PCA
Last updated: 2025-02-04
Checks: 6 1
Knit directory: sapphire/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240923) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/sapphire/data/serum_vit_D_study_with_lab_results.xlsx | data/serum_vit_D_study_with_lab_results.xlsx |
| ~/sapphire/data/sun_expos_data/sun_expos_long.csv | data/sun_expos_data/sun_expos_long.csv |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 564ea55. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Untracked files:
Untracked: .lock_gPLINK
Untracked: .metafile_gPLINK
Untracked: SAfrADMIX/
Untracked: analysis/genotype.R
Untracked: analysis/lilyanalysis.Rmd
Untracked: data/dist_pops.csv
Untracked: data/summer_winter.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/pca.Rmd) and HTML
(docs/pca.html) files. If you’ve configured a remote Git
repository (see ?wflow_git_remote), click on the hyperlinks
in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 564ea55 | Lily Heald | 2025-02-04 | Build site. |
| Rmd | d9fb6da | Lily Heald | 2025-02-04 | wflow |
| html | d9fb6da | Lily Heald | 2025-02-04 | wflow |
| Rmd | 517cf8d | Lily Heald | 2025-02-04 | More analyses |
| Rmd | 131b1d2 | Lily Heald | 2025-01-22 | initial PCAs |
file_path <- "~/sapphire/data/serum_vit_D_study_with_lab_results.xlsx"
data_summer <- read_excel(file_path, sheet = "ScreeningDataCollectionSummer")
data_winter <- read_excel(file_path, sheet = "ScreeningDataCollectionWinter")
data_6weeks <- read_excel(file_path, sheet = "ScreeningDataCollection6Weeks")
sun_expos <- read.csv("~/sapphire/data/sun_expos_data/sun_expos_long.csv")
sun_expos_summer <- sun_expos[sun_expos$collection_period == 'Summer', ]
sun_expos_winter <- sun_expos[sun_expos$collection_period == 'Winter', ]
sun_expos_6Weeks <- sun_expos[sun_expos$collection_period == '6Weeks', ]part 0: cleaning
summer_data <- left_join(data_summer, sun_expos_summer,
by = c("ParticipantCentreID" = "participant_centre_id"))
winter_data <- left_join(data_winter, sun_expos_winter,
by = c("ParticipantCentreID" = "participant_centre_id"))
six_week_data <- left_join(data_6weeks, sun_expos_6Weeks,
by = c("ParticipantCentreID" = "participant_centre_id"))summer_data = subset(summer_data, select = -c(Supplements, Medications,
EthnicitySpecifyOther, SmokingComments,
x9if_apply_sunscreen_spf_used))
winter_data = subset(winter_data, select = -c(Supplements, Medications,
EthnicitySpecifyOther, SmokingComments,
ContinuedInStudy, IfNotContinuedInStudyReason,
x9if_apply_sunscreen_spf_used))
six_week_data = subset(six_week_data, select = -c(Supplements, Medications,
EthnicitySpecifyOther, SmokingComments,
ContinuedInStudy, IfNotContinuedInStudyReason,
x9if_apply_sunscreen_spf_used))
six_week_data = six_week_data[,!grepl("IfNoReasonForExclusion:",names(six_week_data))]
winter_data = winter_data[,!grepl("IfNoReasonForExclusion:",names(winter_data))]
summer_data = summer_data[,!grepl("IfNoReasonForExclusion:",names(summer_data))]
six_week_data = six_week_data[,!grepl("Req Num",names(six_week_data))]
winter_data = winter_data[,!grepl("Req Num",names(winter_data))]
summer_data = summer_data[,!grepl("Req Num",names(summer_data))]
six_week_data <- data_6weeks
summer_data <- data_summer
winter_data <- data_winter# taking the median of three measurements
sites <- c("Forehead", "RightUpperInnerArm", "LeftUpperInnerArm")
metrics <- c("E", "M", "R", "G", "B", "L\\*", "a\\*", "b\\*")
seasons <- c("six_week_data", "summer_data", "winter_data")
for(site in sites) {
for(metric in metrics) {
six_week_data <- six_week_data %>%
rowwise() %>%
mutate(!!paste0("Median", site, metric) := median(c_across(matches(paste0("SkinReflectance", site, metric, "[123]"))), na.rm = TRUE)) %>%
ungroup()
}
}
for(site in sites) {
for(metric in metrics) {
summer_data <- summer_data %>%
rowwise() %>%
mutate(!!paste0("Median", site, metric) := median(c_across(matches(paste0("SkinReflectance", site, metric, "[123]"))), na.rm = TRUE)) %>%
ungroup()
}
}
for(site in sites) {
for(metric in metrics) {
winter_data <- winter_data %>%
rowwise() %>%
mutate(!!paste0("Median", site, metric) := median(c_across(matches(paste0("SkinReflectance", site, metric, "[123]"))), na.rm = TRUE)) %>%
ungroup()
}
}
winter_data <- winter_data %>%
