Recovery RNAseq DOX Data
Emma M Pfortmiller
2025-02-10
Last updated: 2025-04-18
Checks: 6 1
Knit directory: Recovery_5FU/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20250217) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/ensembl_backup_dox.RDS | data/ensembl_backup_dox.RDS |
| C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/ind_num_dox.RDS | data/ind_num_dox.RDS |
| C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/annot_dox.RDS | data/annot_dox.RDS |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 62c3340. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/figure/
Ignored: data/CDKN1A_geneplot_Dox.RDS
Ignored: data/Cormotif_prob_gene_list.RDS
Ignored: data/Cormotif_prob_gene_list_doxonly.RDS
Ignored: data/DMSO_TNN13_plot.RDS
Ignored: data/DOX_TNN13_plot.RDS
Ignored: data/DOXgeneplots.RDS
Ignored: data/MDM2_geneplot_Dox.RDS
Ignored: data/SIRT1_geneplot_Dox.RDS
Ignored: data/annot_dox.RDS
Ignored: data/cormotifARclust_pp.RDS
Ignored: data/counts_DE_df_dox.RDS
Ignored: data/ensembl_backup_dox.RDS
Ignored: data/filt_gene_list_dox.RDS
Ignored: data/gene_clustlike_motif.RDS
Ignored: data/gene_postprob_motif.RDS
Ignored: data/genematrix_dox.RDS
Ignored: data/heartgenes.csv
Ignored: data/heartgenes_dox.csv
Ignored: data/ind_num_dox.RDS
Ignored: data/initial_cormotif.RDS
Ignored: data/initial_cormotif_dox.RDS
Ignored: data/plot_leg_d.RDS
Ignored: data/plot_leg_d_horizontal.RDS
Ignored: data/plot_leg_d_vertical.RDS
Ignored: data/tableED_GOBP.RDS
Ignored: data/tableESR_GOBP_postprob.RDS
Ignored: data/tableLD_GOBP.RDS
Ignored: data/tableLR_GOBP_postprob.RDS
Ignored: data/tableNR_GOBP.RDS
Ignored: data/tableNR_GOBP_postprob.RDS
Ignored: data/top.table_V.D144r_dox.RDS
Ignored: data/top.table_V.D24_dox.RDS
Ignored: data/top.table_V.D24r_dox.RDS
Untracked files:
Untracked: NBL1_motif1_examplegene_1000x600_EMP_250416.png
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Recovery_DOX.Rmd) and HTML
(docs/Recovery_DOX.html) files. If you’ve configured a
remote Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 62c3340 | emmapfort | 2025-04-18 | Updated analysis 25/04/18 |
| Rmd | 424626d | emmapfort | 2025-04-15 | Add In DOX only set |
| html | be23fa9 | emmapfort | 2025-04-15 | Build site. |
| Rmd | ff22325 | emmapfort | 2025-04-15 | workflowr::wflow_publish(files = c("analysis/Recovery_DOX.Rmd")) |
| Rmd | 82840fa | emmapfort | 2025-04-15 | Updating website |
| html | 84d5599 | emmapfort | 2025-04-14 | Build site. |
| Rmd | 3db2eb6 | emmapfort | 2025-04-14 | DOX only page |
I want to separate out my dataset for this to only include the DOX and DMSO vehicle samples
fC_Matrix_Full_cpm_filter <- readRDS("data/fC_Matrix_Full_cpm_filter.RDS")
fC_Matrix_Full_cpm_filter_dox <- as.data.frame(fC_Matrix_Full_cpm_filter) %>% dplyr::select(-(contains("FLUO")))
#this dataset is the original log2cpm transformed and rowMeans filtered dataset which I need for heatmaps
counts_DE_df <- readRDS("data/counts_DE_df.RDS")
#this dataframe is pre-filtered and only contains genes (rows) that were present after log2cpm transformation and filtering by rowMeans > 0
DOX_counts_DE_df <- counts_DE_df %>% dplyr::select(-(contains("FLUO")))
dim(DOX_counts_DE_df)[1] 14170 42#this should have 42 variables if we remove the 21 variables from 5FUcol_tx_large <- rep(c("#499FBD" , "#63666D"), 21)
col_tx_large_2 <- c(rep("#499FBD" , 3), rep("#63666D", 3), 21)
ind_col <- c("#003F5C", "#45AE91", "#58508D", "#BC4099", "#8B3E9B", "#FF6361", "#FF2362")
tx_col <- c("#499FBD","#63666D")
time_col <- c("#fbb4b9", "#f768a1", "#ae017e")
##Add columns with more information to each gene I pull out##
ind_names <- c(rep("Ind1", 6), rep("Ind2", 6), rep("Ind3", 6), rep("Ind4", 6), rep("Ind5", 6), rep("Ind6", 6), rep("Ind6REP", 6))
time_names <- c(rep("24", 2), rep("24rec", 2), rep("144rec", 2))
time_names2 <- c("24", "24rec", "144rec")
time_names <- c(rep(time_names, 7))
time_names2 <- c(rep(time_names2, 7))
tx_names <- c("DOX", "DMSO")
tx_names <- c(rep(tx_names, 21))
tx_names2 <- c(rep("DOX", 3), rep("DMSO", 3))
tx_names2 <- c(rep(tx_names2, 21))
txtime_names <- c("DOX_24", "DMSO_24", "DOX_24rec", "DMSO_24rec", "DOX_144rec", "DMSO_144rec")
txtime_names <- c(rep(txtime_names, 7))
txtime_names2 <- c("DOX_24", "DOX_24rec", "DOX_144rec", "DMSO_24", "DMSO_24rec", "DMSO_144rec")
txtime_names2 <- c(rep(txtime_names2, 7))
genematrix_dox <- fC_Matrix_Full_cpm_filter_dox %>% rownames_to_column(var = "entrezgene_id")
#saveRDS(genematrix_dox, "data/genematrix_dox.RDS")
###CDKN1A - 1026###
CDKN1A_d <- genematrix_dox %>% filter(entrezgene_id=="1026")
CDKN1A_new_d <- as.data.frame(CDKN1A_d) %>% dplyr::select("entrezgene_id", "DOX_24_Ind1", "DOX_24rec_Ind1", "DOX_144rec_Ind1", "DMSO_24_Ind1", "DMSO_24rec_Ind1", "DMSO_144rec_Ind1","DOX_24_Ind2", "DOX_24rec_Ind2", "DOX_144rec_Ind2", "DMSO_24_Ind2", "DMSO_24rec_Ind2", "DMSO_144rec_Ind2", "DOX_24_Ind3", "DOX_24rec_Ind3", "DOX_144rec_Ind3", "DMSO_24_Ind3", "DMSO_24rec_Ind3", "DMSO_144rec_Ind3", "DOX_24_Ind4", "DOX_24rec_Ind4", "DOX_144rec_Ind4", "DMSO_24_Ind4", "DMSO_24rec_Ind4", "DMSO_144rec_Ind4", "DOX_24_Ind5", "DOX_24rec_Ind5", "DOX_144rec_Ind5", "DMSO_24_Ind5", "DMSO_24rec_Ind5", "DMSO_144rec_Ind5", "DOX_24_Ind6", "DOX_24rec_Ind6", "DOX_144rec_Ind6", "DMSO_24_Ind6", "DMSO_24rec_Ind6", "DMSO_144rec_Ind6", "DOX_24_Ind6REP", "DOX_24rec_Ind6REP", "DOX_144rec_Ind6REP", "DMSO_24_Ind6REP", "DMSO_24rec_Ind6REP", "DMSO_144rec_Ind6REP")
CDKN1A_melt_d <- melt(CDKN1A_d, variable.name = "sample")Using entrezgene_id as id variablesCDKN1A_melt_new_d <- melt(CDKN1A_new_d, variable.name = "sample")Using entrezgene_id as id variablesCDKN1A_melt_df_d <- data.frame(tx = factor(tx_names, levels = unique(tx_names)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names, levels = unique(txtime_names)),
time = factor(time_names, levels = unique(time_names)))
CDKN1A_melt_df2_d <- data.frame(tx = factor(tx_names2, levels = unique(tx_names2)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names2, levels = unique(txtime_names2)),
time = factor(time_names2, levels = unique(time_names2)))
CDKN1A_melt_df_all_d <- cbind(CDKN1A_melt_d, CDKN1A_melt_df_d)
CDKN1A_melt_df_all2_d <- cbind(CDKN1A_melt_new_d, CDKN1A_melt_df2_d)
####CDKN1A####
CDKN1A_melt_df_all2_d %>% ggplot(aes(x = txtime, y = value))+
geom_boxplot(aes(fill = tx))+
geom_point(aes(color = ind)) +
labs(title = "CDKN1A")+
theme_bw(base_size = 16)+
scale_fill_manual(values = c(tx_col))+
scale_color_manual(values = c(ind_col))+
xlab("Conditions")+
ylab("log2cpm")+
theme(plot.title = element_text(face = "italic"))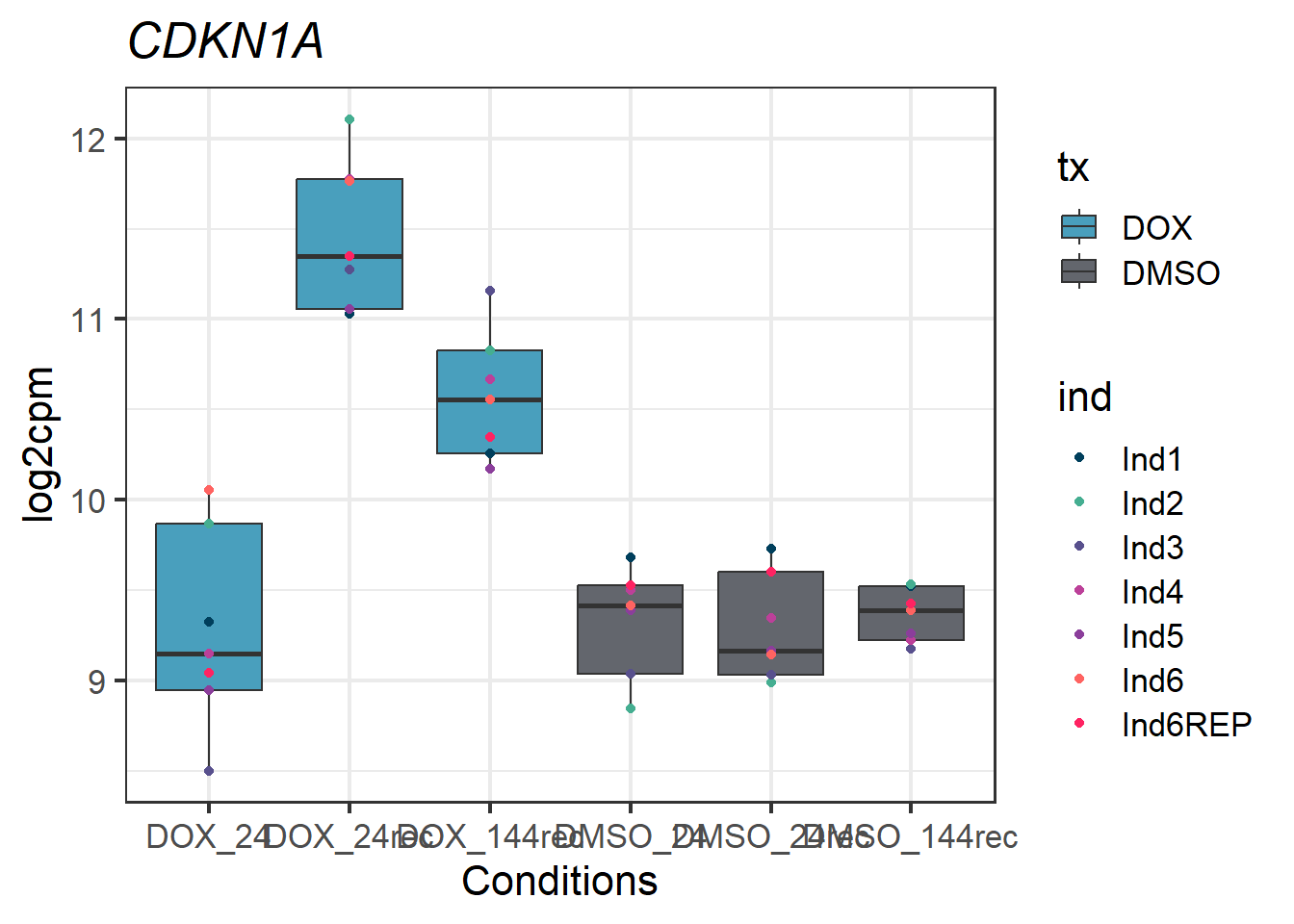
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#saveRDS(CDKN1A_geneplot_Dox, "data/CDKN1A_geneplot_Dox.RDS")
###MDM2 - 4193###
MDM2_d <- genematrix_dox %>% filter(entrezgene_id=="4193")
MDM2_new_d <- as.data.frame(MDM2_d) %>% dplyr::select("entrezgene_id", "DOX_24_Ind1", "DOX_24rec_Ind1", "DOX_144rec_Ind1", "DMSO_24_Ind1", "DMSO_24rec_Ind1", "DMSO_144rec_Ind1","DOX_24_Ind2", "DOX_24rec_Ind2", "DOX_144rec_Ind2", "DMSO_24_Ind2", "DMSO_24rec_Ind2", "DMSO_144rec_Ind2", "DOX_24_Ind3", "DOX_24rec_Ind3", "DOX_144rec_Ind3", "DMSO_24_Ind3", "DMSO_24rec_Ind3", "DMSO_144rec_Ind3", "DOX_24_Ind4", "DOX_24rec_Ind4", "DOX_144rec_Ind4", "DMSO_24_Ind4", "DMSO_24rec_Ind4", "DMSO_144rec_Ind4", "DOX_24_Ind5", "DOX_24rec_Ind5", "DOX_144rec_Ind5", "DMSO_24_Ind5", "DMSO_24rec_Ind5", "DMSO_144rec_Ind5", "DOX_24_Ind6", "DOX_24rec_Ind6", "DOX_144rec_Ind6", "DMSO_24_Ind6", "DMSO_24rec_Ind6", "DMSO_144rec_Ind6", "DOX_24_Ind6REP", "DOX_24rec_Ind6REP", "DOX_144rec_Ind6REP", "DMSO_24_Ind6REP", "DMSO_24rec_Ind6REP", "DMSO_144rec_Ind6REP")
MDM2_melt_d <- melt(MDM2_d, variable.name = "sample")Using entrezgene_id as id variablesMDM2_melt_new_d <- melt(MDM2_new_d, variable.name = "sample")Using entrezgene_id as id variablesMDM2_melt_df_d <- data.frame(tx = factor(tx_names, levels = unique(tx_names)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names, levels = unique(txtime_names)),
time = factor(time_names, levels = unique(time_names)))
MDM2_melt_df2_d <- data.frame(tx = factor(tx_names2, levels = unique(tx_names2)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names2, levels = unique(txtime_names2)),
time = factor(time_names2, levels = unique(time_names2)))
MDM2_melt_df_all_d <- cbind(MDM2_melt_d, MDM2_melt_df_d)
MDM2_melt_df_all2_d <- cbind(MDM2_melt_new_d, MDM2_melt_df2_d)
####MDM2####
MDM2_melt_df_all2_d %>% ggplot(aes(x = txtime, y = value))+
geom_boxplot(aes(fill = tx))+
geom_point(aes(color = ind)) +
labs(title = "MDM2")+
theme_bw(base_size = 16)+
scale_fill_manual(values = c(tx_col))+
scale_color_manual(values = c(ind_col))+
xlab("Conditions")+
ylab("log2cpm")+
theme(plot.title = element_text(face = "italic"))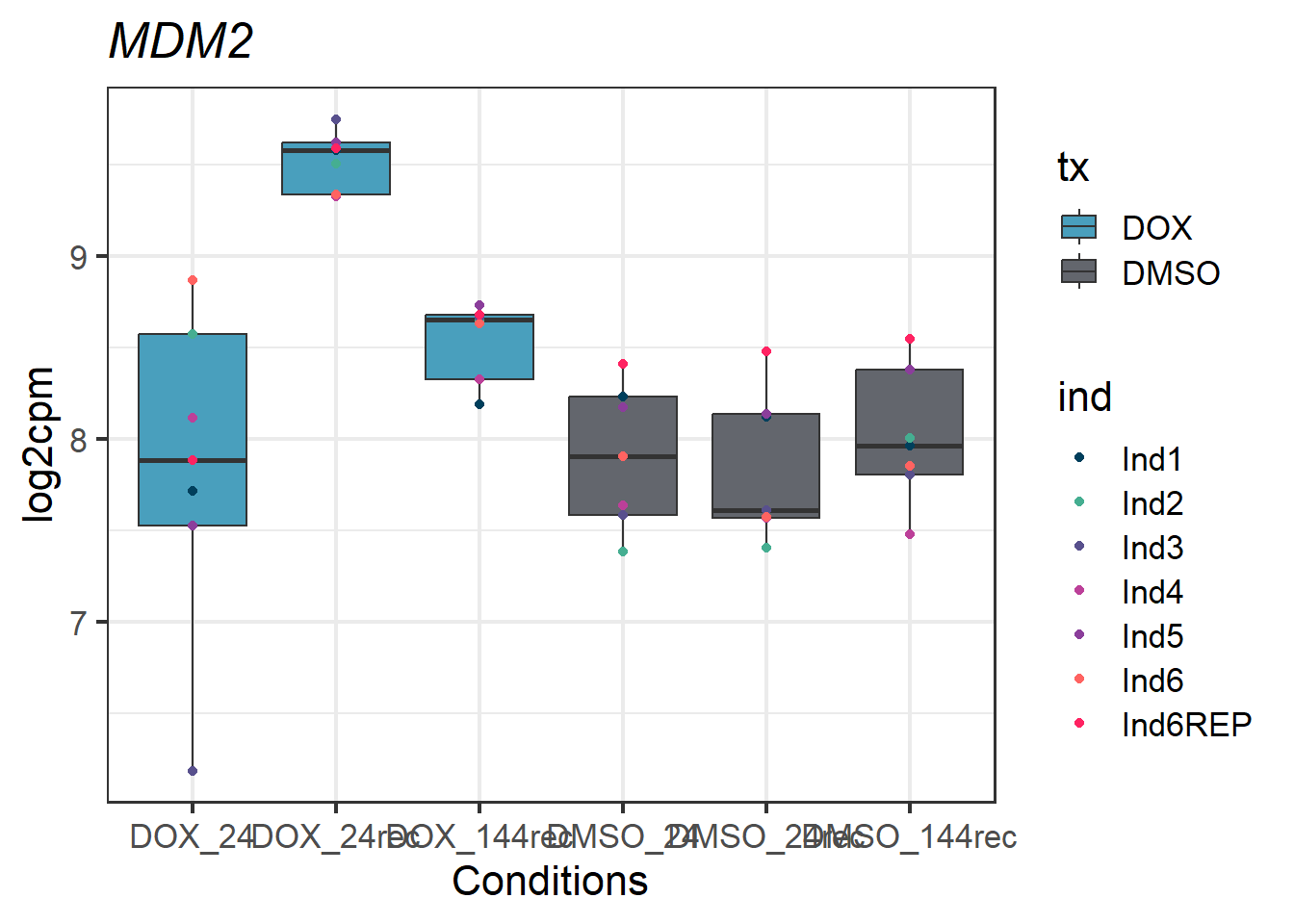
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#saveRDS(MDM2_geneplot_Dox, "data/MDM2_geneplot_Dox.RDS")
###SIRT1 - 23411###
SIRT1_d <- genematrix_dox %>% filter(entrezgene_id=="23411")
SIRT1_new_d <- as.data.frame(SIRT1_d) %>% dplyr::select("entrezgene_id", "DOX_24_Ind1", "DOX_24rec_Ind1", "DOX_144rec_Ind1", "DMSO_24_Ind1", "DMSO_24rec_Ind1", "DMSO_144rec_Ind1","DOX_24_Ind2", "DOX_24rec_Ind2", "DOX_144rec_Ind2", "DMSO_24_Ind2", "DMSO_24rec_Ind2", "DMSO_144rec_Ind2", "DOX_24_Ind3", "DOX_24rec_Ind3", "DOX_144rec_Ind3", "DMSO_24_Ind3", "DMSO_24rec_Ind3", "DMSO_144rec_Ind3", "DOX_24_Ind4", "DOX_24rec_Ind4", "DOX_144rec_Ind4", "DMSO_24_Ind4", "DMSO_24rec_Ind4", "DMSO_144rec_Ind4", "DOX_24_Ind5", "DOX_24rec_Ind5", "DOX_144rec_Ind5", "DMSO_24_Ind5", "DMSO_24rec_Ind5", "DMSO_144rec_Ind5", "DOX_24_Ind6", "DOX_24rec_Ind6", "DOX_144rec_Ind6", "DMSO_24_Ind6", "DMSO_24rec_Ind6", "DMSO_144rec_Ind6", "DOX_24_Ind6REP", "DOX_24rec_Ind6REP", "DOX_144rec_Ind6REP", "DMSO_24_Ind6REP", "DMSO_24rec_Ind6REP", "DMSO_144rec_Ind6REP")
SIRT1_melt_d <- melt(SIRT1_d, variable.name = "sample")Using entrezgene_id as id variablesSIRT1_melt_new_d <- melt(SIRT1_new_d, variable.name = "sample")Using entrezgene_id as id variablesSIRT1_melt_df_d <- data.frame(tx = factor(tx_names, levels = unique(tx_names)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names, levels = unique(txtime_names)),
time = factor(time_names, levels = unique(time_names)))
SIRT1_melt_df2_d <- data.frame(tx = factor(tx_names2, levels = unique(tx_names2)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names2, levels = unique(txtime_names2)),
time = factor(time_names2, levels = unique(time_names2)))
SIRT1_melt_df_all_d <- cbind(SIRT1_melt_d, SIRT1_melt_df_d)
SIRT1_melt_df_all2_d <- cbind(SIRT1_melt_new_d, SIRT1_melt_df2_d)
####SIRT1####
SIRT1_vertplot <- SIRT1_melt_df_all2_d %>% ggplot(aes(x = txtime, y = value))+
geom_boxplot(aes(fill = tx))+
geom_point(aes(color = ind)) +
labs(title = "SIRT1")+
theme_bw(base_size = 16)+
scale_fill_manual(values = c(tx_col))+
scale_color_manual(values = c(ind_col))+
xlab("Conditions")+
ylab("log2cpm")+
theme(plot.title = element_text(face = "italic"))
# get_legend_func <- function(plot) {
# legends <- cowplot::get_plot_component(plot, "guide-box", return_all = TRUE)
# nonzero <- vapply(plot_legend_d, \(x) !inherits(x, "zeroGrob"), TRUE)
# idx <- which(nonzero)
# #this will return the first nonzero legend since it's not in the standard spot
# if (length(idx) >0) {
# return(legends[[idx[1]]])
# } else {
# return(legends[[1]])
# }
# }
# plot_leg_d_hor <- get_legend_func(plot = SIRT1_vertplot + guides(color = guide_legend(nrow=1))+
# theme(legend.position = "bottom")
# )
plot_leg_d_ver <- cowplot::get_legend(SIRT1_vertplot)Warning in get_plot_component(plot, "guide-box"): Multiple components found;
returning the first one. To return all, use `return_all = TRUE`.# ggdraw(plot_leg_d_hor)
ggdraw(plot_leg_d_ver)
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#saveRDS(plot_leg_d_hor, "data/plot_leg_d_horizontal.RDS")
#saveRDS(plot_leg_d_ver, "data/plot_leg_d_vertical.RDS")
#saveRDS(SIRT1_geneplot_Dox, "data/SIRT1_geneplot_Dox.RDS")
#I saved these with no legend
#plot all of these genes together in one set
CDKN1A_geneplot_Dox <- readRDS("data/CDKN1A_geneplot_Dox.RDS")
MDM2_geneplot_Dox <- readRDS("data/MDM2_geneplot_Dox.RDS")
SIRT1_geneplot_Dox <- readRDS("data/SIRT1_geneplot_Dox.RDS")
plot_legend_d_ver <- readRDS("data/plot_leg_d_vertical.RDS")
plot_legend_d_hor <- readRDS("data/plot_leg_d_horizontal.RDS")
DOXgeneplots <- plot_grid(CDKN1A_geneplot_Dox,
MDM2_geneplot_Dox,
SIRT1_geneplot_Dox, nrow = 1, ncol = 3)
DOXgeneplots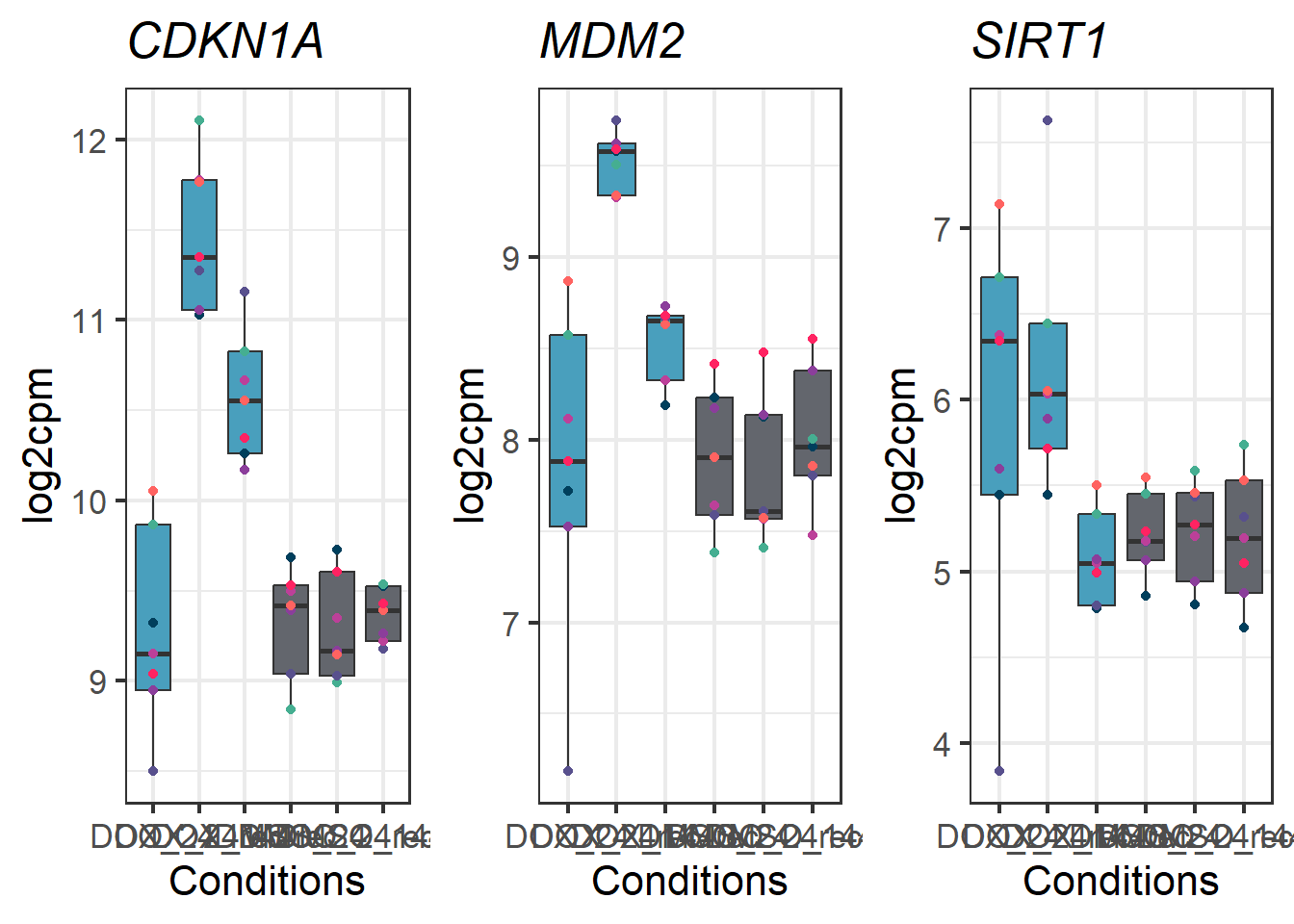
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#saveRDS(DOXgeneplots, "data/DOXgeneplots.RDS")genes_dox <- as.data.frame(fC_Matrix_Full_cpm_filter_dox)
genes_dox_df <- rownames_to_column(fC_Matrix_Full_cpm_filter_dox, var = "entrezgene_id")
###now let's pull some classic cardiac genes expressed in iPSC-CMs###
genecardiccheck_dox <- c("MYH7", "TNNT2","MYH6","ACTN2","BMP3","TNNI3","RYR2","CACNA1C","KCNQ1", "HCN1", "ADRB1", "ADRB2")
#ensembl_dox <- useMart("ensembl", dataset="hsapiens_gene_ensembl")
#saveRDS(ensembl_dox, "data/ensembl_backup_dox.RDS")
ensembl_dox <- readRDS("C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/ensembl_backup_dox.RDS")
my_chr_dox <- c(1:22, 'M', 'X', 'Y') ## creates a filter for each database
my_attributes_dox <- c('entrezgene_id', 'ensembl_gene_id', 'hgnc_symbol')
heartgenes_dox <- getBM(attributes=my_attributes_dox,filters ='hgnc_symbol',
values = genecardiccheck_dox, mart = ensembl_dox)
write.csv(heartgenes_dox, "data/heartgenes_dox.csv")
heartgenes_dox <-read.csv("data/heartgenes_dox.csv")
fungraph_dox <- as.data.frame(fC_Matrix_Full_cpm_filter_dox[rownames(fC_Matrix_Full_cpm_filter_dox) %in% heartgenes_dox$entrezgene_id,])
fungraph_dox %>%
rownames_to_column("entrezgene_id") %>%
pivot_longer(-entrezgene_id, names_to = "samples",values_to = "counts") %>%
mutate(gene = case_match(entrezgene_id,"88"~"ACTN2","153"~"ADRB1",
"154"~"ADRB2","651"~"BMP3","775"~"CACNA1C", "100874369"~"CACNA1C","348980"~"HCN1",
"3784"~"KCNQ1", "4624"~"MYH6","4625"~"MYH7","6262"~"RYR2",
"7137"~"TNNI3","7139"~"TNNT2",.default = entrezgene_id)) %>%
ggplot(., aes(x=reorder(gene,counts,decreasing=TRUE), y=counts))+
geom_boxplot()+
ggtitle(expression("Expression of typical cardiac tissue genes"))+
xlab("")+
ylim(c(0,20))+
ylab(expression("log"[2]~"cpm"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size =10, color = "black", angle = 0),
strip.text.y = element_text(color = "white"))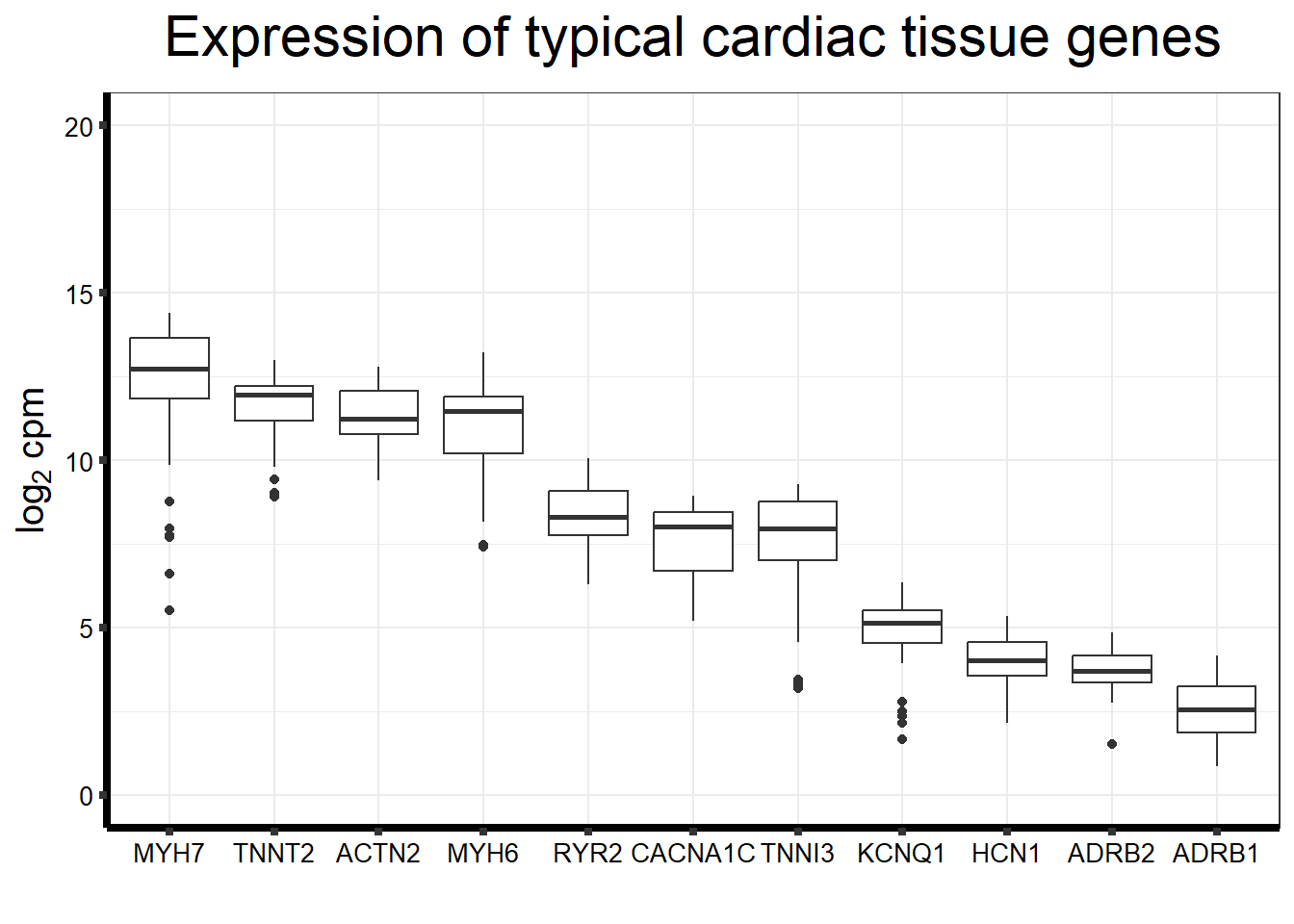
| Version | Author | Date |
|---|---|---|
| be23fa9 | emmapfort | 2025-04-15 |
#Now I want to plot these with DOX vs Veh to see if there are any changes
#I'll choose a subset of these genes - TNN13, TNNT2, NPPB, MYH7, ACTN2
fungraph_dox_only <- fungraph_dox %>% dplyr::select("DOX_24_Ind1", "DOX_24rec_Ind1", "DOX_144rec_Ind1", "DOX_24_Ind2", "DOX_24rec_Ind2", "DOX_144rec_Ind2", "DOX_24_Ind3", "DOX_24rec_Ind3", "DOX_144rec_Ind3", "DOX_24_Ind4", "DOX_24rec_Ind4", "DOX_144rec_Ind4", "DOX_24_Ind5", "DOX_24rec_Ind5", "DOX_144rec_Ind5", "DOX_24_Ind6", "DOX_24rec_Ind6", "DOX_144rec_Ind6", "DOX_24_Ind6REP", "DOX_24rec_Ind6REP", "DOX_144rec_Ind6REP")
fungraph_dmso_only <- fungraph_dox %>% dplyr::select("DMSO_24_Ind1", "DMSO_24rec_Ind1", "DMSO_144rec_Ind1", "DMSO_24_Ind2", "DMSO_24rec_Ind2", "DMSO_144rec_Ind2", "DMSO_24_Ind3", "DMSO_24rec_Ind3", "DMSO_144rec_Ind3", "DMSO_24_Ind4", "DMSO_24rec_Ind4", "DMSO_144rec_Ind4", "DMSO_24_Ind5", "DMSO_24rec_Ind5", "DMSO_144rec_Ind5", "DMSO_24_Ind6", "DMSO_24rec_Ind6", "DMSO_144rec_Ind6", "DMSO_24_Ind6REP", "DMSO_24rec_Ind6REP", "DMSO_144rec_Ind6REP")
fungraph_dox_only %>%
rownames_to_column("entrezgene_id") %>%
pivot_longer(-entrezgene_id, names_to = "samples",values_to = "counts") %>%
mutate(gene = case_match(entrezgene_id,"88"~"ACTN2","153"~"ADRB1",
"154"~"ADRB2","651"~"BMP3","775"~"CACNA1C", "100874369"~"CACNA1C","348980"~"HCN1",
"3784"~"KCNQ1", "4624"~"MYH6","4625"~"MYH7","6262"~"RYR2",
"7137"~"TNNI3","7139"~"TNNT2",.default = entrezgene_id)) %>%
ggplot(., aes(x=reorder(gene,counts,decreasing=TRUE), y=counts))+
geom_boxplot()+
ggtitle(expression("Expression Cardiac Genes DOX Only"))+
xlab("")+
ylim(c(0,20))+
ylab(expression("log"[2]~"cpm"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size =10, color = "black", angle = 0),
strip.text.y = element_text(color = "white"))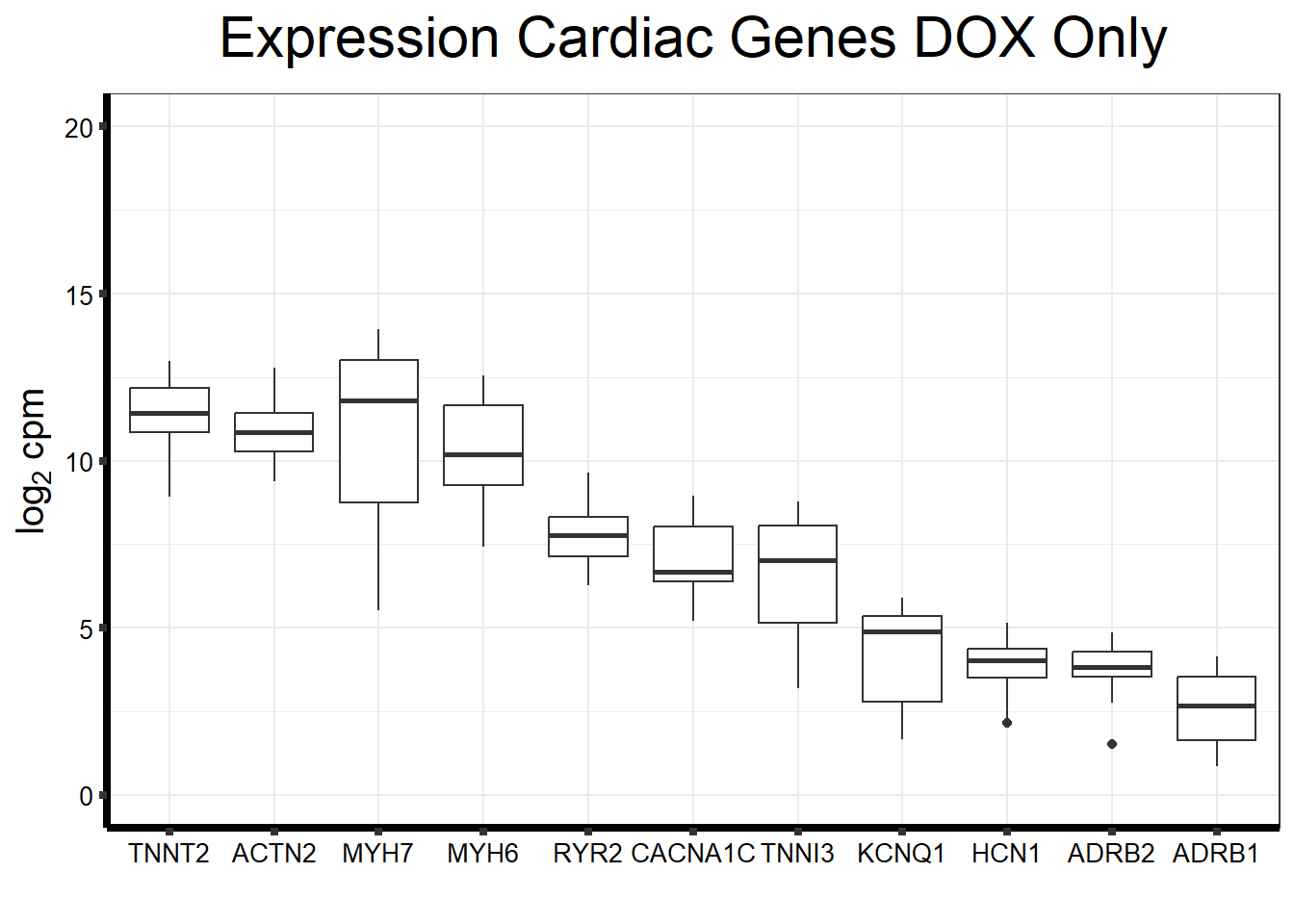
| Version | Author | Date |
|---|---|---|
| be23fa9 | emmapfort | 2025-04-15 |
fungraph_dmso_only %>%
rownames_to_column("entrezgene_id") %>%
pivot_longer(-entrezgene_id, names_to = "samples",values_to = "counts") %>%
mutate(gene = case_match(entrezgene_id,"88"~"ACTN2","153"~"ADRB1",
"154"~"ADRB2","651"~"BMP3","775"~"CACNA1C", "100874369"~"CACNA1C","348980"~"HCN1",
"3784"~"KCNQ1", "4624"~"MYH6","4625"~"MYH7","6262"~"RYR2",
"7137"~"TNNI3","7139"~"TNNT2",.default = entrezgene_id)) %>%
ggplot(., aes(x=reorder(gene,counts,decreasing=TRUE), y=counts))+
geom_boxplot()+
ggtitle(expression("Expression Cardiac Genes Vehicle Only"))+
xlab("")+
ylim(c(0,20))+
ylab(expression("log"[2]~"cpm"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size =10, color = "black", angle = 0),
strip.text.y = element_text(color = "white"))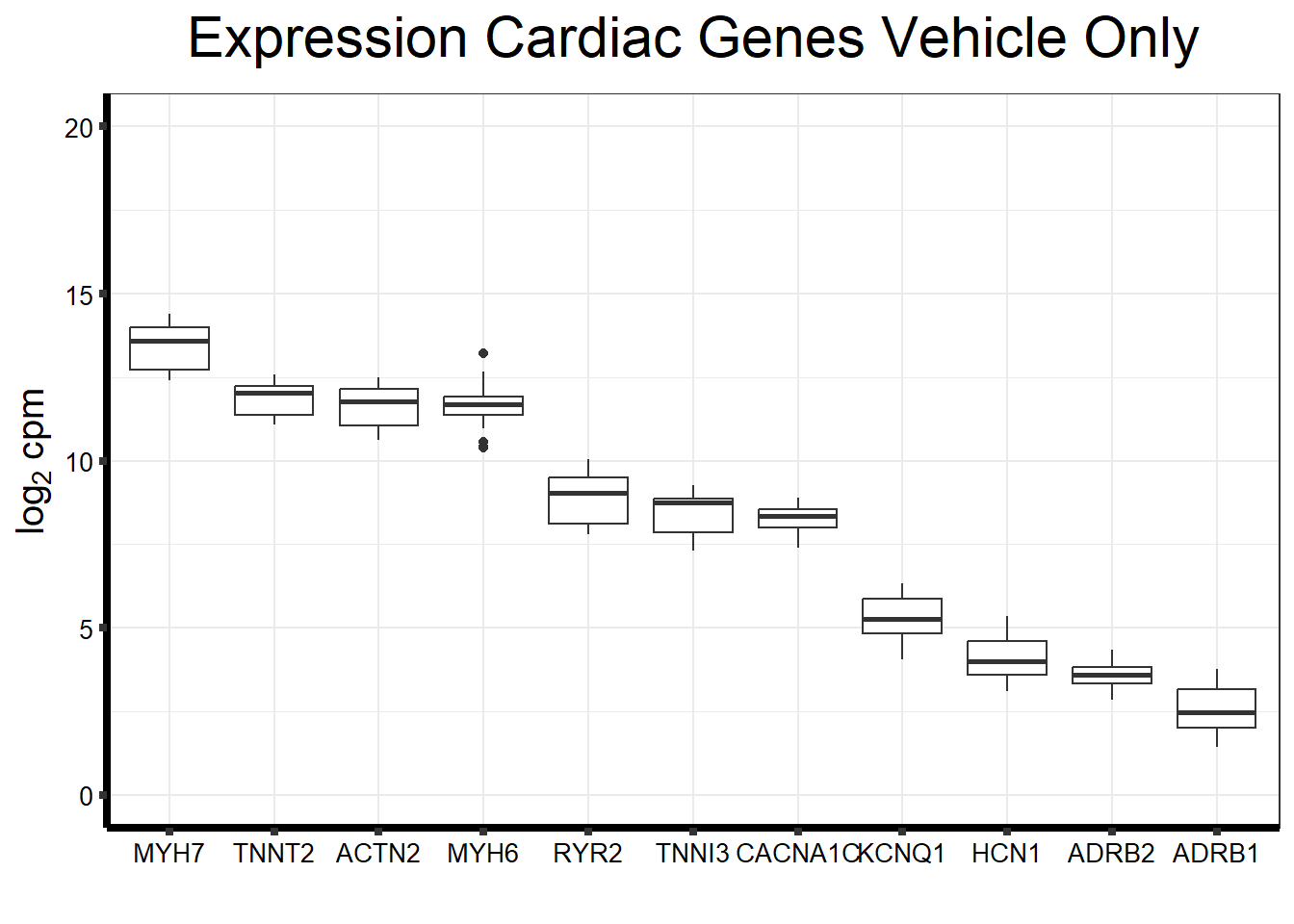
| Version | Author | Date |
|---|---|---|
| be23fa9 | emmapfort | 2025-04-15 |
#I want to just plot TNNI3 now
fungraph_dox_only_TNNI3 <- fungraph_dox_only %>%
rownames_to_column("entrezgene_id") %>%
pivot_longer(-entrezgene_id, names_to = "samples",values_to = "counts") %>%
filter(entrezgene_id == "7137") %>%
mutate(gene = case_match(entrezgene_id,"7137"~"TNNI3",.default = entrezgene_id)) %>%
ggplot(., aes(x=reorder(gene,counts,decreasing=TRUE), y=counts))+
geom_boxplot()+
ggtitle(expression("TNNI3 DOX"))+
xlab("")+
ylim(c(0,10))+
ylab(expression("log"[2]~"cpm"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size =10, color = "black", angle = 0),
strip.text.y = element_text(color = "white"))
#saveRDS(fungraph_dox_only_TNNI3, "data/DOX_TNN13_plot.RDS")
fungraph_dmso_only_TNNI3 <- fungraph_dmso_only %>%
rownames_to_column("entrezgene_id") %>%
pivot_longer(-entrezgene_id, names_to = "samples",values_to = "counts") %>%
filter(entrezgene_id == "7137") %>%
mutate(gene = case_match(entrezgene_id,"7137"~"TNNI3",.default = entrezgene_id)) %>%
ggplot(., aes(x=reorder(gene,counts,decreasing=TRUE), y=counts))+
geom_boxplot()+
ggtitle(expression("TNNI3 VEH"))+
xlab("")+
ylim(c(0,10))+
ylab(expression("log"[2]~"cpm"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size =10, color = "black", angle = 0),
strip.text.y = element_text(color = "white"))
#saveRDS(fungraph_dmso_only_TNNI3, "data/DMSO_TNN13_plot.RDS")
fungraph_dox_only_TNNI3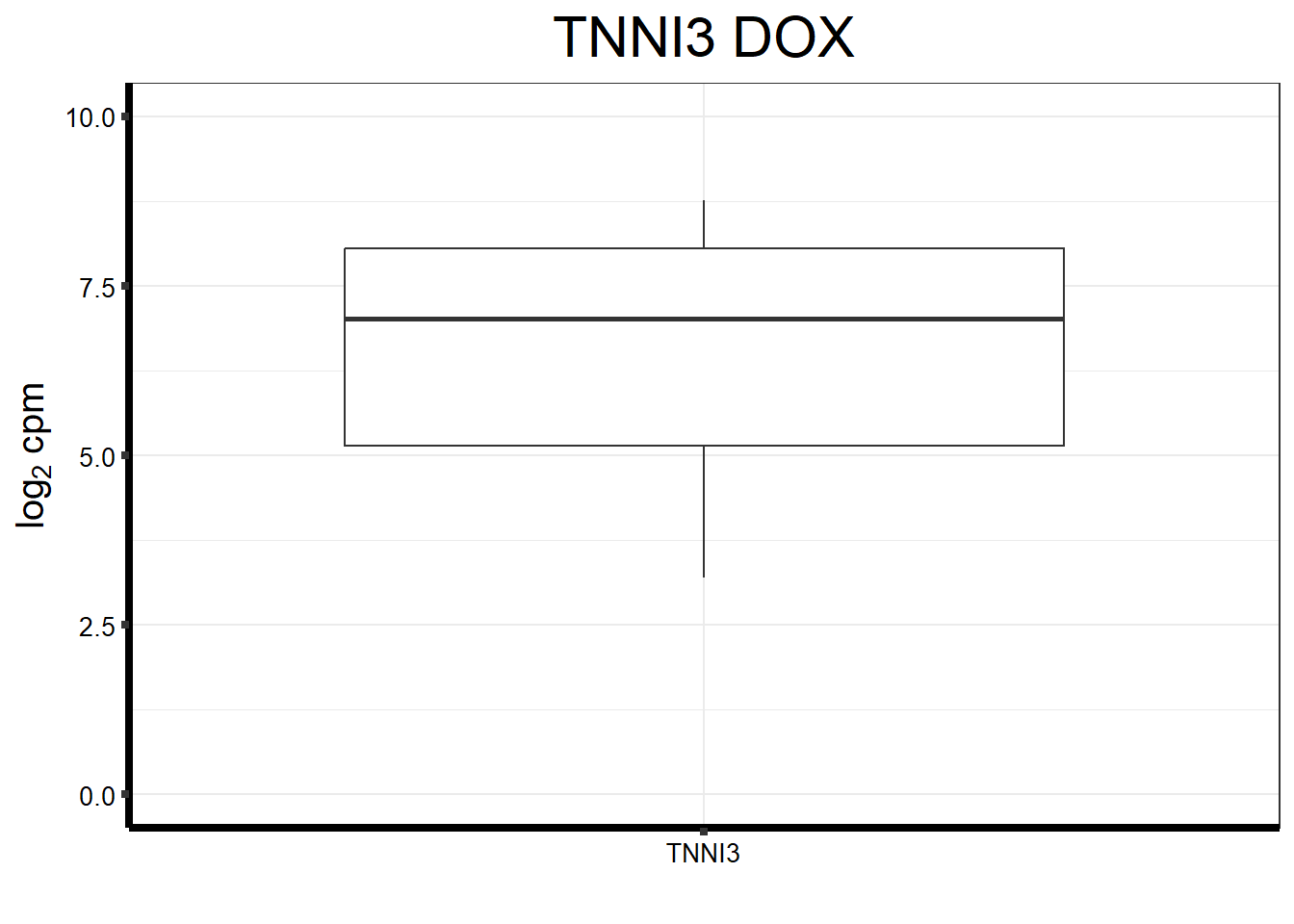
| Version | Author | Date |
|---|---|---|
| be23fa9 | emmapfort | 2025-04-15 |
fungraph_dmso_only_TNNI3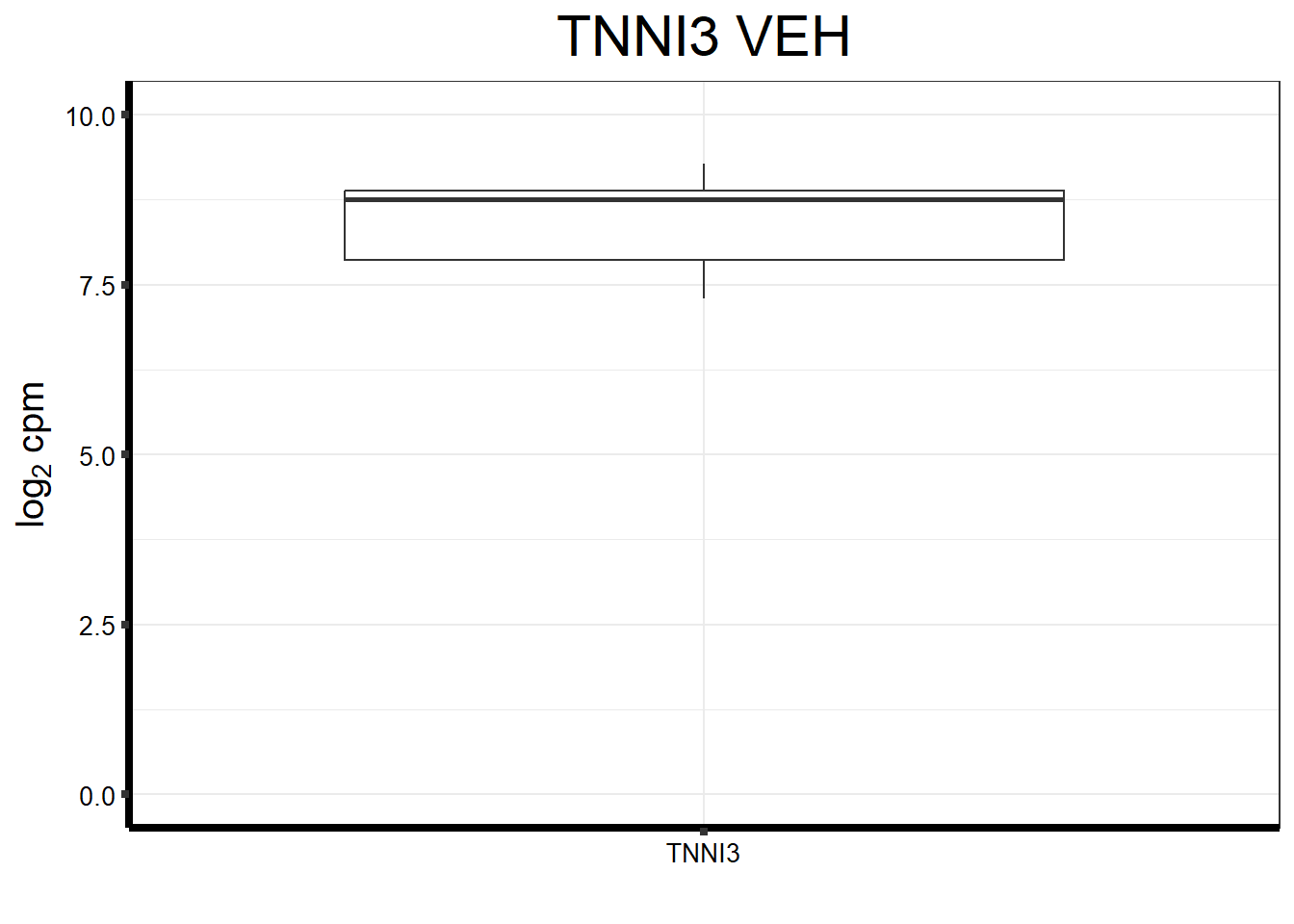
| Version | Author | Date |
|---|---|---|
| be23fa9 | emmapfort | 2025-04-15 |
plot_grid(fungraph_dox_only_TNNI3,
fungraph_dmso_only_TNNI3,
nrow = 1, ncol = 2)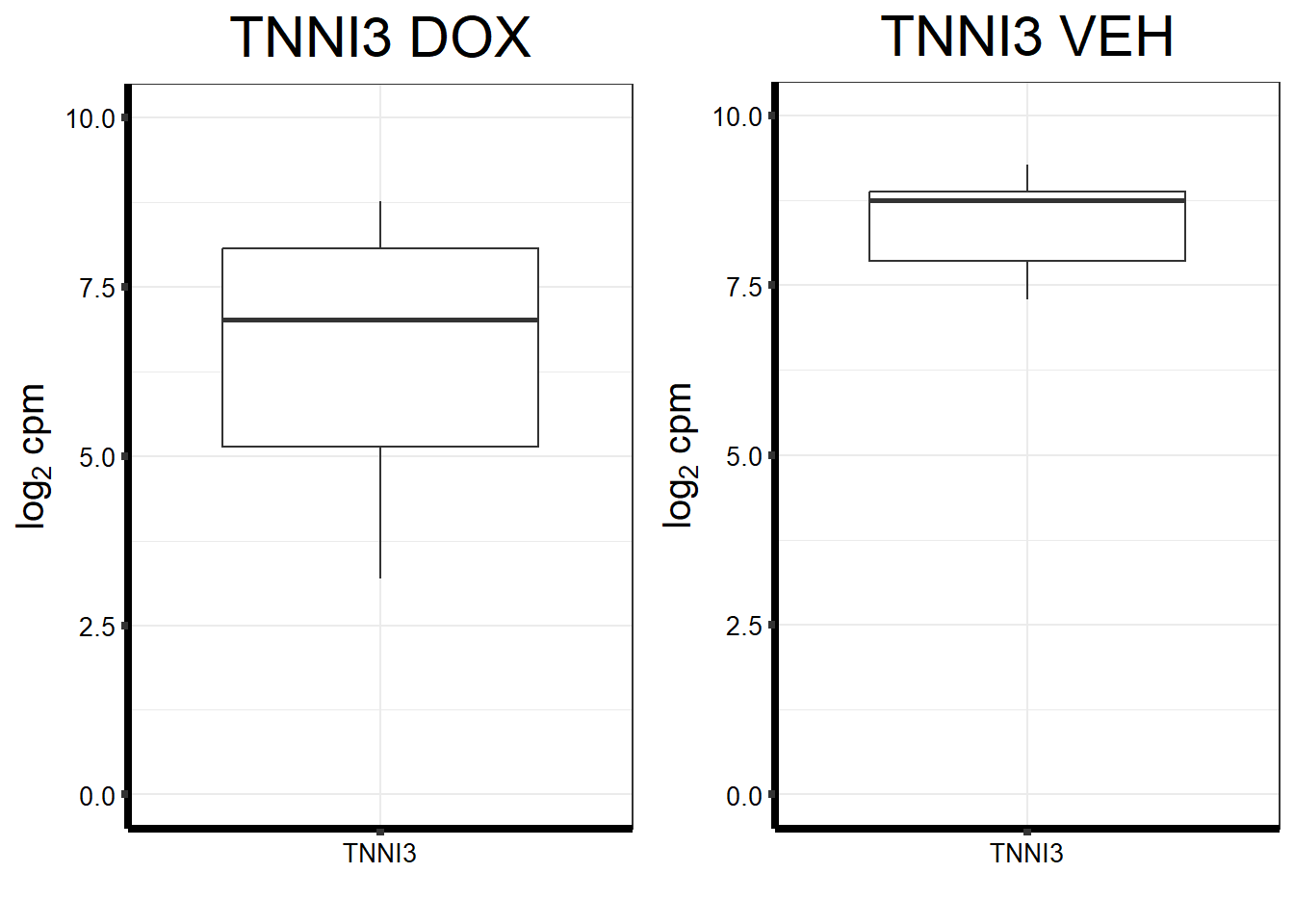
| Version | Author | Date |
|---|---|---|
| be23fa9 | emmapfort | 2025-04-15 |
####SPEARMAN FILTERED####
fC_Matrix_Full_cpm_filter_spearmancor_dox <-
cor(
fC_Matrix_Full_cpm_filter_dox,
y = NULL,
use = "everything",
method = "spearman"
)
Individual <- as.factor(c(rep("Ind1", 6), rep("Ind2", 6), rep("Ind3", 6), rep("Ind4", 6), rep("Ind5", 6), rep("Ind6", 6), rep("Ind6REP", 6)))
#Factor 2 - Treatment
tx_factor <- c("DOX", "DMSO")
Tx <- as.factor(c(rep(tx_factor, 21)))
#view(Treatment)
#Factor 3 - Timepoint
time_factor <- c(rep("24", 2), rep("24rec", 2), rep("144rec", 2))
Time <- as.factor(c(rep(time_factor, 7)))
####annotation for colors####
annot_col_hm = list(Tx = c(DOX = "blue", DMSO = "black"),
Ind = c(Ind1 = "#66E2A5", Ind2 = "#FC8D62", Ind3 = "#1F78B4", Ind4 = "#EFEDA3", Ind5 = "#A6D854", Ind6 = "#FFD92A", Ind6REP = "#8B3E9B"),
Time = c("24" = "#096F38", "24rec" = "#0050B5", "144rec" = "#B725AD"))
####annotation for values####
annot_list_hm <- data.frame(Individual = as.factor(c(rep("Ind1", 6), rep("Ind2", 6), rep("Ind3", 6), rep("Ind4", 6), rep("Ind5", 6), rep("Ind6", 6), rep("Ind6REP", 6))),
Tx = as.factor(c(rep(tx_factor, 21))),
Time = as.factor(c(rep(time_factor, 7))))
##add in the annotations from above into the dataframe
row.names(annot_list_hm) <- colnames(fC_Matrix_Full_cpm_filter_spearmancor_dox)
####ANNOTATED HEATMAPS####
pheatmap(fC_Matrix_Full_cpm_filter_spearmancor_dox, border_color = "black", legend = TRUE, angle_col = 90, display_numbers = FALSE, number_color = "black", fontsize = 10, fontsize_number = 5, annotation_col = annot_list_hm, annotation_colors = annot_col_hm)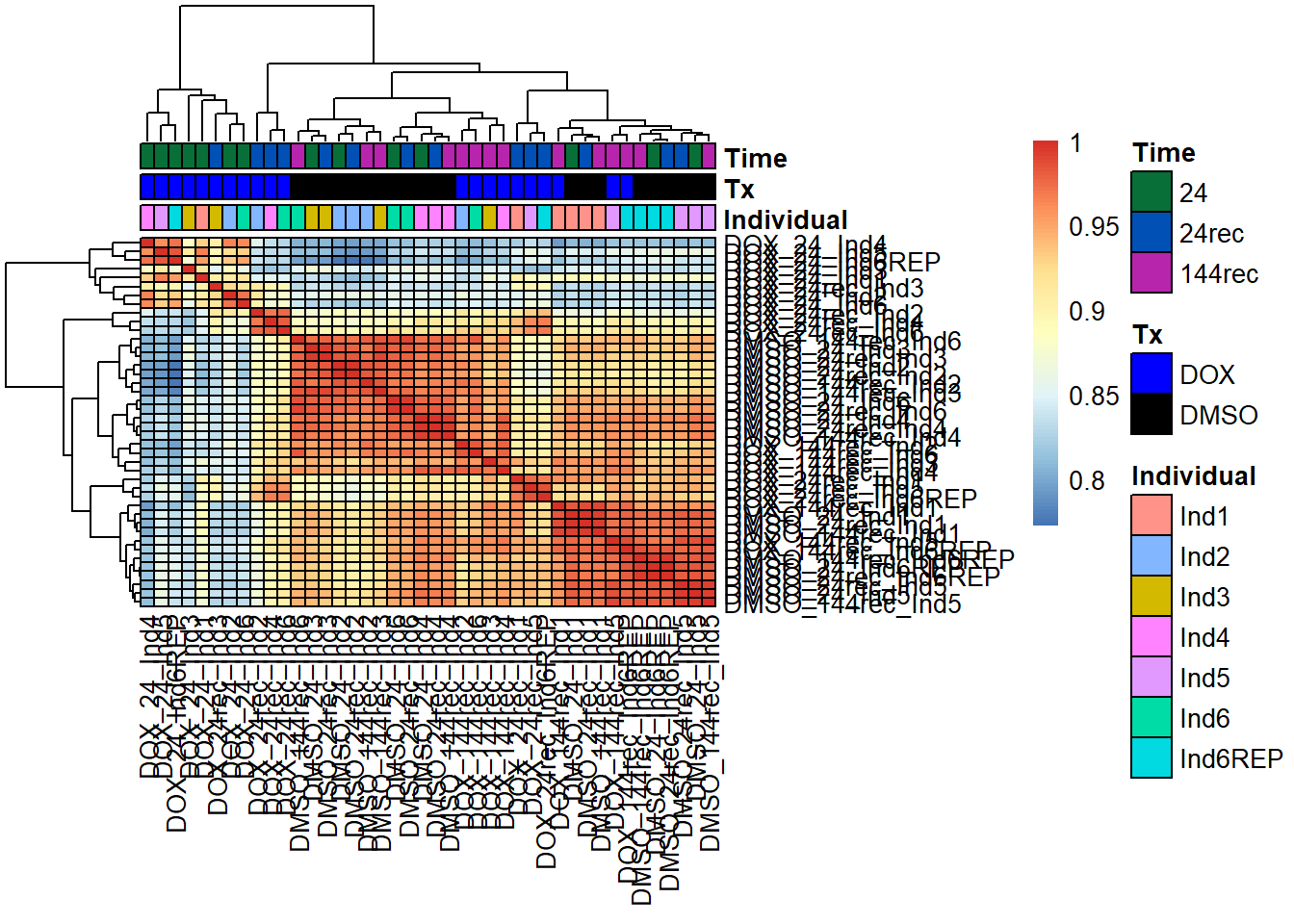
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
Now that I’ve put together my data, let’s begin DE!
