Regime changes 2.0
Francisco Bischoff
on October 04, 2023
Last updated: 2023-10-04
Checks: 7 0
Knit directory:
develop/docs/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20201020) was run prior to running the code in the R Markdown file.
Setting a seed ensures that any results that rely on randomness, e.g.
subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d34d2ab. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the
analysis have been committed to Git prior to generating the results (you can
use wflow_publish or wflow_git_commit). workflowr only
checks the R Markdown file, but you know if there are other scripts or data
files that it depends on. Below is the status of the Git repository when the
results were generated:
Ignored files:
Ignored: .Renviron
Ignored: .Rhistory
Ignored: .docker/
Ignored: .luarc.json
Ignored: analysis/regime_optimize_2.knit.md
Ignored: dev/
Ignored: inst/extdata/
Ignored: output/dbarts_fitted.bak.rds
Ignored: output/dbarts_fitted2.bak.rds
Ignored: output/dbarts_fitted2_lmk.bak.rds
Ignored: output/dbarts_fitted_lmk.bak.rds
Ignored: output/dbarts_fitted_mvds.bak.rds
Ignored: output/dbarts_fitted_vtds.bak.rds
Ignored: output/importances.bak.rds
Ignored: output/importances2.bak.rds
Ignored: output/importances2_lmk.bak.rds
Ignored: output/importances_lmk.bak.rds
Ignored: output/importances_mvds.bak.rds
Ignored: output/importances_vtds.bak.rds
Ignored: renv/staging/
Ignored: tmp/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made
to the R Markdown (analysis/regime_optimize_2.Rmd) and HTML (docs/regime_optimize_2.html)
files. If you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table below to
view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d34d2ab | Francisco Bischoff | 2023-10-04 | revised floss score |
| Rmd | ca252b0 | Francisco Bischoff | 2023-09-08 | main changes |
| html | d06c64f | Francisco Bischoff | 2022-10-09 | Squashed commit of the following: |
| html | f9f551d | Francisco Bischoff | 2022-10-06 | Build site. |
| Rmd | 6b7a6c7 | Francisco Bischoff | 2022-10-06 | PR done |
| Rmd | dbbd1d6 | Francisco Bischoff | 2022-08-22 | Squashed commit of the following: |
| html | dbbd1d6 | Francisco Bischoff | 2022-08-22 | Squashed commit of the following: |
| Rmd | de21180 | Francisco Bischoff | 2022-08-21 | Squashed commit of the following: |
| html | de21180 | Francisco Bischoff | 2022-08-21 | Squashed commit of the following: |
1 Regime changes optimization (continuation)
Following the previous article, this article aims to complete the analysis of the regime change optimization. Here we add the regime_landmark parameter to the optimization.
1.1 Current pipeline
Figure 1.1: FLOSS pipeline.
1.2 Tuning process
As we have seen previously, the FLOSS algorithm is built on top of the Matrix Profile (MP). Thus, we have proposed several parameters that may or not impact the FLOSS prediction performance.
The variables for building the MP are:
mp_threshold: the minimum similarity value to be considered for 1-NN.time_constraint: the maximum distance to look for the nearest neighbor.window_size: the default parameter always used to build an MP.
Later, the FLOSS algorithm also has a parameter that needs tuning to optimize the prediction:
regime_threshold: the threshold below which a regime change is considered.regime_landmark: the point in time where the regime threshold is applied.
Using the tidymodels framework, we performed a basic grid search on all these parameters, now limiting the exploration
on the MP and focusing on the FLOSS parameters.
The workflow is as follows:
- From the original set of 229 records, a subset of 137 records was selected for the grid search.
- The MP parameters were explored using the following values:
mp_threshold: 0.0, 0.4, 0.6 and 0.8;time_constraint: 0, 800 and 1500;window_size: 25, 50, 75, 100, 125 and 150;
- The FLOSS parameters were explored using the following values:
regime_threshold: 0.05 to 0.90, by 0.05 steps;regime_landmark: 1 to 10, by 0.5 steps.
The results were then combined with the previous optimization and deduplicated.
