The Methylome in Female Adolescent Conduct Disorder: Neural Pathomechanisms and Environmental Risk Factors
Post-hoc Analyses
AG Chiocchetti
29 Dezember 2020
Last updated: 2021-09-17
Checks: 7 0
Knit directory: femNATCD_MethSeq/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210128) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2047ac2. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.Rhistory
Ignored: code/.Rhistory
Ignored: data/Epicounts.csv
Ignored: data/Epimeta.csv
Ignored: data/Epitpm.csv
Ignored: data/KangUnivers.txt
Ignored: data/Kang_DataPreprocessing.RData
Ignored: data/Kang_dataset_genesMod_version2.txt
Ignored: data/PatMeta.csv
Ignored: data/ProcessedData.RData
Ignored: data/RTrawdata/
Ignored: data/SNPCommonFilt.csv
Ignored: data/femNAT_PC20.txt
Ignored: output/BrainMod_Enrichemnt.pdf
Ignored: output/Brain_Module_Heatmap.pdf
Ignored: output/DMR_Results.csv
Ignored: output/GOres.xlsx
Ignored: output/LME_GOplot.pdf
Ignored: output/LME_Results.csv
Ignored: output/LME_Results_Sig.csv
Ignored: output/LME_tophit.svg
Ignored: output/ProcessedData.RData
Ignored: output/RNAvsMETplots.pdf
Ignored: output/Regions_GOplot.pdf
Ignored: output/ResultsgroupComp.txt
Ignored: output/SEM_summary_groupEpi_M15.txt
Ignored: output/SEM_summary_groupEpi_M2.txt
Ignored: output/SEM_summary_groupEpi_M_all.txt
Ignored: output/SEM_summary_groupEpi_TopHit.txt
Ignored: output/SEM_summary_groupEpi_all.txt
Ignored: output/SEMplot_Epi_M15.html
Ignored: output/SEMplot_Epi_M15.png
Ignored: output/SEMplot_Epi_M15_files/
Ignored: output/SEMplot_Epi_M2.html
Ignored: output/SEMplot_Epi_M2.png
Ignored: output/SEMplot_Epi_M2_files/
Ignored: output/SEMplot_Epi_M_all.html
Ignored: output/SEMplot_Epi_M_all.png
Ignored: output/SEMplot_Epi_M_all_files/
Ignored: output/SEMplot_Epi_TopHit.html
Ignored: output/SEMplot_Epi_TopHit.png
Ignored: output/SEMplot_Epi_TopHit_files/
Ignored: output/SEMplot_Epi_all.html
Ignored: output/SEMplot_Epi_all.png
Ignored: output/SEMplot_Epi_all_files/
Ignored: output/barplots.pdf
Ignored: output/circos_DMR_tags.svg
Ignored: output/circos_LME_tags.svg
Ignored: output/clinFact.RData
Ignored: output/dds_filt_analyzed.RData
Ignored: output/designh0.RData
Ignored: output/designh1.RData
Ignored: output/envFact.RData
Ignored: output/functional_Enrichemnt.pdf
Ignored: output/gostres.pdf
Ignored: output/modelFact.RData
Ignored: output/resdmr.RData
Ignored: output/resultsdmr_table.RData
Ignored: output/table1_filtered.Rmd
Ignored: output/table1_filtered.docx
Ignored: output/table1_unfiltered.Rmd
Ignored: output/table1_unfiltered.docx
Ignored: setup_built.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/03_01_Posthoc_analyses_Gene_Enrichment.Rmd) and HTML (docs/03_01_Posthoc_analyses_Gene_Enrichment.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2047ac2 | achiocch | 2021-09-17 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| Rmd | fd80c2d | achiocch | 2021-09-16 | adds SEM improvments |
| html | fd80c2d | achiocch | 2021-09-16 | adds SEM improvments |
| html | ccbc9e4 | achiocch | 2021-08-10 | Build site. |
| Rmd | 723a1aa | achiocch | 2021-08-09 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | 70cd649 | achiocch | 2021-08-06 | Build site. |
| Rmd | 611ca24 | achiocch | 2021-08-06 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | 2a53a87 | achiocch | 2021-08-06 | Build site. |
| Rmd | e4425b8 | achiocch | 2021-08-06 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | e3a9ae3 | achiocch | 2021-08-04 | Build site. |
| Rmd | 3979990 | achiocch | 2021-08-04 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | f6bbdc0 | achiocch | 2021-08-04 | Build site. |
| Rmd | 1a30b73 | achiocch | 2021-08-04 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | 1a30b73 | achiocch | 2021-08-04 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | 710904a | achiocch | 2021-08-03 | Build site. |
| Rmd | 16112c3 | achiocch | 2021-08-03 | wflow_publish(c(“analysis/", "code/”, “docs/”)) |
| html | 16112c3 | achiocch | 2021-08-03 | wflow_publish(c(“analysis/", "code/”, “docs/”)) |
| html | cde8384 | achiocch | 2021-08-03 | Build site. |
| Rmd | d3629d5 | achiocch | 2021-08-03 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | d3629d5 | achiocch | 2021-08-03 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | d761be4 | achiocch | 2021-07-31 | Build site. |
| Rmd | b452d2f | achiocch | 2021-07-30 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | b452d2f | achiocch | 2021-07-30 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| Rmd | 1a9f36f | achiocch | 2021-07-30 | reviewed analysis |
| html | 2734c4e | achiocch | 2021-05-08 | Build site. |
| html | a847823 | achiocch | 2021-05-08 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | 9cc52f7 | achiocch | 2021-05-08 | Build site. |
| html | 158d0b4 | achiocch | 2021-05-08 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | 0f262d1 | achiocch | 2021-05-07 | Build site. |
| html | 5167b90 | achiocch | 2021-05-07 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | 05aac7f | achiocch | 2021-04-23 | Build site. |
| Rmd | 5f070a5 | achiocch | 2021-04-23 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | f5c5265 | achiocch | 2021-04-19 | Build site. |
| Rmd | dc9e069 | achiocch | 2021-04-19 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | dc9e069 | achiocch | 2021-04-19 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | 17f1eec | achiocch | 2021-04-10 | Build site. |
| Rmd | 4dee231 | achiocch | 2021-04-10 | wflow_publish(c(“analysis/", "code/”, "docs/*")) |
| html | 91de221 | achiocch | 2021-04-05 | Build site. |
| Rmd | b6c6b33 | achiocch | 2021-04-05 | updated GO function, and model def |
| html | 4ea1bba | achiocch | 2021-02-25 | Build site. |
| Rmd | 6c21638 | achiocch | 2021-02-25 | wflow_publish(c(“analysis/", "code/”, "docs/*"), update = F) |
| html | 6c21638 | achiocch | 2021-02-25 | wflow_publish(c(“analysis/", "code/”, "docs/*"), update = F) |
Home = getwd()Sensitivity Analyses
Sensitivity EWAS
including SES
collector=data.frame(originalP=results_Deseq$pvalue,
originall2FC=results_Deseq$log2FoldChange)
rownames(collector)=paste0("Epi", 1:nrow(collector))
parm="EduPar"
workingcopy = dds_filt
workingcopy=workingcopy[,as.vector(!is.na(colData(dds_filt)[parm]))]
modelpar=as.character(design(dds_filt))[2]
tmpmod=gsub("0", paste0("~ 0 +",parm), modelpar)
tmpmod=gsub("int_dis \\+", "", tmpmod)
modelpar=as.formula(tmpmod)
design(workingcopy) = modelpar
workingcopy = DESeq(workingcopy, parallel = T)
parmres=results(workingcopy)
collector[,paste0(parm,"P")] = parmres$pvalue
collector[,paste0(parm,"l2FC")] = parmres$log2FoldChange
idx=collector$originalP<=thresholdp
idx=collector[,paste0(parm,"P")]<=thresholdp
table(collector$originalP<=thresholdp, collector[paste0(parm,"P")]<=thresholdp) %>% fisher.test()
cor.test(collector$originall2FC[idx],collector[,paste0(parm,"l2FC")][idx],
method = "spearman")
qqplot(y=-log10(collector[,paste0(parm,"P")]),
x = -log(runif(nrow(collector))), xlim=c(0,12),ylim=c(0,12),
col="gray", ylab="", xlab="")
par(new=T)
qqplot(y=-log10(collector$originalP),
x = -log(runif(nrow(collector))),xlim=c(0,12),ylim=c(0,12),
xlab="expected",ylab="observed")
abline(0,1,col="red")
legend("topleft", pch=1, col=c("black", "gray"), legend=c("original", parm))
plot(collector$originall2FC[idx],collector[,paste0(parm,"l2FC")][idx], pch=16,
main="log 2 foldchange", ylab=parm, xlab="original")excluding int_dis
### excluding int_dist
modelpar=as.character(design(dds_filt))[2]
modelpar=as.formula(paste("~",gsub("int_dis +", "", modelpar)))
design(workingcopy) = modelpar
workingcopy = DESeq(workingcopy, parallel=T)
parmres=results(workingcopy)
parm="wo.int.dis"
collector[,paste0(parm,"P")] = parmres$pvalue
collector[,paste0(parm,"l2FC")] = parmres$log2FoldChange
idx=collector$originalP<=thresholdp
idx=collector[,paste0(parm,"P")]<=thresholdp
table(collector$originalP<=thresholdp, collector[paste0(parm,"P")]<=thresholdp) %>% fisher.test()
cor.test(collector$originall2FC[idx],collector[,paste0(parm,"l2FC")][idx],
method = "spearman")
qqplot(y=-log10(collector[,paste0(parm,"P")]),
x = -log(runif(nrow(collector))), xlim=c(0,12),ylim=c(0,12),
col="gray", ylab="", xlab="")
par(new=T)
qqplot(y=-log10(collector$originalP),
x = -log(runif(nrow(collector))),xlim=c(0,12),ylim=c(0,12),
xlab="expected",ylab="observed")
abline(0,1,col="red")
legend("topleft", pch=1, col=c("black", "gray"), legend=c("original", parm))
plot(collector$originall2FC[idx],collector[,paste0(parm,"l2FC")][idx], pch=16,
main="log 2 foldchange", ylab=parm, xlab="original")Sensitivity main hit
For the most significant tag of interest (5’ of the SLITRK5 gene), we tested if the group effect is stable if correcting for Ethnicity (PC1-PC4) or CD associated environmental risk factors.
tophit=which.min(results_Deseq$padj)
methdata=log2_cpm[tophit,]
Probdat=as.data.frame(colData(dds_filt))
Probdat$topHit=methdata[rownames(Probdat)]
model0=as.character(design(dds_filt))[2]
model0=as.formula(gsub("0 +", "topHit ~ 0 + ", model0))
lmres=lm(model0, data=Probdat)
lmrescoeff = as.data.frame(coefficients(summary(lmres)))
totestpar=c("site","PC_1", "PC_2", "PC_3", "PC_4", envFact)
ressens=data.frame(matrix(nrow = length(totestpar)+1, ncol=c(3)))
colnames(ressens) = c("beta", "se", "p.value")
rownames(ressens) = c("original", totestpar)
ressens["original",] = lmrescoeff["groupCD", c("Estimate", "Std. Error", "Pr(>|t|)")]
for( parm in totestpar){
modelpar=as.character(design(dds_filt))[2]
modelpar=as.formula(gsub("0", paste0("topHit ~ 0 +",parm), modelpar))
lmres=lm(modelpar, data=Probdat)
lmrescoeff = as.data.frame(coefficients(summary(lmres)))
ressens[parm,] = lmrescoeff["groupCD", c("Estimate", "Std. Error", "Pr(>|t|)")]
}
modelpar=as.character(design(dds_filt))[2]
modelpar=as.formula(gsub("int_dis +", "", gsub("0", "topHit ~ 0", modelpar)))
lmres=lm(modelpar, data=Probdat)
lmrescoeff = as.data.frame(coefficients(summary(lmres)))
ressens["w/o_int_dis",] = lmrescoeff["groupCD", c("Estimate", "Std. Error", "Pr(>|t|)")]
a = barplot(height = ressens$beta,
ylim=rev(range(c(0,ressens$beta-ressens$se)))*1.3,
names.arg = rownames(ressens), col=Set3, border = NA, las=3,
ylab="beta[se]", main="Effect sensitvity analysis")
arrows(a,ressens$beta, a, ressens$beta+ressens$se, angle = 90, length = 0.1)
arrows(a,ressens$beta, a, ressens$beta-ressens$se, angle = 90, length = 0.1)
text(a, min(ressens$beta-ressens$se)*1.15,
formatC(ressens$p.value), cex=0.6, srt=90)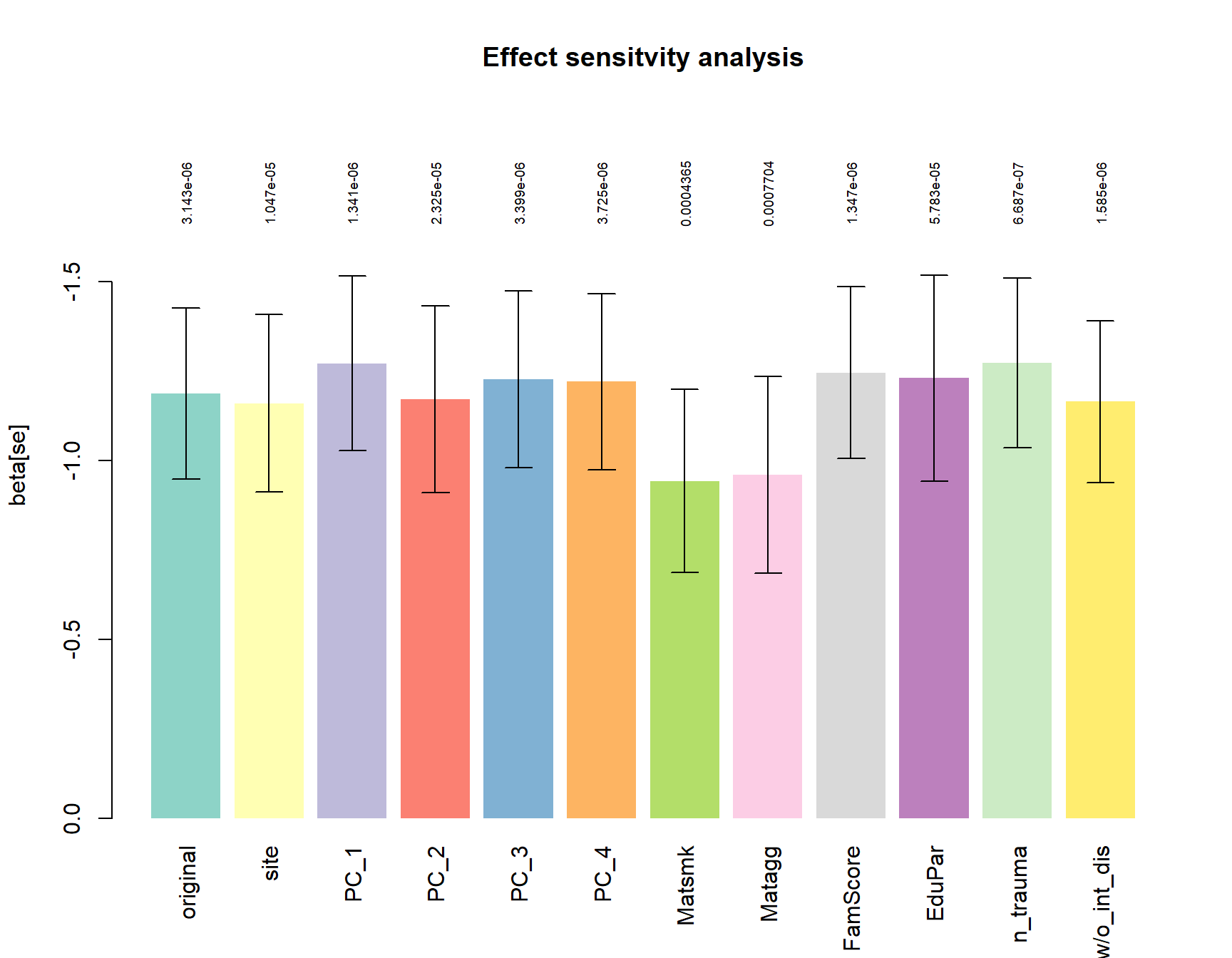
All models are corrected for:
site, Age, Pubstat, int_dis, medication, contraceptives, cigday_1,
site is included as random effect.
original: model defined as 0 + +Age + int_dis + medication + contraceptives + cigday_1 + V8 + group
all other models represent the original model + the variable of interest
Real-time PCR validation
Data loading and parsing
RefGenes = c("GUSB")
Targets_of_Int = c("SLITRK5", "MIR4500HG")
nreplicates = 3
flagscore=Inf #replication quality error
SamplesMeta=read_xlsx(paste0(Home,"/data/RTrawdata/ZelllinienRNA_femNAT.xlsx"))
as.data.frame(SamplesMeta) -> SamplesMeta
SamplesMeta$Pou=paste("POU", SamplesMeta$Pou)
rownames(SamplesMeta)=SamplesMeta$Pou
SamplesMeta$Group = dds_filt$group[match(SamplesMeta$femNATID, dds_filt$ID_femNAT)]
Files=list.files(paste0(Home,"/data/RTrawdata/"), full.names = T)
Files=Files[grepl("_data",Files)]
Sets=unique(substr(basename(Files), 1,8))
Targets_all=vector()
Samples_all=vector()
geoMean=function(x){
x=x[!is.na(x)]
if(length(x)==0)
return(NA)
else
return((prod(x))^(1/length(x)))}
for (Set in Sets){
Setfiles=Files[grep(Set, Files)]
for( i in 1:length(Setfiles)){
tmp=read.table(Setfiles[i], skip=8, header=T, sep="\t", comment.char = "", fill=T)[1:96,]
tmp=tmp[,c("Sample.Name", "Target.Name","CÑ.")]
colnames(tmp)=c("Sample.Name", "Target.Name", "CT")
tmp$Target.Name=gsub("SLITRK5_L", "SLITRK5_", tmp$Target.Name)
tmp$Target.Name=gsub("VD_", "", tmp$Target.Name)
tmp$Target.Name=gsub("_", "", tmp$Target.Name)
tmp$Target.Name=substr(tmp$Target.Name,1, regexpr("#", tmp$Target.Name)-1)
tmp$CT=as.numeric(tmp$CT)
# set bad replicates to NA
tmpmu = tapply(tmp$CT, paste0(tmp$Sample.Name,"_",tmp$Target.Name), mean, na.rm=T)
tmpsd = tapply(tmp$CT, paste0(tmp$Sample.Name,"_",tmp$Target.Name), sd, na.rm=T)
for (corr in which(tmpsd>flagscore)){
index=unlist(strsplit(names(tmpmu)[corr], "_"))
tmp[which(tmp$Sample.Name==index[1] & tmp$Target.Name==index[2]),"CT"] = NA
}
assign(paste0("tmp_",Set,"_",i),tmp)
}
tmp=do.call("rbind", mget(apropos(paste0("tmp_",Set))))
tmp=tmp[which(!(tmp$Sample.Name==""|is.na(tmp$Sample.Name))), ]
tmp=tmp[!tmp$Sample.Name=="NTC",]
Samples=unique(tmp$Sample.Name)
Targets=unique(tmp$Target.Name)
Samples_all=unique(c(Samples_all, Samples))
Targets_all=unique(c(Targets_all, Targets))
Reform=data.frame(matrix(NA, nrow=length(Samples), ncol=length(Targets)*nreplicates))
colnames(Reform)=paste0(rep(Targets, each=3), letters[1:nreplicates])
rownames(Reform)=Samples
for (i in Samples) {
#print(i)
for (j in Targets){
Reform[i,grep(j, colnames(Reform))]=tmp[tmp$Sample.Name==i & tmp$Target.Name==j,"CT"]
}
}
HK=colnames(Reform)[grep(paste0(RefGenes, collapse="|"),colnames(Reform))]
GMHK=apply(Reform[,HK], 1, geoMean)
tmp2=Reform-GMHK
assign(paste0(Set,"_dCT"), tmp2)
rm(list=c(apropos("tmp"), "Reform", "GMHK"))
}Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugtWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'get_java_tmp_dir' nicht gefundenWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'set_java_tmp_dir' nicht gefundenWarning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugtWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'get_java_tmp_dir' nicht gefundenWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'set_java_tmp_dir' nicht gefundenWarning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugtWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'get_java_tmp_dir' nicht gefundenWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'set_java_tmp_dir' nicht gefundenWarning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugtWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'get_java_tmp_dir' nicht gefundenWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'set_java_tmp_dir' nicht gefundenWarning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugtWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'get_java_tmp_dir' nicht gefundenWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'set_java_tmp_dir' nicht gefundenWarning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugtWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'get_java_tmp_dir' nicht gefundenWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'set_java_tmp_dir' nicht gefundenWarning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugtWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'get_java_tmp_dir' nicht gefundenWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'set_java_tmp_dir' nicht gefundenWarning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugt
Warning: NAs durch Umwandlung erzeugtWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'get_java_tmp_dir' nicht gefundenWarning in rm(list = c(apropos("tmp"), "Reform", "GMHK")): Objekt
'set_java_tmp_dir' nicht gefundenSamples_all=unique(Samples_all)
Targets_all = unique(Targets_all)
mergedCTtable=data.frame(matrix(NA,ncol=length(Targets_all)*nreplicates, nrow=length(Samples_all)))
colnames(mergedCTtable)=paste0(rep(unique(Targets_all), each=nreplicates), letters[1:nreplicates])
rownames(mergedCTtable)=Samples_all
CTobj=apropos("_dCT")
for( obj in CTobj){
DF=get(obj)
for(k in colnames(DF)){
for(l in rownames(DF)){
mergedCTtable[l,k]=DF[l,k]
}
}
}
CTmeans=colMeans(mergedCTtable, na.rm = T)
meanvec=tapply(CTmeans,gsub(paste0(letters[1:nreplicates],collapse="|"),"",names(CTmeans)), mean, na.rm=T)
meanvec = rep(meanvec, each=nreplicates)
names(meanvec) = paste0(names(meanvec), letters[1:nreplicates])
meanvec=meanvec[colnames(mergedCTtable)]
ddCT=apply(mergedCTtable,1, function(x){x-meanvec})
FC=2^-ddCT
SamplesMeta$inset=F
SamplesMeta$inset[SamplesMeta$Pou %in% colnames(FC)]=T
SamplesMeta=SamplesMeta[SamplesMeta$inset,]
CTRLCASEsorter=c(which(SamplesMeta$Group=="CTRL"),which(SamplesMeta$Group=="CD"))
SamplesMeta = SamplesMeta[CTRLCASEsorter, ]
searcher=paste0(Targets_of_Int, collapse = "|")
FC = FC[grepl(searcher, rownames(FC)),SamplesMeta$Pou]
MuFC=apply(FC, 2, function(x){tapply(log2(x), gsub("a|b|c","",rownames(FC)), mean, na.rm=T)})
SDFC=apply(FC, 2, function(x){tapply(log2(x), gsub("a|b|c","",rownames(FC)), sd, na.rm=T)})plot relative expression by samples
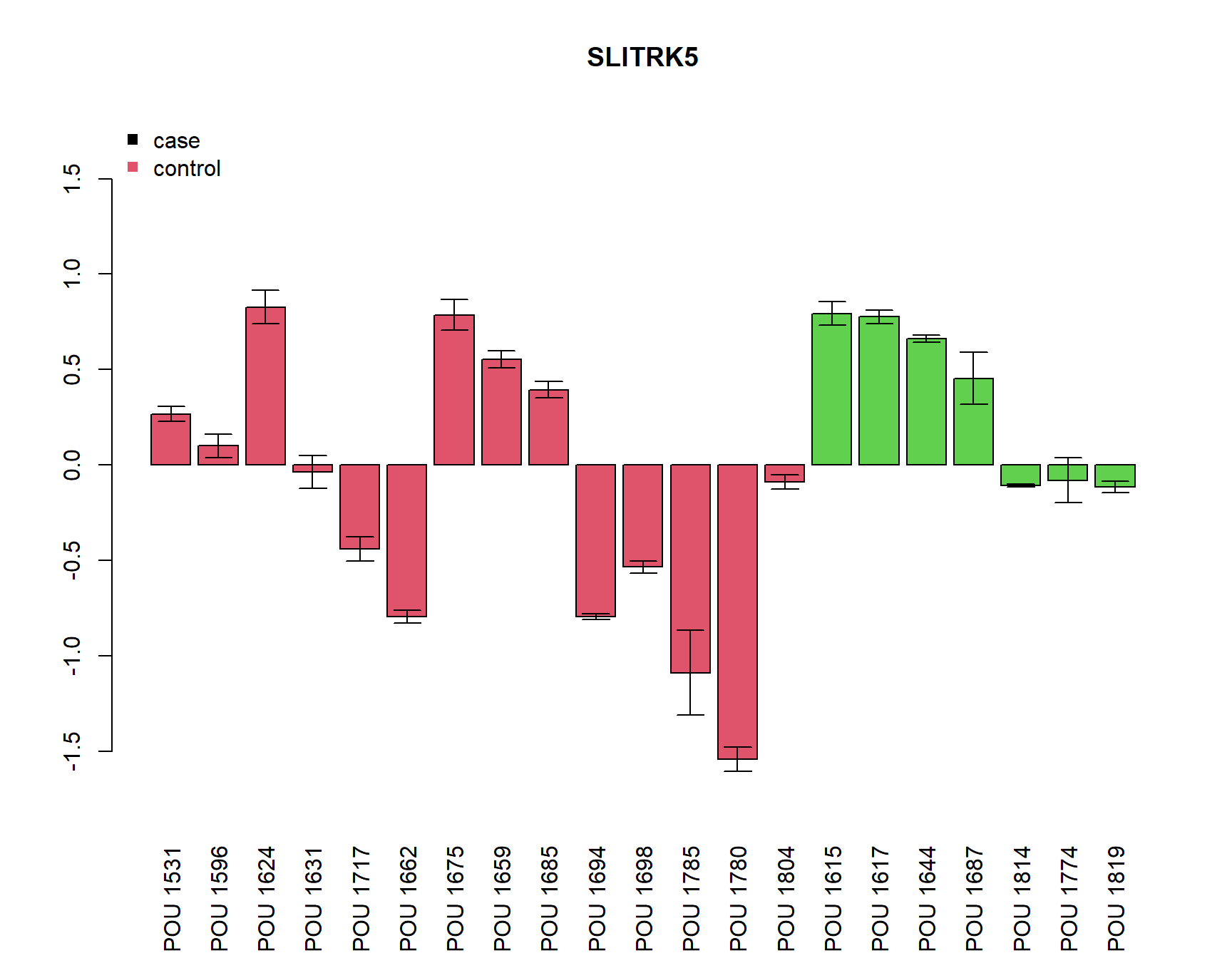
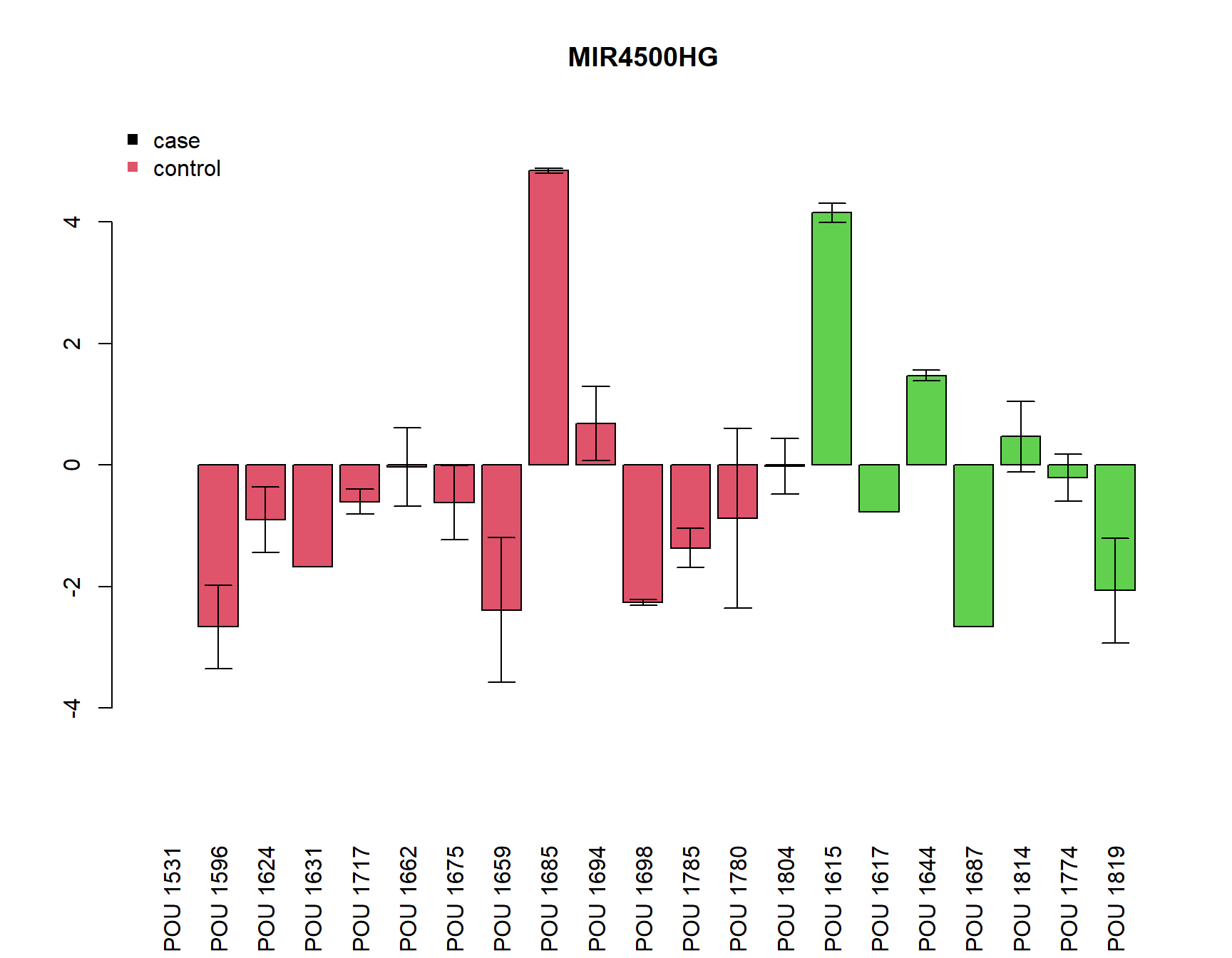
pdf(paste0(Home, "/output/barplots.pdf"))
for(i in Targets_of_Int){
if(any(!is.na(MuFC[i,]))){
a=barplot(unlist(MuFC[i,]), col=as.numeric(SamplesMeta[colnames(MuFC),"Group"])+1, main=i, las=3,
ylim=c(-max(abs(MuFC[i,])*1.2, na.rm=T), (max(abs(MuFC[i,])*1.2, na.rm=T))))
arrows(a, MuFC[i,], a, MuFC[i,]+SDFC[i,], angle = 90, length = 0.1)
arrows(a, MuFC[i,], a, MuFC[i,]-SDFC[i,], angle = 90, length = 0.1)
legend("topleft", c("case", "control"), col=c(1,2), pch=15, bty="n")
} else {
plot(0,0, type="n", main=paste(i, "not detected"))
}
}
dev.off()png
2 for(i in Targets_of_Int){
if(any(!is.na(MuFC[i,]))){
a=barplot(unlist(MuFC[i,]), col=as.numeric(SamplesMeta[colnames(MuFC),"Group"])+1, main=i, las=3,
ylim=c(-max(abs(MuFC[i,])*1.2, na.rm=T), (max(abs(MuFC[i,])*1.2, na.rm=T))))
arrows(a, MuFC[i,], a, MuFC[i,]+SDFC[i,], angle = 90, length = 0.1)
arrows(a, MuFC[i,], a, MuFC[i,]-SDFC[i,], angle = 90, length = 0.1)
legend("topleft", c("case", "control"), col=c(1,2), pch=15, bty="n")
} else {
plot(0,0, type="n", main=paste(i, "not detected"))
}
}
compare across groups
sink(paste0(Home, "/output/ResultsgroupComp.txt"))
Group=SamplesMeta$Group
for(i in Targets_of_Int){
print(i)
print(summary(try(lm(unlist(MuFC[i,])~Group))))
print(t.test(unlist(MuFC[i,])~Group))
}[1] "SLITRK5"
Call:
lm(formula = unlist(MuFC[i, ]) ~ Group)
Residuals:
Min 1Q Median 3Q Max
-1.3710 -0.4489 0.1142 0.4382 0.9983
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.1703 0.1718 -0.991 0.334
GroupCD 0.5109 0.2975 1.717 0.102
Residual standard error: 0.6427 on 19 degrees of freedom
Multiple R-squared: 0.1343, Adjusted R-squared: 0.08878
F-statistic: 2.949 on 1 and 19 DF, p-value: 0.1022
Welch Two Sample t-test
data: unlist(MuFC[i, ]) by Group
t = -2.0316, df = 18.197, p-value = 0.05706
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-1.03871726 0.01701686
sample estimates:
mean in group CTRL mean in group CD
-0.1702834 0.3405668
[1] "MIR4500HG"
Call:
lm(formula = unlist(MuFC[i, ]) ~ Group)
Residuals:
Min 1Q Median 3Q Max
-2.7172 -1.2116 -0.2660 0.5749 5.4474
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.6053 0.5683 -1.065 0.301
GroupCD 0.6614 0.9607 0.688 0.500
Residual standard error: 2.049 on 18 degrees of freedom
(1 observation deleted due to missingness)
Multiple R-squared: 0.02566, Adjusted R-squared: -0.02847
F-statistic: 0.474 on 1 and 18 DF, p-value: 0.4999
Welch Two Sample t-test
data: unlist(MuFC[i, ]) by Group
t = -0.65047, df = 10.602, p-value = 0.5292
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-2.909646 1.586853
sample estimates:
mean in group CTRL mean in group CD
-0.60525003 0.05614653 sink()
SamplesMeta$femNATID2=paste0("ID_",gsub("-","_",SamplesMeta$femNATID))
SamplesMeta=SamplesMeta[SamplesMeta$Pou %in% colnames(MuFC),]
MuFC=MuFC[,SamplesMeta$Pou]
TPM4RNA=selEpitpm[,SamplesMeta$femNATID2]
colnames(TPM4RNA)=SamplesMeta$Pou
tags=list()
Targets=Targets_of_Int
sigtags=which(restab$padj<=0.05)
tagsOI=grep(paste0(Targets, collapse = "|"),selEpiMeta$gene)
sigtagsOI = tagsOI[tagsOI %in% sigtags]
fintagsOI=data.frame(tags=sigtagsOI, gene=selEpiMeta[sigtagsOI,"gene"])
#Targ=Targets[1]
#tag=tags[1]compare against methylation level
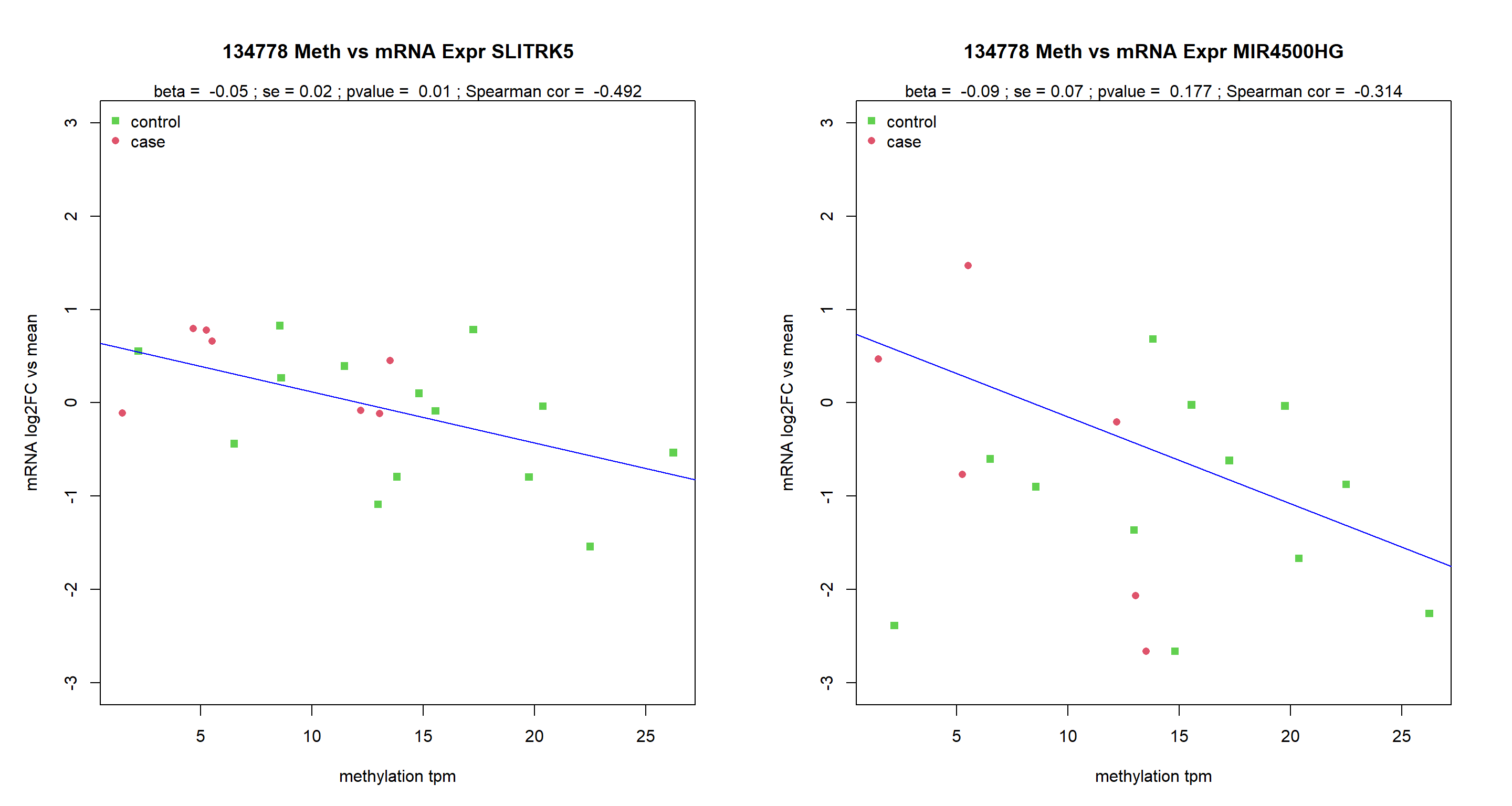
pdf(paste0(Home,"/output/RNAvsMETplots.pdf"), width = 15, height = 8)
MuFC=as.data.frame(MuFC)
MuFCsel=MuFC[Targets,]
par(mar=c(5,5,5,3), mfrow=c(1,2))
for (Targ in Targets){
tags=fintagsOI$tags[grep(Targ, fintagsOI$gene)]
for (tag in tags){
data=data.frame(tpm=unlist(TPM4RNA[tag,SamplesMeta$Pou]) ,
RT=unlist(MuFCsel[grep(Targ, rownames(MuFCsel)),SamplesMeta$Pou]))
plot(data$tpm,data$RT,
xlab="methylation tpm",
ylab = "mRNA log2FC vs mean",
ylim=c(-3,3),
col=4-as.numeric(SamplesMeta$Group),
pch=as.numeric(SamplesMeta$Group)+14,
main=paste(tag, "Meth vs mRNA Expr", Targ))
legend("topleft", c("control", "case"), pch=c(15,16), col=c(3,2), bty="n")
a=lm(RT~tpm, data)
b=summary(a)
abline(a, col="blue")
SperCor=cor(data$RT,data$tpm,use = "c", method = "spearman")
mtext(3, text = paste("beta = ", round(coefficients(a)[2],2),
"; se =", round(b$coefficients[2,2],2),
"; pvalue = ", round(b$coefficients[2,4],3),
"; sperman cor = ", round(SperCor,3)))
}
}
dev.off()png
2 MuFC=as.data.frame(MuFC)
MuFCsel=MuFC[Targets,]
par(mar=c(5,5,5,3), mfrow=c(1,2))
for (Targ in Targets){
tags=fintagsOI$tags[grep(Targ, fintagsOI$gene)]
for (tag in tags){
data=data.frame(tpm=unlist(TPM4RNA[tag,SamplesMeta$Pou]) ,
RT=unlist(MuFCsel[grep(Targ, rownames(MuFCsel)),SamplesMeta$Pou]))
plot(data$tpm,data$RT,
xlab="methylation tpm",
ylab = "mRNA log2FC vs mean",
ylim=c(-3,3),
col=4-as.numeric(SamplesMeta$Group),
pch=as.numeric(SamplesMeta$Group)+14,
main=paste(tag, "Meth vs mRNA Expr", Targ))
legend("topleft", c("control", "case"), pch=c(15,16), col=c(3,2), bty="n")
a=lm(RT~tpm, data)
b=summary(a)
abline(a, col="blue")
SperCor=cor(data$RT,data$tpm,use = "c", method = "spearman")
mtext(3, text = paste("beta = ", round(coefficients(a)[2],2),
"; se =", round(b$coefficients[2,2],2),
"; pvalue = ", round(b$coefficients[2,4],3),
"; Spearman cor = ", round(SperCor,3)))
}
}
Systems Analysis
Genomic feature enrichment
Significant loci with a p-value <= 0.01 and a absolute log2 fold-change lager 0.5 were tested for enrichment in annotated genomic feature using fisher exact test.
Ranges=rowData(dds_filt)
TotTagsofInterest=sum(Ranges$WaldPvalue_groupCD<=thresholdp & abs(Ranges$groupCD)>thresholdLFC)
Resall=data.frame()
index = Ranges$WaldPvalue_groupCD<=thresholdp& abs(Ranges$groupCD)>thresholdLFC
for (feat in unique(Ranges$feature)){
tmp=table(Ranges$feature == feat, signif=index)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resall = rbind(Resall, res)
}
tmp=table(Ranges$tf_binding!="", signif=index)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resall = rbind(Resall, res)
tmp=table(Ranges$cpg=="cpg", signif=index)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resall = rbind(Resall, res)
colnames(Resall)=c("OR", "CI95L", "CI95U", "P")
rownames(Resall)=c(unique(Ranges$feature), "TF-binding", "CpG-island")
Resall$Beta = log(Resall$OR)
Resall$SE = (log(Resall$OR)-log(Resall$CI95L))/1.96
Resall$Padj=p.adjust(Resall$P, method = "bonferroni")
Resdown=data.frame()
index = Ranges$WaldPvalue_groupCD<=thresholdp & Ranges$groupCD<thresholdLFC
for (feat in unique(Ranges$feature)){
tmp=table(Ranges$feature == feat, signif=index)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resdown = rbind(Resdown, res)
}
tmp=table(Ranges$tf_binding!="", signif=index)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resdown = rbind(Resdown, res)
tmp=table(Ranges$cpg=="cpg", signif=index)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resdown = rbind(Resdown, res)
colnames(Resdown)=c("OR", "CI95L", "CI95U", "P")
rownames(Resdown)=c(unique(Ranges$feature), "TF-binding", "CpG-island")
Resdown$Beta = log(Resdown$OR)
Resdown$SE = (log(Resdown$OR)-log(Resdown$CI95L))/1.96
Resdown$Padj=p.adjust(Resdown$P, method = "bonferroni")
Resup=data.frame()
index = Ranges$WaldPvalue_groupCD<=thresholdp & Ranges$groupCD>thresholdLFC
for (feat in unique(Ranges$feature)){
tmp=table(Ranges$feature == feat, signif=index)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resup = rbind(Resup, res)
}
tmp=table(Ranges$tf_binding!="", signif=index)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resup = rbind(Resup, res)
tmp=table(Ranges$cpg=="cpg", signif=index)
resfish=fisher.test(tmp)
res = c(resfish$estimate, unlist(resfish$conf.int), resfish$p.value)
Resup = rbind(Resup, res)
colnames(Resup)=c("OR", "CI95L", "CI95U", "P")
rownames(Resup)=c(unique(Ranges$feature), "TF-binding", "CpG-island")
Resup$Beta = log(Resup$OR)
Resup$SE = (log(Resup$OR)-log(Resup$CI95L))/1.96
Resup$Padj=p.adjust(Resup$P, method = "bonferroni")
multiORplot(Resall, Pval = "P", Padj = "Padj", beta="Beta",SE = "SE", pheno="All diff. methylated loci")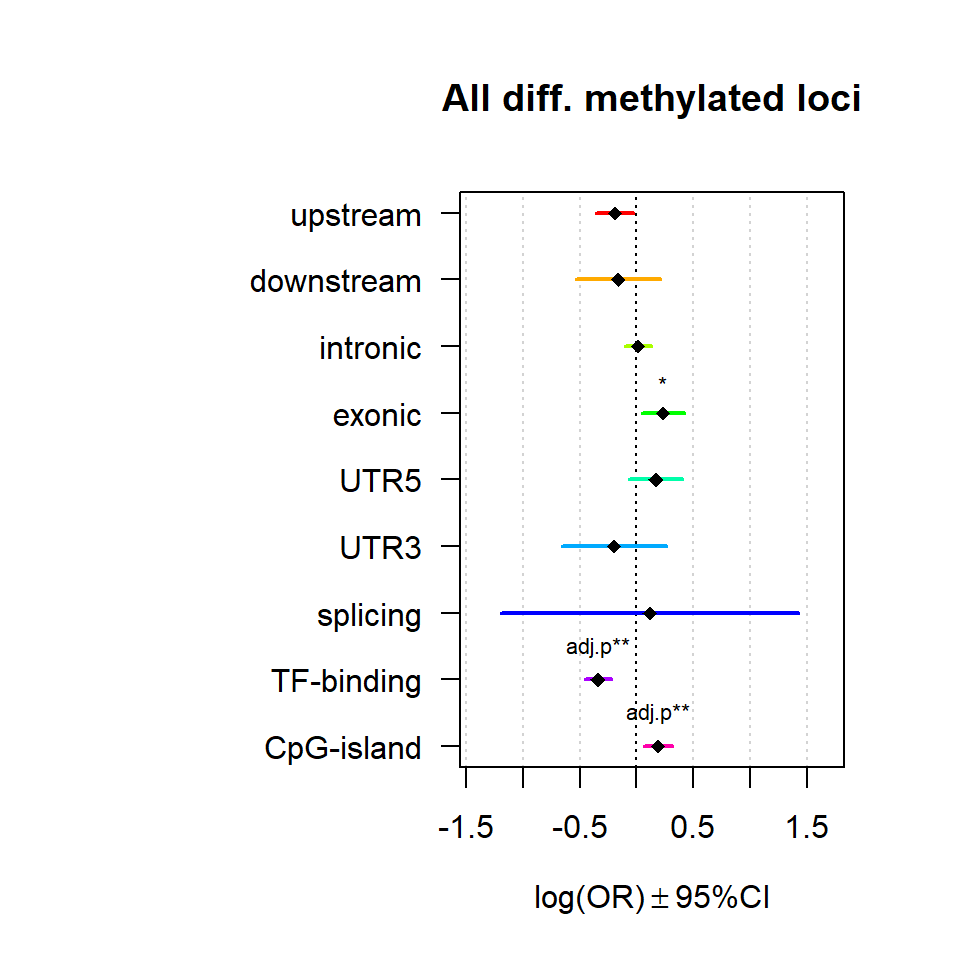
multiORplot(Resup, Pval = "P", Padj = "Padj", beta="Beta",SE = "SE", pheno="hypomethylated loci")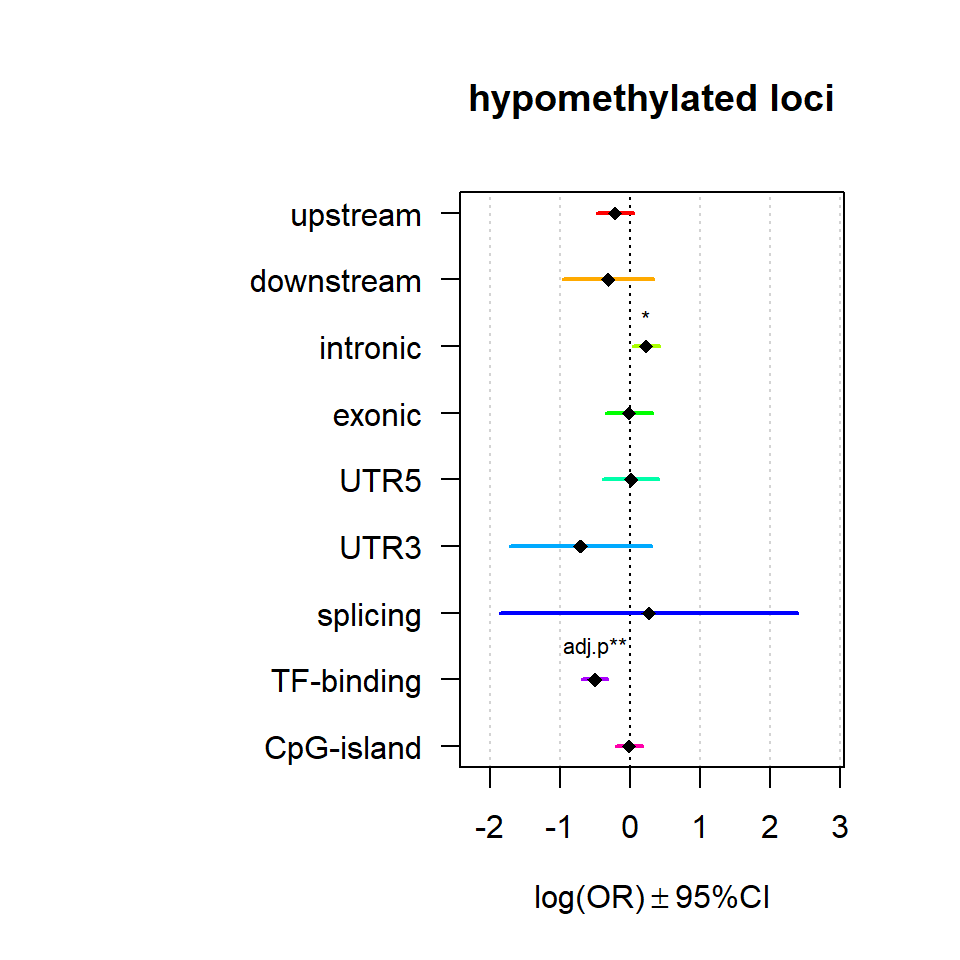
multiORplot(Resdown, Pval = "P", Padj = "Padj", beta="Beta",SE = "SE", pheno="Hypermethylated loci")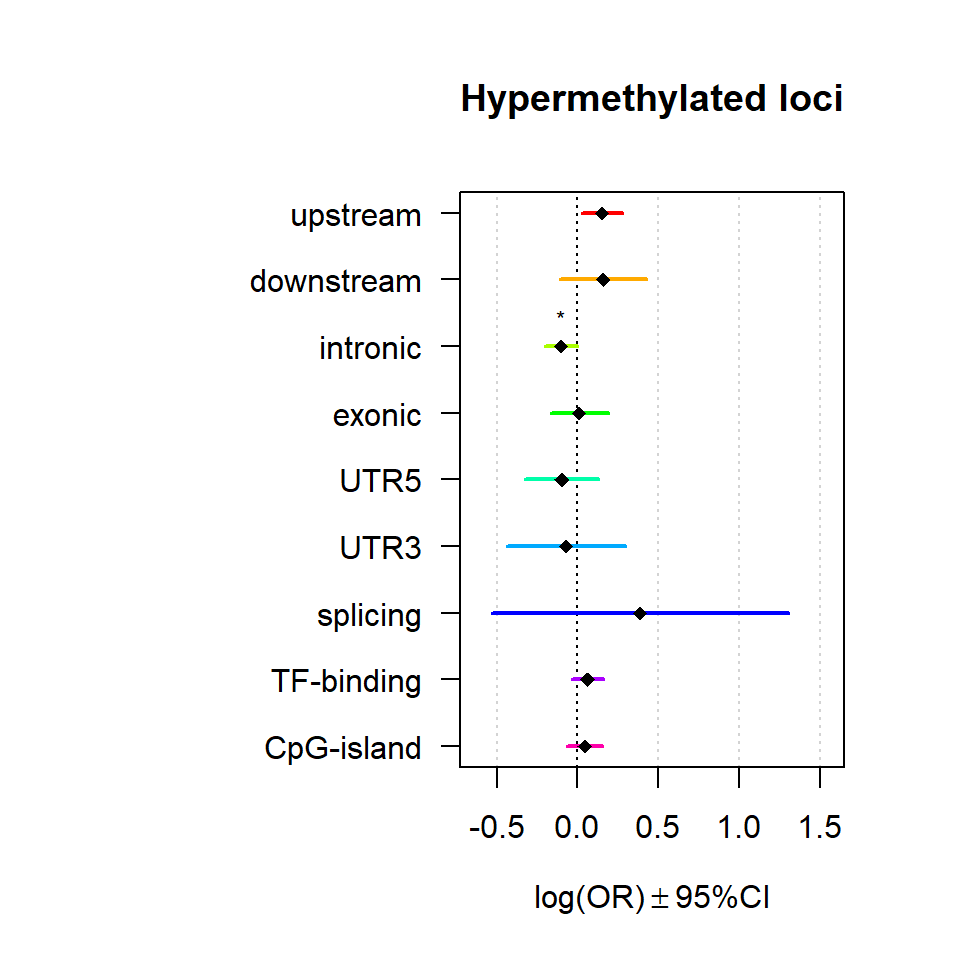
pdf(paste0(Home, "/output/functional_Enrichemnt.pdf"))
multiORplot(Resall, Pval = "P", Padj = "Padj", beta="Beta",SE = "SE", pheno="All diff. methylated loci")
multiORplot(Resup, Pval = "P", Padj = "Padj", beta="Beta",SE = "SE", pheno="hypomethylated loci")
multiORplot(Resdown, Pval = "P", Padj = "Padj", beta="Beta",SE = "SE", pheno="Hypermethylated loci")
dev.off()png
2 GO-term Enrichment
Significant loci and differentially methylated regions with a p-value <= 0.01 and an absolute log2 fold-change lager 0.5 were tested for enrichment among GO-terms Molecular Function, Cellular Compartment and Biological Processes, KEGG pathways, Transcription factor Binding sites, Human Protein Atlas Tissue Expression, Human Phenotypes.
getGOresults = function(geneset, genereference){
resgo = gost(geneset, organism = "hsapiens",
correction_method = "g_SCS",
domain_scope = "custom",
sources = c("GO:BP", "GO:MF", "GO:CC"),
custom_bg = genereference)
if(length(resgo) != 0){
return(resgo)
} else {
print("no significant results")
return(NULL)
}
}
gene_univers = getuniquegenes(as.data.frame(rowRanges(dds_filt))$gene)
idx = (results_Deseq$pvalue <= thresholdp &
(abs(results_Deseq$log2FoldChange) > thresholdLFC))
genes_reg = getuniquegenes(as.data.frame(rowRanges(dds_filt))$gene[idx])
dmr_genes = unique(resultsdmr_table$name[resultsdmr_table$p.value<=thresholdp &
abs(resultsdmr_table$value)>=thresholdLFC])
Genes_of_interset = list("01_dmregions" = dmr_genes,
"02_dmtag" = genes_reg
)
gostres = getGOresults(Genes_of_interset, gene_univers)
gostplot(gostres, capped = TRUE, interactive = T)p = gostplot(gostres, capped = TRUE, interactive = F)
toptab = gostres$result
pp = publish_gostplot(p, filename = paste0(Home,"/output/gostres.pdf"))The image is saved to C:/Users/chiocchetti/Projects/femNATCD_MethSeq/output/gostres.pdfwrite.xlsx2(toptab, file = paste0(Home,"/output/GOres.xlsx"), sheetName = "GO_enrichment")Brain Developmental Processes Enrichment tests
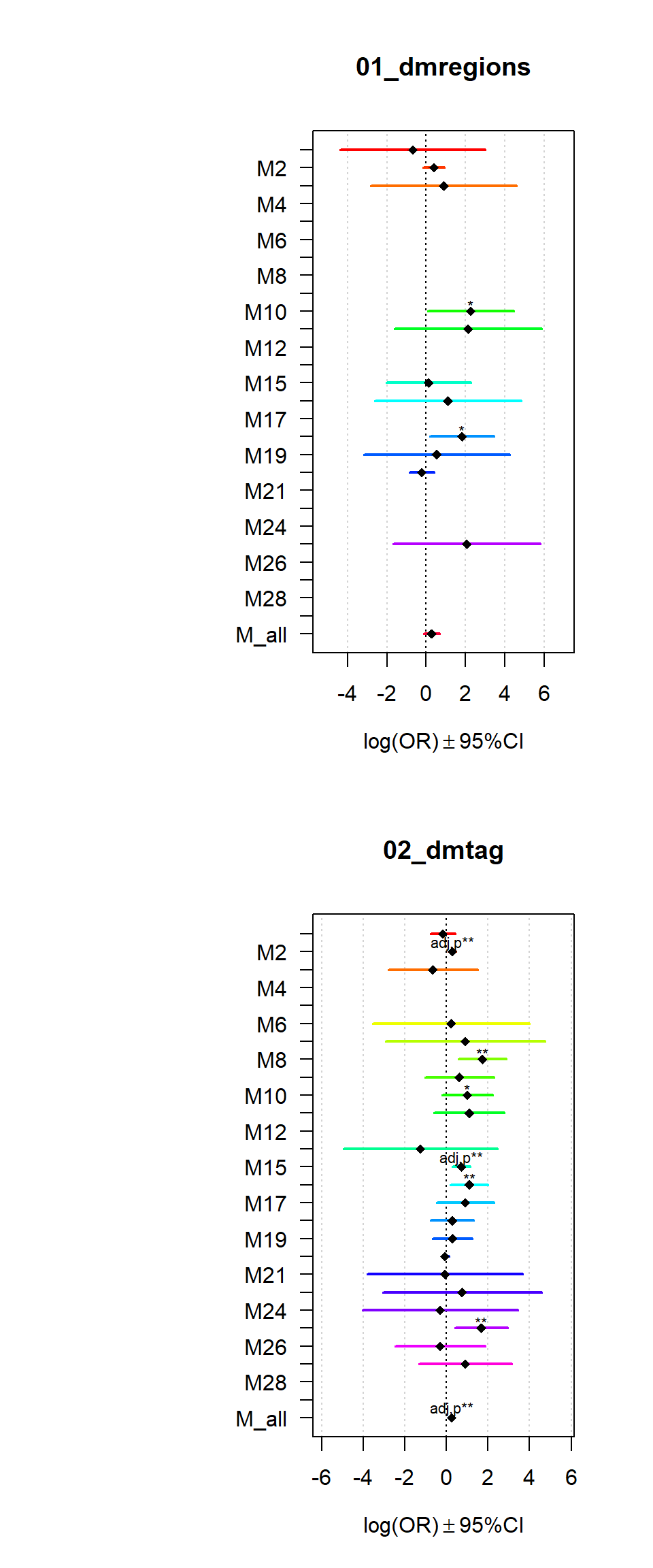
Gene sets identified to be deferentially methylated with a p-value <= 0.01 and an absolute log2 fold-change larger 0.5 were tested for enrichment among gene-modules coregulated during Brain expression.
