Running the MCMC
Last updated: 2020-04-24
Checks: 6 1
Knit directory: lieb/
This reproducible R Markdown analysis was created with workflowr (version 1.6.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190717) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 717e762. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/.Rhistory
Ignored: analysis/pairwise_fitting_cache/
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Ignored: output/chip_shiny/.DS_Store
Ignored: output/chip_shiny/Data/.DS_Store
Ignored: output/vision_shiny/.DS_Store
Ignored: output/vision_shiny/.Rhistory
Ignored: output/vision_shiny/Data/.DS_Store
Unstaged changes:
Modified: analysis/pairwise_fitting.Rmd
Modified: analysis/preprocessing.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/running_mcmc.Rmd) and HTML (docs/running_mcmc.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 828e725 | hillarykoch | 2020-04-23 | add a preprocessing section |
| html | c5fd6fc | hillarykoch | 2020-04-23 | change nav bar to accomodate a menu |
| html | 5d5c3dd | hillarykoch | 2020-03-07 | Build site. |
| Rmd | 2a75de4 | hillarykoch | 2020-02-20 | remove too-big header |
| html | 2a75de4 | hillarykoch | 2020-02-20 | remove too-big header |
| html | 3a9bf9d | hillarykoch | 2020-02-20 | Build site. |
| html | 72e6fec | hillarykoch | 2020-02-20 | Build site. |
| Rmd | 1c27b8e | hillarykoch | 2020-02-20 | add description of mcmc output |
| html | 1c27b8e | hillarykoch | 2020-02-20 | add description of mcmc output |
| Rmd | 862bc02 | hillarykoch | 2020-02-07 | big updates for mcmc processors |
| html | 862bc02 | hillarykoch | 2020-02-07 | big updates for mcmc processors |
| html | 8b3e556 | hillarykoch | 2019-12-03 | Build site. |
| html | 39383ac | hillarykoch | 2019-12-03 | Build site. |
| Rmd | 645d408 | hillarykoch | 2019-12-03 | workflowr::wflow_publish(files = “*“) |
| html | e467a51 | hillarykoch | 2019-08-16 | different shiny location |
| html | d58e1a6 | hillarykoch | 2019-08-16 | resource data |
| html | 38fb1c0 | hillarykoch | 2019-08-16 | edit shiny |
| html | d6d897c | hillarykoch | 2019-07-18 | reorder mcmc section |
| Rmd | 330ef60 | hillarykoch | 2019-07-18 | reorder mcmc section |
| html | 330ef60 | hillarykoch | 2019-07-18 | reorder mcmc section |
| Rmd | a8d87b4 | hillarykoch | 2019-07-18 | updat mcmc section |
| html | a8d87b4 | hillarykoch | 2019-07-18 | updat mcmc section |
| Rmd | 4991356 | hillarykoch | 2019-07-18 | updat mcmc section |
| html | 4991356 | hillarykoch | 2019-07-18 | updat mcmc section |
| html | 228123d | hillarykoch | 2019-07-17 | update up to obtaining the hyperparameters |
| html | c1dc0c1 | hillarykoch | 2019-07-17 | update up to obtaining the hyperparameters |
| html | 674120c | hillarykoch | 2019-07-17 | update about page |
| html | e67a3a2 | hillarykoch | 2019-07-17 | update about page |
| html | 2faf446 | hillarykoch | 2019-07-17 | update about page |
| html | 2177dfa | hillarykoch | 2019-07-17 | update about page |
| html | 8b30694 | hillarykoch | 2019-07-17 | update about page |
| html | e36b267 | hillarykoch | 2019-07-17 | update about page |
| html | a991668 | hillarykoch | 2019-07-17 | update about page |
| html | a36d893 | hillarykoch | 2019-07-17 | update about page |
| html | e8e54b7 | hillarykoch | 2019-07-17 | update about page |
| html | f47c013 | hillarykoch | 2019-07-17 | update about page |
| Rmd | 50cf23e | hillarykoch | 2019-07-17 | make skeleton |
| html | 50cf23e | hillarykoch | 2019-07-17 | make skeleton |
—This is likely the most time-consuming step of CLIMB analysis—
The final step of CLIMB involves doing inference on the parsimonious Gaussian mixture using MCMC. MCMC is an iterative method, and thus the user needs to specify how many iterations to use. We recommend running a quick pilot analysis–say, for 10 iterations. This pilot analysis will give a good idea of how long an analysis will need to run for a given larger number of iterations (say, 20,000 iterations).
You can run an mcmc simply with the function run_mcmc(). This function calls a script written in Julia, and executes everything at the default settings in the CLIMB methodology. The user needs to provide 4 arguments:
dat: the input data you’ve been using throughout the analysishyp: the hyperparameter values estimated in the previous stepnstep: number of MCMC iterations to runretained_classes: the parsimonious list of candidate latent classes, after finally filtering out by prior weights as done in the previous step
First, we load in our data, list of candidate latent classes, and estimated hyperparameters.