1.3 Parameters analysis
As before, we started by computing the importance of each parameter1. We used the same approach using the Bayesian Additive Regression Trees (BART) model to fit the tuning parameters as predictors of the FLOSS score.
1.3.1 Interactions
Before starting the parameter importance analysis, we need to consider the parameter interactions since this is usually the weak spot of the analysis techniques.
The first BART model was fitted using the following parameters:
\[\begin{equation} \begin{aligned} E( score ) &= \alpha + time\_constraint\\ &\quad + mp\_threshold + window\_size\\ &\quad + regime\_threshold + regime\_landmark \end{aligned} \tag{1.1} \end{equation}\]
After checking the interactions, this is the refitted model:
\[\begin{equation} \begin{aligned} E( score ) &= \alpha + time\_constraint\\ &\quad + mp\_threshold + window\_size\\ &\quad + regime\_threshold + regime\_landmark\\ &\quad + \left(regime\_threshold \times regime\_landmark\right)\\ &\quad + \left(mp\_threshold \times regime\_landmark\right)\\ &\quad + \left(mp\_threshold \times window\_size\right) \end{aligned} \tag{1.2} \end{equation}\]
Fig. 1.2 shows the variable interaction strength between pairs of variables. That allows us to
verify if there are any significant interactions between the variables. Using the information from the first model fit,
equation (1.1), we see that regime_threshold interacts strongly with regime_landmark. This interaction
was already expected, and we see that even after refitting the model, equation (1.2), this interaction is
still strong.
This is not a problem per se but a signal we must be aware of when exploring the parameters.
Figure 1.2: Variable interactions strength using feature importance ranking measure (FIRM) approach2. A) Shows strong interaction between regime_threshold and regime_landmark, mp_threshold and window_size, mp_threshold and regime_landmark. B) Refitting the model with these interactions taken into account, the strength is substantially reduced, except for the first, showing that indeed there is a strong correlation between those variables.
1.3.2 Importance
After evaluating the interactions, we can then perform the analysis of the variable importance. The goal is to understand how the FLOSS score behaves when we change the parameters.
The techniques for evaluating the variable importances were described in the previous article.
1.3.3 Importance analysis
Using the three techniques simultaneously allows a broad comparison of the model behavior3. All three methods are model-agnostic (separates interpretation from the model), but as we have seen, each method has its advantages and disadvantages4.
Fig. 1.3 then shows the variable importance using three methods: Feature Importance Ranking Measure
(FIRM) using Individual Conditional Expectation (ICE), Permutation-based, and Shapley Additive explanations (SHAP). The
first line of this figure shows an interesting result that probably comes from the main disadvantage of the FIRM method:
the method does not take into account interactions. We see that FIRM is the only one that disagrees with the other
two methods, giving much importance to window_size.
In the second line, taking into account the interactions, we see that all methods somewhat agree with each other,
accentuating the importance of regime_threshold, which makes sense as it is the most evident parameter we need to set
to determine if the Arc Counts are low enough to indicate a regime change.
Figure 1.3: Variables importances using three different methods. A) Feature Importance Ranking Measure using ICE curves. B) Permutation method. C) SHAP (400 iterations). Line 1 refers to the original fit, and line 2 to the re-fit, taking into account the interactions between variables (Fig. 1.2).
Fig. 1.4 and 1.5 show the effect of each feature on the FLOSS score. The
more evident difference is the shape of the effect of time_constraint that initially suggested better results with
larger values. However, removing the interactions seems to be a flat line.
