Prepare core temperature data and vertically align to climatology
David Stappard & Jens Daniel Müller
14 December, 2023
Last updated: 2023-12-14
Checks: 7 0
Knit directory: bgc_argo_r_argodata/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211008) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 64fd104. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: output/
Untracked files:
Untracked: code/doxy_vertical_align.Rmd
Untracked: code/nitrate_vertical_align.Rmd
Untracked: nitrate_align_climatology.Rmd
Unstaged changes:
Modified: analysis/_site.yml
Deleted: analysis/doxy_vertical_align.Rmd
Deleted: analysis/nitrate_vertical_align.Rmd
Deleted: code/doxy_align_climatology.Rmd
Deleted: code/load_clim_doxy_woa.Rmd
Modified: code/start_background_job.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/temp_core_align_climatology.Rmd) and HTML
(docs/temp_core_align_climatology.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 64fd104 | ds2n19 | 2023-12-14 | revised coverage analysis and SO focused cluster analysis. |
| html | f110b74 | ds2n19 | 2023-12-13 | Build site. |
| Rmd | acb6523 | ds2n19 | 2023-12-12 | Added documentation added to tasks section at start of each script. |
| html | e60ebd2 | ds2n19 | 2023-12-07 | Build site. |
| Rmd | 59f5cc4 | ds2n19 | 2023-11-23 | Moved spatiotemporal analysis to use aligned profiles. |
| html | 80c16c2 | ds2n19 | 2023-11-15 | Build site. |
| Rmd | 3eba518 | ds2n19 | 2023-11-15 | Introduction of vertical alignment and cluster analysis to github website. |
Task
This markdown file reads previously created temperature climatology file and uses that as the definition of the depth levels that the Argo temperature data should be aligned to. As the climatology is based on pressure it is first aligned to standard depths. Previously created annual data files (yyyy_core_data_temp.rds), metadata (yyyy_core_metadata.rds) and fileid (core_fileid.rds) files are loaded from the core preprocessed folder.
Base data qc flags are checked to ensure that the float position, pressure measurements and temperature measurements are good. Pressure is used to derive the depth of each measurement. The temperature profile is checked to ensure that significant gaps (specified by the opt_gap_limit, opt_gap_min_depth and opt_gap_max_depth) do not exist. Profiles are assigned a profile_range field that identifies the depth 1 = 600 m, 2 = 1200 m and 3 = 1500 m.
The float temperature profiles are then aligned using the spline function to match the depth levels of the climatology resulting in data frame temp_core_va. An anomaly file is then created from the vertically aligned profiles and climatology.
Dependencies
yyyy_core_data_temp.rds, yyyy_core_metadata.rds, core_fileid.rds - core preprocessed folder created by load_argo_core.
temp_clim_va.rds - BGC preprocessed folder created by temp_align_climatology.
Outputs (Core preprocessed folder)
temp_core_va.rds – vertically aligned temperature profiles.
temp_clim_va.rds – climatology temperature profiles.
temp_anomaly_va.rds – anomaly temperature profiles.
Set directories
location of pre-prepared data
Set options
Define options that are used to determine profiles that we will us in the ongoing analysis
# Options
# opt_profile_depth_range
# The profile must have at least one temperature reading at a depth <= opt_profile_depth_range[1, ]
# The profile must have at least one temperature reading at a depth >= opt_profile_depth_range[2, ].
# In addition if the profile depth does not exceed the min(opt_profile_depth_range[2, ]) (i.e. 600) it will be removed.
profile_range <- c(1, 2, 3)
min_depth <- c(5.0, 5.0, 5.0)
max_depth <- c(600, 1200, 1500)
opt_profile_depth_range <- data.frame(profile_range, min_depth, max_depth)
# The profile should not have a gap greater that opt_gap_limit within the range defined by opt_gap_min_depth and opt_gap_max_depth
opt_gap_limit <- c(28, 55, 110)
opt_gap_min_depth <- c(0, 400, 1000)
opt_gap_max_depth <- c(400, 1000, 1500)
# year to be refreshed are set by opt_min_year and opt_max_year
opt_min_year = 2013
opt_max_year = 2023
# opt_measure_label, opt_xlim and opt_xbreaks are associated with formatting
opt_measure_label <- "temperature anomaly (°C)"
opt_xlim <- c(-4.5, 4.5)
opt_xbreaks <- c(-4, -2, 0, 2, 4)
# opt_exclude_shallower
# This option will exclude depths from the climatology and subsequent vertically aligned data that are shallower than opt_exclude_shallower.
