Differential Splicing
Briana Mittleman
11/11/2019
Last updated: 2019-12-18
Checks: 7 0
Knit directory: Comparative_APA/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.5.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190902) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/chimp_log/
Ignored: code/human_log/
Ignored: data/.DS_Store
Ignored: data/metadata_HCpanel.txt.sb-a5794dd2-i594qs/
Untracked files:
Untracked: ._.DS_Store
Untracked: Chimp/
Untracked: Human/
Untracked: analysis/assessReadQual.Rmd
Untracked: analysis/diffExpressionPantro6.Rmd
Untracked: code/._ClassifyLeafviz.sh
Untracked: code/._Config_chimp.yaml
Untracked: code/._Config_chimp_full.yaml
Untracked: code/._Config_human.yaml
Untracked: code/._ConvertJunc2Bed.sh
Untracked: code/._CountNucleotides.py
Untracked: code/._CrossMapChimpRNA.sh
Untracked: code/._DiffSplice.sh
Untracked: code/._DiffSplicePlots.sh
Untracked: code/._DiffSplicePlots_gencode.sh
Untracked: code/._DiffSplice_gencode.sh
Untracked: code/._DiffSplice_removebad.sh
Untracked: code/._GetMAPQscore.py
Untracked: code/._GetSecondaryMap.py
Untracked: code/._LiftFinalChimpJunc2Human.sh
Untracked: code/._LiftOrthoPAS2chimp.sh
Untracked: code/._MapBadSamples.sh
Untracked: code/._QuantMergedClusters.sh
Untracked: code/._ReverseLiftFilter.R
Untracked: code/._RunFixLeafCluster.sh
Untracked: code/._Snakefile
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilePASfilt
Untracked: code/._SortIndexBadSamples.sh
Untracked: code/._bed215upbed.py
Untracked: code/._bed2SAF_gen.py
Untracked: code/._buildIndecpantro5
Untracked: code/._buildIndecpantro5.sh
Untracked: code/._buildLeafviz.sh
Untracked: code/._buildLeafviz_leadAnno.sh
Untracked: code/._buildStarIndex.sh
Untracked: code/._chimpChromprder.sh
Untracked: code/._cleanbed2saf.py
Untracked: code/._cluster.json
Untracked: code/._cluster2bed.py
Untracked: code/._clusterLiftReverse.sh
Untracked: code/._clusterLiftReverse_removebad.sh
Untracked: code/._clusterLiftprimary.sh
Untracked: code/._clusterLiftprimary_removebad.sh
Untracked: code/._converBam2Junc.sh
Untracked: code/._converBam2Junc_removeBad.sh
Untracked: code/._extraSnakefiltpas
Untracked: code/._filter5percPAS.py
Untracked: code/._filterNumChroms.py
Untracked: code/._filterPASforMP.py
Untracked: code/._filterPostLift.py
Untracked: code/._fixExonFC.py
Untracked: code/._fixLeafCluster.py
Untracked: code/._fixLiftedJunc.py
Untracked: code/._fixUTRexonanno.py
Untracked: code/._formathg38Anno.py
Untracked: code/._formatpantro6Anno.py
Untracked: code/._getRNAseqMapStats.sh
Untracked: code/._hg19MapStats.sh
Untracked: code/._humanChromorder.sh
Untracked: code/._intersectLiftedPAS.sh
Untracked: code/._liftJunctionFiles.sh
Untracked: code/._liftPAS19to38.sh
Untracked: code/._liftedchimpJunc2human.sh
Untracked: code/._makeSamplyGroupsHuman_TvN.py
Untracked: code/._mapRNAseqhg19.sh
Untracked: code/._mapRNAseqhg19_newPipeline.sh
Untracked: code/._maphg19.sh
Untracked: code/._maphg19_subjunc.sh
Untracked: code/._mergedBam2BW.sh
Untracked: code/._nameClusters.py
Untracked: code/._numMultimap.py
Untracked: code/._overlapapaQTLPAS.sh
Untracked: code/._prepareCleanLiftedFC_5perc4LC.py
Untracked: code/._prepareLeafvizAnno.sh
Untracked: code/._preparePAS4lift.py
Untracked: code/._primaryLift.sh
Untracked: code/._processhg38exons.py
Untracked: code/._quantJunc.sh
Untracked: code/._quantJunc_TEST.sh
Untracked: code/._quantJunc_removeBad.sh
Untracked: code/._quantMerged_seperatly.sh
Untracked: code/._recLiftchim2human.sh
Untracked: code/._revLiftPAShg38to19.sh
Untracked: code/._reverseLift.sh
Untracked: code/._runCheckReverseLift.sh
Untracked: code/._runChimpDiffIso.sh
Untracked: code/._runCountNucleotides.sh
Untracked: code/._runFilterNumChroms.sh
Untracked: code/._runHumanDiffIso.sh
Untracked: code/._runNuclearDifffIso.sh
Untracked: code/._run_chimpverifybam.sh
Untracked: code/._run_verifyBam.sh
Untracked: code/._snakemake.batch
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakePASchimp.batch
Untracked: code/._snakemakePAShuman.batch
Untracked: code/._snakemake_chimp.batch
Untracked: code/._snakemake_human.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._snakemakefiltPAS_chimp
Untracked: code/._snakemakefiltPAS_chimp.sh
Untracked: code/._snakemakefiltPAS_human.sh
Untracked: code/._submit-snakemake-chimp.sh
Untracked: code/._submit-snakemake-human.sh
Untracked: code/._submit-snakemakePAS-chimp.sh
Untracked: code/._submit-snakemakePAS-human.sh
Untracked: code/._submit-snakemakefiltPAS-chimp.sh
Untracked: code/._submit-snakemakefiltPAS-human.sh
Untracked: code/._subset_diffisopheno_Nuclear_HvC.py
Untracked: code/._transcriptDTplotsNuclear.sh
Untracked: code/._verifyBam4973.sh
Untracked: code/._verifyBam4973inHuman.sh
Untracked: code/._wrap_chimpverifybam.sh
Untracked: code/._wrap_verifyBam.sh
Untracked: code/._writeMergecode.py
Untracked: code/.snakemake/
Untracked: code/ClassifyLeafviz.sh
Untracked: code/Config_chimp.yaml
Untracked: code/Config_chimp_full.yaml
Untracked: code/Config_human.yaml
Untracked: code/ConvertJunc2Bed.err
Untracked: code/ConvertJunc2Bed.out
Untracked: code/ConvertJunc2Bed.sh
Untracked: code/CountNucleotides.py
Untracked: code/CrossMapChimpRNA.sh
Untracked: code/CrossmapChimpRNA.err
Untracked: code/CrossmapChimpRNA.out
Untracked: code/DiffSplice.err
Untracked: code/DiffSplice.out
Untracked: code/DiffSplice.sh
Untracked: code/DiffSplicePlots.err
