Column inventories
Jens Daniel Müller
09 April, 2022
Last updated: 2022-04-09
Checks: 7 0
Knit directory: emlr_obs_analysis/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210412) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 46091ea. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/
Ignored: output/other/
Ignored: output/publication/
Unstaged changes:
Modified: analysis/_site.yml
Modified: code/Workflowr_project_managment.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/classic_column_inventories.Rmd) and HTML (docs/classic_column_inventories.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | ae5f326 | jens-daniel-mueller | 2022-04-08 | Build site. |
| html | 2839d72 | jens-daniel-mueller | 2022-03-15 | Build site. |
| html | c3a6238 | jens-daniel-mueller | 2022-03-08 | Build site. |
| html | de557de | jens-daniel-mueller | 2022-01-28 | Build site. |
| html | 6c2e89a | jens-daniel-mueller | 2022-01-27 | Build site. |
| html | 9753eb8 | jens-daniel-mueller | 2022-01-26 | Build site. |
| html | f347cd7 | jens-daniel-mueller | 2022-01-18 | Build site. |
| html | 513630f | jens-daniel-mueller | 2022-01-18 | Build site. |
| html | d7dfc7c | jens-daniel-mueller | 2022-01-18 | Build site. |
| html | 2c8371d | jens-daniel-mueller | 2022-01-17 | Build site. |
| Rmd | 53f06ba | jens-daniel-mueller | 2022-01-17 | removed data cleaning with CANYONB or MLR residuals |
| html | 269809e | jens-daniel-mueller | 2022-01-12 | Build site. |
| html | 1696b98 | jens-daniel-mueller | 2022-01-11 | Build site. |
| html | 570e738 | jens-daniel-mueller | 2022-01-10 | Build site. |
| Rmd | d3903e6 | jens-daniel-mueller | 2022-01-10 | rebuild with child docs |
| html | 9bf6789 | jens-daniel-mueller | 2022-01-10 | Build site. |
| html | 23068f5 | jens-daniel-mueller | 2022-01-10 | Build site. |
| Rmd | a79fb02 | jens-daniel-mueller | 2022-01-10 | split child documents |
| html | ccc57bf | jens-daniel-mueller | 2022-01-10 | Build site. |
| Rmd | 8f07aad | jens-daniel-mueller | 2022-01-10 | split child documents |
| html | 33f190b | jens-daniel-mueller | 2022-01-09 | Build site. |
| Rmd | 053a459 | jens-daniel-mueller | 2022-01-09 | test with MLR_target runs |
| html | 88c47ca | jens-daniel-mueller | 2022-01-07 | Build site. |
| Rmd | 22e83d8 | jens-daniel-mueller | 2022-01-07 | test with MLR_target runs |
| html | a1b4fac | jens-daniel-mueller | 2022-01-07 | Build site. |
| html | 69b058c | jens-daniel-mueller | 2022-01-07 | Build site. |
| html | 296acd8 | jens-daniel-mueller | 2022-01-07 | Build site. |
| Rmd | 71c6a60 | jens-daniel-mueller | 2022-01-06 | created child documents |
| html | 030836b | jens-daniel-mueller | 2022-01-06 | Build site. |
| html | bc2c182 | jens-daniel-mueller | 2021-12-23 | Build site. |
| html | e55f868 | jens-daniel-mueller | 2021-12-23 | Build site. |
| html | db09d70 | jens-daniel-mueller | 2021-12-10 | Build site. |
| html | 7e0a36b | jens-daniel-mueller | 2021-11-21 | Build site. |
| html | e505a4b | jens-daniel-mueller | 2021-11-09 | Build site. |
| html | 66ec048 | jens-daniel-mueller | 2021-11-04 | Build site. |
| Rmd | 3eaddfc | jens-daniel-mueller | 2021-11-04 | compared G19 and this study directly |
| html | 3f7b649 | jens-daniel-mueller | 2021-11-03 | Build site. |
| Rmd | d675dd9 | jens-daniel-mueller | 2021-11-03 | updated plots |
| html | f7c3da2 | jens-daniel-mueller | 2021-11-03 | Build site. |
| html | e534f51 | jens-daniel-mueller | 2021-11-02 | Build site. |
| Rmd | 31cbfae | jens-daniel-mueller | 2021-11-02 | rebuild with revised IO tests |
| html | aea138c | jens-daniel-mueller | 2021-11-01 | Build site. |
| Rmd | 133bbab | jens-daniel-mueller | 2021-11-01 | tried to align plots |
| html | 57cfc36 | jens-daniel-mueller | 2021-11-01 | Build site. |
| html | 4331a22 | jens-daniel-mueller | 2021-10-29 | Build site. |
| html | a162b7e | jens-daniel-mueller | 2021-10-28 | Build site. |
| Rmd | ebaf3bd | jens-daniel-mueller | 2021-10-28 | cleaned output |
| html | ae5ae64 | jens-daniel-mueller | 2021-10-26 | Build site. |
| Rmd | 059b2bf | jens-daniel-mueller | 2021-10-26 | rebuild after /docs error |
| html | 059b2bf | jens-daniel-mueller | 2021-10-26 | rebuild after /docs error |
| html | 8b4e334 | jens-daniel-mueller | 2021-10-26 | Build site. |
| Rmd | 09c80f9 | jens-daniel-mueller | 2021-10-26 | revised composite plots |
| html | e753cfc | jens-daniel-mueller | 2021-10-25 | Build site. |
| Rmd | 78f1dbc | jens-daniel-mueller | 2021-10-25 | revised composite plot |
| html | 7ae4bdd | jens-daniel-mueller | 2021-10-25 | Build site. |
| Rmd | a7bcc04 | jens-daniel-mueller | 2021-10-25 | started composite plot |
| html | a8576a8 | jens-daniel-mueller | 2021-10-25 | Build site. |
| Rmd | 8cbaf95 | jens-daniel-mueller | 2021-10-25 | ensemble lat-lon grid budgets added |
| html | 287324c | jens-daniel-mueller | 2021-10-25 | Build site. |
| Rmd | c99a911 | jens-daniel-mueller | 2021-10-25 | lat-lon grid budgets added |
| html | 8c9fa17 | jens-daniel-mueller | 2021-10-22 | Build site. |
| html | 968fe94 | jens-daniel-mueller | 2021-10-18 | Build site. |
| html | 581baa0 | jens-daniel-mueller | 2021-10-07 | Build site. |
| html | a7af62f | jens-daniel-mueller | 2021-10-06 | Build site. |
| html | f9b4f93 | jens-daniel-mueller | 2021-10-05 | Build site. |
| html | 54c2b1e | jens-daniel-mueller | 2021-09-30 | Build site. |
| Rmd | 4efa83e | jens-daniel-mueller | 2021-09-30 | maps added |
| html | 8a01fb2 | jens-daniel-mueller | 2021-09-30 | Build site. |
| Rmd | 9a115bf | jens-daniel-mueller | 2021-09-30 | maps added |
| html | c225f90 | jens-daniel-mueller | 2021-09-30 | Build site. |
| Rmd | 67edc71 | jens-daniel-mueller | 2021-09-30 | maps added |