# e.g. set to 4.5 to ensure that the top depth of 0.0 m is excluded
# Set to 0.0 to ensure no depths are excluded.
opt_exclude_shallower <- 4.5read climatology
Temperature climatology has been prepared during BGC analysis
temp_clim_va <- read_rds(file = paste0(path_argo_preprocessed, "/temp_clim_va.rds"))
target_depth_levels <- unique(temp_clim_va[c("depth")])
target_depth_levels <- target_depth_levels %>% select(target_depth = depth)
# Exclude 0.0 m depth from the climatology and target_depth_levels
target_depth_levels <- target_depth_levels %>% filter(target_depth > opt_exclude_shallower)Prepare years
Read annual core temperature and metadata file. Validate profile against limits and align to target_depth_range
for (target_year in opt_min_year:opt_max_year) {
#target_year = 2023
# --------------------------------------------------------------------------------
# Read data
# --------------------------------------------------------------------------------
# base data and associated metadata
core_data <- read_rds(file = paste0(path_core_preprocessed, "/", target_year, "_core_data_temp.rds"))
core_metadata <- read_rds(file = paste0(path_core_preprocessed, "/", target_year, "_core_metadata.rds"))
core_fileid <- read_rds(file = paste0(path_core_preprocessed, "/core_fileid.rds"))
#Replace file with file_id
core_metadata <- full_join(core_metadata, core_fileid)
core_metadata <- core_metadata %>%
select(-c(file))
core_data <- full_join(core_data, core_fileid)
core_data <- core_data %>%
select(-c(file))
# Select relevant field from metadata ready to join to core_data
core_metadata_select <- core_metadata %>%
filter (position_qc == 1) %>%
select(file_id,
date,
lat,
lon) %>%
mutate(year = year(date),
month = month(date),
.after = date)
# we only want pressure and temperature data
# conditions
# !is.na(depth) - pressure value must be present
# !is.na(temp_adjusted) - temperature value must be present
# pres_adjusted_qc %in% c(1, 8), temp_adjusted_qc %in% c(1, 8) and n_prof == 1 has been applied in load process
core_data_temp <- core_data %>%
filter(!is.na(depth) & !is.na(temp_adjusted)) %>%
select(file_id,
depth,
temp_adjusted)
# # join with metadata information and calculate depth field
# core_data_temp <-
# inner_join(core_metadata_select %>% select(file_id, lat),
# core_data_temp) %>%
# select(-c(lat, pres_adjusted))
# # ensure we have a depth, and temp_adjusted for all rows in bgc_data_temp
# core_data_temp <- core_data_temp %>%
# filter(!is.na(depth) & !is.na(temp_adjusted))
# clean up working tables
rm(core_data, core_metadata)
gc()
# --------------------------------------------------------------------------------
# Profile limits
# --------------------------------------------------------------------------------
# Apply the rules that are determined by options set in set_options.
# Profile must cover a set range and not contain gaps.
# Determine profile min and max depths
core_profile_limits <- core_data_temp %>%
group_by(file_id) %>%
summarise(
min_depth = min(depth),
