eMLR - data preparation
Jens Daniel Müller
19 August, 2020
Last updated: 2020-08-19
Checks: 7 0
Knit directory: Cant_eMLR/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200707) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 94f9375. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rproj.user/
Ignored: data/GLODAPv1_1/
Ignored: data/GLODAPv2_2016b_MappedClimatologies/
Ignored: data/GLODAPv2_2020/
Ignored: data/Gruber_2019/
Ignored: data/WOCE/
Ignored: data/World_Ocean_Atlas_2018/
Ignored: data/dclement/
Ignored: data/eMLR/
Ignored: data/mapping/
Ignored: data/pCO2_atmosphere/
Ignored: dump/
Ignored: figure/
Unstaged changes:
Modified: analysis/_site.yml
Deleted: analysis/eMLR.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/eMLR_data_preparation.Rmd) and HTML (docs/eMLR_data_preparation.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 94f9375 | jens-daniel-mueller | 2020-08-19 | split emlr into data preparation, assumption testing, and model fitting |
| html | 828f44e | jens-daniel-mueller | 2020-08-19 | Build site. |
| Rmd | 77b2dc1 | jens-daniel-mueller | 2020-08-19 | split emlr into data preparation, assumption testing, and model fitting |
library(tidyverse)
library(lubridate)
library(patchwork)1 Required data
Required are:
- GLODAPv2.2020
- cleaned data file
- horizontal grid of sampling coordinates
- Cant from GLODAPv2_2016b_MappedClimatologies
- annual mean atmospheric pCO2
GLODAP <-
read_csv(
here::here(
"data/GLODAPv2_2020/_summarized_data_files",
"GLODAPv2.2020_clean.csv"
)
)
GLODAP_obs_grid <-
read_csv(
here::here(
"data/GLODAPv2_2020/_summarized_data_files",
"GLODAPv2.2020_clean_obs_grid.csv"
)
)
# Cant_clim <-
# read_csv(here::here("data/GLODAPv1_1/_summarized_files",
# "Cant_94.csv"))
Cant_clim <-
read_csv(
here::here(
"data/GLODAPv2_2016b_MappedClimatologies/_summarized_files",
"Cant.csv"
)
)
co2_atm <-
read_csv(here::here(
"data/pCO2_atmosphere/_summarized_data_files",
"co2_atm.csv"
))Cant_clim <- Cant_clim %>%
rename(cant = Cant)2 C*
C* serves as a conservative tracer of anthropogenic CO2 uptake. It is derived from measured DIC by removing the impact of
- organic matter formation and respiration
- calcification and calcium carbonate dissolution
Contributions of those processes are estimated from phosphate and alkalinity concentrations.
2.1 Stoichiometric ratios
rCP <- 117
rNP <- 16The stoichiometric nutrient ratios for the production and mineralization of organic matter were set to:
- C/P: 117
- N/P: 16
2.2 Calculation
C* is calculated as:
print("Cstar = tco2 + rCP_phosphate + talk_05 + rNP_phosphate_05")[1] "Cstar = tco2 + rCP_phosphate + talk_05 + rNP_phosphate_05"GLODAP <- GLODAP %>%
mutate(rCP_phosphate = -rCP * phosphate,
talk_05 = -0.5 * talk,
rNP_phosphate_05 = -0.5 * rNP * phosphate,
Cstar = tco2 + rCP_phosphate + talk_05 + rNP_phosphate_05)
rm(rCP, rNP)3 PO4* calculation
Currently, the predictor PO4* is calculated according to Clement and Gruber (2018), ie based on oxygen rather than nitrate, as claimed in Gruber et al (2019).
GLODAP <- GLODAP %>%
mutate(phosphate_star_oxy_corr = (oxygen / 170) - 1.95,
phosphate_star_oxy = phosphate + phosphate_star_oxy_corr)GLODAP <- GLODAP %>%
mutate(phosphate_star_nit_corr = - nitrate/16 + 2.9,
phosphate_star_nit = phosphate + phosphate_star_nit_corr)3.1 Comparison of approaches
GLODAP %>%
ggplot(aes(oxygen,
nitrate)) +
geom_bin2d() +
scale_fill_viridis_c() +
facet_wrap(~basin)
GLODAP %>%
ggplot(aes(phosphate,
nitrate)) +
geom_bin2d() +
scale_fill_viridis_c() +
facet_wrap(~basin)
GLODAP %>%
ggplot(aes(phosphate,
oxygen)) +
geom_bin2d() +
scale_fill_viridis_c() +
facet_wrap(~basin)
GLODAP %>%
ggplot(aes(phosphate_star_oxy_corr,
phosphate_star_nit_corr)) +
geom_bin2d() +
scale_fill_viridis_c() +
facet_wrap(~basin)
GLODAP %>%
ggplot(aes(phosphate_star_oxy, phosphate_star_nit)) +
geom_bin2d() +
scale_fill_viridis_c() +
facet_wrap(~basin)
GLODAP <- GLODAP %>%
select(-c(
phosphate_star_nit,
phosphate_star_oxy_corr)) %>%
rename(phosphate_star = phosphate_star_oxy)4 Reference year adjustment
The reference year adjustment relies on an apriori estimate of Cant at a given location and depth, which is used as a scaling factor for the concurrent change in atmospheric CO2. The underlying assumption is a transient steady state for the oceanic Cant uptake. Here, Cant from the GLODAP mapped Climatology was used.
Note that eq. 6 in Clement and Gruber (2018) misses pCO2 pre-industrial in the denominator. Here we use the equation published in Gruber et al. (2019).
