GLODAPv2_2016: Mapped Climatologies
Jens Daniel Müller
15 December, 2020
Last updated: 2020-12-15
Checks: 7 0
Knit directory: emlr_obs_preprocessing/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200707) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 53a6197. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/
Unstaged changes:
Modified: code/Workflowr_project_managment.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/read_GLODAPv2_2016_MappedClimatologies.Rmd) and HTML (docs/read_GLODAPv2_2016_MappedClimatologies.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | f049b99 | jens-daniel-mueller | 2020-12-15 | Build site. |
| Rmd | f58a773 | jens-daniel-mueller | 2020-12-15 | code and output cleaning |
| html | 2fdd2a7 | jens-daniel-mueller | 2020-12-14 | Build site. |
| Rmd | 0a3cc05 | jens-daniel-mueller | 2020-12-14 | rebuild with new root folder and basinmask |
| html | 474daac | jens-daniel-mueller | 2020-12-12 | Build site. |
| Rmd | 53d8356 | jens-daniel-mueller | 2020-12-12 | removed basinmask issues |
| html | 5c773fa | jens-daniel-mueller | 2020-12-11 | Build site. |
| html | 999cd9d | jens-daniel-mueller | 2020-12-02 | Build site. |
| html | e28cc90 | jens-daniel-mueller | 2020-11-30 | Build site. |
| Rmd | cd676e8 | jens-daniel-mueller | 2020-11-30 | created global parameterization file params_global.rds |
| html | 825309e | jens-daniel-mueller | 2020-11-27 | Build site. |
| html | 58359ac | jens-daniel-mueller | 2020-11-27 | Build site. |
| Rmd | 2f37595 | jens-daniel-mueller | 2020-11-27 | first rebuild after splitting the preprocessing part |
| Rmd | 92e10aa | Jens Müller | 2020-11-27 | Initial commit |
| html | 92e10aa | Jens Müller | 2020-11-27 | Initial commit |
1 Read source files
Data source: Globally mapped climatologies from Lauvset et al. (2016) downloaded in June 2020 from glodap.info.
Following files were used:
file_list <- c(
paste(path_glodapv2_2016b, "GLODAPv2.2016b.Cant.nc", sep = ""),
paste(path_glodapv2_2016b, "GLODAPv2.2016b.NO3.nc", sep = ""),
paste(path_glodapv2_2016b, "GLODAPv2.2016b.oxygen.nc", sep = ""),
paste(path_glodapv2_2016b, "GLODAPv2.2016b.PO4.nc", sep = ""),
paste(path_glodapv2_2016b, "GLODAPv2.2016b.salinity.nc", sep = ""),
paste(path_glodapv2_2016b, "GLODAPv2.2016b.silicate.nc", sep = ""),
paste(path_glodapv2_2016b, "GLODAPv2.2016b.TAlk.nc", sep = ""),
paste(path_glodapv2_2016b, "GLODAPv2.2016b.TCO2.nc", sep = ""),
paste(path_glodapv2_2016b, "GLODAPv2.2016b.temperature.nc", sep = "")
)
print(file_list)[1] "/nfs/kryo/work/updata/glodapv2.2016b/GLODAPv2.2016b.Cant.nc"
[2] "/nfs/kryo/work/updata/glodapv2.2016b/GLODAPv2.2016b.NO3.nc"
[3] "/nfs/kryo/work/updata/glodapv2.2016b/GLODAPv2.2016b.oxygen.nc"
[4] "/nfs/kryo/work/updata/glodapv2.2016b/GLODAPv2.2016b.PO4.nc"
[5] "/nfs/kryo/work/updata/glodapv2.2016b/GLODAPv2.2016b.salinity.nc"
[6] "/nfs/kryo/work/updata/glodapv2.2016b/GLODAPv2.2016b.silicate.nc"
[7] "/nfs/kryo/work/updata/glodapv2.2016b/GLODAPv2.2016b.TAlk.nc"
[8] "/nfs/kryo/work/updata/glodapv2.2016b/GLODAPv2.2016b.TCO2.nc"
[9] "/nfs/kryo/work/updata/glodapv2.2016b/GLODAPv2.2016b.temperature.nc"# use only three basin to assign general basin mask
# ie this is not specific to the MLR fitting
basinmask <- basinmask %>%
filter(MLR_basins == "2") %>%
select(lat, lon, basin_AIP)2 Plot data and write csv
Below, subsets of the climatologies are plotted. For all relevant parameters, the plots show:
- maps at depth levels
- concentration along global section
The global section path is indicated as white line in maps.
