Marine Heat Waves anomaly profiles
Marguerite Larriere & Jens Daniel Müller
20 May, 2024
Last updated: 2024-05-20
Checks: 7 0
Knit directory:
bgc_argo_r_argodata/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211008) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5304024. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rproj.user/
Ignored: analysis/figure/
Untracked files:
Untracked: analysis/draft.Rmd
Untracked: load_argo_core_output.txt
Unstaged changes:
Modified: analysis/CESM_comparison.Rmd
Deleted: analysis/MHWs_categorisation.Rmd
Modified: analysis/_site.yml
Modified: analysis/anomaly_SST_2023.Rmd
Modified: analysis/child/cluster_analysis_base.Rmd
Modified: analysis/coverage_maps_North_Atlantic.Rmd
Modified: analysis/load_broullon_DIC_TA_clim.Rmd
Modified: code/Workflowr_project_managment.R
Modified: code/start_background_job.R
Modified: code/start_background_job_load.R
Modified: code/start_background_job_partial.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/MHWs_vertical_anomaly.Rmd)
and HTML (docs/MHWs_vertical_anomaly.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5304024 | mlarriere | 2024-05-20 | choosing extent |
| html | b0fb164 | mlarriere | 2024-05-16 | Build site. |
| Rmd | fbf0c64 | mlarriere | 2024-05-16 | latitude, longitude and day of the month graphs |
| html | 22e0206 | mlarriere | 2024-05-16 | Build site. |
| Rmd | d0f58b6 | mlarriere | 2024-05-16 | latitude, longitude and day of the month graphs |
| html | 96d4b76 | mlarriere | 2024-05-14 | Build site. |
| Rmd | 91e6028 | mlarriere | 2024-05-13 | adding subsection CESM comparison |
| html | af6594f | mlarriere | 2024-05-13 | Build site. |
| Rmd | 30f9250 | mlarriere | 2024-05-13 | Adding CESM subsection |
Task
Focusing on 2023 Only core Argo - focus on temperature anomalies
Dependencies
temp_core_va.rds - temperature of core argo floats after vertical alignment.
core_metadata.rds - File with metadata concerning the floats such as platform number, cycle number, date, lat, lon and quality control results.
temp_anomaly_va.rds - file containing the temperature anomalies (temp core - climatology).
2023_mhw_raw.csv - CSV file containing the categorization of surface marine heatwaves, in 2023 and in a 0.25°x0.25° grid.
Outputs
2023_surface_mhws_1x1.rds - file containing the categorization of surface marine heatwaves in a 1°x1° grid, in 2023.
path_emlr_utilities <- "/nfs/kryo/work/jenmueller/emlr_cant/utilities/files/"