group_1d <- rep(c("DOX_24",
"DMSO_24",
"DOX_24rec",
"DMSO_24rec",
"DOX_144rec",
"DMSO_144rec"), 6)
group_2d <- rep(c("DOX_24",
"DMSO_24",
"DOX_24rec",
"DMSO_24rec",
"DOX_144rec",
"DMSO_144rec"), 7)
dge_d <- DGEList.data.frame(counts = DOX_counts_DE_df, group = group_2d, genes = row.names(DOX_counts_DE_df))
#calculate the normalization factors with method TMM
dged_calc <- calcNormFactors(dge_d, method = "TMM")
#Pull out factors
snames_d <- data.frame("samples" = colnames(dged_calc)) %>% separate_wider_delim(., cols = samples, names = c("Treatment", "Time", "Individual"), delim = "_", cols_remove = FALSE)
snames_time_d <- snames_d$Time
snames_tx_d <- snames_d$Treatment
snames_ind_d <- snames_d$Individual
#Create my model matrix
mm_r_d <- model.matrix(~0 + group_2d)
p_d <- voom(dged_calc$counts, mm_r_d, plot = TRUE)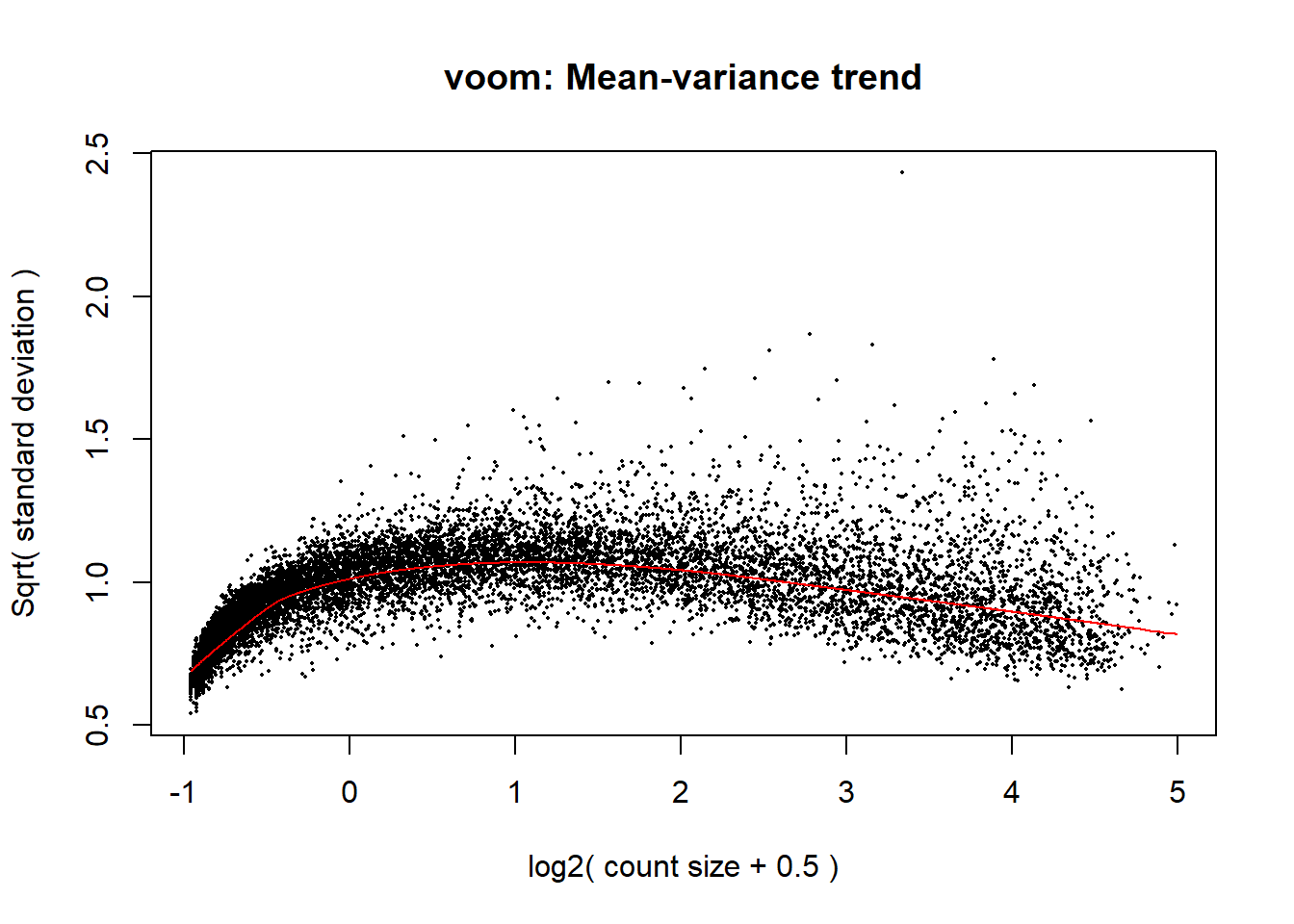
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
corfit_d <- duplicateCorrelation(p_d, mm_r_d, block = snames_ind_d)
v_d <- voom(dged_calc$counts, mm_r_d, block = snames_ind_d, correlation = corfit_d$consensus)
fit_d <- lmFit(v_d, mm_r_d, block = snames_ind_d, correlation = corfit_d$consensus)
#make sure to check which order the columns are in - otherwise they won't match right (it was moved into alphabetical and number order)
colnames(mm_r_d) <- c("DMSO_144rec","DMSO_24","DMSO_24rec","DOX_144rec","DOX_24","DOX_24rec")
cm_r_d <- makeContrasts(
V.D24 = DOX_24 - DMSO_24,
V.D24r = DOX_24rec - DMSO_24rec,
V.D144r = DOX_144rec - DMSO_144rec,
levels = mm_r_d
)
vfit_r_d <- lmFit(p_d, mm_r_d)
vfit_r_d <- contrasts.fit(vfit_r_d, contrasts = cm_r_d)
efit2_d <- eBayes(vfit_r_d)
results_d = decideTests(efit2_d)
summary(results_d)
V.D24 V.D24r V.D144r
Down 9709 6869 31
NotSig 3276 6379 13680
Up 1185 922 459
# # V.D24 V.D24r V.D144r
# Down 9709 6869 31
# NotSig 3276 6379 13680
# Up 1185 922 459
####plot your voom####
voom_plot_d <- voom(dged_calc, mm_r_d, plot = TRUE)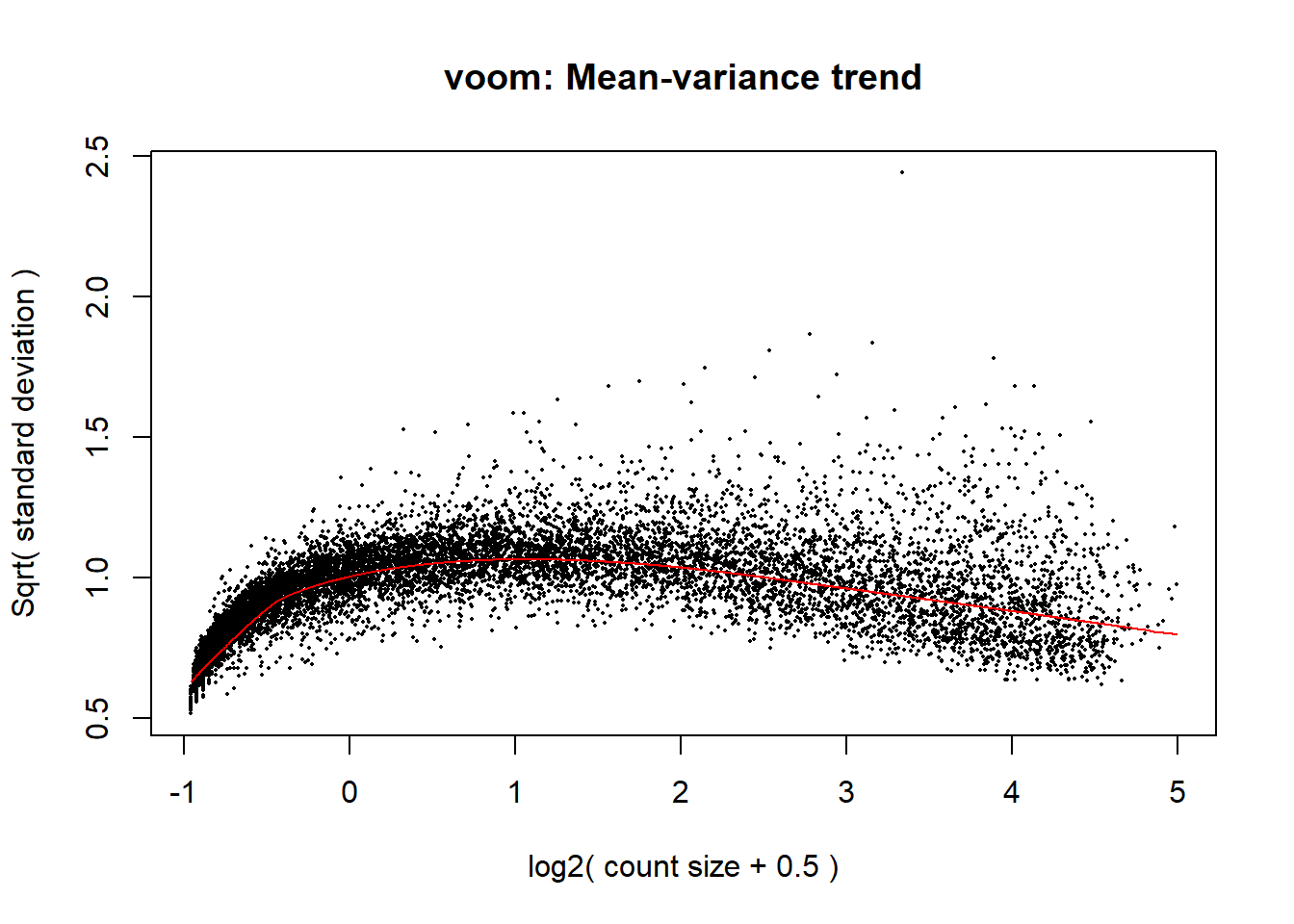
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
top.table_V.D24_dox <- topTable(fit = efit2_d, coef = "V.D24", number = nrow(dged_calc), adjust.method = "BH", p.value = 1, sort.by = "none")
head(top.table_V.D24_dox) logFC AveExpr t P.Value adj.P.Val B
100287102 -1.804419 5.673476 -1.741845 8.886085e-02 1.088296e-01 -4.985757
102466751 -1.386392 6.598993 -2.215141 3.223163e-02 4.236756e-02 -4.208131
100302278 2.117211 4.110796 5.136780 6.832114e-06 1.543790e-05 3.484472
645520 -1.060785 3.557889 -2.870907 6.387858e-03 9.406209e-03 -3.056843
79501 -1.851133 3.191793 -7.289085 5.649161e-09 1.915955e-08 10.231400
102725121 -2.392951 5.717312 -2.447792 1.863753e-02 2.550153e-02 -3.658644#saveRDS(top.table_V.D24_dox, "data/top.table_V.D24_dox.RDS")
top.table_V.D24r_dox <- topTable(fit = efit2_d, coef = "V.D24r", number = nrow(dged_calc), adjust.method = "BH", p.value = 1, sort.by = "none")
head(top.table_V.D24r_dox) logFC AveExpr t P.Value adj.P.Val B
100287102 0.2141767 5.673476 0.2255907 0.82261472 0.86440123 -6.218826
102466751 -0.8094950 6.598993 -1.3328291 0.18977853 0.26317888 -5.413117
100302278 -0.8734257 4.110796 -1.7817528 0.08202618 0.12937566 -4.863398
645520 -0.1192432 3.557889 -0.2881042 0.77468499 0.82587957 -6.419091
79501 -0.6825584 3.191793 -2.3475219 0.02368945 0.04379952 -4.150543
102725121 0.3419483 5.717312 0.3858246 0.70157538 0.76548265 -6.184783#saveRDS(top.table_V.D24r_dox, "data/top.table_V.D24r_dox.RDS")
top.table_V.D144r_dox <- topTable(fit = efit2_d, coef = "V.D144r", number = nrow(dged_calc), adjust.method = "BH", p.value = 1, sort.by = "none")
head(top.table_V.D144r_dox) logFC AveExpr t P.Value adj.P.Val B
100287102 -0.42467259 5.673476 -0.4139649 0.68100778 0.9656919 -6.176321
102466751 -0.04911403 6.598993 -0.0842465 0.93326102 0.9658168 -6.400185
100302278 0.32459251 4.110796 1.0815817 0.28561401 0.8618346 -6.375234
645520 -0.46815849 3.557889 -1.2874535 0.20499081 0.7713611 -5.861459
79501 0.56678196 3.191793 1.9032026 0.06388777 0.4666442 -5.076972
102725121 0.18759316 5.717312 0.1938705 0.84721256 0.9656919 -6.246426#saveRDS(top.table_V.D144r_dox, "data/top.table_V.D144r_dox.RDS")
#plot the top 5 most DE genes for each condition
#i sorted the toptable by the top 5 most DE genes for each + found the corresponding gene name
#####24hr Tx####
topDEG_dox24 <- top.table_V.D24_dox %>%
dplyr::slice_min(., n=5, order_by=P.Value) %>%
rownames_to_column(var = "GeneID")
Gene_Name <- c("FAM163A","SLC26A8","THBS-AS1","FOXD3-AS1","GRIK3")
topDEG_dox24 %>% mutate(GeneID = factor(GeneID, levels = c("148753", "116369", "101929523", "100996301", "2899"))) %>%
cbind(Gene_Name) GeneID logFC AveExpr t P.Value adj.P.Val B
1 148753 6.274101 5.578715 22.98007 2.128816e-25 3.016533e-21 47.14980
2 116369 5.789178 6.348094 20.56787 1.542184e-23 1.092637e-19 43.01677
3 101929523 -6.034510 6.305356 -19.08546 2.647316e-22 1.004705e-18 39.91738
4 100996301 5.149384 5.289969 18.98930 3.202808e-22 1.004705e-18 40.23872
5 2899 5.508418 5.862853 18.93819 3.545183e-22 1.004705e-18 40.05644
Gene_Name
1 FAM163A
2 SLC26A8
3 THBS-AS1
4 FOXD3-AS1
5 GRIK3#top 5 DE genes based on p-value and adj p value
ggplot(topDEG_dox24, aes(x=Gene_Name, y=logFC))+
geom_boxplot(aes(colour = adj.P.Val))+
ggtitle(expression("Top 5 DE Genes DOX 24"))+
xlab("")+
ylim(c(-10, 10))+
ylab(expression("logFC"))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5)),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size =10, color = "black", angle = 0),
strip.text.y = element_text(color = "white"))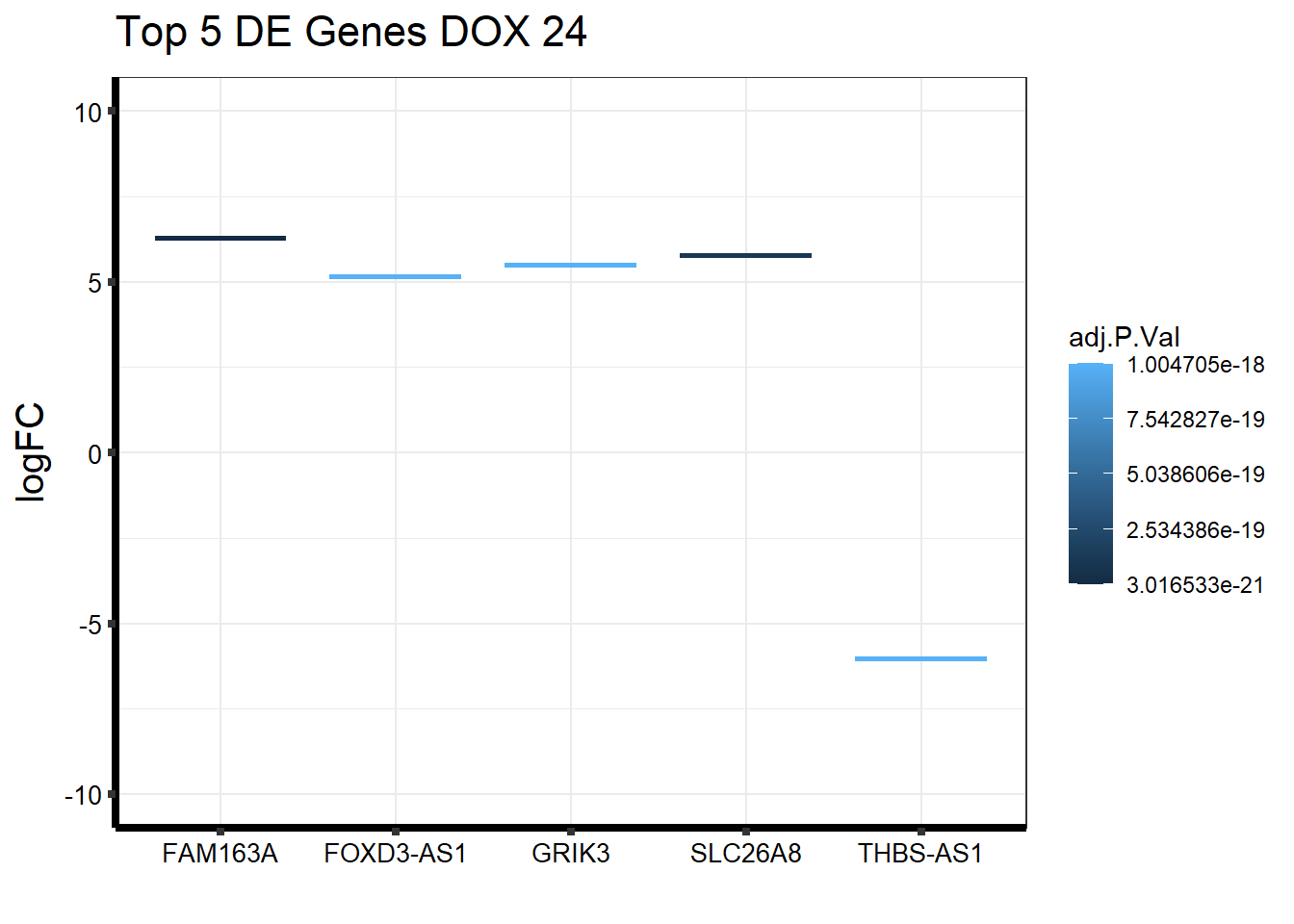
####24hr Recovery####
topDEG_dox24R <- top.table_V.D24r_dox %>%
dplyr::slice_min(., n=5, order_by=P.Value) %>%
rownames_to_column(var = "GeneID")
Gene_Name1 <- c("KCNK10", "CSTA", "LCE1B", "OTP", "KRTAP19-1")
topDEG_dox24R %>% mutate(GeneID = factor(GeneID, levels = c("54207", "1475", "353132", "23440", "337882"))) %>%
cbind(Gene_Name1) GeneID logFC AveExpr t P.Value adj.P.Val B
1 54207 4.949406 6.530192 15.82299 2.695651e-19 2.698663e-15 33.02274
2 1475 5.603936 5.360183 15.67190 3.808981e-19 2.698663e-15 32.95920
3 353132 4.839166 4.461296 14.34492 8.816170e-18 4.164171e-14 30.06752
4 23440 5.863845 6.216741 13.53171 6.670038e-17 2.362861e-13 27.67891
5 337882 6.367767 5.675415 13.17894 1.644191e-16 4.659637e-13 27.24446
Gene_Name1
1 KCNK10
2 CSTA
3 LCE1B
4 OTP
5 KRTAP19-1ggplot(topDEG_dox24R, aes(x=Gene_Name1, y=logFC))+
geom_boxplot(aes(colour = adj.P.Val))+
ggtitle(expression("Top 5 DE Genes DOX 24 Recovery"))+
xlab("")+
ylim(c(-10,10))+
ylab(expression("logFC"))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5)),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size =10, color = "black", angle = 0),
strip.text.y = element_text(color = "white"))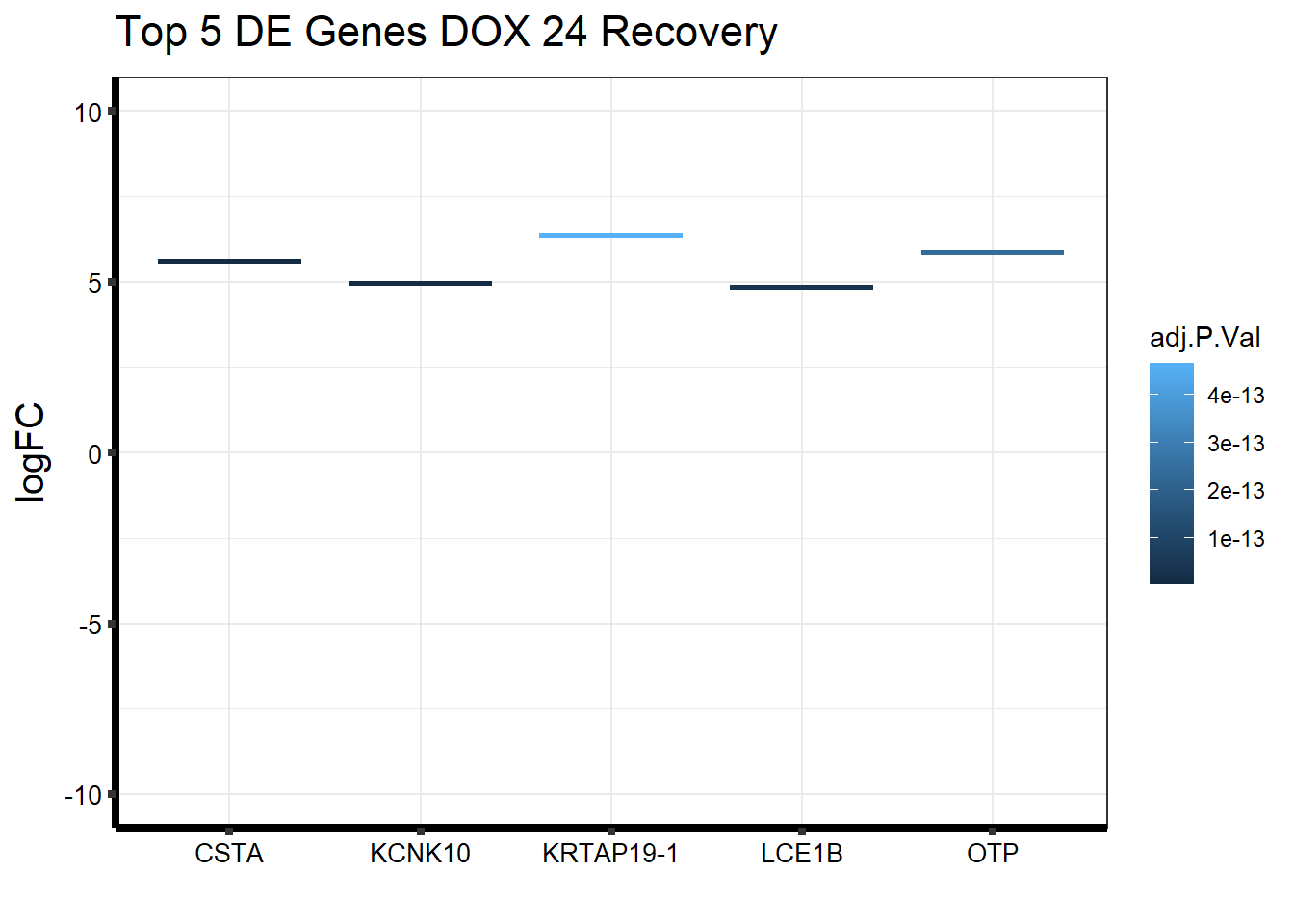
####144R####
topDEG_dox144R <- top.table_V.D144r_dox %>%
dplyr::slice_min(., n=5, order_by=P.Value) %>%
rownames_to_column(var = "GeneID")
Gene_Name2 <- c("LINC03025", "SLC4A10", "VIT", "PSMA8", "MYO3B")
topDEG_dox144R %>% mutate(GeneID = factor(GeneID, levels = c("440896", "57282", "5212", "143471", "140469"))) %>%
cbind(Gene_Name2) GeneID logFC AveExpr t P.Value adj.P.Val B
1 440896 4.681589 4.937852 13.65775 4.849711e-17 6.872040e-13 28.36219
2 57282 4.710121 5.295198 12.59160 7.637380e-16 5.411084e-12 25.77519
3 5212 3.923330 4.773412 12.10946 2.782463e-15 1.314250e-11 24.40702
4 143471 3.521809 4.241896 11.71091 8.284121e-15 2.643174e-11 23.33360
5 140469 4.647516 6.057544 11.66805 9.326656e-15 2.643174e-11 23.23254
Gene_Name2
1 LINC03025
2 SLC4A10
3 VIT
4 PSMA8
5 MYO3Bggplot(topDEG_dox144R, aes(x=Gene_Name2, y=logFC))+
geom_boxplot(aes(colour = adj.P.Val))+
ggtitle(expression("Top 5 DE Genes DOX 144 Recovery"))+
xlab("")+
ylim(c(-10,10))+
ylab(expression("logFC"))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5)),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size =10, color = "black", angle = 0),
strip.text.y = element_text(color = "white"))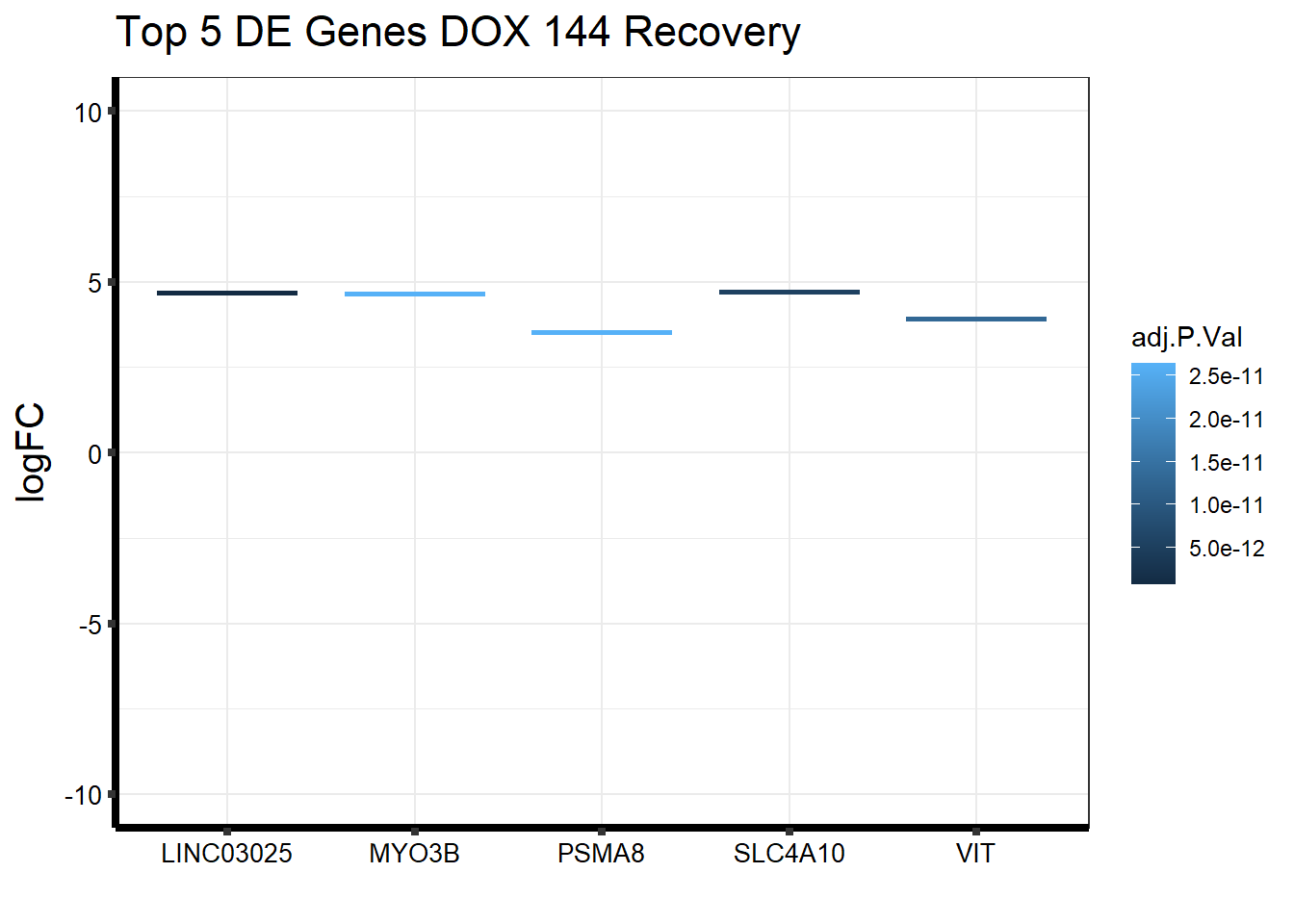
generate_volcano_plot <- function(toptable, title) {
#make significance labels
toptable$Significance <- "Not Significant"
toptable$Significance[toptable$logFC > 0 & toptable$adj.P.Val < 0.05] <- "Upregulated"
toptable$Significance[toptable$logFC < 0 & toptable$adj.P.Val < 0.05] <- "Downregulated"
#add number of genes for each significance label
upgenes <- toptable %>% filter(Significance == "Upregulated") %>% nrow()
downgenes <- toptable %>% filter(Significance == "Downregulated") %>% nrow()
nsgenes <- toptable %>% filter(Significance == "Not Significant") %>% nrow()
#make legend labels for no of genes
legend_lab <- c(
str_c('Upregulated: ', upgenes),
str_c('Not Significant: ', nsgenes),
str_c('Downregulated: ', downgenes)
)
#generate volcano plot w/ legend
ggplot(toptable, aes(x = logFC, y = -log10(P.Value), color = Significance)) +
geom_point(alpha = 0.4, size = 2) +
scale_color_manual(values = c("Upregulated" = "blue", "Downregulated" = "red", "Not Significant" = "gray"), labels = legend_lab) +
xlim(-5, 5) +
labs(title = title, x = expression(x = "log"[2]*"FC"), y = expression(y = "-log"[10]*"P-value")) +
theme(legend.position = "none",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25))) +
theme_bw()
}
#now that I've made a function, I can make volcano plots for each of my comparisons (6 total)
volcano_plots <- list(
"V.D24" = generate_volcano_plot(top.table_V.D24_dox, "Volcano Plot DOX 24hr (adj P-val<0.05)"),
"V.D24r" = generate_volcano_plot(top.table_V.D24r_dox, "Volcano Plot DOX 24hr Recovery (adj P-val<0.05)"),
"V.D144r" = generate_volcano_plot(top.table_V.D144r_dox, "Volcano Plot DOX 144hr Recovery (adj P-val<0.05)")
)
# Display each volcano plot
for (plot_name in names(volcano_plots)) {
print(volcano_plots[[plot_name]])
}Warning: Removed 90 rows containing missing values or values outside the scale range
(`geom_point()`).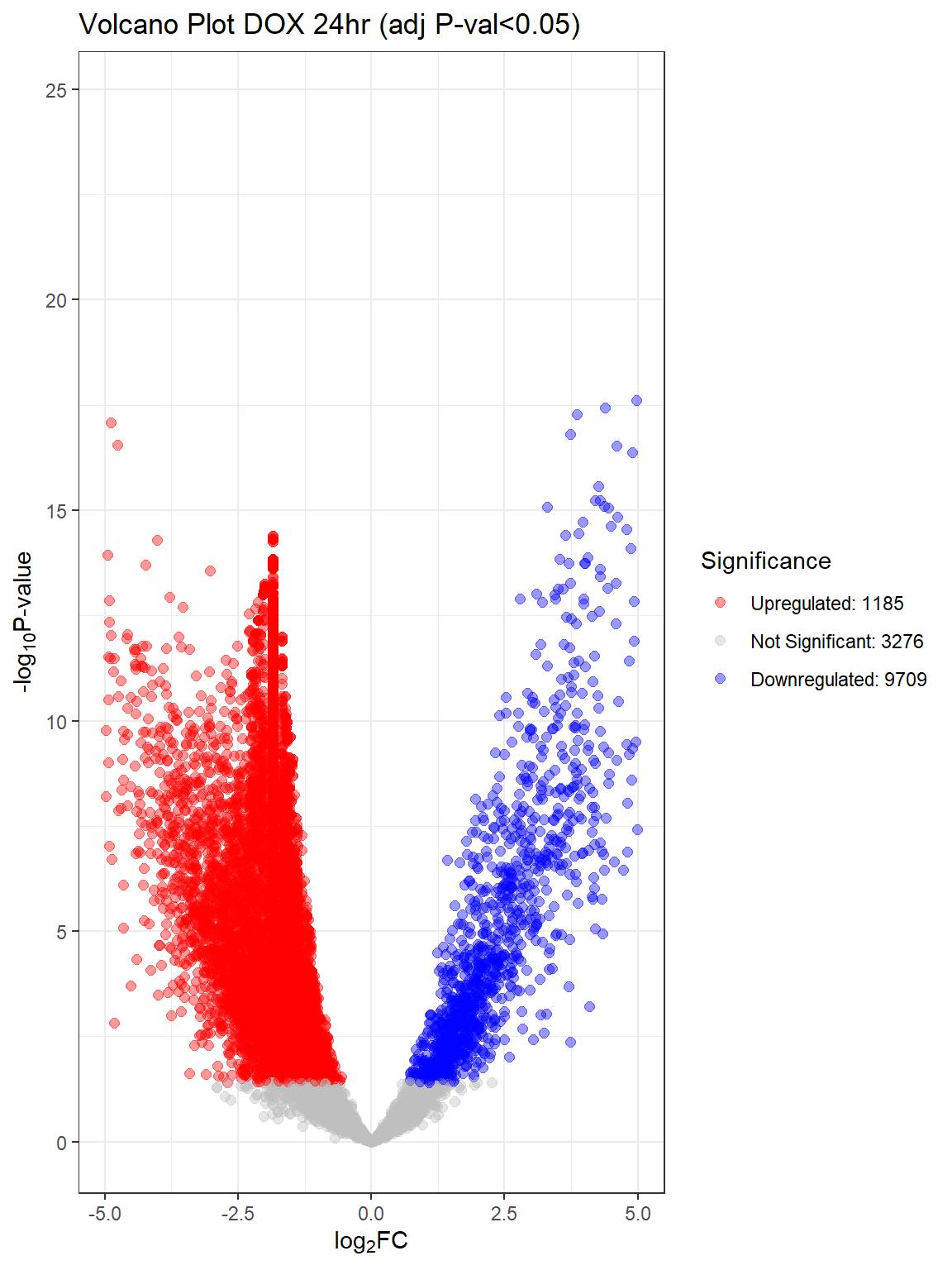
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
Warning: Removed 11 rows containing missing values or values outside the scale range
(`geom_point()`).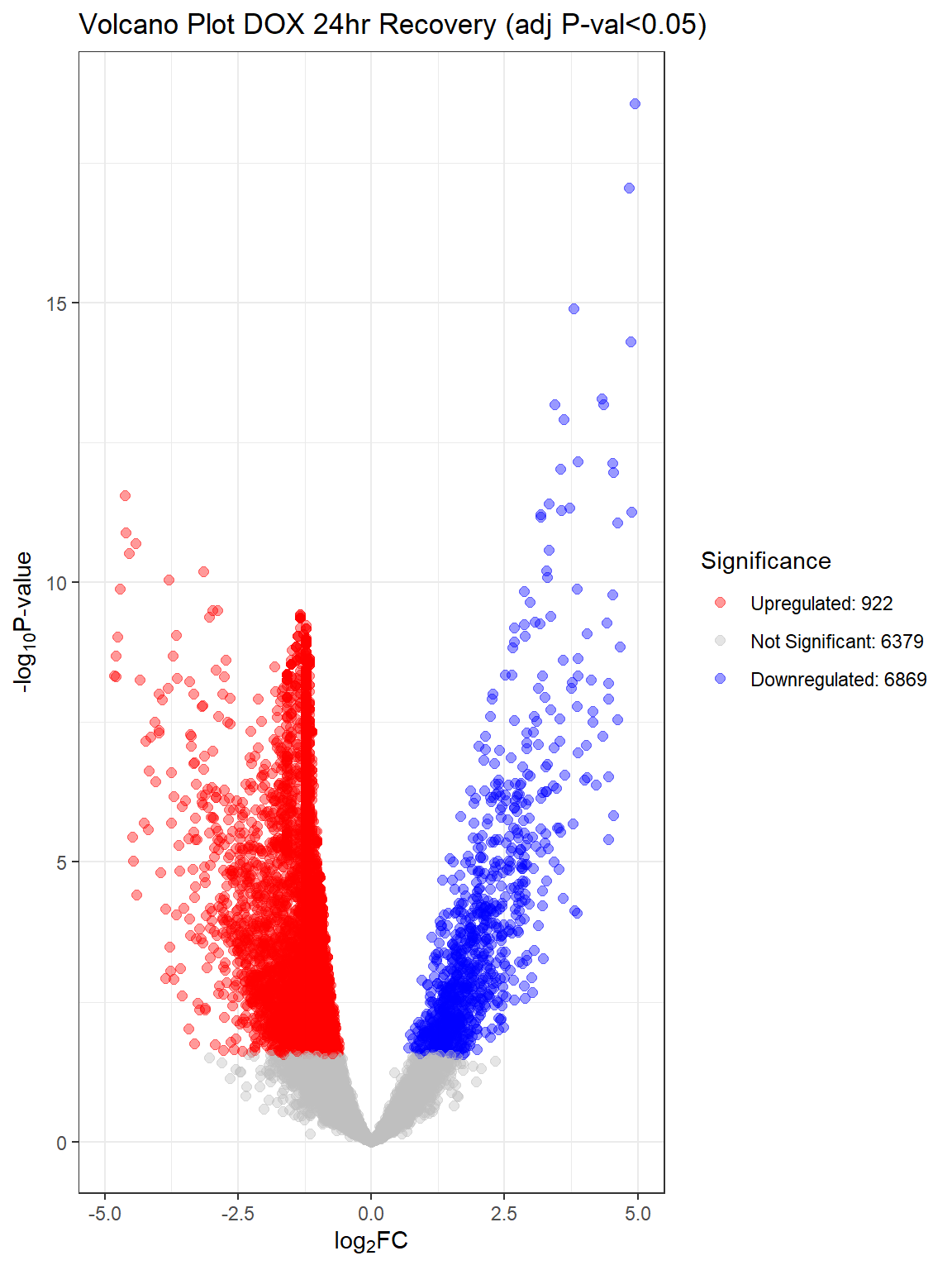
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
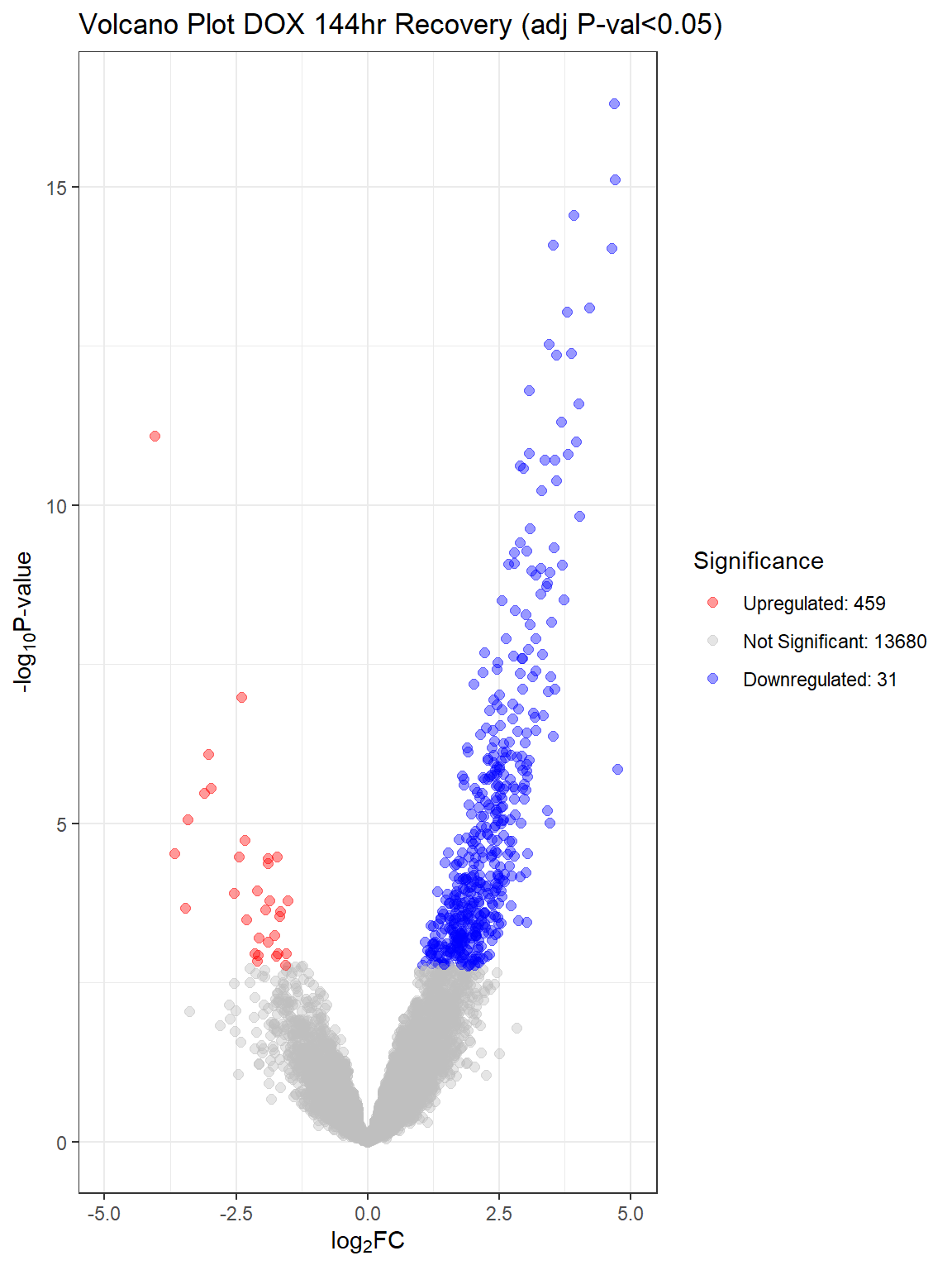
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#I want to add the number of DEG onto this for each
#make each volcano plot into an object so I can see how many rows it has
# Alternative function from Renee to combine them all into one image
# volcanosig <- function(df, psig.lvl,topg) {
# df <- df %>%
# mutate(threshold = ifelse(adj.P.Val > psig.lvl, "A", ifelse(adj.P.Val <= psig.lvl & logFC<=0,"B","C")))
#
# ggplot(df, aes(x=logFC, y=-log10(adj.P.Val))) +
# geom_point(aes(color=threshold))+
# xlab(expression("Log"[2]*" FC"))+
# ylim(0,30)+
# ylab(expression("-log"[10]*"P Value"))+
# scale_color_manual(values = c("black", "red","blue"))+
# theme_cowplot()+
# theme(legend.position = "none",
# plot.title = element_text(size = rel(0.8), hjust = 0.5),
# axis.title = element_text(size = rel(0.8)))
# }
#
# vol1 <- volcanosig(top.table_V.D24_dox, 0.01,0)+ ggtitle("Doxorubicin \n 24 hour")
# vol2 <- volcanosig(top.table_V.D24r_dox, 0.01,0)+ ggtitle("Doxorubicin \n 24 hour Recovery")
# vol3 <- volcanosig(top.table_V.D144r_dox, 0.01,0)+ ggtitle("Doxorubicin\n 144 hour Recovery")+ylab("")
#
#
# Volcanoplots <- plot_grid(vol1,vol2,vol3, nrow = 1, ncol = 3)
# Volcanoplots#now like the hypoxia paper, I'd like to put together comparisons between samples and conditions to check if they are more similar than othersPCA_data_d <- fC_Matrix_Full_cpm_filter_dox %>%
prcomp(.) %>%
