Sexual selection and sexual size dimorphism: a meta-analysis of comparative studies
Main analyses
Lennart Winkler1, Robert P
Freckleton2, Tamas Szekely34 & Tim
Janicke1,5
1Applied Zoology, Technical
University Dresden 2Department of Zoology, University of
Oxford, South Parks Road, Oxford OX1 3PS, UK
3Milner
Centre for Evolution, University of Bath, Bath,
UK84Department of Evolutionary Zoology and Human
Behaviour, University of Debrecen, Debrecen, Hungary
5Centre d’Écologie Fonctionnelle et Évolutive, UMR 5175,
CNRS, Université de Montpellier
Last updated: 2023-04-30
Checks: 7 0
Knit directory:
SSD_and_sexual_selection_2023/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230430) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1a5561c. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Untracked files:
Untracked: analysis/index2.Rmd
Untracked: data/Supplement4_SexSelSSD_V01.csv
Untracked: data/Supplement6_SexSelSSD_V01.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/index.Rmd) and HTML
(docs/index.html) files. If you’ve configured a remote Git
repository (see ?wflow_git_remote), click on the hyperlinks
in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1a5561c | LennartWinkler | 2023-04-30 | wflow_publish(all = T) |
| Rmd | 4482c88 | LennartWinkler | 2023-04-30 | Start workflowr project. |
Supplementary material reporting R code for the manuscript ‘Sexual selection and sexual size dimorphism: a meta-analysis of comparative studies’. Additional analyses excluding studies that did not control for phylogenetic non-independence (Supplement 2) can be found at:
# Load and prepare data Before we started the analyses, we loaded all necessary packages and data.
rm(list = ls()) # Clear work environment
# Load R-packages ####
list_of_packages=cbind('ape','matrixcalc','metafor','Matrix','MASS','pwr','psych','multcomp','data.table','ggplot2','RColorBrewer','MCMCglmm','ggdist','cowplot','PupillometryR','dplyr','wesanderson')
lapply(list_of_packages, require, character.only = TRUE)
# Load data set ####
MetaData <- read.csv("./data/Supplement4_SexSelSSD_V01.csv", sep=";", header=TRUE) # Load data set
N_Studies <- length(summary(as.factor(MetaData$Study_ID))) # Number of included primary studies
Tree<- read.tree("./data/Supplement6_SexSelSSD_V01.txt") # Load phylogenetic tree
# Prune phylogenetic tree
MetaData_Class_Data <- unique(MetaData$Class)
Tree_Class<-drop.tip(Tree, Tree$tip.label[-na.omit(match(MetaData_Class_Data, Tree$tip.label))])
forcedC_Moderators <- as.matrix(forceSymmetric(vcv(Tree_Class, corr=TRUE)))
# Order moderator levels
MetaData$SexSel_Mode=as.factor(MetaData$SexSel_Mode)
MetaData$SexSel_Mode=relevel(MetaData$SexSel_Mode,c("post-copulatory"))
MetaData$SexSel_Mode=relevel(MetaData$SexSel_Mode,c("pre-copulatory"))
MetaData$SexSel_Sex=as.factor(MetaData$SexSel_Sex)
MetaData$SexSel_Sex=relevel(MetaData$SexSel_Sex,c("Male"))
# Set figure theme and colors
theme=theme(panel.border = element_blank(),