4.1 Merge data sets
4.1.1 GLODAP + Cant
Cant_clim <- Cant_clim %>%
drop_na()
Cant_clim_obs <- left_join(GLODAP_obs_grid, Cant_clim) %>%
select(-n)
# Cant_clim_obs_nr <- Cant_clim_obs %>%
# group_by(lon, lat) %>%
# summarise(n_cant = n()) %>%
# ungroup()
# Cant_clim_obs %>%
# filter(n < 1) %>%
# ggplot(aes(lon,lat)) +
# geom_point(data = GLODAP_obs_grid, aes(lon, lat)) +
# geom_point(col = "red")
rm(Cant_clim, GLODAP_obs_grid)
GLODAP_Cant_obs <- full_join(GLODAP, Cant_clim_obs)
rm(GLODAP, Cant_clim_obs)The mapped Cant product was merged with GLODAP observation by:
- using an identical 1x1° horizontal grid
- linear interpolation of Cant from standard to sampling depth
# GLODAP_Cant_obs <- full_join(GLODAP_Cant_obs, Cant_clim_obs_nr)
GLODAP_Cant_obs <- GLODAP_Cant_obs %>%
# filter(n_cant > 1) %>%
group_by(lat, lon) %>%
arrange(depth) %>%
mutate(cant_int = approxfun(depth, cant, rule = 2)(depth)) %>%
ungroup()
# GLODAP_Cant_obs_set <- GLODAP_Cant_obs %>%
# filter(n_cant == 1) %>%
# group_by(lat, lon) %>%
# arrange(depth) %>%
# mutate(cant_int = mean(cant, na.rm = TRUE)) %>%
# ungroup()
ggplot() +
geom_path(
data = GLODAP_Cant_obs %>%
filter(lat == 48.5, lon == 165.5,!is.na(cant)) %>%
arrange(depth),
aes(cant, depth, col = "mapped")
) +
geom_point(
data = GLODAP_Cant_obs %>%
filter(lat == 48.5, lon == 165.5,!is.na(cant)) %>%
arrange(depth),
aes(cant, depth, col = "mapped")
) +
geom_point(
data = GLODAP_Cant_obs %>%
filter(lat == 48.5, lon == 165.5, date == ymd("2018-06-27")),
aes(cant_int, depth, col = "interpolated")
) +
scale_y_reverse() +
scale_color_brewer(palette = "Dark2", name = "") +
labs(title = "Cant interpolation to sampling depth - example profile")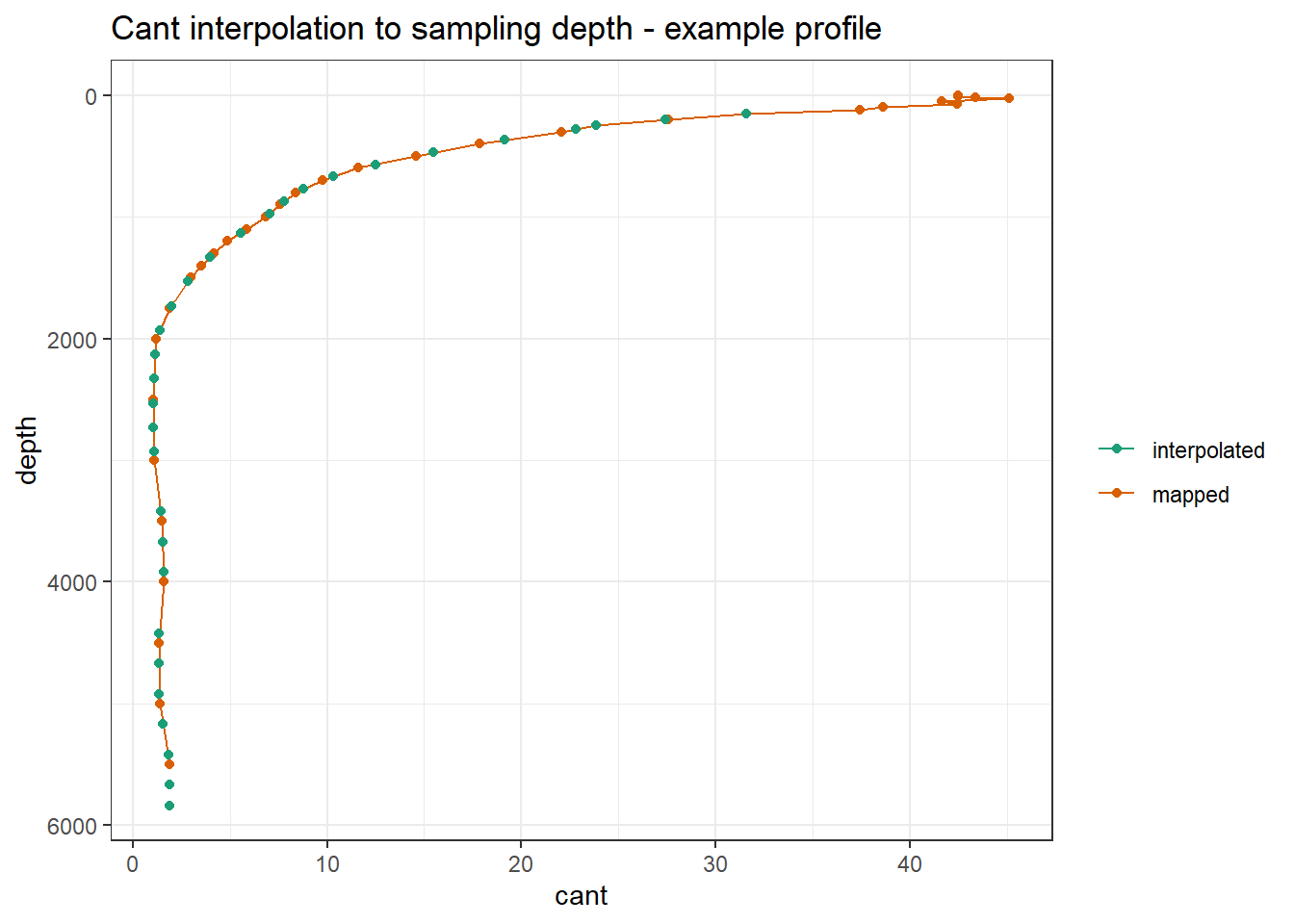
# remove cant data at grid cells without observations
GLODAP <- GLODAP_Cant_obs %>%
filter(!is.na(Cstar)) %>%
mutate(cant = cant_int) %>%
select(-cant_int)
rm(GLODAP_Cant_obs)4.1.2 GLODAP + atm. pCO2
GLODAP observations were merged with mean annual atmospheric pCO2 levels by year.