| html | 318dbe4 | jens-daniel-mueller | 2021-09-30 | Build site. |
| Rmd | 9d7d257 | jens-daniel-mueller | 2021-09-30 | maps added |
| html | a6689de | jens-daniel-mueller | 2021-09-29 | Build site. |
| Rmd | d635f71 | jens-daniel-mueller | 2021-09-29 | column inventory plots for presentation |
| html | 960d158 | jens-daniel-mueller | 2021-09-29 | Build site. |
| html | 0573621 | jens-daniel-mueller | 2021-09-29 | Build site. |
| html | f51dcb9 | jens-daniel-mueller | 2021-09-28 | Build site. |
| Rmd | 056c790 | jens-daniel-mueller | 2021-09-28 | new MLR_basin added |
| html | 55d2059 | jens-daniel-mueller | 2021-09-28 | Build site. |
| Rmd | 9307c46 | jens-daniel-mueller | 2021-09-28 | new MLR_basin added |
| html | 5d5d6f7 | jens-daniel-mueller | 2021-09-28 | Build site. |
| Rmd | ae39df2 | jens-daniel-mueller | 2021-09-28 | update density distribution |
| html | 83fb5b3 | jens-daniel-mueller | 2021-09-28 | Build site. |
| Rmd | 2c64a26 | jens-daniel-mueller | 2021-09-28 | update map color scale range |
| html | aa8bd1f | jens-daniel-mueller | 2021-09-28 | Build site. |
| Rmd | 134da12 | jens-daniel-mueller | 2021-09-28 | update map color scale range |
| html | d0dbf3e | jens-daniel-mueller | 2021-09-27 | Build site. |
| Rmd | f8ae9b3 | jens-daniel-mueller | 2021-09-27 | filter MLR basins with both criteria |
| html | 09002a7 | jens-daniel-mueller | 2021-09-26 | Build site. |
| html | 0c6be7e | jens-daniel-mueller | 2021-09-24 | Build site. |
| Rmd | 6a79fd0 | jens-daniel-mueller | 2021-09-24 | consider ensemble selection across all analysis |
| html | f54d5db | jens-daniel-mueller | 2021-09-24 | Build site. |
| Rmd | 76c942b | jens-daniel-mueller | 2021-09-24 | 1990 as start year 1 for classic runs |
| html | 31c33cb | jens-daniel-mueller | 2021-09-23 | Build site. |
| Rmd | 9b1df4d | jens-daniel-mueller | 2021-09-23 | 1994 vs 2014 added |
| html | ec4f702 | jens-daniel-mueller | 2021-09-22 | Build site. |
| html | 6525680 | jens-daniel-mueller | 2021-09-21 | Build site. |
| html | 6bf38c9 | jens-daniel-mueller | 2021-09-21 | Build site. |
| html | 63911d0 | jens-daniel-mueller | 2021-09-21 | Build site. |
| html | 499c9d5 | jens-daniel-mueller | 2021-09-21 | Build site. |
| html | 271a3ac | jens-daniel-mueller | 2021-09-21 | Build site. |
| html | 35ad8b5 | jens-daniel-mueller | 2021-09-20 | Build site. |
| Rmd | 919f74b | jens-daniel-mueller | 2021-09-20 | rebuildt with canyon b cleaning analysis |
version_id_pattern <- "1"
config <- "MLR_basins"1 Read files
# identify required version IDs
Version_IDs_1 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_1", "1"))
Version_IDs_2 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_2", "1"))
Version_IDs_3 <- list.files(path = "/nfs/kryo/work/jenmueller/emlr_cant/observations",
pattern = paste0("v_3", "1"))
Version_IDs <- c(Version_IDs_1, Version_IDs_2, Version_IDs_3)
print(Version_IDs) [1] "v_1101" "v_1102" "v_1103" "v_1104" "v_1105" "v_1106" "v_2101" "v_2102"
[9] "v_2103" "v_2104" "v_2105" "v_2106" "v_3101" "v_3102" "v_3103" "v_3104"
[17] "v_3105" "v_3106"for (i_Version_IDs in Version_IDs) {
# i_Version_IDs <- Version_IDs[1]
print(i_Version_IDs)
path_version_data <-
paste(path_observations,
i_Version_IDs,
"/data/",
sep = "")
# load and join data files
dcant_inv <-
read_csv(paste(path_version_data,
"dcant_inv.csv",
sep = ""))
dcant_inv_mod_truth <-
read_csv(paste(path_version_data,
"dcant_inv_mod_truth.csv",
sep = "")) %>%
filter(method == "total") %>%
select(-method)
dcant_inv_bias <-
read_csv(paste(path_version_data,
"dcant_inv_bias.csv",
sep = "")) %>%
mutate(Version_ID = i_Version_IDs)
dcant_inv <- bind_rows(dcant_inv,
dcant_inv_mod_truth) %>%
mutate(Version_ID = i_Version_IDs)
dcant_budget_lat_grid <-
read_csv(paste(path_version_data,
"dcant_budget_lat_grid.csv",
sep = "")) %>%
mutate(Version_ID = i_Version_IDs)
dcant_budget_lon_grid <-
read_csv(paste(path_version_data,
"dcant_budget_lon_grid.csv",
sep = "")) %>%
mutate(Version_ID = i_Version_IDs)
params_local <-
read_rds(paste(path_version_data,
"params_local.rds",
sep = ""))
params_local <- bind_cols(
Version_ID = i_Version_IDs,
MLR_basins := str_c(params_local$MLR_basins, collapse = "|"),
tref1 = params_local$tref1,
tref2 = params_local$tref2)
tref <- read_csv(paste(path_version_data,
"tref.csv",
sep = ""))
params_local <- params_local %>%
mutate(
median_year_1 = sort(tref$median_year)[1],
median_year_2 = sort(tref$median_year)[2],
duration = median_year_2 - median_year_1,
period = paste(median_year_1, "-", median_year_2)
)
if (exists("dcant_inv_all")) {
dcant_inv_all <- bind_rows(dcant_inv_all, dcant_inv)
}
if (!exists("dcant_inv_all")) {
dcant_inv_all <- dcant_inv
}
if (exists("dcant_inv_bias_all")) {
dcant_inv_bias_all <- bind_rows(dcant_inv_bias_all, dcant_inv_bias)
}
if (!exists("dcant_inv_bias_all")) {
dcant_inv_bias_all <- dcant_inv_bias
}
if (exists("dcant_budget_lat_grid_all")) {
dcant_budget_lat_grid_all <- bind_rows(dcant_budget_lat_grid_all, dcant_budget_lat_grid)
}
if (!exists("dcant_budget_lat_grid_all")) {
dcant_budget_lat_grid_all <- dcant_budget_lat_grid
}
if (exists("dcant_budget_lon_grid_all")) {
dcant_budget_lon_grid_all <- bind_rows(dcant_budget_lon_grid_all, dcant_budget_lon_grid)
}
if (!exists("dcant_budget_lon_grid_all")) {
dcant_budget_lon_grid_all <- dcant_budget_lon_grid
}
if (exists("params_local_all")) {
params_local_all <- bind_rows(params_local_all, params_local)
}
if (!exists("params_local_all")) {
params_local_all <- params_local
}
}[1] "v_1101"
[1] "v_1102"
[1] "v_1103"
[1] "v_1104"
[1] "v_1105"
[1] "v_1106"
[1] "v_2101"
[1] "v_2102"
[1] "v_2103"
[1] "v_2104"
[1] "v_2105"
[1] "v_2106"