Based on Figures 1.3 and 1.5 we can infer that:
regime_threshold: is the most important feature, has an optimal value to be set, and since the high interaction with theregime_landmark, both must be tuned simultaneously. In this setting, high thresholds significantly impact the score, probably due to an increase in false positives starting on >0.65 the overall impact is mostly negative.regime_landmark: is not as important as theregime_threshold,but since there is a high interaction, it must not be underestimated. It is known that the Arc Counts have more uncertainty as we approach the margin of the streaming, and this becomes evident looking at how the score is negatively affected for values below 3.5s.window_size: has a near zero impact on the score when correctly set. Nevertheless, for higher window values, the score is negatively affected. This high value probably depends on the data domain. In this setting, the model is being tuned towards the changes from atrial fibrillation/non-fibrillation; thus, the “shape of interest” is small compared to the whole heartbeat waveform. Window sizes smaller than 150 are more suitable in this case. As Beyer et al. noted, “as dimensionality increases, the distance to the nearest data point approaches the distance to the farthest data point”5, which means that the bigger the window size, the smaller will be the contrast between different regimes.mp_threshold: has a fair impact on the score, but primarily by not using it. We start to see a negative impact on the score with values above 0.60, while a constant positive impact with lower values.time_constraint: is a parameter that must be interpreted cautiously. The 0 (zero) value means no constraint, which is equivalent to the size of the FLOSS history buffer (in our setting, 5000). We can see that this parameter’s impact throughout the possible values is constantly near zero.
In short, for the MP computation, the parameter that is worth tuning is the window_size, while for the FLOSS
computation, both regime_threshold (mainly) and regime_landmark shall be tuned.
Figure 1.4: This shows the effect each variable has on the FLOSS score. This plot doesn’t take into account the variable interactions.
Figure 1.5: This shows the effect each variable has on the FLOSS score, taking into account the interactions.
According to the FLOSS paper6, the window_size is indeed a feature that can be tuned; nevertheless,
the results appear to be similar in a reasonably wide range of window sizes, up to a limit, consistent with our findings.
1.4 Visualizing the predictions
At this point, the grid search tested a total of 23,389 models with resulting (individual) scores from 0.0002 to 1669.83 (Q25: 0.9838, Q50: 1.8093, Q75: 3.3890).
1.4.1 By recording
First, we will visualize how the models (in general) performed throughout the individual recordings.
Fig. 1.6 shows a violin plot of equal areas clipped to the minimum value. The blue color indicates the recordings with a small IQR (interquartile range) of model scores. We see on the left half 10% of the recordings with the worst minimum score, and on the right half, 10% of the recordings with the best minimum score.
Next, we will visualize some of these predictions to understand why some recordings were difficult to segment. For us to have a simple baseline: a recording with just one regime change, and the model predicts exactly one regime change, but far from the truth, the score will be roughly 1.
Figure 1.6: Violin plot showing the distribution of the FLOSS score achieved by all tested models by recording. The left half shows the recordings that were difficult to predict (10% overall), whereas the right half shows the recordings that at least one model could achieve a good prediction (10% overall). The recordings are sorted (left-right) by the minimum (best) score achieved in descending order, and ties are sorted by the median of all recording scores. The blue color highlights recordings where models had an IQR variability of less than one. As a simple example, a recording with just one regime change, and the model predicts exactly one change, far from the truth, the score will be roughly 1.
Fig. 1.7 shows the best effort in predicting the most complex recordings. One information not declared before is that if the model does not predict any change, it will put a mark on the zero position. On the other side, the truth markers positioned at the beginning and the end of the recording were removed, as these locations lack information and do not represent a streaming setting. Compared to the corresponding figure in the previous article, we can see that even the most complex recordings had better predictions.
