Load Core-Argo Data
Marguerite Larriere, David Stappard, Pasqualina Vonlanthen & Jens Daniel Müller
04 June, 2024
Last updated: 2024-06-04
Checks: 7 0
Knit directory:
bgc_argo_r_argodata/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211008) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e14743e. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/draft.Rmd
Untracked: load_argo_core_output.txt
Untracked: poster_profile_argo.png
Unstaged changes:
Modified: analysis/CESM_comparison.Rmd
Deleted: analysis/MHWs_categorisation.Rmd
Modified: analysis/MHWs_vertical_anomaly.Rmd
Modified: analysis/_site.yml
Modified: analysis/anomaly_SST_2023.Rmd
Modified: analysis/child/cluster_analysis_base.Rmd
Modified: analysis/coverage_maps_North_Atlantic.Rmd
Modified: analysis/load_broullon_DIC_TA_clim.Rmd
Modified: analysis/temp_align_climatology.Rmd
Modified: analysis/temp_core_align_climatology.Rmd
Modified: code/Workflowr_project_managment.R
Modified: code/start_background_job.R
Modified: code/start_background_job_load.R
Modified: code/start_background_job_partial.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/load_argo_core.Rmd) and
HTML (docs/load_argo_core.html) files. If you’ve configured
a remote Git repository (see ?wflow_git_remote), click on
the hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | e14743e | mlarriere | 2024-06-04 | duplicates 2022 fixed |
| html | 8cb6d7d | mlarriere | 2024-06-03 | Build site. |
| Rmd | f2bcbb6 | mlarriere | 2024-06-03 | clean code |
| html | 96d4b76 | mlarriere | 2024-05-14 | Build site. |
| html | af6594f | mlarriere | 2024-05-13 | Build site. |
| Rmd | 30f9250 | mlarriere | 2024-05-13 | Adding CESM subsection |
| html | d2c67b8 | mlarriere | 2024-05-13 | Build site. |
| Rmd | 037926c | mlarriere | 2024-05-13 | adding realtime files to the dataset |
| html | af44bcd | mlarriere | 2024-04-26 | Build site. |
| Rmd | f37a13f | mlarriere | 2024-04-26 | adding subsection SST anomaly |
| html | 780276c | mlarriere | 2024-04-19 | Build site. |
| Rmd | c4eb5b4 | mlarriere | 2024-04-19 | Adding SST anomaly subsection |
| Rmd | cec0b43 | mlarriere | 2024-04-19 | Adding SST anomaly subsection |
| html | bc9a34e | mlarriere | 2024-04-12 | Build site. |
| Rmd | 1800745 | mlarriere | 2024-04-12 | build load_data section |
| html | c076fba | mlarriere | 2024-04-12 | Build site. |
| html | 91f08a6 | mlarriere | 2024-04-07 | Build site. |
| html | db21f55 | mlarriere | 2024-04-06 | Build site. |
| Rmd | 5627ad6 | mlarriere | 2024-04-04 | until coverage_maps_North_Atlantic building test |
| html | c6dfd99 | mlarriere | 2024-03-31 | Build site. |
| Rmd | 6af4dc0 | mlarriere | 2024-03-31 | load_argo_core building test |
| Rmd | de0401f | mlarriere | 2024-03-27 | running test. |
| html | b82a3be | mlarriere | 2024-03-26 | Build site. |
| Rmd | 4ee65d7 | mlarriere | 2024-03-26 | core dataset refresh - adding realtime mode files in 2023. |
| Rmd | 825f6aa | mlarriere | 2024-03-25 | core dataset refresh - adding realtime mode files in 2023. |
| Rmd | 500bc32 | mlarriere | 2024-03-15 | core dataset refresh. |
| Rmd | 33ab76a | mlarriere | 2024-03-14 | core dataset refresh. |
| Rmd | 26fb454 | mlarriere | 2024-03-11 | core dataset refresh. |
| Rmd | 7c8bd44 | mlarriere | 2024-03-08 | core dataset refresh. |
| Rmd | 5ffa0da | mlarriere | 2024-03-07 | core dataset refresh. |
| Rmd | 2593007 | mlarriere | 2024-03-06 | core dataset refresh. |
| Rmd | d638c85 | mlarriere | 2024-03-05 | core dataset refresh. |
| Rmd | e3fa964 | mlarriere | 2024-03-01 | core dataset refresh. |
| html | f9de50e | ds2n19 | 2024-01-01 | Build site. |
| Rmd | 76ebe5b | ds2n19 | 2024-01-01 | load refresh end Dec 2023 |
| Rmd | 38a1c4f | ds2n19 | 2023-12-21 | core dataset refresh. |
| Rmd | 17dfa44 | ds2n19 | 2023-12-20 | builing generic cluster analysis. |
| html | 4eb3da7 | ds2n19 | 2023-12-20 | Build site. |
| Rmd | fe896a1 | ds2n19 | 2023-12-20 | builing generic cluster analysis. |
| html | 07d4eb8 | ds2n19 | 2023-12-20 | Build site. |
| html | fa6cf38 | ds2n19 | 2023-12-14 | Build site. |
| html | f110b74 | ds2n19 | 2023-12-13 | Build site. |
| Rmd | acb6523 | ds2n19 | 2023-12-12 | Added documentation added to tasks section at start of each script. |
| html | e60ebd2 | ds2n19 | 2023-12-07 | Build site. |
| html | 7b0dde9 | ds2n19 | 2023-11-27 | Build site. |
| Rmd | 48f510c | ds2n19 | 2023-11-27 | Cluster under surface extreme. |
| html | b8a2611 | ds2n19 | 2023-11-27 | Build site. |
| Rmd | 6c81f96 | ds2n19 | 2023-11-27 | Cluster under surface extreme. |
| html | 80c16c2 | ds2n19 | 2023-11-15 | Build site. |
| Rmd | 3eba518 | ds2n19 | 2023-11-15 | Introduction of vertical alignment and cluster analysis to github website. |
| Rmd | e07ffb5 | ds2n19 | 2023-11-11 | Updates after code reviewand additional documentation. |
| Rmd | b18136a | jens-daniel-mueller | 2023-11-09 | code review |
| Rmd | 4fcef97 | ds2n19 | 2023-11-01 | vertical alignment of bgc and core profiles and then cluster analysis. |
| Rmd | 8edb9f4 | ds2n19 | 2023-10-17 | BGC load process aligned to core load. Associated changes to pH and oxygen analysis. |
| html | c7c6b3c | ds2n19 | 2023-10-14 | Build site. |
| Rmd | 806687a | ds2n19 | 2023-10-14 | Added load qc metrics |
| html | 7227bdb | ds2n19 | 2023-10-14 | Build site. |
| Rmd | d6e0aca | ds2n19 | 2023-10-14 | Added load qc metrics |
| html | 12d26f4 | ds2n19 | 2023-10-14 | Build site. |
| Rmd | 311a3ad | ds2n19 | 2023-10-14 | Added load qc metrics |
| html | c1dc2c4 | ds2n19 | 2023-10-14 | Build site. |
| Rmd | 7b9cd9b | ds2n19 | 2023-10-14 | Added load qc metrics |
| html | d6fe78d | ds2n19 | 2023-10-14 | Build site. |
| Rmd | cc09ea4 | ds2n19 | 2023-10-14 | Added load qc metrics |
| html | eec64f7 | ds2n19 | 2023-10-13 | Build site. |
| Rmd | 2ca768d | ds2n19 | 2023-10-13 | Added load qc metrics |
| html | 35d0933 | ds2n19 | 2023-10-13 | Build site. |
| Rmd | 3f8f36e | ds2n19 | 2023-10-13 | Added load qc metrics |
| Rmd | 6d38b59 | ds2n19 | 2023-10-12 | minor changes after JDM code review |
| html | 2e78f0a | ds2n19 | 2023-10-12 | Build site. |
| Rmd | d1405fb | ds2n19 | 2023-10-11 | Run core Argo after code review 2013 - 2023 |
| Rmd | 72f849f | jens-daniel-mueller | 2023-10-11 | code review |
| Rmd | 5bf13a5 | ds2n19 | 2023-10-11 | load core conflicts resolved |
