mr.mash issue with larger number of conditions
Fabio Morgante
April 13, 2021
Last updated: 2021-04-13
Checks: 7 0
Knit directory: mr_mash_test/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200328) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bf711b8. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .sos/
Ignored: analysis/.sos/
Ignored: code/.sos/
Ignored: code/assess_mrmash_speed.2835257.err
Ignored: code/assess_mrmash_speed.2835257.out
Ignored: code/assess_mrmash_speed.2914525.err
Ignored: code/assess_mrmash_speed.2914525.out
Ignored: code/fit_mr_mash.66662433.err
Ignored: code/fit_mr_mash.66662433.out
Ignored: data/cis_eqtl_analysis_ready
Ignored: data/gtex-v8-ids.txt
Ignored: data/gtex-v8-manifest-full-X-10tissues-missing-rate.txt
Ignored: data/gtex-v8-manifest-full-X.txt
Ignored: data/gtex-v8-manifest.txt
Ignored: dsc/.sos/
Ignored: dsc/dsc_mvreg_07_29_20.3339807.err
Ignored: dsc/dsc_mvreg_07_29_20.3339807.out
Ignored: dsc/dsc_mvreg_07_29_20.3340460.err
Ignored: dsc/dsc_mvreg_07_29_20.3340460.out
Ignored: dsc/dsc_mvreg_07_29_20.3341113.err
Ignored: dsc/dsc_mvreg_07_29_20.3341113.out
Ignored: dsc/dsc_mvreg_07_29_20.3352804.err
Ignored: dsc/dsc_mvreg_07_29_20.3352804.out
Ignored: dsc/functions/__pycache__/
Ignored: dsc/mvreg_test/rep1/
Ignored: dsc/mvreg_test/rep2/
Ignored: dsc/outfiles/
Ignored: output/dsc.html
Ignored: output/dsc/
Ignored: output/dsc_05_18_20.html
Ignored: output/dsc_05_18_20/
Ignored: output/dsc_07_29_20.html
Ignored: output/dsc_07_29_20/
Ignored: output/dsc_OLD.html
Ignored: output/dsc_OLD/
Ignored: output/dsc_test.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10.html
Ignored: output/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10/
Ignored: output/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_inter/
Ignored: output/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10.html
Ignored: output/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10/
Ignored: output/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_inter/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_inter/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_inter/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_inter/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_test.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_inter/
Ignored: output/test_dense_issue.rds
Ignored: output/test_sparse_issue.rds
Ignored: scripts/.sos/
Ignored: scripts/extract_gtex_ids.10523031.err
Ignored: scripts/extract_gtex_ids.10523031.out
Ignored: scripts/get_full_X_gtex_v8_manifest.10520041.err
Ignored: scripts/get_full_X_gtex_v8_manifest.10520041.out
Ignored: scripts/get_gtex_v8_manifest.10520039.err
Ignored: scripts/get_gtex_v8_manifest.10520039.out
Ignored: scripts/get_missing_Y_rate_10tissues_full_X_gtex_v8_manifest.6515875.err
Ignored: scripts/get_missing_Y_rate_10tissues_full_X_gtex_v8_manifest.6515875.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6297709.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6297709.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6297720.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6297720.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6300278.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6300278.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6300959.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6300959.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6312239.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6312239.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6030933.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6030933.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6030949.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6030949.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6034734.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6034734.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6035003.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6035003.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6190207.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6190207.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6318547.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6318547.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6318567.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6318567.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6322126.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6322126.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6322461.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6322461.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6401917.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6401917.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6433374.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6433374.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6449097.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6449097.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6450584.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6450584.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6192054.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6192054.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6192082.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6192082.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6193059.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6193059.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6194214.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6194214.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6219626.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6219626.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE_pipeline.6295843.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE_pipeline.6295843.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6400836.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6400836.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6400839.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6400839.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6401592.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6401592.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6401599.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6401599.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955005.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955005.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955011.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955011.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955473.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955473.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955517.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955517.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6342504.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6342504.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6342513.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6342513.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6350805.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6350805.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6350813.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6350813.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6355751.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6355751.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6017919.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6017919.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6017922.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6017922.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6019026.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6019026.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6019566.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6019566.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6372840.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6372840.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6372856.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6372856.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6373601.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6373601.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6373615.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6373615.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6382984.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6382984.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6400810.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6400810.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6225970.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6225970.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6226041.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6226041.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6228446.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6228446.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6228985.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6228985.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5297178.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5297178.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5297184.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5297184.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5299552.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5299552.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5300596.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5300596.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5300796.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5300796.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5305725.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5305725.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5305727.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5305727.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5308354.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5308354.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5309181.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5309181.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5310380.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5310380.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5867284.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5867284.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5867300.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5867300.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5870492.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5870492.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5870684.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5870684.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5876670.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5876670.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5845435.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5845435.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5845459.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5845459.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5850880.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5850880.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5851060.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5851060.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5290364.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5290364.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5290365.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5290365.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5293048.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5293048.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5294079.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5294079.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5295637.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5295637.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5297100.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5297100.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5315542.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5315542.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5315575.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5315575.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5318936.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5318936.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5374687.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5374687.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5374902.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5374902.out
Ignored: scripts/pred_HHB_imputed.6496220.err
Ignored: scripts/pred_HHB_imputed.6496220.out
Ignored: scripts/pred_fullV_HHB_imputed.6503400.err
Ignored: scripts/pred_fullV_HHB_imputed.6503400.out
Untracked files:
Untracked: analysis/dense_issue_investigation2/
Untracked: code/20200502_Prepare_ED_prior.ipynb
Untracked: code/plot_test.R
Untracked: code/pred_fullV_LNP1.R
Untracked: dsc/mvreg_all_genes_prior_09_11_20_COPY.dsc
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/mrmash_larger_r_issue.Rmd) and HTML (docs/mrmash_larger_r_issue.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bf711b8 | fmorgante | 2021-04-13 | Add null data lfsr |
| html | f3b730a | fmorgante | 2021-04-13 | Build site. |
| Rmd | c544bf2 | fmorgante | 2021-04-13 | Add null data results |
| html | 2034558 | fmorgante | 2021-04-13 | Build site. |
| Rmd | 0091cf6 | fmorgante | 2021-04-13 | Add more mash results |
| html | 446489f | fmorgante | 2021-04-12 | Build site. |
| Rmd | 0fe4f50 | fmorgante | 2021-04-12 | Add mash results |
| html | b2c3824 | fmorgante | 2021-03-25 | Build site. |
| Rmd | 5361380 | fmorgante | 2021-03-25 | Add a few things |
| html | cde23ac | fmorgante | 2021-03-24 | Build site. |
| Rmd | d5846a6 | fmorgante | 2021-03-24 | Add investigation on the issue with larger r |
library(mr.mash.alpha)
library(glmnet)
library(mashr)
options(stringsAsFactors = FALSE)
###Functions to compute g-lasso coefficients
compute_coefficients_glasso <- function(X, Y, standardize, nthreads, version=c("Rcpp", "R")){
version <- match.arg(version)
n <- nrow(X)
p <- ncol(X)
r <- ncol(Y)
Y_has_missing <- any(is.na(Y))
if(Y_has_missing){
###Extract per-individual Y missingness patterns
Y_miss_patterns <- mr.mash.alpha:::extract_missing_Y_pattern(Y)
###Initialize missing Ys
muy <- colMeans(Y, na.rm=TRUE)
for(l in 1:r){
Y[is.na(Y[, l]), l] <- muy[l]
}
###Compute V and its inverse
V <- mr.mash.alpha:::compute_V_init(X, Y, matrix(0, p, r), method="flash")
Vinv <- chol2inv(chol(V))
###Compute expected Y (assuming B=0)
mu <- matrix(rep(muy, each=n), n, r)
###Impute missing Ys
Y <- mr.mash.alpha:::impute_missing_Y(Y=Y, mu=mu, Vinv=Vinv, miss=Y_miss_patterns$miss, non_miss=Y_miss_patterns$non_miss,
version=version)$Y
}
##Fit group-lasso
if(nthreads>1){
doMC::registerDoMC(nthreads)
paral <- TRUE
} else {
paral <- FALSE
}
cvfit_glmnet <- glmnet::cv.glmnet(x=X, y=Y, family="mgaussian", alpha=1, standardize=standardize, parallel=paral)
coeff_glmnet <- coef(cvfit_glmnet, s="lambda.min")
##Build matrix of initial estimates for mr.mash
B <- matrix(as.numeric(NA), nrow=p, ncol=r)
for(i in 1:length(coeff_glmnet)){
B[, i] <- as.vector(coeff_glmnet[[i]])[-1]
}
return(list(B=B, Y=Y))
}In this website, we want to investigate an issue that happens with larger number of conditions than the 10 conditions we have used in the other investigations. The issue is that mr.mash fails to shrink to 0 coefficients that are actually 0. As we will see, some of them are pretty badly misestimated.