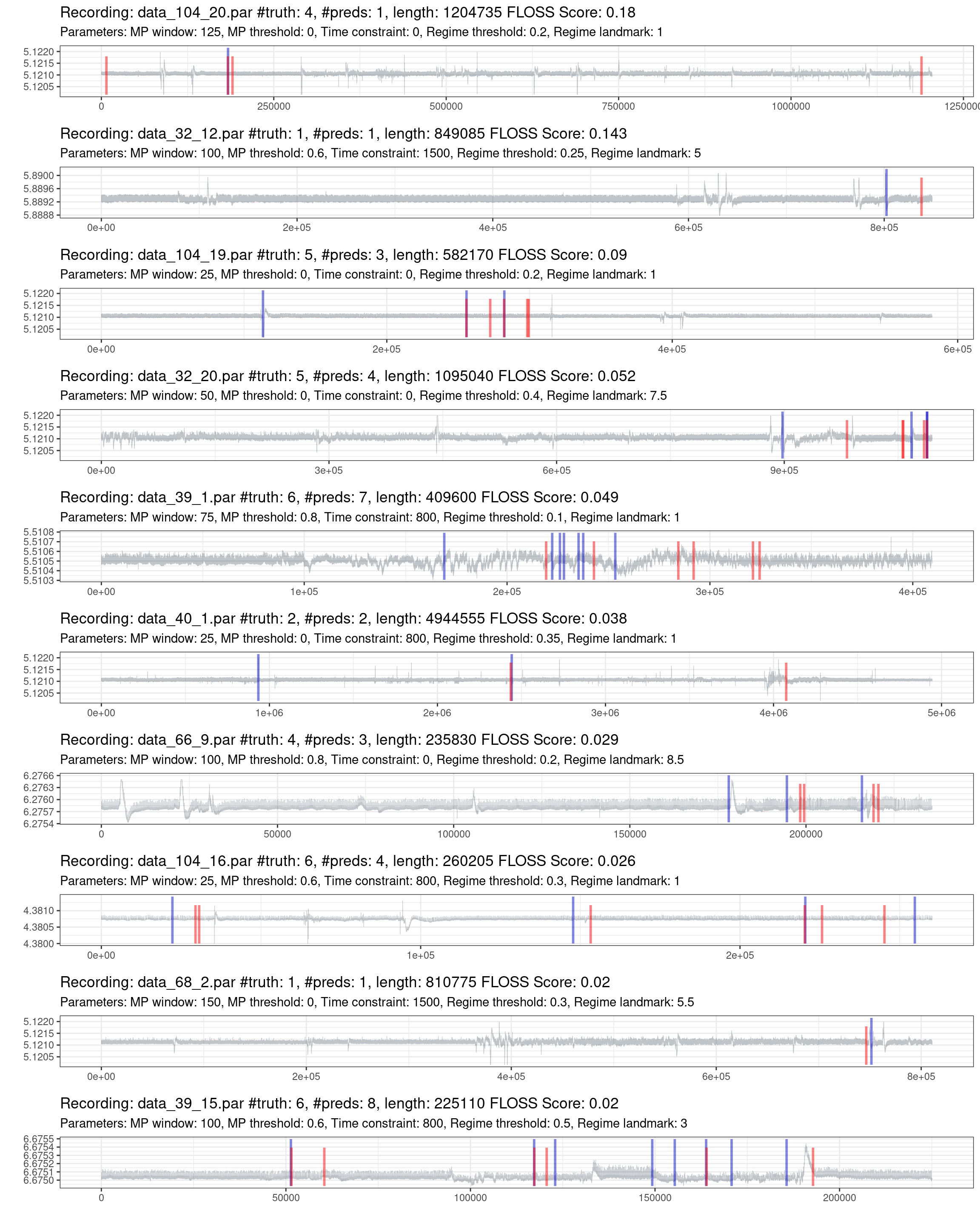
Figure 1.7: Prediction of the worst 10% of recordings (red is the truth, blue are the predictions).
Fig. 1.8 shows the best performances of the best recordings. Notice that there are recordings with a significant duration and few regime changes, making it hard for a “trivial model” to predict randomly.

Figure 1.8: Prediction of the best 10% of recordings (red is the truth, blue are the predictions).
An online interactive version of all the datasets and predictions can be accessed at Shiny app.
1.4.2 By model
Fig. 1.9 shows the distribution of the FLOSS score of the 10% worst (left side) and 10% best models across the recordings (right side). The bluish color highlights the models with SD below 3 and IQR below 1.
Here again, we can compare with the previous article and see an improvement in the performance, as the models present lower SD and IQR.
Figure 1.9: Violin plot showing the distribution of the FLOSS score achieved by all tested models during the inner ressample. The left half shows the models with the worst performances (10% overall), whereas the right half shows the models with the best performances (10% overall). The models are sorted (left-right) by the mean score (top) and by the median (below). Ties are sorted by the SD and IQR, respectively. The bluish colors highlights models with an SD below 3 and IQR below 1.
Fig. 1.10 the performance of the six best models. They are ordered from left to right, from the worst record to the best record. The top model is the one with the lowest mean across the scores. The blue line indicates the mean score, and the red line the median score. The scores above 3 are squished in the plot and colored according to the scale in the legend. Notice the improvement on the blue and red lines compared to the previous article.
