Response time correction of Contros HydroC pCO2 data
Jens Daniel Müller
08 November, 2019
Last updated: 2019-11-08
Checks: 7 0
Knit directory: BloomSail/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191021) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/TinaV/
Ignored: data/_merged_data_files/
Ignored: data/_summarized_data_files/
Unstaged changes:
Modified: output/Plots/response_time/RT_determination.pdf
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | aa27fd4 | jens-daniel-mueller | 2019-11-08 | Finalized RT determination |
| html | 4256bcf | jens-daniel-mueller | 2019-11-08 | Build site. |
| Rmd | 7f52e66 | jens-daniel-mueller | 2019-11-08 | response_time pdf updated |
| html | 72687ee | jens-daniel-mueller | 2019-11-08 | Build site. |
| Rmd | 74632d6 | jens-daniel-mueller | 2019-11-08 | response_time pdf updated |
| html | 74212a6 | jens-daniel-mueller | 2019-11-08 | Build site. |
| Rmd | 6cb1935 | jens-daniel-mueller | 2019-11-08 | response_time updated |
| html | 33e3659 | jens-daniel-mueller | 2019-10-22 | Build site. |
| Rmd | efcafd1 | jens-daniel-mueller | 2019-10-22 | Added data base, merging, and RT determination |
| html | 1595fe9 | jens-daniel-mueller | 2019-10-21 | Build site. |
| html | a059c41 | jens-daniel-mueller | 2019-10-21 | Build site. |
| Rmd | eff54ce | jens-daniel-mueller | 2019-10-21 | Added CTD read-in |
| html | 32ec4f7 | jens-daniel-mueller | 2019-10-21 | Build site. |
| Rmd | b2d2bbb | jens-daniel-mueller | 2019-10-21 | Structured data base and response time Rmd |
| html | bafa88f | jens-daniel-mueller | 2019-10-21 | Build site. |
