Last updated: 2020-12-02
Checks:
Knit directory: emlr_obs_v_XXX/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200707) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 9eb4281 . See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Unstaged changes:
Modified: README.md
Modified: analysis/_site.yml
Deleted: analysis/config_nomenclature.Rmd
Modified: code/Workflowr_project_managment.R
Modified: data/auxillary/params_local.rds
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/analysis_this_study.Rmd) and HTML (docs/analysis_this_study.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
File
Version
Author
Date
Message
html
9eb4281
jens-daniel-mueller
2020-12-02
Build site.
html
478f74d
jens-daniel-mueller
2020-12-02
Build site.
html
902f65a
jens-daniel-mueller
2020-12-02
Build site.
html
ee9bf6d
jens-daniel-mueller
2020-12-01
Build site.
Rmd
e2f207c
jens-daniel-mueller
2020-12-01
corrected slab definition for plot
html
22d0127
jens-daniel-mueller
2020-12-01
Build site.
html
0ff728b
jens-daniel-mueller
2020-12-01
Build site.
html
92edddb
jens-daniel-mueller
2020-12-01
Build site.
Rmd
4843275
jens-daniel-mueller
2020-12-01
auto eras naming
html
cf19652
jens-daniel-mueller
2020-11-30
Build site.
html
196be51
jens-daniel-mueller
2020-11-30
Build site.
Rmd
7a4b015
jens-daniel-mueller
2020-11-30
first rebuild on ETH server
Rmd
bc61ce3
Jens Müller
2020-11-30
Initial commit
html
bc61ce3
Jens Müller
2020-11-30
Initial commit
path_functions <- "/nfs/kryo/work/updata/emlr_cant/utilities/functions/"
path_files <- "/nfs/kryo/work/updata/emlr_cant/utilities/files/"path_preprocessing <-
"/nfs/kryo/work/updata/emlr_cant/observations/preprocessing/"
path_version_data <-
paste(
"/nfs/kryo/work/updata/emlr_cant/observations/",
params_local$Version_ID,
"/data/",
sep = ""
)
path_version_figures <-
paste(
"/nfs/kryo/work/updata/emlr_cant/observations/",
params_local$Version_ID,
"/figures/",
sep = ""
)
Libraries
Loading libraries specific to the the analysis performed in this section.
library(scales)
library(marelac)
library(kableExtra)
Data sources
cant estimates from this study:
Mean and SD per grid cell (lat, lon, depth)
Zonal mean and SD (basin, lat, depth)
Inventories (lat, lon)
cant_3d <-
read_csv(paste(path_version_data,
"cant_3d.csv",
sep = ""))
cant_zonal <-
read_csv(paste(path_version_data,
"cant_zonal.csv",
sep = ""))
cant_predictor_zonal <-
read_csv(paste(path_version_data,
"cant_predictor_zonal.csv",
sep = ""))
cant_inv <-
read_csv(paste(path_version_data,
"cant_inv.csv",
sep = ""))C* estimates from this study:
Mean and SD per grid cell (lat, lon, depth)
Zonal mean and SD (basin, lat, depth)
cstar_3d <-
read_csv(paste(path_version_data,
"cstar_3d.csv",
sep = ""))
cstar_zonal <-
read_csv(paste(path_version_data,
"cstar_zonal.csv",
sep = ""))Cleaned GLODAPv2_2020 file as used in this study
GLODAP <-
read_csv(paste(
path_version_data,
"GLODAPv2.2020_MLR_fitting_ready.csv",
sep = ""
))
Cant budgets
Global Cant inventories were estimated in units of Pg C. Please note that here we only added positive Cant values in the upper 3000m and do not apply additional corrections for areas not covered.
cant_inv_budget <- cant_inv %>%
mutate(surface_area = earth_surf(lat, lon),
cant_inv_grid = cant_inv*surface_area) %>%
group_by(eras, basin_AIP) %>%
summarise(cant_total = sum(cant_inv_grid)*12*1e-15,
cant_total = round(cant_total,1)) %>%
ungroup() %>%
arrange(desc(eras)) %>%
pivot_wider(values_from = cant_total, names_from = basin_AIP) %>%
mutate(total = Atlantic + Indian + Pacific)
cant_inv_budget %>%
kableExtra::kable() %>%
add_header_above() %>%
kable_styling(full_width = FALSE)
eras
Atlantic
Indian
Pacific
total
(1999,2012] –> (2012,Inf]
7.3
3.8
14.7
25.8
(1981,1999] –> (1999,2012]
10.9
10.1
17.4
38.4
rm(cant_inv_budget)
Cant - positive
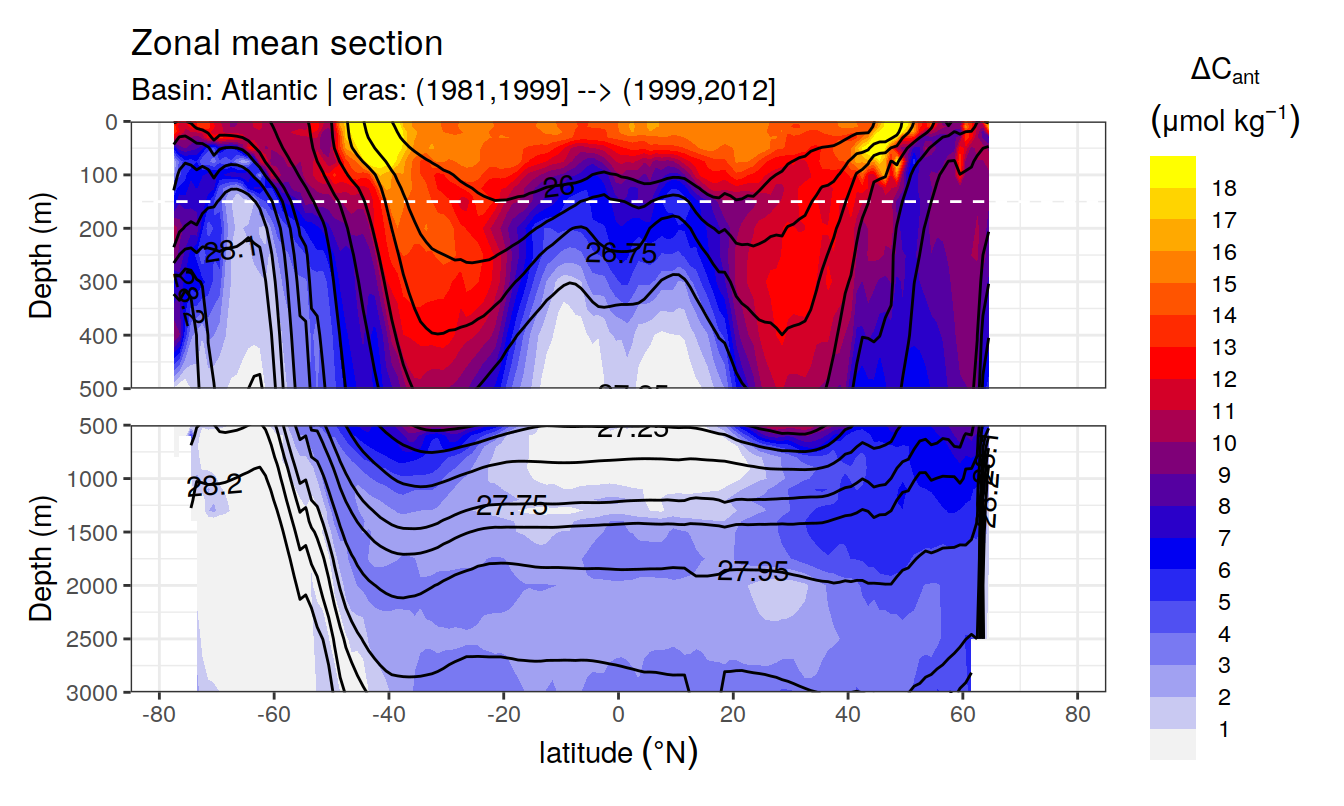
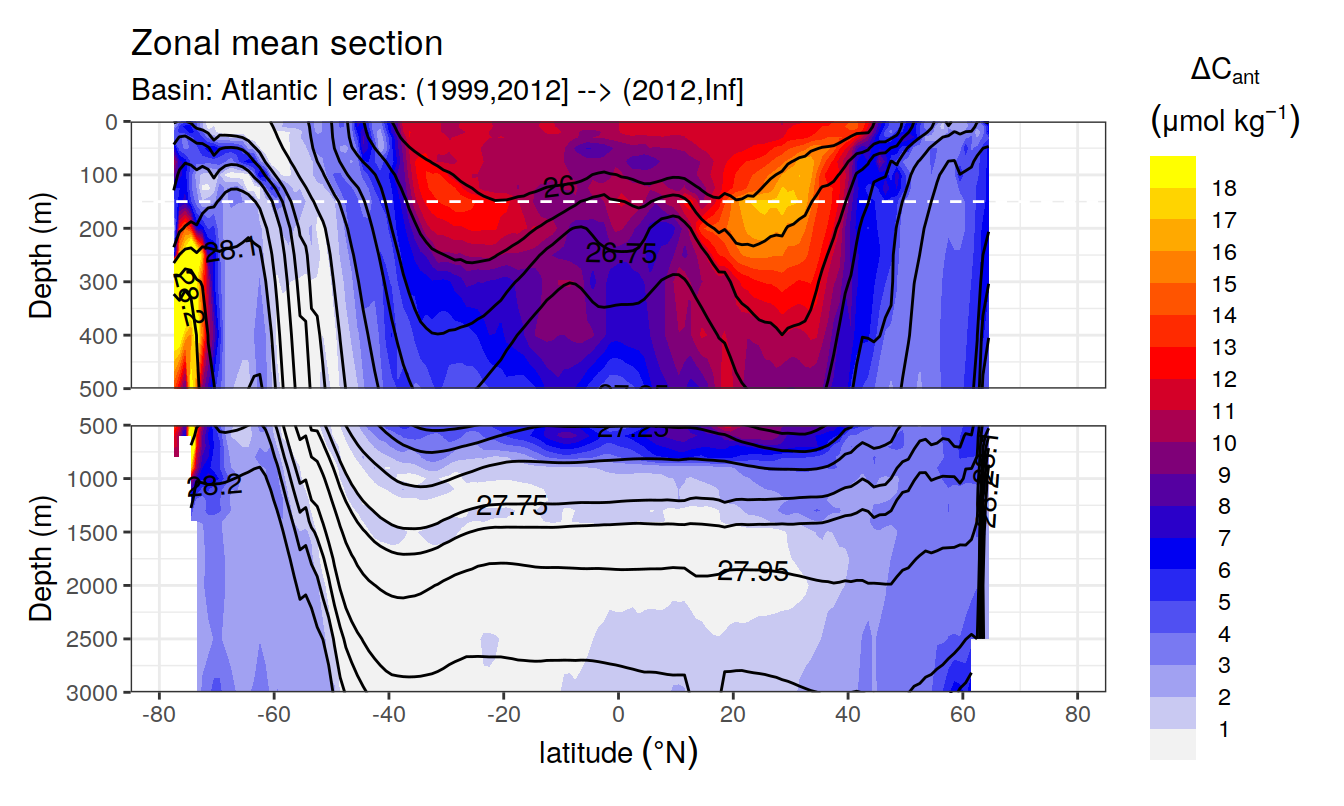
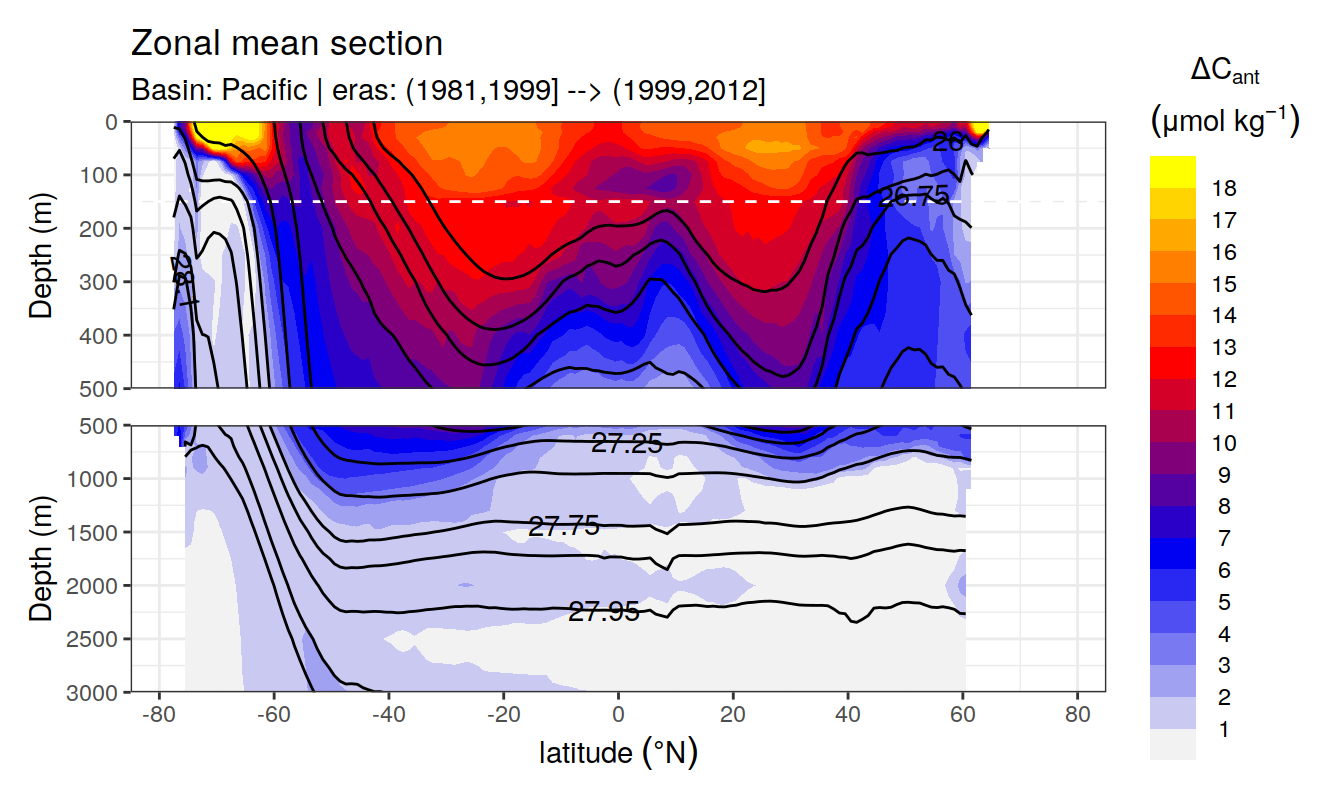
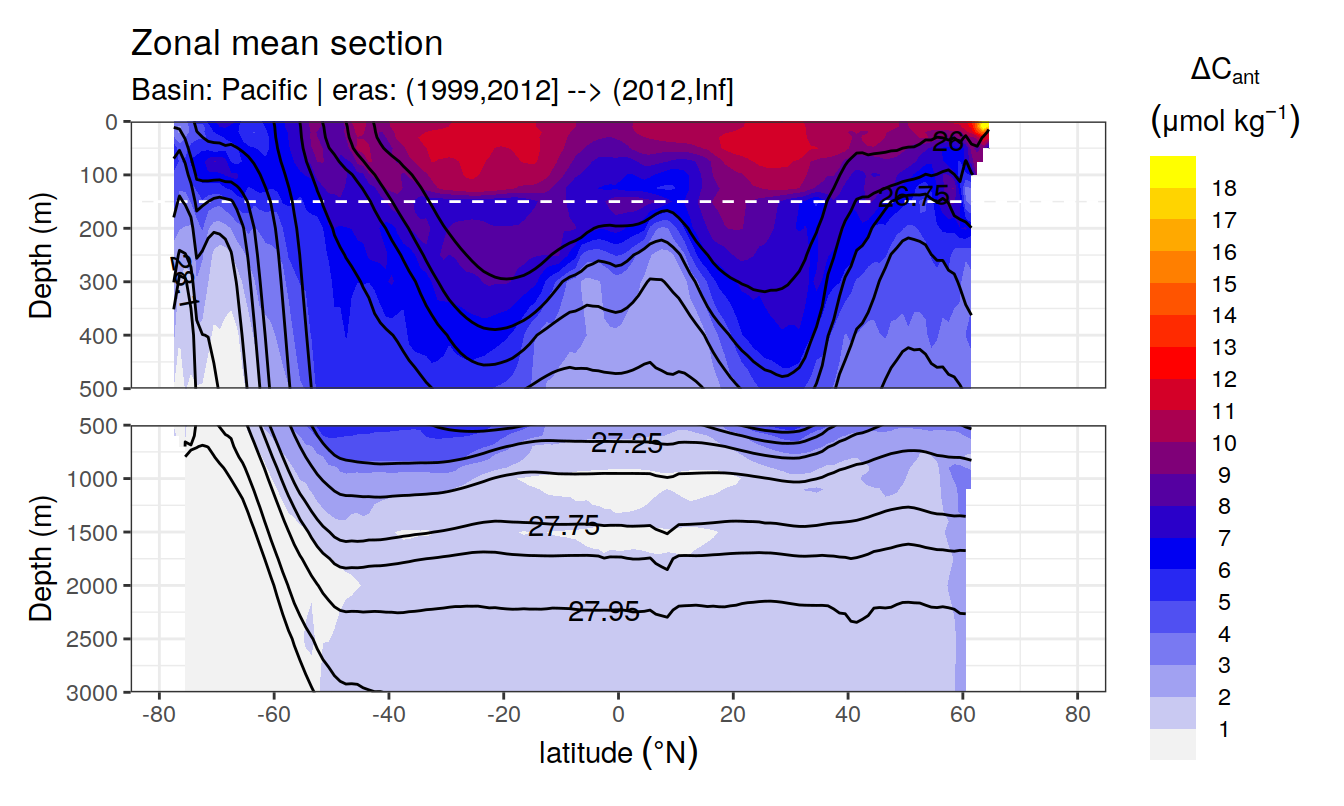
In a first series of plots we explore the distribution of cant, taking only positive estimates into account (positive here refers to the mean cant estimate across 10 eMLR model predictions available for each grid cell). Negative values were set to zero before calculating mean sections and inventories.