select(-matches(".*[EMRGBL\\*a\\*b\\*]\\d$"))
summer_data <- summer_data %>%
select(-matches(".*[EMRGBL\\*a\\*b\\*]\\d$"))
six_week_data <- six_week_data %>%
select(-matches(".*[EMRGBL\\*a\\*b\\*]\\d$"))ethnicity <- function(EthnicityAfricanBlack, EthnicityColoured, EthnicityWhite,
EthnicityIndianAsian) {
case_when(
EthnicityAfricanBlack == TRUE &
EthnicityColoured == FALSE &
EthnicityWhite == FALSE &
EthnicityIndianAsian == FALSE ~ "Xhosa",
EthnicityAfricanBlack == FALSE &
EthnicityColoured == TRUE &
EthnicityWhite == FALSE &
EthnicityIndianAsian == FALSE ~ "Cape_colored",
TRUE ~ NA_character_
)
}
summer_data <- summer_data %>%
mutate(Ethnicity = ethnicity(EthnicityAfricanBlack, EthnicityColoured,
EthnicityWhite, EthnicityIndianAsian))
summer_data = subset(summer_data, select = -c(EthnicityAfricanBlack,
EthnicityColoured, EthnicityWhite,
EthnicityIndianAsian))
winter_data <- winter_data %>%
mutate(Ethnicity = ethnicity(EthnicityAfricanBlack, EthnicityColoured,
EthnicityWhite, EthnicityIndianAsian))
winter_data = subset(winter_data, select = -c(EthnicityAfricanBlack,
EthnicityColoured, EthnicityWhite,
EthnicityIndianAsian))
six_week_data <- six_week_data %>%
mutate(Ethnicity = ethnicity(EthnicityAfricanBlack, EthnicityColoured,
EthnicityWhite, EthnicityIndianAsian))
six_week_data = subset(six_week_data, select = -c(EthnicityAfricanBlack,
EthnicityColoured, EthnicityWhite,
EthnicityIndianAsian))ggplot(six_week_data, aes(x = VitDResult, y = MedianForeheadM, color = Ethnicity)) +
geom_jitter() +
theme(legend.position = "right")Warning: Removed 1 row containing missing values or values outside the scale range
(`geom_point()`).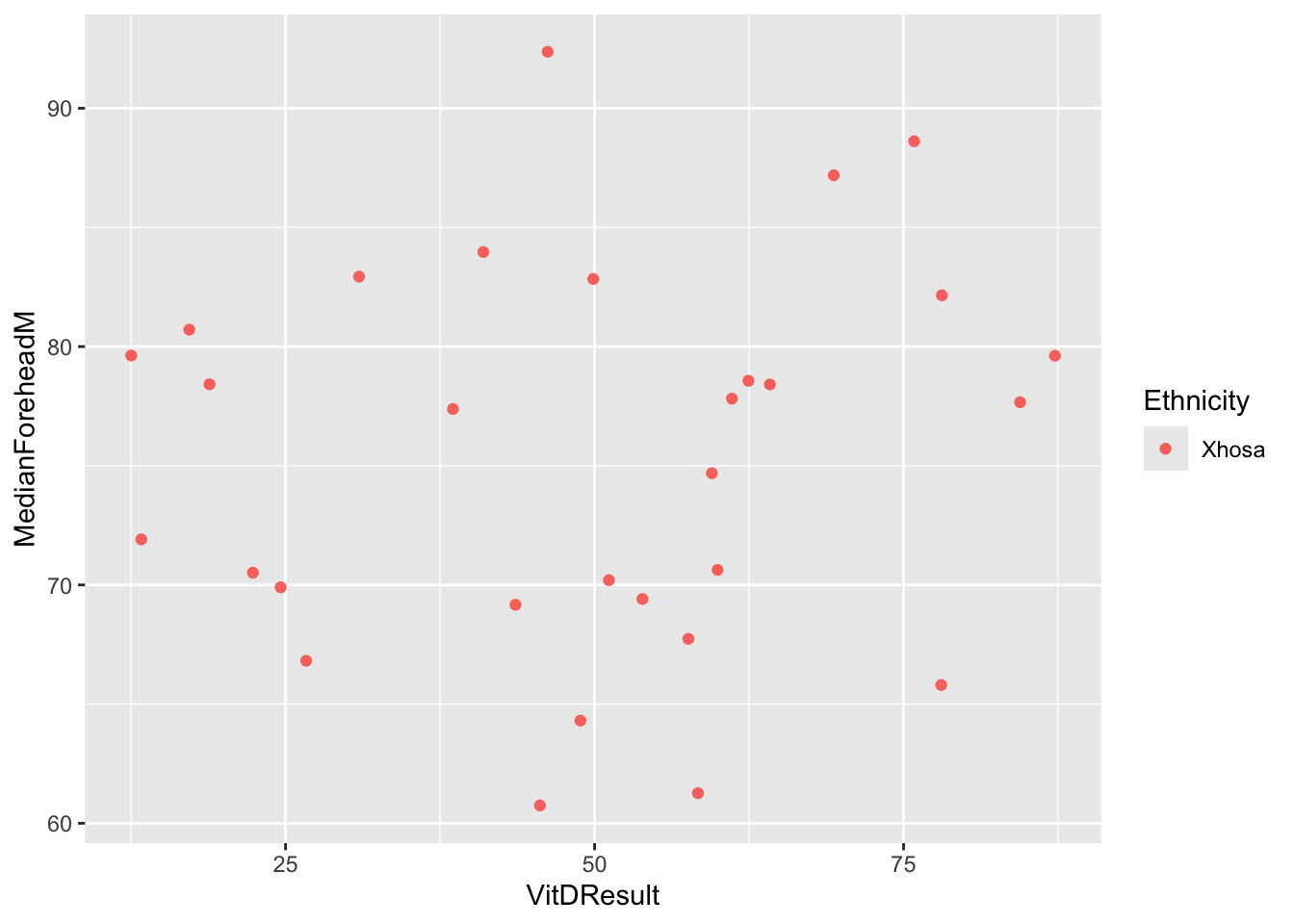
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
ggplot(six_week_data, aes(x = Ethnicity, y = MedianForeheadM, color = Ethnicity, fill = Ethnicity)) +
geom_violin()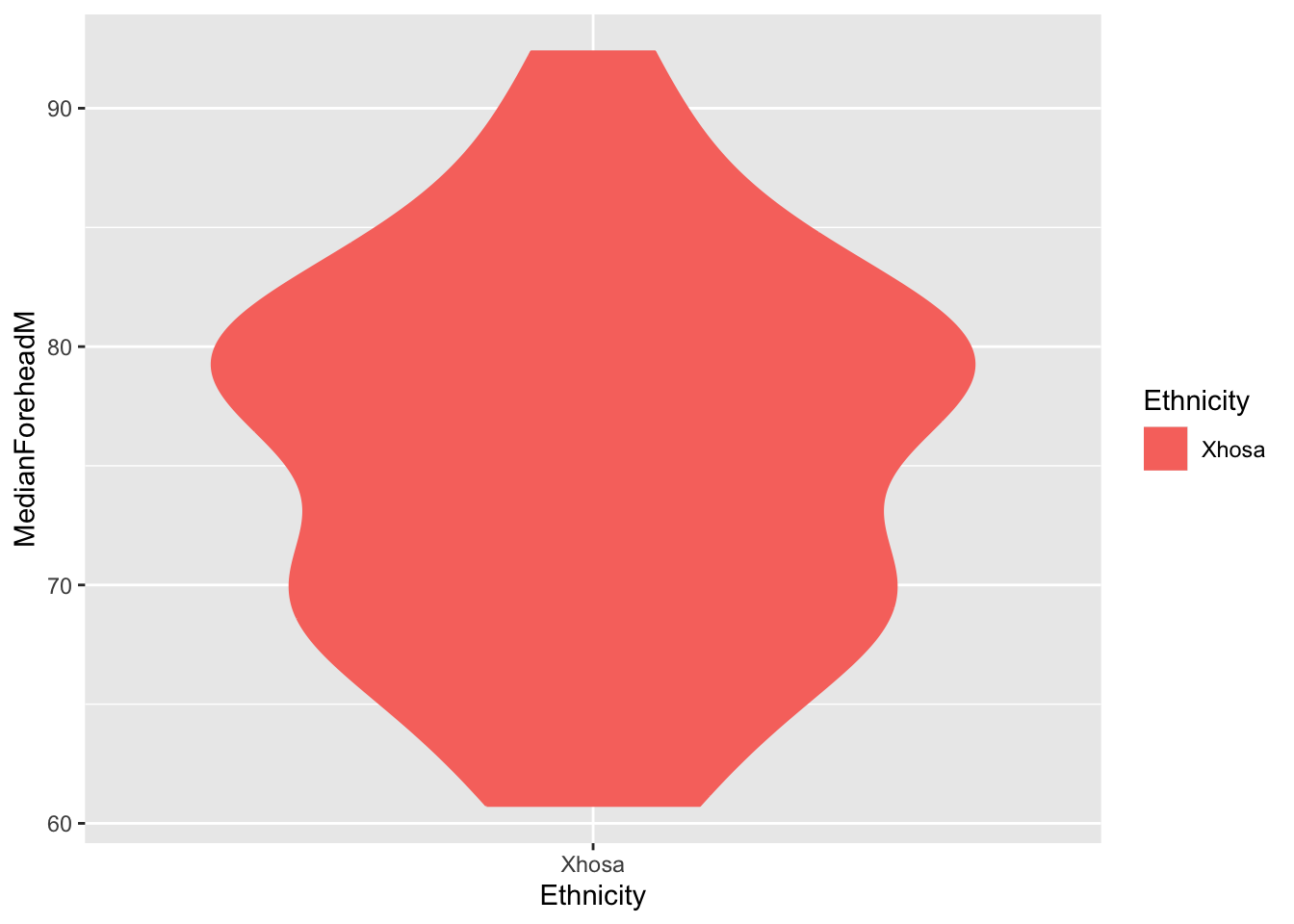
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
ggplot(summer_data, aes(x = VitDResult, y = MedianForeheadM, color = Ethnicity)) +
geom_jitter() +
theme(legend.position = "right")Warning: Removed 1 row containing missing values or values outside the scale range
(`geom_point()`).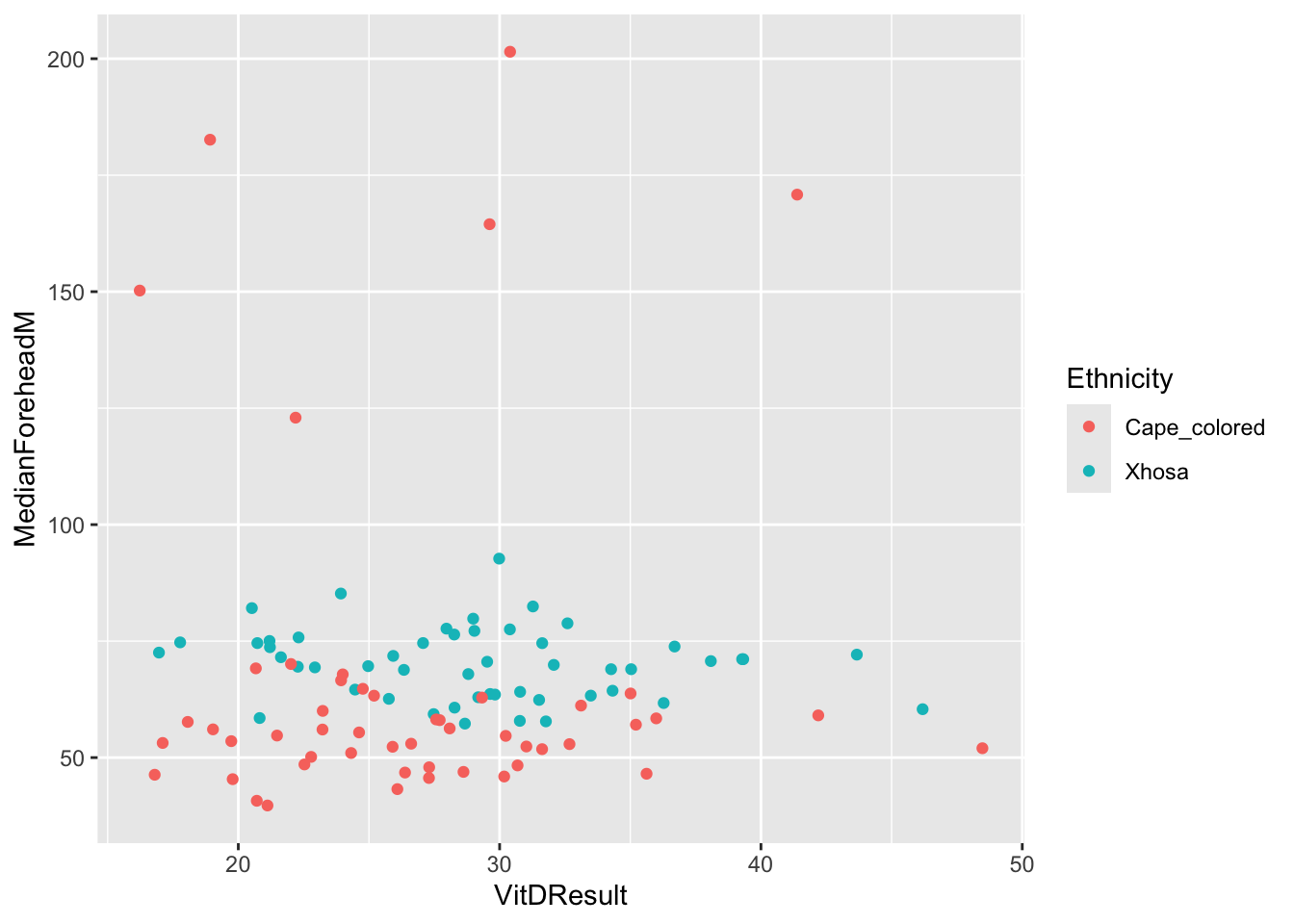
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
ggplot(summer_data, aes(x = Ethnicity, y = MedianForeheadM, color = Ethnicity, fill = Ethnicity)) +
geom_boxplot()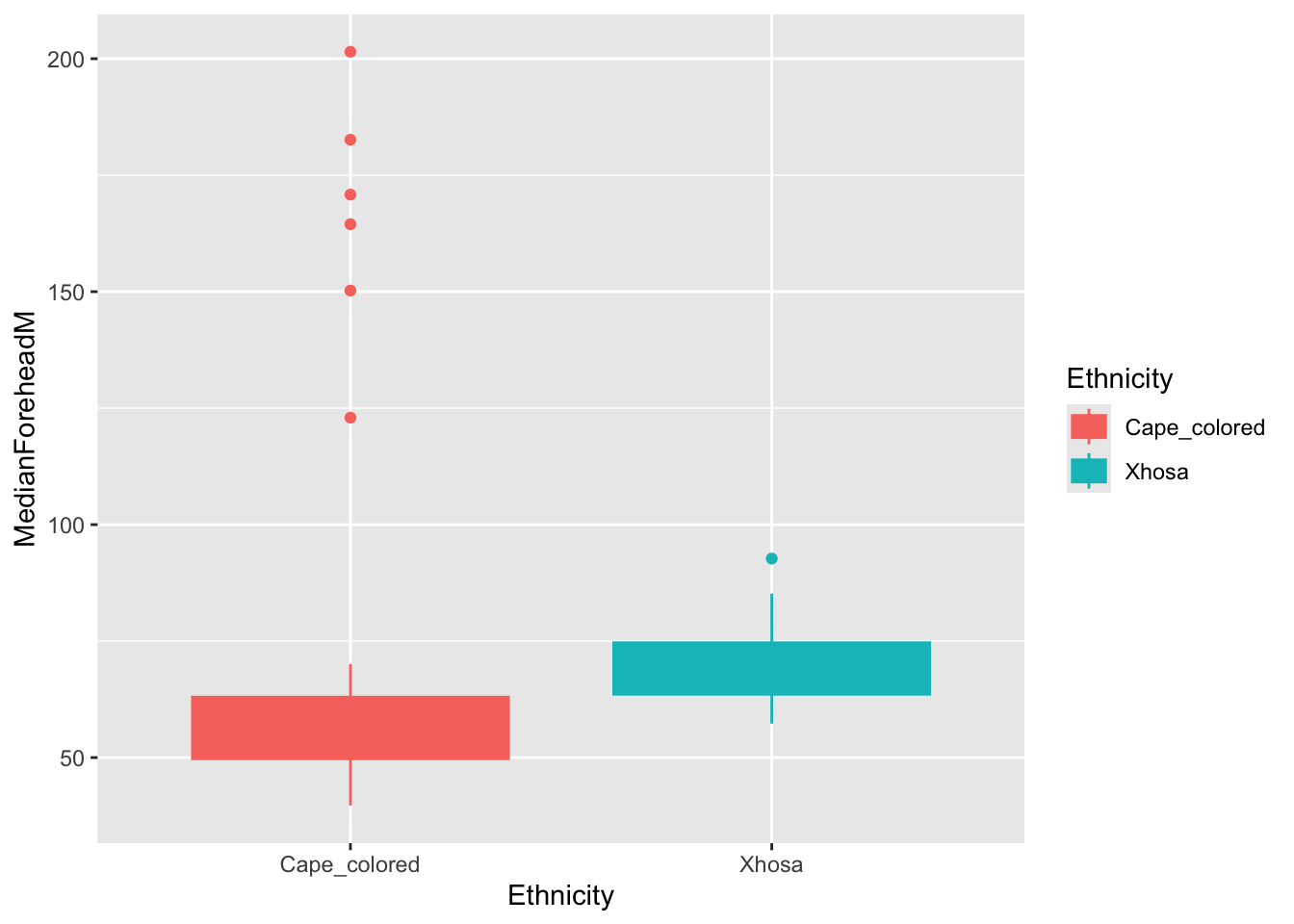
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
# there are still outliers even when taking the median
for(site in sites) {
for(metric in metrics) {
column_name <- paste0("Median", site, metric)
iqr <- IQR(winter_data[[column_name]], na.rm = TRUE)
Q <- quantile(winter_data[[column_name]], probs = c(0.25, 0.75), na.rm = TRUE)
up <- Q[2] + 1.5 * iqr
low <- Q[1] - 1.5 * iqr
winter_data <- winter_data %>%
filter(!!sym(column_name) > low & !!sym(column_name) < up)
}
}
for(site in sites) {
for(metric in metrics) {
column_name <- paste0("Median", site, metric)
iqr <- IQR(summer_data[[column_name]], na.rm = TRUE)
Q <- quantile(summer_data[[column_name]], probs = c(0.25, 0.75), na.rm = TRUE)
up <- Q[2] + 1.5 * iqr
low <- Q[1] - 1.5 * iqr
summer_data <- summer_data %>%
filter(!!sym(column_name) > low & !!sym(column_name) < up)
}
}
for(site in sites) {
for(metric in metrics) {
column_name <- paste0("Median", site, metric)
iqr <- IQR(winter_data[[column_name]], na.rm = TRUE)
Q <- quantile(winter_data[[column_name]], probs = c(0.25, 0.75), na.rm = TRUE)
up <- Q[2] + 1.5 * iqr
low <- Q[1] - 1.5 * iqr
six_week_data <- six_week_data %>%
filter(!!sym(column_name) > low & !!sym(column_name) < up)
}
}ggplot(six_week_data, aes(x = VitDResult, y = MedianForeheadM, color = Ethnicity)) +
geom_jitter() +
theme(legend.position = "right")Warning: Removed 1 row containing missing values or values outside the scale range
(`geom_point()`).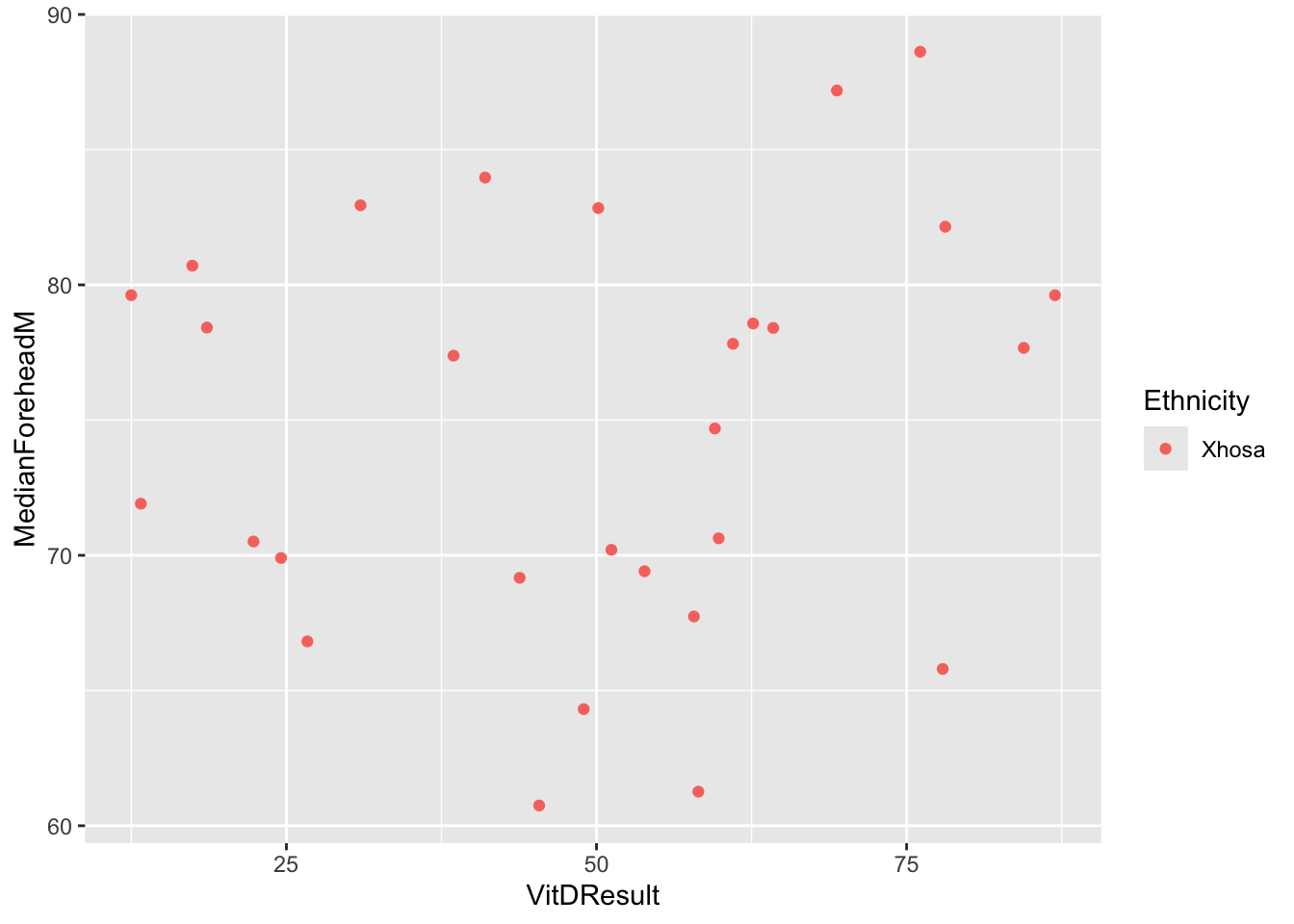
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
ggplot(six_week_data, aes(x = Ethnicity, y = MedianForeheadM, color = Ethnicity, fill = Ethnicity)) +
geom_violin()
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
ggplot(summer_data, aes(x = VitDResult, y = MedianForeheadM, color = Ethnicity)) +
geom_jitter() +