| Rmd | 53ad162 | jens-daniel-mueller | 2019-10-21 | Structured data base and response time Rmd |
| html | 076a36b | jens-daniel-mueller | 2019-10-21 | Build site. |
| Rmd | 3e8a32e | jens-daniel-mueller | 2019-10-21 | Structured data base and response time Rmd |
| html | b2d0164 | jens-daniel-mueller | 2019-10-21 | Build site. |
| Rmd | 53ae361 | jens-daniel-mueller | 2019-10-21 | Added data base and response time Rmd |
library(tidyverse)
library(seacarb)
library(data.table)
library(broom)
library(lubridate)Sensitivity considerations
A change in DIC of 1 µmol kg-1 corresponds to a change in pCO2 of around 1 µatm, in the Central Baltic Sea at a pCO2 of around 100 µatm (summertime conditions).
df <- data.frame(cbind(
(c(1720)),
(c(7))))
Tem <- seq(5,25,5)
pCO2<-seq(50,500,20)
df<-merge(df, Tem)
names(df) <- c("AT", "S", "Tem")
df<-merge(df, pCO2)
names(df) <- c("AT", "S", "Tem", "pCO2")
df<-data.table(df)
df$AT<-df$AT*1e-6
df$DIC<-carb(flag=24, var1=df$pCO2, var2=df$AT, S=df$S, T=df$Tem, k1k2="m10", kf="dg", pHscale="T")[,16]
df$pCO2.corr<-carb(flag=15, var1=df$AT, var2=df$DIC, S=df$S, T=df$Tem, k1k2="m10", kf="dg", pHscale="T")[,9]
df$pCO2.2<-df$pCO2.corr + 25
df$DIC.2<-carb(flag=24, var1=df$pCO2.2, var2=df$AT, S=df$S, T=df$Tem, k1k2="m10", kf="dg", pHscale="T")[,16]
df$ratio<-(df$pCO2.2-df$pCO2.corr)/(df$DIC.2*1e6-df$DIC*1e6)
df %>%
ggplot(aes(pCO2, ratio, col=as.factor(Tem)))+
geom_line()+
scale_color_viridis_d(option = "C",name="Tem [°C]")+
labs(x=expression(pCO[2]~(µatm)), y=expression(Delta~pCO[2]~"/"~Delta~DIC~(µatm~µmol^{-1}~kg)))+
scale_y_continuous(limits = c(0,8), breaks = seq(0,10,1))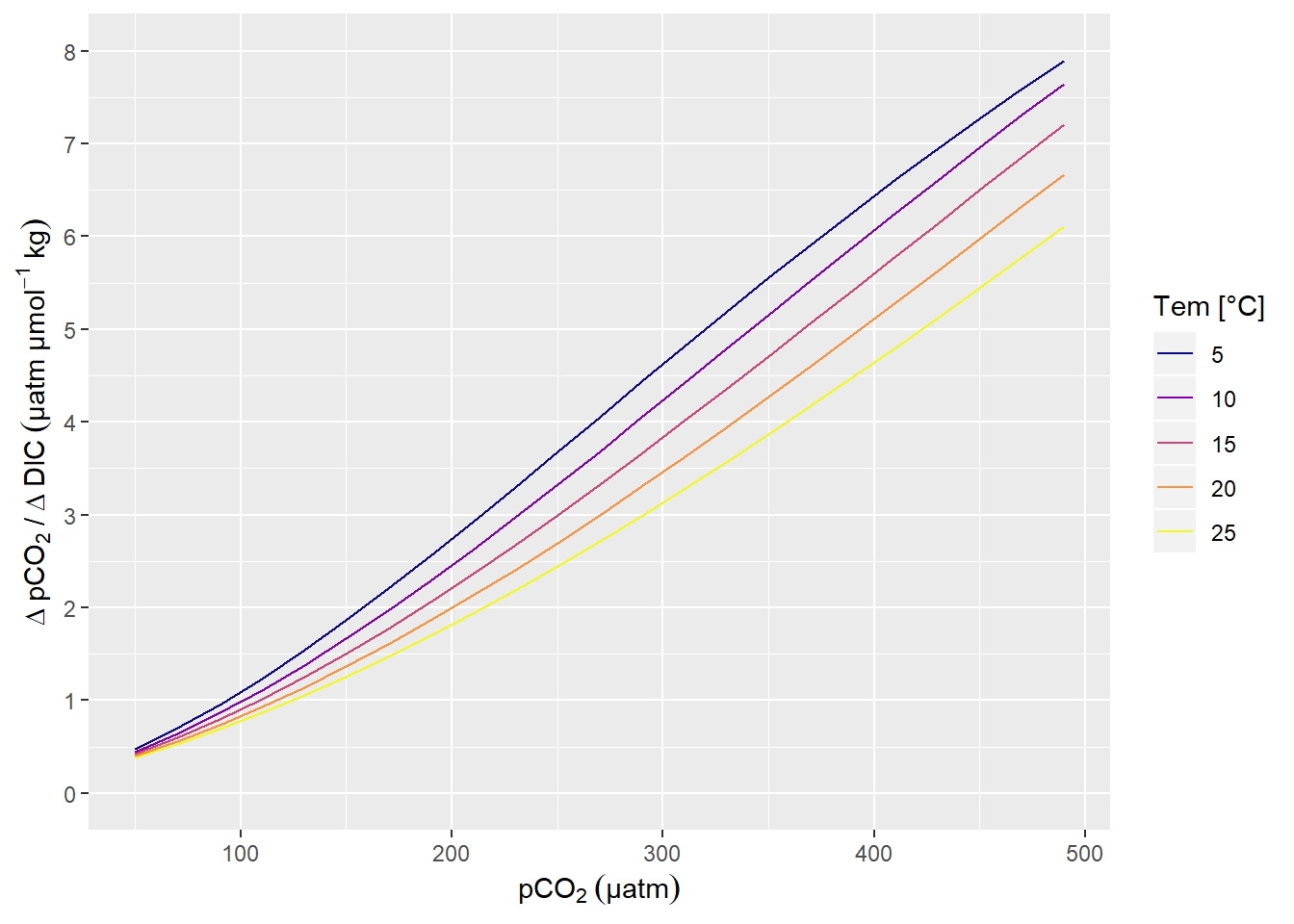
pCO2 sensitivity to changes in DIC.
