Demographic Data
Tina Lasisi
2024-03-03 20:03:14
Last updated: 2024-03-03
Checks: 7 0
Knit directory: PODFRIDGE/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230302) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2596546. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: output/.DS_Store
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/demo-data.Rmd) and HTML
(docs/demo-data.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2596546 | Tina Lasisi | 2024-03-03 | wflow_publish("analysis/*", republish = TRUE, all = TRUE, verbose = TRUE) |
| html | 48acb9f | Tina Lasisi | 2024-03-02 | Build site. |
| Rmd | 5352065 | Tina Lasisi | 2024-03-02 | workflowr::wflow_publish(files = "analysis/*", all = TRUE, update = TRUE, |
| html | aa3ff5c | Tina Lasisi | 2024-03-01 | Build site. |
| html | 9e71347 | Tina Lasisi | 2024-01-22 | Build site. |
| html | c280b6f | Tina Lasisi | 2023-04-16 | Build site. |
| html | f89a90f | Tina Lasisi | 2023-04-16 | Build site. |
| html | 5f805fe | Tina Lasisi | 2023-03-06 | Build site. |
| Rmd | 5106bab | Tina Lasisi | 2023-03-06 | add analyses |
| html | c3948af | Tina Lasisi | 2023-03-04 | Build site. |
| html | f02bc38 | Tina Lasisi | 2023-03-03 | Build site. |
| html | c9130d5 | Tina Lasisi | 2023-03-03 | wflow_git_commit(all = TRUE) |
| html | a4a7d45 | Tina Lasisi | 2023-03-03 | Build site. |
| html | 00073fd | Tina Lasisi | 2023-03-03 | Build site. |
| html | 51ed5a6 | Tina Lasisi | 2023-03-02 | Build site. |
| Rmd | 13ed9ae | Tina Lasisi | 2023-03-02 | Publishing POPFORGE |
| html | 13ed9ae | Tina Lasisi | 2023-03-02 | Publishing POPFORGE |
# Load necessary packages
library(wesanderson) # for color palettes
library(RColorBrewer)
library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.4.4 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.0
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorslibrary(patchwork)
knitr::opts_knit$set(root.dir = "..")
knitr::opts_chunk$set(eval = TRUE, echo = FALSE, warning = FALSE, fig.width = 7, fig.height = 6)Demographic data
The population size data (as well as number of children) in our analyses is based on the US Census data hosted at IPUMS USA.
Detailed demographic data
The website here has some figures from an exploratory analysis of birth rate-related data from IPUMS. Due to the terms of agreement for using this data, we cannot share the full dataset but our repo contains the subset that was used to calculate the mean number of offspring and variance.
“STANDARD REDISTRIBUTION TERM You will not redistribute the data without permission. You may publish a subset of the data to meet journal requirements for accessing data related to a particular publication. Contact us for permission for any other redistribution; we will consider requests for free and commercial redistribution.”
citation: Steven Ruggles, Sarah Flood, Matthew Sobek, Daniel Backman, Annie Chen, Grace Cooper, Stephanie Richards, Renae Rogers, and Megan Schouweiler. IPUMS USA: Version 14.0 [dataset]. Minneapolis, MN: IPUMS, 2023. https://doi.org/10.18128/D010.V14.0
Number of children
We start by loading our pre-processed dataset which focuses on the number of children per family from the US Census data.
Attaching package: 'data.table'The following objects are masked from 'package:lubridate':
hour, isoweek, mday, minute, month, quarter, second, wday, week,
yday, yearThe following objects are masked from 'package:dplyr':
between, first, lastThe following object is masked from 'package:purrr':
transposeNext, we identify the unique values of the number of children to understand the range of family sizes.