[1] "v_3101"
[1] "v_3102"
[1] "v_3103"
[1] "v_3104"
[1] "v_3105"
[1] "v_3106"rm(dcant_inv,
dcant_inv_bias,
dcant_inv_mod_truth,
dcant_budget_lat_grid,
dcant_budget_lon_grid,
params_local,
tref)
# params_local_all <-
# params_local_all %>%
# mutate(period = factor(period, c("1994 - 2004", "2004 - 2014", "1994 - 2014")))dcant_inv_all <- dcant_inv_all %>%
filter(inv_depth == params_global$inventory_depth_standard)
dcant_budget_lat_grid_all <- dcant_budget_lat_grid_all %>%
filter(inv_depth == params_global$inventory_depth_standard)
dcant_budget_lon_grid_all <- dcant_budget_lon_grid_all %>%
filter(inv_depth == params_global$inventory_depth_standard)all_predictors <- c("saltempaouoxygenphosphatenitratesilicate")
params_local_all <- params_local_all %>%
mutate(MLR_predictors = str_remove_all(all_predictors,
MLR_predictors))dcant_budget_lat_grid_all <- dcant_budget_lat_grid_all %>%
pivot_wider(names_from = estimate,
values_from = value) %>%
filter(period != "1994 - 2014",
method == "total")
dcant_budget_lon_grid_all <- dcant_budget_lon_grid_all %>%
pivot_wider(names_from = estimate,
values_from = value) %>%
filter(period != "1994 - 2014",
method == "total")2 Uncertainty limit
sd_uncertainty_limit <- 43 Individual cases
3.1 Absoulte values
dcant_inv_all %>%
filter(data_source %in% c("mod", "obs"),
period != "1994 - 2014") %>%
group_by(data_source) %>%
group_split() %>%
# head(1) %>%
map(
~ p_map_cant_inv(df = .x,
var = "dcant",
subtitle_text = paste("data_source:",
unique(.x$data_source))) +
facet_grid(MLR_basins ~ period) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())
)[[1]]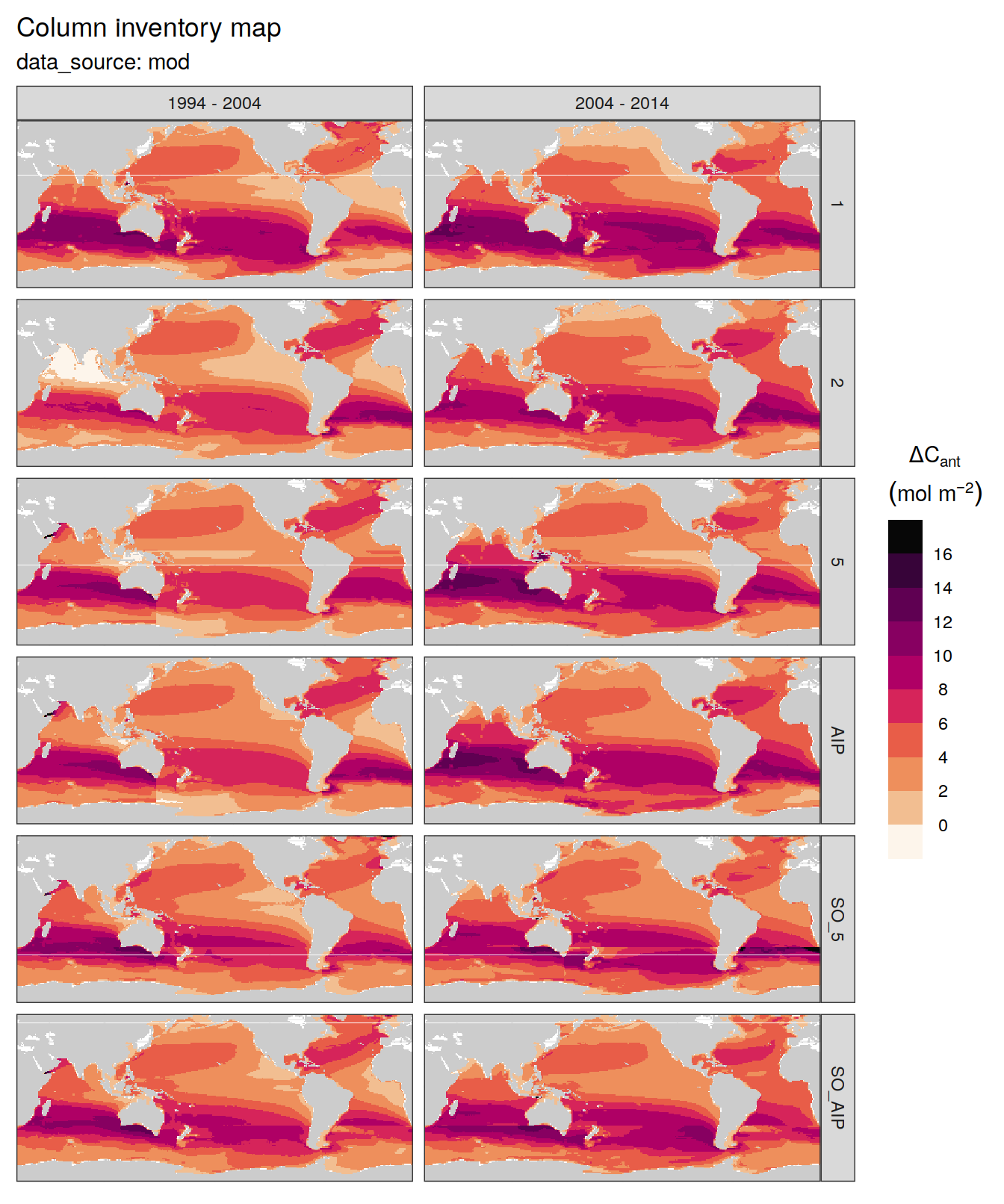
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| 83fb5b3 | jens-daniel-mueller | 2021-09-28 |
| aa8bd1f | jens-daniel-mueller | 2021-09-28 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
[[2]]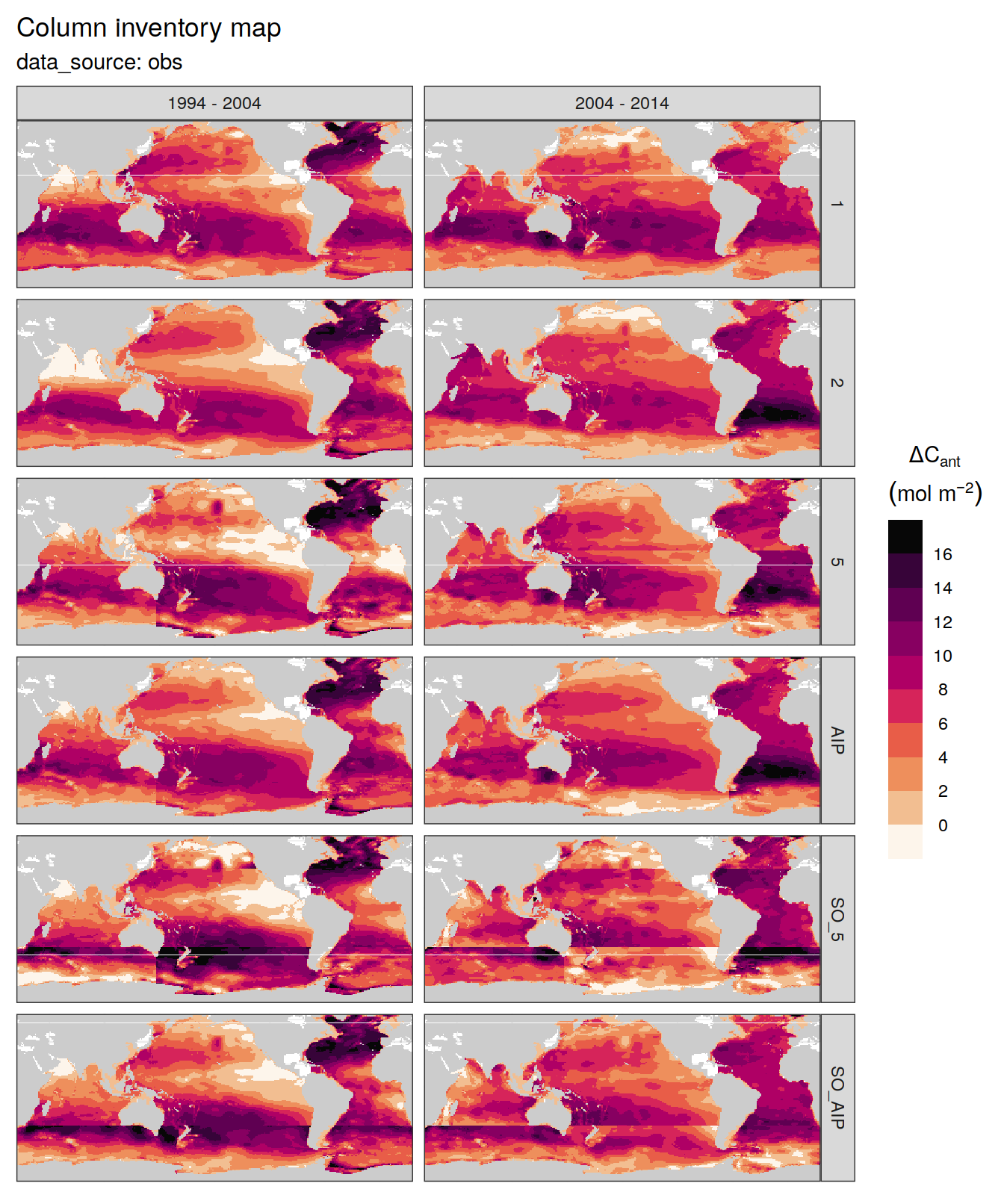
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| 83fb5b3 | jens-daniel-mueller | 2021-09-28 |
| aa8bd1f | jens-daniel-mueller | 2021-09-28 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 31c33cb | jens-daniel-mueller | 2021-09-23 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
p_dcant_inv_all_1994_2004 <-
dcant_inv_all %>%
filter(data_source %in% c("obs"),
period == "1994 - 2004") %>%
mutate(period = recode(period,
"1994 - 2004" = "Observation-based")) %>%
p_map_cant_inv(var = "dcant",
title_text = "1994 - 2004") +
facet_grid(MLR_basins ~ period) +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
strip.background.y = element_blank(),
strip.text.y = element_blank(),
legend.position = "left"
)3.2 Biases
dcant_inv_bias_all %>%
filter(period != "1994 - 2014") %>%
p_map_cant_inv(var = "dcant_bias",
col = "bias",
subtitle_text = "data_source: mod - mod_truth") +
facet_grid(MLR_basins ~ period)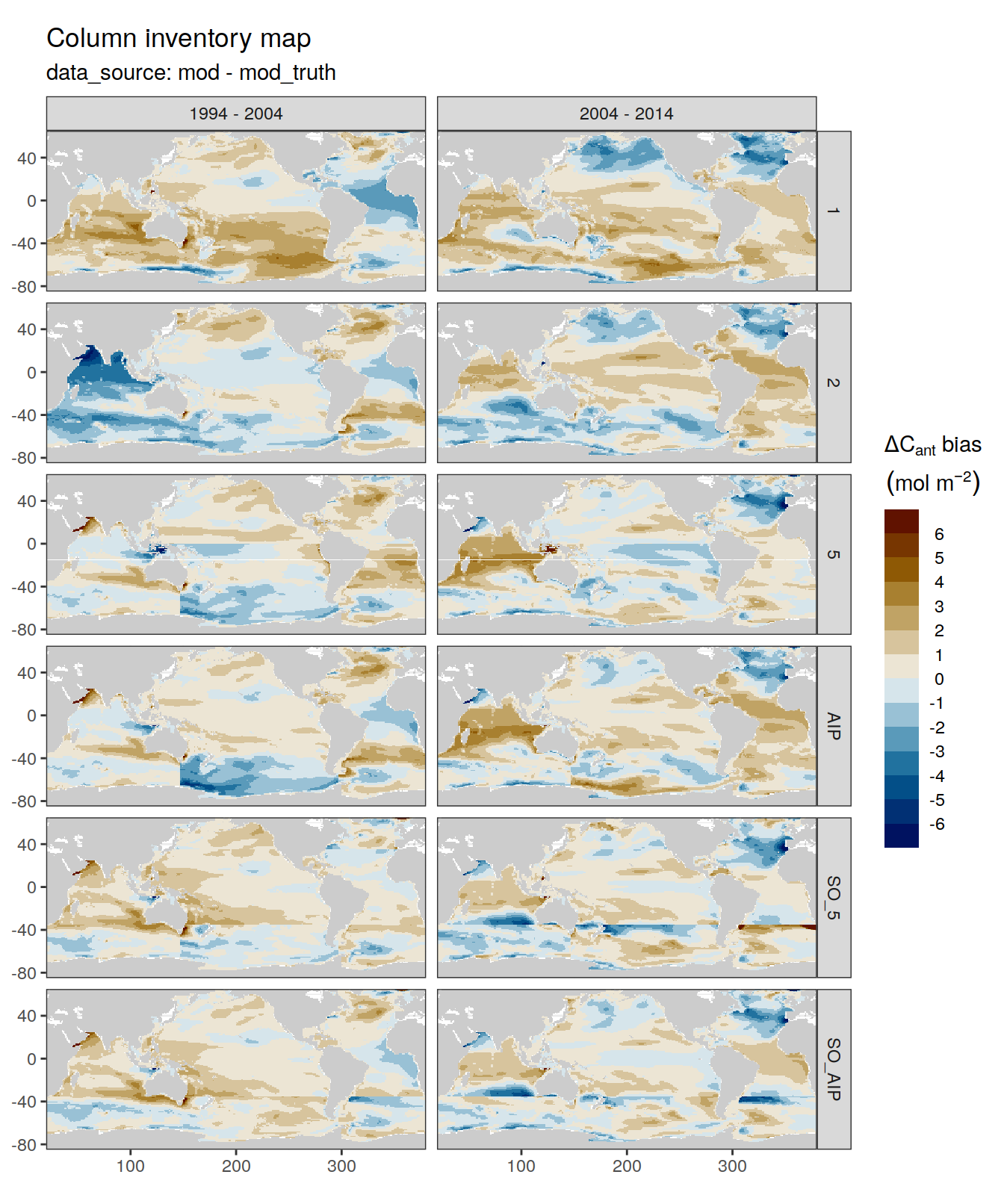
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| 5d5d6f7 | jens-daniel-mueller | 2021-09-28 |
| 83fb5b3 | jens-daniel-mueller | 2021-09-28 |
| aa8bd1f | jens-daniel-mueller | 2021-09-28 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 31c33cb | jens-daniel-mueller | 2021-09-23 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
dcant_inv_bias_all %>%
filter(period == "1994 - 2014") %>%
p_map_cant_inv(var = "dcant_bias",
col = "bias",
subtitle_text = "data_source: mod - mod_truth") +
facet_grid(MLR_basins ~ period)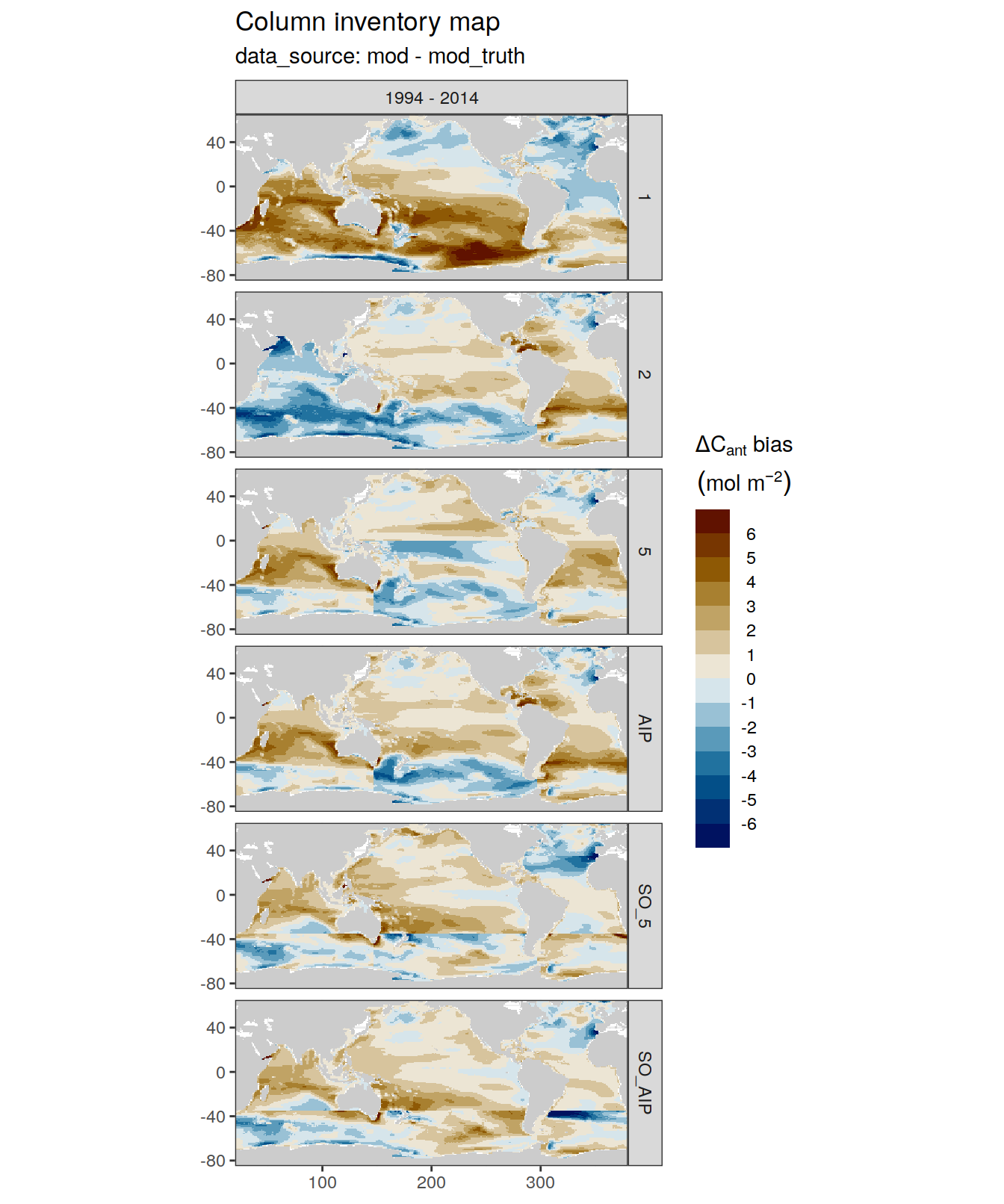
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| 5d5d6f7 | jens-daniel-mueller | 2021-09-28 |
| 83fb5b3 | jens-daniel-mueller | 2021-09-28 |
p_dcant_inv_bias_all_1994_2004 <-
dcant_inv_bias_all %>%
filter(period == "1994 - 2004") %>%
mutate(period = recode(period,
"1994 - 2004" = "Model-based")) %>%
p_map_cant_inv(var = "dcant_bias",
col = "bias",
title_text = "Models") +
facet_grid(MLR_basins ~ period) +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
plot.title = element_blank()
)p_dcant_1994_2004 <-
p_dcant_inv_all_1994_2004 +
p_dcant_inv_bias_all_1994_2004
p_dcant_1994_2004
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
# ggsave(plot = p_dcant_1994_2004,
# path = "output/other",
# filename = "inv_dcant_1994_2004_abs_bias.png",
# height = 8,
# width = 8)3.2.1 Density distribution
dcant_inv_bias_all %>%
filter(abs(dcant_bias) < 10) %>%
ggplot(aes(dcant_bias, col = MLR_basins)) +
scale_color_brewer(palette = "Dark2") +
geom_vline(xintercept = 0) +
geom_density() +
facet_grid(period ~.)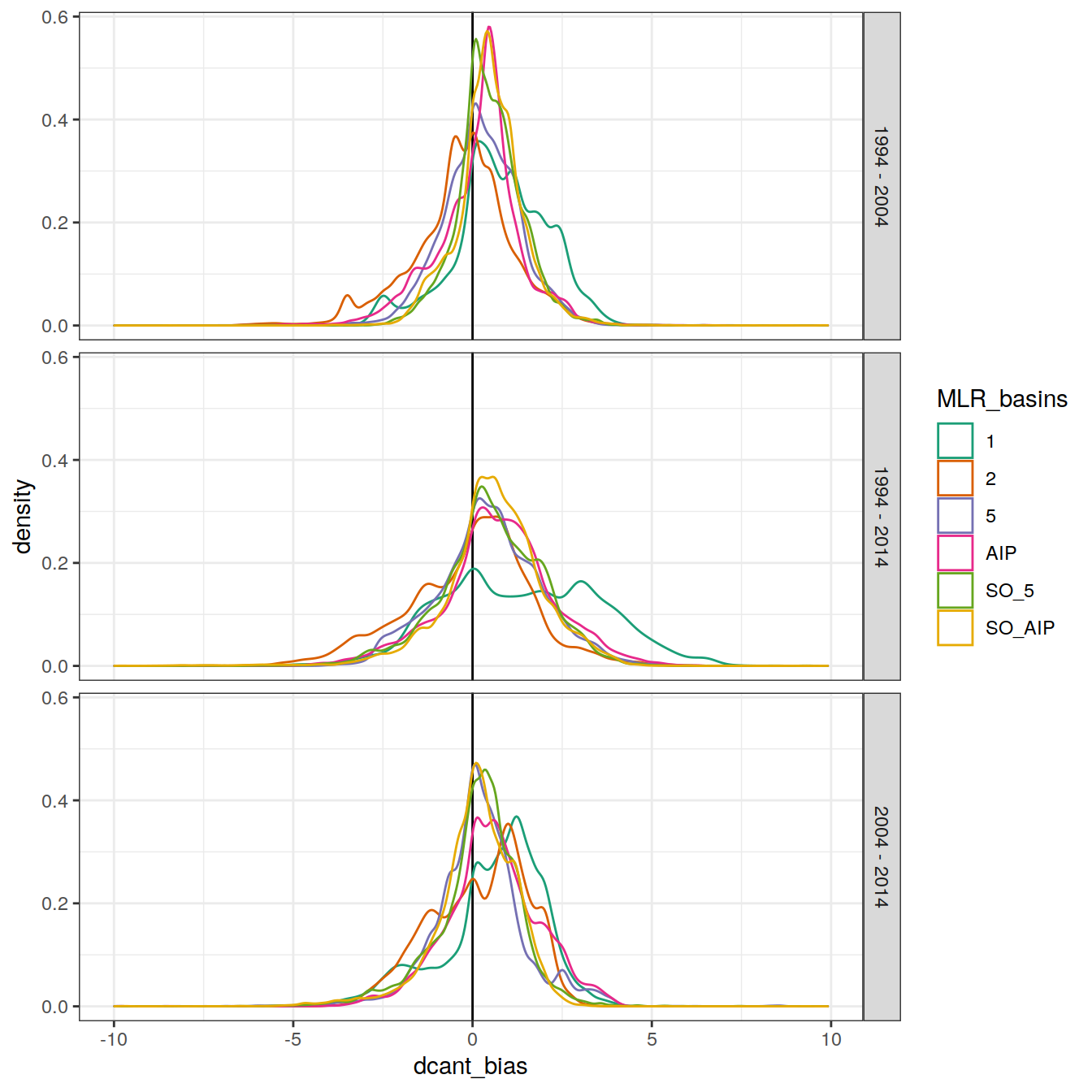
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| 66ec048 | jens-daniel-mueller | 2021-11-04 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
3.3 Lat grid budgets
dcant_budget_lat_grid_all %>%
group_split(data_source) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(lat_grid, dcant, fill = MLR_basins)) +
geom_hline(yintercept = 0) +
geom_col(position = "dodge") +
coord_flip() +
scale_fill_brewer(palette = "Dark2") +
labs(title = paste("data_source:", unique(.x$data_source))) +
facet_grid(basin_AIP ~ period)
)[[1]]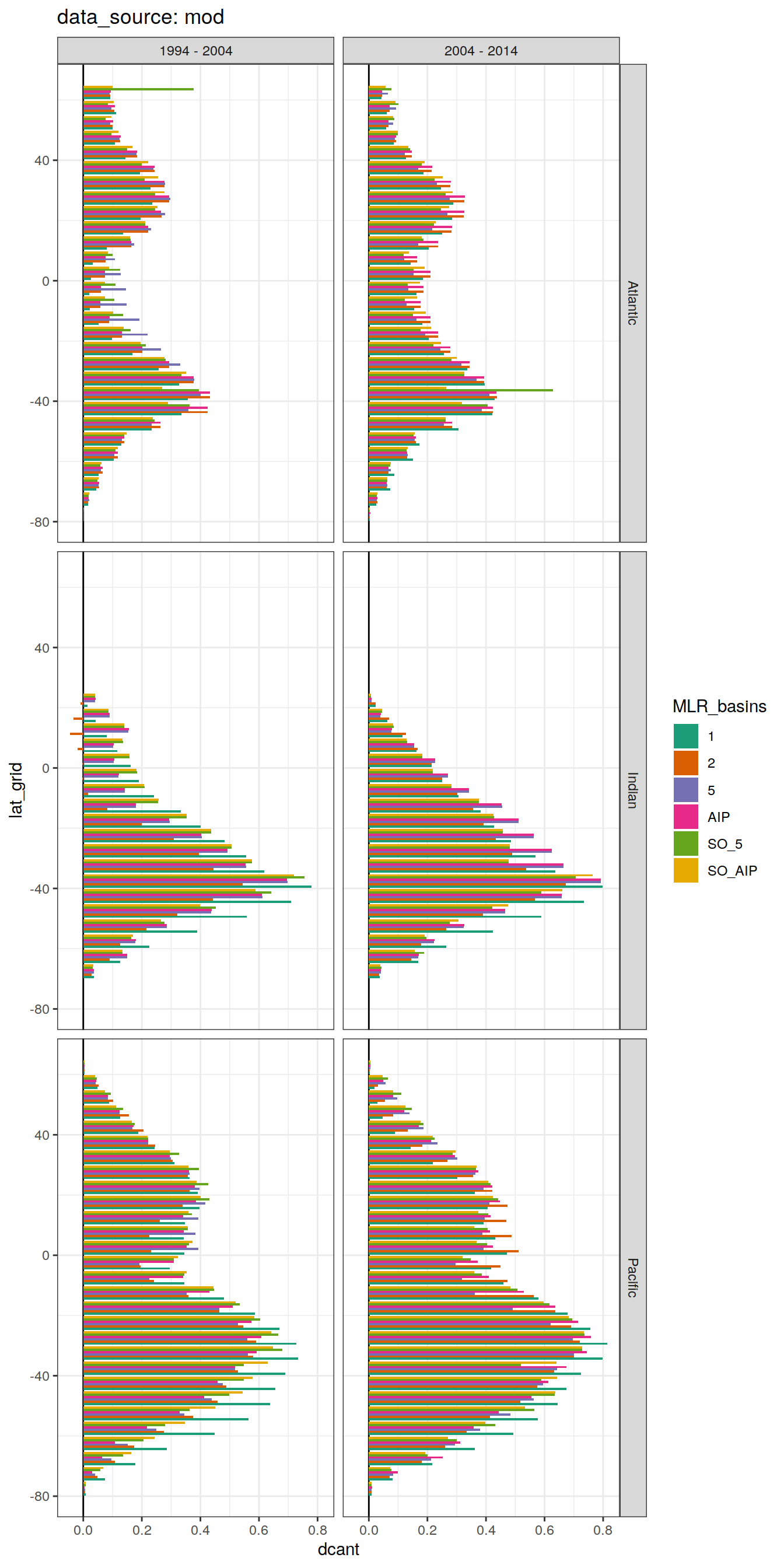
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 287324c | jens-daniel-mueller | 2021-10-25 |
[[2]]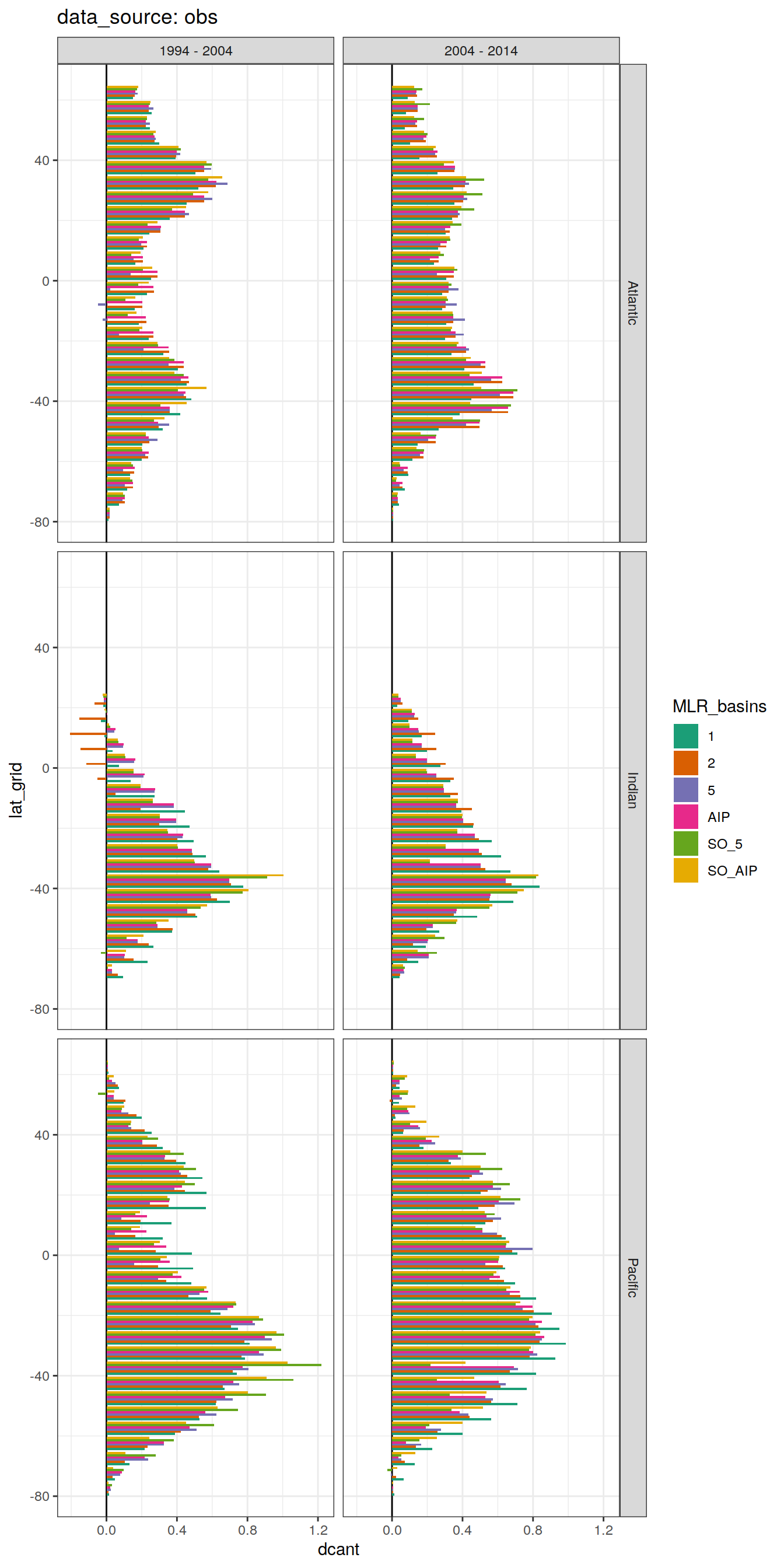
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 287324c | jens-daniel-mueller | 2021-10-25 |
3.4 Lon grid budgets
dcant_budget_lon_grid_all %>%
group_split(data_source, period) %>%
# head(1) %>%
map(
~ ggplot(data = .x,
aes(lon_grid, dcant, fill = MLR_basins)) +
geom_col(position = "dodge") +
scale_fill_brewer(palette = "Dark2") +
labs(title = paste(
"data_source:",
unique(.x$data_source),
"| period:",
unique(.x$period)
)) +
facet_grid(basin_AIP ~ .)