| Rmd | 1ae81b3 | ds2n19 | 2023-10-11 | reworked core load process to work initially by year and then finally create consolidated all years files. |
| html | f4c8d6f | ds2n19 | 2023-10-11 | Build site. |
| Rmd | f5edbe3 | ds2n19 | 2023-10-10 | Create simplified data sets and run for 2013 - 2023 |
| html | 7394ba8 | ds2n19 | 2023-10-10 | Build site. |
| Rmd | a5002da | ds2n19 | 2023-10-10 | Create simplified data sets and run for 2013 - 2023 |
| Rmd | aac59da | ds2n19 | 2023-10-09 | Create targetted data sets and run for 2013 - 2023 |
| html | 26f85f0 | ds2n19 | 2023-10-06 | Build site. |
| Rmd | 2bd702c | ds2n19 | 2023-10-06 | Changed core Argo location folders and run for 2022 |
| html | c3381d0 | ds2n19 | 2023-10-06 | Build site. |
| Rmd | bc8d46d | ds2n19 | 2023-10-06 | Changed core Argo location folders and run for 2022 |
| html | 8d5c853 | pasqualina-vonlanthendinenna | 2022-08-30 | Build site. |
| Rmd | 9bde106 | pasqualina-vonlanthendinenna | 2022-08-30 | added 6 months of core data (still have to fix the dates |
| html | 7b3d8c5 | pasqualina-vonlanthendinenna | 2022-08-29 | Build site. |
| Rmd | 8e81570 | pasqualina-vonlanthendinenna | 2022-08-29 | load and add in core-argo data (1 month) |
Task
Makes use of argodata libraries to load argo profile related data, stages load index, data and metadata files. Cache files (saved in /nfs/kryo/work/datasets/ungridded/3d/ocean/floats/core_argo_r_argodata and dac subdirectory) are used, set option opt_refresh_cache to TRUE to force a refresh – This process takes considerable time.
The load process described in this paragraph is carried out for each year and the files are saved to the core preprocessed. core_index data frame is created based on delayed mode files and a supplied date range. The index file is then used to load profile data into core_data and associated meta data into core_metadata. core_data is segregated into core_data_temp and core_data_psal. core_data_temp, core_data_psal and core_metadata are filtered by n_prof == opt_n_prof_sel. Description of n_prof usage is provided at https://argo.ucsd.edu/data/data-faq/version-3-profile-files/ the following is from that page. The main Argo CTD profile is stored in N_PROF=1. All other parameters (including biogeochemical parameters) that are measured with the same vertical sampling scheme and at the same location and time as the main Argo CTD profile are also stored in N_PROF=1.
On completion combined all year core_data_temp, core_data_psal and core_metadata that incorporate all years. core_data_temp, core_data_psal and core_metadata initially contain a string field file that uniquely identifies the profile and links the two data frames. An additional data from core_fileid is created with a unique list file fields along with a numeric file_id field. The file fields in core_data and core_metadata are then replaced with file_id.
core_temp_flag_A and core_temp_flag_AB are created that support prior analysis. In addition, a data frame core_measure_summary is created that is used for load level reporting figures.
Files are written to the core preprocessed folder for ongoing analysis.
Dependencies
Cache files - /nfs/kryo/work/datasets/ungridded/3d/ocean/floats/core_argo_r_argodata_2024-03-13
Outputs (in core preprocessed folder)
core_index.rds – Copy of the index file that was used in the selection of data and meta data files.
core_data_temp.rds – The main core temperature profile data.
core_data_psal.rds – The main core salinity profile data.
core_metadata.rds – The associated meta data information.
core_fileid.rds – A lookup from file_id to file.
Each of the below files are used in prior analysis and maintained to support that analysis.
core_temp_flag_A.rds
core_temp_flag_AB.rds
Set load options
Determine if files are refreshed from dac or cache directory is used. Are metadata, temperature and salinity year files renewed? Are the consolidated all year files created from the individual year files?
# opt_refresh_cache
# FALSE = do not refresh cache.
# TRUE = refresh cache. (any none zero value will force a refresh)
opt_refresh_cache = FALSE
# opt_refresh_years_temp, opt_refresh_years_psal, opt_refresh_years_metadata
# FALSE = do not refresh the yearly files. (any value <> 1 will omit annual refresh)
# TRUE = refresh yearly files for given parameter.
# year to be refreshed are set by opt_min_year and opt_max_year
opt_refresh_years_temp = TRUE
opt_refresh_years_psal = TRUE
opt_refresh_years_metadata = TRUE
opt_min_year = 2013
opt_max_year = 2024
# opt_consolidate_temp, opt_consolidate_psal, opt_consolidate_metadata
# Yearly files must have already been created!
# FALSE = do not build consolidated file from previously written yearly files. (any value <> 1 will omit consolidation)
# TRUE = build consolidated file from previously written yearly files for given parameter.
# year to be included in the consolidation are set by opt_min_year and opt_max_year
opt_consolidate_temp = TRUE
opt_consolidate_psal = TRUE
opt_consolidate_metadata = TRUE
# opt_A_AB_files
# consolidated temp files must have already been created!
# FALSE = do not build temp_A and temp_AB file from previously written consolidated files. (any value <> 1 will omit A and AB files)
# TRUE = build temp_A and temp_AB file from previously written consolidated files.
opt_A_AB_files = TRUE
# opt_review_mode
# if set (TRUE) the processing will take place in a sub-directory opt_review_dir and only process 10 days of profiles per year to reduce size
# of output and processing time
opt_review_mode = FALSE
opt_review_dir = "/review_mode"
#if (opt_review_mode) {
# path_argo_core_preprocessed <- paste0(path_argo_core, "/preprocessed_core_data", opt_review_dir)
#}
# opt_qc_only
# Avoids reprocessing files and ensures qc summary plots are created from a previous run!
# FALSE = carry out reprocessing based on options set above and create QC summaries.
# TRUE = do NOT reprocessing files and just create QC summaries from previous loads.
opt_qc_only = FALSE
# opt_n_prof_sel
# The selection criteria that is used against n_prof, here set to 1
# Description of n_prof usage is provided at https://argo.ucsd.edu/data/data-faq/version-3-profile-files/ the next two lines are from that page.