path_basin_mask <- "/nfs/kryo/work/datasets/gridded/ocean/interior/reccap2/supplementary/"
path_argo <- '/nfs/kryo/work/datasets/ungridded/3d/ocean/floats/bgc_argo'
path_argo_core <- '/nfs/kryo/work/datasets/ungridded/3d/ocean/floats/core_argo_r_argodata_2024-03-13'
path_argo_core_preprocessed <- paste0(path_argo_core, "/preprocessed_core_data")
path_mhw<- '/net/kryo/work/datasets/gridded/ocean/2d/obs/mhw'#Area of interest: North Atlantic north west - lat:(60,30), lon:(-70,-30), North Atlantic east - lat:(0,40), lon:(-30,0)
chosen_extent <- list(
lat_min = 0, #30
lat_max = 40, #60
lon_min = -30, #-70
lon_max = 0 #-30
)
name_extent<- "East" #NorthwestLoad data
Load biomes
Load Argo floats data
Load SST anomaly
#Read SST anomaly, computed using climatology:2009-2019 (from anomaly_SST_2023.Rmd)
sst_anomaly_northAtlantic<- read_rds(paste(path_argo_core_preprocessed, "/SST_anomaly2023_NorthAtlantic_clim2004-2019.rds", sep = ""))Argo floats
Distribution over North Atlantic basin
#Histogram -- number of floats per month and biome
unique_platforms_per_month <- core_anomaly_with_platform_2023 %>%
group_by(month) %>%
filter(lat>0, lat<70, lon>-80, lon<0) %>%
summarize(unique_platforms = n_distinct(platform_number))
hist <- ggplot(unique_platforms_per_month, aes(x = factor(month), y = unique_platforms)) +
geom_bar(stat = "identity", fill = "darkred") +
labs(title = "Amount of platform per month, in North Atlantic, 2023",
x = "Months", y = "Number of argo floats", fill = "Biomes") +
theme_minimal() +
guides(fill = FALSE)
# scale_fill_manual(values = c("1" = "#1034A6", "2" = "#f59c04", "3" = "darkred"),
# labels = c("1" = "SPSS", "2" = "STSS", "3" = "STPS")) +
# theme_minimal()
print(hist)
#Number of cycle per platform
# cycle_counts <- core_anomaly_with_platform_2023 %>%
# group_by(platform_number) %>%
# summarise(cycle_count = n_distinct(cycle_number))
# print(cycle_counts)#Join the SST anomaly with the anomaly profiles
core_anomaly_with_platform_2023<-core_anomaly_with_platform_2023 %>%
filter(!is.na(depth)) %>%
select(-profile_range, -day_of_year)
complete_anomaly_profile<- core_anomaly_with_platform_2023 %>%
inner_join(sst_anomaly_northAtlantic, by = c("lat", "lon", "month"))
#---Chosen subset
core_anomaly_2023_natlantic_subset<- complete_anomaly_profile %>%
filter(lat > chosen_extent$lat_min, lat < chosen_extent$lat_max,
lon > chosen_extent$lon_min, lon < chosen_extent$lon_max) %>%
select(-interpolated_temp)
cycles_per_platform <-core_anomaly_2023_natlantic_subset %>%
