Prepare doxy data and vertically align to same depths as (pH) climatology
David Stappard & Jens Daniel Müller
06 December, 2023
Last updated: 2023-12-06
Checks: 7 0
Knit directory: bgc_argo_r_argodata/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211008) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 83269d5. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: output/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/doxy_vertical_align.Rmd)
and HTML (docs/doxy_vertical_align.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 59f5cc4 | ds2n19 | 2023-11-23 | Moved spatiotemporal analysis to use aligned profiles. |
| html | 80c16c2 | ds2n19 | 2023-11-15 | Build site. |
| Rmd | 3eba518 | ds2n19 | 2023-11-15 | Introduction of vertical alignment and cluster analysis to github website. |
Task
This markdown file reads previously created pH climatology file and uses that as the definition of the depth levels that the Argo dissolved oxygen data should be aligned to. Previously created BGC data (bgc_data.rds) and metadata (bgc_metadata.rds) are loaded from the BGC preprocessed folder.
Base data qc flags are checked to ensure that the float position, pressure measurements and dissolved oxygen measurements are good. Pressure is used to derive the depth of each measurement. The dissolved oxygen profile is checked to ensure that significant gaps (specified by the opt_gap_limit, opt_gap_min_depth and opt_gap_max_depth) do not exist. Profiles are assigned a profile_range field that identifies the depth 1 = 614 m, 2 = 1225 m and 3 = 1600 m.
The float dissolved oxygen profiles are then aligned using the spline function to match the depth levels of the climatology resulting in data frame bgc_data_doxy_interpolated_clean.
Set directories
location of pre-prepared data
Set options
Define options that are used to determine profiles that we will us in the ongoing analysis
# Options
# opt_profile_depth_range
# The profile must have at least one doxy reading at a depth <= opt_profile_depth_range[1, ]
# The profile must have at least one doxy reading at a depth >= opt_profile_depth_range[2, ].
# In addition if the profile depth does not exceed the min(opt_profile_depth_range[2, ]) (i.e. 600) it will be removed.
profile_range <- c(1, 2, 3)
min_depth <- c(10, 10, 10)
max_depth <- c(614, 1225, 1600)
opt_profile_depth_range <- data.frame(profile_range, min_depth, max_depth)
# opt_gap...
# The profile should not have a gap greater that opt_gap_limit within the range defined by opt_gap_min_depth and opt_gap_max_depth
opt_gap_limit <- c(28, 55, 110)
opt_gap_min_depth <- c(0, 400, 1000)
opt_gap_max_depth <- c(400, 1000, 1600)
# opt_measure_label, opt_xlim and opt_xbreaks are associated formatting
opt_measure_label <- expression("dissolved oxygen ( µmol kg"^"-1"~")")
opt_xlim <- c(50, 350)
opt_xbreaks <- c(50, 100, 150, 200, 250, 300, 350)
# opt_n_prof_sel
# The selection criteria that is used against n_prof, here set to 1
# Description of n_prof usage is provided at https://argo.ucsd.edu/data/data-faq/version-3-profile-files/ the next two lines are from that page.
# The main Argo CTD profile is stored in N_PROF=1. All other parameters (including biogeochemical parameters) that are measured
# with the same vertical sampling scheme and at the same location and time as the main Argo CTD profile are also stored in N_PROF=1.
opt_n_prof_sel = 1read climatology
read pH climatology, values are provided at set depths
# climatology values (pH_clim_va) available for lat, lon, month and depth
pH_clim_va <- read_rds(file = paste0(path_argo_preprocessed, "/pH_clim_va.rds"))
# What is the max depth we are interested in
opt_profile_max_depth <- max(opt_profile_depth_range$max_depth)
# existing depth levels that we will align to
target_depth_levels <- pH_clim_va %>%
filter(depth <= opt_profile_max_depth) %>%
rename(target_depth = depth) %>%
distinct(target_depth)
rm(pH_clim_va)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1276535 68.2 2476715 132.3 2476715 132.3
Vcells 2192738 16.8 91952148 701.6 92803266 708.1read doxy data
read doxy profile and carry out basic checks, good data.