Untracked: code/DiffSplicePlots.out
Untracked: code/DiffSplicePlots.sh
Untracked: code/DiffSplicePlots_gencode.sh
Untracked: code/DiffSplice_gencode.sh
Untracked: code/DiffSplice_removebad.err
Untracked: code/DiffSplice_removebad.out
Untracked: code/DiffSplice_removebad.sh
Untracked: code/FilterReverseLift.err
Untracked: code/FilterReverseLift.out
Untracked: code/GencodeDiffSplice.err
Untracked: code/GencodeDiffSplice.out
Untracked: code/GetMAPQscore.py
Untracked: code/GetSecondaryMap.py
Untracked: code/HchromOrder.err
Untracked: code/HchromOrder.out
Untracked: code/JunctionLift.err
Untracked: code/JunctionLift.out
Untracked: code/JunctionLiftFinalChimp.err
Untracked: code/JunctionLiftFinalChimp.out
Untracked: code/LiftClustersFirst.err
Untracked: code/LiftClustersFirst.out
Untracked: code/LiftClustersFirst_remove.err
Untracked: code/LiftClustersFirst_remove.out
Untracked: code/LiftClustersSecond.err
Untracked: code/LiftClustersSecond.out
Untracked: code/LiftClustersSecond_remove.err
Untracked: code/LiftClustersSecond_remove.out
Untracked: code/LiftFinalChimpJunc2Human.sh
Untracked: code/LiftOrthoPAS2chimp.sh
Untracked: code/LiftorthoPAS.err
Untracked: code/LiftorthoPASt.out
Untracked: code/Log.out
Untracked: code/MapBadSamples.err
Untracked: code/MapBadSamples.out
Untracked: code/MapBadSamples.sh
Untracked: code/MapStats.err
Untracked: code/MapStats.out
Untracked: code/MergeClusters.err
Untracked: code/MergeClusters.out
Untracked: code/MergeClusters.sh
Untracked: code/QuantMergeClusters
Untracked: code/QuantMergeClusters.err
Untracked: code/QuantMergeClusters.out
Untracked: code/QuantMergedClusters.sh
Untracked: code/Rev_liftoverPAShg19to38.err
Untracked: code/Rev_liftoverPAShg19to38.out
Untracked: code/ReverseLiftFilter.R
Untracked: code/RunFixCluster.err
Untracked: code/RunFixCluster.out
Untracked: code/RunFixLeafCluster.sh
Untracked: code/SAF215upbed_gen.py
Untracked: code/Snakefile
Untracked: code/SnakefilePAS
Untracked: code/SnakefilePASfilt
Untracked: code/SortIndexBadSamples.err
Untracked: code/SortIndexBadSamples.out
Untracked: code/SortIndexBadSamples.sh
Untracked: code/TotalTranscriptDTplot.err
Untracked: code/TotalTranscriptDTplot.out
Untracked: code/Upstream10Bases_general.py
Untracked: code/apaQTLsnake.err
Untracked: code/apaQTLsnake.out
Untracked: code/apaQTLsnakePAS.err
Untracked: code/apaQTLsnakePAS.out
Untracked: code/apaQTLsnakePAShuman.err
Untracked: code/bam2junc.err
Untracked: code/bam2junc.out
Untracked: code/bam2junc_remove.err
Untracked: code/bam2junc_remove.out
Untracked: code/bed215upbed.py
Untracked: code/bed2SAF_gen.py
Untracked: code/bed2saf.py
Untracked: code/bg_to_cov.py
Untracked: code/buildIndecpantro5
Untracked: code/buildIndecpantro5.sh
Untracked: code/buildLeafviz.err
Untracked: code/buildLeafviz.out
Untracked: code/buildLeafviz.sh
Untracked: code/buildLeafviz_leadAnno.sh
Untracked: code/buildLeafviz_leafanno.err
Untracked: code/buildLeafviz_leafanno.out
Untracked: code/buildStarIndex.sh
Untracked: code/callPeaksYL.py
Untracked: code/chimpChromprder.sh
Untracked: code/chooseAnno2Bed.py
Untracked: code/chooseAnno2SAF.py
Untracked: code/chromOrder.err
Untracked: code/chromOrder.out
Untracked: code/classifyLeafviz.err
Untracked: code/classifyLeafviz.out
Untracked: code/cleanbed2saf.py
Untracked: code/cluster.json
Untracked: code/cluster2bed.py
Untracked: code/clusterLiftReverse.sh
Untracked: code/clusterLiftReverse_removebad.sh
Untracked: code/clusterLiftprimary.sh
Untracked: code/clusterLiftprimary_removebad.sh
Untracked: code/clusterPAS.json
Untracked: code/clusterfiltPAS.json
Untracked: code/comands2Mege.sh
Untracked: code/converBam2Junc.sh
Untracked: code/converBam2Junc_removeBad.sh
Untracked: code/convertNumeric.py
Untracked: code/environment.yaml
Untracked: code/extraSnakefiltpas
Untracked: code/filter5perc.R
Untracked: code/filter5percPAS.py
Untracked: code/filter5percPheno.py
Untracked: code/filterBamforMP.pysam2_gen.py
Untracked: code/filterJuncChroms.err
Untracked: code/filterJuncChroms.out
Untracked: code/filterMissprimingInNuc10_gen.py
Untracked: code/filterNumChroms.py
Untracked: code/filterPASforMP.py
Untracked: code/filterPostLift.py
Untracked: code/filterSAFforMP_gen.py
Untracked: code/filterSortBedbyCleanedBed_gen.R
Untracked: code/filterpeaks.py
Untracked: code/fixExonFC.py
Untracked: code/fixFChead.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixLeafCluster.py
Untracked: code/fixLiftedJunc.py
Untracked: code/fixUTRexonanno.py
Untracked: code/formathg38Anno.py
Untracked: code/generateStarIndex.err
Untracked: code/generateStarIndex.out
Untracked: code/generateStarIndexHuman.err
Untracked: code/generateStarIndexHuman.out
Untracked: code/getRNAseqMapStats.sh
Untracked: code/hg19MapStats.err
Untracked: code/hg19MapStats.out
Untracked: code/hg19MapStats.sh
Untracked: code/humanChromorder.sh
Untracked: code/humanFiles
Untracked: code/intersectAnno.err
Untracked: code/intersectAnno.out
Untracked: code/intersectLiftedPAS.sh
Untracked: code/leafcutter_merge_regtools_redo.py
Untracked: code/liftJunctionFiles.sh
Untracked: code/liftPAS19to38.sh
Untracked: code/liftoverPAShg19to38.err
Untracked: code/liftoverPAShg19to38.out
Untracked: code/log/
Untracked: code/make5percPeakbed.py
Untracked: code/makeFileID.py
Untracked: code/makePheno.py
Untracked: code/makeSamplyGroupsChimp_TvN.py
Untracked: code/makeSamplyGroupsHuman_TvN.py
Untracked: code/mapRNAseqhg19.sh