max_depth = max(depth),
) %>%
ungroup()
# The profile much match at least one of teh range criteria
force_min <- min(opt_profile_depth_range$min_depth)
force_max <- min(opt_profile_depth_range$max_depth)
# Apply profile min and max restrictions
core_apply_limits <- core_profile_limits %>%
filter(
min_depth <= force_min &
max_depth >= force_max
)
# Ensure working data set only contains profiles that have confrormed to the range test
core_data_temp <- right_join(core_data_temp,
core_apply_limits %>% select(file_id))
# Add profile type field and set all to 1.
# All profile that meet the minimum requirements are profile_range = 1
core_data_temp <- core_data_temp %>%
mutate(profile_range = 1)
for (i in 2:nrow(opt_profile_depth_range)) {
#i = 3
range_min <- opt_profile_depth_range[i,'min_depth']
range_max <- opt_profile_depth_range[i,'max_depth']
# Apply profile min and max restrictions
core_apply_limits <- core_profile_limits %>%
filter(min_depth <= range_min &
max_depth >= range_max) %>%
select(file_id) %>%
mutate (profile_range = i)
# Update profile range to i for these profiles
# core_data_temp <- full_join(core_data_temp, core_apply_limits) %>%
# filter(!is.na(min_depth))
core_data_temp <-
core_data_temp %>% rows_update(core_apply_limits, by = "file_id")
}
# Find the gaps within the profiles
profile_gaps <- full_join(core_data_temp,
opt_profile_depth_range) %>%
filter(depth >= min_depth &
depth <= max_depth) %>%
select(file_id,
depth) %>%
arrange(file_id, depth) %>%
group_by(file_id) %>%
mutate(gap = depth - lag(depth, default = 0)) %>%
ungroup()
# Ensure we do not have gaps in the profile that invalidate it
for (i_gap in opt_gap_limit) {
# The limits to be applied in that pass of for loop
# i_gap <- opt_gap_limit[3]
i_gap_min = opt_gap_min_depth[which(opt_gap_limit == i_gap)]
i_gap_max = opt_gap_max_depth[which(opt_gap_limit == i_gap)]
# Which gaps are greater than i_gap
profile_gaps_remove <- profile_gaps %>%
filter(gap > i_gap) %>%
filter(depth >= i_gap_min & depth <= i_gap_max) %>%
distinct(file_id) %>%
pull()
# Remonve gap-containing profiles from working data set
core_data_temp <- core_data_temp %>%
filter(!file_id %in% profile_gaps_remove)
}
# clean up working tables
rm(core_apply_limits, core_profile_limits, profile_gaps, profile_gaps_remove)
gc()
# create df that contains the observations prior to vertical alignment
core_data_temp_full <- left_join(core_data_temp, core_metadata_select)
# --------------------------------------------------------------------------------
# vertical alignment
# --------------------------------------------------------------------------------
# We have a set of temperature profiles that match our criteria we now need to align that data set to match the
# depth that are in target_depth_range, this will match the range of climatology values in temp_clim_va
# create unique combinations of file_id and profile ranges
profile_range_file_id <-
core_data_temp %>%
distinct(file_id, profile_range)
# select variable of interest and prepare target_depth field
core_data_temp_clean <- core_data_temp %>%