theme(legend.position = "right")Warning: Removed 1 row containing missing values or values outside the scale range
(`geom_point()`).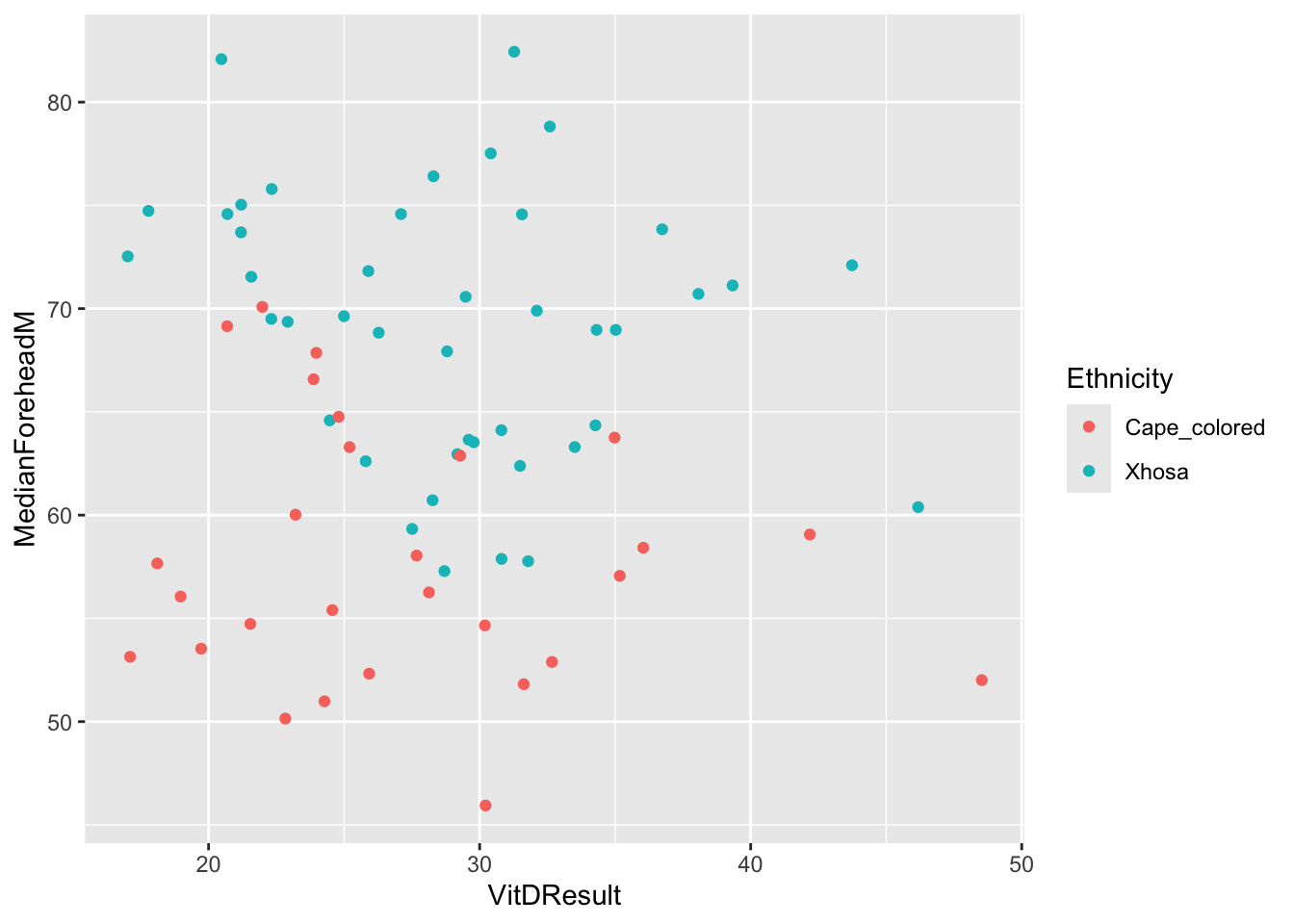
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
ggplot(summer_data, aes(x = Ethnicity, y = MedianForeheadM, color = Ethnicity, fill = Ethnicity)) +
geom_boxplot()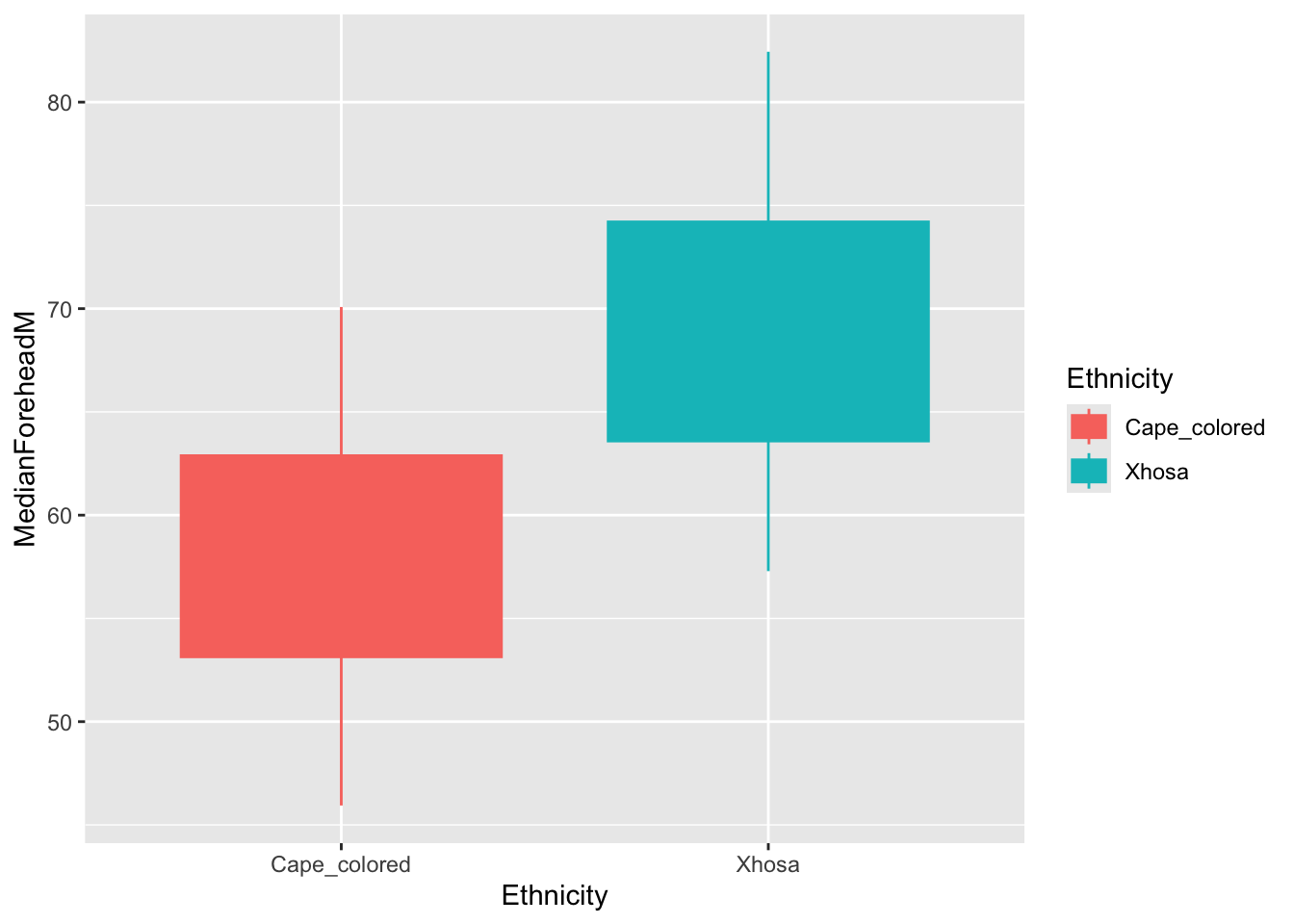
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
# mean left and right inner arm
for (metric in metrics) {
summer_data <- summer_data %>%
mutate(!!paste0("InnerArm", metric) := rowMeans(
select(., starts_with(paste0("MedianLeftInnerArm", metric)),
starts_with(paste0("MedianRightUpperInnerArm", metric))),
na.rm = TRUE
))
}
for (metric in metrics) {
winter_data <- winter_data %>%
mutate(!!paste0("InnerArm", metric) := rowMeans(
select(., starts_with(paste0("MedianLeftInnerArm", metric)),
starts_with(paste0("MedianRightUpperInnerArm", metric))),
na.rm = TRUE
))
}
for (metric in metrics) {
six_week_data <- six_week_data %>%
mutate(!!paste0("InnerArm", metric) := rowMeans(
select(., starts_with(paste0("MedianLeftInnerArm", metric)),
starts_with(paste0("MedianRightUpperInnerArm", metric))),
na.rm = TRUE
))
}
winter_data <- winter_data %>%
select(-matches("Left|Right"))
summer_data <- summer_data %>%
select(-matches("Left|Right"))
six_week_data <- six_week_data %>%
select(-matches("Left|Right"))for (metric in metrics) {
six_week_data <- six_week_data %>%
mutate(!!paste0(metric, "Difference") :=
.[[paste0("MedianForehead", metric)]] -
.[[paste0("InnerArm", metric)]])
}
for (metric in metrics) {
summer_data <- summer_data %>%
mutate(!!paste0(metric, "Difference") :=
.[[paste0("MedianForehead", metric)]] -
.[[paste0("InnerArm", metric)]])
}
for (metric in metrics) {
winter_data <- winter_data %>%
mutate(!!paste0(metric, "Difference") :=
.[[paste0("MedianForehead", metric)]] -
.[[paste0("InnerArm", metric)]])
}six_week_rename <- six_week_data %>%
rename_with(~ paste0(., "_sixweeks"), -ParticipantCentreID)
joinone <- left_join(summer_data, winter_data,
by = "ParticipantCentreID",
suffix = c("_summer", "_winter"))
joined_data <- left_join(joinone, six_week_rename,
by = "ParticipantCentreID")
head(joined_data)# A tibble: 6 × 197
ParticipantCentreID InterviewerName_summer TodayDate_summer AgeYears_summer
<chr> <chr> <dttm> <dbl>
1 VDKH001 Betty 2013-02-11 00:00:00 20
2 VDKH002 Betty 2013-02-11 00:00:00 23
3 VDKH003 Betty 2013-02-11 00:00:00 23
4 VDKH005 Betty 2013-02-12 00:00:00 19
5 VDKH006 Betty 2013-02-12 00:00:00 21
6 VDKH007 Betty 2013-02-12 00:00:00 22
# ℹ 193 more variables: DateOfBirth_summer <dttm>, Gender_summer <dbl>,
# EthnicityOther_summer <lgl>, EthnicitySpecifyOther_summer <lgl>,
# Ethnicity_summer <chr>, RefuseToAnswer_summer <lgl>,
# AvgWeight_summer <dbl>, AvgHeight_summer <dbl>, BMI_summer <dbl>,
# SoreThroatYes_summer <lgl>, SoreThroatNo_summer <lgl>,
# RunnyNoseYes_summer <lgl>, RunnyNoseNo_summer <lgl>, CoughYes_summer <lgl>,
# CoughNo_summer <lgl>, FeverYes_summer <lgl>, FeverNo_summer <lgl>, …part 1: PCAs
PCAs to run: - run wideform pca - run pigmentation subset pca for each season - run RGB subset for each season - run ME subset for each season - run CIElab subset for each season
wideform
summer_data$Ethnicity <- as.factor (summer_data$Ethnicity)
names(summer_data) <- gsub("\\\\", "", names(summer_data))
names(summer_data) <- gsub("\\*", "", names(summer_data))
#Xhosa is value 2
winter_data$Ethnicity <- as.factor (winter_data$Ethnicity)
names(winter_data) <- gsub("\\\\", "", names(winter_data))
names(winter_data) <- gsub("\\*", "", names(winter_data))
summer_winter <- left_join(summer_data, winter_data,
by = "ParticipantCentreID",
suffix = c("_summer", "_winter"))
summer_winter <- summer_winter %>%
select(-(matches("EthnicitySpecifyOther")))
clean_sw <- summer_winter %>%
select(
contains("MedianForehead"),
contains("InnerArm"),
contains("ParticipantCentreID"),
contains("Ethnicity"),
contains("Difference")
)summer_winter <- summer_winter %>%
select(matches("MedianForehead|InnerArm|VitD|ParticipantCentreID|Ethnicity"))
summer_winter_clean <- na.omit(summer_winter)
reflectance_metrics_ws <- summer_winter_clean %>%
select(matches("MedianForehead|InnerArm"))
reflectance_metrics_ws# A tibble: 52 × 32
