1.05819 | 93.755 | 0.29269 | 0.77376 |
| motC; chemotaxis protein MotC | 0.00021 | 0.00009 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00032 | 0.00000 | 0.00014 | 93 | 0.00015 | -1.93830 | 96.789 | 0.05550 | 0.70577 |
| mcbA; MqsR-controlled colanic acid and biofilm protein A | 0.00021 | 0.00008 | 0.00018 | 0.00000 | 0.00008 | 65 | 0.00022 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.30464 | 144.655 | 0.76107 | 0.93806 |
| hblC; hemolysin BL lytic component L2 | 0.00020 | 0.00014 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00033 | 0.00001 | 0.00024 | 93 | 0.00024 | -1.31871 | 92.798 | 0.19051 | 0.77376 |
| hblD; hemolysin BL lytic component L1 | 0.00020 | 0.00014 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00033 | 0.00001 | 0.00024 | 93 | 0.00024 | -1.31871 | 92.798 | 0.19051 | 0.77376 |
| dhaB; glycerol dehydratase large subunit [EC:4.2.1.30] | 0.00020 | 0.00017 | 0.00044 | 0.00001 | 0.00042 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00042 | 0.93618 | 64.727 | 0.35266 | 0.79194 |
| dhbC; glycerol dehydratase medium subunit [EC:4.2.1.30] | 0.00020 | 0.00017 | 0.00044 | 0.00001 | 0.00042 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00042 | 0.93618 | 64.727 | 0.35266 | 0.79194 |
| gumH, aceA; alpha-1,3-mannosyltransferase [EC:2.4.1.252] | 0.00020 | 0.00007 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00021 | 0.00000 | 0.00009 | 93 | 0.00015 | -0.15296 | 132.510 | 0.87866 | 0.96759 |
| wcaD; putative colanic acid polymerase | 0.00020 | 0.00008 | 0.00016 | 0.00000 | 0.00007 | 65 | 0.00024 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.54694 | 140.456 | 0.58529 | 0.87720 |
| wcaE; putative colanic acid biosynthesis glycosyltransferase [EC:2.4.-.-] | 0.00020 | 0.00008 | 0.00016 | 0.00000 | 0.00007 | 65 | 0.00024 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.54694 | 140.456 | 0.58529 | 0.87720 |
| nanT; MFS transporter, SHS family, sialic acid transporter | 0.00020 | 0.00008 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00030 | 0.00000 | 0.00014 | 93 | 0.00014 | -1.61130 | 102.160 | 0.11020 | 0.77376 |
| mngR, farR; GntR family transcriptional regulator, mannosyl-D-glycerate transport/metabolism system repressor | 0.00020 | 0.00008 | 0.00021 | 0.00000 | 0.00009 | 65 | 0.00020 | 0.00000 | 0.00012 | 93 | 0.00015 | 0.10022 | 151.852 | 0.92030 | 0.97905 |
| cofF; gamma-F420-2:alpha-L-glutamate ligase [EC:6.3.2.32] | 0.00020 | 0.00011 | 0.00012 | 0.00000 | 0.00009 | 65 | 0.00026 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.70293 | 135.199 | 0.48331 | 0.84354 |
| acdA; acetate—CoA ligase (ADP-forming) subunit alpha [EC:6.2.1.13] | 0.00020 | 0.00015 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00033 | 0.00001 | 0.00025 | 93 | 0.00025 | -1.25892 | 92.374 | 0.21123 | 0.77376 |
| tetP_A, tet40; MFS transporter, DHA3 family, tetracycline resistance protein | 0.00020 | 0.00015 | 0.00035 | 0.00001 | 0.00035 | 65 | 0.00009 | 0.00000 | 0.00009 | 93 | 0.00037 | 0.71434 | 71.928 | 0.47733 | 0.84209 |
| K07494; putative transposase | 0.00020 | 0.00005 | 0.00026 | 0.00000 | 0.00012 | 65 | 0.00016 | 0.00000 | 0.00003 | 93 | 0.00012 | 0.87312 | 72.075 | 0.38550 | 0.80258 |
| cotM; spore coat protein M | 0.00020 | 0.00009 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00032 | 0.00000 | 0.00016 | 93 | 0.00016 | -1.81027 | 95.667 | 0.07339 | 0.74128 |
| K16901; anthranilate 3-monooxygenase (FAD) / 4-hydroxyphenylacetate 3-monooxygenase [EC:1.14.14.8 1.14.14.9] | 0.00020 | 0.00010 | 0.00018 | 0.00000 | 0.00016 | 65 | 0.00021 | 0.00000 | 0.00012 | 93 | 0.00020 | -0.16694 | 126.597 | 0.86768 | 0.96354 |
| torS; two-component system, OmpR family, sensor histidine kinase TorS [EC:2.7.13.3] | 0.00020 | 0.00008 | 0.00016 | 0.00000 | 0.00006 | 65 | 0.00023 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.45848 | 130.146 | 0.64737 | 0.90344 |
| ramA; AraC family of transcriptional regulator, multidrug resistance transcriptional activator | 0.00020 | 0.00007 | 0.00029 | 0.00000 | 0.00016 | 65 | 0.00014 | 0.00000 | 0.00006 | 93 | 0.00017 | 0.87703 | 82.971 | 0.38300 | 0.80064 |
| lysK, argE; LysW-gamma-L-lysine/LysW-L-ornithine carboxypeptidase | 0.00020 | 0.00011 | 0.00011 | 0.00000 | 0.00009 | 65 | 0.00026 | 0.00000 | 0.00018 | 93 | 0.00020 | -0.73701 | 132.504 | 0.46242 | 0.83654 |
| hpgT, nocG; (S)-3,5-dihydroxyphenylglycine transaminase [EC:2.6.1.103] | 0.00020 | 0.00009 | 0.00018 | 0.00000 | 0.00010 | 65 | 0.00021 | 0.00000 | 0.00014 | 93 | 0.00018 | -0.19258 | 153.460 | 0.84754 | 0.95682 |
| phlD; phloroglucinol synthase [EC:2.3.1.253] | 0.00020 | 0.00009 | 0.00018 | 0.00000 | 0.00010 | 65 | 0.00021 | 0.00000 | 0.00014 | 93 | 0.00018 | -0.19258 | 153.460 | 0.84754 | 0.95682 |
| ydjG; methylglyoxal reductase [EC:1.1.1.-] | 0.00020 | 0.00008 | 0.00011 | 0.00000 | 0.00005 | 65 | 0.00026 | 0.00000 | 0.00013 | 93 | 0.00014 | -1.05077 | 114.109 | 0.29559 | 0.77376 |
| flu; antigen 43 | 0.00020 | 0.00014 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00031 | 0.00001 | 0.00024 | 93 | 0.00024 | -1.09774 | 93.896 | 0.27512 | 0.77376 |
| vanJ; vancomycin resistance protein VanJ | 0.00020 | 0.00012 | 0.00017 | 0.00000 | 0.00017 | 65 | 0.00022 | 0.00000 | 0.00017 | 93 | 0.00024 | -0.21073 | 152.617 | 0.83338 | 0.95506 |
| acfC; accessory colonization factor AcfC | 0.00019 | 0.00011 | 0.00011 | 0.00000 | 0.00004 | 65 | 0.00025 | 0.00000 | 0.00018 | 93 | 0.00019 | -0.76172 | 102.114 | 0.44798 | 0.83372 |
| blaOXA-213; beta-lactamase class D OXA-213 [EC:3.5.2.6] | 0.00019 | 0.00011 | 0.00011 | 0.00000 | 0.00004 | 65 | 0.00025 | 0.00000 | 0.00018 | 93 | 0.00019 | -0.76172 | 102.114 | 0.44798 | 0.83372 |
| blaOXA-51; beta-lactamase class D OXA-51 [EC:3.5.2.6] | 0.00019 | 0.00011 | 0.00011 | 0.00000 | 0.00004 | 65 | 0.00025 | 0.00000 | 0.00018 | 93 | 0.00019 | -0.76172 | 102.114 | 0.44798 | 0.83372 |
| PAT, AAT; bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase [EC:2.6.1.1 2.6.1.78 2.6.1.79] | 0.00019 | 0.00011 | 0.00011 | 0.00000 | 0.00004 | 65 | 0.00025 | 0.00000 | 0.00018 | 93 | 0.00019 | -0.76172 | 102.114 | 0.44798 | 0.83372 |
| galD; 4-oxalomesaconate tautomerase [EC:5.3.2.8] | 0.00019 | 0.00006 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00030 | 0.00000 | 0.00010 | 93 | 0.00010 | -2.58278 | 101.128 | 0.01123 | 0.58859 |
| wcaC; putative colanic acid biosynthesis glycosyltransferase [EC:2.4.-.-] | 0.00019 | 0.00008 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00022 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.50243 | 141.020 | 0.61615 | 0.88882 |
| wcaM; colanic acid biosynthesis protein WcaM | 0.00019 | 0.00008 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00022 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.50243 | 141.020 | 0.61615 | 0.88882 |
| wzxC; lipopolysaccharide exporter | 0.00019 | 0.00008 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00022 | 0.00000 | 0.00013 | 93 | 0.00015 | -0.50243 | 141.020 | 0.61615 | 0.88882 |
| E2.1.3.1-5S; methylmalonyl-CoA carboxyltransferase 5S subunit [EC:2.1.3.1] | 0.00019 | 0.00006 | 0.00022 | 0.00000 | 0.00012 | 65 | 0.00017 | 0.00000 | 0.00007 | 93 | 0.00014 | 0.39184 | 103.374 | 0.69599 | 0.91772 |
| mcbR; GntR family transcriptional regulator, colanic acid and biofilm gene transcriptional regulator | 0.00019 | 0.00008 | 0.00018 | 0.00000 | 0.00008 | 65 | 0.00020 | 0.00000 | 0.00012 | 93 | 0.00014 | -0.14424 | 145.557 | 0.88551 | 0.97001 |
| DAO, aao; D-amino-acid oxidase [EC:1.4.3.3] | 0.00019 | 0.00007 | 0.00018 | 0.00000 | 0.00012 | 65 | 0.00020 | 0.00000 | 0.00009 | 93 | 0.00015 | -0.14713 | 131.617 | 0.88326 | 0.96974 |
| RAD52; DNA repair and recombination protein RAD52 | 0.00019 | 0.00009 | 0.00017 | 0.00000 | 0.00015 | 65 | 0.00021 | 0.00000 | 0.00012 | 93 | 0.00020 | -0.19720 | 132.334 | 0.84398 | 0.95530 |
| yihQ; sulfoquinovosidase [EC:3.2.1.199] | 0.00019 | 0.00008 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00029 | 0.00000 | 0.00014 | 93 | 0.00014 | -1.84195 | 96.149 | 0.06856 | 0.73473 |
| mshK; MSHA biogenesis protein MshK | 0.00019 | 0.00009 | 0.00027 | 0.00000 | 0.00016 | 65 | 0.00013 | 0.00000 | 0.00010 | 93 | 0.00019 | 0.73231 | 107.259 | 0.46558 | 0.83786 |
| vexE; membrane fusion protein, multidrug efflux system | 0.00019 | 0.00009 | 0.00019 | 0.00000 | 0.00018 | 65 | 0.00018 | 0.00000 | 0.00009 | 93 | 0.00020 | 0.05679 | 94.685 | 0.95483 | 0.98792 |
| vexF; multidrug efflux pump | 0.00019 | 0.00009 | 0.00019 | 0.00000 | 0.00018 | 65 | 0.00018 | 0.00000 | 0.00009 | 93 | 0.00020 | 0.05679 | 94.685 | 0.95483 | 0.98792 |
| gfrR; sigma-54 dependent transcriptional regulator, gfr operon transcriptional activator | 0.00019 | 0.00006 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00026 | 0.00000 | 0.00010 | 93 | 0.00011 | -1.59470 | 115.482 | 0.11351 | 0.77376 |
| caiB; L-carnitine CoA-transferase [EC:2.8.3.21] | 0.00018 | 0.00007 | 0.00015 | 0.00000 | 0.00005 | 65 | 0.00021 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.38590 | 121.393 | 0.70025 | 0.91953 |
| oraE; D-ornithine 4,5-aminomutase subunit beta [EC:5.4.3.5] | 0.00018 | 0.00005 | 0.00029 | 0.00000 | 0.00010 | 65 | 0.00011 | 0.00000 | 0.00004 | 93 | 0.00010 | 1.79048 | 86.685 | 0.07687 | 0.75063 |
| oraS; D-ornithine 4,5-aminomutase subunit alpha [EC:5.4.3.5] | 0.00018 | 0.00005 | 0.00029 | 0.00000 | 0.00010 | 65 | 0.00011 | 0.00000 | 0.00004 | 93 | 0.00010 | 1.79048 | 86.685 | 0.07687 | 0.75063 |
| csm5; CRISPR-associated protein Csm5 | 0.00018 | 0.00006 | 0.00020 | 0.00000 | 0.00009 | 65 | 0.00017 | 0.00000 | 0.00009 | 93 | 0.00012 | 0.19814 | 153.709 | 0.84320 | 0.95519 |
| waaR, waaT, rfaJ; UDP-glucose/galactose:(glucosyl)LPS alpha-1,2-glucosyl/galactosyltransferase [EC:2.4.1.-] | 0.00018 | 0.00011 | 0.00022 | 0.00000 | 0.00020 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00023 | 0.26599 | 110.166 | 0.79074 | 0.94658 |
| agd31B; oligosaccharide 4-alpha-D-glucosyltransferase [EC:2.4.1.161] | 0.00018 | 0.00015 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00025 | 0.00001 | 0.00024 | 93 | 0.00025 | -0.65562 | 100.539 | 0.51357 | 0.85202 |
| entB; probable enterotoxin B | 0.00018 | 0.00017 | 0.00041 | 0.00001 | 0.00040 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00041 | 0.93997 | 64.300 | 0.35075 | 0.79120 |
| frlC; fructoselysine 3-epimerase [EC:5.1.3.41] | 0.00018 | 0.00008 | 0.00011 | 0.00000 | 0.00004 | 65 | 0.00023 | 0.00000 | 0.00013 | 93 | 0.00013 | -0.94818 | 111.287 | 0.34509 | 0.78739 |
| ASPH; aspartate beta-hydroxylase [EC:1.14.11.16] | 0.00018 | 0.00014 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00027 | 0.00000 | 0.00023 | 93 | 0.00023 | -0.92764 | 96.648 | 0.35590 | 0.79399 |
| rtxB, fitA; ATP-binding cassette, subfamily B, bacterial RtxB | 0.00018 | 0.00012 | 0.00019 | 0.00000 | 0.00017 | 65 | 0.00017 | 0.00000 | 0.00016 | 93 | 0.00023 | 0.06717 | 150.743 | 0.94653 | 0.98674 |
| rtxD, fitB; membrane fusion protein, RTX toxin transport system | 0.00018 | 0.00012 | 0.00019 | 0.00000 | 0.00017 | 65 | 0.00017 | 0.00000 | 0.00016 | 93 | 0.00023 | 0.06717 | 150.743 | 0.94653 | 0.98674 |
| rtxE, fitC; ATP-binding cassette, subfamily B, bacterial RtxE | 0.00018 | 0.00012 | 0.00019 | 0.00000 | 0.00017 | 65 | 0.00017 | 0.00000 | 0.00016 | 93 | 0.00023 | 0.06717 | 150.743 | 0.94653 | 0.98674 |
| adh1; NAD+-dependent secondary alcohol dehydrogenase Adh1 [EC:1.1.1.-] | 0.00018 | 0.00006 | 0.00025 | 0.00000 | 0.00013 | 65 | 0.00013 | 0.00000 | 0.00006 | 93 | 0.00014 | 0.86268 | 89.075 | 0.39063 | 0.80384 |
| cmr; MFS transporter, DHA3 family, multidrug efflux protein | 0.00018 | 0.00009 | 0.00020 | 0.00000 | 0.00019 | 65 | 0.00016 | 0.00000 | 0.00007 | 93 | 0.00020 | 0.16976 | 81.934 | 0.86562 | 0.96270 |
| tet35; tetracycline resistance efflux pump | 0.00017 | 0.00009 | 0.00024 | 0.00000 | 0.00018 | 65 | 0.00013 | 0.00000 | 0.00008 | 93 | 0.00020 | 0.53227 | 90.267 | 0.59585 | 0.88005 |
| ccrA; crotonyl-CoA reductase [EC:1.3.1.86] | 0.00017 | 0.00006 | 0.00028 | 0.00000 | 0.00014 | 65 | 0.00010 | 0.00000 | 0.00004 | 93 | 0.00015 | 1.26271 | 74.870 | 0.21061 | 0.77376 |
| sodN; nickel superoxide dismutase [EC:1.15.1.1] | 0.00017 | 0.00006 | 0.00031 | 0.00000 | 0.00015 | 65 | 0.00008 | 0.00000 | 0.00003 | 93 | 0.00015 | 1.56203 | 69.114 | 0.12285 | 0.77376 |
| cmr6; CRISPR-associated protein Cmr6 | 0.00017 | 0.00007 | 0.00015 | 0.00000 | 0.00008 | 65 | 0.00018 | 0.00000 | 0.00010 | 93 | 0.00013 | -0.23508 | 155.831 | 0.81445 | 0.95409 |
| nfrA; bacteriophage N4 adsorption protein A | 0.00017 | 0.00008 | 0.00015 | 0.00000 | 0.00009 | 65 | 0.00019 | 0.00000 | 0.00012 | 93 | 0.00015 | -0.24873 | 151.280 | 0.80391 | 0.94931 |
| caiA; crotonobetainyl-CoA dehydrogenase [EC:1.3.8.13] | 0.00017 | 0.00007 | 0.00014 | 0.00000 | 0.00005 | 65 | 0.00019 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.43209 | 121.104 | 0.66644 | 0.91126 |
| eutE; aldehyde dehydrogenase | 0.00017 | 0.00008 | 0.00012 | 0.00000 | 0.00005 | 65 | 0.00021 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.67326 | 117.628 | 0.50210 | 0.84942 |
| mbtM; long-chain-fatty-acid–[acyl-carrier-protein] ligase [EC:6.2.1.20] | 0.00017 | 0.00007 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00028 | 0.00000 | 0.00012 | 93 | 0.00012 | -2.18098 | 92.942 | 0.03171 | 0.61842 |
| yihU; 4-hydroxybutyrate dehydrogenase / sulfolactaldehyde 3-reductase [EC:1.1.1.61 1.1.1.373] | 0.00017 | 0.00008 | 0.00010 | 0.00000 | 0.00004 | 65 | 0.00022 | 0.00000 | 0.00013 | 93 | 0.00013 | -0.94540 | 111.566 | 0.34650 | 0.78921 |
| aac2-I; aminoglycoside 2’-N-acetyltransferase I [EC:2.3.1.59] | 0.00017 | 0.00006 | 0.00016 | 0.00000 | 0.00012 | 65 | 0.00017 | 0.00000 | 0.00006 | 93 | 0.00014 | -0.10011 | 101.027 | 0.92045 | 0.97905 |
| plc, cpa; phospholipase C / alpha-toxin [EC:3.1.4.3] | 0.00017 | 0.00017 | 0.00041 | 0.00001 | 0.00040 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00041 | 0.99395 | 64.002 | 0.32399 | 0.77376 |
| fimE; type 1 fimbriae regulatory protein FimE | 0.00017 | 0.00009 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00023 | 0.00000 | 0.00014 | 93 | 0.00015 | -0.98798 | 112.868 | 0.32528 | 0.77376 |
| ndhE; NAD(P)H-quinone oxidoreductase subunit 4L [EC:1.6.5.3] | 0.00017 | 0.00009 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00021 | 0.00000 | 0.00014 | 93 | 0.00015 | -0.69718 | 126.397 | 0.48697 | 0.84553 |
| garP; MFS transporter, ACS family, probable galactarate transporter | 0.00017 | 0.00008 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00023 | 0.00000 | 0.00013 | 93 | 0.00014 | -1.04949 | 108.406 | 0.29629 | 0.77376 |
| eutK; ethanolamine utilization protein EutK | 0.00017 | 0.00008 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00021 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.65275 | 130.132 | 0.51507 | 0.85216 |
| nodT, ameC; outer membrane protein, multidrug efflux system | 0.00017 | 0.00009 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00028 | 0.00000 | 0.00016 | 93 | 0.00016 | -1.67904 | 93.249 | 0.09649 | 0.76667 |
| cloSI; clostripain [EC:3.4.22.8] | 0.00017 | 0.00017 | 0.00041 | 0.00001 | 0.00040 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00040 | 0.99886 | 64.000 | 0.32162 | 0.77376 |
| entA; probable enterotoxin A | 0.00017 | 0.00017 | 0.00041 | 0.00001 | 0.00040 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00040 | 0.99886 | 64.000 | 0.32162 | 0.77376 |
| K09128; uncharacterized protein | 0.00017 | 0.00008 | 0.00022 | 0.00000 | 0.00018 | 65 | 0.00013 | 0.00000 | 0.00007 | 93 | 0.00019 | 0.43060 | 86.103 | 0.66784 | 0.91150 |
| sapZ; predicted membrane protein | 0.00017 | 0.00006 | 0.00016 | 0.00000 | 0.00008 | 65 | 0.00017 | 0.00000 | 0.00009 | 93 | 0.00012 | -0.14274 | 155.169 | 0.88668 | 0.97020 |
| hyaF; hydrogenase-1 operon protein HyaF | 0.00017 | 0.00008 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00024 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.44373 | 102.243 | 0.15187 | 0.77376 |
| crtW; beta-carotene ketolase (CrtW type) | 0.00017 | 0.00008 | 0.00021 | 0.00000 | 0.00011 | 65 | 0.00014 | 0.00000 | 0.00011 | 93 | 0.00016 | 0.45594 | 148.534 | 0.64910 | 0.90350 |
| E1.1.1.251, gatD; galactitol-1-phosphate 5-dehydrogenase [EC:1.1.1.251] | 0.00016 | 0.00008 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.66137 | 130.594 | 0.50954 | 0.84956 |
| TC.HME; heavy-metal exporter, HME family | 0.00016 | 0.00006 | 0.00013 | 0.00000 | 0.00006 | 65 | 0.00019 | 0.00000 | 0.00009 | 93 | 0.00011 | -0.58719 | 145.991 | 0.55798 | 0.86813 |
| epsN; pyridoxal phosphate-dependent aminotransferase EpsN [EC:2.6.1.-] | 0.00016 | 0.00008 | 0.00010 | 0.00000 | 0.00009 | 65 | 0.00021 | 0.00000 | 0.00012 | 93 | 0.00015 | -0.74772 | 154.707 | 0.45576 | 0.83438 |
| blaTEM; beta-lactamase class A TEM [EC:3.5.2.6] | 0.00016 | 0.00008 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00022 | 0.00000 | 0.00013 | 93 | 0.00014 | -1.02340 | 108.540 | 0.30839 | 0.77376 |
| TYR; tyrosinase [EC:1.14.18.1] | 0.00016 | 0.00006 | 0.00019 | 0.00000 | 0.00012 | 65 | 0.00015 | 0.00000 | 0.00006 | 93 | 0.00013 | 0.31268 | 97.502 | 0.75519 | 0.93665 |
| psuT; putative pseudouridine transporter | 0.00016 | 0.00008 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00021 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.87266 | 126.236 | 0.38451 | 0.80197 |
| gci; D-galactarolactone cycloisomerase [EC:5.5.1.27] | 0.00016 | 0.00007 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00022 | 0.00000 | 0.00011 | 93 | 0.00012 | -1.10849 | 133.095 | 0.26965 | 0.77376 |
| rap; autoaggregation protein RapA/B/C | 0.00016 | 0.00012 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00023 | 0.00000 | 0.00020 | 93 | 0.00021 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| TC.GBP; general bacterial porin, GBP family | 0.00016 | 0.00013 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00024 | 0.00000 | 0.00023 | 93 | 0.00023 | -0.82117 | 96.697 | 0.41357 | 0.81655 |
| waaH; heptose III glucuronosyltransferase [EC:2.4.1.-] | 0.00016 | 0.00008 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00021 | 0.00000 | 0.00013 | 93 | 0.00013 | -0.96736 | 108.799 | 0.33551 | 0.78186 |
| egtE; hercynylcysteine S-oxide lyase [EC:4.4.1.36] | 0.00016 | 0.00006 | 0.00015 | 0.00000 | 0.00012 | 65 | 0.00017 | 0.00000 | 0.00006 | 93 | 0.00013 | -0.17211 | 101.677 | 0.86369 | 0.96263 |
| MGD; 1,2-diacylglycerol 3-beta-galactosyltransferase [EC:2.4.1.46] | 0.00016 | 0.00006 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00023 | 0.00000 | 0.00010 | 93 | 0.00011 | -1.67219 | 104.128 | 0.09749 | 0.76673 |
| whiB6; WhiB family transcriptional regulator, redox-sensing transcriptional regulator | 0.00016 | 0.00007 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00026 | 0.00000 | 0.00012 | 93 | 0.00012 | -2.12878 | 93.045 | 0.03591 | 0.62411 |
| tasA, cotN; spore coat-associated protein N | 0.00016 | 0.00010 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00024 | 0.00000 | 0.00016 | 93 | 0.00016 | -1.26289 | 98.501 | 0.20961 | 0.77376 |
| hepC; heparan-sulfate lyase [EC:4.2.2.8] | 0.00016 | 0.00008 | 0.00010 | 0.00000 | 0.00006 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.75992 | 127.906 | 0.44870 | 0.83406 |
| caiE; carnitine operon protein CaiE | 0.00016 | 0.00007 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00021 | 0.00000 | 0.00012 | 93 | 0.00013 | -1.02445 | 107.923 | 0.30791 | 0.77376 |
| aksD; methanogen homoaconitase large subunit [EC:4.2.1.114] | 0.00016 | 0.00011 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00020 | 0.00000 | 0.00017 | 93 | 0.00019 | -0.56472 | 133.245 | 0.57321 | 0.87292 |
| mbtG; mycobactin lysine-N-oxygenase | 0.00016 | 0.00005 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00025 | 0.00000 | 0.00008 | 93 | 0.00008 | -2.79859 | 96.599 | 0.00620 | 0.54022 |
| QPCT; glutaminyl-peptide cyclotransferase [EC:2.3.2.5] | 0.00016 | 0.00010 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00026 | 0.00000 | 0.00018 | 93 | 0.00018 | -1.42059 | 92.237 | 0.15881 | 0.77376 |
| eutD; phosphotransacetylase | 0.00016 | 0.00008 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00021 | 0.00000 | 0.00013 | 93 | 0.00013 | -0.90136 | 109.125 | 0.36938 | 0.79587 |
| eutG; alcohol dehydrogenase | 0.00016 | 0.00008 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00021 | 0.00000 | 0.00013 | 93 | 0.00013 | -0.90136 | 109.125 | 0.36938 | 0.79587 |
| zraP; zinc resistance-associated protein | 0.00016 | 0.00008 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00021 | 0.00000 | 0.00013 | 93 | 0.00013 | -0.90136 | 109.125 | 0.36938 | 0.79587 |
| crtU; isorenieratene synthase | 0.00016 | 0.00005 | 0.00007 | 0.00000 | 0.00005 | 65 | 0.00021 | 0.00000 | 0.00008 | 93 | 0.00009 | -1.53991 | 144.098 | 0.12577 | 0.77376 |
| mop; aldehyde oxidoreductase [EC:1.2.99.7] | 0.00016 | 0.00009 | 0.00018 | 0.00000 | 0.00017 | 65 | 0.00014 | 0.00000 | 0.00008 | 93 | 0.00019 | 0.17909 | 93.378 | 0.85825 | 0.96053 |
| rhmA; 2-dehydro-3-deoxy-L-rhamnonate aldolase [EC:4.1.2.53] | 0.00016 | 0.00008 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00013 | -0.88970 | 109.129 | 0.37559 | 0.79810 |
| nheBC; non-hemolytic enterotoxin B/C | 0.00015 | 0.00009 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00024 | 0.00000 | 0.00016 | 93 | 0.00016 | -1.32607 | 95.488 | 0.18798 | 0.77376 |
| araQ; arabinosaccharide transport system permease protein | 0.00015 | 0.00006 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00023 | 0.00000 | 0.00009 | 93 | 0.00010 | -1.88962 | 108.325 | 0.06148 | 0.73001 |
| APA1_2; ATP adenylyltransferase [EC:2.7.7.53] | 0.00015 | 0.00007 | 0.00021 | 0.00000 | 0.00012 | 65 | 0.00012 | 0.00000 | 0.00009 | 93 | 0.00015 | 0.61213 | 120.370 | 0.54161 | 0.86207 |
| pimE; alpha-1,2-mannosyltransferase [EC:2.4.1.-] | 0.00015 | 0.00005 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00025 | 0.00000 | 0.00008 | 93 | 0.00008 | -2.79965 | 95.008 | 0.00620 | 0.54022 |
| K09129; uncharacterized protein | 0.00015 | 0.00007 | 0.00014 | 0.00000 | 0.00009 | 65 | 0.00016 | 0.00000 | 0.00010 | 93 | 0.00013 | -0.13497 | 152.932 | 0.89281 | 0.97159 |
| nifV; homocitrate synthase NifV [EC:2.3.3.14] | 0.00015 | 0.00004 | 0.00011 | 0.00000 | 0.00004 | 65 | 0.00018 | 0.00000 | 0.00006 | 93 | 0.00007 | -1.03829 | 147.257 | 0.30084 | 0.77376 |
| amyM; maltogenic alpha-amylase [EC:3.2.1.133] | 0.00015 | 0.00006 | 0.00022 | 0.00000 | 0.00013 | 65 | 0.00010 | 0.00000 | 0.00005 | 93 | 0.00014 | 0.89131 | 79.908 | 0.37544 | 0.79810 |
| IRPC; inward rectifier potassium channel | 0.00015 | 0.00006 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00020 | 0.00000 | 0.00009 | 93 | 0.00011 | -0.98258 | 148.823 | 0.32741 | 0.77376 |
| ethA; monooxygenase [EC:1.14.13.-] | 0.00015 | 0.00005 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00023 | 0.00000 | 0.00008 | 93 | 0.00008 | -2.39530 | 103.101 | 0.01841 | 0.59095 |
| mph; macrolide phosphotransferase | 0.00015 | 0.00008 | 0.00015 | 0.00000 | 0.00012 | 65 | 0.00015 | 0.00000 | 0.00011 | 93 | 0.00016 | -0.00106 | 148.013 | 0.99916 | 0.99957 |
| E3.5.1.41; chitin deacetylase [EC:3.5.1.41] | 0.00015 | 0.00007 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00025 | 0.00000 | 0.00013 | 93 | 0.00013 | -2.03396 | 92.000 | 0.04484 | 0.65981 |
| tpa; taurine-pyruvate aminotransferase [EC:2.6.1.77] | 0.00015 | 0.00007 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00025 | 0.00000 | 0.00013 | 93 | 0.00013 | -2.03396 | 92.000 | 0.04484 | 0.65981 |
| K09744; uncharacterized protein | 0.00015 | 0.00010 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00023 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.10121 | 97.851 | 0.27351 | 0.77376 |
| 2PGK; 2-phosphoglycerate kinase [EC:2.7.2.-] | 0.00015 | 0.00010 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00023 | 0.00000 | 0.00017 | 93 | 0.00018 | -1.12196 | 97.234 | 0.26464 | 0.77376 |
| K09133; uncharacterized protein | 0.00015 | 0.00010 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00025 | 0.00000 | 0.00016 | 93 | 0.00016 | -1.54315 | 92.000 | 0.12623 | 0.77376 |
| entC; probable enterotoxin C | 0.00015 | 0.00015 | 0.00035 | 0.00001 | 0.00035 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00035 | 0.99309 | 64.003 | 0.32441 | 0.77376 |
| xdhD; putative selenate reductase molybdopterin-binding subunit | 0.00015 | 0.00009 | 0.00014 | 0.00000 | 0.00012 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00017 | -0.11079 | 152.855 | 0.91192 | 0.97664 |
| GATM; glycine amidinotransferase [EC:2.1.4.1] | 0.00015 | 0.00004 | 0.00017 | 0.00000 | 0.00005 | 65 | 0.00013 | 0.00000 | 0.00005 | 93 | 0.00007 | 0.49309 | 154.461 | 0.62265 | 0.89150 |
| vanC, vanE, vanG; D-alanine—D-serine ligase [EC:6.3.2.35] | 0.00015 | 0.00009 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00025 | 0.00000 | 0.00015 | 93 | 0.00015 | -1.69915 | 92.000 | 0.09267 | 0.76567 |
| cpe; enterotoxin Cpe | 0.00015 | 0.00015 | 0.00035 | 0.00001 | 0.00035 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00035 | 0.99870 | 64.000 | 0.32170 | 0.77376 |
| entD; probable enterotoxin D | 0.00015 | 0.00015 | 0.00035 | 0.00001 | 0.00035 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00035 | 0.99870 | 64.000 | 0.32170 | 0.77376 |
| aguG; alpha-1,4-digalacturonate transport system permease protein | 0.00015 | 0.00007 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00023 | 0.00000 | 0.00011 | 93 | 0.00012 | -1.74268 | 101.240 | 0.08443 | 0.76567 |
| cobR; cob(II)yrinic acid a,c-diamide reductase [EC:1.16.8.1] | 0.00015 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00023 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.89261 | 94.593 | 0.06147 | 0.73001 |
| yihT; sulfofructosephosphate aldolase [EC:4.1.2.57] | 0.00015 | 0.00007 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00021 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.14254 | 102.683 | 0.25589 | 0.77376 |
| hokB; protein HokB | 0.00014 | 0.00008 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.09515 | 101.937 | 0.27603 | 0.77376 |
| ceo; N5-(carboxyethyl)ornithine synthase [EC:1.5.1.24] | 0.00014 | 0.00007 | 0.00017 | 0.00000 | 0.00013 | 65 | 0.00013 | 0.00000 | 0.00007 | 93 | 0.00014 | 0.27204 | 105.448 | 0.78612 | 0.94458 |
| pmrD; signal transduction protein PmrD | 0.00014 | 0.00007 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.11490 | 102.677 | 0.26750 | 0.77376 |
| ypdF; aminopeptidase [EC:3.4.11.-] | 0.00014 | 0.00007 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.11490 | 102.677 | 0.26750 | 0.77376 |
| K09726; uncharacterized protein | 0.00014 | 0.00007 | 0.00010 | 0.00000 | 0.00006 | 65 | 0.00017 | 0.00000 | 0.00011 | 93 | 0.00013 | -0.53809 | 130.686 | 0.59143 | 0.87896 |
| MEP; peptidyl-Lys metalloendopeptidase [EC:3.4.24.20] | 0.00014 | 0.00007 | 0.00022 | 0.00000 | 0.00015 | 65 | 0.00008 | 0.00000 | 0.00005 | 93 | 0.00015 | 0.88940 | 78.381 | 0.37651 | 0.79810 |
| traA; conjugal transfer pilus assembly protein TraA | 0.00014 | 0.00009 | 0.00014 | 0.00000 | 0.00012 | 65 | 0.00014 | 0.00000 | 0.00013 | 93 | 0.00018 | -0.01305 | 153.218 | 0.98961 | 0.99801 |
| ITGB8; integrin beta 8 | 0.00014 | 0.00006 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00017 | 0.00000 | 0.00010 | 93 | 0.00011 | -0.68129 | 138.265 | 0.49683 | 0.84797 |
| kinD; two-component system, sporulation sensor kinase D [EC:2.7.13.3] | 0.00014 | 0.00008 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.11940 | 102.764 | 0.26558 | 0.77376 |
| slcC; (S)-sulfolactate dehydrogenase [EC:1.1.1.310] | 0.00014 | 0.00005 | 0.00015 | 0.00000 | 0.00007 | 65 | 0.00013 | 0.00000 | 0.00006 | 93 | 0.00009 | 0.24262 | 132.653 | 0.80868 | 0.95002 |
| K11325; L-cysteine/cystine lyase | 0.00014 | 0.00007 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00020 | 0.00000 | 0.00012 | 93 | 0.00013 | -1.14316 | 117.323 | 0.25530 | 0.77376 |
| epsH; glycosyltransferase EpsH [EC:2.4.-.-] | 0.00014 | 0.00007 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00023 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.81178 | 93.676 | 0.07322 | 0.74128 |
| irp2, HMWP2; yersiniabactin nonribosomal peptide synthetase | 0.00014 | 0.00008 | 0.00010 | 0.00000 | 0.00006 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00014 | -0.43603 | 135.265 | 0.66351 | 0.91006 |
| AOX1, AOX2; ubiquinol oxidase [EC:1.10.3.11] | 0.00014 | 0.00008 | 0.00018 | 0.00000 | 0.00018 | 65 | 0.00011 | 0.00000 | 0.00007 | 93 | 0.00019 | 0.37426 | 81.933 | 0.70918 | 0.92004 |
| coxA; spore cortex protein | 0.00014 | 0.00008 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.10288 | 102.762 | 0.27265 | 0.77376 |
| comC; (2R)-3-sulfolactate dehydrogenase (NADP+) [EC:1.1.1.338] | 0.00014 | 0.00006 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00010 | 93 | 0.00010 | -1.56856 | 104.987 | 0.11976 | 0.77376 |
| dhpH; 2,6-dihydroxypyridine 3-monooxygenase [EC:1.14.13.10] | 0.00014 | 0.00006 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00015 | 0.00000 | 0.00007 | 93 | 0.00013 | -0.28423 | 108.579 | 0.77678 | 0.94269 |
| yscF, sctF, ssaG, prgI; type III secretion protein F | 0.00014 | 0.00008 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00021 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.47369 | 93.796 | 0.14391 | 0.77376 |
| csh1; CRISPR-associated protein Csh1 | 0.00014 | 0.00003 | 0.00015 | 0.00000 | 0.00005 | 65 | 0.00012 | 0.00000 | 0.00005 | 93 | 0.00007 | 0.45256 | 138.485 | 0.65158 | 0.90504 |
| frlB; fructoselysine 6-phosphate deglycase [EC:3.5.-.-] | 0.00014 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00012 | 93 | 0.00013 | -1.15500 | 102.972 | 0.25076 | 0.77376 |
| frlR; GntR family transcriptional regulator, frlABCD operon transcriptional regulator | 0.00014 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00012 | 93 | 0.00013 | -1.15500 | 102.972 | 0.25076 | 0.77376 |
| E3.2.1.15; polygalacturonase [EC:3.2.1.15] | 0.00013 | 0.00007 | 0.00011 | 0.00000 | 0.00007 | 65 | 0.00016 | 0.00000 | 0.00010 | 93 | 0.00012 | -0.39680 | 151.667 | 0.69207 | 0.91676 |
| ynfF; Tat-targeted selenate reductase subunit YnfF [EC:1.97.1.9] | 0.00013 | 0.00007 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00018 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.85150 | 103.787 | 0.39645 | 0.80459 |
| irp5, ybtE; yersiniabactin salicyl-AMP ligase [EC:6.3.2.-] | 0.00013 | 0.00008 | 0.00010 | 0.00000 | 0.00006 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00014 | -0.39747 | 135.266 | 0.69165 | 0.91676 |
| ttrC; tetrathionate reductase subunit C | 0.00013 | 0.00004 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00015 | 0.00000 | 0.00006 | 93 | 0.00009 | -0.43651 | 150.465 | 0.66309 | 0.91006 |
| caiT; L-carnitine/gamma-butyrobetaine antiporter | 0.00013 | 0.00008 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.64273 | 128.274 | 0.52154 | 0.85613 |
| epsJ; glycosyltransferase EpsJ [EC:2.4.-.-] | 0.00013 | 0.00003 | 0.00013 | 0.00000 | 0.00005 | 65 | 0.00013 | 0.00000 | 0.00004 | 93 | 0.00006 | 0.02612 | 139.434 | 0.97920 | 0.99657 |
| waaW; (galactosyl)LPS 1,2-glucosyltransferase [EC:2.4.1.-] | 0.00013 | 0.00008 | 0.00010 | 0.00000 | 0.00008 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00015 | -0.33128 | 152.189 | 0.74089 | 0.93000 |
| irtA; ATP-binding cassette, subfamily B, bacterial IrtA [EC:3.6.3.-] | 0.00013 | 0.00006 | 0.00013 | 0.00000 | 0.00012 | 65 | 0.00013 | 0.00000 | 0.00006 | 93 | 0.00014 | -0.00289 | 95.491 | 0.99770 | 0.99955 |
| argHA; argininosuccinate lyase / amino-acid N-acetyltransferase [EC:4.3.2.1 2.3.1.1] | 0.00013 | 0.00008 | 0.00024 | 0.00000 | 0.00018 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00019 | 0.90741 | 78.269 | 0.36697 | 0.79587 |
| NAALAD; N-acetylated-alpha-linked acidic dipeptidase [EC:3.4.17.21] | 0.00013 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00021 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.62464 | 94.168 | 0.10758 | 0.77376 |
| waaZ, rfaZ; KDO transferase III [EC:2.4.99.-] | 0.00013 | 0.00008 | 0.00026 | 0.00000 | 0.00018 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00018 | 1.20160 | 67.987 | 0.23369 | 0.77376 |
| aksE; methanogen homoaconitase small subunit [EC:4.2.1.114] | 0.00013 | 0.00010 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00017 | 93 | 0.00017 | -0.99374 | 97.228 | 0.32282 | 0.77376 |
| K09785; uncharacterized protein | 0.00013 | 0.00010 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00017 | 93 | 0.00017 | -0.99374 | 97.228 | 0.32282 | 0.77376 |
| LYS12; homoisocitrate dehydrogenase [EC:1.1.1.87] | 0.00013 | 0.00010 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00017 | 93 | 0.00017 | -0.99374 | 97.228 | 0.32282 | 0.77376 |
| LYS21, LYS20; homocitrate synthase [EC:2.3.3.14] | 0.00013 | 0.00010 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00017 | 93 | 0.00017 | -0.99374 | 97.228 | 0.32282 | 0.77376 |
| narC; cytochrome b-561 | 0.00013 | 0.00010 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00017 | 93 | 0.00017 | -0.99374 | 97.228 | 0.32282 | 0.77376 |
| gumE; putative polymerase | 0.00013 | 0.00005 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00016 | 0.00000 | 0.00008 | 93 | 0.00010 | -0.71586 | 153.110 | 0.47517 | 0.84095 |
| trbC; conjugal transfer pilus assembly protein TrbC | 0.00013 | 0.00004 | 0.00015 | 0.00000 | 0.00009 | 65 | 0.00011 | 0.00000 | 0.00004 | 93 | 0.00010 | 0.42461 | 92.398 | 0.67211 | 0.91184 |
| gtfB, gtfE; vancomycin aglycone glucosyltransferase [EC:2.4.1.310] | 0.00013 | 0.00006 | 0.00015 | 0.00000 | 0.00012 | 65 | 0.00012 | 0.00000 | 0.00006 | 93 | 0.00013 | 0.28141 | 95.284 | 0.77900 | 0.94269 |
| acfD; accessory colonization factor AcfD | 0.00013 | 0.00007 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.46980 | 118.527 | 0.63936 | 0.89853 |
| HMGCR; hydroxymethylglutaryl-CoA reductase (NADPH) [EC:1.1.1.34] | 0.00013 | 0.00005 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00016 | 0.00000 | 0.00007 | 93 | 0.00009 | -0.98942 | 146.429 | 0.32409 | 0.77376 |
| exoP, vpsO; polysaccharide biosynthesis transport protein | 0.00013 | 0.00006 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00020 | 0.00000 | 0.00009 | 93 | 0.00010 | -1.90820 | 96.252 | 0.05935 | 0.72498 |
| mdcF; malonate transporter | 0.00013 | 0.00005 | 0.00015 | 0.00000 | 0.00009 | 65 | 0.00011 | 0.00000 | 0.00005 | 93 | 0.00010 | 0.39461 | 104.240 | 0.69394 | 0.91701 |
| E1.12.7.2G; ferredoxin hydrogenase gamma subunit [EC:1.12.7.2] | 0.00013 | 0.00009 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00022 | 0.00000 | 0.00016 | 93 | 0.00016 | -1.36617 | 92.000 | 0.17521 | 0.77376 |
| fdhB; formate dehydrogenase (NADP+) beta subunit [EC:1.17.1.10] | 0.00013 | 0.00009 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00022 | 0.00000 | 0.00016 | 93 | 0.00016 | -1.36617 | 92.000 | 0.17521 | 0.77376 |
| epsL; sugar transferase EpsL [EC:2.-.-.-] | 0.00013 | 0.00007 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00021 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.68501 | 93.700 | 0.09531 | 0.76609 |
| cqsA; CAI-1 autoinducer synthase [EC:2.3.-.-] | 0.00013 | 0.00006 | 0.00016 | 0.00000 | 0.00008 | 65 | 0.00011 | 0.00000 | 0.00009 | 93 | 0.00012 | 0.42189 | 155.366 | 0.67369 | 0.91184 |
| ybtX, irp8; MFS transporter, putative signal transducer | 0.00013 | 0.00007 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.92120 | 104.504 | 0.35907 | 0.79496 |
| PTS-Gam-EIIB, agaB; PTS system, galactosamine-specific IIB component [EC:2.7.1.-] | 0.00013 | 0.00007 | 0.00009 | 0.00000 | 0.00004 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.51700 | 115.589 | 0.60615 | 0.88536 |
| aguE; alpha-1,4-digalacturonate transport system substrate-binding protein | 0.00013 | 0.00007 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.51567 | 101.934 | 0.13270 | 0.77376 |
| aguF; alpha-1,4-digalacturonate transport system permease protein | 0.00013 | 0.00007 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00020 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.51567 | 101.934 | 0.13270 | 0.77376 |
| pcpB; pentachlorophenol monooxygenase [EC:1.14.13.50] | 0.00013 | 0.00006 | 0.00018 | 0.00000 | 0.00012 | 65 | 0.00009 | 0.00000 | 0.00006 | 93 | 0.00013 | 0.67612 | 89.910 | 0.50070 | 0.84900 |
| fbp-SEBP; fructose-1,6-bisphosphatase I / sedoheptulose-1,7-bisphosphatase [EC:3.1.3.11 3.1.3.37] | 0.00013 | 0.00006 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00016 | 0.00000 | 0.00009 | 93 | 0.00010 | -0.83559 | 139.280 | 0.40482 | 0.81147 |
| csxA; exo-1,4-beta-D-glucosaminidase [EC:3.2.1.165] | 0.00013 | 0.00005 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00020 | 0.00000 | 0.00008 | 93 | 0.00008 | -2.25511 | 96.002 | 0.02640 | 0.60812 |
| ahlD, aiiA, attM, blcC; N-acyl homoserine lactone hydrolase [EC:3.1.1.81] | 0.00013 | 0.00008 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.41914 | 93.705 | 0.15918 | 0.77376 |
| devR; two-component system, NarL family, response regulator DevR | 0.00013 | 0.00004 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00020 | 0.00000 | 0.00007 | 93 | 0.00007 | -2.59459 | 96.119 | 0.01095 | 0.58284 |
| chiA; bifunctional chitinase/lysozyme [EC:3.2.1.14 3.2.1.17] | 0.00013 | 0.00007 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.80763 | 106.099 | 0.42111 | 0.82292 |
| prlF, sohA; antitoxin PrlF | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00018 | 0.00000 | 0.00012 | 93 | 0.00013 | -1.02607 | 103.123 | 0.30726 | 0.77376 |
| PTS-Gam-EIIC, agaC; PTS system, galactosamine-specific IIC component | 0.00012 | 0.00007 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.56927 | 115.481 | 0.57028 | 0.87135 |
| PTS-Gam-EIID, agaD; PTS system, galactosamine-specific IID component | 0.00012 | 0.00007 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.56927 | 115.481 | 0.57028 | 0.87135 |
| ompT; omptin [EC:3.4.23.49] | 0.00012 | 0.00007 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.66416 | 109.909 | 0.50798 | 0.84956 |
| K07092; uncharacterized protein | 0.00012 | 0.00005 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00016 | 0.00000 | 0.00008 | 93 | 0.00009 | -0.88712 | 140.776 | 0.37653 | 0.79810 |
| azr; azobenzene reductase [EC:1.7.1.6] | 0.00012 | 0.00008 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00019 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.26925 | 96.636 | 0.20740 | 0.77376 |
| exoQ; exopolysaccharide production protein ExoQ | 0.00012 | 0.00006 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00020 | 0.00000 | 0.00009 | 93 | 0.00010 | -1.92213 | 95.525 | 0.05757 | 0.71337 |
| lprG; lipoprotein LprG | 0.00012 | 0.00004 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00018 | 0.00000 | 0.00006 | 93 | 0.00007 | -2.00594 | 115.024 | 0.04721 | 0.66255 |
| phsB; thiosulfate reductase electron transport protein | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.92561 | 103.371 | 0.35680 | 0.79399 |
| phsC; thiosulfate reductase cytochrome b subunit | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.92561 | 103.371 | 0.35680 | 0.79399 |
| nprB; neutral peptidase B [EC:3.4.24.-] | 0.00012 | 0.00008 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00020 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.38567 | 93.711 | 0.16914 | 0.77376 |
| yeiL; CRP/FNR family transcriptional regulator, putaive post-exponential-phase nitrogen-starvation regulator | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.99975 | 103.236 | 0.31977 | 0.77376 |
| caiF; transcriptional activator CaiF | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.91332 | 103.377 | 0.36320 | 0.79496 |
| scpB, mmcD; methylmalonyl-CoA decarboxylase [EC:4.1.1.41] | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.91332 | 103.377 | 0.36320 | 0.79496 |
| yahN; amino acid exporter | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.91332 | 103.377 | 0.36320 | 0.79496 |
| ydiB; quinate/shikimate dehydrogenase [EC:1.1.1.282] | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.91332 | 103.377 | 0.36320 | 0.79496 |
| ydiF; acetate CoA-transferase [EC:2.8.3.8] | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.91332 | 103.377 | 0.36320 | 0.79496 |
| yhdJ; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.91332 | 103.377 | 0.36320 | 0.79496 |
| K08677; kumamolisin | 0.00012 | 0.00004 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00018 | 0.00000 | 0.00007 | 93 | 0.00007 | -2.01834 | 107.088 | 0.04606 | 0.66255 |
| acsE; 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase [EC:2.1.1.258] | 0.00012 | 0.00006 | 0.00007 | 0.00000 | 0.00006 | 65 | 0.00016 | 0.00000 | 0.00009 | 93 | 0.00011 | -0.79621 | 148.313 | 0.42718 | 0.82661 |
| cdhE, acsC; acetyl-CoA decarbonylase/synthase complex subunit gamma [EC:2.1.1.245] | 0.00012 | 0.00006 | 0.00007 | 0.00000 | 0.00006 | 65 | 0.00016 | 0.00000 | 0.00009 | 93 | 0.00011 | -0.79621 | 148.313 | 0.42718 | 0.82661 |
| pgi-pmi; glucose/mannose-6-phosphate isomerase [EC:5.3.1.9 5.3.1.8] | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.91041 | 119.964 | 0.36443 | 0.79587 |
| K15878, narB; rieske iron-sulphur protein | 0.00012 | 0.00010 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00020 | 0.00000 | 0.00017 | 93 | 0.00017 | -1.14392 | 92.195 | 0.25562 | 0.77376 |
| allS; LysR family transcriptional regulator, transcriptional activator of the allD operon | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.90080 | 103.380 | 0.36979 | 0.79587 |
| pgtP; MFS transporter, OPA family, phosphoglycerate transporter protein | 0.00012 | 0.00007 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00018 | 0.00000 | 0.00011 | 93 | 0.00012 | -1.26053 | 98.052 | 0.21047 | 0.77376 |
| K09120; uncharacterized protein | 0.00012 | 0.00010 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00020 | 0.00000 | 0.00017 | 93 | 0.00017 | -1.19542 | 92.000 | 0.23499 | 0.77376 |
| agaI; galactosamine-6-phosphate isomerase [EC:5.3.1.-] | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.97254 | 103.431 | 0.33305 | 0.77955 |
| yogA; zinc-binding alcohol dehydrogenase/oxidoreductase | 0.00012 | 0.00006 | 0.00013 | 0.00000 | 0.00008 | 65 | 0.00012 | 0.00000 | 0.00008 | 93 | 0.00011 | 0.09898 | 145.551 | 0.92129 | 0.97905 |
| K16654; spore-specific protein | 0.00012 | 0.00008 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00019 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.34486 | 93.711 | 0.18192 | 0.77376 |
| L2HGDH; 2-hydroxyglutarate dehydrogenase [EC:1.1.99.2] | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.97470 | 118.033 | 0.33170 | 0.77825 |
| enr; 2-enoate reductase [EC:1.3.1.31] | 0.00012 | 0.00005 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00017 | 0.00000 | 0.00008 | 93 | 0.00009 | -1.33287 | 125.908 | 0.18498 | 0.77376 |
| K07096; uncharacterized protein | 0.00012 | 0.00005 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00014 | 0.00000 | 0.00008 | 93 | 0.00009 | -0.54512 | 143.198 | 0.58652 | 0.87731 |
| pchF; 4-cresol dehydrogenase (hydroxylating) flavoprotein subunit [EC:1.17.99.1] | 0.00012 | 0.00008 | 0.00028 | 0.00000 | 0.00020 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00020 | 1.34564 | 64.093 | 0.18316 | 0.77376 |
| K06951; uncharacterized protein | 0.00012 | 0.00010 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00020 | 0.00000 | 0.00017 | 93 | 0.00017 | -1.17706 | 92.000 | 0.24221 | 0.77376 |
| elaD, sseL; deubiquitinase [EC:3.4.22.-] | 0.00012 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.88172 | 103.361 | 0.37997 | 0.79852 |
| gspS; general secretion pathway protein S | 0.00012 | 0.00008 | 0.00018 | 0.00000 | 0.00017 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00018 | 0.60860 | 75.305 | 0.54462 | 0.86388 |
| mbtI, irp9, ybtS; salicylate synthetase [EC:5.4.4.2 4.2.99.21] | 0.00012 | 0.00007 | 0.00003 | 0.00000 | 0.00001 | 65 | 0.00018 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.26555 | 94.347 | 0.20879 | 0.77376 |
| glcA; glycolate permease | 0.00012 | 0.00007 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.79791 | 103.923 | 0.42674 | 0.82661 |
| cmtC, dhbA; 2,3-dihydroxy-p-cumate/2,3-dihydroxybenzoate 3,4-dioxygenase [EC:1.13.11.- 1.13.11.14] | 0.00012 | 0.00006 | 0.00016 | 0.00000 | 0.00012 | 65 | 0.00009 | 0.00000 | 0.00006 | 93 | 0.00013 | 0.52664 | 98.060 | 0.59963 | 0.88240 |
| esp, sigA, sepA; serine protease autotransporter [EC:3.4.21.-] | 0.00011 | 0.00007 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00017 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.03467 | 96.255 | 0.30342 | 0.77376 |
| acsB; acetyl-CoA synthase [EC:2.3.1.169] | 0.00011 | 0.00006 | 0.00007 | 0.00000 | 0.00006 | 65 | 0.00015 | 0.00000 | 0.00009 | 93 | 0.00011 | -0.70165 | 148.502 | 0.48400 | 0.84354 |
| cdhD, acsD; acetyl-CoA decarbonylase/synthase complex subunit delta [EC:2.1.1.245] | 0.00011 | 0.00006 | 0.00007 | 0.00000 | 0.00006 | 65 | 0.00015 | 0.00000 | 0.00009 | 93 | 0.00011 | -0.70165 | 148.502 | 0.48400 | 0.84354 |
| cansdh; carboxynorspermidine synthase [EC:1.5.1.43] | 0.00011 | 0.00008 | 0.00019 | 0.00000 | 0.00018 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00019 | 0.68356 | 78.760 | 0.49626 | 0.84797 |
| esxA, esat6; 6 kDa early secretory antigenic target | 0.00011 | 0.00004 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00017 | 0.00000 | 0.00006 | 93 | 0.00007 | -2.11629 | 106.773 | 0.03664 | 0.62578 |
| SUOX; sulfite oxidase [EC:1.8.3.1] | 0.00011 | 0.00004 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00011 | 0.00000 | 0.00005 | 93 | 0.00008 | -0.03857 | 131.841 | 0.96930 | 0.99287 |
| phnA; phosphonoacetate hydrolase [EC:3.11.1.2] | 0.00011 | 0.00005 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00014 | 0.00000 | 0.00008 | 93 | 0.00009 | -0.60847 | 153.014 | 0.54378 | 0.86313 |
| vgb; virginiamycin B lyase [EC:4.2.99.-] | 0.00011 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00018 | 0.00000 | 0.00007 | 93 | 0.00007 | -2.52587 | 95.686 | 0.01318 | 0.59095 |
| ctpI; cation-transporting P-type ATPase I [EC:3.6.3.-] | 0.00011 | 0.00009 | 0.00025 | 0.00000 | 0.00023 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00023 | 1.00281 | 64.330 | 0.31971 | 0.77376 |
| mdtE; membrane fusion protein, multidrug efflux system | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.86587 | 103.469 | 0.38856 | 0.80262 |
| mdtF; multidrug efflux pump | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.86587 | 103.469 | 0.38856 | 0.80262 |
| sfmF; fimbrial-like protein | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00016 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.86587 | 103.469 | 0.38856 | 0.80262 |
| ndx1; diadenosine hexaphosphate hydrolase (ATP-forming) [EC:3.6.1.61] | 0.00011 | 0.00010 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00017 | 0.00000 | 0.00017 | 93 | 0.00017 | -0.81776 | 97.305 | 0.41549 | 0.81730 |
| allR; IclR family transcriptional regulator, negative regulator of allantoin and glyoxylate utilization operons | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| dos; c-di-GMP-specific phosphodiesterase [EC:3.1.4.52] | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| emrK; multidrug resistance protein K | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| emrY; MFS transporter, DHA2 family, multidrug resistance protein | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| focB; formate transporter | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| frlA; fructoselysine transporter | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| frvR; putative frv operon regulatory protein | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| hyfR; hydrogenase-4 transcriptional activator | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| pphB; serine/threonine protein phosphatase 2 [EC:3.1.3.16] | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| sfmD; outer membrane usher protein | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| tnaB; low affinity tryptophan permease | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| uidC, gusC; putative glucuronide porin | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| yfaL; autotransporter family porin | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| ygeU, xdhC; xanthine dehydrogenase iron-sulfur-binding subunit | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| ygfM; putative selenate reductase FAD-binding subunit | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.85325 | 103.472 | 0.39549 | 0.80459 |
| echA; ech hydrogenase subunit A | 0.00011 | 0.00008 | 0.00005 | 0.00000 | 0.00002 | 65 | 0.00016 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.81139 | 96.412 | 0.41914 | 0.81980 |
| echC; ech hydrogenase subunit C | 0.00011 | 0.00008 | 0.00005 | 0.00000 | 0.00002 | 65 | 0.00016 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.81139 | 96.412 | 0.41914 | 0.81980 |
| echE; ech hydrogenase subunit E | 0.00011 | 0.00008 | 0.00005 | 0.00000 | 0.00002 | 65 | 0.00016 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.81139 | 96.412 | 0.41914 | 0.81980 |
| E3.4.21.66; thermitase [EC:3.4.21.66] | 0.00011 | 0.00004 | 0.00010 | 0.00000 | 0.00005 | 65 | 0.00012 | 0.00000 | 0.00006 | 93 | 0.00008 | -0.23508 | 154.410 | 0.81446 | 0.95409 |
| gntP; high-affinity gluconate transporter | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.84618 | 103.492 | 0.39940 | 0.80691 |
| phcB; extracellular factor (EF) 3-hydroxypalmitic acid methyl ester biosynthesis protein | 0.00011 | 0.00009 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00018 | 0.00000 | 0.00015 | 93 | 0.00015 | -1.11154 | 92.287 | 0.26922 | 0.77376 |
| phcQ; two-component system, probable response regulator PhcQ | 0.00011 | 0.00009 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00018 | 0.00000 | 0.00015 | 93 | 0.00015 | -1.11154 | 92.287 | 0.26922 | 0.77376 |
| phcS; two-component system, sensor histidine kinase PhcS [EC:2.7.13.3] | 0.00011 | 0.00009 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00018 | 0.00000 | 0.00015 | 93 | 0.00015 | -1.11154 | 92.287 | 0.26922 | 0.77376 |
| nodD; LysR family transcriptional regulator, nod-box dependent transcriptional activator | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.77084 | 109.975 | 0.44246 | 0.83186 |
| envY; AraC family transcriptional regulator | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.83356 | 103.495 | 0.40645 | 0.81189 |
| nupX; nucleoside permease | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.83356 | 103.495 | 0.40645 | 0.81189 |
| waaV; UDP-glucose:(glucosyl)LPS beta-1,3-glucosyltransferase [EC:2.4.1.-] | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.83356 | 103.495 | 0.40645 | 0.81189 |
| yahA; c-di-GMP-specific phosphodiesterase [EC:3.1.4.52] | 0.00011 | 0.00007 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -0.83356 | 103.495 | 0.40645 | 0.81189 |
| pks5; polyketide synthase 5 | 0.00011 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00018 | 0.00000 | 0.00007 | 93 | 0.00007 | -2.47646 | 95.307 | 0.01503 | 0.59095 |
| tetD; AraC family transcriptional regulator, transposon Tn10 TetD protein | 0.00011 | 0.00004 | 0.00016 | 0.00000 | 0.00009 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00009 | 0.90873 | 87.627 | 0.36598 | 0.79587 |
| NTPCR; nucleoside-triphosphatase [EC:3.6.1.15] | 0.00011 | 0.00006 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00014 | 0.00000 | 0.00009 | 93 | 0.00011 | -0.79088 | 148.280 | 0.43028 | 0.82749 |
| mer; 5,10-methylenetetrahydromethanopterin reductase [EC:1.5.98.2] | 0.00011 | 0.00009 | 0.00023 | 0.00000 | 0.00023 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00023 | 0.90901 | 64.358 | 0.36674 | 0.79587 |
| SC5DL, ERG3; Delta7-sterol 5-desaturase [EC:1.14.19.20] | 0.00011 | 0.00006 | 0.00018 | 0.00000 | 0.00014 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00014 | 0.83909 | 75.399 | 0.40407 | 0.81062 |
| oleC5; oleandomycin transport system permease protein | 0.00010 | 0.00004 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00010 | 0.00000 | 0.00004 | 93 | 0.00008 | 0.20148 | 115.004 | 0.84068 | 0.95506 |
| embA; arabinosyltransferase A [EC:2.4.2.-] | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00017 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.44944 | 95.531 | 0.01613 | 0.59095 |
| COMT; catechol O-methyltransferase [EC:2.1.1.6] | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00017 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.39968 | 95.535 | 0.01835 | 0.59095 |
| mbtB; mycobactin phenyloxazoline synthetase | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00017 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.39968 | 95.535 | 0.01835 | 0.59095 |
| hicA; mRNA interferase HicA [EC:3.1.-.-] | 0.00010 | 0.00003 | 0.00010 | 0.00000 | 0.00004 | 65 | 0.00010 | 0.00000 | 0.00004 | 93 | 0.00005 | 0.01258 | 153.630 | 0.98998 | 0.99801 |
| hoxH; NAD-reducing hydrogenase large subunit [EC:1.12.1.2] | 0.00010 | 0.00004 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00012 | 0.00000 | 0.00006 | 93 | 0.00007 | -0.76785 | 152.500 | 0.44376 | 0.83186 |
| nasB; assimilatory nitrate reductase electron transfer subunit [EC:1.7.99.-] | 0.00010 | 0.00004 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00010 | 0.00000 | 0.00004 | 93 | 0.00007 | 0.15305 | 126.867 | 0.87860 | 0.96759 |
| hoxF; [NiFe] hydrogenase diaphorase moiety large subunit [EC:1.12.1.2] | 0.00010 | 0.00004 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00011 | 0.00000 | 0.00006 | 93 | 0.00008 | -0.18598 | 149.899 | 0.85271 | 0.95848 |
| desA2; acyl-[acyl-carrier-protein] desaturase [EC:1.14.19.2] | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.41354 | 95.660 | 0.01770 | 0.59095 |
| E2.5.1.86; trans,polycis-decaprenyl diphosphate synthase [EC:2.5.1.86] | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.41354 | 95.660 | 0.01770 | 0.59095 |
| fadD21; fatty acid CoA ligase FadD21 | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.41354 | 95.660 | 0.01770 | 0.59095 |
| hsd; 3beta-hydroxy-Delta5-steroid dehydrogenase / steroid Delta-isomerase [EC:1.1.1.145 5.3.3.1] | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.41354 | 95.660 | 0.01770 | 0.59095 |
| PTS-Sor-EIIB, sorB; PTS system, sorbose-specific IIB component [EC:2.7.1.206] | 0.00010 | 0.00004 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00013 | 0.00000 | 0.00006 | 93 | 0.00007 | -0.86597 | 140.259 | 0.38798 | 0.80262 |
| exoY; exopolysaccharide production protein ExoY | 0.00010 | 0.00005 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.71220 | 96.553 | 0.09007 | 0.76567 |
| pmmS; 2-phosphinomethylmalate synthase [EC:2.3.3.18] | 0.00010 | 0.00010 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00017 | 0.00000 | 0.00017 | 93 | 0.00017 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| aph3-III; aminoglycoside 3’-phosphotransferase III [EC:2.7.1.95] | 0.00010 | 0.00007 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00017 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.47493 | 92.142 | 0.14364 | 0.77376 |
| ydaV; putative replication protein | 0.00010 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.11335 | 93.899 | 0.26840 | 0.77376 |
| mbtA; mycobactin salicyl-AMP ligase [EC:6.3.2.-] | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.35670 | 95.660 | 0.02048 | 0.59095 |
| mbtE; mycobactin peptide synthetase MbtE | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.35670 | 95.660 | 0.02048 | 0.59095 |
| mbtF; mycobactin peptide synthetase MbtF | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.35670 | 95.660 | 0.02048 | 0.59095 |
| PLD1_2; phospholipase D1/2 [EC:3.1.4.4] | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00007 | 93 | 0.00007 | -2.05890 | 98.195 | 0.04215 | 0.65026 |
| exoO; succinoglycan biosynthesis protein ExoO [EC:2.4.-.-] | 0.00010 | 0.00005 | 0.00001 | 0.00000 | 0.00000 | 65 | 0.00016 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.82288 | 92.338 | 0.07156 | 0.73962 |
| K15016; enoyl-CoA hydratase / 3-hydroxyacyl-CoA dehydrogenase [EC:4.2.1.17 1.1.1.35] | 0.00010 | 0.00007 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00014 | 0.00000 | 0.00011 | 93 | 0.00012 | -0.72387 | 120.391 | 0.47055 | 0.83905 |
| xanB2; chorismate lyase / 3-hydroxybenzoate synthase [EC:4.1.3.40 4.1.3.45] | 0.00010 | 0.00004 | 0.00010 | 0.00000 | 0.00006 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00008 | 0.13302 | 136.422 | 0.89437 | 0.97180 |
| med; transcriptional activator of comK gene | 0.00010 | 0.00005 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00014 | 0.00000 | 0.00008 | 93 | 0.00009 | -1.27081 | 109.912 | 0.20648 | 0.77376 |
| fyuA; pesticin/yersiniabactin receptor | 0.00010 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.08772 | 93.898 | 0.27950 | 0.77376 |
| irp1, HMWP1; yersiniabactin nonribosomal peptide/polyketide synthase | 0.00010 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.08772 | 93.898 | 0.27950 | 0.77376 |
| irp3, ybtU; yersiniabactin synthetase, thiazolinyl reductase component | 0.00010 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.08772 | 93.898 | 0.27950 | 0.77376 |
| irp4, ybtT; yersiniabactin synthetase, thioesterase component | 0.00010 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.08772 | 93.898 | 0.27950 | 0.77376 |
| sfmH; fimbrial protein | 0.00010 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.08772 | 93.898 | 0.27950 | 0.77376 |
| tdcR; threonine dehydratase operon activator protein | 0.00010 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.08772 | 93.898 | 0.27950 | 0.77376 |
| ybtA; AraC family transcriptional regulator | 0.00010 | 0.00007 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00012 | 93 | 0.00012 | -1.08772 | 93.898 | 0.27950 | 0.77376 |
| plyA; polysaccharidase protein | 0.00010 | 0.00007 | 0.00004 | 0.00000 | 0.00004 | 65 | 0.00014 | 0.00000 | 0.00012 | 93 | 0.00013 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| ccsA; citryl-CoA synthetase large subunit [EC:6.2.1.18] | 0.00010 | 0.00007 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00013 | 0.00000 | 0.00011 | 93 | 0.00012 | -0.69837 | 120.393 | 0.48629 | 0.84538 |
| mptA; GTP cyclohydrolase IV [EC:3.5.4.39] | 0.00010 | 0.00007 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00013 | 0.00000 | 0.00011 | 93 | 0.00012 | -0.69837 | 120.393 | 0.48629 | 0.84538 |
| pchA; salicylate biosynthesis isochorismate synthase [EC:5.4.4.2] | 0.00010 | 0.00007 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00013 | 0.00000 | 0.00011 | 93 | 0.00012 | -0.69837 | 120.393 | 0.48629 | 0.84538 |
| NOP2; 25S rRNA (cytosine2870-C5)-methyltransferase [EC:2.1.1.310] | 0.00010 | 0.00008 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00014 | 0.00000 | 0.00013 | 93 | 0.00014 | -0.85742 | 95.919 | 0.39335 | 0.80459 |
| ADE5; phosphoribosylamine–glycine ligase / phosphoribosylformylglycinamidine cyclo-ligase [EC:6.3.4.13 6.3.3.1] | 0.00010 | 0.00008 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00016 | 0.00000 | 0.00013 | 93 | 0.00013 | -1.16452 | 92.461 | 0.24721 | 0.77376 |
| tetV; MFS transporter, DHA3 family, tetracycline resistance protein | 0.00010 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.27140 | 95.727 | 0.02536 | 0.59941 |
| LYS9; saccharopine dehydrogenase (NADP+, L-glutamate forming) [EC:1.5.1.10] | 0.00009 | 0.00008 | 0.00017 | 0.00000 | 0.00017 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00018 | 0.75691 | 70.477 | 0.45163 | 0.83438 |
| kdnB; 3-deoxy-alpha-D-manno-octulosonate 8-oxidase [EC:1.1.3.48] | 0.00009 | 0.00006 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00010 | 0.00000 | 0.00008 | 93 | 0.00012 | -0.10072 | 146.106 | 0.91991 | 0.97880 |
| fimY; fimbrial protein FimY | 0.00009 | 0.00004 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00008 | 0.74118 | 100.878 | 0.46030 | 0.83465 |
| SMARCAD1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 [EC:3.6.4.12] | 0.00009 | 0.00009 | 0.00023 | 0.00000 | 0.00023 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00023 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| uxaA1; altronate dehydratase small subunit [EC:4.2.1.7] | 0.00009 | 0.00007 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00010 | 0.00000 | 0.00010 | 93 | 0.00013 | -0.07997 | 155.289 | 0.93636 | 0.98459 |
| hoxU; [NiFe] hydrogenase diaphorase moiety small subunit [EC:1.12.1.2] | 0.00009 | 0.00004 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00009 | 0.00000 | 0.00006 | 93 | 0.00008 | -0.01495 | 148.269 | 0.98809 | 0.99801 |
| mbtC; mycobactin polyketide synthetase MbtC | 0.00009 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.21007 | 95.735 | 0.02948 | 0.61334 |
| mbtD; mycobactin polyketide synthetase MbtD | 0.00009 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.21007 | 95.735 | 0.02948 | 0.61334 |
| nat; arylamine N-acetyltransferase [EC:2.3.1.5] | 0.00009 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.21007 | 95.735 | 0.02948 | 0.61334 |
| 4CL; 4-coumarate–CoA ligase [EC:6.2.1.12] | 0.00009 | 0.00004 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00012 | 0.00000 | 0.00006 | 93 | 0.00007 | -0.86475 | 153.098 | 0.38853 | 0.80262 |
| hoxY; NAD-reducing hydrogenase small subunit [EC:1.12.1.2] | 0.00009 | 0.00004 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00011 | 0.00000 | 0.00006 | 93 | 0.00007 | -0.55822 | 153.188 | 0.57751 | 0.87480 |
| gpuA; guanidinopropionase [EC:3.5.3.17] | 0.00009 | 0.00005 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00015 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.69316 | 93.308 | 0.09376 | 0.76609 |
| exoA; succinoglycan biosynthesis protein ExoA [EC:2.4.-.-] | 0.00009 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00015 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.74647 | 92.178 | 0.08406 | 0.76567 |
| exoF; polysaccharide biosynthesis/export protein ExoF | 0.00009 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00015 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.74647 | 92.178 | 0.08406 | 0.76567 |
| exoK; endo-1,3-1,4-beta-glycanase ExoK [EC:3.2.1.-] | 0.00009 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00015 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.74647 | 92.178 | 0.08406 | 0.76567 |
| K16929; energy-coupling factor transport system substrate-specific component | 0.00009 | 0.00008 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00015 | 0.00000 | 0.00014 | 93 | 0.00014 | -1.06766 | 92.000 | 0.28847 | 0.77376 |
| iucA; N2-citryl-N6-acetyl-N6-hydroxylysine synthase [EC:6.3.2.38] | 0.00009 | 0.00004 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00006 | 0.00000 | 0.00003 | 93 | 0.00008 | 0.83325 | 93.488 | 0.40683 | 0.81189 |
| iucC; aerobactin synthase [EC:6.3.2.39] | 0.00009 | 0.00004 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00006 | 0.00000 | 0.00003 | 93 | 0.00008 | 0.83325 | 93.488 | 0.40683 | 0.81189 |
| trcR; two-component system, OmpR family, response regulator TrcR | 0.00009 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00015 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.23970 | 92.874 | 0.02750 | 0.61334 |
| phzE; 2-amino-4-deoxychorismate synthase [EC:2.6.1.86] | 0.00009 | 0.00004 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00008 | 0.05319 | 146.066 | 0.95765 | 0.98896 |
| aph3-I; aminoglycoside 3’-phosphotransferase I [EC:2.7.1.95] | 0.00009 | 0.00004 | 0.00012 | 0.00000 | 0.00006 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00008 | 0.68546 | 121.382 | 0.49436 | 0.84797 |
| exoL; succinoglycan biosynthesis protein ExoL [EC:2.-.-.-] | 0.00009 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00015 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.69473 | 92.178 | 0.09350 | 0.76599 |
| ntdA; 3-dehydro-glucose-6-phosphate—glutamate transaminase [EC:2.6.1.104] | 0.00009 | 0.00006 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00014 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.16346 | 93.903 | 0.24759 | 0.77376 |
| ntdB; kanosamine-6-phosphate phosphatase [EC:3.1.3.92] | 0.00009 | 0.00006 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00014 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.16346 | 93.903 | 0.24759 | 0.77376 |
| sinI; antagonist of SinR | 0.00009 | 0.00006 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00014 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.16346 | 93.903 | 0.24759 | 0.77376 |
| EPHX1; microsomal epoxide hydrolase [EC:3.3.2.9] | 0.00009 | 0.00005 | 0.00017 | 0.00000 | 0.00012 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00012 | 1.19986 | 66.624 | 0.23444 | 0.77376 |
| exoV; succinoglycan biosynthesis protein ExoV | 0.00009 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00015 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.65921 | 92.256 | 0.10047 | 0.77236 |
| SHPK; sedoheptulokinase [EC:2.7.1.14] | 0.00009 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00015 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.70848 | 92.273 | 0.09091 | 0.76567 |
| dhaa; 3-hydroxy-D-aspartate aldolase [EC:4.1.3.41] | 0.00009 | 0.00005 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00012 | 0.00000 | 0.00008 | 93 | 0.00009 | -0.79182 | 126.210 | 0.42995 | 0.82749 |
| porD; pyruvate ferredoxin oxidoreductase delta subunit [EC:1.2.7.1] | 0.00009 | 0.00005 | 0.00012 | 0.00000 | 0.00009 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00011 | 0.59229 | 110.457 | 0.55486 | 0.86806 |
| nifZ; nitrogen fixation protein NifZ | 0.00009 | 0.00005 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00014 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.42725 | 96.637 | 0.15673 | 0.77376 |
| gli; D-galactarolactone isomerase [EC:5.4.1.4] | 0.00009 | 0.00005 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00011 | 0.00000 | 0.00007 | 93 | 0.00008 | -0.66294 | 131.938 | 0.50853 | 0.84956 |
| K07499; putative transposase | 0.00009 | 0.00005 | 0.00013 | 0.00000 | 0.00011 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00012 | 0.62316 | 80.992 | 0.53493 | 0.86207 |
| LEU1; 3-isopropylmalate dehydratase [EC:4.2.1.33] | 0.00009 | 0.00004 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00013 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.66493 | 100.935 | 0.09903 | 0.76942 |
| dfrD, dhfr; dihydrofolate reductase (trimethoprim resistance protein) [EC:1.5.1.3] | 0.00008 | 0.00006 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00014 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.24714 | 92.149 | 0.21551 | 0.77376 |
| IDE, ide; insulysin [EC:3.4.24.56] | 0.00008 | 0.00006 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00014 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.24714 | 92.149 | 0.21551 | 0.77376 |
| ituB, mycB, bmyB; iturin family lipopeptide synthetase B | 0.00008 | 0.00006 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00014 | 0.00000 | 0.00011 | 93 | 0.00011 | -1.24714 | 92.149 | 0.21551 | 0.77376 |
| togA; oligogalacturonide transport system ATP-binding protein | 0.00008 | 0.00004 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00012 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.34505 | 112.056 | 0.18132 | 0.77376 |
| siaA, neuC1; UDP-N-acetylglucosamine 2-epimerase (hydrolysing) [EC:3.2.1.183] | 0.00008 | 0.00005 | 0.00016 | 0.00000 | 0.00012 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00012 | 1.12435 | 69.180 | 0.26475 | 0.77376 |
| URE; urease [EC:3.5.1.5] | 0.00008 | 0.00004 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00013 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.71134 | 98.033 | 0.09018 | 0.76567 |
| E2.1.3.1-1.3S; methylmalonyl-CoA carboxyltransferase 1.3S subunit [EC:2.1.3.1] | 0.00008 | 0.00004 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00010 | 0.00000 | 0.00006 | 93 | 0.00007 | -0.74443 | 149.868 | 0.45778 | 0.83438 |
| toxR; cholera toxin transcriptional activator | 0.00008 | 0.00004 | 0.00016 | 0.00000 | 0.00009 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00009 | 1.33634 | 78.997 | 0.18527 | 0.77376 |
| irtB; ATP-binding cassette, subfamily B, bacterial IrtB [EC:3.6.3.-] | 0.00008 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00013 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.99435 | 95.923 | 0.04895 | 0.67403 |
| E2.3.1.7; carnitine O-acetyltransferase [EC:2.3.1.7] | 0.00008 | 0.00004 | 0.00007 | 0.00000 | 0.00005 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00007 | -0.32925 | 155.926 | 0.74241 | 0.93050 |
| ramA; (R)-amidase [EC:3.5.1.100] | 0.00008 | 0.00004 | 0.00011 | 0.00000 | 0.00008 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00009 | 0.59019 | 96.796 | 0.55644 | 0.86813 |
| hypBA1; non-reducing end beta-L-arabinofuranosidase [EC:3.2.1.185] | 0.00008 | 0.00005 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00008 | 0.00000 | 0.00006 | 93 | 0.00010 | 0.09907 | 112.620 | 0.92126 | 0.97905 |
| mcl2; (3S)-malyl-CoA thioesterase [EC:3.1.2.30] | 0.00008 | 0.00006 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00014 | 0.00000 | 0.00010 | 93 | 0.00010 | -1.43172 | 92.000 | 0.15561 | 0.77376 |
| K14728; phthiodiolone/phenolphthiodiolone dimycocerosates ketoreductase [EC:1.2.-.-] | 0.00008 | 0.00003 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00012 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.80205 | 113.017 | 0.07420 | 0.74128 |
| cpnA; cyclopentanol dehydrogenase [EC:1.1.1.163] | 0.00008 | 0.00004 | 0.00013 | 0.00000 | 0.00008 | 65 | 0.00005 | 0.00000 | 0.00002 | 93 | 0.00009 | 0.95944 | 75.001 | 0.34042 | 0.78475 |
| nifK; nitrogenase molybdenum-iron protein beta chain [EC:1.18.6.1] | 0.00008 | 0.00003 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00012 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.71822 | 103.121 | 0.08876 | 0.76567 |
| chlH, bchH; magnesium chelatase subunit H [EC:6.6.1.1] | 0.00008 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00013 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.81420 | 96.254 | 0.07276 | 0.74128 |
| iatA; inositol transport system ATP-binding protein | 0.00008 | 0.00003 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00008 | 1.04103 | 80.568 | 0.30097 | 0.77376 |
| iatP; inositol transport system permease protein | 0.00008 | 0.00003 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00008 | 1.04103 | 80.568 | 0.30097 | 0.77376 |
| K17208, ibpA; inositol transport system substrate-binding protein | 0.00008 | 0.00003 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00008 | 1.04103 | 80.568 | 0.30097 | 0.77376 |
| xsc; sulfoacetaldehyde acetyltransferase [EC:2.3.3.15] | 0.00008 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00013 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.58124 | 92.194 | 0.11725 | 0.77376 |
| xynD; arabinoxylan arabinofuranohydrolase [EC:3.2.1.55] | 0.00008 | 0.00005 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00013 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.33301 | 94.725 | 0.18573 | 0.77376 |
| PTS-Sor-EIIC, sorA; PTS system, sorbose-specific IIC component | 0.00008 | 0.00003 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00006 | -0.42164 | 154.254 | 0.67388 | 0.91184 |
| dgaE; D-glucosaminate-6-phosphate ammonia-lyase [EC:4.3.1.29] | 0.00008 | 0.00005 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00013 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.33808 | 93.477 | 0.18412 | 0.77376 |
| dgaF; 2-dehydro-3-deoxy-phosphogluconate aldolase [EC:4.1.2.14] | 0.00008 | 0.00005 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00013 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.33808 | 93.477 | 0.18412 | 0.77376 |
| cmtD, dhbB; HCOMODA/2-hydroxy-3-carboxy-muconic semialdehyde decarboxylase [EC:4.1.1.-] | 0.00008 | 0.00003 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00007 | 0.08417 | 119.281 | 0.93307 | 0.98428 |
| K16191, arfA; peptidoglycan-binding protein ArfA | 0.00008 | 0.00004 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00011 | 0.00000 | 0.00006 | 93 | 0.00007 | -1.08442 | 142.666 | 0.28001 | 0.77376 |
| E3.5.1.56; N,N-dimethylformamidase [EC:3.5.1.56] | 0.00008 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00013 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.02769 | 92.343 | 0.04547 | 0.65981 |
| tetX; tetracycline 11a-monooxygenase, tetracycline resistance protein [EC:1.14.13.231] | 0.00008 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00012 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.60769 | 98.204 | 0.11111 | 0.77376 |
| PTS-Sor-EIID, sorM; PTS system, sorbose-specific IID component | 0.00008 | 0.00003 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00006 | -0.35132 | 154.319 | 0.72583 | 0.92177 |
| flbB; flagellar protein FlbB | 0.00008 | 0.00004 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00009 | 0.00000 | 0.00007 | 93 | 0.00007 | -0.63088 | 117.829 | 0.52934 | 0.86094 |
| kgp; gingipain K [EC:3.4.22.47] | 0.00007 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00013 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.80148 | 92.000 | 0.07490 | 0.74200 |
| SMUG1; single-strand selective monofunctional uracil DNA glycosylase [EC:3.2.2.-] | 0.00007 | 0.00004 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00009 | 0.00000 | 0.00006 | 93 | 0.00007 | -0.66966 | 151.617 | 0.50409 | 0.84956 |
| flK; fluoroacetyl-CoA thioesterase [EC:3.1.2.29] | 0.00007 | 0.00004 | 0.00015 | 0.00000 | 0.00010 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00010 | 1.26501 | 69.554 | 0.21009 | 0.77376 |
| E3.1.4.12; sphingomyelin phosphodiesterase [EC:3.1.4.12] | 0.00007 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00012 | 0.00000 | 0.00006 | 93 | 0.00007 | -1.62807 | 96.007 | 0.10679 | 0.77376 |
| POMT, pmt; dolichyl-phosphate-mannose-protein mannosyltransferase [EC:2.4.1.109] | 0.00007 | 0.00002 | 0.00007 | 0.00000 | 0.00003 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.06724 | 154.241 | 0.94648 | 0.98674 |
| frhB; coenzyme F420 hydrogenase subunit beta [EC:1.12.98.1] | 0.00007 | 0.00003 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00011 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.68911 | 111.840 | 0.09398 | 0.76609 |
| ACADSB; short/branched chain acyl-CoA dehydrogenase [EC:1.3.99.12] | 0.00007 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00012 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.57887 | 93.273 | 0.11775 | 0.77376 |
| blaACT_MIR; beta-lactamase class C ACT/MIR [EC:3.5.2.6] | 0.00007 | 0.00003 | 0.00013 | 0.00000 | 0.00007 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00007 | 1.23989 | 73.737 | 0.21895 | 0.77376 |
| prmB; propane monooxygenase reductase component [EC:1.18.1.-] | 0.00007 | 0.00003 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00007 | 0.83270 | 83.314 | 0.40739 | 0.81200 |
| prmC; propane 2-monooxygenase small subunit [EC:1.14.13.227] | 0.00007 | 0.00003 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00007 | 0.83270 | 83.314 | 0.40739 | 0.81200 |
| kdnA; 8-amino-3,8-dideoxy-alpha-D-manno-octulosonate transaminase [EC:2.6.1.109] | 0.00007 | 0.00004 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00010 | 0.27770 | 96.625 | 0.78183 | 0.94310 |
| trcS; two-component system, OmpR family, sensor histidine kinase TrcS [EC:2.7.13.3] | 0.00007 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00012 | 0.00000 | 0.00006 | 93 | 0.00006 | -2.04025 | 92.000 | 0.04419 | 0.65981 |
| E4.1.1.64; 2,2-dialkylglycine decarboxylase (pyruvate) [EC:4.1.1.64] | 0.00007 | 0.00003 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00008 | 0.00000 | 0.00005 | 93 | 0.00007 | -0.25867 | 151.695 | 0.79624 | 0.94791 |
| gppmt; geranyl diphosphate 2-C-methyltransferase [EC:2.1.1.255] | 0.00007 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00011 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.31963 | 96.801 | 0.19007 | 0.77376 |
| E1.2.1.5; aldehyde dehydrogenase (NAD(P)+) [EC:1.2.1.5] | 0.00007 | 0.00003 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.06936 | 125.025 | 0.28697 | 0.77376 |
| gumK; 2-beta-glucuronyltransferase [EC:2.4.1.264] | 0.00007 | 0.00003 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00007 | 0.50109 | 106.850 | 0.61734 | 0.88903 |
| K16905; fluoroquinolone transport system permease protein | 0.00007 | 0.00002 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00007 | 0.00000 | 0.00003 | 93 | 0.00005 | 0.12116 | 141.182 | 0.90373 | 0.97641 |
| K16906; fluoroquinolone transport system permease protein | 0.00007 | 0.00002 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00007 | 0.00000 | 0.00003 | 93 | 0.00005 | 0.12116 | 141.182 | 0.90373 | 0.97641 |
| K16907; fluoroquinolone transport system ATP-binding protein [EC:3.6.3.-] | 0.00007 | 0.00002 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00007 | 0.00000 | 0.00003 | 93 | 0.00005 | 0.12116 | 141.182 | 0.90373 | 0.97641 |
| prmA; propane 2-monooxygenase large subunit [EC:1.14.13.227] | 0.00007 | 0.00003 | 0.00011 | 0.00000 | 0.00006 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00007 | 0.95078 | 78.750 | 0.34462 | 0.78711 |
| nifN; nitrogenase molybdenum-iron protein NifN | 0.00007 | 0.00003 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00010 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.51325 | 102.341 | 0.13330 | 0.77376 |
| fexA_B; MFS transporter, DHA2 family, florfenicol/chloramphenicol resistance protein | 0.00007 | 0.00004 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00009 | 0.00000 | 0.00007 | 93 | 0.00007 | -0.87063 | 118.393 | 0.38572 | 0.80258 |
| rhlB; rhamnosyltransferase subunit B [EC:2.4.1.-] | 0.00007 | 0.00004 | 0.00013 | 0.00000 | 0.00010 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00010 | 1.09146 | 69.723 | 0.27883 | 0.77376 |
| lasB; pseudolysin [EC:3.4.24.26] | 0.00007 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00011 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.87428 | 92.000 | 0.06406 | 0.73334 |
| fae; 5,6,7,8-tetrahydromethanopterin hydro-lyase [EC:4.2.1.147] | 0.00007 | 0.00003 | 0.00010 | 0.00000 | 0.00005 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00006 | 0.90131 | 94.164 | 0.36973 | 0.79587 |
| nifQ; nitrogen fixation protein NifQ | 0.00007 | 0.00004 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00010 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.34340 | 100.254 | 0.18217 | 0.77376 |
| mauB; methylamine dehydrogenase heavy chain [EC:1.4.9.1] | 0.00007 | 0.00003 | 0.00006 | 0.00000 | 0.00003 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.03189 | 154.351 | 0.97460 | 0.99546 |
| andAd; anthranilate 1,2-dioxygenase small subunit [EC:1.14.12.1] | 0.00007 | 0.00004 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00009 | 0.00000 | 0.00007 | 93 | 0.00007 | -0.69747 | 122.734 | 0.48683 | 0.84553 |
| CDIPT; CDP-diacylglycerol–inositol 3-phosphatidyltransferase [EC:2.7.8.11] | 0.00007 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00010 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.61841 | 98.195 | 0.10878 | 0.77376 |
| K07065; uncharacterized protein | 0.00007 | 0.00004 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00009 | 0.00000 | 0.00007 | 93 | 0.00007 | -0.79247 | 124.975 | 0.42959 | 0.82749 |
| fliR-flhB; flagellar biosynthetic protein FliR/FlhB | 0.00007 | 0.00005 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00010 | 0.00000 | 0.00009 | 93 | 0.00009 | -0.90329 | 96.470 | 0.36862 | 0.79587 |
| tlh; thermolabile hemolysin | 0.00006 | 0.00005 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00008 | 0.00000 | 0.00008 | 93 | 0.00009 | -0.38274 | 128.727 | 0.70255 | 0.92004 |
| ATX, chlB1, mdpB; 6-methylsalicylic acid synthase [EC:2.3.1.165] | 0.00006 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00011 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.88854 | 92.000 | 0.06210 | 0.73004 |
| K09116; uncharacterized protein | 0.00006 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00011 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.86249 | 92.000 | 0.06573 | 0.73334 |
| IDO, INDO; indoleamine 2,3-dioxygenase [EC:1.13.11.52] | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00005 | 0.00000 | 0.00002 | 93 | 0.00008 | 0.48840 | 76.027 | 0.62667 | 0.89255 |
| exoW; succinoglycan biosynthesis protein ExoW [EC:2.4.-.-] | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00011 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.39710 | 92.359 | 0.16573 | 0.77376 |
| K07123; uncharacterized protein | 0.00006 | 0.00003 | 0.00013 | 0.00000 | 0.00006 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00006 | 1.76734 | 70.653 | 0.08149 | 0.76350 |
| exoU; succinoglycan biosynthesis protein ExoU [EC:2.4.-.-] | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00011 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.41794 | 92.250 | 0.15958 | 0.77376 |
| dndE; DNA sulfur modification protein DndE | 0.00006 | 0.00003 | 0.00007 | 0.00000 | 0.00005 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00007 | 0.20795 | 127.458 | 0.83560 | 0.95506 |
| gesA, mexP; membrane fusion protein, gold/copper resistance efflux system | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00009 | 0.61531 | 82.976 | 0.54003 | 0.86207 |
| gesB, mexQ; gold/copper resistance efflux pump | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00009 | 0.61531 | 82.976 | 0.54003 | 0.86207 |
| luxO; two-component system, repressor protein LuxO | 0.00006 | 0.00003 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00006 | 0.00000 | 0.00005 | 93 | 0.00006 | -0.03025 | 155.393 | 0.97590 | 0.99571 |
| pksL; polyketide synthase PksL | 0.00006 | 0.00003 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00007 | 0.76467 | 118.282 | 0.44599 | 0.83357 |
| K09942; uncharacterized protein | 0.00006 | 0.00003 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00008 | 0.00000 | 0.00005 | 93 | 0.00006 | -0.86333 | 146.498 | 0.38937 | 0.80347 |
| nifT; nitrogen fixation protein NifT | 0.00006 | 0.00003 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00006 | -1.34537 | 102.550 | 0.18147 | 0.77376 |
| nifW; nitrogenase-stabilizing/protective protein | 0.00006 | 0.00003 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00006 | -1.34537 | 102.550 | 0.18147 | 0.77376 |
| nifX; nitrogen fixation protein NifX | 0.00006 | 0.00003 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00009 | 0.00000 | 0.00005 | 93 | 0.00006 | -1.34537 | 102.550 | 0.18147 | 0.77376 |
| K07453; putative restriction endonuclease | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00008 | 0.56966 | 75.305 | 0.57060 | 0.87146 |
| vanT; serine/alanine racemase [EC:5.1.1.18 5.1.1.1] | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00010 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.54595 | 92.000 | 0.12555 | 0.77376 |
| HOGA1; 4-hydroxy-2-oxoglutarate aldolase [EC:4.1.3.16] | 0.00006 | 0.00004 | 0.00011 | 0.00000 | 0.00009 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00009 | 0.97766 | 67.534 | 0.33173 | 0.77825 |
| E2.1.1.113; site-specific DNA-methyltransferase (cytosine-N4-specific) [EC:2.1.1.113] | 0.00006 | 0.00005 | 0.00013 | 0.00000 | 0.00012 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00012 | 0.96117 | 64.195 | 0.34007 | 0.78467 |
| kaiB; circadian clock protein KaiB | 0.00006 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00010 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.25215 | 92.000 | 0.21369 | 0.77376 |
| araN; arabinosaccharide transport system substrate-binding protein | 0.00006 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00009 | 0.00000 | 0.00004 | 93 | 0.00005 | -1.84937 | 98.676 | 0.06740 | 0.73334 |
| araP; arabinosaccharide transport system permease protein | 0.00006 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00009 | 0.00000 | 0.00004 | 93 | 0.00005 | -1.84937 | 98.676 | 0.06740 | 0.73334 |
| etbAa; ethylbenzene dioxygenase subunit alpha [EC:1.14.12.-] | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00008 | 0.61995 | 75.024 | 0.53717 | 0.86207 |
| etbAb; ethylbenzene dioxygenase subunit beta [EC:1.14.12.-] | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00008 | 0.61995 | 75.024 | 0.53717 | 0.86207 |
| etbD; 2-hydroxy-6-oxo-octa-2,4-dienoate hydrolase [EC:3.7.1.-] | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00008 | 0.61995 | 75.024 | 0.53717 | 0.86207 |
| phdE; cis-3,4-dihydrophenanthrene-3,4-diol dehydrogenase [EC:1.3.1.49] | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00008 | 0.61995 | 75.024 | 0.53717 | 0.86207 |
| PRMT9; type II protein arginine methyltransferase [EC:2.1.1.320] | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00008 | 0.61995 | 75.024 | 0.53717 | 0.86207 |
| rfbV; abequosyltransferase [EC:2.4.1.60] | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00008 | 0.61995 | 75.024 | 0.53717 | 0.86207 |
| oleC4; oleandomycin transport system ATP-binding protein | 0.00006 | 0.00003 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00006 | 0.00000 | 0.00003 | 93 | 0.00006 | 0.06267 | 126.177 | 0.95013 | 0.98774 |
| ATPeV1G, ATP6G; V-type H+-transporting ATPase subunit G | 0.00006 | 0.00004 | 0.00004 | 0.00000 | 0.00004 | 65 | 0.00007 | 0.00000 | 0.00007 | 93 | 0.00008 | -0.29791 | 147.757 | 0.76619 | 0.93900 |
| ctpD; cobalt/nickel-transporting P-type ATPase D [EC:3.6.3.-] | 0.00006 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00010 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.69697 | 92.000 | 0.09308 | 0.76567 |
| lfrR; TetR/AcrR family transcriptional regulator, repressor for lfrA | 0.00006 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00010 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.69697 | 92.000 | 0.09308 | 0.76567 |
| PTS-Sor-EIIA, sorF; PTS system, sorbose-specific IIA component [EC:2.7.1.206] | 0.00006 | 0.00003 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00005 | -0.76798 | 126.201 | 0.44393 | 0.83186 |
| pfkC; ADP-dependent phosphofructokinase/glucokinase [EC:2.7.1.146 2.7.1.147] | 0.00006 | 0.00002 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00005 | 0.52042 | 132.993 | 0.60364 | 0.88434 |
| ligM; vanillate/3-O-methylgallate O-demethylase [EC:2.1.1.341] | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00009 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.21247 | 92.118 | 0.22843 | 0.77376 |
| traR; LuxR family transcriptional regulator, activator of conjugal transfer of Ti plasmids | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00009 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.25897 | 92.254 | 0.21122 | 0.77376 |
| K16327; putative LysE/RhtB family amino acid efflux pump | 0.00006 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00009 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.31814 | 92.000 | 0.19073 | 0.77376 |
| PTGS2, COX2; prostaglandin-endoperoxide synthase 2 [EC:1.14.99.1] | 0.00006 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00012 | 0.86607 | 65.350 | 0.38962 | 0.80353 |
| hepA; heparin lyase [EC:4.2.2.7] | 0.00006 | 0.00003 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.78325 | 127.624 | 0.43493 | 0.82874 |
| opmE; outer membrane protein, multidrug efflux system | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00009 | 0.72178 | 82.132 | 0.47248 | 0.84050 |
| waaS, rfaS; 1,5-rhamnosyltransferase [EC:2.4.1.-] | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00008 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00009 | 0.72178 | 82.132 | 0.47248 | 0.84050 |
| E2.4.1.14; sucrose-phosphate synthase [EC:2.4.1.14] | 0.00006 | 0.00003 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00006 | 0.84528 | 87.635 | 0.40026 | 0.80762 |
| liaG; lia operon protein LiaG | 0.00006 | 0.00003 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.78474 | 146.841 | 0.43387 | 0.82874 |
| tfrA; fumarate reductase (CoM/CoB) subunit A [EC:1.3.4.1] | 0.00006 | 0.00004 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00009 | 0.58706 | 81.978 | 0.55878 | 0.86813 |
| migA; alpha-1,6-rhamnosyltransferase [EC:2.4.1.-] | 0.00005 | 0.00003 | 0.00013 | 0.00000 | 0.00008 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00008 | 1.54077 | 64.149 | 0.12829 | 0.77376 |
| exoH; succinoglycan biosynthesis protein ExoH | 0.00005 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00009 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.29701 | 92.000 | 0.19787 | 0.77376 |
| pgtA; two-component system, NtrC family, phosphoglycerate transport system response regulator PgtA | 0.00005 | 0.00003 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00005 | 0.69310 | 128.133 | 0.48950 | 0.84797 |
| pgtB; two-component system, NtrC family, phosphoglycerate transport system sensor histidine kinase PgtB [EC:2.7.13.3] | 0.00005 | 0.00003 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00005 | 0.69310 | 128.133 | 0.48950 | 0.84797 |
| atzB; hydroxydechloroatrazine ethylaminohydrolase [EC:3.5.4.43] | 0.00005 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00008 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.30538 | 102.974 | 0.19467 | 0.77376 |
| waaB, rfaB; UDP-D-galactose:(glucosyl)LPS alpha-1,6-D-galactosyltransferase [EC:2.4.1.-] | 0.00005 | 0.00003 | 0.00007 | 0.00000 | 0.00004 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00005 | 0.42248 | 149.242 | 0.67328 | 0.91184 |
| tuaH; teichuronic acid biosynthesis glycosyltransferase TuaH [EC:2.4.-.-] | 0.00005 | 0.00005 | 0.00012 | 0.00000 | 0.00011 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00011 | 1.02839 | 64.244 | 0.30762 | 0.77376 |
| pksD; bacillaene synthase trans-acting acyltransferase | 0.00005 | 0.00005 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00008 | 0.00000 | 0.00008 | 93 | 0.00008 | -0.79000 | 97.982 | 0.43143 | 0.82801 |
| exoT; succinoglycan exporter | 0.00005 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00009 | 0.00000 | 0.00007 | 93 | 0.00007 | -1.23576 | 92.000 | 0.21969 | 0.77376 |
| hemQ; Fe-coproporphyrin III decarboxylase [EC:1.11.1.-] | 0.00005 | 0.00003 | 0.00010 | 0.00000 | 0.00007 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00008 | 1.09948 | 68.242 | 0.27542 | 0.77376 |
| tisB; small toxic protein TisB | 0.00005 | 0.00002 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00006 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.21793 | 155.795 | 0.82777 | 0.95506 |
| hepB; heparin/heparan-sulfate lyase [EC:4.2.2.7 4.2.2.8] | 0.00005 | 0.00003 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.49381 | 145.940 | 0.62218 | 0.89150 |
| nox1; NADH oxidase (H2O2-forming) [EC:1.6.3.3] | 0.00005 | 0.00002 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00005 | 0.90741 | 80.825 | 0.36689 | 0.79587 |
| ftr; formylmethanofuran–tetrahydromethanopterin N-formyltransferase [EC:2.3.1.101] | 0.00005 | 0.00002 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 1.21173 | 75.942 | 0.22937 | 0.77376 |
| fwdA, fmdA; formylmethanofuran dehydrogenase subunit A [EC:1.2.7.12] | 0.00005 | 0.00002 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 1.21173 | 75.942 | 0.22937 | 0.77376 |
| fwdB, fmdB; formylmethanofuran dehydrogenase subunit B [EC:1.2.7.12] | 0.00005 | 0.00002 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 1.21173 | 75.942 | 0.22937 | 0.77376 |
| fwdC, fmdC; formylmethanofuran dehydrogenase subunit C [EC:1.2.7.12] | 0.00005 | 0.00002 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 1.21173 | 75.942 | 0.22937 | 0.77376 |
| K09154; uncharacterized protein | 0.00005 | 0.00002 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 1.21173 | 75.942 | 0.22937 | 0.77376 |
| mch; methenyltetrahydromethanopterin cyclohydrolase [EC:3.5.4.27] | 0.00005 | 0.00002 | 0.00009 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 1.21173 | 75.942 | 0.22937 | 0.77376 |
| chlB; light-independent protochlorophyllide reductase subunit B [EC:1.3.7.7] | 0.00005 | 0.00003 | 0.00001 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.70263 | 94.218 | 0.09194 | 0.76567 |
| chlN; light-independent protochlorophyllide reductase subunit N [EC:1.3.7.7] | 0.00005 | 0.00003 | 0.00001 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.70263 | 94.218 | 0.09194 | 0.76567 |
| gck, gckA, GLYCTK; glycerate 2-kinase [EC:2.7.1.165] | 0.00005 | 0.00004 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00009 | 0.70573 | 71.457 | 0.48265 | 0.84354 |
| whiEII; putative monooxygenase | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00011 | 0.95533 | 64.245 | 0.34299 | 0.78567 |
| whiEIII; minimal PKS ketosynthase (KS/KS alpha) [EC:2.3.1.-] | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00011 | 0.95533 | 64.245 | 0.34299 | 0.78567 |
| whiEIV; minimal PKS chain-length factor (CLF/KS beta) [EC:2.3.1.-] | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00011 | 0.95533 | 64.245 | 0.34299 | 0.78567 |
| whiEVI; aromatase | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00011 | 0.95533 | 64.245 | 0.34299 | 0.78567 |
| whiEVII; cyclase | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00011 | 0.95533 | 64.245 | 0.34299 | 0.78567 |
| cmr4; CRISPR-associated protein Cmr4 | 0.00005 | 0.00003 | 0.00004 | 0.00000 | 0.00004 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.17355 | 145.388 | 0.86246 | 0.96263 |
| cmr5; CRISPR-associated protein Cmr5 | 0.00005 | 0.00003 | 0.00004 | 0.00000 | 0.00004 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.17355 | 145.388 | 0.86246 | 0.96263 |
| yraG; similar to spore coat protein | 0.00005 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00008 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.66254 | 99.835 | 0.09954 | 0.76942 |
| tfdB; 2,4-dichlorophenol 6-monooxygenase [EC:1.14.13.20] | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00011 | 0.95987 | 64.199 | 0.34072 | 0.78492 |
| impD; type VI secretion system protein ImpD | 0.00005 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.13812 | 103.431 | 0.25770 | 0.77376 |
| creS; crescentin | 0.00005 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00007 | 0.00000 | 0.00006 | 93 | 0.00006 | -0.90595 | 102.232 | 0.36709 | 0.79587 |
| EPT1; ethanolaminephosphotransferase [EC:2.7.8.1] | 0.00005 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.16452 | 92.461 | 0.24721 | 0.77376 |
| pyrBI; aspartate carbamoyltransferase [EC:2.1.3.2] | 0.00005 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.16452 | 92.461 | 0.24721 | 0.77376 |
| PPIL1; peptidyl-prolyl cis-trans isomerase-like 1 [EC:5.2.1.8] | 0.00005 | 0.00004 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00009 | 0.75691 | 70.477 | 0.45163 | 0.83438 |
| cpt; carboxypeptidase T [EC:3.4.17.18] | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00011 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| mlhB, chnC; epsilon-lactone hydrolase [EC:3.1.1.83] | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00011 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| rdmC; aclacinomycin methylesterase [EC:3.1.1.95] | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00011 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| whiEV; minimal PKS acyl carrier protein | 0.00005 | 0.00005 | 0.00011 | 0.00000 | 0.00011 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00011 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| pchR; AraC family transcriptional regulator, transcriptional activator of the genes for pyochelin and ferripyochelin receptors | 0.00005 | 0.00002 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.30916 | 155.975 | 0.75761 | 0.93719 |
| pucC; MFS transporter, BCD family, chlorophyll transporter | 0.00005 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.44734 | 92.000 | 0.15120 | 0.77376 |
| bxlE; xylobiose transport system substrate-binding protein | 0.00005 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.44103 | 92.000 | 0.15297 | 0.77376 |
| bxlF; xylobiose transport system permease protein | 0.00005 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.44103 | 92.000 | 0.15297 | 0.77376 |
| bxlG; xylobiose transport system permease protein | 0.00005 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.44103 | 92.000 | 0.15297 | 0.77376 |
| xynC; glucuronoarabinoxylan endo-1,4-beta-xylanase [EC:3.2.1.136] | 0.00005 | 0.00002 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.15179 | 155.013 | 0.87955 | 0.96779 |
| aphB; LysR family transcriptional regulator, transcriptional activator AphB | 0.00005 | 0.00003 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00005 | 0.30742 | 151.803 | 0.75894 | 0.93719 |
| E3.4.21.100; pseudomonalisin [EC:3.4.21.100] | 0.00005 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| E3.4.21.101; xanthomonalisin [EC:3.4.21.101] | 0.00005 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00008 | 93 | 0.00008 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hap, nprV; vibriolysin [EC:3.4.24.25] | 0.00005 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.47377 | 93.999 | 0.14389 | 0.77376 |
| mtdB; methylene-tetrahydromethanopterin dehydrogenase [EC:1.5.1.-] | 0.00005 | 0.00003 | 0.00008 | 0.00000 | 0.00006 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 1.01605 | 74.916 | 0.31288 | 0.77376 |
| codA; choline oxidase [EC:1.1.3.17] | 0.00005 | 0.00003 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00006 | 0.91809 | 81.203 | 0.36129 | 0.79496 |
| PTS-Dga-EIIB, dgaB; PTS system, D-glucosaminate-specific IIB component [EC:2.7.1.203] | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00008 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.73747 | 92.014 | 0.08565 | 0.76567 |
| hypBA2; beta-L-arabinobiosidase [EC:3.2.1.187] | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.35421 | 92.000 | 0.17899 | 0.77376 |
| bglK; beta-glucoside kinase [EC:2.7.1.85] | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.22339 | 150.536 | 0.82353 | 0.95506 |
| actIV; cyclase [EC:4.-.-.-] | 0.00004 | 0.00002 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00005 | 0.52508 | 88.701 | 0.60084 | 0.88272 |
| gbd; 4-hydroxybutyrate dehydrogenase [EC:1.1.1.61] | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.50538 | 92.000 | 0.13565 | 0.77376 |
| E3.4.11.24; aminopeptidase S [EC:3.4.11.24] | 0.00004 | 0.00002 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00005 | 0.56884 | 87.785 | 0.57092 | 0.87172 |
| desB, galA; gallate dioxygenase [EC:1.13.11.57] | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.30529 | 148.806 | 0.76057 | 0.93791 |
| galR; LysR family transcriptional regulator, regulator for genes of the gallate degradation pathway | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.30529 | 148.806 | 0.76057 | 0.93791 |
| IDUA; L-iduronidase [EC:3.2.1.76] | 0.00004 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00005 | -1.06370 | 107.040 | 0.28986 | 0.77376 |
| hupR, hoxA; two-component system, NtrC family, response regulator HupR/HoxA | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.62754 | 92.000 | 0.10704 | 0.77376 |
| hupT, hoxJ; two-component system, NtrC family, sensor histidine kinase HupT/HoxJ [EC:2.7.13.3] | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.62754 | 92.000 | 0.10704 | 0.77376 |
| sct; succinyl-CoA—D-citramalate CoA-transferase [EC:2.8.3.20] | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.38785 | 93.759 | 0.16847 | 0.77376 |
| mxaG; cytochrome c-L | 0.00004 | 0.00003 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00006 | 1.23776 | 67.480 | 0.22010 | 0.77376 |
| aac6-Ib; aminoglycoside 6’-N-acetyltransferase Ib [EC:2.3.1.82] | 0.00004 | 0.00002 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00004 | 0.21465 | 155.915 | 0.83032 | 0.95506 |
| sprC; streptogrisin C [EC:3.4.21.-] | 0.00004 | 0.00002 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00005 | 0.62747 | 87.390 | 0.53199 | 0.86182 |
| mxaA; mxaA protein | 0.00004 | 0.00003 | 0.00008 | 0.00000 | 0.00007 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00007 | 1.09115 | 66.825 | 0.27912 | 0.77376 |
| mxaL; mxaL protein | 0.00004 | 0.00003 | 0.00008 | 0.00000 | 0.00007 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00007 | 1.09115 | 66.825 | 0.27912 | 0.77376 |
| andAb; anthranilate 1,2-dioxygenase ferredoxin component | 0.00004 | 0.00004 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00006 | -0.86809 | 94.871 | 0.38754 | 0.80262 |
| cmr2, cas10; CRISPR-associated protein Cmr2 | 0.00004 | 0.00003 | 0.00004 | 0.00000 | 0.00004 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00005 | 0.12007 | 143.519 | 0.90460 | 0.97658 |
| narB; ferredoxin-nitrate reductase [EC:1.7.7.2] | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.52558 | 92.000 | 0.13055 | 0.77376 |
| K00183; prokaryotic molybdopterin-containing oxidoreductase family, molybdopterin binding subunit | 0.00004 | 0.00001 | 0.00005 | 0.00000 | 0.00002 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00003 | 0.44601 | 146.329 | 0.65625 | 0.90679 |
| K06931; uncharacterized protein | 0.00004 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00007 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.67991 | 92.000 | 0.09637 | 0.76667 |
| crtM; 4,4’-diapophytoene synthase [EC:2.5.1.96] | 0.00004 | 0.00002 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.88649 | 145.137 | 0.37682 | 0.79810 |
| graR; two-component system, OmpR family, response regulator protein GraR | 0.00004 | 0.00002 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.88649 | 145.137 | 0.37682 | 0.79810 |
| vraF; cationic antimicrobial peptide transport system ATP-binding protein | 0.00004 | 0.00002 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.88649 | 145.137 | 0.37682 | 0.79810 |
| cotSA; spore coat protein SA | 0.00004 | 0.00002 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00004 | 0.61231 | 133.102 | 0.54138 | 0.86207 |
| fadD28; long-chain fatty acid adenylyltransferase FadD28 [EC:6.2.1.49] | 0.00004 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00006 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.67243 | 111.655 | 0.09724 | 0.76673 |
| prmD; propane monooxygenase coupling protein | 0.00004 | 0.00002 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00005 | 0.71777 | 77.661 | 0.47505 | 0.84095 |
| cotY; spore coat protein Y | 0.00004 | 0.00003 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.63735 | 120.146 | 0.52511 | 0.85728 |
| glnT; putative sodium/glutamine symporter | 0.00004 | 0.00003 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.63735 | 120.146 | 0.52511 | 0.85728 |
| rapG; response regulator aspartate phosphatase G [EC:3.1.-.-] | 0.00004 | 0.00003 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.63735 | 120.146 | 0.52511 | 0.85728 |
| E3.1.30.1; nuclease S1 [EC:3.1.30.1] | 0.00004 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.27008 | 95.926 | 0.20713 | 0.77376 |
| lhpI; 1-piperideine-2-carboxylate/1-pyrroline-2-carboxylate reductase [NAD(P)H] [EC:1.5.1.1] | 0.00004 | 0.00003 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.58236 | 132.362 | 0.56131 | 0.86813 |
| chiS; two-component system, sensor histidine kinase ChiS | 0.00004 | 0.00002 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00005 | 1.17342 | 71.025 | 0.24455 | 0.77376 |
| E1.3.3.5; bilirubin oxidase [EC:1.3.3.5] | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.31650 | 92.638 | 0.19126 | 0.77376 |
| fccA; cytochrome subunit of sulfide dehydrogenase | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| fccB; sulfide dehydrogenase [flavocytochrome c] flavoprotein chain [EC:1.8.2.3] | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| nalC; TetR/AcrR family transcriptional regulator, transcriptional repressor NalC | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpaA; two-component system, OmpR family, response regulator RpaA | 0.00004 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| blaSHV; beta-lactamase class A SHV [EC:3.5.2.6] | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00004 | 0.02042 | 151.394 | 0.98373 | 0.99706 |
| sul1; dihydropteroate synthase type 1 [EC:2.5.1.15] | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00004 | 0.02042 | 151.394 | 0.98373 | 0.99706 |
| E4.1.3.46, ccl; (R)-citramalyl-CoA lyase [EC:4.1.3.46] | 0.00004 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00005 | 0.42326 | 101.144 | 0.67300 | 0.91184 |
| pla; plasminogen activator [EC:3.4.23.48] | 0.00004 | 0.00002 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00004 | 1.58986 | 67.537 | 0.11654 | 0.77376 |
| RSAD2; radical S-adenosyl methionine domain-containing protein 2 | 0.00004 | 0.00003 | 0.00009 | 0.00000 | 0.00007 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00007 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| suyA; (2R)-sulfolactate sulfo-lyase subunit alpha [EC:4.4.1.24] | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00003 | 0.02497 | 130.774 | 0.98012 | 0.99657 |
| suyB; (2R)-sulfolactate sulfo-lyase subunit beta [EC:4.4.1.24] | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00003 | 0.02497 | 130.774 | 0.98012 | 0.99657 |
| mdlB; (S)-mandelate dehydrogenase [EC:1.1.99.31] | 0.00004 | 0.00002 | 0.00008 | 0.00000 | 0.00004 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.58912 | 65.472 | 0.11685 | 0.77376 |
| nodA; nodulation protein A [EC:2.3.1.-] | 0.00004 | 0.00002 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.81289 | 110.250 | 0.41803 | 0.81941 |
| nodC; N-acetylglucosaminyltransferase [EC:2.4.1.-] | 0.00004 | 0.00002 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.81289 | 110.250 | 0.41803 | 0.81941 |
| PTS-Glc-EIIB, ptsG; PTS system, glucose-specific IIB component [EC:2.7.1.199] | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.05991 | 149.460 | 0.95230 | 0.98792 |
| K15527; cysteate synthase [EC:2.5.1.76] | 0.00004 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.32595 | 92.000 | 0.18814 | 0.77376 |
| DHRS13; dehydrogenase/reductase SDR family member 13 [EC:1.1.-.-] | 0.00004 | 0.00004 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00009 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| hcrB, hbaD; 4-hydroxybenzoyl-CoA reductase subunit beta [EC:1.3.7.9] | 0.00004 | 0.00004 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00009 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| aac6-I; aminoglycoside 6’-N-acetyltransferase I [EC:2.3.1.82] | 0.00004 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00004 | -1.04988 | 122.200 | 0.29584 | 0.77376 |
| mexZ; TetR/AcrR family transcriptional regulator, mexXY operon repressor | 0.00004 | 0.00003 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.44764 | 143.290 | 0.65509 | 0.90679 |
| adh3; alcohol dehydrogenase [EC:1.1.1.-] | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00005 | 0.20527 | 145.023 | 0.83765 | 0.95506 |
| pduW; propionate kinase [EC:2.7.2.15] | 0.00004 | 0.00002 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.23148 | 148.192 | 0.81726 | 0.95487 |
| pgtC; phosphoglycerate transport regulatory protein PgtC | 0.00004 | 0.00002 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.23148 | 148.192 | 0.81726 | 0.95487 |
| kdxD; 2-dehydro-3-deoxy-D-arabinonate dehydratase [EC:4.2.1.141] | 0.00004 | 0.00002 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00004 | 0.25752 | 121.117 | 0.79721 | 0.94810 |
| nodB; chitooligosaccharide deacetylase [EC:3.5.1.-] | 0.00004 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.88671 | 109.417 | 0.37718 | 0.79815 |
| K15667, ppsD, fenA; fengycin family lipopeptide synthetase D | 0.00004 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.86167 | 100.564 | 0.39092 | 0.80384 |
| nheA; non-hemolytic enterotoxin A | 0.00004 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00005 | -0.86167 | 100.564 | 0.39092 | 0.80384 |
| ameR, rmiR; TetR/AcrR family transcriptional regulator, repressor of the ameABC operon | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.87032 | 109.345 | 0.38603 | 0.80258 |
| csn; chitosanase [EC:3.2.1.132] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.41559 | 92.000 | 0.16027 | 0.77376 |
| E1.14.13.81, acsF, chlE; magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase [EC:1.14.13.81] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.54425 | 92.522 | 0.12594 | 0.77376 |
| SLC34A, NPT, nptA; solute carrier family 34 (sodium-dependent phosphate cotransporter) | 0.00003 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 1.15760 | 77.769 | 0.25057 | 0.77376 |
| CDO1; cysteine dioxygenase [EC:1.13.11.20] | 0.00003 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.34765 | 92.000 | 0.18108 | 0.77376 |
| camD; 5-exo-hydroxycamphor dehydrogenase [EC:1.1.1.327] | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.31963 | 96.801 | 0.19007 | 0.77376 |
| kdhA; 6-hydroxypseudooxynicotine dehydrogenase subunit alpha [EC:1.5.99.14] | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.31963 | 96.801 | 0.19007 | 0.77376 |
| thnE; carboxymethylproline synthase [EC:2.3.1.226] | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.31963 | 96.801 | 0.19007 | 0.77376 |
| blaKPC; beta-lactamase class A KPC [EC:3.5.2.6] | 0.00003 | 0.00002 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.19032 | 148.266 | 0.84932 | 0.95712 |
| ptxD; phosphonate dehydrogenase [EC:1.20.1.1] | 0.00003 | 0.00002 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.19032 | 148.266 | 0.84932 | 0.95712 |
| E4.99.1.2; alkylmercury lyase [EC:4.99.1.2] | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.79143 | 83.406 | 0.43094 | 0.82763 |
| waaI, rfaI; UDP-D-galactose:(glucosyl)LPS alpha-1,3-D-galactosyltransferase [EC:2.4.1.44] | 0.00003 | 0.00002 | 0.00007 | 0.00000 | 0.00006 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00006 | 0.93696 | 64.654 | 0.35227 | 0.79159 |
| hmfA; 2-furoyl-CoA dehydrogenase large subunit [EC:1.3.99.8] | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.86730 | 82.562 | 0.38829 | 0.80262 |
| hmfB; 2-furoyl-CoA dehydrogenase FAD binding subunit [EC:1.3.99.8] | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.86730 | 82.562 | 0.38829 | 0.80262 |
| hmfC; 2-furoyl-CoA dehydrogenase 2Fe-2S iron sulfur subunit [EC:1.3.99.8] | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.86730 | 82.562 | 0.38829 | 0.80262 |
| hmfD; 2-furoate—CoA ligase [EC:6.2.1.31] | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.86730 | 82.562 | 0.38829 | 0.80262 |
| hmfE; 2-oxoglutaroyl-CoA hydrolase | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.86730 | 82.562 | 0.38829 | 0.80262 |
| pvadh; polyvinyl alcohol dehydrogenase (cytochrome) [EC:1.1.2.6] | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.32100 | 118.944 | 0.18904 | 0.77376 |
| dptF; DNA phosphorothioation-dependent restriction protein DptF | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00005 | 0.53490 | 83.341 | 0.59414 | 0.88005 |
| SDC1, CD138; syndecan 1 | 0.00003 | 0.00002 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00005 | 0.90639 | 74.654 | 0.36765 | 0.79587 |
| gumL; pyruvyltransferase | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.66288 | 94.859 | 0.09964 | 0.76942 |
| alsA; D-allose transport system ATP-binding protein [EC:3.6.3.17] | 0.00003 | 0.00002 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00004 | -0.16388 | 155.851 | 0.87004 | 0.96495 |
| epsK; membrane protein EpsK | 0.00003 | 0.00003 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.93923 | 99.157 | 0.34990 | 0.79120 |
| epsE; glycosyltransferase EpsE [EC:2.4.-.-] | 0.00003 | 0.00002 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00005 | 0.85218 | 72.002 | 0.39694 | 0.80459 |
| pks12; polyketide synthase 12 | 0.00003 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.20039 | 92.000 | 0.23307 | 0.77376 |
| E4.2.2.17; inulin fructotransferase (DFA-I-forming) [EC:4.2.2.17] | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| impN; type VI secretion system protein ImpN [EC:2.7.11.1] | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| inoE; inositol-phosphate transport system substrate-binding protein | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| inoF; inositol-phosphate transport system permease protein | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| inoG; inositol-phosphate transport system permease protein | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| inoK; inositol-phosphate transport system ATP-binding protein | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| lpxQ; lipid A oxidase | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| prsD; ATP-binding cassette, subfamily C, bacterial PrsD | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| prsE; membrane fusion protein | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.76912 | 109.501 | 0.44348 | 0.83186 |
| amzA, AMZ2, AMZ1; archaemetzincin [EC:3.4.-.-] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.30087 | 92.000 | 0.19655 | 0.77376 |
| pksN; polyketide synthase PksN | 0.00003 | 0.00003 | 0.00008 | 0.00000 | 0.00007 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00007 | 1.08860 | 64.000 | 0.28041 | 0.77376 |
| bchC; bacteriochlorophyllide a dehydrogenase [EC:1.1.1.396] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| bchX; 3,8-divinyl chlorophyllide a/chlorophyllide a reductase subunit X [EC:1.3.7.14 1.3.7.15] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| bchY; 3,8-divinyl chlorophyllide a/chlorophyllide a reductase subunit Y [EC:1.3.7.14 1.3.7.15] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| bchZ; 3,8-divinyl chlorophyllide a/chlorophyllide a reductase subunit Z [EC:1.3.7.14 1.3.7.15] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| chlL; light-independent protochlorophyllide reductase subunit L [EC:1.3.7.7] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| crtF; demethylspheroidene O-methyltransferase [EC:2.1.1.210] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| pufA; light-harvesting complex 1 alpha chain | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| pufB; light-harvesting complex 1 beta chain | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| pufL; photosynthetic reaction center L subunit | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| pufM; photosynthetic reaction center M subunit | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.40642 | 92.525 | 0.16295 | 0.77376 |
| tuaE; teichuronic acid biosynthesis protein TuaE | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.82111 | 110.540 | 0.41335 | 0.81655 |
| bchF; 3-vinyl bacteriochlorophyllide hydratase [EC:4.2.1.165] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.46187 | 92.000 | 0.14718 | 0.77376 |
| bchO; magnesium chelatase accessory protein | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.46187 | 92.000 | 0.14718 | 0.77376 |
| chlG, bchG; chlorophyll/bacteriochlorophyll a synthase [EC:2.5.1.62 2.5.1.133] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.46187 | 92.000 | 0.14718 | 0.77376 |
| chlP, bchP; geranylgeranyl diphosphate/geranylgeranyl-bacteriochlorophyllide a reductase [EC:1.3.1.83 1.3.1.111] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.46187 | 92.000 | 0.14718 | 0.77376 |
| crtD; 1-hydroxycarotenoid 3,4-desaturase [EC:1.3.99.27] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.46187 | 92.000 | 0.14718 | 0.77376 |
| puhA; photosynthetic reaction center H subunit | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.46187 | 92.000 | 0.14718 | 0.77376 |
| AKR1A1, adh; alcohol dehydrogenase (NADP+) [EC:1.1.1.2] | 0.00003 | 0.00001 | 0.00005 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 1.14358 | 69.399 | 0.25673 | 0.77376 |
| baiH; NAD+-dependent 7beta-hydroxy-3-oxo bile acid-CoA-ester 4-dehydrogenase | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.49453 | 92.000 | 0.13846 | 0.77376 |
| RPR2, RPP21; ribonuclease P protein subunit RPR2 [EC:3.1.26.5] | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00005 | 0.54947 | 80.272 | 0.58421 | 0.87720 |
| SDR16C5; all-trans-retinol dehydrogenase (NAD+) [EC:1.1.1.105] | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00005 | 0.51860 | 88.264 | 0.60534 | 0.88512 |
| petC; cytochrome b6-f complex iron-sulfur subunit [EC:1.10.9.1] | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.31150 | 108.097 | 0.19247 | 0.77376 |
| acpK; polyketide biosynthesis acyl carrier protein | 0.00003 | 0.00002 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.56950 | 114.880 | 0.57013 | 0.87135 |
| pksF; malonyl-ACP decarboxylase | 0.00003 | 0.00002 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.56950 | 114.880 | 0.57013 | 0.87135 |
| pksG; polyketide biosynthesis 3-hydroxy-3-methylglutaryl-CoA synthase-like enzyme PksG | 0.00003 | 0.00002 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.56950 | 114.880 | 0.57013 | 0.87135 |
| pksH; polyketide biosynthesis enoyl-CoA hydratase PksH | 0.00003 | 0.00002 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.56950 | 114.880 | 0.57013 | 0.87135 |
| cefD; isopenicillin-N epimerase [EC:5.1.1.17] | 0.00003 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.05353 | 92.000 | 0.29486 | 0.77376 |
| alsK; allose kinase [EC:2.7.1.55] | 0.00003 | 0.00002 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00004 | 0.00232 | 144.662 | 0.99815 | 0.99957 |
| DNA2; DNA replication ATP-dependent helicase Dna2 [EC:3.6.4.12] | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00004 | 0.79455 | 87.425 | 0.42903 | 0.82749 |
| NPEPPS; puromycin-sensitive aminopeptidase [EC:3.4.11.-] | 0.00003 | 0.00002 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.60220 | 135.097 | 0.54805 | 0.86551 |
| K09141; uncharacterized protein | 0.00003 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.68208 | 84.895 | 0.49705 | 0.84797 |
| K15019; 3-hydroxypropionyl-coenzyme A dehydratase [EC:4.2.1.116] | 0.00003 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00003 | 0.62341 | 93.810 | 0.53453 | 0.86207 |
| FUK; fucokinase [EC:2.7.1.52] | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.77655 | 92.000 | 0.07895 | 0.75800 |
| lipL; lipoyl amidotransferase [EC:2.3.1.200] | 0.00003 | 0.00003 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00006 | 0.98393 | 64.182 | 0.32885 | 0.77419 |
| K11646; 3-dehydroquinate synthase II [EC:1.4.1.24] | 0.00003 | 0.00003 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00006 | 0.99409 | 64.338 | 0.32390 | 0.77376 |
| mxaC; mxaC protein | 0.00003 | 0.00002 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00005 | 1.22230 | 65.558 | 0.22597 | 0.77376 |
| mxaK; mxaK protein | 0.00003 | 0.00002 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00005 | 1.22230 | 65.558 | 0.22597 | 0.77376 |
| CTBP; C-terminal binding protein | 0.00003 | 0.00002 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00004 | 0.12567 | 137.891 | 0.90018 | 0.97503 |
| MIOX; inositol oxygenase [EC:1.13.99.1] | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -0.69639 | 105.796 | 0.48771 | 0.84656 |
| K09144; uncharacterized protein | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.43790 | 94.268 | 0.15377 | 0.77376 |
| gplH; glycopeptidolipid biosynthesis protein | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.72701 | 92.000 | 0.08752 | 0.76567 |
| add; aminodeoxyfutalosine deaminase [EC:3.5.4.40] | 0.00003 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00002 | -0.32986 | 155.367 | 0.74195 | 0.93049 |
| K06922; uncharacterized protein | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.21668 | 101.869 | 0.22654 | 0.77376 |
| MANEA; glycoprotein endo-alpha-1,2-mannosidase [EC:3.2.1.130] | 0.00003 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.21668 | 101.869 | 0.22654 | 0.77376 |
| andAc; anthranilate 1,2-dioxygenase large subunit [EC:1.14.12.1] | 0.00003 | 0.00002 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00004 | 0.12168 | 138.389 | 0.90333 | 0.97641 |
| tphA2; terephthalate 1,2-dioxygenase oxygenase component alpha subunit [EC:1.14.12.15] | 0.00003 | 0.00002 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00004 | 0.12168 | 138.389 | 0.90333 | 0.97641 |
| tphB; 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate dehydrogenase [EC:1.3.1.53] | 0.00003 | 0.00002 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00004 | 0.12168 | 138.389 | 0.90333 | 0.97641 |
| PTS-Dga-EIIC, dgaC; PTS system, D-glucosaminate-specific IIC component | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.47411 | 92.032 | 0.14387 | 0.77376 |
| PTS-Dga-EIID, dgaD; PTS system, D-glucosaminate-specific IID component | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.47411 | 92.032 | 0.14387 | 0.77376 |
| hapR, luxR, litR; TetR/AcrR family transcriptional regulator, hemagglutinin/protease regulatory protein | 0.00003 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.57307 | 64.000 | 0.12063 | 0.77376 |
| mshB; MSHA pilin protein MshB | 0.00003 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.57307 | 64.000 | 0.12063 | 0.77376 |
| ETF1, ERF1; peptide chain release factor subunit 1 | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.11289 | 92.000 | 0.26865 | 0.77376 |
| K07477; translin | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.11289 | 92.000 | 0.26865 | 0.77376 |
| rifG, asm47; 5-deoxy-5-amino-3-dehydroquinate synthase | 0.00002 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.05590 | 109.583 | 0.29334 | 0.77376 |
| hutM; histidine permease | 0.00002 | 0.00002 | 0.00006 | 0.00000 | 0.00005 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00005 | 1.15398 | 64.000 | 0.25280 | 0.77376 |
| hmfF; 2,5-furandicarboxylate decarboxylase 1 | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.36239 | 64.718 | 0.17780 | 0.77376 |
| PRSS33; serine protease 33 [EC:3.4.21.-] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.36239 | 64.718 | 0.17780 | 0.77376 |
| yscY, sctY; type III secretion protein Y | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.36239 | 64.718 | 0.17780 | 0.77376 |
| K07495; putative transposase | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| gumI; beta-1,4-mannosyltransferase [EC:2.4.1.251] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.56174 | 92.000 | 0.12178 | 0.77376 |
| dndB; DNA sulfur modification protein DndB | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00005 | 0.92118 | 64.715 | 0.36038 | 0.79496 |
| petF; ferredoxin | 0.00002 | 0.00002 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.35097 | 140.269 | 0.72613 | 0.92177 |
| actVI1, RED1; ketoreductase RED1 [EC:1.1.1.-] | 0.00002 | 0.00002 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.46544 | 83.533 | 0.64283 | 0.90098 |
| bgtB; arginine/lysine/histidine/glutamine transport system substrate-binding and permease protein | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.42160 | 92.000 | 0.15852 | 0.77376 |
| cmlR, cmx; MFS transporter, DHA1 family, chloramphenicol resistance protein | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.30440 | 92.000 | 0.19535 | 0.77376 |
| K07041; uncharacterized protein | 0.00002 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 0.80377 | 81.500 | 0.42387 | 0.82432 |
| repA; regulatory protein RepA | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.52351 | 95.758 | 0.13093 | 0.77376 |
| ehbQ; energy-converting hydrogenase B subunit Q | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.22552 | 97.293 | 0.22334 | 0.77376 |
| iolX; scyllo-inositol 2-dehydrogenase (NAD+) [EC:1.1.1.370] | 0.00002 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 0.85182 | 89.862 | 0.39658 | 0.80459 |
| qhpA; quinohemoprotein amine dehydrogenase [EC:1.4.9.1] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.19300 | 92.630 | 0.23591 | 0.77376 |
| K16192, arfB; uncharacterized membrane protein ArfB | 0.00002 | 0.00002 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.51788 | 77.653 | 0.60601 | 0.88535 |
| aknOx; aclacinomycin-N/aclacinomycin-A oxidase [EC:1.1.3.45 1.3.3.14] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| blaVEB; beta-lactamase class A VEB [EC:3.5.2.6] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| DEGS; sphingolipid 4-desaturase/C4-monooxygenase [EC:1.14.19.17 1.14.18.5] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EDEM1; ER degradation enhancer, mannosidase alpha-like 1 | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EDEM2; ER degradation enhancer, mannosidase alpha-like 2 | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ILR1; IAA-amino acid hydrolase [EC:3.5.1.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| PIGB; phosphatidylinositol glycan, class B [EC:2.4.1.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| pksI; polyketide biosynthesis enoyl-CoA hydratase PksI | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rhiF; rhizoxin biosynthesis, polyketide synthase RhiF | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TOP2; DNA topoisomerase II [EC:5.99.1.3] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| tycC; tyrocidine synthetase III | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| wbyK; mannosyltransferase [EC:2.4.1.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| aphA; PadR family transcriptional regulator, regulatory protein AphA | 0.00002 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.45431 | 64.000 | 0.15075 | 0.77376 |
| chbP; N,N’-diacetylchitobiose phosphorylase [EC:2.4.1.280] | 0.00002 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.45431 | 64.000 | 0.15075 | 0.77376 |
| feoC; putative ferrous iron transport protein C | 0.00002 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.45431 | 64.000 | 0.15075 | 0.77376 |
| luxU; two-component system, phosphorelay protein LuxU | 0.00002 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.45431 | 64.000 | 0.15075 | 0.77376 |
| toxS; transmembrane regulatory protein ToxS | 0.00002 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.45431 | 64.000 | 0.15075 | 0.77376 |
| trmY; tRNA (pseudouridine54-N1)-methyltransferase [EC:2.1.1.257] | 0.00002 | 0.00002 | 0.00006 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.45431 | 64.000 | 0.15075 | 0.77376 |
| ASPG; 60kDa lysophospholipase [EC:3.1.1.5 3.1.1.47 3.5.1.1] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| lasA; LasA protease [EC:3.4.24.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| sipB, ipaB, bipB; invasin B | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| sipC, ipaC, bipC; invasin C | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| sipD, ipaD, bipD; invasin D | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| PAL; phenylalanine ammonia-lyase [EC:4.3.1.24] | 0.00002 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 1.20273 | 74.401 | 0.23290 | 0.77376 |
| ADHFE1; hydroxyacid-oxoacid transhydrogenase [EC:1.1.99.24] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.03280 | 92.000 | 0.30440 | 0.77376 |
| DPH4, DNAJC24; diphthamide biosynthesis protein 4 | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.03280 | 92.000 | 0.30440 | 0.77376 |
| blaCMY-2; beta-lactamase class C CMY-2 [EC:3.5.2.6] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.93927 | 92.915 | 0.35003 | 0.79120 |
| dptG; DNA phosphorothioation-dependent restriction protein DptG | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00005 | 0.95873 | 64.200 | 0.34129 | 0.78526 |
| dptH; DNA phosphorothioation-dependent restriction protein DptH | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00005 | 0.95873 | 64.200 | 0.34129 | 0.78526 |
| K18336; 2,4-diketo-3-deoxy-L-fuconate hydrolase [EC:3.7.1.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.31665 | 93.000 | 0.19119 | 0.77376 |
| uidR; TetR/AcrR family transcriptional regulator, repressor for uid operon | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.02306 | 93.356 | 0.30892 | 0.77376 |
| ctcP, cts4, prnC; tetracycline 7-halogenase / FADH2 O2-dependent halogenase [EC:1.14.19.49 1.14.19.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.24047 | 92.081 | 0.21795 | 0.77376 |
| wbqR; UDP-perosamine 4-acetyltransferase [EC:2.3.1.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.32173 | 92.000 | 0.18954 | 0.77376 |
| nifHD1, nifI1; nitrogen regulatory protein PII 1 | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00003 | 0.00000 | 0.00001 | 93 | 0.00002 | -0.50679 | 147.030 | 0.61306 | 0.88788 |
| nifHD2, nifI2; nitrogen regulatory protein PII 2 | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00003 | 0.00000 | 0.00001 | 93 | 0.00002 | -0.50679 | 147.030 | 0.61306 | 0.88788 |
| ipct; 1L-myo-inositol 1-phosphate cytidylyltransferase [EC:2.7.7.74] | 0.00002 | 0.00002 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.43441 | 130.471 | 0.66471 | 0.91100 |
| gumC; GumC protein | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.39398 | 92.000 | 0.16668 | 0.77376 |
| gumD; undecaprenyl-phosphate glucose phosphotransferase [EC:2.7.8.31] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.39398 | 92.000 | 0.16668 | 0.77376 |
| gumF; acyltransferase [EC:2.3.1.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.39398 | 92.000 | 0.16668 | 0.77376 |
| gumM; beta-1,4-glucosyltransferase [EC:2.4.1.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.39398 | 92.000 | 0.16668 | 0.77376 |
| ICMT, STE14; protein-S-isoprenylcysteine O-methyltransferase [EC:2.1.1.100] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.39398 | 92.000 | 0.16668 | 0.77376 |
| RYR2; ryanodine receptor 2 | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.31265 | 131.132 | 0.75504 | 0.93665 |
| nox2; NADH oxidase (H2O-forming) [EC:1.6.3.4] | 0.00002 | 0.00001 | 0.00004 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.65998 | 68.570 | 0.10149 | 0.77376 |
| draT; NAD+—dinitrogen-reductase ADP-D-ribosyltransferase [EC:2.4.2.37] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00004 | 93 | 0.00004 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mxaD; mxaD protein | 0.00002 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 1.23776 | 67.480 | 0.22010 | 0.77376 |
| mxaJ; mxaJ protein | 0.00002 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 1.23776 | 67.480 | 0.22010 | 0.77376 |
| CBH2, cbhA; cellulose 1,4-beta-cellobiosidase [EC:3.2.1.91] | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00003 | 0.50929 | 125.262 | 0.61145 | 0.88722 |
| E1.12.7.2S; ferredoxin hydrogenase small subunit [EC:1.12.7.2] | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.38111 | 92.000 | 0.17059 | 0.77376 |
| araD; D-arabinonate dehydratase [EC:4.2.1.5] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.91496 | 93.234 | 0.36257 | 0.79496 |
| hpaC; type III secretion control protein HpaP | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00003 | -0.14666 | 155.884 | 0.88359 | 0.96974 |
| pimF; putative glycosyltransferase [EC:2.4.-.-] | 0.00002 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00003 | -0.74798 | 116.369 | 0.45598 | 0.83438 |
| arfC; uncharacterized membrane protein ArfC | 0.00002 | 0.00002 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00004 | 0.67104 | 75.574 | 0.50424 | 0.84956 |
| PAM, PHM; peptidylglycine monooxygenase [EC:1.14.17.3] | 0.00002 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 1.04443 | 74.818 | 0.29965 | 0.77376 |
| fadD26; long chain fatty acid CoA FadD26 | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.22821 | 92.000 | 0.22250 | 0.77376 |
| wbtD; galacturonosyltransferase [EC:2.4.1.-] | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 0.59479 | 92.117 | 0.55344 | 0.86788 |
| yafO; mRNA interferase YafO [EC:3.1.-.-] | 0.00002 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.26286 | 64.525 | 0.21118 | 0.77376 |
| cmaB; non-haem Fe2+, alpha-ketoglutarate-dependent halogenase | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.14752 | 64.000 | 0.25544 | 0.77376 |
| hopM1; effector protein HopM1 | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.14752 | 64.000 | 0.25544 | 0.77376 |
| ospF, mkaD, spvC; phosphothreonine lyase [EC:4.2.3.-] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.14752 | 64.000 | 0.25544 | 0.77376 |
| syrB1; Syringomycin synthetase protein SyrB1 | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.14752 | 64.000 | 0.25544 | 0.77376 |
| ARSA; arylsulfatase A [EC:3.1.6.8] | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 0.62797 | 71.181 | 0.53203 | 0.86182 |
| aliA; cyclohexanecarboxylate-CoA ligase [EC:6.2.1.-] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00005 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| vanB, vanA, vanD; D-alanine—(R)-lactate ligase [EC:6.1.2.1] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00005 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| vanH; D-specific alpha-keto acid dehydrogenase [EC:1.1.1.-] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00005 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00005 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| acfA; accessory colonization factor AcfA | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| ainS, luxM; acyl homoserine lactone synthase [EC:2.3.1.184] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| ASPA, aspA; aspartoacylase [EC:3.5.1.15] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| blaCARB-17; beta-lactamase class A CARB-17 [EC:3.5.2.6] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| dnk; deoxynucleoside kinase [EC:2.7.1.145] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| GNS; N-acetylglucosamine-6-sulfatase [EC:3.1.6.14] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| luxA; alkanal monooxygenase alpha chain [EC:1.14.14.3] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| luxB; alkanal monooxygenase beta chain [EC:1.14.14.3] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| luxC; long-chain-fatty-acyl-CoA reductase [EC:1.2.1.50] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| luxD; acyl transferase [EC:2.3.1.-] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| luxE; long-chain-fatty-acid—luciferin-component ligase [EC:6.2.1.19] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| luxN; two-component system, autoinducer 1 sensor kinase/phosphatase LuxN [EC:2.7.13.3 3.1.3.-] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| vieA; c-di-GMP phosphodiesterase [EC:3.1.4.52] | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| yadB_C; adhesin YadB/C | 0.00002 | 0.00002 | 0.00005 | 0.00000 | 0.00004 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00004 | 1.21433 | 64.000 | 0.22908 | 0.77376 |
| ACSM; medium-chain acyl-CoA synthetase [EC:6.2.1.2] | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.93545 | 97.604 | 0.35186 | 0.79120 |
| CPGS; cyclic 2,3-diphosphoglycerate synthetase [EC:4.6.1.-] | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.93545 | 97.604 | 0.35186 | 0.79120 |
| K07244; mgtE-like transporter | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.93545 | 97.604 | 0.35186 | 0.79120 |
| ndhG; NAD(P)H-quinone oxidoreductase subunit 6 [EC:1.6.5.3] | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.93545 | 97.604 | 0.35186 | 0.79120 |
| wbbJ; lipopolysaccharide O-acetyltransferase [EC:2.3.1.-] | 0.00002 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00002 | -0.85250 | 104.337 | 0.39589 | 0.80459 |
| fimW; fimbrial protein FimW | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.05273 | 92.032 | 0.29522 | 0.77376 |
| mbhJ; membrane-bound hydrogenase subunit mbhJ [EC:1.12.7.2] | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.30817 | 142.133 | 0.75840 | 0.93719 |
| mbhK; membrane-bound hydrogenase subunit beta [EC:1.12.7.2] | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.30817 | 142.133 | 0.75840 | 0.93719 |
| mbhL; membrane-bound hydrogenase subunit alpha [EC:1.12.7.2] | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.30817 | 142.133 | 0.75840 | 0.93719 |
| tuaB; teichuronic acid exporter | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.45952 | 69.312 | 0.14894 | 0.77376 |
| rppA; 1,3,6,8-tetrahydroxynaphthalene synthase [EC:2.3.1.233] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| sprD; streptogrisin D [EC:3.4.21.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| crtC; carotenoid 1,2-hydratase [EC:4.2.1.131] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.98161 | 92.938 | 0.32884 | 0.77419 |
| oprJ; outer membrane protein, multidrug efflux system | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00003 | 0.62610 | 78.823 | 0.53306 | 0.86207 |
| amsD; amylovoran biosynthesis glycosyltransferase AmsD [EC:2.4.-.-] | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ANK; ankyrin | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K01622; fructose 1,6-bisphosphate aldolase/phosphatase [EC:4.1.2.13 3.1.3.11] | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.70493 | 67.532 | 0.48328 | 0.84354 |
| pufC; photosynthetic reaction center cytochrome c subunit | 0.00002 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00003 | -0.92619 | 92.938 | 0.35675 | 0.79399 |
| phcR; two-component system, response regulator PhcR | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.16902 | 92.000 | 0.24541 | 0.77376 |
| BCS1; mitochondrial chaperone BCS1 | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.19075 | 92.000 | 0.23682 | 0.77376 |
| UBLCP1; ubiquitin-like domain-containing CTD phosphatase 1 [EC:3.1.3.16] | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.19075 | 92.000 | 0.23682 | 0.77376 |
| ABO; histo-blood group ABO system transferase [EC:2.4.1.40 2.4.1.37] | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.23334 | 93.662 | 0.22053 | 0.77376 |
| NANS, SAS; sialic acid synthase [EC:2.5.1.56 2.5.1.57 2.5.1.132] | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.23334 | 93.662 | 0.22053 | 0.77376 |
| bchM, chlM; magnesium-protoporphyrin O-methyltransferase [EC:2.1.1.11] | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00002 | -0.97912 | 93.161 | 0.33006 | 0.77623 |
| mtkA; malate-CoA ligase subunit beta [EC:6.2.1.9] | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.50562 | 102.238 | 0.61421 | 0.88788 |
| mtkB; malate-CoA ligase subunit alpha [EC:6.2.1.9] | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.50562 | 102.238 | 0.61421 | 0.88788 |
| nodF; nodulation protein F [EC:2.3.1.-] | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.50562 | 102.238 | 0.61421 | 0.88788 |
| cfa; cAMP factor | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.09693 | 97.401 | 0.27538 | 0.77376 |
| bpr; bacillopeptidase F [EC:3.4.21.-] | 0.00002 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.17294 | 65.326 | 0.24508 | 0.77376 |
| mexY, amrB; multidrug efflux pump | 0.00002 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.59657 | 71.621 | 0.55267 | 0.86788 |
| sul2; dihydropteroate synthase type 2 [EC:2.5.1.15] | 0.00001 | 0.00001 | 0.00004 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.23946 | 64.000 | 0.21970 | 0.77376 |
| strA; streptomycin 3"-kinase [EC:2.7.1.87] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.80077 | 67.234 | 0.42608 | 0.82652 |
| ctpA_B; cation-transporting P-type ATPase A/B [EC:3.6.3.-] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.75202 | 92.000 | 0.08310 | 0.76567 |
| K06944; uncharacterized protein | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.89361 | 65.328 | 0.37481 | 0.79810 |
| aac3-I; aminoglycoside 3-N-acetyltransferase I [EC:2.3.1.60] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.41606 | 83.632 | 0.67844 | 0.91311 |
| hpdB; 4-hydroxyphenylacetate decarboxylase large subunit [EC:4.1.1.83] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.32700 | 73.864 | 0.18860 | 0.77376 |
| alsB; D-allose transport system substrate-binding protein | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.90779 | 64.643 | 0.36736 | 0.79587 |
| alsC; D-allose transport system permease protein | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.90779 | 64.643 | 0.36736 | 0.79587 |
| yafN; antitoxin YafN | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.90779 | 64.643 | 0.36736 | 0.79587 |
| ARSB; arylsulfatase B [EC:3.1.6.12] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.11318 | 70.631 | 0.26940 | 0.77376 |
| LOXL2_3_4; lysyl oxidase-like protein 2/3/4 [EC:1.4.3.-] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.10349 | 98.936 | 0.27249 | 0.77376 |
| mfnB; (5-formylfuran-3-yl)methyl phosphate synthase [EC:4.2.3.153] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.92877 | 64.610 | 0.35647 | 0.79399 |
| mfnF; (4-(4-[2-(gamma-L-glutamylamino)ethyl]phenoxymethyl)furan-2-yl)methanamine synthase [EC:2.5.1.131] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.92877 | 64.610 | 0.35647 | 0.79399 |
| RABGGTB; geranylgeranyl transferase type-2 subunit beta [EC:2.5.1.60] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.92877 | 64.610 | 0.35647 | 0.79399 |
| aliB; cyclohexanecarboxyl-CoA dehydrogenase [EC:1.3.99.-] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K14165; atypical dual specificity phosphatase [EC:3.1.3.16 3.1.3.48] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 0.88171 | 65.629 | 0.38115 | 0.79920 |
| aprE; subtilisin [EC:3.4.21.62] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.57524 | 64.000 | 0.12013 | 0.77376 |
| gumG; acyltransferase [EC:2.3.1.-] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hpa1; type III secretion harpin protein Hpa1 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hpa2; lysozyme-related protein Hpa2 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hpaA; type III secretion regulatory protein HpaA | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hpaB; type III secretion control protein HpaB | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hrpB1; type III secretion protein HrpB1 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hrpB2; type III secretion inner rod protein HrpB2 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hrpE; type III secretion hrp pilus HrpE | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hrpF; type III secretion translocon protein HrpF | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mexX, amrA; membrane fusion protein, multidrug efflux system | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | 0.77074 | 69.790 | 0.44346 | 0.83186 |
| natE; neutral amino acid transport system ATP-binding protein | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.49279 | 92.000 | 0.13891 | 0.77376 |
| top6A; DNA topoisomerase VI subunit A [EC:5.99.1.3] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.16053 | 92.000 | 0.24883 | 0.77376 |
| top6B; DNA topoisomerase VI subunit B [EC:5.99.1.3] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.16053 | 92.000 | 0.24883 | 0.77376 |
| CYP134A, cypX; pulcherriminic acid synthase [EC:1.14.15.13] | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | -0.36908 | 140.702 | 0.71263 | 0.92004 |
| FMN2; formin 2 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07128; uncharacterized protein | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| K08980; putative membrane protein | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| MGME1, DDK1; mitochondrial genome maintenance exonuclease 1 [EC:3.1.-.-] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| mphR; TetR/AcrR family transcriptional regulator, macrolide resistance operon repressor | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | 0.43253 | 124.172 | 0.66611 | 0.91126 |
| mrx; macrolide resistance protein | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | 0.43253 | 124.172 | 0.66611 | 0.91126 |
| sfmC; fimbrial chaperone protein | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| DECR2; peroxisomal 2,4-dienoyl-CoA reductase [EC:1.3.1.34] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| SKI3, TTC37; superkiller protein 3 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TH; tyrosine 3-monooxygenase [EC:1.14.16.2] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| E2.4.1.217; mannosyl-3-phosphoglycerate synthase [EC:2.4.1.217] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 0.85853 | 66.298 | 0.39369 | 0.80459 |
| HYDIN; hydrocephalus-inducing protein | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| tphA1; terephthalate 1,2-dioxygenase reductase component [EC:1.18.1.-] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| tphA3; terephthalate 1,2-dioxygenase oxygenase component beta subunit [EC:1.14.12.15] | 0.00001 | 0.00001 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00003 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| asm11; 4,5-epoxidase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.33129 | 92.000 | 0.18638 | 0.77376 |
| NUDT2; bis(5’-nucleosidyl)-tetraphosphatase [EC:3.6.1.17] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 0.89864 | 65.209 | 0.37215 | 0.79738 |
| NDUFAF7; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 7 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -0.90636 | 129.570 | 0.36643 | 0.79587 |
| blaIND; metallo-beta-lactamase class B IND [EC:3.5.2.6] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -0.80310 | 98.140 | 0.42386 | 0.82432 |
| mmoX; methane monooxygenase component A alpha chain [EC:1.14.13.25] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mmoY; methane monooxygenase component A beta chain [EC:1.14.13.25] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ACAA2; acetyl-CoA acyltransferase 2 [EC:2.3.1.16] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K09139; uncharacterized protein | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| tcmG, elmG; tetracenomycin A2 monooxygenase-dioxygenase [EC:1.14.13.200] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -0.94760 | 99.635 | 0.34563 | 0.78802 |
| clrA, serA; complex iron-sulfur molybdoenzyme family reductase subunit alpha | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| clrb, serB; complex iron-sulfur molybdoenzyme family reductase subunit beta | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| clrC, serC; complex iron-sulfur molybdoenzyme family reductase subunit gamma | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ddhA; dimethylsulfide dehydrogenase subunit alpha [EC:1.8.2.4] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ddhB; dimethylsulfide dehydrogenase subunit beta | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ddhC; dimethylsulfide dehydrogenase subunit gamma | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| dmd-tmd; dimethylamine/trimethylamine dehydrogenase [EC:1.5.8.1 1.5.8.2] | 0.00001 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.67713 | 101.917 | 0.49986 | 0.84896 |
| ppsA; phthiocerol/phenolphthiocerol synthesis type-I polyketide synthase A | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.22821 | 92.000 | 0.22250 | 0.77376 |
| ppsB; phthiocerol/phenolphthiocerol synthesis type-I polyketide synthase B | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.22821 | 92.000 | 0.22250 | 0.77376 |
| ppsD; phthiocerol/phenolphthiocerol synthesis type-I polyketide synthase D | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.22821 | 92.000 | 0.22250 | 0.77376 |
| ppsE; phthiocerol/phenolphthiocerol synthesis type-I polyketide synthase E | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.22821 | 92.000 | 0.22250 | 0.77376 |
| K09143; uncharacterized protein | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 0.82939 | 67.290 | 0.40982 | 0.81416 |
| whiEVIII; putative polyketide hydroxylase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00002 | -0.80761 | 97.877 | 0.42127 | 0.82295 |
| aoxB; arsenite oxidase large subunit [EC:1.20.2.1 1.20.9.1] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| bpsA; N4-bis(aminopropyl)spermidine synthase [EC:2.5.1.128] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| waaK, rfaK; UDP-N-acetylglucosamine:(glucosyl)LPS alpha-1,2-N-acetylglucosaminyltransferase [EC:2.4.1.56] | 0.00001 | 0.00000 | 0.00001 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.42337 | 151.323 | 0.15669 | 0.77376 |
| snbC; pristinamycin I synthase 2 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.17483 | 92.000 | 0.24310 | 0.77376 |
| sfa3; sigma-54 dependent transcriptional regulator | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| cmr3; CRISPR-associated protein Cmr3 | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.80148 | 92.000 | 0.07490 | 0.74200 |
| tpr; thiol protease [EC:3.4.22.-] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.80148 | 92.000 | 0.07490 | 0.74200 |
| K06913; uncharacterized protein | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TFIIB, GTF2B, SUA7, tfb; transcription initiation factor TFIIB | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TTN; titin [EC:2.7.11.1] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.41774 | 92.000 | 0.15964 | 0.77376 |
| exoX; exopolysaccharide production repressor protein | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00002 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.31194 | 92.000 | 0.19281 | 0.77376 |
| vanK; vancomycin resistance protein VanK | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | 0.22929 | 98.602 | 0.81912 | 0.95506 |
| asnO; L-asparagine oxygenase [EC:1.14.11.39] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| cmmT, thnT; putative pantetheine hydrolase [EC:3.5.1.-] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| cyc2; germacradienol/geosmin synthase [EC:4.2.3.22 4.2.3.75 4.1.99.16] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| CYP105D; pentalenic acid synthase [EC:1.14.15.11] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| E2.3.2.21; cyclo(L-tyrosyl-L-tyrosyl) synthase [EC:2.3.2.21] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| pchC; pyochelin biosynthetic protein PchC | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ptlI, CYP183A; pentalenene oxygenase [EC:1.14.13.133] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| snpA; snapalysin [EC:3.4.24.77] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| sprB; streptogrisin B [EC:3.4.21.81] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| thnD; putative oxidoreductase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| wtpA; molybdate/tungstate transport system substrate-binding protein | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.35559 | 93.349 | 0.17850 | 0.77376 |
| dipps; CDP-L-myo-inositol myo-inositolphosphotransferase [EC:2.7.8.34] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rsaF; outer membrane protein, S-layer protein transport system | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.24728 | 92.000 | 0.21546 | 0.77376 |
| crtR; beta-carotene hydroxylase [EC:1.14.13.-] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| DHRS12; dehydrogenase/reductase SDR family member 12 [EC:1.1.-.-] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| PRDX5; peroxiredoxin 5, atypical 2-Cys peroxiredoxin [EC:1.11.1.15] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| dfrA12, dhfr; dihydrofolate reductase (trimethoprim resistance protein) [EC:1.5.1.3] | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -0.23220 | 154.477 | 0.81669 | 0.95487 |
| dmrA; dihydromethanopterin reductase [EC:1.5.1.47] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| mdh1, mxaF; methanol dehydrogenase (cytochrome c) subunit 1 [EC:1.1.2.7] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| mdh2, mxaI; methanol dehydrogenase (cytochrome c) subunit 2 [EC:1.1.2.7] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| PRMT1; type I protein arginine methyltransferase [EC:2.1.1.319] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| traI; conjugal transfer pilus assembly protein TraI | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 0.97828 | 66.103 | 0.33150 | 0.77825 |
| glcP; MFS transporter, FHS family, glucose/mannose:H+ symporter | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.41718 | 64.000 | 0.16128 | 0.77376 |
| pelC; pectate lyase C [EC:4.2.2.2 4.2.2.10] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.41718 | 64.000 | 0.16128 | 0.77376 |
| ydfI; two-component system, NarL family, response regulator YdfI | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.41718 | 64.000 | 0.16128 | 0.77376 |
| E2.4.1.213; glucosylglycerol-phosphate synthase [EC:2.4.1.213] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.14427 | 64.297 | 0.25675 | 0.77376 |
| C5AP, scpA, scpB; C5a peptidase [EC:3.4.21.110] | 0.00001 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.13670 | 120.702 | 0.89150 | 0.97141 |
| vasH; sigma-54 dependent transcriptional regulator | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -0.42648 | 134.288 | 0.67044 | 0.91184 |
| NAPEPLD; N-acyl-phosphatidylethanolamine-hydrolysing phospholipase D [EC:3.1.4.54] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| yscI, sctI; type III secretion protein I | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09884 | 92.000 | 0.27470 | 0.77376 |
| yscO, sctO; type III secretion protein O | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09884 | 92.000 | 0.27470 | 0.77376 |
| yscX, sctX; type III secretion protein X | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09884 | 92.000 | 0.27470 | 0.77376 |
| srfAA, lchAA; surfactin family lipopeptide synthetase A | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| cymC; p-cumic aldehyde dehydrogenase | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.14614 | 102.324 | 0.25441 | 0.77376 |
| ITGB3, CD61; integrin beta 3 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.14614 | 102.324 | 0.25441 | 0.77376 |
| snoaF, dnrE, dauE, aknU; nogalaviketone/aklaviketone reductase [EC:1.1.1.- 1.1.1.362] | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.14614 | 102.324 | 0.25441 | 0.77376 |
| dsrA; dissimilatory sulfite reductase alpha subunit [EC:1.8.99.5] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09140 | 93.486 | 0.27790 | 0.77376 |
| dsrB; dissimilatory sulfite reductase beta subunit [EC:1.8.99.5] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09140 | 93.486 | 0.27790 | 0.77376 |
| K09126; uncharacterized protein | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09140 | 93.486 | 0.27790 | 0.77376 |
| NSF, SEC18; vesicle-fusing ATPase [EC:3.6.4.6] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09140 | 93.486 | 0.27790 | 0.77376 |
| qmoA; quinone-modifying oxidoreductase, subunit QmoA | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09140 | 93.486 | 0.27790 | 0.77376 |
| qmoB; quinone-modifying oxidoreductase, subunit QmoB | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09140 | 93.486 | 0.27790 | 0.77376 |
| qmoC; quinone-modifying oxidoreductase, subunit QmoC | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.09140 | 93.486 | 0.27790 | 0.77376 |
| MAN1; mannosyl-oligosaccharide alpha-1,2-mannosidase [EC:3.2.1.113] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rsaA; S-layer protein | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rsaD; ATP-binding cassette, subfamily C, bacterial RsaD | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rsaE; membrane fusion protein, S-layer protein transport system | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| wbqL; O-antigen biosynthesis protein WbqL | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| penM, pntM, CYP161C; pentalenolactone synthase [EC:1.14.19.8] | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| E2.1.1.103, NMT; phosphoethanolamine N-methyltransferase [EC:2.1.1.103] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| vanXY; zinc D-Ala-D-Ala dipeptidase/carboxypeptidase [EC:3.4.13.22 3.4.17.14] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ciaX; regulatory peptide CiaX | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.08860 | 64.000 | 0.28041 | 0.77376 |
| ska; streptokinase A [EC:3.4.-.-] | 0.00001 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.43212 | 109.570 | 0.66650 | 0.91126 |
| K18611; 4-pyridoxate dehydrogenase [EC:1.1.1.-] | 0.00001 | 0.00001 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.19572 | 64.000 | 0.23622 | 0.77376 |
| devS; two-component system, NarL family, sensor histidine kinase DevS [EC:2.7.13.3] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| diox1; all-trans-8’-apo-beta-carotenal 15,15’-oxygenase [EC:1.13.11.75] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07450; putative resolvase | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| KDM8, JMJD5; lysine-specific demethylase 8 [EC:1.14.11.27] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| whiB5; WhiB family transcriptional regulator, redox-sensing transcriptional regulator | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| pmoC-amoC; methane/ammonia monooxygenase subunit C | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.37089 | 92.000 | 0.17375 | 0.77376 |
| opmD; outer membrane protein, multidrug efflux system | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| radA; DNA repair protein RadA | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| glyQS; glycyl-tRNA synthetase [EC:6.1.1.14] | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| loxA; arachidonate 15-lipoxygenase [EC:1.13.11.33] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| GLCAK; glucuronokinase [EC:2.7.1.43] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| GYS; glycogen synthase [EC:2.4.1.11] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mepR; MarR family transcriptional regulator, repressor for mepA | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ADT, PDT; arogenate/prephenate dehydratase [EC:4.2.1.91 4.2.1.51] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.27135 | 92.000 | 0.20681 | 0.77376 |
| amsB; amylovoran biosynthesis glycosyltransferase AmsB [EC:2.4.-.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 0.05563 | 146.421 | 0.95571 | 0.98792 |
| amsC; amylovoran biosynthesis protein AmsC | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 0.05563 | 146.421 | 0.95571 | 0.98792 |
| amsF; amylovoran biosynthesis protein AmsF | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 0.05563 | 146.421 | 0.95571 | 0.98792 |
| amsG; UDP-galactose-lipid carrier transferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 0.05563 | 146.421 | 0.95571 | 0.98792 |
| amsL; exopolysaccharide (amylovoran) exporter | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 0.05563 | 146.421 | 0.95571 | 0.98792 |
| idnT; Gnt-II system L-idonate transporter | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 0.05563 | 146.421 | 0.95571 | 0.98792 |
| pagO; putative membrane protein PagO | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 0.05563 | 146.421 | 0.95571 | 0.98792 |
| bphD; 2,6-dioxo-6-phenylhexa-3-enoate hydrolase [EC:3.7.1.8] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| carAa; carbazole 1,9a-dioxygenase [EC:1.14.12.22] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| carAc; carbazole 1,9a-dioxygenase ferredoxin component | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| carAd; carbazole 1,9a-dioxygenase ferredoxin reductase component | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| carBa; 2’-aminobiphenyl-2,3-diol 1,2-dioxygenase, small subunit [EC:1.13.11.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| carBb; 2’-aminobiphenyl-2,3-diol 1,2-dioxygenase, large subunit [EC:1.13.11.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| carC; 2-hydroxy-6-oxo-6-(2’-aminophenyl)hexa-2,4-dienoate hydrolase [EC:3.7.1.13] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| E1.1.1.387; L-serine 3-dehydrogenase (NAD+) [EC:1.1.1.387] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| HSD17B4; 3-hydroxyacyl-CoA dehydrogenase / 3a,7a,12a-trihydroxy-5b-cholest-24-enoyl-CoA hydratase / enoyl-CoA hydratase 2 [EC:1.1.1.35 4.2.1.107 4.2.1.119] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| tmoF, tbuC, touF; toluene monooxygenase electron transfer component [EC:1.18.1.3] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.92118 | 73.357 | 0.35998 | 0.79496 |
| flaI; flagellar rod protein FlaI | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| luxP; autoinducer 2-binding periplasmic protein LuxP | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| luxQ; two-component system, autoinducer 2 sensor kinase/phosphatase LuxQ [EC:2.7.13.3 3.1.3.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| mshF; MSHA biogenesis protein MshF | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| tfoS; AraC family transcriptional regulator, chitin signaling transcriptional activator | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| PIP5K; 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| ZDS, crtQ; zeta-carotene desaturase [EC:1.3.5.6] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| epsI; pyruvyl transferase EpsI [EC:2.-.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.88101 | 65.522 | 0.38153 | 0.79976 |
| crtISO, crtH; prolycopene isomerase [EC:5.2.1.13] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.37573 | 155.883 | 0.70763 | 0.92004 |
| cruC; chlorobactene glucosyltransferase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.37573 | 155.883 | 0.70763 | 0.92004 |
| E1.1.1.102; 3-dehydrosphinganine reductase [EC:1.1.1.102] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.37573 | 155.883 | 0.70763 | 0.92004 |
| ydfH; two-component system, NarL family, sensor histidine kinase YdfH [EC:2.7.13.3] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.18733 | 64.000 | 0.23949 | 0.77376 |
| GABRE; gamma-aminobutyric acid receptor subunit epsilon | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.97904 | 72.899 | 0.33080 | 0.77745 |
| luxR, vanR; LuxR family transcriptional regulator, transcriptional activator of the bioluminescence operon | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| pmoA-amoA; methane/ammonia monooxygenase subunit A [EC:1.14.18.3 1.14.99.39] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.41872 | 92.000 | 0.15936 | 0.77376 |
| pmoB-amoB; methane/ammonia monooxygenase subunit B | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.41872 | 92.000 | 0.15936 | 0.77376 |
| AKR7; aflatoxin B1 aldehyde reductase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.12153 | 92.000 | 0.26498 | 0.77376 |
| UAH; ureidoglycolate amidohydrolase [EC:3.5.1.116] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.29892 | 114.457 | 0.76554 | 0.93900 |
| helS; helicase [EC:3.6.4.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 0.30301 | 113.904 | 0.76244 | 0.93813 |
| cdc6A; archaeal cell division control protein 6 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| endA; tRNA-intron endonuclease, archaea type [EC:4.6.1.16] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ginS; DNA replication factor GINS | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07159; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07463; archaea-specific RecJ-like exonuclease | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| purP; 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5’-monophosphate synthetase [EC:6.3.4.23] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RFA1, RPA1, rpa; replication factor A1 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RUXX; small nuclear ribonucleoprotein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ssh10b; archaea-specific DNA-binding protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| bioA, bioK; lysine—8-amino-7-oxononanoate aminotransferase [EC:2.6.1.105] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| bioI, CYP107H; pimeloyl-[acyl-carrier protein] synthase [EC:1.14.14.46] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| cgeE; spore maturation protein CgeE | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| cotV; spore coat protein V | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| cotW; spore coat protein W | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| cwlS; peptidoglycan DL-endopeptidase CwlS [EC:3.4.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| dppE; dipeptide transport system substrate-binding protein | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| epsA; protein tyrosine kinase EpsB modulator | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| epsC; polysaccharide biosynthesis protein EpsC | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| gbsB; choline dehydrogenase [EC:1.1.1.1] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| gerAA; spore germination protein AA | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| gerAB; spore germination protein AB | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| gerAC; spore germination protein AC | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| iolS; myo-inositol catabolism protein IolS [EC:1.1.1.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| liaH; lia operon protein LiaH | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| ntdC; glucose-6-phosphate 3-dehydrogenase [EC:1.1.1.361] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| phrE; phosphatase RapE regulator | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| PTS-Fru1-EIIA, levD; PTS system, fructose-specific IIA component [EC:2.7.1.202] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| PTS-Fru1-EIIB, levE; PTS system, fructose-specific IIB component [EC:2.7.1.202] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| rapA, spo0L; response regulator aspartate phosphatase A (stage 0 sporulation protein L) [EC:3.1.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| rapC; response regulator aspartate phosphatase C [EC:3.1.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| rapD; response regulator aspartate phosphatase D [EC:3.1.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| rapE; response regulator aspartate phosphatase E [EC:3.1.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| rapH; response regulator aspartate phosphatase H [EC:3.1.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| rapI; response regulator aspartate phosphatase I [EC:3.1.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| rapK; response regulator aspartate phosphatase K [EC:3.1.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| slrA; anti-repressor of SlrR | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| slrR; HTH-type transcriptional regulator, biofilm formation regulator | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| spoIISA; stage II sporulation protein SA | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| spoIISB; stage II sporulation protein SB | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| srfAB, lchAB; surfactin family lipopeptide synthetase B | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| srfAC, lchAC; surfactin family lipopeptide synthetase C | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| tapA; TasA anchoring/assembly protein | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| tuaC; teichuronic acid biosynthesis glycosyltransferase TuaC [EC:2.4.-.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| tuaF; teichuronic acid biosynthesis protein TuaF | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| wprA; cell wall-associated protease [EC:3.4.21.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| ydfJ; membrane protein YdfJ | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| yesX; rhamnogalacturonan exolyase [EC:4.2.2.24] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| yknT; sigma-E controlled sporulation protein | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| ytrB; acetoin utilization transport system ATP-binding protein | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| ytrC_D; acetoin utilization transport system permease protein | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| yvmC; cyclo(L-leucyl-L-leucyl) synthase [EC:2.3.2.22] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| yxdJ; two-component system, OmpR family, response regulator YxdJ | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| yxdK; two-component system, OmpR family, sensor histidine kinase YxdK [EC:2.7.13.3] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| inlB; internalin B | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.43420 | 92.000 | 0.15491 | 0.77376 |
| E4.3.1.23; tyrosine ammonia-lyase [EC:4.3.1.23] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.82903 | 92.000 | 0.07063 | 0.73788 |
| aac6-II; aminoglycoside 6’-N-acetyltransferase II [EC:2.3.1.82] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| blaDHA; beta-lactamase class C DHA [EC:3.5.2.6] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| blaGES; beta-lactamase class A GES [EC:3.5.2.6] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| blaOXA-10; beta-lactamase class D OXA-10 [EC:3.5.2.6] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| dfrA1, dhfr; dihydrofolate reductase (trimethoprim resistance protein) [EC:1.5.1.3] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| tauY; taurine dehydrogenase large subunit [EC:1.4.2.-] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.01506 | 73.383 | 0.31341 | 0.77376 |
| ligX; 5,5’-dehydrodivanillate O-demethylase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.41798 | 92.000 | 0.15958 | 0.77376 |
| fadD10; long chain fatty acid CoA ligase FadD10 [EC:6.2.1.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.19227 | 92.000 | 0.23622 | 0.77376 |
| K11954, natB; neutral amino acid transport system substrate-binding protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.19227 | 92.000 | 0.23622 | 0.77376 |
| K11957, natA; neutral amino acid transport system ATP-binding protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.19227 | 92.000 | 0.23622 | 0.77376 |
| mps2; glycopeptidolipid biosynthesis protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.19227 | 92.000 | 0.23622 | 0.77376 |
| natD; neutral amino acid transport system permease protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.19227 | 92.000 | 0.23622 | 0.77376 |
| nprE; bacillolysin [EC:3.4.24.28] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.19227 | 92.000 | 0.23622 | 0.77376 |
| phdF; extradiol dioxygenase [EC:1.13.11.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.19227 | 92.000 | 0.23622 | 0.77376 |
| SCRN; secernin | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.19227 | 92.000 | 0.23622 | 0.77376 |
| cynD; cyanide dihydratase [EC:3.5.5.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.22593 | 92.000 | 0.22335 | 0.77376 |
| pchF; pyochelin synthetase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.22593 | 92.000 | 0.22335 | 0.77376 |
| E1.1.1.374; UDP-N-acetylglucosamine 3-dehydrogenase [EC:1.1.1.374] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.48822 | 92.000 | 0.14011 | 0.77376 |
| bxlA; beta-xylosidase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| cd, ma, nplT; cyclomaltodextrinase / maltogenic alpha-amylase / neopullulanase [EC:3.2.1.54 3.2.1.133 3.2.1.135] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| jadI; cyclase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| NIP; aquaporin NIP | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| qodI; quercetin 2,3-dioxygenase [EC:1.13.11.24] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| cgt; cyclomaltodextrin glucanotransferase [EC:2.4.1.19] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| dcsG; O-ureido-D-serine cyclo-ligase [EC:6.3.3.5] | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| gap2; glyceraldehyde-3-phosphate dehydrogenase (NAD(P)) [EC:1.2.1.59] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.27151 | 92.000 | 0.20675 | 0.77376 |
| phcA; LysR family transcriptional regulator, virulence genes transcriptional regulator | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.42185 | 92.000 | 0.15845 | 0.77376 |
| spoVM; stage V sporulation protein M | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.67608 | 107.549 | 0.50044 | 0.84899 |
| K07445; putative DNA methylase | 0.00000 | 0.00000 | 0.00001 | 0.00000 | 0.00001 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00001 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| bbsF; benzylsuccinate CoA-transferase BbsF subunit [EC:2.8.3.15] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.15996 | 155.670 | 0.87312 | 0.96576 |
| desA; syringate O-demethylase [EC:2.1.1.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.15996 | 155.670 | 0.87312 | 0.96576 |
| POLRMT, RPO41; DNA-directed RNA polymerase, mitochondrial [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.15996 | 155.670 | 0.87312 | 0.96576 |
| ctpF; cation-transporting P-type ATPase F [EC:3.6.3.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mmoB; methane monooxygenase regulatory protein B | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mmoC; methane monooxygenase component C [EC:1.14.13.25] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mmoZ; methane monooxygenase component A gamma chain [EC:1.14.13.25] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RIMS2, RIM2; regulating synaptic membrane exocytosis protein 2 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| vioO; nonribosomal peptide synthetase protein VioO | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| WIPF; WAS/WASL-interacting protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| blaCMY-1; beta-lactamase class C CMY-1 [EC:3.5.2.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| blaOXA-12; beta-lactamase class D OXA-12 [EC:3.5.2.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| lldR; LysR family transcriptional regulator, L-lactate utilization regulator | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| phaJ; enoyl-CoA hydratase [EC:4.2.1.119] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| E1.10.3.3; L-ascorbate oxidase [EC:1.10.3.3] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| tmoC, tbuB, touC; toluene monooxygenase system ferredoxin subunit | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ABCE1, Rli1; ATP-binding cassette, sub-family E, member 1 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| AK6, FAP7; adenylate kinase [EC:2.7.4.3] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ALG7; UDP-N-acetylglucosamine–dolichyl-phosphate N-acetylglucosaminephosphotransferase [EC:2.7.8.15] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| atrm56; tRNA (cytidine56-2’-O)-methyltransferase [EC:2.1.1.206] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| cobY; adenosylcobinamide-phosphate guanylyltransferase [EC:2.7.7.62] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| cofH; FO synthase subunit 2 [EC:2.5.1.77] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| CSL4, EXOSC1; exosome complex component CSL4 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| DKC1, NOLA4, CBF5; H/ACA ribonucleoprotein complex subunit 4 [EC:5.4.99.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| DOHH; deoxyhypusine monooxygenase [EC:1.14.99.29] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| DPH1, dph2; 2-(3-amino-3-carboxypropyl)histidine synthase [EC:2.5.1.108] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| DPH5; diphthine methyl ester synthase [EC:2.1.1.314] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| dtdA, GEK1; D-aminoacyl-tRNA deacylase [EC:3.1.1.96] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| E2.5.1.41; phosphoglycerol geranylgeranyltransferase [EC:2.5.1.41] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| E2.5.1.42; geranylgeranylglycerol-phosphate geranylgeranyltransferase [EC:2.5.1.42] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| E2.7.7.3B; pantetheine-phosphate adenylyltransferase [EC:2.7.7.3] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| E2.7.8.39; archaetidylinositol phosphate synthase [EC:2.7.8.39] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EEF1A; elongation factor 1-alpha | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EEF1B; elongation factor 1-beta | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EEF2; elongation factor 2 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EGD2, NACA; nascent polypeptide-associated complex subunit alpha | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EIF1A; translation initiation factor 1A | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EIF2S1; translation initiation factor 2 subunit 1 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EIF2S2; translation initiation factor 2 subunit 2 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EIF2S3; translation initiation factor 2 subunit 3 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EIF5A; translation initiation factor 5A | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EIF5B; translation initiation factor 5B | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EIF6; translation initiation factor 6 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| EMG1, NEP1; rRNA small subunit pseudouridine methyltransferase Nep1 [EC:2.1.1.260] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| FEN1, RAD2; flap endonuclease-1 [EC:3.-.-.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| flpA; fibrillarin-like pre-rRNA processing protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| gar1; RNA-binding protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| gatD; glutamyl-tRNA(Gln) amidotransferase subunit D [EC:6.3.5.7] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| gatE; glutamyl-tRNA(Gln) amidotransferase subunit E [EC:6.3.5.7] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| gch3; GTP cyclohydrolase IIa [EC:3.5.4.29] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| GGR; digeranylgeranylglycerophospholipid reductase [EC:1.3.1.101 1.3.7.11] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| GNPNAT1, GNA1; glucosamine-phosphate N-acetyltransferase [EC:2.3.1.4] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| GRC3, NOL9; polynucleotide 5’-hydroxyl-kinase GRC3/NOL9 [EC:2.7.1.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| hjr; holliday junction resolvase Hjr [EC:3.1.22.4] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ipk; isopentenyl phosphate kinase [EC:2.7.4.26] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K06865; ATPase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K06869; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07108; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07158; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07176; putative serine/threonine protein kinase | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07398; conserved protein with predicted RNA binding PUA domain | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07558, cca; tRNA nucleotidyltransferase (CCA-adding enzyme) [EC:2.7.7.72] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07572; putative nucleotide binding protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07575; PUA domain protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07580; Zn-ribbon RNA-binding protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K07581; RNA-binding protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K08975; putative membrane protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K08979; putative membrane protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K09142; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K09148; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K09152; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K09721; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K09722, pps; 4-phosphopantoate—beta-alanine ligase [EC:6.3.2.36] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K09735; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K09736; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K09738; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K16306; fructose-bisphosphate aldolase / 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate synthase [EC:4.1.2.13 2.2.1.10] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K18593; 4-hydroxybutyryl-CoA synthetase (ADP-forming) [EC:6.2.1.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K18594; 3-hydroxypropionyl-CoA synthetase (ADP-forming) [EC:6.2.1.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K18601; aldehyde dehydrogenase [EC:1.2.1.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K18602; malonic semialdehyde reductase [EC:1.1.1.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K18603; acetyl-CoA/propionyl-CoA carboxylase [EC:6.4.1.2 6.4.1.3] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K18604; acetyl-CoA/propionyl-CoA carboxylase [EC:6.4.1.2 6.4.1.3 2.1.3.15] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K18605; biotin carboxyl carrier protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| KRR1; ribosomal RNA assembly protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| MBF1; putative transcription factor | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mcm, cdc21; replicative DNA helicase Mcm [EC:3.6.4.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mptE; 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase [EC:2.7.6.3] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| mtrA; tetrahydromethanopterin S-methyltransferase subunit A [EC:2.1.1.86] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| nob1; endoribonuclease Nob1 [EC:3.1.-.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| NOP10, NOLA3; H/ACA ribonucleoprotein complex subunit 3 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| NOP56; nucleolar protein 56 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| PCNA; proliferating cell nuclear antigen | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| PDCD5, TFAR19; programmed cell death protein 5 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| PELO, DOM34, pelA; protein pelota | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| pfdA, PFDN5; prefoldin alpha subunit | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| pfdB, PFDN6; prefoldin beta subunit | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| pok; pantoate kinase [EC:2.7.1.169] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| pol; DNA polymerase, archaea type [EC:2.7.7.7] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| polB; DNA polymerase II small subunit [EC:2.7.7.7] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| polC; DNA polymerase II large subunit [EC:2.7.7.7] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| POP4, RPP29; ribonuclease P protein subunit POP4 [EC:3.1.26.5] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| priL, pri2, priB; DNA primase large subunit [EC:2.7.7.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| priS, pri1, priA; DNA primase small subunit [EC:2.7.7.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| PUS10; tRNA pseudouridine synthase 10 [EC:5.4.99.25] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RAD51; DNA repair protein RAD51 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rfcL; replication factor C large subunit | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rfcS; replication factor C small subunit | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rfk; riboflavin kinase, archaea type [EC:2.7.1.161] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RIB7, arfC; 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5’-phosphate reductase [EC:1.1.1.302] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| ribL; FAD synthetase [EC:2.7.7.2] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L10e, RPL10; large subunit ribosomal protein L10e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L12, rpl12; large subunit ribosomal protein L12 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L15e, RPL15; large subunit ribosomal protein L15e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L18e, RPL18; large subunit ribosomal protein L18e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L19e, RPL19; large subunit ribosomal protein L19e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L21e, RPL21; large subunit ribosomal protein L21e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L24e, RPL24; large subunit ribosomal protein L24e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L30e, RPL30; large subunit ribosomal protein L30e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L31e, RPL31; large subunit ribosomal protein L31e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L32e, RPL32; large subunit ribosomal protein L32e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L37Ae, RPL37A; large subunit ribosomal protein L37Ae | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L39e, RPL39; large subunit ribosomal protein L39e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L44e, RPL44; large subunit ribosomal protein L44e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L4e, RPL4; large subunit ribosomal protein L4e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-L7Ae, RPL7A; large subunit ribosomal protein L7Ae | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S17e, RPS17; small subunit ribosomal protein S17e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S19e, RPS19; small subunit ribosomal protein S19e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S24e, RPS24; small subunit ribosomal protein S24e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S25e, RPS25; small subunit ribosomal protein S25e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S26e, RPS26; small subunit ribosomal protein S26e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S27Ae, RPS27A; small subunit ribosomal protein S27Ae | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S27e, RPS27; small subunit ribosomal protein S27e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S28e, RPS28; small subunit ribosomal protein S28e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S30e, RPS30; small subunit ribosomal protein S30e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S3Ae, RPS3A; small subunit ribosomal protein S3Ae | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S4e, RPS4; small subunit ribosomal protein S4e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S6e, RPS6; small subunit ribosomal protein S6e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RP-S8e, RPS8; small subunit ribosomal protein S8e | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RPB6, POLR2F; DNA-directed RNA polymerases I, II, and III subunit RPABC2 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoA1; DNA-directed RNA polymerase subunit A’ [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoB; DNA-directed RNA polymerase subunit B [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoD; DNA-directed RNA polymerase subunit D [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoE1; DNA-directed RNA polymerase subunit E’ [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoE2; DNA-directed RNA polymerase subunit E" [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoF; DNA-directed RNA polymerase subunit F [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoH; DNA-directed RNA polymerase subunit H [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoL; DNA-directed RNA polymerase subunit L [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoN; DNA-directed RNA polymerase subunit N [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| rpoP; DNA-directed RNA polymerase subunit P [EC:2.7.7.6] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RRP4, EXOSC2; exosome complex component RRP4 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RRP41, EXOSC4, SKI6; exosome complex component RRP41 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| RRP42, EXOSC7; exosome complex component RRP42 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| SDO1, SBDS; ribosome maturation protein SDO1 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| SEC61A; protein transport protein SEC61 subunit alpha | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| SEC61G, SSS1, secE; protein transport protein SEC61 subunit gamma and related proteins | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| SRP19; signal recognition particle subunit SRP19 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| STT3; dolichyl-diphosphooligosaccharide—protein glycosyltransferase [EC:2.4.99.18] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TAN1, THUMPD1; tRNA acetyltransferase TAN1 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TBP, tbp; transcription initiation factor TFIID TATA-box-binding protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TFIIE1, GTF2E1, TFA1, tfe; transcription initiation factor TFIIE subunit alpha | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| tfs; transcription termination factor TFS | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| tgtA; 7-cyano-7-deazaguanine tRNA-ribosyltransferase [EC:2.4.2.48] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| tiaS; tRNA(Ile2)-agmatinylcytidine synthase [EC:6.3.4.22] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TOP1; DNA topoisomerase I [EC:5.99.1.2] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TP53RK, PRPK, BUD32; TP53 regulating kinase and related kinases [EC:2.7.11.1] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TRM5, TRMT5; tRNA (guanine37-N1)-methyltransferase [EC:2.1.1.228] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TRMT1, trm1; tRNA (guanine26-N2/guanine27-N2)-dimethyltransferase [EC:2.1.1.215 2.1.1.216] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TSR3; pre-rRNA-processing protein TSR3 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| TYW1; tRNA wybutosine-synthesizing protein 1 [EC:4.1.3.44] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| uppS, cpdS; tritrans,polycis-undecaprenyl-diphosphate synthase [geranylgeranyl-diphosphate specific] [EC:2.5.1.89] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| wtpB; molybdate/tungstate transport system permease protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| wtpC; molybdate/tungstate transport system ATP-binding protein [EC:3.6.3.- 3.6.3.55] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| yfkT; spore germination protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| pucA; light-harvesting protein B-800-850 alpha chain | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.37193 | 92.000 | 0.17342 | 0.77376 |
| amnA; 2-aminophenol/2-amino-5-chlorophenol 1,6-dioxygenase subunit alpha | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| amnB; 2-aminophenol/2-amino-5-chlorophenol 1,6-dioxygenase subunit beta [EC:1.13.11.74 1.13.11.76] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| BHMT; betaine-homocysteine S-methyltransferase [EC:2.1.1.5] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| desZ; 3-O-methylgallate 3,4-dioxygenase [EC:1.13.11.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| nga; NAD+ glycohydrolase [EC:3.2.2.5] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| sfb1; fibronectin-binding protein 1 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| dgaR; sigma-54 dependent transcriptional regulator, dga operon transcriptional activator | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.78906 | 98.990 | 0.43196 | 0.82810 |
| idnR, gntH; LacI family transcriptional regulator, gluconate utilization system Gnt-II transcriptional activator | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.78906 | 98.990 | 0.43196 | 0.82810 |
| PTS-Dga-EIIA, dgaA; PTS system, D-glucosaminate-specific IIA component [EC:2.7.1.203] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.78906 | 98.990 | 0.43196 | 0.82810 |
| tutB; tyrosine permease | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -0.78906 | 98.990 | 0.43196 | 0.82810 |
| hsf; adhesin Hsf | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.41964 | 92.000 | 0.15909 | 0.77376 |
| K18132, porA; major outer membrane protein P.IA | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.41964 | 92.000 | 0.15909 | 0.77376 |
| sphB1, nalP, ausP; autotransporter serine protease [EC:3.4.21.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.41964 | 92.000 | 0.15909 | 0.77376 |
| xopD; type III effector protein XopD | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| glmU; UDP-N-acetylglucosamine pyrophosphorylase [EC:2.7.7.23] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| IDS; iduronate 2-sulfatase [EC:3.1.6.13] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| sfmA; type 1 fimbrial protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| boNT; botulinum neurotoxin [EC:3.4.24.69] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.41575 | 92.000 | 0.16023 | 0.77376 |
| ntnH; botulinum neurotoxin type non-toxic component | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.41575 | 92.000 | 0.16023 | 0.77376 |
| actA; actin-assembly inducing protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.39323 | 92.000 | 0.16691 | 0.77376 |
| mpl; zinc metalloproteinase [EC:3.4.24.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.39323 | 92.000 | 0.16691 | 0.77376 |
| bacC; dihydroanticapsin dehydrogenase [EC:1.1.1.385] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| bbsH; E-phenylitaconyl-CoA hydratase [EC:4.2.1.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| hph; hygromycin-B 4-O-kinase [EC:2.7.1.163] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| K18612; 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylate dehydrogenase [EC:1.2.1.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| K18613; 3-hydroxy-2-methylpyridine-4,5-dicarboxylate 4-decarboxylase [EC:4.1.1.51] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| K18614; 2-(acetamidomethylene)succinate hydrolase [EC:3.5.1.29] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| mhpco; 2-methyl-3-hydroxypyridine 5-carboxylic acid dioxygenase [EC:1.14.12.4] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| pdla; 4-pyridoxolactonase [EC:3.1.1.27] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| pldh; pyridoxal 4-dehydrogenase [EC:1.1.1.107] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| pno; pyridoxine 4-oxidase [EC:1.1.3.12] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| ppaT; pyridoxamine—pyruvate transaminase [EC:2.6.1.30] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| SUGCT; succinate—hydroxymethylglutarate CoA-transferase [EC:2.8.3.13] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| K07063; uncharacterized protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 0.48642 | 91.550 | 0.62783 | 0.89365 |
| bchJ; divinyl protochlorophyllide a 8-vinyl-reductase [EC:1.-.-.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.37193 | 92.000 | 0.17342 | 0.77376 |
| crtA; spheroidene monooxygenase [EC:1.14.15.9] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.37193 | 92.000 | 0.17342 | 0.77376 |
| dddL; dimethylpropiothetin dethiomethylase [EC:4.4.1.3] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.37193 | 92.000 | 0.17342 | 0.77376 |
| DVR; divinyl chlorophyllide a 8-vinyl-reductase [EC:1.3.1.75] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.37193 | 92.000 | 0.17342 | 0.77376 |
| E3.6.3.17; monosaccharide-transporting ATPase [EC:3.6.3.17] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.37193 | 92.000 | 0.17342 | 0.77376 |
| pucB; light-harvesting protein B-800-850 beta chain | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.37193 | 92.000 | 0.17342 | 0.77376 |
| pufX; photosynthetic reaction center PufX protein | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.37193 | 92.000 | 0.17342 | 0.77376 |
| tauX; taurine dehydrogenase small subunit [EC:1.4.2.-] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.37193 | 92.000 | 0.17342 | 0.77376 |
| E3.2.1.73; licheninase [EC:3.2.1.73] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| K18237; ribose 1,5-bisphosphate isomerase [EC:5.3.1.29] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | 1.00000 | 64.000 | 0.32108 | 0.77376 |
| PDC, pdc; pyruvate decarboxylase [EC:4.1.1.1] | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
| K14486, ARF; auxin response factor | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00000 | -1.00000 | 92.000 | 0.31993 | 0.77376 |
plot.dat <- tb.ra %>%
arrange(desc(`Overall Mean`)) %>%
slice_head(n=50)%>%
mutate(
description = fct_reorder(description, `Overall Mean`),
description = factor(description, levels = levels(description), ordered=T)
) %>%
arrange(description) %>%
mutate(
id = 1:n(),
step = ifelse(id%%2 == 0, 1, 0),
Mean_diff = `Tumor Mean` - `Non-Tumor Mean`,
diff_se = SEpooled,
Mean_diff_ll = Mean_diff - qt(0.975, df)*diff_se,
Mean_diff_ul = Mean_diff + qt(0.975, df)*diff_se
) %>%
pivot_longer(
cols=contains("Mean"),
names_to = "group",
values_to = "mean"
)
p1.d <- plot.dat %>%
filter(group %in% c("Tumor Mean","Non-Tumor Mean")) %>%
mutate(
group = ifelse(group == "Tumor Mean", "Tumor", "Non-Tumor"),
col = ifelse(step == 1, "grey90", "white"),
h=1, w=Inf
)
p1 <- ggplot()+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_bar(data=p1.d,
aes(x=mean, y=description,
group=group, color=group,
fill=group),
stat="identity",position = "dodge",
alpha = 1)+
labs(x="Mean Proportion (%)")+
theme_classic()+
theme(
legend.position = "bottom",
plot.margin = unit(c(1,0,1,1), "lines")
)
p2.d <- plot.dat %>%
filter(group %in% c("Mean_diff", "Mean_diff_ll", "Mean_diff_ul")) %>%
pivot_wider(
names_from = group,
values_from = mean
) %>%
mutate(
group = ifelse(Mean_diff > 0, "Tumor", "Non-Tumor"),
p = sprintf("%.3f", round(p,3)),
ll = min(Mean_diff_ll)-0.01,
ul = max(Mean_diff_ul)+0.01
)
p2<-ggplot(p2.d, aes(x=Mean_diff, y=description))+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_vline(xintercept = 0, linetype="dashed", alpha=0.5)+
geom_segment(aes(x=Mean_diff_ll, y=description, xend=Mean_diff_ul, yend=description))+
geom_point(aes(fill=group, color=group))+
geom_text(aes(label=p, x=unique(ul)+0.1))+
coord_cartesian(xlim = c(unique(p2.d$ll), unique(p2.d$ul)),
clip = 'off') +
annotate("text", x=unique(p2.d$ul)+0.2,y = 25,
angle=90,
label="p-value (uncorrected)")+
labs(x="Mean Difference in Proportions")+
theme_classic()+
theme(
legend.position = "bottom",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
plot.margin = unit(c(1,4,1,0), "lines")
)
# plot
p1 + p2+
plot_annotation(title="KO Data: 50 most abundant descriptions")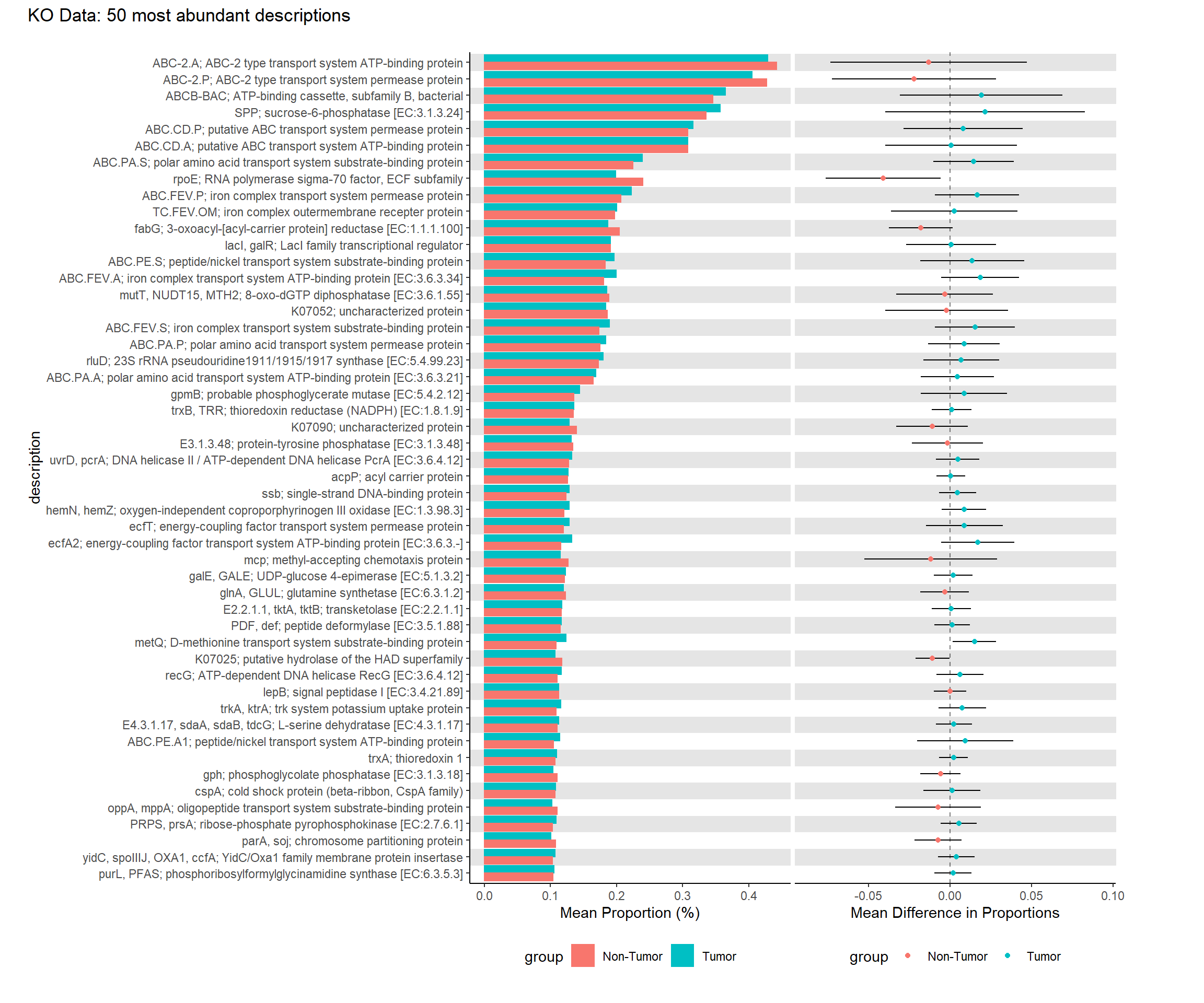
Modeling Difference between Tumor and Non-Tumor
For the modeling, we used a generalized linear mixed model (GLMM).