select(-profile_range) %>%
mutate(target_depth = depth, .after = depth)
rm(core_data_temp)
gc()
# create all possible combinations of location, month and depth levels for interpolation
target_depth_grid <-
expand_grid(
target_depth_levels,
profile_range_file_id
)
# Constrain target_depth_grid to profile depth range
target_depth_grid <-
left_join(target_depth_grid, opt_profile_depth_range) %>%
filter(target_depth <= max_depth)
target_depth_grid <- target_depth_grid %>%
select(target_depth,
file_id)
# extend temp depth vectors with target depths
core_data_temp_extended <-
full_join(core_data_temp_clean, target_depth_grid) %>%
arrange(file_id, target_depth)
rm(core_data_temp_clean)
gc()
# predict spline interpolation on adjusted depth grid for temp location and month
core_data_temp_interpolated <-
core_data_temp_extended %>%
group_by(file_id) %>%
mutate(temp_spline = spline(target_depth, temp_adjusted,
method = "natural",
xout = target_depth)$y) %>%
ungroup()
rm(core_data_temp_extended)
gc()
# subset interpolated values on target depth range
core_data_temp_interpolated_clean <-
inner_join(target_depth_levels, core_data_temp_interpolated)
rm(core_data_temp_interpolated)
gc()
# select columns and rename to initial names
core_data_temp_interpolated_clean <-
core_data_temp_interpolated_clean %>%
select(file_id,
depth = target_depth,
temp = temp_spline)
# merge with profile range
core_data_temp_interpolated_clean <-
full_join(core_data_temp_interpolated_clean,
profile_range_file_id)
# merge with meta data
core_data_temp_interpolated_clean <-
left_join(core_data_temp_interpolated_clean,
core_metadata_select)
# --------------------------------------------------------------------------------
# Create anomaly profiles
# --------------------------------------------------------------------------------
# Create anomaly profiles as observed - climatology
# Create core_temp_anomaly, but only where we have an climatology temperature
core_temp_anomaly <-
inner_join(core_data_temp_interpolated_clean,
temp_clim_va)
# Calculate the anomaly temperature
core_temp_anomaly <- core_temp_anomaly %>%
mutate(anomaly = temp - clim_temp)
# -----------------------------------------------------------------------------
# Climatology check
# -----------------------------------------------------------------------------
# It is possible even though we have observational measures to full depth the climatology may not matach all depth.
# These profiles will be removed from both temp_core_va and temp_anomaly_va
# Anomaly max depth
core_temp_anomaly_depth_check <- core_temp_anomaly %>%
group_by(file_id,
profile_range) %>%
summarise(min_pdepth = min(depth),
max_pdepth = max(depth)) %>%
ungroup()
# Add the required min depth and max depth
core_temp_anomaly_depth_check <-
left_join(core_temp_anomaly_depth_check, opt_profile_depth_range)