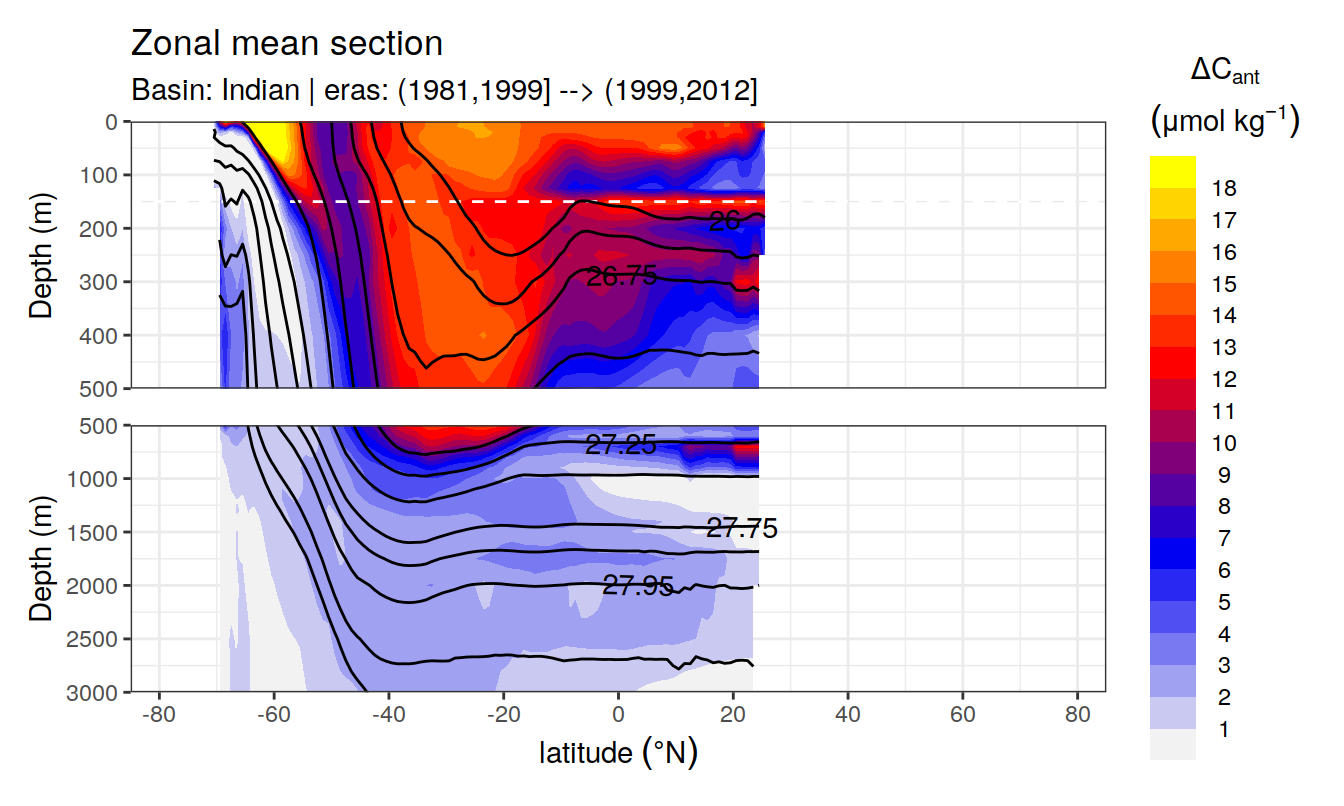
Zonal mean sections
# i_basin_AIP <- unique(cant_zonal$basin_AIP)[2]
# i_eras <- unique(cant_zonal$eras)[1]
for (i_basin_AIP in unique(cant_zonal$basin_AIP)) {
for (i_eras in unique(cant_zonal$eras)) {
print(
p_section_zonal(
df = cant_zonal %>%
filter(basin_AIP == i_basin_AIP,
eras == i_eras),
var = "cant_pos_mean",
subtitle_text =
paste("Basin:", i_basin_AIP, "| eras:", i_eras))
)
}
}
Past versions of cant_pos_zonal_mean_sections-1.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_zonal_mean_sections-2.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_zonal_mean_sections-3.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_zonal_mean_sections-4.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_zonal_mean_sections-5.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_zonal_mean_sections-6.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
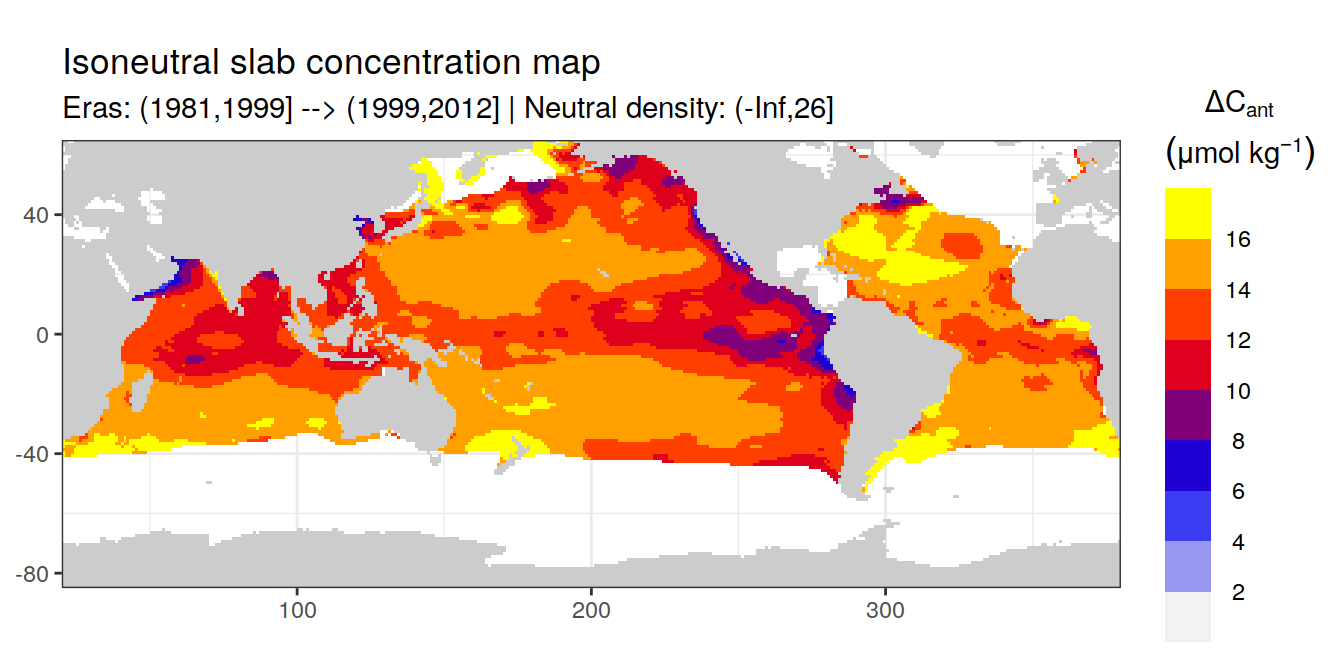
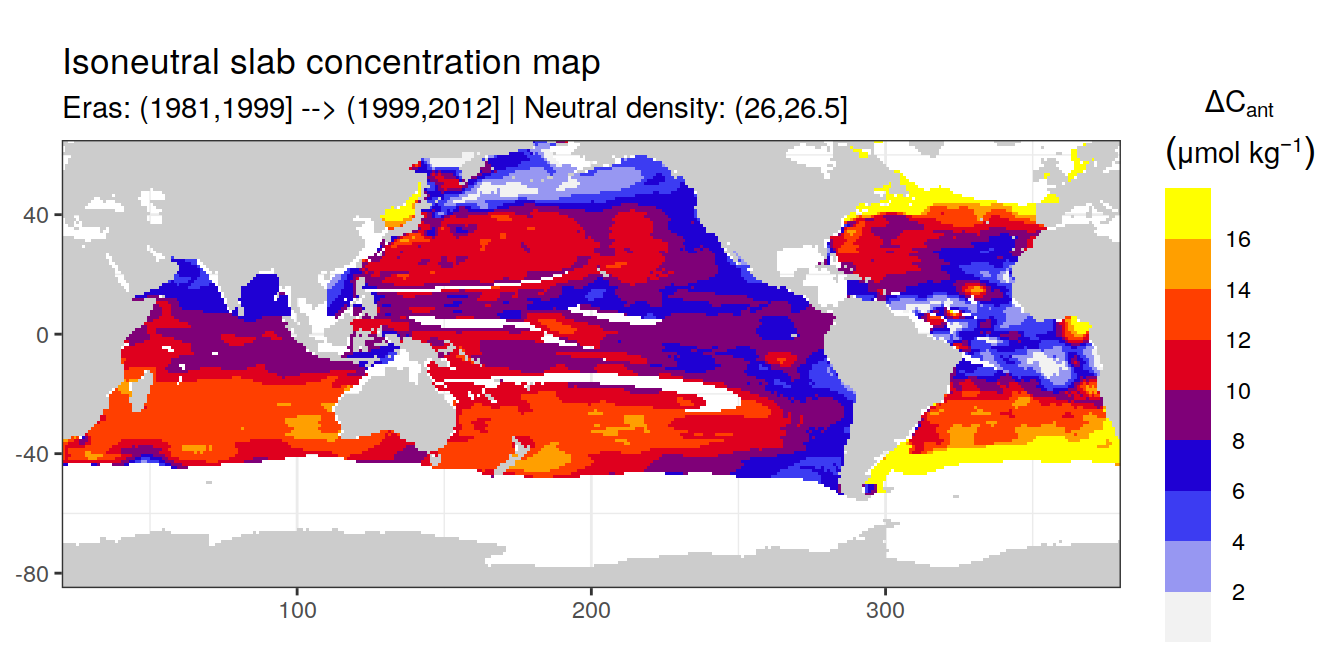
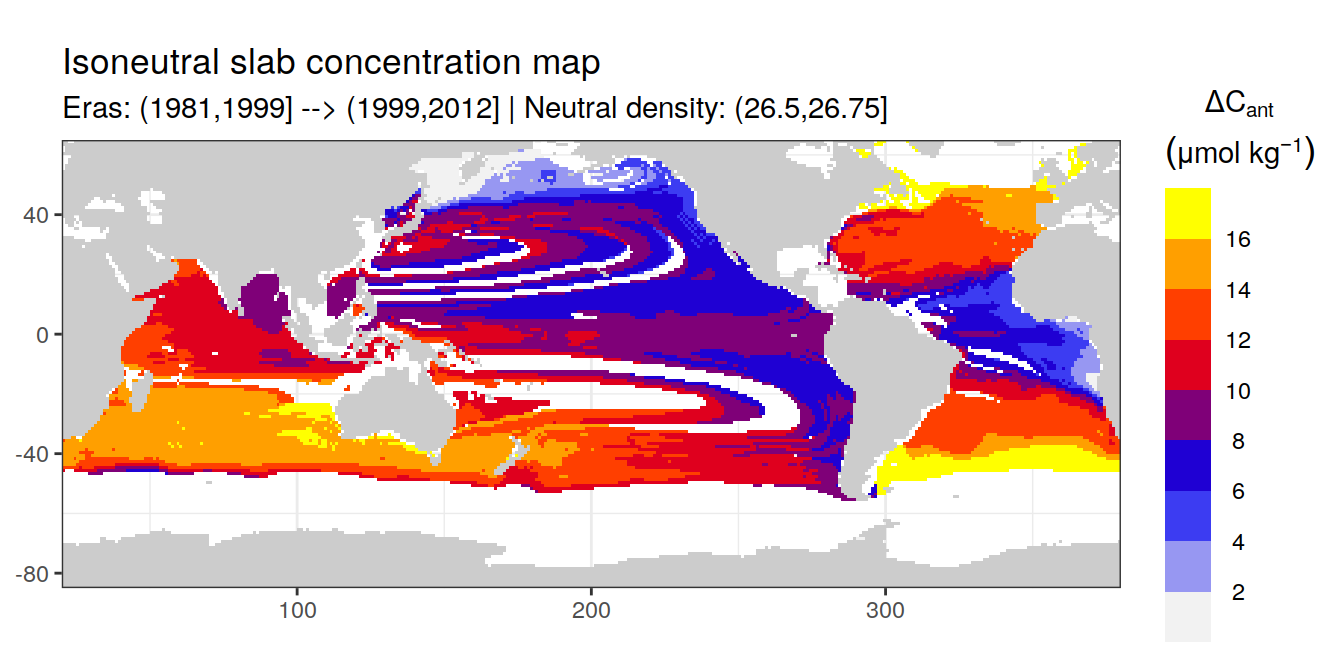
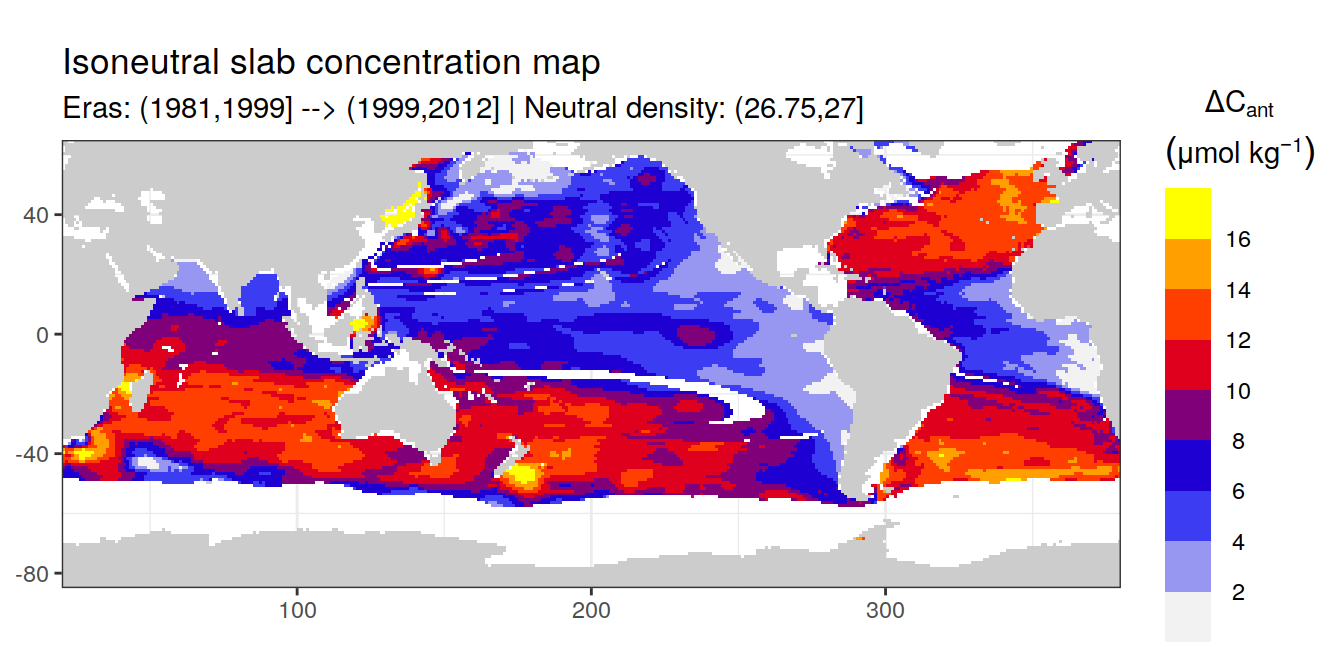
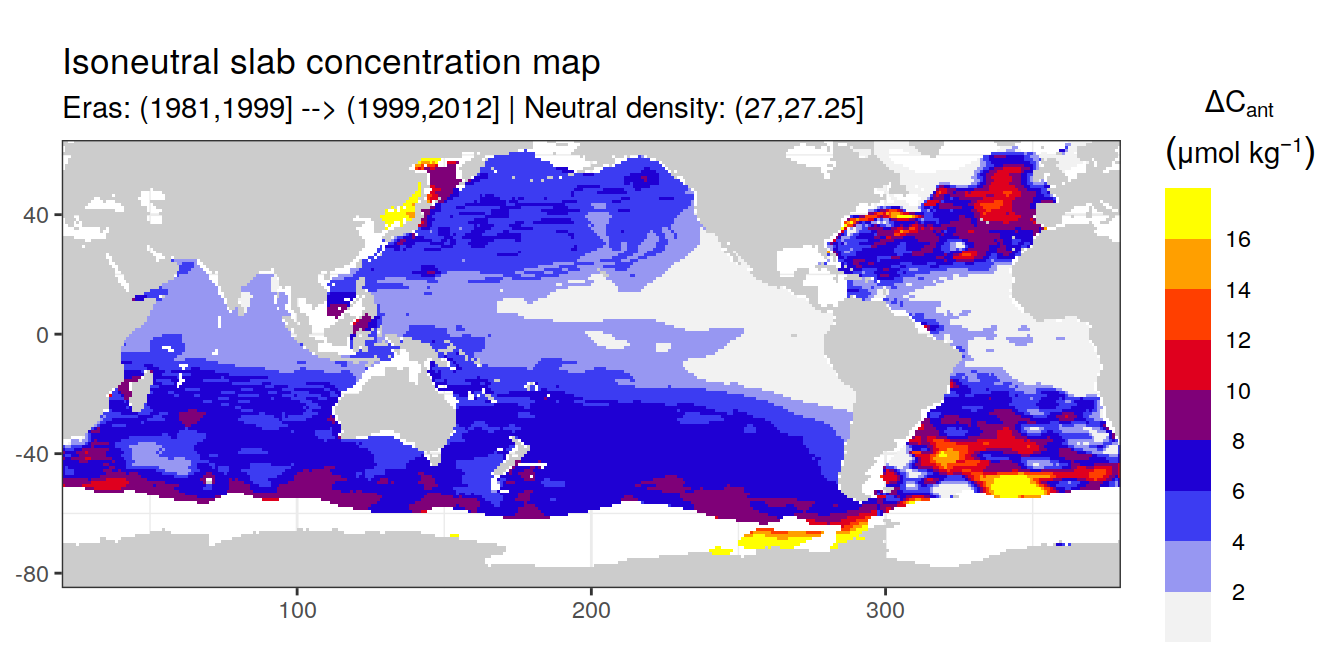
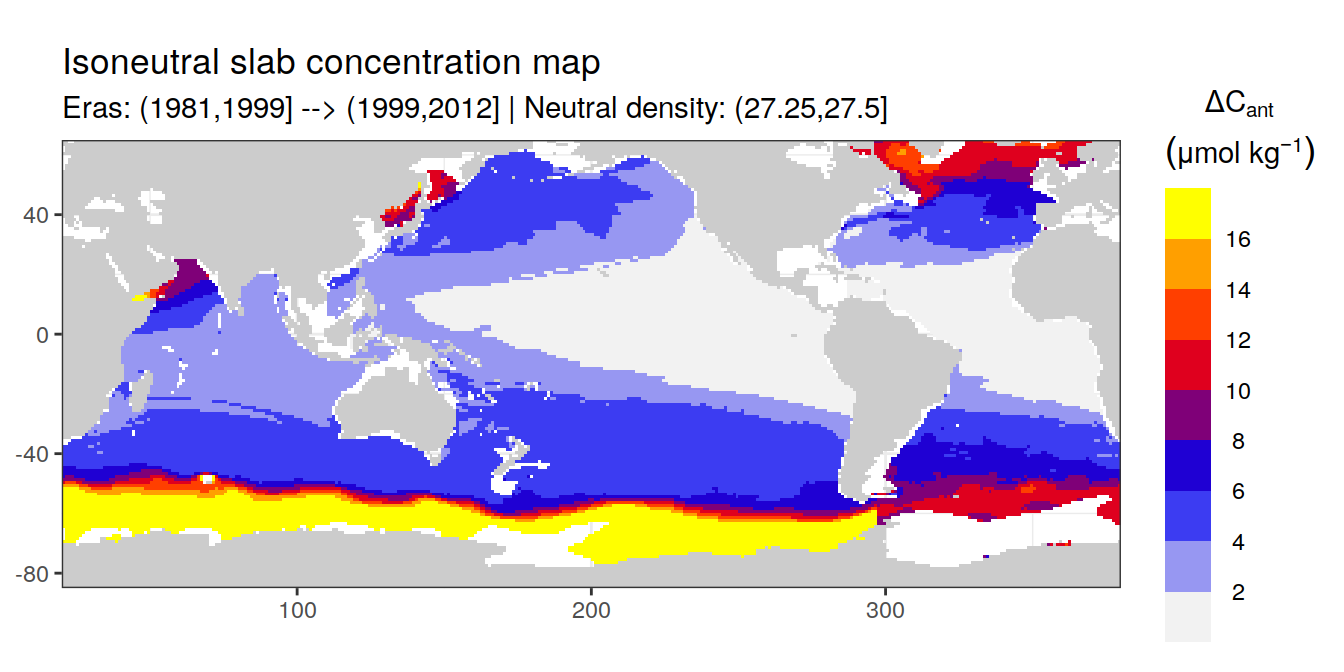
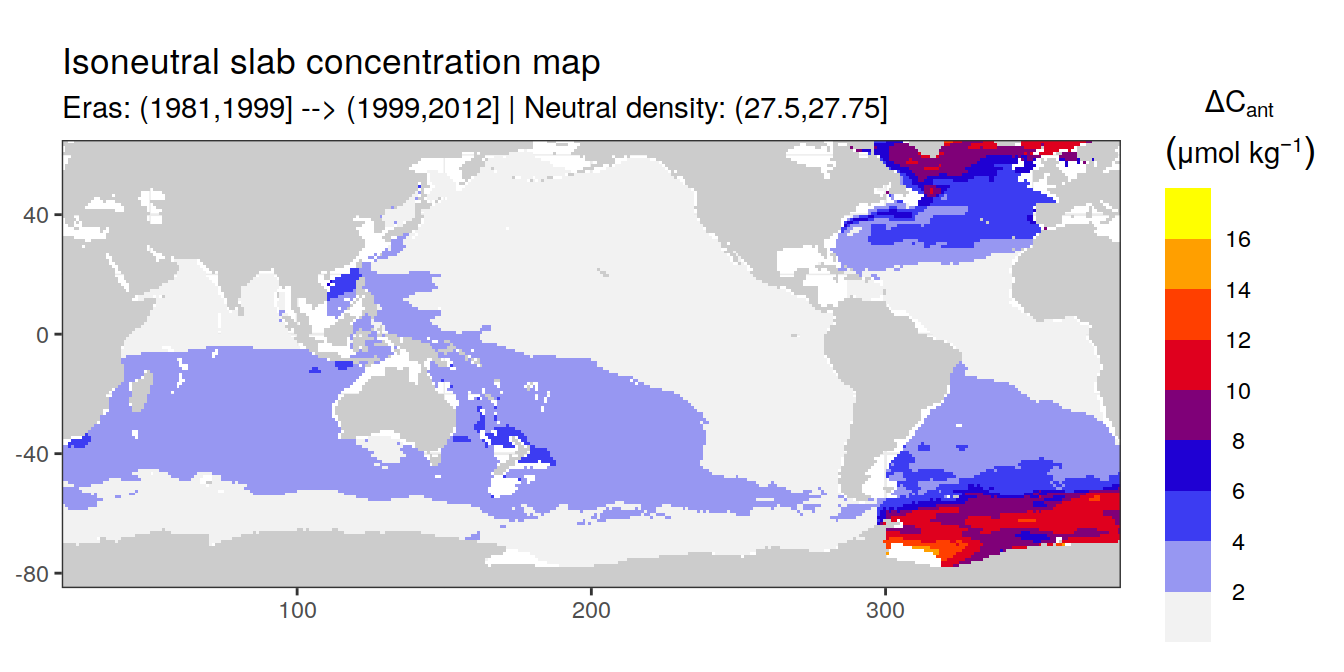
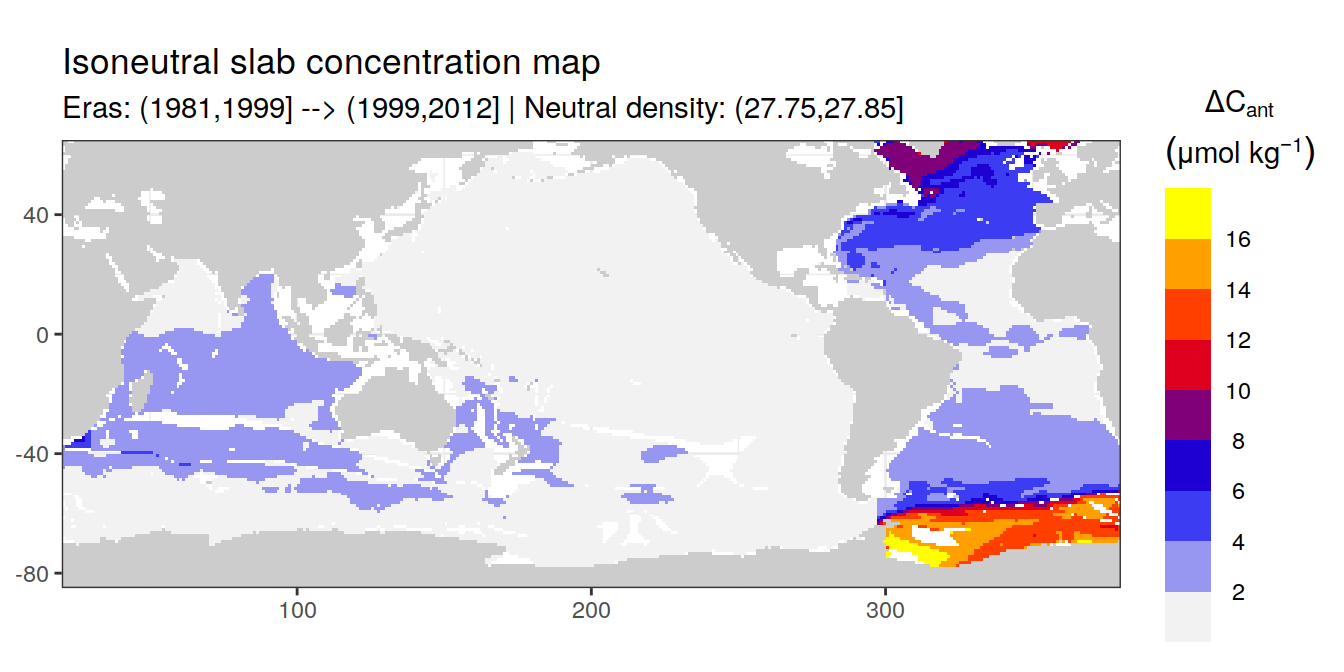