Let’s look at an example with n=150, p=400, r=25, PVE=0.2, p_causal=5, independent residuals, independent predictors, and equal effects across conditions from a single multivariate normal with variance 1.
set.seed(12)
n <- 200
ntest <- 50
p <- 400
p_causal <- 5
r <- 25
X_cor <- 0
pve <- 0.2
r_causal <- list(1:r)
B_cor <- 1
B_scale <- 1
w <- 1
###Simulate V, B, X and Y
out <- simulate_mr_mash_data(n, p, p_causal, r, r_causal, intercepts = rep(1, r),
pve=pve, B_cor=B_cor, B_scale=B_scale, w=w,
X_cor=X_cor, X_scale=1, V_cor=0)
colnames(out$Y) <- paste0("Y", seq(1, r))
rownames(out$Y) <- paste0("N", seq(1, n))
colnames(out$X) <- paste0("X", seq(1, p))
rownames(out$X) <- paste0("N", seq(1, n))
###Split the data in training and test sets
test_set <- sort(sample(x=c(1:n), size=ntest, replace=FALSE))
Ytrain <- out$Y[-test_set, ]
Xtrain <- out$X[-test_set, ]
Ytest <- out$Y[test_set, ]
Xtest <- out$X[test_set, ]mr.mash is run standardizing X and dropping mixture components with weight less than \(1e-8\). However, the results look the same without dropping any component. The mixture prior consists only of canonical matrices, scaled by a grid computed as in the mash paper. group-LASSO is used to initialize mr.mash and as a comparison.
standardize <- TRUE
nthreads <- 4
w0_threshold <- 1e-8
S0_can <- compute_canonical_covs(ncol(Ytrain), singletons=TRUE, hetgrid=c(0, 0.25, 0.5, 0.75, 1))
####Complete data####
###Compute grid of variances
univ_sumstats <- compute_univariate_sumstats(Xtrain, Ytrain, standardize=standardize,
standardize.response=FALSE, mc.cores=nthreads)
grid <- autoselect.mixsd(univ_sumstats, mult=sqrt(2))^2
###Compute prior with only canonical matrices
S0 <- expand_covs(S0_can, grid, zeromat=TRUE)
###Fit mr.mash
out_coeff <- compute_coefficients_glasso(Xtrain, Ytrain, standardize=standardize, nthreads=nthreads, version="Rcpp")
prop_nonzero_glmnet <- sum(out_coeff$B[, 1]!=0)/p
w0 <- c((1-prop_nonzero_glmnet), rep(prop_nonzero_glmnet/(length(S0)-1), (length(S0)-1)))
fit_mrmash <- mr.mash(Xtrain, Ytrain, S0, w0=w0, update_w0=TRUE, update_w0_method="EM", tol=1e-2,
convergence_criterion="ELBO", compute_ELBO=TRUE, standardize=standardize,
verbose=FALSE, update_V=TRUE, update_V_method="diagonal", e=1e-8,
w0_threshold=w0_threshold, nthreads=nthreads, mu1_init=out_coeff$B)
#####Coefficients plots######
layout(matrix(c(1, 1, 2, 2,
1, 1, 2, 2,
0, 3, 3, 0,
0, 3, 3, 0), 4, 4, byrow = TRUE))
###Plot estimated vs true coeffcients
##mr.mash
plot(out$B, fit_mrmash$mu1, main="mr.mash", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
##g-lasso
plot(out$B, out_coeff$B, main="g-lasso", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
###Plot mr.mash vs g-lasso estimated coeffcients
colorz <- matrix("black", nrow=p, ncol=r)
zeros <- apply(out$B, 2, function(x) x==0)
for(i in 1:ncol(colorz)){
colorz[zeros[, i], i] <- "red"
}
plot(fit_mrmash$mu1, out_coeff$B, main="mr.mash vs g-lasso",
xlab="mr.mash coefficients", ylab="g-lasso coefficients",
col=colorz, cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
legend("topleft",
legend = c("Non-zero", "Zero"),
col = c("black", "red"),
pch = c(1, 1),
horiz = FALSE,
cex=2) 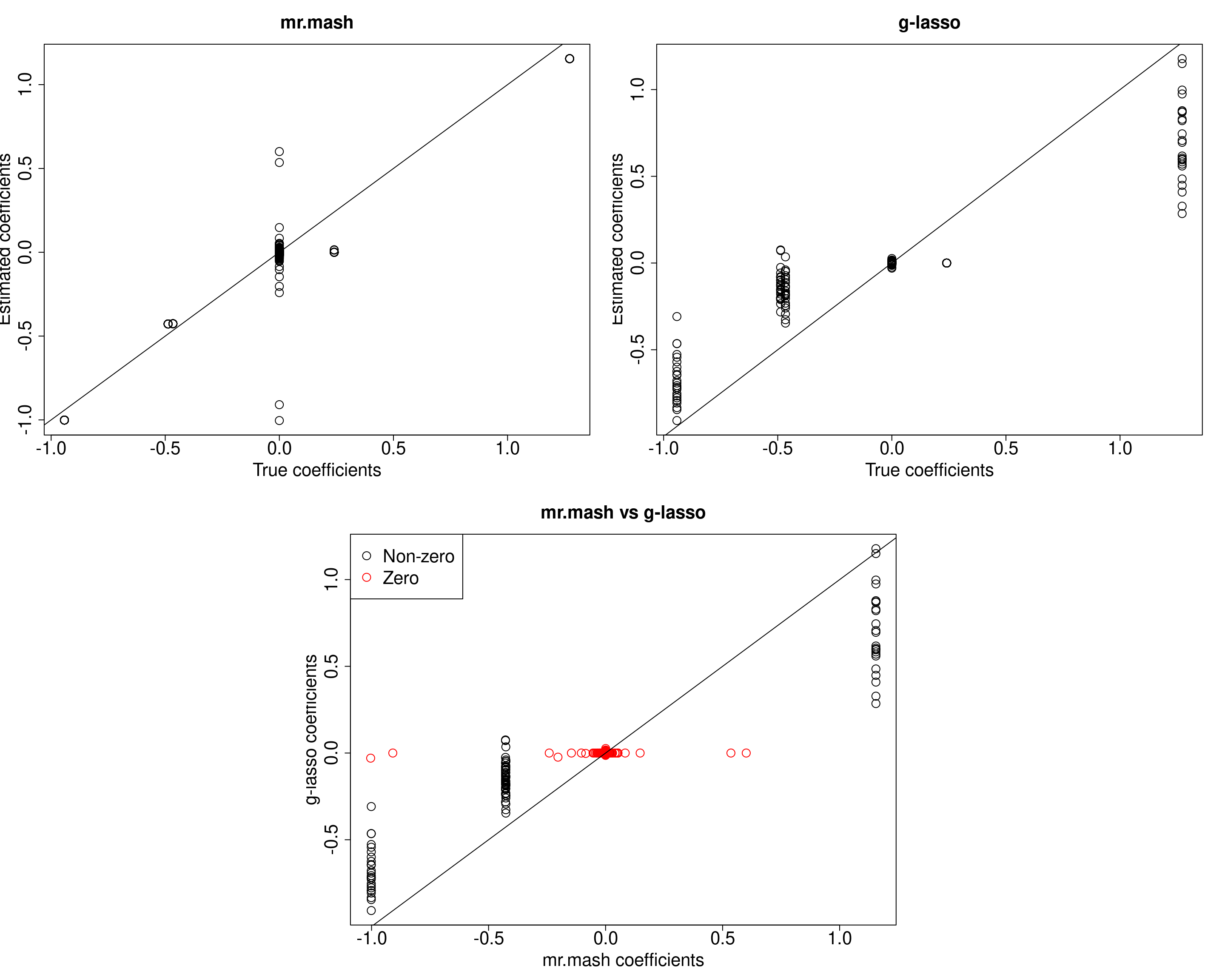
###Identify badly estimated variables
bad_est_1 <- which(out$B==0 & fit_mrmash$mu1 < -0.5, arr.ind = TRUE)
bad_est_2 <- which(out$B==0 & fit_mrmash$mu1 > 0.5, arr.ind = TRUE)
bad_est <- rbind(bad_est_1, bad_est_2)
cat("Bad variables:\n")Bad variables:bad_est row col
X274 274 10
X347 347 15
X183 183 10
X72 72 13mr.mash is much better than group-LASSO at correctly estimating the non-zero coefficients. However, it does much worse at shrinking the true null coefficients to 0. In particular, there are a few coeffcients that are pretty badly misestimated. We are going to look at those cases more closely.
