Figure 6 Analysis
Last updated: 2026-01-06
Checks: 7 0
Knit directory: DXR_website/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20251201) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e4705f6. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: DXR_website/data/
Ignored: code/Flow_Purity_Stressor_Boxplots_EMP_250226.R
Ignored: data/CDKN1A_geneplot_Dox.RDS
Ignored: data/Cormotif_dox.RDS
Ignored: data/Cormotif_prob_gene_list.RDS
Ignored: data/Cormotif_prob_gene_list_doxonly.RDS
Ignored: data/Counts_RNA_EMPfortmiller.txt
Ignored: data/DMSO_TNN13_plot.RDS
Ignored: data/DOX_TNN13_plot.RDS
Ignored: data/DOXgeneplots.RDS
Ignored: data/ExpressionMatrix_EMP.csv
Ignored: data/Ind1_DOX_Spearman_plot.RDS
Ignored: data/Ind6REP_Spearman_list.csv
Ignored: data/Ind6REP_Spearman_plot.RDS
Ignored: data/Ind6REP_Spearman_set.csv
Ignored: data/Ind6_Spearman_plot.RDS
Ignored: data/MDM2_geneplot_Dox.RDS
Ignored: data/Master_summary_75-1_EMP_250811_final.csv
Ignored: data/Master_summary_87-1_EMP_250811_final.csv
Ignored: data/Metadata.csv
Ignored: data/Metadata_2_norep_update.RDS
Ignored: data/Metadata_update.csv
Ignored: data/QC/
Ignored: data/Recovery_flow_purity.csv
Ignored: data/SIRT1_geneplot_Dox.RDS
Ignored: data/Sample_annotated.csv
Ignored: data/SpearmanHeatmapMatrix_EMP
Ignored: data/VolcanoPlot_D144R_original_EMP_250623.png
Ignored: data/VolcanoPlot_D24R_original_EMP_250623.png
Ignored: data/VolcanoPlot_D24T_original_EMP_250623.png
Ignored: data/annot_dox.RDS
Ignored: data/annot_list_hm.csv
Ignored: data/cormotif_dxr_1.RDS
Ignored: data/cormotif_dxr_2.RDS
Ignored: data/counts/
Ignored: data/counts_DE_df_dox.RDS
Ignored: data/counts_DE_raw_data.RDS
Ignored: data/counts_raw_filt.RDS
Ignored: data/counts_raw_matrix.RDS
Ignored: data/counts_raw_matrix_EMP_250514.csv
Ignored: data/d24_Spearman_plot.RDS
Ignored: data/dge_calc_dxr.RDS
Ignored: data/dge_calc_matrix.RDS
Ignored: data/ensembl_backup_dox.RDS
Ignored: data/fC_AllCounts.RDS
Ignored: data/fC_DOXCounts.RDS
Ignored: data/featureCounts_Concat_Matrix_DOXSamples_EMP_250430.csv
Ignored: data/featureCounts_DOXdata_updatedind.RDS
Ignored: data/filcpm_colnames_matrix.csv
Ignored: data/filcpm_matrix.csv
Ignored: data/filt_gene_list_dox.RDS
Ignored: data/filter_gene_list_final.RDS
Ignored: data/final_data/
Ignored: data/gene_clustlike_motif.RDS
Ignored: data/gene_postprob_motif.RDS
Ignored: data/genedf_dxr.RDS
Ignored: data/genematrix_dox.RDS
Ignored: data/genematrix_dxr.RDS
Ignored: data/heartgenes.csv
Ignored: data/heartgenes_dox.csv
Ignored: data/image_intensity_stats_all_conditions.csv
Ignored: data/image_level_stats_all_conditions.csv
Ignored: data/image_level_summary_yH2AX_quant.csv
Ignored: data/ind_num_dox.RDS
Ignored: data/ind_num_dxr.RDS
Ignored: data/initial_cormotif.RDS
Ignored: data/initial_cormotif_dox.RDS
Ignored: data/low_nuclei_samples.csv
Ignored: data/low_veh_samples.csv
Ignored: data/new/
Ignored: data/new_cormotif_dox.RDS
Ignored: data/perc_mean_all_10X.png
Ignored: data/perc_mean_all_20X.png
Ignored: data/perc_mean_all_40X.png
Ignored: data/perc_mean_consistent_10X.png
Ignored: data/perc_mean_consistent_20X.png
Ignored: data/perc_mean_consistent_40X.png
Ignored: data/perc_median_all_10X.png
Ignored: data/perc_median_all_20X.png
Ignored: data/perc_median_all_40X.png
Ignored: data/perc_median_consistent_10X.png
Ignored: data/perc_median_consistent_20X.png
Ignored: data/perc_median_consistent_40X.png
Ignored: data/plot_leg_d.RDS
Ignored: data/plot_leg_d_horizontal.RDS
Ignored: data/plot_leg_d_vertical.RDS
Ignored: data/plot_theme_custom.RDS
Ignored: data/process_gene_data_funct.RDS
Ignored: data/raw_mean_all_10X.png
Ignored: data/raw_mean_all_20X.png
Ignored: data/raw_mean_all_40X.png
Ignored: data/raw_mean_consistent_10X.png
Ignored: data/raw_mean_consistent_20X.png
Ignored: data/raw_mean_consistent_40X.png
Ignored: data/raw_median_all_10X.png
Ignored: data/raw_median_all_20X.png
Ignored: data/raw_median_all_40X.png
Ignored: data/raw_median_consistent_10X.png
Ignored: data/raw_median_consistent_20X.png
Ignored: data/raw_median_consistent_40X.png
Ignored: data/ruv/
Ignored: data/summary_all_IF_ROI.RDS
Ignored: data/tableED_GOBP.RDS
Ignored: data/tableESR_GOBP_postprob.RDS
Ignored: data/tableLD_GOBP.RDS
Ignored: data/tableLR_GOBP_postprob.RDS
Ignored: data/tableNR_GOBP.RDS
Ignored: data/tableNR_GOBP_postprob.RDS
Ignored: data/table_incsus_genes_GOBP_RUV.RDS
Ignored: data/table_motif1_GOBP_d.RDS
Ignored: data/table_motif1_genes_GOBP.RDS
Ignored: data/table_motif2_GOBP_d.RDS
Ignored: data/table_motif3_genes_GOBP.RDS
Ignored: data/thing.R
Ignored: data/top.table_V.D144r_dox.RDS
Ignored: data/top.table_V.D24_dox.RDS
Ignored: data/top.table_V.D24r_dox.RDS
Ignored: data/yH2AX_75-1_EMP_250811.csv
Ignored: data/yH2AX_87-1_EMP_250811.csv
Ignored: data/yH2AX_Boxplots_MeanIntensity_10X_EMP_250811.png
Ignored: data/yH2AX_Boxplots_MeanIntensity_10X_NormtoMax_EMP_250811.png
Ignored: data/yH2AX_Boxplots_MeanIntensity_20X_EMP_250811.png
Ignored: data/yH2AX_Boxplots_MeanIntensity_20X_NormtoMax_EMP_250811.png
Ignored: data/yH2AX_Boxplots_MedianIntensity_10X_NormtoMax_EMP_250811.png
Ignored: data/yH2AX_Boxplots_MedianIntensity_20X_NormtoMax_EMP_250811.png
Ignored: data/yH2AX_Boxplots_MedianIntensity_NormtoMax_EMP_250811.png
Ignored: data/yH2AX_Boxplots_Quant_10X_EMP_250817.png
Ignored: data/yH2AX_Boxplots_Quant_20X_EMP_250817.png
Ignored: data/yh2ax_all_df_EMP_250812.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (DXR_website/analysis/Figure6.Rmd)
and HTML (DXR_website/docs/Figure6.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | c382b38 | emmapfort | 2026-01-06 | Build site. |
| Rmd | cd535e9 | emmapfort | 2026-01-06 | Final Website Touches 01/06/25 EMP |
| html | 7f22593 | emmapfort | 2026-01-05 | Build site. |
| Rmd | 49fa865 | emmapfort | 2026-01-05 | Final Updates to Website EMP 01/05/26 |
Libraries
Load necessary libraries for analysis.
library(tidyverse)
library(readxl)
library(readr)
library(reshape2)
library(circlize)
library(grid)
library(stringr)
library(org.Hs.eg.db)
library(AnnotationDbi)
library(gprofiler2)
library(purrr)Define my custom plot theme
# Define the custom theme
# plot_theme_custom <- function() {
# theme_minimal() +
# theme(
# #line for x and y axis
# axis.line = element_line(linewidth = 1,
# color = "black"),
#
# #axis ticks only on x and y, length standard
# axis.ticks.x = element_line(color = "black",
# linewidth = 1),
# axis.ticks.y = element_line(color = "black",
# linewidth = 1),
# axis.ticks.length = unit(0.05, "in"),
#
# #text and font
# axis.text = element_text(color = "black",
# family = "Arial",
# size = 7),
# axis.title = element_text(color = "black",
# family = "Arial",
# size = 9),
# legend.text = element_text(color = "black",
# family = "Arial",
# size = 7),
# legend.title = element_text(color = "black",
# family = "Arial",
# size = 9),
# plot.title = element_text(color = "black",
# family = "Arial",
# size = 10),
#
# #blank background and border
# panel.background = element_blank(),
# panel.border = element_blank(),
#
# #gridlines for alignment
# panel.grid.major = element_line(color = "grey80", linewidth = 0.5), #grey major grid for align in illus
# panel.grid.minor = element_line(color = "grey90", linewidth = 0.5) #grey minor grid for align in illus
# )
# }
# saveRDS(plot_theme_custom, "data/plot_theme_custom.RDS")
theme_custom <- readRDS("data/plot_theme_custom.RDS")Define a function to save plots as .pdf and .png
save_plot <- function(plot, filename,
folder = ".",
width = 8,
height = 6,
units = "in",
dpi = 300,
add_date = TRUE) {
if (missing(filename)) stop("Please provide a filename (without extension) for the plot.")
date_str <- if (add_date) paste0("_", format(Sys.Date(), "%y%m%d")) else ""
pdf_file <- file.path(folder, paste0(filename, date_str, ".pdf"))
png_file <- file.path(folder, paste0(filename, date_str, ".png"))
ggsave(filename = pdf_file, plot = plot, device = cairo_pdf, width = width, height = height, units = units, bg = "transparent")
ggsave(filename = png_file, plot = plot, device = "png", width = width, height = height, units = units, dpi = dpi, bg = "transparent")
message("Saved plot as Cairo PDF: ", pdf_file)
message("Saved plot as PNG: ", png_file)
}
output_folder <- "C:/Users/emmap/OneDrive/Desktop/DXR Manuscript Materials/Fig pdfs"
#save plot function created
#to use: just define the plot name, filename_base, width, heightCharacterize the Chronic response
Identify genes that are progressively Chronic over time
We became interested in our Chronic gene cluster, and wanted to identify those genes that had an absolute fold change which increased over time (progressively Chronic: absFC tx+0 < tx+24 < tx+144).
#read in my gene set for my Chronic cluster
final_genes_2_RUV <- readRDS("data/Fig3/final_genes_2_RUV.RDS")
#read in my toptables
DOX_24T_R <- read.csv("data/DE/Toptable_RUV_24T_final.csv") %>%
dplyr::select(-(X))
DOX_24R_R <- read.csv("data/DE/Toptable_RUV_24R_final.csv") %>%
dplyr::select(-(X))
DOX_144R_R <- read.csv("data/DE/Toptable_RUV_144R_final.csv") %>%
dplyr::select(-(X))
#add in the absolute fold change for each timepoint so I can define my set of progressively Chronic genes
##we use absolute fold change to select genes that are progressively different over time regardless of response direction, so they may be increasingly upregulated or downregulated
combined_toptables_RUV_dxr <- list(
"t0" = DOX_24T_R %>% mutate(absFC = abs(logFC)),
"t24" = DOX_24R_R %>% mutate(absFC = abs(logFC)),
"t144" = DOX_144R_R %>% mutate(absFC = abs(logFC))
)
#break these up by timepoint
# t0_proChronic <- combined_toptables_RUV_dxr[["t0"]] %>%
# mutate(
# logFC_t0 = logFC,
# absFC_t0 = absFC,
# adjP_t0 = adj.P.Val,
# Sig_t0 = ifelse(adj.P.Val < 0.05, "DE", "nonDE")
# ) %>%
# dplyr::select(Entrez_ID, logFC_t0, absFC_t0, adjP_t0, Sig_t0)
# t24_proChronic <- combined_toptables_RUV_dxr[["t24"]] %>%
# mutate(
# logFC_t24 = logFC,
# absFC_t24 = absFC,
# adjP_t24 = adj.P.Val,
# Sig_t24 = ifelse(adj.P.Val < 0.05, "DE", "nonDE")
# ) %>%
# dplyr::select(Entrez_ID, logFC_t24, absFC_t24, adjP_t24, Sig_t24)
# t144_proChronic <- combined_toptables_RUV_dxr[["t144"]] %>%
# mutate(
# logFC_t144 = logFC,
# absFC_t144 = absFC,
# adjP_t144 = adj.P.Val,
# Sig_t144 = ifelse(adj.P.Val < 0.05, "DE", "nonDE")
# ) %>%
# dplyr::select(Entrez_ID, logFC_t144, absFC_t144, adjP_t144, Sig_t144)
#merge into one central dataframe
# proChronic_df <- t0_proChronic %>%
# dplyr::select(Entrez_ID, logFC_t0, absFC_t0, adjP_t0, Sig_t0) %>%
# left_join(t24_proChronic %>%
# dplyr::select(Entrez_ID, logFC_t24, absFC_t24, adjP_t24, Sig_t24), by = "Entrez_ID") %>%
# left_join(t144_proChronic %>%
# dplyr::select(Entrez_ID, logFC_t144, absFC_t144, adjP_t144, Sig_t144), by = "Entrez_ID")
#classify genes by their absFC response
# proChronic_df <- proChronic_df %>%
# dplyr::filter(Entrez_ID %in% final_genes_2_RUV) %>%
# mutate(
# Cluster = "Chronic",
# Response = ifelse(absFC_t0 < absFC_t24 & absFC_t24 < absFC_t144,
# "ProChronic",
# "non-ProChronic")
# )
#I used the absFC to determine whether they were Progressively Chronic
#ie absFC t0 < t24 & t24 < t144 = ProChronic
#all others are non-ProChronic (of the 501 Chronic genes)
# saveRDS(proChronic_df, "data/Fig6/proChronic_df.RDS")
#now I want to add in a SYMBOL category so I can see the names of the genes
# symbol_map <- AnnotationDbi::select(
# org.Hs.eg.db,
# keys = as.character(proChronic_df$Entrez_ID),
# columns = c("SYMBOL"),
# keytype = "ENTREZID"
# )
#ensure that they're the same type
# proChronic_df$Entrez_ID <- as.character(proChronic_df$Entrez_ID)
#remove any duplicates (if any)
# symbol_map <- symbol_map[!duplicated(symbol_map$ENTREZID), ]
# proChronic_df <- proChronic_df %>%
# left_join(symbol_map, by = c("Entrez_ID" = "ENTREZID"))
#reorder columns so Entrez_ID and SYMBOL are together
# proChronic_df <- proChronic_df %>%
# dplyr::select(
# Entrez_ID,
# SYMBOL,
# Cluster,
# Response,
# everything()
# )
#save a final version of this dataframe
# saveRDS(proChronic_df, "data/Fig6/Chronic_proChron_dataframe_symbolentrez.RDS")
proChronic_df <- readRDS("data/Fig6/Chronic_proChron_dataframe_symbolentrez.RDS")
#ensure numeric values
proChronic_df_all <- proChronic_df %>%
mutate(
absFC_t0 = as.numeric(absFC_t0),
absFC_t24 = as.numeric(absFC_t24),
absFC_t144 = as.numeric(absFC_t144)
)
proChronic_df_all <- proChronic_df_all %>%
mutate(
Category_final = case_when(
absFC_t0 < absFC_t24 & absFC_t24 < absFC_t144 ~ "ProChronic",
!is.na(absFC_t0) & !is.na(absFC_t24) & !is.na(absFC_t144) ~ "non-ProChronic",
TRUE ~ NA_character_ #keep genes missing any FC as NA
)
)
table(proChronic_df_all$Category_final, useNA = "ifany")
non-ProChronic ProChronic
338 163 #make my final proChronic gene list
proChronic_gene_list <- proChronic_df_all %>%
filter(Category_final == "ProChronic") %>%
pull(Entrez_ID) %>%
unique()
#make my final non-ProChronic gene list
nonproChronic_gene_list <- proChronic_df_all %>%
filter(Category_final == "non-ProChronic") %>%
pull(Entrez_ID) %>%
unique()
length(proChronic_gene_list)[1] 163length(nonproChronic_gene_list)[1] 338# saveRDS(proChronic_gene_list, "data/Fig6/proChronic_gene_list.RDS")
# saveRDS(nonproChronic_gene_list, "data/Fig6/non-proChronic_gene_list.RDS")Figure 6A - Progressively Chronic genes line plot
proChronic_gene_list [1] "79180" "1870" "79000" "63967" "55143" "991"
[7] "11004" "6491" "4998" "55635" "8317" "343099"
[13] "29899" "128239" "83540" "460" "9928" "4751"
[19] "51514" "1063" "9156" "55388" "56952" "11130"
[25] "983" "219790" "9585" "55165" "3070" "4288"
[31] "64787" "283120" "3481" "6240" "81930" "374393"
[37] "113130" "53340" "2305" "10635" "83461" "10024"
[43] "29127" "25840" "9700" "100128191" "283431" "55010"
[49] "221150" "675" "79866" "1033" "11169" "9787"
[55] "9824" "57082" "5888" "11339" "51203" "79968"
[61] "9133" "9768" "55055" "57214" "55215" "90381"
[67] "641" "9055" "9088" "116028" "79801" "51659"
[73] "81620" "83903" "54478" "9212" "6470" "4239"
[79] "10615" "146857" "990" "7153" "672" "78995"
[85] "146909" "146956" "55771" "7083" "332" "10403"
[91] "220134" "29128" "147841" "10535" "6241" "1058"
[97] "23397" "150468" "57405" "83879" "50940" "151246"
[103] "55355" "994" "9837" "22974" "1869" "81610"
[109] "4605" "11065" "8208" "8318" "79019" "2192"
[115] "51512" "2177" "151648" "25886" "55799" "57650"
[121] "10721" "4171" "5984" "55214" "64151" "4085"
[127] "890" "84057" "1363" "79682" "9319" "375444"
[133] "55789" "64105" "64946" "1393" "4001" "10112"
[139] "995" "3161" "6941" "1302" "3833" "442213"
[145] "55166" "7272" "253714" "26271" "5982" "4176"
[151] "43" "7516" "54892" "157313" "157570" "55872"
[157] "8836" "79075" "29028" "26147" "89958" "24137"
[163] "54821" nonproChronic_gene_list [1] "64856" "26038" "3604" "1188" "57822" "3925"
[7] "84958" "5453" "26508" "1263" "1031" "163782"
[13] "55283" "2634" "57535" "84722" "3737" "4803"
[19] "8458" "653820" "554282" "728833" "23632" "8991"
[25] "6275" "1163" "8407" "339416" "57795" "5997"
[31] "259266" "2494" "23046" "29089" "55220" "729533"
[37] "10637" "7044" "414149" "8516" "84930" "2658"
[43] "118738" "100144748" "355" "3832" "55118" "150"
[49] "153" "399827" "338707" "6786" "10346" "7762"
[55] "51700" "272" "57586" "8534" "29015" "710"
[61] "220001" "2237" "3619" "80150" "23649" "283131"
[67] "51083" "9633" "55107" "10714" "220042" "53826"
[73] "3014" "1111" "2113" "9918" "397" "441631"
[79] "55089" "79962" "11226" "269" "6490" "1017"
[85] "8914" "5557" "23306" "8091" "55592" "121601"
[91] "55332" "11113" "92558" "9735" "5426" "55835"
[97] "5983" "1910" "1880" "650669" "55320" "5427"
[103] "54331" "5228" "145508" "7443" "1396" "100288637"
[109] "701" "90417" "619189" "22995" "388121" "145773"
[115] "51285" "84465" "9493" "10620" "1138" "11173"
[121] "145864" "2242" "56963" "899" "124222" "9074"
[127] "9235" "5347" "728741" "3835" "112755" "23594"
[133] "23090" "4501" "55715" "64785" "1006" "23491"
[139] "3931" "2175" "359845" "5176" "342527" "100507131"
[145] "29893" "83990" "9902" "3838" "9120" "5608"
[151] "3759" "2905" "2232" "10610" "84733" "7298"
[157] "284217" "3613" "5932" "162681" "23526" "92840"
[163] "10036" "126282" "55723" "3780" "9518" "7643"
[169] "9745" "333" "284403" "163255" "54922" "27338"
[175] "129607" "3754" "4613" "6382" "79172" "109"
[181] "3795" "151056" "822" "30818" "699" "200373"
[187] "4175" "3800" "130940" "115677" "5163" "26154"
[193] "9427" "1293" "23178" "343637" "56265" "83959"
[199] "79980" "55959" "6790" "30811" "54069" "54058"
[205] "11200" "129080" "3976" "4174" "102800317" "57633"
[211] "6854" "80852" "56992" "29122" "993" "10293"
[217] "1795" "7348" "7200" "92369" "4071" "10051"
[223] "1894" "8646" "116832" "5648" "80235" "10460"
[229] "2261" "5522" "3815" "1633" "51191" "3015"
[235] "1062" "8492" "10733" "3148" "10309" "11174"
[241] "891" "1719" "1404" "4888" "9499" "6444"
[247] "9232" "54908" "9945" "100130357" "221687" "51053"
[253] "6890" "3159" "2289" "1026" "105375039" "266727"
[259] "2740" "860" "221391" "4172" "1310" "9096"
[265] "202" "387103" "2037" "113115" "2852" "4521"
[271] "55698" "10457" "29887" "54443" "7378" "222183"
[277] "27445" "10926" "4885" "79690" "375611" "4897"
[283] "1129" "27147" "3792" "2146" "83648" "55790"
[289] "1960" "8794" "8797" "51435" "84296" "4173"
[295] "254778" "4603" "23213" "5569" "23462" "25788"
[301] "9134" "284" "5820" "9401" "81704" "158326"
[307] "347240" "2189" "9833" "55071" "1164" "401541"
[313] "195828" "10592" "54566" "3371" "57000" "286204"
[319] "29941" "10319" "84628" "9719" "1757" "441478"
[325] "10742" "9468" "6839" "60401" "9452" "2491"
[331] "11013" "55859" "9363" "51765" "347475" "644538"
[337] "5365" "57595" #simplify the line plot
proChronic_df_all Entrez_ID SYMBOL Cluster Response logFC_t0 absFC_t0
1 64856 VWA1 Chronic non-ProChronic -0.1790655602 0.1790655602
2 26038 CHD5 Chronic non-ProChronic 3.0173128205 3.0173128205
3 3604 TNFRSF9 Chronic non-ProChronic -1.1811544525 1.1811544525
4 79180 EFHD2 Chronic ProChronic 0.0449537348 0.0449537348
5 1188 CLCNKB Chronic non-ProChronic -0.9670272054 0.9670272054
6 1870 E2F2 Chronic ProChronic 0.9067155249 0.9067155249
7 57822 GRHL3 Chronic non-ProChronic 3.5117220467 3.5117220467
8 79000 AUNIP Chronic ProChronic -0.0339386917 0.0339386917
9 3925 STMN1 Chronic non-ProChronic 0.4575116918 0.4575116918
10 84958 SYTL1 Chronic non-ProChronic 0.6513254703 0.6513254703
11 63967 CLSPN Chronic ProChronic 0.4344069282 0.4344069282
12 55143 CDCA8 Chronic ProChronic -0.0217210339 0.0217210339
13 5453 POU3F1 Chronic non-ProChronic 8.6287638175 8.6287638175
14 26508 HEYL Chronic non-ProChronic -1.6454138189 1.6454138189
15 991 CDC20 Chronic ProChronic -0.7947951384 0.7947951384
16 11004 KIF2C Chronic ProChronic -0.6668487346 0.6668487346
17 1263 PLK3 Chronic non-ProChronic 1.5917610696 1.5917610696
18 6491 STIL Chronic ProChronic -0.1528496481 0.1528496481
19 1031 CDKN2C Chronic non-ProChronic 2.2583366100 2.2583366100
20 4998 ORC1 Chronic ProChronic 0.3244009724 0.3244009724
21 163782 KANK4 Chronic non-ProChronic -0.5564019636 0.5564019636
22 55635 DEPDC1 Chronic ProChronic -1.9856317499 1.9856317499
23 55283 MCOLN3 Chronic non-ProChronic 4.6313840275 4.6313840275
24 2634 GBP2 Chronic non-ProChronic -1.6431769341 1.6431769341
25 8317 CDC7 Chronic ProChronic 0.3058329348 0.3058329348
26 343099 CCDC18 Chronic ProChronic 0.4358016819 0.4358016819
27 29899 GPSM2 Chronic ProChronic -1.2646002969 1.2646002969
28 57535 ELAPOR1 Chronic non-ProChronic 4.4746039473 4.4746039473
29 84722 PSRC1 Chronic non-ProChronic -1.9774803520 1.9774803520
30 3737 KCNA2 Chronic non-ProChronic 5.2080867614 5.2080867614
31 4803 NGF Chronic non-ProChronic 1.3921755232 1.3921755232
32 8458 TTF2 Chronic non-ProChronic 0.7981825768 0.7981825768
33 653820 FAM72B Chronic non-ProChronic 2.2344741015 2.2344741015
34 554282 FAM72C Chronic non-ProChronic 2.4350367942 2.4350367942
35 728833 FAM72D Chronic non-ProChronic 2.3359339007 2.3359339007
36 23632 CA14 Chronic non-ProChronic -0.2365023251 0.2365023251
37 8991 SELENBP1 Chronic non-ProChronic -0.2840422801 0.2840422801
38 6275 S100A4 Chronic non-ProChronic -0.0418515172 0.0418515172
39 1163 CKS1B Chronic non-ProChronic 0.4199373952 0.4199373952
40 128239 IQGAP3 Chronic ProChronic -0.5843640109 0.5843640109
41 8407 TAGLN2 Chronic non-ProChronic -0.4400625978 0.4400625978
42 83540 NUF2 Chronic ProChronic -2.5965484211 2.5965484211
43 339416 ANKRD45 Chronic non-ProChronic 0.2621226961 0.2621226961
44 460 ASTN1 Chronic ProChronic 0.2813923699 0.2813923699
45 57795 BRINP2 Chronic non-ProChronic 1.4155088616 1.4155088616
46 5997 RGS2 Chronic non-ProChronic 0.5463391682 0.5463391682
47 259266 ASPM Chronic non-ProChronic -0.8529704561 0.8529704561
48 2494 NR5A2 Chronic non-ProChronic 0.6192769413 0.6192769413
49 9928 KIF14 Chronic ProChronic -1.4348245203 1.4348245203
50 23046 KIF21B Chronic non-ProChronic -0.2226322164 0.2226322164
51 29089 UBE2T Chronic non-ProChronic -1.6038186300 1.6038186300
52 55220 KLHDC8A Chronic non-ProChronic -0.9195248965 0.9195248965
53 729533 FAM72A Chronic non-ProChronic 2.2859127819 2.2859127819
54 4751 NEK2 Chronic ProChronic -1.5477990700 1.5477990700
55 51514 DTL Chronic ProChronic -0.6815649561 0.6815649561
56 1063 CENPF Chronic ProChronic -1.7915168080 1.7915168080
57 10637 LEFTY1 Chronic non-ProChronic 0.5991522243 0.5991522243
58 7044 LEFTY2 Chronic non-ProChronic -0.0123746427 0.0123746427
59 9156 EXO1 Chronic ProChronic -0.8134403890 0.8134403890
60 55388 MCM10 Chronic ProChronic -0.8400720570 0.8400720570
61 414149 ACBD7 Chronic non-ProChronic 2.0101829420 2.0101829420