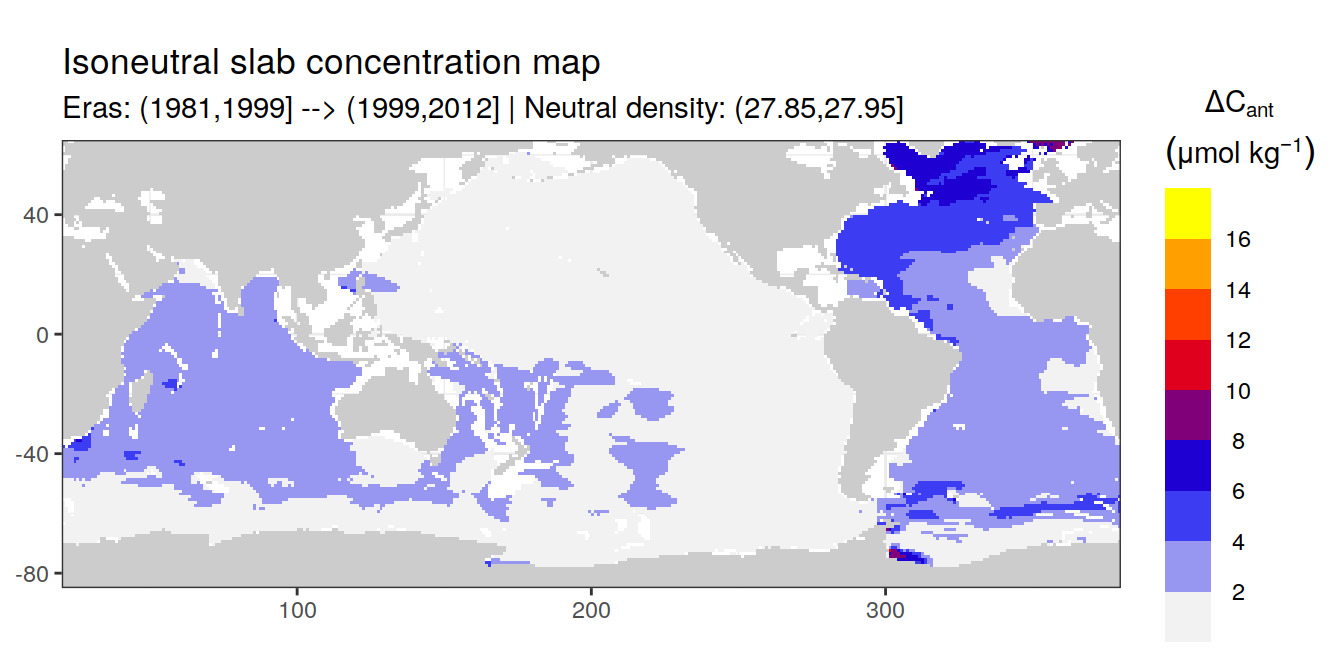
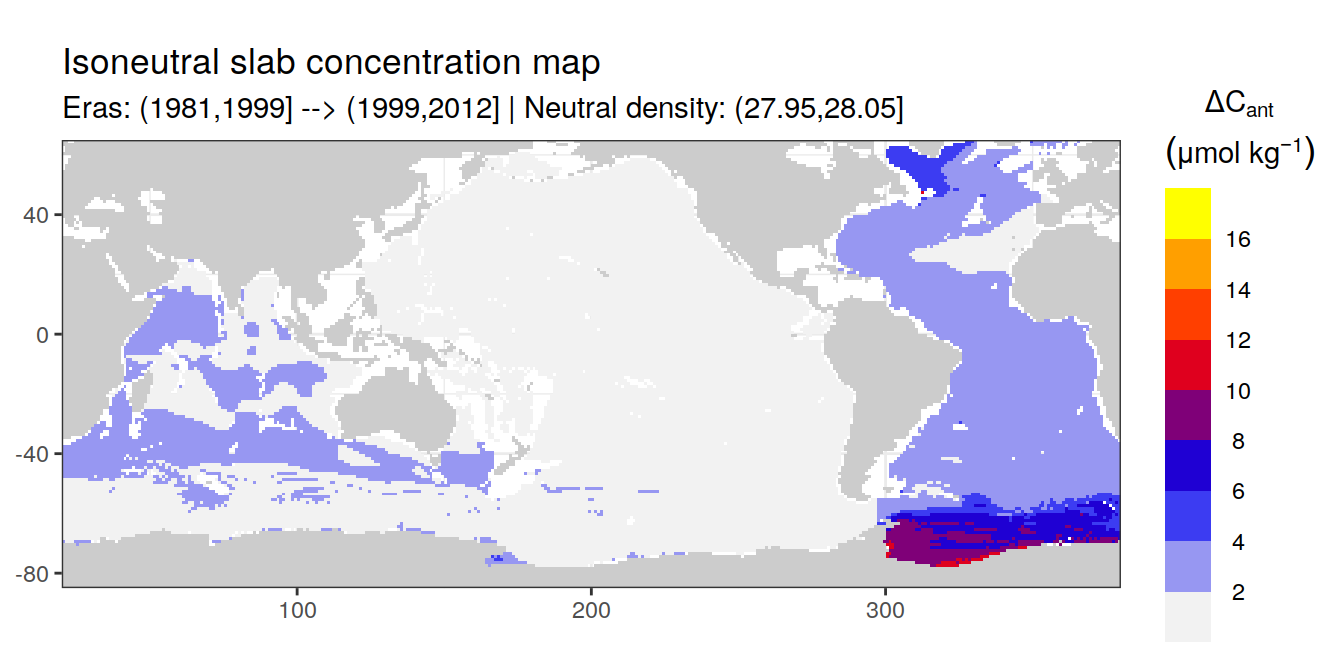
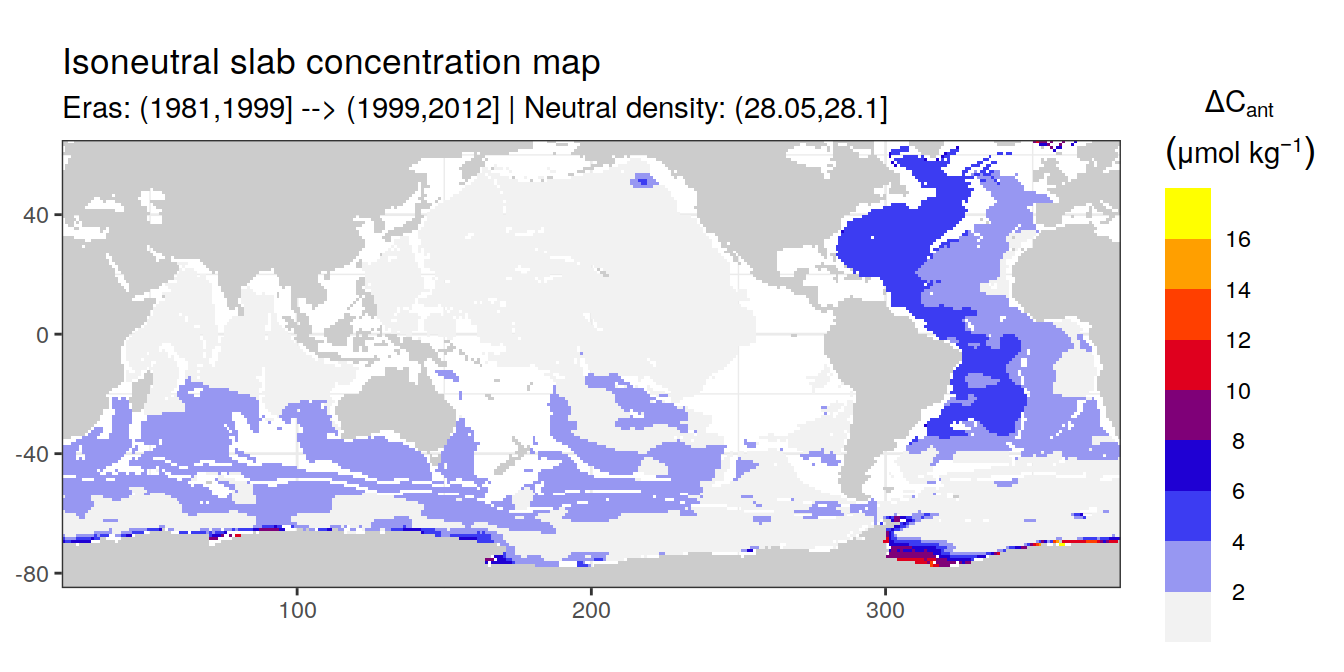
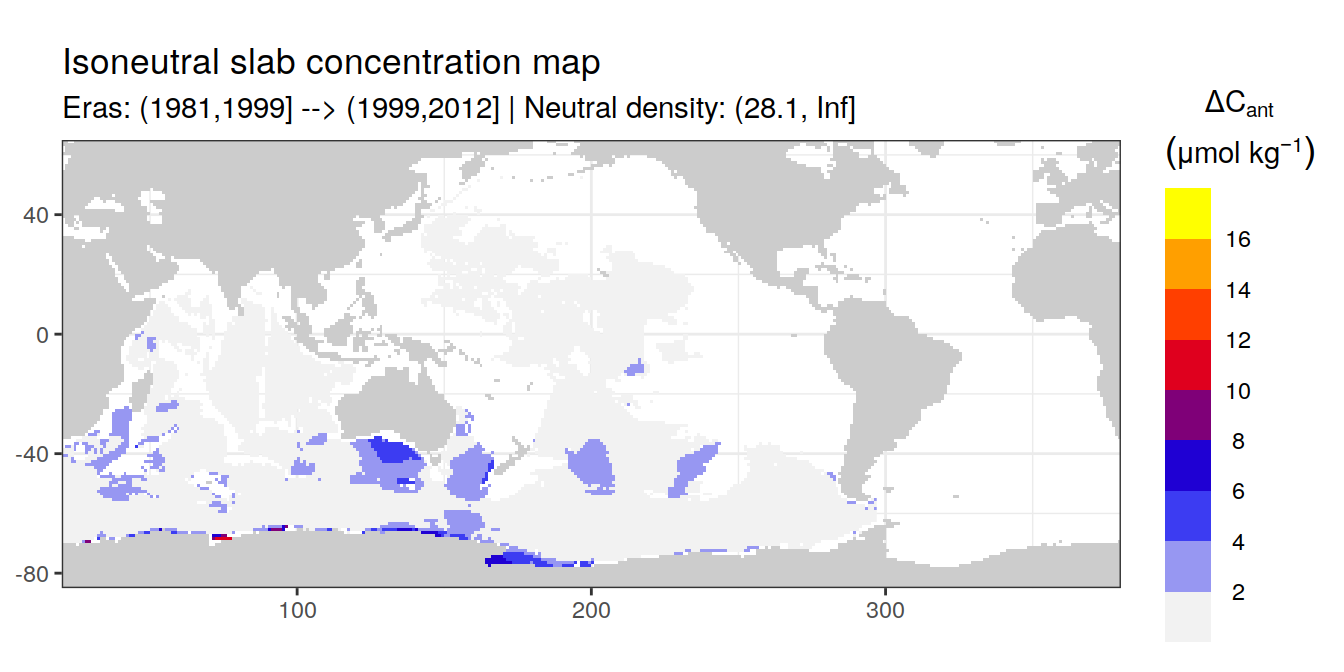
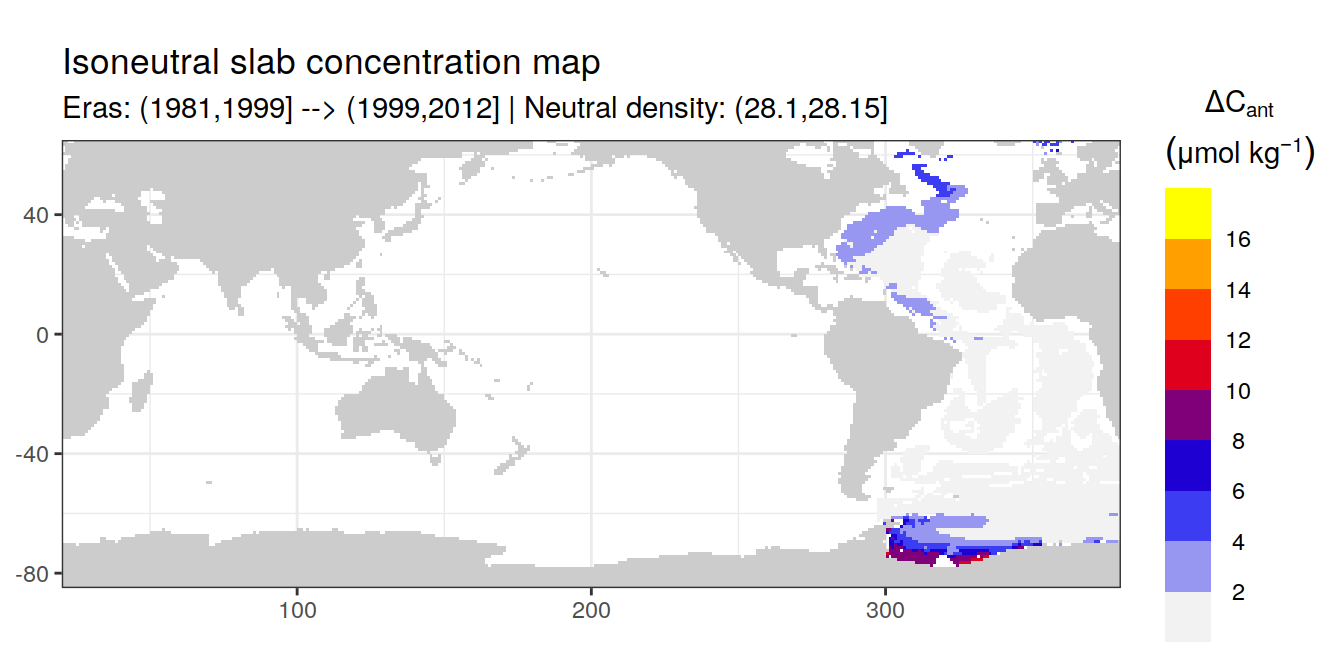
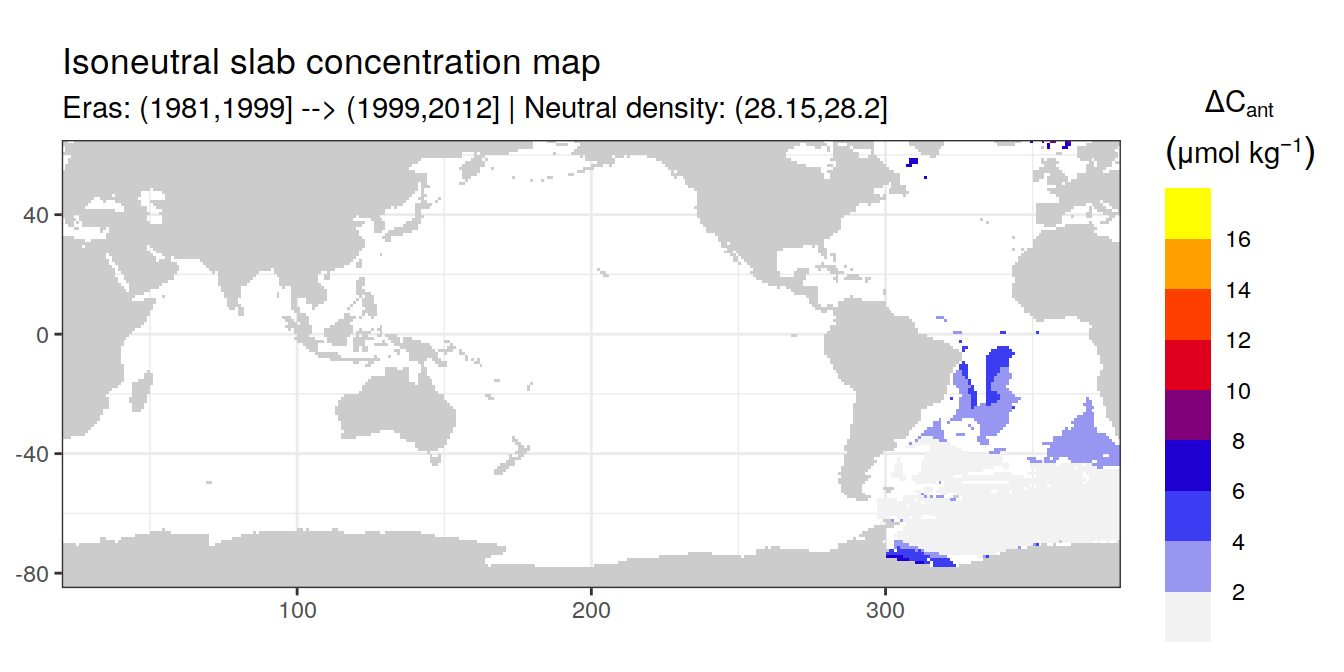
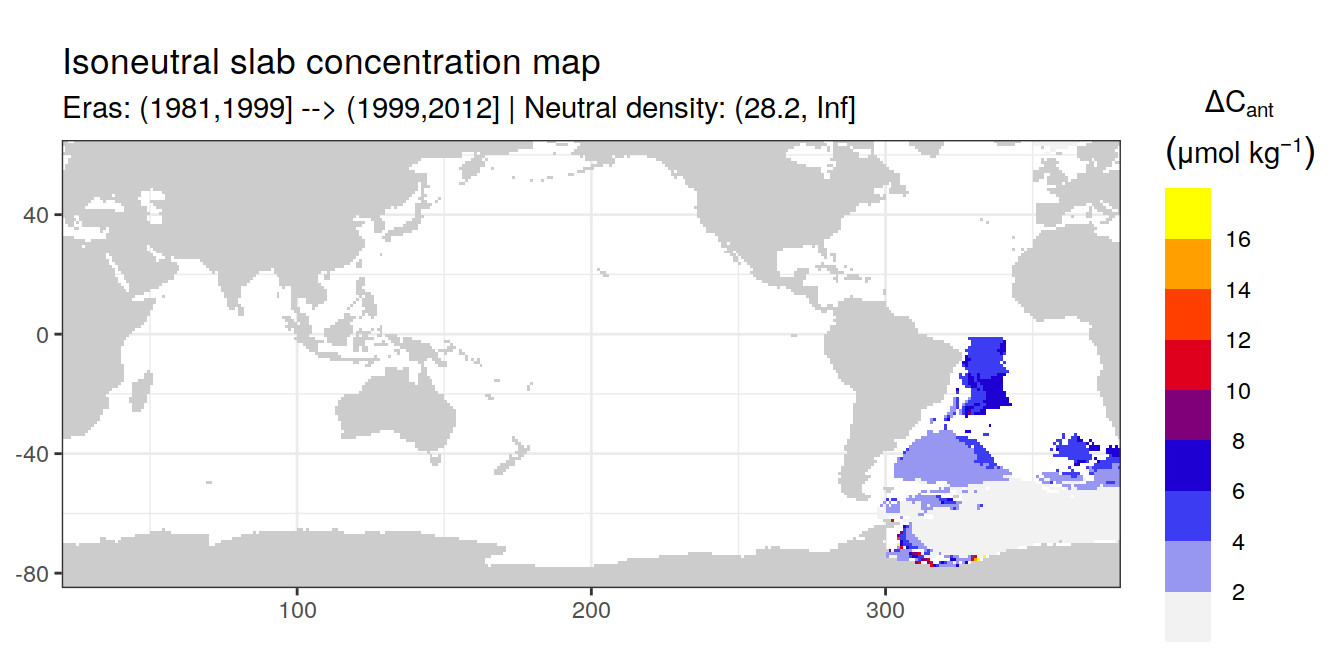
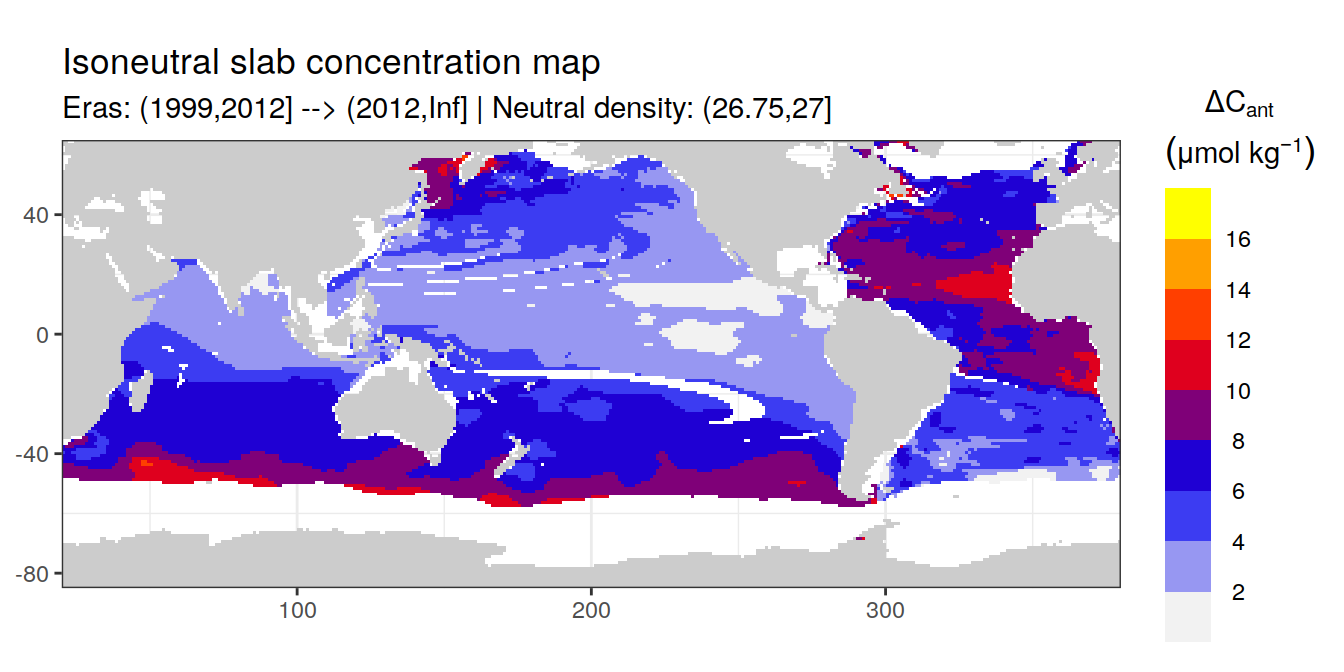
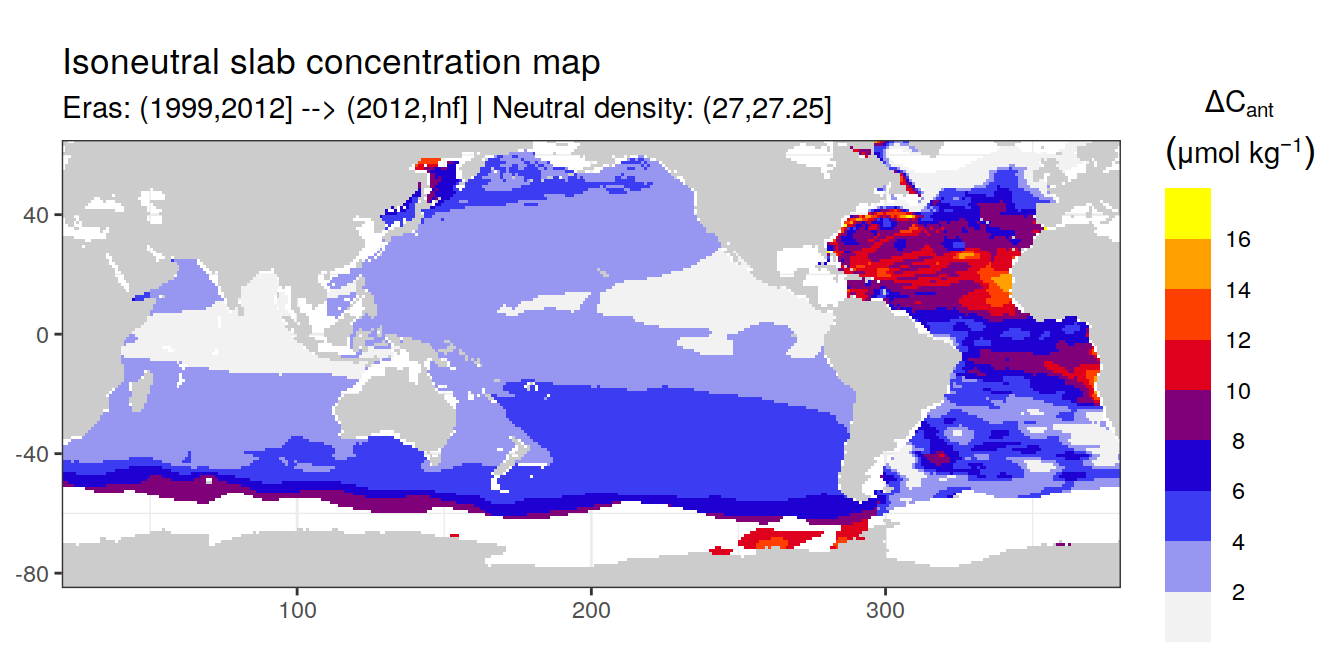
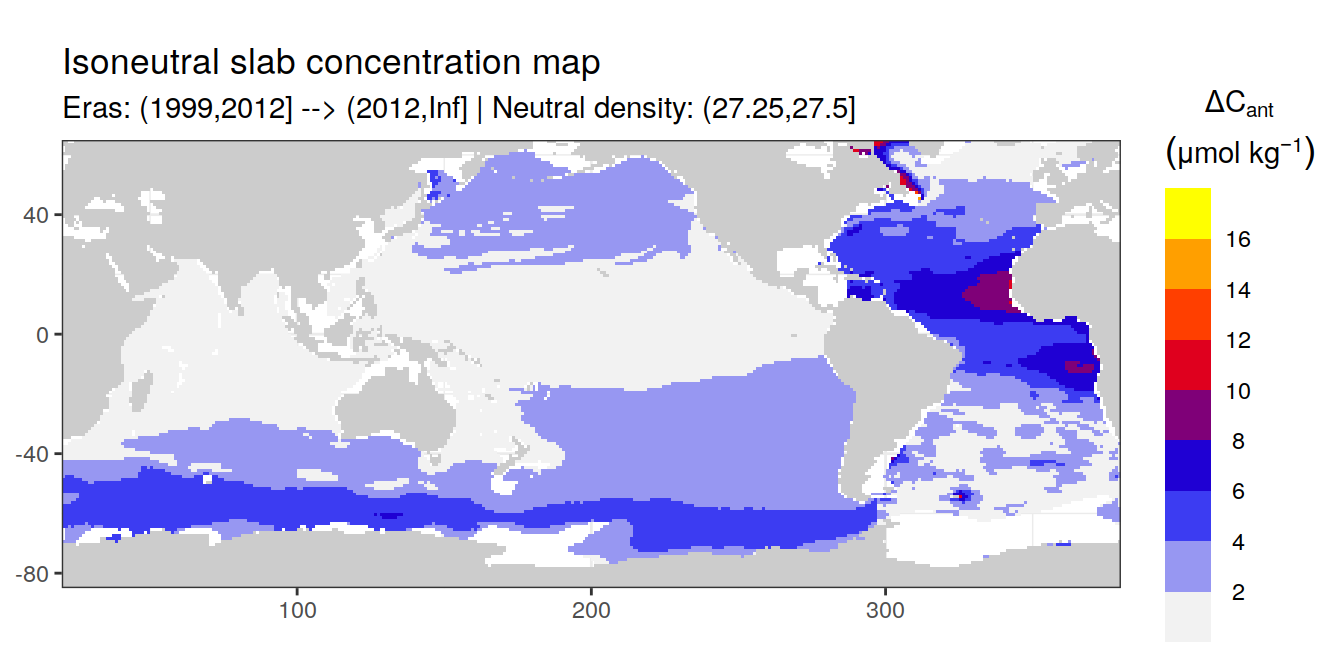
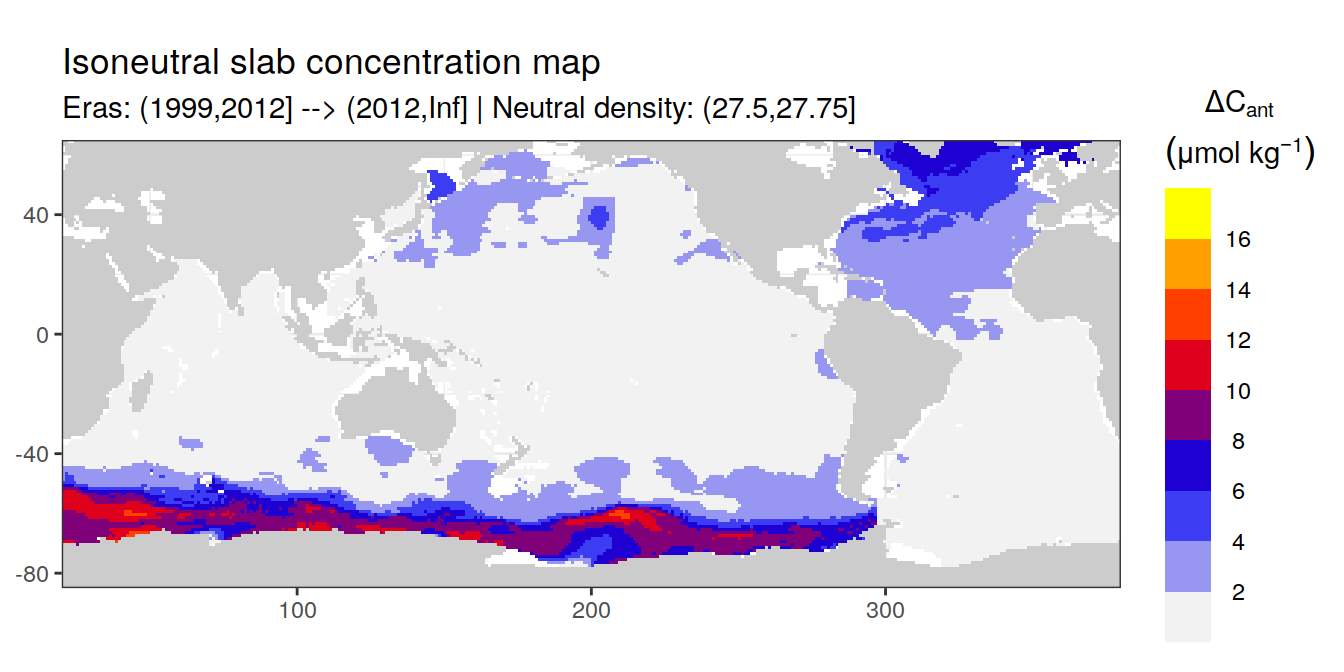
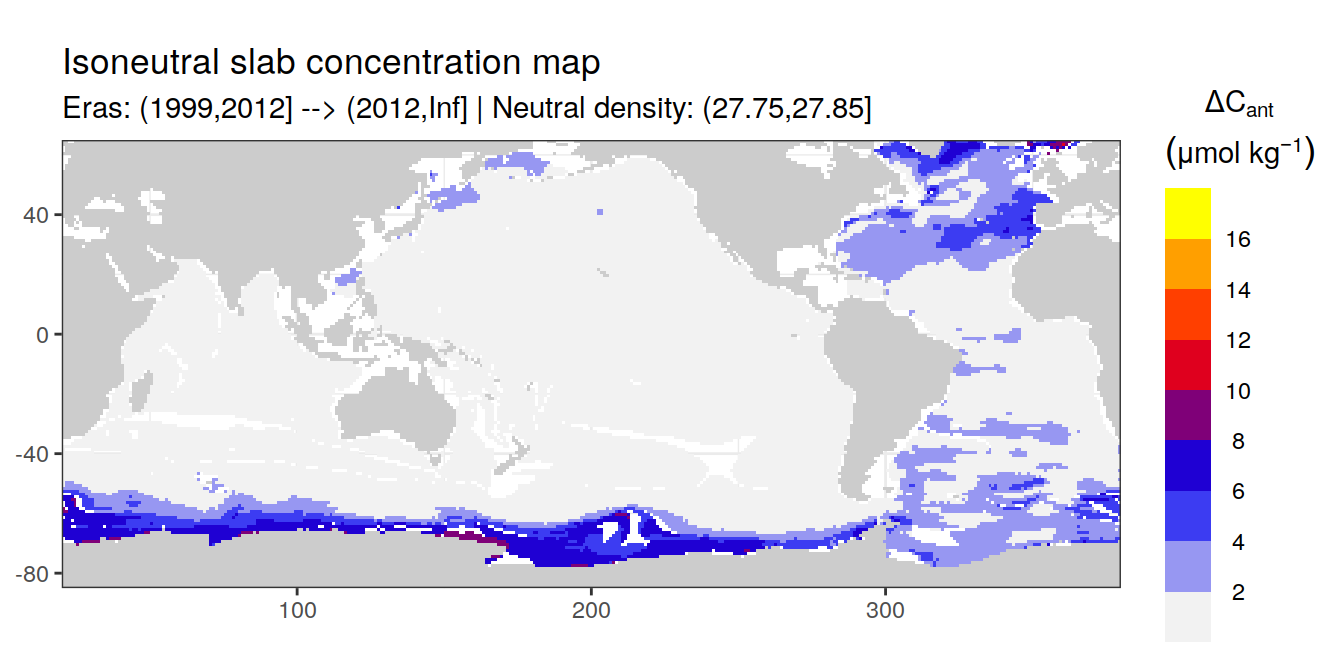
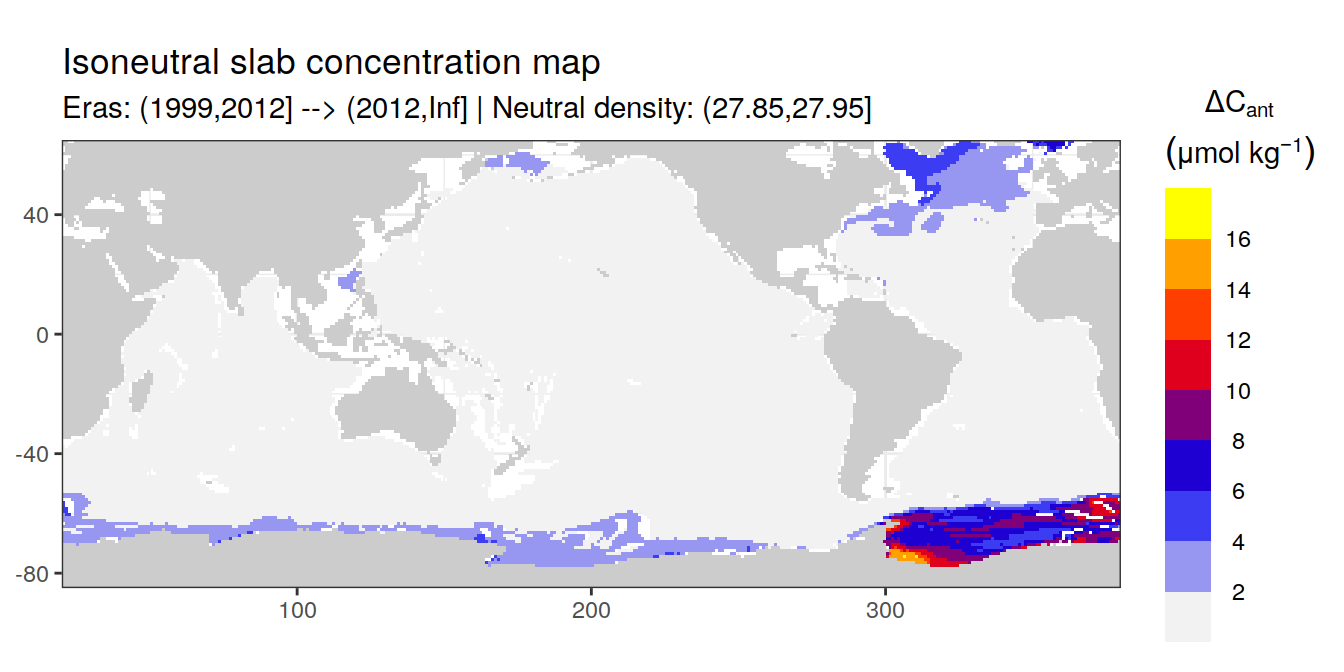
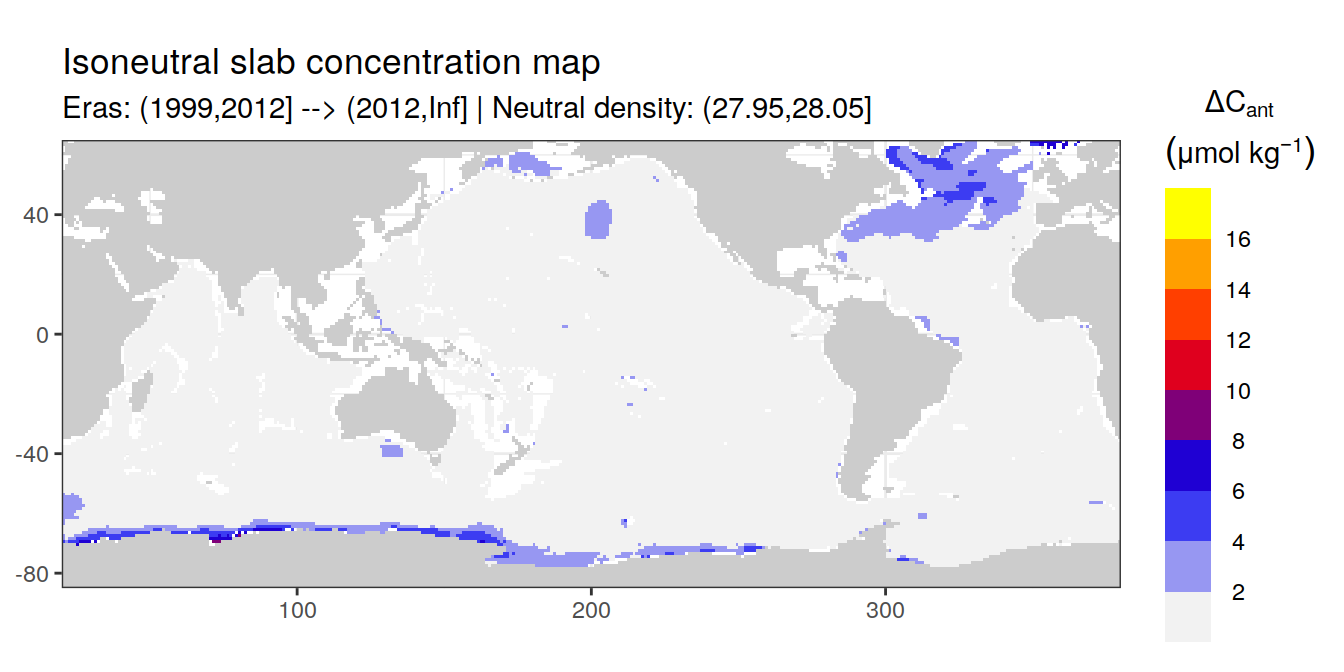
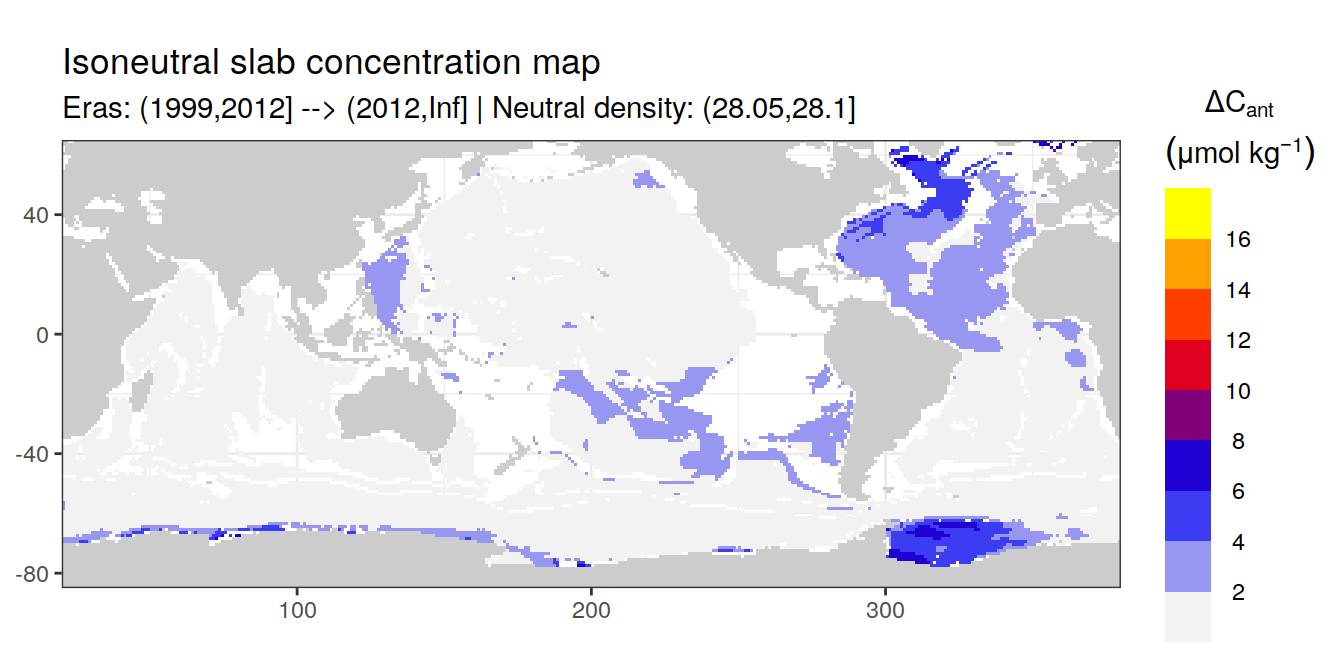
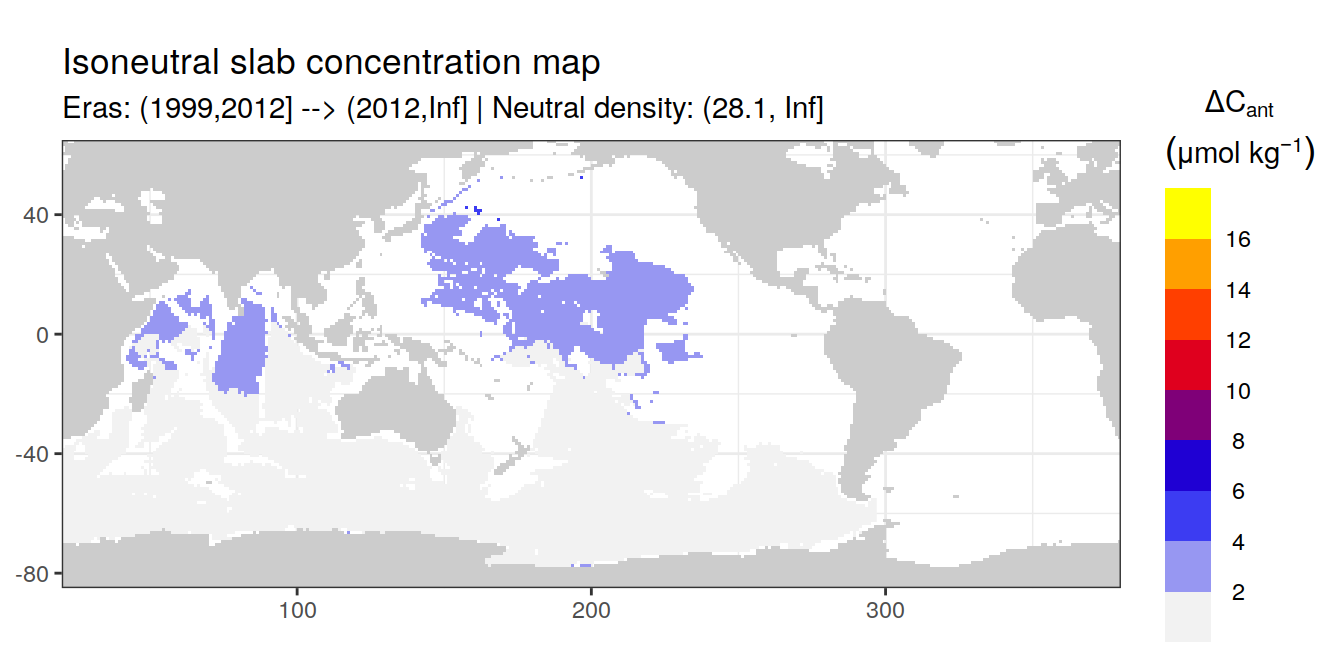
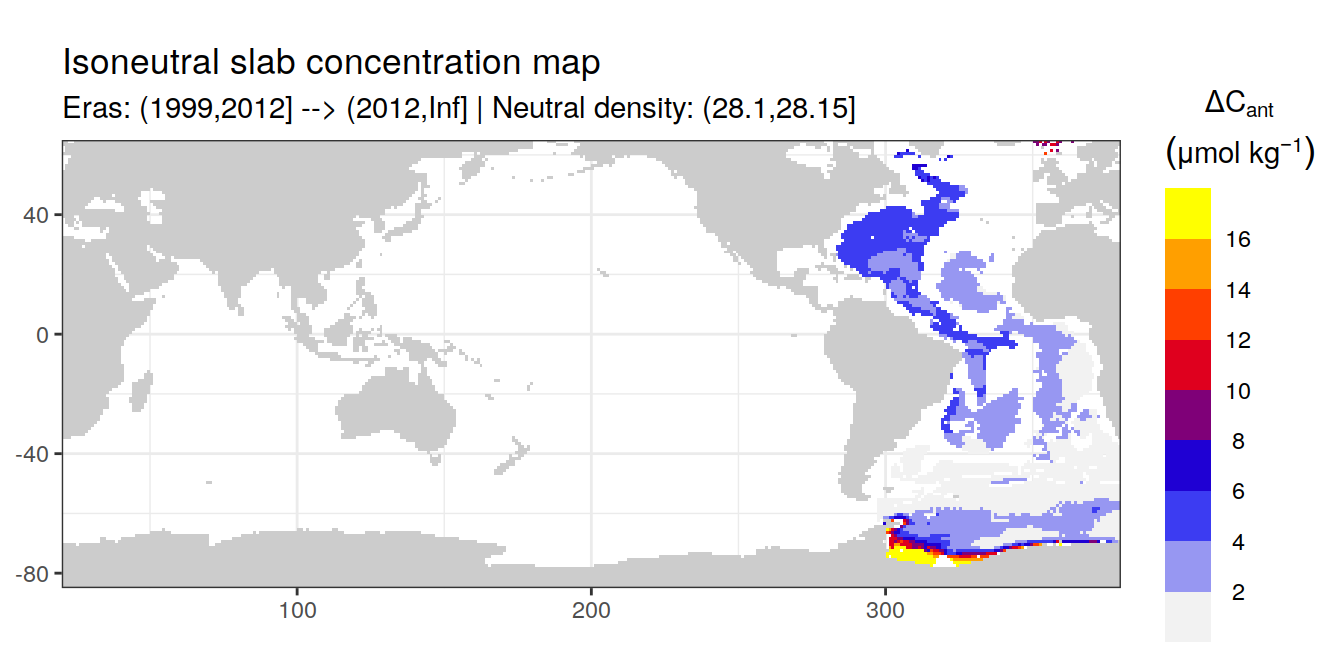
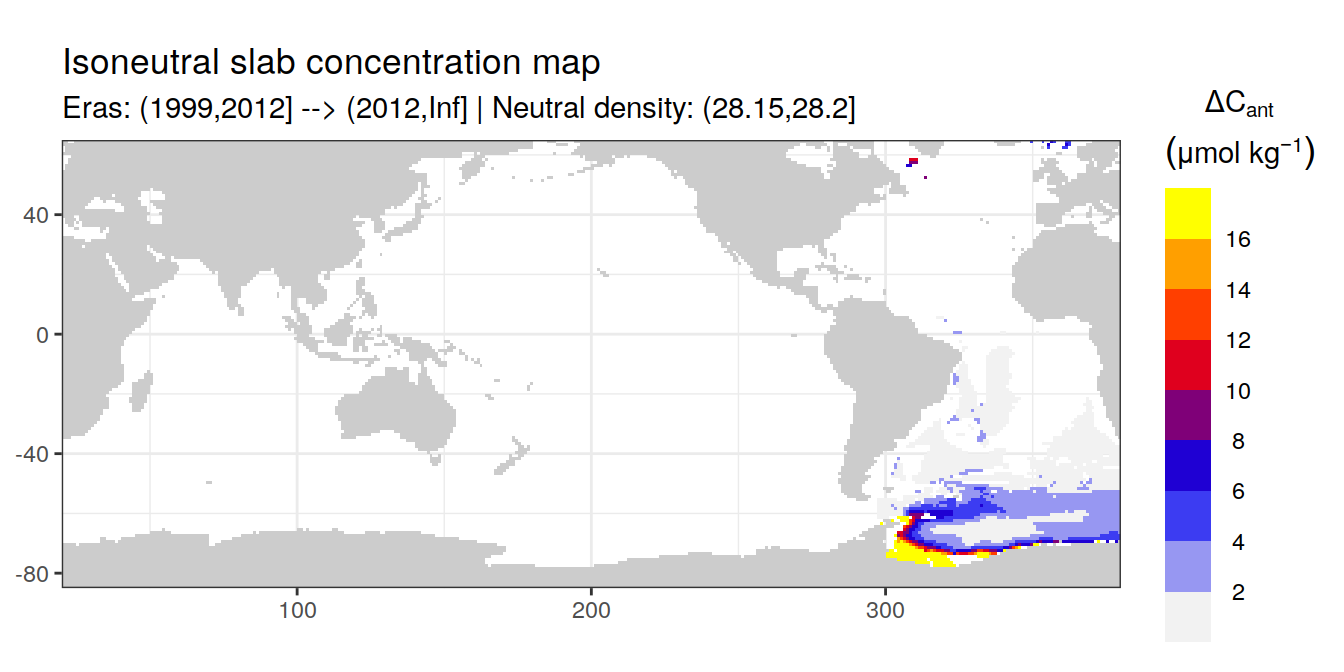
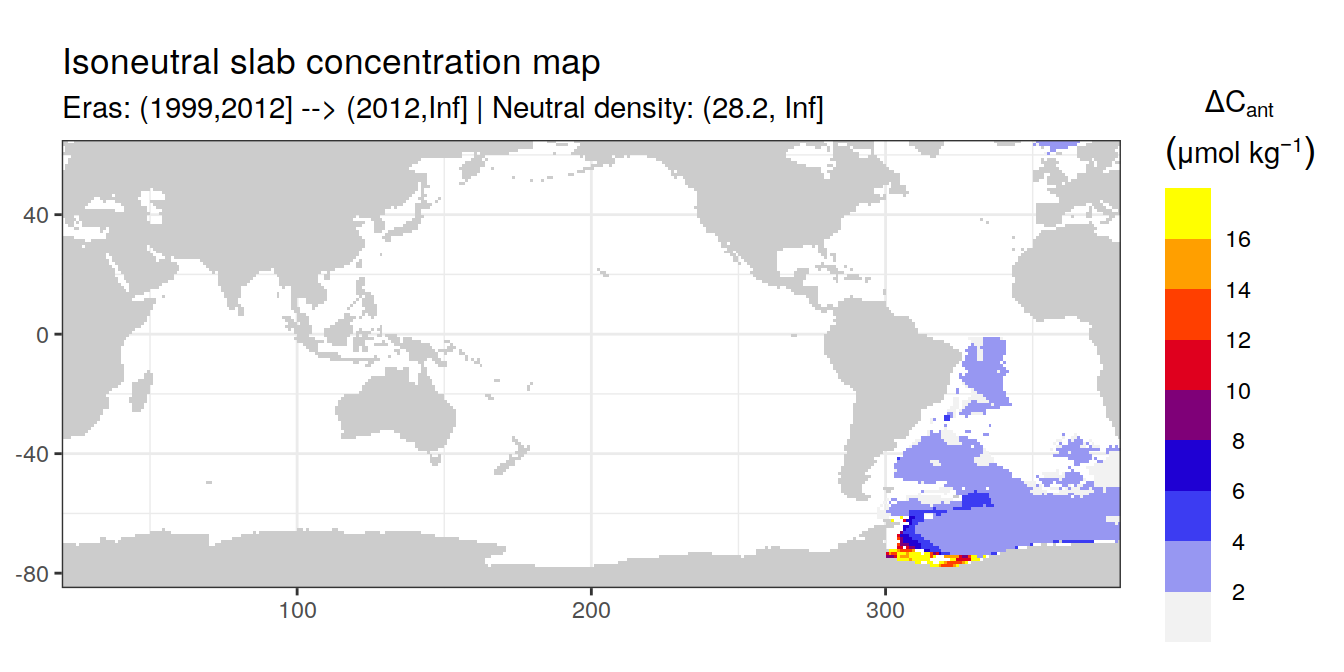
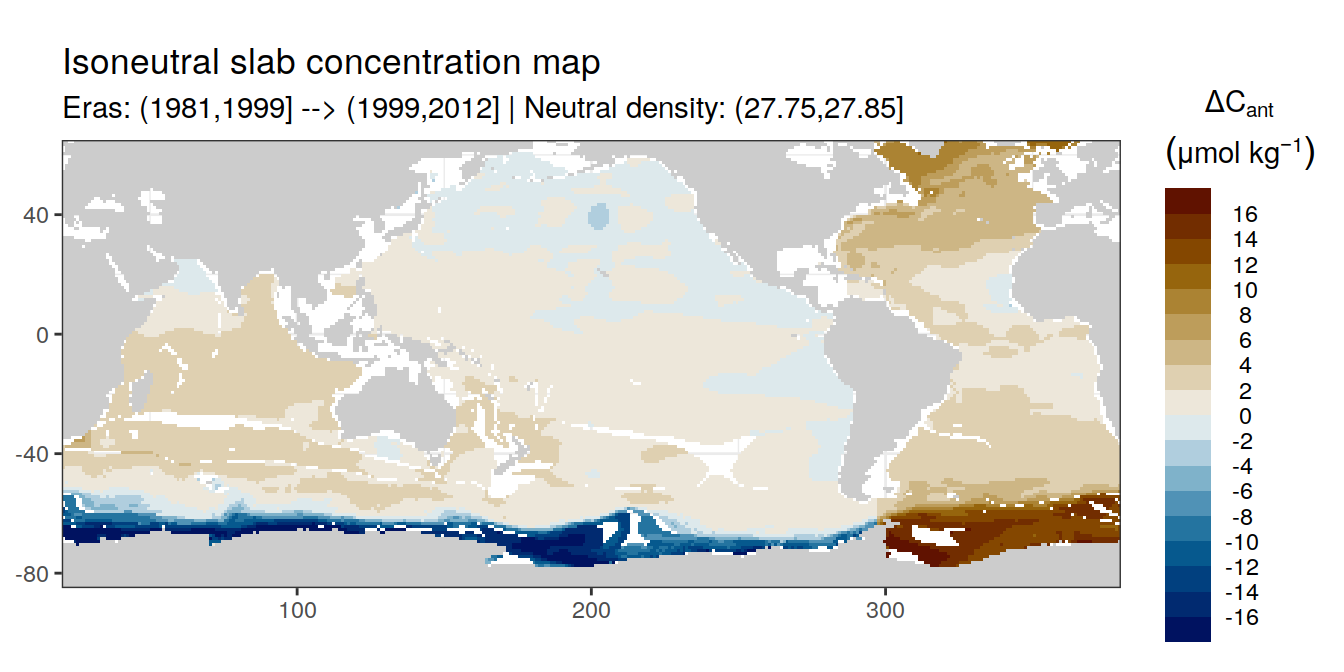
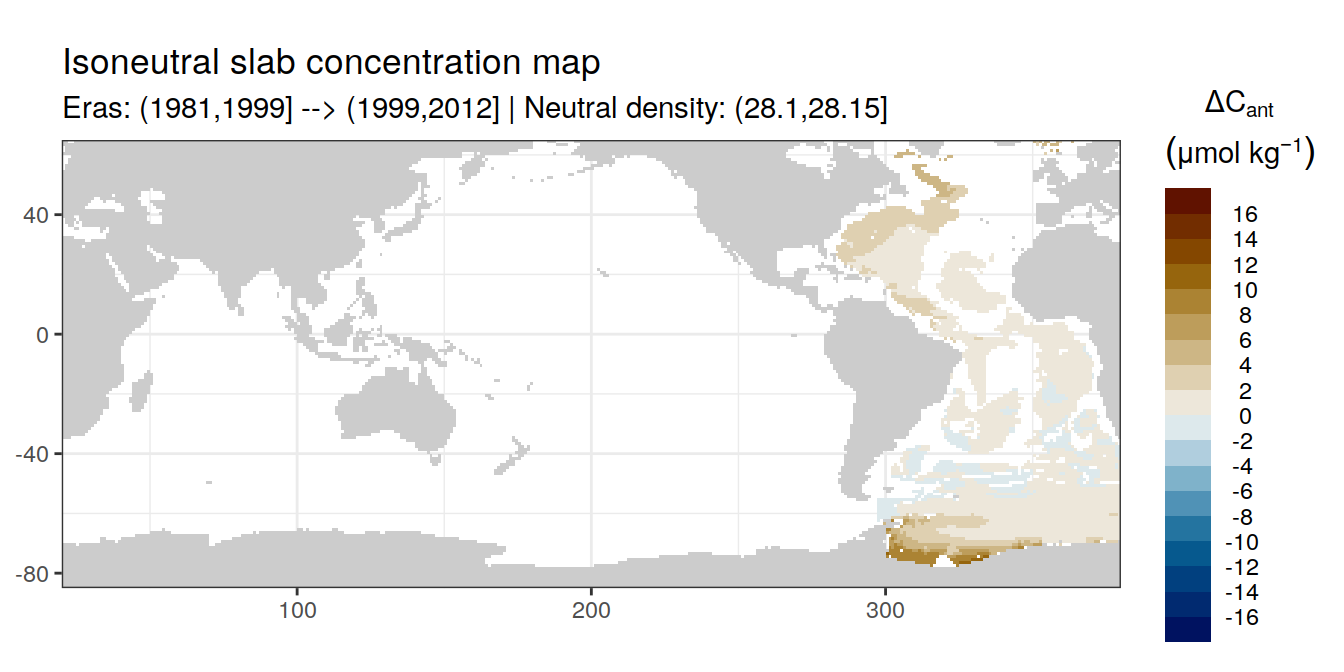
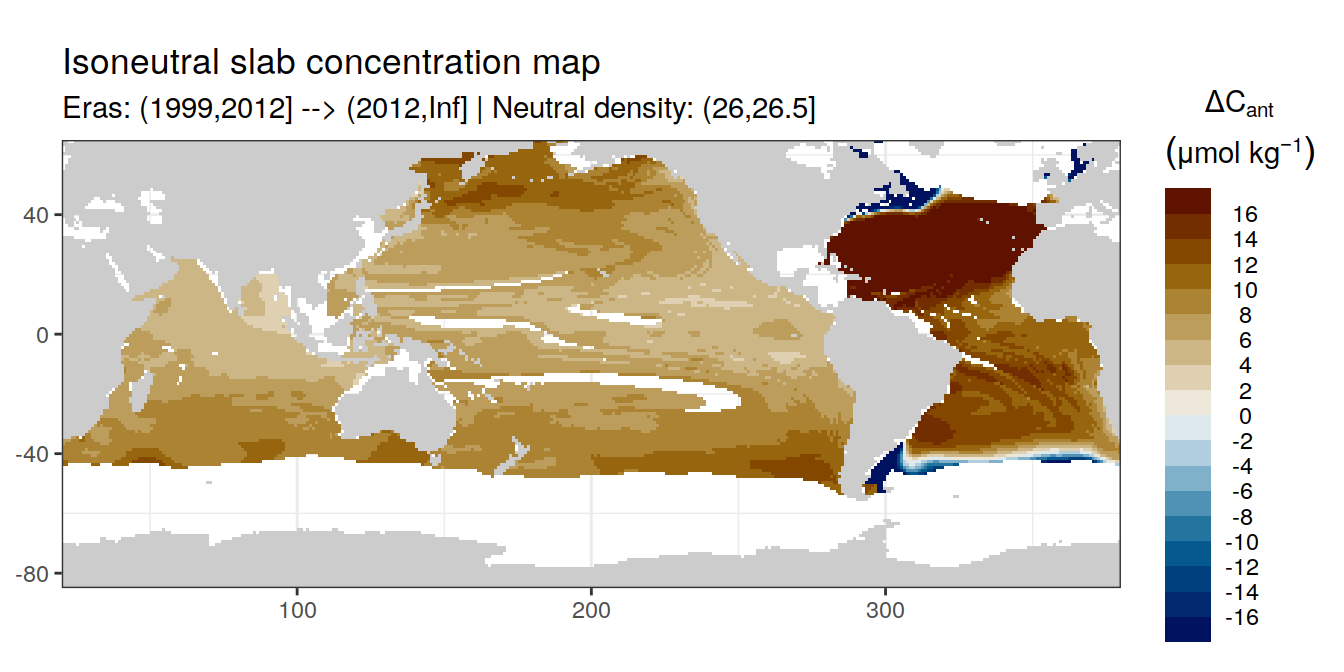
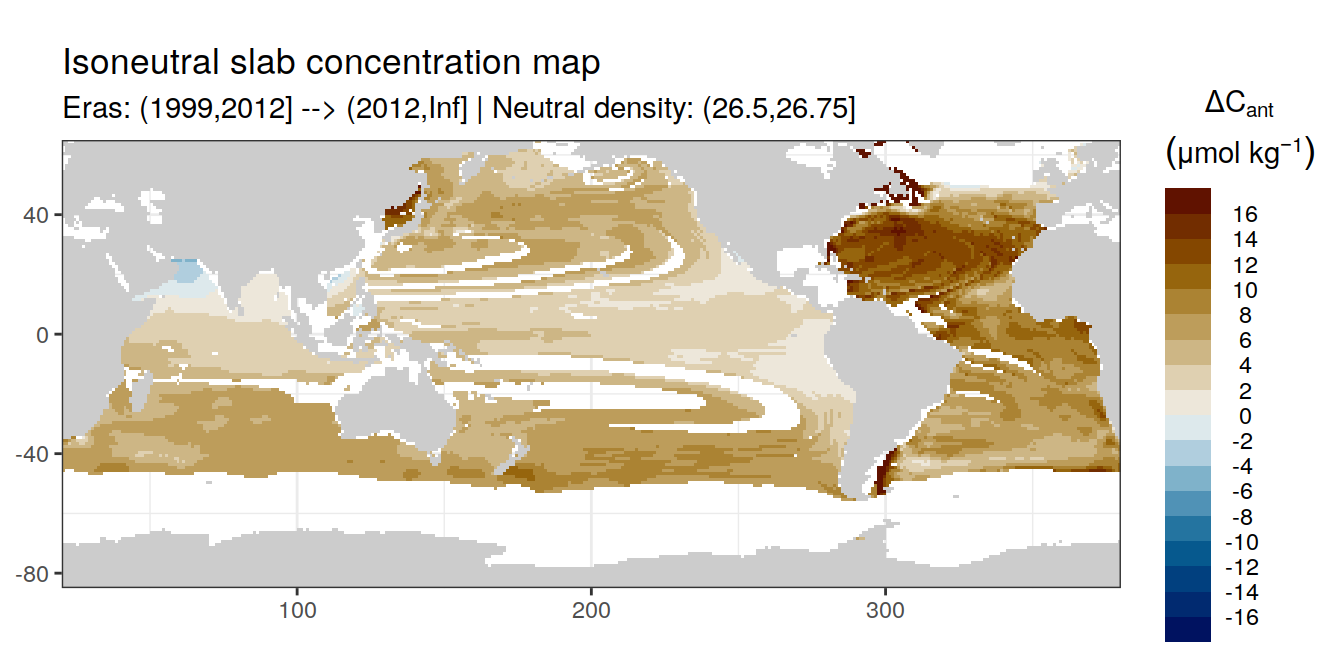
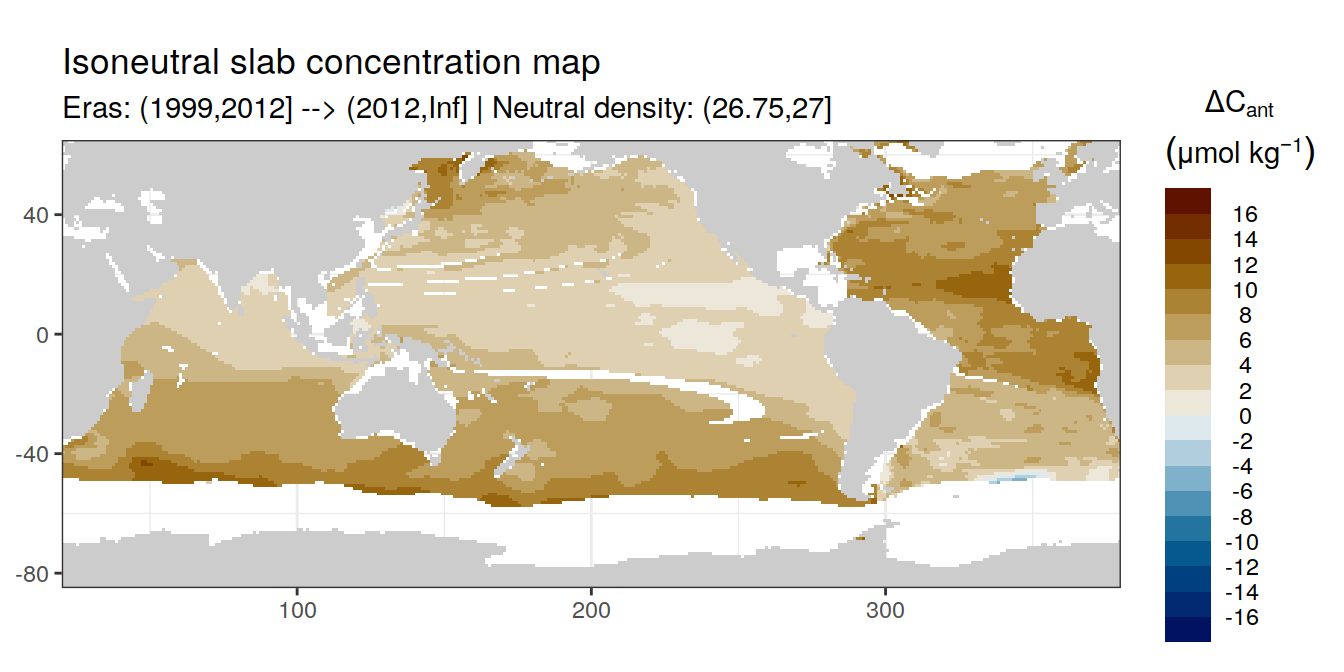
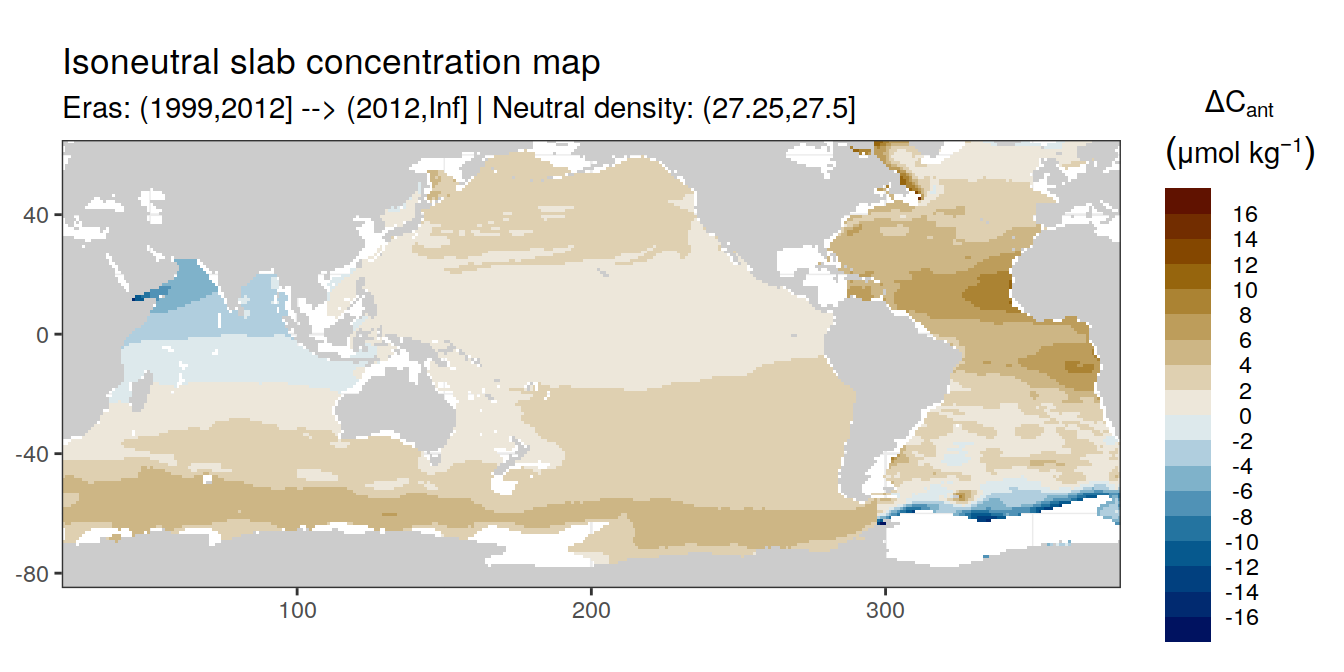
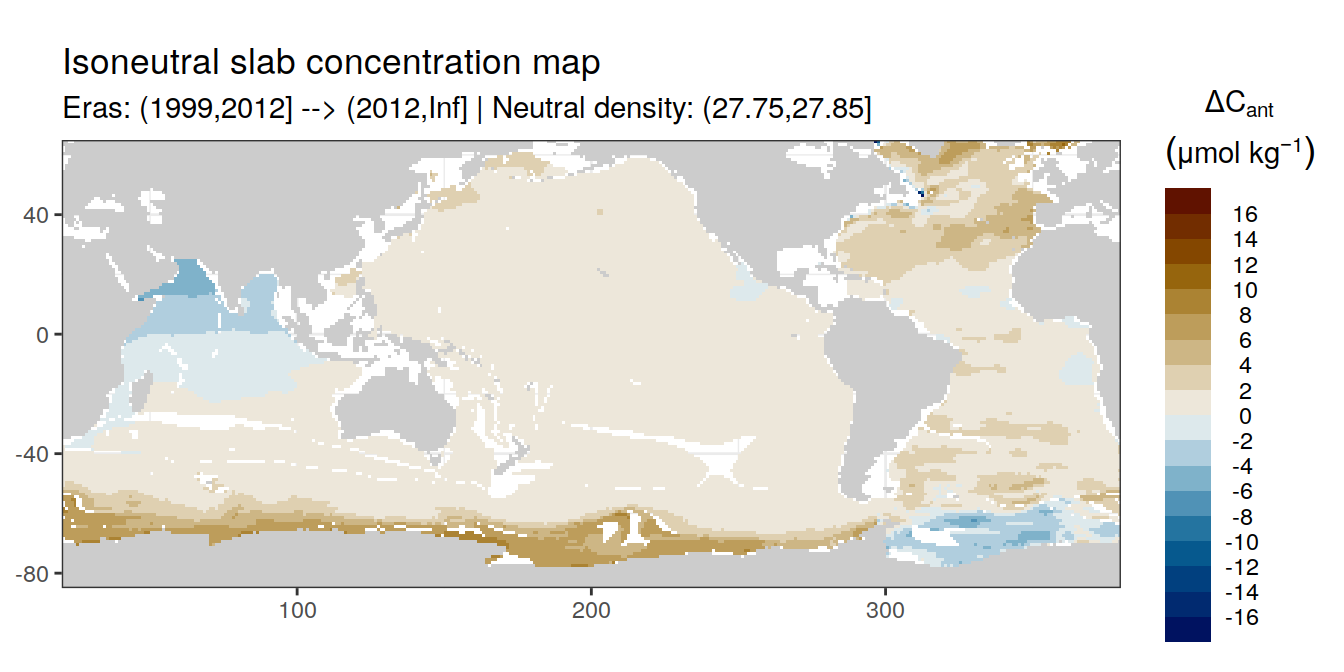
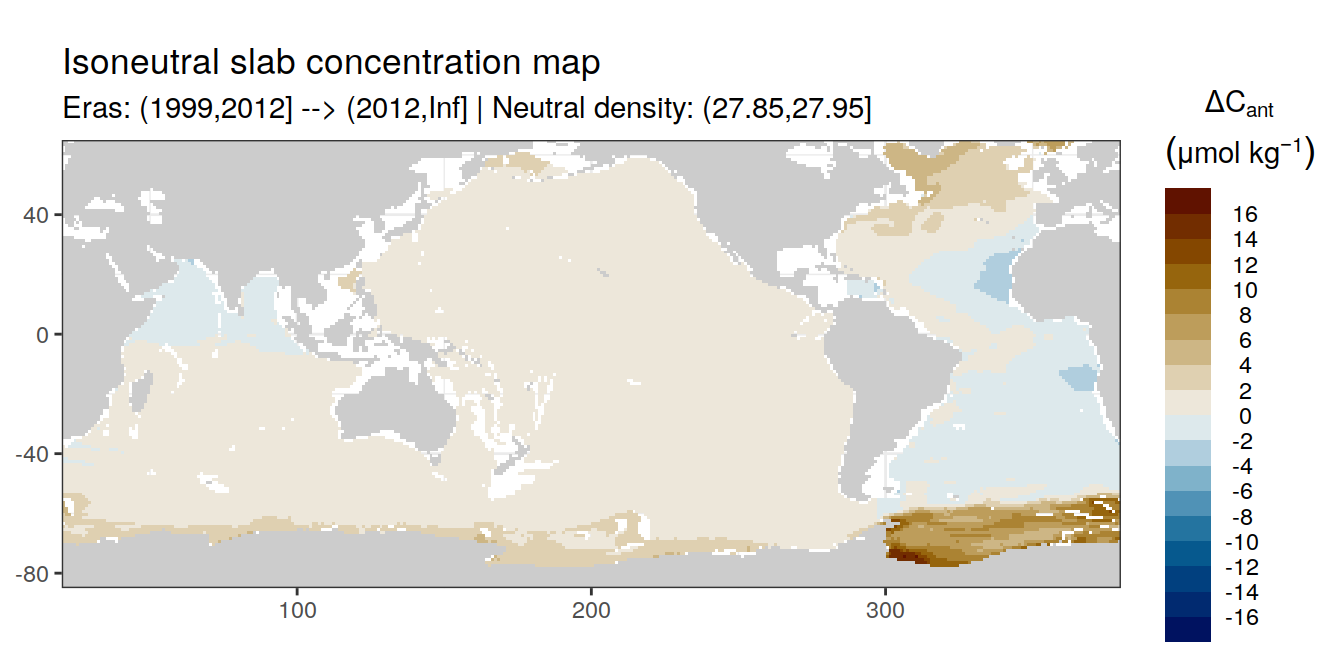
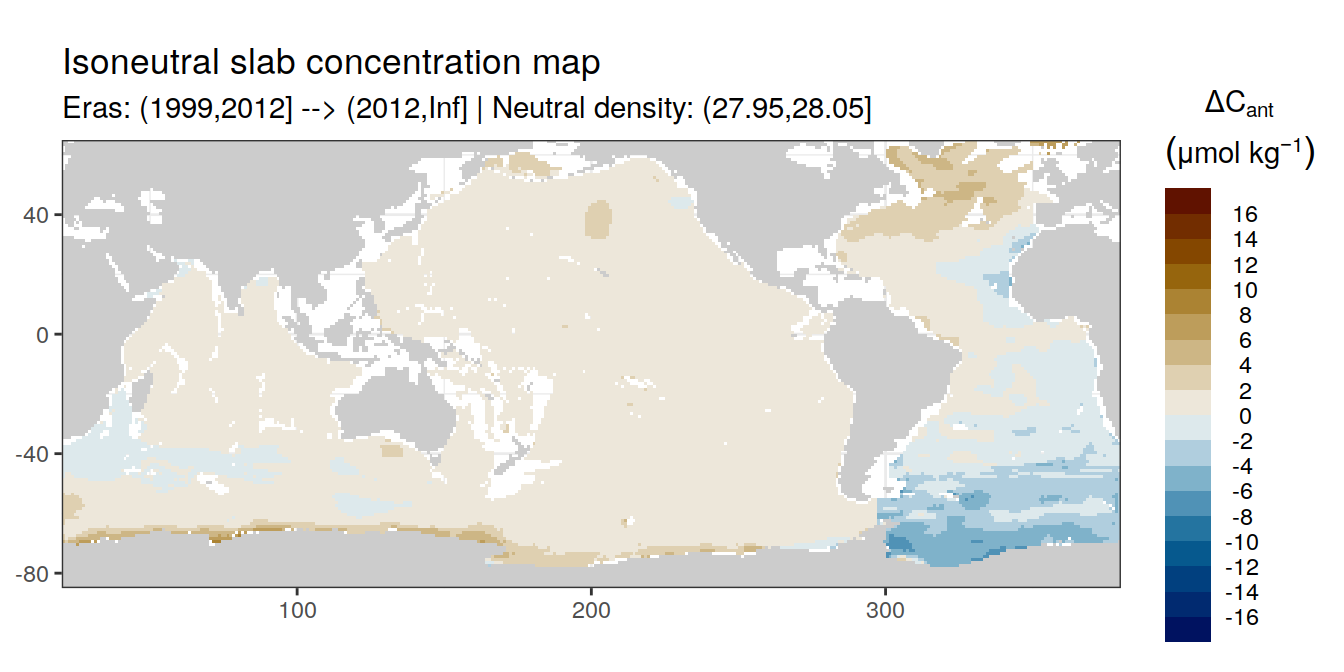
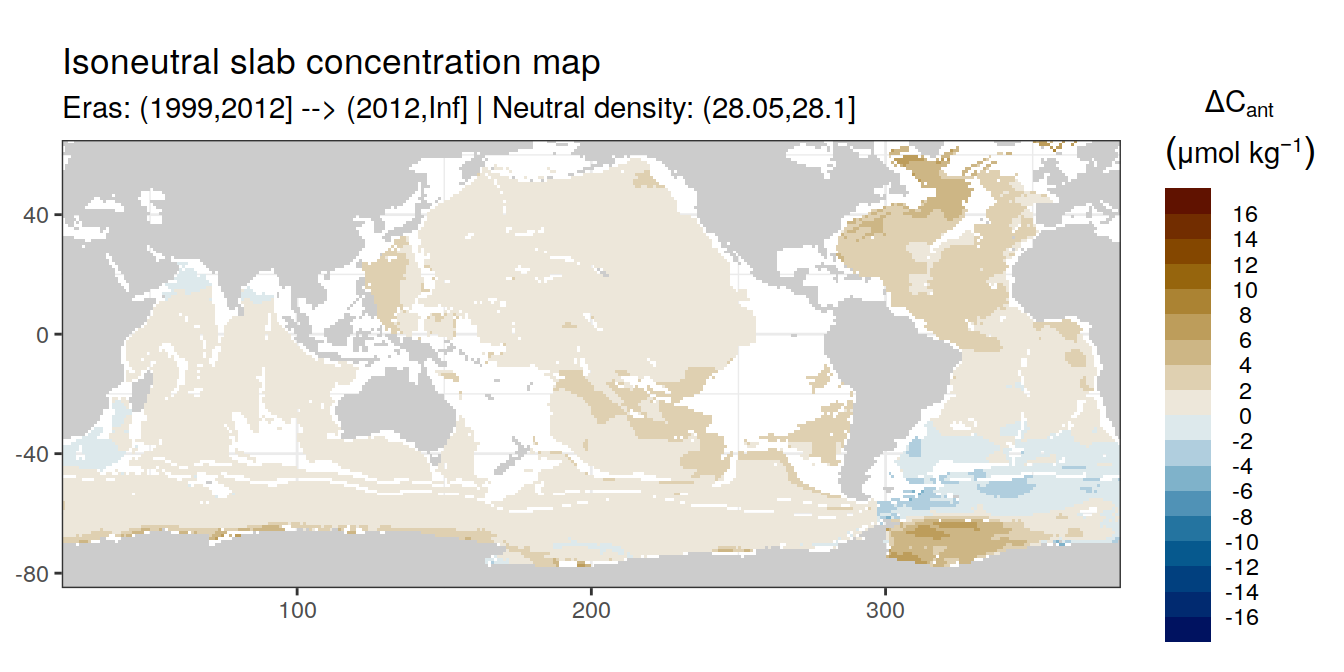
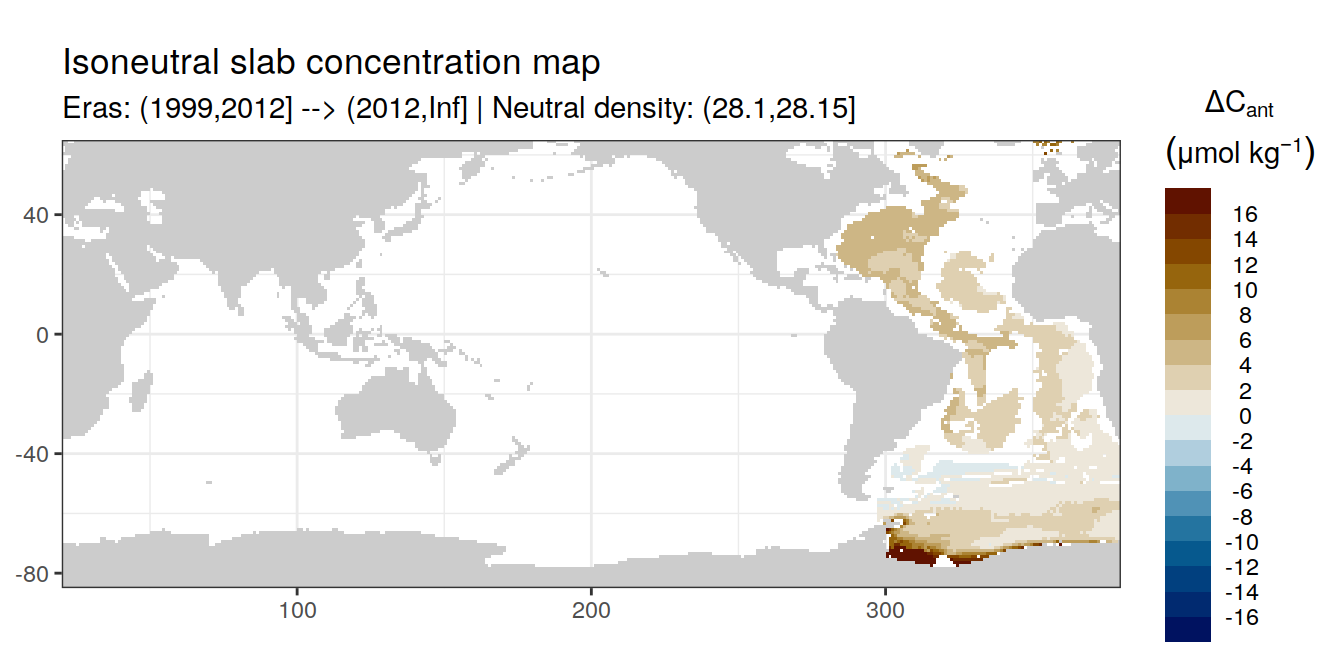