# base data and associated metadata
bgc_data <- read_rds(file = paste0(path_argo_preprocessed, '/bgc_data.rds'))
bgc_metadata <- read_rds(file = paste0(path_argo_preprocessed, '/bgc_metadata.rds'))
# Select relevant field from metadata ready to join to bgc_data
bgc_metadata_select <- bgc_metadata %>%
filter(position_qc == 1) %>%
select(file_id,
date,
lat,
lon) %>%
mutate(year = year(date),
month = month(date),
.after = date)
# we drive alignment from pressure and doxy data
# conditions
# n_prof == 1
# pres_adjusted_qc %in% c(1, 8) - pressure data marked as good
# doxy_adjusted_qc %in% c(1, 8) - doxy data marked as good
# !is.na(pres_adjusted) - pressure value must be present
# !is.na(doxy_adjusted) - doxy value must be present
bgc_data_doxy <- bgc_data %>%
filter(
pres_adjusted_qc %in% c(1, 8) &
doxy_adjusted_qc %in% c(1, 8) &
n_prof == opt_n_prof_sel &
!is.na(pres_adjusted) &
!is.na(doxy_adjusted)
) %>%
select(file_id,
pres_adjusted,
doxy_adjusted)
# join with metadata information and calculate depth field
bgc_data_doxy <- inner_join(bgc_metadata_select %>% select(file_id, lat),
bgc_data_doxy) %>%
mutate(depth = gsw_z_from_p(pres_adjusted, latitude = lat) * -1.0,
.before = pres_adjusted) %>%
select(-c(lat, pres_adjusted))
# ensure we have a depth, and doxy_adjusted for all rows in bgc_data_doxy
bgc_data_doxy <- bgc_data_doxy %>%
filter(!is.na(depth) & !is.na(doxy_adjusted))
# clean up working tables
rm(bgc_data, bgc_metadata)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1313120 70.2 2476715 132.3 2476715 132.3
Vcells 71918833 548.7 3356060008 25604.8 3484793727 26586.9Profile limits
Apply the rules that are determined by options set in set_options. Profile must cover a set range and not contain gaps.
# Determine profile min and max depths
bgc_profile_limits <- bgc_data_doxy %>%
group_by(file_id) %>%
summarise(
min_depth = min(depth),
max_depth = max(depth),
) %>%
ungroup()
# The profile much match at least one of the range criteria
force_min <- min(opt_profile_depth_range$min_depth)
force_max <- min(opt_profile_depth_range$max_depth)
# Apply profile min and max restrictions
bgc_apply_limits <- bgc_profile_limits %>%
filter(
min_depth <= force_min &
max_depth >= force_max
)
# Ensure working data set only contains profiles that have confrormed to the range test
bgc_data_doxy <- right_join(bgc_data_doxy,
bgc_apply_limits %>% select(file_id))
# Add profile type field and set all to 1.
# All profile that meet the minimum requirements are profile_range = 1
bgc_data_doxy <- bgc_data_doxy %>%
mutate(profile_range = 1)
for (i in 2:nrow(opt_profile_depth_range)) {
#i = 3
range_min <- opt_profile_depth_range[i,'min_depth']
range_max <- opt_profile_depth_range[i,'max_depth']
# Apply profile min and max restrictions
bgc_apply_limits <- bgc_profile_limits %>%
filter(min_depth <= range_min &
max_depth >= range_max) %>%
select(file_id) %>%
mutate (profile_range = i)
# Update profile range to i for these profiles
# bgc_data_temp <- full_join(bgc_data_temp, bgc_apply_limits) %>%
# filter(!is.na(min_depth))
bgc_data_doxy <-
bgc_data_doxy %>% rows_update(bgc_apply_limits, by = "file_id")
}
# Find the gaps within the profiles
profile_gaps <- full_join(bgc_data_doxy,
opt_profile_depth_range) %>%
filter(depth >= min_depth &
depth <= max_depth) %>%
select(file_id,
depth) %>%
arrange(file_id, depth) %>%
group_by(file_id) %>%
mutate(gap = depth - lag(depth, default = 0)) %>%
ungroup()
# Ensure we do not have gaps in the profile that invalidate it
for (i_gap in opt_gap_limit) {
# The limits to be applied in that pass of for loop
# i_gap <- opt_gap_limit[3]
i_gap_min = opt_gap_min_depth[which(opt_gap_limit == i_gap)]
i_gap_max = opt_gap_max_depth[which(opt_gap_limit == i_gap)]