62 8516 ITGA8 Chronic non-ProChronic 0.6092595059 0.6092595059
63 56952 PRTFDC1 Chronic ProChronic -0.3036136997 0.3036136997
64 84930 MASTL Chronic non-ProChronic 1.7196748010 1.7196748010
65 2658 GDF2 Chronic non-ProChronic -3.2705253708 3.2705253708
66 118738 ZNF488 Chronic non-ProChronic 4.9867576997 4.9867576997
67 11130 ZWINT Chronic ProChronic -0.6199000529 0.6199000529
68 983 CDK1 Chronic ProChronic 0.4750771745 0.4750771745
69 219790 RTKN2 Chronic ProChronic -0.1900966591 0.1900966591
70 100144748 KLLN Chronic non-ProChronic 4.6982900846 4.6982900846
71 355 FAS Chronic non-ProChronic 1.9821375884 1.9821375884
72 9585 KIF20B Chronic ProChronic -0.9654855886 0.9654855886
73 3832 KIF11 Chronic non-ProChronic -0.9267187568 0.9267187568
74 55165 CEP55 Chronic ProChronic -0.9353547388 0.9353547388
75 3070 HELLS Chronic ProChronic -0.3775414607 0.3775414607
76 55118 CRTAC1 Chronic non-ProChronic 7.4226195629 7.4226195629
77 150 ADRA2A Chronic non-ProChronic 5.0620934604 5.0620934604
78 153 ADRB1 Chronic non-ProChronic 1.7114657666 1.7114657666
79 4288 MKI67 Chronic ProChronic -0.6858450854 0.6858450854
80 399827 LINC01164 Chronic non-ProChronic 4.7966667200 4.7966667200
81 338707 B4GALNT4 Chronic non-ProChronic -1.2482075763 1.2482075763
82 64787 EPS8L2 Chronic ProChronic -0.3254681706 0.3254681706
83 283120 H19 Chronic ProChronic 0.3273789715 0.3273789715
84 3481 IGF2 Chronic ProChronic 0.4863761574 0.4863761574
85 6786 STIM1 Chronic non-ProChronic 1.6288593218 1.6288593218
86 6240 RRM1 Chronic ProChronic 0.3205654022 0.3205654022
87 10346 TRIM22 Chronic non-ProChronic 1.4540317052 1.4540317052
88 7762 ZNF215 Chronic non-ProChronic -1.8769432132 1.8769432132
89 51700 CYB5R2 Chronic non-ProChronic 0.5336124717 0.5336124717
90 272 AMPD3 Chronic non-ProChronic 0.7957323497 0.7957323497
91 81930 KIF18A Chronic ProChronic -1.6677368228 1.6677368228
92 57586 SYT13 Chronic non-ProChronic -1.1257412548 1.1257412548
93 8534 CHST1 Chronic non-ProChronic 2.1475600115 2.1475600115
94 29015 SLC43A3 Chronic non-ProChronic -0.8801436690 0.8801436690
95 710 SERPING1 Chronic non-ProChronic 0.0847401743 0.0847401743
96 374393 FAM111B Chronic ProChronic -0.5871795773 0.5871795773
97 220001 VWCE Chronic non-ProChronic 2.3012737762 2.3012737762
98 2237 FEN1 Chronic non-ProChronic 1.0555613470 1.0555613470
99 3619 INCENP Chronic non-ProChronic -1.1693393140 1.1693393140
100 80150 ASRGL1 Chronic non-ProChronic 0.6600732048 0.6600732048
101 113130 CDCA5 Chronic ProChronic 1.0096718605 1.0096718605
102 23649 POLA2 Chronic non-ProChronic -0.2167647471 0.2167647471
103 283131 NEAT1 Chronic non-ProChronic -1.9652715372 1.9652715372
104 51083 GAL Chronic non-ProChronic 1.2534032436 1.2534032436
105 9633 TESMIN Chronic non-ProChronic -2.7838108318 2.7838108318
106 55107 ANO1 Chronic non-ProChronic -0.0129147028 0.0129147028
107 10714 POLD3 Chronic non-ProChronic -1.5041415589 1.5041415589
108 220042 DDIAS Chronic non-ProChronic 1.1928462323 1.1928462323
109 53826 FXYD6 Chronic non-ProChronic 0.0078673304 0.0078673304
110 3014 H2AX Chronic non-ProChronic -1.0313928001 1.0313928001
111 53340 SPA17 Chronic ProChronic -0.1096071864 0.1096071864
112 1111 CHEK1 Chronic non-ProChronic -0.7486688480 0.7486688480
113 2113 ETS1 Chronic non-ProChronic -1.3981265368 1.3981265368
114 2305 FOXM1 Chronic ProChronic 0.3655872026 0.3655872026
115 10635 RAD51AP1 Chronic ProChronic -0.3894265996 0.3894265996
116 9918 NCAPD2 Chronic non-ProChronic 0.6818634813 0.6818634813
117 83461 CDCA3 Chronic ProChronic 0.7825873469 0.7825873469
118 397 ARHGDIB Chronic non-ProChronic -1.9908748177 1.9908748177
119 441631 TSPAN11 Chronic non-ProChronic 3.0951662011 3.0951662011
120 55089 SLC38A4 Chronic non-ProChronic 1.0009645389 1.0009645389
121 10024 TROAP Chronic ProChronic -0.6705157913 0.6705157913
122 79962 DNAJC22 Chronic non-ProChronic 1.7913766521 1.7913766521
123 29127 RACGAP1 Chronic ProChronic -1.1383474539 1.1383474539
124 25840 TMT1A Chronic ProChronic -0.5563528161 0.5563528161
125 11226 GALNT6 Chronic non-ProChronic 0.2798889264 0.2798889264
126 9700 ESPL1 Chronic ProChronic 0.0859603646 0.0859603646
127 269 AMHR2 Chronic non-ProChronic -2.9633599427 2.9633599427
128 6490 PMEL Chronic non-ProChronic 0.9018113457 0.9018113457
129 1017 CDK2 Chronic non-ProChronic 0.8923691023 0.8923691023
130 8914 TIMELESS Chronic non-ProChronic -1.0306258478 1.0306258478
131 5557 PRIM1 Chronic non-ProChronic -1.3268893800 1.3268893800
132 23306 NEMP1 Chronic non-ProChronic 1.5711453922 1.5711453922
133 8091 HMGA2 Chronic non-ProChronic -0.0538344631 0.0538344631
134 100128191 TMPO-AS1 Chronic ProChronic 0.9912381488 0.9912381488
135 55592 GOLGA2P5 Chronic non-ProChronic 0.9151973320 0.9151973320
136 283431 GAS2L3 Chronic ProChronic 0.1276668542 0.1276668542
137 121601 ANO4 Chronic non-ProChronic -1.7844951409 1.7844951409
138 55332 DRAM1 Chronic non-ProChronic 1.8357370525 1.8357370525
139 55010 PARPBP Chronic ProChronic -1.2452384250 1.2452384250
140 11113 CIT Chronic non-ProChronic -1.1300315503 1.1300315503
141 92558 BICDL1 Chronic non-ProChronic 2.1974295888 2.1974295888
142 9735 KNTC1 Chronic non-ProChronic 1.0066557284 1.0066557284
143 5426 POLE Chronic non-ProChronic 0.6139253184 0.6139253184
144 221150 SKA3 Chronic ProChronic -1.3308598956 1.3308598956
145 55835 CENPJ Chronic non-ProChronic -0.4345572023 0.4345572023
146 675 BRCA2 Chronic ProChronic 0.1867923156 0.1867923156
147 5983 RFC3 Chronic non-ProChronic -2.0694683881 2.0694683881
148 79866 BORA Chronic ProChronic -0.6756767084 0.6756767084
149 1910 EDNRB Chronic non-ProChronic 0.2661809815 0.2661809815
150 1880 GPR183 Chronic non-ProChronic -4.9913001864 4.9913001864
151 650669 GAS6-AS1 Chronic non-ProChronic -0.4669403965 0.4669403965
152 55320 MIS18BP1 Chronic non-ProChronic -2.7130556199 2.7130556199
153 5427 POLE2 Chronic non-ProChronic -1.1874213729 1.1874213729
154 54331 GNG2 Chronic non-ProChronic -2.9979880047 2.9979880047
155 1033 CDKN3 Chronic ProChronic -1.2913713797 1.2913713797
156 11169 WDHD1 Chronic ProChronic -0.0641235214 0.0641235214
157 9787 DLGAP5 Chronic ProChronic -1.5072527435 1.5072527435
158 5228 PGF Chronic non-ProChronic 2.3736788732 2.3736788732
159 145508 CEP128 Chronic non-ProChronic -1.7262619436 1.7262619436
160 7443 VRK1 Chronic non-ProChronic 0.8861875627 0.8861875627
161 1396 CRIP1 Chronic non-ProChronic -0.2747159939 0.2747159939
162 100288637 LOC100288637 Chronic non-ProChronic -0.5739656484 0.5739656484
163 9824 ARHGAP11A Chronic ProChronic -1.7836008962 1.7836008962
164 701 BUB1B Chronic non-ProChronic -2.7010118622 2.7010118622
165 90417 KNSTRN Chronic non-ProChronic -0.4915228400 0.4915228400
166 57082 KNL1 Chronic ProChronic -0.5871089447 0.5871089447
167 5888 RAD51 Chronic ProChronic -1.4596037220 1.4596037220
168 11339 OIP5 Chronic ProChronic -0.0914251518 0.0914251518
169 51203 NUSAP1 Chronic ProChronic 0.6072271181 0.6072271181
170 619189 SERINC4 Chronic non-ProChronic 1.6840133045 1.6840133045
171 79968 WDR76 Chronic ProChronic -1.5585436893 1.5585436893
172 22995 CEP152 Chronic non-ProChronic -0.6477730141 0.6477730141
173 388121 TNFAIP8L3 Chronic non-ProChronic 0.8949152590 0.8949152590
174 9133 CCNB2 Chronic ProChronic -1.3712764394 1.3712764394
175 145773 FAM81A Chronic non-ProChronic 0.6790254253 0.6790254253
176 9768 PCLAF Chronic ProChronic 0.0943528453 0.0943528453
177 51285 RASL12 Chronic non-ProChronic -1.5394690509 1.5394690509
178 84465 MEGF11 Chronic non-ProChronic 3.6374970466 3.6374970466
179 55055 ZWILCH Chronic ProChronic -0.1559319733 0.1559319733
180 9493 KIF23 Chronic non-ProChronic -1.2534992340 1.2534992340
181 10620 ARID3B Chronic non-ProChronic 1.2503570683 1.2503570683
182 1138 CHRNA5 Chronic non-ProChronic 1.5653078035 1.5653078035
183 11173 ADAMTS7 Chronic non-ProChronic 2.0456415444 2.0456415444
184 57214 CEMIP Chronic ProChronic 0.6428023142 0.6428023142
185 145864 HAPLN3 Chronic non-ProChronic 2.4841519350 2.4841519350
186 55215 FANCI Chronic ProChronic -0.0467358271 0.0467358271
187 90381 TICRR Chronic ProChronic 0.0502445076 0.0502445076
188 641 BLM Chronic ProChronic -1.4488503254 1.4488503254
189 2242 FES Chronic non-ProChronic 1.4186150093 1.4186150093
190 9055 PRC1 Chronic ProChronic -0.5505839709 0.5505839709
191 56963 RGMA Chronic non-ProChronic 6.5180569781 6.5180569781
192 899 CCNF Chronic non-ProChronic 0.2697298434 0.2697298434
193 124222 PAQR4 Chronic non-ProChronic 0.8090473501 0.8090473501
194 9088 PKMYT1 Chronic ProChronic 0.0572455038 0.0572455038
195 9074 CLDN6 Chronic non-ProChronic -0.2383223503 0.2383223503
196 9235 IL32 Chronic non-ProChronic -0.5409395671 0.5409395671
197 116028 RMI2 Chronic ProChronic -0.2767979987 0.2767979987
198 5347 PLK1 Chronic non-ProChronic -1.7690828331 1.7690828331
199 728741 NPIPB6 Chronic non-ProChronic 1.4877517976 1.4877517976
200 3835 KIF22 Chronic non-ProChronic -0.4595427171 0.4595427171
201 112755 STX1B Chronic non-ProChronic 0.7050951760 0.7050951760
202 79801 SHCBP1 Chronic ProChronic 0.0394646223 0.0394646223
203 23594 ORC6 Chronic non-ProChronic 0.5307798329 0.5307798329
204 23090 ZNF423 Chronic non-ProChronic 3.9122970084 3.9122970084
205 4501 MT1X Chronic non-ProChronic -0.9393658413 0.9393658413
206 55715 DOK4 Chronic non-ProChronic -0.5280069399 0.5280069399
207 64785 GINS3 Chronic non-ProChronic 0.9571939480 0.9571939480
208 1006 CDH8 Chronic non-ProChronic 0.8304663043 0.8304663043
209 23491 CES3 Chronic non-ProChronic 3.6063255694 3.6063255694
210 3931 LCAT Chronic non-ProChronic -0.2422521379 0.2422521379
211 51659 GINS2 Chronic ProChronic 0.0561536410 0.0561536410
212 81620 CDT1 Chronic ProChronic -0.3009427249 0.3009427249
213 2175 FANCA Chronic non-ProChronic 0.4184914075 0.4184914075
214 359845 RFLNB Chronic non-ProChronic 4.0930746863 4.0930746863
215 5176 SERPINF1 Chronic non-ProChronic 0.7082216663 0.7082216663
216 83903 HASPIN Chronic ProChronic 1.2764499194 1.2764499194
217 342527 SMTNL2 Chronic non-ProChronic -0.2950930454 0.2950930454
218 54478 PIMREG Chronic ProChronic 0.9722987570 0.9722987570
219 9212 AURKB Chronic ProChronic -1.9272541928 1.9272541928
220 100507131 <NA> Chronic non-ProChronic -1.7853121834 1.7853121834
221 6470 SHMT1 Chronic ProChronic -0.0002336503 0.0002336503
222 4239 MFAP4 Chronic ProChronic 0.3554046242 0.3554046242
223 10615 SPAG5 Chronic ProChronic -0.8295177973 0.8295177973
224 146857 SLFN13 Chronic ProChronic 0.0945600293 0.0945600293
225 990 CDC6 Chronic ProChronic -0.8427347972 0.8427347972
226 7153 TOP2A Chronic ProChronic -0.5384586323 0.5384586323
227 29893 PSMC3IP Chronic non-ProChronic 1.7012823220 1.7012823220
228 672 BRCA1 Chronic ProChronic -0.8783838599 0.8783838599
229 78995 HROB Chronic ProChronic 0.4070622642 0.4070622642
230 146909 KIF18B Chronic ProChronic -1.6434557030 1.6434557030
231 146956 EME1 Chronic ProChronic -0.1833507206 0.1833507206
232 55771 PRR11 Chronic ProChronic -0.2143900597 0.2143900597
233 83990 BRIP1 Chronic non-ProChronic -0.8870532821 0.8870532821
234 9902 MRC2 Chronic non-ProChronic 0.3357883018 0.3357883018
235 3838 KPNA2 Chronic non-ProChronic -0.3461608473 0.3461608473
236 9120 SLC16A6 Chronic non-ProChronic 4.4974432130 4.4974432130
237 5608 MAP2K6 Chronic non-ProChronic -2.2623503559 2.2623503559
238 3759 KCNJ2 Chronic non-ProChronic 2.6965582616 2.6965582616
239 2905 GRIN2C Chronic non-ProChronic 3.8164025874 3.8164025874
240 2232 FDXR Chronic non-ProChronic 2.1937384998 2.1937384998
241 10610 ST6GALNAC2 Chronic non-ProChronic 4.3065228080 4.3065228080
242 7083 TK1 Chronic ProChronic 0.2695282081 0.2695282081
243 332 BIRC5 Chronic ProChronic -0.3249127652 0.3249127652
244 84733 CBX2 Chronic non-ProChronic 1.1448895831 1.1448895831
245 7298 TYMS Chronic non-ProChronic 1.1989460399 1.1989460399
246 10403 NDC80 Chronic ProChronic -1.9703958906 1.9703958906
247 284217 LAMA1 Chronic non-ProChronic 0.6913162057 0.6913162057
248 3613 IMPA2 Chronic non-ProChronic 2.7648081552 2.7648081552
249 5932 RBBP8 Chronic non-ProChronic 0.6126905615 0.6126905615
250 220134 SKA1 Chronic ProChronic -2.7409234865 2.7409234865
251 162681 C18orf54 Chronic non-ProChronic -1.4051352660 1.4051352660
252 23526 ARHGAP45 Chronic non-ProChronic -0.7009356222 0.7009356222
253 92840 REEP6 Chronic non-ProChronic -0.3298074521 0.3298074521
254 10036 CHAF1A Chronic non-ProChronic -0.8076872688 0.8076872688
255 126282 TNFAIP8L1 Chronic non-ProChronic -1.5299092498 1.5299092498
256 29128 UHRF1 Chronic ProChronic 0.0968097483 0.0968097483
257 147841 SPC24 Chronic ProChronic -1.7990513261 1.7990513261
258 10535 RNASEH2A Chronic ProChronic -0.2611937437 0.2611937437
259 55723 ASF1B Chronic non-ProChronic 3.1946217418 3.1946217418
260 3780 KCNN1 Chronic non-ProChronic -0.4974848853 0.4974848853
261 9518 GDF15 Chronic non-ProChronic 1.5030267937 1.5030267937
262 7643 ZNF90 Chronic non-ProChronic 0.5620211673 0.5620211673
263 9745 ZNF536 Chronic non-ProChronic 3.3446029394 3.3446029394
264 333 APLP1 Chronic non-ProChronic 0.8144991227 0.8144991227
265 284403 WDR62 Chronic non-ProChronic -0.1689598319 0.1689598319
266 163255 ZNF540 Chronic non-ProChronic -0.1088767955 0.1088767955
267 54922 RASIP1 Chronic non-ProChronic 1.4677989893 1.4677989893
268 27338 UBE2S Chronic non-ProChronic -1.2770037180 1.2770037180
269 129607 CMPK2 Chronic non-ProChronic 3.7041637567 3.7041637567
270 6241 RRM2 Chronic ProChronic -0.5951215411 0.5951215411
271 3754 KCNF1 Chronic non-ProChronic 2.6114211031 2.6114211031
272 4613 MYCN Chronic non-ProChronic 0.2927066391 0.2927066391
273 6382 SDC1 Chronic non-ProChronic 3.5644437413 3.5644437413
274 79172 CENPO Chronic non-ProChronic -0.5945742726 0.5945742726
275 109 ADCY3 Chronic non-ProChronic 2.6650529121 2.6650529121
276 1058 CENPA Chronic ProChronic -1.1011412579 1.1011412579
277 3795 KHK Chronic non-ProChronic -0.5655516495 0.5655516495
278 151056 PLB1 Chronic non-ProChronic 1.2239167098 1.2239167098
279 822 CAPG Chronic non-ProChronic -0.4363272286 0.4363272286
280 30818 KCNIP3 Chronic non-ProChronic 4.0142103598 4.0142103598
281 23397 NCAPH Chronic ProChronic -1.9739138451 1.9739138451
282 699 BUB1 Chronic non-ProChronic -0.7703993683 0.7703993683
283 150468 CKAP2L Chronic ProChronic -1.0740970054 1.0740970054
284 200373 CFAP221 Chronic non-ProChronic 0.5213887822 0.5213887822
285 4175 MCM6 Chronic non-ProChronic 0.0365924396 0.0365924396
286 3800 KIF5C Chronic non-ProChronic 3.0701571024 3.0701571024
287 130940 CCDC148 Chronic non-ProChronic 1.5454662353 1.5454662353
288 115677 NOSTRIN Chronic non-ProChronic -1.8022549141 1.8022549141
289 57405 SPC25 Chronic ProChronic -2.5879852429 2.5879852429
290 5163 PDK1 Chronic non-ProChronic 0.2805017352 0.2805017352
291 83879 CDCA7 Chronic ProChronic 0.3431916890 0.3431916890
292 50940 PDE11A Chronic ProChronic 0.6453385356 0.6453385356
293 151246 SGO2 Chronic ProChronic 0.4267702456 0.4267702456
294 26154 ABCA12 Chronic non-ProChronic 2.5490176806 2.5490176806
295 9427 ECEL1 Chronic non-ProChronic 3.2715078136 3.2715078136
296 55355 HJURP Chronic ProChronic -1.0635362786 1.0635362786
297 1293 COL6A3 Chronic non-ProChronic -1.2326938429 1.2326938429
298 23178 PASK Chronic non-ProChronic 1.1745891469 1.1745891469
299 343637 RSPO4 Chronic non-ProChronic 1.1215249160 1.1215249160
300 56265 CPXM1 Chronic non-ProChronic 0.0313000354 0.0313000354
301 83959 SLC4A11 Chronic non-ProChronic 5.9272703752 5.9272703752
302 994 CDC25B Chronic ProChronic 0.0681008129 0.0681008129
303 9837 GINS1 Chronic ProChronic -0.4471030337 0.4471030337
304 22974 TPX2 Chronic ProChronic -1.0530089125 1.0530089125
305 1869 E2F1 Chronic ProChronic 1.2157320479 1.2157320479
306 79980 DSN1 Chronic non-ProChronic -1.0094632132 1.0094632132
307 81610 FAM83D Chronic ProChronic 1.4412587120 1.4412587120
308 4605 MYBL2 Chronic ProChronic -1.1500223697 1.1500223697
309 11065 UBE2C Chronic ProChronic -1.2995793934 1.2995793934
310 55959 SULF2 Chronic non-ProChronic 1.9766963638 1.9766963638
311 6790 AURKA Chronic non-ProChronic 0.7363017280 0.7363017280
312 30811 HUNK Chronic non-ProChronic 0.8635791073 0.8635791073
313 54069 MIS18A Chronic non-ProChronic -2.1638940678 2.1638940678
314 8208 CHAF1B Chronic ProChronic -0.3822638264 0.3822638264
315 54058 C21orf58 Chronic non-ProChronic 0.9355189347 0.9355189347
316 8318 CDC45 Chronic ProChronic -0.2916154690 0.2916154690
317 11200 CHEK2 Chronic non-ProChronic -1.1746215478 1.1746215478
318 129080 EMID1 Chronic non-ProChronic 0.5720408079 0.5720408079
319 3976 LIF Chronic non-ProChronic 1.6534928349 1.6534928349
320 4174 MCM5 Chronic non-ProChronic -0.4910090645 0.4910090645
321 102800317 TPTEP2-CSNK1E Chronic non-ProChronic 1.4187125590 1.4187125590
322 79019 CENPM Chronic ProChronic -1.1762109876 1.1762109876
323 2192 FBLN1 Chronic ProChronic 0.3969246580 0.3969246580
324 51512 GTSE1 Chronic ProChronic -0.7656164726 0.7656164726
325 57633 LRRN1 Chronic non-ProChronic 7.2469860247 7.2469860247
326 2177 FANCD2 Chronic ProChronic -1.2548540233 1.2548540233
327 6854 SYN2 Chronic non-ProChronic 5.5886599049 5.5886599049
328 80852 GRIP2 Chronic non-ProChronic 2.5361184300 2.5361184300
329 151648 SGO1 Chronic ProChronic -0.5117484961 0.5117484961
330 56992 KIF15 Chronic non-ProChronic 0.1706259061 0.1706259061
331 29122 PRSS50 Chronic non-ProChronic -0.0575312356 0.0575312356
332 993 CDC25A Chronic non-ProChronic 0.7529371228 0.7529371228
333 10293 TRAIP Chronic non-ProChronic -0.7842837949 0.7842837949
334 1795 DOCK3 Chronic non-ProChronic 1.4771694641 1.4771694641
335 25886 POC1A Chronic ProChronic 0.7571337541 0.7571337541
336 55799 CACNA2D3 Chronic ProChronic 0.8776787117 0.8776787117
337 57650 CIP2A Chronic ProChronic -0.0065562200 0.0065562200
338 7348 UPK1B Chronic non-ProChronic -0.7180315956 0.7180315956
339 10721 POLQ Chronic ProChronic -1.0817468009 1.0817468009
340 4171 MCM2 Chronic ProChronic -1.0971695098 1.0971695098
341 7200 TRH Chronic non-ProChronic 0.5900660443 0.5900660443
342 92369 SPSB4 Chronic non-ProChronic 0.0698217803 0.0698217803
343 4071 TM4SF1 Chronic non-ProChronic -1.4717489690 1.4717489690
344 10051 SMC4 Chronic non-ProChronic -0.6037557564 0.6037557564
345 1894 ECT2 Chronic non-ProChronic -1.0957077120 1.0957077120
346 8646 CHRD Chronic non-ProChronic 5.9242487260 5.9242487260
347 5984 RFC4 Chronic ProChronic 0.0340837340 0.0340837340
348 116832 RPL39L Chronic non-ProChronic -1.5414019099 1.5414019099
349 5648 MASP1 Chronic non-ProChronic 0.1966341880 0.1966341880
350 55214 P3H2 Chronic ProChronic 0.0448335432 0.0448335432
351 80235 PIGZ Chronic non-ProChronic 2.7667297544 2.7667297544
352 10460 TACC3 Chronic non-ProChronic 0.2041157142 0.2041157142
353 2261 FGFR3 Chronic non-ProChronic 4.3990940861 4.3990940861
354 5522 PPP2R2C Chronic non-ProChronic 3.3020827002 3.3020827002
355 64151 NCAPG Chronic ProChronic -0.2266053338 0.2266053338
356 3815 KIT Chronic non-ProChronic 0.6088591920 0.6088591920
357 1633 DCK Chronic non-ProChronic -0.3372869665 0.3372869665
358 51191 HERC5 Chronic non-ProChronic 3.9649566774 3.9649566774
359 3015 H2AZ1 Chronic non-ProChronic 0.5552216893 0.5552216893
360 1062 CENPE Chronic non-ProChronic -2.7160882851 2.7160882851
361 8492 PRSS12 Chronic non-ProChronic 4.4110971584 4.4110971584
362 4085 MAD2L1 Chronic ProChronic -1.1455927709 1.1455927709
363 890 CCNA2 Chronic ProChronic -0.1779116288 0.1779116288
364 10733 PLK4 Chronic non-ProChronic -0.5784570770 0.5784570770
365 84057 MND1 Chronic ProChronic -1.0005900506 1.0005900506
366 1363 CPE Chronic ProChronic 1.0664542599 1.0664542599
367 3148 HMGB2 Chronic non-ProChronic 0.7444766455 0.7444766455
368 79682 CENPU Chronic ProChronic -0.1709207449 0.1709207449
369 9319 TRIP13 Chronic ProChronic -0.0873659287 0.0873659287
370 375444 C5orf34 Chronic ProChronic -0.3230447096 0.3230447096
371 10309 CCNO Chronic non-ProChronic 4.9771928722 4.9771928722
372 55789 DEPDC1B Chronic ProChronic 0.2409855411 0.2409855411
373 11174 ADAMTS6 Chronic non-ProChronic -2.6092721161 2.6092721161
374 64105 CENPK Chronic ProChronic -1.2275965940 1.2275965940
375 891 CCNB1 Chronic non-ProChronic -1.6246902814 1.6246902814
376 64946 CENPH Chronic ProChronic 0.0487045123 0.0487045123
377 1393 CRHBP Chronic ProChronic -0.3362015742 0.3362015742
378 1719 DHFR Chronic non-ProChronic 0.4973710301 0.4973710301
379 1404 HAPLN1 Chronic non-ProChronic -0.8145323703 0.8145323703
380 4001 LMNB1 Chronic ProChronic -0.3563513201 0.3563513201
381 4888 NPY6R Chronic non-ProChronic -3.6019167704 3.6019167704
382 9499 MYOT Chronic non-ProChronic 0.3167201860 0.3167201860
383 10112 KIF20A Chronic ProChronic -0.9851112692 0.9851112692
384 995 CDC25C Chronic ProChronic 0.6508613563 0.6508613563
385 6444 SGCD Chronic non-ProChronic -1.3055424492 1.3055424492
386 9232 PTTG1 Chronic non-ProChronic 0.8987346155 0.8987346155
387 3161 HMMR Chronic ProChronic -0.0595088279 0.0595088279
388 54908 SPDL1 Chronic non-ProChronic -0.8485704947 0.8485704947
389 9945 GFPT2 Chronic non-ProChronic 1.7994707254 1.7994707254
390 100130357 LOC100130357 Chronic non-ProChronic -1.0347649066 1.0347649066
391 221687 RNF182 Chronic non-ProChronic 4.3988492364 4.3988492364
392 51053 GMNN Chronic non-ProChronic -0.4428899525 0.4428899525
393 6941 TCF19 Chronic ProChronic -1.8386154737 1.8386154737
394 6890 TAP1 Chronic non-ProChronic 2.6207158626 2.6207158626
395 1302 COL11A2 Chronic ProChronic 0.1765950717 0.1765950717
396 3833 KIFC1 Chronic ProChronic -0.0840252340 0.0840252340
397 3159 HMGA1 Chronic non-ProChronic 0.5619840055 0.5619840055
398 2289 FKBP5 Chronic non-ProChronic 2.2291838892 2.2291838892