GLODAP <- left_join(GLODAP, co2_atm)4.2 Calculate adjustment
GLODAP <- GLODAP %>%
group_by(era) %>%
mutate(tref = median(year)) %>%
ungroup()
tref <- GLODAP %>%
group_by(era) %>%
summarise(year = median(year)) %>%
ungroup()
co2_atm_tref <- right_join(co2_atm, tref) %>%
select(-year) %>%
rename(pCO2_tref = pCO2)
GLODAP <- full_join(GLODAP, co2_atm_tref)
rm(co2_atm, co2_atm_tref, tref)
GLODAP <- GLODAP %>%
mutate(Cstar_tref_delta =
((pCO2 - pCO2_tref) / (pCO2_tref - 280)) * cant,
Cstar_tref = Cstar - Cstar_tref_delta)4.3 Control plots
4.3.1 Histogram
GLODAP %>%
ggplot(aes(Cstar_tref_delta)) +
geom_histogram()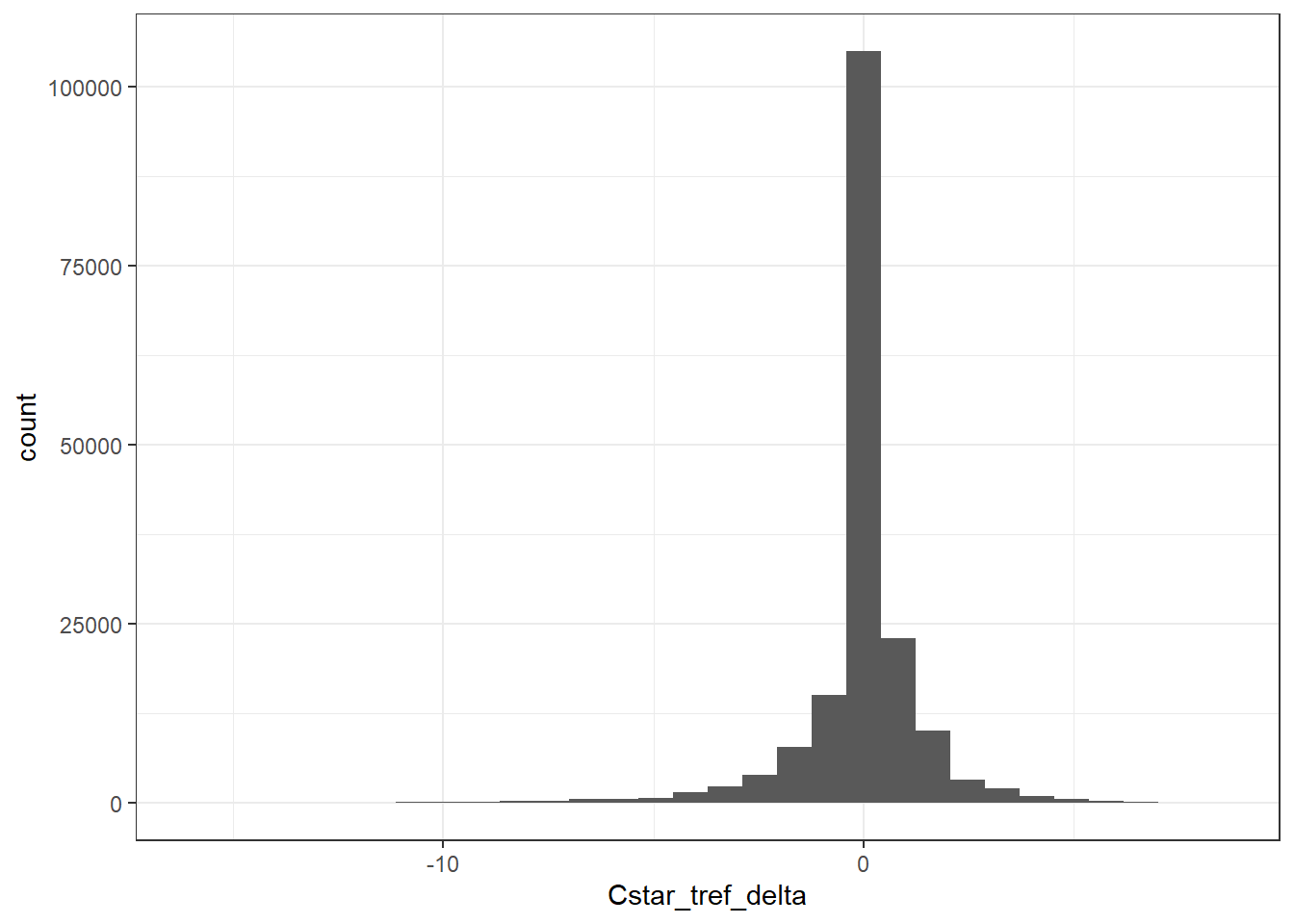
4.3.2 Time series
GLODAP %>%
sample_n(1e4) %>%
ggplot(aes(year, Cstar_tref_delta, col = cant)) +
geom_point() +
scale_color_viridis_c() +
labs(title = "random subsample 1e4")
5 Selected section plots
A selected section is plotted to demonstrate the magnitude of various parameters and corrections relevant to C*.