Please note that NA values in the climatologies were filled with neighbouring values on the longitudinal axis.
# file <- file_list[2]
for (file in file_list) {
# open file
clim <- read_stars(file,
quiet = TRUE)
# extract parameter name
parameter <- str_split(file, pattern = "16b/GLODAPv2.2016b.", simplify = TRUE)[2]
parameter <- str_split(parameter, pattern = ".nc", simplify = TRUE)[1]
print(parameter)
# extract parameter
clim <- clim %>% select(all_of(parameter))
#convert to table
clim_tibble <- clim %>%
as_tibble()
# harmonize column names
clim_tibble <- clim_tibble %>%
rename(lat = y,
lon = x,
depth = depth_surface)
# join with basin mask and remove data outside basin mask
clim_tibble <- inner_join(clim_tibble, basinmask)
# determine bottom depth
bottom_depth <- clim_tibble %>%
filter(!is.na(!!sym(parameter))) %>%
group_by(lon, lat) %>%
summarise(bottom_depth = max(depth)) %>%
ungroup()
# remove data below bottom depth
clim_tibble <- left_join(clim_tibble, bottom_depth)
rm(bottom_depth)
clim_tibble <- clim_tibble %>%
filter(depth <= bottom_depth) %>%
select(-bottom_depth)
# fill NAs with closest value along longitude
clim_tibble <- clim_tibble %>%
group_by(lat, depth, basin_AIP) %>%
arrange(lon) %>%
fill(!!sym(parameter), .direction = "downup") %>%
ungroup()
# # plot column NA inventory
#
# clim_tibble_grid <- clim_tibble %>%
# filter(!is.na(!!sym(parameter))) %>%
# group_by(lat, lon) %>%
# summarise(depth_max = max(depth)) %>%
# ungroup()
#
# clim_tibble_grid <- full_join(clim_tibble, clim_tibble_grid)
#
# clim_tibble_grid <- clim_tibble_grid %>%
# filter(depth <= depth_max) %>%
# select(-depth_max)
#
# clim_tibble_grid <- clim_tibble_grid %>%
# filter(is.na(!!sym(parameter)),
# depth <= params_global$inventory_depth) %>%
# count(lat, lon)
#
# # plot NA map
# print(
#
# map +
# geom_raster(data = clim_tibble_grid,
# aes(lon, lat, fill = n)) +
# scale_fill_viridis_c() +
# theme(axis.title = element_blank()) +
# labs(title = paste(parameter, "colum NA"))
#
# )
#
# rm(clim_tibble_grid)
# remove NAs
clim_tibble <- clim_tibble %>%
drop_na()
# plot maps
print(
p_map_climatology(
df = clim_tibble,