| Version | Author | Date |
|---|---|---|
| 33e3659 | jens-daniel-mueller | 2019-10-22 |
rm(df, Tem, pCO2)Response time determination
HydroC sensor settings
The sensor was first run with a low power pump (1W), later and for most parts of the expedition with a stronger (8W) pump. Pumps were switched between recordings (data file: SD_datafile_20180718_170417CO2-0618-001.txt):
- 2018-07-17;13:08:34
- 2018-07-17;13:08:35
Logging frequency for all measurement modes (Measure, Zero, Flush) was set to:
10 sec for all recordings including SD_datafile_20180714_073641CO2-0618-001.txt
Increase to 2 sec in SD_datafile_20180717_121052CO2-0618-001.txt at:
- 2018-07-14;07:52:02
- 2018-07-14;07:52:12
- 2018-07-14;07:52:14
Increase to 2 sec in SD_datafile_20180718_170417CO2-0618-001 at:
- 2018-07-17;12:27:25
- 2018-07-17;12:27:27
- 2018-07-17;12:27:28
Model fitting
Response times were determined by fitting a nonlinear least-squares model with the nls function as described here by Douglas Watson.
- Flush period length: variable
- Flush period restricted to equilibration phase, avoiding initial gas mixing effects occuring at the start of each Flush period
- only completet Flush periods (duration > 500 sec) included
df <- read_csv(here::here("data/_merged_data_files",
"BloomSail_CTD_HydroC_Contros_clean.csv"),
col_types = cols(ID = col_character(),
pCO2_analog = col_double(),
pCO2 = col_double(),
Zero = col_factor(),
Flush = col_factor(),
Zero_ID = col_integer(),
duration = col_double(),
mixing = col_character()))
df <- df %>%
select(date_time, ID, dep, tem, Flush, pCO2, Zero_ID, duration, mixing)
df <- df %>%
filter(Flush == 1, mixing == "equilibration")
df <- df %>%
group_by(Zero_ID) %>%
mutate(duration = duration- min(duration),
max_duration = max(duration)) %>%
ungroup() %>%
filter(max_duration >= 500) %>%
select(-max_duration)An example plot for a nls model fitted to pCO2 observations during a Flush phase is shown below.
i <- 95
df_ID <- df %>%
filter(Zero_ID == i, duration <= 300)
fit <-
df_ID %>%
nls(pCO2 ~ SSasymp(duration, yf, y0, log_alpha), data = .)
tau <- as.numeric(exp(-tidy(fit)[3,2]))
pCO2_end <- as.numeric(tidy(fit)[1,2])
pCO2_start <- as.numeric(tidy(fit)[2,2])
dpCO2 = pCO2_end - pCO2_start
mean_abs_resid <- mean(abs(resid(fit)))
augment(fit) %>%
ggplot(aes(duration, pCO2))+
geom_point()+
geom_line(aes(y = .fitted))+
geom_vline(xintercept = tau)+
geom_hline(yintercept = pCO2_start + 0.63 *(dpCO2))+
labs(y=expression(pCO[2]~(µatm)), x="Duration of Flush period (s)")
Example response time determination by non-linear least squares fit to the pCO2 recovery signal after zeroing. The vertical line indicates the determined response time tau.