t()
PCA_data_test_d <- (prcomp(t(fC_Matrix_Full_cpm_filter_dox), scale. = TRUE))
#ind_num_dox <- c("1", "1", "1", "1", "1", "1", "2", "2", "2", "2", "2", "2", "3", "3", "3", "3", "3", "3", "4", "4", "4", "4", "4", "4","5", "5", "5", "5", "5", "5", "6", "6", "6", "6", "6", "6", "6R", "6R", "6R", "6R", "6R", "6R")
#saveRDS(ind_num_dox, "data/ind_num_dox.RDS")
ind_num_dox <- readRDS("C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/ind_num_dox.RDS")
####now make an annotation for my PCA####
annot_d <- data.frame("samples" = colnames(fC_Matrix_Full_cpm_filter_dox)) %>% separate_wider_delim(., cols = samples, names = c("Tx", "Time", "Ind"), delim = "_", cols_remove = FALSE) %>% unite(., col = "Tx_Time", Tx, Time, sep = "_", remove = FALSE) %>% cbind(., ind_num_dox)
#saveRDS(annot_d, "C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/annot_dox.RDS")
#combine the prcomp matrix and annotation
annot_PCA_matrix_d <- PCA_data_test_d$x %>% cbind(., annot_d)
#now I can make a graph where I have filled values for individual! I have seven colors for seven individuals
# I have three fill values for three timepoints
#using annotation matrix above as well as annotated PCA matrix #(annot_PCA_matrix)
#extra info for colors in the graph (fill parameter)
fill_col_ind <- c("#66C2A5", "#FC8D62", "#1F78B4", "#E78AC3", "#A6D854", "#FFD92A", "#8B3E9B")
fill_col_ind_dark <- c("#003F5C", "#45AE91", "#58508D", "#BC4099", "#8B3E9B", "#FF6361", "#FF2362")
fill_col_tx_dox <- c("#63666D", "#499FBD")
fill_col_txtime_d <- c("#45AE91", "#58508D", "#BC4099", "#FF2362", "#A6D854", "#FC8D62")
#I want the color parameter to be treatment, the shape as time, and individual as number
####PC1/PC2####
annot_PCA_matrix_d %>% ggplot(., aes(x=PC1, y=PC2, size=10)) +
geom_point(aes(color = Tx, shape = Time)) +
scale_color_manual(values = c(fill_col_tx_dox))+
scale_shape_manual(values = c(15, 19, 17))+
geom_text_repel(aes(label = ind_num_dox))+
theme_bw(base_size = 10)+
ggtitle(expression("PCA of filtered log"[2]*"cpm"))+
guides(size="none")Warning: ggrepel: 27 unlabeled data points (too many overlaps). Consider
increasing max.overlaps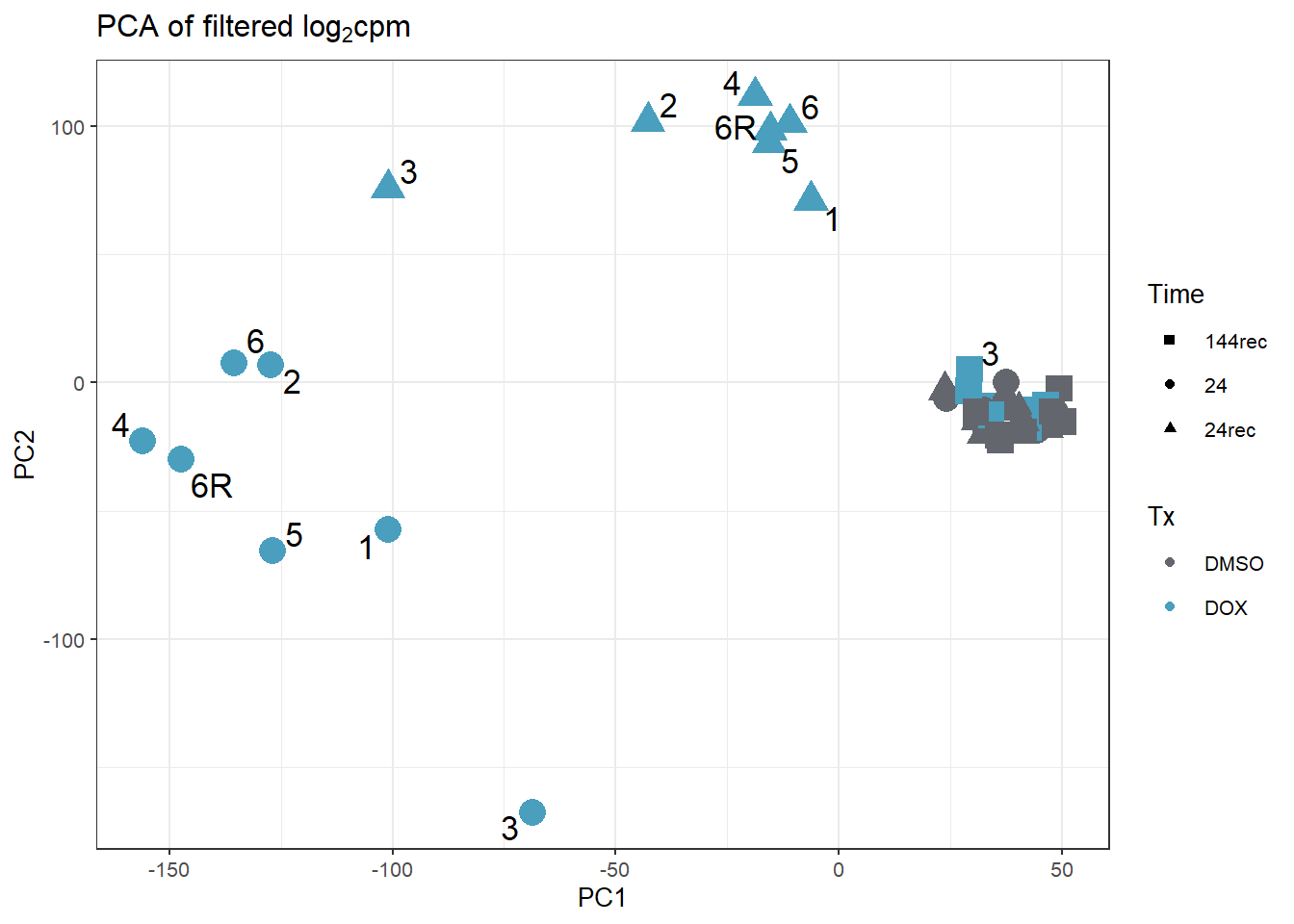
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
####PC1/PC2 autoplot####
prcomp_res_d <- prcomp(t(fC_Matrix_Full_cpm_filter_dox), scale. = FALSE, center = TRUE)
annot_prcomp_res_d <- prcomp_res_d$x %>% cbind(., annot_d)
ggplot2::autoplot(prcomp_res_d, data = annot_d, colour = "Tx_Time", size =4)+
theme_bw()+
scale_color_manual(values = c(fill_col_txtime_d))+
ggrepel::geom_text_repel(label= ind_num_dox)+
ggtitle("PCA of DOX filtered log2cpm")Warning: ggrepel: 8 unlabeled data points (too many overlaps). Consider
increasing max.overlaps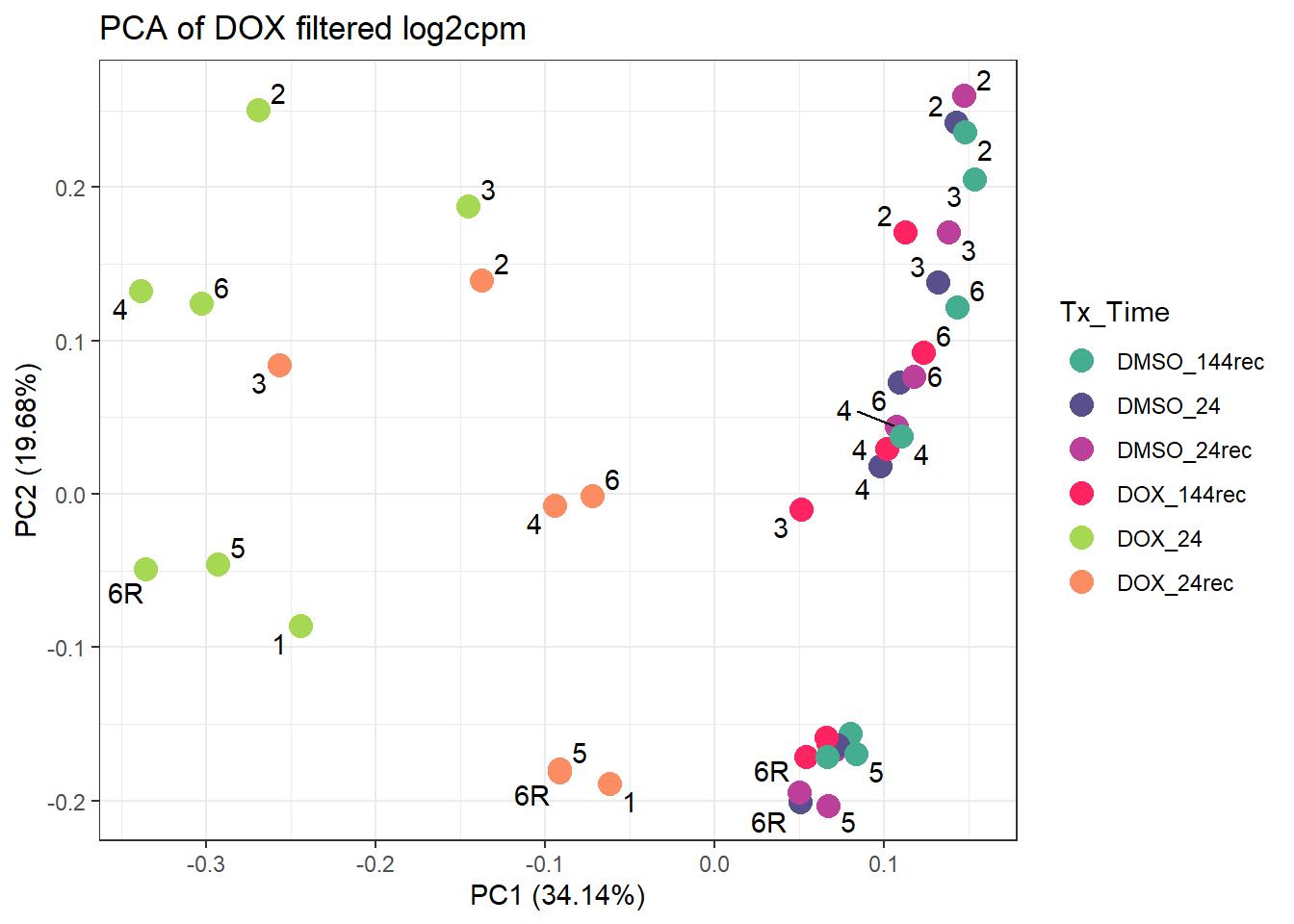
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
####PC2/PC3####
annot_PCA_matrix_d %>% ggplot(., aes(x=PC2, y=PC3, size=10)) +
geom_point(aes(color = Tx, shape = Time)) +
scale_color_manual(values = c(fill_col_tx_dox))+
scale_shape_manual(values = c(15, 19, 17))+
geom_text_repel(aes(label = ind_num_dox))+
theme_bw(base_size = 10)+
ggtitle(expression("PCA of filtered log"[2]*"cpm"))+
guides(size="none")Warning: ggrepel: 7 unlabeled data points (too many overlaps). Consider
increasing max.overlaps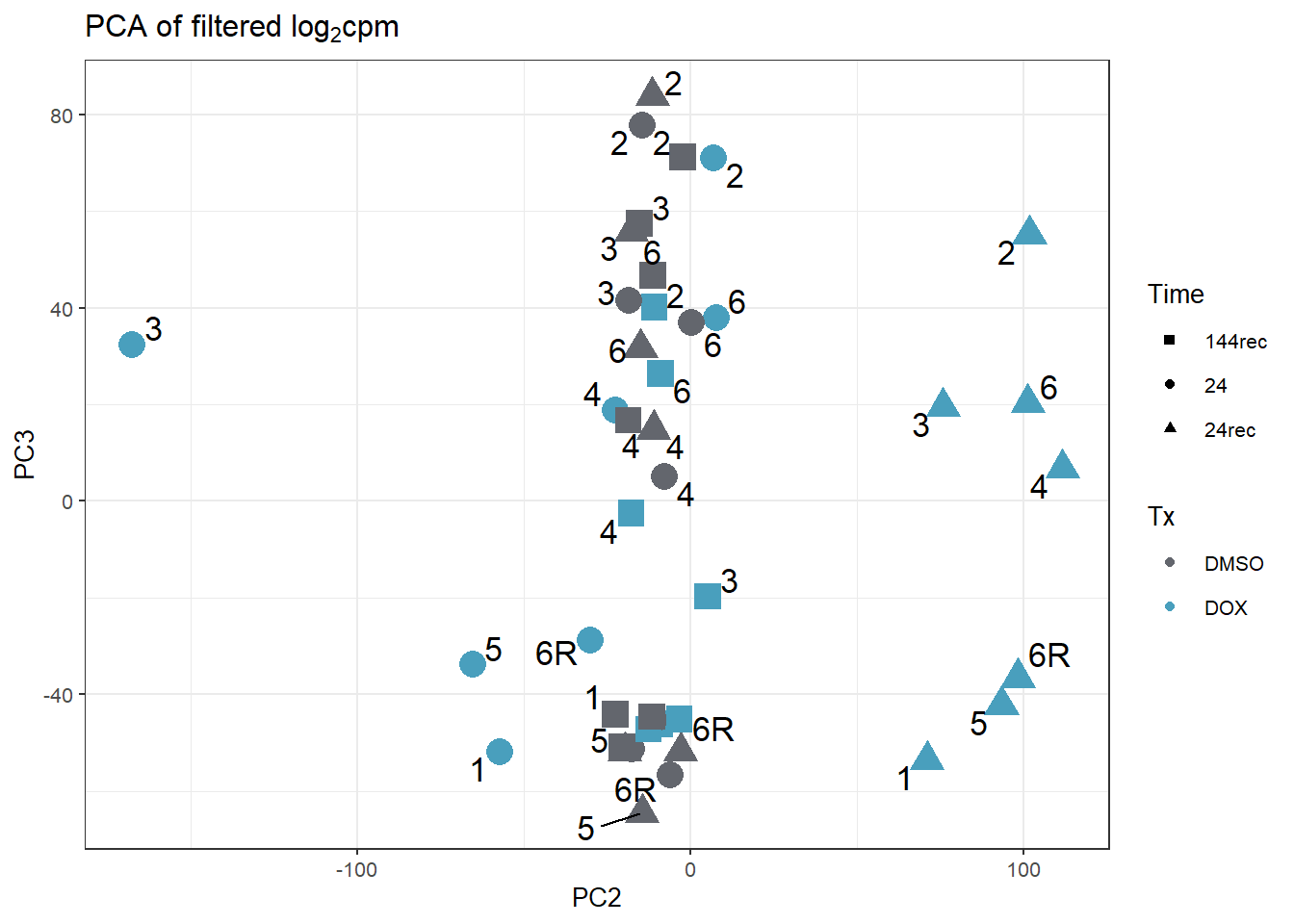
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
####PC3/PC4####
annot_PCA_matrix_d %>% ggplot(., aes(x=PC3, y=PC4, size=10)) +
geom_point(aes(color = Tx, shape = Time)) +
scale_color_manual(values = c(fill_col_tx_dox))+
scale_shape_manual(values = c(15, 19, 17))+
geom_text_repel(aes(label = ind_num_dox))+
theme_bw(base_size = 10)+
ggtitle(expression("PCA of filtered log"[2]*"cpm"))+
guides(size="none")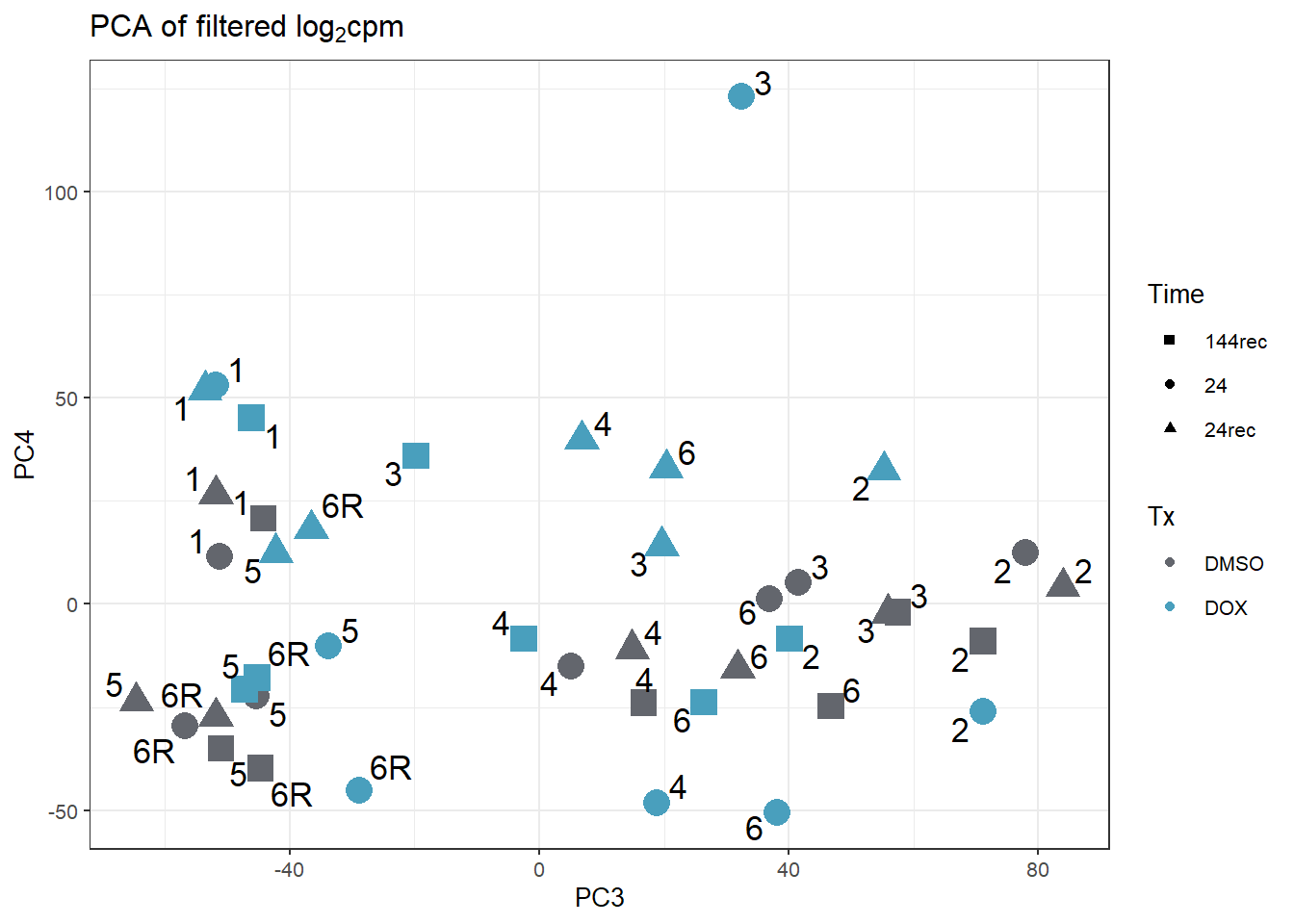
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
fC_Matrix_Full <- readRDS("data/fC_Matrix_Full.RDS")
fC_Matrix_Full_dox <- as.data.frame(fC_Matrix_Full) %>% dplyr::select(-(contains("FLUO")))
Counts_Full_df_dox <- data.frame(fC_Matrix_Full_dox)
#filter this dataframe by the filtered gene list I have
filt_gene_list_dox <- row.names(fC_Matrix_Full_cpm_filter_dox)
#saveRDS(filt_gene_list_dox, "data/filt_gene_list_dox.RDS")
counts_DE_df_dox <- Counts_Full_df_dox[! (rownames(Counts_Full_df_dox) %in% filt_gene_list_dox), ]
#saveRDS(counts_DE_df_dox, "data/counts_DE_df_dox.RDS")
RUV_filt_counts_dox <- fC_Matrix_Full_dox %>%
as.data.frame() %>%
dplyr::filter(., row.names(.)%in% filt_gene_list_dox)
#add in the annotation files
ind_num_dox <- readRDS("C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/ind_num_dox.RDS")
annot_d <- readRDS("C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/annot_dox.RDS")
# counts need to be integer values and in a numeric matrix
# note: the log transformation needs to be accounted for in the isLog argument in RUVs function.
counts_d <- as.matrix(RUV_filt_counts_dox)
dim(counts_d)[1] 14225 42#14225 genes
# Create a DataFrame for the phenoData
phenoData_d <- DataFrame(annot_d)
# Now create the RangedSummarizedExperiment necessary for RUVs input
# looks like it did need both the phenodata and the counts.
set_dox <- SummarizedExperiment(assays = counts_d, metadata = phenoData_d)
# Generate a background matrix
# The column "Cond" holds the comparisons that you actually want to make. DOX_24, DMSO_24,5FU_24, DOX_3,etc.
scIdx_d <-RUVSeq::makeGroups(phenoData_d$Tx_Time)
scIdx_d [,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 6 12 18 24 30 36 42
[2,] 2 8 14 20 26 32 38
[3,] 4 10 16 22 28 34 40
[4,] 5 11 17 23 29 35 41
[5,] 1 7 13 19 25 31 37
[6,] 3 9 15 21 27 33 39#now I've made all of the data I need for this - they are located in each section for k values
#DO NOT USE THESE COUNTS FOR LINEAR MODELING
#colors for all of the plots
fill_col_ind <- c("#66C2A5", "#FC8D62", "#1F78B4", "#E78AC3", "#A6D854", "#FFD92A", "#8B3E9B")
fill_col_ind_dark <- c("#003F5C", "#45AE91", "#58508D", "#BC4099", "#8B3E9B", "#FF6361", "#FF2362")
fill_col_tx_dox <- c("#63666D", "#499FBD")
fill_col_txtime_dox <- c("#45AE91", "#58508D", "#BC4099", "#FF2362", "#A6D854", "#FC8D62")
# before ruv
prcomp_res_d <- prcomp(t(counts_d), scale. = FALSE, center = TRUE)
ggplot2::autoplot(prcomp_res_d, data = annot_d, colour = "Tx_Time", size =4)+
theme_bw()+
scale_color_manual(values = c(fill_col_txtime_dox))+
ggrepel::geom_text_repel(label= ind_num_dox)+
ggtitle("No RUV")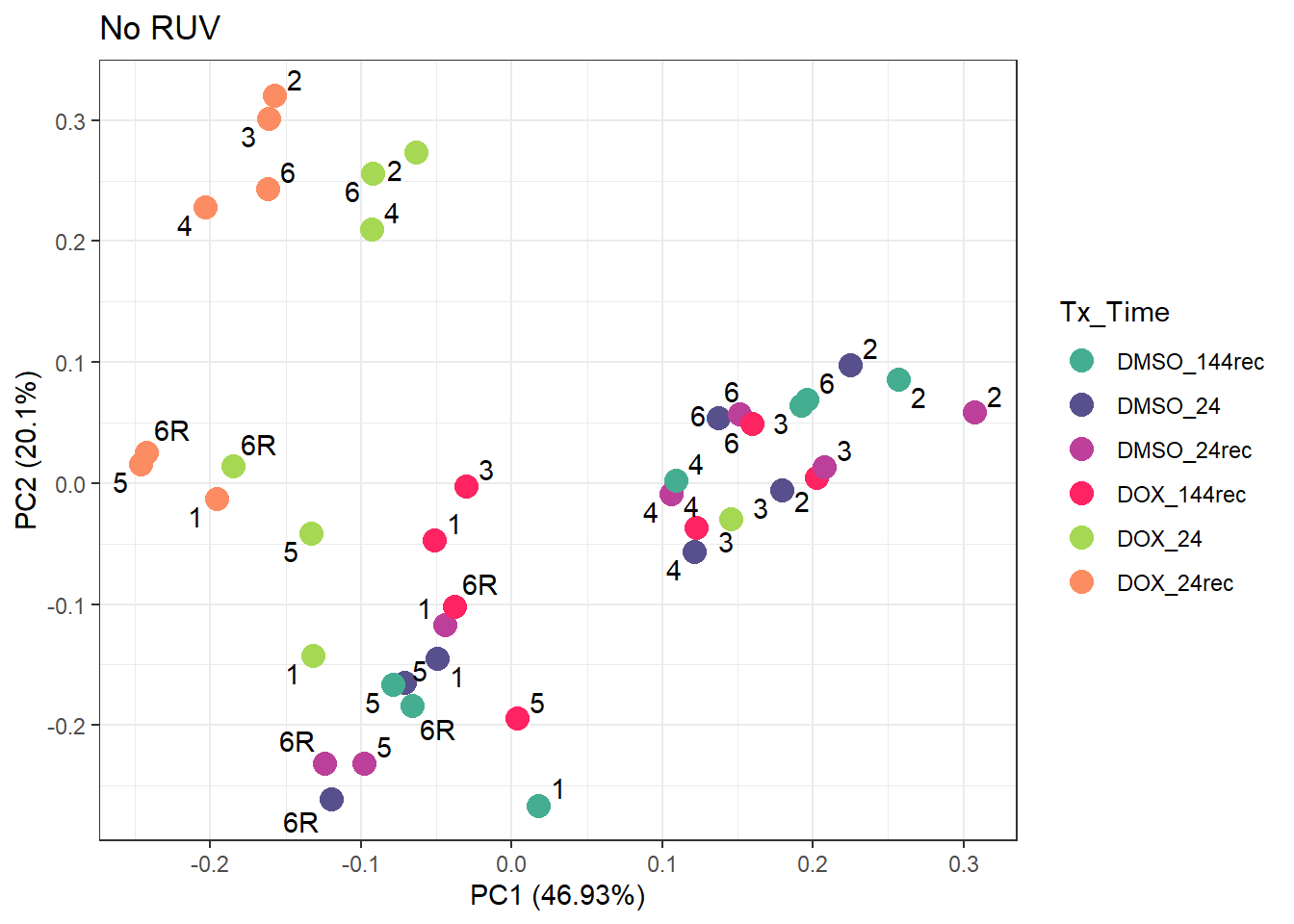
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
####new PCA plots no correction####
#PCA plots for each value of k attached in each section
prcomp_res_d <- prcomp(t(counts_d), scale. = FALSE, center = TRUE)
annot_prcomp_res_d <- prcomp_res_d$x %>% cbind(., annot_d)
group_2d <- rep(c("DOX_24",
"DMSO_24",
"DOX_24rec",
"DMSO_24rec",
"DOX_144rec",
"DMSO_144rec"), 7)
dge1_d <- DGEList.data.frame(counts = fC_Matrix_Full_dox, group = group_2d, genes = row.names(fC_Matrix_Full_dox))
#calculate the normalization factors with method TMM
dge1_calc_d <- calcNormFactors(dge1_d, method = "TMM")
#Pull out factors
snames1_d <- data.frame("samples" = colnames(dge1_calc_d)) %>% separate_wider_delim(., cols = samples, names = c("Treatment", "Time", "Individual"), delim = "_", cols_remove = FALSE)
snames1_ind_d <- snames1_d$Individual#Apply RUVs function from RUVSeq
#"k" will be iteratively adjusted over time depending on your PCA.
set_d <- RUVSeq::RUVs(x = counts_d, k =1, scIdx = scIdx_d, isLog = FALSE)
#get the ruv weights to put into the linear model. n weights = k.
#k=1
RUV_df_d <- set_d$W %>% as.data.frame()
RUV_df_d$Names <- rownames(RUV_df_d)
#Check that the names match
#k=1
RUV_df_rm_d <- RUV_df_d[RUV_df_d$Names %in% annot_d$samples, ]
RUV_1_d <- RUV_df_rm_d$W_1
# after ruv k=1
#PCA checks
#k=1
prcomp_res_1_d <- prcomp(t(set_d$normalizedCounts), scale. = FALSE, center = TRUE)
annot_prcomp_res_1_d <- prcomp_res_1_d$x %>% cbind(., annot_d)
ggplot2::autoplot(prcomp_res_1_d, data = annot_d, colour = "Tx_Time", size =4)+
theme_bw()+
scale_color_manual(values = c(fill_col_txtime_dox))+
ggrepel::geom_text_repel(label= ind_num_dox)+
ggtitle("RUVs Correction k=1")Warning: ggrepel: 5 unlabeled data points (too many overlaps). Consider
increasing max.overlaps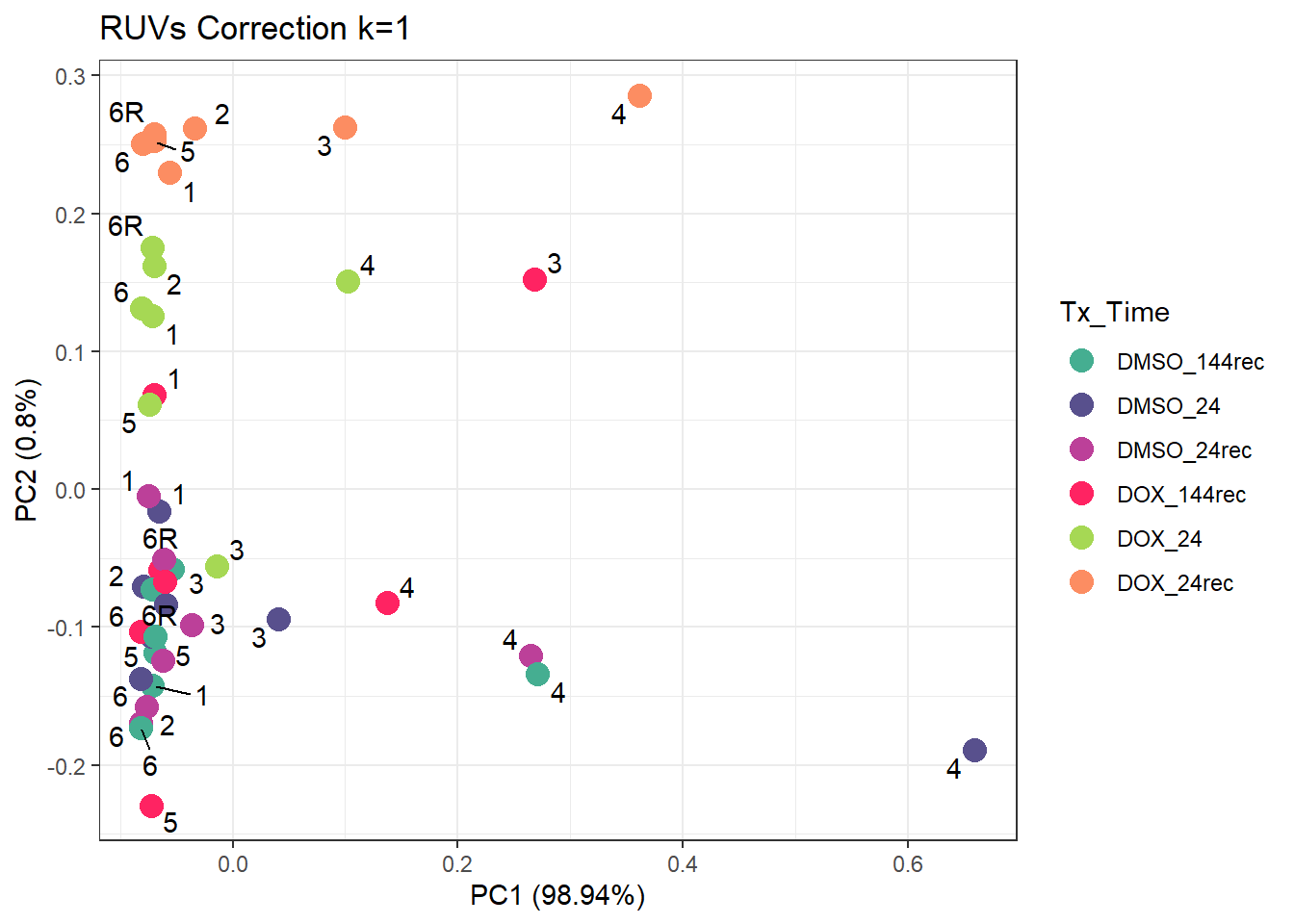
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#k=1
annot_d$samples == RUV_df_rm_d$Names [1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[16] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[31] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUEannot_d$RUV_1_d <- RUV_df_rm_d$W_1
#Create my model matrix
#k=1
mm_r1_d <- model.matrix(~0 + group_2d + RUV_1_d, data = annot_d)
p1_d <- voom(dge1_calc_d$counts, mm_r1_d, plot = TRUE)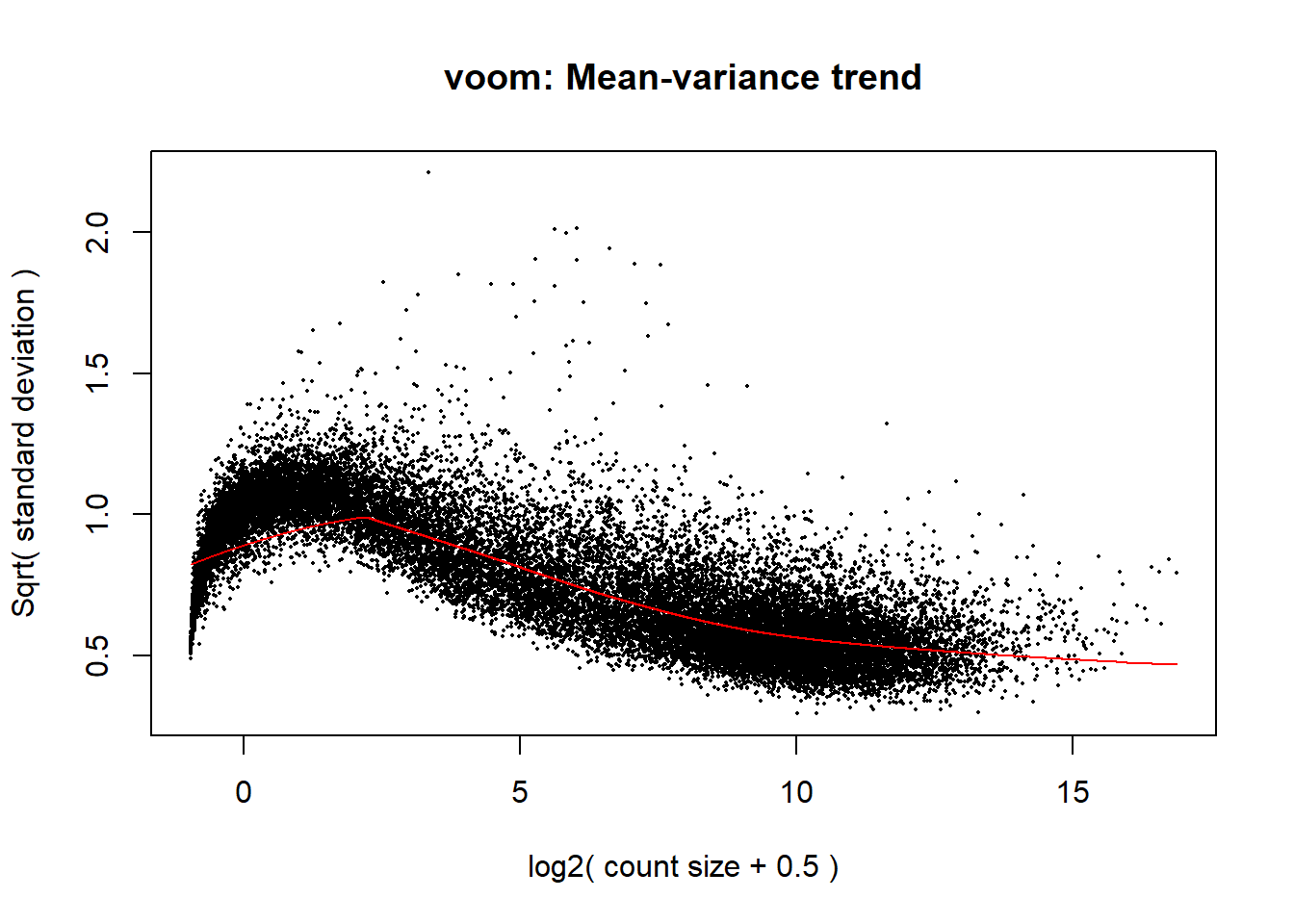
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
corfit1_d <- duplicateCorrelation(p1_d, mm_r1_d, block = snames1_ind_d)
v1_d <- voom(dge1_calc_d$counts, mm_r1_d, block = snames1_ind_d, correlation = corfit1_d$consensus)
fit1_d <- lmFit(v1_d, mm_r1_d, block = snames1_ind_d, correlation = corfit1_d$consensus)
#k=1
mm_r1_names_d <- str_replace(string = colnames(mm_r1_d), pattern = "group_2d", replacement = "")
design_d <- model.matrix(~ group_2d + RUV_1_d , annot_d)
colnames(mm_r1_d) <- mm_r1_names_d
#k=1
cm_r1_d <- makeContrasts(
V.D24 = DOX_24 - DMSO_24,
V.D24r = DOX_24rec - DMSO_24rec,
V.D144r = DOX_144rec - DMSO_144rec,
RUV_1_24 = RUV_1_d - DMSO_24,
RUV_1_24r= RUV_1_d - DMSO_24rec,
RUV_1_144r = RUV_1_d - DMSO_144rec,
levels = mm_r1_d
)
#k=1
vfit_r1_d <- lmFit(p1_d, mm_r1_d)
vfit_r1_d <- contrasts.fit(vfit_r1_d, contrasts = cm_r1_d)
#k=1
efit1_d <- eBayes(vfit_r1_d)
#k=1
results1_d = decideTests(efit1_d)
summary(results1_d) V.D24 V.D24r V.D144r RUV_1_24 RUV_1_24r RUV_1_144r
Down 5222 2650 486 13532 13539 13491
NotSig 14886 16841 26921 1645 1641 1721
Up 8287 8904 988 13218 13215 13183# V.D24 V.D24r V.D144r RUV_1_24 RUV_1_24r RUV_1_144r
# Down 5222 2650 486 13532 13539 13491
# NotSig 14886 16841 26921 1645 1641 1721
# Up 8287 8904 988 13218 13215 13183
#k=1
toptable_Dupcor_DOX_d <- topTable(efit1_d, coef = "V.D24", number = nrow(dge1_calc_d$counts), p.value = 1)
toptable_Dupcor_DOXrec_d <- topTable(efit1_d, coef = "V.D24r", number = nrow(dge1_calc_d$counts), p.value = 1)
toptable_Dupcor_DOX144_d <- topTable(efit1_d, coef = "V.D144r", number = nrow(dge1_calc_d$counts), p.value = 1)
#k=1 plots
toptable_Dupcor_DOX_d$logFC %>% hist(, main= "RUVs k=1 DOX 24hr Toptable")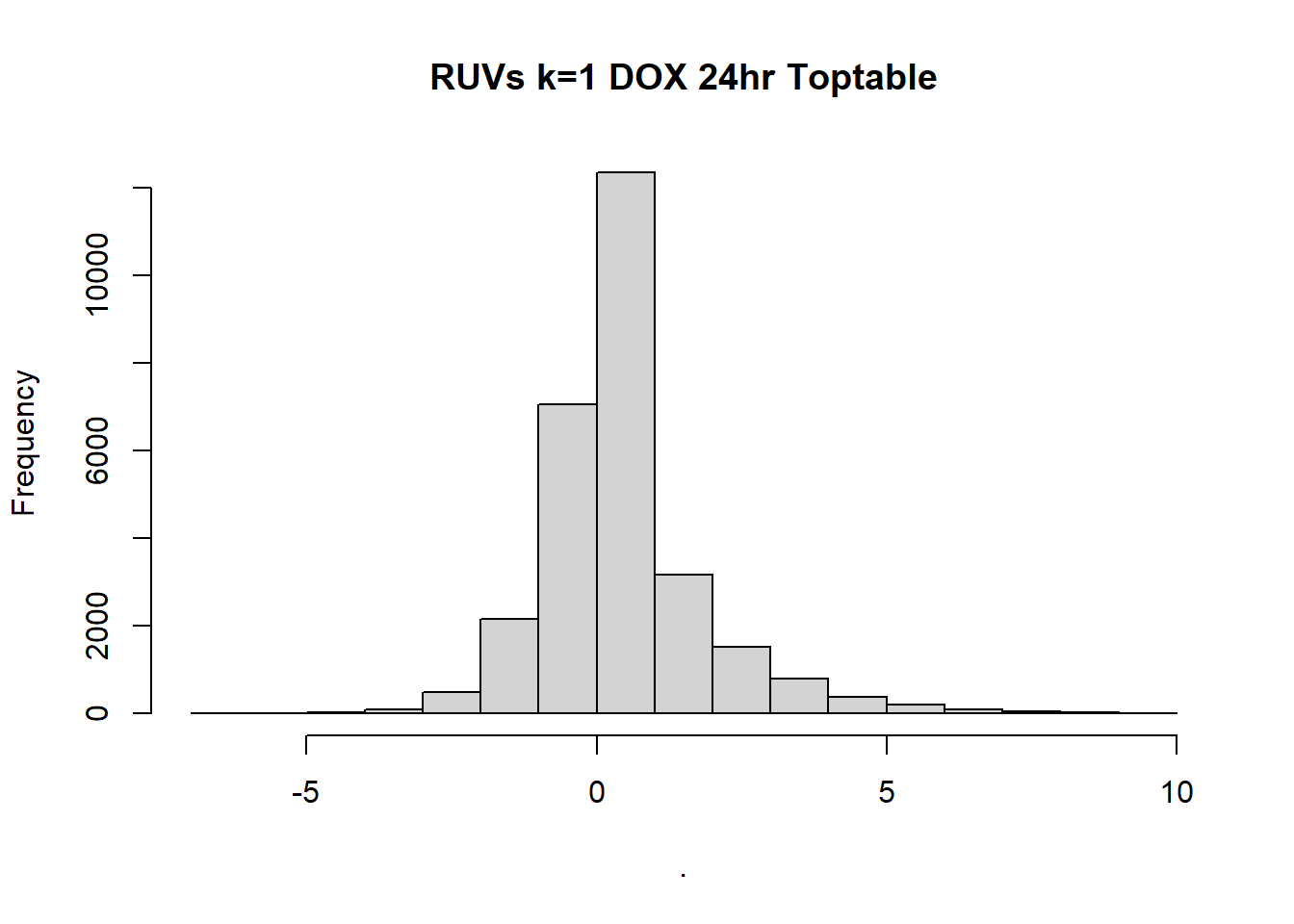
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
toptable_Dupcor_DOXrec_d$logFC %>% hist(, main = "RUVs k=1 DOX 24Rec Toptable")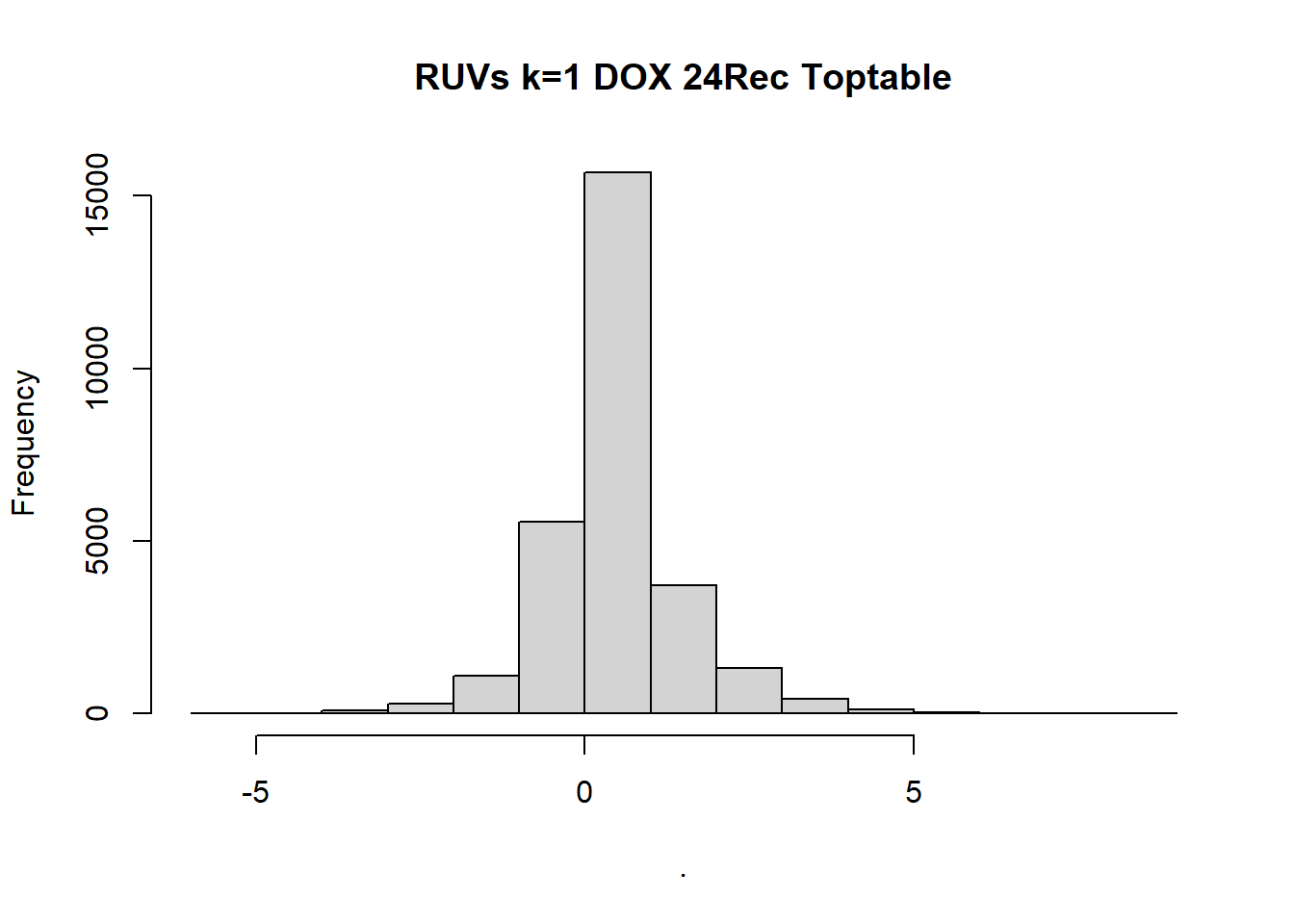
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
toptable_Dupcor_DOX144_d$logFC %>% hist(, main = "RUVs k=1 DOX 144Rec Toptable")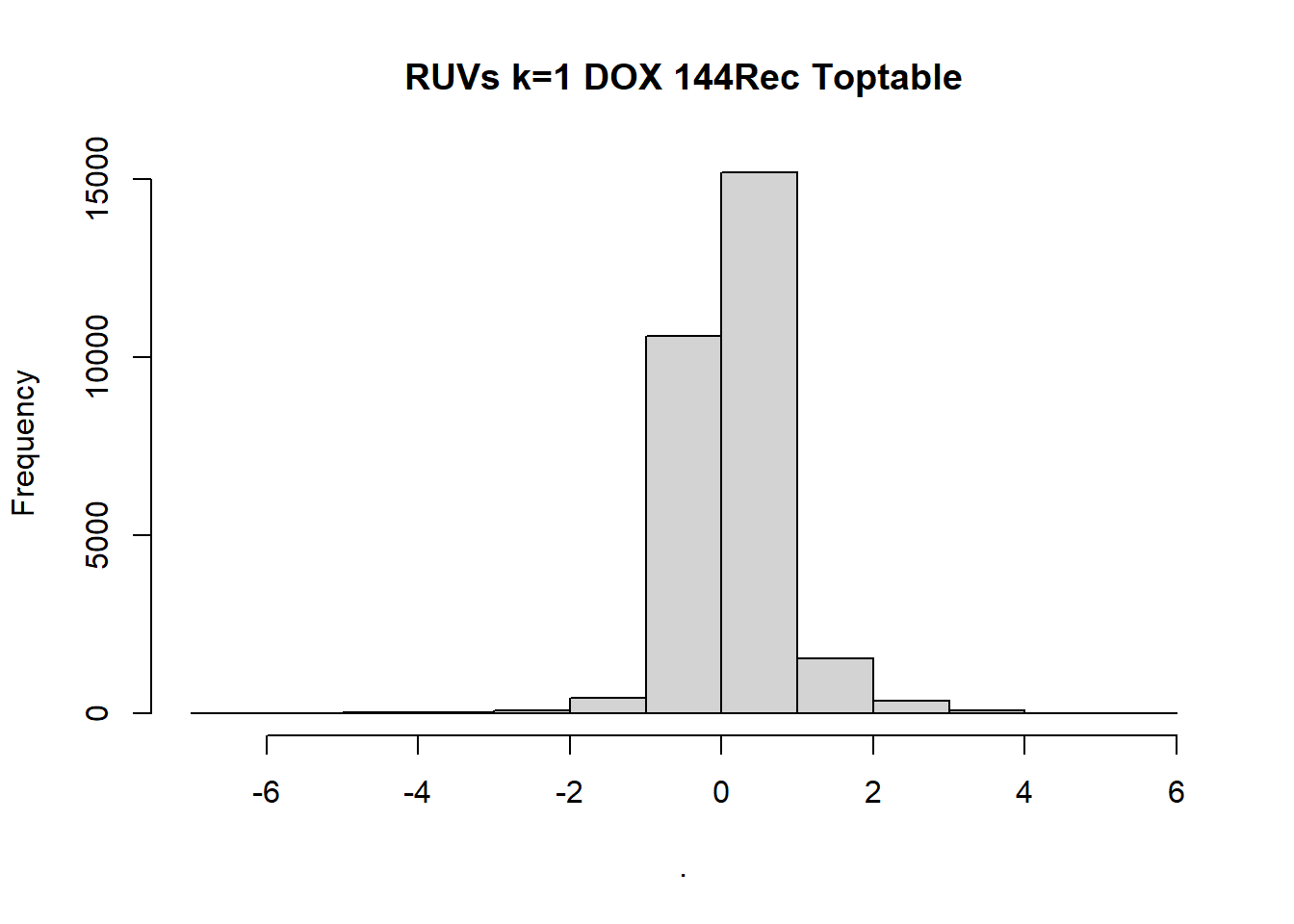
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#function is on the previous
annot_data_d <- annot_d %>% dplyr::select("Tx_Time", "Time", "Tx", "Ind", "samples")
Cormotif_d <- counts_DE_df_dox %>% cpm(., log = TRUE)
Cormotif_df_d <- as.data.frame(Cormotif_d)
groupid_d <- rep(c(1, 2, 3, 4, 5, 6), 7)
compid_d <- data.frame(c1 = c(1, 3, 5), c2 = c(2, 4, 6))#set.seed(12345)
# initial_cormotif_dox <- cormotiffit(exprs = Cormotif_d,
# groupid = groupid_d,
# compid = compid_d,
# K=1:8 , max.iter = 500, runtype = "logCPM")
#only need to run this once!
