eMLR - model fitting
Jens Daniel Müller and Donghe Zhu
23 January, 2021
Last updated: 2021-01-23
Checks: 7 0
Knit directory: emlr_mod_v_XXX/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200707) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0819197. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rproj.user/
Unstaged changes:
Modified: data/auxillary/params_local.rds
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/eMLR_model_fitting.Rmd) and HTML (docs/eMLR_model_fitting.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 28509fc | Donghe-Zhu | 2021-01-23 | Build site. |
| html | 4c28e4a | Donghe-Zhu | 2021-01-22 | Build site. |
| html | 24cc264 | jens-daniel-mueller | 2021-01-22 | cleaned /docs before creating copies |
| html | 88eb28f | Donghe-Zhu | 2021-01-21 | Build site. |
| html | 2679490 | Donghe-Zhu | 2021-01-21 | Build site. |
| html | 7891955 | Donghe-Zhu | 2021-01-21 | Build site. |
| html | d4cf1cb | Donghe-Zhu | 2021-01-21 | Build site. |
| Rmd | 167eeec | Donghe-Zhu | 2021-01-21 | surface DIC calculation with atmospheric equilibrium option |
| html | 1f3e5b6 | jens-daniel-mueller | 2021-01-20 | Build site. |
| html | 0e7bdf1 | jens-daniel-mueller | 2021-01-15 | cleaning template repository |
| html | 73cbef3 | jens-daniel-mueller | 2021-01-15 | Build site. |
| html | 4571843 | jens-daniel-mueller | 2021-01-14 | revision and html deleted for template copying |
| html | 23151cd | jens-daniel-mueller | 2021-01-14 | Build site. |
| html | b3564aa | jens-daniel-mueller | 2021-01-14 | Build site. |
| html | 8d032c3 | jens-daniel-mueller | 2021-01-14 | Build site. |
| html | 022871c | Donghe-Zhu | 2021-01-13 | Build site. |
| Rmd | d44f36f | Donghe-Zhu | 2021-01-13 | reorder analysis final |
| html | 17dee1d | jens-daniel-mueller | 2021-01-13 | Build site. |
| Rmd | 9e04fd7 | jens-daniel-mueller | 2021-01-13 | local rebuild after revision |
| html | a076226 | Donghe-Zhu | 2021-01-11 | Build site. |
| Rmd | 52eff18 | Donghe-Zhu | 2021-01-09 | Implemet model_run and subsetting |
| html | 7cdea0c | jens-daniel-mueller | 2021-01-06 | Build site. |
| Rmd | b5934dd | jens-daniel-mueller | 2021-01-06 | local rebuild after revision |
| html | fa85b93 | jens-daniel-mueller | 2021-01-06 | Build site. |
| html | e5cb81a | Donghe-Zhu | 2021-01-05 | Build site. |
| Rmd | 608cc45 | Donghe-Zhu | 2021-01-05 | modification of analysis |
| html | a499f10 | Donghe-Zhu | 2021-01-05 | Build site. |
| Rmd | 715bdb4 | Donghe-Zhu | 2021-01-02 | model modification |
| html | fb8a752 | Donghe-Zhu | 2020-12-23 | Build site. |
| Rmd | 82e3c9c | Donghe-Zhu | 2020-12-23 | first build after creating model template |
| html | 8fae0b2 | Donghe-Zhu | 2020-12-21 | Build site. |
| Rmd | 00a1322 | Donghe-Zhu | 2020-12-21 | first build after creating model template |
| Rmd | d73ae35 | Donghe-Zhu | 2020-12-21 | first version with lm error |
| html | c8b76b3 | jens-daniel-mueller | 2020-12-19 | Build site. |
| Rmd | b5fedce | jens-daniel-mueller | 2020-12-19 | first build after creating model template |
| Rmd | 8e8abf5 | Jens Müller | 2020-12-18 | Initial commit |
1 Required data
Required are:
- cleaned and prepared GLODAP-based synthetic model subsetting file
GLODAP <-
read_csv(paste(path_version_data,
"GLODAPv2.2020_MLR_fitting_ready.csv",
sep = ""))2 Predictor combinations
Find all possible combinations of following considered predictor variables:
- sal, temp, aou, nitrate, silicate, phosphate, phosphate_star
# the following code is a workaround to find all predictor combinations
# using the olsrr package and fit all models for one era, slab, and basin
i_basin <- unique(GLODAP$basin)[1]
i_era <- unique(GLODAP$era)[1]
# subset one basin and era for fitting
GLODAP_basin_era <- GLODAP %>%
filter(basin == i_basin, era == i_era)
i_gamma_slab <- unique(GLODAP_basin_era$gamma_slab)[1]
print(i_gamma_slab)
# subset one gamma slab
GLODAP_basin_era_slab <- GLODAP_basin_era %>%
filter(gamma_slab == i_gamma_slab)
# fit the full linear model, i.e. all predictor combinations
lm_full <- lm(paste(
params_local$MLR_target,
paste(params_local$MLR_predictors, collapse = " + "),
sep = " ~ "
),
data = GLODAP_basin_era_slab)
# fit linear models for all possible predictor combinations
# unfortunately, this functions does not provide model coefficients (yet)
lm_all <- ols_step_all_possible(lm_full)
# convert to tibble
lm_all <- as_tibble(lm_all)
# extract relevant columns and format model formula
lm_all <- lm_all %>%
select(n, predictors) %>%
mutate(lm_coeff = str_replace_all(predictors, " ", " + "),
lm_coeff = paste(params_local$MLR_target, "~", lm_coeff))
# remove helper objects
rm(i_gamma_slab,
i_era,
i_basin,
GLODAP_basin_era,
GLODAP_basin_era_slab,
lm_full)3 Apply predictor threshold
Select combinations with a total number of predictors in the range:
- Minimum: 2
- Maximum: 5
lm_all <- lm_all %>%
filter(n >= params_local$MLR_predictors_min,
n <= params_local$MLR_predictors_max)This results in a total number of MLR models of:
- 112
4 Fit all models
Individual linear regression models were fitted for the chosen target variable:
- cstar_tref
as a function of each predictor combination. Fitting was performed separately within each basin, era, and slab. Model diagnostics, such as the root mean squared error (RMSE), were calculated for each fitted model.
# loop across all basins, era, gamma slabs, and MLRs
# fit all MLR models
for (i_basin in unique(GLODAP$basin)) {
for (i_era in unique(GLODAP$era)) {
# i_basin <- unique(GLODAP$basin)[1]
# i_era <- unique(GLODAP$era)[1]
print(i_basin)
print(i_era)
GLODAP_basin_era <- GLODAP %>%
filter(basin == i_basin, era == i_era)
for (i_gamma_slab in unique(GLODAP_basin_era$gamma_slab)) {
# i_gamma_slab <- unique(GLODAP_basin_era$gamma_slab)[1]
print(i_gamma_slab)
GLODAP_basin_era_slab <- GLODAP_basin_era %>%
filter(gamma_slab == i_gamma_slab)
for (i_predictors in unique(lm_all$predictors)) {
# i_predictors <- unique(lm_all$predictors)[110]
# extract one model definition
i_lm <- lm_all %>%
filter(predictors == i_predictors) %>%
select(lm_coeff) %>%
pull()
# extract number of predictors
i_n_predictors <- lm_all %>%
filter(predictors == i_predictors) %>%
select(n) %>%
pull()
# fit model
i_lm_fit <- lm(as.formula(i_lm),
data = GLODAP_basin_era_slab)
# find max predictor correlation
i_cor_max <- GLODAP_basin_era_slab %>%
select(!!!syms(str_split(i_predictors, " ",
simplify = TRUE))) %>%
correlate(quiet = TRUE) %>%
select(-term) %>%
abs() %>%
max(na.rm = TRUE)
# calculate root mean squared error
i_rmse <- sqrt(
c(crossprod(i_lm_fit$residuals)) /
length(i_lm_fit$residuals)
)
# calculate maximum residual
i_resid_max <- max(abs(i_lm_fit$residuals))
# calculate Akaike information criterion aic
i_aic <- AIC(i_lm_fit)
# collect model coefficients and diagnostics
coefficients <- tidy(i_lm_fit)
coefficients <- coefficients %>%
mutate(
basin = i_basin,
era = i_era,
gamma_slab = i_gamma_slab,
model = i_lm,
rmse = i_rmse,
aic = i_aic,
resid_max = i_resid_max,
n_predictors = i_n_predictors,
na_predictor = anyNA(coefficients$estimate),
cor_max = i_cor_max
)
if (exists("lm_all_fitted")) {
lm_all_fitted <- bind_rows(lm_all_fitted, coefficients)
}
if (!exists("lm_all_fitted")) {
lm_all_fitted <- coefficients
}
# # plot model diagnostics, if activated
# if (params_local$plot_all_figures == "y") {
# p_model <- ggnostic(
# i_lm_fit,
# columnsY = c(params_local$MLR_target, ".fitted", ".resid"),
# title = paste(
# "| era:",
# i_era,
# "| basin:",
# i_basin,
# "| gamma slab:",
# i_gamma_slab,
# "| predictors:",
# i_predictors
# )
# )
#
# ggsave(
# plot = p_model,
# path = paste(path_version_figures, "eMLR_diagnostics/", sep = ""),
# filename = paste(
# "MLR_residuals",
# i_era,
# i_basin,
# i_gamma_slab,
# i_predictors,
# "predictors.png",
# sep = "_"
# ),
# width = 14,
# height = 8
# )
#
# rm(p_model)
#
# }
}
}
}
}
rm(i_lm_fit, coefficients, i_rmse,
GLODAP_basin_era, GLODAP_basin_era_slab,
i_lm,
i_basin, i_era, i_gamma_slab, i_predictors,
lm_all,
i_aic, i_n_predictors, i_resid_max)5 Prepare coeffcients
Coefficients are prepared for the mapping of Cant and the chosen target variable.
5.1 Formatting
# select relevant columns
lm_all_fitted <- lm_all_fitted %>%
select(basin, gamma_slab, era, model, n_predictors,
term, estimate,
rmse, aic, resid_max, na_predictor, cor_max)
# set coefficient to zero if not fitted (=NA)
lm_all_fitted <- lm_all_fitted %>%
mutate(estimate = if_else(is.na(estimate), 0, estimate))
# Prepare model coefficients for mapping of target variable
lm_all_fitted_wide <- lm_all_fitted %>%
pivot_wider(values_from = estimate,
names_from = term,
names_prefix = "coeff_",
values_fill = 0)5.2 Predictor selection
Within each basin and slab, the following number of best linear regression models was selected:
- 10
The criterion used to select the best models was:
- rmse
The criterion was summed up for two adjacent eras, and the models with lowest summed values were selected.
Please note, that currently the lm() function produces NAs for some predictors. It is not yet entirely clear when this happens, but presumably it is caused by some form of collinearity between predictors, such that including another predictor does not help to explain the target variable any better. The issues also expresses as exactly identical rmse values of different models. As an interim solution, models with fitted NA predictors were not included.