| Version | Author | Date |
|---|---|---|
| 74212a6 | jens-daniel-mueller | 2019-11-08 |
rm(df_ID, fit, i, tau, dpCO2, pCO2_end, pCO2_start)duration_intervals <- seq(150,500,50)As there was some speculation about the dependence of determined response times (\(\tau\)) on the chosen duration of the fit interval, \(\tau\) was determined for all zeroings and for total durations of:
150, 200, 250, 300, 350, 400, 450, 500 secs
# Plot all individual Flush periods with exponential fit ----------------------
pdf(file=here::here("output/Plots/response_time",
"RT_determination.pdf"), onefile = TRUE, width = 7, height = 4)
for (i in unique(df$Zero_ID)) {
for (max_duration in duration_intervals) {
df_ID <- df %>%
filter(Zero_ID == i, duration <= max_duration)
fit <-
try(
df_ID %>%
nls(pCO2 ~ SSasymp(duration, yf, y0, log_alpha), data = .),
TRUE)
if (class(fit) == "nls"){
tau <- as.numeric(exp(-tidy(fit)[3,2]))
pCO2_end <- as.numeric(tidy(fit)[1,2])
pCO2_start <- as.numeric(tidy(fit)[2,2])
dpCO2 = pCO2_end - pCO2_start
mean_abs_resid <- mean(abs(resid(fit))/pCO2_end)*100
temp <- as_tibble(bind_cols(Zero_ID = i, duration = max_duration, tau = tau, resid = mean_abs_resid))
if (exists("tau_df")){tau_df <- bind_rows(tau_df, temp)}
else {tau_df <- temp}
print(
augment(fit) %>%
ggplot(aes(duration, pCO2))+
geom_point()+
geom_line(aes(y = .fitted))+
geom_vline(xintercept = tau)+
geom_hline(yintercept = pCO2_start + 0.63 *(dpCO2))+
labs(y=expression(pCO[2]~(µatm)), x="Duration of Flush period (s)",
title = paste("Zero_ID: ", i, "Mean absolute residual (%): ", round(mean_abs_resid*100, 2)))+
xlim(0,600)
)
}
}
}
dev.off()
rm(df_ID, fit, i, tau, dpCO2, pCO2_end, pCO2_start, temp, max_duration, mean_abs_resid)
tau_df %>%
write_csv(here::here("data/_summarized_data_files", "Tina_V_HydroC_response_times_all.csv"))
# Plot individual Flush periods with linearized response variable --------
# for (i in unique(df$Zero_ID)) {
#
# #i <- 50
# df_ID <- df %>%
# filter(Zero_ID == i,
# mixing == "equilibration")
#
# mean_pCO2 <- df_ID %>%
# slice((n()-4) : n()) %>%
# summarise(mean_pCO2 = mean(pCO2))
#
# df_ID <- full_join(df_ID, mean_pCO2) %>%
# mutate(dpCO2 = max(pCO2) - pCO2,
# ln_dpCO2 = log(dpCO2))
#
#
# df_ID %>%
# ggplot(aes(duration_equi, ln_dpCO2))+
# geom_point()+
# geom_smooth(method = "lm")+
# theme_bw()
#
# # augment(fit) %>%
# # ggplot(aes(duration_equi, pCO2))+
# # geom_point()+
# # geom_line(aes(y = .fitted))+
# # geom_vline(xintercept = tau)
#
# ggsave(here::here("/Plots/TinaV/Sensor/HydroC_diagnostics/Response_time_fits",
# paste(i,"_Zero_ID_HydroC_RT_linear.jpg", sep="")),
# width = 10, height = 4)
# }A pdf with plots of all individual response time fits can be accessed here
tau_df <- read_csv(here::here("data/_summarized_data_files", "Tina_V_HydroC_response_times_all.csv"))
# define subsetting parameters
tau_high <- 120
tau_low <- 20
resid_limit <- 1
# subset determined tau values
tau_resid <- tau_df %>%
filter(resid < resid_limit)
tau_resid_min_max <- tau_resid %>%
filter(tau > tau_low, tau < tau_high)
# calculate some metrics for the subsetting
n_Zero_IDs <- df %>%
group_by(Zero_ID) %>%
n_groups()
n_duration_intervals <- length(duration_intervals)
n_tau_max <- n_Zero_IDs * length(duration_intervals)
n_tau_total <- nrow(tau_df)
n_tau_resid <- nrow(tau_resid)
n_tau_resid_min_max <- nrow(tau_resid_min_max)Outcome
Estimated \(\tau\) values were restricted to * mean absolute fit residual above 1 % of the final equilibrium pCO2 * \(\tau\) falling between 20 and 120 seconds
Metrics to characterize the fitting procedure include the number of:
- Flush periods: 55
- Duration intervals: 8
- Exercised response time fits: 440
- Succesful response times determinations: 417 (94.8)%
- \(\tau\)’s after removing fits with high absolute fit residual: 375 (89.9 %)
- \(\tau\)’s after restriction to \(\tau\) thresholds: 365 (87.5 %)
tau_df %>%
ggplot(aes(resid))+
geom_histogram()+
facet_wrap(~duration, labeller = label_both)+
geom_vline(xintercept = resid_limit)+
labs(x=expression(Mean~absolute~residuals~("%"~of~equilibrium~pCO[2])))
Histogram of residuals from fit displayed for the investigate durations of the fit interval. Vertical line represents the chosen threshold.