#save this to an object so I can retrieve it as needed
#saveRDS(initial_cormotif_dox, "data/initial_cormotif_dox.RDS")
initial_cormotif_dox <- readRDS("data/initial_cormotif_dox.RDS")plotIC(initial_cormotif_dox)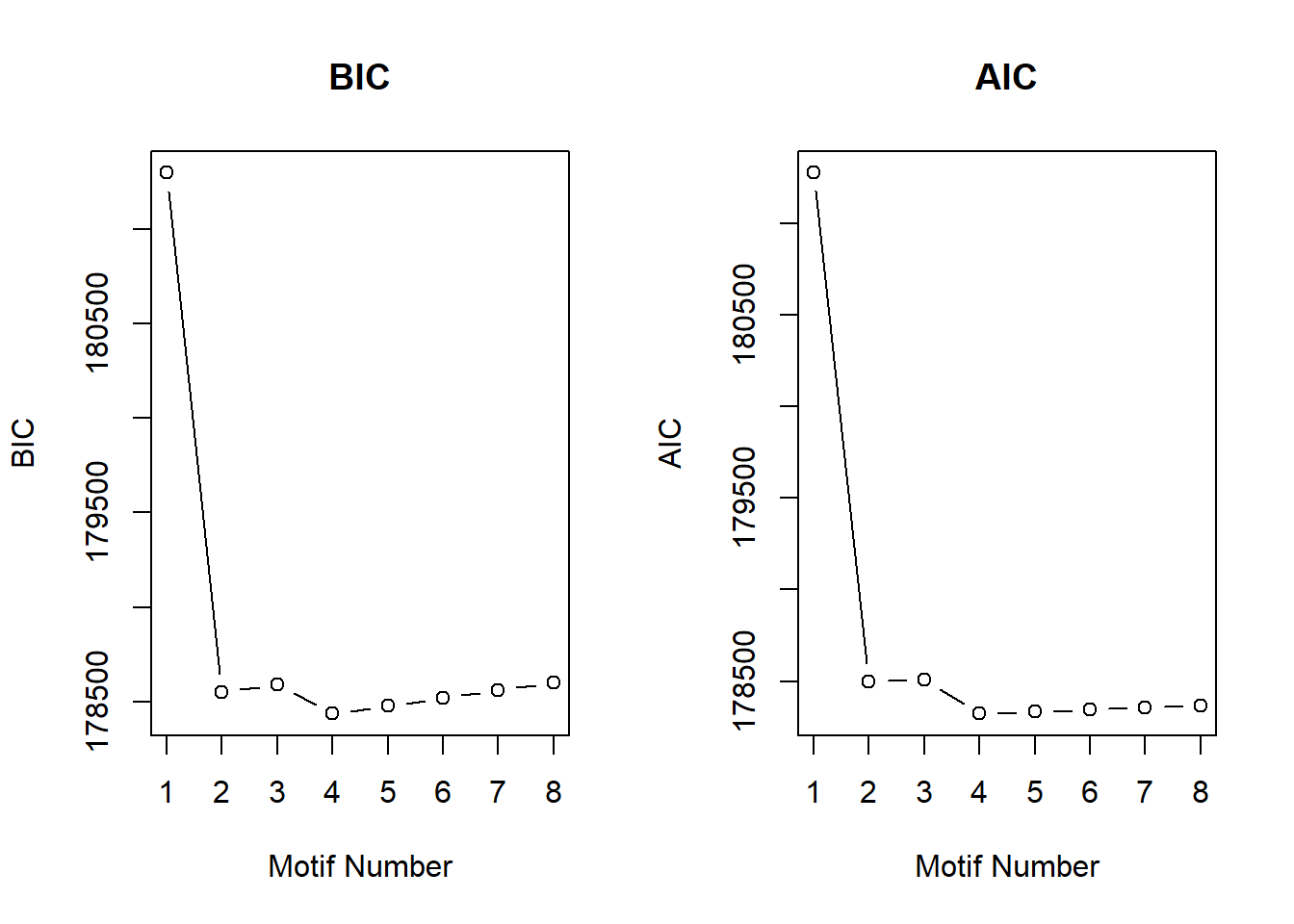
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
plotMotif(initial_cormotif_dox)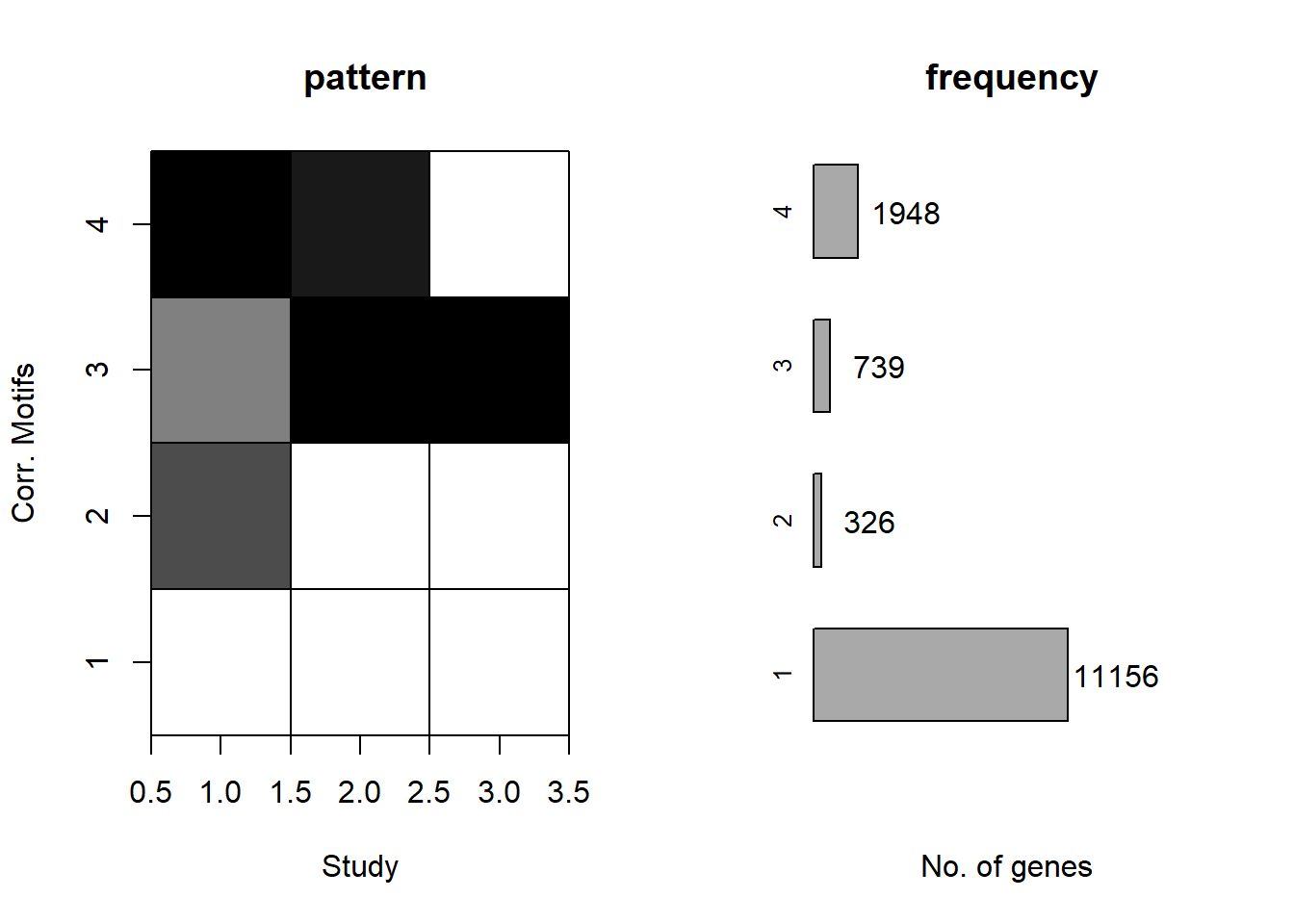
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
myColors <- rev(c("#FFFFFF", "#E6E6E6" ,"#CCCCCC", "#B3B3B3", "#999999", "#808080", "#666666","#4C4C4C", "#333333", "#191919","#000000"))
plot.new()
legend('bottomleft',fill=myColors, legend =rev(c("0", "0.1", "0.2", "0.3", "0.4", "0.5", "0.6", "0.7", "0.8","0.9", "1")), box.col="white",title = "Probability\nlegend", horiz=FALSE,title.cex=.8)
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
topgenelist_d <- generank(initial_cormotif_dox$bestmotif$p.post)
rownames(topgenelist_d) <- rownames(Cormotif_df_d)
motif_prob_d <- initial_cormotif_dox$bestmotif$clustlike
rownames(motif_prob_d) <- rownames(topgenelist_d)
#saveRDS(motif_prob_d, "data/Cormotif_prob_gene_list_doxonly.RDS")
#Define the gene probability groups - I have 4
clust1_d <- motif_prob_d %>%
as.data.frame() %>%
filter(V1>0.57) %>%
rownames
#using a filter of >0.57 I get 11130 genes compared to 11156 above
length(clust1_d)[1] 11130#11482 > 0.5
#11130 > 0.57
#is this clust1 a non-response?
clust1_d_df <- as.data.frame(clust1_d)
#example gene - UID 4681 - NBL1
NBL1_motif1 <- Cormotif_df_d %>%
rownames_to_column(var = "entrezgene_id") %>%
dplyr::filter(entrezgene_id == "4681")
NBL1_motif1_long <- melt(NBL1_motif1,
id.vars = c("entrezgene_id"),
variable.name = "Sample",
value.name = "log2cpm")
#now add in my factors like time, tx, tx_time, and ind by breaking up the Sample column
NBL1_motif1_long_df <- data.frame(tx = factor(tx_names2, levels = unique(tx_names2)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names2, levels = unique(txtime_names2)),
time = factor(time_names2, levels = unique(time_names2)))
NBL1_motif1_long_factors <- cbind(NBL1_motif1_long_df, NBL1_motif1_long)
NBL1_motif1_long_factors %>% ggplot(aes(x = txtime, y = log2cpm))+
geom_boxplot(aes(fill = tx))+
geom_point(aes(color = ind)) +
labs(title = "NBL1")+
theme_bw(base_size = 16)+
scale_fill_manual(values = c(tx_col))+
scale_color_manual(values = c(ind_col))+
xlab("Conditions")+
ylab("log2cpm")+
ylim(0,10)+
theme(plot.title = element_text(face = "italic"))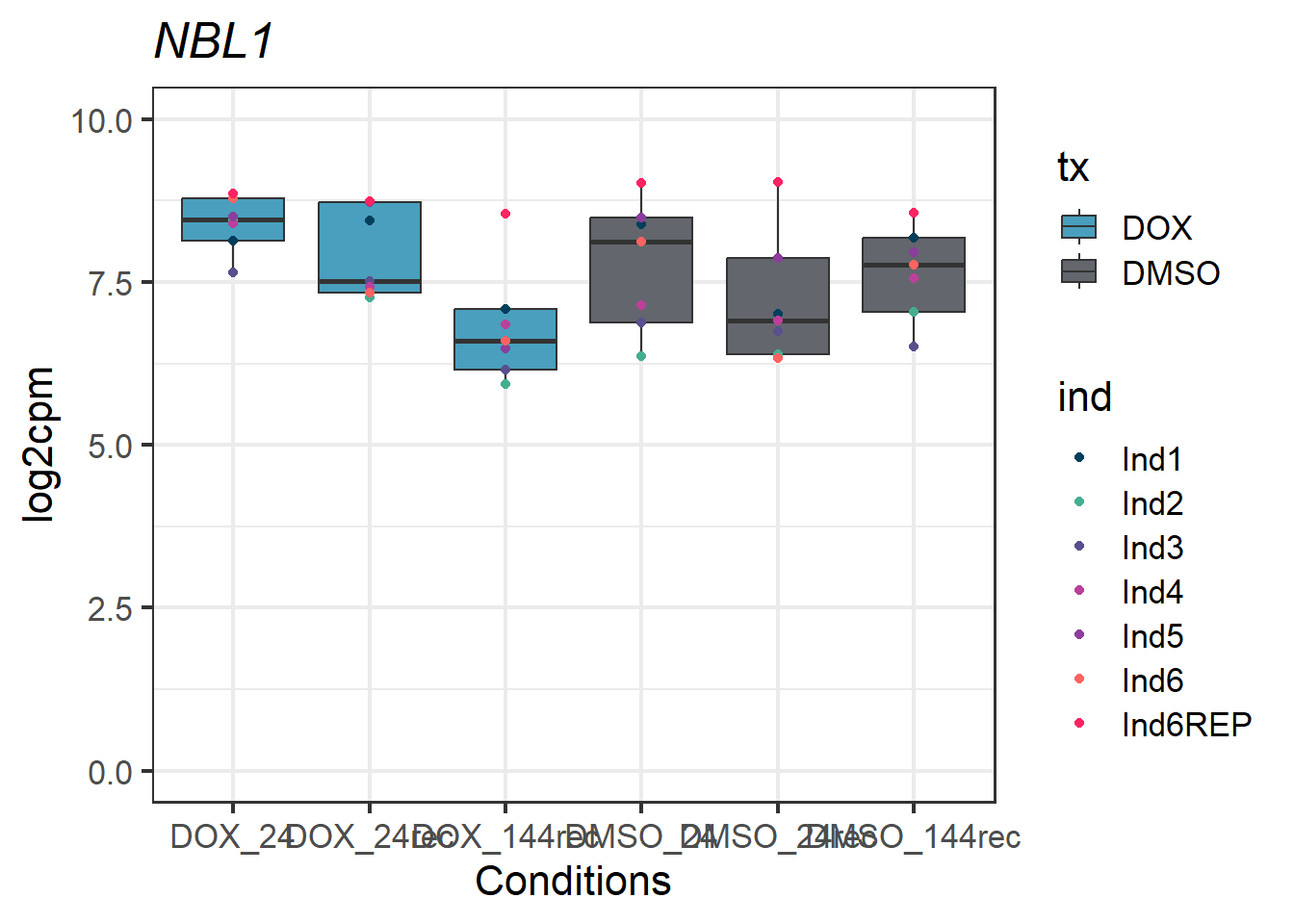
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#saveRDS(CDKN1A_geneplot_Dox, "data/CDKN1A_geneplot_Dox.RDS")
clust2_d <- motif_prob_d %>%
as.data.frame() %>%
filter(V2>0.132) %>%
rownames
#using a filter of >0.132 I get the closest - 326 original vs 327 genes here
length(clust2_d)[1] 327#0 > 0.5
#326 > 0.132
#is clust2 a dox early response?
clust2_d_df <- as.data.frame(clust2_d)
#example gene - 597 - BCL2A1
BCL2A1_motif1 <- Cormotif_df_d %>%
rownames_to_column(var = "entrezgene_id") %>%
dplyr::filter(entrezgene_id == "597")
BCL2A1_motif1_long <- melt(BCL2A1_motif1,
id.vars = c("entrezgene_id"),
variable.name = "Sample",
value.name = "log2cpm")
#now add in my factors like time, tx, tx_time, and ind by breaking up the Sample column
BCL2A1_motif1_long_df <- data.frame(tx = factor(tx_names2, levels = unique(tx_names2)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names2, levels = unique(txtime_names2)),
time = factor(time_names2, levels = unique(time_names2)))
BCL2A1_motif1_long_factors <- cbind(BCL2A1_motif1_long_df, BCL2A1_motif1_long)
BCL2A1_motif1_long_factors %>% ggplot(aes(x = txtime, y = log2cpm))+
geom_boxplot(aes(fill = tx))+
geom_point(aes(color = ind)) +
labs(title = "BCL2A1")+
theme_bw(base_size = 16)+
scale_fill_manual(values = c(tx_col))+
scale_color_manual(values = c(ind_col))+
xlab("Conditions")+
ylab("log2cpm")+
ylim(0,10)+
theme(plot.title = element_text(face = "italic"))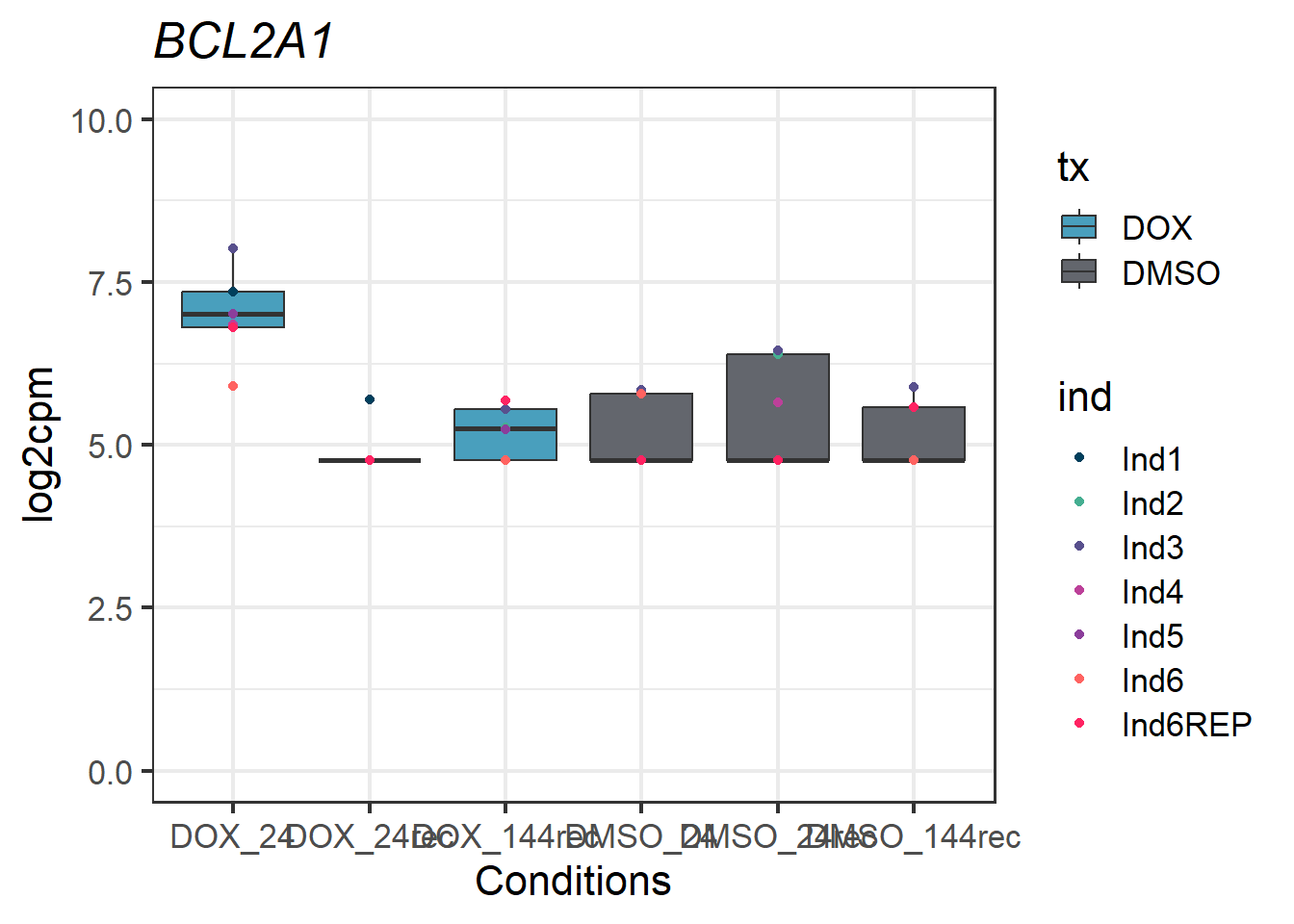
clust3_d <- motif_prob_d %>%
as.data.frame() %>%
filter(V3>0.3) %>%
rownames
#using a filter of >0.3 I get 740 genes vs original 739 genes
length(clust3_d)[1] 740#538 > 0.5
#740 > 0.3
#is clust 3 a late response?
clust3_d_df <- as.data.frame(clust3_d)
#example gene - 8856 - NR1I2
NR1I2_motif1 <- Cormotif_df_d %>%
rownames_to_column(var = "entrezgene_id") %>%
dplyr::filter(entrezgene_id == "8856")
NR1I2_motif1_long <- melt(NR1I2_motif1,
id.vars = c("entrezgene_id"),
variable.name = "Sample",
value.name = "log2cpm")
#now add in my factors like time, tx, tx_time, and ind by breaking up the Sample column
NR1I2_motif1_long_df <- data.frame(tx = factor(tx_names2, levels = unique(tx_names2)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names2, levels = unique(txtime_names2)),
time = factor(time_names2, levels = unique(time_names2)))
NR1I2_motif1_long_factors <- cbind(NR1I2_motif1_long_df, NR1I2_motif1_long)
NR1I2_motif1_long_factors %>% ggplot(aes(x = txtime, y = log2cpm))+
geom_boxplot(aes(fill = tx))+
geom_point(aes(color = ind)) +
labs(title = "NR1I2")+
theme_bw(base_size = 16)+
scale_fill_manual(values = c(tx_col))+
scale_color_manual(values = c(ind_col))+
xlab("Conditions")+
ylab("log2cpm")+
ylim(0,10)+
theme(plot.title = element_text(face = "italic"))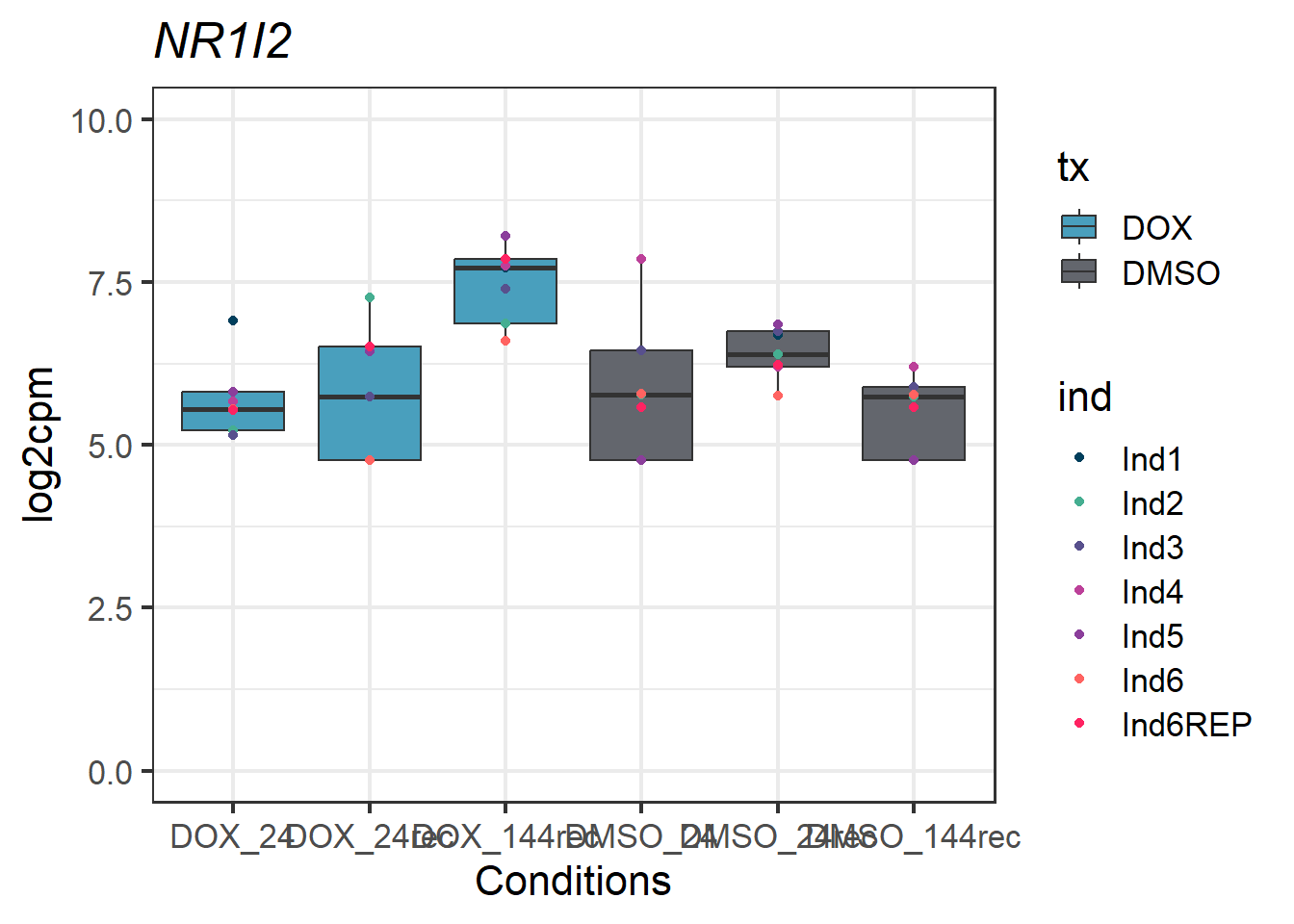
#cluster 4
clust4_d <- motif_prob_d %>%
as.data.frame() %>%
filter(V4>0.4) %>%
rownames
#using >0.4 as my cutoff, I get 1938 genes as compared to original 1948 genes
length(clust4_d)[1] 1938#1576 > 0.5
#1938 > 0.4
#is clust4 a DOX late response?
clust4_d_df <- as.data.frame(clust4_d)
#example gene - 6725 - SRMS
SRMS_motif1 <- Cormotif_df_d %>%
rownames_to_column(var = "entrezgene_id") %>%
dplyr::filter(entrezgene_id == "6725")
SRMS_motif1_long <- melt(SRMS_motif1,
id.vars = c("entrezgene_id"),
variable.name = "Sample",
value.name = "log2cpm")
#now add in my factors like time, tx, tx_time, and ind by breaking up the Sample column
SRMS_motif1_long_df <- data.frame(tx = factor(tx_names2, levels = unique(tx_names2)),
ind = factor(ind_names, levels = unique(ind_names)),
txtime = factor(txtime_names2, levels = unique(txtime_names2)),
time = factor(time_names2, levels = unique(time_names2)))
SRMS_motif1_long_factors <- cbind(SRMS_motif1_long_df, SRMS_motif1_long)
SRMS_motif1_long_factors %>% ggplot(aes(x = txtime, y = log2cpm))+
geom_boxplot(aes(fill = tx))+
geom_point(aes(color = ind)) +
labs(title = "SRMS")+
theme_bw(base_size = 16)+
scale_fill_manual(values = c(tx_col))+
scale_color_manual(values = c(ind_col))+
xlab("Conditions")+
ylab("log2cpm")+
ylim(0,10)+
theme(plot.title = element_text(face = "italic"))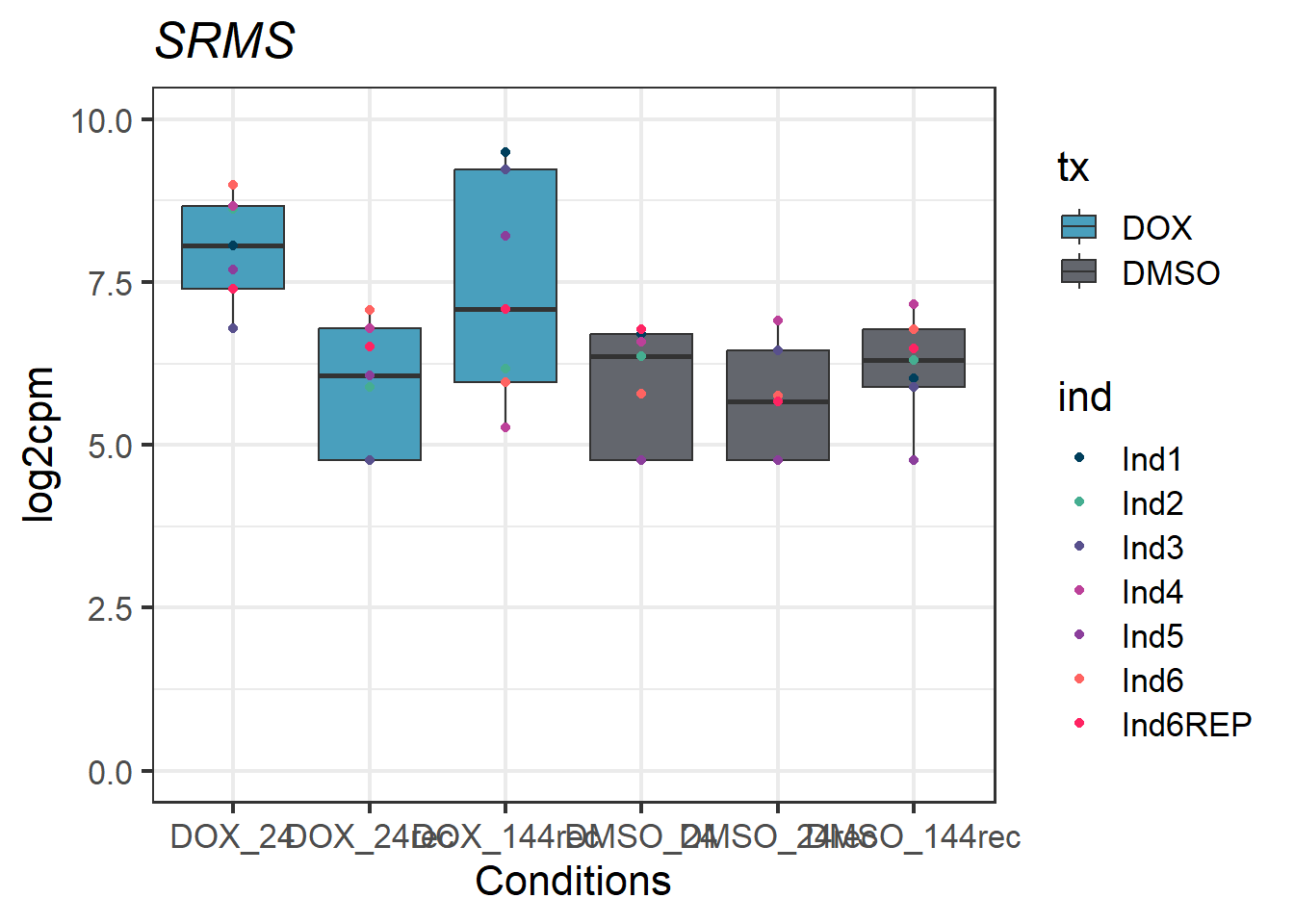
#now let's make a pie chart with the above number of genes for each motif
clusterdata_d_adj <- data.frame(
Category = c("No Response","Acute Response", "Late Response", "Early Sustained Response"),
Value = c(11130, 327, 740, 1938)
)
piecolors <- c("No Response" = "#007896",
"Acute Response" = "#079255",
"Late Response" = "#58508D",
"Early Sustained Response" = "#BC5090")
#make a piechart of these distributions
clusterdata_d_adj %>% ggplot(aes(x = "", y = Value, fill = Category))+
geom_bar(width = 1, stat = "identity")+
coord_polar("y", start = 0)+
geom_text(aes(label = Value),
position = position_stack(vjust = 0.5),
size = 4, color = "black")+
labs(title = "Distribution of Gene Clusters Identified By Cormotif", x = NULL, y = NULL)+
theme_void()+
scale_fill_manual(values = piecolors)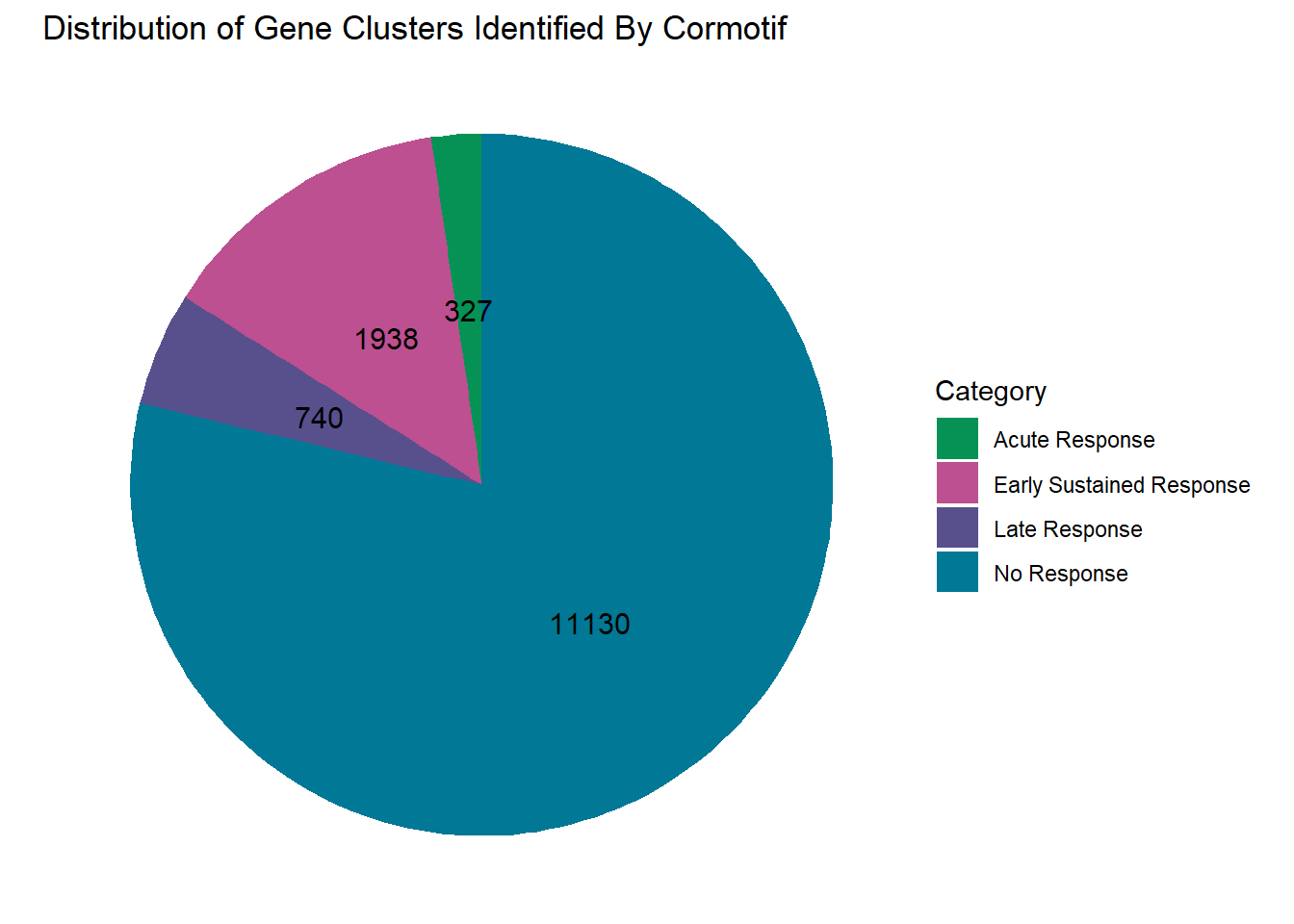
#now let's look at each of these motifs by study with the posterior probability
#changing the cutoffs based on the look of each motif - if it's very dark = > 0.5
#if white < 0.5
#if grey try a less stringent cutoff > 0.1
#ideally each of these are similar to the number of genes in the above plot
#gene_postprob_motif <- initial_cormotif_dox$bestmotif$p.post
#rownames(gene_postprob_motif) <- rownames(Cormotif_df_d)
#saveRDS(gene_postprob_motif, "data/gene_postprob_motif.RDS")
gene_postprob_motif <- readRDS("data/gene_postprob_motif.RDS")
gene_postprob_motif_df <- as.data.frame(gene_postprob_motif)
#motif 1 no response p.prob
prob_motif_1 <- rownames(gene_postprob_motif_df[(gene_postprob_motif_df[,1] < 0.62
& gene_postprob_motif_df[,2] < 0.62
& gene_postprob_motif_df[,3] < 0.62),])
length(prob_motif_1)[1] 11157#10903 genes post prob
#gene example: 943 - TNFRSF8
TNFRF8_gene <-
#10903 genes post prob - close to value of 11156 on cormotif pattern graph
#gene example: 11009 - IL24
IL24_gene_motif1 <-
#motif 2 acute response
prob_motif_2 <- rownames(gene_postprob_motif_df[(gene_postprob_motif_df[,1] > 0.99
& gene_postprob_motif_df[,2] < 0.5
& gene_postprob_motif_df[,3] < 0.5),])
length(prob_motif_2)[1] 277#2586 genes post prob
#motif 3 late response
prob_motif_3 <- rownames(gene_postprob_motif_df[(gene_postprob_motif_df[,1] > 0.1
& gene_postprob_motif_df[,2] > 0.5
& gene_postprob_motif_df[,3] > 0.5),])
length(prob_motif_3)[1] 451#451 genes post prob
#motif 4 early sustained response
prob_motif_4 <- rownames(gene_postprob_motif_df[(gene_postprob_motif_df[,1] > 0.5
& gene_postprob_motif_df[,2] > 0.1
& gene_postprob_motif_df[,3] < 0.5),])
length(prob_motif_4)[1] 2600#2600 genes post prob
#this totals 16540clusterdata_postprob <- data.frame(
Category = c("No Response", "Acute Response", "Late Response", "Early Sustained Response"),
Value = c(10903, 2586, 451, 2600)
)
piecolors_2 <- c("No Response" = "#007896",
"Acute Response" = "#58508D",
"Late Response" = "#ABCC59",
"Early Sustained Response" = "#BC5090")
#make a piechart of these distributions
clusterdata_postprob %>% ggplot(aes(x = "", y = Value, fill = Category))+
geom_bar(width = 1, stat = "identity")+
coord_polar("y", start = 0)+
geom_text(aes(label = Value),
position = position_stack(vjust = 0.5),
size = 4, color = "black")+
labs(title = "Distribution of Gene Clusters Identified By Cormotif", x = NULL, y = NULL)+
theme_void()+
scale_fill_manual(values = piecolors_2)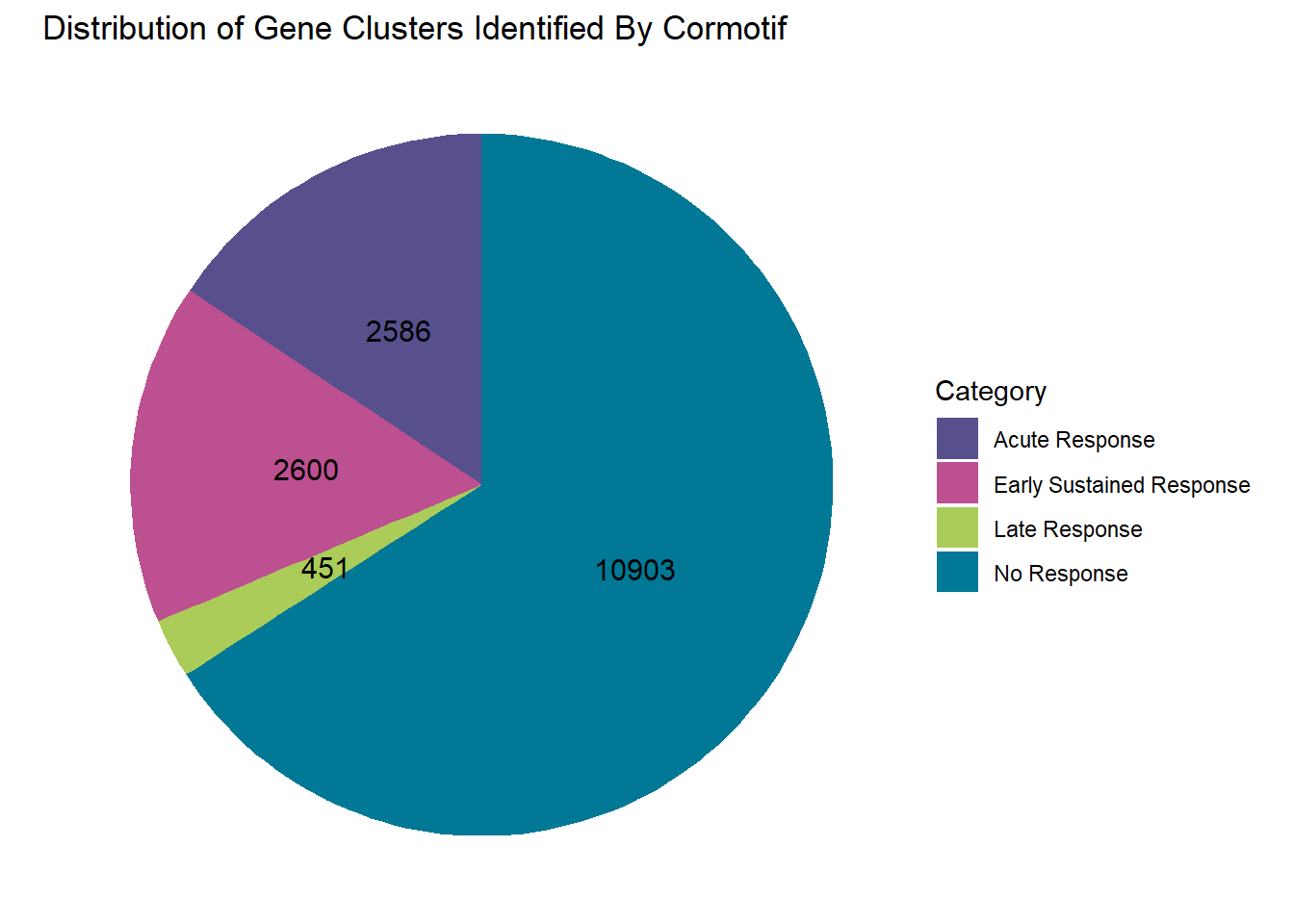
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
Now we will use Gene Ontology analysis to discover biological relevance to the genes that are assigned to this cluster This set is for the No Response motif (motif 1) with clustlike
motif_NR_d <- clust1_d
NRmotif_genes_d <- gost(query = motif_NR_d,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
cormotifNFclust_d <- gostplot(NRmotif_genes_d, capped = FALSE, interactive = TRUE)
cormotifNFclust_dtableNR_d <- NRmotif_genes_d$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
tableNR_d %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0050907 | detection of chemical stimulus involved in sensory perception | 391 | 443 | 1.264e-213 |
| GO:BP | GO:0035195 | miRNA-mediated post-transcriptional gene silencing | 472 | 612 | 3.524e-208 |
| GO:BP | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell | 360 | 397 | 3.627e-206 |
| GO:BP | GO:0007606 | sensory perception of chemical stimulus | 414 | 497 | 3.627e-206 |
| GO:BP | GO:0035194 | regulatory ncRNA-mediated post-transcriptional gene silencing | 473 | 624 | 2.926e-203 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 474 | 630 | 2.423e-201 |
| GO:BP | GO:0009593 | detection of chemical stimulus | 400 | 480 | 2.737e-199 |
| GO:BP | GO:0007608 | sensory perception of smell | 367 | 423 | 4.500e-195 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 414 | 515 | 8.464e-195 |
| GO:BP | GO:0031047 | regulatory ncRNA-mediated gene silencing | 480 | 686 | 1.791e-181 |
| GO:BP | GO:0051606 | detection of stimulus | 451 | 643 | 1.107e-170 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 688 | 1275 | 1.722e-169 |
| GO:BP | GO:0007600 | sensory perception | 546 | 955 | 4.494e-147 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 511 | 1087 | 9.743e-93 |
| GO:BP | GO:0050877 | nervous system process | 632 | 1489 | 3.079e-92 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 625 | 1640 | 2.152e-68 |
| GO:BP | GO:0003008 | system process | 784 | 2266 | 1.973e-65 |
| GO:BP | GO:0042221 | response to chemical | 1079 | 3867 | 4.895e-38 |
| GO:BP | GO:0065007 | biological regulation | 2878 | 12671 | 2.495e-34 |
| GO:BP | GO:0050789 | regulation of biological process | 2791 | 12278 | 5.075e-32 |
| GO:BP | GO:0042742 | defense response to bacterium | 163 | 326 | 8.829e-32 |
| GO:BP | GO:0006959 | humoral immune response | 138 | 254 | 1.143e-31 |
| GO:BP | GO:0006952 | defense response | 562 | 1809 | 9.256e-30 |
| GO:BP | GO:0050794 | regulation of cellular process | 2701 | 11876 | 9.256e-30 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 787 | 2772 | 1.709e-28 |
| GO:BP | GO:0032501 | multicellular organismal process | 1760 | 7234 | 3.177e-28 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 795 | 2838 | 8.308e-27 |
| GO:BP | GO:0009617 | response to bacterium | 270 | 721 | 3.102e-26 |
| GO:BP | GO:0031424 | keratinization | 62 | 82 | 6.445e-25 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 82 | 131 | 4.159e-24 |
| GO:BP | GO:0098542 | defense response to other organism | 397 | 1228 | 6.247e-24 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 467 | 1558 | 1.238e-20 |
| GO:BP | GO:0051707 | response to other organism | 465 | 1555 | 2.567e-20 |
| GO:BP | GO:0009607 | response to biotic stimulus | 474 | 1593 | 2.791e-20 |
| GO:BP | GO:0140546 | defense response to symbiont | 358 | 1125 | 4.440e-20 |
| GO:BP | GO:0007165 | signal transduction | 1439 | 5979 | 9.834e-19 |
| GO:BP | GO:0141060 | disruption of anatomical structure in another organism | 61 | 98 | 1.442e-17 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 829 | 3197 | 3.109e-17 |
| GO:BP | GO:0001906 | cell killing | 102 | 219 | 8.951e-17 |
| GO:BP | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 54 | 83 | 9.352e-17 |
| GO:BP | GO:0031640 | killing of cells of another organism | 57 | 91 | 1.355e-16 |
| GO:BP | GO:0141061 | disruption of cell in another organism | 57 | 91 | 1.355e-16 |
| GO:BP | GO:0009605 | response to external stimulus | 638 | 2367 | 1.659e-16 |
| GO:BP | GO:0006955 | immune response | 553 | 2008 | 5.299e-16 |
| GO:BP | GO:0006954 | inflammatory response | 272 | 847 | 1.527e-15 |
| GO:BP | GO:0045109 | intermediate filament organization | 48 | 73 | 3.954e-15 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 482 | 1724 | 5.590e-15 |
| GO:BP | GO:0007154 | cell communication | 1520 | 6496 | 6.072e-15 |
| GO:BP | GO:0023052 | signaling | 1514 | 6471 | 7.621e-15 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 860 | 3418 | 2.574e-14 |
| GO:BP | GO:0045087 | innate immune response | 300 | 977 | 2.609e-14 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 83 | 176 | 5.360e-14 |
| GO:BP | GO:0050830 | defense response to Gram-positive bacterium | 63 | 120 | 3.260e-13 |
| GO:BP | GO:0050829 | defense response to Gram-negative bacterium | 52 | 91 | 8.895e-13 |
| GO:BP | GO:0097530 | granulocyte migration | 74 | 156 | 1.383e-12 |
| GO:BP | GO:0002252 | immune effector process | 225 | 703 | 1.451e-12 |
| GO:BP | GO:1990266 | neutrophil migration | 65 | 129 | 1.483e-12 |
| GO:BP | GO:0050896 | response to stimulus | 2011 | 8993 | 1.588e-12 |
| GO:BP | GO:0050909 | sensory perception of taste | 42 | 67 | 3.801e-12 |
| GO:BP | GO:0030593 | neutrophil chemotaxis | 56 | 106 | 7.292e-12 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 63 | 129 | 2.269e-11 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 100 | 250 | 2.693e-11 |
| GO:BP | GO:0007586 | digestion | 65 | 136 | 3.273e-11 |
| GO:BP | GO:0050913 | sensory perception of bitter taste | 30 | 42 | 1.067e-10 |
| GO:BP | GO:0002697 | regulation of immune effector process | 135 | 381 | 1.068e-10 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 94 | 237 | 2.386e-10 |
| GO:BP | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste | 30 | 43 | 2.686e-10 |
| GO:BP | GO:0019731 | antibacterial humoral response | 40 | 68 | 2.686e-10 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 49 | 94 | 4.602e-10 |
| GO:BP | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste | 27 | 37 | 6.090e-10 |
| GO:BP | GO:0045103 | intermediate filament-based process | 49 | 95 | 7.457e-10 |
| GO:BP | GO:0002323 | natural killer cell activation involved in immune response | 26 | 35 | 7.504e-10 |
| GO:BP | GO:0002376 | immune system process | 693 | 2796 | 9.320e-10 |
| GO:BP | GO:0050900 | leukocyte migration | 136 | 396 | 1.032e-09 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 125 | 355 | 1.062e-09 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 92 | 237 | 1.473e-09 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1642 | 7320 | 2.062e-09 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 46 | 90 | 4.511e-09 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 87 | 224 | 4.907e-09 |
| GO:BP | GO:0008544 | epidermis development | 133 | 394 | 6.241e-09 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 110 | 308 | 6.493e-09 |
| GO:BP | GO:0042100 | B cell proliferation | 51 | 106 | 6.852e-09 |
| GO:BP | GO:0070661 | leukocyte proliferation | 121 | 351 | 9.824e-09 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 52 | 110 | 9.948e-09 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 117 | 337 | 1.186e-08 |
| GO:BP | GO:0007338 | single fertilization | 70 | 169 | 1.246e-08 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 111 | 315 | 1.258e-08 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 83 | 214 | 1.313e-08 |
| GO:BP | GO:0060326 | cell chemotaxis | 112 | 319 | 1.313e-08 |
| GO:BP | GO:0052695 | cellular glucuronidation | 18 | 21 | 1.679e-08 |
| GO:BP | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein | 18 | 21 | 1.679e-08 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 159 | 503 | 2.248e-08 |
| GO:BP | GO:0007283 | spermatogenesis | 192 | 637 | 2.801e-08 |
| GO:BP | GO:0045321 | leukocyte activation | 267 | 951 | 3.624e-08 |
| GO:BP | GO:0001775 | cell activation | 301 | 1098 | 4.438e-08 |
| GO:BP | GO:0009566 | fertilization | 81 | 212 | 4.742e-08 |
| GO:BP | GO:0048232 | male gamete generation | 195 | 654 | 5.249e-08 |
| GO:BP | GO:0002526 | acute inflammatory response | 50 | 109 | 7.298e-08 |
| GO:BP | GO:0002548 | monocyte chemotaxis | 36 | 68 | 1.324e-07 |
| GO:BP | GO:0048245 | eosinophil chemotaxis | 20 | 27 | 1.736e-07 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 94 | 265 | 1.895e-07 |
| GO:BP | GO:1990869 | cellular response to chemokine | 46 | 99 | 1.952e-07 |
| GO:BP | GO:1990868 | response to chemokine | 46 | 99 | 1.952e-07 |
| GO:BP | GO:0002682 | regulation of immune system process | 395 | 1526 | 2.069e-07 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 104 | 305 | 3.139e-07 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 105 | 309 | 3.244e-07 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 89 | 251 | 4.844e-07 |
| GO:BP | GO:0071715 | icosanoid transport | 34 | 65 | 5.205e-07 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 267 | 980 | 7.291e-07 |
| GO:BP | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 18 | 24 | 7.541e-07 |
| GO:BP | GO:0072677 | eosinophil migration | 21 | 31 | 8.249e-07 |
| GO:BP | GO:0006953 | acute-phase response | 28 | 49 | 8.250e-07 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 85 | 240 | 1.064e-06 |
| GO:BP | GO:0022600 | digestive system process | 46 | 105 | 1.752e-06 |
| GO:BP | GO:0050832 | defense response to fungus | 31 | 59 | 1.977e-06 |
| GO:BP | GO:0042501 | serine phosphorylation of STAT protein | 18 | 25 | 2.076e-06 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 64 | 167 | 2.152e-06 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 83 | 236 | 2.156e-06 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 84 | 240 | 2.271e-06 |
| GO:BP | GO:0097722 | sperm motility | 58 | 147 | 2.747e-06 |
| GO:BP | GO:0030317 | flagellated sperm motility | 58 | 147 | 2.747e-06 |
| GO:BP | GO:0022414 | reproductive process | 394 | 1556 | 2.902e-06 |
| GO:BP | GO:0048247 | lymphocyte chemotaxis | 32 | 63 | 3.165e-06 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 87 | 253 | 3.165e-06 |
| GO:BP | GO:0043588 | skin development | 105 | 322 | 3.203e-06 |
| GO:BP | GO:0050776 | regulation of immune response | 244 | 897 | 3.229e-06 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 49 | 117 | 3.229e-06 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 84 | 243 | 3.983e-06 |
| GO:BP | GO:0032609 | type II interferon production | 49 | 118 | 4.392e-06 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 282 | 1066 | 4.680e-06 |
| GO:BP | GO:0006063 | uronic acid metabolic process | 18 | 26 | 4.819e-06 |
| GO:BP | GO:0019585 | glucuronate metabolic process | 18 | 26 | 4.819e-06 |
| GO:BP | GO:0140975 | disruption of cellular anatomical structure in another organism | 10 | 10 | 6.564e-06 |
| GO:BP | GO:0031341 | regulation of cell killing | 45 | 106 | 6.564e-06 |
| GO:BP | GO:0002775 | antimicrobial peptide production | 10 | 10 | 6.564e-06 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 64 | 172 | 6.684e-06 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 64 | 172 | 6.684e-06 |
| GO:BP | GO:0002922 | positive regulation of humoral immune response | 16 | 22 | 8.534e-06 |
| GO:BP | GO:0042330 | taxis | 139 | 465 | 9.224e-06 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 84 | 248 | 1.005e-05 |
| GO:BP | GO:0030101 | natural killer cell activation | 41 | 95 | 1.337e-05 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 24 | 43 | 1.355e-05 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 124 | 408 | 1.568e-05 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 137 | 461 | 1.585e-05 |
| GO:BP | GO:0007342 | fusion of sperm to egg plasma membrane involved in single fertilization | 19 | 30 | 1.624e-05 |
| GO:BP | GO:0006935 | chemotaxis | 137 | 463 | 2.051e-05 |
| GO:BP | GO:0007276 | gamete generation | 222 | 822 | 2.055e-05 |
| GO:BP | GO:0019953 | sexual reproduction | 286 | 1103 | 2.287e-05 |
| GO:BP | GO:0072676 | lymphocyte migration | 49 | 124 | 2.346e-05 |
| GO:BP | GO:0001909 | leukocyte mediated cytotoxicity | 52 | 135 | 2.690e-05 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 65 | 182 | 2.751e-05 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 64 | 179 | 3.148e-05 |
| GO:BP | GO:0007259 | cell surface receptor signaling pathway via JAK-STAT | 61 | 168 | 3.148e-05 |
| GO:BP | GO:0002385 | mucosal immune response | 22 | 39 | 3.148e-05 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 116 | 381 | 3.157e-05 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 48 | 122 | 3.413e-05 |
| GO:BP | GO:0032689 | negative regulation of type II interferon production | 23 | 42 | 3.479e-05 |
| GO:BP | GO:0097696 | cell surface receptor signaling pathway via STAT | 63 | 176 | 3.580e-05 |
| GO:BP | GO:0010468 | regulation of gene expression | 1223 | 5515 | 3.696e-05 |
| GO:BP | GO:0051873 | killing by host of symbiont cells | 18 | 29 | 4.703e-05 |
| GO:BP | GO:0015732 | prostaglandin transport | 18 | 29 | 4.703e-05 |
| GO:BP | GO:0007631 | feeding behavior | 43 | 106 | 4.721e-05 |
| GO:BP | GO:0009620 | response to fungus | 32 | 70 | 5.214e-05 |
| GO:BP | GO:0009584 | detection of visible light | 19 | 32 | 6.033e-05 |
| GO:BP | GO:0001817 | regulation of cytokine production | 206 | 765 | 6.062e-05 |
| GO:BP | GO:0050865 | regulation of cell activation | 171 | 616 | 6.744e-05 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 140 | 486 | 7.081e-05 |
| GO:BP | GO:0001816 | cytokine production | 207 | 772 | 7.722e-05 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 63 | 180 | 8.120e-05 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 72 | 214 | 8.125e-05 |
| GO:BP | GO:0007286 | spermatid development | 72 | 214 | 8.125e-05 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 60 | 169 | 8.216e-05 |
| GO:BP | GO:0036230 | granulocyte activation | 25 | 50 | 9.827e-05 |
| GO:BP | GO:0050778 | positive regulation of immune response | 197 | 732 | 1.026e-04 |
| GO:BP | GO:0048240 | sperm capacitation | 19 | 33 | 1.084e-04 |
| GO:BP | GO:0071674 | mononuclear cell migration | 77 | 235 | 1.105e-04 |
| GO:BP | GO:0002784 | regulation of antimicrobial peptide production | 8 | 8 | 1.253e-04 |
| GO:BP | GO:0002778 | antibacterial peptide production | 8 | 8 | 1.253e-04 |
| GO:BP | GO:0002760 | positive regulation of antimicrobial humoral response | 8 | 8 | 1.253e-04 |
| GO:BP | GO:0046649 | lymphocyte activation | 209 | 787 | 1.311e-04 |
| GO:BP | GO:0042119 | neutrophil activation | 22 | 42 | 1.434e-04 |
| GO:BP | GO:0002920 | regulation of humoral immune response | 23 | 45 | 1.479e-04 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 156 | 561 | 1.593e-04 |
| GO:BP | GO:0048515 | spermatid differentiation | 73 | 222 | 1.659e-04 |
| GO:BP | GO:0015908 | fatty acid transport | 45 | 118 | 1.732e-04 |
| GO:BP | GO:0045026 | plasma membrane fusion | 19 | 34 | 1.879e-04 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 57 | 163 | 2.332e-04 |
| GO:BP | GO:0098586 | cellular response to virus | 33 | 78 | 2.535e-04 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 37 | 92 | 2.921e-04 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 31 | 72 | 3.105e-04 |
| GO:BP | GO:0032642 | regulation of chemokine production | 38 | 96 | 3.379e-04 |
| GO:BP | GO:0032602 | chemokine production | 38 | 96 | 3.379e-04 |
| GO:BP | GO:0002673 | regulation of acute inflammatory response | 23 | 47 | 3.600e-04 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 139 | 497 | 3.677e-04 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 46 | 125 | 3.901e-04 |
| GO:BP | GO:0006805 | xenobiotic metabolic process | 46 | 125 | 3.901e-04 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 28 | 63 | 3.982e-04 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 55 | 159 | 4.619e-04 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 195 | 741 | 4.718e-04 |
| GO:BP | GO:0007602 | phototransduction | 23 | 48 | 5.364e-04 |
| GO:BP | GO:0032309 | icosanoid secretion | 23 | 48 | 5.364e-04 |
| GO:BP | GO:0015670 | carbon dioxide transport | 11 | 15 | 5.364e-04 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 116 | 404 | 5.371e-04 |
| GO:BP | GO:0052697 | xenobiotic glucuronidation | 7 | 7 | 5.500e-04 |
| GO:BP | GO:0002225 | positive regulation of antimicrobial peptide production | 7 | 7 | 5.500e-04 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1231 | 5633 | 5.528e-04 |
| GO:BP | GO:0019755 | one-carbon compound transport | 15 | 25 | 5.573e-04 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 43 | 116 | 5.784e-04 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 71 | 222 | 5.798e-04 |
| GO:BP | GO:0042113 | B cell activation | 86 | 282 | 6.050e-04 |
| GO:BP | GO:0042110 | T cell activation | 152 | 558 | 6.140e-04 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 39 | 102 | 6.140e-04 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 44 | 120 | 6.204e-04 |
| GO:BP | GO:0002227 | innate immune response in mucosa | 16 | 28 | 6.551e-04 |
| GO:BP | GO:0007601 | visual perception | 70 | 219 | 6.599e-04 |
| GO:BP | GO:0009583 | detection of light stimulus | 29 | 68 | 6.711e-04 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 40 | 106 | 6.728e-04 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 38 | 99 | 6.881e-04 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 50 | 143 | 7.433e-04 |
| GO:BP | GO:0002759 | regulation of antimicrobial humoral response | 9 | 11 | 7.517e-04 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 61 | 185 | 7.716e-04 |
| GO:BP | GO:0034341 | response to type II interferon | 51 | 147 | 7.754e-04 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 45 | 125 | 8.165e-04 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 67 | 210 | 1.009e-03 |
| GO:BP | GO:0032637 | interleukin-8 production | 33 | 83 | 1.009e-03 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 33 | 83 | 1.009e-03 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 72 | 230 | 1.044e-03 |
| GO:BP | GO:0032757 | positive regulation of interleukin-8 production | 27 | 63 | 1.110e-03 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 105 | 366 | 1.252e-03 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 50 | 146 | 1.331e-03 |
| GO:BP | GO:0050863 | regulation of T cell activation | 107 | 375 | 1.343e-03 |
| GO:BP | GO:0003341 | cilium movement | 67 | 212 | 1.372e-03 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 30 | 74 | 1.454e-03 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 34 | 88 | 1.494e-03 |
| GO:BP | GO:0032612 | interleukin-1 production | 40 | 110 | 1.672e-03 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 40 | 110 | 1.672e-03 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 47 | 136 | 1.691e-03 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 44 | 125 | 1.762e-03 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1256 | 5795 | 1.818e-03 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 36 | 96 | 1.846e-03 |
| GO:BP | GO:0007281 | germ cell development | 105 | 370 | 1.929e-03 |
| GO:BP | GO:0006968 | cellular defense response | 23 | 52 | 2.223e-03 |
| GO:BP | GO:0042363 | fat-soluble vitamin catabolic process | 9 | 12 | 2.237e-03 |
| GO:BP | GO:1901317 | regulation of flagellated sperm motility | 13 | 22 | 2.284e-03 |
| GO:BP | GO:0002786 | regulation of antibacterial peptide production | 6 | 6 | 2.295e-03 |
| GO:BP | GO:0051673 | disruption of plasma membrane integrity in another organism | 6 | 6 | 2.295e-03 |
| GO:BP | GO:0051552 | flavone metabolic process | 6 | 6 | 2.295e-03 |
| GO:BP | GO:0002687 | positive regulation of leukocyte migration | 51 | 153 | 2.325e-03 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 213 | 843 | 2.388e-03 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 30 | 76 | 2.454e-03 |
| GO:BP | GO:0032653 | regulation of interleukin-10 production | 25 | 59 | 2.472e-03 |
| GO:BP | GO:0032613 | interleukin-10 production | 25 | 59 | 2.472e-03 |
| GO:BP | GO:0008228 | opsonization | 11 | 17 | 2.592e-03 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 27 | 66 | 2.665e-03 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 20 | 43 | 2.665e-03 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 33 | 87 | 2.704e-03 |
| GO:BP | GO:0042098 | T cell proliferation | 66 | 213 | 2.758e-03 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 52 | 158 | 2.776e-03 |
| GO:BP | GO:0060295 | regulation of cilium movement involved in cell motility | 16 | 31 | 2.887e-03 |
| GO:BP | GO:1902019 | regulation of cilium-dependent cell motility | 16 | 31 | 2.887e-03 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 38 | 106 | 3.287e-03 |
| GO:BP | GO:0030183 | B cell differentiation | 52 | 159 | 3.287e-03 |
| GO:BP | GO:0002544 | chronic inflammatory response | 12 | 20 | 3.343e-03 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 64 | 207 | 3.667e-03 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1265 | 5865 | 3.699e-03 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 40 | 114 | 3.699e-03 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 251 | 1022 | 3.867e-03 |
| GO:BP | GO:0015669 | gas transport | 13 | 23 | 3.897e-03 |
| GO:BP | GO:0030277 | maintenance of gastrointestinal epithelium | 13 | 23 | 3.897e-03 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 101 | 361 | 4.256e-03 |
| GO:BP | GO:0042755 | eating behavior | 18 | 38 | 4.256e-03 |
| GO:BP | GO:0060259 | regulation of feeding behavior | 14 | 26 | 4.256e-03 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 36 | 100 | 4.315e-03 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 16 | 32 | 4.436e-03 |
| GO:BP | GO:1903963 | arachidonate transport | 16 | 32 | 4.436e-03 |
| GO:BP | GO:0050482 | arachidonate secretion | 16 | 32 | 4.436e-03 |
| GO:BP | GO:0009581 | detection of external stimulus | 47 | 142 | 4.779e-03 |
| GO:BP | GO:0031347 | regulation of defense response | 197 | 783 | 5.095e-03 |
| GO:BP | GO:0002707 | negative regulation of lymphocyte mediated immunity | 23 | 55 | 5.477e-03 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 94 | 334 | 5.515e-03 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 83 | 288 | 5.529e-03 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 48 | 147 | 5.710e-03 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 25 | 62 | 5.710e-03 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 48 | 147 | 5.710e-03 |
| GO:BP | GO:0032615 | interleukin-12 production | 25 | 62 | 5.710e-03 |
| GO:BP | GO:0002228 | natural killer cell mediated immunity | 29 | 76 | 5.710e-03 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 260 | 1071 | 6.032e-03 |
| GO:BP | GO:0044058 | regulation of digestive system process | 18 | 39 | 6.059e-03 |
| GO:BP | GO:0070555 | response to interleukin-1 | 44 | 132 | 6.132e-03 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 36 | 102 | 6.407e-03 |
| GO:BP | GO:0042832 | defense response to protozoan | 14 | 27 | 6.752e-03 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 37 | 106 | 6.757e-03 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 34 | 95 | 6.944e-03 |
| GO:BP | GO:1903027 | regulation of opsonization | 8 | 11 | 6.986e-03 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 52 | 164 | 6.986e-03 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 52 | 164 | 6.986e-03 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 62 | 204 | 7.018e-03 |