Untracked: code/mapRNAseqhg19_newPipeline.sh
Untracked: code/maphg19.err
Untracked: code/maphg19.out
Untracked: code/maphg19.sh
Untracked: code/maphg19_new.err
Untracked: code/maphg19_new.out
Untracked: code/maphg19_sub.err
Untracked: code/maphg19_sub.out
Untracked: code/maphg19_subjunc.sh
Untracked: code/merge.err
Untracked: code/merge_leafcutter_clusters_redo.py
Untracked: code/mergedBam2BW.sh
Untracked: code/mergedbam2bw.err
Untracked: code/mergedbam2bw.out
Untracked: code/nameClusters.py
Untracked: code/namePeaks.py
Untracked: code/nuclearTranscriptDTplot.err
Untracked: code/nuclearTranscriptDTplot.out
Untracked: code/numMultimap.py
Untracked: code/overlapPAS.err
Untracked: code/overlapPAS.out
Untracked: code/overlapapaQTLPAS.sh
Untracked: code/peak2PAS.py
Untracked: code/pheno2countonly.R
Untracked: code/prepareAnnoLeafviz.err
Untracked: code/prepareAnnoLeafviz.out
Untracked: code/prepareCleanLiftedFC_5perc4LC.py
Untracked: code/prepareLeafvizAnno.sh
Untracked: code/preparePAS4lift.py
Untracked: code/prepare_phenotype_table.py
Untracked: code/primaryLift.err
Untracked: code/primaryLift.out
Untracked: code/primaryLift.sh
Untracked: code/processhg38exons.py
Untracked: code/quantJunc.sh
Untracked: code/quantJunc_TEST.sh
Untracked: code/quantJunc_removeBad.sh
Untracked: code/quantLiftedPAS.err
Untracked: code/quantLiftedPAS.out
Untracked: code/quantLiftedPAS.sh
Untracked: code/quatJunc.err
Untracked: code/quatJunc.out
Untracked: code/recChimpback2Human.err
Untracked: code/recChimpback2Human.out
Untracked: code/recLiftchim2human.sh
Untracked: code/revLift.err
Untracked: code/revLift.out
Untracked: code/revLiftPAShg38to19.sh
Untracked: code/reverseLift.sh
Untracked: code/runCheckReverseLift.sh
Untracked: code/runChimpDiffIso.sh
Untracked: code/runCountNucleotides.err
Untracked: code/runCountNucleotides.out
Untracked: code/runCountNucleotides.sh
Untracked: code/runCountNucleotidesPantro6.err
Untracked: code/runCountNucleotidesPantro6.out
Untracked: code/runCountNucleotides_pantro6.sh
Untracked: code/runFilterNumChroms.sh
Untracked: code/runHumanDiffIso.sh
Untracked: code/runNuclearDifffIso.sh
Untracked: code/run_Chimpleafcutter_ds.err
Untracked: code/run_Chimpleafcutter_ds.out
Untracked: code/run_Chimpverifybam.err
Untracked: code/run_Chimpverifybam.out
Untracked: code/run_Humanleafcutter_ds.err
Untracked: code/run_Humanleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_ds.err
Untracked: code/run_Nuclearleafcutter_ds.out
Untracked: code/run_chimpverifybam.sh
Untracked: code/run_verifyBam.sh
Untracked: code/run_verifybam.err
Untracked: code/run_verifybam.out
Untracked: code/slurm-62824013.out
Untracked: code/slurm-62825841.out
Untracked: code/slurm-62826116.out
Untracked: code/slurm-64108209.out
Untracked: code/slurm-64108521.out
Untracked: code/slurm-64108557.out
Untracked: code/snakePASChimp.err
Untracked: code/snakePASChimp.out
Untracked: code/snakePAShuman.out
Untracked: code/snakemake.batch
Untracked: code/snakemakeChimp.err
Untracked: code/snakemakeChimp.out
Untracked: code/snakemakeHuman.err
Untracked: code/snakemakeHuman.out
Untracked: code/snakemakePAS.batch
Untracked: code/snakemakePASFiltChimp.err
Untracked: code/snakemakePASFiltChimp.out
Untracked: code/snakemakePASFiltHuman.err
Untracked: code/snakemakePASFiltHuman.out
Untracked: code/snakemakePASchimp.batch
Untracked: code/snakemakePAShuman.batch
Untracked: code/snakemake_chimp.batch
Untracked: code/snakemake_human.batch
Untracked: code/snakemakefiltPAS.batch
Untracked: code/snakemakefiltPAS_chimp.sh
Untracked: code/snakemakefiltPAS_human.sh
Untracked: code/submit-snakemake-chimp.sh
Untracked: code/submit-snakemake-human.sh
Untracked: code/submit-snakemakePAS-chimp.sh
Untracked: code/submit-snakemakePAS-human.sh
Untracked: code/submit-snakemakefiltPAS-chimp.sh
Untracked: code/submit-snakemakefiltPAS-human.sh
Untracked: code/subset_diffisopheno.py
Untracked: code/subset_diffisopheno_Chimp_tvN.py
Untracked: code/subset_diffisopheno_Huma_tvN.py
Untracked: code/subset_diffisopheno_Nuclear_HvC.py
Untracked: code/test
Untracked: code/transcriptDTplotsNuclear.sh
Untracked: code/transcriptDTplotsTotal.sh
Untracked: code/verifyBam4973.sh
Untracked: code/verifyBam4973inHuman.sh
Untracked: code/verifybam4973.err
Untracked: code/verifybam4973.out
Untracked: code/verifybam4973HumanMap.err
Untracked: code/verifybam4973HumanMap.out
Untracked: code/wrap_Chimpverifybam.err
Untracked: code/wrap_Chimpverifybam.out
Untracked: code/wrap_chimpverifybam.sh
Untracked: code/wrap_verifyBam.sh
Untracked: code/wrap_verifybam.err
Untracked: code/wrap_verifybam.out
Untracked: code/writeMergecode.py
Untracked: data/._.DS_Store
Untracked: data/._HC_filenames.txt
Untracked: data/._HC_filenames.txt.sb-4426323c-IKIs0S
Untracked: data/._HC_filenames.xlsx
Untracked: data/._MapPantro6_meta.txt
Untracked: data/._MapPantro6_meta.txt.sb-a5794dd2-Cskmlm
Untracked: data/._MapPantro6_meta.xlsx
Untracked: data/._OppositeSpeciesMap.txt
Untracked: data/._OppositeSpeciesMap.txt.sb-a5794dd2-mayWJf
Untracked: data/._OppositeSpeciesMap.xlsx
Untracked: data/._RNASEQ_metadata.txt
Untracked: data/._RNASEQ_metadata.txt.sb-4426323c-TE4ns3
Untracked: data/._RNASEQ_metadata.txt.sb-51f67ae1-HXp7Gq
Untracked: data/._RNASEQ_metadata_2Removed.txt
Untracked: data/._RNASEQ_metadata_2Removed.txt.sb-4426323c-a4lBwx
Untracked: data/._RNASEQ_metadata_2Removed.xlsx
Untracked: data/._RNASEQ_metadata_stranded.txt