First, we looked at the biserial correlation between the abundance of each description and the tumor status.
tb <- mydata %>%
group_by(description)%>%
summarise(
r = cor(tumor, Abundance)
) %>%
mutate(
M=mean(r)
)
ggplot(tb, aes(x=r))+
geom_density()+
geom_vline(aes(xintercept = M))+
labs(x="Biserial Correlation",title="Relationship between description abundance and tumor (tumor vs. non-tumor)")+
theme(panel.grid = element_blank())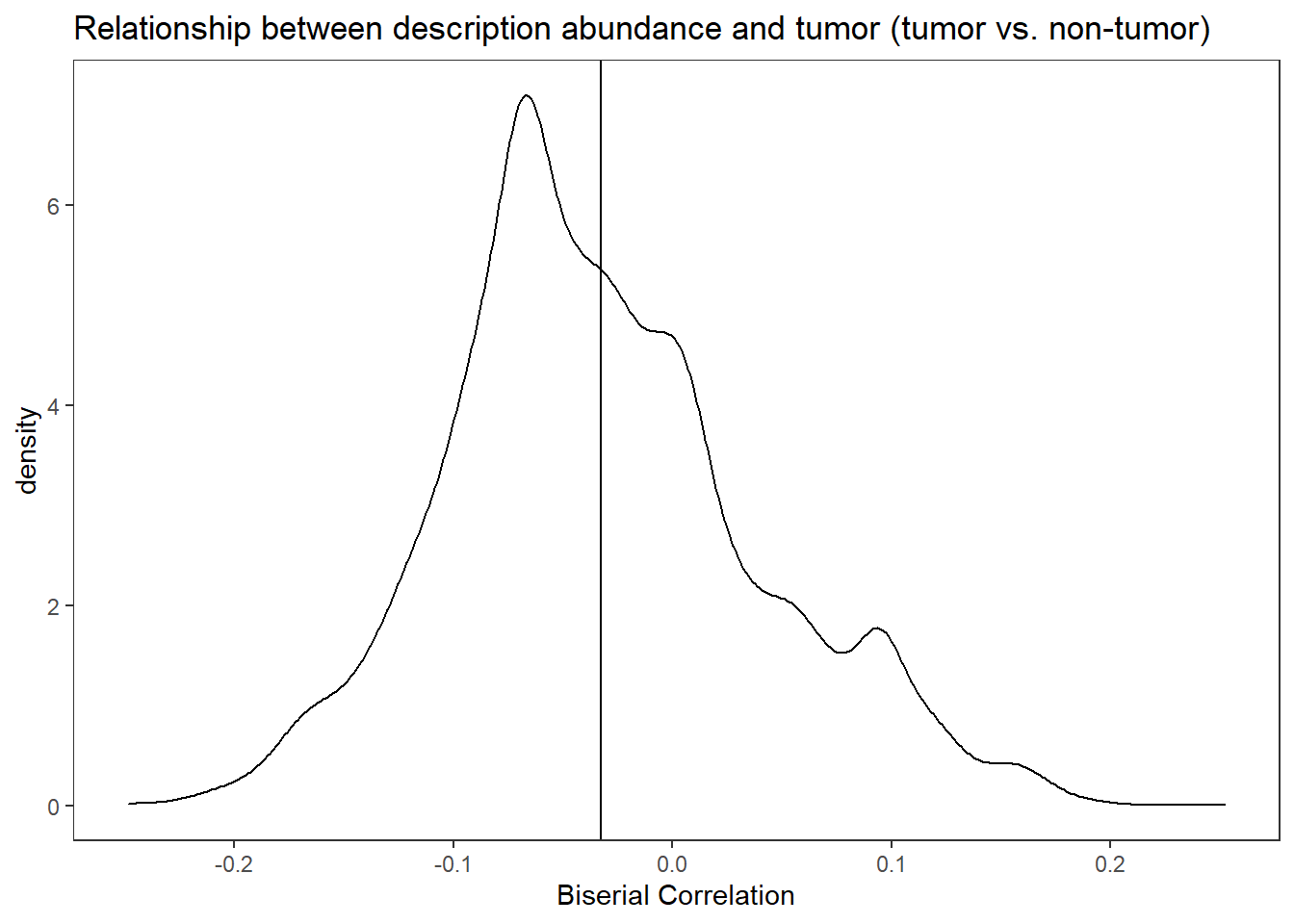
Next, we need the data to be on an interpretable scale. First, let’s use the raw abundance like scale.
p <- ggplot(mydata, aes(x=Abundance))+
geom_density()
p
mydata <- mydata %>%
mutate(Abundance.dich=ifelse(Abundance==0, 0, 1))
table(mydata$Abundance.dich)
0 1
484856 599814 Let’s first run models by description so that we can avoid the nesting issue initially. We will come back to this to conduct the final model (it will be more powerful).
DESCRIPTIONS <- unique(mydata$description)
i<-1
dat0 <- mydata %>% filter(description==DESCRIPTIONS[i])
# quasipossion (approximately negative binom) give an approx. answer.
fit0 <- glm(
Abundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
summary(fit0)
Call:
glm(formula = Abundance ~ 1 + tumor, family = quasipoisson(link = "log"),
data = dat0)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.389 -1.389 -1.389 -0.656 21.089
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.0365 0.6350 -0.06 0.95
tumor -1.4989 1.7279 -0.87 0.39
(Dispersion parameter for quasipoisson family taken to be 36.153)
Null deviance: 805.22 on 157 degrees of freedom
Residual deviance: 767.38 on 156 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 8NExt, let’s model the percent relative abundance. This will allow us to make inference about the difference average relative abundance. Which is a simpler interpretation than trying to model differences in the log relative abundance which will need to be interpreted as the multiplicative relative change.
results.out <- as.data.frame(matrix(ncol=4, nrow=length(DESCRIPTIONS)))
colnames(results.out) <- c("description", "Est", "SE", "p")
#results.out$description <-DESCRIPTIONS
i <- 1
for(i in 1:length(DESCRIPTIONS)){
#for(i in 1:5){
dat0 <- mydata %>%
filter(description == DESCRIPTIONS[i])%>%
mutate(RelAbundance= RelAbundance*100)
fit0 <- glm(
RelAbundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
fit.sum <- summary(fit0)
results.out[i, 1] <- DESCRIPTIONS[i]
results.out[i, 2:4] <- fit.sum$coefficients[2, c(1,2,4)]
}
results.out$fdr_p <- p.adjust(results.out$p, method="fdr")
kable(results.out, format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="600px")| description | Est | SE | p | fdr_p |
|---|---|---|---|---|
| 2PGK; 2-phosphoglycerate kinase [EC:2.7.2.-] | -1.929 | 1.989 | 0.334 | 0.978 |
| 4CL; 4-coumarate–CoA ligase [EC:6.2.1.12] | -0.780 | 0.986 | 0.430 | 0.978 |
| 4hbD, abfH; 4-hydroxybutyrate dehydrogenase [EC:1.1.1.61] | 0.079 | 0.466 | 0.865 | 0.999 |
| aac2-I; aminoglycoside 2’-N-acetyltransferase I [EC:2.3.1.59] | -0.081 | 0.760 | 0.915 | 0.999 |
| aac3-I; aminoglycoside 3-N-acetyltransferase I [EC:2.3.1.60] | 0.592 | 1.206 | 0.624 | 0.999 |
| aac6-I, aacA7; aminoglycoside 6’-N-acetyltransferase I [EC:2.3.1.82] | 0.199 | 0.468 | 0.671 | 0.999 |
| aac6-I; aminoglycoside 6’-N-acetyltransferase I [EC:2.3.1.82] | -1.307 | 1.417 | 0.357 | 0.978 |
| aac6-Ib; aminoglycoside 6’-N-acetyltransferase Ib [EC:2.3.1.82] | 0.210 | 1.071 | 0.845 | 0.999 |
| aac6-II; aminoglycoside 6’-N-acetyltransferase II [EC:2.3.1.82] | 18.165 | 2857.430 | 0.995 | 0.999 |
| aacA; aminoglycoside 6’-N-acetyltransferase [EC:2.3.1.82] | -1.286 | 0.818 | 0.118 | 0.917 |
| aacC; aminoglycoside 3-N-acetyltransferase [EC:2.3.1.81] | 0.162 | 0.354 | 0.648 | 0.999 |
| AACS, acsA; acetoacetyl-CoA synthetase [EC:6.2.1.16] | -0.313 | 0.287 | 0.277 | 0.978 |
| aadA; streptomycin 3"-adenylyltransferase [EC:2.7.7.47] | -0.327 | 0.425 | 0.443 | 0.978 |
| aadK; aminoglycoside 6-adenylyltransferase [EC:2.7.7.-] | -0.045 | 0.167 | 0.789 | 0.999 |
| aaeA; p-hydroxybenzoic acid efflux pump subunit AaeA | 0.134 | 0.621 | 0.829 | 0.999 |
| aaeB; p-hydroxybenzoic acid efflux pump subunit AaeB | 0.134 | 0.621 | 0.829 | 0.999 |
| aapJ, bztA; general L-amino acid transport system substrate-binding protein | -0.331 | 0.283 | 0.244 | 0.978 |
| aapM, bztC; general L-amino acid transport system permease protein | -0.089 | 0.266 | 0.740 | 0.999 |
| aapP, bztD; general L-amino acid transport system ATP-binding protein [EC:3.6.3.-] | -0.125 | 0.291 | 0.668 | 0.999 |
| aapQ, bztB; general L-amino acid transport system permease protein | -0.090 | 0.267 | 0.738 | 0.999 |
| aarC, cat1; succinyl-CoA:acetate CoA-transferase [EC:2.8.3.18] | -0.018 | 0.139 | 0.897 | 0.999 |
| AARS, alaS; alanyl-tRNA synthetase [EC:6.1.1.7] | 0.050 | 0.040 | 0.205 | 0.972 |
| AARSD1, ALAX; misacylated tRNA(Ala) deacylase [EC:3.1.1.-] | -1.207 | 0.555 | 0.031 | 0.839 |
| aas; acyl-[acyl-carrier-protein]-phospholipid O-acyltransferase / long-chain-fatty-acid–[acyl-carrier-protein] ligase [EC:2.3.1.40 6.2.1.20] | 0.533 | 0.264 | 0.045 | 0.839 |
| aat; leucyl/phenylalanyl-tRNA—protein transferase [EC:2.3.2.6] | -0.117 | 0.151 | 0.442 | 0.978 |
| aauR; two-component system, response regulator AauR | 0.235 | 0.343 | 0.495 | 0.981 |
| aauS; two-component system, sensor histidine kinase AauS [EC:2.7.13.3] | 0.235 | 0.343 | 0.495 | 0.981 |
| ABC-2.A; ABC-2 type transport system ATP-binding protein | -0.030 | 0.071 | 0.669 | 0.999 |
| ABC-2.CPSE.A; capsular polysaccharide transport system ATP-binding protein [EC:3.6.3.38] | -0.138 | 0.379 | 0.715 | 0.999 |
| ABC-2.CPSE.P; capsular polysaccharide transport system permease protein | -0.041 | 0.382 | 0.914 | 0.999 |
| ABC-2.CPSE.P1; capsular polysaccharide transport system permease protein | -0.147 | 0.376 | 0.696 | 0.999 |
| ABC-2.CYL.A, cylA; multidrug/hemolysin transport system ATP-binding protein | -0.013 | 0.168 | 0.938 | 0.999 |
| ABC-2.CYL.P, cylB; multidrug/hemolysin transport system permease protein | -0.014 | 0.168 | 0.935 | 0.999 |
| ABC-2.LPSE.A; lipopolysaccharide transport system ATP-binding protein | -0.164 | 0.252 | 0.517 | 0.986 |
| ABC-2.LPSE.P; lipopolysaccharide transport system permease protein | -0.045 | 0.206 | 0.829 | 0.999 |
| ABC-2.OM, wza; polysaccharide biosynthesis/export protein | -0.116 | 0.111 | 0.299 | 0.978 |
| ABC-2.P; ABC-2 type transport system permease protein | -0.053 | 0.062 | 0.394 | 0.978 |
| ABC-2.TX; HlyD family secretion protein | -0.149 | 0.118 | 0.208 | 0.976 |
| ABC.ARG.S, argT; lysine/arginine/ornithine transport system substrate-binding protein | -0.091 | 0.490 | 0.853 | 0.999 |
| ABC.CD.A; putative ABC transport system ATP-binding protein | 0.002 | 0.069 | 0.978 | 0.999 |
| ABC.CD.P; putative ABC transport system permease protein | 0.026 | 0.061 | 0.675 | 0.999 |
| ABC.CD.TX; HlyD family secretion protein | -0.108 | 0.092 | 0.242 | 0.978 |
| ABC.CYST.A; cystine transport system ATP-binding protein [EC:3.6.3.-] | 0.146 | 0.150 | 0.333 | 0.978 |
| ABC.CYST.P; cystine transport system permease protein | 0.172 | 0.135 | 0.205 | 0.972 |
| ABC.FEV.A; iron complex transport system ATP-binding protein [EC:3.6.3.34] | 0.098 | 0.060 | 0.103 | 0.904 |
| ABC.FEV.P; iron complex transport system permease protein | 0.077 | 0.058 | 0.188 | 0.966 |
| ABC.FEV.S; iron complex transport system substrate-binding protein | 0.084 | 0.065 | 0.196 | 0.970 |
| ABC.GGU.A, gguA; putative multiple sugar transport system ATP-binding protein [EC:3.6.3.17] | -0.205 | 0.292 | 0.485 | 0.979 |
| ABC.GGU.P, gguB; putative multiple sugar transport system permease protein | -0.051 | 0.261 | 0.845 | 0.999 |
| ABC.GGU.S, chvE; putative multiple sugar transport system substrate-binding protein | -0.247 | 0.303 | 0.417 | 0.978 |
| ABC.GLN1.A; putative glutamine transport system ATP-binding protein [EC:3.6.3.-] | 0.051 | 0.168 | 0.760 | 0.999 |
| ABC.GLN1.P; putative glutamine transport system permease protein | 0.044 | 0.162 | 0.789 | 0.999 |
| ABC.GLN1.S; putative glutamine transport system substrate-binding protein | 0.063 | 0.172 | 0.713 | 0.999 |
| ABC.MN.A; manganese/iron transport system ATP-binding protein | 0.129 | 0.343 | 0.706 | 0.999 |
| ABC.MN.P; manganese/iron transport system permease protein | 0.120 | 0.289 | 0.678 | 0.999 |
| ABC.MN.S; manganese/iron transport system substrate-binding protein | -1.025 | 0.742 | 0.169 | 0.961 |
| ABC.MR.TX; HlyD family secretion protein | -0.947 | 0.476 | 0.049 | 0.839 |
| ABC.MR; putative ABC transport system ATP-binding protein | -0.359 | 0.627 | 0.568 | 0.995 |
| ABC.MS.P; multiple sugar transport system permease protein | -0.146 | 0.115 | 0.206 | 0.974 |
| ABC.MS.P1; multiple sugar transport system permease protein | -0.089 | 0.125 | 0.478 | 0.979 |
| ABC.MS.S; multiple sugar transport system substrate-binding protein | -0.051 | 0.112 | 0.650 | 0.999 |
| ABC.NGC.P; N-acetylglucosamine transport system permease protein | -0.072 | 0.763 | 0.925 | 0.999 |
| ABC.NGC.P1; N-acetylglucosamine transport system permease protein | 0.325 | 0.407 | 0.426 | 0.978 |
| ABC.NGC.S; N-acetylglucosamine transport system substrate-binding protein | -0.072 | 0.763 | 0.925 | 0.999 |
| ABC.PA.A; polar amino acid transport system ATP-binding protein [EC:3.6.3.21] | 0.027 | 0.070 | 0.705 | 0.999 |
| ABC.PA.P; polar amino acid transport system permease protein | 0.048 | 0.064 | 0.462 | 0.979 |
| ABC.PA.S; polar amino acid transport system substrate-binding protein | 0.062 | 0.056 | 0.268 | 0.978 |
| ABC.PE.A; peptide/nickel transport system ATP-binding protein | 0.066 | 0.154 | 0.670 | 0.999 |
| ABC.PE.A1; peptide/nickel transport system ATP-binding protein | 0.085 | 0.127 | 0.507 | 0.981 |
| ABC.PE.P; peptide/nickel transport system permease protein | 0.051 | 0.124 | 0.683 | 0.999 |
| ABC.PE.P1; peptide/nickel transport system permease protein | 0.049 | 0.122 | 0.691 | 0.999 |
| ABC.PE.S; peptide/nickel transport system substrate-binding protein | 0.071 | 0.081 | 0.380 | 0.978 |
| ABC.SN.A; NitT/TauT family transport system ATP-binding protein | -0.247 | 0.127 | 0.053 | 0.839 |
| ABC.SN.P; NitT/TauT family transport system permease protein | -0.204 | 0.141 | 0.150 | 0.954 |
| ABC.SN.S; NitT/TauT family transport system substrate-binding protein | -0.229 | 0.133 | 0.087 | 0.880 |
| ABC.SP.A; putative spermidine/putrescine transport system ATP-binding protein | -0.191 | 0.268 | 0.478 | 0.979 |
| ABC.SP.P; putative spermidine/putrescine transport system permease protein | -0.226 | 0.281 | 0.423 | 0.978 |
| ABC.SP.P1; putative spermidine/putrescine transport system permease protein | -0.206 | 0.280 | 0.464 | 0.979 |
| ABC.SP.S; putative spermidine/putrescine transport system substrate-binding protein | -0.127 | 0.267 | 0.635 | 0.999 |
| ABC.SS.A; simple sugar transport system ATP-binding protein [EC:3.6.3.17] | -0.021 | 0.085 | 0.804 | 0.999 |
| ABC.SS.P; simple sugar transport system permease protein | 0.010 | 0.086 | 0.909 | 0.999 |
| ABC.SS.S; simple sugar transport system substrate-binding protein | -0.251 | 0.207 | 0.229 | 0.978 |
| ABC.VB12.A, btuD; vitamin B12 transport system ATP-binding protein [EC:3.6.3.33] | 0.218 | 0.576 | 0.706 | 0.999 |
| ABC.VB12.P, btuC; vitamin B12 transport system permease protein | 0.218 | 0.576 | 0.706 | 0.999 |
| ABC.VB12.S1, btuF; vitamin B12 transport system substrate-binding protein | 0.172 | 0.298 | 0.564 | 0.994 |
| ABC.VB1X.A; putative thiamine transport system ATP-binding protein | 0.067 | 0.570 | 0.906 | 0.999 |
| ABC.VB1X.P; putative thiamine transport system permease protein | 0.067 | 0.570 | 0.906 | 0.999 |
| ABC.VB1X.S; putative thiamine transport system substrate-binding protein | 0.067 | 0.570 | 0.906 | 0.999 |
| ABC.X2.A; putative ABC transport system ATP-binding protein | -0.061 | 0.200 | 0.759 | 0.999 |
| ABC.X2.P; putative ABC transport system permease protein | -0.117 | 0.174 | 0.502 | 0.981 |
| ABC.X4.A; putative ABC transport system ATP-binding protein | 0.085 | 0.097 | 0.383 | 0.978 |
| ABC.X4.P; putative ABC transport system permease protein | 0.083 | 0.098 | 0.399 | 0.978 |
| ABC.X4.S; putative ABC transport system substrate-binding protein | 0.106 | 0.093 | 0.255 | 0.978 |
| ABC.ZM.A; zinc/manganese transport system ATP-binding protein | 0.048 | 0.216 | 0.824 | 0.999 |
| ABC.ZM.P; zinc/manganese transport system permease protein | 0.046 | 0.203 | 0.820 | 0.999 |
| ABC.ZM.S; zinc/manganese transport system substrate-binding protein | 0.212 | 0.202 | 0.297 | 0.978 |
| abcA, bmrA; ATP-binding cassette, subfamily B, bacterial AbcA/BmrA [EC:3.6.3.44] | -0.199 | 0.349 | 0.569 | 0.995 |
| ABCB-BAC; ATP-binding cassette, subfamily B, bacterial | 0.054 | 0.071 | 0.450 | 0.978 |
| ABCC-BAC; ATP-binding cassette, subfamily C, bacterial | 0.025 | 0.100 | 0.804 | 0.999 |
| ABCE1, Rli1; ATP-binding cassette, sub-family E, member 1 | -17.234 | 3077.514 | 0.996 | 0.999 |
| ABCF3; ATP-binding cassette, subfamily F, member 3 | 0.022 | 0.042 | 0.595 | 0.999 |
| abfA; alpha-N-arabinofuranosidase [EC:3.2.1.55] | -0.838 | 0.353 | 0.019 | 0.839 |
| abfD; 4-hydroxybutyryl-CoA dehydratase / vinylacetyl-CoA-Delta-isomerase [EC:4.2.1.120 5.3.3.3] | -0.540 | 0.437 | 0.218 | 0.978 |
| abgA; aminobenzoyl-glutamate utilization protein A | 0.076 | 0.261 | 0.773 | 0.999 |
| abgB; aminobenzoyl-glutamate utilization protein B | -0.545 | 0.361 | 0.133 | 0.942 |
| abgR; LysR family transcriptional regulator, regulator of abg operon | 0.233 | 0.698 | 0.738 | 0.999 |
| abgT; aminobenzoyl-glutamate transport protein | 0.404 | 0.258 | 0.120 | 0.917 |
| abiQ; protein AbiQ | 0.560 | 0.390 | 0.153 | 0.954 |
| abmG; 2-aminobenzoate-CoA ligase [EC:6.2.1.32] | -0.961 | 0.592 | 0.106 | 0.911 |
| abnA; arabinan endo-1,5-alpha-L-arabinosidase [EC:3.2.1.99] | -0.677 | 0.441 | 0.126 | 0.927 |
| ABO; histo-blood group ABO system transferase [EC:2.4.1.40 2.4.1.37] | -2.641 | 2.722 | 0.333 | 0.978 |
| abrB; transcriptional pleiotropic regulator of transition state genes | -0.659 | 0.425 | 0.123 | 0.919 |
| ACAA2; acetyl-CoA acyltransferase 2 [EC:2.3.1.16] | -17.970 | 4446.494 | 0.997 | 0.999 |
| ACADL; long-chain-acyl-CoA dehydrogenase [EC:1.3.8.8] | -0.460 | 0.423 | 0.278 | 0.978 |
| ACADM, acd; acyl-CoA dehydrogenase [EC:1.3.8.7] | -0.528 | 0.255 | 0.040 | 0.839 |
| ACADS, bcd; butyryl-CoA dehydrogenase [EC:1.3.8.1] | 0.493 | 0.245 | 0.046 | 0.839 |
| ACADSB; short/branched chain acyl-CoA dehydrogenase [EC:1.3.99.12] | -2.997 | 2.591 | 0.249 | 0.978 |
| accA; acetyl-CoA carboxylase carboxyl transferase subunit alpha [EC:6.4.1.2 2.1.3.15] | 0.035 | 0.055 | 0.528 | 0.989 |
| accB, bccP; acetyl-CoA carboxylase biotin carboxyl carrier protein | -0.032 | 0.059 | 0.591 | 0.999 |
| accC; acetyl-CoA carboxylase, biotin carboxylase subunit [EC:6.4.1.2 6.3.4.14] | -0.035 | 0.057 | 0.539 | 0.990 |
| accD; acetyl-CoA carboxylase carboxyl transferase subunit beta [EC:6.4.1.2 2.1.3.15] | 0.047 | 0.050 | 0.358 | 0.978 |
| accD6; acetyl-CoA/propionyl-CoA carboxylase carboxyl transferase subunit [EC:6.4.1.2 6.4.1.3 2.1.3.15] | -0.957 | 0.903 | 0.291 | 0.978 |
| acd; glutaryl-CoA dehydrogenase (non-decarboxylating) [EC:1.3.99.32] | -2.766 | 1.309 | 0.036 | 0.839 |
| acdA; acetate—CoA ligase (ADP-forming) subunit alpha [EC:6.2.1.13] | -2.857 | 2.948 | 0.334 | 0.978 |
| ACE, CD143; peptidyl-dipeptidase A [EC:3.4.15.1] | -0.216 | 0.394 | 0.585 | 0.999 |
| aceB, glcB; malate synthase [EC:2.3.3.9] | -0.196 | 0.174 | 0.263 | 0.978 |
| aceE; pyruvate dehydrogenase E1 component [EC:1.2.4.1] | -0.072 | 0.155 | 0.642 | 0.999 |
| aceK; isocitrate dehydrogenase kinase/phosphatase [EC:2.7.11.5 3.1.3.-] | 0.105 | 0.219 | 0.632 | 0.999 |
| acfA; accessory colonization factor AcfA | 18.902 | 3402.727 | 0.996 | 0.999 |
| acfC; accessory colonization factor AcfC | -0.816 | 1.161 | 0.483 | 0.979 |
| acfD; accessory colonization factor AcfD | -0.499 | 1.160 | 0.668 | 0.999 |
| ackA; acetate kinase [EC:2.7.2.1] | 0.073 | 0.065 | 0.263 | 0.978 |
| acm; lysozyme | -0.109 | 0.140 | 0.440 | 0.978 |
| acmB; methyl acetate hydrolase [EC:3.1.1.-] | -1.889 | 2.270 | 0.407 | 0.978 |
| ACMSD; aminocarboxymuconate-semialdehyde decarboxylase [EC:4.1.1.45] | -1.252 | 0.627 | 0.047 | 0.839 |
| acnB; aconitate hydratase 2 / 2-methylisocitrate dehydratase [EC:4.2.1.3 4.2.1.99] | -0.015 | 0.191 | 0.936 | 0.999 |
| ACO, acnA; aconitate hydratase [EC:4.2.1.3] | -0.139 | 0.080 | 0.083 | 0.867 |
| acpD, azoR; FMN-dependent NADH-azoreductase [EC:1.7.1.17] | -0.086 | 0.142 | 0.548 | 0.991 |
| acpH; acyl carrier protein phosphodiesterase [EC:3.1.4.14] | 0.276 | 0.549 | 0.616 | 0.999 |
| acpK; polyketide biosynthesis acyl carrier protein | -0.931 | 1.781 | 0.602 | 0.999 |
| acpP; acyl carrier protein | 0.003 | 0.035 | 0.939 | 0.999 |
| acpS; holo-[acyl-carrier protein] synthase [EC:2.7.8.7] | 0.072 | 0.073 | 0.325 | 0.978 |
| ACR3, arsB; arsenite transporter | -0.050 | 0.170 | 0.770 | 0.999 |
| acrA, mexA, adeI, smeD, mtrC, cmeA; membrane fusion protein, multidrug efflux system | -0.104 | 0.120 | 0.387 | 0.978 |
| acrB, mexB, adeJ, smeE, mtrD, cmeB; multidrug efflux pump | -0.076 | 0.185 | 0.681 | 0.999 |
| acrD; multidrug efflux pump | 0.134 | 0.621 | 0.829 | 0.999 |
| acrE; membrane fusion protein, multidrug efflux system | -0.243 | 0.717 | 0.735 | 0.999 |
| acrF; multidrug efflux pump | -0.243 | 0.717 | 0.735 | 0.999 |
| acrR, smeT; TetR/AcrR family transcriptional regulator, acrAB operon repressor | -0.003 | 0.122 | 0.982 | 0.999 |
| acrR; TetR/AcrR family transcriptional regulator, multidrug resistance operon repressor | 0.248 | 0.349 | 0.478 | 0.979 |
| acsB; acetyl-CoA synthase [EC:2.3.1.169] | -0.727 | 1.126 | 0.520 | 0.988 |
| acsE; 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase [EC:2.1.1.258] | -0.789 | 1.083 | 0.468 | 0.979 |
| ACSL, fadD; long-chain acyl-CoA synthetase [EC:6.2.1.3] | -0.098 | 0.089 | 0.275 | 0.978 |
| ACSM; medium-chain acyl-CoA synthetase [EC:6.2.1.2] | -1.859 | 2.280 | 0.416 | 0.978 |
| ACSS, acs; acetyl-CoA synthetase [EC:6.2.1.1] | -0.290 | 0.135 | 0.033 | 0.839 |
| actA; actin-assembly inducing protein | -16.755 | 1738.673 | 0.992 | 0.999 |
| actIV; cyclase [EC:4.-.-.-] | 0.590 | 0.979 | 0.548 | 0.991 |
| actP; cation/acetate symporter | -0.236 | 0.213 | 0.269 | 0.978 |
| actVI1, RED1; ketoreductase RED1 [EC:1.1.1.-] | 0.692 | 1.263 | 0.585 | 0.999 |
| acuA; acetoin utilization protein AcuA [EC:2.3.1.-] | -0.946 | 0.502 | 0.061 | 0.839 |
| acuB; acetoin utilization protein AcuB | -0.053 | 0.145 | 0.717 | 0.999 |
| acuC; acetoin utilization protein AcuC | -0.967 | 0.400 | 0.017 | 0.839 |
| acuI; acrylyl-CoA reductase (NADPH) [EC:1.3.1.-] | -0.116 | 0.281 | 0.681 | 0.999 |
| acxC; acetone carboxylase, gamma subunit [EC:6.4.1.6] | -1.386 | 1.551 | 0.373 | 0.978 |
| acyP; acylphosphatase [EC:3.6.1.7] | 0.064 | 0.088 | 0.472 | 0.979 |
| ada-alkA; AraC family transcriptional regulator, regulatory protein of adaptative response / DNA-3-methyladenine glycosylase II [EC:3.2.2.21] | -0.254 | 0.328 | 0.439 | 0.978 |
| ada; AraC family transcriptional regulator, regulatory protein of adaptative response / methylated-DNA-[protein]-cysteine methyltransferase [EC:2.1.1.63] | -0.217 | 0.164 | 0.189 | 0.966 |
| adaA; AraC family transcriptional regulator, regulatory protein of adaptative response / methylphosphotriester-DNA alkyltransferase methyltransferase [EC:2.1.1.-] | -0.785 | 1.127 | 0.487 | 0.979 |
| adaB; methylated-DNA-[protein]-cysteine S-methyltransferase [EC:2.1.1.63] | 0.015 | 0.477 | 0.975 | 0.999 |
| adc; acetoacetate decarboxylase [EC:4.1.1.4] | -1.294 | 0.758 | 0.090 | 0.882 |
| add, ADA; adenosine deaminase [EC:3.5.4.4] | 0.037 | 0.137 | 0.790 | 0.999 |
| add; aminodeoxyfutalosine deaminase [EC:3.5.4.40] | -0.299 | 0.948 | 0.753 | 0.999 |
| addA; ATP-dependent helicase/nuclease subunit A [EC:3.1.-.- 3.6.4.12] | -0.010 | 0.095 | 0.920 | 0.999 |
| addB; ATP-dependent helicase/nuclease subunit B [EC:3.1.-.- 3.6.4.12] | 0.033 | 0.096 | 0.734 | 0.999 |
| ade; adenine deaminase [EC:3.5.4.2] | -0.259 | 0.263 | 0.327 | 0.978 |
| ADE5; phosphoribosylamine–glycine ligase / phosphoribosylformylglycinamidine cyclo-ligase [EC:6.3.4.13 6.3.3.1] | -3.190 | 3.831 | 0.406 | 0.978 |
| adeA; membrane fusion protein, multidrug efflux system | -0.058 | 0.392 | 0.883 | 0.999 |
| adeB; multidrug efflux pump | -0.031 | 0.378 | 0.936 | 0.999 |
| adeC; outer membrane protein, multidrug efflux system | -0.126 | 0.434 | 0.771 | 0.999 |
| adeN; TetR/AcrR family transcriptional regulator, repressor of the adeIJK operon | -0.267 | 0.454 | 0.557 | 0.992 |
| adeR; two-component system, OmpR family, response regulator AdeR | -0.155 | 0.372 | 0.678 | 0.999 |
| adeS; two-component system, OmpR family, sensor histidine kinase AdeS [EC:2.7.13.3] | -0.129 | 0.383 | 0.738 | 0.999 |
| adh1; NAD+-dependent secondary alcohol dehydrogenase Adh1 [EC:1.1.1.-] | 0.681 | 0.693 | 0.327 | 0.978 |
| adh2; alcohol dehydrogenase [EC:1.1.1.-] | -0.147 | 0.182 | 0.419 | 0.978 |
| adh3; alcohol dehydrogenase [EC:1.1.1.-] | 0.266 | 1.338 | 0.843 | 0.999 |
| adhE; acetaldehyde dehydrogenase / alcohol dehydrogenase [EC:1.2.1.10 1.1.1.1] | 0.108 | 0.109 | 0.326 | 0.978 |
| ADHFE1; hydroxyacid-oxoacid transhydrogenase [EC:1.1.99.24] | -17.756 | 3869.767 | 0.996 | 0.999 |
| adhP; alcohol dehydrogenase, propanol-preferring [EC:1.1.1.1] | -0.019 | 0.107 | 0.862 | 0.999 |
| adiA; arginine decarboxylase [EC:4.1.1.19] | -0.016 | 0.241 | 0.946 | 0.999 |
| adiC; arginine:agmatine antiporter | -0.170 | 0.435 | 0.697 | 0.999 |
| adiY; AraC family transcriptional regulator, transcriptional activator of adiA | -1.145 | 1.438 | 0.427 | 0.978 |
| adk, AK; adenylate kinase [EC:2.7.4.3] | 0.024 | 0.033 | 0.467 | 0.979 |
| adrA; diguanylate cyclase [EC:2.7.7.65] | -0.170 | 0.342 | 0.621 | 0.999 |
| ADT, PDT; arogenate/prephenate dehydratase [EC:4.2.1.91 4.2.1.51] | -17.211 | 2393.329 | 0.994 | 0.999 |
| aefR; TetR/AcrR family transcriptional regulator, regulator of autoinduction and epiphytic fitness | -0.361 | 0.348 | 0.300 | 0.978 |
| aepZ; 2-aminoethylphosphonate-pyruvate transaminase | 0.133 | 0.547 | 0.808 | 0.999 |
| aer; aerotaxis receptor | 0.021 | 0.270 | 0.938 | 0.999 |
| aes; acetyl esterase [EC:3.1.1.-] | -0.330 | 0.220 | 0.135 | 0.944 |
| AFMID; arylformamidase [EC:3.5.1.9] | -0.789 | 0.484 | 0.106 | 0.911 |
| afr; 1,5-anhydro-D-fructose reductase (1,5-anhydro-D-mannitol-forming) [EC:1.1.1.292] | -1.267 | 1.091 | 0.248 | 0.978 |
| aftA; galactan 5-O-arabinofuranosyltransferase [EC:2.4.2.46] | -0.821 | 0.702 | 0.244 | 0.978 |
| aftB; arabinofuranosyltransferase [EC:2.4.2.-] | -0.801 | 0.731 | 0.275 | 0.978 |
| aftC; arabinofuranan 3-O-arabinosyltransferase [EC:2.4.2.47] | -0.796 | 0.696 | 0.255 | 0.978 |
| aftD; arabinofuranan 3-O-arabinosyltransferase [EC:2.4.2.-] | -0.677 | 0.706 | 0.339 | 0.978 |
| afuA, fbpA; iron(III) transport system substrate-binding protein | 0.071 | 0.082 | 0.391 | 0.978 |
| afuB, fbpB; iron(III) transport system permease protein | 0.085 | 0.105 | 0.418 | 0.978 |
| afuC, fbpC; iron(III) transport system ATP-binding protein [EC:3.6.3.30] | 0.057 | 0.097 | 0.555 | 0.992 |
| AGA, aspG; N4-(beta-N-acetylglucosaminyl)-L-asparaginase [EC:3.5.1.26] | -0.132 | 0.369 | 0.721 | 0.999 |
| agaA; N-acetylgalactosamine-6-phosphate deacetylase [EC:3.5.1.25] | -0.005 | 0.732 | 0.995 | 0.999 |
| agaI; galactosamine-6-phosphate isomerase [EC:5.3.1.-] | -1.246 | 1.403 | 0.376 | 0.978 |
| agaR; DeoR family transcriptional regulator, aga operon transcriptional repressor | -0.151 | 0.325 | 0.642 | 0.999 |
| agaS; tagatose-6-phosphate ketose/aldose isomerase [EC:5.-.-.-] | 0.064 | 0.127 | 0.613 | 0.999 |
| agd31B; oligosaccharide 4-alpha-D-glucosyltransferase [EC:2.4.1.161] | -1.060 | 1.749 | 0.545 | 0.990 |
| aglE, ggtB; alpha-glucoside transport system substrate-binding protein | -0.371 | 0.446 | 0.407 | 0.978 |
| aglF, ggtC; alpha-glucoside transport system permease protein | -0.427 | 0.624 | 0.494 | 0.981 |
| aglG, ggtD; alpha-glucoside transport system permease protein | -0.427 | 0.624 | 0.494 | 0.981 |
| aglK; alpha-glucoside transport system ATP-binding protein | -3.373 | 2.075 | 0.106 | 0.911 |
| agp; glucose-1-phosphatase [EC:3.1.3.10] | 0.069 | 0.221 | 0.755 | 0.999 |
| AGPS, agpS; alkyldihydroxyacetonephosphate synthase [EC:2.5.1.26] | -0.997 | 0.646 | 0.125 | 0.923 |
| agrA, blpR, fsrA; two-component system, LytTR family, response regulator AgrA | -0.123 | 0.195 | 0.528 | 0.989 |
| agrB; accessory gene regulator B | 0.008 | 0.430 | 0.984 | 0.999 |
| agrC, blpH, fsrC; two-component system, LytTR family, sensor histidine kinase AgrC [EC:2.7.13.3] | -0.113 | 0.210 | 0.590 | 0.999 |
| agrD; AgrD protein | -0.467 | 0.685 | 0.496 | 0.981 |
| aguA; agmatine deiminase [EC:3.5.3.12] | -0.126 | 0.136 | 0.354 | 0.978 |
| aguA; alpha-glucuronidase [EC:3.2.1.139] | -0.682 | 0.397 | 0.088 | 0.880 |
| aguB; N-carbamoylputrescine amidase [EC:3.5.1.53] | -0.214 | 0.116 | 0.065 | 0.839 |
| aguE; alpha-1,4-digalacturonate transport system substrate-binding protein | -2.034 | 1.626 | 0.213 | 0.978 |
| aguF; alpha-1,4-digalacturonate transport system permease protein | -2.034 | 1.626 | 0.213 | 0.978 |
| aguG; alpha-1,4-digalacturonate transport system permease protein | -2.188 | 1.564 | 0.164 | 0.957 |
| AGXT; alanine-glyoxylate transaminase / serine-glyoxylate transaminase / serine-pyruvate transaminase [EC:2.6.1.44 2.6.1.45 2.6.1.51] | -1.185 | 0.645 | 0.068 | 0.839 |
| ahlD, aiiA, attM, blcC; N-acyl homoserine lactone hydrolase [EC:3.1.1.81] | -2.503 | 2.184 | 0.254 | 0.978 |
| ahpD; alkyl hydroperoxide reductase subunit D | -0.113 | 0.334 | 0.736 | 0.999 |
| ahpF; alkyl hydroperoxide reductase subunit F [EC:1.6.4.-] | -0.040 | 0.104 | 0.698 | 0.999 |
| AIBP, nnrE; NAD(P)H-hydrate epimerase [EC:5.1.99.6] | 0.006 | 0.105 | 0.954 | 0.999 |
| aidA-I, misL; autotransporter family porin | -0.081 | 0.458 | 0.859 | 0.999 |
| aidB; putative acyl-CoA dehydrogenase | -0.144 | 0.210 | 0.494 | 0.981 |
| ail; attachment invasion locus protein | -0.446 | 1.068 | 0.677 | 0.999 |
| ainS, luxM; acyl homoserine lactone synthase [EC:2.3.1.184] | 18.902 | 3402.727 | 0.996 | 0.999 |
| AK6, FAP7; adenylate kinase [EC:2.7.4.3] | -17.234 | 3077.514 | 0.996 | 0.999 |
| aknOx; aclacinomycin-N/aclacinomycin-A oxidase [EC:1.1.3.45 1.3.3.14] | -17.761 | 4005.858 | 0.996 | 0.999 |
| AKR1A1, adh; alcohol dehydrogenase (NADP+) [EC:1.1.1.2] | 1.278 | 0.848 | 0.134 | 0.942 |
| AKR7; aflatoxin B1 aldehyde reductase | -17.989 | 4004.197 | 0.996 | 0.999 |
| aksD; methanogen homoaconitase large subunit [EC:4.2.1.114] | -0.774 | 1.492 | 0.605 | 0.999 |
| aksE; methanogen homoaconitase small subunit [EC:4.2.1.114] | -1.940 | 2.261 | 0.392 | 0.978 |
| ala; alanine dehydrogenase [EC:1.4.1.1] | 0.206 | 0.724 | 0.777 | 0.999 |
| alaA; alanine-synthesizing transaminase [EC:2.6.1.66 2.6.1.2] | -0.017 | 0.086 | 0.842 | 0.999 |
| alaC; alanine-synthesizing transaminase [EC:2.6.1.-] | -0.078 | 0.230 | 0.736 | 0.999 |
| alc, ALLC; allantoicase [EC:3.5.3.4] | -0.733 | 0.403 | 0.071 | 0.841 |
| ald; alanine dehydrogenase [EC:1.4.1.1] | -0.324 | 0.115 | 0.005 | 0.839 |
| aldA; lactaldehyde dehydrogenase / glycolaldehyde dehydrogenase [EC:1.2.1.22 1.2.1.21] | 0.091 | 0.389 | 0.816 | 0.999 |
| aldB; aldehyde dehydrogenase [EC:1.2.1.-] | -0.229 | 0.218 | 0.293 | 0.978 |
| ALDH; aldehyde dehydrogenase (NAD+) [EC:1.2.1.3] | -0.384 | 0.156 | 0.015 | 0.839 |
| aldH; NADP-dependent aldehyde dehydrogenase [EC:1.2.1.4] | -0.046 | 0.300 | 0.880 | 0.999 |
| ALDO; fructose-bisphosphate aldolase, class I [EC:4.1.2.13] | 0.298 | 0.227 | 0.192 | 0.970 |
| alg44; mannuronan synthase [EC:2.4.1.33] | 0.255 | 0.342 | 0.458 | 0.979 |
| ALG7; UDP-N-acetylglucosamine–dolichyl-phosphate N-acetylglucosaminephosphotransferase [EC:2.7.8.15] | -17.234 | 3077.514 | 0.996 | 0.999 |
| alg8; mannuronan synthase [EC:2.4.1.33] | 0.253 | 0.342 | 0.460 | 0.979 |
| algA, xanB, rfbA, wbpW, pslB; mannose-1-phosphate guanylyltransferase / mannose-6-phosphate isomerase [EC:2.7.7.13 5.3.1.8] | -0.143 | 0.258 | 0.580 | 0.997 |
| algB; two-component system, NtrC family, response regulator AlgB | 0.229 | 0.340 | 0.502 | 0.981 |
| algD; GDP-mannose 6-dehydrogenase [EC:1.1.1.132] | 0.279 | 0.351 | 0.428 | 0.978 |
| algE; alginate production protein | 0.255 | 0.342 | 0.458 | 0.979 |
| algF; alginate O-acetyltransferase complex protein AlgF | 0.263 | 0.341 | 0.441 | 0.978 |
| algG; mannuronan 5-epimerase [EC:5.1.3.37] | 0.364 | 0.362 | 0.317 | 0.978 |
| algH; putative transcriptional regulator | -0.091 | 0.135 | 0.503 | 0.981 |
| algI; alginate O-acetyltransferase complex protein AlgI | 0.164 | 0.239 | 0.494 | 0.981 |
| algJ; alginate O-acetyltransferase complex protein AlgJ | 0.081 | 0.367 | 0.825 | 0.999 |
| algK; alginate biosynthesis protein AlgK | 0.255 | 0.342 | 0.458 | 0.979 |
| algL; poly(beta-D-mannuronate) lyase [EC:4.2.2.3] | 0.065 | 0.307 | 0.833 | 0.999 |
| algR; two-component system, LytTR family, response regulator AlgR | -0.015 | 0.207 | 0.941 | 0.999 |
| algX; alginate biosynthesis protein AlgX | 0.253 | 0.342 | 0.460 | 0.979 |
| algZ; two-component system, LytTR family, sensor histidine kinase AlgZ [EC:2.7.13.3] | 0.032 | 0.228 | 0.888 | 0.999 |
| aliA; cyclohexanecarboxylate-CoA ligase [EC:6.2.1.-] | 17.929 | 2539.480 | 0.994 | 0.999 |
| aliB; cyclohexanecarboxyl-CoA dehydrogenase [EC:1.3.99.-] | -17.243 | 3092.681 | 0.996 | 0.999 |
| alkA; DNA-3-methyladenine glycosylase II [EC:3.2.2.21] | -0.387 | 0.210 | 0.067 | 0.839 |
| alkB; DNA oxidative demethylase [EC:1.14.11.33] | -0.366 | 0.268 | 0.174 | 0.961 |
| alkB1_2, alkM; alkane 1-monooxygenase [EC:1.14.15.3] | -0.529 | 0.459 | 0.251 | 0.978 |
| allA; ureidoglycolate lyase [EC:4.3.2.3] | -0.199 | 0.265 | 0.454 | 0.978 |
| allB; allantoinase [EC:3.5.2.5] | -0.292 | 0.449 | 0.516 | 0.985 |
| allC; allantoate deiminase [EC:3.5.3.9] | -0.592 | 0.448 | 0.188 | 0.966 |
| allD; ureidoglycolate dehydrogenase (NAD+) [EC:1.1.1.350] | -1.030 | 0.945 | 0.277 | 0.978 |
| allP; allantoin permease | -0.727 | 1.189 | 0.542 | 0.990 |
| allR; IclR family transcriptional regulator, negative regulator of allantoin and glyoxylate utilization operons | -1.153 | 1.472 | 0.435 | 0.978 |
| allS; LysR family transcriptional regulator, transcriptional activator of the allD operon | -1.125 | 1.359 | 0.409 | 0.978 |
| alpA; prophage regulatory protein | 0.124 | 0.213 | 0.561 | 0.992 |
| alr; alanine racemase [EC:5.1.1.1] | 0.081 | 0.037 | 0.032 | 0.839 |
| alsA; D-allose transport system ATP-binding protein [EC:3.6.3.17] | -0.216 | 1.383 | 0.876 | 0.999 |
| alsB; D-allose transport system substrate-binding protein | 2.069 | 1.635 | 0.208 | 0.976 |
| alsC; D-allose transport system permease protein | 2.069 | 1.635 | 0.208 | 0.976 |
| alsD, budA, aldC; acetolactate decarboxylase [EC:4.1.1.5] | 0.074 | 0.135 | 0.583 | 0.999 |
| alsE; D-allulose-6-phosphate 3-epimerase [EC:5.1.3.-] | 0.221 | 0.800 | 0.783 | 0.999 |
| alsK; allose kinase [EC:2.7.1.55] | 0.003 | 1.318 | 0.998 | 1.000 |
| ameR, rmiR; TetR/AcrR family transcriptional regulator, repressor of the ameABC operon | -1.366 | 1.756 | 0.438 | 0.978 |
| amgK; N-acetylmuramate 1-kinase [EC:2.7.1.221] | -0.128 | 0.179 | 0.474 | 0.979 |
| amhX; amidohydrolase [EC:3.5.1.-] | -1.202 | 0.930 | 0.198 | 0.970 |
| amiABC; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | -0.009 | 0.074 | 0.908 | 0.999 |
| amn; AMP nucleosidase [EC:3.2.2.4] | -0.177 | 0.189 | 0.351 | 0.978 |
| amnA; 2-aminophenol/2-amino-5-chlorophenol 1,6-dioxygenase subunit alpha | -17.155 | 2959.362 | 0.995 | 0.999 |
| amnB; 2-aminophenol/2-amino-5-chlorophenol 1,6-dioxygenase subunit beta [EC:1.13.11.74 1.13.11.76] | -17.155 | 2959.362 | 0.995 | 0.999 |
| amnD; 2-aminomuconate deaminase [EC:3.5.99.5] | -1.395 | 1.817 | 0.444 | 0.978 |
| ampC; beta-lactamase class C [EC:3.5.2.6] | -0.005 | 0.111 | 0.963 | 0.999 |
| ampD; N-acetyl-anhydromuramoyl-L-alanine amidase [EC:3.5.1.28] | 0.081 | 0.171 | 0.635 | 0.999 |
| ampE; AmpE protein | 0.101 | 0.259 | 0.697 | 0.999 |
| ampG; MFS transporter, PAT family, beta-lactamase induction signal transducer AmpG | -0.117 | 0.124 | 0.345 | 0.978 |
| ampH; serine-type D-Ala-D-Ala carboxypeptidase/endopeptidase [EC:3.4.16.4 3.4.21.-] | -0.295 | 0.639 | 0.645 | 0.999 |
| ampR; LysR family transcriptional regulator, regulator of gene expression of beta-lactamase | -0.394 | 0.344 | 0.254 | 0.978 |
| ampS, pepS, ampT; aminopeptidase [EC:3.4.11.-] | -0.056 | 0.109 | 0.609 | 0.999 |
| amsB; amylovoran biosynthesis glycosyltransferase AmsB [EC:2.4.-.-] | 0.055 | 1.009 | 0.957 | 0.999 |
| amsC; amylovoran biosynthesis protein AmsC | 0.055 | 1.009 | 0.957 | 0.999 |
| amsD; amylovoran biosynthesis glycosyltransferase AmsD [EC:2.4.-.-] | -17.444 | 3418.523 | 0.996 | 0.999 |
| amsF; amylovoran biosynthesis protein AmsF | 0.055 | 1.009 | 0.957 | 0.999 |
| amsG; UDP-galactose-lipid carrier transferase | 0.055 | 1.009 | 0.957 | 0.999 |
| amsL; exopolysaccharide (amylovoran) exporter | 0.055 | 1.009 | 0.957 | 0.999 |
| amt, AMT, MEP; ammonium transporter, Amt family | -0.119 | 0.107 | 0.268 | 0.978 |
| AMY, amyA, malS; alpha-amylase [EC:3.2.1.1] | -0.069 | 0.121 | 0.569 | 0.995 |
| amyM; maltogenic alpha-amylase [EC:3.2.1.133] | 0.779 | 0.726 | 0.285 | 0.978 |
| amzA, AMZ2, AMZ1; archaemetzincin [EC:3.4.-.-] | -18.092 | 3633.323 | 0.996 | 0.999 |
| andAb; anthranilate 1,2-dioxygenase ferredoxin component | -2.078 | 2.789 | 0.457 | 0.979 |
| andAc; anthranilate 1,2-dioxygenase large subunit [EC:1.14.12.1] | 0.173 | 1.432 | 0.904 | 0.999 |
| andAd; anthranilate 1,2-dioxygenase small subunit [EC:1.14.12.1] | -0.888 | 1.390 | 0.524 | 0.989 |
| ANK; ankyrin | -17.444 | 3418.523 | 0.996 | 0.999 |
| anmK; anhydro-N-acetylmuramic acid kinase [EC:2.7.1.170] | -0.055 | 0.147 | 0.710 | 0.999 |
| ansP; L-asparagine permease | -0.113 | 0.341 | 0.740 | 0.999 |
| antA; anthranilate 1,2-dioxygenase (deaminating, decarboxylating) large subunit [EC:1.14.12.1] | -0.662 | 0.600 | 0.271 | 0.978 |
| antB; anthranilate 1,2-dioxygenase (deaminating, decarboxylating) small subunit [EC:1.14.12.1] | -0.683 | 0.608 | 0.263 | 0.978 |
| antB; anti-repressor protein | -0.038 | 0.511 | 0.941 | 0.999 |
| antC; anthranilate 1,2-dioxygenase reductase component [EC:1.18.1.-] | -0.683 | 0.608 | 0.263 | 0.978 |
| AOC3, AOC2, tynA; primary-amine oxidase [EC:1.4.3.21] | -0.264 | 0.421 | 0.531 | 0.990 |
| aor; aldehyde:ferredoxin oxidoreductase [EC:1.2.7.5] | 0.190 | 0.716 | 0.791 | 0.999 |
| aotJ; arginine/ornithine transport system substrate-binding protein | 0.088 | 0.330 | 0.790 | 0.999 |
| aotM; arginine/ornithine transport system permease protein | 0.155 | 0.366 | 0.673 | 0.999 |
| aotP; arginine/ornithine transport system ATP-binding protein [EC:3.6.3.-] | 0.276 | 0.327 | 0.400 | 0.978 |
| aotQ; arginine/ornithine transport system permease protein | 0.211 | 0.340 | 0.537 | 0.990 |
| AOX1, AOX2; ubiquinol oxidase [EC:1.10.3.11] | 0.510 | 1.145 | 0.657 | 0.999 |
| aoxB; arsenite oxidase large subunit [EC:1.20.2.1 1.20.9.1] | 18.245 | 2974.604 | 0.995 | 0.999 |
| APA1_2; ATP adenylyltransferase [EC:2.7.7.53] | 0.588 | 0.957 | 0.539 | 0.990 |
| apaG; ApaG protein | -0.114 | 0.186 | 0.539 | 0.990 |
| apaH; bis(5’-nucleosyl)-tetraphosphatase (symmetrical) [EC:3.6.1.41] | 0.063 | 0.155 | 0.686 | 0.999 |
| apbE; FAD:protein FMN transferase [EC:2.7.1.180] | 0.114 | 0.060 | 0.061 | 0.839 |
| apeE, estA, lip-1; outer membrane lipase/esterase | 0.146 | 0.282 | 0.607 | 0.999 |
| APEH; acylaminoacyl-peptidase [EC:3.4.19.1] | 0.610 | 0.389 | 0.119 | 0.917 |
| APEX1; AP endonuclease 1 [EC:4.2.99.18] | 0.190 | 0.356 | 0.594 | 0.999 |
| apgM; 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [EC:5.4.2.12] | -0.068 | 0.344 | 0.843 | 0.999 |
| aph, spcN; spectinomycin phosphotransferase | -0.054 | 0.487 | 0.912 | 0.999 |
| aph3-I; aminoglycoside 3’-phosphotransferase I [EC:2.7.1.95] | 0.583 | 0.849 | 0.493 | 0.981 |
| aph3-II; aminoglycoside 3’-phosphotransferase II [EC:2.7.1.95] | -0.165 | 0.465 | 0.722 | 0.999 |
| aph3-III; aminoglycoside 3’-phosphotransferase III [EC:2.7.1.95] | -3.992 | 4.568 | 0.383 | 0.978 |
| aphA; acid phosphatase (class B) [EC:3.1.3.2] | 0.252 | 0.278 | 0.365 | 0.978 |
| aphA; kanamycin kinase [EC:2.7.1.95] | 0.083 | 0.162 | 0.610 | 0.999 |
| aphA; PadR family transcriptional regulator, regulatory protein AphA | 18.119 | 1920.327 | 0.992 | 0.999 |
| aphB; LysR family transcriptional regulator, transcriptional activator AphB | 0.359 | 1.252 | 0.775 | 0.999 |
| aphD; aminoglycoside 2’’-phosphotransferase [EC:2.7.1.190] | -0.924 | 1.156 | 0.425 | 0.978 |
| APOD; apolipoprotein D and lipocalin family protein | -0.057 | 0.250 | 0.821 | 0.999 |
| appA; 4-phytase / acid phosphatase [EC:3.1.3.26 3.1.3.2] | -0.132 | 0.528 | 0.803 | 0.999 |
| aprA; adenylylsulfate reductase, subunit A [EC:1.8.99.2] | 0.018 | 0.503 | 0.971 | 0.999 |
| aprB; adenylylsulfate reductase, subunit B [EC:1.8.99.2] | 0.018 | 0.503 | 0.971 | 0.999 |
| aprE; subtilisin [EC:3.4.21.62] | 18.553 | 2202.522 | 0.993 | 0.999 |
| APRT, apt; adenine phosphoribosyltransferase [EC:2.4.2.7] | 0.090 | 0.051 | 0.079 | 0.859 |
| aprX; serine protease AprX [EC:3.4.21.-] | -1.870 | 0.972 | 0.056 | 0.839 |
| aqpZ; aquaporin Z | -0.236 | 0.107 | 0.030 | 0.839 |
| araA; L-arabinose 1-dehydrogenase [EC:1.1.1.376] | -2.135 | 1.044 | 0.043 | 0.839 |
| araA; L-arabinose isomerase [EC:5.3.1.4] | 0.143 | 0.286 | 0.619 | 0.999 |
| araB; L-ribulokinase [EC:2.7.1.16] | -0.358 | 0.355 | 0.315 | 0.978 |
| araC; AraC family transcriptional regulator, arabinose operon regulatory protein | -0.105 | 0.446 | 0.813 | 0.999 |
| araD, ulaF, sgaE, sgbE; L-ribulose-5-phosphate 4-epimerase [EC:5.1.3.4] | -0.570 | 0.365 | 0.120 | 0.917 |
| araD; 2-keto-3-deoxy-L-arabinonate dehydratase [EC:4.2.1.43] | -2.481 | 2.614 | 0.344 | 0.978 |
| araD; D-arabinonate dehydratase [EC:4.2.1.5] | -2.501 | 3.364 | 0.458 | 0.979 |
| araE; MFS transporter, SP family, arabinose:H+ symporter | -0.475 | 0.662 | 0.474 | 0.979 |
| araF; L-arabinose transport system substrate-binding protein | -0.507 | 0.558 | 0.366 | 0.978 |
| araG; L-arabinose transport system ATP-binding protein [EC:3.6.3.17] | -0.513 | 0.559 | 0.360 | 0.978 |
| araH; L-arabinose transport system permease protein | -0.505 | 0.559 | 0.368 | 0.978 |
| araJ; MFS transporter, DHA1 family, arabinose polymer utilization protein | -0.447 | 0.349 | 0.202 | 0.972 |
| araL; arabinose operon protein AraL | -2.599 | 2.089 | 0.215 | 0.978 |
| araM, egsA; glycerol-1-phosphate dehydrogenase [NAD(P)+] [EC:1.1.1.261] | -0.185 | 0.341 | 0.587 | 0.999 |
| araN; arabinosaccharide transport system substrate-binding protein | -2.383 | 1.643 | 0.149 | 0.954 |
| araP; arabinosaccharide transport system permease protein | -2.383 | 1.643 | 0.149 | 0.954 |
| araQ; arabinosaccharide transport system permease protein | -1.613 | 0.985 | 0.104 | 0.905 |
| araR; GntR family transcriptional regulator, arabinose operon transcriptional repressor | 0.280 | 0.416 | 0.501 | 0.981 |
| arcA; arginine deiminase [EC:3.5.3.6] | 0.142 | 0.107 | 0.189 | 0.966 |
| arcA; two-component system, OmpR family, aerobic respiration control protein ArcA | 0.226 | 0.307 | 0.463 | 0.979 |
| arcB; two-component system, OmpR family, aerobic respiration control sensor histidine kinase ArcB [EC:2.7.13.3] | 0.217 | 0.299 | 0.468 | 0.979 |
| arcC; carbamate kinase [EC:2.7.2.2] | 0.130 | 0.116 | 0.263 | 0.978 |
| arcD, lysl, lysP; arginine:ornithine antiporter / lysine permease | 0.167 | 0.184 | 0.366 | 0.978 |
| arfA; alternative ribosome-rescue factor | 0.196 | 0.216 | 0.365 | 0.978 |
| arfC; uncharacterized membrane protein ArfC | 1.206 | 1.487 | 0.419 | 0.978 |
| argA; amino-acid N-acetyltransferase [EC:2.3.1.1] | -0.143 | 0.312 | 0.646 | 0.999 |
| argAB; amino-acid N-acetyltransferase [EC:2.3.1.1] | 0.109 | 0.176 | 0.537 | 0.990 |
| argB; acetylglutamate kinase [EC:2.7.2.8] | -0.074 | 0.056 | 0.189 | 0.966 |
| argC; N-acetyl-gamma-glutamyl-phosphate reductase [EC:1.2.1.38] | -0.071 | 0.062 | 0.254 | 0.978 |
| argD; acetylornithine/N-succinyldiaminopimelate aminotransferase [EC:2.6.1.11 2.6.1.17] | 0.048 | 0.107 | 0.654 | 0.999 |
| argE; acetylornithine deacetylase [EC:3.5.1.16] | -0.050 | 0.154 | 0.744 | 0.999 |
| argF; N-acetylornithine carbamoyltransferase [EC:2.1.3.9] | -0.095 | 0.459 | 0.836 | 0.999 |
| argF; N-succinyl-L-ornithine transcarbamylase [EC:2.1.3.11] | -0.596 | 0.413 | 0.151 | 0.954 |
| argG, ASS1; argininosuccinate synthase [EC:6.3.4.5] | -0.003 | 0.053 | 0.955 | 0.999 |
| argH, ASL; argininosuccinate lyase [EC:4.3.2.1] | -0.050 | 0.051 | 0.330 | 0.978 |
| argHA; argininosuccinate lyase / amino-acid N-acetyltransferase [EC:4.3.2.1 2.3.1.1] | 1.355 | 1.313 | 0.304 | 0.978 |
| argJ; glutamate N-acetyltransferase / amino-acid N-acetyltransferase [EC:2.3.1.35 2.3.1.1] | -0.125 | 0.104 | 0.233 | 0.978 |
| argK; LAO/AO transport system kinase [EC:2.7.-.-] | -0.082 | 0.180 | 0.649 | 0.999 |
| argR, ahrC; transcriptional regulator of arginine metabolism | 0.039 | 0.103 | 0.705 | 0.999 |
| arlR; two-component system, OmpR family, response regulator ArlR | -0.320 | 0.688 | 0.642 | 0.999 |
| arlS; two-component system, OmpR family, sensor histidine kinase ArlS [EC:2.7.13.3] | -0.349 | 0.702 | 0.620 | 0.999 |
| arnA, pmrI; UDP-4-amino-4-deoxy-L-arabinose formyltransferase / UDP-glucuronic acid dehydrogenase (UDP-4-keto-hexauronic acid decarboxylating) [EC:2.1.2.13 1.1.1.305] | -0.221 | 0.462 | 0.633 | 0.999 |
| arnB, pmrH; UDP-4-amino-4-deoxy-L-arabinose-oxoglutarate aminotransferase [EC:2.6.1.87] | 0.093 | 0.462 | 0.841 | 0.999 |
| arnC, pmrF; undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase [EC:2.4.2.53] | -0.165 | 0.387 | 0.670 | 0.999 |
| arnD; undecaprenyl phosphate-alpha-L-ara4FN deformylase [EC:3.5.1.-] | -0.221 | 0.462 | 0.633 | 0.999 |
| arnE; undecaprenyl phosphate-alpha-L-ara4N flippase subunit ArnE | -0.389 | 0.339 | 0.253 | 0.978 |
| arnF; undecaprenyl phosphate-alpha-L-ara4N flippase subunit ArnF | -0.222 | 0.462 | 0.631 | 0.999 |
| arnT, pmrK; 4-amino-4-deoxy-L-arabinose transferase [EC:2.4.2.43] | -0.384 | 0.529 | 0.469 | 0.979 |
| aroA; 3-phosphoshikimate 1-carboxyvinyltransferase [EC:2.5.1.19] | 0.009 | 0.038 | 0.813 | 0.999 |
| AROA1, aroA; chorismate mutase [EC:5.4.99.5] | -0.036 | 0.126 | 0.777 | 0.999 |
| AROA2, aroA; 3-deoxy-7-phosphoheptulonate synthase [EC:2.5.1.54] | 0.374 | 0.191 | 0.052 | 0.839 |
| aroB; 3-dehydroquinate synthase [EC:4.2.3.4] | -0.040 | 0.046 | 0.392 | 0.978 |
| aroC; chorismate synthase [EC:4.2.3.5] | 0.014 | 0.041 | 0.732 | 0.999 |
| aroD; 3-dehydroquinate dehydratase I [EC:4.2.1.10] | -0.057 | 0.110 | 0.605 | 0.999 |
| aroDE, DHQ-SDH; 3-dehydroquinate dehydratase / shikimate dehydrogenase [EC:4.2.1.10 1.1.1.25] | 0.338 | 0.821 | 0.681 | 0.999 |
| aroE; shikimate dehydrogenase [EC:1.1.1.25] | 0.085 | 0.059 | 0.155 | 0.954 |
| aroG, aroA; 3-deoxy-7-phosphoheptulonate synthase / chorismate mutase [EC:2.5.1.54 5.4.99.5] | -0.960 | 0.463 | 0.040 | 0.839 |
| aroH; chorismate mutase [EC:5.4.99.5] | -0.227 | 0.375 | 0.546 | 0.990 |
| aroKB; shikimate kinase / 3-dehydroquinate synthase [EC:2.7.1.71 4.2.3.4] | -0.036 | 0.355 | 0.919 | 0.999 |
| aroM; protein AroM | -0.496 | 0.850 | 0.561 | 0.992 |
| aroP; aromatic amino acid permease | 0.116 | 0.410 | 0.778 | 0.999 |
| aroP; aromatic amino acid transport protein AroP | 0.026 | 0.179 | 0.882 | 0.999 |
| aroQ, qutE; 3-dehydroquinate dehydratase II [EC:4.2.1.10] | 0.017 | 0.069 | 0.804 | 0.999 |
| arr; rifampin ADP-ribosylating transferase | -0.031 | 0.418 | 0.940 | 0.999 |
| arsA, ASNA1, GET3; arsenite/tail-anchored protein-transporting ATPase [EC:3.6.3.16 3.6.3.-] | 0.578 | 0.430 | 0.180 | 0.961 |
| ARSA; arylsulfatase A [EC:3.1.6.8] | 1.036 | 1.270 | 0.416 | 0.978 |
| arsB; arsenical pump membrane protein | -0.219 | 0.199 | 0.272 | 0.978 |
| ARSB; arylsulfatase B [EC:3.1.6.12] | 1.360 | 0.956 | 0.157 | 0.954 |
| arsC; arsenate-mycothiol transferase [EC:2.8.4.2] | -0.564 | 0.759 | 0.459 | 0.979 |
| ARSC1, arsC; arsenate reductase [EC:1.20.4.1] | 0.048 | 0.040 | 0.240 | 0.978 |
| ARSC2, arsC; arsenate reductase [EC:1.20.4.1] | -0.051 | 0.124 | 0.683 | 0.999 |
| arsH; arsenical resistance protein ArsH | -0.117 | 0.249 | 0.640 | 0.999 |
| arsR; ArsR family transcriptional regulator, arsenate/arsenite/antimonite-responsive transcriptional repressor | 0.008 | 0.077 | 0.914 | 0.999 |
| artI; arginine transport system substrate-binding protein | 0.225 | 0.305 | 0.463 | 0.979 |
| artJ; arginine transport system substrate-binding protein | 0.106 | 0.608 | 0.862 | 0.999 |
| artM; arginine transport system permease protein | 0.205 | 0.312 | 0.513 | 0.982 |
| artP; arginine transport system ATP-binding protein [EC:3.6.3.-] | 0.205 | 0.312 | 0.513 | 0.982 |
| artQ; arginine transport system permease protein | 0.205 | 0.312 | 0.513 | 0.982 |
| aruH; arginine:pyruvate transaminase [EC:2.6.1.84] | -0.077 | 0.441 | 0.862 | 0.999 |
| aruI; 5-guanidino-2-oxopentanoate decarboxylase [EC:4.1.1.75] | 0.778 | 0.610 | 0.204 | 0.972 |
| ASAH2; neutral ceramidase [EC:3.5.1.23] | 0.068 | 0.358 | 0.850 | 0.999 |
| asbF; 3-dehydroshikimate dehydratase [EC:4.2.1.118] | -0.682 | 1.317 | 0.605 | 0.999 |
| ascC, ddhC, rfbH; CDP-4-dehydro-6-deoxyglucose reductase, E1 [EC:1.17.1.1] | 0.186 | 0.341 | 0.587 | 0.999 |
| ascD, ddhD, rfbI; CDP-4-dehydro-6-deoxyglucose reductase, E3 [EC:1.17.1.1] | 0.067 | 0.226 | 0.766 | 0.999 |
| ascG; LacI family transcriptional regulator, asc operon repressor | 0.577 | 0.507 | 0.257 | 0.978 |
| asd; aspartate-semialdehyde dehydrogenase [EC:1.2.1.11] | -0.004 | 0.039 | 0.928 | 0.999 |
| asdA; aspartate 4-decarboxylase [EC:4.1.1.12] | 0.297 | 0.427 | 0.488 | 0.979 |
| asl; D-aspartate ligase [EC:6.3.1.12] | 0.130 | 0.363 | 0.721 | 0.999 |
| asm11; 4,5-epoxidase | -18.019 | 3423.353 | 0.996 | 0.999 |
| asmA; AsmA protein | -0.084 | 0.391 | 0.830 | 0.999 |
| asnA; aspartate–ammonia ligase [EC:6.3.1.1] | 0.143 | 0.120 | 0.234 | 0.978 |
| asnB, ASNS; asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4] | -0.164 | 0.152 | 0.281 | 0.978 |
| asnC; Lrp/AsnC family transcriptional regulator, regulator for asnA, asnC and gidA | -0.122 | 0.186 | 0.513 | 0.982 |
| asnO; L-asparagine oxygenase [EC:1.14.11.39] | -17.794 | 4072.025 | 0.997 | 0.999 |
| asp1; accessory secretory protein Asp1 | -0.146 | 0.186 | 0.434 | 0.978 |
| asp2; accessory secretory protein Asp2 | -0.145 | 0.186 | 0.436 | 0.978 |
| asp3; accessory secretory protein Asp3 | -0.183 | 0.187 | 0.331 | 0.978 |
| ASPA, aspA; aspartoacylase [EC:3.5.1.15] | 18.902 | 3402.727 | 0.996 | 0.999 |
| aspA; aspartate ammonia-lyase [EC:4.3.1.1] | -0.046 | 0.106 | 0.665 | 0.999 |
| aspB; aspartate aminotransferase [EC:2.6.1.1] | -0.212 | 0.141 | 0.135 | 0.946 |
| aspC, aspS; nondiscriminating aspartyl-tRNA synthetase [EC:6.1.1.23] | -0.411 | 0.778 | 0.598 | 0.999 |
| aspC; aspartate aminotransferase [EC:2.6.1.1] | 0.226 | 0.284 | 0.427 | 0.978 |
| ASPG; 60kDa lysophospholipase [EC:3.1.1.5 3.1.1.47 3.5.1.1] | -17.760 | 4004.372 | 0.996 | 0.999 |
| ASPH; aspartate beta-hydroxylase [EC:1.14.11.16] | -1.636 | 1.961 | 0.405 | 0.978 |
| aspQ, ansB, ansA; glutamin-(asparagin-)ase [EC:3.5.1.38] | -0.347 | 0.339 | 0.308 | 0.978 |
| aspS; aspartyl-tRNA synthetase [EC:6.1.1.12] | 0.043 | 0.045 | 0.344 | 0.978 |
| asrA; anaerobic sulfite reductase subunit A | 0.916 | 0.464 | 0.050 | 0.839 |
| asrB; anaerobic sulfite reductase subunit B | 0.916 | 0.464 | 0.050 | 0.839 |
| asrC; anaerobic sulfite reductase subunit C | 0.916 | 0.464 | 0.050 | 0.839 |
| assT; arylsulfate sulfotransferase [EC:2.8.2.22] | -0.949 | 0.951 | 0.320 | 0.978 |
| astA; arginine N-succinyltransferase [EC:2.3.1.109] | 0.068 | 0.266 | 0.800 | 0.999 |
| astB; succinylarginine dihydrolase [EC:3.5.3.23] | 0.044 | 0.238 | 0.853 | 0.999 |
| astC; succinylornithine aminotransferase [EC:2.6.1.81] | -0.338 | 0.393 | 0.391 | 0.978 |
| astD; succinylglutamic semialdehyde dehydrogenase [EC:1.2.1.71] | 0.012 | 0.260 | 0.964 | 0.999 |
| astE; succinylglutamate desuccinylase [EC:3.5.1.96] | 0.088 | 0.255 | 0.730 | 0.999 |
| ATE1; arginyl-tRNA—protein transferase [EC:2.3.2.8] | -0.131 | 0.196 | 0.505 | 0.981 |
| atl; bifunctional autolysin [EC:3.5.1.28 3.2.1.96] | 0.659 | 0.773 | 0.395 | 0.978 |
| atoA; acetate CoA/acetoacetate CoA-transferase beta subunit [EC:2.8.3.8 2.8.3.9] | 0.825 | 0.378 | 0.031 | 0.839 |
| atoC; two-component system, NtrC family, response regulator AtoC | -0.308 | 0.815 | 0.706 | 0.999 |
| atoD; acetate CoA/acetoacetate CoA-transferase alpha subunit [EC:2.8.3.8 2.8.3.9] | 0.811 | 0.378 | 0.033 | 0.839 |
| atoE; short-chain fatty acids transporter | 0.230 | 0.198 | 0.247 | 0.978 |
| atoS; two-component system, NtrC family, sensor histidine kinase AtoS [EC:2.7.13.3] | -1.548 | 1.264 | 0.223 | 0.978 |
| ATOX1, ATX1, copZ, golB; copper chaperone | -0.135 | 0.112 | 0.227 | 0.978 |
| ATPeV1G, ATP6G; V-type H+-transporting ATPase subunit G | -0.438 | 1.579 | 0.782 | 0.999 |
| ATPF0A, atpB; F-type H+-transporting ATPase subunit a | 0.031 | 0.036 | 0.388 | 0.978 |
| ATPF0B, atpF; F-type H+-transporting ATPase subunit b | 0.004 | 0.037 | 0.911 | 0.999 |
| ATPF0C, atpE; F-type H+-transporting ATPase subunit c | 0.031 | 0.036 | 0.399 | 0.978 |
| ATPF1A, atpA; F-type H+-transporting ATPase subunit alpha [EC:3.6.3.14] | 0.031 | 0.036 | 0.394 | 0.978 |
| ATPF1B, atpD; F-type H+-transporting ATPase subunit beta [EC:3.6.3.14] | 0.029 | 0.033 | 0.380 | 0.978 |
| ATPF1D, atpH; F-type H+-transporting ATPase subunit delta | 0.038 | 0.037 | 0.315 | 0.978 |
| ATPF1E, atpC; F-type H+-transporting ATPase subunit epsilon | 0.032 | 0.036 | 0.388 | 0.978 |
| ATPF1G, atpG; F-type H+-transporting ATPase subunit gamma | 0.031 | 0.036 | 0.394 | 0.978 |
| atpI; ATP synthase protein I | 0.039 | 0.127 | 0.761 | 0.999 |
| ATPVA, ntpA, atpA; V/A-type H+/Na+-transporting ATPase subunit A [EC:3.6.3.14 3.6.3.15] | 0.314 | 0.150 | 0.038 | 0.839 |
| ATPVB, ntpB, atpB; V/A-type H+/Na+-transporting ATPase subunit B | 0.311 | 0.150 | 0.040 | 0.839 |
| ATPVC, ntpC, atpC; V/A-type H+/Na+-transporting ATPase subunit C | 0.397 | 0.159 | 0.014 | 0.839 |
| ATPVD, ntpD, atpD; V/A-type H+/Na+-transporting ATPase subunit D | 0.318 | 0.150 | 0.036 | 0.839 |
| ATPVE, ntpE, atpE; V/A-type H+/Na+-transporting ATPase subunit E | 0.213 | 0.172 | 0.216 | 0.978 |
| ATPVF, ntpF, atpF; V/A-type H+/Na+-transporting ATPase subunit F | 0.373 | 0.159 | 0.020 | 0.839 |
| ATPVG, ahaH, atpH; V/A-type H+/Na+-transporting ATPase subunit G/H | 0.909 | 0.363 | 0.013 | 0.839 |
| ATPVI, ntpI, atpI; V/A-type H+/Na+-transporting ATPase subunit I | 0.310 | 0.150 | 0.040 | 0.839 |
| ATPVK, ntpK, atpK; V/A-type H+/Na+-transporting ATPase subunit K | 0.312 | 0.150 | 0.040 | 0.839 |
| atrm56; tRNA (cytidine56-2’-O)-methyltransferase [EC:2.1.1.206] | -17.234 | 3077.514 | 0.996 | 0.999 |
| attA1; mannopine transport system ATP-binding protein | -2.174 | 2.005 | 0.280 | 0.978 |
| attA2; mannopine transport system permease protein | -2.174 | 2.005 | 0.280 | 0.978 |
| attC; mannopine transport system substrate-binding protein | -2.174 | 2.005 | 0.280 | 0.978 |
| atuB; citronellol/citronellal dehydrogenase | -0.345 | 0.333 | 0.302 | 0.978 |
| atuC; geranyl-CoA carboxylase beta subunit [EC:6.4.1.5] | -0.310 | 0.329 | 0.348 | 0.978 |
| atuD; citronellyl-CoA dehydrogenase [EC:1.3.99.-] | -0.288 | 0.334 | 0.390 | 0.978 |
| atuE; isohexenylglutaconyl-CoA hydratase [EC:4.2.1.57] | -0.176 | 0.370 | 0.634 | 0.999 |
| atuF; geranyl-CoA carboxylase alpha subunit [EC:6.4.1.5] | -0.280 | 0.340 | 0.410 | 0.978 |
| atuG; citronellol/citronellal dehydrogenase | 0.086 | 0.334 | 0.798 | 0.999 |
| atuH; citronellyl-CoA synthetase [EC:6.2.1.-] | -0.431 | 0.378 | 0.256 | 0.978 |
| ATX, chlB1, mdpB; 6-methylsalicylic acid synthase [EC:2.3.1.165] | -17.790 | 2152.624 | 0.993 | 0.999 |
| atzB; hydroxydechloroatrazine ethylaminohydrolase [EC:3.5.4.43] | -1.865 | 1.685 | 0.270 | 0.978 |
| atzD; cyanuric acid amidohydrolase [EC:3.5.2.15] | -0.999 | 0.882 | 0.259 | 0.978 |
| atzF; allophanate hydrolase [EC:3.5.1.54] | -0.127 | 0.298 | 0.670 | 0.999 |
| AUP1; ancient ubiquitous protein 1 | -0.976 | 0.998 | 0.329 | 0.978 |
| aur; aureolysin [EC:3.4.24.29] | -0.467 | 0.685 | 0.496 | 0.981 |
| avtA; valine–pyruvate aminotransferase [EC:2.6.1.66] | 0.335 | 0.356 | 0.349 | 0.978 |
| AXY8, FUC95A, afcA; alpha-L-fucosidase 2 [EC:3.2.1.51] | -0.275 | 0.194 | 0.160 | 0.954 |
| azr; azobenzene reductase [EC:1.7.1.6] | -2.035 | 1.879 | 0.280 | 0.978 |
| babA; outer membrane protein BabA | -1.386 | 1.551 | 0.373 | 0.978 |
| bacA; undecaprenyl-diphosphatase [EC:3.6.1.27] | 0.001 | 0.044 | 0.989 | 0.999 |
| bacA; vitamin B12/bleomycin/antimicrobial peptide transport system ATP-binding/permease protein | -0.123 | 0.187 | 0.514 | 0.983 |
| bacC; dihydroanticapsin dehydrogenase [EC:1.1.1.385] | 18.076 | 2733.666 | 0.995 | 0.999 |
| badA; benzoate-CoA ligase [EC:6.2.1.25] | -2.028 | 1.566 | 0.197 | 0.970 |
| badH; 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase [EC:1.1.1.-] | -0.879 | 0.559 | 0.118 | 0.917 |
| badI; 2-ketocyclohexanecarboxyl-CoA hydrolase [EC:3.1.2.-] | -1.983 | 1.811 | 0.275 | 0.978 |
| baeR, smeR; two-component system, OmpR family, response regulator BaeR | -0.262 | 0.267 | 0.328 | 0.978 |
| baeS, smeS; two-component system, OmpR family, sensor histidine kinase BaeS [EC:2.7.13.3] | -0.301 | 0.268 | 0.262 | 0.978 |
| baiB; bile acid-coenzyme A ligase [EC:6.2.1.7] | -0.872 | 0.696 | 0.212 | 0.978 |
| baiH; NAD+-dependent 7beta-hydroxy-3-oxo bile acid-CoA-ester 4-dehydrogenase | -18.040 | 3081.666 | 0.995 | 0.999 |
| bamB; outer membrane protein assembly factor BamB | 0.024 | 0.215 | 0.911 | 0.999 |
| bamC; outer membrane protein assembly factor BamC | 0.223 | 0.191 | 0.246 | 0.978 |
| bamD; outer membrane protein assembly factor BamD | -0.108 | 0.101 | 0.286 | 0.978 |
| bamE, smpA; outer membrane protein assembly factor BamE | 0.071 | 0.156 | 0.650 | 0.999 |
| barA, gacS, varS; two-component system, NarL family, sensor histidine kinase BarA [EC:2.7.13.3] | 0.108 | 0.271 | 0.689 | 0.999 |
| basR; two-component system, OmpR family, response regulator BasR | -0.198 | 0.560 | 0.724 | 0.999 |
| basS; two-component system, OmpR family, sensor histidine kinase BasS [EC:2.7.13.3] | 0.134 | 0.621 | 0.829 | 0.999 |
| bax; Bax protein | 0.927 | 0.447 | 0.040 | 0.839 |
| bbsF; benzylsuccinate CoA-transferase BbsF subunit [EC:2.8.3.15] | -0.229 | 1.513 | 0.880 | 0.999 |
| bbsH; E-phenylitaconyl-CoA hydratase [EC:4.2.1.-] | 18.076 | 2733.666 | 0.995 | 0.999 |
| bccA, pccA; acetyl-CoA/propionyl-CoA carboxylase, biotin carboxylase, biotin carboxyl carrier protein [EC:6.4.1.2 6.4.1.3 6.3.4.14] | -0.119 | 0.227 | 0.602 | 0.999 |
| bceA, vraD; bacitracin transport system ATP-binding protein | -1.232 | 0.669 | 0.067 | 0.839 |
| bceB, vraE; bacitracin transport system permease protein | -1.131 | 0.665 | 0.091 | 0.885 |
| bceR; two-component system, OmpR family, bacitracin resistance response regulator BceR | -1.150 | 0.694 | 0.100 | 0.896 |
| bceS; two-component system, OmpR family, bacitracin resistance sensor histidine kinase BceS [EC:2.7.13.3] | -1.001 | 0.831 | 0.230 | 0.978 |
| bchC; bacteriochlorophyllide a dehydrogenase [EC:1.1.1.396] | -3.309 | 3.390 | 0.331 | 0.978 |
| bchE; anaerobic magnesium-protoporphyrin IX monomethyl ester cyclase [EC:1.21.98.3] | 0.616 | 1.054 | 0.560 | 0.992 |
| bchF; 3-vinyl bacteriochlorophyllide hydratase [EC:4.2.1.165] | -18.056 | 3176.254 | 0.995 | 0.999 |
| bchJ; divinyl protochlorophyllide a 8-vinyl-reductase [EC:1.-.-.-] | -17.500 | 2562.559 | 0.995 | 0.999 |
| bchM, chlM; magnesium-protoporphyrin O-methyltransferase [EC:2.1.1.11] | -2.592 | 3.309 | 0.435 | 0.978 |
| bchO; magnesium chelatase accessory protein | -18.056 | 3176.254 | 0.995 | 0.999 |
| bchX; 3,8-divinyl chlorophyllide a/chlorophyllide a reductase subunit X [EC:1.3.7.14 1.3.7.15] | -3.309 | 3.390 | 0.331 | 0.978 |
| bchY; 3,8-divinyl chlorophyllide a/chlorophyllide a reductase subunit Y [EC:1.3.7.14 1.3.7.15] | -3.309 | 3.390 | 0.331 | 0.978 |
| bchZ; 3,8-divinyl chlorophyllide a/chlorophyllide a reductase subunit Z [EC:1.3.7.14 1.3.7.15] | -3.309 | 3.390 | 0.331 | 0.978 |
| BCKDHA, bkdA1; 2-oxoisovalerate dehydrogenase E1 component alpha subunit [EC:1.2.4.4] | -0.565 | 0.245 | 0.022 | 0.839 |
| BCKDHB, bkdA2; 2-oxoisovalerate dehydrogenase E1 component beta subunit [EC:1.2.4.4] | -0.511 | 0.259 | 0.050 | 0.839 |
| BCP, PRXQ, DOT5; peroxiredoxin Q/BCP [EC:1.11.1.15] | -0.125 | 0.066 | 0.060 | 0.839 |
| bcpA; oxaloacetate decarboxylase [EC:4.1.1.3] | 0.144 | 0.326 | 0.658 | 0.999 |
| bcr, tcaB; MFS transporter, DHA1 family, multidrug resistance protein | -0.084 | 0.098 | 0.388 | 0.978 |
| bcrA; bacitracin transport system ATP-binding protein | -1.238 | 0.807 | 0.127 | 0.927 |
| bcrB; bacitracin transport system permease protein | -1.640 | 0.955 | 0.088 | 0.880 |
| bcrC; undecaprenyl-diphosphatase [EC:3.6.1.27] | -0.093 | 0.048 | 0.055 | 0.839 |
| BCS1; mitochondrial chaperone BCS1 | -17.404 | 2813.939 | 0.995 | 0.999 |
| bcsA; cellulose synthase (UDP-forming) [EC:2.4.1.12] | -0.123 | 0.269 | 0.647 | 0.999 |
| bdcA; cyclic-di-GMP-binding biofilm dispersal mediator protein | -0.330 | 0.726 | 0.650 | 0.999 |
| bdcR; TetR/AcrR family transcriptional regulator, repressor for divergent bdcA | -0.157 | 0.790 | 0.842 | 0.999 |
| BDH, butB; (R,R)-butanediol dehydrogenase / meso-butanediol dehydrogenase / diacetyl reductase [EC:1.1.1.4 1.1.1.- 1.1.1.303] | -0.054 | 0.151 | 0.723 | 0.999 |
| bdhAB; butanol dehydrogenase [EC:1.1.1.-] | -0.018 | 0.123 | 0.885 | 0.999 |
| benA-xylX; benzoate/toluate 1,2-dioxygenase subunit alpha [EC:1.14.12.10 1.14.12.-] | 0.204 | 0.322 | 0.528 | 0.989 |
| benB-xylY; benzoate/toluate 1,2-dioxygenase subunit beta [EC:1.14.12.10 1.14.12.-] | 0.206 | 0.321 | 0.521 | 0.988 |
| benC-xylZ; benzoate/toluate 1,2-dioxygenase reductase component [EC:1.18.1.-] | 0.209 | 0.315 | 0.509 | 0.981 |
| benD-xylL; dihydroxycyclohexadiene carboxylate dehydrogenase [EC:1.3.1.25 1.3.1.-] | 0.220 | 0.307 | 0.473 | 0.979 |
| benE; benzoate membrane transport protein | -0.123 | 0.202 | 0.544 | 0.990 |
| benK; MFS transporter, AAHS family, benzoate transport protein | -0.017 | 0.326 | 0.960 | 0.999 |
| betA, CHDH; choline dehydrogenase [EC:1.1.99.1] | -0.167 | 0.199 | 0.403 | 0.978 |
| betB, gbsA; betaine-aldehyde dehydrogenase [EC:1.2.1.8] | -0.199 | 0.188 | 0.291 | 0.978 |
| betC; choline-sulfatase [EC:3.1.6.6] | 0.095 | 0.323 | 0.769 | 0.999 |
| betI; TetR/AcrR family transcriptional regulator, transcriptional repressor of bet genes | -0.047 | 0.214 | 0.827 | 0.999 |
| betT, betS; choline/glycine/proline betaine transport protein | 0.047 | 0.176 | 0.790 | 0.999 |
| bfd; bacterioferritin-associated ferredoxin | 0.014 | 0.176 | 0.936 | 0.999 |
| bfr; bacterioferritin [EC:1.16.3.1] | -0.040 | 0.185 | 0.830 | 0.999 |
| bgaB, lacA; beta-galactosidase [EC:3.2.1.23] | -0.162 | 0.140 | 0.248 | 0.978 |
| bglB; beta-glucosidase [EC:3.2.1.21] | -0.190 | 0.313 | 0.546 | 0.990 |
| bglG1; transcriptional antiterminator | -0.027 | 0.197 | 0.889 | 0.999 |
| bglH; carbohydrate-specific outer membrane porin | -0.338 | 1.004 | 0.737 | 0.999 |
| bglK; beta-glucoside kinase [EC:2.7.1.85] | -0.202 | 0.975 | 0.836 | 0.999 |
| bglX; beta-glucosidase [EC:3.2.1.21] | -0.195 | 0.144 | 0.178 | 0.961 |
| bgtB; arginine/lysine/histidine/glutamine transport system substrate-binding and permease protein | -17.795 | 2866.649 | 0.995 | 0.999 |
| BHMT; betaine-homocysteine S-methyltransferase [EC:2.1.1.5] | -17.155 | 2959.362 | 0.995 | 0.999 |
| bhsA; multiple stress resistance protein BhsA | 0.011 | 0.621 | 0.986 | 0.999 |
| bigA; putative surface-exposed virulence protein | 1.203 | 0.476 | 0.013 | 0.839 |
| bioA, bioK; lysine—8-amino-7-oxononanoate aminotransferase [EC:2.6.1.105] | 18.239 | 2966.075 | 0.995 | 0.999 |
| bioA; adenosylmethionine—8-amino-7-oxononanoate aminotransferase [EC:2.6.1.62] | 0.010 | 0.088 | 0.908 | 0.999 |
| bioB; biotin synthase [EC:2.8.1.6] | 0.052 | 0.083 | 0.534 | 0.990 |
| bioC; malonyl-CoA O-methyltransferase [EC:2.1.1.197] | 0.158 | 0.136 | 0.246 | 0.978 |
| bioD; dethiobiotin synthetase [EC:6.3.3.3] | 0.064 | 0.095 | 0.502 | 0.981 |
| bioF; 8-amino-7-oxononanoate synthase [EC:2.3.1.47] | -0.041 | 0.119 | 0.730 | 0.999 |
| bioG; pimeloyl-[acyl-carrier protein] methyl ester esterase [EC:3.1.1.85] | 0.386 | 0.259 | 0.138 | 0.949 |
| bioH; pimeloyl-[acyl-carrier protein] methyl ester esterase [EC:3.1.1.85] | 0.078 | 0.194 | 0.689 | 0.999 |
| bioI, CYP107H; pimeloyl-[acyl-carrier protein] synthase [EC:1.14.14.46] | 18.239 | 2966.075 | 0.995 | 0.999 |
| bioM; biotin transport system ATP-binding protein [EC:3.6.3.-] | -0.071 | 0.282 | 0.803 | 0.999 |
| bioN; biotin transport system permease protein | -0.048 | 0.317 | 0.879 | 0.999 |
| bioW; 6-carboxyhexanoate–CoA ligase [EC:6.2.1.14] | 0.071 | 0.251 | 0.779 | 0.999 |
| bioY; biotin transport system substrate-specific component | 0.066 | 0.087 | 0.451 | 0.978 |
| birA-coaX; biotin—[acetyl-CoA-carboxylase] ligase / type III pantothenate kinase [EC:6.3.4.15 2.7.1.33] | 0.291 | 0.478 | 0.543 | 0.990 |
| birA; BirA family transcriptional regulator, biotin operon repressor / biotin—[acetyl-CoA-carboxylase] ligase [EC:6.3.4.15] | 0.037 | 0.039 | 0.349 | 0.978 |
| bisC; biotin/methionine sulfoxide reductase [EC:1.-.-.-] | -0.714 | 0.485 | 0.143 | 0.951 |
| bjaI, rpaI, braI, rhiI; acyl-homoserine lactone synthase [EC:2.3.1.228 2.3.1.229 2.3.1.-] | -0.574 | 0.684 | 0.403 | 0.978 |
| bjaR1, rpaR, rhiR; LuxR family transcriptional regulator, quorum-sensing system regulator BjaR1 | -2.618 | 1.591 | 0.102 | 0.897 |
| bkdA; 2-oxoisovalerate dehydrogenase E1 component [EC:1.2.4.4] | -0.150 | 0.300 | 0.618 | 0.999 |
| bla2, blm, ccrA, blaB; metallo-beta-lactamase class B [EC:3.5.2.6] | -0.190 | 0.361 | 0.599 | 0.999 |
| blaACT_MIR; beta-lactamase class C ACT/MIR [EC:3.5.2.6] | 1.305 | 0.861 | 0.132 | 0.941 |
| blaCARB-17; beta-lactamase class A CARB-17 [EC:3.5.2.6] | 18.902 | 3402.727 | 0.996 | 0.999 |
| blaCMY-1; beta-lactamase class C CMY-1 [EC:3.5.2.6] | 17.680 | 2242.142 | 0.994 | 0.999 |
| blaCMY-2; beta-lactamase class C CMY-2 [EC:3.5.2.6] | -2.157 | 2.664 | 0.419 | 0.978 |
| blaDHA; beta-lactamase class C DHA [EC:3.5.2.6] | 18.165 | 2857.430 | 0.995 | 0.999 |
| blaGES; beta-lactamase class A GES [EC:3.5.2.6] | 18.165 | 2857.430 | 0.995 | 0.999 |
| blaI; BlaI family transcriptional regulator, penicillinase repressor | -0.770 | 0.560 | 0.171 | 0.961 |
| blaIND; metallo-beta-lactamase class B IND [EC:3.5.2.6] | -1.696 | 2.377 | 0.477 | 0.979 |
| blaKPC; beta-lactamase class A KPC [EC:3.5.2.6] | -0.213 | 1.210 | 0.860 | 0.999 |
| blaOXA-10; beta-lactamase class D OXA-10 [EC:3.5.2.6] | 18.165 | 2857.430 | 0.995 | 0.999 |
| blaOXA-12; beta-lactamase class D OXA-12 [EC:3.5.2.6] | 17.680 | 2242.142 | 0.994 | 0.999 |
| blaOXA-213; beta-lactamase class D OXA-213 [EC:3.5.2.6] | -0.816 | 1.161 | 0.483 | 0.979 |
| blaOXA-51; beta-lactamase class D OXA-51 [EC:3.5.2.6] | -0.816 | 1.161 | 0.483 | 0.979 |
| blaR1; bla regulator protein blaR1 | -0.744 | 0.598 | 0.215 | 0.978 |
| blaSHV; beta-lactamase class A SHV [EC:3.5.2.6] | 0.021 | 1.115 | 0.985 | 0.999 |
| blaTEM; beta-lactamase class A TEM [EC:3.5.2.6] | -1.000 | 1.063 | 0.348 | 0.978 |
| blaVEB; beta-lactamase class A VEB [EC:3.5.2.6] | -17.761 | 4005.858 | 0.996 | 0.999 |
| blaZ; beta-lactamase class A BlaZ [EC:3.5.2.6] | -0.457 | 0.700 | 0.514 | 0.983 |
| BLMH, pepC; bleomycin hydrolase [EC:3.4.22.40] | -0.004 | 0.115 | 0.973 | 0.999 |
| blt; MFS transporter, DHA1 family, multidrug resistance protein | 0.294 | 0.456 | 0.520 | 0.988 |
| bluB; 5,6-dimethylbenzimidazole synthase [EC:1.13.11.79] | -0.163 | 0.258 | 0.528 | 0.989 |
| bmaC; fibronectin-binding autotransporter adhesin | -1.934 | 1.573 | 0.221 | 0.978 |
| bmpA, bmpB, tmpC; basic membrane protein A and related proteins | -0.043 | 0.100 | 0.667 | 0.999 |
| bmr; MFS transporter, DHA1 family, multidrug resistance protein | -0.912 | 1.119 | 0.417 | 0.978 |
| bmrR; MerR family transcriptional regulator, activator of bmr gene | -0.635 | 1.401 | 0.651 | 0.999 |
| bofA; inhibitor of the pro-sigma K processing machinery | -0.260 | 0.338 | 0.443 | 0.978 |
| bofC; forespore regulator of the sigma-K checkpoint | -2.072 | 0.971 | 0.034 | 0.839 |
| bolA; BolA family transcriptional regulator, general stress-responsive regulator | -0.029 | 0.146 | 0.840 | 0.999 |
| boNT; botulinum neurotoxin [EC:3.4.24.69] | -16.780 | 1732.243 | 0.992 | 0.999 |
| boxA; benzoyl-CoA 2,3-epoxidase subunit A [EC:1.14.13.208] | -2.792 | 2.260 | 0.218 | 0.978 |
| boxB; benzoyl-CoA 2,3-epoxidase subunit B [EC:1.14.13.208] | -1.730 | 0.934 | 0.066 | 0.839 |
| boxC; benzoyl-CoA-dihydrodiol lyase [EC:4.1.2.44] | -1.730 | 0.934 | 0.066 | 0.839 |
| boxD; 3,4-dehydroadipyl-CoA semialdehyde dehydrogenase [EC:1.2.1.77] | -3.265 | 3.528 | 0.356 | 0.978 |
| boxR, bzdR; XRE family transcriptional regulator, aerobic/anaerobic benzoate catabolism transcriptional regulator | -1.796 | 0.891 | 0.045 | 0.839 |
| bpeE; membrane fusion protein, multidrug efflux system | -0.760 | 0.417 | 0.070 | 0.841 |
| bpeF; multidrug efflux pump | -0.690 | 0.447 | 0.125 | 0.923 |
| bpeT; LysR family transcriptional regulator, regulator for bpeEF and oprC | -0.298 | 0.651 | 0.648 | 0.999 |
| bphD; 2,6-dioxo-6-phenylhexa-3-enoate hydrolase [EC:3.7.1.8] | 1.513 | 1.385 | 0.276 | 0.978 |
| bphH, xylJ, tesE; 2-oxopent-4-enoate/cis-2-oxohex-4-enoate hydratase [EC:4.2.1.80 4.2.1.132] | 0.639 | 0.465 | 0.172 | 0.961 |
| bphI, xylK, nahM, tesG; 4-hydroxy-2-oxovalerate/4-hydroxy-2-oxohexanoate aldolase [EC:4.1.3.39 4.1.3.43] | -0.058 | 0.514 | 0.910 | 0.999 |
| bphJ, xylQ, nahO, tesF; acetaldehyde/propanal dehydrogenase [EC:1.2.1.10 1.2.1.87] | -0.058 | 0.514 | 0.910 | 0.999 |
| bpr; bacillopeptidase F [EC:3.4.21.-] | 2.533 | 1.763 | 0.153 | 0.954 |
| bpsA, srsA, pks11, pks10; alkylresorcinol/alkylpyrone synthase | -1.154 | 0.787 | 0.145 | 0.951 |
| bpsA; N4-bis(aminopropyl)spermidine synthase [EC:2.5.1.128] | 18.245 | 2974.604 | 0.995 | 0.999 |
| bpsB, srsB; methyltransferase | -1.449 | 0.786 | 0.067 | 0.839 |
| braD, bceA; bacitracin transport system ATP-binding protein | -0.467 | 0.685 | 0.496 | 0.981 |
| braE, bceB; bacitracin transport system permease protein | -0.394 | 0.621 | 0.527 | 0.989 |
| braR, bceR; two-component system, OmpR family, response regulator protein BraR/BceR | 0.087 | 0.436 | 0.842 | 0.999 |
| braS, bceS; two-component system, OmpR family, sensor histidine kinase BraS/BceS [EC:2.7.13.3] | 0.087 | 0.436 | 0.842 | 0.999 |
| bshA; L-malate glycosyltransferase [EC:2.4.1.-] | 0.383 | 0.215 | 0.077 | 0.856 |
| bssR; biofilm regulator BssR | -0.243 | 0.717 | 0.735 | 0.999 |
| bssS; biofilm regulator BssS | 0.119 | 0.601 | 0.843 | 0.999 |
| btaA; S-adenosylmethionine-diacylglycerol 3-amino-3-carboxypropyl transferase | 0.109 | 0.323 | 0.735 | 0.999 |
| btaB; S-adenosylmethionine-diacylgycerolhomoserine-N-methlytransferase | -1.153 | 0.548 | 0.037 | 0.839 |
| btaE; hyaluronate-binding autotransporter adhesin | -2.174 | 2.005 | 0.280 | 0.978 |
| btaF; ECM component-binding autotransporter adhesin | -2.174 | 2.005 | 0.280 | 0.978 |
| btuB; vitamin B12 transporter | 0.011 | 0.127 | 0.930 | 0.999 |
| budC; meso-butanediol dehydrogenase / (S,S)-butanediol dehydrogenase / diacetyl reductase [EC:1.1.1.- 1.1.1.76 1.1.1.304] | 0.606 | 0.604 | 0.317 | 0.978 |
| buk; butyrate kinase [EC:2.7.2.7] | -0.237 | 0.237 | 0.319 | 0.978 |
| butA, budC; meso-butanediol dehydrogenase / (S,S)-butanediol dehydrogenase / diacetyl reductase [EC:1.1.1.- 1.1.1.76 1.1.1.304] | -0.116 | 0.192 | 0.545 | 0.990 |
| bxlA; beta-xylosidase | -17.703 | 3891.582 | 0.996 | 0.999 |
| bxlE; xylobiose transport system substrate-binding protein | -17.470 | 2403.378 | 0.994 | 0.999 |
| bxlF; xylobiose transport system permease protein | -17.470 | 2403.378 | 0.994 | 0.999 |
| bxlG; xylobiose transport system permease protein | -17.470 | 2403.378 | 0.994 | 0.999 |
| C5AP, scpA, scpB; C5a peptidase [EC:3.4.21.110] | 0.143 | 1.011 | 0.887 | 0.999 |
| cadB; cadaverine:lysine antiporter | 0.088 | 0.766 | 0.909 | 0.999 |
| cadC; transcriptional activator of cad operon | -0.074 | 0.827 | 0.929 | 0.999 |
| cag1; cag pathogenicity island protein 1 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag10; cag pathogenicity island protein 10 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag11; cag pathogenicity island protein 11 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag12; cag pathogenicity island protein 12 | -0.684 | 1.205 | 0.571 | 0.995 |
| cag13; cag pathogenicity island protein 13 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag14; cag pathogenicity island protein 14 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag15; cag pathogenicity island protein 15 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag16; cag pathogenicity island protein 16 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag17; cag pathogenicity island protein 17 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag18; cag pathogenicity island protein 18 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag19; cag pathogenicity island protein 19 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag2; cag pathogenicity island protein 2 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag20; cag pathogenicity island protein 20 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag21; cag pathogenicity island protein 21 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag22; cag pathogenicity island protein 22 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag23; cag pathogenicity island protein 23 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag24; cag pathogenicity island protein 24 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag25; cag pathogenicity island protein 25 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag3; cag pathogenicity island protein 3 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag4; cag pathogenicity island protein 4 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag5; cag pathogenicity island protein 5 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag6; cag pathogenicity island protein 6 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag7; cag pathogenicity island protein 7 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag8; cag pathogenicity island protein 8 | -1.386 | 1.551 | 0.373 | 0.978 |
| cag9; cag pathogenicity island protein 9 | -1.386 | 1.551 | 0.373 | 0.978 |
| cagA; cytotoxicity-associated immunodominant antigen | -1.386 | 1.551 | 0.373 | 0.978 |
| cah; carbonic anhydrase [EC:4.2.1.1] | 0.211 | 0.296 | 0.477 | 0.979 |
| cah; cephalosporin-C deacetylase [EC:3.1.1.41] | -0.015 | 0.195 | 0.939 | 0.999 |
| caiA; crotonobetainyl-CoA dehydrogenase [EC:1.3.8.13] | -0.348 | 0.888 | 0.696 | 0.999 |
| caiB; L-carnitine CoA-transferase [EC:2.8.3.21] | -0.285 | 0.818 | 0.728 | 0.999 |
| caiC; carnitine-CoA ligase [EC:6.2.1.48] | -0.662 | 0.498 | 0.185 | 0.966 |
| caiD; crotonobetainyl-CoA hydratase [EC:4.2.1.149] | -1.726 | 0.568 | 0.003 | 0.839 |
| caiE; carnitine operon protein CaiE | -0.982 | 1.042 | 0.347 | 0.978 |
| caiF; transcriptional activator CaiF | -1.130 | 1.346 | 0.402 | 0.978 |
| caiT; L-carnitine/gamma-butyrobetaine antiporter | -0.715 | 1.208 | 0.555 | 0.992 |
| camD; 5-exo-hydroxycamphor dehydrogenase [EC:1.1.1.327] | -2.224 | 2.044 | 0.278 | 0.978 |
| cansdh; carboxynorspermidine synthase [EC:1.5.1.43] | 1.142 | 1.423 | 0.423 | 0.978 |
| capA, pgsA; gamma-polyglutamate biosynthesis protein CapA | -0.074 | 0.119 | 0.533 | 0.990 |
| capD; UDP-glucose 4-epimerase [EC:5.1.3.2] | -0.584 | 0.371 | 0.118 | 0.917 |
| carA, CPA1; carbamoyl-phosphate synthase small subunit [EC:6.3.5.5] | 0.010 | 0.054 | 0.854 | 0.999 |
| carAa; carbazole 1,9a-dioxygenase [EC:1.14.12.22] | 1.513 | 1.385 | 0.276 | 0.978 |
| carAc; carbazole 1,9a-dioxygenase ferredoxin component | 1.513 | 1.385 | 0.276 | 0.978 |
| carAd; carbazole 1,9a-dioxygenase ferredoxin reductase component | 1.513 | 1.385 | 0.276 | 0.978 |
| carB, CPA2; carbamoyl-phosphate synthase large subunit [EC:6.3.5.5] | 0.018 | 0.053 | 0.732 | 0.999 |
| carBa; 2’-aminobiphenyl-2,3-diol 1,2-dioxygenase, small subunit [EC:1.13.11.-] | 1.513 | 1.385 | 0.276 | 0.978 |
| carBb; 2’-aminobiphenyl-2,3-diol 1,2-dioxygenase, large subunit [EC:1.13.11.-] | 1.513 | 1.385 | 0.276 | 0.978 |
| carC; 2-hydroxy-6-oxo-6-(2’-aminophenyl)hexa-2,4-dienoate hydrolase [EC:3.7.1.13] | 1.513 | 1.385 | 0.276 | 0.978 |
| carD; CarD family transcriptional regulator | -0.289 | 0.197 | 0.145 | 0.951 |
| CARP, pepA; leucyl aminopeptidase [EC:3.4.11.1] | -0.083 | 0.093 | 0.374 | 0.978 |
| CARS, cysS; cysteinyl-tRNA synthetase [EC:6.1.1.16] | 0.035 | 0.032 | 0.279 | 0.978 |
| cas1; CRISP-associated protein Cas1 | 0.109 | 0.096 | 0.258 | 0.978 |
| cas2; CRISPR-associated protein Cas2 | 0.166 | 0.100 | 0.099 | 0.896 |
| cas3; CRISPR-associated endonuclease/helicase Cas3 [EC:3.1.-.- 3.6.4.-] | 0.249 | 0.132 | 0.061 | 0.839 |
| cas4; CRISPR-associated exonuclease Cas4 [EC:3.1.12.1] | 0.180 | 0.179 | 0.316 | 0.978 |
| cas5d; CRISPR-associated protein Cas5d | -0.222 | 0.319 | 0.488 | 0.979 |
| cas5h; CRISPR-associated protein Cas5h | 0.144 | 0.628 | 0.819 | 0.999 |
| cas5t; CRISPR-associated protein Cas5t | 1.060 | 0.567 | 0.064 | 0.839 |
| cas6; CRISPR-associated endoribonuclease Cas6 [EC:3.1.-.-] | 0.214 | 0.193 | 0.267 | 0.978 |
| casA, cse1; CRISPR system Cascade subunit CasA | 0.140 | 0.395 | 0.723 | 0.999 |
| casB, cse2; CRISPR system Cascade subunit CasB | 0.086 | 0.414 | 0.836 | 0.999 |
| casC, cse4; CRISPR system Cascade subunit CasC | 0.198 | 0.299 | 0.509 | 0.981 |
| casD, cse5; CRISPR system Cascade subunit CasD | 0.155 | 0.392 | 0.694 | 0.999 |
| casE, cse3; CRISPR system Cascade subunit CasE | 0.141 | 0.395 | 0.722 | 0.999 |
| cat2, abfT; 4-hydroxybutyrate CoA-transferase [EC:2.8.3.-] | 0.491 | 0.351 | 0.164 | 0.957 |
| catA; catechol 1,2-dioxygenase [EC:1.13.11.1] | -0.107 | 0.256 | 0.677 | 0.999 |
| catA; chloramphenicol O-acetyltransferase type A [EC:2.3.1.28] | -0.877 | 0.531 | 0.101 | 0.896 |
| catB; chloramphenicol O-acetyltransferase type B [EC:2.3.1.28] | -0.311 | 0.415 | 0.455 | 0.978 |
| catB; muconate cycloisomerase [EC:5.5.1.1] | 0.016 | 0.272 | 0.954 | 0.999 |
| catC; muconolactone D-isomerase [EC:5.3.3.4] | 0.036 | 0.273 | 0.894 | 0.999 |
| catE; catechol 2,3-dioxygenase [EC:1.13.11.2] | -0.134 | 0.147 | 0.363 | 0.978 |
| cbe, mbe; cellobiose epimerase [EC:5.1.3.11] | -0.960 | 0.463 | 0.040 | 0.839 |
| cbeA; cytoskeleton bundling-enhancing protein CbeA and related proteins | -1.818 | 1.796 | 0.313 | 0.978 |
| cbf, cbf1; 3’-5’ exoribonuclease [EC:3.1.-.-] | 0.002 | 0.120 | 0.985 | 0.999 |
| CBH2, cbhA; cellulose 1,4-beta-cellobiosidase [EC:3.2.1.91] | 0.609 | 1.214 | 0.617 | 0.999 |
| cbiB, cobD; adenosylcobinamide-phosphate synthase [EC:6.3.1.10] | 0.099 | 0.112 | 0.379 | 0.978 |
| cbiD; cobalt-precorrin-5B (C1)-methyltransferase [EC:2.1.1.195] | 0.290 | 0.146 | 0.048 | 0.839 |
| cbiE; cobalt-precorrin-7 (C5)-methyltransferase [EC:2.1.1.289] | 0.539 | 0.215 | 0.013 | 0.839 |
| cbiG; cobalt-precorrin 5A hydrolase [EC:3.7.1.12] | 0.297 | 0.135 | 0.030 | 0.839 |
| cbiGH-cobJ; cobalt-precorrin 5A hydrolase / precorrin-3B C17-methyltransferase [EC:3.7.1.12 2.1.1.131] | 0.121 | 0.345 | 0.727 | 0.999 |
| cbiK; nickel transport protein | 0.272 | 0.653 | 0.677 | 0.999 |
| cbiK; sirohydrochlorin cobaltochelatase [EC:4.99.1.3] | 0.367 | 0.188 | 0.053 | 0.839 |
| cbiL; nickel transport protein | 0.042 | 0.323 | 0.896 | 0.999 |
| cbiM; cobalt/nickel transport system permease protein | 0.097 | 0.243 | 0.689 | 0.999 |
| cbiN; cobalt/nickel transport protein | 0.668 | 0.310 | 0.033 | 0.839 |
| cbiO; cobalt/nickel transport system ATP-binding protein | 0.092 | 0.240 | 0.701 | 0.999 |
| cbiQ; cobalt/nickel transport system permease protein | 0.205 | 0.231 | 0.378 | 0.978 |
| cbiT; cobalt-precorrin-6B (C15)-methyltransferase [EC:2.1.1.196] | 0.561 | 0.218 | 0.011 | 0.839 |
| cbiX; sirohydrochlorin cobaltochelatase [EC:4.99.1.3] | -0.834 | 0.568 | 0.144 | 0.951 |
| cbl; LysR family transcriptional regulator, cys regulon transcriptional activator | -0.312 | 0.343 | 0.365 | 0.978 |
| CBP3, UQCC; cytochrome b pre-mRNA-processing protein 3 | -1.109 | 0.434 | 0.012 | 0.839 |
| cbpA; curved DNA-binding protein | -0.135 | 0.131 | 0.301 | 0.978 |
| cbpM; chaperone modulatory protein CbpM | 0.032 | 0.232 | 0.891 | 0.999 |
| cbrT; energy-coupling factor transport system substrate-specific component | -0.874 | 0.473 | 0.067 | 0.839 |
| CBS; cystathionine beta-synthase [EC:4.2.1.22] | -0.194 | 0.223 | 0.385 | 0.978 |
| cbtA; cytoskeleton-binding toxin CbtA and related proteins | -0.735 | 1.149 | 0.523 | 0.989 |
| cca; tRNA nucleotidyltransferase (CCA-adding enzyme) [EC:2.7.7.72 3.1.3.- 3.1.4.-] | 0.111 | 0.051 | 0.031 | 0.839 |
| cccA; cytochrome c550 | -1.518 | 0.736 | 0.041 | 0.839 |
| cccB; cytochrome c551 | -1.475 | 0.759 | 0.054 | 0.839 |
| ccdA; antitoxin CcdA | 0.076 | 0.596 | 0.899 | 0.999 |
| ccdA; cytochrome c-type biogenesis protein | -0.026 | 0.124 | 0.835 | 0.999 |
| ccdB; toxin CcdB | 0.160 | 0.637 | 0.801 | 0.999 |
| cciR; LuxR family transcriptional regulator, quorum-sensing system regulator CciR | -1.019 | 0.711 | 0.154 | 0.954 |
| cckA; two-component system, cell cycle sensor histidine kinase and response regulator CckA [EC:2.7.13.3] | -1.110 | 0.434 | 0.011 | 0.839 |
| ccmA; heme exporter protein A [EC:3.6.3.41] | -0.012 | 0.148 | 0.937 | 0.999 |
| ccmB; heme exporter protein B | -0.010 | 0.147 | 0.948 | 0.999 |
| ccmC; heme exporter protein C | -0.016 | 0.147 | 0.916 | 0.999 |
| ccmD; heme exporter protein D | 0.057 | 0.178 | 0.750 | 0.999 |
| ccmE; cytochrome c-type biogenesis protein CcmE | -0.017 | 0.147 | 0.907 | 0.999 |
| ccmF; cytochrome c-type biogenesis protein CcmF | -0.033 | 0.147 | 0.821 | 0.999 |
| ccmG, dsbE; cytochrome c biogenesis protein CcmG, thiol:disulfide interchange protein DsbE | -0.012 | 0.172 | 0.947 | 0.999 |
| ccmH; cytochrome c-type biogenesis protein CcmH | -0.037 | 0.172 | 0.832 | 0.999 |
| ccoN; cytochrome c oxidase cbb3-type subunit I [EC:1.9.3.1] | -0.060 | 0.265 | 0.820 | 0.999 |
| ccoNO; cytochrome c oxidase cbb3-type subunit I/II [EC:1.9.3.1] | -0.186 | 0.512 | 0.717 | 0.999 |
| ccoO; cytochrome c oxidase cbb3-type subunit II | -0.007 | 0.224 | 0.976 | 0.999 |
| ccoP; cytochrome c oxidase cbb3-type subunit III | -0.042 | 0.236 | 0.861 | 0.999 |
| ccoQ; cytochrome c oxidase cbb3-type subunit IV | 0.009 | 0.228 | 0.968 | 0.999 |
| ccr; crotonyl-CoA carboxylase/reductase [EC:1.3.1.85] | -1.101 | 0.945 | 0.246 | 0.978 |
| ccrA; crotonyl-CoA reductase [EC:1.3.1.86] | 1.083 | 0.694 | 0.120 | 0.917 |
| ccrM; modification methylase [EC:2.1.1.72] | -0.894 | 0.322 | 0.006 | 0.839 |
| ccsA; citryl-CoA synthetase large subunit [EC:6.2.1.18] | -1.047 | 1.651 | 0.527 | 0.989 |
| cd, ma, nplT; cyclomaltodextrinase / maltogenic alpha-amylase / neopullulanase [EC:3.2.1.54 3.2.1.133 3.2.1.135] | -17.703 | 3891.582 | 0.996 | 0.999 |
| cdaR; carbohydrate diacid regulator | 0.194 | 0.227 | 0.395 | 0.978 |
| cdc6A; archaeal cell division control protein 6 | -16.927 | 2639.780 | 0.995 | 0.999 |
| cdd, CDA; cytidine deaminase [EC:3.5.4.5] | -0.021 | 0.082 | 0.797 | 0.999 |
| cdgJ; c-di-GMP phosphodiesterase [EC:3.1.4.52] | -0.431 | 0.507 | 0.397 | 0.978 |
| cdh; CDP-diacylglycerol pyrophosphatase [EC:3.6.1.26] | -0.100 | 0.369 | 0.786 | 0.999 |
| cdhD, acsD; acetyl-CoA decarbonylase/synthase complex subunit delta [EC:2.1.1.245] | -0.727 | 1.126 | 0.520 | 0.988 |
| cdhE, acsC; acetyl-CoA decarbonylase/synthase complex subunit gamma [EC:2.1.1.245] | -0.789 | 1.083 | 0.468 | 0.979 |
| cdhR; AraC family transcriptional regulator, carnitine catabolism transcriptional activator | 0.186 | 0.370 | 0.616 | 0.999 |
| CDIPT; CDP-diacylglycerol–inositol 3-phosphatidyltransferase [EC:2.7.8.11] | -2.295 | 1.765 | 0.195 | 0.970 |
| CDO1; cysteine dioxygenase [EC:1.13.11.20] | -18.166 | 3639.630 | 0.996 | 0.999 |
| cdr; CoA-disulfide reductase [EC:1.8.1.14] | -0.198 | 0.832 | 0.812 | 0.999 |
| cebE; cellobiose transport system substrate-binding protein | 0.106 | 0.588 | 0.857 | 0.999 |
| cebF; cellobiose transport system permease protein | -0.077 | 0.639 | 0.904 | 0.999 |
| cebG; cellobiose transport system permease protein | -0.077 | 0.639 | 0.904 | 0.999 |
| CECR1, ADA2; adenosine deaminase CECR1 [EC:3.5.4.4] | -0.261 | 1.196 | 0.828 | 0.999 |
| cedA; cell division activator | -0.131 | 0.760 | 0.863 | 0.999 |
| cefD; isopenicillin-N epimerase [EC:5.1.1.17] | -17.999 | 4282.978 | 0.997 | 0.999 |
| ceo; N5-(carboxyethyl)ornithine synthase [EC:1.5.1.24] | 0.270 | 0.918 | 0.769 | 0.999 |
| cfa; cAMP factor | -2.011 | 2.150 | 0.351 | 0.978 |
| cfa; cyclopropane-fatty-acyl-phospholipid synthase [EC:2.1.1.79] | -0.048 | 0.135 | 0.719 | 0.999 |
| cgeB; spore maturation protein CgeB | -0.081 | 0.372 | 0.829 | 0.999 |
| cgeE; spore maturation protein CgeE | 18.239 | 2966.075 | 0.995 | 0.999 |
| cggR; central glycolytic genes regulator | 0.008 | 0.158 | 0.960 | 0.999 |
| cgt; cyclomaltodextrin glucanotransferase [EC:2.4.1.19] | 17.922 | 2530.829 | 0.994 | 0.999 |
| cgtA; beta-1,4-N-acetylgalactosaminyltransferase [EC:2.4.1.-] | -1.386 | 1.551 | 0.373 | 0.978 |
| chaA, CAX; Ca2+:H+ antiporter | -0.727 | 0.323 | 0.026 | 0.839 |
| chaB; cation transport regulator | 0.167 | 0.620 | 0.788 | 0.999 |
| CHAC, chaC; glutathione-specific gamma-glutamylcyclotransferase [EC:4.3.2.7] | -0.455 | 0.285 | 0.112 | 0.917 |
| chbG; chitin disaccharide deacetylase [EC:3.5.1.105] | -0.287 | 0.363 | 0.430 | 0.978 |
| chbP; N,N’-diacetylchitobiose phosphorylase [EC:2.4.1.280] | 18.119 | 1920.327 | 0.992 | 0.999 |
| chbR, celD; AraC family transcriptional regulator, dual regulator of chb operon | -0.067 | 0.598 | 0.911 | 0.999 |
| cheA; two-component system, chemotaxis family, sensor kinase CheA [EC:2.7.13.3] | -0.164 | 0.176 | 0.354 | 0.978 |
| cheB; two-component system, chemotaxis family, protein-glutamate methylesterase/glutaminase [EC:3.1.1.61 3.5.1.44] | -0.107 | 0.184 | 0.562 | 0.992 |
| cheBR; two-component system, chemotaxis family, CheB/CheR fusion protein [EC:2.1.1.80 3.1.1.61] | -0.209 | 0.386 | 0.588 | 0.999 |
| cheC; chemotaxis protein CheC | -0.316 | 0.351 | 0.370 | 0.978 |
| cheD; chemotaxis protein CheD [EC:3.5.1.44] | -0.206 | 0.190 | 0.278 | 0.978 |
| cheR; chemotaxis protein methyltransferase CheR [EC:2.1.1.80] | -0.096 | 0.182 | 0.598 | 0.999 |
| cheV; two-component system, chemotaxis family, chemotaxis protein CheV | -0.022 | 0.229 | 0.922 | 0.999 |
| cheW; purine-binding chemotaxis protein CheW | -0.157 | 0.174 | 0.369 | 0.978 |
| cheX; chemotaxis protein CheX | 0.028 | 0.403 | 0.945 | 0.999 |
| cheY; two-component system, chemotaxis family, chemotaxis protein CheY | -0.182 | 0.150 | 0.228 | 0.978 |
| cheZ; chemotaxis protein CheZ | 0.005 | 0.227 | 0.984 | 0.999 |
| chiA; bifunctional chitinase/lysozyme [EC:3.2.1.14 3.2.1.17] | -0.931 | 1.250 | 0.458 | 0.979 |
| chiE; putative chitobiose transport system substrate-binding protein | -1.528 | 1.228 | 0.215 | 0.978 |
| chiF; putative chitobiose transport system permease protein | -1.533 | 1.224 | 0.212 | 0.978 |
| chiG; putative chitobiose transport system permease protein | -1.483 | 1.248 | 0.237 | 0.978 |
| chiS; two-component system, sensor histidine kinase ChiS | 1.818 | 1.333 | 0.175 | 0.961 |
| chlB; light-independent protochlorophyllide reductase subunit B [EC:1.3.7.7] | -2.547 | 1.881 | 0.178 | 0.961 |
| chlD, bchD; magnesium chelatase subunit D [EC:6.6.1.1] | 0.126 | 0.222 | 0.571 | 0.995 |
| chlG, bchG; chlorophyll/bacteriochlorophyll a synthase [EC:2.5.1.62 2.5.1.133] | -18.056 | 3176.254 | 0.995 | 0.999 |
| chlH, bchH; magnesium chelatase subunit H [EC:6.6.1.1] | -2.415 | 1.668 | 0.150 | 0.954 |
| chlI, bchI; magnesium chelatase subunit I [EC:6.6.1.1] | 0.115 | 0.165 | 0.487 | 0.979 |
| chlL; light-independent protochlorophyllide reductase subunit L [EC:1.3.7.7] | -3.309 | 3.390 | 0.331 | 0.978 |
| chlN; light-independent protochlorophyllide reductase subunit N [EC:1.3.7.7] | -2.547 | 1.881 | 0.178 | 0.961 |
| chlP, bchP; geranylgeranyl diphosphate/geranylgeranyl-bacteriochlorophyllide a reductase [EC:1.3.1.83 1.3.1.111] | -18.056 | 3176.254 | 0.995 | 0.999 |
| chnB; cyclohexanone monooxygenase [EC:1.14.13.22] | -0.863 | 0.502 | 0.088 | 0.880 |
| cho; excinuclease Cho [EC:3.1.25.-] | 0.041 | 0.560 | 0.942 | 0.999 |
| CHO1, pssA; CDP-diacylglycerol—serine O-phosphatidyltransferase [EC:2.7.8.8] | -0.050 | 0.071 | 0.477 | 0.979 |
| choD; cholesterol oxidase [EC:1.1.3.6] | -1.088 | 0.900 | 0.229 | 0.978 |
| chpA; chemosensory pili system protein ChpA (sensor histidine kinase/response regulator) | -0.068 | 0.298 | 0.819 | 0.999 |
| chpB, chpBK; mRNA interferase ChpB [EC:3.1.-.-] | -0.873 | 0.999 | 0.384 | 0.978 |
| chpB; chemosensory pili system protein ChpB (putative protein-glutamate methylesterase) | 0.212 | 0.337 | 0.529 | 0.989 |
| chpC; chemosensory pili system protein ChpC | 0.118 | 0.271 | 0.664 | 0.999 |
| chpD; AraC family transcriptional regulator, chemosensory pili system protein ChpD | -0.976 | 0.998 | 0.329 | 0.978 |
| chpE; chemosensory pili system protein ChpE | -0.942 | 0.967 | 0.332 | 0.978 |
| chpS, chpBI; antitoxin ChpS | -0.033 | 0.795 | 0.967 | 0.999 |
| chpT; histidine phosphotransferase ChpT | -1.210 | 0.442 | 0.007 | 0.839 |
| chqB; hydroxyquinol 1,2-dioxygenase [EC:1.13.11.37] | -0.393 | 0.707 | 0.579 | 0.997 |
| chr, crh; catabolite repression HPr-like protein | -0.159 | 0.359 | 0.659 | 0.999 |
| chrA; chromate transporter | -0.042 | 0.117 | 0.718 | 0.999 |
| chrR, NQR; chromate reductase, NAD(P)H dehydrogenase (quinone) | -0.061 | 0.233 | 0.796 | 0.999 |
| chrR; putative transcriptional regulator | -0.241 | 0.398 | 0.546 | 0.990 |
| chuX; heme iron utilization protein | -0.447 | 0.479 | 0.352 | 0.978 |
| chvB, cgs, ndvB; cyclic beta-1,2-glucan synthetase [EC:2.4.1.-] | -2.205 | 1.237 | 0.076 | 0.856 |
| chvG; two-component system, OmpR family, sensor histidine kinase ChvG [EC:2.7.13.3] | -1.064 | 0.432 | 0.015 | 0.839 |
| chvI; two-component system, OmpR family, response regulator ChvI | -1.064 | 0.432 | 0.015 | 0.839 |
| ciaH; two-component system, OmpR family, sensor histidine kinase CiaH [EC:2.7.13.3] | -0.015 | 0.172 | 0.930 | 0.999 |
| ciaR; two-component system, OmpR family, response regulator CiaR | -0.018 | 0.171 | 0.916 | 0.999 |
| ciaX; regulatory peptide CiaX | 18.830 | 3661.468 | 0.996 | 0.999 |
| cidA; holin-like protein | 0.069 | 0.079 | 0.386 | 0.978 |
| cimA; (R)-citramalate synthase [EC:2.3.1.182] | -0.593 | 0.574 | 0.303 | 0.978 |
| citA, tcuC; MFS transporter, MHS family, citrate/tricarballylate:H+ symporter | -0.346 | 0.485 | 0.477 | 0.979 |
| citB, tcuB; citrate/tricarballylate utilization protein | -0.855 | 0.547 | 0.120 | 0.917 |
| citC; [citrate (pro-3S)-lyase] ligase [EC:6.2.1.22] | 0.767 | 0.297 | 0.011 | 0.839 |
| citD; citrate lyase subunit gamma (acyl carrier protein) | 0.497 | 0.190 | 0.010 | 0.839 |
| citE; citrate lyase subunit beta / citryl-CoA lyase [EC:4.1.3.34] | 0.013 | 0.140 | 0.929 | 0.999 |
| citF; citrate lyase subunit alpha / citrate CoA-transferase [EC:2.8.3.10] | 0.493 | 0.188 | 0.010 | 0.839 |
| citG; triphosphoribosyl-dephospho-CoA synthase [EC:2.4.2.52] | 1.003 | 0.421 | 0.018 | 0.839 |
| citR; LysR family transcriptional regulator, repressor for citA | -0.635 | 0.832 | 0.446 | 0.978 |
| citS; two-component system, CitB family, sensor histidine kinase CitS [EC:2.7.13.3] | -2.116 | 1.208 | 0.082 | 0.865 |
| citT; citrate:succinate antiporter | -0.042 | 0.268 | 0.876 | 0.999 |
| citX; holo-ACP synthase [EC:2.7.7.61] | 1.060 | 0.437 | 0.017 | 0.839 |
| citXG; holo-ACP synthase / triphosphoribosyl-dephospho-CoA synthase [EC:2.7.7.61 2.4.2.52] | 0.424 | 0.521 | 0.417 | 0.978 |
| cld; chlorite dismutase [EC:1.13.11.49] | -0.180 | 0.180 | 0.319 | 0.978 |
| clfA; clumping factor A | -1.670 | 1.245 | 0.182 | 0.961 |
| clfB; clumping factor B | -1.670 | 1.245 | 0.182 | 0.961 |
| cloSI; clostripain [EC:3.4.22.8] | 6.328 | 12.689 | 0.619 | 0.999 |
| clpA; ATP-dependent Clp protease ATP-binding subunit ClpA | -0.170 | 0.160 | 0.290 | 0.978 |
| clpB; ATP-dependent Clp protease ATP-binding subunit ClpB | 0.009 | 0.058 | 0.878 | 0.999 |
| clpC; ATP-dependent Clp protease ATP-binding subunit ClpC | 0.005 | 0.082 | 0.956 | 0.999 |
| clpE; ATP-dependent Clp protease ATP-binding subunit ClpE | 0.085 | 0.155 | 0.584 | 0.999 |
| clpL; ATP-dependent Clp protease ATP-binding subunit ClpL | 0.019 | 0.160 | 0.906 | 0.999 |
| clpP, CLPP; ATP-dependent Clp protease, protease subunit [EC:3.4.21.92] | 0.015 | 0.044 | 0.739 | 0.999 |
| clpS; ATP-dependent Clp protease adaptor protein ClpS | -0.143 | 0.127 | 0.263 | 0.978 |
| clpX, CLPX; ATP-dependent Clp protease ATP-binding subunit ClpX | 0.050 | 0.041 | 0.226 | 0.978 |
| clrA, serA; complex iron-sulfur molybdoenzyme family reductase subunit alpha | -17.931 | 4360.659 | 0.997 | 0.999 |
| clrb, serB; complex iron-sulfur molybdoenzyme family reductase subunit beta | -17.931 | 4360.659 | 0.997 | 0.999 |
| clrC, serC; complex iron-sulfur molybdoenzyme family reductase subunit gamma | -17.931 | 4360.659 | 0.997 | 0.999 |
| clsA_B; cardiolipin synthase A/B [EC:2.7.8.-] | 0.040 | 0.050 | 0.427 | 0.978 |
| clsC; cardiolipin synthase C [EC:2.7.8.-] | 0.071 | 0.198 | 0.722 | 0.999 |
| cmaB; non-haem Fe2+, alpha-ketoglutarate-dependent halogenase | 18.932 | 3654.080 | 0.996 | 0.999 |
| cmeR; TetR/AcrR family transcriptional regulator, cmeABC operon repressor | 0.936 | 0.710 | 0.189 | 0.966 |
| cmk; CMP/dCMP kinase [EC:2.7.4.25] | 0.050 | 0.044 | 0.251 | 0.978 |
| cmlA, cmlB, floR; MFS transporter, DHA1 family, florfenicol/chloramphenicol resistance protein | -0.062 | 0.452 | 0.892 | 0.999 |
| cmlR, cmx; MFS transporter, DHA1 family, chloramphenicol resistance protein | -17.792 | 3118.911 | 0.995 | 0.999 |
| cmmT, thnT; putative pantetheine hydrolase [EC:3.5.1.-] | -17.794 | 4072.025 | 0.997 | 0.999 |
| CMO; choline monooxygenase [EC:1.14.15.7] | 0.140 | 0.455 | 0.758 | 0.999 |
| cmoA; tRNA (cmo5U34)-methyltransferase [EC:2.1.1.-] | 0.089 | 0.123 | 0.471 | 0.979 |
| cmoB; tRNA (mo5U34)-methyltransferase [EC:2.1.1.-] | 0.123 | 0.214 | 0.565 | 0.994 |
| cmr; MFS transporter, DHA3 family, multidrug efflux protein | 0.193 | 0.978 | 0.844 | 0.999 |
| cmr2, cas10; CRISPR-associated protein Cmr2 | 0.158 | 1.340 | 0.906 | 0.999 |
| cmr3; CRISPR-associated protein Cmr3 | -17.860 | 2336.427 | 0.994 | 0.999 |
| cmr4; CRISPR-associated protein Cmr4 | -0.193 | 1.135 | 0.865 | 0.999 |
| cmr5; CRISPR-associated protein Cmr5 | -0.193 | 1.135 | 0.865 | 0.999 |
| cmr6; CRISPR-associated protein Cmr6 | -0.175 | 0.787 | 0.824 | 0.999 |
| cmtC, dhbA; 2,3-dihydroxy-p-cumate/2,3-dihydroxybenzoate 3,4-dioxygenase [EC:1.13.11.- 1.13.11.14] | 0.589 | 1.020 | 0.565 | 0.994 |
| cmtD, dhbB; HCOMODA/2-hydroxy-3-carboxy-muconic semialdehyde decarboxylase [EC:4.1.1.-] | 0.077 | 0.877 | 0.930 | 0.999 |
| CNP; 2’,3’-cyclic-nucleotide 3’-phosphodiesterase [EC:3.1.4.37] | 1.286 | 0.611 | 0.037 | 0.839 |
| coaA; type I pantothenate kinase [EC:2.7.1.33] | 0.093 | 0.113 | 0.413 | 0.978 |
| coaBC, dfp; phosphopantothenoylcysteine decarboxylase / phosphopantothenate—cysteine ligase [EC:4.1.1.36 6.3.2.5] | 0.051 | 0.049 | 0.305 | 0.978 |
| coaE; dephospho-CoA kinase [EC:2.7.1.24] | 0.031 | 0.044 | 0.486 | 0.979 |
| coaW; type II pantothenate kinase [EC:2.7.1.33] | -0.376 | 0.241 | 0.121 | 0.919 |
| coaX; type III pantothenate kinase [EC:2.7.1.33] | 0.010 | 0.082 | 0.905 | 0.999 |
| cobA-hemD; uroporphyrinogen III methyltransferase / synthase [EC:2.1.1.107 4.2.1.75] | 0.288 | 0.161 | 0.076 | 0.855 |
| cobA, btuR; cob(I)alamin adenosyltransferase [EC:2.5.1.17] | 0.129 | 0.120 | 0.284 | 0.978 |
| cobA; uroporphyrin-III C-methyltransferase [EC:2.1.1.107] | -0.327 | 0.190 | 0.087 | 0.880 |
| cobB-cbiA; cobyrinic acid a,c-diamide synthase [EC:6.3.5.9 6.3.5.11] | 0.131 | 0.116 | 0.260 | 0.978 |
| cobC, phpB; alpha-ribazole phosphatase [EC:3.1.3.73] | 0.051 | 0.106 | 0.633 | 0.999 |
| cobC1, cobC; cobalamin biosynthetic protein CobC | -0.226 | 0.272 | 0.407 | 0.978 |
| cobD; threonine-phosphate decarboxylase [EC:4.1.1.81] | 0.282 | 0.172 | 0.103 | 0.904 |
| cobF; precorrin-6A synthase [EC:2.1.1.152] | -0.209 | 0.292 | 0.476 | 0.979 |
| cobG; precorrin-3B synthase [EC:1.14.13.83] | -0.099 | 0.294 | 0.737 | 0.999 |
| cobH-cbiC; precorrin-8X/cobalt-precorrin-8 methylmutase [EC:5.4.99.61 5.4.99.60] | 0.228 | 0.130 | 0.082 | 0.865 |
| cobI-cbiL; precorrin-2/cobalt-factor-2 C20-methyltransferase [EC:2.1.1.130 2.1.1.151] | 0.250 | 0.134 | 0.065 | 0.839 |
| cobIJ; precorrin-2 C20-methyltransferase / precorrin-3B C17-methyltransferase [EC:2.1.1.130 2.1.1.131] | -0.654 | 0.610 | 0.286 | 0.978 |
| cobK-cbiJ; precorrin-6A/cobalt-precorrin-6A reductase [EC:1.3.1.54 1.3.1.106] | 0.278 | 0.132 | 0.036 | 0.839 |
| cobL; precorrin-6Y C5,15-methyltransferase (decarboxylating) [EC:2.1.1.132] | -0.055 | 0.198 | 0.782 | 0.999 |
| cobM, cbiF; precorrin-4/cobalt-precorrin-4 C11-methyltransferase [EC:2.1.1.133 2.1.1.271] | 0.229 | 0.130 | 0.079 | 0.859 |
| cobN; cobaltochelatase CobN [EC:6.6.1.2] | -0.061 | 0.166 | 0.712 | 0.999 |
| cobP, cobU; adenosylcobinamide kinase / adenosylcobinamide-phosphate guanylyltransferase [EC:2.7.1.156 2.7.7.62] | 0.044 | 0.110 | 0.687 | 0.999 |
| cobQ, cbiP; adenosylcobyric acid synthase [EC:6.3.5.10] | 0.236 | 0.135 | 0.083 | 0.870 |
| cobR; cob(II)yrinic acid a,c-diamide reductase [EC:1.16.8.1] | -2.528 | 1.681 | 0.135 | 0.944 |
| cobS; cobaltochelatase CobS [EC:6.6.1.2] | -1.063 | 0.415 | 0.011 | 0.839 |
| cobT; cobaltochelatase CobT [EC:6.6.1.2] | -1.109 | 0.435 | 0.012 | 0.839 |
| cobW; cobalamin biosynthesis protein CobW | -0.143 | 0.316 | 0.652 | 0.999 |
| cobY; adenosylcobinamide-phosphate guanylyltransferase [EC:2.7.7.62] | -17.234 | 3077.514 | 0.996 | 0.999 |
| cobZ, tcuA; tricarballylate dehydrogenase | -1.023 | 0.497 | 0.041 | 0.839 |
| codA; choline oxidase [EC:1.1.3.17] | 1.196 | 1.148 | 0.299 | 0.978 |
| codA; cytosine deaminase [EC:3.5.4.1] | -0.083 | 0.194 | 0.670 | 0.999 |
| codB; cytosine permease | -0.115 | 0.311 | 0.713 | 0.999 |
| codY; transcriptional pleiotropic repressor | 0.027 | 0.125 | 0.828 | 0.999 |
| cof; HMP-PP phosphatase [EC:3.6.1.-] | -0.320 | 0.646 | 0.621 | 0.999 |
| cofC; 2-phospho-L-lactate guanylyltransferase [EC:2.7.7.68] | -0.759 | 0.566 | 0.182 | 0.961 |
| cofD; LPPG:FO 2-phospho-L-lactate transferase [EC:2.7.8.28] | -0.752 | 0.620 | 0.227 | 0.978 |
| cofE, fbiB; coenzyme F420-0:L-glutamate ligase / coenzyme F420-1:gamma-L-glutamate ligase [EC:6.3.2.31 6.3.2.34] | -0.440 | 0.538 | 0.415 | 0.978 |
| cofF; gamma-F420-2:alpha-L-glutamate ligase [EC:6.3.2.32] | -0.786 | 1.219 | 0.520 | 0.988 |
| cofH; FO synthase subunit 2 [EC:2.5.1.77] | -17.234 | 3077.514 | 0.996 | 0.999 |
| coiA; competence protein CoiA | 0.081 | 0.142 | 0.567 | 0.995 |
| colA; microbial collagenase [EC:3.4.24.3] | -0.506 | 0.753 | 0.502 | 0.981 |
| comA; ATP-binding cassette, subfamily C, bacterial, competence factor transporting protein [EC:3.4.22.-] | -0.038 | 0.211 | 0.858 | 0.999 |
| comA; phosphosulfolactate synthase [EC:4.4.1.19] | -1.102 | 0.800 | 0.170 | 0.961 |
| comA; two-component system, NarL family, competent response regulator ComA | -0.524 | 1.324 | 0.693 | 0.999 |
| comB; 2-phosphosulfolactate phosphatase [EC:3.1.3.71] | -0.551 | 0.347 | 0.114 | 0.917 |
| comB; competence factor transport accessory protein ComB | -0.041 | 0.212 | 0.847 | 0.999 |
| comB10; ComB10 competence protein | -1.386 | 1.551 | 0.373 | 0.978 |
| comB4; ComB4 competence protein | -0.613 | 1.039 | 0.556 | 0.992 |
| comB6; ComB6 competence protein | -1.386 | 1.551 | 0.373 | 0.978 |
| comB8; ComB8 competence protein | -0.260 | 0.900 | 0.773 | 0.999 |
| comB9; ComB9 competence protein | -0.260 | 0.900 | 0.773 | 0.999 |
| comC; (2R)-3-sulfolactate dehydrogenase (NADP+) [EC:1.1.1.338] | -1.631 | 1.190 | 0.173 | 0.961 |
| comC; leader peptidase (prepilin peptidase) / N-methyltransferase [EC:3.4.23.43 2.1.1.-] | 0.035 | 0.134 | 0.793 | 0.999 |
| comD; two-component system, LytTR family, sensor histidine kinase ComD [EC:2.7.13.3] | -0.192 | 0.205 | 0.351 | 0.978 |
| comE; two-component system, LytTR family, response regulator ComE | -0.088 | 0.185 | 0.636 | 0.999 |
| comEA; competence protein ComEA | 0.120 | 0.044 | 0.007 | 0.839 |
| comEB; dCMP deaminase [EC:3.5.4.12] | 0.073 | 0.089 | 0.412 | 0.978 |
| comEC; competence protein ComEC | 0.074 | 0.045 | 0.102 | 0.899 |
| comER; competence protein ComER | -2.661 | 1.051 | 0.012 | 0.839 |
| comFA; competence protein ComFA | 0.000 | 0.133 | 0.999 | 1.000 |
| comFB; competence protein ComFB | 0.103 | 0.521 | 0.844 | 0.999 |
| comFC; competence protein ComFC | 0.137 | 0.119 | 0.252 | 0.978 |
| comGA; competence protein ComGA | 0.000 | 0.133 | 0.999 | 1.000 |
| comGB; competence protein ComGB | 0.003 | 0.134 | 0.983 | 0.999 |
| comGC; competence protein ComGC | 0.000 | 0.133 | 0.999 | 1.000 |
| comGD; competence protein ComGD | 0.025 | 0.141 | 0.861 | 0.999 |
| comGE; competence protein ComGE | -3.408 | 2.625 | 0.196 | 0.970 |
| comGF; competence protein ComGF | 0.065 | 0.141 | 0.646 | 0.999 |
| comGG; competence protein ComGG | -3.418 | 2.305 | 0.140 | 0.949 |
| comK; competence protein ComK | -0.918 | 0.515 | 0.077 | 0.856 |
| comM; magnesium chelatase family protein | 0.088 | 0.081 | 0.276 | 0.978 |
| comP; two-component system, NarL family, sensor histidine kinase ComP [EC:2.7.13.3] | -0.627 | 1.390 | 0.653 | 0.999 |
| comQ; competence protein ComQ | -0.612 | 1.369 | 0.656 | 0.999 |
| COMT; catechol O-methyltransferase [EC:2.1.1.6] | -2.635 | 1.432 | 0.068 | 0.839 |
| comX; competence protein ComX | -0.622 | 1.388 | 0.655 | 0.999 |
| comX1_2; competence protein ComX | -0.084 | 0.183 | 0.646 | 0.999 |
| comZ; competence protein ComZ | -2.682 | 1.755 | 0.128 | 0.931 |
| cooC; CO dehydrogenase maturation factor | -0.682 | 0.939 | 0.469 | 0.979 |
| cooF; anaerobic carbon-monoxide dehydrogenase iron sulfur subunit | 0.752 | 0.968 | 0.438 | 0.978 |
| cooS, acsA; anaerobic carbon-monoxide dehydrogenase catalytic subunit [EC:1.2.7.4] | 0.313 | 0.241 | 0.196 | 0.970 |
| copA, ATP7; Cu+-exporting ATPase [EC:3.6.3.54] | 0.030 | 0.034 | 0.390 | 0.978 |
| copB; Cu2+-exporting ATPase [EC:3.6.3.4] | 0.000 | 0.093 | 0.996 | 0.999 |
| copC, pcoC; copper resistance protein C | -0.275 | 0.254 | 0.280 | 0.978 |
| COQ10; coenzyme Q-binding protein COQ10 | -1.205 | 0.444 | 0.007 | 0.839 |
| COQ4; ubiquinone biosynthesis protein COQ4 | -0.622 | 0.535 | 0.247 | 0.978 |
| COQ7; ubiquinone biosynthesis monooxygenase Coq7 [EC:1.14.13.-] | -0.051 | 0.209 | 0.809 | 0.999 |
| COQ9; ubiquinone biosynthesis protein COQ9 | -0.865 | 0.505 | 0.089 | 0.880 |
| corA; magnesium transporter | -0.031 | 0.039 | 0.429 | 0.978 |
| corC; magnesium and cobalt transporter | 0.046 | 0.154 | 0.768 | 0.999 |
| cotA; spore coat protein A, manganese oxidase [EC:1.16.3.3] | -1.211 | 1.014 | 0.234 | 0.978 |
| cotB; spore coat protein B | -2.453 | 2.237 | 0.275 | 0.978 |
| cotD; spore coat protein D | -1.553 | 0.763 | 0.043 | 0.839 |
| cotE; spore coat protein E | -1.431 | 0.741 | 0.055 | 0.839 |
| cotF; spore coat protein F | -3.213 | 1.928 | 0.098 | 0.896 |
| cotH; spore coat protein H | 0.059 | 0.323 | 0.855 | 0.999 |
| cotI; spore coat protein I | 1.182 | 1.331 | 0.376 | 0.978 |
| cotJA; spore coat protein JA | -1.601 | 0.839 | 0.058 | 0.839 |
| cotJB; spore coat protein JB | -1.079 | 0.543 | 0.049 | 0.839 |
| cotJC; spore coat protein JC | -1.140 | 0.557 | 0.042 | 0.839 |
| cotM; spore coat protein M | -2.263 | 1.511 | 0.136 | 0.949 |
| cotSA; spore coat protein SA | 0.577 | 0.978 | 0.556 | 0.992 |
| cotV; spore coat protein V | 18.239 | 2966.075 | 0.995 | 0.999 |
| cotW; spore coat protein W | 18.239 | 2966.075 | 0.995 | 0.999 |
| cotX; spore coat protein X | -2.290 | 1.313 | 0.083 | 0.869 |
| cotY; spore coat protein Y | -0.904 | 1.547 | 0.560 | 0.992 |
| cotZ; spore coat protein Z | -0.809 | 0.999 | 0.419 | 0.978 |
| COX11, ctaG; cytochrome c oxidase assembly protein subunit 11 | -0.171 | 0.227 | 0.451 | 0.978 |
| COX15, ctaA; cytochrome c oxidase assembly protein subunit 15 | -0.378 | 0.183 | 0.041 | 0.839 |
| coxA, ctaD; cytochrome c oxidase subunit I [EC:1.9.3.1] | -0.474 | 0.212 | 0.026 | 0.839 |
| coxA; spore cortex protein | -1.366 | 1.367 | 0.319 | 0.978 |
| coxAC; cytochrome c oxidase subunit I+III [EC:1.9.3.1] | -0.815 | 0.475 | 0.088 | 0.880 |
| coxB, ctaC; cytochrome c oxidase subunit II [EC:1.9.3.1] | -0.482 | 0.202 | 0.018 | 0.839 |
| coxC, ctaE; cytochrome c oxidase subunit III [EC:1.9.3.1] | -0.361 | 0.192 | 0.062 | 0.839 |
| coxD, ctaF; cytochrome c oxidase subunit IV [EC:1.9.3.1] | -1.053 | 0.568 | 0.066 | 0.839 |
| coxL, cutL; aerobic carbon-monoxide dehydrogenase large subunit [EC:1.2.5.3] | -1.072 | 0.581 | 0.067 | 0.839 |
| coxM, cutM; aerobic carbon-monoxide dehydrogenase medium subunit [EC:1.2.5.3] | -0.662 | 0.503 | 0.190 | 0.968 |
| coxS; aerobic carbon-monoxide dehydrogenase small subunit [EC:1.2.5.3] | -0.207 | 0.211 | 0.327 | 0.978 |
| cpaA, tadV; prepilin peptidase CpaA [EC:3.4.23.43] | -0.130 | 0.229 | 0.571 | 0.995 |
| cpaB, rcpC; pilus assembly protein CpaB | -0.329 | 0.228 | 0.151 | 0.954 |
| cpaC, rcpA; pilus assembly protein CpaC | -0.322 | 0.244 | 0.189 | 0.966 |
| cpaD; pilus assembly protein CpaD | -0.935 | 0.449 | 0.039 | 0.839 |
| cpaE, tadZ; pilus assembly protein CpaE | -0.357 | 0.240 | 0.140 | 0.949 |
| cpaF, tadA; pilus assembly protein CpaF | -0.023 | 0.154 | 0.882 | 0.999 |
| cpbD; chitin-binding protein | -0.116 | 0.400 | 0.772 | 0.999 |
| cpdA; 3’,5’-cyclic-AMP phosphodiesterase [EC:3.1.4.53] | 0.140 | 0.192 | 0.465 | 0.979 |
| cpdB; 2’,3’-cyclic-nucleotide 2’-phosphodiesterase / 3’-nucleotidase [EC:3.1.4.16 3.1.3.6] | -0.097 | 0.119 | 0.413 | 0.978 |
| cpdP; 3’,5’-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17] | -0.019 | 0.651 | 0.976 | 0.999 |
| cpdR; two-component system, cell cycle response regulator CpdR | -1.071 | 0.429 | 0.014 | 0.839 |
| cpe; enterotoxin Cpe | 6.196 | 11.877 | 0.603 | 0.999 |
| cpg; glutamate carboxypeptidase [EC:3.4.17.11] | -0.271 | 0.186 | 0.147 | 0.954 |
| CPGS; cyclic 2,3-diphosphoglycerate synthetase [EC:4.6.1.-] | -1.859 | 2.280 | 0.416 | 0.978 |
| cphA; cyanophycin synthetase [EC:6.3.2.29 6.3.2.30] | -0.243 | 0.500 | 0.628 | 0.999 |
| cphB; cyanophycinase [EC:3.4.15.6] | -0.378 | 0.593 | 0.525 | 0.989 |
| cpnA; cyclopentanol dehydrogenase [EC:1.1.1.163] | 1.027 | 0.862 | 0.236 | 0.978 |
| cpo; non-heme chloroperoxidase [EC:1.11.1.10] | -0.150 | 0.256 | 0.559 | 0.992 |
| cpoA; 1,2-diacylglycerol-3-alpha-glucose alpha-1,2-galactosyltransferase [EC:2.4.1.-] | 0.124 | 0.216 | 0.569 | 0.995 |
| CPOX, hemF; coproporphyrinogen III oxidase [EC:1.3.3.3] | -0.085 | 0.178 | 0.632 | 0.999 |
| cpt; carboxypeptidase T [EC:3.4.17.18] | 18.832 | 3989.590 | 0.996 | 0.999 |
| cpt; chloramphenicol 3-O phosphotransferase [EC:2.7.1.-] | -1.812 | 1.125 | 0.109 | 0.917 |
| cptA; toxin CptA | 0.218 | 0.258 | 0.400 | 0.978 |
| cptB; antitoxin CptB | -0.033 | 0.146 | 0.821 | 0.999 |
| cpxA; two-component system, OmpR family, sensor histidine kinase CpxA [EC:2.7.13.3] | 0.304 | 0.325 | 0.352 | 0.978 |
| cpxP; periplasmic protein CpxP | 0.210 | 0.578 | 0.716 | 0.999 |
| cpxR; two-component system, OmpR family, response regulator CpxR | 0.018 | 0.264 | 0.947 | 0.999 |
| cqsA; CAI-1 autoinducer synthase [EC:2.3.-.-] | 0.400 | 1.051 | 0.704 | 0.999 |
| cqsS; two-component system, CAI-1 autoinducer sensor kinase/phosphatase CqsS [EC:2.7.13.3 3.1.3.-] | -0.436 | 1.356 | 0.748 | 0.999 |
| crcB, FEX; fluoride exporter | -0.051 | 0.066 | 0.439 | 0.978 |
| creA; CreA protein | -0.031 | 0.209 | 0.882 | 0.999 |
| creB; two-component system, OmpR family, catabolic regulation response regulator CreB | -0.028 | 0.243 | 0.909 | 0.999 |
| creC; two-component system, OmpR family, sensor histidine kinase CreC [EC:2.7.13.3] | -0.043 | 0.240 | 0.859 | 0.999 |
| creD; inner membrane protein | 0.291 | 0.207 | 0.162 | 0.957 |
| creS; crescentin | -1.592 | 1.984 | 0.423 | 0.978 |
| crl; sigma factor-binding protein Crl | 0.352 | 0.531 | 0.508 | 0.981 |
| CRLS; cardiolipin synthase (CMP-forming) [EC:2.7.8.41] | -0.626 | 0.257 | 0.016 | 0.839 |
| croR; 3-hydroxybutyryl-CoA dehydratase [EC:4.2.1.55] | -0.451 | 0.330 | 0.174 | 0.961 |
| crp; CRP/FNR family transcriptional regulator, cyclic AMP receptor protein | 0.106 | 0.167 | 0.526 | 0.989 |