| GO:BP | GO:0015671 | oxygen transport | 10 | 16 | 7.200e-03 |
| GO:BP | GO:0060046 | regulation of acrosome reaction | 10 | 16 | 7.200e-03 |
| GO:BP | GO:0034374 | low-density lipoprotein particle remodeling | 10 | 16 | 7.200e-03 |
| GO:BP | GO:0008037 | cell recognition | 50 | 157 | 7.939e-03 |
| GO:BP | GO:0035036 | sperm-egg recognition | 22 | 53 | 7.939e-03 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 53 | 169 | 8.073e-03 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 53 | 169 | 8.073e-03 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 58 | 189 | 8.083e-03 |
| GO:BP | GO:0002715 | regulation of natural killer cell mediated immunity | 21 | 50 | 8.818e-03 |
| GO:BP | GO:0034097 | response to cytokine | 229 | 937 | 8.945e-03 |
| GO:BP | GO:1903028 | positive regulation of opsonization | 7 | 9 | 8.998e-03 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 59 | 194 | 9.181e-03 |
| GO:BP | GO:0052696 | flavonoid glucuronidation | 5 | 5 | 9.181e-03 |
| GO:BP | GO:0002803 | positive regulation of antibacterial peptide production | 5 | 5 | 9.181e-03 |
| GO:BP | GO:0046456 | icosanoid biosynthetic process | 23 | 57 | 9.213e-03 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 158 | 619 | 9.711e-03 |
| GO:BP | GO:0009988 | cell-cell recognition | 28 | 75 | 1.018e-02 |
| GO:BP | GO:0002710 | negative regulation of T cell mediated immunity | 14 | 28 | 1.020e-02 |
| GO:BP | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway | 6 | 7 | 1.042e-02 |
| GO:BP | GO:0070942 | neutrophil mediated cytotoxicity | 9 | 14 | 1.053e-02 |
| GO:BP | GO:0009111 | vitamin catabolic process | 9 | 14 | 1.053e-02 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 24 | 61 | 1.055e-02 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 19 | 44 | 1.059e-02 |
| GO:BP | GO:1901652 | response to peptide | 231 | 950 | 1.097e-02 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 21 | 51 | 1.160e-02 |
| GO:BP | GO:0032733 | positive regulation of interleukin-10 production | 18 | 41 | 1.163e-02 |
| GO:BP | GO:0001523 | retinoid metabolic process | 30 | 83 | 1.181e-02 |
| GO:BP | GO:0050766 | positive regulation of phagocytosis | 26 | 69 | 1.334e-02 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 46 | 145 | 1.392e-02 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 50 | 161 | 1.416e-02 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 27 | 73 | 1.464e-02 |
| GO:BP | GO:0002825 | regulation of T-helper 1 type immune response | 15 | 32 | 1.464e-02 |
| GO:BP | GO:0043299 | leukocyte degranulation | 30 | 84 | 1.464e-02 |
| GO:BP | GO:0042445 | hormone metabolic process | 70 | 243 | 1.468e-02 |
| GO:BP | GO:0006910 | phagocytosis, recognition | 14 | 29 | 1.514e-02 |
| GO:BP | GO:0009812 | flavonoid metabolic process | 8 | 12 | 1.514e-02 |
| GO:BP | GO:0016056 | G protein-coupled opsin signaling pathway | 8 | 12 | 1.514e-02 |
| GO:BP | GO:0070943 | neutrophil-mediated killing of symbiont cell | 8 | 12 | 1.514e-02 |
| GO:BP | GO:0018149 | peptide cross-linking | 14 | 29 | 1.514e-02 |
| GO:BP | GO:0032661 | regulation of interleukin-18 production | 8 | 12 | 1.514e-02 |
| GO:BP | GO:0032621 | interleukin-18 production | 8 | 12 | 1.514e-02 |
| GO:BP | GO:0016101 | diterpenoid metabolic process | 31 | 88 | 1.523e-02 |
| GO:BP | GO:0098581 | detection of external biotic stimulus | 13 | 26 | 1.535e-02 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 40 | 123 | 1.736e-02 |
| GO:BP | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 22 | 56 | 1.739e-02 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 22 | 56 | 1.739e-02 |
| GO:BP | GO:0072678 | T cell migration | 27 | 74 | 1.786e-02 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 32 | 93 | 1.956e-02 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 32 | 93 | 1.956e-02 |
| GO:BP | GO:0141005 | transposable element silencing by heterochromatin formation | 9 | 15 | 1.985e-02 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 56 | 188 | 2.010e-02 |
| GO:BP | GO:0002456 | T cell mediated immunity | 40 | 124 | 2.030e-02 |
| GO:BP | GO:0042573 | retinoic acid metabolic process | 15 | 33 | 2.064e-02 |
| GO:BP | GO:0035747 | natural killer cell chemotaxis | 7 | 10 | 2.159e-02 |
| GO:BP | GO:0008343 | adult feeding behavior | 7 | 10 | 2.159e-02 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 7 | 10 | 2.159e-02 |
| GO:BP | GO:0002765 | immune response-inhibiting signal transduction | 7 | 10 | 2.159e-02 |
| GO:BP | GO:0001562 | response to protozoan | 14 | 30 | 2.194e-02 |
| GO:BP | GO:0007603 | phototransduction, visible light | 10 | 18 | 2.238e-02 |
| GO:BP | GO:0002675 | positive regulation of acute inflammatory response | 13 | 27 | 2.308e-02 |
| GO:BP | GO:0002577 | regulation of antigen processing and presentation | 11 | 21 | 2.355e-02 |
| GO:BP | GO:0002523 | leukocyte migration involved in inflammatory response | 11 | 21 | 2.355e-02 |
| GO:BP | GO:0055078 | sodium ion homeostasis | 19 | 47 | 2.522e-02 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 123 | 478 | 2.636e-02 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 27 | 76 | 2.716e-02 |
| GO:BP | GO:2000501 | regulation of natural killer cell chemotaxis | 6 | 8 | 2.976e-02 |
| GO:BP | GO:0032741 | positive regulation of interleukin-18 production | 6 | 8 | 2.976e-02 |
| GO:BP | GO:0070268 | cornification | 6 | 8 | 2.976e-02 |
| GO:BP | GO:0060456 | positive regulation of digestive system process | 8 | 13 | 2.976e-02 |
| GO:BP | GO:0031349 | positive regulation of defense response | 124 | 484 | 2.976e-02 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1203 | 5646 | 2.976e-02 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 61 | 212 | 2.976e-02 |
| GO:BP | GO:0046942 | carboxylic acid transport | 92 | 344 | 2.986e-02 |
| GO:BP | GO:0042116 | macrophage activation | 35 | 107 | 2.986e-02 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 128 | 502 | 2.986e-02 |
| GO:BP | GO:0042267 | natural killer cell mediated cytotoxicity | 26 | 73 | 3.098e-02 |
| GO:BP | GO:0009072 | aromatic amino acid metabolic process | 17 | 41 | 3.163e-02 |
| GO:BP | GO:0006775 | fat-soluble vitamin metabolic process | 19 | 48 | 3.258e-02 |
| GO:BP | GO:0015849 | organic acid transport | 92 | 345 | 3.258e-02 |
| GO:BP | GO:0010817 | regulation of hormone levels | 138 | 548 | 3.277e-02 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 24 | 66 | 3.324e-02 |
| GO:BP | GO:1902221 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process | 13 | 28 | 3.324e-02 |
| GO:BP | GO:0051607 | defense response to virus | 84 | 311 | 3.403e-02 |
| GO:BP | GO:0035821 | modulation of process of another organism | 9 | 16 | 3.403e-02 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 98 | 372 | 3.480e-02 |
| GO:BP | GO:0019370 | leukotriene biosynthetic process | 10 | 19 | 3.571e-02 |
| GO:BP | GO:0030431 | sleep | 11 | 22 | 3.571e-02 |
| GO:BP | GO:0009253 | peptidoglycan catabolic process | 5 | 6 | 3.571e-02 |
| GO:BP | GO:0002325 | natural killer cell differentiation involved in immune response | 4 | 4 | 3.571e-02 |
| GO:BP | GO:0032635 | interleukin-6 production | 46 | 152 | 3.571e-02 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 46 | 152 | 3.571e-02 |
| GO:BP | GO:0098543 | detection of other organism | 10 | 19 | 3.571e-02 |
| GO:BP | GO:0000270 | peptidoglycan metabolic process | 5 | 6 | 3.571e-02 |
| GO:BP | GO:0044278 | disruption of cell wall in another organism | 4 | 4 | 3.571e-02 |
| GO:BP | GO:0034382 | chylomicron remnant clearance | 5 | 6 | 3.571e-02 |
| GO:BP | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production | 10 | 19 | 3.571e-02 |
| GO:BP | GO:0097272 | ammonium homeostasis | 5 | 6 | 3.571e-02 |
| GO:BP | GO:0060480 | lung goblet cell differentiation | 4 | 4 | 3.571e-02 |
| GO:BP | GO:2000503 | positive regulation of natural killer cell chemotaxis | 5 | 6 | 3.571e-02 |
| GO:BP | GO:2000851 | positive regulation of glucocorticoid secretion | 4 | 4 | 3.571e-02 |
| GO:BP | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep | 4 | 4 | 3.571e-02 |
| GO:BP | GO:0051464 | positive regulation of cortisol secretion | 4 | 4 | 3.571e-02 |
| GO:BP | GO:0032826 | regulation of natural killer cell differentiation involved in immune response | 4 | 4 | 3.571e-02 |
| GO:BP | GO:0042747 | circadian sleep/wake cycle, REM sleep | 4 | 4 | 3.571e-02 |
| GO:BP | GO:0006869 | lipid transport | 111 | 431 | 3.906e-02 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 58 | 203 | 4.221e-02 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 25 | 71 | 4.221e-02 |
| GO:BP | GO:0032372 | negative regulation of sterol transport | 7 | 11 | 4.274e-02 |
| GO:BP | GO:0032375 | negative regulation of cholesterol transport | 7 | 11 | 4.274e-02 |
| GO:BP | GO:0070944 | neutrophil-mediated killing of bacterium | 7 | 11 | 4.274e-02 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 62 | 220 | 4.285e-02 |
| GO:BP | GO:0007340 | acrosome reaction | 16 | 39 | 4.545e-02 |
| GO:BP | GO:0015721 | bile acid and bile salt transport | 13 | 29 | 4.563e-02 |
| GO:BP | GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 18 | 46 | 4.607e-02 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 112 | 438 | 4.710e-02 |
| GO:BP | GO:0019369 | arachidonate metabolic process | 21 | 57 | 4.919e-02 |
| KEGG | KEGG:04740 | Olfactory transduction | 367 | 428 | 1.117e-182 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 152 | 291 | 3.555e-28 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 169 | 365 | 9.506e-24 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 147 | 310 | 7.052e-22 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 58 | 86 | 1.316e-17 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 62 | 98 | 8.388e-17 |
| KEGG | KEGG:00830 | Retinol metabolism | 40 | 68 | 2.581e-09 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 32 | 49 | 4.064e-09 |
| KEGG | KEGG:04742 | Taste transduction | 45 | 85 | 1.597e-08 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 40 | 74 | 5.987e-08 |
| KEGG | KEGG:05310 | Asthma | 20 | 27 | 3.423e-07 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 45 | 92 | 3.423e-07 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 36 | 67 | 3.698e-07 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 26 | 44 | 2.759e-06 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 34 | 68 | 8.022e-06 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 63 | 162 | 1.691e-05 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 40 | 88 | 1.700e-05 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 19 | 30 | 2.543e-05 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 52 | 132 | 7.983e-05 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 21 | 37 | 7.983e-05 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 29 | 62 | 2.206e-04 |
| KEGG | KEGG:04976 | Bile secretion | 37 | 89 | 4.124e-04 |
| KEGG | KEGG:05144 | Malaria | 24 | 49 | 4.179e-04 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 28 | 61 | 4.179e-04 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 17 | 30 | 5.363e-04 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 28 | 62 | 5.568e-04 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 19 | 36 | 6.668e-04 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 15 | 26 | 1.038e-03 |
| KEGG | KEGG:05330 | Allograft rejection | 17 | 34 | 3.520e-03 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 20 | 43 | 3.520e-03 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 20 | 43 | 3.520e-03 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 44 | 124 | 4.374e-03 |
| KEGG | KEGG:05152 | Tuberculosis | 57 | 175 | 8.150e-03 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 30 | 79 | 9.433e-03 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 18 | 40 | 9.990e-03 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 17 | 37 | 9.995e-03 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 31 | 86 | 1.923e-02 |
| KEGG | KEGG:05164 | Influenza A | 53 | 167 | 1.925e-02 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 16 | 36 | 1.925e-02 |
| KEGG | KEGG:05143 | African trypanosomiasis | 16 | 36 | 1.925e-02 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 36 | 106 | 2.611e-02 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 57 | 188 | 3.938e-02 |
| KEGG | KEGG:04744 | Phototransduction | 13 | 29 | 4.150e-02 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 12 | 26 | 4.202e-02 |
| KEGG | KEGG:04972 | Pancreatic secretion | 34 | 102 | 4.245e-02 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 31 | 92 | 4.935e-02 |
#write.csv(tableNR, "output/table_NRmotif.csv")
#GO:BP
tableNR_GOBP_d <- tableNR_d %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(tableNR_GOBP, "data/tableNR_GOBP.RDS")
tableNR_GOBP_d %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("No Response Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))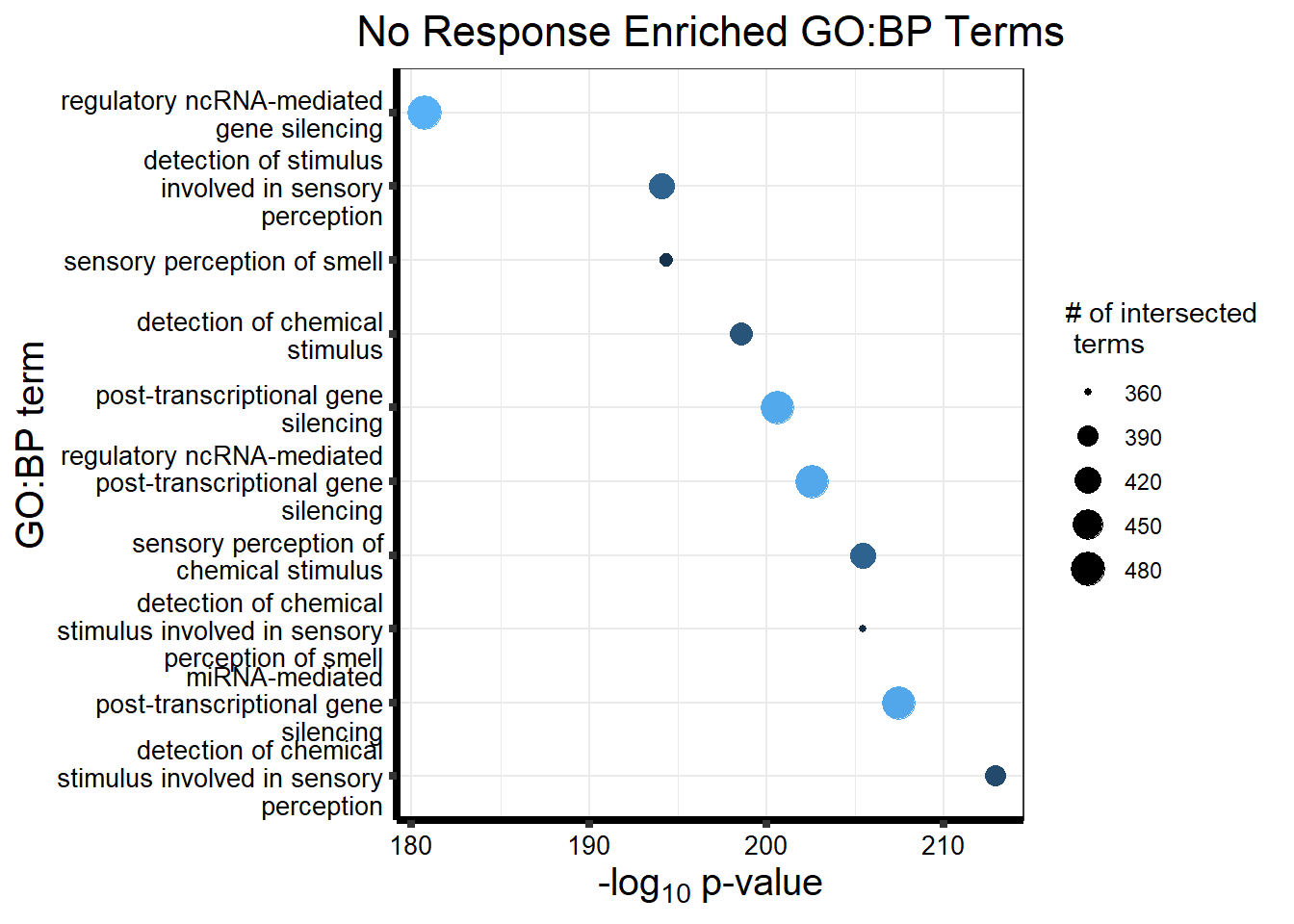
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#KEGG
tableNR_KEGG_d <- tableNR_d %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
tableNR_KEGG_d %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("No Response Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))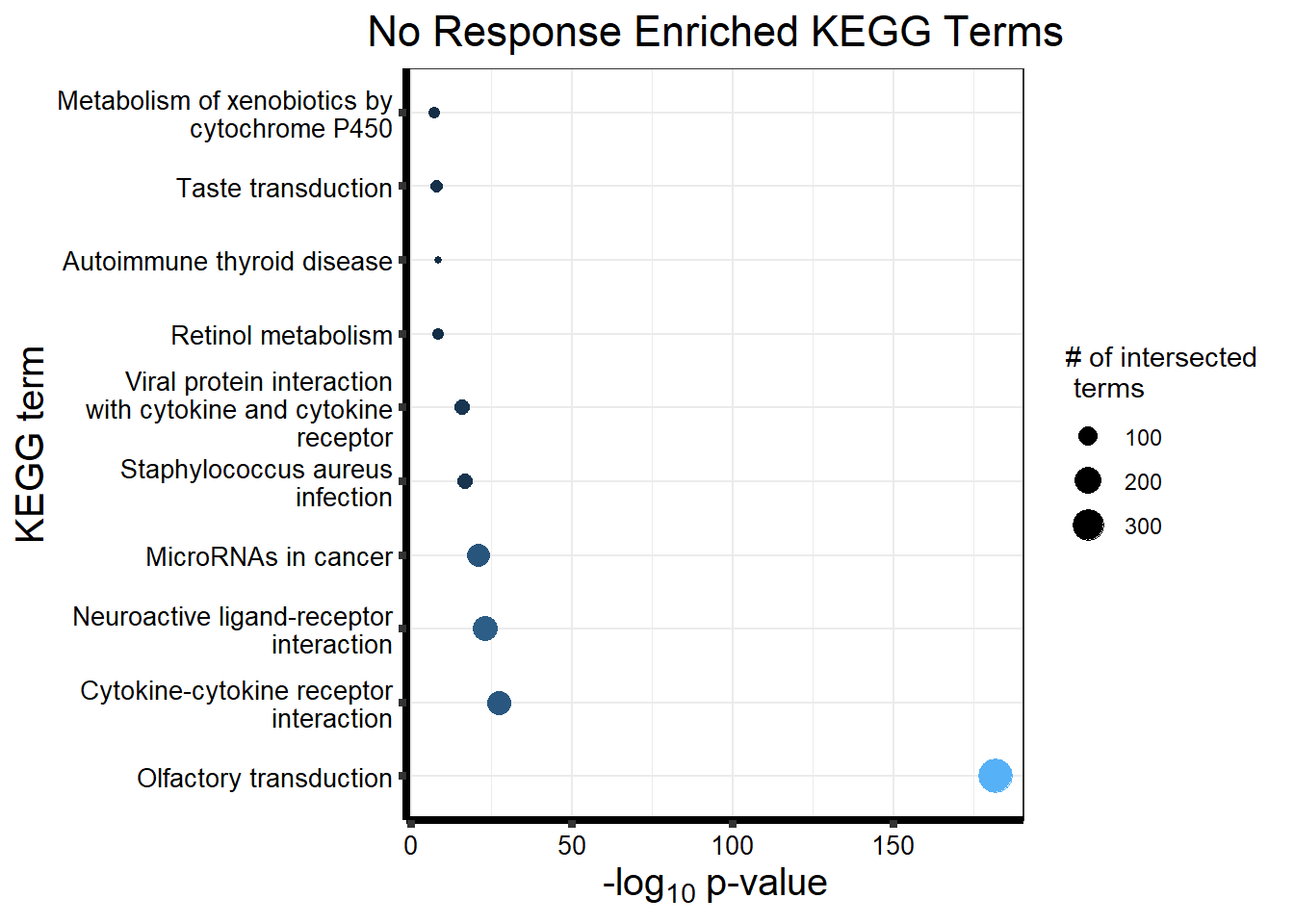
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
Now let’s do the same thing but with the posterior probability rather than the clustlike
motif_NR_pp <- prob_motif_1
NRmotif_genes_pp <- gost(query = motif_NR_pp,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
cormotifNFclust_pp <- gostplot(NRmotif_genes_pp, capped = FALSE, interactive = TRUE)
cormotifNFclust_pptableNR_pp <- NRmotif_genes_pp$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
tableNR_pp %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0050907 | detection of chemical stimulus involved in sensory perception | 390 | 443 | 1.287e-211 |
| GO:BP | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell | 361 | 397 | 3.880e-207 |
| GO:BP | GO:0035195 | miRNA-mediated post-transcriptional gene silencing | 470 | 612 | 1.992e-205 |
| GO:BP | GO:0007606 | sensory perception of chemical stimulus | 413 | 497 | 2.606e-204 |
| GO:BP | GO:0035194 | regulatory ncRNA-mediated post-transcriptional gene silencing | 471 | 624 | 2.134e-200 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 472 | 630 | 1.663e-198 |
| GO:BP | GO:0009593 | detection of chemical stimulus | 399 | 480 | 1.882e-197 |
| GO:BP | GO:0007608 | sensory perception of smell | 368 | 423 | 4.525e-196 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 412 | 515 | 8.753e-192 |
| GO:BP | GO:0031047 | regulatory ncRNA-mediated gene silencing | 478 | 686 | 7.125e-179 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 691 | 1275 | 6.531e-171 |
| GO:BP | GO:0051606 | detection of stimulus | 450 | 643 | 3.612e-169 |
| GO:BP | GO:0007600 | sensory perception | 550 | 955 | 1.330e-149 |
| GO:BP | GO:0050877 | nervous system process | 640 | 1489 | 7.974e-96 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 507 | 1087 | 6.512e-90 |
| GO:BP | GO:0003008 | system process | 792 | 2266 | 7.511e-68 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 623 | 1640 | 4.964e-67 |
| GO:BP | GO:0042221 | response to chemical | 1081 | 3867 | 6.538e-38 |
| GO:BP | GO:0065007 | biological regulation | 2890 | 12671 | 3.299e-35 |
| GO:BP | GO:0050789 | regulation of biological process | 2803 | 12278 | 6.612e-33 |
| GO:BP | GO:0042742 | defense response to bacterium | 163 | 326 | 1.270e-31 |
| GO:BP | GO:0006952 | defense response | 567 | 1809 | 7.997e-31 |
| GO:BP | GO:0032501 | multicellular organismal process | 1775 | 7234 | 4.261e-30 |
| GO:BP | GO:0050794 | regulation of cellular process | 2710 | 11876 | 4.261e-30 |
| GO:BP | GO:0006959 | humoral immune response | 135 | 254 | 1.532e-29 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 784 | 2772 | 2.288e-27 |
| GO:BP | GO:0009617 | response to bacterium | 270 | 721 | 5.192e-26 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 792 | 2838 | 1.031e-25 |
| GO:BP | GO:0098542 | defense response to other organism | 401 | 1228 | 7.042e-25 |
| GO:BP | GO:0031424 | keratinization | 62 | 82 | 7.370e-25 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 81 | 131 | 3.323e-23 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 472 | 1558 | 1.127e-21 |
| GO:BP | GO:0051707 | response to other organism | 470 | 1555 | 2.367e-21 |
| GO:BP | GO:0140546 | defense response to symbiont | 363 | 1125 | 2.502e-21 |
| GO:BP | GO:0009607 | response to biotic stimulus | 479 | 1593 | 2.604e-21 |
| GO:BP | GO:0007165 | signal transduction | 1438 | 5979 | 5.050e-18 |
| GO:BP | GO:0009605 | response to external stimulus | 645 | 2367 | 1.527e-17 |
| GO:BP | GO:0141060 | disruption of anatomical structure in another organism | 60 | 98 | 1.074e-16 |
| GO:BP | GO:0001906 | cell killing | 102 | 219 | 1.111e-16 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 826 | 3197 | 2.318e-16 |
| GO:BP | GO:0006954 | inflammatory response | 275 | 847 | 3.167e-16 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 489 | 1724 | 3.212e-16 |
| GO:BP | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 53 | 83 | 7.347e-16 |
| GO:BP | GO:0031640 | killing of cells of another organism | 56 | 91 | 9.719e-16 |
| GO:BP | GO:0141061 | disruption of cell in another organism | 56 | 91 | 9.719e-16 |
| GO:BP | GO:0045087 | innate immune response | 305 | 977 | 1.891e-15 |
| GO:BP | GO:0045109 | intermediate filament organization | 48 | 73 | 4.381e-15 |
| GO:BP | GO:0006955 | immune response | 549 | 2008 | 5.955e-15 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 84 | 176 | 1.835e-14 |
| GO:BP | GO:0007154 | cell communication | 1520 | 6496 | 1.967e-14 |
| GO:BP | GO:0023052 | signaling | 1513 | 6471 | 3.321e-14 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 859 | 3418 | 7.863e-14 |
| GO:BP | GO:0097530 | granulocyte migration | 74 | 156 | 1.684e-12 |
| GO:BP | GO:1990266 | neutrophil migration | 65 | 129 | 1.812e-12 |
| GO:BP | GO:0050896 | response to stimulus | 2016 | 8993 | 1.856e-12 |
| GO:BP | GO:0050829 | defense response to Gram-negative bacterium | 51 | 91 | 5.170e-12 |
| GO:BP | GO:0050830 | defense response to Gram-positive bacterium | 61 | 120 | 6.701e-12 |
| GO:BP | GO:0030593 | neutrophil chemotaxis | 56 | 106 | 8.589e-12 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 101 | 250 | 1.235e-11 |
| GO:BP | GO:0002252 | immune effector process | 221 | 703 | 2.576e-11 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 63 | 129 | 2.607e-11 |
| GO:BP | GO:0007586 | digestion | 65 | 136 | 3.831e-11 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 95 | 237 | 1.092e-10 |
| GO:BP | GO:0050900 | leukocyte migration | 139 | 396 | 1.402e-10 |
| GO:BP | GO:0050909 | sensory perception of taste | 40 | 67 | 1.572e-10 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 127 | 355 | 2.877e-10 |
| GO:BP | GO:0002697 | regulation of immune effector process | 134 | 381 | 2.911e-10 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 49 | 94 | 5.229e-10 |
| GO:BP | GO:0002376 | immune system process | 696 | 2796 | 6.460e-10 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 93 | 237 | 7.182e-10 |
| GO:BP | GO:0002323 | natural killer cell activation involved in immune response | 26 | 35 | 8.175e-10 |
| GO:BP | GO:0045103 | intermediate filament-based process | 49 | 95 | 8.229e-10 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 54 | 110 | 8.825e-10 |
| GO:BP | GO:0019731 | antibacterial humoral response | 39 | 68 | 1.527e-09 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 88 | 224 | 2.298e-09 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 119 | 337 | 3.248e-09 |
| GO:BP | GO:0008544 | epidermis development | 134 | 394 | 3.770e-09 |
| GO:BP | GO:0070661 | leukocyte proliferation | 122 | 351 | 5.808e-09 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 84 | 214 | 6.541e-09 |
| GO:BP | GO:0050913 | sensory perception of bitter taste | 28 | 42 | 7.382e-09 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 110 | 308 | 7.797e-09 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1639 | 7320 | 1.120e-08 |
| GO:BP | GO:0007283 | spermatogenesis | 194 | 637 | 1.246e-08 |
| GO:BP | GO:0007338 | single fertilization | 70 | 169 | 1.462e-08 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 111 | 315 | 1.544e-08 |
| GO:BP | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste | 28 | 43 | 1.592e-08 |
| GO:BP | GO:0052695 | cellular glucuronidation | 18 | 21 | 1.831e-08 |
| GO:BP | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein | 18 | 21 | 1.831e-08 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 45 | 90 | 1.838e-08 |
| GO:BP | GO:0048232 | male gamete generation | 197 | 654 | 2.315e-08 |
| GO:BP | GO:0042100 | B cell proliferation | 50 | 106 | 2.576e-08 |
| GO:BP | GO:0071715 | icosanoid transport | 36 | 65 | 2.911e-08 |
| GO:BP | GO:0060326 | cell chemotaxis | 111 | 319 | 3.324e-08 |
| GO:BP | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste | 25 | 37 | 4.338e-08 |
| GO:BP | GO:0009566 | fertilization | 81 | 212 | 5.545e-08 |
| GO:BP | GO:0002682 | regulation of immune system process | 399 | 1526 | 7.018e-08 |
| GO:BP | GO:0045321 | leukocyte activation | 265 | 951 | 1.229e-07 |
| GO:BP | GO:0002548 | monocyte chemotaxis | 36 | 68 | 1.442e-07 |
| GO:BP | GO:0001775 | cell activation | 299 | 1098 | 1.442e-07 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 156 | 503 | 1.723e-07 |
| GO:BP | GO:0048245 | eosinophil chemotaxis | 20 | 27 | 1.818e-07 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 94 | 265 | 2.192e-07 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 90 | 251 | 2.555e-07 |
| GO:BP | GO:0002526 | acute inflammatory response | 49 | 109 | 2.614e-07 |
| GO:BP | GO:0022414 | reproductive process | 401 | 1556 | 3.920e-07 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 269 | 980 | 4.080e-07 |
| GO:BP | GO:0022600 | digestive system process | 47 | 105 | 6.278e-07 |
| GO:BP | GO:1990869 | cellular response to chemokine | 45 | 99 | 7.116e-07 |
| GO:BP | GO:1990868 | response to chemokine | 45 | 99 | 7.116e-07 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 103 | 305 | 7.469e-07 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 104 | 309 | 7.689e-07 |
| GO:BP | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 18 | 24 | 7.793e-07 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 89 | 253 | 8.236e-07 |
| GO:BP | GO:0072677 | eosinophil migration | 21 | 31 | 8.501e-07 |
| GO:BP | GO:0006953 | acute-phase response | 28 | 49 | 8.612e-07 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 85 | 240 | 1.193e-06 |
| GO:BP | GO:0050832 | defense response to fungus | 31 | 59 | 2.090e-06 |
| GO:BP | GO:0042501 | serine phosphorylation of STAT protein | 18 | 25 | 2.145e-06 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 64 | 167 | 2.377e-06 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 83 | 236 | 2.430e-06 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 84 | 240 | 2.562e-06 |
| GO:BP | GO:0031341 | regulation of cell killing | 46 | 106 | 2.568e-06 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 284 | 1066 | 2.902e-06 |
| GO:BP | GO:0030317 | flagellated sperm motility | 58 | 147 | 2.965e-06 |
| GO:BP | GO:0097722 | sperm motility | 58 | 147 | 2.965e-06 |
| GO:BP | GO:0048247 | lymphocyte chemotaxis | 32 | 63 | 3.354e-06 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 49 | 117 | 3.559e-06 |
| GO:BP | GO:0043588 | skin development | 105 | 322 | 3.654e-06 |
| GO:BP | GO:0032609 | type II interferon production | 49 | 118 | 4.838e-06 |
| GO:BP | GO:0019585 | glucuronate metabolic process | 18 | 26 | 5.102e-06 |
| GO:BP | GO:0006063 | uronic acid metabolic process | 18 | 26 | 5.102e-06 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 85 | 248 | 5.705e-06 |
| GO:BP | GO:0050776 | regulation of immune response | 243 | 897 | 6.122e-06 |
| GO:BP | GO:0002775 | antimicrobial peptide production | 10 | 10 | 6.776e-06 |
| GO:BP | GO:0140975 | disruption of cellular anatomical structure in another organism | 10 | 10 | 6.776e-06 |
| GO:BP | GO:0019953 | sexual reproduction | 290 | 1103 | 7.052e-06 |
| GO:BP | GO:0007631 | feeding behavior | 45 | 106 | 7.082e-06 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 64 | 172 | 7.376e-06 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 64 | 172 | 7.376e-06 |
| GO:BP | GO:0002922 | positive regulation of humoral immune response | 16 | 22 | 8.801e-06 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 83 | 243 | 8.934e-06 |
| GO:BP | GO:0072676 | lymphocyte migration | 50 | 124 | 9.854e-06 |
| GO:BP | GO:0030101 | natural killer cell activation | 41 | 95 | 1.434e-05 |
| GO:BP | GO:0007259 | cell surface receptor signaling pathway via JAK-STAT | 62 | 168 | 1.566e-05 |
| GO:BP | GO:0010468 | regulation of gene expression | 1231 | 5515 | 1.601e-05 |
| GO:BP | GO:0007342 | fusion of sperm to egg plasma membrane involved in single fertilization | 19 | 30 | 1.685e-05 |
| GO:BP | GO:0007276 | gamete generation | 223 | 822 | 1.685e-05 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 124 | 408 | 1.784e-05 |
| GO:BP | GO:0097696 | cell surface receptor signaling pathway via STAT | 64 | 176 | 1.813e-05 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 117 | 381 | 2.093e-05 |
| GO:BP | GO:0048240 | sperm capacitation | 20 | 33 | 2.241e-05 |
| GO:BP | GO:0001909 | leukocyte mediated cytotoxicity | 52 | 135 | 2.922e-05 |
| GO:BP | GO:0042330 | taxis | 137 | 465 | 3.037e-05 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 142 | 486 | 3.187e-05 |
| GO:BP | GO:0015908 | fatty acid transport | 47 | 118 | 3.220e-05 |
| GO:BP | GO:0071674 | mononuclear cell migration | 79 | 235 | 3.327e-05 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 64 | 179 | 3.440e-05 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 48 | 122 | 3.689e-05 |
| GO:BP | GO:0032309 | icosanoid secretion | 25 | 48 | 4.224e-05 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 64 | 180 | 4.227e-05 |
| GO:BP | GO:0007286 | spermatid development | 73 | 214 | 4.564e-05 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 46 | 116 | 4.695e-05 |
| GO:BP | GO:0015732 | prostaglandin transport | 18 | 29 | 4.842e-05 |
| GO:BP | GO:0051873 | killing by host of symbiont cells | 18 | 29 | 4.842e-05 |
| GO:BP | GO:0001817 | regulation of cytokine production | 207 | 765 | 4.892e-05 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 121 | 404 | 5.096e-05 |
| GO:BP | GO:0009620 | response to fungus | 32 | 70 | 5.455e-05 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 23 | 43 | 5.998e-05 |
| GO:BP | GO:0001816 | cytokine production | 208 | 772 | 6.150e-05 |
| GO:BP | GO:0006935 | chemotaxis | 135 | 463 | 6.150e-05 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 64 | 182 | 6.150e-05 |
| GO:BP | GO:0050865 | regulation of cell activation | 171 | 616 | 7.851e-05 |
| GO:BP | GO:0050778 | positive regulation of immune response | 198 | 732 | 8.254e-05 |
| GO:BP | GO:0034341 | response to type II interferon | 54 | 147 | 8.849e-05 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 72 | 214 | 8.885e-05 |
| GO:BP | GO:0098586 | cellular response to virus | 34 | 78 | 9.303e-05 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 74 | 222 | 9.367e-05 |
| GO:BP | GO:0048515 | spermatid differentiation | 74 | 222 | 9.367e-05 |
| GO:BP | GO:0036230 | granulocyte activation | 25 | 50 | 1.007e-04 |
| GO:BP | GO:0007601 | visual perception | 73 | 219 | 1.084e-04 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 57 | 159 | 1.093e-04 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 38 | 92 | 1.145e-04 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 157 | 561 | 1.184e-04 |
| GO:BP | GO:0002760 | positive regulation of antimicrobial humoral response | 8 | 8 | 1.237e-04 |
| GO:BP | GO:0002778 | antibacterial peptide production | 8 | 8 | 1.237e-04 |
| GO:BP | GO:0002784 | regulation of antimicrobial peptide production | 8 | 8 | 1.237e-04 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 133 | 461 | 1.239e-04 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 29 | 63 | 1.296e-04 |
| GO:BP | GO:0002385 | mucosal immune response | 21 | 39 | 1.355e-04 |
| GO:BP | GO:0042119 | neutrophil activation | 22 | 42 | 1.433e-04 |
| GO:BP | GO:0032689 | negative regulation of type II interferon production | 22 | 42 | 1.433e-04 |
| GO:BP | GO:0002920 | regulation of humoral immune response | 23 | 45 | 1.480e-04 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 141 | 497 | 1.646e-04 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 47 | 125 | 1.725e-04 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 47 | 125 | 1.725e-04 |
| GO:BP | GO:0006805 | xenobiotic metabolic process | 47 | 125 | 1.725e-04 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 59 | 169 | 1.783e-04 |
| GO:BP | GO:0045026 | plasma membrane fusion | 19 | 34 | 1.852e-04 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 109 | 366 | 1.921e-04 |
| GO:BP | GO:0042363 | fat-soluble vitamin catabolic process | 10 | 12 | 2.025e-04 |
| GO:BP | GO:0046649 | lymphocyte activation | 208 | 787 | 2.146e-04 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 57 | 163 | 2.373e-04 |
| GO:BP | GO:0009584 | detection of visible light | 18 | 32 | 2.865e-04 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1238 | 5633 | 2.971e-04 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 31 | 72 | 3.070e-04 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 69 | 210 | 3.102e-04 |
| GO:BP | GO:0032642 | regulation of chemokine production | 38 | 96 | 3.356e-04 |
| GO:BP | GO:0032602 | chemokine production | 38 | 96 | 3.356e-04 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 74 | 230 | 3.391e-04 |
| GO:BP | GO:0050863 | regulation of T cell activation | 110 | 375 | 3.516e-04 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 46 | 125 | 3.931e-04 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 49 | 136 | 3.991e-04 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 62 | 185 | 4.263e-04 |
| GO:BP | GO:0003341 | cilium movement | 69 | 212 | 4.284e-04 |
| GO:BP | GO:0042110 | T cell activation | 153 | 558 | 4.571e-04 |
| GO:BP | GO:0015670 | carbon dioxide transport | 11 | 15 | 5.153e-04 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 195 | 741 | 5.213e-04 |
| GO:BP | GO:0002225 | positive regulation of antimicrobial peptide production | 7 | 7 | 5.269e-04 |
| GO:BP | GO:0052697 | xenobiotic glucuronidation | 7 | 7 | 5.269e-04 |
| GO:BP | GO:0019755 | one-carbon compound transport | 15 | 25 | 5.456e-04 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 31 | 74 | 5.586e-04 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 35 | 88 | 6.260e-04 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 39 | 102 | 6.260e-04 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 40 | 106 | 7.014e-04 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 38 | 99 | 7.157e-04 |
| GO:BP | GO:0002759 | regulation of antimicrobial humoral response | 9 | 11 | 7.523e-04 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 54 | 158 | 7.761e-04 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 50 | 143 | 7.761e-04 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 37 | 96 | 8.104e-04 |
| GO:BP | GO:0030277 | maintenance of gastrointestinal epithelium | 14 | 23 | 8.118e-04 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 257 | 1022 | 8.286e-04 |
| GO:BP | GO:0032613 | interleukin-10 production | 26 | 59 | 8.894e-04 |
| GO:BP | GO:0032653 | regulation of interleukin-10 production | 26 | 59 | 8.894e-04 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 31 | 76 | 9.966e-04 |
| GO:BP | GO:0032637 | interleukin-8 production | 33 | 83 | 1.030e-03 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 33 | 83 | 1.030e-03 |
| GO:BP | GO:0032757 | positive regulation of interleukin-8 production | 27 | 63 | 1.130e-03 |
| GO:BP | GO:0002673 | regulation of acute inflammatory response | 22 | 47 | 1.171e-03 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 34 | 87 | 1.182e-03 |
| GO:BP | GO:0050482 | arachidonate secretion | 17 | 32 | 1.192e-03 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 17 | 32 | 1.192e-03 |
| GO:BP | GO:1903963 | arachidonate transport | 17 | 32 | 1.192e-03 |
| GO:BP | GO:0002687 | positive regulation of leukocyte migration | 52 | 153 | 1.220e-03 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 50 | 146 | 1.361e-03 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1261 | 5795 | 1.384e-03 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 50 | 147 | 1.653e-03 |
| GO:BP | GO:0042098 | T cell proliferation | 67 | 213 | 1.665e-03 |
| GO:BP | GO:0009111 | vitamin catabolic process | 10 | 14 | 1.665e-03 |
| GO:BP | GO:0007602 | phototransduction | 22 | 48 | 1.679e-03 |
| GO:BP | GO:0009583 | detection of light stimulus | 28 | 68 | 1.847e-03 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 62 | 194 | 1.847e-03 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1273 | 5865 | 1.948e-03 |
| GO:BP | GO:0007281 | germ cell development | 105 | 370 | 2.077e-03 |
| GO:BP | GO:0051552 | flavone metabolic process | 6 | 6 | 2.251e-03 |
| GO:BP | GO:1901317 | regulation of flagellated sperm motility | 13 | 22 | 2.251e-03 |
| GO:BP | GO:0002786 | regulation of antibacterial peptide production | 6 | 6 | 2.251e-03 |