MedianForeheadE_summer MedianForeheadM_summer MedianForeheadR_summer
<dbl> <dbl> <dbl>
1 18.6 70.6 50
2 17.4 74.7 45
3 14.3 69.6 51
4 19.7 63.0 57
5 15.3 82.1 38
6 23.8 69.0 51
7 18.9 59.3 66
8 18.6 64.6 59
9 15.5 75.0 44
10 19.3 64.1 65
# ℹ 42 more rows
# ℹ 29 more variables: MedianForeheadG_summer <dbl>,
# MedianForeheadB_summer <dbl>, MedianForeheadL_summer <dbl>,
# MedianForeheada_summer <dbl>, MedianForeheadb_summer <dbl>,
# InnerArmE_summer <dbl>, InnerArmM_summer <dbl>, InnerArmR_summer <dbl>,
# InnerArmG_summer <dbl>, InnerArmB_summer <dbl>, InnerArmL_summer <dbl>,
# InnerArma_summer <dbl>, InnerArmb_summer <dbl>, …reflectance3 <- scale(reflectance_metrics_ws)
reflectancepcaws <- prcomp(reflectance3)
summary(reflectancepcaws)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 4.8004 1.8402 1.35595 1.00167 0.83059 0.78552 0.61637
Proportion of Variance 0.7201 0.1058 0.05746 0.03135 0.02156 0.01928 0.01187
Cumulative Proportion 0.7201 0.8259 0.88341 0.91477 0.93632 0.95561 0.96748
PC8 PC9 PC10 PC11 PC12 PC13 PC14
Standard deviation 0.5905 0.4868 0.38054 0.28874 0.23220 0.20548 0.16805
Proportion of Variance 0.0109 0.0074 0.00453 0.00261 0.00168 0.00132 0.00088
Cumulative Proportion 0.9784 0.9858 0.99031 0.99291 0.99460 0.99591 0.99680
PC15 PC16 PC17 PC18 PC19 PC20 PC21
Standard deviation 0.16165 0.14597 0.11881 0.09832 0.09465 0.07705 0.06204
Proportion of Variance 0.00082 0.00067 0.00044 0.00030 0.00028 0.00019 0.00012
Cumulative Proportion 0.99761 0.99828 0.99872 0.99902 0.99930 0.99949 0.99961
PC22 PC23 PC24 PC25 PC26 PC27 PC28
Standard deviation 0.05768 0.04873 0.04403 0.03387 0.03195 0.02696 0.02514
Proportion of Variance 0.00010 0.00007 0.00006 0.00004 0.00003 0.00002 0.00002
Cumulative Proportion 0.99971 0.99979 0.99985 0.99988 0.99992 0.99994 0.99996
PC29 PC30 PC31 PC32
Standard deviation 0.02301 0.02118 0.01825 0.006431
Proportion of Variance 0.00002 0.00001 0.00001 0.000000
Cumulative Proportion 0.99997 0.99999 1.00000 1.000000reflectancepcaws$loadings[, 1:2]NULLfviz_eig(reflectancepcaws, addlabels = TRUE)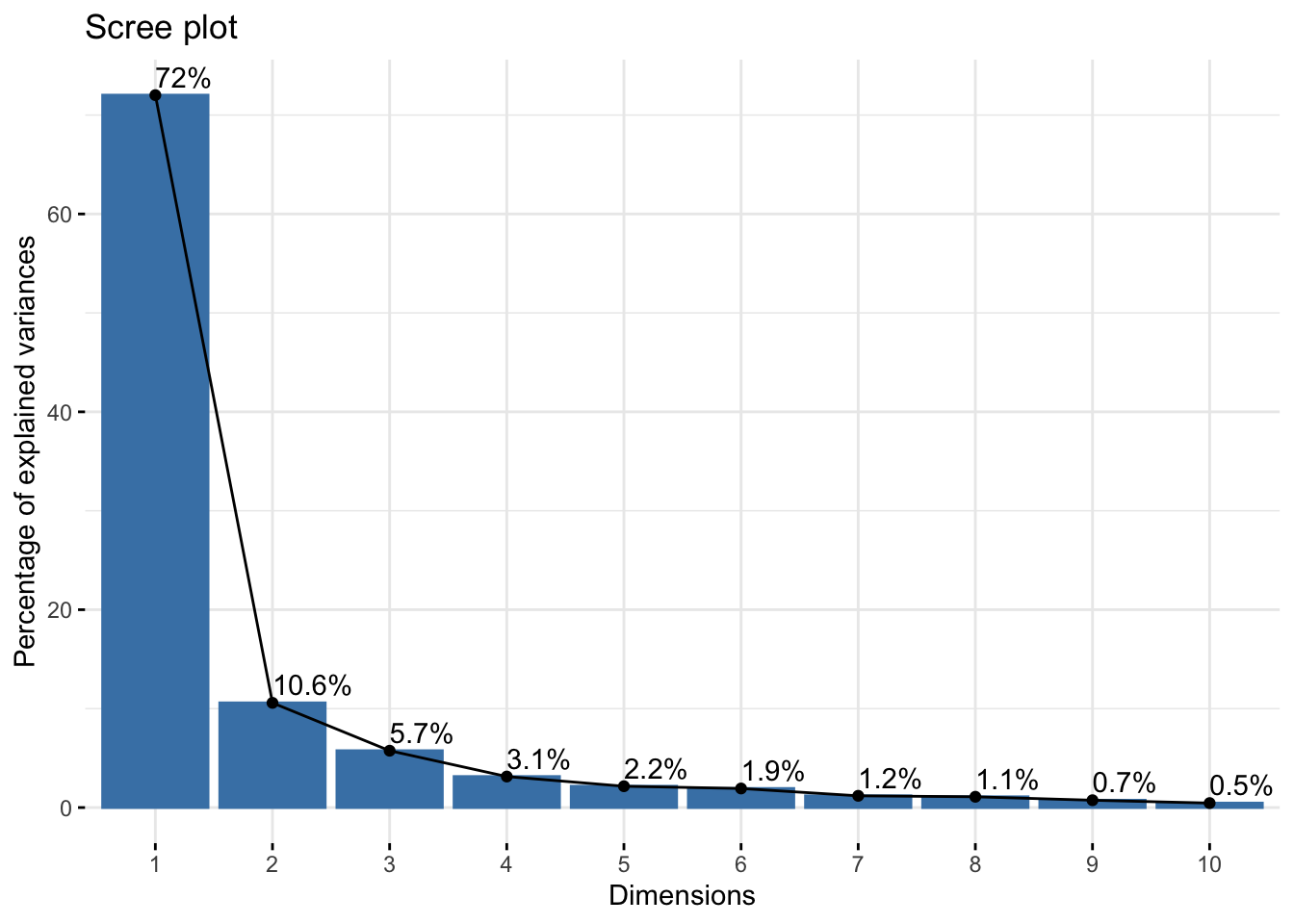
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(reflectancepcaws, col.var = "black")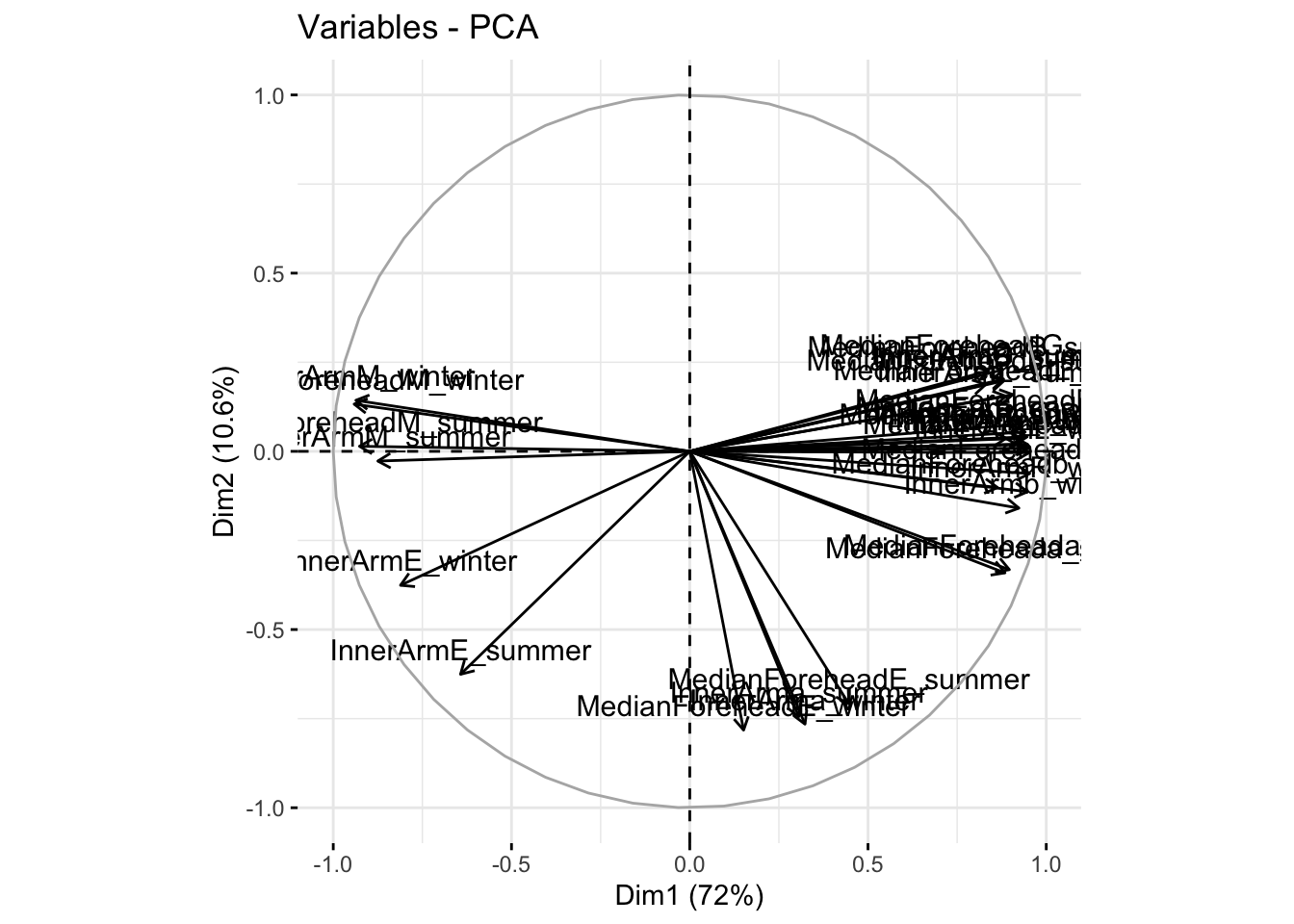
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(reflectancepcaws, choice = "var", axes = 1:2)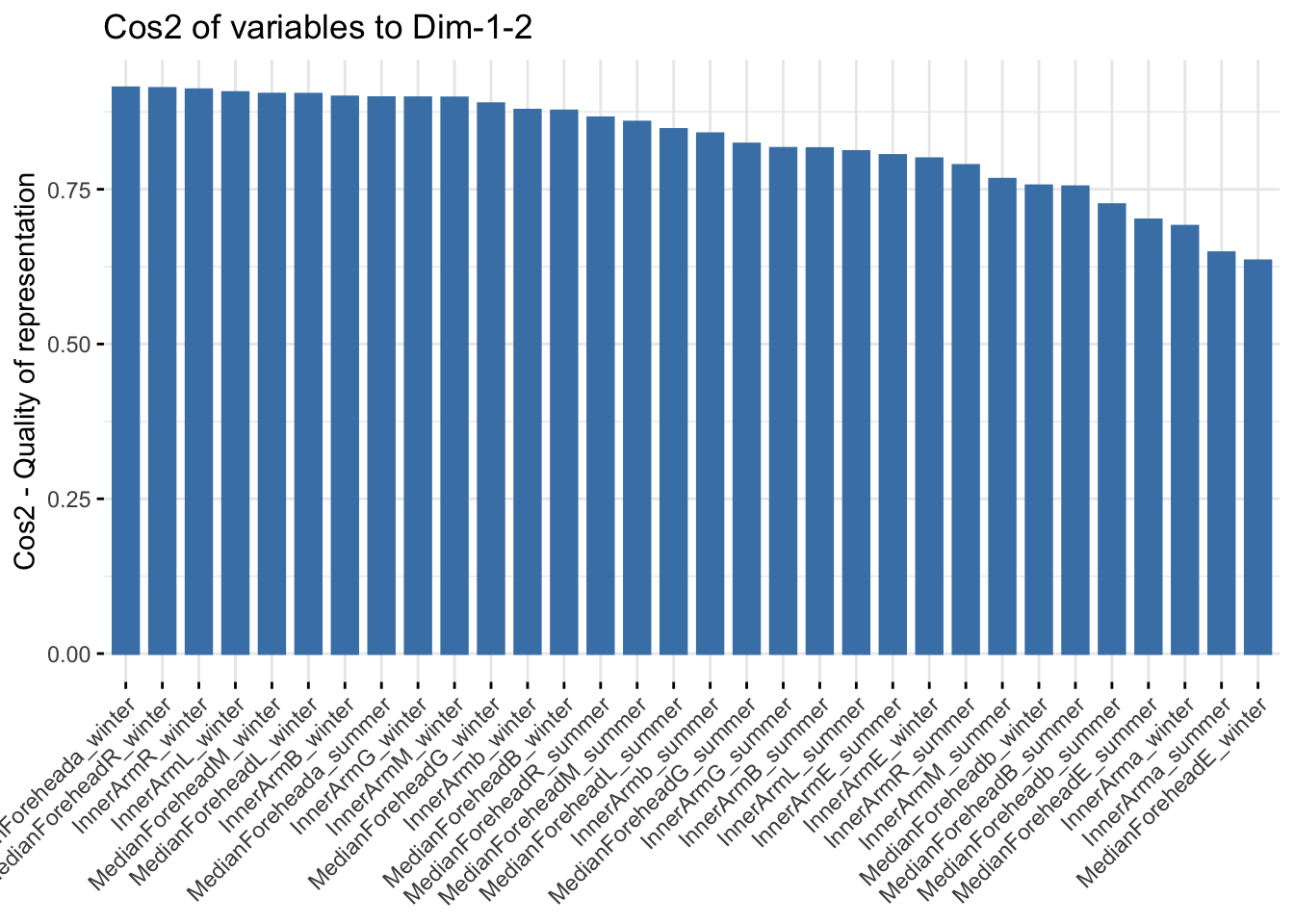
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
wscomps <- as.data.frame(reflectancepcaws$x)
reflectance_ws <- cbind(summer_winter_clean,wscomps[,c(1,2)])
reflectance_ws <- reflectance_ws %>%
select(-matches("SkinReflectance"))
reflectance_ws <- reflectance_ws %>%
select(-"Ethnicity_winter")
names(reflectance_ws)[names(reflectance_ws) == 'Ethnicity_summer'] <- 'Ethnicity'
ggplot(reflectance_ws, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Wide Pigmentation PCA")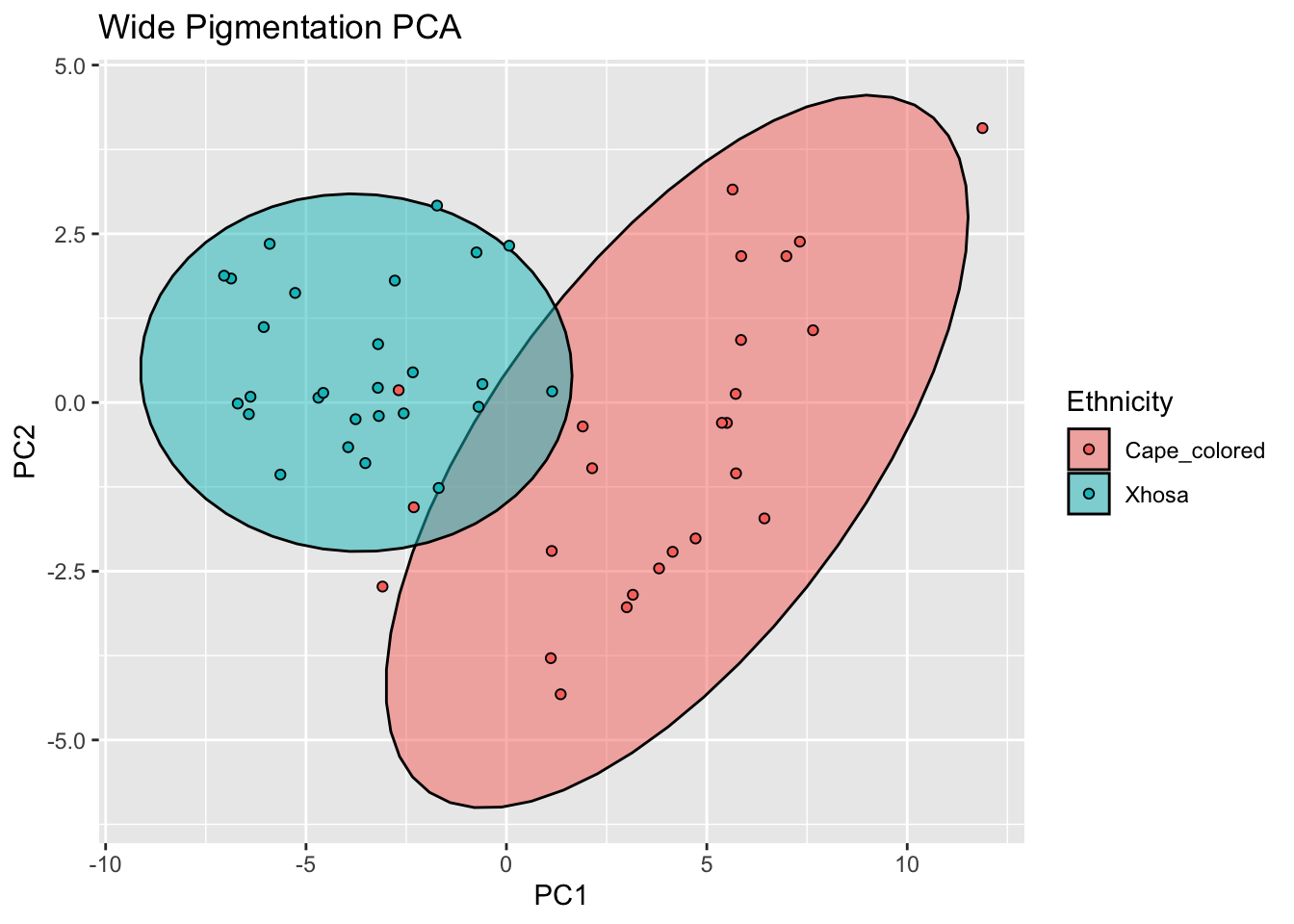
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(reflectancepcaws, choice = "var", axes = 1, top = 20)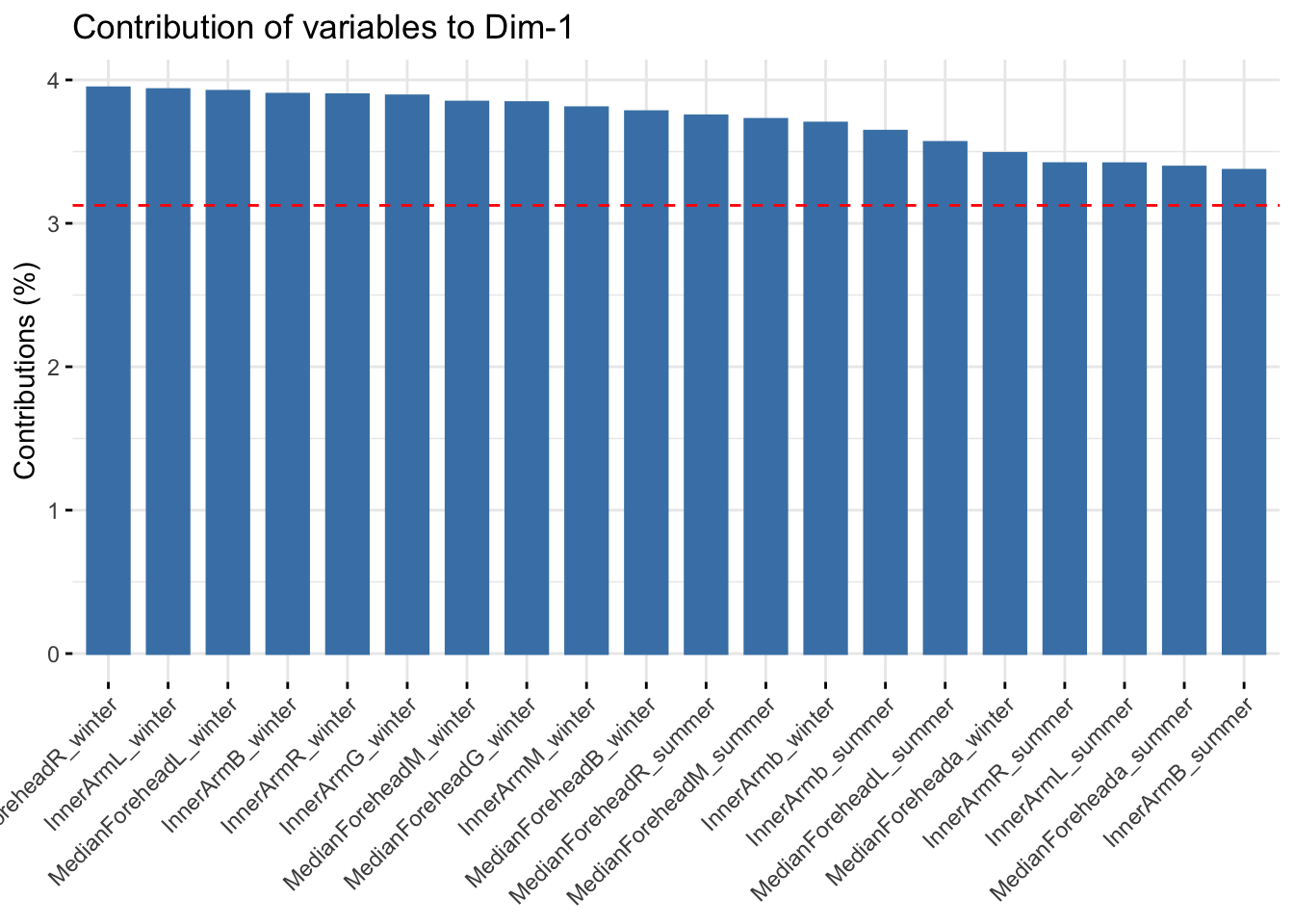
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(reflectancepcaws, choice = "var", axes = 2, top = 20)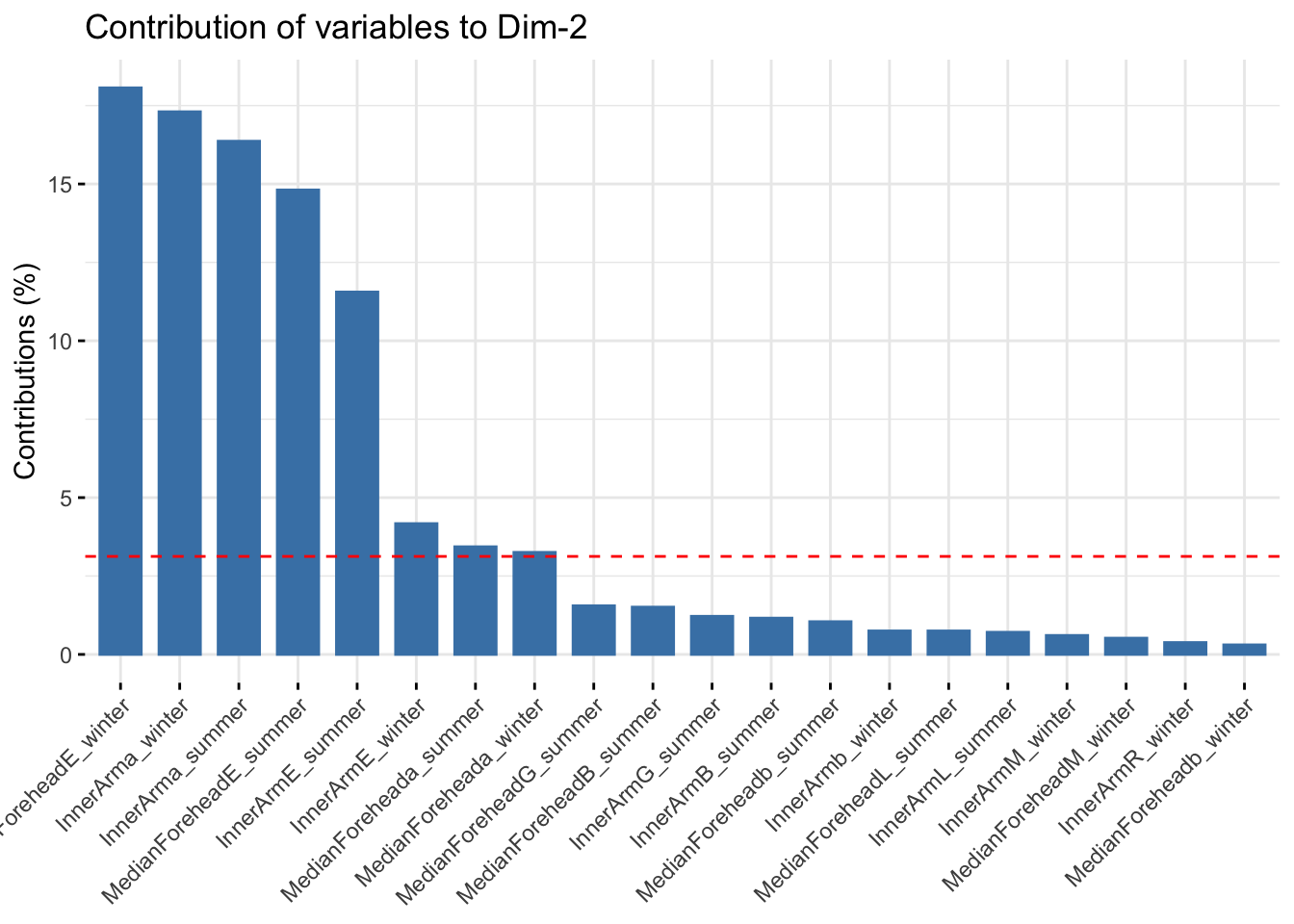
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
pigmentation subset
summer_refl_metrics <- summer_data %>%
select(matches("InnerArm|MedianForehead"))
summer_refl_metrics <- na.omit(summer_refl_metrics)
winter_refl_metrics <- winter_data %>%
select(matches("InnerArm|MedianForehead"))
winter_refl_metrics <- na.omit(winter_refl_metrics)
six_refl_metrics <- six_week_data %>%
select(matches("InnerArm|MedianForehead"))
six_refl_metrics <- na.omit(six_refl_metrics)six_refl <- scale(six_refl_metrics)
six_refl_pca <- prcomp(six_refl)
summary(six_refl_pca)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 3.237 1.6206 1.06822 0.99730 0.54065 0.43093 0.36791
Proportion of Variance 0.655 0.1641 0.07132 0.06216 0.01827 0.01161 0.00846
Cumulative Proportion 0.655 0.8191 0.89045 0.95261 0.97088 0.98248 0.99094
PC8 PC9 PC10 PC11 PC12 PC13 PC14
Standard deviation 0.2467 0.19478 0.13699 0.10300 0.08596 0.06440 0.04847
Proportion of Variance 0.0038 0.00237 0.00117 0.00066 0.00046 0.00026 0.00015
Cumulative Proportion 0.9948 0.99712 0.99829 0.99895 0.99942 0.99967 0.99982