)[[1]]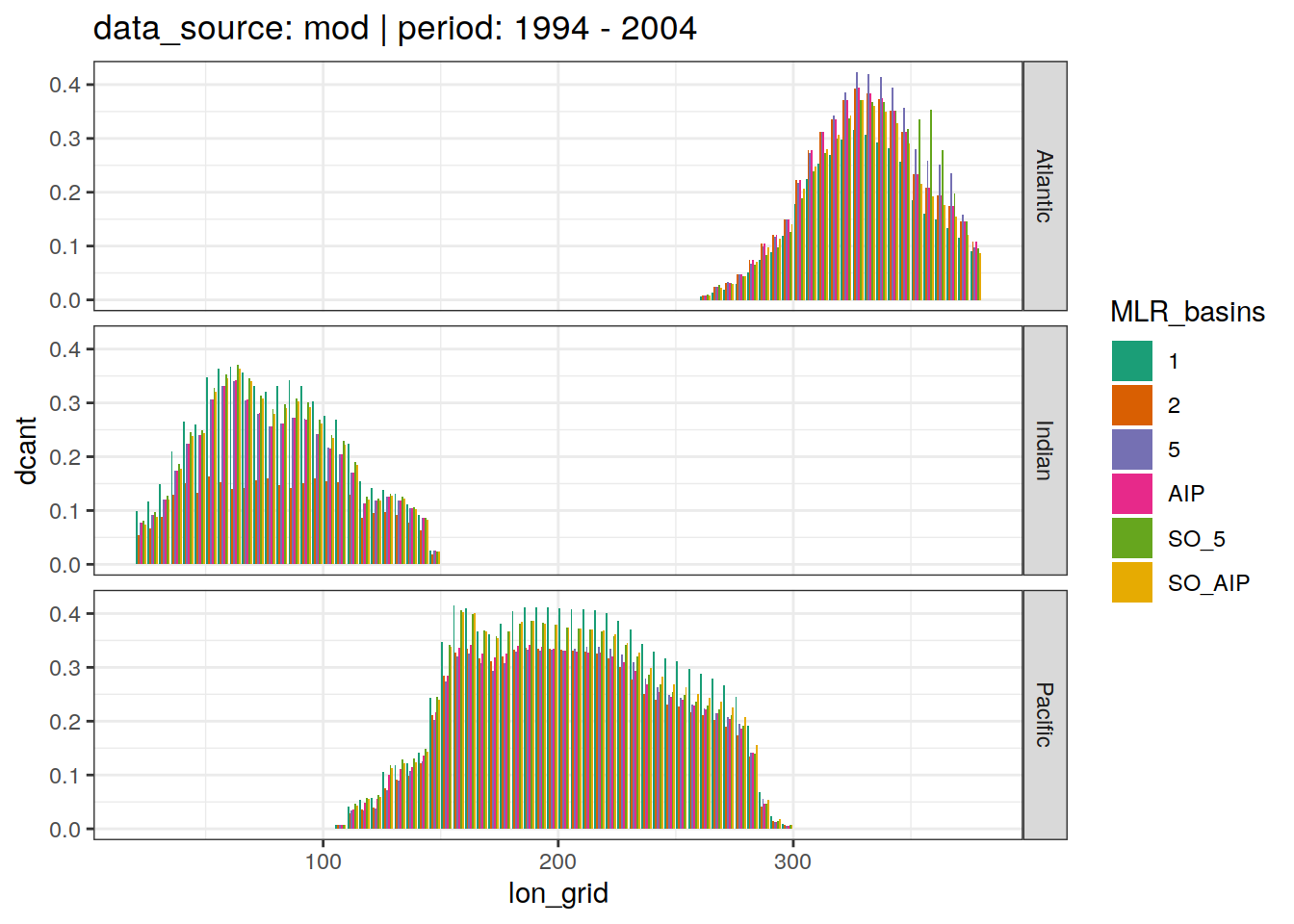
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 287324c | jens-daniel-mueller | 2021-10-25 |
[[2]]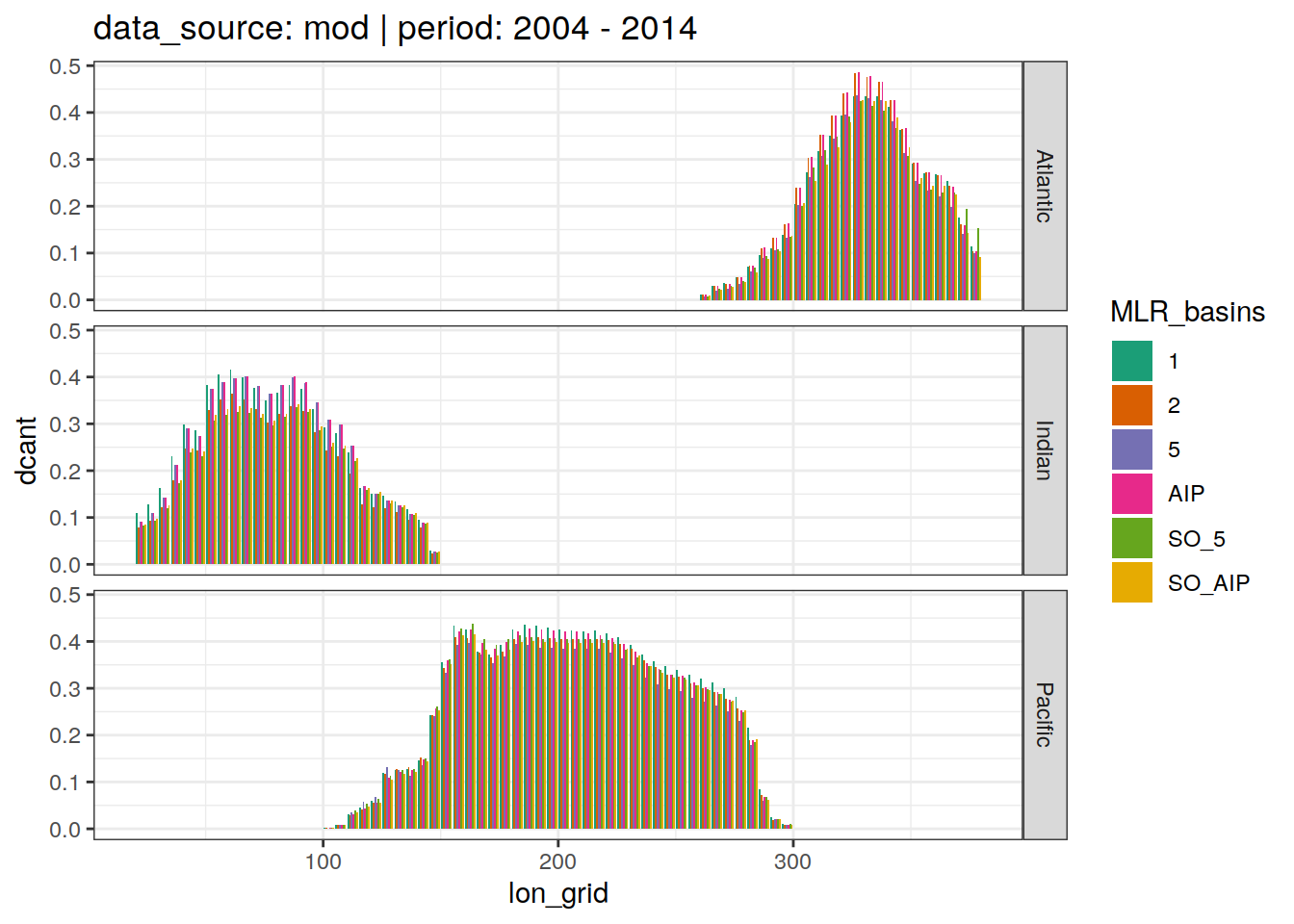
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 287324c | jens-daniel-mueller | 2021-10-25 |
[[3]]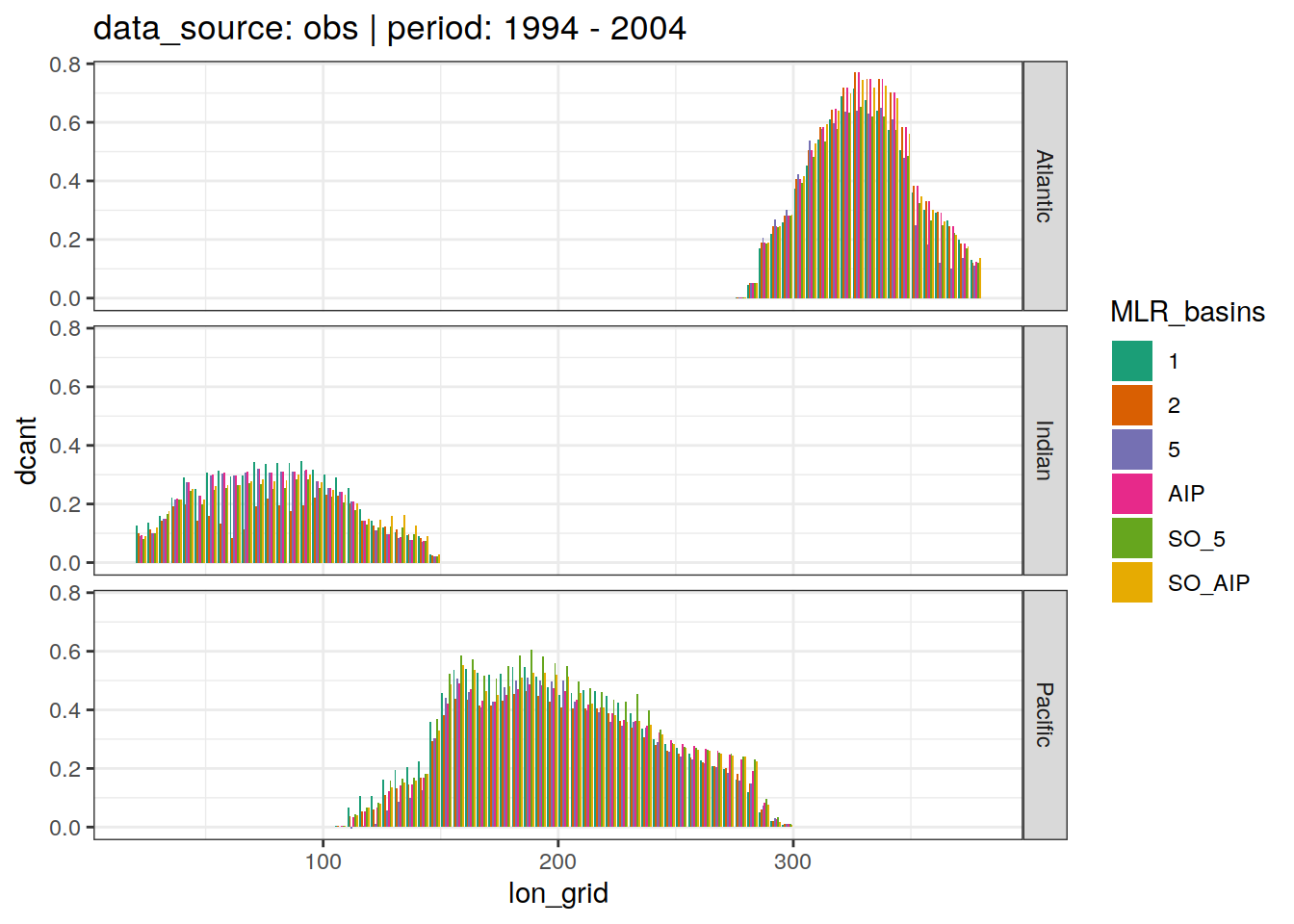
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 287324c | jens-daniel-mueller | 2021-10-25 |
[[4]]
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 287324c | jens-daniel-mueller | 2021-10-25 |
4 Ensemble
dcant_inv_ensemble <- dcant_inv_all %>%
filter(data_source %in% c("mod", "obs")) %>%
group_by(lat, lon, data_source, period) %>%
summarise(dcant_mean = mean(dcant),
dcant_sd = sd(dcant),
dcant_range = max(dcant)- min(dcant)) %>%
ungroup()`summarise()` has grouped output by 'lat', 'lon', 'data_source'. You can override using the `.groups` argument.dcant_lat_grid_ensemble <- dcant_budget_lat_grid_all %>%
filter(data_source %in% c("mod", "obs")) %>%
group_by(lat_grid, data_source, period, MLR_basins) %>%
summarise(dcant = sum(dcant, na.rm = TRUE)) %>%
ungroup() %>%
group_by(lat_grid, data_source, period) %>%
summarise(dcant_mean = mean(dcant),
dcant_sd = sd(dcant),
dcant_max = max(dcant),
dcant_min = min(dcant)) %>%
ungroup()`summarise()` has grouped output by 'lat_grid', 'data_source', 'period'. You can override using the `.groups` argument.`summarise()` has grouped output by 'lat_grid', 'data_source'. You can override using the `.groups` argument.dcant_lon_grid_ensemble <- dcant_budget_lon_grid_all %>%
filter(data_source %in% c("mod", "obs")) %>%
group_by(lon_grid, data_source, period, MLR_basins) %>%
summarise(dcant = sum(dcant, na.rm = TRUE)) %>%
ungroup() %>%
group_by(lon_grid, data_source, period) %>%
summarise(dcant_mean = mean(dcant),
dcant_sd = sd(dcant),
dcant_max = max(dcant),
dcant_min = min(dcant)) %>%
ungroup()`summarise()` has grouped output by 'lon_grid', 'data_source', 'period'. You can override using the `.groups` argument.`summarise()` has grouped output by 'lon_grid', 'data_source'. You can override using the `.groups` argument.4.1 Mean
p_map_cant_inv(
df = dcant_inv_ensemble %>%
filter(period != "1994 - 2014",
data_source == "obs"),
var = "dcant_mean",
subtitle_text = paste("Ensemble mean")
) +
facet_grid(period ~ .) +
theme(axis.text = element_blank(),
axis.ticks = element_blank())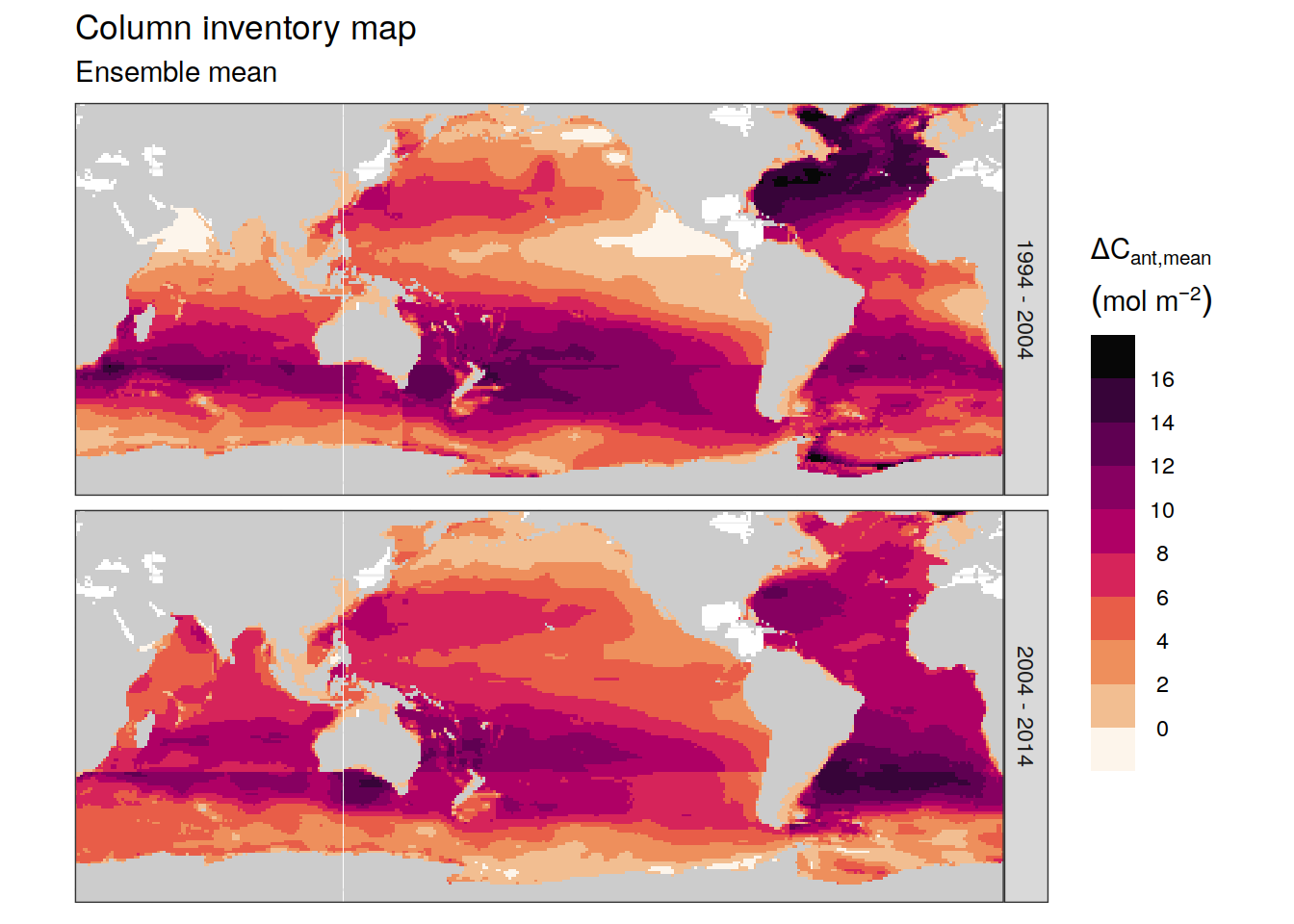
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| a6689de | jens-daniel-mueller | 2021-09-29 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| 83fb5b3 | jens-daniel-mueller | 2021-09-28 |
| aa8bd1f | jens-daniel-mueller | 2021-09-28 |
| d0dbf3e | jens-daniel-mueller | 2021-09-27 |
| 09002a7 | jens-daniel-mueller | 2021-09-26 |
| 0c6be7e | jens-daniel-mueller | 2021-09-24 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 31c33cb | jens-daniel-mueller | 2021-09-23 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
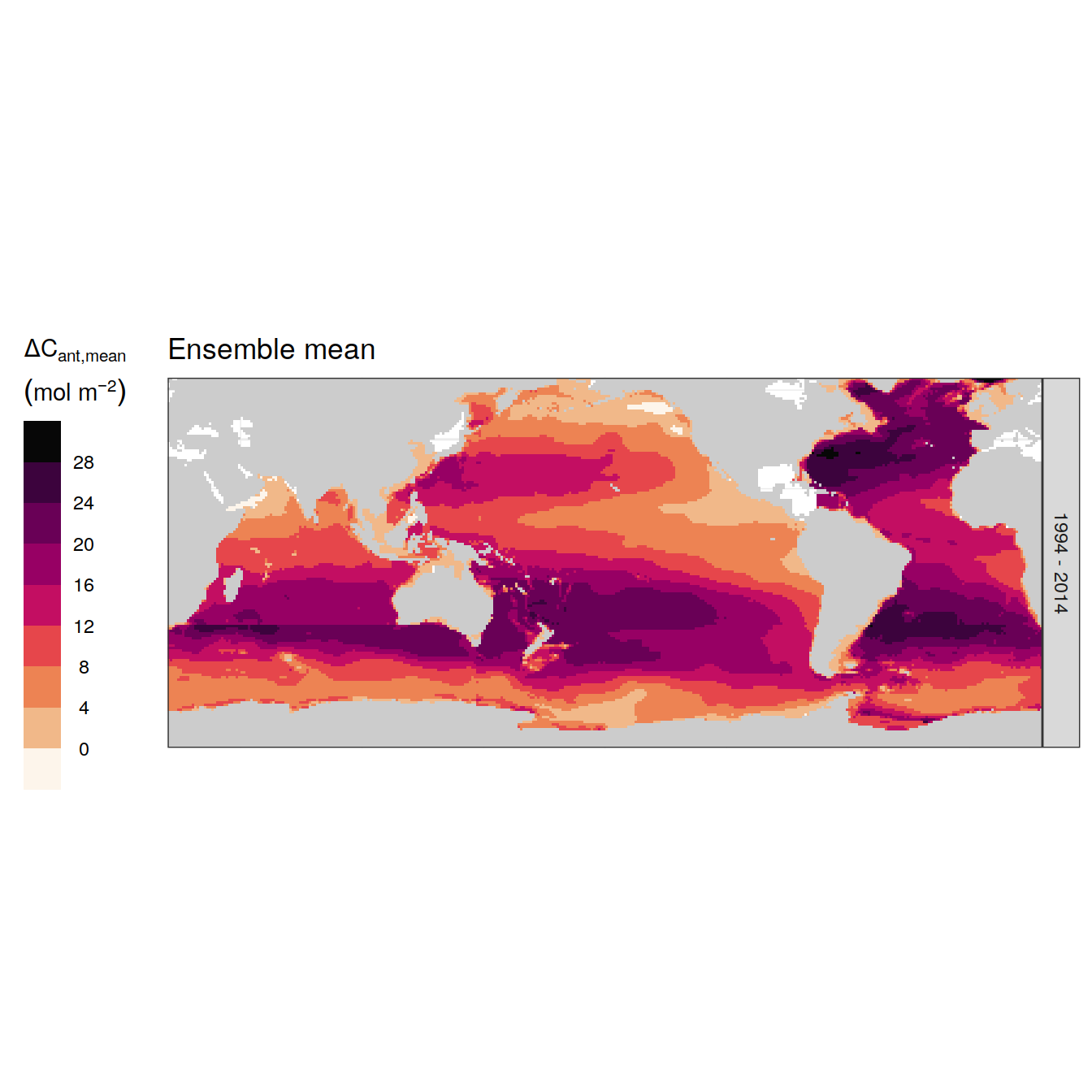
p_map_cant_inv(
df = dcant_inv_ensemble %>%
filter(data_source == "obs",
period == "1994 - 2014"),
var = "dcant_mean",
title_text = paste("Ensemble mean"),
breaks = c(-Inf, seq(0, 28, 4), Inf)
) +
facet_grid(period ~ .) +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
legend.position = "left"
)
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
4.2 Mean bias
dcant_inv_ensemble_bias <- full_join(
dcant_inv_ensemble %>%
filter(data_source == "mod") %>%
select(lat, lon, period, dcant_mean, dcant_sd),
dcant_inv_all %>%
filter(data_source == "mod_truth",
MLR_basins == unique(dcant_inv_all$MLR_basins)[1]) %>%
select(lat, lon, period, dcant)
)Joining, by = c("lat", "lon", "period")dcant_inv_ensemble_bias <- dcant_inv_ensemble_bias %>%
mutate(dcant_mean_bias = dcant_mean - dcant)
dcant_inv_ensemble_bias %>%