Figure 1.10: Performances of the best 6 models across all inner resample of recordings. The recordings are ordered by score, from the worst to the best. Each plot shows one model, starting from the best one. The red line indicates the median score of the model. The blue line indicates the mean score of the model. The gray line limits the zero-score region. The plot is limited on the “y” axis, and the scores above this limit are shown in color.
We can see that some records (namely #19, #41, #93, #100, #107) are contained in the set of “difficult” records shown in Fig. 1.6.
2 Current status
The current status of the project shows that FLOSS is up to the task of signaling possible regime changes.
After introducing the regime_landmark feature, the performance improves significantly, and we can narrow down the tuning space to a small number of parameters.
In parallel, another score measure is being developed based on the concept of Precision and Recall, but for time-series7. It is expected that such a score measure will help to choose the best final model where most of the significant regime changes are detected, keeping a reasonable amount of false positives that will be ruled out further by the classification algorithm.
Further evaluation will be performed in more datasets; later, the results will be presented here.
References
─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.1 (2023-06-16)
os Ubuntu 22.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Europe/Lisbon
date 2023-10-04
pandoc 2.17.0.1 @ /usr/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
askpass 1.1 2019-01-13 [1] CRAN (R 4.3.0)
backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.1)
base64url 1.4 2018-05-14 [1] CRAN (R 4.3.0)
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
bookdown 0.35.1 2023-08-13 [1] Github (rstudio/bookdown@661567e)
bslib 0.5.1 2023-08-11 [1] CRAN (R 4.3.1)
cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
callr 3.7.3 2022-11-02 [1] CRAN (R 4.3.1)
checkmate 2.2.0 2023-04-27 [1] CRAN (R 4.3.0)
class 7.3-22 2023-05-03 [2] CRAN (R 4.3.1)
cli 3.6.1 2023-03-23 [1] CRAN (R 4.3.1)
codetools 0.2-19 2023-02-01 [2] CRAN (R 4.3.0)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.1)
credentials 1.3.2 2021-11-29 [1] CRAN (R 4.3.0)
data.table 1.14.8 2023-02-17 [1] CRAN (R 4.3.0)
dbarts 0.9-23 2023-01-23 [1] CRAN (R 4.3.0)
debugme 1.1.0 2017-10-22 [1] CRAN (R 4.3.0)
devtools 2.4.5 2022-10-11 [1] CRAN (R 4.3.0)
dials 1.2.0 2023-04-03 [1] CRAN (R 4.3.0)
DiceDesign 1.9 2021-02-13 [1] CRAN (R 4.3.0)
digest 0.6.33 2023-07-07 [1] CRAN (R 4.3.1)
dplyr 1.1.3 2023-09-03 [1] CRAN (R 4.3.1)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
evaluate 0.21 2023-05-05 [1] CRAN (R 4.3.0)
fansi 1.0.4 2023-01-22 [1] CRAN (R 4.3.0)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
fastshap 0.0.7 2021-12-06 [1] CRAN (R 4.3.0)
forcats 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
foreach 1.5.2 2022-02-02 [1] CRAN (R 4.3.0)
fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.1)
furrr 0.3.1 2022-08-15 [1] CRAN (R 4.3.0)
future 1.33.0 2023-07-01 [1] CRAN (R 4.3.1)
future.apply 1.11.0 2023-05-21 [1] CRAN (R 4.3.1)
future.callr 0.8.2 2023-08-09 [1] CRAN (R 4.3.1)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
gert 1.9.3 2023-08-07 [1] CRAN (R 4.3.1)
getPass 0.2-2 2017-07-21 [1] CRAN (R 4.3.0)
ggplot2 * 3.4.3 2023-08-14 [1] CRAN (R 4.3.1)
git2r 0.32.0.9000 2023-06-30 [1] Github (ropensci/git2r@9c42d41)
gittargets * 0.0.6.9000 2023-05-05 [1] Github (wlandau/gittargets@2d448ff)
globals 0.16.2 2022-11-21 [1] CRAN (R 4.3.0)
glue * 1.6.2 2022-02-24 [1] CRAN (R 4.3.1)
gower 1.0.1 2022-12-22 [1] CRAN (R 4.3.0)
GPfit 1.0-8 2019-02-08 [1] CRAN (R 4.3.0)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.0)
gtable 0.3.3 2023-03-21 [1] CRAN (R 4.3.0)
hardhat 1.3.0 2023-03-30 [1] CRAN (R 4.3.0)
here * 1.0.1 2020-12-13 [1] CRAN (R 4.3.0)
highr 0.10 2022-12-22 [1] CRAN (R 4.3.1)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
htmltools 0.5.6 2023-08-10 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.2 2023-03-17 [1] CRAN (R 4.3.0)
httpuv 1.6.11 2023-05-11 [1] CRAN (R 4.3.1)
httr 1.4.6 2023-05-08 [1] CRAN (R 4.3.1)
igraph 1.5.1 2023-08-10 [1] CRAN (R 4.3.1)
ipred 0.9-14 2023-03-09 [1] CRAN (R 4.3.0)
iterators 1.0.14 2022-02-05 [1] CRAN (R 4.3.0)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
jsonlite 1.8.7 2023-06-29 [1] CRAN (R 4.3.0)
kableExtra * 1.3.4 2021-02-20 [1] CRAN (R 4.3.0)
knitr 1.43 2023-05-25 [1] CRAN (R 4.3.0)
labeling 0.4.2 2020-10-20 [1] CRAN (R 4.3.0)
later 1.3.1 2023-05-02 [1] CRAN (R 4.3.1)
lattice 0.21-8 2023-04-05 [2] CRAN (R 4.3.0)
lava 1.7.2.1 2023-02-27 [1] CRAN (R 4.3.0)
lhs 1.1.6 2022-12-17 [1] CRAN (R 4.3.0)
lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.3.1)
listenv 0.9.0 2022-12-16 [1] CRAN (R 4.3.0)
lubridate 1.9.2 2023-02-10 [1] CRAN (R 4.3.0)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.1)
MASS 7.3-60 2023-05-04 [2] CRAN (R 4.3.1)
Matrix 1.6-0 2023-07-08 [2] CRAN (R 4.3.1)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
mgcv 1.9-0 2023-07-11 [2] CRAN (R 4.3.1)
mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.0)
modelenv 0.1.1 2023-03-08 [1] CRAN (R 4.3.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
nlme 3.1-163 2023-08-09 [1] CRAN (R 4.3.1)
nnet 7.3-19 2023-05-03 [2] CRAN (R 4.3.1)
openssl 2.1.0 2023-07-15 [1] CRAN (R 4.3.1)
parallelly 1.36.0 2023-05-26 [1] CRAN (R 4.3.1)
parsnip 1.1.0 2023-04-12 [1] CRAN (R 4.3.0)
patchwork * 1.1.2 2022-08-19 [1] CRAN (R 4.3.0)
pdp 0.8.1 2023-06-22 [1] Github (bgreenwell/pdp@4f22141)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgbuild 1.4.2 2023-06-26 [1] CRAN (R 4.3.1)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
pkgload 1.3.2.1 2023-07-08 [1] CRAN (R 4.3.1)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.3.0)
processx 3.8.2 2023-06-30 [1] CRAN (R 4.3.1)
prodlim 2023.03.31 2023-04-02 [1] CRAN (R 4.3.0)
profvis 0.3.8 2023-05-02 [1] CRAN (R 4.3.1)
