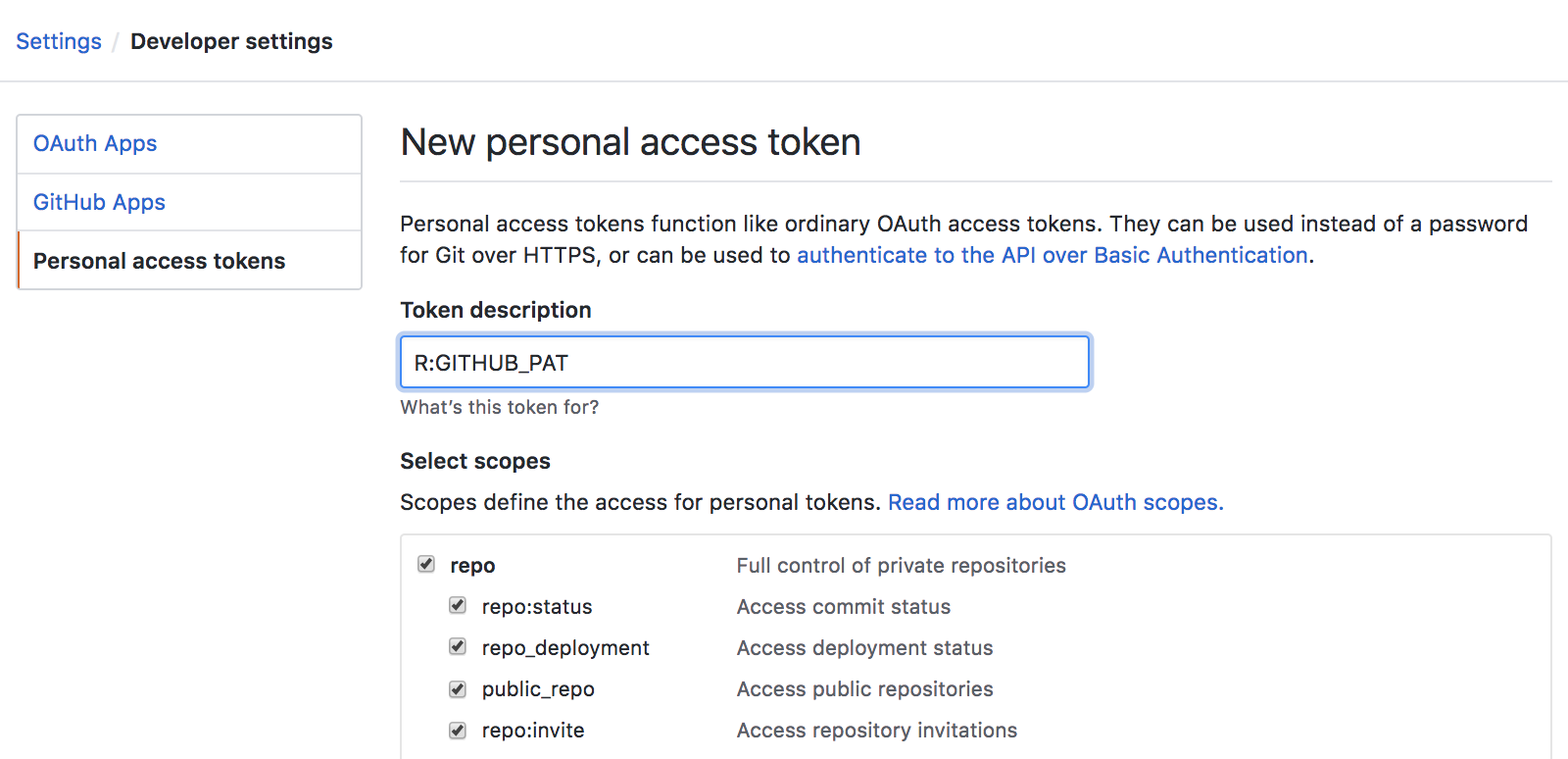
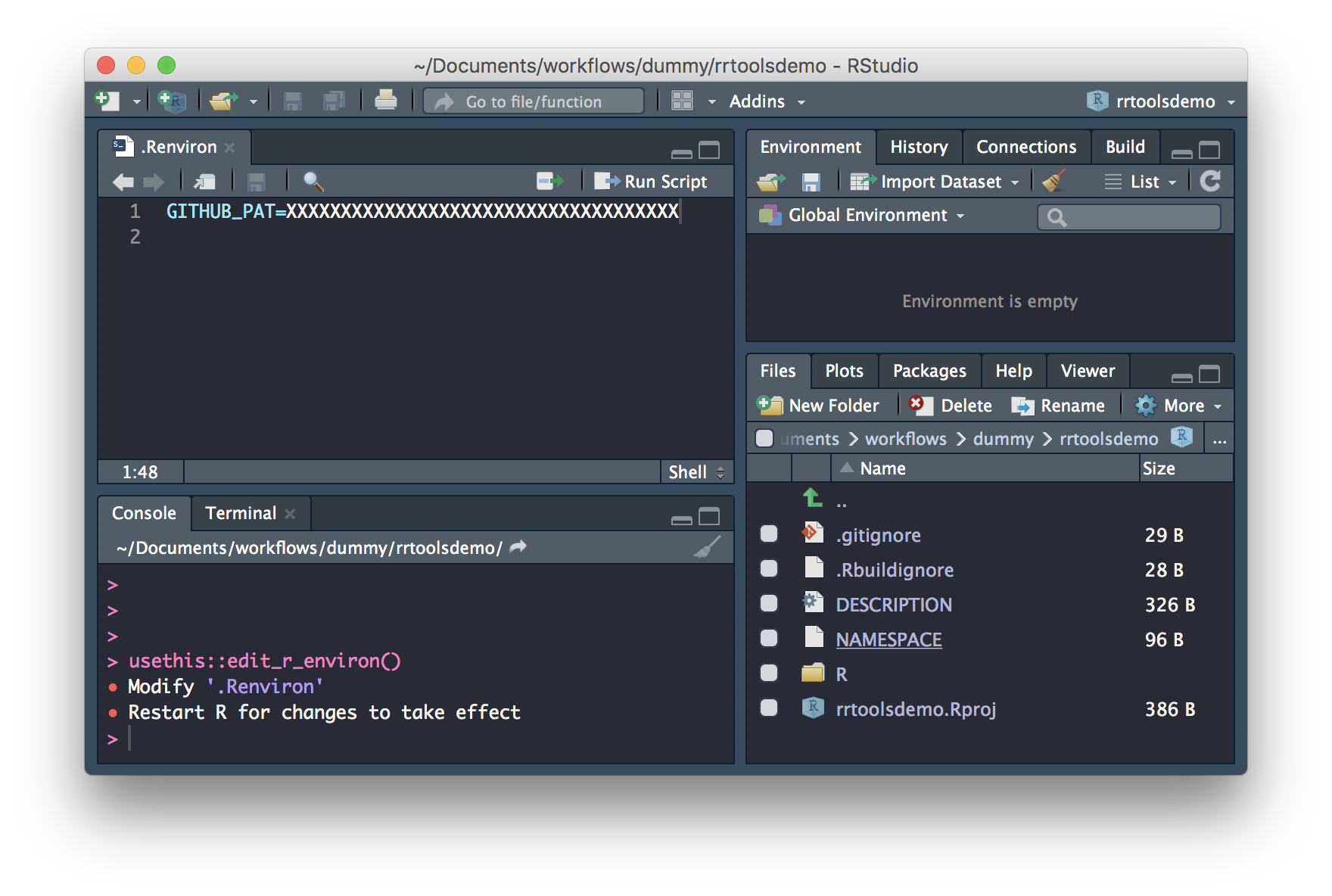
Create a Research Compendium
Last updated: 2018-10-31
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20181015)The command
set.seed(20181015)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 4d17ca9
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: analysis/.DS_Store Ignored: analysis/data/ Ignored: analysis/package.Rmd Ignored: assets/ Ignored: docs/.DS_Store Untracked files: Untracked: docs/assets/Boettiger-2018-Ecology_Letters.pdf Untracked: docs/assets/Packaging-Data-Analytical Work-Reproducibly-Using-R-and-Friends.pdf Untracked: docs/css/ Untracked: libs/ Unstaged changes: Modified: analysis/_site.yml Modified: analysis/index.Rmd
Expand here to see past versions:
Workshop setup
Launch Rstudio
Let’s begin by launching Rstudio
Install packages
Next, let’s install the packages we’ll need, starting with rrtools (if you haven’t got devtools installed, you’ll need to before you can install rrtools from GitHub).
Installing rrtools imports many of the packages we’ll need today (eg, have a look at the imports section of the DESCRIPTION file).