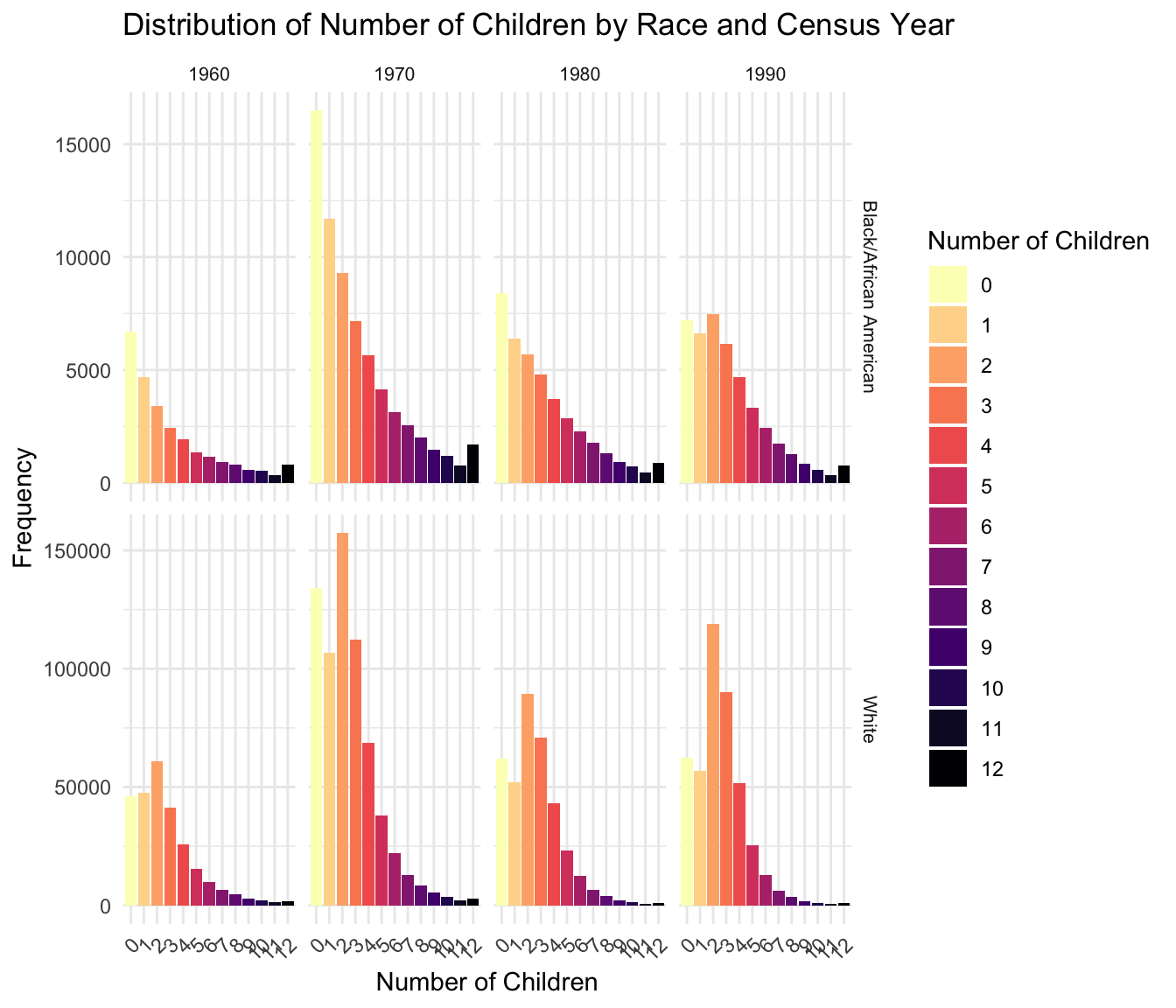
[1] 0 1 2 3 4 5 6 7 8 9 10 11 12We then create a frequency table to analyze the distribution of family sizes across race and census years.