| crt; enoyl-CoA hydratase [EC:4.2.1.17] | 0.279 | 0.194 | 0.151 | 0.954 |
| crtA; spheroidene monooxygenase [EC:1.14.15.9] | -17.500 | 2562.559 | 0.995 | 0.999 |
| crtB; 15-cis-phytoene synthase [EC:2.5.1.32] | -0.465 | 0.226 | 0.041 | 0.839 |
| crtC; carotenoid 1,2-hydratase [EC:4.2.1.131] | -2.693 | 3.485 | 0.441 | 0.978 |
| crtD; 1-hydroxycarotenoid 3,4-desaturase [EC:1.3.99.27] | -18.056 | 3176.254 | 0.995 | 0.999 |
| crtF; demethylspheroidene O-methyltransferase [EC:2.1.1.210] | -3.309 | 3.390 | 0.331 | 0.978 |
| crtI; phytoene desaturase [EC:1.3.99.26 1.3.99.28 1.3.99.29 1.3.99.31] | -0.189 | 0.379 | 0.618 | 0.999 |
| crtISO, crtH; prolycopene isomerase [EC:5.2.1.13] | -0.456 | 1.281 | 0.723 | 0.999 |
| crtM; 4,4’-diapophytoene synthase [EC:2.5.1.96] | -0.811 | 1.001 | 0.419 | 0.978 |
| crtN; 4,4’-diapophytoene desaturase [EC:1.3.8.2] | -1.554 | 1.110 | 0.164 | 0.957 |
| crtP; diapolycopene oxygenase [EC:1.14.99.44] | -1.554 | 1.110 | 0.164 | 0.957 |
| crtQ; 4,4’-diaponeurosporenoate glycosyltransferase [EC:2.4.1.-] | -1.551 | 1.058 | 0.145 | 0.951 |
| crtR; beta-carotene hydroxylase [EC:1.14.13.-] | -17.770 | 4023.440 | 0.996 | 0.999 |
| crtU; isorenieratene synthase | -1.061 | 0.778 | 0.175 | 0.961 |
| crtW; beta-carotene ketolase (CrtW type) | 0.424 | 0.994 | 0.670 | 0.999 |
| crtX; zeaxanthin glucosyltransferase [EC:2.4.1.276] | -0.124 | 0.669 | 0.853 | 0.999 |
| crtZ; beta-carotene 3-hydroxylase [EC:1.14.15.24] | 0.048 | 0.333 | 0.886 | 0.999 |
| cruC; chlorobactene glucosyltransferase | -0.456 | 1.281 | 0.723 | 0.999 |
| cruF; bisanhydrobacterioruberin hydratase [EC:4.2.1.161] | -1.094 | 0.877 | 0.214 | 0.978 |
| CS, gltA; citrate synthase [EC:2.3.3.1] | -0.240 | 0.083 | 0.005 | 0.839 |
| csb1; CRISPR-associated protein Csb1 | 0.298 | 0.433 | 0.492 | 0.981 |
| csb2; CRISPR-associated protein Csb2 | 0.298 | 0.433 | 0.492 | 0.981 |
| csb3; CRISPR-associated protein Csb3 | 0.288 | 0.435 | 0.509 | 0.981 |
| csd1, cas8c; CRISPR-associated protein Csd1 | -0.213 | 0.319 | 0.505 | 0.981 |
| csd2, cas7; CRISPR-associated protein Csd2 | -0.222 | 0.319 | 0.488 | 0.979 |
| csdA; cysteine sulfinate desulfinase [EC:4.4.1.-] | 0.187 | 0.217 | 0.389 | 0.978 |
| csgA; major curlin subunit | -0.329 | 0.769 | 0.670 | 0.999 |
| csgB; minor curlin subunit | 0.452 | 0.365 | 0.218 | 0.978 |
| csgC; curli production protein | -0.329 | 0.769 | 0.670 | 0.999 |
| csgD; LuxR family transcriptional regulator, csgAB operon transcriptional regulatory protein | -0.261 | 0.701 | 0.710 | 0.999 |
| csgE; curli production assembly/transport component CsgE | 0.607 | 0.365 | 0.098 | 0.896 |
| csgF; curli production assembly/transport component CsgF | 0.545 | 0.347 | 0.118 | 0.917 |
| csgG; curli production assembly/transport component CsgG | 0.399 | 0.327 | 0.225 | 0.978 |
| csh1; CRISPR-associated protein Csh1 | 0.230 | 0.514 | 0.655 | 0.999 |
| csh2; CRISPR-associated protein Csh2 | 0.144 | 0.628 | 0.819 | 0.999 |
| cshB; ATP-dependent RNA helicase CshB [EC:3.6.4.13] | 0.000 | 0.133 | 0.999 | 1.000 |
| csiD; protein CsiD | 0.049 | 0.519 | 0.924 | 0.999 |
| csiR; GntR family transcriptional regulator, carbon starvation induced regulator | 0.277 | 0.557 | 0.620 | 0.999 |
| CSL4, EXOSC1; exosome complex component CSL4 | -17.234 | 3077.514 | 0.996 | 0.999 |
| cslA; chondroitin AC lyase [EC:4.2.2.5] | -0.274 | 0.652 | 0.675 | 0.999 |
| csm1, cas10; CRISPR-associated protein Csm1 | -0.262 | 0.678 | 0.699 | 0.999 |
| csm2; CRISPR-associated protein Csm2 | -0.735 | 0.874 | 0.402 | 0.978 |
| csm3; CRISPR-associated protein Csm3 | -0.213 | 0.577 | 0.712 | 0.999 |
| csm4; CRISPR-associated protein Csm4 | 0.611 | 0.488 | 0.213 | 0.978 |
| csm5; CRISPR-associated protein Csm5 | 0.133 | 0.706 | 0.851 | 0.999 |
| csn; chitosanase [EC:3.2.1.132] | -18.175 | 3481.021 | 0.996 | 0.999 |
| csn1, cas9; CRISPR-associated endonuclease Csn1 [EC:3.1.-.-] | 0.000 | 0.135 | 0.997 | 0.999 |
| csn2; CRISPR-associated protein Csn2 | 0.398 | 0.370 | 0.284 | 0.978 |
| cspA; cold shock protein (beta-ribbon, CspA family) | 0.010 | 0.083 | 0.902 | 0.999 |
| csrA; carbon storage regulator | -0.032 | 0.168 | 0.850 | 0.999 |
| csrD; RNase E specificity factor CsrD | 0.444 | 0.544 | 0.416 | 0.978 |
| cssR; two-component system, OmpR family, response regulator CssR | -0.304 | 0.509 | 0.552 | 0.992 |
| cssS; two-component system, OmpR family, sensor histidine kinase CssS [EC:2.7.13.3] | -0.664 | 0.549 | 0.228 | 0.978 |
| cst1, cas8a; CRISPR-associated protein Cst1 | 1.077 | 0.574 | 0.062 | 0.839 |
| cst2, cas7; CRISPR-associated protein Cst2 | 1.133 | 0.554 | 0.043 | 0.839 |
| cstA; carbon starvation protein | -0.074 | 0.115 | 0.523 | 0.989 |
| csx16; CRISPR-associated protein Csx16 | -0.086 | 0.592 | 0.884 | 0.999 |
| csxA; exo-1,4-beta-D-glucosaminidase [EC:3.2.1.165] | -2.352 | 1.287 | 0.069 | 0.841 |
| csy1; CRISPR-associated protein Csy1 | 0.301 | 0.531 | 0.572 | 0.995 |
| csy2; CRISPR-associated protein Csy2 | 0.431 | 0.513 | 0.401 | 0.978 |
| csy3; CRISPR-associated protein Csy3 | 0.249 | 0.472 | 0.598 | 0.999 |
| csy4, cas6f; CRISPR-associated endonuclease Csy4 [EC:3.1.-.-] | 0.431 | 0.513 | 0.401 | 0.978 |
| ctaG; putative membrane protein | -1.574 | 0.739 | 0.035 | 0.839 |
| CTBP; C-terminal binding protein | 0.179 | 1.431 | 0.901 | 0.999 |
| ctcP, cts4, prnC; tetracycline 7-halogenase / FADH2 O2-dependent halogenase [EC:1.14.19.49 1.14.19.-] | -4.099 | 5.721 | 0.475 | 0.979 |
| CTDSP; carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase [EC:3.1.3.16] | -0.367 | 0.918 | 0.690 | 0.999 |
| CTDSPL2; CTD small phosphatase-like protein 2 [EC:3.1.3.-] | -0.259 | 0.965 | 0.788 | 0.999 |
| CTH; cystathionine gamma-lyase [EC:4.4.1.1] | -0.102 | 0.260 | 0.694 | 0.999 |
| ctpA_B; cation-transporting P-type ATPase A/B [EC:3.6.3.-] | -18.303 | 2998.425 | 0.995 | 0.999 |
| ctpC; manganese/zinc-transporting P-type ATPase C [EC:3.6.3.-] | 0.827 | 0.360 | 0.023 | 0.839 |
| ctpD; cobalt/nickel-transporting P-type ATPase D [EC:3.6.3.-] | -17.661 | 2245.180 | 0.994 | 0.999 |
| ctpE; cation-transporting P-type ATPase E [EC:3.6.3.-] | 0.041 | 0.129 | 0.750 | 0.999 |
| ctpF; cation-transporting P-type ATPase F [EC:3.6.3.-] | -17.368 | 3290.703 | 0.996 | 0.999 |
| ctpG; cation-transporting P-type ATPase G [EC:3.6.3.-] | -0.594 | 1.011 | 0.558 | 0.992 |
| ctpI; cation-transporting P-type ATPase I [EC:3.6.3.-] | 2.619 | 1.993 | 0.191 | 0.970 |
| ctrA; two-component system, cell cycle response regulator CtrA | -1.219 | 0.446 | 0.007 | 0.839 |
| ctsR; transcriptional regulator of stress and heat shock response | 0.050 | 0.112 | 0.655 | 0.999 |
| cueO; blue copper oxidase | -0.500 | 0.681 | 0.464 | 0.979 |
| cueR; MerR family transcriptional regulator, copper efflux regulator | 0.092 | 0.271 | 0.735 | 0.999 |
| cusA, silA; Cu(I)/Ag(I) efflux system membrane protein CusA/SilA | -0.075 | 0.218 | 0.730 | 0.999 |
| cusB, silB; membrane fusion protein, Cu(I)/Ag(I) efflux system | -0.030 | 0.223 | 0.892 | 0.999 |
| cusC, silC; outer membrane protein, Cu(I)/Ag(I) efflux system | -0.207 | 0.708 | 0.770 | 0.999 |
| cusF; Cu(I)/Ag(I) efflux system periplasmic protein CusF | 0.447 | 0.457 | 0.329 | 0.978 |
| cusR, copR, silR; two-component system, OmpR family, copper resistance phosphate regulon response regulator CusR | 0.013 | 0.256 | 0.961 | 0.999 |
| cusS, copS, silS; two-component system, OmpR family, heavy metal sensor histidine kinase CusS [EC:2.7.13.3] | 0.000 | 0.256 | 1.000 | 1.000 |
| cutA; periplasmic divalent cation tolerance protein | -0.063 | 0.259 | 0.808 | 0.999 |
| cutC; copper homeostasis protein | 0.049 | 0.086 | 0.569 | 0.995 |
| cutF, nlpE; copper homeostasis protein (lipoprotein) | -0.181 | 0.426 | 0.671 | 0.999 |
| cuyA; L-cysteate sulfo-lyase [EC:4.4.1.25] | -0.469 | 0.804 | 0.560 | 0.992 |
| cvpA; membrane protein required for colicin V production | 0.090 | 0.118 | 0.445 | 0.978 |
| cvrA, nhaP2; cell volume regulation protein A | 0.326 | 0.174 | 0.064 | 0.839 |
| cwlJ, sleB; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | -0.771 | 0.433 | 0.077 | 0.856 |
| cwlK; peptidoglycan LD-endopeptidase CwlK [EC:3.4.-.-] | -0.279 | 0.340 | 0.412 | 0.978 |
| cwlS; peptidoglycan DL-endopeptidase CwlS [EC:3.4.-.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| cyaA; adenylate cyclase, class 1 [EC:4.6.1.1] | 0.180 | 0.231 | 0.435 | 0.978 |
| cyaC, hlyC, rtxC; cytolysin-activating lysine-acyltransferase [EC:2.3.1.-] | -0.199 | 0.687 | 0.772 | 0.999 |
| cyaY; CyaY protein | 0.137 | 0.181 | 0.451 | 0.978 |
| cybB; cytochrome b561 | -0.059 | 0.208 | 0.777 | 0.999 |
| cybC; soluble cytochrome b562 | -0.310 | 0.368 | 0.402 | 0.978 |
| CYC; cytochrome c | -0.445 | 0.233 | 0.058 | 0.839 |
| CYC1, CYT1, petC; ubiquinol-cytochrome c reductase cytochrome c1 subunit | -0.089 | 0.184 | 0.629 | 0.999 |
| cyc2; germacradienol/geosmin synthase [EC:4.2.3.22 4.2.3.75 4.1.99.16] | -17.794 | 4072.025 | 0.997 | 0.999 |
| cycA; D-serine/D-alanine/glycine transporter | -0.086 | 0.145 | 0.555 | 0.992 |
| cycB, ganO; arabinogalactan oligomer / maltooligosaccharide transport system substrate-binding protein | 0.033 | 0.117 | 0.781 | 0.999 |
| cydA; cytochrome bd ubiquinol oxidase subunit I [EC:1.10.3.14] | -0.006 | 0.067 | 0.927 | 0.999 |
| cydB; cytochrome bd ubiquinol oxidase subunit II [EC:1.10.3.14] | -0.034 | 0.066 | 0.605 | 0.999 |
| cydC; ATP-binding cassette, subfamily C, bacterial CydC | 0.080 | 0.137 | 0.557 | 0.992 |
| cydCD; ATP-binding cassette, subfamily C, bacterial CydCD | 0.213 | 0.204 | 0.300 | 0.978 |
| cydD; ATP-binding cassette, subfamily C, bacterial CydD | 0.083 | 0.137 | 0.546 | 0.990 |
| cymC; p-cumic aldehyde dehydrogenase | -1.780 | 1.801 | 0.324 | 0.978 |
| cymR; Rrf2 family transcriptional regulator, cysteine metabolism repressor | -0.821 | 0.533 | 0.126 | 0.927 |
| cynD; cyanide dihydratase [EC:3.5.5.-] | -17.750 | 3249.062 | 0.996 | 0.999 |
| cynR; LysR family transcriptional regulator, cyn operon transcriptional activator | -0.525 | 0.309 | 0.092 | 0.891 |
| cynS; cyanate lyase [EC:4.2.1.104] | -0.258 | 0.366 | 0.483 | 0.979 |
| cynT, can; carbonic anhydrase [EC:4.2.1.1] | -0.094 | 0.059 | 0.115 | 0.917 |
| cyoA; cytochrome o ubiquinol oxidase subunit II [EC:1.10.3.10] | -0.105 | 0.222 | 0.636 | 0.999 |
| cyoB; cytochrome o ubiquinol oxidase subunit I [EC:1.10.3.10] | -0.134 | 0.224 | 0.552 | 0.992 |
| cyoC; cytochrome o ubiquinol oxidase subunit III | -0.139 | 0.212 | 0.513 | 0.982 |
| cyoD; cytochrome o ubiquinol oxidase subunit IV | -0.135 | 0.223 | 0.545 | 0.990 |
| CYP105D; pentalenic acid synthase [EC:1.14.15.11] | -17.794 | 4072.025 | 0.997 | 0.999 |
| CYP125A; cholest-4-en-3-one 26-monooxygenase [EC:1.14.13.141] | -2.070 | 1.521 | 0.175 | 0.961 |
| CYP134A, cypX; pulcherriminic acid synthase [EC:1.14.15.13] | -0.554 | 1.623 | 0.733 | 0.999 |
| CYP142; cholest-4-en-3-one 26-monooxygenase [EC:1.14.13.221] | -2.024 | 1.667 | 0.227 | 0.978 |
| CYP152A; fatty-acid peroxygenase [EC:1.11.2.4] | 0.570 | 0.620 | 0.359 | 0.978 |
| CYP51; sterol 14-demethylase [EC:1.14.13.70] | -2.129 | 1.604 | 0.186 | 0.966 |
| CYP81F; indol-3-yl-methylglucosinolate hydroxylase [EC:1.14.-.-] | -0.512 | 0.338 | 0.132 | 0.941 |
| cypD_E, CYP102A2_3; cytochrome P450 / NADPH-cytochrome P450 reductase [EC:1.14.14.1 1.6.2.4] | -0.554 | 0.668 | 0.408 | 0.978 |
| cysA; sulfate transport system ATP-binding protein [EC:3.6.3.25] | -0.141 | 0.169 | 0.407 | 0.978 |
| cysB; LysR family transcriptional regulator, cys regulon transcriptional activator | 0.162 | 0.182 | 0.376 | 0.978 |
| cysC; adenylylsulfate kinase [EC:2.7.1.25] | -0.618 | 0.282 | 0.030 | 0.839 |
| cysD; sulfate adenylyltransferase subunit 2 [EC:2.7.7.4] | -0.227 | 0.168 | 0.178 | 0.961 |
| cysE; serine O-acetyltransferase [EC:2.3.1.30] | -0.024 | 0.047 | 0.602 | 0.999 |
| cysG; uroporphyrin-III C-methyltransferase / precorrin-2 dehydrogenase / sirohydrochlorin ferrochelatase [EC:2.1.1.107 1.3.1.76 4.99.1.4] | 0.063 | 0.168 | 0.709 | 0.999 |
| cysH; phosphoadenosine phosphosulfate reductase [EC:1.8.4.8 1.8.4.10] | -0.277 | 0.152 | 0.070 | 0.841 |
| cysI; sulfite reductase (NADPH) hemoprotein beta-component [EC:1.8.1.2] | -0.138 | 0.162 | 0.395 | 0.978 |
| cysJ; sulfite reductase (NADPH) flavoprotein alpha-component [EC:1.8.1.2] | -0.046 | 0.166 | 0.781 | 0.999 |
| cysK; cysteine synthase A [EC:2.5.1.47] | 0.023 | 0.032 | 0.472 | 0.979 |
| cysM; cysteine synthase B [EC:2.5.1.47] | -0.030 | 0.222 | 0.891 | 0.999 |
| cysN; sulfate adenylyltransferase subunit 1 [EC:2.7.7.4] | -0.167 | 0.176 | 0.344 | 0.978 |
| cysNC; bifunctional enzyme CysN/CysC [EC:2.7.7.4 2.7.1.25] | -0.266 | 0.279 | 0.343 | 0.978 |
| cysP, sbp; sulfate transport system substrate-binding protein | -0.112 | 0.206 | 0.586 | 0.999 |
| cysQ, MET22, BPNT1; 3’(2’), 5’-bisphosphate nucleotidase [EC:3.1.3.7] | -0.105 | 0.139 | 0.451 | 0.978 |
| cysS1; cysteinyl-tRNA synthetase, unknown class [EC:6.1.1.16] | 1.189 | 0.538 | 0.029 | 0.839 |
| cysU; sulfate transport system permease protein | -0.176 | 0.174 | 0.314 | 0.978 |
| cysW; sulfate transport system permease protein | -0.176 | 0.174 | 0.314 | 0.978 |
| cysZ; CysZ protein | 0.125 | 0.194 | 0.518 | 0.987 |
| CYTB, petB; ubiquinol-cytochrome c reductase cytochrome b subunit | -0.112 | 0.180 | 0.535 | 0.990 |
| cytR; LacI family transcriptional regulator, repressor for deo operon, udp, cdd, tsx, nupC, and nupG | -0.324 | 0.299 | 0.281 | 0.978 |
| czcA; cobalt-zinc-cadmium resistance protein CzcA | -0.100 | 0.262 | 0.702 | 0.999 |
| czcB; membrane fusion protein, cobalt-zinc-cadmium efflux system | -0.086 | 0.289 | 0.766 | 0.999 |
| czcC; outer membrane protein, cobalt-zinc-cadmium efflux system | -0.077 | 0.313 | 0.806 | 0.999 |
| czcD, zitB; cobalt-zinc-cadmium efflux system protein | -0.086 | 0.077 | 0.268 | 0.978 |
| dacA; diadenylate cyclase [EC:2.7.7.85] | 0.049 | 0.102 | 0.631 | 0.999 |
| dacB; serine-type D-Ala-D-Ala carboxypeptidase/endopeptidase (penicillin-binding protein 4) [EC:3.4.16.4 3.4.21.-] | -0.099 | 0.103 | 0.338 | 0.978 |
| dacC, dacA, dacD; serine-type D-Ala-D-Ala carboxypeptidase (penicillin-binding protein 5/6) [EC:3.4.16.4] | 0.021 | 0.067 | 0.756 | 0.999 |
| dadA; D-amino-acid dehydrogenase [EC:1.4.5.1] | -0.245 | 0.190 | 0.200 | 0.971 |
| dagK; diacylglycerol kinase (ATP) [EC:2.7.1.107] | 0.091 | 0.139 | 0.513 | 0.982 |
| DAK, TKFC; triose/dihydroxyacetone kinase / FAD-AMP lyase (cyclizing) [EC:2.7.1.28 2.7.1.29 4.6.1.15] | -0.251 | 0.198 | 0.209 | 0.977 |
| dalD; D-arabinitol 4-dehydrogenase [EC:1.1.1.11] | -1.947 | 1.738 | 0.264 | 0.978 |
| dam; DNA adenine methylase [EC:2.1.1.72] | 0.064 | 0.156 | 0.684 | 0.999 |
| damX; DamX protein | 0.221 | 0.302 | 0.466 | 0.979 |
| DAO, aao; D-amino-acid oxidase [EC:1.4.3.3] | -0.119 | 0.802 | 0.882 | 0.999 |
| dapA; 4-hydroxy-tetrahydrodipicolinate synthase [EC:4.3.3.7] | -0.046 | 0.029 | 0.119 | 0.917 |
| dapB; 4-hydroxy-tetrahydrodipicolinate reductase [EC:1.17.1.8] | -0.033 | 0.048 | 0.485 | 0.979 |
| dapC; N-succinyldiaminopimelate aminotransferase [EC:2.6.1.17] | -0.043 | 0.216 | 0.841 | 0.999 |
| dapD; 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase [EC:2.3.1.117] | -0.027 | 0.073 | 0.711 | 0.999 |
| dapdh; diaminopimelate dehydrogenase [EC:1.4.1.16] | 0.009 | 0.181 | 0.959 | 0.999 |
| dapE; succinyl-diaminopimelate desuccinylase [EC:3.5.1.18] | 0.114 | 0.081 | 0.163 | 0.957 |
| dapF; diaminopimelate epimerase [EC:5.1.1.7] | 0.016 | 0.058 | 0.784 | 0.999 |
| dapH, dapD; tetrahydrodipicolinate N-acetyltransferase [EC:2.3.1.89] | 0.011 | 0.201 | 0.957 | 0.999 |
| dapL; N-acetyldiaminopimelate deacetylase [EC:3.5.1.47] | 0.055 | 0.145 | 0.707 | 0.999 |
| dasA; N,N’-diacetylchitobiose transport system substrate-binding protein | 1.075 | 0.640 | 0.095 | 0.896 |
| dasB; N,N’-diacetylchitobiose transport system permease protein | 0.788 | 0.636 | 0.217 | 0.978 |
| dasC; N,N’-diacetylchitobiose transport system permease protein | 0.936 | 0.688 | 0.175 | 0.961 |
| dat; D-alanine transaminase [EC:2.6.1.21] | -0.176 | 0.177 | 0.321 | 0.978 |
| dauA; D-arginine dehydrogenase [EC:1.4.99.6] | 0.119 | 0.351 | 0.736 | 0.999 |
| dauB; L-arginine dehydrogenase [EC:1.4.1.25] | 0.113 | 0.859 | 0.896 | 0.999 |
| davT, gabT; 5-aminovalerate/4-aminobutyrate aminotransferase [EC:2.6.1.48 2.6.1.19] | 0.221 | 0.340 | 0.517 | 0.986 |
| dbh; DNA polymerase IV (archaeal DinB-like DNA polymerase) [EC:2.7.7.7] | 0.933 | 0.703 | 0.186 | 0.966 |
| dbpA; ATP-independent RNA helicase DbpA [EC:3.6.4.13] | 0.062 | 0.220 | 0.779 | 0.999 |
| DBT, bkdB; 2-oxoisovalerate dehydrogenase E2 component (dihydrolipoyl transacylase) [EC:2.3.1.168] | -0.464 | 0.264 | 0.081 | 0.864 |
| DCAA; acyl-CoA dehydrogenase [EC:1.3.99.-] | -1.296 | 0.601 | 0.033 | 0.839 |
| dcd; dCTP deaminase [EC:3.5.4.13] | -0.060 | 0.109 | 0.581 | 0.999 |
| dck; deoxyadenosine/deoxycytidine kinase [EC:2.7.1.76 2.7.1.74] | -0.746 | 0.499 | 0.137 | 0.949 |
| dcp; peptidyl-dipeptidase Dcp [EC:3.4.15.5] | -0.239 | 0.158 | 0.131 | 0.941 |
| dcsG; O-ureido-D-serine cyclo-ligase [EC:6.3.3.5] | 17.922 | 2530.829 | 0.994 | 0.999 |
| dctA; aerobic C4-dicarboxylate transport protein | -0.263 | 0.204 | 0.199 | 0.970 |
| dctB; two-component system, NtrC family, C4-dicarboxylate transport sensor histidine kinase DctB [EC:2.7.13.3] | 0.043 | 0.309 | 0.890 | 0.999 |
| dctD; two-component system, NtrC family, C4-dicarboxylate transport response regulator DctD | 0.030 | 0.329 | 0.927 | 0.999 |
| dctM; C4-dicarboxylate transporter, DctM subunit | 0.363 | 0.211 | 0.088 | 0.880 |
| dctP; C4-dicarboxylate-binding protein DctP | 0.035 | 0.303 | 0.908 | 0.999 |
| dctQ; C4-dicarboxylate transporter, DctQ subunit | -0.063 | 0.313 | 0.841 | 0.999 |
| dctR; two-component system, LuxR family, response regulator DctR | -1.105 | 0.662 | 0.097 | 0.896 |
| dctS; two-component system, LuxR family, sensor histidine kinase DctS [EC:2.7.13.3] | -0.586 | 0.578 | 0.313 | 0.978 |
| dcuA; anaerobic C4-dicarboxylate transporter DcuA | -0.172 | 0.201 | 0.393 | 0.978 |
| dcuB; anaerobic C4-dicarboxylate transporter DcuB | 0.049 | 0.232 | 0.831 | 0.999 |
| dcuR; two-component system, CitB family, response regulator DcuR | 0.064 | 0.603 | 0.915 | 0.999 |
| dcuS; two-component system, CitB family, sensor histidine kinase DcuS [EC:2.7.13.3] | 0.134 | 0.621 | 0.829 | 0.999 |
| dcyD; D-cysteine desulfhydrase [EC:4.4.1.15] | -0.067 | 0.311 | 0.829 | 0.999 |
| DDAH, ddaH; dimethylargininase [EC:3.5.3.18] | 0.026 | 0.569 | 0.964 | 0.999 |
| DDC, TDC; aromatic-L-amino-acid/L-tryptophan decarboxylase [EC:4.1.1.28 4.1.1.105] | 1.008 | 0.512 | 0.051 | 0.839 |
| ddc; L-2,4-diaminobutyrate decarboxylase [EC:4.1.1.86] | -0.260 | 0.321 | 0.419 | 0.978 |
| dddL; dimethylpropiothetin dethiomethylase [EC:4.4.1.3] | -17.500 | 2562.559 | 0.995 | 0.999 |
| ddhA; dimethylsulfide dehydrogenase subunit alpha [EC:1.8.2.4] | -17.931 | 4360.659 | 0.997 | 0.999 |
| ddhB; dimethylsulfide dehydrogenase subunit beta | -17.931 | 4360.659 | 0.997 | 0.999 |
| ddhC; dimethylsulfide dehydrogenase subunit gamma | -17.931 | 4360.659 | 0.997 | 0.999 |
| ddl; D-alanine-D-alanine ligase [EC:6.3.2.4] | 0.007 | 0.034 | 0.832 | 0.999 |
| deaD, cshA; ATP-dependent RNA helicase DeaD [EC:3.6.4.13] | 0.021 | 0.050 | 0.682 | 0.999 |
| DECR2; peroxisomal 2,4-dienoyl-CoA reductase [EC:1.3.1.34] | -17.068 | 2832.569 | 0.995 | 0.999 |
| dedA; membrane-associated protein | 0.093 | 0.152 | 0.542 | 0.990 |
| dedD; DedD protein | 0.132 | 0.237 | 0.577 | 0.996 |
| degP, htrA; serine protease Do [EC:3.4.21.107] | -0.045 | 0.054 | 0.401 | 0.978 |
| degQ, hhoA; serine protease DegQ [EC:3.4.21.-] | 0.166 | 0.212 | 0.436 | 0.978 |
| DEGS; sphingolipid 4-desaturase/C4-monooxygenase [EC:1.14.19.17 1.14.18.5] | -17.761 | 4005.858 | 0.996 | 0.999 |
| degS; two-component system, NarL family, sensor histidine kinase DegS [EC:2.7.13.3] | -0.290 | 0.427 | 0.498 | 0.981 |
| degU; two-component system, NarL family, response regulator DegU | -1.241 | 0.742 | 0.096 | 0.896 |
| dehH; haloacetate dehalogenase [EC:3.8.1.3] | -0.101 | 0.225 | 0.653 | 0.999 |
| deoA, TYMP; thymidine phosphorylase [EC:2.4.2.4] | -0.427 | 0.258 | 0.100 | 0.896 |
| deoB; phosphopentomutase [EC:5.4.2.7] | 0.010 | 0.105 | 0.924 | 0.999 |
| deoC, DERA; deoxyribose-phosphate aldolase [EC:4.1.2.4] | 0.016 | 0.076 | 0.835 | 0.999 |
| deoD; purine-nucleoside phosphorylase [EC:2.4.2.1] | 0.084 | 0.096 | 0.380 | 0.978 |
| deoR; DeoR family transcriptional regulator, deoxyribose operon repressor | 0.148 | 0.599 | 0.805 | 0.999 |
| deoR; deoxyribonucleoside regulator | 0.355 | 0.437 | 0.417 | 0.978 |
| desA; syringate O-demethylase [EC:2.1.1.-] | -0.229 | 1.513 | 0.880 | 0.999 |
| desA2; acyl-[acyl-carrier-protein] desaturase [EC:1.14.19.2] | -2.624 | 1.417 | 0.066 | 0.839 |
| desB, galA; gallate dioxygenase [EC:1.13.11.57] | -0.283 | 0.997 | 0.777 | 0.999 |
| desK; two-component system, NarL family, sensor histidine kinase DesK [EC:2.7.13.3] | -0.127 | 0.127 | 0.318 | 0.978 |
| desR; two-component system, NarL family, response regulator DesR | -0.214 | 0.237 | 0.368 | 0.978 |
| desZ; 3-O-methylgallate 3,4-dioxygenase [EC:1.13.11.-] | -17.155 | 2959.362 | 0.995 | 0.999 |
| devR; two-component system, NarL family, response regulator DevR | -2.428 | 1.174 | 0.040 | 0.839 |
| devS; two-component system, NarL family, sensor histidine kinase DevS [EC:2.7.13.3] | -17.416 | 3370.607 | 0.996 | 0.999 |
| dexA; dextranase [EC:3.2.1.11] | 0.033 | 0.473 | 0.944 | 0.999 |
| dexB; glucan 1,6-alpha-glucosidase [EC:3.2.1.70] | 0.044 | 0.170 | 0.797 | 0.999 |
| dfrA1, dhfr; dihydrofolate reductase (trimethoprim resistance protein) [EC:1.5.1.3] | 18.165 | 2857.430 | 0.995 | 0.999 |
| dfrA12, dhfr; dihydrofolate reductase (trimethoprim resistance protein) [EC:1.5.1.3] | -0.295 | 1.323 | 0.824 | 0.999 |
| dfrD, dhfr; dihydrofolate reductase (trimethoprim resistance protein) [EC:1.5.1.3] | -3.465 | 4.101 | 0.400 | 0.978 |
| dfx; superoxide reductase [EC:1.15.1.2] | 0.115 | 0.196 | 0.559 | 0.992 |
| dgaE; D-glucosaminate-6-phosphate ammonia-lyase [EC:4.3.1.29] | -2.728 | 2.632 | 0.302 | 0.978 |
| dgaF; 2-dehydro-3-deoxy-phosphogluconate aldolase [EC:4.1.2.14] | -2.728 | 2.632 | 0.302 | 0.978 |
| dgaR; sigma-54 dependent transcriptional regulator, dga operon transcriptional activator | -1.630 | 2.316 | 0.483 | 0.979 |
| dgcB; diguanylate cyclase [EC:2.7.7.65] | -0.085 | 0.292 | 0.772 | 0.999 |
| dge1; diguanylate cyclase [EC:2.7.7.65] | -1.341 | 1.026 | 0.193 | 0.970 |
| dgk; deoxyguanosine kinase [EC:2.7.1.113] | -0.718 | 0.512 | 0.163 | 0.957 |
| dgkA, DGK; diacylglycerol kinase (ATP) [EC:2.7.1.107] | -0.001 | 0.122 | 0.996 | 0.999 |
| dgkA; undecaprenol kinase [EC:2.7.1.66] | 0.049 | 0.137 | 0.723 | 0.999 |
| dgoA; 2-dehydro-3-deoxyphosphogalactonate aldolase [EC:4.1.2.21] | -0.902 | 0.356 | 0.012 | 0.839 |
| dgoD; galactonate dehydratase [EC:4.2.1.6] | -0.714 | 0.380 | 0.062 | 0.839 |
| dgoK; 2-dehydro-3-deoxygalactonokinase [EC:2.7.1.58] | -0.892 | 0.350 | 0.012 | 0.839 |
| dgoR; GntR family transcriptional regulator, galactonate operon transcriptional repressor | -0.196 | 0.598 | 0.744 | 0.999 |
| dgoT; MFS transporter, ACS family, D-galactonate transporter | 0.020 | 0.395 | 0.961 | 0.999 |
| dgs, bgsA; 1,2-diacylglycerol-3-alpha-glucose alpha-1,2-glucosyltransferase [EC:2.4.1.208] | 0.175 | 0.170 | 0.304 | 0.978 |
| dgt; dGTPase [EC:3.1.5.1] | -0.022 | 0.087 | 0.803 | 0.999 |
| dhaa; 3-hydroxy-D-aspartate aldolase [EC:4.1.3.41] | -0.946 | 1.312 | 0.472 | 0.979 |
| dhaA; haloalkane dehalogenase [EC:3.8.1.5] | -0.132 | 0.381 | 0.731 | 0.999 |
| dhaB; glycerol dehydratase large subunit [EC:4.2.1.30] | 2.329 | 1.863 | 0.213 | 0.978 |
| dhaK; phosphoenolpyruvate—glycerone phosphotransferase subunit DhaK [EC:2.7.1.121] | 0.235 | 0.124 | 0.059 | 0.839 |
| dhaL; phosphoenolpyruvate—glycerone phosphotransferase subunit DhaL [EC:2.7.1.121] | 0.140 | 0.115 | 0.228 | 0.978 |
| dhaM; phosphoenolpyruvate—glycerone phosphotransferase subunit DhaM [EC:2.7.1.121] | -0.088 | 0.299 | 0.768 | 0.999 |
| dhaR; transcriptional activator for dhaKLM operon | -0.105 | 0.784 | 0.893 | 0.999 |
| dhaT; 1,3-propanediol dehydrogenase [EC:1.1.1.202] | 0.338 | 0.536 | 0.529 | 0.989 |
| dhbC; glycerol dehydratase medium subunit [EC:4.2.1.30] | 2.329 | 1.863 | 0.213 | 0.978 |
| DHBD; 2,3-dihydroxybenzoate decarboxylase [EC:4.1.1.46] | -1.005 | 0.947 | 0.290 | 0.978 |
| dhbE; glycerol dehydratase small subunit [EC:4.2.1.30] | 2.414 | 1.814 | 0.185 | 0.966 |
| dhbF; nonribosomal peptide synthetase DhbF | -0.321 | 0.659 | 0.627 | 0.999 |
| dhcR; LysR family transcriptional regulator, carnitine catabolism transcriptional activator | 0.082 | 0.289 | 0.777 | 0.999 |
| DHFR, folA; dihydrofolate reductase [EC:1.5.1.3] | 0.039 | 0.039 | 0.319 | 0.978 |
| DHODH, pyrD; dihydroorotate dehydrogenase [EC:1.3.5.2] | -0.065 | 0.117 | 0.579 | 0.997 |
| dhpH; 2,6-dihydroxypyridine 3-monooxygenase [EC:1.14.13.10] | -0.286 | 0.987 | 0.772 | 0.999 |
| DHPS, dys; deoxyhypusine synthase [EC:2.5.1.46] | -0.243 | 0.326 | 0.457 | 0.979 |
| DHRS12; dehydrogenase/reductase SDR family member 12 [EC:1.1.-.-] | -17.770 | 4023.440 | 0.996 | 0.999 |
| DHRS13; dehydrogenase/reductase SDR family member 13 [EC:1.1.-.-] | 18.563 | 3487.927 | 0.996 | 0.999 |
| diaA; DnaA initiator-associating protein | 0.352 | 0.531 | 0.508 | 0.981 |
| dinB; DNA polymerase IV [EC:2.7.7.7] | -0.013 | 0.040 | 0.750 | 0.999 |
| dinD; DNA-damage-inducible protein D | -0.509 | 0.396 | 0.201 | 0.972 |
| dinG; ATP-dependent DNA helicase DinG [EC:3.6.4.12] | 0.069 | 0.056 | 0.220 | 0.978 |
| dinI; DNA-damage-inducible protein I | 0.015 | 0.615 | 0.981 | 0.999 |
| dinJ; DNA-damage-inducible protein J | 0.311 | 0.176 | 0.079 | 0.859 |
| diox1; all-trans-8’-apo-beta-carotenal 15,15’-oxygenase [EC:1.13.11.75] | -17.416 | 3370.607 | 0.996 | 0.999 |
| dipps; CDP-L-myo-inositol myo-inositolphosphotransferase [EC:2.7.8.34] | -17.779 | 4041.596 | 0.996 | 0.999 |
| DIS3, RRP44; exosome complex exonuclease DIS3/RRP44 [EC:3.1.13.-] | 0.295 | 0.433 | 0.497 | 0.981 |
| disA; diadenylate cyclase [EC:2.7.7.85] | 0.349 | 0.229 | 0.130 | 0.934 |
| divIC, divA; cell division protein DivIC | 0.057 | 0.119 | 0.631 | 0.999 |
| divIVA; cell division initiation protein | 0.058 | 0.111 | 0.605 | 0.999 |
| divJ; two-component system, cell cycle sensor histidine kinase DivJ [EC:2.7.13.3] | -1.479 | 0.650 | 0.024 | 0.839 |
| divK; two-component system, cell cycle response regulator DivK | -1.062 | 0.396 | 0.008 | 0.839 |
| djlA; DnaJ like chaperone protein | -0.004 | 0.149 | 0.980 | 0.999 |
| DKC1, NOLA4, CBF5; H/ACA ribonucleoprotein complex subunit 4 [EC:5.4.99.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| dkgA; 2,5-diketo-D-gluconate reductase A [EC:1.1.1.346] | -0.038 | 0.220 | 0.862 | 0.999 |
| dkgB; 2,5-diketo-D-gluconate reductase B [EC:1.1.1.346] | -0.008 | 0.229 | 0.972 | 0.999 |
| dksA; DnaK suppressor protein | -0.040 | 0.164 | 0.806 | 0.999 |
| DLAT, aceF, pdhC; pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12] | -0.052 | 0.070 | 0.457 | 0.979 |
| DLD, lpd, pdhD; dihydrolipoamide dehydrogenase [EC:1.8.1.4] | -0.093 | 0.056 | 0.096 | 0.896 |
| dld; D-lactate dehydrogenase | 0.112 | 0.168 | 0.508 | 0.981 |
| dld; D-lactate dehydrogenase (quinone) [EC:1.1.5.12] | -0.002 | 0.229 | 0.993 | 0.999 |
| dlgD; 3-dehydro-L-gulonate 2-dehydrogenase [EC:1.1.1.130] | 0.506 | 0.545 | 0.355 | 0.978 |
| DLST, sucB; 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide succinyltransferase) [EC:2.3.1.61] | -0.130 | 0.109 | 0.236 | 0.978 |
| dltA; D-alanine–poly(phosphoribitol) ligase subunit 1 [EC:6.1.1.13] | 0.024 | 0.136 | 0.863 | 0.999 |
| dltB; membrane protein involved in D-alanine export | 0.023 | 0.139 | 0.871 | 0.999 |
| dltC; D-alanine–poly(phosphoribitol) ligase subunit 2 [EC:6.1.1.13] | 0.049 | 0.142 | 0.731 | 0.999 |
| dltD; D-alanine transfer protein | 0.055 | 0.144 | 0.701 | 0.999 |
| dltE; uncharacterized oxidoreductase [EC:1.-.-.-] | -0.665 | 0.441 | 0.134 | 0.942 |
| dmd-tmd; dimethylamine/trimethylamine dehydrogenase [EC:1.5.8.1 1.5.8.2] | 0.663 | 0.915 | 0.470 | 0.979 |
| DMGDH; dimethylglycine dehydrogenase [EC:1.5.8.4] | -1.655 | 0.931 | 0.077 | 0.856 |
| dmlR; LysR family transcriptional regulator, transcriptional activator for dmlA | -0.691 | 0.459 | 0.134 | 0.942 |
| dmpA, dap; D-aminopeptidase [EC:3.4.11.19] | -0.989 | 0.437 | 0.025 | 0.839 |
| dmpB, xylE; catechol 2,3-dioxygenase [EC:1.13.11.2] | -1.134 | 0.536 | 0.036 | 0.839 |
| dmpC, xylG, praB; aminomuconate-semialdehyde/2-hydroxymuconate-6-semialdehyde dehydrogenase [EC:1.2.1.32 1.2.1.85] | -0.501 | 0.699 | 0.474 | 0.979 |
| dmpD, xylF; 2-hydroxymuconate-semialdehyde hydrolase [EC:3.7.1.9] | -0.673 | 0.937 | 0.474 | 0.979 |
| dmpH, xylI, nahK; 2-oxo-3-hexenedioate decarboxylase [EC:4.1.1.77] | -1.003 | 0.705 | 0.157 | 0.954 |
| dmpK, poxA; phenol hydroxylase P0 protein | 0.272 | 0.524 | 0.605 | 0.999 |
| dmpL, poxC; Phenol hydroxylase P1 protein | 0.140 | 0.492 | 0.776 | 0.999 |
| dmpM, poxB; phenol hydroxylase P2 protein | 0.140 | 0.492 | 0.776 | 0.999 |
| dmpN, poxD; phenol hydroxylase P3 protein [EC:1.14.13.-] | 0.140 | 0.492 | 0.776 | 0.999 |
| dmpO, poxE; phenol hydroxylase P4 protein | 0.140 | 0.492 | 0.776 | 0.999 |
| dmpP, poxF; phenol hydroxylase P5 protein | 0.140 | 0.492 | 0.776 | 0.999 |
| dmrA; dihydromethanopterin reductase [EC:1.5.1.47] | 18.066 | 2719.856 | 0.995 | 0.999 |
| dmsA; anaerobic dimethyl sulfoxide reductase subunit A [EC:1.8.5.3] | -0.930 | 0.750 | 0.217 | 0.978 |
| dmsB; anaerobic dimethyl sulfoxide reductase subunit B (DMSO reductase iron- sulfur subunit) | -0.911 | 0.639 | 0.156 | 0.954 |
| dmsC; anaerobic dimethyl sulfoxide reductase subunit C (DMSO reductase anchor subunit) | -0.163 | 0.426 | 0.703 | 0.999 |
| DNA2; DNA replication ATP-dependent helicase Dna2 [EC:3.6.4.12] | 1.107 | 1.279 | 0.388 | 0.978 |
| dnaA; chromosomal replication initiator protein | -0.007 | 0.037 | 0.850 | 0.999 |
| dnaB; replication initiation and membrane attachment protein | 0.002 | 0.134 | 0.985 | 0.999 |
| dnaB; replicative DNA helicase [EC:3.6.4.12] | 0.021 | 0.029 | 0.472 | 0.979 |
| dnaC; DNA replication protein DnaC | 0.121 | 0.214 | 0.572 | 0.995 |
| dnaD; DNA replication protein | 0.001 | 0.133 | 0.992 | 0.999 |
| dnaE; DNA polymerase III subunit alpha [EC:2.7.7.7] | 0.034 | 0.032 | 0.292 | 0.978 |
| dnaE2; error-prone DNA polymerase [EC:2.7.7.7] | -0.173 | 0.192 | 0.368 | 0.978 |
| dnaG; DNA primase [EC:2.7.7.-] | 0.035 | 0.035 | 0.328 | 0.978 |
| dnaI; primosomal protein DnaI | 0.001 | 0.132 | 0.996 | 0.999 |
| dnaJ; molecular chaperone DnaJ | 0.024 | 0.037 | 0.507 | 0.981 |
| dnaK, HSPA9; molecular chaperone DnaK | 0.041 | 0.035 | 0.245 | 0.978 |
| dnaN; DNA polymerase III subunit beta [EC:2.7.7.7] | 0.075 | 0.042 | 0.077 | 0.856 |
| dnaQ; DNA polymerase III subunit epsilon [EC:2.7.7.7] | -0.026 | 0.045 | 0.570 | 0.995 |
| dnaT; DNA replication protein DnaT | -0.243 | 0.717 | 0.735 | 0.999 |
| dnaX; DNA polymerase III subunit gamma/tau [EC:2.7.7.7] | 0.034 | 0.032 | 0.295 | 0.978 |
| dndB; DNA sulfur modification protein DndB | 2.301 | 1.860 | 0.218 | 0.978 |
| dndC; DNA sulfur modification protein DndC | 0.215 | 0.502 | 0.669 | 0.999 |
| dndD; DNA sulfur modification protein DndD | 0.154 | 0.512 | 0.764 | 0.999 |
| dndE; DNA sulfur modification protein DndE | 0.219 | 1.032 | 0.832 | 0.999 |
| dnk; deoxynucleoside kinase [EC:2.7.1.145] | 18.902 | 3402.727 | 0.996 | 0.999 |
| DNMT1, dcm; DNA (cytosine-5)-methyltransferase 1 [EC:2.1.1.37] | -0.065 | 0.096 | 0.500 | 0.981 |
| DNPEP; aspartyl aminopeptidase [EC:3.4.11.21] | 0.287 | 0.121 | 0.019 | 0.839 |
| doc; death on curing protein | -0.042 | 0.147 | 0.776 | 0.999 |
| doeA; ectoine hydrolase [EC:3.5.4.44] | -1.042 | 0.807 | 0.198 | 0.970 |
| doeB; N2-acetyl-L-2,4-diaminobutanoate deacetylase [EC:3.5.1.125] | -0.605 | 0.735 | 0.411 | 0.978 |
| doeC; aspartate-semialdehyde dehydrogenase [EC:1.2.1.-] | -0.878 | 0.747 | 0.242 | 0.978 |
| doeD; L-2,4-diaminobutyrate transaminase [EC:2.6.1.76] | -1.030 | 0.802 | 0.201 | 0.972 |
| doeX; Lrp/AsnC family transcriptional regulator, regulator of ectoine-degradation genes | -1.135 | 0.827 | 0.172 | 0.961 |
| DOHH; deoxyhypusine monooxygenase [EC:1.14.99.29] | -17.234 | 3077.514 | 0.996 | 0.999 |
| DOPA; 4,5-DOPA dioxygenase extradiol [EC:1.13.11.-] | 0.002 | 0.132 | 0.990 | 0.999 |
| dos; c-di-GMP-specific phosphodiesterase [EC:3.1.4.52] | -1.153 | 1.472 | 0.435 | 0.978 |
| dotB, traJ; defect in organelle trafficking protein DotB | -0.259 | 0.965 | 0.788 | 0.999 |
| dotC, traI; defect in organelle trafficking protein DotC | -0.259 | 0.965 | 0.788 | 0.999 |
| doxD; thiosulfate dehydrogenase [quinone] large subunit [EC:1.8.5.2] | -0.916 | 0.428 | 0.034 | 0.839 |
| dpe, lre; D-psicose/D-tagatose/L-ribulose 3-epimerase [EC:5.1.3.30 5.1.3.31] | -1.629 | 0.699 | 0.021 | 0.839 |
| DPEP; membrane dipeptidase [EC:3.4.13.19] | -0.071 | 0.120 | 0.553 | 0.992 |
| DPH1, dph2; 2-(3-amino-3-carboxypropyl)histidine synthase [EC:2.5.1.108] | -17.234 | 3077.514 | 0.996 | 0.999 |
| DPH4, DNAJC24; diphthamide biosynthesis protein 4 | -17.756 | 3869.767 | 0.996 | 0.999 |
| DPH5; diphthine methyl ester synthase [EC:2.1.1.314] | -17.234 | 3077.514 | 0.996 | 0.999 |
| dpiA, citB; two-component system, CitB family, response regulator CitB | 0.410 | 0.621 | 0.510 | 0.981 |
| dpiB, citA; two-component system, CitB family, cit operon sensor histidine kinase CitA [EC:2.7.13.3] | 0.280 | 0.570 | 0.624 | 0.999 |
| dpkA, lhpD; delta1-piperideine-2-carboxylate reductase [EC:1.5.1.21] | -0.055 | 0.408 | 0.892 | 0.999 |
| DPM1; dolichol-phosphate mannosyltransferase [EC:2.4.1.83] | 0.004 | 0.087 | 0.965 | 0.999 |
| dpo; DNA polymerase bacteriophage-type [EC:2.7.7.7] | 0.009 | 0.130 | 0.946 | 0.999 |
| DPP3; dipeptidyl-peptidase III [EC:3.4.14.4] | -0.252 | 0.245 | 0.305 | 0.978 |
| DPP4, CD26; dipeptidyl-peptidase 4 [EC:3.4.14.5] | -0.210 | 0.173 | 0.226 | 0.978 |
| dppA; dipeptide transport system substrate-binding protein | -0.035 | 0.244 | 0.887 | 0.999 |
| dppA1; D-amino peptidase [EC:3.4.11.-] | -0.948 | 0.658 | 0.152 | 0.954 |
| dppB; dipeptide transport system permease protein | 0.003 | 0.211 | 0.989 | 0.999 |
| dppB1; dipeptide transport system permease protein | -1.804 | 1.242 | 0.149 | 0.954 |
| dppC; dipeptide transport system permease protein | -0.037 | 0.204 | 0.857 | 0.999 |
| dppD; dipeptide transport system ATP-binding protein | 0.028 | 0.154 | 0.856 | 0.999 |
| dppE; dipeptide transport system substrate-binding protein | 18.239 | 2966.075 | 0.995 | 0.999 |
| dppF; dipeptide transport system ATP-binding protein | 0.006 | 0.211 | 0.976 | 0.999 |
| dprE1; decaprenylphospho-beta-D-ribofuranose 2-oxidase [EC:1.1.98.3] | -0.412 | 0.588 | 0.484 | 0.979 |
| dprE2; decaprenylphospho-beta-D-erythro-pentofuranosid-2-ulose 2-reductase [EC:1.1.1.333] | -0.356 | 0.574 | 0.535 | 0.990 |
| dps; starvation-inducible DNA-binding protein | 0.020 | 0.044 | 0.657 | 0.999 |
| dptF; DNA phosphorothioation-dependent restriction protein DptF | 0.842 | 1.349 | 0.533 | 0.990 |
| dptG; DNA phosphorothioation-dependent restriction protein DptG | 3.041 | 2.596 | 0.243 | 0.978 |
| dptH; DNA phosphorothioation-dependent restriction protein DptH | 3.041 | 2.596 | 0.243 | 0.978 |
| DPYS, dht, hydA; dihydropyrimidinase [EC:3.5.2.2] | -0.326 | 0.230 | 0.159 | 0.954 |
| draG; ADP-ribosyl-[dinitrogen reductase] hydrolase [EC:3.2.2.24] | 0.285 | 0.485 | 0.557 | 0.992 |
| draT; NAD+—dinitrogen-reductase ADP-D-ribosyltransferase [EC:2.4.2.37] | -17.665 | 3819.214 | 0.996 | 0.999 |
| drp35; lactonase [EC:3.1.1.-] | -1.395 | 0.969 | 0.152 | 0.954 |
| dsbA; thiol:disulfide interchange protein DsbA | 0.048 | 0.164 | 0.768 | 0.999 |
| dsbB; disulfide bond formation protein DsbB | -0.075 | 0.145 | 0.604 | 0.999 |
| dsbC; thiol:disulfide interchange protein DsbC [EC:5.3.4.1] | 0.041 | 0.161 | 0.800 | 0.999 |
| dsbD; thiol:disulfide interchange protein DsbD [EC:1.8.1.8] | -0.044 | 0.109 | 0.684 | 0.999 |
| dsbG; thiol:disulfide interchange protein DsbG | 0.070 | 0.265 | 0.792 | 0.999 |
| dsdA; D-serine dehydratase [EC:4.3.1.18] | 0.344 | 0.257 | 0.182 | 0.961 |
| dsdC; LysR family transcriptional regulator, D-serine deaminase activator | 0.653 | 0.517 | 0.208 | 0.977 |
| dsdX; D-serine transporter | -0.371 | 0.685 | 0.589 | 0.999 |
| dsrA; dissimilatory sulfite reductase alpha subunit [EC:1.8.99.5] | -2.579 | 2.960 | 0.385 | 0.978 |
| dsrB; dissimilatory sulfite reductase beta subunit [EC:1.8.99.5] | -2.579 | 2.960 | 0.385 | 0.978 |
| dszB; 2’-hydroxybiphenyl-2-sulfinate desulfinase [EC:3.13.1.3] | -1.076 | 1.026 | 0.296 | 0.978 |
| dtd, DTD1; D-tyrosyl-tRNA(Tyr) deacylase [EC:3.1.-.-] | 0.062 | 0.041 | 0.138 | 0.949 |
| dtdA, GEK1; D-aminoacyl-tRNA deacylase [EC:3.1.1.96] | -17.234 | 3077.514 | 0.996 | 0.999 |
| dusA; tRNA-dihydrouridine synthase A [EC:1.-.-.-] | -0.003 | 0.136 | 0.981 | 0.999 |
| dusB; tRNA-dihydrouridine synthase B [EC:1.-.-.-] | -0.010 | 0.138 | 0.944 | 0.999 |
| dusC; tRNA-dihydrouridine synthase C [EC:1.-.-.-] | 0.095 | 0.177 | 0.590 | 0.999 |
| dut, DUT; dUTP pyrophosphatase [EC:3.6.1.23] | 0.058 | 0.037 | 0.122 | 0.919 |
| DVR; divinyl chlorophyllide a 8-vinyl-reductase [EC:1.3.1.75] | -17.500 | 2562.559 | 0.995 | 0.999 |
| dxr; 1-deoxy-D-xylulose-5-phosphate reductoisomerase [EC:1.1.1.267] | -0.006 | 0.062 | 0.921 | 0.999 |
| dxs; 1-deoxy-D-xylulose-5-phosphate synthase [EC:2.2.1.7] | 0.073 | 0.070 | 0.300 | 0.978 |
| E1.1.1.1, adh; alcohol dehydrogenase [EC:1.1.1.1] | -0.222 | 0.138 | 0.110 | 0.917 |
| E1.1.1.102; 3-dehydrosphinganine reductase [EC:1.1.1.102] | -0.456 | 1.281 | 0.723 | 0.999 |
| E1.1.1.122; D-threo-aldose 1-dehydrogenase [EC:1.1.1.122] | -0.693 | 0.383 | 0.072 | 0.845 |
| E1.1.1.219; dihydroflavonol-4-reductase [EC:1.1.1.219] | -0.434 | 0.337 | 0.199 | 0.970 |
| E1.1.1.251, gatD; galactitol-1-phosphate 5-dehydrogenase [EC:1.1.1.251] | -0.620 | 1.017 | 0.543 | 0.990 |
| E1.1.1.275; (+)-trans-carveol dehydrogenase [EC:1.1.1.275] | 0.042 | 0.899 | 0.963 | 0.999 |
| E1.1.1.3; homoserine dehydrogenase [EC:1.1.1.3] | -0.026 | 0.050 | 0.600 | 0.999 |
| E1.1.1.30, bdh; 3-hydroxybutyrate dehydrogenase [EC:1.1.1.30] | -0.322 | 0.207 | 0.122 | 0.919 |
| E1.1.1.306; S-(hydroxymethyl)mycothiol dehydrogenase [EC:1.1.1.306] | -0.065 | 0.338 | 0.848 | 0.999 |
| E1.1.1.374; UDP-N-acetylglucosamine 3-dehydrogenase [EC:1.1.1.374] | -17.716 | 2631.659 | 0.995 | 0.999 |
| E1.1.1.387; L-serine 3-dehydrogenase (NAD+) [EC:1.1.1.387] | 1.513 | 1.385 | 0.276 | 0.978 |
| E1.1.1.40, maeB; malate dehydrogenase (oxaloacetate-decarboxylating)(NADP+) [EC:1.1.1.40] | -0.176 | 0.104 | 0.093 | 0.893 |
| E1.1.1.43; phosphogluconate 2-dehydrogenase [EC:1.1.1.43] | -0.010 | 0.435 | 0.981 | 0.999 |
| E1.1.1.51; 3(or 17)beta-hydroxysteroid dehydrogenase [EC:1.1.1.51] | 0.029 | 0.779 | 0.970 | 0.999 |
| E1.1.1.53; 3alpha(or 20beta)-hydroxysteroid dehydrogenase [EC:1.1.1.53] | -2.054 | 0.957 | 0.033 | 0.839 |
| E1.1.1.65; pyridoxine 4-dehydrogenase [EC:1.1.1.65] | -1.421 | 1.156 | 0.221 | 0.978 |
| E1.1.1.67, mtlK; mannitol 2-dehydrogenase [EC:1.1.1.67] | -0.217 | 0.342 | 0.526 | 0.989 |
| E1.1.1.9; D-xylulose reductase [EC:1.1.1.9] | -2.185 | 1.070 | 0.043 | 0.839 |
| E1.1.1.90; aryl-alcohol dehydrogenase [EC:1.1.1.90] | 0.063 | 0.328 | 0.849 | 0.999 |
| E1.1.1.91; aryl-alcohol dehydrogenase (NADP+) [EC:1.1.1.91] | -1.220 | 0.679 | 0.074 | 0.847 |
| E1.1.3.21; alpha-glycerophosphate oxidase [EC:1.1.3.21] | -0.153 | 0.197 | 0.438 | 0.978 |
| E1.1.99.21; D-sorbitol dehydrogenase (acceptor) [EC:1.1.99.21] | -0.349 | 0.648 | 0.591 | 0.999 |
| E1.1.99.3A; gluconate 2-dehydrogenase alpha chain [EC:1.1.99.3] | -0.155 | 0.443 | 0.726 | 0.999 |
| E1.1.99.3G; gluconate 2-dehydrogenase gamma chain [EC:1.1.99.3] | -0.124 | 0.404 | 0.759 | 0.999 |
| E1.10.3.3; L-ascorbate oxidase [EC:1.10.3.3] | -17.298 | 3177.464 | 0.996 | 0.999 |
| E1.11.1.19; dye decolorizing peroxidase [EC:1.11.1.19] | -0.764 | 0.546 | 0.164 | 0.958 |
| E1.11.1.5; cytochrome c peroxidase [EC:1.11.1.5] | 0.029 | 0.196 | 0.884 | 0.999 |
| E1.12.7.2; ferredoxin hydrogenase [EC:1.12.7.2] | 1.855 | 1.562 | 0.237 | 0.978 |
| E1.12.7.2G; ferredoxin hydrogenase gamma subunit [EC:1.12.7.2] | -17.478 | 2545.196 | 0.995 | 0.999 |
| E1.12.7.2L; ferredoxin hydrogenase large subunit [EC:1.12.7.2] | -2.495 | 1.789 | 0.165 | 0.960 |
| E1.12.7.2S; ferredoxin hydrogenase small subunit [EC:1.12.7.2] | -17.650 | 2743.732 | 0.995 | 0.999 |
| E1.13.11.4; gentisate 1,2-dioxygenase [EC:1.13.11.4] | -1.268 | 0.661 | 0.057 | 0.839 |
| E1.13.12.4; lactate 2-monooxygenase [EC:1.13.12.4] | -2.096 | 1.174 | 0.076 | 0.855 |
| E1.14.11.1; gamma-butyrobetaine dioxygenase [EC:1.14.11.1] | -0.016 | 0.627 | 0.980 | 0.999 |
| E1.14.13.1; salicylate hydroxylase [EC:1.14.13.1] | -1.133 | 0.541 | 0.038 | 0.839 |
| E1.14.13.40; anthraniloyl-CoA monooxygenase [EC:1.14.13.40] | -0.707 | 0.492 | 0.153 | 0.954 |
| E1.14.13.7; phenol 2-monooxygenase [EC:1.14.13.7] | -0.003 | 0.739 | 0.996 | 0.999 |
| E1.14.13.81, acsF, chlE; magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase [EC:1.14.13.81] | -3.402 | 3.248 | 0.296 | 0.978 |
| E1.14.14.1; unspecific monooxygenase [EC:1.14.14.1] | -2.036 | 1.058 | 0.056 | 0.839 |
| E1.14.19.3; linoleoyl-CoA desaturase [EC:1.14.19.3] | -0.340 | 0.319 | 0.289 | 0.978 |
| E1.17.4.1A, nrdA, nrdE; ribonucleoside-diphosphate reductase alpha chain [EC:1.17.4.1] | -0.014 | 0.041 | 0.726 | 0.999 |
| E1.17.4.1B, nrdB, nrdF; ribonucleoside-diphosphate reductase beta chain [EC:1.17.4.1] | 0.013 | 0.046 | 0.777 | 0.999 |
| E1.2.1.10; acetaldehyde dehydrogenase (acetylating) [EC:1.2.1.10] | 0.321 | 0.893 | 0.719 | 0.999 |
| E1.2.1.5; aldehyde dehydrogenase (NAD(P)+) [EC:1.2.1.5] | -0.936 | 0.959 | 0.331 | 0.978 |
| E1.2.1.68; coniferyl-aldehyde dehydrogenase [EC:1.2.1.68] | -0.145 | 0.220 | 0.512 | 0.982 |
| E1.2.1.88; 1-pyrroline-5-carboxylate dehydrogenase [EC:1.2.1.88] | -0.551 | 0.376 | 0.145 | 0.951 |
| E1.2.3.3, poxL; pyruvate oxidase [EC:1.2.3.3] | -0.044 | 0.160 | 0.783 | 0.999 |
| E1.2.7.8; indolepyruvate ferredoxin oxidoreductase [EC:1.2.7.8] | -0.180 | 0.259 | 0.488 | 0.979 |
| E1.3.1.12; prephenate dehydrogenase [EC:1.3.1.12] | -0.967 | 0.603 | 0.111 | 0.917 |
| E1.3.1.32; maleylacetate reductase [EC:1.3.1.32] | -1.446 | 1.084 | 0.184 | 0.965 |
| E1.3.1.45; 2’-hydroxyisoflavone reductase [EC:1.3.1.45] | -2.603 | 1.798 | 0.150 | 0.954 |
| E1.3.1.74; 2-alkenal reductase [EC:1.3.1.74] | -1.643 | 1.075 | 0.129 | 0.931 |
| E1.3.3.5; bilirubin oxidase [EC:1.3.3.5] | -3.152 | 3.336 | 0.346 | 0.978 |
| E1.3.3.6, ACOX1, ACOX3; acyl-CoA oxidase [EC:1.3.3.6] | -0.755 | 0.479 | 0.117 | 0.917 |
| E1.4.1.4, gdhA; glutamate dehydrogenase (NADP+) [EC:1.4.1.4] | -0.065 | 0.082 | 0.429 | 0.978 |
| E1.4.1.9; leucine dehydrogenase [EC:1.4.1.9] | -0.215 | 0.208 | 0.302 | 0.978 |
| E1.4.7.1; glutamate synthase (ferredoxin) [EC:1.4.7.1] | -0.228 | 0.219 | 0.301 | 0.978 |
| E1.5.3.1; sarcosine oxidase [EC:1.5.3.1] | -0.719 | 0.518 | 0.167 | 0.961 |
| E1.6.99.1; NADPH2 dehydrogenase [EC:1.6.99.1] | -0.981 | 0.448 | 0.030 | 0.839 |
| E1.6.99.3; NADH dehydrogenase [EC:1.6.99.3] | -0.854 | 0.331 | 0.011 | 0.839 |
| E1.7.1.7, guaC; GMP reductase [EC:1.7.1.7] | 0.020 | 0.120 | 0.870 | 0.999 |
| E1.8.2.1; sulfite dehydrogenase [EC:1.8.2.1] | -2.235 | 1.776 | 0.210 | 0.978 |
| E2.1.1.103, NMT; phosphoethanolamine N-methyltransferase [EC:2.1.1.103] | -17.477 | 3476.545 | 0.996 | 0.999 |
| E2.1.1.104; caffeoyl-CoA O-methyltransferase [EC:2.1.1.104] | 0.939 | 0.441 | 0.035 | 0.839 |
| E2.1.1.113; site-specific DNA-methyltransferase (cytosine-N4-specific) [EC:2.1.1.113] | 2.392 | 1.808 | 0.188 | 0.966 |
| E2.1.1.131, cobJ, cbiH; precorrin-3B C17-methyltransferase [EC:2.1.1.131] | 0.300 | 0.182 | 0.102 | 0.896 |
| E2.1.1.72; site-specific DNA-methyltransferase (adenine-specific) [EC:2.1.1.72] | -0.117 | 0.127 | 0.358 | 0.978 |
| E2.1.1.77, pcm; protein-L-isoaspartate(D-aspartate) O-methyltransferase [EC:2.1.1.77] | -0.160 | 0.169 | 0.346 | 0.978 |
| E2.1.3.-; carbamoyltransferase [EC:2.1.3.-] | 0.032 | 0.359 | 0.929 | 0.999 |
| E2.1.3.1-1.3S; methylmalonyl-CoA carboxyltransferase 1.3S subunit [EC:2.1.3.1] | -0.655 | 0.951 | 0.492 | 0.981 |
| E2.1.3.1-12S; methylmalonyl-CoA carboxyltransferase 12S subunit [EC:2.1.3.1] | 0.429 | 0.582 | 0.462 | 0.979 |
| E2.1.3.1-5S; methylmalonyl-CoA carboxyltransferase 5S subunit [EC:2.1.3.1] | 0.269 | 0.633 | 0.671 | 0.999 |
| E2.2.1.1, tktA, tktB; transketolase [EC:2.2.1.1] | 0.006 | 0.052 | 0.912 | 0.999 |
| E2.2.1.2, talA, talB; transaldolase [EC:2.2.1.2] | -0.079 | 0.063 | 0.213 | 0.978 |
| E2.2.1.6L, ilvB, ilvG, ilvI; acetolactate synthase I/II/III large subunit [EC:2.2.1.6] | 0.000 | 0.040 | 0.992 | 0.999 |
| E2.2.1.6S, ilvH, ilvN; acetolactate synthase I/III small subunit [EC:2.2.1.6] | -0.026 | 0.067 | 0.704 | 0.999 |
| E2.3.1.20; diacylglycerol O-acyltransferase [EC:2.3.1.20] | -0.432 | 0.394 | 0.275 | 0.978 |
| E2.3.1.207; beta-ketodecanoyl-[acyl-carrier-protein] synthase [EC:2.3.1.207] | 0.038 | 0.258 | 0.884 | 0.999 |
| E2.3.1.37, ALAS; 5-aminolevulinate synthase [EC:2.3.1.37] | -1.243 | 0.452 | 0.007 | 0.839 |
| E2.3.1.54, pflD; formate C-acetyltransferase [EC:2.3.1.54] | 0.082 | 0.097 | 0.397 | 0.978 |
| E2.3.1.7; carnitine O-acetyltransferase [EC:2.3.1.7] | -0.297 | 0.946 | 0.754 | 0.999 |
| E2.3.1.8, pta; phosphate acetyltransferase [EC:2.3.1.8] | 0.048 | 0.088 | 0.590 | 0.999 |
| E2.3.1.9, atoB; acetyl-CoA C-acetyltransferase [EC:2.3.1.9] | -0.169 | 0.112 | 0.133 | 0.942 |
| E2.3.2.21; cyclo(L-tyrosyl-L-tyrosyl) synthase [EC:2.3.2.21] | -17.794 | 4072.025 | 0.997 | 0.999 |
| E2.3.3.10; hydroxymethylglutaryl-CoA synthase [EC:2.3.3.10] | 0.113 | 0.135 | 0.406 | 0.978 |
| E2.4.-.-; glycosyltransferase [EC:2.4.-.-] | -0.036 | 0.185 | 0.847 | 0.999 |
| E2.4.1.14; sucrose-phosphate synthase [EC:2.4.1.14] | 0.948 | 1.004 | 0.346 | 0.978 |
| E2.4.1.20; cellobiose phosphorylase [EC:2.4.1.20] | -0.415 | 0.682 | 0.543 | 0.990 |
| E2.4.1.213; glucosylglycerol-phosphate synthase [EC:2.4.1.213] | 3.210 | 2.428 | 0.188 | 0.966 |
| E2.4.1.217; mannosyl-3-phosphoglycerate synthase [EC:2.4.1.217] | 2.011 | 1.804 | 0.267 | 0.978 |
| E2.4.1.321; cellobionic acid phosphorylase [EC:2.4.1.321] | -0.745 | 1.362 | 0.585 | 0.999 |
| E2.4.1.4; amylosucrase [EC:2.4.1.4] | -0.287 | 0.334 | 0.391 | 0.978 |
| E2.4.1.5; dextransucrase [EC:2.4.1.5] | 0.017 | 0.344 | 0.961 | 0.999 |
| E2.4.1.64; alpha,alpha-trehalose phosphorylase [EC:2.4.1.64] | 0.445 | 0.504 | 0.379 | 0.978 |
| E2.4.1.7; sucrose phosphorylase [EC:2.4.1.7] | 0.082 | 0.181 | 0.653 | 0.999 |
| E2.4.2.21, cobU, cobT; nicotinate-nucleotide–dimethylbenzimidazole phosphoribosyltransferase [EC:2.4.2.21] | 0.076 | 0.112 | 0.495 | 0.981 |
| E2.4.2.6; nucleoside deoxyribosyltransferase [EC:2.4.2.6] | 0.399 | 0.539 | 0.461 | 0.979 |
| E2.5.1.41; phosphoglycerol geranylgeranyltransferase [EC:2.5.1.41] | -17.234 | 3077.514 | 0.996 | 0.999 |
| E2.5.1.42; geranylgeranylglycerol-phosphate geranylgeranyltransferase [EC:2.5.1.42] | -17.234 | 3077.514 | 0.996 | 0.999 |
| E2.5.1.54, aroF, aroG, aroH; 3-deoxy-7-phosphoheptulonate synthase [EC:2.5.1.54] | -0.021 | 0.076 | 0.786 | 0.999 |
| E2.5.1.56, neuB; N-acetylneuraminate synthase [EC:2.5.1.56] | 0.029 | 0.237 | 0.903 | 0.999 |
| E2.5.1.68; short-chain Z-isoprenyl diphosphate synthase [EC:2.5.1.68] | -0.008 | 0.292 | 0.980 | 0.999 |
| E2.5.1.86; trans,polycis-decaprenyl diphosphate synthase [EC:2.5.1.86] | -2.624 | 1.417 | 0.066 | 0.839 |
| E2.6.1.-E, patB; aminotransferase [EC:2.6.1.-] | 0.226 | 0.513 | 0.660 | 0.999 |
| E2.6.1.11, argD; acetylornithine aminotransferase [EC:2.6.1.11] | -0.277 | 0.123 | 0.026 | 0.839 |
| E2.6.1.18; beta-alanine–pyruvate transaminase [EC:2.6.1.18] | -0.129 | 0.226 | 0.569 | 0.995 |
| E2.6.1.42, ilvE; branched-chain amino acid aminotransferase [EC:2.6.1.42] | -0.044 | 0.047 | 0.347 | 0.978 |
| E2.6.1.83; LL-diaminopimelate aminotransferase [EC:2.6.1.83] | -0.003 | 0.208 | 0.989 | 0.999 |
| E2.7.1.-; kinase [EC:2.7.1.-] | -0.363 | 0.324 | 0.265 | 0.978 |
| E2.7.1.12, gntK, idnK; gluconokinase [EC:2.7.1.12] | 0.069 | 0.126 | 0.586 | 0.999 |
| E2.7.1.20, ADK; adenosine kinase [EC:2.7.1.20] | -0.305 | 0.300 | 0.312 | 0.978 |
| E2.7.1.36, MVK, mvaK1; mevalonate kinase [EC:2.7.1.36] | 0.121 | 0.135 | 0.371 | 0.978 |
| E2.7.1.37; protein kinase [EC:2.7.1.37] | 1.231 | 0.567 | 0.032 | 0.839 |
| E2.7.1.4, scrK; fructokinase [EC:2.7.1.4] | -0.006 | 0.058 | 0.913 | 0.999 |
| E2.7.1.47; D-ribulokinase [EC:2.7.1.47] | -1.522 | 1.449 | 0.295 | 0.978 |
| E2.7.1.71, aroK, aroL; shikimate kinase [EC:2.7.1.71] | 0.039 | 0.049 | 0.436 | 0.978 |
| E2.7.1.76, dak; deoxyadenosine kinase [EC:2.7.1.76] | 0.027 | 0.524 | 0.958 | 0.999 |
| E2.7.10.1; receptor protein-tyrosine kinase [EC:2.7.10.1] | -1.209 | 0.853 | 0.159 | 0.954 |
| E2.7.10.2; non-specific protein-tyrosine kinase [EC:2.7.10.2] | -2.283 | 1.626 | 0.162 | 0.957 |
| E2.7.11.1; non-specific serine/threonine protein kinase [EC:2.7.11.1] | -0.397 | 0.524 | 0.449 | 0.978 |
| E2.7.13.1; protein-histidine pros-kinase [EC:2.7.13.1] | -1.102 | 0.824 | 0.183 | 0.964 |
| E2.7.13.3; histidine kinase [EC:2.7.13.3] | -0.198 | 0.172 | 0.252 | 0.978 |
| E2.7.4.2, mvaK2; phosphomevalonate kinase [EC:2.7.4.2] | 0.114 | 0.139 | 0.412 | 0.978 |
| E2.7.4.8, gmk; guanylate kinase [EC:2.7.4.8] | 0.074 | 0.049 | 0.137 | 0.949 |
| E2.7.6.5X; putative GTP pyrophosphokinase [EC:2.7.6.5] | -0.011 | 0.117 | 0.922 | 0.999 |
| E2.7.7.24, rfbA, rffH; glucose-1-phosphate thymidylyltransferase [EC:2.7.7.24] | 0.029 | 0.062 | 0.637 | 0.999 |
| E2.7.7.3A, coaD, kdtB; pantetheine-phosphate adenylyltransferase [EC:2.7.7.3] | 0.033 | 0.036 | 0.357 | 0.978 |
| E2.7.7.3B; pantetheine-phosphate adenylyltransferase [EC:2.7.7.3] | -17.234 | 3077.514 | 0.996 | 0.999 |
| E2.7.7.41, CDS1, CDS2, cdsA; phosphatidate cytidylyltransferase [EC:2.7.7.41] | 0.062 | 0.029 | 0.038 | 0.839 |
| E2.7.7.53; ATP adenylyltransferase [EC:2.7.7.53] | 0.042 | 0.241 | 0.863 | 0.999 |
| E2.7.7.6; DNA-directed RNA polymerase [EC:2.7.7.6] | -2.149 | 1.058 | 0.044 | 0.839 |
| E2.7.7.65; diguanylate cyclase [EC:2.7.7.65] | -0.079 | 0.380 | 0.836 | 0.999 |
| E2.7.8.26, cobS, cobV; adenosylcobinamide-GDP ribazoletransferase [EC:2.7.8.26] | 0.053 | 0.106 | 0.617 | 0.999 |
| E2.7.8.39; archaetidylinositol phosphate synthase [EC:2.7.8.39] | -17.234 | 3077.514 | 0.996 | 0.999 |
| E2.8.3.5A, scoA; 3-oxoacid CoA-transferase subunit A [EC:2.8.3.5] | -0.258 | 0.189 | 0.174 | 0.961 |
| E2.8.3.5B, scoB; 3-oxoacid CoA-transferase subunit B [EC:2.8.3.5] | -0.257 | 0.189 | 0.175 | 0.961 |
| E3.1.1.11; pectinesterase [EC:3.1.1.11] | -0.863 | 0.398 | 0.032 | 0.839 |
| E3.1.1.17, gnl, RGN; gluconolactonase [EC:3.1.1.17] | -0.720 | 0.311 | 0.022 | 0.839 |
| E3.1.1.22; hydroxybutyrate-dimer hydrolase [EC:3.1.1.22] | -1.795 | 1.203 | 0.138 | 0.949 |
| E3.1.1.45; carboxymethylenebutenolidase [EC:3.1.1.45] | -0.390 | 0.224 | 0.084 | 0.871 |
| E3.1.1.74; cutinase [EC:3.1.1.74] | -2.782 | 1.601 | 0.084 | 0.873 |
| E3.1.11.2, xthA; exodeoxyribonuclease III [EC:3.1.11.2] | 0.035 | 0.033 | 0.290 | 0.978 |
| E3.1.11.5; exodeoxyribonuclease V [EC:3.1.11.5] | -0.615 | 0.294 | 0.038 | 0.839 |
| E3.1.2.1, ACH1; acetyl-CoA hydrolase [EC:3.1.2.1] | -1.626 | 2.053 | 0.429 | 0.978 |
| E3.1.2.20; acyl-CoA hydrolase [EC:3.1.2.20] | -0.158 | 0.571 | 0.782 | 0.999 |
| E3.1.2.23; 4-hydroxybenzoyl-CoA thioesterase [EC:3.1.2.23] | -0.029 | 0.284 | 0.919 | 0.999 |
| E3.1.21.4; type II restriction enzyme [EC:3.1.21.4] | -0.443 | 0.377 | 0.243 | 0.978 |
| E3.1.21.7, nfi; deoxyribonuclease V [EC:3.1.21.7] | -0.082 | 0.316 | 0.796 | 0.999 |
| E3.1.27.1; ribonuclease T2 [EC:3.1.27.1] | -0.282 | 0.209 | 0.178 | 0.961 |
| E3.1.27.3; ribonuclease T1 [EC:3.1.27.3] | -0.829 | 0.686 | 0.229 | 0.978 |
| E3.1.3.1, phoA, phoB; alkaline phosphatase [EC:3.1.3.1] | -0.412 | 0.196 | 0.038 | 0.839 |
| E3.1.3.15B; histidinol-phosphatase (PHP family) [EC:3.1.3.15] | 0.197 | 0.154 | 0.204 | 0.972 |
| E3.1.3.16; protein phosphatase [EC:3.1.3.16] | -0.042 | 0.084 | 0.620 | 0.999 |
| E3.1.3.25, IMPA, suhB; myo-inositol-1(or 4)-monophosphatase [EC:3.1.3.25] | -0.102 | 0.071 | 0.152 | 0.954 |
| E3.1.3.41; 4-nitrophenyl phosphatase [EC:3.1.3.41] | 0.148 | 0.130 | 0.257 | 0.978 |
| E3.1.3.48; protein-tyrosine phosphatase [EC:3.1.3.48] | -0.012 | 0.080 | 0.877 | 0.999 |
| E3.1.3.5; 5’-nucleotidase [EC:3.1.3.5] | -0.166 | 0.089 | 0.063 | 0.839 |
| E3.1.3.70; mannosyl-3-phosphoglycerate phosphatase [EC:3.1.3.70] | -0.456 | 0.714 | 0.524 | 0.989 |
| E3.1.3.8; 3-phytase [EC:3.1.3.8] | -0.362 | 0.330 | 0.274 | 0.978 |
| E3.1.3.97; 3’,5’-nucleoside bisphosphate phosphatase [EC:3.1.3.97] | 0.079 | 0.098 | 0.420 | 0.978 |
| E3.1.30.1; nuclease S1 [EC:3.1.30.1] | -2.279 | 2.181 | 0.298 | 0.978 |
| E3.1.4.12; sphingomyelin phosphodiesterase [EC:3.1.4.12] | -2.282 | 1.706 | 0.183 | 0.964 |
| E3.1.4.46, glpQ, ugpQ; glycerophosphoryl diester phosphodiesterase [EC:3.1.4.46] | 0.023 | 0.063 | 0.716 | 0.999 |
| E3.1.6.1, aslA; arylsulfatase [EC:3.1.6.1] | -0.486 | 0.241 | 0.045 | 0.839 |
| E3.2.1.123; endoglycosylceramidase [EC:3.2.1.123] | -1.900 | 2.241 | 0.398 | 0.978 |
| E3.2.1.14; chitinase [EC:3.2.1.14] | -0.198 | 0.356 | 0.579 | 0.997 |
| E3.2.1.15; polygalacturonase [EC:3.2.1.15] | -0.383 | 1.032 | 0.711 | 0.999 |
| E3.2.1.17; lysozyme [EC:3.2.1.17] | -0.026 | 0.235 | 0.912 | 0.999 |
| E3.2.1.1A; alpha-amylase [EC:3.2.1.1] | -0.233 | 0.239 | 0.331 | 0.978 |
| E3.2.1.22B, galA, rafA; alpha-galactosidase [EC:3.2.1.22] | 0.022 | 0.146 | 0.880 | 0.999 |
| E3.2.1.25, MANBA, manB; beta-mannosidase [EC:3.2.1.25] | -0.424 | 0.289 | 0.144 | 0.951 |
| E3.2.1.4; endoglucanase [EC:3.2.1.4] | -0.551 | 0.223 | 0.015 | 0.839 |
| E3.2.1.58; glucan 1,3-beta-glucosidase [EC:3.2.1.58] | -0.299 | 0.372 | 0.422 | 0.978 |
| E3.2.1.73; licheninase [EC:3.2.1.73] | 17.747 | 2319.587 | 0.994 | 0.999 |
| E3.2.1.8, xynA; endo-1,4-beta-xylanase [EC:3.2.1.8] | -0.669 | 0.358 | 0.064 | 0.839 |
| E3.2.1.85, lacG; 6-phospho-beta-galactosidase [EC:3.2.1.85] | -0.020 | 0.155 | 0.898 | 0.999 |
| E3.2.1.86A, celF; 6-phospho-beta-glucosidase [EC:3.2.1.86] | 0.050 | 0.372 | 0.893 | 0.999 |
| E3.2.1.86B, bglA; 6-phospho-beta-glucosidase [EC:3.2.1.86] | 0.082 | 0.121 | 0.498 | 0.981 |
| E3.2.1.89; arabinogalactan endo-1,4-beta-galactosidase [EC:3.2.1.89] | -0.567 | 0.333 | 0.090 | 0.882 |
| E3.2.1.96; mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase [EC:3.2.1.96] | -0.189 | 0.225 | 0.402 | 0.978 |
| E3.3.1.1, ahcY; adenosylhomocysteinase [EC:3.3.1.1] | -0.068 | 0.120 | 0.574 | 0.995 |
| E3.3.2.8; limonene-1,2-epoxide hydrolase [EC:3.3.2.8] | -2.062 | 2.006 | 0.306 | 0.978 |
| E3.4.11.-; aminopeptidase [EC:3.4.11.-] | 0.866 | 0.467 | 0.065 | 0.839 |
| E3.4.11.10; bacterial leucyl aminopeptidase [EC:3.4.11.10] | -0.958 | 1.043 | 0.360 | 0.978 |
| E3.4.11.14; cytosol alanyl aminopeptidase [EC:3.4.11.14] | 0.002 | 0.524 | 0.997 | 0.999 |
| E3.4.11.24; aminopeptidase S [EC:3.4.11.24] | 0.656 | 1.004 | 0.514 | 0.983 |
| E3.4.13.-; D-alanyl-D-alanine dipeptidase [EC:3.4.13.-] | -0.213 | 0.201 | 0.290 | 0.978 |
| E3.4.16.4; D-alanyl-D-alanine carboxypeptidase [EC:3.4.16.4] | -0.129 | 0.171 | 0.453 | 0.978 |
| E3.4.17.19; carboxypeptidase Taq [EC:3.4.17.19] | 0.220 | 0.189 | 0.247 | 0.978 |
| E3.4.21.100; pseudomonalisin [EC:3.4.21.100] | -17.453 | 3434.806 | 0.996 | 0.999 |
| E3.4.21.101; xanthomonalisin [EC:3.4.21.101] | -17.453 | 3434.806 | 0.996 | 0.999 |
| E3.4.21.102, prc, ctpA; carboxyl-terminal processing protease [EC:3.4.21.102] | -0.049 | 0.068 | 0.472 | 0.979 |
| E3.4.21.66; thermitase [EC:3.4.21.66] | -0.163 | 0.723 | 0.822 | 0.999 |
| E3.4.21.96; lactocepin [EC:3.4.21.96] | 0.048 | 0.179 | 0.790 | 0.999 |
| E3.5.1.1, ansA, ansB; L-asparaginase [EC:3.5.1.1] | 0.040 | 0.044 | 0.366 | 0.978 |
| E3.5.1.11; penicillin amidase [EC:3.5.1.11] | -0.160 | 0.228 | 0.484 | 0.979 |
| E3.5.1.24; choloylglycine hydrolase [EC:3.5.1.24] | 0.070 | 0.259 | 0.786 | 0.999 |
| E3.5.1.28D, amiD; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | 0.532 | 0.231 | 0.023 | 0.839 |
| E3.5.1.4, amiE; amidase [EC:3.5.1.4] | -0.182 | 0.163 | 0.266 | 0.978 |
| E3.5.1.41; chitin deacetylase [EC:3.5.1.41] | -17.631 | 1845.307 | 0.992 | 0.999 |
| E3.5.1.49; formamidase [EC:3.5.1.49] | -0.553 | 0.417 | 0.187 | 0.966 |
| E3.5.1.56; N,N-dimethylformamidase [EC:3.5.1.56] | -3.871 | 3.159 | 0.222 | 0.978 |
| E3.5.1.77; N-carbamoyl-D-amino-acid hydrolase [EC:3.5.1.77] | -1.391 | 0.982 | 0.159 | 0.954 |
| E3.5.1.81; N-acyl-D-amino-acid deacylase [EC:3.5.1.81] | -0.278 | 0.308 | 0.367 | 0.978 |
| E3.5.1.82; N-acyl-D-glutamate deacylase [EC:3.5.1.82] | -0.437 | 0.814 | 0.592 | 0.999 |
| E3.5.2.10; creatinine amidohydrolase [EC:3.5.2.10] | -0.903 | 0.370 | 0.016 | 0.839 |
| E3.5.3.1, rocF, arg; arginase [EC:3.5.3.1] | -0.247 | 0.179 | 0.170 | 0.961 |
| E3.5.3.3; creatinase [EC:3.5.3.3] | 0.537 | 0.553 | 0.332 | 0.978 |
| E3.5.4.3, guaD; guanine deaminase [EC:3.5.4.3] | -0.124 | 0.190 | 0.515 | 0.983 |
| E3.5.4.32; 8-oxoguanine deaminase [EC:3.5.4.32] | -0.084 | 0.429 | 0.844 | 0.999 |
| E3.5.5.1; nitrilase [EC:3.5.5.1] | -0.226 | 0.246 | 0.360 | 0.978 |
| E3.5.5.7; aliphatic nitrilase [EC:3.5.5.7] | -0.645 | 0.732 | 0.379 | 0.978 |
| E3.5.99.7; 1-aminocyclopropane-1-carboxylate deaminase [EC:3.5.99.7] | 0.123 | 0.269 | 0.649 | 0.999 |
| E3.6.1.22, NUDT12, nudC; NAD+ diphosphatase [EC:3.6.1.22] | 0.014 | 0.088 | 0.879 | 0.999 |
| E3.6.3.17; monosaccharide-transporting ATPase [EC:3.6.3.17] | -17.500 | 2562.559 | 0.995 | 0.999 |
| E3.6.3.25; sulfate-transporting ATPase [EC:3.6.3.25] | -0.144 | 0.324 | 0.659 | 0.999 |
| E3.6.3.8; Ca2+-transporting ATPase [EC:3.6.3.8] | 0.038 | 0.113 | 0.740 | 0.999 |
| E3.8.1.2; 2-haloacid dehalogenase [EC:3.8.1.2] | -0.367 | 0.170 | 0.032 | 0.839 |
| E4.1.1.15, gadB, gadA, GAD; glutamate decarboxylase [EC:4.1.1.15] | -0.073 | 0.440 | 0.868 | 0.999 |
| E4.1.1.17, ODC1, speC, speF; ornithine decarboxylase [EC:4.1.1.17] | 0.112 | 0.137 | 0.413 | 0.978 |
| E4.1.1.18, ldcC, cadA; lysine decarboxylase [EC:4.1.1.18] | -0.352 | 0.176 | 0.047 | 0.839 |
| E4.1.1.19; arginine decarboxylase [EC:4.1.1.19] | -0.230 | 0.231 | 0.322 | 0.978 |
| E4.1.1.32, pckA, PCK; phosphoenolpyruvate carboxykinase (GTP) [EC:4.1.1.32] | -0.160 | 0.213 | 0.454 | 0.978 |
| E4.1.1.49, pckA; phosphoenolpyruvate carboxykinase (ATP) [EC:4.1.1.49] | 0.061 | 0.078 | 0.440 | 0.978 |
| E4.1.1.64; 2,2-dialkylglycine decarboxylase (pyruvate) [EC:4.1.1.64] | -0.252 | 1.005 | 0.803 | 0.999 |
| E4.1.1.82; phosphonopyruvate decarboxylase [EC:4.1.1.82] | -0.372 | 0.491 | 0.450 | 0.978 |
| E4.1.3.1, aceA; isocitrate lyase [EC:4.1.3.1] | -0.271 | 0.173 | 0.120 | 0.917 |
| E4.1.3.25; (S)-citramalyl-CoA lyase [EC:4.1.3.25] | 0.096 | 0.375 | 0.798 | 0.999 |
| E4.1.3.3, nanA, NPL; N-acetylneuraminate lyase [EC:4.1.3.3] | 0.089 | 0.114 | 0.436 | 0.978 |
| E4.1.3.4, HMGCL, hmgL; hydroxymethylglutaryl-CoA lyase [EC:4.1.3.4] | -0.114 | 0.187 | 0.541 | 0.990 |
| E4.1.3.46, ccl; (R)-citramalyl-CoA lyase [EC:4.1.3.46] | 0.582 | 1.270 | 0.648 | 0.999 |
| E4.1.99.2; tyrosine phenol-lyase [EC:4.1.99.2] | 0.983 | 0.495 | 0.049 | 0.839 |
| E4.2.1.2A, fumA, fumB; fumarate hydratase, class I [EC:4.2.1.2] | -0.183 | 0.108 | 0.093 | 0.892 |
| E4.2.1.2AA, fumA; fumarate hydratase subunit alpha [EC:4.2.1.2] | 0.088 | 0.204 | 0.667 | 0.999 |
| E4.2.1.2AB, fumB; fumarate hydratase subunit beta [EC:4.2.1.2] | 0.088 | 0.203 | 0.666 | 0.999 |
| E4.2.1.2B, fumC, FH; fumarate hydratase, class II [EC:4.2.1.2] | -0.019 | 0.103 | 0.854 | 0.999 |
| E4.2.1.46, rfbB, rffG; dTDP-glucose 4,6-dehydratase [EC:4.2.1.46] | 0.157 | 0.059 | 0.008 | 0.839 |
| E4.2.2.10; pectin lyase [EC:4.2.2.10] | -0.835 | 0.918 | 0.364 | 0.978 |
| E4.2.2.17; inulin fructotransferase (DFA-I-forming) [EC:4.2.2.17] | -1.272 | 1.833 | 0.489 | 0.979 |
| E4.3.1.12, ocd; ornithine cyclodeaminase [EC:4.3.1.12] | -0.269 | 0.163 | 0.101 | 0.896 |
| E4.3.1.15; diaminopropionate ammonia-lyase [EC:4.3.1.15] | 0.180 | 0.200 | 0.370 | 0.978 |
| E4.3.1.17, sdaA, sdaB, tdcG; L-serine dehydratase [EC:4.3.1.17] | 0.020 | 0.050 | 0.682 | 0.999 |
| E4.3.1.19, ilvA, tdcB; threonine dehydratase [EC:4.3.1.19] | -0.060 | 0.049 | 0.228 | 0.978 |
| E4.3.1.23; tyrosine ammonia-lyase [EC:4.3.1.23] | -17.816 | 2251.882 | 0.994 | 0.999 |
| E4.3.1.4; formiminotetrahydrofolate cyclodeaminase [EC:4.3.1.4] | 0.374 | 0.195 | 0.057 | 0.839 |
| E4.4.1.11; methionine-gamma-lyase [EC:4.4.1.11] | 0.121 | 0.151 | 0.423 | 0.978 |
| E4.6.1.1; adenylate cyclase [EC:4.6.1.1] | -0.495 | 0.257 | 0.056 | 0.839 |
| E4.6.1.1B, cyaB; adenylate cyclase, class 2 [EC:4.6.1.1] | -0.050 | 0.218 | 0.820 | 0.999 |
| E4.6.1.2; guanylate cyclase, other [EC:4.6.1.2] | -1.465 | 1.176 | 0.215 | 0.978 |
| E4.99.1.2; alkylmercury lyase [EC:4.99.1.2] | 0.932 | 1.018 | 0.362 | 0.978 |
| E5.1.3.15; glucose-6-phosphate 1-epimerase [EC:5.1.3.15] | 0.110 | 0.180 | 0.542 | 0.990 |
| E5.1.3.6; UDP-glucuronate 4-epimerase [EC:5.1.3.6] | -0.206 | 0.203 | 0.313 | 0.978 |
| E5.1.99.4, AMACR, mcr; alpha-methylacyl-CoA racemase [EC:5.1.99.4] | -0.959 | 0.471 | 0.043 | 0.839 |
| E5.2.1.8; peptidylprolyl isomerase [EC:5.2.1.8] | 0.041 | 0.127 | 0.748 | 0.999 |
| E5.3.1.26, lacA, lacB; galactose-6-phosphate isomerase [EC:5.3.1.26] | -0.055 | 0.155 | 0.726 | 0.999 |
| E5.3.3.1; steroid Delta-isomerase [EC:5.3.3.1] | -1.585 | 1.445 | 0.274 | 0.978 |
| E5.3.4.1; protein disulfide-isomerase [EC:5.3.4.1] | -0.099 | 0.471 | 0.833 | 0.999 |
| E5.4.99.2A, mcmA1; methylmalonyl-CoA mutase, N-terminal domain [EC:5.4.99.2] | 0.131 | 0.223 | 0.557 | 0.992 |
| E5.4.99.2B, mcmA2; methylmalonyl-CoA mutase, C-terminal domain [EC:5.4.99.2] | 0.179 | 0.221 | 0.420 | 0.978 |
| E6.3.4.6; urea carboxylase [EC:6.3.4.6] | -0.232 | 0.317 | 0.464 | 0.979 |
| E6.3.5.1, NADSYN1, QNS1, nadE; NAD+ synthase (glutamine-hydrolysing) [EC:6.3.5.1] | -0.101 | 0.125 | 0.417 | 0.978 |
| E6.4.1.4A; 3-methylcrotonyl-CoA carboxylase alpha subunit [EC:6.4.1.4] | -0.240 | 0.220 | 0.277 | 0.978 |
| E6.4.1.4B; 3-methylcrotonyl-CoA carboxylase beta subunit [EC:6.4.1.4] | -0.217 | 0.221 | 0.329 | 0.978 |
| E6.5.1.2, ligA, ligB; DNA ligase (NAD+) [EC:6.5.1.2] | 0.037 | 0.024 | 0.130 | 0.934 |
| eamA; O-acetylserine/cysteine efflux transporter | -0.248 | 0.312 | 0.429 | 0.978 |
| eamB; cysteine/O-acetylserine efflux protein | -0.237 | 0.733 | 0.747 | 0.999 |
| eap, map; protein Map | -1.670 | 1.245 | 0.182 | 0.961 |
| EARS, gltX; glutamyl-tRNA synthetase [EC:6.1.1.17] | 0.002 | 0.073 | 0.984 | 0.999 |
| eat, eutP; ethanolamine permease | -0.167 | 0.294 | 0.573 | 0.995 |
| ebgA; evolved beta-galactosidase subunit alpha [EC:3.2.1.23] | 0.715 | 0.525 | 0.175 | 0.961 |
| ebgC; evolved beta-galactosidase subunit beta | 0.264 | 0.709 | 0.710 | 0.999 |
| ebgR; LacI family transcriptional regulator, ebg operon repressor | -0.014 | 0.767 | 0.986 | 0.999 |
| ebr, qacEdelta1; small multidrug resistance pump | 0.237 | 0.542 | 0.663 | 0.999 |
| ebrA; multidrug resistance protein EbrA | -0.653 | 1.290 | 0.614 | 0.999 |
| ebrB; multidrug resistance protein EbrB | -0.653 | 1.290 | 0.614 | 0.999 |
| ECE; endothelin-converting enzyme [EC:3.4.24.71] | -0.431 | 0.554 | 0.438 | 0.978 |
| ecfA1; energy-coupling factor transport system ATP-binding protein [EC:3.6.3.-] | 0.124 | 0.097 | 0.203 | 0.972 |
| ecfA2; energy-coupling factor transport system ATP-binding protein [EC:3.6.3.-] | 0.137 | 0.091 | 0.133 | 0.942 |
| ecfT; energy-coupling factor transport system permease protein | 0.071 | 0.097 | 0.470 | 0.979 |
| echA; ech hydrogenase subunit A | -1.229 | 1.638 | 0.454 | 0.978 |
| echC; ech hydrogenase subunit C | -1.229 | 1.638 | 0.454 | 0.978 |
| echE; ech hydrogenase subunit E | -1.229 | 1.638 | 0.454 | 0.978 |
| ecm; ethylmalonyl-CoA mutase [EC:5.4.99.63] | -0.313 | 0.603 | 0.605 | 0.999 |
| ECM4, yqjG; glutathionyl-hydroquinone reductase [EC:1.8.5.7] | -0.003 | 0.237 | 0.990 | 0.999 |
| ecnA; entericidin A | -0.303 | 0.643 | 0.638 | 0.999 |
| ecnB; entericidin B | 0.111 | 0.624 | 0.859 | 0.999 |
| eco; ecotin | 0.159 | 0.296 | 0.592 | 0.999 |
| ecpD; chaperone protein EcpD | -0.399 | 0.608 | 0.513 | 0.982 |
| ectA; L-2,4-diaminobutyric acid acetyltransferase [EC:2.3.1.178] | -0.756 | 0.523 | 0.151 | 0.954 |
| ectB, dat; diaminobutyrate-2-oxoglutarate transaminase [EC:2.6.1.76] | -0.089 | 0.254 | 0.728 | 0.999 |
| ectC; L-ectoine synthase [EC:4.2.1.108] | -0.689 | 0.485 | 0.157 | 0.954 |
| ectD; ectoine hydroxylase [EC:1.14.11.55] | -0.712 | 0.460 | 0.124 | 0.919 |
| eda; 2-dehydro-3-deoxyphosphogluconate aldolase / (4S)-4-hydroxy-2-oxoglutarate aldolase [EC:4.1.2.14 4.1.3.42] | 0.005 | 0.109 | 0.963 | 0.999 |
| edd; phosphogluconate dehydratase [EC:4.2.1.12] | -0.109 | 0.212 | 0.609 | 0.999 |
| EDEM1; ER degradation enhancer, mannosidase alpha-like 1 | -17.761 | 4005.858 | 0.996 | 0.999 |
| EDEM2; ER degradation enhancer, mannosidase alpha-like 2 | -17.761 | 4005.858 | 0.996 | 0.999 |
| EEF1A; elongation factor 1-alpha | -17.234 | 3077.514 | 0.996 | 0.999 |
| EEF1B; elongation factor 1-beta | -17.234 | 3077.514 | 0.996 | 0.999 |
| EEF2; elongation factor 2 | -17.234 | 3077.514 | 0.996 | 0.999 |
| eexD; ATP-binding cassette, subfamily C, bacterial EexD | 0.679 | 0.662 | 0.306 | 0.978 |
| eexE; membrane fusion protein, epimerase transport system | 0.828 | 0.677 | 0.223 | 0.978 |
| efeB; deferrochelatase/peroxidase EfeB [EC:1.11.1.-] | -0.138 | 0.142 | 0.333 | 0.978 |
| efeO; iron uptake system component EfeO | -0.138 | 0.140 | 0.327 | 0.978 |
| efp; elongation factor P | 0.034 | 0.038 | 0.373 | 0.978 |
| efrA; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.104 | 0.184 | 0.575 | 0.995 |
| efrB; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.104 | 0.184 | 0.575 | 0.995 |
| EGD2, NACA; nascent polypeptide-associated complex subunit alpha | -17.234 | 3077.514 | 0.996 | 0.999 |
| egtB; gamma-glutamyl hercynylcysteine S-oxide synthase [EC:1.14.99.50] | -0.956 | 0.805 | 0.237 | 0.978 |
| egtC; gamma-glutamyl hercynylcysteine S-oxide hydrolase [EC:3.5.1.118] | -0.023 | 0.172 | 0.892 | 0.999 |
| egtD; L-histidine Nalpha-methyltransferase [EC:2.1.1.44] | -0.927 | 0.747 | 0.217 | 0.978 |
| egtE; hercynylcysteine S-oxide lyase [EC:4.4.1.36] | -0.145 | 0.798 | 0.856 | 0.999 |
| ehbQ; energy-converting hydrogenase B subunit Q | -2.116 | 2.060 | 0.306 | 0.978 |
| EIF1, SUI1; translation initiation factor 1 | 0.036 | 0.173 | 0.837 | 0.999 |
| EIF1A; translation initiation factor 1A | -17.234 | 3077.514 | 0.996 | 0.999 |
| EIF2S1; translation initiation factor 2 subunit 1 | -17.234 | 3077.514 | 0.996 | 0.999 |
| EIF2S2; translation initiation factor 2 subunit 2 | -17.234 | 3077.514 | 0.996 | 0.999 |
| EIF2S3; translation initiation factor 2 subunit 3 | -17.234 | 3077.514 | 0.996 | 0.999 |
| EIF5A; translation initiation factor 5A | -17.234 | 3077.514 | 0.996 | 0.999 |
| EIF5B; translation initiation factor 5B | -17.234 | 3077.514 | 0.996 | 0.999 |
| EIF6; translation initiation factor 6 | -17.234 | 3077.514 | 0.996 | 0.999 |
| elaA; ElaA protein | 0.141 | 0.136 | 0.299 | 0.978 |
| elaB; ElaB protein | 0.169 | 0.657 | 0.798 | 0.999 |
| elaD, sseL; deubiquitinase [EC:3.4.22.-] | -1.125 | 1.387 | 0.419 | 0.978 |
| ELP3, KAT9; elongator complex protein 3 [EC:2.3.1.48] | -0.417 | 0.662 | 0.529 | 0.989 |
| embA; arabinosyltransferase A [EC:2.4.2.-] | -2.654 | 1.420 | 0.063 | 0.839 |
| embB; arabinosyltransferase B [EC:2.4.2.-] | -1.615 | 1.113 | 0.149 | 0.954 |
| embC; arabinosyltransferase C [EC:2.4.2.-] | -0.901 | 0.888 | 0.312 | 0.978 |
| EMG1, NEP1; rRNA small subunit pseudouridine methyltransferase Nep1 [EC:2.1.1.260] | -17.234 | 3077.514 | 0.996 | 0.999 |
| emrA; membrane fusion protein, multidrug efflux system | -0.056 | 0.176 | 0.750 | 0.999 |
| emrB; MFS transporter, DHA2 family, multidrug resistance protein | -0.120 | 0.202 | 0.553 | 0.992 |
| emrD; MFS transporter, DHA1 family, 2-module integral membrane pump EmrD | 0.285 | 0.565 | 0.614 | 0.999 |
| emrE, qac, mmr, smr; small multidrug resistance pump | -0.075 | 0.152 | 0.621 | 0.999 |
| emrK; multidrug resistance protein K | -1.153 | 1.472 | 0.435 | 0.978 |
| emrR, mprA; MarR family transcriptional regulator, negative regulator of the multidrug operon emrRAB | 0.126 | 0.383 | 0.742 | 0.999 |
| emrY; MFS transporter, DHA2 family, multidrug resistance protein | -1.153 | 1.472 | 0.435 | 0.978 |
| ena; enamidase [EC:3.5.2.18] | -2.122 | 1.292 | 0.102 | 0.899 |
| endA; deoxyribonuclease I [EC:3.1.21.1] | 0.336 | 0.299 | 0.263 | 0.978 |
| endA; DNA-entry nuclease | 0.112 | 0.162 | 0.491 | 0.980 |
| endA; tRNA-intron endonuclease, archaea type [EC:4.6.1.16] | -16.927 | 2639.780 | 0.995 | 0.999 |
| ENDOG; endonuclease G, mitochondrial | -0.116 | 0.166 | 0.488 | 0.979 |
| engA, der; GTPase | 0.035 | 0.036 | 0.325 | 0.978 |
| engB; GTP-binding protein | 0.045 | 0.042 | 0.289 | 0.978 |
| engCP, engBF, endoEF; endo-alpha-N-acetylgalactosaminidase [EC:3.2.1.97] | 0.967 | 0.925 | 0.298 | 0.978 |
| ENO, eno; enolase [EC:4.2.1.11] | 0.038 | 0.056 | 0.498 | 0.981 |
| enr; 2-enoate reductase [EC:1.3.1.31] | -1.250 | 1.064 | 0.242 | 0.978 |
| entA; 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase [EC:1.3.1.28] | -0.118 | 0.268 | 0.659 | 0.999 |
| entA; probable enterotoxin A | 6.328 | 12.689 | 0.619 | 0.999 |
| entB, dhbB, vibB, mxcF; bifunctional isochorismate lyase / aryl carrier protein [EC:3.3.2.1 6.3.2.14] | -0.157 | 0.274 | 0.568 | 0.995 |
| entB; probable enterotoxin B | 2.821 | 2.375 | 0.237 | 0.978 |
| entC; isochorismate synthase [EC:5.4.4.2] | -0.186 | 0.153 | 0.225 | 0.978 |
| entC; probable enterotoxin C | 4.874 | 6.172 | 0.431 | 0.978 |
| entD; enterobactin synthetase component D [EC:6.3.2.14 2.7.8.-] | 0.100 | 0.313 | 0.750 | 0.999 |
| entD; probable enterotoxin D | 6.196 | 11.877 | 0.603 | 0.999 |
| entE, dhbE, vibE, mxcE; 2,3-dihydroxybenzoate-AMP ligase [EC:6.3.2.14 2.7.7.58] | -0.109 | 0.255 | 0.669 | 0.999 |
| entF; enterobactin synthetase component F [EC:6.3.2.14] | -0.038 | 0.278 | 0.891 | 0.999 |
| entS; MFS transporter, ENTS family, enterobactin (siderophore) exporter | -0.273 | 0.586 | 0.642 | 0.999 |
| envR, acrS; TetR/AcrR family transcriptional regulator, acrEF/envCD operon repressor | -0.243 | 0.717 | 0.735 | 0.999 |
| envY; AraC family transcriptional regulator | -1.154 | 1.507 | 0.445 | 0.978 |
| envZ; two-component system, OmpR family, osmolarity sensor histidine kinase EnvZ [EC:2.7.13.3] | -0.174 | 0.223 | 0.436 | 0.978 |
| epd; D-erythrose 4-phosphate dehydrogenase [EC:1.2.1.72] | 0.107 | 0.268 | 0.690 | 0.999 |
| EPHX1; microsomal epoxide hydrolase [EC:3.3.2.9] | 1.838 | 1.160 | 0.115 | 0.917 |
| epmA, poxA; elongation factor P–(R)-beta-lysine ligase [EC:6.3.1.-] | -0.055 | 0.202 | 0.787 | 0.999 |
| epmC; elongation factor P hydroxylase [EC:1.14.-.-] | 0.102 | 0.257 | 0.692 | 0.999 |
| epr; minor extracellular protease Epr [EC:3.4.21.-] | -1.144 | 1.057 | 0.281 | 0.978 |
| EPS15; epidermal growth factor receptor substrate 15 | -0.144 | 0.503 | 0.775 | 0.999 |
| epsA; protein tyrosine kinase EpsB modulator | 18.239 | 2966.075 | 0.995 | 0.999 |
| epsC; polysaccharide biosynthesis protein EpsC | 18.239 | 2966.075 | 0.995 | 0.999 |
| epsD; glycosyltransferase EpsD [EC:2.4.-.-] | -0.166 | 0.421 | 0.694 | 0.999 |
| epsE; glycosyltransferase EpsE [EC:2.4.-.-] | 1.425 | 1.354 | 0.294 | 0.978 |
| epsF; glycosyltransferase EpsF [EC:2.4.-.-] | -0.454 | 1.283 | 0.724 | 0.999 |
| epsG; transmembrane protein EpsG | -1.684 | 1.115 | 0.133 | 0.942 |
| epsH; glycosyltransferase EpsH [EC:2.4.-.-] | -2.855 | 2.098 | 0.176 | 0.961 |
| epsI; pyruvyl transferase EpsI [EC:2.-.-.-] | 1.929 | 1.610 | 0.232 | 0.978 |
| epsJ; glycosyltransferase EpsJ [EC:2.4.-.-] | 0.012 | 0.446 | 0.979 | 0.999 |
| epsK; membrane protein EpsK | -1.763 | 2.141 | 0.411 | 0.978 |
| epsL; sugar transferase EpsL [EC:2.-.-.-] | -2.780 | 2.162 | 0.200 | 0.971 |
| epsM; acetyltransferase EpsM [EC:2.3.1.-] | -0.221 | 0.665 | 0.740 | 0.999 |
| epsN; pyridoxal phosphate-dependent aminotransferase EpsN [EC:2.6.1.-] | -0.734 | 1.068 | 0.493 | 0.981 |
| epsO; pyruvyl transferase EpsO [EC:2.-.-.-] | 0.268 | 0.371 | 0.471 | 0.979 |
| EPT1; ethanolaminephosphotransferase [EC:2.7.8.1] | -3.190 | 3.828 | 0.406 | 0.978 |
| eptA, pmrC; lipid A ethanolaminephosphotransferase [EC:2.7.8.43] | -0.135 | 0.237 | 0.571 | 0.995 |
| eptB; KDO II ethanolaminephosphotransferase [EC:2.7.8.42] | 0.056 | 0.619 | 0.928 | 0.999 |
| eptC; heptose-I-phosphate ethanolaminephosphotransferase [EC:2.7.8.-] | 0.025 | 0.176 | 0.887 | 0.999 |
| era, ERAL1; GTPase | 0.038 | 0.033 | 0.253 | 0.978 |
| ERCC2, XPD; DNA excision repair protein ERCC-2 [EC:3.6.4.12] | 1.708 | 0.931 | 0.068 | 0.839 |
| ERCC3, XPB; DNA excision repair protein ERCC-3 [EC:3.6.4.12] | -0.232 | 0.353 | 0.512 | 0.982 |
| ERCC4, XPF; DNA excision repair protein ERCC-4 [EC:3.1.-.-] | -0.272 | 0.958 | 0.777 | 0.999 |
| ereA_B; erythromycin esterase [EC:3.1.1.-] | -0.750 | 1.127 | 0.507 | 0.981 |
| erfK; L,D-transpeptidase ErfK/SrfK | 0.203 | 0.319 | 0.526 | 0.989 |
| ermC, ermA; 23S rRNA (adenine-N6)-dimethyltransferase [EC:2.1.1.184] | 0.229 | 0.322 | 0.478 | 0.979 |
| erpA; iron-sulfur cluster insertion protein | 0.034 | 0.152 | 0.826 | 0.999 |
| eryA; erythritol kinase (D-erythritol 1-phosphate-forming) [EC:2.7.1.215] | -2.827 | 1.713 | 0.101 | 0.896 |
| eryE; erythritol transport system ATP-binding protein | -2.415 | 1.241 | 0.053 | 0.839 |
| esp, sigA, sepA; serine protease autotransporter [EC:3.4.21.-] | -1.432 | 1.511 | 0.345 | 0.978 |
| estA; putative tributyrin esterase [EC:3.1.1.-] | -0.019 | 0.173 | 0.915 | 0.999 |
| esxA, esat6; 6 kDa early secretory antigenic target | -1.749 | 0.971 | 0.073 | 0.845 |
| eta; exfoliative toxin A/B | -0.016 | 0.168 | 0.924 | 0.999 |
| etbAa; ethylbenzene dioxygenase subunit alpha [EC:1.14.12.-] | 0.884 | 1.140 | 0.439 | 0.978 |
| etbAb; ethylbenzene dioxygenase subunit beta [EC:1.14.12.-] | 0.884 | 1.140 | 0.439 | 0.978 |
| etbD; 2-hydroxy-6-oxo-octa-2,4-dienoate hydrolase [EC:3.7.1.-] | 0.884 | 1.140 | 0.439 | 0.978 |
| ETF1, ERF1; peptide chain release factor subunit 1 | -17.833 | 3732.576 | 0.996 | 0.999 |
| ETFDH; electron-transferring-flavoprotein dehydrogenase [EC:1.5.5.1] | -0.168 | 0.185 | 0.365 | 0.978 |
| ethA; monooxygenase [EC:1.14.13.-] | -1.874 | 0.925 | 0.044 | 0.839 |
| etk-wzc; tyrosine-protein kinase Etk/Wzc [EC:2.7.10.-] | -0.027 | 0.267 | 0.920 | 0.999 |
| eutA; ethanolamine utilization protein EutA | 0.959 | 0.486 | 0.050 | 0.839 |
| eutB; ethanolamine ammonia-lyase large subunit [EC:4.3.1.7] | 0.329 | 0.230 | 0.155 | 0.954 |
| eutC; ethanolamine ammonia-lyase small subunit [EC:4.3.1.7] | 0.339 | 0.232 | 0.146 | 0.954 |
| eutD; phosphotransacetylase | -0.890 | 1.071 | 0.408 | 0.978 |
| eutE; aldehyde dehydrogenase | -0.590 | 0.955 | 0.537 | 0.990 |
| eutG; alcohol dehydrogenase | -0.890 | 1.071 | 0.408 | 0.978 |
| eutH; ethanolamine transporter | 0.883 | 0.465 | 0.059 | 0.839 |
| eutJ; ethanolamine utilization protein EutJ | 1.011 | 0.454 | 0.027 | 0.839 |
| eutK; ethanolamine utilization protein EutK | -0.603 | 1.003 | 0.548 | 0.991 |
| eutL; ethanolamine utilization protein EutL | 0.979 | 0.493 | 0.049 | 0.839 |
| eutM; ethanolamine utilization protein EutM | 1.230 | 0.534 | 0.023 | 0.839 |
| eutN; ethanolamine utilization protein EutN | 0.574 | 0.362 | 0.114 | 0.917 |
| eutP; ethanolamine utilization protein EutP | 0.513 | 0.227 | 0.025 | 0.839 |
| eutQ; ethanolamine utilization protein EutQ | 1.105 | 0.499 | 0.028 | 0.839 |
| eutR; AraC family transcriptional regulator, ethanolamine operon transcriptional activator | -0.248 | 0.346 | 0.473 | 0.979 |
| eutS; ethanolamine utilization protein EutS | 0.489 | 0.225 | 0.031 | 0.839 |
| eutT; ethanolamine utilization cobalamin adenosyltransferase [EC:2.5.1.17] | 1.226 | 0.546 | 0.026 | 0.839 |
| evgA, bvgA; two-component system, NarL family, response regulator EvgA | -0.023 | 0.392 | 0.953 | 0.999 |
| evgS, bvgS; two-component system, NarL family, sensor histidine kinase EvgS [EC:2.7.13.3] | -0.034 | 0.306 | 0.912 | 0.999 |
| exaA; alcohol dehydrogenase (cytochrome c) [EC:1.1.2.8] | 0.421 | 0.340 | 0.218 | 0.978 |
| exbB; biopolymer transport protein ExbB | 0.024 | 0.094 | 0.802 | 0.999 |
| exbD; biopolymer transport protein ExbD | 0.020 | 0.118 | 0.864 | 0.999 |
| exoA; succinoglycan biosynthesis protein ExoA [EC:2.4.-.-] | -4.046 | 3.981 | 0.311 | 0.978 |
| exoF; polysaccharide biosynthesis/export protein ExoF | -4.046 | 3.981 | 0.311 | 0.978 |
| exoH; succinoglycan biosynthesis protein ExoH | -17.628 | 2890.370 | 0.995 | 0.999 |
| exoK; endo-1,3-1,4-beta-glycanase ExoK [EC:3.2.1.-] | -4.046 | 3.981 | 0.311 | 0.978 |
| exoL; succinoglycan biosynthesis protein ExoL [EC:2.-.-.-] | -4.016 | 4.039 | 0.322 | 0.978 |
| exoM; succinoglycan biosynthesis protein ExoM [EC:2.4.-.-] | -1.443 | 1.225 | 0.241 | 0.978 |
| exoO; succinoglycan biosynthesis protein ExoO [EC:2.4.-.-] | -3.243 | 2.510 | 0.198 | 0.970 |
| exoP, vpsO; polysaccharide biosynthesis transport protein | -2.239 | 1.419 | 0.117 | 0.917 |
| exoQ; exopolysaccharide production protein ExoQ | -2.549 | 1.696 | 0.135 | 0.944 |
| exoT; succinoglycan exporter | -17.579 | 2959.469 | 0.995 | 0.999 |
| exoU; succinoglycan biosynthesis protein ExoU [EC:2.4.-.-] | -3.677 | 4.053 | 0.366 | 0.978 |
| exoV; succinoglycan biosynthesis protein ExoV | -3.493 | 3.143 | 0.268 | 0.978 |
| exoW; succinoglycan biosynthesis protein ExoW [EC:2.4.-.-] | -3.164 | 3.139 | 0.315 | 0.978 |
| exoX; exodeoxyribonuclease X [EC:3.1.11.-] | -0.287 | 0.337 | 0.395 | 0.978 |
| exoX; exopolysaccharide production repressor protein | -17.825 | 3152.083 | 0.995 | 0.999 |
| exoY; exopolysaccharide production protein ExoY | -2.328 | 1.679 | 0.167 | 0.961 |
| exoZ; exopolysaccharide production protein ExoZ | -1.789 | 0.898 | 0.048 | 0.839 |
| expR; LuxR family transcriptional regulator, quorum-sensing system regulator ExpR | 1.333 | 0.980 | 0.176 | 0.961 |
| exuR; GntR family transcriptional regulator, hexuronate regulon transcriptional repressor | 0.135 | 0.629 | 0.830 | 0.999 |
| exuT; MFS transporter, ACS family, hexuronate transporter | -0.174 | 0.217 | 0.425 | 0.978 |
| ezrA; septation ring formation regulator | 0.030 | 0.140 | 0.830 | 0.999 |
| faaH; fumarylacetoacetate (FAA) hydrolase [EC:3.7.1.2] | -0.091 | 0.274 | 0.741 | 0.999 |
| FAB2, SSI2, desA1; acyl-[acyl-carrier-protein] desaturase [EC:1.14.19.2 1.14.19.11 1.14.19.26] | -1.679 | 0.724 | 0.022 | 0.839 |
| fabA; 3-hydroxyacyl-[acyl-carrier protein] dehydratase / trans-2-decenoyl-[acyl-carrier protein] isomerase [EC:4.2.1.59 5.3.3.14] | -0.027 | 0.165 | 0.869 | 0.999 |
| fabB; 3-oxoacyl-[acyl-carrier-protein] synthase I [EC:2.3.1.41] | -0.120 | 0.135 | 0.377 | 0.978 |
| fabD; [acyl-carrier-protein] S-malonyltransferase [EC:2.3.1.39] | 0.016 | 0.032 | 0.623 | 0.999 |
| fabF; 3-oxoacyl-[acyl-carrier-protein] synthase II [EC:2.3.1.179] | 0.004 | 0.031 | 0.891 | 0.999 |
| fabG; 3-oxoacyl-[acyl-carrier protein] reductase [EC:1.1.1.100] | -0.092 | 0.051 | 0.072 | 0.845 |
| fabH; 3-oxoacyl-[acyl-carrier-protein] synthase III [EC:2.3.1.180] | 0.049 | 0.045 | 0.276 | 0.978 |
| fabI; enoyl-[acyl-carrier protein] reductase I [EC:1.3.1.9 1.3.1.10] | -0.139 | 0.105 | 0.188 | 0.966 |
| fabK; enoyl-[acyl-carrier protein] reductase II [EC:1.3.1.9] | 0.130 | 0.098 | 0.186 | 0.966 |
| fabL; enoyl-[acyl-carrier protein] reductase III [EC:1.3.1.104] | -1.976 | 0.924 | 0.034 | 0.839 |
| fabM; trans-2-decenoyl-[acyl-carrier protein] isomerase [EC:5.3.3.14] | 0.008 | 0.152 | 0.958 | 0.999 |
| fabV, ter; enoyl-[acyl-carrier protein] reductase / trans-2-enoyl-CoA reductase (NAD+) [EC:1.3.1.9 1.3.1.44] | 0.093 | 0.231 | 0.688 | 0.999 |
| fabY; acetoacetyl-[acyl-carrier protein] synthase [EC:2.3.1.180] | 0.790 | 0.468 | 0.093 | 0.892 |
| fabZ; 3-hydroxyacyl-[acyl-carrier-protein] dehydratase [EC:4.2.1.59] | 0.073 | 0.046 | 0.116 | 0.917 |
| FAD6, desA; acyl-lipid omega-6 desaturase (Delta-12 desaturase) [EC:1.14.19.23 1.14.19.45] | -1.235 | 0.632 | 0.052 | 0.839 |
| fadA, fadI; acetyl-CoA acyltransferase [EC:2.3.1.16] | -0.208 | 0.172 | 0.229 | 0.978 |
| fadB; 3-hydroxyacyl-CoA dehydrogenase / enoyl-CoA hydratase / 3-hydroxybutyryl-CoA epimerase / enoyl-CoA isomerase [EC:1.1.1.35 4.2.1.17 5.1.2.3 5.3.3.8] | 0.101 | 0.251 | 0.687 | 0.999 |
| fadB; enoyl-CoA hydratase [EC:4.2.1.17] | -1.302 | 0.910 | 0.155 | 0.954 |
| fadD10; long chain fatty acid CoA ligase FadD10 [EC:6.2.1.-] | -17.763 | 3362.856 | 0.996 | 0.999 |
| fadD21; fatty acid CoA ligase FadD21 | -2.624 | 1.417 | 0.066 | 0.839 |
| fadD26; long chain fatty acid CoA FadD26 | -17.615 | 3031.381 | 0.995 | 0.999 |
| fadD28; long-chain fatty acid adenylyltransferase FadD28 [EC:6.2.1.49] | -1.593 | 1.107 | 0.152 | 0.954 |
| fadD32; fatty acid CoA ligase FadD32 | -0.698 | 0.752 | 0.355 | 0.978 |
| fadD36; fatty acid CoA ligase FadD36 | -1.655 | 1.561 | 0.291 | 0.978 |
| fadD9; fatty acid CoA ligase FadD9 | -2.953 | 1.583 | 0.064 | 0.839 |
| fadE; acyl-CoA dehydrogenase [EC:1.3.99.-] | -0.083 | 0.246 | 0.735 | 0.999 |
| fadH; 2,4-dienoyl-CoA reductase (NADPH2) [EC:1.3.1.34] | -0.086 | 0.210 | 0.681 | 0.999 |
| fadJ; 3-hydroxyacyl-CoA dehydrogenase / enoyl-CoA hydratase / 3-hydroxybutyryl-CoA epimerase [EC:1.1.1.35 4.2.1.17 5.1.2.3] | -0.214 | 0.176 | 0.225 | 0.978 |
| fadK; acyl-CoA synthetase [EC:6.2.1.-] | -0.272 | 0.632 | 0.667 | 0.999 |
| fadL; long-chain fatty acid transport protein | 0.056 | 0.141 | 0.695 | 0.999 |
| fadN; 3-hydroxyacyl-CoA dehydrogenase [EC:1.1.1.35] | -0.469 | 0.267 | 0.080 | 0.864 |
| fadR; GntR family transcriptional regulator, negative regulator for fad regulon and positive regulator of fabA | 0.231 | 0.308 | 0.455 | 0.978 |
| fae; 5,6,7,8-tetrahydromethanopterin hydro-lyase [EC:4.2.1.147] | 0.810 | 0.822 | 0.327 | 0.978 |
| FAEB; feruloyl esterase [EC:3.1.1.73] | -1.085 | 0.762 | 0.157 | 0.954 |
| FAH, fahA; fumarylacetoacetase [EC:3.7.1.2] | -0.062 | 0.247 | 0.802 | 0.999 |
| FANCM; fanconi anemia group M protein | -0.283 | 0.927 | 0.760 | 0.999 |
| FARSA, pheS; phenylalanyl-tRNA synthetase alpha chain [EC:6.1.1.20] | 0.037 | 0.036 | 0.306 | 0.978 |
| FARSB, pheT; phenylalanyl-tRNA synthetase beta chain [EC:6.1.1.20] | 0.031 | 0.031 | 0.318 | 0.978 |
| fas; fatty acid synthase, bacteria type [EC:2.3.1.-] | -0.113 | 0.284 | 0.692 | 0.999 |
| FBA, fbaA; fructose-bisphosphate aldolase, class II [EC:4.1.2.13] | 0.030 | 0.054 | 0.585 | 0.999 |
| fbaB; fructose-bisphosphate aldolase, class I [EC:4.1.2.13] | -0.570 | 0.381 | 0.136 | 0.949 |
| fbcH; ubiquinol-cytochrome c reductase cytochrome b/c1 subunit | -1.305 | 0.900 | 0.149 | 0.954 |
| fbiC; FO synthase [EC:2.5.1.77] | -0.737 | 0.622 | 0.238 | 0.978 |
| fbp-SEBP; fructose-1,6-bisphosphatase I / sedoheptulose-1,7-bisphosphatase [EC:3.1.3.11 3.1.3.37] | -0.735 | 0.957 | 0.443 | 0.978 |
| FBP, fbp; fructose-1,6-bisphosphatase I [EC:3.1.3.11] | -0.013 | 0.141 | 0.927 | 0.999 |
| fbp; diacylglycerol O-acyltransferase / trehalose O-mycolyltransferase [EC:2.3.1.20 2.3.1.122] | -1.796 | 1.084 | 0.100 | 0.896 |
| fbp3; fructose-1,6-bisphosphatase III [EC:3.1.3.11] | 0.059 | 0.137 | 0.667 | 0.999 |
| fccA; cytochrome subunit of sulfide dehydrogenase | -17.256 | 3111.566 | 0.996 | 0.999 |
| fccB; sulfide dehydrogenase [flavocytochrome c] flavoprotein chain [EC:1.8.2.3] | -17.256 | 3111.566 | 0.996 | 0.999 |
| fcs; feruloyl-CoA synthase [EC:6.2.1.34] | -1.018 | 0.456 | 0.027 | 0.839 |
| fctD; glutamate formiminotransferase [EC:2.1.2.5] | 0.354 | 0.188 | 0.062 | 0.839 |
| FDFT1; farnesyl-diphosphate farnesyltransferase [EC:2.5.1.21] | -0.730 | 1.221 | 0.551 | 0.992 |
| FDH; formate dehydrogenase [EC:1.17.1.9] | -1.397 | 0.682 | 0.042 | 0.839 |
| fdhA; formate dehydrogenase (NADP+) alpha subunit [EC:1.17.1.10] | 0.118 | 0.468 | 0.801 | 0.999 |
| fdhA; glutathione-independent formaldehyde dehydrogenase [EC:1.2.1.46] | 0.151 | 0.345 | 0.663 | 0.999 |
| fdhB; formate dehydrogenase (NADP+) beta subunit [EC:1.17.1.10] | -17.478 | 2545.196 | 0.995 | 0.999 |
| fdhD; FdhD protein | -0.099 | 0.095 | 0.302 | 0.978 |
| fdhE; FdhE protein | 0.155 | 0.191 | 0.420 | 0.978 |
| fdnG; formate dehydrogenase-N, alpha subunit [EC:1.17.5.3] | 0.062 | 0.630 | 0.921 | 0.999 |
| fdnH; formate dehydrogenase-N, beta subunit | 0.313 | 0.638 | 0.625 | 0.999 |
| fdnI; formate dehydrogenase-N, gamma subunit | 0.313 | 0.638 | 0.625 | 0.999 |
| fdoG, fdhF, fdwA; formate dehydrogenase major subunit [EC:1.17.1.9] | 0.042 | 0.120 | 0.725 | 0.999 |
| fdoH, fdsB; formate dehydrogenase iron-sulfur subunit | 0.113 | 0.160 | 0.482 | 0.979 |
| fdoI, fdsG; formate dehydrogenase subunit gamma | 0.140 | 0.155 | 0.369 | 0.978 |
| fdrA; FdrA protein | -0.309 | 0.946 | 0.744 | 0.999 |
| fdsD; formate dehydrogenase subunit delta [EC:1.17.1.9] | -0.261 | 0.323 | 0.420 | 0.978 |
| fdx; ferredoxin, 2Fe-2S | -0.058 | 0.133 | 0.663 | 0.999 |
| fdxA; ferredoxin | -0.144 | 0.171 | 0.401 | 0.978 |
| feaB; phenylacetaldehyde dehydrogenase [EC:1.2.1.39] | 0.007 | 0.354 | 0.984 | 0.999 |
| feaR; AraC family transcriptional regulator, positive regulator of tynA and feaB | 0.179 | 0.636 | 0.779 | 0.999 |
| fecA; Fe(3+) dicitrate transport protein | -0.332 | 0.298 | 0.267 | 0.978 |
| fecR; transmembrane sensor | -0.045 | 0.268 | 0.867 | 0.999 |
| femA; peptidoglycan pentaglycine glycine transferase (the second and third glycine) [EC:2.3.2.17] | -0.220 | 0.810 | 0.786 | 0.999 |
| femB; peptidoglycan pentaglycine glycine transferase (the fourth and fifth glycine) [EC:2.3.2.18] | -0.220 | 0.810 | 0.786 | 0.999 |
| femX, fmhB; peptidoglycan pentaglycine glycine transferase (the first glycine) [EC:2.3.2.16] | -0.140 | 0.624 | 0.823 | 0.999 |
| FEN1, RAD2; flap endonuclease-1 [EC:3.-.-.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| feoA; ferrous iron transport protein A | 0.098 | 0.134 | 0.466 | 0.979 |
| feoB; ferrous iron transport protein B | 0.003 | 0.109 | 0.976 | 0.999 |
| feoC; ferrous iron transport protein C | 0.120 | 0.603 | 0.842 | 0.999 |
| feoC; putative ferrous iron transport protein C | 18.119 | 1920.327 | 0.992 | 0.999 |
| fepA, pfeA, iroN, pirA; ferric enterobactin receptor | 0.031 | 0.270 | 0.908 | 0.999 |
| fer; ferredoxin | -0.085 | 0.122 | 0.485 | 0.979 |
| fes; enterochelin esterase and related enzymes | -0.176 | 0.208 | 0.397 | 0.978 |
| fexA_B; MFS transporter, DHA2 family, florfenicol/chloramphenicol resistance protein | -1.119 | 1.422 | 0.432 | 0.978 |
| fgd1; coenzyme F420-dependent glucose-6-phosphate dehydrogenase [EC:1.1.98.2] | -0.993 | 0.998 | 0.322 | 0.978 |
| fha1; type VI secretion system protein | -0.395 | 0.579 | 0.496 | 0.981 |
| fhaB; filamentous hemagglutinin | 0.515 | 0.290 | 0.077 | 0.856 |
| fhaC; hemolysin activation/secretion protein | -0.516 | 0.625 | 0.410 | 0.978 |
| fhlA; formate hydrogenlyase transcriptional activator | -0.180 | 0.641 | 0.779 | 0.999 |
| fhs; formate–tetrahydrofolate ligase [EC:6.3.4.3] | 0.063 | 0.083 | 0.453 | 0.978 |
| fhuF; ferric iron reductase protein FhuF | -0.311 | 0.480 | 0.518 | 0.986 |
| fic; cell filamentation protein | -0.084 | 0.157 | 0.594 | 0.999 |
| fieF; ferrous-iron efflux pump FieF | 0.213 | 0.299 | 0.476 | 0.979 |
| fimA; major type 1 subunit fimbrin (pilin) | -0.242 | 0.289 | 0.404 | 0.978 |
| fimB; type 1 fimbriae regulatory protein FimB | -0.156 | 0.728 | 0.831 | 0.999 |
| fimC; fimbrial chaperone protein | -0.033 | 0.276 | 0.906 | 0.999 |
| fimD, fimC, mrkC, htrE, cssD; outer membrane usher protein | -0.109 | 0.236 | 0.644 | 0.999 |
| fimE; type 1 fimbriae regulatory protein FimE | -1.040 | 1.152 | 0.368 | 0.978 |
| fimF; minor fimbrial subunit | -0.595 | 0.909 | 0.513 | 0.982 |
| fimG; minor fimbrial subunit | -0.421 | 0.507 | 0.407 | 0.978 |
| fimH; minor fimbrial subunit | -0.626 | 0.936 | 0.504 | 0.981 |
| fimI; fimbrial protein | -0.151 | 0.664 | 0.820 | 0.999 |
| fimT; type IV fimbrial biogenesis protein FimT | 0.027 | 0.207 | 0.898 | 0.999 |
| fimU; type IV fimbrial biogenesis protein FimU | 0.516 | 0.465 | 0.269 | 0.978 |
| fimV; pilus assembly protein FimV | 0.123 | 0.240 | 0.610 | 0.999 |
| fimW; fimbrial protein FimW | -4.388 | 7.789 | 0.574 | 0.995 |
| fimY; fimbrial protein FimY | 0.630 | 0.788 | 0.425 | 0.978 |
| fimZ; two-component system, NarL family, response regulator, fimbrial Z protein, FimZ | -0.267 | 0.694 | 0.701 | 0.999 |
| fis; Fis family transcriptional regulator, factor for inversion stimulation protein | 0.082 | 0.156 | 0.601 | 0.999 |
| fitB; toxin FitB [EC:3.1.-.-] | -0.264 | 0.214 | 0.219 | 0.978 |
| fitD, mcf; insecticidal toxin | -0.241 | 0.840 | 0.775 | 0.999 |
| fiu; catecholate siderophore receptor | -0.100 | 0.234 | 0.669 | 0.999 |
| fixA, etfB; electron transfer flavoprotein beta subunit | 0.089 | 0.093 | 0.337 | 0.978 |
| fixB, etfA; electron transfer flavoprotein alpha subunit | 0.094 | 0.093 | 0.310 | 0.978 |
| fixC; electron transfer flavoprotein-quinone oxidoreductase [EC:1.5.5.-] | 0.434 | 0.230 | 0.061 | 0.839 |
| fixJ; two-component system, LuxR family, response regulator FixJ | -0.941 | 0.471 | 0.048 | 0.839 |
| fixK; CRP/FNR family transcriptional regulator, nitrogen fixation regulation protein | -1.279 | 0.882 | 0.149 | 0.954 |
| fixL; two-component system, LuxR family, sensor kinase FixL [EC:2.7.13.3] | -1.110 | 0.493 | 0.026 | 0.839 |
| fixX; ferredoxin like protein | 0.101 | 0.258 | 0.695 | 0.999 |
| fklB; FKBP-type peptidyl-prolyl cis-trans isomerase FklB [EC:5.2.1.8] | -0.066 | 0.145 | 0.649 | 0.999 |
| fkpA; FKBP-type peptidyl-prolyl cis-trans isomerase FkpA [EC:5.2.1.8] | -0.058 | 0.199 | 0.771 | 0.999 |
| flA; adenosyl-fluoride synthase [EC:2.5.1.63] | -0.039 | 0.146 | 0.789 | 0.999 |
| flaF; flagellar protein FlaF | -1.417 | 0.489 | 0.004 | 0.839 |
| flaG; flagellar protein FlaG | -0.076 | 0.213 | 0.722 | 0.999 |
| flaI; flagellar rod protein FlaI | 18.482 | 3349.575 | 0.996 | 0.999 |
| flbB; flagellar protein FlbB | -0.671 | 1.155 | 0.562 | 0.992 |
| flbD; flagellar protein FlbD | -0.119 | 0.361 | 0.741 | 0.999 |
| flbT; flagellar protein FlbT | -1.448 | 0.508 | 0.005 | 0.839 |
| fldA, nifF, isiB; flavodoxin I | 0.255 | 0.211 | 0.229 | 0.978 |
| fldA; cinnamoyl-CoA:phenyllactate CoA-transferase [EC:2.8.3.17] | 0.902 | 0.687 | 0.191 | 0.970 |
| fldB; flavodoxin II | 0.329 | 0.522 | 0.530 | 0.989 |
| fleQ, flrA; sigma-54 dependent transcriptional regulator, flagellar regulatory protein | 0.144 | 0.267 | 0.591 | 0.999 |
| flgA; flagella basal body P-ring formation protein FlgA | -0.072 | 0.180 | 0.692 | 0.999 |
| flgB; flagellar basal-body rod protein FlgB | -0.189 | 0.153 | 0.216 | 0.978 |
| flgC; flagellar basal-body rod protein FlgC | -0.214 | 0.150 | 0.156 | 0.954 |
| flgD; flagellar basal-body rod modification protein FlgD | -0.185 | 0.155 | 0.236 | 0.978 |
| flgE; flagellar hook protein FlgE | -0.211 | 0.158 | 0.182 | 0.961 |
| flgF; flagellar basal-body rod protein FlgF | -0.105 | 0.211 | 0.618 | 0.999 |
| flgG; flagellar basal-body rod protein FlgG | -0.276 | 0.164 | 0.094 | 0.894 |
| flgH; flagellar L-ring protein precursor FlgH | -0.092 | 0.173 | 0.597 | 0.999 |
| flgI; flagellar P-ring protein precursor FlgI | -0.086 | 0.174 | 0.619 | 0.999 |
| flgJ; flagellar protein FlgJ | 0.028 | 0.194 | 0.884 | 0.999 |
| flgK; flagellar hook-associated protein 1 FlgK | -0.182 | 0.166 | 0.273 | 0.978 |
| flgL; flagellar hook-associated protein 3 FlgL | -0.145 | 0.156 | 0.356 | 0.978 |
| flgM; negative regulator of flagellin synthesis FlgM | -0.047 | 0.175 | 0.791 | 0.999 |
| flgN; flagella synthesis protein FlgN | 0.176 | 0.289 | 0.545 | 0.990 |
| flhA; flagellar biosynthesis protein FlhA | -0.224 | 0.152 | 0.142 | 0.951 |
| flhB; flagellar biosynthetic protein FlhB | -0.191 | 0.151 | 0.209 | 0.977 |
| flhB2; flagellar biosynthesis protein | -0.184 | 0.204 | 0.369 | 0.978 |
| flhC; flagellar transcriptional activator FlhC | -0.134 | 0.398 | 0.737 | 0.999 |
| flhD; flagellar transcriptional activator FlhD | -0.208 | 0.402 | 0.604 | 0.999 |
| flhE; flagellar protein FlhE | 0.298 | 0.612 | 0.627 | 0.999 |
| flhF; flagellar biosynthesis protein FlhF | -0.099 | 0.181 | 0.584 | 0.999 |
| flhG, fleN; flagellar biosynthesis protein FlhG | -0.118 | 0.186 | 0.527 | 0.989 |
| fliA; RNA polymerase sigma factor for flagellar operon FliA | -0.102 | 0.157 | 0.517 | 0.986 |
| fliB; lysine-N-methylase [EC:2.1.1.-] | 0.458 | 0.536 | 0.394 | 0.978 |
| fliC; flagellin | -0.140 | 0.178 | 0.433 | 0.978 |
| fliD; flagellar hook-associated protein 2 | -0.042 | 0.170 | 0.805 | 0.999 |
| fliE; flagellar hook-basal body complex protein FliE | -0.232 | 0.153 | 0.133 | 0.942 |
| fliF; flagellar M-ring protein FliF | -0.168 | 0.153 | 0.274 | 0.978 |
| fliG; flagellar motor switch protein FliG | -0.194 | 0.151 | 0.200 | 0.971 |
| fliH; flagellar assembly protein FliH | -0.120 | 0.160 | 0.454 | 0.978 |
| fliI; flagellum-specific ATP synthase [EC:3.6.3.14] | -0.207 | 0.149 | 0.166 | 0.961 |
| fliJ; flagellar FliJ protein | -0.040 | 0.175 | 0.821 | 0.999 |
| fliK; flagellar hook-length control protein FliK | 0.017 | 0.188 | 0.928 | 0.999 |
| fliL; flagellar FliL protein | -0.069 | 0.173 | 0.690 | 0.999 |
| fliM; flagellar motor switch protein FliM | -0.196 | 0.152 | 0.201 | 0.971 |
| fliNY, fliN; flagellar motor switch protein FliN/FliY | -0.259 | 0.156 | 0.099 | 0.896 |
| fliOZ, fliO; flagellar protein FliO/FliZ | -0.051 | 0.173 | 0.770 | 0.999 |
| fliP; flagellar biosynthetic protein FliP | -0.198 | 0.153 | 0.197 | 0.970 |
| fliQ; flagellar biosynthetic protein FliQ | -0.201 | 0.147 | 0.172 | 0.961 |
| fliR-flhB; flagellar biosynthetic protein FliR/FlhB | -1.706 | 2.114 | 0.421 | 0.978 |
| fliR; flagellar biosynthetic protein FliR | -0.186 | 0.151 | 0.219 | 0.978 |
| fliS; flagellar protein FliS | -0.170 | 0.162 | 0.296 | 0.978 |
| fliT; flagellar protein FliT | -0.580 | 0.392 | 0.141 | 0.949 |
| fliW; flagellar assembly factor FliW | -0.368 | 0.315 | 0.245 | 0.978 |
| fliY; cystine transport system substrate-binding protein | 0.225 | 0.125 | 0.073 | 0.845 |
| fliZ; regulator of sigma S factor FliZ | 0.141 | 0.630 | 0.823 | 0.999 |
| flK; fluoroacetyl-CoA thioesterase [EC:3.1.2.29] | 1.977 | 1.345 | 0.144 | 0.951 |
| FLOT; flotillin | -0.251 | 0.156 | 0.109 | 0.917 |
| flp, pilA; pilus assembly protein Flp/PilA | -0.416 | 0.235 | 0.079 | 0.859 |
| flpA; fibrillarin-like pre-rRNA processing protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| flr, flipr; FPRL1 inhibitory protein | -1.670 | 1.245 | 0.182 | 0.961 |
| flrB, fleS; two-component system, sensor histidine kinase FlrB [EC:2.7.13.3] | 0.233 | 0.327 | 0.477 | 0.979 |
| flrC, fleR; two-component system, response regulator FlrC | 0.117 | 0.334 | 0.727 | 0.999 |
| flu; antigen 43 | -2.027 | 2.120 | 0.341 | 0.978 |
| FMN2; formin 2 | -17.128 | 2919.746 | 0.995 | 0.999 |
| fnbA; fibronectin-binding protein A | -0.042 | 0.532 | 0.937 | 0.999 |
| fnbB; fibronectin-binding protein B | -1.670 | 1.245 | 0.182 | 0.961 |
| fno; 8-hydroxy-5-deazaflavin:NADPH oxidoreductase [EC:1.5.1.40] | -0.173 | 0.341 | 0.613 | 0.999 |
| fnr; CRP/FNR family transcriptional regulator, anaerobic regulatory protein | 0.066 | 0.112 | 0.553 | 0.992 |
| focA; formate transporter | 0.207 | 0.312 | 0.508 | 0.981 |
| focB; formate transporter | -1.153 | 1.472 | 0.435 | 0.978 |
| folB; 7,8-dihydroneopterin aldolase/epimerase/oxygenase [EC:4.1.2.25 5.1.99.8 1.13.11.81] | -0.041 | 0.059 | 0.487 | 0.979 |
| folC; dihydrofolate synthase / folylpolyglutamate synthase [EC:6.3.2.12 6.3.2.17] | 0.035 | 0.053 | 0.512 | 0.982 |
| folD; methylenetetrahydrofolate dehydrogenase (NADP+) / methenyltetrahydrofolate cyclohydrolase [EC:1.5.1.5 3.5.4.9] | 0.009 | 0.030 | 0.757 | 0.999 |
| folE2; GTP cyclohydrolase IB [EC:3.5.4.16] | 0.106 | 0.192 | 0.583 | 0.999 |
| folK; 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase [EC:2.7.6.3] | -0.062 | 0.058 | 0.289 | 0.978 |
| folM; dihydromonapterin reductase / dihydrofolate reductase [EC:1.5.1.50 1.5.1.3] | 0.218 | 0.315 | 0.491 | 0.980 |
| folP; dihydropteroate synthase [EC:2.5.1.15] | -0.016 | 0.043 | 0.713 | 0.999 |
| folX; D-erythro-7,8-dihydroneopterin triphosphate epimerase [EC:5.1.99.7] | 0.077 | 0.342 | 0.821 | 0.999 |
| fosB; metallothiol transferase [EC:2.5.1.-] | -1.331 | 0.715 | 0.065 | 0.839 |
| fpr; ferredoxin/flavodoxin—NADP+ reductase [EC:1.18.1.2 1.19.1.1] | -0.022 | 0.114 | 0.848 | 0.999 |
| frc; formyl-CoA transferase [EC:2.8.3.16] | -0.268 | 0.269 | 0.321 | 0.978 |
| frcA; fructose transport system ATP-binding protein | -1.797 | 0.995 | 0.073 | 0.845 |
| frcB; fructose transport system substrate-binding protein | -1.677 | 0.993 | 0.093 | 0.892 |
| frcC; fructose transport system permease protein | -1.677 | 0.993 | 0.093 | 0.892 |
| frdA; fumarate reductase flavoprotein subunit [EC:1.3.5.4] | 0.302 | 0.226 | 0.185 | 0.966 |
| frdB; fumarate reductase iron-sulfur subunit [EC:1.3.5.4] | 0.054 | 0.288 | 0.852 | 0.999 |
| frdC; fumarate reductase subunit C | 0.054 | 0.288 | 0.852 | 0.999 |
| frdD; fumarate reductase subunit D | 0.211 | 0.304 | 0.488 | 0.979 |
| fre, ubiB; aquacobalamin reductase / NAD(P)H-flavin reductase [EC:1.16.1.3 1.5.1.41] | 0.405 | 0.522 | 0.439 | 0.978 |
| frhB; coenzyme F420 hydrogenase subunit beta [EC:1.12.98.1] | -1.448 | 0.975 | 0.139 | 0.949 |
| frlA; fructoselysine transporter | -1.153 | 1.472 | 0.435 | 0.978 |
| frlB; fructoselysine 6-phosphate deglycase [EC:3.5.-.-] | -1.386 | 1.327 | 0.298 | 0.978 |
| frlC; fructoselysine 3-epimerase [EC:5.1.3.41] | -0.789 | 0.903 | 0.384 | 0.978 |
| frlD; fructoselysine 6-kinase [EC:2.7.1.218] | -1.044 | 0.869 | 0.232 | 0.978 |
| frlR; GntR family transcriptional regulator, frlABCD operon transcriptional regulator | -1.386 | 1.327 | 0.298 | 0.978 |
| frmA, ADH5, adhC; S-(hydroxymethyl)glutathione dehydrogenase / alcohol dehydrogenase [EC:1.1.1.284 1.1.1.1] | -0.352 | 0.201 | 0.082 | 0.865 |
| frmB, ESD, fghA; S-formylglutathione hydrolase [EC:3.1.2.12] | -0.119 | 0.171 | 0.487 | 0.979 |
| frr, MRRF, RRF; ribosome recycling factor | 0.036 | 0.036 | 0.321 | 0.978 |
| frsA; esterase FrsA [EC:3.1.-.-] | -0.015 | 0.590 | 0.980 | 0.999 |
| fruA; fructan beta-fructosidase [EC:3.2.1.80] | -0.195 | 0.161 | 0.227 | 0.978 |
| fruK; 1-phosphofructokinase [EC:2.7.1.56] | 0.093 | 0.067 | 0.170 | 0.961 |
| fruR1, fruR; LacI family transcriptional regulator, fructose operon transcriptional repressor | 0.157 | 0.271 | 0.563 | 0.993 |
| fruR2, fruR; DeoR family transcriptional regulator, fructose operon transcriptional repressor | 0.053 | 0.098 | 0.588 | 0.999 |
| frvR; putative frv operon regulatory protein | -1.153 | 1.472 | 0.435 | 0.978 |
| frvX; putative aminopeptidase FrvX [EC:3.4.11.-] | 0.557 | 0.641 | 0.386 | 0.978 |
| fsaA, mipB; fructose-6-phosphate aldolase 1 [EC:4.1.2.-] | 0.162 | 0.287 | 0.574 | 0.995 |
| fsaB, talC; fructose-6-phosphate aldolase 2 [EC:4.1.2.-] | 0.288 | 0.287 | 0.318 | 0.978 |
| fsr; MFS transporter, FSR family, fosmidomycin resistance protein | -0.057 | 0.130 | 0.661 | 0.999 |
| FTCD; glutamate formiminotransferase / formiminotetrahydrofolate cyclodeaminase [EC:2.1.2.5 4.3.1.4] | -0.035 | 0.286 | 0.903 | 0.999 |
| ftnA, ftn; ferritin [EC:1.16.3.2] | -0.006 | 0.147 | 0.965 | 0.999 |
| ftnB; ferritin-like protein 2 | -0.243 | 0.717 | 0.735 | 0.999 |
| FTR, FTH1, efeU; high-affinity iron transporter | 0.114 | 0.088 | 0.198 | 0.970 |
| ftr; formylmethanofuran–tetrahydromethanopterin N-formyltransferase [EC:2.3.1.101] | 1.390 | 0.984 | 0.160 | 0.954 |
| ftrA; AraC family transcriptional regulator, transcriptional activator FtrA | -0.135 | 0.399 | 0.735 | 0.999 |
| ftrB; CRP/FNR family transcriptional regulator, transcriptional activator FtrB | -2.525 | 1.880 | 0.181 | 0.961 |
| ftsA; cell division protein FtsA | 0.001 | 0.047 | 0.980 | 0.999 |
| ftsB; cell division protein FtsB | 0.124 | 0.143 | 0.386 | 0.978 |
| ftsE; cell division transport system ATP-binding protein | -0.034 | 0.059 | 0.562 | 0.992 |
| ftsH, hflB; cell division protease FtsH [EC:3.4.24.-] | 0.030 | 0.043 | 0.485 | 0.979 |
| ftsI; cell division protein FtsI (penicillin-binding protein 3) [EC:3.4.16.4] | 0.024 | 0.078 | 0.758 | 0.999 |
| ftsK, spoIIIE; DNA segregation ATPase FtsK/SpoIIIE, S-DNA-T family | -0.032 | 0.053 | 0.544 | 0.990 |
| ftsL; cell division protein FtsL | 0.095 | 0.161 | 0.555 | 0.992 |
| ftsN; cell division protein FtsN | 0.194 | 0.314 | 0.537 | 0.990 |
| ftsQ; cell division protein FtsQ | 0.057 | 0.037 | 0.126 | 0.927 |
| ftsW, spoVE; cell division protein FtsW | 0.002 | 0.035 | 0.963 | 0.999 |
| ftsX; cell division transport system permease protein | 0.016 | 0.054 | 0.773 | 0.999 |
| ftsY; fused signal recognition particle receptor | 0.036 | 0.036 | 0.319 | 0.978 |
| ftsZ; cell division protein FtsZ | 0.057 | 0.043 | 0.189 | 0.966 |
| FUCA; alpha-L-fucosidase [EC:3.2.1.51] | -0.197 | 0.148 | 0.185 | 0.966 |
| fucA; L-fuculose-phosphate aldolase [EC:4.1.2.17] | 0.286 | 0.159 | 0.074 | 0.845 |
| fucD; L-fuconate dehydratase [EC:4.2.1.68] | -0.121 | 0.340 | 0.722 | 0.999 |
| fucI; L-fucose/D-arabinose isomerase [EC:5.3.1.25 5.3.1.3] | 0.178 | 0.281 | 0.528 | 0.989 |
| fucK; L-fuculokinase [EC:2.7.1.51] | 0.465 | 0.444 | 0.296 | 0.978 |
| fucO; lactaldehyde reductase [EC:1.1.1.77] | 0.494 | 0.247 | 0.047 | 0.839 |
| fucP; MFS transporter, FHS family, L-fucose permease | -0.070 | 0.122 | 0.567 | 0.995 |
| fucR; DeoR family transcriptional regulator, L-fucose operon activator | -0.643 | 0.591 | 0.278 | 0.978 |
| fucU, FUOM; L-fucose mutarotase [EC:5.1.3.29] | 0.012 | 0.243 | 0.962 | 0.999 |
| FUK; fucokinase [EC:2.7.1.52] | -17.986 | 2523.431 | 0.994 | 0.999 |
| fur, zur, furB; Fur family transcriptional regulator, ferric uptake regulator | -0.019 | 0.067 | 0.779 | 0.999 |
| fusA, GFM, EFG; elongation factor G | 0.056 | 0.051 | 0.278 | 0.978 |
| fwdA, fmdA; formylmethanofuran dehydrogenase subunit A [EC:1.2.7.12] | 1.390 | 0.984 | 0.160 | 0.954 |
| fwdB, fmdB; formylmethanofuran dehydrogenase subunit B [EC:1.2.7.12] | 1.390 | 0.984 | 0.160 | 0.954 |
| fwdC, fmdC; formylmethanofuran dehydrogenase subunit C [EC:1.2.7.12] | 1.390 | 0.984 | 0.160 | 0.954 |
| fwdE, fmdE; formylmethanofuran dehydrogenase subunit E [EC:1.2.7.12] | 1.250 | 0.606 | 0.041 | 0.839 |
| fxsA; UPF0716 protein FxsA | -0.002 | 0.153 | 0.988 | 0.999 |
| fyuA; pesticin/yersiniabactin receptor | -2.044 | 2.163 | 0.346 | 0.978 |
| G6PD, zwf; glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49 1.1.1.363] | -0.014 | 0.060 | 0.814 | 0.999 |
| gabD; succinate-semialdehyde dehydrogenase / glutarate-semialdehyde dehydrogenase [EC:1.2.1.16 1.2.1.79 1.2.1.20] | -0.077 | 0.138 | 0.578 | 0.997 |
| gabP; GABA permease | -0.003 | 0.323 | 0.991 | 0.999 |
| GABRE; gamma-aminobutyric acid receptor subunit epsilon | 1.580 | 1.368 | 0.250 | 0.978 |
| gabT; 4-aminobutyrate aminotransferase / (S)-3-amino-2-methylpropionate transaminase [EC:2.6.1.19 2.6.1.22] | -0.800 | 0.357 | 0.027 | 0.839 |
| gal; D-galactose 1-dehydrogenase [EC:1.1.1.48] | -1.047 | 0.447 | 0.020 | 0.839 |
| galB; 4-oxalomesaconate hydratase [EC:4.2.1.83] | -1.679 | 0.743 | 0.025 | 0.839 |
| galD; 4-oxalomesaconate tautomerase [EC:5.3.2.8] | -1.895 | 0.862 | 0.029 | 0.839 |
| GalDH; L-galactose dehydrogenase [EC:1.1.1.316] | -1.432 | 1.197 | 0.234 | 0.978 |
| galE, GALE; UDP-glucose 4-epimerase [EC:5.1.3.2] | 0.016 | 0.049 | 0.747 | 0.999 |
| galK; galactokinase [EC:2.7.1.6] | 0.098 | 0.080 | 0.219 | 0.978 |
| galM, GALM; aldose 1-epimerase [EC:5.1.3.3] | 0.000 | 0.066 | 0.999 | 1.000 |
| galP; MFS transporter, SP family, galactose:H+ symporter | -0.012 | 0.648 | 0.986 | 0.999 |
| galR; LysR family transcriptional regulator, regulator for genes of the gallate degradation pathway | -0.283 | 0.997 | 0.777 | 0.999 |
| galT, GALT; UDPglucose–hexose-1-phosphate uridylyltransferase [EC:2.7.7.12] | 0.171 | 0.084 | 0.045 | 0.839 |
| ganP; arabinogalactan oligomer / maltooligosaccharide transport system permease protein | -0.066 | 0.134 | 0.622 | 0.999 |
| ganQ; arabinogalactan oligomer / maltooligosaccharide transport system permease protein | -0.025 | 0.122 | 0.840 | 0.999 |
| gap2; glyceraldehyde-3-phosphate dehydrogenase (NAD(P)) [EC:1.2.1.59] | -17.524 | 2799.236 | 0.995 | 0.999 |
| GAPDH, gapA; glyceraldehyde 3-phosphate dehydrogenase [EC:1.2.1.12] | 0.005 | 0.031 | 0.858 | 0.999 |
| gapN; glyceraldehyde-3-phosphate dehydrogenase (NADP+) [EC:1.2.1.9] | 0.050 | 0.142 | 0.724 | 0.999 |
| gar1; RNA-binding protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| garD; galactarate dehydratase [EC:4.2.1.42] | -0.121 | 0.277 | 0.664 | 0.999 |
| garL; 2-dehydro-3-deoxyglucarate aldolase [EC:4.1.2.20] | 0.383 | 0.542 | 0.481 | 0.979 |
| garP; MFS transporter, ACS family, probable galactarate transporter | -1.006 | 1.043 | 0.336 | 0.978 |