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-D659m2
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-ImNMoY
Untracked: data/._RNASEQ_metadata_stranded.xlsx
Untracked: data/._metadata_HCpanel.txt
Untracked: data/._metadata_HCpanel.txt.sb-a3d92a2d-b9cYoF
Untracked: data/._metadata_HCpanel.txt.sb-a5794dd2-i594qs
Untracked: data/._metadata_HCpanel.txt.sb-f4823d1e-qihGek
Untracked: data/._metadata_HCpanel.xlsx
Untracked: data/._metadata_HCpanel_frompantro5.xlsx
Untracked: data/._~$RNASEQ_metadata.xlsx
Untracked: data/._~$metadata_HCpanel.xlsx
Untracked: data/._.xlsx
Untracked: data/CompapaQTLpas/
Untracked: data/DTmatrix/
Untracked: data/DiffExpression/
Untracked: data/DiffIso_Nuclear/
Untracked: data/DiffSplice/
Untracked: data/DiffSplice_liftedJunc/
Untracked: data/DiffSplice_removeBad/
Untracked: data/EvalPantro5/
Untracked: data/HC_filenames.txt
Untracked: data/HC_filenames.xlsx
Untracked: data/MapPantro6_meta.txt
Untracked: data/MapPantro6_meta.xlsx
Untracked: data/MapStats/
Untracked: data/NuclearHvC/
Untracked: data/OppositeSpeciesMap.txt
Untracked: data/OppositeSpeciesMap.xlsx
Untracked: data/Peaks_5perc/
Untracked: data/Pheno_5perc/
Untracked: data/Pheno_5perc_nuclear/
Untracked: data/Pheno_5perc_total/
Untracked: data/RNASEQ_metadata.txt
Untracked: data/RNASEQ_metadata_2Removed.txt
Untracked: data/RNASEQ_metadata_2Removed.xlsx
Untracked: data/RNASEQ_metadata_stranded.txt
Untracked: data/RNASEQ_metadata_stranded.xlsx
Untracked: data/TwoBadSampleAnalysis/
Untracked: data/chainFiles/
Untracked: data/cleanPeaks_anno/
Untracked: data/cleanPeaks_byspecies/
Untracked: data/cleanPeaks_lifted/
Untracked: data/leafviz/
Untracked: data/liftover_files/
Untracked: data/metadata_HCpanel.txt
Untracked: data/metadata_HCpanel.xlsx
Untracked: data/metadata_HCpanel_frompantro5.txt
Untracked: data/metadata_HCpanel_frompantro5.xlsx
Untracked: data/primaryLift/
Untracked: data/reverseLift/
Untracked: data/~$RNASEQ_metadata.xlsx
Untracked: data/~$metadata_HCpanel.xlsx
Untracked: data/.xlsx
Untracked: output/dtPlots/
Untracked: projectNotes.Rmd
Unstaged changes:
Modified: analysis/OppositeMap.Rmd
Modified: analysis/annotationInfo.Rmd
Modified: analysis/diffExpression.Rmd
Modified: analysis/investigatePantro5.Rmd
Modified: analysis/multiMap.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | ac06540 | brimittleman | 2019-12-18 | update pantro6 |
| html | 5ac753a | brimittleman | 2019-12-11 | Build site. |
| Rmd | 76142de | brimittleman | 2019-12-11 | update reciprocal lift |
| html | d7f04d9 | brimittleman | 2019-12-11 | Build site. |
| Rmd | 6b8b23c | brimittleman | 2019-12-11 | fix redundancy in splicing genes |
| html | 8ae44d5 | brimittleman | 2019-12-10 | Build site. |
| Rmd | 28c77be | brimittleman | 2019-12-10 | add examples and anno code |
| html | 174fba1 | brimittleman | 2019-12-07 | Build site. |
| Rmd | 81cbc5f | brimittleman | 2019-12-07 | add res |
| html | e9cfec8 | brimittleman | 2019-12-07 | Build site. |
| Rmd | 1d92e9f | brimittleman | 2019-12-07 | add res |
| html | a288a29 | brimittleman | 2019-12-04 | Build site. |
| Rmd | 558b39f | brimittleman | 2019-12-04 | add current error for splice and write out DE genes |
| html | 2fbf2d4 | brimittleman | 2019-12-02 | Build site. |
| Rmd | b5df813 | brimittleman | 2019-12-02 | initial results no overlap |
| html | 12a7fcb | brimittleman | 2019-11-20 | Build site. |
| Rmd | 6d6e7d5 | brimittleman | 2019-11-20 | fix label |
| html | 9b73cff | brimittleman | 2019-11-18 | Build site. |
| Rmd | 3c2512b | brimittleman | 2019-11-18 | wflow_publish(“analysis/diffSplicing.Rmd”) |
| html | 7770d55 | brimittleman | 2019-11-18 | Build site. |
| Rmd | fa2c075 | brimittleman | 2019-11-18 | code for diff splice |
| html | 106f3c1 | brimittleman | 2019-11-15 | Build site. |
| Rmd | 4ad7bce | brimittleman | 2019-11-15 | look at results of cluster lift |
| html | dc91b0a | brimittleman | 2019-11-11 | Build site. |
| Rmd | b5ba82e | brimittleman | 2019-11-11 | add diff expression and diff splicing |
library(tidyverse)── Attaching packages ──────────────────────────────────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.1.1 ✔ purrr 0.3.2
✔ tibble 2.1.1 ✔ dplyr 0.8.0.1
✔ tidyr 0.8.3 ✔ stringr 1.3.1
✔ readr 1.3.1 ✔ forcats 0.3.0 ── Conflicts ─────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(reshape2)
Attaching package: 'reshape2'The following object is masked from 'package:tidyr':
smithsI want to use the RNA seq I collected to also perform a differential splicing analysis with leafcutter. I will follow the pipeline found at http://davidaknowles.github.io/leafcutter/articles/Usage.html. For a first pass I will use the bam files from the snakemake and differential expression analysis pipeline.
I will get clusters in both species then perform reciprocal liftover. I can use a liftover pipeline similar to the one I used for the differnetial PAS analysis.
Pipeline from example on leafcutter github.
for bamfile in `ls run/geuvadis/*chr1.bam`; do
echo Converting $bamfile to $bamfile.junc
samtools index $bamfile
regtools junctions extract -a 8 -m 50 -M 500000 $bamfile -o $bamfile.junc
echo $bamfile.junc >> test_juncfiles.txt
done
python ../clustering/leafcutter_cluster_regtools.py -j test_juncfiles.txt -m 50 -o testYRIvsEU -l 500000At this point I will be able to liftover the junctions. I can use the human corrdinates for the differential splicing step.