# The main Argo CTD profile is stored in N_PROF=1. All other parameters (including biogeochemical parameters) that are measured
# with the same vertical sampling scheme and at the same location and time as the main Argo CTD profile are also stored in N_PROF=1.
opt_n_prof_sel = 1Set cache directory
Directory where the core-Argo profile files are stored. Either use the cached files or force a refresh from dac (long process)
# argodata package: https://rdrr.io/github/ArgoCanada/argodata/man/
if (!opt_qc_only) {
# print("Defining settings")
# set cache directory
argo_set_cache_dir(cache_dir = path_argo_core)
# check cache directory
argo_cache_dir()
# Server options are available.
# https://data-argo.ifremer.fr - This is the default. Downloads are quick but when carrying out a refresh in Dec
# 2023 the process would halt part way through and not continue. -- SAME in March 2024
# ftp://ftp.ifremer.fr/ifremer/argo - Progress does not seem as fast as the HTTPS server but as yet the halting
# issue has not occurred.
# https://usgodae.org/pub/outgoing/argo - alternative HTTPS
argo_set_mirror("https://usgodae.org/pub/outgoing/argo")
# check argo mirror
argo_mirror()
# age argument: age of the cached files to update in hours (Inf means always use the cached file, and -Inf means always download from the server)
# ex: max_global_cache_age = 5 updates files that have been in the cache for more than 5 hours, max_global_cache_age = 0.5 updates
# files that have been in the cache for more than 30 minutes, etc.
if (opt_refresh_cache){
print("Updating files from website -- in progress")
# Terminal command for synchronization with dynamic ARGO archive -- about 12h of computation
command <- 'rsync -avzh --delete vdmzrs.ifremer.fr::argo/ /nfs/kryo/work/datasets/ungridded/3d/ocean/floats/core_argo_r_argodata_2024-13-03/dac'
system(command)
# argodata function to update data -- facing some writing permission issues as the entire database has to be redownload each time (dynamic archive)
# argo_update_global(max_global_cache_age = -Inf)
# argo_update_data(max_data_cache_age = -Inf)
} else {
print("Retreiving files from local server")
argo_update_global(max_global_cache_age = Inf)
argo_update_data(max_data_cache_age = Inf)
}
}Load by year
Builds yearly files for temperature, salinity and metadata that can be consolidated in the next code chunk (consolidate_into_allyears)
#------------------------------------------------------------------------------
# Important - file are loaded for the given year processed and the files written to disk.
#------------------------------------------------------------------------------
if (!opt_qc_only) {
for (target_year in opt_min_year:opt_max_year) {
# for manual testing of the loop
# target_year <- 2022
cat("year being processed:", target_year, "\n")
# if updating any year files it will be based on the initial index file core_index
if (opt_refresh_years_temp | opt_refresh_years_psal | opt_refresh_years_metadata){
print("Reading file on online server")
# if working in review mode only consider first 10 days of the year
if (opt_review_mode) {
core_index <- argo_global_prof() %>%
argo_filter_data_mode(data_mode = 'delayed') %>%
argo_filter_date(
date_min = paste0(target_year, "-01-01"),
date_max = paste0(target_year, "-01-05")
)
} else {
# We select the delayed mode files, and add the realtime-mode files, in order to have good data coverage
#Download delayed-mode files
delayed_files <- argo_global_prof() %>%
argo_filter_data_mode(data_mode = 'delayed') %>%
argo_filter_date(
date_min = paste0(target_year, "-01-01"),
date_max = paste0(target_year, "-12-31")
)
#Download realtime files
realtime_files <- argo_global_prof() %>%
argo_filter_data_mode(data_mode = 'realtime') %>%
argo_filter_date(
date_min = paste0(target_year, "-01-01"),
date_max = paste0(target_year, "-12-31")
)
}
}
# if temp or psal are being updated get the profile data
if (opt_refresh_years_temp | opt_refresh_years_psal){
#Reading profiles (takes a while)
#---------------- Realtime files ----------------
# see: http://www.argodatamgt.org/content/download/20685/142877/file/argo-quality-control-manual_version2.9.pdf
# when PARAM_ADJUSTED and PARAM_ADJUSTED_QC are available for Real-time files -> DATA_MODE = ‘A’, i.e. real-time adjustments are available for one or more parameters.
# If we use PARAM_ADJUSTED and PARAM_ADJUSTED_QC, we loose some data (about 2 time less), so we select all files
realtime_float<- argo_prof_levels(
path = realtime_files,
vars = c( 'PRES', # variables for realtime files
'PRES_QC',
'PSAL',
'PSAL_QC',
'TEMP',
'TEMP_QC'),
quiet = TRUE) # contains qc flags= 1, 2, 3, 4, 9, " "
#Remove rows containing NA
realtime_float_filtered <- realtime_float %>%
filter(!is.na(pres), !is.na(temp), !is.na(psal)) %>%
mutate( pres_adjusted = pres, #same structure than delayed files
pres_adjusted_qc = pres_qc,
psal_adjusted = psal,
psal_adjusted_qc = psal_qc,
temp_adjusted = temp,
temp_adjusted_qc = temp_qc
) %>%
select(-pres, -pres_qc, -psal, -psal_qc, -temp, -temp_qc )
#---------------- Delayed files ----------------
delayed_float <- argo_prof_levels(
path = delayed_files,
vars = c('PRES_ADJUSTED', #variables for delayed files
'PRES_ADJUSTED_QC',
'PSAL_ADJUSTED',
'PSAL_ADJUSTED_QC',
'TEMP_ADJUSTED',
'TEMP_ADJUSTED_QC'),
quiet = TRUE) # contains qc flags= 1, 2, 3, 4, 9, " "
#Remove rows containing NA
delayed_float_filtered <- delayed_float %>%
filter(!is.na(pres_adjusted), !is.na(temp_adjusted), !is.na(psal_adjusted)) %>%
distinct(file, pres_adjusted, .keep_all = TRUE)
#Combining datasets
core_data_yr <- bind_rows(delayed_float_filtered, realtime_float_filtered)
# see option section above to know why we want n_prof = 1 profiles
core_data_yr <- core_data_yr %>%
filter(n_prof == opt_n_prof_sel)