panel.background = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.2,0.5),
legend.title = element_blank(),
legend.text = element_text(colour="black", size=12),
axis.line.x = element_line(colour = "black", size = 1),
axis.line.y = element_line(colour = "black", size = 1),
axis.text.x = element_text(face="plain", color="black", size=16, angle=0),
axis.text.y = element_text(face="plain", color="black", size=16, angle=0),
axis.title.x = element_text(size=16,face="plain", margin = margin(r=0,10,0,0)),
axis.title.y = element_text(size=16,face="plain", margin = margin(r=10,0,0,0)),
axis.ticks = element_line(size = 1),
axis.ticks.length = unit(.3, "cm"))
colpal=c("#4DAF4A","#377EB8","#E41A1C")
colpal2=brewer.pal(7, 'Dark2')
colpal4=c("grey50","grey65")
colpal4=wes_palette('FantasticFox1', 9, type = c("continuous"))
Meta_col=c('grey85','grey50','grey20','black')
# Global models ####
# Phylogenetic Model
Model_REML_Null = rma.mv(r ~ 1, V=Var_r, data = MetaData, random = c(~ 1 | Study_ID,~ 1 | Index, ~ 1 | Class), R = list(Class = forcedC_Moderators), method = "REML")
summary(Model_REML_Null)
# Non-phylogenetic Model
Model_cREML_Null = rma.mv(r ~ 1, V=Var_r, data = MetaData, random = c(~ 1 | Study_ID,~ 1 | Index), method = "REML")
summary(Model_cREML_Null)Global models
We began the analysis by running global models without additional moderators. First, we ran a global model including the phylogeny:
Model_REML_Null = rma.mv(r ~ 1, V=Var_r, data = MetaData, random = c(~ 1 | Study_ID,~ 1 | Index, ~ 1 | Class), R = list(Class = forcedC_Moderators), method = "REML")
summary(Model_REML_Null)
Multivariate Meta-Analysis Model (k = 85; method: REML)
logLik Deviance AIC BIC AICc
-23.9950 47.9900 55.9900 65.7133 56.4964
Variance Components:
estim sqrt nlvls fixed factor R
sigma^2.1 0.0252 0.1586 51 no Study_ID no
sigma^2.2 0.0617 0.2485 85 no Index no
sigma^2.3 0.0161 0.1269 9 no Class yes
Test for Heterogeneity:
Q(df = 84) = 1447.8890, p-val < .0001
Model Results:
estimate se zval pval ci.lb ci.ub
0.2898 0.0875 3.3109 0.0009 0.1182 0.4613 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Second, we ran a global model without the phylogeny:
Model_cREML_Null = rma.mv(r ~ 1, V=Var_r, data = MetaData, random = c(~ 1 | Study_ID,~ 1 | Index), method = "REML")
summary(Model_cREML_Null)
Multivariate Meta-Analysis Model (k = 85; method: REML)
logLik Deviance AIC BIC AICc
-24.9810 49.9619 55.9619 63.2544 56.2619
Variance Components:
estim sqrt nlvls fixed factor
sigma^2.1 0.0336 0.1834 51 no Study_ID
sigma^2.2 0.0622 0.2494 85 no Index
Test for Heterogeneity:
Q(df = 84) = 1447.8890, p-val < .0001
Model Results:
estimate se zval pval ci.lb ci.ub
0.2823 0.0410 6.8804 <.0001 0.2019 0.3627 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Moderator tests for phylogenetic models
Next, we ran a series of models that test the effect of different moderators. Again we started with models including the phylogeny.