../scripts/leafcutter_ds.R --num_threads 4 ../example_data/testYRIvsEU_perind_numers.counts.gz example_geuvadis/groups_file.txt
I now have my RNA seq for each species. I can write a script that runs the junctions for each species.
mkdir /project2/gilad/briana/Comparative_APA/Human/data/RNAseq/DiffSplice/
mkdir /project2/gilad/briana/Comparative_APA/Chimp/data/RNAseq/DiffSplice/
touch /project2/gilad/briana/Comparative_APA/Human/data/RNAseq/DiffSplice/human_juncfiles.txt
touch /project2/gilad/briana/Comparative_APA/Chimp/data/RNAseq/DiffSplice/chimp_juncfiles.txt
sbatch converBam2Junc.shCreate a script that only keeps the number chromosomes (2A and 2B for chimp). This means I will not have any of the chimp contigs.
I should lift first then filter
mkdir ../data/DiffSplice_liftedJunc
sbatch liftJunctionFiles.sh
Assess the number that liftover: Resiprocal must map back to the same spot.
First I will look at how many pass the original lift.
#Original:
touch /project2/gilad/briana/Comparative_APA/data/DiffSplice_liftedJunc/JuncNums.txt
for i in $(ls /project2/gilad/briana/Comparative_APA/Human/data/RNAseq/sort/*.bam.junc)
do
describer=$(echo ${i} cut -d/ -f 9 | cut -d- -f 4)
number=$(wc -l < $i)
echo -e "\t" ${describer} ${number} "original" "Human" >> /project2/gilad/briana/Comparative_APA/data/DiffSplice_liftedJunc/JuncNums.txt
done
#reverse
for i in $(ls /project2/gilad/briana/Comparative_APA/Human/data/RNAseq/sort/*.bam.junc.2Chimp)
do
describer=$(echo ${i} cut -d/ -f 9 | cut -d- -f 4)
number=$(wc -l < $i)
echo -e "\t" ${describer} ${number} "ForwardLift" "Human" >> /project2/gilad/briana/Comparative_APA/data/DiffSplice_liftedJunc/JuncNums.txt
done
#chimp files
#original
for i in $(ls /project2/gilad/briana/Comparative_APA/Chimp/data/RNAseq/sort/*.bam.junc)
do
describer=$(echo ${i} cut -d/ -f 9 | cut -d- -f 4)
number=$(wc -l < $i)
echo -e "\t" ${describer} ${number} "original" "Chimp" >> /project2/gilad/briana/Comparative_APA/data/DiffSplice_liftedJunc/JuncNums.txt
done
#reverse
for i in $(ls /project2/gilad/briana/Comparative_APA/Chimp/data/RNAseq/sort/*.bam.junc.2Human)
do