Imports: devtools, git2r, whisker, rstudioapi, rmarkdown, knitr,
bookdown, curl, RCurl, jsonlite, methods, httr, usethis, clisymbols,
crayon, glue, readr (>= 1.1.1)Now, install some additional packages we’ll need for the workshop.
install.packages(c(
# source paper analysis
"dplyr", "ggplot2", "ggthemes", "here",
# bibliographic / publishing
"citr", "rticles",
# documentation
"roxygen2",
# graphics
"Cairo"))Get workshop materials
Today we’ll be working with a subset of materials from the published compendium of code, data, and author’s manuscript:
Carl Boettiger. (2018, April 17). cboettig/noise-phenomena: Supplement to: “From noise to knowledge: how randomness generates novel phenomena and reveals information” (Version revision-2). Zenodo. http://doi.org/10.5281/zenodo.1219780
accompanying the publication:
Carl Boettiger  . From noise to knowledge: how randomness generates novel phenomena and reveals information. Published in Ecology Letters, 22 May 2018 https://doi.org/10.1111/ele.13085
. From noise to knowledge: how randomness generates novel phenomena and reveals information. Published in Ecology Letters, 22 May 2018 https://doi.org/10.1111/ele.13085

You can download the materials using usethis::use_course() and supplying a path to a destination folder to argument destdir:
This will download everything we need from a GitHub repository as a .zip file, unzip it and launch it in a new Rstudio session for us to explore.
Inspect materials
├── README.md <- .......................repo README
├── analysis.R <- ......................analysis underlying paper
├── gillespie.csv <- ...................data
├── paper.pdf <- .......................LaTex pdf of the paper
├── paper.txt <- .......................text body of the paper
├── refs.bib <- ........................bibtex bibliographic file
└── rrtools-wkshp-materials.Rproj <- ...rstudio project fileIn this workshop we’ll attempt a partial reproduction of the original paper using the materials we’ve just downloaded.
We’ll use this as an opportunity to create a new research compendium using rrtools and friends! 🎊
Create compendium
Now that we’ve got all the materials we need, let’s start by *creating a blank research compendium for us to work in.
load library
First we need to load rrtools
This performs a quick check to confirm you have Git installed and configured
If you do, you should see the following output in the console.
configure git
If your git configuration hasn’t been set yet, you can use usethis::use_git_config()
Set git configuration:
Check git configuration:
$user.name
[1] "Jane"
$user.email
[1] "jane@example.org"create compendium
Now we’re ready to create our compendium. We use function rrtools::use_compendium and supply it with a path at which our compendium will be created. The final part of our path becomes the compendium name. Because the function effectively creates a package, only a single string of lowercase alpha characters is accepted as a name. so let’s go for rrcompendium as the final part of our path.
To create rrcompendium in a directory called Documents/workflows/ I use:
Go ahead and create a compendium at a location of your choice. Stick with compendium name rrcompendium for ease of following the materials. If the call was successfull you should see the following console output:
✔ Setting active project to '/Users/Anna/Documents/workflows/rrcompendium'
✔ Creating 'R/'
✔ Creating 'man/'
✔ Writing 'DESCRIPTION'
✔ Writing 'NAMESPACE'
✔ Writing 'rrcompendium.Rproj'
✔ Adding '.Rproj.user' to '.gitignore'
✔ Adding '^rrcompendium\\.Rproj$', '^\\.Rproj\\.user$' to '.Rbuildignore'
✔ Opening new project 'rrcompendium' in RStudio
✔ The package rrcompendium has been created
✔ Opening the new compendium in a new RStudio session...