filter(period != "1994 - 2014") %>%
p_map_cant_inv(var = "dcant_mean_bias",
col = "bias",
subtitle_text = "Ensemble mean - mod_truth") +
facet_grid(period ~ .)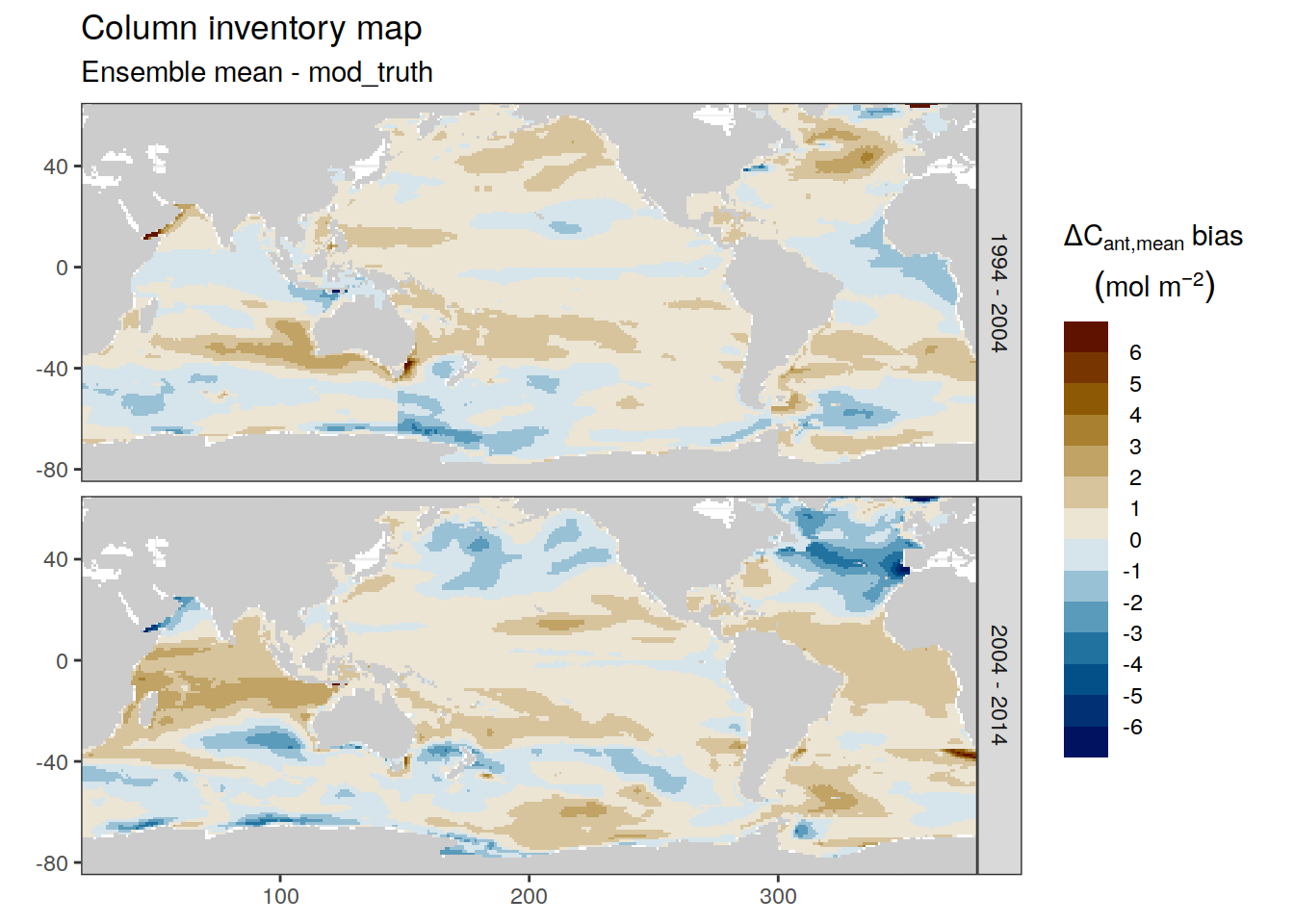
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| 5d5d6f7 | jens-daniel-mueller | 2021-09-28 |
| 83fb5b3 | jens-daniel-mueller | 2021-09-28 |
| aa8bd1f | jens-daniel-mueller | 2021-09-28 |
| d0dbf3e | jens-daniel-mueller | 2021-09-27 |
| 09002a7 | jens-daniel-mueller | 2021-09-26 |
| 0c6be7e | jens-daniel-mueller | 2021-09-24 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 31c33cb | jens-daniel-mueller | 2021-09-23 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
4.2.1 Density distribution
dcant_inv_bias_all %>%
ggplot() +
scale_color_manual(values = c("red", "grey")) +
geom_vline(xintercept = 0) +
geom_density(aes(dcant_bias, group = MLR_basins, col = "Individual")) +
geom_density(data = dcant_inv_ensemble_bias,
aes(dcant_mean_bias, col = "Ensemble")) +
facet_grid(period ~.) +
coord_cartesian(xlim = c(-10, 10))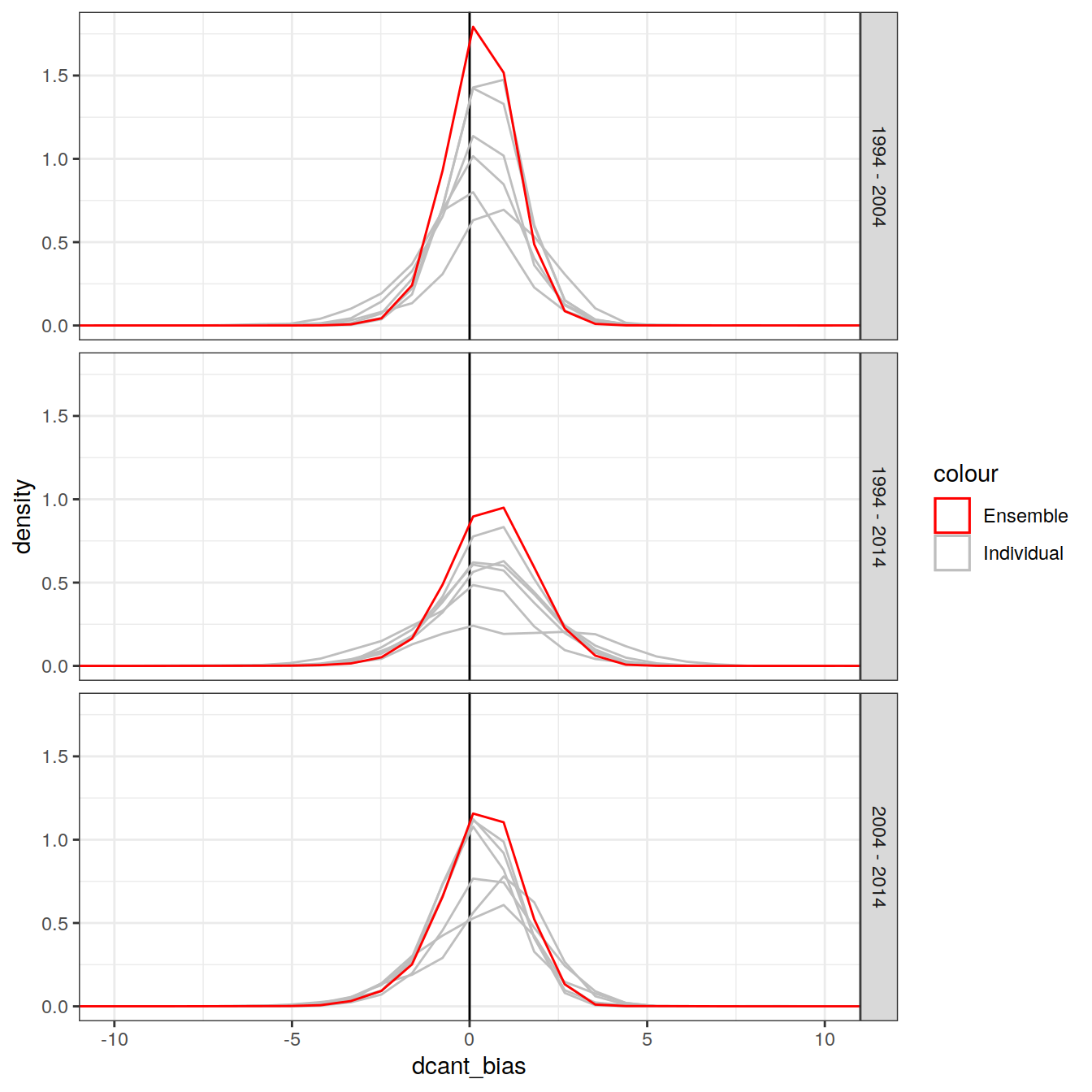
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
4.3 Mean lat grid budgets
dcant_lat_grid_ensemble %>%
ggplot(aes(lat_grid, dcant_mean)) +
geom_hline(yintercept = 0) +
geom_col(position = "dodge",
fill = "grey80",
col = "grey20") +
geom_errorbar(aes(
ymin = dcant_min,
ymax = dcant_max
),
col = "grey20",
width = 0) +
scale_color_brewer(palette = "Set1") +
coord_flip() +
scale_fill_brewer(palette = "Dark2") +
facet_grid(data_source ~ period)
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| a8576a8 | jens-daniel-mueller | 2021-10-25 |
4.4 Mean lon grid budgets
dcant_lon_grid_ensemble %>%
ggplot(aes(lon_grid, dcant_mean)) +
geom_col(position = "dodge",
fill = "grey80",
col = "grey20") +
geom_errorbar(aes(
ymin = dcant_min,
ymax = dcant_max
),
col = "grey20",
width = 0) +
facet_grid(data_source ~ period)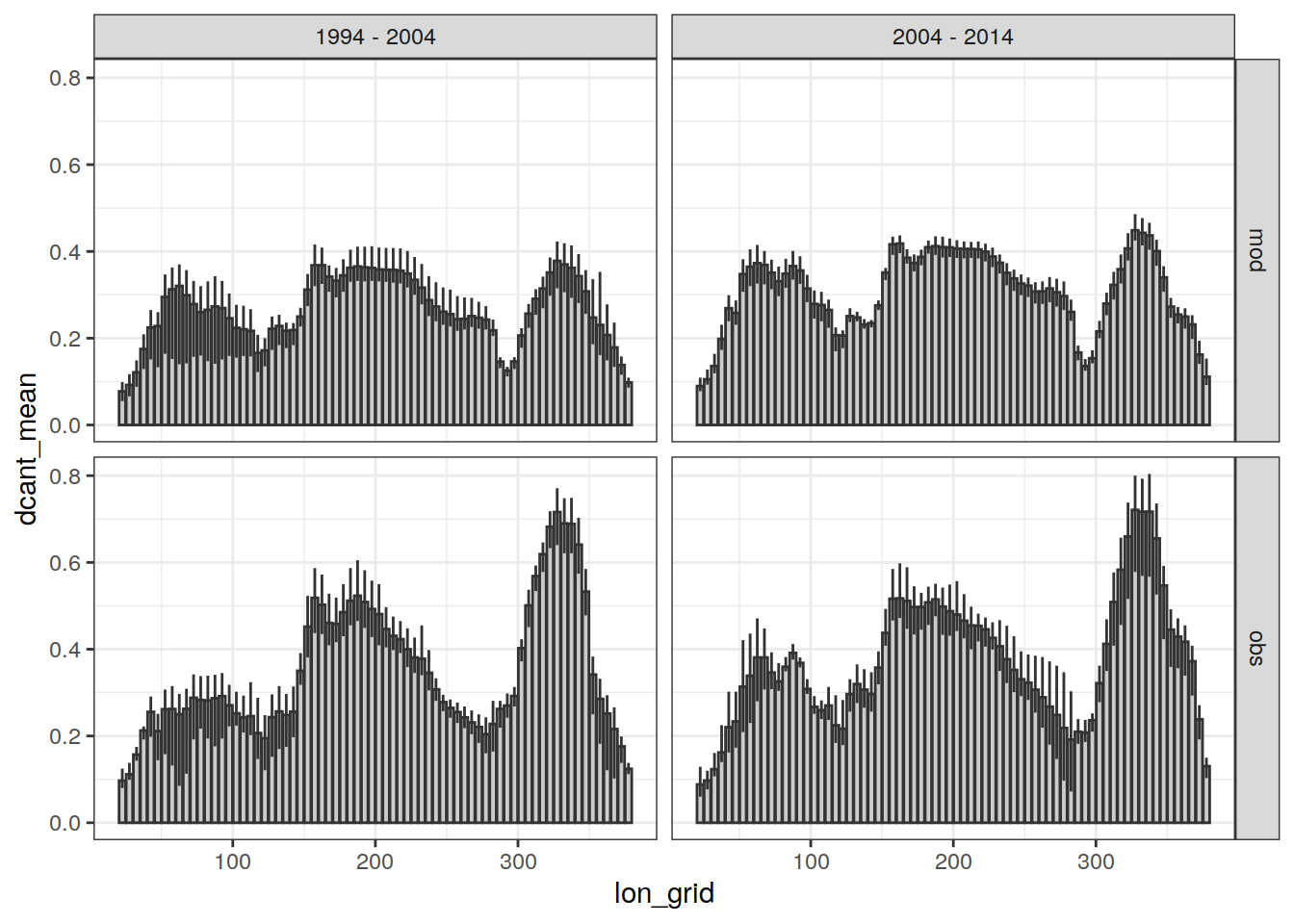
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 7ae4bdd | jens-daniel-mueller | 2021-10-25 |
| a8576a8 | jens-daniel-mueller | 2021-10-25 |
4.5 Standard deviation
p_map_cant_inv(
df = dcant_inv_ensemble,
var = "dcant_sd",
breaks = c(seq(0, 4, 0.4), Inf),
subtitle_text = paste("Ensemble SD")
) +
facet_grid(period ~ data_source)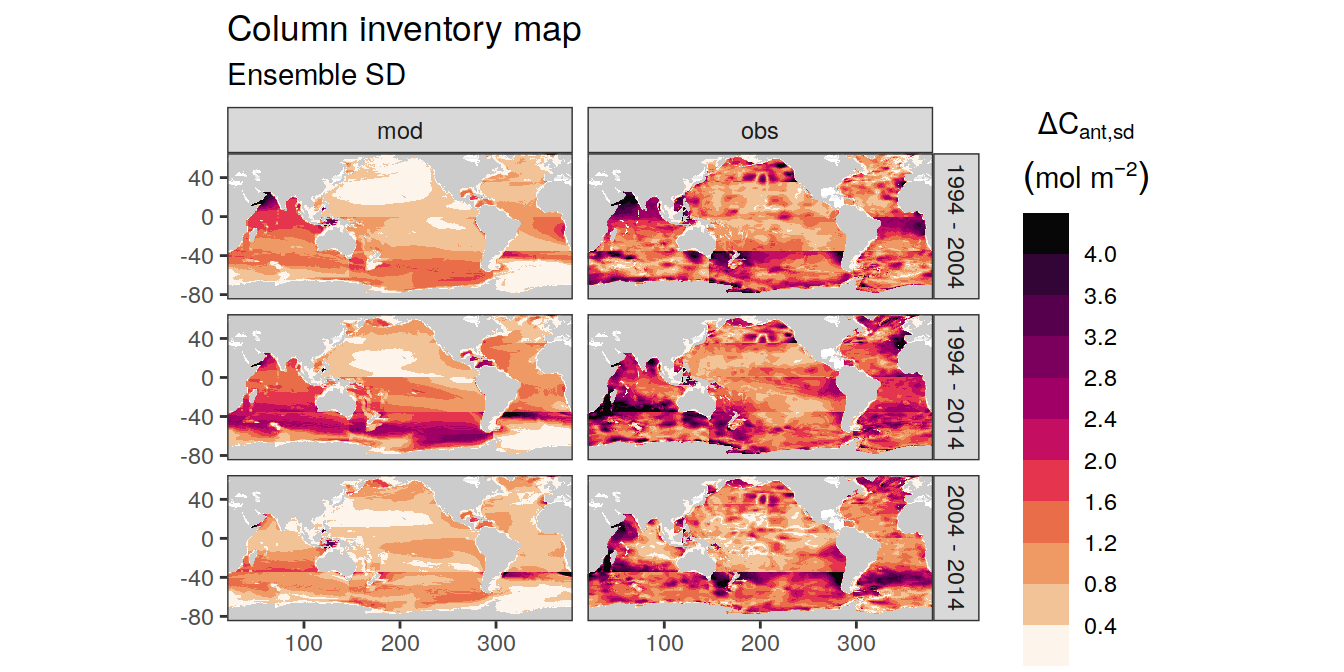
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| c225f90 | jens-daniel-mueller | 2021-09-30 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| aa8bd1f | jens-daniel-mueller | 2021-09-28 |
| d0dbf3e | jens-daniel-mueller | 2021-09-27 |
| 09002a7 | jens-daniel-mueller | 2021-09-26 |
| 0c6be7e | jens-daniel-mueller | 2021-09-24 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 31c33cb | jens-daniel-mueller | 2021-09-23 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
4.6 SD as uncertainty
uncertainty_grid <- dcant_inv_ensemble %>%
filter(dcant_sd > sd_uncertainty_limit) %>%
distinct(lon, lat, data_source, period)
uncertainty_grid <- m_grid_horizontal_coarse(uncertainty_grid) %>%
distinct(lon_grid, lat_grid, data_source, period)
map +
geom_point(data =
uncertainty_grid,
aes(lon_grid, lat_grid),
shape = 3) +
facet_grid(period ~ data_source)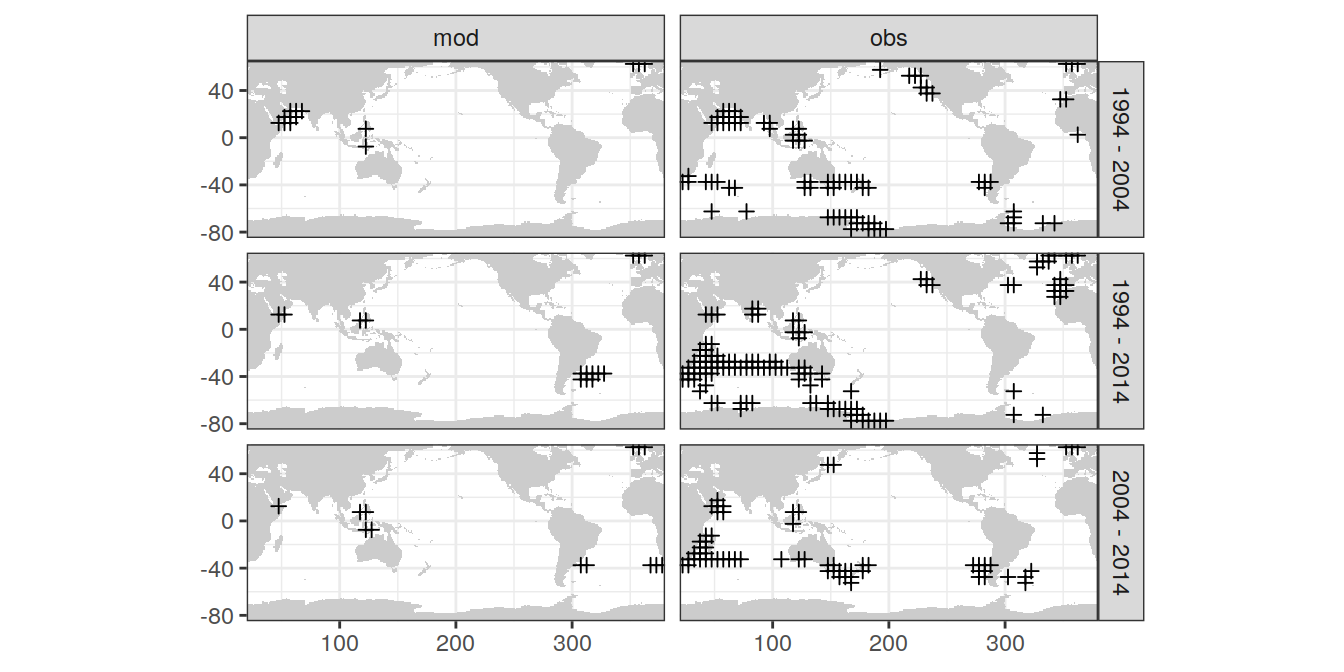
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
4.7 SD vs abs bias
4.7.1 2D bin
dcant_inv_ensemble_bias %>%
ggplot(aes(abs(dcant_mean_bias), dcant_sd)) +
geom_bin2d() +
scale_fill_viridis_c() +
facet_grid(. ~ period)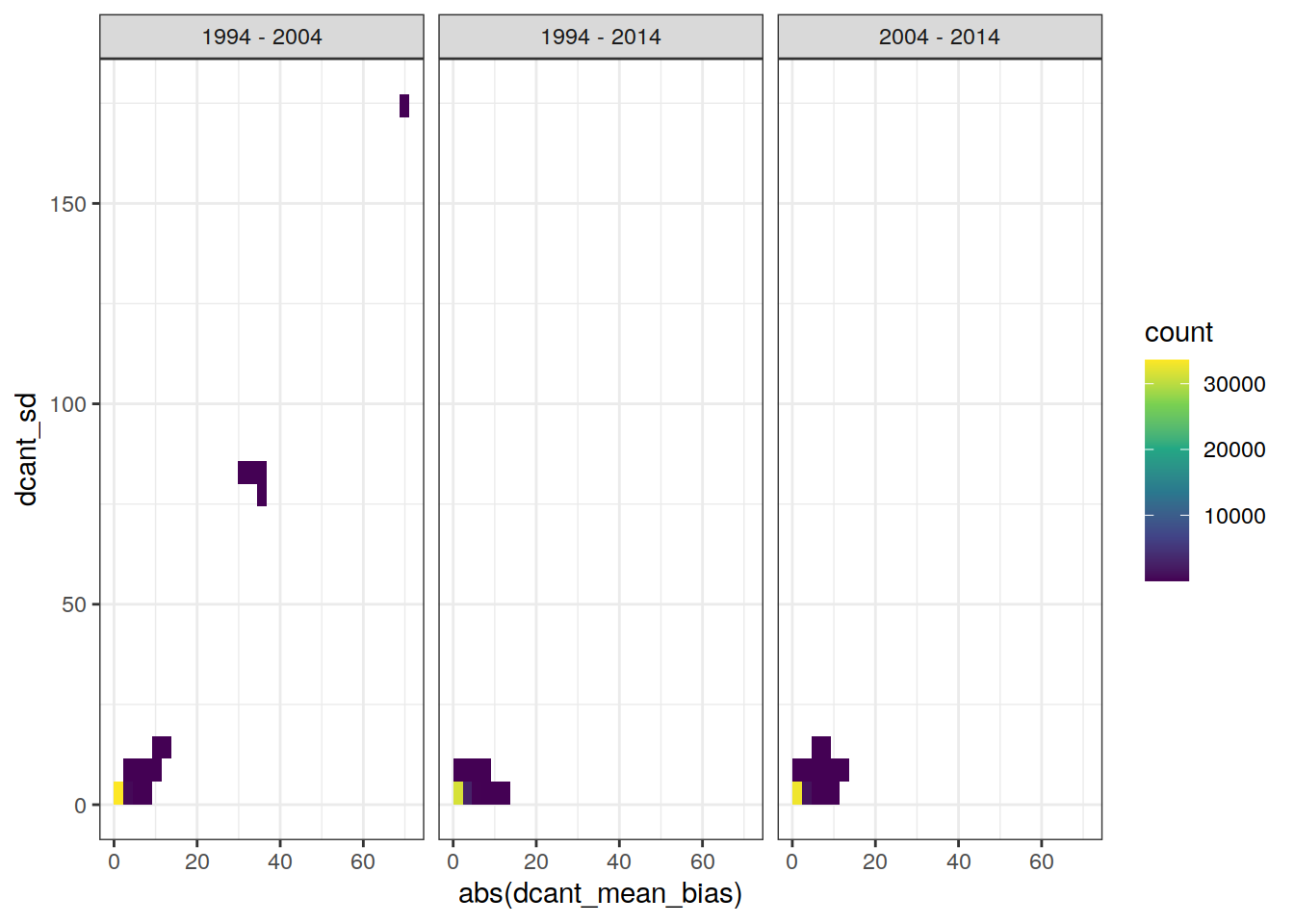
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| c225f90 | jens-daniel-mueller | 2021-09-30 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| aa8bd1f | jens-daniel-mueller | 2021-09-28 |
| d0dbf3e | jens-daniel-mueller | 2021-09-27 |
| 09002a7 | jens-daniel-mueller | 2021-09-26 |