describer=$(echo ${i} cut -d/ -f 9 | cut -d- -f 4)
number=$(wc -l < $i)
echo -e "\t" ${describer} ${number} "ForwardLift" "Chimp" >> /project2/gilad/briana/Comparative_APA/data/DiffSplice_liftedJunc/JuncNums.txt
done
Next I need to make sure everything lifts back to the same place.
I will write an R script that takes both files and inner joins to the same locations. I need the original and the final lift.
sbatch runCheckReverseLift.shEvaluate results:
for i in $(ls /project2/gilad/briana/Comparative_APA/Human/data/RNAseq/sort/*.SamePlace)
do
describer=$(echo ${i} cut -d/ -f 9 | cut -d- -f 4)
number=$(wc -l < $i)
echo -e "\t" ${describer} ${number} "SamePlace" "Human" >> /project2/gilad/briana/Comparative_APA/data/DiffSplice_liftedJunc/JuncNums.txt
done
for i in $(ls /project2/gilad/briana/Comparative_APA/Chimp/data/RNAseq/sort/*.SamePlace)
do
describer=$(echo ${i} cut -d/ -f 9 | cut -d- -f 4)
number=$(wc -l < $i)
echo -e "\t" ${describer} ${number} "SamePlace" "Chimp" >> /project2/gilad/briana/Comparative_APA/data/DiffSplice_liftedJunc/JuncNums.txt
done
liftStats=read.table("../data/DiffSplice_liftedJunc/JuncNums.txt", col.names = c("line", "Njunc","File", "Species"),stringsAsFactors = F)
#spread by line
liftStatsSpread= liftStats %>% spread(key="File", value="Njunc") %>% mutate(PercLift=ForwardLift/original, PercSame=SamePlace/original)
ggplot(liftStatsSpread, aes(x=line, fill=Species, y=PercLift)) + geom_bar(stat="identity")+geom_text(aes(label=round(PercLift,3)), vjust=1.6, color="black") + labs(title="Proportion of Junctions lifting first")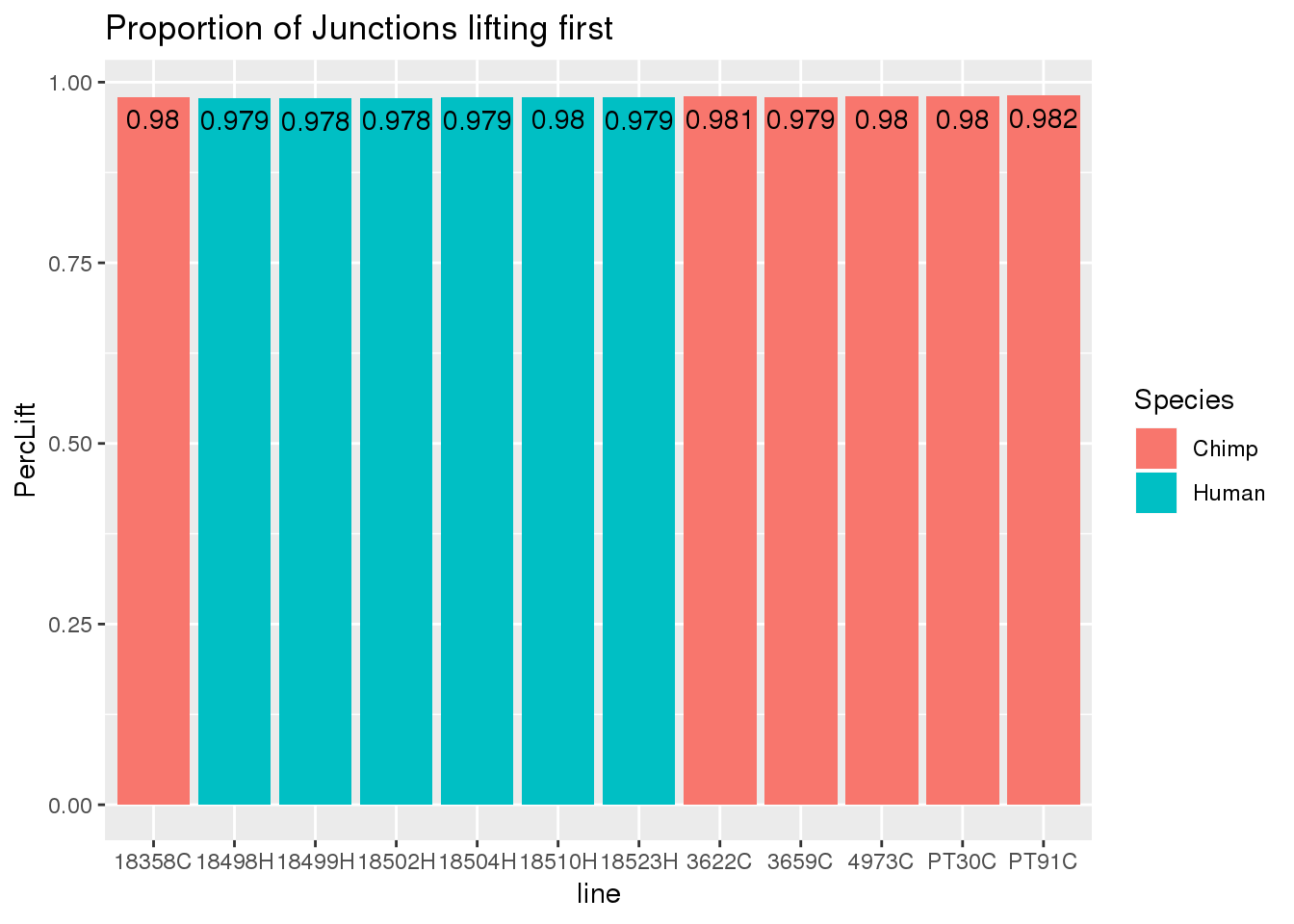
| Version | Author | Date |
|---|---|---|
| 5ac753a | brimittleman | 2019-12-11 |
ggplot(liftStatsSpread, aes(x=line, fill=Species, y=PercSame)) + geom_bar(stat="identity")+geom_text(aes(label=round(PercSame,3)), vjust=1.6, color="black") + labs(title="Proportion of Junctions Passing reciprocal liftover")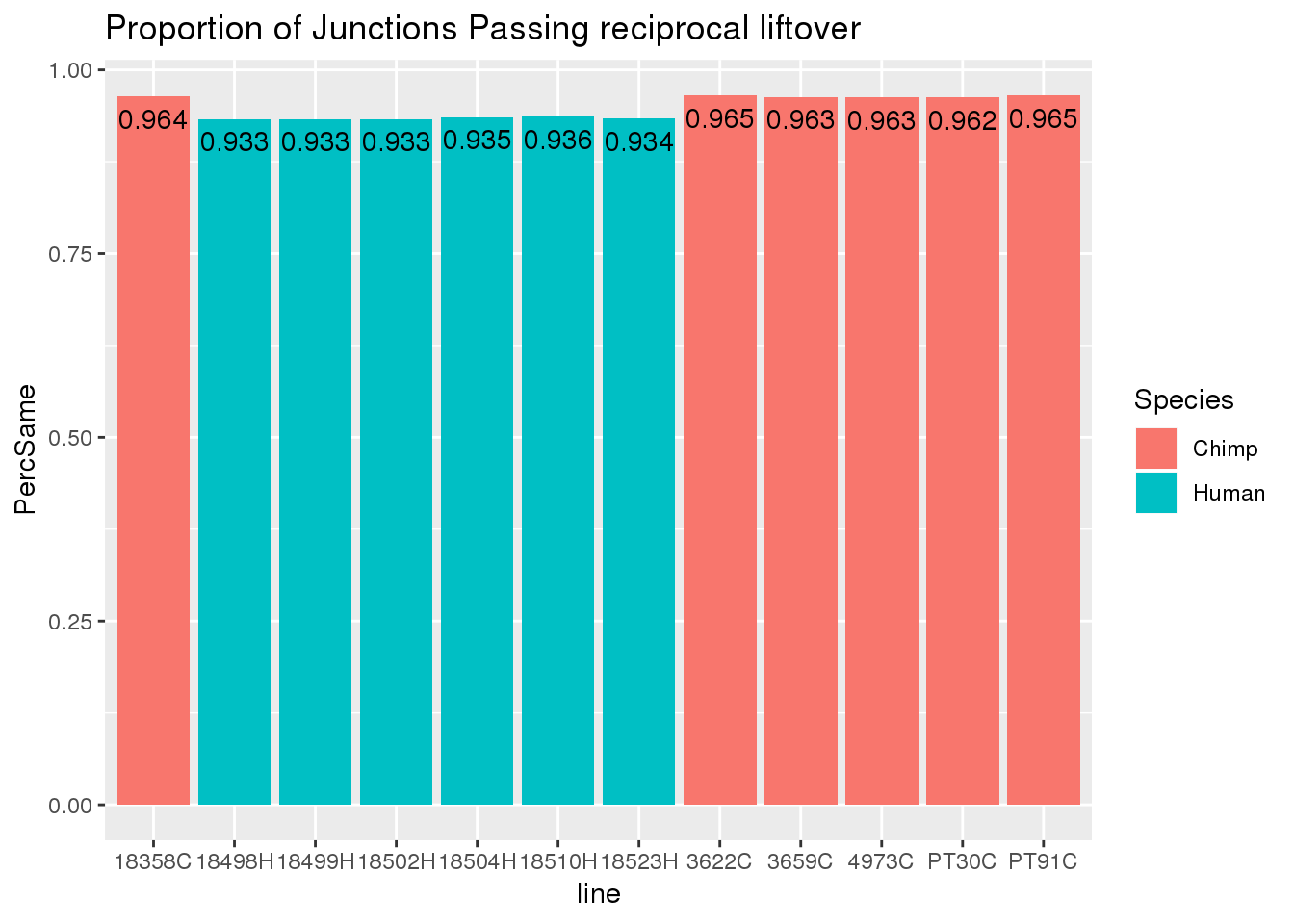
Lift the passing chimps to human:
sbatch LiftFinalChimpJunc2Human.shI need to write a script that fixes the lifted files. I need to make column 9 255,0,0 and remove the commas from the last 2 columns.