###Estimated coefficients by mr.mash for the bad variables
cat("mr.mash estimated coefficients:\n")mr.mash estimated coefficients:fit_mrmash$mu1[bad_est[, 1], ] Y1 Y2 Y3 Y4 Y5
X274 -2.642841e-05 -2.663867e-05 -2.335354e-05 -2.653733e-05 -2.613332e-05
X347 6.491175e-07 2.916934e-06 1.739985e-06 2.150244e-06 2.321851e-06
X183 1.133417e-04 1.284499e-04 1.203046e-04 1.115160e-04 1.203380e-04
X72 9.529781e-06 -4.624202e-06 1.235475e-06 1.068643e-06 2.780234e-06
Y6 Y7 Y8 Y9 Y10
X274 -2.233829e-05 -2.511605e-05 -2.896884e-05 -2.870533e-05 -9.092047e-01
X347 1.451095e-06 6.299573e-06 3.501019e-06 6.391154e-07 6.646096e-05
X183 1.200349e-04 1.184511e-04 1.168999e-04 9.990242e-05 5.354676e-01
X72 1.060682e-05 1.326606e-05 7.372047e-06 -2.390740e-06 -6.613187e-04
Y11 Y12 Y13 Y14 Y15
X274 -2.773517e-05 -2.442013e-05 8.829146e-05 -3.148464e-05 6.064451e-05
X347 5.231139e-06 -3.038223e-05 -2.331779e-04 1.595482e-06 -1.003837e+00
X183 1.290409e-04 1.330501e-04 3.121302e-04 1.422738e-04 3.496837e-04
X72 9.218040e-06 -3.617465e-05 6.008830e-01 -6.688684e-06 1.265758e-03
Y16 Y17 Y18 Y19 Y20
X274 -2.554938e-05 -2.579409e-05 -2.871577e-05 -2.471882e-05 -2.455460e-05
X347 4.505555e-06 2.149654e-06 1.773809e-06 2.081464e-06 -8.069752e-08
X183 1.223263e-04 1.269347e-04 1.225920e-04 1.272094e-04 1.296489e-04
X72 5.131390e-06 3.996038e-06 2.568455e-06 4.814783e-06 9.623803e-06
Y21 Y22 Y23 Y24 Y25
X274 -2.592537e-05 4.360263e-04 -2.732129e-05 -1.867228e-05 -2.871367e-05
X347 9.000980e-06 -9.735566e-07 2.812928e-06 3.894336e-06 8.448670e-07
X183 1.170115e-04 -1.310602e-04 1.225248e-04 1.238062e-04 9.362710e-05
X72 -1.681922e-05 3.996730e-04 4.902352e-06 9.337151e-06 1.645090e-05We can see that the estimated coefficients for each one of those variables look like a singleton structure, where there is an effect in one condition and nowhere else. When we look at the prior weights at convergence, we can see that those singleton matrices get relatively large weight (see below).
###Estimated prior weights by mr.mash for the bad variables
cat("mr.mash estimated prior weights:\n")mr.mash estimated prior weights:head(sort(fit_mrmash$w0, decreasing=TRUE), 20) null shared1_grid11 singleton13_grid10
0.9346759513 0.0086745867 0.0069429060
singleton10_grid10 singleton22_grid10 singleton10_grid11
0.0065330516 0.0052718769 0.0051348432
singleton15_grid11 singleton15_grid10 shared1_grid10
0.0049436750 0.0047741792 0.0034326125
singleton22_grid9 singleton13_grid9 singleton13_grid11
0.0030192287 0.0018686026 0.0014915698
singleton15_grid9 singleton10_grid9 singleton22_grid8
0.0004987422 0.0004802332 0.0003878599
singleton13_grid8 singleton12_grid8 singleton24_grid8
0.0003738464 0.0002453590 0.0001796324
singleton12_grid7 singleton22_grid11
0.0001623117 0.0001445351 To see why that is happening, the next step will be to look at the data. While the best thing to do would be to look at the summary statistics from a simple multivariate regression with the same residual covariance matrix as in mr.mash, we are going to look at the univariate summary statistics that are readily available.
###Estimated coefficients and Z-scores by univariate regression for the bad variables
rownames(univ_sumstats$Bhat) <- colnames(out$X)
colnames(univ_sumstats$Bhat) <- colnames(out$Y)
cat("SLR estimated coefficients:\n")SLR estimated coefficients:univ_sumstats$Bhat[bad_est[, 1], ] Y1 Y2 Y3 Y4 Y5 Y6
X274 -0.1325482 -0.09893288 0.61508065 -0.1165136 -0.1069066 0.35151305
X347 -0.2427989 0.28123165 0.06309551 0.1649556 0.3030400 0.02980773
X183 -0.2428503 0.49439918 0.20907556 -0.2292677 0.2192696 0.17487806
X72 0.3468332 -0.38077183 -0.36862902 -0.2403712 -0.2716020 0.18512322
Y7 Y8 Y9 Y10 Y11 Y12
X274 0.1024990 -0.454970612 -0.25909065 -1.0881426 -0.2781424 0.0916439
X347 0.4888237 0.446450157 -0.08708036 0.2033015 0.6209967 -0.5022021
X183 0.1270435 0.003659657 -0.29134310 1.1836731 0.5654748 0.3028926
X72 0.2467956 0.150784203 -0.25086360 -0.1810409 0.2310031 -0.3425984
Y13 Y14 Y15 Y16 Y17 Y18
X274 0.2435822 -0.39061436 0.2271793 0.071133592 0.01441381 -0.58419877
X347 -0.3747620 0.04851022 -1.1350130 0.640749645 0.19264130 0.07550262
X183 0.3618534 0.62647779 0.4556610 0.337172801 0.62552105 0.39018457
X72 1.1717008 -0.33024120 0.5026009 0.009030604 -0.08879668 -0.23306137
Y19 Y20 Y21 Y22 Y23
X274 0.23015756 0.1516004 0.02275088 0.52279901 -0.226039596
X347 0.15321483 -0.1375850 0.51024287 0.07540335 0.296716744
X183 0.54839476 0.4148681 0.10061303 -0.04286237 0.308742629
X72 -0.01456058 0.1413060 -0.37970711 0.21641262 -0.006988736
Y24 Y25
X274 0.32427352 -0.3004417
X347 0.19882637 -0.0776774
X183 0.22366030 -0.4267544
X72 0.06677394 0.3970696###Estimated Z-scores by univariate regression for the bad variables
Zscores <- univ_sumstats$Bhat/univ_sumstats$Shat
cat("SLR estimated Z-scores:\n")SLR estimated Z-scores:Zscores[bad_est[, 1], ] Y1 Y2 Y3 Y4 Y5 Y6
X274 -0.4365736 -0.3252120 1.7394660 -0.3690672 -0.3517424 1.11406779
X347 -0.8009137 0.9267963 0.1766698 0.5227513 0.9999831 0.09408288
X183 -0.8010841 1.6391949 0.5860324 -0.7271807 0.7224003 0.55252082
X72 1.1466597 -1.2578544 -1.0357761 -0.7625325 -0.8956515 0.58496230
Y7 Y8 Y9 Y10 Y11 Y12
X274 0.3267747 -1.44776342 -0.8270740 -3.3784413 -0.8473222 0.2965010
X347 1.5706941 1.42027984 -0.2774153 0.6090926 1.9102079 -1.6389006
X183 0.4051023 0.01156436 -0.9305960 3.7011173 1.7357935 0.9828482
X72 0.7881596 0.47683468 -0.8006966 -0.5422600 0.7031937 -1.1126963
Y13 Y14 Y15 Y16 Y17 Y18
X274 0.732031 -1.1812598 0.696065 0.23658439 0.04131068 -1.9612099
X347 -1.129041 0.1460284 -3.621566 2.16389997 0.55268195 0.2503124
X183 1.089836 1.9086398 1.403033 1.12595782 1.81241787 1.3006204
X72 3.670433 -0.9973537 1.549786 0.03002948 -0.25454953 -0.7740532
Y19 Y20 Y21 Y22 Y23 Y24
X274 0.71527253 0.4970719 0.0678337 1.7837366 -0.74185722 1.0726073
X347 0.47569908 -0.4510520 1.5332637 0.2546203 0.97512125 0.6560840
X183 1.71812707 1.3676601 0.3000724 -0.1447153 1.01491063 0.7383136
X72 -0.04517348 0.4632679 -1.1370125 0.7319304 -0.02289468 0.2200577
Y25
X274 -0.932783
X347 -0.240511
X183 -1.328901
X72 1.235480plot(fit_mrmash$mu1[bad_est[, 1], ], univ_sumstats$Bhat[bad_est[, 1], ], main="mr.mash vs SLR -- bad variables",
xlab="mr.mash coefficients", ylab="SLR coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)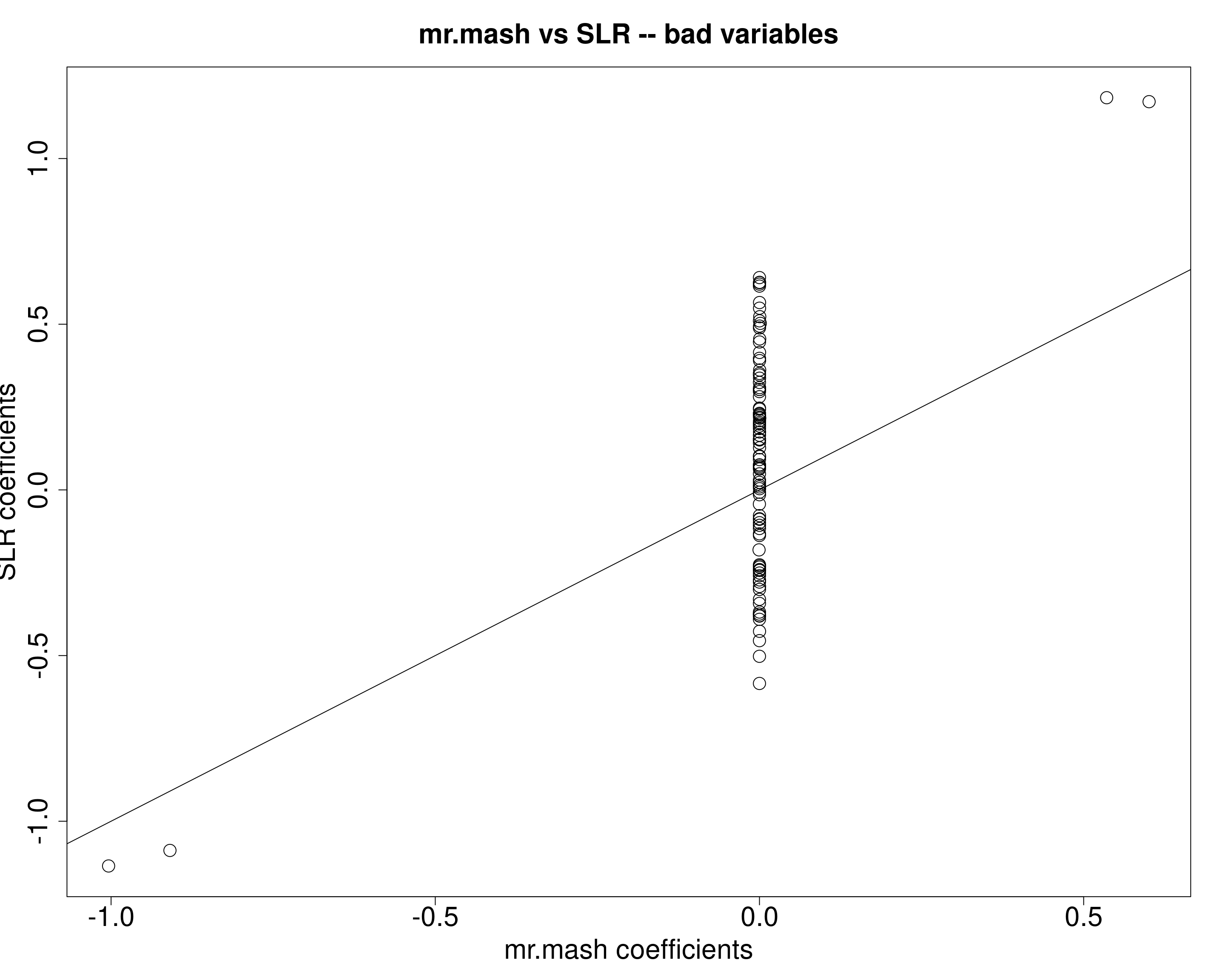
The data (wrongly) supports the singleton structure (i.e., large effect in one condition and smaller effects in the other conditions). mr.mash then estimates the coefficients accordingly, by shrinking to 0 the coefficients in the conditions with smaller effects and leaving the coefficent in the one condition almost unshrunken for two variables and a little shrunken for the other two variables.