Sexual selection mode
The first model explores the effect of the sexual selection mode (i.e. pre-copulatory, post-copulatory or both):
MetaData$SexSel_Mode=relevel(MetaData$SexSel_Mode,c("pre-copulatory"))
Model_REML_by_SexSelMode = rma.mv(r ~ factor(SexSel_Mode), V=Var_r, data = MetaData, random = c(~ 1 | Study_ID,~ 1 | Index, ~ 1 | Class), R = list(Class = forcedC_Moderators), method = "REML")
summary(Model_REML_by_SexSelMode)
Multivariate Meta-Analysis Model (k = 85; method: REML)
logLik Deviance AIC BIC AICc
-11.4578 22.9157 34.9157 49.3560 36.0357
Variance Components:
estim sqrt nlvls fixed factor R
sigma^2.1 0.0169 0.1300 51 no Study_ID no
sigma^2.2 0.0438 0.2093 85 no Index no
sigma^2.3 0.0164 0.1281 9 no Class yes
Test for Residual Heterogeneity:
QE(df = 82) = 1230.9203, p-val < .0001
Test of Moderators (coefficients 2:3):
QM(df = 2) = 30.1226, p-val < .0001
Model Results:
estimate se zval pval ci.lb
intrcpt 0.2671 0.0897 2.9790 0.0029 0.0914
factor(SexSel_Mode)post-copulatory -0.4664 0.1109 -4.2062 <.0001 -0.6837
factor(SexSel_Mode)both 0.1505 0.0654 2.3026 0.0213 0.0224
ci.ub
intrcpt 0.4429 **
factor(SexSel_Mode)post-copulatory -0.2491 ***
factor(SexSel_Mode)both 0.2786 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1We then re-leveled the model for post-hoc comparisons:
MetaData$SexSel_Mode=relevel(MetaData$SexSel_Mode,c("post-copulatory"))
Model_REML_by_SexSelMode2 = rma.mv(r ~ factor(SexSel_Mode), V=Var_r, data = MetaData, random = c(~ 1 | Study_ID,~ 1 | Index, ~ 1 | Class), R = list(Class = forcedC_Moderators), method = "REML")
summary(Model_REML_by_SexSelMode2)
MetaData$SexSel_Mode=relevel(MetaData$SexSel_Mode,c("both"))
Model_REML_by_SexSelMode3 = rma.mv(r ~ factor(SexSel_Mode), V=Var_r, data = MetaData, random = c(~ 1 | Study_ID,~ 1 | Index, ~ 1 | Class), R = list(Class = forcedC_Moderators), method = "REML")
summary(Model_REML_by_SexSelMode3)Finally, we computed FDR corrected p-values:
tab2=as.data.frame(round(p.adjust(c(0.0029, 0.1203, .0001), method = 'fdr'),digit=3),row.names=cbind("Pre-copulatory","Post-copulatory","Both"))
colnames(tab2)<-cbind('P-value')
tab2 P-value
Pre-copulatory 0.004
Post-copulatory 0.120
Both 0.000Plot sexual selection mode (Figure 2)