399 1026 CDKN1A Chronic non-ProChronic 0.0075017212 0.0075017212
400 105375039 <NA> Chronic non-ProChronic 2.7704861241 2.7704861241
401 266727 MDGA1 Chronic non-ProChronic 3.0511657074 3.0511657074
402 2740 GLP1R Chronic non-ProChronic -0.2106183770 0.2106183770
403 860 RUNX2 Chronic non-ProChronic 3.8907660197 3.8907660197
404 221391 OPN5 Chronic non-ProChronic -1.7620509005 1.7620509005
405 442213 PTCHD4 Chronic ProChronic -0.4129355964 0.4129355964
406 55166 CENPQ Chronic ProChronic -0.2223360617 0.2223360617
407 4172 MCM3 Chronic non-ProChronic -1.1780619235 1.1780619235
408 1310 COL19A1 Chronic non-ProChronic 0.5643606013 0.5643606013
409 7272 TTK Chronic ProChronic -1.6458190111 1.6458190111
410 9096 TBX18 Chronic non-ProChronic 0.0347079453 0.0347079453
411 253714 MMS22L Chronic ProChronic 0.6790601183 0.6790601183
412 202 CRYBG1 Chronic non-ProChronic 3.8279822118 3.8279822118
413 387103 CENPW Chronic non-ProChronic -1.3377741188 1.3377741188
414 2037 EPB41L2 Chronic non-ProChronic 1.8202388087 1.8202388087
415 113115 MTFR2 Chronic non-ProChronic 1.8838277463 1.8838277463
416 26271 FBXO5 Chronic ProChronic 0.2246730017 0.2246730017
417 2852 GPER1 Chronic non-ProChronic -2.0121477927 2.0121477927
418 4521 NUDT1 Chronic non-ProChronic 0.5166207343 0.5166207343
419 55698 RADIL Chronic non-ProChronic -1.2875797227 1.2875797227
420 10457 GPNMB Chronic non-ProChronic 0.4378714834 0.4378714834
421 29887 SNX10 Chronic non-ProChronic -1.4571321008 1.4571321008
422 54443 ANLN Chronic non-ProChronic -1.8075819799 1.8075819799
423 7378 UPP1 Chronic non-ProChronic 0.6550447242 0.6550447242
424 5982 RFC2 Chronic ProChronic 0.2013439009 0.2013439009
425 222183 SRRM3 Chronic non-ProChronic 2.5417453313 2.5417453313
426 27445 PCLO Chronic non-ProChronic 1.1991433587 1.1991433587
427 10926 DBF4 Chronic non-ProChronic -0.5394016421 0.5394016421
428 4885 NPTX2 Chronic non-ProChronic 8.4023554274 8.4023554274
429 4176 MCM7 Chronic ProChronic 0.2022672582 0.2022672582
430 79690 GAL3ST4 Chronic non-ProChronic 0.9364593206 0.9364593206
431 43 ACHE Chronic ProChronic 2.3791708823 2.3791708823
432 375611 SLC26A5 Chronic non-ProChronic 3.9244415879 3.9244415879
433 4897 NRCAM Chronic non-ProChronic 2.2679562332 2.2679562332
434 1129 CHRM2 Chronic non-ProChronic -2.9496438163 2.9496438163
435 27147 DENND2A Chronic non-ProChronic 0.6145981925 0.6145981925
436 3792 KEL Chronic non-ProChronic -0.2171832006 0.2171832006
437 2146 EZH2 Chronic non-ProChronic 2.2198697845 2.2198697845
438 7516 XRCC2 Chronic ProChronic -0.6399268747 0.6399268747
439 54892 NCAPG2 Chronic ProChronic -0.7788121744 0.7788121744
440 83648 FAM167A Chronic non-ProChronic 3.5590368443 3.5590368443
441 55790 CSGALNACT1 Chronic non-ProChronic 3.0947795623 3.0947795623
442 1960 EGR3 Chronic non-ProChronic 3.6280357677 3.6280357677
443 8794 TNFRSF10C Chronic non-ProChronic 2.2178890274 2.2178890274
444 8797 TNFRSF10A Chronic non-ProChronic 3.9919491191 3.9919491191
445 157313 CDCA2 Chronic ProChronic 1.1084300798 1.1084300798
446 51435 SCARA3 Chronic non-ProChronic -0.6076031455 0.6076031455
447 157570 ESCO2 Chronic ProChronic 0.6652723908 0.6652723908
448 55872 PBK Chronic ProChronic -1.3228002708 1.3228002708
449 84296 GINS4 Chronic non-ProChronic 1.3546479759 1.3546479759
450 4173 MCM4 Chronic non-ProChronic 0.9116463039 0.9116463039
451 8836 GGH Chronic ProChronic 0.8684528779 0.8684528779
452 254778 VXN Chronic non-ProChronic 2.5077898825 2.5077898825
453 4603 MYBL1 Chronic non-ProChronic 2.0199982032 2.0199982032
454 23213 SULF1 Chronic non-ProChronic -1.3281603035 1.3281603035
455 5569 PKIA Chronic non-ProChronic -0.6792923077 0.6792923077
456 23462 HEY1 Chronic non-ProChronic 0.3212571454 0.3212571454
457 25788 RAD54B Chronic non-ProChronic -0.4281616609 0.4281616609
458 9134 CCNE2 Chronic non-ProChronic 2.5864263271 2.5864263271
459 284 ANGPT1 Chronic non-ProChronic -2.2320702547 2.2320702547
460 79075 DSCC1 Chronic ProChronic -0.5309637138 0.5309637138
461 29028 ATAD2 Chronic ProChronic -0.4940570783 0.4940570783
462 5820 PVT1 Chronic non-ProChronic 0.7425173695 0.7425173695
463 9401 RECQL4 Chronic non-ProChronic 1.4025553898 1.4025553898
464 81704 DOCK8 Chronic non-ProChronic 4.6037921787 4.6037921787
465 158326 FREM1 Chronic non-ProChronic -0.7724442601 0.7724442601
466 347240 KIF24 Chronic non-ProChronic -0.7758952924 0.7758952924
467 2189 FANCG Chronic non-ProChronic -0.6821684093 0.6821684093
468 9833 MELK Chronic non-ProChronic -1.9026265753 1.9026265753
469 55071 C9orf40 Chronic non-ProChronic 1.1001665679 1.1001665679
470 1164 CKS2 Chronic non-ProChronic 2.0263054664 2.0263054664
471 401541 CENPP Chronic non-ProChronic 0.6305869124 0.6305869124
472 195828 ZNF367 Chronic non-ProChronic 2.7496194644 2.7496194644
473 10592 SMC2 Chronic non-ProChronic -0.6846039585 0.6846039585
474 54566 EPB41L4B Chronic non-ProChronic 5.7864669136 5.7864669136
475 3371 TNC Chronic non-ProChronic -0.6783941530 0.6783941530
476 26147 PHF19 Chronic ProChronic -0.7028339761 0.7028339761
477 57000 GSN-AS1 Chronic non-ProChronic -0.1220834800 0.1220834800
478 286204 CRB2 Chronic non-ProChronic -2.4021211874 2.4021211874
479 29941 PKN3 Chronic non-ProChronic 0.5911060512 0.5911060512
480 10319 LAMC3 Chronic non-ProChronic 0.0975949392 0.0975949392
481 84628 NTNG2 Chronic non-ProChronic 4.6060999799 4.6060999799
482 9719 ADAMTSL2 Chronic non-ProChronic -0.7090768498 0.7090768498
483 1757 SARDH Chronic non-ProChronic -1.2916657386 1.2916657386
484 89958 SAPCD2 Chronic ProChronic 0.7876663015 0.7876663015
485 441478 NRARP Chronic non-ProChronic 1.9395866870 1.9395866870
486 10742 RAI2 Chronic non-ProChronic 2.7387876543 2.7387876543
487 9468 PCYT1B Chronic non-ProChronic -2.8879610923 2.8879610923
488 6839 SUV39H1 Chronic non-ProChronic -1.1501622155 1.1501622155
489 60401 EDA2R Chronic non-ProChronic 0.2434850060 0.2434850060
490 24137 KIF4A Chronic ProChronic -1.2423815078 1.2423815078
491 54821 ERCC6L Chronic ProChronic -0.8429381920 0.8429381920
492 9452 ITM2A Chronic non-ProChronic 1.5472992291 1.5472992291
493 2491 CENPI Chronic non-ProChronic -3.1521897724 3.1521897724
494 11013 TMSB15A Chronic non-ProChronic 1.3810626063 1.3810626063
495 55859 BEX1 Chronic non-ProChronic -0.5650655054 0.5650655054
496 9363 RAB33A Chronic non-ProChronic -1.6454270162 1.6454270162
497 51765 STK26 Chronic non-ProChronic 0.9039809792 0.9039809792
498 347475 CCDC160 Chronic non-ProChronic -2.5210308584 2.5210308584
499 644538 SMIM10 Chronic non-ProChronic -1.9988186402 1.9988186402
500 5365 PLXNB3 Chronic non-ProChronic -0.8926253789 0.8926253789
501 57595 PDZD4 Chronic non-ProChronic 2.7666221466 2.7666221466
adjP_t0 Sig_t0 logFC_t24 absFC_t24 adjP_t24 Sig_t24
1 6.250208e-01 nonDE -1.414973198 1.414973198 4.822452e-04 DE
2 5.913300e-07 DE 2.279923132 2.279923132 1.053718e-04 DE
3 3.667021e-02 DE -2.484967549 2.484967549 1.880482e-04 DE
4 8.552949e-01 nonDE -0.379180547 0.379180547 1.209064e-01 nonDE
5 3.271422e-04 DE -1.821637350 1.821637350 9.933969e-08 DE
6 4.250464e-02 DE -2.617233890 2.617233890 7.143744e-06 DE
7 1.708866e-06 DE 3.739638759 3.739638759 1.338741e-06 DE
8 9.094122e-01 nonDE -0.445088746 0.445088746 1.242555e-01 nonDE
9 1.340109e-01 nonDE -0.756676355 0.756676355 2.062751e-02 DE
10 2.299780e-02 DE 2.203116032 2.203116032 8.647200e-09 DE
11 2.242321e-01 nonDE -2.450698333 2.450698333 4.922357e-07 DE
12 9.675077e-01 nonDE -3.292741124 3.292741124 5.035305e-07 DE
13 1.189710e-10 DE 6.777394620 6.777394620 4.916767e-08 DE
14 2.406624e-06 DE -1.369485669 1.369485669 8.650872e-05 DE
15 1.001819e-01 nonDE -3.338876308 3.338876308 4.021547e-07 DE
16 1.894204e-02 DE -3.123179542 3.123179542 5.993740e-11 DE
17 2.873788e-03 DE 2.248452185 2.248452185 1.264282e-04 DE
18 6.482785e-01 nonDE -1.372625426 1.372625426 2.183280e-04 DE
19 3.905571e-04 DE -1.269516917 1.269516917 6.230180e-02 nonDE
20 4.568904e-01 nonDE -1.585570217 1.585570217 1.181633e-03 DE
21 2.062193e-01 nonDE -1.868819299 1.868819299 3.208626e-04 DE
22 8.310248e-06 DE -3.864142813 3.864142813 6.483884e-10 DE
23 2.729515e-09 DE 4.071315688 4.071315688 1.347329e-07 DE
24 8.935719e-04 DE 0.515936029 0.515936029 2.965991e-01 nonDE
25 5.121943e-02 nonDE -0.642475132 0.642475132 3.521833e-04 DE
26 4.400805e-02 DE -0.591685185 0.591685185 1.164955e-02 DE
27 9.899696e-05 DE -1.439440826 1.439440826 4.101402e-05 DE
28 1.318906e-11 DE 2.319334405 2.319334405 1.325850e-05 DE
29 1.368045e-03 DE -3.230591543 3.230591543 9.400314e-06 DE
30 7.705738e-10 DE 1.226746208 1.226746208 7.268767e-02 nonDE
31 1.380929e-02 DE 1.878384343 1.878384343 1.838266e-03 DE
32 2.000231e-03 DE -0.112867412 0.112867412 6.967008e-01 nonDE
33 1.058017e-08 DE -1.178941210 1.178941210 8.578950e-04 DE
34 7.231313e-08 DE -1.168752373 1.168752373 4.738244e-03 DE
35 1.171819e-07 DE -1.310957909 1.310957909 1.640425e-03 DE
36 5.358591e-01 nonDE -2.629669313 2.629669313 3.307341e-07 DE
37 4.084764e-01 nonDE -1.196818245 1.196818245 1.627019e-03 DE
38 9.024567e-01 nonDE -1.408083837 1.408083837 1.308750e-04 DE
39 1.830844e-01 nonDE -1.085121908 1.085121908 2.188973e-03 DE
40 5.542866e-02 nonDE -3.865960872 3.865960872 8.908983e-12 DE
41 3.303452e-02 DE -0.142992017 0.142992017 5.342565e-01 nonDE
42 1.960837e-08 DE -3.936781145 3.936781145 7.554244e-11 DE
43 5.854543e-01 nonDE 1.687209158 1.687209158 7.707936e-04 DE
44 3.957396e-01 nonDE 0.456766669 0.456766669 1.924709e-01 nonDE
45 2.569697e-02 DE 3.720432379 3.720432379 8.362818e-07 DE
46 2.812491e-01 nonDE 2.167689077 2.167689077 1.315802e-04 DE
47 2.274381e-02 DE -4.331129320 4.331129320 5.194121e-11 DE
48 2.783292e-01 nonDE 2.395668644 2.395668644 1.639182e-04 DE
49 1.179194e-04 DE -3.179504867 3.179504867 1.449383e-09 DE
50 5.661754e-01 nonDE -0.070418394 0.070418394 8.749022e-01 nonDE
51 6.467254e-07 DE -2.113035022 2.113035022 1.953063e-08 DE
52 6.013056e-02 nonDE 0.165381913 0.165381913 7.704738e-01 nonDE
53 2.568602e-08 DE -0.629796474 0.629796474 8.019097e-02 nonDE
54 1.617231e-03 DE -3.416991022 3.416991022 1.588570e-07 DE
55 7.229339e-02 nonDE -2.747312897 2.747312897 1.815688e-07 DE
56 3.979934e-05 DE -3.915385108 3.915385108 3.750054e-10 DE
57 2.645581e-01 nonDE -2.078396969 2.078396969 7.166634e-04 DE
58 9.833620e-01 nonDE -2.359630074 2.359630074 1.319362e-04 DE
59 2.878984e-02 DE -2.353111447 2.353111447 1.117013e-06 DE
60 9.473857e-02 nonDE -3.268422872 3.268422872 1.434963e-06 DE
61 2.089094e-05 DE -0.558198489 0.558198489 2.413500e-01 nonDE
62 3.154620e-01 nonDE -2.066610421 2.066610421 2.881749e-03 DE
63 3.088258e-01 nonDE -0.377704576 0.377704576 2.408494e-01 nonDE
64 3.973227e-09 DE -0.086780595 0.086780595 7.442249e-01 nonDE
65 2.547814e-07 DE -1.032493924 1.032493924 5.087501e-02 nonDE
66 2.481572e-08 DE 1.986095594 1.986095594 1.014054e-02 DE
67 1.115979e-01 nonDE -2.454327881 2.454327881 1.288192e-06 DE
68 2.005402e-01 nonDE -2.567125509 2.567125509 3.153546e-07 DE
69 5.425142e-01 nonDE -1.034204242 1.034204242 2.032046e-03 DE
70 3.401782e-14 DE 3.603689004 3.603689004 5.877674e-11 DE
71 1.304020e-05 DE 1.509441145 1.509441145 8.112115e-04 DE
72 1.166016e-03 DE -1.867024906 1.867024906 4.440795e-07 DE
73 4.098021e-03 DE -3.260535951 3.260535951 2.200035e-10 DE
74 1.289357e-02 DE -3.772261363 3.772261363 4.391275e-10 DE
75 1.638124e-01 nonDE -0.899842212 0.899842212 2.800600e-03 DE
76 3.401782e-14 DE 8.664023345 8.664023345 7.260759e-15 DE
77 3.162688e-09 DE 2.787904933 2.787904933 1.789419e-04 DE
78 3.602064e-07 DE -0.822789533 0.822789533 9.126128e-03 DE
79 8.213147e-02 nonDE -4.428636299 4.428636299 7.554315e-11 DE
80 1.054269e-05 DE 7.440620202 7.440620202 9.785520e-09 DE
81 8.229094e-04 DE -2.460131566 2.460131566 1.985573e-07 DE
82 1.382195e-01 nonDE 0.499164973 0.499164973 3.108440e-02 DE
83 4.952827e-01 nonDE -1.468024007 1.468024007 3.650563e-03 DE
84 1.943004e-01 nonDE -0.973159197 0.973159197 1.756216e-02 DE
85 3.143208e-09 DE 0.145725014 0.145725014 5.407010e-01 nonDE
86 4.154369e-02 DE -0.665409702 0.665409702 2.015266e-04 DE
87 1.208607e-05 DE 2.136803238 2.136803238 3.986373e-08 DE
88 3.143898e-07 DE 0.005815022 0.005815022 9.881655e-01 nonDE
89 7.272788e-02 nonDE -0.936866708 0.936866708 4.200880e-03 DE
90 7.793632e-04 DE -0.154132183 0.154132183 5.507748e-01 nonDE
91 2.718244e-06 DE -2.868259662 2.868259662 9.278987e-10 DE
92 6.271803e-04 DE -0.396426850 0.396426850 2.426978e-01 nonDE
93 4.747246e-05 DE -0.536841329 0.536841329 3.222056e-01 nonDE
94 8.504296e-03 DE -0.351471879 0.351471879 3.256546e-01 nonDE
95 7.954894e-01 nonDE -1.284240148 1.284240148 2.717834e-04 DE
96 1.770103e-01 nonDE -3.841409732 3.841409732 1.477691e-08 DE
97 1.479886e-08 DE 2.957287568 2.957287568 3.532046e-10 DE
98 1.177956e-03 DE -0.857718881 0.857718881 1.247713e-02 DE
99 2.283021e-04 DE -1.629004081 1.629004081 6.108004e-06 DE
100 2.680087e-03 DE -0.601880754 0.601880754 9.945325e-03 DE
101 3.402390e-02 DE -1.646236117 1.646236117 2.540016e-03 DE
102 4.424822e-01 nonDE -1.322978207 1.322978207 5.389365e-05 DE
103 4.222530e-10 DE 2.328151781 2.328151781 1.009823e-10 DE
104 7.815057e-03 DE 2.375082877 2.375082877 1.363673e-05 DE
105 2.418970e-08 DE -1.936802147 1.936802147 2.762217e-05 DE
106 9.797456e-01 nonDE -1.195836799 1.195836799 1.504251e-02 DE
107 2.251291e-09 DE -0.664330748 0.664330748 1.356797e-03 DE
108 2.760165e-03 DE -1.016046368 1.016046368 1.809959e-02 DE
109 9.803673e-01 nonDE -1.225560893 1.225560893 1.880482e-04 DE
110 2.167837e-05 DE -0.885100441 0.885100441 3.170218e-04 DE
111 7.369916e-01 nonDE -0.198082374 0.198082374 5.682485e-01 nonDE
112 1.220012e-03 DE -1.561051185 1.561051185 1.250860e-07 DE
113 2.857758e-05 DE -0.096452976 0.096452976 7.811491e-01 nonDE
114 3.094512e-01 nonDE -2.576723379 2.576723379 1.634504e-07 DE
115 2.976502e-01 nonDE -2.257469660 2.257469660 2.357905e-06 DE
116 1.372963e-03 DE -0.405679964 0.405679964 6.475336e-02 nonDE
117 6.140592e-03 DE -2.057641026 2.057641026 1.095341e-07 DE
118 1.194148e-04 DE -1.274791826 1.274791826 1.232257e-02 DE
119 2.255078e-08 DE 2.643597864 2.643597864 1.289630e-06 DE
120 2.211930e-02 DE 0.677978557 0.677978557 1.382408e-01 nonDE
121 1.220383e-01 nonDE -4.232203820 4.232203820 1.443149e-09 DE
122 1.023077e-03 DE 1.040931642 1.040931642 6.185319e-02 nonDE
123 2.420006e-04 DE -2.211214356 2.211214356 3.353711e-08 DE
124 4.560456e-02 DE -0.601961390 0.601961390 4.367357e-02 DE
125 4.810506e-01 nonDE -0.318395133 0.318395133 4.628804e-01 nonDE
126 8.492192e-01 nonDE -2.899040687 2.899040687 4.496599e-07 DE
127 3.192909e-09 DE -0.531943238 0.531943238 1.644601e-01 nonDE
128 1.757225e-02 DE -0.360513862 0.360513862 3.950148e-01 nonDE
129 1.457335e-06 DE -0.547040769 0.547040769 2.103155e-03 DE
130 2.291156e-04 DE -1.452891535 1.452891535 5.489832e-06 DE
131 9.335959e-07 DE -2.093183463 2.093183463 1.211525e-09 DE
132 3.905782e-09 DE -0.622246326 0.622246326 5.971427e-03 DE
133 8.054416e-01 nonDE -0.530276499 0.530276499 1.572948e-02 DE
134 3.964507e-03 DE -1.152789621 1.152789621 2.530192e-03 DE
135 4.160835e-03 DE -0.673478817 0.673478817 4.824908e-02 DE
136 6.653279e-01 nonDE -1.211949434 1.211949434 2.162235e-04 DE
137 1.648503e-06 DE -2.388162466 2.388162466 4.336499e-08 DE
138 4.804075e-07 DE 2.114780616 2.114780616 1.031431e-07 DE
139 6.605227e-05 DE -1.711706658 1.711706658 1.663064e-06 DE
140 3.518146e-04 DE -3.145110618 3.145110618 1.318201e-10 DE
141 1.853005e-06 DE 0.717701990 0.717701990 9.111965e-02 nonDE
142 2.420006e-04 DE -1.089133601 1.089133601 2.889931e-04 DE
143 1.033685e-02 DE -1.476562236 1.476562236 1.082216e-06 DE
144 2.340527e-03 DE -2.873195871 2.873195871 3.958574e-07 DE
145 4.895329e-02 DE -1.283755084 1.283755084 2.837631e-06 DE
146 5.574500e-01 nonDE -1.663242084 1.663242084 1.220179e-05 DE
147 1.630912e-10 DE -1.148786667 1.148786667 1.798719e-05 DE
148 3.437176e-02 DE -1.181210484 1.181210484 8.683131e-04 DE
149 5.633817e-01 nonDE 0.082189181 0.082189181 8.830171e-01 nonDE
150 1.227487e-12 DE -2.342952626 2.342952626 1.104583e-05 DE
151 1.044516e-01 nonDE 1.664564527 1.664564527 1.437969e-06 DE
152 4.608653e-13 DE -2.266940957 2.266940957 1.009823e-10 DE
153 1.453079e-03 DE -2.259601311 2.259601311 1.017200e-06 DE
154 3.550184e-07 DE -1.393186788 1.393186788 6.688484e-03 DE
155 4.897827e-04 DE -2.533842136 2.533842136 1.029740e-07 DE
156 8.010863e-01 nonDE -0.882295499 0.882295499 9.708502e-04 DE
157 1.380056e-06 DE -4.131691236 4.131691236 8.258916e-14 DE
158 6.007786e-04 DE 2.750587955 2.750587955 2.070549e-04 DE
159 1.460485e-06 DE -1.417964551 1.417964551 6.515616e-05 DE
160 2.470967e-04 DE -0.639581397 0.639581397 1.040627e-02 DE
161 3.862953e-01 nonDE -2.309745009 2.309745009 1.040620e-07 DE
162 9.722524e-02 nonDE -2.742840875 2.742840875 2.341384e-08 DE
163 5.246775e-07 DE -2.886337156 2.886337156 4.354853e-10 DE
164 3.836630e-12 DE -3.610414045 3.610414045 9.991335e-14 DE
165 2.706397e-02 DE -1.297065235 1.297065235 2.397524e-06 DE
166 4.843933e-02 DE -2.867955034 2.867955034 6.833389e-10 DE
167 4.787595e-06 DE -2.358549610 2.358549610 5.494461e-09 DE
168 7.591661e-01 nonDE -2.221215310 2.221215310 2.341384e-08 DE
169 1.483819e-01 nonDE -2.856469315 2.856469315 5.084987e-07 DE
170 2.825651e-07 DE 0.247001036 0.247001036 4.172312e-01 nonDE
171 1.420384e-06 DE -2.722372036 2.722372036 4.315401e-10 DE
172 1.684201e-03 DE -0.129702876 0.129702876 5.589246e-01 nonDE
173 1.553076e-02 DE 0.713715872 0.713715872 6.830683e-02 nonDE
174 1.238344e-04 DE -2.205575075 2.205575075 3.191334e-07 DE
175 1.523872e-02 DE 0.450642111 0.450642111 1.353009e-01 nonDE
176 8.306467e-01 nonDE -2.251863970 2.251863970 1.351859e-05 DE
177 1.024748e-05 DE -0.730496072 0.730496072 2.673892e-02 DE
178 5.032757e-09 DE 2.391626181 2.391626181 2.257643e-05 DE
179 3.503138e-01 nonDE -0.252559423 0.252559423 1.510480e-01 nonDE
180 8.641408e-05 DE -3.229870843 3.229870843 4.661895e-11 DE
181 9.300189e-08 DE 0.640495260 0.640495260 2.499754e-03 DE
182 9.690863e-06 DE -0.493036976 0.493036976 1.522362e-01 nonDE
183 1.571560e-08 DE 1.673790890 1.673790890 2.031771e-06 DE
184 1.697676e-01 nonDE 1.025362848 1.025362848 3.544566e-02 DE
185 4.137465e-07 DE 2.043435302 2.043435302 2.366703e-05 DE
186 8.560342e-01 nonDE -1.675887031 1.675887031 1.405107e-07 DE
187 8.938714e-01 nonDE -2.393762691 2.393762691 2.918467e-07 DE
188 2.711338e-05 DE -1.740888376 1.740888376 5.628814e-06 DE
189 2.047481e-05 DE -0.332313853 0.332313853 3.200919e-01 nonDE
190 7.049384e-02 nonDE -3.159092533 3.159092533 1.545632e-10 DE
191 3.417708e-13 DE 6.112840605 6.112840605 9.919541e-12 DE
192 4.066095e-01 nonDE -1.202757012 1.202757012 8.971090e-04 DE
193 3.420014e-03 DE -1.207610663 1.207610663 1.543586e-04 DE
194 8.860324e-01 nonDE -2.313369442 2.313369442 1.448054e-06 DE
195 4.375639e-01 nonDE -1.110840558 1.110840558 9.251446e-04 DE
196 1.656404e-01 nonDE -1.237639562 1.237639562 3.885216e-03 DE
197 2.987555e-01 nonDE -1.247255363 1.247255363 7.791906e-05 DE
198 1.348618e-06 DE -3.355852298 3.355852298 7.636302e-11 DE
199 4.039096e-06 DE 1.570492317 1.570492317 4.070514e-06 DE
200 9.851165e-02 nonDE -1.531658748 1.531658748 7.010185e-06 DE
201 6.513072e-03 DE -0.657712543 0.657712543 1.880208e-02 DE
202 9.111169e-01 nonDE -2.792696736 2.792696736 5.135317e-09 DE
203 1.274117e-01 nonDE -1.586959711 1.586959711 1.202372e-04 DE
204 3.435422e-10 DE 2.058452329 2.058452329 9.250127e-05 DE
205 7.052551e-03 DE -1.312767349 1.312767349 6.837984e-04 DE
206 4.623241e-04 DE -0.776148269 0.776148269 6.866692e-06 DE
207 2.981318e-02 DE -0.770530202 0.770530202 1.109828e-01 nonDE
208 1.899013e-02 DE 0.332775362 0.332775362 3.923469e-01 nonDE
209 2.713878e-07 DE 2.556792543 2.556792543 1.168678e-04 DE
210 1.926618e-01 nonDE 0.224675413 0.224675413 2.561750e-01 nonDE
211 8.682145e-01 nonDE -2.236620661 2.236620661 1.791818e-07 DE
212 4.522944e-01 nonDE -1.904092999 1.904092999 5.906777e-05 DE
213 2.491730e-01 nonDE -1.890617716 1.890617716 2.153313e-05 DE
214 3.097286e-13 DE 0.654557870 0.654557870 8.013205e-02 nonDE
215 1.583176e-02 DE 0.313296532 0.313296532 3.304471e-01 nonDE
216 1.510239e-02 DE -1.955724292 1.955724292 1.238391e-03 DE
217 4.026843e-01 nonDE -0.671700472 0.671700472 7.341742e-02 nonDE
218 5.131210e-02 nonDE -3.549265351 3.549265351 4.645872e-07 DE
219 1.534595e-05 DE -3.584215330 3.584215330 2.618179e-09 DE
220 1.957127e-09 DE -1.360550592 1.360550592 8.594608e-07 DE
221 9.994906e-01 nonDE -1.193035364 1.193035364 1.319284e-04 DE
222 3.553919e-01 nonDE -1.480808342 1.480808342 7.088032e-04 DE
223 9.923679e-03 DE -2.423093325 2.423093325 3.913518e-08 DE
224 7.417823e-01 nonDE -0.775608754 0.775608754 9.104097e-03 DE
225 4.915260e-02 DE -2.570530397 2.570530397 2.811415e-06 DE
226 1.684338e-01 nonDE -3.792652865 3.792652865 4.391275e-10 DE
227 3.292690e-05 DE -0.264772329 0.264772329 5.378485e-01 nonDE
228 3.551607e-05 DE -1.034651267 1.034651267 8.865667e-06 DE
229 2.616694e-01 nonDE -0.831019257 0.831019257 3.404678e-02 DE
230 3.281637e-04 DE -4.025435385 4.025435385 2.029300e-09 DE
231 6.101791e-01 nonDE -1.416664455 1.416664455 3.904684e-04 DE
232 5.408194e-01 nonDE -3.240592393 3.240592393 1.216771e-09 DE
233 6.638307e-03 DE -2.929261728 2.929261728 1.799119e-09 DE
234 1.134799e-01 nonDE -0.208955552 0.208955552 3.705142e-01 nonDE
235 3.491703e-01 nonDE -1.041576983 1.041576983 8.311321e-03 DE
236 7.558860e-12 DE 0.602012321 0.602012321 2.084754e-01 nonDE
237 1.191241e-07 DE -1.588578159 1.588578159 8.615671e-05 DE
238 6.180963e-05 DE 1.744353720 1.744353720 8.336927e-03 DE
239 2.722480e-09 DE 3.538629630 3.538629630 4.381887e-08 DE
240 1.574143e-10 DE 2.622646088 2.622646088 3.494125e-11 DE