group_by(platform_number) %>%
summarize(unique_cycles = toString(unique(cycle_number)))
months_per_float <- core_anomaly_2023_natlantic_subset %>%
group_by(platform_number) %>%
summarize(unique_months = toString(unique(month)))Distribution over East-North Atlantic
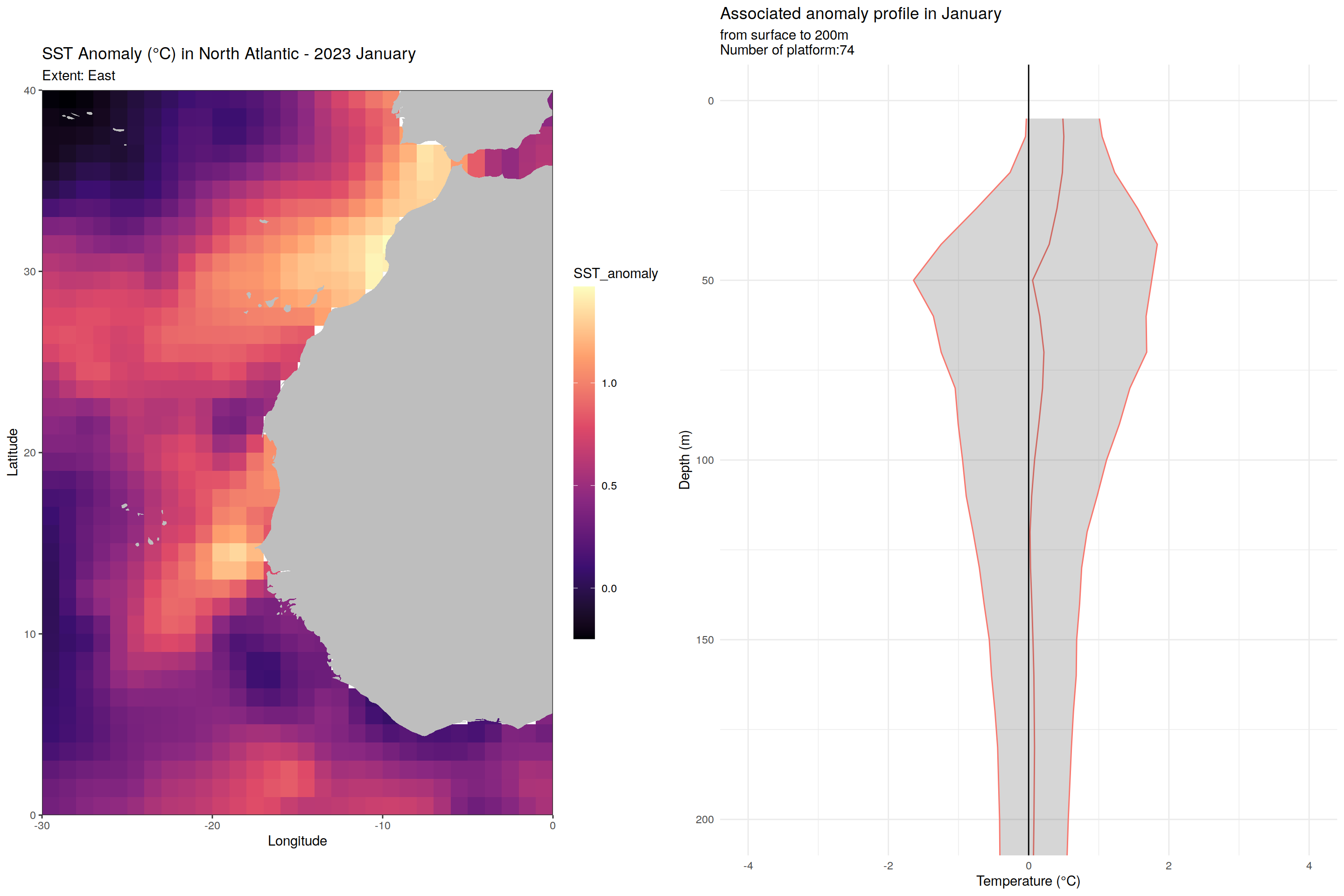
Using the SST map computed in “anomaly_SST_2023.Rmd” and ClimateReanalyser, we identify 2 areas of particular interest for SST anomalies in 2023 in the North Atlantic Ocean:
Northeast, near the Canada/USA coast. SST anomaly particularly strong in summer and autumn (JJA and SON).
East coast of the North Atlantic Ocean. Here, SST anomalies are high on an annual basis, with a sharp increase from June onwards.
#PLatfrom in the east north atlantic over 2023
platform_counts <- aggregate(platform_number ~ month, data = core_anomaly_2023_natlantic_subset, FUN = function(x) length(unique(x)))
cycle_count_per_platform_month <- core_anomaly_2023_natlantic_subset %>%
group_by(month, platform_number) %>%
summarise(cycle_count = n_distinct(cycle_number))
custom_labeller <- function(variable, value) {
month_count <- platform_counts[platform_counts$month == value, "platform_number"]
return(paste("Month:", value, "\nNumber of floats:", month_count))
}
#PLot
sst_anomaly_northAtlantic_subset<-sst_anomaly_northAtlantic %>%
filter(lat > chosen_extent$lat_min, lat < chosen_extent$lat_max,
lon > chosen_extent$lon_min, lon < chosen_extent$lon_max)
ggplot() +
geom_tile(data=sst_anomaly_northAtlantic_subset, aes(x = lon, y = lat, fill = SST_anomaly)) + #tile with SST
geom_point(data = core_anomaly_2023_natlantic_subset, aes(x = lon, y = lat), color = "black") + # Point for float position
geom_map(data = world_coordinates, map = world_coordinates, aes(long, lat, map_id = region), fill = "grey") + #base map
lims(x = c(chosen_extent$lon_min, chosen_extent$lon_max), y=c(chosen_extent$lat_min, chosen_extent$lat_max)) +
coord_quickmap(expand = 0) +
scale_fill_viridis_c(option = "magma") +
labs(title = "Platform Locations",
subtitle = "Resolution: 1°x1°, SST from SOM",
color = "Months with float") +
theme(plot.title = element_text(size = 16),
plot.subtitle = element_text(size = 12),
legend.text = element_text(size = 10),
legend.title = element_text(size = 12, face = "bold"),
legend.key.width = unit(0.5, "cm"),
legend.key.height = unit(2, "cm"))+
facet_wrap(~month, ncol = 3, labeller = custom_labeller)
Anomaly profiles