Here we plot the sexual selection mode moderator:
MetaData$SexSel_Mode=factor(MetaData$SexSel_Mode, levels = c("both","post-copulatory" ,"pre-copulatory"))
ggplot(MetaData, aes(x=SexSel_Mode, y=r, fill = SexSel_Mode, colour = SexSel_Mode)) +
geom_hline(yintercept=0, linetype="longdash", color = "black", linewidth=1)+
geom_flat_violin(position = position_nudge(x = 0.25, y = 0),adjust =1, trim = F,alpha=0.6)+
geom_point(position = position_jitter(width = .1), size = 2.5,alpha=0.6,stroke=0,shape=19)+
geom_point(inherit.aes = F,mapping = aes(y=Model_REML_by_SexSelMode$b[1,1], x=3.25), size = 3.5,alpha=1,stroke=0,shape=19,color='grey30')+
geom_point(inherit.aes = F,mapping = aes(y=Model_REML_by_SexSelMode2$b[1,1], x=2.25), size = 3.5,alpha=1,stroke=0,shape=19,color='grey30')+
geom_point(inherit.aes = F,mapping = aes(y=Model_REML_by_SexSelMode3$b[1,1], x=1.25), size = 3.5,alpha=1,stroke=0,shape=19,color='grey30')+
geom_segment(inherit.aes = F,mapping = aes(y=Model_REML_by_SexSelMode$ci.lb[1], x=3.25, xend= 3.25, yend= Model_REML_by_SexSelMode$ci.ub[1]), alpha=1,linewidth=1,color='grey30')+
geom_segment(inherit.aes = F,mapping = aes(y=Model_REML_by_SexSelMode2$ci.lb[1], x=2.25, xend= 2.25, yend= Model_REML_by_SexSelMode2$ci.ub[1]), alpha=1,linewidth=1,color='grey30')+
geom_segment(inherit.aes = F,mapping = aes(y=Model_REML_by_SexSelMode3$ci.lb[1], x=1.25, xend= 1.25, yend= Model_REML_by_SexSelMode3$ci.ub[1]), alpha=1,linewidth=1,color='grey30')+
ylab(expression(paste("Effect size (", italic("r"),')')))+xlab('Sexual selection mode')+coord_flip()+guides(fill = FALSE, colour = FALSE) +
scale_color_manual(values =colpal)+
scale_fill_manual(values =colpal)+
scale_x_discrete(labels=c("Both","Post-copulatory" ,"Pre-copulatory"),expand=c(.1,0))+
annotate("text", x=1, y=1.2, label= "n = 37",size=4.5) +
annotate("text", x=2, y=1.2, label= "n = 9",size=4.5) +
annotate("text", x=3, y=1.2, label= "n = 39",size=4.5) + theme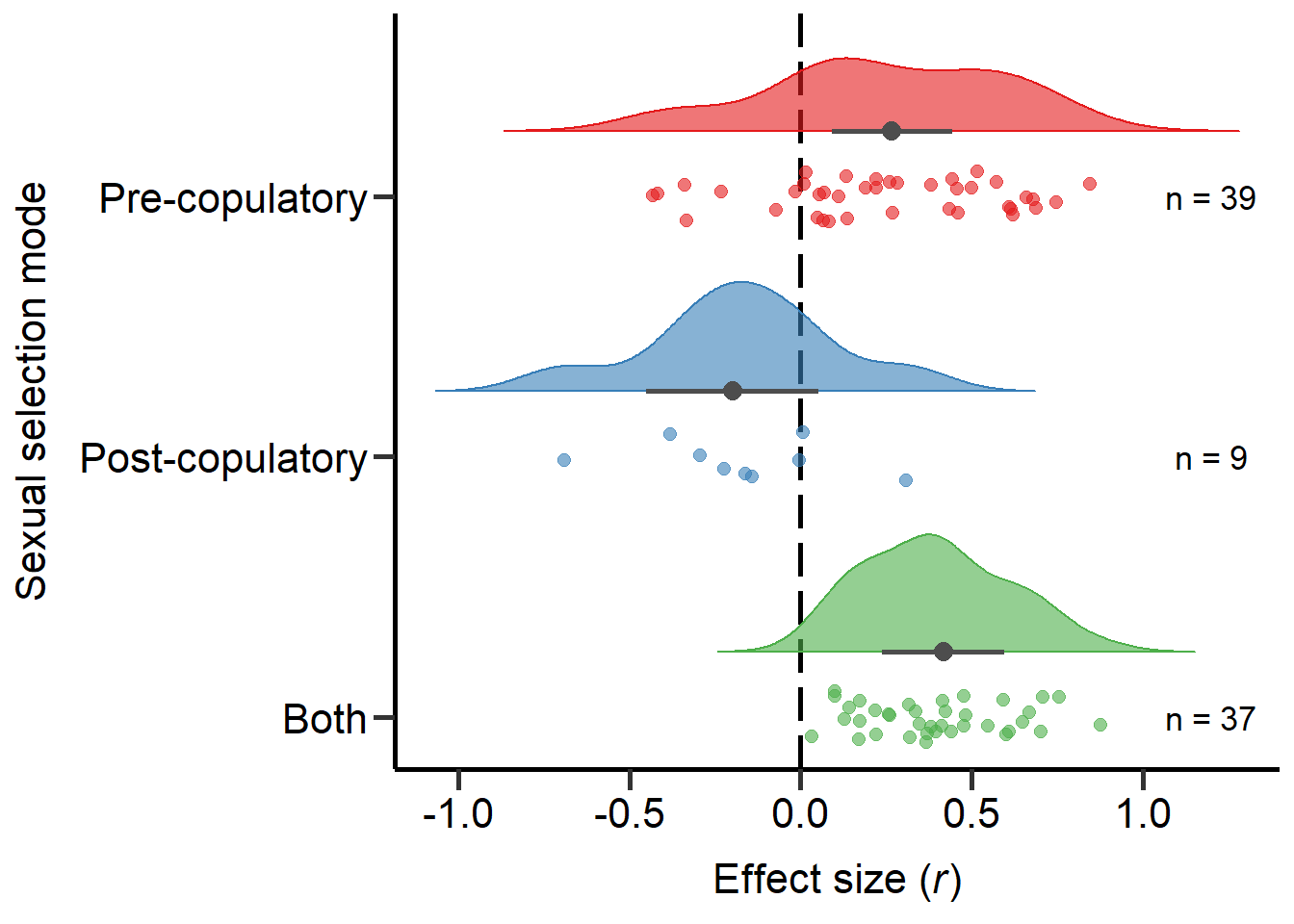
Figure 2: Raincloud plot of correlation coefficients between SSD and the modes of sexual selection proxies (i.e. pre-copulatory, post-copulatory or both) including sample sizes and estimates with 95%CI from phylogenetic model.