# remove models with predictors fitted as NA
lm_all_fitted_wide <- lm_all_fitted_wide %>%
filter(na_predictor == FALSE)# calculate RMSE sum for adjacent eras
lm_all_fitted_wide_eras <- lm_all_fitted_wide %>%
select(basin, gamma_slab, model, era, rmse, aic, resid_max) %>%
arrange(era) %>%
group_by(basin, gamma_slab, model) %>%
mutate(eras = paste(lag(era), era, sep = " --> "),
rmse_sum = rmse + lag(rmse),
aic_sum = aic + lag(aic)
) %>%
ungroup() %>%
select(-c(era)) %>%
drop_na()
# subset models with lowest summed criterion
# chose which criterion is applied
if (params_local$MLR_criterion == "aic") {
lm_best <- lm_all_fitted_wide_eras %>%
group_by(basin, gamma_slab, eras) %>%
slice_min(order_by = aic_sum,
with_ties = FALSE,
n = params_local$MLR_number) %>%
ungroup() %>%
arrange(basin, gamma_slab, eras, model)
} else {
lm_best <- lm_all_fitted_wide_eras %>%
group_by(basin, gamma_slab, eras) %>%
slice_min(order_by = rmse_sum,
with_ties = FALSE,
n = params_local$MLR_number) %>%
ungroup() %>%
arrange(basin, gamma_slab, eras, model)
}
# print table
lm_best %>%
kable() %>%
add_header_above() %>%
kable_styling() %>%
scroll_box(width = "100%", height = "400px")| basin | gamma_slab | model | rmse | aic | resid_max | eras | rmse_sum | aic_sum |
|---|---|---|---|---|---|---|---|---|
| Atlantic | (-Inf,26] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 1.0529313 | 508.0936 | 4.021817 | 1982-1999 –> 2000-2012 | 2.3845411 | 1286.0801 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + aou + phosphate + phosphate_star | 1.0669064 | 510.5238 | 4.506279 | 1982-1999 –> 2000-2012 | 2.3985898 | 1286.5351 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 1.0650698 | 511.9449 | 4.426016 | 1982-1999 –> 2000-2012 | 2.3967507 | 1289.9554 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 1.0574420 | 509.5299 | 4.227758 | 1982-1999 –> 2000-2012 | 2.3903131 | 1287.9406 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 1.0589440 | 510.0068 | 4.281729 | 1982-1999 –> 2000-2012 | 2.3921140 | 1288.5180 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + phosphate | 1.0709739 | 511.8023 | 4.694811 | 1982-1999 –> 2000-2012 | 2.4040133 | 1288.2696 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 1.0646493 | 511.8122 | 4.234683 | 1982-1999 –> 2000-2012 | 2.3962400 | 1289.7923 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 1.0694684 | 513.3297 | 4.624413 | 1982-1999 –> 2000-2012 | 2.4024546 | 1291.7791 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 1.0708017 | 513.7483 | 4.671370 | 1982-1999 –> 2000-2012 | 2.4040952 | 1292.3010 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 1.0605961 | 510.5306 | 4.333869 | 1982-1999 –> 2000-2012 | 2.3945656 | 1289.3103 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 1.1285639 | 254.2220 | 2.798637 | 2000-2012 –> 2013-2019 | 2.1814952 | 762.3155 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 1.1819575 | 261.4332 | 3.306338 | 2000-2012 –> 2013-2019 | 2.2470273 | 773.3781 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 1.1357044 | 255.2059 | 2.761390 | 2000-2012 –> 2013-2019 | 2.1931464 | 764.7358 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 1.1385676 | 255.5987 | 2.813203 | 2000-2012 –> 2013-2019 | 2.1975116 | 765.6055 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + nitrate + silicate | 1.1929038 | 262.8713 | 2.724893 | 2000-2012 –> 2013-2019 | 2.2980531 | 787.2281 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 1.2437146 | 269.3784 | 3.455189 | 2000-2012 –> 2013-2019 | 2.3083639 | 781.1906 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 1.1901456 | 262.5102 | 3.453009 | 2000-2012 –> 2013-2019 | 2.2596141 | 775.8399 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 1.1931661 | 262.9056 | 3.497480 | 2000-2012 –> 2013-2019 | 2.2639678 | 776.6539 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 1.1417526 | 256.0345 | 2.864096 | 2000-2012 –> 2013-2019 | 2.2023487 | 766.5651 |
| Atlantic | (-Inf,26] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 1.1950624 | 263.1534 | 3.519214 | 2000-2012 –> 2013-2019 | 2.2672406 | 777.3333 |
| Atlantic | (26,26.8] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 3.8890327 | 14988.1178 | 17.924664 | 1982-1999 –> 2000-2012 | 7.7302552 | 35875.8233 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 3.8093271 | 14876.4608 | 35.094315 | 1982-1999 –> 2000-2012 | 7.5543650 | 35572.7063 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 3.9527677 | 15075.7677 | 12.077204 | 1982-1999 –> 2000-2012 | 7.6801708 | 35736.3775 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 3.9016580 | 15005.5940 | 34.479878 | 1982-1999 –> 2000-2012 | 7.7462780 | 35899.9743 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 3.9178675 | 15027.9487 | 34.551996 | 1982-1999 –> 2000-2012 | 7.7812613 | 35959.1070 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 4.0773464 | 15243.0835 | 12.235786 | 1982-1999 –> 2000-2012 | 7.9517081 | 36195.6453 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 3.9571830 | 15081.7873 | 34.091338 | 1982-1999 –> 2000-2012 | 7.8588609 | 36087.3938 |
| Atlantic | (26,26.8] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate | 3.9171389 | 15026.9458 | 19.265703 | 1982-1999 –> 2000-2012 | 7.7858304 | 35968.4500 |
| Atlantic | (26,26.8] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate_star | 3.9335417 | 15049.4774 | 20.037089 | 1982-1999 –> 2000-2012 | 7.8206681 | 36026.8731 |
| Atlantic | (26,26.8] | cstar_tref ~ temp + nitrate + silicate + phosphate + phosphate_star | 3.9537134 | 15077.0576 | 19.792862 | 1982-1999 –> 2000-2012 | 7.8585277 | 36088.7307 |
| Atlantic | (26,26.8] | cstar_tref ~ aou + nitrate + phosphate + phosphate_star | 3.9880919 | 7594.8924 | 11.221907 | 2000-2012 –> 2013-2019 | 8.0029119 | 22752.6485 |
| Atlantic | (26,26.8] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 3.9766030 | 7589.0857 | 10.789211 | 2000-2012 –> 2013-2019 | 7.8656357 | 22577.2035 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 3.6938392 | 7389.4868 | 10.426509 | 2000-2012 –> 2013-2019 | 7.5031663 | 22265.9476 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 3.7903241 | 7459.2615 | 10.303387 | 2000-2012 –> 2013-2019 | 7.6919822 | 22464.8555 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 3.8027479 | 7468.1166 | 10.282146 | 2000-2012 –> 2013-2019 | 7.7206154 | 22496.0653 |
| Atlantic | (26,26.8] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 3.8477109 | 7499.9242 | 10.304416 | 2000-2012 –> 2013-2019 | 7.8048939 | 22581.7116 |
| Atlantic | (26,26.8] | cstar_tref ~ temp + aou + nitrate + phosphate + phosphate_star | 3.9836624 | 7593.8852 | 10.957736 | 2000-2012 –> 2013-2019 | 7.9973746 | 22752.1534 |
| Atlantic | (26,26.8] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate | 3.9827716 | 7593.2800 | 10.489451 | 2000-2012 –> 2013-2019 | 7.8999104 | 22620.2258 |
| Atlantic | (26,26.8] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate_star | 3.9885200 | 7597.1828 | 10.434835 | 2000-2012 –> 2013-2019 | 7.9220617 | 22646.6603 |
| Atlantic | (26,26.8] | cstar_tref ~ temp + nitrate + silicate + phosphate + phosphate_star | 4.0053015 | 7608.5443 | 10.301486 | 2000-2012 –> 2013-2019 | 7.9590149 | 22685.6019 |
| Atlantic | (26.8,27.2] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 2.6049278 | 23706.1447 | 12.821629 | 1982-1999 –> 2000-2012 | 4.7227741 | 52194.8778 |
| Atlantic | (26.8,27.2] | cstar_tref ~ aou + silicate + phosphate + phosphate_star | 2.7988071 | 24419.8743 | 15.180973 | 1982-1999 –> 2000-2012 | 5.0446380 | 53676.7874 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 2.7087789 | 24095.9017 | 12.446419 | 1982-1999 –> 2000-2012 | 5.2917970 | 55190.9183 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 2.4952938 | 23277.4496 | 12.395986 | 1982-1999 –> 2000-2012 | 4.5472903 | 51351.5796 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 2.5926602 | 23659.0811 | 11.819352 | 1982-1999 –> 2000-2012 | 4.7711050 | 52518.1196 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 2.6956406 | 24047.4269 | 16.030288 | 1982-1999 –> 2000-2012 | 4.9917184 | 53596.7763 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 2.7291979 | 24170.7744 | 18.335328 | 1982-1999 –> 2000-2012 | 5.0677790 | 53960.8811 |
| Atlantic | (26.8,27.2] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate | 2.8307542 | 24535.0326 | 13.200750 | 1982-1999 –> 2000-2012 | 5.1366416 | 54140.3412 |
| Atlantic | (26.8,27.2] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate_star | 2.9021450 | 24783.3550 | 15.154767 | 1982-1999 –> 2000-2012 | 5.3019287 | 54912.5616 |
| Atlantic | (26.8,27.2] | cstar_tref ~ temp + aou + silicate + phosphate + phosphate_star | 2.6069906 | 23714.0366 | 43.074769 | 1982-1999 –> 2000-2012 | 4.8105108 | 52723.3009 |
| Atlantic | (26.8,27.2] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 3.1662044 | 13894.8061 | 17.068745 | 2000-2012 –> 2013-2019 | 5.7711322 | 37600.9508 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 3.0839375 | 13752.6967 | 17.006646 | 2000-2012 –> 2013-2019 | 5.7927164 | 37848.5984 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + aou + silicate + phosphate | 3.1017187 | 13781.7308 | 19.605376 | 2000-2012 –> 2013-2019 | 5.8105956 | 37875.9932 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 3.0569199 | 13705.1978 | 21.445281 | 2000-2012 –> 2013-2019 | 5.5522137 | 36982.6474 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 3.0954121 | 13772.7440 | 20.313104 | 2000-2012 –> 2013-2019 | 5.6880723 | 37431.8252 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 3.1772794 | 13913.6548 | 20.412323 | 2000-2012 –> 2013-2019 | 5.8729200 | 37961.0816 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 3.3115817 | 14137.1353 | 23.434895 | 2000-2012 –> 2013-2019 | 6.1922401 | 38846.4009 |
| Atlantic | (26.8,27.2] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 3.2014731 | 13954.6027 | 20.350140 | 2000-2012 –> 2013-2019 | 5.9306710 | 38125.3770 |
| Atlantic | (26.8,27.2] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate | 3.3844672 | 14254.6528 | 18.021890 | 2000-2012 –> 2013-2019 | 6.2152213 | 38789.6854 |
| Atlantic | (26.8,27.2] | cstar_tref ~ temp + aou + silicate + phosphate + phosphate_star | 2.8811712 | 13385.5769 | 24.655222 | 2000-2012 –> 2013-2019 | 5.4881618 | 37099.6135 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 3.1757893 | 32231.2271 | 20.869331 | 1982-1999 –> 2000-2012 | 5.7146447 | 73000.8298 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + aou + phosphate + phosphate_star | 3.1759618 | 32229.9070 | 21.151860 | 1982-1999 –> 2000-2012 | 5.7369712 | 73148.1435 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 2.9166117 | 31165.8648 | 21.944476 | 1982-1999 –> 2000-2012 | 5.3273116 | 71037.4244 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 3.2032099 | 32338.8125 | 22.469231 | 1982-1999 –> 2000-2012 | 5.7568597 | 73209.1532 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + aou + nitrate + silicate | 3.1110548 | 31973.5090 | 14.366119 | 1982-1999 –> 2000-2012 | 5.5786023 | 72249.1763 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 3.1200544 | 32009.6572 | 19.335570 | 1982-1999 –> 2000-2012 | 5.6799906 | 72922.6269 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 2.9646699 | 31370.3827 | 22.962120 | 1982-1999 –> 2000-2012 | 5.4033109 | 71441.7419 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 3.1602554 | 32169.8665 | 24.980092 | 1982-1999 –> 2000-2012 | 5.7574391 | 73333.2896 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 3.1435113 | 32103.3869 | 24.153462 | 1982-1999 –> 2000-2012 | 5.7512899 | 73337.3942 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 3.1484843 | 32123.1682 | 24.748967 | 1982-1999 –> 2000-2012 | 5.7503303 | 73317.6875 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 4.0138129 | 18916.4175 | 24.683522 | 2000-2012 –> 2013-2019 | 7.1896022 | 51147.6445 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 3.7123766 | 18391.0106 | 26.346727 | 2000-2012 –> 2013-2019 | 6.7824309 | 50198.5018 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + aou + silicate + phosphate | 3.7338343 | 18427.7983 | 28.528346 | 2000-2012 –> 2013-2019 | 6.8169993 | 50286.6170 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 3.7204750 | 18405.6760 | 27.646487 | 2000-2012 –> 2013-2019 | 6.6370867 | 49571.5407 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + aou + nitrate + silicate | 3.7919939 | 18533.8194 | 17.331054 | 2000-2012 –> 2013-2019 | 6.9030487 | 50507.3284 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 3.8425282 | 18622.9148 | 22.550614 | 2000-2012 –> 2013-2019 | 6.9625826 | 50632.5719 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 3.7335455 | 18429.2778 | 28.438479 | 2000-2012 –> 2013-2019 | 6.6982154 | 49799.6606 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 3.9472747 | 18803.9173 | 30.746162 | 2000-2012 –> 2013-2019 | 7.1075301 | 50973.7838 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 3.9021439 | 18726.5271 | 28.929716 | 2000-2012 –> 2013-2019 | 7.0456552 | 50829.9140 |