GLODAP_cruise <- GLODAP %>%
filter(cruise %in% parameters$cruises_meridional)GLODAP_cruise %>%
arrange(date) %>%
ggplot(aes(lon, lat)) +
geom_raster(data = landmask %>% filter(region == "land"),
aes(lon, lat), fill = "grey80") +
geom_path() +
geom_point(aes(col = date)) +
coord_quickmap(expand = 0) +
scale_color_viridis_c(trans = "date") +
labs(title = paste("Cruise year:", mean(GLODAP_cruise$year))) +
theme(legend.position = "bottom")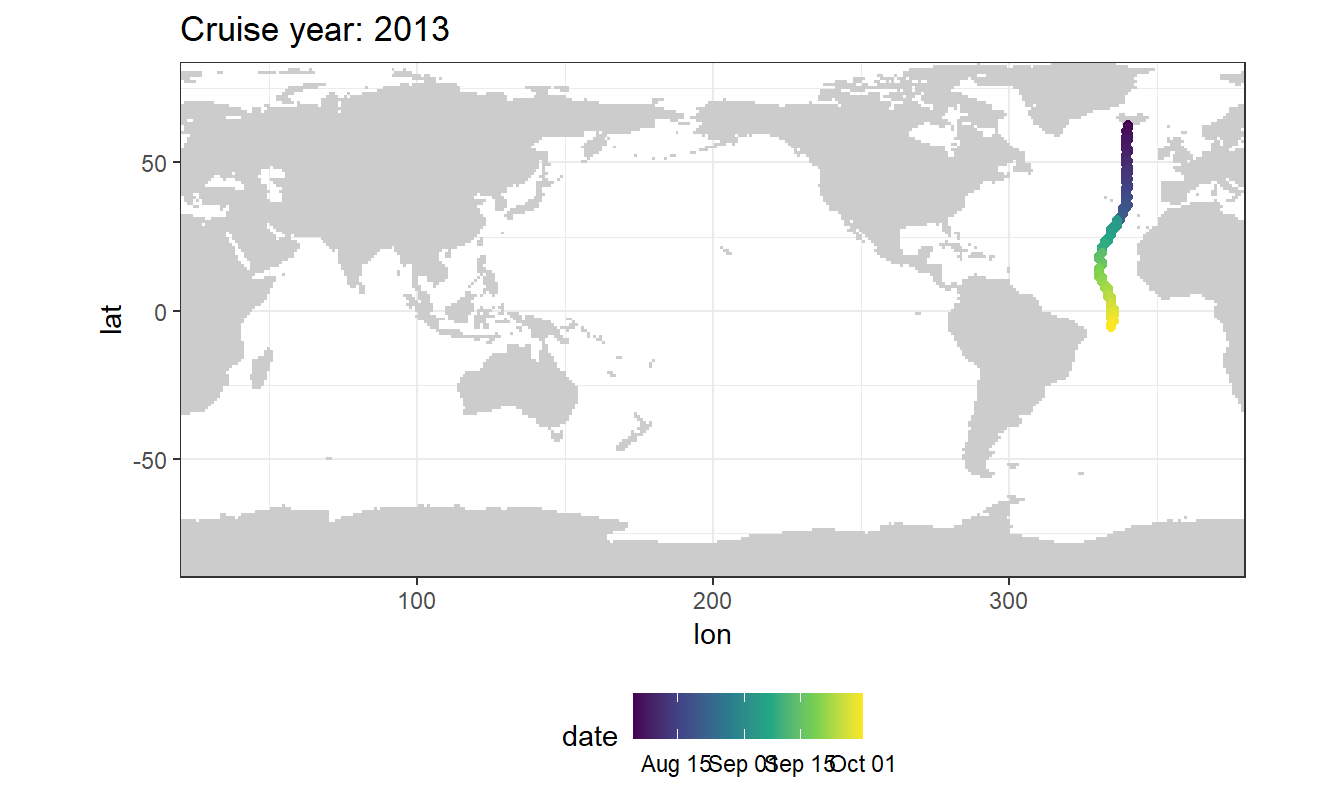