# This profiles do not match the depth required by the profile_range
# min_depth_check <- min(target_depth_grid$target_depth)
# Please double check if the criterion should be
# each profile is checked against the min depth specified in min_depth
# max depth specified in max_depth.
remove_profiles <- core_temp_anomaly_depth_check %>%
filter((max_pdepth < max_depth) | (min_pdepth > min_depth)) %>%
distinct(file_id)
# remove from both core_data_temp_interpolated_clean and core_temp_anomaly
core_data_temp_interpolated_clean <-
anti_join(core_data_temp_interpolated_clean, remove_profiles)
core_temp_anomaly <- anti_join(core_temp_anomaly, remove_profiles)
# --------------------------------------------------------------------------------
# Write files
# --------------------------------------------------------------------------------
# Write the climatology that maps onto depth levels, interpolated temperature profiles that map onto depth levels and resulting anomaly files.
core_data_temp_interpolated_clean %>%
write_rds(file = paste0(path_core_preprocessed, "/", target_year, "_temp_core_va.rds"))
core_temp_anomaly %>%
write_rds(file = paste0(path_core_preprocessed, "/", target_year, "_temp_anomaly_va.rds"))
core_data_temp_full %>%
select(file_id, lat, lon, date, year, month, depth, temp_adjusted) %>%
write_rds(file = paste0(path_core_preprocessed, "/", target_year, "_temp_core_observed.rds"))
rm(core_data_temp_interpolated_clean, core_temp_anomaly, core_data_temp_full)
}Consolidate years
This process create three files in the path_argo_core_preprocessed directory that will be used for further analysis
# ------------------------------------------------------------------------------
# Process temperature file
# ------------------------------------------------------------------------------
consolidated_created = 0
for (target_year in opt_min_year:opt_max_year) {
# read the yearly file based on target_year
temp_core_va_yr <-
read_rds(file = paste0(path_core_preprocessed, "/", target_year, "_temp_core_va.rds"))
# Combine into a consolidated all years file
if (consolidated_created == 0) {
temp_core_va <- temp_core_va_yr
consolidated_created = 1
} else {
temp_core_va <- rbind(temp_core_va, temp_core_va_yr)
}
}
# write consolidated files
temp_core_va %>%
write_rds(file = paste0(path_core_preprocessed, "/temp_core_va.rds"))
# remove files to free space
rm(temp_core_va)
rm(temp_core_va_yr)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 3630535 193.9 7049188 376.5 5588982 298.5
Vcells 2429128466 18532.8 4273545849 32604.6 4273545849 32604.6# ------------------------------------------------------------------------------
# Process anomaly file
# ------------------------------------------------------------------------------
consolidated_created = 0
for (target_year in opt_min_year:opt_max_year) {
# read the yearly file based on target_year
temp_anomaly_va_yr <-
read_rds(file = paste0(path_core_preprocessed, "/", target_year, "_temp_anomaly_va.rds"))
# Combine into a consolidated all years file
if (consolidated_created == 0) {
temp_anomaly_va <- temp_anomaly_va_yr
consolidated_created = 1
} else {