| Atlantic | (27.2,27.8] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 3.9191116 | 18755.7277 | 30.301004 | 2000-2012 –> 2013-2019 | 7.0675959 | 50878.8959 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 3.0476531 | 12868.0168 | 81.780590 | 1982-1999 –> 2000-2012 | 5.4914003 | 28542.0711 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + aou + phosphate + phosphate_star | 3.1183048 | 12982.3012 | 73.984632 | 1982-1999 –> 2000-2012 | 5.6425583 | 28873.8548 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 3.0218764 | 12824.9189 | 79.766840 | 1982-1999 –> 2000-2012 | 5.5111994 | 28624.1076 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 3.1640789 | 13058.2419 | 89.496383 | 1982-1999 –> 2000-2012 | 5.6267083 | 28784.4207 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + phosphate | 3.2406006 | 13177.4937 | 80.912266 | 1982-1999 –> 2000-2012 | 5.8188571 | 29212.3973 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 2.5738885 | 12010.7440 | 32.175994 | 1982-1999 –> 2000-2012 | 5.0229280 | 27699.4484 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 3.1570872 | 13047.0173 | 87.178381 | 1982-1999 –> 2000-2012 | 5.6929871 | 28971.7441 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 3.3141969 | 13293.4387 | 93.760615 | 1982-1999 –> 2000-2012 | 5.9197878 | 29401.7604 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 3.2960446 | 13265.5714 | 95.572250 | 1982-1999 –> 2000-2012 | 5.7925628 | 29084.3057 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 3.3120399 | 13290.1354 | 92.752085 | 1982-1999 –> 2000-2012 | 5.9084877 | 29374.6519 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 2.7163855 | 6610.9605 | 24.468703 | 2000-2012 –> 2013-2019 | 5.7640386 | 19478.9773 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 2.8361089 | 6728.6215 | 27.328764 | 2000-2012 –> 2013-2019 | 5.8579853 | 19553.5405 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 2.7301970 | 6624.7960 | 24.747403 | 2000-2012 –> 2013-2019 | 5.8942760 | 19683.0379 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 2.8537508 | 6745.5384 | 26.191802 | 2000-2012 –> 2013-2019 | 6.2596436 | 20177.4555 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 2.8265818 | 6719.4422 | 23.123666 | 2000-2012 –> 2013-2019 | 5.4004703 | 18730.1862 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 2.8894257 | 6779.4298 | 27.966117 | 2000-2012 –> 2013-2019 | 6.0465128 | 19826.4472 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 2.9696416 | 6854.1321 | 28.539746 | 2000-2012 –> 2013-2019 | 6.2838384 | 20147.5707 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 2.7566583 | 6651.1087 | 25.015350 | 2000-2012 –> 2013-2019 | 6.0527029 | 19916.6800 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 2.9580732 | 6843.4843 | 28.659117 | 2000-2012 –> 2013-2019 | 6.2701132 | 20133.6197 |
| Atlantic | (27.8,27.9] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 2.9761188 | 6860.0758 | 28.456667 | 2000-2012 –> 2013-2019 | 6.2861048 | 20147.0636 |
| Atlantic | (27.9,28.1] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 3.0353096 | 15816.7662 | 27.753840 | 1982-1999 –> 2000-2012 | 5.9985122 | 37094.9816 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 2.9530508 | 15645.1048 | 28.365399 | 1982-1999 –> 2000-2012 | 5.7793567 | 36521.8369 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + nitrate + silicate + phosphate | 3.0307699 | 15805.4146 | 30.200762 | 1982-1999 –> 2000-2012 | 6.0340957 | 37195.7905 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + nitrate + silicate + phosphate + phosphate_star | 2.9853149 | 15712.9983 | 28.583363 | 1982-1999 –> 2000-2012 | 5.8488618 | 36700.8426 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 2.9182477 | 15571.0319 | 25.339640 | 1982-1999 –> 2000-2012 | 5.9356573 | 37003.1184 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 2.9580799 | 15655.7362 | 26.215211 | 1982-1999 –> 2000-2012 | 6.0384664 | 37263.1540 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate | 3.0172724 | 15779.5271 | 30.280490 | 1982-1999 –> 2000-2012 | 6.0161720 | 37159.3844 |
| Atlantic | (27.9,28.1] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate | 3.0318909 | 15809.7251 | 27.198793 | 1982-1999 –> 2000-2012 | 5.9844675 | 37057.4479 |
| Atlantic | (27.9,28.1] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate_star | 3.0451837 | 15837.0585 | 25.723841 | 1982-1999 –> 2000-2012 | 5.9839591 | 37045.0131 |
| Atlantic | (27.9,28.1] | cstar_tref ~ temp + nitrate + silicate + phosphate + phosphate_star | 3.0317310 | 15809.3955 | 27.104322 | 1982-1999 –> 2000-2012 | 5.9821775 | 37050.9927 |
| Atlantic | (27.9,28.1] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 2.9459116 | 8396.8428 | 26.617676 | 2000-2012 –> 2013-2019 | 5.9812212 | 24213.6091 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 2.7824213 | 8205.3402 | 26.236983 | 2000-2012 –> 2013-2019 | 5.7755555 | 23934.6822 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 2.6535975 | 8046.3431 | 28.239775 | 2000-2012 –> 2013-2019 | 5.6066483 | 23691.4478 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + nitrate + silicate + phosphate | 2.6733422 | 8069.2069 | 29.469179 | 2000-2012 –> 2013-2019 | 5.7041121 | 23874.6214 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + nitrate + silicate + phosphate + phosphate_star | 2.6655502 | 8061.4167 | 28.790431 | 2000-2012 –> 2013-2019 | 5.6508651 | 23774.4150 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 2.7619183 | 8180.5338 | 25.763326 | 2000-2012 –> 2013-2019 | 5.7285528 | 23854.3129 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 2.7184662 | 8127.3473 | 24.745497 | 2000-2012 –> 2013-2019 | 5.6367139 | 23698.3792 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 2.7562851 | 8173.6860 | 25.631013 | 2000-2012 –> 2013-2019 | 5.7143649 | 23829.4222 |
| Atlantic | (27.9,28.1] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate | 2.6475444 | 8038.6835 | 29.634210 | 2000-2012 –> 2013-2019 | 5.6648168 | 23818.2106 |
| Atlantic | (27.9,28.1] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate | 2.9588859 | 8411.5821 | 26.124471 | 2000-2012 –> 2013-2019 | 5.9907769 | 24221.3072 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 0.8439233 | 8536.3485 | 7.593862 | 1982-1999 –> 2000-2012 | 1.5746245 | 19036.3713 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + aou + nitrate + phosphate_star | 0.8861600 | 8867.5073 | 6.460803 | 1982-1999 –> 2000-2012 | 1.6606546 | 19917.7882 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate_star | 0.8664944 | 8716.4085 | 7.542880 | 1982-1999 –> 2000-2012 | 1.6308643 | 19643.8388 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + aou + nitrate | 0.8504911 | 8587.2347 | 7.418466 | 1982-1999 –> 2000-2012 | 1.5896611 | 19194.5910 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 0.8504032 | 8588.5299 | 7.510925 | 1982-1999 –> 2000-2012 | 1.5878841 | 19176.1792 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 0.8504903 | 8589.2286 | 7.426875 | 1982-1999 –> 2000-2012 | 1.5886548 | 19185.6683 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + aou + nitrate + silicate | 0.8501907 | 8586.8249 | 7.612598 | 1982-1999 –> 2000-2012 | 1.5889554 | 19190.9765 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 0.8563499 | 8636.0690 | 7.491839 | 1982-1999 –> 2000-2012 | 1.6020058 | 19328.3136 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 0.8859848 | 8868.1583 | 8.112323 | 1982-1999 –> 2000-2012 | 1.6617056 | 19935.4496 |
| Atlantic | (28.1,28.2] | cstar_tref ~ temp + aou + nitrate + phosphate + phosphate_star | 0.8231615 | 8366.4182 | 13.869518 | 1982-1999 –> 2000-2012 | 1.4598735 | 17560.0637 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 0.9482691 | 4903.6413 | 8.416903 | 2000-2012 –> 2013-2019 | 1.7921924 | 13439.9898 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + aou + nitrate + phosphate_star | 0.9843129 | 5035.1948 | 7.395627 | 2000-2012 –> 2013-2019 | 1.8704729 | 13902.7021 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate_star | 0.9627538 | 4957.9118 | 8.657656 | 2000-2012 –> 2013-2019 | 1.8292482 | 13674.3202 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + aou + nitrate | 0.9560785 | 4931.0032 | 8.225577 | 2000-2012 –> 2013-2019 | 1.8065695 | 13518.2379 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 0.9559603 | 4932.5607 | 8.338042 | 2000-2012 –> 2013-2019 | 1.8063635 | 13521.0906 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 0.9560784 | 4933.0029 | 8.222789 | 2000-2012 –> 2013-2019 | 1.8065687 | 13522.2316 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + aou + nitrate + silicate | 0.9552180 | 4929.7799 | 8.585688 | 2000-2012 –> 2013-2019 | 1.8054087 | 13516.6049 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 0.9617924 | 4954.3351 | 8.348077 | 2000-2012 –> 2013-2019 | 1.8181423 | 13590.4041 |
| Atlantic | (28.1,28.2] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 0.9823514 | 5030.0538 | 8.939639 | 2000-2012 –> 2013-2019 | 1.8683362 | 13898.2120 |
| Atlantic | (28.1,28.2] | cstar_tref ~ temp + aou + nitrate + phosphate + phosphate_star | 0.8670608 | 4583.1265 | 15.561133 | 2000-2012 –> 2013-2019 | 1.6902223 | 12949.5447 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + phosphate | 0.3429184 | 2958.3108 | 2.037539 | 1982-1999 –> 2000-2012 | 0.6478106 | 5713.4989 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 0.3391792 | 2867.6653 | 2.211193 | 1982-1999 –> 2000-2012 | 0.6440319 | 5623.3136 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + phosphate_star | 0.3501288 | 3134.1416 | 1.949898 | 1982-1999 –> 2000-2012 | 0.6565772 | 5949.7518 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + silicate | 0.3534411 | 3213.7066 | 1.988553 | 1982-1999 –> 2000-2012 | 0.6596790 | 6021.1606 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 0.3255418 | 2520.8956 | 2.143799 | 1982-1999 –> 2000-2012 | 0.6303960 | 5276.6069 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate_star | 0.3490030 | 3108.9287 | 2.067840 | 1982-1999 –> 2000-2012 | 0.6550932 | 5912.6573 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 0.3393669 | 2872.3409 | 2.206547 | 1982-1999 –> 2000-2012 | 0.6442495 | 5629.1521 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 0.3395940 | 2877.9930 | 2.205103 | 1982-1999 –> 2000-2012 | 0.6447888 | 5646.9529 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + temp + aou + nitrate + silicate | 0.3509290 | 3155.4335 | 2.134668 | 1982-1999 –> 2000-2012 | 0.6570626 | 5960.8427 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 0.3379706 | 2837.5014 | 2.209241 | 1982-1999 –> 2000-2012 | 0.6389390 | 5440.9641 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + phosphate | 0.3950700 | 2362.2400 | 2.495027 | 2000-2012 –> 2013-2019 | 0.7379884 | 5320.5508 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 0.3783565 | 2157.0140 | 2.771724 | 2000-2012 –> 2013-2019 | 0.7175357 | 5024.6793 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + phosphate_star | 0.4135856 | 2581.8126 | 2.572196 | 2000-2012 –> 2013-2019 | 0.7637143 | 5715.9542 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + silicate | 0.4165997 | 2616.6231 | 2.435991 | 2000-2012 –> 2013-2019 | 0.7700408 | 5830.3298 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 0.3619309 | 1944.2398 | 2.679695 | 2000-2012 –> 2013-2019 | 0.6874727 | 4465.1354 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate_star | 0.4134611 | 2582.3694 | 2.655486 | 2000-2012 –> 2013-2019 | 0.7624641 | 5691.2982 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 0.3786330 | 2160.5167 | 2.768969 | 2000-2012 –> 2013-2019 | 0.7179999 | 5032.8576 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 0.3788945 | 2163.8268 | 2.767863 | 2000-2012 –> 2013-2019 | 0.7184885 | 5041.8198 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + temp + aou + nitrate + silicate | 0.4031930 | 2461.8100 | 2.685125 | 2000-2012 –> 2013-2019 | 0.7541221 | 5617.2435 |