PC15 PC16
Standard deviation 0.04121 0.03399
Proportion of Variance 0.00011 0.00007
Cumulative Proportion 0.99993 1.00000six_refl_pca$loadings[, 1:2]NULLfviz_eig(six_refl_pca, addlabels = TRUE)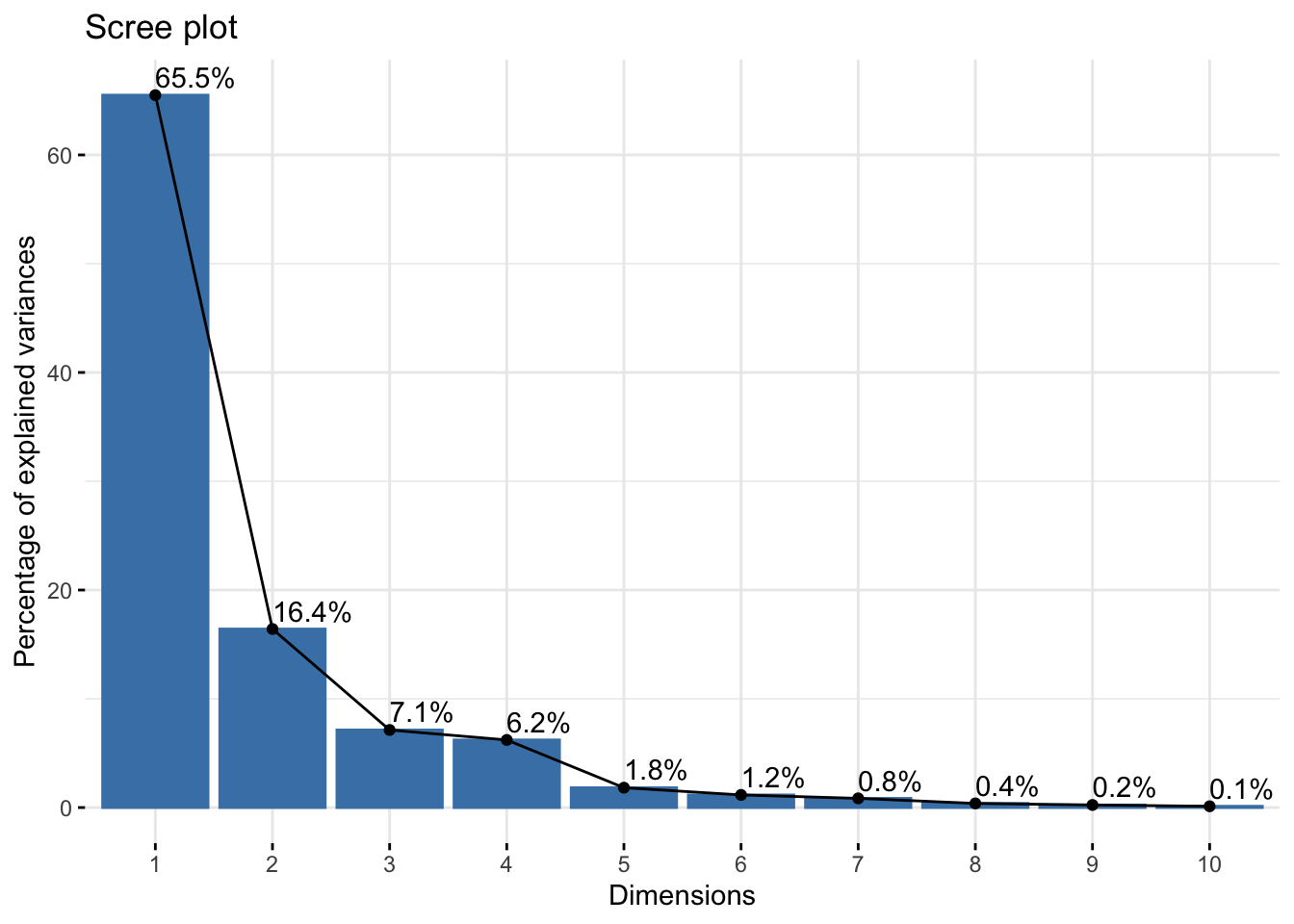
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(six_refl_pca, col.var = "black")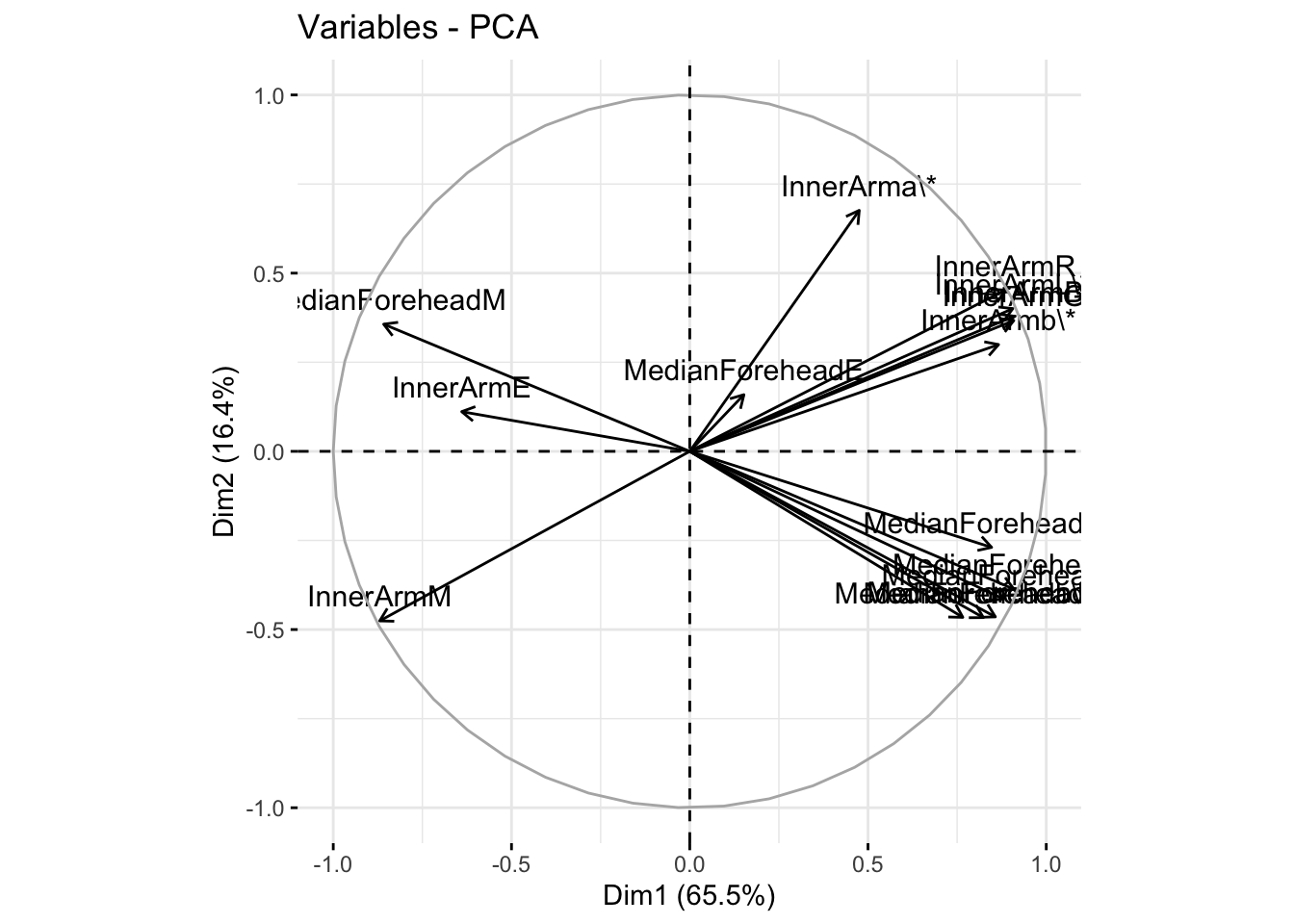
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(six_refl_pca, choice = "var", axes = 1:2)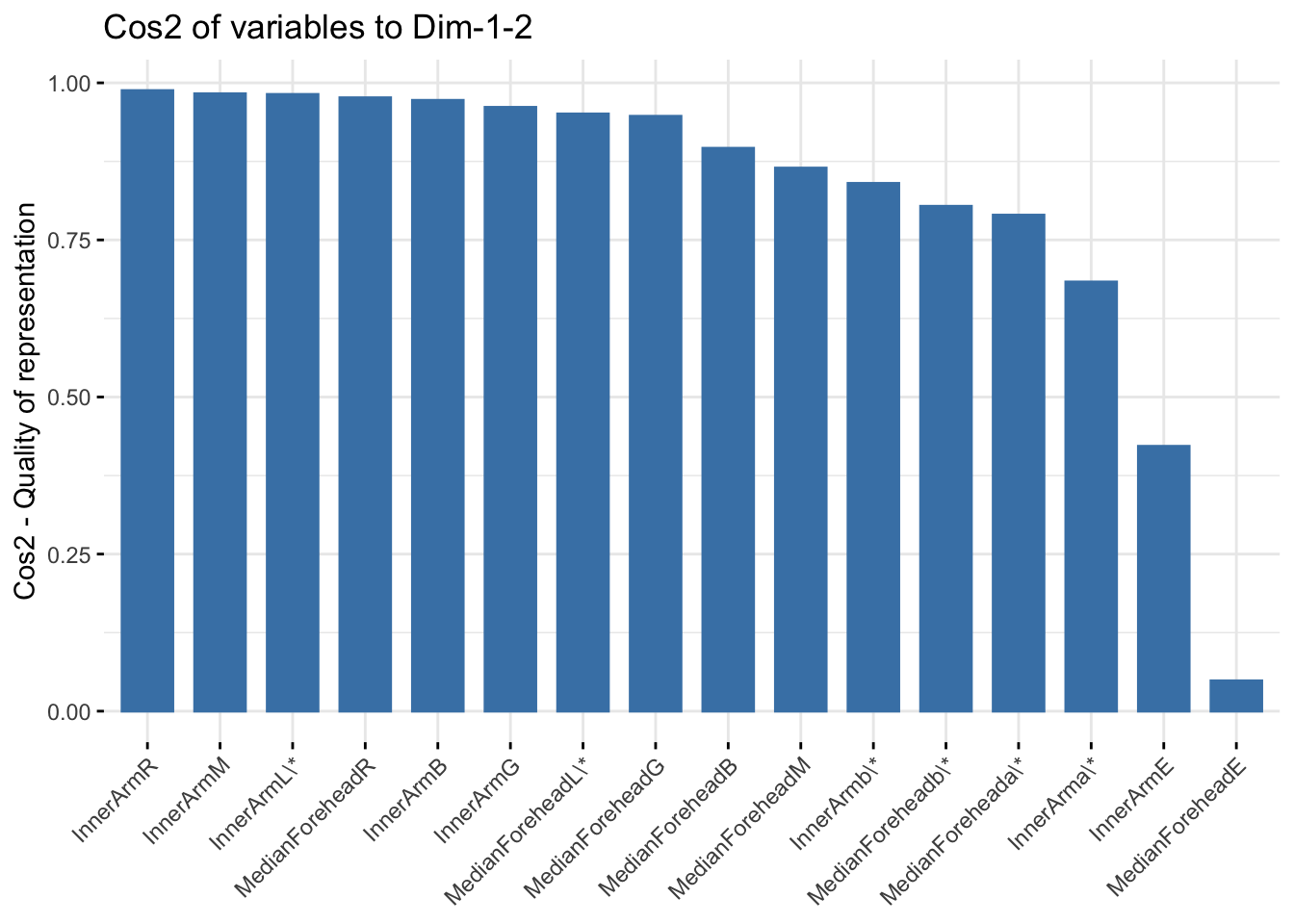
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
six_pigm_comps <- as.data.frame(six_refl_pca$x)
six_pigment <- cbind(six_week_data,six_pigm_comps[,c(1,2)])
ggplot(six_pigment, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Six Week Pigmentation PCA")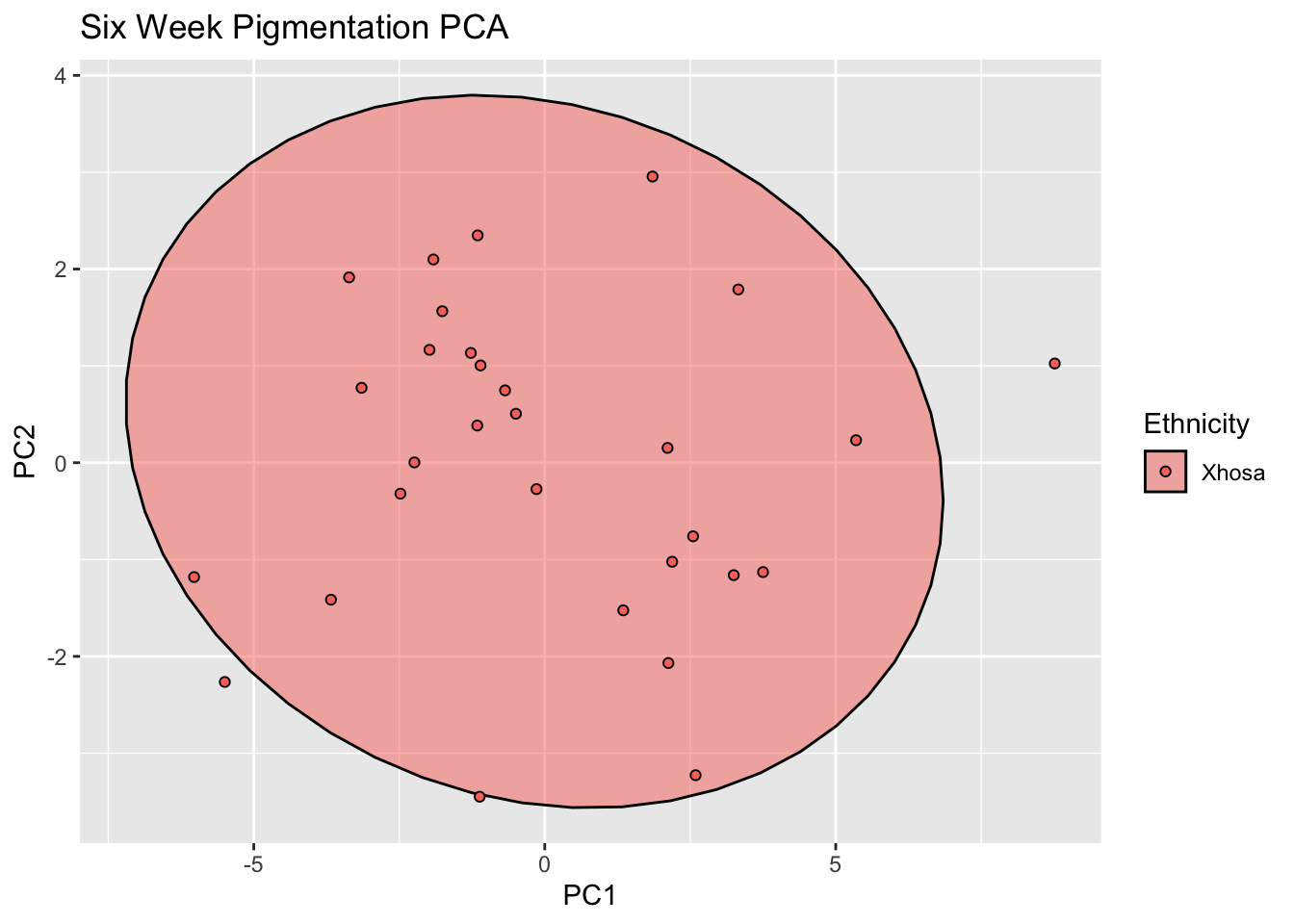
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(six_refl_pca, choice = "var", axes = 1, top = 10)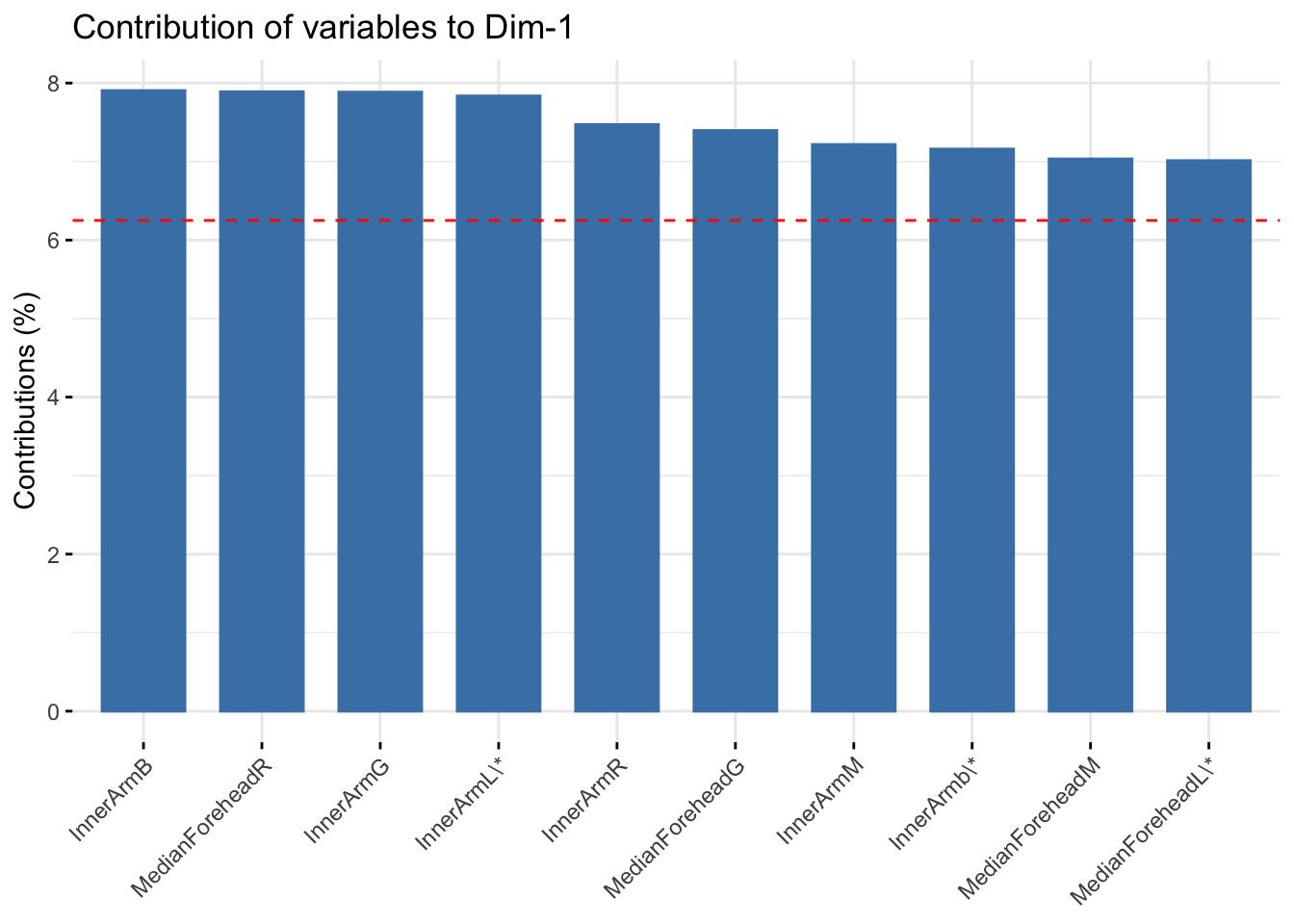
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(six_refl_pca, choice = "var", axes = 2, top = 10)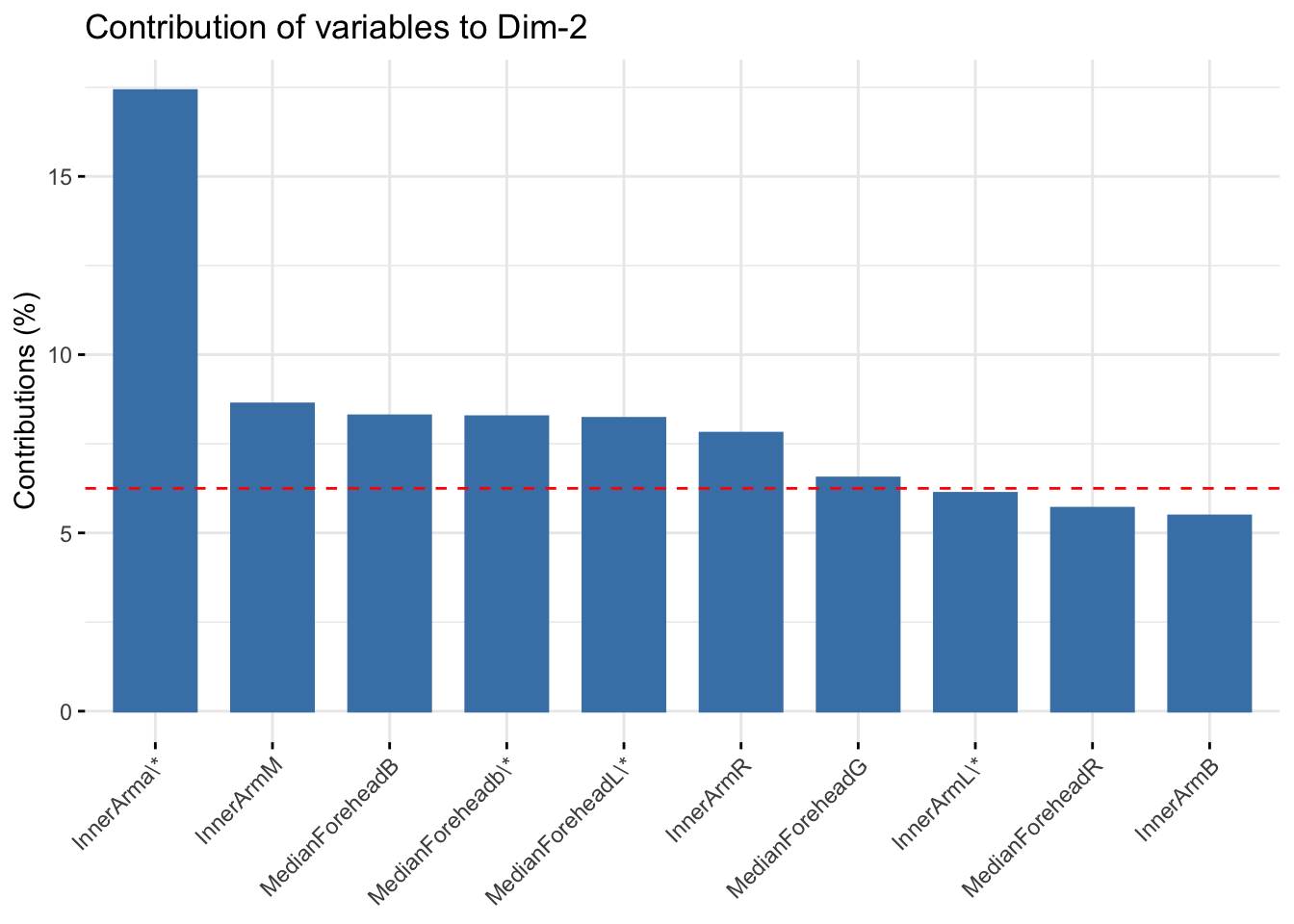
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
summer_refl <- scale(summer_refl_metrics)
summer_refl_pca <- prcomp(summer_refl)
summary(summer_refl_pca)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 3.4053 1.3280 1.23143 0.78241 0.52805 0.3175 0.20528
Proportion of Variance 0.7248 0.1102 0.09478 0.03826 0.01743 0.0063 0.00263
Cumulative Proportion 0.7248 0.8350 0.92975 0.96801 0.98543 0.9917 0.99437
PC8 PC9 PC10 PC11 PC12 PC13 PC14
Standard deviation 0.17041 0.15034 0.11867 0.09491 0.08545 0.05918 0.04667
Proportion of Variance 0.00182 0.00141 0.00088 0.00056 0.00046 0.00022 0.00014
Cumulative Proportion 0.99618 0.99760 0.99848 0.99904 0.99949 0.99971 0.99985
PC15 PC16
Standard deviation 0.03646 0.03273
Proportion of Variance 0.00008 0.00007
Cumulative Proportion 0.99993 1.00000summer_refl_pca$loadings[, 1:2]NULLfviz_eig(summer_refl_pca, addlabels = TRUE)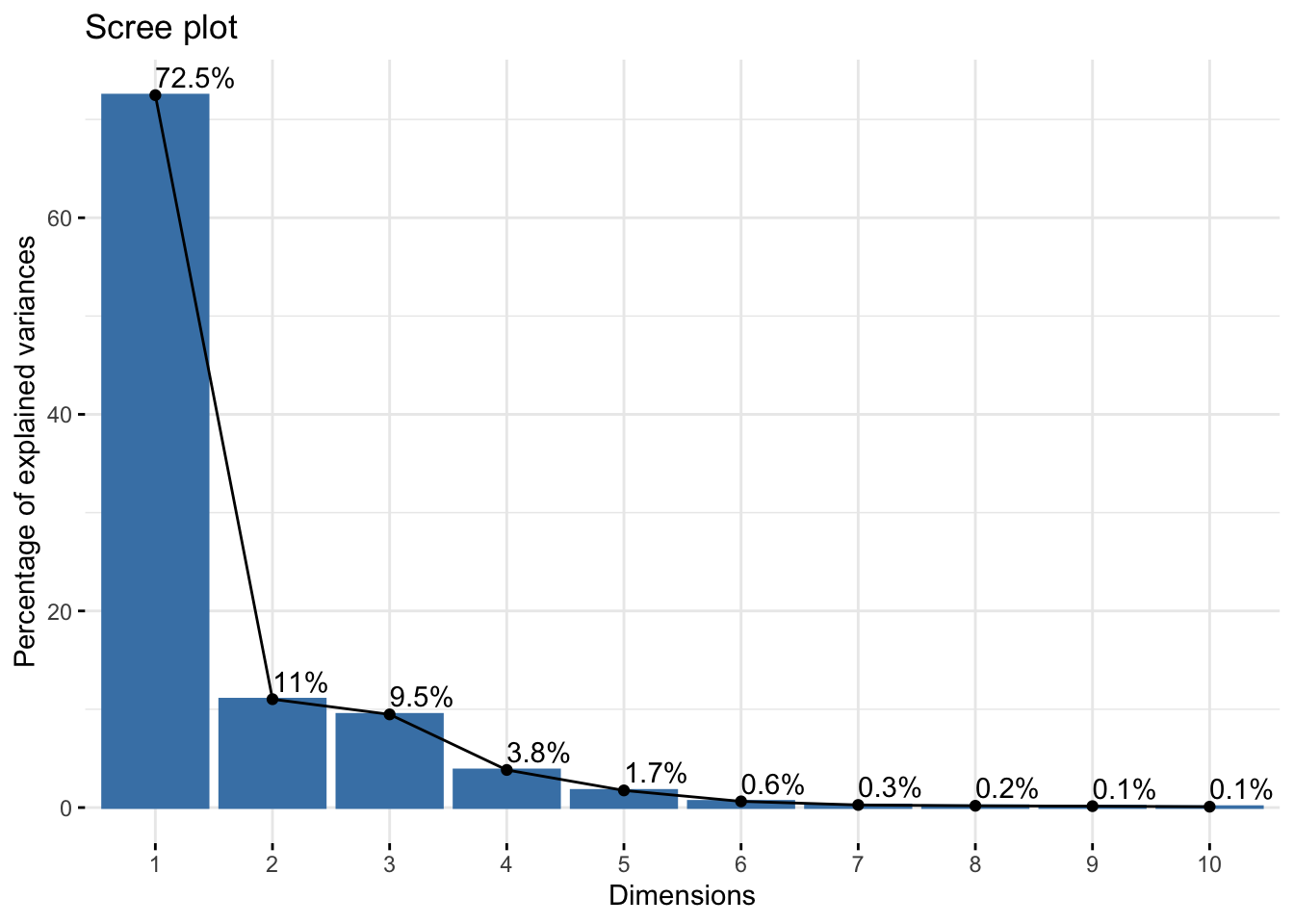
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(summer_refl_pca, col.var = "black")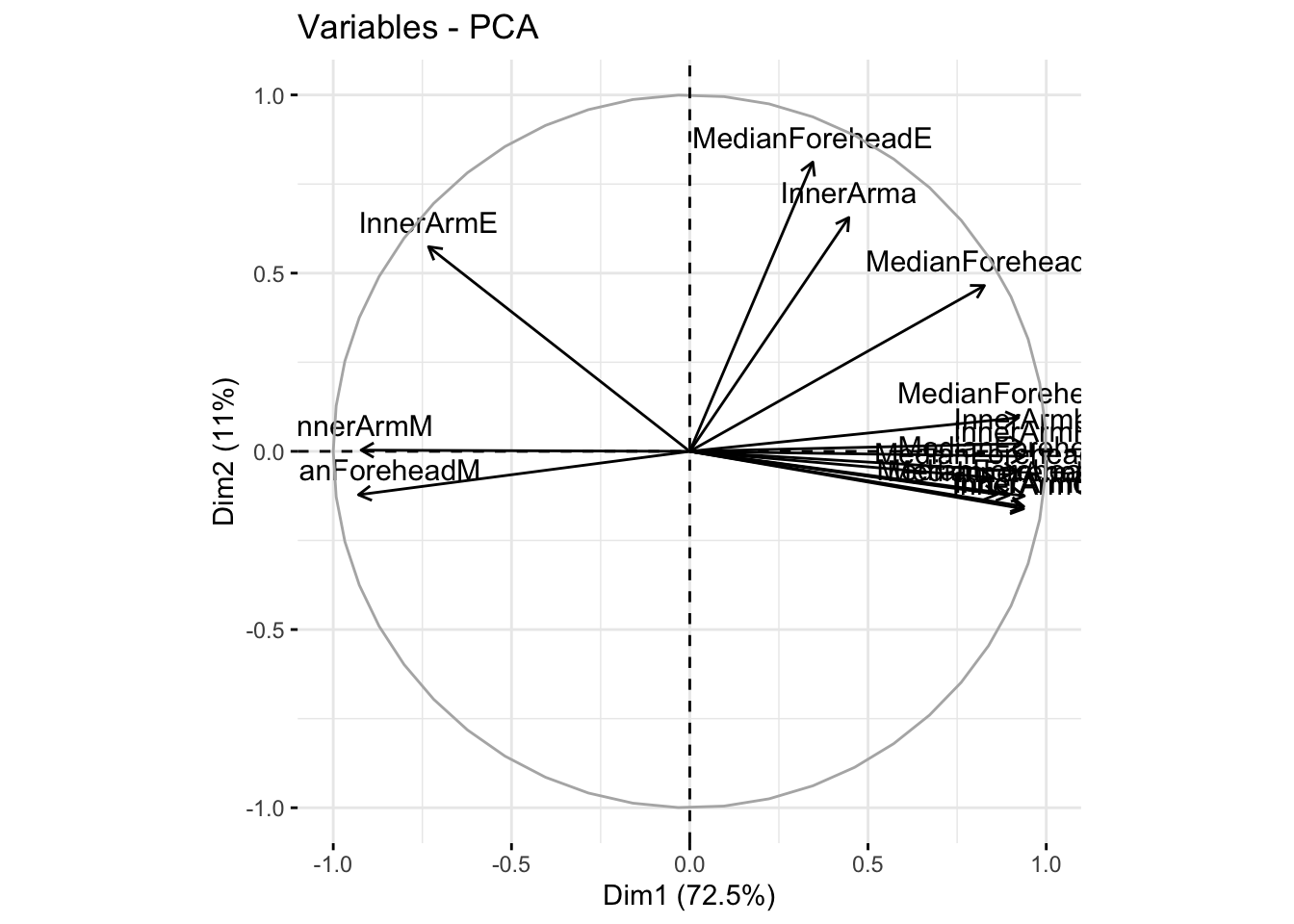
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(summer_refl_pca, choice = "var", axes = 1:2)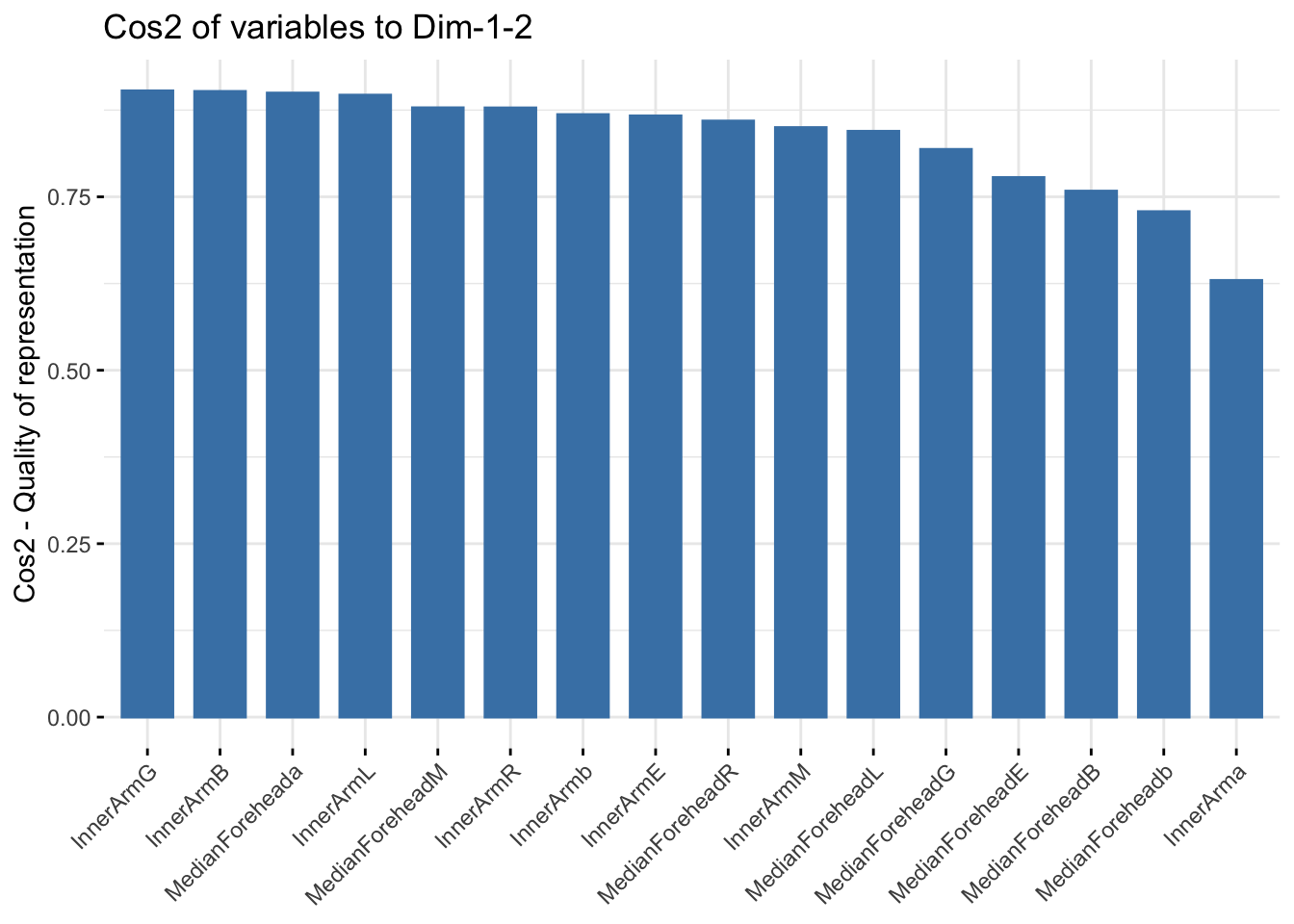
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
summer_pigm_comps <- as.data.frame(summer_refl_pca$x)
summer_pigment <- cbind(summer_data,summer_pigm_comps[,c(1,2)])
ggplot(summer_pigment, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Summer Pigmentation PCA")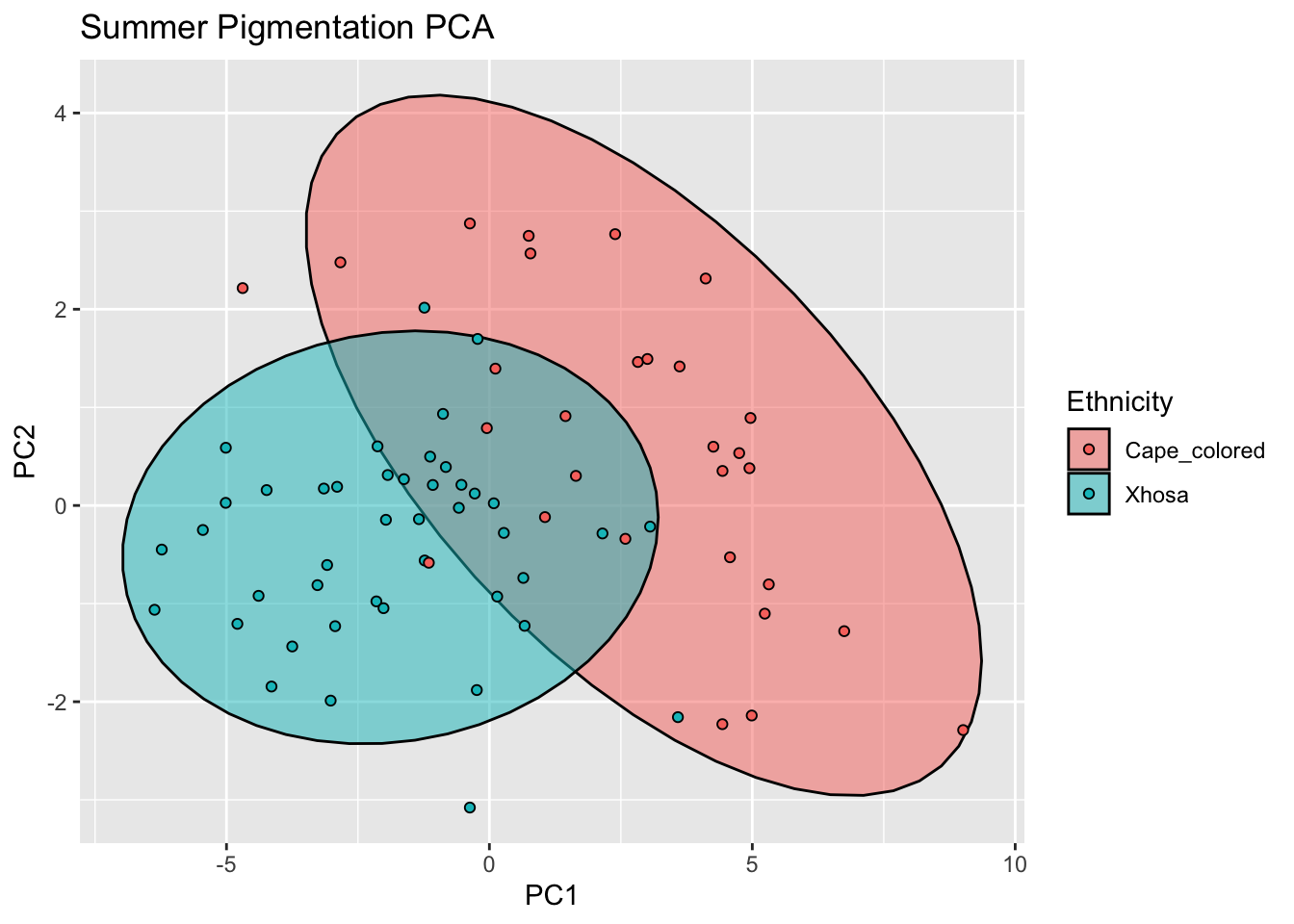
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(summer_refl_pca, choice = "var", axes = 1, top = 10)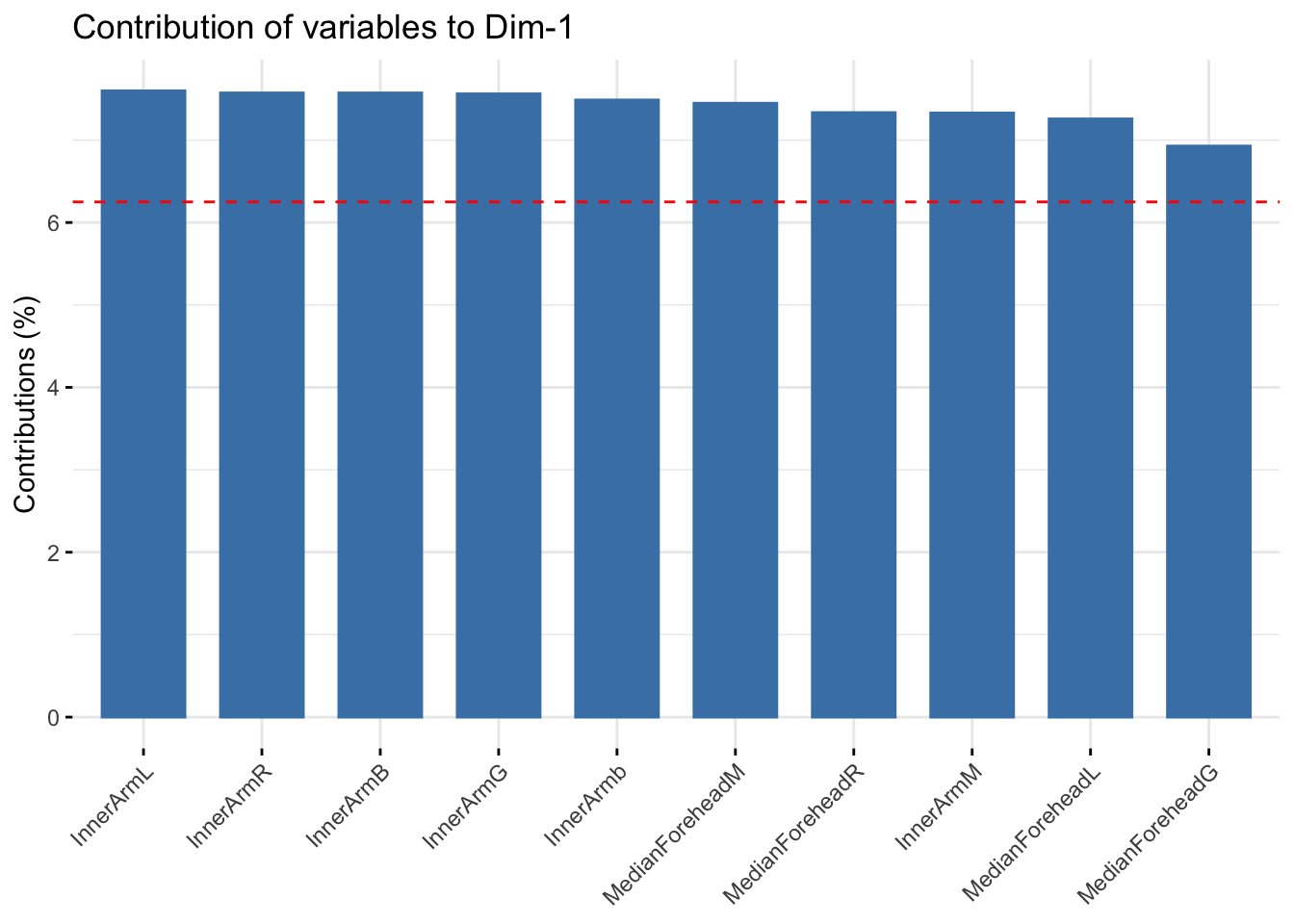
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(summer_refl_pca, choice = "var", axes = 2, top = 10)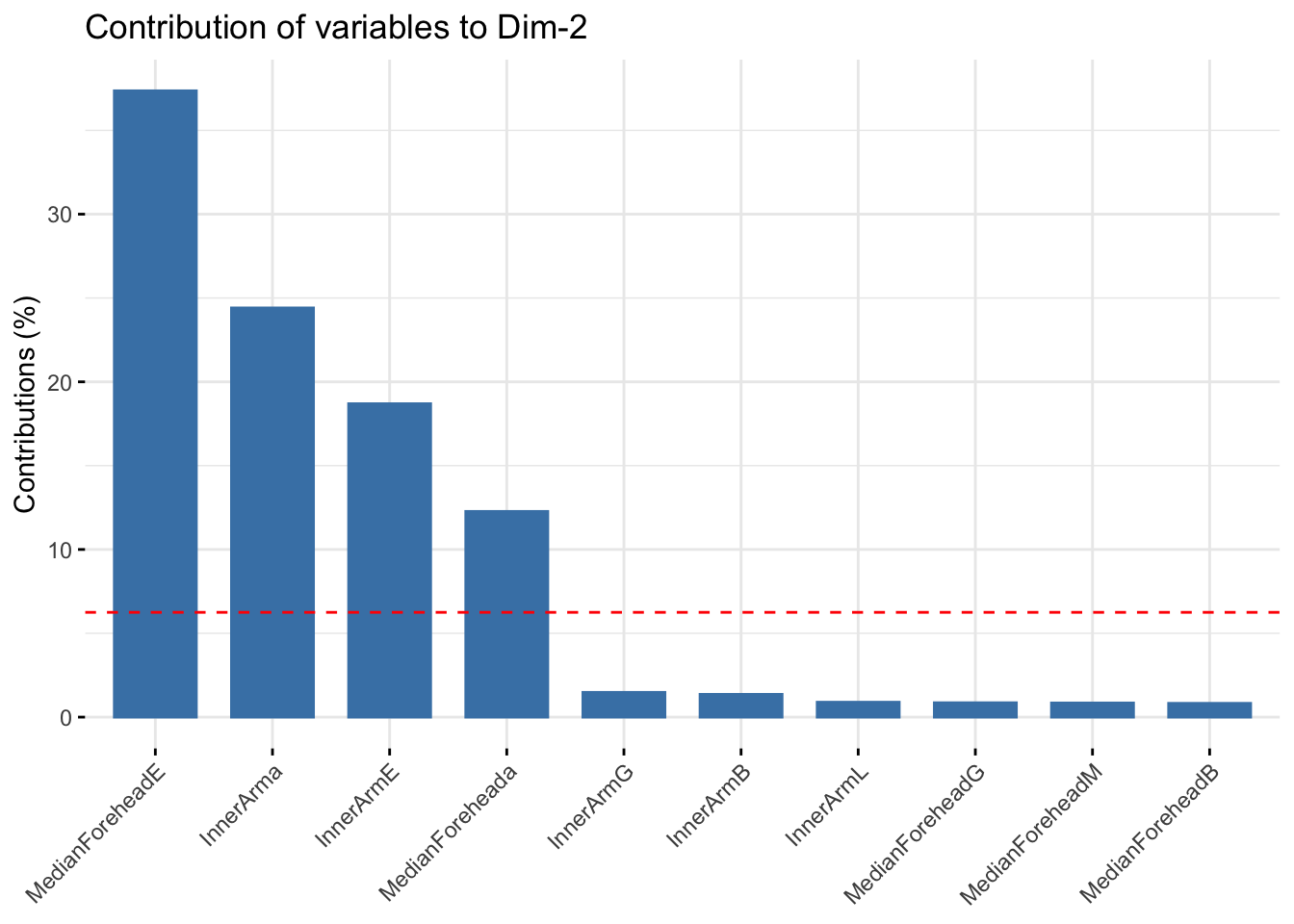
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
winter_refl <- scale(winter_refl_metrics)
winter_refl_pca <- prcomp(winter_refl)
summary(winter_refl_pca)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 3.5551 1.3839 0.80737 0.64068 0.48775 0.26632 0.15397
Proportion of Variance 0.7899 0.1197 0.04074 0.02565 0.01487 0.00443 0.00148
Cumulative Proportion 0.7899 0.9096 0.95038 0.97603 0.99090 0.99534 0.99682
PC8 PC9 PC10 PC11 PC12 PC13 PC14
Standard deviation 0.14316 0.12764 0.07986 0.06160 0.03646 0.03546 0.02830
Proportion of Variance 0.00128 0.00102 0.00040 0.00024 0.00008 0.00008 0.00005
Cumulative Proportion 0.99810 0.99912 0.99952 0.99975 0.99984 0.99991 0.99996
PC15 PC16
Standard deviation 0.01758 0.01615
Proportion of Variance 0.00002 0.00002
Cumulative Proportion 0.99998 1.00000winter_refl_pca$loadings[, 1:2]NULLfviz_eig(winter_refl_pca, addlabels = TRUE)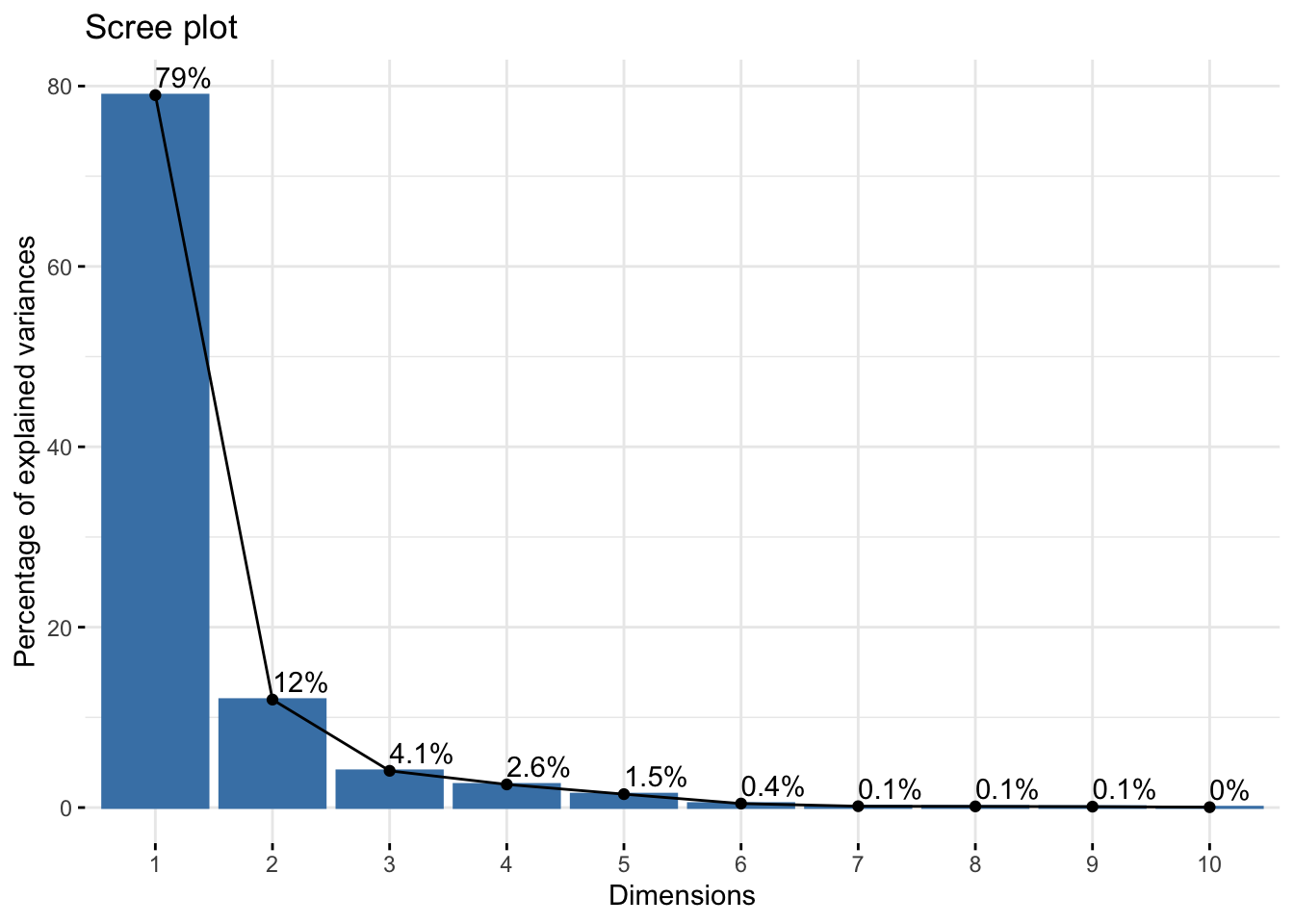
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(winter_refl_pca, col.var = "black")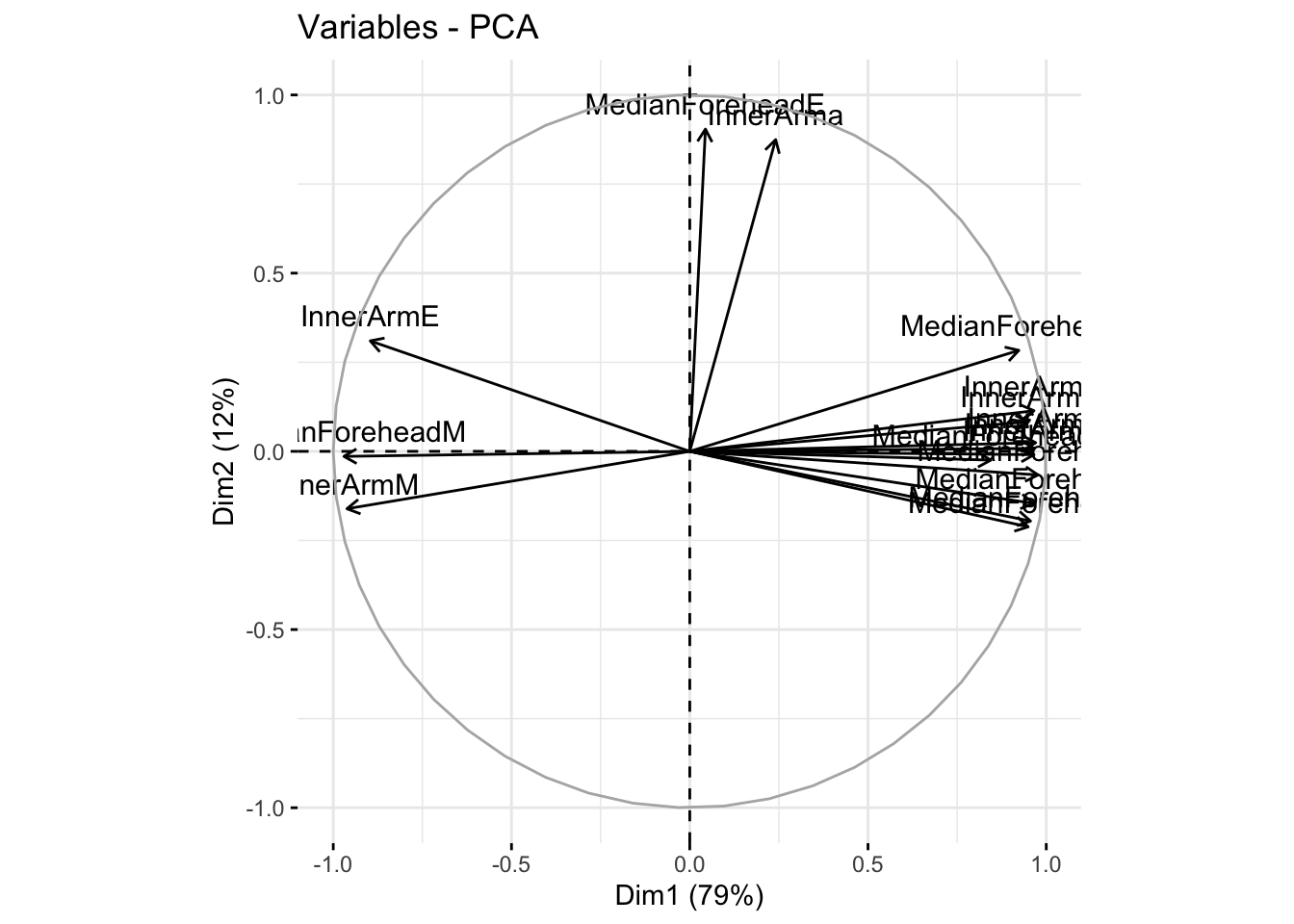
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(winter_refl_pca, choice = "var", axes = 1:2)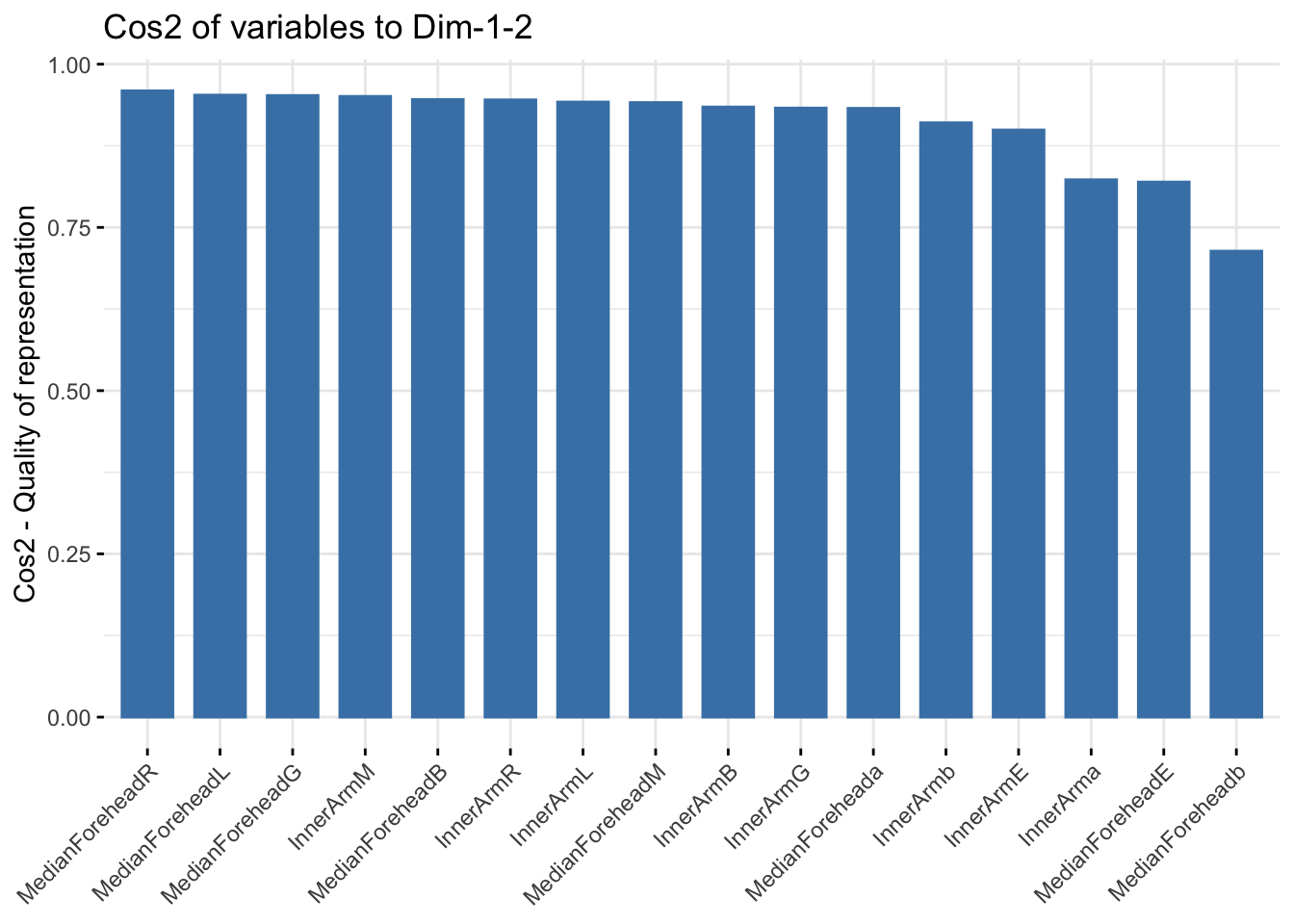
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
winter_pigm_comps <- as.data.frame(winter_refl_pca$x)
winter_pigment <- cbind(winter_data,winter_pigm_comps[,c(1,2)])