Kang Modules
# define Reference Universe
KangUnivers<- read.table(paste0(Home,"/data/KangUnivers.txt"), sep="\t", header=T)
colnames(KangUnivers)<-c("EntrezId","Symbol")
Kang_genes<-read.table(paste0(Home,"/data/Kang_dataset_genesMod_version2.txt"),sep="\t",header=TRUE)
#3)Generate Gene universe to be used for single gene lists
tmp=merge(KangUnivers,Kang_genes,by.y="EntrezGene",by.x="EntrezId",all=TRUE) #18826
KangUni_Final<-tmp[duplicated(tmp$EntrezId)==FALSE,] #18675
# Local analysis gene universe
Annotation_list<-data.frame(Symbol = gene_univers)
# match modules
Annotation_list$Module = Kang_genes$Module[match(Annotation_list$Symbol,Kang_genes$symbol)]
# check if overlapping in gene universes
Annotation_list$univers = Annotation_list$Symbol %in% KangUni_Final$Symbol
# drop duplicates
Annotation_list = Annotation_list[duplicated(Annotation_list$Symbol)==FALSE,]
# selct only genes that have been detected on both datasets
Annotation_list = Annotation_list[Annotation_list$univers==T,]
# final reference
UniversalGeneset=Annotation_list$Symbol
# define Gene lists to test
# sort and order Modules to be tested
Modules=unique(Annotation_list$Module)
Modules = Modules[! Modules %in% c(NA, "")]
Modules = Modules[order(as.numeric(gsub("M","",Modules)))]
GL_all=list()
for(i in Modules){
GL_all[[i]]=Annotation_list$Symbol[Annotation_list$Module%in%i]
}
GL_all[["M_all"]]=Kang_genes$symbol[Kang_genes$Module %in% Modules]
GOI1 = Genes_of_interset
Resultsall=list()
for(j in names(GOI1)){
Res = data.frame()
for(i in names(GL_all)){
Modulegene=GL_all[[i]]
Factorgene=GOI1[[j]]
Testframe<-fisher.test(table(factor(UniversalGeneset %in% Factorgene,levels=c("TRUE","FALSE")),
factor(UniversalGeneset %in% Modulegene,levels=c("TRUE","FALSE"))))
beta=log(Testframe$estimate)
Res[i, "beta"] =beta
Res[i, "SE"]=abs(beta-log(Testframe$conf.int[1]))/1.96
Res[i, "Pval"]=Testframe$p.value
Res[i, "OR"]=(Testframe$estimate)
Res[i, "ORL"]=(Testframe$conf.int[1])
Res[i, "ORU"]=(Testframe$conf.int[2])
}
Res$Padj = p.adjust(Res$Pval, method = "bonferroni")
Resultsall[[j]] = Res
}
par(mfrow = c(2,1))
for (i in names(Resultsall)){
multiORplot(datatoplot = Resultsall[[i]], pheno=i)
}
par(mfrow = c(1,1))
pdf(paste0(Home, "/output/BrainMod_Enrichemnt.pdf"))
for (i in names(Resultsall)){
multiORplot(datatoplot = Resultsall[[i]], pheno=i)
}
dev.off()png
2 Modsig = c()
for(r in names(Resultsall)){
a=rownames(Resultsall[[r]])[Resultsall[[r]]$Padj<=0.05]
Modsig = c(Modsig,a)
}Brain espresseion heatmaps
# show brains and expression
Modsig2=unique(Modsig[Modsig!="M_all"])
load(paste0(Home,"/data/Kang_DataPreprocessing.RData")) #Load the Kang expression data of all genes
datExprPlot=matriz #Expression data of Kang loaded as Rdata object DataPreprocessing.RData
Genes = GL_all[names(GL_all)!="M_all"]
Genes_expression<-list()
pcatest<-list()
for (i in names(Genes)){
Genes_expression[[i]]<-matriz[,which(colnames(matriz) %in% Genes[[i]])]
pcatest[[i]]=prcomp(t(as.matrix(Genes_expression[[i]])),retx=TRUE)
}
# PCA test
PCA<-data.frame(pcatest[[1]]$rotation)
PCA$donor_name<-rownames(PCA)
PC1<-data.frame(PCA[,c(1,ncol(PCA))])
#Combining the age with expression data
list <- strsplit(sampleInfo$age, " ")
library("plyr")------------------------------------------------------------------------------You have loaded plyr after dplyr - this is likely to cause problems.
If you need functions from both plyr and dplyr, please load plyr first, then dplyr:
library(plyr); library(dplyr)------------------------------------------------------------------------------
Attache Paket: 'plyr'The following object is masked from 'package:matrixStats':
countThe following object is masked from 'package:IRanges':
descThe following object is masked from 'package:S4Vectors':
renameThe following objects are masked from 'package:dplyr':
arrange, count, desc, failwith, id, mutate, rename, summarise,
summarizeThe following object is masked from 'package:purrr':
compactdf <- ldply(list)
colnames(df) <- c("Age", "time")
sampleInfo<-cbind(sampleInfo[,1:9],df)
sampleInfo$Age<-as.numeric(sampleInfo$Age)
sampleInfo$period<-ifelse(sampleInfo$time=="pcw",sampleInfo$Age*7,ifelse(sampleInfo$time=="yrs",sampleInfo$Age*365+270,ifelse(sampleInfo$time=="mos",sampleInfo$Age*30+270,NA)))
#We need it just for the donor names
PCA_matrix<-merge.with.order(PC1,sampleInfo,by.y="SampleID",by.x="donor_name",keep_order=1)
#Select which have phenotype info present
matriz2<-matriz[which(rownames(matriz) %in% PCA_matrix$donor_name),]
FactorGenes_expression<-list()
#Factors here mean modules
for (i in names(Genes)){
FactorGenes_expression[[i]]<-matriz2[,which(colnames(matriz2) %in% Genes[[i]])]
}
FactorseGE<-list()
for (i in names(Genes)){
FactorseGE[[i]]<-FactorGenes_expression[[i]]
}
allModgenes=NULL
colors=vector()
for ( i in names(Genes)){
allModgenes=cbind(allModgenes,FactorseGE[[i]])
colors=c(colors, rep(i, ncol(FactorseGE[[i]])))
}
lengths=unlist(lapply(FactorGenes_expression, ncol), use.names = F)
MEorig=moduleEigengenes(allModgenes, colors)
PCA_matrixfreeze=PCA_matrix
index=!PCA_matrix$structure_acronym %in% c("URL", "DTH", "CGE","LGE", "MGE", "Ocx", "PCx", "M1C-S1C","DIE", "TCx", "CB")
PCA_matrix=PCA_matrix[index,]
ME = MEorig$eigengenes[index,]
matsel = matriz2[index,]
colnames(ME) = gsub("ME", "", colnames(ME))
timepoints=seq(56,15000, length.out=1000)
matrix(c("CB", "THA", "CBC", "MD"), ncol=2 ) -> cnm
brainheatmap=function(Module){
MEmod=ME[,Module]
toplot=data.frame(matrix(NA, nrow=length(table(PCA_matrix$structure_acronym)), ncol=998))
rownames(toplot)=unique(PCA_matrix$structure_acronym)
target <- c("OFC", "DFC", "VFC", "MFC","M1C","S1C","IPC","A1C","STC","ITC","V1C","HIP","AMY","STR","MD","CBC")
toplot<-toplot[c(6,2,8,5,11,12,10,9,7,4,14,3,1,13,16,15),]
for ( i in unique(PCA_matrix$structure_acronym)){
index=PCA_matrix$structure_acronym==i
LOESS=loess(MEmod[index]~PCA_matrix$period[index])
toplot[i,]=predict(LOESS,newdata = round(exp(seq(log(56),log(15000), length.out=998)),2))
colnames(toplot)[c(1,77,282,392,640,803,996)]<-
c("1pcw","21pcw","Birth","1.3years","5.4years","13.6years","40.7years")
}
cols=viridis(100)
labvec <- c(rep(NA, 1000))
labvec[c(1,77,282,392,640,803,996)] <- c("1pcw","21pcw","Birth","1.3years","5.4years","13.6years","40.7years")
toplot<-toplot[,1:998]
date<-c(1:998)
dateY<-paste0(round(date/365,2),"_Years")
names(toplot)<-dateY
par(xpd=FALSE)
heatmap.2(as.matrix(toplot), col = cols,
main=Module,
trace = "none",
na.color = "grey",
Colv = F, Rowv = F,
labCol = labvec,
#breaks = seq(-0.1,0.1, length.out=101),
symkey = T,
scale = "row",
key.title = "",
dendrogram = "none",
key.xlab = "eigengene",
density.info = "none",
#main=paste("Module",1),
srtCol=90,
tracecol = "none",
cexRow = 1,
add.expr=eval.parent(abline(v=282),
axis(1,at=c(1,77,282,392,640,803,996),
labels =FALSE)),cexCol = 1)
}
brainheatmap_gene=function(Genename){
MEmod=matsel[,Genename]
toplot=data.frame(matrix(NA, nrow=length(table(PCA_matrix$structure_acronym)), ncol=998))
rownames(toplot)=unique(PCA_matrix$structure_acronym)
target <- c("OFC", "DFC", "VFC", "MFC","M1C","S1C","IPC","A1C","STC","ITC","V1C","HIP","AMY","STR","MD","CBC")
toplot<-toplot[c(6,2,8,5,11,12,10,9,7,4,14,3,1,13,16,15),]
for ( i in unique(PCA_matrix$structure_acronym)){
index=PCA_matrix$structure_acronym==i
LOESS=loess(MEmod[index]~PCA_matrix$period[index])
toplot[i,]=predict(LOESS,newdata = round(exp(seq(log(56),log(15000), length.out=998)),2))
colnames(toplot)[c(1,77,282,392,640,803,996)]<-
c("1pcw","21pcw","Birth","1.3years","5.4years","13.6years","40.7years")
}
cols=viridis(100)
labvec <- c(rep(NA, 1000))
labvec[c(1,77,282,392,640,803,996)] <- c("1pcw","21pcw","Birth","1.3years","5.4years","13.6years","40.7years")
toplot<-toplot[,1:998]
date<-c(1:998)
dateY<-paste0(round(date/365,2),"_Years")
names(toplot)<-dateY
par(xpd=FALSE)
heatmap.2(as.matrix(toplot), col = cols,
main=Genename,
trace = "none",
na.color = "grey",
Colv = F, Rowv = F,
labCol = labvec,
#breaks = seq(-0.1,0.1, length.out=101),
symkey = F,
scale = "none",
key.title = "",
dendrogram = "none",
key.xlab = "eigengene",
density.info = "none",
#main=paste("Module",1),
#srtCol=90,
tracecol = "none",
cexRow = 1,
add.expr=eval.parent(abline(v=282),
axis(1,at=c(1,77,282,392,640,803,996),
labels =FALSE))
,cexCol = 1)
}
brainheatmap_gene("SLITRK5")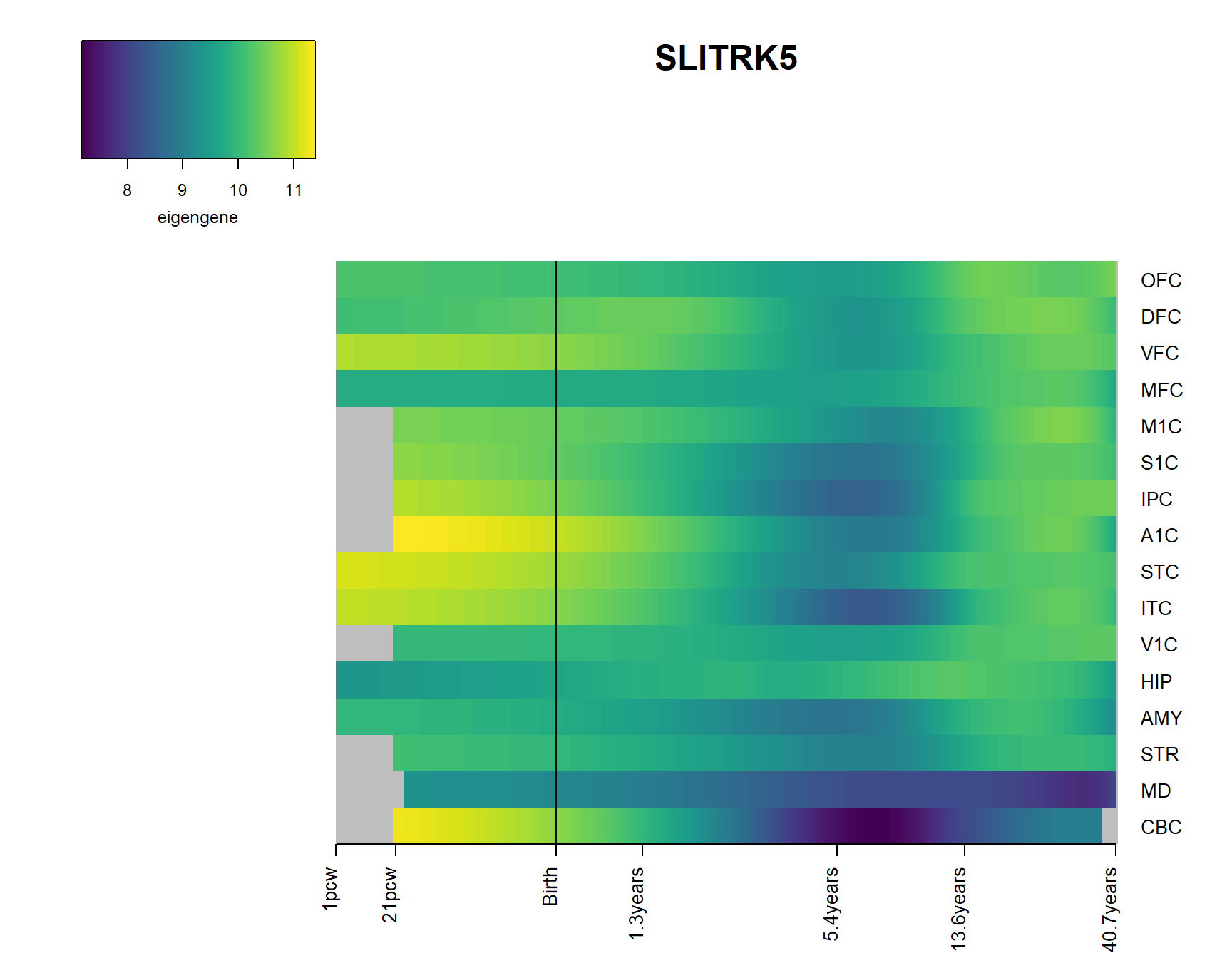
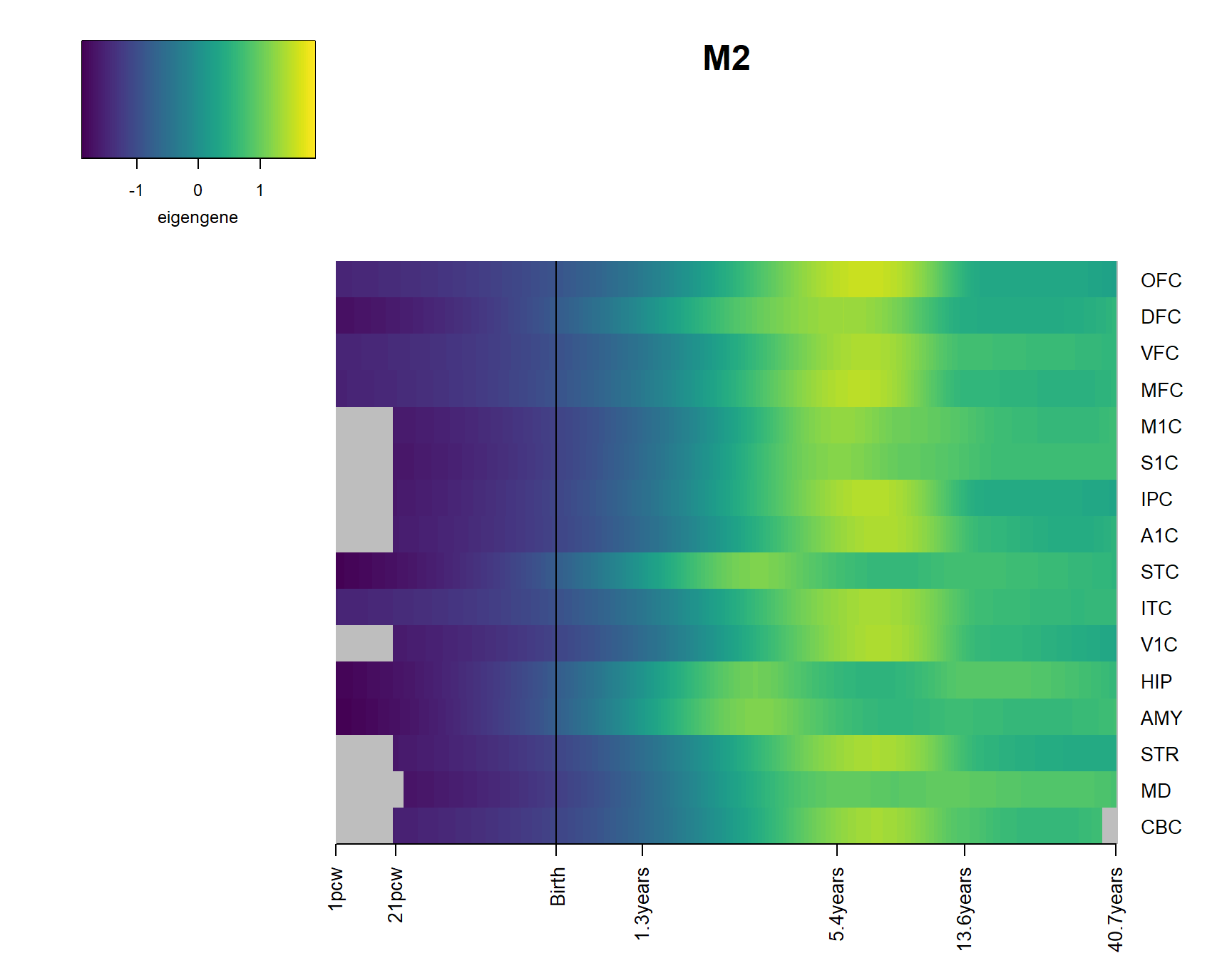
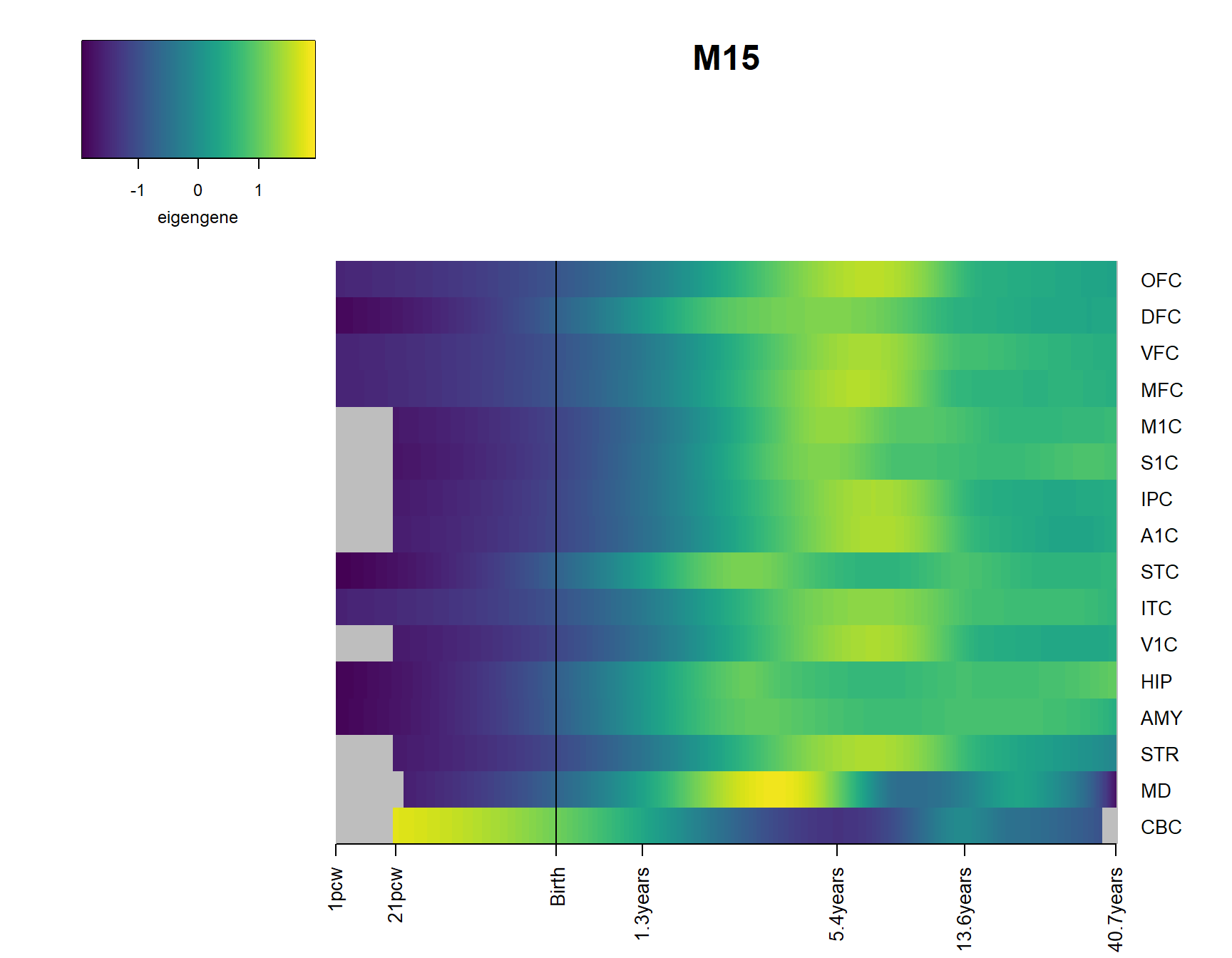
for(Module in Modsig2){
brainheatmap(Module)
}
pdf(paste0(Home, "/output/Brain_Module_Heatmap.pdf"))
brainheatmap_gene("SLITRK5")
for(Module in Modsig2){
brainheatmap(Module)
}
dev.off()png
2 Risk Factor Mediation Analysis
Risk factor loading and correlation plots
dropfact=c("site", "0", "group")
modelFact=strsplit(as.character(design(dds_filt))[2], " \\+ ")[[1]]
Patdata=as.data.frame(colData(dds_filt))
load(paste0(Home, "/output/envFact.RData"))
envFact=envFact[!envFact %in% dropfact]
modelFact=modelFact[!modelFact %in% dropfact]
EpiMarker = c()
# TopHit
Patdata$Epi_TopHit=log2_cpm[base::which.min(results_Deseq$pvalue),]
# 1PC of all diff met
tmp=glmpca(log2_cpm[base::which(results_Deseq$pvalue<=thresholdp),], 1)
Patdata$Epi_all= tmp$factors$dim1
EpiMarker = c(EpiMarker, "Epi_TopHit", "Epi_all")
#Brain Modules
Epitestset=GL_all[Modsig]
for(n in names(Epitestset)){
index=gettaglistforgenelist(genelist = Epitestset[[n]], dds_filt)
index = base::intersect(index, base::which(results_Deseq$pvalue<=thresholdp))
# get eigenvalue
epiname=paste0("Epi_",n)
tmp=glmpca(log2_cpm[index,], 1)
Patdata[,epiname]= tmp$factors$dim1
EpiMarker = c(EpiMarker, epiname)
}
cormat = cor(apply(Patdata[,c("group", envFact, modelFact, EpiMarker)] %>% mutate_all(as.numeric), 2, minmax_scaling),
use = "pairwise.complete.obs")
par(mfrow=c(1,2))
corrplot(cormat, main="correlations")
corrplot(cormat, order = "hclust", main="correlations ordered")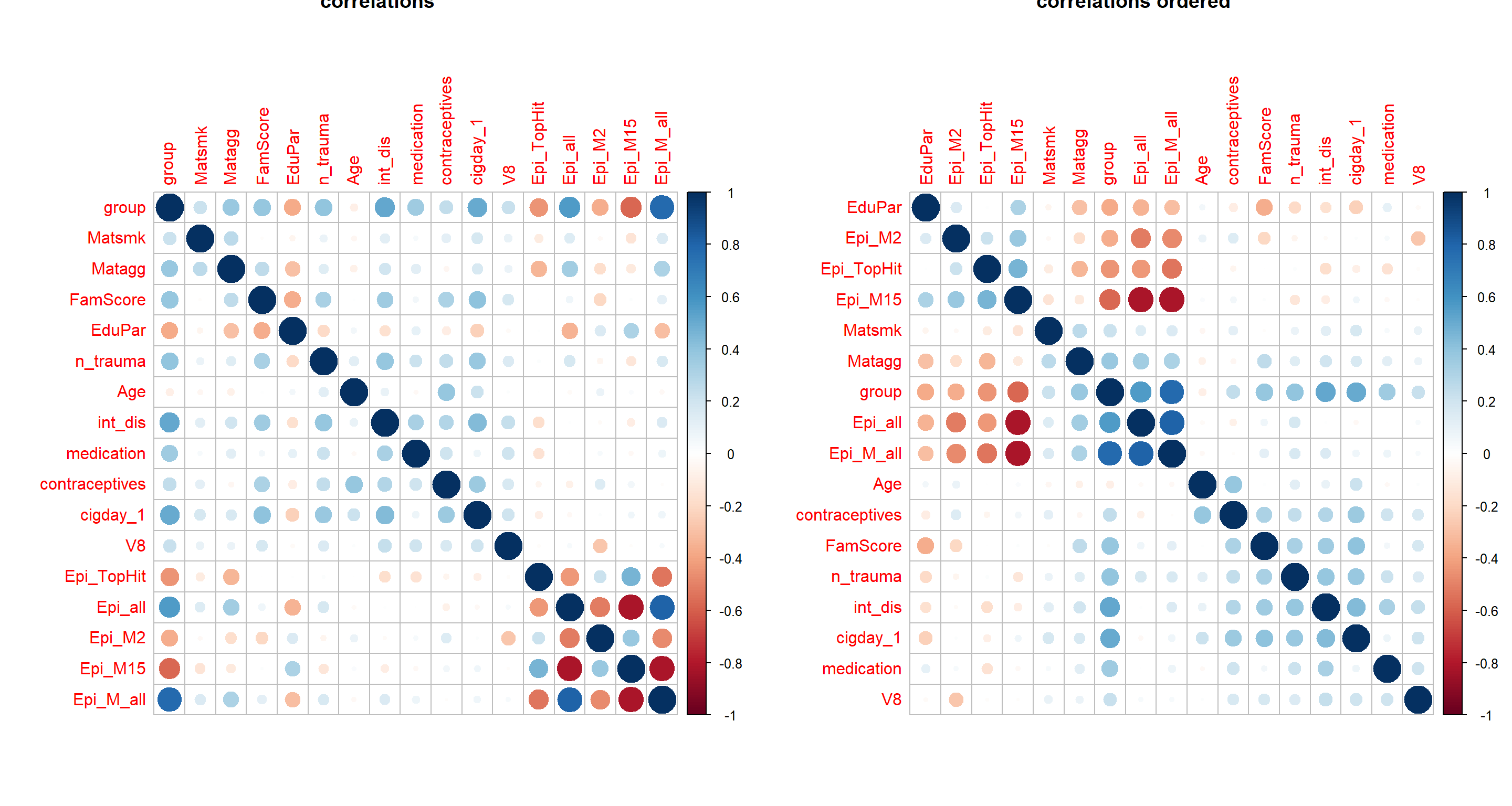
SEM analysis
fullmodEnv=paste(unique(envFact,modelFact), sep = "+", collapse = "+")
Dataset = Patdata[,c("group", envFact, modelFact,EpiMarker)]
model = "
Epi~a*Matsmk+b*Matagg+c*FamScore+d*EduPar+e*n_trauma+Age+int_dis+medication+contraceptives+V8+cigday_1
group~f*Matsmk+g*Matagg+h*FamScore+i*EduPar+j*n_trauma+Age+int_dis+medication+contraceptives+V8+cigday_1+z*Epi
#direct
directMatsmk := f
directMatagg := g
directFamScore := h
directEduPar := i
directn_trauma := j
#indirect
EpiMatsmk := a*z
EpiMatagg := b*z
EpiFamScore := c*z
EpiEduPar := d*z
Epin_trauma := e*z
total := f + g + h + i + j + (a*z)+(b*z)+(c*z)+(d*z)+(e*z)
"
model0="group~1+Matsmk+Matagg+FamScore+EduPar+n_trauma+Age+int_dis+medication+contraceptives+V8+Epi"
Netlist = list()
for (marker in EpiMarker) {
Dataset$Epi = Dataset[,marker]
Datasetscaled = Dataset %>% mutate_if(is.numeric, minmax_scaling)
Datasetscaled = Datasetscaled %>% mutate_if(is.factor, ~ as.numeric(.)-1)
Datasetscaled$group = ordered(Datasetscaled$group)
fit<-lavaan(model,data=Datasetscaled)
sink(paste0(Home,"/output/SEM_summary_group",marker,".txt"))
summary(fit)
print(fitMeasures(fit))
print(parameterEstimates(fit))
sink()
cat("############################\n")
cat("############################\n")
cat(marker, "\n")
cat("############################\n")
cat("############################\n")
cat("##Mediation Model ##\n")
summary(fit)
cat("\n")
print(fitMeasures(fit))
cat("\n")
print(parameterEstimates(fit))
cat("\n")
#SOURCE FOR PLOT https://stackoverflow.com/questions/51270032/how-can-i-display-only-significant-path-lines-on-a-path-diagram-r-lavaan-sem
restab=lavaan::standardizedSolution(fit) %>% dplyr::filter(!is.na(pvalue)) %>%
arrange(desc(pvalue)) %>% mutate_if("is.numeric","round",3) %>%
dplyr::select(-ci.lower,-ci.upper,-z)
pvalue_cutoff <- 0.05
obj <- semPlot:::semPlotModel(fit)
original_Pars <- obj@Pars
print(original_Pars)
check_Pars <- obj@Pars %>% dplyr:::filter(!(edge %in% c("int","<->") | lhs == rhs)) # this is the list of parameter to sift thru
keep_Pars <- obj@Pars %>% dplyr:::filter(edge %in% c("int","<->") | lhs == rhs) # this is the list of parameter to keep asis
test_against <- lavaan::standardizedSolution(fit) %>% dplyr::filter(pvalue < pvalue_cutoff, rhs != lhs)
# for some reason, the rhs and lhs are reversed in the standardizedSolution() output, for some of the values
# I'll have to reverse it myself, and test against both orders
test_against_rev <- test_against %>% dplyr::rename(rhs2 = lhs, lhs = rhs) %>% dplyr::rename(rhs = rhs2)
checked_Pars <-
check_Pars %>% semi_join(test_against, by = c("lhs", "rhs")) %>% bind_rows(
check_Pars %>% semi_join(test_against_rev, by = c("lhs", "rhs"))
)
obj@Pars <- keep_Pars %>% bind_rows(checked_Pars) %>%
mutate_if("is.numeric","round",3) %>%
mutate_at(c("lhs","rhs"),~gsub("Epi", marker,.))