| Atlantic | (28.2, Inf] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 0.3777333 | 2149.1120 | 2.771424 | 2000-2012 –> 2013-2019 | 0.7157039 | 4986.6135 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 5.9368924 | 35970.5975 | 33.206006 | 1982-1999 –> 2000-2012 | 12.1836597 | 85952.5079 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + aou + phosphate + phosphate_star | 6.2801597 | 36600.1682 | 29.818984 | 1982-1999 –> 2000-2012 | 12.8626803 | 87384.7542 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 6.2388929 | 36528.0930 | 29.366420 | 1982-1999 –> 2000-2012 | 12.7725705 | 87200.2068 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 6.0579613 | 36197.4241 | 33.818877 | 1982-1999 –> 2000-2012 | 12.4174769 | 86454.2748 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 6.1136918 | 36300.3175 | 34.053133 | 1982-1999 –> 2000-2012 | 12.5238257 | 86679.0210 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + phosphate | 6.4254929 | 36857.2246 | 30.208689 | 1982-1999 –> 2000-2012 | 13.1429671 | 87953.7368 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 6.2733617 | 36589.9990 | 29.675295 | 1982-1999 –> 2000-2012 | 12.8523383 | 87368.3078 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 6.3773992 | 36774.8088 | 29.718986 | 1982-1999 –> 2000-2012 | 13.0347184 | 87735.0625 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 6.4461499 | 36895.2887 | 29.845366 | 1982-1999 –> 2000-2012 | 13.1621308 | 87990.3838 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 6.1369641 | 36343.0072 | 34.054990 | 1982-1999 –> 2000-2012 | 12.5721086 | 86781.5633 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 5.6641426 | 18718.1540 | 28.268543 | 2000-2012 –> 2013-2019 | 11.6010350 | 54688.7514 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + aou + phosphate + phosphate_star | 5.9966384 | 19054.5361 | 24.645119 | 2000-2012 –> 2013-2019 | 12.2767981 | 55654.7043 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 5.9243606 | 18984.6030 | 23.875073 | 2000-2012 –> 2013-2019 | 12.1632535 | 55512.6960 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 5.7682183 | 18826.1621 | 29.166503 | 2000-2012 –> 2013-2019 | 11.8261796 | 55023.5862 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 5.8220861 | 18881.3024 | 29.808032 | 2000-2012 –> 2013-2019 | 11.9357779 | 55181.6200 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 5.9924378 | 19052.3794 | 24.457831 | 2000-2012 –> 2013-2019 | 12.2657995 | 55642.3784 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 6.0470335 | 19106.1797 | 24.650519 | 2000-2012 –> 2013-2019 | 12.4244327 | 55880.9886 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 5.8363044 | 18895.7715 | 29.341398 | 2000-2012 –> 2013-2019 | 11.9732685 | 55238.7787 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + nitrate + phosphate_star | 5.9978940 | 19055.7781 | 27.345614 | 2000-2012 –> 2013-2019 | 12.4545857 | 55967.4267 |
| Indo-Pacific | (-Inf,26] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 5.9955754 | 19055.4845 | 27.200018 | 2000-2012 –> 2013-2019 | 12.4477444 | 55961.2600 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 4.6199548 | 64645.4743 | 37.089849 | 1982-1999 –> 2000-2012 | 9.0638277 | 153579.9948 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 4.8242499 | 65593.7003 | 56.356823 | 1982-1999 –> 2000-2012 | 9.3833055 | 155310.0229 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 4.8681188 | 65792.0728 | 56.308779 | 1982-1999 –> 2000-2012 | 9.4419727 | 155607.4044 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 4.6498108 | 64786.6354 | 35.212652 | 1982-1999 –> 2000-2012 | 9.0460965 | 153392.2260 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 4.7432544 | 65222.6575 | 34.972147 | 1982-1999 –> 2000-2012 | 9.2002384 | 154247.1855 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 4.8704518 | 65802.5724 | 56.209203 | 1982-1999 –> 2000-2012 | 9.4490013 | 155649.2531 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + nitrate + phosphate_star | 4.8917582 | 65896.2291 | 54.417884 | 1982-1999 –> 2000-2012 | 9.6943665 | 157200.5895 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 4.7457183 | 65234.0380 | 40.259916 | 1982-1999 –> 2000-2012 | 9.5072391 | 156277.8943 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 4.7474245 | 65241.9151 | 35.164825 | 1982-1999 –> 2000-2012 | 9.2147567 | 154337.2958 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + silicate + phosphate_star | 4.7747259 | 65365.5765 | 33.757204 | 1982-1999 –> 2000-2012 | 9.5399697 | 156431.3124 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 4.7381641 | 34888.0729 | 37.416197 | 2000-2012 –> 2013-2019 | 9.3581189 | 99533.5472 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 4.9385239 | 35373.6429 | 43.798277 | 2000-2012 –> 2013-2019 | 9.7627738 | 100967.3432 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 5.0112464 | 35545.0263 | 42.147309 | 2000-2012 –> 2013-2019 | 9.8793651 | 101337.0991 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 4.8112061 | 35067.4275 | 35.106247 | 2000-2012 –> 2013-2019 | 9.4610169 | 99854.0629 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 4.9254596 | 35342.5872 | 41.789883 | 2000-2012 –> 2013-2019 | 9.6687140 | 100565.2447 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 5.0111174 | 35544.7247 | 42.003616 | 2000-2012 –> 2013-2019 | 9.8815692 | 101347.2971 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + nitrate + phosphate_star | 5.0112471 | 35543.0280 | 42.156824 | 2000-2012 –> 2013-2019 | 9.9030053 | 101439.2572 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 4.8028438 | 35047.0323 | 39.334231 | 2000-2012 –> 2013-2019 | 9.5485621 | 100281.0702 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 4.9239888 | 35339.0857 | 41.490198 | 2000-2012 –> 2013-2019 | 9.6714133 | 100581.0008 |
| Indo-Pacific | (26,26.8] | cstar_tref ~ sal + temp + silicate + phosphate_star | 4.9275027 | 35345.4493 | 42.358810 | 2000-2012 –> 2013-2019 | 9.7022286 | 100711.0258 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 4.5795336 | 81813.8219 | 34.801865 | 1982-1999 –> 2000-2012 | 8.5516150 | 189873.0313 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ aou + silicate + phosphate + phosphate_star | 4.7886149 | 83053.7301 | 33.349845 | 1982-1999 –> 2000-2012 | 8.9291782 | 192714.9439 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 4.8295980 | 83292.7959 | 45.368888 | 1982-1999 –> 2000-2012 | 8.9093005 | 192384.2516 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ sal + aou + phosphate + phosphate_star | 4.9176703 | 83793.5132 | 43.210781 | 1982-1999 –> 2000-2012 | 9.0702458 | 193566.5823 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 4.7371486 | 82755.1342 | 36.785071 | 1982-1999 –> 2000-2012 | 8.7534986 | 191242.2911 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 4.6565455 | 82277.7345 | 40.769086 | 1982-1999 –> 2000-2012 | 8.6285536 | 190336.2311 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ temp + aou + nitrate + phosphate + phosphate_star | 4.7350846 | 82743.0111 | 38.131037 | 1982-1999 –> 2000-2012 | 8.8754037 | 192403.9473 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate | 4.7380874 | 82760.6466 | 33.937656 | 1982-1999 –> 2000-2012 | 8.8483849 | 192140.5852 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ temp + aou + phosphate + phosphate_star | 4.7560441 | 82863.8732 | 36.970104 | 1982-1999 –> 2000-2012 | 8.9114400 | 192663.1588 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ temp + aou + silicate + phosphate + phosphate_star | 4.5137641 | 81411.4136 | 31.307713 | 1982-1999 –> 2000-2012 | 8.4468954 | 189090.1265 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 5.1129226 | 45414.6620 | 38.741744 | 2000-2012 –> 2013-2019 | 9.6924562 | 127228.4838 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ aou + silicate + phosphate + phosphate_star | 5.3461176 | 46076.3907 | 36.840632 | 2000-2012 –> 2013-2019 | 10.1347325 | 129130.1208 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 5.4336533 | 46320.0907 | 48.460676 | 2000-2012 –> 2013-2019 | 10.2632513 | 129612.8866 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 5.3224960 | 46012.4896 | 39.131883 | 2000-2012 –> 2013-2019 | 10.0596447 | 128767.6238 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 5.2455588 | 45795.7988 | 42.954185 | 2000-2012 –> 2013-2019 | 9.9021043 | 128073.5333 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ temp + aou + nitrate + phosphate + phosphate_star | 5.2853539 | 45908.2742 | 41.361839 | 2000-2012 –> 2013-2019 | 10.0204386 | 128651.2852 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate | 5.2749045 | 45878.8224 | 37.966053 | 2000-2012 –> 2013-2019 | 10.0129919 | 128639.4689 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ temp + aou + phosphate + phosphate_star | 5.3051466 | 45961.9005 | 40.029218 | 2000-2012 –> 2013-2019 | 10.0611907 | 128825.7737 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ temp + aou + silicate + phosphate + phosphate_star | 5.0451120 | 45215.9674 | 34.042217 | 2000-2012 –> 2013-2019 | 9.5588760 | 126627.3810 |
| Indo-Pacific | (26.8,27.2] | cstar_tref ~ temp + nitrate + silicate + phosphate + phosphate_star | 5.3883644 | 46195.5309 | 37.019791 | 2000-2012 –> 2013-2019 | 10.2500015 | 129672.2592 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 3.6311940 | 82054.4668 | 35.438607 | 1982-1999 –> 2000-2012 | 6.4340072 | 182995.3260 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate_star | 3.4872184 | 80829.0205 | 31.145000 | 1982-1999 –> 2000-2012 | 6.2166685 | 180677.0600 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 3.6724121 | 82396.3547 | 32.077068 | 1982-1999 –> 2000-2012 | 6.5157353 | 183928.4591 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + aou + silicate + phosphate_star | 3.7333729 | 82893.0309 | 30.468524 | 1982-1999 –> 2000-2012 | 6.5781398 | 184444.0506 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + nitrate + silicate + phosphate + phosphate_star | 3.5110881 | 81035.6466 | 30.468186 | 1982-1999 –> 2000-2012 | 6.2335009 | 180777.3192 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 3.5551878 | 81413.7232 | 32.908179 | 1982-1999 –> 2000-2012 | 6.3364189 | 182036.0933 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 3.5989799 | 81784.5499 | 33.006624 | 1982-1999 –> 2000-2012 | 6.4275883 | 183102.8720 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate | 3.4841970 | 80802.7655 | 34.064284 | 1982-1999 –> 2000-2012 | 6.4875823 | 184591.3607 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + silicate + phosphate | 3.5678878 | 81519.7339 | 33.667524 | 1982-1999 –> 2000-2012 | 6.5791832 | 185414.7023 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 3.5618307 | 81470.2678 | 33.134929 | 1982-1999 –> 2000-2012 | 6.3680145 | 182460.6458 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 4.2684165 | 46155.0368 | 39.014604 | 2000-2012 –> 2013-2019 | 7.8996105 | 128209.5036 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate_star | 4.0818001 | 45436.3622 | 29.917429 | 2000-2012 –> 2013-2019 | 7.5690185 | 126265.3827 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 4.2806890 | 46201.1922 | 32.074435 | 2000-2012 –> 2013-2019 | 7.9531011 | 128597.5469 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + aou + silicate + phosphate_star | 4.4049957 | 46659.3727 | 29.189160 | 2000-2012 –> 2013-2019 | 8.1383686 | 129552.4037 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + nitrate + silicate + phosphate + phosphate_star | 4.1297449 | 45624.0905 | 31.749891 | 2000-2012 –> 2013-2019 | 7.6408330 | 126659.7371 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 4.1330172 | 45636.8236 | 33.253329 | 2000-2012 –> 2013-2019 | 7.6882050 | 127050.5468 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 4.1728802 | 45791.1337 | 33.395820 | 2000-2012 –> 2013-2019 | 7.7718600 | 127575.6836 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate | 3.9857466 | 45053.5372 | 33.202756 | 2000-2012 –> 2013-2019 | 7.4699436 | 125856.3026 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + silicate + phosphate | 4.1369757 | 45650.2135 | 32.731002 | 2000-2012 –> 2013-2019 | 7.7048636 | 127169.9474 |