RACE YEAR chborn_num n
1 Black/African American 1960 0 6697
2 Black/African American 1960 1 4698
3 Black/African American 1960 2 3411
4 Black/African American 1960 3 2445
5 Black/African American 1960 4 1949
6 Black/African American 1960 5 1361
7 Black/African American 1960 6 1162
8 Black/African American 1960 7 932
9 Black/African American 1960 8 810
10 Black/African American 1960 9 588
11 Black/African American 1960 10 549
12 Black/African American 1960 11 326
13 Black/African American 1960 12 821
14 Black/African American 1970 0 16490
15 Black/African American 1970 1 11686
16 Black/African American 1970 2 9275
17 Black/African American 1970 3 7161
18 Black/African American 1970 4 5659
19 Black/African American 1970 5 4147
20 Black/African American 1970 6 3147
21 Black/African American 1970 7 2542
22 Black/African American 1970 8 2012
23 Black/African American 1970 9 1473
24 Black/African American 1970 10 1194
25 Black/African American 1970 11 784
26 Black/African American 1970 12 1682
27 Black/African American 1980 0 8417
28 Black/African American 1980 1 6383
29 Black/African American 1980 2 5681
30 Black/African American 1980 3 4797
31 Black/African American 1980 4 3732
32 Black/African American 1980 5 2870
33 Black/African American 1980 6 2264
34 Black/African American 1980 7 1782
35 Black/African American 1980 8 1310
36 Black/African American 1980 9 943
37 Black/African American 1980 10 717
38 Black/African American 1980 11 452
39 Black/African American 1980 12 870
40 Black/African American 1990 0 7193