241 3.657921e-08 DE 3.602290792 3.602290792 2.361724e-06 DE
242 4.327129e-01 nonDE -2.222078510 2.222078510 7.153739e-07 DE
243 4.554538e-01 nonDE -3.851255320 3.851255320 6.986471e-09 DE
244 2.999962e-04 DE -0.615751536 0.615751536 5.963148e-02 nonDE
245 9.593220e-08 DE 0.424067032 0.424067032 3.211862e-02 DE
246 1.541320e-07 DE -3.862927156 3.862927156 7.826764e-12 DE
247 2.487224e-02 DE 0.992524343 0.992524343 3.106250e-03 DE
248 6.788946e-08 DE -0.038029653 0.038029653 9.434575e-01 nonDE
249 9.743340e-05 DE 0.217394260 0.217394260 1.675967e-01 nonDE
250 1.020297e-08 DE -2.846095378 2.846095378 2.544862e-08 DE
251 9.889126e-06 DE -1.542597548 1.542597548 6.776552e-06 DE
252 6.426349e-03 DE -1.599583123 1.599583123 7.073217e-07 DE
253 1.254582e-01 nonDE -1.819978750 1.819978750 6.719097e-09 DE
254 7.331256e-03 DE -1.737961708 1.737961708 2.796756e-06 DE
255 1.866220e-06 DE -0.141406629 0.141406629 6.445933e-01 nonDE
256 8.449746e-01 nonDE -2.270130433 2.270130433 5.365666e-05 DE
257 3.218887e-06 DE -2.692185728 2.692185728 1.685051e-08 DE
258 2.544798e-01 nonDE -0.901344423 0.901344423 4.771412e-04 DE
259 3.960124e-06 DE -1.144939232 1.144939232 9.208412e-02 nonDE
260 2.980778e-01 nonDE -2.839340435 2.839340435 4.971436e-06 DE
261 8.817658e-03 DE 4.448682075 4.448682075 8.448875e-09 DE
262 1.890603e-01 nonDE -0.546717659 0.546717659 2.465262e-01 nonDE
263 2.352033e-07 DE -0.518487643 0.518487643 3.904286e-01 nonDE
264 8.840771e-04 DE 0.359814253 0.359814253 1.542051e-01 nonDE
265 5.985313e-01 nonDE -2.275165738 2.275165738 9.583337e-08 DE
266 6.548956e-01 nonDE 1.259349383 1.259349383 4.510816e-06 DE
267 6.159630e-04 DE 0.656089192 0.656089192 1.375457e-01 nonDE
268 2.592065e-07 DE -0.417977535 0.417977535 5.158818e-02 nonDE
269 4.425311e-07 DE 0.638365973 0.638365973 3.587569e-01 nonDE
270 1.620348e-01 nonDE -2.699148471 2.699148471 1.239798e-06 DE
271 4.978336e-05 DE 0.566701846 0.566701846 3.928471e-01 nonDE
272 2.095626e-01 nonDE -2.557253901 2.557253901 3.953460e-11 DE
273 6.096643e-13 DE 2.485674542 2.485674542 7.057737e-09 DE
274 5.335314e-04 DE -0.519409357 0.519409357 3.341360e-03 DE
275 1.378245e-08 DE -0.448553454 0.448553454 2.778722e-01 nonDE
276 1.683343e-02 DE -3.582976228 3.582976228 2.469021e-08 DE
277 9.033398e-02 nonDE -1.326425733 1.326425733 4.790668e-04 DE
278 6.625468e-06 DE 1.690652990 1.690652990 7.192147e-08 DE
279 2.488745e-01 nonDE 0.182861221 0.182861221 6.659295e-01 nonDE
280 2.341657e-10 DE 2.214938829 2.214938829 4.153175e-05 DE
281 9.407956e-07 DE -3.685390661 3.685390661 9.211214e-11 DE
282 7.520005e-03 DE -3.126605768 3.126605768 4.238670e-11 DE
283 1.733641e-02 DE -3.637974057 3.637974057 3.161799e-08 DE
284 2.264329e-02 DE -1.402413270 1.402413270 1.764276e-06 DE
285 8.925414e-01 nonDE -2.125490034 2.125490034 3.422839e-09 DE
286 1.674316e-10 DE 3.028511031 3.028511031 1.126296e-09 DE
287 3.105924e-03 DE 3.363501752 3.363501752 1.531610e-07 DE
288 1.014165e-06 DE -0.484682000 0.484682000 1.378024e-01 nonDE
289 1.645349e-08 DE -3.305149627 3.305149627 9.062902e-10 DE
290 4.169273e-01 nonDE -0.941766724 0.941766724 9.382442e-03 DE
291 1.784552e-01 nonDE -0.838368393 0.838368393 3.201256e-03 DE
292 6.810953e-02 nonDE 1.524827546 1.524827546 1.338489e-04 DE
293 1.938490e-01 nonDE -1.346614219 1.346614219 3.600105e-04 DE
294 8.663685e-05 DE 4.407155994 4.407155994 2.295870e-08 DE
295 2.788608e-06 DE 3.747669300 3.747669300 6.897724e-07 DE
296 2.340165e-02 DE -3.804485344 3.804485344 3.256078e-08 DE
297 2.071735e-02 DE -1.767760822 1.767760822 2.634683e-03 DE
298 2.171547e-03 DE -1.614373304 1.614373304 2.451943e-04 DE
299 4.188185e-03 DE -0.691077725 0.691077725 9.647498e-02 nonDE
300 9.356741e-01 nonDE -1.706651136 1.706651136 4.409785e-05 DE
301 7.712616e-14 DE 4.133645832 4.133645832 7.520349e-10 DE
302 7.565880e-01 nonDE -0.594061339 0.594061339 7.999672e-03 DE
303 8.425525e-02 nonDE -1.468886033 1.468886033 5.165306e-06 DE
304 4.176451e-03 DE -3.269235046 3.269235046 1.507086e-09 DE
305 8.253555e-05 DE -1.461074283 1.461074283 2.997026e-05 DE
306 8.797238e-04 DE -1.595685064 1.595685064 7.062724e-06 DE
307 3.526379e-03 DE -1.799148121 1.799148121 1.282079e-03 DE
308 5.330261e-03 DE -3.095131475 3.095131475 6.044801e-08 DE
309 1.438138e-02 DE -2.954011102 2.954011102 7.339560e-06 DE
310 1.019889e-07 DE 2.231655745 2.231655745 4.968744e-08 DE
311 5.696914e-03 DE -1.330780332 1.330780332 2.222645e-05 DE
312 6.328899e-03 DE -0.853831799 0.853831799 1.277447e-02 DE
313 1.933475e-13 DE -0.596654288 0.596654288 1.524695e-03 DE
314 1.301827e-01 nonDE -1.342176840 1.342176840 1.365397e-05 DE
315 1.462835e-03 DE -2.043565180 2.043565180 1.624371e-07 DE
316 4.738529e-01 nonDE -2.392564957 2.392564957 3.227566e-06 DE
317 5.350922e-05 DE -1.542839654 1.542839654 2.757682e-06 DE
318 7.464572e-02 nonDE -0.264891934 0.264891934 4.683686e-01 nonDE
319 1.371346e-05 DE 1.749634093 1.749634093 1.263690e-05 DE
320 1.487937e-01 nonDE -2.235622740 2.235622740 5.735238e-07 DE
321 8.383496e-06 DE 0.349565969 0.349565969 2.586490e-01 nonDE
322 1.100171e-02 DE -2.955199885 2.955199885 9.608116e-07 DE
323 1.629603e-01 nonDE -1.269492457 1.269492457 1.213302e-04 DE
324 2.105734e-02 DE -3.519678133 3.519678133 1.677261e-10 DE
325 2.464254e-11 DE 3.335106761 3.335106761 9.773640e-05 DE
326 4.350759e-05 DE -1.853530897 1.853530897 3.675394e-07 DE
327 6.757094e-12 DE 2.579945583 2.579945583 4.609161e-05 DE
328 5.857508e-06 DE 2.170857177 2.170857177 1.107641e-04 DE
329 1.305913e-01 nonDE -2.908013764 2.908013764 7.087294e-09 DE
330 7.283654e-01 nonDE -2.809656337 2.809656337 3.791458e-06 DE
331 8.781756e-01 nonDE -2.313665923 2.313665923 3.865659e-07 DE
332 2.767415e-02 DE -1.590701810 1.590701810 1.000249e-04 DE
333 2.937134e-02 DE -1.994893977 1.994893977 6.480093e-06 DE
334 7.762668e-05 DE 1.643701009 1.643701009 3.939310e-05 DE
335 4.938144e-03 DE -0.950441909 0.950441909 1.402333e-03 DE
336 2.765338e-02 DE 1.243247466 1.243247466 3.913063e-03 DE
337 9.776151e-01 nonDE -1.220614927 1.220614927 2.206562e-06 DE
338 4.336896e-02 DE -1.963551735 1.963551735 8.240564e-06 DE
339 1.929418e-05 DE -2.720038038 2.720038038 9.919541e-12 DE
340 1.733423e-04 DE -1.500326172 1.500326172 6.558977e-06 DE
341 8.235308e-02 nonDE -1.117104713 1.117104713 3.471194e-03 DE
342 7.394879e-01 nonDE -0.920274977 0.920274977 7.546887e-05 DE
343 1.380823e-05 DE -0.878513303 0.878513303 6.852556e-03 DE
344 1.376916e-02 DE -1.740632053 1.740632053 6.663580e-08 DE
345 4.851305e-07 DE -1.656471084 1.656471084 8.856808e-10 DE
346 4.983116e-10 DE 5.114499563 5.114499563 3.269820e-08 DE
347 8.758823e-01 nonDE -0.551843480 0.551843480 1.144807e-02 DE
348 2.130446e-06 DE -0.513183163 0.513183163 8.077906e-02 nonDE
349 5.481909e-01 nonDE -1.135380841 1.135380841 1.169347e-03 DE
350 8.863051e-01 nonDE 0.224551045 0.224551045 4.865665e-01 nonDE
351 1.586572e-10 DE -0.781166026 0.781166026 2.410707e-02 DE
352 3.149670e-01 nonDE -1.211282212 1.211282212 1.589014e-06 DE
353 1.431075e-09 DE 1.458732395 1.458732395 1.372043e-02 DE
354 8.253442e-08 DE 1.378445415 1.378445415 1.139866e-02 DE
355 5.191858e-01 nonDE -3.366364426 3.366364426 8.606526e-10 DE
356 1.050736e-01 nonDE 1.245600303 1.245600303 1.915395e-03 DE
357 2.130860e-02 DE -0.047512813 0.047512813 7.798272e-01 nonDE
358 1.343836e-12 DE 4.399973516 4.399973516 1.227736e-12 DE
359 7.644193e-02 nonDE 0.184372954 0.184372954 6.055630e-01 nonDE
360 8.648186e-11 DE -3.809642304 3.809642304 1.230858e-12 DE
361 1.045124e-09 DE 3.351577634 3.351577634 5.986227e-07 DE
362 7.194024e-04 DE -2.529332287 2.529332287 2.751402e-08 DE
363 6.416467e-01 nonDE -2.598502288 2.598502288 2.031891e-07 DE
364 9.382668e-02 nonDE -2.290328147 2.290328147 5.104373e-07 DE
365 8.376021e-03 DE -2.578905688 2.578905688 2.635959e-07 DE
366 2.511993e-04 DE 1.081828630 1.081828630 4.792899e-04 DE
367 3.204512e-02 DE -0.635520553 0.635520553 8.712570e-02 nonDE
368 5.646058e-01 nonDE -1.784471915 1.784471915 1.530623e-06 DE
369 7.457719e-01 nonDE -1.644674402 1.644674402 5.836430e-07 DE
370 3.846660e-01 nonDE -1.534138744 1.534138744 2.512554e-04 DE
371 1.268179e-11 DE 2.496336377 2.496336377 2.208774e-05 DE
372 6.690827e-01 nonDE -1.956724930 1.956724930 1.488161e-03 DE
373 7.866750e-08 DE -1.969345427 1.969345427 1.743946e-05 DE
374 1.455172e-03 DE -2.191188338 2.191188338 2.831956e-06 DE
375 1.569469e-04 DE -2.609657854 2.609657854 4.563200e-07 DE
376 8.921874e-01 nonDE -1.459898701 1.459898701 1.675970e-04 DE
377 7.220902e-01 nonDE -1.823962173 1.823962173 5.863394e-02 nonDE
378 4.433189e-03 DE -0.165604165 0.165604165 3.798128e-01 nonDE
379 8.248356e-02 nonDE -2.391196971 2.391196971 2.021974e-05 DE
380 2.090678e-01 nonDE -0.463884771 0.463884771 1.260672e-01 nonDE
381 2.047775e-11 DE -0.317899949 0.317899949 4.075026e-01 nonDE
382 4.728409e-01 nonDE 0.924098724 0.924098724 4.019655e-02 DE
383 3.602155e-03 DE -4.408636339 4.408636339 4.284642e-12 DE
384 1.530220e-01 nonDE -2.753506927 2.753506927 3.604119e-06 DE
385 1.377279e-03 DE -2.175614448 2.175614448 6.192821e-06 DE
386 3.217590e-03 DE -0.303940697 0.303940697 3.564500e-01 nonDE
387 8.751969e-01 nonDE -2.805824149 2.805824149 2.734848e-08 DE
388 2.014082e-05 DE -0.273689473 0.273689473 1.499235e-01 nonDE
389 1.049348e-07 DE 0.799866416 0.799866416 7.765939e-03 DE
390 3.933000e-04 DE 0.648811931 0.648811931 2.335803e-02 DE
391 1.184234e-08 DE 5.259699194 5.259699194 1.152077e-09 DE
392 2.915727e-02 DE -0.607870787 0.607870787 5.904156e-03 DE
393 5.726046e-07 DE -2.379281586 2.379281586 2.804427e-08 DE
394 1.689897e-09 DE 1.183575427 1.183575427 1.125477e-03 DE
395 6.090569e-01 nonDE -1.641582949 1.641582949 3.233610e-05 DE
396 8.492662e-01 nonDE -2.387676034 2.387676034 5.001016e-06 DE
397 3.467146e-03 DE 0.336511587 0.336511587 9.351268e-02 nonDE
398 8.837397e-09 DE 1.040164286 1.040164286 1.866831e-03 DE
399 9.750165e-01 nonDE 2.019330389 2.019330389 4.391275e-10 DE
400 3.471391e-07 DE 2.235246411 2.235246411 2.614765e-05 DE
401 5.211649e-09 DE 1.421010388 1.421010388 1.375906e-03 DE
402 6.301520e-01 nonDE -3.382681946 3.382681946 4.017781e-08 DE
403 2.930252e-09 DE 2.508859472 2.508859472 2.088820e-05 DE
404 1.091848e-04 DE -0.942958356 0.942958356 3.342332e-02 DE
405 6.984090e-02 nonDE 0.777009636 0.777009636 1.560009e-03 DE
406 5.453706e-01 nonDE -0.823516613 0.823516613 3.256129e-02 DE
407 4.784310e-07 DE -1.407425530 1.407425530 7.527454e-08 DE
408 8.151509e-03 DE -0.896684125 0.896684125 2.495954e-04 DE
409 1.151773e-04 DE -3.544840554 3.544840554 3.505997e-09 DE
410 9.413966e-01 nonDE -1.363474353 1.363474353 3.702493e-03 DE
411 8.464397e-03 DE -0.752408657 0.752408657 7.417444e-03 DE
412 3.295739e-09 DE 2.181948731 2.181948731 1.218876e-04 DE
413 2.864207e-04 DE -0.870429563 0.870429563 1.764333e-02 DE
414 1.655329e-06 DE -0.271888469 0.271888469 4.651246e-01 nonDE
415 1.578377e-03 DE -0.542499210 0.542499210 4.047805e-01 nonDE
416 6.272812e-01 nonDE -0.422492197 0.422492197 3.932595e-01 nonDE
417 9.832937e-05 DE -0.775756716 0.775756716 1.258222e-01 nonDE
418 5.670911e-02 nonDE -0.162685957 0.162685957 5.987474e-01 nonDE
419 2.025189e-05 DE -3.226701323 3.226701323 2.898532e-11 DE
420 4.552468e-01 nonDE 3.066140107 3.066140107 5.041754e-06 DE
421 3.474171e-06 DE -1.293662252 1.293662252 4.597302e-05 DE
422 6.046217e-05 DE -3.889383230 3.889383230 4.747170e-10 DE
423 1.011971e-02 DE 1.587006632 1.587006632 5.330483e-07 DE
424 2.531967e-01 nonDE -0.587865481 0.587865481 2.492190e-03 DE
425 1.541320e-07 DE 1.782544216 1.782544216 1.117828e-04 DE
426 6.735486e-05 DE 1.771043622 1.771043622 3.604521e-07 DE
427 3.949908e-02 DE -0.998856988 0.998856988 6.709556e-04 DE
428 1.405553e-09 DE 5.971957393 5.971957393 2.203809e-06 DE
429 5.286567e-01 nonDE -1.069322339 1.069322339 1.922175e-03 DE
430 7.822047e-03 DE 1.971234918 1.971234918 2.077287e-06 DE
431 1.091179e-07 DE 2.448891933 2.448891933 1.984459e-07 DE
432 1.594405e-11 DE 1.265956557 1.265956557 3.755251e-03 DE
433 1.149943e-06 DE 3.183046853 3.183046853 6.986471e-09 DE
434 4.846711e-11 DE -1.265450234 1.265450234 2.884638e-04 DE
435 1.270857e-01 nonDE -1.059391080 1.059391080 1.780285e-02 DE
436 6.225819e-01 nonDE 1.012650062 1.012650062 2.543422e-02 DE
437 1.227100e-07 DE 0.379751801 0.379751801 3.185400e-01 nonDE
438 6.100635e-02 nonDE -2.958298422 2.958298422 5.689240e-09 DE
439 1.823835e-03 DE -1.519133629 1.519133629 7.741858e-07 DE
440 1.723248e-10 DE 0.697622316 0.697622316 1.104782e-01 nonDE
441 4.916734e-08 DE 1.589643252 1.589643252 1.615697e-03 DE
442 7.523388e-08 DE 2.414506110 2.414506110 1.408333e-04 DE
443 1.011135e-06 DE 2.668670068 2.668670068 1.275609e-07 DE
444 2.247366e-09 DE 3.333427782 3.333427782 2.444370e-07 DE
445 3.609127e-02 DE -1.339613977 1.339613977 2.256905e-02 DE
446 1.748623e-01 nonDE -2.764767300 2.764767300 1.867117e-06 DE
447 1.608876e-01 nonDE -2.326716799 2.326716799 7.618673e-05 DE
448 1.801575e-03 DE -4.168145193 4.168145193 6.557790e-10 DE
449 6.645646e-05 DE -0.354350401 0.354350401 3.077907e-01 nonDE
450 1.144186e-04 DE -0.287021264 0.287021264 2.251307e-01 nonDE
451 2.683702e-03 DE -1.024033159 1.024033159 1.224669e-03 DE
452 2.533462e-09 DE -0.488126393 0.488126393 1.681556e-01 nonDE
453 3.820719e-07 DE -0.718150785 0.718150785 5.075986e-02 nonDE
454 3.810901e-03 DE -1.960602696 1.960602696 1.335709e-04 DE
455 1.742068e-02 DE 0.430033268 0.430033268 1.574537e-01 nonDE
456 2.782045e-01 nonDE -0.643240439 0.643240439 4.240881e-02 DE
457 8.924912e-02 nonDE -2.060933367 2.060933367 1.049612e-08 DE
458 1.081783e-07 DE -0.795550843 0.795550843 7.265658e-02 nonDE
459 8.766799e-06 DE -2.346729258 2.346729258 1.314952e-05 DE
460 1.001729e-01 nonDE -1.661999217 1.661999217 2.366703e-05 DE
461 2.451825e-02 DE -1.025509668 1.025509668 5.735198e-05 DE
462 1.237012e-02 DE 2.060106305 2.060106305 5.806578e-08 DE
463 1.389876e-06 DE -0.579996207 0.579996207 3.361298e-02 DE
464 3.153840e-10 DE 2.617315892 2.617315892 2.908479e-05 DE
465 3.283017e-02 DE -2.126348950 2.126348950 2.295750e-06 DE
466 9.541304e-03 DE -1.476525236 1.476525236 2.548879e-05 DE
467 3.317138e-03 DE -1.384527917 1.384527917 1.212259e-06 DE
468 1.436261e-07 DE -2.762086962 2.762086962 8.298610e-10 DE
469 1.335758e-04 DE 0.175443183 0.175443183 5.660536e-01 nonDE
470 1.240790e-04 DE -0.398983429 0.398983429 4.734022e-01 nonDE
471 5.866227e-02 nonDE 0.332412908 0.332412908 3.664256e-01 nonDE
472 1.553265e-08 DE -0.218305537 0.218305537 6.257916e-01 nonDE
473 7.080648e-03 DE -1.669514262 1.669514262 2.621514e-07 DE
474 5.096618e-11 DE 3.106164769 3.106164769 1.363673e-05 DE
475 1.742829e-01 nonDE -0.368506691 0.368506691 5.085002e-01 nonDE
476 2.219978e-03 DE -1.196289580 1.196289580 8.991871e-06 DE
477 7.729011e-01 nonDE 0.547717405 0.547717405 1.851632e-01 nonDE
478 2.301032e-07 DE -3.542478461 3.542478461 1.215861e-09 DE
479 3.122570e-02 DE -0.696822395 0.696822395 1.985894e-02 DE
480 7.556743e-01 nonDE -1.284949892 1.284949892 1.809326e-04 DE
481 1.185507e-12 DE 2.751059962 2.751059962 2.197817e-07 DE
482 2.350883e-03 DE -1.720212689 1.720212689 2.634679e-08 DE
483 3.254671e-06 DE -1.154472109 1.154472109 4.327833e-05 DE
484 4.449507e-02 DE -3.020276919 3.020276919 8.274300e-08 DE
485 1.518904e-03 DE -0.174083152 0.174083152 8.067255e-01 nonDE
486 3.118804e-08 DE 2.107659133 2.107659133 8.556705e-06 DE
487 4.410341e-09 DE -0.619679440 0.619679440 1.060107e-01 nonDE
488 1.721876e-04 DE -1.661418599 1.661418599 2.870317e-06 DE
489 1.283480e-01 nonDE 1.162937819 1.162937819 5.207807e-08 DE
490 4.170056e-06 DE -3.711586741 3.711586741 3.732951e-14 DE
491 4.583087e-03 DE -1.557368006 1.557368006 1.061001e-05 DE
492 1.466150e-04 DE 2.198227005 2.198227005 2.053515e-06 DE
493 2.268861e-11 DE -3.143146034 3.143146034 1.799624e-10 DE
494 8.611354e-04 DE -1.074943988 1.074943988 1.455368e-02 DE
495 3.355460e-02 DE 0.662635800 0.662635800 1.971388e-02 DE
496 6.463698e-07 DE -0.471774805 0.471774805 1.047971e-01 nonDE
497 2.010550e-04 DE 0.858359361 0.858359361 7.514693e-04 DE
498 1.963149e-09 DE -0.941133384 0.941133384 4.949867e-03 DE
499 1.068121e-06 DE -0.751093440 0.751093440 3.941029e-02 DE
500 4.738535e-03 DE -1.422474365 1.422474365 6.323139e-05 DE
501 4.387167e-10 DE 0.831545684 0.831545684 1.756216e-02 DE
logFC_t144 absFC_t144 adjP_t144 Sig_t144 Category_final
1 -0.46802552 0.46802552 5.651783e-01 nonDE non-ProChronic
2 0.66340150 0.66340150 5.738298e-01 nonDE non-ProChronic
3 -0.41964519 0.41964519 8.160922e-01 nonDE non-ProChronic
4 -0.40374758 0.40374758 3.609532e-01 nonDE ProChronic
5 -0.59242118 0.59242118 1.490759e-01 nonDE non-ProChronic
6 -4.34718859 4.34718859 1.804382e-09 DE ProChronic
7 2.05698541 2.05698541 2.668150e-02 DE non-ProChronic
8 -0.87167987 0.87167987 2.850392e-02 DE ProChronic
9 -0.57989063 0.57989063 2.821062e-01 nonDE non-ProChronic
10 1.16459633 1.16459633 2.607669e-03 DE non-ProChronic
11 -3.35262838 3.35262838 1.951258e-08 DE ProChronic
12 -4.47252730 4.47252730 1.548214e-08 DE ProChronic
13 3.30797025 3.30797025 1.856586e-02 DE non-ProChronic
14 -1.07316672 1.07316672 1.300373e-02 DE non-ProChronic
15 -3.81540132 3.81540132 9.470028e-07 DE ProChronic
16 -3.85833272 3.85833272 8.134679e-12 DE ProChronic
17 1.15723305 1.15723305 1.869362e-01 nonDE non-ProChronic
18 -1.60230685 1.60230685 3.836822e-04 DE ProChronic
19 -3.55140900 3.55140900 4.368427e-05 DE non-ProChronic
20 -2.48201255 2.48201255 4.117201e-05 DE ProChronic
21 -0.82380041 0.82380041 3.218154e-01 nonDE non-ProChronic
22 -5.09078479 5.09078479 1.192141e-10 DE ProChronic
23 1.77850477 1.77850477 4.723081e-02 DE non-ProChronic
24 -0.11985977 0.11985977 9.546234e-01 nonDE non-ProChronic
25 -0.87619089 0.87619089 4.550893e-05 DE ProChronic
26 -0.68459714 0.68459714 3.437800e-02 DE ProChronic
27 -1.70722498 1.70722498 2.930035e-05 DE ProChronic
28 1.37362040 1.37362040 4.230766e-02 DE non-ProChronic
29 -2.81682892 2.81682892 5.726584e-04 DE non-ProChronic
30 1.26086264 1.26086264 2.701616e-01 nonDE non-ProChronic
31 1.34977549 1.34977549 1.372703e-01 nonDE non-ProChronic
32 -0.44309912 0.44309912 3.678451e-01 nonDE non-ProChronic
33 -1.54145384 1.54145384 4.569858e-04 DE non-ProChronic
34 -1.40236927 1.40236927 1.098649e-02 DE non-ProChronic
35 -1.42237612 1.42237612 8.612141e-03 DE non-ProChronic
36 -1.67428167 1.67428167 1.170746e-03 DE non-ProChronic
37 -0.60452627 0.60452627 3.476608e-01 nonDE non-ProChronic
38 -0.16368204 0.16368204 8.853056e-01 nonDE non-ProChronic
39 -0.95809603 0.95809603 4.590955e-02 DE non-ProChronic
40 -4.56642936 4.56642936 3.929771e-12 DE ProChronic
41 -0.08001974 0.08001974 9.218257e-01 nonDE non-ProChronic
42 -4.24731112 4.24731112 3.055318e-11 DE ProChronic
43 1.36554155 1.36554155 4.194150e-02 DE non-ProChronic
44 0.86098644 0.86098644 7.429738e-02 nonDE ProChronic
45 0.95866095 0.95866095 4.279408e-01 nonDE non-ProChronic
46 0.05320048 0.05320048 9.808329e-01 nonDE non-ProChronic
47 -4.12641689 4.12641689 4.796769e-10 DE non-ProChronic
48 1.80527385 1.80527385 2.668150e-02 DE non-ProChronic
49 -4.01236151 4.01236151 1.788282e-10 DE ProChronic
50 -0.58773376 0.58773376 4.514928e-01 nonDE non-ProChronic
51 -1.40075568 1.40075568 1.266897e-04 DE non-ProChronic
52 -0.02665710 0.02665710 9.911057e-01 nonDE non-ProChronic
53 -1.06360148 1.06360148 3.579080e-02 DE non-ProChronic
54 -5.43029198 5.43029198 6.895231e-11 DE ProChronic
55 -4.51593183 4.51593183 3.085996e-10 DE ProChronic
56 -4.96788892 4.96788892 4.876545e-11 DE ProChronic
57 -1.65050534 1.65050534 4.945900e-02 DE non-ProChronic
58 -1.87374513 1.87374513 1.708804e-02 DE non-ProChronic
59 -3.33464386 3.33464386 1.607607e-08 DE ProChronic
60 -5.55401696 5.55401696 2.174408e-10 DE ProChronic
61 -2.36891080 2.36891080 1.079727e-04 DE non-ProChronic
62 -1.44621195 1.44621195 1.624734e-01 nonDE non-ProChronic
63 -1.25207222 1.25207222 2.484002e-03 DE ProChronic
64 -0.76148203 0.76148203 2.297902e-02 DE non-ProChronic
65 1.63289970 1.63289970 1.880202e-02 DE non-ProChronic
66 1.07405197 1.07405197 4.961472e-01 nonDE non-ProChronic
67 -2.54905816 2.54905816 8.323847e-06 DE ProChronic
68 -5.39630241 5.39630241 2.059789e-11 DE ProChronic
69 -1.72097928 1.72097928 4.743664e-05 DE ProChronic
70 0.76065852 0.76065852 2.213483e-01 nonDE non-ProChronic
71 1.12641980 1.12641980 6.776060e-02 nonDE non-ProChronic
72 -1.91189411 1.91189411 2.237342e-06 DE ProChronic
73 -3.23733646 3.23733646 1.014375e-09 DE non-ProChronic
74 -4.64860314 4.64860314 3.362220e-11 DE ProChronic
75 -1.62028185 1.62028185 1.958850e-05 DE ProChronic
76 5.61933556 5.61933556 2.869240e-10 DE non-ProChronic
77 2.38501504 2.38501504 1.168519e-02 DE non-ProChronic
78 -0.97029450 0.97029450 1.573603e-02 DE non-ProChronic
79 -6.09226460 6.09226460 1.065307e-11 DE ProChronic
80 4.76779931 4.76779931 2.324545e-04 DE non-ProChronic
81 -1.01872474 1.01872474 5.828374e-02 nonDE non-ProChronic
82 0.94195642 0.94195642 1.256584e-03 DE ProChronic
83 -2.62866990 2.62866990 2.324174e-05 DE ProChronic
84 -2.05319521 2.05319521 6.855368e-05 DE ProChronic
85 0.54353315 0.54353315 9.386867e-02 nonDE non-ProChronic
86 -0.82695490 0.82695490 8.250599e-05 DE ProChronic
87 0.70807396 0.70807396 1.407537e-01 nonDE non-ProChronic
88 -1.01417380 1.01417380 2.168737e-02 DE non-ProChronic
89 -0.13276014 0.13276014 9.080254e-01 nonDE non-ProChronic
90 -0.44342870 0.44342870 2.888582e-01 nonDE non-ProChronic
91 -3.05478760 3.05478760 1.080149e-09 DE ProChronic
92 -1.69728492 1.69728492 4.884471e-05 DE non-ProChronic
93 -0.21988213 0.21988213 9.077996e-01 nonDE non-ProChronic
94 -0.42503631 0.42503631 5.715710e-01 nonDE non-ProChronic