ggplot(winter_pigment, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Winter Pigmentation PCA")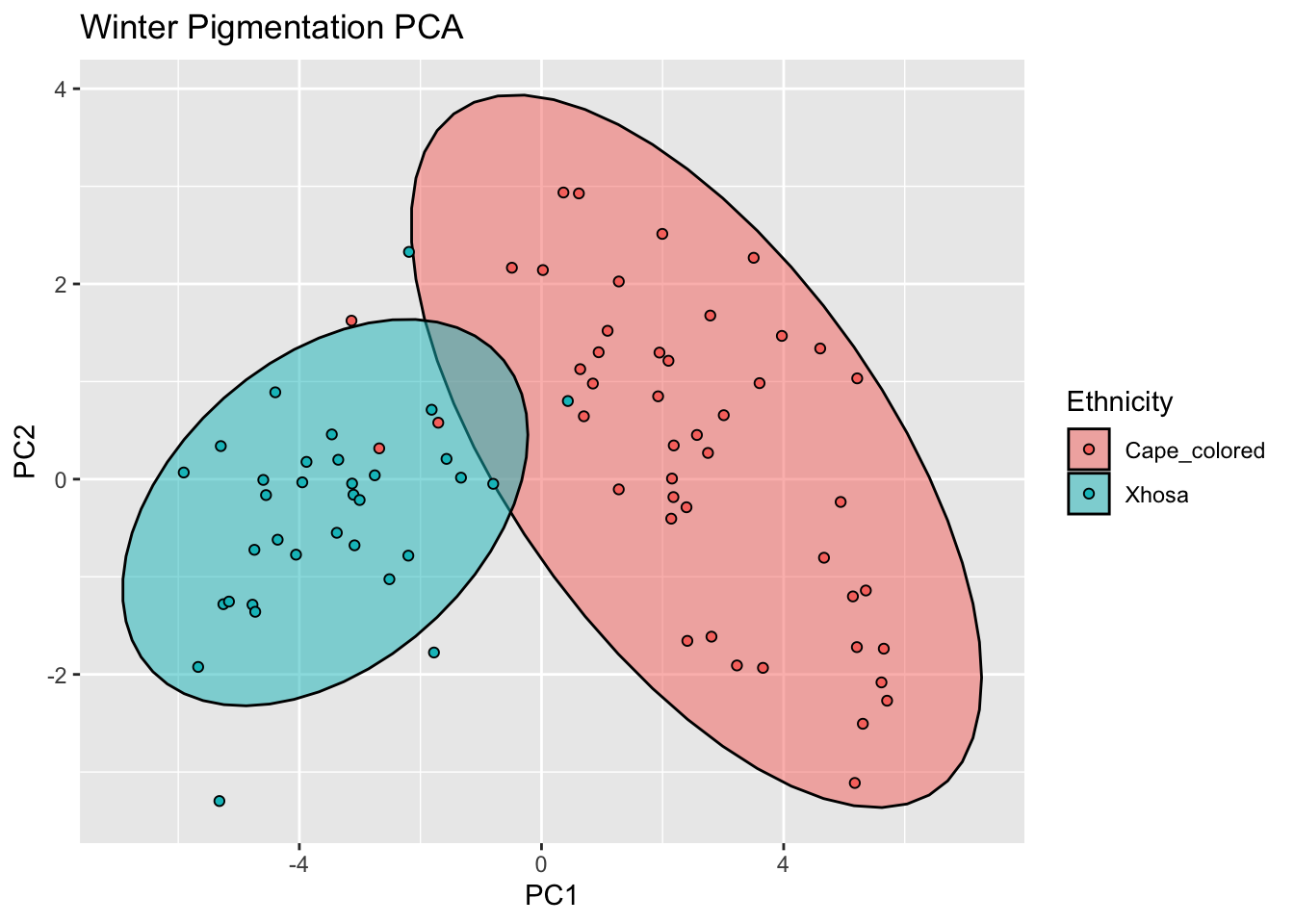
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(winter_refl_pca, choice = "var", axes = 1, top = 10)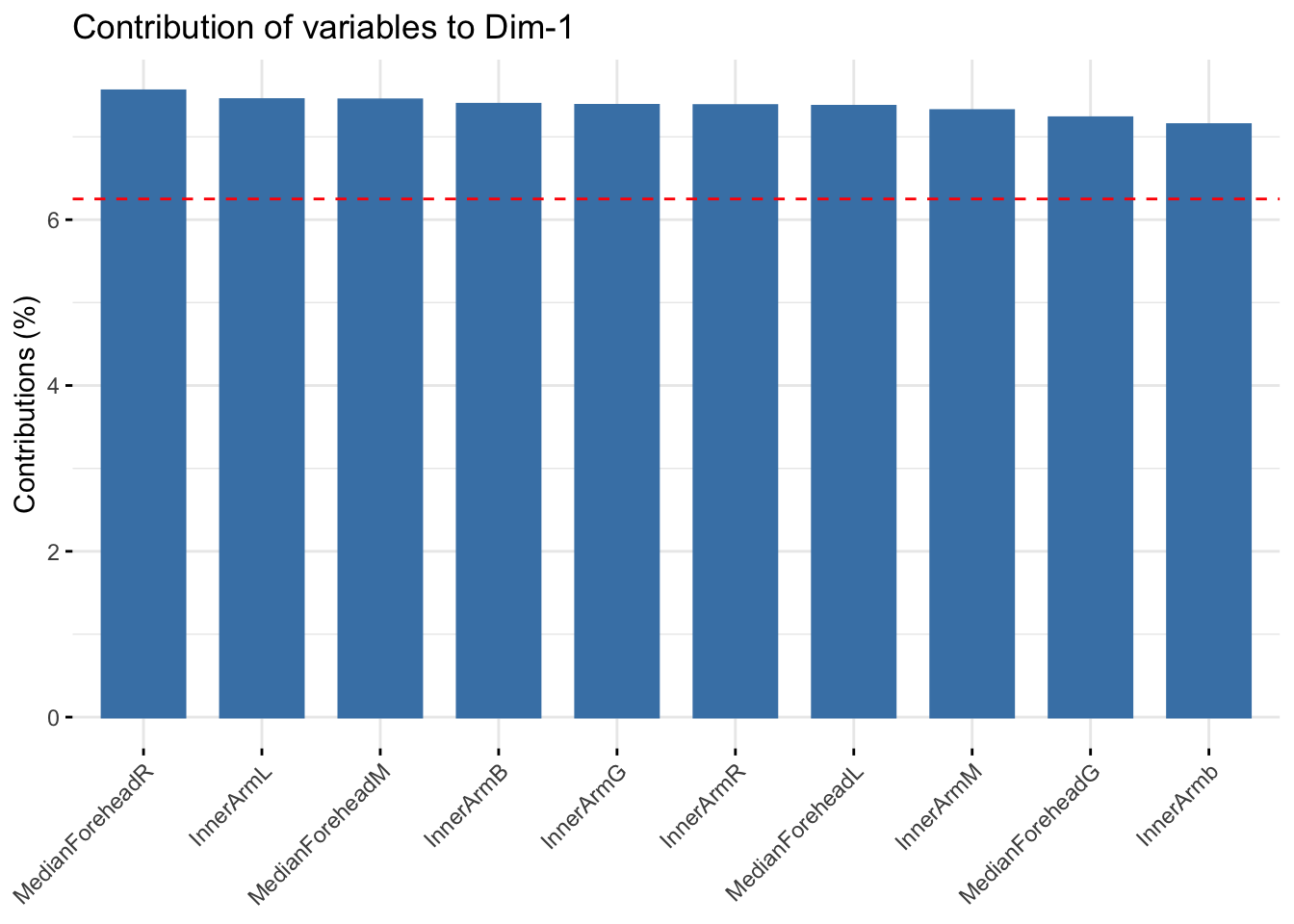
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(winter_refl_pca, choice = "var", axes = 2, top = 10)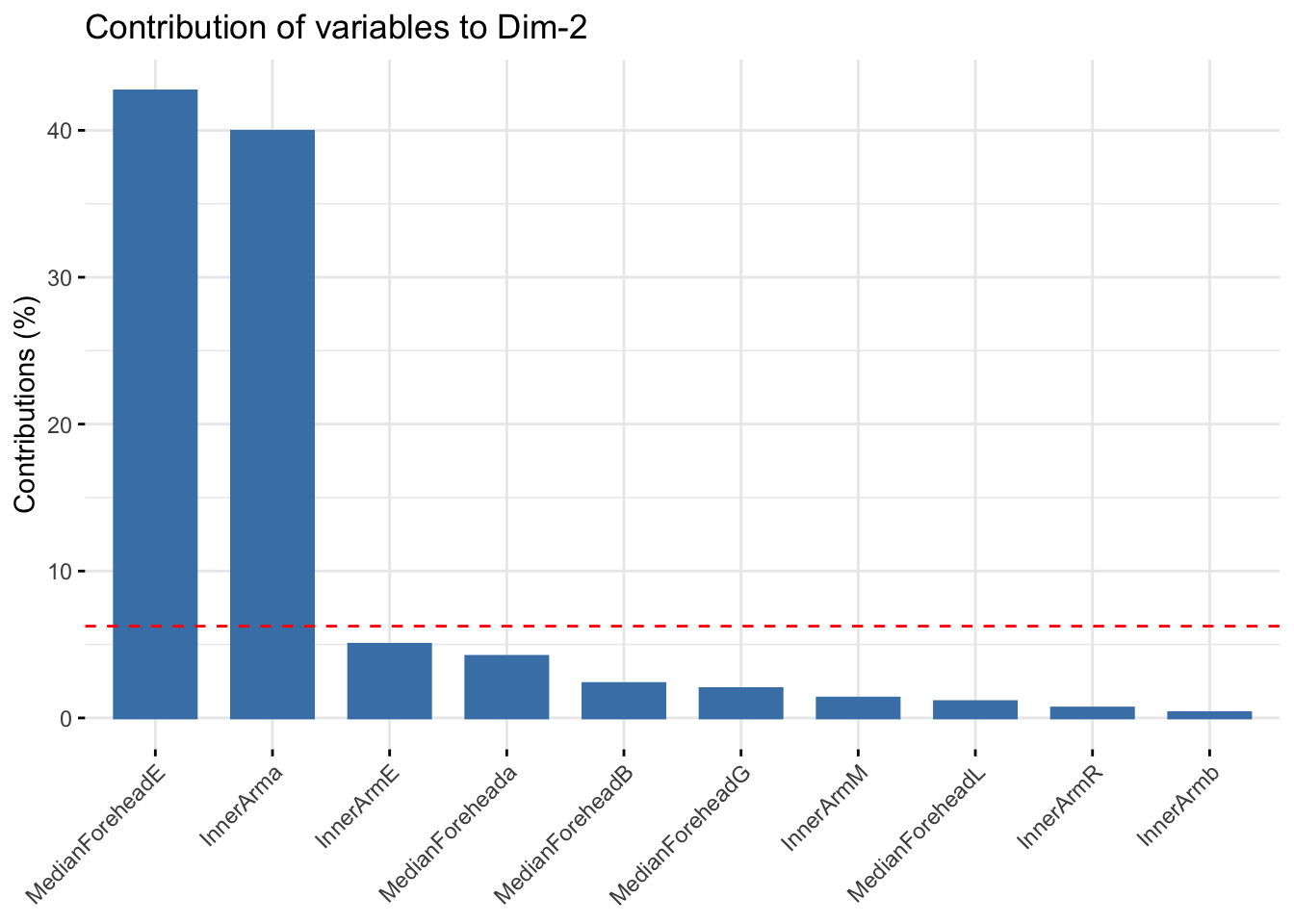
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
RGB subset
summer_winter_clean <- na.omit(summer_winter)
reflectance_rgb_ws <- summer_winter_clean %>%
select(matches("MedianForeheadR|MedianForeheadG|MedianForeheadB|InnerArmR|InnerArmG|InnerArmB", ignore.case = FALSE))
reflectance4 <- scale(reflectance_rgb_ws)
reflectancergbws <- prcomp(reflectance4)
summary(reflectancergbws)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 3.1878 0.94496 0.72698 0.51980 0.2593 0.19373 0.12847
Proportion of Variance 0.8468 0.07441 0.04404 0.02252 0.0056 0.00313 0.00138
Cumulative Proportion 0.8468 0.92126 0.96530 0.98782 0.9934 0.99655 0.99792
PC8 PC9 PC10 PC11 PC12
Standard deviation 0.10024 0.08260 0.07502 0.04244 0.02558
Proportion of Variance 0.00084 0.00057 0.00047 0.00015 0.00005
Cumulative Proportion 0.99876 0.99933 0.99980 0.99995 1.00000reflectancergbws$loadings[, 1:2]NULLfviz_eig(reflectancergbws, addlabels = TRUE)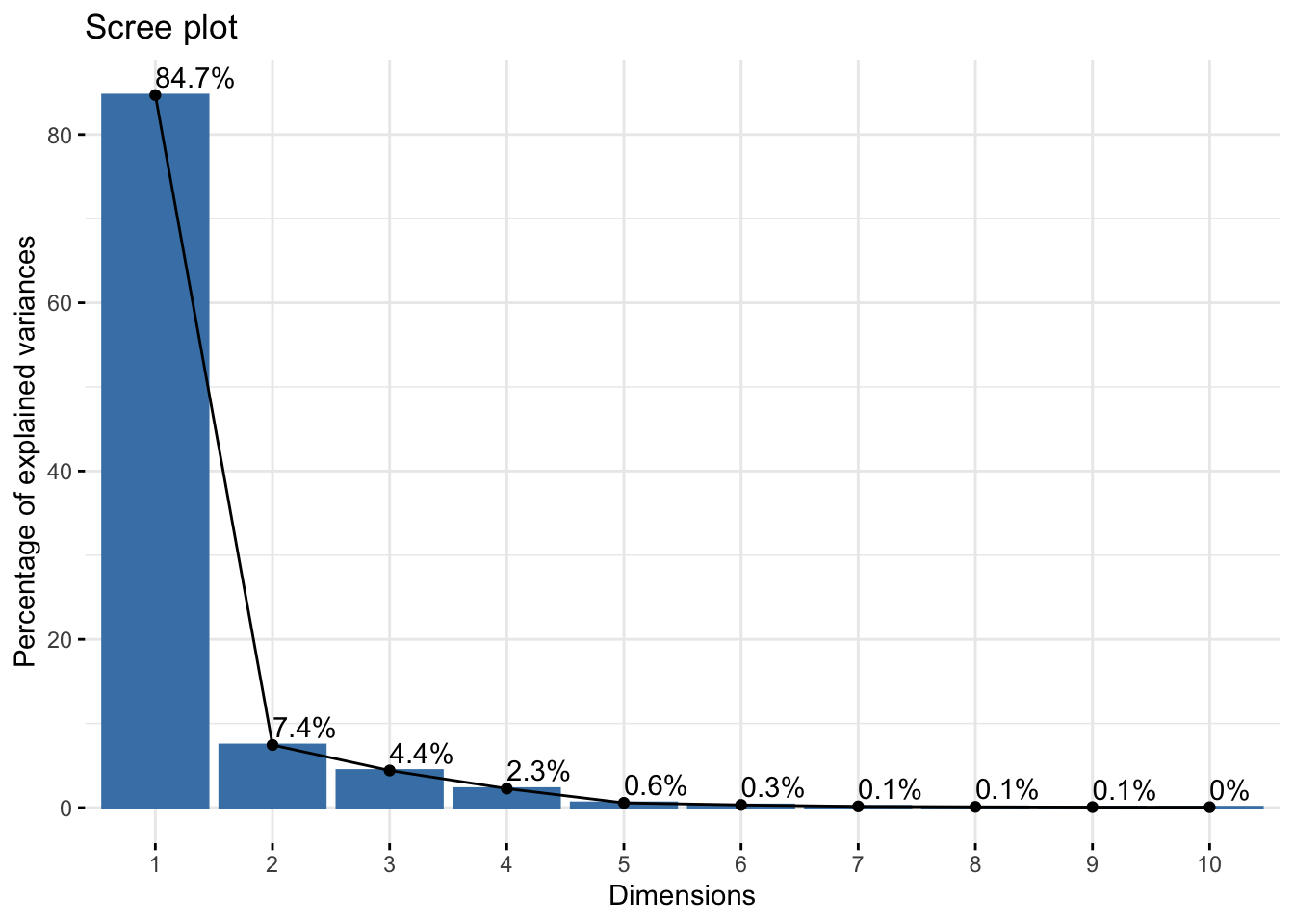
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(reflectancergbws, col.var = "black")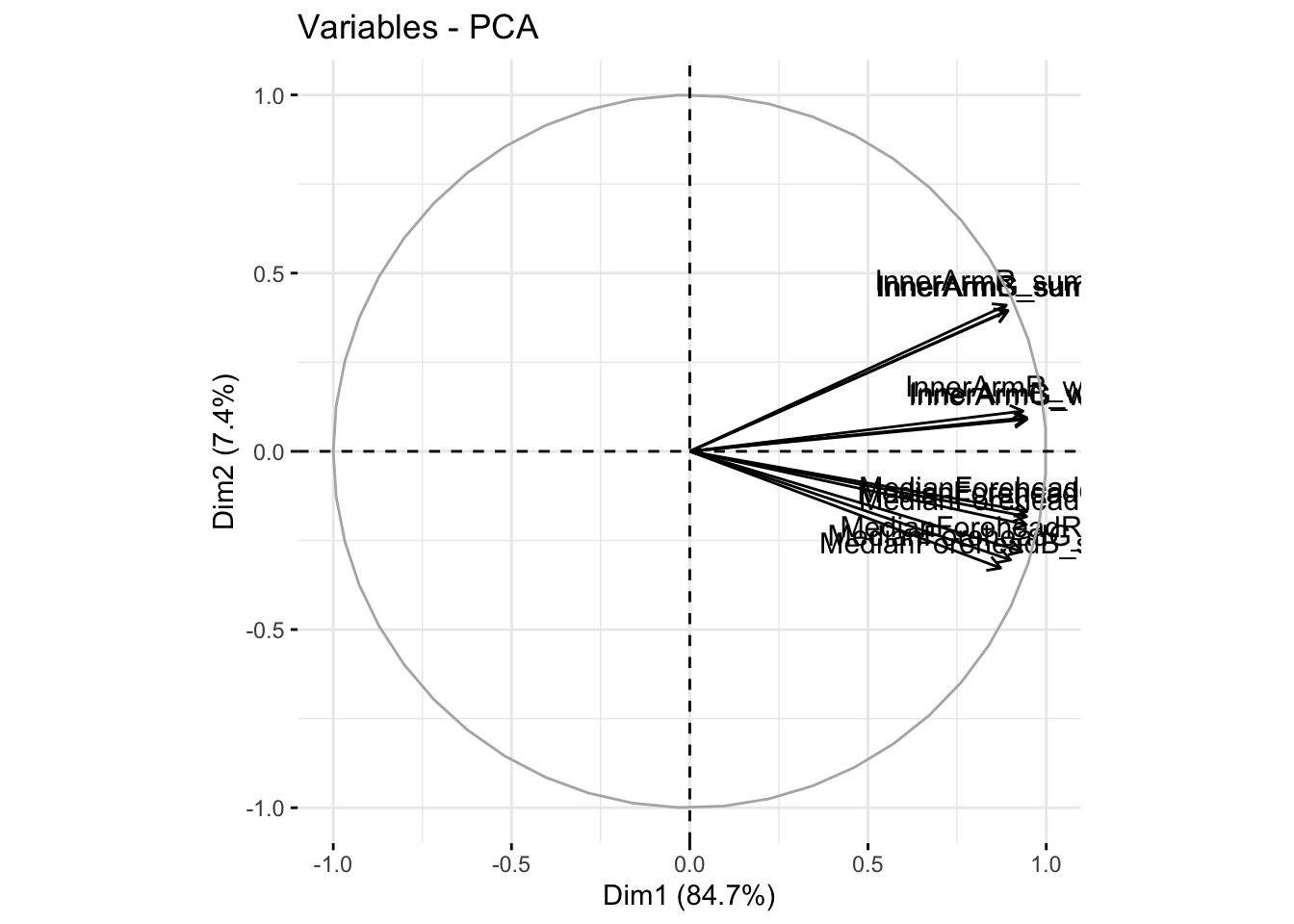
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(reflectancergbws, choice = "var", axes = 1:2)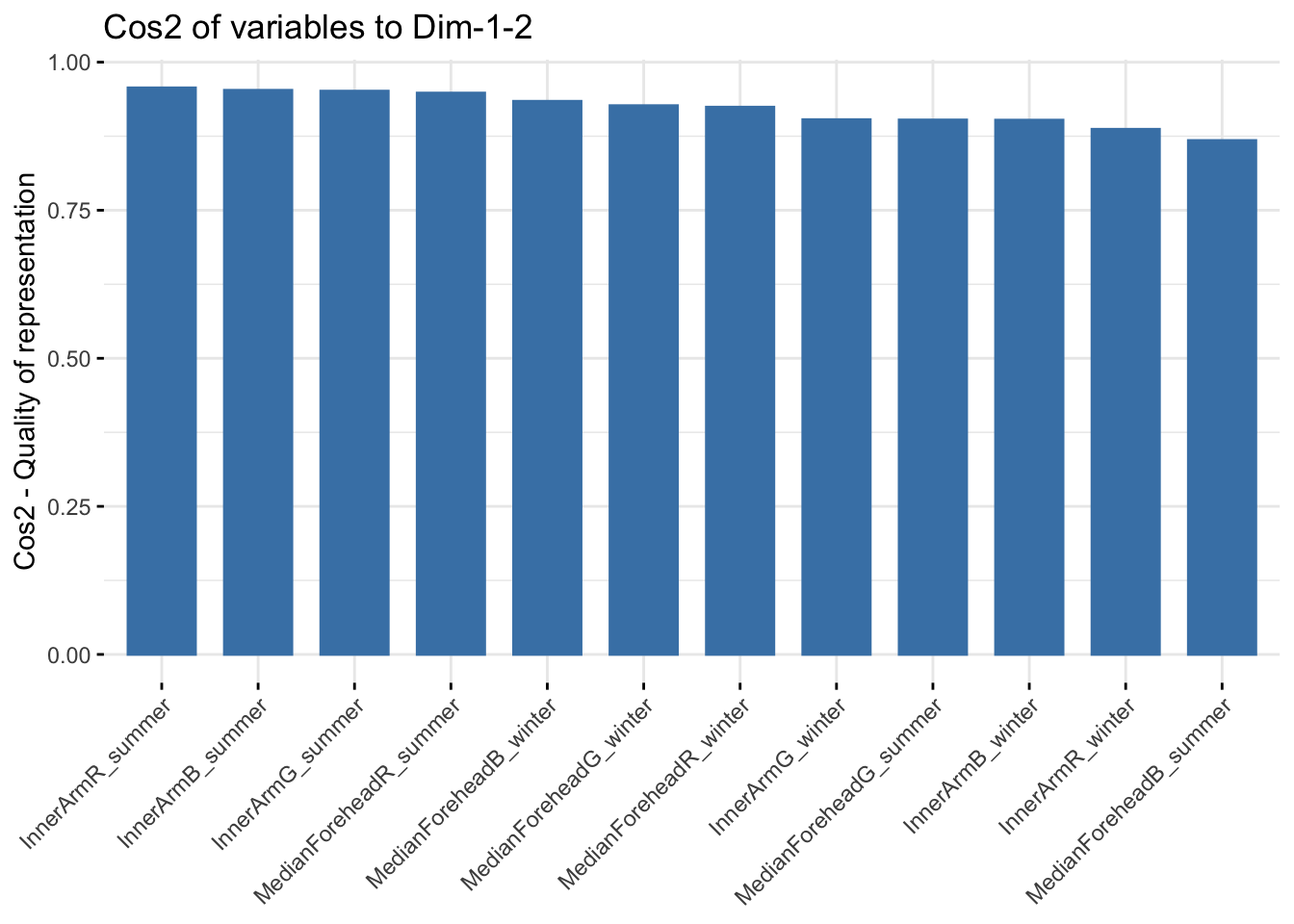
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
wsrgbcomps <- as.data.frame(reflectancergbws$x)
rgb_ws_bind <- cbind(summer_winter_clean,wsrgbcomps[,c(1,2)])
rgb_ws_bind <- rgb_ws_bind %>%
select(-matches("SkinReflectance"))
rgb_ws_bind <- rgb_ws_bind %>%
select(-"Ethnicity_winter")
names(rgb_ws_bind)[names(rgb_ws_bind) == 'Ethnicity_summer'] <- 'Ethnicity'
ggplot(rgb_ws_bind, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Wide Pigmentation PCA")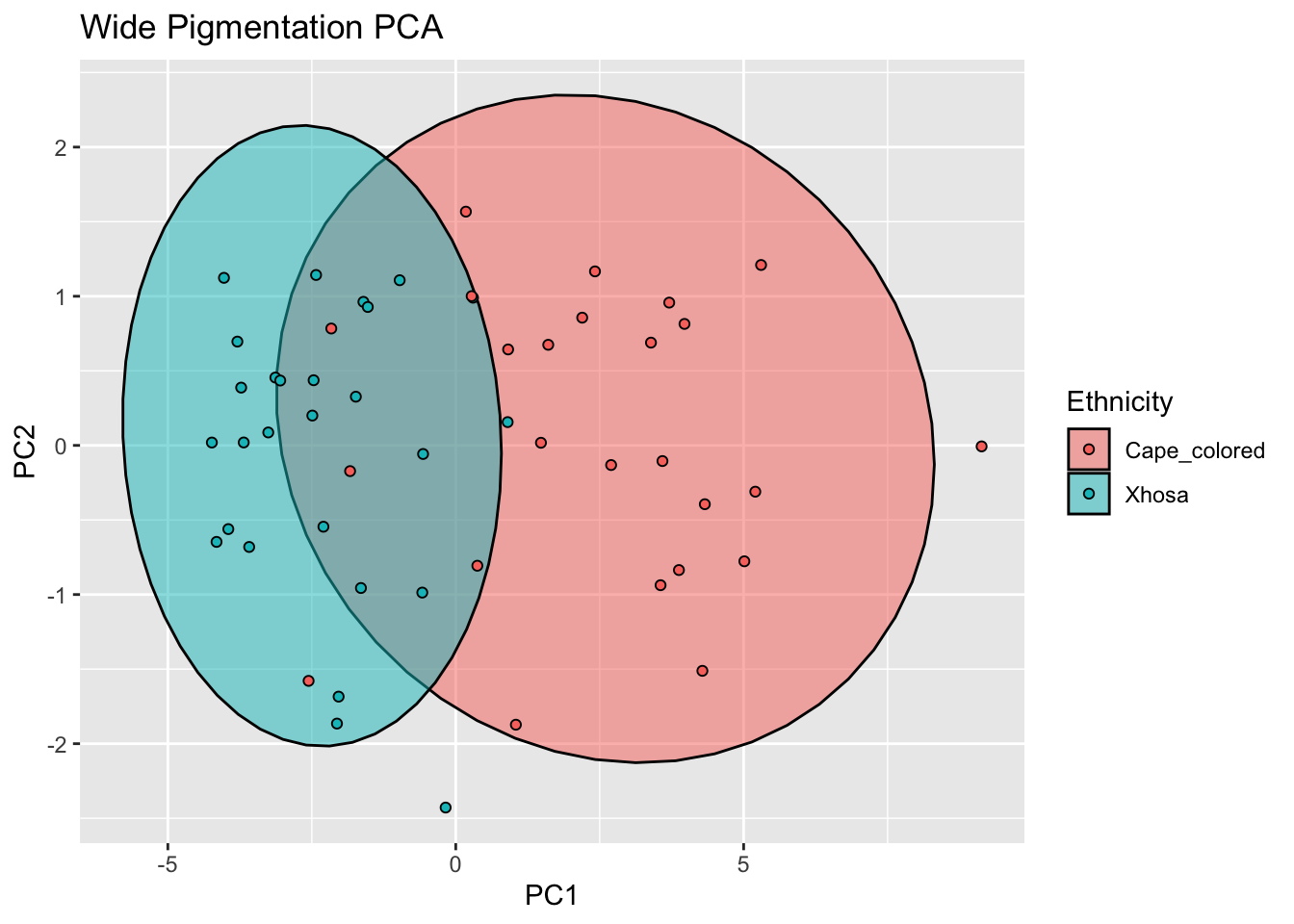
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(reflectancergbws, choice = "var", axes = 1, top = 20)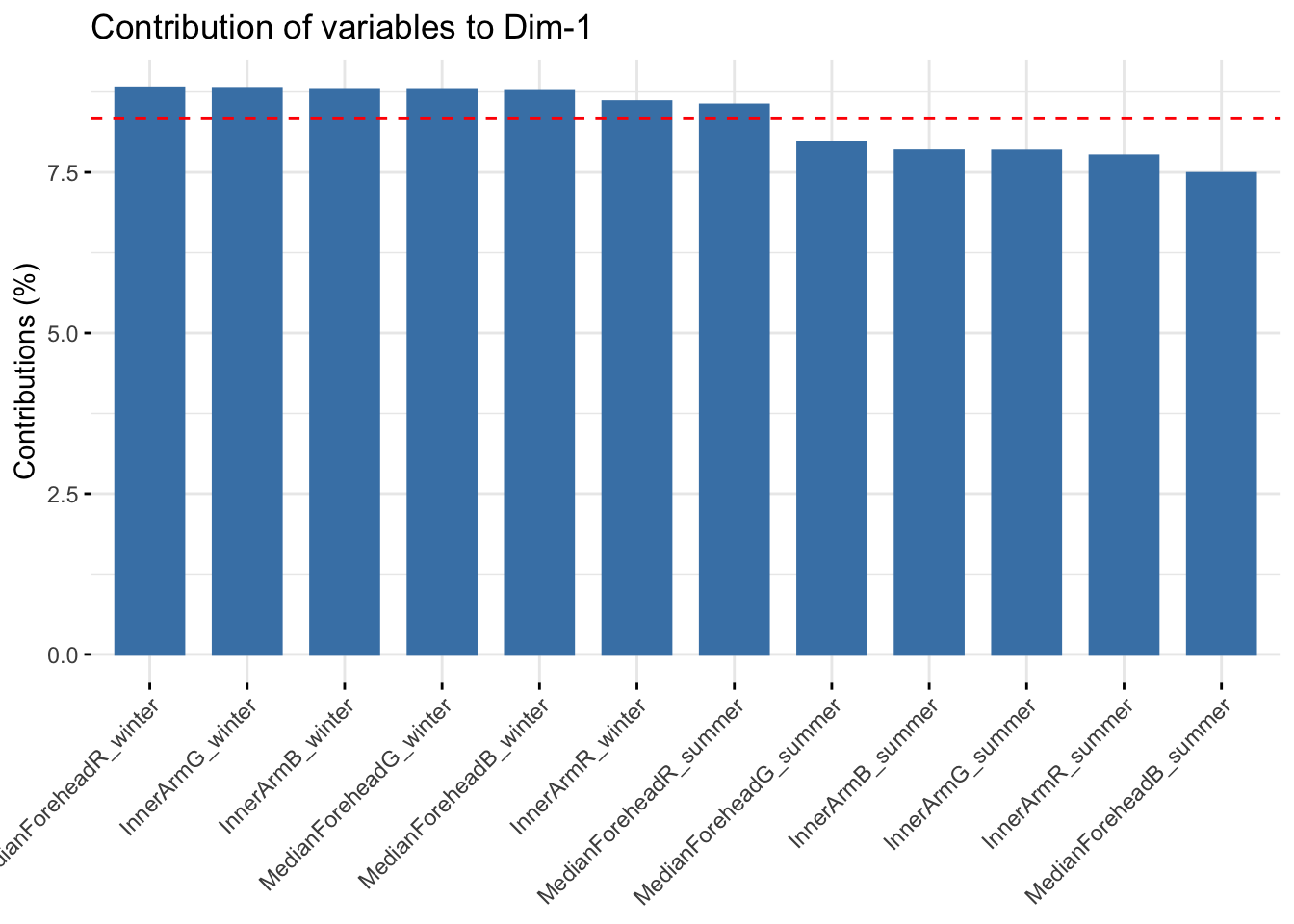
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(reflectancergbws, choice = "var", axes = 2, top = 20)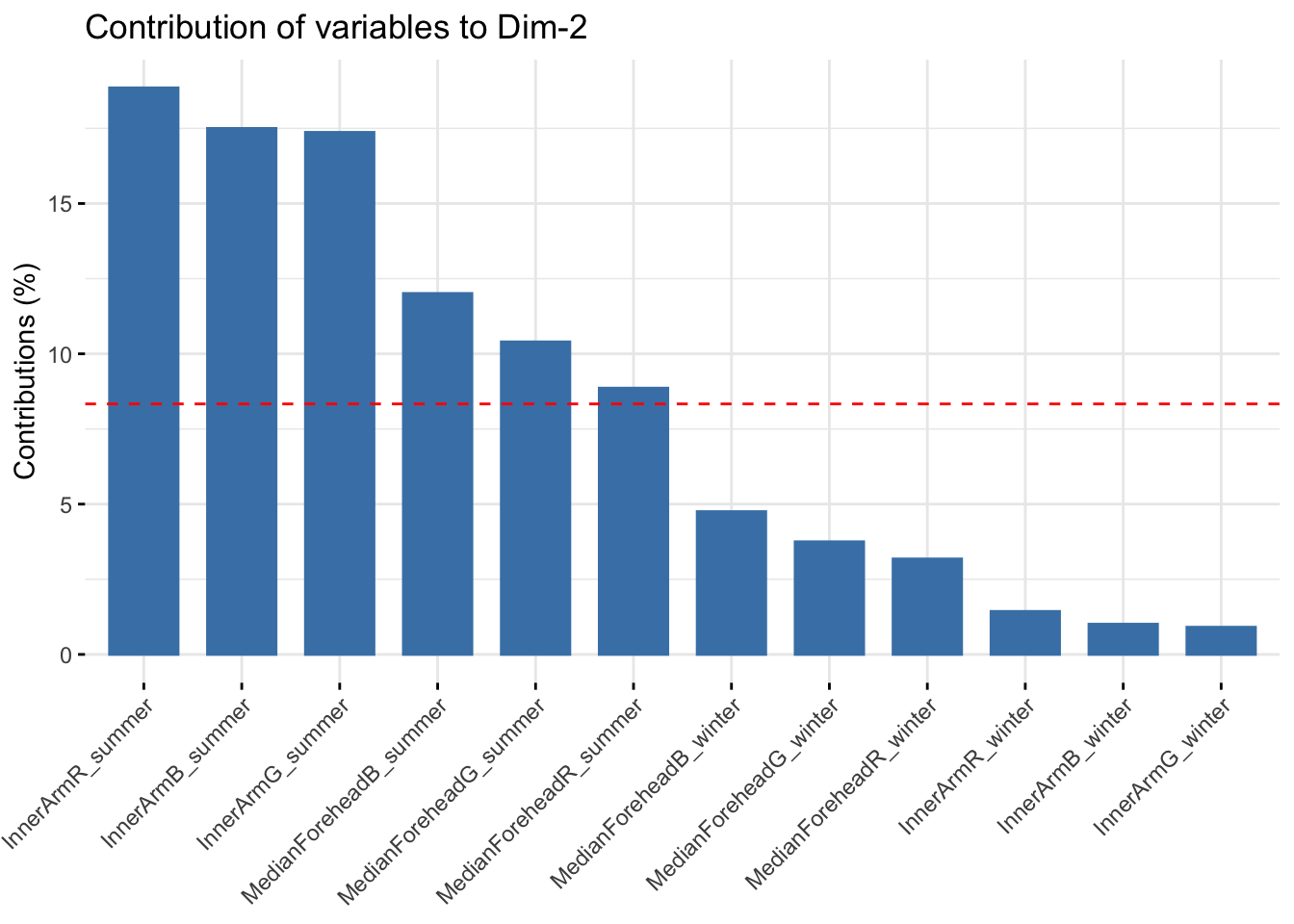
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
summer_rgb <- summer_data %>%
select(matches("ForeheadR|ForeheadG|ForeheadB|InnerArmR|InnerArmG|InnerArmB", ignore.case = FALSE))
summer_rgb <- na.omit(summer_rgb)
winter_rgb <- winter_data %>%
select(matches("ForeheadR|ForeheadG|ForeheadB|InnerArmR|InnerArmG|InnerArmB", ignore.case = FALSE))
winter_rgb <- na.omit(winter_rgb)
six_rgb <- six_week_data %>%
select(matches("ForeheadR|ForeheadG|ForeheadB|InnerArmR|InnerArmG|InnerArmB", ignore.case = FALSE))
six_rgb <- na.omit(six_rgb)winter_rgb_scale <- scale(winter_rgb)
winter_rgb_pca <- prcomp(winter_rgb_scale)
summary(winter_rgb_pca)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6
Standard deviation 2.3720 0.58112 0.15130 0.08075 0.07684 0.01941
Proportion of Variance 0.9378 0.05628 0.00382 0.00109 0.00098 0.00006
Cumulative Proportion 0.9378 0.99405 0.99787 0.99895 0.99994 1.00000winter_rgb_pca$loadings[, 1:2]NULLfviz_eig(winter_rgb_pca, addlabels = TRUE)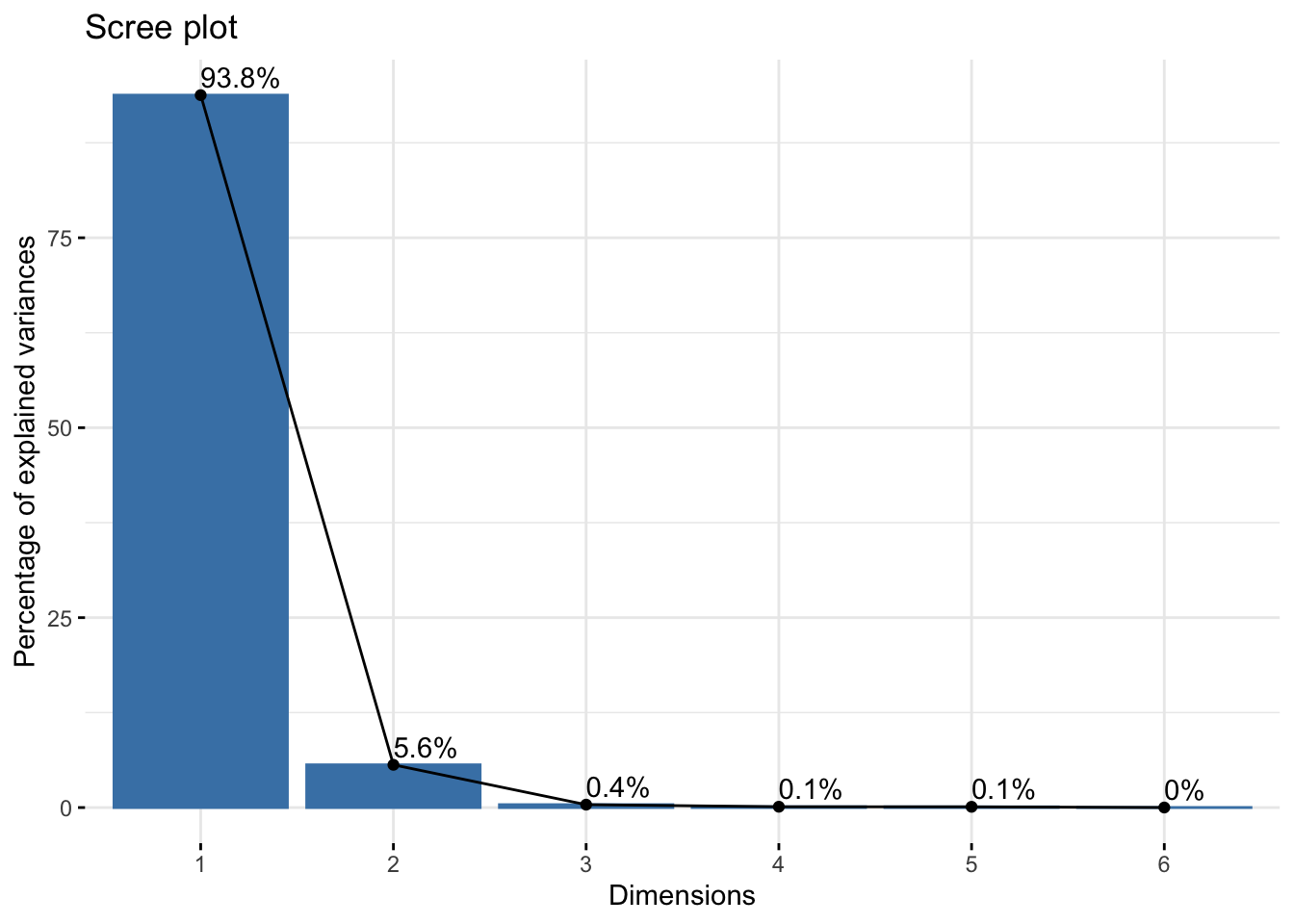
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(winter_rgb_pca, col.var = "black")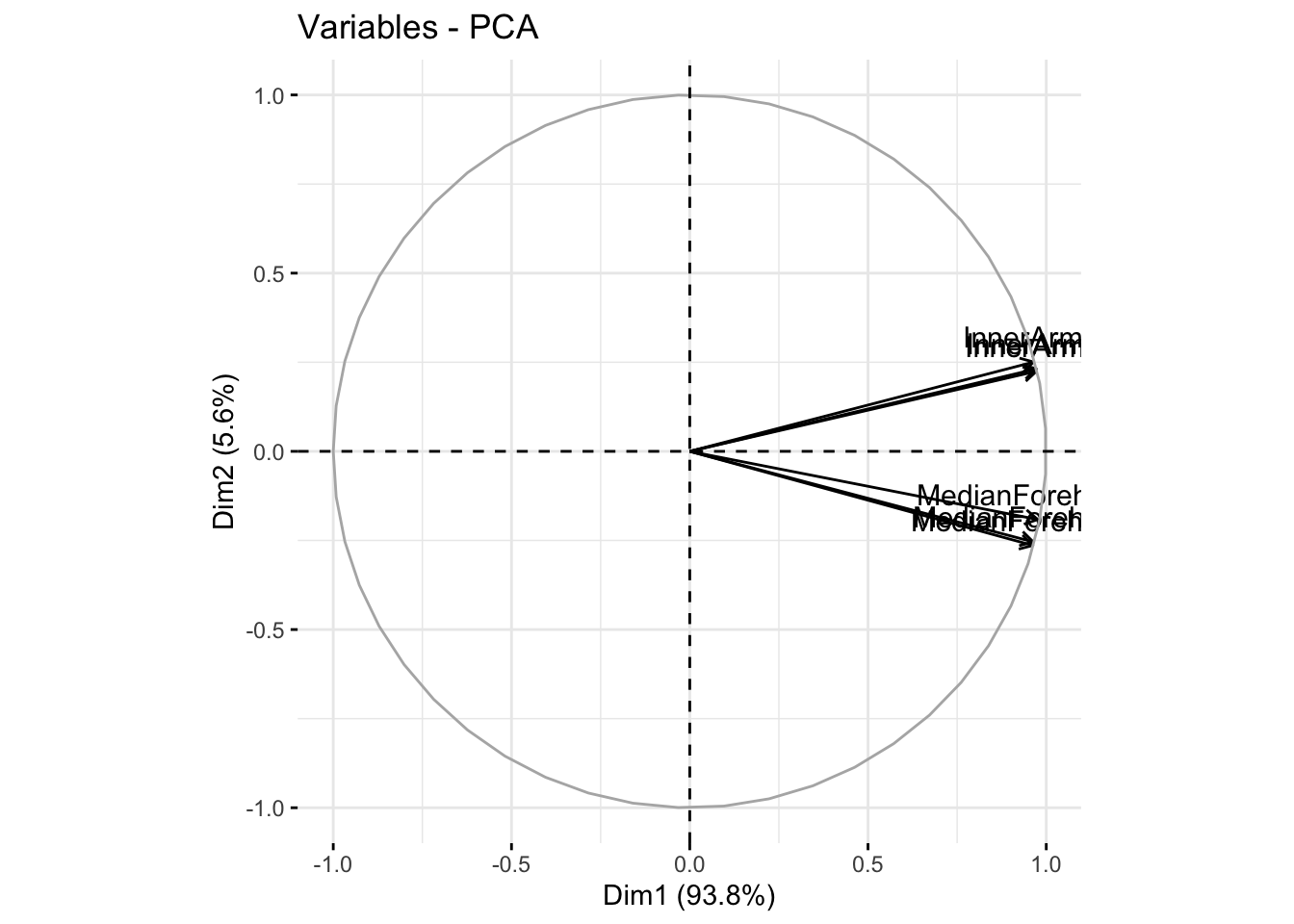
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(winter_rgb_pca, choice = "var", axes = 1:2)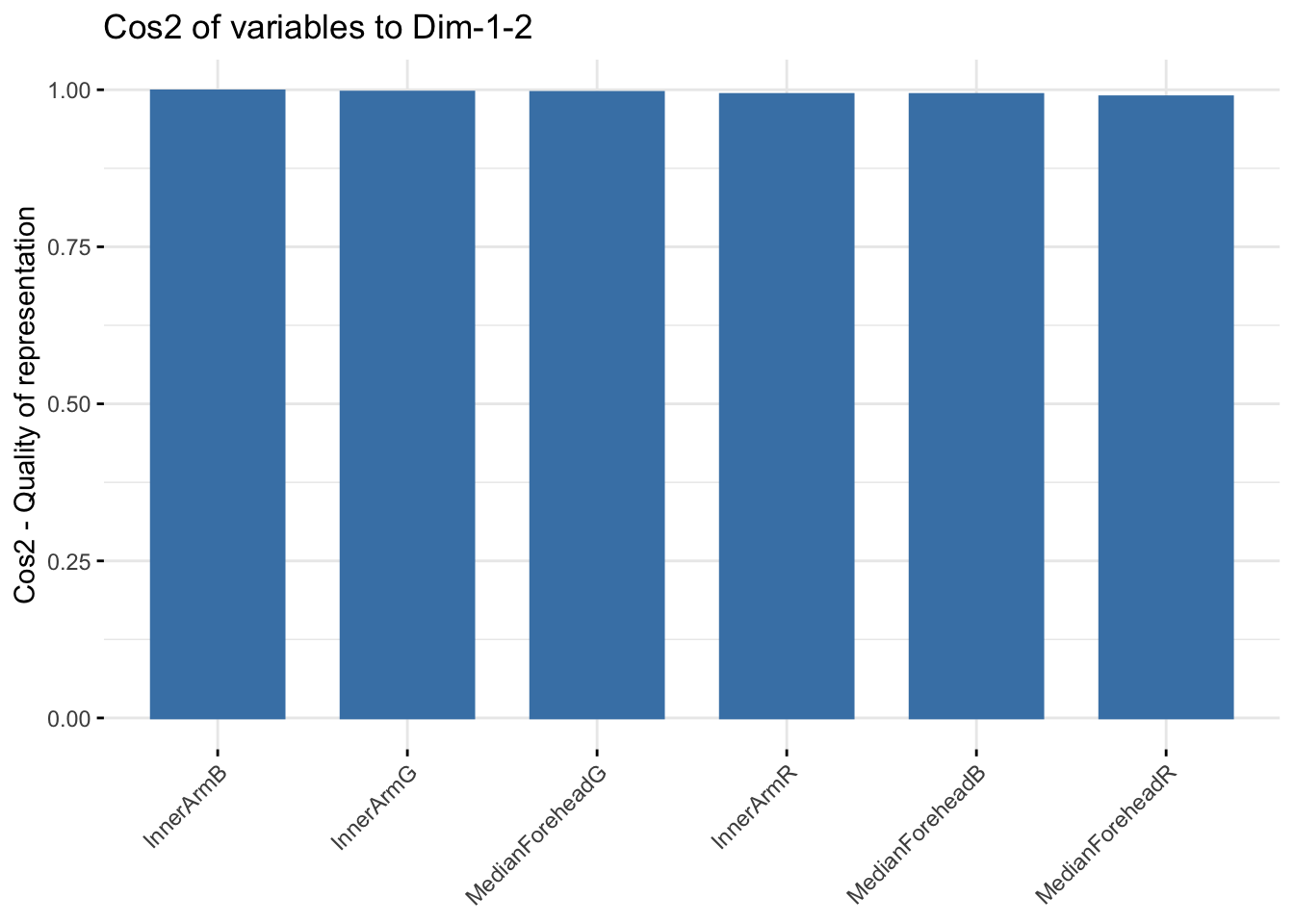
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
winter_rgb_comps <- as.data.frame(winter_rgb_pca$x)
winter_rgb_new <- cbind(winter_data,winter_rgb_comps[,c(1,2)])
ggplot(winter_rgb_new, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Winter RGB PCA")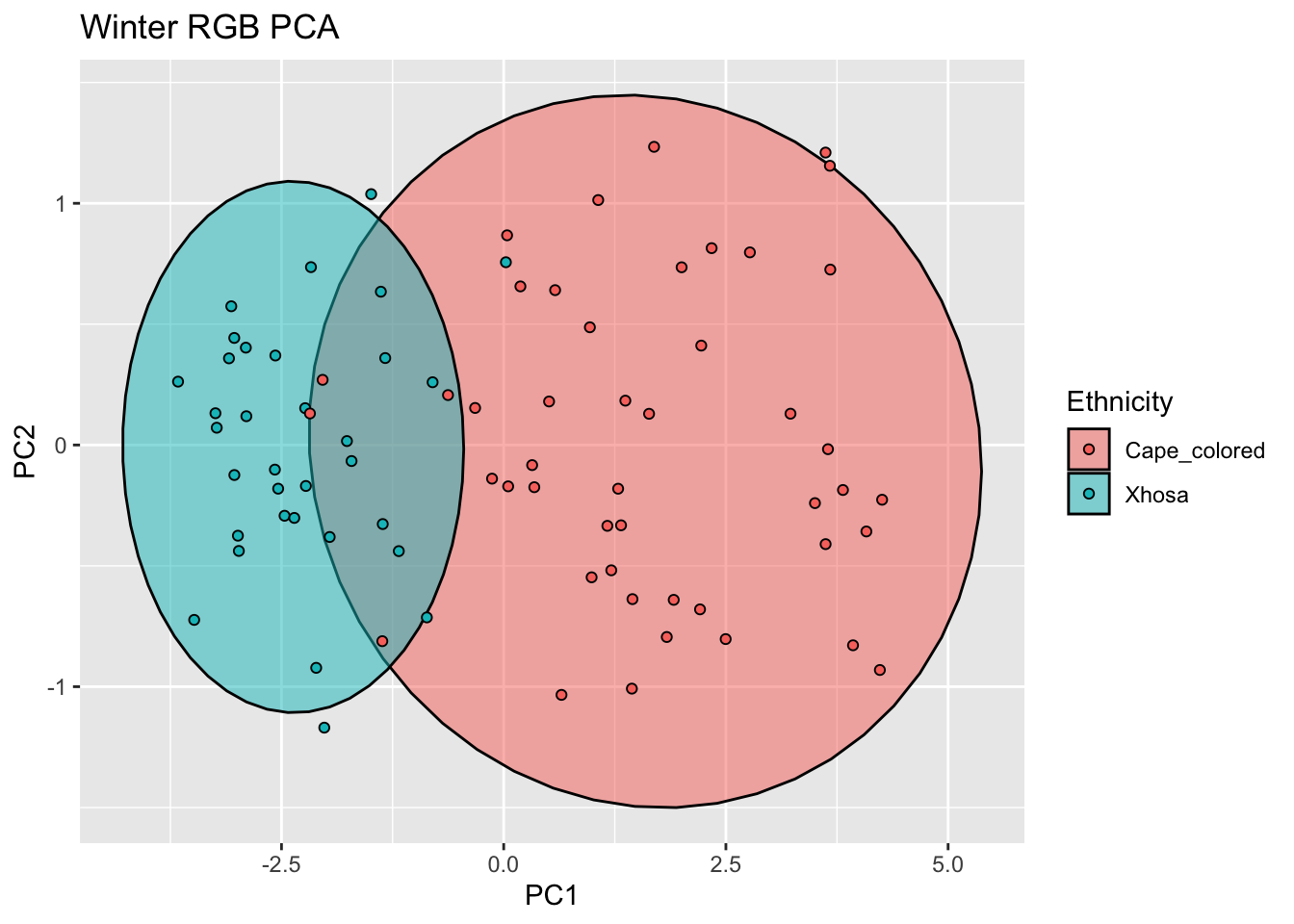
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(winter_rgb_pca, choice = "var", axes = 1, top = 10)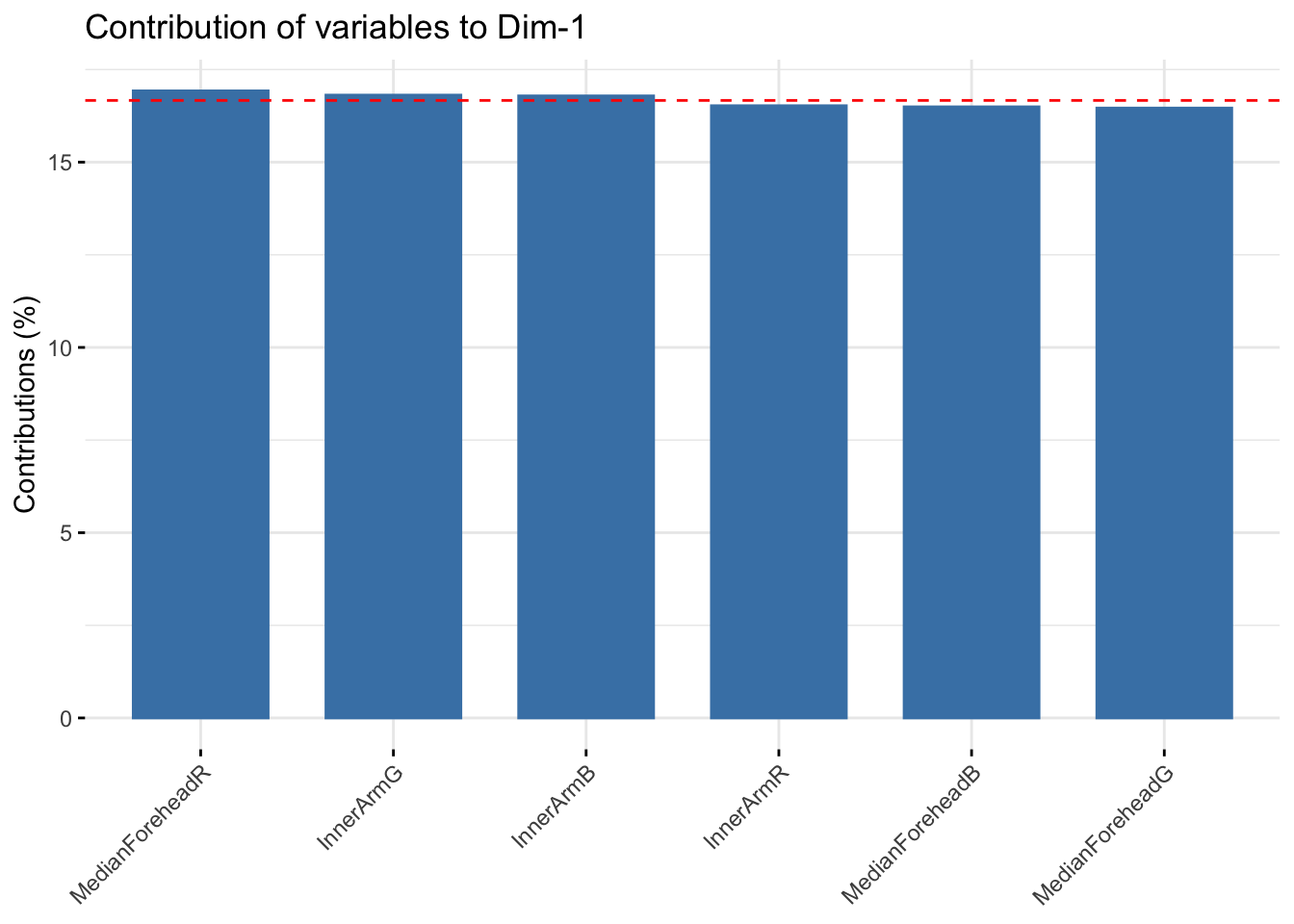