| Indo-Pacific | (27.2,27.8] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 4.1270636 | 45613.6495 | 33.571282 | 2000-2012 –> 2013-2019 | 7.6888944 | 127083.9174 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + aou + nitrate + phosphate + phosphate_star | 2.8482817 | 36170.3565 | 34.472962 | 1982-1999 –> 2000-2012 | 5.0269370 | 80264.7475 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 2.8354017 | 36103.8955 | 36.169899 | 1982-1999 –> 2000-2012 | 4.9934882 | 80008.0179 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + aou + nitrate + phosphate | 2.8494127 | 36176.1785 | 34.292463 | 1982-1999 –> 2000-2012 | 5.0349683 | 80333.9972 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 2.8201792 | 36024.9560 | 36.836244 | 1982-1999 –> 2000-2012 | 4.9998907 | 80129.0692 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 2.8358069 | 36105.9905 | 36.038293 | 1982-1999 –> 2000-2012 | 5.0018154 | 80083.6091 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 2.8333838 | 36093.4557 | 35.912394 | 1982-1999 –> 2000-2012 | 5.0034524 | 80108.6361 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 2.8335276 | 36094.1996 | 35.912551 | 1982-1999 –> 2000-2012 | 5.0075779 | 80146.1495 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 2.8332168 | 36092.5910 | 35.893479 | 1982-1999 –> 2000-2012 | 5.0046659 | 80120.5281 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + silicate + phosphate_star | 2.8335702 | 36092.4202 | 35.966529 | 1982-1999 –> 2000-2012 | 5.0286896 | 80335.8196 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ temp + aou + silicate + phosphate + phosphate_star | 2.8288658 | 36070.0540 | 35.663023 | 1982-1999 –> 2000-2012 | 4.9609466 | 79731.0031 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 3.2619062 | 20683.5347 | 42.052061 | 2000-2012 –> 2013-2019 | 6.0962975 | 56782.2037 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 3.2562933 | 20669.8499 | 39.928836 | 2000-2012 –> 2013-2019 | 6.0916950 | 56773.7454 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + aou + nitrate + phosphate_star | 3.2496245 | 20653.5600 | 39.978779 | 2000-2012 –> 2013-2019 | 6.0971681 | 56820.1163 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + aou + phosphate + phosphate_star | 3.1692189 | 20454.4785 | 40.940090 | 2000-2012 –> 2013-2019 | 5.9893980 | 56479.4345 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 3.2508193 | 20656.4811 | 39.746171 | 2000-2012 –> 2013-2019 | 6.0866262 | 56762.4715 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 3.2445752 | 20641.2039 | 39.567399 | 2000-2012 –> 2013-2019 | 6.0779590 | 56734.6596 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + nitrate + phosphate + phosphate_star | 3.2502667 | 20655.1301 | 39.767367 | 2000-2012 –> 2013-2019 | 6.0975419 | 56820.3040 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 3.2423743 | 20635.8121 | 40.235128 | 2000-2012 –> 2013-2019 | 6.0759019 | 56730.0118 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 3.2440939 | 20640.0250 | 39.539922 | 2000-2012 –> 2013-2019 | 6.0773106 | 56732.6159 |
| Indo-Pacific | (27.8,27.9] | cstar_tref ~ sal + temp + silicate + phosphate_star | 3.2457468 | 20642.0726 | 39.732509 | 2000-2012 –> 2013-2019 | 6.0793170 | 56734.4929 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 2.1497875 | 28763.8559 | 9.090062 | 1982-1999 –> 2000-2012 | 4.0365372 | 67315.2492 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + aou + nitrate + silicate | 2.1452171 | 28735.8437 | 9.090571 | 1982-1999 –> 2000-2012 | 4.0411647 | 67378.4894 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 2.1542792 | 28791.3276 | 9.110385 | 1982-1999 –> 2000-2012 | 4.0444431 | 67376.6450 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 2.1543768 | 28791.9239 | 9.105887 | 1982-1999 –> 2000-2012 | 4.0455476 | 67387.2340 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + nitrate + silicate | 2.1455127 | 28735.6571 | 9.069301 | 1982-1999 –> 2000-2012 | 4.0738926 | 67694.5677 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate | 2.1279997 | 28629.7803 | 8.688730 | 1982-1999 –> 2000-2012 | 4.0494159 | 67522.8074 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 2.1439932 | 28728.3326 | 8.974954 | 1982-1999 –> 2000-2012 | 4.0335617 | 67307.7384 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 2.1540557 | 28789.9619 | 9.104070 | 1982-1999 –> 2000-2012 | 4.0449001 | 67382.0337 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + silicate + phosphate_star | 2.1607357 | 28828.7155 | 9.138491 | 1982-1999 –> 2000-2012 | 4.0619310 | 67521.2255 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ temp + aou + silicate + phosphate + phosphate_star | 2.1490185 | 28759.1465 | 8.506487 | 1982-1999 –> 2000-2012 | 4.0341289 | 67294.2300 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate_star | 2.3308618 | 16676.6391 | 8.794806 | 2000-2012 –> 2013-2019 | 4.5047731 | 45587.3695 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 2.3484670 | 16731.9910 | 9.300589 | 2000-2012 –> 2013-2019 | 4.4982546 | 45495.8469 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + aou + nitrate + silicate | 2.3705243 | 16800.7576 | 9.489054 | 2000-2012 –> 2013-2019 | 4.5157414 | 45536.6013 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 2.3536974 | 16748.3557 | 9.325255 | 2000-2012 –> 2013-2019 | 4.5079767 | 45539.6833 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 2.3528743 | 16745.7827 | 9.319013 | 2000-2012 –> 2013-2019 | 4.5072512 | 45537.7066 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate | 2.3261398 | 16661.7216 | 8.688527 | 2000-2012 –> 2013-2019 | 4.4541395 | 45291.5019 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 2.3403400 | 16706.4907 | 9.157466 | 2000-2012 –> 2013-2019 | 4.4843332 | 45434.8233 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 2.3525686 | 16744.8270 | 9.317052 | 2000-2012 –> 2013-2019 | 4.5066244 | 45534.7889 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ sal + temp + silicate + phosphate_star | 2.3576049 | 16758.5576 | 9.351008 | 2000-2012 –> 2013-2019 | 4.5183406 | 45587.2731 |
| Indo-Pacific | (27.9,28.1] | cstar_tref ~ temp + aou + silicate + phosphate + phosphate_star | 2.3589130 | 16764.6377 | 8.470486 | 2000-2012 –> 2013-2019 | 4.5079314 | 45523.7842 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ aou + nitrate + silicate + phosphate | 1.5361773 | 73634.6174 | 8.733378 | 1982-1999 –> 2000-2012 | 2.8079755 | 165389.8842 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 1.5089135 | 72923.3001 | 10.415983 | 1982-1999 –> 2000-2012 | 2.7513833 | 163390.6644 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 1.3948961 | 69793.5465 | 10.490447 | 1982-1999 –> 2000-2012 | 2.5772322 | 157518.1298 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + nitrate + silicate + phosphate + phosphate_star | 1.4354153 | 70934.1635 | 8.449386 | 1982-1999 –> 2000-2012 | 2.6823797 | 161601.1701 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 1.5314928 | 73514.9585 | 14.185354 | 1982-1999 –> 2000-2012 | 2.8636804 | 167837.0700 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 1.5281199 | 73427.1334 | 14.378394 | 1982-1999 –> 2000-2012 | 2.8513606 | 167376.6806 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 1.5314910 | 73514.9130 | 14.185270 | 1982-1999 –> 2000-2012 | 2.8636761 | 167836.9173 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate | 1.5143319 | 73066.0856 | 10.220413 | 1982-1999 –> 2000-2012 | 2.7611966 | 163728.6687 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ temp + aou + nitrate + silicate + phosphate_star | 1.5308210 | 73497.4825 | 10.138151 | 1982-1999 –> 2000-2012 | 2.7926269 | 164818.6463 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ temp + nitrate + silicate + phosphate + phosphate_star | 1.5184453 | 73174.1397 | 10.078924 | 1982-1999 –> 2000-2012 | 2.7707584 | 164077.7898 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ aou + nitrate + silicate + phosphate + phosphate_star | 1.7716198 | 42649.6756 | 20.936949 | 2000-2012 –> 2013-2019 | 3.2805333 | 115572.9757 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + aou + nitrate + silicate + phosphate | 1.6072486 | 40564.3886 | 17.629482 | 2000-2012 –> 2013-2019 | 3.0021447 | 110357.9351 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + aou + silicate + phosphate + phosphate_star | 1.7314864 | 42158.9485 | 16.581456 | 2000-2012 –> 2013-2019 | 3.2626842 | 115666.2325 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + nitrate + silicate + phosphate + phosphate_star | 1.6318563 | 40889.7931 | 17.025774 | 2000-2012 –> 2013-2019 | 3.0672716 | 111823.9566 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + temp + aou + silicate + phosphate | 1.7317874 | 42162.6704 | 16.576870 | 2000-2012 –> 2013-2019 | 3.2630874 | 115672.6150 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + temp + aou + silicate + phosphate_star | 1.7322153 | 42167.9616 | 16.575055 | 2000-2012 –> 2013-2019 | 3.2637081 | 115682.9201 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate | 1.7038406 | 41814.2505 | 16.464704 | 2000-2012 –> 2013-2019 | 3.2660027 | 116119.0319 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + temp + nitrate + silicate + phosphate_star | 1.7344727 | 42195.8521 | 17.075855 | 2000-2012 –> 2013-2019 | 3.2625926 | 115622.9856 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + temp + silicate + phosphate + phosphate_star | 1.7322017 | 42167.7935 | 16.574226 | 2000-2012 –> 2013-2019 | 3.2636927 | 115682.7065 |
| Indo-Pacific | (28.1, Inf] | cstar_tref ~ sal + temp + silicate + phosphate_star | 1.7349951 | 42200.3016 | 16.913655 | 2000-2012 –> 2013-2019 | 3.2667617 | 115720.3823 |
5.3 Target variable coefficients
A data frame to map the target variable is prepared.
# create table with two era belonging to one eras
eras_forward <- lm_all_fitted_wide %>%
arrange(era) %>%
group_by(basin, gamma_slab, model) %>%
mutate(eras = paste(era, lead(era), sep = " --> ")) %>%
ungroup() %>%
select(era, eras) %>%
unique()
eras_backward <- lm_all_fitted_wide %>%
arrange(era) %>%
group_by(basin, gamma_slab, model) %>%
mutate(eras = paste(lag(era), era, sep = " --> ")) %>%
ungroup() %>%
select(era, eras) %>%
unique()
eras_era <- full_join(eras_backward, eras_forward) %>%
filter(str_detect(eras, "NA") == FALSE)
# extend best model selection from eras to era
lm_best_target <- full_join(
lm_best %>% select(basin, gamma_slab, model, eras),
eras_era)
lm_best_target <- left_join(lm_best_target, lm_all_fitted_wide)
rm(eras_era, eras_forward, eras_backward,
lm_all_fitted)5.4 Cant coeffcients
A data frame of coefficient offsets is prepared to facilitate the direct mapping of Cant.
# pivot long format
lm_best_long <- lm_best_target %>%
pivot_longer(cols = starts_with("coeff_"),
names_to = "term",
values_to = "estimate",
names_prefix = "coeff_")
# subtract coefficients of adjacent era
lm_best_long <- lm_best_long %>%
arrange(era) %>%
group_by(basin, gamma_slab, eras, model, term) %>%
mutate(delta_coeff = estimate - lag(estimate)) %>%
ungroup() %>%
arrange(basin, gamma_slab, model, term, eras) %>%
drop_na() %>%
select(-c(era,estimate))
# pivot back to wide format
lm_best_cant <- lm_best_long %>%
pivot_wider(values_from = delta_coeff,
names_from = term,
names_prefix = "delta_coeff_",
values_fill = 0)5.5 Write files
lm_best_target %>%
select(
basin,
gamma_slab,
model,
eras,
era,
starts_with("coeff_")
) %>%
write_csv(paste(path_version_data,
"lm_best_target.csv",
sep = ""))
lm_best_cant %>%
select(
basin,
gamma_slab,
model,
eras,
starts_with("delta_coeff_")
) %>%
write_csv(paste(path_version_data,
"lm_best_cant.csv",
sep = ""))6 Model diagnotics
6.1 Selection criterion vs predictors
The selection criterion (rmse) was plotted against the number of predictors (limited to 2 - 5).