lat_section <-
GLODAP_cruise %>%
ggplot(aes(lat, depth)) +
scale_y_reverse() +
scale_color_viridis_c() +
theme(legend.position = "bottom")
lat_section +
geom_point(aes(col = tco2))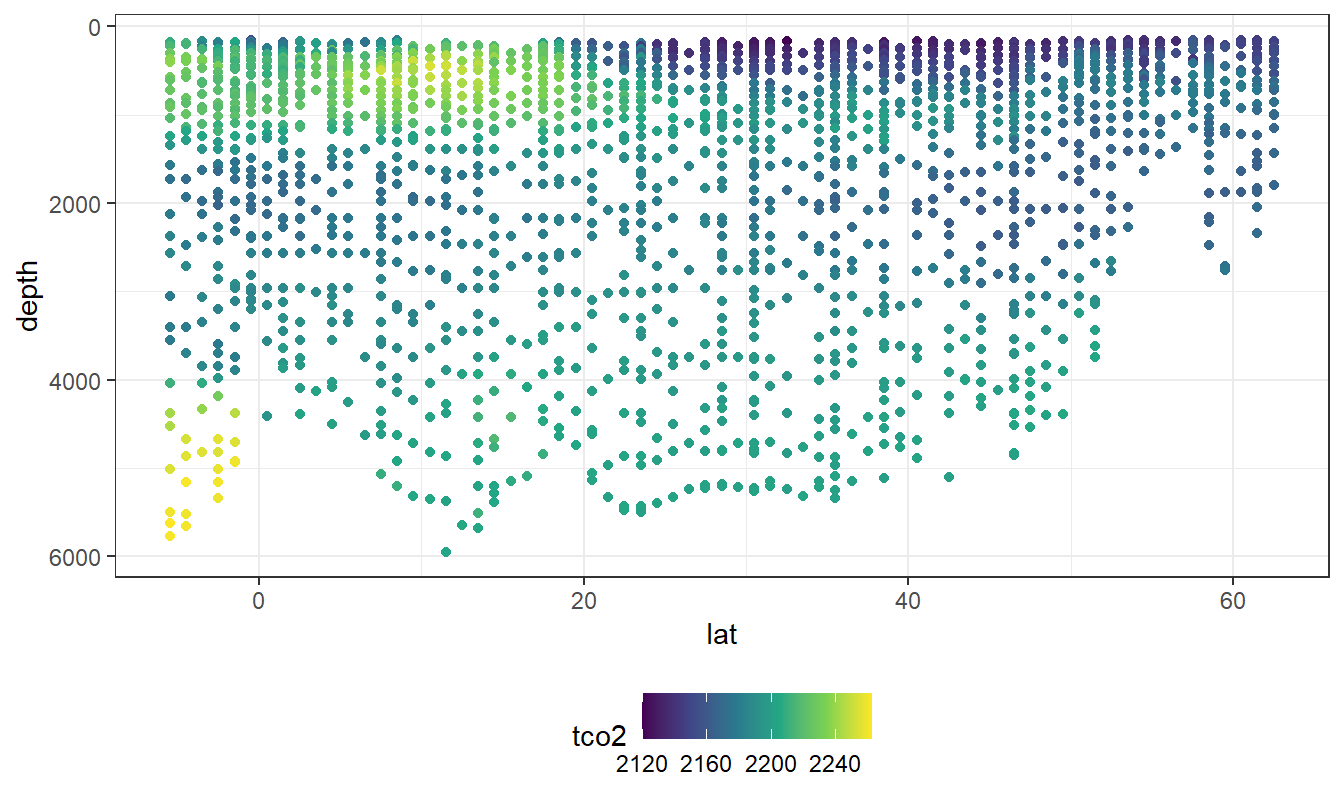
lat_section +
geom_point(aes(col = rCP_phosphate))
lat_section +
geom_point(aes(col = talk_05))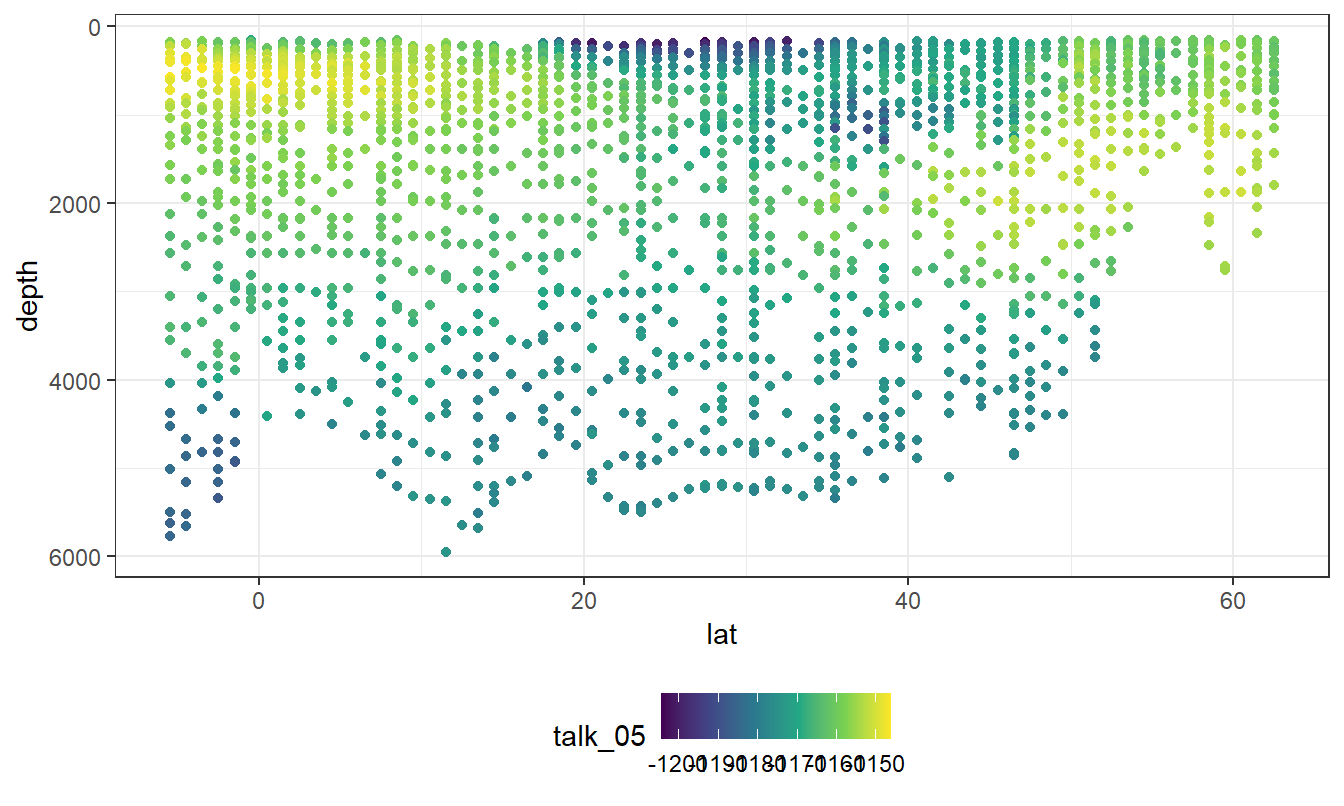
lat_section +
geom_point(aes(col = rNP_phosphate_05))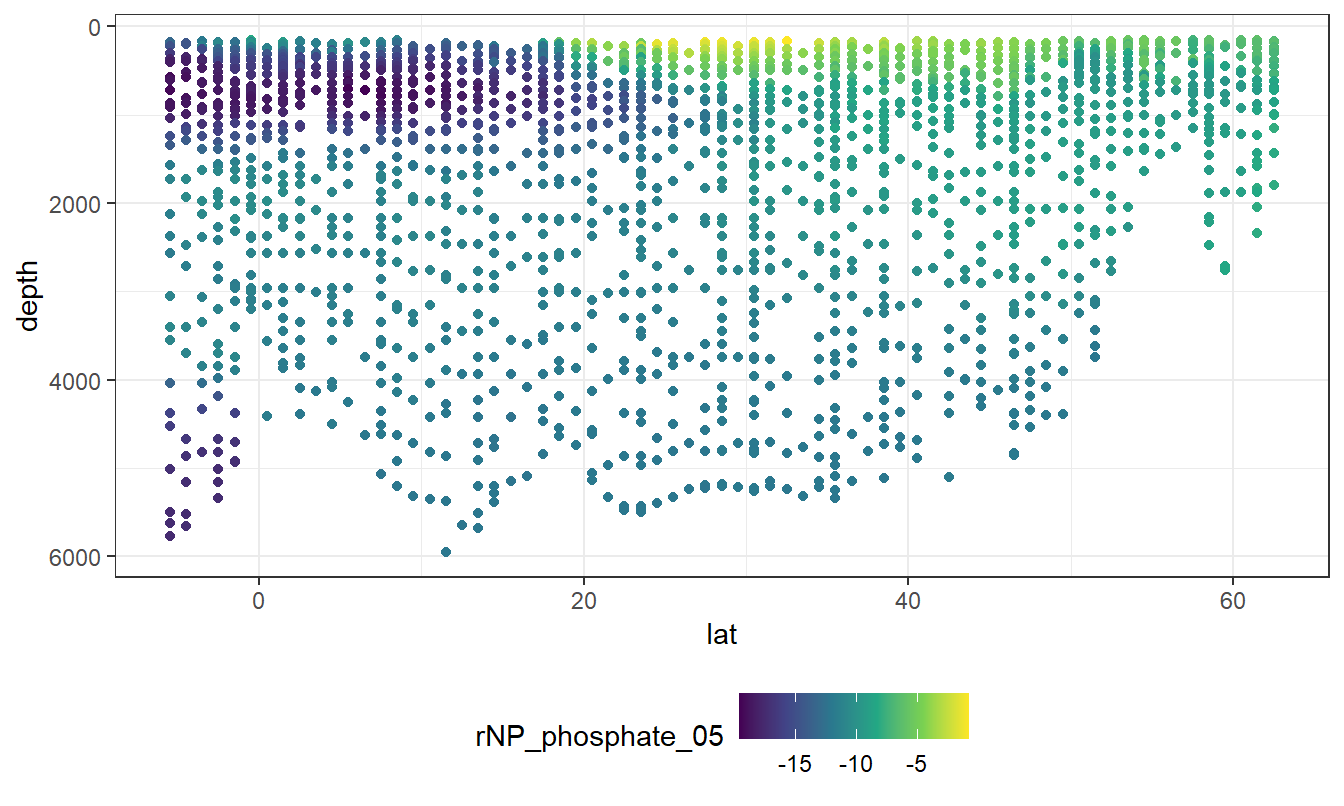
lat_section +