If we do not include the singleton matrices in the prior, this behavior does not appear.
S0_can_nosing <- compute_canonical_covs(ncol(Ytrain), singletons=FALSE, hetgrid=c(0, 0.25, 0.5, 0.75, 1))
###Compute prior with only canonical matrices without singletons
S0_nosing <- expand_covs(S0_can_nosing, grid, zeromat=TRUE)
###Fit mr.mash
w0_nosing <- c((1-prop_nonzero_glmnet), rep(prop_nonzero_glmnet/(length(S0_nosing)-1), (length(S0_nosing)-1)))
fit_mrmash_nosing <- mr.mash(Xtrain, Ytrain, S0_nosing, w0=w0_nosing, update_w0=TRUE, update_w0_method="EM", tol=1e-2,
convergence_criterion="ELBO", compute_ELBO=TRUE, standardize=standardize,
verbose=FALSE, update_V=TRUE, update_V_method="diagonal", e=1e-8,
w0_threshold=w0_threshold, nthreads=nthreads, mu1_init=out_coeff$B)
#####Coefficients plots######
layout(matrix(c(1, 1, 2, 2,
1, 1, 2, 2,
0, 3, 3, 0,
0, 3, 3, 0), 4, 4, byrow = TRUE))
###Plot estimated vs true coeffcients
##mr.mash
plot(out$B, fit_mrmash_nosing$mu1, main="mr.mash", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
##g-lasso
plot(out$B, out_coeff$B, main="g-lasso", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
###Plot mr.mash vs g-lasso estimated coeffcients
colorz <- matrix("black", nrow=p, ncol=r)
zeros <- apply(out$B, 2, function(x) x==0)
for(i in 1:ncol(colorz)){
colorz[zeros[, i], i] <- "red"
}
plot(fit_mrmash_nosing$mu1, out_coeff$B, main="mr.mash vs g-lasso",
xlab="mr.mash coefficients", ylab="g-lasso coefficients",
col=colorz, cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
legend("topleft",
legend = c("Non-zero", "Zero"),
col = c("black", "red"),
pch = c(1, 1),
horiz = FALSE,
cex=2)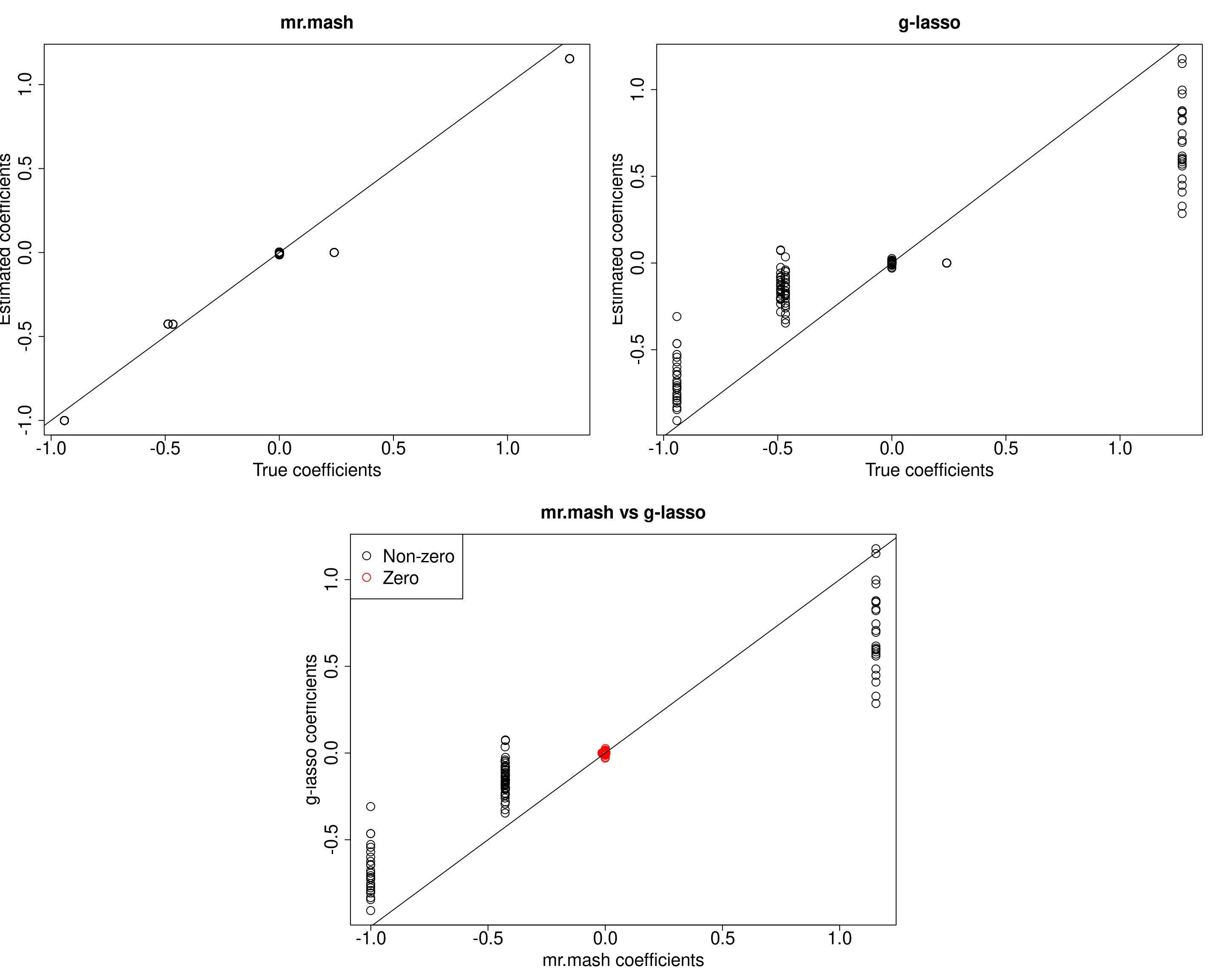
When comparing the ELBO of the two models, we can see that the ELBO for the model with the singletons is a little larger.