95 -0.80020128 0.80020128 1.017758e-01 nonDE non-ProChronic
96 -6.23058289 6.23058289 3.705805e-12 DE ProChronic
97 1.36149144 1.36149144 2.093031e-03 DE non-ProChronic
98 -1.22555802 1.22555802 6.309583e-03 DE non-ProChronic
99 -0.84636260 0.84636260 5.670668e-02 nonDE non-ProChronic
100 -0.41999997 0.41999997 2.816388e-01 nonDE non-ProChronic
101 -2.84305362 2.84305362 9.026665e-05 DE ProChronic
102 -1.17408167 1.17408167 2.373926e-03 DE non-ProChronic
103 0.75113944 0.75113944 2.198429e-02 DE non-ProChronic
104 0.06222601 0.06222601 9.754005e-01 nonDE non-ProChronic
105 -0.87941621 0.87941621 1.618356e-01 nonDE non-ProChronic
106 -0.90741291 0.90741291 2.615384e-01 nonDE non-ProChronic
107 -0.78399555 0.78399555 2.050738e-03 DE non-ProChronic
108 -2.46175680 2.46175680 2.104391e-05 DE non-ProChronic
109 -0.78584716 0.78584716 6.651461e-02 nonDE non-ProChronic
110 -0.83663636 0.83663636 5.998489e-03 DE non-ProChronic
111 -0.65803608 0.65803608 2.206829e-01 nonDE ProChronic
112 -0.62418681 0.62418681 6.541060e-02 nonDE non-ProChronic
113 -0.28588984 0.28588984 7.260248e-01 nonDE non-ProChronic
114 -4.23334815 4.23334815 5.363195e-10 DE ProChronic
115 -3.20659338 3.20659338 4.984524e-08 DE ProChronic
116 -1.01404260 1.01404260 2.481767e-04 DE non-ProChronic
117 -2.14742369 2.14742369 3.895356e-07 DE ProChronic
118 -0.98110619 0.98110619 2.464561e-01 nonDE non-ProChronic
119 0.87010395 0.87010395 2.644940e-01 nonDE non-ProChronic
120 0.46213783 0.46213783 6.689534e-01 nonDE non-ProChronic
121 -6.16294840 6.16294840 1.019023e-12 DE ProChronic
122 -0.81090493 0.81090493 4.876257e-01 nonDE non-ProChronic
123 -2.29112506 2.29112506 1.032314e-07 DE ProChronic
124 -1.16599538 1.16599538 2.359483e-03 DE ProChronic
125 -0.28200623 0.28200623 8.284591e-01 nonDE non-ProChronic
126 -4.88028853 4.88028853 9.933066e-11 DE ProChronic
127 -1.02766935 1.02766935 5.640285e-02 nonDE non-ProChronic
128 -1.16738940 1.16738940 4.921840e-02 DE non-ProChronic
129 -0.79598359 0.79598359 2.554913e-04 DE non-ProChronic
130 -1.09002933 1.09002933 2.466237e-03 DE non-ProChronic
131 -1.23164385 1.23164385 6.920156e-05 DE non-ProChronic
132 -0.91122262 0.91122262 1.513007e-03 DE non-ProChronic
133 -0.16991974 0.16991974 7.780165e-01 nonDE non-ProChronic
134 -1.40013386 1.40013386 3.990928e-03 DE ProChronic
135 -0.72442073 0.72442073 1.669306e-01 nonDE non-ProChronic
136 -1.79667046 1.79667046 7.057750e-06 DE ProChronic
137 -0.72117113 0.72117113 1.635704e-01 nonDE non-ProChronic
138 0.74806314 0.74806314 1.326865e-01 nonDE non-ProChronic
139 -2.05334813 2.05334813 7.043344e-07 DE ProChronic
140 -3.00999481 3.00999481 1.121903e-09 DE non-ProChronic
141 1.21195500 1.21195500 4.379402e-02 DE non-ProChronic
142 -1.08357999 1.08357999 2.904747e-03 DE non-ProChronic
143 -1.07749019 1.07749019 9.235720e-04 DE non-ProChronic
144 -4.10798031 4.10798031 1.057617e-09 DE ProChronic
145 -0.70191479 0.70191479 2.844883e-02 DE non-ProChronic
146 -2.70325274 2.70325274 2.477132e-08 DE ProChronic
147 -1.16278570 1.16278570 1.268383e-04 DE non-ProChronic
148 -1.27212848 1.27212848 4.198517e-03 DE ProChronic
149 -1.20828195 1.20828195 9.150687e-02 nonDE non-ProChronic
150 -0.74966161 0.74966161 3.582831e-01 nonDE non-ProChronic
151 1.40277436 1.40277436 1.392248e-04 DE non-ProChronic
152 -1.29917210 1.29917210 1.393969e-05 DE non-ProChronic
153 -1.88513579 1.88513579 1.266897e-04 DE non-ProChronic
154 -1.33302406 1.33302406 6.904049e-02 nonDE non-ProChronic
155 -2.78537690 2.78537690 1.181049e-07 DE ProChronic
156 -1.17189751 1.17189751 3.622907e-04 DE ProChronic
157 -5.05147856 5.05147856 1.787870e-14 DE ProChronic
158 0.08762880 0.08762880 9.768575e-01 nonDE non-ProChronic
159 -1.58780160 1.58780160 1.266897e-04 DE non-ProChronic
160 -0.84362528 0.84362528 1.112777e-02 DE non-ProChronic
161 -1.17894123 1.17894123 8.469135e-03 DE non-ProChronic
162 -2.02086722 2.02086722 2.974852e-05 DE non-ProChronic
163 -3.39473029 3.39473029 8.908485e-11 DE ProChronic
164 -2.79042256 2.79042256 3.982353e-11 DE non-ProChronic
165 -0.88674785 0.88674785 3.831091e-03 DE non-ProChronic
166 -3.88436911 3.88436911 1.621713e-11 DE ProChronic
167 -2.93910234 2.93910234 2.865574e-10 DE ProChronic
168 -2.39036972 2.39036972 2.456925e-08 DE ProChronic
169 -4.71894789 4.71894789 2.771433e-09 DE ProChronic
170 0.52448500 0.52448500 2.850481e-01 nonDE non-ProChronic
171 -3.02484321 3.02484321 1.352828e-10 DE ProChronic
172 -0.80629425 0.80629425 3.166939e-03 DE non-ProChronic
173 0.07076314 0.07076314 9.681139e-01 nonDE non-ProChronic
174 -3.56221211 3.56221211 5.680696e-10 DE ProChronic
175 0.06252024 0.06252024 9.619882e-01 nonDE non-ProChronic
176 -4.17662645 4.17662645 2.283340e-09 DE ProChronic
177 -0.81359769 0.81359769 8.715618e-02 nonDE non-ProChronic
178 1.30911200 1.30911200 7.840766e-02 nonDE non-ProChronic
179 -0.59527610 0.59527610 1.145229e-02 DE ProChronic
180 -1.67973370 1.67973370 2.974852e-05 DE non-ProChronic
181 -0.11412609 0.11412609 8.661936e-01 nonDE non-ProChronic
182 -0.65761620 0.65761620 2.576823e-01 nonDE non-ProChronic
183 0.69979248 0.69979248 1.225407e-01 nonDE non-ProChronic
184 1.21862544 1.21862544 9.092404e-02 nonDE ProChronic
185 1.31835074 1.31835074 2.929423e-02 DE non-ProChronic
186 -2.35617356 2.35617356 1.080149e-09 DE ProChronic
187 -2.54221092 2.54221092 7.705245e-07 DE ProChronic
188 -2.97714774 2.97714774 2.126892e-09 DE ProChronic
189 -0.10552414 0.10552414 9.350832e-01 nonDE non-ProChronic
190 -3.48776101 3.48776101 1.192141e-10 DE ProChronic
191 2.42362410 2.42362410 1.487530e-03 DE non-ProChronic
192 -1.01728563 1.01728563 3.339750e-02 DE non-ProChronic
193 -0.74664913 0.74664913 7.650896e-02 nonDE non-ProChronic
194 -3.41459710 3.41459710 1.526134e-08 DE ProChronic
195 -0.61313818 0.61313818 2.436109e-01 nonDE non-ProChronic
196 -1.11536438 1.11536438 7.054263e-02 nonDE non-ProChronic
197 -1.79698931 1.79698931 2.239543e-06 DE ProChronic
198 -3.24209712 3.24209712 6.595496e-10 DE non-ProChronic
199 0.96820759 0.96820759 1.573855e-02 DE non-ProChronic
200 -0.85890176 0.85890176 3.608275e-02 DE non-ProChronic
201 -0.44062999 0.44062999 3.814741e-01 nonDE non-ProChronic
202 -3.32951656 3.32951656 1.516840e-09 DE ProChronic
203 -0.98561508 0.98561508 6.776060e-02 nonDE non-ProChronic
204 0.93622600 0.93622600 2.436663e-01 nonDE non-ProChronic
205 -0.40327811 0.40327811 6.240733e-01 nonDE non-ProChronic
206 -0.05288817 0.05288817 9.253543e-01 nonDE non-ProChronic
207 -1.32821864 1.32821864 5.546433e-02 nonDE non-ProChronic
208 1.09960996 1.09960996 2.929423e-02 DE non-ProChronic
209 0.77559981 0.77559981 5.575423e-01 nonDE non-ProChronic
210 0.61064718 0.61064718 1.858038e-02 DE non-ProChronic
211 -3.65068129 3.65068129 8.127478e-11 DE ProChronic
212 -3.11552484 3.11552484 3.060867e-07 DE ProChronic
213 -1.59486697 1.59486697 1.975999e-03 DE non-ProChronic
214 -0.52479373 0.52479373 5.100438e-01 nonDE non-ProChronic
215 0.04671336 0.04671336 9.724847e-01 nonDE non-ProChronic
216 -2.53463892 2.53463892 6.656283e-04 DE ProChronic
217 -0.42243598 0.42243598 5.986976e-01 nonDE non-ProChronic
218 -4.21624255 4.21624255 9.289155e-08 DE ProChronic
219 -4.92930596 4.92930596 9.531625e-12 DE ProChronic
220 0.24468328 0.24468328 6.252051e-01 nonDE non-ProChronic
221 -1.31179916 1.31179916 3.624888e-04 DE ProChronic
222 -2.49815511 2.49815511 2.296140e-06 DE ProChronic
223 -2.77401633 2.77401633 2.456925e-08 DE ProChronic
224 -1.17344761 1.17344761 1.940143e-03 DE ProChronic
225 -3.02542151 3.02542151 2.595718e-06 DE ProChronic
226 -6.01905927 6.01905927 3.082490e-12 DE ProChronic
227 -1.11377453 1.11377453 5.670668e-02 nonDE non-ProChronic
228 -1.11884013 1.11884013 1.999260e-05 DE ProChronic
229 -1.13951424 1.13951424 3.740323e-02 DE ProChronic
230 -5.22774362 5.22774362 1.541413e-11 DE ProChronic
231 -2.02980315 2.02980315 3.393709e-05 DE ProChronic
232 -3.31426894 3.31426894 6.541116e-09 DE ProChronic
233 -2.87158172 2.87158172 1.355830e-08 DE non-ProChronic
234 -0.57660772 0.57660772 7.840766e-02 nonDE non-ProChronic
235 -0.87489766 0.87489766 1.322260e-01 nonDE non-ProChronic
236 0.71009518 0.71009518 4.340002e-01 nonDE non-ProChronic
237 -1.09116835 1.09116835 3.040295e-02 DE non-ProChronic
238 -0.17123449 0.17123449 9.511976e-01 nonDE non-ProChronic
239 2.01818954 2.01818954 2.821398e-03 DE non-ProChronic
240 1.53993195 1.53993195 4.224644e-06 DE non-ProChronic
241 1.22874033 1.22874033 2.699735e-01 nonDE non-ProChronic
242 -4.07589463 4.07589463 2.377675e-10 DE ProChronic
243 -5.45076166 5.45076166 1.426368e-10 DE ProChronic
244 -0.82780747 0.82780747 8.089485e-02 nonDE non-ProChronic
245 -0.37391950 0.37391950 2.500714e-01 nonDE non-ProChronic
246 -4.69118426 4.69118426 1.019023e-12 DE ProChronic
247 0.45080862 0.45080862 4.940334e-01 nonDE non-ProChronic
248 -0.72590851 0.72590851 4.045751e-01 nonDE non-ProChronic
249 -0.58123650 0.58123650 4.058392e-03 DE non-ProChronic
250 -3.92245234 3.92245234 1.146149e-10 DE ProChronic
251 -1.52976069 1.52976069 6.868768e-05 DE non-ProChronic
252 -0.88151708 0.88151708 1.391016e-02 DE non-ProChronic
253 -0.10093873 0.10093873 8.996800e-01 nonDE non-ProChronic
254 -1.59222944 1.59222944 9.213175e-05 DE non-ProChronic
255 -1.05410713 1.05410713 6.262286e-03 DE non-ProChronic
256 -2.46078408 2.46078408 2.764791e-04 DE ProChronic
257 -4.15421195 4.15421195 8.134679e-12 DE ProChronic
258 -1.20069646 1.20069646 1.272148e-04 DE ProChronic
259 -4.17215476 4.17215476 1.543439e-05 DE non-ProChronic
260 -1.59691765 1.59691765 2.166930e-02 DE non-ProChronic
261 2.17066449 2.17066449 5.350542e-03 DE non-ProChronic
262 -1.42734896 1.42734896 2.340903e-02 DE non-ProChronic
263 0.87011787 0.87011787 4.225072e-01 nonDE non-ProChronic
264 1.23809991 1.23809991 7.424294e-05 DE non-ProChronic
265 -0.76506288 0.76506288 1.246191e-01 nonDE non-ProChronic
266 0.38286064 0.38286064 3.817089e-01 nonDE non-ProChronic
267 -0.74173873 0.74173873 3.570222e-01 nonDE non-ProChronic
268 -0.58066020 0.58066020 5.765008e-02 nonDE non-ProChronic
269 -0.69617811 0.69617811 6.693289e-01 nonDE non-ProChronic
270 -6.89630519 6.89630519 3.082490e-12 DE ProChronic
271 2.31618294 2.31618294 4.697801e-03 DE non-ProChronic
272 -0.50944619 0.50944619 1.990865e-01 nonDE non-ProChronic
273 1.23214786 1.23214786 4.427533e-03 DE non-ProChronic
274 -0.87520876 0.87520876 7.720169e-05 DE non-ProChronic
275 -0.71208001 0.71208001 3.310629e-01 nonDE non-ProChronic
276 -4.45888056 4.45888056 5.051713e-10 DE ProChronic
277 -0.53878044 0.53878044 4.228104e-01 nonDE non-ProChronic
278 1.22961316 1.22961316 1.380027e-04 DE non-ProChronic
279 -0.58567992 0.58567992 4.568227e-01 nonDE non-ProChronic
280 0.95652501 0.95652501 2.394494e-01 nonDE non-ProChronic
281 -4.49487252 4.49487252 4.592356e-12 DE ProChronic
282 -2.98187111 2.98187111 2.865574e-10 DE non-ProChronic
283 -5.10156093 5.10156093 1.835786e-10 DE ProChronic
284 -1.38373909 1.38373909 1.241833e-05 DE non-ProChronic
285 -1.59423916 1.59423916 3.369770e-06 DE non-ProChronic
286 1.91739345 1.91739345 4.932211e-05 DE non-ProChronic
287 1.19346493 1.19346493 1.485685e-01 nonDE non-ProChronic
288 -0.64370871 0.64370871 2.325851e-01 nonDE non-ProChronic
289 -5.69520864 5.69520864 8.380099e-15 DE ProChronic
290 -0.88964165 0.88964165 8.379172e-02 nonDE non-ProChronic
291 -1.24059130 1.24059130 4.672194e-04 DE ProChronic
292 1.79296957 1.79296957 1.115615e-04 DE ProChronic
293 -1.98719562 1.98719562 2.140564e-05 DE ProChronic
294 1.76467392 1.76467392 5.153030e-02 nonDE non-ProChronic
295 1.95420958 1.95420958 2.720816e-02 DE non-ProChronic
296 -4.77100688 4.77100688 2.011720e-09 DE ProChronic
297 -0.97481687 0.97481687 3.333562e-01 nonDE non-ProChronic
298 -0.76286938 0.76286938 2.615384e-01 nonDE non-ProChronic
299 -0.31836436 0.31836436 7.855158e-01 nonDE non-ProChronic
300 -0.93320491 0.93320491 1.011275e-01 nonDE non-ProChronic
301 2.16657551 2.16657551 6.488932e-04 DE non-ProChronic
302 -1.21641243 1.21641243 1.841065e-05 DE ProChronic
303 -1.62680277 1.62680277 8.682981e-06 DE ProChronic
304 -3.59840619 3.59840619 1.308475e-09 DE ProChronic
305 -1.83861928 1.83861928 8.178421e-06 DE ProChronic
306 -1.41305871 1.41305871 3.657691e-04 DE non-ProChronic
307 -3.55075694 3.55075694 5.460553e-06 DE ProChronic
308 -5.83218065 5.83218065 3.050739e-12 DE ProChronic
309 -5.74848569 5.74848569 4.796769e-10 DE ProChronic
310 1.26663423 1.26663423 2.063968e-03 DE non-ProChronic
311 -1.02181275 1.02181275 5.519279e-03 DE non-ProChronic
312 -0.85564592 0.85564592 7.453143e-02 nonDE non-ProChronic
313 -0.64080531 0.64080531 7.155605e-03 DE non-ProChronic
314 -1.88071496 1.88071496 3.846424e-07 DE ProChronic
315 -1.61272335 1.61272335 5.684836e-05 DE non-ProChronic
316 -4.04625595 4.04625595 7.975705e-10 DE ProChronic
317 -0.90603871 0.90603871 1.823791e-02 DE non-ProChronic
318 -1.24885422 1.24885422 6.834068e-03 DE non-ProChronic
319 0.77754018 0.77754018 1.735466e-01 nonDE non-ProChronic
320 -2.18872268 2.18872268 5.442750e-06 DE non-ProChronic
321 0.42114344 0.42114344 4.811096e-01 nonDE non-ProChronic
322 -4.90263104 4.90263104 1.146149e-10 DE ProChronic
323 -2.22843826 2.22843826 2.127249e-07 DE ProChronic
324 -4.41747440 4.41747440 2.059789e-11 DE ProChronic
325 0.95748196 0.95748196 5.903767e-01 nonDE non-ProChronic
326 -2.48850554 2.48850554 1.105052e-08 DE ProChronic
327 1.40330617 1.40330617 1.141104e-01 nonDE non-ProChronic
328 1.93665774 1.93665774 5.196003e-03 DE non-ProChronic
329 -3.24541205 3.24541205 1.740243e-09 DE ProChronic
330 -2.34497139 2.34497139 5.392109e-04 DE non-ProChronic
331 1.39255664 1.39255664 2.521912e-03 DE non-ProChronic
332 -1.12629410 1.12629410 2.857754e-02 DE non-ProChronic
333 -1.93760326 1.93760326 9.182008e-05 DE non-ProChronic
334 0.89020801 0.89020801 9.617854e-02 nonDE non-ProChronic
335 -1.38478402 1.38478402 1.931908e-04 DE ProChronic
336 1.28552642 1.28552642 2.149333e-02 DE ProChronic
337 -1.45518063 1.45518063 8.253106e-07 DE ProChronic
338 -0.25867604 0.25867604 8.187765e-01 nonDE non-ProChronic
339 -3.03638114 3.03638114 3.082490e-12 DE ProChronic
340 -2.19764011 2.19764011 4.480330e-08 DE ProChronic
341 -0.70543673 0.70543673 2.578141e-01 nonDE non-ProChronic
342 -0.36835009 0.36835009 3.049616e-01 nonDE non-ProChronic
343 0.08552028 0.08552028 9.511269e-01 nonDE non-ProChronic
344 -1.41301616 1.41301616 1.543439e-05 DE non-ProChronic
345 -1.39222376 1.39222376 1.032314e-07 DE non-ProChronic
346 2.41921366 2.41921366 1.824526e-02 DE non-ProChronic
347 -0.79958922 0.79958922 4.541815e-03 DE ProChronic
348 -0.83934483 0.83934483 3.848651e-02 DE non-ProChronic
349 1.03178111 1.03178111 2.473436e-02 DE non-ProChronic
350 1.22881903 1.22881903 1.638038e-03 DE ProChronic
351 0.95577026 0.95577026 3.461089e-02 DE non-ProChronic
352 -1.05448261 1.05448261 1.260134e-04 DE non-ProChronic
353 0.08047217 0.08047217 9.744926e-01 nonDE non-ProChronic
354 0.99204529 0.99204529 2.725950e-01 nonDE non-ProChronic
355 -4.91037307 4.91037307 8.134679e-12 DE ProChronic
356 0.39182848 0.39182848 6.737863e-01 nonDE non-ProChronic
357 -0.65293545 0.65293545 9.682713e-04 DE non-ProChronic
358 1.25142890 1.25142890 1.637191e-02 DE non-ProChronic
359 -0.67161783 0.67161783 2.087220e-01 nonDE non-ProChronic
360 -3.24550406 3.24550406 7.569559e-11 DE non-ProChronic
361 0.90770601 0.90770601 3.875661e-01 nonDE non-ProChronic
362 -2.73919515 2.73919515 4.218919e-08 DE ProChronic
363 -2.99174529 2.99174529 1.937116e-07 DE ProChronic
364 -2.16365097 2.16365097 1.164457e-05 DE non-ProChronic
365 -2.98257498 2.98257498 5.328235e-08 DE ProChronic
366 1.46046104 1.46046104 6.529596e-05 DE ProChronic
367 -1.49582700 1.49582700 1.637624e-03 DE non-ProChronic
368 -2.15871190 2.15871190 5.121989e-07 DE ProChronic
369 -2.22575610 2.22575610 1.963370e-08 DE ProChronic
370 -1.72355710 1.72355710 6.925522e-04 DE ProChronic
371 1.45894735 1.45894735 5.599953e-02 nonDE non-ProChronic
372 -2.99571185 2.99571185 7.993501e-05 DE ProChronic
373 -1.02213644 1.02213644 7.387320e-02 nonDE non-ProChronic
374 -3.15168851 3.15168851 5.093205e-08 DE ProChronic
375 -2.56751813 2.56751813 6.758053e-06 DE non-ProChronic
376 -2.35563201 2.35563201 1.552599e-06 DE ProChronic
377 -2.77172083 2.77172083 5.414828e-02 nonDE ProChronic
378 -1.12488439 1.12488439 1.522978e-06 DE non-ProChronic
379 -1.29389381 1.29389381 7.032091e-02 nonDE non-ProChronic
380 -0.48863969 0.48863969 3.669844e-01 nonDE ProChronic
381 0.39923884 0.39923884 6.278682e-01 nonDE non-ProChronic
382 0.89175979 0.89175979 2.157717e-01 nonDE non-ProChronic
383 -5.22855118 5.22855118 3.082490e-12 DE ProChronic
384 -2.79570395 2.79570395 1.970675e-05 DE ProChronic
385 -0.45840939 0.45840939 5.903767e-01 nonDE non-ProChronic
386 -3.52508311 3.52508311 1.872359e-10 DE non-ProChronic
387 -3.09430245 3.09430245 4.262605e-08 DE ProChronic
388 -0.56132311 0.56132311 3.047581e-02 DE non-ProChronic
389 -0.13946126 0.13946126 8.951532e-01 nonDE non-ProChronic
390 0.13920160 0.13920160 8.852159e-01 nonDE non-ProChronic
391 2.51757290 2.51757290 2.732010e-03 DE non-ProChronic
392 -0.56683966 0.56683966 6.549850e-02 nonDE non-ProChronic
393 -2.42150179 2.42150179 9.741064e-08 DE ProChronic
394 0.61621691 0.61621691 3.042320e-01 nonDE non-ProChronic
395 -2.01895145 2.01895145 8.191038e-06 DE ProChronic
396 -4.43548864 4.43548864 2.658438e-09 DE ProChronic
397 -0.14157591 0.14157591 8.080578e-01 nonDE non-ProChronic
398 -0.46120680 0.46120680 4.848395e-01 nonDE non-ProChronic
399 1.28304276 1.28304276 9.224978e-06 DE non-ProChronic
400 1.17412625 1.17412625 9.912776e-02 nonDE non-ProChronic
401 0.86194692 0.86194692 2.171428e-01 nonDE non-ProChronic
402 -2.07834568 2.07834568 2.696234e-04 DE non-ProChronic
403 0.74925892 0.74925892 4.888348e-01 nonDE non-ProChronic
404 0.82714125 0.82714125 2.209336e-01 nonDE non-ProChronic
405 0.81604230 0.81604230 7.890101e-03 DE ProChronic
406 -1.22437992 1.22437992 1.823791e-02 DE ProChronic
407 -0.89414246 0.89414246 7.484681e-04 DE non-ProChronic
408 0.31579324 0.31579324 4.486660e-01 nonDE non-ProChronic
409 -3.58169403 3.58169403 2.124786e-08 DE ProChronic
410 -1.09185858 1.09185858 1.069388e-01 nonDE non-ProChronic
411 -0.99481401 0.99481401 5.995313e-03 DE ProChronic
412 1.05224009 1.05224009 2.245354e-01 nonDE non-ProChronic
413 -2.29409888 2.29409888 2.122531e-06 DE non-ProChronic
414 -0.78678677 0.78678677 1.609017e-01 nonDE non-ProChronic
415 -1.75583146 1.75583146 5.035574e-02 nonDE non-ProChronic
416 -1.20444199 1.20444199 9.093401e-02 nonDE ProChronic
417 -0.75576749 0.75576749 4.371955e-01 nonDE non-ProChronic
418 -0.99793507 0.99793507 1.145063e-02 DE non-ProChronic
419 -0.83203397 0.83203397 3.604055e-02 DE non-ProChronic
420 0.64370707 0.64370707 6.387120e-01 nonDE non-ProChronic
421 -0.27452327 0.27452327 6.982417e-01 nonDE non-ProChronic
422 -2.10062614 2.10062614 1.416678e-04 DE non-ProChronic
423 1.07428829 1.07428829 1.487530e-03 DE non-ProChronic
424 -0.96675213 0.96675213 5.962356e-05 DE ProChronic
425 1.14413125 1.14413125 5.730193e-02 nonDE non-ProChronic
426 0.67843166 0.67843166 1.200644e-01 nonDE non-ProChronic
427 -0.86142167 0.86142167 2.374746e-02 DE non-ProChronic
428 2.30760216 2.30760216 2.191838e-01 nonDE non-ProChronic
429 -1.09624392 1.09624392 1.319365e-02 DE ProChronic
430 1.12033209 1.12033209 2.562862e-02 DE non-ProChronic
431 2.53435584 2.53435584 6.289011e-07 DE ProChronic
432 0.58255581 0.58255581 4.961472e-01 nonDE non-ProChronic
433 1.76852460 1.76852460 1.068587e-03 DE non-ProChronic
434 0.32865730 0.32865730 6.716667e-01 nonDE non-ProChronic
435 -0.55684974 0.55684974 5.416847e-01 nonDE non-ProChronic
436 0.04407872 0.04407872 9.806753e-01 nonDE non-ProChronic
437 -0.56443134 0.56443134 4.299133e-01 nonDE non-ProChronic
438 -3.03631656 3.03631656 7.176867e-09 DE ProChronic
439 -1.67660236 1.67660236 1.033584e-06 DE ProChronic
440 -0.73943127 0.73943127 3.596082e-01 nonDE non-ProChronic
441 1.02552497 1.02552497 1.790693e-01 nonDE non-ProChronic
442 -0.53247152 0.53247152 7.443536e-01 nonDE non-ProChronic
443 1.28088770 1.28088770 2.155175e-02 DE non-ProChronic
444 1.20245018 1.20245018 1.519940e-01 nonDE non-ProChronic
445 -3.54956282 3.54956282 2.440372e-05 DE ProChronic
446 -1.30759010 1.30759010 5.882855e-02 nonDE non-ProChronic
447 -3.57785934 3.57785934 9.453682e-07 DE ProChronic
448 -5.89529613 5.89529613 8.134679e-12 DE ProChronic
449 -1.81376537 1.81376537 7.424294e-05 DE non-ProChronic
450 -0.91388851 0.91388851 2.043543e-03 DE non-ProChronic
451 -1.22383851 1.22383851 2.315482e-03 DE ProChronic
452 0.48536417 0.48536417 4.672941e-01 nonDE non-ProChronic
453 -1.33048463 1.33048463 6.919518e-03 DE non-ProChronic
454 -1.44346056 1.44346056 2.432878e-02 DE non-ProChronic
455 -0.62787094 0.62787094 1.797299e-01 nonDE non-ProChronic
456 -0.36643647 0.36643647 6.022705e-01 nonDE non-ProChronic
457 -1.38370944 1.38370944 6.700596e-05 DE non-ProChronic
458 -1.72622171 1.72622171 3.769892e-03 DE non-ProChronic
459 -0.70088361 0.70088361 4.233303e-01 nonDE non-ProChronic
460 -2.42578989 2.42578989 3.991646e-07 DE ProChronic
461 -1.28187665 1.28187665 1.543439e-05 DE ProChronic
462 0.80344632 0.80344632 7.032091e-02 nonDE non-ProChronic
463 -0.79338435 0.79338435 3.764457e-02 DE non-ProChronic
464 1.78135114 1.78135114 2.374746e-02 DE non-ProChronic
465 0.98226322 0.98226322 7.455424e-02 nonDE non-ProChronic
466 -1.35386932 1.35386932 7.634781e-04 DE non-ProChronic
467 -0.83572995 0.83572995 8.987915e-03 DE non-ProChronic
468 -2.69076676 2.69076676 6.630922e-09 DE non-ProChronic