var = parameter)
)
# plot sections
print(
p_section_global(
df = clim_tibble,
var = parameter,
title_text = "GLODAPv2_2016_Mapped_Climatology")
)
# write csv file
clim_tibble %>%
write_csv(paste(
path_preprocessing,
paste("GLODAPv2_2016_MappedClimatology_",
parameter,
".csv",
sep = ""),
sep = ""
))
}[1] "Cant"

[1] "NO3"

[1] "oxygen"

[1] "PO4"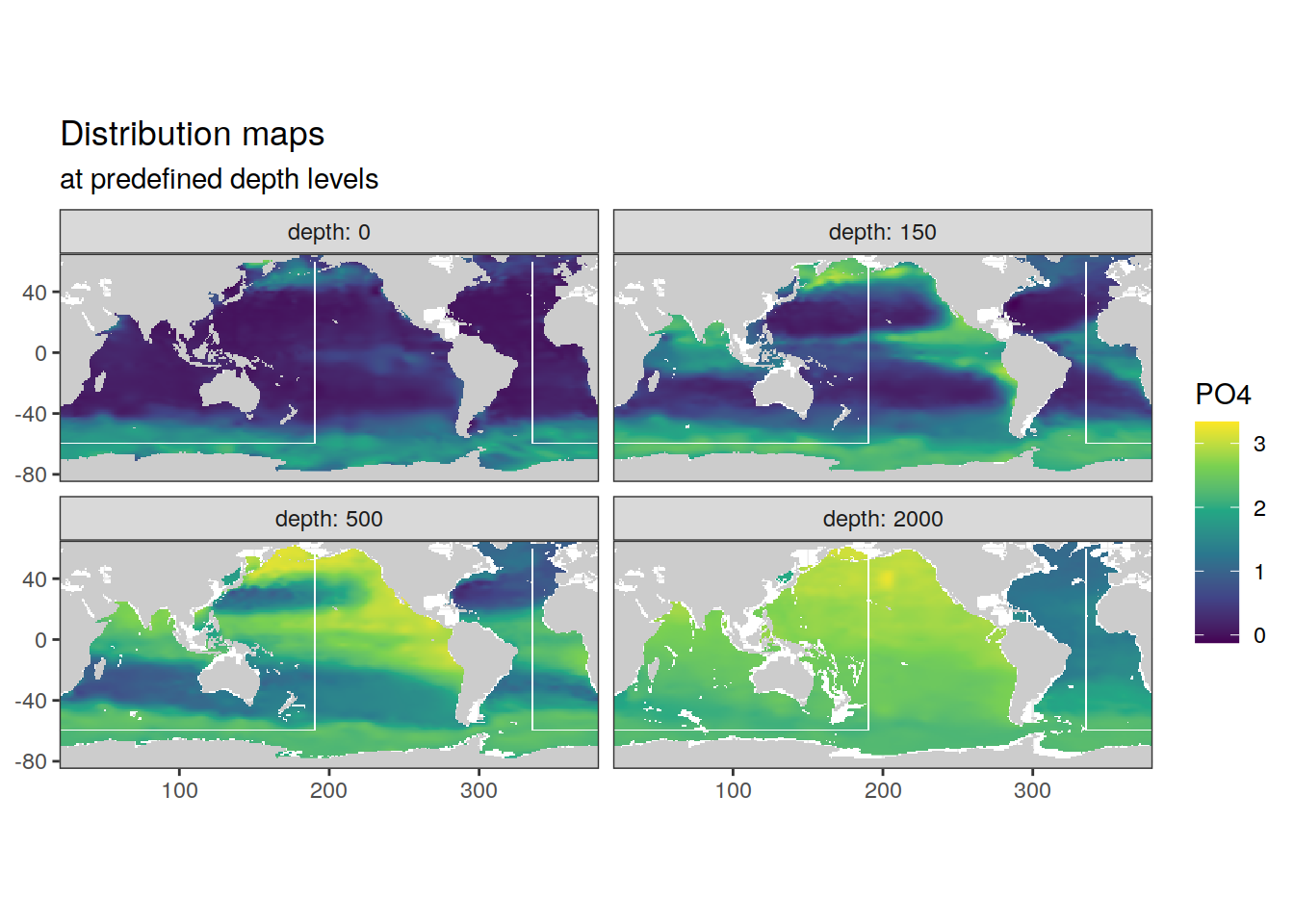

[1] "salinity"

[1] "silicate"

[1] "TAlk"
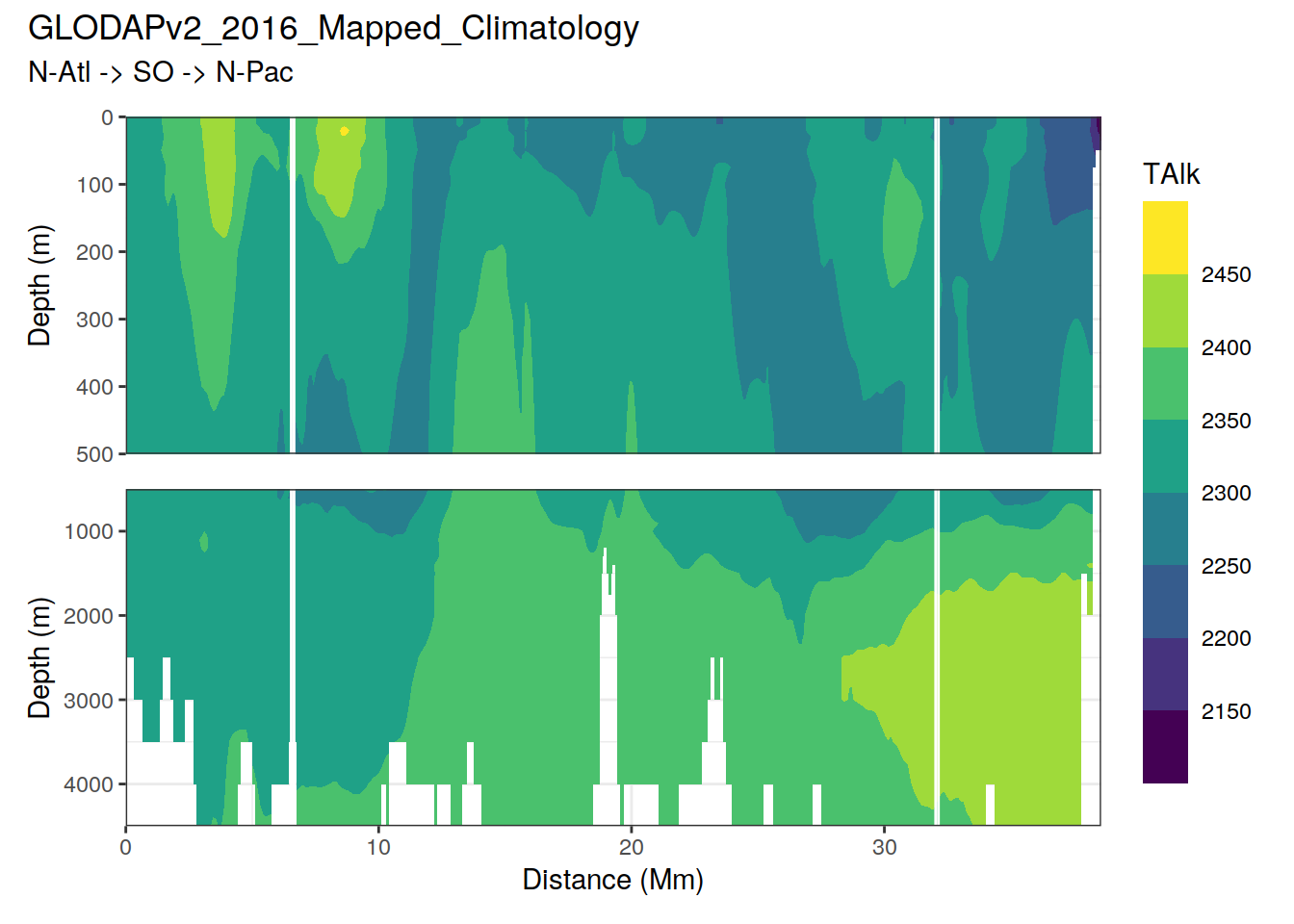
[1] "TCO2"

[1] "temperature"
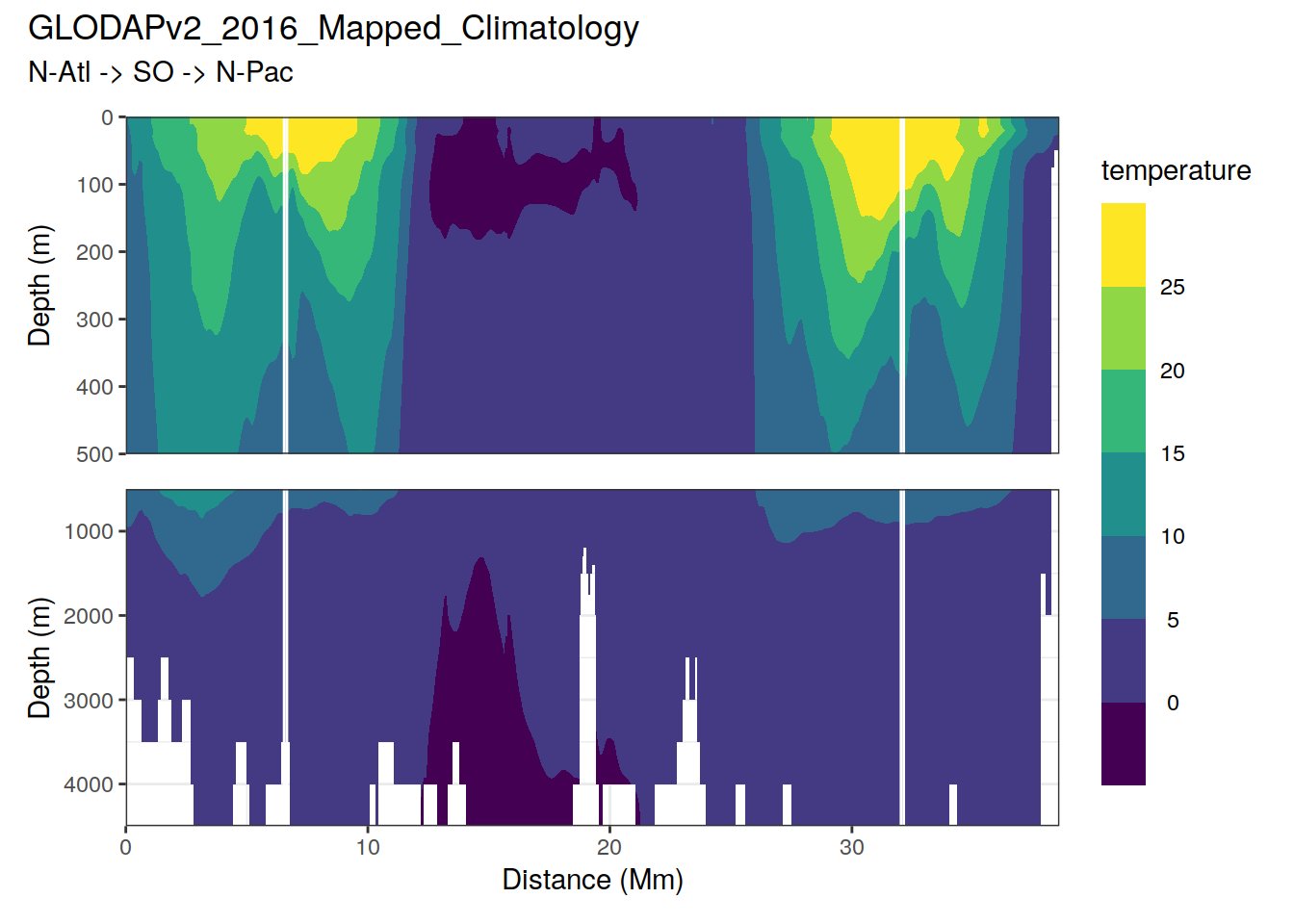
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.1
Matrix products: default
BLAS: /usr/local/R-4.0.3/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.0.3/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] stars_0.4-3 sf_0.9-6 abind_1.4-5 metR_0.9.0
[5] scico_1.2.0 patchwork_1.1.0 collapse_1.4.2 forcats_0.5.0
[9] stringr_1.4.0 dplyr_1.0.2 purrr_0.3.4 readr_1.4.0
[13] tidyr_1.1.2 tibble_3.0.4 ggplot2_3.3.2 tidyverse_1.3.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.2 viridisLite_0.3.0 jsonlite_1.7.1