###ELBO with singletons
cat("mr.mash ELBO with singletons:\n")mr.mash ELBO with singletons:fit_mrmash$ELBO[1] -10083.11###ELBO with singletons
cat("mr.mash ELBO without singletons:\n")mr.mash ELBO without singletons:fit_mrmash_nosing$ELBO[1] -10085.82However, at least in this example, the variable/condition combinations displaying this problem are not so many. Interestingly, a few conditions (10, 13, 15, 22) are the only ones creating this issue.
badd <- which(abs(fit_mrmash$mu1 - out$B) > 0.05, arr.ind = TRUE)
unique_vars <- unique(badd[,1])
unique_vars <- unique_vars[-unique(which(out$B[unique_vars, ] !=0, arr.ind = TRUE)[, 1])]
unique_vars_bad_y <- badd[which(badd[, 1] %in% unique_vars), ]
unique_vars_bad_y row col
X141 141 10
X183 183 10
X274 274 10
X72 72 13
X342 342 13
X357 357 13
X142 142 15
X147 147 15
X332 332 15
X347 347 15
X187 187 22
X192 192 22
X194 194 22
X232 232 22
X327 327 22plot(out$B[unique_vars_bad_y], fit_mrmash$mu1[unique_vars_bad_y], xlab="True coefficents", ylab="Estimated coefficients", main="mr.mash vs truth\nmost bad variables/conditions")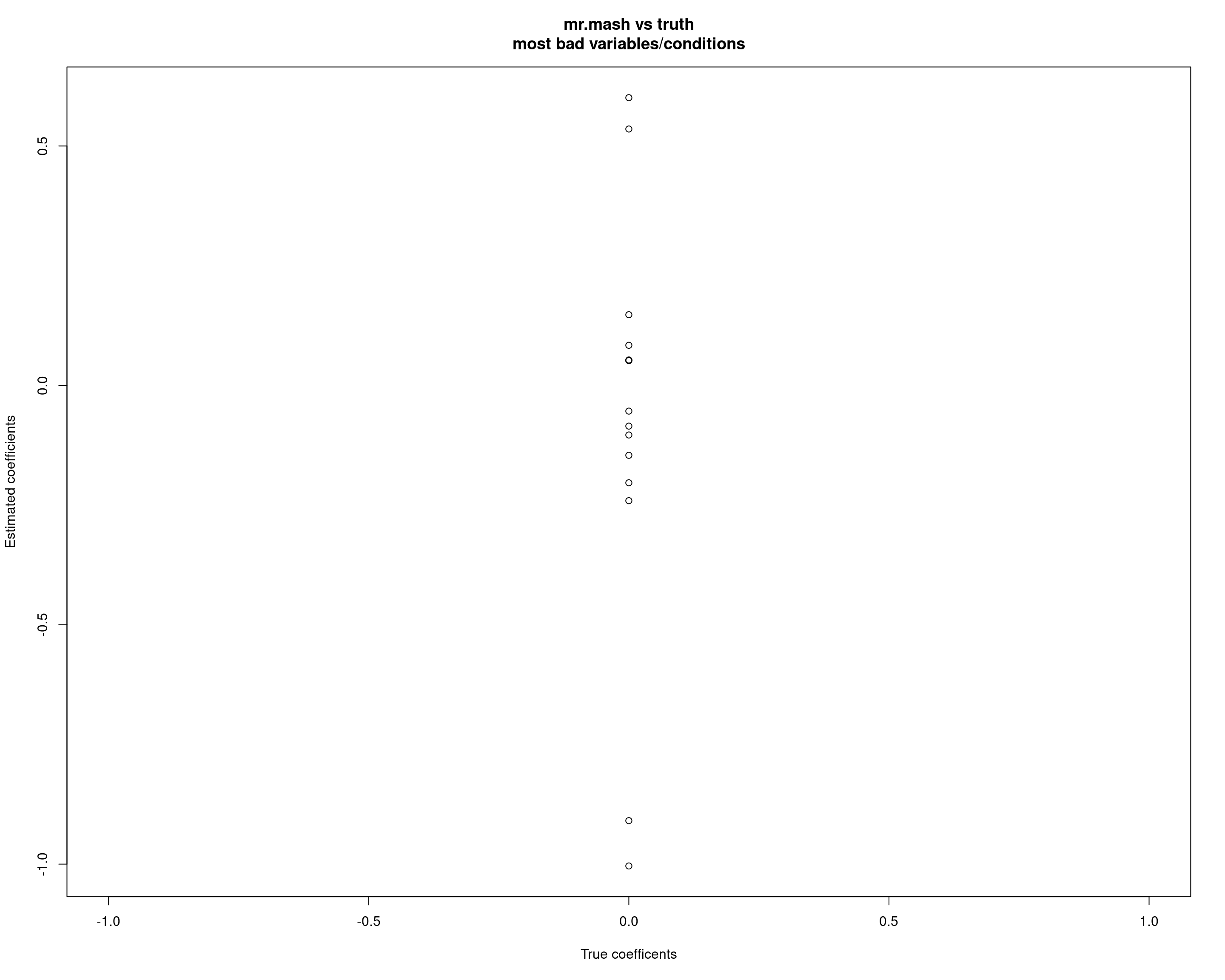
Let’s what happens if we run mash on the summary statistics. Since predictors are independent, so we should roughly get the same results as mr.mash.
dat_mash <- mash_set_data(univ_sumstats$Bhat, univ_sumstats$Shat)
mash_obj <- mash(dat_mash, S0_can) - Computing 400 x 451 likelihood matrix.
- Likelihood calculations took 1.44 seconds.
- Fitting model with 451 mixture components.
- Model fitting took 3.03 seconds.
- Computing posterior matrices.
- Computation allocated took 0.09 seconds.mash_pm <- get_pm(mash_obj)
cat("mash estimated coefficients:\n")mash estimated coefficients:mash_pm[bad_est[, 1], ] Y1 Y2 Y3 Y4 Y5
X274 -0.023650792 -0.023650792 -0.023650792 -0.023650792 -0.023650792
X347 0.057239661 0.057239661 0.057239661 0.057239661 0.057239661
X183 0.213057913 0.213057913 0.213057913 0.213057913 0.213057913
X72 0.006652372 0.006652372 0.006652372 0.006652372 0.006652372
Y6 Y7 Y8 Y9 Y10
X274 -0.023650792 -0.023650792 -0.023650792 -0.023650792 -0.023650792
X347 0.057239661 0.057239661 0.057239661 0.057239661 0.057239661
X183 0.213057913 0.213057913 0.213057913 0.213057913 0.213057913
X72 0.006652372 0.006652372 0.006652372 0.006652372 0.006652372
Y11 Y12 Y13 Y14 Y15
X274 -0.023650792 -0.023650792 -0.02250090 -0.023650792 -0.023650792
X347 0.057239661 0.057239661 0.05573754 0.057239661 0.057239661
X183 0.213057913 0.213057913 0.21306298 0.213057913 0.213057913
X72 0.006652372 0.006652372 0.45452574 0.006652372 0.006652372
Y16 Y17 Y18 Y19 Y20
X274 -0.023650792 -0.023650792 -0.023650792 -0.023650792 -0.023650792
X347 0.057239661 0.057239661 0.057239661 0.057239661 0.057239661
X183 0.213057913 0.213057913 0.213057913 0.213057913 0.213057913
X72 0.006652372 0.006652372 0.006652372 0.006652372 0.006652372
Y21 Y22 Y23 Y24 Y25
X274 -0.023642208 -0.023650792 -0.023650792 -0.023650792 -0.023650792
X347 0.057577792 0.057239661 0.057239661 0.057239661 0.057239661
X183 0.213058005 0.213057913 0.213057913 0.213057913 0.213057913
X72 0.006514678 0.006652372 0.006652372 0.006652372 0.006652372mash seems to have a different problem – it estimates that the effects of variables X274, X347, and X183 to be equal across conditions. However, similar behavior to mr.mash is shown for variable X72. Checking the prior weights, we can see that mash wrongly puts a lot of weight on equal effects components.
###Estimated prior weights by mr.mash for the bad variables
cat("mr.mash estimated prior weights:\n")mr.mash estimated prior weights:head(sort(get_estimated_pi(mash_obj, "all"), decreasing=TRUE), 20) shared1.6 null shared1.10 singleton13.10 shared1.11
0.7600049178 0.2209967909 0.0074711314 0.0061764252 0.0046510891
singleton21.11 singleton1.1 singleton2.1 singleton3.1 singleton4.1
0.0006996457 0.0000000000 0.0000000000 0.0000000000 0.0000000000
singleton5.1 singleton6.1 singleton7.1 singleton8.1 singleton9.1
0.0000000000 0.0000000000 0.0000000000 0.0000000000 0.0000000000
singleton10.1 singleton11.1 singleton12.1 singleton13.1 singleton14.1
0.0000000000 0.0000000000 0.0000000000 0.0000000000 0.0000000000 Let’s now look at the estimated effects from mash, and compare them to those from mr.mash.