| garR, glxR; 2-hydroxy-3-oxopropionate reductase [EC:1.1.1.60] | -0.308 | 0.207 | 0.139 | 0.949 |
| GARS, glyS1; glycyl-tRNA synthetase [EC:6.1.1.14] | -0.193 | 0.124 | 0.122 | 0.919 |
| GART; phosphoribosylamine–glycine ligase / phosphoribosylglycinamide formyltransferase / phosphoribosylformylglycinamidine cyclo-ligase [EC:6.3.4.13 2.1.2.2 6.3.3.1] | -0.878 | 0.664 | 0.188 | 0.966 |
| gatA, QRSL1; aspartyl-tRNA(Asn)/glutamyl-tRNA(Gln) amidotransferase subunit A [EC:6.3.5.6 6.3.5.7] | -0.061 | 0.053 | 0.254 | 0.978 |
| gatB, PET112; aspartyl-tRNA(Asn)/glutamyl-tRNA(Gln) amidotransferase subunit B [EC:6.3.5.6 6.3.5.7] | 0.037 | 0.053 | 0.491 | 0.980 |
| gatC, GATC; aspartyl-tRNA(Asn)/glutamyl-tRNA(Gln) amidotransferase subunit C [EC:6.3.5.6 6.3.5.7] | 0.035 | 0.054 | 0.510 | 0.981 |
| gatD; glutamyl-tRNA(Gln) amidotransferase subunit D [EC:6.3.5.7] | -17.234 | 3077.514 | 0.996 | 0.999 |
| gatE; glutamyl-tRNA(Gln) amidotransferase subunit E [EC:6.3.5.7] | -17.234 | 3077.514 | 0.996 | 0.999 |
| GATM; glycine amidinotransferase [EC:2.1.4.1] | 0.228 | 0.493 | 0.645 | 0.999 |
| gatR; DeoR family transcriptional regulator, galactitol utilization operon repressor | 0.105 | 0.355 | 0.767 | 0.999 |
| gatY-kbaY; tagatose 1,6-diphosphate aldolase GatY/KbaY [EC:4.1.2.40] | -0.205 | 0.487 | 0.675 | 0.999 |
| gatZ-kbaZ; D-tagatose-1,6-bisphosphate aldolase subunit GatZ/KbaZ | -0.179 | 0.364 | 0.625 | 0.999 |
| GBA, srfJ; glucosylceramidase [EC:3.2.1.45] | -0.058 | 0.319 | 0.856 | 0.999 |
| gbcA; glycine betaine catabolism A | 0.016 | 0.304 | 0.957 | 0.999 |
| gbd; 4-hydroxybutyrate dehydrogenase [EC:1.1.1.61] | -17.398 | 2219.101 | 0.994 | 0.999 |
| GBE1, glgB; 1,4-alpha-glucan branching enzyme [EC:2.4.1.18] | 0.032 | 0.071 | 0.652 | 0.999 |
| gbsB; choline dehydrogenase [EC:1.1.1.1] | 18.239 | 2966.075 | 0.995 | 0.999 |
| gbuA; guanidinobutyrase [EC:3.5.3.7] | 0.203 | 0.334 | 0.543 | 0.990 |
| gcd; quinoprotein glucose dehydrogenase [EC:1.1.5.2] | -0.088 | 0.234 | 0.707 | 0.999 |
| gcdA; glutaconyl-CoA decarboxylase [EC:4.1.1.70] | 0.344 | 0.206 | 0.098 | 0.896 |
| GCDH, gcdH; glutaryl-CoA dehydrogenase [EC:1.3.8.6] | -0.297 | 0.204 | 0.147 | 0.954 |
| GCH1, folE; GTP cyclohydrolase IA [EC:3.5.4.16] | 0.014 | 0.042 | 0.737 | 0.999 |
| gch3; GTP cyclohydrolase IIa [EC:3.5.4.29] | -17.234 | 3077.514 | 0.996 | 0.999 |
| gci; D-galactarolactone cycloisomerase [EC:5.5.1.27] | -0.957 | 0.953 | 0.317 | 0.978 |
| gck, gckA, GLYCTK; glycerate 2-kinase [EC:2.7.1.165] | 1.294 | 1.445 | 0.372 | 0.978 |
| gcl; tartronate-semialdehyde synthase [EC:4.1.1.47] | -0.035 | 0.324 | 0.915 | 0.999 |
| gcpE, ispG; (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase [EC:1.17.7.1 1.17.7.3] | -0.011 | 0.060 | 0.855 | 0.999 |
| gcrA; GcrA cell cycle regulator | -1.113 | 0.473 | 0.020 | 0.839 |
| gctA; glutaconate CoA-transferase, subunit A [EC:2.8.3.12] | 0.427 | 0.238 | 0.075 | 0.855 |
| gctB; glutaconate CoA-transferase, subunit B [EC:2.8.3.12] | 0.400 | 0.254 | 0.118 | 0.917 |
| gcvA; LysR family transcriptional regulator, glycine cleavage system transcriptional activator | -0.094 | 0.193 | 0.629 | 0.999 |
| gcvH, GCSH; glycine cleavage system H protein | -0.038 | 0.078 | 0.631 | 0.999 |
| gcvPA; glycine dehydrogenase subunit 1 [EC:1.4.4.2] | -0.331 | 0.201 | 0.102 | 0.899 |
| gcvPB; glycine dehydrogenase subunit 2 [EC:1.4.4.2] | -0.338 | 0.202 | 0.097 | 0.896 |
| gcvR; glycine cleavage system transcriptional repressor | 0.118 | 0.249 | 0.636 | 0.999 |
| gcvT, AMT; aminomethyltransferase [EC:2.1.2.10] | -0.172 | 0.109 | 0.116 | 0.917 |
| gdh; glucose 1-dehydrogenase [EC:1.1.1.47] | -0.523 | 0.390 | 0.182 | 0.961 |
| GDH2; glutamate dehydrogenase [EC:1.4.1.2] | -0.084 | 0.208 | 0.687 | 0.999 |
| gerAA; spore germination protein AA | 18.239 | 2966.075 | 0.995 | 0.999 |
| gerAB; spore germination protein AB | 18.239 | 2966.075 | 0.995 | 0.999 |
| gerAC; spore germination protein AC | 18.239 | 2966.075 | 0.995 | 0.999 |
| gerD; spore germination protein D | -1.426 | 0.741 | 0.056 | 0.839 |
| gerE; LuxR family transcriptional regulator, transcriptional regulator of spore coat protein | -1.431 | 0.741 | 0.055 | 0.839 |
| gerKA; spore germination protein KA | -1.212 | 0.647 | 0.063 | 0.839 |
| gerKB; spore germination protein KB | -1.509 | 0.743 | 0.044 | 0.839 |
| gerKC; spore germination protein KC | -1.320 | 0.819 | 0.109 | 0.917 |
| gerM; germination protein M | -0.183 | 0.404 | 0.651 | 0.999 |
| gerPA; spore germination protein PA | -2.631 | 1.091 | 0.017 | 0.839 |
| gerPB; spore germination protein PB | -2.721 | 1.132 | 0.017 | 0.839 |
| gerPC; spore germination protein PC | -2.721 | 1.132 | 0.017 | 0.839 |
| gerPD; spore germination protein PD | -2.930 | 1.399 | 0.038 | 0.839 |
| gerPE; spore germination protein PE | -2.721 | 1.132 | 0.017 | 0.839 |
| gerPF; spore germination protein PF | -3.219 | 1.604 | 0.047 | 0.839 |
| gerQ; spore germination protein Q | -1.431 | 0.741 | 0.055 | 0.839 |
| gesA, mexP; membrane fusion protein, gold/copper resistance efflux system | 0.835 | 1.158 | 0.472 | 0.979 |
| gesB, mexQ; gold/copper resistance efflux pump | 0.835 | 1.158 | 0.472 | 0.979 |
| gfa; S-(hydroxymethyl)glutathione synthase [EC:4.4.1.22] | -1.027 | 0.720 | 0.156 | 0.954 |
| gfo; glucose-fructose oxidoreductase [EC:1.1.99.28] | -1.424 | 0.662 | 0.033 | 0.839 |
| gfrE; glucoselysine-6-phosphate deglycase | 1.258 | 0.621 | 0.044 | 0.839 |
| gfrF; fructoselysine-6-phosphate deglycase | 1.019 | 0.542 | 0.062 | 0.839 |
| gfrR; sigma-54 dependent transcriptional regulator, gfr operon transcriptional activator | -1.152 | 0.798 | 0.151 | 0.954 |
| GGPS; geranylgeranyl diphosphate synthase, type II [EC:2.5.1.1 2.5.1.10 2.5.1.29] | 0.078 | 0.065 | 0.231 | 0.978 |
| GGR; digeranylgeranylglycerophospholipid reductase [EC:1.3.1.101 1.3.7.11] | -17.234 | 3077.514 | 0.996 | 0.999 |
| ggt; gamma-glutamyltranspeptidase / glutathione hydrolase [EC:2.3.2.2 3.4.19.13] | -0.063 | 0.162 | 0.698 | 0.999 |
| ghrA; glyoxylate/hydroxypyruvatereductase [EC:1.1.1.79 1.1.1.81] | -0.501 | 0.284 | 0.080 | 0.864 |
| ghrB; glyoxylate/hydroxypyruvate/2-ketogluconate reductase [EC:1.1.1.79 1.1.1.81 1.1.1.215] | -0.040 | 0.229 | 0.863 | 0.999 |
| gidA, mnmG, MTO1; tRNA uridine 5-carboxymethylaminomethyl modification enzyme | 0.086 | 0.048 | 0.072 | 0.845 |
| gidB, rsmG; 16S rRNA (guanine527-N7)-methyltransferase [EC:2.1.1.170] | 0.038 | 0.036 | 0.285 | 0.978 |
| ginS; DNA replication factor GINS | -16.927 | 2639.780 | 0.995 | 0.999 |
| glbN; hemoglobin | -0.471 | 0.189 | 0.014 | 0.839 |
| glcA; glycolate permease | -1.002 | 1.362 | 0.463 | 0.979 |
| GLCAK; glucuronokinase [EC:2.7.1.43] | -17.214 | 3047.217 | 0.995 | 0.999 |
| glcC; GntR family transcriptional regulator, glc operon transcriptional activator | 0.589 | 0.361 | 0.104 | 0.907 |
| glcD; glycolate oxidase [EC:1.1.3.15] | -0.020 | 0.192 | 0.919 | 0.999 |
| glcE; glycolate oxidase FAD binding subunit | -0.134 | 0.272 | 0.621 | 0.999 |
| glcF; glycolate oxidase iron-sulfur subunit | -0.312 | 0.250 | 0.214 | 0.978 |
| glcG; glc operon protein GlcG | 0.568 | 0.349 | 0.106 | 0.911 |
| glcP; MFS transporter, FHS family, glucose/mannose:H+ symporter | 18.045 | 1898.825 | 0.992 | 0.999 |
| glcT; transcriptional antiterminator | -0.743 | 0.435 | 0.090 | 0.882 |
| glcU; glucose uptake protein | 0.257 | 0.350 | 0.465 | 0.979 |
| gldA; glycerol dehydrogenase [EC:1.1.1.6] | 0.179 | 0.124 | 0.152 | 0.954 |
| GLDC, gcvP; glycine dehydrogenase [EC:1.4.4.2] | -0.047 | 0.146 | 0.746 | 0.999 |
| glf; UDP-galactopyranose mutase [EC:5.4.99.9] | 0.088 | 0.167 | 0.600 | 0.999 |
| glft1; rhamnopyranosyl-N-acetylglucosaminyl-diphospho-decaprenol beta-1,3/1,4-galactofuranosyltransferase [EC:2.4.1.287] | -0.370 | 0.599 | 0.538 | 0.990 |
| glft2; galactofuranosylgalactofuranosylrhamnosyl-N-acetylglucosaminyl-diphospho-decaprenol beta-1,5/1,6-galactofuranosyltransferase [EC:2.4.1.288] | 0.025 | 0.344 | 0.942 | 0.999 |
| glgA; starch synthase [EC:2.4.1.21] | 0.033 | 0.076 | 0.664 | 0.999 |
| glgC; glucose-1-phosphate adenylyltransferase [EC:2.7.7.27] | 0.072 | 0.103 | 0.482 | 0.979 |
| glgE; starch synthase (maltosyl-transferring) [EC:2.4.99.16] | 0.031 | 0.242 | 0.897 | 0.999 |
| glgM; alpha-maltose-1-phosphate synthase [EC:2.4.1.342] | -0.081 | 0.258 | 0.754 | 0.999 |
| glgX; glycogen debranching enzyme [EC:3.2.1.196] | 0.157 | 0.122 | 0.199 | 0.970 |
| gli; D-galactarolactone isomerase [EC:5.4.1.4] | -0.697 | 1.141 | 0.542 | 0.990 |
| glk; glucokinase [EC:2.7.1.2] | -0.009 | 0.045 | 0.837 | 0.999 |
| glmE, mutE, mamB; methylaspartate mutase epsilon subunit [EC:5.4.99.1] | 0.808 | 0.439 | 0.068 | 0.839 |
| glmM; phosphoglucosamine mutase [EC:5.4.2.10] | 0.067 | 0.037 | 0.073 | 0.845 |
| glmS, GFPT; glucosamine—fructose-6-phosphate aminotransferase (isomerizing) [EC:2.6.1.16] | 0.002 | 0.028 | 0.951 | 0.999 |
| glmS, mutS, mamA; methylaspartate mutase sigma subunit [EC:5.4.99.1] | 0.736 | 0.419 | 0.081 | 0.864 |
| glmU; bifunctional UDP-N-acetylglucosamine pyrophosphorylase / Glucosamine-1-phosphate N-acetyltransferase [EC:2.7.7.23 2.3.1.157] | 0.053 | 0.037 | 0.159 | 0.954 |
| glmU; UDP-N-acetylglucosamine pyrophosphorylase [EC:2.7.7.23] | -16.845 | 2534.319 | 0.995 | 0.999 |
| glnA, GLUL; glutamine synthetase [EC:6.3.1.2] | -0.028 | 0.061 | 0.649 | 0.999 |
| glnB; nitrogen regulatory protein P-II 1 | -0.098 | 0.127 | 0.444 | 0.978 |
| glnD; [protein-PII] uridylyltransferase [EC:2.7.7.59] | -0.065 | 0.136 | 0.632 | 0.999 |
| glnE; [glutamine synthetase] adenylyltransferase / [glutamine synthetase]-adenylyl-L-tyrosine phosphorylase [EC:2.7.7.42 2.7.7.89] | -0.032 | 0.117 | 0.784 | 0.999 |
| glnG, ntrC; two-component system, NtrC family, nitrogen regulation response regulator GlnG | 0.099 | 0.168 | 0.557 | 0.992 |
| glnH; glutamine transport system substrate-binding protein | 0.179 | 0.480 | 0.710 | 0.999 |
| glnK; nitrogen regulatory protein P-II 2 | -0.142 | 0.210 | 0.501 | 0.981 |
| glnL, ntrB; two-component system, NtrC family, nitrogen regulation sensor histidine kinase GlnL [EC:2.7.13.3] | -0.148 | 0.199 | 0.457 | 0.979 |
| glnP; glutamine transport system permease protein | 0.202 | 0.506 | 0.691 | 0.999 |
| glnQ; glutamine transport system ATP-binding protein [EC:3.6.3.-] | 0.185 | 0.429 | 0.668 | 0.999 |
| glnR; MerR family transcriptional regulator, glutamine synthetase repressor | -0.065 | 0.146 | 0.660 | 0.999 |
| glnT; putative sodium/glutamine symporter | -0.904 | 1.547 | 0.560 | 0.992 |
| GLO1, gloA; lactoylglutathione lyase [EC:4.4.1.5] | 0.092 | 0.055 | 0.099 | 0.896 |
| gloB, HAGH; hydroxyacylglutathione hydrolase [EC:3.1.2.6] | 0.081 | 0.123 | 0.510 | 0.981 |
| glpA, glpD; glycerol-3-phosphate dehydrogenase [EC:1.1.5.3] | -0.030 | 0.082 | 0.715 | 0.999 |
| glpB; glycerol-3-phosphate dehydrogenase subunit B [EC:1.1.5.3] | 0.003 | 0.201 | 0.989 | 0.999 |
| glpC; glycerol-3-phosphate dehydrogenase subunit C [EC:1.1.5.3] | -0.001 | 0.201 | 0.997 | 0.999 |
| glpE; thiosulfate sulfurtransferase [EC:2.8.1.1] | 0.204 | 0.210 | 0.334 | 0.978 |
| GLPF; glycerol uptake facilitator protein | 0.084 | 0.090 | 0.354 | 0.978 |
| glpG; GlpG protein | 0.548 | 0.202 | 0.007 | 0.839 |
| glpK, GK; glycerol kinase [EC:2.7.1.30] | 0.009 | 0.035 | 0.806 | 0.999 |
| glpM; membrane protein GlpM | -0.569 | 0.523 | 0.278 | 0.978 |
| glpP; glycerol uptake operon antiterminator | 0.175 | 0.294 | 0.552 | 0.992 |
| glpQ; glycerol transport system permease protein | -0.622 | 0.391 | 0.114 | 0.917 |
| glpR; DeoR family transcriptional regulator, glycerol-3-phosphate regulon repressor | 0.078 | 0.137 | 0.569 | 0.995 |
| glpS; glycerol transport system ATP-binding protein | -0.627 | 0.390 | 0.110 | 0.917 |
| glpT; MFS transporter, OPA family, glycerol-3-phosphate transporter | 0.021 | 0.165 | 0.897 | 0.999 |
| glpV; glycerol transport system substrate-binding protein | -0.622 | 0.391 | 0.114 | 0.917 |
| glpX-SEBP; fructose-1,6-bisphosphatase II / sedoheptulose-1,7-bisphosphatase [EC:3.1.3.11 3.1.3.37] | -1.374 | 0.475 | 0.004 | 0.839 |
| glpX; fructose-1,6-bisphosphatase II [EC:3.1.3.11] | 0.144 | 0.138 | 0.298 | 0.978 |
| glrK, qseE; two-component system, NtrC family, sensor histidine kinase GlrK [EC:2.7.13.3] | -0.864 | 0.596 | 0.149 | 0.954 |
| glrR, qseF; two-component system, NtrC family, response regulator GlrR | -0.789 | 0.616 | 0.202 | 0.972 |
| glsA, GLS; glutaminase [EC:3.5.1.2] | 0.213 | 0.155 | 0.171 | 0.961 |
| gltB; glutamate synthase (NADPH/NADH) large chain [EC:1.4.1.13 1.4.1.14] | -0.255 | 0.149 | 0.089 | 0.882 |
| gltC; LysR family transcriptional regulator, transcription activator of glutamate synthase operon | -1.508 | 0.781 | 0.055 | 0.839 |
| gltD; glutamate synthase (NADPH/NADH) small chain [EC:1.4.1.13 1.4.1.14] | -0.147 | 0.087 | 0.095 | 0.896 |
| gltI, aatJ; glutamate/aspartate transport system substrate-binding protein | -0.291 | 0.279 | 0.299 | 0.978 |
| gltJ, aatQ; glutamate/aspartate transport system permease protein | -0.031 | 0.235 | 0.893 | 0.999 |
| gltK, aatM; glutamate/aspartate transport system permease protein | -0.031 | 0.235 | 0.893 | 0.999 |
| gltL, aatP; glutamate/aspartate transport system ATP-binding protein [EC:3.6.3.-] | -0.008 | 0.234 | 0.973 | 0.999 |
| gltP, gltT; proton glutamate symport protein | -0.172 | 0.166 | 0.302 | 0.978 |
| gltS; glutamate:Na+ symporter, ESS family | 0.352 | 0.156 | 0.025 | 0.839 |
| gltX; nondiscriminating glutamyl-tRNA synthetase [EC:6.1.1.24] | 0.019 | 0.117 | 0.872 | 0.999 |
| gluA; glutamate transport system ATP-binding protein [EC:3.6.3.-] | -0.037 | 0.272 | 0.891 | 0.999 |
| gluB; glutamate transport system substrate-binding protein | -0.011 | 0.268 | 0.966 | 0.999 |
| gluC; glutamate transport system permease protein | -0.027 | 0.277 | 0.921 | 0.999 |
| gluD; glutamate transport system permease protein | -0.030 | 0.270 | 0.912 | 0.999 |
| GLUD1_2, gdhA; glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3] | -0.062 | 0.181 | 0.731 | 0.999 |
| gluP; rhomboid protease GluP [EC:3.4.21.105] | 0.281 | 0.262 | 0.285 | 0.978 |
| gluQ; glutamyl-Q tRNA(Asp) synthetase [EC:6.1.1.-] | -0.104 | 0.168 | 0.536 | 0.990 |
| glvA; maltose-6’-phosphate glucosidase [EC:3.2.1.122] | 0.181 | 0.307 | 0.556 | 0.992 |
| glvR; RpiR family transcriptional regulator, glv operon transcriptional regulator | 0.071 | 0.266 | 0.789 | 0.999 |
| glxK, garK; glycerate 2-kinase [EC:2.7.1.165] | 0.010 | 0.067 | 0.884 | 0.999 |
| glyA, SHMT; glycine hydroxymethyltransferase [EC:2.1.2.1] | 0.016 | 0.034 | 0.650 | 0.999 |
| GLYK; D-glycerate 3-kinase [EC:2.7.1.31] | -0.079 | 0.430 | 0.855 | 0.999 |
| glyQ; glycyl-tRNA synthetase alpha chain [EC:6.1.1.14] | 0.136 | 0.053 | 0.011 | 0.839 |
| glyQS; glycyl-tRNA synthetase [EC:6.1.1.14] | -17.259 | 3117.043 | 0.996 | 0.999 |
| glyS; glycyl-tRNA synthetase beta chain [EC:6.1.1.14] | 0.133 | 0.051 | 0.010 | 0.839 |
| gmd, GMDS; GDPmannose 4,6-dehydratase [EC:4.2.1.47] | -0.285 | 0.145 | 0.051 | 0.839 |
| gmhA, lpcA; D-sedoheptulose 7-phosphate isomerase [EC:5.3.1.28] | 0.293 | 0.119 | 0.015 | 0.839 |
| gmhB; D-glycero-D-manno-heptose 1,7-bisphosphate phosphatase [EC:3.1.3.82 3.1.3.83] | 0.084 | 0.109 | 0.445 | 0.978 |
| gmhC, hldE, waaE, rfaE; D-beta-D-heptose 7-phosphate kinase / D-beta-D-heptose 1-phosphate adenosyltransferase [EC:2.7.1.167 2.7.7.70] | 0.107 | 0.129 | 0.406 | 0.978 |
| gmhD, rfaD; ADP-L-glycero-D-manno-heptose 6-epimerase [EC:5.1.3.20] | 0.278 | 0.150 | 0.065 | 0.839 |
| GMPP; mannose-1-phosphate guanylyltransferase [EC:2.7.7.13] | -0.826 | 0.416 | 0.049 | 0.839 |
| gmr; c-di-GMP phosphodiesterase Gmr [EC:3.1.4.52] | -0.406 | 0.675 | 0.548 | 0.991 |
| gmuG; mannan endo-1,4-beta-mannosidase [EC:3.2.1.78] | -0.693 | 0.417 | 0.099 | 0.896 |
| GNPNAT1, GNA1; glucosamine-phosphate N-acetyltransferase [EC:2.3.1.4] | -17.234 | 3077.514 | 0.996 | 0.999 |
| GNS; N-acetylglucosamine-6-sulfatase [EC:3.1.6.14] | 18.902 | 3402.727 | 0.996 | 0.999 |
| gntP; high-affinity gluconate transporter | -1.158 | 1.491 | 0.439 | 0.978 |
| gntR; LacI family transcriptional regulator, gluconate utilization system Gnt-I transcriptional repressor | -0.192 | 0.282 | 0.498 | 0.981 |
| gntT; Gnt-I system high-affinity gluconate transporter | 0.109 | 0.252 | 0.666 | 0.999 |
| gntU; Gnt-I system low-affinity gluconate transporter | 0.118 | 0.601 | 0.844 | 0.999 |
| golS; MerR family transcriptional regulator, gold-responsive activator of gol and ges genes | -0.141 | 0.415 | 0.735 | 0.999 |
| golT; Au+-exporting ATPase [EC:3.6.1.-] | -0.207 | 0.417 | 0.620 | 0.999 |
| gpgS; glucosyl-3-phosphoglycerate synthase [EC:2.4.1.266] | -0.321 | 0.579 | 0.580 | 0.997 |
| gph; phosphoglycolate phosphatase [EC:3.1.3.18] | -0.056 | 0.057 | 0.328 | 0.978 |
| GPI, pgi; glucose-6-phosphate isomerase [EC:5.3.1.9] | 0.042 | 0.046 | 0.369 | 0.978 |
| gplH; glycopeptidolipid biosynthesis protein | -17.885 | 2468.597 | 0.994 | 0.999 |
| gpmB; probable phosphoglycerate mutase [EC:5.4.2.12] | 0.061 | 0.094 | 0.518 | 0.986 |
| gpmI; 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [EC:5.4.2.12] | -0.094 | 0.099 | 0.343 | 0.978 |
| gppmt; geranyl diphosphate 2-C-methyltransferase [EC:2.1.1.255] | -2.224 | 2.043 | 0.278 | 0.978 |
| gpr; L-glyceraldehyde 3-phosphate reductase [EC:1.1.1.-] | -0.149 | 0.213 | 0.486 | 0.979 |
| gpr; spore protease [EC:3.4.24.78] | -0.960 | 0.501 | 0.057 | 0.839 |
| gpsA; glycerol-3-phosphate dehydrogenase (NAD(P)+) [EC:1.1.1.94] | 0.033 | 0.036 | 0.356 | 0.978 |
| gpt; xanthine phosphoribosyltransferase [EC:2.4.2.22] | -0.141 | 0.251 | 0.576 | 0.996 |
| gpuA; guanidinopropionase [EC:3.5.3.17] | -2.860 | 2.236 | 0.203 | 0.972 |
| gpx; glutathione peroxidase [EC:1.11.1.9] | -0.005 | 0.066 | 0.938 | 0.999 |
| graR; two-component system, OmpR family, response regulator protein GraR | -0.811 | 1.001 | 0.419 | 0.978 |
| graS; two-component system, OmpR family, sensor histidine kinase GraS [EC:2.7.13.3] | -1.554 | 1.110 | 0.164 | 0.957 |
| GRC3, NOL9; polynucleotide 5’-hydroxyl-kinase GRC3/NOL9 [EC:2.7.1.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| grcA; autonomous glycyl radical cofactor | 0.210 | 0.304 | 0.490 | 0.979 |
| grdA; glycine/sarcosine/betaine reductase complex component A [EC:1.21.4.2 1.21.4.3 1.21.4.4] | 0.469 | 0.260 | 0.073 | 0.845 |
| grdB; glycine reductase complex component B subunit gamma [EC:1.21.4.2] | 0.136 | 0.448 | 0.762 | 0.999 |
| greA; transcription elongation factor GreA | 0.013 | 0.055 | 0.815 | 0.999 |
| greB; transcription elongation factor GreB | 0.044 | 0.153 | 0.774 | 0.999 |
| groEL, HSPD1; chaperonin GroEL | 0.026 | 0.038 | 0.502 | 0.981 |
| groES, HSPE1; chaperonin GroES | 0.030 | 0.035 | 0.388 | 0.978 |
| GRPE; molecular chaperone GrpE | 0.036 | 0.036 | 0.313 | 0.978 |
| grxA; glutaredoxin 1 | 0.207 | 0.312 | 0.508 | 0.981 |
| grxB; glutaredoxin 2 | 0.414 | 0.398 | 0.300 | 0.978 |
| grxC, GLRX, GLRX2; glutaredoxin 3 | -0.158 | 0.175 | 0.366 | 0.978 |
| grxD, GLRX5; monothiol glutaredoxin | -0.040 | 0.163 | 0.807 | 0.999 |
| gshA, ybdK; glutamate—cysteine ligase / carboxylate-amine ligase [EC:6.3.2.2 6.3.-.-] | -0.059 | 0.230 | 0.797 | 0.999 |
| gshA; glutamate–cysteine ligase [EC:6.3.2.2] | 0.134 | 0.112 | 0.234 | 0.978 |
| gshB; glutathione synthase [EC:6.3.2.3] | -0.085 | 0.172 | 0.619 | 0.999 |
| gsiA; glutathione transport system ATP-binding protein | -0.466 | 0.496 | 0.349 | 0.978 |
| gsiB; glutathione transport system substrate-binding protein | -0.357 | 0.703 | 0.612 | 0.999 |
| gsiC; glutathione transport system permease protein | -0.410 | 0.656 | 0.533 | 0.990 |
| gsiD; glutathione transport system permease protein | -0.527 | 0.667 | 0.431 | 0.978 |
| gsk; inosine kinase [EC:2.7.1.73] | 0.418 | 0.520 | 0.423 | 0.978 |
| gsmt; glycine/sarcosine N-methyltransferase [EC:2.1.1.156] | -0.937 | 0.976 | 0.338 | 0.978 |
| gsp; glutathionylspermidine amidase/synthetase [EC:3.5.1.78 6.3.1.8] | 0.777 | 0.390 | 0.048 | 0.839 |
| GSP13; general stress protein 13 | -0.008 | 0.134 | 0.952 | 0.999 |
| gspA; general secretion pathway protein A | -0.398 | 0.554 | 0.473 | 0.979 |
| gspB; general secretion pathway protein B | -0.443 | 0.632 | 0.484 | 0.979 |
| gspC; general secretion pathway protein C | -0.115 | 0.225 | 0.611 | 0.999 |
| gspD; general secretion pathway protein D | 0.063 | 0.144 | 0.663 | 0.999 |
| gspE; general secretion pathway protein E | 0.063 | 0.142 | 0.659 | 0.999 |
| gspF; general secretion pathway protein F | 0.074 | 0.173 | 0.669 | 0.999 |
| gspG; general secretion pathway protein G | 0.113 | 0.158 | 0.475 | 0.979 |
| gspH; general secretion pathway protein H | -0.148 | 0.264 | 0.575 | 0.995 |
| gspI; general secretion pathway protein I | -0.162 | 0.239 | 0.500 | 0.981 |
| gspJ; general secretion pathway protein J | -0.144 | 0.238 | 0.545 | 0.990 |
| gspK; general secretion pathway protein K | -0.145 | 0.238 | 0.543 | 0.990 |
| gspK; glucosamine kinase [EC:2.7.1.8] | -1.341 | 0.612 | 0.030 | 0.839 |
| gspL; general secretion pathway protein L | -0.149 | 0.240 | 0.535 | 0.990 |
| gspM; general secretion pathway protein M | -0.208 | 0.256 | 0.417 | 0.978 |
| gspN; general secretion pathway protein N | -0.144 | 0.247 | 0.560 | 0.992 |
| gspO; general secretion pathway protein O [EC:3.4.23.43 2.1.1.-] | -0.089 | 0.796 | 0.911 | 0.999 |
| gspS; general secretion pathway protein S | 0.920 | 1.213 | 0.450 | 0.978 |
| GSR, gor; glutathione reductase (NADPH) [EC:1.8.1.7] | 0.110 | 0.093 | 0.235 | 0.978 |
| GST, gst; glutathione S-transferase [EC:2.5.1.18] | -0.179 | 0.194 | 0.358 | 0.978 |
| gtfB, gtfE; vancomycin aglycone glucosyltransferase [EC:2.4.1.310] | 0.277 | 0.884 | 0.754 | 0.999 |
| gtsA, glcE; glucose/mannose transport system substrate-binding protein | -0.263 | 0.265 | 0.322 | 0.978 |
| gtsB, glcF; glucose/mannose transport system permease protein | -0.315 | 0.272 | 0.248 | 0.978 |
| gtsC, glcG; glucose/mannose transport system permease protein | -0.332 | 0.269 | 0.220 | 0.978 |
| guaA, GMPS; GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2] | 0.004 | 0.033 | 0.896 | 0.999 |
| gudB, rocG; glutamate dehydrogenase [EC:1.4.1.2] | 0.076 | 0.165 | 0.645 | 0.999 |
| gudD; glucarate dehydratase [EC:4.2.1.40] | -0.074 | 0.286 | 0.795 | 0.999 |
| gudP; MFS transporter, ACS family, glucarate transporter | 0.069 | 0.261 | 0.791 | 0.999 |
| gudX; glucarate dehydratase-related protein | 0.176 | 0.678 | 0.796 | 0.999 |
| GULO; L-gulonolactone oxidase [EC:1.1.3.8] | -1.121 | 1.071 | 0.297 | 0.978 |
| gumC; GumC protein | -17.687 | 2769.090 | 0.995 | 0.999 |
| gumD; undecaprenyl-phosphate glucose phosphotransferase [EC:2.7.8.31] | -17.687 | 2769.090 | 0.995 | 0.999 |
| gumE; putative polymerase | -0.583 | 0.874 | 0.506 | 0.981 |
| gumF; acyltransferase [EC:2.3.1.-] | -17.687 | 2769.090 | 0.995 | 0.999 |
| gumG; acyltransferase [EC:2.3.1.-] | -17.175 | 2988.990 | 0.995 | 0.999 |
| gumH, aceA; alpha-1,3-mannosyltransferase [EC:2.4.1.252] | -0.112 | 0.728 | 0.878 | 0.999 |
| gumI; beta-1,4-mannosyltransferase [EC:2.4.1.251] | -17.816 | 2636.156 | 0.995 | 0.999 |
| gumK; 2-beta-glucuronyltransferase [EC:2.4.1.264] | 0.482 | 0.901 | 0.594 | 0.999 |
| gumL; pyruvyltransferase | -2.368 | 1.742 | 0.176 | 0.961 |
| gumM; beta-1,4-glucosyltransferase [EC:2.4.1.-] | -17.687 | 2769.090 | 0.995 | 0.999 |
| gutA, gutP; probable glucitol transport protein GutA | -2.015 | 1.481 | 0.176 | 0.961 |
| gutM; glucitol operon activator protein | 0.118 | 0.597 | 0.844 | 0.999 |
| gutQ; arabinose 5-phosphate isomerase [EC:5.3.1.13] | 0.161 | 0.632 | 0.799 | 0.999 |
| gyaR, GOR1; glyoxylate reductase [EC:1.1.1.26] | -0.283 | 0.208 | 0.175 | 0.961 |
| gyrA; DNA gyrase subunit A [EC:5.99.1.3] | 0.039 | 0.041 | 0.340 | 0.978 |
| gyrB; DNA gyrase subunit B [EC:5.99.1.3] | 0.039 | 0.044 | 0.381 | 0.978 |
| GYS; glycogen synthase [EC:2.4.1.11] | -17.214 | 3047.217 | 0.995 | 0.999 |
| HAAO; 3-hydroxyanthranilate 3,4-dioxygenase [EC:1.13.11.6] | -0.162 | 0.441 | 0.714 | 0.999 |
| hap, nprV; vibriolysin [EC:3.4.24.25] | -2.721 | 2.400 | 0.259 | 0.978 |
| hapE; 4-hydroxyacetophenone monooxygenase [EC:1.14.13.84] | -1.946 | 0.836 | 0.021 | 0.839 |
| hapR, luxR, litR; TetR/AcrR family transcriptional regulator, hemagglutinin/protease regulatory protein | 18.202 | 1851.167 | 0.992 | 0.999 |
| HARS, hisS; histidyl-tRNA synthetase [EC:6.1.1.21] | 0.028 | 0.035 | 0.434 | 0.978 |
| hasA; heme acquisition protein HasA | -0.500 | 0.646 | 0.440 | 0.978 |
| hasA; hyaluronan synthase [EC:2.4.1.212] | 0.229 | 0.562 | 0.684 | 0.999 |
| hasD, prtD, aprD; ATP-binding cassette, subfamily C, bacterial exporter for protease/lipase | -0.329 | 0.460 | 0.476 | 0.979 |
| hasE, prtE; membrane fusion protein, protease secretion system | -0.280 | 0.502 | 0.578 | 0.997 |
| hasF, prtF; outer membrane protein, protease secretion system | -0.256 | 0.524 | 0.626 | 0.999 |
| hasR; heme acquisition protein HasR | -0.372 | 0.640 | 0.562 | 0.992 |
| hbaA; 4-hydroxybenzoate-CoA ligase [EC:6.2.1.27 6.2.1.25] | -1.407 | 1.166 | 0.229 | 0.978 |
| hbhA; heparin binding hemagglutinin HbhA | -0.843 | 0.908 | 0.355 | 0.978 |
| hblAB; hemolysin BL binding component | -2.586 | 2.420 | 0.287 | 0.978 |
| hblC; hemolysin BL lytic component L2 | -2.767 | 2.698 | 0.307 | 0.978 |
| hblD; hemolysin BL lytic component L1 | -2.767 | 2.698 | 0.307 | 0.978 |
| hcaB; 2,3-dihydroxy-2,3-dihydrophenylpropionate dehydrogenase [EC:1.3.1.87] | -0.934 | 0.927 | 0.316 | 0.978 |
| hcaC; 3-phenylpropionate/trans-cinnamate dioxygenase ferredoxin component | -0.364 | 0.297 | 0.223 | 0.978 |
| hcaD; 3-phenylpropionate/trans-cinnamate dioxygenase ferredoxin reductase component [EC:1.18.1.3] | -0.890 | 0.300 | 0.003 | 0.839 |
| hcaE, hcaA1; 3-phenylpropionate/trans-cinnamate dioxygenase subunit alpha [EC:1.14.12.19] | -0.934 | 0.927 | 0.316 | 0.978 |
| hcaF, hcaA2; 3-phenylpropionate/trans-cinnamate dioxygenase subunit beta [EC:1.14.12.19] | -0.934 | 0.927 | 0.316 | 0.978 |
| hcaR; LysR family transcriptional regulator, hca operon transcriptional activator | -0.417 | 0.482 | 0.388 | 0.978 |
| hcaT; MFS transporter, PPP family, 3-phenylpropionic acid transporter | 0.021 | 0.142 | 0.883 | 0.999 |
| hchA; D-lactate dehydratase / protein deglycase [EC:4.2.1.130 3.5.1.124] | -0.287 | 0.298 | 0.338 | 0.978 |
| hcnA; hydrogen cyanide synthase HcnA [EC:1.4.99.5] | -0.121 | 0.577 | 0.834 | 0.999 |
| hcnB; hydrogen cyanide synthase HcnB [EC:1.4.99.5] | -0.121 | 0.577 | 0.834 | 0.999 |
| hcnC; hydrogen cyanide synthase HcnC [EC:1.4.99.5] | -0.131 | 0.627 | 0.835 | 0.999 |
| hcp; hydroxylamine reductase [EC:1.7.99.1] | 0.210 | 0.176 | 0.233 | 0.978 |
| hcp; type VI secretion system secreted protein Hcp | 0.231 | 0.249 | 0.355 | 0.978 |
| hcr; NADH oxidoreductase Hcr [EC:1.-.-.-] | 0.147 | 0.599 | 0.807 | 0.999 |
| hcrB, hbaD; 4-hydroxybenzoyl-CoA reductase subunit beta [EC:1.3.7.9] | 18.563 | 3486.631 | 0.996 | 0.999 |
| hda; DnaA-homolog protein | 0.064 | 0.156 | 0.683 | 0.999 |
| hdc, HDC; histidine decarboxylase [EC:4.1.1.22] | 0.616 | 1.082 | 0.570 | 0.995 |
| hddA; D-glycero-alpha-D-manno-heptose-7-phosphate kinase [EC:2.7.1.168] | 0.607 | 0.465 | 0.194 | 0.970 |
| hddC; D-glycero-alpha-D-manno-heptose 1-phosphate guanylyltransferase [EC:2.7.7.71] | -0.613 | 0.543 | 0.260 | 0.978 |
| hdeA; acid stress chaperone HdeA | -1.965 | 1.067 | 0.067 | 0.839 |
| hdeB; acid stress chaperone HdeB | -1.186 | 0.785 | 0.133 | 0.942 |
| hdhA; 7-alpha-hydroxysteroid dehydrogenase [EC:1.1.1.159] | -0.491 | 0.458 | 0.285 | 0.978 |
| HDHD1; pseudouridine 5’-phosphatase [EC:3.1.3.96] | -0.189 | 0.933 | 0.840 | 0.999 |
| hdrA2; heterodisulfide reductase subunit A2 [EC:1.8.7.3 1.8.98.4 1.8.98.5 1.8.98.6] | -0.100 | 0.477 | 0.834 | 0.999 |
| hdrB2; heterodisulfide reductase subunit B2 [EC:1.8.7.3 1.8.98.4 1.8.98.5 1.8.98.6] | 0.032 | 0.479 | 0.946 | 0.999 |
| hdrC2; heterodisulfide reductase subunit C2 [EC:1.8.7.3 1.8.98.4 1.8.98.5 1.8.98.6] | 0.058 | 0.485 | 0.905 | 0.999 |
| helD; DNA helicase IV [EC:3.6.4.12] | 0.008 | 0.384 | 0.984 | 0.999 |
| helS; helicase [EC:3.6.4.-] | 0.446 | 1.410 | 0.752 | 0.999 |
| helY; ATP-dependent RNA helicase HelY [EC:3.6.4.-] | 0.038 | 0.249 | 0.878 | 0.999 |
| hemA; glutamyl-tRNA reductase [EC:1.2.1.70] | 0.125 | 0.070 | 0.079 | 0.859 |
| hemAT; heam-based aerotactic trancducer | -0.817 | 1.168 | 0.485 | 0.979 |
| hemB, ALAD; porphobilinogen synthase [EC:4.2.1.24] | 0.060 | 0.067 | 0.373 | 0.978 |
| hemC, HMBS; hydroxymethylbilane synthase [EC:2.5.1.61] | 0.066 | 0.067 | 0.322 | 0.978 |
| hemD, UROS; uroporphyrinogen-III synthase [EC:4.2.1.75] | -0.195 | 0.088 | 0.027 | 0.839 |
| hemDX; uroporphyrinogen III methyltransferase / synthase [EC:2.1.1.107 4.2.1.75] | -0.151 | 0.718 | 0.834 | 0.999 |
| hemE, UROD; uroporphyrinogen decarboxylase [EC:4.1.1.37] | -0.121 | 0.075 | 0.111 | 0.917 |
| hemG; menaquinone-dependent protoporphyrinogen oxidase [EC:1.3.5.3] | 0.149 | 0.261 | 0.569 | 0.995 |
| hemH, FECH; protoporphyrin/coproporphyrin ferrochelatase [EC:4.99.1.1 4.99.1.9] | -0.090 | 0.070 | 0.204 | 0.972 |
| hemK, prmC, HEMK; release factor glutamine methyltransferase [EC:2.1.1.297] | 0.012 | 0.033 | 0.706 | 0.999 |
| hemL; glutamate-1-semialdehyde 2,1-aminomutase [EC:5.4.3.8] | 0.045 | 0.074 | 0.545 | 0.990 |
| hemN, hemZ; oxygen-independent coproporphyrinogen III oxidase [EC:1.3.98.3] | 0.068 | 0.053 | 0.199 | 0.970 |
| hemQ; Fe-coproporphyrin III decarboxylase [EC:1.11.1.-] | 1.758 | 1.259 | 0.165 | 0.960 |
| hemX; HemX protein | -0.865 | 0.546 | 0.115 | 0.917 |
| hemX; uroporphyrin-III C-methyltransferase [EC:2.1.1.107] | 0.128 | 0.171 | 0.454 | 0.978 |
| hemY; HemY protein | 0.009 | 0.162 | 0.957 | 0.999 |
| hepA; ATP-dependent helicase HepA [EC:3.6.4.-] | 0.125 | 0.205 | 0.541 | 0.990 |
| hepA; heparin lyase [EC:4.2.2.7] | -0.768 | 1.066 | 0.473 | 0.979 |
| hepB; heparin/heparan-sulfate lyase [EC:4.2.2.7 4.2.2.8] | -0.468 | 1.020 | 0.647 | 0.999 |
| hepC; heparan-sulfate lyase [EC:4.2.2.8] | -0.748 | 1.071 | 0.486 | 0.979 |
| hepST; heptaprenyl diphosphate synthase [EC:2.5.1.30] | 0.277 | 0.126 | 0.029 | 0.839 |
| HEXA_B; hexosaminidase [EC:3.2.1.52] | -0.103 | 0.128 | 0.424 | 0.978 |
| hexR; RpiR family transcriptional regulator, carbohydrate utilization regulator | 0.178 | 0.257 | 0.489 | 0.979 |
| hfaA; holdfast attachment protein HfaA | -0.550 | 0.876 | 0.531 | 0.990 |
| hfaB; holdfast attachment protein HfaB | -0.550 | 0.876 | 0.531 | 0.990 |
| hflC; membrane protease subunit HflC [EC:3.4.-.-] | -0.129 | 0.158 | 0.417 | 0.978 |
| hflD; high frequency lysogenization protein | 0.095 | 0.180 | 0.598 | 0.999 |
| hflK; membrane protease subunit HflK [EC:3.4.-.-] | -0.101 | 0.169 | 0.552 | 0.992 |
| hflX; GTPase | 0.029 | 0.037 | 0.436 | 0.978 |
| hfq; host factor-I protein | -0.095 | 0.116 | 0.415 | 0.978 |
| HGD, hmgA; homogentisate 1,2-dioxygenase [EC:1.13.11.5] | -0.117 | 0.213 | 0.584 | 0.999 |
| hha; haemolysin expression modulating protein | -0.095 | 0.626 | 0.879 | 0.999 |
| hhoB, degS; serine protease DegS [EC:3.4.21.-] | 0.144 | 0.191 | 0.451 | 0.978 |
| HIBCH; 3-hydroxyisobutyryl-CoA hydrolase [EC:3.1.2.4] | -0.898 | 0.984 | 0.363 | 0.978 |
| hicA; mRNA interferase HicA [EC:3.1.-.-] | 0.007 | 0.554 | 0.990 | 0.999 |
| hicB; antitoxin HicB | -0.795 | 0.654 | 0.226 | 0.978 |
| higA; HTH-type transcriptional regulator / antitoxin HigA | -0.266 | 0.269 | 0.325 | 0.978 |
| higB-1; toxin HigB-1 | -0.013 | 0.182 | 0.943 | 0.999 |
| higB; mRNA interferase HigB [EC:3.1.-.-] | -0.566 | 0.528 | 0.286 | 0.978 |
| HINT1, hinT, hit; histidine triad (HIT) family protein | 0.032 | 0.045 | 0.475 | 0.979 |
| hipA; serine/threonine-protein kinase HipA [EC:2.7.11.1] | -0.075 | 0.146 | 0.608 | 0.999 |
| hipB; HTH-type transcriptional regulator / antitoxin HipB | 0.384 | 0.640 | 0.550 | 0.992 |
| hipO; hippurate hydrolase [EC:3.5.1.32] | -0.393 | 0.236 | 0.098 | 0.896 |
| hisA; phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase [EC:5.3.1.16] | -0.060 | 0.057 | 0.289 | 0.978 |
| hisB; imidazoleglycerol-phosphate dehydratase / histidinol-phosphatase [EC:4.2.1.19 3.1.3.15] | -0.005 | 0.208 | 0.981 | 0.999 |
| hisB; imidazoleglycerol-phosphate dehydratase [EC:4.2.1.19] | -0.117 | 0.074 | 0.115 | 0.917 |
| hisC; histidinol-phosphate aminotransferase [EC:2.6.1.9] | -0.124 | 0.057 | 0.032 | 0.839 |
| hisD; histidinol dehydrogenase [EC:1.1.1.23] | -0.098 | 0.064 | 0.126 | 0.927 |
| hisE; phosphoribosyl-ATP pyrophosphohydrolase [EC:3.6.1.31] | -0.132 | 0.115 | 0.254 | 0.978 |
| hisF; cyclase [EC:4.1.3.-] | -0.094 | 0.065 | 0.153 | 0.954 |
| hisG; ATP phosphoribosyltransferase [EC:2.4.2.17] | -0.089 | 0.059 | 0.134 | 0.942 |
| hisH; glutamine amidotransferase [EC:2.4.2.-] | -0.080 | 0.062 | 0.193 | 0.970 |
| hisI; phosphoribosyl-AMP cyclohydrolase [EC:3.5.4.19] | -0.159 | 0.120 | 0.187 | 0.966 |
| hisIE; phosphoribosyl-ATP pyrophosphohydrolase / phosphoribosyl-AMP cyclohydrolase [EC:3.6.1.31 3.5.4.19] | -0.084 | 0.137 | 0.539 | 0.990 |
| hisJ; histidine transport system substrate-binding protein | -0.336 | 0.459 | 0.466 | 0.979 |
| hisM; histidine transport system permease protein | -0.276 | 0.431 | 0.522 | 0.988 |
| hisN; histidinol-phosphatase [EC:3.1.3.15] | 0.065 | 0.272 | 0.813 | 0.999 |
| hisP; histidine transport system ATP-binding protein [EC:3.6.3.21] | -0.291 | 0.427 | 0.496 | 0.981 |
| hisQ; histidine transport system permease protein | -0.263 | 0.425 | 0.536 | 0.990 |
| hisZ; ATP phosphoribosyltransferase regulatory subunit | -0.116 | 0.088 | 0.190 | 0.969 |
| hjr; holliday junction resolvase Hjr [EC:3.1.22.4] | -17.234 | 3077.514 | 0.996 | 0.999 |
| HK; hexokinase [EC:2.7.1.1] | 0.302 | 0.487 | 0.536 | 0.990 |
| hld; delta-hemolysin | -1.670 | 1.245 | 0.182 | 0.961 |
| hlg, luk; leukocidin/hemolysin toxin family protein | -1.670 | 1.245 | 0.182 | 0.961 |
| hlpA, ompH; outer membrane protein | -0.001 | 0.142 | 0.996 | 0.999 |
| hlyB, cyaB; ATP-binding cassette, subfamily B, bacterial HlyB/CyaB | -0.437 | 0.445 | 0.328 | 0.978 |
| hlyD, cyaD; hemolysin D | -0.513 | 0.480 | 0.287 | 0.978 |
| hlyII; hemolysin II | -1.658 | 1.181 | 0.162 | 0.957 |
| hlyIII; hemolysin III | 0.042 | 0.052 | 0.415 | 0.978 |
| hmfA; 2-furoyl-CoA dehydrogenase large subunit [EC:1.3.99.8] | 0.995 | 0.993 | 0.318 | 0.978 |
| hmfB; 2-furoyl-CoA dehydrogenase FAD binding subunit [EC:1.3.99.8] | 0.995 | 0.993 | 0.318 | 0.978 |
| hmfC; 2-furoyl-CoA dehydrogenase 2Fe-2S iron sulfur subunit [EC:1.3.99.8] | 0.995 | 0.993 | 0.318 | 0.978 |
| hmfD; 2-furoate—CoA ligase [EC:6.2.1.31] | 0.995 | 0.993 | 0.318 | 0.978 |
| hmfE; 2-oxoglutaroyl-CoA hydrolase | 0.995 | 0.993 | 0.318 | 0.978 |
| hmfF; 2,5-furandicarboxylate decarboxylase 1 | 2.635 | 1.537 | 0.088 | 0.880 |
| hmfG; 2,5-furandicarboxylate decarboxylase 2 | -2.174 | 2.005 | 0.280 | 0.978 |
| hmfH; 5-(hydroxymethyl)furfural/furfural oxidase [EC:1.1.3.47 1.1.3.-] | -0.553 | 0.830 | 0.507 | 0.981 |
| HMGCR; hydroxymethylglutaryl-CoA reductase (NADPH) [EC:1.1.1.34] | -0.732 | 0.804 | 0.364 | 0.978 |
| hmo, nocN; 4-hydroxymandelate oxidase [EC:1.1.3.46] | -0.360 | 0.791 | 0.650 | 0.999 |
| HMOX1; heme oxygenase 1 [EC:1.14.14.18] | 0.179 | 0.358 | 0.618 | 0.999 |
| hmp, YHB1; nitric oxide dioxygenase [EC:1.14.12.17] | -0.156 | 0.173 | 0.368 | 0.978 |
| hmuS; putative hemin transport protein | -0.194 | 0.442 | 0.662 | 0.999 |
| hndA; NADP-reducing hydrogenase subunit HndA [EC:1.12.1.3] | -0.496 | 0.544 | 0.363 | 0.978 |
| hndB; NADP-reducing hydrogenase subunit HndB [EC:1.12.1.3] | -0.553 | 0.722 | 0.445 | 0.978 |
| hndC; NADP-reducing hydrogenase subunit HndC [EC:1.12.1.3] | -0.336 | 0.489 | 0.492 | 0.981 |
| hndD; NADP-reducing hydrogenase subunit HndD [EC:1.12.1.3] | 0.173 | 0.451 | 0.703 | 0.999 |
| hns; DNA-binding protein H-NS | -0.107 | 0.248 | 0.668 | 0.999 |
| hofB; protein transport protein HofB | 0.193 | 0.314 | 0.539 | 0.990 |
| hofC; protein transport protein HofC | 0.193 | 0.314 | 0.539 | 0.990 |
| hofD, hopD; leader peptidase HopD [EC:3.4.23.43] | 0.211 | 0.345 | 0.542 | 0.990 |
| hofM; pilus assembly protein HofM | 0.119 | 0.601 | 0.843 | 0.999 |
| hofN; pilus assembly protein HofN | 0.119 | 0.601 | 0.843 | 0.999 |
| hofO; pilus assembly protein HofO | -0.353 | 0.718 | 0.623 | 0.999 |
| hofP; pilus assembly protein HofP | 0.207 | 0.679 | 0.761 | 0.999 |
| hofQ; protein transport protein HofQ | 0.164 | 0.213 | 0.442 | 0.978 |
| HOGA1; 4-hydroxy-2-oxoglutarate aldolase [EC:4.1.3.16] | 1.551 | 1.188 | 0.194 | 0.970 |
| hokA; protein HokA | -1.266 | 1.272 | 0.321 | 0.978 |
| hokB; protein HokB | -1.237 | 1.233 | 0.317 | 0.978 |
| hokC_D; protein HokC/D | -1.662 | 1.552 | 0.286 | 0.978 |
| hokE; protein HokE | -1.001 | 1.258 | 0.428 | 0.978 |
| holA; DNA polymerase III subunit delta [EC:2.7.7.7] | 0.035 | 0.036 | 0.331 | 0.978 |
| holB; DNA polymerase III subunit delta’ [EC:2.7.7.7] | 0.040 | 0.036 | 0.262 | 0.978 |
| holC; DNA polymerase III subunit chi [EC:2.7.7.7] | -0.045 | 0.146 | 0.759 | 0.999 |
| holD; DNA polymerase III subunit psi [EC:2.7.7.7] | 0.237 | 0.307 | 0.441 | 0.978 |
| holE; DNA polymerase III subunit theta [EC:2.7.7.7] | 0.260 | 0.598 | 0.664 | 0.999 |
| hopB, alpB; outer membrane protein HopB/AlpB | -1.386 | 1.551 | 0.373 | 0.978 |
| hopC, alpA; outer membrane protein HopC/AlpA | -1.386 | 1.551 | 0.373 | 0.978 |
| hopM1; effector protein HopM1 | 18.932 | 3654.080 | 0.996 | 0.999 |
| hopZ; outer membrane protein HopZ | -1.386 | 1.551 | 0.373 | 0.978 |
| hoxF; [NiFe] hydrogenase diaphorase moiety large subunit [EC:1.12.1.2] | -0.152 | 0.837 | 0.856 | 0.999 |
| hoxH; NAD-reducing hydrogenase large subunit [EC:1.12.1.2] | -0.577 | 0.807 | 0.476 | 0.979 |
| hoxU; [NiFe] hydrogenase diaphorase moiety small subunit [EC:1.12.1.2] | -0.013 | 0.884 | 0.988 | 0.999 |
| hoxY; NAD-reducing hydrogenase small subunit [EC:1.12.1.2] | -0.445 | 0.851 | 0.602 | 0.999 |
| hpa1; type III secretion harpin protein Hpa1 | -17.175 | 2988.990 | 0.995 | 0.999 |
| hpa2; lysozyme-related protein Hpa2 | -17.175 | 2988.990 | 0.995 | 0.999 |
| hpaA; AraC family transcriptional regulator, 4-hydroxyphenylacetate 3-monooxygenase operon regulatory protein | -0.305 | 0.541 | 0.573 | 0.995 |
| hpaA; neuraminyllactose-binding hemagglutinin | -1.386 | 1.551 | 0.373 | 0.978 |
| hpaA; type III secretion regulatory protein HpaA | -17.175 | 2988.990 | 0.995 | 0.999 |
| hpaB; 4-hydroxyphenylacetate 3-monooxygenase [EC:1.14.14.9] | -0.832 | 0.570 | 0.146 | 0.954 |
| hpaB; type III secretion control protein HpaB | -17.175 | 2988.990 | 0.995 | 0.999 |
| hpaC; flavin reductase (NADH) [EC:1.5.1.36] | 0.453 | 0.393 | 0.250 | 0.978 |
| hpaC; type III secretion control protein HpaP | -0.209 | 1.508 | 0.890 | 0.999 |
| hpaD, hpcB; 3,4-dihydroxyphenylacetate 2,3-dioxygenase [EC:1.13.11.15] | -0.736 | 0.576 | 0.203 | 0.972 |
| hpaE, hpcC; 5-carboxymethyl-2-hydroxymuconic-semialdehyde dehydrogenase [EC:1.2.1.60] | -0.929 | 0.391 | 0.019 | 0.839 |
| hpaF, hpcD; 5-carboxymethyl-2-hydroxymuconate isomerase [EC:5.3.3.10] | -0.544 | 0.342 | 0.113 | 0.917 |
| hpaG; 5-oxopent-3-ene-1,2,5-tricarboxylate decarboxylase / 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase [EC:4.1.1.68 5.3.3.-] | -0.449 | 0.459 | 0.329 | 0.978 |
| hpaH; 2-oxo-hept-3-ene-1,7-dioate hydratase [EC:4.2.1.-] | 0.124 | 0.421 | 0.768 | 0.999 |
| hpaI, hpcH; 4-hydroxy-2-oxoheptanedioate aldolase [EC:4.1.2.52] | -0.762 | 0.331 | 0.023 | 0.839 |
| hpaX; MFS transporter, ACS family, 4-hydroxyphenylacetate permease | 0.667 | 0.617 | 0.281 | 0.978 |
| HPD, hppD; 4-hydroxyphenylpyruvate dioxygenase [EC:1.13.11.27] | -0.113 | 0.208 | 0.587 | 0.999 |
| hpdB; 4-hydroxyphenylacetate decarboxylase large subunit [EC:4.1.1.83] | 1.521 | 0.977 | 0.122 | 0.919 |
| hpgT, nocG; (S)-3,5-dihydroxyphenylglycine transaminase [EC:2.6.1.103] | -0.173 | 0.963 | 0.858 | 0.999 |
| hph; hygromycin-B 4-O-kinase [EC:2.7.1.163] | 18.076 | 2733.666 | 0.995 | 0.999 |
| hppA; K(+)-stimulated pyrophosphate-energized sodium pump [EC:3.6.1.1] | -0.032 | 0.152 | 0.832 | 0.999 |
| hpr; MarR family transcriptional regulator, protease production regulatory protein HPr | -1.586 | 0.806 | 0.051 | 0.839 |
| hprA; glycerate dehydrogenase [EC:1.1.1.29] | 0.179 | 0.119 | 0.134 | 0.942 |
| hprK, ptsK; HPr kinase/phosphorylase [EC:2.7.11.- 2.7.4.-] | 0.083 | 0.087 | 0.340 | 0.978 |
| hprT, hpt, HPRT1; hypoxanthine phosphoribosyltransferase [EC:2.4.2.8] | 0.078 | 0.049 | 0.111 | 0.917 |
| hpsN; sulfopropanediol 3-dehydrogenase [EC:1.1.1.308] | -0.933 | 0.878 | 0.290 | 0.978 |
| hpxA; allantoin racemase [EC:5.1.99.3] | -0.354 | 0.361 | 0.327 | 0.978 |
| hpxB; allantoinase [EC:3.5.2.5] | -0.557 | 0.565 | 0.326 | 0.978 |
| hpxO; FAD-dependent urate hydroxylase [EC:1.14.13.113] | -0.670 | 0.520 | 0.200 | 0.971 |
| hpxQ; 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase [EC:4.1.1.97] | -0.833 | 0.491 | 0.092 | 0.892 |
| hrcA; heat-inducible transcriptional repressor | 0.050 | 0.067 | 0.459 | 0.979 |
| hrpA; ATP-dependent helicase HrpA [EC:3.6.4.13] | 0.066 | 0.124 | 0.594 | 0.999 |
| hrpB; ATP-dependent helicase HrpB [EC:3.6.4.13] | -0.091 | 0.189 | 0.632 | 0.999 |
| hrpB1; type III secretion protein HrpB1 | -17.175 | 2988.990 | 0.995 | 0.999 |
| hrpB2; type III secretion inner rod protein HrpB2 | -17.175 | 2988.990 | 0.995 | 0.999 |
| hrpE; type III secretion hrp pilus HrpE | -17.175 | 2988.990 | 0.995 | 0.999 |
| hrpF; type III secretion translocon protein HrpF | -17.175 | 2988.990 | 0.995 | 0.999 |
| hrtA; hemin transport system ATP-binding protein [EC:3.6.3.-] | -0.467 | 0.685 | 0.496 | 0.981 |
| hrtB; hemin transport system permease protein | -0.160 | 0.828 | 0.847 | 0.999 |
| hsaA; 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione monooxygenase [EC:1.14.14.12] | -1.016 | 0.789 | 0.200 | 0.971 |
| hsaB; 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione monooxygenase reductase component [EC:1.5.1.-] | -0.679 | 0.691 | 0.327 | 0.978 |
| hsaC; 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione 4,5-dioxygenase [EC:1.13.11.25] | -0.838 | 0.724 | 0.248 | 0.978 |
| hsaD; 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase [EC:3.7.1.17] | -0.882 | 0.709 | 0.215 | 0.978 |
| hscA; molecular chaperone HscA | 0.089 | 0.171 | 0.601 | 0.999 |
| hscB, HSCB, HSC20; molecular chaperone HscB | 0.089 | 0.171 | 0.601 | 0.999 |
| hscC; molecular chaperone HscC | -0.062 | 0.337 | 0.855 | 0.999 |
| hsd; 3beta-hydroxy-Delta5-steroid dehydrogenase / steroid Delta-isomerase [EC:1.1.1.145 5.3.3.1] | -2.624 | 1.417 | 0.066 | 0.839 |
| HSD17B4; 3-hydroxyacyl-CoA dehydrogenase / 3a,7a,12a-trihydroxy-5b-cholest-24-enoyl-CoA hydratase / enoyl-CoA hydratase 2 [EC:1.1.1.35 4.2.1.107 4.2.1.119] | 1.513 | 1.385 | 0.276 | 0.978 |
| hsdM; type I restriction enzyme M protein [EC:2.1.1.72] | -0.010 | 0.071 | 0.892 | 0.999 |
| hsdR; type I restriction enzyme, R subunit [EC:3.1.21.3] | 0.032 | 0.084 | 0.701 | 0.999 |
| hsdS; type I restriction enzyme, S subunit [EC:3.1.21.3] | -0.017 | 0.091 | 0.852 | 0.999 |
| hsf; adhesin Hsf | -16.880 | 1816.243 | 0.993 | 0.999 |
| hsiF3; type VI secretion system lysozyme-related protein | -0.189 | 0.933 | 0.840 | 0.999 |
| hslJ; heat shock protein HslJ | 0.143 | 0.297 | 0.631 | 0.999 |
| hslO; molecular chaperone Hsp33 | 0.077 | 0.042 | 0.069 | 0.840 |
| hslR; ribosome-associated heat shock protein Hsp15 | -0.082 | 0.088 | 0.353 | 0.978 |
| hslU; ATP-dependent HslUV protease ATP-binding subunit HslU | 0.020 | 0.090 | 0.826 | 0.999 |
| hslV, clpQ; ATP-dependent HslUV protease, peptidase subunit HslV [EC:3.4.25.2] | 0.017 | 0.090 | 0.847 | 0.999 |
| HSP20; HSP20 family protein | -0.103 | 0.177 | 0.562 | 0.992 |
| HSP90A, htpG; molecular chaperone HtpG | 0.023 | 0.107 | 0.830 | 0.999 |
| hspQ; heat shock protein HspQ | -0.780 | 0.356 | 0.030 | 0.839 |
| hspR; MerR family transcriptional regulator, heat shock protein HspR | -0.051 | 0.228 | 0.823 | 0.999 |
| hss; homospermidine synthase [EC:2.5.1.44] | -1.940 | 0.811 | 0.018 | 0.839 |
| htpX; heat shock protein HtpX [EC:3.4.24.-] | 0.100 | 0.054 | 0.063 | 0.839 |
| htsT; energy-coupling factor transport system substrate-specific component | 0.038 | 0.144 | 0.793 | 0.999 |
| hugZ, hutZ; heme iron utilization protein | -0.164 | 0.234 | 0.486 | 0.979 |
| hupA; DNA-binding protein HU-alpha | 0.213 | 0.212 | 0.316 | 0.978 |
| hupB; DNA-binding protein HU-beta | -0.024 | 0.052 | 0.651 | 0.999 |
| hupR, hoxA; two-component system, NtrC family, response regulator HupR/HoxA | -17.351 | 2005.003 | 0.993 | 0.999 |
| hupT, hoxJ; two-component system, NtrC family, sensor histidine kinase HupT/HoxJ [EC:2.7.13.3] | -17.351 | 2005.003 | 0.993 | 0.999 |
| hutC; GntR family transcriptional regulator, histidine utilization repressor | -0.150 | 0.212 | 0.480 | 0.979 |
| hutF; formimidoylglutamate deiminase [EC:3.5.3.13] | -0.176 | 0.328 | 0.592 | 0.999 |
| hutG; formiminoglutamase [EC:3.5.3.8] | -0.029 | 0.115 | 0.801 | 0.999 |
| hutG; N-formylglutamate deformylase [EC:3.5.1.68] | -0.739 | 0.630 | 0.243 | 0.978 |
| hutH, HAL; histidine ammonia-lyase [EC:4.3.1.3] | -0.019 | 0.097 | 0.844 | 0.999 |
| hutI, AMDHD1; imidazolonepropionase [EC:3.5.2.7] | 0.048 | 0.090 | 0.594 | 0.999 |
| hutM; histidine permease | 18.183 | 2499.687 | 0.994 | 0.999 |
| hutP; hut operon positive regulatory protein | -2.614 | 1.445 | 0.072 | 0.845 |
| hutT; histidine transporter | -0.066 | 0.225 | 0.769 | 0.999 |
| hutU, UROC1; urocanate hydratase [EC:4.2.1.49] | 0.049 | 0.089 | 0.585 | 0.999 |
| hxlA; 3-hexulose-6-phosphate synthase [EC:4.1.2.43] | -1.500 | 0.672 | 0.027 | 0.839 |
| hxlB; 6-phospho-3-hexuloisomerase [EC:5.3.1.27] | -1.048 | 0.480 | 0.031 | 0.839 |
| HXT; MFS transporter, SP family, sugar:H+ symporter | -0.652 | 0.520 | 0.212 | 0.978 |
| hya; hyaluronoglucosaminidase [EC:3.2.1.35] | 0.004 | 0.323 | 0.989 | 0.999 |
| hyaA, hybO; hydrogenase small subunit [EC:1.12.99.6] | 0.126 | 0.223 | 0.572 | 0.995 |
| hyaB, hybC; hydrogenase large subunit [EC:1.12.99.6] | 0.113 | 0.222 | 0.613 | 0.999 |
| hyaC; Ni/Fe-hydrogenase 1 B-type cytochrome subunit | 0.088 | 0.232 | 0.706 | 0.999 |
| hyaD, hybD; hydrogenase maturation protease [EC:3.4.23.-] | 0.048 | 0.210 | 0.820 | 0.999 |
| hyaE; hydrogenase-1 operon protein HyaE | -2.046 | 1.315 | 0.122 | 0.919 |
| hyaF; hydrogenase-1 operon protein HyaF | -1.500 | 1.160 | 0.198 | 0.970 |
| hycA; formate hydrogenlyase regulatory protein HycA | -0.371 | 0.685 | 0.589 | 0.999 |
| hycB; formate hydrogenlyase subunit 2 | -0.371 | 0.685 | 0.589 | 0.999 |
| hycC; formate hydrogenlyase subunit 3 | -0.371 | 0.685 | 0.589 | 0.999 |
| hycD; formate hydrogenlyase subunit 4 | -0.371 | 0.685 | 0.589 | 0.999 |
| hycE; formate hydrogenlyase subunit 5 | -0.158 | 0.634 | 0.804 | 0.999 |
| hycF; formate hydrogenlyase subunit 6 | -0.371 | 0.685 | 0.589 | 0.999 |
| hycG; formate hydrogenlyase subunit 7 | -0.344 | 0.676 | 0.612 | 0.999 |
| hycH; formate hydrogenlyase maturation protein HycH | -0.051 | 0.450 | 0.910 | 0.999 |
| hycI; hydrogenase 3 maturation protease [EC:3.4.23.51] | 0.332 | 0.427 | 0.437 | 0.978 |
| hydA; quinone-reactive Ni/Fe-hydrogenase small subunit [EC:1.12.5.1] | -0.366 | 0.782 | 0.641 | 0.999 |
| hydB; quinone-reactive Ni/Fe-hydrogenase large subunit [EC:1.12.5.1] | -0.366 | 0.782 | 0.641 | 0.999 |
| HYDIN; hydrocephalus-inducing protein | -17.059 | 2820.383 | 0.995 | 0.999 |
| hydN; electron transport protein HydN | 0.167 | 0.257 | 0.517 | 0.986 |
| hyfA; hydrogenase-4 component A [EC:1.-.-.-] | 0.662 | 0.632 | 0.297 | 0.978 |
| hyfB; hydrogenase-4 component B [EC:1.-.-.-] | 0.292 | 0.500 | 0.561 | 0.992 |
| hyfC; hydrogenase-4 component C [EC:1.-.-.-] | 0.428 | 0.535 | 0.425 | 0.978 |
| hyfD; hydrogenase-4 component D [EC:1.-.-.-] | 0.942 | 0.928 | 0.312 | 0.978 |
| hyfE; hydrogenase-4 component E [EC:1.-.-.-] | 0.235 | 0.483 | 0.627 | 0.999 |
| hyfF; hydrogenase-4 component F [EC:1.-.-.-] | 0.235 | 0.483 | 0.627 | 0.999 |
| hyfG; hydrogenase-4 component G [EC:1.-.-.-] | 0.723 | 0.834 | 0.387 | 0.978 |
| hyfH; hydrogenase-4 component H | 0.609 | 0.765 | 0.427 | 0.978 |
| hyfI; hydrogenase-4 component I [EC:1.-.-.-] | 0.633 | 0.869 | 0.468 | 0.979 |
| hyfJ; hydrogenase-4 component J [EC:1.-.-.-] | 0.633 | 0.869 | 0.468 | 0.979 |
| hyfR; hydrogenase-4 transcriptional activator | -1.153 | 1.472 | 0.435 | 0.978 |
| hyg; hygromycin-B 7’’-O-kinase [EC:2.7.1.119] | -1.407 | 1.166 | 0.229 | 0.978 |
| hyi, gip; hydroxypyruvate isomerase [EC:5.3.1.22] | -0.268 | 0.210 | 0.205 | 0.972 |
| hypA, hybF; hydrogenase nickel incorporation protein HypA/HybF | 0.016 | 0.191 | 0.934 | 0.999 |
| hypB; hydrogenase nickel incorporation protein HypB | 0.027 | 0.194 | 0.892 | 0.999 |
| hypBA1; non-reducing end beta-L-arabinofuranosidase [EC:3.2.1.185] | 0.125 | 1.190 | 0.917 | 0.999 |
| hypBA2; beta-L-arabinobiosidase [EC:3.2.1.187] | -17.405 | 2475.781 | 0.994 | 0.999 |
| hypC; hydrogenase expression/formation protein HypC | -0.061 | 0.190 | 0.747 | 0.999 |
| hypD; hydrogenase expression/formation protein HypD | 0.045 | 0.195 | 0.816 | 0.999 |
| hypE; hydrogenase expression/formation protein HypE | 0.017 | 0.196 | 0.929 | 0.999 |
| hypF; hydrogenase maturation protein HypF | -0.003 | 0.193 | 0.986 | 0.999 |
| hypX, hoxX; putative two-component system protein, hydrogenase maturation factor HypX/HoxX | -2.100 | 2.239 | 0.350 | 0.978 |
| hysA, hylA, hylB; hyaluronate lyase [EC:4.2.2.1] | 0.517 | 0.452 | 0.255 | 0.978 |
| hyuA; N-methylhydantoinase A [EC:3.5.2.14] | -0.556 | 0.407 | 0.174 | 0.961 |
| hyuB; N-methylhydantoinase B [EC:3.5.2.14] | -0.537 | 0.402 | 0.184 | 0.965 |
| iaaA, ASRGL1; beta-aspartyl-peptidase (threonine type) [EC:3.4.19.5] | -0.313 | 0.302 | 0.302 | 0.978 |
| iaaM; tryptophan 2-monooxygenase [EC:1.13.12.3] | -0.159 | 0.438 | 0.717 | 0.999 |
| iadA; beta-aspartyl-dipeptidase (metallo-type) [EC:3.4.19.-] | -0.221 | 0.453 | 0.626 | 0.999 |
| IAL; isopenicillin-N N-acyltransferase like protein | -0.227 | 0.655 | 0.729 | 0.999 |
| iap; alkaline phosphatase isozyme conversion protein [EC:3.4.11.-] | 0.134 | 0.621 | 0.829 | 0.999 |
| IARS, ileS; isoleucyl-tRNA synthetase [EC:6.1.1.5] | 0.029 | 0.032 | 0.377 | 0.978 |
| iatA; inositol transport system ATP-binding protein | 0.983 | 0.801 | 0.222 | 0.978 |
| iatP; inositol transport system permease protein | 0.983 | 0.801 | 0.222 | 0.978 |
| ibpA; molecular chaperone IbpA | -0.215 | 0.234 | 0.361 | 0.978 |
| ibpB; molecular chaperone IbpB | 0.135 | 0.629 | 0.830 | 0.999 |
| ich-P; itaconyl-CoA hydratase / mesaconyl-C4 CoA hydratase [EC:4.2.1.56 4.2.1.-] | 0.082 | 0.403 | 0.839 | 0.999 |
| iciA; LysR family transcriptional regulator, chromosome initiation inhibitor | -0.067 | 0.200 | 0.738 | 0.999 |
| iclR; IclR family transcriptional regulator, acetate operon repressor | -0.744 | 0.379 | 0.052 | 0.839 |
| icmB, dotO; intracellular multiplication protein IcmB | -0.342 | 0.934 | 0.714 | 0.999 |
| icmF; isobutyryl-CoA mutase [EC:5.4.99.13] | -0.418 | 0.234 | 0.075 | 0.855 |
| icmJ, dotN; intracellular multiplication protein IcmJ | -0.259 | 0.965 | 0.788 | 0.999 |
| icmL, traM, dotI; intracellular multiplication protein IcmL | -0.259 | 0.965 | 0.788 | 0.999 |
| icmO, trbC, dotL; intracellular multiplication protein IcmO | -0.259 | 0.965 | 0.788 | 0.999 |
| ICMT, STE14; protein-S-isoprenylcysteine O-methyltransferase [EC:2.1.1.100] | -17.687 | 2769.090 | 0.995 | 0.999 |
| ICP; inhibitor of cysteine peptidase | 0.229 | 0.339 | 0.501 | 0.981 |
| ict-P; itaconate CoA-transferase [EC:2.8.3.- 2.8.3.22] | 0.152 | 0.433 | 0.726 | 0.999 |
| ict-Y; itaconate CoA-transferase [EC:2.8.3.-] | -0.543 | 0.889 | 0.542 | 0.990 |
| ictB; putative inorganic carbon (hco3(-)) transporter | 0.479 | 0.624 | 0.444 | 0.978 |
| IDE, ide; insulysin [EC:3.4.24.56] | -3.465 | 4.101 | 0.400 | 0.978 |
| IDH1, IDH2, icd; isocitrate dehydrogenase [EC:1.1.1.42] | -0.144 | 0.091 | 0.117 | 0.917 |
| IDH3; isocitrate dehydrogenase (NAD+) [EC:1.1.1.41] | 0.517 | 0.291 | 0.078 | 0.859 |
| idi, IDI; isopentenyl-diphosphate Delta-isomerase [EC:5.3.3.2] | 0.027 | 0.108 | 0.801 | 0.999 |
| idnD; L-idonate 5-dehydrogenase [EC:1.1.1.264] | -0.610 | 0.498 | 0.222 | 0.978 |
| idnO; gluconate 5-dehydrogenase [EC:1.1.1.69] | 0.088 | 0.199 | 0.660 | 0.999 |
| idnR, gntH; LacI family transcriptional regulator, gluconate utilization system Gnt-II transcriptional activator | -1.630 | 2.316 | 0.483 | 0.979 |
| idnT; Gnt-II system L-idonate transporter | 0.055 | 1.009 | 0.957 | 0.999 |
| IDO, INDO; indoleamine 2,3-dioxygenase [EC:1.13.11.52] | 0.624 | 1.029 | 0.545 | 0.990 |
| IDS; iduronate 2-sulfatase [EC:3.1.6.13] | -16.845 | 2534.319 | 0.995 | 0.999 |
| idsA; geranylgeranyl diphosphate synthase, type I [EC:2.5.1.1 2.5.1.10 2.5.1.29] | -0.011 | 0.239 | 0.965 | 0.999 |
| IDUA; L-iduronidase [EC:3.2.1.76] | -1.572 | 1.689 | 0.353 | 0.978 |
| iga; IgA-specific metalloendopeptidase [EC:3.4.24.13] | -0.032 | 0.687 | 0.963 | 0.999 |
| iga; IgA-specific serine endopeptidase [EC:3.4.21.72] | 0.261 | 0.357 | 0.465 | 0.979 |
| ihfA, himA; integration host factor subunit alpha | -0.071 | 0.132 | 0.594 | 0.999 |
| ihfB, himD; integration host factor subunit beta | -0.049 | 0.144 | 0.734 | 0.999 |
| ihk; two-component system, OmpR family, sensor kinase Ihk [EC:2.7.13.3] | 0.077 | 0.182 | 0.672 | 0.999 |
| ILR1; IAA-amino acid hydrolase [EC:3.5.1.-] | -17.761 | 4005.858 | 0.996 | 0.999 |
| ilvC; ketol-acid reductoisomerase [EC:1.1.1.86] | -0.061 | 0.052 | 0.249 | 0.978 |
| ilvD; dihydroxy-acid dehydratase [EC:4.2.1.9] | -0.144 | 0.060 | 0.017 | 0.839 |
| ilvM; acetolactate synthase II small subunit [EC:2.2.1.6] | 0.161 | 0.257 | 0.532 | 0.990 |
| ilvY; LysR family transcriptional regulator, positive regulator for ilvC | 0.232 | 0.307 | 0.451 | 0.978 |
| IMA, malL; oligo-1,6-glucosidase [EC:3.2.1.10] | 0.149 | 0.223 | 0.505 | 0.981 |
| impA; type VI secretion system protein ImpA | -0.160 | 0.366 | 0.662 | 0.999 |
| impB; type VI secretion system protein ImpB | 0.094 | 0.266 | 0.723 | 0.999 |
| impC; type VI secretion system protein ImpC | 0.095 | 0.239 | 0.691 | 0.999 |
| impD; type VI secretion system protein ImpD | -1.734 | 1.761 | 0.326 | 0.978 |
| IMPDH, guaB; IMP dehydrogenase [EC:1.1.1.205] | 0.022 | 0.037 | 0.555 | 0.992 |
| impE; type VI secretion system protein ImpE | -1.023 | 0.780 | 0.191 | 0.970 |
| impF; type VI secretion system protein ImpF | -0.176 | 0.363 | 0.629 | 0.999 |
| impG, vasA; type VI secretion system protein ImpG | 0.146 | 0.256 | 0.570 | 0.995 |
| impH, vasB; type VI secretion system protein ImpH | 0.153 | 0.250 | 0.543 | 0.990 |
| impI, vasC; type VI secretion system protein ImpI | 0.056 | 0.332 | 0.865 | 0.999 |
| impJ, vasE; type VI secretion system protein ImpJ | 0.131 | 0.263 | 0.619 | 0.999 |
| impK, ompA, vasF, dotU; type VI secretion system protein ImpK | 0.138 | 0.240 | 0.567 | 0.995 |
| impL, vasK, icmF; type VI secretion system protein ImpL | 0.164 | 0.303 | 0.590 | 0.999 |
| impM; type VI secretion system protein ImpM | -0.231 | 0.366 | 0.529 | 0.989 |
| impN; type VI secretion system protein ImpN [EC:2.7.11.1] | -1.272 | 1.833 | 0.489 | 0.979 |
| imuA; protein ImuA | -0.743 | 0.347 | 0.034 | 0.839 |
| imuB; protein ImuB | -0.144 | 0.190 | 0.449 | 0.978 |
| ina; immune inhibitor A [EC:3.4.24.-] | -0.411 | 0.295 | 0.166 | 0.960 |
| infA; translation initiation factor IF-1 | 0.037 | 0.035 | 0.285 | 0.978 |
| infB, MTIF2; translation initiation factor IF-2 | 0.038 | 0.037 | 0.305 | 0.978 |
| infC, MTIF3; translation initiation factor IF-3 | 0.036 | 0.036 | 0.323 | 0.978 |
| inhA; cyclohexyl-isocyanide hydratase [EC:4.2.1.103] | -1.074 | 0.538 | 0.048 | 0.839 |
| inhA; enoyl ACP reductase [EC:1.3.1.9] | -2.137 | 1.493 | 0.154 | 0.954 |
| inlA; internalin A | -0.128 | 0.461 | 0.782 | 0.999 |
| inlB; internalin B | -17.821 | 2878.738 | 0.995 | 0.999 |
| INO1, ISYNA1; myo-inositol-1-phosphate synthase [EC:5.5.1.4] | -0.200 | 0.213 | 0.348 | 0.978 |
| inoE; inositol-phosphate transport system substrate-binding protein | -1.272 | 1.833 | 0.489 | 0.979 |
| inoF; inositol-phosphate transport system permease protein | -1.272 | 1.833 | 0.489 | 0.979 |
| inoG; inositol-phosphate transport system permease protein | -1.272 | 1.833 | 0.489 | 0.979 |
| inoK; inositol-phosphate transport system ATP-binding protein | -1.272 | 1.833 | 0.489 | 0.979 |
| insB; insertion element IS1 protein InsB | -1.011 | 1.306 | 0.440 | 0.978 |
| int; integrase | -0.115 | 0.230 | 0.617 | 0.999 |
| INV, sacA; beta-fructofuranosidase [EC:3.2.1.26] | 0.139 | 0.103 | 0.182 | 0.961 |
| iolB; 5-deoxy-glucuronate isomerase [EC:5.3.1.30] | -0.354 | 0.271 | 0.194 | 0.970 |
| iolC; 5-dehydro-2-deoxygluconokinase [EC:2.7.1.92] | -0.337 | 0.271 | 0.215 | 0.978 |
| iolD; 3D-(3,5/4)-trihydroxycyclohexane-1,2-dione acylhydrolase (decyclizing) [EC:3.7.1.22] | -0.314 | 0.272 | 0.250 | 0.978 |
| iolE; inosose dehydratase [EC:4.2.1.44] | -0.307 | 0.283 | 0.280 | 0.978 |
| iolF; MFS transporter, SP family, inositol transporter | -0.717 | 0.458 | 0.120 | 0.917 |
| iolG; myo-inositol 2-dehydrogenase / D-chiro-inositol 1-dehydrogenase [EC:1.1.1.18 1.1.1.369] | -0.416 | 0.344 | 0.228 | 0.978 |
| iolH; myo-inositol catabolism protein IolH | -0.123 | 0.472 | 0.794 | 0.999 |
| iolI; 2-keto-myo-inositol isomerase [EC:5.3.99.11] | -0.511 | 0.568 | 0.370 | 0.978 |
| iolJ; 6-phospho-5-dehydro-2-deoxy-D-gluconate aldolase [EC:4.1.2.29] | 0.070 | 0.758 | 0.927 | 0.999 |
| iolR; DeoR family transcriptional regulator, myo-inositol catabolism operon repressor | 2.127 | 1.621 | 0.191 | 0.970 |
| iolS; myo-inositol catabolism protein IolS [EC:1.1.1.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| iolT; MFS transporter, SP family, major inositol transporter | 0.274 | 0.336 | 0.416 | 0.978 |
| iolW; scyllo-inositol 2-dehydrogenase (NADP+) [EC:1.1.1.371] | -0.656 | 0.669 | 0.329 | 0.978 |
| iolX; scyllo-inositol 2-dehydrogenase (NAD+) [EC:1.1.1.370] | 1.125 | 1.241 | 0.366 | 0.978 |
| iorA; indolepyruvate ferredoxin oxidoreductase, alpha subunit [EC:1.2.7.8] | -0.539 | 0.368 | 0.145 | 0.951 |
| iorA; isoquinoline 1-oxidoreductase subunit alpha [EC:1.3.99.16] | -0.312 | 0.298 | 0.297 | 0.978 |
| iorB; indolepyruvate ferredoxin oxidoreductase, beta subunit [EC:1.2.7.8] | -0.537 | 0.369 | 0.148 | 0.954 |
| iorB; isoquinoline 1-oxidoreductase subunit beta [EC:1.3.99.16] | -0.322 | 0.316 | 0.310 | 0.978 |
| ipct; 1L-myo-inositol 1-phosphate cytidylyltransferase [EC:2.7.7.74] | -0.623 | 1.557 | 0.689 | 0.999 |
| ipdC; indolepyruvate decarboxylase [EC:4.1.1.74] | -0.426 | 0.425 | 0.318 | 0.978 |
| ipk; isopentenyl phosphate kinase [EC:2.7.4.26] | -17.234 | 3077.514 | 0.996 | 0.999 |
| irp1, HMWP1; yersiniabactin nonribosomal peptide/polyketide synthase | -2.044 | 2.163 | 0.346 | 0.978 |
| irp2, HMWP2; yersiniabactin nonribosomal peptide synthetase | -0.456 | 1.133 | 0.688 | 0.999 |
| irp3, ybtU; yersiniabactin synthetase, thiazolinyl reductase component | -2.044 | 2.163 | 0.346 | 0.978 |
| irp4, ybtT; yersiniabactin synthetase, thioesterase component | -2.044 | 2.163 | 0.346 | 0.978 |
| irp5, ybtE; yersiniabactin salicyl-AMP ligase [EC:6.3.2.-] | -0.423 | 1.154 | 0.715 | 0.999 |
| IRPC; inward rectifier potassium channel | -0.783 | 0.871 | 0.370 | 0.978 |
| irr; Fur family transcriptional regulator, iron response regulator | -1.497 | 0.628 | 0.018 | 0.839 |
| irr; two-component system, OmpR family, response regulator Irr | 0.080 | 0.182 | 0.662 | 0.999 |
| irtA; ATP-binding cassette, subfamily B, bacterial IrtA [EC:3.6.3.-] | -0.003 | 0.934 | 0.997 | 0.999 |
| irtB; ATP-binding cassette, subfamily B, bacterial IrtB [EC:3.6.3.-] | -2.417 | 1.514 | 0.112 | 0.917 |
| IS15, IS26; transposase, IS6 family | -0.753 | 0.718 | 0.296 | 0.978 |
| iscA; iron-sulfur cluster assembly protein | -0.199 | 0.114 | 0.082 | 0.865 |
| iscR; Rrf2 family transcriptional regulator, iron-sulfur cluster assembly transcription factor | 0.044 | 0.134 | 0.745 | 0.999 |
| iscS, NFS1; cysteine desulfurase [EC:2.8.1.7] | 0.019 | 0.061 | 0.754 | 0.999 |
| iscU, nifU; nitrogen fixation protein NifU and related proteins | 0.094 | 0.048 | 0.052 | 0.839 |
| isdG, isdI; heme oxygenase (staphylobilin-producing) [EC:1.14.99.48] | -0.671 | 0.630 | 0.288 | 0.978 |
| isfD; sulfoacetaldehyde reductase [EC:1.1.1.313] | 0.000 | 0.766 | 1.000 | 1.000 |
| isp; major intracellular serine protease [EC:3.4.21.-] | -1.298 | 1.153 | 0.262 | 0.978 |
| ispA; farnesyl diphosphate synthase [EC:2.5.1.1 2.5.1.10] | -0.157 | 0.168 | 0.350 | 0.978 |
| ispB; octaprenyl-diphosphate synthase [EC:2.5.1.90] | -0.083 | 0.091 | 0.361 | 0.978 |
| ispD; 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [EC:2.7.7.60] | -0.013 | 0.063 | 0.834 | 0.999 |
| ispDF; 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase / 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [EC:2.7.7.60 4.6.1.12] | -0.118 | 0.176 | 0.505 | 0.981 |
| ispE; 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase [EC:2.7.1.148] | 0.036 | 0.050 | 0.469 | 0.979 |
| ispF; 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [EC:4.6.1.12] | 0.036 | 0.069 | 0.606 | 0.999 |
| ispH, lytB; 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase [EC:1.17.7.4] | 0.017 | 0.067 | 0.803 | 0.999 |
| ispZ; intracellular septation protein | -0.031 | 0.157 | 0.843 | 0.999 |
| ITGB3, CD61; integrin beta 3 | -1.780 | 1.801 | 0.324 | 0.978 |
| ITGB8; integrin beta 8 | -0.596 | 0.946 | 0.530 | 0.989 |
| ituB, mycB, bmyB; iturin family lipopeptide synthetase B | -3.465 | 4.101 | 0.400 | 0.978 |
| iucA; N2-citryl-N6-acetyl-N6-hydroxylysine synthase [EC:6.3.2.38] | 0.719 | 0.779 | 0.357 | 0.978 |
| iucB; acetyl CoA:N6-hydroxylysine acetyl transferase [EC:2.3.1.102] | 0.709 | 0.605 | 0.243 | 0.978 |
| iucC; aerobactin synthase [EC:6.3.2.39] | 0.719 | 0.779 | 0.357 | 0.978 |
| iucD; lysine N6-hydroxylase [EC:1.14.13.59] | -0.323 | 0.408 | 0.430 | 0.978 |
| iunH; purine nucleosidase [EC:3.2.2.1] | -0.028 | 0.186 | 0.882 | 0.999 |
| IVD, ivd; isovaleryl-CoA dehydrogenase [EC:1.3.8.4] | -0.198 | 0.210 | 0.348 | 0.978 |
| jadI; cyclase | -17.703 | 3891.582 | 0.996 | 0.999 |
| jag; spoIIIJ-associated protein | 0.119 | 0.087 | 0.171 | 0.961 |
| JEN; MFS transporter, SHS family, lactate transporter | 0.035 | 0.738 | 0.963 | 0.999 |
| K00183; prokaryotic molybdopterin-containing oxidoreductase family, molybdopterin binding subunit | 0.301 | 0.703 | 0.669 | 0.999 |
| K00184; prokaryotic molybdopterin-containing oxidoreductase family, iron-sulfur binding subunit | -0.302 | 0.470 | 0.521 | 0.988 |
| K00185; prokaryotic molybdopterin-containing oxidoreductase family, membrane subunit | -0.047 | 0.442 | 0.915 | 0.999 |
| K00243; uncharacterized protein | 0.108 | 0.073 | 0.140 | 0.949 |
| K00375; GntR family transcriptional regulator / MocR family aminotransferase | -0.050 | 0.129 | 0.697 | 0.999 |
| K00666; fatty-acyl-CoA synthase [EC:6.2.1.-] | -0.397 | 0.196 | 0.045 | 0.839 |
| K01138; uncharacterized sulfatase [EC:3.1.6.-] | 0.335 | 0.521 | 0.521 | 0.988 |
| K01163; uncharacterized protein | 0.236 | 0.170 | 0.168 | 0.961 |
| K01436; amidohydrolase [EC:3.5.1.-] | 0.057 | 0.115 | 0.620 | 0.999 |
| K01622; fructose 1,6-bisphosphate aldolase/phosphatase [EC:4.1.2.13 3.1.3.11] | 1.254 | 1.302 | 0.337 | 0.978 |
| K02351; putative membrane protein | -0.438 | 0.471 | 0.354 | 0.978 |
| K02475; two-component system, CitB family, response regulator | -0.502 | 0.707 | 0.478 | 0.979 |
| K02476; two-component system, CitB family, sensor kinase [EC:2.7.13.3] | -0.637 | 0.579 | 0.273 | 0.978 |
| K02477; two-component system, LytTR family, response regulator | 0.035 | 0.217 | 0.874 | 0.999 |
| K02478; two-component system, LytTR family, sensor kinase [EC:2.7.13.3] | 0.604 | 0.342 | 0.080 | 0.861 |
| K02479; two-component system, NarL family, response regulator | -0.315 | 0.453 | 0.487 | 0.979 |
| K02480; two-component system, NarL family, sensor kinase [EC:2.7.13.3] | -0.786 | 0.531 | 0.141 | 0.949 |
| K02481; two-component system, NtrC family, response regulator | 0.120 | 0.294 | 0.683 | 0.999 |
| K02482; two-component system, NtrC family, sensor kinase [EC:2.7.13.3] | -0.814 | 0.570 | 0.155 | 0.954 |
| K02483; two-component system, OmpR family, response regulator | -0.206 | 0.173 | 0.236 | 0.978 |
| K02484; two-component system, OmpR family, sensor kinase [EC:2.7.13.3] | -0.107 | 0.157 | 0.497 | 0.981 |
| K03653; N-glycosylase/DNA lyase [EC:3.2.2.- 4.2.99.18] | 0.914 | 0.445 | 0.041 | 0.839 |
| K03710; GntR family transcriptional regulator | -0.074 | 0.110 | 0.499 | 0.981 |
| K03791; putative chitinase | -0.287 | 0.365 | 0.433 | 0.978 |
| K03822; putative long chain acyl-CoA synthase [EC:6.2.1.-] | -2.137 | 1.493 | 0.154 | 0.954 |
| K05303; O-methyltransferase [EC:2.1.1.-] | -0.972 | 0.731 | 0.186 | 0.966 |
| K05558; pyridoxamine 5’-phosphate oxidase family protein | -1.482 | 1.416 | 0.297 | 0.978 |
| K05937; uncharacterized protein | -0.091 | 0.824 | 0.912 | 0.999 |
| K05952; uncharacterized protein | 0.295 | 0.233 | 0.207 | 0.976 |
| K05967; uncharacterized protein | -1.071 | 0.837 | 0.203 | 0.972 |
| K06860; putative heme uptake system protein | 0.016 | 0.288 | 0.954 | 0.999 |
| K06865; ATPase | -17.234 | 3077.514 | 0.996 | 0.999 |
| K06867; uncharacterized protein | 0.244 | 0.226 | 0.282 | 0.978 |
| K06869; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K06871; uncharacterized protein | -0.018 | 0.155 | 0.907 | 0.999 |
| K06872; uncharacterized protein | -0.127 | 0.116 | 0.275 | 0.978 |
| K06876; deoxyribodipyrimidine photolyase-related protein | 0.242 | 0.281 | 0.391 | 0.978 |
| K06877; DEAD/DEAH box helicase domain-containing protein | -0.227 | 0.226 | 0.318 | 0.978 |
| K06878; tRNA-binding protein | -0.024 | 0.116 | 0.834 | 0.999 |
| K06882; uncharacterized protein | -0.324 | 0.725 | 0.656 | 0.999 |
| K06883; uncharacterized protein | -2.378 | 1.422 | 0.096 | 0.896 |
| K06884; uncharacterized protein | 0.272 | 0.292 | 0.353 | 0.978 |
| K06885; uncharacterized protein | 0.043 | 0.108 | 0.689 | 0.999 |
| K06887; uncharacterized protein | -0.163 | 0.641 | 0.800 | 0.999 |
| K06888; uncharacterized protein | -0.659 | 0.303 | 0.031 | 0.839 |
| K06889; uncharacterized protein | 0.033 | 0.087 | 0.708 | 0.999 |
| K06890; uncharacterized protein | 0.044 | 0.101 | 0.662 | 0.999 |
| K06893; uncharacterized protein | 0.028 | 0.263 | 0.915 | 0.999 |
| K06894; uncharacterized protein | 0.121 | 0.191 | 0.528 | 0.989 |
| K06897; 7,8-dihydropterin-6-yl-methyl-4-(beta-D-ribofuranosyl)aminobenzene 5’-phosphate synthase [EC:2.5.1.105] | -0.200 | 0.483 | 0.679 | 0.999 |
| K06903; uncharacterized protein | -0.299 | 0.307 | 0.331 | 0.978 |
| K06904; uncharacterized protein | -0.829 | 0.322 | 0.011 | 0.839 |
| K06905; uncharacterized protein | -0.244 | 0.315 | 0.440 | 0.978 |
| K06906; uncharacterized protein | -0.399 | 0.426 | 0.350 | 0.978 |
| K06907; uncharacterized protein | -0.200 | 0.311 | 0.522 | 0.988 |
| K06908; uncharacterized protein | -0.194 | 0.313 | 0.537 | 0.990 |
| K06910; uncharacterized protein | 0.050 | 0.124 | 0.686 | 0.999 |
| K06911; uncharacterized protein | -0.109 | 0.148 | 0.464 | 0.979 |
| K06913; uncharacterized protein | -17.845 | 4178.817 | 0.997 | 0.999 |
| K06915; uncharacterized protein | 0.127 | 0.110 | 0.248 | 0.978 |
| K06918; uncharacterized protein | -0.089 | 0.269 | 0.741 | 0.999 |
| K06919; putative DNA primase/helicase | 0.241 | 0.233 | 0.303 | 0.978 |
| K06921; uncharacterized protein | 0.280 | 0.326 | 0.391 | 0.978 |
| K06922; uncharacterized protein | -1.849 | 1.778 | 0.300 | 0.978 |
| K06923; uncharacterized protein | 0.014 | 0.158 | 0.930 | 0.999 |
| K06926; uncharacterized protein | 0.527 | 0.303 | 0.084 | 0.872 |
| K06929; uncharacterized protein | -0.035 | 0.125 | 0.781 | 0.999 |
| K06931; uncharacterized protein | -18.313 | 3142.249 | 0.995 | 0.999 |
| K06934; uncharacterized protein | -0.426 | 0.394 | 0.282 | 0.978 |
| K06937; 7,8-dihydro-6-hydroxymethylpterin dimethyltransferase [EC:2.1.1.-] | -1.874 | 0.840 | 0.027 | 0.839 |
| K06938; uncharacterized protein | -0.068 | 0.222 | 0.759 | 0.999 |
| K06940; uncharacterized protein | -0.049 | 0.122 | 0.686 | 0.999 |
| K06944; uncharacterized protein | 2.285 | 1.976 | 0.249 | 0.978 |
| K06945; uncharacterized protein | -0.138 | 0.257 | 0.592 | 0.999 |
| K06946; uncharacterized protein | -1.289 | 1.180 | 0.277 | 0.978 |
| K06950; uncharacterized protein | 0.043 | 0.089 | 0.628 | 0.999 |
| K06951; uncharacterized protein | -17.398 | 2838.871 | 0.995 | 0.999 |
| K06952; uncharacterized protein | -1.040 | 0.449 | 0.022 | 0.839 |
| K06953; uncharacterized protein | -0.424 | 0.352 | 0.230 | 0.978 |
| K06954; uncharacterized protein | -0.117 | 0.341 | 0.733 | 0.999 |
| K06955; uncharacterized protein | -0.050 | 0.320 | 0.877 | 0.999 |
| K06956; uncharacterized protein | 0.073 | 0.128 | 0.568 | 0.995 |
| K06960; uncharacterized protein | 0.116 | 0.087 | 0.184 | 0.965 |
| K06962; uncharacterized protein | 0.069 | 0.121 | 0.566 | 0.995 |
| K06971; uncharacterized protein | -1.425 | 0.705 | 0.045 | 0.839 |
| K06973; uncharacterized protein | -0.056 | 0.161 | 0.730 | 0.999 |
| K06975; uncharacterized protein | 0.093 | 0.081 | 0.253 | 0.978 |
| K06976; uncharacterized protein | -0.377 | 0.298 | 0.207 | 0.976 |
| K06977; uncharacterized protein | -0.032 | 0.461 | 0.944 | 0.999 |
| K06978; uncharacterized protein | -0.424 | 0.277 | 0.128 | 0.927 |
| K06983; uncharacterized protein | -0.028 | 0.466 | 0.953 | 0.999 |
| K06985; aspartyl protease family protein | -0.006 | 0.262 | 0.981 | 0.999 |
| K06986; uncharacterized protein | 0.174 | 0.302 | 0.564 | 0.994 |
| K06987; uncharacterized protein | -0.106 | 0.304 | 0.728 | 0.999 |
| K06991; uncharacterized protein | -0.113 | 0.307 | 0.713 | 0.999 |
| K06992; uncharacterized protein | -0.058 | 0.236 | 0.806 | 0.999 |
| K06994; putative drug exporter of the RND superfamily | -0.503 | 0.278 | 0.073 | 0.845 |
| K06995; uncharacterized protein | -0.015 | 0.295 | 0.961 | 0.999 |
| K06996; uncharacterized protein | -0.175 | 0.246 | 0.479 | 0.979 |
| K06999; phospholipase/carboxylesterase | -0.041 | 0.153 | 0.790 | 0.999 |
| K07000; uncharacterized protein | -0.025 | 0.145 | 0.862 | 0.999 |
| K07001; NTE family protein | -0.135 | 0.114 | 0.236 | 0.978 |
| K07002; uncharacterized protein | -0.237 | 0.202 | 0.243 | 0.978 |
| K07003; uncharacterized protein | 0.019 | 0.181 | 0.917 | 0.999 |
| K07004; uncharacterized protein | -0.109 | 0.146 | 0.458 | 0.979 |
| K07005; uncharacterized protein | 0.241 | 0.195 | 0.217 | 0.978 |
| K07006; uncharacterized protein | -0.088 | 0.205 | 0.670 | 0.999 |
| K07007; uncharacterized protein | -0.029 | 0.049 | 0.557 | 0.992 |
| K07009; uncharacterized protein | 0.033 | 0.124 | 0.791 | 0.999 |
| K07010; putative glutamine amidotransferase | 0.138 | 0.076 | 0.070 | 0.841 |
| K07011; uncharacterized protein | -0.183 | 0.144 | 0.204 | 0.972 |
| K07014; uncharacterized protein | 0.110 | 0.244 | 0.654 | 0.999 |
| K07015; uncharacterized protein | 0.004 | 0.120 | 0.976 | 0.999 |
| K07017; uncharacterized protein | 0.038 | 0.233 | 0.869 | 0.999 |
| K07018; uncharacterized protein | -0.060 | 0.158 | 0.706 | 0.999 |
| K07019; uncharacterized protein | -0.001 | 0.176 | 0.996 | 0.999 |
| K07020; uncharacterized protein | 0.092 | 0.280 | 0.741 | 0.999 |
| K07023; putative hydrolases of HD superfamily | -0.161 | 0.175 | 0.360 | 0.978 |
| K07025; putative hydrolase of the HAD superfamily | -0.097 | 0.046 | 0.039 | 0.839 |
| K07027; glycosyltransferase 2 family protein | -0.043 | 0.110 | 0.696 | 0.999 |
| K07028; uncharacterized protein | -0.053 | 0.290 | 0.856 | 0.999 |
| K07030; uncharacterized protein | 0.111 | 0.093 | 0.233 | 0.978 |
| K07032; uncharacterized protein | -0.033 | 0.195 | 0.865 | 0.999 |
| K07033; uncharacterized protein | -0.522 | 0.528 | 0.324 | 0.978 |
| K07034; uncharacterized protein | 0.147 | 0.276 | 0.595 | 0.999 |
| K07035; uncharacterized protein | 0.333 | 0.174 | 0.058 | 0.839 |
| K07037; uncharacterized protein | 0.125 | 0.185 | 0.499 | 0.981 |
| K07038; inner membrane protein | -0.013 | 0.177 | 0.940 | 0.999 |
| K07039; uncharacterized protein | 0.022 | 0.232 | 0.923 | 0.999 |
| K07040; uncharacterized protein | 0.025 | 0.054 | 0.639 | 0.999 |
| K07041; uncharacterized protein | 0.973 | 1.035 | 0.349 | 0.978 |
| K07043; uncharacterized protein | 0.012 | 0.084 | 0.887 | 0.999 |
| K07044; uncharacterized protein | -0.439 | 0.396 | 0.270 | 0.978 |
| K07045; uncharacterized protein | -0.324 | 0.264 | 0.221 | 0.978 |
| K07046; L-fuconolactonase [EC:3.1.1.-] | -0.260 | 0.353 | 0.463 | 0.979 |
| K07051; uncharacterized protein | -0.457 | 0.619 | 0.461 | 0.979 |
| K07052; uncharacterized protein | -0.012 | 0.106 | 0.912 | 0.999 |
| K07054; uncharacterized protein | -0.022 | 0.117 | 0.851 | 0.999 |
| K07058; membrane protein | -0.022 | 0.041 | 0.591 | 0.999 |
| K07063; uncharacterized protein | 0.768 | 1.418 | 0.589 | 0.999 |
| K07064; uncharacterized protein | 0.450 | 0.526 | 0.394 | 0.978 |
| K07065; uncharacterized protein | -1.016 | 1.414 | 0.473 | 0.979 |
| K07068; uncharacterized protein | -0.856 | 0.480 | 0.076 | 0.855 |
| K07069; uncharacterized protein | 0.986 | 0.446 | 0.029 | 0.839 |
| K07070; uncharacterized protein | 0.080 | 0.241 | 0.739 | 0.999 |
| K07071; uncharacterized protein | -0.165 | 0.155 | 0.287 | 0.978 |
| K07074; uncharacterized protein | 0.244 | 0.399 | 0.542 | 0.990 |
| K07075; uncharacterized protein | 0.067 | 0.230 | 0.769 | 0.999 |
| K07076; uncharacterized protein | 0.216 | 0.733 | 0.769 | 0.999 |
| K07077; uncharacterized protein | -0.925 | 0.428 | 0.032 | 0.839 |
| K07078; uncharacterized protein | 0.184 | 0.210 | 0.382 | 0.978 |
| K07079; uncharacterized protein | -0.507 | 0.552 | 0.360 | 0.978 |
| K07080; uncharacterized protein | -0.143 | 0.213 | 0.503 | 0.981 |
| K07082; UPF0755 protein | 0.027 | 0.040 | 0.488 | 0.979 |
| K07085; putative transport protein | 0.099 | 0.161 | 0.542 | 0.990 |
| K07086; uncharacterized protein | -1.904 | 1.211 | 0.118 | 0.917 |
| K07088; uncharacterized protein | 0.135 | 0.073 | 0.068 | 0.839 |
| K07089; uncharacterized protein | -0.019 | 0.111 | 0.865 | 0.999 |
| K07090; uncharacterized protein | -0.083 | 0.083 | 0.320 | 0.978 |
| K07092; uncharacterized protein | -0.732 | 0.897 | 0.416 | 0.978 |
| K07093; uncharacterized protein | -0.146 | 0.249 | 0.558 | 0.992 |
| K07095; uncharacterized protein | 0.071 | 0.092 | 0.447 | 0.978 |
| K07096; uncharacterized protein | -0.442 | 0.875 | 0.614 | 0.999 |
| K07097; uncharacterized protein | 1.116 | 0.499 | 0.027 | 0.839 |
| K07098; uncharacterized protein | -0.109 | 0.105 | 0.300 | 0.978 |
| K07099; uncharacterized protein | 0.036 | 0.221 | 0.870 | 0.999 |
| K07100; putative phosphoribosyl transferase | -0.599 | 0.623 | 0.338 | 0.978 |
| K07101; uncharacterized protein | 0.018 | 0.220 | 0.934 | 0.999 |
| K07105; uncharacterized protein | 0.057 | 0.111 | 0.611 | 0.999 |
| K07108; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K07109; uncharacterized protein | 0.170 | 0.307 | 0.580 | 0.997 |
| K07112; uncharacterized protein | -0.072 | 0.120 | 0.547 | 0.991 |
| K07117; uncharacterized protein | -0.314 | 0.337 | 0.352 | 0.978 |
| K07118; uncharacterized protein | 0.130 | 0.169 | 0.444 | 0.978 |
| K07119; uncharacterized protein | -0.300 | 0.208 | 0.151 | 0.954 |
| K07120; uncharacterized protein | 0.012 | 0.198 | 0.952 | 0.999 |
| K07121; uncharacterized protein | 0.111 | 0.173 | 0.520 | 0.988 |
| K07123; uncharacterized protein | 1.864 | 0.909 | 0.042 | 0.839 |
| K07124; uncharacterized protein | 0.062 | 0.065 | 0.341 | 0.978 |
| K07125; uncharacterized protein | 0.326 | 0.421 | 0.440 | 0.978 |
| K07126; uncharacterized protein | -0.069 | 0.164 | 0.673 | 0.999 |
| K07128; uncharacterized protein | 18.458 | 3309.834 | 0.996 | 0.999 |
| K07131; uncharacterized protein | -0.218 | 0.261 | 0.405 | 0.978 |
| K07133; uncharacterized protein | 0.165 | 0.157 | 0.296 | 0.978 |
| K07136; uncharacterized protein | -0.476 | 0.363 | 0.192 | 0.970 |
| K07137; uncharacterized protein | 0.141 | 0.143 | 0.326 | 0.978 |
| K07138; uncharacterized protein | 0.094 | 0.231 | 0.685 | 0.999 |
| K07139; uncharacterized protein | 0.045 | 0.100 | 0.652 | 0.999 |
| K07140; uncharacterized protein | -0.051 | 0.241 | 0.834 | 0.999 |
| K07146; UPF0176 protein | -0.065 | 0.097 | 0.505 | 0.981 |
| K07148; uncharacterized protein | -0.537 | 0.317 | 0.093 | 0.892 |
| K07149; uncharacterized protein | -0.278 | 0.326 | 0.396 | 0.978 |
| K07150; uncharacterized protein | 0.113 | 0.148 | 0.448 | 0.978 |
| K07157; uncharacterized protein | -0.187 | 0.212 | 0.378 | 0.978 |
| K07158; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K07159; uncharacterized protein | -16.927 | 2639.780 | 0.995 | 0.999 |
| K07160; UPF0271 protein | 0.005 | 0.129 | 0.969 | 0.999 |
| K07161; uncharacterized protein | -0.977 | 0.529 | 0.067 | 0.839 |
| K07164; uncharacterized protein | -0.252 | 0.172 | 0.145 | 0.952 |
| K07166; ACT domain-containing protein | 0.087 | 0.108 | 0.425 | 0.978 |
| K07168; CBS domain-containing membrane protein | -0.084 | 0.243 | 0.730 | 0.999 |
| K07169; FHA domain-containing protein | -0.839 | 0.851 | 0.326 | 0.978 |
| K07176; putative serine/threonine protein kinase | -17.234 | 3077.514 | 0.996 | 0.999 |
| K07177; Lon-like protease | 0.032 | 0.124 | 0.795 | 0.999 |
| K07182; CBS domain-containing protein | 0.032 | 0.276 | 0.909 | 0.999 |
| K07216; hemerythrin | -0.302 | 0.261 | 0.250 | 0.978 |
| K07217; Mn-containing catalase | -0.480 | 0.387 | 0.217 | 0.978 |
| K07219; putative molybdopterin biosynthesis protein | -1.375 | 0.650 | 0.036 | 0.839 |
| K07220; uncharacterized protein | -0.067 | 0.109 | 0.541 | 0.990 |
| K07222; putative flavoprotein involved in K+ transport | -0.164 | 0.248 | 0.508 | 0.981 |
| K07223; putative iron-dependent peroxidase | 0.063 | 0.147 | 0.667 | 0.999 |
| K07228; TrkA domain protein | -0.657 | 0.778 | 0.400 | 0.978 |
| K07231; putative iron-regulated protein | 0.126 | 0.318 | 0.693 | 0.999 |
| K07234; uncharacterized protein involved in response to NO | 0.025 | 0.224 | 0.912 | 0.999 |
| K07244; mgtE-like transporter | -1.859 | 2.280 | 0.416 | 0.978 |
| K07270; glycosyl transferase, family 25 | -0.015 | 0.256 | 0.953 | 0.999 |
| K07276; uncharacterized protein | -0.149 | 0.378 | 0.694 | 0.999 |
| K07280; outer membrane protein | 0.065 | 0.537 | 0.903 | 0.999 |
| K07317; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | -0.492 | 0.629 | 0.435 | 0.978 |
| K07318; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | 0.408 | 0.325 | 0.211 | 0.978 |
| K07336; PKHD-type hydroxylase [EC:1.14.11.-] | -0.025 | 0.222 | 0.912 | 0.999 |
| K07337; penicillin-binding protein activator | -0.502 | 0.396 | 0.207 | 0.976 |
| K07338; uncharacterized protein | 0.099 | 0.325 | 0.760 | 0.999 |
| K07387; putative metalloprotease [EC:3.4.24.-] | -0.299 | 0.233 | 0.201 | 0.972 |
| K07394; SM-20-related protein | 0.076 | 0.269 | 0.779 | 0.999 |
| K07395; putative proteasome-type protease | -0.068 | 0.245 | 0.781 | 0.999 |
| K07396; putative protein-disulfide isomerase | 0.048 | 0.306 | 0.874 | 0.999 |
| K07398; conserved protein with predicted RNA binding PUA domain | -17.234 | 3077.514 | 0.996 | 0.999 |
| K07401; selenoprotein W-related protein | 0.057 | 0.298 | 0.848 | 0.999 |
| K07445; putative DNA methylase | 17.810 | 2393.050 | 0.994 | 0.999 |
| K07449; similar to archaeal holliday junction resolvase and Mrr protein | -1.386 | 1.551 | 0.373 | 0.978 |
| K07450; putative resolvase | -17.416 | 3370.607 | 0.996 | 0.999 |
| K07453; putative restriction endonuclease | 0.776 | 1.090 | 0.477 | 0.979 |
| K07454; putative restriction endonuclease | 0.142 | 0.541 | 0.793 | 0.999 |
| K07457; endonuclease III related protein | -0.089 | 0.289 | 0.759 | 0.999 |
| K07461; putative endonuclease | 0.106 | 0.057 | 0.064 | 0.839 |
| K07463; archaea-specific RecJ-like exonuclease | -16.927 | 2639.780 | 0.995 | 0.999 |
| K07465; putative RecB family exonuclease | -0.036 | 0.311 | 0.907 | 0.999 |
| K07477; translin | -17.833 | 3732.576 | 0.996 | 0.999 |
| K07481; transposase, IS5 family | -0.094 | 0.215 | 0.663 | 0.999 |
| K07482; transposase, IS30 family | -0.094 | 0.459 | 0.839 | 0.999 |
| K07483; transposase | 0.158 | 0.087 | 0.071 | 0.841 |
| K07484; transposase | 0.091 | 0.390 | 0.816 | 0.999 |
| K07485; transposase | 0.570 | 0.277 | 0.041 | 0.839 |
| K07486; transposase | 0.711 | 0.427 | 0.098 | 0.896 |
| K07487; transposase | 1.158 | 0.590 | 0.051 | 0.839 |
| K07488; transposase | 0.053 | 0.366 | 0.885 | 0.999 |
| K07491; putative transposase | 0.070 | 0.137 | 0.610 | 0.999 |
| K07492; putative transposase | -1.171 | 0.957 | 0.223 | 0.978 |
| K07493; putative transposase | -0.854 | 0.891 | 0.339 | 0.978 |
| K07494; putative transposase | 0.506 | 0.458 | 0.270 | 0.978 |
| K07495; putative transposase | -17.817 | 4120.281 | 0.997 | 0.999 |
| K07496; putative transposase | 0.011 | 0.416 | 0.979 | 0.999 |
| K07497; putative transposase | 0.050 | 0.131 | 0.703 | 0.999 |
| K07498; putative transposase | -0.994 | 0.584 | 0.091 | 0.884 |
| K07499; putative transposase | 0.878 | 1.188 | 0.461 | 0.979 |
| K07501; 3’-5’ exonuclease | -0.069 | 0.247 | 0.780 | 0.999 |
| K07504; predicted type IV restriction endonuclease | 0.031 | 0.331 | 0.925 | 0.999 |
| K07506; AraC family transcriptional regulator | -0.015 | 0.245 | 0.951 | 0.999 |
| K07558, cca; tRNA nucleotidyltransferase (CCA-adding enzyme) [EC:2.7.7.72] | -17.234 | 3077.514 | 0.996 | 0.999 |
| K07571; S1 RNA binding domain protein | 0.091 | 0.146 | 0.533 | 0.990 |
| K07572; putative nucleotide binding protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K07575; PUA domain protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K07576; metallo-beta-lactamase family protein | 0.224 | 0.191 | 0.243 | 0.978 |
| K07577; putative mRNA 3-end processing factor | -0.200 | 0.229 | 0.384 | 0.978 |
| K07580; Zn-ribbon RNA-binding protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K07581; RNA-binding protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K07726; putative transcriptional regulator | -0.266 | 0.249 | 0.286 | 0.978 |
| K07727; putative transcriptional regulator | -0.150 | 0.133 | 0.262 | 0.978 |
| K07729; putative transcriptional regulator | -0.083 | 0.130 | 0.526 | 0.989 |
| K07807; uncharacterized protein | 0.799 | 0.419 | 0.059 | 0.839 |
| K07814; putative two-component system response regulator | -0.521 | 0.430 | 0.228 | 0.978 |
| K08303; putative protease [EC:3.4.-.-] | 0.047 | 0.070 | 0.507 | 0.981 |
| K08677; kumamolisin | -1.665 | 0.956 | 0.084 | 0.871 |
| K08884; serine/threonine protein kinase, bacterial [EC:2.7.11.1] | 0.026 | 0.090 | 0.770 | 0.999 |
| K08961; chondroitin-sulfate-ABC endolyase/exolyase [EC:4.2.2.20 4.2.2.21] | -0.668 | 0.517 | 0.199 | 0.970 |
| K08972; putative membrane protein | 0.017 | 0.174 | 0.924 | 0.999 |
| K08973; putative membrane protein | -0.143 | 0.166 | 0.390 | 0.978 |
| K08974; putative membrane protein | 0.101 | 0.130 | 0.436 | 0.978 |
| K08975; putative membrane protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K08976; putative membrane protein | -0.976 | 0.456 | 0.034 | 0.839 |
| K08979; putative membrane protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K08980; putative membrane protein | 18.458 | 3309.834 | 0.996 | 0.999 |
| K08981; putative membrane protein | -0.132 | 0.205 | 0.520 | 0.988 |
| K08982; putative membrane protein | -1.277 | 0.723 | 0.079 | 0.860 |
| K08983; putative membrane protein | 0.184 | 0.395 | 0.643 | 0.999 |
| K08985; putative lipoprotein | 0.247 | 0.336 | 0.464 | 0.979 |
| K08987; putative membrane protein | 0.058 | 0.094 | 0.541 | 0.990 |
| K08988; putative membrane protein | -0.309 | 0.303 | 0.310 | 0.978 |
| K08989; putative membrane protein | -0.111 | 0.172 | 0.521 | 0.988 |
| K08995; putative membrane protein | -0.203 | 0.265 | 0.446 | 0.978 |
| K08998; uncharacterized protein | 0.039 | 0.059 | 0.507 | 0.981 |
| K08999; uncharacterized protein | -0.271 | 0.186 | 0.147 | 0.954 |
| K09003; uncharacterized protein | -1.161 | 0.713 | 0.106 | 0.911 |
| K09004; uncharacterized protein | -0.309 | 0.522 | 0.555 | 0.992 |
| K09005; uncharacterized protein | -0.331 | 0.253 | 0.192 | 0.970 |
| K09009; uncharacterized protein | 0.045 | 0.252 | 0.858 | 0.999 |
| K09116; uncharacterized protein | -17.790 | 2181.954 | 0.994 | 0.999 |
| K09117; uncharacterized protein | -0.029 | 0.105 | 0.782 | 0.999 |
| K09118; uncharacterized protein | 0.112 | 0.252 | 0.656 | 0.999 |
| K09120; uncharacterized protein | -17.414 | 2816.885 | 0.995 | 0.999 |
| K09122; uncharacterized protein | -0.342 | 0.764 | 0.655 | 0.999 |
| K09124; uncharacterized protein | -1.555 | 0.964 | 0.109 | 0.917 |
| K09125; uncharacterized protein | 0.030 | 0.103 | 0.769 | 0.999 |
| K09126; uncharacterized protein | -2.579 | 2.960 | 0.385 | 0.978 |
| K09128; uncharacterized protein | 0.480 | 0.959 | 0.617 | 0.999 |
| K09129; uncharacterized protein | -0.119 | 0.909 | 0.896 | 0.999 |
| K09131; uncharacterized protein | -0.078 | 0.233 | 0.737 | 0.999 |
| K09133; uncharacterized protein | -17.617 | 2415.867 | 0.994 | 0.999 |
| K09137; uncharacterized protein | 0.195 | 0.527 | 0.712 | 0.999 |
| K09138; uncharacterized protein | -0.085 | 0.430 | 0.844 | 0.999 |
| K09139; uncharacterized protein | -17.970 | 4446.494 | 0.997 | 0.999 |
| K09141; uncharacterized protein | 0.930 | 1.193 | 0.437 | 0.978 |
| K09142; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K09143; uncharacterized protein | 1.832 | 1.711 | 0.286 | 0.978 |
| K09144; uncharacterized protein | -2.640 | 2.355 | 0.264 | 0.978 |
| K09145; uncharacterized protein | -0.469 | 0.734 | 0.524 | 0.989 |
| K09146; uncharacterized protein | -0.818 | 0.662 | 0.219 | 0.978 |
| K09148; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K09152; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K09153; uncharacterized protein | 0.115 | 0.147 | 0.436 | 0.978 |
| K09154; uncharacterized protein | 1.390 | 0.984 | 0.160 | 0.954 |
| K09155; uncharacterized protein | 0.166 | 0.113 | 0.142 | 0.951 |
| K09157; uncharacterized protein | 0.108 | 0.101 | 0.284 | 0.978 |
| K09158; uncharacterized protein | 0.073 | 0.152 | 0.631 | 0.999 |
| K09160; uncharacterized protein | -0.027 | 0.173 | 0.876 | 0.999 |
| K09161; uncharacterized protein | 0.364 | 0.455 | 0.425 | 0.978 |
| K09163; uncharacterized protein | -0.937 | 0.698 | 0.181 | 0.961 |
| K09164; uncharacterized protein | 0.001 | 0.342 | 0.999 | 1.000 |
| K09165; uncharacterized protein | -0.043 | 0.241 | 0.859 | 0.999 |
| K09166; uncharacterized protein | -0.606 | 0.752 | 0.422 | 0.978 |
| K09167; uncharacterized protein | -0.097 | 0.201 | 0.630 | 0.999 |
| K09190; uncharacterized protein | -0.153 | 0.376 | 0.684 | 0.999 |
| K09384; uncharacterized protein | 0.240 | 0.436 | 0.582 | 0.999 |
| K09386; uncharacterized protein | -1.794 | 0.634 | 0.005 | 0.839 |
| K09388; uncharacterized protein | -2.099 | 0.942 | 0.027 | 0.839 |
| K09700; uncharacterized protein | 0.466 | 0.837 | 0.579 | 0.997 |
| K09701; uncharacterized protein | -0.095 | 0.212 | 0.653 | 0.999 |
| K09702; uncharacterized protein | -0.536 | 0.447 | 0.232 | 0.978 |
| K09703; uncharacterized protein | -0.838 | 0.909 | 0.358 | 0.978 |
| K09704; uncharacterized protein | -0.096 | 0.144 | 0.506 | 0.981 |
| K09705; uncharacterized protein | -0.266 | 0.304 | 0.383 | 0.978 |
| K09706; uncharacterized protein | -1.886 | 0.944 | 0.047 | 0.839 |
| K09707; uncharacterized protein | -3.092 | 1.855 | 0.098 | 0.896 |
| K09712; uncharacterized protein | 0.077 | 0.244 | 0.753 | 0.999 |
| K09721; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K09722, pps; 4-phosphopantoate—beta-alanine ligase [EC:6.3.2.36] | -17.234 | 3077.514 | 0.996 | 0.999 |
| K09726; uncharacterized protein | -0.516 | 1.041 | 0.621 | 0.999 |
| K09729; uncharacterized protein | 0.484 | 0.225 | 0.033 | 0.839 |
| K09735; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K09736; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K09738; uncharacterized protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K09740; uncharacterized protein | -1.471 | 1.002 | 0.144 | 0.951 |
| K09744; uncharacterized protein | -1.873 | 1.957 | 0.340 | 0.978 |
| K09747; uncharacterized protein | 0.063 | 0.039 | 0.108 | 0.917 |
| K09749; uncharacterized protein | -0.084 | 0.417 | 0.841 | 0.999 |
| K09762; uncharacterized protein | 0.124 | 0.085 | 0.148 | 0.954 |
| K09763; uncharacterized protein | -0.838 | 0.488 | 0.088 | 0.880 |
| K09764; uncharacterized protein | -0.352 | 0.416 | 0.399 | 0.978 |
| K09766; uncharacterized protein | -0.154 | 0.503 | 0.759 | 0.999 |
| K09768; uncharacterized protein | -0.090 | 0.155 | 0.563 | 0.992 |
| K09769; uncharacterized protein | 0.101 | 0.172 | 0.557 | 0.992 |
| K09770; uncharacterized protein | -0.302 | 0.408 | 0.460 | 0.979 |
| K09775; uncharacterized protein | -0.102 | 0.193 | 0.598 | 0.999 |
| K09776; uncharacterized protein | -2.159 | 0.933 | 0.022 | 0.839 |
| K09777; uncharacterized protein | -0.014 | 0.208 | 0.946 | 0.999 |
| K09778; uncharacterized protein | 0.350 | 0.305 | 0.253 | 0.978 |
| K09779; uncharacterized protein | -0.585 | 0.424 | 0.170 | 0.961 |
| K09780; uncharacterized protein | 0.173 | 0.116 | 0.136 | 0.947 |
| K09781; uncharacterized protein | -0.023 | 0.140 | 0.871 | 0.999 |
| K09785; uncharacterized protein | -1.940 | 2.261 | 0.392 | 0.978 |
| K09786; uncharacterized protein | -0.160 | 0.226 | 0.479 | 0.979 |
| K09787; uncharacterized protein | 0.146 | 0.095 | 0.125 | 0.923 |
| K09790; uncharacterized protein | 0.066 | 0.071 | 0.354 | 0.978 |
| K09791; uncharacterized protein | 0.033 | 0.162 | 0.838 | 0.999 |
| K09792; uncharacterized protein | 0.025 | 0.228 | 0.914 | 0.999 |
| K09793; uncharacterized protein | -0.359 | 0.241 | 0.139 | 0.949 |
| K09794; uncharacterized protein | 0.168 | 0.196 | 0.393 | 0.978 |
| K09795; uncharacterized protein | -0.295 | 0.298 | 0.324 | 0.978 |
| K09797; uncharacterized protein | -0.201 | 0.201 | 0.319 | 0.978 |
| K09798; uncharacterized protein | 0.080 | 0.167 | 0.631 | 0.999 |
| K09799; uncharacterized protein | -0.470 | 0.355 | 0.187 | 0.966 |
| K09801; uncharacterized protein | 0.035 | 0.162 | 0.829 | 0.999 |
| K09802; uncharacterized protein | 0.581 | 0.261 | 0.028 | 0.839 |
| K09803; uncharacterized protein | -0.514 | 0.418 | 0.221 | 0.978 |
| K09804; uncharacterized protein | -0.412 | 0.797 | 0.605 | 0.999 |
| K09805; uncharacterized protein | -0.830 | 0.560 | 0.141 | 0.949 |
| K09806; uncharacterized protein | 0.088 | 0.161 | 0.583 | 0.999 |
| K09807; uncharacterized protein | -0.153 | 0.163 | 0.350 | 0.978 |
| K09822; uncharacterized protein | -0.688 | 0.464 | 0.140 | 0.949 |
| K09857; uncharacterized protein | 0.026 | 0.280 | 0.926 | 0.999 |
| K09858; SEC-C motif domain protein | 0.054 | 0.126 | 0.670 | 0.999 |
| K09859; uncharacterized protein | -0.670 | 0.484 | 0.169 | 0.961 |
| K09860; uncharacterized protein | 0.334 | 0.556 | 0.549 | 0.991 |
| K09861; uncharacterized protein | 0.099 | 0.062 | 0.115 | 0.917 |
| K09862; uncharacterized protein | -0.049 | 0.159 | 0.759 | 0.999 |
| K09891; uncharacterized protein | 0.170 | 0.272 | 0.532 | 0.990 |
| K09894; uncharacterized protein | 0.231 | 0.308 | 0.455 | 0.978 |
| K09895; uncharacterized protein | 0.108 | 0.172 | 0.532 | 0.990 |
| K09896; uncharacterized protein | 0.231 | 0.308 | 0.455 | 0.978 |
| K09897; uncharacterized protein | 0.207 | 0.312 | 0.508 | 0.981 |
| K09898; uncharacterized protein | 0.226 | 0.212 | 0.289 | 0.978 |
| K09899; uncharacterized protein | 0.231 | 0.308 | 0.455 | 0.978 |
| K09900; uncharacterized protein | 0.253 | 0.334 | 0.450 | 0.978 |
| K09901; uncharacterized protein | 0.208 | 0.312 | 0.505 | 0.981 |
| K09902; uncharacterized protein | 0.109 | 0.172 | 0.528 | 0.989 |
| K09904; uncharacterized protein | 0.231 | 0.308 | 0.455 | 0.978 |
| K09907; uncharacterized protein | 0.207 | 0.312 | 0.508 | 0.981 |
| K09908; uncharacterized protein | 0.215 | 0.213 | 0.314 | 0.978 |
| K09909; uncharacterized protein | 0.119 | 0.262 | 0.649 | 0.999 |
| K09910; uncharacterized protein | 0.231 | 0.308 | 0.455 | 0.978 |
| K09911; uncharacterized protein | 0.206 | 0.312 | 0.510 | 0.981 |
| K09912; uncharacterized protein | 0.066 | 0.221 | 0.766 | 0.999 |
| K09914; putative lipoprotein | -0.352 | 0.330 | 0.287 | 0.978 |
| K09915; uncharacterized protein | 0.037 | 0.235 | 0.875 | 0.999 |
| K09916; uncharacterized protein | 0.120 | 0.249 | 0.630 | 0.999 |
| K09917; uncharacterized protein | 0.206 | 0.312 | 0.510 | 0.981 |
| K09918; uncharacterized protein | 0.246 | 0.310 | 0.428 | 0.978 |
| K09919; uncharacterized protein | -0.149 | 0.214 | 0.489 | 0.979 |
| K09920; uncharacterized protein | 0.125 | 0.255 | 0.624 | 0.999 |
| K09921; uncharacterized protein | 0.137 | 0.184 | 0.458 | 0.979 |
| K09922; uncharacterized protein | -0.812 | 0.285 | 0.005 | 0.839 |
| K09923; uncharacterized protein | 0.215 | 0.214 | 0.317 | 0.978 |
| K09924; uncharacterized protein | -0.089 | 0.197 | 0.651 | 0.999 |
| K09925; uncharacterized protein | -0.094 | 0.332 | 0.778 | 0.999 |
| K09926; uncharacterized protein | 0.112 | 0.233 | 0.633 | 0.999 |
| K09927; uncharacterized protein | -0.176 | 0.232 | 0.450 | 0.978 |
| K09928; uncharacterized protein | -0.098 | 0.237 | 0.679 | 0.999 |
| K09929; uncharacterized protein | 0.144 | 0.252 | 0.569 | 0.995 |
| K09930; uncharacterized protein | 0.002 | 0.192 | 0.992 | 0.999 |
| K09931; uncharacterized protein | -0.371 | 0.441 | 0.402 | 0.978 |
| K09932; uncharacterized protein | 0.177 | 0.309 | 0.569 | 0.995 |
| K09934; uncharacterized protein | -0.482 | 0.770 | 0.532 | 0.990 |
| K09935; uncharacterized protein | 0.083 | 0.330 | 0.801 | 0.999 |
| K09937; uncharacterized protein | -0.071 | 0.284 | 0.802 | 0.999 |
| K09938; uncharacterized protein | 0.141 | 0.267 | 0.598 | 0.999 |
| K09939; uncharacterized protein | -0.204 | 0.302 | 0.502 | 0.981 |
| K09940; uncharacterized protein | -0.022 | 0.203 | 0.916 | 0.999 |
| K09941; uncharacterized protein | -0.012 | 0.292 | 0.968 | 0.999 |
| K09942; uncharacterized protein | -0.945 | 1.218 | 0.439 | 0.978 |
| K09943; uncharacterized protein | -0.083 | 0.310 | 0.790 | 0.999 |
| K09944; uncharacterized protein | -0.428 | 0.802 | 0.595 | 0.999 |
| K09945; uncharacterized protein | 0.157 | 0.295 | 0.595 | 0.999 |
| K09946; uncharacterized protein | 0.027 | 0.218 | 0.900 | 0.999 |
| K09947; uncharacterized protein | 0.071 | 0.315 | 0.823 | 0.999 |
| K09948; uncharacterized protein | -0.013 | 0.286 | 0.962 | 0.999 |
| K09949; uncharacterized protein | 0.299 | 0.197 | 0.130 | 0.934 |
| K09950; uncharacterized protein | 0.089 | 0.268 | 0.740 | 0.999 |
| K09954; uncharacterized protein | 0.107 | 0.250 | 0.670 | 0.999 |
| K09955; uncharacterized protein | -1.083 | 0.471 | 0.023 | 0.839 |
| K09956; uncharacterized protein | -0.167 | 0.270 | 0.537 | 0.990 |
| K09957; uncharacterized protein | -0.628 | 0.612 | 0.306 | 0.978 |
| K09958; uncharacterized protein | -0.043 | 0.234 | 0.853 | 0.999 |
| K09959; uncharacterized protein | -1.161 | 0.400 | 0.004 | 0.839 |
| K09960; uncharacterized protein | -0.575 | 0.741 | 0.439 | 0.978 |
| K09961; uncharacterized protein | -0.355 | 0.710 | 0.618 | 0.999 |
| K09962; uncharacterized protein | -0.119 | 0.137 | 0.387 | 0.978 |
| K09963; uncharacterized protein | 0.221 | 0.225 | 0.329 | 0.978 |
| K09964; uncharacterized protein | -0.006 | 0.303 | 0.985 | 0.999 |
| K09965; uncharacterized protein | -0.244 | 0.339 | 0.473 | 0.979 |
| K09966; uncharacterized protein | -0.403 | 0.349 | 0.250 | 0.978 |
| K09967; uncharacterized protein | -0.113 | 0.269 | 0.675 | 0.999 |
| K09968; uncharacterized protein | -1.850 | 1.129 | 0.103 | 0.904 |
| K09973; uncharacterized protein | -0.312 | 0.246 | 0.207 | 0.976 |
| K09974; uncharacterized protein | 0.158 | 0.280 | 0.574 | 0.995 |
| K09975; uncharacterized protein | -0.097 | 0.165 | 0.555 | 0.992 |
| K09976; uncharacterized protein | 0.000 | 0.133 | 0.998 | 1.000 |
| K09977; uncharacterized protein | 0.154 | 0.229 | 0.504 | 0.981 |
| K09978; uncharacterized protein | 0.219 | 0.315 | 0.488 | 0.979 |
| K09979; uncharacterized protein | 0.195 | 0.266 | 0.466 | 0.979 |
| K09980; uncharacterized protein | 0.206 | 0.508 | 0.686 | 0.999 |
| K09981; uncharacterized protein | -0.009 | 0.395 | 0.982 | 0.999 |
| K09982; uncharacterized protein | 0.028 | 0.411 | 0.946 | 0.999 |
| K09983; uncharacterized protein | -0.804 | 0.378 | 0.035 | 0.839 |
| K09984; uncharacterized protein | 0.071 | 0.300 | 0.814 | 0.999 |
| K09985; uncharacterized protein | -1.205 | 0.444 | 0.007 | 0.839 |
| K09986; uncharacterized protein | -0.168 | 0.241 | 0.487 | 0.979 |
| K09987; uncharacterized protein | -1.205 | 0.444 | 0.007 | 0.839 |
| K09989; uncharacterized protein | -0.149 | 0.226 | 0.512 | 0.982 |
| K09990; uncharacterized protein | -0.998 | 0.542 | 0.068 | 0.839 |
| K09991; uncharacterized protein | -1.096 | 0.432 | 0.012 | 0.839 |
| K09992; uncharacterized protein | -0.725 | 0.445 | 0.105 | 0.910 |
| K10120, msmE; fructooligosaccharide transport system substrate-binding protein | 0.789 | 0.408 | 0.055 | 0.839 |
| K10121, msmF; fructooligosaccharide transport system permease protein | 0.811 | 0.413 | 0.051 | 0.839 |
| K10122, msmG; fructooligosaccharide transport system permease protein | 0.811 | 0.413 | 0.051 | 0.839 |
| K10212, crtO; glycosyl-4,4’-diaponeurosporenoate acyltransferase [EC:2.3.1.-] | -1.551 | 1.058 | 0.145 | 0.951 |
| K10253; DOPA 4,5-dioxygenase [EC:1.14.99.-] | 0.108 | 0.283 | 0.703 | 0.999 |
| K10907; aminotransferase [EC:2.6.1.-] | 0.102 | 0.197 | 0.607 | 0.999 |
| K11145; ribonuclease III family protein [EC:3.1.26.-] | 0.124 | 0.095 | 0.195 | 0.970 |
| K11159; carotenoid cleavage dioxygenase | -0.939 | 0.405 | 0.022 | 0.839 |
| K11312; cupin 2 domain-containing protein | 0.687 | 0.312 | 0.029 | 0.839 |
| K11325; L-cysteine/cystine lyase | -1.368 | 1.361 | 0.316 | 0.978 |
| K11442; putative uridylyltransferase [EC:2.7.7.-] | -0.198 | 0.832 | 0.812 | 0.999 |
| K11476, gntR; GntR family transcriptional regulator, gluconate operon transcriptional repressor | -0.657 | 1.216 | 0.590 | 0.999 |
| K11527; two-component system, sensor histidine kinase and response regulator [EC:2.7.13.3] | -1.104 | 0.594 | 0.065 | 0.839 |
| K11638, citT; two-component system, CitB family, response regulator CitT | -2.116 | 1.208 | 0.082 | 0.865 |
| K11646; 3-dehydroquinate synthase II [EC:1.4.1.24] | 3.014 | 2.517 | 0.233 | 0.978 |
| K11691, dctS; two-component system, CitB family, sensor histidine kinase DctS [EC:2.7.13.3] | -1.135 | 1.118 | 0.312 | 0.978 |
| K11692, dctR; two-component system, CitB family, response regulator DctR | -0.507 | 0.741 | 0.495 | 0.981 |
| K11905; type VI secretion system protein | 0.369 | 0.250 | 0.142 | 0.951 |
| K11954, natB; neutral amino acid transport system substrate-binding protein | -17.763 | 3362.856 | 0.996 | 0.999 |
| K11957, natA; neutral amino acid transport system ATP-binding protein | -17.763 | 3362.856 | 0.996 | 0.999 |
| K12055, parA; chromosome partitioning related protein ParA | -0.045 | 0.613 | 0.941 | 0.999 |
| K13522, nadM; bifunctional NMN adenylyltransferase/nudix hydrolase [EC:2.7.7.1 3.6.1.-] | -0.445 | 0.423 | 0.294 | 0.978 |
| K13580; magnesium chelatase subunit ChlD-like protein | 0.247 | 0.337 | 0.464 | 0.979 |
| K13652; AraC family transcriptional regulator | -0.741 | 0.439 | 0.093 | 0.892 |
| K13653; AraC family transcriptional regulator | 0.111 | 0.107 | 0.303 | 0.978 |
| K13671; alpha-1,2-mannosyltransferase [EC:2.4.1.-] | -0.054 | 0.356 | 0.880 | 0.999 |
| K13819; NifU-like protein | -0.447 | 0.766 | 0.561 | 0.992 |
| K13874, araB; L-arabinonolactonase [EC:3.1.1.15] | -0.755 | 1.314 | 0.566 | 0.995 |
| K13875, araC; L-arabonate dehydrase [EC:4.2.1.25] | -1.441 | 1.293 | 0.267 | 0.978 |
| K14136; decaprenyl-phosphate phosphoribosyltransferase [EC:2.4.2.45] | -0.412 | 0.588 | 0.484 | 0.979 |
| K14165; atypical dual specificity phosphatase [EC:3.1.3.16 3.1.3.48] | 2.183 | 1.908 | 0.254 | 0.978 |
| K14331; fatty aldehyde decarbonylase [EC:4.1.99.5] | -0.976 | 0.998 | 0.329 | 0.978 |
| K14340; mannosyltransferase [EC:2.4.1.-] | 1.102 | 0.729 | 0.133 | 0.942 |
| K14486, ARF; auxin response factor | -16.380 | 2008.024 | 0.994 | 0.999 |
| K14645; serine protease [EC:3.4.21.-] | 0.090 | 0.308 | 0.771 | 0.999 |
| K14728; phthiodiolone/phenolphthiodiolone dimycocerosates ketoreductase [EC:1.2.-.-] | -1.627 | 1.060 | 0.127 | 0.927 |
| K15016; enoyl-CoA hydratase / 3-hydroxyacyl-CoA dehydrogenase [EC:4.2.1.17 1.1.1.35] | -1.070 | 1.632 | 0.513 | 0.982 |
| K15019; 3-hydroxypropionyl-coenzyme A dehydratase [EC:4.2.1.116] | 0.636 | 0.916 | 0.488 | 0.979 |
| K15024; putative phosphotransacetylase [EC:2.3.1.8] | -0.791 | 0.633 | 0.213 | 0.978 |
| K15383; MtN3 and saliva related transmembrane protein | 0.063 | 0.369 | 0.864 | 0.999 |
| K15527; cysteate synthase [EC:2.5.1.76] | -18.213 | 3786.487 | 0.996 | 0.999 |
| K15533; 1,3-beta-galactosyl-N-acetylhexosamine phosphorylase [EC:2.4.1.211] | 0.275 | 0.427 | 0.521 | 0.988 |
| K15640, phoE; uncharacterized phosphatase | -2.077 | 0.911 | 0.024 | 0.839 |
| K15667, ppsD, fenA; fengycin family lipopeptide synthetase D | -1.438 | 1.843 | 0.437 | 0.978 |
| K15878, narB; rieske iron-sulphur protein | -3.588 | 4.783 | 0.454 | 0.978 |
| K15975; glyoxalase family protein | -0.370 | 0.371 | 0.321 | 0.978 |
| K15976; putative NAD(P)H nitroreductase [EC:1.-.-.-] | -0.676 | 1.197 | 0.573 | 0.995 |
| K15977; putative oxidoreductase | -0.095 | 0.144 | 0.509 | 0.981 |
| K16149; 1,4-alpha-glucan branching enzyme [EC:2.4.1.18] | -0.204 | 0.395 | 0.607 | 0.999 |
| K16150; glycogen synthase [EC:2.4.1.11] | -1.418 | 0.992 | 0.155 | 0.954 |
| K16153; glycogen phosphorylase/synthase [EC:2.4.1.1 2.4.1.11] | 0.074 | 0.437 | 0.866 | 0.999 |
| K16163; maleylpyruvate isomerase [EC:5.2.1.4] | -0.568 | 0.959 | 0.554 | 0.992 |
| K16164; acylpyruvate hydrolase [EC:3.7.1.5] | 0.203 | 0.376 | 0.590 | 0.999 |
| K16191, arfA; peptidoglycan-binding protein ArfA | -1.140 | 1.201 | 0.344 | 0.978 |
| K16192, arfB; uncharacterized membrane protein ArfB | 0.787 | 1.240 | 0.527 | 0.989 |
| K16306; fructose-bisphosphate aldolase / 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate synthase [EC:4.1.2.13 2.2.1.10] | -17.234 | 3077.514 | 0.996 | 0.999 |
| K16327; putative LysE/RhtB family amino acid efflux pump | -17.644 | 2866.856 | 0.995 | 0.999 |
| K16637, exoY; adenylate cyclase ExoY | -0.976 | 0.998 | 0.329 | 0.978 |
| K16638, exoU; exoenzyme U | -0.976 | 0.998 | 0.329 | 0.978 |
| K16654; spore-specific protein | -2.453 | 2.237 | 0.275 | 0.978 |
| K16881; mannose-1-phosphate guanylyltransferase / phosphomannomutase [EC:2.7.7.13 5.4.2.8] | -2.257 | 0.984 | 0.023 | 0.839 |
| K16901; anthranilate 3-monooxygenase (FAD) / 4-hydroxyphenylacetate 3-monooxygenase [EC:1.14.14.8 1.14.14.9] | -0.169 | 0.999 | 0.866 | 0.999 |
| K16905; fluoroquinolone transport system permease protein | 0.088 | 0.729 | 0.905 | 0.999 |
| K16906; fluoroquinolone transport system permease protein | 0.088 | 0.729 | 0.905 | 0.999 |
| K16907; fluoroquinolone transport system ATP-binding protein [EC:3.6.3.-] | 0.088 | 0.729 | 0.905 | 0.999 |
| K16929; energy-coupling factor transport system substrate-specific component | -18.129 | 4509.872 | 0.997 | 0.999 |
| K17076, lysY; putative lysine transport system ATP-binding protein [EC:3.6.3.-] | 0.040 | 0.134 | 0.764 | 0.999 |
| K17202, eryG; erythritol transport system substrate-binding protein | -2.415 | 1.241 | 0.053 | 0.839 |
| K17203, eryF; erythritol transport system permease protein | -2.420 | 1.237 | 0.052 | 0.839 |
| K17208, ibpA; inositol transport system substrate-binding protein | 0.983 | 0.801 | 0.222 | 0.978 |
| K17213; inositol transport system substrate-binding protein | -0.777 | 0.496 | 0.119 | 0.917 |
| K17214; inositol transport system permease protein | -0.813 | 0.537 | 0.132 | 0.941 |
| K17215; inositol transport system ATP-binding protein | -0.948 | 0.532 | 0.077 | 0.856 |
| K17318, lplA; putative aldouronate transport system substrate-binding protein | 0.107 | 0.154 | 0.488 | 0.979 |
| K17322, glpP; glycerol transport system permease protein | -0.622 | 0.391 | 0.114 | 0.917 |
| K17325, glpT; glycerol transport system ATP-binding protein | -0.629 | 0.390 | 0.109 | 0.917 |
| K18132, porA; major outer membrane protein P.IA | -16.880 | 1816.243 | 0.993 | 0.999 |
| K18133, porB; major outer membrane protein P.IB | 0.177 | 0.456 | 0.699 | 0.999 |
| K18237; ribose 1,5-bisphosphate isomerase [EC:5.3.1.29] | 17.675 | 2236.882 | 0.994 | 0.999 |
| K18284; adenosylhomocysteine/aminodeoxyfutalosine nucleosidase [EC:3.2.2.9 3.2.2.30] | -0.428 | 0.802 | 0.595 | 0.999 |
| K18333; L-fucose dehydrogenase | 0.114 | 0.257 | 0.660 | 0.999 |
| K18335; 2-keto-3-deoxy-L-fuconate dehydrogenase [EC:1.1.1.-] | -0.893 | 0.417 | 0.034 | 0.839 |
| K18336; 2,4-diketo-3-deoxy-L-fuconate hydrolase [EC:3.7.1.-] | -2.939 | 2.989 | 0.327 | 0.978 |