| 0c6be7e | jens-daniel-mueller | 2021-09-24 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 31c33cb | jens-daniel-mueller | 2021-09-23 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
4.7.2 Density distributions
legend_title = expression(Delta * C[ant]~(mol~m^2))
dcant_density_distribution_bias <- dcant_inv_ensemble_bias %>%
select(dcant_mean_bias, period) %>%
pivot_longer(dcant_mean_bias,
names_to = "estimate",
values_to = "value") %>%
mutate(data_source = "mod")
dcant_density_distribution <- dcant_inv_ensemble %>%
select(dcant_mean, dcant_sd, period, data_source) %>%
pivot_longer(dcant_mean:dcant_sd,
names_to = "estimate",
values_to = "value")
dcant_density_distribution <- bind_rows(dcant_density_distribution,
dcant_density_distribution_bias) %>%
mutate(period = factor(period, c("1994 - 2004", "2004 - 2014", "1994 - 2014")))
p_ensemble_distribution <-
dcant_density_distribution %>%
filter(period != "1994 - 2014") %>%
mutate(
data_source = recode(data_source,
mod = "Model",
obs = "Observations"),
estimate = recode(
estimate,
dcant_mean = "Mean",
dcant_mean_bias = "Bias",
dcant_sd = "SD"
)
) %>%
ggplot(aes(value, fill = estimate, col = estimate)) +
scale_color_brewer(palette = "Dark2") +
scale_fill_brewer(palette = "Dark2") +
geom_density(alpha = 0.2) +
facet_grid(period ~ data_source) +
labs(title = "Density distributions of column inventory ensemble properties",
x = legend_title,
y = "Density of 1x1° grid cells") +
coord_cartesian(ylim = c(0, 0.5),
xlim = c(-5, 32))
# ggsave(plot = p_ensemble_distribution,
# path = "output/other",
# filename = "inv_ensemble_distribution.png",
# height = 4,
# width = 8)
p_ensemble_distribution <-
dcant_density_distribution %>%
filter(data_source == "obs") %>%
mutate(
data_source = recode(data_source,
mod = "Model",
obs = "Observations"),
estimate = recode(
estimate,
dcant_mean = "Mean",
dcant_mean_bias = "Bias",
dcant_sd = "SD"
)
) %>%
ggplot(aes(value, fill = estimate, col = estimate)) +
scale_color_brewer(palette = "Dark2") +
scale_fill_brewer(palette = "Dark2") +
geom_density(alpha = 0.2) +
facet_grid(period ~ data_source) +
labs(title = "Density distributions of column inventory ensemble properties",
x = legend_title,
y = "Density of 1x1° grid cells") +
coord_cartesian(ylim = c(0, 0.5),
xlim = c(-5, 32))
p_ensemble_distribution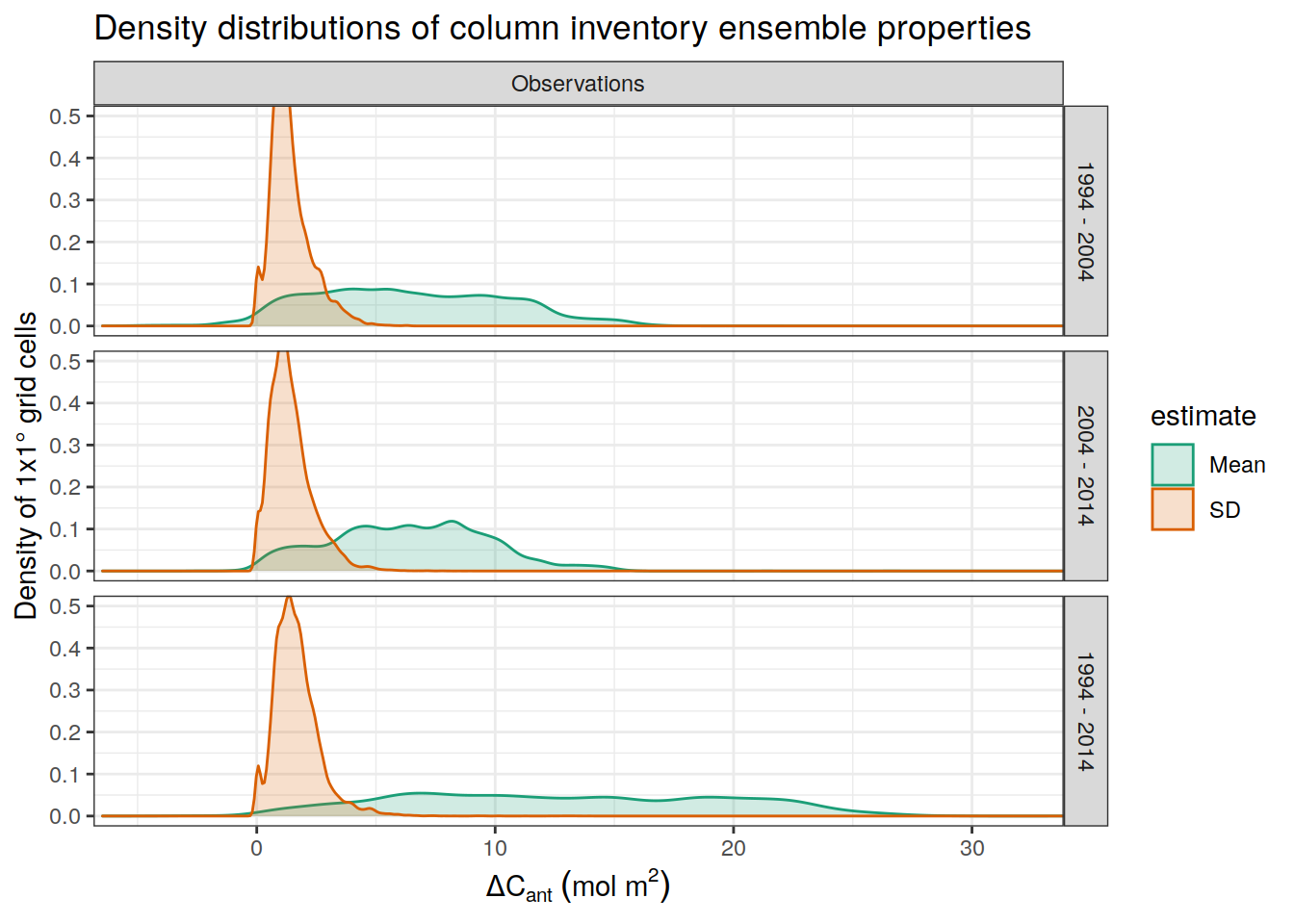
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
# ggsave(plot = p_ensemble_distribution,
# path = "output/other",
# filename = "inv_ensemble_distribution_all.png",
# height = 6,
# width = 5)4.8 Range
p_map_cant_inv(
df = dcant_inv_ensemble,
var = "dcant_range",
breaks = c(seq(0,8,0.8), Inf),
subtitle_text = paste("Ensemble range")
) +
facet_grid(period ~ data_source)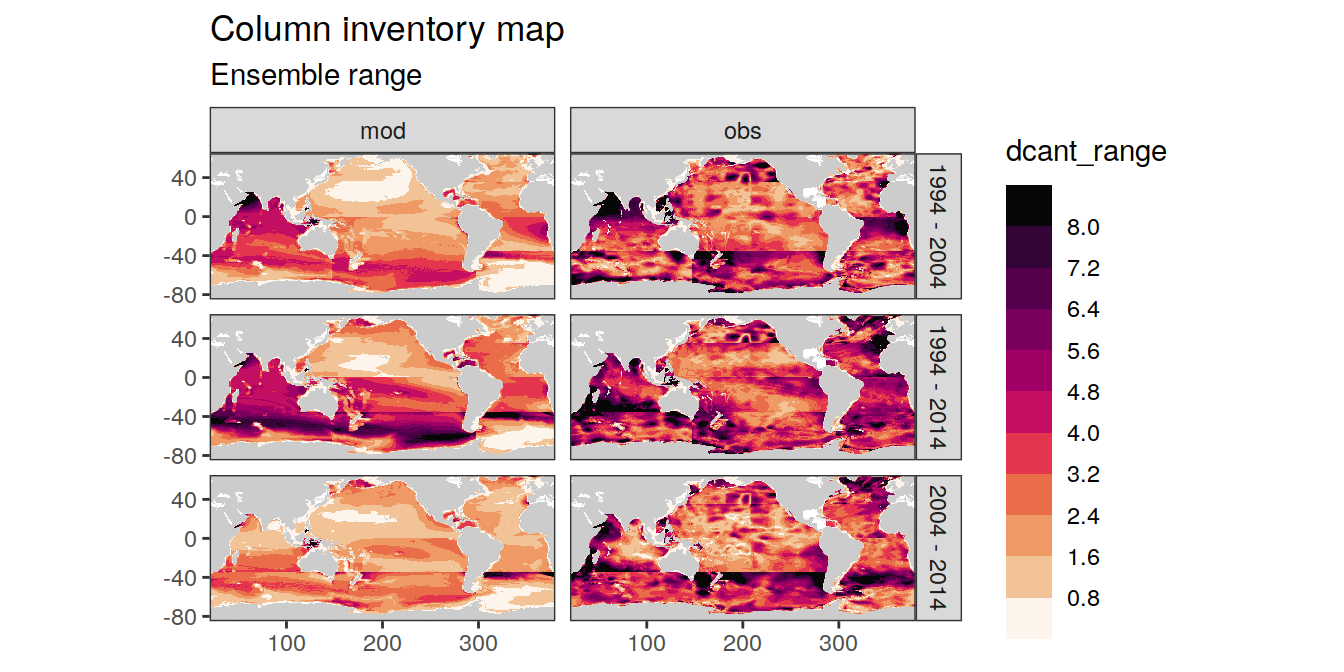
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| c225f90 | jens-daniel-mueller | 2021-09-30 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| aa8bd1f | jens-daniel-mueller | 2021-09-28 |
| d0dbf3e | jens-daniel-mueller | 2021-09-27 |
| 09002a7 | jens-daniel-mueller | 2021-09-26 |
| 0c6be7e | jens-daniel-mueller | 2021-09-24 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 31c33cb | jens-daniel-mueller | 2021-09-23 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
4.9 Composed figure
uncertainty_grid <- uncertainty_grid %>%
filter(data_source == "obs")
p_map_94 <- p_map_cant_inv(
df = dcant_inv_ensemble %>%
filter(period == "1994 - 2004",
data_source == "obs"),
var = "dcant_mean",
subtitle_text = NULL,
title_text = NULL
) +
geom_point(
data =
uncertainty_grid %>% filter(period == "1994 - 2004"),
aes(lon_grid, lat_grid),
shape = 3,
col = "red"
) +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
legend.position = "left"
) +
facet_grid(period ~ .,
switch = "y")
p_map_04 <- p_map_cant_inv(
df = dcant_inv_ensemble %>%
filter(period == "2004 - 2014",
data_source == "obs"),
var = "dcant_mean",