tau_resid %>%
group_by(duration) %>%
mutate(n_tau = n()) %>%
ggplot(aes(duration, n_tau))+
geom_point()+
labs(x="Fit interval duration (sec)", y="Number of tau values")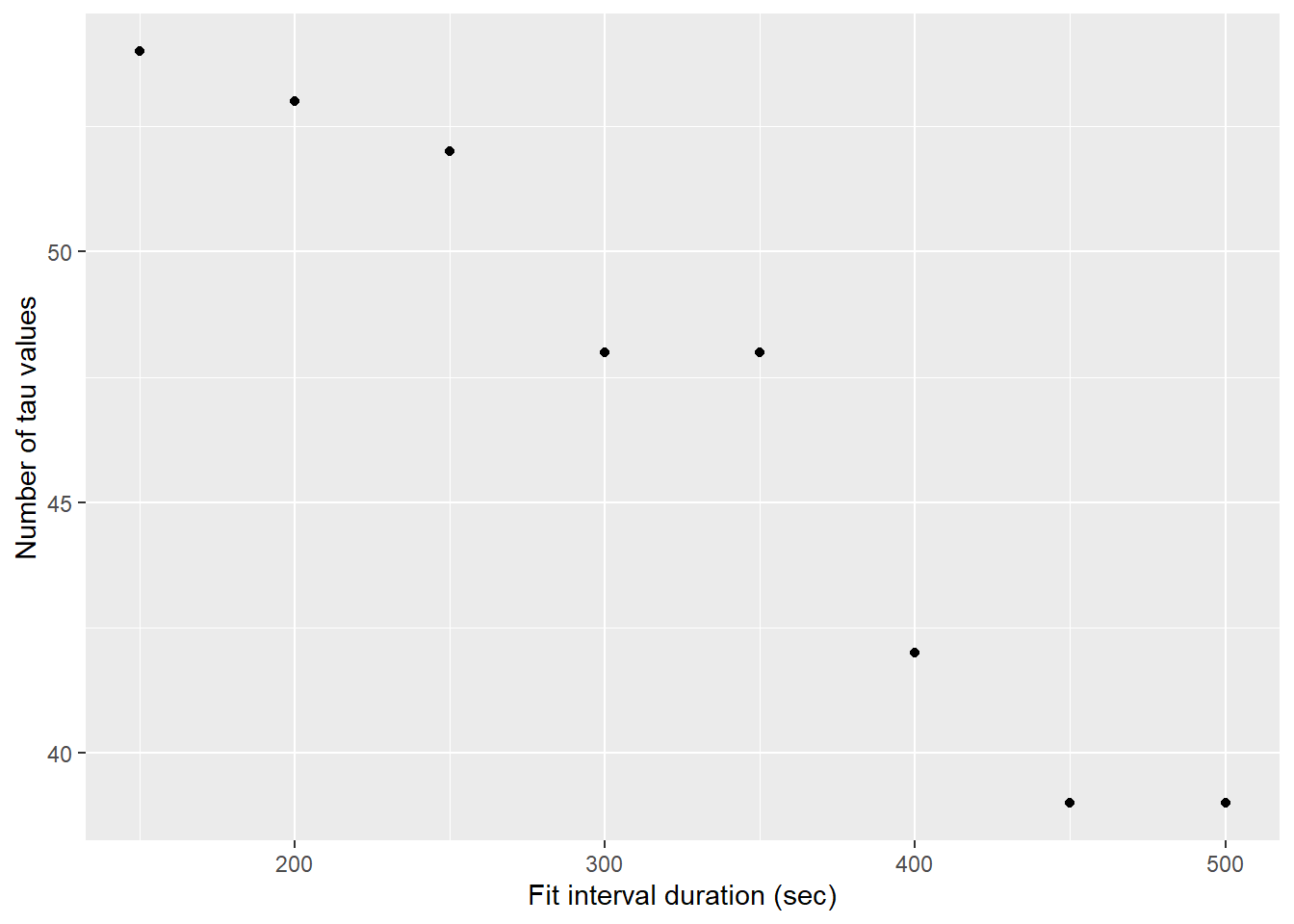
Number of determined tau values as a function of the fit interval length, after applying a fit residual threshold
tau_resid_min_max %>%
ggplot(aes(Zero_ID, tau, col=duration))+
geom_point()+
scale_color_viridis_c(name="Duration (sec)")+
labs(y="Tau (sec)")
Tau for all Zeroings with color representing the fit interval duration.