geom_point(aes(col = Cstar))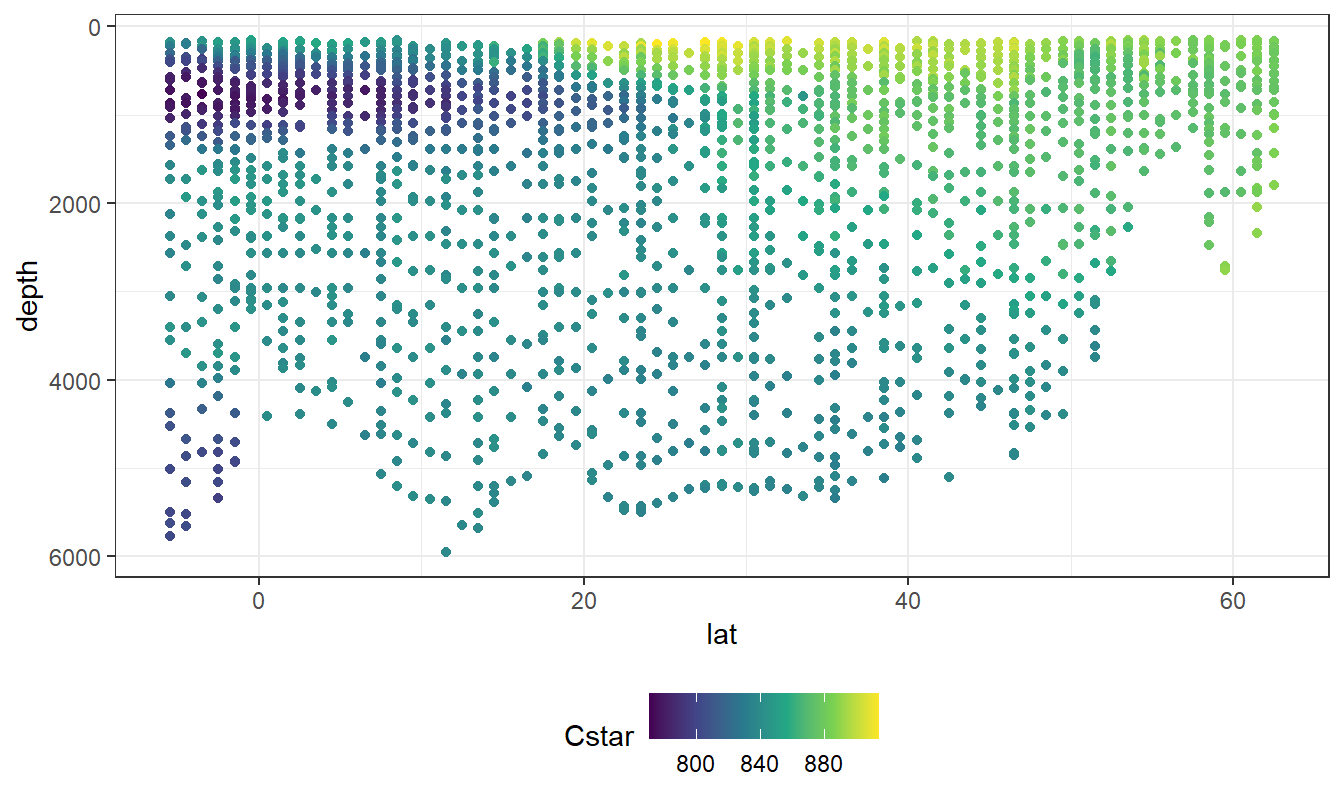
lat_section +
geom_point(aes(col = -Cstar_tref_delta))
rm(lat_section, GLODAP_cruise)6 Isoneutral slabs
The following boundaries for isoneutral slabs were defined:
- Atlantic: -, 26, 26.5, 26.75, 27, 27.25, 27.5, 27.75, 27.85, 27.95, 28.05, 28.1, 28.15, 28.2,
- Indo-Pacific: -, 26, 26.5, 26.75, 27, 27.25, 27.5, 27.75, 27.85, 27.95, 28.05, 28.1,
Continuous neutral densities (gamma) values from GLODAP are grouped into isoneutral slabs.