sessionInfo()R version 4.2.0 (2022-04-22 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=German_Germany.utf8 LC_CTYPE=German_Germany.utf8
[3] LC_MONETARY=German_Germany.utf8 LC_NUMERIC=C
[5] LC_TIME=German_Germany.utf8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] wesanderson_0.3.6 PupillometryR_0.0.4 rlang_1.0.6
[4] dplyr_1.1.0 cowplot_1.1.1 ggdist_3.2.1
[7] MCMCglmm_2.34 coda_0.19-4 RColorBrewer_1.1-3
[10] ggplot2_3.4.1 data.table_1.14.8 multcomp_1.4-23
[13] TH.data_1.1-1 survival_3.3-1 mvtnorm_1.1-3
[16] psych_2.2.9 pwr_1.3-0 MASS_7.3-56
[19] metafor_3.8-1 metadat_1.2-0 Matrix_1.5-3
[22] matrixcalc_1.0-6 ape_5.7-1 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] httr_1.4.5 sass_0.4.5 jsonlite_1.8.4
[4] splines_4.2.0 bslib_0.4.2 getPass_0.2-2
[7] distributional_0.3.1 highr_0.10 tensorA_0.36.2
[10] yaml_2.3.7 pillar_1.8.1 lattice_0.20-45
[13] glue_1.6.2 digest_0.6.31 promises_1.2.0.1
[16] colorspace_2.1-0 sandwich_3.0-2 htmltools_0.5.4
[19] httpuv_1.6.9 pkgconfig_2.0.3 corpcor_1.6.10
[22] scales_1.2.1 processx_3.8.0 whisker_0.4.1
[25] later_1.3.0 cubature_2.0.4.6 git2r_0.31.0
[28] tibble_3.2.0 generics_0.1.3 farver_2.1.1
[31] cachem_1.0.7 withr_2.5.0 cli_3.6.1
[34] mnormt_2.1.1 magrittr_2.0.3 evaluate_0.20
[37] ps_1.7.2 fs_1.6.1 fansi_1.0.4
[40] nlme_3.1-157 tools_4.2.0 lifecycle_1.0.3
[43] stringr_1.5.0 munsell_0.5.0 callr_3.7.3
[46] compiler_4.2.0 jquerylib_0.1.4 grid_4.2.0
[49] rstudioapi_0.14 labeling_0.4.2 rmarkdown_2.20
[52] gtable_0.3.1 codetools_0.2-18 R6_2.5.1
[55] zoo_1.8-11 knitr_1.42 fastmap_1.1.1
[58] utf8_1.2.3 mathjaxr_1.6-0 rprojroot_2.0.3
[61] stringi_1.7.12 parallel_4.2.0 Rcpp_1.0.10
[64] vctrs_0.5.2 tidyselect_1.2.0 xfun_0.37