obj@Vars = obj@Vars %>% mutate_at(c("name"),~gsub("Epi", marker,.))
DF = obj@Pars
DF = DF[DF$lhs!=DF$rhs,]
DF = DF[abs(DF$est)>0.1,]
DF = DF[DF$edge == "~>",] # only include directly modelled effects in figure
nodes <- data.frame(id=obj@Vars$name, label = obj@Vars$name)
nodes$color<-Dark8[8]
nodes$color[nodes$label == "group"] = Dark8[3]
nodes$color[nodes$label == marker] = Dark8[4]
nodes$color[nodes$label %in% envFact] = Dark8[5]
edges <- data.frame(from = DF$lhs,
to = DF$rhs,
width=abs(DF$est),
arrows ="to")
edges$dashes = F
edges$label = DF$est
edges$color=c("firebrick", "forestgreen")[1:2][factor(sign(DF$est), levels=c(-1,0,1),labels=c(1,2,2))]
edges$width=2
cexlab = 18
plotnet<- visNetwork(nodes, edges,
main=list(text=marker,
style="font-family:arial;font-size:20px;text-align:center"),
submain=list(text="significant paths",
style="font-family:arial;text-align:center")) %>%
visEdges(arrows =list(to = list(enabled = TRUE, scaleFactor = 0.7)),
font=list(size=cexlab, style="font-family:arial;text-align:center")) %>%
visNodes(font=list(size=cexlab, style="font-family:arial;text-align:center")) %>%
visPhysics(enabled = T, solver = "forceAtlas2Based")
Netlist[[marker]] = plotnet
htmlfile = paste0(Home,"/output/SEMplot_",marker,".html")
visSave(plotnet, htmlfile)
webshot(paste0(Home,"/output/SEMplot_",marker,".html"), zoom = 1,
file = paste0(Home,"/output/SEMplot_",marker,".png"))
}Warning in lav_bvmix_cor_twostep_fit(fit.y1 = UNI[[j]], fit.y2 = UNI[[i]], : lavaan WARNING: estimation polyserial correlation did not converge for
variables Epi and groupWarning in lav_samplestats_step2(UNI = FIT, wt = wt, ov.names = ov.names, :
lavaan WARNING: correlation between variables group and Epi is (nearly) 1.0Warning in lav_object_post_check(object): lavaan WARNING: some estimated ov
variances are negative############################
############################
Epi_TopHit
############################
############################
##Mediation Model ##
lavaan 0.6-7 ended normally after 108 iterations
Estimator DWLS
Optimization method NLMINB
Number of free parameters 23
Used Total
Number of observations 80 99
Model Test User Model:
Standard Robust
Test Statistic 148.312 132.700
Degrees of freedom 3 3
P-value (Chi-square) 0.000 0.000
Scaling correction factor 1.120
Shift parameter 0.322
simple second-order correction
Parameter Estimates:
Standard errors Robust.sem
Information Expected
Information saturated (h1) model Unstructured
Regressions:
Estimate Std.Err z-value P(>|z|)
Epi ~
Matsmk (a) -0.033 0.050 -0.657 0.511
Matagg (b) -0.014 0.072 -0.202 0.840
FamScore (c) 0.056 0.076 0.739 0.460
EduPar (d) -0.038 0.108 -0.358 0.721
n_trauma (e) 0.085 0.111 0.767 0.443
Age -0.090 0.097 -0.929 0.353
int_dis -0.074 0.057 -1.310 0.190
medication -0.044 0.059 -0.759 0.448
contrcptvs -0.017 0.049 -0.341 0.733
V8 -0.089 0.568 -0.156 0.876
cigday_1 -0.111 0.136 -0.815 0.415
group ~
Matsmk (f) 0.194 1.123 0.172 0.863
Matagg (g) 1.502 2.429 0.618 0.536
FamScore (h) 0.182 2.078 0.088 0.930
EduPar (i) -3.511 2.216 -1.584 0.113
n_trauma (j) 2.563 1.399 1.832 0.067
Age -3.056 2.476 -1.234 0.217
int_dis 1.143 0.802 1.426 0.154
medication 1.079 0.834 1.294 0.196
contrcptvs 0.108 0.798 0.135 0.892
V8 13.100 16.569 0.791 0.429
cigday_1 10.186 7.161 1.422 0.155
Epi (z) -1.921 0.014 -133.612 0.000
Intercepts:
Estimate Std.Err z-value P(>|z|)
.Epi 0.000
.group 0.000
Thresholds:
Estimate Std.Err z-value P(>|z|)
group|t1 5.748
Variances:
Estimate Std.Err z-value P(>|z|)
.Epi 1.000
.group -2.692
Scales y*:
Estimate Std.Err z-value P(>|z|)
group 1.000
Defined Parameters:
Estimate Std.Err z-value P(>|z|)
directMatsmk 0.194 1.123 0.172 0.863
directMatagg 1.502 2.429 0.618 0.536
directFamScore 0.182 2.078 0.088 0.930
directEduPar -3.511 2.216 -1.584 0.113
directn_trauma 2.563 1.399 1.832 0.067
EpiMatsmk 0.063 0.095 0.656 0.512
EpiMatagg 0.028 0.138 0.202 0.840
EpiFamScore -0.108 0.147 -0.738 0.460
EpiEduPar 0.074 0.206 0.358 0.720
Epin_trauma -0.163 0.213 -0.767 0.443
total 0.823 4.116 0.200 0.842
npar fmin
23.000 0.927
chisq df
148.312 3.000
pvalue chisq.scaled
0.000 132.700
df.scaled pvalue.scaled
3.000 0.000
chisq.scaling.factor baseline.chisq
1.120 111.273
baseline.df baseline.pvalue
1.000 0.000
baseline.chisq.scaled baseline.df.scaled
111.273 1.000
baseline.pvalue.scaled baseline.chisq.scaling.factor
0.000 1.000
cfi tli
0.000 0.561
nnfi rfi
0.561 NA
nfi pnfi
NA -0.999
ifi rni
-0.342 -0.318
cfi.scaled tli.scaled
0.000 0.608
cfi.robust tli.robust
NA NA
nnfi.scaled nnfi.robust
0.608 NA
rfi.scaled nfi.scaled
NA NA
ifi.scaled rni.scaled
-0.198 -0.176
rni.robust rmsea
NA 0.783
rmsea.ci.lower rmsea.ci.upper
0.678 0.893
rmsea.pvalue rmsea.scaled
0.000 0.740
rmsea.ci.lower.scaled rmsea.ci.upper.scaled
0.635 0.850
rmsea.pvalue.scaled rmsea.robust
0.000 NA
rmsea.ci.lower.robust rmsea.ci.upper.robust
NA NA
rmsea.pvalue.robust rmr
NA 0.980
rmr_nomean srmr
1.167 25.068
srmr_bentler srmr_bentler_nomean
19.574 25.068
crmr crmr_nomean
3.232 0.929
srmr_mplus srmr_mplus_nomean
NA NA
cn_05 cn_01
5.163 7.043
gfi agfi
-401.552 -3487.786
pgfi mfi
-46.333 0.399
lhs op rhs label
1 Epi ~ Matsmk a
2 Epi ~ Matagg b
3 Epi ~ FamScore c
4 Epi ~ EduPar d
5 Epi ~ n_trauma e
6 Epi ~ Age
7 Epi ~ int_dis
8 Epi ~ medication
9 Epi ~ contraceptives
10 Epi ~ V8
11 Epi ~ cigday_1
12 group ~ Matsmk f
13 group ~ Matagg g
14 group ~ FamScore h
15 group ~ EduPar i
16 group ~ n_trauma j
17 group ~ Age
18 group ~ int_dis
19 group ~ medication
20 group ~ contraceptives
21 group ~ V8
22 group ~ cigday_1
23 group ~ Epi z
24 group | t1
25 Epi ~~ Epi
26 group ~~ group
27 Matsmk ~~ Matsmk
28 Matsmk ~~ Matagg
29 Matsmk ~~ FamScore
30 Matsmk ~~ EduPar
31 Matsmk ~~ n_trauma
32 Matsmk ~~ Age
33 Matsmk ~~ int_dis
34 Matsmk ~~ medication
35 Matsmk ~~ contraceptives
36 Matsmk ~~ V8
37 Matsmk ~~ cigday_1
38 Matagg ~~ Matagg
39 Matagg ~~ FamScore
40 Matagg ~~ EduPar
41 Matagg ~~ n_trauma
42 Matagg ~~ Age
43 Matagg ~~ int_dis
44 Matagg ~~ medication
45 Matagg ~~ contraceptives
46 Matagg ~~ V8
47 Matagg ~~ cigday_1
48 FamScore ~~ FamScore
49 FamScore ~~ EduPar
50 FamScore ~~ n_trauma
51 FamScore ~~ Age
52 FamScore ~~ int_dis
53 FamScore ~~ medication
54 FamScore ~~ contraceptives
55 FamScore ~~ V8
56 FamScore ~~ cigday_1
57 EduPar ~~ EduPar
58 EduPar ~~ n_trauma
59 EduPar ~~ Age
60 EduPar ~~ int_dis
61 EduPar ~~ medication
62 EduPar ~~ contraceptives
63 EduPar ~~ V8
64 EduPar ~~ cigday_1
65 n_trauma ~~ n_trauma
66 n_trauma ~~ Age
67 n_trauma ~~ int_dis
68 n_trauma ~~ medication
69 n_trauma ~~ contraceptives
70 n_trauma ~~ V8
71 n_trauma ~~ cigday_1
72 Age ~~ Age
73 Age ~~ int_dis
74 Age ~~ medication
75 Age ~~ contraceptives
76 Age ~~ V8
77 Age ~~ cigday_1
78 int_dis ~~ int_dis
79 int_dis ~~ medication
80 int_dis ~~ contraceptives
81 int_dis ~~ V8
82 int_dis ~~ cigday_1
83 medication ~~ medication
84 medication ~~ contraceptives
85 medication ~~ V8
86 medication ~~ cigday_1
87 contraceptives ~~ contraceptives
88 contraceptives ~~ V8
89 contraceptives ~~ cigday_1
90 V8 ~~ V8
91 V8 ~~ cigday_1
92 cigday_1 ~~ cigday_1
93 group ~*~ group
94 Epi ~1
95 group ~1
96 Matsmk ~1
97 Matagg ~1
98 FamScore ~1
99 EduPar ~1
100 n_trauma ~1
101 Age ~1
102 int_dis ~1
103 medication ~1
104 contraceptives ~1
105 V8 ~1
106 cigday_1 ~1
107 directMatsmk := f directMatsmk
108 directMatagg := g directMatagg
109 directFamScore := h directFamScore
110 directEduPar := i directEduPar
111 directn_trauma := j directn_trauma
112 EpiMatsmk := a*z EpiMatsmk
113 EpiMatagg := b*z EpiMatagg
114 EpiFamScore := c*z EpiFamScore
115 EpiEduPar := d*z EpiEduPar
116 Epin_trauma := e*z Epin_trauma
117 total := f+g+h+i+j+(a*z)+(b*z)+(c*z)+(d*z)+(e*z) total
est se z pvalue ci.lower ci.upper
1 -0.033 0.050 -0.657 0.511 -0.130 0.065
2 -0.014 0.072 -0.202 0.840 -0.155 0.126
3 0.056 0.076 0.739 0.460 -0.093 0.206
4 -0.038 0.108 -0.358 0.721 -0.249 0.172
5 0.085 0.111 0.767 0.443 -0.132 0.302
6 -0.090 0.097 -0.929 0.353 -0.280 0.100
7 -0.074 0.057 -1.310 0.190 -0.186 0.037
8 -0.044 0.059 -0.759 0.448 -0.159 0.070
9 -0.017 0.049 -0.341 0.733 -0.113 0.080
10 -0.089 0.568 -0.156 0.876 -1.202 1.025
11 -0.111 0.136 -0.815 0.415 -0.378 0.156
12 0.194 1.123 0.172 0.863 -2.007 2.395
13 1.502 2.429 0.618 0.536 -3.259 6.264
14 0.182 2.078 0.088 0.930 -3.891 4.255
15 -3.511 2.216 -1.584 0.113 -7.855 0.833
16 2.563 1.399 1.832 0.067 -0.179 5.306
17 -3.056 2.476 -1.234 0.217 -7.908 1.797
18 1.143 0.802 1.426 0.154 -0.429 2.715
19 1.079 0.834 1.294 0.196 -0.555 2.712
20 0.108 0.798 0.135 0.892 -1.456 1.672
21 13.100 16.569 0.791 0.429 -19.375 45.576
22 10.186 7.161 1.422 0.155 -3.850 24.222
23 -1.921 0.014 -133.612 0.000 -1.950 -1.893
24 5.748 0.000 NA NA 5.748 5.748
25 1.000 0.000 NA NA 1.000 1.000
26 -2.692 0.000 NA NA -2.692 -2.692
27 0.196 0.000 NA NA 0.196 0.196
28 0.049 0.000 NA NA 0.049 0.049
29 -0.009 0.000 NA NA -0.009 -0.009
30 -0.006 0.000 NA NA -0.006 -0.006
31 0.007 0.000 NA NA 0.007 0.007
32 -0.003 0.000 NA NA -0.003 -0.003
33 0.022 0.000 NA NA 0.022 0.022
34 0.004 0.000 NA NA 0.004 0.004
35 0.022 0.000 NA NA 0.022 0.022
36 0.003 0.000 NA NA 0.003 0.003
37 0.017 0.000 NA NA 0.017 0.017
38 0.091 0.000 NA NA 0.091 0.091
39 0.034 0.000 NA NA 0.034 0.034
40 -0.017 0.000 NA NA -0.017 -0.017
41 0.007 0.000 NA NA 0.007 0.007
42 0.000 0.000 NA NA 0.000 0.000
43 0.033 0.000 NA NA 0.033 0.033
44 0.008 0.000 NA NA 0.008 0.008
45 0.008 0.000 NA NA 0.008 0.008
46 0.001 0.000 NA NA 0.001 0.001
47 0.010 0.000 NA NA 0.010 0.010
48 0.132 0.000 NA NA 0.132 0.132
49 -0.029 0.000 NA NA -0.029 -0.029
50 0.027 0.000 NA NA 0.027 0.027
51 0.004 0.000 NA NA 0.004 0.004
52 0.065 0.000 NA NA 0.065 0.065
53 0.004 0.000 NA NA 0.004 0.004
54 0.058 0.000 NA NA 0.058 0.058
55 0.001 0.000 NA NA 0.001 0.001
56 0.044 0.000 NA NA 0.044 0.044
57 0.054 0.000 NA NA 0.054 0.054
58 -0.008 0.000 NA NA -0.008 -0.008
59 0.003 0.000 NA NA 0.003 0.003
60 -0.019 0.000 NA NA -0.019 -0.019
61 0.010 0.000 NA NA 0.010 0.010
62 -0.013 0.000 NA NA -0.013 -0.013
63 0.000 0.000 NA NA 0.000 0.000
64 -0.015 0.000 NA NA -0.015 -0.015
65 0.051 0.000 NA NA 0.051 0.051
66 0.002 0.000 NA NA 0.002 0.002
67 0.042 0.000 NA NA 0.042 0.042
68 0.018 0.000 NA NA 0.018 0.018
69 0.018 0.000 NA NA 0.018 0.018
70 0.000 0.000 NA NA 0.000 0.000
71 0.021 0.000 NA NA 0.021 0.021
72 0.047 0.000 NA NA 0.047 0.047
73 0.008 0.000 NA NA 0.008 0.008
74 -0.002 0.000 NA NA -0.002 -0.002
75 0.035 0.000 NA NA 0.035 0.035
76 -0.001 0.000 NA NA -0.001 -0.001
77 0.009 0.000 NA NA 0.009 0.009
78 0.213 0.000 NA NA 0.213 0.213
79 0.061 0.000 NA NA 0.061 0.061
80 0.061 0.000 NA NA 0.061 0.061
81 0.006 0.000 NA NA 0.006 0.006
82 0.044 0.000 NA NA 0.044 0.044
83 0.146 0.000 NA NA 0.146 0.146
84 0.035 0.000 NA NA 0.035 0.035
85 0.002 0.000 NA NA 0.002 0.002
86 0.003 0.000 NA NA 0.003 0.003
87 0.213 0.000 NA NA 0.213 0.213
88 0.003 0.000 NA NA 0.003 0.003
89 0.049 0.000 NA NA 0.049 0.049
90 0.005 0.000 NA NA 0.005 0.005
91 0.002 0.000 NA NA 0.002 0.002
92 0.062 0.000 NA NA 0.062 0.062
93 1.000 0.000 NA NA 1.000 1.000
94 0.000 0.000 NA NA 0.000 0.000
95 0.000 0.000 NA NA 0.000 0.000
96 0.262 0.000 NA NA 0.262 0.262
97 0.100 0.000 NA NA 0.100 0.100
98 0.225 0.000 NA NA 0.225 0.225
99 0.606 0.000 NA NA 0.606 0.606
100 0.196 0.000 NA NA 0.196 0.196
101 0.562 0.000 NA NA 0.562 0.562
102 0.300 0.000 NA NA 0.300 0.300
103 0.175 0.000 NA NA 0.175 0.175
104 0.300 0.000 NA NA 0.300 0.300
105 0.529 0.000 NA NA 0.529 0.529
106 0.124 0.000 NA NA 0.124 0.124
107 0.194 1.123 0.172 0.863 -2.007 2.395
108 1.502 2.429 0.618 0.536 -3.259 6.264
109 0.182 2.078 0.088 0.930 -3.891 4.255
110 -3.511 2.216 -1.584 0.113 -7.855 0.833
111 2.563 1.399 1.832 0.067 -0.179 5.306
112 0.063 0.095 0.656 0.512 -0.124 0.249
113 0.028 0.138 0.202 0.840 -0.243 0.298
114 -0.108 0.147 -0.738 0.460 -0.396 0.179
115 0.074 0.206 0.358 0.720 -0.331 0.478
116 -0.163 0.213 -0.767 0.443 -0.580 0.254
117 0.823 4.116 0.200 0.842 -7.244 8.889
label lhs edge rhs est std group
1 a Matsmk ~> Epi -3.256589e-02 -0.0143904486
2 b Matagg ~> Epi -1.446988e-02 -0.0043596534
3 c FamScore ~> Epi 5.638498e-02 0.0204664407
4 d EduPar ~> Epi -3.844427e-02 -0.0088987514
5 e n_trauma ~> Epi 8.486733e-02 0.0191918495
6 Age ~> Epi -9.011774e-02 -0.0195992540
7 int_dis ~> Epi -7.447596e-02 -0.0342761092
8 medication ~> Epi -4.445645e-02 -0.0169647132
9 contraceptives ~> Epi -1.677936e-02 -0.0077223731
10 V8 ~> Epi -8.888122e-02 -0.0060402510
11 cigday_1 ~> Epi -1.109172e-01 -0.0274559411
12 f Matsmk ~> group 1.936833e-01 0.0213188605
13 g Matagg ~> group 1.502114e+00 0.1127328103
14 h FamScore ~> group 1.819577e-01 0.0164516638
15 i EduPar ~> group -3.511101e+00 -0.2024423075
16 j n_trauma ~> group 2.563273e+00 0.1443881567
17 Age ~> group -3.055589e+00 -0.1655330454
18 int_dis ~> group 1.143258e+00 0.1310630080
19 medication ~> group 1.078602e+00 0.1025259005
20 contraceptives ~> group 1.080677e-01 0.0123888729
21 V8 ~> group 1.310041e+01 0.2217637334
22 cigday_1 ~> group 1.018628e+01 0.6280782422
23 z Epi ~> group -1.921485e+00 -0.4786273469
25 Epi <-> Epi 1.000000e+00 0.9960212963
26 group <-> group -2.692105e+00 -0.1663725243
27 Matsmk <-> Matsmk 1.960443e-01 1.0000000000
28 Matsmk <-> Matagg 4.936709e-02 0.3693241433
29 Matsmk <-> FamScore -9.177215e-03 -0.0569887592
30 Matsmk <-> EduPar -5.564346e-03 -0.0541843459
31 Matsmk <-> n_trauma 7.459313e-03 0.0743497863
32 Matsmk <-> Age -3.217300e-03 -0.0333441329
33 Matsmk <-> int_dis 2.151899e-02 0.1053910232
34 Matsmk <-> medication 4.113924e-03 0.0242997446
35 Matsmk <-> contraceptives 2.151899e-02 0.1053910232
36 Matsmk <-> V8 2.928139e-03 0.0971189320
37 Matsmk <-> cigday_1 1.693829e-02 0.1542372547
38 Matagg <-> Matagg 9.113924e-02 1.0000000000
39 Matagg <-> FamScore 3.417722e-02 0.3112715087
40 Matagg <-> EduPar -1.656118e-02 -0.2365241196
41 Matagg <-> n_trauma 7.233273e-03 0.1057402114
42 Matagg <-> Age 3.118694e-04 0.0047405101
43 Matagg <-> int_dis 3.291139e-02 0.2364027144
44 Matagg <-> medication 7.594937e-03 0.0657951695
45 Matagg <-> contraceptives 7.594937e-03 0.0545544726
46 Matagg <-> V8 8.272067e-04 0.0402393217
47 Matagg <-> cigday_1 1.018987e-02 0.1360858260
48 FamScore <-> FamScore 1.322785e-01 1.0000000000
49 FamScore <-> EduPar -2.948312e-02 -0.3495149022
50 FamScore <-> n_trauma 2.667269e-02 0.3236534989
51 FamScore <-> Age 3.636947e-03 0.0458878230
52 FamScore <-> int_dis 6.455696e-02 0.3849084009
53 FamScore <-> medication 4.430380e-03 0.0318580293
54 FamScore <-> contraceptives 5.822785e-02 0.3471722832
55 FamScore <-> V8 7.814844e-04 0.0315547719
56 FamScore <-> cigday_1 4.381329e-02 0.4856887960
57 EduPar <-> EduPar 5.379307e-02 1.0000000000
58 EduPar <-> n_trauma -8.024412e-03 -0.1526891136
59 EduPar <-> Age 2.762108e-03 0.0546490350
60 EduPar <-> int_dis -1.909283e-02 -0.1785114035
61 EduPar <-> medication 1.017932e-02 0.1147832062
62 EduPar <-> contraceptives -1.329114e-02 -0.1242676068
63 EduPar <-> V8 -8.860887e-06 -0.0005610523
64 EduPar <-> cigday_1 -1.493803e-02 -0.2596730517
65 n_trauma <-> n_trauma 5.134332e-02 1.0000000000
66 n_trauma <-> Age 1.582278e-03 0.0320439451
67 n_trauma <-> int_dis 4.159132e-02 0.3980335009
68 n_trauma <-> medication 1.763110e-02 0.2034979577
69 n_trauma <-> contraceptives 1.808318e-02 0.1730580439
70 n_trauma <-> V8 -4.694340e-04 -0.0304243917
71 n_trauma <-> cigday_1 2.128165e-02 0.3786692420
72 Age <-> Age 4.748866e-02 1.0000000000
73 Age <-> int_dis 8.090259e-03 0.0805056484
74 Age <-> medication -1.655660e-03 -0.0198700345
75 Age <-> contraceptives 3.524124e-02 0.3506833348
76 Age <-> V8 -1.333633e-03 -0.0898732659
77 Age <-> cigday_1 8.542355e-03 0.1580445206
78 int_dis <-> int_dis 2.126582e-01 1.0000000000
79 int_dis <-> medication 6.075949e-02 0.3445843938
80 int_dis <-> contraceptives 6.075949e-02 0.2857142857
81 int_dis <-> V8 5.645344e-03 0.1797788722
82 int_dis <-> cigday_1 4.449367e-02 0.3890038953
83 medication <-> medication 1.462025e-01 1.0000000000
84 medication <-> contraceptives 3.544304e-02 0.2010075631
85 medication <-> V8 2.084232e-03 0.0800493604
86 medication <-> cigday_1 3.275316e-03 0.0345360471
87 contraceptives <-> contraceptives 2.126582e-01 1.0000000000
88 contraceptives <-> V8 2.688833e-03 0.0856272484
89 contraceptives <-> cigday_1 4.892405e-02 0.4277382803
90 V8 <-> V8 4.636832e-03 1.0000000000
91 V8 <-> cigday_1 1.509276e-03 0.0893623867
92 cigday_1 <-> cigday_1 6.151859e-02 1.0000000000
93 group <-> group 1.000000e+00 1.0000000000
94 int Epi 0.000000e+00 0.0000000000
95 int group 0.000000e+00 0.0000000000
96 int Matsmk 2.625000e-01 0.5928600601
97 int Matagg 1.000000e-01 0.3312434486
98 int FamScore 2.250000e-01 0.6186398880
99 int EduPar 6.062500e-01 2.6138976225
100 int n_trauma 1.964286e-01 0.8668873691
101 int Age 5.621377e-01 2.5795724974
102 int int_dis 3.000000e-01 0.6505492185
103 int medication 1.750000e-01 0.4576785957
104 int contraceptives 3.000000e-01 0.6505492185
105 int V8 5.286908e-01 7.7640983340
106 int cigday_1 1.243750e-01 0.5014526157
fixed par
1 FALSE 1
2 FALSE 2
3 FALSE 3
4 FALSE 4
5 FALSE 5
6 FALSE 6
7 FALSE 7
8 FALSE 8
9 FALSE 9
10 FALSE 10
11 FALSE 11
12 FALSE 12
13 FALSE 13
14 FALSE 14
15 FALSE 15
16 FALSE 16
17 FALSE 17
18 FALSE 18
19 FALSE 19
20 FALSE 20
21 FALSE 21
22 FALSE 22
23 FALSE 23
25 TRUE 0
26 TRUE 0
27 TRUE 0
28 TRUE 0
29 TRUE 0
30 TRUE 0
31 TRUE 0
32 TRUE 0
33 TRUE 0
34 TRUE 0
35 TRUE 0
36 TRUE 0
37 TRUE 0
38 TRUE 0
39 TRUE 0
40 TRUE 0
41 TRUE 0
42 TRUE 0
43 TRUE 0
44 TRUE 0
45 TRUE 0
46 TRUE 0
47 TRUE 0
48 TRUE 0
49 TRUE 0
50 TRUE 0
51 TRUE 0
52 TRUE 0
53 TRUE 0
54 TRUE 0
55 TRUE 0
56 TRUE 0
57 TRUE 0
58 TRUE 0
59 TRUE 0