#####Coefficients plots######
layout(matrix(c(1, 1, 2, 2,
1, 1, 2, 2), 2, 4, byrow = TRUE))
##mash
plot(out$B, mash_pm, main="mash", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
##mr.mash vs mash
plot(fit_mrmash$mu1, mash_pm, main="mr.mash vs mash -- all variables",
xlab="mr.mash coefficients", ylab="mash coefficients",
col=colorz, cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
legend("topleft",
legend = c("Non-zero", "Zero"),
col = c("black", "red"),
pch = c(1, 1),
horiz = FALSE,
cex=2)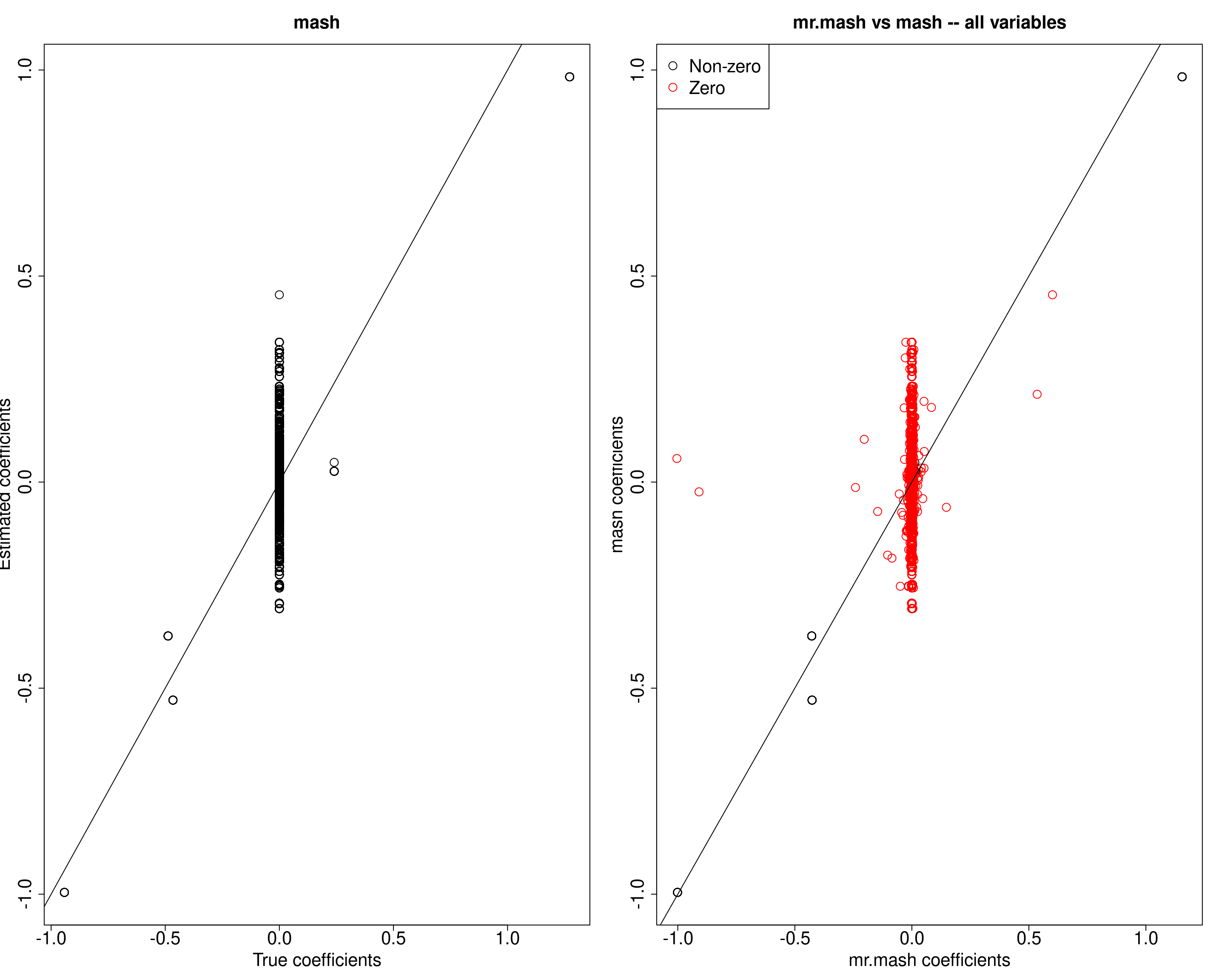
In general, mash seems to have more difficulty to shrink true 0 coefficients.
Let’s now compare the two methods on completely null data.
nreps <- 4
mrmash_null_coeff <- vector("list", nreps)
mash_obj_null <- vector("list", nreps)
for(i in 1:nreps){
###Simulate null phenotypes
Ytrain_null <- matrix(rnorm((n-ntest)*r), nrow=(n-ntest), ncol=r)
###Compute grid of variances
univ_sumstats_null <- compute_univariate_sumstats(Xtrain, Ytrain_null, standardize=standardize,
standardize.response=FALSE, mc.cores=nthreads)
grid_null <- autoselect.mixsd(univ_sumstats_null, mult=sqrt(2))^2
###Compute prior with only canonical matrices
S0_null <- expand_covs(S0_can, grid_null, zeromat=TRUE)
###Fit mr.mash
out_coeff_null <- compute_coefficients_glasso(Xtrain, Ytrain_null, standardize=standardize, nthreads=nthreads, version="Rcpp")
prop_nonzero_glmnet_null <- sum(out_coeff_null$B[, 1]!=0)/p
w0_null <- c((1-prop_nonzero_glmnet_null), rep(prop_nonzero_glmnet_null/(length(S0_null)-1), (length(S0_null)-1)))
fit_mrmash_null <- mr.mash(Xtrain, Ytrain_null, S0_null, w0=w0_null, update_w0=TRUE, update_w0_method="EM", tol=1e-2,
convergence_criterion="ELBO", compute_ELBO=TRUE, standardize=standardize,
verbose=FALSE, update_V=TRUE, update_V_method="diagonal", e=1e-8,
w0_threshold=w0_threshold, nthreads=nthreads, mu1_init=out_coeff_null$B)
mrmash_null_coeff[[i]] <- fit_mrmash_null
###Fit mash
dat_mash_null <- mash_set_data(univ_sumstats_null$Bhat, univ_sumstats_null$Shat)
mash_obj_null[[i]] <- mash(dat_mash_null, S0_can)
} - Computing 400 x 451 likelihood matrix.
- Likelihood calculations took 1.41 seconds.
- Fitting model with 451 mixture components.
- Model fitting took 4.28 seconds.
- Computing posterior matrices.
- Computation allocated took 0.17 seconds.
- Computing 400 x 451 likelihood matrix.
- Likelihood calculations took 1.45 seconds.
- Fitting model with 451 mixture components.
- Model fitting took 3.14 seconds.
- Computing posterior matrices.
- Computation allocated took 0.13 seconds.
- Computing 400 x 451 likelihood matrix.
- Likelihood calculations took 1.41 seconds.
- Fitting model with 451 mixture components.
- Model fitting took 3.70 seconds.
- Computing posterior matrices.
- Computation allocated took 0.13 seconds.
- Computing 400 x 451 likelihood matrix.
- Likelihood calculations took 1.42 seconds.
- Fitting model with 451 mixture components.
- Model fitting took 3.78 seconds.
- Computing posterior matrices.
- Computation allocated took 0.13 seconds.#####Coefficients plots######
layout(matrix(c(1, 1, 2, 2,
1, 1, 2, 2,
3, 3, 4, 4,
3, 3, 4, 4), 4, 4, byrow = TRUE))
for(i in 1:nreps){
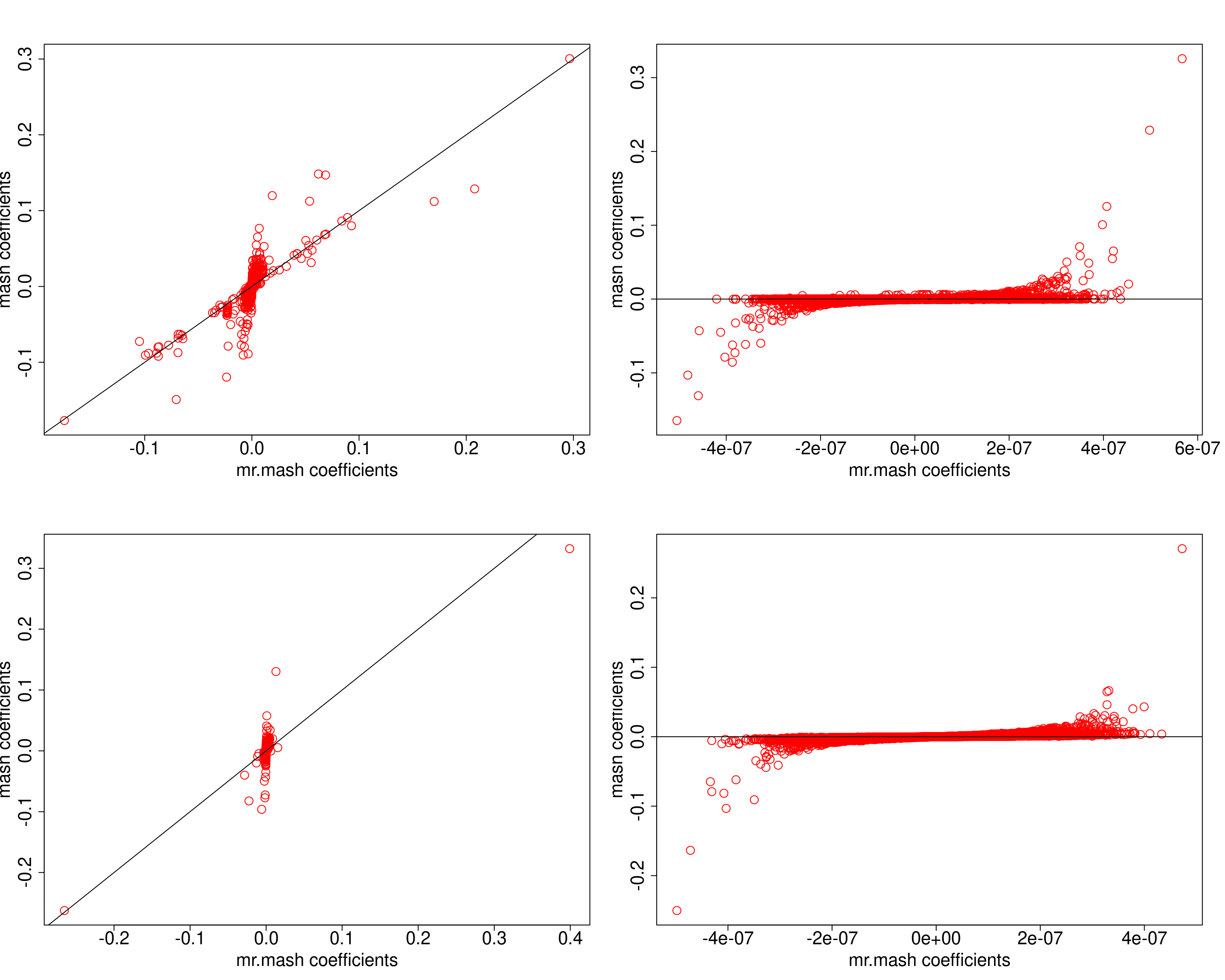
##mr.mash vs mash
plot(mrmash_null_coeff[[i]][[1]], get_pm(mash_obj_null[[i]]),
xlab="mr.mash coefficients", ylab="mash coefficients",
col="red", cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
}
| Version | Author | Date |
|---|---|---|
| f3b730a | fmorgante | 2021-04-13 |
It turns out that mr.mash does not seem to give more false pasitives than mash – it is actually the opposite. However, the lfsr might be informative about actual false positives in mash.