I can impliment the fix in the filter file.
sbatch runFilterNumChroms.shMake clusters:
juncfiles=read.table("../data/DiffSplice_liftedJunc/BothSpec_juncfiles.txt", header = F)
humanFiles=juncfiles %>% slice(1:6)
write.table(humanFiles, "../data/DiffSplice_liftedJunc/Human_juncfiles.txt", quote = F, col.names = F, row.names = F)
chimpFiles=juncfiles %>% slice(7:12)
write.table(chimpFiles, "../data/DiffSplice_liftedJunc/Chimp_juncfiles.txt", quote = F, col.names = F, row.names = F)sbatch quantJunc.shNow I can merge all of the culsters with: /project2/yangili1/yangili/leafcutter_scripts/merge_leafcutter_clusters.py
sbatch MergeClusters.sh
sbatch QuantMergedClusters.sh
gunzip ../data/DiffSplice_liftedJunc/MergeCombined_perind_numers.counts.gzMake the sample list:
combinedCounts=read.table("../data/DiffSplice_liftedJunc/MergeCombined_perind_numers.counts", header=T)
x=colnames(combinedCounts)
#YG-BM-S8-18499H-Total_S8_R1_001-sort.bam
indiv=as.data.frame(x) %>% separate(x, into=c("yg", "bm","lane", "sample", "total", "sort", "bam"), sep="[.]") %>% mutate(sample=paste(yg, "-", bm, "-", lane, "-", sample, "-", total, "-", sort, ".", bam, sep="")) %>% dplyr::select(sample) %>% mutate(Species=ifelse(grepl("H",sample), "Human", "Chimp"))
write.table(indiv, "../data/DiffSplice_liftedJunc/groups_file.txt", quote = F, col.names = F, row.names = F, sep = "\t")fix to -Total instead of .Total
counts=read.table('../data/DiffSplice_liftedJunc/MergeCombined_perind_numers.counts', header=T, check.names = F)
meta=read.table("../data/DiffSplice_liftedJunc/groups_file.txt", header=F, stringsAsFactors = F)
colnames(meta)[1:2]=c("sample","group")
counts=counts[,meta$sample]
#rownames(counts)gzip ../data/DiffSplice_liftedJunc/MergeCombined_perind_numers.countsI am going to write a python work around to change the cluster format. This will take the unziped version of the counts file. When I run it, I can unzip and zip the results in the bash script.
sbatch RunFixLeafCluster.sh
sbatch DiffSplice.sh
results=read.table("../data/DiffSplice_liftedJunc/MergedRes_cluster_significance.txt",stringsAsFactors = F, header = T, sep="\t") %>% separate(cluster, into=c("chrom", "clus"),sep=":") %>% filter(status=="Success")
results$p.adjust=as.numeric(as.character(results$p.adjust))
nrow(results)[1] 6014results %>% filter(p.adjust < .05 ) %>% nrow()[1] 2274results_sig= results %>% filter(p.adjust < .05 )qqplot(-log10(runif(nrow(results))), -log10(results$p.adjust),ylab="-log10 Total Adjusted Leafcutter pvalue", xlab="-log 10 Uniform expectation", main="Human Chimp differential splicing")