60 TRUE 0
61 TRUE 0
62 TRUE 0
63 TRUE 0
64 TRUE 0
65 TRUE 0
66 TRUE 0
67 TRUE 0
68 TRUE 0
69 TRUE 0
70 TRUE 0
71 TRUE 0
72 TRUE 0
73 TRUE 0
74 TRUE 0
75 TRUE 0
76 TRUE 0
77 TRUE 0
78 TRUE 0
79 TRUE 0
80 TRUE 0
81 TRUE 0
82 TRUE 0
83 TRUE 0
84 TRUE 0
85 TRUE 0
86 TRUE 0
87 TRUE 0
88 TRUE 0
89 TRUE 0
90 TRUE 0
91 TRUE 0
92 TRUE 0
93 TRUE 0
94 TRUE 0
95 TRUE 0
96 TRUE 0
97 TRUE 0
98 TRUE 0
99 TRUE 0
100 TRUE 0
101 TRUE 0
102 TRUE 0
103 TRUE 0
104 TRUE 0
105 TRUE 0
106 TRUE 0Warning in lav_samplestats_step2(UNI = FIT, wt = wt, ov.names = ov.names, :
lavaan WARNING: correlation between variables group and Epi is (nearly) 1.0
Warning in lav_samplestats_step2(UNI = FIT, wt = wt, ov.names = ov.names, :
lavaan WARNING: some estimated ov variances are negative############################
############################
Epi_all
############################
############################
##Mediation Model ##
lavaan 0.6-7 ended normally after 114 iterations
Estimator DWLS
Optimization method NLMINB
Number of free parameters 23
Used Total
Number of observations 80 99
Model Test User Model:
Standard Robust
Test Statistic 27.282 24.091
Degrees of freedom 3 3
P-value (Chi-square) 0.000 0.000
Scaling correction factor 1.151
Shift parameter 0.394
simple second-order correction
Parameter Estimates:
Standard errors Robust.sem
Information Expected
Information saturated (h1) model Unstructured
Regressions:
Estimate Std.Err z-value P(>|z|)
Epi ~
Matsmk (a) 0.014 0.025 0.573 0.567
Matagg (b) 0.023 0.038 0.590 0.555
FamScore (c) -0.037 0.042 -0.871 0.384
EduPar (d) -0.090 0.060 -1.514 0.130
n_trauma (e) 0.025 0.048 0.523 0.601
Age 0.034 0.064 0.534 0.593
int_dis 0.023 0.024 0.934 0.350
medication 0.007 0.030 0.248 0.804
contrcptvs -0.016 0.027 -0.583 0.560
V8 0.060 0.306 0.197 0.844
cigday_1 0.022 0.052 0.425 0.671
group ~
Matsmk (f) 0.236 1.139 0.207 0.836
Matagg (g) 1.498 2.470 0.607 0.544
FamScore (h) 0.125 2.069 0.060 0.952
EduPar (i) -3.311 2.205 -1.502 0.133
n_trauma (j) 2.365 1.396 1.694 0.090
Age -2.930 2.463 -1.190 0.234
int_dis 1.254 0.801 1.566 0.117
medication 1.154 0.840 1.373 0.170
contrcptvs 0.162 0.774 0.210 0.834
V8 13.187 16.629 0.793 0.428
cigday_1 10.369 7.189 1.442 0.149
Epi (z) 1.393 0.134 10.435 0.000
Intercepts:
Estimate Std.Err z-value P(>|z|)
.Epi 0.000
.group 0.000
Thresholds:
Estimate Std.Err z-value P(>|z|)
group|t1 5.748
Variances:
Estimate Std.Err z-value P(>|z|)
.Epi 1.000
.group -0.942
Scales y*:
Estimate Std.Err z-value P(>|z|)
group 1.000
Defined Parameters:
Estimate Std.Err z-value P(>|z|)
directMatsmk 0.236 1.139 0.207 0.836
directMatagg 1.498 2.470 0.607 0.544
directFamScore 0.125 2.069 0.060 0.952
directEduPar -3.311 2.205 -1.502 0.133
directn_trauma 2.365 1.396 1.694 0.090
EpiMatsmk 0.020 0.035 0.572 0.567
EpiMatagg 0.032 0.054 0.590 0.556
EpiFamScore -0.051 0.059 -0.868 0.385
EpiEduPar -0.126 0.084 -1.501 0.133
Epin_trauma 0.035 0.067 0.522 0.602
total 0.823 4.116 0.200 0.842
npar fmin
23.000 0.171
chisq df
27.282 3.000
pvalue chisq.scaled
0.000 24.091
df.scaled pvalue.scaled
3.000 0.000
chisq.scaling.factor baseline.chisq
1.151 0.358
baseline.df baseline.pvalue
1.000 0.549
baseline.chisq.scaled baseline.df.scaled
0.358 1.000
baseline.pvalue.scaled baseline.chisq.scaling.factor
0.549 1.000
cfi tli
0.000 13.613
nnfi rfi
13.613 NA
nfi pnfi
NA -225.435
ifi rni
1.000 38.839
cfi.scaled tli.scaled
0.000 11.956
cfi.robust tli.robust
NA NA
nnfi.scaled nnfi.robust
11.956 NA
rfi.scaled nfi.scaled
NA NA
ifi.scaled rni.scaled
1.000 33.867
rni.robust rmsea
NA 0.320
rmsea.ci.lower rmsea.ci.upper
0.217 0.435
rmsea.pvalue rmsea.scaled
0.000 0.298
rmsea.ci.lower.scaled rmsea.ci.upper.scaled
0.195 0.414
rmsea.pvalue.scaled rmsea.robust
0.000 NA
rmsea.ci.lower.robust rmsea.ci.upper.robust
NA NA
rmsea.pvalue.robust rmr
NA 0.739
rmr_nomean srmr
0.951 89.392
srmr_bentler srmr_bentler_nomean
69.246 89.392
crmr crmr_nomean
0.990 0.398
srmr_mplus srmr_mplus_nomean
NA NA
cn_05 cn_01
23.629 33.852
gfi agfi
-13172.670 -114170.808
pgfi mfi
-1519.923 0.858
lhs op rhs label
1 Epi ~ Matsmk a
2 Epi ~ Matagg b
3 Epi ~ FamScore c
4 Epi ~ EduPar d
5 Epi ~ n_trauma e
6 Epi ~ Age
7 Epi ~ int_dis
8 Epi ~ medication
9 Epi ~ contraceptives
10 Epi ~ V8
11 Epi ~ cigday_1
12 group ~ Matsmk f
13 group ~ Matagg g
14 group ~ FamScore h
15 group ~ EduPar i
16 group ~ n_trauma j
17 group ~ Age
18 group ~ int_dis
19 group ~ medication
20 group ~ contraceptives
21 group ~ V8
22 group ~ cigday_1
23 group ~ Epi z
24 group | t1
25 Epi ~~ Epi
26 group ~~ group
27 Matsmk ~~ Matsmk
28 Matsmk ~~ Matagg
29 Matsmk ~~ FamScore
30 Matsmk ~~ EduPar
31 Matsmk ~~ n_trauma
32 Matsmk ~~ Age
33 Matsmk ~~ int_dis
34 Matsmk ~~ medication
35 Matsmk ~~ contraceptives
36 Matsmk ~~ V8
37 Matsmk ~~ cigday_1
38 Matagg ~~ Matagg
39 Matagg ~~ FamScore
40 Matagg ~~ EduPar
41 Matagg ~~ n_trauma
42 Matagg ~~ Age
43 Matagg ~~ int_dis
44 Matagg ~~ medication
45 Matagg ~~ contraceptives
46 Matagg ~~ V8
47 Matagg ~~ cigday_1
48 FamScore ~~ FamScore
49 FamScore ~~ EduPar
50 FamScore ~~ n_trauma
51 FamScore ~~ Age
52 FamScore ~~ int_dis
53 FamScore ~~ medication
54 FamScore ~~ contraceptives
55 FamScore ~~ V8
56 FamScore ~~ cigday_1
57 EduPar ~~ EduPar
58 EduPar ~~ n_trauma
59 EduPar ~~ Age
60 EduPar ~~ int_dis
61 EduPar ~~ medication
62 EduPar ~~ contraceptives
63 EduPar ~~ V8
64 EduPar ~~ cigday_1
65 n_trauma ~~ n_trauma
66 n_trauma ~~ Age
67 n_trauma ~~ int_dis
68 n_trauma ~~ medication
69 n_trauma ~~ contraceptives
70 n_trauma ~~ V8
71 n_trauma ~~ cigday_1
72 Age ~~ Age
73 Age ~~ int_dis
74 Age ~~ medication
75 Age ~~ contraceptives
76 Age ~~ V8
77 Age ~~ cigday_1
78 int_dis ~~ int_dis
79 int_dis ~~ medication
80 int_dis ~~ contraceptives
81 int_dis ~~ V8
82 int_dis ~~ cigday_1
83 medication ~~ medication
84 medication ~~ contraceptives
85 medication ~~ V8
86 medication ~~ cigday_1
87 contraceptives ~~ contraceptives
88 contraceptives ~~ V8
89 contraceptives ~~ cigday_1
90 V8 ~~ V8
91 V8 ~~ cigday_1
92 cigday_1 ~~ cigday_1
93 group ~*~ group
94 Epi ~1
95 group ~1
96 Matsmk ~1
97 Matagg ~1
98 FamScore ~1
99 EduPar ~1
100 n_trauma ~1
101 Age ~1
102 int_dis ~1
103 medication ~1
104 contraceptives ~1
105 V8 ~1
106 cigday_1 ~1
107 directMatsmk := f directMatsmk
108 directMatagg := g directMatagg
109 directFamScore := h directFamScore
110 directEduPar := i directEduPar
111 directn_trauma := j directn_trauma
112 EpiMatsmk := a*z EpiMatsmk
113 EpiMatagg := b*z EpiMatagg
114 EpiFamScore := c*z EpiFamScore
115 EpiEduPar := d*z EpiEduPar
116 Epin_trauma := e*z Epin_trauma
117 total := f+g+h+i+j+(a*z)+(b*z)+(c*z)+(d*z)+(e*z) total
est se z pvalue ci.lower ci.upper
1 0.014 0.025 0.573 0.567 -0.035 0.064
2 0.023 0.038 0.590 0.555 -0.053 0.098
3 -0.037 0.042 -0.871 0.384 -0.119 0.046
4 -0.090 0.060 -1.514 0.130 -0.208 0.027
5 0.025 0.048 0.523 0.601 -0.069 0.119
6 0.034 0.064 0.534 0.593 -0.091 0.160
7 0.023 0.024 0.934 0.350 -0.025 0.071
8 0.007 0.030 0.248 0.804 -0.051 0.066
9 -0.016 0.027 -0.583 0.560 -0.069 0.037
10 0.060 0.306 0.197 0.844 -0.540 0.660
11 0.022 0.052 0.425 0.671 -0.079 0.123
12 0.236 1.139 0.207 0.836 -1.997 2.469
13 1.498 2.470 0.607 0.544 -3.343 6.339
14 0.125 2.069 0.060 0.952 -3.930 4.179
15 -3.311 2.205 -1.502 0.133 -7.632 1.010
16 2.365 1.396 1.694 0.090 -0.371 5.101
17 -2.930 2.463 -1.190 0.234 -7.757 1.897
18 1.254 0.801 1.566 0.117 -0.316 2.825
19 1.154 0.840 1.373 0.170 -0.493 2.801
20 0.162 0.774 0.210 0.834 -1.356 1.680
21 13.187 16.629 0.793 0.428 -19.405 45.780
22 10.369 7.189 1.442 0.149 -3.721 24.458
23 1.393 0.134 10.435 0.000 1.132 1.655
24 5.748 0.000 NA NA 5.748 5.748
25 1.000 0.000 NA NA 1.000 1.000
26 -0.942 0.000 NA NA -0.942 -0.942
27 0.196 0.000 NA NA 0.196 0.196
28 0.049 0.000 NA NA 0.049 0.049
29 -0.009 0.000 NA NA -0.009 -0.009
30 -0.006 0.000 NA NA -0.006 -0.006
31 0.007 0.000 NA NA 0.007 0.007
32 -0.003 0.000 NA NA -0.003 -0.003
33 0.022 0.000 NA NA 0.022 0.022
34 0.004 0.000 NA NA 0.004 0.004
35 0.022 0.000 NA NA 0.022 0.022
36 0.003 0.000 NA NA 0.003 0.003
37 0.017 0.000 NA NA 0.017 0.017
38 0.091 0.000 NA NA 0.091 0.091
39 0.034 0.000 NA NA 0.034 0.034
40 -0.017 0.000 NA NA -0.017 -0.017
41 0.007 0.000 NA NA 0.007 0.007
42 0.000 0.000 NA NA 0.000 0.000
43 0.033 0.000 NA NA 0.033 0.033
44 0.008 0.000 NA NA 0.008 0.008
45 0.008 0.000 NA NA 0.008 0.008
46 0.001 0.000 NA NA 0.001 0.001
47 0.010 0.000 NA NA 0.010 0.010
48 0.132 0.000 NA NA 0.132 0.132
49 -0.029 0.000 NA NA -0.029 -0.029
50 0.027 0.000 NA NA 0.027 0.027
51 0.004 0.000 NA NA 0.004 0.004
52 0.065 0.000 NA NA 0.065 0.065
53 0.004 0.000 NA NA 0.004 0.004
54 0.058 0.000 NA NA 0.058 0.058
55 0.001 0.000 NA NA 0.001 0.001
56 0.044 0.000 NA NA 0.044 0.044
57 0.054 0.000 NA NA 0.054 0.054
58 -0.008 0.000 NA NA -0.008 -0.008
59 0.003 0.000 NA NA 0.003 0.003
60 -0.019 0.000 NA NA -0.019 -0.019
61 0.010 0.000 NA NA 0.010 0.010
62 -0.013 0.000 NA NA -0.013 -0.013
63 0.000 0.000 NA NA 0.000 0.000
64 -0.015 0.000 NA NA -0.015 -0.015
65 0.051 0.000 NA NA 0.051 0.051
66 0.002 0.000 NA NA 0.002 0.002
67 0.042 0.000 NA NA 0.042 0.042
68 0.018 0.000 NA NA 0.018 0.018
69 0.018 0.000 NA NA 0.018 0.018
70 0.000 0.000 NA NA 0.000 0.000
71 0.021 0.000 NA NA 0.021 0.021
72 0.047 0.000 NA NA 0.047 0.047
73 0.008 0.000 NA NA 0.008 0.008
74 -0.002 0.000 NA NA -0.002 -0.002
75 0.035 0.000 NA NA 0.035 0.035
76 -0.001 0.000 NA NA -0.001 -0.001
77 0.009 0.000 NA NA 0.009 0.009
78 0.213 0.000 NA NA 0.213 0.213
79 0.061 0.000 NA NA 0.061 0.061
80 0.061 0.000 NA NA 0.061 0.061
81 0.006 0.000 NA NA 0.006 0.006
82 0.044 0.000 NA NA 0.044 0.044
83 0.146 0.000 NA NA 0.146 0.146
84 0.035 0.000 NA NA 0.035 0.035
85 0.002 0.000 NA NA 0.002 0.002
86 0.003 0.000 NA NA 0.003 0.003
87 0.213 0.000 NA NA 0.213 0.213
88 0.003 0.000 NA NA 0.003 0.003
89 0.049 0.000 NA NA 0.049 0.049
90 0.005 0.000 NA NA 0.005 0.005
91 0.002 0.000 NA NA 0.002 0.002
92 0.062 0.000 NA NA 0.062 0.062
93 1.000 0.000 NA NA 1.000 1.000
94 0.000 0.000 NA NA 0.000 0.000
95 0.000 0.000 NA NA 0.000 0.000
96 0.262 0.000 NA NA 0.262 0.262
97 0.100 0.000 NA NA 0.100 0.100
98 0.225 0.000 NA NA 0.225 0.225
99 0.606 0.000 NA NA 0.606 0.606
100 0.196 0.000 NA NA 0.196 0.196
101 0.562 0.000 NA NA 0.562 0.562
102 0.300 0.000 NA NA 0.300 0.300
103 0.175 0.000 NA NA 0.175 0.175
104 0.300 0.000 NA NA 0.300 0.300
105 0.529 0.000 NA NA 0.529 0.529
106 0.124 0.000 NA NA 0.124 0.124
107 0.236 1.139 0.207 0.836 -1.997 2.469
108 1.498 2.470 0.607 0.544 -3.343 6.339
109 0.125 2.069 0.060 0.952 -3.930 4.179
110 -3.311 2.205 -1.502 0.133 -7.632 1.010
111 2.365 1.396 1.694 0.090 -0.371 5.101
112 0.020 0.035 0.572 0.567 -0.049 0.089
113 0.032 0.054 0.590 0.556 -0.073 0.137
114 -0.051 0.059 -0.868 0.385 -0.166 0.064
115 -0.126 0.084 -1.501 0.133 -0.291 0.039
116 0.035 0.067 0.522 0.602 -0.096 0.166
117 0.823 4.116 0.200 0.842 -7.244 8.889
label lhs edge rhs est std group
1 a Matsmk ~> Epi 1.438673e-02 0.0063670252
2 b Matagg ~> Epi 2.267876e-02 0.0068433600
3 c FamScore ~> Epi -3.652791e-02 -0.0132790452
4 d EduPar ~> Epi -9.048835e-02 -0.0209774780
5 e n_trauma ~> Epi 2.508125e-02 0.0056805267
6 Age ~> Epi 3.417168e-02 0.0074431841
7 int_dis ~> Epi 2.287155e-02 0.0105422718
8 medication ~> Epi 7.364844e-03 0.0028147411
9 contraceptives ~> Epi -1.577718e-02 -0.0072722355
10 V8 ~> Epi 6.028764e-02 0.0041033295
11 cigday_1 ~> Epi 2.200542e-02 0.0054554452
12 f Matsmk ~> group 2.362105e-01 0.0259998632
13 g Matagg ~> group 1.498314e+00 0.1124475905
14 h FamScore ~> group 1.245177e-01 0.0112582378
15 i EduPar ~> group -3.311137e+00 -0.1909128192
16 j n_trauma ~> group 2.365251e+00 0.1332336693
17 Age ~> group -2.930048e+00 -0.1587319634
18 int_dis ~> group 1.254491e+00 0.1438147824
19 medication ~> group 1.153762e+00 0.1096701467
20 contraceptives ~> group 1.622942e-01 0.0186053926
21 V8 ~> group 1.318719e+01 0.2232327021
22 cigday_1 ~> group 1.036875e+01 0.6393286206
23 z Epi ~> group 1.393481e+00 0.3465760226
25 Epi <-> Epi 1.000000e+00 0.9990675738
26 group <-> group -9.417906e-01 -0.0582027947
27 Matsmk <-> Matsmk 1.960443e-01 1.0000000000
28 Matsmk <-> Matagg 4.936709e-02 0.3693241433
29 Matsmk <-> FamScore -9.177215e-03 -0.0569887592
30 Matsmk <-> EduPar -5.564346e-03 -0.0541843459
31 Matsmk <-> n_trauma 7.459313e-03 0.0743497863
32 Matsmk <-> Age -3.217300e-03 -0.0333441329
33 Matsmk <-> int_dis 2.151899e-02 0.1053910232
34 Matsmk <-> medication 4.113924e-03 0.0242997446
35 Matsmk <-> contraceptives 2.151899e-02 0.1053910232
36 Matsmk <-> V8 2.928139e-03 0.0971189320
37 Matsmk <-> cigday_1 1.693829e-02 0.1542372547
38 Matagg <-> Matagg 9.113924e-02 1.0000000000
39 Matagg <-> FamScore 3.417722e-02 0.3112715087
40 Matagg <-> EduPar -1.656118e-02 -0.2365241196
41 Matagg <-> n_trauma 7.233273e-03 0.1057402114
42 Matagg <-> Age 3.118694e-04 0.0047405101
43 Matagg <-> int_dis 3.291139e-02 0.2364027144
44 Matagg <-> medication 7.594937e-03 0.0657951695
45 Matagg <-> contraceptives 7.594937e-03 0.0545544726
46 Matagg <-> V8 8.272067e-04 0.0402393217
47 Matagg <-> cigday_1 1.018987e-02 0.1360858260
48 FamScore <-> FamScore 1.322785e-01 1.0000000000
49 FamScore <-> EduPar -2.948312e-02 -0.3495149022
50 FamScore <-> n_trauma 2.667269e-02 0.3236534989
51 FamScore <-> Age 3.636947e-03 0.0458878230
52 FamScore <-> int_dis 6.455696e-02 0.3849084009
53 FamScore <-> medication 4.430380e-03 0.0318580293
54 FamScore <-> contraceptives 5.822785e-02 0.3471722832
55 FamScore <-> V8 7.814844e-04 0.0315547719
56 FamScore <-> cigday_1 4.381329e-02 0.4856887960
57 EduPar <-> EduPar 5.379307e-02 1.0000000000
58 EduPar <-> n_trauma -8.024412e-03 -0.1526891136
59 EduPar <-> Age 2.762108e-03 0.0546490350
60 EduPar <-> int_dis -1.909283e-02 -0.1785114035
61 EduPar <-> medication 1.017932e-02 0.1147832062
62 EduPar <-> contraceptives -1.329114e-02 -0.1242676068
63 EduPar <-> V8 -8.860887e-06 -0.0005610523
64 EduPar <-> cigday_1 -1.493803e-02 -0.2596730517
65 n_trauma <-> n_trauma 5.134332e-02 1.0000000000
66 n_trauma <-> Age 1.582278e-03 0.0320439451
67 n_trauma <-> int_dis 4.159132e-02 0.3980335009
68 n_trauma <-> medication 1.763110e-02 0.2034979577
69 n_trauma <-> contraceptives 1.808318e-02 0.1730580439
70 n_trauma <-> V8 -4.694340e-04 -0.0304243917
71 n_trauma <-> cigday_1 2.128165e-02 0.3786692420
72 Age <-> Age 4.748866e-02 1.0000000000
73 Age <-> int_dis 8.090259e-03 0.0805056484
74 Age <-> medication -1.655660e-03 -0.0198700345
75 Age <-> contraceptives 3.524124e-02 0.3506833348
76 Age <-> V8 -1.333633e-03 -0.0898732659
77 Age <-> cigday_1 8.542355e-03 0.1580445206
78 int_dis <-> int_dis 2.126582e-01 1.0000000000
79 int_dis <-> medication 6.075949e-02 0.3445843938
80 int_dis <-> contraceptives 6.075949e-02 0.2857142857
81 int_dis <-> V8 5.645344e-03 0.1797788722
82 int_dis <-> cigday_1 4.449367e-02 0.3890038953
83 medication <-> medication 1.462025e-01 1.0000000000
84 medication <-> contraceptives 3.544304e-02 0.2010075631
85 medication <-> V8 2.084232e-03 0.0800493604
86 medication <-> cigday_1 3.275316e-03 0.0345360471
87 contraceptives <-> contraceptives 2.126582e-01 1.0000000000
88 contraceptives <-> V8 2.688833e-03 0.0856272484
89 contraceptives <-> cigday_1 4.892405e-02 0.4277382803
90 V8 <-> V8 4.636832e-03 1.0000000000
91 V8 <-> cigday_1 1.509276e-03 0.0893623867
92 cigday_1 <-> cigday_1 6.151859e-02 1.0000000000
93 group <-> group 1.000000e+00 1.0000000000
94 int Epi 0.000000e+00 0.0000000000
95 int group 0.000000e+00 0.0000000000
96 int Matsmk 2.625000e-01 0.5928600601
97 int Matagg 1.000000e-01 0.3312434486
98 int FamScore 2.250000e-01 0.6186398880
99 int EduPar 6.062500e-01 2.6138976225
100 int n_trauma 1.964286e-01 0.8668873691
101 int Age 5.621377e-01 2.5795724974
102 int int_dis 3.000000e-01 0.6505492185
103 int medication 1.750000e-01 0.4576785957
104 int contraceptives 3.000000e-01 0.6505492185
105 int V8 5.286908e-01 7.7640983340
106 int cigday_1 1.243750e-01 0.5014526157
fixed par
1 FALSE 1
2 FALSE 2
3 FALSE 3
4 FALSE 4
5 FALSE 5
6 FALSE 6
7 FALSE 7
8 FALSE 8
9 FALSE 9
10 FALSE 10
11 FALSE 11
12 FALSE 12
13 FALSE 13
14 FALSE 14
15 FALSE 15
16 FALSE 16
17 FALSE 17
18 FALSE 18
19 FALSE 19
20 FALSE 20
21 FALSE 21
22 FALSE 22
23 FALSE 23
25 TRUE 0
26 TRUE 0
27 TRUE 0
28 TRUE 0
29 TRUE 0
30 TRUE 0
31 TRUE 0
32 TRUE 0
33 TRUE 0
34 TRUE 0
35 TRUE 0
36 TRUE 0
37 TRUE 0
38 TRUE 0
39 TRUE 0
40 TRUE 0
41 TRUE 0
42 TRUE 0
43 TRUE 0
44 TRUE 0
45 TRUE 0
46 TRUE 0
47 TRUE 0
48 TRUE 0
49 TRUE 0
50 TRUE 0
51 TRUE 0
52 TRUE 0
53 TRUE 0
54 TRUE 0
55 TRUE 0
56 TRUE 0
57 TRUE 0
58 TRUE 0
59 TRUE 0
60 TRUE 0
61 TRUE 0
62 TRUE 0
63 TRUE 0
64 TRUE 0
65 TRUE 0
66 TRUE 0
67 TRUE 0
68 TRUE 0
69 TRUE 0
70 TRUE 0
71 TRUE 0
72 TRUE 0
73 TRUE 0
74 TRUE 0
75 TRUE 0
76 TRUE 0
77 TRUE 0
78 TRUE 0
79 TRUE 0
80 TRUE 0
81 TRUE 0
82 TRUE 0
83 TRUE 0
84 TRUE 0
85 TRUE 0
86 TRUE 0
87 TRUE 0
88 TRUE 0
89 TRUE 0
90 TRUE 0
91 TRUE 0
92 TRUE 0
93 TRUE 0
94 TRUE 0
95 TRUE 0
96 TRUE 0
97 TRUE 0
98 TRUE 0
99 TRUE 0
100 TRUE 0
101 TRUE 0
102 TRUE 0
103 TRUE 0
104 TRUE 0
105 TRUE 0
106 TRUE 0Warning in lav_object_post_check(object): lavaan WARNING: some estimated ov
variances are negative############################
############################
Epi_M2
############################
############################
##Mediation Model ##
lavaan 0.6-7 ended normally after 108 iterations
Estimator DWLS
Optimization method NLMINB
Number of free parameters 23
Used Total
Number of observations 80 99
Model Test User Model:
Standard Robust
Test Statistic 257.012 248.644
Degrees of freedom 3 3
P-value (Chi-square) 0.000 0.000
Scaling correction factor 1.034
Shift parameter 0.099
simple second-order correction
Parameter Estimates:
Standard errors Robust.sem
Information Expected
Information saturated (h1) model Unstructured
Regressions:
Estimate Std.Err z-value P(>|z|)
Epi ~
Matsmk (a) 0.009 0.042 0.217 0.829
Matagg (b) -0.017 0.064 -0.261 0.794
FamScore (c) -0.054 0.063 -0.853 0.393
EduPar (d) -0.010 0.092 -0.109 0.913
n_trauma (e) 0.018 0.096 0.188 0.851
Age -0.042 0.101 -0.416 0.677
int_dis -0.010 0.038 -0.255 0.799
medication 0.018 0.039 0.453 0.651
contrcptvs 0.074 0.053 1.390 0.165
V8 -0.522 0.239 -2.187 0.029
cigday_1 -0.058 0.091 -0.641 0.522
group ~
Matsmk (f) 0.308 1.115 0.276 0.782
Matagg (g) 1.435 2.312 0.621 0.535
FamScore (h) -0.234 1.945 -0.120 0.904
EduPar (i) -3.495 2.188 -1.597 0.110
n_trauma (j) 2.505 1.290 1.942 0.052
Age -3.123 2.418 -1.292 0.196
int_dis 1.231 0.767 1.605 0.109
medication 1.267 0.812 1.560 0.119
contrcptvs 0.566 0.713 0.794 0.427
V8 10.274 16.690 0.616 0.538
cigday_1 10.065 7.155 1.407 0.160
Epi (z) -5.741 0.009 -669.813 0.000
Intercepts:
Estimate Std.Err z-value P(>|z|)
.Epi 0.000
.group 0.000
Thresholds:
Estimate Std.Err z-value P(>|z|)
group|t1 5.748
Variances:
Estimate Std.Err z-value P(>|z|)
.Epi 1.000
.group -31.957
Scales y*:
Estimate Std.Err z-value P(>|z|)
group 1.000
Defined Parameters:
Estimate Std.Err z-value P(>|z|)
directMatsmk 0.308 1.115 0.276 0.782
directMatagg 1.435 2.312 0.621 0.535
directFamScore -0.234 1.945 -0.120 0.904
directEduPar -3.495 2.188 -1.597 0.110
directn_trauma 2.505 1.290 1.942 0.052
EpiMatsmk -0.052 0.239 -0.217 0.829
EpiMatagg 0.095 0.365 0.261 0.794
EpiFamScore 0.308 0.361 0.853 0.393
EpiEduPar 0.058 0.526 0.110 0.913
Epin_trauma -0.104 0.554 -0.188 0.851
total 0.823 4.116 0.200 0.842
npar fmin
23.000 1.606
chisq df
257.012 3.000
pvalue chisq.scaled
0.000 248.644
df.scaled pvalue.scaled
3.000 0.000
chisq.scaling.factor baseline.chisq
1.034 171.816
baseline.df baseline.pvalue
1.000 0.000
baseline.chisq.scaled baseline.df.scaled
171.816 1.000
baseline.pvalue.scaled baseline.chisq.scaling.factor
0.000 1.000
cfi tli
0.000 0.504
nnfi rfi
0.504 NA
nfi pnfi
NA -1.488
ifi rni
-0.505 -0.487
cfi.scaled tli.scaled
0.000 0.521
cfi.robust tli.robust
NA NA
nnfi.scaled nnfi.robust
0.521 NA
rfi.scaled nfi.scaled
NA NA
ifi.scaled rni.scaled
-0.455 -0.438
rni.robust rmsea
NA 1.035
rmsea.ci.lower rmsea.ci.upper
0.930 1.144
rmsea.pvalue rmsea.scaled
0.000 1.018
rmsea.ci.lower.scaled rmsea.ci.upper.scaled
0.913 1.127
rmsea.pvalue.scaled rmsea.robust
0.000 NA
rmsea.ci.lower.robust rmsea.ci.upper.robust
NA NA
rmsea.pvalue.robust rmr
NA 2.586
rmr_nomean srmr
3.299 52.490
srmr_bentler srmr_bentler_nomean
40.808 52.490
crmr crmr_nomean
5.267 4.753
srmr_mplus srmr_mplus_nomean
NA NA
cn_05 cn_01
3.402 4.487
gfi agfi
-2488.184 -21571.926
pgfi mfi
-287.098 0.200
lhs op rhs label
1 Epi ~ Matsmk a
2 Epi ~ Matagg b
3 Epi ~ FamScore c
4 Epi ~ EduPar d
5 Epi ~ n_trauma e
6 Epi ~ Age
7 Epi ~ int_dis
8 Epi ~ medication
9 Epi ~ contraceptives
10 Epi ~ V8
11 Epi ~ cigday_1
12 group ~ Matsmk f
13 group ~ Matagg g
14 group ~ FamScore h
15 group ~ EduPar i
16 group ~ n_trauma j
17 group ~ Age
18 group ~ int_dis
19 group ~ medication
20 group ~ contraceptives
21 group ~ V8
22 group ~ cigday_1
23 group ~ Epi z
24 group | t1
25 Epi ~~ Epi
26 group ~~ group
27 Matsmk ~~ Matsmk
28 Matsmk ~~ Matagg
29 Matsmk ~~ FamScore
30 Matsmk ~~ EduPar
31 Matsmk ~~ n_trauma
32 Matsmk ~~ Age
33 Matsmk ~~ int_dis
34 Matsmk ~~ medication
35 Matsmk ~~ contraceptives
36 Matsmk ~~ V8
37 Matsmk ~~ cigday_1
38 Matagg ~~ Matagg
39 Matagg ~~ FamScore
40 Matagg ~~ EduPar
41 Matagg ~~ n_trauma
42 Matagg ~~ Age
43 Matagg ~~ int_dis
44 Matagg ~~ medication
45 Matagg ~~ contraceptives
46 Matagg ~~ V8
47 Matagg ~~ cigday_1
48 FamScore ~~ FamScore
49 FamScore ~~ EduPar
50 FamScore ~~ n_trauma
51 FamScore ~~ Age
52 FamScore ~~ int_dis
53 FamScore ~~ medication
54 FamScore ~~ contraceptives
55 FamScore ~~ V8
56 FamScore ~~ cigday_1
57 EduPar ~~ EduPar
58 EduPar ~~ n_trauma
59 EduPar ~~ Age
60 EduPar ~~ int_dis
61 EduPar ~~ medication
62 EduPar ~~ contraceptives
63 EduPar ~~ V8
64 EduPar ~~ cigday_1
65 n_trauma ~~ n_trauma
66 n_trauma ~~ Age
67 n_trauma ~~ int_dis
68 n_trauma ~~ medication
69 n_trauma ~~ contraceptives
70 n_trauma ~~ V8
71 n_trauma ~~ cigday_1
72 Age ~~ Age
73 Age ~~ int_dis
74 Age ~~ medication
75 Age ~~ contraceptives
76 Age ~~ V8
77 Age ~~ cigday_1
78 int_dis ~~ int_dis
79 int_dis ~~ medication
80 int_dis ~~ contraceptives
81 int_dis ~~ V8
82 int_dis ~~ cigday_1
83 medication ~~ medication
84 medication ~~ contraceptives
85 medication ~~ V8
86 medication ~~ cigday_1
87 contraceptives ~~ contraceptives
88 contraceptives ~~ V8
89 contraceptives ~~ cigday_1
90 V8 ~~ V8
91 V8 ~~ cigday_1
92 cigday_1 ~~ cigday_1
93 group ~*~ group
94 Epi ~1
95 group ~1
96 Matsmk ~1
97 Matagg ~1
98 FamScore ~1
99 EduPar ~1
100 n_trauma ~1
101 Age ~1
102 int_dis ~1
103 medication ~1
104 contraceptives ~1
105 V8 ~1
106 cigday_1 ~1
107 directMatsmk := f directMatsmk
108 directMatagg := g directMatagg
109 directFamScore := h directFamScore
110 directEduPar := i directEduPar
111 directn_trauma := j directn_trauma
112 EpiMatsmk := a*z EpiMatsmk
113 EpiMatagg := b*z EpiMatagg
114 EpiFamScore := c*z EpiFamScore
115 EpiEduPar := d*z EpiEduPar
116 Epin_trauma := e*z Epin_trauma
117 total := f+g+h+i+j+(a*z)+(b*z)+(c*z)+(d*z)+(e*z) total
est se z pvalue ci.lower ci.upper
1 0.009 0.042 0.217 0.829 -0.073 0.091
2 -0.017 0.064 -0.261 0.794 -0.141 0.108
3 -0.054 0.063 -0.853 0.393 -0.177 0.070
4 -0.010 0.092 -0.109 0.913 -0.190 0.170
5 0.018 0.096 0.188 0.851 -0.171 0.207
6 -0.042 0.101 -0.416 0.677 -0.239 0.155
7 -0.010 0.038 -0.255 0.799 -0.083 0.064
8 0.018 0.039 0.453 0.651 -0.060 0.095
9 0.074 0.053 1.390 0.165 -0.030 0.179
10 -0.522 0.239 -2.187 0.029 -0.990 -0.054
11 -0.058 0.091 -0.641 0.522 -0.236 0.120
12 0.308 1.115 0.276 0.782 -1.878 2.494
13 1.435 2.312 0.621 0.535 -3.096 5.966
14 -0.234 1.945 -0.120 0.904 -4.046 3.578
15 -3.495 2.188 -1.597 0.110 -7.783 0.794
16 2.505 1.290 1.942 0.052 -0.023 5.032
17 -3.123 2.418 -1.292 0.196 -7.861 1.616
18 1.231 0.767 1.605 0.109 -0.273 2.735
19 1.267 0.812 1.560 0.119 -0.325 2.858
20 0.566 0.713 0.794 0.427 -0.831 1.963
21 10.274 16.690 0.616 0.538 -22.437 42.986
22 10.065 7.155 1.407 0.160 -3.958 24.089
23 -5.741 0.009 -669.813 0.000 -5.758 -5.724
24 5.748 0.000 NA NA 5.748 5.748
25 1.000 0.000 NA NA 1.000 1.000
26 -31.957 0.000 NA NA -31.957 -31.957
27 0.196 0.000 NA NA 0.196 0.196
28 0.049 0.000 NA NA 0.049 0.049
29 -0.009 0.000 NA NA -0.009 -0.009
30 -0.006 0.000 NA NA -0.006 -0.006
31 0.007 0.000 NA NA 0.007 0.007
32 -0.003 0.000 NA NA -0.003 -0.003
33 0.022 0.000 NA NA 0.022 0.022
34 0.004 0.000 NA NA 0.004 0.004
35 0.022 0.000 NA NA 0.022 0.022
36 0.003 0.000 NA NA 0.003 0.003
37 0.017 0.000 NA NA 0.017 0.017
38 0.091 0.000 NA NA 0.091 0.091
39 0.034 0.000 NA NA 0.034 0.034
40 -0.017 0.000 NA NA -0.017 -0.017
41 0.007 0.000 NA NA 0.007 0.007
42 0.000 0.000 NA NA 0.000 0.000
43 0.033 0.000 NA NA 0.033 0.033
44 0.008 0.000 NA NA 0.008 0.008
45 0.008 0.000 NA NA 0.008 0.008
46 0.001 0.000 NA NA 0.001 0.001
47 0.010 0.000 NA NA 0.010 0.010
48 0.132 0.000 NA NA 0.132 0.132
49 -0.029 0.000 NA NA -0.029 -0.029
50 0.027 0.000 NA NA 0.027 0.027
51 0.004 0.000 NA NA 0.004 0.004
52 0.065 0.000 NA NA 0.065 0.065
53 0.004 0.000 NA NA 0.004 0.004
54 0.058 0.000 NA NA 0.058 0.058
55 0.001 0.000 NA NA 0.001 0.001
56 0.044 0.000 NA NA 0.044 0.044
57 0.054 0.000 NA NA 0.054 0.054
58 -0.008 0.000 NA NA -0.008 -0.008
59 0.003 0.000 NA NA 0.003 0.003
60 -0.019 0.000 NA NA -0.019 -0.019
61 0.010 0.000 NA NA 0.010 0.010
62 -0.013 0.000 NA NA -0.013 -0.013
63 0.000 0.000 NA NA 0.000 0.000
64 -0.015 0.000 NA NA -0.015 -0.015
65 0.051 0.000 NA NA 0.051 0.051
66 0.002 0.000 NA NA 0.002 0.002
67 0.042 0.000 NA NA 0.042 0.042
68 0.018 0.000 NA NA 0.018 0.018
69 0.018 0.000 NA NA 0.018 0.018
70 0.000 0.000 NA NA 0.000 0.000
71 0.021 0.000 NA NA 0.021 0.021
72 0.047 0.000 NA NA 0.047 0.047
73 0.008 0.000 NA NA 0.008 0.008
74 -0.002 0.000 NA NA -0.002 -0.002
75 0.035 0.000 NA NA 0.035 0.035
76 -0.001 0.000 NA NA -0.001 -0.001
77 0.009 0.000 NA NA 0.009 0.009
78 0.213 0.000 NA NA 0.213 0.213
79 0.061 0.000 NA NA 0.061 0.061
80 0.061 0.000 NA NA 0.061 0.061
81 0.006 0.000 NA NA 0.006 0.006
82 0.044 0.000 NA NA 0.044 0.044
83 0.146 0.000 NA NA 0.146 0.146
84 0.035 0.000 NA NA 0.035 0.035
85 0.002 0.000 NA NA 0.002 0.002
86 0.003 0.000 NA NA 0.003 0.003
87 0.213 0.000 NA NA 0.213 0.213
88 0.003 0.000 NA NA 0.003 0.003
89 0.049 0.000 NA NA 0.049 0.049
90 0.005 0.000 NA NA 0.005 0.005
91 0.002 0.000 NA NA 0.002 0.002
92 0.062 0.000 NA NA 0.062 0.062
93 1.000 0.000 NA NA 1.000 1.000
94 0.000 0.000 NA NA 0.000 0.000
95 0.000 0.000 NA NA 0.000 0.000
96 0.262 0.000 NA NA 0.262 0.262
97 0.100 0.000 NA NA 0.100 0.100
98 0.225 0.000 NA NA 0.225 0.225
99 0.606 0.000 NA NA 0.606 0.606
100 0.196 0.000 NA NA 0.196 0.196
101 0.562 0.000 NA NA 0.562 0.562
102 0.300 0.000 NA NA 0.300 0.300
103 0.175 0.000 NA NA 0.175 0.175
104 0.300 0.000 NA NA 0.300 0.300
105 0.529 0.000 NA NA 0.529 0.529
106 0.124 0.000 NA NA 0.124 0.124
107 0.308 1.115 0.276 0.782 -1.878 2.494
108 1.435 2.312 0.621 0.535 -3.096 5.966
109 -0.234 1.945 -0.120 0.904 -4.046 3.578
110 -3.495 2.188 -1.597 0.110 -7.783 0.794
111 2.505 1.290 1.942 0.052 -0.023 5.032
112 -0.052 0.239 -0.217 0.829 -0.520 0.417
113 0.095 0.365 0.261 0.794 -0.620 0.810
114 0.308 0.361 0.853 0.393 -0.399 1.015
115 0.058 0.526 0.110 0.913 -0.973 1.088
116 -0.104 0.554 -0.188 0.851 -1.190 0.981
117 0.823 4.116 0.200 0.842 -7.244 8.889
label lhs edge rhs est std group
1 a Matsmk ~> Epi 9.021717e-03 0.0039897063
2 b Matagg ~> Epi -1.656139e-02 -0.0049937195
3 c FamScore ~> Epi -5.363066e-02 -0.0194819461
4 d EduPar ~> Epi -1.003054e-02 -0.0023236029
5 e n_trauma ~> Epi 1.817152e-02 0.0041125181
6 Age ~> Epi -4.183870e-02 -0.0091064217
7 int_dis ~> Epi -9.609870e-03 -0.0044262219
8 medication ~> Epi 1.786833e-02 0.0068239536
9 contraceptives ~> Epi 7.415666e-02 0.0341559075
10 V8 ~> Epi -5.220008e-01 -0.0355023001
11 cigday_1 ~> Epi -5.823695e-02 -0.0144270114
12 f Matsmk ~> group 3.080501e-01 0.0339073245
13 g Matagg ~> group 1.434825e+00 0.1076829249
14 h FamScore ~> group -2.342742e-01 -0.0211818628
15 i EduPar ~> group -3.494814e+00 -0.2015033907
16 j n_trauma ~> group 2.504516e+00 0.1410785375
17 Age ~> group -3.122626e+00 -0.1691648443
18 int_dis ~> group 1.231194e+00 0.1411440745
19 medication ~> group 1.266603e+00 0.1203963282
20 contraceptives ~> group 5.660290e-01 0.0648895666
21 V8 ~> group 1.027449e+01 0.1739266853
22 cigday_1 ~> group 1.006508e+01 0.6206054481
23 z Epi ~> group -5.740817e+00 -1.4288750028
25 Epi <-> Epi 1.000000e+00 0.9975833547
26 group <-> group -3.195698e+01 -1.9749494979
27 Matsmk <-> Matsmk 1.960443e-01 1.0000000000
28 Matsmk <-> Matagg 4.936709e-02 0.3693241433
29 Matsmk <-> FamScore -9.177215e-03 -0.0569887592
30 Matsmk <-> EduPar -5.564346e-03 -0.0541843459
31 Matsmk <-> n_trauma 7.459313e-03 0.0743497863
32 Matsmk <-> Age -3.217300e-03 -0.0333441329
33 Matsmk <-> int_dis 2.151899e-02 0.1053910232
34 Matsmk <-> medication 4.113924e-03 0.0242997446
35 Matsmk <-> contraceptives 2.151899e-02 0.1053910232
36 Matsmk <-> V8 2.928139e-03 0.0971189320
37 Matsmk <-> cigday_1 1.693829e-02 0.1542372547
38 Matagg <-> Matagg 9.113924e-02 1.0000000000
39 Matagg <-> FamScore 3.417722e-02 0.3112715087
40 Matagg <-> EduPar -1.656118e-02 -0.2365241196
41 Matagg <-> n_trauma 7.233273e-03 0.1057402114
42 Matagg <-> Age 3.118694e-04 0.0047405101
43 Matagg <-> int_dis 3.291139e-02 0.2364027144
44 Matagg <-> medication 7.594937e-03 0.0657951695
45 Matagg <-> contraceptives 7.594937e-03 0.0545544726
46 Matagg <-> V8 8.272067e-04 0.0402393217
47 Matagg <-> cigday_1 1.018987e-02 0.1360858260
48 FamScore <-> FamScore 1.322785e-01 1.0000000000
49 FamScore <-> EduPar -2.948312e-02 -0.3495149022
50 FamScore <-> n_trauma 2.667269e-02 0.3236534989
51 FamScore <-> Age 3.636947e-03 0.0458878230
52 FamScore <-> int_dis 6.455696e-02 0.3849084009
53 FamScore <-> medication 4.430380e-03 0.0318580293
54 FamScore <-> contraceptives 5.822785e-02 0.3471722832
55 FamScore <-> V8 7.814844e-04 0.0315547719
56 FamScore <-> cigday_1 4.381329e-02 0.4856887960
57 EduPar <-> EduPar 5.379307e-02 1.0000000000
58 EduPar <-> n_trauma -8.024412e-03 -0.1526891136
59 EduPar <-> Age 2.762108e-03 0.0546490350
60 EduPar <-> int_dis -1.909283e-02 -0.1785114035
61 EduPar <-> medication 1.017932e-02 0.1147832062
62 EduPar <-> contraceptives -1.329114e-02 -0.1242676068
63 EduPar <-> V8 -8.860887e-06 -0.0005610523
64 EduPar <-> cigday_1 -1.493803e-02 -0.2596730517
65 n_trauma <-> n_trauma 5.134332e-02 1.0000000000
66 n_trauma <-> Age 1.582278e-03 0.0320439451
67 n_trauma <-> int_dis 4.159132e-02 0.3980335009
68 n_trauma <-> medication 1.763110e-02 0.2034979577
69 n_trauma <-> contraceptives 1.808318e-02 0.1730580439
70 n_trauma <-> V8 -4.694340e-04 -0.0304243917
71 n_trauma <-> cigday_1 2.128165e-02 0.3786692420
72 Age <-> Age 4.748866e-02 1.0000000000
73 Age <-> int_dis 8.090259e-03 0.0805056484
74 Age <-> medication -1.655660e-03 -0.0198700345
75 Age <-> contraceptives 3.524124e-02 0.3506833348
76 Age <-> V8 -1.333633e-03 -0.0898732659
77 Age <-> cigday_1 8.542355e-03 0.1580445206
78 int_dis <-> int_dis 2.126582e-01 1.0000000000
79 int_dis <-> medication 6.075949e-02 0.3445843938
80 int_dis <-> contraceptives 6.075949e-02 0.2857142857
81 int_dis <-> V8 5.645344e-03 0.1797788722
82 int_dis <-> cigday_1 4.449367e-02 0.3890038953
83 medication <-> medication 1.462025e-01 1.0000000000
84 medication <-> contraceptives 3.544304e-02 0.2010075631
85 medication <-> V8 2.084232e-03 0.0800493604
86 medication <-> cigday_1 3.275316e-03 0.0345360471
87 contraceptives <-> contraceptives 2.126582e-01 1.0000000000
88 contraceptives <-> V8 2.688833e-03 0.0856272484
89 contraceptives <-> cigday_1 4.892405e-02 0.4277382803
90 V8 <-> V8 4.636832e-03 1.0000000000
91 V8 <-> cigday_1 1.509276e-03 0.0893623867
92 cigday_1 <-> cigday_1 6.151859e-02 1.0000000000
93 group <-> group 1.000000e+00 1.0000000000
94 int Epi 0.000000e+00 0.0000000000
95 int group 0.000000e+00 0.0000000000
96 int Matsmk 2.625000e-01 0.5928600601
97 int Matagg 1.000000e-01 0.3312434486
98 int FamScore 2.250000e-01 0.6186398880
99 int EduPar 6.062500e-01 2.6138976225
100 int n_trauma 1.964286e-01 0.8668873691
101 int Age 5.621377e-01 2.5795724974
102 int int_dis 3.000000e-01 0.6505492185
103 int medication 1.750000e-01 0.4576785957
104 int contraceptives 3.000000e-01 0.6505492185
105 int V8 5.286908e-01 7.7640983340
106 int cigday_1 1.243750e-01 0.5014526157
fixed par
1 FALSE 1
2 FALSE 2
3 FALSE 3
4 FALSE 4
5 FALSE 5
6 FALSE 6
7 FALSE 7
8 FALSE 8
9 FALSE 9
10 FALSE 10
11 FALSE 11
12 FALSE 12
13 FALSE 13
14 FALSE 14
15 FALSE 15
16 FALSE 16
17 FALSE 17
18 FALSE 18
19 FALSE 19
20 FALSE 20
21 FALSE 21
22 FALSE 22
23 FALSE 23
25 TRUE 0
26 TRUE 0
27 TRUE 0
28 TRUE 0
29 TRUE 0
30 TRUE 0
31 TRUE 0
32 TRUE 0
33 TRUE 0
34 TRUE 0
35 TRUE 0
36 TRUE 0
37 TRUE 0
38 TRUE 0
39 TRUE 0
40 TRUE 0
41 TRUE 0
42 TRUE 0
43 TRUE 0
44 TRUE 0
45 TRUE 0
46 TRUE 0
47 TRUE 0
48 TRUE 0
49 TRUE 0
50 TRUE 0
51 TRUE 0
52 TRUE 0
53 TRUE 0
54 TRUE 0
55 TRUE 0
56 TRUE 0
57 TRUE 0
58 TRUE 0
59 TRUE 0
60 TRUE 0
61 TRUE 0
62 TRUE 0
63 TRUE 0
64 TRUE 0
65 TRUE 0
66 TRUE 0
67 TRUE 0
68 TRUE 0
69 TRUE 0
70 TRUE 0
71 TRUE 0
72 TRUE 0
73 TRUE 0
74 TRUE 0
75 TRUE 0
76 TRUE 0
77 TRUE 0
78 TRUE 0
79 TRUE 0
80 TRUE 0
81 TRUE 0
82 TRUE 0
83 TRUE 0
84 TRUE 0
85 TRUE 0
86 TRUE 0
87 TRUE 0
88 TRUE 0
89 TRUE 0
90 TRUE 0
91 TRUE 0
92 TRUE 0
93 TRUE 0
94 TRUE 0
95 TRUE 0
96 TRUE 0
97 TRUE 0
98 TRUE 0
99 TRUE 0
100 TRUE 0
101 TRUE 0
102 TRUE 0
103 TRUE 0
104 TRUE 0
105 TRUE 0
106 TRUE 0Warning in lav_object_post_check(object): lavaan WARNING: some estimated ov
variances are negative############################
############################
Epi_M15
############################
############################
##Mediation Model ##
lavaan 0.6-7 ended normally after 107 iterations
Estimator DWLS
Optimization method NLMINB
Number of free parameters 23
Used Total
Number of observations 80 99
Model Test User Model:
Standard Robust
Test Statistic 73.074 70.375
Degrees of freedom 3 3
P-value (Chi-square) 0.000 0.000
Scaling correction factor 1.040
Shift parameter 0.116
simple second-order correction
Parameter Estimates:
Standard errors Robust.sem
Information Expected
Information saturated (h1) model Unstructured
Regressions:
Estimate Std.Err z-value P(>|z|)
Epi ~
Matsmk (a) -0.055 0.057 -0.964 0.335
Matagg (b) 0.074 0.106 0.695 0.487
FamScore (c) 0.113 0.082 1.378 0.168
EduPar (d) 0.229 0.104 2.213 0.027
n_trauma (e) -0.062 0.089 -0.701 0.483
Age -0.088 0.121 -0.727 0.467
int_dis -0.065 0.047 -1.378 0.168
medication -0.024 0.056 -0.433 0.665
contrcptvs 0.032 0.055 0.579 0.562
V8 0.007 0.271 0.026 0.979
cigday_1 -0.052 0.113 -0.458 0.647
group ~
Matsmk (f) 0.193 1.131 0.171 0.864
Matagg (g) 1.614 2.461 0.656 0.512
FamScore (h) 0.202 2.055 0.098 0.922
EduPar (i) -3.176 2.214 -1.435 0.151
n_trauma (j) 2.329 1.390 1.676 0.094
Age -2.983 2.470 -1.208 0.227
int_dis 1.212 0.799 1.517 0.129
medication 1.137 0.833 1.365 0.172
contrcptvs 0.177 0.770 0.230 0.818
V8 13.279 16.848 0.788 0.431
cigday_1 10.340 7.173 1.442 0.149
Epi (z) -1.141 0.027 -41.523 0.000
Intercepts:
Estimate Std.Err z-value P(>|z|)
.Epi 0.000
.group 0.000
Thresholds:
Estimate Std.Err z-value P(>|z|)
group|t1 5.748
Variances:
Estimate Std.Err z-value P(>|z|)
.Epi 1.000
.group -0.302
Scales y*:
Estimate Std.Err z-value P(>|z|)
group 1.000
Defined Parameters:
Estimate Std.Err z-value P(>|z|)