Next, you need to: ↓ ↓ ↓
● Edit the DESCRIPTION file
● Use other 'rrtools' functions to add components to the compendiumand a new Rstudio session launched for the compendium:
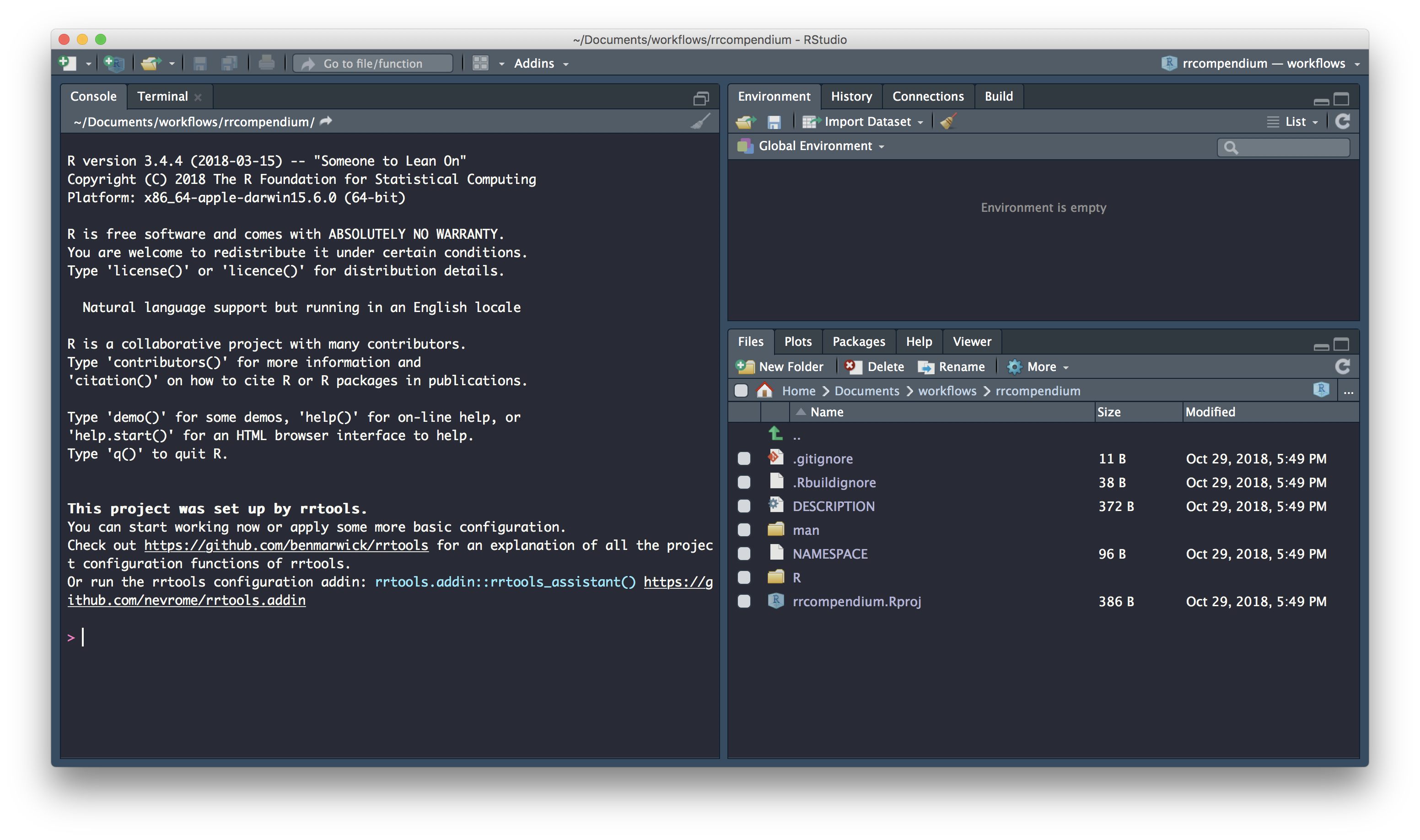
Initiate git
We can initialise our compendium with .git using:
N.B. Beware, you may have ended up with two Rstudio sessions of rrcompendium. Make sure to only have one session of a single project at one time to avoid problems.
Inspect templates
.
├── DESCRIPTION <- .............................package metadata
| dependency management
├── NAMESPACE <- ...............................AUTO-GENERATED on build
├── R <- .......................................folder for functions
├── man <- .....................................AUTO-GENERATED on build
└── rrcompendium.Rproj <- ......................rstudio project filerrtools::use_compendium() creates the bare backbone of infrastructure required for a research compendium. At this point it provides facilities to store general metadata about our compendium (eg bibliographic details to create a citation) and manage dependencies in the DESCRIPTION file and store and document functions in the R/ folder. Together these allow us to manage, install and share functionality associated with our project.
update description file
Let’s update some basic details in the DESCRIPTION file:
Package: rrcompendium
Title: What the Package Does (One Line, Title Case)
Version: 0.0.0.9000
Authors@R:
person(given = "First",
family = "Last",
role = c("aut", "cre"),
email = "first.last@example.com")
Description: What the package does (one paragraph)
License: What license it uses
ByteCompile: true
Encoding: UTF-8
LazyData: true
Title
Let’s start with giving our compendium a descriptive title:
Title: Partial Reproduction of Boettiger Ecology Letters 2018;21:1255–1267
with rrtoolsVersion
We don’t need to change the version now but using semantic versioning for our compendium can be a really useful way to track versions. In general, versions below 0.0.1 are in development, hence the DESCRIPTION file defaults to 0.0.0.9000.