temp_anomaly_va <- rbind(temp_anomaly_va, temp_anomaly_va_yr)
}
}
# write consolidated files
temp_anomaly_va %>%
write_rds(file = paste0(path_core_preprocessed, "/temp_anomaly_va.rds"))
# remove files to free space
rm(temp_anomaly_va)
rm(temp_anomaly_va_yr)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 3630807 194.0 7049188 376.5 5588982 298.5
Vcells 2429128928 18532.8 5128335018 39126.1 4273545849 32604.6# ------------------------------------------------------------------------------
# Process observed file
# ------------------------------------------------------------------------------
consolidated_created = 0
for (target_year in opt_min_year:opt_max_year) {
# read the yearly file based on target_year
temp_core_observed_yr <-
read_rds(file = paste0(path_core_preprocessed, "/", target_year, "_temp_core_observed.rds"))
# Combine into a consolidated all years file
if (consolidated_created == 0) {
temp_core_observed <- temp_core_observed_yr
consolidated_created = 1
} else {
temp_core_observed <- rbind(temp_core_observed, temp_core_observed_yr)
}
}
# write consolidated files
temp_core_observed %>%
write_rds(file = paste0(path_core_preprocessed, "/temp_core_observed.rds"))
# remove files to free space
rm(temp_core_observed)
rm(temp_core_observed_yr)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 3630900 194.0 7049188 376.5 5588982 298.5
Vcells 2429129015 18532.8 12826199373 97856.2 16032749216 122320.2read files
Read files that were previously created ready for analysis
# read files
temp_core_va <- read_rds(file = paste0(path_core_preprocessed, "/temp_core_va.rds"))
temp_clim_va <- read_rds(file = paste0(path_argo_preprocessed, "/temp_clim_va.rds"))
temp_anomaly_va <- read_rds(file = paste0(path_core_preprocessed, "/temp_anomaly_va.rds"))Temperature anomaly
Details of mean anomaly over analysis period
max_depth_1 <- opt_profile_depth_range[1, "max_depth"]
max_depth_2 <- opt_profile_depth_range[2, "max_depth"]
max_depth_3 <- opt_profile_depth_range[3, "max_depth"]
# Profiles to 600m
anomaly_overall_mean_1 <- temp_anomaly_va %>%
filter(profile_range %in% c(1, 2, 3) & depth <= max_depth_1) %>%
group_by(depth) %>%
summarise(temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE),
temp_anomaly_sd = sd(anomaly, na.rm = TRUE))
anomaly_year_mean_1 <- temp_anomaly_va %>%
filter(profile_range %in% c(1, 2, 3) & depth <= max_depth_1) %>%
group_by(year, depth) %>%
summarise(temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE),
temp_anomaly_sd = sd(anomaly, na.rm = TRUE))
# Profiles to 1200m
anomaly_overall_mean_2 <- temp_anomaly_va %>%
filter(profile_range %in% c(2, 3) & depth <= max_depth_2) %>%
group_by(depth) %>%
summarise(temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE),
temp_anomaly_sd = sd(anomaly, na.rm = TRUE))
anomaly_year_mean_2 <- temp_anomaly_va %>%
filter(profile_range %in% c(2, 3) & depth <= max_depth_2) %>%
group_by(year, depth) %>%
summarise(temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE),
temp_anomaly_sd = sd(anomaly, na.rm = TRUE))