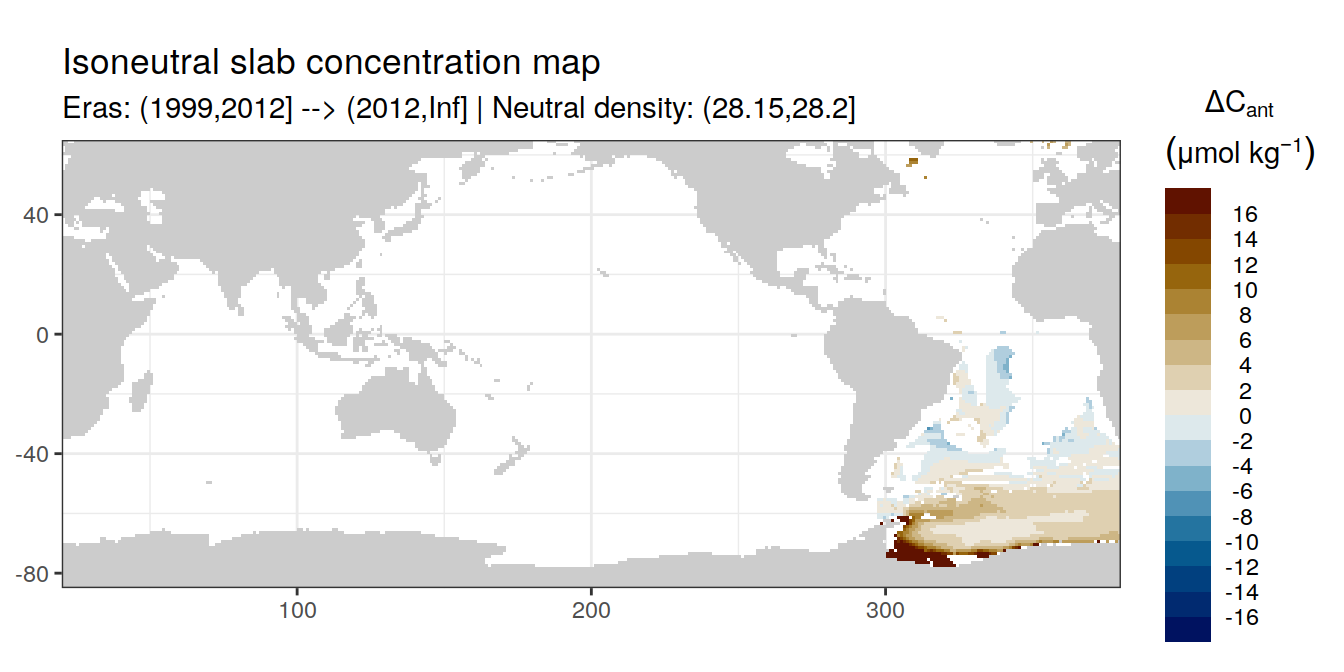
Isoneutral slab distribution
Mean of positive cant within each horizontal grid cell (lon x lat) per isoneutral slab.
Please note that:
density slabs covering values >28.1 occur by definition only either in the Atlantic or Indo-Pacific basin
gaps in the maps represent areas where (thin) density layers fit between discrete depth levels used for mapping
cant_gamma_maps <- m_cant_slab(cant_3d)
cant_gamma_maps <- cant_gamma_maps %>%
arrange(gamma_slab, eras)# i_eras <- unique(cant_gamma_maps$eras)[1]
# i_gamma_slab <- unique(cant_gamma_maps$gamma_slab)[1]
for (i_eras in unique(cant_gamma_maps$eras)) {
for (i_gamma_slab in unique(cant_gamma_maps$gamma_slab)) {
print(
p_map_cant_slab(
df = cant_gamma_maps %>%
filter(eras == i_eras,
gamma_slab == i_gamma_slab),
subtitle_text = paste(
"Eras:", i_eras,
"| Neutral density:", i_gamma_slab)
)
)
}
}
Past versions of cant_pos_gamma_slab_maps-1.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-2.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-3.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-4.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-5.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-6.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-7.