# Which gaps are greater than i_gap
profile_gaps_remove <- profile_gaps %>%
filter(gap > i_gap) %>%
filter(depth >= i_gap_min & depth <= i_gap_max) %>%
distinct(file_id) %>%
pull()
# Remonve these profiles from working data set
bgc_data_doxy <- bgc_data_doxy %>%
filter(!file_id %in% profile_gaps_remove)
}
# clean up working tables
rm(bgc_profile_limits, profile_gaps, profile_gaps_remove, bgc_apply_limits)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1328238 71.0 2476715 132.3 2476715 132.3
Vcells 92807295 708.1 2684848007 20483.8 3484793727 26586.9# Write file that are observations prior to vertical alignment
bgc_data_doxy_full <- left_join(bgc_data_doxy, bgc_metadata_select)
bgc_data_doxy_full %>%
select(file_id, lat, lon, date, year, month, depth, doxy_adjusted) %>%
write_rds(file = paste0(path_argo_preprocessed, "/doxy_bgc_observed.rds"))
rm(bgc_data_doxy_full)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1328614 71.0 2476715 132.3 2476715 132.3
Vcells 92808056 708.1 2147878406 16387.1 3484793727 26586.9Vertical align doxy
We have a set of doxy profiles that match our criteria we now need to align that data set to match the depth that are in target_depth_range, this will match the range of climatology values in ucsd_clim
# create unique combinations of file_id and profile ranges
profile_range_file_id <-
bgc_data_doxy %>%
distinct(file_id, profile_range)
# select variable of interest and prepare target_depth field
bgc_data_doxy_clean <- bgc_data_doxy %>%
select(-profile_range) %>%
mutate(target_depth = depth, .after = depth)
rm(bgc_data_doxy)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1328258 71 2476715 132.3 2476715 132.3
Vcells 67367051 514 1718302725 13109.7 3484793727 26586.9# create all possible combinations of location, month and depth levels for interpolation
target_depth_grid <-
expand_grid(
target_depth_levels,
profile_range_file_id
)
# Constrain target_depth_grid to profile depth range
target_depth_grid <-
left_join(target_depth_grid, opt_profile_depth_range) %>%
filter(target_depth <= max_depth)
target_depth_grid <- target_depth_grid %>%
select(target_depth,
file_id)
# extend doxy depth vectors with target depths
bgc_data_doxy_extended <-
full_join(bgc_data_doxy_clean, target_depth_grid) %>%
arrange(file_id, target_depth)
rm(bgc_data_doxy_clean)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1330427 71.1 2476715 132.3 2476715 132.3
Vcells 112806447 860.7 1374642180 10487.7 3484793727 26586.9# predict spline interpolation on adjusted depth grid for doxy location and month
bgc_data_doxy_interpolated <-
bgc_data_doxy_extended %>%
group_by(file_id) %>%
mutate(doxy_spline = spline(target_depth, doxy_adjusted,
method = "natural",
xout = target_depth)$y) %>%
ungroup()
rm(bgc_data_doxy_extended)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1332199 71.2 3119584 166.7 3119584 166.7
Vcells 142379849 1086.3 879770996 6712.2 3484793727 26586.9# subset interpolated values on target depth range
bgc_data_doxy_interpolated_clean <-
inner_join(target_depth_levels, bgc_data_doxy_interpolated)
rm(bgc_data_doxy_interpolated)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1332246 71.2 3119584 166.7 3119584 166.7
Vcells 27168616 207.3 703816797 5369.7 3484793727 26586.9# select columns and rename to initial names
bgc_data_doxy_interpolated_clean <-
bgc_data_doxy_interpolated_clean %>%
select(file_id,
depth = target_depth,
doxy = doxy_spline)
# merge with profile range
bgc_data_doxy_interpolated_clean <-
full_join(bgc_data_doxy_interpolated_clean,
profile_range_file_id)
# merge with meta data
bgc_data_doxy_interpolated_clean <-
left_join(bgc_data_doxy_interpolated_clean,
bgc_metadata_select)Write files
Write the interpolated doxy profiles that map onto depth levels.