# Profiles to 1500m
anomaly_overall_mean_3 <- temp_anomaly_va %>%
filter(profile_range %in% c(3) & depth <= max_depth_3) %>%
group_by(depth) %>%
summarise(temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE),
temp_anomaly_sd = sd(anomaly, na.rm = TRUE))
anomaly_year_mean_3 <- temp_anomaly_va %>%
filter(profile_range %in% c(3) & depth <= max_depth_3) %>%
group_by(year, depth) %>%
summarise(temp_count = n(),
temp_anomaly_mean = mean(anomaly, na.rm = TRUE),
temp_anomaly_sd = sd(anomaly, na.rm = TRUE))
# All years anomaly
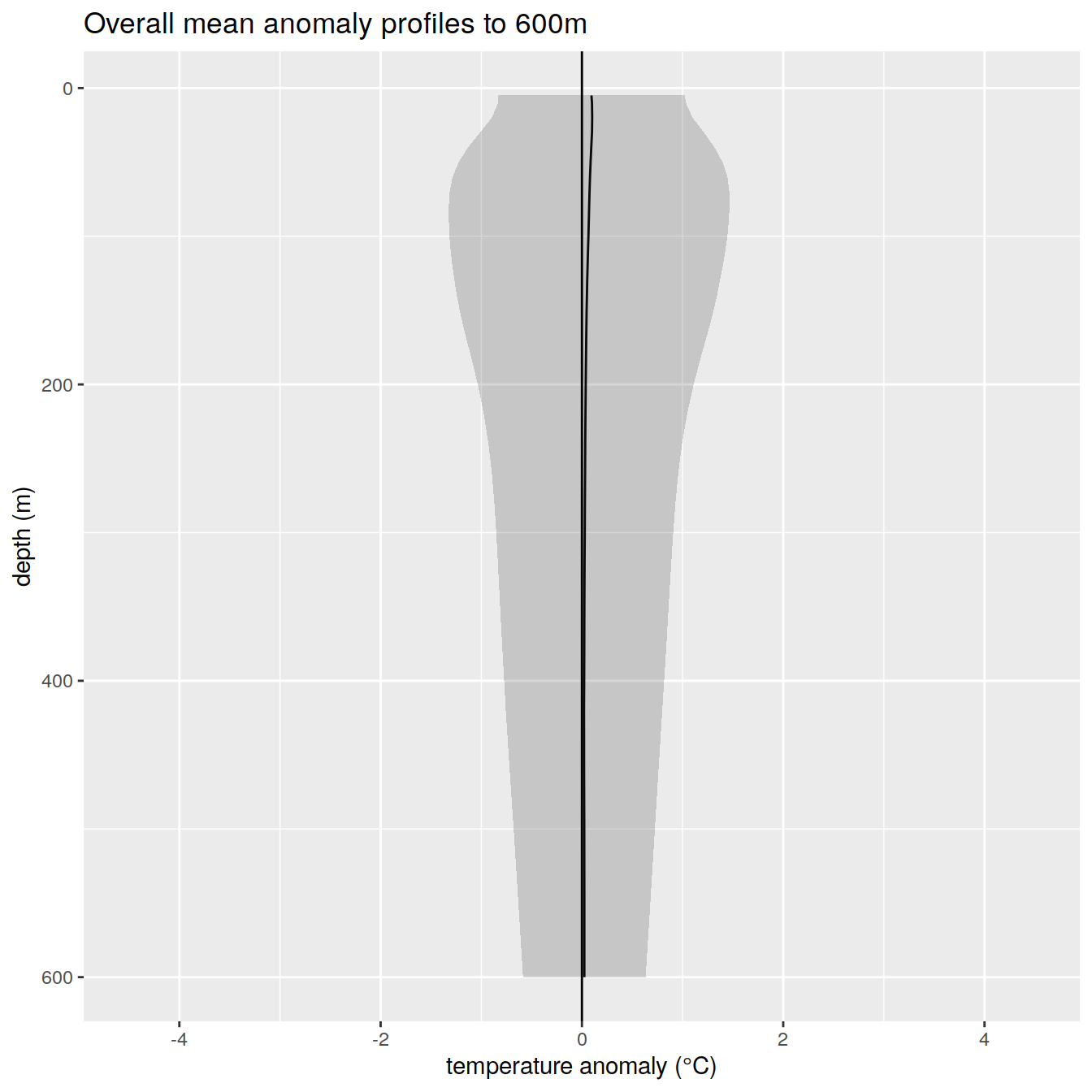
anomaly_overall_mean_1 %>%
ggplot()+
geom_path(aes(x = temp_anomaly_mean,
y = depth))+
geom_ribbon(aes(xmax = temp_anomaly_mean + temp_anomaly_sd,
xmin = temp_anomaly_mean - temp_anomaly_sd,
y = depth),
alpha = 0.2)+
geom_vline(xintercept = 0)+
scale_y_reverse() +
coord_cartesian(xlim = opt_xlim) +
scale_x_continuous(breaks = opt_xbreaks) +
labs(
title = paste0('Overall mean anomaly profiles to ', max_depth_1, 'm'),
x = opt_measure_label,
y = 'depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
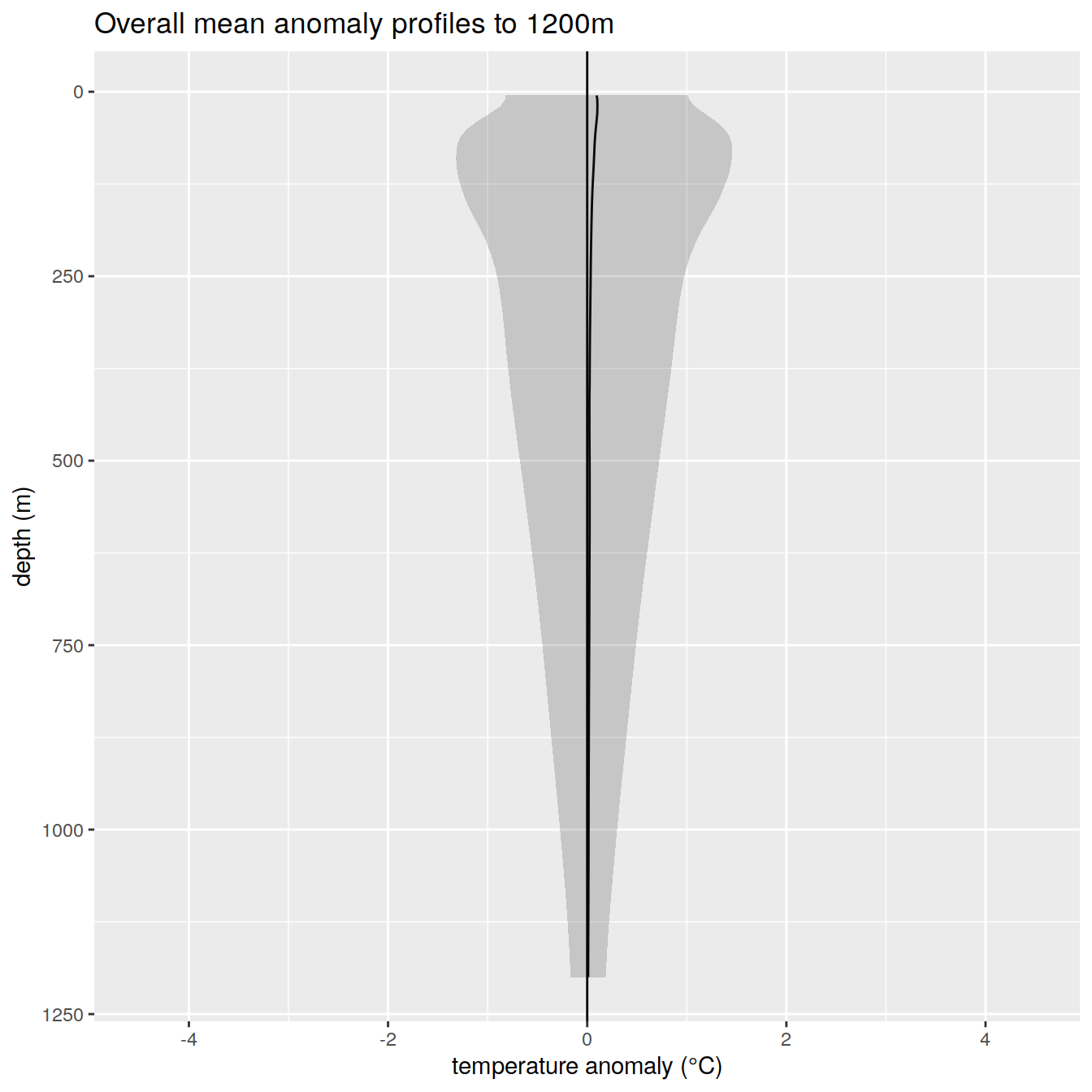
anomaly_overall_mean_2 %>%
ggplot()+
geom_path(aes(x = temp_anomaly_mean,
y = depth))+
geom_ribbon(aes(xmax = temp_anomaly_mean + temp_anomaly_sd,
xmin = temp_anomaly_mean - temp_anomaly_sd,
y = depth),
alpha = 0.2)+
geom_vline(xintercept = 0)+
scale_y_reverse() +
coord_cartesian(xlim = opt_xlim) +
scale_x_continuous(breaks = opt_xbreaks) +
labs(
title = paste0('Overall mean anomaly profiles to ', max_depth_2, 'm'),
x = opt_measure_label,
y = 'depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
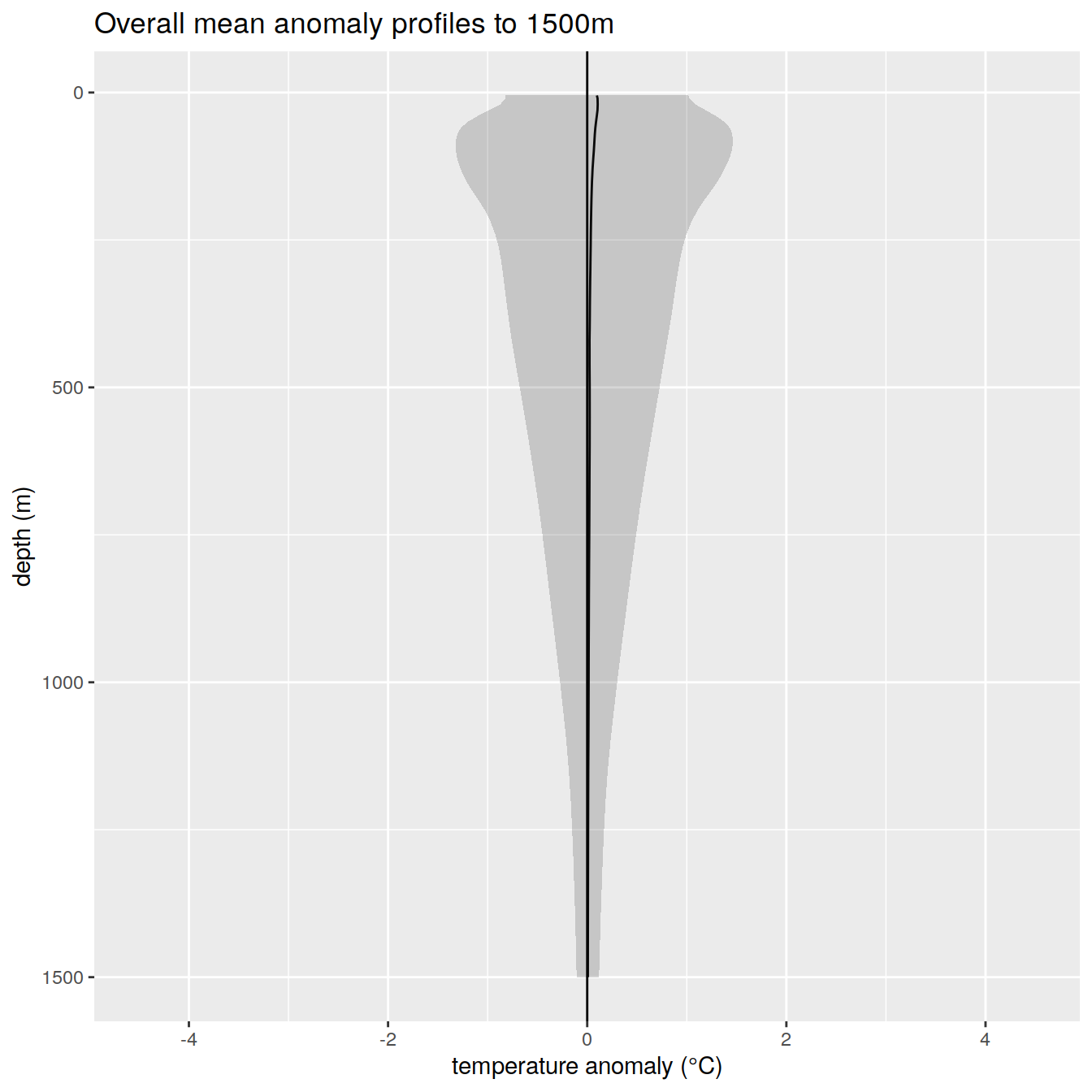
anomaly_overall_mean_3 %>%
ggplot()+
geom_path(aes(x = temp_anomaly_mean,
y = depth))+
geom_ribbon(aes(xmax = temp_anomaly_mean + temp_anomaly_sd,
xmin = temp_anomaly_mean - temp_anomaly_sd,
y = depth),
alpha = 0.2)+
geom_vline(xintercept = 0)+
scale_y_reverse() +
coord_cartesian(xlim = opt_xlim) +
scale_x_continuous(breaks = opt_xbreaks) +
labs(
title = paste0('Overall mean anomaly profiles to ', max_depth_3, 'm'),
x = opt_measure_label,
y = 'depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
# yearly anomaly
anomaly_year_mean_1 %>%
ggplot()+
geom_path(aes(x = temp_anomaly_mean,
y = depth))+
geom_ribbon(aes(xmax = temp_anomaly_mean + temp_anomaly_sd,
xmin = temp_anomaly_mean - temp_anomaly_sd,
y = depth),
alpha = 0.2)+
geom_vline(xintercept = 0)+
scale_y_reverse() +
facet_wrap(~year)+
coord_cartesian(xlim = opt_xlim) +
scale_x_continuous(breaks = opt_xbreaks) +
labs(
title = paste0('Yearly mean anomaly profiles to ', max_depth_1, 'm'),
x = opt_measure_label,
y = 'depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
anomaly_year_mean_2 %>%
ggplot()+
geom_path(aes(x = temp_anomaly_mean,
y = depth))+
geom_ribbon(aes(xmax = temp_anomaly_mean + temp_anomaly_sd,
xmin = temp_anomaly_mean - temp_anomaly_sd,
y = depth),
alpha = 0.2)+
geom_vline(xintercept = 0)+
scale_y_reverse() +
facet_wrap(~year)+
coord_cartesian(xlim = opt_xlim) +
scale_x_continuous(breaks = opt_xbreaks) +