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(winter_rgb_pca, choice = "var", axes = 2, top = 10)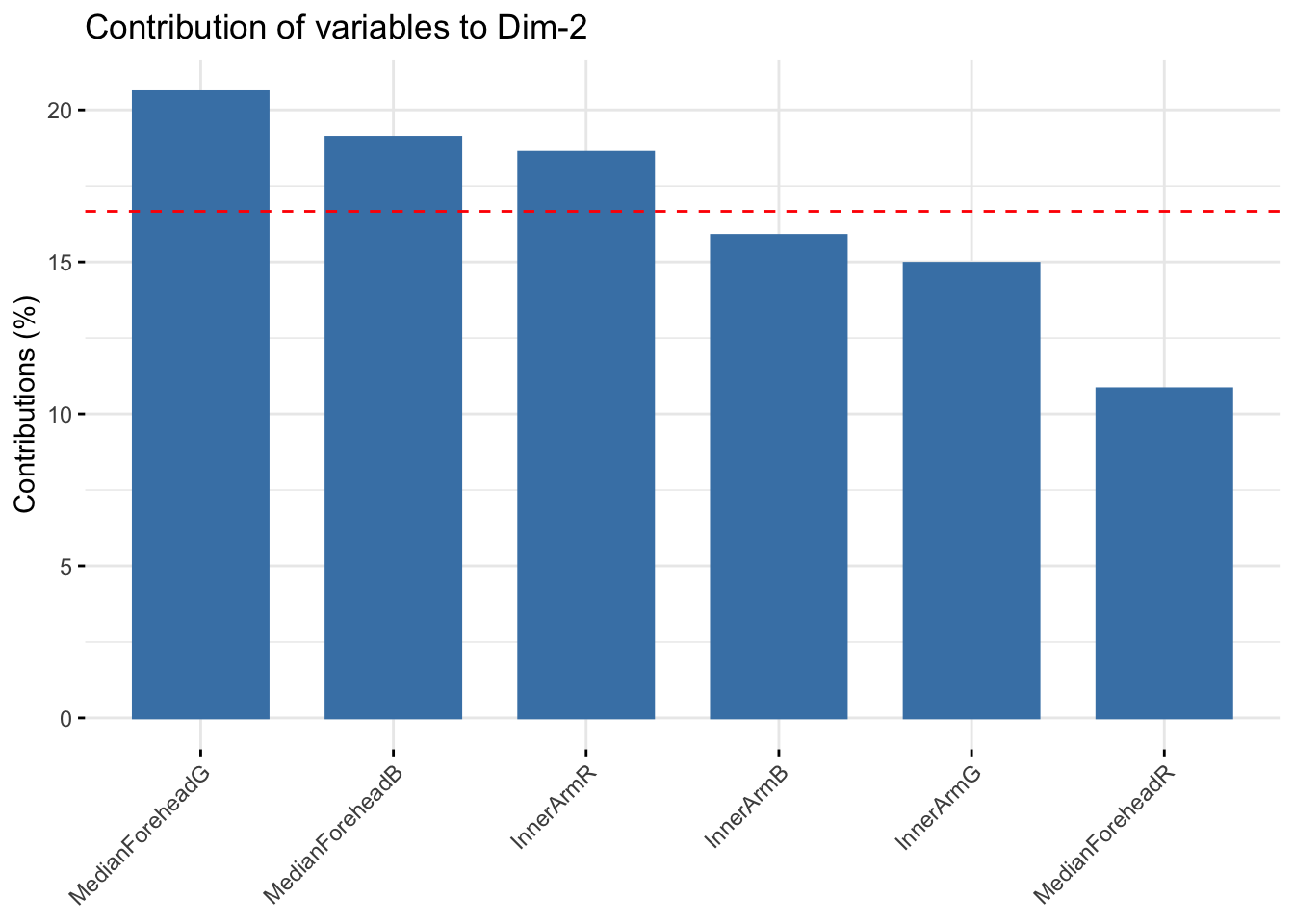
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
summer_rgb_scale <- scale(summer_rgb)
summer_rgb_pca <- prcomp(summer_rgb_scale)
summary(summer_rgb_pca)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6
Standard deviation 2.2617 0.8751 0.26995 0.17017 0.11981 0.05129
Proportion of Variance 0.8526 0.1277 0.01215 0.00483 0.00239 0.00044
Cumulative Proportion 0.8526 0.9802 0.99234 0.99717 0.99956 1.00000summer_rgb_pca$loadings[, 1:2]NULLfviz_eig(summer_rgb_pca, addlabels = TRUE)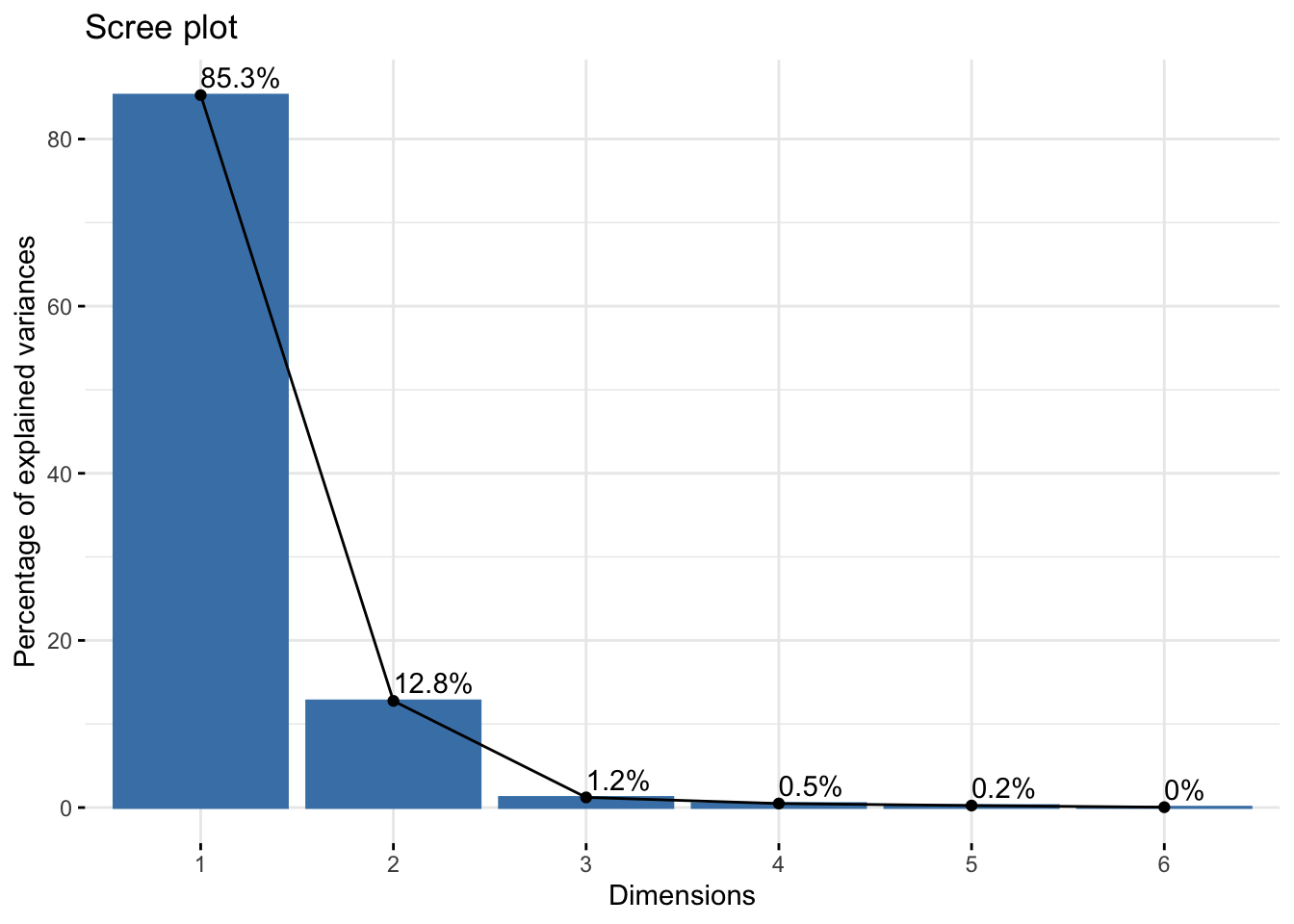
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(summer_rgb_pca, col.var = "black")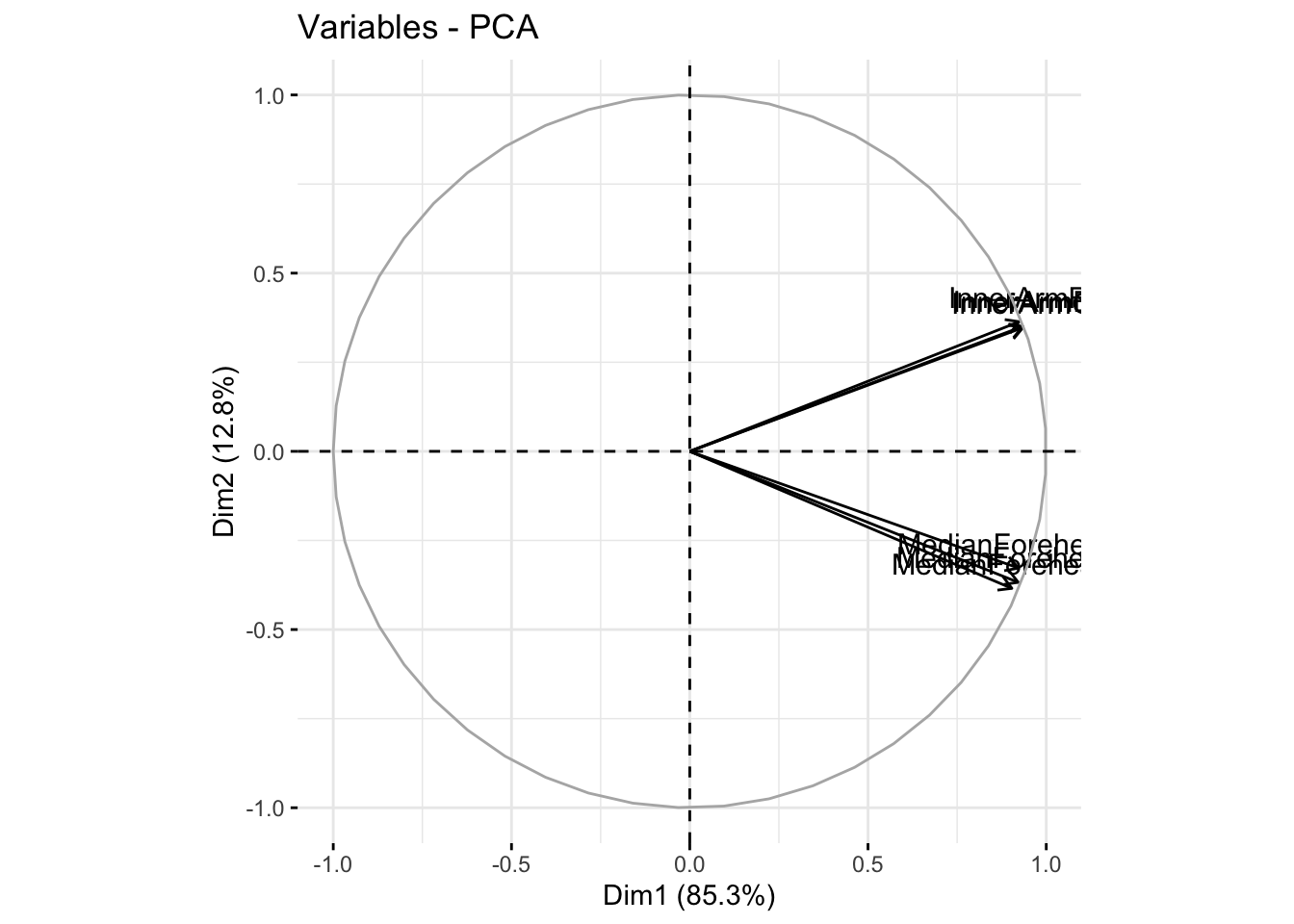
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(summer_rgb_pca, choice = "var", axes = 1:2)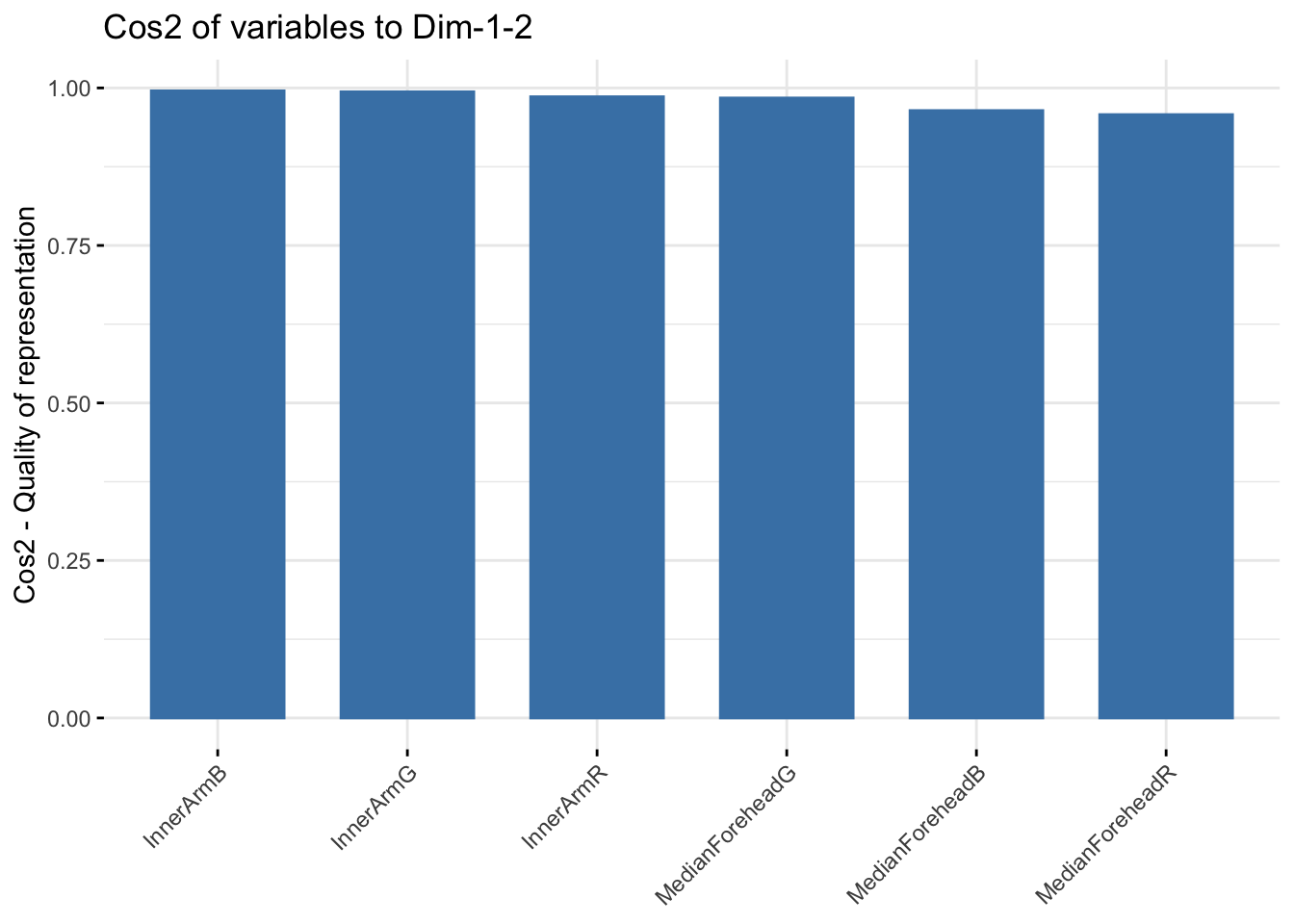
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
summer_rgb_comps <- as.data.frame(summer_rgb_pca$x)
summer_rgb_new <- cbind(summer_data,summer_rgb_comps[,c(1,2)])
ggplot(summer_rgb_new, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Summer RGB PCA")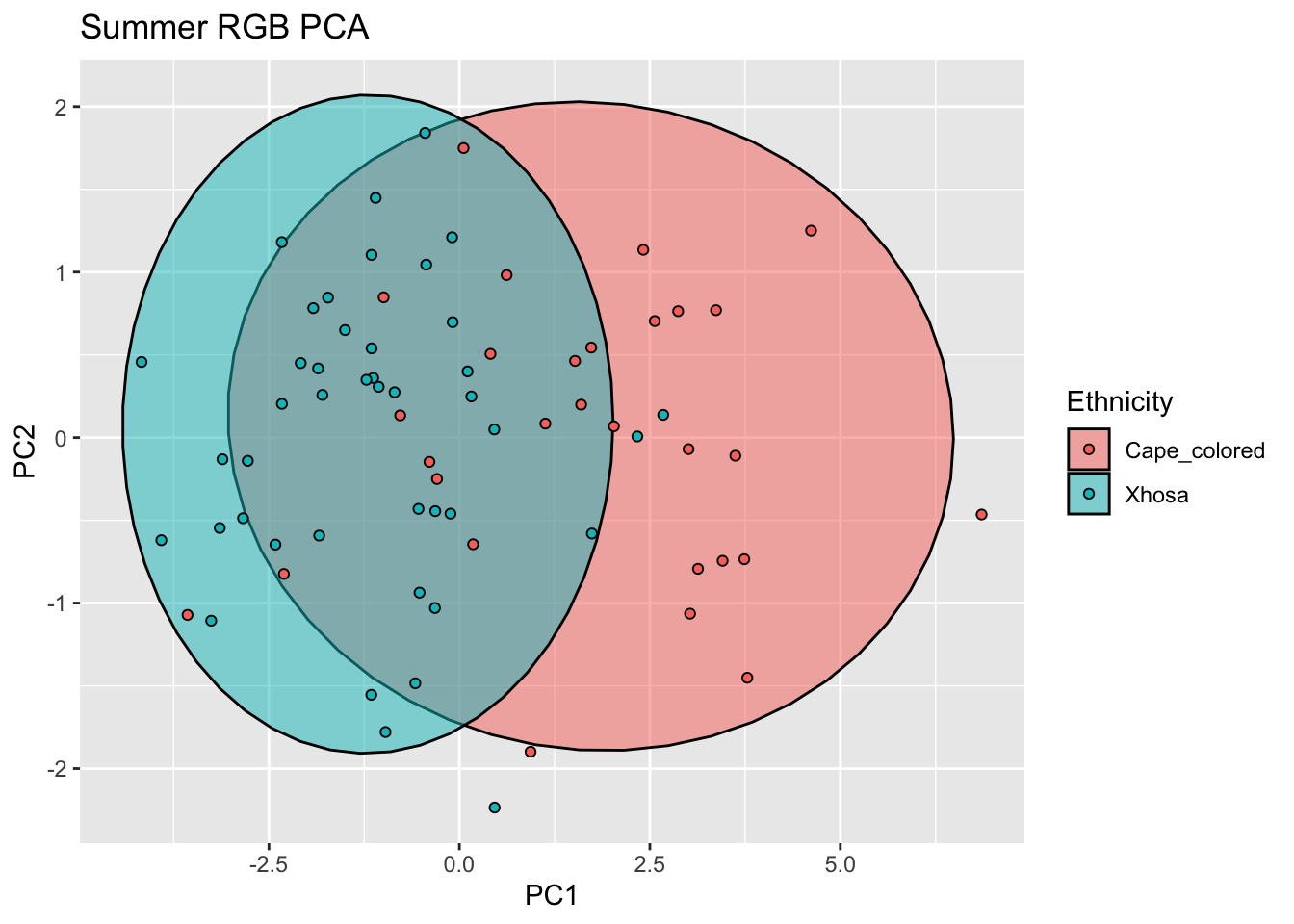
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(summer_rgb_pca, choice = "var", axes = 1, top = 10)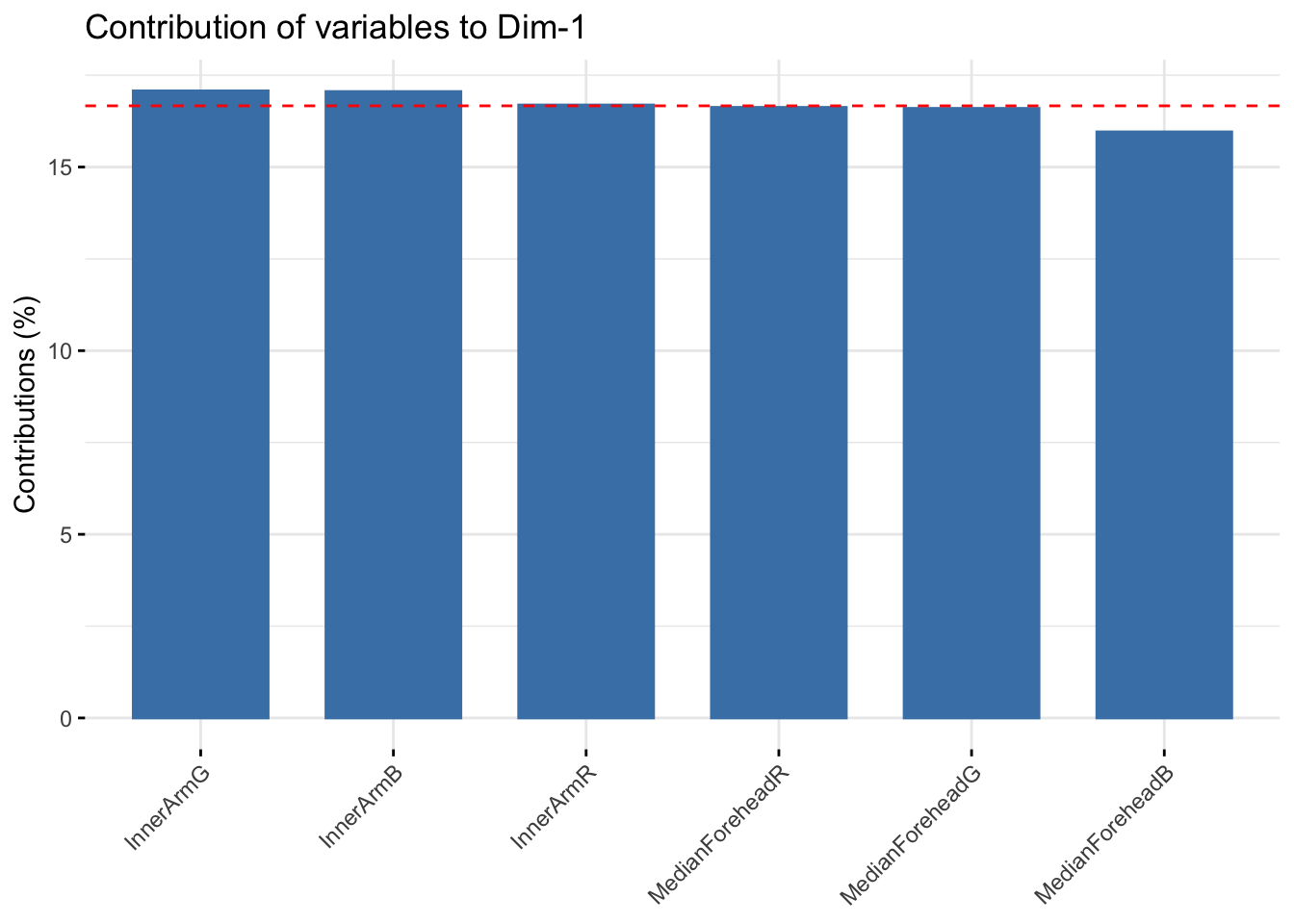
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(summer_rgb_pca, choice = "var", axes = 2, top = 10)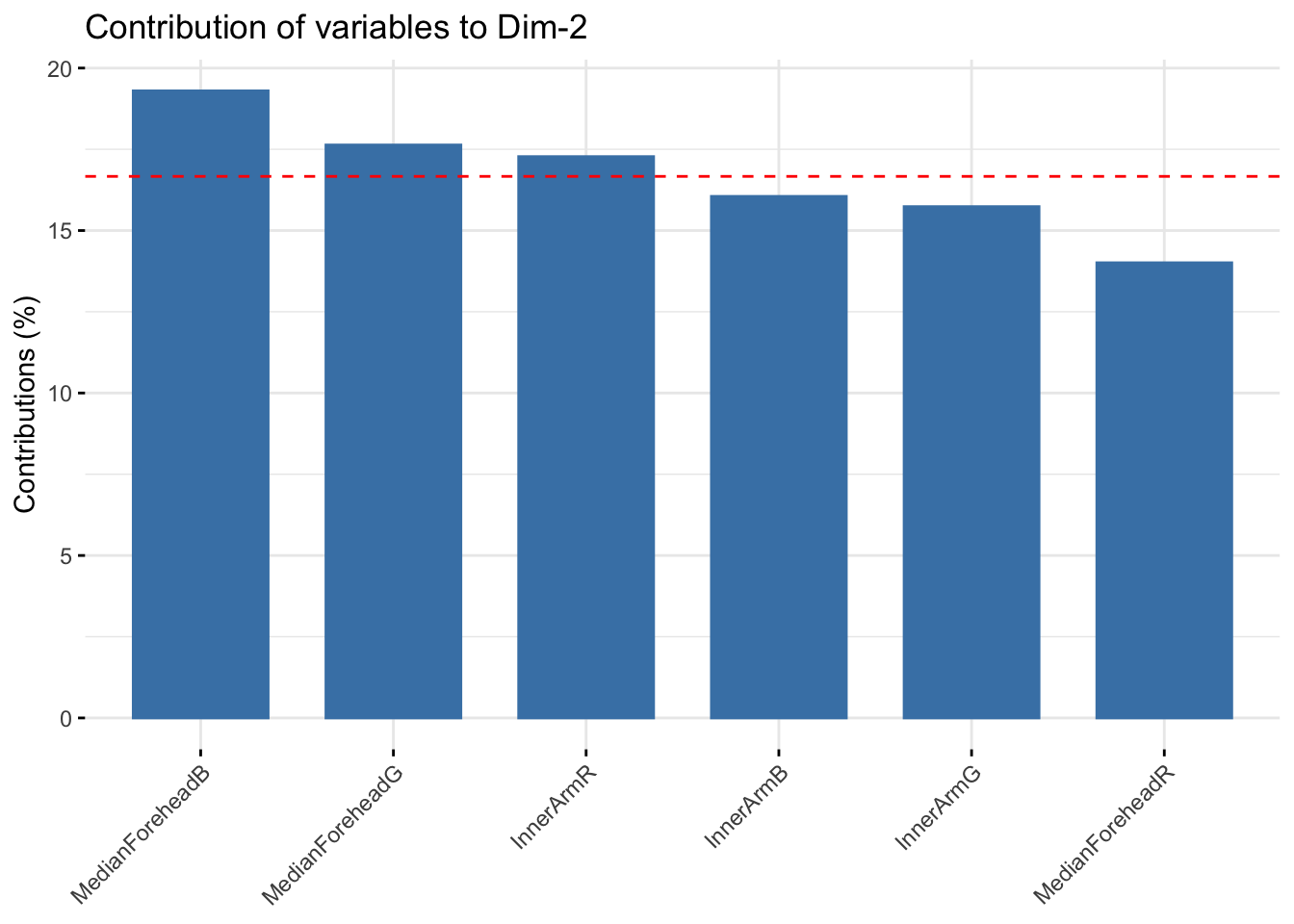
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
six_rgb_scale <- scale(six_rgb)
six_rgb_pca <- prcomp(six_rgb_scale)
summary(six_rgb_pca)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6
Standard deviation 2.1876 1.0663 0.18148 0.14888 0.13270 0.06947
Proportion of Variance 0.7976 0.1895 0.00549 0.00369 0.00293 0.00080
Cumulative Proportion 0.7976 0.9871 0.99257 0.99626 0.99920 1.00000six_rgb_pca$loadings[, 1:2]NULLfviz_eig(six_rgb_pca, addlabels = TRUE)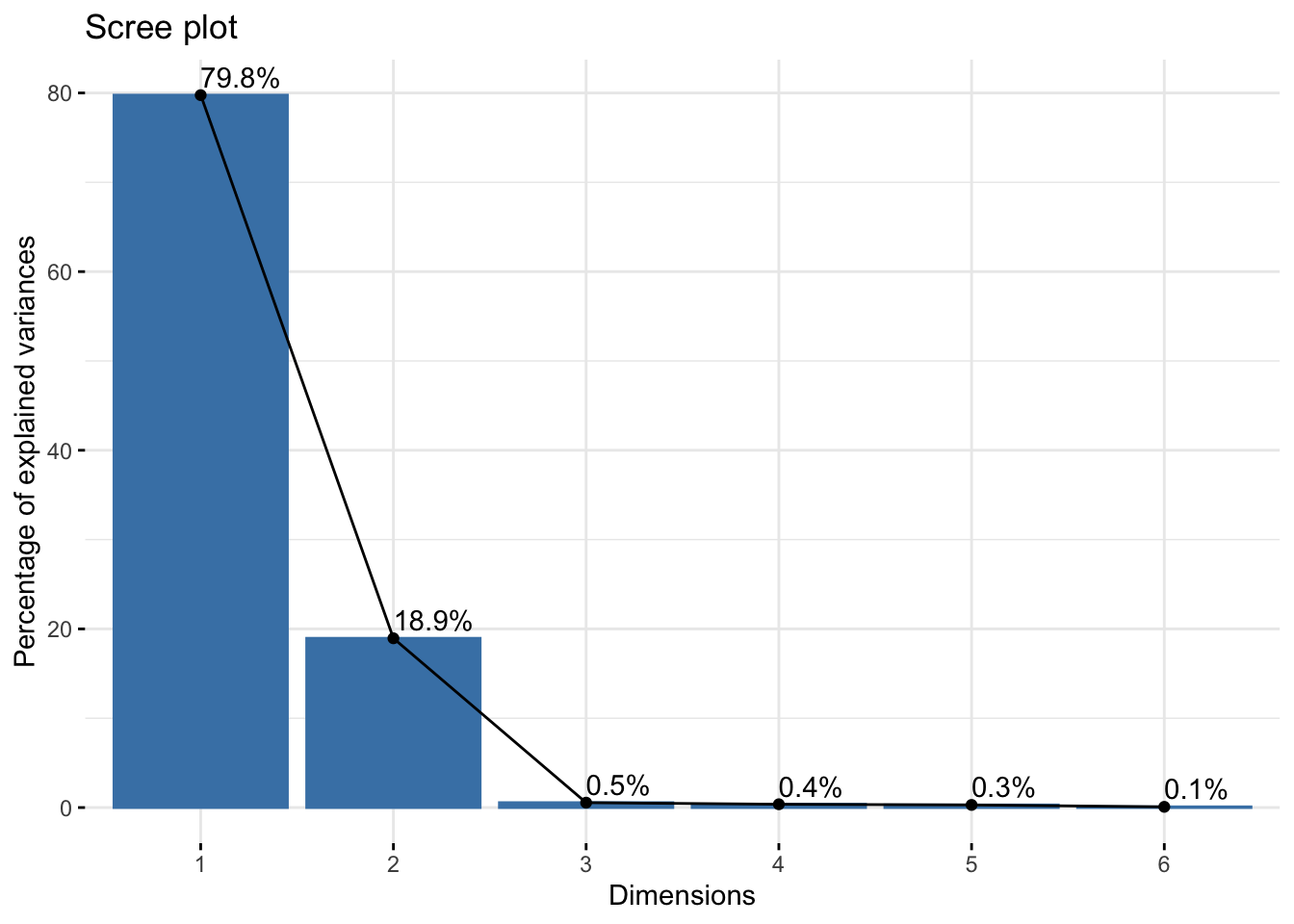
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(six_rgb_pca, col.var = "black")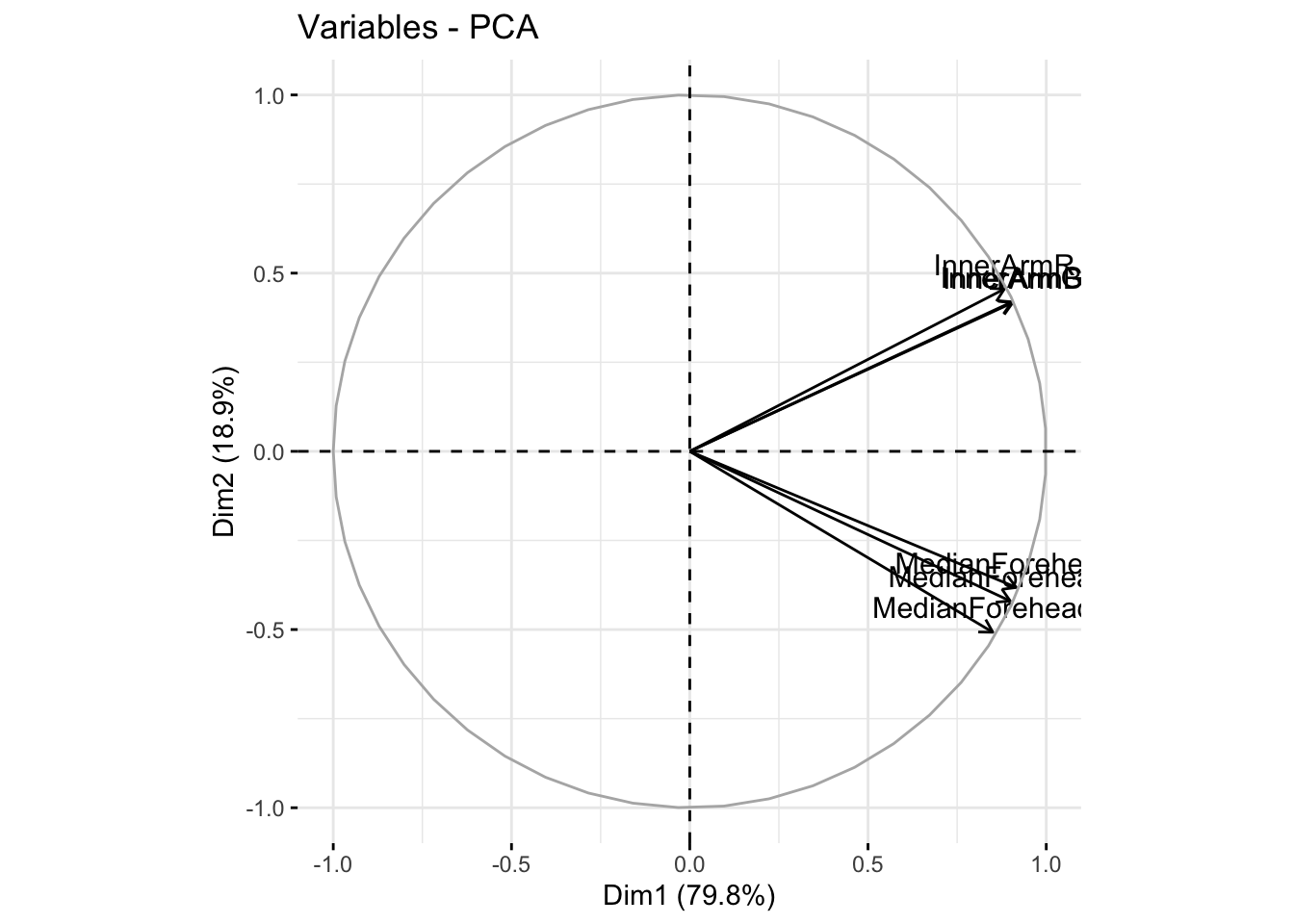
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(six_rgb_pca, choice = "var", axes = 1:2)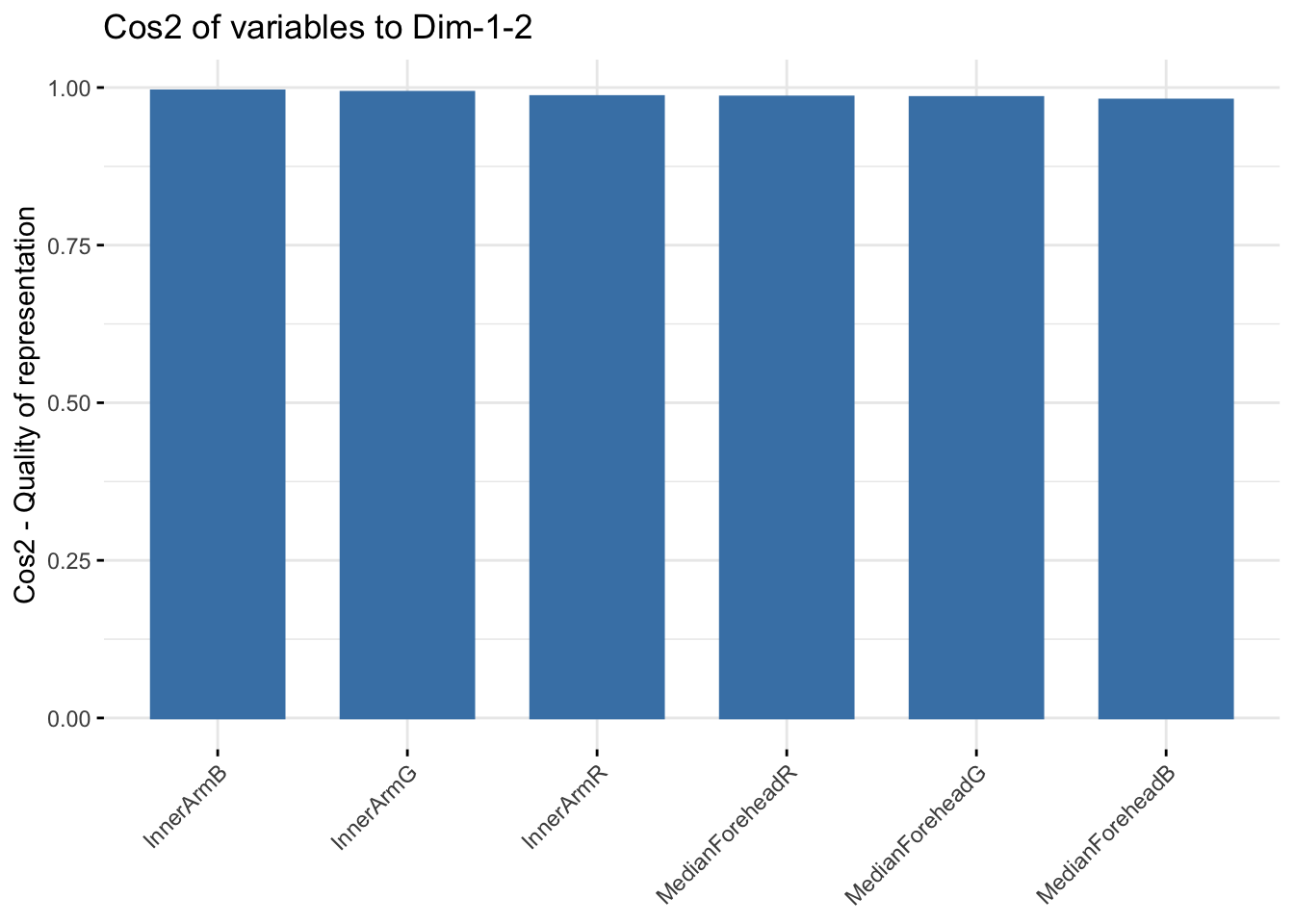
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
six_rgb_comps <- as.data.frame(six_rgb_pca$x)
six_rgb_new <- cbind(six_week_data,six_rgb_comps[,c(1,2)])
ggplot(six_rgb_new, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Six Week RGB PCA")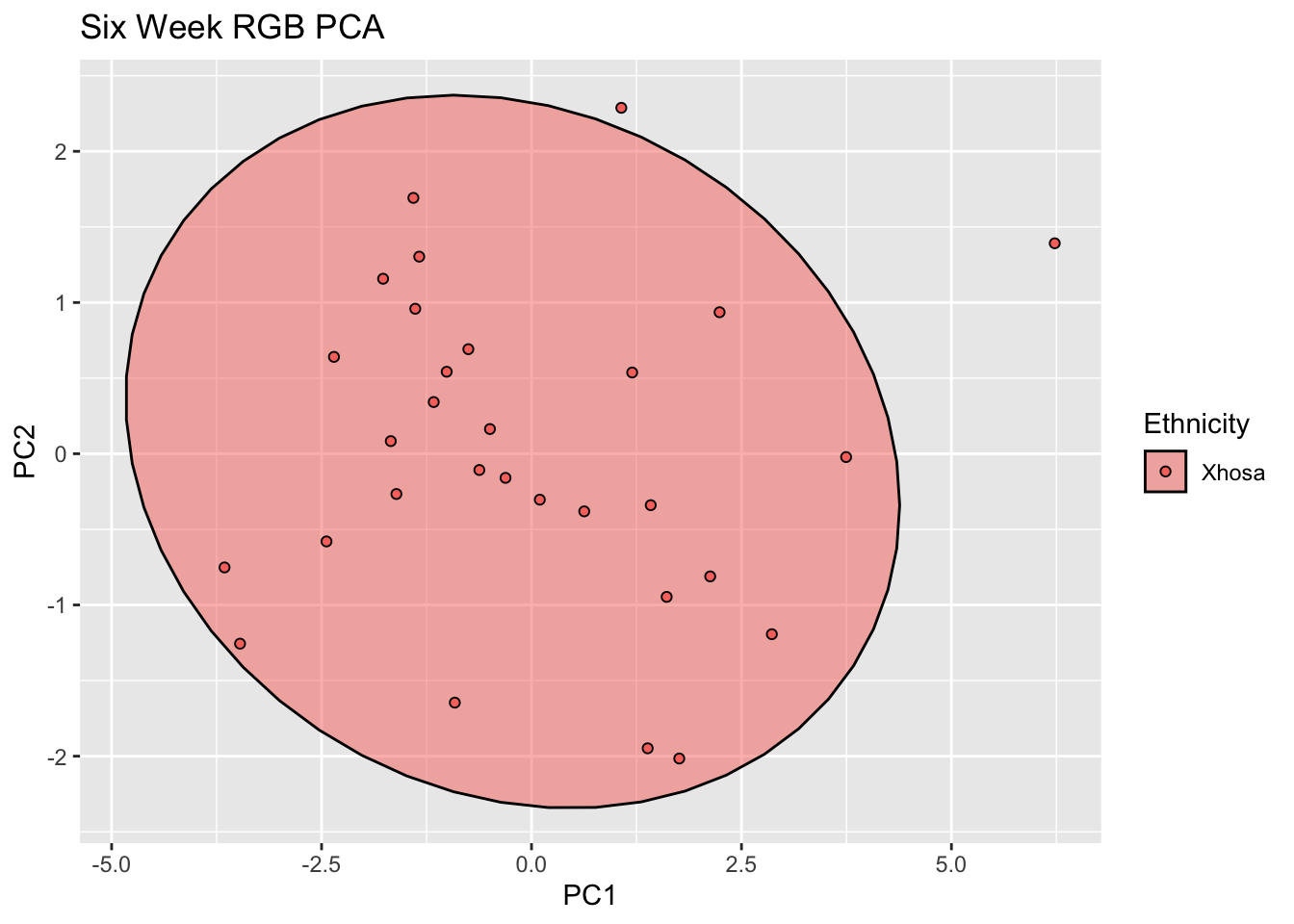
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(six_rgb_pca, choice = "var", axes = 1, top = 10)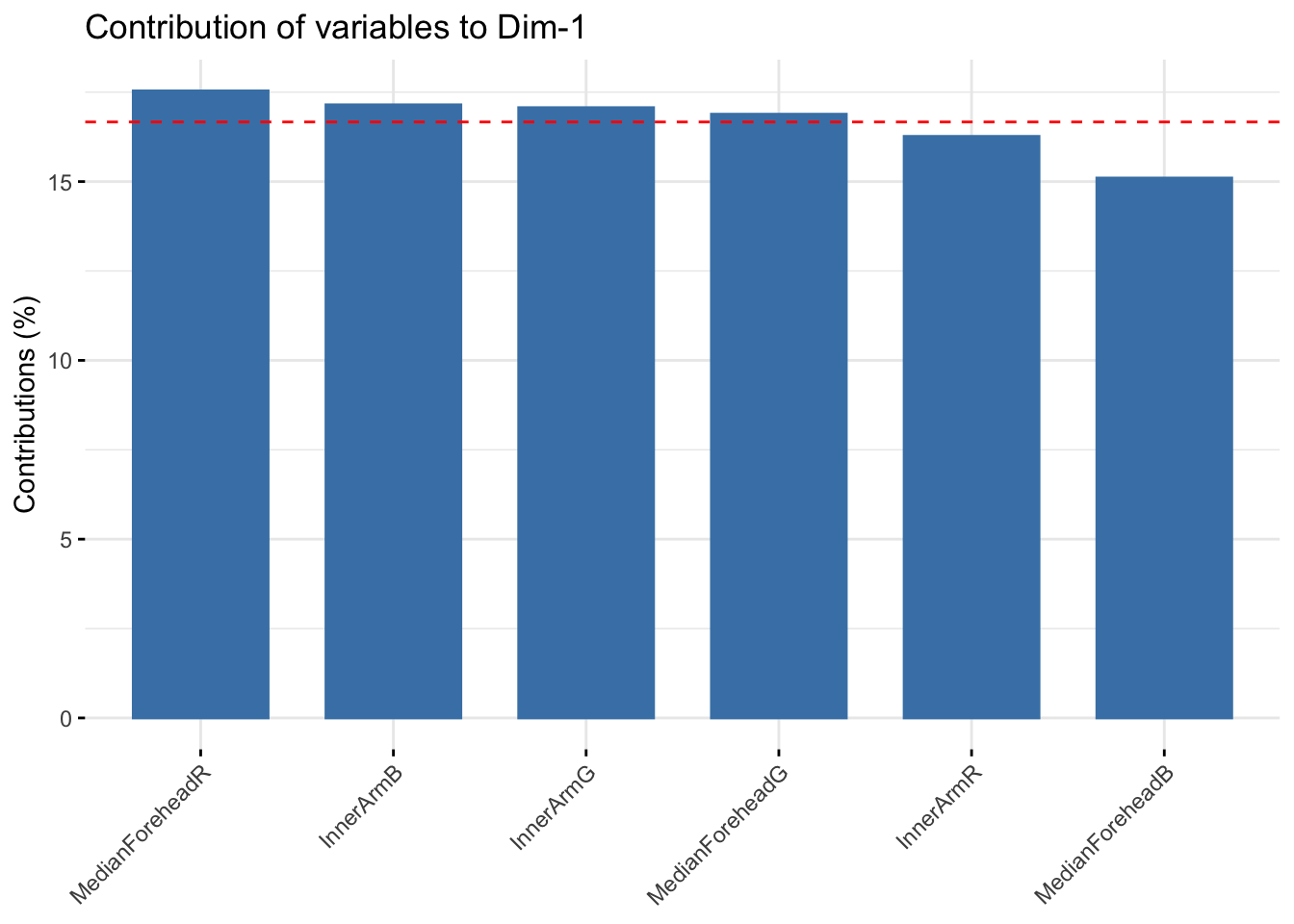
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(six_rgb_pca, choice = "var", axes = 2, top = 10)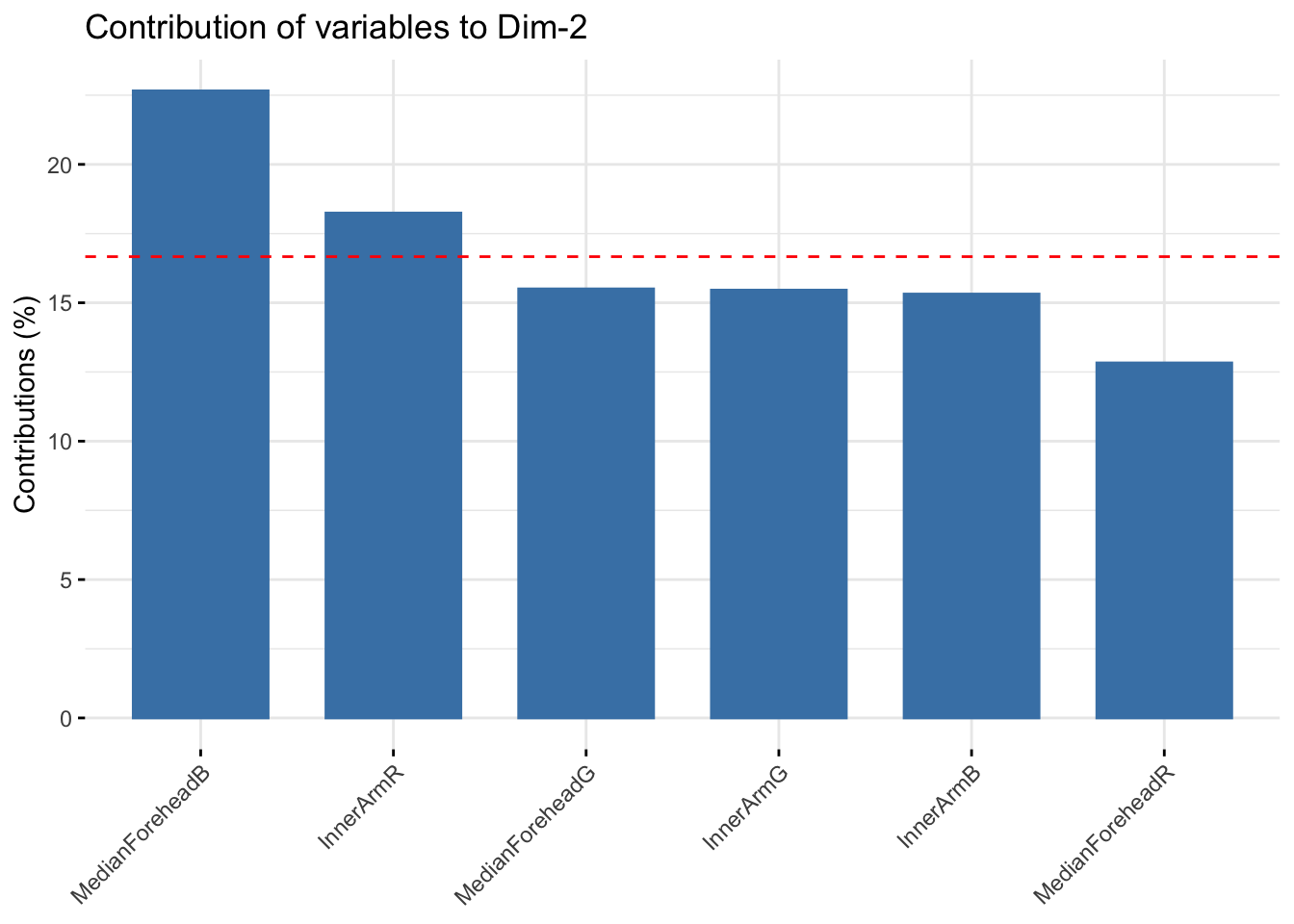
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
CIElab subset
summer_winter_clean <- na.omit(summer_winter)
reflectance_cie_ws <- summer_winter_clean %>%
select(matches("ForeheadL|Foreheada|Foreheadb|InnerArmL|InnerArma|InnerArmb", ignore.case = FALSE))
reflectance5 <- scale(reflectance_cie_ws)
reflectanceciews <- prcomp(reflectance5)
summary(reflectanceciews)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 2.9138 1.2773 0.72925 0.60171 0.55023 0.46879 0.42674
Proportion of Variance 0.7075 0.1360 0.04432 0.03017 0.02523 0.01831 0.01518
Cumulative Proportion 0.7075 0.8435 0.88778 0.91795 0.94318 0.96149 0.97667
PC8 PC9 PC10 PC11 PC12
Standard deviation 0.31641 0.26634 0.23849 0.16406 0.15854
Proportion of Variance 0.00834 0.00591 0.00474 0.00224 0.00209
Cumulative Proportion 0.98501 0.99092 0.99566 0.99791 1.00000reflectanceciews$loadings[, 1:2]NULLfviz_eig(reflectanceciews, addlabels = TRUE)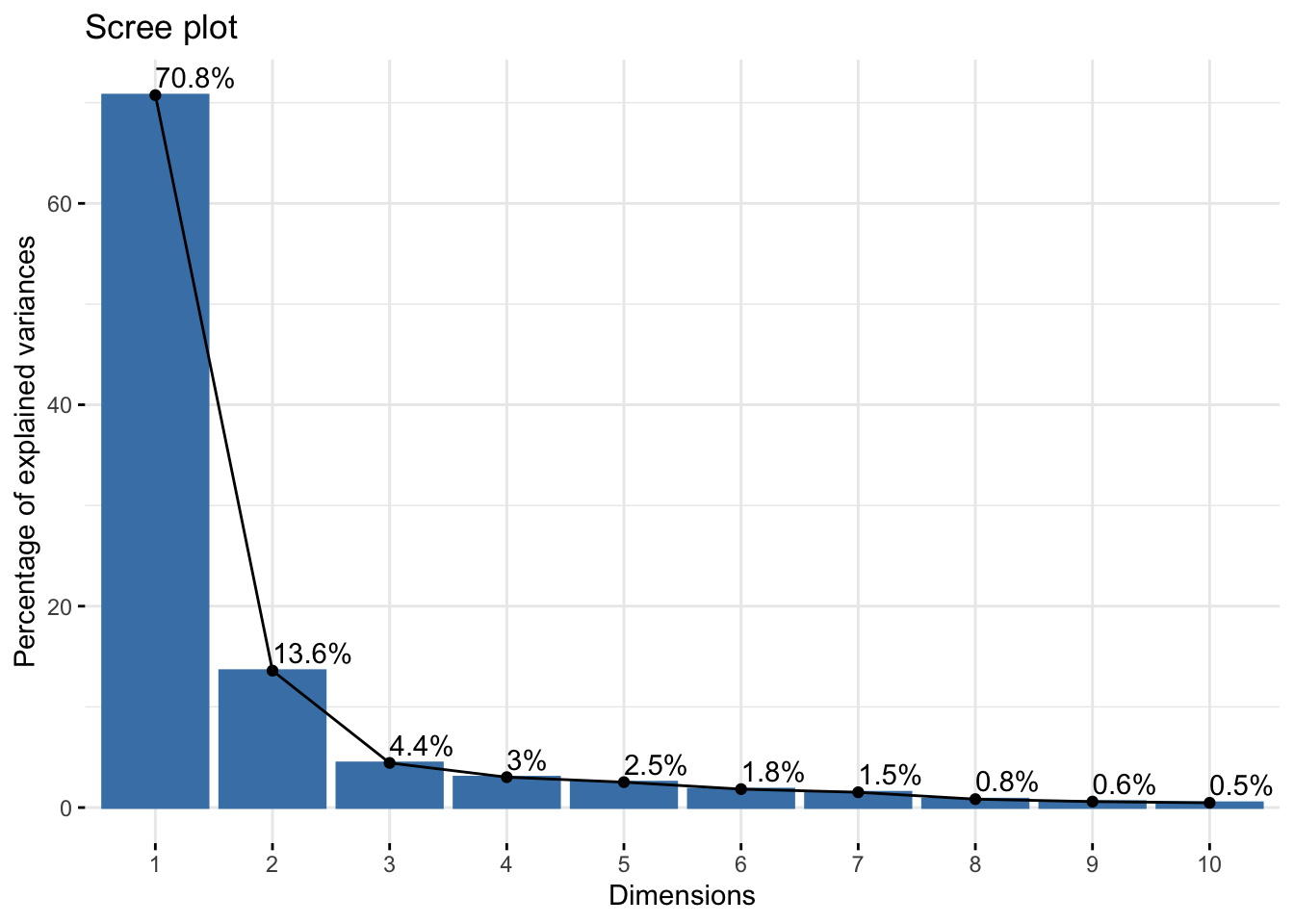
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(reflectanceciews, col.var = "black")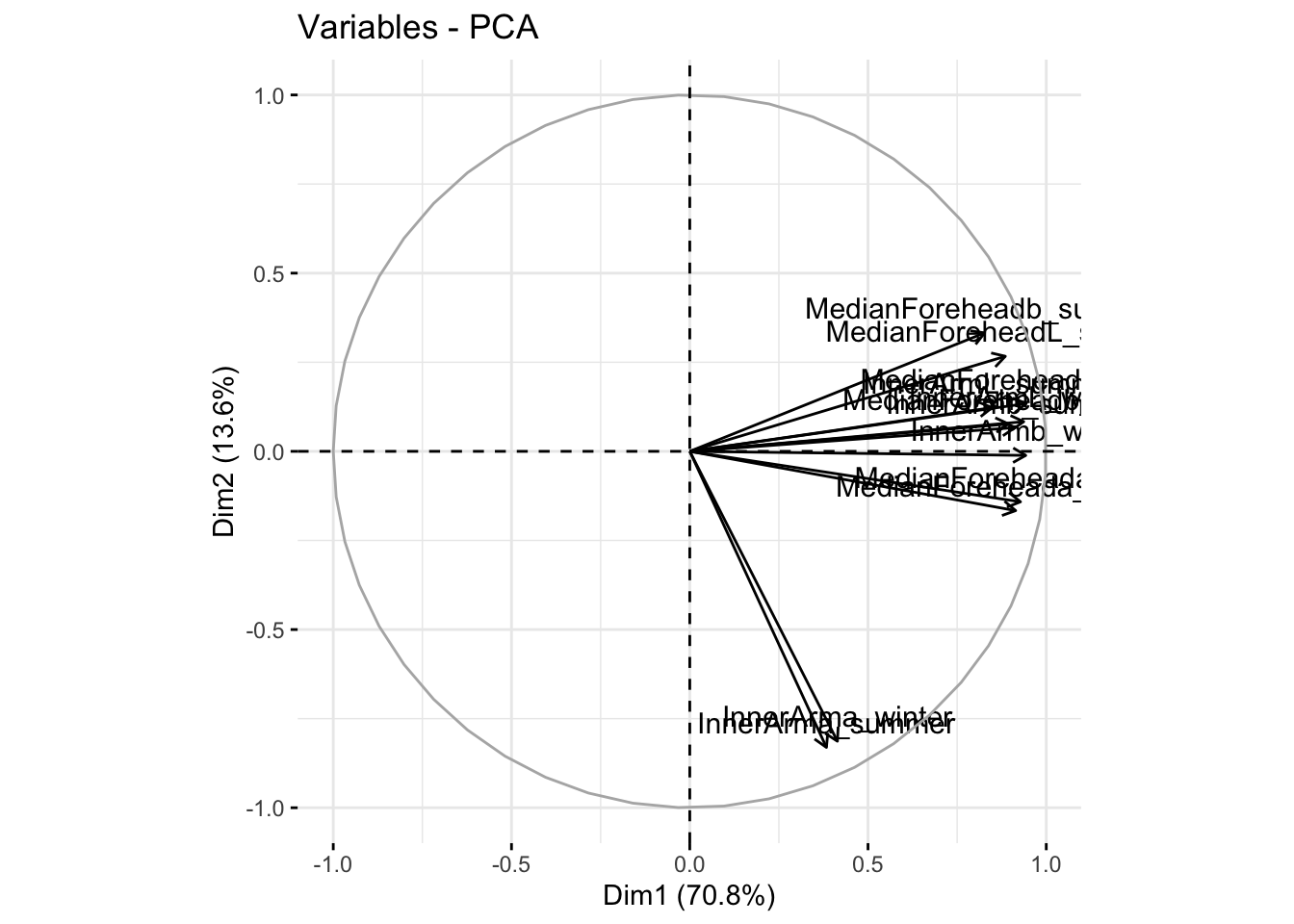
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(reflectanceciews, choice = "var", axes = 1:2)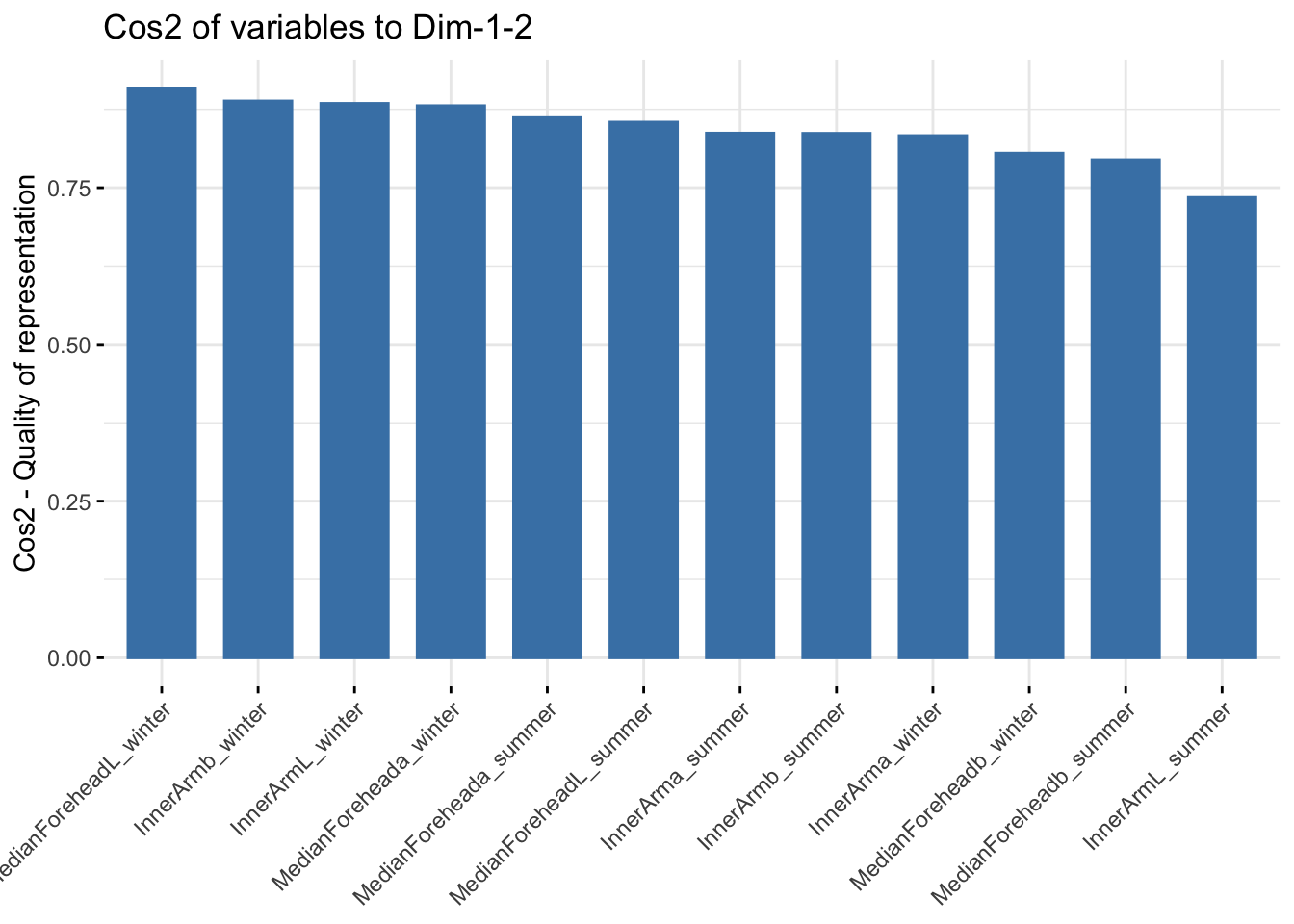
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
wsciecomps <- as.data.frame(reflectanceciews$x)
cie_ws_bind <- cbind(summer_winter_clean,wsciecomps[,c(1,2)])