tau_resid_min_max %>%
group_by(Zero_ID) %>%
mutate(d_tau = tau - mean(tau)) %>%
ggplot(aes(duration, d_tau))+
geom_hline(yintercept = 0)+
geom_smooth()+
geom_point()+
facet_wrap(~Zero_ID, ncol = 4, labeller = label_both)+
labs(x="Duration (sec)", y="Deviation from mean tau (sec)")
Determined tau values as a function of the fit interval duration, displayed individually for each flush period.
tau_resid_min_max %>%
group_by(Zero_ID) %>%
mutate(d_tau = tau - mean(tau)) %>%
ggplot(aes(duration, d_tau))+
geom_violin(aes(group=duration))+
geom_point()+
labs(x="Duration (sec)", y="Deviation from mean tau (sec)")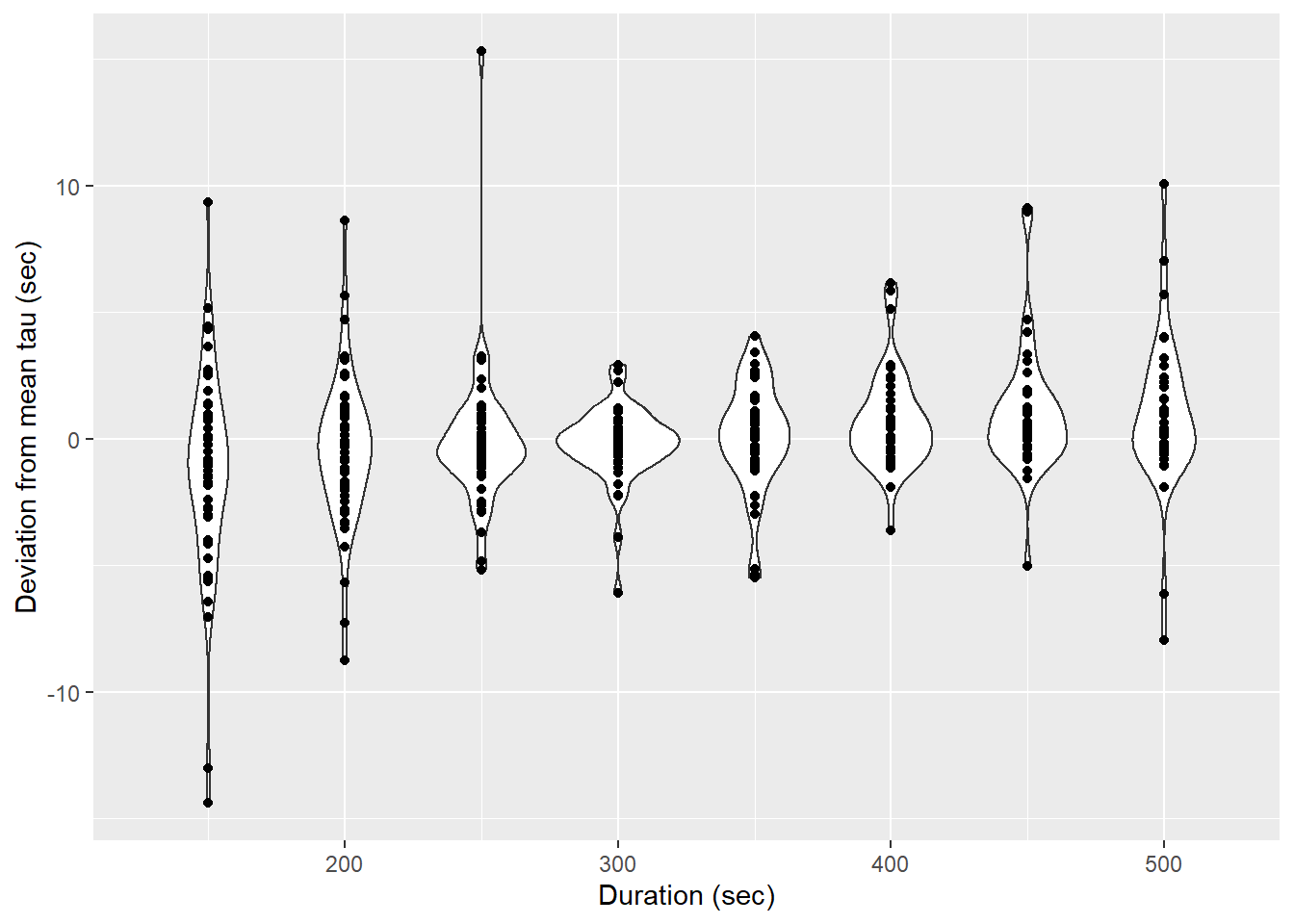
Determined tau values as a function of the fit interval duration, pooled for all flush period.
Pre-smoothing
Response time correction
Post-smoothing
Response time optimization
Open tasks / questions
- Compare Contros and own response time estimates
- Compare differnt response time correction methods (Bittig vs. Fiedler, Miloshevich, Fietzek)
- Test impact of duration for response time estimation on final mean response time
- Test impact of selection criterion for “good” response time estimates on final mean response time
- Check results from field response time experiment (high zeroing frequency)