data("sim")
load("output/hyperparameters.Rdata")
retained_classes <- readr::read_tsv("output/retained_classes.txt", col_names = FALSE)Now we are ready to launch an MCMC:
results <- run_mcmc(sim$data, hyp = hyp, nstep = 100, retained_classes = retained_classes)
out <- extract_chains(results)Running the MCMC...100%|████████████████████████████████| Time: 0:00:33The object results contains 3 objects:
chain: the estimate parameters over the course ofnstepiterationsacceptane_rate_chain: an \(M\times\)nstepmatrix of the acceptance rates for each cluster covariance. The proposals for each cluster are adaptively tuned such that the acceptance rates converge to about 0.3tune_df_chain: the tuning degrees of freedom across the chain, adjusted to yield optimal acceptance rates
results is effectively a Julia object, so the first thing you should do with this object is to extract the data for R’s use:
out will contain all the different chains from the MCMC. For example, you can check the MCMC trace plots. Here is the trace plot of the mean from the first cluster in the third dimension:
plot(out$mu_chains[[1]][,3], type = "l", xlab = "iteration", ylab = expression(mu[3]))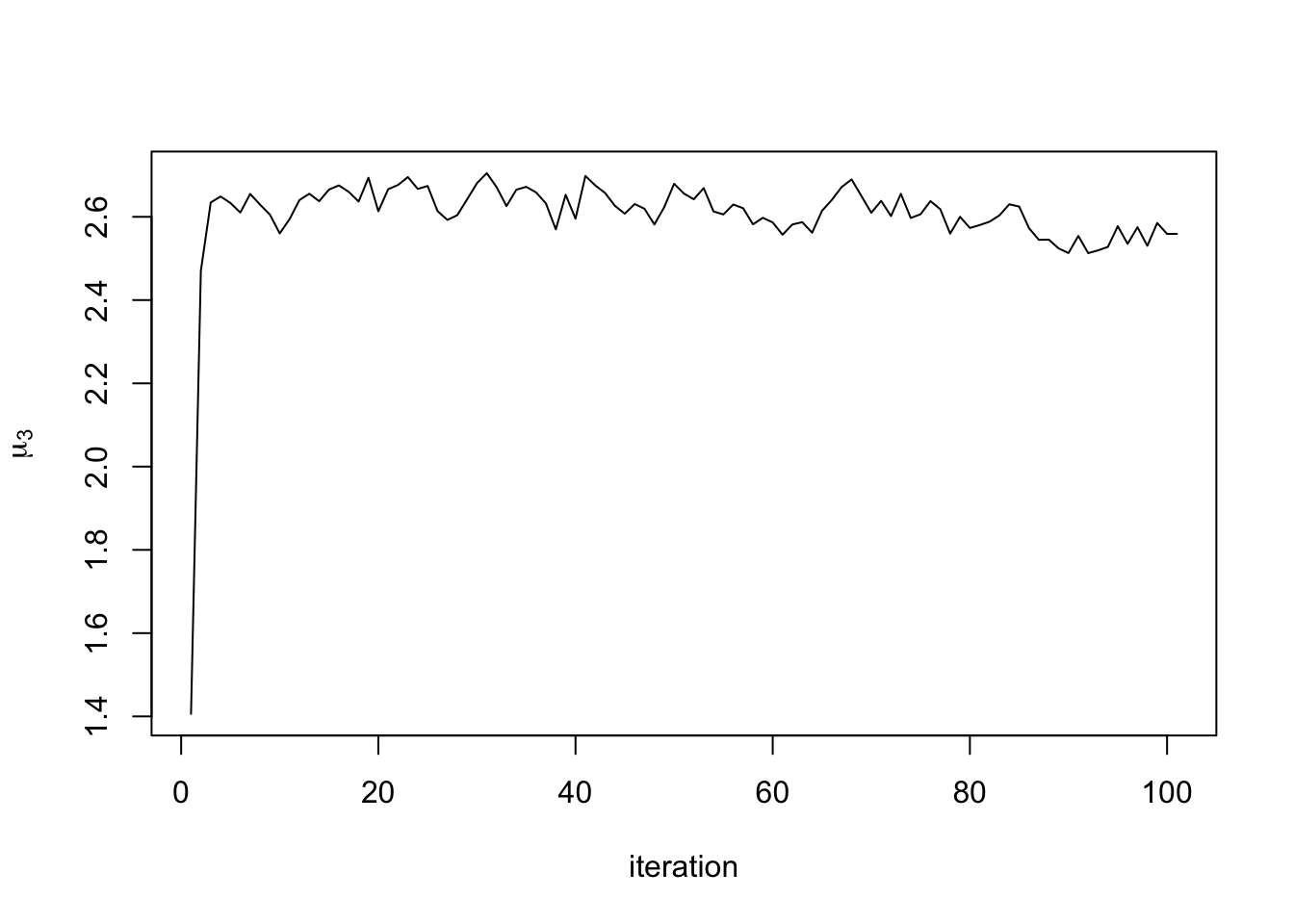
| Version | Author | Date |
|---|---|---|
| 862bc02 | hillarykoch | 2020-02-07 |
More specifically, extract_chains() returns a list with 4 elements. First, recall that M is the number of input classes, D is the dimension of the data, and let iterations be nstep+1.
mu_chains: a list withMelements, each element a matrix of dimensioniterations x D.mu_chains[[i]]is the MCMC samples for the mean vector of clusteri.Sigma_chains: a list withMelements, each element an array of dimensionD x D x iterations.Sigma_chains[[i]]is the MCMC samples for the covariance matrix of clusteri.prop_chain: A matrix of dimensionM x iterations, containing the MCMC samples for the mixing proportions of each class.z_chain: A matrix of dimensionn x iterations, containing the MCMC samples for the class labels of each observation. These labels correspond to the row indices ofretained_classes(above).