| K18383; trans-feruloyl-CoA hydratase / vanillin synthase [EC:4.2.1.101 4.1.2.41] | -0.442 | 0.526 | 0.403 | 0.978 |
| K18593; 4-hydroxybutyryl-CoA synthetase (ADP-forming) [EC:6.2.1.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| K18594; 3-hydroxypropionyl-CoA synthetase (ADP-forming) [EC:6.2.1.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| K18601; aldehyde dehydrogenase [EC:1.2.1.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| K18602; malonic semialdehyde reductase [EC:1.1.1.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| K18603; acetyl-CoA/propionyl-CoA carboxylase [EC:6.4.1.2 6.4.1.3] | -17.234 | 3077.514 | 0.996 | 0.999 |
| K18604; acetyl-CoA/propionyl-CoA carboxylase [EC:6.4.1.2 6.4.1.3 2.1.3.15] | -17.234 | 3077.514 | 0.996 | 0.999 |
| K18605; biotin carboxyl carrier protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| K18611; 4-pyridoxate dehydrogenase [EC:1.1.1.-] | 18.788 | 3263.010 | 0.995 | 0.999 |
| K18612; 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylate dehydrogenase [EC:1.2.1.-] | 18.076 | 2733.666 | 0.995 | 0.999 |
| K18613; 3-hydroxy-2-methylpyridine-4,5-dicarboxylate 4-decarboxylase [EC:4.1.1.51] | 18.076 | 2733.666 | 0.995 | 0.999 |
| K18614; 2-(acetamidomethylene)succinate hydrolase [EC:3.5.1.29] | 18.076 | 2733.666 | 0.995 | 0.999 |
| KAE1, tsaD, QRI7; N6-L-threonylcarbamoyladenine synthase [EC:2.3.1.234] | 0.058 | 0.047 | 0.215 | 0.978 |
| kaiB; circadian clock protein KaiB | -17.699 | 3101.861 | 0.995 | 0.999 |
| kaiC; circadian clock protein KaiC | -0.194 | 0.349 | 0.579 | 0.997 |
| kal; 3-aminobutyryl-CoA ammonia-lyase [EC:4.3.1.14] | 0.871 | 0.432 | 0.045 | 0.839 |
| kamA; lysine 2,3-aminomutase [EC:5.4.3.2] | 0.240 | 0.317 | 0.449 | 0.978 |
| kamD; beta-lysine 5,6-aminomutase alpha subunit [EC:5.4.3.3] | 0.954 | 0.451 | 0.036 | 0.839 |
| kamE; beta-lysine 5,6-aminomutase beta subunit [EC:5.4.3.3] | 0.954 | 0.451 | 0.036 | 0.839 |
| kapB; kinase-associated protein B | -0.669 | 0.631 | 0.291 | 0.978 |
| kapD; sporulation inhibitor KapD | -2.853 | 1.593 | 0.075 | 0.855 |
| KARS, lysS; lysyl-tRNA synthetase, class II [EC:6.1.1.6] | 0.054 | 0.040 | 0.174 | 0.961 |
| kas; beta-ketoacyl ACP synthase [EC:2.3.1.-] | -2.272 | 1.310 | 0.085 | 0.873 |
| katE, CAT, catB, srpA; catalase [EC:1.11.1.6] | -0.188 | 0.157 | 0.232 | 0.978 |
| katG; catalase-peroxidase [EC:1.11.1.21] | -0.206 | 0.231 | 0.372 | 0.978 |
| kauB; 4-guanidinobutyraldehyde dehydrogenase / NAD-dependent aldehyde dehydrogenase [EC:1.2.1.54 1.2.1.-] | 0.204 | 0.318 | 0.522 | 0.988 |
| kbaA; KinB signaling pathway activation protein | -1.543 | 0.758 | 0.044 | 0.839 |
| kbl, GCAT; glycine C-acetyltransferase [EC:2.3.1.29] | -0.171 | 0.138 | 0.215 | 0.978 |
| kce; 3-keto-5-aminohexanoate cleavage enzyme [EC:2.3.1.247] | 0.943 | 0.450 | 0.038 | 0.839 |
| kch, trkA, mthK, pch; voltage-gated potassium channel | -0.083 | 0.083 | 0.321 | 0.978 |
| kdd; L-erythro-3,5-diaminohexanoate dehydrogenase [EC:1.4.1.11] | 0.941 | 0.446 | 0.036 | 0.839 |
| kdgD; 5-dehydro-4-deoxyglucarate dehydratase [EC:4.2.1.41] | -0.182 | 0.353 | 0.607 | 0.999 |
| kdgK; 2-dehydro-3-deoxygluconokinase [EC:2.7.1.45] | -0.098 | 0.139 | 0.481 | 0.979 |
| kdgR; IclR family transcriptional regulator, KDG regulon repressor | 0.350 | 0.514 | 0.497 | 0.981 |
| kdgR; LacI family transcriptional regulator, kdg operon repressor | 0.146 | 0.440 | 0.741 | 0.999 |
| kdgT; 2-keto-3-deoxygluconate permease | -0.140 | 0.355 | 0.695 | 0.999 |
| kdhA; 6-hydroxypseudooxynicotine dehydrogenase subunit alpha [EC:1.5.99.14] | -2.224 | 2.044 | 0.278 | 0.978 |
| kdkA; 3-deoxy-D-manno-octulosonic acid kinase [EC:2.7.1.166] | 0.129 | 0.259 | 0.620 | 0.999 |
| KDM8, JMJD5; lysine-specific demethylase 8 [EC:1.14.11.27] | -17.416 | 3370.607 | 0.996 | 0.999 |
| kdnA; 8-amino-3,8-dideoxy-alpha-D-manno-octulosonate transaminase [EC:2.6.1.109] | 0.373 | 1.208 | 0.758 | 0.999 |
| kdnB; 3-deoxy-alpha-D-manno-octulosonate 8-oxidase [EC:1.1.3.48] | -0.129 | 1.300 | 0.921 | 0.999 |
| kdpA; K+-transporting ATPase ATPase A chain [EC:3.6.3.12] | -0.241 | 0.174 | 0.168 | 0.961 |
| kdpB; K+-transporting ATPase ATPase B chain [EC:3.6.3.12] | -0.224 | 0.186 | 0.230 | 0.978 |
| kdpC; K+-transporting ATPase ATPase C chain [EC:3.6.3.12] | -0.243 | 0.174 | 0.165 | 0.960 |
| kdpD; two-component system, OmpR family, sensor histidine kinase KdpD [EC:2.7.13.3] | -0.323 | 0.195 | 0.101 | 0.896 |
| kdpE; two-component system, OmpR family, KDP operon response regulator KdpE | -0.249 | 0.175 | 0.157 | 0.954 |
| kdpF; K+-transporting ATPase ATPase F chain | -0.047 | 0.440 | 0.915 | 0.999 |
| kdsA; 2-dehydro-3-deoxyphosphooctonate aldolase (KDO 8-P synthase) [EC:2.5.1.55] | 0.076 | 0.090 | 0.399 | 0.978 |
| kdsB; 3-deoxy-manno-octulosonate cytidylyltransferase (CMP-KDO synthetase) [EC:2.7.7.38] | 0.066 | 0.090 | 0.465 | 0.979 |
| kdsC; 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (KDO 8-P phosphatase) [EC:3.1.3.45] | 0.107 | 0.096 | 0.265 | 0.978 |
| kdsD, kpsF; arabinose-5-phosphate isomerase [EC:5.3.1.13] | 0.078 | 0.090 | 0.386 | 0.978 |
| kdtA, waaA; 3-deoxy-D-manno-octulosonic-acid transferase [EC:2.4.99.12 2.4.99.13 2.4.99.14 2.4.99.15] | -0.032 | 0.090 | 0.718 | 0.999 |
| kduD; 2-dehydro-3-deoxy-D-gluconate 5-dehydrogenase [EC:1.1.1.127] | -0.407 | 0.340 | 0.234 | 0.978 |
| kduI; 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase [EC:5.3.1.17] | 0.111 | 0.264 | 0.676 | 0.999 |
| kdxD; 2-dehydro-3-deoxy-D-arabinonate dehydratase [EC:4.2.1.141] | 0.280 | 1.055 | 0.791 | 0.999 |
| kefB; glutathione-regulated potassium-efflux system protein KefB | -0.050 | 0.185 | 0.788 | 0.999 |
| kefC; glutathione-regulated potassium-efflux system ancillary protein KefC | -0.402 | 0.448 | 0.371 | 0.978 |
| kefF; glutathione-regulated potassium-efflux system ancillary protein KefF | -0.122 | 0.402 | 0.761 | 0.999 |
| kefG; glutathione-regulated potassium-efflux system ancillary protein KefG | -0.130 | 0.364 | 0.721 | 0.999 |
| kgd; 2-oxoglutarate decarboxylase [EC:4.1.1.71] | -0.384 | 0.602 | 0.525 | 0.989 |
| kgp; gingipain K [EC:3.4.22.47] | -17.939 | 2431.100 | 0.994 | 0.999 |
| kgtP; MFS transporter, MHS family, alpha-ketoglutarate permease | -0.231 | 0.223 | 0.303 | 0.978 |
| kguK; dehydrogluconokinase [EC:2.7.1.13] | 0.011 | 0.432 | 0.979 | 0.999 |
| kinA; two-component system, sporulation sensor kinase A [EC:2.7.13.3] | -1.829 | 1.029 | 0.077 | 0.856 |
| kinB; two-component system, NtrC family, sensor histidine kinase KinB [EC:2.7.13.3] | 0.318 | 0.354 | 0.369 | 0.978 |
| kinB; two-component system, sporulation sensor kinase B [EC:2.7.13.3] | -2.788 | 1.210 | 0.023 | 0.839 |
| kinC; two-component system, sporulation sensor kinase C [EC:2.7.13.3] | -2.412 | 1.515 | 0.113 | 0.917 |
| kinD; two-component system, sporulation sensor kinase D [EC:2.7.13.3] | -1.364 | 1.344 | 0.312 | 0.978 |
| kinE; two-component system, sporulation sensor kinase E [EC:2.7.13.3] | -1.440 | 0.897 | 0.110 | 0.917 |
| kipA; antagonist of KipI | -2.198 | 0.720 | 0.003 | 0.839 |
| kipI; inhibitor of KinA | -2.376 | 0.866 | 0.007 | 0.839 |
| KMO; kynurenine 3-monooxygenase [EC:1.14.13.9] | -0.038 | 0.444 | 0.933 | 0.999 |
| kojP; kojibiose phosphorylase [EC:2.4.1.230] | 0.902 | 0.693 | 0.195 | 0.970 |
| korA, oorA, oforA; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit alpha [EC:1.2.7.3 1.2.7.11] | -0.215 | 0.155 | 0.167 | 0.961 |
| korB, oorB, oforB; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit beta [EC:1.2.7.3 1.2.7.11] | -0.221 | 0.153 | 0.152 | 0.954 |
| korC, oorC; 2-oxoglutarate ferredoxin oxidoreductase subunit gamma [EC:1.2.7.3] | -0.146 | 0.232 | 0.530 | 0.989 |
| korD, oorD; 2-oxoglutarate ferredoxin oxidoreductase subunit delta [EC:1.2.7.3] | -0.144 | 0.229 | 0.530 | 0.989 |
| kpsC, lipA; capsular polysaccharide export protein | 0.114 | 0.362 | 0.753 | 0.999 |
| kpsS, lipB; capsular polysaccharide export protein | -0.060 | 0.383 | 0.876 | 0.999 |
| kptA; putative RNA 2’-phosphotransferase [EC:2.7.1.-] | 0.778 | 0.311 | 0.013 | 0.839 |
| KRR1; ribosomal RNA assembly protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| ksgA; 16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase [EC:2.1.1.182] | 0.037 | 0.036 | 0.308 | 0.978 |
| kshA; 3-ketosteroid 9alpha-monooxygenase subunit A [EC:1.14.13.142] | -1.369 | 1.252 | 0.276 | 0.978 |
| kshB; 3-ketosteroid 9alpha-monooxygenase subunit B [EC:1.14.13.142] | -1.058 | 0.641 | 0.101 | 0.896 |
| kstD; 3-oxosteroid 1-dehydrogenase [EC:1.3.99.4] | -0.967 | 0.852 | 0.258 | 0.978 |
| ku; DNA end-binding protein Ku | -0.260 | 0.207 | 0.209 | 0.978 |
| kup; KUP system potassium uptake protein | -0.093 | 0.134 | 0.489 | 0.979 |
| kynB; arylformamidase [EC:3.5.1.9] | -1.221 | 0.450 | 0.007 | 0.839 |
| KYNU, kynU; kynureninase [EC:3.7.1.3] | -0.332 | 0.280 | 0.237 | 0.978 |
| L2HGDH; 2-hydroxyglutarate dehydrogenase [EC:1.1.99.2] | -1.306 | 1.512 | 0.389 | 0.978 |
| L3HYPDH; trans-L-3-hydroxyproline dehydratase [EC:4.2.1.77] | -0.054 | 0.828 | 0.948 | 0.999 |
| laaA; L-proline amide hydrolase [EC:3.5.1.101] | -0.537 | 0.648 | 0.408 | 0.978 |
| lacA; galactoside O-acetyltransferase [EC:2.3.1.18] | -0.109 | 0.257 | 0.672 | 0.999 |
| lacC; tagatose 6-phosphate kinase [EC:2.7.1.144] | 0.068 | 0.151 | 0.652 | 0.999 |
| lacD; tagatose 1,6-diphosphate aldolase [EC:4.1.2.40] | 0.111 | 0.142 | 0.435 | 0.978 |
| lacE, araN; lactose/L-arabinose transport system substrate-binding protein | 0.389 | 0.540 | 0.472 | 0.979 |
| lacF, araP; lactose/L-arabinose transport system permease protein | 0.326 | 0.540 | 0.547 | 0.991 |
| lacG, araQ; lactose/L-arabinose transport system permease protein | 0.341 | 0.541 | 0.529 | 0.989 |
| lacI, galR; LacI family transcriptional regulator | 0.003 | 0.069 | 0.969 | 0.999 |
| lacK; lactose/L-arabinose transport system ATP-binding protein | -1.601 | 0.942 | 0.091 | 0.885 |
| lacR; DeoR family transcriptional regulator, lactose phosphotransferase system repressor | -0.050 | 0.152 | 0.742 | 0.999 |
| lacS, galP, rafP; lactose/raffinose/galactose permease | 0.466 | 0.328 | 0.158 | 0.954 |
| lacT; transcriptional antiterminator | 0.005 | 0.167 | 0.976 | 0.999 |
| lacY; MFS transporter, OHS family, lactose permease | -0.005 | 0.175 | 0.977 | 0.999 |
| lacZ; beta-galactosidase [EC:3.2.1.23] | -0.023 | 0.135 | 0.866 | 0.999 |
| lamB; maltoporin | -0.304 | 0.305 | 0.320 | 0.978 |
| lapA; lipopolysaccharide assembly protein A | 0.128 | 0.262 | 0.626 | 0.999 |
| lapA; surface adhesion protein | -0.150 | 0.534 | 0.779 | 0.999 |
| lapB; ATP-binding cassette, subfamily C, bacterial LapB | 0.163 | 0.253 | 0.519 | 0.988 |
| lapC; membrane fusion protein, adhesin transport system | 0.100 | 0.298 | 0.736 | 0.999 |
| lapE; outer membrane protein, adhesin transport system | 0.195 | 0.227 | 0.392 | 0.978 |
| larB; pyridinium-3,5-biscarboxylic acid mononucleotide synthase [EC:2.5.1.143] | 0.097 | 0.206 | 0.638 | 0.999 |
| larC; pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickel chelatase [EC:4.99.1.12] | 0.146 | 0.212 | 0.490 | 0.980 |
| larE; pyridinium-3,5-biscarboxylic acid mononucleotide sulfurtransferase | 0.077 | 0.199 | 0.701 | 0.999 |
| LARS, leuS; leucyl-tRNA synthetase [EC:6.1.1.4] | 0.025 | 0.039 | 0.522 | 0.989 |
| lasA; LasA protease [EC:3.4.24.-] | -17.760 | 4004.372 | 0.996 | 0.999 |
| lasB; pseudolysin [EC:3.4.24.26] | -17.824 | 2205.841 | 0.994 | 0.999 |
| lasI, luxI; acyl homoserine lactone synthase [EC:2.3.1.184] | -0.105 | 0.455 | 0.818 | 0.999 |
| lasR; LuxR family transcriptional regulator, quorum-sensing system regulator LasR | -0.347 | 0.595 | 0.561 | 0.992 |
| lasT; tRNA/rRNA methyltransferase [EC:2.1.1.-] | -0.458 | 0.232 | 0.050 | 0.839 |
| lat; L-lysine 6-transaminase [EC:2.6.1.36] | -0.880 | 0.726 | 0.227 | 0.978 |
| lcdH, cdhA; carnitine 3-dehydrogenase [EC:1.1.1.108] | 0.077 | 0.352 | 0.827 | 0.999 |
| lctB; potassium channel LctB | -2.295 | 0.912 | 0.013 | 0.839 |
| lctO; L-lactate oxidase [EC:1.1.3.2] | -0.117 | 0.229 | 0.609 | 0.999 |
| lctP; lactate permease | 0.009 | 0.122 | 0.942 | 0.999 |
| lcyB, crtL1, crtY; lycopene beta-cyclase [EC:5.5.1.19] | -0.426 | 0.440 | 0.333 | 0.978 |
| ldcA; muramoyltetrapeptide carboxypeptidase [EC:3.4.17.13] | 0.052 | 0.134 | 0.701 | 0.999 |
| LDH, ldh; L-lactate dehydrogenase [EC:1.1.1.27] | 0.208 | 0.150 | 0.168 | 0.961 |
| ldhA; D-lactate dehydrogenase [EC:1.1.1.28] | 0.345 | 0.177 | 0.053 | 0.839 |
| LDHD, dld; D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4] | -1.232 | 0.345 | 0.000 | 0.839 |
| ldrA_B_C_D; small toxic polypeptide LdrA/B/C/D | -0.974 | 1.366 | 0.477 | 0.979 |
| legF, ptmB; CMP-N,N’-diacetyllegionaminic acid synthase [EC:2.7.7.82] | -0.431 | 0.647 | 0.506 | 0.981 |
| legG, neuC2; GDP/UDP-N,N’-diacetylbacillosamine 2-epimerase (hydrolysing) [EC:3.2.1.184] | -0.482 | 0.763 | 0.529 | 0.989 |
| legI, neuB2; N,N’-diacetyllegionaminate synthase [EC:2.5.1.101] | -0.375 | 0.668 | 0.575 | 0.995 |
| lemA; LemA protein | 0.048 | 0.054 | 0.371 | 0.978 |
| lepA; GTP-binding protein LepA | 0.036 | 0.036 | 0.311 | 0.978 |
| lepB; signal peptidase I [EC:3.4.21.89] | -0.001 | 0.044 | 0.978 | 0.999 |
| LEU1; 3-isopropylmalate dehydratase [EC:4.2.1.33] | -1.992 | 1.429 | 0.165 | 0.960 |
| leuA, IMS; 2-isopropylmalate synthase [EC:2.3.3.13] | -0.070 | 0.056 | 0.214 | 0.978 |
| leuB, IMDH; 3-isopropylmalate dehydrogenase [EC:1.1.1.85] | -0.051 | 0.056 | 0.363 | 0.978 |
| leuC, IPMI-L; 3-isopropylmalate/(R)-2-methylmalate dehydratase large subunit [EC:4.2.1.33 4.2.1.35] | -0.077 | 0.056 | 0.167 | 0.961 |
| leuD, IPMI-S; 3-isopropylmalate/(R)-2-methylmalate dehydratase small subunit [EC:4.2.1.33 4.2.1.35] | -0.069 | 0.054 | 0.201 | 0.971 |
| leuE; leucine efflux protein | -0.090 | 0.337 | 0.789 | 0.999 |
| leuO; LysR family transcriptional regulator, transcriptional activator for leuABCD operon | 0.212 | 0.583 | 0.717 | 0.999 |
| lexA; repressor LexA [EC:3.4.21.88] | 0.084 | 0.059 | 0.157 | 0.954 |
| lf2; levanbiose-producing levanase [EC:3.2.1.64] | 0.126 | 0.576 | 0.827 | 0.999 |
| lfrR; TetR/AcrR family transcriptional regulator, repressor for lfrA | -17.661 | 2245.180 | 0.994 | 0.999 |
| lgt, umpA; phosphatidylglycerol:prolipoprotein diacylglycerol transferase [EC:2.-.-.-] | 0.074 | 0.040 | 0.065 | 0.839 |
| lhgO; L-2-hydroxyglutarate oxidase [EC:1.1.3.-] | -0.159 | 0.482 | 0.742 | 0.999 |
| lhpA; 4-hydroxyproline epimerase [EC:5.1.1.8] | -0.265 | 0.302 | 0.382 | 0.978 |
| lhpI; 1-piperideine-2-carboxylate/1-pyrroline-2-carboxylate reductase [NAD(P)H] [EC:1.5.1.1] | -0.831 | 1.558 | 0.595 | 0.999 |
| lhpI; cis-L-3-hydroxyproline dehydratase [EC:4.2.1.171] | 0.123 | 0.543 | 0.821 | 0.999 |
| lhr; ATP-dependent helicase Lhr and Lhr-like helicase [EC:3.6.4.-] | -0.068 | 0.177 | 0.702 | 0.999 |
| liaF; lia operon protein LiaF | -1.427 | 0.697 | 0.042 | 0.839 |
| liaG; lia operon protein LiaG | -0.762 | 1.059 | 0.473 | 0.979 |
| liaH; lia operon protein LiaH | 18.239 | 2966.075 | 0.995 | 0.999 |
| liaI; lia operon protein LiaI | -1.388 | 0.807 | 0.088 | 0.880 |
| liaR; two-component system, NarL family, response regulator LiaR | -0.005 | 0.150 | 0.975 | 0.999 |
| liaS; two-component system, NarL family, sensor histidine kinase LiaS [EC:2.7.13.3] | 0.003 | 0.154 | 0.982 | 0.999 |
| licD; lipopolysaccharide cholinephosphotransferase [EC:2.7.8.-] | 0.050 | 0.132 | 0.703 | 0.999 |
| licR; lichenan operon transcriptional antiterminator | -0.124 | 0.162 | 0.445 | 0.978 |
| licT, bglG; beta-glucoside operon transcriptional antiterminator | 0.067 | 0.133 | 0.619 | 0.999 |
| LIG1; DNA ligase 1 [EC:6.5.1.1 6.5.1.6 6.5.1.7] | -0.040 | 0.162 | 0.806 | 0.999 |
| ligA; protocatechuate 4,5-dioxygenase, alpha chain [EC:1.13.11.8] | -1.927 | 1.476 | 0.194 | 0.970 |
| ligB; protocatechuate 4,5-dioxygenase, beta chain [EC:1.13.11.8] | -1.869 | 1.144 | 0.104 | 0.908 |
| ligC; 2-hydroxy-4-carboxymuconate semialdehyde hemiacetal dehydrogenase [EC:1.1.1.312] | -2.126 | 1.538 | 0.169 | 0.961 |
| ligD; bifunctional non-homologous end joining protein LigD [EC:6.5.1.1] | -0.368 | 0.222 | 0.100 | 0.896 |
| ligI; 2-pyrone-4,6-dicarboxylate lactonase [EC:3.1.1.57] | -2.223 | 1.497 | 0.140 | 0.949 |
| ligJ; 4-oxalmesaconate hydratase [EC:4.2.1.83] | -1.323 | 0.762 | 0.084 | 0.873 |
| ligK, galC; 4-hydroxy-4-methyl-2-oxoglutarate aldolase [EC:4.1.3.17] | -0.923 | 0.504 | 0.069 | 0.840 |
| ligM; vanillate/3-O-methylgallate O-demethylase [EC:2.1.1.341] | -3.890 | 5.265 | 0.461 | 0.979 |
| ligX; 5,5’-dehydrodivanillate O-demethylase | -17.782 | 2854.684 | 0.995 | 0.999 |
| limB; limonene 1,2-monooxygenase [EC:1.14.13.107] | -0.896 | 0.658 | 0.176 | 0.961 |
| linN; cholesterol transport system auxiliary component | -0.135 | 0.231 | 0.561 | 0.992 |
| lip, TGL2; triacylglycerol lipase [EC:3.1.1.3] | -0.066 | 0.203 | 0.746 | 0.999 |
| lip3; type VI secretion system protein | -0.189 | 0.933 | 0.840 | 0.999 |
| lipA; lipoyl synthase [EC:2.8.1.8] | -0.126 | 0.085 | 0.140 | 0.949 |
| lipB; lipoyl(octanoyl) transferase [EC:2.3.1.181] | -0.057 | 0.119 | 0.633 | 0.999 |
| lipL; lipoyl amidotransferase [EC:2.3.1.200] | 2.823 | 2.239 | 0.209 | 0.977 |
| lipL; octanoyl-[GcvH]:protein N-octanoyltransferase [EC:2.3.1.204] | -0.131 | 0.256 | 0.610 | 0.999 |
| lipV; lipase [EC:3.1.1.-] | -1.419 | 1.138 | 0.214 | 0.978 |
| liuC; methylglutaconyl-CoA hydratase [EC:4.2.1.18] | -0.123 | 0.198 | 0.535 | 0.990 |
| livF; branched-chain amino acid transport system ATP-binding protein | -0.185 | 0.117 | 0.117 | 0.917 |
| livG; branched-chain amino acid transport system ATP-binding protein | -0.182 | 0.116 | 0.117 | 0.917 |
| livH; branched-chain amino acid transport system permease protein | -0.191 | 0.116 | 0.100 | 0.896 |
| livK; branched-chain amino acid transport system substrate-binding protein | -0.192 | 0.120 | 0.111 | 0.917 |
| livM; branched-chain amino acid transport system permease protein | -0.185 | 0.117 | 0.116 | 0.917 |
| lldD; L-lactate dehydrogenase (cytochrome) [EC:1.1.2.3] | -0.144 | 0.151 | 0.341 | 0.978 |
| lldE; L-lactate dehydrogenase complex protein LldE | -0.024 | 0.120 | 0.840 | 0.999 |
| lldF; L-lactate dehydrogenase complex protein LldF | -0.029 | 0.119 | 0.809 | 0.999 |
| lldG; L-lactate dehydrogenase complex protein LldG | 0.100 | 0.118 | 0.399 | 0.978 |
| lldP, lctP; L-lactate permease | -0.190 | 0.353 | 0.591 | 0.999 |
| lldR; GntR family transcriptional regulator, L-lactate dehydrogenase operon regulator | -0.182 | 0.353 | 0.608 | 0.999 |
| lldR; LysR family transcriptional regulator, L-lactate utilization regulator | 17.680 | 2242.142 | 0.994 | 0.999 |
| lmrA, yxaF; TetR/AcrR family transcriptional regulator, lmrAB and yxaGH operons repressor | -0.924 | 0.975 | 0.345 | 0.978 |
| lmrB; MFS transporter, DHA2 family, lincomycin resistance protein | 0.171 | 0.313 | 0.587 | 0.999 |
| lmrP; MFS transporter, DHA1 family, multidrug resistance protein B | -0.545 | 0.724 | 0.453 | 0.978 |
| lmrS; MFS transporter, DHA2 family, multidrug resistance protein | -0.268 | 0.571 | 0.639 | 0.999 |
| lnt; apolipoprotein N-acyltransferase [EC:2.3.1.-] | -0.100 | 0.136 | 0.463 | 0.979 |
| lnuA_C_D_E, lin; lincosamide nucleotidyltransferase A/C/D/E | 0.898 | 1.130 | 0.428 | 0.978 |
| lolA; outer membrane lipoprotein carrier protein | 0.007 | 0.142 | 0.961 | 0.999 |
| lolB; outer membrane lipoprotein LolB | 0.066 | 0.160 | 0.681 | 0.999 |
| lolC_E; lipoprotein-releasing system permease protein | 0.007 | 0.103 | 0.946 | 0.999 |
| lolD; lipoprotein-releasing system ATP-binding protein [EC:3.6.3.-] | 0.012 | 0.100 | 0.901 | 0.999 |
| lon; ATP-dependent Lon protease [EC:3.4.21.53] | 0.099 | 0.086 | 0.253 | 0.978 |
| lonB; ATP-dependent Lon protease [EC:3.4.21.53] | -1.153 | 0.740 | 0.121 | 0.919 |
| lonH; Lon-like ATP-dependent protease [EC:3.4.21.-] | 0.231 | 0.308 | 0.455 | 0.978 |
| loxA; arachidonate 15-lipoxygenase [EC:1.13.11.33] | 18.588 | 3531.830 | 0.996 | 0.999 |
| LOXL2_3_4; lysyl oxidase-like protein 2/3/4 [EC:1.4.3.-] | -1.911 | 2.017 | 0.345 | 0.978 |
| lpcC; mannosyltransferase [EC:2.4.1.-] | -2.179 | 0.935 | 0.021 | 0.839 |
| lplA, lplJ; lipoate—protein ligase [EC:6.3.1.20] | -0.001 | 0.124 | 0.995 | 0.999 |
| lplB; putative aldouronate transport system permease protein | 0.063 | 0.151 | 0.677 | 0.999 |
| lplC; putative aldouronate transport system permease protein | 0.060 | 0.149 | 0.686 | 0.999 |
| lplT; MFS transporter, LPLT family, lysophospholipid transporter | 0.119 | 0.601 | 0.843 | 0.999 |
| lpp; murein lipoprotein | -0.022 | 0.389 | 0.955 | 0.999 |
| lpqC; polyhydroxybutyrate depolymerase | -0.314 | 0.411 | 0.446 | 0.978 |
| lpqH; ipoprotein LpqH | -2.357 | 1.491 | 0.116 | 0.917 |
| lprG; lipoprotein LprG | -1.457 | 0.833 | 0.082 | 0.865 |
| lptA; lipopolysaccharide export system protein LptA | 0.003 | 0.101 | 0.975 | 0.999 |
| lptB; lipopolysaccharide export system ATP-binding protein [EC:3.6.3.-] | 0.050 | 0.087 | 0.563 | 0.993 |
| lptC; lipopolysaccharide export system protein LptC | 0.018 | 0.116 | 0.878 | 0.999 |
| lptD, imp, ostA; LPS-assembly protein | 0.002 | 0.101 | 0.980 | 0.999 |
| lptE, rlpB; LPS-assembly lipoprotein | -0.043 | 0.146 | 0.769 | 0.999 |
| lptF; lipopolysaccharide export system permease protein | 0.010 | 0.099 | 0.924 | 0.999 |
| lptG; lipopolysaccharide export system permease protein | 0.043 | 0.087 | 0.620 | 0.999 |
| lpxA; UDP-N-acetylglucosamine acyltransferase [EC:2.3.1.129] | 0.012 | 0.102 | 0.909 | 0.999 |
| lpxB; lipid-A-disaccharide synthase [EC:2.4.1.182] | 0.072 | 0.090 | 0.423 | 0.978 |
| lpxC-fabZ; UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase / 3-hydroxyacyl-[acyl-carrier-protein] dehydratase [EC:3.5.1.108 4.2.1.59] | -0.107 | 0.221 | 0.630 | 0.999 |
| lpxC; UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase [EC:3.5.1.108] | 0.157 | 0.093 | 0.093 | 0.892 |
| lpxD; UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase [EC:2.3.1.191] | 0.132 | 0.102 | 0.197 | 0.970 |
| lpxE; lipid A 1-phosphatase [EC:3.1.3.-] | -1.386 | 1.551 | 0.373 | 0.978 |
| lpxH; UDP-2,3-diacylglucosamine hydrolase [EC:3.6.1.54] | -0.053 | 0.107 | 0.619 | 0.999 |
| lpxK; tetraacyldisaccharide 4’-kinase [EC:2.7.1.130] | 0.050 | 0.091 | 0.581 | 0.998 |
| lpxL, htrB; Kdo2-lipid IVA lauroyltransferase [EC:2.3.1.241] | 0.072 | 0.082 | 0.377 | 0.978 |
| lpxM, msbB; lauroyl-Kdo2-lipid IVA myristoyltransferase [EC:2.3.1.243] | 0.232 | 0.308 | 0.453 | 0.978 |
| lpxO; beta-hydroxylase [EC:1.14.11.-] | -0.119 | 0.243 | 0.626 | 0.999 |
| lpxP; KDO2-lipid IV(A) palmitoleoyltransferase [EC:2.3.1.242] | 0.119 | 0.601 | 0.843 | 0.999 |
| lpxQ; lipid A oxidase | -1.272 | 1.833 | 0.489 | 0.979 |
| LRA1; L-rhamnose 1-dehydrogenase [EC:1.1.1.378 1.1.1.377 1.1.1.173] | -0.972 | 0.627 | 0.123 | 0.919 |
| LRA3, yfaW; L-rhamnonate dehydratase [EC:4.2.1.90] | 0.904 | 0.479 | 0.061 | 0.839 |
| lrgA; holin-like protein | -0.114 | 0.538 | 0.833 | 0.999 |
| lrgB; holin-like protein LrgB | -0.114 | 0.538 | 0.833 | 0.999 |
| lrp; Lrp/AsnC family transcriptional regulator, leucine-responsive regulatory protein | -0.114 | 0.134 | 0.396 | 0.978 |
| lsa; lincosamide and streptogramin A transport system ATP-binding/permease protein | 0.477 | 0.292 | 0.104 | 0.907 |
| lspA; signal peptidase II [EC:3.4.23.36] | 0.042 | 0.038 | 0.267 | 0.978 |
| lsrA, ego; AI-2 transport system ATP-binding protein | -0.624 | 0.598 | 0.298 | 0.978 |
| lsrB; AI-2 transport system substrate-binding protein | -0.624 | 0.598 | 0.298 | 0.978 |
| lsrC; AI-2 transport system permease protein | -0.614 | 0.595 | 0.304 | 0.978 |
| lsrD; AI-2 transport system permease protein | -0.624 | 0.598 | 0.298 | 0.978 |
| lsrF; 3-hydroxy-5-phosphonooxypentane-2,4-dione thiolase [EC:2.3.1.245] | -0.603 | 0.540 | 0.266 | 0.978 |
| lsrG; (4S)-4-hydroxy-5-phosphonooxypentane-2,3-dione isomerase [EC:5.3.1.32] | 0.120 | 0.625 | 0.847 | 0.999 |
| lsrK; autoinducer-2 kinase [EC:2.7.1.189] | -0.172 | 0.521 | 0.742 | 0.999 |
| lsrR; lsr operon transcriptional repressor | -0.102 | 0.657 | 0.876 | 0.999 |
| lst; beta-galactosamide-alpha-2,3-sialyltransferase [EC:2.4.99.-] | -0.321 | 0.480 | 0.504 | 0.981 |
| ltaE; threonine aldolase [EC:4.1.2.48] | 0.024 | 0.123 | 0.846 | 0.999 |
| ltaS; lipoteichoic acid synthase [EC:2.7.8.20] | 0.201 | 0.192 | 0.296 | 0.978 |
| ltnD; L-threonate 2-dehydrogenase [EC:1.1.1.411] | -0.553 | 0.613 | 0.368 | 0.978 |
| ltrA; RNA-directed DNA polymerase [EC:2.7.7.49] | 0.268 | 0.521 | 0.608 | 0.999 |
| luxA; alkanal monooxygenase alpha chain [EC:1.14.14.3] | 18.902 | 3402.727 | 0.996 | 0.999 |
| luxB; alkanal monooxygenase beta chain [EC:1.14.14.3] | 18.902 | 3402.727 | 0.996 | 0.999 |
| luxC; long-chain-fatty-acyl-CoA reductase [EC:1.2.1.50] | 18.902 | 3402.727 | 0.996 | 0.999 |
| luxD; acyl transferase [EC:2.3.1.-] | 18.902 | 3402.727 | 0.996 | 0.999 |
| luxE; long-chain-fatty-acid—luciferin-component ligase [EC:6.2.1.19] | 18.902 | 3402.727 | 0.996 | 0.999 |
| luxN; two-component system, autoinducer 1 sensor kinase/phosphatase LuxN [EC:2.7.13.3 3.1.3.-] | 18.902 | 3402.727 | 0.996 | 0.999 |
| luxO; two-component system, repressor protein LuxO | -0.031 | 1.089 | 0.978 | 0.999 |
| luxP; autoinducer 2-binding periplasmic protein LuxP | 18.482 | 3349.575 | 0.996 | 0.999 |
| luxQ; two-component system, autoinducer 2 sensor kinase/phosphatase LuxQ [EC:2.7.13.3 3.1.3.-] | 18.482 | 3349.575 | 0.996 | 0.999 |
| luxR, vanR; LuxR family transcriptional regulator, transcriptional activator of the bioluminescence operon | 18.373 | 3170.868 | 0.995 | 0.999 |
| luxS; S-ribosylhomocysteine lyase [EC:4.4.1.21] | 0.029 | 0.087 | 0.739 | 0.999 |
| luxU; two-component system, phosphorelay protein LuxU | 18.119 | 1920.327 | 0.992 | 0.999 |
| LYS1; saccharopine dehydrogenase (NAD+, L-lysine forming) [EC:1.5.1.7] | -0.223 | 0.154 | 0.150 | 0.954 |
| LYS12; homoisocitrate dehydrogenase [EC:1.1.1.87] | -1.940 | 2.261 | 0.392 | 0.978 |
| LYS21, LYS20; homocitrate synthase [EC:2.3.3.14] | -1.940 | 2.261 | 0.392 | 0.978 |
| LYS5, acpT; 4’-phosphopantetheinyl transferase [EC:2.7.8.-] | 0.000 | 0.156 | 0.998 | 1.000 |
| LYS9; saccharopine dehydrogenase (NADP+, L-glutamate forming) [EC:1.5.1.10] | 1.495 | 1.571 | 0.343 | 0.978 |
| lysA; diaminopimelate decarboxylase [EC:4.1.1.20] | -0.023 | 0.049 | 0.646 | 0.999 |
| lysAC; bifunctional diaminopimelate decarboxylase / aspartate kinase [EC:4.1.1.20 2.7.2.4] | -0.046 | 0.470 | 0.923 | 0.999 |
| lysC; aspartate kinase [EC:2.7.2.4] | -0.039 | 0.050 | 0.439 | 0.978 |
| lysDH; lysine 6-dehydrogenase [EC:1.4.1.18] | -1.374 | 1.003 | 0.173 | 0.961 |
| lysE, argO; L-lysine exporter family protein LysE/ArgO | 0.086 | 0.102 | 0.402 | 0.978 |
| lysJ, argD; LysW-gamma-L-lysine/LysW-L-ornithine aminotransferase [EC:2.6.1.-] | -0.631 | 1.117 | 0.573 | 0.995 |
| lysK, argE; LysW-gamma-L-lysine/LysW-L-ornithine carboxypeptidase | -0.817 | 1.210 | 0.500 | 0.981 |
| lysK; lysyl-tRNA synthetase, class I [EC:6.1.1.6] | -1.110 | 0.399 | 0.006 | 0.839 |
| LYSN; 2-aminoadipate transaminase [EC:2.6.1.-] | 0.069 | 0.205 | 0.735 | 0.999 |
| lysP; lysine-specific permease | -0.008 | 0.147 | 0.957 | 0.999 |
| lysW; alpha-aminoadipate/glutamate carrier protein LysW | -0.639 | 1.036 | 0.538 | 0.990 |
| lysX; [lysine-biosynthesis-protein LysW]—L-2-aminoadipate ligase [EC:6.3.2.43] | -1.103 | 1.184 | 0.353 | 0.978 |
| lysX1; putative lysine transport system substrate-binding protein | -0.790 | 0.717 | 0.273 | 0.978 |
| lysX2; putative lysine transport system permease protein | 0.033 | 0.133 | 0.803 | 0.999 |
| lysY, argC; LysW-gamma-L-alpha-aminoadipyl-6-phosphate/LysW-L-glutamyl-5-phosphate reductase [EC:1.2.1.-] | -0.631 | 1.117 | 0.573 | 0.995 |
| lysZ, argB; LysW-gamma-L-alpha-aminoadipate/LysW-L-glutamate kinase [EC:2.7.2.-] | -0.631 | 1.117 | 0.573 | 0.995 |
| lytE, cwlF; peptidoglycan DL-endopeptidase LytE [EC:3.4.-.-] | -0.776 | 0.642 | 0.229 | 0.978 |
| lytF, cwlE; peptidoglycan DL-endopeptidase LytF [EC:3.4.-.-] | -0.635 | 1.401 | 0.651 | 0.999 |
| lytM; lysostaphin [EC:3.4.24.75] | -1.554 | 1.110 | 0.164 | 0.957 |
| lytS; two-component system, LytTR family, sensor histidine kinase LytS [EC:2.7.13.3] | 0.054 | 0.216 | 0.804 | 0.999 |
| lytT, lytR; two-component system, LytTR family, response regulator LytT | -0.236 | 0.351 | 0.503 | 0.981 |
| lyxA; D-lyxose ketol-isomerase [EC:5.3.1.15] | 0.275 | 0.537 | 0.609 | 0.999 |
| lyxK; L-xylulokinase [EC:2.7.1.53] | -0.279 | 0.379 | 0.463 | 0.979 |
| maa; maltose O-acetyltransferase [EC:2.3.1.79] | 0.051 | 0.123 | 0.679 | 0.999 |
| mabA; beta-ketoacyl ACP reductase [EC:1.1.1.100] | -2.137 | 1.493 | 0.154 | 0.954 |
| mabO; 4-methylaminobutanoate oxidase (formaldehyde-forming) [EC:1.5.3.19] | -1.405 | 0.793 | 0.078 | 0.859 |
| macA; membrane fusion protein, macrolide-specific efflux system | 0.062 | 0.147 | 0.671 | 0.999 |
| macB; macrolide transport system ATP-binding/permease protein [EC:3.6.3.-] | -0.009 | 0.184 | 0.960 | 0.999 |
| maeN; malate:Na+ symporter | -0.019 | 0.577 | 0.974 | 0.999 |
| maf; septum formation protein | -0.057 | 0.072 | 0.430 | 0.978 |
| maiA, GSTZ1; maleylacetoacetate isomerase [EC:5.2.1.2] | -0.113 | 0.215 | 0.599 | 0.999 |
| mal; methylaspartate ammonia-lyase [EC:4.3.1.2] | 0.003 | 0.480 | 0.996 | 0.999 |
| malE; maltose/maltodextrin transport system substrate-binding protein | -0.440 | 0.420 | 0.296 | 0.978 |
| malF; maltose/maltodextrin transport system permease protein | -0.288 | 0.457 | 0.529 | 0.989 |
| malG; maltose/maltodextrin transport system permease protein | -0.288 | 0.457 | 0.529 | 0.989 |
| malI; LacI family transcriptional regulator, maltose regulon regulatory protein | 0.397 | 0.551 | 0.473 | 0.979 |
| malK, mtlK, thuK; multiple sugar transport system ATP-binding protein [EC:3.6.3.-] | -0.502 | 0.252 | 0.048 | 0.839 |
| malM; maltose operon periplasmic protein | -0.340 | 0.514 | 0.510 | 0.981 |
| malQ; 4-alpha-glucanotransferase [EC:2.4.1.25] | 0.076 | 0.086 | 0.376 | 0.978 |
| malR; two-component system, CitB family, response regulator MalR | 0.179 | 0.459 | 0.697 | 0.999 |
| malT; LuxR family transcriptional regulator, maltose regulon positive regulatory protein | 0.191 | 0.280 | 0.495 | 0.981 |
| malY, malT; maltose/moltooligosaccharide transporter | 0.072 | 0.214 | 0.738 | 0.999 |
| malZ; alpha-glucosidase [EC:3.2.1.20] | -0.024 | 0.128 | 0.854 | 0.999 |
| MAN; mannan endo-1,4-beta-mannosidase [EC:3.2.1.78] | -0.201 | 0.251 | 0.426 | 0.978 |
| MAN1; mannosyl-oligosaccharide alpha-1,2-mannosidase [EC:3.2.1.113] | -17.510 | 3533.982 | 0.996 | 0.999 |
| MAN2C1; alpha-mannosidase [EC:3.2.1.24] | 0.038 | 0.183 | 0.835 | 0.999 |
| manA, MPI; mannose-6-phosphate isomerase [EC:5.3.1.8] | -0.021 | 0.072 | 0.766 | 0.999 |
| manB; phosphomannomutase [EC:5.4.2.8] | -0.136 | 0.117 | 0.247 | 0.978 |
| manC, cpsB; mannose-1-phosphate guanylyltransferase [EC:2.7.7.13] | -0.202 | 0.128 | 0.116 | 0.917 |
| MANEA; glycoprotein endo-alpha-1,2-mannosidase [EC:3.2.1.130] | -1.849 | 1.778 | 0.300 | 0.978 |
| manR; activator of the mannose operon, transcriptional antiterminator | -1.067 | 0.899 | 0.237 | 0.978 |
| MAO, aofH; monoamine oxidase [EC:1.4.3.4] | -0.324 | 0.280 | 0.250 | 0.978 |
| map; methionyl aminopeptidase [EC:3.4.11.18] | -0.018 | 0.029 | 0.540 | 0.990 |
| mapA; maltose phosphorylase [EC:2.4.1.8] | 0.407 | 0.337 | 0.230 | 0.978 |
| mapP; maltose 6’-phosphate phosphatase [EC:3.1.3.90] | 0.072 | 0.153 | 0.639 | 0.999 |
| marA; AraC family transcriptional regulator, mar-sox-rob regulon activator | -0.243 | 0.717 | 0.735 | 0.999 |
| marB; multiple antibiotic resistance protein MarB | -0.243 | 0.717 | 0.735 | 0.999 |
| marC; multiple antibiotic resistance protein | -0.081 | 0.138 | 0.558 | 0.992 |
| marR; MarR family transcriptional regulator, multiple antibiotic resistance protein MarR | -0.243 | 0.717 | 0.735 | 0.999 |
| MARS, metG; methionyl-tRNA synthetase [EC:6.1.1.10] | 0.040 | 0.037 | 0.280 | 0.978 |
| mas; Mce-associated membrane protein | -1.971 | 1.422 | 0.168 | 0.961 |
| matB; malonyl-CoA/methylmalonyl-CoA synthetase [EC:6.2.1.-] | -0.738 | 0.518 | 0.156 | 0.954 |
| mauA; methylamine dehydrogenase light chain [EC:1.4.9.1] | -1.203 | 1.540 | 0.436 | 0.978 |
| mauB; methylamine dehydrogenase heavy chain [EC:1.4.9.1] | -0.024 | 0.778 | 0.976 | 0.999 |
| mazE, chpAI; antitoxin MazE | -0.408 | 0.471 | 0.388 | 0.978 |
| mazF, ndoA, chpA; mRNA interferase MazF [EC:3.1.-.-] | -0.069 | 0.191 | 0.720 | 0.999 |
| mazG; nucleoside triphosphate diphosphatase [EC:3.6.1.9] | -0.025 | 0.173 | 0.887 | 0.999 |
| MBF1; putative transcription factor | -17.234 | 3077.514 | 0.996 | 0.999 |
| mbhJ; membrane-bound hydrogenase subunit mbhJ [EC:1.12.7.2] | 0.340 | 1.138 | 0.766 | 0.999 |
| mbhK; membrane-bound hydrogenase subunit beta [EC:1.12.7.2] | 0.340 | 1.138 | 0.766 | 0.999 |
| mbhL; membrane-bound hydrogenase subunit alpha [EC:1.12.7.2] | 0.340 | 1.138 | 0.766 | 0.999 |
| mbtA; mycobactin salicyl-AMP ligase [EC:6.3.2.-] | -2.602 | 1.431 | 0.071 | 0.841 |
| mbtB; mycobactin phenyloxazoline synthetase | -2.635 | 1.432 | 0.068 | 0.839 |
| mbtC; mycobactin polyketide synthetase MbtC | -2.534 | 1.465 | 0.086 | 0.878 |
| mbtD; mycobactin polyketide synthetase MbtD | -2.534 | 1.465 | 0.086 | 0.878 |
| mbtE; mycobactin peptide synthetase MbtE | -2.602 | 1.431 | 0.071 | 0.841 |
| mbtF; mycobactin peptide synthetase MbtF | -2.602 | 1.431 | 0.071 | 0.841 |
| mbtG; mycobactin lysine-N-oxygenase | -2.462 | 1.118 | 0.029 | 0.839 |
| mbtH, nocI; MbtH protein | -0.016 | 0.316 | 0.960 | 0.999 |
| mbtI, irp9, ybtS; salicylate synthetase [EC:5.4.4.2 4.2.99.21] | -1.935 | 1.740 | 0.268 | 0.978 |
| mbtM; long-chain-fatty-acid–[acyl-carrier-protein] ligase [EC:6.2.1.20] | -3.164 | 2.043 | 0.124 | 0.919 |
| mbtN, fadE14; acyl-ACP dehydrogenase [EC:1.3.99.-] | -0.276 | 0.188 | 0.143 | 0.951 |
| mca; mycothiol S-conjugate amidase [EC:3.5.1.115] | -0.027 | 0.318 | 0.933 | 0.999 |
| mcbA; MqsR-controlled colanic acid and biofilm protein A | -0.224 | 0.799 | 0.779 | 0.999 |
| mcbR; GntR family transcriptional regulator, colanic acid and biofilm gene transcriptional regulator | -0.109 | 0.831 | 0.895 | 0.999 |
| mccA; cystathionine beta-synthase (O-acetyl-L-serine) [EC:2.5.1.134] | -0.963 | 0.518 | 0.065 | 0.839 |
| mccB; cystathionine gamma-lyase / homocysteine desulfhydrase [EC:4.4.1.1 4.4.1.2] | -0.647 | 0.444 | 0.147 | 0.954 |
| mcd; (2S)-methylsuccinyl-CoA dehydrogenase [EC:1.3.8.12] | -1.138 | 0.757 | 0.135 | 0.944 |
| MCEE, epi; methylmalonyl-CoA/ethylmalonyl-CoA epimerase [EC:5.1.99.1] | -0.270 | 0.161 | 0.095 | 0.896 |
| mch, mcd; 2-methylfumaryl-CoA hydratase [EC:4.2.1.148] | -1.609 | 1.137 | 0.159 | 0.954 |
| MCH; medium-chain acyl-[acyl-carrier-protein] hydrolase [EC:3.1.2.21] | 0.134 | 0.147 | 0.362 | 0.978 |
| mch; methenyltetrahydromethanopterin cyclohydrolase [EC:3.5.4.27] | 1.390 | 0.984 | 0.160 | 0.954 |
| mcl; malyl-CoA/(S)-citramalyl-CoA lyase [EC:4.1.3.24 4.1.3.25] | -1.353 | 0.883 | 0.128 | 0.927 |
| mcl2; (3S)-malyl-CoA thioesterase [EC:3.1.2.30] | -18.019 | 3183.927 | 0.995 | 0.999 |
| mcm, cdc21; replicative DNA helicase Mcm [EC:3.6.4.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| mcp; methyl-accepting chemotaxis protein | -0.099 | 0.166 | 0.554 | 0.992 |
| mcrA; 5-methylcytosine-specific restriction enzyme A [EC:3.1.21.-] | -0.136 | 0.265 | 0.608 | 0.999 |
| mcrB; 5-methylcytosine-specific restriction enzyme B [EC:3.1.21.-] | 0.624 | 0.378 | 0.100 | 0.896 |
| mcrC; 5-methylcytosine-specific restriction enzyme subunit McrC | 0.379 | 0.302 | 0.212 | 0.978 |
| mcsA; protein arginine kinase activator | 0.042 | 0.186 | 0.820 | 0.999 |
| mcsB; protein arginine kinase [EC:2.7.14.1] | 0.073 | 0.191 | 0.702 | 0.999 |
| mct; 2-methylfumaryl-CoA isomerase [EC:5.4.1.3] | -1.235 | 0.941 | 0.191 | 0.970 |
| mcyA; microcystin synthetase protein McyA | 0.445 | 0.951 | 0.640 | 0.999 |
| mdaB; modulator of drug activity B | 0.126 | 0.165 | 0.448 | 0.978 |
| mdcA; malonate decarboxylase alpha subunit [EC:2.3.1.187] | 0.054 | 0.259 | 0.834 | 0.999 |
| mdcB; triphosphoribosyl-dephospho-CoA synthase [EC:2.4.2.52] | 0.031 | 0.274 | 0.910 | 0.999 |
| mdcC; malonate decarboxylase delta subunit | 0.034 | 0.274 | 0.902 | 0.999 |
| mdcD; malonate decarboxylase beta subunit [EC:4.1.1.87] | 0.016 | 0.298 | 0.957 | 0.999 |
| mdcE; malonate decarboxylase gamma subunit [EC:4.1.1.87] | 0.054 | 0.259 | 0.834 | 0.999 |
| mdcF; malonate transporter | 0.298 | 0.698 | 0.670 | 0.999 |
| mdcG; phosphoribosyl-dephospho-CoA transferase [EC:2.7.7.66] | 0.082 | 0.265 | 0.758 | 0.999 |
| mdcH; malonate decarboxylase epsilon subunit [EC:2.3.1.39] | 0.110 | 0.342 | 0.748 | 0.999 |
| mdcR; LysR family transcriptional regulator, malonate utilization transcriptional regulator | 0.100 | 0.338 | 0.767 | 0.999 |
| mdeA; MFS transporter, DHA2 family, multidrug resistance protein | -0.433 | 0.673 | 0.521 | 0.988 |
| mdfA, cmr; MFS transporter, DHA1 family, multidrug/chloramphenicol efflux transport protein | 0.072 | 0.282 | 0.797 | 0.999 |
| mdh; malate dehydrogenase [EC:1.1.1.37] | -0.143 | 0.094 | 0.128 | 0.927 |
| mdh1, mxaF; methanol dehydrogenase (cytochrome c) subunit 1 [EC:1.1.2.7] | 18.066 | 2719.856 | 0.995 | 0.999 |
| MDH1; malate dehydrogenase [EC:1.1.1.37] | 0.072 | 0.539 | 0.894 | 0.999 |
| mdh2, mxaI; methanol dehydrogenase (cytochrome c) subunit 2 [EC:1.1.2.7] | 18.066 | 2719.856 | 0.995 | 0.999 |
| mdlA, smdA; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.069 | 0.519 | 0.894 | 0.999 |
| mdlA; mandelate racemase [EC:5.1.2.2] | -1.407 | 1.166 | 0.229 | 0.978 |
| mdlB, smdB; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.069 | 0.519 | 0.894 | 0.999 |
| mdlB; (S)-mandelate dehydrogenase [EC:1.1.99.31] | 2.148 | 1.026 | 0.038 | 0.839 |
| mdlC; benzoylformate decarboxylase [EC:4.1.1.7] | -0.974 | 0.719 | 0.178 | 0.961 |
| mdlY; mandelamide amidase [EC:3.5.1.86] | -0.514 | 0.952 | 0.590 | 0.999 |
| mdo; formaldehyde dismutase / methanol dehydrogenase [EC:1.2.98.1 1.1.99.37] | -1.099 | 1.173 | 0.350 | 0.978 |
| mdoB; phosphoglycerol transferase [EC:2.7.8.20] | -0.086 | 0.357 | 0.810 | 0.999 |
| mdoC; glucans biosynthesis protein C [EC:2.1.-.-] | -0.085 | 0.575 | 0.882 | 0.999 |
| mdoG; periplasmic glucans biosynthesis protein | -0.090 | 0.248 | 0.717 | 0.999 |
| mdoH; membrane glycosyltransferase [EC:2.4.1.-] | -0.105 | 0.266 | 0.693 | 0.999 |
| mdtA; membrane fusion protein, multidrug efflux system | -0.032 | 0.220 | 0.884 | 0.999 |
| mdtB; multidrug efflux pump | -0.016 | 0.236 | 0.946 | 0.999 |
| mdtC; multidrug efflux pump | 0.010 | 0.228 | 0.964 | 0.999 |
| mdtD; MFS transporter, DHA2 family, multidrug resistance protein | 0.134 | 0.621 | 0.829 | 0.999 |
| mdtE; membrane fusion protein, multidrug efflux system | -1.158 | 1.457 | 0.428 | 0.978 |
| mdtF; multidrug efflux pump | -1.158 | 1.457 | 0.428 | 0.978 |
| mdtG; MFS transporter, DHA1 family, multidrug resistance protein | 0.258 | 0.287 | 0.370 | 0.978 |
| mdtH; MFS transporter, DHA1 family, multidrug resistance protein | -0.217 | 0.582 | 0.709 | 0.999 |
| mdtI; spermidine export protein MdtI | -0.005 | 0.390 | 0.989 | 0.999 |
| mdtJ; spermidine export protein MdtJ | -0.005 | 0.390 | 0.989 | 0.999 |
| mdtL; MFS transporter, DHA1 family, multidrug resistance protein | -0.252 | 0.698 | 0.718 | 0.999 |
| mdtN; membrane fusion protein, multidrug efflux system | -0.226 | 0.626 | 0.719 | 0.999 |
| mdtO; multidrug resistance protein MdtO | -0.226 | 0.626 | 0.719 | 0.999 |
| mdtP; outer membrane protein, multidrug efflux system | -1.292 | 1.211 | 0.288 | 0.978 |
| ME2, sfcA, maeA; malate dehydrogenase (oxaloacetate-decarboxylating) [EC:1.1.1.38] | 0.187 | 0.117 | 0.113 | 0.917 |
| mecA; penicillin-binding protein 2 prime [EC:3.4.16.4] | -1.670 | 1.245 | 0.182 | 0.961 |
| mecA1_2; adapter protein MecA 1/2 | 0.010 | 0.123 | 0.935 | 0.999 |
| mecR1; methicillin resistance protein | -1.670 | 1.245 | 0.182 | 0.961 |
| med; transcriptional activator of comK gene | -1.431 | 1.272 | 0.262 | 0.978 |
| mef; MFS transporter, DHA3 family, macrolide efflux protein | -0.181 | 0.339 | 0.595 | 0.999 |
| meh; 3-methylfumaryl-CoA hydratase [EC:4.2.1.153] | -1.103 | 0.571 | 0.055 | 0.839 |
| melA; alpha-galactosidase [EC:3.2.1.22] | -1.759 | 0.569 | 0.002 | 0.839 |
| melB; melibiose permease | -0.779 | 0.545 | 0.155 | 0.954 |
| MEMO1; MEMO1 family protein | 0.350 | 0.498 | 0.484 | 0.979 |
| menA; 1,4-dihydroxy-2-naphthoate octaprenyltransferase [EC:2.5.1.74 2.5.1.-] | 0.080 | 0.103 | 0.436 | 0.978 |
| menB; naphthoate synthase [EC:4.1.3.36] | 0.021 | 0.110 | 0.852 | 0.999 |
| menC; O-succinylbenzoate synthase [EC:4.2.1.113] | 0.000 | 0.149 | 0.997 | 0.999 |
| menD; 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase [EC:2.2.1.9] | -0.016 | 0.120 | 0.896 | 0.999 |
| menE; O-succinylbenzoic acid—CoA ligase [EC:6.2.1.26] | 0.026 | 0.111 | 0.814 | 0.999 |
| menF; menaquinone-specific isochorismate synthase [EC:5.4.4.2] | 0.089 | 0.166 | 0.593 | 0.999 |
| menH; 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase [EC:4.2.99.20] | 0.080 | 0.183 | 0.661 | 0.999 |
| menI; 1,4-dihydroxy-2-naphthoyl-CoA hydrolase [EC:3.1.2.28] | -0.148 | 0.426 | 0.729 | 0.999 |
| MEP; peptidyl-Lys metalloendopeptidase [EC:3.4.24.20] | 0.969 | 0.905 | 0.286 | 0.978 |
| mepA; multidrug efflux pump | -1.161 | 0.945 | 0.221 | 0.978 |
| mepA; penicillin-insensitive murein DD-endopeptidase [EC:3.4.24.-] | -0.151 | 0.281 | 0.592 | 0.999 |
| mepH; murein DD-endopeptidase [EC:3.4.-.-] | 0.134 | 0.621 | 0.829 | 0.999 |
| mepM; murein DD-endopeptidase [EC:3.4.24.-] | 0.195 | 0.476 | 0.683 | 0.999 |
| mepR; MarR family transcriptional regulator, repressor for mepA | -17.214 | 3047.217 | 0.995 | 0.999 |
| mepS, spr; murein DD-endopeptidase / murein LD-carboxypeptidase [EC:3.4.-.- 3.4.17.13] | 0.170 | 0.223 | 0.447 | 0.978 |
| mer; 5,10-methylenetetrahydromethanopterin reductase [EC:1.5.98.2] | 2.411 | 1.959 | 0.220 | 0.978 |
| merA; mercuric reductase [EC:1.16.1.1] | 0.067 | 0.415 | 0.872 | 0.999 |
| merC; mercuric ion transport protein | 0.331 | 0.696 | 0.635 | 0.999 |
| merD; MerR family transcriptional regulator, mercuric resistance operon regulatory protein | 0.754 | 0.982 | 0.444 | 0.978 |
| merE; mercuric ion transport protein | -1.844 | 1.522 | 0.227 | 0.978 |
| merP; periplasmic mercuric ion binding protein | 0.140 | 0.235 | 0.552 | 0.992 |
| merR; MerR family transcriptional regulator, mercuric resistance operon regulatory protein | -0.058 | 0.306 | 0.850 | 0.999 |
| merT; mercuric ion transport protein | 0.136 | 0.238 | 0.568 | 0.995 |
| MET8; precorrin-2 dehydrogenase / sirohydrochlorin ferrochelatase [EC:1.3.1.76 4.99.1.4] | 0.220 | 0.162 | 0.178 | 0.961 |
| metA; homoserine O-succinyltransferase/O-acetyltransferase [EC:2.3.1.46 2.3.1.31] | 0.002 | 0.103 | 0.988 | 0.999 |
| metB; cystathionine gamma-synthase [EC:2.5.1.48] | 0.080 | 0.075 | 0.292 | 0.978 |
| metC; cystathionine beta-lyase [EC:4.4.1.8] | 0.136 | 0.126 | 0.283 | 0.978 |
| metE; 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase [EC:2.1.1.14] | 0.050 | 0.065 | 0.446 | 0.978 |
| metF, MTHFR; methylenetetrahydrofolate reductase (NADPH) [EC:1.5.1.20] | -0.036 | 0.059 | 0.540 | 0.990 |
| metH, MTR; 5-methyltetrahydrofolate–homocysteine methyltransferase [EC:2.1.1.13] | -0.035 | 0.091 | 0.698 | 0.999 |
| metI; D-methionine transport system permease protein | 0.070 | 0.045 | 0.122 | 0.919 |
| metJ; MetJ family transcriptional regulator, methionine regulon repressor | 0.231 | 0.308 | 0.455 | 0.978 |
| metK; S-adenosylmethionine synthetase [EC:2.5.1.6] | 0.032 | 0.036 | 0.381 | 0.978 |
| metL; bifunctional aspartokinase / homoserine dehydrogenase 2 [EC:2.7.2.4 1.1.1.3] | 0.218 | 0.576 | 0.706 | 0.999 |
| metN; D-methionine transport system ATP-binding protein | 0.090 | 0.039 | 0.024 | 0.839 |
| metQ; D-methionine transport system substrate-binding protein | 0.129 | 0.055 | 0.020 | 0.839 |
| metR; LysR family transcriptional regulator, regulator for metE and metH | 0.062 | 0.154 | 0.689 | 0.999 |
| metX; homoserine O-acetyltransferase/O-succinyltransferase [EC:2.3.1.31 2.3.1.46] | -0.097 | 0.128 | 0.449 | 0.978 |
| metY; O-acetylhomoserine (thiol)-lyase [EC:2.5.1.49] | -0.109 | 0.078 | 0.162 | 0.957 |
| metZ; O-succinylhomoserine sulfhydrylase [EC:2.5.1.-] | -0.140 | 0.195 | 0.474 | 0.979 |
| mexC; membrane fusion protein, multidrug efflux system | -0.793 | 0.893 | 0.376 | 0.978 |
| mexD; multidrug efflux pump | -0.793 | 0.893 | 0.376 | 0.978 |
| mexE; membrane fusion protein, multidrug efflux system | 0.153 | 0.368 | 0.678 | 0.999 |
| mexF; multidrug efflux pump | 0.245 | 0.336 | 0.467 | 0.979 |
| mexG; transmembrane protein | -0.071 | 0.484 | 0.884 | 0.999 |
| mexH; membrane fusion protein, multidrug efflux system | -0.290 | 0.569 | 0.611 | 0.999 |
| mexI; multidrug efflux pump | -0.203 | 0.649 | 0.754 | 0.999 |
| mexJ; membrane fusion protein, multidrug efflux system | 0.053 | 0.249 | 0.832 | 0.999 |
| mexK; multidrug efflux pump | 0.064 | 0.230 | 0.782 | 0.999 |
| mexL; TetR/AcrR family transcriptional regulator, mexJK operon transcriptional repressor | 0.010 | 0.235 | 0.966 | 0.999 |
| mexT; LysR family transcriptional regulator, mexEF-oprN operon transcriptional activator | 0.128 | 0.366 | 0.728 | 0.999 |
| mexX, amrA; membrane fusion protein, multidrug efflux system | 1.551 | 1.591 | 0.331 | 0.978 |
| mexY, amrB; multidrug efflux pump | 0.952 | 1.231 | 0.441 | 0.978 |
| mexZ; TetR/AcrR family transcriptional regulator, mexXY operon repressor | -0.627 | 1.514 | 0.679 | 0.999 |
| mfd; transcription-repair coupling factor (superfamily II helicase) [EC:3.6.4.-] | 0.030 | 0.035 | 0.384 | 0.978 |
| mfnB; (5-formylfuran-3-yl)methyl phosphate synthase [EC:4.2.3.153] | 2.673 | 2.283 | 0.243 | 0.978 |
| mfnF; (4-(4-[2-(gamma-L-glutamylamino)ethyl]phenoxymethyl)furan-2-yl)methanamine synthase [EC:2.5.1.131] | 2.673 | 2.283 | 0.243 | 0.978 |
| MFS.CP; MFS transporter, CP family, cyanate transporter | -0.065 | 0.102 | 0.523 | 0.989 |
| MFS.SET; MFS transporter, SET family, sugar efflux transporter | -0.360 | 0.654 | 0.583 | 0.999 |
| MFS.SP; MFS transporter, SP family, sugar porter, other | -1.609 | 0.993 | 0.107 | 0.913 |
| MGD; 1,2-diacylglycerol 3-beta-galactosyltransferase [EC:2.4.1.46] | -1.459 | 0.975 | 0.136 | 0.949 |
| mglA; methyl-galactoside transport system ATP-binding protein [EC:3.6.3.17] | 0.392 | 0.170 | 0.023 | 0.839 |
| mglB; methyl-galactoside transport system substrate-binding protein | 0.396 | 0.183 | 0.032 | 0.839 |
| mglC; methyl-galactoside transport system permease protein | 0.391 | 0.171 | 0.023 | 0.839 |
| MGME1, DDK1; mitochondrial genome maintenance exonuclease 1 [EC:3.1.-.-] | 18.458 | 3309.834 | 0.996 | 0.999 |
| mgp; 4-O-beta-D-mannosyl-D-glucose phosphorylase [EC:2.4.1.281] | -0.963 | 0.443 | 0.031 | 0.839 |
| mgrA; MarR family transcriptional regulator, multiple gene regulator MgrA | -0.171 | 0.819 | 0.835 | 0.999 |
| mgs, bgsB; 1,2-diacylglycerol 3-alpha-glucosyltransferase [EC:2.4.1.337] | 0.139 | 0.140 | 0.322 | 0.978 |
| mgsA; methylglyoxal synthase [EC:4.2.3.3] | 0.150 | 0.188 | 0.426 | 0.978 |
| mgtA, mgtB; Mg2+-importing ATPase [EC:3.6.3.2] | -0.017 | 0.107 | 0.876 | 0.999 |
| mgtA; phosphatidylinositol alpha 1,6-mannosyltransferase [EC:2.4.1.-] | -0.402 | 0.438 | 0.360 | 0.978 |
| mgtC; putative Mg2+ transporter-C (MgtC) family protein | 0.071 | 0.103 | 0.489 | 0.979 |
| mgtE; magnesium transporter | 0.141 | 0.103 | 0.174 | 0.961 |
| mhpA; 3-(3-hydroxy-phenyl)propionate hydroxylase [EC:1.14.13.127] | -0.784 | 0.377 | 0.039 | 0.839 |
| mhpB; 2,3-dihydroxyphenylpropionate 1,2-dioxygenase [EC:1.13.11.16] | -0.815 | 0.653 | 0.214 | 0.978 |
| mhpC; 2-hydroxy-6-oxonona-2,4-dienedioate hydrolase [EC:3.7.1.14] | -1.148 | 0.806 | 0.156 | 0.954 |
| mhpco; 2-methyl-3-hydroxypyridine 5-carboxylic acid dioxygenase [EC:1.14.12.4] | 18.076 | 2733.666 | 0.995 | 0.999 |
| mhpD; 2-keto-4-pentenoate hydratase [EC:4.2.1.80] | -0.662 | 0.342 | 0.055 | 0.839 |
| mhpE; 4-hydroxy 2-oxovalerate aldolase [EC:4.1.3.39] | -1.155 | 0.628 | 0.067 | 0.839 |
| mhpF; acetaldehyde dehydrogenase [EC:1.2.1.10] | -1.101 | 0.703 | 0.119 | 0.917 |
| mhpR; IclR family transcriptional regulator, mhp operon transcriptional activator | -0.880 | 0.590 | 0.138 | 0.949 |
| mhpT; MFS transporter, AAHS family, 3-hydroxyphenylpropionic acid transporter | -0.116 | 0.352 | 0.742 | 0.999 |
| mhqR; MarR family transcriptional regulator, 2-MHQ and catechol-resistance regulon repressor | -0.763 | 0.321 | 0.019 | 0.839 |
| miaA, TRIT1; tRNA dimethylallyltransferase [EC:2.5.1.75] | 0.010 | 0.046 | 0.826 | 0.999 |
| miaB; tRNA-2-methylthio-N6-dimethylallyladenosine synthase [EC:2.8.4.3] | -0.014 | 0.067 | 0.828 | 0.999 |
| miaE; tRNA-(ms[2]io[6]A)-hydroxylase [EC:1.-.-.-] | 0.123 | 0.251 | 0.626 | 0.999 |
| migA; alpha-1,6-rhamnosyltransferase [EC:2.4.1.-] | 3.499 | 2.055 | 0.091 | 0.884 |
| minC; septum site-determining protein MinC | 0.096 | 0.122 | 0.434 | 0.978 |
| minD; septum site-determining protein MinD | 0.050 | 0.095 | 0.596 | 0.999 |
| minE; cell division topological specificity factor | 0.173 | 0.135 | 0.202 | 0.972 |
| mioC; MioC protein | 0.215 | 0.212 | 0.311 | 0.978 |
| MIOX; inositol oxygenase [EC:1.13.99.1] | -1.283 | 2.030 | 0.528 | 0.989 |
| mipA, ompV; MipA family protein | 0.428 | 0.269 | 0.114 | 0.917 |
| MKS1; Meckel syndrome type 1 protein | 0.159 | 0.783 | 0.840 | 0.999 |
| mlaA, vacJ; phospholipid-binding lipoprotein MlaA | 0.182 | 0.143 | 0.205 | 0.974 |
| mlaB; phospholipid transport system transporter-binding protein | 0.098 | 0.163 | 0.549 | 0.992 |
| mlaC; phospholipid transport system substrate-binding protein | 0.065 | 0.152 | 0.671 | 0.999 |
| mlaD, linM; phospholipid/cholesterol/gamma-HCH transport system substrate-binding protein | -0.162 | 0.119 | 0.173 | 0.961 |
| mlaE, linK; phospholipid/cholesterol/gamma-HCH transport system permease protein | -0.180 | 0.115 | 0.118 | 0.917 |
| mlaF, linL, mkl; phospholipid/cholesterol/gamma-HCH transport system ATP-binding protein | -0.068 | 0.091 | 0.456 | 0.979 |
| mlc; transcriptional regulator of PTS gene | 0.298 | 0.330 | 0.368 | 0.978 |
| mlhB, chnC; epsilon-lactone hydrolase [EC:3.1.1.83] | 18.832 | 3989.590 | 0.996 | 0.999 |
| mltA; membrane-bound lytic murein transglycosylase A [EC:4.2.2.-] | -0.100 | 0.173 | 0.562 | 0.992 |
| mltB; membrane-bound lytic murein transglycosylase B [EC:4.2.2.-] | 0.063 | 0.169 | 0.709 | 0.999 |
| mltC; membrane-bound lytic murein transglycosylase C [EC:4.2.2.-] | 0.231 | 0.308 | 0.455 | 0.978 |
| mltD, dniR; membrane-bound lytic murein transglycosylase D [EC:4.2.2.-] | -0.073 | 0.132 | 0.583 | 0.999 |
| mltE, emtA; membrane-bound lytic murein transglycosylase E [EC:4.2.2.-] | 0.050 | 0.594 | 0.933 | 0.999 |
| mltF; membrane-bound lytic murein transglycosylase F [EC:4.2.2.-] | 0.017 | 0.176 | 0.925 | 0.999 |
| MLYCD; malonyl-CoA decarboxylase [EC:4.1.1.9] | -1.210 | 0.641 | 0.061 | 0.839 |
| MMAB, pduO; cob(I)alamin adenosyltransferase [EC:2.5.1.17] | 0.041 | 0.091 | 0.652 | 0.999 |
| MME, CD10; neprilysin [EC:3.4.24.11] | -2.231 | 1.889 | 0.239 | 0.978 |
| mmoB; methane monooxygenase regulatory protein B | -17.368 | 3290.703 | 0.996 | 0.999 |
| mmoC; methane monooxygenase component C [EC:1.14.13.25] | -17.368 | 3290.703 | 0.996 | 0.999 |
| mmoX; methane monooxygenase component A alpha chain [EC:1.14.13.25] | -17.977 | 4462.995 | 0.997 | 0.999 |
| mmoY; methane monooxygenase component A beta chain [EC:1.14.13.25] | -17.977 | 4462.995 | 0.997 | 0.999 |
| mmoZ; methane monooxygenase component A gamma chain [EC:1.14.13.25] | -17.368 | 3290.703 | 0.996 | 0.999 |
| mmr; MFS transporter, DHA2 family, methylenomycin A resistance protein | -0.302 | 0.220 | 0.172 | 0.961 |
| mmsA, iolA, ALDH6A1; malonate-semialdehyde dehydrogenase (acetylating) / methylmalonate-semialdehyde dehydrogenase [EC:1.2.1.18 1.2.1.27] | -0.274 | 0.212 | 0.197 | 0.970 |
| mmsB, HIBADH; 3-hydroxyisobutyrate dehydrogenase [EC:1.1.1.31] | -0.178 | 0.129 | 0.170 | 0.961 |
| mmuM, BHMT2; homocysteine S-methyltransferase [EC:2.1.1.10] | -0.092 | 0.149 | 0.537 | 0.990 |
| mmuP; S-methylmethionine transporter | 0.011 | 0.252 | 0.965 | 0.999 |
| mngB; mannosylglycerate hydrolase [EC:3.2.1.170] | 0.052 | 0.617 | 0.933 | 0.999 |
| mngR, farR; GntR family transcriptional regulator, mannosyl-D-glycerate transport/metabolism system repressor | 0.073 | 0.804 | 0.928 | 0.999 |
| mnhA, mrpA; multicomponent Na+:H+ antiporter subunit A | -0.589 | 0.286 | 0.041 | 0.839 |
| mnhB, mrpB; multicomponent Na+:H+ antiporter subunit B | -0.488 | 0.319 | 0.129 | 0.932 |
| mnhC, mrpC; multicomponent Na+:H+ antiporter subunit C | -0.451 | 0.263 | 0.088 | 0.880 |
| mnhD, mrpD; multicomponent Na+:H+ antiporter subunit D | -0.473 | 0.258 | 0.069 | 0.840 |
| mnhE, mrpE; multicomponent Na+:H+ antiporter subunit E | -0.455 | 0.264 | 0.087 | 0.880 |
| mnhF, mrpF; multicomponent Na+:H+ antiporter subunit F | -0.500 | 0.355 | 0.161 | 0.957 |
| mnhG, mrpG; multicomponent Na+:H+ antiporter subunit G | -0.453 | 0.264 | 0.089 | 0.880 |
| mnmA, trmU; tRNA-uridine 2-sulfurtransferase [EC:2.8.1.13] | 0.097 | 0.057 | 0.088 | 0.880 |
| mnmC; tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein [EC:2.1.1.61 1.5.-.-] | 0.052 | 0.194 | 0.790 | 0.999 |
| mnmE, trmE, MSS1; tRNA modification GTPase [EC:3.6.-.-] | 0.041 | 0.042 | 0.330 | 0.978 |
| mnoP; high affinity Mn2+ porin | -0.411 | 0.812 | 0.614 | 0.999 |
| mntA; manganese transport system ATP-binding protein | -0.091 | 0.359 | 0.800 | 0.999 |
| mntB; manganese transport system permease protein | -0.011 | 0.369 | 0.976 | 0.999 |
| mntC; manganese transport system substrate-binding protein | 0.031 | 0.378 | 0.935 | 0.999 |
| mntH; manganese transport protein | -0.094 | 0.094 | 0.318 | 0.978 |
| mntR; DtxR family transcriptional regulator, manganese transport regulator | -0.025 | 0.406 | 0.951 | 0.999 |
| moaA, CNX2; GTP 3’,8-cyclase [EC:4.1.99.22] | -0.003 | 0.074 | 0.970 | 0.999 |
| moaB; molybdopterin adenylyltransferase [EC:2.7.7.75] | -0.032 | 0.139 | 0.818 | 0.999 |
| moaC, CNX3; cyclic pyranopterin monophosphate synthase [EC:4.6.1.17] | 0.012 | 0.077 | 0.881 | 0.999 |
| moaD, cysO; sulfur-carrier protein | -0.163 | 0.128 | 0.204 | 0.972 |
| mobA; molybdenum cofactor guanylyltransferase [EC:2.7.7.77] | -0.040 | 0.096 | 0.680 | 0.999 |
| mobAB; molybdopterin-guanine dinucleotide biosynthesis protein [EC:2.7.7.77] | 0.204 | 0.248 | 0.410 | 0.978 |
| mobB; molybdopterin-guanine dinucleotide biosynthesis adapter protein | -0.127 | 0.191 | 0.508 | 0.981 |
| mocA; molybdenum cofactor cytidylyltransferase [EC:2.7.7.76] | -0.077 | 0.128 | 0.549 | 0.992 |
| MOCS2B, moaE; molybdopterin synthase catalytic subunit [EC:2.8.1.12] | -0.156 | 0.113 | 0.167 | 0.961 |
| MOCS3, UBA4; adenylyltransferase and sulfurtransferase [EC:2.7.7.80 2.8.1.11] | -0.141 | 0.124 | 0.257 | 0.978 |
| mod; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | -0.176 | 0.300 | 0.559 | 0.992 |
| modA; molybdate transport system substrate-binding protein | -0.103 | 0.086 | 0.236 | 0.978 |
| modB; molybdate transport system permease protein | -0.035 | 0.079 | 0.660 | 0.999 |
| modC; molybdate transport system ATP-binding protein [EC:3.6.3.29] | -0.022 | 0.094 | 0.813 | 0.999 |
| modD; molybdenum transport protein [EC:2.4.2.-] | 0.173 | 0.492 | 0.725 | 0.999 |
| modE; molybdate transport system regulatory protein | 0.086 | 0.140 | 0.542 | 0.990 |
| modF; molybdate transport system ATP-binding protein | 0.052 | 0.300 | 0.862 | 0.999 |
| moeA; molybdopterin molybdotransferase [EC:2.10.1.1] | -0.054 | 0.089 | 0.545 | 0.990 |
| mogA; molybdopterin adenylyltransferase [EC:2.7.7.75] | -0.084 | 0.240 | 0.728 | 0.999 |
| mop; aldehyde oxidoreductase [EC:1.2.99.7] | 0.220 | 1.099 | 0.841 | 0.999 |
| motA; chemotaxis protein MotA | -0.079 | 0.169 | 0.641 | 0.999 |
| motB; chemotaxis protein MotB | -0.023 | 0.118 | 0.843 | 0.999 |
| motC; chemotaxis protein MotC | -2.135 | 1.313 | 0.106 | 0.911 |
| motD; chemotaxis protein MotD | -2.294 | 1.439 | 0.113 | 0.917 |
| moxR; MoxR-like ATPase [EC:3.6.3.-] | -0.140 | 0.119 | 0.241 | 0.978 |
| mp2; beta-1,4-mannooligosaccharide/beta-1,4-mannosyl-N-acetylglucosamine phosphorylase [EC:2.4.1.319 2.4.1.320] | -0.333 | 0.234 | 0.156 | 0.954 |
| mpa; proteasome-associated ATPase | 0.008 | 0.257 | 0.976 | 0.999 |
| mpaA; protein MpaA | 0.085 | 0.609 | 0.889 | 0.999 |
| MPG; DNA-3-methyladenine glycosylase [EC:3.2.2.21] | 0.115 | 0.154 | 0.458 | 0.979 |
| mph; macrolide phosphotransferase | -0.001 | 1.078 | 0.999 | 1.000 |
| mphR; TetR/AcrR family transcriptional regulator, macrolide resistance operon repressor | 0.469 | 1.077 | 0.664 | 0.999 |
| mpl; UDP-N-acetylmuramate: L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase [EC:6.3.2.45] | 0.077 | 0.155 | 0.621 | 0.999 |
| mpl; zinc metalloproteinase [EC:3.4.24.-] | -16.755 | 1738.673 | 0.992 | 0.999 |
| mprA; two-component system, OmpR family, response regulator MprA | -0.374 | 0.478 | 0.435 | 0.978 |
| mprB; two-component system, OmpR family, sensor histidine kinase MprB [EC:2.7.13.3] | -0.678 | 0.803 | 0.400 | 0.978 |
| mprF, fmtC; phosphatidylglycerol lysyltransferase [EC:2.3.2.3] | 0.145 | 0.134 | 0.281 | 0.978 |
| mps2; glycopeptidolipid biosynthesis protein | -17.763 | 3362.856 | 0.996 | 0.999 |
| mptA; alpha-1,6-mannosyltransferase [EC:2.4.1.-] | -0.538 | 0.597 | 0.370 | 0.978 |
| mptA; GTP cyclohydrolase IV [EC:3.5.4.39] | -1.047 | 1.651 | 0.527 | 0.989 |
| mptB; alpha-1,6-mannosyltransferase [EC:2.4.1.-] | -0.490 | 0.617 | 0.428 | 0.978 |
| mptE; 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase [EC:2.7.6.3] | -17.234 | 3077.514 | 0.996 | 0.999 |
| MQCRA, qcrA, bfcA, petC; menaquinol-cytochrome c reductase iron-sulfur subunit [EC:1.10.2.-] | -1.543 | 0.758 | 0.044 | 0.839 |
| MQCRB, qcrB, bfcB, petB; menaquinol-cytochrome c reductase cytochrome b subunit | -1.510 | 0.698 | 0.032 | 0.839 |
| MQCRC, qcrC, bfcC, petD; menaquinol-cytochrome c reductase cytochrome b/c subunit | -1.510 | 0.698 | 0.032 | 0.839 |
| mqnA; chorismate dehydratase [EC:4.2.1.151] | 0.021 | 0.429 | 0.961 | 0.999 |
| mqnB; futalosine hydrolase [EC:3.2.2.26] | -2.184 | 1.575 | 0.168 | 0.961 |
| mqnC; cyclic dehypoxanthinyl futalosine synthase [EC:1.21.98.1] | 0.117 | 0.443 | 0.791 | 0.999 |
| mqnD; 1,4-dihydroxy-6-naphthoate synthase [EC:1.14.-.-] | -0.427 | 0.591 | 0.472 | 0.979 |
| mqnE; aminodeoxyfutalosine synthase [EC:2.5.1.120] | 0.202 | 0.466 | 0.665 | 0.999 |
| mqo; malate dehydrogenase (quinone) [EC:1.1.5.4] | -0.090 | 0.169 | 0.594 | 0.999 |
| mqsA; HTH-type transcriptional regulator / antitoxin MqsA | -0.632 | 0.526 | 0.232 | 0.978 |
| mqsR; motility quorum-sensing regulator / GCU-specific mRNA interferase toxin | -0.541 | 0.541 | 0.319 | 0.978 |
| mraW, rsmH; 16S rRNA (cytosine1402-N4)-methyltransferase [EC:2.1.1.199] | 0.034 | 0.037 | 0.360 | 0.978 |
| mraY; phospho-N-acetylmuramoyl-pentapeptide-transferase [EC:2.7.8.13] | 0.031 | 0.035 | 0.377 | 0.978 |
| mraZ; MraZ protein | 0.013 | 0.062 | 0.830 | 0.999 |
| mrcA; penicillin-binding protein 1A [EC:2.4.1.129 3.4.16.4] | -0.023 | 0.051 | 0.654 | 0.999 |
| mrcB; penicillin-binding protein 1B [EC:2.4.1.129 3.4.16.4] | 0.111 | 0.178 | 0.536 | 0.990 |
| mrdA; penicillin-binding protein 2 [EC:3.4.16.4] | 0.063 | 0.089 | 0.476 | 0.979 |
| mreB; rod shape-determining protein MreB and related proteins | 0.129 | 0.085 | 0.132 | 0.941 |
| mreC; rod shape-determining protein MreC | 0.060 | 0.044 | 0.175 | 0.961 |
| mreD; rod shape-determining protein MreD | 0.077 | 0.054 | 0.157 | 0.954 |
| mrp, NUBPL; ATP-binding protein involved in chromosome partitioning | -0.140 | 0.076 | 0.067 | 0.839 |
| mrr; restriction system protein | -0.023 | 0.156 | 0.881 | 0.999 |
| mrx; macrolide resistance protein | 0.469 | 1.077 | 0.664 | 0.999 |
| mrx1; mycoredoxin [EC:1.20.4.3] | -0.508 | 0.522 | 0.333 | 0.978 |
| msbA; ATP-binding cassette, subfamily B, bacterial MsbA [EC:3.6.3.-] | 0.011 | 0.082 | 0.892 | 0.999 |
| mscK, kefA, aefA; potassium-dependent mechanosensitive channel | 0.183 | 0.208 | 0.379 | 0.978 |
| mscL; large conductance mechanosensitive channel | 0.044 | 0.036 | 0.222 | 0.978 |
| mscS; small conductance mechanosensitive channel | 0.096 | 0.056 | 0.087 | 0.880 |
| mshA; D-inositol-3-phosphate glycosyltransferase [EC:2.4.1.250] | -0.035 | 0.316 | 0.913 | 0.999 |
| mshA; MSHA pilin protein MshA | 0.535 | 0.848 | 0.529 | 0.989 |
| mshB; MSHA pilin protein MshB | 18.202 | 1851.167 | 0.992 | 0.999 |
| mshB; N-acetyl-1-D-myo-inositol-2-amino-2-deoxy-alpha-D-glucopyranoside deacetylase [EC:3.5.1.103] | -0.023 | 0.357 | 0.948 | 0.999 |
| mshC; L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase [EC:6.3.1.13] | -0.037 | 0.316 | 0.908 | 0.999 |
| mshC; MSHA pilin protein MshC | 0.207 | 0.875 | 0.813 | 0.999 |
| mshD; MSHA pilin protein MshD | 0.207 | 0.875 | 0.813 | 0.999 |
| mshD; mycothiol synthase [EC:2.3.1.189] | -0.027 | 0.318 | 0.933 | 0.999 |
| mshE; MSHA biogenesis protein MshE | 0.207 | 0.875 | 0.813 | 0.999 |
| mshF; MSHA biogenesis protein MshF | 18.482 | 3349.575 | 0.996 | 0.999 |
| mshG; MSHA biogenesis protein MshG | 0.207 | 0.875 | 0.813 | 0.999 |
| mshI; MSHA biogenesis protein MshI | 0.207 | 0.875 | 0.813 | 0.999 |
| mshJ; MSHA biogenesis protein MshJ | 0.015 | 1.131 | 0.989 | 0.999 |
| mshK; MSHA biogenesis protein MshK | 0.713 | 0.938 | 0.448 | 0.978 |
| mshL; MSHA biogenesis protein MshL | 0.207 | 0.875 | 0.813 | 0.999 |
| mshM; MSHA biogenesis protein MshM | 0.305 | 0.924 | 0.741 | 0.999 |
| mshN; MSHA biogenesis protein MshN | 0.015 | 1.131 | 0.989 | 0.999 |
| mshO; MSHA biogenesis protein MshO | 0.207 | 0.875 | 0.813 | 0.999 |
| mshP; MSHA biogenesis protein MshP | 0.154 | 0.892 | 0.863 | 0.999 |
| mshQ; MSHA biogenesis protein MshQ | -0.359 | 1.296 | 0.782 | 0.999 |
| msmE; raffinose/stachyose/melibiose transport system substrate-binding protein | -0.059 | 0.204 | 0.771 | 0.999 |
| msmF; raffinose/stachyose/melibiose transport system permease protein | -0.048 | 0.198 | 0.809 | 0.999 |
| msmG; raffinose/stachyose/melibiose transport system permease protein | -0.037 | 0.204 | 0.856 | 0.999 |
| msmX, msmK, malK, sugC, ggtA, msiK; multiple sugar transport system ATP-binding protein | -0.115 | 0.075 | 0.127 | 0.927 |
| msrA, vmlR; macrolide transport system ATP-binding/permease protein | -0.024 | 0.325 | 0.942 | 0.999 |
| msrA; peptide-methionine (S)-S-oxide reductase [EC:1.8.4.11] | -0.099 | 0.113 | 0.381 | 0.978 |
| msrAB; peptide methionine sulfoxide reductase msrA/msrB [EC:1.8.4.11 1.8.4.12] | 0.103 | 0.108 | 0.342 | 0.978 |
| msrB; peptide-methionine (R)-S-oxide reductase [EC:1.8.4.12] | -0.002 | 0.105 | 0.988 | 0.999 |
| msrP; methionine sulfoxide reductase catalytic subunit [EC:1.8.-.-] | -0.097 | 0.268 | 0.719 | 0.999 |
| msrQ; methionine sulfoxide reductase heme-binding subunit | -0.105 | 0.247 | 0.672 | 0.999 |
| msyB; acidic protein MsyB | 0.134 | 0.621 | 0.829 | 0.999 |
| mtaB; threonylcarbamoyladenosine tRNA methylthiotransferase MtaB [EC:2.8.4.5] | -0.037 | 0.131 | 0.777 | 0.999 |
| mtaD; 5-methylthioadenosine/S-adenosylhomocysteine deaminase [EC:3.5.4.31 3.5.4.28] | -0.026 | 0.093 | 0.782 | 0.999 |
| mtaP, MTAP; 5’-methylthioadenosine phosphorylase [EC:2.4.2.28] | -0.005 | 0.182 | 0.979 | 0.999 |
| mtdB; methylene-tetrahydromethanopterin dehydrogenase [EC:1.5.1.-] | 1.357 | 1.122 | 0.228 | 0.978 |
| mtfA; MtfA peptidase | 0.003 | 0.229 | 0.991 | 0.999 |
| mtfabH; beta-ketoacyl-[acyl-carrier-protein] synthase III [EC:2.3.1.180] | -2.005 | 1.868 | 0.285 | 0.978 |
| MTFMT, fmt; methionyl-tRNA formyltransferase [EC:2.1.2.9] | 0.031 | 0.032 | 0.338 | 0.978 |
| mtgA; monofunctional glycosyltransferase [EC:2.4.1.129] | -0.084 | 0.101 | 0.410 | 0.978 |
| MTHFS; 5-formyltetrahydrofolate cyclo-ligase [EC:6.3.3.2] | 0.070 | 0.043 | 0.108 | 0.915 |
| mtiP; 5’-methylthioinosine phosphorylase [EC:2.4.2.44] | 0.207 | 0.344 | 0.548 | 0.991 |
| mtkA; malate-CoA ligase subunit beta [EC:6.2.1.9] | 0.717 | 1.336 | 0.592 | 0.999 |
| mtkB; malate-CoA ligase subunit alpha [EC:6.2.1.9] | 0.717 | 1.336 | 0.592 | 0.999 |
| mtlD; mannitol-1-phosphate 5-dehydrogenase [EC:1.1.1.17] | 0.063 | 0.210 | 0.763 | 0.999 |
| mtlR; mannitol operon repressor | 0.158 | 0.565 | 0.780 | 0.999 |
| mtlR; mannitol operon transcriptional antiterminator | 0.226 | 0.287 | 0.432 | 0.978 |
| mtnA; methylthioribose-1-phosphate isomerase [EC:5.3.1.23] | 0.235 | 0.125 | 0.062 | 0.839 |
| mtnB; methylthioribulose-1-phosphate dehydratase [EC:4.2.1.109] | -0.025 | 0.294 | 0.931 | 0.999 |
| mtnC, ENOPH1; enolase-phosphatase E1 [EC:3.1.3.77] | 0.180 | 0.318 | 0.572 | 0.995 |
| mtnD, mtnZ, ADI1; 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase [EC:1.13.11.53 1.13.11.54] | -0.035 | 0.241 | 0.886 | 0.999 |
| mtnE, mtnV; aminotransferase [EC:2.6.1.-] | -0.066 | 0.326 | 0.839 | 0.999 |
| mtnK; 5-methylthioribose kinase [EC:2.7.1.100] | 0.614 | 0.336 | 0.070 | 0.841 |
| mtnN, mtn, pfs; adenosylhomocysteine nucleosidase [EC:3.2.2.9] | 0.029 | 0.063 | 0.646 | 0.999 |
| mtnW; 2,3-diketo-5-methylthiopentyl-1-phosphate enolase [EC:5.3.2.5] | -2.767 | 1.444 | 0.057 | 0.839 |
| mtnX; 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase [EC:3.1.3.87] | -1.438 | 0.929 | 0.124 | 0.919 |
| mtr; mycothione reductase [EC:1.8.1.15] | -0.067 | 0.333 | 0.840 | 0.999 |
| mtr; tryptophan-specific transport protein | 0.253 | 0.271 | 0.352 | 0.978 |
| mtrA; AraC family transcriptional regulator, activator of mtrCDE | 0.140 | 0.518 | 0.787 | 0.999 |
| mtrA; tetrahydromethanopterin S-methyltransferase subunit A [EC:2.1.1.86] | -17.234 | 3077.514 | 0.996 | 0.999 |
| mtrA; two-component system, OmpR family, response regulator MtrA | 0.008 | 0.268 | 0.976 | 0.999 |
| mtrB; transcription attenuation protein (tryptophan RNA-binding attenuator protein) | -1.221 | 0.708 | 0.086 | 0.880 |
| mtrB; two-component system, OmpR family, sensor histidine kinase MtrB [EC:2.7.13.3] | -0.036 | 0.286 | 0.899 | 0.999 |
| mtsA; iron/zinc/manganese/copper transport system substrate-binding protein | -0.012 | 0.156 | 0.938 | 0.999 |
| mtsB; iron/zinc/manganese/copper transport system ATP-binding protein | -0.012 | 0.156 | 0.937 | 0.999 |
| mtsC; iron/zinc/manganese/copper transport system permease protein | -0.012 | 0.156 | 0.939 | 0.999 |
| mtsT; energy-coupling factor transport system substrate-specific component | -0.001 | 0.141 | 0.995 | 0.999 |
| mttB; trimethylamine—corrinoid protein Co-methyltransferase [EC:2.1.1.250] | -2.132 | 1.532 | 0.166 | 0.961 |
| MuB; ATP-dependent target DNA activator [EC:3.6.1.3] | -0.276 | 0.454 | 0.545 | 0.990 |
| mucK; MFS transporter, AAHS family, cis,cis-muconate transporter | -0.677 | 1.048 | 0.519 | 0.988 |
| mug; double-stranded uracil-DNA glycosylase [EC:3.2.2.28] | -0.756 | 0.369 | 0.042 | 0.839 |
| mukB; chromosome partition protein MukB | 0.207 | 0.312 | 0.508 | 0.981 |
| mukE; chromosome partition protein MukE | 0.207 | 0.312 | 0.508 | 0.981 |
| mukF; chromosome partition protein MukF | 0.207 | 0.312 | 0.508 | 0.981 |
| murA; UDP-N-acetylglucosamine 1-carboxyvinyltransferase [EC:2.5.1.7] | 0.030 | 0.050 | 0.552 | 0.992 |
| murB; UDP-N-acetylmuramate dehydrogenase [EC:1.3.1.98] | 0.032 | 0.037 | 0.376 | 0.978 |
| murC; UDP-N-acetylmuramate–alanine ligase [EC:6.3.2.8] | 0.040 | 0.039 | 0.303 | 0.978 |
| murD; UDP-N-acetylmuramoylalanine–D-glutamate ligase [EC:6.3.2.9] | 0.036 | 0.036 | 0.321 | 0.978 |
| murE; UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–2,6-diaminopimelate ligase [EC:6.3.2.13] | 0.061 | 0.055 | 0.274 | 0.978 |
| murE; UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-L-lysine ligase [EC:6.3.2.7] | -0.039 | 0.161 | 0.810 | 0.999 |
| murEF; murE/murF fusion protein [EC:6.3.2.13 6.3.2.10] | 0.345 | 0.558 | 0.537 | 0.990 |
| murF; UDP-N-acetylmuramoyl-tripeptide–D-alanyl-D-alanine ligase [EC:6.3.2.10] | 0.023 | 0.041 | 0.584 | 0.999 |
| murG; UDP-N-acetylglucosamine–N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase [EC:2.4.1.227] | 0.030 | 0.033 | 0.353 | 0.978 |
| murI; glutamate racemase [EC:5.1.1.3] | 0.034 | 0.046 | 0.454 | 0.978 |
| murJ, mviN; putative peptidoglycan lipid II flippase | 0.142 | 0.107 | 0.189 | 0.966 |
| murM; serine/alanine adding enzyme [EC:2.3.2.10] | -0.026 | 0.171 | 0.880 | 0.999 |
| murN; alanine adding enzyme [EC:2.3.2.-] | -0.070 | 0.171 | 0.680 | 0.999 |
| murQ; N-acetylmuramic acid 6-phosphate etherase [EC:4.2.1.126] | -0.020 | 0.123 | 0.870 | 0.999 |
| murR; RpiR family transcriptional regulator, murPQ operon repressor | -0.993 | 0.976 | 0.311 | 0.978 |
| murU; N-acetyl-alpha-D-muramate 1-phosphate uridylyltransferase [EC:2.7.7.99] | -0.136 | 0.181 | 0.453 | 0.978 |
| MUT; methylmalonyl-CoA mutase [EC:5.4.99.2] | -0.219 | 0.167 | 0.192 | 0.970 |
| mutH; DNA mismatch repair protein MutH | 0.231 | 0.308 | 0.455 | 0.978 |
| mutL; DNA mismatch repair protein MutL | 0.042 | 0.041 | 0.310 | 0.978 |
| mutM, fpg; formamidopyrimidine-DNA glycosylase [EC:3.2.2.23 4.2.99.18] | 0.003 | 0.060 | 0.960 | 0.999 |
| mutS; DNA mismatch repair protein MutS | 0.042 | 0.038 | 0.274 | 0.978 |
| mutS2; DNA mismatch repair protein MutS2 | 0.067 | 0.088 | 0.443 | 0.978 |
| mutT, NUDT15, MTH2; 8-oxo-dGTP diphosphatase [EC:3.6.1.55] | -0.018 | 0.082 | 0.828 | 0.999 |
| mutY; A/G-specific adenine glycosylase [EC:3.2.2.31] | -0.055 | 0.050 | 0.274 | 0.978 |
| mvaA; hydroxymethylglutaryl-CoA reductase [EC:1.1.1.88] | 0.103 | 0.132 | 0.435 | 0.978 |
| MVD, mvaD; diphosphomevalonate decarboxylase [EC:4.1.1.33] | 0.122 | 0.134 | 0.364 | 0.978 |
| mviM; virulence factor | 0.077 | 0.237 | 0.746 | 0.999 |
| mvpA, vapC; tRNA(fMet)-specific endonuclease VapC [EC:3.1.-.-] | -0.186 | 0.225 | 0.410 | 0.978 |
| mvpT, vapB; antitoxin VapB | 0.009 | 0.317 | 0.978 | 0.999 |
| mxaA; mxaA protein | 2.133 | 1.580 | 0.179 | 0.961 |
| mxaC; mxaC protein | 2.498 | 1.682 | 0.140 | 0.949 |
| mxaD; mxaD protein | 2.154 | 1.456 | 0.141 | 0.949 |
| mxaG; cytochrome c-L | 2.154 | 1.456 | 0.141 | 0.949 |
| mxaJ; mxaJ protein | 2.154 | 1.456 | 0.141 | 0.949 |
| mxaK; mxaK protein | 2.498 | 1.682 | 0.140 | 0.949 |
| mxaL; mxaL protein | 2.133 | 1.580 | 0.179 | 0.961 |
| mycP; membrane-anchored mycosin MYCP [EC:3.4.21.-] | -0.982 | 0.591 | 0.099 | 0.896 |
| NAALAD; N-acetylated-alpha-linked acidic dipeptidase [EC:3.4.17.21] | -2.301 | 1.700 | 0.178 | 0.961 |
| nac; LysR family transcriptional regulator, nitrogen assimilation regulatory protein | -0.502 | 0.339 | 0.141 | 0.949 |
| nadA; quinolinate synthase [EC:2.5.1.72] | -0.065 | 0.105 | 0.538 | 0.990 |
| nadB; L-aspartate oxidase [EC:1.4.3.16] | -0.040 | 0.107 | 0.707 | 0.999 |
| nadC, QPRT; nicotinate-nucleotide pyrophosphorylase (carboxylating) [EC:2.4.2.19] | -0.079 | 0.095 | 0.411 | 0.978 |
| nadD; nicotinate-nucleotide adenylyltransferase [EC:2.7.7.18] | 0.026 | 0.043 | 0.543 | 0.990 |
| nadE; NAD+ synthase [EC:6.3.1.5] | 0.070 | 0.067 | 0.293 | 0.978 |
| nadM; nicotinamide-nucleotide adenylyltransferase [EC:2.7.7.1] | 0.793 | 0.576 | 0.171 | 0.961 |
| nadR; HTH-type transcriptional regulator, transcriptional repressor of NAD biosynthesis genes [EC:2.7.7.1 2.7.1.22] | 0.348 | 0.252 | 0.170 | 0.961 |
| nadX, ASPDH; aspartate dehydrogenase [EC:1.4.1.21] | -0.604 | 0.424 | 0.156 | 0.954 |
| nagA, AMDHD2; N-acetylglucosamine-6-phosphate deacetylase [EC:3.5.1.25] | 0.002 | 0.065 | 0.971 | 0.999 |
| nagB, GNPDA; glucosamine-6-phosphate deaminase [EC:3.5.99.6] | 0.027 | 0.088 | 0.756 | 0.999 |
| nagC; N-acetylglucosamine repressor | 0.469 | 0.353 | 0.186 | 0.966 |
| nagD; NagD protein | 0.424 | 0.262 | 0.108 | 0.916 |
| nagG; salicylate 5-hydroxylase large subunit [EC:1.14.13.172] | -3.045 | 2.774 | 0.274 | 0.978 |
| nagH; salicylate 5-hydroxylase small subunit [EC:1.14.13.172] | -3.053 | 2.762 | 0.271 | 0.978 |
| NAGK, nagK; N-acetylglucosamine kinase [EC:2.7.1.59] | 0.055 | 0.243 | 0.820 | 0.999 |
| nagK; fumarylpyruvate hydrolase [EC:3.7.1.20] | -0.886 | 0.359 | 0.015 | 0.839 |
| nagL; maleylpyruvate isomerase [EC:5.2.1.4] | -0.659 | 0.361 | 0.070 | 0.841 |
| NAGLU; alpha-N-acetylglucosaminidase [EC:3.2.1.50] | -0.960 | 0.591 | 0.107 | 0.912 |
| nagZ; beta-N-acetylhexosaminidase [EC:3.2.1.52] | -0.084 | 0.105 | 0.424 | 0.978 |
| nahAa, nagAa, ndoR, nbzAa, dntAa; naphthalene 1,2-dioxygenase ferredoxin reductase component [EC:1.18.1.7] | -1.511 | 1.419 | 0.289 | 0.978 |
| nahAb, nagAb, ndoA, nbzAb, dntAb; naphthalene 1,2-dioxygenase ferredoxin component | -3.045 | 2.774 | 0.274 | 0.978 |
| nahK, lnpB; N-acetylhexosamine 1-kinase [EC:2.7.1.162] | -0.068 | 0.450 | 0.879 | 0.999 |
| nalC; TetR/AcrR family transcriptional regulator, transcriptional repressor NalC | -17.256 | 3111.566 | 0.996 | 0.999 |
| nalD; TetR/AcrR family transcriptional regulator, repressor of the mexAB-oprM multidrug resistance operon | 0.634 | 0.449 | 0.160 | 0.954 |
| namH; UDP-MurNAc hydroxylase | -1.723 | 1.212 | 0.157 | 0.954 |
| NAMPT; nicotinamide phosphoribosyltransferase [EC:2.4.2.12] | -0.033 | 0.331 | 0.920 | 0.999 |
| nanE; N-acylglucosamine-6-phosphate 2-epimerase [EC:5.1.3.9] | 0.144 | 0.101 | 0.157 | 0.954 |
| nanEK; N-acetylmannosamine-6-phosphate 2-epimerase / N-acetylmannosamine kinase [EC:5.1.3.9 2.7.1.60] | -2.185 | 2.024 | 0.282 | 0.978 |
| nanK; N-acylmannosamine kinase [EC:2.7.1.60] | 0.107 | 0.335 | 0.749 | 0.999 |
| nanM; N-acetylneuraminate epimerase [EC:5.1.3.24] | -0.296 | 0.565 | 0.602 | 0.999 |
| NANS, SAS; sialic acid synthase [EC:2.5.1.56 2.5.1.57 2.5.1.132] | -2.641 | 2.722 | 0.333 | 0.978 |
| nanT; MFS transporter, SHS family, sialic acid transporter | -1.421 | 0.976 | 0.148 | 0.954 |
| napA; periplasmic nitrate reductase NapA [EC:1.7.99.-] | 0.222 | 0.235 | 0.348 | 0.978 |
| napB; cytochrome c-type protein NapB | 0.266 | 0.239 | 0.266 | 0.978 |
| napC; cytochrome c-type protein NapC | 0.180 | 0.247 | 0.468 | 0.979 |
| napD; periplasmic nitrate reductase NapD | 0.223 | 0.235 | 0.346 | 0.978 |
| napE; periplasmic nitrate reductase NapE | 0.825 | 0.419 | 0.051 | 0.839 |
| NAPEPLD; N-acyl-phosphatidylethanolamine-hydrolysing phospholipase D [EC:3.1.4.54] | 17.966 | 2587.320 | 0.994 | 0.999 |
| napF; ferredoxin-type protein NapF | 0.150 | 0.285 | 0.598 | 0.999 |
| napG; ferredoxin-type protein NapG | 0.079 | 0.285 | 0.781 | 0.999 |
| napH; ferredoxin-type protein NapH | 0.111 | 0.283 | 0.696 | 0.999 |
| narB; ferredoxin-nitrate reductase [EC:1.7.7.2] | -17.319 | 2105.764 | 0.993 | 0.999 |
| narC; cytochrome b-561 | -1.940 | 2.261 | 0.392 | 0.978 |
| narG, narZ, nxrA; nitrate reductase / nitrite oxidoreductase, alpha subunit [EC:1.7.5.1 1.7.99.-] | 0.080 | 0.126 | 0.529 | 0.989 |
| narH, narY, nxrB; nitrate reductase / nitrite oxidoreductase, beta subunit [EC:1.7.5.1 1.7.99.-] | 0.080 | 0.125 | 0.520 | 0.988 |
| narI, narV; nitrate reductase gamma subunit [EC:1.7.5.1 1.7.99.-] | 0.080 | 0.126 | 0.526 | 0.989 |
| narJ, narW; nitrate reductase molybdenum cofactor assembly chaperone NarJ/NarW | 0.081 | 0.128 | 0.531 | 0.989 |
| narL; two-component system, NarL family, nitrate/nitrite response regulator NarL | -0.395 | 0.266 | 0.141 | 0.949 |
| narP; two-component system, NarL family, nitrate/nitrite response regulator NarP | 0.229 | 0.317 | 0.472 | 0.979 |
| narQ; two-component system, NarL family, nitrate/nitrite sensor histidine kinase NarQ [EC:2.7.13.3] | 0.243 | 0.322 | 0.452 | 0.978 |
| NARS, asnS; asparaginyl-tRNA synthetase [EC:6.1.1.22] | 0.069 | 0.077 | 0.371 | 0.978 |
| narT; MFS transporter, NNP family, putative nitrate transporter | -0.469 | 0.677 | 0.489 | 0.979 |
| narX; two-component system, NarL family, nitrate/nitrite sensor histidine kinase NarX [EC:2.7.13.3] | -0.236 | 0.274 | 0.391 | 0.978 |
| nasA; assimilatory nitrate reductase catalytic subunit [EC:1.7.99.-] | -0.210 | 0.249 | 0.401 | 0.978 |
| nasB; assimilatory nitrate reductase electron transfer subunit [EC:1.7.99.-] | 0.110 | 0.700 | 0.876 | 0.999 |
| nasT; two-component system, response regulator / RNA-binding antiterminator | 0.144 | 0.109 | 0.187 | 0.966 |
| nat; arylamine N-acetyltransferase [EC:2.3.1.5] | -2.534 | 1.465 | 0.086 | 0.878 |
| nat; isonocardicin synthase [EC:2.5.1.38] | -0.145 | 0.723 | 0.842 | 0.999 |
| natA; sodium transport system ATP-binding protein [EC:3.6.3.7] | -0.114 | 0.418 | 0.786 | 0.999 |
| natB; sodium transport system permease protein | -0.110 | 0.419 | 0.794 | 0.999 |
| natD; neutral amino acid transport system permease protein | -17.763 | 3362.856 | 0.996 | 0.999 |
| natE; neutral amino acid transport system ATP-binding protein | -18.156 | 3269.627 | 0.996 | 0.999 |
| nboR; nicotine blue oxidoreductase [EC:1.1.1.328] | -0.078 | 0.495 | 0.875 | 0.999 |
| ncd2, npd; nitronate monooxygenase [EC:1.13.12.16] | -0.222 | 0.161 | 0.170 | 0.961 |
| ndh; NADH dehydrogenase [EC:1.6.99.3] | -0.088 | 0.074 | 0.233 | 0.978 |
| ndhE; NAD(P)H-quinone oxidoreductase subunit 4L [EC:1.6.5.3] | -0.701 | 1.092 | 0.522 | 0.988 |
| ndhF; NAD(P)H-quinone oxidoreductase subunit 5 [EC:1.6.5.3] | -0.644 | 0.470 | 0.173 | 0.961 |
| ndhG; NAD(P)H-quinone oxidoreductase subunit 6 [EC:1.6.5.3] | -1.859 | 2.280 | 0.416 | 0.978 |
| ndk, NME; nucleoside-diphosphate kinase [EC:2.7.4.6] | 0.008 | 0.050 | 0.865 | 0.999 |
| ndoAI; CopG family transcriptional regulator / antitoxin EndoAI | -0.802 | 0.506 | 0.115 | 0.917 |
| ndpA; nucleoid-associated protein | 0.244 | 0.211 | 0.249 | 0.978 |
| NDUFAF7; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 7 | -1.128 | 1.396 | 0.420 | 0.978 |
| ndx1; diadenosine hexaphosphate hydrolase (ATP-forming) [EC:3.6.1.61] | -1.769 | 2.449 | 0.471 | 0.979 |
| nei; endonuclease VIII [EC:3.2.2.- 4.2.99.18] | -0.065 | 0.238 | 0.784 | 0.999 |
| nemA; N-ethylmaleimide reductase [EC:1.-.-.-] | -0.350 | 0.255 | 0.172 | 0.961 |
| nemR; TetR/AcrR family transcriptional regulator, transcriptional repressor for nem operon | -0.381 | 0.237 | 0.109 | 0.917 |
| nepI; MFS transporter, DHA1 family, purine ribonucleoside efflux pump | 0.512 | 0.385 | 0.186 | 0.966 |
| ner, nlp, sfsB; Ner family transcriptional regulator | 0.085 | 0.392 | 0.829 | 0.999 |
| NEU1; sialidase-1 [EC:3.2.1.18] | -0.110 | 0.156 | 0.481 | 0.979 |
| neuA; N-acylneuraminate cytidylyltransferase [EC:2.7.7.43] | -0.123 | 0.278 | 0.659 | 0.999 |
| nfdA; N-substituted formamide deformylase [EC:3.5.1.91] | 0.036 | 0.138 | 0.795 | 0.999 |
| nfeD; membrane-bound serine protease (ClpP class) | 0.124 | 0.177 | 0.487 | 0.979 |
| nfnB, nfsB; nitroreductase / dihydropteridine reductase [EC:1.-.-.- 1.5.1.34] | 0.134 | 0.233 | 0.566 | 0.995 |
| nfo; deoxyribonuclease IV [EC:3.1.21.2] | 0.035 | 0.123 | 0.773 | 0.999 |
| nfrA; bacteriophage N4 adsorption protein A | -0.223 | 0.965 | 0.817 | 0.999 |
| nfrA1; FMN reductase (NADPH) [EC:1.5.1.38] | -0.643 | 0.275 | 0.021 | 0.839 |
| nfrA2; FMN reductase [NAD(P)H] [EC:1.5.1.39] | -0.123 | 0.248 | 0.620 | 0.999 |
| nfrB; bacteriophage N4 adsorption protein B | -0.087 | 0.378 | 0.818 | 0.999 |
| nfsA; nitroreductase [EC:1.-.-.-] | 0.206 | 0.346 | 0.552 | 0.992 |
| nfuA; Fe/S biogenesis protein NfuA | 0.106 | 0.172 | 0.538 | 0.990 |
| nfxB; TetR/AcrR family transcriptional regulator, mexCD-oprJ operon repressor | -0.025 | 0.479 | 0.959 | 0.999 |
| nga; NAD+ glycohydrolase [EC:3.2.2.5] | -16.913 | 2621.982 | 0.995 | 0.999 |
| nhaA; Na+:H+ antiporter, NhaA family | 0.008 | 0.103 | 0.941 | 0.999 |
| nhaB; Na+:H+ antiporter, NhaB family | 0.220 | 0.265 | 0.408 | 0.978 |
| nhaC; Na+:H+ antiporter, NhaC family | 0.578 | 0.216 | 0.008 | 0.839 |
| nhaR; LysR family transcriptional regulator, transcriptional activator of nhaA | -0.151 | 0.307 | 0.623 | 0.999 |
| nheA; non-hemolytic enterotoxin A | -1.438 | 1.843 | 0.437 | 0.978 |
| nheBC; non-hemolytic enterotoxin B/C | -2.138 | 1.905 | 0.263 | 0.978 |
| nhoA; N-hydroxyarylamine O-acetyltransferase [EC:2.3.1.118] | -0.398 | 0.307 | 0.197 | 0.970 |
| niaX; niacin transporter | 0.059 | 0.158 | 0.710 | 0.999 |
| nicA; nicotinate dehydrogenase subunit A [EC:1.17.2.1] | -0.136 | 0.414 | 0.743 | 0.999 |
| nicB; nicotinate dehydrogenase subunit B [EC:1.17.2.1] | -1.262 | 0.758 | 0.098 | 0.896 |
| nicC; 6-hydroxynicotinate 3-monooxygenase [EC:1.14.13.114] | -1.020 | 0.779 | 0.193 | 0.970 |
| nicD; N-formylmaleamate deformylase [EC:3.5.1.106] | -0.896 | 0.878 | 0.309 | 0.978 |
| nicE, maiA; maleate isomerase [EC:5.2.1.1] | 0.122 | 0.395 | 0.759 | 0.999 |
| nicF; maleamate amidohydrolase [EC:3.5.1.107] | -0.763 | 0.779 | 0.329 | 0.978 |
| nicX; 2,5-dihydroxypyridine 5,6-dioxygenase [EC:1.13.11.9] | -0.975 | 0.881 | 0.270 | 0.978 |
| nifA; Nif-specific regulatory protein | 0.059 | 0.712 | 0.934 | 0.999 |
| nifB; nitrogen fixation protein NifB | -0.386 | 0.573 | 0.502 | 0.981 |
| nifD; nitrogenase molybdenum-iron protein alpha chain [EC:1.18.6.1] | 0.361 | 0.562 | 0.522 | 0.988 |
| nifE; nitrogenase molybdenum-cofactor synthesis protein NifE | 0.149 | 0.409 | 0.715 | 0.999 |
| nifH; nitrogenase iron protein NifH [EC:1.18.6.1] | 0.681 | 0.329 | 0.040 | 0.839 |
| nifHD1, nifI1; nitrogen regulatory protein PII 1 | -0.506 | 1.046 | 0.629 | 0.999 |
| nifHD2, nifI2; nitrogen regulatory protein PII 2 | -0.506 | 1.046 | 0.629 | 0.999 |
| nifK; nitrogenase molybdenum-iron protein beta chain [EC:1.18.6.1] | -1.721 | 1.155 | 0.138 | 0.949 |
| nifN; nitrogenase molybdenum-iron protein NifN | -1.810 | 1.393 | 0.196 | 0.970 |
| nifQ; nitrogen fixation protein NifQ | -1.844 | 1.592 | 0.249 | 0.978 |
| nifT; nitrogen fixation protein NifT | -1.746 | 1.498 | 0.246 | 0.978 |
| nifV; homocitrate synthase NifV [EC:2.3.3.14] | -0.512 | 0.530 | 0.336 | 0.978 |
| nifW; nitrogenase-stabilizing/protective protein | -1.746 | 1.498 | 0.246 | 0.978 |
| nifX; nitrogen fixation protein NifX | -1.746 | 1.498 | 0.246 | 0.978 |
| nifZ; nitrogen fixation protein NifZ | -2.140 | 1.787 | 0.233 | 0.978 |
| nikA; nickel transport system substrate-binding protein | 0.488 | 0.365 | 0.184 | 0.965 |
| nikB; nickel transport system permease protein | 0.513 | 0.370 | 0.168 | 0.961 |
| nikC; nickel transport system permease protein | 0.328 | 0.220 | 0.138 | 0.949 |
| nikD; nickel transport system ATP-binding protein [EC:3.6.3.24] | 0.576 | 0.376 | 0.127 | 0.927 |
| nikE; nickel transport system ATP-binding protein [EC:3.6.3.24] | 0.538 | 0.371 | 0.149 | 0.954 |
| nikR; CopG family transcriptional regulator, nickel-responsive regulator | -0.617 | 0.371 | 0.099 | 0.896 |
| NIP; aquaporin NIP | -17.703 | 3891.582 | 0.996 | 0.999 |
| nirA; ferredoxin-nitrite reductase [EC:1.7.7.1] | 0.062 | 0.234 | 0.792 | 0.999 |
| nirB; nitrite reductase (NADH) large subunit [EC:1.7.1.15] | -0.186 | 0.174 | 0.287 | 0.978 |
| nirC; cytochrome c55X | -1.146 | 0.826 | 0.167 | 0.961 |
| nirC; nitrite transporter | 0.087 | 0.407 | 0.832 | 0.999 |
| nirD; nitrite reductase (NADH) small subunit [EC:1.7.1.15] | -0.186 | 0.179 | 0.301 | 0.978 |
| nirF; protein NirF | -1.146 | 0.826 | 0.167 | 0.961 |
| nirK; nitrite reductase (NO-forming) [EC:1.7.2.1] | 0.108 | 0.216 | 0.617 | 0.999 |
| nirS; nitrite reductase (NO-forming) / hydroxylamine reductase [EC:1.7.2.1 1.7.99.1] | -1.082 | 0.784 | 0.170 | 0.961 |
| nixA; high-affinity nickel-transport protein | -1.029 | 0.567 | 0.071 | 0.843 |
| nlpC; probable lipoprotein NlpC | 0.158 | 0.305 | 0.605 | 0.999 |
| nlpD; lipoprotein NlpD | 0.064 | 0.154 | 0.680 | 0.999 |
| nlpI; lipoprotein NlpI | 0.224 | 0.306 | 0.466 | 0.979 |
| nmpC, ompD; outer membrane porin protein LC | -0.644 | 0.975 | 0.510 | 0.981 |
| nnr; ADP-dependent NAD(P)H-hydrate dehydratase [EC:4.2.1.136] | 0.382 | 0.467 | 0.415 | 0.978 |
| nob1; endoribonuclease Nob1 [EC:3.1.-.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| nodA; nodulation protein A [EC:2.3.1.-] | -1.162 | 1.572 | 0.461 | 0.979 |
| nodB; chitooligosaccharide deacetylase [EC:3.5.1.-] | -1.378 | 1.742 | 0.430 | 0.978 |
| nodC; N-acetylglucosaminyltransferase [EC:2.4.1.-] | -1.162 | 1.572 | 0.461 | 0.979 |
| nodD; LysR family transcriptional regulator, nod-box dependent transcriptional activator | -1.091 | 1.548 | 0.482 | 0.979 |
| nodE; nodulation protein E [EC:2.3.1.-] | -0.513 | 0.625 | 0.413 | 0.978 |
| nodF; nodulation protein F [EC:2.3.1.-] | 0.717 | 1.336 | 0.592 | 0.999 |
| nodI; lipooligosaccharide transport system ATP-binding protein | -0.547 | 0.424 | 0.198 | 0.970 |
| nodJ; lipooligosaccharide transport system permease protein | -0.702 | 0.488 | 0.152 | 0.954 |
| nodT, ameC; outer membrane protein, multidrug efflux system | -3.065 | 2.530 | 0.228 | 0.978 |
| NOP10, NOLA3; H/ACA ribonucleoprotein complex subunit 3 | -17.234 | 3077.514 | 0.996 | 0.999 |
| NOP2; 25S rRNA (cytosine2870-C5)-methyltransferase [EC:2.1.1.310] | -1.647 | 2.132 | 0.441 | 0.978 |
| NOP56; nucleolar protein 56 | -17.234 | 3077.514 | 0.996 | 0.999 |
| norA; MFS transporter, DHA1 family, quinolone resistance protein | -1.554 | 1.110 | 0.164 | 0.957 |
| norB, norC; MFS transporter, DHA2 family, multidrug resistance protein | -0.570 | 0.646 | 0.379 | 0.978 |
| norB; nitric oxide reductase subunit B [EC:1.7.2.5] | -0.035 | 0.214 | 0.872 | 0.999 |
| norC; nitric oxide reductase subunit C | -1.269 | 0.648 | 0.052 | 0.839 |
| norD; nitric oxide reductase NorD protein | -1.301 | 0.654 | 0.048 | 0.839 |
| norE; nitric oxide reductase NorE protein | -1.340 | 0.639 | 0.038 | 0.839 |
| norG; GntR family transcriptional regulator, regulator for abcA and norABC | -1.279 | 0.729 | 0.081 | 0.865 |
| norQ; nitric oxide reductase NorQ protein | -1.605 | 0.485 | 0.001 | 0.839 |
| norR; anaerobic nitric oxide reductase transcription regulator | 0.151 | 0.296 | 0.610 | 0.999 |
| norV; anaerobic nitric oxide reductase flavorubredoxin | 0.453 | 0.567 | 0.425 | 0.978 |
| norW; nitric oxide reductase FlRd-NAD(+) reductase [EC:1.18.1.-] | 0.467 | 0.563 | 0.407 | 0.978 |
| nos; nitric-oxide synthase, bacterial [EC:1.14.14.47] | -0.962 | 0.473 | 0.044 | 0.839 |
| nosD; nitrous oxidase accessory protein | -0.524 | 0.327 | 0.111 | 0.917 |
| nosF; Cu-processing system ATP-binding protein | -0.480 | 0.349 | 0.171 | 0.961 |
| nosL; copper chaperone NosL | -0.552 | 0.347 | 0.113 | 0.917 |
| nosR; NosR/NirI family transcriptional regulator, nitrous oxide reductase regulator | -0.644 | 0.449 | 0.153 | 0.954 |
| nosY; Cu-processing system permease protein | -0.644 | 0.387 | 0.098 | 0.896 |
| nosZ; nitrous-oxide reductase [EC:1.7.2.4] | -0.412 | 0.313 | 0.190 | 0.968 |
| nox1; NADH oxidase (H2O2-forming) [EC:1.6.3.3] | 0.954 | 0.891 | 0.286 | 0.978 |
| nox2; NADH oxidase (H2O-forming) [EC:1.6.3.4] | 2.006 | 1.015 | 0.050 | 0.839 |
| npdA; NAD-dependent deacetylase [EC:3.5.1.-] | 0.080 | 0.072 | 0.269 | 0.978 |
| NPEPPS; puromycin-sensitive aminopeptidase [EC:3.4.11.-] | -0.651 | 1.172 | 0.579 | 0.997 |
| npr; NADH peroxidase [EC:1.11.1.1] | 0.501 | 0.402 | 0.215 | 0.978 |
| npr; thermolysin [EC:3.4.24.27] | -3.737 | 2.743 | 0.175 | 0.961 |
| nprB; neutral peptidase B [EC:3.4.24.-] | -2.480 | 2.207 | 0.263 | 0.978 |
| nprE; bacillolysin [EC:3.4.24.28] | -17.763 | 3362.856 | 0.996 | 0.999 |
| NQO1; NAD(P)H dehydrogenase (quinone) [EC:1.6.5.2] | 0.017 | 0.117 | 0.884 | 0.999 |
| nqrA; Na+-transporting NADH:ubiquinone oxidoreductase subunit A [EC:1.6.5.8] | -0.003 | 0.165 | 0.983 | 0.999 |
| nqrB; Na+-transporting NADH:ubiquinone oxidoreductase subunit B [EC:1.6.5.8] | -0.003 | 0.165 | 0.985 | 0.999 |
| nqrC; Na+-transporting NADH:ubiquinone oxidoreductase subunit C [EC:1.6.5.8] | -0.003 | 0.165 | 0.984 | 0.999 |
| nqrD; Na+-transporting NADH:ubiquinone oxidoreductase subunit D [EC:1.6.5.8] | -0.003 | 0.165 | 0.985 | 0.999 |
| nqrE; Na+-transporting NADH:ubiquinone oxidoreductase subunit E [EC:1.6.5.8] | -0.003 | 0.165 | 0.986 | 0.999 |
| nqrF; Na+-transporting NADH:ubiquinone oxidoreductase subunit F [EC:1.6.5.8] | -0.003 | 0.165 | 0.985 | 0.999 |
| nrdG; anaerobic ribonucleoside-triphosphate reductase activating protein [EC:1.97.1.4] | 0.102 | 0.085 | 0.229 | 0.978 |
| nrdH; glutaredoxin-like protein NrdH | 0.069 | 0.122 | 0.574 | 0.995 |
| nrdI; protein involved in ribonucleotide reduction | 0.030 | 0.121 | 0.801 | 0.999 |
| nrdR; transcriptional repressor NrdR | 0.084 | 0.036 | 0.022 | 0.839 |
| nreA; nitrogen regulatory protein A | -0.143 | 0.545 | 0.794 | 0.999 |
| nreB; two-component system, NarL family, sensor histidine kinase NreB [EC:2.7.13.3] | 0.075 | 0.499 | 0.881 | 0.999 |
| nreC; two-component system, NarL family, response regulator NreC | 0.362 | 0.476 | 0.449 | 0.978 |
| nrfA; nitrite reductase (cytochrome c-552) [EC:1.7.2.2] | 0.054 | 0.207 | 0.794 | 0.999 |
| nrfB; cytochrome c-type protein NrfB | 0.167 | 0.346 | 0.629 | 0.999 |
| nrfC; protein NrfC | 0.024 | 0.314 | 0.939 | 0.999 |
| nrfD; protein NrfD | 0.095 | 0.326 | 0.772 | 0.999 |
| nrfE; cytochrome c-type biogenesis protein NrfE | 0.166 | 0.346 | 0.632 | 0.999 |
| nrfF; formate-dependent nitrite reductase complex subunit NrfF | -0.523 | 0.648 | 0.421 | 0.978 |
| nrfG; formate-dependent nitrite reductase complex subunit NrfG | -0.516 | 0.609 | 0.398 | 0.978 |
| nrfH; cytochrome c nitrite reductase small subunit | 0.005 | 0.268 | 0.984 | 0.999 |
| nrnA; bifunctional oligoribonuclease and PAP phosphatase NrnA [EC:3.1.3.7 3.1.13.3] | 0.061 | 0.085 | 0.472 | 0.979 |
| NRT, narK, nrtP, nasA; MFS transporter, NNP family, nitrate/nitrite transporter | -0.017 | 0.124 | 0.889 | 0.999 |
| nrtA, nasF, cynA; nitrate/nitrite transport system substrate-binding protein | -0.070 | 0.256 | 0.784 | 0.999 |
| nrtB, nasE, cynB; nitrate/nitrite transport system permease protein | -0.160 | 0.306 | 0.602 | 0.999 |
| nrtC, nasD; nitrate/nitrite transport system ATP-binding protein [EC:3.6.3.-] | -0.160 | 0.306 | 0.602 | 0.999 |
| NSF, SEC18; vesicle-fusing ATPase [EC:3.6.4.6] | -2.579 | 2.960 | 0.385 | 0.978 |
| nspC; carboxynorspermidine decarboxylase [EC:4.1.1.96] | -0.221 | 0.121 | 0.069 | 0.840 |
| nsrR; Rrf2 family transcriptional regulator, nitric oxide-sensitive transcriptional repressor | -0.346 | 0.197 | 0.081 | 0.864 |
| ntdA; 3-dehydro-glucose-6-phosphate—glutamate transaminase [EC:2.6.1.104] | -2.386 | 2.492 | 0.340 | 0.978 |
| ntdB; kanosamine-6-phosphate phosphatase [EC:3.1.3.92] | -2.386 | 2.492 | 0.340 | 0.978 |
| ntdC; glucose-6-phosphate 3-dehydrogenase [EC:1.1.1.361] | 18.239 | 2966.075 | 0.995 | 0.999 |
| NTH; endonuclease III [EC:4.2.99.18] | 0.024 | 0.038 | 0.529 | 0.989 |
| nthA; nitrile hydratase subunit alpha [EC:4.2.1.84] | -0.716 | 0.463 | 0.124 | 0.919 |
| ntnH; botulinum neurotoxin type non-toxic component | -16.780 | 1732.243 | 0.992 | 0.999 |
| NTPCR; nucleoside-triphosphatase [EC:3.6.1.15] | -0.896 | 1.253 | 0.476 | 0.979 |
| ntrX; two-component system, NtrC family, nitrogen regulation response regulator NtrX | -0.850 | 0.398 | 0.034 | 0.839 |
| ntrY; two-component system, NtrC family, nitrogen regulation sensor histidine kinase NtrY [EC:2.7.13.3] | -0.914 | 0.424 | 0.033 | 0.839 |
| nuc; micrococcal nuclease [EC:3.1.31.1] | -0.323 | 0.286 | 0.260 | 0.978 |
| nucS; endonuclease [EC:3.1.-.-] | -0.009 | 0.263 | 0.974 | 0.999 |
| nudB, ntpA; dihydroneopterin triphosphate diphosphatase [EC:3.6.1.67] | 0.021 | 0.157 | 0.895 | 0.999 |
| nudE; ADP-ribose diphosphatase [EC:3.6.1.-] | 0.186 | 0.193 | 0.336 | 0.978 |
| nudF; ADP-ribose pyrophosphatase [EC:3.6.1.13] | 0.132 | 0.051 | 0.010 | 0.839 |
| nudG; (d)CTP diphosphatase [EC:3.6.1.65] | 0.243 | 0.601 | 0.686 | 0.999 |
| nudH; putative (di)nucleoside polyphosphate hydrolase [EC:3.6.1.-] | -0.062 | 0.141 | 0.661 | 0.999 |
| nudI; nucleoside triphosphatase [EC:3.6.1.-] | 0.270 | 0.651 | 0.679 | 0.999 |
| nudJ; phosphatase NudJ [EC:3.6.1.-] | 0.119 | 0.601 | 0.843 | 0.999 |
| nudK; GDP-mannose pyrophosphatase NudK [EC:3.6.1.-] | 0.196 | 0.645 | 0.761 | 0.999 |
| NUDT14; UDP-sugar diphosphatase [EC:3.6.1.45] | -0.428 | 0.802 | 0.595 | 0.999 |
| NUDT2; bis(5’-nucleosidyl)-tetraphosphatase [EC:3.6.1.17] | 2.332 | 2.008 | 0.247 | 0.978 |
| nuoA; NADH-quinone oxidoreductase subunit A [EC:1.6.5.3] | -0.201 | 0.101 | 0.048 | 0.839 |
| nuoB; NADH-quinone oxidoreductase subunit B [EC:1.6.5.3] | -0.215 | 0.099 | 0.031 | 0.839 |
| nuoC; NADH-quinone oxidoreductase subunit C [EC:1.6.5.3] | -0.324 | 0.186 | 0.083 | 0.867 |
| nuoCD; NADH-quinone oxidoreductase subunit C/D [EC:1.6.5.3] | -0.119 | 0.152 | 0.435 | 0.978 |
| nuoD; NADH-quinone oxidoreductase subunit D [EC:1.6.5.3] | -0.353 | 0.183 | 0.055 | 0.839 |
| nuoE; NADH-quinone oxidoreductase subunit E [EC:1.6.5.3] | -0.126 | 0.118 | 0.287 | 0.978 |
| nuoF; NADH-quinone oxidoreductase subunit F [EC:1.6.5.3] | -0.097 | 0.129 | 0.453 | 0.978 |
| nuoG; NADH-quinone oxidoreductase subunit G [EC:1.6.5.3] | -0.008 | 0.099 | 0.937 | 0.999 |
| nuoH; NADH-quinone oxidoreductase subunit H [EC:1.6.5.3] | -0.213 | 0.100 | 0.036 | 0.839 |
| nuoI; NADH-quinone oxidoreductase subunit I [EC:1.6.5.3] | -0.191 | 0.099 | 0.056 | 0.839 |
| nuoJ; NADH-quinone oxidoreductase subunit J [EC:1.6.5.3] | -0.208 | 0.099 | 0.037 | 0.839 |
| nuoK; NADH-quinone oxidoreductase subunit K [EC:1.6.5.3] | -0.195 | 0.103 | 0.060 | 0.839 |
| nuoL; NADH-quinone oxidoreductase subunit L [EC:1.6.5.3] | -0.217 | 0.101 | 0.034 | 0.839 |
| nuoM; NADH-quinone oxidoreductase subunit M [EC:1.6.5.3] | -0.211 | 0.100 | 0.038 | 0.839 |
| nuoN; NADH-quinone oxidoreductase subunit N [EC:1.6.5.3] | -0.209 | 0.100 | 0.038 | 0.839 |
| nupC; nucleoside transport protein | -0.492 | 0.455 | 0.281 | 0.978 |
| nupG; MFS transporter, NHS family, nucleoside permease | -0.178 | 0.263 | 0.499 | 0.981 |
| nupX; nucleoside permease | -1.154 | 1.507 | 0.445 | 0.978 |
| nusA; N utilization substance protein A | 0.036 | 0.036 | 0.323 | 0.978 |
| nusB; N utilization substance protein B | 0.029 | 0.037 | 0.431 | 0.978 |
| nusG; transcriptional antiterminator NusG | 0.029 | 0.036 | 0.416 | 0.978 |
| nylA; 6-aminohexanoate-cyclic-dimer hydrolase [EC:3.5.2.12] | -1.596 | 1.095 | 0.147 | 0.954 |
| nylB; 6-aminohexanoate-oligomer exohydrolase [EC:3.5.1.46] | -0.966 | 0.454 | 0.035 | 0.839 |
| oadA; oxaloacetate decarboxylase, alpha subunit [EC:4.1.1.3] | 0.302 | 0.172 | 0.081 | 0.864 |
| oadB; oxaloacetate decarboxylase, beta subunit [EC:4.1.1.3] | 0.257 | 0.187 | 0.172 | 0.961 |
| oadG; oxaloacetate decarboxylase, gamma subunit [EC:4.1.1.3] | -0.286 | 0.437 | 0.513 | 0.982 |
| oapA; opacity associated protein | 0.205 | 0.358 | 0.567 | 0.995 |
| obgE, cgtA; GTPase [EC:3.6.5.-] | 0.035 | 0.033 | 0.290 | 0.978 |
| occM, nocM; octopine/nopaline transport system permease protein | -0.721 | 0.837 | 0.390 | 0.978 |
| occP, nocP; octopine/nopaline transport system ATP-binding protein [EC:3.6.3.-] | -0.711 | 0.818 | 0.386 | 0.978 |
| occQ, nocQ; octopine/nopaline transport system permease protein | -0.670 | 0.869 | 0.442 | 0.978 |
| occT, nocT; octopine/nopaline transport system substrate-binding protein | -1.924 | 1.625 | 0.238 | 0.978 |
| odh; opine dehydrogenase [EC:1.5.1.28] | -0.323 | 0.612 | 0.599 | 0.999 |
| ofaA, arfA; arthrofactin-type cyclic lipopeptide synthetase A | -0.274 | 0.663 | 0.680 | 0.999 |
| ofaB, arfB; arthrofactin-type cyclic lipopeptide synthetase B | -0.224 | 0.682 | 0.743 | 0.999 |
| ofaC, arfC; arthrofactin-type cyclic lipopeptide synthetase C | -0.202 | 0.692 | 0.771 | 0.999 |
| OGDH, sucA; 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2] | -0.096 | 0.112 | 0.396 | 0.978 |
| OGG1; N-glycosylase/DNA lyase [EC:3.2.2.- 4.2.99.18] | 0.327 | 0.342 | 0.340 | 0.978 |
| ogl; oligogalacturonide lyase [EC:4.2.2.6] | -0.978 | 0.757 | 0.198 | 0.970 |
| ogt, MGMT; methylated-DNA-[protein]-cysteine S-methyltransferase [EC:2.1.1.63] | 0.017 | 0.042 | 0.688 | 0.999 |
| ohyA, sph; oleate hydratase [EC:4.2.1.53] | 0.140 | 0.243 | 0.566 | 0.995 |
| oleC4; oleandomycin transport system ATP-binding protein | 0.062 | 0.971 | 0.949 | 0.999 |
| oleC5; oleandomycin transport system permease protein | 0.148 | 0.697 | 0.833 | 0.999 |
| omp31; outer membrane immunogenic protein | -0.987 | 0.416 | 0.019 | 0.839 |
| ompC; outer membrane pore protein C | 0.129 | 0.613 | 0.834 | 0.999 |
| ompF; outer membrane pore protein F | 0.129 | 0.597 | 0.829 | 0.999 |
| ompG; outer membrane protein G | -0.285 | 0.754 | 0.707 | 0.999 |
| ompN; outer membrane protein N | 0.108 | 0.641 | 0.866 | 0.999 |
| ompR; two-component system, OmpR family, phosphate regulon response regulator OmpR | -0.181 | 0.248 | 0.466 | 0.979 |
| ompT; omptin [EC:3.4.23.49] | -0.752 | 1.230 | 0.542 | 0.990 |
| ompU; outer membrane protein OmpU | 0.066 | 0.819 | 0.936 | 0.999 |
| ompW; outer membrane protein | -0.112 | 0.211 | 0.598 | 0.999 |
| ompX; outer membrane protein X | -0.141 | 0.276 | 0.610 | 0.999 |
| OPLAH, OXP1, oplAH; 5-oxoprolinase (ATP-hydrolysing) [EC:3.5.2.9] | -1.424 | 0.725 | 0.051 | 0.839 |
| opmD; outer membrane protein, multidrug efflux system | -17.335 | 3236.897 | 0.996 | 0.999 |
| opmE; outer membrane protein, multidrug efflux system | 1.117 | 1.359 | 0.412 | 0.978 |
| oppA, mppA; oligopeptide transport system substrate-binding protein | -0.071 | 0.130 | 0.584 | 0.999 |
| oppB; oligopeptide transport system permease protein | -0.022 | 0.101 | 0.827 | 0.999 |
| oppC; oligopeptide transport system permease protein | -0.027 | 0.101 | 0.787 | 0.999 |
| oppD; oligopeptide transport system ATP-binding protein | 0.009 | 0.098 | 0.926 | 0.999 |
| oppF; oligopeptide transport system ATP-binding protein | 0.001 | 0.129 | 0.994 | 0.999 |
| oprB; porin | -0.122 | 0.272 | 0.654 | 0.999 |
| oprC, opcM; outer membrane protein, multidrug efflux system | -0.479 | 0.499 | 0.339 | 0.978 |
| oprD; imipenem/basic amino acid-specific outer membrane pore [EC:3.4.21.-] | 0.462 | 0.330 | 0.163 | 0.957 |
| oprJ; outer membrane protein, multidrug efflux system | 1.082 | 1.460 | 0.460 | 0.979 |
| oprM, emhC, ttgC, cusC, adeK, smeF, mtrE, cmeC, gesC; outer membrane protein, multidrug efflux system | -0.092 | 0.208 | 0.659 | 0.999 |
| oprN; outer membrane protein, multidrug efflux system | 0.204 | 0.337 | 0.545 | 0.990 |
| oprO_P; phosphate-selective porin OprO and OprP | 0.001 | 0.254 | 0.996 | 0.999 |
| opsX; heptosyltransferase I [EC:2.4.-.-] | 0.125 | 0.268 | 0.643 | 0.999 |
| opuA; osmoprotectant transport system ATP-binding protein | 0.078 | 0.077 | 0.311 | 0.978 |
| opuBD; osmoprotectant transport system permease protein | 0.053 | 0.077 | 0.488 | 0.979 |
| opuC; osmoprotectant transport system substrate-binding protein | 0.055 | 0.171 | 0.747 | 0.999 |
| opuD, betL; glycine betaine transporter | -0.214 | 0.313 | 0.494 | 0.981 |
| oqxA; membrane fusion protein, multidrug efflux system | 0.048 | 0.426 | 0.910 | 0.999 |
| oqxB; multidrug efflux pump | 0.028 | 0.431 | 0.949 | 0.999 |
| oqxR; Rrf2 family transcriptional regulator, repressor of oqxAB | 0.594 | 0.664 | 0.372 | 0.978 |
| oraE; D-ornithine 4,5-aminomutase subunit beta [EC:5.4.3.5] | 1.010 | 0.506 | 0.048 | 0.839 |
| oraS; D-ornithine 4,5-aminomutase subunit alpha [EC:5.4.3.5] | 1.010 | 0.506 | 0.048 | 0.839 |
| orn, REX2, REXO2; oligoribonuclease [EC:3.1.-.-] | 0.058 | 0.120 | 0.631 | 0.999 |
| osmB; osmotically inducible lipoprotein OsmB | 0.195 | 0.269 | 0.471 | 0.979 |
| osmC; osmotically inducible protein OsmC | -0.099 | 0.219 | 0.653 | 0.999 |
| osmE; osmotically inducible lipoprotein OsmE | 0.434 | 0.310 | 0.164 | 0.957 |
| osmY; hyperosmotically inducible periplasmic protein | 0.091 | 0.228 | 0.689 | 0.999 |
| ospF, mkaD, spvC; phosphothreonine lyase [EC:4.2.3.-] | 18.932 | 3654.080 | 0.996 | 0.999 |
| OTC, argF, argI; ornithine carbamoyltransferase [EC:2.1.3.3] | 0.085 | 0.061 | 0.167 | 0.961 |
| otsA; trehalose 6-phosphate synthase [EC:2.4.1.15 2.4.1.347] | -0.374 | 0.254 | 0.144 | 0.951 |
| otsB; trehalose 6-phosphate phosphatase [EC:3.1.3.12] | -0.382 | 0.253 | 0.134 | 0.942 |
| oxa; beta-lactamase class D [EC:3.5.2.6] | -1.951 | 0.853 | 0.024 | 0.839 |
| oxc; oxalyl-CoA decarboxylase [EC:4.1.1.8] | 0.469 | 0.679 | 0.491 | 0.980 |
| OXCT; 3-oxoacid CoA-transferase [EC:2.8.3.5] | -0.584 | 0.736 | 0.428 | 0.978 |
| oxdA; aldoxime dehydratase [EC:4.99.1.5] | -1.122 | 0.876 | 0.202 | 0.972 |
| oxdD; oxalate decarboxylase [EC:4.1.1.2] | -0.761 | 0.844 | 0.368 | 0.978 |
| oxlT; MFS transporter, OFA family, oxalate/formate antiporter | -0.055 | 0.188 | 0.770 | 0.999 |
| oxyR; LysR family transcriptional regulator, hydrogen peroxide-inducible genes activator | -0.125 | 0.099 | 0.208 | 0.977 |
| p19, ftrA; periplasmic iron binding protein | 0.718 | 0.216 | 0.001 | 0.839 |
| P4HA; prolyl 4-hydroxylase [EC:1.14.11.2] | -0.815 | 0.437 | 0.064 | 0.839 |
| paaA; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaA [EC:1.14.13.149] | -0.284 | 0.280 | 0.312 | 0.978 |
| paaB; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaB | -0.256 | 0.283 | 0.366 | 0.978 |
| paaC; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaC [EC:1.14.13.149] | -0.238 | 0.281 | 0.398 | 0.978 |
| paaD; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaD | -0.256 | 0.283 | 0.366 | 0.978 |
| paaE; ring-1,2-phenylacetyl-CoA epoxidase subunit PaaE | -0.062 | 0.251 | 0.806 | 0.999 |
| paaF, echA; enoyl-CoA hydratase [EC:4.2.1.17] | -0.384 | 0.243 | 0.116 | 0.917 |
| paaG; 2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA isomerase [EC:5.3.3.18] | -0.588 | 0.253 | 0.021 | 0.839 |
| paaH, hbd, fadB, mmgB; 3-hydroxybutyryl-CoA dehydrogenase [EC:1.1.1.157] | -0.061 | 0.161 | 0.705 | 0.999 |
| paaI; acyl-CoA thioesterase [EC:3.1.2.-] | 0.019 | 0.097 | 0.849 | 0.999 |
| paaJ; 3-oxo-5,6-didehydrosuberyl-CoA/3-oxoadipyl-CoA thiolase [EC:2.3.1.223 2.3.1.174] | -0.063 | 0.582 | 0.915 | 0.999 |
| paaK; phenylacetate-CoA ligase [EC:6.2.1.30] | -0.381 | 0.216 | 0.079 | 0.859 |
| paaX; phenylacetic acid degradation operon negative regulatory protein | -0.387 | 0.307 | 0.208 | 0.977 |
| paaY; phenylacetic acid degradation protein | 0.118 | 0.363 | 0.746 | 0.999 |
| paaZ; oxepin-CoA hydrolase / 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase [EC:3.3.2.12 1.2.1.91] | -0.108 | 0.321 | 0.737 | 0.999 |
| pabA; para-aminobenzoate synthetase component II [EC:2.6.1.85] | 0.181 | 0.159 | 0.255 | 0.978 |
| pabAB; para-aminobenzoate synthetase [EC:2.6.1.85] | -0.132 | 0.309 | 0.670 | 0.999 |
| pabB; para-aminobenzoate synthetase component I [EC:2.6.1.85] | 0.023 | 0.118 | 0.843 | 0.999 |
| pabBC; para-aminobenzoate synthetase / 4-amino-4-deoxychorismate lyase [EC:2.6.1.85 4.1.3.38] | -0.174 | 0.168 | 0.302 | 0.978 |
| pabC; 4-amino-4-deoxychorismate lyase [EC:4.1.3.38] | 0.062 | 0.104 | 0.553 | 0.992 |
| padR; PadR family transcriptional regulator, regulatory protein PadR | -0.091 | 0.118 | 0.442 | 0.978 |
| pafA; proteasome accessory factor A [EC:6.3.1.19] | -0.002 | 0.259 | 0.994 | 0.999 |
| pafB; proteasome accessory factor B | 0.078 | 0.292 | 0.790 | 0.999 |
| pafC; proteasome accessory factor C | -0.097 | 0.277 | 0.725 | 0.999 |
| pagC; putatice virulence related protein PagC | -0.534 | 1.027 | 0.604 | 0.999 |
| pagL; lipid A 3-O-deacylase [EC:3.1.-.-] | 0.288 | 0.324 | 0.374 | 0.978 |
| pagO; putative membrane protein PagO | 0.055 | 1.009 | 0.957 | 0.999 |
| pagP, crcA; lipid IVA palmitoyltransferase [EC:2.3.1.251] | 0.134 | 0.529 | 0.800 | 0.999 |
| paiB; transcriptional regulator | -0.230 | 0.207 | 0.268 | 0.978 |
| pal; peptidoglycan-associated lipoprotein | -0.089 | 0.146 | 0.540 | 0.990 |
| PAL; phenylalanine ammonia-lyase [EC:4.3.1.24] | 1.419 | 0.994 | 0.155 | 0.954 |
| PAM, PHM; peptidylglycine monooxygenase [EC:1.14.17.3] | 1.559 | 1.305 | 0.234 | 0.978 |
| panB; 3-methyl-2-oxobutanoate hydroxymethyltransferase [EC:2.1.2.11] | -0.199 | 0.083 | 0.017 | 0.839 |
| panC; pantoate–beta-alanine ligase [EC:6.3.2.1] | -0.121 | 0.072 | 0.094 | 0.894 |
| panD; aspartate 1-decarboxylase [EC:4.1.1.11] | -0.237 | 0.111 | 0.035 | 0.839 |
| panE, apbA; 2-dehydropantoate 2-reductase [EC:1.1.1.169] | 0.044 | 0.090 | 0.621 | 0.999 |
| panF; sodium/pantothenate symporter | 0.326 | 0.215 | 0.131 | 0.941 |
| parA, soj; chromosome partitioning protein | -0.071 | 0.070 | 0.313 | 0.978 |
| parB, spo0J; chromosome partitioning protein, ParB family | -0.080 | 0.064 | 0.213 | 0.978 |
| parC; topoisomerase IV subunit A [EC:5.99.1.-] | -0.046 | 0.054 | 0.396 | 0.978 |
| parD1_3_4; antitoxin ParD1/3/4 | -0.585 | 0.399 | 0.145 | 0.951 |
| parE; topoisomerase IV subunit B [EC:5.99.1.-] | -0.043 | 0.054 | 0.423 | 0.978 |
| parE1_3_4; toxin ParE1/3/4 | -0.613 | 0.365 | 0.095 | 0.896 |
| parM; plasmid segregation protein ParM | -0.991 | 0.496 | 0.047 | 0.839 |
| PARP; poly [ADP-ribose] polymerase [EC:2.4.2.30] | -0.259 | 0.965 | 0.788 | 0.999 |
| parR; two-component system, OmpR family, response regulator ParR | -0.229 | 0.402 | 0.570 | 0.995 |
| PARS, proS; prolyl-tRNA synthetase [EC:6.1.1.15] | 0.027 | 0.032 | 0.406 | 0.978 |
| parS; two-component system, OmpR family, sensor kinase ParS [EC:2.7.13.3] | -0.229 | 0.402 | 0.570 | 0.995 |
| PAT, AAT; bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase [EC:2.6.1.1 2.6.1.78 2.6.1.79] | -0.816 | 1.161 | 0.483 | 0.979 |