# Write files
bgc_data_doxy_interpolated_clean %>%
write_rds(file = paste0(path_argo_preprocessed, "/doxy_bgc_va.rds"))
# Rename so that names match if just reading existing files
doxy_bgc_va <- bgc_data_doxy_interpolated_clean
rm(bgc_data_doxy_interpolated_clean)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1331875 71.2 3119584 166.7 3119584 166.7
Vcells 43032647 328.4 563053438 4295.8 3484793727 26586.9read files
Read files that were previously created ready for analysis
# read files
doxy_bgc_va <- read_rds(file = paste0(path_argo_preprocessed, "/doxy_bgc_va.rds"))Analysis
doxy mean profile
max_depth_1 <- opt_profile_depth_range[1, "max_depth"]
max_depth_2 <- opt_profile_depth_range[2, "max_depth"]
max_depth_3 <- opt_profile_depth_range[3, "max_depth"]
# Profiles to 600m
doxy_overall_mean_1 <- doxy_bgc_va %>%
filter(profile_range %in% c(1, 2, 3) & depth <= max_depth_1) %>%
group_by(depth) %>%
summarise(count_measures = n(),
doxy_mean = mean(doxy, na.rm = TRUE),
doxy_sd = sd(doxy, na.rm = TRUE))
doxy_year_mean_1 <- doxy_bgc_va %>%
filter(profile_range %in% c(1, 2, 3) & depth <= max_depth_1) %>%
group_by(year, depth) %>%
summarise(count_measures = n(),
doxy_mean = mean(doxy, na.rm = TRUE),
doxy_sd = sd(doxy, na.rm = TRUE))
# Profiles to 1200m
doxy_overall_mean_2 <- doxy_bgc_va %>%
filter(profile_range %in% c(2, 3) & depth <= max_depth_2) %>%
group_by(depth) %>%
summarise(count_measures = n(),
doxy_mean = mean(doxy, na.rm = TRUE),
doxy_sd = sd(doxy, na.rm = TRUE))
doxy_year_mean_2 <- doxy_bgc_va %>%
filter(profile_range %in% c(2, 3) & depth <= max_depth_2) %>%
group_by(year, depth) %>%
summarise(count_measures = n(),
doxy_mean = mean(doxy, na.rm = TRUE),
doxy_sd = sd(doxy, na.rm = TRUE))
# Profiles to 1500m
doxy_overall_mean_3 <- doxy_bgc_va %>%
filter(profile_range %in% c(3) & depth <= max_depth_3) %>%
group_by(depth) %>%
summarise(count_measures = n(),
doxy_mean = mean(doxy, na.rm = TRUE),
doxy_sd = sd(doxy, na.rm = TRUE))
doxy_year_mean_3 <- doxy_bgc_va %>%
filter(profile_range %in% c(3) & depth <= max_depth_3) %>%
group_by(year, depth) %>%
summarise(count_measures = n(),
doxy_mean = mean(doxy, na.rm = TRUE),
doxy_sd = sd(doxy, na.rm = TRUE))
# All years
doxy_overall_mean_1 %>%
ggplot()+
geom_path(aes(x = doxy_mean,
y = depth))+
geom_ribbon(aes(
xmax = doxy_mean + doxy_sd,
xmin = doxy_mean - doxy_sd,
y = depth