# if necessary make summary data frame
if (!exists("core_measure_summary")){
core_measure_summary <-
tibble("year" = numeric(),
"measure" = character(),
"measure_order" = numeric(),
"measure_qc" = numeric(),
"count_measures" = numeric()
)
}
# ---------------- Pressure ----------------
# Default QC counts for measurements - Pressure
qc_defaults <-
tibble(
year = rep(target_year, 8),
measure = rep("Pressure",8),
measure_order = rep(1, 8), #1st measure in the dataset
measure_qc = c('0', '1', '2', '3', '4', '5', '8', '9'),
count_measures = rep(0, 8)
)
core_measure_summary = rbind(core_measure_summary, qc_defaults)
# Build summary of qc flags for pressure and update core_measure_summary
agg_tbl <- core_data_yr %>%
group_by(
year = target_year,
measure = "Pressure",
measure_order = 1,
measure_qc = pres_adjusted_qc
) %>%
summarise(count_measures = n())
core_measure_summary <- rows_update(core_measure_summary,
agg_tbl,
by = c('year', 'measure_order', 'measure_qc'))
# ---------------- Temperature ----------------
# Default qc counts for measurements - temperature
qc_defaults <-
data.frame(
year = c( target_year,
target_year,
target_year,
target_year,
target_year,
target_year,
target_year,
target_year
),
measure = c( "Temperature",
"Temperature",
"Temperature",
"Temperature",
"Temperature",
"Temperature",
"Temperature",
"Temperature"
),
measure_order = c(2, 2, 2, 2, 2, 2, 2, 2), #2nd measure in the dataset
measure_qc = c('0', '1', '2', '3', '4', '5', '8', '9'),
count_measures = c(0, 0, 0, 0, 0, 0, 0, 0)
)
core_measure_summary = rbind(core_measure_summary, qc_defaults)
# Build summary of qc flags for temperature and update core_measure_summary
agg_tbl <- core_data_yr %>%
group_by(
year = target_year,
measure = "Temperature",
measure_order = 2,
measure_qc = temp_adjusted_qc
) %>%
summarise(count_measures = n())
core_measure_summary <- rows_update(core_measure_summary,
agg_tbl,
by = c('year', 'measure_order', 'measure_qc'))
# ---------------- Salinity ----------------
# Default qc counts for measurements - salinity
qc_defaults <-
data.frame(
year = c( target_year,
target_year,
target_year,
target_year,
target_year,
target_year,
target_year,
target_year
),
measure = c( "Salinity",
"Salinity",
"Salinity",
"Salinity",
"Salinity",
"Salinity",
"Salinity",
"Salinity"
),
measure_order = c(3, 3, 3, 3, 3, 3, 3, 3), #3rd measure in the dataset
measure_qc = c('0', '1', '2', '3', '4', '5', '8', '9'),
count_measures = c(0, 0, 0, 0, 0, 0, 0, 0)
)
core_measure_summary = rbind(core_measure_summary, qc_defaults)
# Build summary of qc flags for salinity and update core_measure_summary
agg_tbl <- core_data_yr %>%
group_by(
year = target_year,
measure = "Salinity",
measure_order = 3,
measure_qc = psal_adjusted_qc
) %>%
summarise(count_measures = n())
core_measure_summary <- rows_update(core_measure_summary,
agg_tbl,
by = c('year', 'measure_order', 'measure_qc'))
rm(agg_tbl)
}
# if updating metadata get the file based on core_index
if (opt_refresh_years_metadata){
# read associated metadata
realtime_metadata <- argo_prof_prof(path = realtime_files)
delayed_metadata <- argo_prof_prof(path = delayed_files)
#combined metadata dataset
core_metadata_yr <- bind_rows(delayed_metadata, realtime_metadata)
# see option section above to know why we want n_prof = 1 profiles
core_metadata_yr <- core_metadata_yr %>%
filter(n_prof == opt_n_prof_sel)
}
# if temp or psal are being updated get the profile data
if (opt_refresh_years_temp | opt_refresh_years_psal)
{
# remove columns that are not needed in merged temperature and salinity files
realtime_files <- realtime_files %>%
select(file,
date,
latitude,
longitude)
delayed_files <- delayed_files %>%
select(file,
date,
latitude,
longitude)
core_index<-bind_rows(delayed_files, realtime_files)
# core_index <- core_index %>%
# select(file,
# date,
# latitude,
# longitude)
# resolve lat and lon
core_index <- core_index %>%
rename(lon = longitude,
lat = latitude) %>%
mutate(lon = if_else(lon < 20, lon + 360, lon)) %>%
mutate(
lat = cut(lat, seq(-90, 90, 1), seq(-89.5, 89.5, 1)),
lat = as.numeric(as.character(lat)),
lon = cut(lon, seq(20, 380, 1), seq(20.5, 379.5, 1)),
lon = as.numeric(as.character(lon))
)
# join to index to incorporate date, lat and lon
core_data_yr <- left_join(core_data_yr, core_index)
# derive depth from the pressure using TEOS=10
core_data_yr <- core_data_yr %>%
mutate(depth = gsw_z_from_p(pres_adjusted, latitude = lat) * -1.0,
.before = pres_adjusted)
core_data_yr <-
core_data_yr %>%
select(-c(n_levels, n_prof, pres_adjusted))
# print("Adding QC flag to realtime file")
#-----------core_data_temp_yr contains coriolis/6903067 (R) and 7900545 (D)
rm(core_index)
}
# ------------------------------------------------------------------------------
# Process temperature file
# ------------------------------------------------------------------------------
if (opt_refresh_years_temp){
# Base temperature data where qc flag = good (1) and estimated value (8)
# Could this cause incomplete profiles to be maintained?
core_data_temp_yr <- core_data_yr %>%
filter(pres_adjusted_qc %in% c(1, 8) & #good and estimated value
temp_adjusted_qc %in% c(1, 8)) %>%
select(-contains(c("_qc", "psal")))
print("Writing temperature file")
#-----------core_data_temp_yr contains coriolis/6903067 (R) and 7900545 (D)
#write file
core_data_temp_yr %>% write_rds(file = paste0(path_argo_core_preprocessed, "/",
target_year, "_core_data_temp.rds"))
}
# ------------------------------------------------------------------------------
# Process salinity file
# ------------------------------------------------------------------------------
if (opt_refresh_years_psal){
# Base salinity data where qc flag = good (1) and and estimated value (8)
core_data_psal_yr <- core_data_yr %>%
filter(pres_adjusted_qc %in% c(1, 8) &