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-8.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-9.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-10.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-11.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-12.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-13.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-14.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-15.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-16.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-17.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-18.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-19.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-20.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-21.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-22.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-23.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-24.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-25.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-26.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-27.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-28.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-29.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_gamma_slab_maps-30.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Inventory map
Column inventory of positive cant between the surface and 3000m water depth per horizontal grid cell (lat x lon).
# i_eras <- unique(cant_inv$eras)[1]
for (i_eras in unique(cant_inv$eras)) {
print(
p_map_cant_inv(
df = cant_inv %>% filter(eras == i_eras),
var = "cant_pos_inv",
subtitle_text = paste("Eras:", i_eras))
)
}
Past versions of cant_pos_inventory_map-1.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_pos_inventory_map-2.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
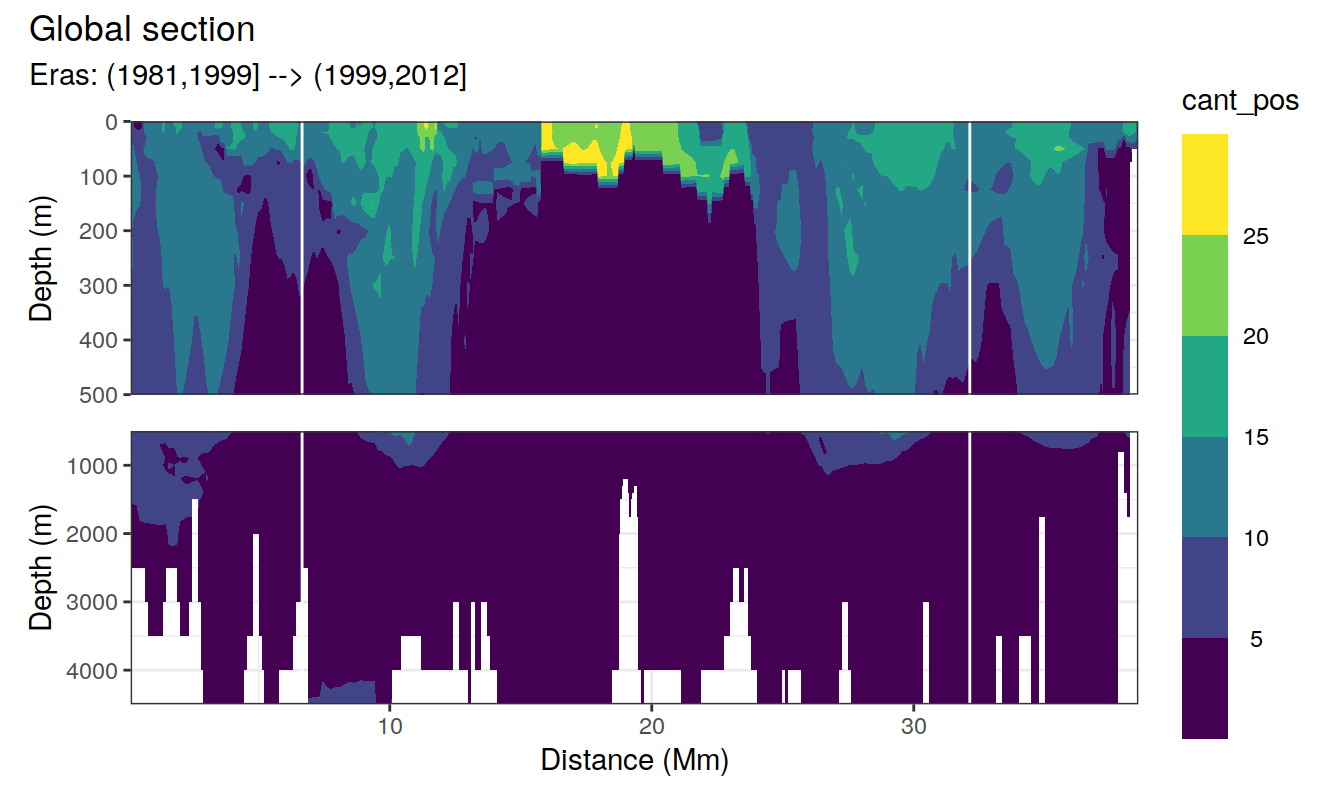
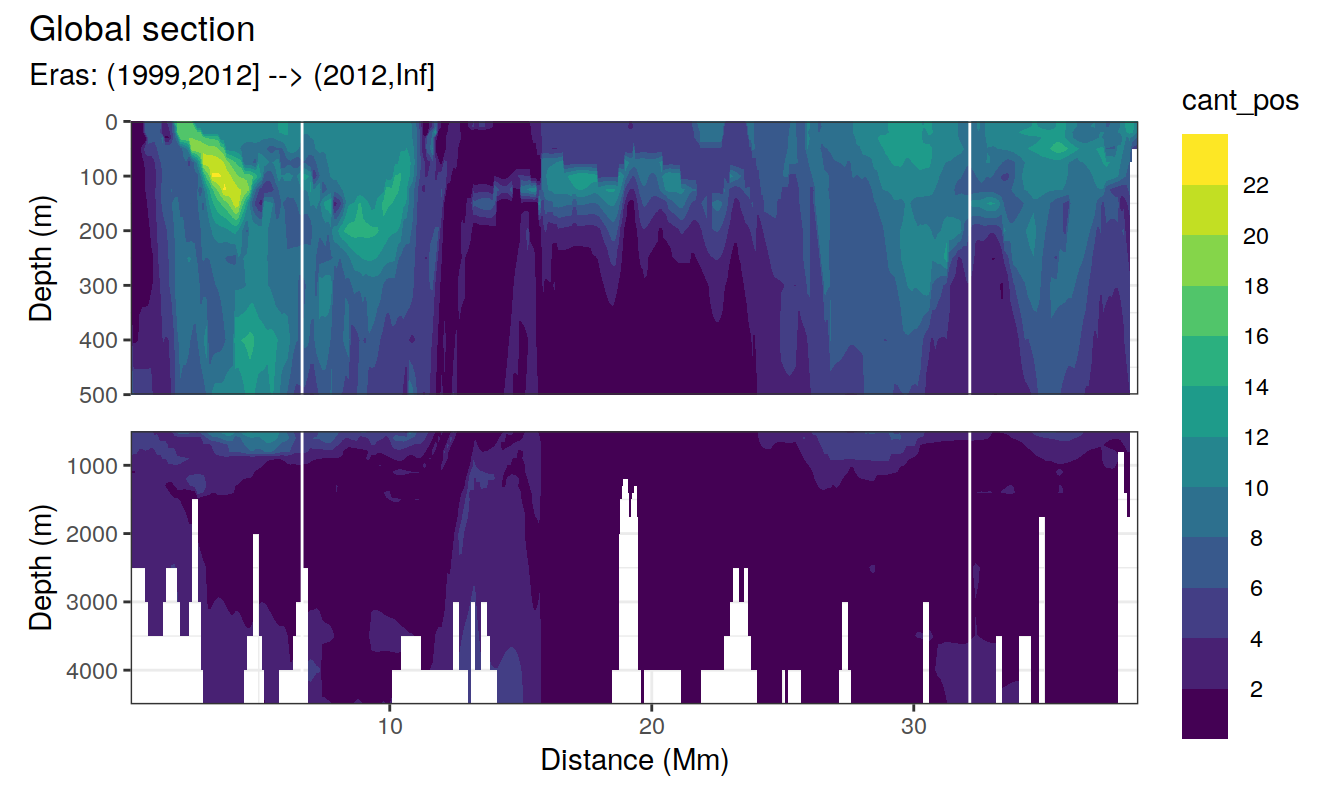
Global sections
for (i_eras in unique(cant_3d$eras)) {
print(
p_section_global(
df = cant_3d %>% filter(eras == i_eras),
var = "cant_pos",
subtitle_text = paste("Eras:", i_eras)
)
)
}
Past versions of cant_sections_positive_mean_one_lon_JGOFS_GO-1.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_positive_mean_one_lon_JGOFS_GO-2.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Cant - all
In a second series of plots we explore the distribution of cant, taking positive and negative estimates into account.