[4] modelr_0.1.8 assertthat_0.2.1 blob_1.2.1
[7] cellranger_1.1.0 yaml_2.2.1 pillar_1.4.7
[10] backports_1.1.10 lattice_0.20-41 glue_1.4.2
[13] RcppEigen_0.3.3.7.0 digest_0.6.27 promises_1.1.1
[16] checkmate_2.0.0 rvest_0.3.6 colorspace_2.0-0
[19] htmltools_0.5.0 httpuv_1.5.4 Matrix_1.2-18
[22] pkgconfig_2.0.3 broom_0.7.2 haven_2.3.1
[25] scales_1.1.1 whisker_0.4 later_1.1.0.1
[28] git2r_0.27.1 farver_2.0.3 generics_0.0.2
[31] ellipsis_0.3.1 withr_2.3.0 cli_2.2.0
[34] magrittr_2.0.1 crayon_1.3.4 readxl_1.3.1
[37] evaluate_0.14 fs_1.5.0 fansi_0.4.1
[40] lwgeom_0.2-5 xml2_1.3.2 RcppArmadillo_0.10.1.2.0
[43] class_7.3-17 tools_4.0.3 data.table_1.13.2
[46] hms_0.5.3 lifecycle_0.2.0 munsell_0.5.0
[49] reprex_0.3.0 isoband_0.2.2 compiler_4.0.3
[52] e1071_1.7-4 rlang_0.4.9 units_0.6-7
[55] classInt_0.4-3 grid_4.0.3 rstudioapi_0.13
[58] labeling_0.4.2 rmarkdown_2.5 gtable_0.3.0
[61] DBI_1.1.0 R6_2.5.0 lubridate_1.7.9
[64] knitr_1.30 rprojroot_2.0.2 KernSmooth_2.23-18
[67] stringi_1.5.3 parallel_4.0.3 Rcpp_1.0.5
[70] vctrs_0.3.5 dbplyr_1.4.4 tidyselect_1.1.0
[73] xfun_0.18