Individual float
unique_platform<-core_anomaly_2023_natlantic_subset %>%
filter(platform_number==6904231)
# ggplot(unique_platform, aes(x = anomaly, y = depth, color = factor(cycle_number))) +
# geom_path() +
# geom_vline(xintercept = 0) +
# scale_y_reverse() +
# coord_cartesian(xlim = c(-6, 6), ylim = c(200, 0)) +
# labs(title = 'Anomaly profiles by month',
# subtitle= paste0('Platform ', unique(unique_platform$platform_number)),
# x = 'Temperature (°C)', y = 'Depth (m)', color = 'Cycle Number') +
# scale_color_viridis(discrete = TRUE) +
# facet_wrap(~month, scales = "free", ncol = 3)
ggplot() +
geom_path(data=unique_platform, aes(x = anomaly, y = depth, color = factor(month), group = cycle_number)) +
geom_vline(xintercept = 0) +
scale_y_reverse() +
coord_cartesian(xlim = c(-6, 6), ylim = c(200, 0)) +
scale_color_manual(values = colorRampPalette(c("#2796A5", "#F3712B", "#880D1E"))(12)) +
labs(title = 'SST anomalies propagation - 1 float',
x = 'Temperature (°C)', y = 'Depth (m)', color = 'Months') +
theme(plot.title = element_text(size = 16),
plot.subtitle = element_text(size = 12),
legend.text = element_text(size = 10),
legend.title = element_text(size = 12, face = "bold"))
# unique_platform_base_map<- base_map + lims(x=c(-30,-10), y=c(0,20))
# unique_coords_with_cycles <- unique_platform %>%
# distinct(lat, lon, .keep_all = TRUE) %>%
# group_by(lat, lon) %>%
# summarise(cycle_numbers = toString(unique(cycle_number)))
# #base map and SST anomaly
# map_anomaly_east <- unique_platform_base_map +
# geom_tile(data = east_sst_anomaly, aes(x = lon, y = lat, fill = SST_anomaly)) +
# scale_fill_gradientn(colors = continuous_palette, limits = scale_limits, breaks = scale_breaks) +
# labs(fill = "SST Anomaly") +
# theme_minimal()
#
# #trajetory
# map_anomaly_east +
# geom_point(data = unique_coords_with_cycles, aes(x = lon, y = lat, color = cycle_numbers), size = 2) +
# scale_color_discrete(name = "Cycle Number", guide = guide_legend(title.position = "top")) +
# labs(title = 'Float position') +
# theme(legend.position = "bottom")+
# guides(fill = guide_colorbar(barwidth = 20, barheight = 1, title.position = "top"))Coloring by lat/lon
#Calculating monthly mean anomaly + std for each lat/lon pair of the area
anomaly_lat_lon <- core_anomaly_2023_natlantic_subset %>%
group_by(lat, lon, depth, month, platform_number, cycle_number) %>%
summarise(
temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE)
)