),
alpha = 0.2) +
scale_y_reverse()+
coord_cartesian(xlim = opt_xlim)+
scale_x_continuous(breaks = opt_xbreaks)+
labs(
title = paste0('Overall mean dissolved oxygen to ', max_depth_1, 'm'),
x=opt_measure_label,
y='depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
doxy_overall_mean_2 %>%
ggplot()+
geom_path(aes(x = doxy_mean,
y = depth))+
geom_ribbon(aes(
xmax = doxy_mean + doxy_sd,
xmin = doxy_mean - doxy_sd,
y = depth
),
alpha = 0.2) +
scale_y_reverse()+
coord_cartesian(xlim = opt_xlim)+
scale_x_continuous(breaks = opt_xbreaks)+
labs(
title = paste0('Overall mean dissolved oxygen to ', max_depth_2, 'm'),
x=opt_measure_label,
y='depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
doxy_overall_mean_3 %>%
ggplot()+
geom_path(aes(x = doxy_mean,
y = depth))+
geom_ribbon(aes(
xmax = doxy_mean + doxy_sd,
xmin = doxy_mean - doxy_sd,
y = depth
),
alpha = 0.2) +
scale_y_reverse()+
coord_cartesian(xlim = opt_xlim)+
scale_x_continuous(breaks = opt_xbreaks)+
labs(
title = paste0('Overall mean dissolved oxygen to ', max_depth_3, 'm'),
x=opt_measure_label,
y='depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
# by years
doxy_year_mean_1 %>%
ggplot()+
geom_path(aes(x = doxy_mean,
y = depth))+
geom_ribbon(aes(
xmax = doxy_mean + doxy_sd,
xmin = doxy_mean - doxy_sd,
y = depth
),
alpha = 0.2) +
scale_y_reverse()+
facet_wrap(~year)+
coord_cartesian(xlim = opt_xlim)+
scale_x_continuous(breaks = opt_xbreaks)+
labs(
title = paste0('Yearly overall mean dissolved oxygen to ', max_depth_1, 'm'),
x = opt_measure_label,
y = 'depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
doxy_year_mean_2 %>%
ggplot()+
geom_path(aes(x = doxy_mean,
y = depth))+
geom_ribbon(aes(
xmax = doxy_mean + doxy_sd,
xmin = doxy_mean - doxy_sd,
y = depth
),
alpha = 0.2) +
scale_y_reverse()+
facet_wrap(~year)+
coord_cartesian(xlim = opt_xlim)+
scale_x_continuous(breaks = opt_xbreaks)+
labs(
title = paste0('Yearly overall mean dissolved oxygen to ', max_depth_2, 'm'),
x = opt_measure_label,
y = 'depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
doxy_year_mean_3 %>%
ggplot()+
geom_path(aes(x = doxy_mean,
y = depth))+
geom_ribbon(aes(
xmax = doxy_mean + doxy_sd,
xmin = doxy_mean - doxy_sd,
y = depth
),
alpha = 0.2) +
scale_y_reverse()+
facet_wrap(~year)+
coord_cartesian(xlim = opt_xlim)+
scale_x_continuous(breaks = opt_xbreaks)+
labs(