GLODAP_Atl <- GLODAP %>%
filter(basin == "Atlantic") %>%
mutate(gamma_slab = cut(gamma, parameters$slabs_Atl))
GLODAP_Ind_Pac <- GLODAP %>%
filter(basin == "Indo-Pacific") %>%
mutate(gamma_slab = cut(gamma, parameters$slabs_Ind_Pac))
GLODAP <- bind_rows(GLODAP_Atl, GLODAP_Ind_Pac)
rm(GLODAP_Atl, GLODAP_Ind_Pac)GLODAP_cruise <- GLODAP %>%
filter(cruise %in% parameters$cruises_meridional)
lat_section <-
GLODAP_cruise %>%
ggplot(aes(lat, depth)) +
scale_y_reverse() +
theme(legend.position = "bottom")
lat_section +
geom_point(aes(col = gamma_slab)) +
scale_color_viridis_d()
rm(lat_section, GLODAP_cruise)GLODAP_cruise <- GLODAP %>%
filter(cruise %in% parameters$cruises_meridional)
library(oce)
GLODAP_cruise <- GLODAP_cruise %>%
mutate(THETA = swTheta(salinity = sal,
temperature = tem,
pressure = depth,
referencePressure = 0,
longitude = lon-180,
latitude = lat))
GLODAP_cruise <- GLODAP_cruise %>%
rename(LATITUDE = lat,
LONGITUDE = lon,
SALNTY = sal,
CTDPRS = depth,
gamma_provided = gamma)
library(reticulate)
source_python(here::here("code/python_scripts",
"Gamma_GLODAP_python.py"))
GLODAP_cruise <- calculate_gamma(GLODAP_cruise)
GLODAP_cruise <- GLODAP_cruise %>%
mutate(gamma_delta = gamma_provided - GAMMA)
lat_section <-
GLODAP_cruise %>%
ggplot(aes(LATITUDE, CTDPRS)) +
scale_y_reverse() +
theme(legend.position = "bottom")
lat_section +
geom_point(aes(col = gamma_delta)) +
scale_color_viridis_c()
GLODAP_cruise %>%
ggplot(aes(gamma_delta))+
geom_histogram()
rm(lat_section, GLODAP_cruise, cruises_meridional)7 Observations coverage
GLODAP <- GLODAP %>%
mutate(era = factor(era, c("JGOFS_WOCE", "GO_SHIP", "new_era"))) %>%
mutate(gamma_slab = factor(gamma_slab),
gamma_slab = factor(gamma_slab, levels = rev(levels(gamma_slab))))
GLODAP %>%
filter(basin == "Atlantic") %>%
ggplot(aes(lat, gamma_slab)) +
geom_bin2d(binwidth = 5) +
scale_fill_viridis_c(option = "magma", direction = -1, trans = "log10",
name = "log10(n)") +
scale_x_continuous(breaks = seq(-100,100,20)) +