subtitle_text = NULL,
title_text = NULL
) +
geom_point(
data =
uncertainty_grid %>% filter(period == "2004 - 2014"),
aes(lon_grid, lat_grid),
shape = 3,
col = "red"
) +
theme(
axis.text = element_blank(),
axis.ticks = element_blank(),
legend.position = "left"
) +
facet_grid(period ~ .,
switch = "y")
p_lon_94 <- dcant_lon_grid_ensemble %>%
filter(data_source == "obs",
period == "1994 - 2004") %>%
ggplot(aes(lon_grid, dcant_mean)) +
geom_col(position = "dodge",
fill = "grey80",
col = "grey20") +
geom_errorbar(aes(ymin = dcant_min,
ymax = dcant_max),
col = "grey20",
width = 0) +
scale_y_continuous(
limits = c(0, 1),
expand = c(0, 0),
name = expression(Delta * C[ant] ~ (PgC)),
position = "right"
) +
scale_x_continuous(name = "Longitude (°E)",
limits = c(20, 380),
expand = c(0, 0)) +
theme(
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank()
)
# coord_fixed(ratio = 100)
p_lon_04 <- dcant_lon_grid_ensemble %>%
filter(data_source == "obs",
period == "2004 - 2014") %>%
ggplot(aes(lon_grid, dcant_mean)) +
geom_col(position = "dodge",
fill = "grey80",
col = "grey20") +
geom_errorbar(aes(
ymin = dcant_min,
ymax = dcant_max
),
col = "grey20",
width = 0) +
scale_y_continuous(limits = c(0,1), expand = c(0,0),
name = expression(Delta*C[ant]~(PgC)),
position = "right") +
scale_x_continuous(name = "Longitude (°E)",
limits = c(20,380), expand = c(0,0)) +
theme(title = element_blank())
# coord_fixed(ratio = 100)
p_lat_94 <- dcant_lat_grid_ensemble %>%
filter(data_source == "obs",
period == "1994 - 2004") %>%
ggplot(aes(dcant_mean, lat_grid)) +
geom_col(
position = "dodge",
fill = "grey80",
col = "grey20",
orientation = "y"
) +
geom_errorbar(aes(xmin = dcant_min,
xmax = dcant_max),
col = "grey20",
width = 0) +
scale_x_continuous(
limits = c(0, 2.8),
expand = c(0, 0),
name = expression(Delta * C[ant] ~ (PgC))
) +
scale_y_continuous(
name = "Latitude (°N)",
limits = c(-80, 65),
expand = c(0, 0),
position = "right"
) +
theme(
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank()
)
# coord_fixed(ratio = 5e-2)
p_lat_04 <- dcant_lat_grid_ensemble %>%
filter(data_source == "obs",
period == "2004 - 2014") %>%
ggplot(aes(dcant_mean, lat_grid)) +
geom_col(position = "dodge",
fill = "grey80",
col = "grey20",
orientation = "y") +
geom_errorbar(aes(xmin = dcant_min,
xmax = dcant_max),
col = "grey20",
width = 0) +
scale_x_continuous(
limits = c(0, 2.8),
expand = c(0, 0),
name = expression(Delta * C[ant] ~ (PgC))
) +
scale_y_continuous(name = "Latitude (°N)",
limits = c(-80, 65),
expand = c(0, 0),
position = "right")
t_94 <- grid::textGrob("1994 -\n2004")
t_04 <- grid::textGrob("2004 -\n2014")
layout <- "
BBB#
AAAD
AAAD
CCCE
CCCE
GGG#
"
wrap_plots(B = p_lon_94, A = p_map_94, D = p_lat_94,
G = p_lon_04, C = p_map_04, E = p_lat_04,
design = layout,
guides = "collect")&
theme(legend.position = "left")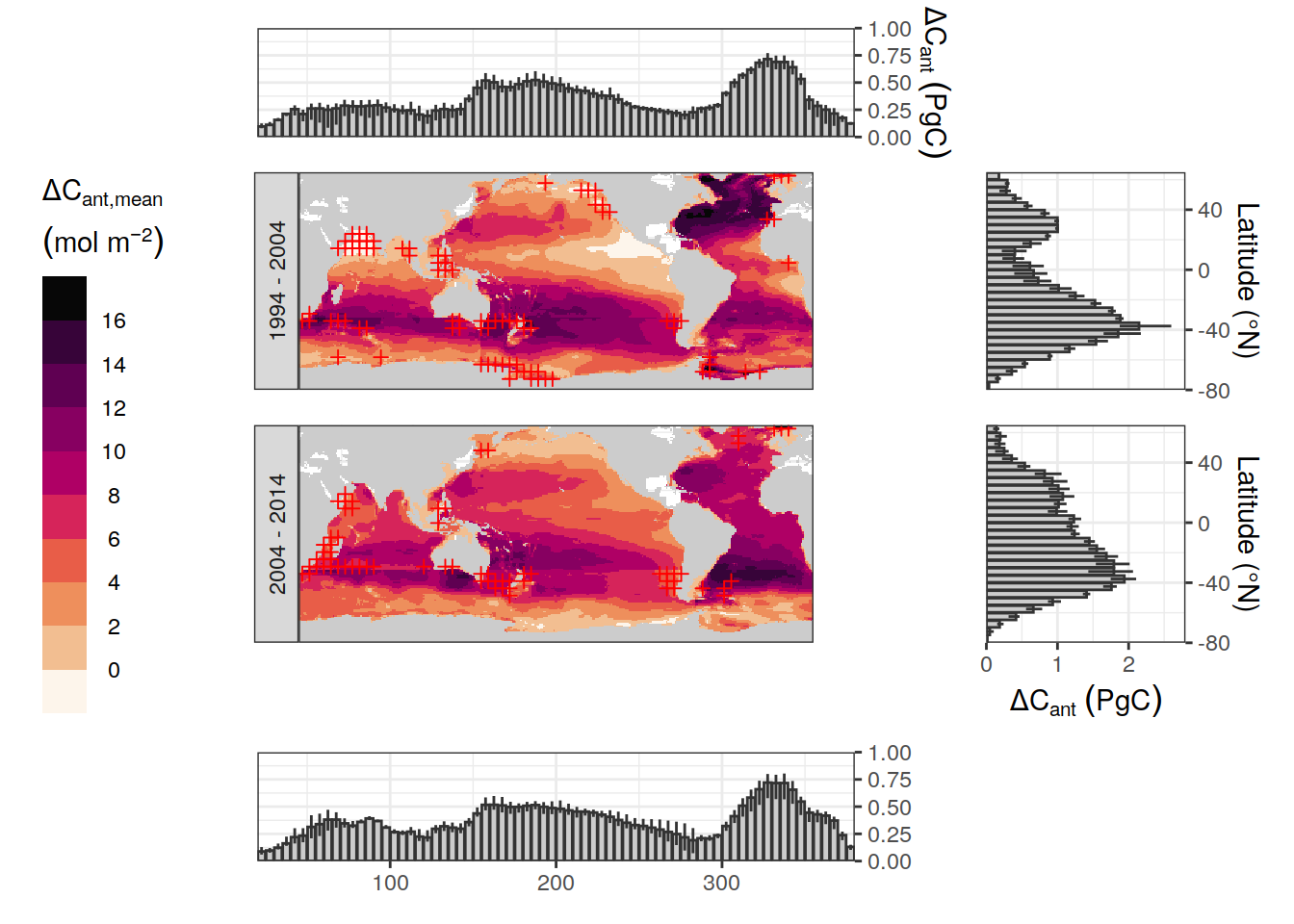
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 059b2bf | jens-daniel-mueller | 2021-10-26 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| e753cfc | jens-daniel-mueller | 2021-10-25 |
| 7ae4bdd | jens-daniel-mueller | 2021-10-25 |
# ggsave("output/publication/Fig_column_inventories.png",
# width=11,
# height=9.27)5 Cases vs ensemble
5.1 Offset from mean
dcant_inv_all <- full_join(dcant_inv_all,
dcant_inv_ensemble)Joining, by = c("data_source", "lon", "lat", "period")dcant_inv_all <- dcant_inv_all %>%
mutate(dcant_offset = dcant - dcant_mean)
dcant_inv_all %>%
filter(data_source %in% c("mod", "obs")) %>%
group_by(period) %>%
group_split() %>%
# head(1) %>%
map(
~ p_map_cant_inv(df = .x,
var = "dcant_offset",
col = "bias",
subtitle_text = paste("period:",
unique(.x$period))) +
facet_grid(MLR_basins ~ data_source)
)[[1]]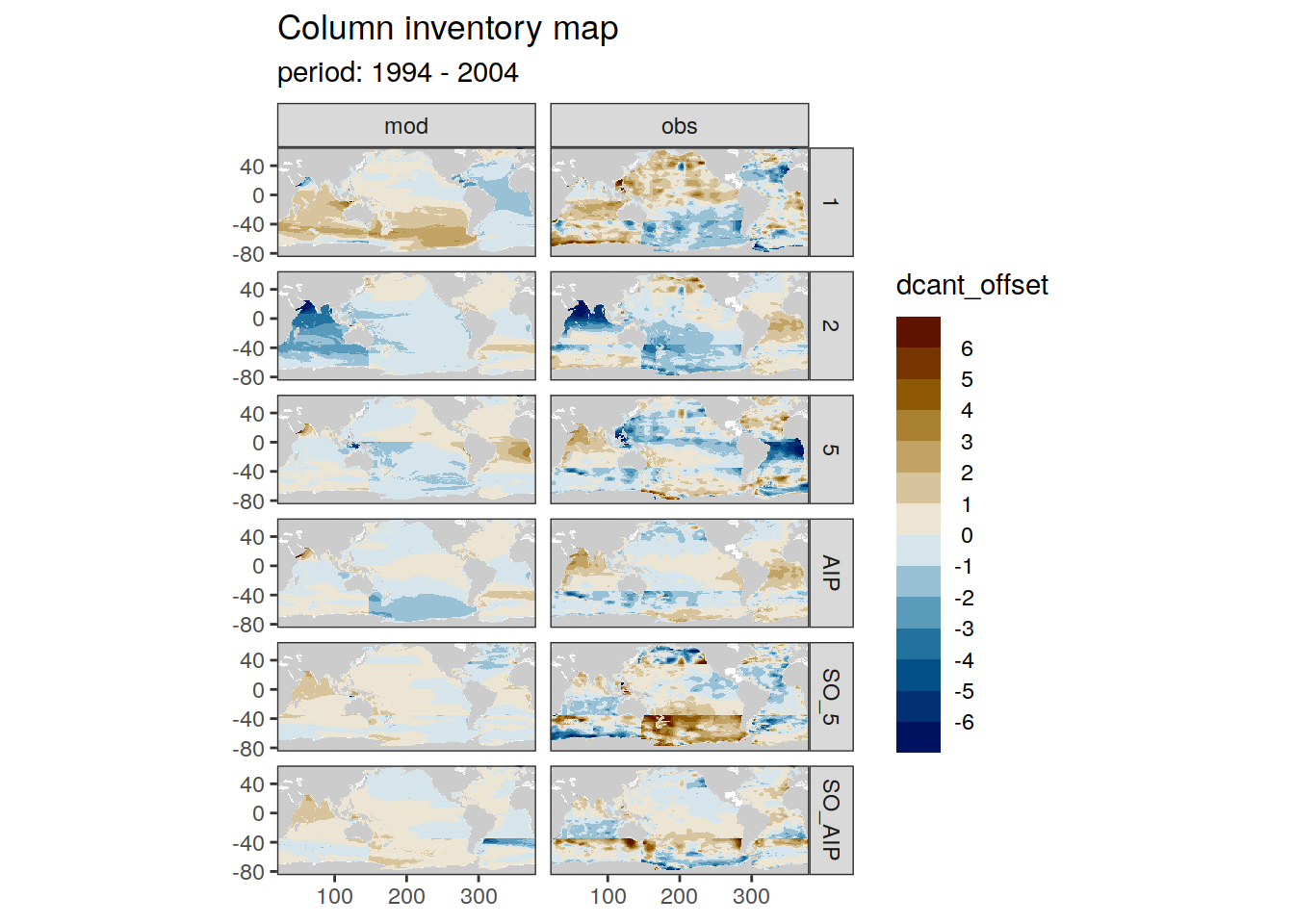
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| aa8bd1f | jens-daniel-mueller | 2021-09-28 |