psal_adjusted_qc %in% c(1, 8)) %>%
select(-contains(c("_qc", "temp")))
#-----------core_data_psal_yr contains coriolis/6903067 (R) and 7900545 (D)
#write file
print("Writing salinity file")
core_data_psal_yr %>% write_rds(file = paste0(path_argo_core_preprocessed, "/",
target_year, "_core_data_psal.rds"))
}
# ------------------------------------------------------------------------------
# Process metadata file
# ------------------------------------------------------------------------------
if (opt_refresh_years_metadata){
# resolve lat and lon so that it is hamonised with data files
core_metadata_yr <- core_metadata_yr %>%
rename(lon = longitude,
lat = latitude) %>%
mutate(lon = if_else(lon < 20, lon + 360, lon)) %>%
mutate(
lat = cut(lat, seq(-90, 90, 1), seq(-89.5, 89.5, 1)),
lat = as.numeric(as.character(lat)),
lon = cut(lon, seq(20, 380, 1), seq(20.5, 379.5, 1)),
lon = as.numeric(as.character(lon))
)
# Select just the columns we are interested in
core_metadata_yr <- core_metadata_yr %>%
select (
file,
date,
lat,
lon,
platform_number,
cycle_number,
position_qc,
profile_pres_qc,
profile_temp_qc,
profile_psal_qc
)
# write file
print("Writing metadata file")
core_metadata_yr %>% write_rds(file = paste0(path_argo_core_preprocessed, "/",
target_year, "_core_metadata.rds"))
}
}
# Write measure summary file
print("Writing summary file")
core_measure_summary %>% write_rds(file = paste0(path_argo_core_preprocessed, "/core_measure_summary.rds"))
rm(core_measure_summary)
}Consolidate years
This process create three files in the path_argo_core_preprocessed directory that will be used for further analysis
core_data_temp.rds
Contains approximately 500 measuring points per profile and only contains those points that are marked as good. Fields listed below fileid - the source file date - date of profile lat - aligned to closest 0.5° lat lon - aligned to closest 0.5° lon depth - calculated from pres_adjusted and latitude pres_adjusted - recorded and adjusted (after qc proccess) pressure temp_adjusted - recorded and adjusted (after qc proccess) temperature
core_data_psal.rds
Contains approximately 500 measuring points per profile and only contains those points that are marked as good. Fields listed below fileid - the source file date - date of profile lat - aligned to closest 0.5° lat lon - aligned to closest 0.5° lon depth - calculated from pres_adjusted and latitude pres_adjusted - recorded and adjusted (after qc proccess) pressure psal_adjusted - recorded and adjusted (after qc proccess) salinity
core_metadata.rds
Contains 1 row per profile. Fields listed below fileid - the source file date - date of profile lat - aligned to closest 0.5° lat lon - aligned to closest 0.5° lon platform_number - identifier of float cycle_number - the profile number for the given float position_qc - qc flag associated with the positioning of the float profile profile_pres_qc - qc flag associated with the pressure readings of the profile (A-F) profile_temp_qc - qc flag associated with the temperature readings of the profile (A-F) profile_psal_qc - qc flag associated with the salinity readings of the profile (A-F)
if (!opt_qc_only) {
# ------------------------------------------------------------------------------
# Process temperature file
# ------------------------------------------------------------------------------
if (opt_consolidate_temp){
print("All years together -- temperature")
consolidated_created = 0
for (target_year in opt_min_year:2023) {
# target_year<-2023
cat("year being processed ", target_year, "\n")
# read the montly files based on target_year
# for (month in sprintf("%02d", 1:12)){
# cat("month being processed ", month, "\n")
core_temp_path <- paste0(path_argo_core_preprocessed, "/", target_year, "_core_data_temp.rds")
if (file.exists(core_temp_path)) {
core_data_temp_yr <- read_rds(file = core_temp_path)
# Combine into a consolidated all years file
if (consolidated_created == 0) {
core_data_temp <- core_data_temp_yr
consolidated_created = 1
} else {
core_data_temp <- rbind(core_data_temp, core_data_temp_yr)
}
}
}
}
# ------------------------------------------------------------------------------
# Process salinity file
# ------------------------------------------------------------------------------
if (opt_consolidate_psal){
print("All years together -- salinity")
consolidated_created = 0
for (target_year in opt_min_year:2023) {
# target_year<-2023
cat("year being processed ", target_year, "\n")
# read the montly files based on target_year
# for (month in sprintf("%02d", 1:12)){
# cat("month being processed ", month, "\n")
core_psal_path <- paste0(path_argo_core_preprocessed, "/", target_year, "_core_data_psal.rds")
if (file.exists(core_psal_path)) {
# read the yearly file based on target_year
core_data_psal_yr <- read_rds(file = core_psal_path)
# Combine into a consolidated all years file
if (consolidated_created == 0) {
core_data_psal <- core_data_psal_yr
consolidated_created = 1
} else {
core_data_psal <- rbind(core_data_psal, core_data_psal_yr)
}
}
}
}
# ------------------------------------------------------------------------------
# Process metadata file
# ------------------------------------------------------------------------------
if (opt_consolidate_metadata){
print("All years together -- metadata")
consolidated_created = 0
for (target_year in opt_min_year:2023) {
# target_year<-2023
cat("year being processed ", target_year, "\n")
# read the montly files based on target_year
# for (month in sprintf("%02d", 1:12)){
# cat("month being processed ", month, "\n")
metadata_path <- paste0(path_argo_core_preprocessed, "/", target_year, "_core_metadata.rds")
if (file.exists(metadata_path)) {
# read the yearly file based on target_year
core_metadata_yr <- read_rds(file = metadata_path)