- Why does nls model failure increase with higher fit duration
sessionInfo()R version 3.5.0 (2018-04-23)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 17763)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] lubridate_1.7.4 broom_0.5.2 data.table_1.12.6
[4] seacarb_3.2.12 oce_1.1-1 gsw_1.0-5
[7] testthat_2.2.1 forcats_0.4.0 stringr_1.4.0
[10] dplyr_0.8.3 purrr_0.3.3 readr_1.3.1
[13] tidyr_1.0.0 tibble_2.1.3 ggplot2_3.2.1
[16] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 xfun_0.10 haven_2.1.1
[4] lattice_0.20-35 colorspace_1.4-1 vctrs_0.2.0
[7] generics_0.0.2 viridisLite_0.3.0 htmltools_0.4.0
[10] yaml_2.2.0 rlang_0.4.1 pillar_1.4.2
[13] glue_1.3.1 withr_2.1.2 modelr_0.1.5
[16] readxl_1.3.1 lifecycle_0.1.0 munsell_0.5.0
[19] gtable_0.3.0 workflowr_1.4.0 cellranger_1.1.0
[22] rvest_0.3.4 evaluate_0.14 labeling_0.3
[25] knitr_1.25 highr_0.8 Rcpp_1.0.2
[28] scales_1.0.0 backports_1.1.5 jsonlite_1.6
[31] fs_1.3.1 hms_0.5.1 digest_0.6.22
[34] stringi_1.4.3 grid_3.5.0 rprojroot_1.3-2
[37] here_0.1 cli_1.1.0 tools_3.5.0
[40] magrittr_1.5 lazyeval_0.2.2 crayon_1.3.4
[43] whisker_0.4 pkgconfig_2.0.3 zeallot_0.1.0
[46] xml2_1.2.2 assertthat_0.2.1 rmarkdown_1.16
[49] httr_1.4.1 rstudioapi_0.10 R6_2.4.0
[52] nlme_3.1-137 git2r_0.26.1 compiler_3.5.0