| pat; phosphinothricin acetyltransferase [EC:2.3.1.183] | -0.036 | 0.053 | 0.496 | 0.981 |
| patA, rscA, lmrC, satA; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.037 | 0.161 | 0.817 | 0.999 |
| patA; aminotransferase [EC:2.6.1.-] | 0.128 | 0.165 | 0.440 | 0.978 |
| patA; putrescine aminotransferase [EC:2.6.1.82] | -0.197 | 0.592 | 0.740 | 0.999 |
| patB, malY; cystathione beta-lyase [EC:4.4.1.8] | 0.097 | 0.094 | 0.304 | 0.978 |
| patB, rscB, lmrC, satB; ATP-binding cassette, subfamily B, multidrug efflux pump | 0.037 | 0.161 | 0.816 | 0.999 |
| pbp1b; penicillin-binding protein 1B | -0.007 | 0.136 | 0.961 | 0.999 |
| pbp2A; penicillin-binding protein 2A [EC:2.4.1.129 3.4.16.4] | 0.088 | 0.144 | 0.542 | 0.990 |
| pbp2B, penA; penicillin-binding protein 2B | -0.041 | 0.170 | 0.811 | 0.999 |
| pbp2X; penicillin-binding protein 2X | -0.019 | 0.173 | 0.915 | 0.999 |
| pbp3; penicillin-binding protein 3 [EC:3.4.-.-] | -0.234 | 0.817 | 0.775 | 0.999 |
| pbp5, pbp4, pbp3; penicillin-binding protein | -0.018 | 0.440 | 0.967 | 0.999 |
| pbpA; penicillin-binding protein 1 [EC:3.4.-.-] | -0.237 | 0.786 | 0.763 | 0.999 |
| pbpA; penicillin-binding protein A | -0.062 | 0.238 | 0.794 | 0.999 |
| pbpB; penicillin-binding protein 2B | 0.056 | 0.248 | 0.823 | 0.999 |
| pbpC; penicillin-binding protein 1C [EC:2.4.1.129] | 0.128 | 0.177 | 0.470 | 0.979 |
| pbpD; penicillin-binding protein 4 [EC:2.4.1.129 3.4.16.4] | -0.707 | 1.201 | 0.557 | 0.992 |
| pbpG; serine-type D-Ala-D-Ala endopeptidase (penicillin-binding protein 7) [EC:3.4.21.-] | 0.066 | 0.184 | 0.721 | 0.999 |
| pbuE; MFS transporter, DHA1 family, purine base/nucleoside efflux pump | -0.859 | 1.033 | 0.407 | 0.978 |
| pbuG; putative MFS transporter, AGZA family, xanthine/uracil permease | 0.162 | 0.058 | 0.006 | 0.839 |
| pbuX; xanthine permease | 0.047 | 0.129 | 0.716 | 0.999 |
| PC, pyc; pyruvate carboxylase [EC:6.4.1.1] | -0.061 | 0.174 | 0.727 | 0.999 |
| pcaB; 3-carboxy-cis,cis-muconate cycloisomerase [EC:5.5.1.2] | -0.187 | 0.347 | 0.592 | 0.999 |
| pcaC; 4-carboxymuconolactone decarboxylase [EC:4.1.1.44] | -0.229 | 0.163 | 0.162 | 0.957 |
| pcaD; 3-oxoadipate enol-lactonase [EC:3.1.1.24] | -0.164 | 0.234 | 0.485 | 0.979 |
| pcaF; 3-oxoadipyl-CoA thiolase [EC:2.3.1.174] | 0.057 | 0.342 | 0.867 | 0.999 |
| pcaG; protocatechuate 3,4-dioxygenase, alpha subunit [EC:1.13.11.3] | -0.100 | 0.253 | 0.693 | 0.999 |
| pcaH; protocatechuate 3,4-dioxygenase, beta subunit [EC:1.13.11.3] | -0.105 | 0.252 | 0.677 | 0.999 |
| pcaI; 3-oxoadipate CoA-transferase, alpha subunit [EC:2.8.3.6] | -0.301 | 0.356 | 0.399 | 0.978 |
| pcaJ; 3-oxoadipate CoA-transferase, beta subunit [EC:2.8.3.6] | -0.308 | 0.355 | 0.386 | 0.978 |
| pcaK; MFS transporter, AAHS family, 4-hydroxybenzoate transporter | -0.038 | 0.292 | 0.897 | 0.999 |
| pcaL; 3-oxoadipate enol-lactonase / 4-carboxymuconolactone decarboxylase [EC:3.1.1.24 4.1.1.44] | -1.117 | 0.625 | 0.076 | 0.855 |
| pcaQ; LysR family transcriptional regulator, pca operon transcriptional activator | 0.248 | 0.325 | 0.447 | 0.978 |
| pcaR; IclR family transcriptional regulator, pca regulon regulatory protein | -0.202 | 0.259 | 0.438 | 0.978 |
| pcaT; MFS transporter, MHS family, dicarboxylic acid transporter PcaT | -0.018 | 0.452 | 0.969 | 0.999 |
| PCBD, phhB; 4a-hydroxytetrahydrobiopterin dehydratase [EC:4.2.1.96] | -0.261 | 0.196 | 0.186 | 0.966 |
| PCCA, pccA; propionyl-CoA carboxylase alpha chain [EC:6.4.1.3] | -1.083 | 0.381 | 0.005 | 0.839 |
| pccA; periplasmic copper chaperone A | -0.129 | 0.196 | 0.513 | 0.982 |
| PCCB, pccB; propionyl-CoA carboxylase beta chain [EC:6.4.1.3 2.1.3.15] | -0.447 | 0.211 | 0.036 | 0.839 |
| pchA; salicylate biosynthesis isochorismate synthase [EC:5.4.4.2] | -1.047 | 1.651 | 0.527 | 0.989 |
| pchB; isochorismate pyruvate lyase [EC:4.2.99.21] | -0.183 | 0.290 | 0.529 | 0.989 |
| pchC; pyochelin biosynthetic protein PchC | -17.794 | 4072.025 | 0.997 | 0.999 |
| pchF; 4-cresol dehydrogenase (hydroxylating) flavoprotein subunit [EC:1.17.99.1] | 3.658 | 2.527 | 0.150 | 0.954 |
| pchF; pyochelin synthetase | -17.750 | 3249.062 | 0.996 | 0.999 |
| pchR; AraC family transcriptional regulator, transcriptional activator of the genes for pyochelin and ferripyochelin receptors | -0.290 | 0.988 | 0.770 | 0.999 |
| PCNA; proliferating cell nuclear antigen | -17.234 | 3077.514 | 0.996 | 0.999 |
| pcnB; poly(A) polymerase [EC:2.7.7.19] | -0.058 | 0.108 | 0.592 | 0.999 |
| pcoB, copB; copper resistance protein B | 0.015 | 0.247 | 0.950 | 0.999 |
| pcoD; copper resistance protein D | -0.326 | 0.254 | 0.200 | 0.971 |
| pcp; pyroglutamyl-peptidase [EC:3.4.19.3] | 0.064 | 0.105 | 0.542 | 0.990 |
| pcpB; pentachlorophenol monooxygenase [EC:1.14.13.50] | 0.704 | 0.921 | 0.445 | 0.978 |
| pcrB; putative glycerol-1-phosphate prenyltransferase [EC:2.5.1.-] | -0.852 | 0.453 | 0.062 | 0.839 |
| pcs; phosphatidylcholine synthase [EC:2.7.8.24] | -0.063 | 0.290 | 0.829 | 0.999 |
| pct; propionate CoA-transferase [EC:2.8.3.1] | -0.272 | 0.331 | 0.412 | 0.978 |
| PCYT1; choline-phosphate cytidylyltransferase [EC:2.7.7.15] | -0.283 | 0.428 | 0.509 | 0.981 |
| pdaA; peptidoglycan-N-acetylmuramic acid deacetylase [EC:3.5.1.-] | -0.996 | 0.535 | 0.065 | 0.839 |
| pdaD; arginine decarboxylase [EC:4.1.1.19] | -0.267 | 0.597 | 0.655 | 0.999 |
| PDC, pdc; pyruvate decarboxylase [EC:4.1.1.1] | -17.148 | 2948.067 | 0.995 | 0.999 |
| pdc; phenolic acid decarboxylase [EC:4.1.1.-] | 0.163 | 0.700 | 0.817 | 0.999 |
| PDCD5, TFAR19; programmed cell death protein 5 | -17.234 | 3077.514 | 0.996 | 0.999 |
| pdeA; c-di-GMP-specific phosphodiesterase [EC:3.1.4.52] | -0.499 | 0.897 | 0.579 | 0.997 |
| PDF, def; peptide deformylase [EC:3.5.1.88] | 0.010 | 0.047 | 0.830 | 0.999 |
| pdh; phenylalanine dehydrogenase [EC:1.4.1.20] | -2.184 | 1.411 | 0.124 | 0.919 |
| PDHA, pdhA; pyruvate dehydrogenase E1 component alpha subunit [EC:1.2.4.1] | -0.075 | 0.092 | 0.415 | 0.978 |
| PDHB, pdhB; pyruvate dehydrogenase E1 component beta subunit [EC:1.2.4.1] | -0.071 | 0.092 | 0.438 | 0.978 |
| pdhR; GntR family transcriptional regulator, transcriptional repressor for pyruvate dehydrogenase complex | -0.083 | 0.203 | 0.683 | 0.999 |
| pdla; 4-pyridoxolactonase [EC:3.1.1.27] | 18.076 | 2733.666 | 0.995 | 0.999 |
| pdp; pyrimidine-nucleoside phosphorylase [EC:2.4.2.2] | 0.034 | 0.109 | 0.758 | 0.999 |
| pdtaS; two-component system, sensor histidine kinase PdtaS [EC:2.7.13.3] | 0.184 | 0.130 | 0.159 | 0.954 |
| pduC; propanediol dehydratase large subunit [EC:4.2.1.28] | 0.268 | 0.250 | 0.286 | 0.978 |
| pduD; propanediol dehydratase medium subunit [EC:4.2.1.28] | 0.295 | 0.251 | 0.242 | 0.978 |
| pduE; propanediol dehydratase small subunit [EC:4.2.1.28] | 0.268 | 0.250 | 0.286 | 0.978 |
| pduL; phosphate propanoyltransferase [EC:2.3.1.222] | 0.264 | 0.264 | 0.319 | 0.978 |
| pduP; propionaldehyde dehydrogenase [EC:1.2.1.87] | 0.263 | 0.244 | 0.282 | 0.978 |
| pduQ; 1-propanol dehydrogenase | 0.265 | 0.261 | 0.310 | 0.978 |
| pduW; propionate kinase [EC:2.7.2.15] | -0.251 | 1.170 | 0.830 | 0.999 |
| pduX; L-threonine kinase [EC:2.7.1.177] | 0.204 | 0.242 | 0.402 | 0.978 |
| pdxA; 4-hydroxythreonine-4-phosphate dehydrogenase [EC:1.1.1.262] | -0.116 | 0.099 | 0.243 | 0.978 |
| pdxB; erythronate-4-phosphate dehydrogenase [EC:1.1.1.290] | -0.002 | 0.191 | 0.993 | 0.999 |
| pdxH, PNPO; pyridoxamine 5’-phosphate oxidase [EC:1.4.3.5] | -0.074 | 0.137 | 0.590 | 0.999 |
| pdxJ; pyridoxine 5-phosphate synthase [EC:2.6.99.2] | -0.170 | 0.139 | 0.222 | 0.978 |
| pdxK, pdxY; pyridoxine kinase [EC:2.7.1.35] | -0.009 | 0.060 | 0.876 | 0.999 |
| pdxS, pdx1; pyridoxal 5’-phosphate synthase pdxS subunit [EC:4.3.3.6] | -0.077 | 0.113 | 0.498 | 0.981 |
| pdxT, pdx2; 5’-phosphate synthase pdxT subunit [EC:4.3.3.6] | -0.233 | 0.117 | 0.049 | 0.839 |
| pecM; probable blue pigment (indigoidine) exporter | -0.785 | 0.468 | 0.096 | 0.896 |
| pehX; exo-poly-alpha-galacturonosidase [EC:3.2.1.82] | -0.976 | 0.998 | 0.329 | 0.978 |
| pel; pectate lyase [EC:4.2.2.2] | -0.783 | 0.643 | 0.225 | 0.978 |
| pelC; pectate lyase C [EC:4.2.2.2 4.2.2.10] | 18.045 | 1898.825 | 0.992 | 0.999 |
| PELO, DOM34, pelA; protein pelota | -17.234 | 3077.514 | 0.996 | 0.999 |
| penM, pntM, CYP161C; pentalenolactone synthase [EC:1.14.19.8] | 17.848 | 2439.579 | 0.994 | 0.999 |
| penP; beta-lactamase class A [EC:3.5.2.6] | 0.020 | 0.148 | 0.894 | 0.999 |
| PEO1; twinkle protein [EC:3.6.4.12] | -1.754 | 1.128 | 0.122 | 0.919 |
| pep2; maltokinase [EC:2.7.1.175] | -0.231 | 0.393 | 0.558 | 0.992 |
| pepA; glutamyl aminopeptidase [EC:3.4.11.7] | 0.011 | 0.153 | 0.942 | 0.999 |
| pepB; PepB aminopeptidase [EC:3.4.11.23] | 0.241 | 0.306 | 0.431 | 0.978 |
| pepD; dipeptidase D [EC:3.4.13.-] | 0.239 | 0.165 | 0.150 | 0.954 |
| pepD; putative serine protease PepD [EC:3.4.21.-] | 0.064 | 0.241 | 0.789 | 0.999 |
| pepDA, pepDB; dipeptidase [EC:3.4.-.-] | 0.255 | 0.304 | 0.402 | 0.978 |
| pepE; dipeptidase E [EC:3.4.13.21] | 0.197 | 0.155 | 0.205 | 0.972 |
| pepF, pepB; oligoendopeptidase F [EC:3.4.24.-] | -0.013 | 0.092 | 0.889 | 0.999 |
| pepM; phosphoenolpyruvate phosphomutase [EC:5.4.2.9] | -0.144 | 0.499 | 0.773 | 0.999 |
| pepN; aminopeptidase N [EC:3.4.11.2] | -0.004 | 0.083 | 0.965 | 0.999 |
| pepO; putative endopeptidase [EC:3.4.24.-] | -0.032 | 0.103 | 0.753 | 0.999 |
| pepP; Xaa-Pro aminopeptidase [EC:3.4.11.9] | 0.041 | 0.043 | 0.347 | 0.978 |
| pepQ; Xaa-Pro dipeptidase [EC:3.4.13.9] | 0.001 | 0.088 | 0.995 | 0.999 |
| pepT; tripeptide aminopeptidase [EC:3.4.11.4] | 0.052 | 0.089 | 0.560 | 0.992 |
| pepX; X-Pro dipeptidyl-peptidase [EC:3.4.14.11] | -0.007 | 0.158 | 0.965 | 0.999 |
| per, rfbE; perosamine synthetase [EC:2.6.1.102] | -0.945 | 0.904 | 0.298 | 0.978 |
| perM; putative permease | 0.212 | 0.212 | 0.320 | 0.978 |
| perR; Fur family transcriptional regulator, peroxide stress response regulator | 0.151 | 0.098 | 0.125 | 0.923 |
| petC; cytochrome b6-f complex iron-sulfur subunit [EC:1.10.9.1] | -1.715 | 1.535 | 0.265 | 0.978 |
| petF; ferredoxin | -0.445 | 1.371 | 0.746 | 0.999 |
| pezA; HTH-type transcriptional regulator / antitoxin PezA | -0.188 | 0.233 | 0.421 | 0.978 |
| pezT; UDP-N-acetylglucosamine kinase [EC:2.7.1.176] | -0.119 | 0.219 | 0.587 | 0.999 |
| pfbA; plasmin and fibronectin-binding protein A | -0.032 | 0.687 | 0.963 | 0.999 |
| pfdA, PFDN5; prefoldin alpha subunit | -17.234 | 3077.514 | 0.996 | 0.999 |
| pfdB, PFDN6; prefoldin beta subunit | -17.234 | 3077.514 | 0.996 | 0.999 |
| pfeR, pirR; two-component system, OmpR family, response regulator PfeR | 0.601 | 0.361 | 0.098 | 0.896 |
| pfeS, pirS; two-component system, OmpR family, sensor histidine kinase PfeS [EC:2.7.13.3] | 0.601 | 0.361 | 0.098 | 0.896 |
| pfkA, PFK; 6-phosphofructokinase 1 [EC:2.7.1.11] | 0.066 | 0.082 | 0.420 | 0.978 |
| pfkB; 6-phosphofructokinase 2 [EC:2.7.1.11] | -1.273 | 0.551 | 0.022 | 0.839 |
| pfkC; ADP-dependent phosphofructokinase/glucokinase [EC:2.7.1.146 2.7.1.147] | 0.426 | 0.829 | 0.609 | 0.999 |
| pflA, pflC, pflE; pyruvate formate lyase activating enzyme [EC:1.97.1.4] | 0.083 | 0.096 | 0.389 | 0.978 |
| pflX; putative pyruvate formate lyase activating enzyme [EC:1.97.1.4] | 0.095 | 0.348 | 0.786 | 0.999 |
| pfp, PFP; diphosphate-dependent phosphofructokinase [EC:2.7.1.90] | -0.222 | 0.215 | 0.303 | 0.978 |
| pfpI; protease I [EC:3.5.1.124] | -0.184 | 0.181 | 0.311 | 0.978 |
| pgaA; biofilm PGA synthesis protein PgaA | -0.212 | 0.355 | 0.552 | 0.992 |
| pgaB; poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase [EC:3.5.1.-] | -0.210 | 0.290 | 0.470 | 0.979 |
| pgaC, icaA; poly-beta-1,6-N-acetyl-D-glucosamine synthase [EC:2.4.1.-] | -0.355 | 0.287 | 0.219 | 0.978 |
| pgaD; biofilm PGA synthesis protein PgaD | -0.212 | 0.384 | 0.582 | 0.999 |
| PGAM, gpmA; 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase [EC:5.4.2.11] | 0.112 | 0.087 | 0.204 | 0.972 |
| PGD, gnd, gntZ; 6-phosphogluconate dehydrogenase [EC:1.1.1.44 1.1.1.343] | -0.017 | 0.061 | 0.782 | 0.999 |
| pgi-pmi; glucose/mannose-6-phosphate isomerase [EC:5.3.1.9 5.3.1.8] | -1.130 | 1.377 | 0.413 | 0.978 |
| pgi1; glucose-6-phosphate isomerase, archaeal [EC:5.3.1.9] | -1.636 | 0.858 | 0.058 | 0.839 |
| PGK, pgk; phosphoglycerate kinase [EC:2.7.2.3] | 0.033 | 0.037 | 0.364 | 0.978 |
| pgl; 6-phosphogluconolactonase [EC:3.1.1.31] | 0.083 | 0.132 | 0.531 | 0.990 |
| pglA; N,N’-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase [EC:2.4.1.290] | 0.480 | 0.618 | 0.438 | 0.978 |
| pglB; undecaprenyl-diphosphooligosaccharide—protein glycotransferase [EC:2.4.99.19] | 0.483 | 0.525 | 0.360 | 0.978 |
| pglC; undecaprenyl phosphate N,N’-diacetylbacillosamine 1-phosphate transferase [EC:2.7.8.36] | 0.475 | 0.616 | 0.442 | 0.978 |
| pglD; UDP-N-acetylbacillosamine N-acetyltransferase [EC:2.3.1.203] | 0.480 | 0.618 | 0.438 | 0.978 |
| pglE; UDP-N-acetylbacillosamine transaminase [EC:2.6.1.34] | 0.480 | 0.618 | 0.438 | 0.978 |
| pglF; UDP-N-acetyl-D-glucosamine 4,6-dehydratase [EC:4.2.1.135] | 0.480 | 0.618 | 0.438 | 0.978 |
| pglH; GalNAc-alpha-(1->4)-GalNAc-alpha-(1->3)-diNAcBac-PP-undecaprenol alpha-1,4-N-acetyl-D-galactosaminyltransferase [EC:2.4.1.292] | 0.439 | 0.516 | 0.397 | 0.978 |
| pglI; GalNAc5-diNAcBac-PP-undecaprenol beta-1,3-glucosyltransferase [EC:2.4.1.293] | 0.051 | 0.670 | 0.939 | 0.999 |
| pglJ; N-acetylgalactosamine-N,N’-diacetylbacillosaminyl-diphospho-undecaprenol 4-alpha-N-acetylgalactosaminyltransferase [EC:2.4.1.291] | 0.498 | 0.586 | 0.396 | 0.978 |
| PGLS, pgl, devB; 6-phosphogluconolactonase [EC:3.1.1.31] | -0.043 | 0.106 | 0.682 | 0.999 |
| pgm; phosphoglucomutase [EC:5.4.2.2] | 0.035 | 0.059 | 0.551 | 0.992 |
| pgmB; beta-phosphoglucomutase [EC:5.4.2.6] | 0.447 | 0.291 | 0.126 | 0.927 |
| pgpA; phosphatidylglycerophosphatase A [EC:3.1.3.27] | 0.245 | 0.147 | 0.098 | 0.896 |
| pgpB; phosphatidylglycerophosphatase B [EC:3.1.3.27 3.1.3.81 3.1.3.4 3.6.1.27] | 0.053 | 0.256 | 0.836 | 0.999 |
| pgpC; phosphatidylglycerophosphatase C [EC:3.1.3.27] | -0.060 | 0.383 | 0.876 | 0.999 |
| PGRP; peptidoglycan recognition protein | -0.766 | 0.729 | 0.295 | 0.978 |
| pgsA, PGS1; CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase [EC:2.7.8.5] | 0.092 | 0.043 | 0.035 | 0.839 |
| pgtA; two-component system, NtrC family, phosphoglycerate transport system response regulator PgtA | 0.657 | 0.981 | 0.504 | 0.981 |
| pgtB; two-component system, NtrC family, phosphoglycerate transport system sensor histidine kinase PgtB [EC:2.7.13.3] | 0.657 | 0.981 | 0.504 | 0.981 |
| pgtC; phosphoglycerate transport regulatory protein PgtC | -0.251 | 1.170 | 0.830 | 0.999 |
| pgtP; MFS transporter, OPA family, phosphoglycerate transporter protein | -1.641 | 1.456 | 0.261 | 0.978 |
| phaA; multicomponent K+:H+ antiporter subunit A | -0.135 | 0.216 | 0.531 | 0.990 |
| phaC; multicomponent K+:H+ antiporter subunit C | -0.115 | 0.217 | 0.596 | 0.999 |
| phaD; multicomponent K+:H+ antiporter subunit D | -0.115 | 0.217 | 0.596 | 0.999 |
| phaE; multicomponent K+:H+ antiporter subunit E | -0.118 | 0.217 | 0.589 | 0.999 |
| phaF; multicomponent K+:H+ antiporter subunit F | -0.176 | 0.213 | 0.411 | 0.978 |
| phaG; multicomponent K+:H+ antiporter subunit G | -0.120 | 0.216 | 0.580 | 0.997 |
| phaJ; enoyl-CoA hydratase [EC:4.2.1.119] | 17.680 | 2242.142 | 0.994 | 0.999 |
| phaZ; poly(3-hydroxybutyrate) depolymerase [EC:3.1.1.75] | -0.332 | 0.301 | 0.272 | 0.978 |
| phbB; acetoacetyl-CoA reductase [EC:1.1.1.36] | -0.234 | 0.326 | 0.474 | 0.979 |
| phbC, phaC; polyhydroxyalkanoate synthase [EC:2.3.1.-] | -0.124 | 0.207 | 0.551 | 0.992 |
| phcA; LysR family transcriptional regulator, virulence genes transcriptional regulator | -17.504 | 2478.325 | 0.994 | 0.999 |
| phcB; extracellular factor (EF) 3-hydroxypalmitic acid methyl ester biosynthesis protein | -3.034 | 3.672 | 0.410 | 0.978 |
| phcQ; two-component system, probable response regulator PhcQ | -3.034 | 3.672 | 0.410 | 0.978 |
| phcR; two-component system, response regulator PhcR | -17.405 | 2868.530 | 0.995 | 0.999 |
| phcS; two-component system, sensor histidine kinase PhcS [EC:2.7.13.3] | -3.034 | 3.672 | 0.410 | 0.978 |
| phd; antitoxin Phd | 0.652 | 0.577 | 0.260 | 0.978 |
| phdE; cis-3,4-dihydrophenanthrene-3,4-diol dehydrogenase [EC:1.3.1.49] | 0.884 | 1.140 | 0.439 | 0.978 |
| phdF; extradiol dioxygenase [EC:1.13.11.-] | -17.763 | 3362.856 | 0.996 | 0.999 |
| pheA; chorismate mutase / prephenate dehydratase [EC:5.4.99.5 4.2.1.51] | 0.150 | 0.111 | 0.179 | 0.961 |
| pheA1; chorismate mutase [EC:5.4.99.5] | -0.094 | 0.301 | 0.756 | 0.999 |
| pheA2; prephenate dehydratase [EC:4.2.1.51] | -0.070 | 0.101 | 0.488 | 0.979 |
| pheB; chorismate mutase [EC:5.4.99.5] | 0.144 | 0.152 | 0.345 | 0.978 |
| pheC; cyclohexadienyl dehydratase [EC:4.2.1.51 4.2.1.91] | -0.899 | 0.479 | 0.062 | 0.839 |
| pheP; phenylalanine-specific permease | -0.241 | 0.701 | 0.732 | 0.999 |
| phhA, PAH; phenylalanine-4-hydroxylase [EC:1.14.16.1] | -0.087 | 0.234 | 0.712 | 0.999 |
| PHKA_B; phosphorylase kinase alpha/beta subunit | -2.599 | 2.089 | 0.215 | 0.978 |
| phlD; phloroglucinol synthase [EC:2.3.1.253] | -0.173 | 0.963 | 0.858 | 0.999 |
| phnA; phosphonoacetate hydrolase [EC:3.11.1.2] | -0.535 | 0.942 | 0.571 | 0.995 |
| phnA; protein PhnA | -0.001 | 0.082 | 0.989 | 0.999 |
| phnB; PhnB protein | -0.070 | 0.127 | 0.583 | 0.999 |
| phnC; phosphonate transport system ATP-binding protein [EC:3.6.3.28] | 0.158 | 0.164 | 0.339 | 0.978 |
| phnD; phosphonate transport system substrate-binding protein | 0.094 | 0.156 | 0.547 | 0.991 |
| phnE; phosphonate transport system permease protein | 0.112 | 0.162 | 0.490 | 0.979 |
| phnF; GntR family transcriptional regulator, phosphonate transport system regulatory protein | -1.056 | 0.445 | 0.019 | 0.839 |
| phnG; alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG [EC:2.7.8.37] | -0.970 | 0.443 | 0.030 | 0.839 |
| phnH; alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH [EC:2.7.8.37] | -0.966 | 0.444 | 0.031 | 0.839 |
| phnI; alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI [EC:2.7.8.37] | -0.970 | 0.443 | 0.030 | 0.839 |
| phnJ; alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase [EC:4.7.1.1] | -0.970 | 0.443 | 0.030 | 0.839 |
| phnK; putative phosphonate transport system ATP-binding protein | -0.970 | 0.443 | 0.030 | 0.839 |
| phnL; alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnL [EC:2.7.8.37] | -0.964 | 0.445 | 0.032 | 0.839 |
| phnM; alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase [EC:3.6.1.63] | -1.074 | 0.403 | 0.009 | 0.839 |
| phnN; ribose 1,5-bisphosphokinase [EC:2.7.4.23] | -0.143 | 0.275 | 0.605 | 0.999 |
| phnO; aminoalkylphosphonate N-acetyltransferase [EC:2.3.1.-] | -0.685 | 0.723 | 0.345 | 0.978 |
| phnP; phosphoribosyl 1,2-cyclic phosphate phosphodiesterase [EC:3.1.4.55] | -0.367 | 0.169 | 0.032 | 0.839 |
| phnS; 2-aminoethylphosphonate transport system substrate-binding protein | -0.464 | 1.200 | 0.700 | 0.999 |
| phnT; 2-aminoethylphosphonate transport system ATP-binding protein | -0.464 | 1.200 | 0.700 | 0.999 |
| phnU; 2-aminoethylphosphonate transport system permease protein | -0.464 | 1.200 | 0.700 | 0.999 |
| phnV; 2-aminoethylphosphonate transport system permease protein | -0.440 | 1.101 | 0.690 | 0.999 |
| phnW; 2-aminoethylphosphonate-pyruvate transaminase [EC:2.6.1.37] | -0.337 | 0.304 | 0.269 | 0.978 |
| phnX; phosphonoacetaldehyde hydrolase [EC:3.11.1.1] | -0.237 | 0.318 | 0.457 | 0.979 |
| PHO; acid phosphatase [EC:3.1.3.2] | -0.203 | 0.374 | 0.587 | 0.999 |
| phoB; two-component system, OmpR family, phosphate regulon response regulator PhoB | -0.181 | 0.179 | 0.314 | 0.978 |
| phoB1, phoP; two-component system, OmpR family, alkaline phosphatase synthesis response regulator PhoP | -0.087 | 0.147 | 0.552 | 0.992 |
| phoD; alkaline phosphatase D [EC:3.1.3.1] | -0.191 | 0.213 | 0.372 | 0.978 |
| phoE; outer membrane pore protein E | 0.107 | 0.627 | 0.865 | 0.999 |
| phoH, phoL; phosphate starvation-inducible protein PhoH and related proteins | -0.008 | 0.042 | 0.842 | 0.999 |
| phoH2; PhoH-like ATPase | -0.146 | 0.187 | 0.437 | 0.978 |
| phoN; acid phosphatase (class A) [EC:3.1.3.2] | -0.055 | 0.228 | 0.811 | 0.999 |
| phoP; two-component system, OmpR family, response regulator PhoP | 0.098 | 0.258 | 0.703 | 0.999 |
| phoQ; two-component system, OmpR family, sensor histidine kinase PhoQ [EC:2.7.13.3] | 0.121 | 0.249 | 0.629 | 0.999 |
| phoR; two-component system, OmpR family, phosphate regulon sensor histidine kinase PhoR [EC:2.7.13.3] | -0.190 | 0.085 | 0.027 | 0.839 |
| phoU; phosphate transport system protein | -0.079 | 0.069 | 0.258 | 0.978 |
| phrB; deoxyribodipyrimidine photo-lyase [EC:4.1.99.3] | -0.153 | 0.173 | 0.377 | 0.978 |
| phrE; phosphatase RapE regulator | 18.239 | 2966.075 | 0.995 | 0.999 |
| phsA, psrA; thiosulfate reductase / polysulfide reductase chain A [EC:1.8.5.5] | 0.494 | 0.590 | 0.403 | 0.978 |
| phsB; thiosulfate reductase electron transport protein | -1.134 | 1.333 | 0.396 | 0.978 |
| phsC; thiosulfate reductase cytochrome b subunit | -1.134 | 1.333 | 0.396 | 0.978 |
| pht5; 4,5-dihydroxyphthalate decarboxylase [EC:4.1.1.55] | -1.618 | 1.105 | 0.145 | 0.951 |
| PHYH; phytanoyl-CoA hydroxylase [EC:1.14.11.18] | -0.282 | 1.106 | 0.799 | 0.999 |
| phzE; 2-amino-4-deoxychorismate synthase [EC:2.6.1.86] | 0.045 | 0.862 | 0.958 | 0.999 |
| phzF; trans-2,3-dihydro-3-hydroxyanthranilate isomerase [EC:5.3.3.17] | -1.240 | 0.497 | 0.014 | 0.839 |
| pigA, hemO; heme oxygenase (biliverdin-IX-beta and delta-forming) [EC:1.14.99.58] | -0.038 | 0.229 | 0.869 | 0.999 |
| PIGB; phosphatidylinositol glycan, class B [EC:2.4.1.-] | -17.761 | 4005.858 | 0.996 | 0.999 |
| pilA; type IV pilus assembly protein PilA | -0.015 | 0.199 | 0.940 | 0.999 |
| pilB; type IV pilus assembly protein PilB | 0.171 | 0.110 | 0.121 | 0.919 |
| pilC; type IV pilus assembly protein PilC | 0.113 | 0.098 | 0.250 | 0.978 |
| pilD, pppA; leader peptidase (prepilin peptidase) / N-methyltransferase [EC:3.4.23.43 2.1.1.-] | 0.163 | 0.083 | 0.052 | 0.839 |
| pilE; type IV pilus assembly protein PilE | 0.011 | 0.190 | 0.953 | 0.999 |
| pilF; type IV pilus assembly protein PilF | 0.085 | 0.161 | 0.598 | 0.999 |
| pilG; twitching motility two-component system response regulator PilG | -0.013 | 0.221 | 0.952 | 0.999 |
| pilH; twitching motility two-component system response regulator PilH | -0.027 | 0.245 | 0.913 | 0.999 |
| pilI; twitching motility protein PilI | -0.010 | 0.221 | 0.964 | 0.999 |
| pilJ; twitching motility protein PilJ | -0.027 | 0.220 | 0.903 | 0.999 |
| pilK; type IV pilus assembly protein PilK | 0.684 | 0.464 | 0.142 | 0.951 |
| pilL; type IV pili sensor histidine kinase and response regulator | -0.130 | 0.561 | 0.818 | 0.999 |
| pilM; type IV pilus assembly protein PilM | -0.067 | 0.173 | 0.700 | 0.999 |
| pilN; type IV pilus assembly protein PilN | 0.018 | 0.188 | 0.924 | 0.999 |
| pilO; type IV pilus assembly protein PilO | 0.000 | 0.176 | 0.998 | 1.000 |
| pilP; type IV pilus assembly protein PilP | 0.146 | 0.200 | 0.466 | 0.979 |
| pilQ; type IV pilus assembly protein PilQ | 0.030 | 0.168 | 0.856 | 0.999 |
| pilR, pehR; two-component system, NtrC family, response regulator PilR | 0.017 | 0.212 | 0.937 | 0.999 |
| pilS, pehS; two-component system, NtrC family, sensor histidine kinase PilS [EC:2.7.13.3] | 0.014 | 0.216 | 0.950 | 0.999 |
| pilT; twitching motility protein PilT | 0.071 | 0.097 | 0.466 | 0.979 |
| pilU; twitching motility protein PilU | -0.019 | 0.234 | 0.936 | 0.999 |
| pilV; type IV pilus assembly protein PilV | 0.023 | 0.214 | 0.915 | 0.999 |
| pilW; type IV pilus assembly protein PilW | 0.033 | 0.200 | 0.869 | 0.999 |
| pilX; type IV pilus assembly protein PilX | 0.084 | 0.222 | 0.705 | 0.999 |
| pilY1; type IV pilus assembly protein PilY1 | -0.017 | 0.246 | 0.945 | 0.999 |
| pilZ; type IV pilus assembly protein PilZ | 0.034 | 0.217 | 0.877 | 0.999 |
| pimA; phosphatidyl-myo-inositol alpha-mannosyltransferase [EC:2.4.1.345] | -0.043 | 0.282 | 0.878 | 0.999 |
| pimB; phosphatidyl-myo-inositol dimannoside synthase [EC:2.4.1.346] | -0.236 | 0.411 | 0.567 | 0.995 |
| pimC; alpha-1,6-mannosyltransferase [EC:2.4.1.-] | 0.176 | 0.276 | 0.526 | 0.989 |
| pimE; alpha-1,2-mannosyltransferase [EC:2.4.1.-] | -2.640 | 1.222 | 0.032 | 0.839 |
| pimF; putative glycosyltransferase [EC:2.4.-.-] | -1.141 | 1.688 | 0.500 | 0.981 |
| pinR; putative DNA-invertase from lambdoid prophage Rac | -0.241 | 0.649 | 0.711 | 0.999 |
| pip; proline iminopeptidase [EC:3.4.11.5] | -0.012 | 0.147 | 0.936 | 0.999 |
| PIP5K; 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68] | 18.478 | 3342.830 | 0.996 | 0.999 |
| pit; low-affinity inorganic phosphate transporter | 0.200 | 0.299 | 0.505 | 0.981 |
| PITRM1, PreP, CYM1; presequence protease [EC:3.4.24.-] | 0.142 | 0.178 | 0.427 | 0.978 |
| PK, pyk; pyruvate kinase [EC:2.7.1.40] | 0.055 | 0.030 | 0.063 | 0.839 |
| pknG; serine/threonine-protein kinase PknG [EC:2.7.11.1] | -0.672 | 0.650 | 0.303 | 0.978 |
| pks12; polyketide synthase 12 | -18.098 | 3949.754 | 0.996 | 0.999 |
| pks13; polyketide synthase 13 | -0.698 | 0.752 | 0.355 | 0.978 |
| pks5; polyketide synthase 5 | -2.695 | 1.434 | 0.062 | 0.839 |
| pksD; bacillaene synthase trans-acting acyltransferase | -1.624 | 2.294 | 0.480 | 0.979 |
| pksE; trans-AT polyketide synthase, acyltransferase and oxidoreductase domains | -0.133 | 0.664 | 0.842 | 0.999 |
| pksF; malonyl-ACP decarboxylase | -0.931 | 1.781 | 0.602 | 0.999 |
| pksG; polyketide biosynthesis 3-hydroxy-3-methylglutaryl-CoA synthase-like enzyme PksG | -0.931 | 1.781 | 0.602 | 0.999 |
| pksH; polyketide biosynthesis enoyl-CoA hydratase PksH | -0.931 | 1.781 | 0.602 | 0.999 |
| pksI; polyketide biosynthesis enoyl-CoA hydratase PksI | -17.761 | 4005.858 | 0.996 | 0.999 |
| pksJ; polyketide synthase PksJ | -0.169 | 0.700 | 0.810 | 0.999 |
| pksL; polyketide synthase PksL | 0.862 | 1.172 | 0.463 | 0.979 |
| pksM; polyketide synthase PksM | -0.145 | 0.723 | 0.842 | 0.999 |
| pksN; polyketide synthase PksN | 18.440 | 3011.936 | 0.995 | 0.999 |
| pksS; cytochrome P450 PksS | -1.768 | 1.095 | 0.108 | 0.917 |
| pla; plasminogen activator [EC:3.4.23.48] | 2.018 | 1.029 | 0.052 | 0.839 |
| plc, cpa; phospholipase C / alpha-toxin [EC:3.1.4.3] | 5.007 | 6.589 | 0.448 | 0.978 |
| plc; 1-phosphatidylinositol phosphodiesterase [EC:4.6.1.13] | -1.530 | 0.813 | 0.062 | 0.839 |
| plc; phospholipase C [EC:3.1.4.3] | -0.321 | 0.293 | 0.275 | 0.978 |
| PLD1_2; phospholipase D1/2 [EC:3.1.4.4] | -2.514 | 1.599 | 0.118 | 0.917 |
| pldA; phospholipase A1/A2 [EC:3.1.1.32 3.1.1.4] | -0.106 | 0.217 | 0.624 | 0.999 |
| pldB; lysophospholipase [EC:3.1.1.5] | -0.484 | 0.242 | 0.047 | 0.839 |
| pldh; pyridoxal 4-dehydrogenase [EC:1.1.1.107] | 18.076 | 2733.666 | 0.995 | 0.999 |
| pleC; two-component system, cell cycle sensor histidine kinase PleC [EC:2.7.13.3] | -1.410 | 0.565 | 0.014 | 0.839 |
| pleD; two-component system, cell cycle response regulator [EC:2.7.7.65] | -1.084 | 0.510 | 0.035 | 0.839 |
| plsB; glycerol-3-phosphate O-acyltransferase [EC:2.3.1.15] | 0.042 | 0.173 | 0.807 | 0.999 |
| plsC; 1-acyl-sn-glycerol-3-phosphate acyltransferase [EC:2.3.1.51] | -0.017 | 0.035 | 0.632 | 0.999 |
| plsX; glycerol-3-phosphate acyltransferase PlsX [EC:2.3.1.15] | 0.087 | 0.054 | 0.110 | 0.917 |
| plsY; glycerol-3-phosphate acyltransferase PlsY [EC:2.3.1.15] | 0.068 | 0.057 | 0.234 | 0.978 |
| plyA; polysaccharidase protein | -1.272 | 1.833 | 0.489 | 0.979 |
| PM20D1; carboxypeptidase PM20D1 [EC:3.4.17.-] | -0.574 | 0.491 | 0.243 | 0.978 |
| PMA1, PMA2; H+-transporting ATPase [EC:3.6.3.6] | -0.022 | 0.858 | 0.979 | 0.999 |
| pmbA; PmbA protein | -0.051 | 0.146 | 0.726 | 0.999 |
| pmm-pgm; phosphomannomutase / phosphoglucomutase [EC:5.4.2.8 5.4.2.2] | -0.058 | 0.201 | 0.774 | 0.999 |
| pmmS; 2-phosphinomethylmalate synthase [EC:2.3.3.18] | -17.228 | 3068.640 | 0.996 | 0.999 |
| pmoA-amoA; methane/ammonia monooxygenase subunit A [EC:1.14.18.3 1.14.99.39] | -17.996 | 3175.764 | 0.995 | 0.999 |
| pmoB-amoB; methane/ammonia monooxygenase subunit B | -17.996 | 3175.764 | 0.995 | 0.999 |
| pmoC-amoC; methane/ammonia monooxygenase subunit C | -17.379 | 2414.057 | 0.994 | 0.999 |
| pmrD; signal transduction protein PmrD | -1.244 | 1.220 | 0.309 | 0.978 |
| pmtA; phosphatidylethanolamine/phosphatidyl-N-methylethanolamine N-methyltransferase [EC:2.1.1.17 2.1.1.71] | -1.322 | 0.651 | 0.044 | 0.839 |
| pnbA; para-nitrobenzyl esterase [EC:3.1.1.-] | -0.709 | 0.334 | 0.035 | 0.839 |
| PNC1; nicotinamidase [EC:3.5.1.19] | 0.282 | 0.536 | 0.600 | 0.999 |
| pncA; nicotinamidase/pyrazinamidase [EC:3.5.1.19 3.5.1.-] | -0.097 | 0.109 | 0.375 | 0.978 |
| pncB, NAPRT1; nicotinate phosphoribosyltransferase [EC:6.3.4.21] | 0.034 | 0.039 | 0.391 | 0.978 |
| pncC; nicotinamide-nucleotide amidase [EC:3.5.1.42] | 0.041 | 0.051 | 0.416 | 0.978 |
| pno; pyridoxine 4-oxidase [EC:1.1.3.12] | 18.076 | 2733.666 | 0.995 | 0.999 |
| pnp, PNPT1; polyribonucleotide nucleotidyltransferase [EC:2.7.7.8] | 0.018 | 0.044 | 0.693 | 0.999 |
| pntA; NAD(P) transhydrogenase subunit alpha [EC:1.6.1.2] | -0.122 | 0.144 | 0.396 | 0.978 |
| pntB; NAD(P) transhydrogenase subunit beta [EC:1.6.1.2] | -0.094 | 0.151 | 0.533 | 0.990 |
| pnuC; nicotinamide mononucleotide transporter | -0.009 | 0.078 | 0.905 | 0.999 |
| pobA; p-hydroxybenzoate 3-monooxygenase [EC:1.14.13.2] | -0.240 | 0.269 | 0.373 | 0.978 |
| pobR; AraC family transcriptional regulator, transcriptional activator of pobA | -0.095 | 0.295 | 0.749 | 0.999 |
| podJ; localization factor PodJL | -1.360 | 0.578 | 0.020 | 0.839 |
| pok; pantoate kinase [EC:2.7.1.169] | -17.234 | 3077.514 | 0.996 | 0.999 |
| pol; DNA polymerase, archaea type [EC:2.7.7.7] | -17.234 | 3077.514 | 0.996 | 0.999 |
| polA; DNA polymerase I [EC:2.7.7.7] | 0.025 | 0.037 | 0.498 | 0.981 |
| polB; DNA polymerase II [EC:2.7.7.7] | 0.212 | 0.288 | 0.462 | 0.979 |
| polB; DNA polymerase II small subunit [EC:2.7.7.7] | -17.234 | 3077.514 | 0.996 | 0.999 |
| polC; DNA polymerase II large subunit [EC:2.7.7.7] | -17.234 | 3077.514 | 0.996 | 0.999 |
| polC; DNA polymerase III subunit alpha, Gram-positive type [EC:2.7.7.7] | 0.178 | 0.099 | 0.076 | 0.855 |
| POLRMT, RPO41; DNA-directed RNA polymerase, mitochondrial [EC:2.7.7.6] | -0.229 | 1.513 | 0.880 | 0.999 |
| polX, dpx; DNA polymerase (family X) | -0.885 | 0.396 | 0.027 | 0.839 |
| POMT, pmt; dolichyl-phosphate-mannose-protein mannosyltransferase [EC:2.4.1.109] | -0.042 | 0.671 | 0.950 | 0.999 |
| POP2; 4-aminobutyrate—pyruvate transaminase [EC:2.6.1.96] | -1.359 | 0.792 | 0.088 | 0.880 |
| POP4, RPP29; ribonuclease P protein subunit POP4 [EC:3.1.26.5] | -17.234 | 3077.514 | 0.996 | 0.999 |
| popA; two-component system, cell cycle response regulator PopA | -0.576 | 0.864 | 0.506 | 0.981 |
| por, nifJ; pyruvate-ferredoxin/flavodoxin oxidoreductase [EC:1.2.7.1 1.2.7.-] | 0.490 | 0.232 | 0.037 | 0.839 |
| porA; pyruvate ferredoxin oxidoreductase alpha subunit [EC:1.2.7.1] | -0.534 | 0.545 | 0.329 | 0.978 |
| porB; pyruvate ferredoxin oxidoreductase beta subunit [EC:1.2.7.1] | -0.480 | 0.598 | 0.424 | 0.978 |
| porD; pyruvate ferredoxin oxidoreductase delta subunit [EC:1.2.7.1] | 0.713 | 1.177 | 0.545 | 0.990 |
| porG; pyruvate ferredoxin oxidoreductase gamma subunit [EC:1.2.7.1] | -0.320 | 0.514 | 0.535 | 0.990 |
| potA; spermidine/putrescine transport system ATP-binding protein [EC:3.6.3.31] | 0.085 | 0.092 | 0.355 | 0.978 |
| potB; spermidine/putrescine transport system permease protein | 0.107 | 0.092 | 0.246 | 0.978 |
| potC; spermidine/putrescine transport system permease protein | 0.123 | 0.098 | 0.212 | 0.978 |
| potD; spermidine/putrescine transport system substrate-binding protein | 0.113 | 0.092 | 0.221 | 0.978 |
| potE; putrescine:ornithine antiporter | 0.050 | 0.236 | 0.833 | 0.999 |
| potF; putrescine transport system substrate-binding protein | 0.181 | 0.259 | 0.485 | 0.979 |
| potG; putrescine transport system ATP-binding protein | -0.024 | 0.204 | 0.906 | 0.999 |
| potH; putrescine transport system permease protein | -0.030 | 0.203 | 0.884 | 0.999 |
| potI; putrescine transport system permease protein | -0.012 | 0.204 | 0.953 | 0.999 |
| poxB; pyruvate dehydrogenase (quinone) [EC:1.2.5.1] | -0.099 | 0.224 | 0.658 | 0.999 |
| ppa; inorganic pyrophosphatase [EC:3.6.1.1] | 0.069 | 0.101 | 0.498 | 0.981 |
| ppaC; manganese-dependent inorganic pyrophosphatase [EC:3.6.1.1] | 0.136 | 0.093 | 0.145 | 0.951 |
| ppaT; pyridoxamine—pyruvate transaminase [EC:2.6.1.30] | 18.076 | 2733.666 | 0.995 | 0.999 |
| ppaX; pyrophosphatase PpaX [EC:3.6.1.1] | -0.762 | 0.582 | 0.192 | 0.970 |
| ppc; phosphoenolpyruvate carboxylase [EC:4.1.1.31] | 0.015 | 0.070 | 0.828 | 0.999 |
| PPCDC, coaC; phosphopantothenoylcysteine decarboxylase [EC:4.1.1.36] | -0.019 | 0.172 | 0.912 | 0.999 |
| PPCS, COAB; phosphopantothenate—cysteine ligase (ATP) [EC:6.3.2.51] | -0.020 | 0.172 | 0.910 | 0.999 |
| ppdA; prepilin peptidase dependent protein A | 0.193 | 0.314 | 0.539 | 0.990 |
| ppdB; prepilin peptidase dependent protein B | 0.182 | 0.312 | 0.560 | 0.992 |
| ppdC; prepilin peptidase dependent protein C | 0.193 | 0.314 | 0.539 | 0.990 |
| ppdD; prepilin peptidase dependent protein D | 0.199 | 0.311 | 0.523 | 0.989 |
| ppdK; pyruvate, orthophosphate dikinase [EC:2.7.9.1] | -0.026 | 0.126 | 0.835 | 0.999 |
| ppgK; polyphosphate glucokinase [EC:2.7.1.63] | 0.026 | 0.255 | 0.918 | 0.999 |
| pphA; serine/threonine protein phosphatase 1 [EC:3.1.3.16] | 0.048 | 0.110 | 0.666 | 0.999 |
| pphB; serine/threonine protein phosphatase 2 [EC:3.1.3.16] | -1.153 | 1.472 | 0.435 | 0.978 |
| PPIA; peptidyl-prolyl cis-trans isomerase A (cyclophilin A) [EC:5.2.1.8] | 0.020 | 0.082 | 0.803 | 0.999 |
| PPIB, ppiB; peptidyl-prolyl cis-trans isomerase B (cyclophilin B) [EC:5.2.1.8] | 0.047 | 0.045 | 0.304 | 0.978 |
| ppiC; peptidyl-prolyl cis-trans isomerase C [EC:5.2.1.8] | 0.025 | 0.177 | 0.887 | 0.999 |
| ppiD; peptidyl-prolyl cis-trans isomerase D [EC:5.2.1.8] | 0.026 | 0.100 | 0.792 | 0.999 |
| PPIL1; peptidyl-prolyl cis-trans isomerase-like 1 [EC:5.2.1.8] | 1.495 | 1.571 | 0.343 | 0.978 |
| ppk; polyphosphate kinase [EC:2.7.4.1] | -0.028 | 0.078 | 0.717 | 0.999 |
| ppkA; serine/threonine-protein kinase PpkA [EC:2.7.11.1] | -0.265 | 0.631 | 0.675 | 0.999 |
| ppnK, NADK; NAD+ kinase [EC:2.7.1.23] | 0.027 | 0.036 | 0.447 | 0.978 |
| ppnN; pyrimidine/purine-5’-nucleotide nucleosidase [EC:3.2.2.10 3.2.2.-] | 0.026 | 0.068 | 0.701 | 0.999 |
| ppnP; purine/pyrimidine-nucleoside phosphorylase [EC:2.4.2.1 2.4.2.2] | 0.020 | 0.234 | 0.931 | 0.999 |
| PPOX, hemY; protoporphyrinogen/coproporphyrinogen III oxidase [EC:1.3.3.4 1.3.3.15] | -0.144 | 0.151 | 0.343 | 0.978 |
| pps, ppsA; pyruvate, water dikinase [EC:2.7.9.2] | 0.022 | 0.087 | 0.797 | 0.999 |
| ppsA; phthiocerol/phenolphthiocerol synthesis type-I polyketide synthase A | -17.921 | 3534.051 | 0.996 | 0.999 |
| ppsB; phthiocerol/phenolphthiocerol synthesis type-I polyketide synthase B | -17.921 | 3534.051 | 0.996 | 0.999 |
| ppsD; phthiocerol/phenolphthiocerol synthesis type-I polyketide synthase D | -17.921 | 3534.051 | 0.996 | 0.999 |
| ppsE; phthiocerol/phenolphthiocerol synthesis type-I polyketide synthase E | -17.921 | 3534.051 | 0.996 | 0.999 |
| ppsR; [pyruvate, water dikinase]-phosphate phosphotransferase / [pyruvate, water dikinase] kinase [EC:2.7.4.28 2.7.11.33] | -0.056 | 0.066 | 0.397 | 0.978 |
| ppx-gppA; exopolyphosphatase / guanosine-5’-triphosphate,3’-diphosphate pyrophosphatase [EC:3.6.1.11 3.6.1.40] | 0.144 | 0.080 | 0.076 | 0.855 |
| pqiA; paraquat-inducible protein A | 0.038 | 0.249 | 0.879 | 0.999 |
| pqiB; paraquat-inducible protein B | -0.012 | 0.270 | 0.966 | 0.999 |
| pqqA; pyrroloquinoline quinone biosynthesis protein A | -0.910 | 0.952 | 0.340 | 0.978 |
| pqqB; pyrroloquinoline quinone biosynthesis protein B | 0.041 | 0.287 | 0.887 | 0.999 |
| pqqC; pyrroloquinoline-quinone synthase [EC:1.3.3.11] | 0.017 | 0.278 | 0.951 | 0.999 |
| pqqD; pyrroloquinoline quinone biosynthesis protein D | 0.130 | 0.281 | 0.646 | 0.999 |
| pqqE; pyrroloquinoline quinone biosynthesis protein E | 0.002 | 0.314 | 0.994 | 0.999 |
| pqqL; zinc protease [EC:3.4.24.-] | -0.107 | 0.137 | 0.435 | 0.978 |
| praC, xylH; 4-oxalocrotonate tautomerase [EC:5.3.2.6] | -0.027 | 0.095 | 0.779 | 0.999 |
| prdA; D-proline reductase (dithiol) PrdA [EC:1.21.4.1] | -0.124 | 0.345 | 0.719 | 0.999 |
| prdB; D-proline reductase (dithiol) PrdB [EC:1.21.4.1] | -0.199 | 0.357 | 0.578 | 0.997 |
| prdD; D-proline reductase (dithiol)-stabilizing protein PrdD | -0.124 | 0.345 | 0.719 | 0.999 |
| prdE; D-proline reductase (dithiol)-stabilizing protein PrdE | -0.124 | 0.345 | 0.719 | 0.999 |
| prdF; proline racemase [EC:5.1.1.4] | -0.248 | 0.540 | 0.647 | 0.999 |
| prdX, proX; Ala-tRNA(Pro) deacylase [EC:3.1.1.-] | 0.189 | 0.115 | 0.101 | 0.896 |
| PRDX2_4, ahpC; peroxiredoxin (alkyl hydroperoxide reductase subunit C) [EC:1.11.1.15] | 0.072 | 0.055 | 0.195 | 0.970 |
| PRDX5; peroxiredoxin 5, atypical 2-Cys peroxiredoxin [EC:1.11.1.15] | -17.770 | 4023.440 | 0.996 | 0.999 |
| preA; dihydropyrimidine dehydrogenase (NAD+) subunit PreA [EC:1.3.1.1] | -0.282 | 0.284 | 0.323 | 0.978 |
| PREP; prolyl oligopeptidase [EC:3.4.21.26] | -0.141 | 0.160 | 0.379 | 0.978 |
| preT; dihydropyrimidine dehydrogenase (NAD+) subunit PreT [EC:1.3.1.1] | 1.003 | 0.698 | 0.153 | 0.954 |
| prfA, MTRF1, MRF1; peptide chain release factor 1 | 0.035 | 0.036 | 0.325 | 0.978 |
| prfB; peptide chain release factor 2 | 0.036 | 0.036 | 0.321 | 0.978 |
| prfC; peptide chain release factor 3 | -0.003 | 0.044 | 0.944 | 0.999 |
| prfH; peptide chain release factor | 0.288 | 0.348 | 0.410 | 0.978 |
| PRHOXNB, URAD; 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase [EC:4.1.1.97] | -0.672 | 0.641 | 0.296 | 0.978 |
| priA; primosomal protein N’ (replication factor Y) (superfamily II helicase) [EC:3.6.4.-] | 0.032 | 0.031 | 0.303 | 0.978 |
| priB; primosomal replication protein N | 0.161 | 0.243 | 0.509 | 0.981 |
| priC; primosomal replication protein N’’ | 0.169 | 0.272 | 0.535 | 0.990 |
| priL, pri2, priB; DNA primase large subunit [EC:2.7.7.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| priS, pri1, priA; DNA primase small subunit [EC:2.7.7.-] | -17.234 | 3077.514 | 0.996 | 0.999 |
| PRK, prkB; phosphoribulokinase [EC:2.7.1.19] | -0.226 | 0.433 | 0.603 | 0.999 |
| prkA; serine protein kinase | -0.151 | 0.246 | 0.540 | 0.990 |
| prlC; oligopeptidase A [EC:3.4.24.70] | 0.022 | 0.158 | 0.889 | 0.999 |
| prlF, sohA; antitoxin PrlF | -1.294 | 1.385 | 0.351 | 0.978 |
| prmA; propane 2-monooxygenase large subunit [EC:1.14.13.227] | 0.954 | 0.835 | 0.255 | 0.978 |
| prmA; ribosomal protein L11 methyltransferase [EC:2.1.1.-] | 0.044 | 0.041 | 0.280 | 0.978 |
| prmB; propane monooxygenase reductase component [EC:1.18.1.-] | 0.791 | 0.811 | 0.330 | 0.978 |
| prmB; ribosomal protein L3 glutamine methyltransferase [EC:2.1.1.298] | 0.052 | 0.154 | 0.737 | 0.999 |
| prmC; propane 2-monooxygenase small subunit [EC:1.14.13.227] | 0.791 | 0.811 | 0.330 | 0.978 |
| prmD; propane monooxygenase coupling protein | 0.883 | 1.008 | 0.383 | 0.978 |
| PRMT1; type I protein arginine methyltransferase [EC:2.1.1.319] | 18.066 | 2719.856 | 0.995 | 0.999 |
| PRMT9; type II protein arginine methyltransferase [EC:2.1.1.320] | 0.884 | 1.140 | 0.439 | 0.978 |
| prnA, rebH, ktzQ; tryptophan 7-halogenase [EC:1.14.19.9] | -0.739 | 0.522 | 0.159 | 0.954 |
| proA; glutamate-5-semialdehyde dehydrogenase [EC:1.2.1.41] | 0.012 | 0.050 | 0.812 | 0.999 |
| proB; glutamate 5-kinase [EC:2.7.2.11] | 0.014 | 0.046 | 0.763 | 0.999 |
| proC; pyrroline-5-carboxylate reductase [EC:1.5.1.2] | -0.011 | 0.046 | 0.819 | 0.999 |
| PRODH; proline dehydrogenase [EC:1.5.-.-] | -0.714 | 0.364 | 0.051 | 0.839 |
| proP; MFS transporter, MHS family, proline/betaine transporter | -0.146 | 0.235 | 0.537 | 0.990 |
| proQ; ProP effector | 0.361 | 0.244 | 0.141 | 0.949 |
| proV; glycine betaine/proline transport system ATP-binding protein [EC:3.6.3.32] | 0.092 | 0.238 | 0.699 | 0.999 |
| proW; glycine betaine/proline transport system permease protein | -0.017 | 0.262 | 0.949 | 0.999 |
| proX; glycine betaine/proline transport system substrate-binding protein | 0.089 | 0.276 | 0.748 | 0.999 |
| proY; proline-specific permease ProY | 0.134 | 0.621 | 0.829 | 0.999 |
| prpB; methylisocitrate lyase [EC:4.1.3.30] | -0.056 | 0.185 | 0.762 | 0.999 |
| prpC; 2-methylcitrate synthase [EC:2.3.3.5] | 0.016 | 0.197 | 0.934 | 0.999 |
| prpD; 2-methylcitrate dehydratase [EC:4.2.1.79] | -0.143 | 0.254 | 0.573 | 0.995 |
| prpE; propionyl-CoA synthetase [EC:6.2.1.17] | -0.293 | 0.266 | 0.273 | 0.978 |
| prpF; 2-methylaconitate isomerase [EC:5.3.3.-] | -0.038 | 0.215 | 0.860 | 0.999 |
| prpR; transcriptional regulator, propionate catabolism operon regulatory protein | -0.133 | 0.426 | 0.755 | 0.999 |
| PRPS, prsA; ribose-phosphate pyrophosphokinase [EC:2.7.6.1] | 0.051 | 0.053 | 0.340 | 0.978 |
| prr; aminobutyraldehyde dehydrogenase [EC:1.2.1.19] | -0.214 | 0.390 | 0.584 | 0.999 |
| prrA; two-component system, OmpR family, response regulator PrrA | -1.593 | 1.633 | 0.331 | 0.978 |
| prrB; two-component system, OmpR family, sensor histidine kinase PrrB [EC:2.7.13.3] | -1.890 | 2.089 | 0.367 | 0.978 |
| prsA; foldase protein PrsA [EC:5.2.1.8] | -0.033 | 0.124 | 0.789 | 0.999 |
| prsD; ATP-binding cassette, subfamily C, bacterial PrsD | -1.272 | 1.833 | 0.489 | 0.979 |
| prsE; membrane fusion protein | -1.272 | 1.833 | 0.489 | 0.979 |
| PRSS15, PIM1; ATP-dependent Lon protease [EC:3.4.21.53] | -0.020 | 0.682 | 0.976 | 0.999 |
| PRSS33; serine protease 33 [EC:3.4.21.-] | 2.635 | 1.537 | 0.088 | 0.880 |
| prtC; serralysin [EC:3.4.24.40] | -0.579 | 0.506 | 0.254 | 0.978 |
| psd, PISD; phosphatidylserine decarboxylase [EC:4.1.1.65] | -0.007 | 0.062 | 0.907 | 0.999 |
| pseB; UDP-N-acetylglucosamine 4,6-dehydratase [EC:4.2.1.115] | -0.628 | 0.606 | 0.301 | 0.978 |
| pseC; UDP-4-amino-4,6-dideoxy-L-N-acetyl-beta-L-altrosamine transaminase [EC:2.6.1.92] | -0.751 | 0.907 | 0.409 | 0.978 |
| pseF; pseudaminic acid cytidylyltransferase [EC:2.7.7.81] | 0.023 | 0.527 | 0.966 | 0.999 |
| pseG; UDP-2,4-diacetamido-2,4,6-trideoxy-beta-L-altropyranose hydrolase [EC:3.6.1.57] | 0.051 | 0.670 | 0.939 | 0.999 |
| pseH; UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine N-acetyltransferase [EC:2.3.1.202] | -0.751 | 0.907 | 0.409 | 0.978 |
| pseI, neuB3; pseudaminic acid synthase [EC:2.5.1.97] | -0.850 | 0.850 | 0.319 | 0.978 |
| psiE; protein PsiE | 0.157 | 0.281 | 0.576 | 0.995 |
| psmA, prcA; proteasome alpha subunit [EC:3.4.25.1] | -0.483 | 0.706 | 0.495 | 0.981 |
| psmB, prcB; proteasome beta subunit [EC:3.4.25.1] | -0.487 | 0.704 | 0.490 | 0.979 |
| pspA; phage shock protein A | -0.118 | 0.201 | 0.557 | 0.992 |
| pspB; phage shock protein B | -0.005 | 0.387 | 0.990 | 0.999 |
| pspC; phage shock protein C | 0.371 | 0.291 | 0.204 | 0.972 |
| pspD; phage shock protein D | 0.154 | 0.615 | 0.803 | 0.999 |
| pspE; phage shock protein E | 0.163 | 0.426 | 0.703 | 0.999 |
| pspF; psp operon transcriptional activator | -0.120 | 0.390 | 0.759 | 0.999 |
| psrB; polysulfide reductase chain B | 0.480 | 0.618 | 0.438 | 0.978 |
| psrC; polysulfide reductase chain C | 0.480 | 0.618 | 0.438 | 0.978 |
| PSRP4, RPS31; 30S ribosomal protein S31 | -0.080 | 0.448 | 0.858 | 0.999 |
| pssA; CDP-diacylglycerol—serine O-phosphatidyltransferase [EC:2.7.8.8] | 0.142 | 0.241 | 0.556 | 0.992 |
| pstA; phosphate transport system permease protein | -0.064 | 0.059 | 0.279 | 0.978 |
| pstB; phosphate transport system ATP-binding protein [EC:3.6.3.27] | -0.037 | 0.079 | 0.643 | 0.999 |
| pstC; phosphate transport system permease protein | -0.058 | 0.061 | 0.341 | 0.978 |
| pstS; phosphate transport system substrate-binding protein | -0.052 | 0.048 | 0.281 | 0.978 |
| psuG; pseudouridylate synthase [EC:4.2.1.70] | -0.711 | 0.304 | 0.021 | 0.839 |
| psuK; pseudouridine kinase [EC:2.7.1.83] | -0.118 | 0.342 | 0.730 | 0.999 |
| psuT; putative pseudouridine transporter | -0.846 | 1.057 | 0.425 | 0.978 |
| pta; phosphate acetyltransferase [EC:2.3.1.8] | 0.051 | 0.136 | 0.711 | 0.999 |
| ptb; phosphate butyryltransferase [EC:2.3.1.19] | -0.200 | 0.235 | 0.397 | 0.978 |
| ptcA; putrescine carbamoyltransferase [EC:2.1.3.6] | 0.110 | 0.740 | 0.882 | 0.999 |
| PTER, php; phosphotriesterase-related protein | -1.156 | 0.570 | 0.044 | 0.839 |
| PTGS2, COX2; prostaglandin-endoperoxide synthase 2 [EC:1.14.99.1] | 2.045 | 1.751 | 0.245 | 0.978 |
| PTH1, pth, spoVC; peptidyl-tRNA hydrolase, PTH1 family [EC:3.1.1.29] | 0.038 | 0.036 | 0.294 | 0.978 |
| PTH2; peptidyl-tRNA hydrolase, PTH2 family [EC:3.1.1.29] | -0.352 | 0.553 | 0.526 | 0.989 |
| ptlF; type IV secretion system protein PtlF | -2.174 | 2.005 | 0.280 | 0.978 |
| ptlI, CYP183A; pentalenene oxygenase [EC:1.14.13.133] | -17.794 | 4072.025 | 0.997 | 0.999 |
| ptpA; Xaa-Xaa-Pro tripeptidyl-peptidase [EC:3.4.14.12] | -0.222 | 0.624 | 0.723 | 0.999 |
| PTR1; pteridine reductase [EC:1.5.1.33] | 0.011 | 0.403 | 0.979 | 0.999 |
| ptrA; protease III [EC:3.4.24.55] | 0.214 | 0.344 | 0.534 | 0.990 |
| ptrB; oligopeptidase B [EC:3.4.21.83] | -0.020 | 0.146 | 0.893 | 0.999 |
| PTS-Aga-EIIA, agaF; PTS system, N-acetylgalactosamine-specific IIA component [EC:2.7.1.-] | -0.035 | 0.161 | 0.829 | 0.999 |
| PTS-Aga-EIIB, agaV; PTS system, N-acetylgalactosamine-specific IIB component [EC:2.7.1.-] | 0.008 | 0.339 | 0.981 | 0.999 |
| PTS-Aga-EIIC, agaW; PTS system, N-acetylgalactosamine-specific IIC component | 0.035 | 0.340 | 0.919 | 0.999 |
| PTS-Aga-EIID, agaE; PTS system, N-acetylgalactosamine-specific IID component | -0.146 | 0.384 | 0.705 | 0.999 |
| PTS-Asc-EIIC, ascF; PTS system, beta-glucoside (arbutin/salicin/cellobiose)-specific IIC component | 0.450 | 0.516 | 0.385 | 0.978 |
| PTS-Bgl-EIIA, bglF, bglP; PTS system, beta-glucoside-specific IIA component [EC:2.7.1.-] | -1.670 | 1.245 | 0.182 | 0.961 |
| PTS-Bgl-EIIC, bglF, bglP; PTS system, beta-glucoside-specific IIC component | 0.308 | 0.170 | 0.072 | 0.845 |
| PTS-Cel-EIIA, celC, chbA; PTS system, cellobiose-specific IIA component [EC:2.7.1.196 2.7.1.205] | 0.243 | 0.162 | 0.136 | 0.949 |
| PTS-Cel-EIIB, celA, chbB; PTS system, cellobiose-specific IIB component [EC:2.7.1.196 2.7.1.205] | 0.269 | 0.171 | 0.118 | 0.917 |
| PTS-Cel-EIIC, celB, chbC; PTS system, cellobiose-specific IIC component | 0.217 | 0.177 | 0.222 | 0.978 |
| PTS-Dga-EIIA, dgaA; PTS system, D-glucosaminate-specific IIA component [EC:2.7.1.203] | -1.630 | 2.316 | 0.483 | 0.979 |
| PTS-Dga-EIIB, dgaB; PTS system, D-glucosaminate-specific IIB component [EC:2.7.1.203] | -5.290 | 7.436 | 0.478 | 0.979 |
| PTS-Dga-EIIC, dgaC; PTS system, D-glucosaminate-specific IIC component | -4.720 | 6.582 | 0.474 | 0.979 |
| PTS-Dga-EIID, dgaD; PTS system, D-glucosaminate-specific IID component | -4.720 | 6.582 | 0.474 | 0.979 |
| PTS-Dgl-EIIA, gamP; PTS system, D-glucosamine-specific IIA component [EC:2.7.1.-] | 0.375 | 0.933 | 0.689 | 0.999 |
| PTS-Dgl-EIIC, gamP; PTS system, D-glucosamine-specific IIC component | -0.108 | 0.179 | 0.548 | 0.991 |
| PTS-EI.PTSI, ptsI; phosphotransferase system, enzyme I, PtsI [EC:2.7.3.9] | 0.112 | 0.068 | 0.099 | 0.896 |
| PTS-EI.PTSP, ptsP; phosphotransferase system, enzyme I, PtsP [EC:2.7.3.9] | -0.111 | 0.219 | 0.614 | 0.999 |
| PTS-Fru-EIIA, fruB; PTS system, fructose-specific IIA component [EC:2.7.1.202] | 0.694 | 0.305 | 0.024 | 0.839 |
| PTS-Fru-EIIB, fruA; PTS system, fructose-specific IIB component [EC:2.7.1.202] | 0.586 | 0.413 | 0.158 | 0.954 |
| PTS-Fru-EIIC, fruA; PTS system, fructose-specific IIC component | 0.123 | 0.072 | 0.088 | 0.880 |
| PTS-Fru1-EIIA, levD; PTS system, fructose-specific IIA component [EC:2.7.1.202] | 18.239 | 2966.075 | 0.995 | 0.999 |
| PTS-Fru1-EIIB, levE; PTS system, fructose-specific IIB component [EC:2.7.1.202] | 18.239 | 2966.075 | 0.995 | 0.999 |
| PTS-Fru1-EIID, levG; PTS system, fructose-specific IID component | -0.216 | 0.209 | 0.303 | 0.978 |
| PTS-Fru2-EIIA; PTS system, fructose-specific IIA-like component [EC:2.7.1.-] | -1.019 | 1.147 | 0.376 | 0.978 |
| PTS-Fru2-EIIB; PTS system, fructose-specific IIB-like component [EC:2.7.1.-] | -0.517 | 0.626 | 0.410 | 0.978 |
| PTS-Fru2-EIIC; PTS system, fructose-specific IIC-like component | -0.167 | 0.597 | 0.780 | 0.999 |
| PTS-Gam-EIIB, agaB; PTS system, galactosamine-specific IIB component [EC:2.7.1.-] | -0.560 | 1.181 | 0.636 | 0.999 |
| PTS-Gam-EIIC, agaC; PTS system, galactosamine-specific IIC component | -0.638 | 1.220 | 0.602 | 0.999 |
| PTS-Gam-EIID, agaD; PTS system, galactosamine-specific IID component | -0.638 | 1.220 | 0.602 | 0.999 |
| PTS-Gat-EIIA, gatA, sgcA; PTS system, galactitol-specific IIA component [EC:2.7.1.200] | 0.052 | 0.169 | 0.758 | 0.999 |
| PTS-Gat-EIIB, gatB, sgcB; PTS system, galactitol-specific IIB component [EC:2.7.1.200] | 0.038 | 0.152 | 0.802 | 0.999 |
| PTS-Gat-EIIC, gatC, sgcC; PTS system, galactitol-specific IIC component | 0.095 | 0.210 | 0.651 | 0.999 |
| PTS-Gfr-EIIA, gfrA; PTS system, fructoselysine/glucoselysine-specific IIA component [EC:2.7.1.-] | 0.713 | 0.423 | 0.094 | 0.894 |
| PTS-Gfr-EIIB, gfrB; PTS system, fructoselysine/glucoselysine-specific IIB component [EC:2.7.1.-] | 0.872 | 0.495 | 0.080 | 0.862 |
| PTS-Gfr-EIIC, gfrC; PTS system, fructoselysine/glucoselysine-specific IIC component | 0.622 | 0.400 | 0.122 | 0.919 |
| PTS-Gfr-EIID, gfrD; PTS system, fructoselysine/glucoselysine-specific IID component | 0.783 | 0.459 | 0.090 | 0.882 |
| PTS-Glc-EIIA, crr; PTS system, sugar-specific IIA component [EC:2.7.1.-] | 0.206 | 0.140 | 0.145 | 0.951 |
| PTS-Glc-EIIB, ptsG; PTS system, glucose-specific IIB component [EC:2.7.1.199] | -0.063 | 1.143 | 0.956 | 0.999 |
| PTS-Glc-EIIC, ptsG; PTS system, glucose-specific IIC component | 0.131 | 0.129 | 0.311 | 0.978 |
| PTS-Glv-EIIC, glvC, malP, aglA; PTS system, alpha-glucoside-specific IIC component | 0.242 | 0.242 | 0.319 | 0.978 |
| PTS-Gut-EIIA, srlB; PTS system, glucitol/sorbitol-specific IIA component [EC:2.7.1.198] | 0.382 | 0.272 | 0.162 | 0.957 |
| PTS-Gut-EIIC, srlA; PTS system, glucitol/sorbitol-specific IIC component | 0.202 | 0.443 | 0.649 | 0.999 |
| PTS-HPR.FRUB, fruB, fpr; phosphocarrier protein FPr | 0.033 | 0.247 | 0.894 | 0.999 |
| PTS-HPR.PTSH, ptsH; phosphocarrier protein HPr | 0.583 | 0.254 | 0.023 | 0.839 |
| PTS-HPR.PTSO, ptsO, npr; phosphocarrier protein NPr | -0.057 | 0.346 | 0.870 | 0.999 |
| PTS-HPR; phosphocarrier protein | 0.037 | 0.047 | 0.433 | 0.978 |
| PTS-Lac-EIIA, lacF; PTS system, lactose-specific IIA component [EC:2.7.1.207] | -0.020 | 0.155 | 0.898 | 0.999 |
| PTS-Lac-EIIC, lacE; PTS system, lactose-specific IIC component | -0.020 | 0.155 | 0.899 | 0.999 |
| PTS-MalGlc-EIIC, malX; PTS system, maltose/glucose-specific IIC component | 0.311 | 0.265 | 0.242 | 0.978 |
| PTS-Man-EIIA, manX; PTS system, mannose-specific IIA component [EC:2.7.1.191] | 0.002 | 0.132 | 0.991 | 0.999 |
| PTS-Man-EIIB, manX; PTS system, mannose-specific IIB component [EC:2.7.1.191] | 0.051 | 0.120 | 0.673 | 0.999 |
| PTS-Man-EIIC, manY; PTS system, mannose-specific IIC component | 0.035 | 0.127 | 0.782 | 0.999 |
| PTS-Man-EIID, manZ; PTS system, mannose-specific IID component | 0.068 | 0.125 | 0.588 | 0.999 |
| PTS-Mng-EIIC, mngA, hrsA; PTS system, 2-O-A-mannosyl-D-glycerate-specific IIC component | -1.392 | 0.946 | 0.143 | 0.951 |
| PTS-Mtl-EIIA, mtlA, cmtB; PTS system, mannitol-specific IIA component [EC:2.7.1.197] | 0.237 | 0.324 | 0.466 | 0.979 |
| PTS-Mtl-EIIC, mtlA, cmtA; PTS system, mannitol-specific IIC component | 0.042 | 0.206 | 0.838 | 0.999 |
| PTS-Mur-EIIC, murP; PTS system, N-acetylmuramic acid-specific IIC component | 0.050 | 0.756 | 0.948 | 0.999 |
| PTS-Nag-EIIA, nagE; PTS system, N-acetylglucosamine-specific IIA component [EC:2.7.1.193] | 0.119 | 0.339 | 0.726 | 0.999 |
| PTS-Nag-EIIB, nagE; PTS system, N-acetylglucosamine-specific IIB component [EC:2.7.1.193] | -0.021 | 0.347 | 0.952 | 0.999 |
| PTS-Nag-EIIC, nagE; PTS system, N-acetylglucosamine-specific IIC component | 0.138 | 0.147 | 0.352 | 0.978 |
| PTS-Ntr-EIIA, ptsN; PTS system, nitrogen regulatory IIA component [EC:2.7.1.-] | 0.338 | 0.145 | 0.021 | 0.839 |
| PTS-Scr-EIIC, scrA, sacP, sacX, ptsS; PTS system, sucrose-specific IIC component | 0.140 | 0.109 | 0.199 | 0.970 |
| PTS-Sor-EIIA, sorF; PTS system, sorbose-specific IIA component [EC:2.7.1.206] | -0.750 | 1.062 | 0.481 | 0.979 |
| PTS-Sor-EIIB, sorB; PTS system, sorbose-specific IIB component [EC:2.7.1.206] | -0.668 | 0.835 | 0.425 | 0.978 |
| PTS-Sor-EIIC, sorA; PTS system, sorbose-specific IIC component | -0.324 | 0.818 | 0.692 | 0.999 |
| PTS-Sor-EIID, sorM; PTS system, sorbose-specific IID component | -0.277 | 0.837 | 0.742 | 0.999 |
| PTS-Tre-EIIC, treB; PTS system, trehalose-specific IIC component | 0.179 | 0.143 | 0.215 | 0.978 |
| PTS-Ula-EIIA, ulaC, sgaA; PTS system, ascorbate-specific IIA component [EC:2.7.1.194] | 0.085 | 0.110 | 0.438 | 0.978 |
| PTS-Ula-EIIB, ulaB, sgaB; PTS system, ascorbate-specific IIB component [EC:2.7.1.194] | 0.086 | 0.148 | 0.562 | 0.992 |
| PTS-Ula-EIIC, ulaA, sgaT; PTS system, ascorbate-specific IIC component | 0.106 | 0.136 | 0.439 | 0.978 |
| ptxD; phosphonate dehydrogenase [EC:1.20.1.1] | -0.213 | 1.210 | 0.860 | 0.999 |
| pucA; light-harvesting protein B-800-850 alpha chain | -17.193 | 2198.071 | 0.994 | 0.999 |
| pucB; light-harvesting protein B-800-850 beta chain | -17.500 | 2562.559 | 0.995 | 0.999 |
| pucC; MFS transporter, BCD family, chlorophyll transporter | -17.473 | 2396.504 | 0.994 | 0.999 |
| pucG; (S)-ureidoglycine—glyoxylate transaminase [EC:2.6.1.112] | 0.308 | 0.672 | 0.647 | 0.999 |
| pucR; purine catabolism regulatory protein | -0.806 | 0.707 | 0.256 | 0.978 |
| pufA; light-harvesting complex 1 alpha chain | -3.309 | 3.390 | 0.331 | 0.978 |
| pufB; light-harvesting complex 1 beta chain | -3.309 | 3.390 | 0.331 | 0.978 |
| pufC; photosynthetic reaction center cytochrome c subunit | -2.639 | 3.582 | 0.462 | 0.979 |
| pufL; photosynthetic reaction center L subunit | -3.309 | 3.390 | 0.331 | 0.978 |
| pufM; photosynthetic reaction center M subunit | -3.309 | 3.390 | 0.331 | 0.978 |
| pufX; photosynthetic reaction center PufX protein | -17.500 | 2562.559 | 0.995 | 0.999 |
| puhA; photosynthetic reaction center H subunit | -18.056 | 3176.254 | 0.995 | 0.999 |
| pulA; pullulanase [EC:3.2.1.41] | -0.107 | 0.138 | 0.442 | 0.978 |
| punA, PNP; purine-nucleoside phosphorylase [EC:2.4.2.1] | -0.066 | 0.104 | 0.529 | 0.989 |
| puo; putrescine oxidase [EC:1.4.3.10] | -0.759 | 0.619 | 0.222 | 0.978 |
| pup; prokaryotic ubiquitin-like protein Pup | -0.002 | 0.259 | 0.994 | 0.999 |
| purA, ADSS; adenylosuccinate synthase [EC:6.3.4.4] | 0.030 | 0.032 | 0.359 | 0.978 |
| purB, ADSL; adenylosuccinate lyase [EC:4.3.2.2] | 0.035 | 0.036 | 0.330 | 0.978 |
| purC; phosphoribosylaminoimidazole-succinocarboxamide synthase [EC:6.3.2.6] | 0.038 | 0.038 | 0.315 | 0.978 |
| purD; phosphoribosylamine—glycine ligase [EC:6.3.4.13] | 0.040 | 0.042 | 0.340 | 0.978 |
| purE; 5-(carboxyamino)imidazole ribonucleotide mutase [EC:5.4.99.18] | 0.044 | 0.038 | 0.253 | 0.978 |
| purF, PPAT; amidophosphoribosyltransferase [EC:2.4.2.14] | 0.017 | 0.038 | 0.647 | 0.999 |
| purH; phosphoribosylaminoimidazolecarboxamide formyltransferase / IMP cyclohydrolase [EC:2.1.2.3 3.5.4.10] | 0.034 | 0.048 | 0.482 | 0.979 |
| purK; 5-(carboxyamino)imidazole ribonucleotide synthase [EC:6.3.4.18] | 0.001 | 0.068 | 0.992 | 0.999 |
| purL, PFAS; phosphoribosylformylglycinamidine synthase [EC:6.3.5.3] | 0.017 | 0.053 | 0.751 | 0.999 |
| purM; phosphoribosylformylglycinamidine cyclo-ligase [EC:6.3.3.1] | 0.035 | 0.040 | 0.387 | 0.978 |
| purN; phosphoribosylglycinamide formyltransferase 1 [EC:2.1.2.2] | 0.098 | 0.043 | 0.025 | 0.839 |
| purNH; phosphoribosylglycinamide/phosphoribosylaminoimidazolecarboxamide formyltransferase [EC:2.1.2.2 2.1.2.3] | -0.903 | 0.549 | 0.102 | 0.896 |
| purP; 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5’-monophosphate synthetase [EC:6.3.4.23] | -16.927 | 2639.780 | 0.995 | 0.999 |
| purR; LacI family transcriptional regulator, purine nucleotide synthesis repressor | 0.017 | 0.261 | 0.948 | 0.999 |
| purR; purine operon repressor | 0.033 | 0.118 | 0.782 | 0.999 |
| purT; phosphoribosylglycinamide formyltransferase 2 [EC:2.1.2.2] | -0.022 | 0.105 | 0.837 | 0.999 |
| purU; formyltetrahydrofolate deformylase [EC:3.5.1.10] | -0.133 | 0.168 | 0.430 | 0.978 |
| PUS10; tRNA pseudouridine synthase 10 [EC:5.4.99.25] | -17.234 | 3077.514 | 0.996 | 0.999 |
| putA; RHH-type transcriptional regulator, proline utilization regulon repressor / proline dehydrogenase / delta 1-pyrroline-5-carboxylate dehydrogenase [EC:1.5.5.2 1.2.1.88] | -0.044 | 0.141 | 0.757 | 0.999 |
| putP; sodium/proline symporter | 0.078 | 0.085 | 0.361 | 0.978 |
| puuA; gamma-glutamylputrescine synthase [EC:6.3.1.11] | -0.104 | 0.386 | 0.789 | 0.999 |
| puuB, ordL; gamma-glutamylputrescine oxidase [EC:1.4.3.-] | -0.030 | 0.206 | 0.884 | 0.999 |
| puuC, aldH; 4-(gamma-glutamylamino)butanal dehydrogenase [EC:1.2.1.99] | -0.111 | 0.277 | 0.689 | 0.999 |
| puuD; gamma-glutamyl-gamma-aminobutyrate hydrolase [EC:3.5.1.94] | 0.136 | 0.641 | 0.832 | 0.999 |
| puuE; 4-aminobutyrate aminotransferase [EC:2.6.1.19] | 0.002 | 0.273 | 0.994 | 0.999 |
| puuP; putrescine importer | 0.436 | 0.383 | 0.256 | 0.978 |
| puuR; HTH-type transcriptional regulator, repressor for puuD | 0.068 | 0.634 | 0.915 | 0.999 |
| pvadh; polyvinyl alcohol dehydrogenase (cytochrome) [EC:1.1.2.6] | -1.523 | 1.350 | 0.261 | 0.978 |
| pvdA, SIDA; L-ornithine N5-monooxygenase [EC:1.14.13.195 1.14.13.196] | -0.048 | 0.402 | 0.905 | 0.999 |
| pvdE; putative pyoverdin transport system ATP-binding/permease protein | -0.200 | 0.463 | 0.665 | 0.999 |
| pvdQ, quiP; acyl-homoserine-lactone acylase [EC:3.5.1.97] | -0.018 | 0.343 | 0.958 | 0.999 |
| pycA; pyruvate carboxylase subunit A [EC:6.4.1.1] | 0.165 | 0.336 | 0.624 | 0.999 |
| pycB; pyruvate carboxylase subunit B [EC:6.4.1.1] | -0.098 | 0.176 | 0.576 | 0.996 |
| pydC; beta-ureidopropionase / N-carbamoyl-L-amino-acid hydrolase [EC:3.5.1.6 3.5.1.87] | -0.203 | 0.170 | 0.235 | 0.978 |
| PYG, glgP; glycogen phosphorylase [EC:2.4.1.1] | 0.075 | 0.078 | 0.338 | 0.978 |
| pyrB, PYR2; aspartate carbamoyltransferase catalytic subunit [EC:2.1.3.2] | 0.031 | 0.039 | 0.425 | 0.978 |
| pyrBI; aspartate carbamoyltransferase [EC:2.1.3.2] | -3.190 | 3.828 | 0.406 | 0.978 |
| pyrD; dihydroorotate dehydrogenase (fumarate) [EC:1.3.98.1] | 0.050 | 0.127 | 0.694 | 0.999 |
| pyrDI; dihydroorotate dehydrogenase (NAD+) catalytic subunit [EC:1.3.1.14] | 0.073 | 0.095 | 0.443 | 0.978 |
| pyrDII; dihydroorotate dehydrogenase electron transfer subunit | 0.043 | 0.099 | 0.663 | 0.999 |
| pyrE; orotate phosphoribosyltransferase [EC:2.4.2.10] | 0.031 | 0.037 | 0.407 | 0.978 |
| pyrF; orotidine-5’-phosphate decarboxylase [EC:4.1.1.23] | 0.047 | 0.038 | 0.219 | 0.978 |
| pyrG, CTPS; CTP synthase [EC:6.3.4.2] | -0.048 | 0.038 | 0.208 | 0.977 |
| pyrH; uridylate kinase [EC:2.7.4.22] | 0.033 | 0.036 | 0.359 | 0.978 |
| pyrI; aspartate carbamoyltransferase regulatory subunit | 0.010 | 0.175 | 0.953 | 0.999 |
| pyrP, uraA; uracil permease | 0.098 | 0.068 | 0.154 | 0.954 |
| pyrR; pyrimidine operon attenuation protein / uracil phosphoribosyltransferase [EC:2.4.2.9] | 0.087 | 0.067 | 0.193 | 0.970 |
| QARS, glnS; glutaminyl-tRNA synthetase [EC:6.1.1.18] | 0.019 | 0.090 | 0.831 | 0.999 |
| qcrA; ubiquinol-cytochrome c reductase iron-sulfur subunit | -0.395 | 0.399 | 0.323 | 0.978 |
| qcrB; ubiquinol-cytochrome c reductase cytochrome b subunit | -0.410 | 0.397 | 0.303 | 0.978 |
| qcrC; ubiquinol-cytochrome c reductase cytochrome c subunit | -0.395 | 0.399 | 0.323 | 0.978 |
| qheDH, qbdA; quinohemoprotein ethanol dehydrogenase [EC:1.1.9.1] | -1.254 | 0.686 | 0.069 | 0.841 |
| qhpA; quinohemoprotein amine dehydrogenase [EC:1.4.9.1] | -3.064 | 3.507 | 0.384 | 0.978 |
| qmoA; quinone-modifying oxidoreductase, subunit QmoA | -2.579 | 2.960 | 0.385 | 0.978 |
| qmoB; quinone-modifying oxidoreductase, subunit QmoB | -2.579 | 2.960 | 0.385 | 0.978 |
| qmoC; quinone-modifying oxidoreductase, subunit QmoC | -2.579 | 2.960 | 0.385 | 0.978 |
| qnr, mcbG; fluoroquinolone resistance protein | 0.031 | 0.450 | 0.946 | 0.999 |
| qodI; quercetin 2,3-dioxygenase [EC:1.13.11.24] | -17.703 | 3891.582 | 0.996 | 0.999 |
| qor, CRYZ; NADPH2:quinone reductase [EC:1.6.5.5] | -0.296 | 0.178 | 0.099 | 0.896 |
| qorB; NAD(P)H dehydrogenase (quinone) [EC:1.6.5.2] | 0.238 | 0.259 | 0.361 | 0.978 |
| qoxA; cytochrome aa3-600 menaquinol oxidase subunit II [EC:1.10.3.12] | -0.816 | 0.612 | 0.184 | 0.965 |
| qoxB; cytochrome aa3-600 menaquinol oxidase subunit I [EC:1.10.3.12] | -0.884 | 0.562 | 0.118 | 0.917 |
| qoxC; cytochrome aa3-600 menaquinol oxidase subunit III [EC:1.10.3.12] | -0.816 | 0.612 | 0.184 | 0.965 |
| qoxD; cytochrome aa3-600 menaquinol oxidase subunit IV [EC:1.10.3.12] | -0.816 | 0.612 | 0.184 | 0.965 |
| QPCT; glutaminyl-peptide cyclotransferase [EC:2.3.2.5] | -3.293 | 3.296 | 0.319 | 0.978 |
| qrtT; energy-coupling factor transport system substrate-specific component | 0.151 | 0.248 | 0.543 | 0.990 |
| qseB; two-component system, OmpR family, response regulator QseB | -0.053 | 0.222 | 0.810 | 0.999 |
| qseC; two-component system, OmpR family, sensor histidine kinase QseC [EC:2.7.13.3] | -0.047 | 0.213 | 0.827 | 0.999 |
| queA; S-adenosylmethionine:tRNA ribosyltransferase-isomerase [EC:2.4.99.17] | -0.012 | 0.057 | 0.834 | 0.999 |
| queC; 7-cyano-7-deazaguanine synthase [EC:6.3.4.20] | -0.108 | 0.064 | 0.094 | 0.895 |
| queD, ptpS, PTS; 6-pyruvoyltetrahydropterin/6-carboxytetrahydropterin synthase [EC:4.2.3.12 4.1.2.50] | -0.117 | 0.066 | 0.080 | 0.864 |
| queE; 7-carboxy-7-deazaguanine synthase [EC:4.3.99.3] | -0.112 | 0.092 | 0.227 | 0.978 |
| queF; 7-cyano-7-deazaguanine reductase [EC:1.7.1.13] | -0.122 | 0.064 | 0.058 | 0.839 |
| queG; epoxyqueuosine reductase [EC:1.17.99.6] | -0.145 | 0.077 | 0.062 | 0.839 |
| queH; epoxyqueuosine reductase [EC:1.17.99.6] | 0.121 | 0.100 | 0.226 | 0.978 |
| quiA; quinate dehydrogenase (quinone) [EC:1.1.5.8] | -0.079 | 0.313 | 0.802 | 0.999 |
| quiC; 3-dehydroshikimate dehydratase [EC:4.2.1.118] | -0.169 | 0.414 | 0.683 | 0.999 |
| RABGGTB; geranylgeranyl transferase type-2 subunit beta [EC:2.5.1.60] | 2.673 | 2.283 | 0.243 | 0.978 |
| racA; chromosome-anchoring protein RacA | -2.958 | 1.587 | 0.064 | 0.839 |
| racD; aspartate racemase [EC:5.1.1.13] | 0.157 | 0.171 | 0.362 | 0.978 |
| RAD51; DNA repair protein RAD51 | -17.234 | 3077.514 | 0.996 | 0.999 |
| RAD52; DNA repair and recombination protein RAD52 | -0.207 | 1.047 | 0.844 | 0.999 |
| radA, sms; DNA repair protein RadA/Sms | 0.033 | 0.037 | 0.377 | 0.978 |
| radA; DNA repair protein RadA | -17.332 | 3233.057 | 0.996 | 0.999 |
| radC; DNA repair protein RadC | 0.039 | 0.033 | 0.243 | 0.978 |
| radD; DNA repair protein RadD | 0.305 | 0.500 | 0.542 | 0.990 |
| raiA; ribosome-associated inhibitor A | 0.231 | 0.308 | 0.455 | 0.978 |
| ramA; (R)-amidase [EC:3.5.1.100] | 0.636 | 0.981 | 0.518 | 0.986 |
| ramA; alpha-L-rhamnosidase [EC:3.2.1.40] | -0.703 | 0.415 | 0.093 | 0.892 |
| ramA; AraC family of transcriptional regulator, multidrug resistance transcriptional activator | 0.725 | 0.700 | 0.302 | 0.978 |
| ramB; XRE family transcriptional regulator, fatty acid utilization regulator | -0.736 | 0.293 | 0.013 | 0.839 |
| rap; autoaggregation protein RapA/B/C | -1.272 | 1.833 | 0.489 | 0.979 |
| rapA, spo0L; response regulator aspartate phosphatase A (stage 0 sporulation protein L) [EC:3.1.-.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| rapC; response regulator aspartate phosphatase C [EC:3.1.-.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| rapD; response regulator aspartate phosphatase D [EC:3.1.-.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| rapE; response regulator aspartate phosphatase E [EC:3.1.-.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| rapG; response regulator aspartate phosphatase G [EC:3.1.-.-] | -0.904 | 1.547 | 0.560 | 0.992 |
| rapH; response regulator aspartate phosphatase H [EC:3.1.-.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| rapI; response regulator aspartate phosphatase I [EC:3.1.-.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| rapK; response regulator aspartate phosphatase K [EC:3.1.-.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| rapZ; RNase adapter protein RapZ | 0.089 | 0.040 | 0.026 | 0.839 |
| rarD; chloramphenicol-sensitive protein RarD | -0.022 | 0.092 | 0.811 | 0.999 |
| RARS, argS; arginyl-tRNA synthetase [EC:6.1.1.19] | 0.024 | 0.036 | 0.510 | 0.981 |
| raxA; membrane fusion protein | -0.412 | 0.576 | 0.476 | 0.979 |
| raxB, cvaB; ATP-binding cassette, subfamily B, bacterial RaxB | -0.219 | 0.581 | 0.707 | 0.999 |
| raxST; sulfotransferase | -0.241 | 0.907 | 0.790 | 0.999 |
| rbbA; ribosome-dependent ATPase | -0.212 | 0.302 | 0.483 | 0.979 |
| rbcL; ribulose-bisphosphate carboxylase large chain [EC:4.1.1.39] | 0.010 | 0.240 | 0.967 | 0.999 |
| rbcS; ribulose-bisphosphate carboxylase small chain [EC:4.1.1.39] | -1.054 | 0.724 | 0.148 | 0.954 |
| rbfA; ribosome-binding factor A | 0.036 | 0.036 | 0.311 | 0.978 |
| rbgA; ribosome biogenesis GTPase A | 0.133 | 0.087 | 0.126 | 0.927 |
| rbsA; ribose transport system ATP-binding protein [EC:3.6.3.17] | -0.037 | 0.203 | 0.854 | 0.999 |
| rbsB; ribose transport system substrate-binding protein | -0.056 | 0.144 | 0.699 | 0.999 |
| rbsC; ribose transport system permease protein | -0.065 | 0.218 | 0.764 | 0.999 |
| rbsD; D-ribose pyranase [EC:5.4.99.62] | 0.180 | 0.164 | 0.275 | 0.978 |
| rbsK, RBKS; ribokinase [EC:2.7.1.15] | 0.033 | 0.112 | 0.768 | 0.999 |
| rbsU; putative ribose uptake protein | -0.090 | 0.511 | 0.861 | 0.999 |
| rbtD; ribitol 2-dehydrogenase [EC:1.1.1.56] | -0.782 | 0.569 | 0.171 | 0.961 |
| rcdA; regulator of CtrA degradation | -1.075 | 0.426 | 0.013 | 0.839 |
| rcnA; nickel/cobalt exporter | 0.049 | 0.456 | 0.915 | 0.999 |
| rcsA; LuxR family transcriptional regulator, capsular biosynthesis positive transcription factor | 0.062 | 0.642 | 0.924 | 0.999 |
| rcsB; two-component system, NarL family, captular synthesis response regulator RcsB | -0.155 | 0.356 | 0.663 | 0.999 |
| rcsC; two-component system, NarL family, capsular synthesis sensor histidine kinase RcsC [EC:2.7.13.3] | -0.142 | 0.424 | 0.739 | 0.999 |
| rcsD; two-component system, NarL family, sensor histidine kinase RcsD [EC:2.7.13.3] | 0.119 | 0.601 | 0.843 | 0.999 |
| rcsF; RcsF protein | 0.318 | 0.541 | 0.558 | 0.992 |
| rdgB; XTP/dITP diphosphohydrolase [EC:3.6.1.66] | -0.018 | 0.055 | 0.749 | 0.999 |
| rdgC; recombination associated protein RdgC | 0.119 | 0.165 | 0.472 | 0.979 |
| rdmC; aclacinomycin methylesterase [EC:3.1.1.95] | 18.832 | 3989.590 | 0.996 | 0.999 |
| recA; recombination protein RecA | 0.026 | 0.032 | 0.427 | 0.978 |
| recB; exodeoxyribonuclease V beta subunit [EC:3.1.11.5] | 0.061 | 0.155 | 0.692 | 0.999 |
| recC; exodeoxyribonuclease V gamma subunit [EC:3.1.11.5] | 0.066 | 0.161 | 0.683 | 0.999 |
| recD; exodeoxyribonuclease V alpha subunit [EC:3.1.11.5] | 0.050 | 0.055 | 0.368 | 0.978 |
| recE; exodeoxyribonuclease VIII [EC:3.1.11.-] | -0.833 | 1.086 | 0.444 | 0.978 |
| recF; DNA replication and repair protein RecF | 0.047 | 0.041 | 0.259 | 0.978 |
| recG; ATP-dependent DNA helicase RecG [EC:3.6.4.12] | 0.053 | 0.064 | 0.411 | 0.978 |
| recJ; single-stranded-DNA-specific exonuclease [EC:3.1.-.-] | 0.081 | 0.049 | 0.101 | 0.896 |
| recN; DNA repair protein RecN (Recombination protein N) | 0.036 | 0.036 | 0.314 | 0.978 |
| recO; DNA repair protein RecO (recombination protein O) | 0.074 | 0.044 | 0.094 | 0.895 |
| recQ; ATP-dependent DNA helicase RecQ [EC:3.6.4.12] | 0.028 | 0.069 | 0.685 | 0.999 |
| recR; recombination protein RecR | 0.036 | 0.036 | 0.315 | 0.978 |
| recT; recombination protein RecT | -0.774 | 0.518 | 0.137 | 0.949 |
| recU; recombination protein U | 0.016 | 0.123 | 0.894 | 0.999 |
| recX; regulatory protein | 0.052 | 0.040 | 0.191 | 0.970 |
| regA, regR, actR; two-component system, response regulator RegA | -0.149 | 0.220 | 0.500 | 0.981 |
| regB, regS, actS; two-component system, sensor histidine kinase RegB [EC:2.7.13.3] | -0.148 | 0.229 | 0.517 | 0.986 |
| regX3; two-component system, OmpR family, response regulator RegX3 | -0.031 | 0.269 | 0.907 | 0.999 |
| relA; GTP pyrophosphokinase [EC:2.7.6.5] | 0.054 | 0.048 | 0.265 | 0.978 |
| relB; RHH-type transcriptional regulator, rel operon repressor / antitoxin RelB | -0.365 | 0.436 | 0.403 | 0.978 |
| relE, stbE; mRNA interferase RelE/StbE | 0.371 | 0.220 | 0.095 | 0.896 |
| RENBP; N-acylglucosamine 2-epimerase [EC:5.1.3.8] | -0.313 | 0.229 | 0.174 | 0.961 |
| rep; ATP-dependent DNA helicase Rep [EC:3.6.4.12] | 0.082 | 0.156 | 0.601 | 0.999 |
| repA; regulatory protein RepA | -2.314 | 1.855 | 0.214 | 0.978 |
| repC; replication initiation protein RepC | -1.734 | 0.905 | 0.057 | 0.839 |
| res; type III restriction enzyme [EC:3.1.21.5] | 0.083 | 0.202 | 0.683 | 0.999 |
| resB, ccs1; cytochrome c biogenesis protein | -0.529 | 0.264 | 0.047 | 0.839 |
| resD; two-component system, OmpR family, response regulator ResD | -0.754 | 0.262 | 0.004 | 0.839 |
| resE; two-component system, OmpR family, sensor histidine kinase ResE [EC:2.7.13.3] | -0.888 | 0.482 | 0.067 | 0.839 |
| RETSAT; all-trans-retinol 13,14-reductase [EC:1.3.99.23] | -0.591 | 0.433 | 0.174 | 0.961 |
| rex; redox-sensing transcriptional repressor | 0.017 | 0.101 | 0.870 | 0.999 |
| rexA; oligosaccharide reducing-end xylanase [EC:3.2.1.156] | -4.847 | 4.929 | 0.327 | 0.978 |
| RFA1, RPA1, rpa; replication factor A1 | -16.927 | 2639.780 | 0.995 | 0.999 |
| rfaH; transcriptional antiterminator RfaH | 0.290 | 0.367 | 0.430 | 0.978 |
| rfbC, rmlC; dTDP-4-dehydrorhamnose 3,5-epimerase [EC:5.1.3.13] | 0.147 | 0.073 | 0.047 | 0.839 |
| rfbD, rmlD; dTDP-4-dehydrorhamnose reductase [EC:1.1.1.133] | 0.082 | 0.061 | 0.179 | 0.961 |
| rfbE; CDP-paratose 2-epimerase [EC:5.1.3.10] | -1.774 | 0.925 | 0.057 | 0.839 |
| rfbF, rhlC; rhamnosyltransferase [EC:2.4.1.-] | 0.222 | 0.529 | 0.676 | 0.999 |
| rfbF; glucose-1-phosphate cytidylyltransferase [EC:2.7.7.33] | -0.032 | 0.270 | 0.905 | 0.999 |
| rfbG; CDP-glucose 4,6-dehydratase [EC:4.2.1.45] | 0.160 | 0.303 | 0.600 | 0.999 |
| rfbG; rhamnosyltransferase [EC:2.4.1.-] | 0.311 | 0.350 | 0.374 | 0.978 |
| rfbN; rhamnosyltransferase [EC:2.4.1.-] | 0.393 | 0.368 | 0.288 | 0.978 |
| rfbP; undecaprenyl-phosphate galactose phosphotransferase [EC:2.7.8.6] | 0.500 | 0.498 | 0.317 | 0.978 |
| rfbV; abequosyltransferase [EC:2.4.1.60] | 0.884 | 1.140 | 0.439 | 0.978 |
| rfcL; replication factor C large subunit | -17.234 | 3077.514 | 0.996 | 0.999 |
| rfcS; replication factor C small subunit | -17.234 | 3077.514 | 0.996 | 0.999 |
| rffC, wecD; dTDP-4-amino-4,6-dideoxy-D-galactose acyltransferase [EC:2.3.1.210] | 0.119 | 0.601 | 0.843 | 0.999 |
| rfk; riboflavin kinase, archaea type [EC:2.7.1.161] | -17.234 | 3077.514 | 0.996 | 0.999 |
| RGL4, rhiE; rhamnogalacturonan endolyase [EC:4.2.2.23] | -0.999 | 0.988 | 0.313 | 0.978 |
| rgpA; rhamnosyltransferase [EC:2.4.1.-] | 0.110 | 0.209 | 0.600 | 0.999 |
| rgpA_B; gingipain R [EC:3.4.22.37] | -0.193 | 0.992 | 0.846 | 0.999 |
| rgpB; rhamnosyltransferase [EC:2.4.1.-] | 0.166 | 0.178 | 0.353 | 0.978 |
| rgpE; glucosyltransferase [EC:2.4.1.-] | 0.038 | 0.534 | 0.944 | 0.999 |
| rgpF; rhamnosyltransferase [EC:2.4.1.-] | 0.108 | 0.184 | 0.560 | 0.992 |
| rgpI; glucosyltransferase [EC:2.4.1.-] | 0.107 | 0.576 | 0.853 | 0.999 |
| rhaA; L-rhamnose isomerase / sugar isomerase [EC:5.3.1.14 5.3.1.-] | -0.280 | 0.500 | 0.577 | 0.996 |
| rhaA; L-rhamnose isomerase [EC:5.3.1.14] | -0.881 | 0.413 | 0.035 | 0.839 |
| rhaB; rhamnulokinase [EC:2.7.1.5] | 0.189 | 0.226 | 0.404 | 0.978 |
| rhaD; rhamnulose-1-phosphate aldolase [EC:4.1.2.19] | 0.078 | 0.352 | 0.825 | 0.999 |
| rhaM; L-rhamnose mutarotase [EC:5.1.3.32] | -0.629 | 0.302 | 0.039 | 0.839 |
| rhaP; rhamnose transport system permease protein | -1.118 | 0.720 | 0.123 | 0.919 |
| rhaQ; rhamnose transport system permease protein | -0.973 | 0.750 | 0.197 | 0.970 |
| rhaR; AraC family transcriptional regulator, L-rhamnose operon transcriptional activator RhaR | -0.097 | 0.787 | 0.902 | 0.999 |
| rhaS; AraC family transcriptional regulator, L-rhamnose operon regulatory protein RhaS | -0.091 | 0.746 | 0.903 | 0.999 |
| rhaS; rhamnose transport system substrate-binding protein | -1.271 | 0.722 | 0.080 | 0.864 |
| rhaT; L-rhamnose-H+ transport protein | -0.938 | 0.485 | 0.055 | 0.839 |
| rhaT; rhamnose transport system ATP-binding protein [EC:3.6.3.17] | -1.070 | 0.594 | 0.074 | 0.845 |
| rhiF; rhizoxin biosynthesis, polyketide synthase RhiF | -17.761 | 4005.858 | 0.996 | 0.999 |
| rhlA; rhamnosyltransferase subunit A [EC:2.4.1.-] | -0.964 | 0.982 | 0.328 | 0.978 |
| rhlB; ATP-dependent RNA helicase RhlB [EC:3.6.4.13] | 0.093 | 0.179 | 0.604 | 0.999 |
| rhlB; rhamnosyltransferase subunit B [EC:2.4.1.-] | 1.839 | 1.408 | 0.193 | 0.970 |
| rhlE; ATP-dependent RNA helicase RhlE [EC:3.6.4.13] | -0.186 | 0.183 | 0.311 | 0.978 |
| rhlI, phzI, solI, cepI, tofI; acyl homoserine lactone synthase [EC:2.3.1.184] | -0.143 | 0.719 | 0.842 | 0.999 |
| rhlR, phzR; LuxR family transcriptional regulator, quorum-sensing system regulator RhlR | -0.145 | 0.723 | 0.842 | 0.999 |
| rhmA; 2-dehydro-3-deoxy-L-rhamnonate aldolase [EC:4.1.2.53] | -0.885 | 1.079 | 0.414 | 0.978 |
| rho; transcription termination factor Rho | 0.020 | 0.060 | 0.739 | 0.999 |
| rhtA; inner membrane transporter RhtA | 0.018 | 0.190 | 0.926 | 0.999 |
| rhtB; homoserine/homoserine lactone efflux protein | -0.070 | 0.251 | 0.781 | 0.999 |
| rhtC; threonine efflux protein | -0.075 | 0.355 | 0.833 | 0.999 |
| RIB7, arfC; 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5’-phosphate reductase [EC:1.1.1.302] | -17.234 | 3077.514 | 0.996 | 0.999 |
| ribA, RIB1; GTP cyclohydrolase II [EC:3.5.4.25] | 0.054 | 0.136 | 0.694 | 0.999 |
| ribB, RIB3; 3,4-dihydroxy 2-butanone 4-phosphate synthase [EC:4.1.99.12] | 0.216 | 0.194 | 0.268 | 0.978 |
| ribBA; 3,4-dihydroxy 2-butanone 4-phosphate synthase / GTP cyclohydrolase II [EC:4.1.99.12 3.5.4.25] | 0.002 | 0.069 | 0.978 | 0.999 |
| ribD; diaminohydroxyphosphoribosylaminopyrimidine deaminase / 5-amino-6-(5-phosphoribosylamino)uracil reductase [EC:3.5.4.26 1.1.1.193] | -0.006 | 0.060 | 0.923 | 0.999 |
| ribD2; 5-amino-6-(5-phosphoribosylamino)uracil reductase [EC:1.1.1.193] | -0.900 | 0.733 | 0.222 | 0.978 |
| ribE, RIB5; riboflavin synthase [EC:2.5.1.9] | 0.008 | 0.065 | 0.898 | 0.999 |
| ribF; riboflavin kinase / FMN adenylyltransferase [EC:2.7.1.26 2.7.7.2] | 0.036 | 0.036 | 0.312 | 0.978 |
| ribH, RIB4; 6,7-dimethyl-8-ribityllumazine synthase [EC:2.5.1.78] | -0.034 | 0.059 | 0.564 | 0.994 |
| ribL; FAD synthetase [EC:2.7.7.2] | -17.234 | 3077.514 | 0.996 | 0.999 |
| ribT; riboflavin biosynthesis RibT protein | 0.248 | 0.334 | 0.459 | 0.979 |
| ridA, tdcF, RIDA; 2-iminobutanoate/2-iminopropanoate deaminase [EC:3.5.99.10] | 0.003 | 0.089 | 0.969 | 0.999 |
| rifG, asm47; 5-deoxy-5-amino-3-dehydroquinate synthase | -1.508 | 1.628 | 0.356 | 0.978 |
| rihA; pyrimidine-specific ribonucleoside hydrolase [EC:3.2.-.-] | 0.399 | 0.505 | 0.431 | 0.978 |
| rihB; ribosylpyrimidine nucleosidase [EC:3.2.2.8] | 0.046 | 0.437 | 0.917 | 0.999 |
| rihC; non-specific riboncleoside hydrolase [EC:3.2.-.-] | 0.213 | 0.444 | 0.632 | 0.999 |
| rimI; [ribosomal protein S18]-alanine N-acetyltransferase [EC:2.3.1.266] | 0.078 | 0.050 | 0.119 | 0.917 |
| rimJ; [ribosomal protein S5]-alanine N-acetyltransferase [EC:2.3.1.267] | -0.008 | 0.137 | 0.954 | 0.999 |
| rimK; ribosomal protein S6–L-glutamate ligase [EC:6.3.2.-] | 0.147 | 0.184 | 0.426 | 0.978 |
| rimL; ribosomal-protein-serine acetyltransferase [EC:2.3.1.-] | -0.265 | 0.222 | 0.236 | 0.978 |
| rimM; 16S rRNA processing protein RimM | 0.037 | 0.036 | 0.304 | 0.978 |
| rimO; ribosomal protein S12 methylthiotransferase [EC:2.8.4.4] | -0.022 | 0.090 | 0.803 | 0.999 |
| rimP; ribosome maturation factor RimP | 0.037 | 0.036 | 0.311 | 0.978 |
| RIMS2, RIM2; regulating synaptic membrane exocytosis protein 2 | -17.368 | 3290.703 | 0.996 | 0.999 |
| RIOK1; RIO kinase 1 [EC:2.7.11.1] | 0.251 | 0.308 | 0.417 | 0.978 |
| rirA; Rrf2 family transcriptional regulator, iron-responsive regulator | -2.411 | 1.438 | 0.096 | 0.896 |
| rlmA1; 23S rRNA (guanine745-N1)-methyltransferase [EC:2.1.1.187] | 0.035 | 0.098 | 0.718 | 0.999 |
| rlmB; 23S rRNA (guanosine2251-2’-O)-methyltransferase [EC:2.1.1.185] | 0.052 | 0.032 | 0.108 | 0.917 |
| rlmE, rrmJ, ftsJ; 23S rRNA (uridine2552-2’-O)-methyltransferase [EC:2.1.1.166] | -0.053 | 0.144 | 0.715 | 0.999 |
| rlmF; 23S rRNA (adenine1618-N6)-methyltransferase [EC:2.1.1.181] | 0.271 | 0.291 | 0.353 | 0.978 |
| rlmG; 23S rRNA (guanine1835-N2)-methyltransferase [EC:2.1.1.174] | -0.014 | 0.652 | 0.983 | 0.999 |
| rlmH; 23S rRNA (pseudouridine1915-N3)-methyltransferase [EC:2.1.1.177] | 0.034 | 0.038 | 0.373 | 0.978 |
| rlmI; 23S rRNA (cytosine1962-C5)-methyltransferase [EC:2.1.1.191] | -0.004 | 0.055 | 0.940 | 0.999 |
| rlmJ; 23S rRNA (adenine2030-N6)-methyltransferase [EC:2.1.1.266] | -0.028 | 0.150 | 0.853 | 0.999 |
| rlmL, rlmK; 23S rRNA (guanine2445-N2)-methyltransferase / 23S rRNA (guanine2069-N7)-methyltransferase [EC:2.1.1.173 2.1.1.264] | 0.082 | 0.174 | 0.639 | 0.999 |
| rlmM; 23S rRNA (cytidine2498-2’-O)-methyltransferase [EC:2.1.1.186] | 0.145 | 0.184 | 0.431 | 0.978 |
| rlmN; 23S rRNA (adenine2503-C2)-methyltransferase [EC:2.1.1.192] | 0.016 | 0.042 | 0.705 | 0.999 |
| rlpA; rare lipoprotein A | -0.087 | 0.105 | 0.413 | 0.978 |
| rluA; tRNA pseudouridine32 synthase / 23S rRNA pseudouridine746 synthase [EC:5.4.99.28 5.4.99.29] | -0.061 | 0.093 | 0.508 | 0.981 |
| rluB; 23S rRNA pseudouridine2605 synthase [EC:5.4.99.22] | 0.020 | 0.035 | 0.576 | 0.996 |
| rluC; 23S rRNA pseudouridine955/2504/2580 synthase [EC:5.4.99.24] | 0.106 | 0.109 | 0.330 | 0.978 |
| rluD; 23S rRNA pseudouridine1911/1915/1917 synthase [EC:5.4.99.23] | 0.039 | 0.066 | 0.557 | 0.992 |
| rluE; 23S rRNA pseudouridine2457 synthase [EC:5.4.99.20] | 0.060 | 0.155 | 0.698 | 0.999 |
| rluF; 23S rRNA pseudouridine2604 synthase [EC:5.4.99.21] | 0.055 | 0.178 | 0.758 | 0.999 |
| rmd; GDP-4-dehydro-6-deoxy-D-mannose reductase [EC:1.1.1.281] | -0.611 | 0.563 | 0.279 | 0.978 |
| rmf; ribosome modulation factor | 0.401 | 0.311 | 0.199 | 0.970 |
| rmuC; DNA recombination protein RmuC | -0.052 | 0.066 | 0.430 | 0.978 |
| rna; ribonuclease I (enterobacter ribonuclease) [EC:3.1.27.6] | -0.377 | 0.687 | 0.584 | 0.999 |
| rnb; exoribonuclease II [EC:3.1.13.1] | -0.004 | 0.237 | 0.986 | 0.999 |
| rnc, DROSHA, RNT1; ribonuclease III [EC:3.1.26.3] | 0.037 | 0.033 | 0.256 | 0.978 |
| rnd; ribonuclease D [EC:3.1.13.5] | -0.097 | 0.124 | 0.435 | 0.978 |
| rne; ribonuclease E [EC:3.1.26.12] | -0.023 | 0.118 | 0.846 | 0.999 |
| rnfA; electron transport complex protein RnfA | 0.318 | 0.169 | 0.062 | 0.839 |
| rnfB; electron transport complex protein RnfB | 0.063 | 0.154 | 0.684 | 0.999 |
| rnfC; electron transport complex protein RnfC | 0.356 | 0.185 | 0.056 | 0.839 |
| rnfD; electron transport complex protein RnfD | 0.399 | 0.169 | 0.020 | 0.839 |
| rnfE; electron transport complex protein RnfE | 0.318 | 0.169 | 0.062 | 0.839 |
| rnfG; electron transport complex protein RnfG | 0.235 | 0.152 | 0.124 | 0.919 |
| rng, cafA; ribonuclease G [EC:3.1.26.-] | 0.080 | 0.088 | 0.362 | 0.978 |
| rnhA-dnaQ; ribonuclease HI / DNA polymerase III subunit epsilon [EC:3.1.26.4 2.7.7.7] | -0.264 | 0.437 | 0.547 | 0.991 |
| rnhA, RNASEH1; ribonuclease HI [EC:3.1.26.4] | 0.000 | 0.062 | 0.997 | 0.999 |
| rnhB; ribonuclease HII [EC:3.1.26.4] | 0.018 | 0.042 | 0.672 | 0.999 |
| rnhC; ribonuclease HIII [EC:3.1.26.4] | -0.060 | 0.142 | 0.676 | 0.999 |
| rnj; ribonuclease J [EC:3.1.-.-] | 0.044 | 0.089 | 0.625 | 0.999 |
| rnk; regulator of nucleoside diphosphate kinase | -0.054 | 0.209 | 0.796 | 0.999 |
| rnmV; ribonuclease M5 [EC:3.1.26.8] | 0.126 | 0.098 | 0.199 | 0.970 |
| rnpA; ribonuclease P protein component [EC:3.1.26.5] | 0.053 | 0.040 | 0.193 | 0.970 |
| rnr, vacB; ribonuclease R [EC:3.1.-.-] | 0.043 | 0.041 | 0.294 | 0.978 |
| rnt; ribonuclease T [EC:3.1.13.-] | 0.093 | 0.171 | 0.586 | 0.999 |
| rny; ribonucrease Y [EC:3.1.-.-] | 0.064 | 0.087 | 0.465 | 0.979 |
| rnz; ribonuclease Z [EC:3.1.26.11] | -0.014 | 0.081 | 0.859 | 0.999 |
| rob; AraC family transcriptional regulator, mar-sox-rob regulon activator | 0.151 | 0.565 | 0.790 | 0.999 |
| rocD, OAT; ornithine–oxo-acid transaminase [EC:2.6.1.13] | -0.776 | 0.306 | 0.012 | 0.839 |
| rocE, rocC; arginine/ornithine permease | -0.535 | 0.708 | 0.451 | 0.978 |
| rocR; arginine utilization regulatory protein | -0.922 | 0.852 | 0.281 | 0.978 |
| rodA, mrdB; rod shape determining protein RodA | 0.066 | 0.041 | 0.113 | 0.917 |
| rodZ; cytoskeleton protein RodZ | 0.101 | 0.173 | 0.560 | 0.992 |
| rof; Rho-binding antiterminator | 0.106 | 0.546 | 0.846 | 0.999 |
| RON2; rhoptry neck protein 2 | 1.239 | 1.292 | 0.339 | 0.978 |
| RP-L1, MRPL1, rplA; large subunit ribosomal protein L1 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-L10, MRPL10, rplJ; large subunit ribosomal protein L10 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-L10e, RPL10; large subunit ribosomal protein L10e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L11, MRPL11, rplK; large subunit ribosomal protein L11 | 0.037 | 0.036 | 0.306 | 0.978 |
| RP-L12, rpl12; large subunit ribosomal protein L12 | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L13, MRPL13, rplM; large subunit ribosomal protein L13 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-L14, MRPL14, rplN; large subunit ribosomal protein L14 | 0.037 | 0.036 | 0.306 | 0.978 |
| RP-L15, MRPL15, rplO; large subunit ribosomal protein L15 | 0.037 | 0.036 | 0.306 | 0.978 |
| RP-L15e, RPL15; large subunit ribosomal protein L15e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L16, MRPL16, rplP; large subunit ribosomal protein L16 | 0.037 | 0.036 | 0.295 | 0.978 |
| RP-L17, MRPL17, rplQ; large subunit ribosomal protein L17 | 0.038 | 0.036 | 0.288 | 0.978 |
| RP-L18, MRPL18, rplR; large subunit ribosomal protein L18 | 0.037 | 0.036 | 0.307 | 0.978 |
| RP-L18e, RPL18; large subunit ribosomal protein L18e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L19, MRPL19, rplS; large subunit ribosomal protein L19 | 0.036 | 0.036 | 0.312 | 0.978 |
| RP-L19e, RPL19; large subunit ribosomal protein L19e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L2, MRPL2, rplB; large subunit ribosomal protein L2 | 0.035 | 0.036 | 0.326 | 0.978 |
| RP-L20, MRPL20, rplT; large subunit ribosomal protein L20 | 0.036 | 0.036 | 0.321 | 0.978 |
| RP-L21, MRPL21, rplU; large subunit ribosomal protein L21 | 0.036 | 0.036 | 0.321 | 0.978 |
| RP-L21e, RPL21; large subunit ribosomal protein L21e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L22, MRPL22, rplV; large subunit ribosomal protein L22 | 0.036 | 0.036 | 0.311 | 0.978 |
| RP-L23, MRPL23, rplW; large subunit ribosomal protein L23 | 0.035 | 0.036 | 0.337 | 0.978 |
| RP-L24, MRPL24, rplX; large subunit ribosomal protein L24 | 0.031 | 0.036 | 0.379 | 0.978 |
| RP-L24e, RPL24; large subunit ribosomal protein L24e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L25, rplY; large subunit ribosomal protein L25 | -0.084 | 0.076 | 0.271 | 0.978 |
| RP-L27, MRPL27, rpmA; large subunit ribosomal protein L27 | 0.036 | 0.036 | 0.321 | 0.978 |
| RP-L28, MRPL28, rpmB; large subunit ribosomal protein L28 | 0.026 | 0.035 | 0.466 | 0.979 |
| RP-L29, rpmC; large subunit ribosomal protein L29 | 0.035 | 0.036 | 0.325 | 0.978 |
| RP-L3, MRPL3, rplC; large subunit ribosomal protein L3 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-L30, MRPL30, rpmD; large subunit ribosomal protein L30 | 0.045 | 0.037 | 0.220 | 0.978 |
| RP-L30e, RPL30; large subunit ribosomal protein L30e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L31, rpmE; large subunit ribosomal protein L31 | 0.032 | 0.032 | 0.320 | 0.978 |
| RP-L31e, RPL31; large subunit ribosomal protein L31e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L32, MRPL32, rpmF; large subunit ribosomal protein L32 | 0.040 | 0.036 | 0.269 | 0.978 |
| RP-L32e, RPL32; large subunit ribosomal protein L32e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L33, MRPL33, rpmG; large subunit ribosomal protein L33 | 0.041 | 0.050 | 0.407 | 0.978 |
| RP-L34, MRPL34, rpmH; large subunit ribosomal protein L34 | 0.075 | 0.044 | 0.090 | 0.884 |
| RP-L35, MRPL35, rpmI; large subunit ribosomal protein L35 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-L36, MRPL36, rpmJ; large subunit ribosomal protein L36 | 0.094 | 0.057 | 0.101 | 0.896 |
| RP-L37Ae, RPL37A; large subunit ribosomal protein L37Ae | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L39e, RPL39; large subunit ribosomal protein L39e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L4, MRPL4, rplD; large subunit ribosomal protein L4 | 0.036 | 0.036 | 0.321 | 0.978 |
| RP-L44e, RPL44; large subunit ribosomal protein L44e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L4e, RPL4; large subunit ribosomal protein L4e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L5, MRPL5, rplE; large subunit ribosomal protein L5 | 0.035 | 0.036 | 0.325 | 0.978 |
| RP-L6, MRPL6, rplF; large subunit ribosomal protein L6 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-L7, MRPL12, rplL; large subunit ribosomal protein L7/L12 | 0.036 | 0.036 | 0.320 | 0.978 |
| RP-L7A, rplGB; large subunit ribosomal protein L7A | -0.073 | 0.199 | 0.715 | 0.999 |
| RP-L7Ae, RPL7A; large subunit ribosomal protein L7Ae | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-L9, MRPL9, rplI; large subunit ribosomal protein L9 | 0.036 | 0.036 | 0.311 | 0.978 |
| RP-S1, rpsA; small subunit ribosomal protein S1 | 0.000 | 0.044 | 0.993 | 0.999 |
| RP-S10, MRPS10, rpsJ; small subunit ribosomal protein S10 | 0.035 | 0.036 | 0.337 | 0.978 |
| RP-S11, MRPS11, rpsK; small subunit ribosomal protein S11 | 0.036 | 0.036 | 0.315 | 0.978 |
| RP-S12, MRPS12, rpsL; small subunit ribosomal protein S12 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-S13, rpsM; small subunit ribosomal protein S13 | 0.037 | 0.036 | 0.305 | 0.978 |
| RP-S14, MRPS14, rpsN; small subunit ribosomal protein S14 | 0.053 | 0.048 | 0.265 | 0.978 |
| RP-S15, MRPS15, rpsO; small subunit ribosomal protein S15 | 0.031 | 0.036 | 0.386 | 0.978 |
| RP-S16, MRPS16, rpsP; small subunit ribosomal protein S16 | 0.036 | 0.036 | 0.320 | 0.978 |
| RP-S17, MRPS17, rpsQ; small subunit ribosomal protein S17 | 0.037 | 0.036 | 0.306 | 0.978 |
| RP-S17e, RPS17; small subunit ribosomal protein S17e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S18, MRPS18, rpsR; small subunit ribosomal protein S18 | 0.030 | 0.035 | 0.399 | 0.978 |
| RP-S19, rpsS; small subunit ribosomal protein S19 | 0.036 | 0.036 | 0.313 | 0.978 |
| RP-S19e, RPS19; small subunit ribosomal protein S19e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S2, MRPS2, rpsB; small subunit ribosomal protein S2 | 0.035 | 0.036 | 0.326 | 0.978 |
| RP-S20, rpsT; small subunit ribosomal protein S20 | 0.036 | 0.036 | 0.324 | 0.978 |
| RP-S21, MRPS21, rpsU; small subunit ribosomal protein S21 | -0.012 | 0.045 | 0.787 | 0.999 |
| RP-S24e, RPS24; small subunit ribosomal protein S24e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S25e, RPS25; small subunit ribosomal protein S25e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S26e, RPS26; small subunit ribosomal protein S26e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S27Ae, RPS27A; small subunit ribosomal protein S27Ae | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S27e, RPS27; small subunit ribosomal protein S27e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S28e, RPS28; small subunit ribosomal protein S28e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S3, rpsC; small subunit ribosomal protein S3 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-S30e, RPS30; small subunit ribosomal protein S30e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S3Ae, RPS3A; small subunit ribosomal protein S3Ae | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S4, rpsD; small subunit ribosomal protein S4 | 0.040 | 0.036 | 0.271 | 0.978 |
| RP-S4e, RPS4; small subunit ribosomal protein S4e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S5, MRPS5, rpsE; small subunit ribosomal protein S5 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-S6, MRPS6, rpsF; small subunit ribosomal protein S6 | 0.036 | 0.036 | 0.320 | 0.978 |
| RP-S6e, RPS6; small subunit ribosomal protein S6e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S7, MRPS7, rpsG; small subunit ribosomal protein S7 | 0.034 | 0.036 | 0.340 | 0.978 |
| RP-S8, rpsH; small subunit ribosomal protein S8 | 0.036 | 0.036 | 0.322 | 0.978 |
| RP-S8e, RPS8; small subunit ribosomal protein S8e | -17.234 | 3077.514 | 0.996 | 0.999 |
| RP-S9, MRPS9, rpsI; small subunit ribosomal protein S9 | 0.035 | 0.036 | 0.325 | 0.978 |
| rpaA; two-component system, OmpR family, response regulator RpaA | -17.256 | 3111.566 | 0.996 | 0.999 |
| rpaB; two-component system, OmpR family, response regulator RpaB | -2.545 | 2.077 | 0.222 | 0.978 |
| RPB6, POLR2F; DNA-directed RNA polymerases I, II, and III subunit RPABC2 | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpe, RPE; ribulose-phosphate 3-epimerase [EC:5.1.3.1] | 0.040 | 0.036 | 0.274 | 0.978 |
| rpfC; two-component system, sensor histidine kinase RpfC [EC:2.7.13.3] | -0.067 | 0.469 | 0.886 | 0.999 |
| rpfF; DSF synthase | -0.048 | 0.440 | 0.913 | 0.999 |
| rpfG; two-component system, response regulator RpfG | -0.052 | 0.473 | 0.913 | 0.999 |
| rph; ribonuclease PH [EC:2.7.7.56] | -0.045 | 0.092 | 0.626 | 0.999 |
| rpiA; ribose 5-phosphate isomerase A [EC:5.3.1.6] | 0.019 | 0.089 | 0.828 | 0.999 |
| rpiB; ribose 5-phosphate isomerase B [EC:5.3.1.6] | 0.043 | 0.113 | 0.707 | 0.999 |
| rpoA; DNA-directed RNA polymerase subunit alpha [EC:2.7.7.6] | 0.035 | 0.036 | 0.336 | 0.978 |
| rpoA1; DNA-directed RNA polymerase subunit A’ [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoB; DNA-directed RNA polymerase subunit B [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoB; DNA-directed RNA polymerase subunit beta [EC:2.7.7.6] | 0.040 | 0.031 | 0.207 | 0.976 |
| rpoBC; DNA-directed RNA polymerase subunit beta-beta’ [EC:2.7.7.6] | -1.386 | 1.551 | 0.373 | 0.978 |
| rpoC; DNA-directed RNA polymerase subunit beta’ [EC:2.7.7.6] | 0.045 | 0.036 | 0.220 | 0.978 |
| rpoD; DNA-directed RNA polymerase subunit D [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoD; RNA polymerase primary sigma factor | 0.024 | 0.057 | 0.677 | 0.999 |
| rpoE; DNA-directed RNA polymerase subunit delta | 0.049 | 0.120 | 0.682 | 0.999 |
| rpoE; RNA polymerase sigma-70 factor, ECF subfamily | -0.188 | 0.085 | 0.028 | 0.839 |
| rpoE1; DNA-directed RNA polymerase subunit E’ [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoE2; DNA-directed RNA polymerase subunit E" [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoF; DNA-directed RNA polymerase subunit F [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoH; DNA-directed RNA polymerase subunit H [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoH; RNA polymerase sigma-32 factor | -0.108 | 0.147 | 0.461 | 0.979 |
| rpoL; DNA-directed RNA polymerase subunit L [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoN; DNA-directed RNA polymerase subunit N [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoN; RNA polymerase sigma-54 factor | -0.089 | 0.097 | 0.360 | 0.978 |
| rpoP; DNA-directed RNA polymerase subunit P [EC:2.7.7.6] | -17.234 | 3077.514 | 0.996 | 0.999 |
| rpoS; RNA polymerase nonessential primary-like sigma factor | 0.118 | 0.254 | 0.642 | 0.999 |
| rpoZ; DNA-directed RNA polymerase subunit omega [EC:2.7.7.6] | 0.077 | 0.038 | 0.044 | 0.839 |
| rppA; 1,3,6,8-tetrahydroxynaphthalene synthase [EC:2.3.1.233] | -17.487 | 3492.836 | 0.996 | 0.999 |
| RPR2, RPP21; ribonuclease P protein subunit RPR2 [EC:3.1.26.5] | 0.875 | 1.335 | 0.513 | 0.982 |
| rraA, menG; regulator of ribonuclease activity A | -0.049 | 0.172 | 0.776 | 0.999 |
| rraB; regulator of ribonuclease activity B | 0.231 | 0.302 | 0.446 | 0.978 |
| RRP4, EXOSC2; exosome complex component RRP4 | -17.234 | 3077.514 | 0.996 | 0.999 |
| RRP41, EXOSC4, SKI6; exosome complex component RRP41 | -17.234 | 3077.514 | 0.996 | 0.999 |
| RRP42, EXOSC7; exosome complex component RRP42 | -17.234 | 3077.514 | 0.996 | 0.999 |
| rsaA; S-layer protein | -17.510 | 3533.982 | 0.996 | 0.999 |
| rsaD; ATP-binding cassette, subfamily C, bacterial RsaD | -17.510 | 3533.982 | 0.996 | 0.999 |
| RSAD2; radical S-adenosyl methionine domain-containing protein 2 | 18.595 | 2918.736 | 0.995 | 0.999 |
| rsaE; membrane fusion protein, S-layer protein transport system | -17.510 | 3533.982 | 0.996 | 0.999 |
| rsaF; outer membrane protein, S-layer protein transport system | -17.771 | 3227.286 | 0.996 | 0.999 |
| rsbQ; sigma-B regulation protein RsbQ | -0.358 | 0.502 | 0.477 | 0.979 |
| rsbR; rsbT co-antagonist protein RsbR | -0.243 | 0.416 | 0.560 | 0.992 |
| rsbS; rsbT antagonist protein RsbS | -0.147 | 0.409 | 0.720 | 0.999 |
| rsbT; serine/threonine-protein kinase RsbT [EC:2.7.11.1] | -0.224 | 0.435 | 0.607 | 0.999 |
| rsbU_P; phosphoserine phosphatase RsbU/P [EC:3.1.3.3] | 0.059 | 0.267 | 0.827 | 0.999 |
| rsbV; anti-sigma B factor antagonist | 0.156 | 0.246 | 0.528 | 0.989 |
| rsbW; serine/threonine-protein kinase RsbW [EC:2.7.11.1] | 0.204 | 0.245 | 0.407 | 0.978 |
| rsbX; phosphoserine phosphatase RsbX [EC:3.1.3.3] | -1.333 | 0.949 | 0.162 | 0.957 |
| rsd; regulator of sigma D | 0.248 | 0.307 | 0.421 | 0.978 |
| rseA; sigma-E factor negative regulatory protein RseA | 0.152 | 0.193 | 0.433 | 0.978 |
| rseB; sigma-E factor negative regulatory protein RseB | 0.145 | 0.195 | 0.459 | 0.979 |
| rseC; sigma-E factor negative regulatory protein RseC | 0.335 | 0.188 | 0.076 | 0.855 |
| rseP; regulator of sigma E protease [EC:3.4.24.-] | 0.035 | 0.041 | 0.388 | 0.978 |
| rsfA; prespore-specific regulator | -1.679 | 0.818 | 0.042 | 0.839 |
| rsgA, engC; ribosome biogenesis GTPase / thiamine phosphate phosphatase [EC:3.6.1.- 3.1.3.100] | 0.059 | 0.042 | 0.161 | 0.957 |
| rsmB, sun; 16S rRNA (cytosine967-C5)-methyltransferase [EC:2.1.1.176] | 0.046 | 0.033 | 0.167 | 0.961 |
| rsmC; 16S rRNA (guanine1207-N2)-methyltransferase [EC:2.1.1.172] | 0.065 | 0.066 | 0.326 | 0.978 |
| rsmD; 16S rRNA (guanine966-N2)-methyltransferase [EC:2.1.1.171] | -0.030 | 0.072 | 0.672 | 0.999 |
| rsmE; 16S rRNA (uracil1498-N3)-methyltransferase [EC:2.1.1.193] | 0.033 | 0.038 | 0.391 | 0.978 |
| rsmF; 16S rRNA (cytosine1407-C5)-methyltransferase [EC:2.1.1.178] | 0.444 | 0.544 | 0.416 | 0.978 |
| rsmI; 16S rRNA (cytidine1402-2’-O)-methyltransferase [EC:2.1.1.198] | 0.019 | 0.036 | 0.597 | 0.999 |
| rsmJ; 16S rRNA (guanine1516-N2)-methyltransferase [EC:2.1.1.242] | 0.130 | 0.176 | 0.462 | 0.979 |
| rspA, manD; mannonate dehydratase [EC:4.2.1.8] | 0.019 | 0.346 | 0.957 | 0.999 |
| rspB; L-gulonate 5-dehydrogenase [EC:1.1.1.380] | 0.180 | 0.575 | 0.755 | 0.999 |
| rssB, hnr; two-component system, response regulator | -0.030 | 0.382 | 0.939 | 0.999 |
| rstA; two-component system, OmpR family, response regulator RstA | 0.031 | 0.224 | 0.889 | 0.999 |
| rstA1; phage replication initiation protein | -0.218 | 0.418 | 0.603 | 0.999 |
| rstB; two-component system, OmpR family, sensor histidine kinase RstB [EC:2.7.13.3] | 0.017 | 0.226 | 0.940 | 0.999 |
| rsuA; 16S rRNA pseudouridine516 synthase [EC:5.4.99.19] | 0.096 | 0.071 | 0.176 | 0.961 |
| RTCA, rtcA; RNA 3’-terminal phosphate cyclase (ATP) [EC:6.5.1.4] | -0.030 | 0.422 | 0.943 | 0.999 |
| RTCB, rtcB; tRNA-splicing ligase RtcB (3’-phosphate/5’-hydroxy nucleic acid ligase) [EC:6.5.1.8] | -0.188 | 0.194 | 0.335 | 0.978 |
| rtcB; release factor H-coupled RctB family protein | -0.132 | 0.433 | 0.761 | 0.999 |
| rtcR; transcriptional regulatory protein RtcR | -0.149 | 0.349 | 0.670 | 0.999 |
| rtpR; ribonucleoside-triphosphate reductase (thioredoxin) [EC:1.17.4.2] | 0.071 | 0.090 | 0.431 | 0.978 |
| rtxB, fitA; ATP-binding cassette, subfamily B, bacterial RtxB | 0.088 | 1.356 | 0.948 | 0.999 |
| rtxD, fitB; membrane fusion protein, RTX toxin transport system | 0.088 | 1.356 | 0.948 | 0.999 |
| rtxE, fitC; ATP-binding cassette, subfamily B, bacterial RtxE | 0.088 | 1.356 | 0.948 | 0.999 |
| rubB, alkT; rubredoxin—NAD+ reductase [EC:1.18.1.1] | 0.177 | 0.258 | 0.494 | 0.981 |
| rumA; 23S rRNA (uracil1939-C5)-methyltransferase [EC:2.1.1.190] | 0.014 | 0.048 | 0.774 | 0.999 |
| rumB; 23S rRNA (uracil747-C5)-methyltransferase [EC:2.1.1.189] | 0.174 | 0.219 | 0.428 | 0.978 |
| rusA; crossover junction endodeoxyribonuclease RusA [EC:3.1.22.4] | -0.280 | 0.799 | 0.727 | 0.999 |
| rutA; pyrimidine oxygenase [EC:1.14.99.46] | -0.005 | 0.489 | 0.992 | 0.999 |
| rutB; ureidoacrylate peracid hydrolase [EC:3.5.1.110] | -0.133 | 0.431 | 0.758 | 0.999 |
| rutC; aminoacrylate peracid reductase | 0.045 | 0.500 | 0.929 | 0.999 |
| rutD; aminoacrylate hydrolase [EC:3.5.1.-] | -0.169 | 0.491 | 0.732 | 0.999 |
| rutE; 3-hydroxypropanoate dehydrogenase [EC:1.1.1.-] | -0.119 | 0.247 | 0.632 | 0.999 |
| rutF; flavin reductase [EC:1.5.1.-] | -0.345 | 0.370 | 0.353 | 0.978 |
| rutG; putative pyrimidine permease RutG | -0.344 | 0.693 | 0.620 | 0.999 |
| rutR; TetR/AcrR family transcriptional regulator | 0.021 | 0.277 | 0.941 | 0.999 |
| ruvA; holliday junction DNA helicase RuvA [EC:3.6.4.12] | 0.035 | 0.036 | 0.329 | 0.978 |
| ruvB; holliday junction DNA helicase RuvB [EC:3.6.4.12] | 0.037 | 0.035 | 0.292 | 0.978 |
| ruvC; crossover junction endodeoxyribonuclease RuvC [EC:3.1.22.4] | 0.037 | 0.073 | 0.615 | 0.999 |
| ruvX; putative holliday junction resolvase [EC:3.1.-.-] | 0.038 | 0.036 | 0.290 | 0.978 |
| RUXX; small nuclear ribonucleoprotein | -16.927 | 2639.780 | 0.995 | 0.999 |
| RYR2; ryanodine receptor 2 | 0.298 | 0.949 | 0.754 | 0.999 |
| rzpD; prophage endopeptidase [EC:3.4.-.-] | -0.195 | 0.947 | 0.838 | 0.999 |
| sabA; outer membrane protein SabA | -1.386 | 1.551 | 0.373 | 0.978 |
| sacB; levansucrase [EC:2.4.1.10] | 0.209 | 0.424 | 0.623 | 0.999 |
| sacC, levB; levanase [EC:3.2.1.65] | -0.203 | 0.304 | 0.506 | 0.981 |
| sad; succinate-semialdehyde dehydrogenase [EC:1.2.1.16 1.2.1.24] | 0.005 | 0.259 | 0.986 | 0.999 |
| saeR; two-component system, OmpR family, response regulator SaeR | -0.091 | 0.181 | 0.614 | 0.999 |
| saeS; two-component system, OmpR family, sensor histidine kinase SaeS [EC:2.7.13.3] | -0.142 | 0.178 | 0.425 | 0.978 |
| safA; morphogenetic protein associated with SpoVID | -1.483 | 0.738 | 0.046 | 0.839 |
| sak; staphylokinase | -1.670 | 1.245 | 0.182 | 0.961 |
| salK; two-component system, NarL family, secretion system sensor histidine kinase SalK | -0.147 | 1.349 | 0.913 | 0.999 |
| salR; two-component system, NarL family, secretion system response regulator SalR | -0.056 | 1.120 | 0.960 | 0.999 |
| sam; S-adenosylmethionine uptake transporter | -0.795 | 0.425 | 0.063 | 0.839 |
| SAM50, TOB55, bamA; outer membrane protein insertion porin family | 0.040 | 0.087 | 0.647 | 0.999 |
| sanA; SanA protein | 0.079 | 0.209 | 0.706 | 0.999 |
| sapA; cationic peptide transport system substrate-binding protein | 0.232 | 0.307 | 0.451 | 0.978 |
| sapB; cationic peptide transport system permease protein | 0.232 | 0.307 | 0.451 | 0.978 |
| sapC; cationic peptide transport system permease protein | 0.232 | 0.307 | 0.451 | 0.978 |
| sapD; cationic peptide transport system ATP-binding protein | 0.231 | 0.308 | 0.455 | 0.978 |
| sapF; cationic peptide transport system ATP-binding protein | 0.231 | 0.308 | 0.455 | 0.978 |
| sapZ; predicted membrane protein | -0.105 | 0.772 | 0.892 | 0.999 |
| SARS, serS; seryl-tRNA synthetase [EC:6.1.1.11] | 0.034 | 0.036 | 0.342 | 0.978 |
| sasG; surface protein G | -0.457 | 0.700 | 0.514 | 0.983 |
| SASP-A, sspA; small acid-soluble spore protein A (major alpha-type SASP) | -1.930 | 1.515 | 0.205 | 0.972 |
| SASP-B, sspB; small acid-soluble spore protein B (major beta-type SASP) | -2.463 | 1.012 | 0.016 | 0.839 |
| sat, met3; sulfate adenylyltransferase [EC:2.7.7.4] | -0.604 | 0.412 | 0.145 | 0.951 |
| sauS; sulfoacetaldehyde dehydrogenase [EC:1.2.1.81] | 1.282 | 0.614 | 0.039 | 0.839 |
| sbcB, exoI; exodeoxyribonuclease I [EC:3.1.11.1] | 0.084 | 0.175 | 0.635 | 0.999 |
| sbcC, rad50; DNA repair protein SbcC/Rad50 | 0.207 | 0.083 | 0.014 | 0.839 |
| sbcD, mre11; DNA repair protein SbcD/Mre11 | 0.097 | 0.080 | 0.226 | 0.978 |
| sbi; immunoglobulin G-binding protein Sbi | -1.670 | 1.245 | 0.182 | 0.961 |
| sbmA, bacA; peptide/bleomycin uptake transporter | -0.501 | 0.329 | 0.130 | 0.934 |
| sbmC; DNA gyrase inhibitor | 0.112 | 0.636 | 0.860 | 0.999 |
| SC5DL, ERG3; Delta7-sterol 5-desaturase [EC:1.14.19.20] | 1.123 | 1.093 | 0.306 | 0.978 |
| SCD, desC; stearoyl-CoA desaturase (Delta-9 desaturase) [EC:1.14.19.1] | -0.190 | 0.287 | 0.509 | 0.981 |
| scn, scin; staphylococcal complement inhibitor | -1.670 | 1.245 | 0.182 | 0.961 |
| SCO1_2; protein SCO1/2 | -0.211 | 0.178 | 0.239 | 0.978 |
| scpA; segregation and condensation protein A | 0.059 | 0.043 | 0.179 | 0.961 |
| scpB, mmcD; methylmalonyl-CoA decarboxylase [EC:4.1.1.41] | -1.130 | 1.346 | 0.402 | 0.978 |
| scpB; segregation and condensation protein B | 0.057 | 0.041 | 0.167 | 0.961 |
| SCRN; secernin | -17.763 | 3362.856 | 0.996 | 0.999 |
| scrR; LacI family transcriptional regulator, sucrose operon repressor | 0.065 | 0.135 | 0.631 | 0.999 |
| scrY; sucrose porin | 0.430 | 0.544 | 0.430 | 0.978 |
| scsB; suppressor for copper-sensitivity B | 0.772 | 0.527 | 0.145 | 0.951 |
| sct; succinyl-CoA—D-citramalate CoA-transferase [EC:2.8.3.20] | -2.724 | 2.543 | 0.286 | 0.978 |
| sda; developmental checkpoint coupling sporulation initiation to replication initiation | -2.109 | 0.983 | 0.033 | 0.839 |
| sdaC; serine transporter | -0.043 | 0.237 | 0.857 | 0.999 |
| SDC1, CD138; syndecan 1 | 1.319 | 1.210 | 0.277 | 0.978 |
| sdh; serine 3-dehydrogenase (NADP+) [EC:1.1.1.276] | -0.191 | 0.415 | 0.646 | 0.999 |
| sdhA, frdA; succinate dehydrogenase / fumarate reductase, flavoprotein subunit [EC:1.3.5.1 1.3.5.4] | -0.125 | 0.073 | 0.090 | 0.883 |
| sdhB, frdB; succinate dehydrogenase / fumarate reductase, iron-sulfur subunit [EC:1.3.5.1 1.3.5.4] | -0.127 | 0.075 | 0.095 | 0.896 |
| sdhC, frdC; succinate dehydrogenase / fumarate reductase, cytochrome b subunit | -0.120 | 0.076 | 0.119 | 0.917 |
| sdhD, frdD; succinate dehydrogenase / fumarate reductase, membrane anchor subunit | -0.104 | 0.152 | 0.496 | 0.981 |
| sdiA; LuxR family transcriptional regulator, quorum-sensing system regulator SdiA | -0.331 | 0.371 | 0.374 | 0.978 |
| sdmt; sarcosine/dimethylglycine N-methyltransferase [EC:2.1.1.157] | -1.020 | 0.981 | 0.300 | 0.978 |
| SDO1, SBDS; ribosome maturation protein SDO1 | -17.234 | 3077.514 | 0.996 | 0.999 |
| SDR16C5; all-trans-retinol dehydrogenase (NAD+) [EC:1.1.1.105] | 0.833 | 1.424 | 0.559 | 0.992 |
| sdrC_D_E; serine-aspartate repeat-containing protein C/D/E | -0.562 | 0.669 | 0.402 | 0.978 |
| sdrM; MFS transporter, DHA2 family, multidrug resistance protein | -0.160 | 0.828 | 0.847 | 0.999 |
| SDS, SDH, CHA1; L-serine/L-threonine ammonia-lyase [EC:4.3.1.17 4.3.1.19] | 0.379 | 0.445 | 0.396 | 0.978 |
| se; staphylococcal enterotoxin | -1.670 | 1.245 | 0.182 | 0.961 |
| seaA; uncharacterized protein | -3.068 | 1.924 | 0.113 | 0.917 |
| SEC11, sipW; signal peptidase I [EC:3.4.21.89] | 0.284 | 0.501 | 0.571 | 0.995 |
| SEC61A; protein transport protein SEC61 subunit alpha | -17.234 | 3077.514 | 0.996 | 0.999 |
| SEC61G, SSS1, secE; protein transport protein SEC61 subunit gamma and related proteins | -17.234 | 3077.514 | 0.996 | 0.999 |
| secA; preprotein translocase subunit SecA | -0.011 | 0.045 | 0.799 | 0.999 |
| secB; preprotein translocase subunit SecB | -0.038 | 0.142 | 0.787 | 0.999 |
| secD; preprotein translocase subunit SecD | 0.140 | 0.077 | 0.071 | 0.841 |
| secDF; SecD/SecF fusion protein | -0.312 | 0.148 | 0.037 | 0.839 |
| secE; preprotein translocase subunit SecE | 0.082 | 0.044 | 0.060 | 0.839 |
| secF; preprotein translocase subunit SecF | 0.141 | 0.077 | 0.068 | 0.839 |
| secG; preprotein translocase subunit SecG | 0.036 | 0.043 | 0.398 | 0.978 |
| secM; secretion monitor | 0.119 | 0.601 | 0.843 | 0.999 |