directMatsmk 0.193 1.131 0.171 0.864
directMatagg 1.614 2.461 0.656 0.512
directFamScore 0.202 2.055 0.098 0.922
directEduPar -3.176 2.214 -1.435 0.151
directn_trauma 2.329 1.390 1.676 0.094
EpiMatsmk 0.063 0.065 0.972 0.331
EpiMatagg -0.084 0.121 -0.698 0.485
EpiFamScore -0.129 0.094 -1.364 0.173
EpiEduPar -0.261 0.119 -2.198 0.028
Epin_trauma 0.071 0.102 0.699 0.485
total 0.823 4.116 0.200 0.842
npar fmin
23.000 0.457
chisq df
73.074 3.000
pvalue chisq.scaled
0.000 70.375
df.scaled pvalue.scaled
3.000 0.000
chisq.scaling.factor baseline.chisq
1.040 30.016
baseline.df baseline.pvalue
1.000 0.000
baseline.chisq.scaled baseline.df.scaled
30.016 1.000
baseline.pvalue.scaled baseline.chisq.scaling.factor
0.000 1.000
cfi tli
0.000 0.195
nnfi rfi
0.195 NA
nfi pnfi
NA -4.303
ifi rni
-1.594 -1.415
cfi.scaled tli.scaled
0.000 0.226
cfi.robust tli.robust
NA NA
nnfi.scaled nnfi.robust
0.226 NA
rfi.scaled nfi.scaled
NA NA
ifi.scaled rni.scaled
-1.494 -1.322
rni.robust rmsea
NA 0.544
rmsea.ci.lower rmsea.ci.upper
0.440 0.655
rmsea.pvalue rmsea.scaled
0.000 0.533
rmsea.ci.lower.scaled rmsea.ci.upper.scaled
0.429 0.645
rmsea.pvalue.scaled rmsea.robust
0.000 NA
rmsea.ci.lower.robust rmsea.ci.upper.robust
NA NA
rmsea.pvalue.robust rmr
NA 0.676
rmr_nomean srmr
0.802 22.585
srmr_bentler srmr_bentler_nomean
17.575 22.585
crmr crmr_nomean
2.171 0.193
srmr_mplus srmr_mplus_nomean
NA NA
cn_05 cn_01
9.448 13.265
gfi agfi
-476.384 -4136.328
pgfi mfi
-54.967 0.642
lhs op rhs label
1 Epi ~ Matsmk a
2 Epi ~ Matagg b
3 Epi ~ FamScore c
4 Epi ~ EduPar d
5 Epi ~ n_trauma e
6 Epi ~ Age
7 Epi ~ int_dis
8 Epi ~ medication
9 Epi ~ contraceptives
10 Epi ~ V8
11 Epi ~ cigday_1
12 group ~ Matsmk f
13 group ~ Matagg g
14 group ~ FamScore h
15 group ~ EduPar i
16 group ~ n_trauma j
17 group ~ Age
18 group ~ int_dis
19 group ~ medication
20 group ~ contraceptives
21 group ~ V8
22 group ~ cigday_1
23 group ~ Epi z
24 group | t1
25 Epi ~~ Epi
26 group ~~ group
27 Matsmk ~~ Matsmk
28 Matsmk ~~ Matagg
29 Matsmk ~~ FamScore
30 Matsmk ~~ EduPar
31 Matsmk ~~ n_trauma
32 Matsmk ~~ Age
33 Matsmk ~~ int_dis
34 Matsmk ~~ medication
35 Matsmk ~~ contraceptives
36 Matsmk ~~ V8
37 Matsmk ~~ cigday_1
38 Matagg ~~ Matagg
39 Matagg ~~ FamScore
40 Matagg ~~ EduPar
41 Matagg ~~ n_trauma
42 Matagg ~~ Age
43 Matagg ~~ int_dis
44 Matagg ~~ medication
45 Matagg ~~ contraceptives
46 Matagg ~~ V8
47 Matagg ~~ cigday_1
48 FamScore ~~ FamScore
49 FamScore ~~ EduPar
50 FamScore ~~ n_trauma
51 FamScore ~~ Age
52 FamScore ~~ int_dis
53 FamScore ~~ medication
54 FamScore ~~ contraceptives
55 FamScore ~~ V8
56 FamScore ~~ cigday_1
57 EduPar ~~ EduPar
58 EduPar ~~ n_trauma
59 EduPar ~~ Age
60 EduPar ~~ int_dis
61 EduPar ~~ medication
62 EduPar ~~ contraceptives
63 EduPar ~~ V8
64 EduPar ~~ cigday_1
65 n_trauma ~~ n_trauma
66 n_trauma ~~ Age
67 n_trauma ~~ int_dis
68 n_trauma ~~ medication
69 n_trauma ~~ contraceptives
70 n_trauma ~~ V8
71 n_trauma ~~ cigday_1
72 Age ~~ Age
73 Age ~~ int_dis
74 Age ~~ medication
75 Age ~~ contraceptives
76 Age ~~ V8
77 Age ~~ cigday_1
78 int_dis ~~ int_dis
79 int_dis ~~ medication
80 int_dis ~~ contraceptives
81 int_dis ~~ V8
82 int_dis ~~ cigday_1
83 medication ~~ medication
84 medication ~~ contraceptives
85 medication ~~ V8
86 medication ~~ cigday_1
87 contraceptives ~~ contraceptives
88 contraceptives ~~ V8
89 contraceptives ~~ cigday_1
90 V8 ~~ V8
91 V8 ~~ cigday_1
92 cigday_1 ~~ cigday_1
93 group ~*~ group
94 Epi ~1
95 group ~1
96 Matsmk ~1
97 Matagg ~1
98 FamScore ~1
99 EduPar ~1
100 n_trauma ~1
101 Age ~1
102 int_dis ~1
103 medication ~1
104 contraceptives ~1
105 V8 ~1
106 cigday_1 ~1
107 directMatsmk := f directMatsmk
108 directMatagg := g directMatagg
109 directFamScore := h directFamScore
110 directEduPar := i directEduPar
111 directn_trauma := j directn_trauma
112 EpiMatsmk := a*z EpiMatsmk
113 EpiMatagg := b*z EpiMatagg
114 EpiFamScore := c*z EpiFamScore
115 EpiEduPar := d*z EpiEduPar
116 Epin_trauma := e*z Epin_trauma
117 total := f+g+h+i+j+(a*z)+(b*z)+(c*z)+(d*z)+(e*z) total
est se z pvalue ci.lower ci.upper
1 -0.055 0.057 -0.964 0.335 -0.168 0.057
2 0.074 0.106 0.695 0.487 -0.134 0.282
3 0.113 0.082 1.378 0.168 -0.048 0.273
4 0.229 0.104 2.213 0.027 0.026 0.432
5 -0.062 0.089 -0.701 0.483 -0.236 0.112
6 -0.088 0.121 -0.727 0.467 -0.325 0.149
7 -0.065 0.047 -1.378 0.168 -0.157 0.027
8 -0.024 0.056 -0.433 0.665 -0.133 0.085
9 0.032 0.055 0.579 0.562 -0.076 0.140
10 0.007 0.271 0.026 0.979 -0.524 0.539
11 -0.052 0.113 -0.458 0.647 -0.274 0.170
12 0.193 1.131 0.171 0.864 -2.023 2.409
13 1.614 2.461 0.656 0.512 -3.208 6.437
14 0.202 2.055 0.098 0.922 -3.825 4.229
15 -3.176 2.214 -1.435 0.151 -7.514 1.163
16 2.329 1.390 1.676 0.094 -0.395 5.054
17 -2.983 2.470 -1.208 0.227 -7.823 1.858
18 1.212 0.799 1.517 0.129 -0.354 2.779
19 1.137 0.833 1.365 0.172 -0.496 2.769
20 0.177 0.770 0.230 0.818 -1.332 1.685
21 13.279 16.848 0.788 0.431 -19.743 46.302
22 10.340 7.173 1.442 0.149 -3.718 24.398
23 -1.141 0.027 -41.523 0.000 -1.195 -1.087
24 5.748 0.000 NA NA 5.748 5.748
25 1.000 0.000 NA NA 1.000 1.000
26 -0.302 0.000 NA NA -0.302 -0.302
27 0.196 0.000 NA NA 0.196 0.196
28 0.049 0.000 NA NA 0.049 0.049
29 -0.009 0.000 NA NA -0.009 -0.009
30 -0.006 0.000 NA NA -0.006 -0.006
31 0.007 0.000 NA NA 0.007 0.007
32 -0.003 0.000 NA NA -0.003 -0.003
33 0.022 0.000 NA NA 0.022 0.022
34 0.004 0.000 NA NA 0.004 0.004
35 0.022 0.000 NA NA 0.022 0.022
36 0.003 0.000 NA NA 0.003 0.003
37 0.017 0.000 NA NA 0.017 0.017
38 0.091 0.000 NA NA 0.091 0.091
39 0.034 0.000 NA NA 0.034 0.034
40 -0.017 0.000 NA NA -0.017 -0.017
41 0.007 0.000 NA NA 0.007 0.007
42 0.000 0.000 NA NA 0.000 0.000
43 0.033 0.000 NA NA 0.033 0.033
44 0.008 0.000 NA NA 0.008 0.008
45 0.008 0.000 NA NA 0.008 0.008
46 0.001 0.000 NA NA 0.001 0.001
47 0.010 0.000 NA NA 0.010 0.010
48 0.132 0.000 NA NA 0.132 0.132
49 -0.029 0.000 NA NA -0.029 -0.029
50 0.027 0.000 NA NA 0.027 0.027
51 0.004 0.000 NA NA 0.004 0.004
52 0.065 0.000 NA NA 0.065 0.065
53 0.004 0.000 NA NA 0.004 0.004
54 0.058 0.000 NA NA 0.058 0.058
55 0.001 0.000 NA NA 0.001 0.001
56 0.044 0.000 NA NA 0.044 0.044
57 0.054 0.000 NA NA 0.054 0.054
58 -0.008 0.000 NA NA -0.008 -0.008
59 0.003 0.000 NA NA 0.003 0.003
60 -0.019 0.000 NA NA -0.019 -0.019
61 0.010 0.000 NA NA 0.010 0.010
62 -0.013 0.000 NA NA -0.013 -0.013
63 0.000 0.000 NA NA 0.000 0.000
64 -0.015 0.000 NA NA -0.015 -0.015
65 0.051 0.000 NA NA 0.051 0.051
66 0.002 0.000 NA NA 0.002 0.002
67 0.042 0.000 NA NA 0.042 0.042
68 0.018 0.000 NA NA 0.018 0.018
69 0.018 0.000 NA NA 0.018 0.018
70 0.000 0.000 NA NA 0.000 0.000
71 0.021 0.000 NA NA 0.021 0.021
72 0.047 0.000 NA NA 0.047 0.047
73 0.008 0.000 NA NA 0.008 0.008
74 -0.002 0.000 NA NA -0.002 -0.002
75 0.035 0.000 NA NA 0.035 0.035
76 -0.001 0.000 NA NA -0.001 -0.001
77 0.009 0.000 NA NA 0.009 0.009
78 0.213 0.000 NA NA 0.213 0.213
79 0.061 0.000 NA NA 0.061 0.061
80 0.061 0.000 NA NA 0.061 0.061
81 0.006 0.000 NA NA 0.006 0.006
82 0.044 0.000 NA NA 0.044 0.044
83 0.146 0.000 NA NA 0.146 0.146
84 0.035 0.000 NA NA 0.035 0.035
85 0.002 0.000 NA NA 0.002 0.002
86 0.003 0.000 NA NA 0.003 0.003
87 0.213 0.000 NA NA 0.213 0.213
88 0.003 0.000 NA NA 0.003 0.003
89 0.049 0.000 NA NA 0.049 0.049
90 0.005 0.000 NA NA 0.005 0.005
91 0.002 0.000 NA NA 0.002 0.002
92 0.062 0.000 NA NA 0.062 0.062
93 1.000 0.000 NA NA 1.000 1.000
94 0.000 0.000 NA NA 0.000 0.000
95 0.000 0.000 NA NA 0.000 0.000
96 0.262 0.000 NA NA 0.262 0.262
97 0.100 0.000 NA NA 0.100 0.100
98 0.225 0.000 NA NA 0.225 0.225
99 0.606 0.000 NA NA 0.606 0.606
100 0.196 0.000 NA NA 0.196 0.196
101 0.562 0.000 NA NA 0.562 0.562
102 0.300 0.000 NA NA 0.300 0.300
103 0.175 0.000 NA NA 0.175 0.175
104 0.300 0.000 NA NA 0.300 0.300
105 0.529 0.000 NA NA 0.529 0.529
106 0.124 0.000 NA NA 0.124 0.124
107 0.193 1.131 0.171 0.864 -2.023 2.409
108 1.614 2.461 0.656 0.512 -3.208 6.437
109 0.202 2.055 0.098 0.922 -3.825 4.229
110 -3.176 2.214 -1.435 0.151 -7.514 1.163
111 2.329 1.390 1.676 0.094 -0.395 5.054
112 0.063 0.065 0.972 0.331 -0.064 0.190
113 -0.084 0.121 -0.698 0.485 -0.321 0.153
114 -0.129 0.094 -1.364 0.173 -0.314 0.056
115 -0.261 0.119 -2.198 0.028 -0.495 -0.028
116 0.071 0.102 0.699 0.485 -0.128 0.270
117 0.823 4.116 0.200 0.842 -7.244 8.889
label lhs edge rhs est std group
1 a Matsmk ~> Epi -5.522110e-02 -0.0243855820
2 b Matagg ~> Epi 7.387035e-02 0.0222419975
3 c FamScore ~> Epi 1.127425e-01 0.0408962143
4 d EduPar ~> Epi 2.291129e-01 0.0529985053
5 e n_trauma ~> Epi -6.219036e-02 -0.0140545193
6 Age ~> Epi -8.796430e-02 -0.0191184353
7 int_dis ~> Epi -6.492649e-02 -0.0298616580
8 medication ~> Epi -2.404003e-02 -0.0091677648
9 contraceptives ~> Epi 3.188491e-02 0.0146648385
10 V8 ~> Epi 7.148733e-03 0.0004855016
11 cigday_1 ~> Epi -5.184015e-02 -0.0128239070
12 f Matsmk ~> group 1.932465e-01 0.0212707840
13 g Matagg ~> group 1.614209e+00 0.1211455200
14 h FamScore ~> group 2.022610e-01 0.0182873823
15 i EduPar ~> group -3.175795e+00 -0.1831093090
16 j n_trauma ~> group 2.329237e+00 0.1312050328
17 Age ~> group -2.982804e+00 -0.1615899743
18 int_dis ~> group 1.212276e+00 0.1389752494
19 medication ~> group 1.136593e+00 0.1080381852
20 contraceptives ~> group 1.766922e-01 0.0202559811
21 V8 ~> group 1.327938e+01 0.2247932563
22 cigday_1 ~> group 1.034025e+01 0.6375719698
23 z Epi ~> group -1.141079e+00 -0.2844197409
25 Epi <-> Epi 1.000000e+00 0.9947223019
26 group <-> group -3.020622e-01 -0.0186674942
27 Matsmk <-> Matsmk 1.960443e-01 1.0000000000
28 Matsmk <-> Matagg 4.936709e-02 0.3693241433
29 Matsmk <-> FamScore -9.177215e-03 -0.0569887592
30 Matsmk <-> EduPar -5.564346e-03 -0.0541843459
31 Matsmk <-> n_trauma 7.459313e-03 0.0743497863
32 Matsmk <-> Age -3.217300e-03 -0.0333441329
33 Matsmk <-> int_dis 2.151899e-02 0.1053910232
34 Matsmk <-> medication 4.113924e-03 0.0242997446
35 Matsmk <-> contraceptives 2.151899e-02 0.1053910232
36 Matsmk <-> V8 2.928139e-03 0.0971189320
37 Matsmk <-> cigday_1 1.693829e-02 0.1542372547
38 Matagg <-> Matagg 9.113924e-02 1.0000000000
39 Matagg <-> FamScore 3.417722e-02 0.3112715087
40 Matagg <-> EduPar -1.656118e-02 -0.2365241196
41 Matagg <-> n_trauma 7.233273e-03 0.1057402114
42 Matagg <-> Age 3.118694e-04 0.0047405101
43 Matagg <-> int_dis 3.291139e-02 0.2364027144
44 Matagg <-> medication 7.594937e-03 0.0657951695
45 Matagg <-> contraceptives 7.594937e-03 0.0545544726
46 Matagg <-> V8 8.272067e-04 0.0402393217
47 Matagg <-> cigday_1 1.018987e-02 0.1360858260
48 FamScore <-> FamScore 1.322785e-01 1.0000000000
49 FamScore <-> EduPar -2.948312e-02 -0.3495149022
50 FamScore <-> n_trauma 2.667269e-02 0.3236534989
51 FamScore <-> Age 3.636947e-03 0.0458878230
52 FamScore <-> int_dis 6.455696e-02 0.3849084009
53 FamScore <-> medication 4.430380e-03 0.0318580293
54 FamScore <-> contraceptives 5.822785e-02 0.3471722832
55 FamScore <-> V8 7.814844e-04 0.0315547719
56 FamScore <-> cigday_1 4.381329e-02 0.4856887960
57 EduPar <-> EduPar 5.379307e-02 1.0000000000
58 EduPar <-> n_trauma -8.024412e-03 -0.1526891136
59 EduPar <-> Age 2.762108e-03 0.0546490350
60 EduPar <-> int_dis -1.909283e-02 -0.1785114035
61 EduPar <-> medication 1.017932e-02 0.1147832062
62 EduPar <-> contraceptives -1.329114e-02 -0.1242676068
63 EduPar <-> V8 -8.860887e-06 -0.0005610523
64 EduPar <-> cigday_1 -1.493803e-02 -0.2596730517
65 n_trauma <-> n_trauma 5.134332e-02 1.0000000000
66 n_trauma <-> Age 1.582278e-03 0.0320439451
67 n_trauma <-> int_dis 4.159132e-02 0.3980335009
68 n_trauma <-> medication 1.763110e-02 0.2034979577
69 n_trauma <-> contraceptives 1.808318e-02 0.1730580439
70 n_trauma <-> V8 -4.694340e-04 -0.0304243917
71 n_trauma <-> cigday_1 2.128165e-02 0.3786692420
72 Age <-> Age 4.748866e-02 1.0000000000
73 Age <-> int_dis 8.090259e-03 0.0805056484
74 Age <-> medication -1.655660e-03 -0.0198700345
75 Age <-> contraceptives 3.524124e-02 0.3506833348
76 Age <-> V8 -1.333633e-03 -0.0898732659
77 Age <-> cigday_1 8.542355e-03 0.1580445206
78 int_dis <-> int_dis 2.126582e-01 1.0000000000
79 int_dis <-> medication 6.075949e-02 0.3445843938
80 int_dis <-> contraceptives 6.075949e-02 0.2857142857
81 int_dis <-> V8 5.645344e-03 0.1797788722
82 int_dis <-> cigday_1 4.449367e-02 0.3890038953
83 medication <-> medication 1.462025e-01 1.0000000000
84 medication <-> contraceptives 3.544304e-02 0.2010075631
85 medication <-> V8 2.084232e-03 0.0800493604
86 medication <-> cigday_1 3.275316e-03 0.0345360471
87 contraceptives <-> contraceptives 2.126582e-01 1.0000000000
88 contraceptives <-> V8 2.688833e-03 0.0856272484
89 contraceptives <-> cigday_1 4.892405e-02 0.4277382803
90 V8 <-> V8 4.636832e-03 1.0000000000
91 V8 <-> cigday_1 1.509276e-03 0.0893623867
92 cigday_1 <-> cigday_1 6.151859e-02 1.0000000000
93 group <-> group 1.000000e+00 1.0000000000
94 int Epi 0.000000e+00 0.0000000000
95 int group 0.000000e+00 0.0000000000
96 int Matsmk 2.625000e-01 0.5928600601
97 int Matagg 1.000000e-01 0.3312434486
98 int FamScore 2.250000e-01 0.6186398880
99 int EduPar 6.062500e-01 2.6138976225
100 int n_trauma 1.964286e-01 0.8668873691
101 int Age 5.621377e-01 2.5795724974
102 int int_dis 3.000000e-01 0.6505492185
103 int medication 1.750000e-01 0.4576785957
104 int contraceptives 3.000000e-01 0.6505492185
105 int V8 5.286908e-01 7.7640983340
106 int cigday_1 1.243750e-01 0.5014526157
fixed par
1 FALSE 1
2 FALSE 2
3 FALSE 3
4 FALSE 4
5 FALSE 5
6 FALSE 6
7 FALSE 7
8 FALSE 8
9 FALSE 9
10 FALSE 10
11 FALSE 11
12 FALSE 12
13 FALSE 13
14 FALSE 14
15 FALSE 15
16 FALSE 16
17 FALSE 17
18 FALSE 18
19 FALSE 19
20 FALSE 20
21 FALSE 21
22 FALSE 22
23 FALSE 23
25 TRUE 0
26 TRUE 0
27 TRUE 0
28 TRUE 0
29 TRUE 0
30 TRUE 0
31 TRUE 0
32 TRUE 0
33 TRUE 0
34 TRUE 0
35 TRUE 0
36 TRUE 0
37 TRUE 0
38 TRUE 0
39 TRUE 0
40 TRUE 0
41 TRUE 0
42 TRUE 0
43 TRUE 0
44 TRUE 0
45 TRUE 0
46 TRUE 0
47 TRUE 0
48 TRUE 0
49 TRUE 0
50 TRUE 0
51 TRUE 0
52 TRUE 0
53 TRUE 0
54 TRUE 0
55 TRUE 0
56 TRUE 0
57 TRUE 0
58 TRUE 0
59 TRUE 0
60 TRUE 0
61 TRUE 0
62 TRUE 0
63 TRUE 0
64 TRUE 0
65 TRUE 0
66 TRUE 0
67 TRUE 0
68 TRUE 0
69 TRUE 0
70 TRUE 0
71 TRUE 0
72 TRUE 0
73 TRUE 0
74 TRUE 0
75 TRUE 0
76 TRUE 0
77 TRUE 0
78 TRUE 0
79 TRUE 0
80 TRUE 0
81 TRUE 0
82 TRUE 0
83 TRUE 0
84 TRUE 0
85 TRUE 0
86 TRUE 0
87 TRUE 0
88 TRUE 0
89 TRUE 0
90 TRUE 0
91 TRUE 0
92 TRUE 0
93 TRUE 0
94 TRUE 0
95 TRUE 0
96 TRUE 0
97 TRUE 0
98 TRUE 0
99 TRUE 0
100 TRUE 0
101 TRUE 0
102 TRUE 0
103 TRUE 0
104 TRUE 0
105 TRUE 0
106 TRUE 0Warning in lav_samplestats_step2(UNI = FIT, wt = wt, ov.names = ov.names, :
lavaan WARNING: correlation between variables group and Epi is (nearly) 1.0
Warning in lav_samplestats_step2(UNI = FIT, wt = wt, ov.names = ov.names, :
lavaan WARNING: some estimated ov variances are negative############################
############################
Epi_M_all
############################
############################
##Mediation Model ##
lavaan 0.6-7 ended normally after 112 iterations
Estimator DWLS
Optimization method NLMINB
Number of free parameters 23
Used Total
Number of observations 80 99
Model Test User Model:
Standard Robust
Test Statistic 19.648 17.218
Degrees of freedom 3 3
P-value (Chi-square) 0.000 0.001
Scaling correction factor 1.171
Shift parameter 0.438
simple second-order correction
Parameter Estimates:
Standard errors Robust.sem
Information Expected
Information saturated (h1) model Unstructured
Regressions:
Estimate Std.Err z-value P(>|z|)
Epi ~
Matsmk (a) 0.025 0.082 0.301 0.764
Matagg (b) 0.132 0.124 1.068 0.285
FamScore (c) -0.102 0.109 -0.934 0.350
EduPar (d) -0.281 0.169 -1.658 0.097
n_trauma (e) 0.074 0.126 0.589 0.556
Age 0.122 0.179 0.679 0.497
int_dis 0.085 0.074 1.148 0.251
medication 0.050 0.080 0.623 0.533
contrcptvs -0.074 0.084 -0.872 0.383
V8 0.285 1.060 0.268 0.788
cigday_1 0.168 0.140 1.205 0.228
group ~
Matsmk (f) 0.095 0.963 0.099 0.921
Matagg (g) 0.666 2.153 0.309 0.757
FamScore (h) 0.738 1.855 0.398 0.691
EduPar (i) -1.600 1.990 -0.804 0.421
n_trauma (j) 1.913 1.223 1.564 0.118
Age -3.678 2.115 -1.739 0.082
int_dis 0.728 0.683 1.066 0.286
medication 0.837 0.699 1.197 0.231
contrcptvs 0.622 0.626 0.994 0.320
V8 11.409 12.905 0.884 0.377
cigday_1 9.298 7.066 1.316 0.188
Epi (z) 6.542 0.104 63.023 0.000
Intercepts:
Estimate Std.Err z-value P(>|z|)
.Epi 0.000
.group 0.000
Thresholds:
Estimate Std.Err z-value P(>|z|)
group|t1 5.748
Variances:
Estimate Std.Err z-value P(>|z|)
.Epi 1.000
.group -41.801
Scales y*:
Estimate Std.Err z-value P(>|z|)
group 1.000
Defined Parameters:
Estimate Std.Err z-value P(>|z|)
directMatsmk 0.095 0.963 0.099 0.921
directMatagg 0.666 2.153 0.309 0.757
directFamScore 0.738 1.855 0.398 0.691
directEduPar -1.600 1.990 -0.804 0.421
directn_trauma 1.913 1.223 1.564 0.118
EpiMatsmk 0.161 0.535 0.301 0.764
EpiMatagg 0.864 0.809 1.068 0.286
EpiFamScore -0.665 0.712 -0.933 0.351
EpiEduPar -1.837 1.108 -1.658 0.097
Epin_trauma 0.487 0.827 0.589 0.556
total 0.823 4.116 0.200 0.842
npar fmin
23.000 0.123
chisq df
19.648 3.000
pvalue chisq.scaled
0.000 17.218
df.scaled pvalue.scaled
3.000 0.001
chisq.scaling.factor baseline.chisq
1.171 5.965
baseline.df baseline.pvalue
1.000 0.015
baseline.chisq.scaled baseline.df.scaled
5.965 1.000
baseline.pvalue.scaled baseline.chisq.scaling.factor
0.015 1.000
cfi tli
0.000 -0.118
nnfi rfi
-0.118 NA
nfi pnfi
NA -6.882
ifi rni
-4.615 -2.353
cfi.scaled tli.scaled
0.000 0.045
cfi.robust tli.robust
NA NA
nnfi.scaled nnfi.robust
0.045 NA
rfi.scaled nfi.scaled
NA NA
ifi.scaled rni.scaled
-3.796 -1.864
rni.robust rmsea
NA 0.265
rmsea.ci.lower rmsea.ci.upper
0.162 0.382
rmsea.pvalue rmsea.scaled
0.001 0.245
rmsea.ci.lower.scaled rmsea.ci.upper.scaled