Description
Let’s add a bit more detail about the contents of the compendium in the Description.
Description: This repository contains the research compendium of the
partial reproduction of Boettiger Ecology Letters 2018;21:1255–1267.
The compendium contains all data, code, and text associated with this sub-section of the analysisLicense
Finally, let’s add a license for the material we create. We’ll use an MIT license. Note however that his only covers the code. We can do this with:
✔ Setting License field in DESCRIPTION to 'MIT + file LICENSE'
✔ Writing 'LICENSE.md'
✔ Adding '^LICENSE\\.md$' to '.Rbuildignore'This creates files LICENSE and LICENSE.md and updates the DESCRIPTION file with details of the license.
License: MIT + file LICENSERecap
We’ve finished updating our DESCRIPTION file! 🎉
It should look a bit like this:
Package: rrcompendium
Title: Partial Reproduction of Boettiger Ecology Letters 2018;21:1255–1267
with rrtools
Version: 0.0.0.9000
Authors@R:
person(given = "Anna",
family = "Krystalli",
role = c("aut", "cre"),
email = "annakrystalli@googlemail.com")
Description: This repository contains the research compendium of the partial
reproduction of Boettiger Ecology Letters 2018;21:1255–1267. The compendium
contains all data, code, and text associated with this sub-section of the
analysis.
License: MIT + file LICENSE
ByteCompile: true
Encoding: UTF-8
LazyData: true
and your project folder should contain:
.
├── DESCRIPTION
├── LICENSE
├── LICENSE.md
├── NAMESPACE
├── R
├── man
└── rrcompendium.RprojLet’s commit our work and move on to preparing our compendium for sharing on GitHub.
Setting up the analysis folder
Create analysis
We now need an analysis folder to contain our analysis and paper. We can do this using function rrtools::use_analysis()
The function has three location = options:
top_levelto create a top-levelanalysis/directoryinstto create aninst/directory (so that all the sub-directories are available after the package is installed)vignettesto create avignettes/directory (and automatically update theDESCRIPTION).
The default is a top-level analysis/.
✔ Adding bookdown to Imports
✔ Creating 'analysis' directory and contents
✔ Creating 'analysis'
✔ Creating 'analysis/paper'
✔ Creating 'analysis/figures'
✔ Creating 'analysis/templates'
✔ Creating 'analysis/data'
✔ Creating 'analysis/data/raw_data'
✔ Creating 'analysis/data/derived_data'
✔ Creating 'references.bib' from template.
✔ Creating 'paper.Rmd' from template.
Next, you need to: ↓ ↓ ↓ ↓
● Write your article/report/thesis, start at the paper.Rmd file
● Add the citation style library file (csl) to replace the default provided here, see https://github.com/citation-style-language/
● Add bibliographic details of cited items to the 'references.bib' file
● For adding captions & cross-referencing in an Rmd, see https://bookdown.org/yihui/bookdown/
● For adding citations & reference lists in an Rmd, see http://rmarkdown.rstudio.com/authoring_bibliographies_and_citations.html
Note that:
⚠ Your data files are tracked by Git and will be pushed to GitHubRegardless for location option, the contents of the created sub-directories are the same:
analysis/
|
├── paper/
│ ├── paper.Rmd # this is the main document to edit
│ └── references.bib # this contains the reference list information
├── figures/ # location of the figures produced by the Rmd
|
├── data/
│ ├── DO-NOT-EDIT-ANY-FILES-IN-HERE-BY-HAND
│ ├── raw_data/ # data obtained from elsewhere
│ └── derived_data/ # data generated during the analysis
|
└── templates
├── journal-of-archaeological-science.csl
| # this sets the style of citations & reference list
├── template.docx # used to style the output of the paper.Rmd
└── template.RmdLet’s inspect
paper.Rmd
paper.Rmd is ready to write in and render with bookdown. It includes:
a YAML header that identifies the
references.bibfile and the suppliedcslfile (Citation Style Language) to style the reference list)a colophon that adds some git commit details to the end of the document. This means that the output file (HTML/PDF/Word) is always traceable to a specific state of the code.
references.bib
The references.bib file has just one item to demonstrate the format. It is ready to insert more reference details.
We can replace the supplied csl file with a different citation style from https://github.com/citation-style-language/
Next, let’s set up functionality as a package!
Session information
This reproducible R Markdown analysis was created with workflowr 1.1.1