for(i in 1:nreps){
print(summary(get_lfsr(mash_obj_null[[i]])))
} V1 V2 V3 V4
Min. :0.1418 Min. :0.1999 Min. :0.1391 Min. :0.1767
1st Qu.:0.9292 1st Qu.:0.9337 1st Qu.:0.9359 1st Qu.:0.9350
Median :0.9520 Median :0.9563 Median :0.9551 Median :0.9571
Mean :0.9244 Mean :0.9293 Mean :0.9294 Mean :0.9296
3rd Qu.:0.9627 3rd Qu.:0.9662 3rd Qu.:0.9670 3rd Qu.:0.9663
Max. :0.9879 Max. :0.9871 Max. :0.9884 Max. :0.9859
V5 V6 V7 V8
Min. :0.08579 Min. :0.2306 Min. :0.1092 Min. :0.2326
1st Qu.:0.88698 1st Qu.:0.9128 1st Qu.:0.9328 1st Qu.:0.9107
Median :0.92359 Median :0.9430 Median :0.9560 Median :0.9398
Mean :0.88916 Mean :0.9135 Mean :0.9307 Mean :0.9112
3rd Qu.:0.94137 3rd Qu.:0.9571 3rd Qu.:0.9672 3rd Qu.:0.9547
Max. :0.98829 Max. :0.9881 Max. :0.9864 Max. :0.9894
V9 V10 V11 V12
Min. :0.1544 Min. :0.1193 Min. :0.08225 Min. :0.2153
1st Qu.:0.9354 1st Qu.:0.9345 1st Qu.:0.93602 1st Qu.:0.9340
Median :0.9555 Median :0.9561 Median :0.95659 Median :0.9553
Mean :0.9299 Mean :0.9294 Mean :0.93056 Mean :0.9286
3rd Qu.:0.9669 3rd Qu.:0.9665 3rd Qu.:0.96674 3rd Qu.:0.9661
Max. :0.9850 Max. :0.9893 Max. :0.98796 Max. :0.9871
V13 V14 V15 V16
Min. :0.2457 Min. :0.1506 Min. :0.02006 Min. :0.07406
1st Qu.:0.9340 1st Qu.:0.8032 1st Qu.:0.93549 1st Qu.:0.92717
Median :0.9563 Median :0.8677 Median :0.95685 Median :0.95107
Mean :0.9305 Mean :0.8252 Mean :0.93087 Mean :0.92229
3rd Qu.:0.9657 3rd Qu.:0.8995 3rd Qu.:0.96693 3rd Qu.:0.96176
Max. :0.9884 Max. :0.9869 Max. :0.98593 Max. :0.98606
V17 V18 V19 V20
Min. :0.2025 Min. :0.03426 Min. :0.2216 Min. :0.2060
1st Qu.:0.9361 1st Qu.:0.92685 1st Qu.:0.9341 1st Qu.:0.9348
Median :0.9562 Median :0.95293 Median :0.9560 Median :0.9581
Mean :0.9314 Mean :0.92342 Mean :0.9310 Mean :0.9307
3rd Qu.:0.9661 3rd Qu.:0.96426 3rd Qu.:0.9671 3rd Qu.:0.9669
Max. :0.9864 Max. :0.98517 Max. :0.9881 Max. :0.9872
V21 V22 V23 V24
Min. :0.2022 Min. :0.2143 Min. :0.1581 Min. :0.1300
1st Qu.:0.9201 1st Qu.:0.9343 1st Qu.:0.9330 1st Qu.:0.9205
Median :0.9442 Median :0.9562 Median :0.9560 Median :0.9474
Mean :0.9144 Mean :0.9308 Mean :0.9299 Mean :0.9165
3rd Qu.:0.9577 3rd Qu.:0.9663 3rd Qu.:0.9664 3rd Qu.:0.9606
Max. :0.9855 Max. :0.9895 Max. :0.9883 Max. :0.9875
V25
Min. :0.2032
1st Qu.:0.9352
Median :0.9559
Mean :0.9300
3rd Qu.:0.9664
Max. :0.9846
V1 V2 V3 V4
Min. :0.0947 Min. :0.8252 Min. :0.6955 Min. :0.8246
1st Qu.:0.9116 1st Qu.:0.9933 1st Qu.:0.9712 1st Qu.:0.9934
Median :0.9430 Median :0.9957 Median :0.9791 Median :0.9957
Mean :0.9147 Mean :0.9930 Mean :0.9697 Mean :0.9929
3rd Qu.:0.9565 3rd Qu.:0.9967 3rd Qu.:0.9844 3rd Qu.:0.9966
Max. :0.9997 Max. :1.0000 Max. :0.9999 Max. :1.0000
V5 V6 V7 V8
Min. :0.8203 Min. :0.8258 Min. :0.8232 Min. :0.8253
1st Qu.:0.9868 1st Qu.:0.9933 1st Qu.:0.9934 1st Qu.:0.9932
Median :0.9915 Median :0.9958 Median :0.9957 Median :0.9957
Mean :0.9872 Mean :0.9929 Mean :0.9929 Mean :0.9929
3rd Qu.:0.9934 3rd Qu.:0.9967 3rd Qu.:0.9966 3rd Qu.:0.9966
Max. :1.0000 Max. :1.0000 Max. :1.0000 Max. :1.0000
V9 V10 V11 V12
Min. :0.8253 Min. :0.8253 Min. :0.8242 Min. :0.7842
1st Qu.:0.9932 1st Qu.:0.9934 1st Qu.:0.9933 1st Qu.:0.9909
Median :0.9957 Median :0.9957 Median :0.9958 Median :0.9939
Mean :0.9929 Mean :0.9929 Mean :0.9930 Mean :0.9904
3rd Qu.:0.9966 3rd Qu.:0.9967 3rd Qu.:0.9967 3rd Qu.:0.9953
Max. :1.0000 Max. :1.0000 Max. :1.0000 Max. :1.0000
V13 V14 V15 V16
Min. :0.07464 Min. :0.8269 Min. :0.8268 Min. :0.8209
1st Qu.:0.96251 1st Qu.:0.9934 1st Qu.:0.9934 1st Qu.:0.9892
Median :0.97618 Median :0.9958 Median :0.9957 Median :0.9929
Mean :0.96119 Mean :0.9930 Mean :0.9929 Mean :0.9895
3rd Qu.:0.98213 3rd Qu.:0.9966 3rd Qu.:0.9967 3rd Qu.:0.9945
Max. :0.99993 Max. :1.0000 Max. :1.0000 Max. :1.0000
V17 V18 V19 V20
Min. :0.8242 Min. :0.8260 Min. :0.8254 Min. :0.8249
1st Qu.:0.9934 1st Qu.:0.9933 1st Qu.:0.9934 1st Qu.:0.9934
Median :0.9957 Median :0.9958 Median :0.9957 Median :0.9957
Mean :0.9929 Mean :0.9930 Mean :0.9929 Mean :0.9930
3rd Qu.:0.9966 3rd Qu.:0.9966 3rd Qu.:0.9966 3rd Qu.:0.9967
Max. :1.0000 Max. :1.0000 Max. :1.0000 Max. :1.0000
V21 V22 V23 V24
Min. :0.002521 Min. :0.8237 Min. :0.8254 Min. :0.8251
1st Qu.:0.973019 1st Qu.:0.9933 1st Qu.:0.9933 1st Qu.:0.9935
Median :0.981149 Median :0.9957 Median :0.9957 Median :0.9957
Mean :0.966744 Mean :0.9929 Mean :0.9929 Mean :0.9930
3rd Qu.:0.986189 3rd Qu.:0.9967 3rd Qu.:0.9966 3rd Qu.:0.9966
Max. :0.996424 Max. :1.0000 Max. :1.0000 Max. :1.0000
V25
Min. :0.8245
1st Qu.:0.9933
Median :0.9958
Mean :0.9929
3rd Qu.:0.9967
Max. :1.0000
V1 V2 V3 V4
Min. :0.4998 Min. :0.008582 Min. :0.0923 Min. :0.5022
1st Qu.:0.9514 1st Qu.:0.939807 1st Qu.:0.9442 1st Qu.:0.9310
Median :0.9658 Median :0.959550 Median :0.9602 Median :0.9478
Mean :0.9524 Mean :0.942474 Mean :0.9460 Mean :0.9327
3rd Qu.:0.9730 3rd Qu.:0.968575 3rd Qu.:0.9701 3rd Qu.:0.9593
Max. :0.9996 Max. :0.994603 Max. :0.9997 Max. :0.9995
V5 V6 V7 V8
Min. :0.4986 Min. :0.5173 Min. :0.4560 Min. :0.4758
1st Qu.:0.9527 1st Qu.:0.9530 1st Qu.:0.9207 1st Qu.:0.9478
Median :0.9660 Median :0.9667 Median :0.9447 Median :0.9639
Mean :0.9530 Mean :0.9538 Mean :0.9260 Mean :0.9501