# Combine into a consolidated all years file
if (consolidated_created == 0) {
core_metadata <- core_metadata_yr
consolidated_created = 1
} else {
core_metadata <- rbind(core_metadata, core_metadata_yr)
}
# }
}
}
}
# ------------------------------------------------------------------------------
# Establish file_id and save files
# ------------------------------------------------------------------------------
# create fileid file ready to update data files
core_fileid <- unique(core_metadata$file)
core_fileid <- tibble(core_fileid)
core_fileid <- core_fileid %>% select (file = core_fileid)
core_fileid <- tibble::rowid_to_column(core_fileid, "file_id")
# Change metadate and data to have file_id
core_metadata <- full_join(core_metadata, core_fileid)
core_metadata <- core_metadata %>%
select(-c(file))
core_data_temp <- full_join(core_data_temp, core_fileid)
core_data_temp <- core_data_temp %>%
select(-c(file))
core_data_psal <- full_join(core_data_psal, core_fileid)
core_data_psal <- core_data_psal %>%
select(-c(file))
# write consolidated files
core_fileid %>%
write_rds(file = paste0(path_argo_core_preprocessed, "/core_fileid.rds"))
core_metadata %>%
write_rds(file = paste0(path_argo_core_preprocessed, "/core_metadata.rds"))
core_data_temp %>%
write_rds(file = paste0(path_argo_core_preprocessed, "/core_data_temp.rds"))
core_data_psal %>%
write_rds(file = paste0(path_argo_core_preprocessed, "/core_data_psal.rds"))
rm(core_metadata_yr, core_data_temp_yr, core_data_psal_yr)
rm(core_metadata, core_data_temp, core_data_psal, core_fileid)
gc()
}A and AB flag files
This process create two additional files in the path_argo_core_preprocessed directory that will be used for further analysis
core_temp_flag_A.rds
only ontains profile where profile_pres_qc and profile_temp_qc both equal A (100%). Fields listed below lat - aligned to closest 0.5° lat lon - aligned to closest 0.5° lon date - date of profile depth - depth of observation temp_adjusted - recorded and adjusted (after qc proccess) temperature platform_number - identifier of float cycle_number - the profile number for the given float
core_temp_flag_A.rds
only ontains profile where profile_pres_qc and profile_temp_qc both either A (100%) or B (> 75%). Fields listed below lat - aligned to closest 0.5° lat lon - aligned to closest 0.5° lon date - date of profile depth - depth of observation temp_adjusted - recorded and adjusted (after qc proccess) temperature platform_number - identifier of float cycle_number - the profile number for the given float
if (!opt_qc_only) {
if (opt_A_AB_files){
# Read temp and meta_data
core_data_temp <- read_rds(file = paste0(path_argo_core_preprocessed, "/core_data_temp.rds"))
core_metadata <- read_rds(file = paste0(path_argo_core_preprocessed, "/core_metadata.rds"))
# Join temp and meta_data to form merge
core_merge <- left_join(x = core_data_temp,
y = core_metadata %>%
select(file_id,
platform_number,
cycle_number,
contains("_qc")))
rm(core_data_temp, core_metadata)
gc()
# Select just A profiles into core_temp_flag_A
core_temp_flag_A <- core_merge %>%
filter(profile_temp_qc == 'A' & profile_pres_qc == 'A') %>%
select(-c(file_id, contains("_qc")))
# write core_temp_flag_A
core_temp_flag_A %>% write_rds(file = paste0(path_argo_core_preprocessed, "/core_temp_flag_A.rds"))
rm(core_temp_flag_A)
gc()
# Select just AB profiles into core_temp_flag_A
core_temp_flag_AB <- core_merge %>%
filter((profile_temp_qc == 'A' | profile_temp_qc == 'B') & (profile_pres_qc == 'A' | profile_pres_qc == 'B')) %>%
select(-c(file_id, contains("_qc")))
# write core_temp_flag_AB
core_temp_flag_AB %>% write_rds(file = paste0(path_argo_core_preprocessed, "/core_temp_flag_AB.rds"))
rm(list = ls(pattern = 'core_'))
gc()
}
} QC summary
profile QC flags (A-F)
Produce a summary of profile QC flags (A-F)
# Read metadata file and create profile summary table with a count for each year, measurement type and qc option
path_argo_core_preprocessed <- paste0(path_argo_core, "/preprocessed_core_data")
core_metadata <-read_rds(file = paste0(path_argo_core_preprocessed, "/core_metadata.rds"))
core_metadata["profile_pres_qc"][is.na(core_metadata["profile_pres_qc"])] <- ""
core_metadata["profile_temp_qc"][is.na(core_metadata["profile_temp_qc"])] <- ""
core_metadata["profile_psal_qc"][is.na(core_metadata["profile_psal_qc"])] <- ""
#Pressure
core_profile_summary <- core_metadata %>%
filter (profile_pres_qc != "") %>%
group_by(year = format(date, "%Y"),
measure = "Pressure",
measure_order = 1,
profile_qc = profile_pres_qc) %>%
summarise(count_profiles = n())
#Adding Temperature
core_profile_summary <- rbind(core_profile_summary, core_metadata %>%
filter (profile_temp_qc != "") %>%
group_by(year = format(date, "%Y"),
measure = "Temperature",
measure_order = 2,
profile_qc = profile_temp_qc) %>%
summarise(count_profiles = n()))
#Adding Salinity
core_profile_summary <- rbind(core_profile_summary, core_metadata %>%
filter (profile_psal_qc != "") %>%
group_by(year = format(date, "%Y"),
measure = "Salinity",
measure_order = 3,
profile_qc = profile_psal_qc) %>%
summarise(count_profiles = n()))
# modify data frame to prepare for plotting
core_profile_summary <- ungroup(core_profile_summary)
core_profile_summary <- core_profile_summary %>% group_by(measure_order)
core_profile_summary <- transform(core_profile_summary, year = as.numeric(year))
year_min <- min(core_profile_summary$year)
year_max <- max(core_profile_summary$year)
facet_label <- as_labeller(c("1"="Pressure", "2"="Temperature", "3"="Salinity"))
# draw plots for the separate parameters
core_profile_summary %>%
ggplot(aes(x = year, y = count_profiles, col = profile_qc, group=profile_qc)) +
geom_point() +
geom_line() +
facet_wrap(~measure_order, labeller = facet_label) +