469 -0.69260470 0.69260470 1.129379e-01 nonDE non-ProChronic
470 -1.09338119 1.09338119 2.202224e-01 nonDE non-ProChronic
471 -1.28523329 1.28523329 7.332753e-03 DE non-ProChronic
472 -2.49408028 2.49408028 1.543439e-05 DE non-ProChronic
473 -0.90355009 0.90355009 9.694701e-03 DE non-ProChronic
474 0.83274227 0.83274227 5.449437e-01 nonDE non-ProChronic
475 -1.15430177 1.15430177 1.654675e-01 nonDE non-ProChronic
476 -1.68220226 1.68220226 1.928708e-07 DE ProChronic
477 0.04661151 0.04661151 9.774330e-01 nonDE non-ProChronic
478 -1.10585837 1.10585837 6.091354e-02 nonDE non-ProChronic
479 -0.60953600 0.60953600 2.001489e-01 nonDE non-ProChronic
480 -1.10839203 1.10839203 9.428051e-03 DE non-ProChronic
481 1.29286444 1.29286444 3.463500e-02 DE non-ProChronic
482 -0.96179131 0.96179131 1.975999e-03 DE non-ProChronic
483 -0.24557544 0.24557544 6.928460e-01 nonDE non-ProChronic
484 -4.12678441 4.12678441 5.297746e-10 DE ProChronic
485 -0.74708576 0.74708576 6.026354e-01 nonDE non-ProChronic
486 1.27815652 1.27815652 2.473436e-02 DE non-ProChronic
487 -0.32654894 0.32654894 7.381934e-01 nonDE non-ProChronic
488 -0.63428949 0.63428949 1.783787e-01 nonDE non-ProChronic
489 0.76457316 0.76457316 3.540057e-04 DE non-ProChronic
490 -4.48837802 4.48837802 7.416522e-15 DE ProChronic
491 -2.33642918 2.33642918 8.517273e-08 DE ProChronic
492 1.53154869 1.53154869 2.821398e-03 DE non-ProChronic
493 -2.04729825 2.04729825 2.084487e-06 DE non-ProChronic
494 -3.32011678 3.32011678 3.028347e-08 DE non-ProChronic
495 -0.13334836 0.13334836 8.898392e-01 nonDE non-ProChronic
496 -0.76460308 0.76460308 7.360826e-02 nonDE non-ProChronic
497 -0.39341612 0.39341612 3.617422e-01 nonDE non-ProChronic
498 -0.46659849 0.46659849 4.464545e-01 nonDE non-ProChronic
499 -0.91481437 0.91481437 7.838924e-02 nonDE non-ProChronic
500 0.90945572 0.90945572 3.982726e-02 DE non-ProChronic
501 1.08065426 1.08065426 2.267469e-02 DE non-ProChronic#read in my combined toptables
combined_toptables_dxr_RUV_df <- readRDS("data/DE/combined_toptables_dxr_RUV_df.RDS")
#create a toptable with all of the genes for proChronic
filt_toptable_proChronic_RUV <- combined_toptables_dxr_RUV_df %>%
mutate(ProChronic = case_when(
Entrez_ID %in% proChronic_gene_list ~ "ProChronic",
TRUE ~ NA_character_
)) %>%
dplyr::filter(!is.na(SYMBOL)) %>%
dplyr::filter(!is.na(ProChronic)) %>%
mutate(Time = factor(Time,
levels = c("t0",
"t24",
"t144"))) %>%
mutate(absFC = abs(logFC))
#this should contain 163 unique genes across timepoints
#calculate medians for ALL proSus genes across timepoints
medians_proChronic_RUV <- filt_toptable_proChronic_RUV %>%
group_by(Time) %>%
summarise(
median_logFC = median(logFC),
median_abslogFC = median(absFC),
.groups = "drop"
)
#label with median values
labs_logFC_proChronic <- paste0(
"ProChronic\nmedian logFC =\n",
paste0(medians_proChronic_RUV$Time, "=",
round(medians_proChronic_RUV$median_logFC, 2),
collapse = "\n")
)
labs_absFC_proChronic <- paste0(
"ProChronic\nmedian abslogFC =\n",
paste0(medians_proChronic_RUV$Time, "=",
round(medians_proChronic_RUV$median_abslogFC, 2),
collapse = "\n")
)
#add a column that labels positive/negative direction
filt_toptable_proChronic_RUV <- filt_toptable_proChronic_RUV %>%
mutate(direction = ifelse(logFC >= 0, "up", "down"))
#label direction
medians_proChronic_RUV <- medians_proChronic_RUV %>%
mutate(direction = "median")
#make a logFC version
plot_proChronic_logFC <- ggplot() +
geom_line(data = filt_toptable_proChronic_RUV,
aes(x = Time,
y = logFC,
group = Entrez_ID),
color = "#A06FA8",
alpha = 0.4,
linewidth = 0.9) +
geom_line(data = medians_proChronic_RUV,
aes(x = Time,
y = median_logFC,
group = 1,
color = direction),
linewidth = 3
) +
scale_color_manual(
breaks = c("median"),
values = c("median" = "#7B4F82"),
labels = c(
median = labs_logFC_proChronic),
name = NULL
) +
labs(title = "Progressive Chronic response genes",
y = "log2FC (DOX vs. VEH)", x = "Timepoint") +
scale_y_continuous(breaks = c(-10, -5, 0, 5, 10),
limits = c(-10, 10),
expand = c(0,0)) +
scale_x_discrete(breaks = c("t0", "t24", "t144"),
expand = c(0, 0)) +
theme_custom() +
theme(legend.position = "right")
#now an absFC version
plot_proChronic_absFC <- ggplot() +
geom_line(data = filt_toptable_proChronic_RUV,
aes(x = Time,
y = absFC,
group = Entrez_ID),
color = "orchid",
alpha = 0.4,
linewidth = 0.9) +
geom_line(data = medians_proChronic_RUV,
aes(x = Time,
y = median_abslogFC,
group = 1,
color = direction),
linewidth = 3
) +
scale_color_manual(
breaks = c("median"),
values = c("median" = "#3D6680"),
labels = c(
median = labs_absFC_proChronic),
name = NULL
) +
labs(title = "abs logFC — ProChronic Genes (n = 163)",
y = "absFC", x = "Timepoint") +
scale_y_continuous(expand = c(0,0),
limits = c(0, 10)) +
scale_x_discrete(breaks = c("t0", "t24", "t144"),
expand = c(0, 0)) +
theme_custom() +
theme(legend.position = "right")
print(plot_proChronic_logFC)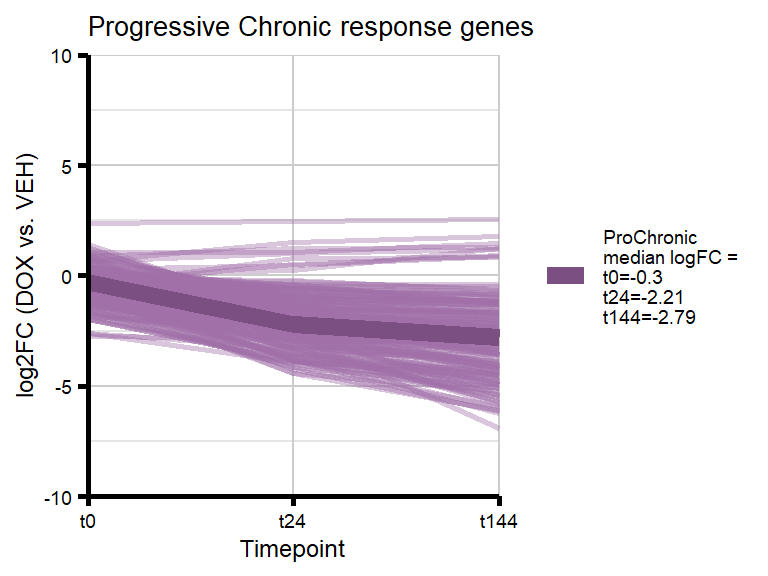
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
print(plot_proChronic_absFC)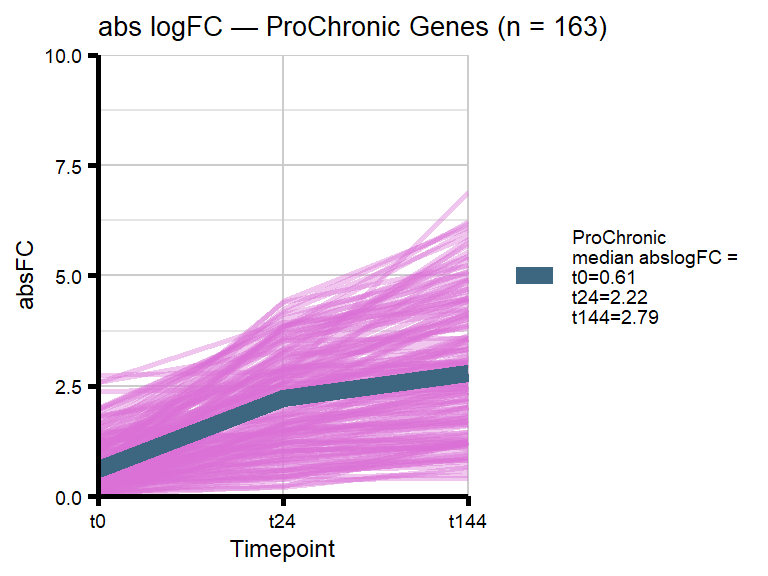
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
#find how many genes are up and how many are down
proChronic_directionality <- filt_toptable_proChronic_RUV %>%
group_by(Time, direction) %>%
summarise(n = n(), .groups = "drop")
# save_plot(
# plot = plot_proChronic_logFC,
# filename = "Lineplot_proChronic_logFC_EMP_251211",
# folder = output_folder,
# height = 4,
# width = 5
# )
# save_plot(
# plot = plot_proChronic_absFC,
# filename = "Lineplot_proChronic_absFC_EMP_251112",
# folder = output_folder,
# height = 4,
# width = 5
# )Figure 6B - Enriched Pathways: ProChronic
#now I would like to look at enriched pathways using GO:BP and KEGG for these progressively Chronic genes
proChronic_gene_list [1] "79180" "1870" "79000" "63967" "55143" "991"
[7] "11004" "6491" "4998" "55635" "8317" "343099"
[13] "29899" "128239" "83540" "460" "9928" "4751"
[19] "51514" "1063" "9156" "55388" "56952" "11130"
[25] "983" "219790" "9585" "55165" "3070" "4288"
[31] "64787" "283120" "3481" "6240" "81930" "374393"
[37] "113130" "53340" "2305" "10635" "83461" "10024"
[43] "29127" "25840" "9700" "100128191" "283431" "55010"
[49] "221150" "675" "79866" "1033" "11169" "9787"
[55] "9824" "57082" "5888" "11339" "51203" "79968"
[61] "9133" "9768" "55055" "57214" "55215" "90381"
[67] "641" "9055" "9088" "116028" "79801" "51659"
[73] "81620" "83903" "54478" "9212" "6470" "4239"
[79] "10615" "146857" "990" "7153" "672" "78995"
[85] "146909" "146956" "55771" "7083" "332" "10403"
[91] "220134" "29128" "147841" "10535" "6241" "1058"
[97] "23397" "150468" "57405" "83879" "50940" "151246"
[103] "55355" "994" "9837" "22974" "1869" "81610"
[109] "4605" "11065" "8208" "8318" "79019" "2192"
[115] "51512" "2177" "151648" "25886" "55799" "57650"
[121] "10721" "4171" "5984" "55214" "64151" "4085"
[127] "890" "84057" "1363" "79682" "9319" "375444"
[133] "55789" "64105" "64946" "1393" "4001" "10112"
[139] "995" "3161" "6941" "1302" "3833" "442213"
[145] "55166" "7272" "253714" "26271" "5982" "4176"
[151] "43" "7516" "54892" "157313" "157570" "55872"
[157] "8836" "79075" "29028" "26147" "89958" "24137"
[163] "54821" #background of all genes
all_genes <- readRDS("data/DE/all_genes.RDS")
length(proChronic_gene_list)[1] 163#163 - don't filter for DEGs
proChronic_GOKEGG_RUV <- gost(query = proChronic_gene_list,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = TRUE,
user_threshold = 0.05,
significant = TRUE,
custom_bg = all_genes,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
proChronic_GOKEGG_genes_RUV <- gostplot(proChronic_GOKEGG_RUV, capped = FALSE, interactive = TRUE)
proChronic_GOKEGG_genes_RUVtable_proChronic_GOKEGG_genes_RUV <- proChronic_GOKEGG_RUV$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_proChronic_GOKEGG_genes_RUV %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0022402 | cell cycle process | 100 | 1094 | 2.092e-65 |
| GO:BP | GO:0007049 | cell cycle | 105 | 1392 | 1.720e-61 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 82 | 687 | 1.521e-60 |
| GO:BP | GO:0000278 | mitotic cell cycle | 85 | 809 | 1.199e-58 |
| GO:BP | GO:0007059 | chromosome segregation | 64 | 380 | 4.075e-55 |
| GO:BP | GO:0000280 | nuclear division | 60 | 358 | 3.595e-51 |
| GO:BP | GO:0048285 | organelle fission | 60 | 403 | 4.948e-48 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 52 | 278 | 2.533e-46 |
| GO:BP | GO:0140014 | mitotic nuclear division | 50 | 258 | 3.138e-45 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 67 | 626 | 6.373e-45 |
| GO:BP | GO:0051276 | chromosome organization | 61 | 485 | 1.388e-44 |
| GO:BP | GO:0000819 | sister chromatid segregation | 46 | 223 | 9.739e-43 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 43 | 185 | 3.548e-42 |
| GO:BP | GO:0051301 | cell division | 62 | 575 | 2.174e-41 |
| GO:BP | GO:0051726 | regulation of cell cycle | 72 | 928 | 2.703e-39 |
| GO:BP | GO:0044770 | cell cycle phase transition | 53 | 483 | 4.600e-35 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 48 | 386 | 5.532e-34 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 50 | 442 | 1.525e-33 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 31 | 106 | 2.523e-33 |
| GO:BP | GO:0051783 | regulation of nuclear division | 32 | 121 | 6.714e-33 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 31 | 125 | 7.157e-31 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 45 | 397 | 5.813e-30 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 34 | 181 | 1.188e-29 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 40 | 303 | 5.438e-29 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 38 | 266 | 9.389e-29 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 36 | 230 | 1.337e-28 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 25 | 74 | 1.538e-28 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 35 | 216 | 2.527e-28 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 27 | 98 | 3.368e-28 |
| GO:BP | GO:0050000 | chromosome localization | 28 | 113 | 7.203e-28 |
| GO:BP | GO:0051304 | chromosome separation | 25 | 80 | 1.310e-27 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 27 | 105 | 2.459e-27 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 22 | 61 | 8.305e-26 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 36 | 278 | 1.022e-25 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 28 | 134 | 1.109e-25 |
| GO:BP | GO:0006260 | DNA replication | 35 | 259 | 1.264e-25 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 22 | 64 | 2.610e-25 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 38 | 335 | 3.842e-25 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 25 | 103 | 1.292e-24 |
| GO:BP | GO:0051321 | meiotic cell cycle | 30 | 195 | 1.692e-23 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 28 | 160 | 1.790e-23 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 20 | 56 | 2.225e-23 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 23 | 91 | 4.681e-23 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 23 | 92 | 6.040e-23 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 19 | 50 | 7.573e-23 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 19 | 50 | 7.573e-23 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 19 | 50 | 7.573e-23 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 19 | 50 | 7.573e-23 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 30 | 208 | 9.880e-23 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 19 | 51 | 1.103e-22 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 19 | 51 | 1.103e-22 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 19 | 51 | 1.103e-22 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 23 | 95 | 1.130e-22 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 23 | 96 | 1.444e-22 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 27 | 158 | 1.968e-22 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 26 | 144 | 3.201e-22 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 43 | 562 | 6.513e-22 |
| GO:BP | GO:0006259 | DNA metabolic process | 52 | 874 | 6.577e-22 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 18 | 48 | 1.375e-21 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 18 | 48 | 1.375e-21 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 18 | 48 | 1.375e-21 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 19 | 58 | 1.686e-21 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 18 | 49 | 2.083e-21 |
| GO:BP | GO:0007051 | spindle organization | 28 | 193 | 2.503e-21 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 19 | 60 | 3.404e-21 |
| GO:BP | GO:0140013 | meiotic nuclear division | 24 | 128 | 6.257e-21 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 18 | 52 | 7.018e-21 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 25 | 151 | 1.931e-20 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 29 | 232 | 2.790e-20 |
| GO:BP | GO:0007017 | microtubule-based process | 47 | 791 | 1.362e-19 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 20 | 93 | 1.233e-18 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 19 | 84 | 3.905e-18 |
| GO:BP | GO:0051656 | establishment of organelle localization | 34 | 424 | 1.021e-17 |
| GO:BP | GO:0007052 | mitotic spindle organization | 21 | 128 | 5.084e-17 |
| GO:BP | GO:0006281 | DNA repair | 37 | 568 | 2.246e-16 |
| GO:BP | GO:0006996 | organelle organization | 85 | 2998 | 2.831e-16 |
| GO:BP | GO:0051225 | spindle assembly | 20 | 126 | 6.545e-16 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 21 | 146 | 8.006e-16 |
| GO:BP | GO:0006974 | DNA damage response | 43 | 826 | 1.179e-15 |
| GO:BP | GO:0051640 | organelle localization | 35 | 543 | 2.633e-15 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 18 | 101 | 3.105e-15 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 17 | 87 | 4.365e-15 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 9 | 11 | 4.741e-15 |
| GO:BP | GO:0007080 | mitotic metaphase chromosome alignment | 15 | 60 | 5.139e-15 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 12 | 30 | 8.436e-15 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 15 | 62 | 8.605e-15 |
| GO:BP | GO:0007127 | meiosis I | 16 | 79 | 1.802e-14 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 19 | 131 | 2.131e-14 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 18 | 115 | 3.125e-14 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 17 | 98 | 3.290e-14 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 13 | 47 | 1.193e-13 |
| GO:BP | GO:0033043 | regulation of organelle organization | 44 | 1002 | 2.046e-13 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 17 | 109 | 2.046e-13 |
| GO:BP | GO:0006310 | DNA recombination | 24 | 283 | 6.537e-13 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 10 | 25 | 2.383e-12 |
| GO:BP | GO:0019953 | sexual reproduction | 35 | 684 | 2.383e-12 |
| GO:BP | GO:0006302 | double-strand break repair | 23 | 291 | 1.013e-11 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 9 | 20 | 1.120e-11 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 10 | 29 | 1.360e-11 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 26 | 391 | 1.451e-11 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 16 | 121 | 1.684e-11 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 10 | 31 | 2.864e-11 |
| GO:BP | GO:0000725 | recombinational repair | 18 | 175 | 4.654e-11 |
| GO:BP | GO:0007010 | cytoskeleton organization | 45 | 1230 | 5.629e-11 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 9 | 24 | 7.814e-11 |
| GO:BP | GO:0000910 | cytokinesis | 17 | 169 | 2.732e-10 |
| GO:BP | GO:0016043 | cellular component organization | 104 | 5231 | 2.794e-10 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 17 | 170 | 2.926e-10 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 22 | 312 | 2.926e-10 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 12 | 71 | 6.363e-10 |
| GO:BP | GO:0022414 | reproductive process | 38 | 988 | 9.513e-10 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 10 | 43 | 9.985e-10 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 6 | 8 | 1.228e-09 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 8 | 22 | 1.584e-09 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 8 | 23 | 2.364e-09 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 8 | 23 | 2.364e-09 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 10 | 47 | 2.479e-09 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 104 | 5431 | 3.328e-09 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 6 | 9 | 3.465e-09 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 24 | 438 | 5.656e-09 |
| GO:BP | GO:0051293 | establishment of spindle localization | 10 | 52 | 6.962e-09 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 6 | 10 | 8.367e-09 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 12 | 89 | 8.787e-09 |
| GO:BP | GO:0033260 | nuclear DNA replication | 9 | 39 | 9.218e-09 |
| GO:BP | GO:0051653 | spindle localization | 10 | 56 | 1.457e-08 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 6 | 11 | 1.765e-08 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 9 | 42 | 1.837e-08 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 5 | 6 | 2.050e-08 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 8 | 30 | 2.365e-08 |
| GO:BP | GO:0051302 | regulation of cell division | 14 | 145 | 2.628e-08 |
| GO:BP | GO:0051231 | spindle elongation | 6 | 12 | 3.337e-08 |
| GO:BP | GO:0051255 | spindle midzone assembly | 6 | 12 | 3.337e-08 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 11 | 80 | 3.534e-08 |
| GO:BP | GO:0035825 | homologous recombination | 9 | 46 | 4.132e-08 |
| GO:BP | GO:0051383 | kinetochore organization | 7 | 21 | 4.310e-08 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 8 | 33 | 5.164e-08 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 6 | 13 | 5.912e-08 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 5 | 7 | 6.593e-08 |
| GO:BP | GO:0006275 | regulation of DNA replication | 12 | 107 | 6.763e-08 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 10 | 66 | 6.964e-08 |
| GO:BP | GO:0006270 | DNA replication initiation | 8 | 35 | 8.276e-08 |
| GO:BP | GO:0033554 | cellular response to stress | 46 | 1609 | 8.718e-08 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 10 | 68 | 9.208e-08 |
| GO:BP | GO:0007292 | female gamete generation | 12 | 111 | 9.973e-08 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 12 | 112 | 1.098e-07 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 12 | 113 | 1.208e-07 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 6 | 15 | 1.565e-07 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 6 | 15 | 1.565e-07 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 8 | 38 | 1.579e-07 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 10 | 73 | 1.783e-07 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 7 | 26 | 2.074e-07 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 6 | 16 | 2.383e-07 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 6 | 16 | 2.383e-07 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 8 | 40 | 2.383e-07 |
| GO:BP | GO:0007144 | female meiosis I | 5 | 9 | 3.455e-07 |
| GO:BP | GO:1902969 | mitotic DNA replication | 6 | 17 | 3.601e-07 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 7 | 29 | 4.566e-07 |
| GO:BP | GO:0034508 | centromere complex assembly | 7 | 29 | 4.566e-07 |
| GO:BP | GO:0034502 | protein localization to chromosome | 11 | 107 | 6.647e-07 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 6 | 19 | 7.501e-07 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 6 | 19 | 7.501e-07 |
| GO:BP | GO:0031297 | replication fork processing | 8 | 48 | 1.014e-06 |
| GO:BP | GO:0000212 | meiotic spindle organization | 6 | 20 | 1.042e-06 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 6 | 20 | 1.042e-06 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 4 | 5 | 1.144e-06 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 4 | 5 | 1.144e-06 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 5 | 11 | 1.147e-06 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 5 | 11 | 1.147e-06 |
| GO:BP | GO:0051781 | positive regulation of cell division | 9 | 69 | 1.337e-06 |
| GO:BP | GO:0030261 | chromosome condensation | 7 | 34 | 1.392e-06 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 13 | 175 | 1.805e-06 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 8 | 52 | 1.830e-06 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 5 | 12 | 1.891e-06 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 7 | 36 | 2.069e-06 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 8 | 54 | 2.437e-06 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 7 | 37 | 2.497e-06 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 8 | 55 | 2.792e-06 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 7 | 38 | 2.980e-06 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 7 | 38 | 2.980e-06 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 4 | 6 | 3.136e-06 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 5 | 14 | 4.457e-06 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 5 | 14 | 4.457e-06 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 23 | 592 | 5.227e-06 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 13 | 194 | 5.528e-06 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 12 | 166 | 6.850e-06 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 48 | 2008 | 7.636e-06 |
| GO:BP | GO:0051716 | cellular response to stimulus | 89 | 4968 | 9.093e-06 |