labs(
title = paste0('Yearly mean anomaly profiles to ', max_depth_2, 'm'),
x = opt_measure_label,
y = 'depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
anomaly_year_mean_3 %>%
ggplot()+
geom_path(aes(x = temp_anomaly_mean,
y = depth))+
geom_ribbon(aes(xmax = temp_anomaly_mean + temp_anomaly_sd,
xmin = temp_anomaly_mean - temp_anomaly_sd,
y = depth),
alpha = 0.2)+
geom_vline(xintercept = 0)+
scale_y_reverse() +
facet_wrap(~year)+
coord_cartesian(xlim = opt_xlim) +
scale_x_continuous(breaks = opt_xbreaks) +
labs(
title = paste0('Yearly mean anomaly profiles to ', max_depth_3, 'm'),
x = opt_measure_label,
y = 'depth (m)'
)
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
#rm(anomaly_overall_mean)Profile counts
Details of the number of profiles and to which depths over the analysis period
temp_histogram <- temp_core_va %>%
group_by(year, profile_range = as.character(profile_range)) %>%
summarise(num_profiles = n_distinct(file_id)) %>%
ungroup()
temp_histogram %>%
ggplot() +
geom_bar(
aes(
x = year,
y = num_profiles,
fill = profile_range,
group = profile_range
),
position = "stack",
stat = "identity"
) +
scale_fill_viridis_d() +
labs(title = "temperature profiles per year and profile range",
x = "year",
y = "profile count",
fill = "profile range")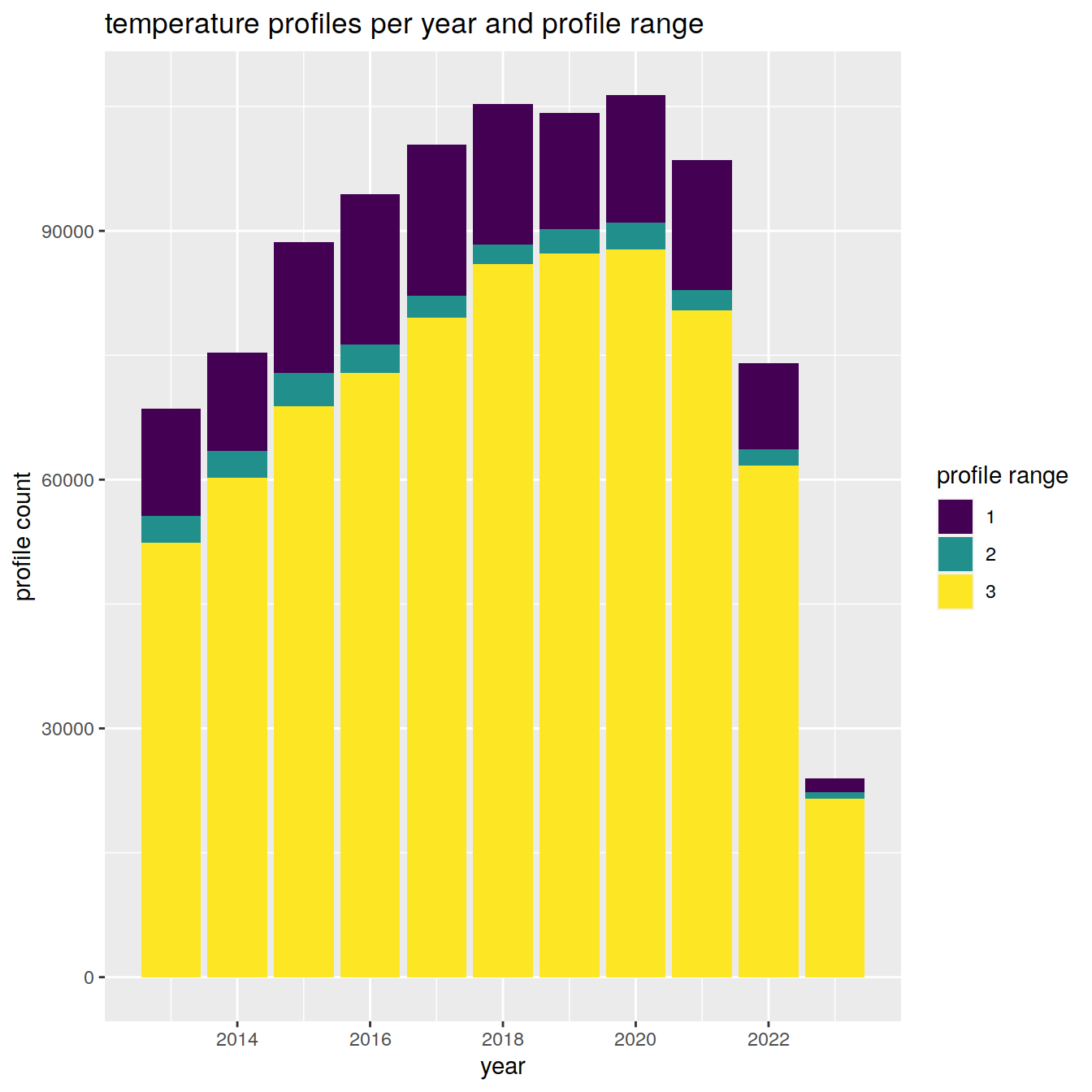
| Version | Author | Date |
|---|---|---|
| 80c16c2 | ds2n19 | 2023-11-15 |
sessionInfo()R version 4.2.2 (2022-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.5
Matrix products: default
BLAS: /usr/local/R-4.2.2/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.2.2/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggforce_0.4.1 gsw_1.1-1 gridExtra_2.3 lubridate_1.9.0
[5] timechange_0.1.1 argodata_0.1.0 forcats_0.5.2 stringr_1.5.0
[9] dplyr_1.1.3 purrr_1.0.2 readr_2.1.3 tidyr_1.3.0
[13] tibble_3.2.1 ggplot2_3.4.4 tidyverse_1.3.2
loaded via a namespace (and not attached):
[1] httr_1.4.4 sass_0.4.4 viridisLite_0.4.1
[4] jsonlite_1.8.3 modelr_0.1.10 bslib_0.4.1
[7] assertthat_0.2.1 highr_0.9 googlesheets4_1.0.1
[10] cellranger_1.1.0 yaml_2.3.6 pillar_1.9.0
[13] backports_1.4.1 glue_1.6.2 digest_0.6.30
[16] promises_1.2.0.1 polyclip_1.10-4 rvest_1.0.3
[19] colorspace_2.0-3 htmltools_0.5.3 httpuv_1.6.6
[22] pkgconfig_2.0.3 broom_1.0.5 haven_2.5.1
[25] scales_1.2.1 tweenr_2.0.2 whisker_0.4
[28] later_1.3.0 tzdb_0.3.0 git2r_0.30.1
[31] googledrive_2.0.0 generics_0.1.3 farver_2.1.1
[34] ellipsis_0.3.2 cachem_1.0.6 withr_2.5.0
[37] cli_3.6.1 magrittr_2.0.3 crayon_1.5.2
[40] readxl_1.4.1 evaluate_0.18 fs_1.5.2
[43] fansi_1.0.3 MASS_7.3-58.1 xml2_1.3.3
[46] tools_4.2.2 hms_1.1.2 gargle_1.2.1
[49] lifecycle_1.0.3 munsell_0.5.0 reprex_2.0.2
[52] compiler_4.2.2 jquerylib_0.1.4 RNetCDF_2.6-1
[55] rlang_1.1.1 grid_4.2.2 rstudioapi_0.15.0
[58] labeling_0.4.2 rmarkdown_2.18 gtable_0.3.1
[61] DBI_1.1.3 R6_2.5.1 knitr_1.41
[64] fastmap_1.1.0 utf8_1.2.2 workflowr_1.7.0
[67] rprojroot_2.0.3 stringi_1.7.8 Rcpp_1.0.10
[70] vctrs_0.6.4 dbplyr_2.2.1 tidyselect_1.2.0
[73] xfun_0.35