scale_x_continuous(breaks = seq(year_min, year_max)) +
labs(x = 'year',
y = 'number of profiles',
col = 'profile QC flag',
title = 'Count of profile qc flags by year') +
theme(axis.text.x =element_text(angle=45))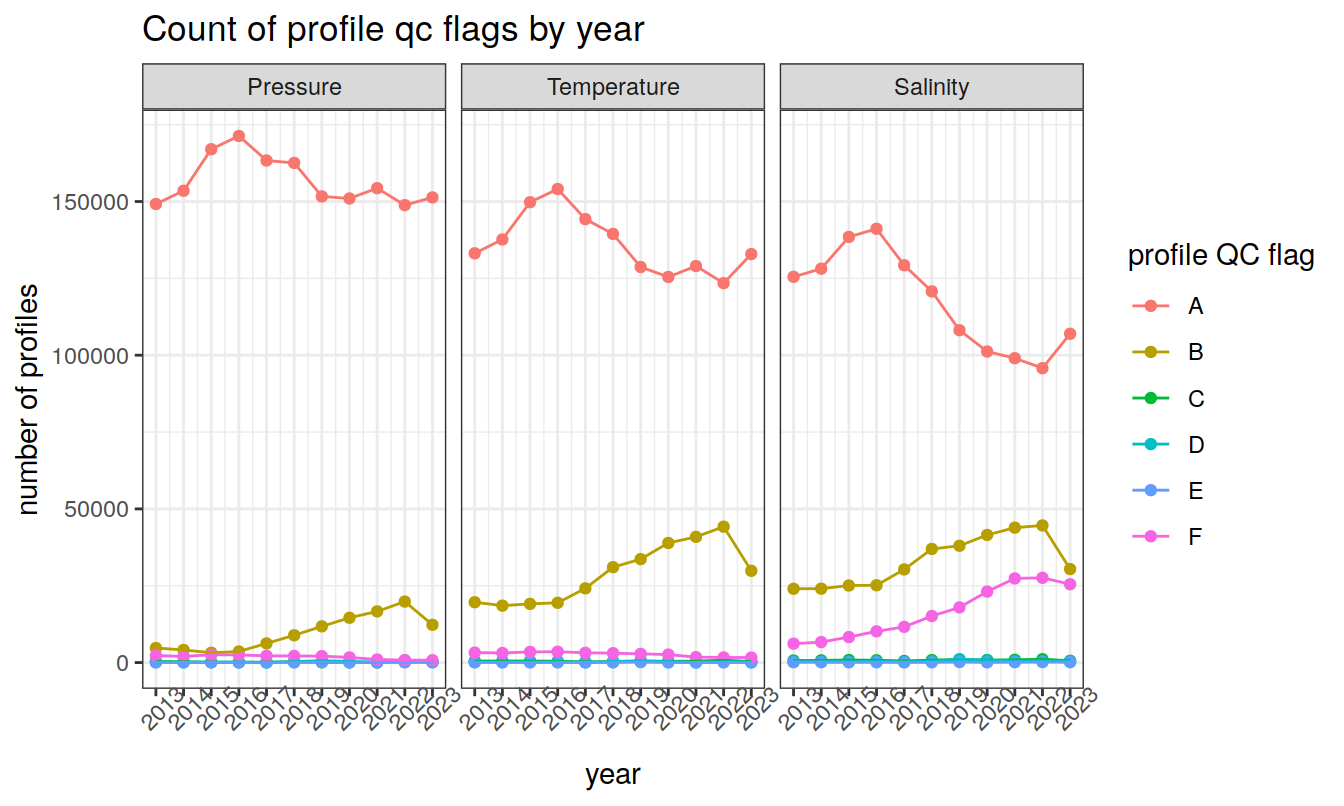
| Version | Author | Date |
|---|---|---|
| 8cb6d7d | mlarriere | 2024-06-03 |
| d2c67b8 | mlarriere | 2024-05-13 |
| bc9a34e | mlarriere | 2024-04-12 |
| c076fba | mlarriere | 2024-04-12 |
| c6dfd99 | mlarriere | 2024-03-31 |
| b82a3be | mlarriere | 2024-03-26 |
| f9de50e | ds2n19 | 2024-01-01 |
| 76ebe5b | ds2n19 | 2024-01-01 |
| 80c16c2 | ds2n19 | 2023-11-15 |
| c7c6b3c | ds2n19 | 2023-10-14 |
| 7227bdb | ds2n19 | 2023-10-14 |
| 12d26f4 | ds2n19 | 2023-10-14 |
| c1dc2c4 | ds2n19 | 2023-10-14 |
measurement QC flags (1-9)
Produce a summary of current measurement QC flags (1-9)
# Read temp and meta_data
core_measure_summary <-
read_rds(file = paste0(path_argo_core_preprocessed, "/core_measure_summary.rds"))
core_measure_summary <- ungroup(core_measure_summary)
year_min <- min(core_measure_summary$year)
year_max <- max(core_measure_summary$year)
# draw plots for the separate parameters
color_palette <- brewer.pal(8, "Dark2") #brewer palette max colors =8
core_measure_summary %>%
filter(measure_qc != " ") %>%
ggplot(aes(x = year, y = count_measures, col = measure_qc, group=measure_qc)) +
geom_point() +
geom_line() +
facet_wrap(~measure_order, labeller = facet_label) +
scale_x_continuous(breaks = seq(year_min, year_max)) +
labs(x = 'Year',
y = 'Number of Measures',
col = 'Measure QC Flag',
title = 'Count of Measure QC Flags by Year',
subtitle = 'Realtime and delayed files') +
scale_color_manual(values = color_palette,
labels = c("0" = "0 (no QC performed)",
"1" = "1 (good data)",
"2" = "2 (probably good data)",
"3" = "3 (bad data potentially correctable)",
"4" = "4 (bad data)",
"5" = "5 (values changed)",
"8" = "8 (estimated value)",
"9"="9 (missing value)"
))+
theme(axis.text.x =element_text(angle=45))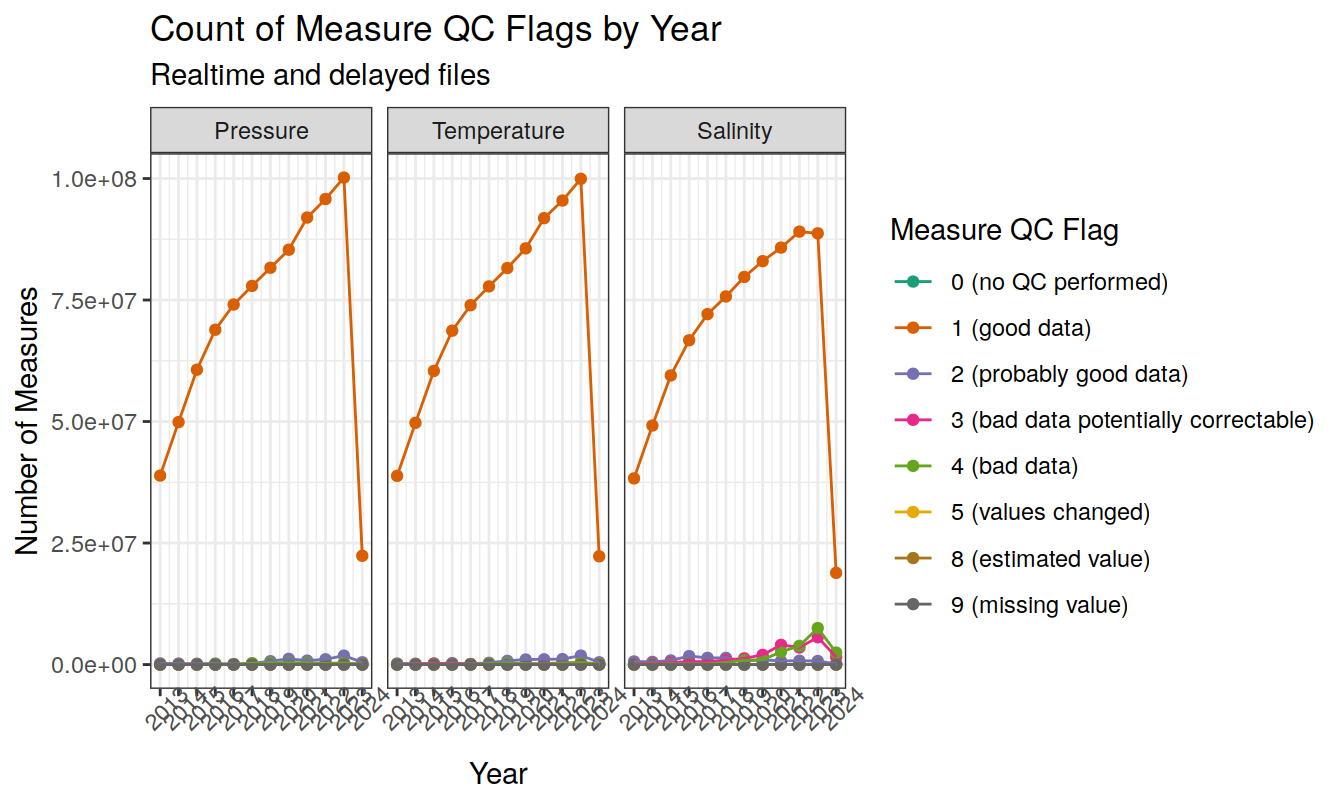
Delayed mode VS Real time mode
delayed_files <- argo_global_prof() %>%
argo_filter_data_mode(data_mode = 'delayed')
realtime_files <- argo_global_prof() %>%
argo_filter_data_mode(data_mode = 'realtime')
#Files difference
realtime_files_count_per_year <- realtime_files %>%
count(year(date)) %>%
rename(realtime_mode = n)%>%
rename(year=`year(date)`)
delayed_files_count_per_year <- delayed_files %>%
count(year(date))%>%
rename(delayed_mode = n) %>%
rename(year=`year(date)`)
files_diff_per_year <- merge(realtime_files_count_per_year, delayed_files_count_per_year, by = "year", all = TRUE) %>%
filter(!is.na(year))
ggplot(files_diff_per_year, aes(x = year)) +
geom_bar(aes(y = realtime_mode , fill = "realtime_mode "), stat = "identity") +
geom_bar(aes(y = delayed_mode, fill = "delayed_mode"), stat = "identity", position = "stack") +
labs(x = "Year", y = "Number of Files", fill = NULL) +
ggtitle("Number of files in delayed and realtime mode per year") +
theme_minimal()+
scale_x_continuous(breaks = files_diff_per_year$year, labels = files_diff_per_year$year) +
scale_fill_manual(values = c("realtime_mode " = "cyan", "delayed_mode" = "orange"),
labels = c("realtime_mode " = "Real Time mode ", "delayed_mode" = "Delayed mode")) +
theme(axis.text.x = element_text(angle = 40, hjust = 1),
legend.position = "right")
# #Difference between "adjusted" anr "realtime" variables
# plot_diff <- function(file_path) {
# # Load data for the given file
# test <- with_argo_example_cache({
# argo_prof_levels(file_path)
# })
#
# # Extract profile ID and file name
# profile_id <- sub(".*/([0-9]+)/profiles/.*", "\\1", file_path)
# file_name <- sub(".*/profiles/(.*)", "\\1", file_path)
#
# # Create individual plots
# temp <- ggplot(test, aes(x = temp, y = temp_adjusted)) +
# geom_point(size = 0.5) +
# labs(x = "Temperature", y = "Adjusted Temperature") +
# ggtitle("Temperature") +
# theme_minimal()
#
# pres <- ggplot(test, aes(x = pres, y = pres_adjusted)) +
# geom_point(size = 0.5) +
# labs(x = "Pressure", y = "Adjusted Pressure") +
# ggtitle("Pressure") +
# theme_minimal()
#
# sal <- ggplot(test, aes(x = psal, y = psal_adjusted)) +
# geom_point(size = 0.5) +
# labs(x = "Salinity", y = "Adjusted Salinity") +
# ggtitle("Salinity") +
# theme_minimal()
#
# main_title <- ggplot() +
# geom_text(aes(label = paste("Profile:", profile_id, ", File:", file_name)),
# x = 0.5, y = 0.5, hjust = 0.5, vjust = 0.5, size = 5) +
# theme_void()
#
# # Arrange plots
# grid.arrange(main_title, temp, pres, sal, ncol = 1, heights = c(0.1, 0.5, 0.5, 0.5))
# }
#
# # List of file paths
# file_paths <- c(
# paste0(path_argo_core,"/dac/aoml/3902561/profiles/D3902561_001.nc"),
# paste0(path_argo_core,"/dac/aoml/3902561/profiles/D3902561_012.nc"),
# paste0(path_argo_core,"/dac/csio/2900313/profiles/D2900313_001.nc")
# )
#
# # Create plots for each file
# for (file_path in file_paths) {
# plot_diff(file_path)
# }#Filter delayed mode files for the year 2023 and 2024
filter_year<-function(files){
#Filter files for the year 2023 and 2024
files_2023<-files %>%
filter(year(date) == 2023)
files_2024<-files %>%
filter(year(date) == 2024)
#Group by month and count the number of delayed mode files
files_month_2023 <- files_2023 %>%
group_by(month = format(date, "%B")) %>%
summarize(files_count = n())
files_month_2024 <- files_2024 %>%
group_by(month = format(date, "%B")) %>%
summarize(files_count = n())
#Order months
month_order <- c("January", "February", "March", "April", "May", "June",
"July", "August", "September", "October", "November", "December")
files_month_2023$month <- factor(files_month_2023$month, levels = month_order)
files_month_2024$month <- factor(files_month_2024$month, levels = month_order)
return(list(files_month_2023 = files_month_2023, files_month_2024 = files_month_2024))