| secY; preprotein translocase subunit SecY | 0.004 | 0.050 | 0.935 | 0.999 |
| selA; L-seryl-tRNA(Ser) seleniumtransferase [EC:2.9.1.1] | 0.163 | 0.165 | 0.324 | 0.978 |
| selB, EEFSEC; selenocysteine-specific elongation factor | 0.284 | 0.166 | 0.088 | 0.880 |
| selD, SEPHS; selenide, water dikinase [EC:2.7.9.3] | 0.177 | 0.109 | 0.106 | 0.911 |
| SELENBP1; selenium-binding protein 1 | -0.998 | 0.740 | 0.179 | 0.961 |
| selU; tRNA 2-selenouridine synthase [EC:2.9.1.-] | 0.180 | 0.218 | 0.409 | 0.978 |
| senX3; two-component system, OmpR family, sensor histidine kinase SenX3 [EC:2.7.13.3] | -0.016 | 0.278 | 0.954 | 0.999 |
| sepF; cell division inhibitor SepF | 0.190 | 0.091 | 0.038 | 0.839 |
| seqA; negative modulator of initiation of replication | 0.231 | 0.308 | 0.455 | 0.978 |
| serA, PHGDH; D-3-phosphoglycerate dehydrogenase / 2-oxoglutarate reductase [EC:1.1.1.95 1.1.1.399] | -0.032 | 0.055 | 0.561 | 0.992 |
| serB-plsC; putative phosphoserine phosphatase / 1-acylglycerol-3-phosphate O-acyltransferase [EC:3.1.3.3 2.3.1.51] | -1.955 | 1.506 | 0.196 | 0.970 |
| serB, PSPH; phosphoserine phosphatase [EC:3.1.3.3] | -0.092 | 0.078 | 0.237 | 0.978 |
| serC, PSAT1; phosphoserine aminotransferase [EC:2.6.1.52] | -0.081 | 0.061 | 0.188 | 0.966 |
| SERPINB; serpin B | -0.145 | 0.291 | 0.618 | 0.999 |
| set; superantigen-like protein | -1.670 | 1.245 | 0.182 | 0.961 |
| sfa2; sigma-54 dependent transcriptional regulator | 0.153 | 0.397 | 0.701 | 0.999 |
| sfa3; sigma-54 dependent transcriptional regulator | 1.513 | 1.386 | 0.276 | 0.978 |
| sfb1; fibronectin-binding protein 1 | -16.913 | 2621.982 | 0.995 | 0.999 |
| sfmA; type 1 fimbrial protein | -16.797 | 2474.383 | 0.995 | 0.999 |
| sfmC; fimbrial chaperone protein | 18.428 | 3259.084 | 0.995 | 0.999 |
| sfmD; outer membrane usher protein | -1.153 | 1.472 | 0.435 | 0.978 |
| sfmF; fimbrial-like protein | -1.158 | 1.457 | 0.428 | 0.978 |
| sfmH; fimbrial protein | -2.044 | 2.163 | 0.346 | 0.978 |
| sfnG; dimethylsulfone monooxygenase [EC:1.14.14.35] | -0.264 | 0.289 | 0.362 | 0.978 |
| sfsA; sugar fermentation stimulation protein A | 0.142 | 0.126 | 0.260 | 0.978 |
| SGA1; glucoamylase [EC:3.2.1.3] | -0.103 | 0.734 | 0.889 | 0.999 |
| sgrR; SgrR family transcriptional regulator | 0.078 | 0.554 | 0.888 | 0.999 |
| sgtA; monofunctional glycosyltransferase [EC:2.4.1.129] | -0.160 | 0.828 | 0.847 | 0.999 |
| sgtB; monofunctional glycosyltransferase [EC:2.4.1.129] | -0.160 | 0.828 | 0.847 | 0.999 |
| shc; squalene-hopene/tetraprenyl-beta-curcumene cyclase [EC:5.4.99.17 4.2.1.129] | -0.226 | 0.580 | 0.697 | 0.999 |
| shiA; MFS transporter, MHS family, shikimate and dehydroshikimate transport protein | 0.107 | 0.625 | 0.864 | 0.999 |
| shlA, hhdA, hpmA; hemolysin | 0.717 | 0.501 | 0.155 | 0.954 |
| shlB, hhdB, hpmB; hemolysin activation/secretion protein | 0.431 | 0.396 | 0.278 | 0.978 |
| SHPK; sedoheptulokinase [EC:2.7.1.14] | -3.815 | 3.621 | 0.294 | 0.978 |
| siaA, neuC1; UDP-N-acetylglucosamine 2-epimerase (hydrolysing) [EC:3.2.1.183] | 1.728 | 1.235 | 0.164 | 0.957 |
| SIAE; sialate O-acetylesterase [EC:3.1.1.53] | -0.382 | 0.224 | 0.090 | 0.882 |
| sigB; RNA polymerase sigma-B factor | -0.562 | 0.328 | 0.089 | 0.880 |
| sigH; RNA polymerase sporulation-specific sigma factor | -0.005 | 0.166 | 0.974 | 0.999 |
| sigI; RNA polymerase sigma factor | -1.590 | 0.823 | 0.055 | 0.839 |
| sinI; antagonist of SinR | -2.386 | 2.492 | 0.340 | 0.978 |
| sinR; XRE family transcriptional regulator, master regulator for biofilm formation | -2.391 | 1.250 | 0.058 | 0.839 |
| sipB, ipaB, bipB; invasin B | -17.760 | 4004.372 | 0.996 | 0.999 |
| sipC, ipaC, bipC; invasin C | -17.760 | 4004.372 | 0.996 | 0.999 |
| sipD, ipaD, bipD; invasin D | -17.760 | 4004.372 | 0.996 | 0.999 |
| sir; sulfite reductase (ferredoxin) [EC:1.8.7.1] | -0.361 | 0.321 | 0.263 | 0.978 |
| sirB; sirohydrochlorin ferrochelatase [EC:4.99.1.4] | -1.552 | 0.893 | 0.084 | 0.873 |
| sitA; manganese/iron transport system substrate-binding protein | 0.120 | 0.296 | 0.686 | 0.999 |
| sitB; manganese/iron transport system ATP-binding protein | 0.095 | 0.290 | 0.743 | 0.999 |
| sitC; manganese/iron transport system permease protein | 0.120 | 0.296 | 0.686 | 0.999 |
| sitD; manganese/iron transport system permease protein | 0.120 | 0.296 | 0.686 | 0.999 |
| sixA; phosphohistidine phosphatase [EC:3.1.3.-] | -0.032 | 0.122 | 0.795 | 0.999 |
| ska; streptokinase A [EC:3.4.-.-] | 0.521 | 1.146 | 0.650 | 0.999 |
| SKI3, TTC37; superkiller protein 3 | -17.068 | 2832.569 | 0.995 | 0.999 |
| SLC10A7, P7; solute carrier family 10 (sodium/bile acid cotransporter), member 7 | 0.013 | 0.159 | 0.934 | 0.999 |
| SLC13A2_3_5; solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2/3/5 | 0.063 | 0.169 | 0.710 | 0.999 |
| SLC34A, NPT, nptA; solute carrier family 34 (sodium-dependent phosphate cotransporter) | 1.403 | 1.070 | 0.192 | 0.970 |
| slcC; (S)-sulfolactate dehydrogenase [EC:1.1.1.310] | 0.164 | 0.669 | 0.807 | 0.999 |
| slmA, ttk; TetR/AcrR family transcriptional regulator | 0.055 | 0.262 | 0.833 | 0.999 |
| slo; thiol-activated cytolysin | -0.181 | 0.201 | 0.368 | 0.978 |
| slp; outer membrane lipoprotein | 0.019 | 0.333 | 0.953 | 0.999 |
| slpA; FKBP-type peptidyl-prolyl cis-trans isomerase SlpA [EC:5.2.1.8] | 0.039 | 0.230 | 0.864 | 0.999 |
| slrA; anti-repressor of SlrR | 18.239 | 2966.075 | 0.995 | 0.999 |
| slrR; HTH-type transcriptional regulator, biofilm formation regulator | 18.239 | 2966.075 | 0.995 | 0.999 |
| slt; soluble lytic murein transglycosylase [EC:4.2.2.-] | -0.023 | 0.095 | 0.811 | 0.999 |
| slyA; MarR family transcriptional regulator, transcriptional regulator for hemolysin | -0.049 | 0.245 | 0.843 | 0.999 |
| slyB; outer membrane lipoprotein SlyB | 0.142 | 0.176 | 0.420 | 0.978 |
| slyD; FKBP-type peptidyl-prolyl cis-trans isomerase SlyD [EC:5.2.1.8] | 0.048 | 0.108 | 0.661 | 0.999 |
| slyX; SlyX protein | -0.002 | 0.152 | 0.988 | 0.999 |
| SMARCAD1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 [EC:3.6.4.12] | 18.525 | 3422.126 | 0.996 | 0.999 |
| smc; chromosome segregation protein | 0.054 | 0.041 | 0.190 | 0.968 |
| smeA; membrane fusion protein, multidrug efflux system | -0.052 | 0.488 | 0.915 | 0.999 |
| smeB; multidrug efflux pump | -0.052 | 0.488 | 0.915 | 0.999 |
| smeC; outer membrane protein, multidrug efflux system | -0.052 | 0.488 | 0.915 | 0.999 |
| smf; DNA processing protein | 0.073 | 0.042 | 0.082 | 0.865 |
| smg; Smg protein | 0.082 | 0.274 | 0.765 | 0.999 |
| smoE, mtlE; sorbitol/mannitol transport system substrate-binding protein | -0.338 | 0.371 | 0.363 | 0.978 |
| smoF, mtlF; sorbitol/mannitol transport system permease protein | -0.302 | 0.336 | 0.370 | 0.978 |
| smoG, mtlG; sorbitol/mannitol transport system permease protein | -0.302 | 0.336 | 0.370 | 0.978 |
| smp; membrane protein | 0.218 | 0.293 | 0.459 | 0.979 |
| smpB; SsrA-binding protein | 0.034 | 0.036 | 0.344 | 0.978 |
| smtA; S-adenosylmethionine-dependent methyltransferase | 0.226 | 0.292 | 0.439 | 0.978 |
| SMUG1; single-strand selective monofunctional uracil DNA glycosylase [EC:3.2.2.-] | -0.661 | 1.067 | 0.536 | 0.990 |
| smvA, qacA, lfrA; MFS transporter, DHA2 family, multidrug resistance protein | -0.147 | 0.221 | 0.508 | 0.981 |
| snbC; pristinamycin I synthase 2 | -17.871 | 3602.141 | 0.996 | 0.999 |
| snoaF, dnrE, dauE, aknU; nogalaviketone/aklaviketone reductase [EC:1.1.1.- 1.1.1.362] | -1.780 | 1.801 | 0.324 | 0.978 |
| snpA; snapalysin [EC:3.4.24.77] | -17.794 | 4072.025 | 0.997 | 0.999 |
| SOD1; superoxide dismutase, Cu-Zn family [EC:1.15.1.1] | -0.138 | 0.178 | 0.440 | 0.978 |
| SOD2; superoxide dismutase, Fe-Mn family [EC:1.15.1.1] | -0.080 | 0.060 | 0.188 | 0.966 |
| sodN; nickel superoxide dismutase [EC:1.15.1.1] | 1.384 | 0.676 | 0.042 | 0.839 |
| sohB; serine protease SohB [EC:3.4.21.-] | 0.114 | 0.188 | 0.546 | 0.990 |
| solA; N-methyl-L-tryptophan oxidase [EC:1.5.3.-] | -0.298 | 0.755 | 0.694 | 0.999 |
| solR, cepR, tofR; LuxR family transcriptional regulator, quorum-sensing system regulator SolR | -2.067 | 1.907 | 0.280 | 0.978 |
| SORD, gutB; L-iditol 2-dehydrogenase [EC:1.1.1.14] | -0.210 | 0.265 | 0.430 | 0.978 |
| sotB; MFS transporter, DHA1 family, L-arabinose/isopropyl-beta-D-thiogalactopyranoside export protein | 0.097 | 0.206 | 0.637 | 0.999 |
| soxA; sarcosine oxidase, subunit alpha [EC:1.5.3.1] | -0.222 | 0.297 | 0.455 | 0.978 |
| soxA; sulfur-oxidizing protein SoxA | -1.335 | 0.667 | 0.047 | 0.839 |
| soxB; sarcosine oxidase, subunit beta [EC:1.5.3.1] | -0.270 | 0.334 | 0.422 | 0.978 |
| soxB; sulfur-oxidizing protein SoxB | -1.085 | 0.603 | 0.074 | 0.845 |
| soxC; sulfane dehydrogenase subunit SoxC | -0.782 | 0.594 | 0.190 | 0.968 |
| soxD; sarcosine oxidase, subunit delta [EC:1.5.3.1] | -0.044 | 0.312 | 0.887 | 0.999 |
| soxG; sarcosine oxidase, subunit gamma [EC:1.5.3.1] | -0.114 | 0.320 | 0.722 | 0.999 |
| soxR; MerR family transcriptional regulator, redox-sensitive transcriptional activator SoxR | -0.053 | 0.234 | 0.821 | 0.999 |
| soxS; AraC family transcriptional regulator, mar-sox-rob regulon activator | -0.223 | 0.684 | 0.744 | 0.999 |
| soxX; sulfur-oxidizing protein SoxX | -1.292 | 0.672 | 0.056 | 0.839 |
| soxY; sulfur-oxidizing protein SoxY | -0.006 | 0.335 | 0.985 | 0.999 |
| soxZ; sulfur-oxidizing protein SoxZ | -0.835 | 0.566 | 0.142 | 0.951 |
| spa; immunoglobulin G-binding protein A | -1.670 | 1.245 | 0.182 | 0.961 |
| spdH; spermidine dehydrogenase [EC:1.5.99.6] | -0.719 | 0.796 | 0.368 | 0.978 |
| speA; arginine decarboxylase [EC:4.1.1.19] | -0.064 | 0.133 | 0.631 | 0.999 |
| speB; agmatinase [EC:3.5.3.11] | -0.100 | 0.181 | 0.580 | 0.997 |
| speD, AMD1; S-adenosylmethionine decarboxylase [EC:4.1.1.50] | 0.051 | 0.140 | 0.717 | 0.999 |
| speE, SRM; spermidine synthase [EC:2.5.1.16] | -0.107 | 0.096 | 0.267 | 0.978 |
| speG, SAT; diamine N-acetyltransferase [EC:2.3.1.57] | -0.076 | 0.122 | 0.535 | 0.990 |
| sphB1, nalP, ausP; autotransporter serine protease [EC:3.4.21.-] | -16.880 | 1816.243 | 0.993 | 0.999 |
| splA; transcriptional regulator of the spore photoproduct lyase operon | -1.305 | 0.884 | 0.142 | 0.951 |
| splB; spore photoproduct lyase [EC:4.1.99.14] | 0.439 | 0.359 | 0.224 | 0.978 |
| spmA; spore maturation protein A | -0.834 | 0.461 | 0.072 | 0.845 |
| spmB; spore maturation protein B | -1.050 | 0.550 | 0.058 | 0.839 |
| spo0A; two-component system, response regulator, stage 0 sporulation protein A | -0.704 | 0.286 | 0.015 | 0.839 |
| spo0B; stage 0 sporulation protein B (sporulation initiation phosphotransferase) [EC:2.7.-.-] | -1.586 | 0.806 | 0.051 | 0.839 |
| spo0E; stage 0 sporulation regulatory protein | -0.708 | 1.237 | 0.568 | 0.995 |
| spo0F; two-component system, response regulator, stage 0 sporulation protein F | -1.421 | 0.741 | 0.057 | 0.839 |
| spo0M; sporulation-control protein | -0.700 | 0.453 | 0.124 | 0.919 |
| spoIIAA; stage II sporulation protein AA (anti-sigma F factor antagonist) | -0.854 | 0.469 | 0.071 | 0.841 |
| spoIIAB; stage II sporulation protein AB (anti-sigma F factor) [EC:2.7.11.1] | -0.960 | 0.501 | 0.057 | 0.839 |
| spoIIB; stage II sporulation protein B | -2.967 | 1.380 | 0.033 | 0.839 |
| spoIID; stage II sporulation protein D | 0.093 | 0.184 | 0.612 | 0.999 |
| spoIIE; stage II sporulation protein E [EC:3.1.3.16] | -0.996 | 0.511 | 0.053 | 0.839 |
| spoIIGA; stage II sporulation protein GA (sporulation sigma-E factor processing peptidase) [EC:3.4.23.-] | -0.893 | 0.514 | 0.085 | 0.873 |
| spoIIIAA; stage III sporulation protein AA | -1.041 | 0.551 | 0.061 | 0.839 |
| spoIIIAB; stage III sporulation protein AB | -0.977 | 0.591 | 0.100 | 0.896 |
| spoIIIAC; stage III sporulation protein AC | -0.957 | 0.501 | 0.058 | 0.839 |
| spoIIIAD; stage III sporulation protein AD | -1.041 | 0.551 | 0.061 | 0.839 |
| spoIIIAE; stage III sporulation protein AE | -0.980 | 0.509 | 0.056 | 0.839 |
| spoIIIAF; stage III sporulation protein AF | -1.058 | 0.586 | 0.073 | 0.845 |
| spoIIIAG; stage III sporulation protein AG | -0.954 | 0.499 | 0.058 | 0.839 |
| spoIIIAH; stage III sporulation protein AH | -0.957 | 0.501 | 0.058 | 0.839 |
| spoIIID; putative DeoR family transcriptional regulator, stage III sporulation protein D | -0.940 | 0.501 | 0.063 | 0.839 |
| spoIIM; stage II sporulation protein M | -1.025 | 0.558 | 0.068 | 0.839 |
| spoIIP; stage II sporulation protein P | -1.015 | 0.487 | 0.039 | 0.839 |
| spoIIQ; stage II sporulation protein Q | -1.487 | 0.738 | 0.046 | 0.839 |
| spoIIR; stage II sporulation protein R | -0.929 | 0.498 | 0.064 | 0.839 |
| spoIISA; stage II sporulation protein SA | 18.239 | 2966.075 | 0.995 | 0.999 |
| spoIISB; stage II sporulation protein SB | 18.239 | 2966.075 | 0.995 | 0.999 |
| spoIVA; stage IV sporulation protein A | -0.960 | 0.501 | 0.057 | 0.839 |
| spoIVB; stage IV sporulation protein B [EC:3.4.21.116] | -0.996 | 0.494 | 0.045 | 0.839 |
| spoIVCA; site-specific DNA recombinase | -0.144 | 0.386 | 0.710 | 0.999 |
| spoIVFA; stage IV sporulation protein FA | -2.174 | 0.860 | 0.012 | 0.839 |
| spoIVFB; stage IV sporulation protein FB [EC:3.4.24.-] | -0.838 | 0.566 | 0.141 | 0.949 |
| spoT; GTP diphosphokinase / guanosine-3’,5’-bis(diphosphate) 3’-diphosphatase [EC:2.7.6.5 3.1.7.2] | 0.199 | 0.139 | 0.153 | 0.954 |
| spoU; RNA methyltransferase, TrmH family | 0.039 | 0.066 | 0.557 | 0.992 |
| spoVAA; stage V sporulation protein AA | -2.374 | 0.831 | 0.005 | 0.839 |
| spoVAB; stage V sporulation protein AB | -2.236 | 0.783 | 0.005 | 0.839 |
| spoVAC; stage V sporulation protein AC | -1.219 | 0.504 | 0.017 | 0.839 |
| spoVAD; stage V sporulation protein AD | -1.202 | 0.504 | 0.018 | 0.839 |
| spoVAE; stage V sporulation protein AE | -1.361 | 0.525 | 0.011 | 0.839 |
| spoVAF; stage V sporulation protein AF | -1.326 | 0.597 | 0.028 | 0.839 |
| spoVB; stage V sporulation protein B | 0.118 | 0.201 | 0.558 | 0.992 |
| spoVD; stage V sporulation protein D (sporulation-specific penicillin-binding protein) | 0.044 | 0.210 | 0.833 | 0.999 |
| spoVFA; dipicolinate synthase subunit A | -1.337 | 0.569 | 0.020 | 0.839 |
| spoVFB; dipicolinate synthase subunit B | -1.337 | 0.569 | 0.020 | 0.839 |
| spoVG; stage V sporulation protein G | 0.226 | 0.235 | 0.339 | 0.978 |
| spoVID; stage VI sporulation protein D | -2.171 | 0.928 | 0.021 | 0.839 |
| spoVK; stage V sporulation protein K | -2.177 | 0.856 | 0.012 | 0.839 |
| spoVM; stage V sporulation protein M | -1.220 | 1.986 | 0.540 | 0.990 |
| spoVR; stage V sporulation protein R | -0.801 | 0.507 | 0.116 | 0.917 |
| spoVS; stage V sporulation protein S | -0.294 | 0.377 | 0.437 | 0.978 |
| spoVT; AbrB family transcriptional regulator, stage V sporulation protein T | -0.966 | 0.503 | 0.057 | 0.839 |
| SPP; sucrose-6-phosphatase [EC:3.1.3.24] | 0.062 | 0.090 | 0.491 | 0.980 |
| sppA; protease IV [EC:3.4.21.-] | 0.072 | 0.101 | 0.475 | 0.979 |
| sprB; streptogrisin B [EC:3.4.21.81] | -17.794 | 4072.025 | 0.997 | 0.999 |
| sprC; streptogrisin C [EC:3.4.21.-] | 0.755 | 1.052 | 0.474 | 0.979 |
| sprD; streptogrisin D [EC:3.4.21.-] | -17.487 | 3492.836 | 0.996 | 0.999 |
| sprL; SprT-like protein | 0.123 | 0.153 | 0.422 | 0.978 |
| sprT; SprT protein | 0.261 | 0.215 | 0.227 | 0.978 |
| spsF; spore coat polysaccharide biosynthesis protein SpsF | -0.329 | 0.665 | 0.622 | 0.999 |
| spuC; putrescine—pyruvate transaminase [EC:2.6.1.113] | 0.116 | 0.328 | 0.724 | 0.999 |
| spxA; regulatory protein spx | -0.012 | 0.137 | 0.928 | 0.999 |
| SQD1, sqdB; UDP-sulfoquinovose synthase [EC:3.13.1.1] | -1.103 | 0.644 | 0.089 | 0.880 |
| sqhC; sporulenol synthase [EC:4.2.1.137] | -2.156 | 1.169 | 0.067 | 0.839 |
| sqr; sulfide:quinone oxidoreductase [EC:1.8.5.4] | -0.132 | 0.206 | 0.523 | 0.989 |
| sra; stationary-phase-induced ribosome-associated protein | -0.281 | 0.729 | 0.700 | 0.999 |
| SRD5A1; 3-oxo-5-alpha-steroid 4-dehydrogenase 1 [EC:1.3.1.22] | -1.103 | 0.669 | 0.101 | 0.896 |
| srfAA, lchAA; surfactin family lipopeptide synthetase A | 17.932 | 2544.192 | 0.994 | 0.999 |
| srfAB, lchAB; surfactin family lipopeptide synthetase B | 18.239 | 2966.075 | 0.995 | 0.999 |
| srfAC, lchAC; surfactin family lipopeptide synthetase C | 18.239 | 2966.075 | 0.995 | 0.999 |
| srfATE, srfAD, lchAD; external thioesterase TEII | -0.607 | 1.382 | 0.661 | 0.999 |
| srlD; sorbitol-6-phosphate 2-dehydrogenase [EC:1.1.1.140] | 0.335 | 0.358 | 0.351 | 0.978 |
| srlR, gutR; DeoR family transcriptional regulator, glucitol operon repressor | 0.129 | 0.619 | 0.835 | 0.999 |
| srmB; ATP-dependent RNA helicase SrmB [EC:3.6.4.13] | 0.231 | 0.308 | 0.455 | 0.978 |
| SRP19; signal recognition particle subunit SRP19 | -17.234 | 3077.514 | 0.996 | 0.999 |
| SRP54, ffh; signal recognition particle subunit SRP54 [EC:3.6.5.4] | 0.035 | 0.036 | 0.326 | 0.978 |
| srtA; sortase A [EC:3.4.22.70] | 0.012 | 0.111 | 0.913 | 0.999 |
| srtB; sortase B [EC:3.4.22.70] | -0.037 | 0.194 | 0.851 | 0.999 |
| ssb; single-strand DNA-binding protein | 0.036 | 0.046 | 0.432 | 0.978 |
| ssh10b; archaea-specific DNA-binding protein | -16.927 | 2639.780 | 0.995 | 0.999 |
| ssp; subtilase-type serine protease [EC:3.4.21.-] | -0.208 | 0.407 | 0.610 | 0.999 |
| sspA, mglA; stringent starvation protein A | 0.061 | 0.155 | 0.696 | 0.999 |
| sspA; glutamyl endopeptidase [EC:3.4.21.19] | -1.676 | 1.105 | 0.131 | 0.941 |
| sspB; staphopain B [EC:3.4.22.-] | -1.670 | 1.245 | 0.182 | 0.961 |
| sspB; stringent starvation protein B | 0.059 | 0.156 | 0.707 | 0.999 |
| sspB2, sspP, scpA; staphopain A [EC:3.4.22.48] | -1.554 | 1.110 | 0.164 | 0.957 |
| sspD; small acid-soluble spore protein D (minor alpha/beta-type SASP) | -2.317 | 1.143 | 0.044 | 0.839 |
| sspE; small acid-soluble spore protein E (minor gamma-type SASP) | -1.247 | 0.767 | 0.106 | 0.911 |
| sspF; small acid-soluble spore protein F (minor alpha/beta-type SASP) | -1.302 | 0.808 | 0.109 | 0.917 |
| sspH; small acid-soluble spore protein H (minor) | -2.107 | 0.917 | 0.023 | 0.839 |
| sspI; small acid-soluble spore protein I (minor) | -1.426 | 0.741 | 0.056 | 0.839 |
| sspK; small acid-soluble spore protein K (minor) | -2.826 | 1.220 | 0.022 | 0.839 |
| sspL; small acid-soluble spore protein L (minor) | -2.930 | 1.399 | 0.038 | 0.839 |
| sspN; small acid-soluble spore protein N (minor) | -2.318 | 1.036 | 0.027 | 0.839 |
| sspO, cotK; small acid-soluble spore protein O (minor) | -2.798 | 1.277 | 0.030 | 0.839 |
| sspP, cotL; small acid-soluble spore protein P (minor) | -2.749 | 1.093 | 0.013 | 0.839 |
| sstT; serine/threonine transporter | 0.039 | 0.091 | 0.667 | 0.999 |
| ssuA; sulfonate transport system substrate-binding protein | -0.137 | 0.228 | 0.550 | 0.992 |
| ssuB; sulfonate transport system ATP-binding protein [EC:3.6.3.-] | -0.094 | 0.173 | 0.586 | 0.999 |
| ssuC; sulfonate transport system permease protein | -0.099 | 0.190 | 0.603 | 0.999 |
| ssuD; alkanesulfonate monooxygenase [EC:1.14.14.5] | -0.338 | 0.265 | 0.204 | 0.972 |
| ssuE; FMN reductase [EC:1.5.1.38] | -0.256 | 0.207 | 0.217 | 0.978 |
| stbD; antitoxin StbD | -0.190 | 0.455 | 0.677 | 0.999 |
| STE24; STE24 endopeptidase [EC:3.4.24.84] | -0.190 | 0.344 | 0.582 | 0.999 |
| steA, tetA46; ATP-binding cassette, subfamily B, tetracycline resistant protein | 0.489 | 0.488 | 0.318 | 0.978 |
| steB, tetB46; ATP-binding cassette, subfamily B, tetracycline resistant protein | 0.489 | 0.488 | 0.318 | 0.978 |
| sthA, udhA; NAD(P) transhydrogenase [EC:1.6.1.1] | 0.033 | 0.236 | 0.888 | 0.999 |
| stk1; serine/threonine-protein kinase Stk1 [EC:2.7.11.-] | -0.308 | 0.470 | 0.513 | 0.982 |
| stp1, pppA; serine/threonine protein phosphatase Stp1 [EC:3.1.3.16] | -0.104 | 0.430 | 0.808 | 0.999 |
| stpA; DNA-binding protein StpA | -0.243 | 0.717 | 0.735 | 0.999 |
| strA; streptomycin 3"-kinase [EC:2.7.1.87] | 1.665 | 1.566 | 0.289 | 0.978 |
| strB; streptomycin 6-kinase [EC:2.7.1.72] | -0.156 | 0.402 | 0.698 | 0.999 |
| STT3; dolichyl-diphosphooligosaccharide—protein glycosyltransferase [EC:2.4.99.18] | -17.234 | 3077.514 | 0.996 | 0.999 |
| sucC; succinyl-CoA synthetase beta subunit [EC:6.2.1.5] | -0.105 | 0.083 | 0.208 | 0.977 |
| sucD; succinate-semialdehyde dehydrogenase [EC:1.2.1.76] | -0.290 | 0.602 | 0.631 | 0.999 |
| sucD; succinyl-CoA synthetase alpha subunit [EC:6.2.1.5] | -0.105 | 0.083 | 0.207 | 0.976 |
| sufA; Fe-S cluster assembly protein SufA | 0.193 | 0.583 | 0.741 | 0.999 |
| sufB; Fe-S cluster assembly protein SufB | -0.030 | 0.075 | 0.690 | 0.999 |
| sufC; Fe-S cluster assembly ATP-binding protein | -0.029 | 0.070 | 0.680 | 0.999 |
| sufD; Fe-S cluster assembly protein SufD | -0.073 | 0.081 | 0.369 | 0.978 |
| sufE; cysteine desulfuration protein SufE | -0.127 | 0.109 | 0.244 | 0.978 |
| sufI; suppressor of ftsI | 0.095 | 0.303 | 0.753 | 0.999 |
| sufS; cysteine desulfurase / selenocysteine lyase [EC:2.8.1.7 4.4.1.16] | -0.024 | 0.067 | 0.717 | 0.999 |
| SUGCT; succinate—hydroxymethylglutarate CoA-transferase [EC:2.8.3.13] | 18.076 | 2733.666 | 0.995 | 0.999 |
| sugE; quaternary ammonium compound-resistance protein SugE | -0.056 | 0.171 | 0.746 | 0.999 |
| sul1; dihydropteroate synthase type 1 [EC:2.5.1.15] | 0.021 | 1.115 | 0.985 | 0.999 |
| sul2; dihydropteroate synthase type 2 [EC:2.5.1.15] | 18.677 | 2978.101 | 0.995 | 0.999 |
| sulA; cell division inhibitor SulA | 0.141 | 0.248 | 0.571 | 0.995 |
| sulD; dihydroneopterin aldolase / 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase [EC:4.1.2.25 2.7.6.3] | 0.083 | 0.141 | 0.560 | 0.992 |
| SUOX; sulfite oxidase [EC:1.8.3.1] | -0.028 | 0.718 | 0.969 | 0.999 |
| SUPV3L1, SUV3; ATP-dependent RNA helicase SUPV3L1/SUV3 [EC:3.6.4.13] | -0.730 | 0.375 | 0.053 | 0.839 |
| surA; peptidyl-prolyl cis-trans isomerase SurA [EC:5.2.1.8] | -0.122 | 0.107 | 0.254 | 0.978 |
| surE; 5’-nucleotidase [EC:3.1.3.5] | -0.054 | 0.087 | 0.538 | 0.990 |
| SURF1, SHY1; surfeit locus 1 family protein | -0.912 | 0.418 | 0.031 | 0.839 |
| suyA; (2R)-sulfolactate sulfo-lyase subunit alpha [EC:4.4.1.24] | 0.023 | 0.915 | 0.980 | 0.999 |
| suyB; (2R)-sulfolactate sulfo-lyase subunit beta [EC:4.4.1.24] | 0.023 | 0.915 | 0.980 | 0.999 |
| syd; SecY interacting protein Syd | 0.352 | 0.531 | 0.508 | 0.981 |
| symE; toxic protein SymE | 0.004 | 0.571 | 0.994 | 0.999 |
| syrB1; Syringomycin synthetase protein SyrB1 | 18.932 | 3654.080 | 0.996 | 0.999 |
| tabA; biofilm protein TabA | 0.245 | 0.608 | 0.688 | 0.999 |
| tadA; tRNA(adenine34) deaminase [EC:3.5.4.33] | -0.015 | 0.074 | 0.836 | 0.999 |
| tadB; tight adherence protein B | -0.009 | 0.155 | 0.953 | 0.999 |
| tadC; tight adherence protein C | -0.085 | 0.156 | 0.586 | 0.999 |
| tag; DNA-3-methyladenine glycosylase I [EC:3.2.2.20] | 0.020 | 0.047 | 0.666 | 0.999 |
| tagA, tarA; N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase [EC:2.4.1.187] | -0.123 | 0.134 | 0.363 | 0.978 |
| tagD; glycerol-3-phosphate cytidylyltransferase [EC:2.7.7.39] | 0.273 | 0.194 | 0.162 | 0.957 |
| tagE; poly(glycerol-phosphate) alpha-glucosyltransferase [EC:2.4.1.52] | 0.094 | 0.245 | 0.701 | 0.999 |
| tagF; CDP-glycerol glycerophosphotransferase [EC:2.7.8.12] | -0.536 | 0.416 | 0.199 | 0.970 |
| tagG; teichoic acid transport system permease protein | 0.174 | 0.312 | 0.577 | 0.997 |
| tagH; teichoic acid transport system ATP-binding protein [EC:3.6.3.40] | 0.276 | 0.312 | 0.378 | 0.978 |
| tal-pgi; transaldolase / glucose-6-phosphate isomerase [EC:2.2.1.2 5.3.1.9] | -0.572 | 0.531 | 0.283 | 0.978 |
| tam; trans-aconitate 2-methyltransferase [EC:2.1.1.144] | -0.151 | 0.266 | 0.570 | 0.995 |
| tamA; translocation and assembly module TamA | -0.033 | 0.151 | 0.828 | 0.999 |
| tamB; translocation and assembly module TamB | 0.030 | 0.132 | 0.822 | 0.999 |
| TAN1, THUMPD1; tRNA acetyltransferase TAN1 | -17.234 | 3077.514 | 0.996 | 0.999 |
| tap; methyl-accepting chemotaxis protein IV, peptide sensor receptor | 0.174 | 0.624 | 0.780 | 0.999 |
| tapA; TasA anchoring/assembly protein | 18.239 | 2966.075 | 0.995 | 0.999 |
| tar; methyl-accepting chemotaxis protein II, aspartate sensor receptor | -0.673 | 0.602 | 0.265 | 0.978 |
| tarJ; ribitol-5-phosphate 2-dehydrogenase (NADP+) [EC:1.1.1.405] | -0.274 | 0.216 | 0.207 | 0.976 |
| tarL; CDP-ribitol ribitolphosphotransferase / teichoic acid ribitol-phosphate polymerase [EC:2.7.8.14 2.7.8.47] | -1.095 | 0.795 | 0.170 | 0.961 |
| TARS, thrS; threonyl-tRNA synthetase [EC:6.1.1.3] | 0.024 | 0.032 | 0.452 | 0.978 |
| tasA, cotN; spore coat-associated protein N | -1.877 | 1.718 | 0.276 | 0.978 |
| tatA; sec-independent protein translocase protein TatA | -0.109 | 0.063 | 0.086 | 0.878 |
| tatB; sec-independent protein translocase protein TatB | -0.068 | 0.120 | 0.570 | 0.995 |
| tatC; sec-independent protein translocase protein TatC | -0.116 | 0.061 | 0.059 | 0.839 |
| tatD; TatD DNase family protein [EC:3.1.21.-] | 0.036 | 0.021 | 0.091 | 0.885 |
| tatE; sec-independent protein translocase protein TatE | 0.130 | 0.620 | 0.834 | 0.999 |
| tauA; taurine transport system substrate-binding protein | -0.212 | 0.291 | 0.469 | 0.979 |
| tauB; taurine transport system ATP-binding protein [EC:3.6.3.36] | -0.219 | 0.260 | 0.401 | 0.978 |
| tauC; taurine transport system permease protein | -0.285 | 0.249 | 0.254 | 0.978 |
| tauD; taurine dioxygenase [EC:1.14.11.17] | -0.171 | 0.278 | 0.539 | 0.990 |
| tauX; taurine dehydrogenase small subunit [EC:1.4.2.-] | -17.500 | 2562.559 | 0.995 | 0.999 |
| tauY; taurine dehydrogenase large subunit [EC:1.4.2.-] | 1.346 | 1.085 | 0.217 | 0.978 |
| TBP, tbp; transcription initiation factor TFIID TATA-box-binding protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| TC.AAA; ATP:ADP antiporter, AAA family | 0.269 | 0.300 | 0.372 | 0.978 |
| TC.AAT; amino acid transporter, AAT family | 0.088 | 0.158 | 0.580 | 0.997 |
| TC.AGCS; alanine or glycine:cation symporter, AGCS family | 0.196 | 0.095 | 0.040 | 0.839 |
| TC.APA; basic amino acid/polyamine antiporter, APA family | 0.065 | 0.127 | 0.608 | 0.999 |
| TC.BASS; bile acid:Na+ symporter, BASS family | -0.112 | 0.090 | 0.218 | 0.978 |
| TC.BAT1; bacterial/archaeal transporter family protein | -0.017 | 0.172 | 0.920 | 0.999 |
| TC.BAT2; bacterial/archaeal transporter family-2 protein | -0.189 | 0.187 | 0.313 | 0.978 |
| TC.BCT; betaine/carnitine transporter, BCCT family | 0.102 | 0.318 | 0.749 | 0.999 |
| TC.CIC; chloride channel protein, CIC family | -0.149 | 0.140 | 0.289 | 0.978 |
| TC.CITMHS; citrate-Mg2+:H+ or citrate-Ca2+:H+ symporter, CitMHS family | 0.127 | 0.148 | 0.392 | 0.978 |
| TC.CNT; concentrative nucleoside transporter, CNT family | 0.052 | 0.184 | 0.777 | 0.999 |
| TC.CPA1; monovalent cation:H+ antiporter, CPA1 family | 0.030 | 0.096 | 0.755 | 0.999 |
| TC.DAACS; dicarboxylate/amino acid:cation (Na+ or H+) symporter, DAACS family | -0.144 | 0.185 | 0.437 | 0.978 |
| TC.DASS; divalent anion:Na+ symporter, DASS family | 0.169 | 0.170 | 0.323 | 0.978 |
| TC.DCUC, dcuC, dcuD; C4-dicarboxylate transporter, DcuC family | 0.107 | 0.240 | 0.657 | 0.999 |
| TC.DME; drug/metabolite transporter, DME family | -0.360 | 0.407 | 0.378 | 0.978 |
| TC.FEV.OM; iron complex outermembrane recepter protein | 0.013 | 0.099 | 0.898 | 0.999 |
| TC.FEV.OM1, fhuE, fpvA, fptA; outer-membrane receptor for ferric coprogen and ferric-rhodotorulic acid | -0.073 | 0.252 | 0.773 | 0.999 |
| TC.FEV.OM2, cirA, cfrA, hmuR; outer membrane receptor for ferrienterochelin and colicins | -0.130 | 0.168 | 0.443 | 0.978 |
| TC.FEV.OM3, tbpA, hemR, lbpA, hpuB, bhuR, hugA, hmbR; hemoglobin/transferrin/lactoferrin receptor protein | 0.052 | 0.185 | 0.781 | 0.999 |
| TC.GBP; general bacterial porin, GBP family | -1.576 | 2.122 | 0.459 | 0.979 |
| TC.GNTP; gluconate:H+ symporter, GntP family | 0.266 | 0.125 | 0.035 | 0.839 |
| TC.GPH; glycoside/pentoside/hexuronide:cation symporter, GPH family | 0.261 | 0.256 | 0.308 | 0.978 |
| TC.HAE1; hydrophobic/amphiphilic exporter-1 (mainly G- bacteria), HAE1 family | -0.188 | 0.101 | 0.065 | 0.839 |
| TC.HME; heavy-metal exporter, HME family | -0.413 | 0.757 | 0.586 | 0.999 |
| TC.KEF; monovalent cation:H+ antiporter-2, CPA2 family | -0.168 | 0.111 | 0.134 | 0.942 |
| TC.LIVCS; branched-chain amino acid:cation transporter, LIVCS family | 0.173 | 0.079 | 0.031 | 0.839 |
| TC.MATE, SLC47A, norM, mdtK, dinF; multidrug resistance protein, MATE family | -0.070 | 0.062 | 0.259 | 0.978 |
| TC.NCS1; nucleobase:cation symporter-1, NCS1 family | -0.141 | 0.307 | 0.648 | 0.999 |
| TC.NCS2; nucleobase:cation symporter-2, NCS2 family | 0.070 | 0.149 | 0.641 | 0.999 |
| TC.NSS; neurotransmitter:Na+ symporter, NSS family | 0.091 | 0.104 | 0.381 | 0.978 |
| TC.OMF; outer membrane factor, OMF family | 1.072 | 0.505 | 0.035 | 0.839 |
| TC.OOP; OmpA-OmpF porin, OOP family | 0.186 | 0.122 | 0.130 | 0.934 |
| TC.PIT; inorganic phosphate transporter, PiT family | -0.098 | 0.097 | 0.317 | 0.978 |
| TC.POT; proton-dependent oligopeptide transporter, POT family | -0.074 | 0.105 | 0.486 | 0.979 |
| TC.PST; polysaccharide transporter, PST family | -0.117 | 0.249 | 0.641 | 0.999 |
| TC.SMR3; small multidrug resistance family-3 protein | 0.019 | 0.144 | 0.894 | 0.999 |
| TC.SSS; solute:Na+ symporter, SSS family | -0.020 | 0.132 | 0.878 | 0.999 |
| TC.SULP; sulfate permease, SulP family | -0.066 | 0.111 | 0.551 | 0.992 |
| TC.ZIP, zupT, ZRT3, ZIP2; zinc transporter, ZIP family | -0.080 | 0.083 | 0.338 | 0.978 |
| tccC; insecticidal toxin complex protein TccC | -0.067 | 0.633 | 0.916 | 0.999 |
| tcmG, elmG; tetracenomycin A2 monooxygenase-dioxygenase [EC:1.14.13.200] | -1.744 | 2.098 | 0.407 | 0.978 |
| tctA; putative tricarboxylic transport membrane protein | -0.031 | 0.192 | 0.872 | 0.999 |
| tctB; putative tricarboxylic transport membrane protein | 0.024 | 0.208 | 0.908 | 0.999 |
| tctC; putative tricarboxylic transport membrane protein | -0.350 | 0.285 | 0.221 | 0.978 |
| tctD; two-component system, OmpR family, response regulator TctD | -0.109 | 0.272 | 0.690 | 0.999 |
| tctE; two-component system, OmpR family, sensor histidine kinase TctE [EC:2.7.13.3] | -0.130 | 0.283 | 0.646 | 0.999 |
| tcuR; LysR family transcriptional regulator, regulatory protein for tcuABC | -0.751 | 0.636 | 0.239 | 0.978 |
| tcyK; L-cystine transport system substrate-binding protein | -0.137 | 0.195 | 0.482 | 0.979 |
| tcyL; L-cystine transport system permease protein | -0.220 | 0.208 | 0.293 | 0.978 |
| tcyM; L-cystine transport system permease protein | -0.190 | 0.200 | 0.344 | 0.978 |
| tcyN; L-cystine transport system ATP-binding protein [EC:3.6.3.-] | -0.171 | 0.190 | 0.370 | 0.978 |
| tdcA; LysR family transcriptional regulator, tdc operon transcriptional activator | -0.243 | 0.717 | 0.735 | 0.999 |
| tdcC; threonine transporter | -0.243 | 0.717 | 0.735 | 0.999 |
| tdcD; propionate kinase [EC:2.7.2.15] | -0.243 | 0.717 | 0.735 | 0.999 |
| tdcR; threonine dehydratase operon activator protein | -2.044 | 2.163 | 0.346 | 0.978 |
| tdh; threonine 3-dehydrogenase [EC:1.1.1.103] | -0.132 | 0.316 | 0.678 | 0.999 |
| tdk, TK; thymidine kinase [EC:2.7.1.21] | -0.029 | 0.077 | 0.707 | 0.999 |
| TDO2, kynA; tryptophan 2,3-dioxygenase [EC:1.13.11.11] | -0.452 | 0.282 | 0.112 | 0.917 |
| tehA; tellurite resistance protein | 0.098 | 0.244 | 0.689 | 0.999 |
| tehB; tellurite methyltransferase [EC:2.1.1.265] | -0.027 | 0.161 | 0.866 | 0.999 |
| tenA; thiaminase (transcriptional activator TenA) [EC:3.5.99.2] | -0.195 | 0.126 | 0.123 | 0.919 |
| tenI; thiazole tautomerase (transcriptional regulator TenI) [EC:5.3.99.10] | -0.863 | 0.607 | 0.157 | 0.954 |
| terA; tellurite resistance protein TerA | -0.421 | 0.628 | 0.504 | 0.981 |
| terB; tellurite resistance protein TerB | -0.258 | 0.795 | 0.746 | 0.999 |
| terC; tellurite resistance protein TerC | -0.024 | 0.146 | 0.871 | 0.999 |
| terD; tellurium resistance protein TerD | 0.119 | 0.386 | 0.758 | 0.999 |
| terZ; tellurium resistance protein TerZ | -0.148 | 0.455 | 0.746 | 0.999 |
| tesA; acyl-CoA thioesterase I [EC:3.1.2.- 3.1.1.5] | -0.146 | 0.199 | 0.463 | 0.979 |
| tesB; acyl-CoA thioesterase II [EC:3.1.2.-] | -0.108 | 0.177 | 0.544 | 0.990 |
| tesC; thioesterase III [EC:3.1.2.-] | 0.214 | 0.374 | 0.568 | 0.995 |
| tesI; 3-oxo-5alpha-steroid 4-dehydrogenase [EC:1.3.99.5] | -1.141 | 1.220 | 0.351 | 0.978 |
| tet35; tetracycline resistance efflux pump | 0.604 | 0.998 | 0.546 | 0.990 |
| tetA; MFS transporter, DHA1 family, tetracycline resistance protein | -0.876 | 0.347 | 0.013 | 0.839 |
| tetB; MFS transporter, DHA2 family, metal-tetracycline-proton antiporter | -0.124 | 0.424 | 0.769 | 0.999 |
| tetD; AraC family transcriptional regulator, transposon Tn10 TetD protein | 0.782 | 0.755 | 0.302 | 0.978 |
| tetM, tetO; ribosomal protection tetracycline resistance protein | 0.041 | 0.221 | 0.854 | 0.999 |
| tetP_A, tet40; MFS transporter, DHA3 family, tetracycline resistance protein | 1.330 | 1.485 | 0.372 | 0.978 |
| tetR; TetR/AcrR family transcriptional regulator, tetracycline repressor protein | -0.305 | 0.802 | 0.704 | 0.999 |
| tetV; MFS transporter, DHA3 family, tetracycline resistance protein | -2.560 | 1.448 | 0.079 | 0.859 |
| tetX; tetracycline 11a-monooxygenase, tetracycline resistance protein [EC:1.14.13.231] | -2.289 | 1.769 | 0.198 | 0.970 |
| tex; protein Tex | 0.061 | 0.038 | 0.107 | 0.913 |
| tfdA; alpha-ketoglutarate-dependent 2,4-dichlorophenoxyacetate dioxygenase [EC:1.14.11.-] | -1.472 | 1.130 | 0.194 | 0.970 |
| tfdB; 2,4-dichlorophenol 6-monooxygenase [EC:1.14.13.20] | 3.234 | 2.883 | 0.264 | 0.978 |
| TFIIB, GTF2B, SUA7, tfb; transcription initiation factor TFIIB | -17.843 | 4173.859 | 0.997 | 0.999 |
| TFIIE1, GTF2E1, TFA1, tfe; transcription initiation factor TFIIE subunit alpha | -17.234 | 3077.514 | 0.996 | 0.999 |
| tfoS; AraC family transcriptional regulator, chitin signaling transcriptional activator | 18.482 | 3349.575 | 0.996 | 0.999 |
| tfoX; DNA transformation protein and related proteins | -0.121 | 0.202 | 0.551 | 0.992 |
| tfrA; fumarate reductase (CoM/CoB) subunit A [EC:1.3.4.1] | 0.985 | 1.443 | 0.496 | 0.981 |
| tfs; transcription termination factor TFS | -17.234 | 3077.514 | 0.996 | 0.999 |
| tgl; protein-glutamine gamma-glutamyltransferase [EC:2.3.2.13] | -2.783 | 1.455 | 0.058 | 0.839 |
| tgt, QTRT1; queuine tRNA-ribosyltransferase [EC:2.4.2.29] | 0.014 | 0.039 | 0.718 | 0.999 |
| tgtA; 7-cyano-7-deazaguanine tRNA-ribosyltransferase [EC:2.4.2.48] | -17.234 | 3077.514 | 0.996 | 0.999 |
| TH; tyrosine 3-monooxygenase [EC:1.14.16.2] | -17.068 | 2832.569 | 0.995 | 0.999 |
| THI4, THI1; cysteine-dependent adenosine diphosphate thiazole synthase [EC:2.4.2.60] | 0.057 | 0.377 | 0.881 | 0.999 |
| thiB, tbpA; thiamine transport system substrate-binding protein | 0.068 | 0.216 | 0.754 | 0.999 |
| thiC; phosphomethylpyrimidine synthase [EC:4.1.99.17] | 0.035 | 0.084 | 0.679 | 0.999 |
| thiD; hydroxymethylpyrimidine/phosphomethylpyrimidine kinase [EC:2.7.1.49 2.7.4.7] | 0.048 | 0.052 | 0.357 | 0.978 |
| thiDE; hydroxymethylpyrimidine kinase / phosphomethylpyrimidine kinase / thiamine-phosphate diphosphorylase [EC:2.7.1.49 2.7.4.7 2.5.1.3] | 0.335 | 0.508 | 0.511 | 0.982 |
| thiE; thiamine-phosphate pyrophosphorylase [EC:2.5.1.3] | 0.031 | 0.057 | 0.583 | 0.999 |
| thiF; sulfur carrier protein ThiS adenylyltransferase [EC:2.7.7.73] | 0.494 | 0.209 | 0.020 | 0.839 |
| thiG; thiazole synthase [EC:2.8.1.10] | 0.034 | 0.078 | 0.667 | 0.999 |
| thiH; 2-iminoacetate synthase [EC:4.1.99.19] | 0.244 | 0.171 | 0.156 | 0.954 |
| thiI; tRNA uracil 4-sulfurtransferase [EC:2.8.1.4] | 0.061 | 0.081 | 0.450 | 0.978 |
| thiJ; protein deglycase [EC:3.5.1.124] | 0.103 | 0.108 | 0.343 | 0.978 |
| thiK; thiamine kinase [EC:2.7.1.89] | 0.128 | 0.599 | 0.832 | 0.999 |
| thiL; thiamine-monophosphate kinase [EC:2.7.4.16] | -0.030 | 0.081 | 0.710 | 0.999 |
| thiM; hydroxyethylthiazole kinase [EC:2.7.1.50] | -0.063 | 0.105 | 0.549 | 0.992 |
| thiN, TPK1, THI80; thiamine pyrophosphokinase [EC:2.7.6.2] | 0.050 | 0.092 | 0.590 | 0.999 |
| thiO; glycine oxidase [EC:1.4.3.19] | -0.182 | 0.167 | 0.279 | 0.978 |
| thiP; thiamine transport system permease protein | 0.044 | 0.232 | 0.851 | 0.999 |
| thiQ; thiamine transport system ATP-binding protein | -0.002 | 0.254 | 0.994 | 0.999 |
| thiS; sulfur carrier protein | 0.026 | 0.081 | 0.754 | 0.999 |
| thiT; thiamine transporter | 0.124 | 0.152 | 0.418 | 0.978 |
| thiX; putative hydroxymethylpyrimidine transport system permease protein | -0.457 | 0.339 | 0.180 | 0.961 |
| thiY; putative hydroxymethylpyrimidine transport system substrate-binding protein | -0.462 | 0.343 | 0.180 | 0.961 |
| thiZ; putative hydroxymethylpyrimidine transport system ATP-binding protein | -0.159 | 0.388 | 0.682 | 0.999 |
| thnD; putative oxidoreductase | -17.794 | 4072.025 | 0.997 | 0.999 |
| thnE; carboxymethylproline synthase [EC:2.3.1.226] | -2.224 | 2.044 | 0.278 | 0.978 |
| THOP1; thimet oligopeptidase [EC:3.4.24.15] | -0.372 | 0.414 | 0.371 | 0.978 |
| thpR; RNA 2’,3’-cyclic 3’-phosphodiesterase [EC:3.1.4.58] | -0.386 | 0.201 | 0.056 | 0.839 |
| thrA; bifunctional aspartokinase / homoserine dehydrogenase 1 [EC:2.7.2.4 1.1.1.3] | -0.046 | 0.153 | 0.761 | 0.999 |
| thrB1; homoserine kinase [EC:2.7.1.39] | 0.016 | 0.078 | 0.838 | 0.999 |
| thrB2; homoserine kinase type II [EC:2.7.1.39] | 0.218 | 0.167 | 0.195 | 0.970 |
| thrC; threonine synthase [EC:4.2.3.1] | -0.015 | 0.044 | 0.743 | 0.999 |
| thrH; phosphoserine / homoserine phosphotransferase [EC:3.1.3.3 2.7.1.39] | -0.046 | 0.256 | 0.857 | 0.999 |
| thuE; trehalose/maltose transport system substrate-binding protein | -1.221 | 0.746 | 0.104 | 0.905 |
| thuF, sugA; trehalose/maltose transport system permease protein | -1.166 | 0.727 | 0.111 | 0.917 |
| thuG, sugB; trehalose/maltose transport system permease protein | 0.118 | 0.520 | 0.821 | 0.999 |
| thyA, TYMS; thymidylate synthase [EC:2.1.1.45] | 0.020 | 0.040 | 0.619 | 0.999 |
| thyX, thy1; thymidylate synthase (FAD) [EC:2.1.1.148] | -0.116 | 0.293 | 0.692 | 0.999 |
| tiaS; tRNA(Ile2)-agmatinylcytidine synthase [EC:6.3.4.22] | -17.234 | 3077.514 | 0.996 | 0.999 |
| tig; trigger factor | 0.042 | 0.040 | 0.284 | 0.978 |
| tilS-hprT; bifunctional protein TilS/HprT [EC:6.3.4.19 2.4.2.8] | 0.621 | 0.477 | 0.195 | 0.970 |
| tilS, mesJ; tRNA(Ile)-lysidine synthase [EC:6.3.4.19] | 0.042 | 0.036 | 0.245 | 0.978 |
| tipF; cyclic-di-GMP phosphodiesterase, flagellum assembly factor TipF | -1.013 | 0.576 | 0.081 | 0.864 |
| tisB; small toxic protein TisB | -0.191 | 0.918 | 0.836 | 0.999 |
| tldD; TldD protein | -0.089 | 0.173 | 0.605 | 0.999 |
| tlh; thermolabile hemolysin | -0.551 | 1.563 | 0.725 | 0.999 |
| tll; dTDP-6-deoxy-L-talose 4-dehydrogenase (NAD+) [EC:1.1.1.339] | 0.378 | 0.283 | 0.184 | 0.965 |
| tlp; small acid-soluble spore protein (thioredoxin-like protein) | -2.516 | 0.971 | 0.010 | 0.839 |
| tlyA; 23S rRNA (cytidine1920-2’-O)/16S rRNA (cytidine1409-2’-O)-methyltransferase [EC:2.1.1.226 2.1.1.227] | 0.079 | 0.078 | 0.312 | 0.978 |
| tlyC; putative hemolysin | 0.124 | 0.080 | 0.123 | 0.919 |
| tmcA; tRNA(Met) cytidine acetyltransferase [EC:2.3.1.193] | 0.227 | 0.307 | 0.460 | 0.979 |
| tmk, DTYMK; dTMP kinase [EC:2.7.4.9] | 0.051 | 0.037 | 0.173 | 0.961 |
| tmm; trimethylamine monooxygenase [EC:1.14.13.148] | -0.723 | 0.638 | 0.258 | 0.978 |
| tmoC, tbuB, touC; toluene monooxygenase system ferredoxin subunit | -17.298 | 3177.464 | 0.996 | 0.999 |
| tmoF, tbuC, touF; toluene monooxygenase electron transfer component [EC:1.18.1.3] | 1.513 | 1.385 | 0.276 | 0.978 |
| tnaA; tryptophanase [EC:4.1.99.1] | 0.529 | 0.281 | 0.062 | 0.839 |
| tnaB; low affinity tryptophan permease | -1.153 | 1.472 | 0.435 | 0.978 |
| toa; taurine—2-oxoglutarate transaminase [EC:2.6.1.55] | -0.145 | 0.455 | 0.751 | 0.999 |
| togA; oligogalacturonide transport system ATP-binding protein | -1.269 | 1.050 | 0.229 | 0.978 |
| togB; oligogalacturonide transport system substrate-binding protein | -1.161 | 0.727 | 0.112 | 0.917 |
| togM; oligogalacturonide transport system permease protein | -1.213 | 0.775 | 0.120 | 0.917 |
| togN; oligogalacturonide transport system permease protein | -1.213 | 0.775 | 0.120 | 0.917 |
| togT, rhiT; oligogalacturonide transporter | 0.181 | 0.601 | 0.764 | 0.999 |
| tolA; colicin import membrane protein | 0.067 | 0.115 | 0.562 | 0.992 |
| tolB; TolB protein | -0.078 | 0.148 | 0.598 | 0.999 |
| tolC; outer membrane protein | -0.227 | 0.143 | 0.114 | 0.917 |
| tolQ; biopolymer transport protein TolQ | -0.106 | 0.159 | 0.508 | 0.981 |
| tolR; biopolymer transport protein TolR | -0.134 | 0.170 | 0.430 | 0.978 |
| tomB; hha toxicity modulator TomB | 0.119 | 0.601 | 0.843 | 0.999 |
| tonB; periplasmic protein TonB | -0.010 | 0.122 | 0.933 | 0.999 |
| TOP1; DNA topoisomerase I [EC:5.99.1.2] | -17.234 | 3077.514 | 0.996 | 0.999 |
| TOP2; DNA topoisomerase II [EC:5.99.1.3] | -17.761 | 4005.858 | 0.996 | 0.999 |
| top6A; DNA topoisomerase VI subunit A [EC:5.99.1.3] | -17.132 | 2520.332 | 0.995 | 0.999 |
| top6B; DNA topoisomerase VI subunit B [EC:5.99.1.3] | -17.132 | 2520.332 | 0.995 | 0.999 |
| topA; DNA topoisomerase I [EC:5.99.1.2] | 0.030 | 0.028 | 0.273 | 0.978 |
| topB; DNA topoisomerase III [EC:5.99.1.2] | -0.057 | 0.120 | 0.635 | 0.999 |
| torA; trimethylamine-N-oxide reductase (cytochrome c) [EC:1.7.2.3] | 0.374 | 0.687 | 0.587 | 0.999 |
| torC; trimethylamine-N-oxide reductase (cytochrome c), cytochrome c-type subunit TorC | 0.391 | 0.425 | 0.360 | 0.978 |
| torD; TorA specific chaperone | 0.164 | 0.714 | 0.819 | 0.999 |
| torR; two-component system, OmpR family, torCAD operon response regulator TorR | -0.429 | 0.762 | 0.574 | 0.995 |
| torS; two-component system, OmpR family, sensor histidine kinase TorS [EC:2.7.13.3] | -0.339 | 0.810 | 0.676 | 0.999 |
| torT; periplasmic protein TorT | -0.636 | 0.760 | 0.404 | 0.978 |
| torY; trimethylamine-N-oxide reductase (cytochrome c), cytochrome c-type subunit TorY | -0.546 | 0.545 | 0.319 | 0.978 |
| torZ; trimethylamine-N-oxide reductase (cytochrome c) [EC:1.7.2.3] | -0.416 | 0.383 | 0.279 | 0.978 |
| toxR; cholera toxin transcriptional activator | 1.589 | 1.119 | 0.158 | 0.954 |
| toxS; transmembrane regulatory protein ToxS | 18.119 | 1920.327 | 0.992 | 0.999 |
| TP53RK, PRPK, BUD32; TP53 regulating kinase and related kinases [EC:2.7.11.1] | -17.234 | 3077.514 | 0.996 | 0.999 |
| tpa; taurine-pyruvate aminotransferase [EC:2.6.1.77] | -17.631 | 1845.307 | 0.992 | 0.999 |
| tphA1; terephthalate 1,2-dioxygenase reductase component [EC:1.18.1.-] | 18.417 | 3241.229 | 0.995 | 0.999 |
| tphA2; terephthalate 1,2-dioxygenase oxygenase component alpha subunit [EC:1.14.12.15] | 0.173 | 1.432 | 0.904 | 0.999 |
| tphA3; terephthalate 1,2-dioxygenase oxygenase component beta subunit [EC:1.14.12.15] | 18.417 | 3241.229 | 0.995 | 0.999 |
| tphB; 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate dehydrogenase [EC:1.3.1.53] | 0.173 | 1.432 | 0.904 | 0.999 |
| TPI, tpiA; triosephosphate isomerase (TIM) [EC:5.3.1.1] | 0.082 | 0.048 | 0.093 | 0.892 |
| TPMT, tpmT; thiopurine S-methyltransferase [EC:2.1.1.67] | 0.195 | 0.307 | 0.525 | 0.989 |
| tpr; thiol protease [EC:3.4.22.-] | -17.860 | 2336.427 | 0.994 | 0.999 |
| TPS; trehalose 6-phosphate synthase/phosphatase [EC:2.4.1.15 3.1.3.12] | -1.397 | 0.774 | 0.073 | 0.845 |
| tpx; thiol peroxidase, atypical 2-Cys peroxiredoxin [EC:1.11.1.15] | 0.030 | 0.081 | 0.712 | 0.999 |
| tqsA; AI-2 transport protein TqsA | 0.124 | 0.483 | 0.798 | 0.999 |
| TR1; tropinone reductase I [EC:1.1.1.206] | -0.046 | 0.470 | 0.923 | 0.999 |
| traA; conjugal transfer pilus assembly protein TraA | -0.017 | 1.322 | 0.990 | 0.999 |
| traB; conjugal transfer pilus assembly protein TraB | -0.053 | 0.667 | 0.936 | 0.999 |
| traC; conjugal transfer ATP-binding protein TraC | -0.314 | 0.637 | 0.623 | 0.999 |
| traD; conjugal transfer pilus assembly protein TraD | -0.731 | 1.247 | 0.559 | 0.992 |
| traE; conjugal transfer pilus assembly protein TraE | -0.043 | 0.603 | 0.943 | 0.999 |
| traF; conjugal transfer pilus assembly protein TraF | 0.212 | 0.687 | 0.758 | 0.999 |
| traG; conjugal transfer mating pair stabilization protein TraG | 0.037 | 0.622 | 0.953 | 0.999 |
| traH; conjugative transfer pilus assembly protein TraH | -0.041 | 0.644 | 0.949 | 0.999 |
| traI; conjugal transfer pilus assembly protein TraI | 2.164 | 1.740 | 0.215 | 0.978 |
| traK; conjugal transfer pilus assembly protein TraK | -0.198 | 0.648 | 0.760 | 0.999 |
| traL; conjugal transfer pilus assembly protein TraL | -0.415 | 0.692 | 0.550 | 0.992 |
| traN; conjugal transfer mating pair stabilization protein TraN | 0.583 | 0.614 | 0.344 | 0.978 |
| traR; LuxR family transcriptional regulator, activator of conjugal transfer of Ti plasmids | -3.552 | 4.274 | 0.407 | 0.978 |
| traU; conjugal transfer pilus assembly protein TraU | -0.422 | 0.650 | 0.517 | 0.986 |
| traV; conjugal transfer pilus assembly protein TraV | -0.313 | 0.629 | 0.619 | 0.999 |
| traW; conjugal transfer pilus assembly protein TraW | -0.192 | 0.660 | 0.772 | 0.999 |
| trbC; conjugal transfer pilus assembly protein TrbC | 0.304 | 0.635 | 0.633 | 0.999 |
| trbI; conjugal transfer pilin signal peptidase TrbI | 0.057 | 0.705 | 0.936 | 0.999 |
| trbL; type IV secretion system protein TrbL | -0.035 | 0.339 | 0.917 | 0.999 |
| trcR; two-component system, OmpR family, response regulator TrcR | -3.513 | 2.411 | 0.147 | 0.954 |
| trcS; two-component system, OmpR family, sensor histidine kinase TrcS [EC:2.7.13.3] | -17.879 | 2083.188 | 0.993 | 0.999 |
| treC; trehalose-6-phosphate hydrolase [EC:3.2.1.93] | 0.106 | 0.124 | 0.396 | 0.978 |
| TREH, treA, treF; alpha,alpha-trehalase [EC:3.2.1.28] | -0.376 | 0.443 | 0.397 | 0.978 |
| treR; LacI family transcriptional regulator, trehalose operon repressor | 0.374 | 0.568 | 0.511 | 0.982 |
| treR2, treR; GntR family transcriptional regulator, trehalose operon transcriptional repressor | 0.109 | 0.117 | 0.352 | 0.978 |
| treS; maltose alpha-D-glucosyltransferase / alpha-amylase [EC:5.4.99.16 3.2.1.1] | -0.066 | 0.268 | 0.806 | 0.999 |
| treT; trehalose synthase [EC:2.4.1.245] | -0.512 | 0.508 | 0.315 | 0.978 |
| treY, glgY; (1->4)-alpha-D-glucan 1-alpha-D-glucosylmutase [EC:5.4.99.15] | -0.122 | 0.258 | 0.638 | 0.999 |
| treZ, glgZ; maltooligosyltrehalose trehalohydrolase [EC:3.2.1.141] | -0.132 | 0.259 | 0.612 | 0.999 |
| trg; methyl-accepting chemotaxis protein III, ribose and galactose sensor receptor | -0.114 | 0.655 | 0.862 | 0.999 |
| tri; tricorn protease [EC:3.4.21.-] | -0.758 | 0.593 | 0.203 | 0.972 |
| trkA, ktrA; trk system potassium uptake protein | 0.066 | 0.066 | 0.316 | 0.978 |
| trkH, trkG, ktrB; trk system potassium uptake protein | 0.104 | 0.075 | 0.165 | 0.960 |
| TRM5, TRMT5; tRNA (guanine37-N1)-methyltransferase [EC:2.1.1.228] | -17.234 | 3077.514 | 0.996 | 0.999 |
| TRM61, GCD14; tRNA (adenine57-N1/adenine58-N1)-methyltransferase catalytic subunit [EC:2.1.1.219 2.1.1.220] | 0.064 | 0.248 | 0.796 | 0.999 |
| trmA; tRNA (uracil-5-)-methyltransferase [EC:2.1.1.35] | 0.053 | 0.160 | 0.742 | 0.999 |
| trmB, METTL1; tRNA (guanine-N7-)-methyltransferase [EC:2.1.1.33] | 0.044 | 0.038 | 0.247 | 0.978 |
| trmD; tRNA (guanine37-N1)-methyltransferase [EC:2.1.1.228] | 0.040 | 0.036 | 0.266 | 0.978 |
| trmFO, gid; methylenetetrahydrofolate–tRNA-(uracil-5-)-methyltransferase [EC:2.1.1.74] | 0.041 | 0.094 | 0.667 | 0.999 |
| trmH; tRNA (guanosine-2’-O-)-methyltransferase [EC:2.1.1.34] | -0.314 | 0.335 | 0.350 | 0.978 |
| trmJ; tRNA (cytidine32/uridine32-2’-O)-methyltransferase [EC:2.1.1.200] | 0.105 | 0.172 | 0.543 | 0.990 |
| trmK; tRNA (adenine22-N1)-methyltransferase [EC:2.1.1.217] | 0.051 | 0.086 | 0.553 | 0.992 |
| trmL, cspR; tRNA (cytidine/uridine-2’-O-)-methyltransferase [EC:2.1.1.207] | 0.075 | 0.035 | 0.036 | 0.839 |
| TRMT1, trm1; tRNA (guanine26-N2/guanine27-N2)-dimethyltransferase [EC:2.1.1.215 2.1.1.216] | -17.234 | 3077.514 | 0.996 | 0.999 |
| trmY; tRNA (pseudouridine54-N1)-methyltransferase [EC:2.1.1.257] | 18.119 | 1920.327 | 0.992 | 0.999 |
| troA, mntA, znuA; manganese/zinc/iron transport system substrate-binding protein | 0.726 | 0.296 | 0.015 | 0.839 |
| troB, mntB, znuC; manganese/zinc/iron transport system ATP- binding protein | 0.630 | 0.287 | 0.030 | 0.839 |
| troC, mntC, znuB; manganese/zinc/iron transport system permease protein | 0.749 | 0.309 | 0.016 | 0.839 |
| troD, mntD, znuB; manganese/zinc/iron transport system permease protein | 0.680 | 0.335 | 0.044 | 0.839 |
| troR; DtxR family transcriptional regulator, Mn-dependent transcriptional regulator | 0.068 | 0.118 | 0.565 | 0.995 |
| TROVE2, SSA2; 60 kDa SS-A/Ro ribonucleoprotein | 0.047 | 0.480 | 0.922 | 0.999 |
| TRP1; anthranilate synthase / indole-3-glycerol phosphate synthase / phosphoribosylanthranilate isomerase [EC:4.1.3.27 4.1.1.48 5.3.1.24] | -2.531 | 2.020 | 0.212 | 0.978 |
| trpA; tryptophan synthase alpha chain [EC:4.2.1.20] | -0.072 | 0.055 | 0.194 | 0.970 |
| trpB; tryptophan synthase beta chain [EC:4.2.1.20] | -0.041 | 0.060 | 0.489 | 0.979 |
| trpC; indole-3-glycerol phosphate synthase [EC:4.1.1.48] | -0.118 | 0.063 | 0.065 | 0.839 |
| trpCF; indole-3-glycerol phosphate synthase / phosphoribosylanthranilate isomerase [EC:4.1.1.48 5.3.1.24] | 0.058 | 0.204 | 0.776 | 0.999 |
| trpD; anthranilate phosphoribosyltransferase [EC:2.4.2.18] | -0.083 | 0.057 | 0.145 | 0.951 |
| trpE; anthranilate synthase component I [EC:4.1.3.27] | -0.075 | 0.056 | 0.178 | 0.961 |
| trpEG; anthranilate synthase [EC:4.1.3.27] | -2.132 | 0.940 | 0.025 | 0.839 |
| trpF; phosphoribosylanthranilate isomerase [EC:5.3.1.24] | -0.112 | 0.073 | 0.128 | 0.927 |
| trpG; anthranilate synthase component II [EC:4.1.3.27] | -0.064 | 0.059 | 0.274 | 0.978 |
| trpGD; anthranilate synthase/phosphoribosyltransferase [EC:4.1.3.27 2.4.2.18] | 0.012 | 0.335 | 0.972 | 0.999 |
| trpR; TrpR family transcriptional regulator, trp operon repressor | 0.205 | 0.302 | 0.498 | 0.981 |
| truA, PUS1; tRNA pseudouridine38-40 synthase [EC:5.4.99.12] | 0.033 | 0.037 | 0.369 | 0.978 |
| truB, PUS4, TRUB1; tRNA pseudouridine55 synthase [EC:5.4.99.25] | 0.043 | 0.033 | 0.196 | 0.970 |
| truC; tRNA pseudouridine65 synthase [EC:5.4.99.26] | 0.106 | 0.215 | 0.622 | 0.999 |
| truD, PUS7; tRNA pseudouridine13 synthase [EC:5.4.99.27] | 0.093 | 0.177 | 0.603 | 0.999 |
| trxA; thioredoxin 1 | 0.019 | 0.040 | 0.630 | 0.999 |
| trxB, TRR; thioredoxin reductase (NADPH) [EC:1.8.1.9] | 0.006 | 0.046 | 0.892 | 0.999 |
| trxC; thioredoxin 2 [EC:1.8.1.8] | -0.007 | 0.229 | 0.974 | 0.999 |
| tsaB; tRNA threonylcarbamoyladenosine biosynthesis protein TsaB | -0.057 | 0.086 | 0.507 | 0.981 |
| tsaC, rimN, SUA5; L-threonylcarbamoyladenylate synthase [EC:2.7.7.87] | 0.037 | 0.041 | 0.362 | 0.978 |
| tsaE; tRNA threonylcarbamoyladenosine biosynthesis protein TsaE | 0.053 | 0.038 | 0.166 | 0.960 |
| tsdA; thiosulfate dehydrogenase [EC:1.8.2.2] | -0.097 | 0.277 | 0.726 | 0.999 |
| tsf, TSFM; elongation factor Ts | 0.036 | 0.036 | 0.321 | 0.978 |
| tsgA; MFS transporter, TsgA protein | 0.232 | 0.577 | 0.688 | 0.999 |
| tspA; uncharacterized membrane protein | 0.044 | 0.667 | 0.948 | 0.999 |
| tsr; methyl-accepting chemotaxis protein I, serine sensor receptor | -0.357 | 0.355 | 0.316 | 0.978 |
| TSR3; pre-rRNA-processing protein TSR3 | -17.234 | 3077.514 | 0.996 | 0.999 |
| TST, MPST, sseA; thiosulfate/3-mercaptopyruvate sulfurtransferase [EC:2.8.1.1 2.8.1.2] | -0.200 | 0.126 | 0.115 | 0.917 |
| TSTA3, fcl; GDP-L-fucose synthase [EC:1.1.1.271] | -0.350 | 0.186 | 0.062 | 0.839 |
| tsx; nucleoside-specific channel-forming protein | -0.161 | 0.470 | 0.733 | 0.999 |
| ttcA; tRNA 2-thiocytidine biosynthesis protein TtcA | -0.072 | 0.155 | 0.641 | 0.999 |
| ttdA; L(+)-tartrate dehydratase alpha subunit [EC:4.2.1.32] | 0.174 | 0.233 | 0.457 | 0.979 |
| ttdB; L(+)-tartrate dehydratase beta subunit [EC:4.2.1.32] | 0.170 | 0.232 | 0.465 | 0.979 |
| ttdT; L-tartrate/succinate antiporter | 0.112 | 0.255 | 0.660 | 0.999 |
| TTN; titin [EC:2.7.11.1] | -17.833 | 2929.773 | 0.995 | 0.999 |
| ttr; acetyltransferase [EC:2.3.1.-] | -0.361 | 0.480 | 0.454 | 0.978 |
| ttrA; tetrathionate reductase subunit A | -0.054 | 0.519 | 0.917 | 0.999 |
| ttrB; tetrathionate reductase subunit B | 0.036 | 0.602 | 0.953 | 0.999 |
| ttrC; tetrathionate reductase subunit C | -0.297 | 0.704 | 0.673 | 0.999 |
| ttrR; two-component system, LuxR family, response regulator TtrR | -2.922 | 2.545 | 0.253 | 0.978 |
| ttrS; two-component system, LuxR family, sensor histidine kinase TtrS [EC:2.7.13.3] | -3.710 | 4.059 | 0.362 | 0.978 |
| ttuB; MFS transporter, ACS family, tartrate transporter | 0.351 | 0.726 | 0.630 | 0.999 |
| ttuC, dmlA; tartrate dehydrogenase/decarboxylase / D-malate dehydrogenase [EC:1.1.1.93 4.1.1.73 1.1.1.83] | -0.750 | 0.373 | 0.046 | 0.839 |
| ttuD; hydroxypyruvate reductase [EC:1.1.1.81] | -0.279 | 0.240 | 0.247 | 0.978 |
| tuaB; teichuronic acid exporter | 1.652 | 0.900 | 0.068 | 0.839 |
| tuaC; teichuronic acid biosynthesis glycosyltransferase TuaC [EC:2.4.-.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| tuaE; teichuronic acid biosynthesis protein TuaE | -1.062 | 1.413 | 0.454 | 0.978 |
| tuaF; teichuronic acid biosynthesis protein TuaF | 18.239 | 2966.075 | 0.995 | 0.999 |
| tuaG; teichuronic acid biosynthesis glycosyltransferase TuaG [EC:2.4.-.-] | -0.725 | 0.464 | 0.121 | 0.919 |
| tuaH; teichuronic acid biosynthesis glycosyltransferase TuaH [EC:2.4.-.-] | 3.201 | 2.664 | 0.231 | 0.978 |
| tuf, TUFM; elongation factor Tu | 0.032 | 0.034 | 0.353 | 0.978 |
| tupA, vupA; tungstate transport system substrate-binding protein | -0.306 | 0.451 | 0.499 | 0.981 |
| tupB, vupB; tungstate transport system permease protein | -0.262 | 0.466 | 0.574 | 0.995 |
| tupC, vupC; tungstate transport system ATP-binding protein [EC:3.6.3.55] | -0.026 | 0.530 | 0.961 | 0.999 |
| tus, tau; DNA replication terminus site-binding protein | 0.141 | 0.595 | 0.813 | 0.999 |
| tusA, sirA; tRNA 2-thiouridine synthesizing protein A [EC:2.8.1.-] | -0.077 | 0.174 | 0.661 | 0.999 |
| tusB, dsrH; tRNA 2-thiouridine synthesizing protein B | 0.234 | 0.218 | 0.285 | 0.978 |
| tusC, dsrF; tRNA 2-thiouridine synthesizing protein C | 0.227 | 0.212 | 0.287 | 0.978 |
| tusD, dsrE; tRNA 2-thiouridine synthesizing protein D [EC:2.8.1.-] | 0.150 | 0.192 | 0.436 | 0.978 |
| tusE, dsrC; tRNA 2-thiouridine synthesizing protein E [EC:2.8.1.-] | 0.149 | 0.192 | 0.439 | 0.978 |
| tutB; tyrosine permease | -1.630 | 2.316 | 0.483 | 0.979 |
| tycC; tyrocidine synthetase III | -17.761 | 4005.858 | 0.996 | 0.999 |
| tylC, oleB, carA, srmB; macrolide transport system ATP-binding/permease protein | -0.229 | 0.485 | 0.637 | 0.999 |
| typA, bipA; GTP-binding protein | 0.037 | 0.036 | 0.310 | 0.978 |
| TYR; tyrosinase [EC:1.14.18.1] | 0.252 | 0.728 | 0.730 | 0.999 |
| tyrA; chorismate mutase / prephenate dehydrogenase [EC:5.4.99.5 1.3.1.12] | 0.238 | 0.308 | 0.440 | 0.978 |
| tyrA1; chorismate mutase [EC:5.4.99.5] | -0.141 | 0.203 | 0.489 | 0.979 |
| tyrA2; prephenate dehydrogenase [EC:1.3.1.12] | -0.028 | 0.076 | 0.714 | 0.999 |
| tyrB; aromatic-amino-acid transaminase [EC:2.6.1.57] | 0.180 | 0.114 | 0.115 | 0.917 |
| tyrC; cyclohexadieny/prephenate dehydrogenase [EC:1.3.1.43 1.3.1.12] | -1.385 | 0.492 | 0.005 | 0.839 |
| tyrP; tyrosine-specific transport protein | 0.218 | 0.328 | 0.508 | 0.981 |
| tyrR; transcriptional regulator of aroF, aroG, tyrA and aromatic amino acid transport | 0.186 | 0.214 | 0.385 | 0.978 |
| TYW1; tRNA wybutosine-synthesizing protein 1 [EC:4.1.3.44] | -17.234 | 3077.514 | 0.996 | 0.999 |
| UAH; ureidoglycolate amidohydrolase [EC:3.5.1.116] | 0.439 | 1.410 | 0.756 | 0.999 |
| UAP1; UDP-N-acetylglucosamine/UDP-N-acetylgalactosamine diphosphorylase [EC:2.7.7.23 2.7.7.83] | -1.564 | 0.892 | 0.082 | 0.865 |
| uaZ; urate oxidase [EC:1.7.3.3] | -1.033 | 0.721 | 0.154 | 0.954 |
| ubiA; 4-hydroxybenzoate polyprenyltransferase [EC:2.5.1.39] | -0.015 | 0.089 | 0.865 | 0.999 |
| ubiB, aarF; ubiquinone biosynthesis protein | -0.049 | 0.115 | 0.671 | 0.999 |
| ubiC; chorismate–pyruvate lyase [EC:4.1.3.40] | 0.198 | 0.214 | 0.356 | 0.978 |
| ubiD; 4-hydroxy-3-polyprenylbenzoate decarboxylase [EC:4.1.1.98] | 0.043 | 0.155 | 0.781 | 0.999 |
| ubiE; demethylmenaquinone methyltransferase / 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase [EC:2.1.1.163 2.1.1.201] | -0.023 | 0.066 | 0.730 | 0.999 |
| ubiF; 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [EC:1.14.13.-] | 0.106 | 0.264 | 0.689 | 0.999 |
| ubiG; 2-polyprenyl-6-hydroxyphenyl methylase / 3-demethylubiquinone-9 3-methyltransferase [EC:2.1.1.222 2.1.1.64] | -0.061 | 0.167 | 0.717 | 0.999 |
| ubiH; 2-octaprenyl-6-methoxyphenol hydroxylase [EC:1.14.13.-] | -0.119 | 0.162 | 0.466 | 0.979 |
| ubiI; 2-octaprenylphenol hydroxylase [EC:1.14.13.-] | 0.192 | 0.242 | 0.429 | 0.978 |
| ubiJ; ubiquinone biosynthesis protein UbiJ | 0.081 | 0.161 | 0.615 | 0.999 |
| ubiX, bsdB, PAD1; flavin prenyltransferase [EC:2.5.1.129] | 0.063 | 0.150 | 0.675 | 0.999 |
| UBLCP1; ubiquitin-like domain-containing CTD phosphatase 1 [EC:3.1.3.16] | -17.404 | 2813.939 | 0.995 | 0.999 |
| uctC; CoA:oxalate CoA-transferase [EC:2.8.3.19] | 0.072 | 0.508 | 0.888 | 0.999 |
| udh; uronate dehydrogenase [EC:1.1.1.203] | -0.186 | 0.329 | 0.572 | 0.995 |
| udk, UCK; uridine kinase [EC:2.7.1.48] | -0.006 | 0.091 | 0.949 | 0.999 |
| udp, UPP; uridine phosphorylase [EC:2.4.2.3] | -0.016 | 0.171 | 0.927 | 0.999 |
| UGCG; ceramide glucosyltransferase [EC:2.4.1.80] | 1.248 | 0.562 | 0.028 | 0.839 |
| UGDH, ugd; UDPglucose 6-dehydrogenase [EC:1.1.1.22] | -0.157 | 0.098 | 0.110 | 0.917 |
| ugl; unsaturated chondroitin disaccharide hydrolase [EC:3.2.1.180] | 0.047 | 0.381 | 0.901 | 0.999 |
| ugl; ureidoglycolate lyase [EC:4.3.2.3] | -0.145 | 0.728 | 0.842 | 0.999 |
| UGP2, galU, galF; UTP–glucose-1-phosphate uridylyltransferase [EC:2.7.7.9] | 0.029 | 0.040 | 0.468 | 0.979 |
| ugpA; sn-glycerol 3-phosphate transport system permease protein | -0.964 | 0.329 | 0.004 | 0.839 |
| ugpB; sn-glycerol 3-phosphate transport system substrate-binding protein | -0.977 | 0.334 | 0.004 | 0.839 |
| ugpC; sn-glycerol 3-phosphate transport system ATP-binding protein [EC:3.6.3.20] | -1.001 | 0.362 | 0.006 | 0.839 |
| ugpE; sn-glycerol 3-phosphate transport system permease protein | -0.965 | 0.331 | 0.004 | 0.839 |
| ugtP; processive 1,2-diacylglycerol beta-glucosyltransferase [EC:2.4.1.315] | 0.072 | 0.195 | 0.713 | 0.999 |
| uhpA; two-component system, NarL family, uhpT operon response regulator UhpA | -0.004 | 0.461 | 0.993 | 0.999 |
| uhpB; two-component system, NarL family, sensor histidine kinase UhpB [EC:2.7.13.3] | 0.125 | 0.336 | 0.711 | 0.999 |
| uhpC; MFS transporter, OPA family, sugar phosphate sensor protein UhpC | 0.529 | 0.459 | 0.252 | 0.978 |
| uhpT; MFS transporter, OPA family, hexose phosphate transport protein UhpT | -0.728 | 0.580 | 0.212 | 0.978 |
| uidA, GUSB; beta-glucuronidase [EC:3.2.1.31] | 0.110 | 0.225 | 0.626 | 0.999 |
| uidB, gusB; glucuronide carrier protein | -1.956 | 1.309 | 0.137 | 0.949 |
| uidC, gusC; putative glucuronide porin | -1.153 | 1.472 | 0.435 | 0.978 |
| uidR; TetR/AcrR family transcriptional regulator, repressor for uid operon | -2.240 | 2.580 | 0.387 | 0.978 |
| ulaD, sgaH, sgbH; 3-dehydro-L-gulonate-6-phosphate decarboxylase [EC:4.1.1.85] | 0.214 | 0.335 | 0.523 | 0.989 |
| ulaE, sgaU, sgbU; L-ribulose-5-phosphate 3-epimerase [EC:5.1.3.22] | 0.428 | 0.378 | 0.260 | 0.978 |
| ulaG; L-ascorbate 6-phosphate lactonase [EC:3.1.1.-] | 0.174 | 0.166 | 0.296 | 0.978 |
| ulaR; DeoR family transcriptional regulator, ulaG and ulaABCDEF operon transcriptional repressor | 0.071 | 0.339 | 0.834 | 0.999 |
| UMF1; MFS transporter, UMF1 family | -0.197 | 0.207 | 0.341 | 0.978 |
| UMF2; MFS transporter, UMF2 family, putative MFS family transporter protein | 0.146 | 0.617 | 0.813 | 0.999 |
| UMPS; uridine monophosphate synthetase [EC:2.4.2.10 4.1.1.23] | -0.571 | 0.758 | 0.452 | 0.978 |
| umuC; DNA polymerase V | -0.005 | 0.084 | 0.951 | 0.999 |
| umuD; DNA polymerase V [EC:3.4.21.-] | 0.017 | 0.261 | 0.949 | 0.999 |
| UNG, UDG; uracil-DNA glycosylase [EC:3.2.2.27] | 0.057 | 0.046 | 0.221 | 0.978 |
| upp, UPRT; uracil phosphoribosyltransferase [EC:2.4.2.9] | 0.044 | 0.038 | 0.247 | 0.978 |
| uppS, cpdS; tritrans,polycis-undecaprenyl-diphosphate synthase [geranylgeranyl-diphosphate specific] [EC:2.5.1.89] | -17.234 | 3077.514 | 0.996 | 0.999 |
| uppS; undecaprenyl diphosphate synthase [EC:2.5.1.31] | 0.043 | 0.036 | 0.232 | 0.978 |
| UQCRFS1, RIP1, petA; ubiquinol-cytochrome c reductase iron-sulfur subunit [EC:1.10.2.2] | -0.126 | 0.180 | 0.484 | 0.979 |
| URA4, pyrC; dihydroorotase [EC:3.5.2.3] | -0.016 | 0.027 | 0.564 | 0.994 |
| uraH, pucM, hiuH; 5-hydroxyisourate hydrolase [EC:3.5.2.17] | -0.229 | 0.236 | 0.333 | 0.978 |
| urdA; urocanate reductase [EC:1.3.99.33] | -0.635 | 0.961 | 0.510 | 0.981 |
| URE; urease [EC:3.5.1.5] | -2.051 | 1.421 | 0.151 | 0.954 |
| ureA; urease subunit gamma [EC:3.5.1.5] | -0.436 | 0.238 | 0.069 | 0.841 |
| ureAB; urease subunit gamma/beta [EC:3.5.1.5] | -0.725 | 0.707 | 0.307 | 0.978 |
| ureB; urease subunit beta [EC:3.5.1.5] | -0.395 | 0.244 | 0.107 | 0.913 |
| ureC; urease subunit alpha [EC:3.5.1.5] | -0.508 | 0.231 | 0.029 | 0.839 |
| ureD, ureH; urease accessory protein | -0.502 | 0.232 | 0.032 | 0.839 |
| ureE; urease accessory protein | -0.497 | 0.236 | 0.037 | 0.839 |
| ureF; urease accessory protein | -0.533 | 0.232 | 0.023 | 0.839 |
| ureG; urease accessory protein | -0.535 | 0.232 | 0.023 | 0.839 |
| ureI; acid-activated urea channel | -1.540 | 1.402 | 0.274 | 0.978 |
| ureJ; urease accessory protein | -0.127 | 0.272 | 0.641 | 0.999 |
| urtA; urea transport system substrate-binding protein | -0.327 | 0.247 | 0.187 | 0.966 |
| urtB; urea transport system permease protein | -0.301 | 0.248 | 0.228 | 0.978 |
| urtC; urea transport system permease protein | -0.301 | 0.248 | 0.227 | 0.978 |
| urtD; urea transport system ATP-binding protein | -0.300 | 0.248 | 0.228 | 0.978 |
| urtE; urea transport system ATP-binding protein | -0.307 | 0.249 | 0.219 | 0.978 |
| ushA; 5’-nucleotidase / UDP-sugar diphosphatase [EC:3.1.3.5 3.6.1.45] | 0.225 | 0.336 | 0.505 | 0.981 |
| uspA; universal stress protein A | 0.199 | 0.209 | 0.343 | 0.978 |
| uspB; universal stress protein B | 0.210 | 0.578 | 0.716 | 0.999 |
| uspC; universal stress protein C | 0.135 | 0.629 | 0.830 | 0.999 |
| uspE; universal stress protein E | 0.281 | 0.337 | 0.406 | 0.978 |
| uspF; universal stress protein F | 0.149 | 0.765 | 0.846 | 0.999 |
| uspG; universal stress protein G | 0.158 | 0.600 | 0.793 | 0.999 |
| utp; urea transporter | -0.213 | 0.282 | 0.452 | 0.978 |
| uup; ABC transport system ATP-binding/permease protein | -0.033 | 0.049 | 0.509 | 0.981 |
| uvrA; excinuclease ABC subunit A | 0.001 | 0.034 | 0.987 | 0.999 |
| uvrB; excinuclease ABC subunit B | 0.033 | 0.033 | 0.309 | 0.978 |
| uvrC; excinuclease ABC subunit C | 0.032 | 0.031 | 0.306 | 0.978 |
| uvrD, pcrA; DNA helicase II / ATP-dependent DNA helicase PcrA [EC:3.6.4.12] | 0.036 | 0.051 | 0.478 | 0.979 |
| uvrY, gacA, varA; two-component system, NarL family, invasion response regulator UvrY | 0.146 | 0.398 | 0.714 | 0.999 |
| uvsE, UVE1; UV DNA damage endonuclease [EC:3.-.-.-] | -0.342 | 0.702 | 0.627 | 0.999 |
| uxaA; altronate hydrolase [EC:4.2.1.7] | -0.063 | 0.228 | 0.783 | 0.999 |
| uxaA1; altronate dehydratase small subunit [EC:4.2.1.7] | -0.113 | 1.484 | 0.939 | 0.999 |
| uxaA2; altronate dehydratase large subunit [EC:4.2.1.7] | 0.169 | 0.270 | 0.533 | 0.990 |
| uxaB; tagaturonate reductase [EC:1.1.1.58] | -0.117 | 0.311 | 0.707 | 0.999 |
| uxaC; glucuronate isomerase [EC:5.3.1.12] | -0.177 | 0.261 | 0.499 | 0.981 |
| UXE, uxe; UDP-arabinose 4-epimerase [EC:5.1.3.5] | -1.276 | 1.013 | 0.210 | 0.978 |
| UXS1, uxs; UDP-glucuronate decarboxylase [EC:4.1.1.35] | -1.224 | 0.337 | 0.000 | 0.839 |
| uxuA; mannonate dehydratase [EC:4.2.1.8] | -0.239 | 0.256 | 0.353 | 0.978 |
| uxuB; fructuronate reductase [EC:1.1.1.57] | -0.019 | 0.268 | 0.943 | 0.999 |
| uxuR; GntR family transcriptional regulator, uxu operon transcriptional repressor | -0.263 | 0.494 | 0.596 | 0.999 |
| vacA; vacuolating cytotoxin | -1.386 | 1.551 | 0.373 | 0.978 |
| vanA; vanillate monooxygenase [EC:1.14.13.82] | -0.340 | 0.309 | 0.273 | 0.978 |
| vanB, vanA, vanD; D-alanine—(R)-lactate ligase [EC:6.1.2.1] | 17.929 | 2539.480 | 0.994 | 0.999 |
| vanB; vanillate monooxygenase ferredoxin subunit | -0.152 | 0.305 | 0.620 | 0.999 |
| vanC, vanE, vanG; D-alanine—D-serine ligase [EC:6.3.2.35] | -17.610 | 2186.533 | 0.994 | 0.999 |
| vanH; D-specific alpha-keto acid dehydrogenase [EC:1.1.1.-] | 17.929 | 2539.480 | 0.994 | 0.999 |
| vanJ; vancomycin resistance protein VanJ | -0.265 | 1.301 | 0.839 | 0.999 |
| vanK; vancomycin resistance protein VanK | 0.299 | 1.184 | 0.801 | 0.999 |
| vanR; GntR family transcriptional regulator, vanillate catabolism transcriptional regulator | -0.421 | 0.358 | 0.241 | 0.978 |
| vanRAc; two-component system, OmpR family, response regulator VanR | -0.004 | 0.523 | 0.994 | 0.999 |
| vanRB, vanR, vanRD; two-component system, OmpR family, response regulator VanR | -0.768 | 0.946 | 0.418 | 0.978 |
| vanRC, vanRE, vanRG; two-component system, OmpR family, response regulator VanR | -0.645 | 0.846 | 0.447 | 0.978 |
| vanSAc; two-component system, OmpR family, sensor histidine kinase VanS [EC:2.7.13.3] | 0.022 | 0.528 | 0.967 | 0.999 |
| vanSB, vanS, vanSD; two-component system, OmpR family, sensor histidine kinase VanS [EC:2.7.13.3] | -0.904 | 1.195 | 0.450 | 0.978 |
| vanSC, vanSE, vanSG; two-component system, OmpR family, sensor histidine kinase VanS [EC:2.7.13.3] | -0.980 | 0.894 | 0.275 | 0.978 |
| vanT; serine/alanine racemase [EC:5.1.1.18 5.1.1.1] | -17.710 | 2526.078 | 0.994 | 0.999 |
| vanW; vancomycin resistance protein VanW | -0.688 | 0.591 | 0.246 | 0.978 |
| vanX; zinc D-Ala-D-Ala dipeptidase [EC:3.4.13.22] | -0.282 | 0.159 | 0.079 | 0.859 |
| vanXY; zinc D-Ala-D-Ala dipeptidase/carboxypeptidase [EC:3.4.13.22 3.4.17.14] | -17.473 | 3469.327 | 0.996 | 0.999 |
| vanY; zinc D-Ala-D-Ala carboxypeptidase [EC:3.4.17.14] | -0.043 | 0.127 | 0.733 | 0.999 |
| vapB; antitoxin VapB | -0.200 | 0.592 | 0.736 | 0.999 |
| vapC; ribonuclease VapC [EC:3.1.-.-] | -0.525 | 0.561 | 0.351 | 0.978 |
| VARS, valS; valyl-tRNA synthetase [EC:6.1.1.9] | 0.026 | 0.030 | 0.392 | 0.978 |
| vasD, lip; type VI secretion system protein VasD | 0.248 | 0.269 | 0.359 | 0.978 |
| vasG, clpV; type VI secretion system protein VasG | 0.121 | 0.258 | 0.641 | 0.999 |
| vasH; sigma-54 dependent transcriptional regulator | -0.655 | 1.665 | 0.695 | 0.999 |
| vasI; type VI secretion system protein VasI | 0.031 | 0.546 | 0.955 | 0.999 |
| vasJ; type VI secretion system protein VasJ | 0.347 | 0.242 | 0.154 | 0.954 |
| vasL; type VI secretion system protein VasL | 0.162 | 0.607 | 0.790 | 0.999 |
| vat; virginiamycin A acetyltransferase [EC:2.3.1.-] | -1.569 | 0.709 | 0.028 | 0.839 |
| vcaM; ATP-binding cassette, subfamily B, multidrug efflux pump | -0.023 | 0.216 | 0.916 | 0.999 |
| VCP, CDC48; transitional endoplasmic reticulum ATPase | -0.651 | 0.404 | 0.109 | 0.917 |
| vdh; valine dehydrogenase (NAD+) [EC:1.4.1.23] | -0.704 | 1.245 | 0.572 | 0.995 |
| vexE; membrane fusion protein, multidrug efflux system | 0.060 | 0.964 | 0.950 | 0.999 |
| vexF; multidrug efflux pump | 0.060 | 0.964 | 0.950 | 0.999 |
| vga; pleuromutilin/lincosamide/streptogramin A transport system ATP-binding/permease protein | 0.591 | 0.952 | 0.535 | 0.990 |
| vgb; virginiamycin B lyase [EC:4.2.99.-] | -2.497 | 1.252 | 0.048 | 0.839 |
| vgrG; type VI secretion system secreted protein VgrG | 0.147 | 0.219 | 0.502 | 0.981 |
| VGSC; voltage-gated sodium channel | -1.439 | 0.812 | 0.078 | 0.859 |
| vicK; two-component system, OmpR family, sensor histidine kinase VicK [EC:2.7.13.3] | -0.005 | 0.132 | 0.967 | 0.999 |
| vicR; two-component system, OmpR family, response regulator VicR | -0.018 | 0.131 | 0.894 | 0.999 |
| vieA; c-di-GMP phosphodiesterase [EC:3.1.4.52] | 18.902 | 3402.727 | 0.996 | 0.999 |
| vioO; nonribosomal peptide synthetase protein VioO | -17.368 | 3290.703 | 0.996 | 0.999 |
| virB1; type IV secretion system protein VirB1 | -0.010 | 0.405 | 0.980 | 0.999 |
| virB10, lvhB10; type IV secretion system protein VirB10 | -0.156 | 0.292 | 0.593 | 0.999 |
| virB11, lvhB11; type IV secretion system protein VirB11 | -0.099 | 0.305 | 0.746 | 0.999 |
| virB2, lvhB2; type IV secretion system protein VirB2 | -0.162 | 0.367 | 0.660 | 0.999 |
| virB3, lvhB3; type IV secretion system protein VirB3 | -0.282 | 0.354 | 0.426 | 0.978 |
| virB4, lvhB4; type IV secretion system protein VirB4 | 0.067 | 0.249 | 0.789 | 0.999 |
| virB5, lvhB5; type IV secretion system protein VirB5 | -0.121 | 0.276 | 0.663 | 0.999 |
| virB6, lvhB6; type IV secretion system protein VirB6 | -0.085 | 0.298 | 0.775 | 0.999 |
| virB7, lvhB7; type IV secretion system protein VirB7 | -1.961 | 1.603 | 0.223 | 0.978 |
| virB8, lvhB8; type IV secretion system protein VirB8 | -0.005 | 0.329 | 0.988 | 0.999 |
| virB9, lvhB9; type IV secretion system protein VirB9 | -0.087 | 0.265 | 0.743 | 0.999 |
| virD4, lvhD4; type IV secretion system protein VirD4 | 0.115 | 0.224 | 0.609 | 0.999 |
| virK; uncharacterized protein | 0.182 | 0.197 | 0.356 | 0.978 |
| vpr; minor extracellular serine protease Vpr [EC:3.4.21.-] | -1.683 | 0.822 | 0.042 | 0.839 |
| vraF; cationic antimicrobial peptide transport system ATP-binding protein | -0.811 | 1.001 | 0.419 | 0.978 |
| vraG; cationic antimicrobial peptide transport system permease protein | -1.554 | 1.110 | 0.164 | 0.957 |
| vraR; two-component system, NarL family, vancomycin resistance associated response regulator VraR | 0.418 | 0.537 | 0.437 | 0.978 |
| vraS; two-component system, NarL family, vancomycin resistance sensor histidine kinase VraS [EC:2.7.13.3] | -0.315 | 0.696 | 0.651 | 0.999 |
| vsr; DNA mismatch endonuclease, patch repair protein [EC:3.1.-.-] | -0.270 | 0.257 | 0.295 | 0.978 |
| waaB, rfaB; UDP-D-galactose:(glucosyl)LPS alpha-1,6-D-galactosyltransferase [EC:2.4.1.-] | 0.416 | 1.053 | 0.694 | 0.999 |
| waaC, rfaC; heptosyltransferase I [EC:2.4.-.-] | 0.138 | 0.135 | 0.308 | 0.978 |
| waaE, kdtX; (heptosyl)LPS beta-1,4-glucosyltransferase [EC:2.4.1.-] | 0.255 | 0.318 | 0.425 | 0.978 |
| waaF, rfaF; heptosyltransferase II [EC:2.4.-.-] | 0.304 | 0.129 | 0.020 | 0.839 |
| waaG, rfaG; UDP-glucose:(heptosyl)LPS alpha-1,3-glucosyltransferase [EC:2.4.1.-] | 0.231 | 0.266 | 0.387 | 0.978 |
| waaH; heptose III glucuronosyltransferase [EC:2.4.1.-] | -0.920 | 1.032 | 0.374 | 0.978 |
| waaI, rfaI; UDP-D-galactose:(glucosyl)LPS alpha-1,3-D-galactosyltransferase [EC:2.4.1.44] | 1.662 | 1.228 | 0.178 | 0.961 |
| waaJ, rfaJ; UDP-glucose:(galactosyl)LPS alpha-1,2-glucosyltransferase [EC:2.4.1.58] | -0.704 | 0.878 | 0.424 | 0.978 |
| waaK, rfaK; UDP-N-acetylglucosamine:(glucosyl)LPS alpha-1,2-N-acetylglucosaminyltransferase [EC:2.4.1.56] | -0.826 | 0.638 | 0.197 | 0.970 |
| waaL, rfaL; O-antigen ligase [EC:2.4.1.-] | 0.289 | 0.213 | 0.178 | 0.961 |
| waaO, rfaI; UDP-glucose:(glucosyl)LPS alpha-1,3-glucosyltransferase [EC:2.4.1.-] | 0.511 | 1.152 | 0.658 | 0.999 |
| waaP, rfaP; heptose I phosphotransferase [EC:2.7.1.-] | 0.213 | 0.327 | 0.517 | 0.986 |
| waaQ, rfaQ; heptosyltransferase III [EC:2.4.-.-] | 0.091 | 0.298 | 0.761 | 0.999 |
| waaR, waaT, rfaJ; UDP-glucose/galactose:(glucosyl)LPS alpha-1,2-glucosyl/galactosyltransferase [EC:2.4.1.-] | 0.327 | 1.156 | 0.778 | 0.999 |
| waaS, rfaS; 1,5-rhamnosyltransferase [EC:2.4.1.-] | 1.117 | 1.359 | 0.412 | 0.978 |
| waaV; UDP-glucose:(glucosyl)LPS beta-1,3-glucosyltransferase [EC:2.4.1.-] | -1.154 | 1.507 | 0.445 | 0.978 |
| waaW; (galactosyl)LPS 1,2-glucosyltransferase [EC:2.4.1.-] | -0.384 | 1.237 | 0.757 | 0.999 |
| waaY, rfaY; heptose II phosphotransferase [EC:2.7.1.-] | 1.102 | 0.497 | 0.028 | 0.839 |
| waaZ, rfaZ; KDO transferase III [EC:2.4.99.-] | 1.834 | 1.209 | 0.131 | 0.941 |
| wapR; alpha-1,3-rhamnosyltransferase [EC:2.4.1.-] | -0.520 | 0.592 | 0.381 | 0.978 |
| WARS, trpS; tryptophanyl-tRNA synthetase [EC:6.1.1.2] | 0.038 | 0.033 | 0.246 | 0.978 |
| wbbJ; lipopolysaccharide O-acetyltransferase [EC:2.3.1.-] | -1.474 | 1.936 | 0.448 | 0.978 |
| wbbL; N-acetylglucosaminyl-diphospho-decaprenol L-rhamnosyltransferase [EC:2.4.1.289] | 0.057 | 0.342 | 0.869 | 0.999 |
| wbdB, wbpY; mannosyl-N-acetyl-alpha-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol 3-alpha-mannosyltransferase / alpha-1,3-rhamnosyltransferase [EC:2.4.1.349 2.4.1.-] | 0.552 | 0.640 | 0.389 | 0.978 |
| wbdC, wbpZ; N-acetyl-alpha-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol 3-alpha-mannosyltransferase / rhamnosyltransferase [EC:2.4.1.348 2.4.1.-] | -0.142 | 0.405 | 0.727 | 0.999 |
| wbdD, wbbD; O-antigen chain-terminating methyltransferase [EC:2.1.1.- 2.1.1.294 2.7.1.181] | 2.766 | 1.217 | 0.024 | 0.839 |
| wbiB; dTDP-L-rhamnose 4-epimerase [EC:5.1.3.25] | -0.679 | 0.553 | 0.221 | 0.978 |
| wbjC; UDP-2-acetamido-2,6-beta-L-arabino-hexul-4-ose reductase [EC:1.1.1.367] | -0.689 | 0.424 | 0.106 | 0.911 |
| wbpA; UDP-N-acetyl-D-glucosamine dehydrogenase [EC:1.1.1.136] | -0.328 | 0.380 | 0.389 | 0.978 |
| wbpB; UDP-N-acetyl-2-amino-2-deoxyglucuronate dehydrogenase [EC:1.1.1.335] | -0.946 | 1.031 | 0.360 | 0.978 |
| wbpD, wlbB; UDP-2-acetamido-3-amino-2,3-dideoxy-glucuronate N-acetyltransferase [EC:2.3.1.201] | -0.104 | 0.445 | 0.816 | 0.999 |
| wbpE, wlbC; UDP-2-acetamido-2-deoxy-ribo-hexuluronate aminotransferase [EC:2.6.1.98] | 0.156 | 0.626 | 0.804 | 0.999 |
| wbpI, wlbD; UDP-GlcNAc3NAcA epimerase [EC:5.1.3.23] | -1.190 | 0.763 | 0.121 | 0.919 |
| wbpL; Fuc2NAc and GlcNAc transferase [EC:2.4.1.-] | 0.260 | 0.365 | 0.478 | 0.979 |
| wbpO; UDP-N-acetyl-D-galactosamine dehydrogenase [EC:1.1.1.-] | 0.191 | 0.170 | 0.262 | 0.978 |
| wbpP; UDP-N-acetylglucosamine 4-epimerase [EC:5.1.3.7] | -0.031 | 0.356 | 0.931 | 0.999 |
| wbpX; alpha-1,2-rhamnosyltransferase [EC:2.4.1.-] | -0.052 | 0.869 | 0.952 | 0.999 |
| wbqL; O-antigen biosynthesis protein WbqL | -17.510 | 3533.982 | 0.996 | 0.999 |
| wbqP; O-antigen biosynthesis protein WbqP | -0.076 | 0.436 | 0.862 | 0.999 |
| wbqR; UDP-perosamine 4-acetyltransferase [EC:2.3.1.-] | -17.702 | 2942.450 | 0.995 | 0.999 |
| wbqV; O-antigen biosynthesis protein WbqV | -1.599 | 0.891 | 0.075 | 0.851 |
| wbtD; galacturonosyltransferase [EC:2.4.1.-] | 0.876 | 1.345 | 0.516 | 0.985 |
| wbyK; mannosyltransferase [EC:2.4.1.-] | -17.761 | 4005.858 | 0.996 | 0.999 |
| wbyL; glycosyltransferase [EC:2.4.1.-] | -1.692 | 1.139 | 0.140 | 0.949 |
| wcaB; putative colanic acid biosynthesis acetyltransferase WcaB [EC:2.3.1.-] | -0.160 | 0.752 | 0.832 | 0.999 |
| wcaC; putative colanic acid biosynthesis glycosyltransferase [EC:2.4.-.-] | -0.395 | 0.850 | 0.643 | 0.999 |
| wcaD; putative colanic acid polymerase | -0.413 | 0.817 | 0.614 | 0.999 |
| wcaE; putative colanic acid biosynthesis glycosyltransferase [EC:2.4.-.-] | -0.413 | 0.817 | 0.614 | 0.999 |
| wcaF; putative colanic acid biosynthesis acetyltransferase WcaF [EC:2.3.1.-] | -1.126 | 0.556 | 0.044 | 0.839 |
| wcaH; colanic acid biosynthesis protein WcaH [EC:3.6.1.-] | -0.796 | 0.716 | 0.268 | 0.978 |
| wcaI; colanic acid biosynthesis glycosyl transferase WcaI | -0.934 | 0.703 | 0.186 | 0.966 |
| wcaJ; putative colanic acid biosysnthesis UDP-glucose lipid carrier transferase | -0.178 | 0.208 | 0.393 | 0.978 |
| wcaK, amsJ; colanic acid/amylovoran biosynthesis protein | -0.248 | 0.710 | 0.728 | 0.999 |
| wcaL, amsK; colanic acid/amylovoran biosynthesis glycosyltransferase [EC:2.4.-.-] | -0.100 | 0.646 | 0.877 | 0.999 |
| wcaM; colanic acid biosynthesis protein WcaM | -0.395 | 0.850 | 0.643 | 0.999 |
| wecA, tagO, rfe; UDP-GlcNAc:undecaprenyl-phosphate/decaprenyl-phosphate GlcNAc-1-phosphate transferase [EC:2.7.8.33 2.7.8.35] | 0.048 | 0.095 | 0.615 | 0.999 |
| wecB; UDP-N-acetylglucosamine 2-epimerase (non-hydrolysing) [EC:5.1.3.14] | -0.029 | 0.075 | 0.702 | 0.999 |
| wecC; UDP-N-acetyl-D-mannosaminuronic acid dehydrogenase [EC:1.1.1.336] | -0.099 | 0.242 | 0.682 | 0.999 |
| wecE, rffA; dTDP-4-amino-4,6-dideoxygalactose transaminase [EC:2.6.1.59] | -0.355 | 0.391 | 0.366 | 0.978 |
| wecF, rffT; dTDP-N-acetylfucosamine:lipid II N-acetylfucosaminyltransferase [EC:2.4.1.325] | 0.119 | 0.601 | 0.843 | 0.999 |
| wecG, rffM; UDP-N-acetyl-D-mannosaminouronate:lipid I N-acetyl-D-mannosaminouronosyltransferase [EC:2.4.1.180] | 0.058 | 0.587 | 0.922 | 0.999 |
| whiB1_2_3_4; WhiB family transcriptional regulator, redox-sensing transcriptional regulator | -0.081 | 0.275 | 0.767 | 0.999 |
| whiB5; WhiB family transcriptional regulator, redox-sensing transcriptional regulator | -17.416 | 3370.607 | 0.996 | 0.999 |
| whiB6; WhiB family transcriptional regulator, redox-sensing transcriptional regulator | -3.091 | 2.016 | 0.127 | 0.927 |
| whiB7; WhiB family transcriptional regulator, redox-sensing transcriptional regulator | -0.520 | 0.756 | 0.492 | 0.981 |
| whiEII; putative monooxygenase | 3.129 | 2.755 | 0.258 | 0.978 |
| whiEIII; minimal PKS ketosynthase (KS/KS alpha) [EC:2.3.1.-] | 3.129 | 2.755 | 0.258 | 0.978 |
| whiEIV; minimal PKS chain-length factor (CLF/KS beta) [EC:2.3.1.-] | 3.129 | 2.755 | 0.258 | 0.978 |
| whiEV; minimal PKS acyl carrier protein | 18.832 | 3989.590 | 0.996 | 0.999 |
| whiEVI; aromatase | 3.129 | 2.755 | 0.258 | 0.978 |
| whiEVII; cyclase | 3.129 | 2.755 | 0.258 | 0.978 |
| whiEVIII; putative polyketide hydroxylase | -1.718 | 2.398 | 0.475 | 0.979 |
| WIPF; WAS/WASL-interacting protein | -17.368 | 3290.703 | 0.996 | 0.999 |
| wlbA, bplA; UDP-N-acetyl-2-amino-2-deoxyglucuronate dehydrogenase [EC:1.1.1.335] | 0.869 | 0.683 | 0.205 | 0.972 |
| wprA; cell wall-associated protease [EC:3.4.21.-] | 18.239 | 2966.075 | 0.995 | 0.999 |
| wrbA; NAD(P)H dehydrogenase (quinone) [EC:1.6.5.2] | -0.138 | 0.182 | 0.447 | 0.978 |
| wspA; methyl-accepting chemotaxis protein WspA | 0.163 | 0.341 | 0.634 | 0.999 |
| wspB; chemotaxis-related protein WspB | 0.228 | 0.329 | 0.489 | 0.979 |
| wspC; chemotaxis protein methyltransferase WspC | 0.234 | 0.329 | 0.478 | 0.979 |
| wspD; chemotaxis-related protein WspD | 0.235 | 0.329 | 0.475 | 0.979 |
| wspE; two-component system, chemotaxis family, sensor histidine kinase and response regulator WspE | 0.126 | 0.388 | 0.746 | 0.999 |
| wspF; two-component system, chemotaxis family, response regulator WspF [EC:3.1.1.61] | 0.225 | 0.329 | 0.496 | 0.981 |
| wspR; two-component system, chemotaxis family, response regulator WspR [EC:2.7.7.65] | 0.228 | 0.329 | 0.489 | 0.979 |
| wtpA; molybdate/tungstate transport system substrate-binding protein | -2.826 | 2.742 | 0.304 | 0.978 |
| wtpB; molybdate/tungstate transport system permease protein | -17.234 | 3077.514 | 0.996 | 0.999 |
| wtpC; molybdate/tungstate transport system ATP-binding protein [EC:3.6.3.- 3.6.3.55] | -17.234 | 3077.514 | 0.996 | 0.999 |
| wzxC; lipopolysaccharide exporter | -0.395 | 0.850 | 0.643 | 0.999 |
| wzxE; enterobacterial common antigen flippase | -0.035 | 0.482 | 0.941 | 0.999 |
| wzy; O-antigen polymerase [EC:2.4.1.-] | 1.132 | 0.878 | 0.199 | 0.970 |
| wzyE, rffT; enterobacterial common antigen polymerase [EC:2.4.1.-] | 0.119 | 0.601 | 0.843 | 0.999 |
| wzzB; chain length determinant protein (polysaccharide antigen chain regulator) | 0.047 | 0.308 | 0.879 | 0.999 |
| wzzE; lipopolysaccharide biosynthesis protein WzzE | 0.186 | 0.350 | 0.596 | 0.999 |
| xanB2; chorismate lyase / 3-hydroxybenzoate synthase [EC:4.1.3.40 4.1.3.45] | 0.107 | 0.805 | 0.894 | 0.999 |
| xanP; xanthine permease XanP | -0.008 | 0.161 | 0.961 | 0.999 |
| xanQ; xanthine permease XanQ | 0.595 | 0.646 | 0.359 | 0.978 |
| xapA; xanthosine phosphorylase [EC:2.4.2.-] | -0.214 | 0.769 | 0.781 | 0.999 |
| xapB; MFS transporter, NHS family, xanthosine permease | -0.245 | 0.278 | 0.379 | 0.978 |
| xdhA; xanthine dehydrogenase small subunit [EC:1.17.1.4] | -0.120 | 0.267 | 0.654 | 0.999 |
| xdhB; xanthine dehydrogenase large subunit [EC:1.17.1.4] | -0.135 | 0.313 | 0.666 | 0.999 |
| xdhC; xanthine dehydrogenase accessory factor | 0.000 | 0.125 | 0.997 | 0.999 |
| xdhD; putative selenate reductase molybdopterin-binding subunit | -0.127 | 1.190 | 0.915 | 0.999 |
| xerC; integrase/recombinase XerC | -0.082 | 0.070 | 0.241 | 0.978 |
| xerD; integrase/recombinase XerD | 0.033 | 0.073 | 0.654 | 0.999 |
| xfp, xpk; xylulose-5-phosphate/fructose-6-phosphate phosphoketolase [EC:4.1.2.9 4.1.2.22] | 0.215 | 0.280 | 0.444 | 0.978 |
| xltA; putative xylitol transport system ATP-binding protein | -1.585 | 1.274 | 0.215 | 0.978 |
| xltB; putative xylitol transport system permease protein | -1.671 | 1.200 | 0.166 | 0.960 |
| xltC; putative xylitol transport system substrate-binding protein | -1.585 | 1.274 | 0.215 | 0.978 |
| xlyAB; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | -0.059 | 0.220 | 0.789 | 0.999 |
| xni; protein Xni | 0.352 | 0.531 | 0.508 | 0.981 |
| xopD; type III effector protein XopD | 18.223 | 2942.586 | 0.995 | 0.999 |
| xpt; xanthine phosphoribosyltransferase [EC:2.4.2.22] | 0.016 | 0.072 | 0.828 | 0.999 |
| xsc; sulfoacetaldehyde acetyltransferase [EC:2.3.3.15] | -3.906 | 4.090 | 0.341 | 0.978 |
| xseA; exodeoxyribonuclease VII large subunit [EC:3.1.11.6] | 0.073 | 0.039 | 0.063 | 0.839 |
| xseB; exodeoxyribonuclease VII small subunit [EC:3.1.11.6] | 0.039 | 0.036 | 0.281 | 0.978 |
| xtmA; phage terminase small subunit | 0.042 | 0.335 | 0.901 | 0.999 |
| xtmB; phage terminase large subunit | -0.611 | 0.503 | 0.226 | 0.978 |
| xylA; xylose isomerase [EC:5.3.1.5] | -0.231 | 0.200 | 0.251 | 0.978 |
| xylB, XYLB; xylulokinase [EC:2.7.1.17] | -0.178 | 0.150 | 0.239 | 0.978 |
| xylC; benzaldehyde dehydrogenase (NAD) [EC:1.2.1.28] | -0.239 | 0.340 | 0.483 | 0.979 |
| xylE; MFS transporter, SP family, xylose:H+ symportor | -0.764 | 0.596 | 0.202 | 0.972 |
| xylF; D-xylose transport system substrate-binding protein | -0.778 | 0.364 | 0.034 | 0.839 |
| xylG; D-xylose transport system ATP-binding protein [EC:3.6.3.17] | -0.468 | 0.294 | 0.113 | 0.917 |
| xylH; D-xylose transport system permease protein | -0.728 | 0.374 | 0.053 | 0.839 |
| xylS, yicI; alpha-D-xyloside xylohydrolase [EC:3.2.1.177] | -0.517 | 0.326 | 0.115 | 0.917 |
| xynB; xylan 1,4-beta-xylosidase [EC:3.2.1.37] | -0.986 | 0.615 | 0.110 | 0.917 |
| xynC; glucuronoarabinoxylan endo-1,4-beta-xylanase [EC:3.2.1.136] | -0.130 | 0.912 | 0.887 | 0.999 |
| xynD; arabinoxylan arabinofuranohydrolase [EC:3.2.1.55] | -2.486 | 2.330 | 0.288 | 0.978 |
| xyoA, aldO; alditol oxidase [EC:1.1.3.41] | 0.757 | 0.738 | 0.307 | 0.978 |
| yaaH; spore germination protein | -1.510 | 0.760 | 0.049 | 0.839 |
| yaaU; MFS transporter, putative metabolite transport protein | 0.184 | 0.435 | 0.672 | 0.999 |
| yabG; spore coat assemly protein | -0.995 | 0.681 | 0.146 | 0.954 |
| yabN; tetrapyrrole methylase family protein / MazG family protein | -0.115 | 0.146 | 0.432 | 0.978 |
| yadB_C; adhesin YadB/C | 18.902 | 3402.727 | 0.996 | 0.999 |
| yaeJ; ribosome-associated protein | -0.117 | 0.195 | 0.550 | 0.992 |
| yaeR; glyoxylase I family protein | -0.098 | 0.125 | 0.433 | 0.978 |
| yafN; antitoxin YafN | 2.069 | 1.635 | 0.208 | 0.976 |
| yafO; mRNA interferase YafO [EC:3.1.-.-] | 2.454 | 1.465 | 0.096 | 0.896 |
| yafP; putative acetyltransferase [EC:2.3.1.-] | -0.746 | 0.414 | 0.073 | 0.845 |
| yafQ; mRNA interferase YafQ [EC:3.1.-.-] | 0.035 | 0.173 | 0.842 | 0.999 |
| yagR; xanthine dehydrogenase YagR molybdenum-binding subunit [EC:1.17.1.4] | -0.125 | 0.251 | 0.618 | 0.999 |
| yagS; xanthine dehydrogenase YagS FAD-binding subunit [EC:1.17.1.4] | -0.133 | 0.248 | 0.593 | 0.999 |
| yagT; xanthine dehydrogenase YagT iron-sulfur-binding subunit | -0.094 | 0.246 | 0.703 | 0.999 |
| yagU; putative membrane protein | 0.201 | 0.237 | 0.397 | 0.978 |
| yahA; c-di-GMP-specific phosphodiesterase [EC:3.1.4.52] | -1.154 | 1.507 | 0.445 | 0.978 |
| yahK; uncharacterized zinc-type alcohol dehydrogenase-like protein [EC:1.-.-.-] | -0.222 | 0.231 | 0.337 | 0.978 |
| yahN; amino acid exporter | -1.130 | 1.346 | 0.402 | 0.978 |
| yajC; preprotein translocase subunit YajC | 0.050 | 0.037 | 0.181 | 0.961 |
| yajG; uncharacterized lipoprotein | 0.218 | 0.203 | 0.284 | 0.978 |
| yajQ; cyclic-di-GMP-binding protein | -0.008 | 0.103 | 0.941 | 0.999 |
| YARS, tyrS; tyrosyl-tRNA synthetase [EC:6.1.1.1] | 0.024 | 0.035 | 0.487 | 0.979 |
| ybaK, ebsC; Cys-tRNA(Pro)/Cys-tRNA(Cys) deacylase [EC:3.1.1.-] | 0.110 | 0.048 | 0.023 | 0.839 |
| ybaO; Lrp/AsnC family transcriptional regulator | -0.245 | 0.255 | 0.338 | 0.978 |
| ybaZ; methylated-DNA-protein-cysteine methyltransferase related protein | -0.028 | 0.095 | 0.771 | 0.999 |
| ybbJ; inner membrane protein | -0.012 | 0.163 | 0.943 | 0.999 |
| ybbN; putative thioredoxin | -0.014 | 0.124 | 0.912 | 0.999 |
| ybcJ; ribosome-associated protein | 0.207 | 0.116 | 0.076 | 0.855 |
| ybcL; MFS transporter, DHA1 family, putative efflux transporter | -0.578 | 1.355 | 0.671 | 0.999 |
| ybdG, mscM; miniconductance mechanosensitive channel | -0.126 | 0.144 | 0.381 | 0.978 |
| ybdH; uncharacterized oxidoreductase [EC:1.1.-.-] | -0.275 | 0.489 | 0.575 | 0.995 |
| ybdL; methionine transaminase [EC:2.6.1.88] | -0.021 | 0.214 | 0.921 | 0.999 |
| ybeB; ribosome-associated protein | -0.010 | 0.040 | 0.803 | 0.999 |
| ybeY, yqfG; probable rRNA maturation factor | 0.067 | 0.041 | 0.105 | 0.910 |
| ybfF; esterase [EC:3.1.-.-] | 0.230 | 0.290 | 0.428 | 0.978 |
| ybgC; acyl-CoA thioester hydrolase [EC:3.1.2.-] | -0.126 | 0.078 | 0.107 | 0.913 |
| ybiC; uncharacterized oxidoreductase [EC:1.1.1.-] | -0.909 | 0.597 | 0.130 | 0.934 |
| ybiS; L,D-transpeptidase YbiS | 0.158 | 0.613 | 0.797 | 0.999 |
| ybiV; sugar-phosphatase [EC:3.1.3.23] | -0.626 | 0.724 | 0.389 | 0.978 |
| ybjD; putative ATP-dependent endonuclease of the OLD family | 0.382 | 0.199 | 0.057 | 0.839 |
| ybtA; AraC family transcriptional regulator | -2.044 | 2.163 | 0.346 | 0.978 |
| ybtX, irp8; MFS transporter, putative signal transducer | -1.077 | 1.271 | 0.398 | 0.978 |
| ycaJ; putative ATPase | 0.037 | 0.045 | 0.415 | 0.978 |
| ycaO; ribosomal protein S12 methylthiotransferase accessory factor | 0.001 | 0.202 | 0.997 | 0.999 |
| ycbA, glnK; two-component system, sensor histidine kinase YcbA [EC:2.7.13.3] | 0.186 | 0.493 | 0.706 | 0.999 |
| ycbB, glnL; two-component system, response regulator YcbB | 0.416 | 0.395 | 0.293 | 0.978 |
| yccA; modulator of FtsH protease | 0.102 | 0.179 | 0.568 | 0.995 |
| ycdX; putative hydrolase | 0.109 | 0.284 | 0.703 | 0.999 |
| ycfD; 50S ribosomal protein L16 3-hydroxylase [EC:1.14.11.47] | 0.074 | 0.162 | 0.648 | 0.999 |
| ycfS; L,D-transpeptidase YcfS | 0.225 | 0.574 | 0.695 | 0.999 |
| ycgQ; putative membrane protein | -0.063 | 0.164 | 0.701 | 0.999 |
| ychF; ribosome-binding ATPase | 0.036 | 0.036 | 0.312 | 0.978 |
| ychN; uncharacterized protein involved in oxidation of intracellular sulfur | 0.296 | 0.323 | 0.360 | 0.978 |
| yciA; acyl-CoA thioesterase YciA [EC:3.1.2.-] | -0.015 | 0.151 | 0.922 | 0.999 |
| ycjF; putative membrane protein | -0.087 | 0.269 | 0.747 | 0.999 |
| ycjT; hypothetical glycosyl hydrolase [EC:3.2.1.-] | 0.739 | 0.609 | 0.227 | 0.978 |
| ycnJ; copper transport protein | -1.106 | 0.523 | 0.036 | 0.839 |
| ydaV; putative replication protein | -2.031 | 2.097 | 0.334 | 0.978 |
| ydfG; 3-hydroxy acid dehydrogenase / malonic semialdehyde reductase [EC:1.1.1.381 1.1.1.-] | 0.207 | 0.278 | 0.458 | 0.979 |
| ydfH; two-component system, NarL family, sensor histidine kinase YdfH [EC:2.7.13.3] | 18.429 | 2747.052 | 0.995 | 0.999 |
| ydfI; two-component system, NarL family, response regulator YdfI | 18.045 | 1898.825 | 0.992 | 0.999 |
| ydfJ; membrane protein YdfJ | 18.239 | 2966.075 | 0.995 | 0.999 |
| ydfJ; MFS transporter, MHS family, metabolite:H+ symporter | 0.193 | 0.574 | 0.736 | 0.999 |
| ydgD; protease YdgD [EC:3.4.21.-] | -0.278 | 0.548 | 0.613 | 0.999 |
| ydhP; MFS transporter, DHA1 family, inner membrane transport protein | -0.119 | 0.218 | 0.588 | 0.999 |
| ydhQ; GntR family transcriptional regulator | 0.423 | 0.295 | 0.154 | 0.954 |
| ydiB; quinate/shikimate dehydrogenase [EC:1.1.1.282] | -1.130 | 1.346 | 0.402 | 0.978 |
| ydiF; acetate CoA-transferase [EC:2.8.3.8] | -1.130 | 1.346 | 0.402 | 0.978 |
| ydiU; uncharacterized protein | -0.170 | 0.327 | 0.604 | 0.999 |
| ydiY; putative salt-induced outer membrane protein | -0.133 | 0.370 | 0.721 | 0.999 |
| ydjE; MFS transporter, putative metabolite:H+ symporter | -0.124 | 0.141 | 0.383 | 0.978 |
| ydjG; methylglyoxal reductase [EC:1.1.1.-] | -0.859 | 0.889 | 0.335 | 0.978 |
| yebQ; MFS transporter, DHA2 family, multidrug resistance protein | -0.648 | 0.290 | 0.027 | 0.839 |
| yedL; putative acetyltransferase [EC:2.3.1.-] | -0.798 | 0.415 | 0.056 | 0.839 |
| yeeJ; adhesin/invasin | -0.355 | 0.839 | 0.673 | 0.999 |
| yefM; antitoxin YefM | -0.157 | 0.164 | 0.340 | 0.978 |
| yegD; hypothetical chaperone protein | -0.104 | 0.209 | 0.617 | 0.999 |
| yeiL; CRP/FNR family transcriptional regulator, putaive post-exponential-phase nitrogen-starvation regulator | -1.272 | 1.395 | 0.363 | 0.978 |
| yejA; microcin C transport system substrate-binding protein | -0.110 | 0.262 | 0.675 | 0.999 |
| yejB; microcin C transport system permease protein | -0.107 | 0.237 | 0.651 | 0.999 |
| yejE; microcin C transport system permease protein | -0.141 | 0.239 | 0.557 | 0.992 |
| yejF; microcin C transport system ATP-binding protein | -0.163 | 0.218 | 0.456 | 0.978 |
| yesM; two-component system, sensor histidine kinase YesM [EC:2.7.13.3] | 0.117 | 0.151 | 0.442 | 0.978 |
| yesN; two-component system, response regulator YesN | 0.128 | 0.146 | 0.380 | 0.978 |
| yesW; rhamnogalacturonan endolyase [EC:4.2.2.23] | -1.266 | 0.768 | 0.101 | 0.896 |
| yesX; rhamnogalacturonan exolyase [EC:4.2.2.24] | 18.239 | 2966.075 | 0.995 | 0.999 |
| yfaE; ferredoxin | 0.271 | 0.315 | 0.392 | 0.978 |
| yfaL; autotransporter family porin | -1.153 | 1.472 | 0.435 | 0.978 |
| yfbK; Ca-activated chloride channel homolog | -0.067 | 0.113 | 0.552 | 0.992 |
| yfbR; 5’-deoxynucleotidase [EC:3.1.3.89] | 0.252 | 0.197 | 0.203 | 0.972 |
| yfbT, yniC; sugar-phosphatase [EC:3.1.3.23] | -0.146 | 0.330 | 0.658 | 0.999 |
| yfiC, trmX; tRNA1Val (adenine37-N6)-methyltransferase [EC:2.1.1.223] | -0.061 | 0.180 | 0.735 | 0.999 |
| yfiF, trmG; RNA methyltransferase, TrmH family [EC:2.1.1.-] | 0.087 | 0.246 | 0.724 | 0.999 |
| yfiH; polyphenol oxidase [EC:1.10.3.-] | 0.012 | 0.060 | 0.839 | 0.999 |
| yfiM; putative lipoprotein | 0.119 | 0.601 | 0.843 | 0.999 |
| yfiP; DTW domain-containing protein | 0.089 | 0.276 | 0.747 | 0.999 |
| yfiQ; acetyltransferase | -0.223 | 0.250 | 0.375 | 0.978 |
| yfkN; 2’,3’-cyclic-nucleotide 2’-phosphodiesterase / 3’-nucleotidase / 5’-nucleotidase [EC:3.1.4.16 3.1.3.6 3.1.3.5] | -0.781 | 1.207 | 0.519 | 0.987 |
| yfkQ; spore germination protein | 0.665 | 1.122 | 0.554 | 0.992 |
| yfkR; spore germination protein | -0.901 | 1.130 | 0.426 | 0.978 |
| yfkT; spore germination protein | -17.198 | 3022.482 | 0.995 | 0.999 |
| ygaC; uncharacterized protein | -0.036 | 0.130 | 0.782 | 0.999 |
| ygeR; lipoprotein YgeR | 0.284 | 0.569 | 0.618 | 0.999 |
| ygeS, xdhA; xanthine dehydrogenase molybdenum-binding subunit [EC:1.17.1.4] | 0.196 | 0.473 | 0.679 | 0.999 |
| ygeT, xdhB; xanthine dehydrogenase FAD-binding subunit [EC:1.17.1.4] | 0.383 | 0.505 | 0.450 | 0.978 |
| ygeU, xdhC; xanthine dehydrogenase iron-sulfur-binding subunit | -1.153 | 1.472 | 0.435 | 0.978 |
| ygfK; putative selenate reductase [EC:1.97.1.9] | 0.400 | 0.489 | 0.414 | 0.978 |
| ygfM; putative selenate reductase FAD-binding subunit | -1.153 | 1.472 | 0.435 | 0.978 |
| ygfZ; tRNA-modifying protein YgfZ | -0.045 | 0.122 | 0.713 | 0.999 |
| yggC; putative kinase | -0.095 | 0.581 | 0.871 | 0.999 |
| yggS, PROSC; PLP dependent protein | 0.017 | 0.037 | 0.646 | 0.999 |
| yggT; YggT family protein | 0.058 | 0.063 | 0.361 | 0.978 |
| yghU, yfcG; GSH-dependent disulfide-bond oxidoreductase [EC:1.8.4.-] | -0.209 | 0.140 | 0.139 | 0.949 |
| ygiF; triphosphatase [EC:3.6.1.25] | 0.354 | 0.530 | 0.505 | 0.981 |
| ygiM; SH3 domain protein | 0.341 | 0.218 | 0.120 | 0.917 |
| ygjK; putative isomerase | -0.453 | 0.664 | 0.496 | 0.981 |
| yhaV; toxin YhaV [EC:3.1.-.-] | -0.580 | 0.775 | 0.456 | 0.978 |
| yhbH; putative sigma-54 modulation protein | 0.068 | 0.065 | 0.294 | 0.978 |
| yhbS; putative acetyltransferase [EC:2.3.1.-] | -0.328 | 0.284 | 0.250 | 0.978 |
| yhbY; RNA-binding protein | 0.137 | 0.051 | 0.008 | 0.839 |
| yhcO; ribonuclease inhibitor | 0.012 | 0.391 | 0.975 | 0.999 |
| yhdJ; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | -1.130 | 1.346 | 0.402 | 0.978 |
| yhdR; aspartate aminotransferase [EC:2.6.1.1] | 0.237 | 0.327 | 0.470 | 0.979 |
| yhfA; putative redox protein | -0.104 | 0.183 | 0.571 | 0.995 |
| yhgE; putative membrane protein | -0.063 | 0.113 | 0.578 | 0.997 |
| yhhL; putative membrane protein | 0.444 | 0.302 | 0.144 | 0.951 |
| yhhY; putative acetyltransferase [EC:2.3.1.-] | -0.213 | 0.379 | 0.575 | 0.995 |
| yhjG; AsmA family protein | -0.022 | 0.232 | 0.925 | 0.999 |
| yiaC; putative acetyltransferase [EC:2.3.1.-] | -0.173 | 0.400 | 0.667 | 0.999 |
| yiaY; alcohol dehydrogenase [EC:1.1.1.1] | -0.284 | 0.262 | 0.281 | 0.978 |
| yibL; ribosome-associated protein | 0.241 | 0.593 | 0.685 | 0.999 |
| yidC, spoIIIJ, OXA1, ccfA; YidC/Oxa1 family membrane protein insertase | 0.037 | 0.054 | 0.492 | 0.981 |
| yidH; putative membrane protein | -0.440 | 0.407 | 0.281 | 0.978 |
| yidP; GntR family transcriptional regulator, glv operon transcriptional regulator | -0.393 | 0.833 | 0.638 | 0.999 |
| yihQ; sulfoquinovosidase [EC:3.2.1.199] | -1.961 | 1.229 | 0.113 | 0.917 |
| yihS; sulfoquinovose isomerase [EC:5.3.1.31] | 0.125 | 0.328 | 0.705 | 0.999 |
| yihT; sulfofructosephosphate aldolase [EC:4.1.2.57] | -1.261 | 1.208 | 0.298 | 0.978 |
| yihU; 4-hydroxybutyrate dehydrogenase / sulfolactaldehyde 3-reductase [EC:1.1.1.61 1.1.1.373] | -0.837 | 0.961 | 0.385 | 0.978 |
| yihV; sulfofructose kinase [EC:2.7.1.184] | -1.643 | 0.985 | 0.097 | 0.896 |
| yitG, ymfD, yfmO; MFS transporter, ACDE family, multidrug resistance protein | -2.266 | 0.864 | 0.010 | 0.839 |
| yjaB; putative acetyltransferase [EC:2.3.1.-] | -0.022 | 0.208 | 0.914 | 0.999 |
| yjbB; phosphate:Na+ symporter | -0.035 | 0.081 | 0.665 | 0.999 |
| yjdF; putative membrane protein | -0.093 | 0.190 | 0.626 | 0.999 |
| yjeH; amino acid efflux transporter | 0.214 | 0.521 | 0.682 | 0.999 |
| yjgA; ribosome-associated protein | 0.066 | 0.156 | 0.670 | 0.999 |
| yjgB; uncharacterized zinc-type alcohol dehydrogenase-like protein [EC:1.-.-.-] | -0.191 | 0.376 | 0.613 | 0.999 |
| yjgM; putative acetyltransferase [EC:2.3.1.-] | -0.172 | 0.198 | 0.387 | 0.978 |
| yjjG; 5’-nucleotidase [EC:3.1.3.5] | -0.117 | 0.686 | 0.865 | 0.999 |
| ykkC; paired small multidrug resistance pump | -0.519 | 0.538 | 0.336 | 0.978 |
| ykkD; paired small multidrug resistance pump | -0.523 | 0.539 | 0.333 | 0.978 |
| yknT; sigma-E controlled sporulation protein | 18.239 | 2966.075 | 0.995 | 0.999 |
| ykoE; energy-coupling factor transport system substrate-specific component | 0.091 | 0.162 | 0.575 | 0.995 |
| ylbA, UGHY; (S)-ureidoglycine aminohydrolase [EC:3.5.3.26] | -0.072 | 0.350 | 0.838 | 0.999 |
| ylnA, cysP; sulfate permease | -0.949 | 0.776 | 0.223 | 0.978 |
| ylxR; uncharacterized protein | 0.086 | 0.084 | 0.308 | 0.978 |
| ynaI, mscMJ; MscS family membrane protein | -0.300 | 0.406 | 0.462 | 0.979 |
| yncG; GST-like protein | -0.470 | 0.475 | 0.323 | 0.978 |
| yndD; spore germination protein | -1.274 | 0.798 | 0.113 | 0.917 |
| yndE; spore germination protein | -1.072 | 1.065 | 0.316 | 0.978 |
| yndF; spore germination protein | -1.069 | 1.066 | 0.318 | 0.978 |
| yneE, BEST; ion channel-forming bestrophin family protein | -0.097 | 0.230 | 0.674 | 0.999 |
| ynfE; Tat-targeted selenate reductase subunit YnfE [EC:1.97.1.9] | -0.569 | 0.642 | 0.377 | 0.978 |
| ynfF; Tat-targeted selenate reductase subunit YnfF [EC:1.97.1.9] | -0.910 | 1.157 | 0.433 | 0.978 |
| ynfH; Tat-targeted selenate reductase subunit YnfH | -0.969 | 0.853 | 0.258 | 0.978 |
| ynfM; MFS transporter, YNFM family, putative membrane transport protein | -0.081 | 0.198 | 0.685 | 0.999 |
| ynhG; L,D-transpeptidase YnhG | -0.243 | 0.717 | 0.735 | 0.999 |
| yoeB; toxin YoeB [EC:3.1.-.-] | 0.391 | 0.201 | 0.054 | 0.839 |
| yogA; zinc-binding alcohol dehydrogenase/oxidoreductase | 0.092 | 0.946 | 0.923 | 0.999 |
| yojI; multidrug/microcin transport system ATP-binding/permease protein | -0.003 | 0.466 | 0.995 | 0.999 |
| ypdF; aminopeptidase [EC:3.4.11.-] | -1.244 | 1.220 | 0.309 | 0.978 |
| ypeB; spore germination protein | -1.426 | 0.741 | 0.056 | 0.839 |
| yprB; uncharacterized protein | -0.071 | 0.265 | 0.791 | 0.999 |
| ypsC; putative N6-adenine-specific DNA methylase [EC:2.1.1.-] | -0.025 | 0.089 | 0.777 | 0.999 |
| yqeH; 30S ribosome assembly GTPase | 0.110 | 0.118 | 0.351 | 0.978 |
| yqfD; similar to stage IV sporulation protein | -0.979 | 0.512 | 0.058 | 0.839 |
| yqgE; MFS transporter, YQGE family, putative transporter | -2.492 | 0.915 | 0.007 | 0.839 |
| yqgT; g-D-glutamyl-meso-diaminopimelate peptidase [EC:3.4.19.11] | -0.095 | 0.405 | 0.814 | 0.999 |
| yqhD; NADP-dependent alcohol dehydrogenase [EC:1.1.-.-] | -0.113 | 0.285 | 0.694 | 0.999 |
| yqjH; ferric-chelate reductase (NADPH) [EC:1.16.1.9] | -0.238 | 0.703 | 0.736 | 0.999 |
| yraD; similar to spore coat protein | -3.216 | 1.601 | 0.046 | 0.839 |
| yraG; similar to spore coat protein | -2.217 | 1.654 | 0.182 | 0.961 |
| yraN; putative endonuclease | 0.063 | 0.071 | 0.378 | 0.978 |
| yrbG; cation:H+ antiporter | -0.044 | 0.128 | 0.734 | 0.999 |
| yrdD; putative DNA topoisomerase | 0.231 | 0.308 | 0.455 | 0.978 |
| yrrT; putative AdoMet-dependent methyltransferase [EC:2.1.1.-] | -1.109 | 0.602 | 0.067 | 0.839 |
| yscC, sctC, ssaC; type III secretion protein C | -0.565 | 0.533 | 0.291 | 0.978 |
| yscD, sctD, ssaD; type III secretion protein D | -0.361 | 0.568 | 0.525 | 0.989 |
| yscF, sctF, ssaG, prgI; type III secretion protein F | -2.151 | 1.705 | 0.209 | 0.977 |
| yscI, sctI; type III secretion protein I | -17.577 | 3325.657 | 0.996 | 0.999 |
| yscJ, sctJ, hrcJ, ssaJ; type III secretion protein J | -0.690 | 0.481 | 0.153 | 0.954 |
| yscL, sctL; type III secretion protein L | -0.328 | 0.544 | 0.548 | 0.991 |
| yscN, sctN, hrcN, ssaN; ATP synthase in type III secretion protein N [EC:3.6.3.14] | -0.407 | 0.482 | 0.400 | 0.978 |
| yscO, sctO; type III secretion protein O | -17.577 | 3325.657 | 0.996 | 0.999 |
| yscQ, sctQ, hrcQ, ssaQ, spaO; type III secretion protein Q | -0.361 | 0.470 | 0.444 | 0.978 |
| yscR, sctR, hrcR, ssaR; type III secretion protein R | -0.491 | 0.441 | 0.268 | 0.978 |
| yscS, sctS, hrcS, ssaS; type III secretion protein S | -0.473 | 0.443 | 0.288 | 0.978 |
| yscT, sctT, hrcT, ssaT; type III secretion protein T | -0.481 | 0.438 | 0.274 | 0.978 |
| yscU, sctU, hrcU, ssaU; type III secretion protein U | -0.560 | 0.439 | 0.204 | 0.972 |
| yscV, sctV, hrcV, ssaV, invA; type III secretion protein V | -0.496 | 0.444 | 0.265 | 0.978 |
| yscW, sctW; type III secretion protein W | -0.314 | 0.548 | 0.567 | 0.995 |
| yscX, sctX; type III secretion protein X | -17.577 | 3325.657 | 0.996 | 0.999 |
| yscY, sctY; type III secretion protein Y | 2.635 | 1.537 | 0.088 | 0.880 |
| ysiA, fadR; TetR/AcrR family transcriptional regulator, fatty acid metabolism regulator protein | -1.616 | 0.769 | 0.037 | 0.839 |
| ysxB; uncharacterized protein | 0.150 | 0.098 | 0.127 | 0.927 |
| yteR, yesR; unsaturated rhamnogalacturonyl hydrolase [EC:3.2.1.172] | -1.136 | 0.553 | 0.042 | 0.839 |
| ytfB; uncharacterized protein | 0.218 | 0.576 | 0.706 | 0.999 |
| ytfE, scdA; regulator of cell morphogenesis and NO signaling | -0.149 | 0.152 | 0.330 | 0.978 |
| ytmI; uncharacterized N-acetyltransferase [EC:2.3.1.-] | 0.528 | 0.256 | 0.040 | 0.839 |
| ytpB; tetraprenyl-beta-curcumene synthase [EC:4.2.3.130] | -2.174 | 0.860 | 0.012 | 0.839 |
| ytrA; GntR family transcriptional regulator | -0.337 | 0.216 | 0.120 | 0.917 |
| ytrB; acetoin utilization transport system ATP-binding protein | 18.239 | 2966.075 | 0.995 | 0.999 |
| ytrC_D; acetoin utilization transport system permease protein | 18.239 | 2966.075 | 0.995 | 0.999 |
| ytrE; acetoin utilization transport system ATP-binding protein | -0.434 | 1.172 | 0.711 | 0.999 |
| ytrF; acetoin utilization transport system permease protein | -0.434 | 1.172 | 0.711 | 0.999 |
| yueD; benzil reductase ((S)-benzoin forming) [EC:1.1.1.320] | -1.021 | 0.950 | 0.284 | 0.978 |
| yufL, malK; two-component system, CitB family, sensor histidine kinase MalK [EC:2.7.13.3] | -1.637 | 0.898 | 0.070 | 0.841 |
| yuiF; putative amino acid transporter | 0.321 | 0.136 | 0.020 | 0.839 |
| yusF; toprim domain protein | -0.867 | 0.524 | 0.100 | 0.896 |
| yvaK; carboxylesterase [EC:3.1.1.1] | -0.062 | 0.203 | 0.760 | 0.999 |
| yvmC; cyclo(L-leucyl-L-leucyl) synthase [EC:2.3.2.22] | 18.239 | 2966.075 | 0.995 | 0.999 |
| ywaD; aminopeptidase YwaD [EC:3.4.11.6 3.4.11.10] | -1.343 | 1.083 | 0.217 | 0.978 |
| yxdJ; two-component system, OmpR family, response regulator YxdJ | 18.239 | 2966.075 | 0.995 | 0.999 |
| yxdK; two-component system, OmpR family, sensor histidine kinase YxdK [EC:2.7.13.3] | 18.239 | 2966.075 | 0.995 | 0.999 |
| yxdL; putative ABC transport system ATP-binding protein | 0.080 | 0.522 | 0.878 | 0.999 |
| yxdM; putative ABC transport system permease protein | -0.218 | 0.564 | 0.699 | 0.999 |
| yxeM; putative amino-acid transport system substrate-binding protein | 0.014 | 0.561 | 0.979 | 0.999 |
| yxeN; putative amino-acid transport system permease protein | 0.726 | 0.644 | 0.261 | 0.978 |
| yxeO; putative amino-acid transport system ATP-binding protein [EC:3.6.3.-] | 0.792 | 0.412 | 0.056 | 0.839 |
| yxjA, nupG; purine nucleoside transport protein | 0.254 | 0.348 | 0.466 | 0.979 |
| yydH; putative peptide zinc metalloprotease protein | -0.261 | 0.425 | 0.540 | 0.990 |
| yydI; putative peptide transport system ATP-binding protein | -1.670 | 1.245 | 0.182 | 0.961 |
| yydJ; putative peptide transport system permease protein | -1.670 | 1.245 | 0.182 | 0.961 |
| yydK; GntR family transcriptional regulator, transcriptional regulator of bglA | -0.072 | 0.510 | 0.888 | 0.999 |
| zapA; cell division protein ZapA | -0.047 | 0.086 | 0.580 | 0.997 |
| zapB; cell division protein ZapB | 0.160 | 0.186 | 0.392 | 0.978 |
| zapC; cell division protein ZapC | 0.352 | 0.531 | 0.508 | 0.981 |
| zapD; cell division protein ZapD | -0.053 | 0.324 | 0.871 | 0.999 |
| zapE; cell division protein ZapE | -0.031 | 0.179 | 0.864 | 0.999 |
| ZDS, crtQ; zeta-carotene desaturase [EC:1.3.5.6] | 18.478 | 3342.830 | 0.996 | 0.999 |
| zipA; cell division protein ZipA | 0.108 | 0.172 | 0.532 | 0.990 |
| zipB; zinc and cadmium transporter | -0.640 | 0.565 | 0.259 | 0.978 |
| zmpB; zinc metalloprotease ZmpB [EC:3.4.24.-] | -0.185 | 0.203 | 0.364 | 0.978 |
| zntA; Cd2+/Zn2+-exporting ATPase [EC:3.6.3.3 3.6.3.5] | 0.107 | 0.091 | 0.241 | 0.978 |
| zntB; zinc transporter | -0.097 | 0.303 | 0.749 | 0.999 |
| zntR; MerR family transcriptional regulator, Zn(II)-responsive regulator of zntA | 0.321 | 0.534 | 0.549 | 0.992 |
| znuA; zinc transport system substrate-binding protein | 0.124 | 0.071 | 0.084 | 0.871 |
| znuB; zinc transport system permease protein | 0.082 | 0.058 | 0.161 | 0.957 |
| znuC; zinc transport system ATP-binding protein [EC:3.6.3.-] | 0.092 | 0.058 | 0.119 | 0.917 |
| zot; zona occludens toxin | -0.423 | 0.534 | 0.430 | 0.978 |
| zraP; zinc resistance-associated protein | -0.890 | 1.071 | 0.408 | 0.978 |
| zraR, hydG; two-component system, NtrC family, response regulator HydG | -0.661 | 0.411 | 0.110 | 0.917 |
| zraS, hydH; two-component system, NtrC family, sensor histidine kinase HydH [EC:2.7.13.3] | -0.647 | 0.721 | 0.371 | 0.978 |
| zur; Fur family transcriptional regulator, zinc uptake regulator | -0.063 | 0.171 | 0.712 | 0.999 |
| zurR, zur; Fur family transcriptional regulator, zinc uptake regulator | 0.037 | 0.227 | 0.870 | 0.999 |
# merge with tb.ra for full results table
full.res <- left_join(tb.ra, results.out, by = "description")
write.csv(full.res, "output/picrust_ko_data_results.csv", row.names = F)Part 3 Pathways Data
pi.dat <- readr::read_tsv("data/PICRUST/path_abun_unstrat_descrip.tsv")
-- Column specification --------------------------------------------------------
cols(
.default = col_double(),
pathway = col_character(),
description = col_character()
)
i Use `spec()` for the full column specifications.# aggregate descriptions to get 1 description per row - unique;y defined.