facet_grid(era~basin)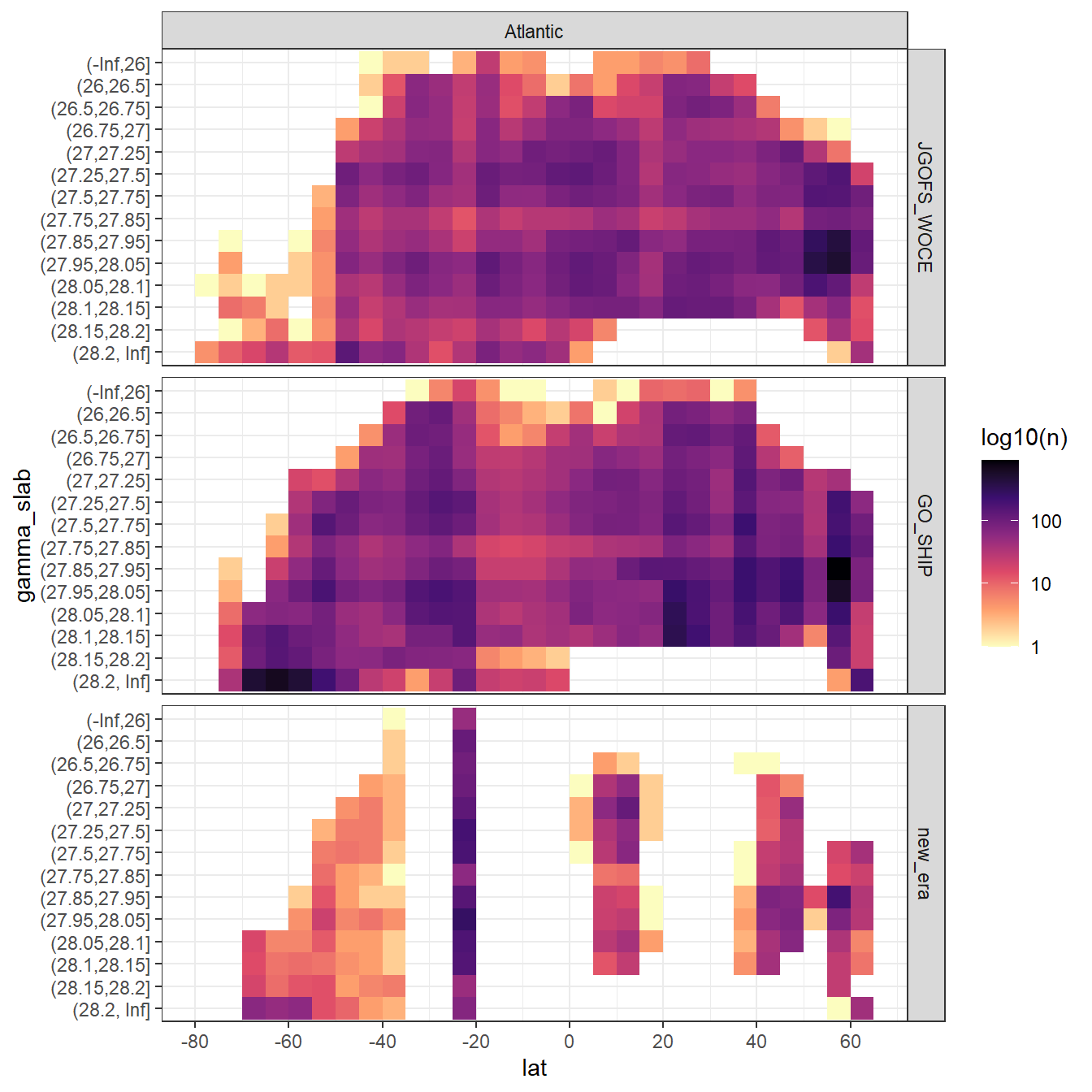
GLODAP %>%
filter(basin == "Indo-Pacific") %>%
ggplot(aes(lat, gamma_slab)) +
geom_bin2d(binwidth = 5) +
scale_fill_viridis_c(option = "magma", direction = -1, trans = "log10",
name = "log10(n)") +
scale_x_continuous(breaks = seq(-100,100,20)) +
facet_grid(era~basin)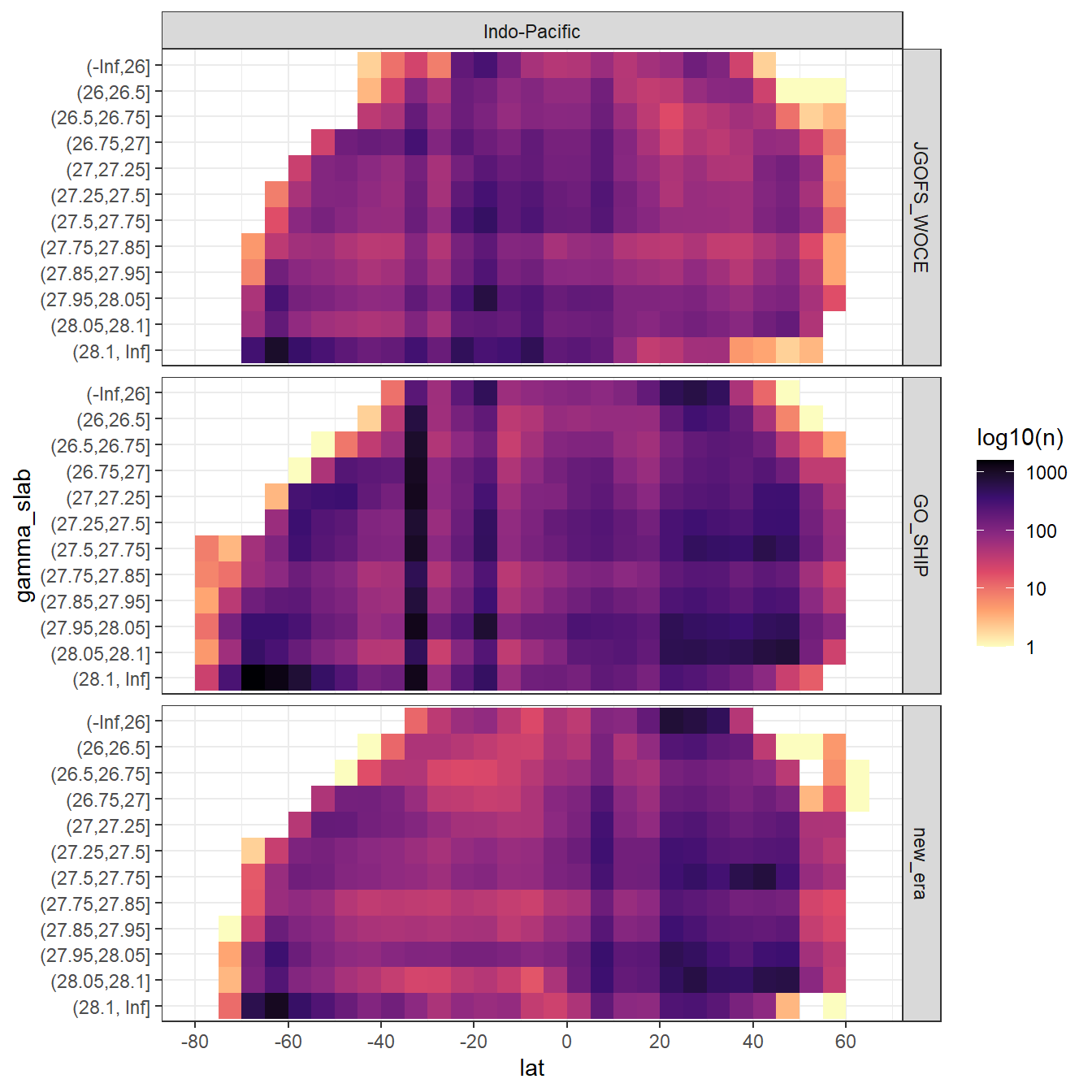
8 Write file
GLODAP %>% write_csv(here::here("data/GLODAPv2_2020/_summarized_data_files",
"GLODAP_MLR_fitting_ready.csv"))9 Open tasks
10 Open questions
- Which PO4* calculation is “correct”?
sessionInfo()R version 4.0.2 (2020-06-22)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 18363)
Matrix products: default
locale:
[1] LC_COLLATE=English_Germany.1252 LC_CTYPE=English_Germany.1252
[3] LC_MONETARY=English_Germany.1252 LC_NUMERIC=C
[5] LC_TIME=English_Germany.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] patchwork_1.0.1 lubridate_1.7.9 forcats_0.5.0 stringr_1.4.0
[5] dplyr_1.0.0 purrr_0.3.4 readr_1.3.1 tidyr_1.1.0
[9] tibble_3.0.3 ggplot2_3.3.2 tidyverse_1.3.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.5 here_0.1 assertthat_0.2.1 rprojroot_1.3-2
[5] digest_0.6.25 R6_2.4.1 cellranger_1.1.0 backports_1.1.8
[9] reprex_0.3.0 evaluate_0.14 httr_1.4.2 pillar_1.4.6
[13] rlang_0.4.7 readxl_1.3.1 rstudioapi_0.11 whisker_0.4
[17] blob_1.2.1 rmarkdown_2.3 labeling_0.3 munsell_0.5.0
[21] broom_0.7.0 compiler_4.0.2 httpuv_1.5.4 modelr_0.1.8
[25] xfun_0.16 pkgconfig_2.0.3 htmltools_0.5.0 tidyselect_1.1.0
[29] fansi_0.4.1 viridisLite_0.3.0 crayon_1.3.4 dbplyr_1.4.4
[33] withr_2.2.0 later_1.1.0.1 grid_4.0.2 jsonlite_1.7.0
[37] gtable_0.3.0 lifecycle_0.2.0 DBI_1.1.0 git2r_0.27.1
[41] magrittr_1.5 scales_1.1.1 cli_2.0.2 stringi_1.4.6
[45] farver_2.0.3 fs_1.4.2 promises_1.1.1 xml2_1.3.2
[49] ellipsis_0.3.1 generics_0.0.2 vctrs_0.3.2 RColorBrewer_1.1-2
[53] tools_4.0.2 glue_1.4.1 hms_0.5.3 yaml_2.2.1
[57] colorspace_1.4-1 rvest_0.3.6 knitr_1.29 haven_2.3.1