41 Black/African American 1990 1 6635
42 Black/African American 1990 2 7485
43 Black/African American 1990 3 6161
44 Black/African American 1990 4 4673
45 Black/African American 1990 5 3333
46 Black/African American 1990 6 2445
47 Black/African American 1990 7 1752
48 Black/African American 1990 8 1263
49 Black/African American 1990 9 853
50 Black/African American 1990 10 590
51 Black/African American 1990 11 348
52 Black/African American 1990 12 772
53 White 1960 0 46202
54 White 1960 1 47433
55 White 1960 2 60732
56 White 1960 3 41272
57 White 1960 4 25666
58 White 1960 5 15327
59 White 1960 6 9697
60 White 1960 7 6347
61 White 1960 8 4518
62 White 1960 9 2990
63 White 1960 10 2148
64 White 1960 11 1280
65 White 1960 12 1747
66 White 1970 0 133940
67 White 1970 1 106663
68 White 1970 2 157405
69 White 1970 3 112397
70 White 1970 4 68603
71 White 1970 5 38000
72 White 1970 6 22023
73 White 1970 7 12927
74 White 1970 8 8534
75 White 1970 9 5342
76 White 1970 10 3593
77 White 1970 11 2082
78 White 1970 12 2858
79 White 1980 0 61909
80 White 1980 1 51856
81 White 1980 2 89551
82 White 1980 3 70716
83 White 1980 4 43190
84 White 1980 5 23170
85 White 1980 6 12556
86 White 1980 7 6589
87 White 1980 8 3874
88 White 1980 9 2254
89 White 1980 10 1368
90 White 1980 11 757
91 White 1980 12 1073
92 White 1990 0 62471
93 White 1990 1 56647
94 White 1990 2 119054
95 White 1990 3 90170
96 White 1990 4 51511
97 White 1990 5 25385
98 White 1990 6 12698
99 White 1990 7 6292
100 White 1990 8 3479