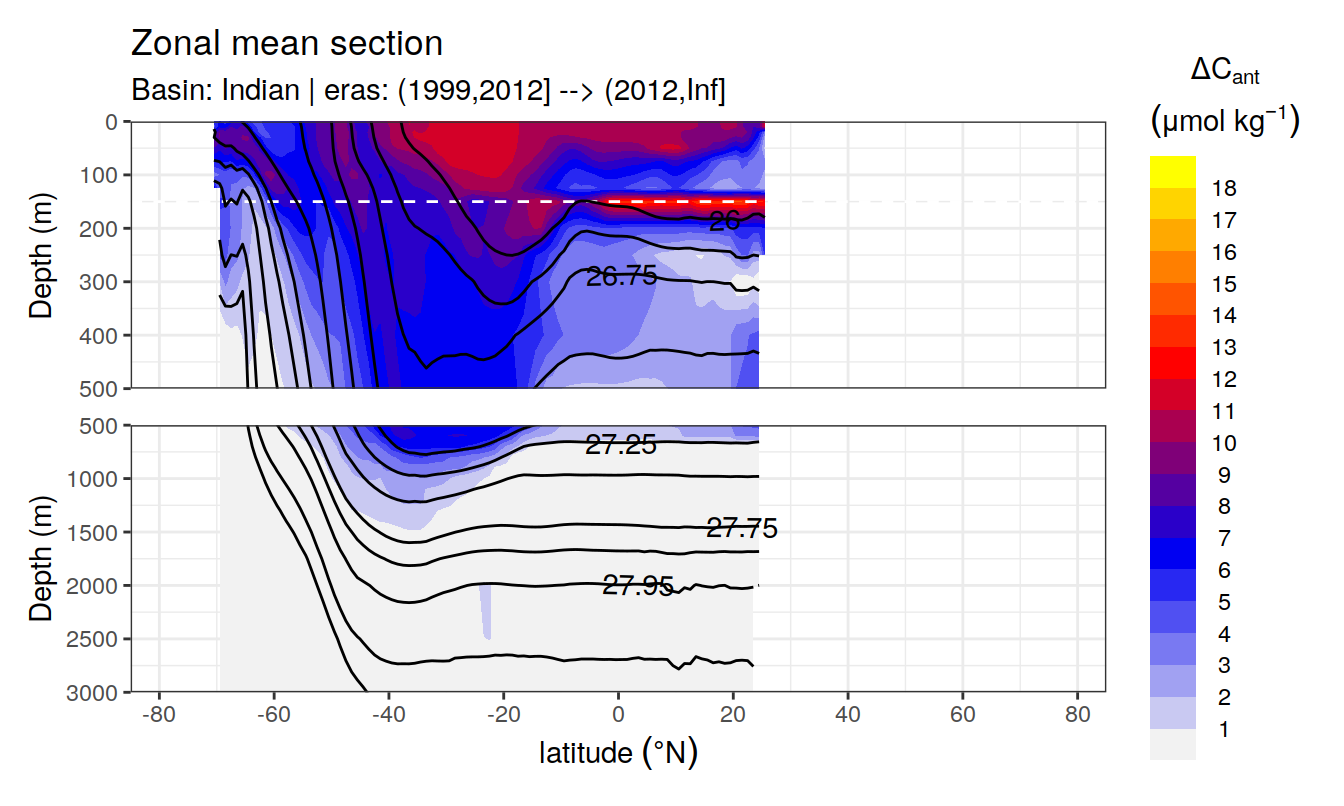
Zonal mean sections
# i_eras <- unique(cant_zonal$eras)[1]
# i_basin_AIP <- unique(cant_zonal$basin_AIP)[1]
for (i_basin_AIP in unique(cant_zonal$basin_AIP)) {
for (i_eras in unique(cant_zonal$eras)) {
print(
p_section_zonal(
df = cant_zonal %>%
filter(basin_AIP == i_basin_AIP,
eras == i_eras),
var = "cant_mean",
gamma = "gamma_mean",
breaks = params_global$breaks_cant,
col = "divergent",
subtitle_text =
paste("Basin:", i_basin_AIP, "| eras:", i_eras))
)
}
}
Past versions of cant_all_zonal_mean_sections-1.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_zonal_mean_sections-2.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_zonal_mean_sections-3.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_zonal_mean_sections-4.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_zonal_mean_sections-5.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_zonal_mean_sections-6.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
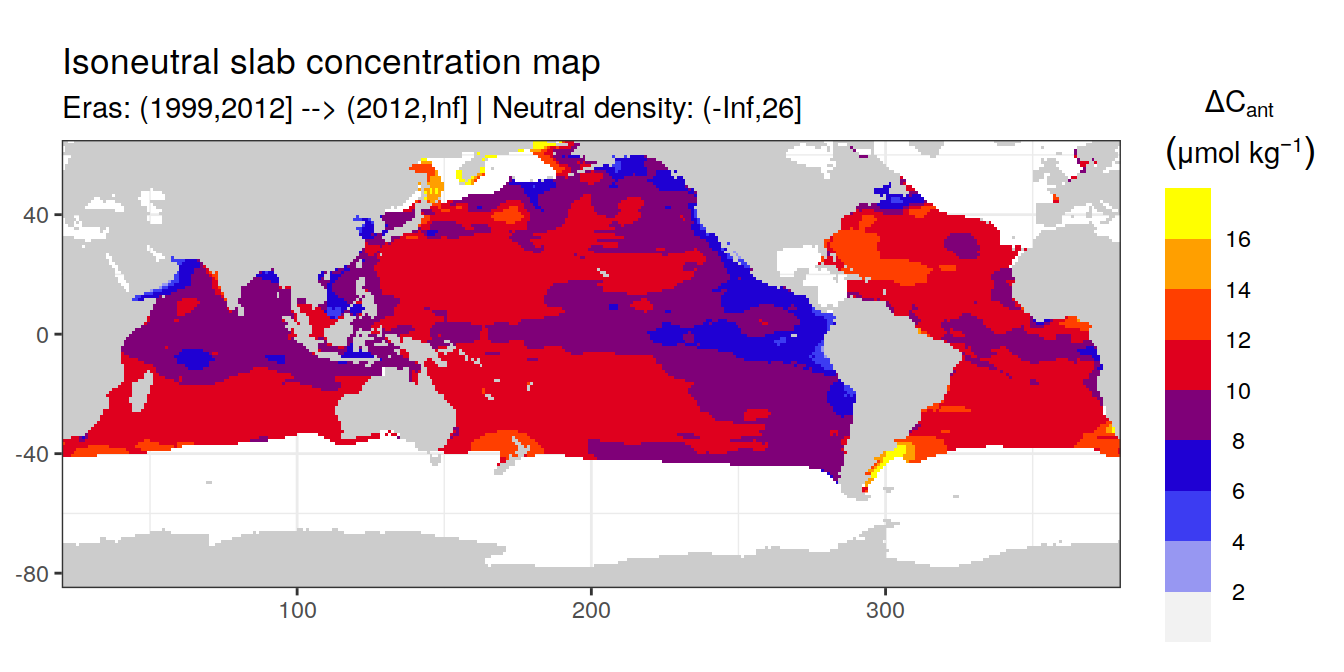
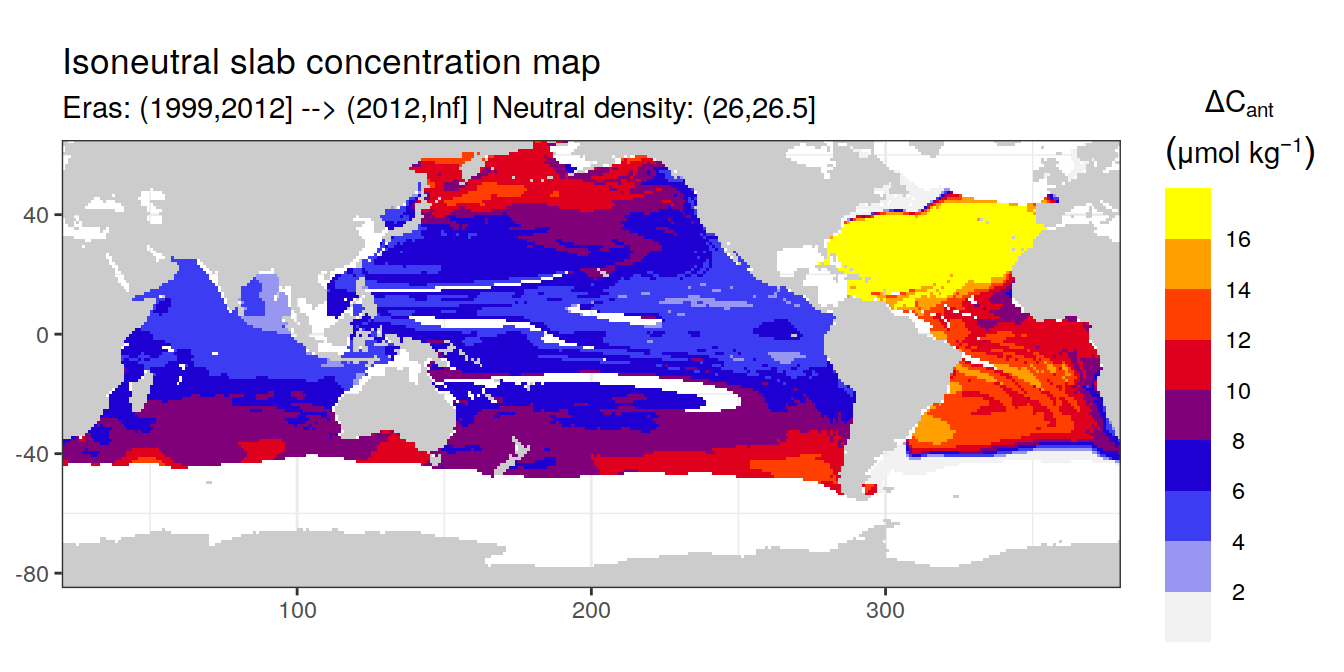
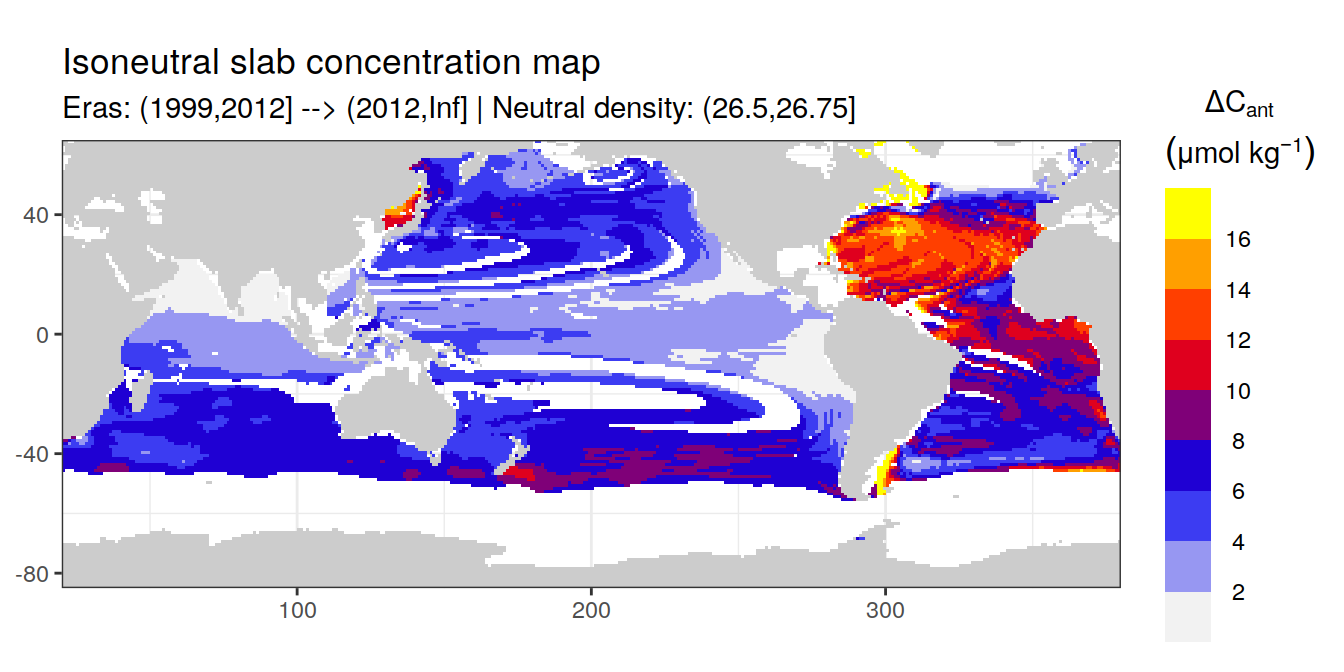
Isoneutral slab distribution
Mean of cant within each horizontal grid cell (lon x lat) per isoneutral slab.
Please note that:
density slabs covering values >28.1 occur by definition only either in the Atlantic or Indo-Pacific basin
gaps in the maps represent areas where (thin) density layers fit between discrete depth levels used for mapping
# i_eras <- unique(cant_gamma_maps$eras)[1]
# i_gamma_slab <- unique(cant_gamma_maps$gamma_slab)[5]
for (i_eras in unique(cant_gamma_maps$eras)) {
for (i_gamma_slab in unique(cant_gamma_maps$gamma_slab)) {
print(
p_map_cant_slab(
df = cant_gamma_maps %>%
filter(eras == i_eras,
gamma_slab == i_gamma_slab),
var = "cant",
col = "divergent",
subtitle_text = paste(
"Eras:", i_eras,
"| Neutral density:", i_gamma_slab))
)
}
}
Past versions of cant_all_gamma_slab_maps-1.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-2.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-3.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-4.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-5.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-6.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-7.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-8.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-9.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-10.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-11.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-12.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-13.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-14.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-15.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-16.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-17.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-18.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-19.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-20.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-21.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-22.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-23.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-24.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-25.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-26.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-27.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-28.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-29.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_gamma_slab_maps-30.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Inventory map
Column inventory of positive cant between the surface and 3000m water depth per horizontal grid cell (lat x lon).
# i_eras <- unique(cant_inv$eras)[1]
for (i_eras in unique(cant_inv$eras)) {
print(
p_map_cant_inv(
df = cant_inv %>% filter(eras == i_eras),
var = "cant_inv",
col = "divergent",
subtitle_text = paste("Eras:", i_eras))
)
}
Past versions of cant_all_inventory_map-1.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_all_inventory_map-2.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Cant - standard deviation
Across models
Standard deviation across Cant from all MLR models was calculate for each grid cell (XYZ). The zonal mean of this standard deviation should reflect the uncertainty associated to the predictor selection within each slab and era.
# i_eras <- unique(cant_zonal$eras)[1]
# i_basin_AIP <- unique(cant_zonal$basin_AIP)[2]
for (i_basin_AIP in unique(cant_zonal$basin_AIP)) {
for (i_eras in unique(cant_zonal$eras)) {
print(
p_section_zonal(
df = cant_zonal %>%
filter(basin_AIP == i_basin_AIP,
eras == i_eras),
var = "cant_sd_mean",
gamma = "gamma_mean",
legend_title = "sd",
title_text = "Zonal mean section of Cant sd between models",
subtitle_text =
paste("Basin:", i_basin_AIP, "| eras:", i_eras))
)
}
}
Past versions of cant_sections_sd_models-1.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_models-2.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_models-3.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_models-4.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_models-5.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_models-6.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Across basins
Standard deviation of mean cant values was calculate across all longitudes. This standard deviation should reflect the zonal variability of cant within the basin and era.
# i_eras <- unique(cant_zonal$eras)[1]
# i_basin_AIP <- unique(cant_zonal$basin_AIP)[2]
for (i_basin_AIP in unique(cant_zonal$basin_AIP)) {
for (i_eras in unique(cant_zonal$eras)) {
print(
p_section_zonal(
df = cant_zonal %>%
filter(basin_AIP == i_basin_AIP,
eras == i_eras),
var = "cant_sd",
gamma = "gamma_mean",
legend_title = "sd",
title_text = "Zonal mean section of sd between model mean Cant",
subtitle_text =
paste("Basin:", i_basin_AIP, "| eras:", i_eras))
)
}
}
Past versions of cant_sections_sd_cant-1.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_cant-2.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_cant-3.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_cant-4.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_cant-5.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_sections_sd_cant-6.png
Version
Author
Date
9eb4281
jens-daniel-mueller
2020-12-02
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Correlation
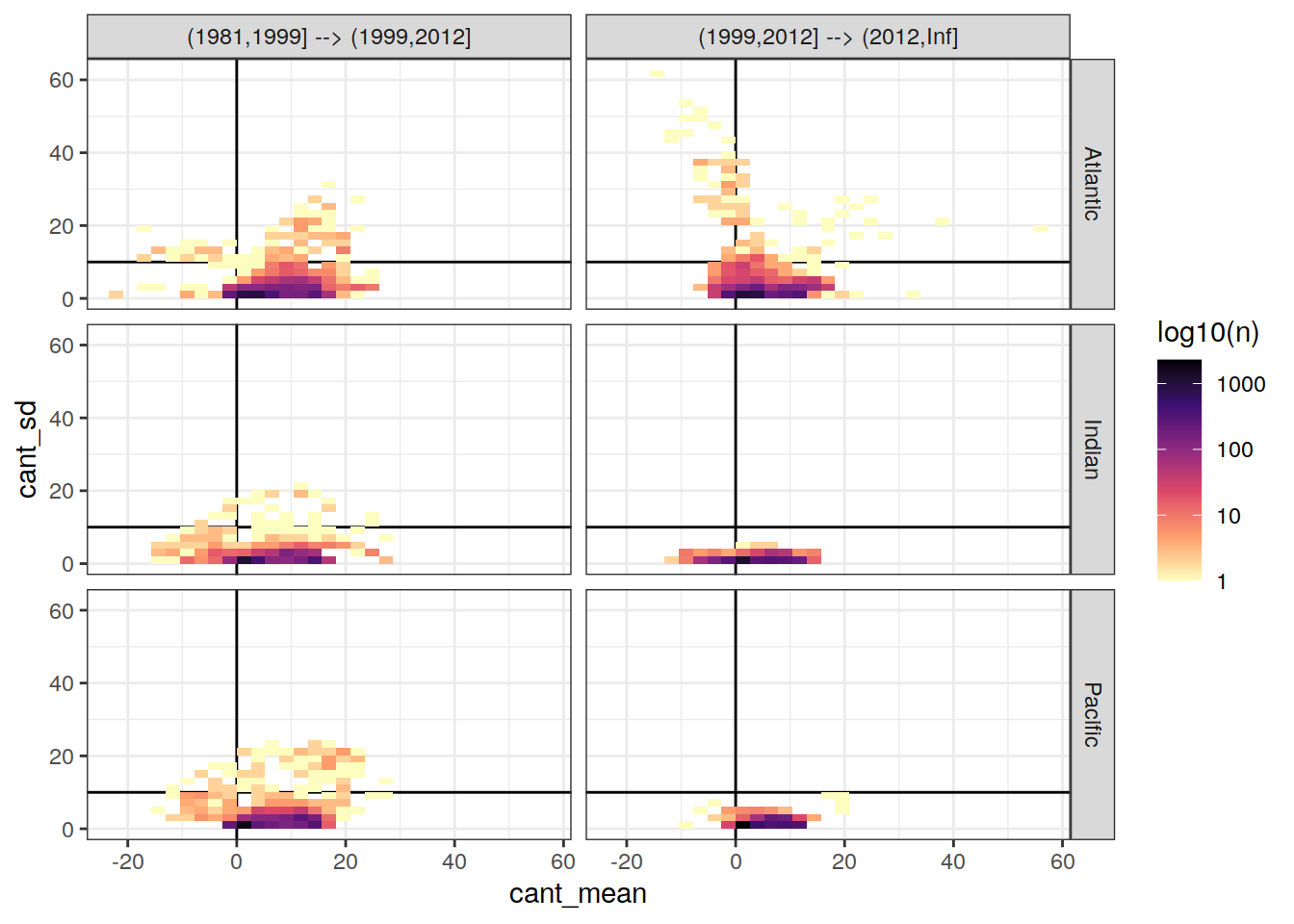
Cant vs model SD
# cant_3d <- cant_3d %>%
# mutate(eras = factor(eras, c("JGOFS_GO", "GO_new")))
cant_3d %>%
ggplot(aes(cant, cant_sd)) +
geom_vline(xintercept = 0) +
geom_hline(yintercept = 10) +
geom_bin2d() +
scale_fill_viridis_c(option = "magma",
direction = -1,
trans = "log10",
name = "log10(n)") +
facet_grid(basin_AIP ~ eras)
Past versions of cant_vs_sd_by_basin_era-1.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
cant_3d %>%
ggplot(aes(cant, cant_sd)) +
geom_vline(xintercept = 0) +
geom_hline(yintercept = 10) +
geom_bin2d() +
scale_fill_viridis_c(option = "magma",
direction = -1,