| GO:BP | GO:0051673 | disruption of plasma membrane integrity in another organism | 6 | 6 | 2.251e-03 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 26 | 62 | 2.262e-03 |
| GO:BP | GO:0032615 | interleukin-12 production | 26 | 62 | 2.262e-03 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 20 | 43 | 2.735e-03 |
| GO:BP | GO:0002227 | innate immune response in mucosa | 15 | 28 | 2.833e-03 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 29 | 73 | 2.833e-03 |
| GO:BP | GO:1902221 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process | 15 | 28 | 2.833e-03 |
| GO:BP | GO:0060295 | regulation of cilium movement involved in cell motility | 16 | 31 | 2.946e-03 |
| GO:BP | GO:1902019 | regulation of cilium-dependent cell motility | 16 | 31 | 2.946e-03 |
| GO:BP | GO:0042113 | B cell activation | 83 | 282 | 2.979e-03 |
| GO:BP | GO:0035036 | sperm-egg recognition | 23 | 53 | 3.039e-03 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 37 | 102 | 3.171e-03 |
| GO:BP | GO:0031347 | regulation of defense response | 199 | 783 | 3.171e-03 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 49 | 147 | 3.178e-03 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 263 | 1071 | 3.242e-03 |
| GO:BP | GO:0002715 | regulation of natural killer cell mediated immunity | 22 | 50 | 3.287e-03 |
| GO:BP | GO:0002544 | chronic inflammatory response | 12 | 20 | 3.327e-03 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 38 | 106 | 3.346e-03 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 38 | 106 | 3.346e-03 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 39 | 110 | 3.566e-03 |
| GO:BP | GO:0032612 | interleukin-1 production | 39 | 110 | 3.566e-03 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 84 | 288 | 3.652e-03 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 64 | 207 | 3.794e-03 |
| GO:BP | GO:0015669 | gas transport | 13 | 23 | 3.873e-03 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 53 | 164 | 4.006e-03 |
| GO:BP | GO:0032733 | positive regulation of interleukin-10 production | 19 | 41 | 4.006e-03 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 53 | 164 | 4.006e-03 |
| GO:BP | GO:0009072 | aromatic amino acid metabolic process | 19 | 41 | 4.006e-03 |
| GO:BP | GO:0060259 | regulation of feeding behavior | 14 | 26 | 4.204e-03 |
| GO:BP | GO:0042755 | eating behavior | 18 | 38 | 4.204e-03 |
| GO:BP | GO:0002825 | regulation of T-helper 1 type immune response | 16 | 32 | 4.434e-03 |
| GO:BP | GO:0009988 | cell-cell recognition | 29 | 75 | 4.561e-03 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 211 | 843 | 4.722e-03 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 54 | 169 | 4.722e-03 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 54 | 169 | 4.722e-03 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 41 | 120 | 5.800e-03 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 160 | 619 | 5.831e-03 |
| GO:BP | GO:0002228 | natural killer cell mediated immunity | 29 | 76 | 5.859e-03 |
| GO:BP | GO:0006968 | cellular defense response | 22 | 52 | 6.103e-03 |
| GO:BP | GO:0044058 | regulation of digestive system process | 18 | 39 | 6.143e-03 |
| GO:BP | GO:0030183 | B cell differentiation | 51 | 159 | 6.256e-03 |
| GO:BP | GO:0070555 | response to interleukin-1 | 44 | 132 | 6.378e-03 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 16 | 33 | 6.736e-03 |
| GO:BP | GO:0042832 | defense response to protozoan | 14 | 27 | 6.782e-03 |
| GO:BP | GO:0015671 | oxygen transport | 10 | 16 | 7.332e-03 |
| GO:BP | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 23 | 56 | 7.332e-03 |
| GO:BP | GO:0034374 | low-density lipoprotein particle remodeling | 10 | 16 | 7.332e-03 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 127 | 478 | 7.559e-03 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 39 | 114 | 7.636e-03 |
| GO:BP | GO:0008037 | cell recognition | 50 | 157 | 8.438e-03 |
| GO:BP | GO:0072678 | T cell migration | 28 | 74 | 8.473e-03 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 51 | 161 | 8.473e-03 |
| GO:BP | GO:0042436 | indole-containing compound catabolic process | 7 | 9 | 9.139e-03 |
| GO:BP | GO:0006569 | tryptophan catabolic process | 7 | 9 | 9.139e-03 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 35 | 100 | 9.235e-03 |
| GO:BP | GO:0002803 | positive regulation of antibacterial peptide production | 5 | 5 | 9.235e-03 |
| GO:BP | GO:0009581 | detection of external stimulus | 46 | 142 | 9.235e-03 |
| GO:BP | GO:0052696 | flavonoid glucuronidation | 5 | 5 | 9.235e-03 |
| GO:BP | GO:0019369 | arachidonate metabolic process | 23 | 57 | 9.561e-03 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 41 | 123 | 9.712e-03 |
| GO:BP | GO:0030431 | sleep | 12 | 22 | 9.851e-03 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 33 | 93 | 9.917e-03 |
| GO:BP | GO:0055078 | sodium ion homeostasis | 20 | 47 | 9.917e-03 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 33 | 93 | 9.917e-03 |
| GO:BP | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway | 6 | 7 | 1.046e-02 |
| GO:BP | GO:0050795 | regulation of behavior | 28 | 75 | 1.046e-02 |
| GO:BP | GO:0010273 | detoxification of copper ion | 9 | 14 | 1.054e-02 |
| GO:BP | GO:0070942 | neutrophil mediated cytotoxicity | 9 | 14 | 1.054e-02 |
| GO:BP | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | 9 | 14 | 1.054e-02 |
| GO:BP | GO:1990169 | stress response to copper ion | 9 | 14 | 1.054e-02 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 21 | 51 | 1.193e-02 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 63 | 212 | 1.215e-02 |
| GO:BP | GO:0001523 | retinoid metabolic process | 30 | 83 | 1.228e-02 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 28 | 76 | 1.312e-02 |
| GO:BP | GO:0034097 | response to cytokine | 228 | 937 | 1.314e-02 |
| GO:BP | GO:0006775 | fat-soluble vitamin metabolic process | 20 | 48 | 1.330e-02 |
| GO:BP | GO:0008228 | opsonization | 10 | 17 | 1.330e-02 |
| GO:BP | GO:0050766 | positive regulation of phagocytosis | 26 | 69 | 1.364e-02 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 33 | 95 | 1.470e-02 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 98 | 361 | 1.513e-02 |
| GO:BP | GO:0032661 | regulation of interleukin-18 production | 8 | 12 | 1.545e-02 |
| GO:BP | GO:0018149 | peptide cross-linking | 14 | 29 | 1.545e-02 |
| GO:BP | GO:0070943 | neutrophil-mediated killing of symbiont cell | 8 | 12 | 1.545e-02 |
| GO:BP | GO:0032621 | interleukin-18 production | 8 | 12 | 1.545e-02 |
| GO:BP | GO:0001912 | positive regulation of leukocyte mediated cytotoxicity | 25 | 66 | 1.549e-02 |
| GO:BP | GO:0042445 | hormone metabolic process | 70 | 243 | 1.549e-02 |
| GO:BP | GO:0098581 | detection of external biotic stimulus | 13 | 26 | 1.571e-02 |
| GO:BP | GO:0016101 | diterpenoid metabolic process | 31 | 88 | 1.584e-02 |
| GO:BP | GO:1901652 | response to peptide | 230 | 950 | 1.603e-02 |
| GO:BP | GO:0042116 | macrophage activation | 36 | 107 | 1.629e-02 |
| GO:BP | GO:0051607 | defense response to virus | 86 | 311 | 1.666e-02 |
| GO:BP | GO:0031349 | positive regulation of defense response | 126 | 484 | 1.670e-02 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 60 | 203 | 1.791e-02 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 100 | 372 | 1.845e-02 |
| GO:BP | GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 19 | 46 | 1.967e-02 |
| GO:BP | GO:0141005 | transposable element silencing by heterochromatin formation | 9 | 15 | 2.007e-02 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 60 | 204 | 2.027e-02 |
| GO:BP | GO:0010669 | epithelial structure maintenance | 15 | 33 | 2.106e-02 |
| GO:BP | GO:0042573 | retinoic acid metabolic process | 15 | 33 | 2.106e-02 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 56 | 188 | 2.117e-02 |
| GO:BP | GO:0008343 | adult feeding behavior | 7 | 10 | 2.169e-02 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 7 | 10 | 2.169e-02 |
| GO:BP | GO:0035747 | natural killer cell chemotaxis | 7 | 10 | 2.169e-02 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1209 | 5646 | 2.169e-02 |
| GO:BP | GO:0001562 | response to protozoan | 14 | 30 | 2.230e-02 |
| GO:BP | GO:0006869 | lipid transport | 113 | 431 | 2.256e-02 |
| GO:BP | GO:0019373 | epoxygenase P450 pathway | 10 | 18 | 2.256e-02 |
| GO:BP | GO:0046456 | icosanoid biosynthetic process | 22 | 57 | 2.269e-02 |
| GO:BP | GO:0002675 | positive regulation of acute inflammatory response | 13 | 27 | 2.330e-02 |
| GO:BP | GO:0002577 | regulation of antigen processing and presentation | 11 | 21 | 2.357e-02 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 56 | 189 | 2.357e-02 |
| GO:BP | GO:0002523 | leukocyte migration involved in inflammatory response | 11 | 21 | 2.357e-02 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 56 | 189 | 2.357e-02 |
| GO:BP | GO:0009074 | aromatic amino acid family catabolic process | 12 | 24 | 2.367e-02 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 40 | 125 | 2.419e-02 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 45 | 145 | 2.498e-02 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 26 | 72 | 2.572e-02 |
| GO:BP | GO:0010817 | regulation of hormone levels | 139 | 548 | 2.650e-02 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 90 | 334 | 2.845e-02 |
| GO:BP | GO:0002690 | positive regulation of leukocyte chemotaxis | 32 | 95 | 2.859e-02 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 18 | 44 | 2.861e-02 |
| GO:BP | GO:0032741 | positive regulation of interleukin-18 production | 6 | 8 | 2.932e-02 |
| GO:BP | GO:0070268 | cornification | 6 | 8 | 2.932e-02 |
| GO:BP | GO:2000501 | regulation of natural killer cell chemotaxis | 6 | 8 | 2.932e-02 |
| GO:BP | GO:0060456 | positive regulation of digestive system process | 8 | 13 | 2.948e-02 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 23 | 62 | 3.141e-02 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 23 | 62 | 3.141e-02 |
| GO:BP | GO:0042267 | natural killer cell mediated cytotoxicity | 26 | 73 | 3.153e-02 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 128 | 502 | 3.217e-02 |
| GO:BP | GO:0060046 | regulation of acrosome reaction | 9 | 16 | 3.408e-02 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 24 | 66 | 3.408e-02 |
| GO:BP | GO:0035821 | modulation of process of another organism | 9 | 16 | 3.408e-02 |
| GO:BP | GO:0072378 | blood coagulation, fibrin clot formation | 9 | 16 | 3.408e-02 |
| GO:BP | GO:0051969 | regulation of transmission of nerve impulse | 9 | 16 | 3.408e-02 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 98 | 371 | 3.419e-02 |
| GO:BP | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0002325 | natural killer cell differentiation involved in immune response | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0048520 | positive regulation of behavior | 12 | 25 | 3.485e-02 |
| GO:BP | GO:0042747 | circadian sleep/wake cycle, REM sleep | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0036101 | leukotriene B4 catabolic process | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0042430 | indole-containing compound metabolic process | 12 | 25 | 3.485e-02 |
| GO:BP | GO:0042377 | vitamin K catabolic process | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0042376 | phylloquinone catabolic process | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0042374 | phylloquinone metabolic process | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0036100 | leukotriene catabolic process | 5 | 6 | 3.485e-02 |
| GO:BP | GO:0044278 | disruption of cell wall in another organism | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0097272 | ammonium homeostasis | 5 | 6 | 3.485e-02 |
| GO:BP | GO:0034382 | chylomicron remnant clearance | 5 | 6 | 3.485e-02 |
| GO:BP | GO:0097501 | stress response to metal ion | 10 | 19 | 3.485e-02 |
| GO:BP | GO:0072376 | protein activation cascade | 10 | 19 | 3.485e-02 |
| GO:BP | GO:0019370 | leukotriene biosynthetic process | 10 | 19 | 3.485e-02 |
| GO:BP | GO:0098543 | detection of other organism | 10 | 19 | 3.485e-02 |
| GO:BP | GO:0019805 | quinolinate biosynthetic process | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production | 10 | 19 | 3.485e-02 |
| GO:BP | GO:0060480 | lung goblet cell differentiation | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0032826 | regulation of natural killer cell differentiation involved in immune response | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0051464 | positive regulation of cortisol secretion | 4 | 4 | 3.485e-02 |
| GO:BP | GO:0002827 | positive regulation of T-helper 1 type immune response | 10 | 19 | 3.485e-02 |
| GO:BP | GO:2000851 | positive regulation of glucocorticoid secretion | 4 | 4 | 3.485e-02 |
| GO:BP | GO:2000503 | positive regulation of natural killer cell chemotaxis | 5 | 6 | 3.485e-02 |
| GO:BP | GO:0002456 | T cell mediated immunity | 39 | 124 | 3.533e-02 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 113 | 438 | 3.570e-02 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 26 | 74 | 3.612e-02 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 94 | 355 | 3.613e-02 |
| GO:BP | GO:0034105 | positive regulation of tissue remodeling | 7 | 11 | 4.159e-02 |
| GO:BP | GO:0021562 | vestibulocochlear nerve development | 7 | 11 | 4.159e-02 |
| GO:BP | GO:0032372 | negative regulation of sterol transport | 7 | 11 | 4.159e-02 |
| GO:BP | GO:0032375 | negative regulation of cholesterol transport | 7 | 11 | 4.159e-02 |
| GO:BP | GO:1903027 | regulation of opsonization | 7 | 11 | 4.159e-02 |
| GO:BP | GO:0070944 | neutrophil-mediated killing of bacterium | 7 | 11 | 4.159e-02 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 25 | 71 | 4.166e-02 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 22 | 60 | 4.209e-02 |
| GO:BP | GO:0046942 | carboxylic acid transport | 91 | 344 | 4.277e-02 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 62 | 220 | 4.368e-02 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 74 | 271 | 4.404e-02 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 16 | 39 | 4.443e-02 |
| GO:BP | GO:0006910 | phagocytosis, recognition | 13 | 29 | 4.443e-02 |
| GO:BP | GO:0015849 | organic acid transport | 91 | 345 | 4.636e-02 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 85 | 319 | 4.654e-02 |
| GO:BP | GO:0002430 | complement receptor mediated signaling pathway | 8 | 14 | 4.950e-02 |
| KEGG | KEGG:04740 | Olfactory transduction | 368 | 428 | 7.954e-184 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 148 | 291 | 9.354e-26 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 173 | 365 | 9.354e-26 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 146 | 310 | 2.799e-21 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 58 | 86 | 1.424e-17 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 59 | 98 | 1.828e-14 |
| KEGG | KEGG:04742 | Taste transduction | 46 | 85 | 4.526e-09 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 32 | 49 | 4.526e-09 |
| KEGG | KEGG:00830 | Retinol metabolism | 39 | 68 | 1.056e-08 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 38 | 74 | 1.027e-06 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 44 | 92 | 1.236e-06 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 35 | 67 | 1.708e-06 |
| KEGG | KEGG:05310 | Asthma | 19 | 27 | 2.846e-06 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 26 | 44 | 2.846e-06 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 34 | 68 | 8.326e-06 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 40 | 88 | 1.881e-05 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 19 | 30 | 2.755e-05 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 62 | 162 | 3.659e-05 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 21 | 37 | 8.608e-05 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 18 | 30 | 1.351e-04 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 29 | 61 | 1.530e-04 |
| KEGG | KEGG:04976 | Bile secretion | 38 | 89 | 1.598e-04 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 51 | 132 | 1.676e-04 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 29 | 62 | 1.909e-04 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 29 | 62 | 1.909e-04 |
| KEGG | KEGG:05144 | Malaria | 24 | 49 | 3.872e-04 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 19 | 36 | 6.806e-04 |
| KEGG | KEGG:05330 | Allograft rejection | 18 | 34 | 9.773e-04 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 15 | 26 | 1.019e-03 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 45 | 124 | 2.337e-03 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 20 | 43 | 3.383e-03 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 19 | 40 | 3.383e-03 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 18 | 37 | 3.383e-03 |
| KEGG | KEGG:05164 | Influenza A | 56 | 167 | 4.074e-03 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 33 | 86 | 4.297e-03 |
| KEGG | KEGG:05152 | Tuberculosis | 58 | 175 | 4.297e-03 |
| KEGG | KEGG:05143 | African trypanosomiasis | 17 | 36 | 6.533e-03 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 17 | 36 | 6.533e-03 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 51 | 153 | 7.350e-03 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 30 | 79 | 8.217e-03 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 19 | 43 | 8.340e-03 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 37 | 106 | 1.339e-02 |
| KEGG | KEGG:04972 | Pancreatic secretion | 35 | 102 | 2.385e-02 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 12 | 26 | 4.247e-02 |
#write.csv(tableNR, "output/table_NRmotif.csv")
#GO:BP
tableNR_GOBP_pp <- tableNR_pp %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(tableNR_GOBP_pp, "data/tableNR_GOBP_postprob.RDS")
tableNR_GOBP_pp %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("No Response Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))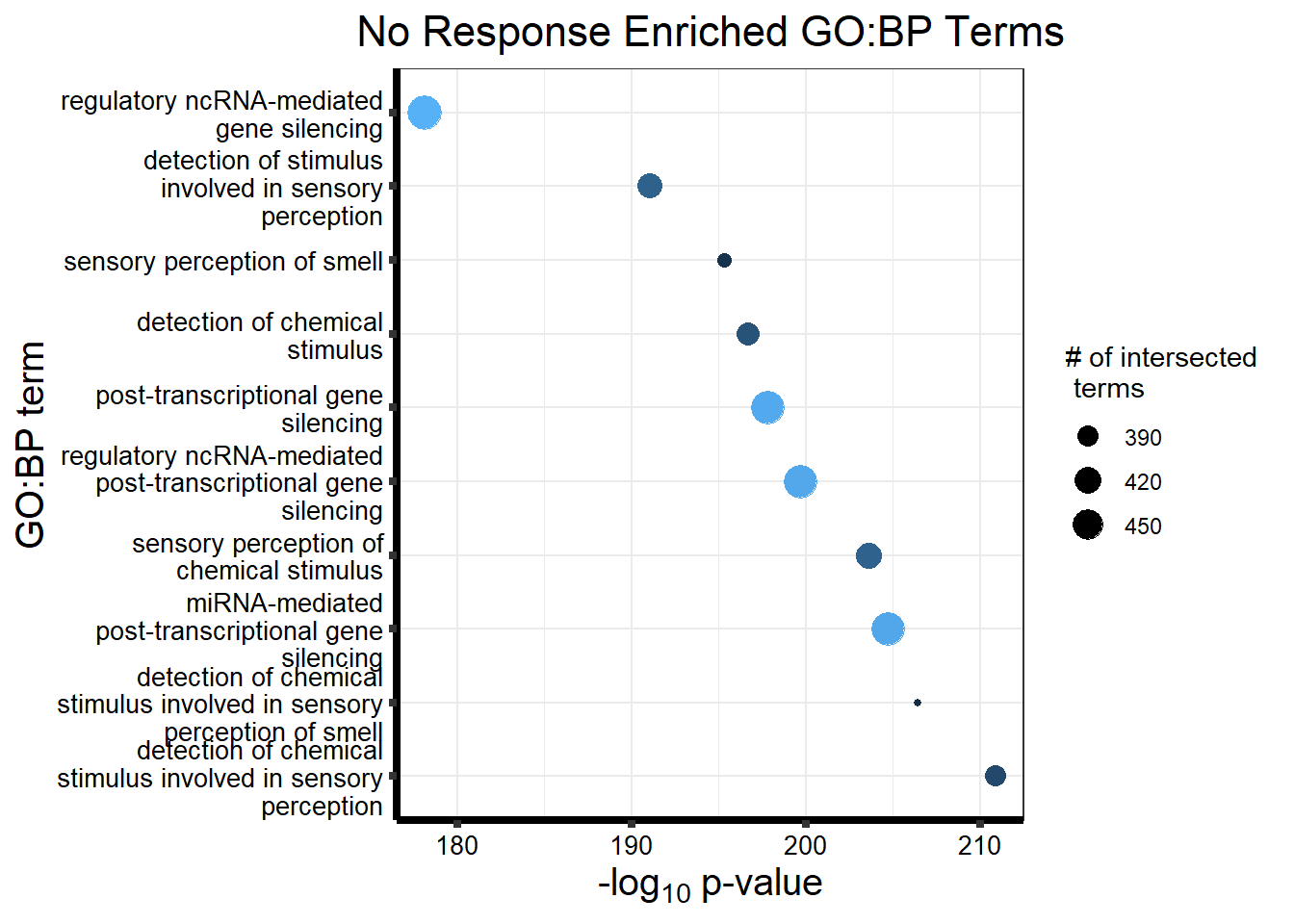
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#KEGG
tableNR_KEGG_pp <- tableNR_pp %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
tableNR_KEGG_pp %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("No Response Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))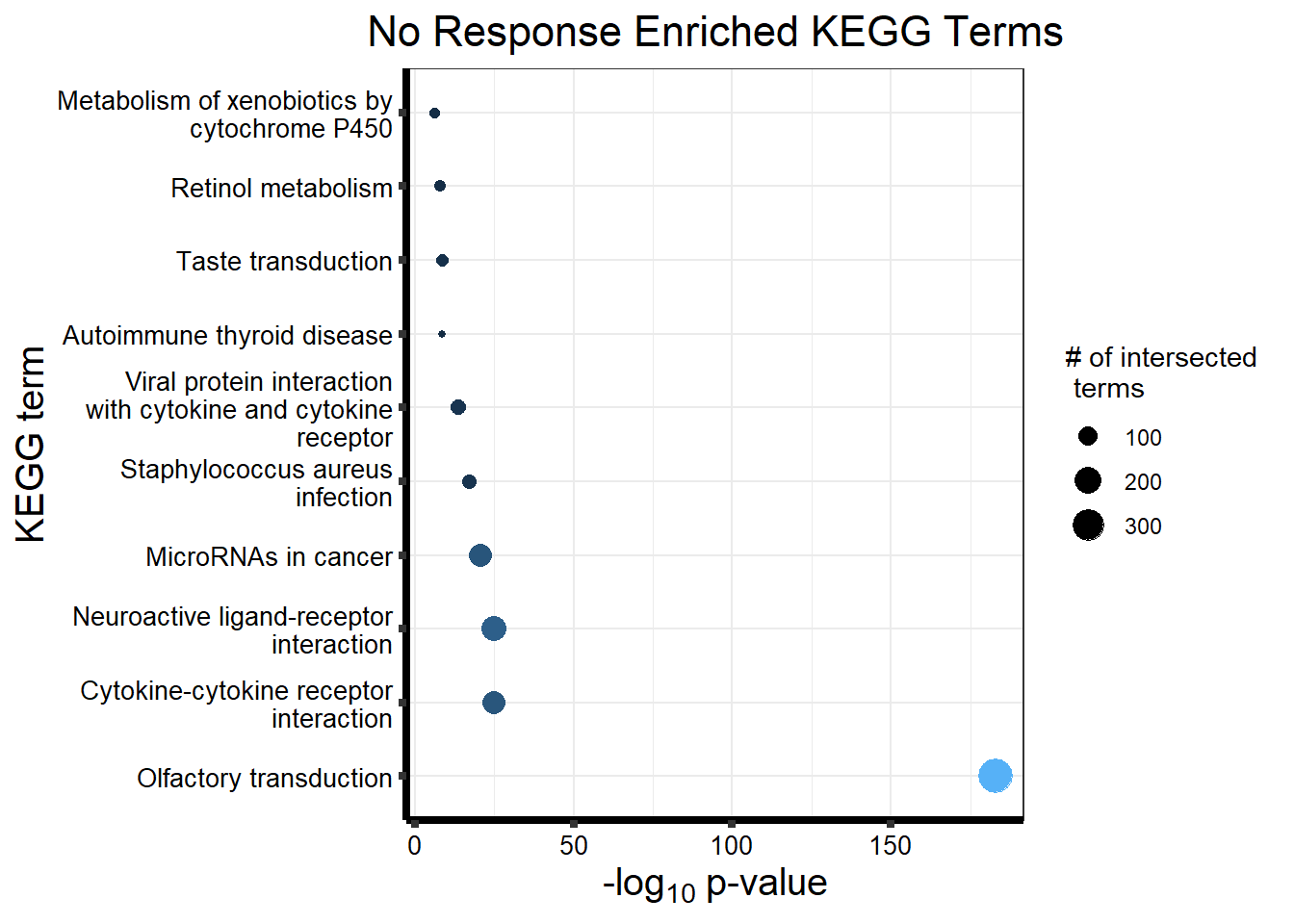
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
The Acute Response had no genes pop up for clustlike at the cutoff of > 0.5 However, when we changed the cutoff and used post prob we got some genes, so let’s use them for GO analysis This is from motif 2, where there is a light gray box in the first study (DOX 24hr) This one doesn’t seem to work, most likely since it’s not really picking up a motif
Next, we’ll look at the Late Response (motif 3)
motif_LD_d <- clust3_d
LDmotif_genes_d <- gost(query = motif_LD_d,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
cormotifLDclust_d <- gostplot(LDmotif_genes_d, capped = FALSE, interactive = TRUE)
cormotifLDclust_dtableLD_d <- LDmotif_genes_d$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
tableLD_d %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006811 | monoatomic ion transport | 90 | 1256 | 4.956e-16 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 75 | 1017 | 9.212e-14 |
| GO:BP | GO:0055085 | transmembrane transport | 95 | 1538 | 2.667e-13 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 68 | 903 | 5.761e-13 |
| GO:BP | GO:0015698 | inorganic anion transport | 26 | 165 | 7.506e-11 |
| GO:BP | GO:0006820 | monoatomic anion transport | 26 | 167 | 8.385e-11 |
| GO:BP | GO:0006812 | monoatomic cation transport | 68 | 1032 | 2.164e-10 |
| GO:BP | GO:0030001 | metal ion transport | 61 | 875 | 3.272e-10 |
| GO:BP | GO:0030154 | cell differentiation | 178 | 4381 | 1.062e-09 |
| GO:BP | GO:0048869 | cellular developmental process | 178 | 4382 | 1.062e-09 |
| GO:BP | GO:0006821 | chloride transport | 21 | 122 | 1.495e-09 |
| GO:BP | GO:0032502 | developmental process | 237 | 6478 | 1.495e-09 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 21 | 127 | 2.871e-09 |
| GO:BP | GO:0006810 | transport | 174 | 4341 | 5.023e-09 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 55 | 832 | 2.061e-08 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 21 | 142 | 2.086e-08 |
| GO:BP | GO:0048468 | cell development | 125 | 2833 | 2.754e-08 |
| GO:BP | GO:0032501 | multicellular organismal process | 252 | 7234 | 2.772e-08 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 18 | 107 | 4.673e-08 |
| GO:BP | GO:0048856 | anatomical structure development | 215 | 5924 | 5.365e-08 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 52 | 811 | 1.500e-07 |
| GO:BP | GO:0050896 | response to stimulus | 293 | 8993 | 3.487e-07 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 25 | 248 | 9.241e-07 |
| GO:BP | GO:0007155 | cell adhesion | 75 | 1511 | 2.123e-06 |
| GO:BP | GO:0002521 | leukocyte differentiation | 42 | 632 | 2.228e-06 |
| GO:BP | GO:0051234 | establishment of localization | 178 | 4869 | 2.720e-06 |
| GO:BP | GO:0006813 | potassium ion transport | 23 | 238 | 7.285e-06 |
| GO:BP | GO:0006968 | cellular defense response | 11 | 52 | 9.549e-06 |
| GO:BP | GO:0065008 | regulation of biological quality | 116 | 2847 | 9.549e-06 |
| GO:BP | GO:0009605 | response to external stimulus | 100 | 2367 | 1.757e-05 |
| GO:BP | GO:0098609 | cell-cell adhesion | 52 | 948 | 1.829e-05 |
| GO:BP | GO:0006814 | sodium ion transport | 22 | 235 | 2.133e-05 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 35 | 529 | 3.275e-05 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 15 | 116 | 3.305e-05 |
| GO:BP | GO:0042110 | T cell activation | 36 | 558 | 3.809e-05 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 18 | 169 | 3.911e-05 |
| GO:BP | GO:0045321 | leukocyte activation | 51 | 951 | 4.086e-05 |
| GO:BP | GO:0051179 | localization | 190 | 5521 | 4.814e-05 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 34 | 521 | 5.698e-05 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 42 | 723 | 5.704e-05 |
| GO:BP | GO:0050957 | equilibrioception | 5 | 8 | 6.531e-05 |
| GO:BP | GO:0050877 | nervous system process | 69 | 1489 | 6.786e-05 |
| GO:BP | GO:0007267 | cell-cell signaling | 63 | 1316 | 7.215e-05 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 12 | 79 | 7.215e-05 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 34 | 530 | 7.306e-05 |
| GO:BP | GO:0001775 | cell activation | 55 | 1098 | 9.547e-05 |
| GO:BP | GO:0015711 | organic anion transport | 30 | 442 | 1.018e-04 |
| GO:BP | GO:0060004 | reflex | 7 | 23 | 1.129e-04 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 36 | 598 | 1.402e-04 |
| GO:BP | GO:0065007 | biological regulation | 369 | 12671 | 1.668e-04 |
| GO:BP | GO:0046649 | lymphocyte activation | 43 | 787 | 1.668e-04 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 18 | 192 | 1.777e-04 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 36 | 608 | 1.876e-04 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 19 | 213 | 1.876e-04 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 29 | 438 | 2.167e-04 |
| GO:BP | GO:0019725 | cellular homeostasis | 44 | 826 | 2.191e-04 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 23 | 306 | 2.958e-04 |
| GO:BP | GO:0048232 | male gamete generation | 37 | 654 | 3.582e-04 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 24 | 334 | 3.751e-04 |
| GO:BP | GO:0007154 | cell communication | 211 | 6496 | 3.751e-04 |
| GO:BP | GO:0030217 | T cell differentiation | 23 | 312 | 3.821e-04 |
| GO:BP | GO:0023052 | signaling | 210 | 6471 | 4.153e-04 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 32 | 531 | 4.166e-04 |
| GO:BP | GO:0048878 | chemical homeostasis | 50 | 1026 | 4.787e-04 |
| GO:BP | GO:0099536 | synaptic signaling | 41 | 783 | 6.366e-04 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 23 | 325 | 6.808e-04 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 22 | 302 | 6.808e-04 |
| GO:BP | GO:0050778 | positive regulation of immune response | 39 | 732 | 6.847e-04 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 23 | 326 | 6.950e-04 |
| GO:BP | GO:0010817 | regulation of hormone levels | 32 | 548 | 7.129e-04 |
| GO:BP | GO:0007283 | spermatogenesis | 35 | 637 | 9.995e-04 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 16 | 181 | 1.010e-03 |
| GO:BP | GO:0007275 | multicellular organism development | 158 | 4658 | 1.033e-03 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 32 | 561 | 1.078e-03 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 57 | 1275 | 1.156e-03 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 27 | 436 | 1.172e-03 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 19 | 248 | 1.189e-03 |
| GO:BP | GO:0042592 | homeostatic process | 71 | 1711 | 1.191e-03 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 24 | 366 | 1.314e-03 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 39 | 759 | 1.322e-03 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 48 | 1022 | 1.520e-03 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 24 | 372 | 1.648e-03 |
| GO:BP | GO:0030097 | hemopoiesis | 46 | 971 | 1.787e-03 |
| GO:BP | GO:0050863 | regulation of T cell activation | 24 | 375 | 1.830e-03 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 21 | 305 | 1.967e-03 |
| GO:BP | GO:0022414 | reproductive process | 65 | 1556 | 2.075e-03 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 23 | 355 | 2.110e-03 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 25 | 404 | 2.110e-03 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 38 | 753 | 2.217e-03 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 38 | 753 | 2.217e-03 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 21 | 309 | 2.223e-03 |
| GO:BP | GO:0050776 | regulation of immune response | 43 | 897 | 2.229e-03 |
| GO:BP | GO:0050865 | regulation of cell activation | 33 | 616 | 2.229e-03 |
| GO:BP | GO:0140013 | meiotic nuclear division | 16 | 198 | 2.339e-03 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 67 | 1632 | 2.405e-03 |
| GO:BP | GO:0042445 | hormone metabolic process | 18 | 243 | 2.449e-03 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 12 | 119 | 2.449e-03 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 17 | 222 | 2.577e-03 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 13 | 139 | 2.588e-03 |
| GO:BP | GO:0002253 | activation of immune response | 30 | 543 | 2.646e-03 |
| GO:BP | GO:0007276 | gamete generation | 40 | 822 | 2.766e-03 |
| GO:BP | GO:0003008 | system process | 86 | 2266 | 2.995e-03 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 19 | 271 | 3.000e-03 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 8 | 55 | 3.073e-03 |
| GO:BP | GO:0006955 | immune response | 78 | 2008 | 3.073e-03 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 15 | 183 | 3.073e-03 |
| GO:BP | GO:0032604 | granulocyte macrophage colony-stimulating factor production | 5 | 18 | 3.073e-03 |
| GO:BP | GO:0032645 | regulation of granulocyte macrophage colony-stimulating factor production | 5 | 18 | 3.073e-03 |
| GO:BP | GO:0046942 | carboxylic acid transport | 22 | 344 | 3.073e-03 |
| GO:BP | GO:0015849 | organic acid transport | 22 | 345 | 3.142e-03 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 48 | 1066 | 3.154e-03 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 9 | 71 | 3.199e-03 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 45 | 980 | 3.354e-03 |
| GO:BP | GO:0007610 | behavior | 34 | 665 | 3.479e-03 |
| GO:BP | GO:0019953 | sexual reproduction | 49 | 1103 | 3.554e-03 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 27 | 477 | 3.601e-03 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 28 | 504 | 3.646e-03 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 27 | 478 | 3.666e-03 |
| GO:BP | GO:0070661 | leukocyte proliferation | 22 | 351 | 3.739e-03 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 15 | 189 | 3.842e-03 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 15 | 189 | 3.842e-03 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 19 | 280 | 3.912e-03 |
| GO:BP | GO:0003341 | cilium movement | 16 | 212 | 4.037e-03 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 66 | 1644 | 4.094e-03 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 6 | 31 | 4.366e-03 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 8 | 59 | 4.435e-03 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 16 | 215 | 4.583e-03 |
| GO:BP | GO:0015747 | urate transport | 4 | 11 | 4.583e-03 |
| GO:BP | GO:0042221 | response to chemical | 131 | 3867 | 4.631e-03 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 8 | 60 | 4.852e-03 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 10 | 94 | 4.942e-03 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 97 | 2692 | 5.112e-03 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 19 | 288 | 5.112e-03 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 16 | 218 | 5.112e-03 |
| GO:BP | GO:0051716 | cellular response to stimulus | 224 | 7320 | 5.112e-03 |
| GO:BP | GO:0002376 | immune system process | 100 | 2796 | 5.137e-03 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 7 | 46 | 5.179e-03 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 18 | 265 | 5.179e-03 |
| GO:BP | GO:0007601 | visual perception | 16 | 219 | 5.179e-03 |
| GO:BP | GO:0001816 | cytokine production | 37 | 772 | 5.179e-03 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 12 | 134 | 5.333e-03 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 16 | 220 | 5.382e-03 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 27 | 497 | 5.739e-03 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 24 | 420 | 6.200e-03 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 5 | 22 | 6.481e-03 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 13 | 158 | 6.481e-03 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 27 | 502 | 6.562e-03 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 14 | 180 | 6.614e-03 |
| GO:BP | GO:0048731 | system development | 133 | 3985 | 6.784e-03 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 103 | 2928 | 6.784e-03 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 14 | 182 | 7.273e-03 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 20 | 324 | 7.429e-03 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 67 | 1724 | 7.478e-03 |
| GO:BP | GO:0060284 | regulation of cell development | 39 | 851 | 7.774e-03 |
| GO:BP | GO:0048513 | animal organ development | 106 | 3047 | 7.901e-03 |
| GO:BP | GO:0001817 | regulation of cytokine production | 36 | 765 | 8.085e-03 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 10 | 102 | 8.130e-03 |
| GO:BP | GO:0006869 | lipid transport | 24 | 431 | 8.289e-03 |
| GO:BP | GO:0006816 | calcium ion transport | 24 | 433 | 8.816e-03 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 5 | 24 | 9.108e-03 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 12 | 145 | 9.774e-03 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 18 | 283 | 9.921e-03 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 65 | 1682 | 1.023e-02 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 16 | 236 | 1.033e-02 |
| GO:BP | GO:0042098 | T cell proliferation | 15 | 213 | 1.046e-02 |
| GO:BP | GO:0031341 | regulation of cell killing | 10 | 106 | 1.052e-02 |
| GO:BP | GO:0050789 | regulation of biological process | 346 | 12278 | 1.059e-02 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 61 | 1558 | 1.096e-02 |
| GO:BP | GO:0002682 | regulation of immune system process | 60 | 1526 | 1.096e-02 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 46 | 1088 | 1.196e-02 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 16 | 240 | 1.196e-02 |
| GO:BP | GO:0090660 | cerebrospinal fluid circulation | 4 | 15 | 1.297e-02 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 7 | 55 | 1.297e-02 |
| GO:BP | GO:0001906 | cell killing | 15 | 219 | 1.332e-02 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 16 | 243 | 1.338e-02 |
| GO:BP | GO:0050900 | leukocyte migration | 22 | 396 | 1.373e-02 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 19 | 319 | 1.408e-02 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 7 | 56 | 1.408e-02 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 10 | 111 | 1.408e-02 |
| GO:BP | GO:0051321 | meiotic cell cycle | 18 | 294 | 1.408e-02 |
| GO:BP | GO:0006952 | defense response | 68 | 1809 | 1.432e-02 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 18 | 295 | 1.449e-02 |
| GO:BP | GO:0045588 | positive regulation of gamma-delta T cell differentiation | 3 | 7 | 1.455e-02 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 10 | 112 | 1.476e-02 |
| GO:BP | GO:0007420 | brain development | 34 | 739 | 1.505e-02 |
| GO:BP | GO:0045058 | T cell selection | 7 | 57 | 1.505e-02 |
| GO:BP | GO:0006957 | complement activation, alternative pathway | 4 | 16 | 1.566e-02 |
| GO:BP | GO:0051707 | response to other organism | 60 | 1555 | 1.586e-02 |
| GO:BP | GO:0015913 | short-chain fatty acid transmembrane transport | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0021630 | olfactory nerve maturation | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0015741 | fumarate transport | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 13 | 179 | 1.610e-02 |
| GO:BP | GO:0038112 | interleukin-8-mediated signaling pathway | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0007600 | sensory perception | 41 | 955 | 1.610e-02 |