# Longitude - gradient north-south
longitude<- ggplot(anomaly_lat_lon, aes(x = temp_anomaly_mean, y = depth, group = interaction(platform_number, cycle_number), color = as.numeric(lon))) +
geom_path() +
geom_vline(xintercept = 0) +
scale_y_reverse(limits = c(200, 0)) +
coord_cartesian(xlim = c(-6, 6)) +
facet_wrap(~ month, ncol = 3) +
labs(title = "Anomaly Profiles for all longitudes",
subtitle = "All floats and cycles included",
x = "Temperature (°C)", y = "Depth (m)", color = "Longitude") +
scale_color_viridis_c()
# Latitude - gradient west-east
latitude<- ggplot(anomaly_lat_lon, aes(x = temp_anomaly_mean, y = depth, group = interaction(platform_number, cycle_number), color = as.numeric(lat))) +
geom_path() +
geom_vline(xintercept = 0) +
scale_y_reverse(limits = c(200, 0)) +
coord_cartesian(xlim = c(-6, 6)) +
facet_wrap(~ month, ncol = 3) +
labs(title = "Anomaly Profile for all latitudes",
subtitle = "All floats and cycles included",
x = "Temperature (°C)", y = "Depth (m)", color = "Latitude") +
scale_color_viridis_c()
combined_plot <- latitude + longitude + plot_layout(ncol = 2)
combined_plot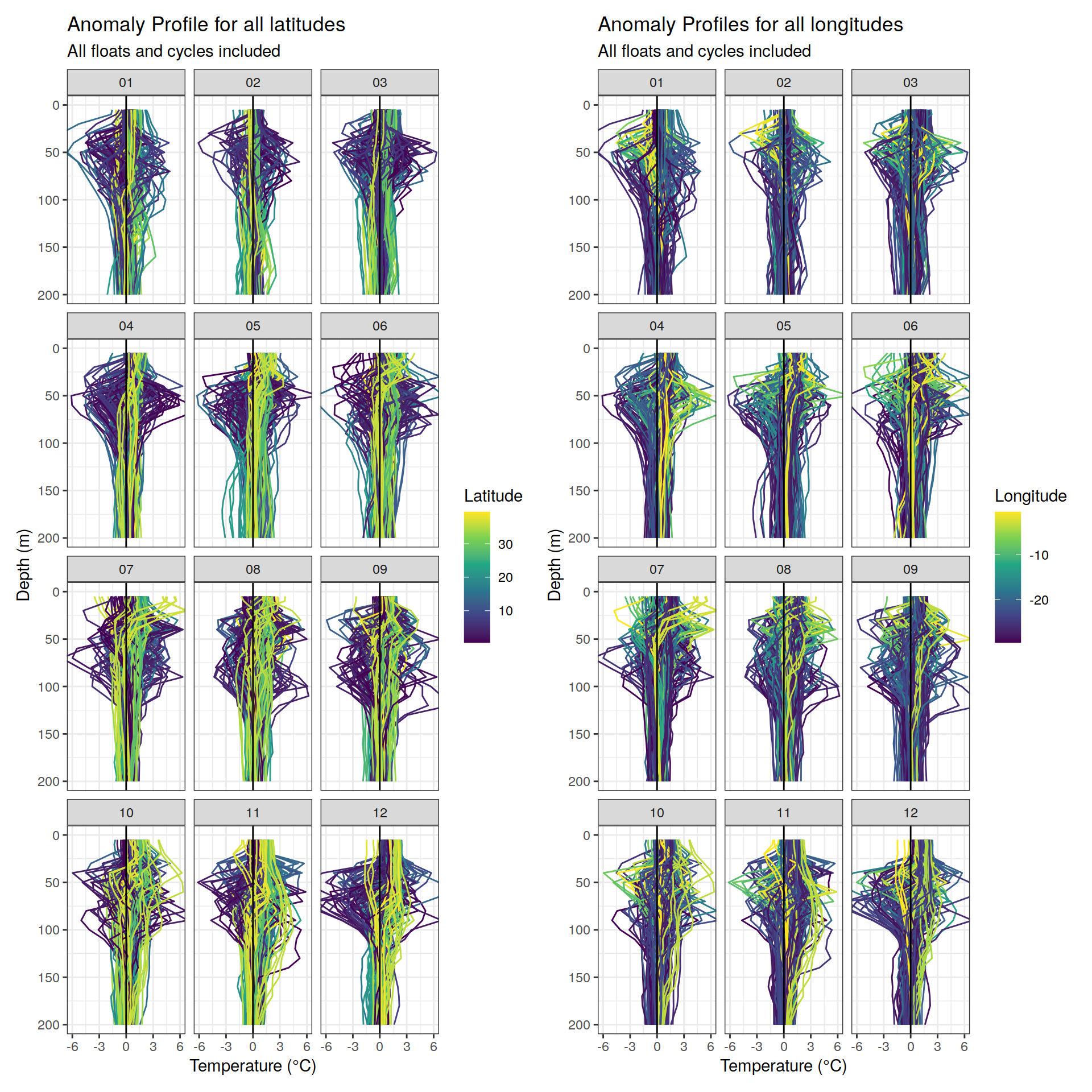
Coloring by SST anomaly
# calculate mean anomaly data
anomaly_summary <- core_anomaly_2023_natlantic_subset %>%
group_by(platform_number, depth, month, cycle_number, SST_anomaly) %>%
summarise(
temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE)
)
#plot for the SST anomalies
ggplot(anomaly_summary, aes(x = temp_anomaly_mean, y = depth, group = interaction(platform_number, cycle_number), color = as.numeric(SST_anomaly))) +
geom_path() +
geom_vline(xintercept = 0) +
scale_y_reverse(limits = c(200, 0)) +
coord_cartesian(xlim = c(-6, 6)) +
facet_wrap(~ month, ncol = 3) +
labs(title = "Mean Anomaly Profile as a function of SST anomaly",
x = "Temperature (°C)", y = "Depth (m)", color = "SST Anomaly") +
scale_color_viridis_c()
Day within the month
# calculate mean anomaly data
anomaly_summary_day <- core_anomaly_2023_natlantic_subset %>%
group_by(platform_number, depth, month, cycle_number, day) %>%
summarise(
temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE)
)
#plot for time during month
ggplot(anomaly_summary_day, aes(x = temp_anomaly_mean, y = depth, group = interaction(platform_number, cycle_number), color = as.numeric(day))) +
geom_path() +
geom_vline(xintercept = 0) +
scale_y_reverse(limits = c(200, 0)) +
coord_cartesian(xlim = c(-6, 6)) +
facet_wrap(~ month, ncol = 3) +
labs(title = "Mean Anomaly Profile",