6.1.1 All models
lm_all_fitted_wide %>%
ggplot(aes(as.factor(n_predictors),
!!sym(params_local$MLR_criterion),
col = basin)) +
geom_hline(yintercept = 10) +
geom_boxplot() +
facet_grid(gamma_slab~era) +
scale_color_brewer(palette = "Set1") +
labs(x="Number of predictors")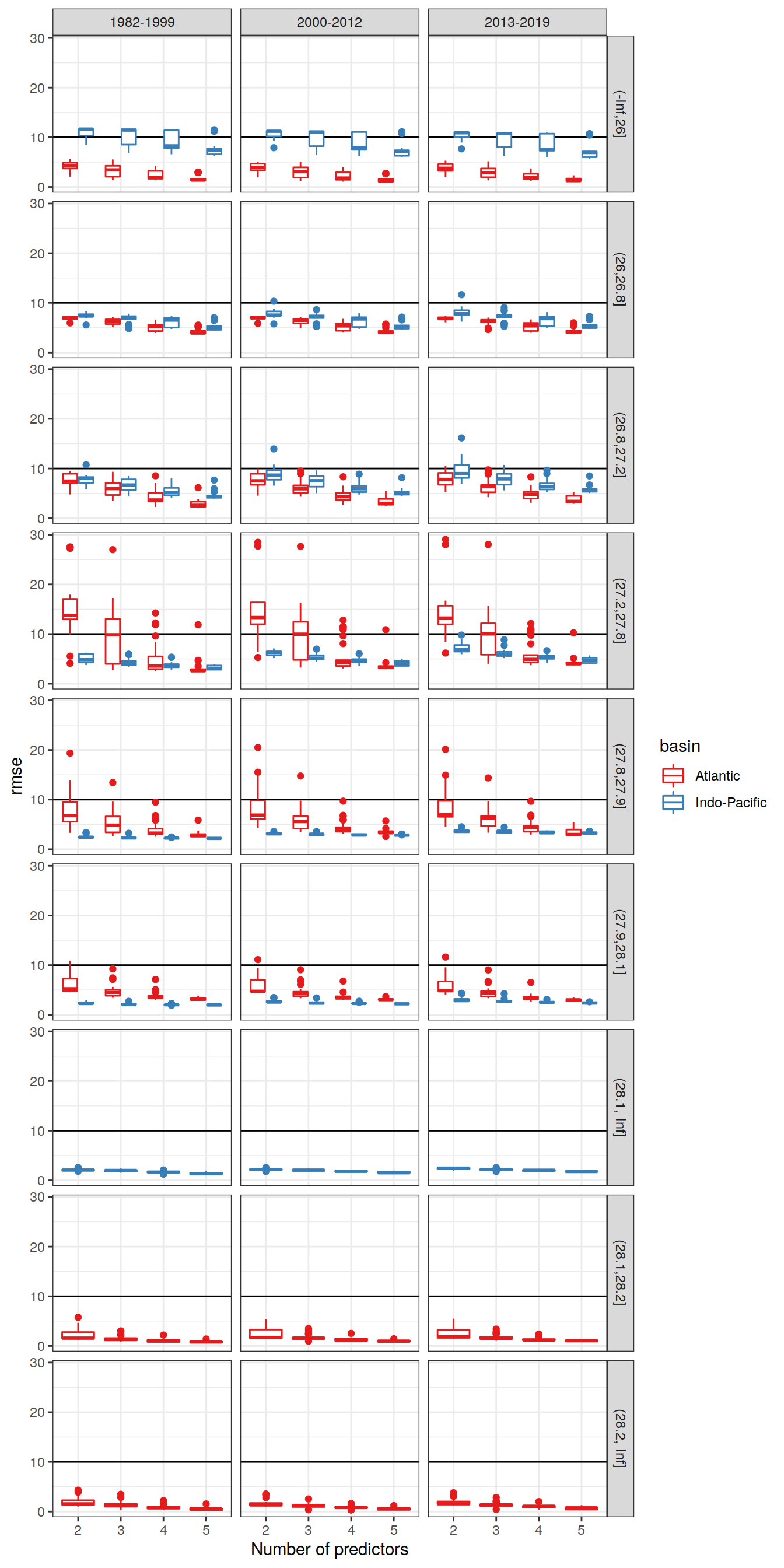
| Version | Author | Date |
|---|---|---|
| 28509fc | Donghe-Zhu | 2021-01-23 |
| 4c28e4a | Donghe-Zhu | 2021-01-22 |
| 24cc264 | jens-daniel-mueller | 2021-01-22 |
| 7891955 | Donghe-Zhu | 2021-01-21 |
| d4cf1cb | Donghe-Zhu | 2021-01-21 |
| 1f3e5b6 | jens-daniel-mueller | 2021-01-20 |
| 0e7bdf1 | jens-daniel-mueller | 2021-01-15 |
| 4571843 | jens-daniel-mueller | 2021-01-14 |
| b3564aa | jens-daniel-mueller | 2021-01-14 |
| 8d032c3 | jens-daniel-mueller | 2021-01-14 |
| 17dee1d | jens-daniel-mueller | 2021-01-13 |
| 7cdea0c | jens-daniel-mueller | 2021-01-06 |
| fa85b93 | jens-daniel-mueller | 2021-01-06 |
| e5cb81a | Donghe-Zhu | 2021-01-05 |
| a499f10 | Donghe-Zhu | 2021-01-05 |
| fb8a752 | Donghe-Zhu | 2020-12-23 |
| 8fae0b2 | Donghe-Zhu | 2020-12-21 |
| c8b76b3 | jens-daniel-mueller | 2020-12-19 |
6.1.2 Best models
lm_best_target %>%
ggplot(aes("",
!!sym(params_local$MLR_criterion),
col = basin)) +
geom_hline(yintercept = 10) +
geom_boxplot() +
facet_grid(gamma_slab~era) +
scale_color_brewer(palette = "Set1") +
labs(x="Number of predictors pooled")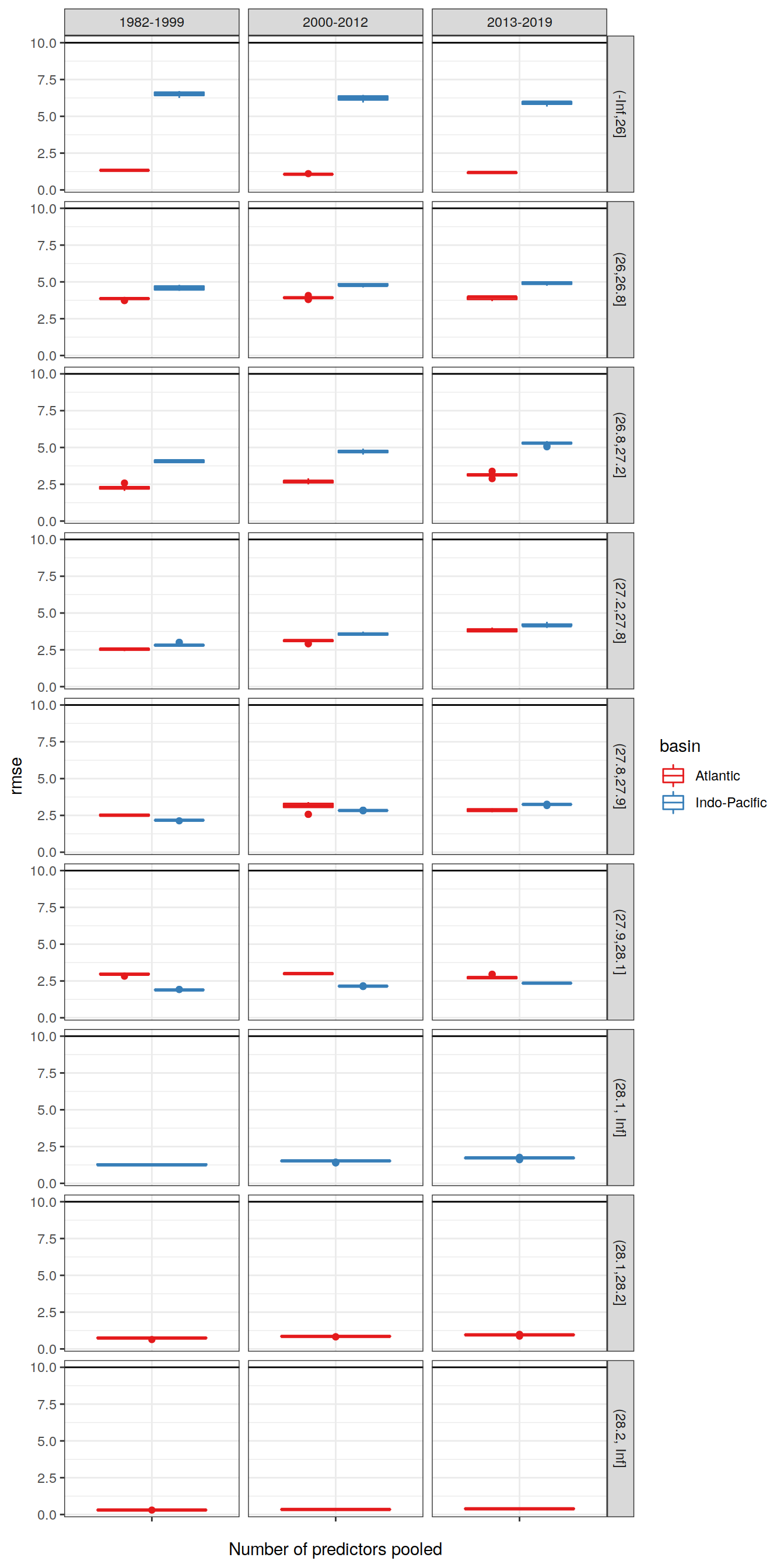
| Version | Author | Date |
|---|---|---|
| 28509fc | Donghe-Zhu | 2021-01-23 |
| 4c28e4a | Donghe-Zhu | 2021-01-22 |
| 24cc264 | jens-daniel-mueller | 2021-01-22 |
| 7891955 | Donghe-Zhu | 2021-01-21 |
| d4cf1cb | Donghe-Zhu | 2021-01-21 |
| 1f3e5b6 | jens-daniel-mueller | 2021-01-20 |
| 0e7bdf1 | jens-daniel-mueller | 2021-01-15 |
| 4571843 | jens-daniel-mueller | 2021-01-14 |
| b3564aa | jens-daniel-mueller | 2021-01-14 |
| 8d032c3 | jens-daniel-mueller | 2021-01-14 |
| 17dee1d | jens-daniel-mueller | 2021-01-13 |
| 7cdea0c | jens-daniel-mueller | 2021-01-06 |
| fa85b93 | jens-daniel-mueller | 2021-01-06 |
| e5cb81a | Donghe-Zhu | 2021-01-05 |
| a499f10 | Donghe-Zhu | 2021-01-05 |
| fb8a752 | Donghe-Zhu | 2020-12-23 |
| 8fae0b2 | Donghe-Zhu | 2020-12-21 |
| c8b76b3 | jens-daniel-mueller | 2020-12-19 |
6.2 RMSE correlation between eras
RMSE was plotted to compare the agreement for one model applied to two adjacent eras (ie check whether the same predictor combination performs equal in both eras).