| d0dbf3e | jens-daniel-mueller | 2021-09-27 |
| 09002a7 | jens-daniel-mueller | 2021-09-26 |
| 0c6be7e | jens-daniel-mueller | 2021-09-24 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
[[2]]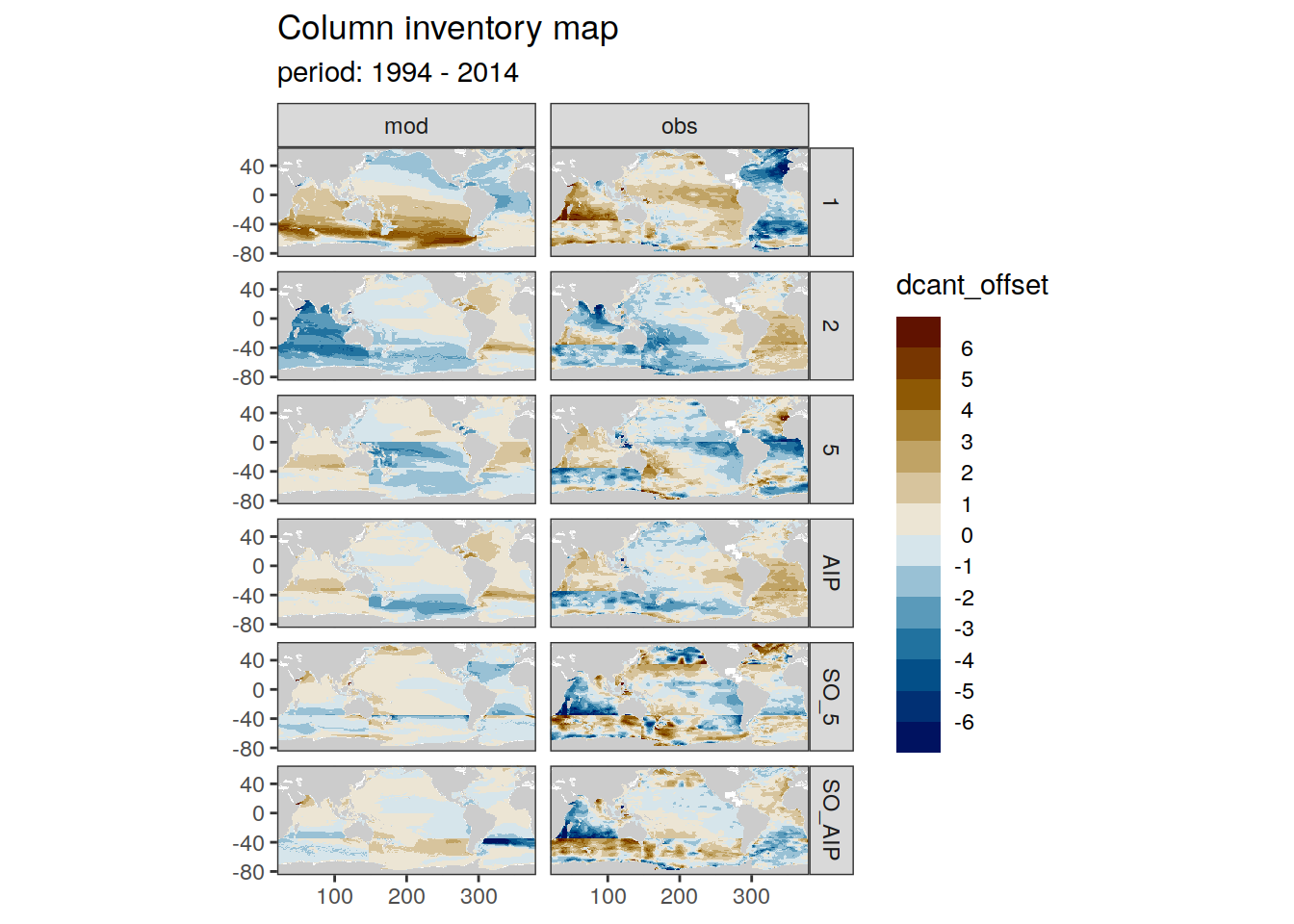
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| c225f90 | jens-daniel-mueller | 2021-09-30 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| d0dbf3e | jens-daniel-mueller | 2021-09-27 |
| 09002a7 | jens-daniel-mueller | 2021-09-26 |
| 0c6be7e | jens-daniel-mueller | 2021-09-24 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 31c33cb | jens-daniel-mueller | 2021-09-23 |
| 35ad8b5 | jens-daniel-mueller | 2021-09-20 |
[[3]]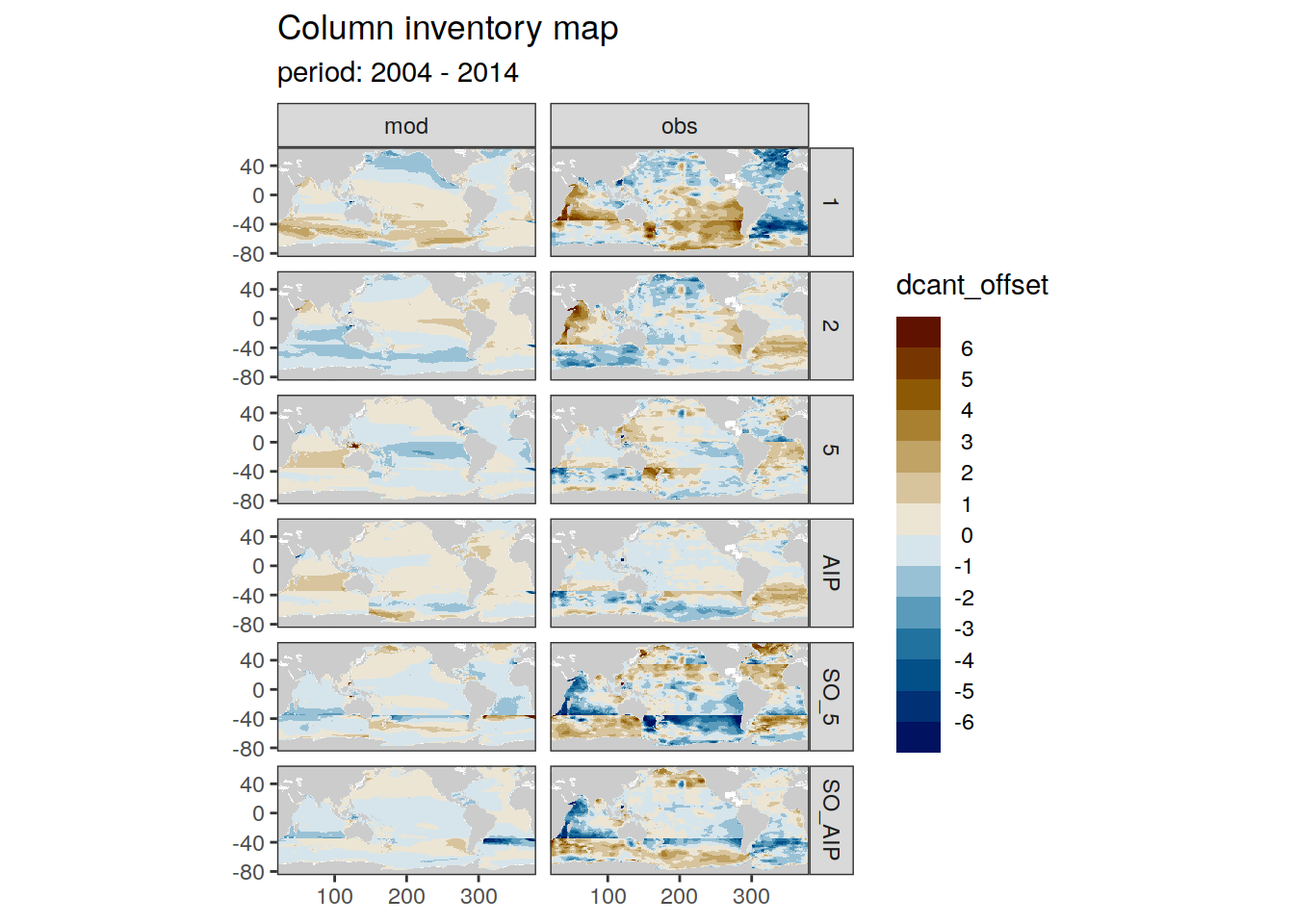
| Version | Author | Date |
|---|---|---|
| ae5f326 | jens-daniel-mueller | 2022-04-08 |
| 2839d72 | jens-daniel-mueller | 2022-03-15 |
| c3a6238 | jens-daniel-mueller | 2022-03-08 |
| 6c2e89a | jens-daniel-mueller | 2022-01-27 |
| 9753eb8 | jens-daniel-mueller | 2022-01-26 |
| 2c8371d | jens-daniel-mueller | 2022-01-17 |
| 570e738 | jens-daniel-mueller | 2022-01-10 |
| ccc57bf | jens-daniel-mueller | 2022-01-10 |
| 33f190b | jens-daniel-mueller | 2022-01-09 |
| 88c47ca | jens-daniel-mueller | 2022-01-07 |
| 296acd8 | jens-daniel-mueller | 2022-01-07 |
| 030836b | jens-daniel-mueller | 2022-01-06 |
| bc2c182 | jens-daniel-mueller | 2021-12-23 |
| e55f868 | jens-daniel-mueller | 2021-12-23 |
| db09d70 | jens-daniel-mueller | 2021-12-10 |
| e534f51 | jens-daniel-mueller | 2021-11-02 |
| a162b7e | jens-daniel-mueller | 2021-10-28 |
| 8b4e334 | jens-daniel-mueller | 2021-10-26 |
| 8c9fa17 | jens-daniel-mueller | 2021-10-22 |
| 968fe94 | jens-daniel-mueller | 2021-10-18 |
| c225f90 | jens-daniel-mueller | 2021-09-30 |
| f51dcb9 | jens-daniel-mueller | 2021-09-28 |
| 55d2059 | jens-daniel-mueller | 2021-09-28 |
| d0dbf3e | jens-daniel-mueller | 2021-09-27 |
| 09002a7 | jens-daniel-mueller | 2021-09-26 |
| 0c6be7e | jens-daniel-mueller | 2021-09-24 |
| f54d5db | jens-daniel-mueller | 2021-09-24 |
| 31c33cb | jens-daniel-mueller | 2021-09-23 |
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.3
Matrix products: default
BLAS: /usr/local/R-4.1.2/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.1.2/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] colorspace_2.0-2 marelac_2.1.10 shape_1.4.6 ggforce_0.3.3
[5] metR_0.11.0 scico_1.3.0 patchwork_1.1.1 collapse_1.7.0
[9] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7 purrr_0.3.4
[13] readr_2.1.1 tidyr_1.1.4 tibble_3.1.6 ggplot2_3.3.5
[17] tidyverse_1.3.1 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] fs_1.5.2 bit64_4.0.5 gsw_1.0-6 lubridate_1.8.0
[5] RColorBrewer_1.1-2 httr_1.4.2 rprojroot_2.0.2 tools_4.1.2
[9] backports_1.4.1 bslib_0.3.1 utf8_1.2.2 R6_2.5.1
[13] DBI_1.1.2 withr_2.4.3 tidyselect_1.1.1 processx_3.5.2
[17] bit_4.0.4 compiler_4.1.2 git2r_0.29.0 cli_3.1.1
[21] rvest_1.0.2 xml2_1.3.3 labeling_0.4.2 sass_0.4.0
[25] scales_1.1.1 checkmate_2.0.0 SolveSAPHE_2.1.0 callr_3.7.0
[29] digest_0.6.29 rmarkdown_2.11 oce_1.5-0 pkgconfig_2.0.3
[33] htmltools_0.5.2 highr_0.9 dbplyr_2.1.1 fastmap_1.1.0
[37] rlang_0.4.12 readxl_1.3.1 rstudioapi_0.13 jquerylib_0.1.4
[41] generics_0.1.1 farver_2.1.0 jsonlite_1.7.3 vroom_1.5.7
[45] magrittr_2.0.1 Rcpp_1.0.8 munsell_0.5.0 fansi_1.0.2
[49] lifecycle_1.0.1 stringi_1.7.6 whisker_0.4 yaml_2.2.1
[53] MASS_7.3-55 grid_4.1.2 parallel_4.1.2 promises_1.2.0.1
[57] crayon_1.4.2 haven_2.4.3 hms_1.1.1 seacarb_3.3.0
[61] knitr_1.37 ps_1.6.0 pillar_1.6.4 reprex_2.0.1
[65] glue_1.6.0 evaluate_0.14 getPass_0.2-2 data.table_1.14.2
[69] modelr_0.1.8 vctrs_0.3.8 tzdb_0.2.0 tweenr_1.0.2
[73] httpuv_1.6.5 cellranger_1.1.0 gtable_0.3.0 polyclip_1.10-0
[77] assertthat_0.2.1 xfun_0.29 broom_0.7.11 later_1.3.0
[81] viridisLite_0.4.0 ellipsis_0.3.2 here_1.0.1