x = "Temperature (°C)", y = "Depth (m)", color = "day within the month") +
scale_color_viridis_c()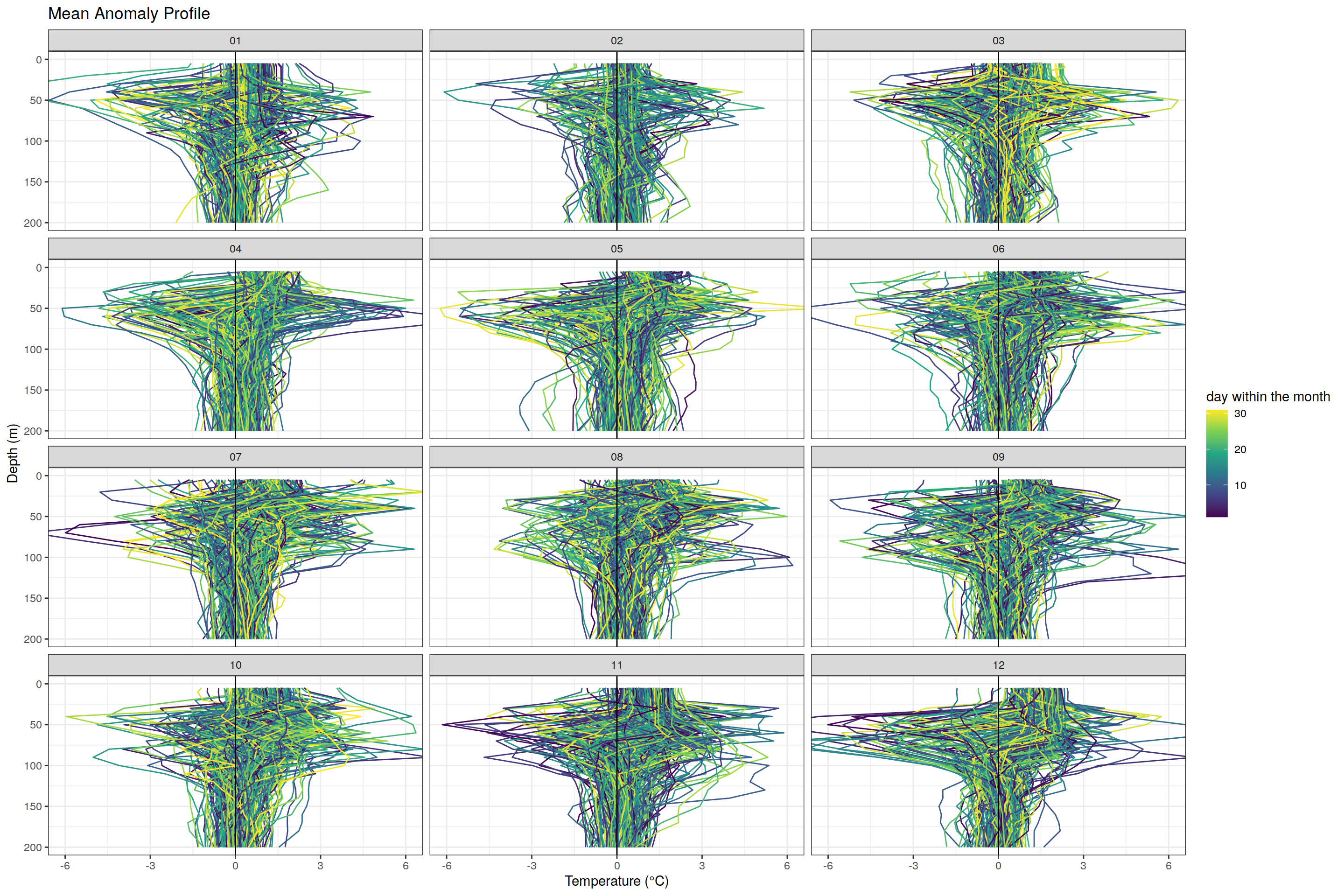
Monthly anomaly profiles
# datsets with the cycle number per float across months
cycles_per_platform_month<- core_anomaly_2023_natlantic_subset %>%
group_by(platform_number, month) %>%
summarize(unique_cycles = toString(unique(cycle_number)))
# Calculating monthly mean anomaly over the east area by averaging the anomaly of each float present
anomaly_mean <- core_anomaly_2023_natlantic_subset %>%
group_by(depth, month) %>%
summarise(temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE),
temp_anomaly_sd = sd(anomaly, na.rm = TRUE))
#For each mont put the SST anomaly next to the anomaly path
for (m in unique(core_anomaly_2023_natlantic_subset$month)) {
# Filter data
map_data <- filter(sst_anomaly_northAtlantic_subset, month == m)
anomaly_data <- filter(anomaly_mean, month == m)
# Plot for temperature anomaly map
map_plot <- ggplot() +
geom_tile(data=map_data, aes(x = lon, y = lat, fill = SST_anomaly)) +
geom_map(data = world_coordinates, map = world_coordinates, aes(long, lat, map_id = region), fill = "grey") + #base map
lims(x = c(chosen_extent$lon_min, chosen_extent$lon_max), y=c(chosen_extent$lat_min, chosen_extent$lat_max)) +
coord_quickmap(expand = 0) +
scale_fill_viridis_c(option = "magma") +
labs(title = paste("SST Anomaly (°C) in North Atlantic - 2023", month.name[as.numeric(m)]),
subtitle = paste0("Extent: ", name_extent),
x = "Longitude", y = "Latitude") +
theme(legend.position = 'right', legend.key.height = unit(2, "cm"))
# Plot for anomaly profiles
anomaly_plot <- ggplot(anomaly_data, aes(x = temp_anomaly_mean, y = depth, color = factor(month))) +
geom_path() +
geom_ribbon(aes(xmax = temp_anomaly_mean + temp_anomaly_sd,
xmin = temp_anomaly_mean - temp_anomaly_sd,
y = depth), alpha = 0.2) +
geom_vline(xintercept = 0) +
scale_y_reverse() +
coord_cartesian(xlim = c(-4, 4), ylim = c(200, 0)) +
labs(title = paste('Associated anomaly profile in', month.name[as.numeric(m)]),
subtitle = paste0("from surface to 200m", "\nNumber of platform:", nrow(filter(cycles_per_platform_month, month==m))),
x = 'Temperature (°C)', y = 'Depth (m)') +
theme_minimal()+
guides(color = FALSE)
#plots side by side
combined_plot <- grid.arrange(map_plot, anomaly_plot, ncol = 2)
print(combined_plot)
}
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]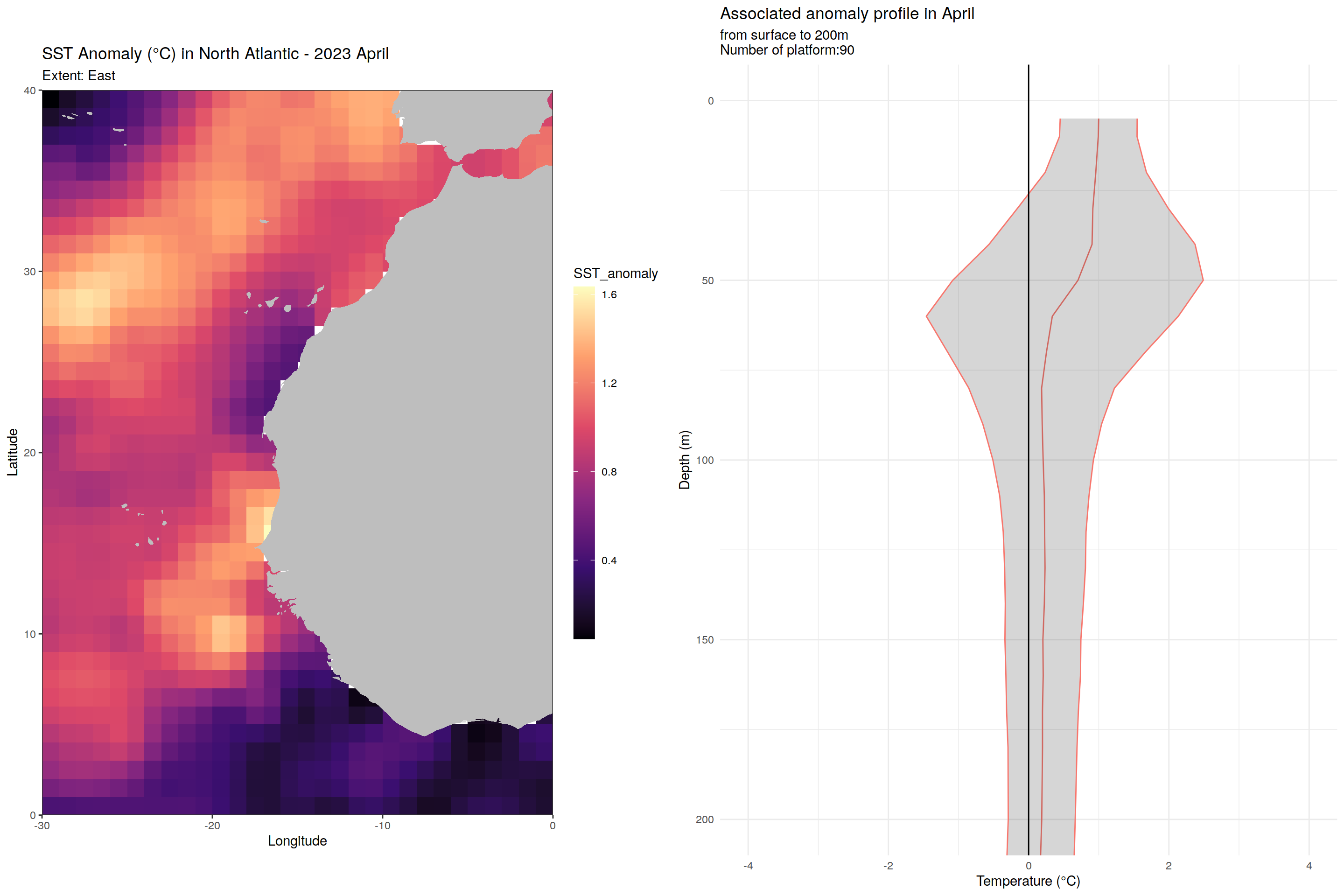