| GO:BP | GO:0030573 | bile acid catabolic process | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0006817 | phosphate ion transport | 5 | 28 | 1.610e-02 |
| GO:BP | GO:2000471 | regulation of hematopoietic stem cell migration | 2 | 2 | 1.610e-02 |
| GO:BP | GO:2000473 | positive regulation of hematopoietic stem cell migration | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0001762 | beta-alanine transport | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0050883 | musculoskeletal movement, spinal reflex action | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 22 | 403 | 1.610e-02 |
| GO:BP | GO:0009607 | response to biotic stimulus | 61 | 1593 | 1.610e-02 |
| GO:BP | GO:0036368 | cone photoresponse recovery | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0033626 | positive regulation of integrin activation by cell surface receptor linked signal transduction | 2 | 2 | 1.610e-02 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 10 | 115 | 1.632e-02 |
| GO:BP | GO:0042391 | regulation of membrane potential | 23 | 433 | 1.634e-02 |
| GO:BP | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste | 6 | 43 | 1.693e-02 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 12 | 159 | 1.741e-02 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 5 | 29 | 1.757e-02 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 18 | 305 | 1.853e-02 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 6 | 44 | 1.883e-02 |
| GO:BP | GO:0008283 | cell population proliferation | 73 | 2009 | 1.945e-02 |
| GO:BP | GO:0046645 | positive regulation of gamma-delta T cell activation | 3 | 8 | 1.945e-02 |
| GO:BP | GO:0120197 | mucociliary clearance | 3 | 8 | 1.945e-02 |
| GO:BP | GO:0051967 | negative regulation of synaptic transmission, glutamatergic | 3 | 8 | 1.945e-02 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 39 | 909 | 1.987e-02 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 18 | 308 | 2.006e-02 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 6 | 45 | 2.059e-02 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 12 | 163 | 2.059e-02 |
| GO:BP | GO:0007165 | signal transduction | 183 | 5979 | 2.067e-02 |
| GO:BP | GO:0032663 | regulation of interleukin-2 production | 7 | 62 | 2.107e-02 |
| GO:BP | GO:0032623 | interleukin-2 production | 7 | 62 | 2.107e-02 |
| GO:BP | GO:0007417 | central nervous system development | 43 | 1038 | 2.119e-02 |
| GO:BP | GO:0002923 | regulation of humoral immune response mediated by circulating immunoglobulin | 4 | 18 | 2.119e-02 |
| GO:BP | GO:0009581 | detection of external stimulus | 11 | 142 | 2.136e-02 |
| GO:BP | GO:0002252 | immune effector process | 32 | 703 | 2.136e-02 |
| GO:BP | GO:0036336 | dendritic cell migration | 5 | 31 | 2.221e-02 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 11 | 143 | 2.243e-02 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 19 | 339 | 2.270e-02 |
| GO:BP | GO:0098657 | import into cell | 39 | 921 | 2.379e-02 |
| GO:BP | GO:0007626 | locomotory behavior | 14 | 214 | 2.404e-02 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 18 | 315 | 2.424e-02 |
| GO:BP | GO:0048518 | positive regulation of biological process | 189 | 6235 | 2.499e-02 |
| GO:BP | GO:0010876 | lipid localization | 24 | 480 | 2.535e-02 |
| GO:BP | GO:0010720 | positive regulation of cell development | 23 | 452 | 2.535e-02 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 15 | 240 | 2.535e-02 |
| GO:BP | GO:0045216 | cell-cell junction organization | 14 | 216 | 2.551e-02 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 11 | 146 | 2.551e-02 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 12 | 169 | 2.562e-02 |
| GO:BP | GO:0030450 | regulation of complement activation, classical pathway | 3 | 9 | 2.562e-02 |
| GO:BP | GO:0045586 | regulation of gamma-delta T cell differentiation | 3 | 9 | 2.562e-02 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 37 | 866 | 2.575e-02 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 18 | 318 | 2.575e-02 |
| GO:BP | GO:0045471 | response to ethanol | 10 | 125 | 2.615e-02 |
| GO:BP | GO:0021543 | pallium development | 13 | 194 | 2.720e-02 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 11 | 148 | 2.752e-02 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 7 | 66 | 2.752e-02 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 24 | 486 | 2.851e-02 |
| GO:BP | GO:0060348 | bone development | 14 | 220 | 2.903e-02 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 179 | 5886 | 2.949e-02 |
| GO:BP | GO:0050909 | sensory perception of taste | 7 | 67 | 2.960e-02 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 10 | 128 | 3.030e-02 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 5 | 34 | 3.068e-02 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 11 | 151 | 3.133e-02 |
| GO:BP | GO:0071695 | anatomical structure maturation | 16 | 274 | 3.268e-02 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 42 | 1037 | 3.268e-02 |
| GO:BP | GO:0060322 | head development | 34 | 789 | 3.304e-02 |
| GO:BP | GO:0023061 | signal release | 24 | 493 | 3.314e-02 |
| GO:BP | GO:2001204 | regulation of osteoclast development | 3 | 10 | 3.319e-02 |
| GO:BP | GO:0046643 | regulation of gamma-delta T cell activation | 3 | 10 | 3.319e-02 |
| GO:BP | GO:0030449 | regulation of complement activation | 4 | 21 | 3.340e-02 |
| GO:BP | GO:0033563 | dorsal/ventral axon guidance | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0021605 | cranial nerve maturation | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0072249 | metanephric podocyte development | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0072312 | metanephric glomerular epithelial cell differentiation | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0071486 | cellular response to high light intensity | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0045957 | negative regulation of complement activation, alternative pathway | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0015912 | short-chain fatty acid transport | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 94 | 2803 | 3.423e-02 |
| GO:BP | GO:0072313 | metanephric glomerular epithelial cell development | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0070543 | response to linoleic acid | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0097195 | pilomotor reflex | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0007127 | meiosis I | 10 | 132 | 3.423e-02 |
| GO:BP | GO:0010482 | regulation of epidermal cell division | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0072248 | metanephric podocyte differentiation | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0072244 | metanephric glomerular epithelium development | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0010481 | epidermal cell division | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0098758 | response to interleukin-8 | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0098759 | cellular response to interleukin-8 | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0006311 | meiotic gene conversion | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0002277 | myeloid dendritic cell activation involved in immune response | 2 | 3 | 3.423e-02 |
| GO:BP | GO:0042471 | ear morphogenesis | 10 | 132 | 3.423e-02 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 9 | 110 | 3.423e-02 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 13 | 203 | 3.521e-02 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 5 | 36 | 3.541e-02 |
| GO:BP | GO:0006959 | humoral immune response | 15 | 254 | 3.625e-02 |
| GO:BP | GO:0070166 | enamel mineralization | 4 | 22 | 3.627e-02 |
| GO:BP | GO:0046629 | gamma-delta T cell activation | 4 | 22 | 3.627e-02 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 76 | 2186 | 3.756e-02 |
| GO:BP | GO:0030900 | forebrain development | 21 | 417 | 3.765e-02 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 58 | 1577 | 3.856e-02 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 8 | 92 | 3.902e-02 |
| GO:BP | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste | 5 | 37 | 3.902e-02 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 3 | 11 | 3.921e-02 |
| GO:BP | GO:0033625 | positive regulation of integrin activation | 3 | 11 | 3.921e-02 |
| GO:BP | GO:1903977 | positive regulation of glial cell migration | 3 | 11 | 3.921e-02 |
| GO:BP | GO:0072015 | podocyte development | 3 | 11 | 3.921e-02 |
| GO:BP | GO:0051898 | negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 6 | 54 | 3.943e-02 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 13 | 207 | 3.943e-02 |
| GO:BP | GO:0048839 | inner ear development | 13 | 207 | 3.943e-02 |
| GO:BP | GO:0007399 | nervous system development | 86 | 2541 | 3.943e-02 |
| GO:BP | GO:0048870 | cell motility | 62 | 1716 | 3.968e-02 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 7 | 73 | 4.027e-02 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 7 | 73 | 4.027e-02 |
| GO:BP | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | 4 | 23 | 4.064e-02 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 34 | 809 | 4.157e-02 |
| GO:BP | GO:0006909 | phagocytosis | 14 | 234 | 4.161e-02 |
| GO:BP | GO:0046164 | alcohol catabolic process | 5 | 38 | 4.161e-02 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 6 | 55 | 4.161e-02 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 6 | 55 | 4.161e-02 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 13 | 209 | 4.161e-02 |
| GO:BP | GO:0046903 | secretion | 39 | 968 | 4.242e-02 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 12 | 185 | 4.242e-02 |
| GO:BP | GO:0071674 | mononuclear cell migration | 14 | 235 | 4.247e-02 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 13 | 210 | 4.271e-02 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 8 | 95 | 4.426e-02 |
| GO:BP | GO:0007281 | germ cell development | 19 | 370 | 4.442e-02 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 6 | 56 | 4.463e-02 |
| GO:BP | GO:0006706 | steroid catabolic process | 4 | 24 | 4.507e-02 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 9 | 117 | 4.507e-02 |
| GO:BP | GO:1903306 | negative regulation of regulated secretory pathway | 4 | 24 | 4.507e-02 |
| GO:BP | GO:0009566 | fertilization | 13 | 212 | 4.507e-02 |
| GO:BP | GO:0009074 | aromatic amino acid family catabolic process | 4 | 24 | 4.507e-02 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 11 | 163 | 4.507e-02 |
| GO:BP | GO:1903818 | positive regulation of voltage-gated potassium channel activity | 3 | 12 | 4.660e-02 |
| GO:BP | GO:0072310 | glomerular epithelial cell development | 3 | 12 | 4.660e-02 |
| GO:BP | GO:0044341 | sodium-dependent phosphate transport | 3 | 12 | 4.660e-02 |
| GO:BP | GO:0032609 | type II interferon production | 9 | 118 | 4.662e-02 |
| GO:BP | GO:0001895 | retina homeostasis | 6 | 57 | 4.708e-02 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 14 | 239 | 4.708e-02 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 10 | 141 | 4.713e-02 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 13 | 214 | 4.769e-02 |
| GO:BP | GO:0009888 | tissue development | 70 | 2012 | 4.842e-02 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 24 | 518 | 4.848e-02 |
| GO:BP | GO:0035270 | endocrine system development | 10 | 142 | 4.900e-02 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 12 | 190 | 4.901e-02 |
| GO:BP | GO:0007605 | sensory perception of sound | 11 | 166 | 4.963e-02 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 37 | 365 | 9.137e-09 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 5 | 16 | 9.906e-03 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 9 | 68 | 1.110e-02 |
| KEGG | KEGG:05033 | Nicotine addiction | 7 | 40 | 1.110e-02 |
#write.csv(tableLD, "output/table_LateDOXmotif.csv")
#GO:BP
tableLD_GOBP_d <- tableLD_d %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(tableLD_GOBP, "data/tableLD_GOBP.RDS")
tableLD_GOBP_d %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Late Response Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))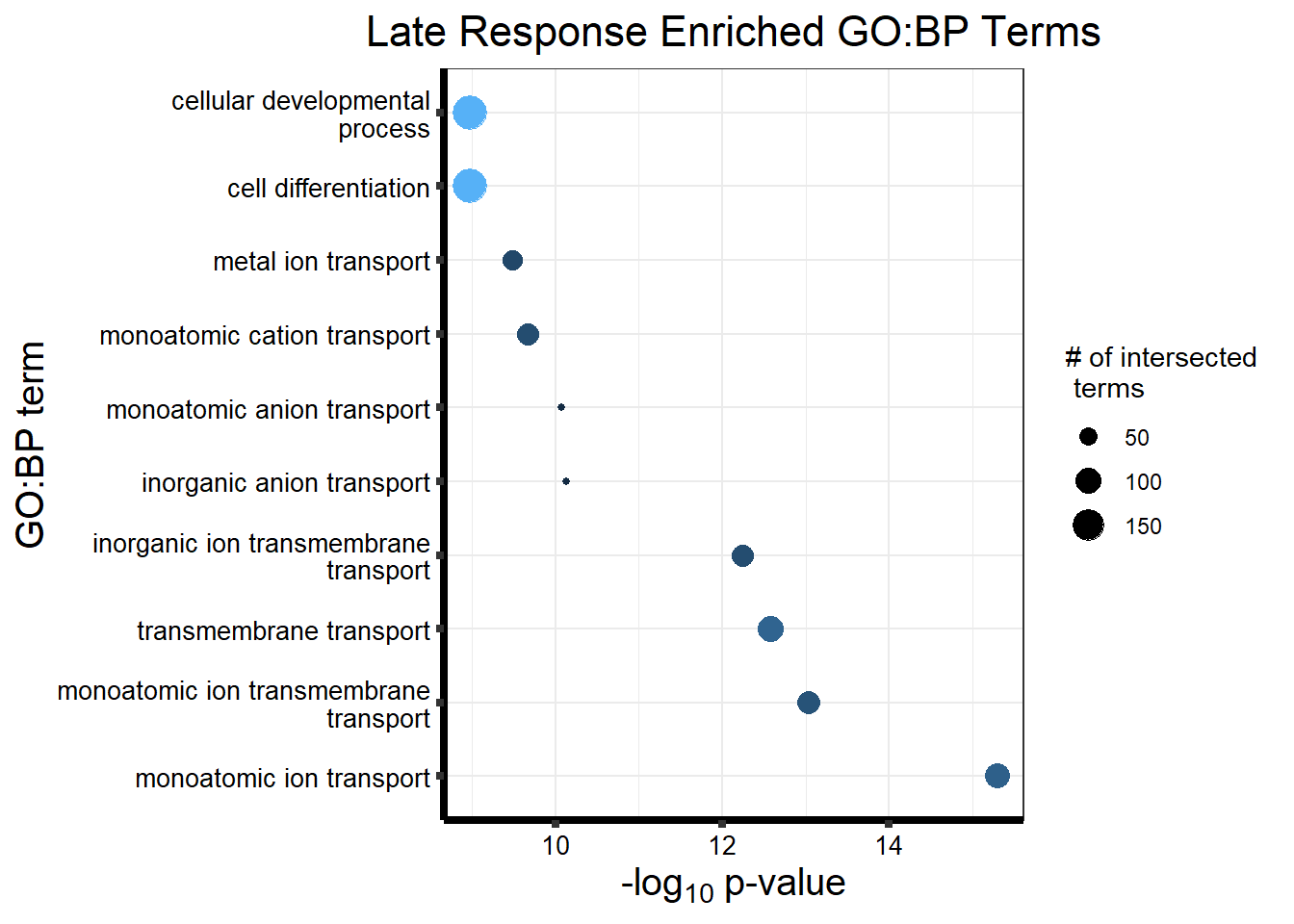
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#KEGG
tableLD_KEGG_d <- tableLD_d %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
tableLD_KEGG_d %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Late Response Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))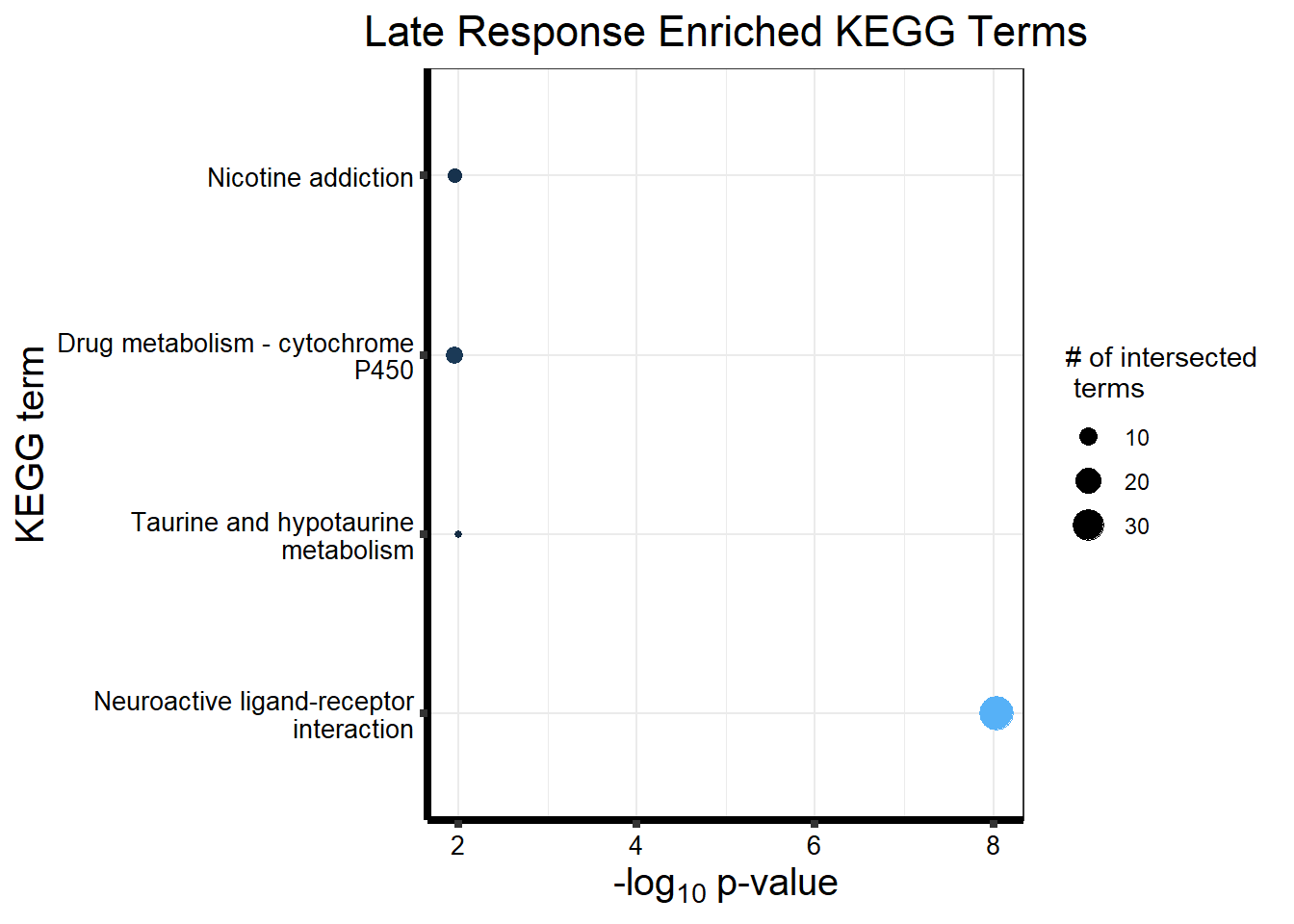
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
Now let’s look at this motif 3 (Late Response) with the posterior probability
motif_LR_pp <- prob_motif_3
LRmotif_genes_pp <- gost(query = motif_LR_pp,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
cormotifLRclust_pp <- gostplot(LRmotif_genes_pp, capped = FALSE, interactive = TRUE)
cormotifLRclust_pptableLR_pp <- LRmotif_genes_pp$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
tableLR_pp %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006811 | monoatomic ion transport | 60 | 1256 | 6.212e-11 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 52 | 1017 | 1.681e-10 |
| GO:BP | GO:0055085 | transmembrane transport | 65 | 1538 | 4.113e-10 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 45 | 903 | 1.161e-08 |
| GO:BP | GO:0015698 | inorganic anion transport | 18 | 165 | 1.375e-07 |
| GO:BP | GO:0006820 | monoatomic anion transport | 18 | 167 | 1.400e-07 |
| GO:BP | GO:0048869 | cellular developmental process | 118 | 4382 | 2.840e-07 |
| GO:BP | GO:0030154 | cell differentiation | 118 | 4381 | 2.840e-07 |
| GO:BP | GO:0006810 | transport | 117 | 4341 | 2.971e-07 |
| GO:BP | GO:0006821 | chloride transport | 15 | 122 | 4.430e-07 |
| GO:BP | GO:0032501 | multicellular organismal process | 167 | 7234 | 1.046e-06 |
| GO:BP | GO:0006812 | monoatomic cation transport | 43 | 1032 | 2.823e-06 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 15 | 142 | 2.844e-06 |
| GO:BP | GO:0032502 | developmental process | 152 | 6478 | 3.014e-06 |
| GO:BP | GO:0048468 | cell development | 83 | 2833 | 4.124e-06 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 13 | 107 | 4.238e-06 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 37 | 832 | 4.606e-06 |
| GO:BP | GO:0050896 | response to stimulus | 193 | 8993 | 4.865e-06 |
| GO:BP | GO:0030001 | metal ion transport | 38 | 875 | 4.881e-06 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 11 | 79 | 1.047e-05 |
| GO:BP | GO:0007267 | cell-cell signaling | 48 | 1316 | 1.298e-05 |
| GO:BP | GO:0051234 | establishment of localization | 120 | 4869 | 1.533e-05 |
| GO:BP | GO:0065008 | regulation of biological quality | 81 | 2847 | 1.552e-05 |
| GO:BP | GO:0065007 | biological regulation | 247 | 12671 | 1.617e-05 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 18 | 248 | 1.781e-05 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 13 | 127 | 2.060e-05 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 34 | 811 | 4.571e-05 |
| GO:BP | GO:0048856 | anatomical structure development | 136 | 5924 | 7.416e-05 |
| GO:BP | GO:0022414 | reproductive process | 51 | 1556 | 9.471e-05 |
| GO:BP | GO:0051179 | localization | 127 | 5521 | 2.049e-04 |
| GO:BP | GO:0006813 | potassium ion transport | 16 | 238 | 2.050e-04 |
| GO:BP | GO:0015711 | organic anion transport | 22 | 442 | 3.627e-04 |
| GO:BP | GO:0023052 | signaling | 142 | 6471 | 4.510e-04 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 8 | 59 | 5.282e-04 |
| GO:BP | GO:0007154 | cell communication | 142 | 6496 | 5.345e-04 |
| GO:BP | GO:0048232 | male gamete generation | 27 | 654 | 8.075e-04 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 36 | 1022 | 8.341e-04 |
| GO:BP | GO:0010817 | regulation of hormone levels | 24 | 548 | 9.563e-04 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 14 | 213 | 9.594e-04 |
| GO:BP | GO:0002521 | leukocyte differentiation | 26 | 632 | 1.150e-03 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 41 | 1275 | 1.504e-03 |
| GO:BP | GO:0050789 | regulation of biological process | 232 | 12278 | 1.750e-03 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 12 | 169 | 1.750e-03 |
| GO:BP | GO:0007275 | multicellular organism development | 107 | 4658 | 1.750e-03 |
| GO:BP | GO:0099536 | synaptic signaling | 29 | 783 | 2.264e-03 |
| GO:BP | GO:0007283 | spermatogenesis | 25 | 637 | 3.208e-03 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 22 | 521 | 3.208e-03 |
| GO:BP | GO:0019953 | sexual reproduction | 36 | 1103 | 3.281e-03 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 27 | 723 | 3.477e-03 |
| GO:BP | GO:0042592 | homeostatic process | 49 | 1711 | 3.593e-03 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 22 | 530 | 3.743e-03 |
| GO:BP | GO:0048731 | system development | 93 | 3985 | 3.743e-03 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 22 | 531 | 3.776e-03 |
| GO:BP | GO:0050877 | nervous system process | 44 | 1489 | 4.173e-03 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 16 | 326 | 5.815e-03 |
| GO:BP | GO:0140013 | meiotic nuclear division | 12 | 198 | 6.435e-03 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 6 | 46 | 6.491e-03 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 27 | 759 | 6.655e-03 |
| GO:BP | GO:0035270 | endocrine system development | 10 | 142 | 6.926e-03 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 23 | 598 | 6.926e-03 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 32 | 980 | 7.045e-03 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 9 | 116 | 7.045e-03 |
| GO:BP | GO:0048878 | chemical homeostasis | 33 | 1026 | 7.079e-03 |
| GO:BP | GO:0006814 | sodium ion transport | 13 | 235 | 7.216e-03 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 15 | 302 | 7.216e-03 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 5 | 31 | 7.902e-03 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 23 | 608 | 7.924e-03 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 15 | 306 | 7.976e-03 |
| GO:BP | GO:0007276 | gamete generation | 28 | 822 | 8.608e-03 |
| GO:BP | GO:0042445 | hormone metabolic process | 13 | 243 | 9.261e-03 |
| GO:BP | GO:0001775 | cell activation | 34 | 1098 | 1.012e-02 |
| GO:BP | GO:0046903 | secretion | 31 | 968 | 1.091e-02 |
| GO:BP | GO:0051716 | cellular response to stimulus | 148 | 7320 | 1.099e-02 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 26 | 753 | 1.099e-02 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 26 | 753 | 1.099e-02 |
| GO:BP | GO:0042221 | response to chemical | 88 | 3867 | 1.099e-02 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 12 | 218 | 1.157e-02 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 45 | 1632 | 1.161e-02 |
| GO:BP | GO:0050957 | equilibrioception | 3 | 8 | 1.165e-02 |
| GO:BP | GO:0060348 | bone development | 12 | 220 | 1.196e-02 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 12 | 220 | 1.196e-02 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 18 | 436 | 1.232e-02 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 6 | 55 | 1.232e-02 |
| GO:BP | GO:0021630 | olfactory nerve maturation | 2 | 2 | 1.261e-02 |
| GO:BP | GO:0042110 | T cell activation | 21 | 558 | 1.261e-02 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 6 | 56 | 1.261e-02 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 18 | 438 | 1.261e-02 |
| GO:BP | GO:0036368 | cone photoresponse recovery | 2 | 2 | 1.261e-02 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 5 | 36 | 1.261e-02 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 11 | 192 | 1.285e-02 |
| GO:BP | GO:0051321 | meiotic cell cycle | 14 | 294 | 1.330e-02 |
| GO:BP | GO:0098609 | cell-cell adhesion | 30 | 948 | 1.330e-02 |
| GO:BP | GO:0007155 | cell adhesion | 42 | 1511 | 1.363e-02 |
| GO:BP | GO:0045321 | leukocyte activation | 30 | 951 | 1.372e-02 |
| GO:BP | GO:0021536 | diencephalon development | 7 | 81 | 1.409e-02 |
| GO:BP | GO:0007600 | sensory perception | 30 | 955 | 1.413e-02 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 15 | 334 | 1.413e-02 |
| GO:BP | GO:0045055 | regulated exocytosis | 12 | 229 | 1.413e-02 |
| GO:BP | GO:0030450 | regulation of complement activation, classical pathway | 3 | 9 | 1.413e-02 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 20 | 529 | 1.475e-02 |
| GO:BP | GO:0019725 | cellular homeostasis | 27 | 826 | 1.487e-02 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 4 | 22 | 1.570e-02 |
| GO:BP | GO:0046649 | lymphocyte activation | 26 | 787 | 1.582e-02 |
| GO:BP | GO:0048732 | gland development | 18 | 454 | 1.582e-02 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 6 | 60 | 1.582e-02 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 14 | 305 | 1.666e-02 |
| GO:BP | GO:0050794 | regulation of cellular process | 219 | 11876 | 1.689e-02 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 11 | 203 | 1.722e-02 |
| GO:BP | GO:0046942 | carboxylic acid transport | 15 | 344 | 1.746e-02 |
| GO:BP | GO:0015849 | organic acid transport | 15 | 345 | 1.783e-02 |
| GO:BP | GO:0009581 | detection of external stimulus | 9 | 142 | 1.795e-02 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 14 | 309 | 1.795e-02 |
| GO:BP | GO:0030217 | T cell differentiation | 14 | 312 | 1.958e-02 |
| GO:BP | GO:1903306 | negative regulation of regulated secretory pathway | 4 | 24 | 1.990e-02 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 9 | 145 | 2.023e-02 |
| GO:BP | GO:0003008 | system process | 56 | 2266 | 2.064e-02 |
| GO:BP | GO:0009888 | tissue development | 51 | 2012 | 2.112e-02 |
| GO:BP | GO:0006869 | lipid transport | 17 | 431 | 2.155e-02 |
| GO:BP | GO:1903977 | positive regulation of glial cell migration | 3 | 11 | 2.180e-02 |
| GO:BP | GO:0015747 | urate transport | 3 | 11 | 2.180e-02 |
| GO:BP | GO:0003341 | cilium movement | 11 | 212 | 2.195e-02 |
| GO:BP | GO:0042391 | regulation of membrane potential | 17 | 433 | 2.195e-02 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 10 | 181 | 2.355e-02 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 7 | 93 | 2.463e-02 |
| GO:BP | GO:0006311 | meiotic gene conversion | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 5 | 45 | 2.463e-02 |
| GO:BP | GO:0010481 | epidermal cell division | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0010482 | regulation of epidermal cell division | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0071486 | cellular response to high light intensity | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0033563 | dorsal/ventral axon guidance | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0021605 | cranial nerve maturation | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 4 | 26 | 2.463e-02 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 14 | 325 | 2.463e-02 |
| GO:BP | GO:0002064 | epithelial cell development | 11 | 217 | 2.463e-02 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 6 | 69 | 2.584e-02 |
| GO:BP | GO:0048513 | animal organ development | 70 | 3047 | 2.597e-02 |
| GO:BP | GO:0030097 | hemopoiesis | 29 | 971 | 2.827e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 10 | 189 | 2.899e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 10 | 189 | 2.899e-02 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 8 | 126 | 2.899e-02 |
| GO:BP | GO:0008283 | cell population proliferation | 50 | 2009 | 2.927e-02 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 4 | 28 | 2.927e-02 |
| GO:BP | GO:0006817 | phosphate ion transport | 4 | 28 | 2.927e-02 |
| GO:BP | GO:0048806 | genitalia development | 5 | 48 | 2.927e-02 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 63 | 2692 | 2.952e-02 |
| GO:BP | GO:1904888 | cranial skeletal system development | 6 | 72 | 2.992e-02 |
| GO:BP | GO:0001501 | skeletal system development | 19 | 536 | 3.011e-02 |
| GO:BP | GO:0043301 | negative regulation of leukocyte degranulation | 3 | 13 | 3.011e-02 |
| GO:BP | GO:0042403 | thyroid hormone metabolic process | 4 | 29 | 3.176e-02 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 4 | 29 | 3.176e-02 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 12 | 264 | 3.206e-02 |
| GO:BP | GO:0001894 | tissue homeostasis | 12 | 264 | 3.206e-02 |
| GO:BP | GO:0030900 | forebrain development | 16 | 417 | 3.206e-02 |
| GO:BP | GO:0060429 | epithelium development | 34 | 1229 | 3.455e-02 |
| GO:BP | GO:0007127 | meiosis I | 8 | 132 | 3.539e-02 |
| GO:BP | GO:0140352 | export from cell | 27 | 901 | 3.547e-02 |
| GO:BP | GO:0035589 | G protein-coupled purinergic nucleotide receptor signaling pathway | 3 | 14 | 3.548e-02 |
| GO:BP | GO:0048518 | positive regulation of biological process | 125 | 6235 | 3.604e-02 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 17 | 465 | 3.669e-02 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 27 | 909 | 3.791e-02 |
| GO:BP | GO:0006968 | cellular defense response | 5 | 52 | 3.791e-02 |
| GO:BP | GO:0046352 | disaccharide catabolic process | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0010845 | positive regulation of reciprocal meiotic recombination | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 9 | 167 | 3.791e-02 |
| GO:BP | GO:0021682 | nerve maturation | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0035822 | gene conversion | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0141006 | transposable element silencing by piRNA-mediated heterochromatin formation | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0140966 | piRNA-mediated heterochromatin formation | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0010520 | regulation of reciprocal meiotic recombination | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0045728 | respiratory burst after phagocytosis | 2 | 4 | 3.791e-02 |
| GO:BP | GO:1990834 | response to odorant | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0009605 | response to external stimulus | 56 | 2367 | 3.880e-02 |
| GO:BP | GO:0071695 | anatomical structure maturation | 12 | 274 | 3.887e-02 |
| GO:BP | GO:0006816 | calcium ion transport | 16 | 433 | 4.134e-02 |
| GO:BP | GO:0007165 | signal transduction | 120 | 5979 | 4.180e-02 |
| GO:BP | GO:0007399 | nervous system development | 59 | 2541 | 4.341e-02 |
| GO:BP | GO:2000241 | regulation of reproductive process | 10 | 206 | 4.341e-02 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 9 | 172 | 4.341e-02 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 9 | 172 | 4.341e-02 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 23 | 741 | 4.442e-02 |
| GO:BP | GO:0032940 | secretion by cell | 25 | 832 | 4.442e-02 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 7 | 110 | 4.452e-02 |
| GO:BP | GO:0006957 | complement activation, alternative pathway | 3 | 16 | 4.452e-02 |
| GO:BP | GO:0007194 | negative regulation of adenylate cyclase activity | 3 | 16 | 4.452e-02 |
| GO:BP | GO:0010876 | lipid localization | 17 | 480 | 4.452e-02 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 8 | 141 | 4.452e-02 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 3 | 16 | 4.452e-02 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 3 | 16 | 4.452e-02 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 118 | 5886 | 4.481e-02 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 19 | 567 | 4.481e-02 |
| GO:BP | GO:0003382 | epithelial cell morphogenesis | 4 | 34 | 4.481e-02 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 10 | 209 | 4.481e-02 |
| GO:BP | GO:0010526 | transposable element silencing | 4 | 34 | 4.481e-02 |
| GO:BP | GO:0015701 | bicarbonate transport | 4 | 34 | 4.481e-02 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 4 | 34 | 4.481e-02 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 11 | 248 | 4.822e-02 |
| GO:BP | GO:0009566 | fertilization | 10 | 212 | 4.830e-02 |
| GO:BP | GO:0045577 | regulation of B cell differentiation | 4 | 35 | 4.848e-02 |
| GO:BP | GO:0051050 | positive regulation of transport | 25 | 844 | 4.848e-02 |
| GO:BP | GO:0043299 | leukocyte degranulation | 6 | 84 | 4.848e-02 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 4 | 35 | 4.848e-02 |
| GO:BP | GO:0002679 | respiratory burst involved in defense response | 3 | 17 | 4.900e-02 |
| GO:BP | GO:0009642 | response to light intensity | 3 | 17 | 4.900e-02 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 31 | 1130 | 4.900e-02 |
| GO:BP | GO:0031280 | negative regulation of cyclase activity | 3 | 17 | 4.900e-02 |
| GO:BP | GO:0045917 | positive regulation of complement activation | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0045920 | negative regulation of exocytosis | 4 | 36 | 4.921e-02 |
| GO:BP | GO:0070995 | NADPH oxidation | 2 | 5 | 4.921e-02 |
| GO:BP | GO:2000611 | positive regulation of thyroid hormone generation | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0032197 | retrotransposition | 4 | 36 | 4.921e-02 |
| GO:BP | GO:0007610 | behavior | 21 | 665 | 4.921e-02 |
| GO:BP | GO:0009644 | response to high light intensity | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 29 | 1037 | 4.921e-02 |
| GO:BP | GO:0030317 | flagellated sperm motility | 8 | 147 | 4.921e-02 |
| GO:BP | GO:0009914 | hormone transport | 13 | 328 | 4.921e-02 |
| GO:BP | GO:0097722 | sperm motility | 8 | 147 | 4.921e-02 |
| GO:BP | GO:0023061 | signal release | 17 | 493 | 4.921e-02 |
| GO:BP | GO:0071484 | cellular response to light intensity | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 42 | 1682 | 4.921e-02 |
| GO:BP | GO:0002434 | immune complex clearance | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0045216 | cell-cell junction organization | 10 | 216 | 4.931e-02 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 25 | 365 | 1.188e-05 |
#write.csv(tableLR_pp, "output/table_LRmotif.csv")
#GO:BP
tableLR_GOBP_pp <- tableLR_pp %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(tableLR_GOBP_pp, "data/tableLR_GOBP_postprob.RDS")
tableLR_GOBP_pp %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("No Response Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))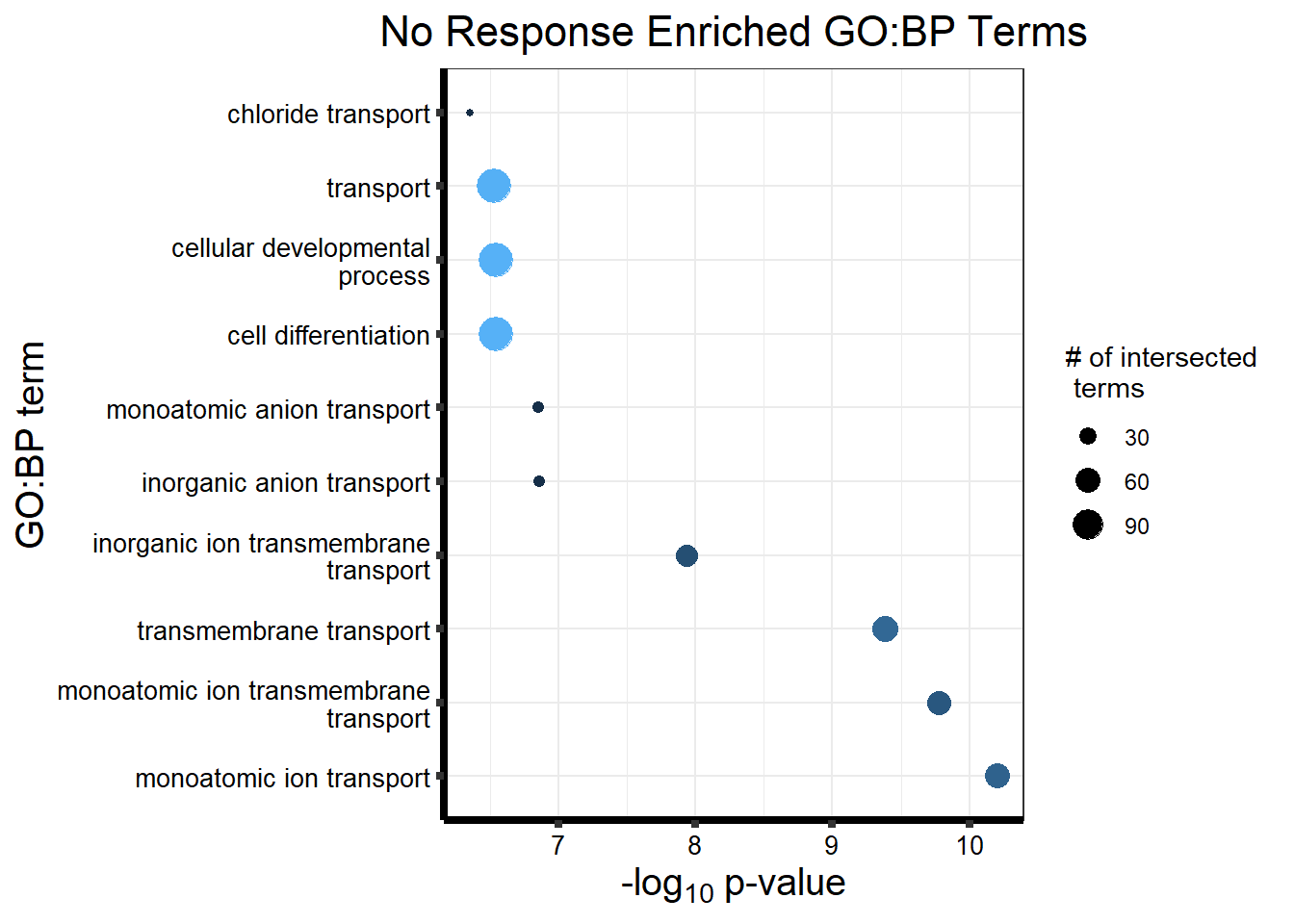
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#KEGG
tableLR_KEGG_pp <- tableLR_pp %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
tableLR_KEGG_pp %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("No Response Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))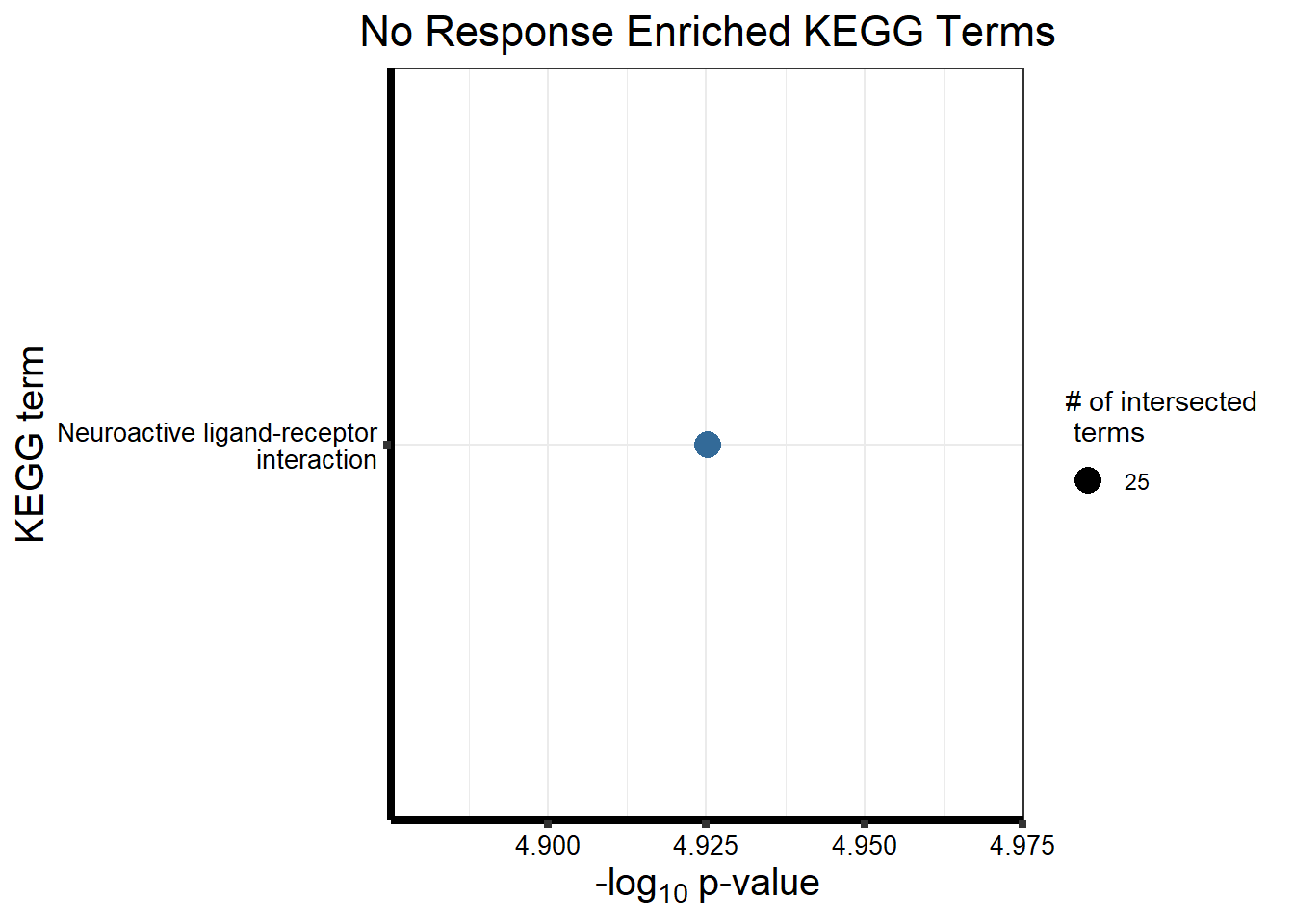
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
Now let’s look at motif 4 - Early Sustained Response
motif_ED_d <- clust4_d
EDmotif_genes_d <- gost(query = motif_ED_d,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
cormotifEDclust_d <- gostplot(EDmotif_genes_d, capped = FALSE, interactive = TRUE)
cormotifEDclust_dtableED_d <- EDmotif_genes_d$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
tableED_d %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0032502 | developmental process | 426 | 6478 | 6.312e-15 |
| GO:BP | GO:0048856 | anatomical structure development | 398 | 5924 | 6.312e-15 |
| GO:BP | GO:0032501 | multicellular organismal process | 453 | 7234 | 1.746e-12 |
| GO:BP | GO:0030154 | cell differentiation | 296 | 4381 | 4.863e-10 |
| GO:BP | GO:0048869 | cellular developmental process | 296 | 4382 | 4.863e-10 |
| GO:BP | GO:0045165 | cell fate commitment | 45 | 291 | 1.087e-09 |
| GO:BP | GO:0007275 | multicellular organism development | 307 | 4658 | 2.268e-09 |
| GO:BP | GO:0048731 | system development | 270 | 3985 | 4.131e-09 |
| GO:BP | GO:0048663 | neuron fate commitment | 19 | 73 | 3.822e-07 |
| GO:BP | GO:0003002 | regionalization | 51 | 434 | 6.431e-07 |
| GO:BP | GO:0007399 | nervous system development | 181 | 2541 | 7.027e-07 |
| GO:BP | GO:0033993 | response to lipid | 84 | 921 | 9.822e-07 |
| GO:BP | GO:0048513 | animal organ development | 208 | 3047 | 9.822e-07 |
| GO:BP | GO:0001708 | cell fate specification | 22 | 113 | 4.095e-06 |
| GO:BP | GO:0007389 | pattern specification process | 52 | 478 | 4.254e-06 |
| GO:BP | GO:0007417 | central nervous system development | 88 | 1038 | 1.055e-05 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 86 | 1017 | 1.619e-05 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 87 | 1037 | 1.814e-05 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 182 | 2692 | 1.819e-05 |
| GO:BP | GO:0060429 | epithelium development | 98 | 1229 | 2.673e-05 |
| GO:BP | GO:0009888 | tissue development | 143 | 2012 | 3.303e-05 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 181 | 2707 | 3.759e-05 |
| GO:BP | GO:0030182 | neuron differentiation | 109 | 1434 | 4.827e-05 |
| GO:BP | GO:0007423 | sensory organ development | 58 | 612 | 5.090e-05 |
| GO:BP | GO:0048665 | neuron fate specification | 11 | 35 | 7.405e-05 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 171 | 2572 | 1.118e-04 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 17 | 90 | 1.595e-04 |
| GO:BP | GO:0048699 | generation of neurons | 111 | 1517 | 1.994e-04 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 24 | 169 | 2.157e-04 |
| GO:BP | GO:0048468 | cell development | 183 | 2833 | 2.404e-04 |
| GO:BP | GO:1901654 | response to ketone | 28 | 221 | 2.767e-04 |
| GO:BP | GO:0006811 | monoatomic ion transport | 95 | 1256 | 2.830e-04 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 19 | 116 | 2.885e-04 |
| GO:BP | GO:0048880 | sensory system development | 42 | 414 | 3.142e-04 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 173 | 2698 | 6.837e-04 |