6.2.1 All models
# find max rmse to scale axis
max_rmse <-
max(c(lm_all_fitted_wide_eras$rmse,
lm_all_fitted_wide_eras$rmse_sum - lm_all_fitted_wide_eras$rmse))
lm_all_fitted_wide_eras %>%
ggplot(aes(rmse, rmse_sum - rmse, col = gamma_slab)) +
geom_point() +
scale_color_viridis_d() +
coord_equal(xlim = c(0,max_rmse),
ylim = c(0,max_rmse)) +
facet_grid(eras ~ basin)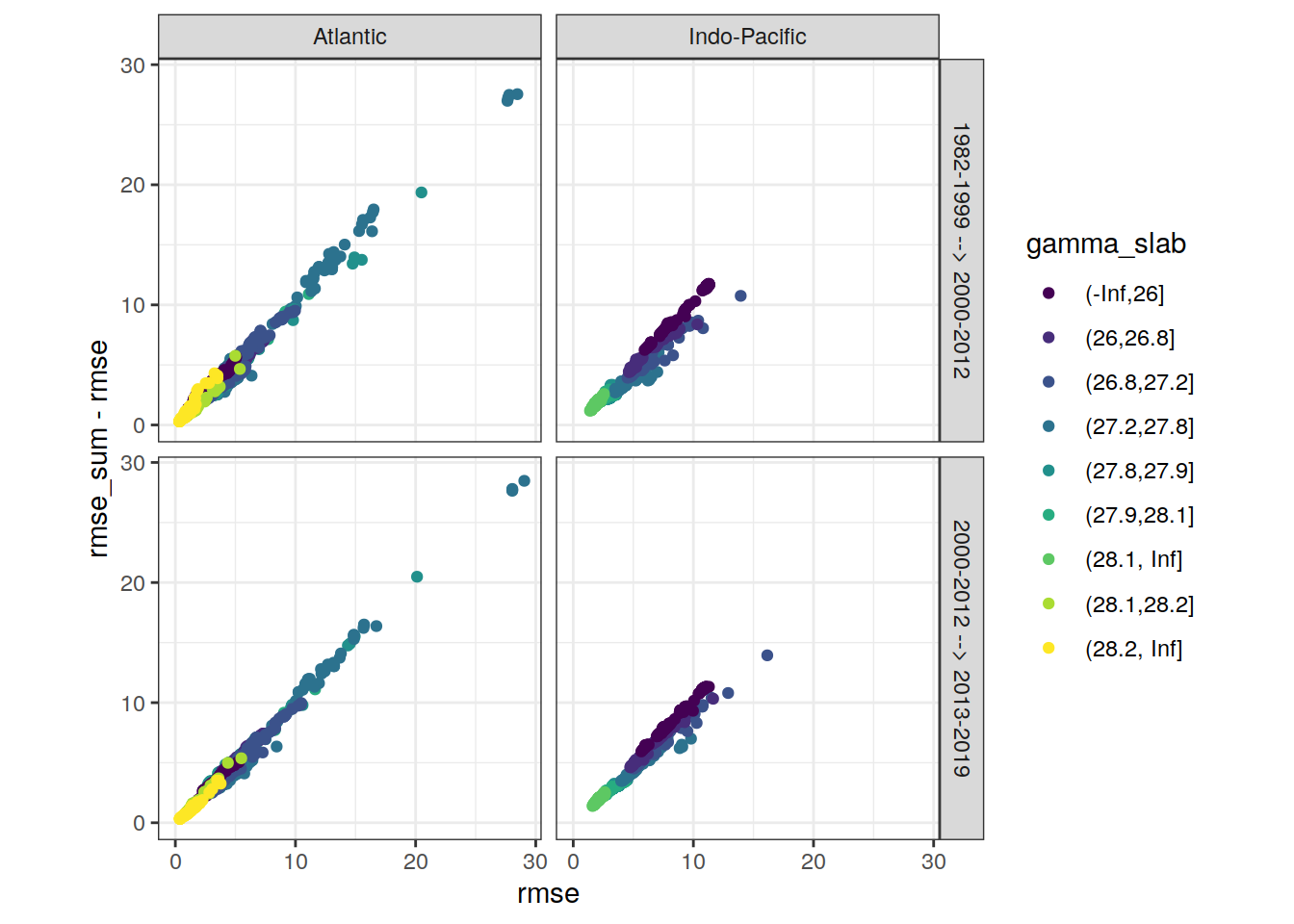
| Version | Author | Date |
|---|---|---|
| 28509fc | Donghe-Zhu | 2021-01-23 |
| 4c28e4a | Donghe-Zhu | 2021-01-22 |
| 24cc264 | jens-daniel-mueller | 2021-01-22 |
| 7891955 | Donghe-Zhu | 2021-01-21 |
| d4cf1cb | Donghe-Zhu | 2021-01-21 |
| 1f3e5b6 | jens-daniel-mueller | 2021-01-20 |
| 0e7bdf1 | jens-daniel-mueller | 2021-01-15 |
| 4571843 | jens-daniel-mueller | 2021-01-14 |
| b3564aa | jens-daniel-mueller | 2021-01-14 |
| 8d032c3 | jens-daniel-mueller | 2021-01-14 |
| 17dee1d | jens-daniel-mueller | 2021-01-13 |
| 7cdea0c | jens-daniel-mueller | 2021-01-06 |
| fa85b93 | jens-daniel-mueller | 2021-01-06 |
| e5cb81a | Donghe-Zhu | 2021-01-05 |
| a499f10 | Donghe-Zhu | 2021-01-05 |
| fb8a752 | Donghe-Zhu | 2020-12-23 |
| 8fae0b2 | Donghe-Zhu | 2020-12-21 |
| c8b76b3 | jens-daniel-mueller | 2020-12-19 |
rm(max_rmse)6.2.2 Best models
# find max rmse to scale axis
max_rmse <-
max(c(lm_best$rmse,
lm_best$rmse_sum - lm_best$rmse))
lm_best %>%
ggplot(aes(rmse, rmse_sum - rmse, col = gamma_slab)) +
geom_point() +
scale_color_viridis_d() +
coord_equal(xlim = c(0,max_rmse),
ylim = c(0,max_rmse)) +
facet_grid(eras ~ basin)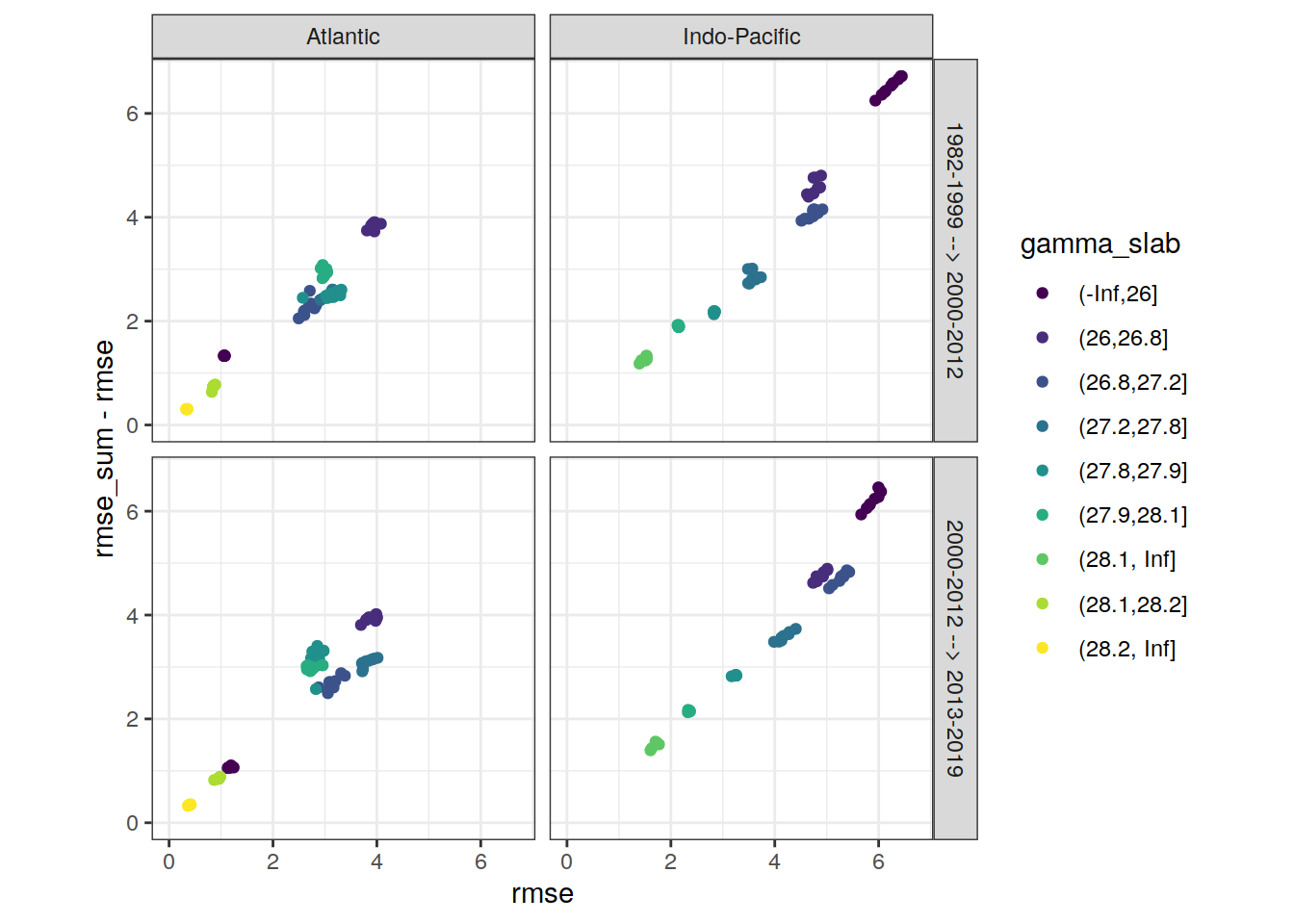
| Version | Author | Date |
|---|---|---|
| 28509fc | Donghe-Zhu | 2021-01-23 |
| 4c28e4a | Donghe-Zhu | 2021-01-22 |
| 24cc264 | jens-daniel-mueller | 2021-01-22 |
| 7891955 | Donghe-Zhu | 2021-01-21 |
| d4cf1cb | Donghe-Zhu | 2021-01-21 |
| 1f3e5b6 | jens-daniel-mueller | 2021-01-20 |
| 0e7bdf1 | jens-daniel-mueller | 2021-01-15 |
| 4571843 | jens-daniel-mueller | 2021-01-14 |
| b3564aa | jens-daniel-mueller | 2021-01-14 |
| 8d032c3 | jens-daniel-mueller | 2021-01-14 |
| 17dee1d | jens-daniel-mueller | 2021-01-13 |
| 7cdea0c | jens-daniel-mueller | 2021-01-06 |
| fa85b93 | jens-daniel-mueller | 2021-01-06 |
| e5cb81a | Donghe-Zhu | 2021-01-05 |
| a499f10 | Donghe-Zhu | 2021-01-05 |
| fb8a752 | Donghe-Zhu | 2020-12-23 |
| 8fae0b2 | Donghe-Zhu | 2020-12-21 |
| c8b76b3 | jens-daniel-mueller | 2020-12-19 |
rm(max_rmse)6.3 Predictor counts
The number of models where a particular predictor was included were counted for each basin, density slab and compared eras
# calculate cases of predictor used
lm_all_stats <- lm_best_long %>%
filter(term != "(Intercept)",
delta_coeff != 0) %>%
group_by(basin, eras, gamma_slab) %>%
count(term) %>%
ungroup() %>%
pivot_wider(values_from = n, names_from = term)
# print table
lm_all_stats %>%
gt(rowname_col = "gamma_slab",
groupname_col = c("basin", "eras")) %>%
summary_rows(
groups = TRUE,
fns = list(total = "sum")
)| aou | nitrate | phosphate | phosphate_star | sal | silicate | temp | |
|---|---|---|---|---|---|---|---|
| Atlantic - 1982-1999 --> 2000-2012 | |||||||
| (-Inf,26] | 9 | 4 | 8 | 7 | 10 | 3 | 7 |
| (26,26.8] | 8 | 8 | 8 | 7 | 6 | 6 | 7 |
| (26.8,27.2] | 9 | 4 | 8 | 7 | 5 | 10 | 6 |
| (27.2,27.8] | 8 | 4 | 7 | 7 | 10 | 6 | 7 |
| (27.8,27.9] | 8 | 4 | 8 | 7 | 10 | 4 | 7 |
| (27.9,28.1] | 5 | 10 | 8 | 6 | 6 | 8 | 6 |
| (28.1,28.2] | 8 | 10 | 4 | 7 | 9 | 3 | 7 |
| (28.2, Inf] | 9 | 10 | 5 | 5 | 10 | 4 | 4 |
| total | 64.00 | 54.00 | 56.00 | 53.00 | 66.00 | 44.00 | 51.00 |
| Atlantic - 2000-2012 --> 2013-2019 | |||||||
| (-Inf,26] | 8 | 5 | 7 | 7 | 10 | 5 | 8 |
| (26,26.8] | 8 | 10 | 8 | 8 | 4 | 4 | 7 |
| (26.8,27.2] | 8 | 4 | 8 | 6 | 7 | 10 | 6 |
| (27.2,27.8] | 8 | 4 | 7 | 6 | 10 | 8 | 6 |
| (27.8,27.9] | 7 | 5 | 7 | 8 | 10 | 5 | 8 |
| (27.9,28.1] | 6 | 10 | 9 | 5 | 8 | 6 | 5 |
| (28.1,28.2] | 8 | 10 | 4 | 7 | 9 | 3 | 7 |
| (28.2, Inf] | 9 | 10 | 5 | 5 | 10 | 4 | 4 |
| total | 62.00 | 58.00 | 55.00 | 52.00 | 68.00 | 45.00 | 51.00 |
| Indo-Pacific - 1982-1999 --> 2000-2012 | |||||||
| (-Inf,26] | 9 | 4 | 8 | 7 | 10 | 3 | 7 |
| (26,26.8] | 5 | 5 | 5 | 8 | 10 | 6 | 9 |
| (26.8,27.2] | 10 | 4 | 10 | 9 | 4 | 5 | 5 |
| (27.2,27.8] | 6 | 4 | 7 | 6 | 10 | 10 | 5 |
| (27.8,27.9] | 7 | 3 | 7 | 8 | 9 | 7 | 8 |
| (27.9,28.1] | 5 | 4 | 5 | 6 | 9 | 10 | 9 |
| (28.1, Inf] | 6 | 8 | 7 | 7 | 5 | 10 | 6 |
| total | 48.00 | 32.00 | 49.00 | 51.00 | 57.00 | 51.00 | 49.00 |
| Indo-Pacific - 2000-2012 --> 2013-2019 | |||||||
| (-Inf,26] | 7 | 6 | 7 | 8 | 10 | 3 | 7 |
| (26,26.8] | 5 | 5 | 5 | 8 | 10 | 6 | 9 |
| (26.8,27.2] | 9 | 5 | 10 | 9 | 3 | 6 | 6 |
| (27.2,27.8] | 6 | 4 | 7 | 6 | 10 | 10 | 5 |
| (27.8,27.9] | 6 | 4 | 6 | 8 | 10 | 7 | 8 |
| (27.9,28.1] | 6 | 4 | 5 | 7 | 9 | 10 | 8 |
| (28.1, Inf] | 5 | 5 | 7 | 7 | 9 | 10 | 6 |
| total | 44.00 | 33.00 | 47.00 | 53.00 | 61.00 | 52.00 | 49.00 |
6.4 RMSE alternatives
AIC is an alternative criterion to RMSE to judge model quality, but not (yet) taken into account.