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]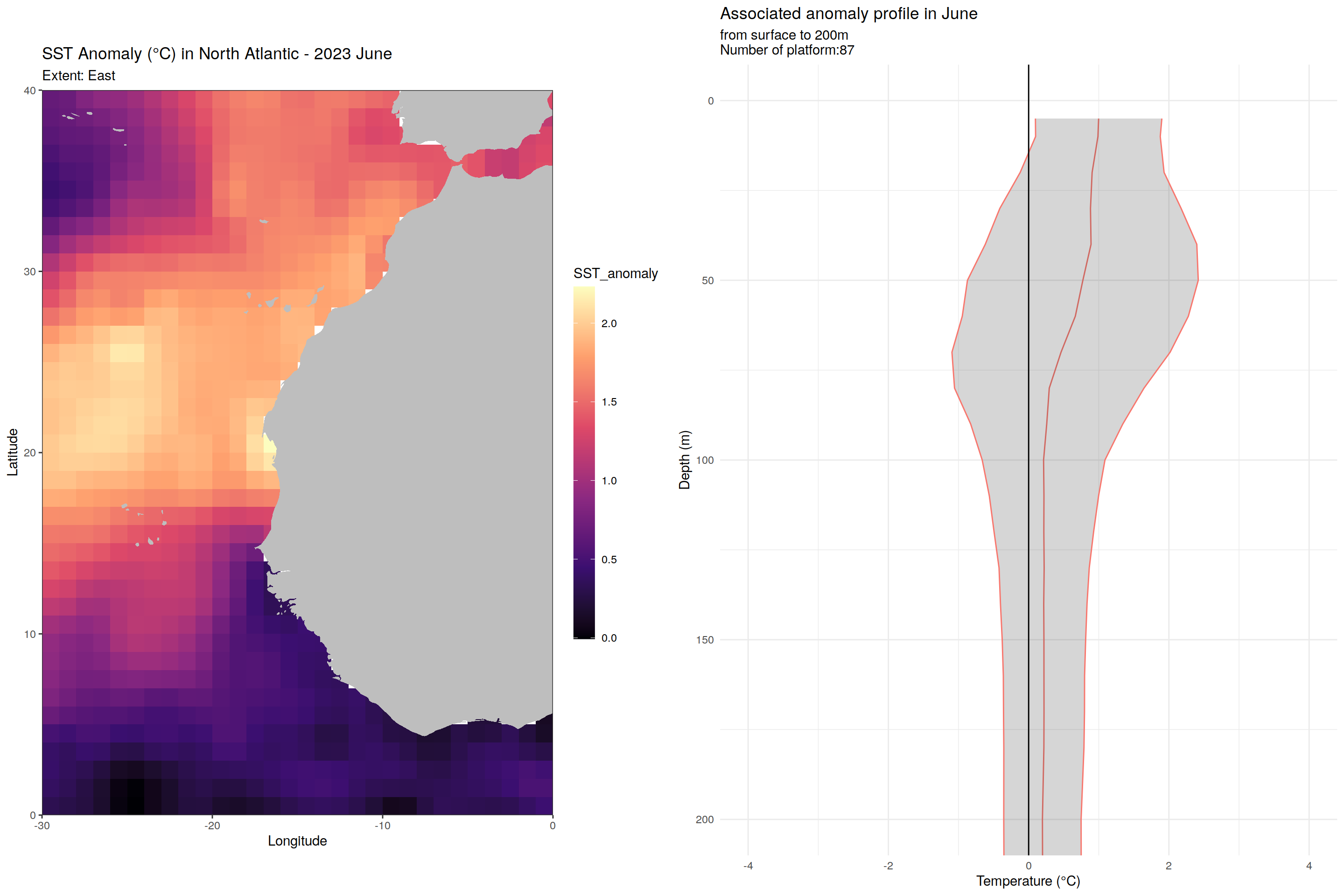
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]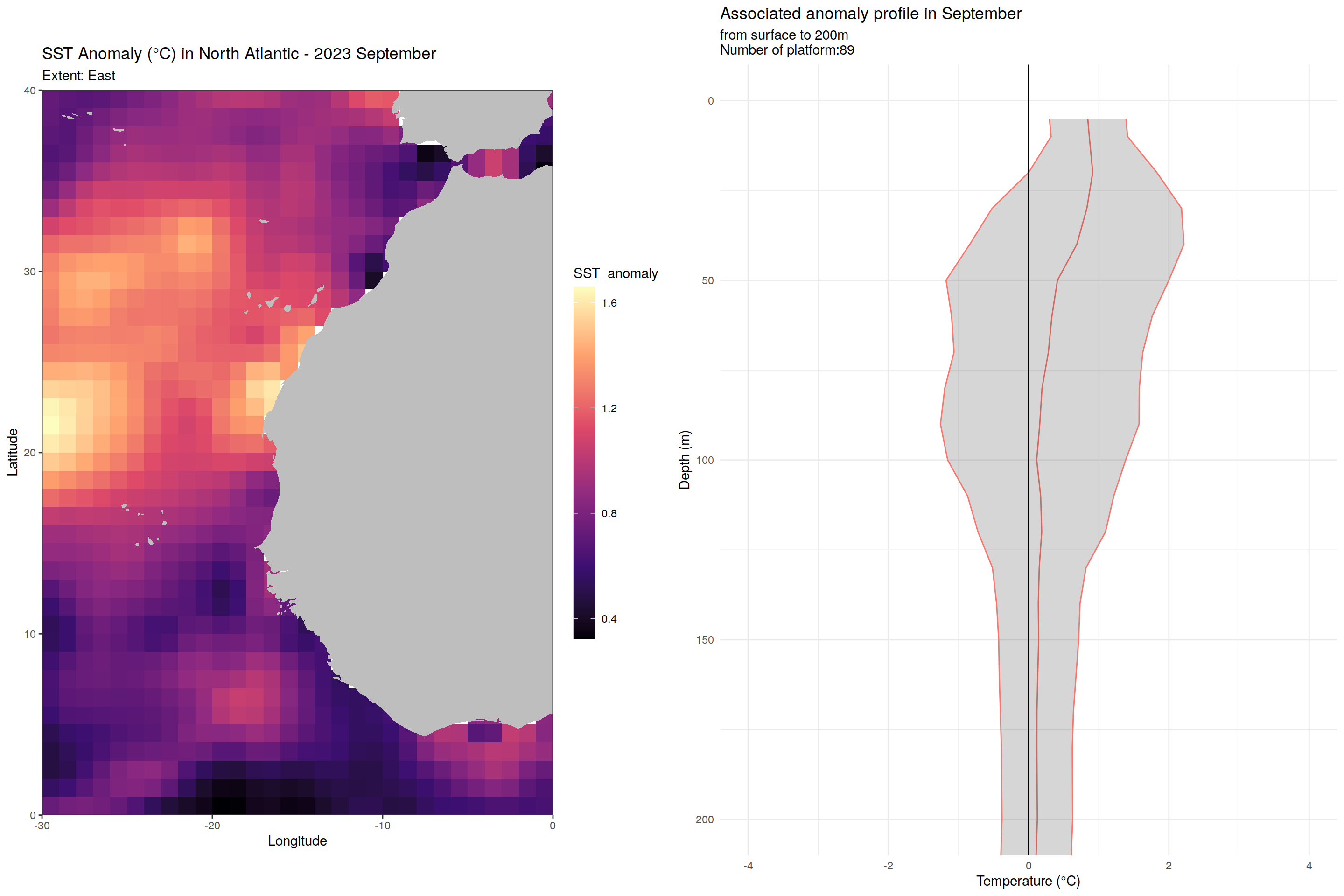
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]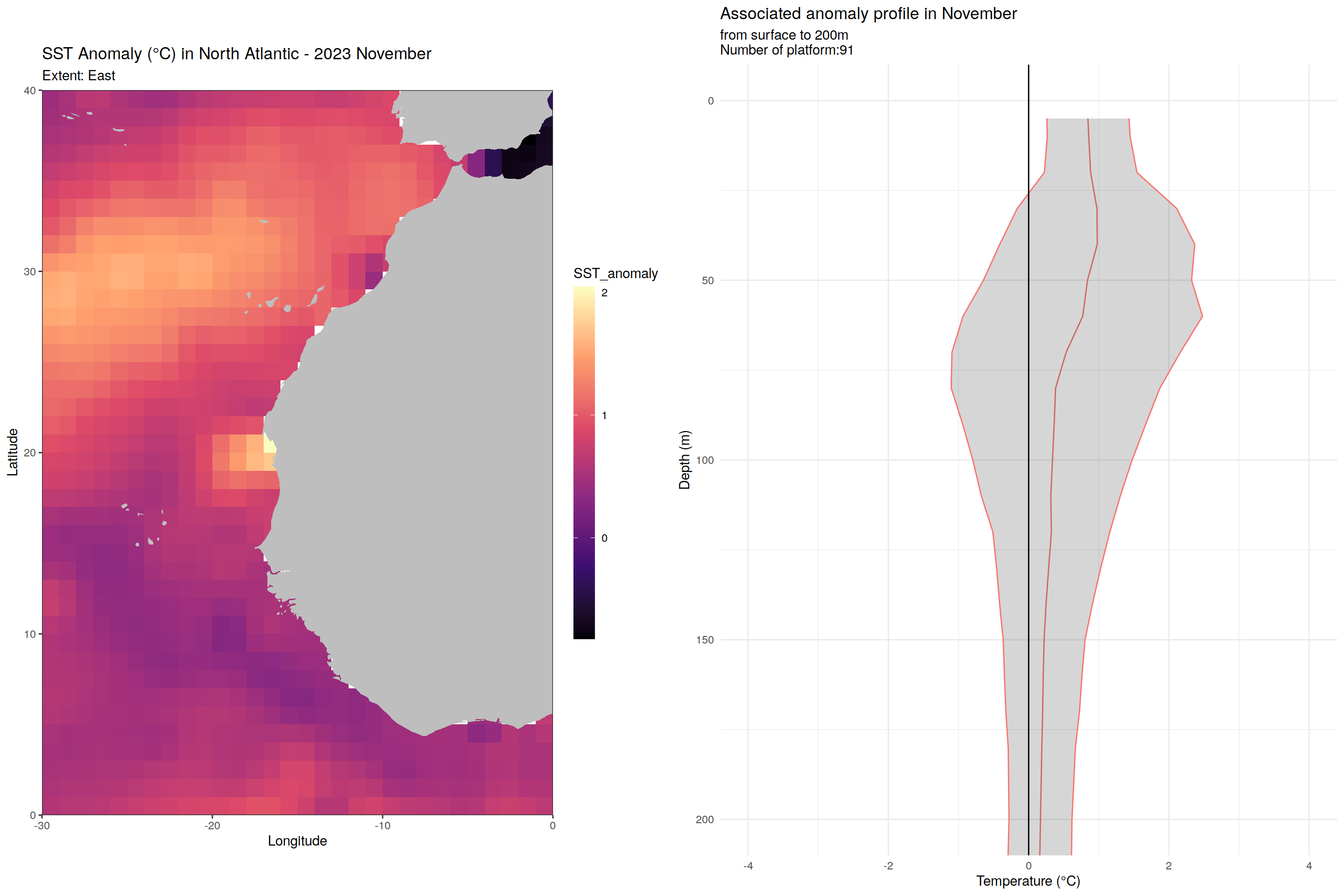
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]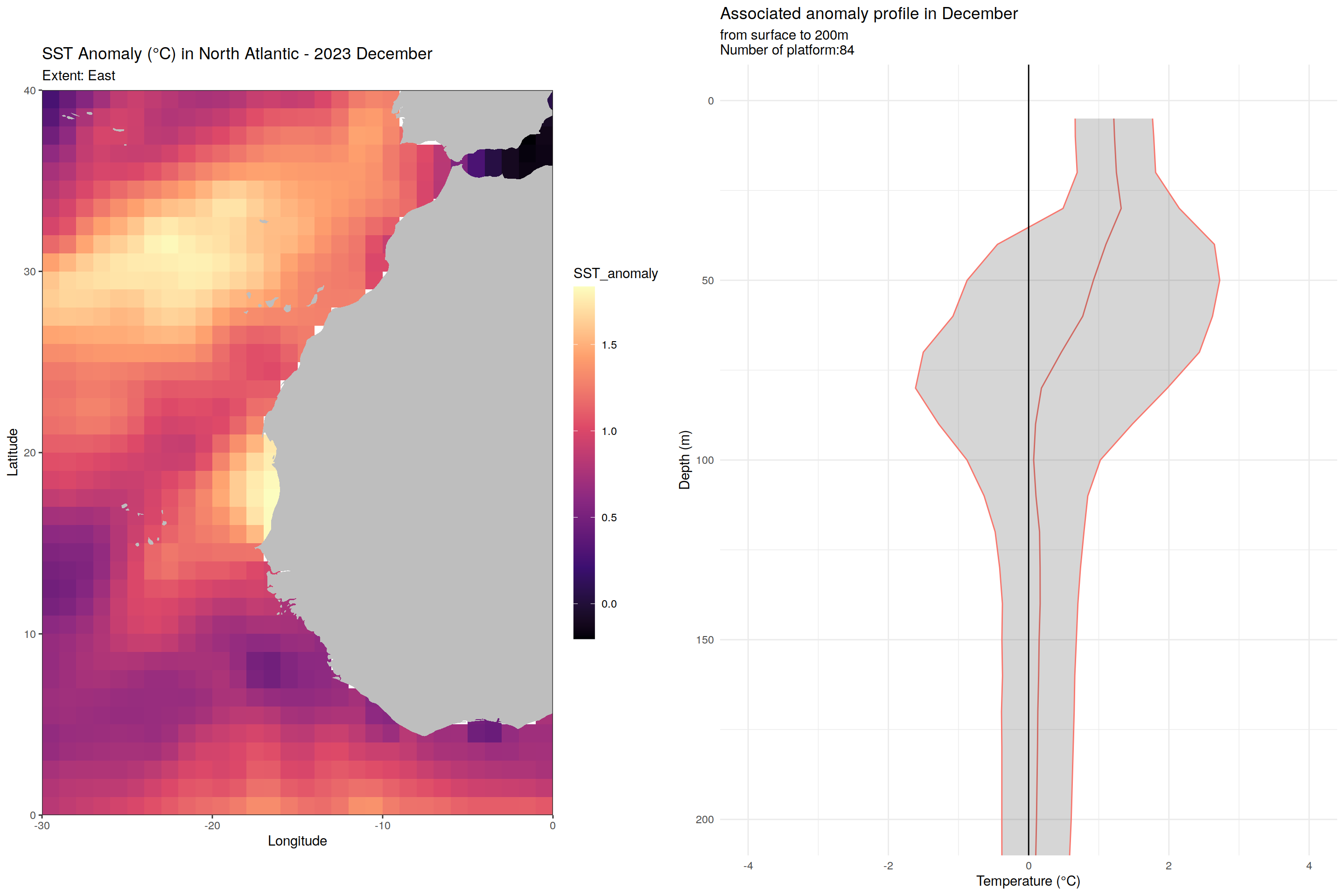
TableGrob (1 x 2) "arrange": 2 grobs
z cells name grob
1 1 (1-1,1-1) arrange gtable[layout]
2 2 (1-1,2-2) arrange gtable[layout]# Vertical anomaly profile for the North Atlantic subset region - monthly
anomaly_plot_monthly <- ggplot(anomaly_mean, aes(x = temp_anomaly_mean, y = depth, color = factor(month))) +
geom_path() +
geom_ribbon(aes(xmax = temp_anomaly_mean + temp_anomaly_sd,
xmin = temp_anomaly_mean - temp_anomaly_sd,
y = depth), alpha = 0.2) +
geom_vline(xintercept = 0) +
scale_y_reverse() +
coord_cartesian(xlim = c(-4, 4), ylim = c(200, 0)) +
labs(title = paste('Monthly anomaly profile'),
subtitle = paste0("Extent: ", name_extent),
x = 'Temperature (°C)', y = 'Depth (m)') +
guides(color = FALSE)+
facet_wrap(~month)
print(anomaly_plot_monthly)
# Calculating monthly mean anomaly over the east area by averaging the anomaly of each float present
anomaly_2months <- core_anomaly_2023_natlantic_subset %>%
mutate(period=(as.numeric(month)+1)%/%2)
anomaly_2months<-anomaly_2months %>%
group_by(depth, period) %>%
summarise(temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE),
temp_anomaly_sd = sd(anomaly, na.rm = TRUE))
anomaly_2023<-core_anomaly_2023_natlantic_subset %>%
group_by(depth) %>%
summarise(temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE),
temp_anomaly_sd = sd(anomaly, na.rm = TRUE))
# Vertical anomaly profile for the North atlatinc east region - monthly
ggplot() +
geom_path(data=anomaly_2months, aes(x = temp_anomaly_mean, y = depth, color = factor(period))) +
geom_ribbon(data=anomaly_2023, aes(xmax = temp_anomaly_mean + temp_anomaly_sd,
xmin = temp_anomaly_mean - temp_anomaly_sd,
y = depth, fill = "spread"), alpha = 0.2) +
geom_vline(xintercept = 0) +
scale_y_reverse() +
coord_cartesian(xlim = c(-4, 4), ylim = c(200, 0)) +
labs(title = paste('Propagation of SST anomalies in the water column'),
subtitle = paste0("Extent: ", name_extent, " North Atlantic bassin in 2023"),
x = 'Temperature (°C)', y = 'Depth (m)') +
scale_color_manual(values = colorRampPalette(c("blue", "orange", "darkred"))(6),
breaks = unique(anomaly_2months$period),