| GO:BP | GO:0002118 | aggressive behavior | 5 | 7 | 7.089e-04 |
| GO:BP | GO:0022008 | neurogenesis | 121 | 1746 | 7.103e-04 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 167 | 2604 | 9.557e-04 |
| GO:BP | GO:0150063 | visual system development | 40 | 408 | 1.104e-03 |
| GO:BP | GO:0006954 | inflammatory response | 68 | 847 | 1.111e-03 |
| GO:BP | GO:0050793 | regulation of developmental process | 158 | 2453 | 1.313e-03 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 71 | 903 | 1.320e-03 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 110 | 1577 | 1.320e-03 |
| GO:BP | GO:0001654 | eye development | 39 | 404 | 1.819e-03 |
| GO:BP | GO:0006952 | defense response | 122 | 1809 | 1.884e-03 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 115 | 1682 | 1.884e-03 |
| GO:BP | GO:0009605 | response to external stimulus | 152 | 2367 | 2.060e-03 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 26 | 225 | 2.246e-03 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 48 | 549 | 2.456e-03 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 8 | 27 | 2.586e-03 |
| GO:BP | GO:0030900 | forebrain development | 39 | 417 | 3.222e-03 |
| GO:BP | GO:0071396 | cellular response to lipid | 51 | 609 | 3.964e-03 |
| GO:BP | GO:0060322 | head development | 62 | 789 | 3.964e-03 |
| GO:BP | GO:0003407 | neural retina development | 14 | 85 | 3.964e-03 |
| GO:BP | GO:0055085 | transmembrane transport | 105 | 1538 | 4.050e-03 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 11 | 55 | 4.056e-03 |
| GO:BP | GO:0043010 | camera-type eye development | 34 | 352 | 4.827e-03 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 89 | 1261 | 4.827e-03 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 29 | 280 | 4.859e-03 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 20 | 159 | 4.945e-03 |
| GO:BP | GO:0007420 | brain development | 58 | 739 | 6.473e-03 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 68 | 909 | 6.718e-03 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 58 | 741 | 6.750e-03 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 9 | 40 | 6.858e-03 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 37 | 408 | 7.153e-03 |
| GO:BP | GO:0060579 | ventral spinal cord interneuron fate commitment | 5 | 11 | 7.153e-03 |
| GO:BP | GO:0006334 | nucleosome assembly | 16 | 115 | 7.153e-03 |
| GO:BP | GO:0060581 | cell fate commitment involved in pattern specification | 5 | 11 | 7.153e-03 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 24 | 218 | 7.153e-03 |
| GO:BP | GO:0007267 | cell-cell signaling | 91 | 1316 | 7.153e-03 |
| GO:BP | GO:0021515 | cell differentiation in spinal cord | 10 | 50 | 7.202e-03 |
| GO:BP | GO:0021536 | diencephalon development | 13 | 81 | 7.238e-03 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 29 | 290 | 7.377e-03 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 63 | 832 | 7.378e-03 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 50 | 617 | 7.654e-03 |
| GO:BP | GO:1901616 | organic hydroxy compound catabolic process | 11 | 61 | 8.104e-03 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 20 | 168 | 8.182e-03 |
| GO:BP | GO:0065007 | biological regulation | 642 | 12671 | 8.182e-03 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 8 | 33 | 8.182e-03 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 9 | 42 | 8.233e-03 |
| GO:BP | GO:0021513 | spinal cord dorsal/ventral patterning | 6 | 18 | 8.579e-03 |
| GO:BP | GO:0008283 | cell population proliferation | 128 | 2009 | 8.587e-03 |
| GO:BP | GO:0048568 | embryonic organ development | 40 | 463 | 8.912e-03 |
| GO:BP | GO:0031960 | response to corticosteroid | 19 | 157 | 8.912e-03 |
| GO:BP | GO:0006812 | monoatomic cation transport | 74 | 1032 | 8.912e-03 |
| GO:BP | GO:0007281 | germ cell development | 34 | 370 | 8.912e-03 |
| GO:BP | GO:0019336 | phenol-containing compound catabolic process | 5 | 12 | 9.104e-03 |
| GO:BP | GO:0098609 | cell-cell adhesion | 69 | 948 | 9.529e-03 |
| GO:BP | GO:0022414 | reproductive process | 103 | 1556 | 9.791e-03 |
| GO:BP | GO:0003008 | system process | 141 | 2266 | 1.008e-02 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 107 | 1632 | 1.021e-02 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 11 | 64 | 1.049e-02 |
| GO:BP | GO:0034728 | nucleosome organization | 17 | 135 | 1.135e-02 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 175 | 2928 | 1.155e-02 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 22 | 203 | 1.261e-02 |
| GO:BP | GO:0021514 | ventral spinal cord interneuron differentiation | 5 | 13 | 1.263e-02 |
| GO:BP | GO:0060732 | positive regulation of inositol phosphate biosynthetic process | 5 | 13 | 1.263e-02 |
| GO:BP | GO:0007501 | mesodermal cell fate specification | 5 | 13 | 1.263e-02 |
| GO:BP | GO:0021511 | spinal cord patterning | 6 | 20 | 1.353e-02 |
| GO:BP | GO:0097305 | response to alcohol | 26 | 262 | 1.383e-02 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 110 | 1710 | 1.534e-02 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 19 | 166 | 1.548e-02 |
| GO:BP | GO:0048545 | response to steroid hormone | 31 | 339 | 1.548e-02 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 110 | 1713 | 1.592e-02 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 23 | 222 | 1.602e-02 |
| GO:BP | GO:0060040 | retinal bipolar neuron differentiation | 4 | 8 | 1.620e-02 |
| GO:BP | GO:0060284 | regulation of cell development | 62 | 851 | 1.643e-02 |
| GO:BP | GO:0042403 | thyroid hormone metabolic process | 7 | 29 | 1.643e-02 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 210 | 3648 | 1.711e-02 |
| GO:BP | GO:0007154 | cell communication | 350 | 6496 | 1.728e-02 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 14 | 104 | 1.728e-02 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 81 | 1189 | 1.728e-02 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 8 | 39 | 1.936e-02 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 93 | 1413 | 1.973e-02 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 19 | 171 | 2.026e-02 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 59 | 811 | 2.212e-02 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 71 | 1022 | 2.233e-02 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 199 | 3454 | 2.257e-02 |
| GO:BP | GO:0010919 | regulation of inositol phosphate biosynthetic process | 5 | 15 | 2.257e-02 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 88 | 1331 | 2.340e-02 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 28 | 305 | 2.406e-02 |
| GO:BP | GO:0042420 | dopamine catabolic process | 4 | 9 | 2.440e-02 |
| GO:BP | GO:0023052 | signaling | 347 | 6471 | 2.464e-02 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 8 | 41 | 2.490e-02 |
| GO:BP | GO:0050789 | regulation of biological process | 618 | 12278 | 2.490e-02 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 11 | 73 | 2.490e-02 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 67 | 958 | 2.490e-02 |
| GO:BP | GO:0006821 | chloride transport | 15 | 122 | 2.509e-02 |
| GO:BP | GO:0051384 | response to glucocorticoid | 16 | 135 | 2.509e-02 |
| GO:BP | GO:0007601 | visual perception | 22 | 219 | 2.621e-02 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 14 | 110 | 2.621e-02 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 24 | 248 | 2.621e-02 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 68 | 980 | 2.673e-02 |
| GO:BP | GO:0050877 | nervous system process | 96 | 1489 | 2.697e-02 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 197 | 3435 | 2.714e-02 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 8 | 42 | 2.729e-02 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 39 | 486 | 2.895e-02 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 16 | 138 | 2.991e-02 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 84 | 1275 | 3.005e-02 |
| GO:BP | GO:0021545 | cranial nerve development | 10 | 64 | 3.005e-02 |
| GO:BP | GO:0042221 | response to chemical | 218 | 3867 | 3.159e-02 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 11 | 76 | 3.160e-02 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 28 | 314 | 3.216e-02 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 61 | 866 | 3.240e-02 |
| GO:BP | GO:0048664 | neuron fate determination | 4 | 10 | 3.252e-02 |
| GO:BP | GO:0019614 | catechol-containing compound catabolic process | 4 | 10 | 3.252e-02 |
| GO:BP | GO:0042424 | catecholamine catabolic process | 4 | 10 | 3.252e-02 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 6 | 25 | 3.392e-02 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 5 | 17 | 3.399e-02 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 31 | 363 | 3.399e-02 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 114 | 1844 | 3.465e-02 |
| GO:BP | GO:0019953 | sexual reproduction | 74 | 1103 | 3.505e-02 |
| GO:BP | GO:0001912 | positive regulation of leukocyte mediated cytotoxicity | 10 | 66 | 3.518e-02 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 32 | 381 | 3.621e-02 |
| GO:BP | GO:0035270 | endocrine system development | 16 | 142 | 3.638e-02 |
| GO:BP | GO:0046069 | cGMP catabolic process | 3 | 5 | 3.688e-02 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 72 | 1071 | 3.761e-02 |
| GO:BP | GO:0042391 | regulation of membrane potential | 35 | 433 | 4.100e-02 |
| GO:BP | GO:1905962 | glutamatergic neuron differentiation | 5 | 18 | 4.245e-02 |
| GO:BP | GO:0001710 | mesodermal cell fate commitment | 5 | 18 | 4.245e-02 |
| GO:BP | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin | 4 | 11 | 4.413e-02 |
| GO:BP | GO:0010455 | positive regulation of cell fate commitment | 4 | 11 | 4.413e-02 |
| GO:BP | GO:0014049 | positive regulation of glutamate secretion | 4 | 11 | 4.413e-02 |
| GO:BP | GO:0010752 | regulation of cGMP-mediated signaling | 4 | 11 | 4.413e-02 |
| GO:BP | GO:0042471 | ear morphogenesis | 15 | 132 | 4.475e-02 |
| GO:BP | GO:0009725 | response to hormone | 62 | 902 | 4.657e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 190 | 3349 | 4.740e-02 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 8 | 47 | 4.799e-02 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 13 | 107 | 4.928e-02 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 49 | 365 | 1.734e-09 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 24 | 132 | 7.521e-07 |
| KEGG | KEGG:05034 | Alcoholism | 25 | 187 | 1.212e-04 |
| KEGG | KEGG:05032 | Morphine addiction | 15 | 89 | 7.783e-04 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 22 | 188 | 2.676e-03 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 29 | 291 | 3.029e-03 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 7 | 26 | 4.479e-03 |
| KEGG | KEGG:04742 | Taste transduction | 13 | 85 | 4.479e-03 |
| KEGG | KEGG:05033 | Nicotine addiction | 8 | 40 | 1.228e-02 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 20 | 192 | 1.417e-02 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 9 | 62 | 4.904e-02 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 11 | 89 | 4.904e-02 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 13 | 114 | 4.904e-02 |
| KEGG | KEGG:04727 | GABAergic synapse | 11 | 89 | 4.904e-02 |
#write.csv(tableED, "output/table_EarlyDOXmotif.csv")
#GO:BP
tableED_GOBP_d <- tableED_d %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(tableED_GOBP, "data/tableED_GOBP.RDS")
tableED_GOBP_d %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Early Sustained Response Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))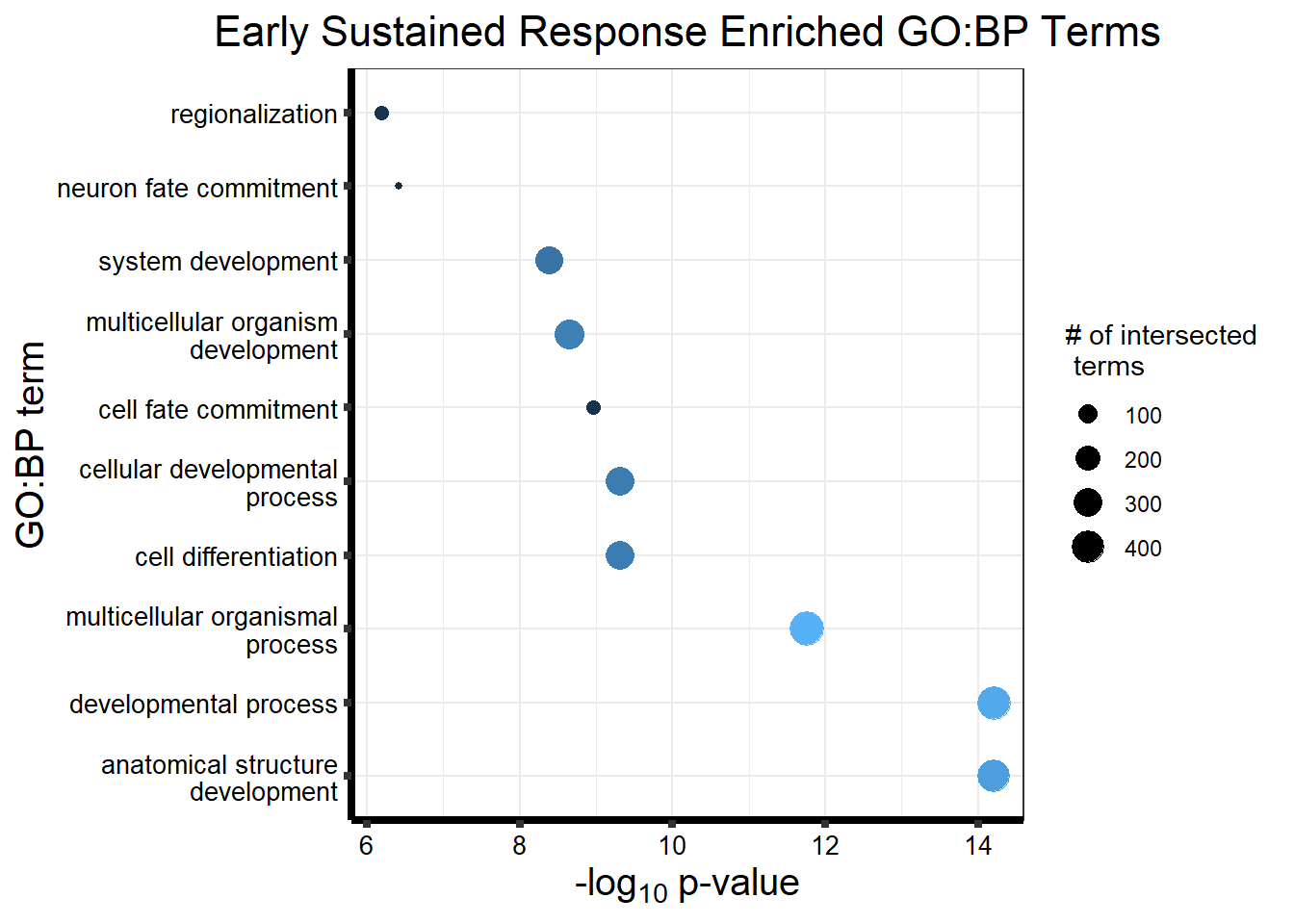
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#KEGG
tableED_KEGG_d <- tableED_d %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
tableED_KEGG_d %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Early Sustained Response Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))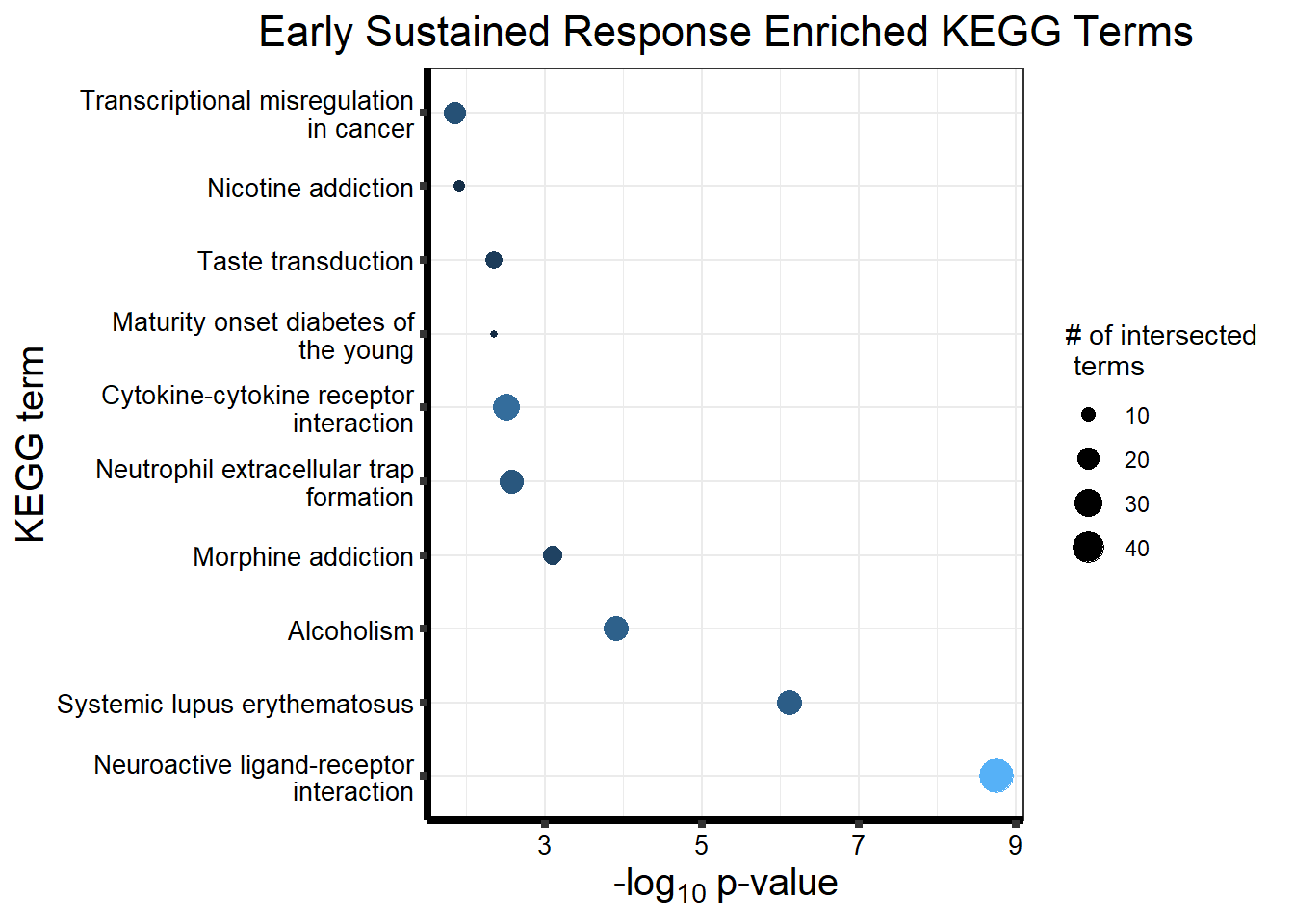
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
Now let’s see this with the posterior probability for the Early Sustained Response motif 4
motif_ESR_pp <- prob_motif_4
ESRmotif_genes_pp <- gost(query = motif_ESR_pp,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
cormotifESRclust_pp <- gostplot(ESRmotif_genes_pp, capped = FALSE, interactive = TRUE)
cormotifESRclust_pptableESR_pp <- LRmotif_genes_pp$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
tableESR_pp %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006811 | monoatomic ion transport | 60 | 1256 | 6.212e-11 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 52 | 1017 | 1.681e-10 |
| GO:BP | GO:0055085 | transmembrane transport | 65 | 1538 | 4.113e-10 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 45 | 903 | 1.161e-08 |
| GO:BP | GO:0015698 | inorganic anion transport | 18 | 165 | 1.375e-07 |
| GO:BP | GO:0006820 | monoatomic anion transport | 18 | 167 | 1.400e-07 |
| GO:BP | GO:0048869 | cellular developmental process | 118 | 4382 | 2.840e-07 |
| GO:BP | GO:0030154 | cell differentiation | 118 | 4381 | 2.840e-07 |
| GO:BP | GO:0006810 | transport | 117 | 4341 | 2.971e-07 |
| GO:BP | GO:0006821 | chloride transport | 15 | 122 | 4.430e-07 |
| GO:BP | GO:0032501 | multicellular organismal process | 167 | 7234 | 1.046e-06 |
| GO:BP | GO:0006812 | monoatomic cation transport | 43 | 1032 | 2.823e-06 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 15 | 142 | 2.844e-06 |
| GO:BP | GO:0032502 | developmental process | 152 | 6478 | 3.014e-06 |
| GO:BP | GO:0048468 | cell development | 83 | 2833 | 4.124e-06 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 13 | 107 | 4.238e-06 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 37 | 832 | 4.606e-06 |
| GO:BP | GO:0050896 | response to stimulus | 193 | 8993 | 4.865e-06 |
| GO:BP | GO:0030001 | metal ion transport | 38 | 875 | 4.881e-06 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 11 | 79 | 1.047e-05 |
| GO:BP | GO:0007267 | cell-cell signaling | 48 | 1316 | 1.298e-05 |
| GO:BP | GO:0051234 | establishment of localization | 120 | 4869 | 1.533e-05 |
| GO:BP | GO:0065008 | regulation of biological quality | 81 | 2847 | 1.552e-05 |
| GO:BP | GO:0065007 | biological regulation | 247 | 12671 | 1.617e-05 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 18 | 248 | 1.781e-05 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 13 | 127 | 2.060e-05 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 34 | 811 | 4.571e-05 |
| GO:BP | GO:0048856 | anatomical structure development | 136 | 5924 | 7.416e-05 |
| GO:BP | GO:0022414 | reproductive process | 51 | 1556 | 9.471e-05 |
| GO:BP | GO:0051179 | localization | 127 | 5521 | 2.049e-04 |
| GO:BP | GO:0006813 | potassium ion transport | 16 | 238 | 2.050e-04 |
| GO:BP | GO:0015711 | organic anion transport | 22 | 442 | 3.627e-04 |
| GO:BP | GO:0023052 | signaling | 142 | 6471 | 4.510e-04 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 8 | 59 | 5.282e-04 |
| GO:BP | GO:0007154 | cell communication | 142 | 6496 | 5.345e-04 |
| GO:BP | GO:0048232 | male gamete generation | 27 | 654 | 8.075e-04 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 36 | 1022 | 8.341e-04 |
| GO:BP | GO:0010817 | regulation of hormone levels | 24 | 548 | 9.563e-04 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 14 | 213 | 9.594e-04 |
| GO:BP | GO:0002521 | leukocyte differentiation | 26 | 632 | 1.150e-03 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 41 | 1275 | 1.504e-03 |
| GO:BP | GO:0050789 | regulation of biological process | 232 | 12278 | 1.750e-03 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 12 | 169 | 1.750e-03 |
| GO:BP | GO:0007275 | multicellular organism development | 107 | 4658 | 1.750e-03 |
| GO:BP | GO:0099536 | synaptic signaling | 29 | 783 | 2.264e-03 |
| GO:BP | GO:0007283 | spermatogenesis | 25 | 637 | 3.208e-03 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 22 | 521 | 3.208e-03 |
| GO:BP | GO:0019953 | sexual reproduction | 36 | 1103 | 3.281e-03 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 27 | 723 | 3.477e-03 |
| GO:BP | GO:0042592 | homeostatic process | 49 | 1711 | 3.593e-03 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 22 | 530 | 3.743e-03 |
| GO:BP | GO:0048731 | system development | 93 | 3985 | 3.743e-03 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 22 | 531 | 3.776e-03 |
| GO:BP | GO:0050877 | nervous system process | 44 | 1489 | 4.173e-03 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 16 | 326 | 5.815e-03 |
| GO:BP | GO:0140013 | meiotic nuclear division | 12 | 198 | 6.435e-03 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 6 | 46 | 6.491e-03 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 27 | 759 | 6.655e-03 |
| GO:BP | GO:0035270 | endocrine system development | 10 | 142 | 6.926e-03 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 23 | 598 | 6.926e-03 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 32 | 980 | 7.045e-03 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 9 | 116 | 7.045e-03 |
| GO:BP | GO:0048878 | chemical homeostasis | 33 | 1026 | 7.079e-03 |
| GO:BP | GO:0006814 | sodium ion transport | 13 | 235 | 7.216e-03 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 15 | 302 | 7.216e-03 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 5 | 31 | 7.902e-03 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 23 | 608 | 7.924e-03 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 15 | 306 | 7.976e-03 |
| GO:BP | GO:0007276 | gamete generation | 28 | 822 | 8.608e-03 |
| GO:BP | GO:0042445 | hormone metabolic process | 13 | 243 | 9.261e-03 |
| GO:BP | GO:0001775 | cell activation | 34 | 1098 | 1.012e-02 |
| GO:BP | GO:0046903 | secretion | 31 | 968 | 1.091e-02 |
| GO:BP | GO:0051716 | cellular response to stimulus | 148 | 7320 | 1.099e-02 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 26 | 753 | 1.099e-02 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 26 | 753 | 1.099e-02 |
| GO:BP | GO:0042221 | response to chemical | 88 | 3867 | 1.099e-02 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 12 | 218 | 1.157e-02 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 45 | 1632 | 1.161e-02 |
| GO:BP | GO:0050957 | equilibrioception | 3 | 8 | 1.165e-02 |
| GO:BP | GO:0060348 | bone development | 12 | 220 | 1.196e-02 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 12 | 220 | 1.196e-02 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 18 | 436 | 1.232e-02 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 6 | 55 | 1.232e-02 |
| GO:BP | GO:0021630 | olfactory nerve maturation | 2 | 2 | 1.261e-02 |
| GO:BP | GO:0042110 | T cell activation | 21 | 558 | 1.261e-02 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 6 | 56 | 1.261e-02 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 18 | 438 | 1.261e-02 |
| GO:BP | GO:0036368 | cone photoresponse recovery | 2 | 2 | 1.261e-02 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 5 | 36 | 1.261e-02 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 11 | 192 | 1.285e-02 |
| GO:BP | GO:0051321 | meiotic cell cycle | 14 | 294 | 1.330e-02 |
| GO:BP | GO:0098609 | cell-cell adhesion | 30 | 948 | 1.330e-02 |
| GO:BP | GO:0007155 | cell adhesion | 42 | 1511 | 1.363e-02 |
| GO:BP | GO:0045321 | leukocyte activation | 30 | 951 | 1.372e-02 |
| GO:BP | GO:0021536 | diencephalon development | 7 | 81 | 1.409e-02 |
| GO:BP | GO:0007600 | sensory perception | 30 | 955 | 1.413e-02 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 15 | 334 | 1.413e-02 |
| GO:BP | GO:0045055 | regulated exocytosis | 12 | 229 | 1.413e-02 |
| GO:BP | GO:0030450 | regulation of complement activation, classical pathway | 3 | 9 | 1.413e-02 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 20 | 529 | 1.475e-02 |
| GO:BP | GO:0019725 | cellular homeostasis | 27 | 826 | 1.487e-02 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 4 | 22 | 1.570e-02 |
| GO:BP | GO:0046649 | lymphocyte activation | 26 | 787 | 1.582e-02 |
| GO:BP | GO:0048732 | gland development | 18 | 454 | 1.582e-02 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 6 | 60 | 1.582e-02 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 14 | 305 | 1.666e-02 |
| GO:BP | GO:0050794 | regulation of cellular process | 219 | 11876 | 1.689e-02 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 11 | 203 | 1.722e-02 |
| GO:BP | GO:0046942 | carboxylic acid transport | 15 | 344 | 1.746e-02 |
| GO:BP | GO:0015849 | organic acid transport | 15 | 345 | 1.783e-02 |
| GO:BP | GO:0009581 | detection of external stimulus | 9 | 142 | 1.795e-02 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 14 | 309 | 1.795e-02 |
| GO:BP | GO:0030217 | T cell differentiation | 14 | 312 | 1.958e-02 |
| GO:BP | GO:1903306 | negative regulation of regulated secretory pathway | 4 | 24 | 1.990e-02 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 9 | 145 | 2.023e-02 |
| GO:BP | GO:0003008 | system process | 56 | 2266 | 2.064e-02 |
| GO:BP | GO:0009888 | tissue development | 51 | 2012 | 2.112e-02 |
| GO:BP | GO:0006869 | lipid transport | 17 | 431 | 2.155e-02 |
| GO:BP | GO:1903977 | positive regulation of glial cell migration | 3 | 11 | 2.180e-02 |
| GO:BP | GO:0015747 | urate transport | 3 | 11 | 2.180e-02 |
| GO:BP | GO:0003341 | cilium movement | 11 | 212 | 2.195e-02 |
| GO:BP | GO:0042391 | regulation of membrane potential | 17 | 433 | 2.195e-02 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 10 | 181 | 2.355e-02 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 7 | 93 | 2.463e-02 |
| GO:BP | GO:0006311 | meiotic gene conversion | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 5 | 45 | 2.463e-02 |
| GO:BP | GO:0010481 | epidermal cell division | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0010482 | regulation of epidermal cell division | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0071486 | cellular response to high light intensity | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0033563 | dorsal/ventral axon guidance | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0021605 | cranial nerve maturation | 2 | 3 | 2.463e-02 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 4 | 26 | 2.463e-02 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 14 | 325 | 2.463e-02 |
| GO:BP | GO:0002064 | epithelial cell development | 11 | 217 | 2.463e-02 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 6 | 69 | 2.584e-02 |
| GO:BP | GO:0048513 | animal organ development | 70 | 3047 | 2.597e-02 |
| GO:BP | GO:0030097 | hemopoiesis | 29 | 971 | 2.827e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 10 | 189 | 2.899e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 10 | 189 | 2.899e-02 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 8 | 126 | 2.899e-02 |
| GO:BP | GO:0008283 | cell population proliferation | 50 | 2009 | 2.927e-02 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 4 | 28 | 2.927e-02 |
| GO:BP | GO:0006817 | phosphate ion transport | 4 | 28 | 2.927e-02 |
| GO:BP | GO:0048806 | genitalia development | 5 | 48 | 2.927e-02 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 63 | 2692 | 2.952e-02 |
| GO:BP | GO:1904888 | cranial skeletal system development | 6 | 72 | 2.992e-02 |
| GO:BP | GO:0001501 | skeletal system development | 19 | 536 | 3.011e-02 |
| GO:BP | GO:0043301 | negative regulation of leukocyte degranulation | 3 | 13 | 3.011e-02 |
| GO:BP | GO:0042403 | thyroid hormone metabolic process | 4 | 29 | 3.176e-02 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 4 | 29 | 3.176e-02 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 12 | 264 | 3.206e-02 |
| GO:BP | GO:0001894 | tissue homeostasis | 12 | 264 | 3.206e-02 |
| GO:BP | GO:0030900 | forebrain development | 16 | 417 | 3.206e-02 |
| GO:BP | GO:0060429 | epithelium development | 34 | 1229 | 3.455e-02 |
| GO:BP | GO:0007127 | meiosis I | 8 | 132 | 3.539e-02 |
| GO:BP | GO:0140352 | export from cell | 27 | 901 | 3.547e-02 |
| GO:BP | GO:0035589 | G protein-coupled purinergic nucleotide receptor signaling pathway | 3 | 14 | 3.548e-02 |
| GO:BP | GO:0048518 | positive regulation of biological process | 125 | 6235 | 3.604e-02 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 17 | 465 | 3.669e-02 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 27 | 909 | 3.791e-02 |
| GO:BP | GO:0006968 | cellular defense response | 5 | 52 | 3.791e-02 |
| GO:BP | GO:0046352 | disaccharide catabolic process | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0010845 | positive regulation of reciprocal meiotic recombination | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 9 | 167 | 3.791e-02 |
| GO:BP | GO:0021682 | nerve maturation | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0035822 | gene conversion | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0141006 | transposable element silencing by piRNA-mediated heterochromatin formation | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0140966 | piRNA-mediated heterochromatin formation | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0010520 | regulation of reciprocal meiotic recombination | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0045728 | respiratory burst after phagocytosis | 2 | 4 | 3.791e-02 |
| GO:BP | GO:1990834 | response to odorant | 2 | 4 | 3.791e-02 |
| GO:BP | GO:0009605 | response to external stimulus | 56 | 2367 | 3.880e-02 |
| GO:BP | GO:0071695 | anatomical structure maturation | 12 | 274 | 3.887e-02 |
| GO:BP | GO:0006816 | calcium ion transport | 16 | 433 | 4.134e-02 |
| GO:BP | GO:0007165 | signal transduction | 120 | 5979 | 4.180e-02 |
| GO:BP | GO:0007399 | nervous system development | 59 | 2541 | 4.341e-02 |
| GO:BP | GO:2000241 | regulation of reproductive process | 10 | 206 | 4.341e-02 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 9 | 172 | 4.341e-02 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 9 | 172 | 4.341e-02 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 23 | 741 | 4.442e-02 |
| GO:BP | GO:0032940 | secretion by cell | 25 | 832 | 4.442e-02 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 7 | 110 | 4.452e-02 |
| GO:BP | GO:0006957 | complement activation, alternative pathway | 3 | 16 | 4.452e-02 |
| GO:BP | GO:0007194 | negative regulation of adenylate cyclase activity | 3 | 16 | 4.452e-02 |
| GO:BP | GO:0010876 | lipid localization | 17 | 480 | 4.452e-02 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 8 | 141 | 4.452e-02 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 3 | 16 | 4.452e-02 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 3 | 16 | 4.452e-02 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 118 | 5886 | 4.481e-02 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 19 | 567 | 4.481e-02 |
| GO:BP | GO:0003382 | epithelial cell morphogenesis | 4 | 34 | 4.481e-02 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 10 | 209 | 4.481e-02 |
| GO:BP | GO:0010526 | transposable element silencing | 4 | 34 | 4.481e-02 |
| GO:BP | GO:0015701 | bicarbonate transport | 4 | 34 | 4.481e-02 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 4 | 34 | 4.481e-02 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 11 | 248 | 4.822e-02 |
| GO:BP | GO:0009566 | fertilization | 10 | 212 | 4.830e-02 |
| GO:BP | GO:0045577 | regulation of B cell differentiation | 4 | 35 | 4.848e-02 |
| GO:BP | GO:0051050 | positive regulation of transport | 25 | 844 | 4.848e-02 |
| GO:BP | GO:0043299 | leukocyte degranulation | 6 | 84 | 4.848e-02 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 4 | 35 | 4.848e-02 |
| GO:BP | GO:0002679 | respiratory burst involved in defense response | 3 | 17 | 4.900e-02 |
| GO:BP | GO:0009642 | response to light intensity | 3 | 17 | 4.900e-02 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 31 | 1130 | 4.900e-02 |
| GO:BP | GO:0031280 | negative regulation of cyclase activity | 3 | 17 | 4.900e-02 |
| GO:BP | GO:0045917 | positive regulation of complement activation | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0045920 | negative regulation of exocytosis | 4 | 36 | 4.921e-02 |
| GO:BP | GO:0070995 | NADPH oxidation | 2 | 5 | 4.921e-02 |
| GO:BP | GO:2000611 | positive regulation of thyroid hormone generation | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0032197 | retrotransposition | 4 | 36 | 4.921e-02 |
| GO:BP | GO:0007610 | behavior | 21 | 665 | 4.921e-02 |
| GO:BP | GO:0009644 | response to high light intensity | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 29 | 1037 | 4.921e-02 |
| GO:BP | GO:0030317 | flagellated sperm motility | 8 | 147 | 4.921e-02 |
| GO:BP | GO:0009914 | hormone transport | 13 | 328 | 4.921e-02 |
| GO:BP | GO:0097722 | sperm motility | 8 | 147 | 4.921e-02 |
| GO:BP | GO:0023061 | signal release | 17 | 493 | 4.921e-02 |
| GO:BP | GO:0071484 | cellular response to light intensity | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 42 | 1682 | 4.921e-02 |
| GO:BP | GO:0002434 | immune complex clearance | 2 | 5 | 4.921e-02 |
| GO:BP | GO:0045216 | cell-cell junction organization | 10 | 216 | 4.931e-02 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 25 | 365 | 1.188e-05 |
write.csv(tableESR_pp, "output/table_ESRmotif.csv")
#GO:BP
tableESR_GOBP_pp <- tableESR_pp %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(tableESR_GOBP_pp, "data/tableESR_GOBP_postprob.RDS")
tableESR_GOBP_pp %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("No Response Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))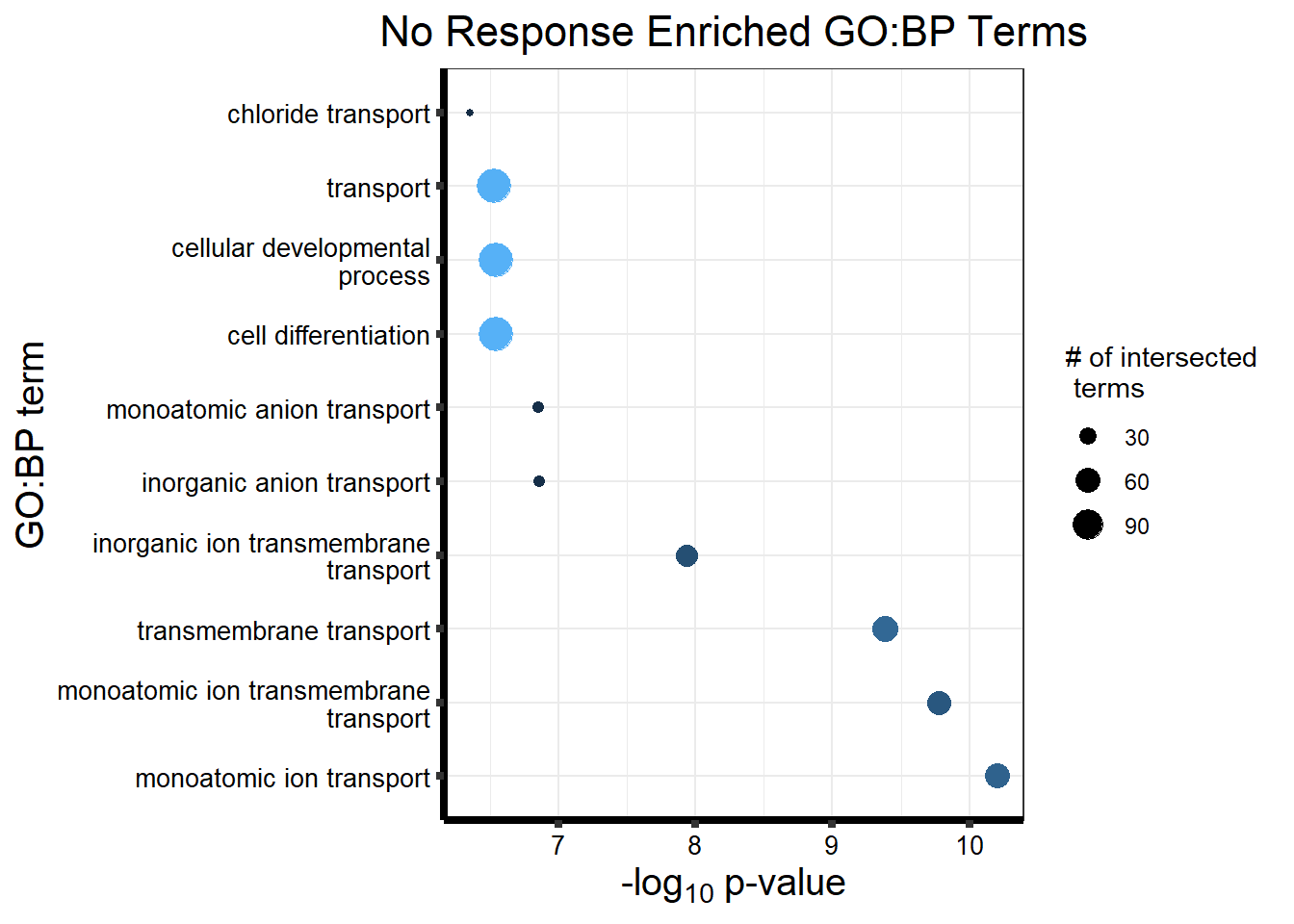
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
#KEGG
tableESR_KEGG_pp <- tableESR_pp %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
tableESR_KEGG_pp %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("No Response Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))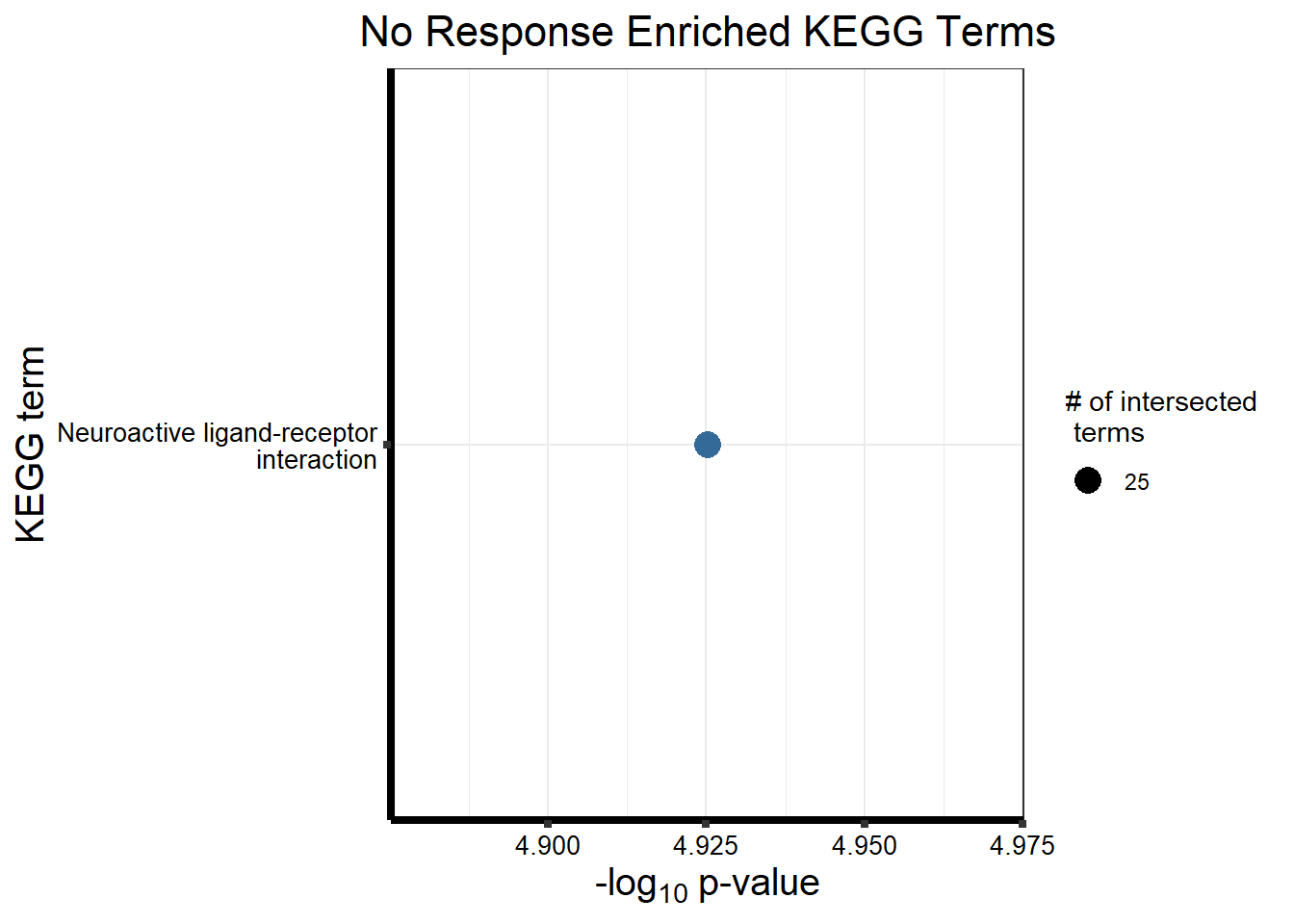
| Version | Author | Date |
|---|---|---|
| 84d5599 | emmapfort | 2025-04-14 |
Currently for the posterior probability code - motif 3 and motif 4 are showing up identically
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 22000)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] gprofiler2_0.2.3 cowplot_1.1.3
[3] Rfast_2.1.5.1 RcppParallel_5.1.10
[5] zigg_0.0.2 Rcpp_1.0.13-1
[7] RUVSeq_1.40.0 EDASeq_2.40.0
[9] ShortRead_1.64.0 GenomicAlignments_1.42.0
[11] SummarizedExperiment_1.36.0 MatrixGenerics_1.18.1
[13] matrixStats_1.4.1 Rsamtools_2.22.0
[15] GenomicRanges_1.58.0 Biostrings_2.74.0
[17] GenomeInfoDb_1.42.3 XVector_0.46.0
[19] IRanges_2.40.0 S4Vectors_0.44.0
[21] BiocParallel_1.40.0 biomaRt_2.62.1
[23] RColorBrewer_1.1-3 Cormotif_1.52.0
[25] affy_1.84.0 Biobase_2.66.0
[27] BiocGenerics_0.52.0 PCAtools_2.18.0
[29] ggfortify_0.4.17 pheatmap_1.0.12
[31] edgeR_4.4.0 limma_3.62.1
[33] readxl_1.4.3 edgebundleR_0.1.4
[35] lubridate_1.9.3 forcats_1.0.0
[37] stringr_1.5.1 dplyr_1.1.4
[39] purrr_1.0.2 readr_2.1.5
[41] tidyr_1.3.1 tidyverse_2.0.0
[43] tibble_3.2.1 hrbrthemes_0.8.7
[45] reshape2_1.4.4 ggrepel_0.9.6
[47] ggplot2_3.5.1 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] later_1.4.1 BiocIO_1.16.0
[3] bitops_1.0-9 filelock_1.0.3
[5] R.oo_1.27.0 cellranger_1.1.0
[7] preprocessCore_1.68.0 XML_3.99-0.18
[9] lifecycle_1.0.4 httr2_1.1.0
[11] pwalign_1.2.0 rprojroot_2.0.4
[13] MASS_7.3-61 processx_3.8.5
[15] lattice_0.22-6 crosstalk_1.2.1
[17] magrittr_2.0.3 plotly_4.10.4
[19] sass_0.4.9 rmarkdown_2.29
[21] jquerylib_0.1.4 yaml_2.3.10
[23] httpuv_1.6.15 DBI_1.2.3
[25] abind_1.4-8 zlibbioc_1.52.0
[27] R.utils_2.12.3 RCurl_1.98-1.16
[29] rappdirs_0.3.3 git2r_0.35.0
[31] gdtools_0.4.1 GenomeInfoDbData_1.2.13
[33] irlba_2.3.5.1 dqrng_0.4.1
[35] svglite_2.1.3 DelayedMatrixStats_1.28.1
[37] codetools_0.2-20 DelayedArray_0.32.0
[39] xml2_1.3.6 tidyselect_1.2.1
[41] farver_2.1.2 UCSC.utils_1.2.0
[43] ScaledMatrix_1.14.0 BiocFileCache_2.14.0
[45] jsonlite_1.8.9 systemfonts_1.1.0
[47] tools_4.4.2 progress_1.2.3
[49] glue_1.8.0 gridExtra_2.3
[51] Rttf2pt1_1.3.12 SparseArray_1.6.0
[53] xfun_0.49 withr_3.0.2
[55] BiocManager_1.30.25 fastmap_1.2.0
[57] latticeExtra_0.6-30 callr_3.7.6
[59] digest_0.6.37 rsvd_1.0.5
[61] timechange_0.3.0 R6_2.6.1
[63] mime_0.12 colorspace_2.1-1
[65] jpeg_0.1-10 RSQLite_2.3.8
[67] R.methodsS3_1.8.2 generics_0.1.3
[69] data.table_1.16.4 fontLiberation_0.1.0
[71] rtracklayer_1.66.0 prettyunits_1.2.0
[73] httr_1.4.7 htmlwidgets_1.6.4
[75] S4Arrays_1.6.0 whisker_0.4.1
[77] pkgconfig_2.0.3 gtable_0.3.6
[79] blob_1.2.4 hwriter_1.3.2.1
[81] htmltools_0.5.8.1 fontBitstreamVera_0.1.1
[83] kableExtra_1.4.0 scales_1.3.0
[85] png_0.1-8 knitr_1.49
[87] rstudioapi_0.17.1 tzdb_0.4.0
[89] rjson_0.2.23 curl_6.0.1
[91] cachem_1.1.0 parallel_4.4.2
[93] extrafont_0.19 AnnotationDbi_1.68.0
[95] restfulr_0.0.15 pillar_1.10.1
[97] grid_4.4.2 vctrs_0.6.5
[99] promises_1.3.2 BiocSingular_1.22.0
[101] dbplyr_2.5.0 beachmat_2.22.0
[103] xtable_1.8-4 extrafontdb_1.0
[105] evaluate_1.0.3 GenomicFeatures_1.58.0
[107] cli_3.6.3 locfit_1.5-9.10
[109] compiler_4.4.2 rlang_1.1.4
[111] crayon_1.5.3 labeling_0.4.3
[113] aroma.light_3.36.0 interp_1.1-6
[115] ps_1.8.1 getPass_0.2-4
[117] plyr_1.8.9 fs_1.6.5
[119] stringi_1.8.4 viridisLite_0.4.2
[121] deldir_2.0-4 munsell_0.5.1
[123] lazyeval_0.2.2 fontquiver_0.2.1
[125] Matrix_1.7-1 hms_1.1.3
[127] sparseMatrixStats_1.18.0 bit64_4.5.2
[129] KEGGREST_1.46.0 statmod_1.5.0
[131] shiny_1.10.0 igraph_2.1.1
[133] memoise_2.0.1 affyio_1.76.0
[135] bslib_0.9.0 bit_4.5.0