promises 1.2.1 2023-08-10 [1] CRAN (R 4.3.1)
ps 1.7.5 2023-04-18 [1] CRAN (R 4.3.1)
purrr 1.0.2 2023-08-10 [1] CRAN (R 4.3.1)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.1)
Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.3.1)
readr 2.1.4 2023-02-10 [1] CRAN (R 4.3.0)
recipes 1.0.7 2023-08-10 [1] CRAN (R 4.3.1)
remotes 2.4.2.1 2023-07-18 [1] CRAN (R 4.3.1)
renv 0.17.3 2023-04-06 [1] CRAN (R 4.3.1)
rlang 1.1.1 2023-04-28 [1] CRAN (R 4.3.0)
rmarkdown 2.23.4 2023-08-13 [1] Github (rstudio/rmarkdown@054d735)
rpart 4.1.19 2022-10-21 [2] CRAN (R 4.3.0)
rprojroot 2.0.3 2022-04-02 [1] CRAN (R 4.3.1)
rsample 1.1.1 2022-12-07 [1] CRAN (R 4.3.0)
rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.1)
rvest 1.0.3 2022-08-19 [1] CRAN (R 4.3.0)
sass 0.4.7 2023-07-15 [1] CRAN (R 4.3.1)
scales 1.2.1 2022-08-20 [1] CRAN (R 4.3.0)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
shapviz 0.9.1 2023-07-18 [1] CRAN (R 4.3.1)
shiny 1.7.5 2023-08-12 [1] CRAN (R 4.3.1)
signal 0.7-7 2021-05-25 [1] CRAN (R 4.3.0)
stringi 1.7.12 2023-01-11 [1] CRAN (R 4.3.1)
stringr 1.5.0 2022-12-02 [1] CRAN (R 4.3.1)
survival 3.5-5 2023-03-12 [2] CRAN (R 4.3.1)
svglite 2.1.1.9000 2023-05-05 [1] Github (r-lib/svglite@6c1d359)
sys 3.4.2 2023-05-23 [1] CRAN (R 4.3.1)
systemfonts 1.0.4 2022-02-11 [1] CRAN (R 4.3.0)
tarchetypes * 0.7.7 2023-06-15 [1] CRAN (R 4.3.1)
targets * 1.2.2 2023-08-10 [1] CRAN (R 4.3.1)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidyr 1.3.0 2023-01-24 [1] CRAN (R 4.3.0)
tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
timechange 0.2.0 2023-01-11 [1] CRAN (R 4.3.0)
timeDate 4022.108 2023-01-07 [1] CRAN (R 4.3.0)
timetk 2.8.3 2023-03-30 [1] CRAN (R 4.3.0)
tune 1.1.1 2023-04-11 [1] CRAN (R 4.3.0)
tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.1)
urlchecker 1.0.1 2021-11-30 [1] CRAN (R 4.3.0)
usethis 2.2.2.9000 2023-07-17 [1] Github (r-lib/usethis@467ff57)
utf8 1.2.3 2023-01-31 [1] CRAN (R 4.3.0)
uuid 1.1-0 2022-04-19 [1] CRAN (R 4.3.0)
vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.3.1)
vip 0.3.2 2020-12-17 [1] CRAN (R 4.3.0)
viridisLite 0.4.2 2023-05-02 [1] CRAN (R 4.3.1)
visNetwork * 2.1.2 2022-09-29 [1] CRAN (R 4.3.0)
vroom 1.6.3 2023-04-28 [1] CRAN (R 4.3.1)
webshot 0.5.5 2023-06-26 [1] CRAN (R 4.3.1)
whisker 0.4.1 2022-12-05 [1] CRAN (R 4.3.0)
withr 2.5.0 2022-03-03 [1] CRAN (R 4.3.1)
workflowr * 1.7.0 2021-12-21 [1] CRAN (R 4.3.0)
workflows 1.1.3 2023-02-22 [1] CRAN (R 4.3.0)
xfun 0.40 2023-08-09 [1] CRAN (R 4.3.1)
xgboost 1.7.5.1 2023-03-30 [1] CRAN (R 4.3.0)
xml2 1.3.5 2023-07-06 [1] CRAN (R 4.3.1)
xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
xts 0.13.1 2023-04-16 [1] CRAN (R 4.3.0)
yaml 2.3.7 2023-01-23 [1] CRAN (R 4.3.1)
yardstick 1.0.0.9000 2023-05-25 [1] Github (tidymodels/yardstick@90ab794)
zoo 1.8-12 2023-04-13 [1] CRAN (R 4.3.0)
[1] /workspace/.cache/R/renv/proj_libs/develop-e2b961e1/R-4.3/x86_64-pc-linux-gnu
[2] /usr/lib/R/library
──────────────────────────────────────────────────────────────────────────────