| GO:BP | GO:0051382 | kinetochore assembly | 5 | 17 | 1.296e-05 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 4 | 8 | 1.361e-05 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 4 | 8 | 1.361e-05 |
| GO:BP | GO:2000241 | regulation of reproductive process | 10 | 118 | 1.394e-05 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 6 | 31 | 1.518e-05 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 8 | 69 | 1.529e-05 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 3 | 3 | 1.705e-05 |
| GO:BP | GO:0070925 | organelle assembly | 28 | 891 | 1.808e-05 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 11 | 154 | 2.170e-05 |
| GO:BP | GO:0035556 | intracellular signal transduction | 52 | 2349 | 2.224e-05 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 4 | 9 | 2.329e-05 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 18 | 431 | 3.467e-05 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 4 | 10 | 3.808e-05 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 6 | 37 | 4.297e-05 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 6 | 37 | 4.297e-05 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 12 | 203 | 5.173e-05 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 3 | 4 | 6.246e-05 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 3 | 4 | 6.246e-05 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 3 | 4 | 6.246e-05 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 3 | 4 | 6.246e-05 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 3 | 4 | 6.246e-05 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 3 | 4 | 6.246e-05 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 3 | 4 | 6.246e-05 |
| GO:BP | GO:0007276 | gamete generation | 19 | 505 | 7.790e-05 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 6 | 43 | 1.013e-04 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 11 | 184 | 1.110e-04 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 4 | 13 | 1.179e-04 |
| GO:BP | GO:0008283 | cell population proliferation | 35 | 1397 | 1.216e-04 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 4 | 14 | 1.612e-04 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 4 | 14 | 1.612e-04 |
| GO:BP | GO:0051338 | regulation of transferase activity | 16 | 398 | 1.831e-04 |
| GO:BP | GO:0032506 | cytokinetic process | 6 | 48 | 1.879e-04 |
| GO:BP | GO:0007098 | centrosome cycle | 9 | 129 | 2.031e-04 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 8 | 100 | 2.181e-04 |
| GO:BP | GO:0010212 | response to ionizing radiation | 9 | 131 | 2.276e-04 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 5 | 32 | 3.081e-04 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 9 | 138 | 3.394e-04 |
| GO:BP | GO:0030010 | establishment of cell polarity | 9 | 138 | 3.394e-04 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 3 | 7 | 4.947e-04 |
| GO:BP | GO:0022607 | cellular component assembly | 51 | 2563 | 5.266e-04 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 8 | 114 | 5.437e-04 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 12 | 261 | 5.599e-04 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 4 | 19 | 5.601e-04 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 4 | 19 | 5.601e-04 |
| GO:BP | GO:0009987 | cellular process | 156 | 11923 | 5.993e-04 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 9 | 150 | 6.273e-04 |
| GO:BP | GO:0009314 | response to radiation | 14 | 351 | 6.273e-04 |
| GO:BP | GO:0050794 | regulation of cellular process | 117 | 7899 | 6.853e-04 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 5 | 38 | 6.853e-04 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 19 | 598 | 7.062e-04 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 3 | 8 | 7.420e-04 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 3 | 8 | 7.420e-04 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 5 | 39 | 7.657e-04 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 5 | 40 | 8.632e-04 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 4 | 22 | 9.774e-04 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 8 | 125 | 9.774e-04 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 3 | 9 | 1.081e-03 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 1.112e-03 |
| GO:BP | GO:0140273 | repair of mitotic kinetochore microtubule attachment defect | 2 | 2 | 1.112e-03 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 1.112e-03 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 1.112e-03 |
| GO:BP | GO:0110029 | negative regulation of meiosis I | 2 | 2 | 1.112e-03 |
| GO:BP | GO:0140274 | repair of kinetochore microtubule attachment defect | 2 | 2 | 1.112e-03 |
| GO:BP | GO:0060631 | regulation of meiosis I | 2 | 2 | 1.112e-03 |
| GO:BP | GO:1905325 | regulation of meiosis I spindle assembly checkpoint | 2 | 2 | 1.112e-03 |
| GO:BP | GO:0031619 | homologous chromosome orientation in meiotic metaphase I | 2 | 2 | 1.112e-03 |
| GO:BP | GO:1905318 | meiosis I spindle assembly checkpoint signaling | 2 | 2 | 1.112e-03 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 10 | 201 | 1.112e-03 |
| GO:BP | GO:0007019 | microtubule depolymerization | 5 | 43 | 1.155e-03 |
| GO:BP | GO:0090224 | regulation of spindle organization | 5 | 44 | 1.286e-03 |
| GO:BP | GO:0001556 | oocyte maturation | 4 | 24 | 1.313e-03 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 3 | 10 | 1.436e-03 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 3 | 10 | 1.436e-03 |
| GO:BP | GO:0050789 | regulation of biological process | 118 | 8147 | 1.896e-03 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 3 | 11 | 1.943e-03 |
| GO:BP | GO:0008150 | biological_process | 157 | 12194 | 2.032e-03 |
| GO:BP | GO:0050896 | response to stimulus | 91 | 5825 | 2.233e-03 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 10 | 222 | 2.357e-03 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 3 | 12 | 2.530e-03 |
| GO:BP | GO:0051649 | establishment of localization in cell | 36 | 1714 | 2.809e-03 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 5 | 53 | 2.992e-03 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 2 | 3 | 3.082e-03 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 2 | 3 | 3.082e-03 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 2 | 3 | 3.082e-03 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 2 | 3 | 3.082e-03 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 2 | 3 | 3.082e-03 |
| GO:BP | GO:0051296 | establishment of meiotic spindle orientation | 2 | 3 | 3.082e-03 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 3 | 13 | 3.131e-03 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 3 | 13 | 3.131e-03 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 3 | 13 | 3.131e-03 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 18 | 629 | 3.350e-03 |
| GO:BP | GO:0071478 | cellular response to radiation | 8 | 153 | 3.388e-03 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 5 | 56 | 3.705e-03 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 3 | 14 | 3.894e-03 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 4 | 33 | 4.244e-03 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 4 | 33 | 4.244e-03 |
| GO:BP | GO:0044085 | cellular component biogenesis | 51 | 2814 | 4.559e-03 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 5 | 59 | 4.637e-03 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 4 | 34 | 4.708e-03 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 3 | 15 | 4.708e-03 |
| GO:BP | GO:0044027 | negative regulation of gene expression via chromosomal CpG island methylation | 3 | 15 | 4.708e-03 |
| GO:BP | GO:0043060 | meiotic metaphase I homologous chromosome alignment | 2 | 4 | 5.644e-03 |
| GO:BP | GO:1901563 | response to camptothecin | 2 | 4 | 5.644e-03 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 2 | 4 | 5.644e-03 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 2 | 4 | 5.644e-03 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 5.644e-03 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 3 | 16 | 5.644e-03 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 2 | 4 | 5.644e-03 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 2 | 4 | 5.644e-03 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 4 | 36 | 5.644e-03 |
| GO:BP | GO:0006301 | postreplication repair | 4 | 36 | 5.644e-03 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 5 | 63 | 5.975e-03 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 65 | 3904 | 5.988e-03 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 4 | 38 | 6.830e-03 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 7 | 133 | 6.830e-03 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 4 | 38 | 6.830e-03 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 19 | 735 | 7.257e-03 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 9 | 218 | 7.608e-03 |
| GO:BP | GO:0006338 | chromatin remodeling | 16 | 570 | 7.788e-03 |
| GO:BP | GO:0048519 | negative regulation of biological process | 66 | 4029 | 8.206e-03 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 5 | 68 | 8.206e-03 |
| GO:BP | GO:0065007 | biological regulation | 118 | 8399 | 8.326e-03 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 2 | 5 | 8.882e-03 |
| GO:BP | GO:0051311 | meiotic metaphase chromosome alignment | 2 | 5 | 8.882e-03 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 4 | 41 | 8.882e-03 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 4 | 42 | 9.666e-03 |
| GO:BP | GO:0001832 | blastocyst growth | 3 | 20 | 1.036e-02 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 4 | 43 | 1.049e-02 |
| GO:BP | GO:0006325 | chromatin organization | 18 | 706 | 1.104e-02 |
| GO:BP | GO:0048599 | oocyte development | 4 | 44 | 1.137e-02 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 5 | 74 | 1.161e-02 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 3 | 21 | 1.179e-02 |
| GO:BP | GO:0009994 | oocyte differentiation | 4 | 45 | 1.224e-02 |
| GO:BP | GO:0006950 | response to stress | 49 | 2807 | 1.251e-02 |
| GO:BP | GO:0042148 | DNA strand invasion | 2 | 6 | 1.273e-02 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 2 | 6 | 1.273e-02 |
| GO:BP | GO:0071139 | resolution of DNA recombination intermediates | 2 | 6 | 1.273e-02 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 4 | 47 | 1.417e-02 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 7 | 154 | 1.470e-02 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 3 | 23 | 1.504e-02 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 6 | 115 | 1.504e-02 |
| GO:BP | GO:0051295 | establishment of meiotic spindle localization | 2 | 7 | 1.741e-02 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 65 | 4082 | 1.840e-02 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 4 | 52 | 2.018e-02 |
| GO:BP | GO:0019985 | translesion synthesis | 3 | 26 | 2.115e-02 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 3 | 26 | 2.115e-02 |
| GO:BP | GO:0006282 | regulation of DNA repair | 8 | 212 | 2.256e-02 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 2 | 8 | 2.256e-02 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 2 | 8 | 2.256e-02 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 3 | 27 | 2.318e-02 |
| GO:BP | GO:0010165 | response to X-ray | 3 | 27 | 2.318e-02 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 3 | 27 | 2.318e-02 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 24 | 1148 | 2.555e-02 |
| GO:BP | GO:0048144 | fibroblast proliferation | 5 | 91 | 2.663e-02 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 2 | 9 | 2.779e-02 |
| GO:BP | GO:0032873 | negative regulation of stress-activated MAPK cascade | 2 | 9 | 2.779e-02 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 2 | 9 | 2.779e-02 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 2 | 9 | 2.779e-02 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 2 | 9 | 2.779e-02 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 2 | 9 | 2.779e-02 |
| GO:BP | GO:0071314 | cellular response to cocaine | 2 | 9 | 2.779e-02 |
| GO:BP | GO:0065009 | regulation of molecular function | 24 | 1174 | 3.286e-02 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 3 | 31 | 3.305e-02 |
| GO:BP | GO:0051642 | centrosome localization | 3 | 31 | 3.305e-02 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 3 | 31 | 3.305e-02 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 6 | 137 | 3.305e-02 |
| GO:BP | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 2 | 10 | 3.350e-02 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 2 | 10 | 3.350e-02 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 2 | 10 | 3.350e-02 |
| GO:BP | GO:0070303 | negative regulation of stress-activated protein kinase signaling cascade | 2 | 10 | 3.350e-02 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 2 | 10 | 3.350e-02 |
| GO:BP | GO:0043933 | protein-containing complex organization | 29 | 1518 | 3.432e-02 |
| GO:BP | GO:0001824 | blastocyst development | 5 | 98 | 3.449e-02 |
| GO:BP | GO:0007018 | microtubule-based movement | 10 | 337 | 3.814e-02 |
| GO:BP | GO:0072710 | response to hydroxyurea | 2 | 11 | 4.019e-02 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 3 | 34 | 4.169e-02 |
| GO:BP | GO:0009411 | response to UV | 6 | 146 | 4.338e-02 |
| GO:BP | GO:0051261 | protein depolymerization | 5 | 104 | 4.355e-02 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 3 | 35 | 4.472e-02 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 3 | 35 | 4.472e-02 |
| GO:BP | GO:0016446 | somatic hypermutation of immunoglobulin genes | 2 | 12 | 4.682e-02 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 2 | 12 | 4.682e-02 |
| GO:BP | GO:0030214 | hyaluronan catabolic process | 2 | 12 | 4.682e-02 |
| GO:BP | GO:0022616 | DNA strand elongation | 3 | 36 | 4.785e-02 |
| GO:BP | GO:0051641 | cellular localization | 47 | 2869 | 4.810e-02 |
| GO:BP | GO:0000723 | telomere maintenance | 6 | 150 | 4.810e-02 |
| GO:BP | GO:0051298 | centrosome duplication | 4 | 69 | 4.888e-02 |
| KEGG | KEGG:04110 | Cell cycle | 28 | 152 | 6.550e-24 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 8 | 49 | 3.891e-06 |
| KEGG | KEGG:03440 | Homologous recombination | 6 | 39 | 1.227e-04 |
| KEGG | KEGG:04114 | Oocyte meiosis | 9 | 110 | 1.227e-04 |
| KEGG | KEGG:03030 | DNA replication | 5 | 35 | 8.619e-04 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 7 | 88 | 1.093e-03 |
| KEGG | KEGG:04814 | Motor proteins | 8 | 150 | 4.736e-03 |
| KEGG | KEGG:04218 | Cellular senescence | 7 | 143 | 1.572e-02 |
| KEGG | KEGG:03430 | Mismatch repair | 3 | 22 | 1.921e-02 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 7 | 177 | 4.136e-02 |
#GO:BP
table_proChronic_genes_GOBP_RUV <- table_proChronic_GOKEGG_genes_RUV %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
proChronic_table_genes_GOBP_plot <- ggplot(table_proChronic_genes_GOBP_RUV, aes(x = log_val, y = reorder(term_name, p_value))) +
geom_point() +
ggtitle("Progressively Chronic Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"adj. p-value"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_custom() +
theme(legend.position = "none")
print(proChronic_table_genes_GOBP_plot)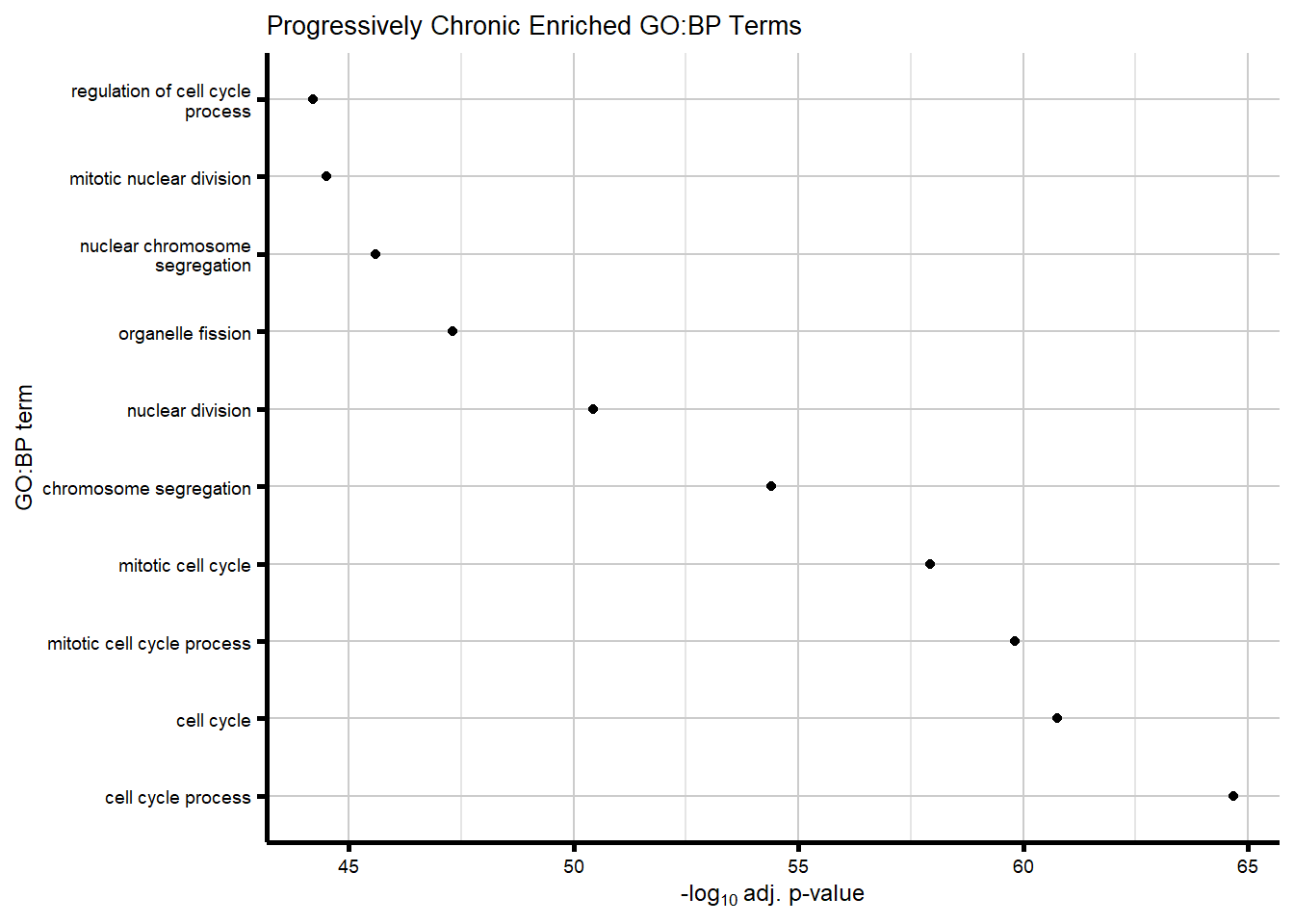
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
# save_plot(
# plot = proChronic_table_genes_GOBP_plot,
# filename = "Chronic_proChronic_GOBP_logval_all_EMP",
# folder = output_folder
# )
#KEGG
table_proChronic_genes_KEGG_RUV <- table_proChronic_GOKEGG_genes_RUV %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
proChronic_table_genes_KEGG_plot <- ggplot(table_proChronic_genes_KEGG_RUV, aes(x = log_val, y = reorder(term_name, p_value))) +
geom_point(aes(size = intersection_size)) +
ggtitle("Enriched Pathways: ProChronic")+
xlab(expression("-log"[10]~"adj. p-value"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_custom() +
theme(legend.position = "none")
print(proChronic_table_genes_KEGG_plot)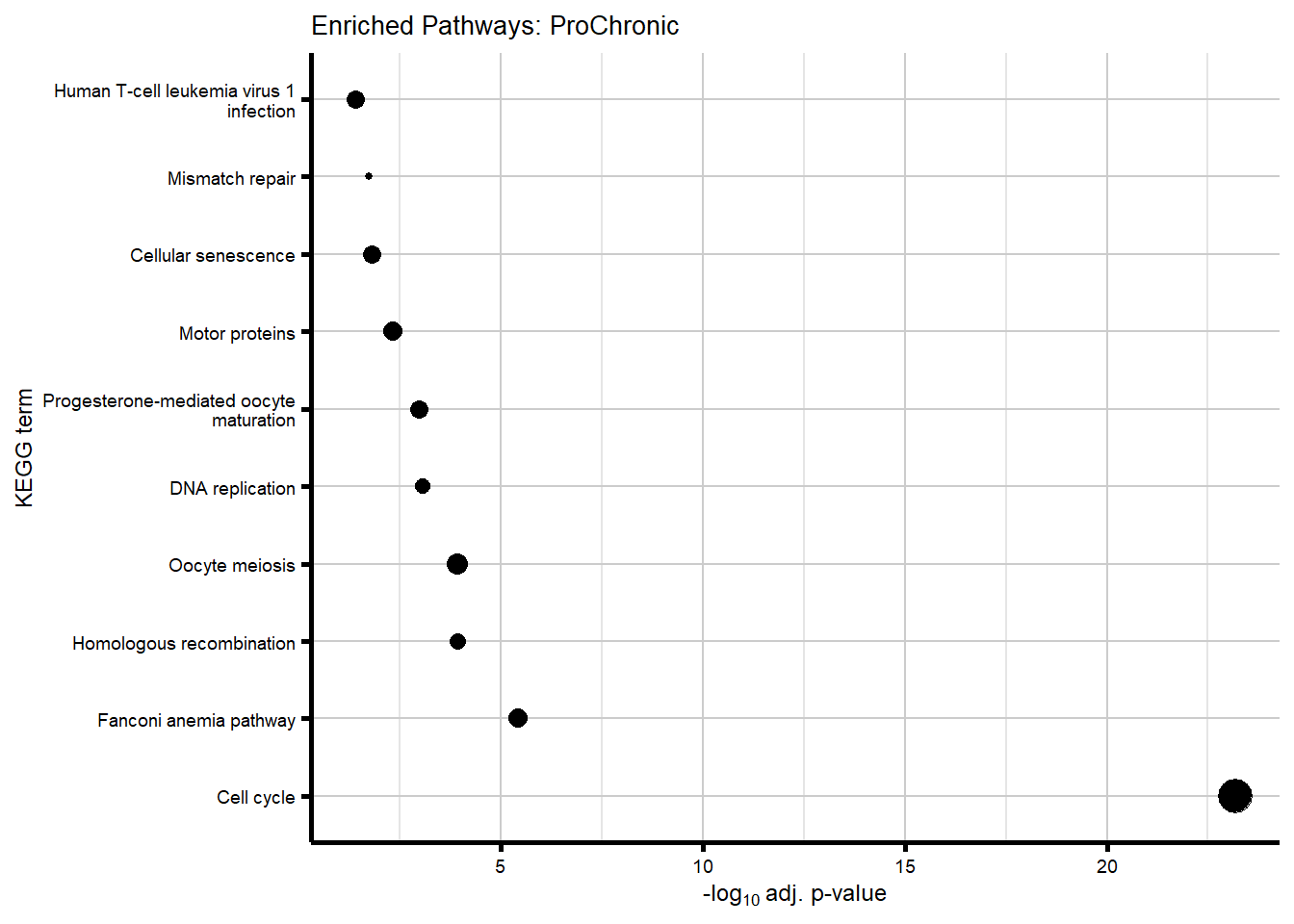
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
# save_plot(
# plot = proChronic_table_genes_KEGG_plot,
# filename = "Chronic_proChronic_KEGG_logval_all_EMP",
# folder = output_folder
# )Figure 6C - Non-progressive Chronic response genes line plot
filt_toptable_nonproChronic_RUV <- combined_toptables_dxr_RUV_df %>%
mutate(nonproChronic = case_when(
Entrez_ID %in% nonproChronic_gene_list ~ "non-ProChronic",
TRUE ~ NA_character_
)) %>%
dplyr::filter(!is.na(SYMBOL)) %>%
dplyr::filter(!is.na(nonproChronic)) %>%
mutate(Time = factor(Time,
levels = c("t0",
"t24",
"t144"))) %>%
mutate(absFC = abs(logFC))
#calculate medians for ALL nonproChronic genes across timepoints
medians_nonproChronic_RUV <- filt_toptable_nonproChronic_RUV %>%
group_by(Time) %>%
summarise(
median_logFC = median(logFC),
median_abslogFC = median(absFC),
.groups = "drop"
)
#label with median values
labs_logFC_nonproChronic <- paste0(
"non-ProChronic\nmedian logFC =\n",
paste0(medians_nonproChronic_RUV$Time, "=",
round(medians_nonproChronic_RUV$median_logFC, 2),
collapse = "\n")
)
labs_absFC_nonproChronic <- paste0(
"non-ProChronic\nmedian absFC =\n",
paste0(medians_nonproChronic_RUV$Time, "=",
round(medians_nonproChronic_RUV$median_abslogFC, 2),
collapse = "\n")
)
#add a column that labels positive/negative direction
filt_toptable_nonproChronic_RUV <- filt_toptable_nonproChronic_RUV %>%
mutate(direction = ifelse(logFC >= 0, "up", "down"))
#label direction
medians_nonproChronic_RUV <- medians_nonproChronic_RUV %>%
mutate(direction = "median")
#logFC version
plot_nonproChronic_logFC <- ggplot() +
geom_line(data = filt_toptable_nonproChronic_RUV,
aes(x = Time,
y = logFC,
group = Entrez_ID),
color = "#E1B9E3",
alpha = 0.4,
linewidth = 0.9) +
geom_line(data = medians_nonproChronic_RUV,
aes(x = Time,
y = median_logFC,
group = 1,
color = direction),
linewidth = 3
) +
scale_color_manual(
breaks = c("median"),
values = c("median" = "#A06FA8"),
labels = c(
median = labs_logFC_nonproChronic),
name = NULL
) +
labs(title = "Non-progressive Chronic response genes",
y = "logFC", x = "Timepoint") +
scale_y_continuous(breaks = c(-10, -5, 0, 5, 10),
limits = c(-10, 10),
expand = c(0,0)) +
scale_x_discrete(breaks = c("t0", "t24", "t144"),
expand = c(0, 0)) +
theme_custom() +
theme(legend.position = "right")
#absFC version
plot_nonproChronic_absFC <- ggplot() +
geom_line(data = filt_toptable_nonproChronic_RUV,
aes(x = Time,
y = absFC,
group = Entrez_ID),
color = "orchid",
alpha = 0.4,
linewidth = 0.9) +
geom_line(data = medians_nonproChronic_RUV,
aes(x = Time,
y = median_abslogFC,
group = 1,
color = direction),
linewidth = 3
) +
scale_color_manual(
breaks = c("median"),
values = c("median" = "#3D6680"),
labels = c(
median = labs_absFC_nonproChronic),
name = NULL
) +
labs(title = "abslogFC — non-ProChronic Genes (n = 338)",
y = "absFC", x = "Timepoint") +
scale_y_continuous(limits = c(0, 10),
expand = c(0, 0)) +
scale_x_discrete(breaks = c("t0", "t24", "t144"),
expand = c(0, 0)) +
theme_custom() +
theme(legend.position = "right")
print(plot_nonproChronic_logFC)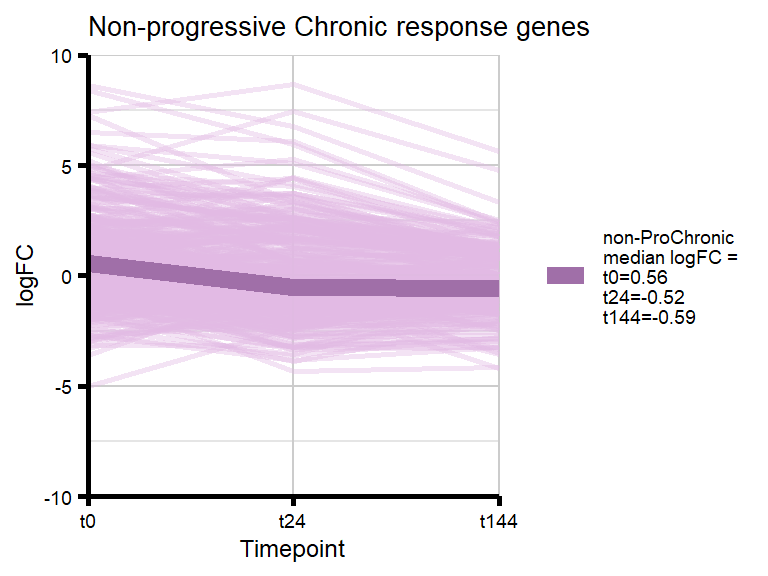
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
print(plot_nonproChronic_absFC)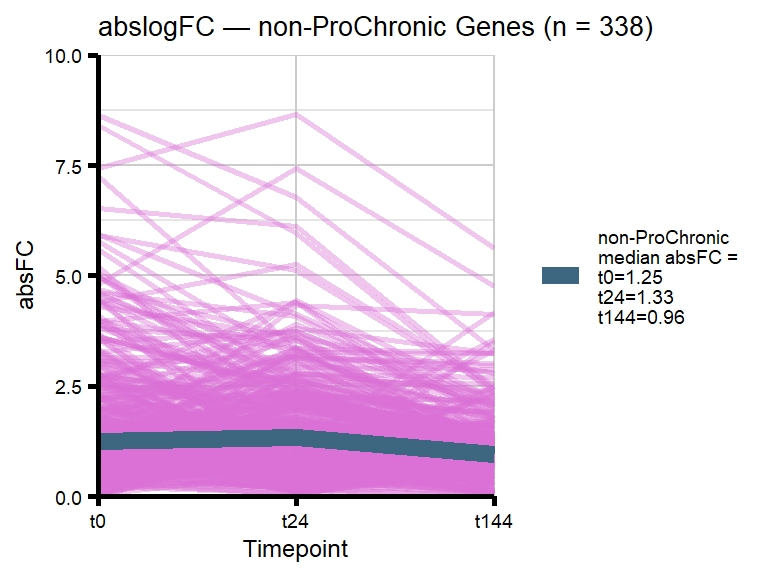
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
# save_plot(
# plot = plot_nonproChronic_logFC,
# filename = "Lineplot_nonproChronic_logFC_EMP_251112",
# folder = output_folder,
# height = 4,
# width = 5
# )
# save_plot(
# plot = plot_nonproChronic_absFC,
# filename = "Lineplot_nonproChronic_absFC_EMP_251112",
# folder = output_folder,
# height = 4,
# width = 5
# )Figure 6D - Enriched Pathways: non-ProChronic
#read in my non-ProChronic gene list
# nonproChronic_gene_list
nonproChronic_GOKEGG_RUV <- gost(query = nonproChronic_gene_list,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = TRUE,
user_threshold = 0.05,
significant = TRUE,
custom_bg = all_genes,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
nonproChronic_GOKEGG_genes_RUV <- gostplot(nonproChronic_GOKEGG_RUV,
capped = FALSE, interactive = TRUE)
nonproChronic_GOKEGG_genes_RUVtable_nonproChronic_GOKEGG_genes_RUV <- nonproChronic_GOKEGG_RUV$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_nonproChronic_GOKEGG_genes_RUV %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0022402 | cell cycle process | 75 | 1094 | 1.548e-13 |
| GO:BP | GO:0007049 | cell cycle | 80 | 1392 | 1.216e-10 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 52 | 687 | 1.384e-10 |
| GO:BP | GO:0051301 | cell division | 47 | 575 | 1.384e-10 |
| GO:BP | GO:0000278 | mitotic cell cycle | 56 | 809 | 4.268e-10 |
| GO:BP | GO:0000280 | nuclear division | 35 | 358 | 1.016e-09 |
| GO:BP | GO:0048285 | organelle fission | 36 | 403 | 5.817e-09 |
| GO:BP | GO:0006260 | DNA replication | 28 | 259 | 1.375e-08 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 21 | 151 | 3.706e-08 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 44 | 626 | 5.347e-08 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 28 | 278 | 5.347e-08 |
| GO:BP | GO:0007059 | chromosome segregation | 33 | 380 | 5.525e-08 |
| GO:BP | GO:0006270 | DNA replication initiation | 11 | 35 | 1.027e-07 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 33 | 397 | 1.480e-07 |
| GO:BP | GO:0051726 | regulation of cell cycle | 54 | 928 | 2.801e-07 |
| GO:BP | GO:0044770 | cell cycle phase transition | 36 | 483 | 3.921e-07 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 19 | 144 | 3.921e-07 |
| GO:BP | GO:0051321 | meiotic cell cycle | 22 | 195 | 3.921e-07 |
| GO:BP | GO:0140013 | meiotic nuclear division | 17 | 128 | 2.340e-06 |