trans = "log10",
name = "log10(n)") +
facet_grid(gamma_slab ~ basin_AIP)
Past versions of cant_vs_sd_by_basin_gamma-1.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
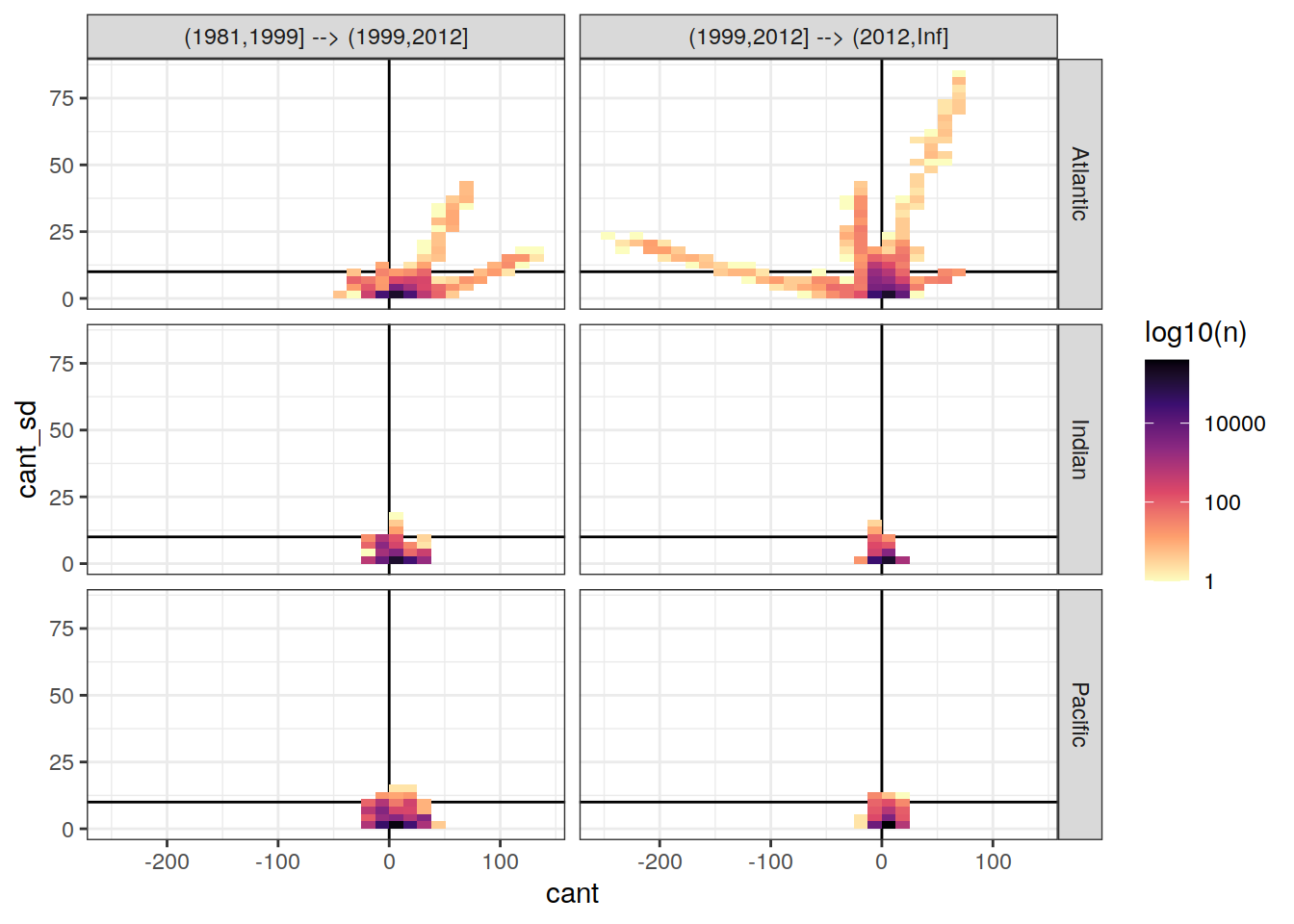
Cant vs regional SD
cant_zonal %>%
ggplot(aes(cant_mean, cant_sd)) +
geom_vline(xintercept = 0) +
geom_hline(yintercept = 10) +
geom_bin2d() +
scale_fill_viridis_c(option = "magma",
direction = -1,
trans = "log10",
name = "log10(n)") +
facet_grid(basin_AIP ~ eras)
Past versions of cant_vs_sd_by_basin_era_zonal-1.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
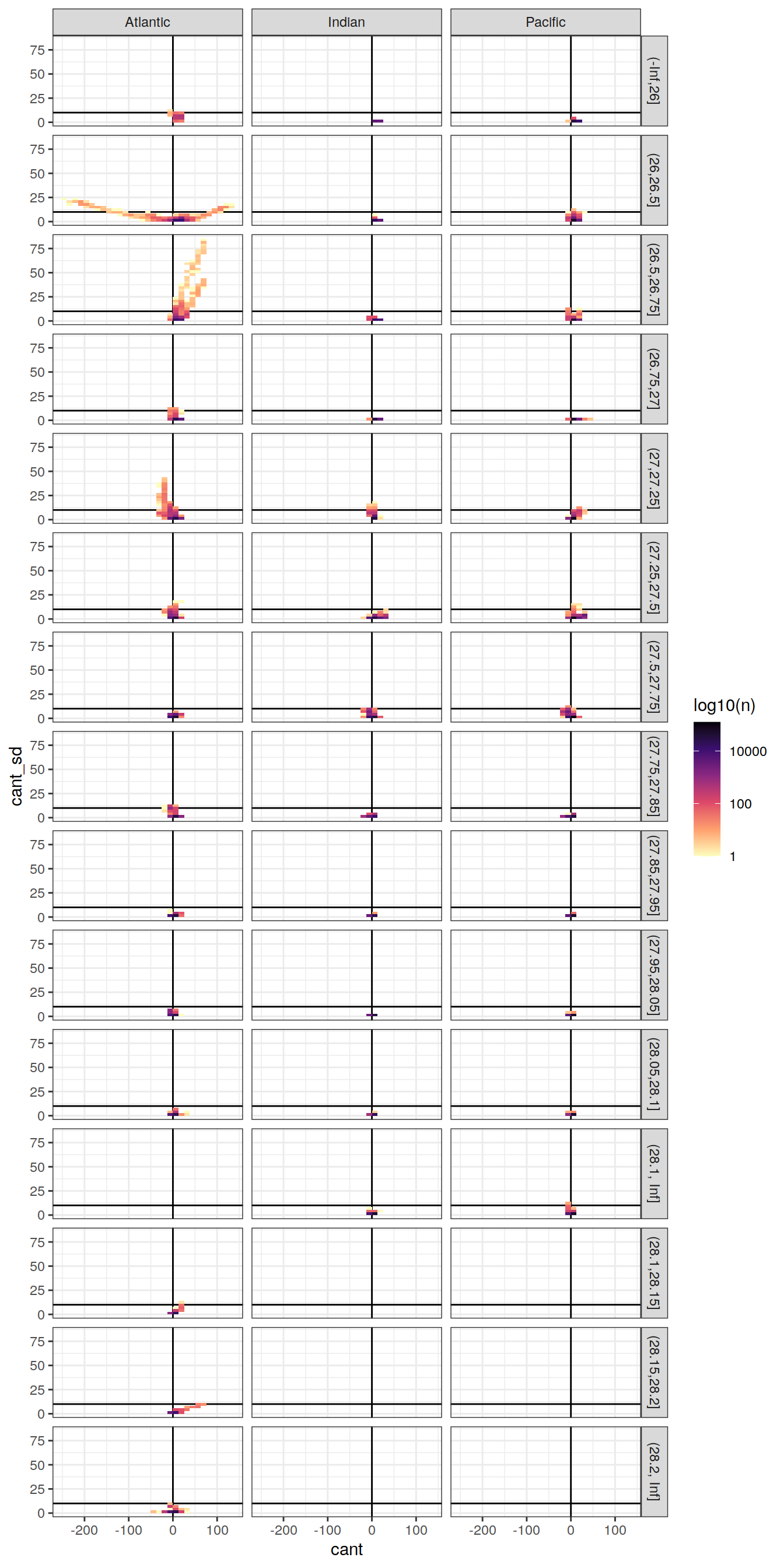
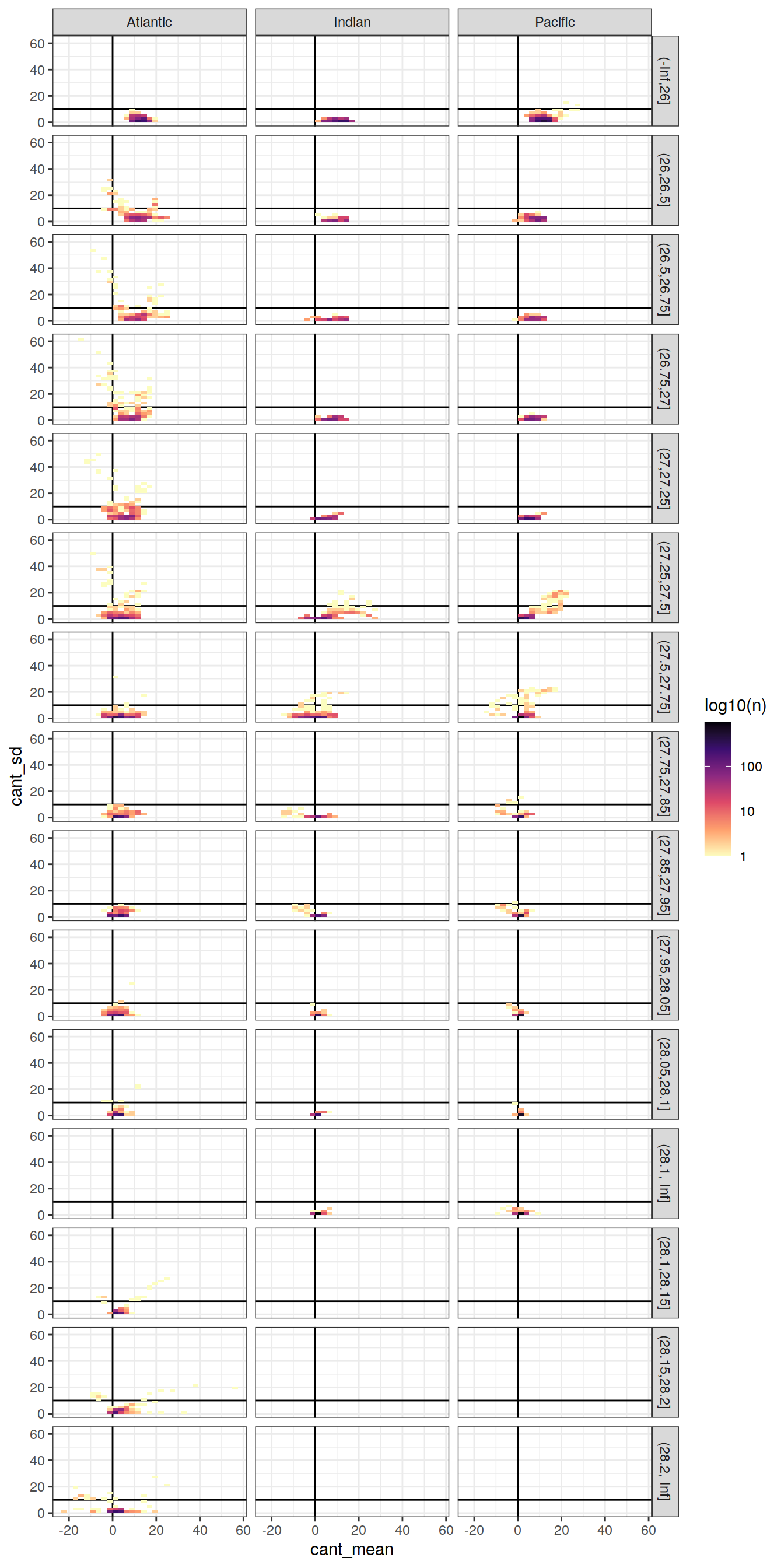
cant_zonal %>%
ggplot(aes(cant_mean, cant_sd)) +
geom_vline(xintercept = 0) +
geom_hline(yintercept = 10) +
geom_bin2d() +
scale_fill_viridis_c(option = "magma",
direction = -1,
trans = "log10",
name = "log10(n)") +
facet_grid(gamma_slab ~ basin_AIP)
Past versions of cant_vs_sd_by_basin_gamma_zonal-1.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Cant - predictor contribution
# cant_predictor_zonal <- cant_predictor_zonal %>%
# mutate(eras = factor(eras, c("JGOFS_GO", "GO_new")))
variable <- "cant_intercept"
for (variable in c(
"cant_intercept",
"cant_aou",
"cant_oxygen",
"cant_phosphate",
"cant_phosphate_star",
"cant_silicate",
"cant_tem",
"cant_sal")) {
print(
p_section_zonal_divergent_gamma_eras_basin(
df = cant_predictor_zonal,
var = variable,
gamma = "gamma")
)
}
Past versions of cant_section_predictor_contribution-1.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_section_predictor_contribution-2.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_section_predictor_contribution-3.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_section_predictor_contribution-4.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_section_predictor_contribution-5.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_section_predictor_contribution-6.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_section_predictor_contribution-7.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
Past versions of cant_section_predictor_contribution-8.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
ee9bf6d
jens-daniel-mueller
2020-12-01
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
rm(variable)
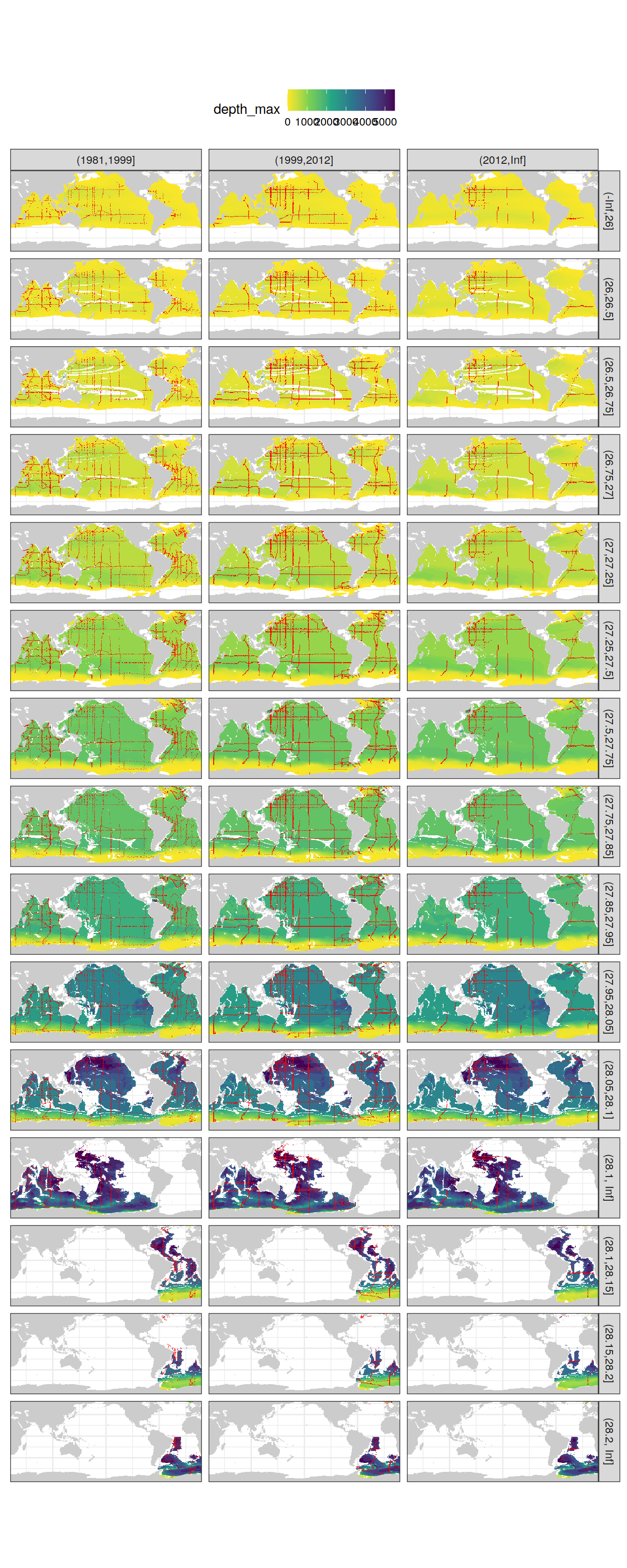
Neutral density
Slab depth
Please note that:
density slabs covering values >28.1 occur by definition only either in the Atlantic or Indo-Pacific basin
predictor density slabs are only shown for the upper 3000m as used for the mapping, whereas GLODAP observations are displayed for the entire water column as used for fitting eMLRs (in both cases shallow waters are excluded at low density)
GLODAP_obs_coverage <- GLODAP %>%
count(lat, lon, gamma_slab, era)
# %>%
# mutate(era = factor(era, c("JGOFS_WOCE", "GO_SHIP", "new_era")))
map +
geom_raster(data = cant_gamma_maps,
aes(lon, lat, fill = depth_max)) +
geom_raster(data = GLODAP_obs_coverage,
aes(lon, lat), fill = "red") +
facet_grid(gamma_slab ~ era) +
scale_fill_viridis_c(direction = -1) +
theme(axis.ticks = element_blank(),
axis.text = element_blank(),
legend.position = "top")
Past versions of gamma_maps-1.png
Version
Author
Date
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
bc61ce3
Jens Müller
2020-11-30
rm(GLODAP_obs_coverage)
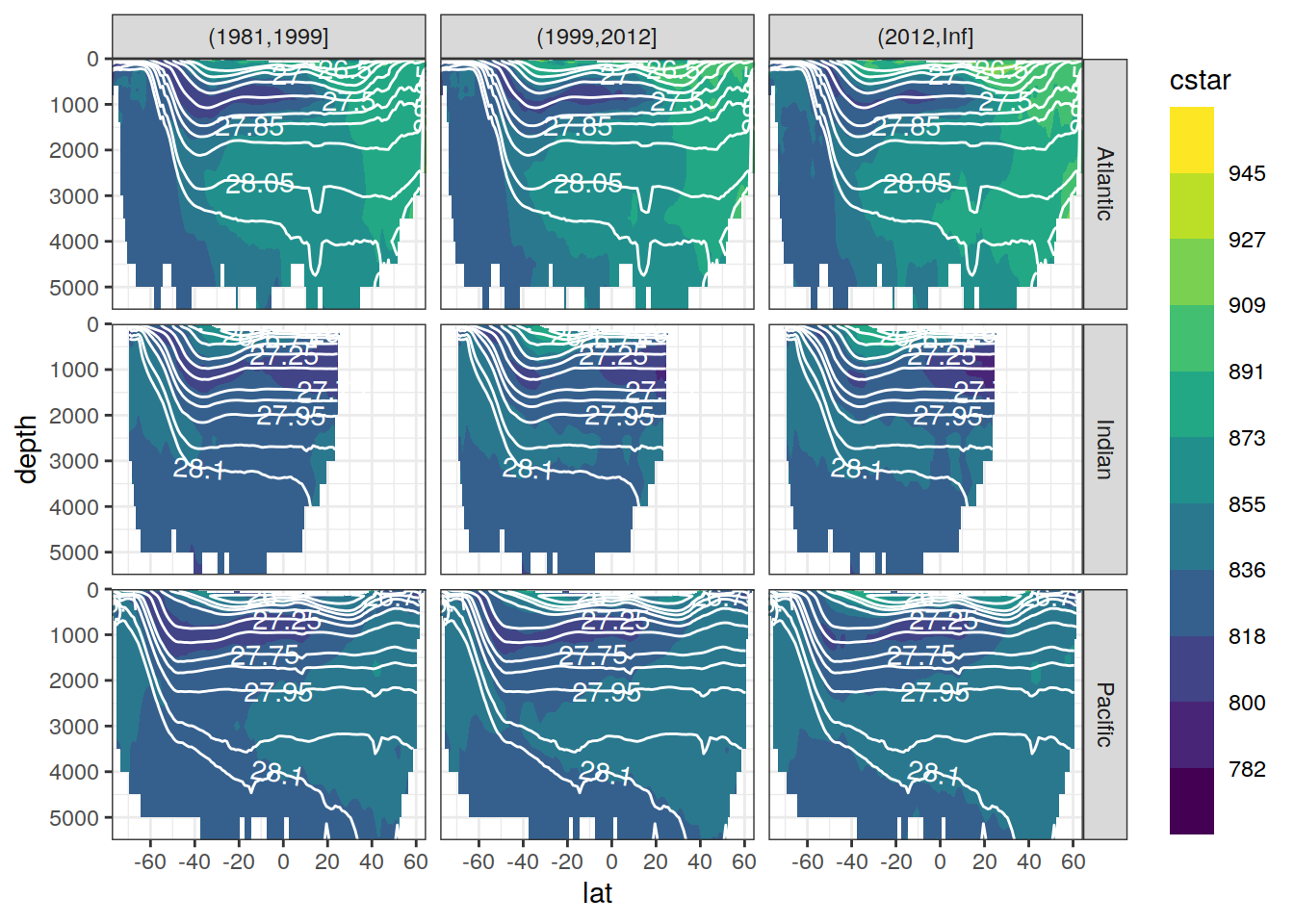
C*
Zonal mean sections
# cstar_zonal <- cstar_zonal %>%
# mutate(era = factor(era, c("JGOFS_WOCE", "GO_SHIP", "new_era")))
slab_breaks <- c(params_local$slabs_Atl[1:12],Inf)
cstar_zonal %>%
ggplot(aes(lat, depth, z = cstar_mean)) +
geom_contour_filled(bins = 11) +
scale_fill_viridis_d(name = "cstar") +
geom_contour(aes(lat, depth, z = gamma_mean),
breaks = slab_breaks,
col = "white") +
geom_text_contour(
aes(lat, depth, z = gamma_mean),
breaks = slab_breaks,
col = "white",
skip = 1
) +
scale_y_reverse() +
scale_x_continuous(breaks = seq(-100,100,20)) +
coord_cartesian(expand = 0) +
guides(fill = guide_colorsteps(barheight = unit(10, "cm"))) +
facet_grid(basin_AIP ~ era)
Past versions of cstar_zonal_mean_era-1.png
Version
Author
Date
902f65a
jens-daniel-mueller
2020-12-02
0ff728b
jens-daniel-mueller
2020-12-01
92edddb
jens-daniel-mueller
2020-12-01
196be51
jens-daniel-mueller
2020-11-30
rm(slab_breaks)
Sections by model
Zonal sections plots are produced for every 20° longitude, each era and for all models individually and can be downloaded here .
Known issues
Deviations between this study and the results by Gruber et al (2019), short G19, for the same period, might be attributable to following known differences in the implementation of the eMLR(C*) method:
GLODAPv2_2020 here vs an extended version of GLODAPv2 in G19
flagging: Here, we accept f flags 0 and 2 (except for tco2, where only 0 is accepted). G19 claim to use 0 throughout, yet have a high coverage of talk observations in the SE Pacific
Neutral density calculation: Here and in GLODAPv2_2020 a polynomial approximation is used, whereas G19 uses the original Matlab code
Predictor climatology: Here we used WOA18, whereas G19 used WOA13
Missing data in the GLODAP mapped climatology, eg NO3 at surface, where not filled in this study
cant on neutral density levels calculate as slab mean, rather than on one surface
Here, surface delta cant were calculated based on Luecker constants, rather than Mehrbach as in G19
Here, pCO2 was calculated from DIC/TA Climatology
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: openSUSE Leap 15.1
Matrix products: default
BLAS: /usr/local/R-4.0.3/lib64/R/lib/libRblas.so
LAPACK: /usr/local/R-4.0.3/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] kableExtra_1.3.1 marelac_2.1.10 shape_1.4.5 scales_1.1.1
[5] metR_0.9.0 scico_1.2.0 patchwork_1.1.0 collapse_1.4.2
[9] forcats_0.5.0 stringr_1.4.0 dplyr_1.0.2 purrr_0.3.4
[13] readr_1.4.0 tidyr_1.1.2 tibble_3.0.4 ggplot2_3.3.2
[17] tidyverse_1.3.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.0 lubridate_1.7.9 gsw_1.0-5
[4] webshot_0.5.2 httr_1.4.2 rprojroot_2.0.2
[7] tools_4.0.3 backports_1.1.10 R6_2.5.0
[10] DBI_1.1.0 colorspace_2.0-0 withr_2.3.0
[13] tidyselect_1.1.0 compiler_4.0.3 git2r_0.27.1
[16] cli_2.2.0 rvest_0.3.6 xml2_1.3.2
[19] isoband_0.2.2 labeling_0.4.2 checkmate_2.0.0
[22] digest_0.6.27 rmarkdown_2.5 oce_1.2-0
[25] pkgconfig_2.0.3 htmltools_0.5.0 dbplyr_1.4.4
[28] highr_0.8 rlang_0.4.9 readxl_1.3.1
[31] rstudioapi_0.13 farver_2.0.3 generics_0.0.2
[34] jsonlite_1.7.1 magrittr_2.0.1 Matrix_1.2-18
[37] Rcpp_1.0.5 munsell_0.5.0 fansi_0.4.1
[40] lifecycle_0.2.0 stringi_1.5.3 whisker_0.4
[43] yaml_2.2.1 plyr_1.8.6 grid_4.0.3
[46] blob_1.2.1 parallel_4.0.3 promises_1.1.1
[49] crayon_1.3.4 lattice_0.20-41 haven_2.3.1
[52] hms_0.5.3 seacarb_3.2.14 knitr_1.30
[55] pillar_1.4.7 reprex_0.3.0 glue_1.4.2
[58] evaluate_0.14 RcppArmadillo_0.10.1.2.0 data.table_1.13.2
[61] modelr_0.1.8 vctrs_0.3.5 httpuv_1.5.4
[64] testthat_3.0.0 cellranger_1.1.0 gtable_0.3.0
[67] assertthat_0.2.1 xfun_0.18 broom_0.7.2
[70] RcppEigen_0.3.3.7.0 later_1.1.0.1 viridisLite_0.3.0
[73] memoise_1.1.0 ellipsis_0.3.1 here_0.1