101 White 1990 9 1876
102 White 1990 10 1132
103 White 1990 11 623
104 White 1990 12 828Loading required package: viridisLite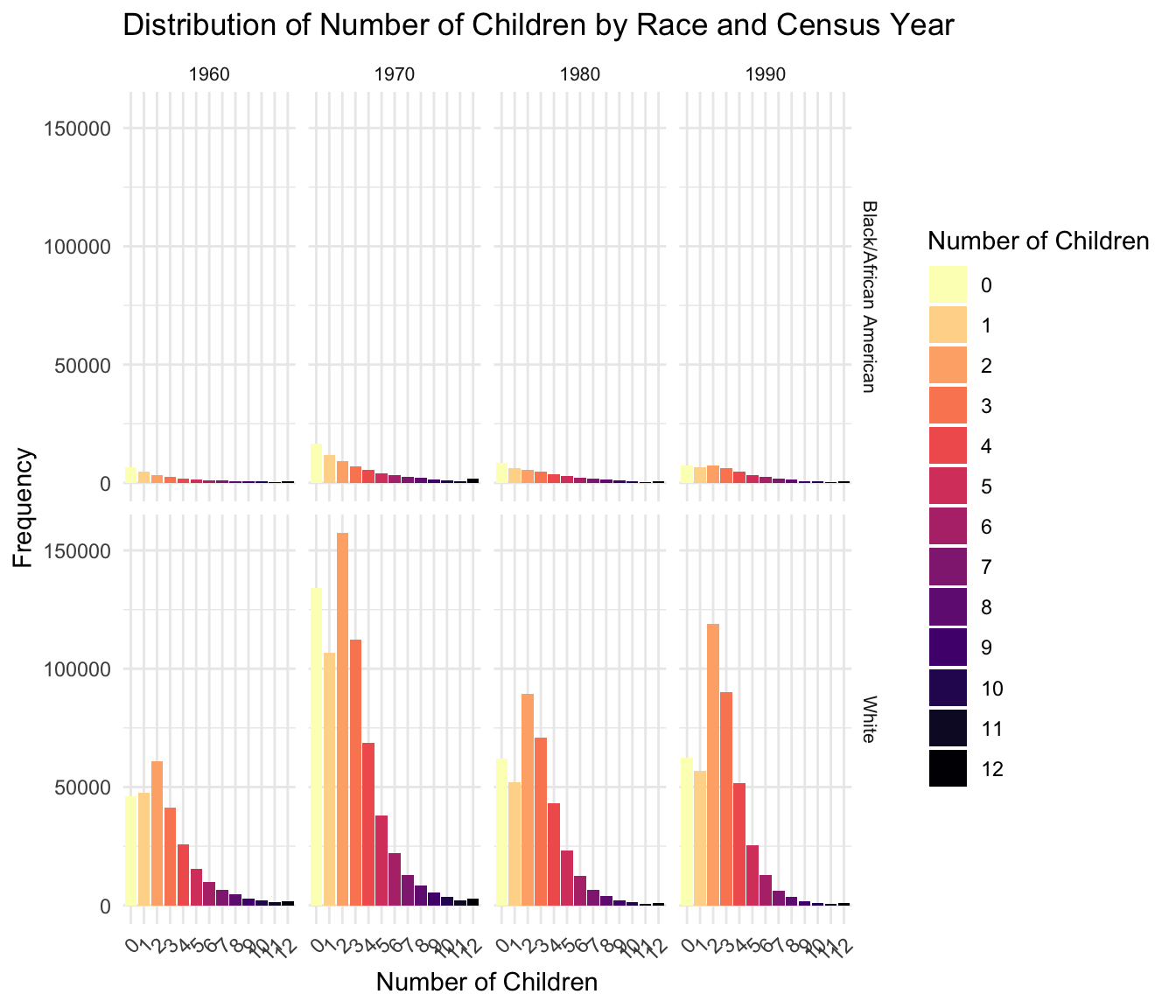
| Version | Author | Date |
|---|---|---|
| 48acb9f | Tina Lasisi | 2024-03-02 |
| Version | Author | Date |
|---|---|---|
| 48acb9f | Tina Lasisi | 2024-03-02 |
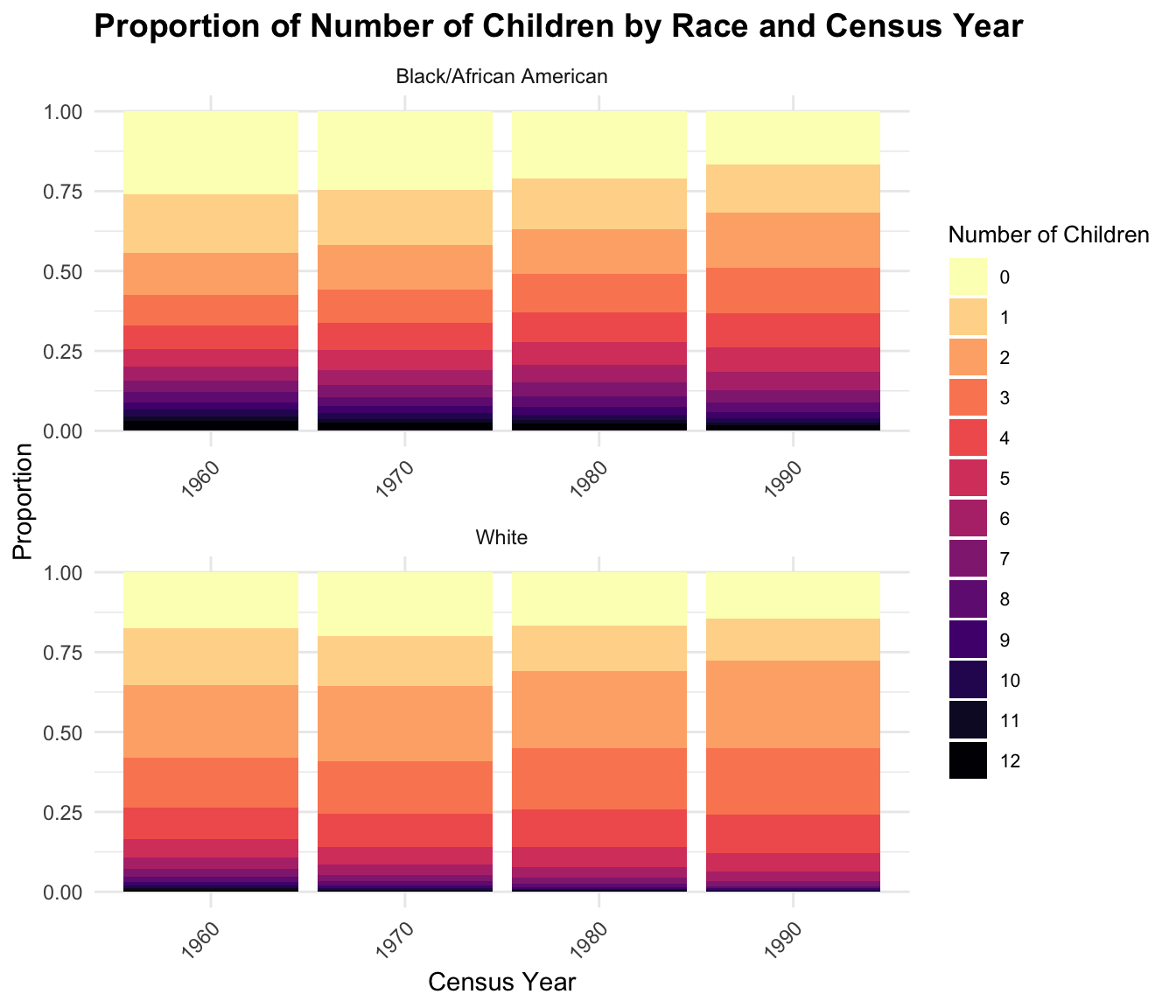
To compare distributions within the context of the total population, we calculate and visualize proportions.
| Version | Author | Date |
|---|---|---|
| 48acb9f | Tina Lasisi | 2024-03-02 |
And we summarize the mean and variance per year
`summarise()` has grouped output by 'RACE'. You can override using the
`.groups` argument.# A tibble: 8 × 4
# Groups: RACE [2]
RACE YEAR Mean Variance
<chr> <int> <dbl> <dbl>
1 Black/African American 1960 3.02 10.5
2 Black/African American 1970 3.00 9.57
3 Black/African American 1980 3.21 9.21
4 Black/African American 1990 3.20 7.84
5 White 1960 2.62 5.47
6 White 1970 2.45 4.69
7 White 1980 2.55 4.10
8 White 1990 2.53 3.46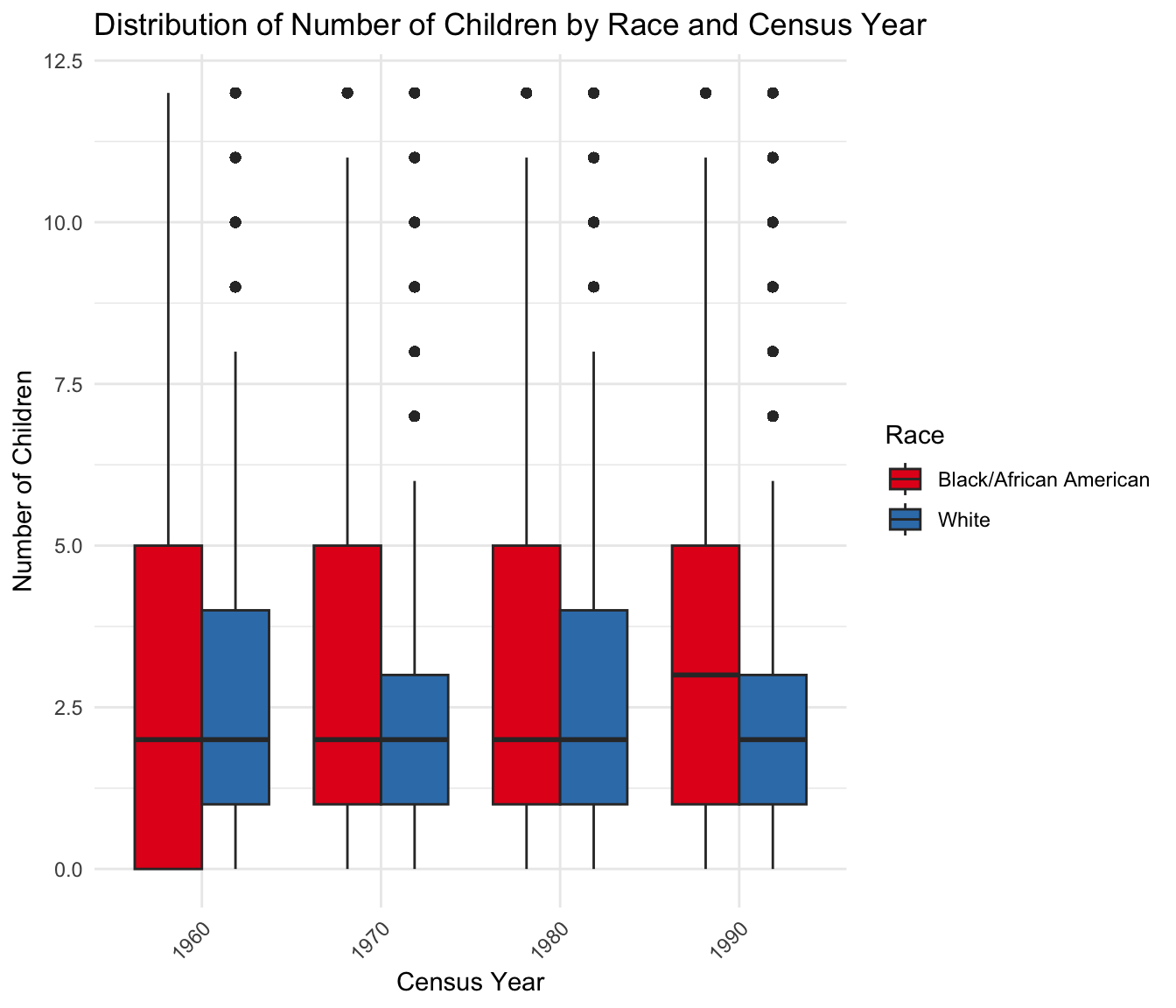
| Version | Author | Date |
|---|---|---|
| 48acb9f | Tina Lasisi | 2024-03-02 |
Database size
What are realistic database sizes for US European- and African American populations? From 23andMe publications it seems that 80% of their customers identify as White (non-Hispanic) and that around 3% of their customers identify as African American or Black (see here where they say that their sample represents their customer database and the US population). This broadly agrees with data seen in a 23andme poster presented in 2011 (see here)