3rd Qu.:0.9722 3rd Qu.:0.9729 3rd Qu.:0.9562 3rd Qu.:0.9711
Max. :0.9997 Max. :0.9996 Max. :0.9995 Max. :0.9996
V9 V10 V11 V12
Min. :0.5261 Min. :0.3935 Min. :0.5017 Min. :0.5258
1st Qu.:0.9511 1st Qu.:0.9399 1st Qu.:0.9505 1st Qu.:0.9499
Median :0.9665 Median :0.9580 Median :0.9664 Median :0.9661
Mean :0.9530 Mean :0.9423 Mean :0.9528 Mean :0.9527
3rd Qu.:0.9729 3rd Qu.:0.9657 3rd Qu.:0.9726 3rd Qu.:0.9733
Max. :0.9997 Max. :0.9995 Max. :0.9997 Max. :0.9996
V13 V14 V15 V16
Min. :0.4916 Min. :0.4014 Min. :0.5037 Min. :0.5218
1st Qu.:0.9500 1st Qu.:0.9282 1st Qu.:0.9497 1st Qu.:0.9516
Median :0.9659 Median :0.9524 Median :0.9657 Median :0.9665
Mean :0.9528 Mean :0.9340 Mean :0.9525 Mean :0.9532
3rd Qu.:0.9729 3rd Qu.:0.9620 3rd Qu.:0.9727 3rd Qu.:0.9731
Max. :0.9996 Max. :0.9995 Max. :0.9996 Max. :0.9997
V17 V18 V19 V20
Min. :0.5375 Min. :0.5035 Min. :0.4859 Min. :0.5072
1st Qu.:0.9475 1st Qu.:0.9501 1st Qu.:0.9502 1st Qu.:0.9503
Median :0.9623 Median :0.9658 Median :0.9662 Median :0.9671
Mean :0.9499 Mean :0.9534 Mean :0.9526 Mean :0.9530
3rd Qu.:0.9712 3rd Qu.:0.9729 3rd Qu.:0.9731 3rd Qu.:0.9728
Max. :0.9997 Max. :0.9997 Max. :0.9996 Max. :0.9996
V21 V22 V23 V24
Min. :0.4975 Min. :0.4823 Min. :0.5121 Min. :0.5127
1st Qu.:0.9510 1st Qu.:0.9498 1st Qu.:0.9513 1st Qu.:0.9504
Median :0.9665 Median :0.9659 Median :0.9656 Median :0.9661
Mean :0.9532 Mean :0.9534 Mean :0.9526 Mean :0.9531
3rd Qu.:0.9733 3rd Qu.:0.9733 3rd Qu.:0.9731 3rd Qu.:0.9725
Max. :0.9997 Max. :0.9996 Max. :0.9996 Max. :0.9997
V25
Min. :0.5104
1st Qu.:0.9488
Median :0.9662
Mean :0.9530
3rd Qu.:0.9731
Max. :0.9996
V1 V2 V3 V4
Min. :0.7763 Min. :0.7834 Min. :0.7880 Min. :0.07595
1st Qu.:0.9137 1st Qu.:0.9197 1st Qu.:0.9167 1st Qu.:0.90631
Median :0.9351 Median :0.9378 Median :0.9354 Median :0.92710
Mean :0.9285 Mean :0.9319 Mean :0.9297 Mean :0.91690
3rd Qu.:0.9496 3rd Qu.:0.9510 3rd Qu.:0.9501 3rd Qu.:0.94315
Max. :0.9839 Max. :0.9819 Max. :0.9818 Max. :0.98285
V5 V6 V7 V8
Min. :0.7734 Min. :0.7562 Min. :0.2977 Min. :0.8238
1st Qu.:0.9110 1st Qu.:0.9143 1st Qu.:0.9129 1st Qu.:0.9162
Median :0.9321 Median :0.9352 Median :0.9337 Median :0.9361
Mean :0.9254 Mean :0.9281 Mean :0.9250 Mean :0.9304
3rd Qu.:0.9470 3rd Qu.:0.9498 3rd Qu.:0.9468 3rd Qu.:0.9503
Max. :0.9847 Max. :0.9816 Max. :0.9856 Max. :0.9850
V9 V10 V11 V12
Min. :0.8145 Min. :0.7682 Min. :0.7521 Min. :0.7331
1st Qu.:0.9169 1st Qu.:0.9143 1st Qu.:0.9162 1st Qu.:0.9187
Median :0.9367 Median :0.9363 Median :0.9349 Median :0.9348
Mean :0.9312 Mean :0.9287 Mean :0.9300 Mean :0.9303
3rd Qu.:0.9494 3rd Qu.:0.9495 3rd Qu.:0.9486 3rd Qu.:0.9499
Max. :0.9860 Max. :0.9856 Max. :0.9827 Max. :0.9855
V13 V14 V15 V16
Min. :0.4343 Min. :0.7583 Min. :0.4274 Min. :0.7685
1st Qu.:0.8873 1st Qu.:0.9167 1st Qu.:0.8618 1st Qu.:0.9177
Median :0.9143 Median :0.9362 Median :0.8942 Median :0.9364
Mean :0.8980 Mean :0.9300 Mean :0.8812 Mean :0.9298
3rd Qu.:0.9327 3rd Qu.:0.9502 3rd Qu.:0.9153 3rd Qu.:0.9497
Max. :0.9824 Max. :0.9858 Max. :0.9820 Max. :0.9860
V17 V18 V19 V20
Min. :0.7392 Min. :0.8013 Min. :0.8169 Min. :0.6024
1st Qu.:0.9170 1st Qu.:0.9184 1st Qu.:0.9173 1st Qu.:0.9069
Median :0.9351 Median :0.9360 Median :0.9372 Median :0.9309
Mean :0.9303 Mean :0.9305 Mean :0.9315 Mean :0.9223
3rd Qu.:0.9494 3rd Qu.:0.9499 3rd Qu.:0.9498 3rd Qu.:0.9452
Max. :0.9825 Max. :0.9842 Max. :0.9835 Max. :0.9826
V21 V22 V23 V24
Min. :0.7855 Min. :0.5693 Min. :0.7650 Min. :0.03912
1st Qu.:0.9183 1st Qu.:0.8637 1st Qu.:0.9188 1st Qu.:0.90507
Median :0.9345 Median :0.8996 Median :0.9364 Median :0.92537
Mean :0.9295 Mean :0.8830 Mean :0.9301 Mean :0.91677
3rd Qu.:0.9498 3rd Qu.:0.9224 3rd Qu.:0.9496 3rd Qu.:0.94356
Max. :0.9817 Max. :0.9803 Max. :0.9829 Max. :0.98370
V25
Min. :0.7727
1st Qu.:0.9164
Median :0.9347
Mean :0.9284
3rd Qu.:0.9482
Max. :0.9849
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] mashr_0.2.41 ashr_2.2-51 glmnet_3.0
[4] Matrix_1.2-18 mr.mash.alpha_0.2-6
loaded via a namespace (and not attached):
[1] foreach_1.4.4 RcppParallel_4.4.3 rmeta_3.0
[4] assertthat_0.2.1 horseshoe_0.2.0 mixsqp_0.3-43
[7] yaml_2.2.1 ebnm_0.1-35 pillar_1.4.2
[10] backports_1.1.4 lattice_0.20-38 quantreg_5.85
[13] glue_1.4.2 digest_0.6.20 promises_1.0.1
[16] colorspace_1.4-1 htmltools_0.3.6 httpuv_1.5.1
[19] plyr_1.8.6 conquer_1.0.2 pkgconfig_2.0.3
[22] invgamma_1.1 SparseM_1.81 purrr_0.3.4
[25] mvtnorm_1.1-1 scales_1.1.0 whisker_0.3-2
[28] later_0.8.0 MatrixModels_0.5-0 git2r_0.26.1
[31] tibble_2.1.3 ggplot2_3.2.1 lazyeval_0.2.2
[34] magrittr_1.5 crayon_1.4.1 mcmc_0.9-7
[37] evaluate_0.14 fs_1.3.1 MASS_7.3-51.4
[40] truncnorm_1.0-8 tools_3.6.1 doMC_1.3.7
[43] softImpute_1.4 lifecycle_1.0.0 matrixStats_0.58.0
[46] stringr_1.4.0 MCMCpack_1.5-0 GIGrvg_0.5
[49] trust_0.1-8 munsell_0.5.0 flashier_0.2.7
[52] MBSP_1.0 irlba_2.3.3 compiler_3.6.1
[55] rlang_0.4.10 grid_3.6.1 iterators_1.0.10
[58] rmarkdown_1.13 gtable_0.3.0 codetools_0.2-16
[61] abind_1.4-5 reshape2_1.4.3 flashr_0.6-7
[64] R6_2.5.0 knitr_1.23 dplyr_0.8.3
[67] workflowr_1.6.2 rprojroot_1.3-2 shape_1.4.4
[70] stringi_1.4.3 parallel_3.6.1 SQUAREM_2021.1
[73] Rcpp_1.0.6 tidyselect_1.1.0 xfun_0.8
[76] coda_0.19-4