pi.dat <- pi.dat %>%
group_by(description) %>%
summarise(across(`1.S37.Jun172016`:`99.D01.S37.Jun232016`,.fns = sum))
# transform to long format
pi.dat <- pi.dat %>%
pivot_longer(
cols=`1.S37.Jun172016`:`99.D01.S37.Jun232016`,
names_to = "ID",
values_to = "Abundance"
)
d <- as.data.frame(dat.16s)
mydata <- full_join(pi.dat, d)Joining, by = "ID"Warning in class(x) <- c(setdiff(subclass, tibble_class), tibble_class): Setting
class(x) to multiple strings ("tbl_df", "tbl", ...); result will no longer be an
S4 objectmydata <- mydata %>%
mutate(
ID.n = as.numeric(as.factor(ID)),
description.n = as.numeric(as.factor(description))
) %>%
group_by(ID) %>%
mutate(
dem = ifelse(sum(Abundance)==0, 1, sum(Abundance)),
RelAbundance = Abundance/dem*100) %>%
ungroup()%>%
group_by(description)%>%
mutate(avgRA = mean(RelAbundance))Relative Abundance
tb.ra1 <- mydata %>%
group_by(description) %>%
summarise(ng = n(),
Overall.M = mean(RelAbundance),
Overall.SE = sd(RelAbundance)/sqrt(ng))
tb.ra2m <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(M = mean(RelAbundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = M)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2se <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(),SE = sd(RelAbundance)/sqrt(ng)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = SE)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2var <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(), VAR = var(RelAbundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = VAR)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2ng <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n()) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = ng)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra <- left_join(tb.ra1, tb.ra2m)Joining, by = "description"tb.ra <- cbind(tb.ra, tb.ra2se[,-1])
tb.ra <- cbind(tb.ra, tb.ra2var[,-1])
tb.ra <- cbind(tb.ra, tb.ra2ng[,-1])
colnames(tb.ra) <- c("description", "ng", "Overall Mean", "Overall SE", "Non-Tumor Mean", "Tumor Mean", "Non-Tumor SE", "Tumor SE","Non-Tumor Var", "Tumor Var", "Non-Tumor Ng", "Tumor Ng")
tb.ra <- tb.ra %>%
arrange(desc(`Overall Mean`))
tb.ra <- tb.ra[, c("description", "Overall Mean", "Overall SE","Tumor Mean","Tumor Var", "Tumor SE","Tumor Ng", "Non-Tumor Mean","Non-Tumor Var", "Non-Tumor SE", "Non-Tumor Ng")]
# compute t-test
tb.ra <- tb.ra %>%
mutate(
SEpooled = sqrt(`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`),
t = (`Tumor Mean` - `Non-Tumor Mean`)/(SEpooled),
df = ((`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`)**2)/(((`Tumor Var`/`Tumor Ng`)**2)/(`Tumor Ng`-1) + ((`Non-Tumor Var`/`Non-Tumor Ng`)**2)/(`Non-Tumor Ng`-1)),
p = pt(q = abs(t), df=df, lower.tail = F)*2,
fdr_p = p.adjust(p, method="fdr")
)
kable(tb.ra, format="html", digits=5, caption="Pathway Data Average Relative Abundance of Each Description (sorted in descending order)") %>%
kable_styling(full_width = T) %>%
scroll_box(width = "100%", height="600px")| description | Overall Mean | Overall SE | Tumor Mean | Tumor Var | Tumor SE | Tumor Ng | Non-Tumor Mean | Non-Tumor Var | Non-Tumor SE | Non-Tumor Ng | SEpooled | t | df | p | fdr_p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| adenosine deoxyribonucleotides de novo biosynthesis II | 0.88690 | 0.02700 | 0.90479 | 0.15154 | 0.04828 | 65 | 0.87439 | 0.09073 | 0.03124 | 93 | 0.05751 | 0.52861 | 114.789 | 0.59810 | 0.87368 |
| guanosine deoxyribonucleotides de novo biosynthesis II | 0.88690 | 0.02700 | 0.90479 | 0.15154 | 0.04828 | 65 | 0.87439 | 0.09073 | 0.03124 | 93 | 0.05751 | 0.52861 | 114.789 | 0.59810 | 0.87368 |
| CDP-diacylglycerol biosynthesis I | 0.88194 | 0.01345 | 0.91170 | 0.03514 | 0.02325 | 65 | 0.86114 | 0.02329 | 0.01582 | 93 | 0.02812 | 1.79786 | 119.225 | 0.07473 | 0.58059 |
| CDP-diacylglycerol biosynthesis II | 0.88194 | 0.01345 | 0.91170 | 0.03514 | 0.02325 | 65 | 0.86114 | 0.02329 | 0.01582 | 93 | 0.02812 | 1.79786 | 119.225 | 0.07473 | 0.58059 |
| gondoate biosynthesis (anaerobic) | 0.87704 | 0.01550 | 0.87629 | 0.04677 | 0.02682 | 65 | 0.87756 | 0.03226 | 0.01862 | 93 | 0.03266 | -0.03889 | 121.016 | 0.96904 | 0.99202 |
| superpathway of adenosine nucleotides de novo biosynthesis I | 0.86393 | 0.01682 | 0.88756 | 0.05826 | 0.02994 | 65 | 0.84740 | 0.03507 | 0.01942 | 93 | 0.03568 | 1.12540 | 115.019 | 0.26276 | 0.70194 |
| superpathway of pyrimidine nucleobases salvage | 0.84874 | 0.01688 | 0.84818 | 0.06365 | 0.03129 | 65 | 0.84914 | 0.03254 | 0.01870 | 93 | 0.03646 | -0.02626 | 108.285 | 0.97909 | 0.99202 |
| pentose phosphate pathway (non-oxidative branch) | 0.83879 | 0.01620 | 0.85375 | 0.05882 | 0.03008 | 65 | 0.82833 | 0.02961 | 0.01784 | 93 | 0.03498 | 0.72667 | 107.682 | 0.46900 | 0.81137 |
| adenosine ribonucleotides de novo biosynthesis | 0.82614 | 0.01420 | 0.85277 | 0.04299 | 0.02572 | 65 | 0.80753 | 0.02365 | 0.01595 | 93 | 0.03026 | 1.49517 | 111.240 | 0.13770 | 0.61876 |
| superpathway of adenosine nucleotides de novo biosynthesis II | 0.82437 | 0.01648 | 0.84462 | 0.05556 | 0.02924 | 65 | 0.81021 | 0.03412 | 0.01915 | 93 | 0.03495 | 0.98446 | 115.876 | 0.32694 | 0.72488 |
| cis-vaccenate biosynthesis | 0.80914 | 0.01221 | 0.82019 | 0.02764 | 0.02062 | 65 | 0.80142 | 0.02083 | 0.01496 | 93 | 0.02548 | 0.73685 | 125.034 | 0.46259 | 0.81137 |
| superpathway of phospholipid biosynthesis I (bacteria) | 0.80733 | 0.01202 | 0.83766 | 0.03348 | 0.02269 | 65 | 0.78613 | 0.01455 | 0.01251 | 93 | 0.02591 | 1.98850 | 102.218 | 0.04943 | 0.58059 |
| pyruvate fermentation to isobutanol (engineered) | 0.79175 | 0.02003 | 0.72829 | 0.06072 | 0.03056 | 65 | 0.83610 | 0.06112 | 0.02564 | 93 | 0.03989 | -2.70248 | 138.160 | 0.00775 | 0.58059 |
| UMP biosynthesis | 0.76402 | 0.01455 | 0.78771 | 0.04413 | 0.02606 | 65 | 0.74747 | 0.02570 | 0.01662 | 93 | 0.03091 | 1.30182 | 113.616 | 0.19561 | 0.63974 |
| guanosine ribonucleotides de novo biosynthesis | 0.75442 | 0.01382 | 0.78105 | 0.03993 | 0.02478 | 65 | 0.73581 | 0.02287 | 0.01568 | 93 | 0.02933 | 1.54265 | 112.905 | 0.12571 | 0.61876 |
| phosphatidylglycerol biosynthesis I (plastidic) | 0.75025 | 0.01308 | 0.78312 | 0.03519 | 0.02327 | 65 | 0.72728 | 0.02034 | 0.01479 | 93 | 0.02757 | 2.02548 | 113.300 | 0.04517 | 0.58059 |
| phosphatidylglycerol biosynthesis II (non-plastidic) | 0.75025 | 0.01308 | 0.78312 | 0.03519 | 0.02327 | 65 | 0.72728 | 0.02034 | 0.01479 | 93 | 0.02757 | 2.02548 | 113.300 | 0.04517 | 0.58059 |
| UDP-N-acetylmuramoyl-pentapeptide biosynthesis I (meso-diaminopimelate containing) | 0.74266 | 0.01450 | 0.76961 | 0.04098 | 0.02511 | 65 | 0.72383 | 0.02731 | 0.01713 | 93 | 0.03040 | 1.50601 | 119.469 | 0.13471 | 0.61876 |
| 5-aminoimidazole ribonucleotide biosynthesis I | 0.73704 | 0.01131 | 0.75812 | 0.02632 | 0.02012 | 65 | 0.72231 | 0.01564 | 0.01297 | 93 | 0.02394 | 1.49556 | 114.476 | 0.13752 | 0.61876 |
| superpathway of aromatic amino acid biosynthesis | 0.73591 | 0.01083 | 0.73848 | 0.02452 | 0.01942 | 65 | 0.73411 | 0.01453 | 0.01250 | 93 | 0.02310 | 0.18902 | 114.351 | 0.85041 | 0.96656 |
| peptidoglycan biosynthesis I (meso-diaminopimelate containing) | 0.73357 | 0.01406 | 0.75986 | 0.03882 | 0.02444 | 65 | 0.71519 | 0.02544 | 0.01654 | 93 | 0.02951 | 1.51388 | 118.724 | 0.13271 | 0.61876 |
| Calvin-Benson-Bassham cycle | 0.73231 | 0.01585 | 0.76008 | 0.06500 | 0.03162 | 65 | 0.71290 | 0.02157 | 0.01523 | 93 | 0.03510 | 1.34428 | 93.632 | 0.18211 | 0.62628 |
| UDP-N-acetylmuramoyl-pentapeptide biosynthesis II (lysine-containing) | 0.72969 | 0.01543 | 0.75467 | 0.04542 | 0.02643 | 65 | 0.71223 | 0.03187 | 0.01851 | 93 | 0.03227 | 1.31518 | 121.783 | 0.19092 | 0.63481 |
| peptidoglycan biosynthesis III (mycobacteria) | 0.72807 | 0.01419 | 0.75401 | 0.03991 | 0.02478 | 65 | 0.70994 | 0.02572 | 0.01663 | 93 | 0.02984 | 1.47648 | 117.985 | 0.14248 | 0.61876 |
| 5-aminoimidazole ribonucleotide biosynthesis II | 0.72310 | 0.01219 | 0.73972 | 0.03015 | 0.02154 | 65 | 0.71148 | 0.01875 | 0.01420 | 93 | 0.02580 | 1.09472 | 116.412 | 0.27590 | 0.70413 |
| superpathway of 5-aminoimidazole ribonucleotide biosynthesis | 0.72310 | 0.01219 | 0.73972 | 0.03015 | 0.02154 | 65 | 0.71148 | 0.01875 | 0.01420 | 93 | 0.02580 | 1.09472 | 116.412 | 0.27590 | 0.70413 |
| L-lysine biosynthesis III | 0.70820 | 0.01052 | 0.70273 | 0.02333 | 0.01895 | 65 | 0.71203 | 0.01357 | 0.01208 | 93 | 0.02247 | -0.41371 | 113.540 | 0.67987 | 0.90725 |
| chorismate biosynthesis I | 0.70514 | 0.01067 | 0.70923 | 0.02424 | 0.01931 | 65 | 0.70227 | 0.01383 | 0.01220 | 93 | 0.02284 | 0.30482 | 112.758 | 0.76107 | 0.92864 |
| chorismate biosynthesis from 3-dehydroquinate | 0.70464 | 0.01214 | 0.72041 | 0.03438 | 0.02300 | 65 | 0.69361 | 0.01549 | 0.01291 | 93 | 0.02637 | 1.01628 | 103.514 | 0.31186 | 0.72280 |
| coenzyme A biosynthesis I | 0.70460 | 0.01292 | 0.72948 | 0.03295 | 0.02251 | 65 | 0.68721 | 0.02134 | 0.01515 | 93 | 0.02714 | 1.55758 | 118.203 | 0.12201 | 0.61876 |
| L-isoleucine biosynthesis II | 0.69810 | 0.01619 | 0.66423 | 0.04890 | 0.02743 | 65 | 0.72178 | 0.03528 | 0.01948 | 93 | 0.03364 | -1.71056 | 123.043 | 0.08968 | 0.60802 |
| inosine-5’-phosphate biosynthesis I | 0.69796 | 0.01228 | 0.72527 | 0.02804 | 0.02077 | 65 | 0.67887 | 0.02025 | 0.01476 | 93 | 0.02548 | 1.82113 | 123.088 | 0.07102 | 0.58059 |
| glycolysis III (from glucose) | 0.69699 | 0.01820 | 0.72401 | 0.06285 | 0.03109 | 65 | 0.67810 | 0.04468 | 0.02192 | 93 | 0.03804 | 1.20678 | 122.382 | 0.22985 | 0.69476 |
| peptidoglycan maturation (meso-diaminopimelate containing) | 0.69402 | 0.02367 | 0.71173 | 0.09817 | 0.03886 | 65 | 0.68165 | 0.08244 | 0.02977 | 93 | 0.04896 | 0.61444 | 130.019 | 0.54000 | 0.85500 |
| glycolysis I (from glucose 6-phosphate) | 0.68809 | 0.01630 | 0.72877 | 0.04859 | 0.02734 | 65 | 0.65966 | 0.03588 | 0.01964 | 93 | 0.03366 | 2.05289 | 124.114 | 0.04218 | 0.58059 |
| superpathway of L-threonine biosynthesis | 0.67764 | 0.01185 | 0.68103 | 0.03005 | 0.02150 | 65 | 0.67526 | 0.01696 | 0.01351 | 93 | 0.02539 | 0.22717 | 112.311 | 0.82070 | 0.95867 |
| L-isoleucine biosynthesis I (from threonine) | 0.66999 | 0.01428 | 0.63175 | 0.03950 | 0.02465 | 65 | 0.69672 | 0.02578 | 0.01665 | 93 | 0.02975 | -2.18411 | 118.545 | 0.03092 | 0.58059 |
| L-valine biosynthesis | 0.66999 | 0.01428 | 0.63175 | 0.03950 | 0.02465 | 65 | 0.69672 | 0.02578 | 0.01665 | 93 | 0.02975 | -2.18411 | 118.545 | 0.03092 | 0.58059 |
| glycolysis II (from fructose 6-phosphate) | 0.66604 | 0.01711 | 0.70667 | 0.05701 | 0.02962 | 65 | 0.63764 | 0.03733 | 0.02003 | 93 | 0.03576 | 1.93063 | 118.688 | 0.05591 | 0.58059 |
| superpathway of purine nucleotides de novo biosynthesis I | 0.66057 | 0.00982 | 0.66589 | 0.02386 | 0.01916 | 65 | 0.65685 | 0.00937 | 0.01003 | 93 | 0.02163 | 0.41802 | 98.759 | 0.67684 | 0.90623 |
| UDP-N-acetyl-D-glucosamine biosynthesis I | 0.65422 | 0.01296 | 0.67813 | 0.03436 | 0.02299 | 65 | 0.63750 | 0.02070 | 0.01492 | 93 | 0.02741 | 1.48219 | 115.046 | 0.14102 | 0.61876 |
| superpathway of pyrimidine ribonucleotides de novo biosynthesis | 0.65400 | 0.01192 | 0.65756 | 0.03607 | 0.02356 | 65 | 0.65151 | 0.01322 | 0.01192 | 93 | 0.02640 | 0.22917 | 96.579 | 0.81922 | 0.95867 |
| superpathway of tetrahydrofolate biosynthesis and salvage | 0.65366 | 0.01210 | 0.65859 | 0.02950 | 0.02130 | 65 | 0.65022 | 0.01894 | 0.01427 | 93 | 0.02564 | 0.32640 | 117.825 | 0.74470 | 0.92580 |
| tRNA charging | 0.65201 | 0.01226 | 0.67270 | 0.02861 | 0.02098 | 65 | 0.63755 | 0.02011 | 0.01471 | 93 | 0.02562 | 1.37219 | 121.870 | 0.17252 | 0.61889 |
| 6-hydroxymethyl-dihydropterin diphosphate biosynthesis I | 0.65198 | 0.01101 | 0.65359 | 0.02559 | 0.01984 | 65 | 0.65085 | 0.01490 | 0.01266 | 93 | 0.02354 | 0.11650 | 113.595 | 0.90746 | 0.97851 |
| adenine and adenosine salvage III | 0.64944 | 0.02406 | 0.67775 | 0.09273 | 0.03777 | 65 | 0.62965 | 0.09063 | 0.03122 | 93 | 0.04900 | 0.98177 | 136.870 | 0.32795 | 0.72488 |
| urate biosynthesis/inosine 5’-phosphate degradation | 0.63801 | 0.01276 | 0.63224 | 0.02800 | 0.02076 | 65 | 0.64205 | 0.02441 | 0.01620 | 93 | 0.02633 | -0.37259 | 131.730 | 0.71005 | 0.91502 |
| superpathway of guanosine nucleotides de novo biosynthesis II | 0.63504 | 0.01282 | 0.64098 | 0.04102 | 0.02512 | 65 | 0.63089 | 0.01572 | 0.01300 | 93 | 0.02829 | 0.35655 | 97.989 | 0.72220 | 0.91502 |
| superpathway of L-isoleucine biosynthesis I | 0.63459 | 0.01127 | 0.60939 | 0.02731 | 0.02050 | 65 | 0.65220 | 0.01448 | 0.01248 | 93 | 0.02400 | -1.78414 | 109.733 | 0.07716 | 0.58059 |
| superpathway of guanosine nucleotides de novo biosynthesis I | 0.62958 | 0.01258 | 0.63605 | 0.04021 | 0.02487 | 65 | 0.62506 | 0.01467 | 0.01256 | 93 | 0.02786 | 0.39426 | 96.441 | 0.69426 | 0.90862 |
| superpathway of branched amino acid biosynthesis | 0.62706 | 0.01378 | 0.59943 | 0.03635 | 0.02365 | 65 | 0.64638 | 0.02501 | 0.01640 | 93 | 0.02878 | -1.63167 | 120.913 | 0.10535 | 0.61876 |
| homolactic fermentation | 0.62203 | 0.01688 | 0.66297 | 0.05689 | 0.02958 | 65 | 0.59342 | 0.03527 | 0.01947 | 93 | 0.03542 | 1.96368 | 116.284 | 0.05195 | 0.58059 |
| superpathway of tetrahydrofolate biosynthesis | 0.62114 | 0.01326 | 0.62219 | 0.03264 | 0.02241 | 65 | 0.62040 | 0.02472 | 0.01630 | 93 | 0.02771 | 0.06471 | 125.268 | 0.94851 | 0.99202 |
| superpathway of purine nucleotides de novo biosynthesis II | 0.61763 | 0.01527 | 0.63888 | 0.03877 | 0.02442 | 65 | 0.60278 | 0.03532 | 0.01949 | 93 | 0.03124 | 1.15562 | 133.723 | 0.24990 | 0.70007 |
| N10-formyl-tetrahydrofolate biosynthesis | 0.61332 | 0.00971 | 0.59836 | 0.01979 | 0.01745 | 65 | 0.62378 | 0.01139 | 0.01107 | 93 | 0.02066 | -1.23057 | 113.101 | 0.22104 | 0.67841 |
| L-isoleucine biosynthesis III | 0.61017 | 0.01344 | 0.58694 | 0.03418 | 0.02293 | 65 | 0.62641 | 0.02426 | 0.01615 | 93 | 0.02805 | -1.40697 | 122.312 | 0.16197 | 0.61876 |
| fatty acid elongation – saturated | 0.60322 | 0.01909 | 0.58223 | 0.06783 | 0.03230 | 65 | 0.61789 | 0.05059 | 0.02332 | 93 | 0.03984 | -0.89510 | 124.568 | 0.37246 | 0.74607 |
| gluconeogenesis I | 0.59898 | 0.01152 | 0.60261 | 0.03408 | 0.02290 | 65 | 0.59645 | 0.01208 | 0.01140 | 93 | 0.02558 | 0.24076 | 95.562 | 0.81026 | 0.95649 |
| pyruvate fermentation to acetate and lactate II | 0.59344 | 0.02637 | 0.64080 | 0.13906 | 0.04625 | 65 | 0.56033 | 0.08804 | 0.03077 | 93 | 0.05555 | 1.44846 | 117.207 | 0.15016 | 0.61876 |
| superpathway of pyrimidine deoxyribonucleotides de novo biosynthesis (E. coli) | 0.59272 | 0.01188 | 0.60045 | 0.03387 | 0.02283 | 65 | 0.58731 | 0.01439 | 0.01244 | 93 | 0.02600 | 0.50551 | 101.431 | 0.61430 | 0.88168 |
| superpathway of L-serine and glycine biosynthesis I | 0.58799 | 0.01284 | 0.56392 | 0.03094 | 0.02182 | 65 | 0.60481 | 0.02226 | 0.01547 | 93 | 0.02675 | -1.52894 | 122.917 | 0.12885 | 0.61876 |
| polyisoprenoid biosynthesis (E. coli) | 0.58795 | 0.01314 | 0.59250 | 0.03389 | 0.02284 | 65 | 0.58477 | 0.02298 | 0.01572 | 93 | 0.02772 | 0.27881 | 120.242 | 0.78087 | 0.93380 |
| 6-hydroxymethyl-dihydropterin diphosphate biosynthesis III (Chlamydia) | 0.58175 | 0.01247 | 0.59572 | 0.03008 | 0.02151 | 65 | 0.57199 | 0.02075 | 0.01494 | 93 | 0.02619 | 0.90594 | 121.012 | 0.36677 | 0.74607 |
| aerobic respiration I (cytochrome c) | 0.57434 | 0.04155 | 0.46362 | 0.24857 | 0.06184 | 65 | 0.65173 | 0.27782 | 0.05466 | 93 | 0.08253 | -2.27921 | 142.537 | 0.02414 | 0.58059 |
| S-adenosyl-L-methionine cycle I | 0.56963 | 0.01684 | 0.58792 | 0.05125 | 0.02808 | 65 | 0.55685 | 0.04043 | 0.02085 | 93 | 0.03497 | 0.88841 | 127.143 | 0.37600 | 0.74607 |
| L-lysine biosynthesis VI | 0.56933 | 0.01090 | 0.57834 | 0.02481 | 0.01954 | 65 | 0.56303 | 0.01468 | 0.01256 | 93 | 0.02323 | 0.65921 | 114.281 | 0.51109 | 0.84266 |
| pyrimidine deoxyribonucleotides de novo biosynthesis I | 0.56568 | 0.01230 | 0.57103 | 0.03756 | 0.02404 | 65 | 0.56194 | 0.01464 | 0.01255 | 93 | 0.02712 | 0.33524 | 98.542 | 0.73815 | 0.92580 |
| (5Z)-dodec-5-enoate biosynthesis | 0.56436 | 0.01589 | 0.54432 | 0.04135 | 0.02522 | 65 | 0.57836 | 0.03884 | 0.02044 | 93 | 0.03246 | -1.04854 | 135.101 | 0.29626 | 0.72280 |
| galactose degradation I (Leloir pathway) | 0.56239 | 0.02324 | 0.61376 | 0.09890 | 0.03901 | 65 | 0.52648 | 0.07362 | 0.02814 | 93 | 0.04809 | 1.81471 | 124.480 | 0.07198 | 0.58059 |
| starch degradation V | 0.56033 | 0.01856 | 0.57691 | 0.05683 | 0.02957 | 65 | 0.54875 | 0.05305 | 0.02388 | 93 | 0.03801 | 0.74084 | 134.829 | 0.46008 | 0.81137 |
| inosine-5’-phosphate biosynthesis III | 0.56024 | 0.01384 | 0.56341 | 0.03918 | 0.02455 | 65 | 0.55801 | 0.02439 | 0.01619 | 93 | 0.02941 | 0.18365 | 116.470 | 0.85460 | 0.96656 |
| L-tryptophan biosynthesis | 0.55463 | 0.01380 | 0.52848 | 0.03318 | 0.02259 | 65 | 0.57291 | 0.02748 | 0.01719 | 93 | 0.02839 | -1.56499 | 129.383 | 0.12003 | 0.61876 |
| sucrose degradation III (sucrose invertase) | 0.54379 | 0.02018 | 0.58081 | 0.07989 | 0.03506 | 65 | 0.51793 | 0.05260 | 0.02378 | 93 | 0.04236 | 1.48428 | 118.932 | 0.14038 | 0.61876 |
| superpathway of S-adenosyl-L-methionine biosynthesis | 0.53781 | 0.01199 | 0.53242 | 0.02622 | 0.02008 | 65 | 0.54157 | 0.02048 | 0.01484 | 93 | 0.02497 | -0.36644 | 126.690 | 0.71464 | 0.91502 |
| L-lysine biosynthesis I | 0.52754 | 0.00996 | 0.53002 | 0.01991 | 0.01750 | 65 | 0.52580 | 0.01291 | 0.01178 | 93 | 0.02110 | 0.19995 | 118.247 | 0.84186 | 0.96656 |
| acetylene degradation | 0.52709 | 0.02706 | 0.55471 | 0.12804 | 0.04438 | 65 | 0.50779 | 0.10739 | 0.03398 | 93 | 0.05590 | 0.83929 | 129.958 | 0.40285 | 0.78408 |
| pyrimidine deoxyribonucleotides de novo biosynthesis II | 0.52654 | 0.01507 | 0.54927 | 0.04080 | 0.02505 | 65 | 0.51065 | 0.03227 | 0.01863 | 93 | 0.03122 | 1.23699 | 127.263 | 0.21837 | 0.67841 |
| thiamin salvage II | 0.52095 | 0.01153 | 0.52137 | 0.02460 | 0.01945 | 65 | 0.52066 | 0.01873 | 0.01419 | 93 | 0.02408 | 0.02934 | 125.518 | 0.97664 | 0.99202 |
| glycogen biosynthesis I (from ADP-D-Glucose) | 0.51514 | 0.01782 | 0.52985 | 0.05455 | 0.02897 | 65 | 0.50486 | 0.04740 | 0.02258 | 93 | 0.03673 | 0.68053 | 131.584 | 0.49736 | 0.82687 |
| flavin biosynthesis I (bacteria and plants) | 0.51351 | 0.01338 | 0.51895 | 0.03616 | 0.02359 | 65 | 0.50970 | 0.02306 | 0.01574 | 93 | 0.02836 | 0.32625 | 117.510 | 0.74482 | 0.92580 |
| O-antigen building blocks biosynthesis (E. coli) | 0.51107 | 0.01275 | 0.53919 | 0.03372 | 0.02278 | 65 | 0.49142 | 0.01940 | 0.01444 | 93 | 0.02697 | 1.77137 | 113.090 | 0.07919 | 0.58059 |
| superpathway of L-alanine biosynthesis | 0.50869 | 0.01622 | 0.50300 | 0.04340 | 0.02584 | 65 | 0.51267 | 0.04074 | 0.02093 | 93 | 0.03325 | -0.29084 | 135.081 | 0.77162 | 0.93296 |
| pyrimidine deoxyribonucleotide phosphorylation | 0.50470 | 0.01084 | 0.51099 | 0.02834 | 0.02088 | 65 | 0.50030 | 0.01191 | 0.01132 | 93 | 0.02375 | 0.44987 | 101.041 | 0.65377 | 0.90427 |
| pentose phosphate pathway | 0.49986 | 0.01287 | 0.48904 | 0.03068 | 0.02173 | 65 | 0.50742 | 0.02315 | 0.01578 | 93 | 0.02685 | -0.68455 | 125.098 | 0.49489 | 0.82687 |
| superpathway of pyrimidine ribonucleosides salvage | 0.49865 | 0.01588 | 0.50149 | 0.04662 | 0.02678 | 65 | 0.49666 | 0.03554 | 0.01955 | 93 | 0.03316 | 0.14562 | 125.574 | 0.88445 | 0.97057 |
| dTDP-L-rhamnose biosynthesis I | 0.49815 | 0.01847 | 0.54236 | 0.08202 | 0.03552 | 65 | 0.46724 | 0.03253 | 0.01870 | 93 | 0.04015 | 1.87105 | 99.106 | 0.06429 | 0.58059 |
| superpathway of geranylgeranyl diphosphate biosynthesis II (via MEP) | 0.49182 | 0.01392 | 0.49777 | 0.03906 | 0.02452 | 65 | 0.48767 | 0.02499 | 0.01639 | 93 | 0.02949 | 0.34261 | 117.669 | 0.73250 | 0.92198 |
| superpathway of L-phenylalanine biosynthesis | 0.48737 | 0.01291 | 0.50505 | 0.02931 | 0.02124 | 65 | 0.47501 | 0.02420 | 0.01613 | 93 | 0.02667 | 1.12621 | 129.238 | 0.26216 | 0.70194 |
| mixed acid fermentation | 0.48706 | 0.01015 | 0.48623 | 0.01758 | 0.01644 | 65 | 0.48763 | 0.01552 | 0.01292 | 93 | 0.02091 | -0.06712 | 132.329 | 0.94659 | 0.99202 |
| methylerythritol phosphate pathway I | 0.48376 | 0.01429 | 0.48681 | 0.04059 | 0.02499 | 65 | 0.48164 | 0.02679 | 0.01697 | 93 | 0.03021 | 0.17112 | 119.048 | 0.86442 | 0.96656 |
| methylerythritol phosphate pathway II | 0.48376 | 0.01429 | 0.48681 | 0.04059 | 0.02499 | 65 | 0.48164 | 0.02679 | 0.01697 | 93 | 0.03021 | 0.17112 | 119.048 | 0.86442 | 0.96656 |
| queuosine biosynthesis | 0.48093 | 0.01234 | 0.46187 | 0.02901 | 0.02113 | 65 | 0.49425 | 0.02042 | 0.01482 | 93 | 0.02580 | -1.25484 | 121.934 | 0.21194 | 0.67308 |
| heme biosynthesis II (anaerobic) | 0.46990 | 0.01526 | 0.44343 | 0.03765 | 0.02407 | 65 | 0.48840 | 0.03572 | 0.01960 | 93 | 0.03104 | -1.44907 | 135.557 | 0.14963 | 0.61876 |
| purine ribonucleosides degradation | 0.46985 | 0.02344 | 0.47520 | 0.08733 | 0.03665 | 65 | 0.46612 | 0.08736 | 0.03065 | 93 | 0.04778 | 0.19001 | 137.889 | 0.84958 | 0.96656 |
| GDP-mannose biosynthesis | 0.46593 | 0.01407 | 0.44276 | 0.03006 | 0.02151 | 65 | 0.48212 | 0.03181 | 0.01849 | 93 | 0.02837 | -1.38755 | 140.290 | 0.16747 | 0.61876 |
| superpathway of L-tyrosine biosynthesis | 0.46268 | 0.01269 | 0.47724 | 0.02899 | 0.02112 | 65 | 0.45250 | 0.02297 | 0.01572 | 93 | 0.02633 | 0.93991 | 127.348 | 0.34904 | 0.73493 |
| L-methionine biosynthesis I | 0.46090 | 0.01288 | 0.45846 | 0.02643 | 0.02017 | 65 | 0.46261 | 0.02631 | 0.01682 | 93 | 0.02626 | -0.15799 | 137.673 | 0.87469 | 0.97057 |
| oleate biosynthesis IV (anaerobic) | 0.45434 | 0.01872 | 0.42978 | 0.05454 | 0.02897 | 65 | 0.47152 | 0.05585 | 0.02451 | 93 | 0.03794 | -1.10011 | 138.899 | 0.27319 | 0.70413 |
| pyrimidine deoxyribonucleosides salvage | 0.45164 | 0.01734 | 0.44054 | 0.04740 | 0.02701 | 65 | 0.45940 | 0.04792 | 0.02270 | 93 | 0.03528 | -0.53469 | 138.342 | 0.59372 | 0.87368 |
| peptidoglycan biosynthesis IV (Enterococcus faecium) | 0.44868 | 0.02141 | 0.46037 | 0.07792 | 0.03462 | 65 | 0.44051 | 0.06921 | 0.02728 | 93 | 0.04408 | 0.45055 | 132.582 | 0.65305 | 0.90427 |
| L-methionine biosynthesis III | 0.44393 | 0.01367 | 0.43527 | 0.03081 | 0.02177 | 65 | 0.44999 | 0.02885 | 0.01761 | 93 | 0.02800 | -0.52569 | 134.969 | 0.59997 | 0.87368 |
| superpathway of L-methionine biosynthesis (transsulfuration) | 0.44205 | 0.01226 | 0.43604 | 0.03024 | 0.02157 | 65 | 0.44626 | 0.01942 | 0.01445 | 93 | 0.02596 | -0.39364 | 117.824 | 0.69456 | 0.90862 |
| superpathway of pyrimidine deoxyribonucleoside salvage | 0.43839 | 0.01363 | 0.43837 | 0.03242 | 0.02233 | 65 | 0.43840 | 0.02757 | 0.01722 | 93 | 0.02820 | -0.00074 | 130.592 | 0.99941 | 0.99941 |
| superpathway of histidine, purine, and pyrimidine biosynthesis | 0.43810 | 0.01336 | 0.43792 | 0.02836 | 0.02089 | 65 | 0.43823 | 0.02836 | 0.01746 | 93 | 0.02723 | -0.01132 | 137.881 | 0.99099 | 0.99849 |
| superpathway of pyrimidine deoxyribonucleotides de novo biosynthesis | 0.43803 | 0.01281 | 0.42563 | 0.02835 | 0.02089 | 65 | 0.44670 | 0.02433 | 0.01617 | 93 | 0.02642 | -0.79752 | 131.002 | 0.42659 | 0.79911 |
| superpathway of L-lysine, L-threonine and L-methionine biosynthesis I | 0.42865 | 0.00988 | 0.42288 | 0.01821 | 0.01674 | 65 | 0.43268 | 0.01359 | 0.01209 | 93 | 0.02065 | -0.47473 | 124.604 | 0.63581 | 0.90147 |
| L-arginine biosynthesis II (acetyl cycle) | 0.42860 | 0.01264 | 0.41588 | 0.03022 | 0.02156 | 65 | 0.43750 | 0.02186 | 0.01533 | 93 | 0.02646 | -0.81715 | 123.181 | 0.41542 | 0.78940 |
| palmitate biosynthesis II (bacteria and plants) | 0.42658 | 0.01745 | 0.42392 | 0.04694 | 0.02687 | 65 | 0.42844 | 0.04946 | 0.02306 | 93 | 0.03541 | -0.12774 | 140.115 | 0.89854 | 0.97689 |
| TCA cycle I (prokaryotic) | 0.41963 | 0.01694 | 0.37898 | 0.04133 | 0.02521 | 65 | 0.44803 | 0.04666 | 0.02240 | 93 | 0.03373 | -2.04752 | 142.941 | 0.04244 | 0.58059 |
| L-arginine biosynthesis I (via L-ornithine) | 0.41934 | 0.01073 | 0.41029 | 0.02241 | 0.01857 | 65 | 0.42567 | 0.01533 | 0.01284 | 93 | 0.02257 | -0.68125 | 120.637 | 0.49702 | 0.82687 |
| pantothenate and coenzyme A biosynthesis I | 0.41791 | 0.01101 | 0.39604 | 0.01926 | 0.01721 | 65 | 0.43319 | 0.01873 | 0.01419 | 93 | 0.02231 | -1.66548 | 136.651 | 0.09811 | 0.61876 |
| L-arginine biosynthesis IV (archaebacteria) | 0.41781 | 0.01067 | 0.40876 | 0.02214 | 0.01845 | 65 | 0.42413 | 0.01520 | 0.01279 | 93 | 0.02245 | -0.68494 | 120.821 | 0.49470 | 0.82687 |
| palmitoleate biosynthesis I (from (5Z)-dodec-5-enoate) | 0.41388 | 0.01918 | 0.38776 | 0.05706 | 0.02963 | 65 | 0.43215 | 0.05868 | 0.02512 | 93 | 0.03884 | -1.14279 | 139.084 | 0.25509 | 0.70193 |
| superpathway of purine deoxyribonucleosides degradation | 0.41048 | 0.01913 | 0.42385 | 0.05950 | 0.03025 | 65 | 0.40114 | 0.05708 | 0.02478 | 93 | 0.03910 | 0.58085 | 136.054 | 0.56230 | 0.86025 |
| glycogen degradation I (bacterial) | 0.41014 | 0.01429 | 0.42046 | 0.03270 | 0.02243 | 65 | 0.40294 | 0.03219 | 0.01861 | 93 | 0.02914 | 0.60113 | 137.189 | 0.54874 | 0.86025 |
| superpathway of N-acetylneuraminate degradation | 0.40919 | 0.01728 | 0.42213 | 0.04022 | 0.02488 | 65 | 0.40015 | 0.05231 | 0.02372 | 93 | 0.03437 | 0.63951 | 148.102 | 0.52348 | 0.84889 |
| stearate biosynthesis II (bacteria and plants) | 0.40897 | 0.01904 | 0.38281 | 0.05618 | 0.02940 | 65 | 0.42725 | 0.05782 | 0.02493 | 93 | 0.03855 | -1.15296 | 139.116 | 0.25090 | 0.70007 |
| mycolate biosynthesis | 0.40880 | 0.02093 | 0.39254 | 0.07254 | 0.03341 | 65 | 0.42016 | 0.06732 | 0.02690 | 93 | 0.04289 | -0.64400 | 134.567 | 0.52067 | 0.84795 |
| TCA cycle VI (obligate autotrophs) | 0.40592 | 0.01047 | 0.38557 | 0.01699 | 0.01617 | 65 | 0.42015 | 0.01724 | 0.01362 | 93 | 0.02114 | -1.63573 | 138.508 | 0.10417 | 0.61876 |
| L-histidine biosynthesis | 0.40527 | 0.01079 | 0.38157 | 0.02088 | 0.01792 | 65 | 0.42184 | 0.01621 | 0.01320 | 93 | 0.02226 | -1.80892 | 126.404 | 0.07284 | 0.58059 |
| L-isoleucine biosynthesis IV | 0.40494 | 0.01641 | 0.41783 | 0.04338 | 0.02583 | 65 | 0.39593 | 0.04225 | 0.02131 | 93 | 0.03349 | 0.65388 | 136.715 | 0.51429 | 0.84445 |
| preQ0 biosynthesis | 0.40307 | 0.01148 | 0.37879 | 0.02158 | 0.01822 | 65 | 0.42005 | 0.01979 | 0.01459 | 93 | 0.02334 | -1.76718 | 134.029 | 0.07947 | 0.58059 |
| superpathway of pyrimidine deoxyribonucleosides degradation | 0.40072 | 0.01762 | 0.41282 | 0.05437 | 0.02892 | 65 | 0.39226 | 0.04569 | 0.02216 | 93 | 0.03644 | 0.56400 | 130.046 | 0.57372 | 0.86383 |
| tetrapyrrole biosynthesis I (from glutamate) | 0.39689 | 0.01357 | 0.41447 | 0.03242 | 0.02233 | 65 | 0.38461 | 0.02673 | 0.01695 | 93 | 0.02804 | 1.06481 | 129.169 | 0.28895 | 0.72280 |
| aspartate superpathway | 0.39573 | 0.00978 | 0.39268 | 0.01814 | 0.01671 | 65 | 0.39787 | 0.01314 | 0.01189 | 93 | 0.02050 | -0.25282 | 123.243 | 0.80083 | 0.94816 |
| superpathway of fatty acid biosynthesis initiation (E. coli) | 0.38795 | 0.01999 | 0.36846 | 0.06370 | 0.03131 | 65 | 0.40157 | 0.06300 | 0.02603 | 93 | 0.04071 | -0.81330 | 137.388 | 0.41745 | 0.78940 |
| superpathway of glycolysis and Entner-Doudoroff | 0.38765 | 0.01110 | 0.37788 | 0.01864 | 0.01693 | 65 | 0.39448 | 0.02014 | 0.01472 | 93 | 0.02243 | -0.73997 | 141.168 | 0.46055 | 0.81137 |
| phosphopantothenate biosynthesis I | 0.38646 | 0.01188 | 0.35778 | 0.02127 | 0.01809 | 65 | 0.40650 | 0.02227 | 0.01547 | 93 | 0.02381 | -2.04628 | 139.837 | 0.04260 | 0.58059 |
| L-ornithine biosynthesis | 0.38421 | 0.01446 | 0.36384 | 0.03550 | 0.02337 | 65 | 0.39845 | 0.03120 | 0.01832 | 93 | 0.02969 | -1.16558 | 132.095 | 0.24588 | 0.70007 |
| tetrapyrrole biosynthesis II (from glycine) | 0.37955 | 0.01311 | 0.39690 | 0.03125 | 0.02193 | 65 | 0.36742 | 0.02426 | 0.01615 | 93 | 0.02723 | 1.08252 | 126.399 | 0.28108 | 0.70982 |
| taxadiene biosynthesis (engineered) | 0.37385 | 0.01091 | 0.38061 | 0.01668 | 0.01602 | 65 | 0.36913 | 0.02043 | 0.01482 | 93 | 0.02182 | 0.52615 | 146.032 | 0.59959 | 0.87368 |
| TCA cycle V (2-oxoglutarate:ferredoxin oxidoreductase) | 0.37375 | 0.01415 | 0.34283 | 0.02364 | 0.01907 | 65 | 0.39537 | 0.03642 | 0.01979 | 93 | 0.02748 | -1.91179 | 152.792 | 0.05778 | 0.58059 |
| incomplete reductive TCA cycle | 0.35902 | 0.01858 | 0.34404 | 0.04805 | 0.02719 | 65 | 0.36950 | 0.05941 | 0.02527 | 93 | 0.03712 | -0.68573 | 146.366 | 0.49397 | 0.82687 |
| L-histidine degradation I | 0.35208 | 0.01866 | 0.36546 | 0.09220 | 0.03766 | 65 | 0.34273 | 0.02956 | 0.01783 | 93 | 0.04167 | 0.54530 | 92.662 | 0.58685 | 0.87368 |
| superpathway of L-aspartate and L-asparagine biosynthesis | 0.35018 | 0.01671 | 0.35696 | 0.05166 | 0.02819 | 65 | 0.34545 | 0.03926 | 0.02055 | 93 | 0.03488 | 0.32989 | 125.435 | 0.74204 | 0.92580 |
| NAD salvage pathway I | 0.33941 | 0.01273 | 0.33777 | 0.02659 | 0.02023 | 65 | 0.34056 | 0.02520 | 0.01646 | 93 | 0.02608 | -0.10725 | 135.501 | 0.91475 | 0.97851 |
| L-lysine biosynthesis II | 0.33611 | 0.01558 | 0.34982 | 0.04429 | 0.02610 | 65 | 0.32653 | 0.03438 | 0.01923 | 93 | 0.03242 | 0.71854 | 126.406 | 0.47375 | 0.81137 |
| lipid IVA biosynthesis | 0.32874 | 0.01601 | 0.34347 | 0.05025 | 0.02780 | 65 | 0.31845 | 0.03391 | 0.01909 | 93 | 0.03373 | 0.74178 | 120.032 | 0.45967 | 0.81137 |
| CMP-3-deoxy-D-manno-octulosonate biosynthesis I | 0.32590 | 0.01525 | 0.34348 | 0.04336 | 0.02583 | 65 | 0.31362 | 0.03217 | 0.01860 | 93 | 0.03183 | 0.93819 | 124.329 | 0.34997 | 0.73493 |
| superpathway of N-acetylglucosamine, N-acetylmannosamine and N-acetylneuraminate degradation | 0.32402 | 0.01732 | 0.34757 | 0.05408 | 0.02884 | 65 | 0.30756 | 0.04257 | 0.02139 | 93 | 0.03591 | 1.11395 | 127.039 | 0.26741 | 0.70194 |
| TCA cycle VII (acetate-producers) | 0.31176 | 0.01707 | 0.28727 | 0.04251 | 0.02557 | 65 | 0.32888 | 0.04832 | 0.02279 | 93 | 0.03426 | -1.21446 | 143.208 | 0.22657 | 0.69009 |
| TCA cycle VIII (helicobacter) | 0.30985 | 0.01850 | 0.27510 | 0.05169 | 0.02820 | 65 | 0.33414 | 0.05487 | 0.02429 | 93 | 0.03722 | -1.58640 | 140.424 | 0.11490 | 0.61876 |
| 8-amino-7-oxononanoate biosynthesis I | 0.30381 | 0.01697 | 0.30676 | 0.04608 | 0.02663 | 65 | 0.30176 | 0.04560 | 0.02214 | 93 | 0.03463 | 0.14435 | 137.420 | 0.88544 | 0.97057 |
| superpathay of heme biosynthesis from glutamate | 0.30306 | 0.01083 | 0.29940 | 0.01994 | 0.01752 | 65 | 0.30561 | 0.01776 | 0.01382 | 93 | 0.02231 | -0.27867 | 132.696 | 0.78093 | 0.93380 |
| biotin biosynthesis I | 0.29403 | 0.01562 | 0.29616 | 0.03903 | 0.02450 | 65 | 0.29255 | 0.03863 | 0.02038 | 93 | 0.03187 | 0.11340 | 137.425 | 0.90988 | 0.97851 |
| fatty acid β-oxidation I | 0.29112 | 0.02296 | 0.25310 | 0.07655 | 0.03432 | 65 | 0.31769 | 0.08714 | 0.03061 | 93 | 0.04598 | -1.40465 | 143.265 | 0.16229 | 0.61876 |
| Kdo transfer to lipid IVA III (Chlamydia) | 0.29002 | 0.01260 | 0.28381 | 0.02263 | 0.01866 | 65 | 0.29436 | 0.02702 | 0.01705 | 93 | 0.02527 | -0.41773 | 145.095 | 0.67676 | 0.90623 |
| anhydromuropeptides recycling | 0.28808 | 0.01435 | 0.26586 | 0.03338 | 0.02266 | 65 | 0.30360 | 0.03174 | 0.01847 | 93 | 0.02924 | -1.29098 | 135.655 | 0.19891 | 0.64524 |
| superpathway of thiamin diphosphate biosynthesis I | 0.28651 | 0.01604 | 0.31376 | 0.05170 | 0.02820 | 65 | 0.26746 | 0.03248 | 0.01869 | 93 | 0.03383 | 1.36868 | 116.865 | 0.17372 | 0.61889 |
| superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate) | 0.28165 | 0.01832 | 0.31326 | 0.06680 | 0.03206 | 65 | 0.25956 | 0.04285 | 0.02147 | 93 | 0.03858 | 1.39167 | 117.784 | 0.16664 | 0.61876 |
| NAD biosynthesis I (from aspartate) | 0.27773 | 0.01317 | 0.27577 | 0.03266 | 0.02242 | 65 | 0.27910 | 0.02405 | 0.01608 | 93 | 0.02759 | -0.12076 | 123.982 | 0.90408 | 0.97851 |
| heme biosynthesis I (aerobic) | 0.27403 | 0.01121 | 0.26179 | 0.02213 | 0.01845 | 65 | 0.28259 | 0.01830 | 0.01403 | 93 | 0.02318 | -0.89725 | 129.312 | 0.37126 | 0.74607 |
| adenosylcobalamin salvage from cobinamide I | 0.26962 | 0.01518 | 0.28336 | 0.04713 | 0.02693 | 65 | 0.26002 | 0.02914 | 0.01770 | 93 | 0.03222 | 0.72437 | 116.170 | 0.47029 | 0.81137 |
| guanosine nucleotides degradation III | 0.26714 | 0.01847 | 0.23739 | 0.04582 | 0.02655 | 65 | 0.28793 | 0.05908 | 0.02521 | 93 | 0.03661 | -1.38041 | 147.815 | 0.16955 | 0.61876 |
| superpathway of sulfate assimilation and cysteine biosynthesis | 0.26083 | 0.01659 | 0.22890 | 0.03879 | 0.02443 | 65 | 0.28315 | 0.04597 | 0.02223 | 93 | 0.03303 | -1.64221 | 144.813 | 0.10272 | 0.61876 |
| colanic acid building blocks biosynthesis | 0.25907 | 0.01271 | 0.23807 | 0.02195 | 0.01838 | 65 | 0.27375 | 0.02778 | 0.01728 | 93 | 0.02523 | -1.41437 | 147.184 | 0.15936 | 0.61876 |
| TCA cycle IV (2-oxoglutarate decarboxylase) | 0.25593 | 0.01691 | 0.21987 | 0.03662 | 0.02374 | 65 | 0.28113 | 0.05004 | 0.02320 | 93 | 0.03319 | -1.84598 | 149.662 | 0.06687 | 0.58059 |
| ppGpp biosynthesis | 0.25490 | 0.01624 | 0.28407 | 0.05689 | 0.02958 | 65 | 0.23451 | 0.03053 | 0.01812 | 93 | 0.03469 | 1.42836 | 110.233 | 0.15601 | 0.61876 |
| fatty acid salvage | 0.25309 | 0.02412 | 0.20325 | 0.07786 | 0.03461 | 65 | 0.28793 | 0.09973 | 0.03275 | 93 | 0.04765 | -1.77718 | 147.595 | 0.07760 | 0.58059 |
| adenosine nucleotides degradation II | 0.25177 | 0.01829 | 0.23210 | 0.05496 | 0.02908 | 65 | 0.26552 | 0.05153 | 0.02354 | 93 | 0.03741 | -0.89338 | 135.024 | 0.37325 | 0.74607 |
| lactose and galactose degradation I | 0.25129 | 0.01776 | 0.25603 | 0.04581 | 0.02655 | 65 | 0.24797 | 0.05319 | 0.02392 | 93 | 0.03573 | 0.22574 | 144.036 | 0.82172 | 0.95867 |
| adenosylcobalamin salvage from cobinamide II | 0.25006 | 0.01445 | 0.26300 | 0.04205 | 0.02543 | 65 | 0.24102 | 0.02687 | 0.01700 | 93 | 0.03059 | 0.71860 | 117.620 | 0.47381 | 0.81137 |
| sucrose degradation IV (sucrose phosphorylase) | 0.24878 | 0.01302 | 0.26472 | 0.03468 | 0.02310 | 65 | 0.23765 | 0.02126 | 0.01512 | 93 | 0.02761 | 0.98061 | 115.807 | 0.32883 | 0.72488 |
| adenosylcobalamin biosynthesis from cobyrinate a,c-diamide I | 0.24877 | 0.01448 | 0.26241 | 0.04263 | 0.02561 | 65 | 0.23923 | 0.02669 | 0.01694 | 93 | 0.03071 | 0.75467 | 116.710 | 0.45197 | 0.81137 |
| superpathway of glycolysis, pyruvate dehydrogenase, TCA, and glyoxylate bypass | 0.24838 | 0.01522 | 0.21919 | 0.03580 | 0.02347 | 65 | 0.26879 | 0.03652 | 0.01982 | 93 | 0.03072 | -1.61471 | 138.728 | 0.10865 | 0.61876 |
| reductive TCA cycle I | 0.24466 | 0.01114 | 0.23547 | 0.01869 | 0.01696 | 65 | 0.25108 | 0.02038 | 0.01480 | 93 | 0.02251 | -0.69341 | 141.537 | 0.48919 | 0.82687 |
| mevalonate pathway I | 0.24035 | 0.01763 | 0.26964 | 0.06584 | 0.03183 | 65 | 0.21987 | 0.03702 | 0.01995 | 93 | 0.03756 | 1.32502 | 112.138 | 0.18786 | 0.63481 |
| superpathway of glyoxylate bypass and TCA | 0.23911 | 0.01791 | 0.20201 | 0.04765 | 0.02708 | 65 | 0.26504 | 0.05171 | 0.02358 | 93 | 0.03590 | -1.75546 | 141.342 | 0.08135 | 0.58059 |
| superpathway of GDP-mannose-derived O-antigen building blocks biosynthesis | 0.23646 | 0.01276 | 0.20918 | 0.01949 | 0.01732 | 65 | 0.25552 | 0.02948 | 0.01780 | 93 | 0.02484 | -1.86590 | 152.381 | 0.06398 | 0.58059 |
| L-arginine biosynthesis III (via N-acetyl-L-citrulline) | 0.23523 | 0.01277 | 0.22903 | 0.02596 | 0.01999 | 65 | 0.23956 | 0.02584 | 0.01667 | 93 | 0.02602 | -0.40464 | 137.663 | 0.68637 | 0.90862 |
| superpathway of L-methionine biosynthesis (by sulfhydrylation) | 0.22836 | 0.01323 | 0.20146 | 0.02416 | 0.01928 | 65 | 0.24717 | 0.02952 | 0.01782 | 93 | 0.02625 | -1.74124 | 145.955 | 0.08375 | 0.58624 |
| glucose and glucose-1-phosphate degradation | 0.22380 | 0.01625 | 0.23099 | 0.04165 | 0.02531 | 65 | 0.21878 | 0.04215 | 0.02129 | 93 | 0.03307 | 0.36924 | 138.391 | 0.71251 | 0.91502 |
| superpathway of hexitol degradation (bacteria) | 0.22096 | 0.01196 | 0.23230 | 0.01813 | 0.01670 | 65 | 0.21304 | 0.02583 | 0.01666 | 93 | 0.02359 | 0.81629 | 150.855 | 0.41562 | 0.78940 |
| superpathway of thiamin diphosphate biosynthesis II | 0.21797 | 0.01297 | 0.22814 | 0.02819 | 0.02082 | 65 | 0.21086 | 0.02561 | 0.01659 | 93 | 0.02663 | 0.64916 | 133.616 | 0.51735 | 0.84599 |
| thiazole biosynthesis I (E. coli) | 0.21770 | 0.01581 | 0.24900 | 0.05222 | 0.02834 | 65 | 0.19582 | 0.02986 | 0.01792 | 93 | 0.03353 | 1.58580 | 112.843 | 0.11558 | 0.61876 |
| ubiquinol-10 biosynthesis (prokaryotic) | 0.20273 | 0.01432 | 0.19263 | 0.03347 | 0.02269 | 65 | 0.20979 | 0.03190 | 0.01852 | 93 | 0.02929 | -0.58585 | 135.756 | 0.55895 | 0.86025 |
| ubiquinol-7 biosynthesis (prokaryotic) | 0.20273 | 0.01432 | 0.19263 | 0.03347 | 0.02269 | 65 | 0.20979 | 0.03190 | 0.01852 | 93 | 0.02929 | -0.58585 | 135.756 | 0.55895 | 0.86025 |
| ubiquinol-8 biosynthesis (prokaryotic) | 0.20273 | 0.01432 | 0.19263 | 0.03347 | 0.02269 | 65 | 0.20979 | 0.03190 | 0.01852 | 93 | 0.02929 | -0.58585 | 135.756 | 0.55895 | 0.86025 |
| ubiquinol-9 biosynthesis (prokaryotic) | 0.20273 | 0.01432 | 0.19263 | 0.03347 | 0.02269 | 65 | 0.20979 | 0.03190 | 0.01852 | 93 | 0.02929 | -0.58585 | 135.756 | 0.55895 | 0.86025 |
| sulfate reduction I (assimilatory) | 0.20163 | 0.01602 | 0.16846 | 0.03127 | 0.02193 | 65 | 0.22482 | 0.04612 | 0.02227 | 93 | 0.03126 | -1.80315 | 151.763 | 0.07335 | 0.58059 |
| superpathway of (R,R)-butanediol biosynthesis | 0.20109 | 0.01608 | 0.19382 | 0.02884 | 0.02106 | 65 | 0.20618 | 0.04956 | 0.02308 | 93 | 0.03125 | -0.39546 | 154.758 | 0.69305 | 0.90862 |
| purine nucleotides degradation II (aerobic) | 0.19865 | 0.01218 | 0.18160 | 0.02300 | 0.01881 | 65 | 0.21057 | 0.02364 | 0.01594 | 93 | 0.02466 | -1.17520 | 139.069 | 0.24192 | 0.70007 |
| pyruvate fermentation to propanoate I | 0.19833 | 0.01311 | 0.16911 | 0.02350 | 0.01901 | 65 | 0.21875 | 0.02898 | 0.01765 | 93 | 0.02595 | -1.91312 | 146.272 | 0.05769 | 0.58059 |
| superpathway of menaquinol-11 biosynthesis | 0.19475 | 0.01165 | 0.18856 | 0.02111 | 0.01802 | 65 | 0.19908 | 0.02187 | 0.01534 | 93 | 0.02366 | -0.44468 | 139.399 | 0.65724 | 0.90427 |
| superpathway of menaquinol-12 biosynthesis | 0.19475 | 0.01165 | 0.18856 | 0.02111 | 0.01802 | 65 | 0.19908 | 0.02187 | 0.01534 | 93 | 0.02366 | -0.44468 | 139.399 | 0.65724 | 0.90427 |
| superpathway of menaquinol-13 biosynthesis | 0.19475 | 0.01165 | 0.18856 | 0.02111 | 0.01802 | 65 | 0.19908 | 0.02187 | 0.01534 | 93 | 0.02366 | -0.44468 | 139.399 | 0.65724 | 0.90427 |
| superpathway of menaquinol-7 biosynthesis | 0.19401 | 0.01141 | 0.18848 | 0.01952 | 0.01733 | 65 | 0.19787 | 0.02147 | 0.01519 | 93 | 0.02305 | -0.40753 | 141.879 | 0.68424 | 0.90862 |
| superpathway of menaquinol-8 biosynthesis I | 0.19303 | 0.01111 | 0.18899 | 0.02050 | 0.01776 | 65 | 0.19585 | 0.01900 | 0.01429 | 93 | 0.02280 | -0.30094 | 134.506 | 0.76392 | 0.92899 |
| urea cycle | 0.19153 | 0.01185 | 0.16526 | 0.02322 | 0.01890 | 65 | 0.20990 | 0.02088 | 0.01498 | 93 | 0.02412 | -1.85081 | 133.135 | 0.06641 | 0.58059 |
| superpathway of UDP-glucose-derived O-antigen building blocks biosynthesis | 0.18482 | 0.01186 | 0.18045 | 0.02352 | 0.01902 | 65 | 0.18787 | 0.02152 | 0.01521 | 93 | 0.02435 | -0.30470 | 133.921 | 0.76106 | 0.92864 |
| Bifidobacterium shunt | 0.18476 | 0.02060 | 0.20452 | 0.09035 | 0.03728 | 65 | 0.17096 | 0.05113 | 0.02345 | 93 | 0.04404 | 0.76192 | 112.410 | 0.44770 | 0.81137 |
| glyoxylate cycle | 0.18378 | 0.01428 | 0.15300 | 0.02883 | 0.02106 | 65 | 0.20529 | 0.03377 | 0.01906 | 93 | 0.02840 | -1.84101 | 144.369 | 0.06767 | 0.58059 |
| superpathway of ubiquinol-8 biosynthesis (prokaryotic) | 0.17836 | 0.01255 | 0.17384 | 0.02597 | 0.01999 | 65 | 0.18151 | 0.02435 | 0.01618 | 93 | 0.02572 | -0.29820 | 135.027 | 0.76601 | 0.92899 |
| L-glutamate and L-glutamine biosynthesis | 0.17801 | 0.01222 | 0.16428 | 0.02170 | 0.01827 | 65 | 0.18761 | 0.02497 | 0.01639 | 93 | 0.02454 | -0.95078 | 143.685 | 0.34331 | 0.72862 |
| superpathway of glucose and xylose degradation | 0.17754 | 0.01064 | 0.16960 | 0.01597 | 0.01568 | 65 | 0.18309 | 0.01931 | 0.01441 | 93 | 0.02129 | -0.63373 | 145.557 | 0.52725 | 0.84889 |
| superpathway of demethylmenaquinol-8 biosynthesis | 0.17266 | 0.01074 | 0.17044 | 0.01986 | 0.01748 | 65 | 0.17422 | 0.01726 | 0.01362 | 93 | 0.02216 | -0.17058 | 131.573 | 0.86481 | 0.96656 |
| ADP-L-glycero-β-D-manno-heptose biosynthesis | 0.16926 | 0.01033 | 0.18069 | 0.01746 | 0.01639 | 65 | 0.16127 | 0.01648 | 0.01331 | 93 | 0.02112 | 0.91999 | 135.309 | 0.35922 | 0.74607 |
| L-1,2-propanediol degradation | 0.16818 | 0.01749 | 0.19385 | 0.06139 | 0.03073 | 65 | 0.15024 | 0.03899 | 0.02047 | 93 | 0.03693 | 1.18092 | 117.341 | 0.24002 | 0.70007 |
| arginine, ornithine and proline interconversion | 0.16658 | 0.01111 | 0.16831 | 0.02069 | 0.01784 | 65 | 0.16538 | 0.01890 | 0.01425 | 93 | 0.02284 | 0.12851 | 133.837 | 0.89794 | 0.97689 |
| tRNA processing | 0.16341 | 0.01128 | 0.16413 | 0.02049 | 0.01776 | 65 | 0.16292 | 0.02005 | 0.01468 | 93 | 0.02304 | 0.05253 | 136.907 | 0.95819 | 0.99202 |
| superpathway of pyridoxal 5’-phosphate biosynthesis and salvage | 0.16302 | 0.01060 | 0.16060 | 0.01747 | 0.01640 | 65 | 0.16471 | 0.01813 | 0.01396 | 93 | 0.02154 | -0.19099 | 139.457 | 0.84881 | 0.96656 |
| heterolactic fermentation | 0.16063 | 0.01913 | 0.18386 | 0.08332 | 0.03580 | 65 | 0.14440 | 0.04004 | 0.02075 | 93 | 0.04138 | 0.95350 | 105.898 | 0.34251 | 0.72862 |
| superpathway of phylloquinol biosynthesis | 0.16006 | 0.01247 | 0.16517 | 0.03435 | 0.02299 | 65 | 0.15649 | 0.01802 | 0.01392 | 93 | 0.02687 | 0.32292 | 109.311 | 0.74737 | 0.92609 |
| 1,4-dihydroxy-2-naphthoate biosynthesis I | 0.15116 | 0.01238 | 0.15676 | 0.03409 | 0.02290 | 65 | 0.14724 | 0.01758 | 0.01375 | 93 | 0.02671 | 0.35620 | 108.622 | 0.72238 | 0.91502 |
| superpathway of arginine and polyamine biosynthesis | 0.15107 | 0.00707 | 0.15080 | 0.00907 | 0.01181 | 65 | 0.15126 | 0.00717 | 0.00878 | 93 | 0.01472 | -0.03165 | 127.283 | 0.97480 | 0.99202 |
| L-leucine degradation I | 0.14411 | 0.01420 | 0.12792 | 0.03155 | 0.02203 | 65 | 0.15543 | 0.03214 | 0.01859 | 93 | 0.02883 | -0.95414 | 138.679 | 0.34167 | 0.72862 |
| thiazole biosynthesis II (Bacillus) | 0.14107 | 0.01281 | 0.16699 | 0.04099 | 0.02511 | 65 | 0.12295 | 0.01496 | 0.01268 | 93 | 0.02813 | 1.56531 | 96.452 | 0.12078 | 0.61876 |
| superpathway of 2,3-butanediol biosynthesis | 0.13765 | 0.01230 | 0.12898 | 0.02313 | 0.01887 | 65 | 0.14371 | 0.02463 | 0.01627 | 93 | 0.02491 | -0.59106 | 140.551 | 0.55543 | 0.86025 |
| octane oxidation | 0.13437 | 0.01759 | 0.12570 | 0.03170 | 0.02209 | 65 | 0.14043 | 0.06132 | 0.02568 | 93 | 0.03387 | -0.43485 | 155.856 | 0.66427 | 0.90545 |
| pyruvate fermentation to acetone | 0.13223 | 0.01912 | 0.17513 | 0.09862 | 0.03895 | 65 | 0.10225 | 0.02777 | 0.01728 | 93 | 0.04261 | 1.71029 | 89.269 | 0.09069 | 0.60802 |
| pyridoxal 5’-phosphate biosynthesis I | 0.13169 | 0.01043 | 0.12583 | 0.01640 | 0.01589 | 65 | 0.13579 | 0.01787 | 0.01386 | 93 | 0.02108 | -0.47273 | 141.500 | 0.63713 | 0.90147 |
| superpathway of heme biosynthesis from uroporphyrinogen-III | 0.12808 | 0.00799 | 0.12173 | 0.00977 | 0.01226 | 65 | 0.13252 | 0.01038 | 0.01056 | 93 | 0.01618 | -0.66673 | 140.441 | 0.50604 | 0.83780 |
| cob(II)yrinate a,c-diamide biosynthesis I (early cobalt insertion) | 0.12719 | 0.01319 | 0.16526 | 0.04324 | 0.02579 | 65 | 0.10059 | 0.01511 | 0.01275 | 93 | 0.02877 | 2.24790 | 95.124 | 0.02689 | 0.58059 |
| protocatechuate degradation II (ortho-cleavage pathway) | 0.12427 | 0.01595 | 0.11461 | 0.03599 | 0.02353 | 65 | 0.13102 | 0.04345 | 0.02162 | 93 | 0.03195 | -0.51344 | 145.507 | 0.60842 | 0.88168 |
| hexitol fermentation to lactate, formate, ethanol and acetate | 0.12391 | 0.00971 | 0.12332 | 0.01662 | 0.01599 | 65 | 0.12432 | 0.01384 | 0.01220 | 93 | 0.02011 | -0.04948 | 129.617 | 0.96061 | 0.99202 |
| L-tyrosine degradation I | 0.12298 | 0.01324 | 0.11465 | 0.02713 | 0.02043 | 65 | 0.12879 | 0.02829 | 0.01744 | 93 | 0.02686 | -0.52638 | 139.667 | 0.59946 | 0.87368 |
| enterobactin biosynthesis | 0.12179 | 0.01104 | 0.11445 | 0.01953 | 0.01734 | 65 | 0.12691 | 0.01923 | 0.01438 | 93 | 0.02252 | -0.55330 | 137.181 | 0.58096 | 0.86817 |
| 4-aminobutanoate degradation V | 0.11762 | 0.01121 | 0.10928 | 0.01997 | 0.01753 | 65 | 0.12344 | 0.01990 | 0.01463 | 93 | 0.02283 | -0.62063 | 137.702 | 0.53587 | 0.85184 |
| superpathway of polyamine biosynthesis I | 0.11591 | 0.00601 | 0.11329 | 0.00576 | 0.00941 | 65 | 0.11775 | 0.00573 | 0.00785 | 93 | 0.01225 | -0.36419 | 137.692 | 0.71627 | 0.91502 |
| superpathway of heme biosynthesis from glycine | 0.11488 | 0.00825 | 0.10403 | 0.00933 | 0.01198 | 65 | 0.12246 | 0.01172 | 0.01122 | 93 | 0.01642 | -1.12257 | 146.908 | 0.26345 | 0.70194 |
| pyrimidine deoxyribonucleotides de novo biosynthesis III | 0.11356 | 0.01087 | 0.11057 | 0.01714 | 0.01624 | 65 | 0.11565 | 0.01990 | 0.01463 | 93 | 0.02186 | -0.23267 | 144.053 | 0.81635 | 0.95867 |
| peptidoglycan biosynthesis V (β-lactam resistance) | 0.11194 | 0.01045 | 0.12950 | 0.02233 | 0.01854 | 65 | 0.09967 | 0.01352 | 0.01206 | 93 | 0.02211 | 1.34887 | 115.273 | 0.18002 | 0.62460 |
| purine nucleobases degradation I (anaerobic) | 0.11137 | 0.01018 | 0.11358 | 0.01794 | 0.01662 | 65 | 0.10982 | 0.01545 | 0.01289 | 93 | 0.02103 | 0.17894 | 131.155 | 0.85826 | 0.96656 |
| 2-methylcitrate cycle II | 0.11082 | 0.01168 | 0.10931 | 0.02118 | 0.01805 | 65 | 0.11187 | 0.02204 | 0.01539 | 93 | 0.02372 | -0.10796 | 139.591 | 0.91418 | 0.97851 |
| D-fructuronate degradation | 0.10899 | 0.01127 | 0.11312 | 0.03400 | 0.02287 | 65 | 0.10611 | 0.01059 | 0.01067 | 93 | 0.02524 | 0.27786 | 91.858 | 0.78174 | 0.93380 |
| NAD salvage pathway II | 0.09570 | 0.00827 | 0.10526 | 0.01051 | 0.01271 | 65 | 0.08902 | 0.01103 | 0.01089 | 93 | 0.01674 | 0.97021 | 139.963 | 0.33362 | 0.72862 |
| superpathway of (Kdo)2-lipid A biosynthesis | 0.09044 | 0.00953 | 0.10204 | 0.01454 | 0.01496 | 65 | 0.08233 | 0.01420 | 0.01236 | 93 | 0.01940 | 1.01594 | 136.855 | 0.31145 | 0.72280 |
| superpathway of polyamine biosynthesis II | 0.09002 | 0.00534 | 0.09284 | 0.00541 | 0.00912 | 65 | 0.08805 | 0.00392 | 0.00649 | 93 | 0.01120 | 0.42781 | 123.255 | 0.66954 | 0.90623 |
| 2-methylcitrate cycle I | 0.08885 | 0.01080 | 0.08659 | 0.01864 | 0.01694 | 65 | 0.09043 | 0.01849 | 0.01410 | 93 | 0.02204 | -0.17411 | 137.524 | 0.86204 | 0.96656 |
| aromatic biogenic amine degradation (bacteria) | 0.08683 | 0.01236 | 0.07124 | 0.01894 | 0.01707 | 65 | 0.09773 | 0.02773 | 0.01727 | 93 | 0.02428 | -1.09086 | 151.580 | 0.27706 | 0.70413 |
| 4-deoxy-L-threo-hex-4-enopyranuronate degradation | 0.08026 | 0.01029 | 0.08820 | 0.03012 | 0.02153 | 65 | 0.07472 | 0.00753 | 0.00900 | 93 | 0.02333 | 0.57784 | 86.479 | 0.56488 | 0.86025 |
| teichoic acid (poly-glycerol) biosynthesis | 0.07638 | 0.00979 | 0.08126 | 0.01944 | 0.01729 | 65 | 0.07297 | 0.01230 | 0.01150 | 93 | 0.02077 | 0.39937 | 117.174 | 0.69035 | 0.90862 |
| L-glutamate degradation V (via hydroxyglutarate) | 0.07595 | 0.01423 | 0.11536 | 0.05963 | 0.03029 | 65 | 0.04841 | 0.01122 | 0.01098 | 93 | 0.03222 | 2.07805 | 80.965 | 0.04087 | 0.58059 |
| glutaryl-CoA degradation | 0.07560 | 0.00857 | 0.07460 | 0.00966 | 0.01219 | 65 | 0.07630 | 0.01310 | 0.01187 | 93 | 0.01701 | -0.10025 | 149.440 | 0.92028 | 0.97918 |
| D-galacturonate degradation I | 0.07061 | 0.00769 | 0.06512 | 0.01267 | 0.01396 | 65 | 0.07445 | 0.00708 | 0.00872 | 93 | 0.01646 | -0.56675 | 111.854 | 0.57202 | 0.86383 |
| cob(II)yrinate a,c-diamide biosynthesis II (late cobalt incorporation) | 0.07057 | 0.00849 | 0.06306 | 0.01044 | 0.01267 | 65 | 0.07582 | 0.01210 | 0.01141 | 93 | 0.01705 | -0.74820 | 143.975 | 0.45556 | 0.81137 |
| acetyl-CoA fermentation to butanoate II | 0.06923 | 0.00754 | 0.05755 | 0.00817 | 0.01121 | 65 | 0.07739 | 0.00946 | 0.01009 | 93 | 0.01508 | -1.31579 | 143.953 | 0.19034 | 0.63481 |
| L-arginine degradation II (AST pathway) | 0.06843 | 0.00967 | 0.07020 | 0.01439 | 0.01488 | 65 | 0.06718 | 0.01520 | 0.01279 | 93 | 0.01962 | 0.15388 | 140.221 | 0.87793 | 0.97057 |
| L-histidine degradation II | 0.06739 | 0.00965 | 0.05722 | 0.01085 | 0.01292 | 65 | 0.07451 | 0.01744 | 0.01370 | 93 | 0.01883 | -0.91819 | 153.658 | 0.35996 | 0.74607 |
| nitrate reduction VI (assimilatory) | 0.06643 | 0.00705 | 0.07043 | 0.00971 | 0.01222 | 65 | 0.06363 | 0.00662 | 0.00844 | 93 | 0.01485 | 0.45813 | 120.514 | 0.64769 | 0.90427 |
| superpathway of β-D-glucuronide and D-glucuronate degradation | 0.06569 | 0.00741 | 0.06425 | 0.01300 | 0.01414 | 65 | 0.06669 | 0.00578 | 0.00788 | 93 | 0.01619 | -0.15080 | 103.023 | 0.88043 | 0.97057 |
| glycerol degradation to butanol | 0.06464 | 0.00646 | 0.07614 | 0.00809 | 0.01116 | 65 | 0.05660 | 0.00546 | 0.00766 | 93 | 0.01353 | 1.44405 | 119.976 | 0.15133 | 0.61876 |
| myo-, chiro- and scillo-inositol degradation | 0.06404 | 0.01065 | 0.04703 | 0.00878 | 0.01162 | 65 | 0.07592 | 0.02416 | 0.01612 | 93 | 0.01987 | -1.45389 | 153.056 | 0.14802 | 0.61876 |
| mono-trans, poly-cis decaprenyl phosphate biosynthesis | 0.06146 | 0.00617 | 0.06364 | 0.00714 | 0.01048 | 65 | 0.05993 | 0.00528 | 0.00754 | 93 | 0.01291 | 0.28679 | 124.179 | 0.77475 | 0.93380 |
| aromatic compounds degradation via β-ketoadipate | 0.05398 | 0.00726 | 0.05454 | 0.00954 | 0.01211 | 65 | 0.05359 | 0.00757 | 0.00902 | 93 | 0.01510 | 0.06271 | 127.423 | 0.95009 | 0.99202 |
| catechol degradation III (ortho-cleavage pathway) | 0.05398 | 0.00726 | 0.05454 | 0.00954 | 0.01211 | 65 | 0.05359 | 0.00757 | 0.00902 | 93 | 0.01510 | 0.06271 | 127.423 | 0.95009 | 0.99202 |
| catechol degradation to β-ketoadipate | 0.05376 | 0.00731 | 0.05539 | 0.01032 | 0.01260 | 65 | 0.05263 | 0.00723 | 0.00882 | 93 | 0.01538 | 0.17964 | 121.694 | 0.85773 | 0.96656 |
| superpathway of hexuronide and hexuronate degradation | 0.05342 | 0.00671 | 0.05189 | 0.01089 | 0.01294 | 65 | 0.05450 | 0.00455 | 0.00699 | 93 | 0.01471 | -0.17774 | 100.841 | 0.85928 | 0.96656 |
| peptidoglycan biosynthesis II (staphylococci) | 0.04940 | 0.00983 | 0.04504 | 0.01763 | 0.01647 | 65 | 0.05245 | 0.01377 | 0.01217 | 93 | 0.02048 | -0.36174 | 126.683 | 0.71815 | 0.91502 |
| L-lysine fermentation to acetate and butanoate | 0.04866 | 0.01191 | 0.07610 | 0.04331 | 0.02581 | 65 | 0.02949 | 0.00723 | 0.00882 | 93 | 0.02728 | 1.70869 | 79.062 | 0.09143 | 0.60802 |
| biotin biosynthesis II | 0.04860 | 0.00510 | 0.04607 | 0.00335 | 0.00718 | 65 | 0.05037 | 0.00467 | 0.00709 | 93 | 0.01009 | -0.42640 | 150.246 | 0.67042 | 0.90623 |
| superpathway of Clostridium acetobutylicum acidogenic fermentation | 0.04387 | 0.00468 | 0.03793 | 0.00233 | 0.00599 | 65 | 0.04802 | 0.00425 | 0.00676 | 93 | 0.00903 | -1.11679 | 155.440 | 0.26581 | 0.70194 |
| superpathway of sulfur oxidation (Acidianus ambivalens) | 0.04215 | 0.00649 | 0.02616 | 0.00305 | 0.00685 | 65 | 0.05332 | 0.00892 | 0.00979 | 93 | 0.01195 | -2.27184 | 151.837 | 0.02450 | 0.58059 |
| toluene degradation I (aerobic) (via o-cresol) | 0.04039 | 0.00562 | 0.03464 | 0.00327 | 0.00709 | 65 | 0.04442 | 0.00621 | 0.00817 | 93 | 0.01082 | -0.90380 | 155.769 | 0.36750 | 0.74607 |
| toluene degradation II (aerobic) (via 4-methylcatechol) | 0.04039 | 0.00562 | 0.03464 | 0.00327 | 0.00709 | 65 | 0.04442 | 0.00621 | 0.00817 | 93 | 0.01082 | -0.90380 | 155.769 | 0.36750 | 0.74607 |
| formaldehyde assimilation II (RuMP Cycle) | 0.03860 | 0.00636 | 0.02074 | 0.00184 | 0.00532 | 65 | 0.05108 | 0.00924 | 0.00997 | 93 | 0.01130 | -2.68537 | 135.992 | 0.00815 | 0.58059 |
| NAD biosynthesis II (from tryptophan) | 0.03801 | 0.00739 | 0.03477 | 0.00889 | 0.01169 | 65 | 0.04028 | 0.00852 | 0.00957 | 93 | 0.01511 | -0.36463 | 136.019 | 0.71595 | 0.91502 |
| myo-inositol degradation I | 0.03719 | 0.00541 | 0.03016 | 0.00311 | 0.00692 | 65 | 0.04211 | 0.00567 | 0.00781 | 93 | 0.01043 | -1.14551 | 155.454 | 0.25376 | 0.70193 |
| mannan degradation | 0.03717 | 0.00571 | 0.02459 | 0.00225 | 0.00588 | 65 | 0.04597 | 0.00702 | 0.00869 | 93 | 0.01049 | -2.03734 | 150.296 | 0.04337 | 0.58059 |
| fucose degradation | 0.03704 | 0.00539 | 0.04418 | 0.00742 | 0.01068 | 65 | 0.03205 | 0.00260 | 0.00529 | 93 | 0.01192 | 1.01781 | 95.270 | 0.31135 | 0.72280 |
| norspermidine biosynthesis | 0.03569 | 0.00455 | 0.03545 | 0.00319 | 0.00701 | 65 | 0.03585 | 0.00335 | 0.00600 | 93 | 0.00923 | -0.04300 | 139.920 | 0.96576 | 0.99202 |
| pyruvate fermentation to butanoate | 0.03568 | 0.00392 | 0.03056 | 0.00161 | 0.00498 | 65 | 0.03927 | 0.00300 | 0.00567 | 93 | 0.00755 | -1.15432 | 155.618 | 0.25014 | 0.70007 |
| mycothiol biosynthesis | 0.03562 | 0.00570 | 0.03565 | 0.00613 | 0.00972 | 65 | 0.03559 | 0.00448 | 0.00694 | 93 | 0.01194 | 0.00453 | 123.617 | 0.99640 | 0.99890 |
| toluene degradation III (aerobic) (via p-cresol) | 0.03483 | 0.00604 | 0.05076 | 0.00981 | 0.01228 | 65 | 0.02369 | 0.00271 | 0.00540 | 93 | 0.01342 | 2.01785 | 88.784 | 0.04663 | 0.58059 |
| superpathway of salicylate degradation | 0.03439 | 0.00558 | 0.04121 | 0.00728 | 0.01058 | 65 | 0.02963 | 0.00329 | 0.00595 | 93 | 0.01214 | 0.95396 | 103.598 | 0.34233 | 0.72862 |
| glucose degradation (oxidative) | 0.03433 | 0.00775 | 0.03301 | 0.00653 | 0.01003 | 65 | 0.03526 | 0.01164 | 0.01119 | 93 | 0.01502 | -0.14936 | 155.208 | 0.88146 | 0.97057 |
| superpathway of chorismate metabolism | 0.03409 | 0.00498 | 0.03709 | 0.00439 | 0.00822 | 65 | 0.03200 | 0.00363 | 0.00625 | 93 | 0.01032 | 0.49375 | 129.347 | 0.62232 | 0.88998 |
| superpathway of menaquinol-8 biosynthesis II | 0.03398 | 0.00505 | 0.03338 | 0.00366 | 0.00750 | 65 | 0.03440 | 0.00432 | 0.00682 | 93 | 0.01014 | -0.10069 | 144.733 | 0.91993 | 0.97918 |
| dTDP-N-acetylthomosamine biosynthesis | 0.03357 | 0.00608 | 0.02749 | 0.00375 | 0.00760 | 65 | 0.03782 | 0.00730 | 0.00886 | 93 | 0.01167 | -0.88472 | 155.879 | 0.37767 | 0.74607 |
| superpathway of menaquinol-10 biosynthesis | 0.03329 | 0.00554 | 0.03344 | 0.00440 | 0.00823 | 65 | 0.03319 | 0.00522 | 0.00749 | 93 | 0.01113 | 0.02251 | 144.833 | 0.98207 | 0.99202 |
| superpathway of menaquinol-6 biosynthesis I | 0.03329 | 0.00554 | 0.03344 | 0.00440 | 0.00823 | 65 | 0.03319 | 0.00522 | 0.00749 | 93 | 0.01113 | 0.02251 | 144.833 | 0.98207 | 0.99202 |
| superpathway of menaquinol-9 biosynthesis | 0.03329 | 0.00554 | 0.03344 | 0.00440 | 0.00823 | 65 | 0.03319 | 0.00522 | 0.00749 | 93 | 0.01113 | 0.02251 | 144.833 | 0.98207 | 0.99202 |
| L-tryptophan degradation to 2-amino-3-carboxymuconate semialdehyde | 0.03229 | 0.00694 | 0.02926 | 0.00741 | 0.01067 | 65 | 0.03440 | 0.00784 | 0.00918 | 93 | 0.01408 | -0.36483 | 140.299 | 0.71578 | 0.91502 |
| methylphosphonate degradation I | 0.03137 | 0.00576 | 0.01533 | 0.00145 | 0.00472 | 65 | 0.04258 | 0.00764 | 0.00906 | 93 | 0.01022 | -2.66728 | 134.406 | 0.00859 | 0.58059 |
| adenosylcobalamin biosynthesis I (early cobalt insertion) | 0.03125 | 0.00360 | 0.03128 | 0.00183 | 0.00531 | 65 | 0.03123 | 0.00222 | 0.00489 | 93 | 0.00721 | 0.00769 | 145.660 | 0.99388 | 0.99889 |
| methyl ketone biosynthesis | 0.03047 | 0.00659 | 0.01797 | 0.00288 | 0.00666 | 65 | 0.03921 | 0.00952 | 0.01012 | 93 | 0.01211 | -1.75402 | 148.807 | 0.08149 | 0.58059 |
| GDP-D-glycero-α-D-manno-heptose biosynthesis | 0.02997 | 0.00430 | 0.03308 | 0.00421 | 0.00805 | 65 | 0.02780 | 0.00205 | 0.00469 | 93 | 0.00932 | 0.56616 | 106.324 | 0.57248 | 0.86383 |
| formaldehyde oxidation I | 0.02950 | 0.00495 | 0.01542 | 0.00101 | 0.00393 | 65 | 0.03934 | 0.00567 | 0.00781 | 93 | 0.00875 | -2.73527 | 132.363 | 0.00709 | 0.58059 |
| 1,4-dihydroxy-6-naphthoate biosynthesis II | 0.02864 | 0.00652 | 0.03165 | 0.00886 | 0.01168 | 65 | 0.02654 | 0.00529 | 0.00754 | 93 | 0.01390 | 0.36751 | 114.611 | 0.71392 | 0.91502 |
| 4-methylcatechol degradation (ortho cleavage) | 0.02736 | 0.00493 | 0.04009 | 0.00688 | 0.01029 | 65 | 0.01846 | 0.00158 | 0.00412 | 93 | 0.01108 | 1.95091 | 84.657 | 0.05438 | 0.58059 |
| photorespiration | 0.02721 | 0.00486 | 0.02318 | 0.00268 | 0.00642 | 65 | 0.03003 | 0.00448 | 0.00694 | 93 | 0.00945 | -0.72470 | 154.369 | 0.46973 | 0.81137 |
| L-rhamnose degradation I | 0.02683 | 0.00357 | 0.01989 | 0.00080 | 0.00351 | 65 | 0.03168 | 0.00282 | 0.00551 | 93 | 0.00653 | -1.80433 | 147.061 | 0.07323 | 0.58059 |
| superpathway of demethylmenaquinol-6 biosynthesis I | 0.02600 | 0.00461 | 0.02584 | 0.00301 | 0.00680 | 65 | 0.02610 | 0.00364 | 0.00626 | 93 | 0.00924 | -0.02809 | 145.655 | 0.97763 | 0.99202 |
| superpathway of demethylmenaquinol-9 biosynthesis | 0.02600 | 0.00461 | 0.02584 | 0.00301 | 0.00680 | 65 | 0.02610 | 0.00364 | 0.00626 | 93 | 0.00924 | -0.02809 | 145.655 | 0.97763 | 0.99202 |
| adenosylcobalamin biosynthesis II (late cobalt incorporation) | 0.02544 | 0.00364 | 0.02185 | 0.00160 | 0.00496 | 65 | 0.02795 | 0.00244 | 0.00513 | 93 | 0.00713 | -0.85510 | 152.610 | 0.39383 | 0.77029 |
| catechol degradation I (meta-cleavage pathway) | 0.02541 | 0.00357 | 0.02337 | 0.00189 | 0.00539 | 65 | 0.02684 | 0.00213 | 0.00478 | 93 | 0.00720 | -0.48157 | 142.889 | 0.63084 | 0.89895 |
| polymyxin resistance | 0.02446 | 0.00604 | 0.01935 | 0.00311 | 0.00692 | 65 | 0.02803 | 0.00765 | 0.00907 | 93 | 0.01141 | -0.76074 | 154.869 | 0.44797 | 0.81137 |
| nitrate reduction I (denitrification) | 0.02372 | 0.00361 | 0.01881 | 0.00098 | 0.00389 | 65 | 0.02715 | 0.00279 | 0.00548 | 93 | 0.00672 | -1.24070 | 152.440 | 0.21662 | 0.67841 |
| 4-hydroxyphenylacetate degradation | 0.02322 | 0.00557 | 0.01511 | 0.00188 | 0.00538 | 65 | 0.02889 | 0.00699 | 0.00867 | 93 | 0.01020 | -1.35067 | 145.437 | 0.17890 | 0.62460 |
| phenylacetate degradation I (aerobic) | 0.02276 | 0.00367 | 0.03036 | 0.00312 | 0.00693 | 65 | 0.01744 | 0.00140 | 0.00388 | 93 | 0.00794 | 1.62803 | 103.280 | 0.10657 | 0.61876 |
| superpathway of UDP-N-acetylglucosamine-derived O-antigen building blocks biosynthesis | 0.01892 | 0.00474 | 0.01181 | 0.00086 | 0.00363 | 65 | 0.02389 | 0.00541 | 0.00763 | 93 | 0.00845 | -1.43081 | 128.854 | 0.15490 | 0.61876 |
| allantoin degradation to glyoxylate III | 0.01844 | 0.00271 | 0.01534 | 0.00080 | 0.00350 | 65 | 0.02061 | 0.00142 | 0.00390 | 93 | 0.00524 | -1.00612 | 155.201 | 0.31592 | 0.72280 |
| CMP-legionaminate biosynthesis I | 0.01755 | 0.00522 | 0.01313 | 0.00192 | 0.00544 | 65 | 0.02064 | 0.00598 | 0.00802 | 93 | 0.00969 | -0.77573 | 150.378 | 0.43913 | 0.81117 |
| superpathway of ornithine degradation | 0.01711 | 0.00406 | 0.01445 | 0.00112 | 0.00415 | 65 | 0.01897 | 0.00365 | 0.00626 | 93 | 0.00751 | -0.60168 | 149.160 | 0.54830 | 0.86025 |
| ectoine biosynthesis | 0.01673 | 0.00322 | 0.01270 | 0.00092 | 0.00376 | 65 | 0.01955 | 0.00214 | 0.00480 | 93 | 0.00610 | -1.12467 | 155.449 | 0.26246 | 0.70194 |
| methanol oxidation to carbon dioxide | 0.01623 | 0.00315 | 0.01793 | 0.00198 | 0.00551 | 65 | 0.01504 | 0.00130 | 0.00374 | 93 | 0.00666 | 0.43423 | 118.931 | 0.66491 | 0.90545 |
| superpathway of phenylethylamine degradation | 0.01276 | 0.00231 | 0.01644 | 0.00121 | 0.00431 | 65 | 0.01019 | 0.00058 | 0.00251 | 93 | 0.00498 | 1.25412 | 106.160 | 0.21255 | 0.67308 |
| glycine betaine degradation I | 0.01245 | 0.00394 | 0.00477 | 0.00083 | 0.00357 | 65 | 0.01781 | 0.00354 | 0.00617 | 93 | 0.00713 | -1.82751 | 141.131 | 0.06974 | 0.58059 |
| 3-phenylpropanoate and 3-(3-hydroxyphenyl)propanoate degradation | 0.01232 | 0.00336 | 0.00735 | 0.00026 | 0.00200 | 65 | 0.01579 | 0.00284 | 0.00552 | 93 | 0.00588 | -1.43485 | 114.937 | 0.15405 | 0.61876 |
| 3-phenylpropanoate degradation | 0.01209 | 0.00377 | 0.00508 | 0.00013 | 0.00143 | 65 | 0.01700 | 0.00368 | 0.00629 | 93 | 0.00645 | -1.84791 | 101.434 | 0.06753 | 0.58059 |
| superpathway of L-arginine and L-ornithine degradation | 0.01187 | 0.00223 | 0.01512 | 0.00093 | 0.00377 | 65 | 0.00959 | 0.00068 | 0.00270 | 93 | 0.00464 | 1.19274 | 123.859 | 0.23525 | 0.70007 |
| superpathway of L-arginine, putrescine, and 4-aminobutanoate degradation | 0.01187 | 0.00223 | 0.01512 | 0.00093 | 0.00377 | 65 | 0.00959 | 0.00068 | 0.00270 | 93 | 0.00464 | 1.19274 | 123.859 | 0.23525 | 0.70007 |
| superpathway of fucose and rhamnose degradation | 0.01163 | 0.00204 | 0.00877 | 0.00024 | 0.00191 | 65 | 0.01363 | 0.00094 | 0.00318 | 93 | 0.00371 | -1.30764 | 143.386 | 0.19309 | 0.63671 |
| succinate fermentation to butanoate | 0.01159 | 0.00228 | 0.00941 | 0.00030 | 0.00215 | 65 | 0.01311 | 0.00119 | 0.00358 | 93 | 0.00417 | -0.88718 | 143.412 | 0.37647 | 0.74607 |
| superpathway of methylglyoxal degradation | 0.01123 | 0.00221 | 0.01003 | 0.00080 | 0.00351 | 65 | 0.01207 | 0.00076 | 0.00286 | 93 | 0.00452 | -0.44971 | 135.631 | 0.65364 | 0.90427 |
| reductive acetyl coenzyme A pathway | 0.01103 | 0.00239 | 0.01014 | 0.00081 | 0.00354 | 65 | 0.01164 | 0.00097 | 0.00323 | 93 | 0.00479 | -0.31264 | 145.012 | 0.75501 | 0.92864 |
| catechol degradation to 2-oxopent-4-enoate II | 0.01091 | 0.00242 | 0.00742 | 0.00036 | 0.00235 | 65 | 0.01335 | 0.00132 | 0.00376 | 93 | 0.00444 | -1.33595 | 145.860 | 0.18365 | 0.62628 |
| isopropanol biosynthesis | 0.01043 | 0.00288 | 0.00592 | 0.00019 | 0.00169 | 65 | 0.01358 | 0.00209 | 0.00474 | 93 | 0.00503 | -1.52311 | 114.228 | 0.13050 | 0.61876 |
| catechol degradation II (meta-cleavage pathway) | 0.01008 | 0.00244 | 0.00959 | 0.00060 | 0.00305 | 65 | 0.01043 | 0.00119 | 0.00357 | 93 | 0.00469 | -0.17879 | 155.926 | 0.85833 | 0.96656 |
| superpathway of demethylmenaquinol-6 biosynthesis II | 0.00991 | 0.00341 | 0.00836 | 0.00062 | 0.00310 | 65 | 0.01100 | 0.00271 | 0.00539 | 93 | 0.00622 | -0.42348 | 140.647 | 0.67260 | 0.90623 |
| superpathway of glycol metabolism and degradation | 0.00975 | 0.00193 | 0.01053 | 0.00080 | 0.00350 | 65 | 0.00921 | 0.00045 | 0.00219 | 93 | 0.00413 | 0.31850 | 111.869 | 0.75070 | 0.92733 |
| glycogen degradation II (eukaryotic) | 0.00953 | 0.00299 | 0.01012 | 0.00141 | 0.00465 | 65 | 0.00913 | 0.00143 | 0.00392 | 93 | 0.00608 | 0.16316 | 138.462 | 0.87063 | 0.97034 |
| nicotinate degradation I | 0.00925 | 0.00370 | 0.00465 | 0.00076 | 0.00343 | 65 | 0.01247 | 0.00314 | 0.00581 | 93 | 0.00674 | -1.15869 | 142.425 | 0.24852 | 0.70007 |
| superpathway of glycerol degradation to 1,3-propanediol | 0.00901 | 0.00285 | 0.01252 | 0.00277 | 0.00652 | 65 | 0.00655 | 0.00025 | 0.00165 | 93 | 0.00673 | 0.88697 | 72.283 | 0.37804 | 0.74607 |
| L-glutamate degradation VIII (to propanoate) | 0.00900 | 0.00244 | 0.01144 | 0.00101 | 0.00395 | 65 | 0.00729 | 0.00090 | 0.00311 | 93 | 0.00503 | 0.82459 | 132.450 | 0.41109 | 0.78940 |
| toluene degradation IV (aerobic) (via catechol) | 0.00885 | 0.00183 | 0.00941 | 0.00056 | 0.00294 | 65 | 0.00845 | 0.00051 | 0.00233 | 93 | 0.00376 | 0.25372 | 133.111 | 0.80010 | 0.94816 |
| superpathway of L-tryptophan biosynthesis | 0.00747 | 0.00281 | 0.00504 | 0.00044 | 0.00259 | 65 | 0.00917 | 0.00182 | 0.00442 | 93 | 0.00512 | -0.80691 | 141.911 | 0.42107 | 0.79248 |
| 3-phenylpropanoate and 3-(3-hydroxyphenyl)propanoate degradation to 2-oxopent-4-enoate | 0.00745 | 0.00229 | 0.00390 | 0.00010 | 0.00125 | 65 | 0.00993 | 0.00132 | 0.00377 | 93 | 0.00397 | -1.51916 | 111.522 | 0.13155 | 0.61876 |
| cinnamate and 3-hydroxycinnamate degradation to 2-oxopent-4-enoate | 0.00745 | 0.00229 | 0.00390 | 0.00010 | 0.00125 | 65 | 0.00993 | 0.00132 | 0.00377 | 93 | 0.00397 | -1.51916 | 111.522 | 0.13155 | 0.61876 |
| ergothioneine biosynthesis I (bacteria) | 0.00739 | 0.00247 | 0.00463 | 0.00027 | 0.00203 | 65 | 0.00931 | 0.00145 | 0.00395 | 93 | 0.00445 | -1.05277 | 133.626 | 0.29435 | 0.72280 |
| creatinine degradation I | 0.00738 | 0.00207 | 0.00716 | 0.00043 | 0.00257 | 65 | 0.00753 | 0.00085 | 0.00303 | 93 | 0.00397 | -0.09198 | 155.964 | 0.92683 | 0.98352 |
| enterobacterial common antigen biosynthesis | 0.00731 | 0.00204 | 0.00779 | 0.00068 | 0.00324 | 65 | 0.00698 | 0.00065 | 0.00265 | 93 | 0.00418 | 0.19341 | 135.778 | 0.84693 | 0.96656 |
| ketogluconate metabolism | 0.00686 | 0.00153 | 0.00694 | 0.00037 | 0.00239 | 65 | 0.00681 | 0.00037 | 0.00200 | 93 | 0.00312 | 0.04324 | 138.324 | 0.96557 | 0.99202 |
| superpathway of L-threonine metabolism | 0.00661 | 0.00137 | 0.00643 | 0.00027 | 0.00203 | 65 | 0.00673 | 0.00032 | 0.00185 | 93 | 0.00275 | -0.10788 | 144.974 | 0.91424 | 0.97851 |
| gallate degradation II | 0.00644 | 0.00253 | 0.00138 | 0.00002 | 0.00050 | 65 | 0.00997 | 0.00168 | 0.00425 | 93 | 0.00428 | -2.00557 | 94.584 | 0.04776 | 0.58059 |
| methylgallate degradation | 0.00643 | 0.00187 | 0.00171 | 0.00003 | 0.00062 | 65 | 0.00973 | 0.00090 | 0.00312 | 93 | 0.00318 | -2.52235 | 99.266 | 0.01325 | 0.58059 |
| mycolyl-arabinogalactan-peptidoglycan complex biosynthesis | 0.00612 | 0.00202 | 0.00361 | 0.00020 | 0.00177 | 65 | 0.00788 | 0.00095 | 0.00320 | 93 | 0.00366 | -1.16593 | 138.469 | 0.24565 | 0.70007 |
| L-methionine salvage cycle III | 0.00610 | 0.00214 | 0.00096 | 0.00002 | 0.00048 | 65 | 0.00970 | 0.00119 | 0.00357 | 93 | 0.00361 | -2.42555 | 95.318 | 0.01717 | 0.58059 |
| S-methyl-5-thio-α-D-ribose 1-phosphate degradation | 0.00604 | 0.00200 | 0.00093 | 0.00002 | 0.00050 | 65 | 0.00962 | 0.00104 | 0.00334 | 93 | 0.00338 | -2.57073 | 96.047 | 0.01168 | 0.58059 |
| D-glucarate degradation I | 0.00554 | 0.00137 | 0.00662 | 0.00039 | 0.00245 | 65 | 0.00479 | 0.00023 | 0.00158 | 93 | 0.00291 | 0.62881 | 114.414 | 0.53072 | 0.84889 |
| D-galactarate degradation I | 0.00552 | 0.00135 | 0.00657 | 0.00037 | 0.00240 | 65 | 0.00478 | 0.00023 | 0.00156 | 93 | 0.00286 | 0.62702 | 115.120 | 0.53189 | 0.84889 |
| superpathway of D-glucarate and D-galactarate degradation | 0.00552 | 0.00135 | 0.00657 | 0.00037 | 0.00240 | 65 | 0.00478 | 0.00023 | 0.00156 | 93 | 0.00286 | 0.62702 | 115.120 | 0.53189 | 0.84889 |
| chlorosalicylate degradation | 0.00525 | 0.00229 | 0.00195 | 0.00004 | 0.00075 | 65 | 0.00756 | 0.00138 | 0.00385 | 93 | 0.00393 | -1.43084 | 98.874 | 0.15563 | 0.61876 |
| chondroitin sulfate degradation I (bacterial) | 0.00521 | 0.00108 | 0.00363 | 0.00004 | 0.00077 | 65 | 0.00632 | 0.00029 | 0.00176 | 93 | 0.00192 | -1.40221 | 124.173 | 0.16335 | 0.61876 |
| protocatechuate degradation I (meta-cleavage pathway) | 0.00485 | 0.00200 | 0.00113 | 0.00002 | 0.00052 | 65 | 0.00745 | 0.00105 | 0.00337 | 93 | 0.00341 | -1.85406 | 96.442 | 0.06679 | 0.58059 |
| superpathway of mycolyl-arabinogalactan-peptidoglycan complex biosynthesis | 0.00425 | 0.00151 | 0.00087 | 0.00003 | 0.00073 | 65 | 0.00661 | 0.00058 | 0.00249 | 93 | 0.00260 | -2.20686 | 107.263 | 0.02945 | 0.58059 |
| L-tryptophan degradation IX | 0.00423 | 0.00220 | 0.00251 | 0.00006 | 0.00099 | 65 | 0.00544 | 0.00126 | 0.00368 | 93 | 0.00381 | -0.76954 | 104.923 | 0.44330 | 0.81137 |
| L-valine degradation I | 0.00411 | 0.00132 | 0.00246 | 0.00011 | 0.00128 | 65 | 0.00527 | 0.00039 | 0.00205 | 93 | 0.00242 | -1.16227 | 146.021 | 0.24702 | 0.70007 |
| methanogenesis from acetate | 0.00389 | 0.00171 | 0.00247 | 0.00025 | 0.00196 | 65 | 0.00487 | 0.00061 | 0.00256 | 93 | 0.00323 | -0.74266 | 154.957 | 0.45881 | 0.81137 |
| 2-amino-3-carboxymuconate semialdehyde degradation to 2-oxopentenoate | 0.00388 | 0.00180 | 0.00187 | 0.00005 | 0.00084 | 65 | 0.00529 | 0.00084 | 0.00300 | 93 | 0.00311 | -1.09833 | 105.892 | 0.27455 | 0.70413 |
| meta cleavage pathway of aromatic compounds | 0.00387 | 0.00110 | 0.00656 | 0.00038 | 0.00243 | 65 | 0.00200 | 0.00005 | 0.00073 | 93 | 0.00253 | 1.80069 | 75.736 | 0.07573 | 0.58059 |
| L-tryptophan degradation XII (Geobacillus) | 0.00384 | 0.00218 | 0.00139 | 0.00003 | 0.00066 | 65 | 0.00555 | 0.00126 | 0.00368 | 93 | 0.00374 | -1.11557 | 97.806 | 0.26734 | 0.70194 |
| protein N-glycosylation (bacterial) | 0.00381 | 0.00119 | 0.00503 | 0.00037 | 0.00238 | 65 | 0.00296 | 0.00013 | 0.00116 | 93 | 0.00265 | 0.77969 | 94.550 | 0.43752 | 0.81117 |
| androstenedione degradation | 0.00373 | 0.00119 | 0.00335 | 0.00017 | 0.00161 | 65 | 0.00399 | 0.00027 | 0.00170 | 93 | 0.00234 | -0.27457 | 153.488 | 0.78402 | 0.93380 |
| 2-nitrobenzoate degradation I | 0.00338 | 0.00178 | 0.00159 | 0.00003 | 0.00066 | 65 | 0.00463 | 0.00083 | 0.00299 | 93 | 0.00306 | -0.99431 | 100.744 | 0.32245 | 0.72280 |
| pyrimidine deoxyribonucleotides biosynthesis from CTP | 0.00334 | 0.00173 | 0.00201 | 0.00026 | 0.00201 | 65 | 0.00427 | 0.00063 | 0.00259 | 93 | 0.00328 | -0.68723 | 155.229 | 0.49296 | 0.82687 |
| isoprene biosynthesis II (engineered) | 0.00320 | 0.00107 | 0.00202 | 0.00007 | 0.00102 | 65 | 0.00402 | 0.00026 | 0.00167 | 93 | 0.00196 | -1.01948 | 144.571 | 0.30968 | 0.72280 |
| allantoin degradation IV (anaerobic) | 0.00297 | 0.00096 | 0.00137 | 0.00002 | 0.00052 | 65 | 0.00409 | 0.00023 | 0.00158 | 93 | 0.00166 | -1.63580 | 110.971 | 0.10471 | 0.61876 |
| CMP-pseudaminate biosynthesis | 0.00293 | 0.00095 | 0.00301 | 0.00015 | 0.00152 | 65 | 0.00288 | 0.00014 | 0.00121 | 93 | 0.00194 | 0.06728 | 133.620 | 0.94646 | 0.99202 |
| ethylmalonyl-CoA pathway | 0.00288 | 0.00128 | 0.00103 | 0.00002 | 0.00052 | 65 | 0.00418 | 0.00043 | 0.00214 | 93 | 0.00220 | -1.42710 | 102.854 | 0.15658 | 0.61876 |
| nylon-6 oligomer degradation | 0.00285 | 0.00109 | 0.00149 | 0.00002 | 0.00054 | 65 | 0.00381 | 0.00030 | 0.00180 | 93 | 0.00188 | -1.23226 | 108.255 | 0.22052 | 0.67841 |
| superpathway of aerobic toluene degradation | 0.00266 | 0.00092 | 0.00316 | 0.00015 | 0.00154 | 65 | 0.00232 | 0.00012 | 0.00114 | 93 | 0.00192 | 0.43848 | 126.623 | 0.66178 | 0.90545 |
| pyrimidine deoxyribonucleotides de novo biosynthesis IV | 0.00266 | 0.00139 | 0.00154 | 0.00015 | 0.00154 | 65 | 0.00344 | 0.00041 | 0.00210 | 93 | 0.00261 | -0.73074 | 153.592 | 0.46605 | 0.81137 |
| 2-aminophenol degradation | 0.00214 | 0.00166 | 0.00076 | 0.00001 | 0.00042 | 65 | 0.00310 | 0.00073 | 0.00281 | 93 | 0.00284 | -0.82442 | 96.137 | 0.41174 | 0.78940 |
| gallate degradation I | 0.00212 | 0.00061 | 0.00053 | 0.00000 | 0.00024 | 65 | 0.00324 | 0.00010 | 0.00102 | 93 | 0.00104 | -2.59259 | 102.240 | 0.01092 | 0.58059 |
| UDP-2,3-diacetamido-2,3-dideoxy-α-D-mannuronate biosynthesis | 0.00208 | 0.00075 | 0.00292 | 0.00017 | 0.00162 | 65 | 0.00149 | 0.00003 | 0.00057 | 93 | 0.00172 | 0.83216 | 80.197 | 0.40779 | 0.78940 |
| 1,5-anhydrofructose degradation | 0.00202 | 0.00092 | 0.00186 | 0.00017 | 0.00161 | 65 | 0.00213 | 0.00011 | 0.00110 | 93 | 0.00195 | -0.13909 | 119.739 | 0.88961 | 0.97248 |
| superpathway of polyamine biosynthesis III | 0.00186 | 0.00127 | 0.00280 | 0.00043 | 0.00257 | 65 | 0.00121 | 0.00014 | 0.00121 | 93 | 0.00284 | 0.55921 | 92.456 | 0.57737 | 0.86605 |
| sucrose biosynthesis I (from photosynthesis) | 0.00165 | 0.00090 | 0.00341 | 0.00030 | 0.00214 | 65 | 0.00042 | 0.00001 | 0.00030 | 93 | 0.00216 | 1.38531 | 66.519 | 0.17059 | 0.61876 |
| coenzyme B biosynthesis | 0.00162 | 0.00129 | 0.00028 | 0.00001 | 0.00028 | 65 | 0.00256 | 0.00044 | 0.00218 | 93 | 0.00220 | -1.03760 | 94.997 | 0.30209 | 0.72280 |
| sulfoglycolysis | 0.00142 | 0.00069 | 0.00068 | 0.00001 | 0.00030 | 65 | 0.00194 | 0.00012 | 0.00116 | 93 | 0.00120 | -1.05252 | 103.970 | 0.29500 | 0.72280 |
| sitosterol degradation to androstenedione | 0.00141 | 0.00050 | 0.00019 | 0.00000 | 0.00015 | 65 | 0.00226 | 0.00007 | 0.00084 | 93 | 0.00085 | -2.41828 | 97.554 | 0.01745 | 0.58059 |
| methylaspartate cycle | 0.00140 | 0.00084 | 0.00144 | 0.00005 | 0.00086 | 65 | 0.00136 | 0.00016 | 0.00130 | 93 | 0.00156 | 0.05152 | 149.028 | 0.95898 | 0.99202 |
| 1,4-dihydroxy-6-naphthoate biosynthesis I | 0.00127 | 0.00051 | 0.00019 | 0.00000 | 0.00014 | 65 | 0.00203 | 0.00007 | 0.00085 | 93 | 0.00086 | -2.12364 | 96.643 | 0.03625 | 0.58059 |
| glycolysis V (Pyrococcus) | 0.00114 | 0.00046 | 0.00143 | 0.00004 | 0.00075 | 65 | 0.00094 | 0.00003 | 0.00059 | 93 | 0.00095 | 0.50793 | 133.132 | 0.61234 | 0.88168 |
| L-arabinose degradation IV | 0.00105 | 0.00054 | 0.00209 | 0.00010 | 0.00121 | 65 | 0.00033 | 0.00001 | 0.00033 | 93 | 0.00126 | 1.40369 | 73.406 | 0.16463 | 0.61876 |
| superpathway of sulfolactate degradation | 0.00102 | 0.00071 | 0.00003 | 0.00000 | 0.00003 | 65 | 0.00171 | 0.00013 | 0.00120 | 93 | 0.00120 | -1.39958 | 92.150 | 0.16500 | 0.61876 |
| vanillin and vanillate degradation II | 0.00091 | 0.00043 | 0.00009 | 0.00000 | 0.00006 | 65 | 0.00148 | 0.00005 | 0.00072 | 93 | 0.00073 | -1.92030 | 93.116 | 0.05788 | 0.58059 |
| superpathway of lipopolysaccharide biosynthesis | 0.00091 | 0.00045 | 0.00138 | 0.00007 | 0.00105 | 65 | 0.00057 | 0.00000 | 0.00019 | 93 | 0.00107 | 0.75687 | 68.210 | 0.45173 | 0.81137 |
| aerobactin biosynthesis | 0.00090 | 0.00035 | 0.00125 | 0.00003 | 0.00065 | 65 | 0.00065 | 0.00001 | 0.00039 | 93 | 0.00076 | 0.77935 | 108.278 | 0.43748 | 0.81117 |
| superpathway of vanillin and vanillate degradation | 0.00087 | 0.00039 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00142 | 0.00004 | 0.00066 | 93 | 0.00067 | -2.00099 | 93.072 | 0.04831 | 0.58059 |
| vanillin and vanillate degradation I | 0.00087 | 0.00039 | 0.00008 | 0.00000 | 0.00005 | 65 | 0.00142 | 0.00004 | 0.00066 | 93 | 0.00067 | -2.00099 | 93.072 | 0.04831 | 0.58059 |
| reductive TCA cycle II | 0.00079 | 0.00054 | 0.00112 | 0.00008 | 0.00112 | 65 | 0.00055 | 0.00002 | 0.00046 | 93 | 0.00122 | 0.46800 | 85.860 | 0.64097 | 0.90370 |
| benzoyl-CoA degradation I (aerobic) | 0.00076 | 0.00036 | 0.00043 | 0.00001 | 0.00031 | 65 | 0.00100 | 0.00003 | 0.00057 | 93 | 0.00065 | -0.88151 | 137.763 | 0.37958 | 0.74607 |
| phospholipases | 0.00073 | 0.00025 | 0.00027 | 0.00000 | 0.00027 | 65 | 0.00105 | 0.00001 | 0.00038 | 93 | 0.00047 | -1.66289 | 152.345 | 0.09839 | 0.61876 |
| superpathway of taurine degradation | 0.00069 | 0.00055 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00118 | 0.00008 | 0.00093 | 93 | 0.00093 | -1.27385 | 92.000 | 0.20593 | 0.66261 |
| formaldehyde assimilation I (serine pathway) | 0.00067 | 0.00048 | 0.00057 | 0.00002 | 0.00052 | 65 | 0.00074 | 0.00005 | 0.00074 | 93 | 0.00091 | -0.19350 | 151.751 | 0.84683 | 0.96656 |
| sucrose biosynthesis III | 0.00056 | 0.00032 | 0.00118 | 0.00004 | 0.00076 | 65 | 0.00013 | 0.00000 | 0.00009 | 93 | 0.00077 | 1.36867 | 65.903 | 0.17575 | 0.62057 |
| chitin derivatives degradation | 0.00042 | 0.00029 | 0.00103 | 0.00003 | 0.00069 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00069 | 1.48998 | 64.000 | 0.14114 | 0.61876 |
| syringate degradation | 0.00039 | 0.00028 | 0.00052 | 0.00002 | 0.00052 | 65 | 0.00031 | 0.00001 | 0.00031 | 93 | 0.00060 | 0.35179 | 107.442 | 0.72569 | 0.91629 |
| chlorophyllide a biosynthesis II (anaerobic) | 0.00036 | 0.00031 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00057 | 0.00003 | 0.00053 | 93 | 0.00053 | -0.96662 | 94.016 | 0.33621 | 0.72862 |
| chlorophyllide a biosynthesis III (aerobic, light independent) | 0.00036 | 0.00031 | 0.00006 | 0.00000 | 0.00006 | 65 | 0.00057 | 0.00003 | 0.00053 | 93 | 0.00053 | -0.96662 | 94.016 | 0.33621 | 0.72862 |
| mandelate degradation to acetyl-CoA | 0.00034 | 0.00024 | 0.00083 | 0.00002 | 0.00058 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00058 | 1.42531 | 64.000 | 0.15893 | 0.61876 |
| mandelate degradation I | 0.00014 | 0.00010 | 0.00033 | 0.00000 | 0.00023 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00023 | 1.42511 | 64.000 | 0.15898 | 0.61876 |
| starch degradation III | 0.00012 | 0.00009 | 0.00016 | 0.00000 | 0.00016 | 65 | 0.00010 | 0.00000 | 0.00010 | 93 | 0.00019 | 0.30942 | 113.039 | 0.75757 | 0.92864 |
| mannosylglycerate biosynthesis I | 0.00012 | 0.00012 | 0.00029 | 0.00001 | 0.00029 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00029 | 1.00000 | 64.000 | 0.32108 | 0.72280 |
| sucrose degradation II (sucrose synthase) | 0.00011 | 0.00011 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00018 | 0.00000 | 0.00018 | 93 | 0.00018 | -1.00000 | 92.000 | 0.31993 | 0.72280 |
| L-rhamnose degradation II | 0.00010 | 0.00006 | 0.00007 | 0.00000 | 0.00007 | 65 | 0.00012 | 0.00000 | 0.00008 | 93 | 0.00011 | -0.51068 | 156.000 | 0.61029 | 0.88168 |
| β-alanine biosynthesis II | 0.00008 | 0.00008 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00014 | 0.00000 | 0.00014 | 93 | 0.00014 | -1.00000 | 92.000 | 0.31993 | 0.72280 |
| superpathway of C1 compounds oxidation to CO2 | 0.00007 | 0.00007 | 0.00016 | 0.00000 | 0.00016 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00016 | 1.00000 | 64.000 | 0.32108 | 0.72280 |
| flavin biosynthesis II (archaea) | 0.00005 | 0.00005 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00009 | 0.00000 | 0.00009 | 93 | 0.00009 | -1.00000 | 92.000 | 0.31993 | 0.72280 |
| 4-coumarate degradation (anaerobic) | 0.00005 | 0.00004 | 0.00009 | 0.00000 | 0.00009 | 65 | 0.00003 | 0.00000 | 0.00003 | 93 | 0.00009 | 0.57833 | 82.726 | 0.56461 | 0.86025 |
| phosphopantothenate biosynthesis III | 0.00003 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00006 | 0.00000 | 0.00006 | 93 | 0.00006 | -1.00000 | 92.000 | 0.31993 | 0.72280 |
| factor 420 biosynthesis | 0.00003 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00005 | 0.00000 | 0.00005 | 93 | 0.00005 | -1.00000 | 92.000 | 0.31993 | 0.72280 |
| superpathway of bacteriochlorophyll a biosynthesis | 0.00003 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00004 | 0.00000 | 0.00003 | 93 | 0.00003 | -1.40047 | 92.000 | 0.16474 | 0.61876 |
| chlorophyllide a biosynthesis I (aerobic, light-dependent) | 0.00002 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00003 | 0.00000 | 0.00002 | 93 | 0.00002 | -1.38405 | 92.000 | 0.16969 | 0.61876 |
| vitamin B6 degradation | 0.00001 | 0.00001 | 0.00002 | 0.00000 | 0.00002 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00002 | 1.00000 | 64.000 | 0.32108 | 0.72280 |
| spirilloxanthin and 2,2’-diketo-spirilloxanthin biosynthesis | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.38433 | 92.000 | 0.16961 | 0.61876 |
| coumarins biosynthesis (engineered) | 0.00001 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.00001 | 0.00000 | 0.00001 | 93 | 0.00001 | -1.00000 | 92.000 | 0.31993 | 0.72280 |
Let’s try a nice bigger plot
plot.dat <- tb.ra %>%
arrange(desc(`Overall Mean`)) %>%
slice_head(n=50)%>%
mutate(
description = fct_reorder(description, `Overall Mean`),
description = factor(description, levels = levels(description), ordered=T)
) %>%
arrange(description) %>%
mutate(
id = 1:n(),
step = ifelse(id%%2 == 0, 1, 0),
Mean_diff = `Tumor Mean` - `Non-Tumor Mean`,
diff_se = SEpooled,
Mean_diff_ll = Mean_diff - qt(0.975, df)*diff_se,
Mean_diff_ul = Mean_diff + qt(0.975, df)*diff_se
) %>%
pivot_longer(
cols=contains("Mean"),
names_to = "group",
values_to = "mean"
)
p1.d <- plot.dat %>%
filter(group %in% c("Tumor Mean","Non-Tumor Mean")) %>%
mutate(
group = ifelse(group == "Tumor Mean", "Tumor", "Non-Tumor"),
col = ifelse(step == 1, "grey90", "white"),
h=1, w=Inf
)
p1 <- ggplot()+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_bar(data=p1.d,
aes(x=mean, y=description,
group=group, color=group,
fill=group),
stat="identity",position = "dodge",
alpha = 1)+
labs(x="Mean Proportion (%)")+
theme_classic()+
theme(
legend.position = "bottom",
plot.margin = unit(c(1,0,1,1), "lines")
)
p2.d <- plot.dat %>%
filter(group %in% c("Mean_diff", "Mean_diff_ll", "Mean_diff_ul")) %>%
pivot_wider(
names_from = group,
values_from = mean
) %>%
mutate(
group = ifelse(Mean_diff > 0, "Tumor", "Non-Tumor"),
p = sprintf("%.3f", round(p,3)),
ll = min(Mean_diff_ll)-0.01,
ul = max(Mean_diff_ul)+0.01
)
p2<-ggplot(p2.d, aes(x=Mean_diff, y=description))+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_vline(xintercept = 0, linetype="dashed", alpha=0.5)+
geom_segment(aes(x=Mean_diff_ll, y=description, xend=Mean_diff_ul, yend=description))+
geom_point(aes(fill=group, color=group))+
geom_text(aes(label=p, x=unique(ul)+0.1))+
coord_cartesian(xlim = c(unique(p2.d$ll), unique(p2.d$ul)),
clip = 'off') +
annotate("text", x=unique(p2.d$ul)+0.2,y = 25,
angle=90,
label="p-value (uncorrected)")+
labs(x="Mean Difference in Proportions")+
theme_classic()+
theme(
legend.position = "bottom",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
plot.margin = unit(c(1,4,1,0), "lines")
)
# plot
p1 + p2+
plot_annotation(title="Path Data: 50 most abundant descriptions")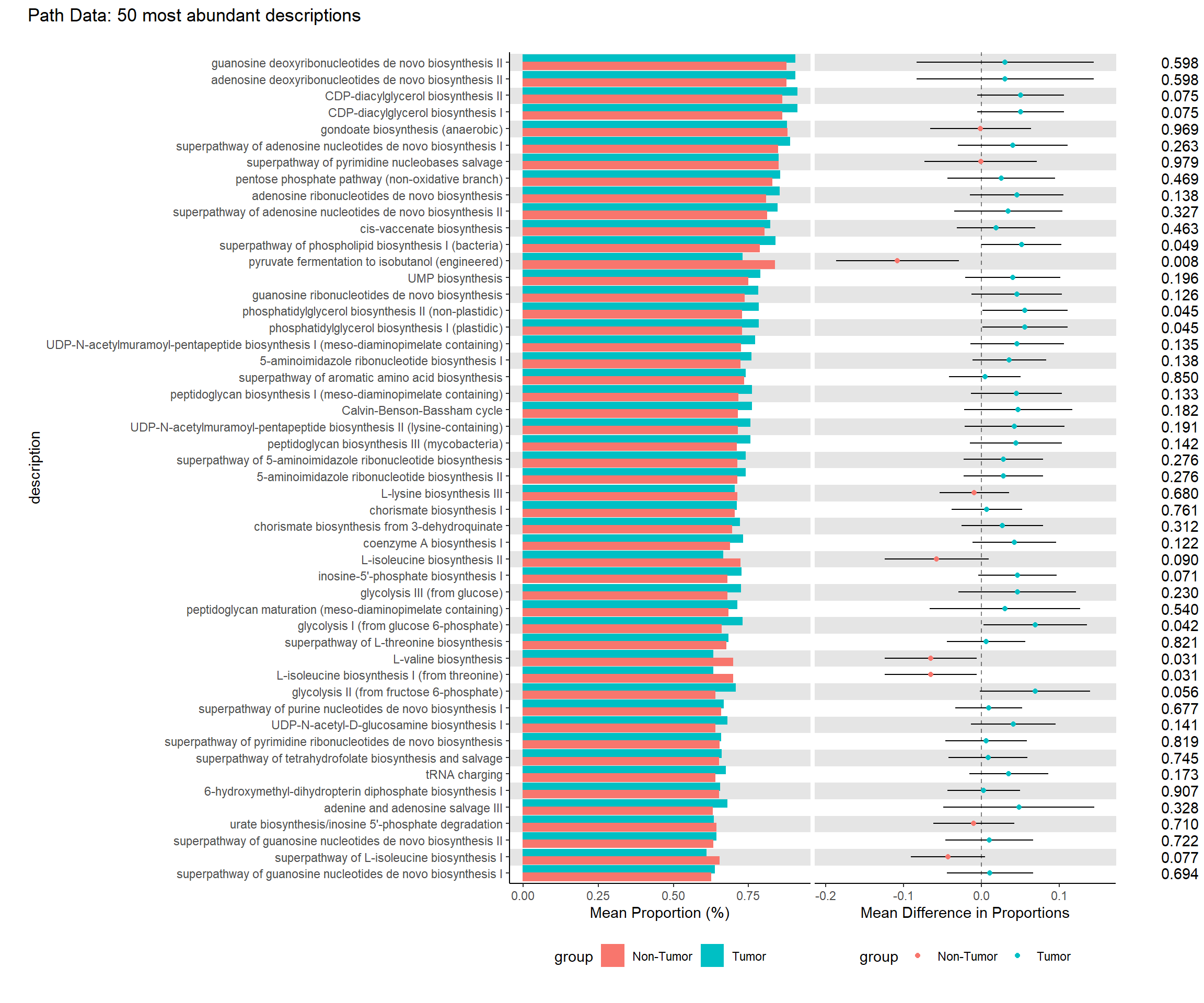
Modeling Difference between Tumor and Non-Tumor
For the modeling, we used a generalized linear mixed model (GLMM).
First, we looked at the biserial correlation between the abundance of each description and the tumor status.
tb <- mydata %>%
group_by(description)%>%
summarise(
r = cor(tumor, Abundance)
) %>%
mutate(
M=mean(r)
)
ggplot(tb, aes(x=r))+
geom_density()+
geom_vline(aes(xintercept = M))+
labs(x="Biserial Correlation",title="Relationship between description abundance and tumor (tumor vs. non-tumor)")+
theme(panel.grid = element_blank())
Next, we need the data to be on an interpretable scale. First, let’s use the raw abundance like scale.
p <- ggplot(mydata, aes(x=Abundance))+
geom_density()
p
mydata <- mydata %>%
mutate(Abundance.dich=ifelse(Abundance==0, 0, 1))
table(mydata$Abundance.dich)
0 1
19644 43398 Let’s first run models by description so that we can avoid the nesting issue initially. We will come back to this to conduct the final model (it will be more powerful).
DESCRIPTIONS <- unique(mydata$description)
i<-1
dat0 <- mydata %>% filter(description==DESCRIPTIONS[i])
# quasipossion (approximately negative binom) give an approx. answer.
fit0 <- glm(
Abundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
summary(fit0)
Call:
glm(formula = Abundance ~ 1 + tumor, family = quasipoisson(link = "log"),
data = dat0)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.34 -0.34 -0.34 0.00 6.16
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.854 0.768 -3.72 0.00028 ***
tumor -17.449 3427.430 -0.01 0.99594
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 3.1611)
Null deviance: 54.273 on 157 degrees of freedom
Residual deviance: 48.591 on 156 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 18NExt, let’s model the percent relative abundance. This will allow us to make inference about the difference average relative abundance. Which is a simpler interpretation than trying to model differences in the log relative abundance which will need to be interpreted as the multiplicative relative change.
results.out <- as.data.frame(matrix(ncol=4, nrow=length(DESCRIPTIONS)))
colnames(results.out) <- c("description", "Est", "SE", "p")
#results.out$description <-DESCRIPTIONS
i <- 1
for(i in 1:length(DESCRIPTIONS)){
#for(i in 1:5){
dat0 <- mydata %>%
filter(description == DESCRIPTIONS[i])%>%
mutate(RelAbundance= RelAbundance*100)
fit0 <- glm(
RelAbundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
fit.sum <- summary(fit0)
results.out[i, 1] <- DESCRIPTIONS[i]
results.out[i, 2:4] <- fit.sum$coefficients[2, c(1,2,4)]
}
results.out$fdr_p <- p.adjust(results.out$p, method="fdr")
kable(results.out, format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="600px")| description | Est | SE | p | fdr_p |
|---|---|---|---|---|
| β-alanine biosynthesis II | -18.045 | 4617.336 | 0.997 | 0.999 |
| (5Z)-dodec-5-enoate biosynthesis | -0.061 | 0.058 | 0.294 | 0.772 |
| 1,4-dihydroxy-2-naphthoate biosynthesis I | 0.063 | 0.165 | 0.705 | 0.952 |
| 1,4-dihydroxy-6-naphthoate biosynthesis I | -2.348 | 1.372 | 0.089 | 0.627 |
| 1,4-dihydroxy-6-naphthoate biosynthesis II | 0.176 | 0.455 | 0.699 | 0.952 |
| 1,5-anhydrofructose degradation | -0.136 | 0.955 | 0.887 | 0.999 |
| 2-amino-3-carboxymuconate semialdehyde degradation to 2-oxopentenoate | -1.041 | 1.031 | 0.314 | 0.778 |
| 2-aminophenol degradation | -1.406 | 1.858 | 0.450 | 0.876 |
| 2-methylcitrate cycle I | -0.043 | 0.249 | 0.862 | 0.999 |
| 2-methylcitrate cycle II | -0.023 | 0.215 | 0.914 | 0.999 |
| 2-nitrobenzoate degradation I | -1.072 | 1.167 | 0.360 | 0.818 |
| 3-phenylpropanoate and 3-(3-hydroxyphenyl)propanoate degradation | -0.764 | 0.578 | 0.188 | 0.688 |
| 3-phenylpropanoate and 3-(3-hydroxyphenyl)propanoate degradation to 2-oxopent-4-enoate | -0.936 | 0.670 | 0.165 | 0.670 |
| 3-phenylpropanoate degradation | -1.208 | 0.712 | 0.092 | 0.627 |
| 4-aminobutanoate degradation V | -0.122 | 0.197 | 0.537 | 0.898 |
| 4-coumarate degradation (anaerobic) | 0.965 | 1.440 | 0.504 | 0.883 |
| 4-deoxy-L-threo-hex-4-enopyranuronate degradation | 0.166 | 0.252 | 0.511 | 0.883 |
| 4-hydroxyphenylacetate degradation | -0.648 | 0.519 | 0.214 | 0.719 |
| 4-methylcatechol degradation (ortho cleavage) | 0.775 | 0.342 | 0.025 | 0.627 |
| 5-aminoimidazole ribonucleotide biosynthesis I | 0.048 | 0.031 | 0.119 | 0.627 |
| 5-aminoimidazole ribonucleotide biosynthesis II | 0.039 | 0.034 | 0.255 | 0.741 |
| 6-hydroxymethyl-dihydropterin diphosphate biosynthesis I | 0.004 | 0.034 | 0.903 | 0.999 |
| 6-hydroxymethyl-dihydropterin diphosphate biosynthesis III (Chlamydia) | 0.041 | 0.043 | 0.350 | 0.807 |
| 8-amino-7-oxononanoate biosynthesis I | 0.016 | 0.114 | 0.885 | 0.999 |
| acetyl-CoA fermentation to butanoate II | -0.296 | 0.230 | 0.200 | 0.688 |
| acetylene degradation | 0.088 | 0.104 | 0.395 | 0.852 |
| adenine and adenosine salvage III | 0.074 | 0.075 | 0.327 | 0.799 |
| adenosine deoxyribonucleotides de novo biosynthesis II | 0.034 | 0.062 | 0.580 | 0.913 |
| adenosine nucleotides degradation II | -0.135 | 0.150 | 0.372 | 0.830 |
| adenosine ribonucleotides de novo biosynthesis | 0.055 | 0.034 | 0.116 | 0.627 |
| adenosylcobalamin biosynthesis from cobyrinate a,c-diamide I | 0.092 | 0.117 | 0.431 | 0.862 |
| adenosylcobalamin biosynthesis I (early cobalt insertion) | 0.002 | 0.235 | 0.994 | 0.999 |
| adenosylcobalamin biosynthesis II (late cobalt incorporation) | -0.246 | 0.298 | 0.410 | 0.853 |
| adenosylcobalamin salvage from cobinamide I | 0.086 | 0.113 | 0.449 | 0.876 |
| adenosylcobalamin salvage from cobinamide II | 0.087 | 0.116 | 0.454 | 0.876 |
| ADP-L-glycero-β-D-manno-heptose biosynthesis | 0.114 | 0.123 | 0.357 | 0.818 |
| aerobactin biosynthesis | 0.644 | 0.792 | 0.417 | 0.853 |
| aerobic respiration I (cytochrome c) | -0.341 | 0.153 | 0.027 | 0.627 |
| allantoin degradation IV (anaerobic) | -1.093 | 0.732 | 0.137 | 0.637 |
| allantoin degradation to glyoxylate III | -0.295 | 0.307 | 0.337 | 0.799 |
| androstenedione degradation | -0.175 | 0.661 | 0.792 | 0.988 |
| anhydromuropeptides recycling | -0.133 | 0.103 | 0.198 | 0.688 |
| arginine, ornithine and proline interconversion | 0.018 | 0.136 | 0.897 | 0.999 |
| aromatic biogenic amine degradation (bacteria) | -0.316 | 0.300 | 0.294 | 0.772 |
| aromatic compounds degradation via β-ketoadipate | 0.018 | 0.274 | 0.949 | 0.999 |
| aspartate superpathway | -0.013 | 0.050 | 0.795 | 0.988 |
| benzoyl-CoA degradation I (aerobic) | -0.850 | 1.056 | 0.422 | 0.853 |
| Bifidobacterium shunt | 0.179 | 0.222 | 0.421 | 0.853 |
| biotin biosynthesis I | 0.012 | 0.108 | 0.910 | 0.999 |
| biotin biosynthesis II | -0.089 | 0.215 | 0.679 | 0.952 |
| Calvin-Benson-Bassham cycle | 0.064 | 0.043 | 0.140 | 0.644 |
| catechol degradation I (meta-cleavage pathway) | -0.138 | 0.291 | 0.635 | 0.946 |
| catechol degradation II (meta-cleavage pathway) | -0.084 | 0.495 | 0.866 | 0.999 |
| catechol degradation III (ortho-cleavage pathway) | 0.018 | 0.274 | 0.949 | 0.999 |
| catechol degradation to β-ketoadipate | 0.051 | 0.275 | 0.853 | 0.999 |
| catechol degradation to 2-oxopent-4-enoate II | -0.587 | 0.474 | 0.217 | 0.723 |
| CDP-diacylglycerol biosynthesis I | 0.057 | 0.031 | 0.064 | 0.627 |
| CDP-diacylglycerol biosynthesis II | 0.057 | 0.031 | 0.064 | 0.627 |
| chitin derivatives degradation | 19.030 | 2956.274 | 0.995 | 0.999 |
| chlorophyllide a biosynthesis I (aerobic, light-dependent) | -17.555 | 2611.080 | 0.995 | 0.999 |
| chlorophyllide a biosynthesis II (anaerobic) | -2.330 | 2.902 | 0.423 | 0.853 |
| chlorophyllide a biosynthesis III (aerobic, light independent) | -2.330 | 2.902 | 0.423 | 0.853 |
| chlorosalicylate degradation | -1.357 | 1.037 | 0.193 | 0.688 |
| chondroitin sulfate degradation I (bacterial) | -0.555 | 0.430 | 0.199 | 0.688 |
| chorismate biosynthesis from 3-dehydroquinate | 0.038 | 0.035 | 0.277 | 0.762 |
| chorismate biosynthesis I | 0.010 | 0.031 | 0.749 | 0.967 |
| cinnamate and 3-hydroxycinnamate degradation to 2-oxopent-4-enoate | -0.936 | 0.670 | 0.165 | 0.670 |
| cis-vaccenate biosynthesis | 0.023 | 0.031 | 0.451 | 0.876 |
| CMP-3-deoxy-D-manno-octulosonate biosynthesis I | 0.091 | 0.094 | 0.336 | 0.799 |
| CMP-legionaminate biosynthesis I | -0.453 | 0.625 | 0.470 | 0.883 |
| CMP-pseudaminate biosynthesis | 0.044 | 0.655 | 0.946 | 0.999 |
| cob(II)yrinate a,c-diamide biosynthesis I (early cobalt insertion) | 0.496 | 0.198 | 0.013 | 0.627 |
| cob(II)yrinate a,c-diamide biosynthesis II (late cobalt incorporation) | -0.184 | 0.250 | 0.462 | 0.876 |
| coenzyme A biosynthesis I | 0.060 | 0.037 | 0.107 | 0.627 |
| coenzyme B biosynthesis | -2.217 | 2.548 | 0.386 | 0.845 |
| colanic acid building blocks biosynthesis | -0.140 | 0.101 | 0.168 | 0.670 |
| coumarins biosynthesis (engineered) | -17.301 | 3182.835 | 0.996 | 0.999 |
| creatinine degradation I | -0.050 | 0.572 | 0.931 | 0.999 |
| D-fructuronate degradation | 0.064 | 0.208 | 0.759 | 0.971 |
| D-galactarate degradation I | 0.319 | 0.485 | 0.512 | 0.883 |
| D-galacturonate degradation I | -0.134 | 0.227 | 0.557 | 0.898 |
| D-glucarate degradation I | 0.324 | 0.490 | 0.510 | 0.883 |
| dTDP-L-rhamnose biosynthesis I | 0.149 | 0.073 | 0.042 | 0.627 |
| dTDP-N-acetylthomosamine biosynthesis | -0.319 | 0.379 | 0.401 | 0.853 |
| ectoine biosynthesis | -0.432 | 0.408 | 0.291 | 0.772 |
| enterobacterial common antigen biosynthesis | 0.110 | 0.565 | 0.846 | 0.999 |
| enterobactin biosynthesis | -0.103 | 0.187 | 0.581 | 0.913 |
| ergothioneine biosynthesis I (bacteria) | -0.698 | 0.720 | 0.334 | 0.799 |
| ethylmalonyl-CoA pathway | -1.396 | 1.083 | 0.199 | 0.688 |
| factor 420 biosynthesis | -18.033 | 4590.844 | 0.997 | 0.999 |
| fatty acid β-oxidation I | -0.227 | 0.164 | 0.169 | 0.670 |
| fatty acid elongation – saturated | -0.059 | 0.065 | 0.361 | 0.818 |
| fatty acid salvage | -0.348 | 0.202 | 0.087 | 0.627 |
| flavin biosynthesis I (bacteria and plants) | 0.018 | 0.053 | 0.735 | 0.957 |
| flavin biosynthesis II (archaea) | -17.624 | 3741.025 | 0.996 | 0.999 |
| formaldehyde assimilation I (serine pathway) | -0.269 | 1.490 | 0.857 | 0.999 |
| formaldehyde assimilation II (RuMP Cycle) | -0.901 | 0.369 | 0.016 | 0.627 |
| formaldehyde oxidation I | -0.937 | 0.377 | 0.014 | 0.627 |
| fucose degradation | 0.321 | 0.283 | 0.258 | 0.741 |
| galactose degradation I (Leloir pathway) | 0.153 | 0.082 | 0.064 | 0.627 |
| gallate degradation I | -1.809 | 0.812 | 0.027 | 0.627 |
| gallate degradation II | -1.975 | 1.128 | 0.082 | 0.627 |
| GDP-D-glycero-α-D-manno-heptose biosynthesis | 0.174 | 0.285 | 0.544 | 0.898 |
| GDP-mannose biosynthesis | -0.085 | 0.062 | 0.170 | 0.670 |
| gluconeogenesis I | 0.010 | 0.039 | 0.793 | 0.988 |
| glucose and glucose-1-phosphate degradation | 0.054 | 0.147 | 0.713 | 0.952 |
| glucose degradation (oxidative) | -0.066 | 0.461 | 0.887 | 0.999 |
| glutaryl-CoA degradation | -0.023 | 0.231 | 0.922 | 0.999 |
| glycerol degradation to butanol | 0.297 | 0.198 | 0.137 | 0.637 |
| glycine betaine degradation I | -1.316 | 0.849 | 0.123 | 0.627 |
| glycogen biosynthesis I (from ADP-D-Glucose) | 0.048 | 0.070 | 0.492 | 0.883 |
| glycogen degradation I (bacterial) | 0.043 | 0.071 | 0.548 | 0.898 |
| glycogen degradation II (eukaryotic) | 0.103 | 0.635 | 0.871 | 0.999 |
| glycolysis I (from glucose 6-phosphate) | 0.100 | 0.047 | 0.036 | 0.627 |
| glycolysis II (from fructose 6-phosphate) | 0.103 | 0.051 | 0.046 | 0.627 |
| glycolysis III (from glucose) | 0.066 | 0.053 | 0.215 | 0.719 |
| glycolysis V (Pyrococcus) | 0.415 | 0.827 | 0.617 | 0.936 |
| glyoxylate cycle | -0.294 | 0.163 | 0.074 | 0.627 |
| gondoate biosynthesis (anaerobic) | -0.001 | 0.036 | 0.968 | 0.999 |
| guanosine deoxyribonucleotides de novo biosynthesis II | 0.034 | 0.062 | 0.580 | 0.913 |
| guanosine nucleotides degradation III | -0.193 | 0.143 | 0.179 | 0.674 |
| guanosine ribonucleotides de novo biosynthesis | 0.060 | 0.037 | 0.106 | 0.627 |
| heme biosynthesis I (aerobic) | -0.076 | 0.084 | 0.364 | 0.818 |
| heme biosynthesis II (anaerobic) | -0.097 | 0.066 | 0.148 | 0.657 |
| heterolactic fermentation | 0.242 | 0.235 | 0.306 | 0.775 |
| hexitol fermentation to lactate, formate, ethanol and acetate | -0.008 | 0.160 | 0.960 | 0.999 |
| homolactic fermentation | 0.111 | 0.054 | 0.041 | 0.627 |
| incomplete reductive TCA cycle | -0.071 | 0.106 | 0.502 | 0.883 |
| inosine-5’-phosphate biosynthesis I | 0.066 | 0.035 | 0.062 | 0.627 |
| inosine-5’-phosphate biosynthesis III | 0.010 | 0.050 | 0.848 | 0.999 |
| isoprene biosynthesis II (engineered) | -0.687 | 0.731 | 0.348 | 0.807 |
| isopropanol biosynthesis | -0.830 | 0.592 | 0.163 | 0.670 |
| Kdo transfer to lipid IVA III (Chlamydia) | -0.037 | 0.089 | 0.681 | 0.952 |
| ketogluconate metabolism | 0.020 | 0.454 | 0.966 | 0.999 |
| L-1,2-propanediol degradation | 0.255 | 0.207 | 0.219 | 0.723 |
| L-arabinose degradation IV | 1.854 | 1.215 | 0.129 | 0.627 |
| L-arginine biosynthesis I (via L-ornithine) | -0.037 | 0.052 | 0.483 | 0.883 |
| L-arginine biosynthesis II (acetyl cycle) | -0.051 | 0.060 | 0.403 | 0.853 |
| L-arginine biosynthesis III (via N-acetyl-L-citrulline) | -0.045 | 0.111 | 0.686 | 0.952 |
| L-arginine biosynthesis IV (archaebacteria) | -0.037 | 0.052 | 0.481 | 0.883 |
| L-arginine degradation II (AST pathway) | 0.044 | 0.287 | 0.879 | 0.999 |
| L-glutamate and L-glutamine biosynthesis | -0.133 | 0.142 | 0.349 | 0.807 |
| L-glutamate degradation V (via hydroxyglutarate) | 0.868 | 0.352 | 0.015 | 0.627 |
| L-glutamate degradation VIII (to propanoate) | 0.450 | 0.555 | 0.418 | 0.853 |
| L-histidine biosynthesis | -0.100 | 0.054 | 0.067 | 0.627 |
| L-histidine degradation I | 0.064 | 0.106 | 0.548 | 0.898 |
| L-histidine degradation II | -0.264 | 0.299 | 0.378 | 0.838 |
| L-isoleucine biosynthesis I (from threonine) | -0.098 | 0.043 | 0.026 | 0.627 |
| L-isoleucine biosynthesis II | -0.083 | 0.047 | 0.082 | 0.627 |
| L-isoleucine biosynthesis III | -0.065 | 0.045 | 0.150 | 0.659 |
| L-isoleucine biosynthesis IV | 0.054 | 0.082 | 0.513 | 0.883 |
| L-leucine degradation I | -0.195 | 0.206 | 0.345 | 0.807 |
| L-lysine biosynthesis I | 0.008 | 0.038 | 0.836 | 0.999 |
| L-lysine biosynthesis II | 0.069 | 0.094 | 0.463 | 0.876 |
| L-lysine biosynthesis III | -0.013 | 0.030 | 0.665 | 0.952 |
| L-lysine biosynthesis VI | 0.027 | 0.039 | 0.490 | 0.883 |
| L-lysine fermentation to acetate and butanoate | 0.948 | 0.463 | 0.042 | 0.627 |
| L-methionine biosynthesis I | -0.009 | 0.057 | 0.875 | 0.999 |
| L-methionine biosynthesis III | -0.033 | 0.063 | 0.598 | 0.925 |
| L-methionine salvage cycle III | -2.317 | 1.162 | 0.048 | 0.627 |
| L-ornithine biosynthesis | -0.091 | 0.077 | 0.241 | 0.741 |
| L-rhamnose degradation I | -0.466 | 0.277 | 0.095 | 0.627 |
| L-rhamnose degradation II | -0.586 | 1.227 | 0.634 | 0.946 |
| L-tryptophan biosynthesis | -0.081 | 0.051 | 0.115 | 0.627 |
| L-tryptophan degradation IX | -0.774 | 1.091 | 0.479 | 0.883 |
| L-tryptophan degradation to 2-amino-3-carboxymuconate semialdehyde | -0.162 | 0.447 | 0.718 | 0.952 |
| L-tryptophan degradation XII (Geobacillus) | -1.389 | 1.361 | 0.309 | 0.775 |
| L-tyrosine degradation I | -0.116 | 0.222 | 0.601 | 0.926 |
| L-valine biosynthesis | -0.098 | 0.043 | 0.026 | 0.627 |
| L-valine degradation I | -0.762 | 0.715 | 0.288 | 0.771 |
| lactose and galactose degradation I | 0.032 | 0.144 | 0.824 | 0.999 |
| lipid IVA biosynthesis | 0.076 | 0.098 | 0.442 | 0.874 |
| mandelate degradation I | 18.887 | 2877.509 | 0.995 | 0.999 |
| mandelate degradation to acetyl-CoA | 18.816 | 2776.770 | 0.995 | 0.999 |
| mannan degradation | -0.626 | 0.331 | 0.061 | 0.627 |
| mannosylglycerate biosynthesis I | 18.776 | 3879.031 | 0.996 | 0.999 |
| meta cleavage pathway of aromatic compounds | 1.189 | 0.546 | 0.031 | 0.627 |
| methanogenesis from acetate | -0.678 | 0.987 | 0.493 | 0.883 |
| methanol oxidation to carbon dioxide | 0.176 | 0.389 | 0.652 | 0.947 |
| methyl ketone biosynthesis | -0.780 | 0.486 | 0.110 | 0.627 |
| methylaspartate cycle | 0.057 | 1.232 | 0.963 | 0.999 |
| methylerythritol phosphate pathway I | 0.011 | 0.060 | 0.859 | 0.999 |
| methylerythritol phosphate pathway II | 0.011 | 0.060 | 0.859 | 0.999 |
| methylgallate degradation | -1.736 | 0.783 | 0.028 | 0.627 |
| methylphosphonate degradation I | -1.021 | 0.426 | 0.018 | 0.627 |
| mevalonate pathway I | 0.204 | 0.145 | 0.162 | 0.670 |
| mixed acid fermentation | -0.003 | 0.042 | 0.946 | 0.999 |
| mono-trans, poly-cis decaprenyl phosphate biosynthesis | 0.060 | 0.203 | 0.768 | 0.973 |
| mycolate biosynthesis | -0.068 | 0.105 | 0.518 | 0.888 |
| mycolyl-arabinogalactan-peptidoglycan complex biosynthesis | -0.780 | 0.729 | 0.286 | 0.771 |
| mycothiol biosynthesis | 0.002 | 0.326 | 0.996 | 0.999 |
| myo-, chiro- and scillo-inositol degradation | -0.479 | 0.352 | 0.176 | 0.671 |
| myo-inositol degradation I | -0.334 | 0.305 | 0.276 | 0.762 |
| N10-formyl-tetrahydrofolate biosynthesis | -0.042 | 0.032 | 0.200 | 0.688 |
| NAD biosynthesis I (from aspartate) | -0.012 | 0.097 | 0.901 | 0.999 |
| NAD biosynthesis II (from tryptophan) | -0.147 | 0.404 | 0.716 | 0.952 |
| NAD salvage pathway I | -0.008 | 0.077 | 0.914 | 0.999 |
| NAD salvage pathway II | 0.168 | 0.174 | 0.338 | 0.799 |
| nicotinate degradation I | -0.986 | 0.948 | 0.300 | 0.775 |
| nitrate reduction I (denitrification) | -0.367 | 0.316 | 0.247 | 0.741 |
| nitrate reduction VI (assimilatory) | 0.102 | 0.214 | 0.635 | 0.946 |
| norspermidine biosynthesis | -0.011 | 0.260 | 0.966 | 0.999 |
| nylon-6 oligomer degradation | -0.941 | 0.829 | 0.258 | 0.741 |
| O-antigen building blocks biosynthesis (E. coli) | 0.093 | 0.050 | 0.064 | 0.627 |
| octane oxidation | -0.111 | 0.268 | 0.680 | 0.952 |
| oleate biosynthesis IV (anaerobic) | -0.093 | 0.085 | 0.275 | 0.762 |
| palmitate biosynthesis II (bacteria and plants) | -0.011 | 0.083 | 0.899 | 0.999 |
| palmitoleate biosynthesis I (from (5Z)-dodec-5-enoate) | -0.108 | 0.095 | 0.257 | 0.741 |
| pantothenate and coenzyme A biosynthesis I | -0.090 | 0.054 | 0.098 | 0.627 |
| pentose phosphate pathway | -0.037 | 0.053 | 0.484 | 0.883 |
| pentose phosphate pathway (non-oxidative branch) | 0.030 | 0.039 | 0.441 | 0.874 |
| peptidoglycan biosynthesis I (meso-diaminopimelate containing) | 0.061 | 0.038 | 0.117 | 0.627 |
| peptidoglycan biosynthesis II (staphylococci) | -0.152 | 0.415 | 0.714 | 0.952 |
| peptidoglycan biosynthesis III (mycobacteria) | 0.060 | 0.039 | 0.126 | 0.627 |
| peptidoglycan biosynthesis IV (Enterococcus faecium) | 0.044 | 0.097 | 0.649 | 0.947 |
| peptidoglycan biosynthesis V (β-lactam resistance) | 0.262 | 0.185 | 0.159 | 0.670 |
| peptidoglycan maturation (meso-diaminopimelate containing) | 0.043 | 0.069 | 0.533 | 0.898 |
| phenylacetate degradation I (aerobic) | 0.555 | 0.317 | 0.082 | 0.627 |
| phosphatidylglycerol biosynthesis I (plastidic) | 0.074 | 0.035 | 0.034 | 0.627 |
| phosphatidylglycerol biosynthesis II (non-plastidic) | 0.074 | 0.035 | 0.034 | 0.627 |
| phospholipases | -1.355 | 0.998 | 0.177 | 0.671 |
| phosphopantothenate biosynthesis I | -0.128 | 0.063 | 0.044 | 0.627 |
| phosphopantothenate biosynthesis III | -17.125 | 2914.120 | 0.995 | 0.999 |
| photorespiration | -0.259 | 0.372 | 0.487 | 0.883 |
| polyisoprenoid biosynthesis (E. coli) | 0.013 | 0.045 | 0.773 | 0.973 |
| polymyxin resistance | -0.371 | 0.517 | 0.475 | 0.883 |
| ppGpp biosynthesis | 0.192 | 0.126 | 0.130 | 0.627 |
| preQ0 biosynthesis | -0.103 | 0.058 | 0.078 | 0.627 |
| protein N-glycosylation (bacterial) | 0.529 | 0.607 | 0.385 | 0.845 |
| protocatechuate degradation I (meta-cleavage pathway) | -1.886 | 1.164 | 0.107 | 0.627 |
| protocatechuate degradation II (ortho-cleavage pathway) | -0.134 | 0.265 | 0.614 | 0.936 |
| purine nucleobases degradation I (anaerobic) | 0.034 | 0.186 | 0.856 | 0.999 |
| purine nucleotides degradation II (aerobic) | -0.148 | 0.127 | 0.244 | 0.741 |
| purine ribonucleosides degradation | 0.019 | 0.102 | 0.850 | 0.999 |
| pyridoxal 5’-phosphate biosynthesis I | -0.076 | 0.163 | 0.640 | 0.947 |
| pyrimidine deoxyribonucleosides salvage | -0.042 | 0.079 | 0.594 | 0.922 |
| pyrimidine deoxyribonucleotide phosphorylation | 0.021 | 0.044 | 0.628 | 0.946 |
| pyrimidine deoxyribonucleotides biosynthesis from CTP | -0.752 | 1.194 | 0.530 | 0.898 |
| pyrimidine deoxyribonucleotides de novo biosynthesis I | 0.016 | 0.044 | 0.717 | 0.952 |
| pyrimidine deoxyribonucleotides de novo biosynthesis II | 0.073 | 0.058 | 0.208 | 0.709 |
| pyrimidine deoxyribonucleotides de novo biosynthesis III | -0.045 | 0.196 | 0.819 | 0.999 |
| pyrimidine deoxyribonucleotides de novo biosynthesis IV | -0.805 | 1.210 | 0.506 | 0.883 |
| pyruvate fermentation to acetate and lactate II | 0.134 | 0.089 | 0.132 | 0.627 |
| pyruvate fermentation to acetone | 0.538 | 0.275 | 0.052 | 0.627 |
| pyruvate fermentation to butanoate | -0.251 | 0.228 | 0.272 | 0.762 |
| pyruvate fermentation to isobutanol (engineered) | -0.138 | 0.051 | 0.008 | 0.627 |
| pyruvate fermentation to propanoate I | -0.257 | 0.138 | 0.063 | 0.627 |
| queuosine biosynthesis | -0.068 | 0.053 | 0.199 | 0.688 |
| reductive acetyl coenzyme A pathway | -0.138 | 0.448 | 0.759 | 0.971 |
| reductive TCA cycle I | -0.064 | 0.093 | 0.492 | 0.883 |
| reductive TCA cycle II | 0.706 | 1.301 | 0.588 | 0.917 |
| S-adenosyl-L-methionine cycle I | 0.054 | 0.060 | 0.365 | 0.818 |
| S-methyl-5-thio-α-D-ribose 1-phosphate degradation | -2.337 | 1.119 | 0.038 | 0.627 |
| sitosterol degradation to androstenedione | -2.452 | 1.299 | 0.061 | 0.627 |
| spirilloxanthin and 2,2’-diketo-spirilloxanthin biosynthesis | -17.719 | 2834.255 | 0.995 | 0.999 |
| starch degradation III | 0.456 | 1.410 | 0.747 | 0.967 |
| starch degradation V | 0.050 | 0.067 | 0.457 | 0.876 |
| stearate biosynthesis II (bacteria and plants) | -0.110 | 0.096 | 0.253 | 0.741 |
| succinate fermentation to butanoate | -0.332 | 0.404 | 0.413 | 0.853 |
| sucrose biosynthesis I (from photosynthesis) | 2.094 | 1.195 | 0.082 | 0.627 |
| sucrose biosynthesis III | 2.210 | 1.269 | 0.084 | 0.627 |
| sucrose degradation II (sucrose synthase) | -17.288 | 3162.774 | 0.996 | 0.999 |
| sucrose degradation III (sucrose invertase) | 0.115 | 0.074 | 0.124 | 0.627 |
| sucrose degradation IV (sucrose phosphorylase) | 0.108 | 0.105 | 0.305 | 0.775 |
| sulfate reduction I (assimilatory) | -0.289 | 0.166 | 0.083 | 0.627 |
| sulfoglycolysis | -1.047 | 1.079 | 0.334 | 0.799 |
| superpathay of heme biosynthesis from glutamate | -0.021 | 0.073 | 0.779 | 0.977 |
| superpathway of β-D-glucuronide and D-glucuronate degradation | -0.037 | 0.232 | 0.872 | 0.999 |
| superpathway of (Kdo)2-lipid A biosynthesis | 0.215 | 0.212 | 0.314 | 0.778 |
| superpathway of (R,R)-butanediol biosynthesis | -0.062 | 0.163 | 0.706 | 0.952 |
| superpathway of 2,3-butanediol biosynthesis | -0.108 | 0.184 | 0.558 | 0.898 |
| superpathway of 5-aminoimidazole ribonucleotide biosynthesis | 0.039 | 0.034 | 0.255 | 0.741 |
| superpathway of adenosine nucleotides de novo biosynthesis I | 0.046 | 0.039 | 0.240 | 0.741 |
| superpathway of adenosine nucleotides de novo biosynthesis II | 0.042 | 0.040 | 0.305 | 0.775 |
| superpathway of aerobic toluene degradation | 0.310 | 0.696 | 0.657 | 0.947 |
| superpathway of arginine and polyamine biosynthesis | -0.003 | 0.095 | 0.974 | 0.999 |
| superpathway of aromatic amino acid biosynthesis | 0.006 | 0.030 | 0.843 | 0.999 |
| superpathway of bacteriochlorophyll a biosynthesis | -17.896 | 3060.915 | 0.995 | 0.999 |
| superpathway of branched amino acid biosynthesis | -0.075 | 0.045 | 0.095 | 0.627 |
| superpathway of C1 compounds oxidation to CO2 | 18.186 | 2888.585 | 0.995 | 0.999 |
| superpathway of chorismate metabolism | 0.148 | 0.294 | 0.616 | 0.936 |
| superpathway of Clostridium acetobutylicum acidogenic fermentation | -0.236 | 0.221 | 0.287 | 0.771 |
| superpathway of D-glucarate and D-galactarate degradation | 0.319 | 0.485 | 0.512 | 0.883 |
| superpathway of demethylmenaquinol-6 biosynthesis I | -0.010 | 0.362 | 0.978 | 0.999 |
| superpathway of demethylmenaquinol-6 biosynthesis II | -0.274 | 0.704 | 0.698 | 0.952 |
| superpathway of demethylmenaquinol-8 biosynthesis | -0.022 | 0.127 | 0.863 | 0.999 |
| superpathway of demethylmenaquinol-9 biosynthesis | -0.010 | 0.362 | 0.978 | 0.999 |
| superpathway of fatty acid biosynthesis initiation (E. coli) | -0.086 | 0.106 | 0.417 | 0.853 |
| superpathway of fucose and rhamnose degradation | -0.441 | 0.363 | 0.227 | 0.740 |
| superpathway of GDP-mannose-derived O-antigen building blocks biosynthesis | -0.200 | 0.111 | 0.073 | 0.627 |
| superpathway of geranylgeranyl diphosphate biosynthesis II (via MEP) | 0.021 | 0.057 | 0.722 | 0.954 |
| superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate) | 0.188 | 0.129 | 0.148 | 0.657 |
| superpathway of glucose and xylose degradation | -0.077 | 0.123 | 0.534 | 0.898 |
| superpathway of glycerol degradation to 1,3-propanediol | 0.647 | 0.571 | 0.258 | 0.741 |
| superpathway of glycol metabolism and degradation | 0.134 | 0.396 | 0.736 | 0.957 |
| superpathway of glycolysis and Entner-Doudoroff | -0.043 | 0.058 | 0.463 | 0.876 |
| superpathway of glycolysis, pyruvate dehydrogenase, TCA, and glyoxylate bypass | -0.204 | 0.127 | 0.111 | 0.627 |
| superpathway of glyoxylate bypass and TCA | -0.272 | 0.157 | 0.086 | 0.627 |
| superpathway of guanosine nucleotides de novo biosynthesis I | 0.017 | 0.041 | 0.668 | 0.952 |
| superpathway of guanosine nucleotides de novo biosynthesis II | 0.016 | 0.041 | 0.699 | 0.952 |
| superpathway of heme biosynthesis from glycine | -0.163 | 0.148 | 0.273 | 0.762 |
| superpathway of heme biosynthesis from uroporphyrinogen-III | -0.085 | 0.128 | 0.508 | 0.883 |
| superpathway of hexitol degradation (bacteria) | 0.087 | 0.110 | 0.432 | 0.862 |
| superpathway of hexuronide and hexuronate degradation | -0.049 | 0.259 | 0.849 | 0.999 |
| superpathway of histidine, purine, and pyrimidine biosynthesis | -0.001 | 0.062 | 0.991 | 0.999 |
| superpathway of L-alanine biosynthesis | -0.019 | 0.065 | 0.770 | 0.973 |
| superpathway of L-arginine and L-ornithine degradation | 0.456 | 0.378 | 0.230 | 0.740 |
| superpathway of L-arginine, putrescine, and 4-aminobutanoate degradation | 0.456 | 0.378 | 0.230 | 0.740 |
| superpathway of L-aspartate and L-asparagine biosynthesis | 0.033 | 0.097 | 0.736 | 0.957 |
| superpathway of L-isoleucine biosynthesis I | -0.068 | 0.036 | 0.063 | 0.627 |
| superpathway of L-lysine, L-threonine and L-methionine biosynthesis I | -0.023 | 0.047 | 0.627 | 0.946 |
| superpathway of L-methionine biosynthesis (by sulfhydrylation) | -0.204 | 0.120 | 0.090 | 0.627 |
| superpathway of L-methionine biosynthesis (transsulfuration) | -0.023 | 0.057 | 0.683 | 0.952 |
| superpathway of L-phenylalanine biosynthesis | 0.061 | 0.053 | 0.253 | 0.741 |
| superpathway of L-serine and glycine biosynthesis I | -0.070 | 0.045 | 0.119 | 0.627 |
| superpathway of L-threonine biosynthesis | 0.009 | 0.036 | 0.812 | 0.999 |
| superpathway of L-threonine metabolism | -0.045 | 0.424 | 0.915 | 0.999 |
| superpathway of L-tryptophan biosynthesis | -0.599 | 0.802 | 0.456 | 0.876 |
| superpathway of L-tyrosine biosynthesis | 0.053 | 0.055 | 0.338 | 0.799 |
| superpathway of lipopolysaccharide biosynthesis | 0.879 | 0.860 | 0.308 | 0.775 |
| superpathway of menaquinol-10 biosynthesis | 0.008 | 0.339 | 0.982 | 0.999 |
| superpathway of menaquinol-11 biosynthesis | -0.054 | 0.123 | 0.658 | 0.947 |
| superpathway of menaquinol-12 biosynthesis | -0.054 | 0.123 | 0.658 | 0.947 |
| superpathway of menaquinol-13 biosynthesis | -0.054 | 0.123 | 0.658 | 0.947 |
| superpathway of menaquinol-6 biosynthesis I | 0.008 | 0.339 | 0.982 | 0.999 |
| superpathway of menaquinol-7 biosynthesis | -0.049 | 0.120 | 0.687 | 0.952 |
| superpathway of menaquinol-8 biosynthesis I | -0.036 | 0.118 | 0.762 | 0.972 |
| superpathway of menaquinol-8 biosynthesis II | -0.030 | 0.304 | 0.921 | 0.999 |
| superpathway of menaquinol-9 biosynthesis | 0.008 | 0.339 | 0.982 | 0.999 |
| superpathway of methylglyoxal degradation | -0.185 | 0.411 | 0.654 | 0.947 |
| superpathway of mycolyl-arabinogalactan-peptidoglycan complex biosynthesis | -2.023 | 1.142 | 0.078 | 0.627 |
| superpathway of N-acetylglucosamine, N-acetylmannosamine and N-acetylneuraminate degradation | 0.122 | 0.107 | 0.256 | 0.741 |
| superpathway of N-acetylneuraminate degradation | 0.053 | 0.086 | 0.534 | 0.898 |
| superpathway of ornithine degradation | -0.272 | 0.487 | 0.577 | 0.913 |
| superpathway of phenylethylamine degradation | 0.478 | 0.357 | 0.182 | 0.678 |
| superpathway of phospholipid biosynthesis I (bacteria) | 0.063 | 0.030 | 0.033 | 0.627 |
| superpathway of phylloquinol biosynthesis | 0.054 | 0.157 | 0.732 | 0.957 |
| superpathway of polyamine biosynthesis I | -0.039 | 0.106 | 0.716 | 0.952 |
| superpathway of polyamine biosynthesis II | 0.053 | 0.120 | 0.660 | 0.947 |
| superpathway of polyamine biosynthesis III | 0.838 | 1.363 | 0.540 | 0.898 |
| superpathway of purine deoxyribonucleosides degradation | 0.055 | 0.094 | 0.561 | 0.899 |
| superpathway of purine nucleotides de novo biosynthesis I | 0.014 | 0.030 | 0.651 | 0.947 |
| superpathway of purine nucleotides de novo biosynthesis II | 0.058 | 0.050 | 0.246 | 0.741 |
| superpathway of pyridoxal 5’-phosphate biosynthesis and salvage | -0.025 | 0.133 | 0.849 | 0.999 |
| superpathway of pyrimidine deoxyribonucleoside salvage | 0.000 | 0.063 | 0.999 | 0.999 |
| superpathway of pyrimidine deoxyribonucleosides degradation | 0.051 | 0.089 | 0.567 | 0.906 |
| superpathway of pyrimidine deoxyribonucleotides de novo biosynthesis | -0.048 | 0.060 | 0.421 | 0.853 |
| superpathway of pyrimidine deoxyribonucleotides de novo biosynthesis (E. coli) | 0.022 | 0.041 | 0.587 | 0.917 |
| superpathway of pyrimidine nucleobases salvage | -0.001 | 0.041 | 0.978 | 0.999 |
| superpathway of pyrimidine ribonucleosides salvage | 0.010 | 0.065 | 0.882 | 0.999 |
| superpathway of pyrimidine ribonucleotides de novo biosynthesis | 0.009 | 0.037 | 0.803 | 0.993 |
| superpathway of S-adenosyl-L-methionine biosynthesis | -0.017 | 0.046 | 0.709 | 0.952 |
| superpathway of salicylate degradation | 0.330 | 0.319 | 0.302 | 0.775 |
| superpathway of sulfate assimilation and cysteine biosynthesis | -0.213 | 0.132 | 0.109 | 0.627 |
| superpathway of sulfolactate degradation | -3.914 | 4.627 | 0.399 | 0.853 |
| superpathway of sulfur oxidation (Acidianus ambivalens) | -0.712 | 0.340 | 0.038 | 0.627 |
| superpathway of taurine degradation | -18.165 | 3849.045 | 0.996 | 0.999 |
| superpathway of tetrahydrofolate biosynthesis | 0.003 | 0.044 | 0.947 | 0.999 |
| superpathway of tetrahydrofolate biosynthesis and salvage | 0.013 | 0.038 | 0.735 | 0.957 |
| superpathway of thiamin diphosphate biosynthesis I | 0.160 | 0.111 | 0.154 | 0.668 |
| superpathway of thiamin diphosphate biosynthesis II | 0.079 | 0.120 | 0.514 | 0.883 |
| superpathway of ubiquinol-8 biosynthesis (prokaryotic) | -0.043 | 0.144 | 0.765 | 0.972 |
| superpathway of UDP-glucose-derived O-antigen building blocks biosynthesis | -0.040 | 0.131 | 0.759 | 0.971 |
| superpathway of UDP-N-acetylglucosamine-derived O-antigen building blocks biosynthesis | -0.705 | 0.535 | 0.190 | 0.688 |
| superpathway of vanillin and vanillate degradation | -2.850 | 1.872 | 0.130 | 0.627 |
| syringate degradation | 0.525 | 1.408 | 0.710 | 0.952 |
| taxadiene biosynthesis (engineered) | 0.031 | 0.059 | 0.606 | 0.931 |
| TCA cycle I (prokaryotic) | -0.167 | 0.083 | 0.045 | 0.627 |
| TCA cycle IV (2-oxoglutarate decarboxylase) | -0.246 | 0.137 | 0.075 | 0.627 |
| TCA cycle V (2-oxoglutarate:ferredoxin oxidoreductase) | -0.143 | 0.077 | 0.067 | 0.627 |
| TCA cycle VI (obligate autotrophs) | -0.086 | 0.053 | 0.105 | 0.627 |
| TCA cycle VII (acetate-producers) | -0.135 | 0.113 | 0.232 | 0.741 |
| TCA cycle VIII (helicobacter) | -0.194 | 0.124 | 0.118 | 0.627 |
| teichoic acid (poly-glycerol) biosynthesis | 0.108 | 0.258 | 0.677 | 0.952 |
| tetrapyrrole biosynthesis I (from glutamate) | 0.075 | 0.069 | 0.280 | 0.765 |
| tetrapyrrole biosynthesis II (from glycine) | 0.077 | 0.070 | 0.269 | 0.762 |
| thiamin salvage II | 0.001 | 0.045 | 0.976 | 0.999 |
| thiazole biosynthesis I (E. coli) | 0.240 | 0.143 | 0.096 | 0.627 |
| thiazole biosynthesis II (Bacillus) | 0.306 | 0.176 | 0.084 | 0.627 |
| toluene degradation I (aerobic) (via o-cresol) | -0.249 | 0.288 | 0.390 | 0.845 |
| toluene degradation II (aerobic) (via 4-methylcatechol) | -0.249 | 0.288 | 0.390 | 0.845 |
| toluene degradation III (aerobic) (via p-cresol) | 0.762 | 0.333 | 0.024 | 0.627 |
| toluene degradation IV (aerobic) (via catechol) | 0.107 | 0.417 | 0.798 | 0.989 |
| tRNA charging | 0.054 | 0.038 | 0.158 | 0.670 |
| tRNA processing | 0.007 | 0.141 | 0.958 | 0.999 |
| ubiquinol-10 biosynthesis (prokaryotic) | -0.085 | 0.145 | 0.558 | 0.898 |
| ubiquinol-7 biosynthesis (prokaryotic) | -0.085 | 0.145 | 0.558 | 0.898 |
| ubiquinol-8 biosynthesis (prokaryotic) | -0.085 | 0.145 | 0.558 | 0.898 |
| ubiquinol-9 biosynthesis (prokaryotic) | -0.085 | 0.145 | 0.558 | 0.898 |
| UDP-2,3-diacetamido-2,3-dideoxy-α-D-mannuronate biosynthesis | 0.671 | 0.670 | 0.318 | 0.783 |
| UDP-N-acetyl-D-glucosamine biosynthesis I | 0.062 | 0.040 | 0.122 | 0.627 |
| UDP-N-acetylmuramoyl-pentapeptide biosynthesis I (meso-diaminopimelate containing) | 0.061 | 0.039 | 0.120 | 0.627 |
| UDP-N-acetylmuramoyl-pentapeptide biosynthesis II (lysine-containing) | 0.058 | 0.043 | 0.176 | 0.671 |
| UMP biosynthesis | 0.052 | 0.038 | 0.173 | 0.671 |
| urate biosynthesis/inosine 5’-phosphate degradation | -0.015 | 0.041 | 0.707 | 0.952 |
| urea cycle | -0.239 | 0.130 | 0.067 | 0.627 |
| vanillin and vanillate degradation I | -2.850 | 1.872 | 0.130 | 0.627 |
| vanillin and vanillate degradation II | -2.796 | 1.894 | 0.142 | 0.644 |
| vitamin B6 degradation | 18.114 | 2786.720 | 0.995 | 0.999 |
# merge with tb.ra for full results table
full.res <- left_join(tb.ra, results.out, by = "description")
#write.csv(full.res, "output/picrust_path_data_results.csv", row.names = F)Misc.
library(brms)
# set up output object
results.out <- as.data.frame(matrix(ncol=7, nrow=length(DESCRIPTIONS)*3))
colnames(results.out) <- c("Description", "Parameter", "Est", "SE", "Q2.5", "Q97.5", "Rhat")
results.out$Description <- rep(DESCRIPTIONS, 3)
# for(i in 1:length(DESCRIPTIONS)){
for(i in 1:5){
dat0 <- mydata %>%
filter(description == DESCRIPTIONS[i])
fit0 <- brms::brm(
Abundance ~ 1 + tumor,
data= dat0,
family=hurdle_lognormal(link="identity"),
control = list(adapt_delta=0.99),
silent = T)
# the resulting "hu" parameter is the model for the proportion of zeros estimated
#summary(fit0)
#plot(fit0)
# save the posterior summaries (intercept, tumor, and hu)
fit.post <- round(posterior_summary(fit0),3)
results.out[results.out$Description == DESCRIPTIONS[i], 2] <- c("b_Intercept", "b_tumor", "hu")
results.out[results.out$Description == DESCRIPTIONS[i], 3:6] <- fit.post[c("b_Intercept", "b_tumor", "hu"),]
results.out[results.out$Description == DESCRIPTIONS[i], 7] <- rhat(fit0)[c("b_Intercept", "b_tumor", "hu")]
cat(".")
}
kable(results.out, format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="600px")Draft of a plots for these results
res.out2 <- left_join(results.out, tb.ra,by=c("Description"="description")) %>% pivot_longer(
cols = c("Tumor Mean", "Non-Tumor Mean"),
names_to = "Tumor", values_to = "RelAbundance"
)
# based on above
dat0 <- res.out2 %>%
mutate(estseR=abs(Est/SE))%>%
na.omit()%>%
filter(Parameter=="b_tumor", estseR >=0.25)
p <- ggplot(dat0, aes(y=Description, x=Est))+
geom_point()+
geom_segment(aes(y=Description, yend=Description, x=Q2.5, xend=Q97.5))
p
p2 <- ggplot(dat0, aes(x=RelAbundance, y=Description, color=Tumor))+
geom_point(position = "dodge")
p2Alternatively, we can model the relative abundance data. For this, transform the RA data to be 0-100 instead of 0-1.
# set up output object
results.out <- as.data.frame(matrix(ncol=7, nrow=length(DESCRIPTIONS)*3))
colnames(results.out) <- c("Description", "Parameter", "Est", "SE", "Q2.5", "Q97.5", "Rhat")
results.out$Description <- rep(DESCRIPTIONS, 3)
# for(i in 1:length(DESCRIPTIONS)){
for(i in 1:5){
dat0 <- mydata %>%
filter(description == DESCRIPTIONS[i])%>%
mutate(RelAbundance= RelAbundance*100)
fit0 <- brms::brm(
RelAbundance ~ 1 + tumor,
data= dat0,
family=hurdle_lognormal(link="identity"),
control = list(adapt_delta=0.99),
silent = T)
# the resulting "hu" parameter is the model for the proportion of zeros estimated
#summary(fit0)
#plot(fit0)
# save the posterior summaries (intercept, tumor, and hu)
fit.post <- round(posterior_summary(fit0),3)
results.out[results.out$Description == DESCRIPTIONS[i], 2] <- c("b_Intercept", "b_tumor", "hu")
results.out[results.out$Description == DESCRIPTIONS[i], 3:6] <- fit.post[c("b_Intercept", "b_tumor", "hu"),]
results.out[results.out$Description == DESCRIPTIONS[i], 7] <- rhat(fit0)[c("b_Intercept", "b_tumor", "hu")]
cat(".")
}
kable(results.out, format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="600px")Draft of a plots for these results
# based on above
p <- results.out %>%
na.omit()%>%
filter(Parameter=="b_tumor")%>%
ggplot(aes(y=Description, x=Est))+
geom_point()+
geom_segment(aes(y=Description, yend=Description, x=Q2.5, xend=Q97.5))
pFor these analyses, we need to use some kind of hurdle/zero-inflated model to account for the large proportion of zeros in SOME of the the descriptions. This creates a programming difficulty because only SOME really need this feature. In the above, I tested two types of approaches. First, we could model each descriptions separately (the quicker - time wise way). Secondly, we could jointly model the descriptions using a multilevel model (2-level) hurdle model. The latter is EXTREMELY computationally intensive.
# random effects model (may be too large)
# let's try on a small subset
dat0 <- mydata %>%
filter(description %in% DESCRIPTIONS[sample(1:length(DESCRIPTIONS),25)])
# priors
priors2 <-c(set_prior("lkj(2)", class = "cor"))
fit0 <- brms::brm(
Abundance ~ 1 + tumor + (1 + tumor | description),
prior = priors2,
data= dat0,
family=hurdle_lognormal(link="identity"),
#iter = 10000, warmup=2000,
save_all_pars = T,
control = list(adapt_delta=0.99))
# the resulting "hu" parameter is the model for the proportion of zeros estimated
summary(fit0)
plot(fit0)
sessionInfo()R version 4.0.5 (2021-03-31)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.1.1 dendextend_1.14.0 ggdendro_0.1.22 reshape2_1.4.4
[5] car_3.0-10 carData_3.0-4 gvlma_1.0.0.3 patchwork_1.1.1
[9] viridis_0.5.1 viridisLite_0.3.0 gridExtra_2.3 xtable_1.8-4
[13] kableExtra_1.3.4 MASS_7.3-53.1 data.table_1.14.0 readxl_1.3.1
[17] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.5 purrr_0.3.4
[21] readr_1.4.0 tidyr_1.1.3 tibble_3.1.0 ggplot2_3.3.3
[25] tidyverse_1.3.0 lmerTest_3.1-3 lme4_1.1-26 Matrix_1.3-2
[29] vegan_2.5-7 lattice_0.20-41 permute_0.9-5 phyloseq_1.34.0
[33] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] minqa_1.2.4 colorspace_2.0-0 rio_0.5.26
[4] ellipsis_0.3.1 rprojroot_2.0.2 XVector_0.30.0
[7] fs_1.5.0 rstudioapi_0.13 farver_2.1.0
[10] fansi_0.4.2 lubridate_1.7.10 xml2_1.3.2
[13] codetools_0.2-18 splines_4.0.5 knitr_1.31
[16] ade4_1.7-16 jsonlite_1.7.2 nloptr_1.2.2.2
[19] broom_0.7.5 cluster_2.1.1 dbplyr_2.1.0
[22] BiocManager_1.30.10 compiler_4.0.5 httr_1.4.2
[25] backports_1.2.1 assertthat_0.2.1 cli_2.3.1
[28] later_1.1.0.1 htmltools_0.5.1.1 prettyunits_1.1.1
[31] tools_4.0.5 igraph_1.2.6 gtable_0.3.0
[34] glue_1.4.2 Rcpp_1.0.6 Biobase_2.50.0
[37] cellranger_1.1.0 jquerylib_0.1.3 vctrs_0.3.6
[40] Biostrings_2.58.0 rhdf5filters_1.2.0 multtest_2.46.0
[43] svglite_2.0.0 ape_5.4-1 nlme_3.1-152
[46] iterators_1.0.13 xfun_0.21 ps_1.6.0
[49] openxlsx_4.2.3 rvest_1.0.0 lifecycle_1.0.0
[52] statmod_1.4.35 zlibbioc_1.36.0 scales_1.1.1
[55] hms_1.0.0 promises_1.2.0.1 parallel_4.0.5
[58] biomformat_1.18.0 rhdf5_2.34.0 curl_4.3
[61] yaml_2.2.1 sass_0.3.1 stringi_1.5.3
[64] highr_0.8 S4Vectors_0.28.1 foreach_1.5.1
[67] BiocGenerics_0.36.0 zip_2.1.1 boot_1.3-27
[70] systemfonts_1.0.1 rlang_0.4.10 pkgconfig_2.0.3
[73] evaluate_0.14 Rhdf5lib_1.12.1 labeling_0.4.2
[76] tidyselect_1.1.0 plyr_1.8.6 magrittr_2.0.1
[79] R6_2.5.0 IRanges_2.24.1 generics_0.1.0
[82] DBI_1.1.1 foreign_0.8-81 pillar_1.5.1
[85] haven_2.3.1 whisker_0.4 withr_2.4.1
[88] mgcv_1.8-34 abind_1.4-5 survival_3.2-10
[91] modelr_0.1.8 crayon_1.4.1 utf8_1.1.4
[94] rmarkdown_2.7 progress_1.2.2 grid_4.0.5
[97] git2r_0.28.0 webshot_0.5.2 reprex_1.0.0
[100] digest_0.6.27 httpuv_1.5.5 numDeriv_2016.8-1.1
[103] stats4_4.0.5 munsell_0.5.0 bslib_0.2.4