For now, the analyses will use an estimate of 80% for White Americans and 5% for Black Americans in the DTC databases.
R version 4.3.2 (2023-10-31)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS Sonoma 14.3.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/Detroit
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] viridis_0.6.5 viridisLite_0.4.2 data.table_1.14.10 patchwork_1.2.0
[5] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4
[9] purrr_1.0.2 readr_2.1.5 tidyr_1.3.0 tibble_3.2.1
[13] ggplot2_3.4.4 tidyverse_2.0.0 RColorBrewer_1.1-3 wesanderson_0.3.7
[17] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] gtable_0.3.4 xfun_0.41 bslib_0.6.1 processx_3.8.3
[5] callr_3.7.3 tzdb_0.4.0 vctrs_0.6.5 tools_4.3.2
[9] ps_1.7.5 generics_0.1.3 fansi_1.0.6 highr_0.10
[13] pkgconfig_2.0.3 lifecycle_1.0.4 compiler_4.3.2 farver_2.1.1
[17] git2r_0.33.0 munsell_0.5.0 getPass_0.2-4 httpuv_1.6.13
[21] htmltools_0.5.7 sass_0.4.8 yaml_2.3.8 later_1.3.2
[25] pillar_1.9.0 jquerylib_0.1.4 whisker_0.4.1 cachem_1.0.8
[29] tidyselect_1.2.0 digest_0.6.34 stringi_1.8.3 labeling_0.4.3
[33] rprojroot_2.0.4 fastmap_1.1.1 grid_4.3.2 colorspace_2.1-0
[37] cli_3.6.2 magrittr_2.0.3 utf8_1.2.4 withr_2.5.2
[41] scales_1.3.0 promises_1.2.1 timechange_0.2.0 rmarkdown_2.25
[45] httr_1.4.7 gridExtra_2.3 hms_1.1.3 evaluate_0.23
[49] knitr_1.45 rlang_1.1.3 Rcpp_1.0.12 glue_1.7.0
[53] rstudioapi_0.15.0 jsonlite_1.8.8 R6_2.5.1 fs_1.6.3