title = paste0('Yearly overall mean dissolved oxygen to ', max_depth_3, 'm'),
x = opt_measure_label,
y = 'depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
Profile counts
Details of the number of profiles and to which depths over the analysis period
doxy_histogram <- doxy_bgc_va %>%
group_by(year, profile_range = as.character(profile_range)) %>%
summarise(num_profiles = n_distinct(file_id)) %>%
ungroup()
doxy_histogram %>%
ggplot() +
geom_bar(
aes(
x = year,
y = num_profiles,
fill = profile_range,
group = profile_range
),
position = "stack",
stat = "identity"
) +
scale_fill_viridis_d() +
labs(title = "dissolved oxygen profiles per year and profile range",
x = "year",
y = "profile count",
fill = "profile range")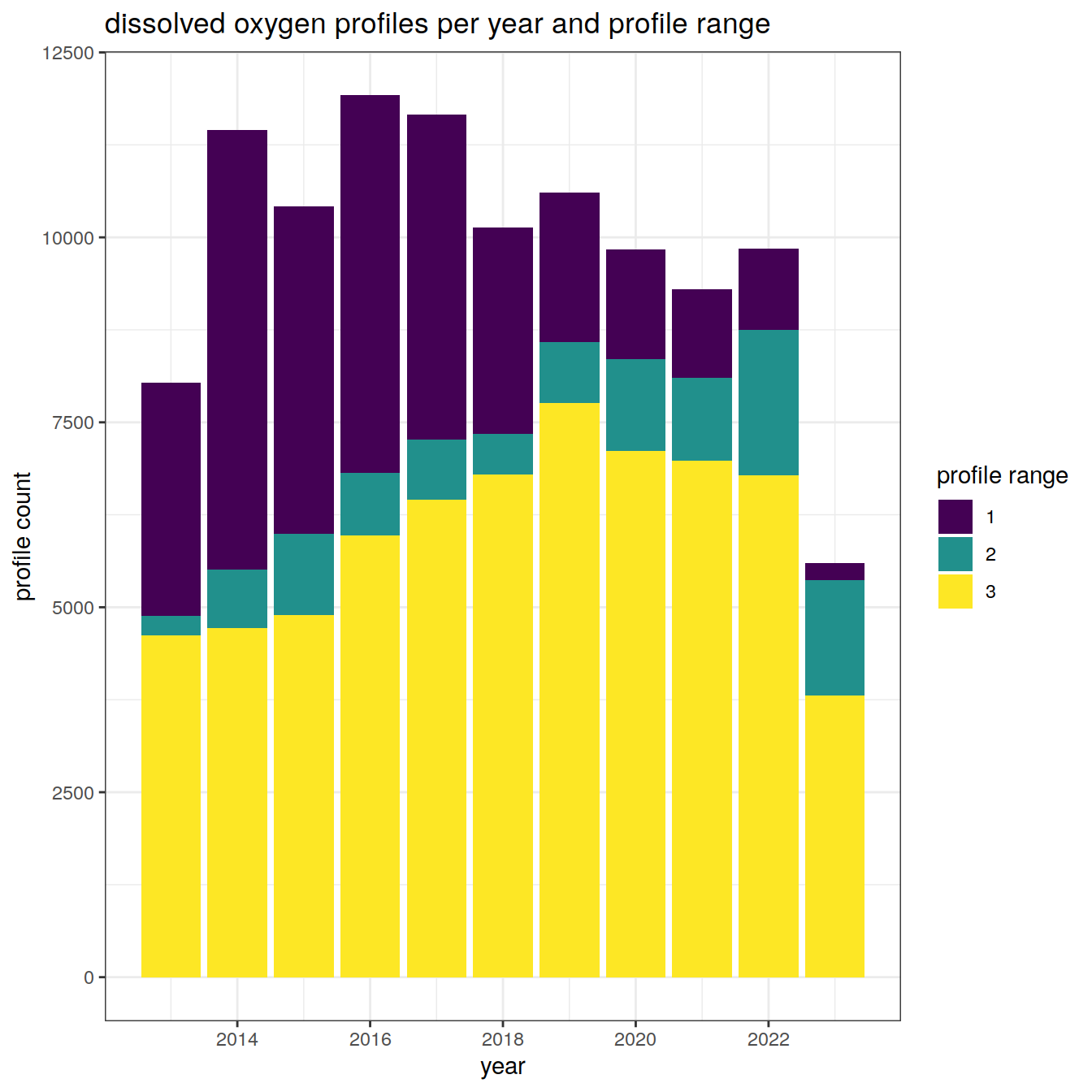
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
sessionInfo()R version 4.2.2 (2022-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.5
Matrix products: default
BLAS: /usr/local/R-4.2.2/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.2.2/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] gsw_1.1-1 gridExtra_2.3 lubridate_1.9.0 timechange_0.1.1
[5] argodata_0.1.0 forcats_0.5.2 stringr_1.5.0 dplyr_1.1.3
[9] purrr_1.0.2 readr_2.1.3 tidyr_1.3.0 tibble_3.2.1
[13] ggplot2_3.4.4 tidyverse_1.3.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.10 assertthat_0.2.1 rprojroot_2.0.3
[4] digest_0.6.30 utf8_1.2.2 R6_2.5.1
[7] cellranger_1.1.0 backports_1.4.1 reprex_2.0.2
[10] evaluate_0.18 highr_0.9 httr_1.4.4
[13] pillar_1.9.0 rlang_1.1.1 googlesheets4_1.0.1
[16] readxl_1.4.1 rstudioapi_0.15.0 whisker_0.4
[19] jquerylib_0.1.4 rmarkdown_2.18 labeling_0.4.2
[22] googledrive_2.0.0 munsell_0.5.0 broom_1.0.5
[25] compiler_4.2.2 httpuv_1.6.6 modelr_0.1.10
[28] xfun_0.35 pkgconfig_2.0.3 htmltools_0.5.3
[31] tidyselect_1.2.0 workflowr_1.7.0 viridisLite_0.4.1
[34] fansi_1.0.3 crayon_1.5.2 withr_2.5.0
[37] tzdb_0.3.0 dbplyr_2.2.1 later_1.3.0
[40] grid_4.2.2 jsonlite_1.8.3 gtable_0.3.1
[43] lifecycle_1.0.3 DBI_1.1.3 git2r_0.30.1
[46] magrittr_2.0.3 scales_1.2.1 cli_3.6.1
[49] stringi_1.7.8 cachem_1.0.6 farver_2.1.1
[52] fs_1.5.2 promises_1.2.0.1 xml2_1.3.3
[55] bslib_0.4.1 ellipsis_0.3.2 generics_0.1.3
[58] vctrs_0.6.4 tools_4.2.2 glue_1.6.2
[61] RNetCDF_2.6-1 hms_1.1.2 fastmap_1.1.0
[64] yaml_2.3.6 colorspace_2.0-3 gargle_1.2.1
[67] rvest_1.0.3 knitr_1.41 haven_2.5.1
[70] sass_0.4.4