0.141 0.363
rmsea.pvalue.scaled rmsea.robust
0.002 NA
rmsea.ci.lower.robust rmsea.ci.upper.robust
NA NA
rmsea.pvalue.robust rmr
NA 2.846
rmr_nomean srmr
3.671 16.430
srmr_bentler srmr_bentler_nomean
12.737 16.430
crmr crmr_nomean
3.270 5.550
srmr_mplus srmr_mplus_nomean
NA NA
cn_05 cn_01
32.422 46.616
gfi agfi
-146.299 -1275.590
pgfi mfi
-16.881 0.900
lhs op rhs label
1 Epi ~ Matsmk a
2 Epi ~ Matagg b
3 Epi ~ FamScore c
4 Epi ~ EduPar d
5 Epi ~ n_trauma e
6 Epi ~ Age
7 Epi ~ int_dis
8 Epi ~ medication
9 Epi ~ contraceptives
10 Epi ~ V8
11 Epi ~ cigday_1
12 group ~ Matsmk f
13 group ~ Matagg g
14 group ~ FamScore h
15 group ~ EduPar i
16 group ~ n_trauma j
17 group ~ Age
18 group ~ int_dis
19 group ~ medication
20 group ~ contraceptives
21 group ~ V8
22 group ~ cigday_1
23 group ~ Epi z
24 group | t1
25 Epi ~~ Epi
26 group ~~ group
27 Matsmk ~~ Matsmk
28 Matsmk ~~ Matagg
29 Matsmk ~~ FamScore
30 Matsmk ~~ EduPar
31 Matsmk ~~ n_trauma
32 Matsmk ~~ Age
33 Matsmk ~~ int_dis
34 Matsmk ~~ medication
35 Matsmk ~~ contraceptives
36 Matsmk ~~ V8
37 Matsmk ~~ cigday_1
38 Matagg ~~ Matagg
39 Matagg ~~ FamScore
40 Matagg ~~ EduPar
41 Matagg ~~ n_trauma
42 Matagg ~~ Age
43 Matagg ~~ int_dis
44 Matagg ~~ medication
45 Matagg ~~ contraceptives
46 Matagg ~~ V8
47 Matagg ~~ cigday_1
48 FamScore ~~ FamScore
49 FamScore ~~ EduPar
50 FamScore ~~ n_trauma
51 FamScore ~~ Age
52 FamScore ~~ int_dis
53 FamScore ~~ medication
54 FamScore ~~ contraceptives
55 FamScore ~~ V8
56 FamScore ~~ cigday_1
57 EduPar ~~ EduPar
58 EduPar ~~ n_trauma
59 EduPar ~~ Age
60 EduPar ~~ int_dis
61 EduPar ~~ medication
62 EduPar ~~ contraceptives
63 EduPar ~~ V8
64 EduPar ~~ cigday_1
65 n_trauma ~~ n_trauma
66 n_trauma ~~ Age
67 n_trauma ~~ int_dis
68 n_trauma ~~ medication
69 n_trauma ~~ contraceptives
70 n_trauma ~~ V8
71 n_trauma ~~ cigday_1
72 Age ~~ Age
73 Age ~~ int_dis
74 Age ~~ medication
75 Age ~~ contraceptives
76 Age ~~ V8
77 Age ~~ cigday_1
78 int_dis ~~ int_dis
79 int_dis ~~ medication
80 int_dis ~~ contraceptives
81 int_dis ~~ V8
82 int_dis ~~ cigday_1
83 medication ~~ medication
84 medication ~~ contraceptives
85 medication ~~ V8
86 medication ~~ cigday_1
87 contraceptives ~~ contraceptives
88 contraceptives ~~ V8
89 contraceptives ~~ cigday_1
90 V8 ~~ V8
91 V8 ~~ cigday_1
92 cigday_1 ~~ cigday_1
93 group ~*~ group
94 Epi ~1
95 group ~1
96 Matsmk ~1
97 Matagg ~1
98 FamScore ~1
99 EduPar ~1
100 n_trauma ~1
101 Age ~1
102 int_dis ~1
103 medication ~1
104 contraceptives ~1
105 V8 ~1
106 cigday_1 ~1
107 directMatsmk := f directMatsmk
108 directMatagg := g directMatagg
109 directFamScore := h directFamScore
110 directEduPar := i directEduPar
111 directn_trauma := j directn_trauma
112 EpiMatsmk := a*z EpiMatsmk
113 EpiMatagg := b*z EpiMatagg
114 EpiFamScore := c*z EpiFamScore
115 EpiEduPar := d*z EpiEduPar
116 Epin_trauma := e*z Epin_trauma
117 total := f+g+h+i+j+(a*z)+(b*z)+(c*z)+(d*z)+(e*z) total
est se z pvalue ci.lower ci.upper
1 0.025 0.082 0.301 0.764 -0.136 0.185
2 0.132 0.124 1.068 0.285 -0.110 0.374
3 -0.102 0.109 -0.934 0.350 -0.315 0.112
4 -0.281 0.169 -1.658 0.097 -0.613 0.051
5 0.074 0.126 0.589 0.556 -0.173 0.322
6 0.122 0.179 0.679 0.497 -0.230 0.473
7 0.085 0.074 1.148 0.251 -0.060 0.231
8 0.050 0.080 0.623 0.533 -0.107 0.207
9 -0.074 0.084 -0.872 0.383 -0.239 0.092
10 0.285 1.060 0.268 0.788 -1.793 2.362
11 0.168 0.140 1.205 0.228 -0.105 0.442
12 0.095 0.963 0.099 0.921 -1.792 1.983
13 0.666 2.153 0.309 0.757 -3.554 4.886
14 0.738 1.855 0.398 0.691 -2.897 4.374
15 -1.600 1.990 -0.804 0.421 -5.501 2.300
16 1.913 1.223 1.564 0.118 -0.484 4.311
17 -3.678 2.115 -1.739 0.082 -7.823 0.467
18 0.728 0.683 1.066 0.286 -0.611 2.068
19 0.837 0.699 1.197 0.231 -0.533 2.206
20 0.622 0.626 0.994 0.320 -0.604 1.848
21 11.409 12.905 0.884 0.377 -13.884 36.702
22 9.298 7.066 1.316 0.188 -4.551 23.147
23 6.542 0.104 63.023 0.000 6.339 6.746
24 5.748 0.000 NA NA 5.748 5.748
25 1.000 0.000 NA NA 1.000 1.000
26 -41.801 0.000 NA NA -41.801 -41.801
27 0.196 0.000 NA NA 0.196 0.196
28 0.049 0.000 NA NA 0.049 0.049
29 -0.009 0.000 NA NA -0.009 -0.009
30 -0.006 0.000 NA NA -0.006 -0.006
31 0.007 0.000 NA NA 0.007 0.007
32 -0.003 0.000 NA NA -0.003 -0.003
33 0.022 0.000 NA NA 0.022 0.022
34 0.004 0.000 NA NA 0.004 0.004
35 0.022 0.000 NA NA 0.022 0.022
36 0.003 0.000 NA NA 0.003 0.003
37 0.017 0.000 NA NA 0.017 0.017
38 0.091 0.000 NA NA 0.091 0.091
39 0.034 0.000 NA NA 0.034 0.034
40 -0.017 0.000 NA NA -0.017 -0.017
41 0.007 0.000 NA NA 0.007 0.007
42 0.000 0.000 NA NA 0.000 0.000
43 0.033 0.000 NA NA 0.033 0.033
44 0.008 0.000 NA NA 0.008 0.008
45 0.008 0.000 NA NA 0.008 0.008
46 0.001 0.000 NA NA 0.001 0.001
47 0.010 0.000 NA NA 0.010 0.010
48 0.132 0.000 NA NA 0.132 0.132
49 -0.029 0.000 NA NA -0.029 -0.029
50 0.027 0.000 NA NA 0.027 0.027
51 0.004 0.000 NA NA 0.004 0.004
52 0.065 0.000 NA NA 0.065 0.065
53 0.004 0.000 NA NA 0.004 0.004
54 0.058 0.000 NA NA 0.058 0.058
55 0.001 0.000 NA NA 0.001 0.001
56 0.044 0.000 NA NA 0.044 0.044
57 0.054 0.000 NA NA 0.054 0.054
58 -0.008 0.000 NA NA -0.008 -0.008
59 0.003 0.000 NA NA 0.003 0.003
60 -0.019 0.000 NA NA -0.019 -0.019
61 0.010 0.000 NA NA 0.010 0.010
62 -0.013 0.000 NA NA -0.013 -0.013
63 0.000 0.000 NA NA 0.000 0.000
64 -0.015 0.000 NA NA -0.015 -0.015
65 0.051 0.000 NA NA 0.051 0.051
66 0.002 0.000 NA NA 0.002 0.002
67 0.042 0.000 NA NA 0.042 0.042
68 0.018 0.000 NA NA 0.018 0.018
69 0.018 0.000 NA NA 0.018 0.018
70 0.000 0.000 NA NA 0.000 0.000
71 0.021 0.000 NA NA 0.021 0.021
72 0.047 0.000 NA NA 0.047 0.047
73 0.008 0.000 NA NA 0.008 0.008
74 -0.002 0.000 NA NA -0.002 -0.002
75 0.035 0.000 NA NA 0.035 0.035
76 -0.001 0.000 NA NA -0.001 -0.001
77 0.009 0.000 NA NA 0.009 0.009
78 0.213 0.000 NA NA 0.213 0.213
79 0.061 0.000 NA NA 0.061 0.061
80 0.061 0.000 NA NA 0.061 0.061
81 0.006 0.000 NA NA 0.006 0.006
82 0.044 0.000 NA NA 0.044 0.044
83 0.146 0.000 NA NA 0.146 0.146
84 0.035 0.000 NA NA 0.035 0.035
85 0.002 0.000 NA NA 0.002 0.002
86 0.003 0.000 NA NA 0.003 0.003
87 0.213 0.000 NA NA 0.213 0.213
88 0.003 0.000 NA NA 0.003 0.003
89 0.049 0.000 NA NA 0.049 0.049
90 0.005 0.000 NA NA 0.005 0.005
91 0.002 0.000 NA NA 0.002 0.002
92 0.062 0.000 NA NA 0.062 0.062
93 1.000 0.000 NA NA 1.000 1.000
94 0.000 0.000 NA NA 0.000 0.000
95 0.000 0.000 NA NA 0.000 0.000
96 0.262 0.000 NA NA 0.262 0.262
97 0.100 0.000 NA NA 0.100 0.100
98 0.225 0.000 NA NA 0.225 0.225
99 0.606 0.000 NA NA 0.606 0.606
100 0.196 0.000 NA NA 0.196 0.196
101 0.562 0.000 NA NA 0.562 0.562
102 0.300 0.000 NA NA 0.300 0.300
103 0.175 0.000 NA NA 0.175 0.175
104 0.300 0.000 NA NA 0.300 0.300
105 0.529 0.000 NA NA 0.529 0.529
106 0.124 0.000 NA NA 0.124 0.124
107 0.095 0.963 0.099 0.921 -1.792 1.983
108 0.666 2.153 0.309 0.757 -3.554 4.886
109 0.738 1.855 0.398 0.691 -2.897 4.374
110 -1.600 1.990 -0.804 0.421 -5.501 2.300
111 1.913 1.223 1.564 0.118 -0.484 4.311
112 0.161 0.535 0.301 0.764 -0.887 1.209
113 0.864 0.809 1.068 0.286 -0.721 2.449
114 -0.665 0.712 -0.933 0.351 -2.061 0.731
115 -1.837 1.108 -1.658 0.097 -4.008 0.334
116 0.487 0.827 0.589 0.556 -1.134 2.108
117 0.823 4.116 0.200 0.842 -7.244 8.889
label lhs edge rhs est std group
1 a Matsmk ~> Epi 2.458188e-02 0.0108076417
2 b Matagg ~> Epi 1.320289e-01 0.0395785961
3 c FamScore ~> Epi -1.016065e-01 -0.0366948417
4 d EduPar ~> Epi -2.807573e-01 -0.0646596116
5 e n_trauma ~> Epi 7.445977e-02 0.0167533797
6 Age ~> Epi 1.216017e-01 0.0263131775
7 int_dis ~> Epi 8.529333e-02 0.0390566463
8 medication ~> Epi 5.004593e-02 0.0190013736
9 contraceptives ~> Epi -7.362493e-02 -0.0337135711
10 V8 ~> Epi 2.846099e-01 0.0192441826
11 cigday_1 ~> Epi 1.683723e-01 0.0414679492
12 f Matsmk ~> group 9.543636e-02 0.0105047481
13 g Matagg ~> group 6.661429e-01 0.0499936408
14 h FamScore ~> group 7.383498e-01 0.0667577132
15 i EduPar ~> group -1.600452e+00 -0.0922784995
16 j n_trauma ~> group 1.913070e+00 0.1077624507
17 Age ~> group -3.677979e+00 -0.1992502675
18 int_dis ~> group 7.283543e-01 0.0834984636
19 medication ~> group 8.366138e-01 0.0795238059
20 contraceptives ~> group 6.219800e-01 0.0713037225
21 V8 ~> group 1.140922e+01 0.1931351079
22 cigday_1 ~> group 9.297883e+00 0.5733000173
23 z Epi ~> group 6.542238e+00 1.6378805741
25 Epi <-> Epi 1.000000e+00 0.9860014692
26 group <-> group -4.180088e+01 -2.5832994533
27 Matsmk <-> Matsmk 1.960443e-01 1.0000000000
28 Matsmk <-> Matagg 4.936709e-02 0.3693241433
29 Matsmk <-> FamScore -9.177215e-03 -0.0569887592
30 Matsmk <-> EduPar -5.564346e-03 -0.0541843459
31 Matsmk <-> n_trauma 7.459313e-03 0.0743497863
32 Matsmk <-> Age -3.217300e-03 -0.0333441329
33 Matsmk <-> int_dis 2.151899e-02 0.1053910232
34 Matsmk <-> medication 4.113924e-03 0.0242997446
35 Matsmk <-> contraceptives 2.151899e-02 0.1053910232
36 Matsmk <-> V8 2.928139e-03 0.0971189320
37 Matsmk <-> cigday_1 1.693829e-02 0.1542372547
38 Matagg <-> Matagg 9.113924e-02 1.0000000000
39 Matagg <-> FamScore 3.417722e-02 0.3112715087
40 Matagg <-> EduPar -1.656118e-02 -0.2365241196
41 Matagg <-> n_trauma 7.233273e-03 0.1057402114
42 Matagg <-> Age 3.118694e-04 0.0047405101
43 Matagg <-> int_dis 3.291139e-02 0.2364027144
44 Matagg <-> medication 7.594937e-03 0.0657951695
45 Matagg <-> contraceptives 7.594937e-03 0.0545544726
46 Matagg <-> V8 8.272067e-04 0.0402393217
47 Matagg <-> cigday_1 1.018987e-02 0.1360858260
48 FamScore <-> FamScore 1.322785e-01 1.0000000000
49 FamScore <-> EduPar -2.948312e-02 -0.3495149022
50 FamScore <-> n_trauma 2.667269e-02 0.3236534989
51 FamScore <-> Age 3.636947e-03 0.0458878230
52 FamScore <-> int_dis 6.455696e-02 0.3849084009
53 FamScore <-> medication 4.430380e-03 0.0318580293
54 FamScore <-> contraceptives 5.822785e-02 0.3471722832
55 FamScore <-> V8 7.814844e-04 0.0315547719
56 FamScore <-> cigday_1 4.381329e-02 0.4856887960
57 EduPar <-> EduPar 5.379307e-02 1.0000000000
58 EduPar <-> n_trauma -8.024412e-03 -0.1526891136
59 EduPar <-> Age 2.762108e-03 0.0546490350
60 EduPar <-> int_dis -1.909283e-02 -0.1785114035
61 EduPar <-> medication 1.017932e-02 0.1147832062
62 EduPar <-> contraceptives -1.329114e-02 -0.1242676068
63 EduPar <-> V8 -8.860887e-06 -0.0005610523
64 EduPar <-> cigday_1 -1.493803e-02 -0.2596730517
65 n_trauma <-> n_trauma 5.134332e-02 1.0000000000
66 n_trauma <-> Age 1.582278e-03 0.0320439451
67 n_trauma <-> int_dis 4.159132e-02 0.3980335009
68 n_trauma <-> medication 1.763110e-02 0.2034979577
69 n_trauma <-> contraceptives 1.808318e-02 0.1730580439
70 n_trauma <-> V8 -4.694340e-04 -0.0304243917
71 n_trauma <-> cigday_1 2.128165e-02 0.3786692420
72 Age <-> Age 4.748866e-02 1.0000000000
73 Age <-> int_dis 8.090259e-03 0.0805056484
74 Age <-> medication -1.655660e-03 -0.0198700345
75 Age <-> contraceptives 3.524124e-02 0.3506833348
76 Age <-> V8 -1.333633e-03 -0.0898732659
77 Age <-> cigday_1 8.542355e-03 0.1580445206
78 int_dis <-> int_dis 2.126582e-01 1.0000000000
79 int_dis <-> medication 6.075949e-02 0.3445843938
80 int_dis <-> contraceptives 6.075949e-02 0.2857142857
81 int_dis <-> V8 5.645344e-03 0.1797788722
82 int_dis <-> cigday_1 4.449367e-02 0.3890038953
83 medication <-> medication 1.462025e-01 1.0000000000
84 medication <-> contraceptives 3.544304e-02 0.2010075631
85 medication <-> V8 2.084232e-03 0.0800493604
86 medication <-> cigday_1 3.275316e-03 0.0345360471
87 contraceptives <-> contraceptives 2.126582e-01 1.0000000000
88 contraceptives <-> V8 2.688833e-03 0.0856272484
89 contraceptives <-> cigday_1 4.892405e-02 0.4277382803
90 V8 <-> V8 4.636832e-03 1.0000000000
91 V8 <-> cigday_1 1.509276e-03 0.0893623867
92 cigday_1 <-> cigday_1 6.151859e-02 1.0000000000
93 group <-> group 1.000000e+00 1.0000000000
94 int Epi 0.000000e+00 0.0000000000
95 int group 0.000000e+00 0.0000000000
96 int Matsmk 2.625000e-01 0.5928600601
97 int Matagg 1.000000e-01 0.3312434486
98 int FamScore 2.250000e-01 0.6186398880
99 int EduPar 6.062500e-01 2.6138976225
100 int n_trauma 1.964286e-01 0.8668873691
101 int Age 5.621377e-01 2.5795724974
102 int int_dis 3.000000e-01 0.6505492185
103 int medication 1.750000e-01 0.4576785957
104 int contraceptives 3.000000e-01 0.6505492185
105 int V8 5.286908e-01 7.7640983340
106 int cigday_1 1.243750e-01 0.5014526157
fixed par
1 FALSE 1
2 FALSE 2
3 FALSE 3
4 FALSE 4
5 FALSE 5
6 FALSE 6
7 FALSE 7
8 FALSE 8
9 FALSE 9
10 FALSE 10
11 FALSE 11
12 FALSE 12
13 FALSE 13
14 FALSE 14
15 FALSE 15
16 FALSE 16
17 FALSE 17
18 FALSE 18
19 FALSE 19
20 FALSE 20
21 FALSE 21
22 FALSE 22
23 FALSE 23
25 TRUE 0
26 TRUE 0
27 TRUE 0
28 TRUE 0
29 TRUE 0
30 TRUE 0
31 TRUE 0
32 TRUE 0
33 TRUE 0
34 TRUE 0
35 TRUE 0
36 TRUE 0
37 TRUE 0
38 TRUE 0
39 TRUE 0
40 TRUE 0
41 TRUE 0
42 TRUE 0
43 TRUE 0
44 TRUE 0
45 TRUE 0
46 TRUE 0
47 TRUE 0
48 TRUE 0
49 TRUE 0
50 TRUE 0
51 TRUE 0
52 TRUE 0
53 TRUE 0
54 TRUE 0
55 TRUE 0
56 TRUE 0
57 TRUE 0
58 TRUE 0
59 TRUE 0
60 TRUE 0
61 TRUE 0
62 TRUE 0
63 TRUE 0
64 TRUE 0
65 TRUE 0
66 TRUE 0
67 TRUE 0
68 TRUE 0
69 TRUE 0
70 TRUE 0
71 TRUE 0
72 TRUE 0
73 TRUE 0
74 TRUE 0
75 TRUE 0
76 TRUE 0
77 TRUE 0
78 TRUE 0
79 TRUE 0
80 TRUE 0
81 TRUE 0
82 TRUE 0
83 TRUE 0
84 TRUE 0
85 TRUE 0
86 TRUE 0
87 TRUE 0
88 TRUE 0
89 TRUE 0
90 TRUE 0
91 TRUE 0
92 TRUE 0
93 TRUE 0
94 TRUE 0
95 TRUE 0
96 TRUE 0
97 TRUE 0
98 TRUE 0
99 TRUE 0
100 TRUE 0
101 TRUE 0
102 TRUE 0
103 TRUE 0
104 TRUE 0
105 TRUE 0
106 TRUE 0rmd_paths <-paste0(tempfile(c(names(Netlist))),".Rmd")
names(rmd_paths) <- names(Netlist)
for (n in names(rmd_paths)) {
sink(file = rmd_paths[n])
cat(" \n",
"```{r, echo = FALSE}",
"Netlist[[n]]",
"```",
sep = " \n")
sink()
}Interactive SEM plots
Only direct effects with a significant standardized effect of p<0.05 are shown.
for (n in names(rmd_paths)) {
cat(knitr::knit_child(rmd_paths[[n]],
quiet= TRUE))
file.remove(rmd_paths[[n]])
}
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
Random number generation:
RNG: Mersenne-Twister
Normal: Inversion
Sample: Rounding
locale:
[1] LC_COLLATE=German_Germany.1252 LC_CTYPE=German_Germany.1252
[3] LC_MONETARY=German_Germany.1252 LC_NUMERIC=C
[5] LC_TIME=German_Germany.1252
attached base packages:
[1] grid parallel stats4 stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] plyr_1.8.6 scales_1.1.1
[3] RCircos_1.2.1 compareGroups_4.4.6
[5] readxl_1.3.1 RRHO_1.28.0
[7] webshot_0.5.2 visNetwork_2.0.9
[9] org.Hs.eg.db_3.12.0 AnnotationDbi_1.52.0
[11] xlsx_0.6.5 gprofiler2_0.2.0
[13] BiocParallel_1.24.1 kableExtra_1.3.1
[15] glmpca_0.2.0 knitr_1.30
[17] DESeq2_1.30.0 SummarizedExperiment_1.20.0
[19] Biobase_2.50.0 MatrixGenerics_1.2.0
[21] matrixStats_0.57.0 GenomicRanges_1.42.0
[23] GenomeInfoDb_1.26.2 IRanges_2.24.1
[25] S4Vectors_0.28.1 BiocGenerics_0.36.0
[27] forcats_0.5.0 stringr_1.4.0
[29] dplyr_1.0.2 purrr_0.3.4
[31] readr_1.4.0 tidyr_1.1.2
[33] tibble_3.0.4 tidyverse_1.3.0
[35] semPlot_1.1.2 lavaan_0.6-7
[37] viridis_0.5.1 viridisLite_0.3.0
[39] WGCNA_1.69 fastcluster_1.1.25
[41] dynamicTreeCut_1.63-1 ggplot2_3.3.3
[43] gplots_3.1.1 corrplot_0.84
[45] RColorBrewer_1.1-2 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] coda_0.19-4 bit64_4.0.5 DelayedArray_0.16.0
[4] data.table_1.13.6 rpart_4.1-15 RCurl_1.98-1.2
[7] doParallel_1.0.16 generics_0.1.0 preprocessCore_1.52.1
[10] callr_3.5.1 lambda.r_1.2.4 RSQLite_2.2.2
[13] mice_3.12.0 chron_2.3-56 bit_4.0.4
[16] xml2_1.3.2 lubridate_1.7.9.2 httpuv_1.5.5
[19] assertthat_0.2.1 d3Network_0.5.2.1 xfun_0.20
[22] hms_1.0.0 rJava_0.9-13 evaluate_0.14
[25] promises_1.1.1 fansi_0.4.1 caTools_1.18.1
[28] dbplyr_2.0.0 igraph_1.2.6 DBI_1.1.1
[31] geneplotter_1.68.0 tmvnsim_1.0-2 Rsolnp_1.16
[34] htmlwidgets_1.5.3 futile.logger_1.4.3 ellipsis_0.3.1
[37] crosstalk_1.1.1 backports_1.2.0 pbivnorm_0.6.0
[40] annotate_1.68.0 vctrs_0.3.6 abind_1.4-5
[43] cachem_1.0.1 withr_2.4.1 HardyWeinberg_1.7.1
[46] checkmate_2.0.0 fdrtool_1.2.16 mnormt_2.0.2
[49] cluster_2.1.0 mi_1.0 lazyeval_0.2.2
[52] crayon_1.3.4 genefilter_1.72.0 pkgconfig_2.0.3
[55] nlme_3.1-151 nnet_7.3-15 rlang_0.4.10
[58] lifecycle_0.2.0 kutils_1.70 modelr_0.1.8
[61] VennDiagram_1.6.20 cellranger_1.1.0 rprojroot_2.0.2
[64] flextable_0.6.2 Matrix_1.2-18 regsem_1.6.2
[67] carData_3.0-4 boot_1.3-26 reprex_1.0.0
[70] base64enc_0.1-3 processx_3.4.5 whisker_0.4
[73] png_0.1-7 rjson_0.2.20 bitops_1.0-6
[76] KernSmooth_2.23-18 blob_1.2.1 arm_1.11-2
[79] jpeg_0.1-8.1 rockchalk_1.8.144 memoise_2.0.0
[82] magrittr_2.0.1 zlibbioc_1.36.0 compiler_4.0.3
[85] lme4_1.1-26 cli_2.2.0 XVector_0.30.0
[88] pbapply_1.4-3 ps_1.5.0 htmlTable_2.1.0
[91] formatR_1.7 Formula_1.2-4 MASS_7.3-53
[94] tidyselect_1.1.0 stringi_1.5.3 lisrelToR_0.1.4
[97] sem_3.1-11 yaml_2.2.1 OpenMx_2.18.1
[100] locfit_1.5-9.4 latticeExtra_0.6-29 tools_4.0.3
[103] matrixcalc_1.0-3 rstudioapi_0.13 uuid_0.1-4
[106] foreach_1.5.1 foreign_0.8-81 git2r_0.28.0
[109] gridExtra_2.3 farver_2.0.3 BDgraph_2.63
[112] digest_0.6.27 shiny_1.6.0 Rcpp_1.0.5
[115] broom_0.7.3 later_1.1.0.1 writexl_1.3.1
[118] gdtools_0.2.3 httr_1.4.2 psych_2.0.12
[121] colorspace_2.0-0 rvest_0.3.6 XML_3.99-0.5
[124] fs_1.5.0 truncnorm_1.0-8 splines_4.0.3
[127] statmod_1.4.35 xlsxjars_0.6.1 systemfonts_0.3.2
[130] plotly_4.9.3 xtable_1.8-4 jsonlite_1.7.2
[133] nloptr_1.2.2.2 futile.options_1.0.1 corpcor_1.6.9
[136] glasso_1.11 R6_2.5.0 Hmisc_4.4-2
[139] mime_0.9 pillar_1.4.7 htmltools_0.5.1.1
[142] glue_1.4.2 fastmap_1.1.0 minqa_1.2.4
[145] codetools_0.2-18 lattice_0.20-41 huge_1.3.4.1
[148] gtools_3.8.2 officer_0.3.16 zip_2.1.1
[151] GO.db_3.12.1 openxlsx_4.2.3 survival_3.2-7
[154] rmarkdown_2.6 qgraph_1.6.5 munsell_0.5.0
[157] GenomeInfoDbData_1.2.4 iterators_1.0.13 impute_1.64.0
[160] haven_2.3.1 reshape2_1.4.4 gtable_0.3.0