}
delayed_files_month_2023<-filter_year(delayed_files)$files_month_2023
delayed_files_month_2024<-filter_year(delayed_files)$files_month_2024
realtime_files_month_2023<-filter_year(realtime_files)$files_month_2023
realtime_files_month_2024<-filter_year(realtime_files)$files_month_2024
#Plots
max_files_count <- max(max(delayed_files_month_2023$files_count), max(realtime_files_month_2023$files_count))
delayed_plot<- ggplot() +
geom_bar(data = delayed_files_month_2023, aes(x = month, y = files_count, fill = "2023"), stat = "identity") +
geom_bar(data = delayed_files_month_2024, aes(x = month, y = files_count, fill = "2024"), stat = "identity") +
scale_fill_manual(values = c("2023" = "skyblue", "2024" = "lightgreen")) +
labs(title = "Number of delayed-mode files per month", x = "Month", y = "Number of files") +
ylim(0, max_files_count) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
realtime_plot<- ggplot() +
geom_bar(data = realtime_files_month_2023, aes(x = month, y = files_count, fill = "2023"), stat = "identity") +
geom_bar(data = realtime_files_month_2024, aes(x = month, y = files_count, fill = "2024"), stat = "identity") +
scale_fill_manual(values = c("2023" = "skyblue", "2024" = "lightgreen")) +
labs(title = "Number of realtime-mode files per month", x = "Month", y = "Number of files") +
ylim(0, max_files_count) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
combined_plot <- delayed_plot + realtime_plot
print(combined_plot)Profiles visualisation
#Read data
read_argo_data <- function(file_path) {
data <- tryCatch({
with_argo_example_cache({argo_prof_levels(file_path)})
}, error = function(e) {
# If an error occurs (e.g., file not found), return NULL
return(NULL)
})
return(data)
}
base_path <- paste0(path_argo_core, "/dac/aoml/3902561/profiles/")
file_names <- sprintf("D3902561_%03d.nc", 1:12)
file_paths <- paste0(base_path, file_names)
all_data <- lapply(file_paths, read_argo_data)
combined_data <- do.call(rbind, all_data)
combined_data$file_index <- as.numeric(factor(combined_data$file))
#---Plots
colors <- rev(brewer.pal(11, "Spectral")) # RdYlBu palette with 11 colors
#T°C vs Pressure
temp_press_plt<-ggplot(combined_data, aes(x = temp_adjusted , y = pres_adjusted, color=file_index, group = file_index)) +
# geom_point() +
geom_path() +
coord_cartesian(xlim=c(8,30), ylim = c(300, 0)) +
labs(x = "Temperature (°C)", y = "Pressure (dbar)",
title = "Temperature measured by 1 Argo float")+
scale_y_continuous(trans = "reverse") +
scale_color_gradientn(colors = colors,
name = "File Index",
breaks = seq(min(combined_data$file_index), max(combined_data$file_index), by = 1)) +
theme(plot.title = element_text(size = 20),
plot.subtitle = element_text(size = 14),
legend.position = "none",
panel.background = element_rect(fill = NA), # Make panel background transparent
plot.background = element_rect(fill = NA), # Make plot background transparent
legend.background = element_rect(fill = NA),
panel.grid = element_blank() # Remove grid lines
)
ggsave("poster_profile_argo.png", plot = temp_press_plt, bg = "transparent", width = 5, height = 8)
#Salinity vs Pressure
sal_press_plt<- ggplot(combined_data, aes(x = psal_adjusted, y = pres_adjusted, color=file_index, group=file_index)) +
geom_point() +
geom_path()+
labs(x = "Salinity (psu)", y = "Pressure (dbar)", title = "Salinity vs Pressure")+
scale_y_continuous(trans = "reverse") +
scale_color_gradientn(colors = colors, name = "File Index", breaks = seq(min(combined_data$file_index), max(combined_data$file_index), by = 1)) +
theme(legend.position = "none")
#T/S Diagram
sal_temp_plt<- ggplot(combined_data, aes(x = psal_adjusted, y = temp_adjusted, color=file_index, group=file_index)) +
geom_point() +
geom_path()+
labs(x = "Salinity (psu)", y = "Temperature (°C)", title = "T/S Diagram")+
scale_color_gradientn(colors = colors, name = "File Index", breaks = seq(min(combined_data$file_index), max(combined_data$file_index), by = 1)) +
theme(legend.position = "right")
(temp_press_plt + sal_press_plt) / sal_temp_plt
sessionInfo()R version 4.2.2 (2022-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.5
Matrix products: default
BLAS: /usr/local/R-4.2.2/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.2.2/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] oce_1.7-10 gsw_1.1-1 sf_1.0-9 patchwork_1.1.2
[5] RColorBrewer_1.1-3 lubridate_1.9.0 timechange_0.1.1 argodata_0.1.0
[9] forcats_0.5.2 stringr_1.5.0 dplyr_1.1.3 purrr_1.0.2
[13] readr_2.1.3 tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.4
[17] tidyverse_1.3.2 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] fs_1.5.2 httr_1.4.4 rprojroot_2.0.3
[4] tools_4.2.2 backports_1.4.1 bslib_0.4.1
[7] utf8_1.2.2 R6_2.5.1 KernSmooth_2.23-20
[10] DBI_1.2.2 colorspace_2.0-3 withr_2.5.0
[13] tidyselect_1.2.0 processx_3.8.0 compiler_4.2.2
[16] git2r_0.30.1 cli_3.6.1 rvest_1.0.3
[19] RNetCDF_2.6-1 xml2_1.3.3 labeling_0.4.2
[22] sass_0.4.4 scales_1.2.1 classInt_0.4-8
[25] callr_3.7.3 proxy_0.4-27 digest_0.6.30
[28] rmarkdown_2.18 pkgconfig_2.0.3 htmltools_0.5.8.1
[31] highr_0.9 dbplyr_2.2.1 fastmap_1.1.0
[34] rlang_1.1.1 readxl_1.4.1 rstudioapi_0.15.0
[37] jquerylib_0.1.4 generics_0.1.3 farver_2.1.1
[40] jsonlite_1.8.3 googlesheets4_1.0.1 magrittr_2.0.3
[43] Rcpp_1.0.10 munsell_0.5.0 fansi_1.0.3
[46] lifecycle_1.0.3 stringi_1.7.8 whisker_0.4
[49] yaml_2.3.6 grid_4.2.2 promises_1.2.0.1
[52] crayon_1.5.2 haven_2.5.1 hms_1.1.2
[55] knitr_1.41 ps_1.7.2 pillar_1.9.0
[58] reprex_2.0.2 glue_1.6.2 evaluate_0.18
[61] getPass_0.2-2 modelr_0.1.10 vctrs_0.6.4
[64] tzdb_0.3.0 httpuv_1.6.6 cellranger_1.1.0
[67] gtable_0.3.1 assertthat_0.2.1 cachem_1.0.6
[70] xfun_0.35 broom_1.0.5 e1071_1.7-12
[73] later_1.3.0 class_7.3-20 googledrive_2.0.0
[76] gargle_1.2.1 units_0.8-0 ellipsis_0.3.2