labels =c("Jan-Feb", "March-April", "May-June", "July-Aug", "Sept-Oct", "Nov-Dec")) +
scale_fill_manual(values = "grey", # Manual fill color setting
labels = "yearly") + # Label for the legend
theme(plot.title = element_text(size = 18),
plot.subtitle = element_text(size = 12),
legend.text = element_text(size = 14)) +
guides(color = guide_legend(title="Period"),
fill = guide_legend(title = "Spread")) # Legend for the ribbon
Hovmoeller plot
sessionInfo()R version 4.2.2 (2022-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.5
Matrix products: default
BLAS: /usr/local/R-4.2.2/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.2.2/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] patchwork_1.1.2 broom_1.0.5 paletteer_1.6.0
[4] cluster_2.1.6 gridExtra_2.3 scatterplot3d_0.3-44
[7] viridis_0.6.2 viridisLite_0.4.1 ggOceanMaps_1.3.4
[10] ggspatial_1.1.7 oce_1.7-10 gsw_1.1-1
[13] lubridate_1.9.0 timechange_0.1.1 forcats_0.5.2
[16] stringr_1.5.0 dplyr_1.1.3 purrr_1.0.2
[19] readr_2.1.3 tidyr_1.3.0 tibble_3.2.1
[22] ggplot2_3.4.4 tidyverse_1.3.2 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] googledrive_2.0.0 colorspace_2.0-3 ellipsis_0.3.2
[4] class_7.3-20 rprojroot_2.0.3 fs_1.5.2
[7] rstudioapi_0.15.0 proxy_0.4-27 farver_2.1.1
[10] fansi_1.0.3 xml2_1.3.3 codetools_0.2-18
[13] cachem_1.0.6 knitr_1.41 jsonlite_1.8.3
[16] dbplyr_2.2.1 rgeos_0.5-9 compiler_4.2.2
[19] httr_1.4.4 backports_1.4.1 assertthat_0.2.1
[22] fastmap_1.1.0 gargle_1.2.1 cli_3.6.1
[25] later_1.3.0 htmltools_0.5.8.1 tools_4.2.2
[28] gtable_0.3.1 glue_1.6.2 maps_3.4.1
[31] Rcpp_1.0.10 cellranger_1.1.0 jquerylib_0.1.4
[34] RNetCDF_2.6-1 raster_3.6-11 vctrs_0.6.4
[37] lwgeom_0.2-10 xfun_0.35 ps_1.7.2
[40] rvest_1.0.3 lifecycle_1.0.3 ncmeta_0.3.5
[43] googlesheets4_1.0.1 terra_1.7-65 getPass_0.2-2
[46] scales_1.2.1 hms_1.1.2 promises_1.2.0.1
[49] parallel_4.2.2 rematch2_2.1.2 yaml_2.3.6
[52] sass_0.4.4 stringi_1.7.8 highr_0.9
[55] e1071_1.7-12 rlang_1.1.1 pkgconfig_2.0.3
[58] evaluate_0.18 lattice_0.20-45 sf_1.0-9
[61] labeling_0.4.2 processx_3.8.0 tidyselect_1.2.0
[64] magrittr_2.0.3 R6_2.5.1 generics_0.1.3
[67] DBI_1.2.2 pillar_1.9.0 haven_2.5.1
[70] whisker_0.4 withr_2.5.0 units_0.8-0
[73] stars_0.6-0 abind_1.4-5 sp_1.5-1
[76] modelr_0.1.10 crayon_1.5.2 KernSmooth_2.23-20
[79] utf8_1.2.2 tzdb_0.3.0 rmarkdown_2.18
[82] grid_4.2.2 readxl_1.4.1 callr_3.7.3
[85] git2r_0.30.1 reprex_2.0.2 digest_0.6.30
[88] classInt_0.4-8 httpuv_1.6.6 munsell_0.5.0
[91] bslib_0.4.1