| GO:BP | GO:0051276 | chromosome organization | 34 | 485 | 4.408e-06 |
| GO:BP | GO:0140014 | mitotic nuclear division | 23 | 258 | 1.297e-05 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 19 | 185 | 1.936e-05 |
| GO:BP | GO:0006259 | DNA metabolic process | 46 | 874 | 7.198e-05 |
| GO:BP | GO:0000819 | sister chromatid segregation | 20 | 223 | 7.244e-05 |
| GO:BP | GO:0007052 | mitotic spindle organization | 15 | 128 | 7.244e-05 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 6 | 14 | 7.379e-05 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 25 | 335 | 8.287e-05 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 27 | 386 | 1.000e-04 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 16 | 158 | 1.827e-04 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 8 | 36 | 2.342e-04 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 21 | 266 | 2.439e-04 |
| GO:BP | GO:0022414 | reproductive process | 48 | 988 | 2.802e-04 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 28 | 442 | 3.710e-04 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 5 | 11 | 3.720e-04 |
| GO:BP | GO:0050896 | response to stimulus | 181 | 5825 | 3.720e-04 |
| GO:BP | GO:0007051 | spindle organization | 17 | 193 | 4.621e-04 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 22 | 303 | 4.621e-04 |
| GO:BP | GO:0034508 | centromere complex assembly | 7 | 29 | 4.622e-04 |
| GO:BP | GO:0006281 | DNA repair | 32 | 568 | 7.246e-04 |
| GO:BP | GO:0032501 | multicellular organismal process | 152 | 4731 | 7.442e-04 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 16 | 181 | 7.645e-04 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 10 | 74 | 1.003e-03 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 18 | 230 | 1.088e-03 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 9 | 60 | 1.119e-03 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 9 | 62 | 1.424e-03 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 13 | 131 | 1.424e-03 |
| GO:BP | GO:0006974 | DNA damage response | 40 | 826 | 1.580e-03 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 17 | 216 | 1.586e-03 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 5 | 15 | 1.595e-03 |
| GO:BP | GO:0051716 | cellular response to stimulus | 156 | 4968 | 1.629e-03 |
| GO:BP | GO:0051304 | chromosome separation | 10 | 80 | 1.634e-03 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 13 | 134 | 1.634e-03 |
| GO:BP | GO:0048856 | anatomical structure development | 137 | 4234 | 1.696e-03 |
| GO:BP | GO:0032502 | developmental process | 146 | 4584 | 1.696e-03 |
| GO:BP | GO:0007275 | multicellular organism development | 117 | 3478 | 1.748e-03 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 14 | 160 | 2.296e-03 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 10 | 84 | 2.296e-03 |
| GO:BP | GO:0033260 | nuclear DNA replication | 7 | 39 | 2.427e-03 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 16 | 208 | 2.978e-03 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 19 | 278 | 2.978e-03 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 12 | 125 | 3.159e-03 |
| GO:BP | GO:0019953 | sexual reproduction | 34 | 684 | 3.166e-03 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 13 | 146 | 3.289e-03 |
| GO:BP | GO:0006275 | regulation of DNA replication | 11 | 107 | 3.313e-03 |
| GO:BP | GO:0006468 | protein phosphorylation | 43 | 963 | 3.734e-03 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 4 | 10 | 3.734e-03 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 4 | 10 | 3.734e-03 |
| GO:BP | GO:0048731 | system development | 102 | 2993 | 3.787e-03 |
| GO:BP | GO:0007098 | centrosome cycle | 12 | 129 | 3.807e-03 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 7 | 43 | 3.861e-03 |
| GO:BP | GO:0006939 | smooth muscle contraction | 9 | 74 | 3.861e-03 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 8 | 58 | 3.903e-03 |
| GO:BP | GO:0048513 | animal organ development | 79 | 2176 | 4.197e-03 |
| GO:BP | GO:0007127 | meiosis I | 9 | 79 | 6.047e-03 |
| GO:BP | GO:0048869 | cellular developmental process | 103 | 3072 | 6.047e-03 |
| GO:BP | GO:0030154 | cell differentiation | 103 | 3071 | 6.047e-03 |
| GO:BP | GO:0051383 | kinetochore organization | 5 | 21 | 6.107e-03 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 10 | 98 | 6.326e-03 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 12 | 138 | 6.382e-03 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 7 | 48 | 6.511e-03 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 7 | 48 | 6.511e-03 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 7 | 48 | 6.511e-03 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 3 | 5 | 6.511e-03 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 3 | 5 | 6.511e-03 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 3 | 5 | 6.511e-03 |
| GO:BP | GO:0090402 | oncogene-induced cell senescence | 3 | 5 | 6.511e-03 |
| GO:BP | GO:0030261 | chromosome condensation | 6 | 34 | 6.579e-03 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 5 | 22 | 6.780e-03 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 7 | 49 | 7.190e-03 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 7 | 50 | 7.838e-03 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 7 | 50 | 7.838e-03 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 7 | 50 | 7.838e-03 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 7 | 50 | 7.838e-03 |
| GO:BP | GO:0016310 | phosphorylation | 44 | 1044 | 7.904e-03 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 7 | 51 | 8.534e-03 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 7 | 51 | 8.534e-03 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 7 | 51 | 8.534e-03 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 10 | 105 | 8.943e-03 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 7 | 52 | 9.465e-03 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 13 | 170 | 9.693e-03 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 3 | 6 | 1.042e-02 |
| GO:BP | GO:0014831 | gastro-intestinal system smooth muscle contraction | 3 | 6 | 1.042e-02 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 6 | 38 | 1.052e-02 |
| GO:BP | GO:0008283 | cell population proliferation | 54 | 1397 | 1.152e-02 |
| GO:BP | GO:0050918 | positive chemotaxis | 6 | 39 | 1.195e-02 |
| GO:BP | GO:0000725 | recombinational repair | 13 | 175 | 1.214e-02 |
| GO:BP | GO:0051299 | centrosome separation | 4 | 15 | 1.382e-02 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 7 | 56 | 1.386e-02 |
| GO:BP | GO:0050000 | chromosome localization | 10 | 113 | 1.461e-02 |
| GO:BP | GO:0007154 | cell communication | 132 | 4292 | 1.587e-02 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 9 | 95 | 1.689e-02 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 9 | 96 | 1.809e-02 |
| GO:BP | GO:0006310 | DNA recombination | 17 | 283 | 1.819e-02 |
| GO:BP | GO:2000241 | regulation of reproductive process | 10 | 118 | 1.974e-02 |
| GO:BP | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway | 2 | 2 | 1.999e-02 |
| GO:BP | GO:0014846 | esophagus smooth muscle contraction | 2 | 2 | 1.999e-02 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 5 | 29 | 1.999e-02 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 9 | 98 | 1.999e-02 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 8 | 79 | 2.068e-02 |
| GO:BP | GO:1902969 | mitotic DNA replication | 4 | 17 | 2.068e-02 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 7 | 61 | 2.100e-02 |
| GO:BP | GO:0051783 | regulation of nuclear division | 10 | 121 | 2.248e-02 |
| GO:BP | GO:0006302 | double-strand break repair | 17 | 291 | 2.275e-02 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 13 | 190 | 2.275e-02 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 3 | 8 | 2.278e-02 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 3 | 8 | 2.278e-02 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 7 | 63 | 2.435e-02 |
| GO:BP | GO:0048608 | reproductive structure development | 14 | 217 | 2.484e-02 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 29 | 640 | 2.528e-02 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 13 | 194 | 2.567e-02 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 7 | 64 | 2.567e-02 |
| GO:BP | GO:0071695 | anatomical structure maturation | 12 | 170 | 2.567e-02 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 9 | 103 | 2.567e-02 |
| GO:BP | GO:0023052 | signaling | 130 | 4280 | 2.567e-02 |
| GO:BP | GO:0006813 | potassium ion transport | 11 | 147 | 2.567e-02 |
| GO:BP | GO:0061458 | reproductive system development | 14 | 220 | 2.672e-02 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 4 | 19 | 2.840e-02 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 9 | 106 | 3.017e-02 |
| GO:BP | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 3 | 9 | 3.042e-02 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 12 | 175 | 3.146e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 10 | 129 | 3.171e-02 |
| GO:BP | GO:0071772 | response to BMP | 10 | 129 | 3.171e-02 |
| GO:BP | GO:0070977 | bone maturation | 4 | 20 | 3.337e-02 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 9 | 109 | 3.526e-02 |
| GO:BP | GO:0048799 | animal organ maturation | 4 | 21 | 3.989e-02 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 14 | 232 | 4.139e-02 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 8 | 91 | 4.273e-02 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 10 | 135 | 4.295e-02 |
| GO:BP | GO:0022616 | DNA strand elongation | 5 | 36 | 4.352e-02 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 17 | 315 | 4.423e-02 |
| GO:BP | GO:0007165 | signal transduction | 119 | 3916 | 4.423e-02 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 12 | 184 | 4.423e-02 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 8 | 92 | 4.423e-02 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 2 | 3 | 4.497e-02 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 5 | 37 | 4.743e-02 |
| KEGG | KEGG:03030 | DNA replication | 11 | 35 | 7.645e-08 |
| KEGG | KEGG:04110 | Cell cycle | 18 | 152 | 2.939e-06 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 11 | 103 | 2.837e-03 |
| KEGG | KEGG:04115 | p53 signaling pathway | 8 | 67 | 9.077e-03 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 17 | 260 | 9.077e-03 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 8 | 88 | 4.822e-02 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 7 | 71 | 4.977e-02 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 9 | 114 | 4.977e-02 |
#GO:BP
table_nonproChronic_genes_GOBP_RUV <- table_nonproChronic_GOKEGG_genes_RUV %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
nonproChronic_table_genes_GOBP_plot <- ggplot(table_nonproChronic_genes_GOBP_RUV, aes(x = log_val, y = reorder(term_name, p_value))) +
geom_point() +
ggtitle("non-ProChronic Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"adj. p-value"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_custom() +
theme(legend.position = "none")
print(nonproChronic_table_genes_GOBP_plot)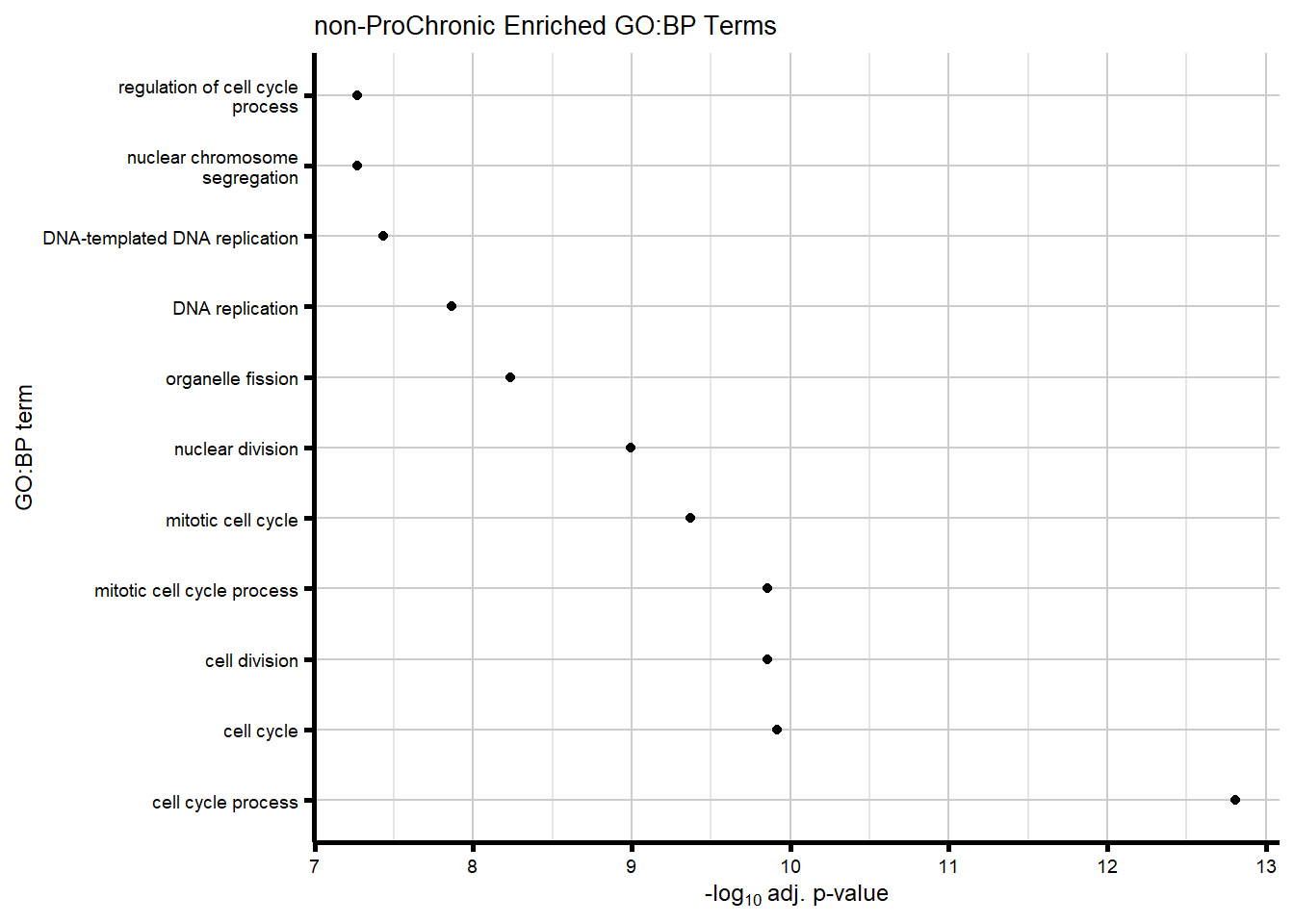
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
# save_plot(
# plot = nonproChronic_table_genes_GOBP_plot,
# filename = "Chronic_nonproChronic_GOBP_logval_all_EMP",
# folder = output_folder
# )
#KEGG
table_nonproChronic_genes_KEGG_RUV <- table_nonproChronic_GOKEGG_genes_RUV %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
nonproChronic_table_genes_KEGG_plot <- ggplot(table_nonproChronic_genes_KEGG_RUV,
aes(x = log_val, y = reorder(term_name, p_value))) +
geom_point() +
ggtitle("Enriched Pathways: non-ProChronic")+
xlab(expression("-log"[10]~"adj. p-value"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_custom() +
theme(legend.position = "none")
print(nonproChronic_table_genes_KEGG_plot)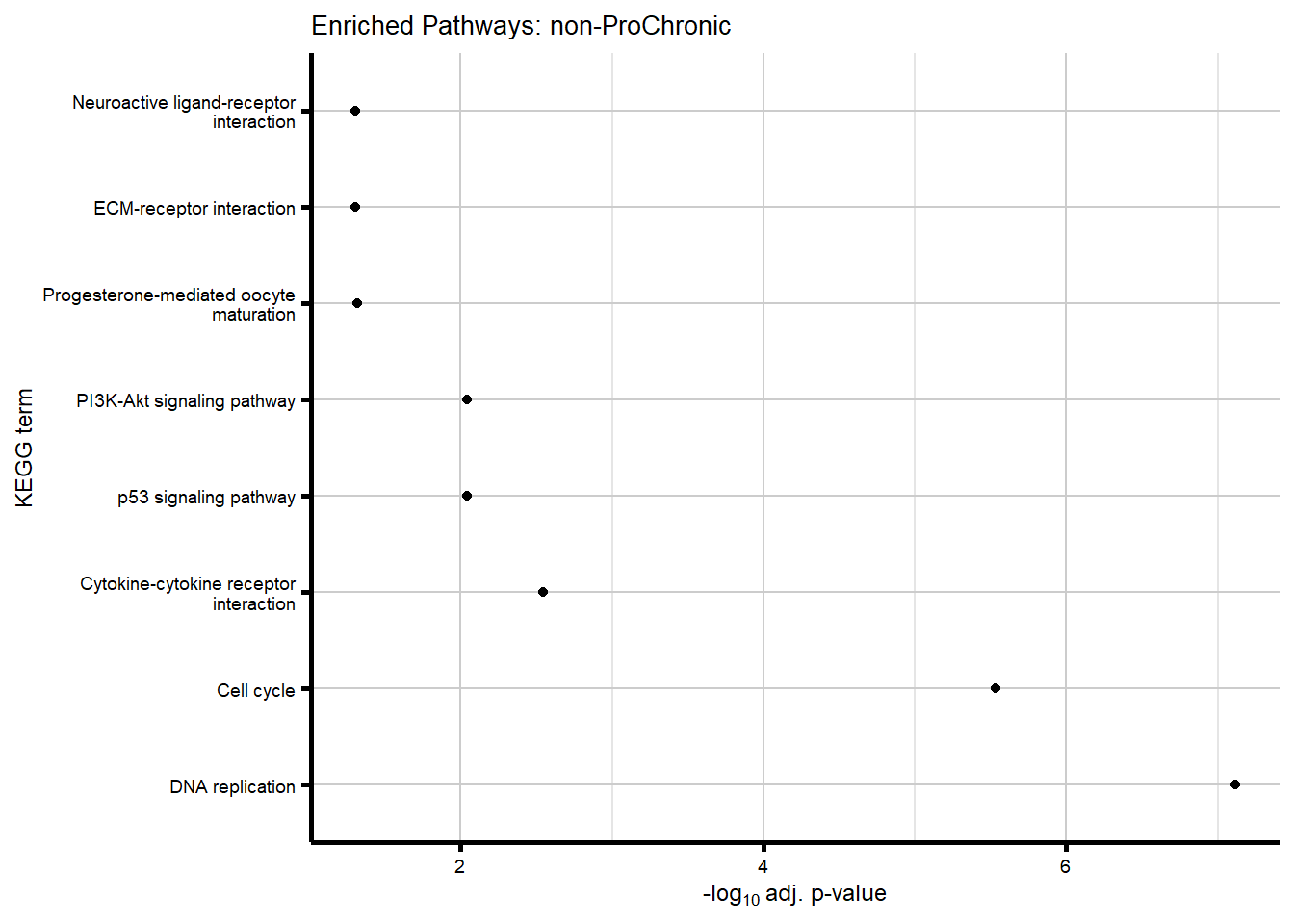
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
# save_plot(
# plot = nonproChronic_table_genes_KEGG_plot,
# filename = "Chronic_nonproChronic_KEGG_logval_all_EMP",
# folder = output_folder
# )Figure 6E - Proportion of ProChronic vs non-ProChronic
proChronic_fin_summary <- proChronic_df %>%
group_by(Cluster, Response) %>%
summarise(Count = n(), .groups = "drop") %>%
mutate(Proportion = Count / sum(Count),
Percent = round(Proportion * 100, 1))
category_colors <- c(
"ProChronic" = "#A06FA8",
"non-ProChronic" = "#E1B9E3"
)
plot_proChronic_fin_summary <- ggplot(proChronic_fin_summary, aes(x = Cluster, y = Percent, fill = Response)) +
geom_bar(stat = "identity") +
geom_text(aes(label = paste0(round(Percent, 1), "\n(n = ", Count, ")")),
position = position_stack(vjust = 0.5),
size = 4, color = "black") +
scale_y_continuous(limits = c(0, 102),
expand = c(0, 0)) +
scale_x_discrete(expand = c(0, 0)) +
scale_fill_manual(values = category_colors) +
theme_custom() +
labs(
y = "Percentage of Chronic Genes (%)",
x = "(n=501)",
fill = "Response Direction"
)
print(plot_proChronic_fin_summary)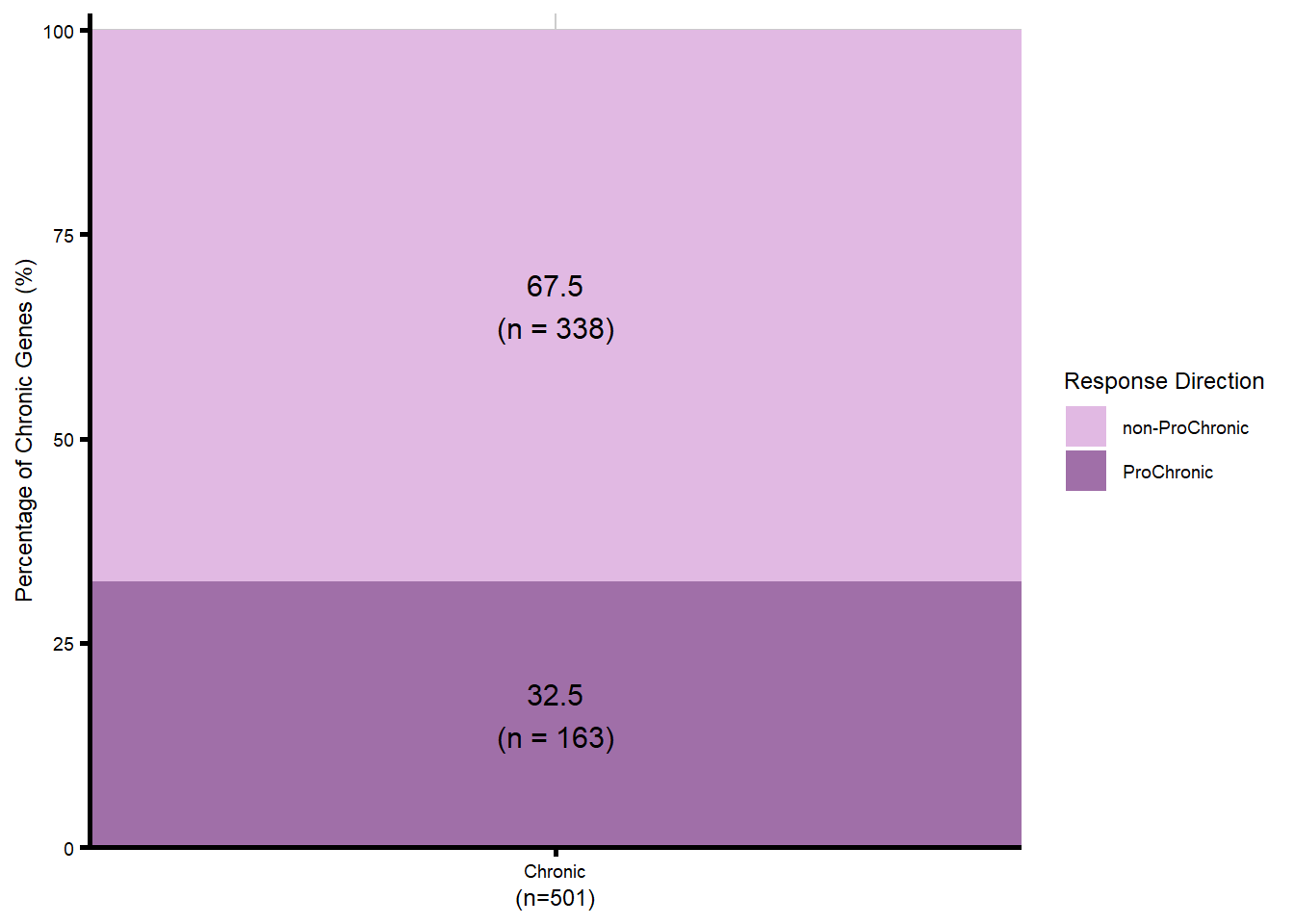
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
#163 genes total
# save_plot(
# plot = plot_proChronic_fin_summary,
# filename = "Sustained_Proportion_Barplot_proChronic_finalset_DEGs_EMP",
# folder = output_folder,
# height = 4,
# width = 5
# )Assess proChronic Genes Directionality
#I want to take my set of proSus genes and stratify by response direction
#pull the proSus genes from the df
proChronic_only_df <- proChronic_df %>%
filter(
Response == "ProChronic"
)
#now find direction
proChronic_only_df <- proChronic_only_df %>%
mutate(Direction = case_when(
!is.na(logFC_t0) & !is.na(logFC_t24) & !is.na(logFC_t144) &
logFC_t0 < logFC_t24 & logFC_t24 < logFC_t144 ~ "Upregulated",
!is.na(logFC_t0) & !is.na(logFC_t24) & !is.na(logFC_t144) &
logFC_t0 > logFC_t24 & logFC_t24 > logFC_t144 ~ "Downregulated",
TRUE ~ "Mixed"
))
table(proChronic_only_df$Direction)
Downregulated Upregulated
154 9 proChronic_up_genes <- proChronic_only_df %>%
dplyr::filter(Direction == "Upregulated") %>%
dplyr::pull(Entrez_ID, SYMBOL)
print(proChronic_up_genes) ASTN1 EPS8L2 CEMIP PDE11A CACNA2D3 P3H2 CPE PTCHD4
"460" "64787" "57214" "50940" "55799" "55214" "1363" "442213"
ACHE
"43" #9 upregulated genes by logFC
#2 of these are DEGs in all timepoints - ACHE & CPE
#CACNA2D3 is a gene of interest
proChronic_down_genes <- proChronic_only_df %>%
dplyr::filter(Direction == "Downregulated") %>%
dplyr::pull(Entrez_ID, SYMBOL)
length(proChronic_down_genes)[1] 154#154 genes downregulated by logFC
#pull out the log2cpm genes that are upregulatedFigure 6F - CACNA2D3 Expression
#there are 9 total genes to investigate that are upregulated in ProChronic
proChronic_up_genes ASTN1 EPS8L2 CEMIP PDE11A CACNA2D3 P3H2 CPE PTCHD4
"460" "64787" "57214" "50940" "55799" "55214" "1363" "442213"
ACHE
"43" dir_cats <- list(
"Upregulated" = proChronic_up_genes
)
proChronic_up_cats <- list(
"Upregulated" = unique(na.omit(proChronic_up_genes))
)
#pull the gene symbol so I can plot log2cpm data
proChronic_up_genes_df <- map_dfr(names(proChronic_up_cats), function(cat) {
tibble(
Entrez_ID = intersect(proChronic_up_genes, proChronic_up_cats[[cat]]),
Category = cat
)
})
dim(proChronic_up_genes_df)[1] 9 2#filter overlaps for non-ProChronic
proChronic_upreg <- proChronic_up_genes_df %>%
filter(Entrez_ID %in% proChronic_gene_list)
#9 genes
#read in my dataframe for plotting log2cpm genes
deg_wide <- readRDS("data/DE/DEGs_overlap_wide_dataframe.RDS")
proChronic_up_degs <- deg_wide %>%
dplyr::filter(Entrez_ID %in% proChronic_upreg$Entrez_ID)
proChronic_upreg <- proChronic_upreg %>%
mutate(
SYMBOL = mapIds(
org.Hs.eg.db,
keys = Entrez_ID,
column = "SYMBOL",
keytype = "ENTREZID",
multiVals = "first"
)
)
#now plot these example genes for each
boxplot_new <- readRDS("data/DE/boxplot_updated.RDS")
#add in colors and factors
txtime_col <- list(
"DOX_t0" = "#263CA5",
"VEH_t0" = "#444343",
"DOX_t24" = "#5B8FD1",
"VEH_t24" = "#757575",
"DOX_t144" = "#89C5E5",
"VEH_t144" = "#AAAAAA"
)
ind_col <- list(
"1" = "#FF9F64",
"2" = "#78EBDA",
"3" = "#ADCD77",
"4" = "#E6CC50",
"5" = "#A68AC0",
"6" = "#F16F90",
"6R" = "#C61E4E"
)
time_col <- list(
"t0" = "#005A4C",
"t24" = "#328477",
"t144" = "#8FB9B1")
tx_col <- list(
"DOX" = "#499FBD",
"VEH" = "#BBBBBC")
#process the cpm data
process_cpm_data_cormotif <- function(gene_id, expr_df) {
gene_data <- expr_df %>% filter(Entrez_ID == gene_id)
long_data <- gene_data %>%
pivot_longer(
cols = -c(Entrez_ID, SYMBOL),
names_to = "Sample",
values_to = "log2CPM"
) %>%
mutate(
Treatment = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("VEH", Sample) ~ "VEH",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("t0", Sample) ~ "t0",
grepl("t24", Sample) ~ "t24",
grepl("t144", Sample) ~ "t144",
TRUE ~ NA_character_
),
Individual = case_when(
grepl("1$", Sample) ~ "1",
grepl("2$", Sample) ~ "2",
grepl("3$", Sample) ~ "3",
grepl("4$", Sample) ~ "4",
grepl("5$", Sample) ~ "5",
grepl("6$", Sample) ~ "6",
grepl("6R$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Treatment, Timepoint, sep = "_")
)
#set factor levels for plotting order
long_data$Condition <- factor(
long_data$Condition,
levels = c("DOX_t0", "VEH_t0",
"DOX_t24", "VEH_t24",
"DOX_t144", "VEH_t144")
)
return(long_data)
}
#function to generate plots for a set of genes
generate_overlap_plots_cormotif <- function(overlap_df, expr_df, comparison_label) {
plots <- list()
for (gene_id in overlap_df$Entrez_ID) {
gene_data <- process_cpm_data_cormotif(gene_id, expr_df)
# Get SYMBOL and all categories
gene_symbol <- unique(gene_data$SYMBOL)
gene_categories <- overlap_df$Category[overlap_df$Entrez_ID == gene_id]
gene_categories <- paste(unique(gene_categories), collapse = ", ")
p <- ggplot(gene_data, aes(x = Condition,
y = log2CPM,
fill = Condition)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Individual), size = 2,
alpha = 0.9,
position = position_identity()) +
scale_fill_manual(values = txtime_col) +
scale_color_manual(values = ind_col) +
ggtitle(paste0(gene_symbol,
" (", comparison_label, " - ", gene_categories, ") ")) +
labs(x = "Condition", y = "log2cpm") +
theme_custom()
#use Entrez_ID + SYMBOL + comparison label for the list name to guarantee uniqueness
plot_name <- paste0(gene_symbol, "_", gene_id, "_", comparison_label)
plots[[plot_name]] <- p
}
return(plots)
}
#generate plots for notproSus overlap genes
proChronic_up_plots <- generate_overlap_plots_cormotif(proChronic_upreg, boxplot_new, "ProChronic")
#preview all plots
for (p in c(proChronic_up_plots)) print(p)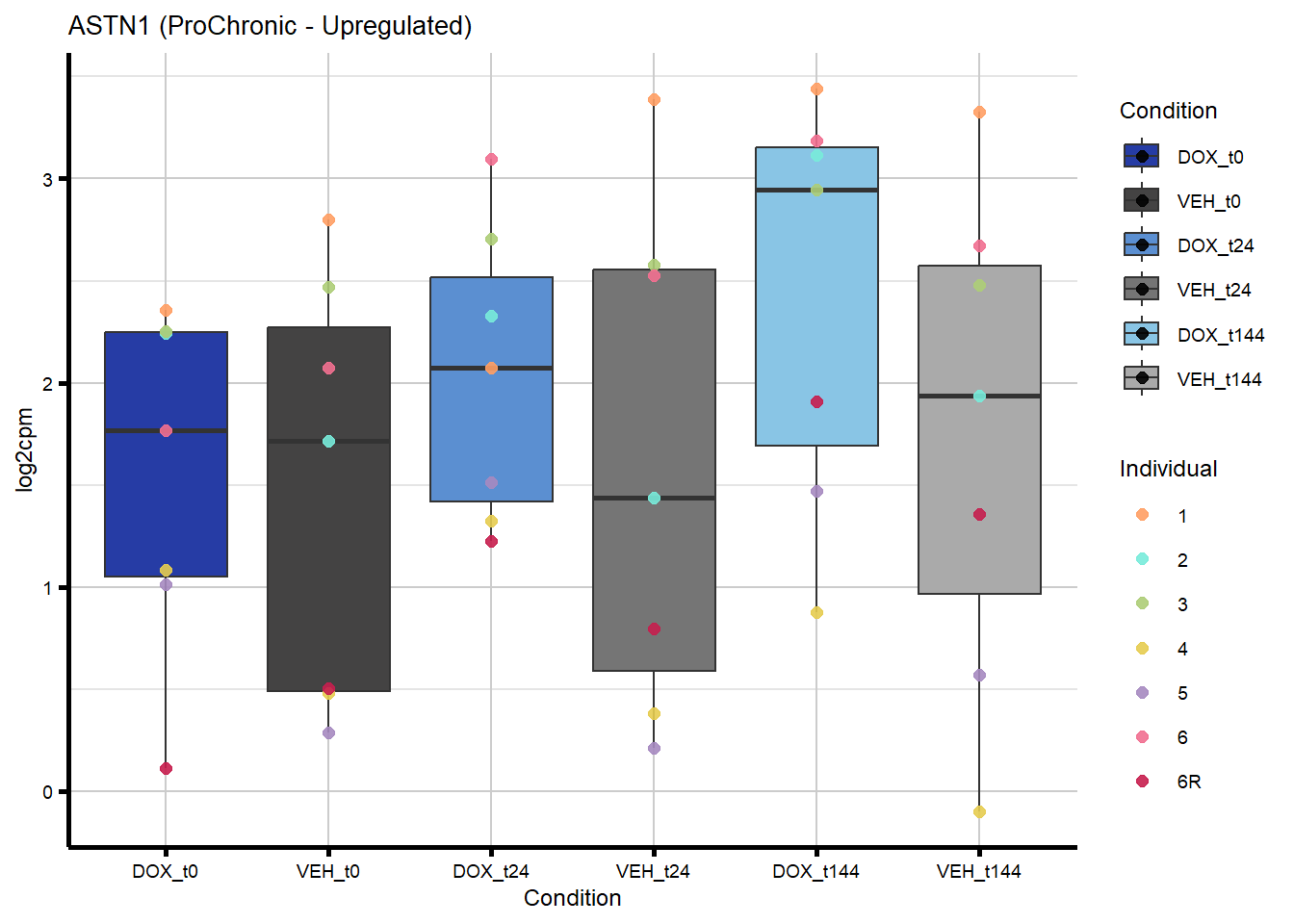
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
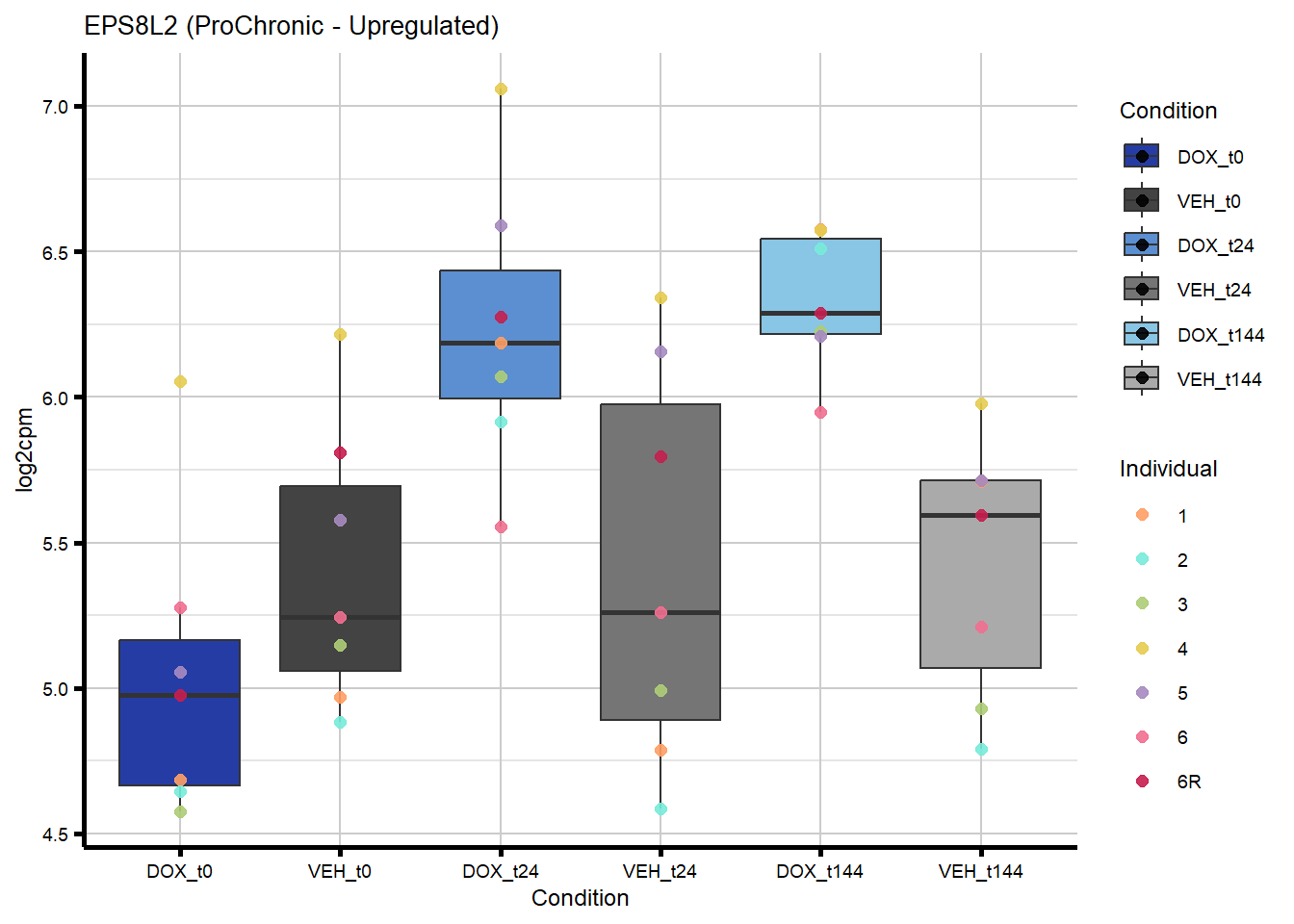
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
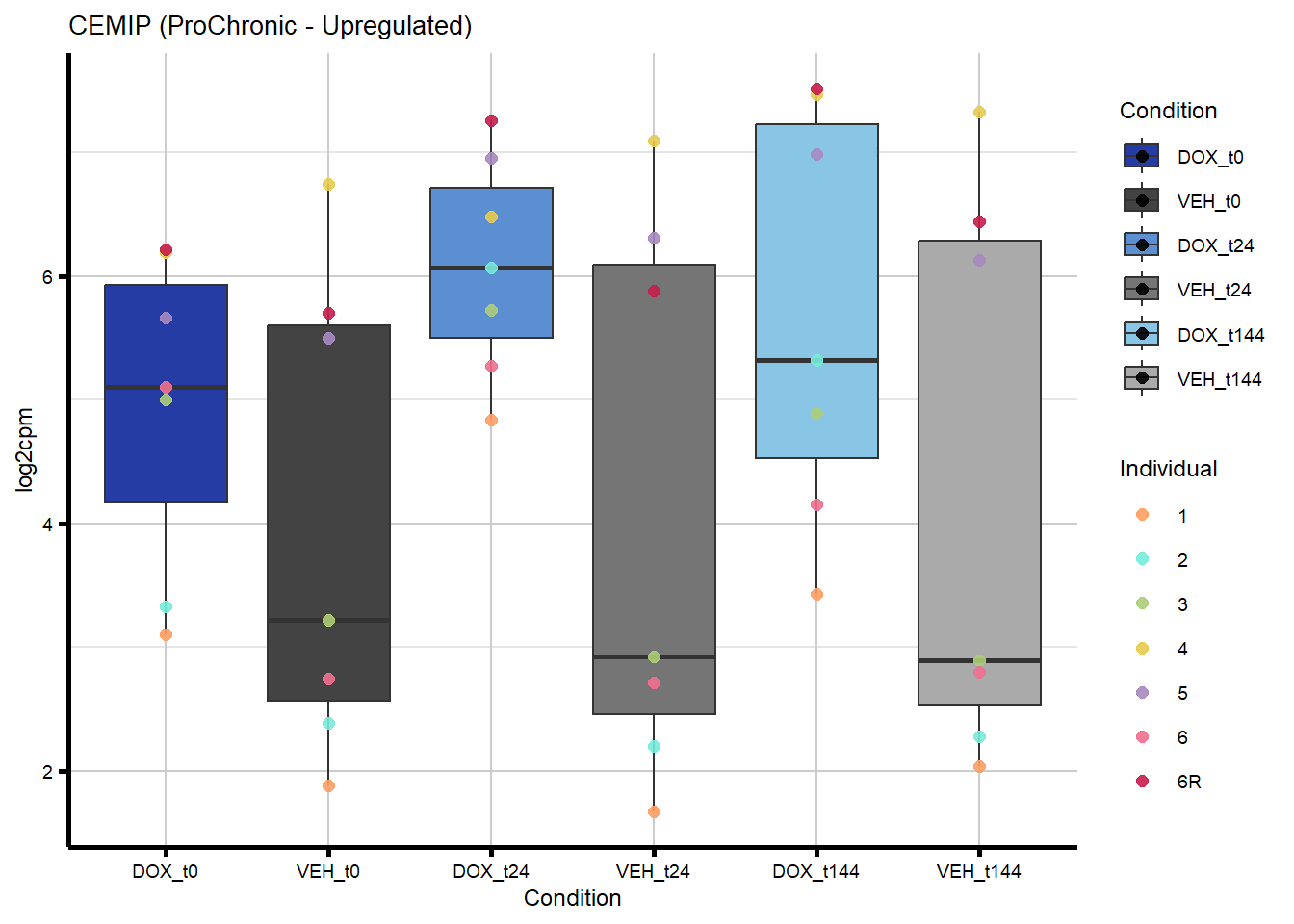
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
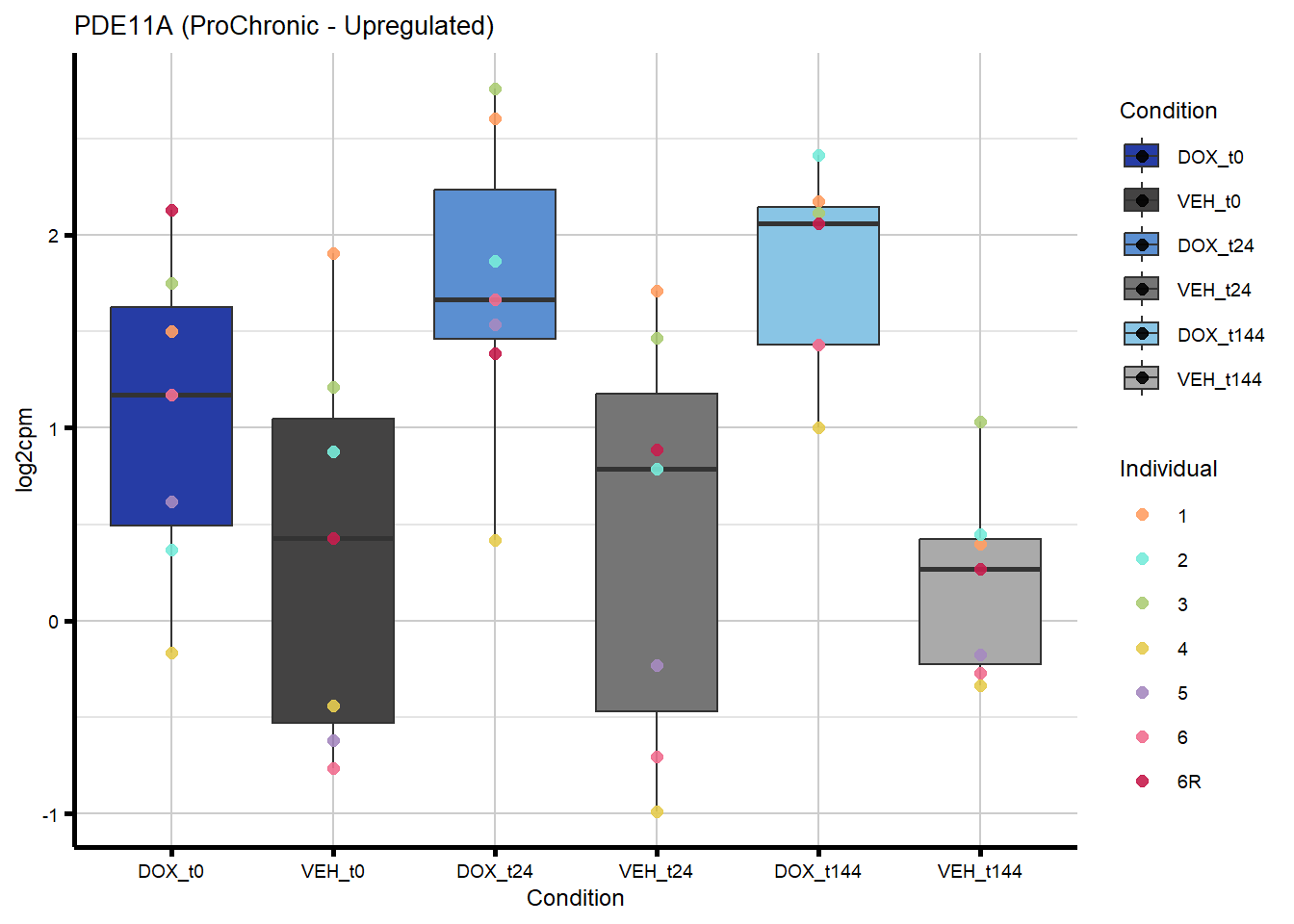
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
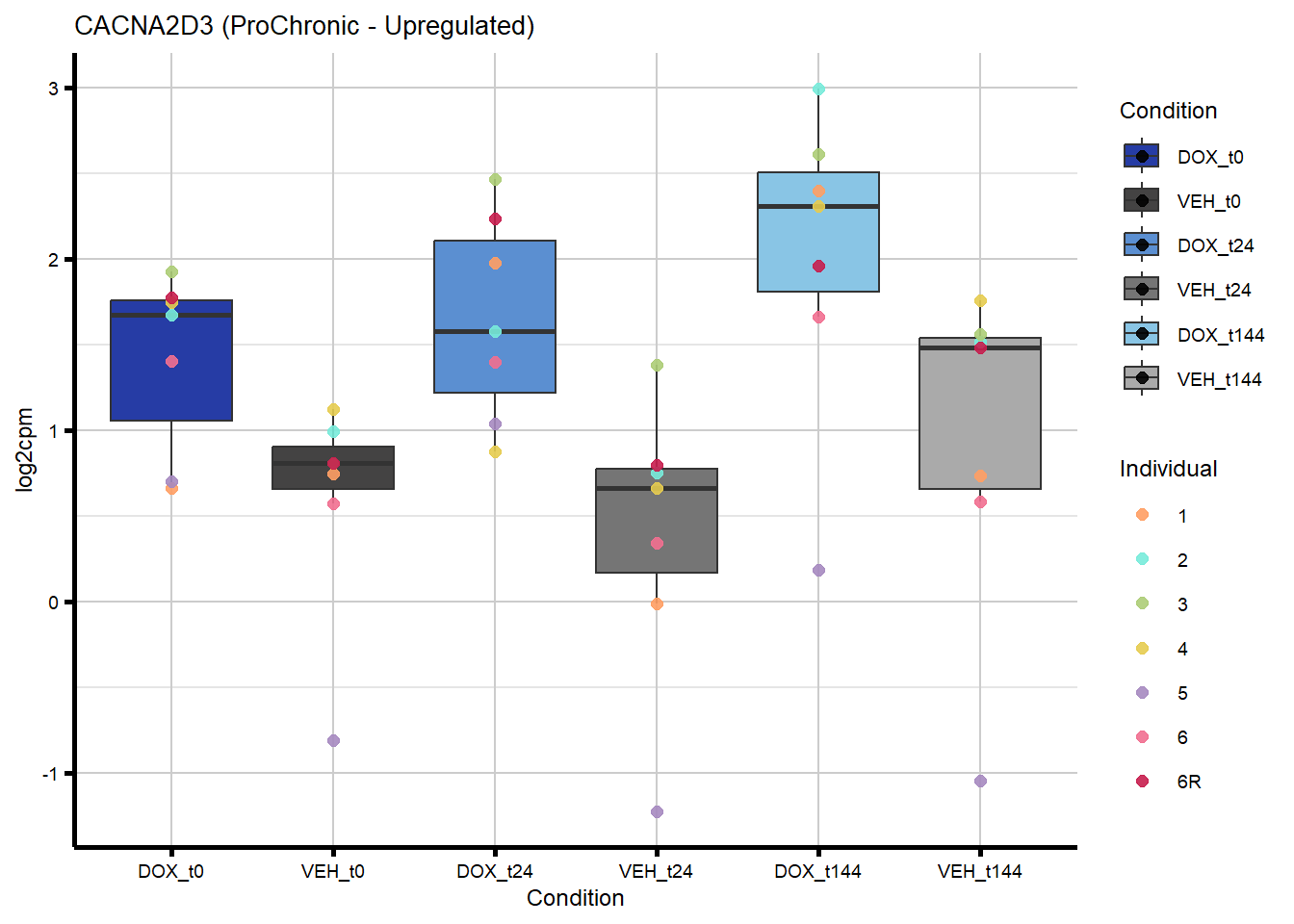
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
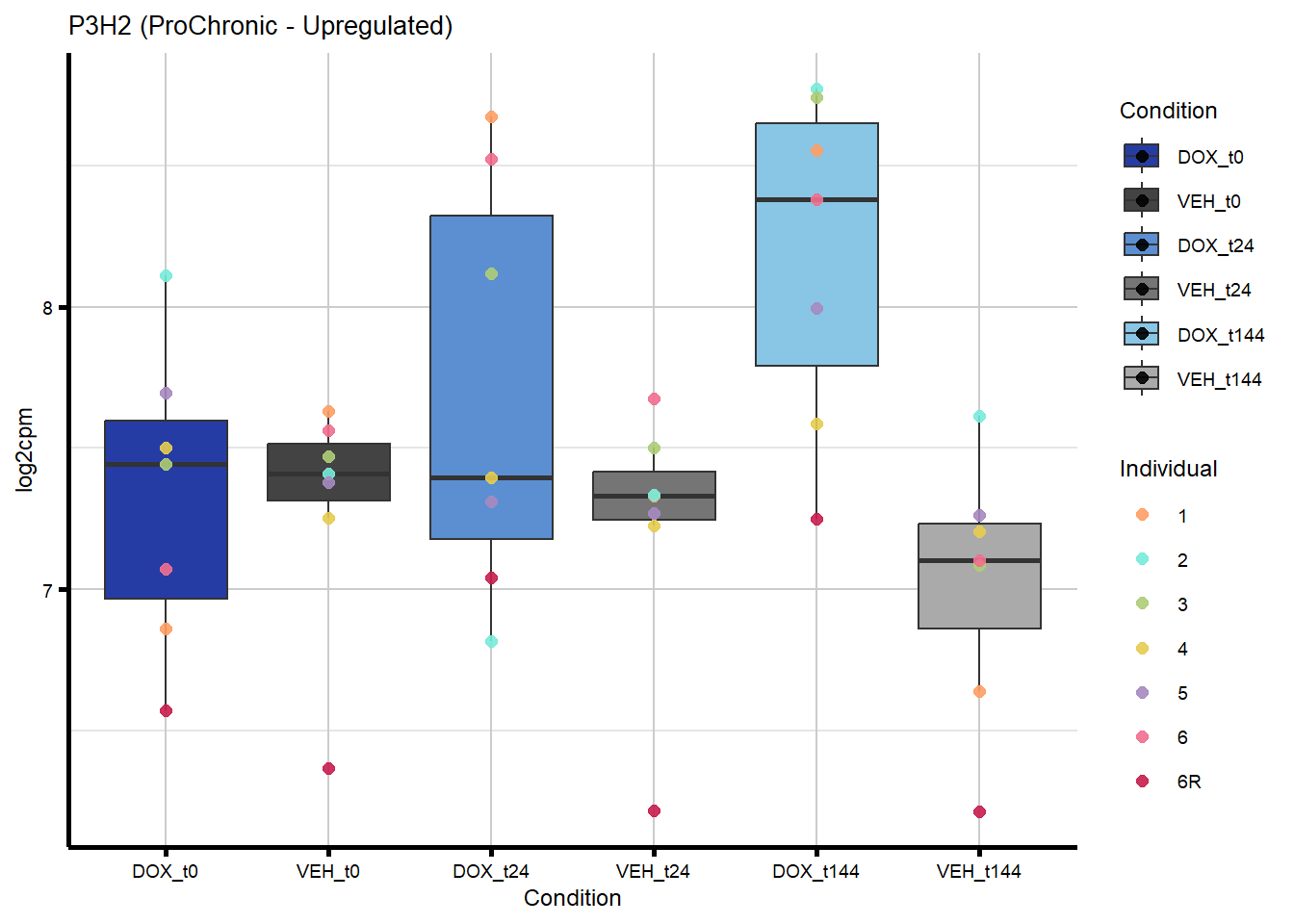
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
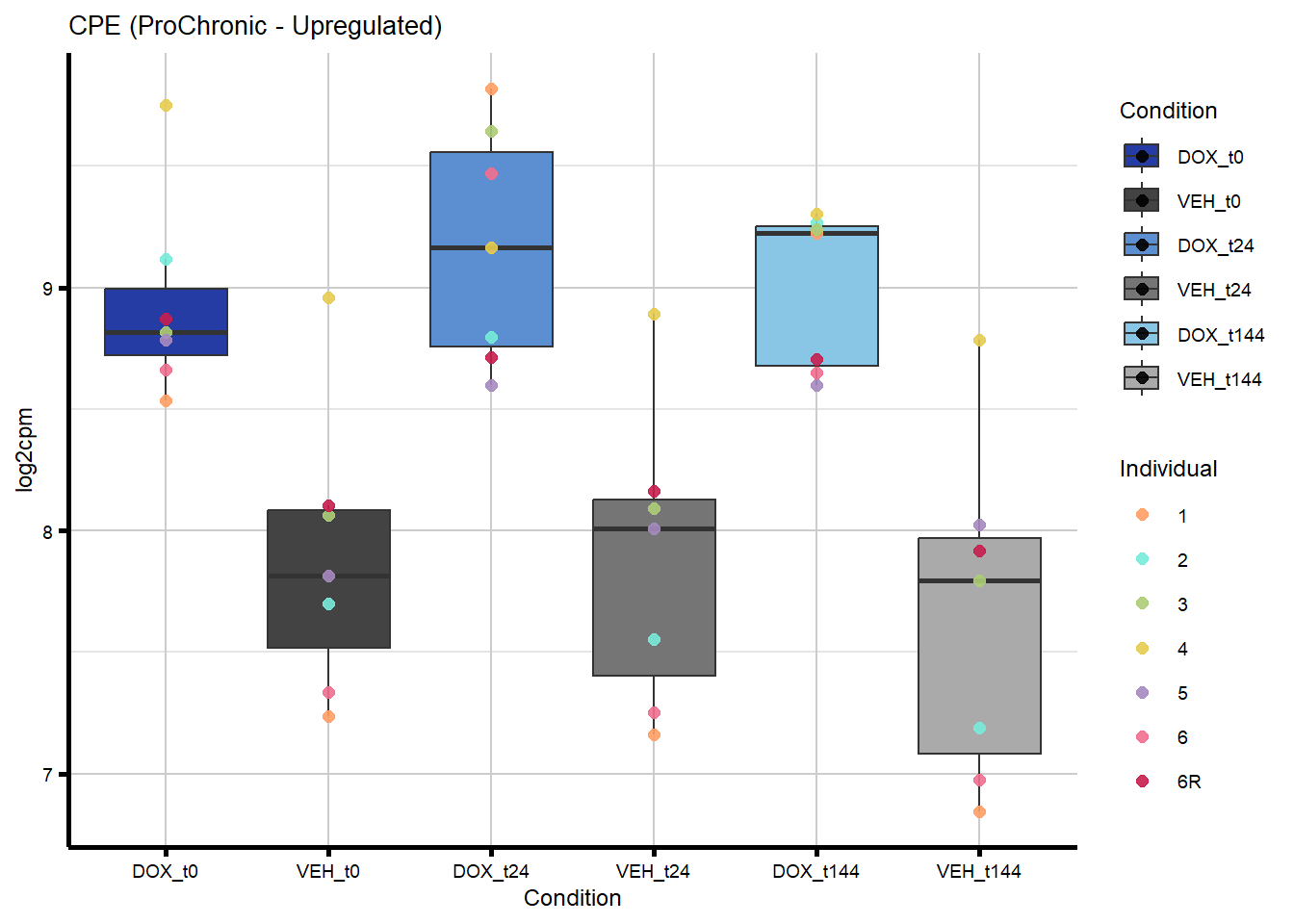
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
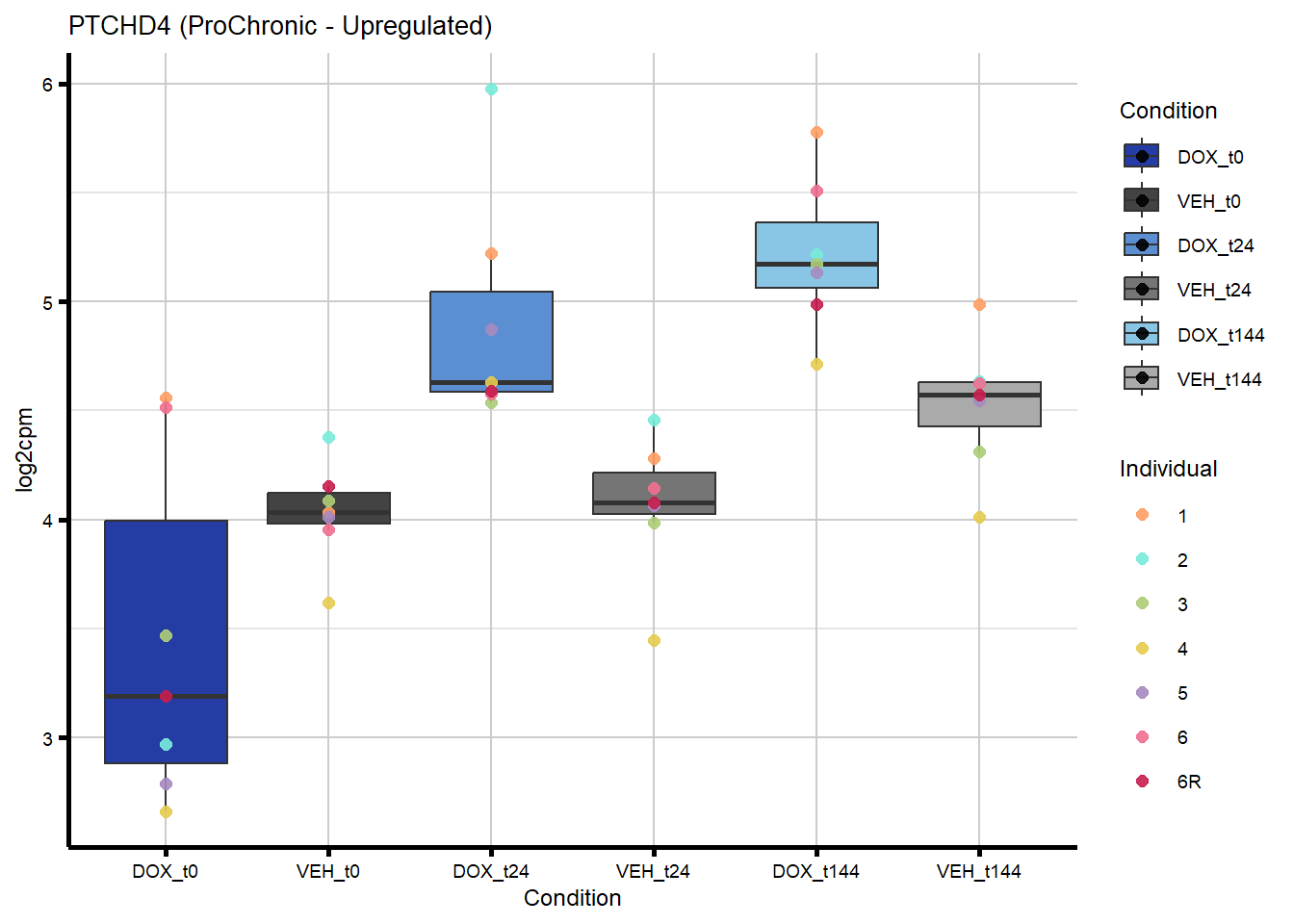
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
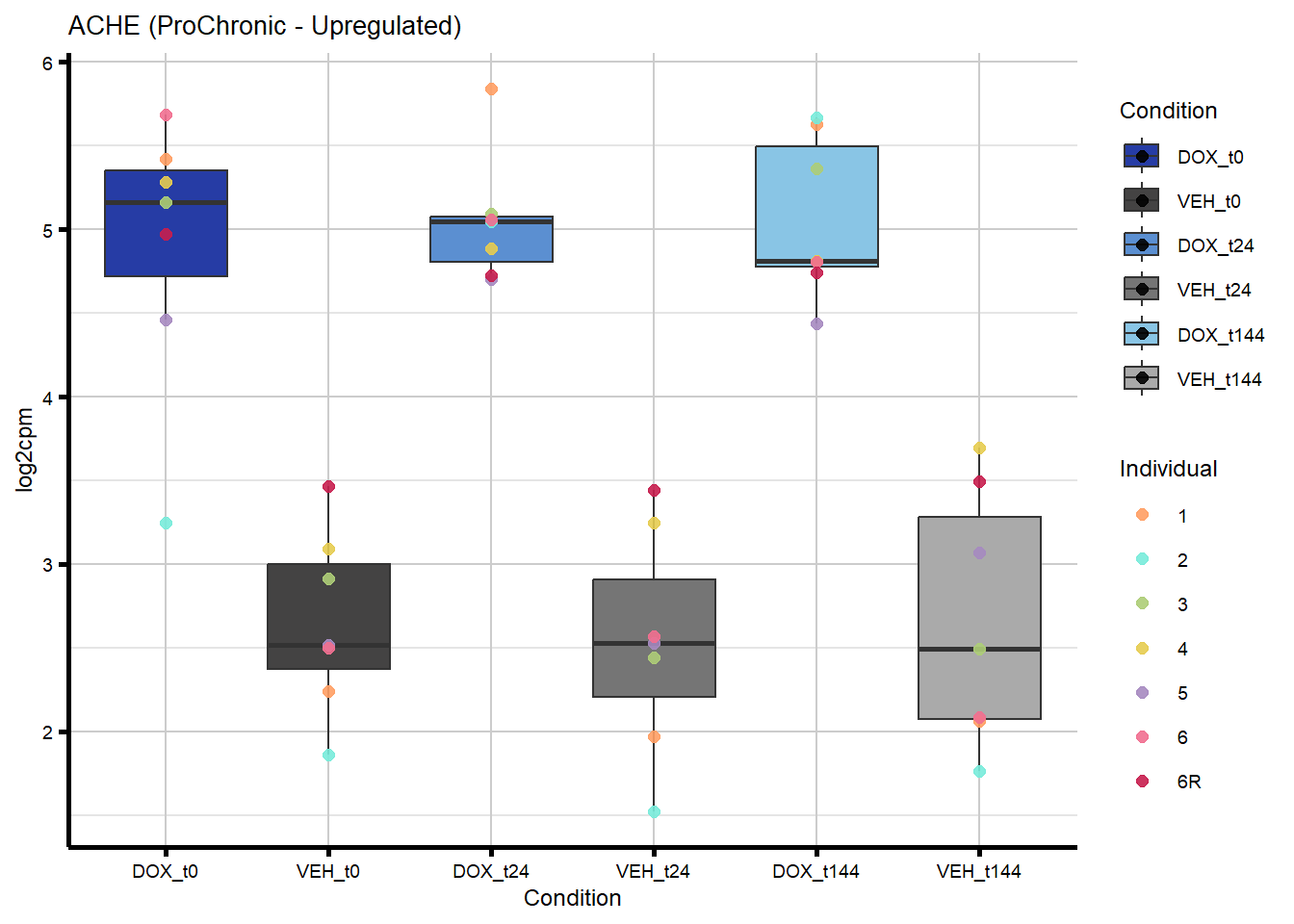
| Version | Author | Date |
|---|---|---|
| 7f22593 | emmapfort | 2026-01-05 |
#I used CACNA2D3 as my example gene for ProChronic in Figure 6F
#save all of these plots with unique names
# for (plot_name in names(proChronic_up_plots)) {
# save_plot(proChronic_up_plots[[plot_name]],
# filename = paste0("ExGene_proChronic_Upreg_", plot_name, "_EMP"),
# folder = output_folder)
# }
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 22000)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats4 grid stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] gprofiler2_0.2.3 org.Hs.eg.db_3.20.0 AnnotationDbi_1.68.0
[4] IRanges_2.40.0 S4Vectors_0.44.0 Biobase_2.66.0
[7] BiocGenerics_0.52.0 circlize_0.4.16 reshape2_1.4.4
[10] readxl_1.4.5 lubridate_1.9.4 forcats_1.0.0
[13] stringr_1.5.1 dplyr_1.1.4 purrr_1.0.4
[16] readr_2.1.5 tidyr_1.3.1 tibble_3.2.1
[19] ggplot2_3.5.2 tidyverse_2.0.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] tidyselect_1.2.1 viridisLite_0.4.2 farver_2.1.2
[4] blob_1.2.4 bitops_1.0-9 Biostrings_2.74.0
[7] RCurl_1.98-1.17 lazyeval_0.2.2 fastmap_1.2.0
[10] promises_1.3.2 digest_0.6.37 mime_0.13
[13] timechange_0.3.0 lifecycle_1.0.4 processx_3.8.6
[16] KEGGREST_1.46.0 RSQLite_2.3.9 magrittr_2.0.3
[19] compiler_4.4.2 rlang_1.1.6 sass_0.4.10
[22] tools_4.4.2 yaml_2.3.10 data.table_1.17.0
[25] knitr_1.50 labeling_0.4.3 htmlwidgets_1.6.4
[28] bit_4.5.0 xml2_1.3.8 plyr_1.8.9
[31] RColorBrewer_1.1-3 withr_3.0.2 git2r_0.36.2
[34] xtable_1.8-4 colorspace_2.1-1 scales_1.4.0
[37] cli_3.6.3 rmarkdown_2.29 crayon_1.5.3
[40] generics_0.1.4 rstudioapi_0.17.1 httr_1.4.7
[43] tzdb_0.5.0 DBI_1.2.3 cachem_1.1.0
[46] zlibbioc_1.52.0 cellranger_1.1.0 XVector_0.46.0
[49] vctrs_0.6.5 jsonlite_2.0.0 callr_3.7.6
[52] hms_1.1.3 bit64_4.5.2 systemfonts_1.2.2
[55] crosstalk_1.2.1 plotly_4.10.4 jquerylib_0.1.4
[58] glue_1.8.0 ps_1.9.1 stringi_1.8.7
[61] shape_1.4.6.1 gtable_0.3.6 later_1.4.2
[64] GenomeInfoDb_1.42.3 UCSC.utils_1.2.0 pillar_1.10.2
[67] htmltools_0.5.8.1 GenomeInfoDbData_1.2.13 R6_2.6.1
[70] rprojroot_2.0.4 kableExtra_1.4.0 shiny_1.10.0
[73] evaluate_1.0.3 png_0.1-8 memoise_2.0.1
[76] httpuv_1.6.16 bslib_0.9.0 Rcpp_1.0.14
[79] svglite_2.1.3 whisker_0.4.1 xfun_0.52
[82] fs_1.6.6 getPass_0.2-4 pkgconfig_2.0.3
[85] GlobalOptions_0.1.2