abline(0,1)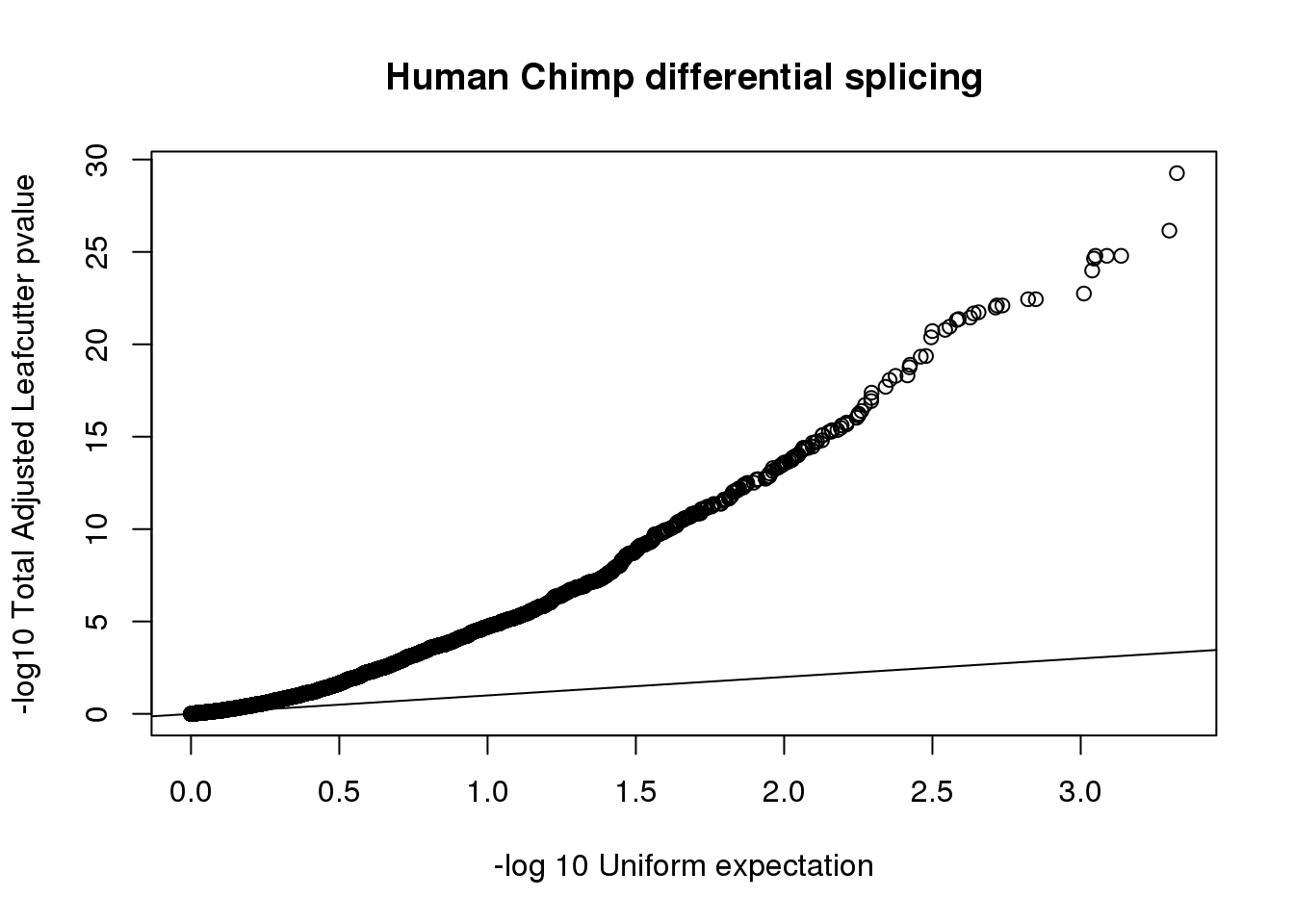
effectsize=read.table("../data/DiffSplice_liftedJunc/MergedRes_effect_sizes.txt", stringsAsFactors = F,header=T)
plot(sort(effectsize$deltapsi), ,main="Human vs Chimp Effect sizes", ylab="Delta PSI", xlab="Cluster Index")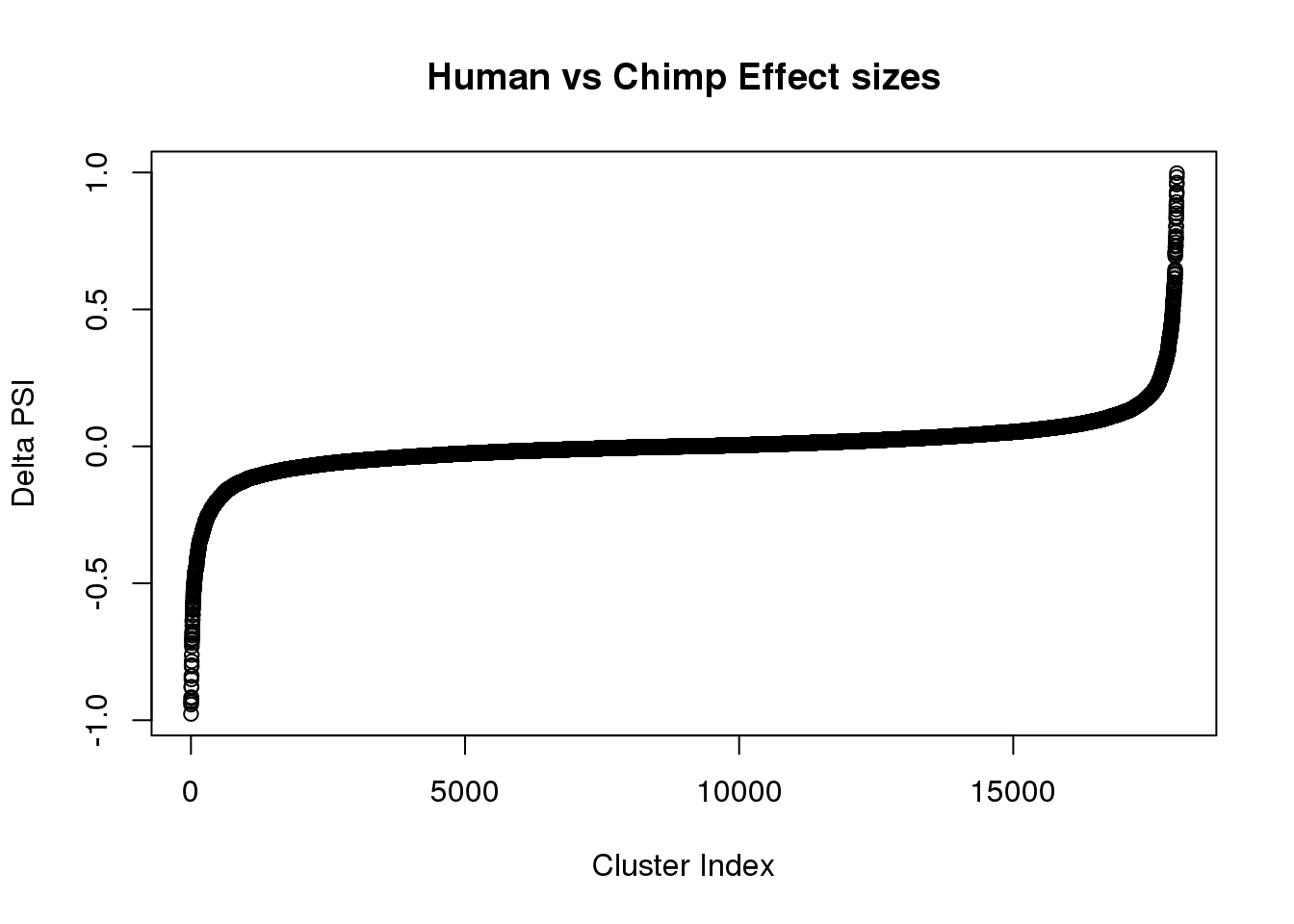
| Version | Author | Date |
|---|---|---|
| 5ac753a | brimittleman | 2019-12-11 |
Use the leafcutter tool to visualize
sbatch DiffSplicePlots.sh prepare genes:
genes=results_sig %>% arrange(p.adjust) %>% dplyr::select(genes) %>% unique()
nrow(genes)[1] 1813write.table(genes, "../data/DiffSplice_liftedJunc/orderedGenelist.txt", col.names = F, row.names = F, quote = F)Fixlist:
tr , '\n' < ../data/DiffSplice_liftedJunc/orderedGenelist.txt > ../data/DiffSplice_liftedJunc/orderedGeneListFixed.txtBuild leafviz
sbatch prepareLeafvizAnno.sh
sbatch buildLeafviz.sh
#with leafcutter annotation (https://github.com/davidaknowles/leafcutter/blob/master/leafviz/download_human_annotation_codes.sh)
sbatch buildLeafviz_leadAnno.sh
#classify clusters
sbatch ClassifyLeafviz.shRun this from leafviz on my own computer
Lift chimp bams to look at in IGV as well.
Pantro6- hg38.
mkdir ../Chimp/data/RNAseq/Sort_hg38
sbatch CrossMapChimpRNA.sh Interesting spots: - WSH3p= alt start -RPL22 (NM_000983.3) -RPL38 - chr3:129171277-129171446, NM_001127192.1, CNBP -chr20:46394503-46406548 - ELMO2, NM_001318253.1 - chimp extra exon chr10:79814716-79826198
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] reshape2_1.4.3 forcats_0.3.0 stringr_1.3.1 dplyr_0.8.0.1
[5] purrr_0.3.2 readr_1.3.1 tidyr_0.8.3 tibble_2.1.1
[9] ggplot2_3.1.1 tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 haven_1.1.2 lattice_0.20-38 colorspace_1.3-2
[5] generics_0.0.2 htmltools_0.3.6 yaml_2.2.0 rlang_0.4.0
[9] later_0.7.5 pillar_1.3.1 glue_1.3.0 withr_2.1.2
[13] modelr_0.1.2 readxl_1.1.0 plyr_1.8.4 munsell_0.5.0
[17] gtable_0.2.0 workflowr_1.5.0 cellranger_1.1.0 rvest_0.3.2
[21] evaluate_0.12 labeling_0.3 knitr_1.20 httpuv_1.4.5
[25] broom_0.5.1 Rcpp_1.0.2 promises_1.0.1 scales_1.0.0
[29] backports_1.1.2 jsonlite_1.6 fs_1.3.1 hms_0.4.2
[33] digest_0.6.18 stringi_1.2.4 grid_3.5.1 rprojroot_1.3-2
[37] cli_1.1.0 tools_3.5.1 magrittr_1.5 lazyeval_0.2.1
[41] crayon_1.3.4 whisker_0.3-2 pkgconfig_2.0.2 xml2_1.2.0
[45] lubridate_1.7.4 assertthat_0.2.0 rmarkdown_1.10 httr_1.3.1
[49] rstudioapi_0.10 R6_2.3.0 nlme_3.1-137 git2r_0.26.1
[53] compiler_3.5.1