lm_all_fitted_wide_eras %>%
ggplot(aes(rmse, aic, col = gamma_slab)) +
geom_point() +
scale_color_viridis_d() +
facet_grid(eras~basin)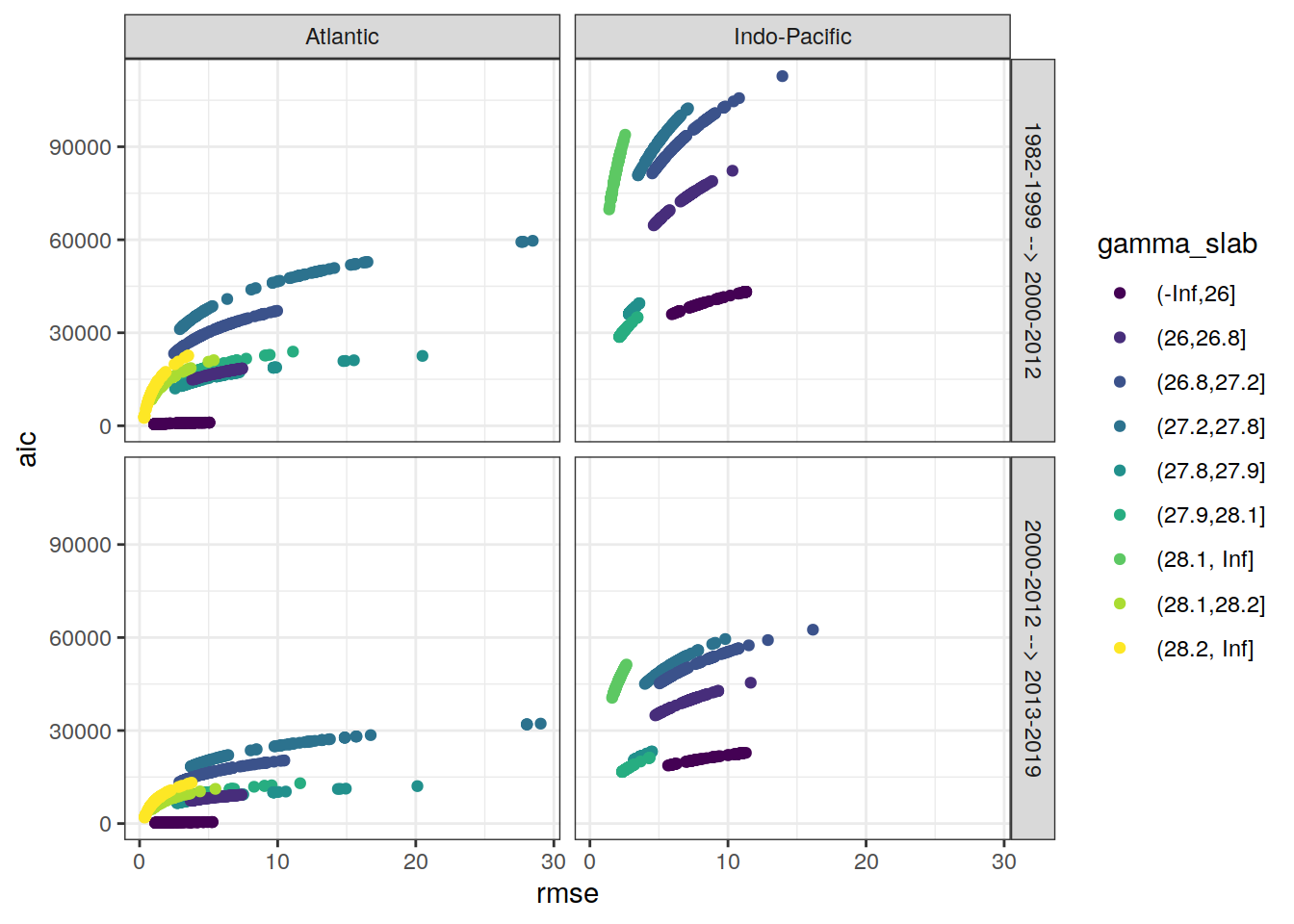
| Version | Author | Date |
|---|---|---|
| 28509fc | Donghe-Zhu | 2021-01-23 |
| 4c28e4a | Donghe-Zhu | 2021-01-22 |
| 24cc264 | jens-daniel-mueller | 2021-01-22 |
| 7891955 | Donghe-Zhu | 2021-01-21 |
| d4cf1cb | Donghe-Zhu | 2021-01-21 |
| 1f3e5b6 | jens-daniel-mueller | 2021-01-20 |
| 0e7bdf1 | jens-daniel-mueller | 2021-01-15 |
| 4571843 | jens-daniel-mueller | 2021-01-14 |
| b3564aa | jens-daniel-mueller | 2021-01-14 |
| 8d032c3 | jens-daniel-mueller | 2021-01-14 |
| 17dee1d | jens-daniel-mueller | 2021-01-13 |
| 7cdea0c | jens-daniel-mueller | 2021-01-06 |
| fa85b93 | jens-daniel-mueller | 2021-01-06 |
| e5cb81a | Donghe-Zhu | 2021-01-05 |
| a499f10 | Donghe-Zhu | 2021-01-05 |
| fb8a752 | Donghe-Zhu | 2020-12-23 |
| 8fae0b2 | Donghe-Zhu | 2020-12-21 |
| c8b76b3 | jens-daniel-mueller | 2020-12-19 |
lm_best %>%
ggplot(aes(rmse, aic, col = gamma_slab)) +
geom_point() +
scale_color_viridis_d() +
facet_grid(eras~basin)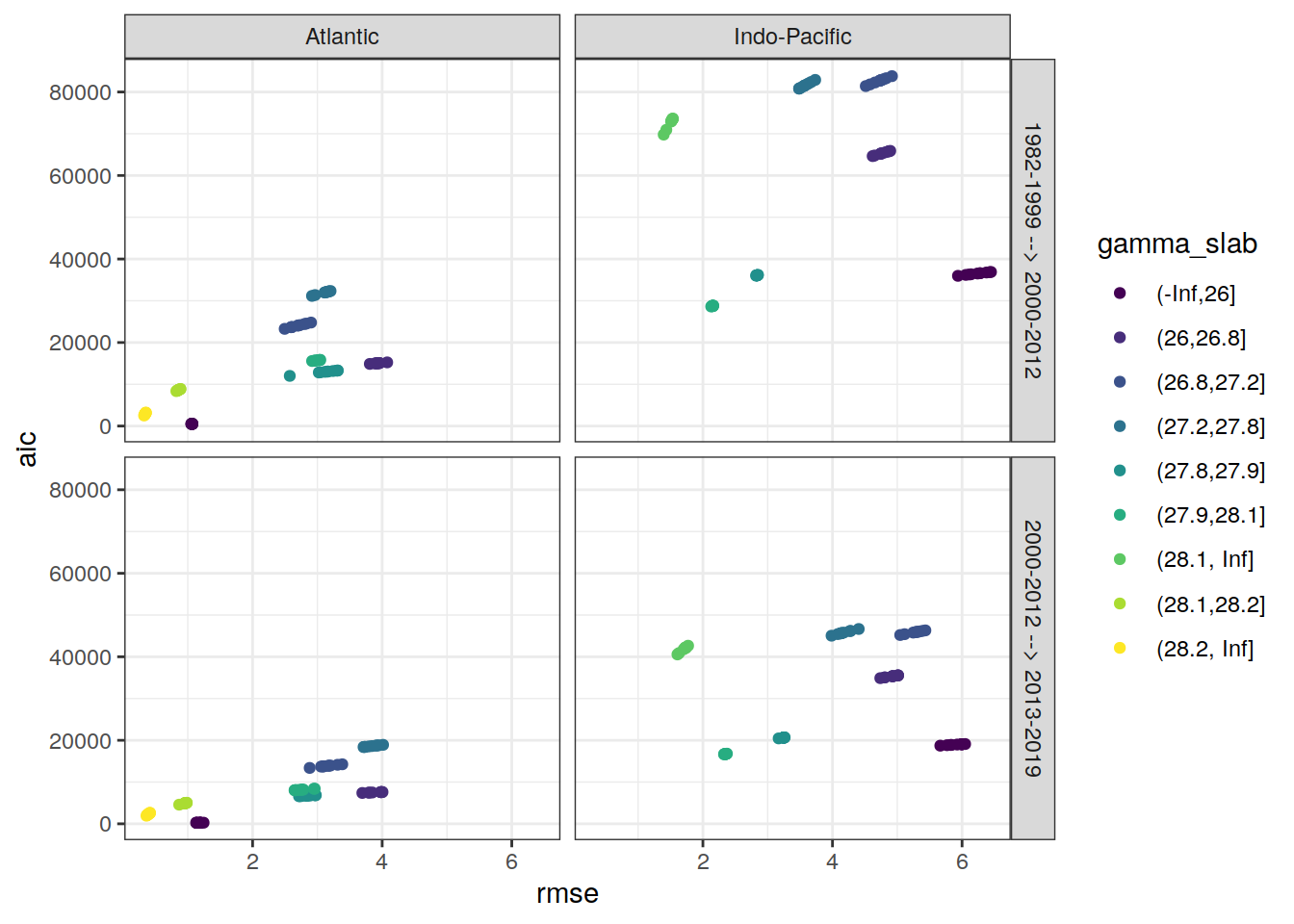
| Version | Author | Date |
|---|---|---|
| 28509fc | Donghe-Zhu | 2021-01-23 |
| 4c28e4a | Donghe-Zhu | 2021-01-22 |
| 24cc264 | jens-daniel-mueller | 2021-01-22 |
| 7891955 | Donghe-Zhu | 2021-01-21 |
| d4cf1cb | Donghe-Zhu | 2021-01-21 |
| 1f3e5b6 | jens-daniel-mueller | 2021-01-20 |
| 0e7bdf1 | jens-daniel-mueller | 2021-01-15 |
| 4571843 | jens-daniel-mueller | 2021-01-14 |
| b3564aa | jens-daniel-mueller | 2021-01-14 |
| 8d032c3 | jens-daniel-mueller | 2021-01-14 |
| 17dee1d | jens-daniel-mueller | 2021-01-13 |
| 7cdea0c | jens-daniel-mueller | 2021-01-06 |
| fa85b93 | jens-daniel-mueller | 2021-01-06 |
| e5cb81a | Donghe-Zhu | 2021-01-05 |
| a499f10 | Donghe-Zhu | 2021-01-05 |
| fb8a752 | Donghe-Zhu | 2020-12-23 |
| 8fae0b2 | Donghe-Zhu | 2020-12-21 |
| c8b76b3 | jens-daniel-mueller | 2020-12-19 |
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.2
Matrix products: default
BLAS: /usr/local/R-4.0.3/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.0.3/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] gt_0.2.2 corrr_0.4.3 broom_0.7.3 kableExtra_1.3.1
[5] knitr_1.30 olsrr_0.5.3 GGally_2.0.0 lubridate_1.7.9
[9] metR_0.9.0 scico_1.2.0 patchwork_1.1.1 collapse_1.5.0
[13] forcats_0.5.0 stringr_1.4.0 dplyr_1.0.2 purrr_0.3.4
[17] readr_1.4.0 tidyr_1.1.2 tibble_3.0.4 ggplot2_3.3.3
[21] tidyverse_1.3.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.0 webshot_0.5.2 RColorBrewer_1.1-2
[4] httr_1.4.2 rprojroot_2.0.2 tools_4.0.3
[7] backports_1.1.10 R6_2.5.0 nortest_1.0-4
[10] DBI_1.1.0 colorspace_2.0-0 withr_2.3.0
[13] gridExtra_2.3 tidyselect_1.1.0 curl_4.3
[16] compiler_4.0.3 git2r_0.27.1 cli_2.2.0
[19] rvest_0.3.6 xml2_1.3.2 sass_0.2.0
[22] labeling_0.4.2 scales_1.1.1 checkmate_2.0.0
[25] goftest_1.2-2 digest_0.6.27 foreign_0.8-80
[28] rmarkdown_2.5 rio_0.5.16 pkgconfig_2.0.3
[31] htmltools_0.5.0 highr_0.8 dbplyr_1.4.4
[34] rlang_0.4.10 readxl_1.3.1 rstudioapi_0.13
[37] farver_2.0.3 generics_0.1.0 jsonlite_1.7.2
[40] zip_2.1.1 car_3.0-10 magrittr_2.0.1
[43] Matrix_1.2-18 Rcpp_1.0.5 munsell_0.5.0
[46] fansi_0.4.1 abind_1.4-5 lifecycle_0.2.0
[49] stringi_1.5.3 whisker_0.4 yaml_2.2.1
[52] carData_3.0-4 plyr_1.8.6 grid_4.0.3
[55] blob_1.2.1 parallel_4.0.3 promises_1.1.1
[58] crayon_1.3.4 lattice_0.20-41 haven_2.3.1
[61] hms_0.5.3 pillar_1.4.7 reprex_0.3.0
[64] glue_1.4.2 evaluate_0.14 RcppArmadillo_0.10.1.2.2
[67] data.table_1.13.6 modelr_0.1.8 vctrs_0.3.6
[70] httpuv_1.5.4 cellranger_1.1.0 gtable_0.3.0
[73] reshape_0.8.8 assertthat_0.2.1 xfun_0.20
[76] openxlsx_4.2.3 RcppEigen_0.3.3.9.1 later_1.1.0.1
[79] viridisLite_0.3.0 ellipsis_0.3.1 here_1.0.1