cie_ws_bind <- rgb_ws_bind %>%
select(-matches("SkinReflectance"))
ggplot(cie_ws_bind, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Wide Pigmentation PCA")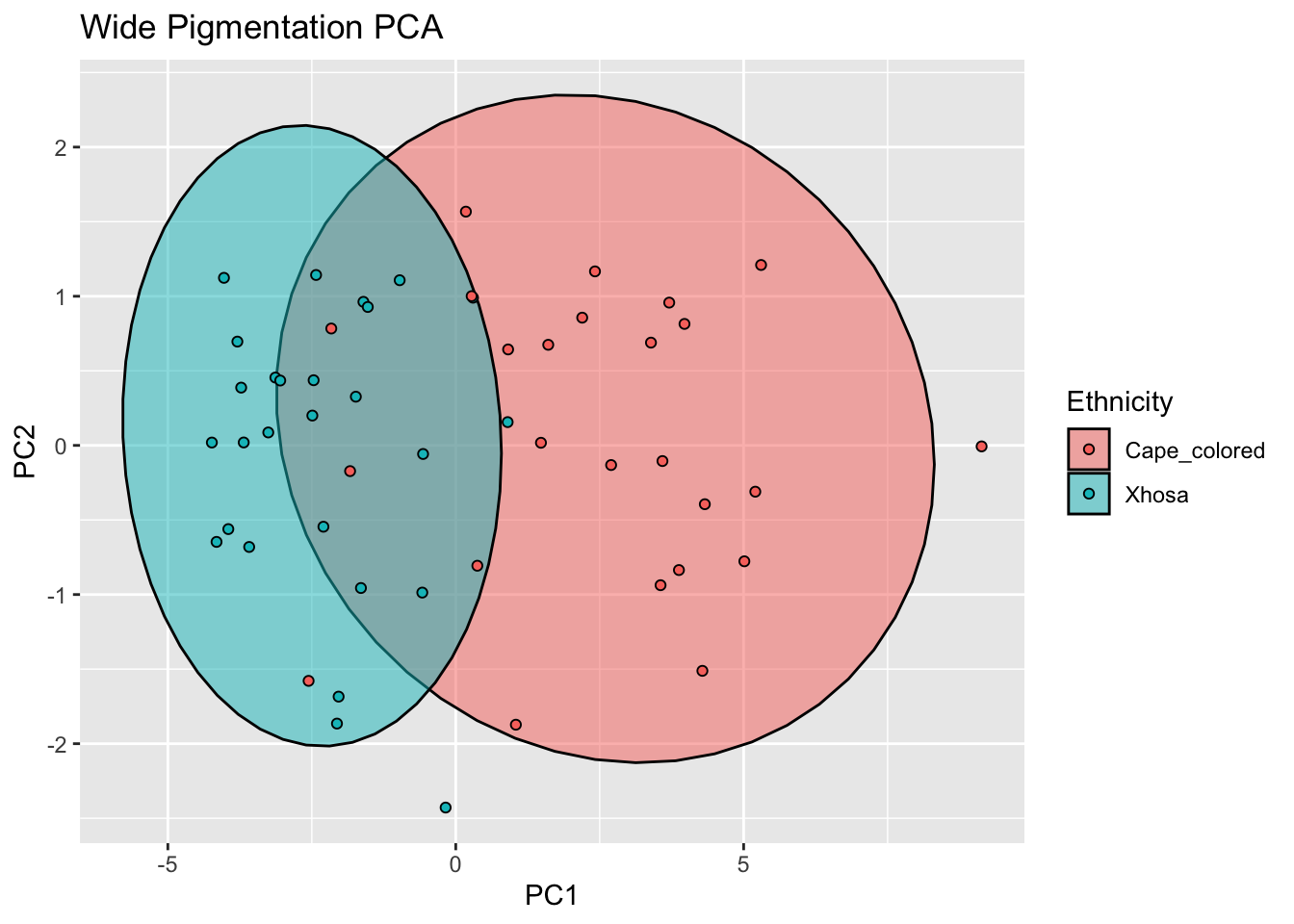
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(reflectanceciews, choice = "var", axes = 1, top = 20)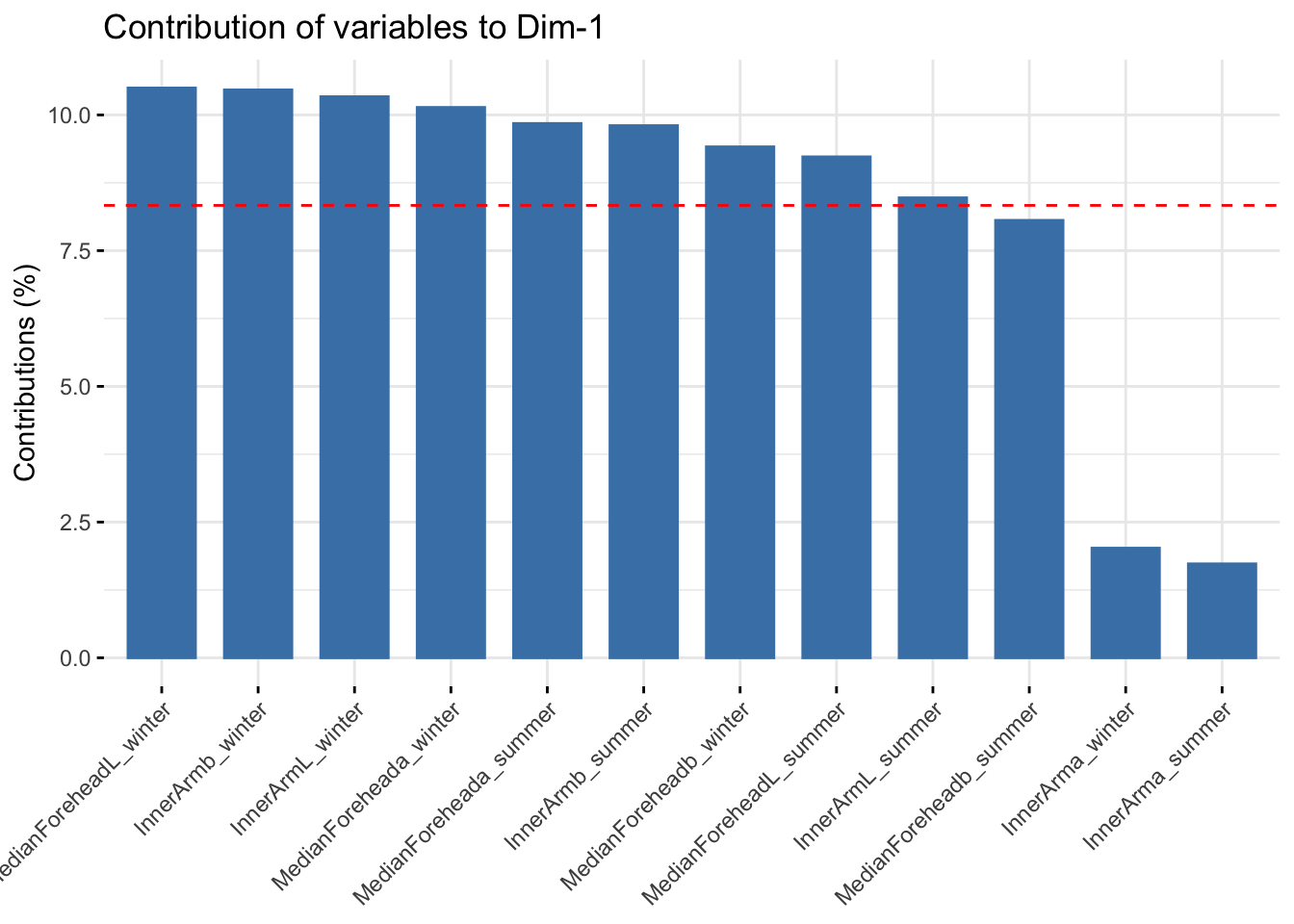
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(reflectanceciews, choice = "var", axes = 2, top = 20)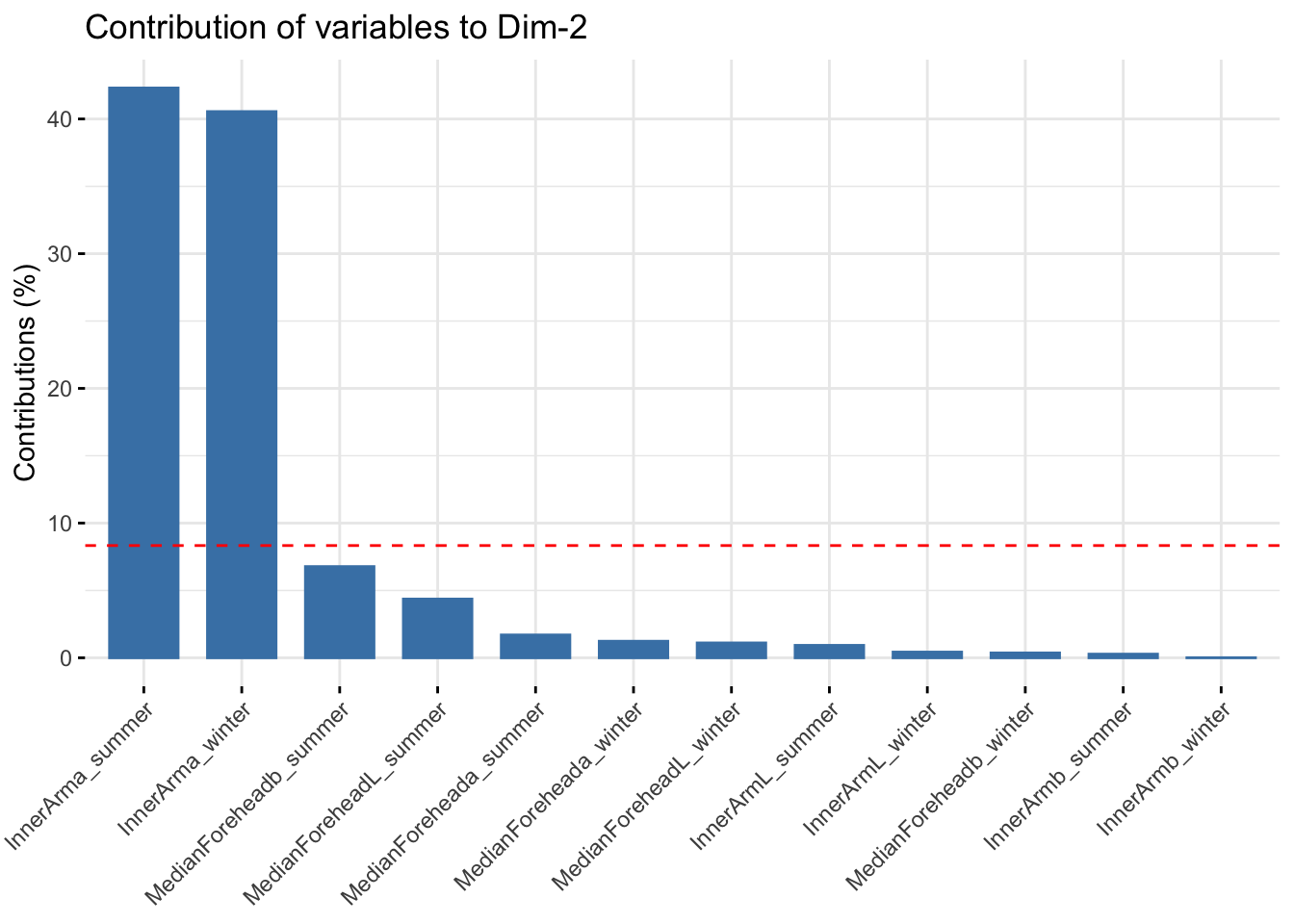
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
summer_cie <- summer_data %>%
select(matches("ForeheadL|Foreheada|Foreheadb|InnerArmL|InnerArma|InnerArmb", ignore.case = FALSE))
summer_cie <- na.omit(summer_cie)
winter_cie <- winter_data %>%
select(matches("ForeheadL|Foreheada|Foreheadb|InnerArmL|InnerArma|InnerArmb", ignore.case = FALSE))
winter_cie <- na.omit(winter_cie)
six_cie <- six_week_data %>%
select(matches("ForeheadL|Foreheada|Foreheadb|InnerArmL|InnerArma|InnerArmb", ignore.case = FALSE))
six_cie <- na.omit(six_cie)winter_cie_scale <- scale(winter_cie)
winter_cie_pca <- prcomp(winter_cie_scale)
summary(winter_cie_pca)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6
Standard deviation 2.1257 0.9808 0.5304 0.36540 0.2486 0.20736
Proportion of Variance 0.7531 0.1603 0.0469 0.02225 0.0103 0.00717
Cumulative Proportion 0.7531 0.9134 0.9603 0.98253 0.9928 1.00000winter_cie_pca$loadings[, 1:2]NULLfviz_eig(winter_cie_pca, addlabels = TRUE)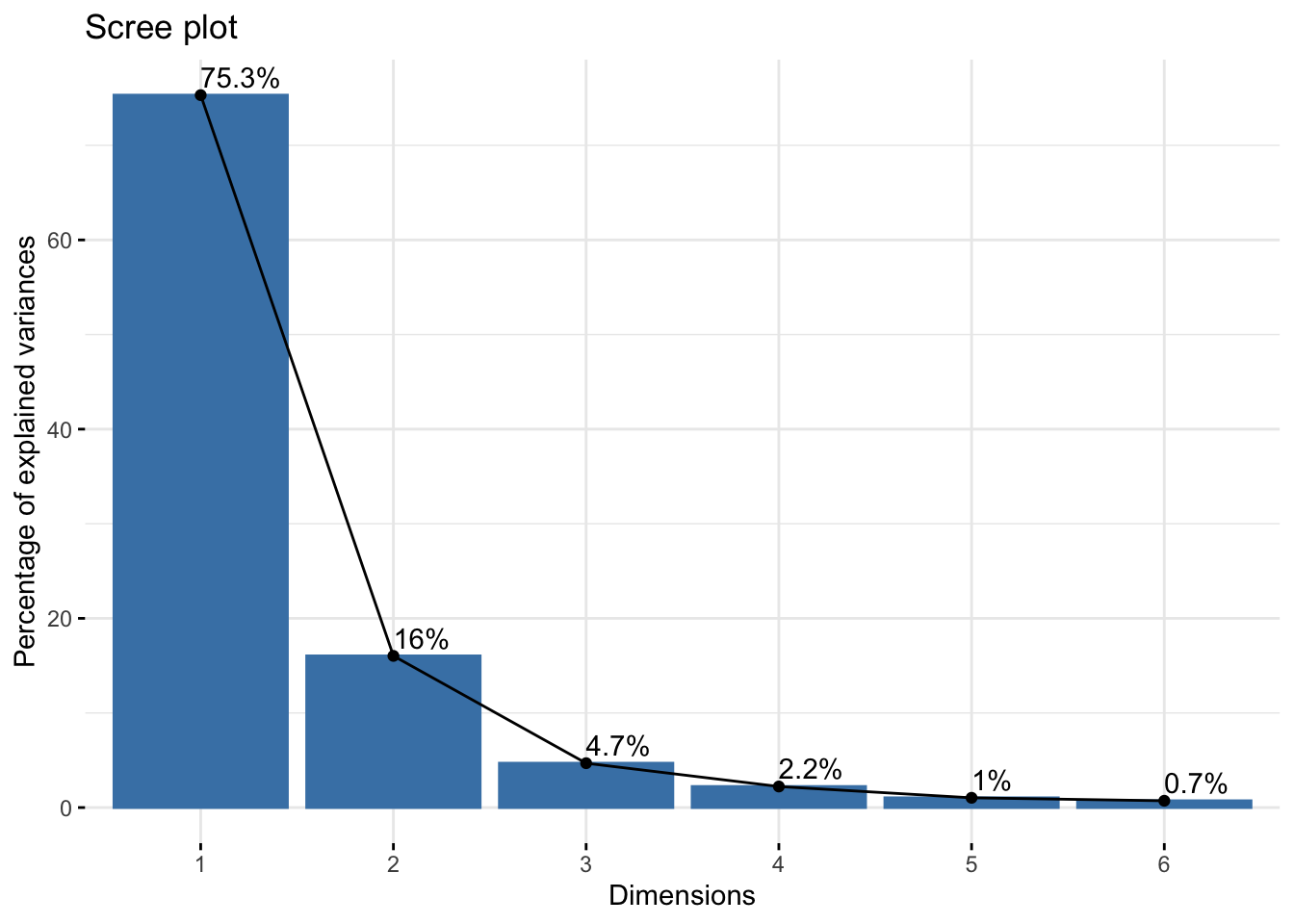
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(winter_cie_pca, col.var = "black")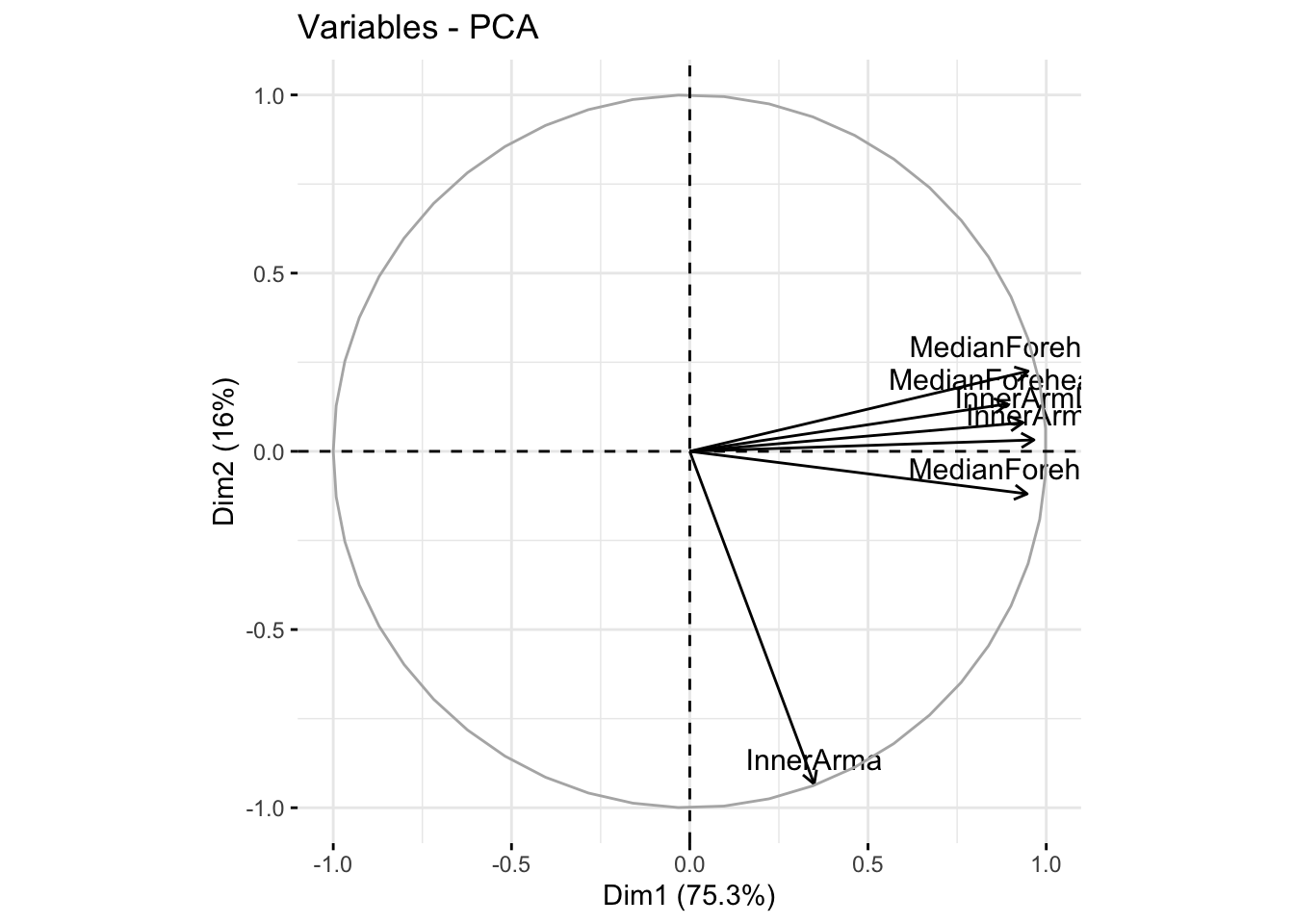
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(winter_cie_pca, choice = "var", axes = 1:2)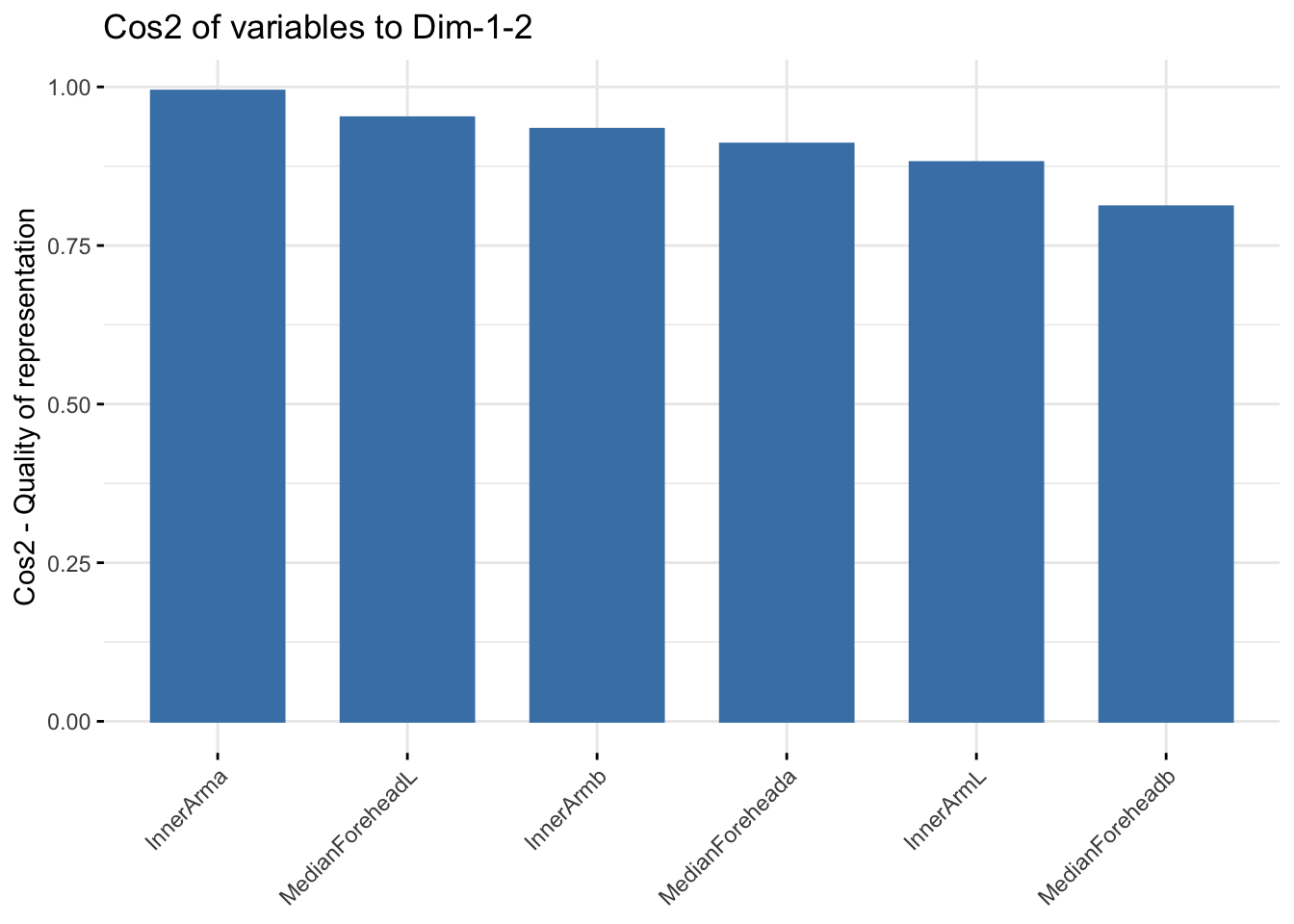
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
winter_cie_comps <- as.data.frame(winter_cie_pca$x)
winter_cie_new <- cbind(winter_data,winter_cie_comps[,c(1,2)])
ggplot(winter_cie_new, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Winter CIELAB PCA")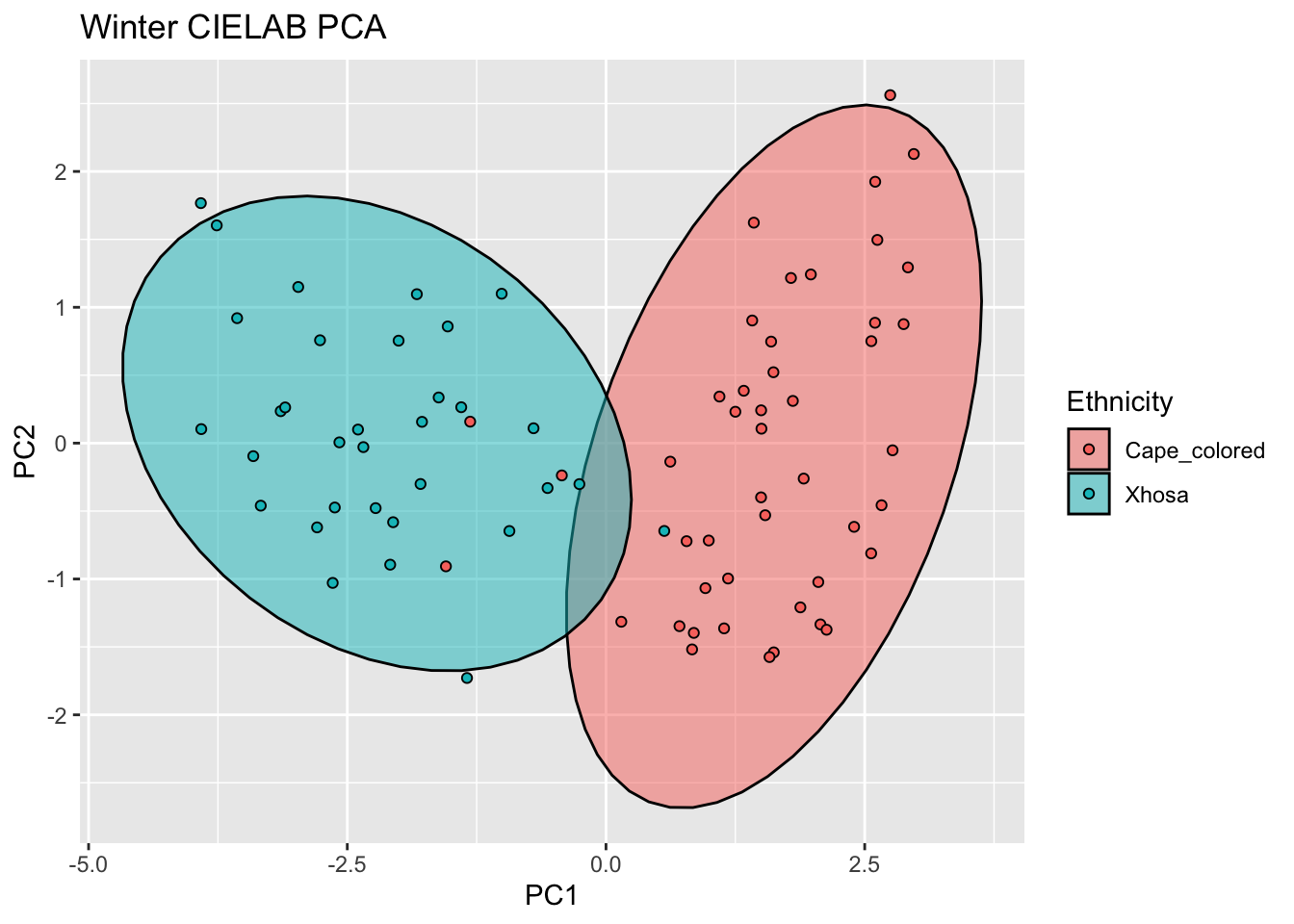
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(winter_cie_pca, choice = "var", axes = 1, top = 10)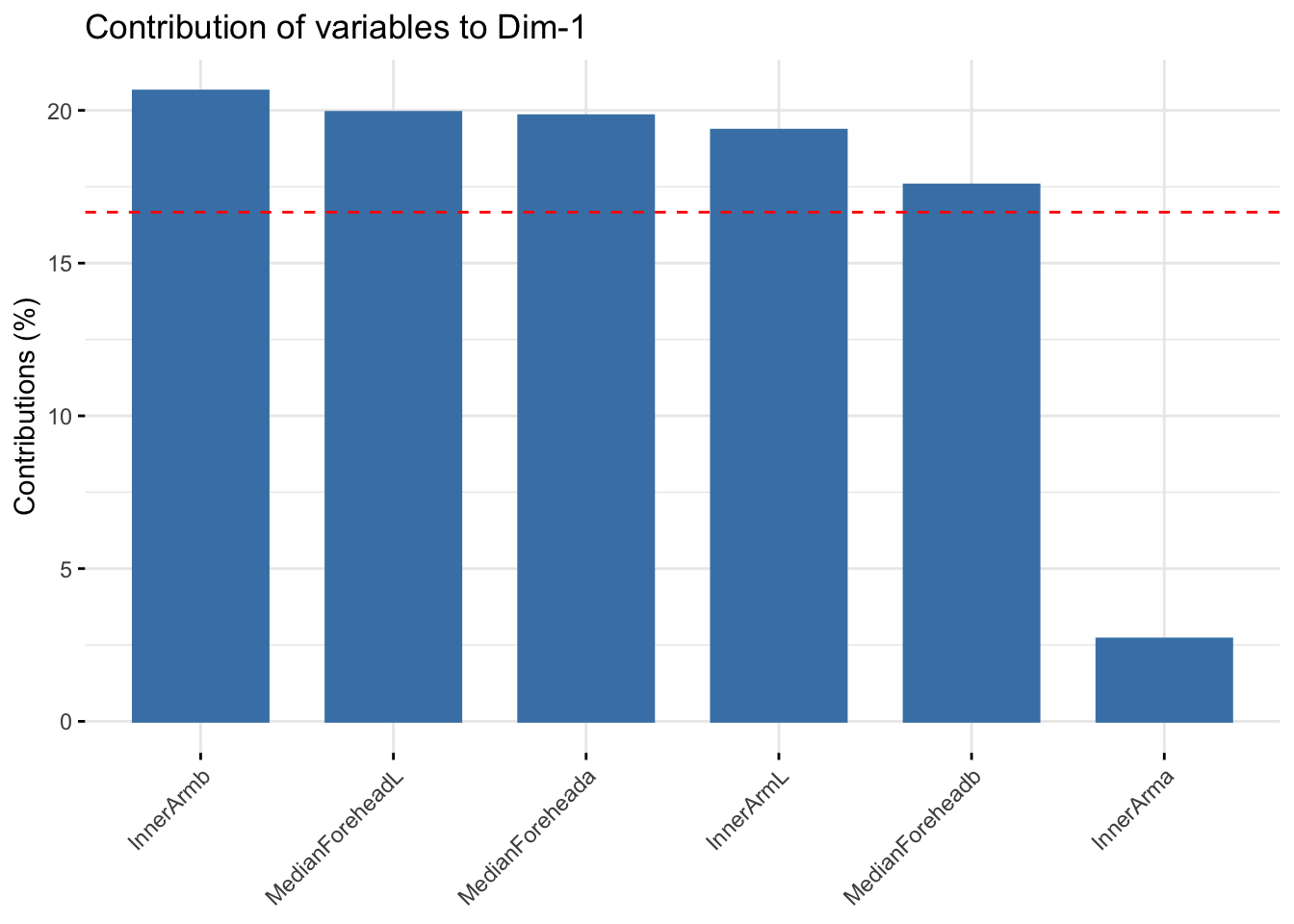
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(winter_cie_pca, choice = "var", axes = 2, top = 10)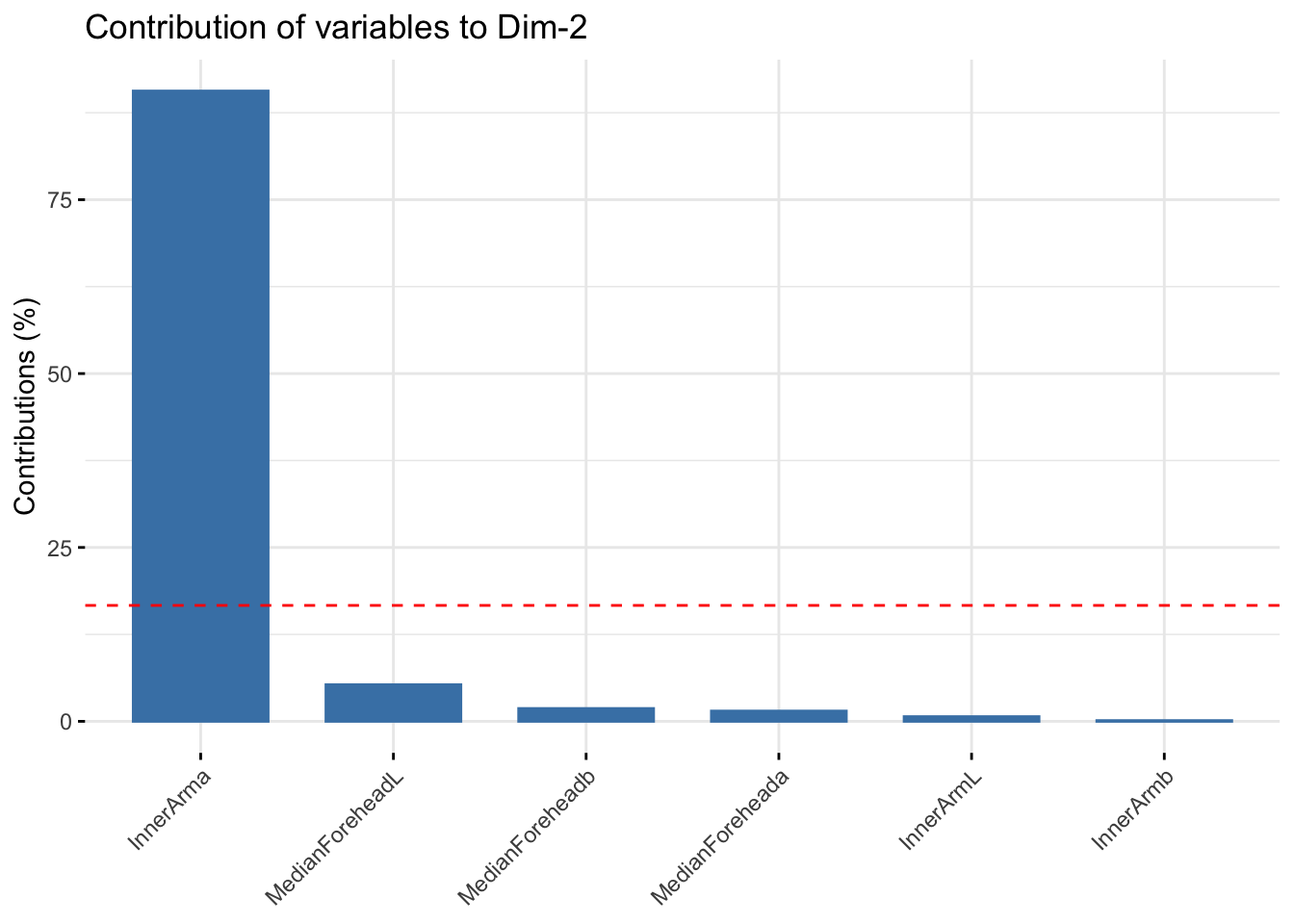
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
summer_cie_scale <- scale(summer_cie)
summer_cie_pca <- prcomp(summer_cie_scale)
summary(summer_cie_pca)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6
Standard deviation 2.0738 0.9392 0.64280 0.4960 0.32130 0.23426
Proportion of Variance 0.7168 0.1470 0.06886 0.0410 0.01721 0.00915
Cumulative Proportion 0.7168 0.8638 0.93265 0.9737 0.99085 1.00000summer_cie_pca$loadings[, 1:2]NULLfviz_eig(summer_cie_pca, addlabels = TRUE)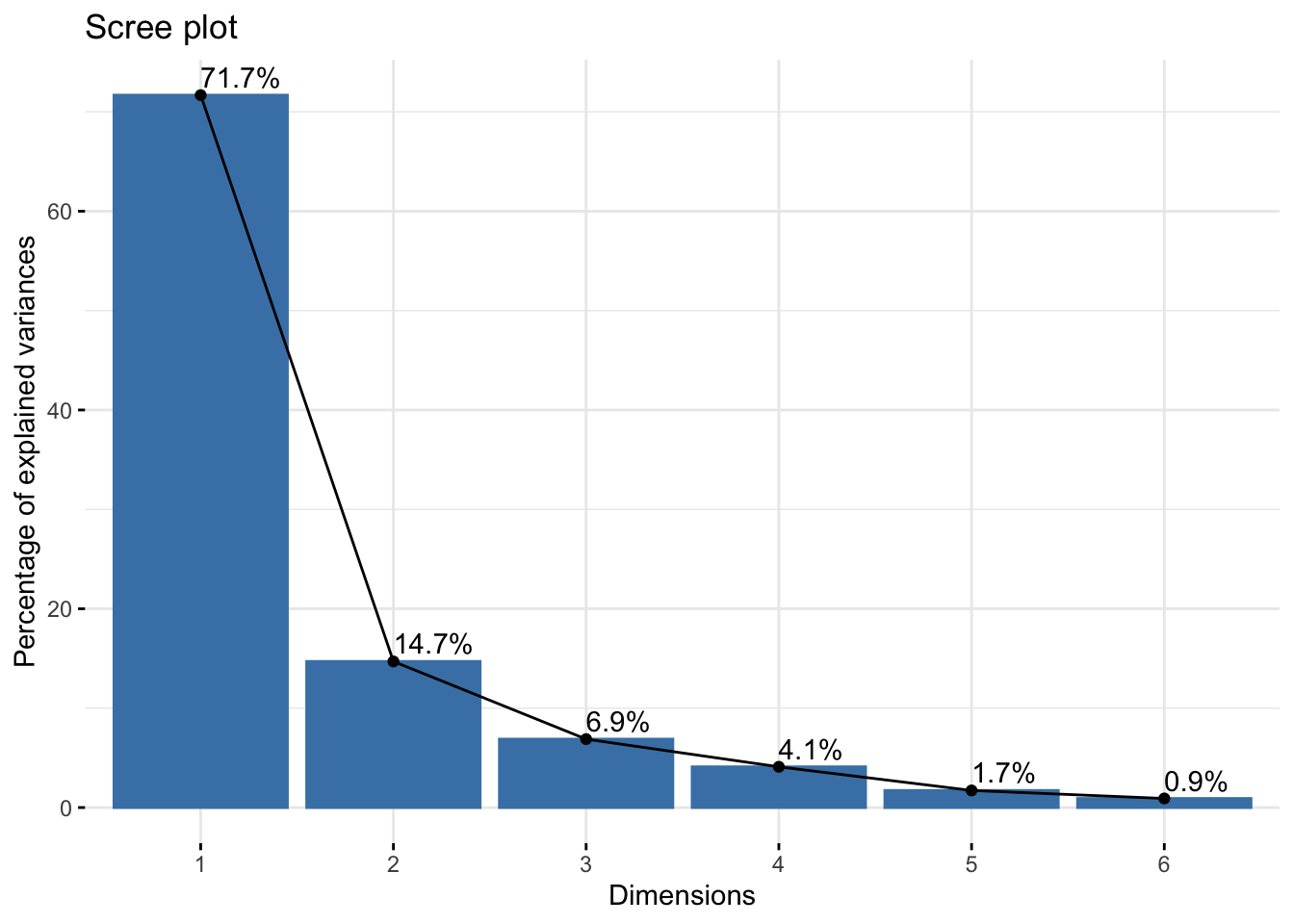
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(summer_cie_pca, col.var = "black")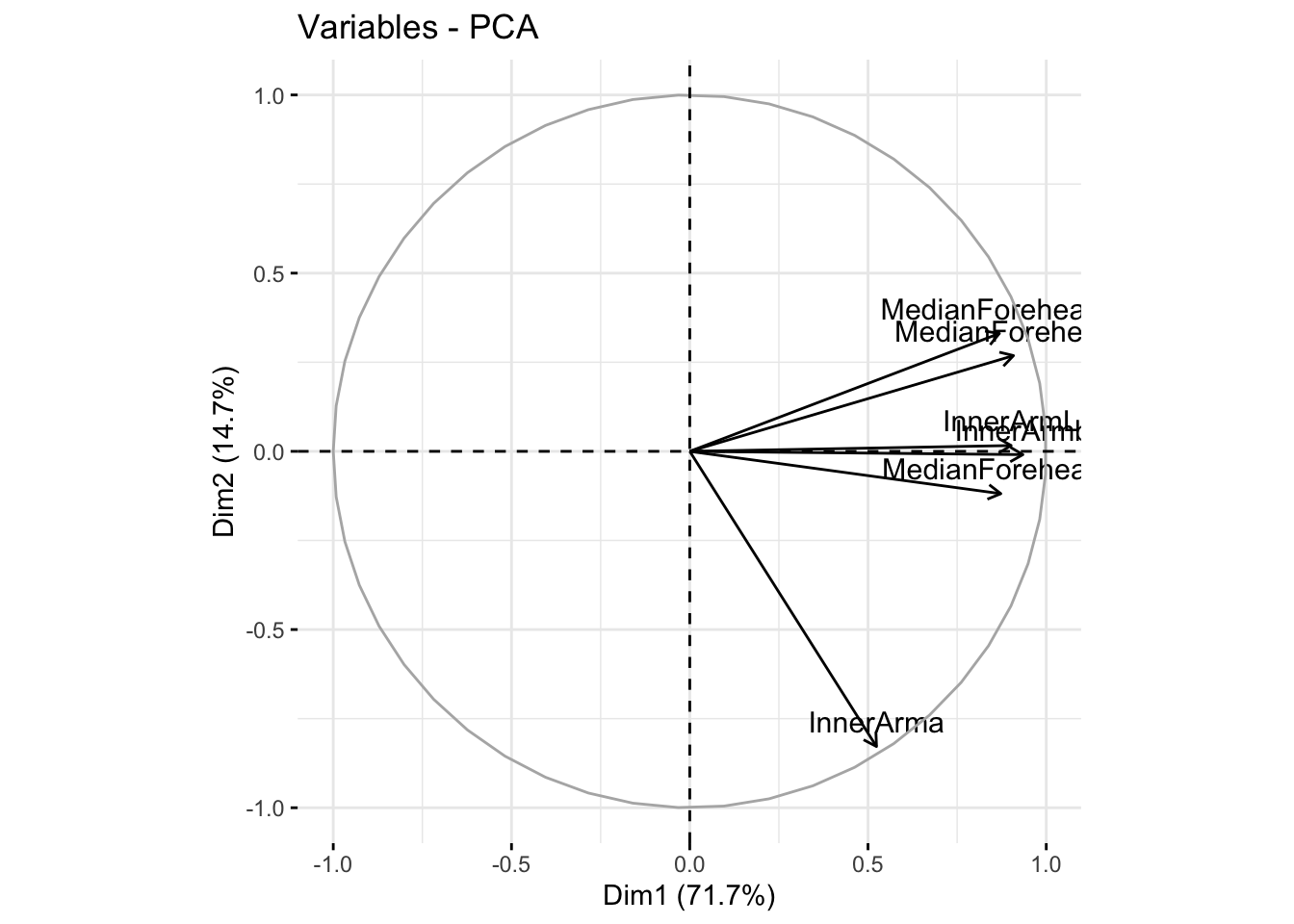
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(summer_cie_pca, choice = "var", axes = 1:2)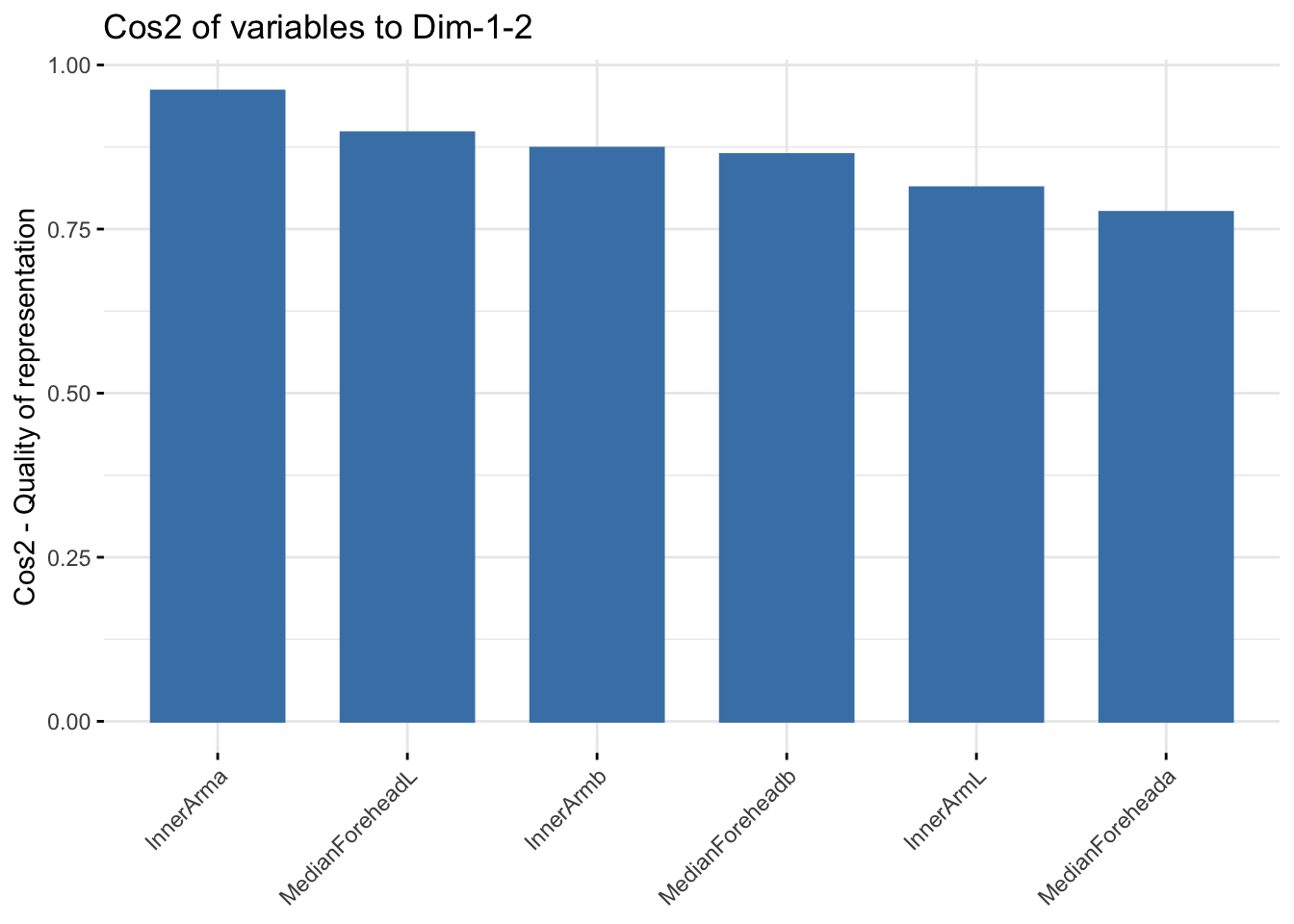
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
summer_cie_comps <- as.data.frame(summer_cie_pca$x)
summer_cie_new <- cbind(summer_data,summer_cie_comps[,c(1,2)])
ggplot(summer_cie_new, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Summer CIELAB PCA")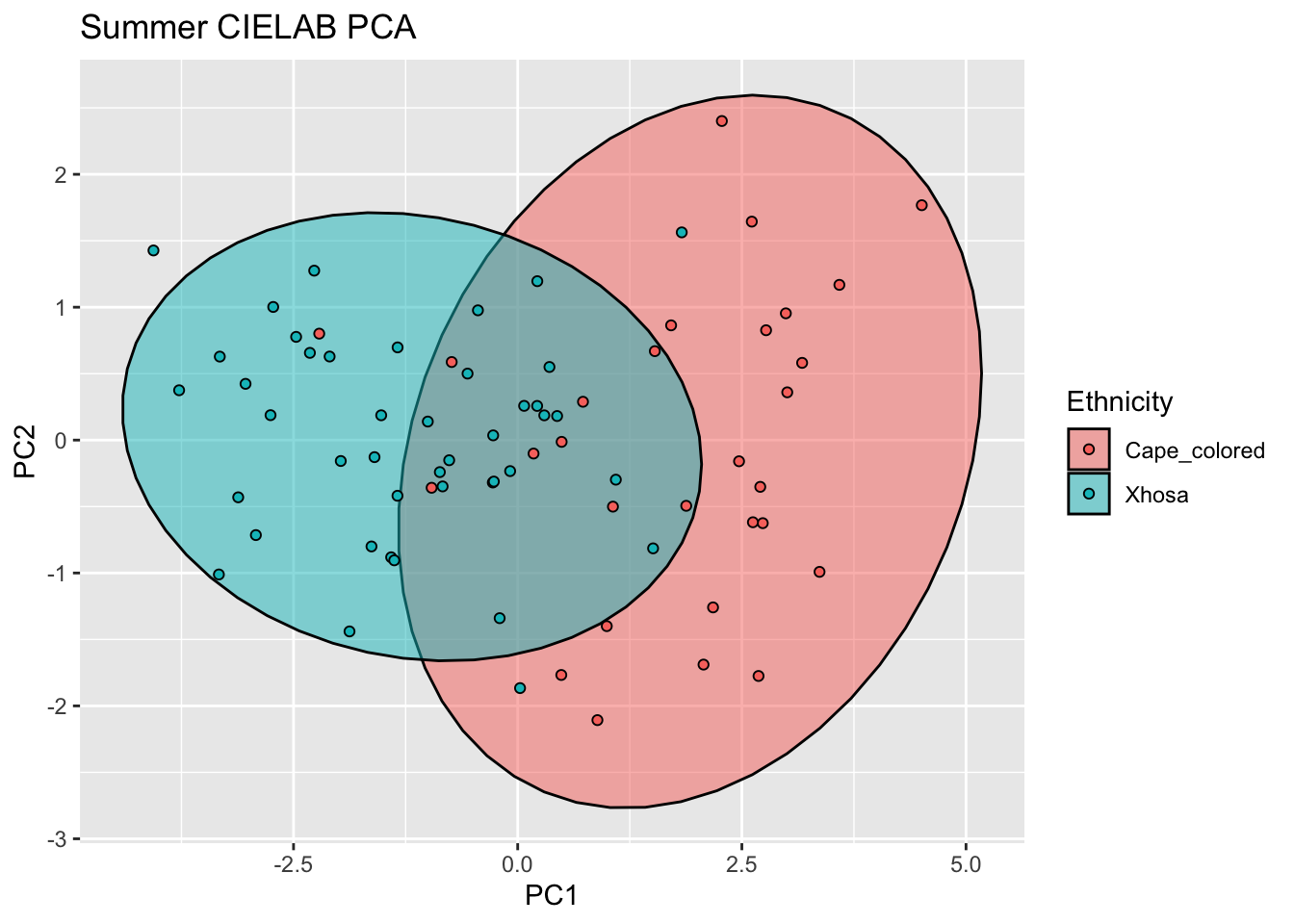
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(summer_cie_pca, choice = "var", axes = 1, top = 10)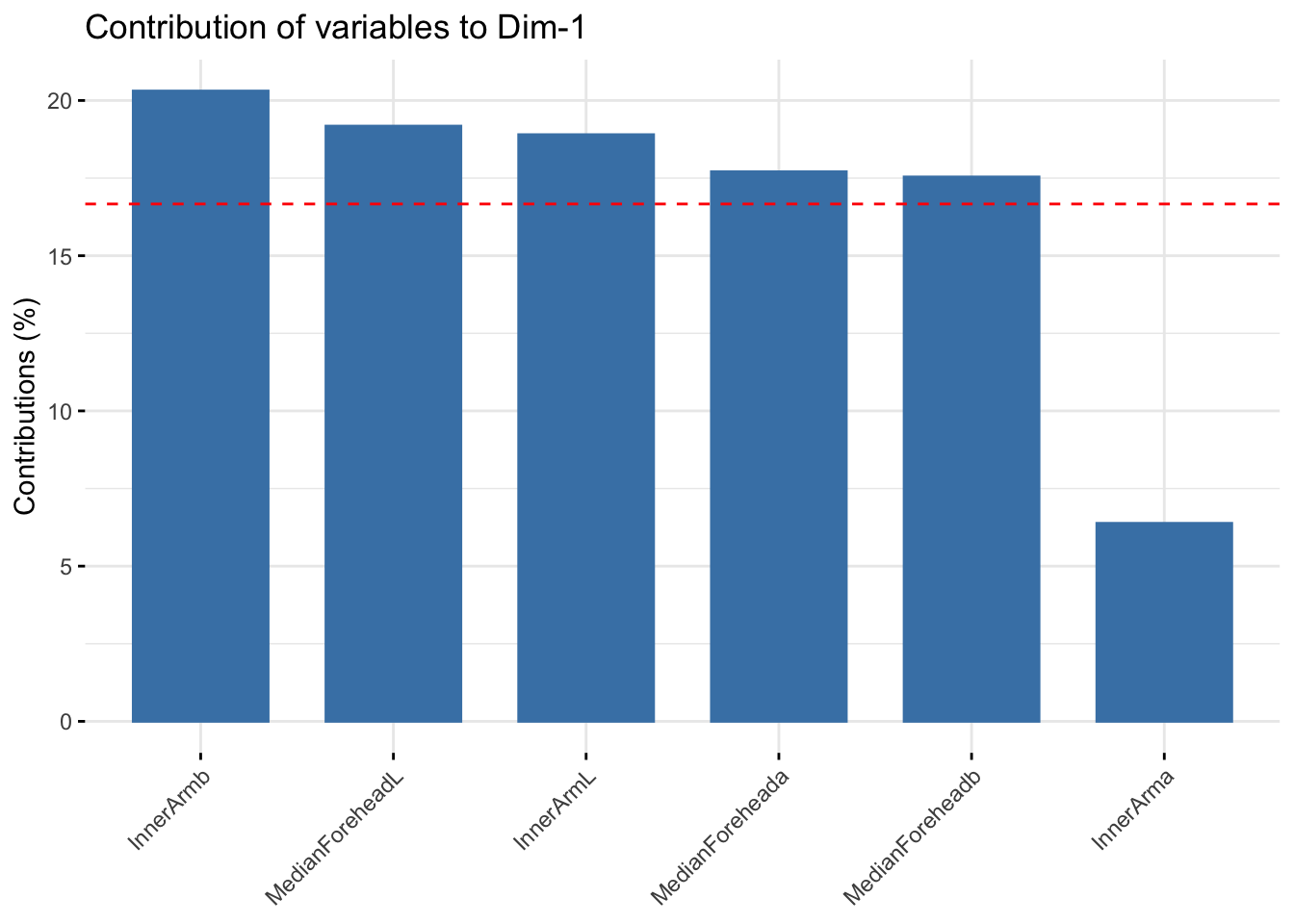
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(summer_cie_pca, choice = "var", axes = 2, top = 10)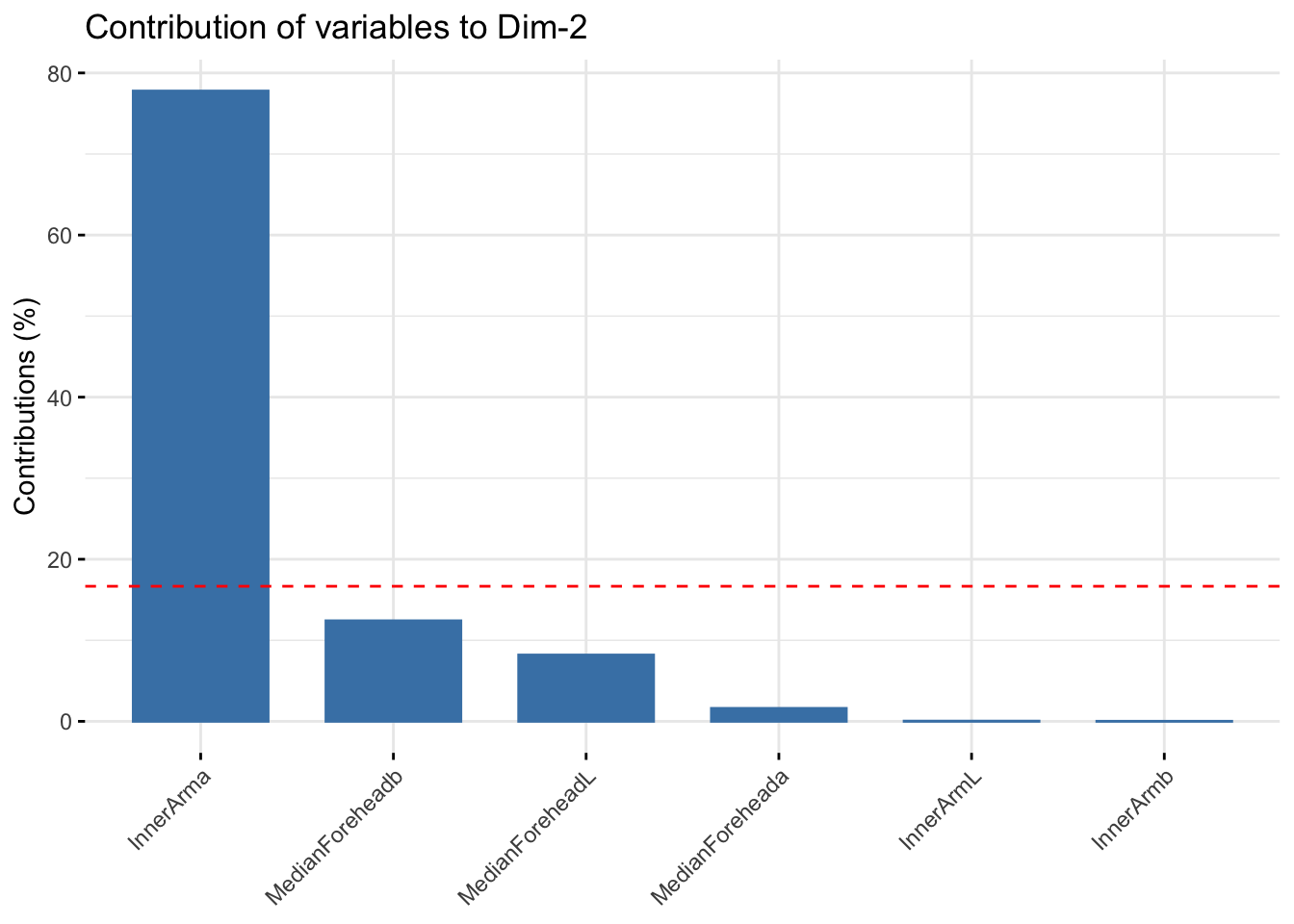
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
six_cie_scale <- scale(six_cie)
six_cie_pca <- prcomp(six_cie_scale)
summary(six_cie_pca)Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6
Standard deviation 1.9741 1.1342 0.62622 0.54034 0.27271 0.24039
Proportion of Variance 0.6495 0.2144 0.06536 0.04866 0.01239 0.00963
Cumulative Proportion 0.6495 0.8639 0.92931 0.97797 0.99037 1.00000six_cie_pca$loadings[, 1:2]NULLfviz_eig(six_cie_pca, addlabels = TRUE)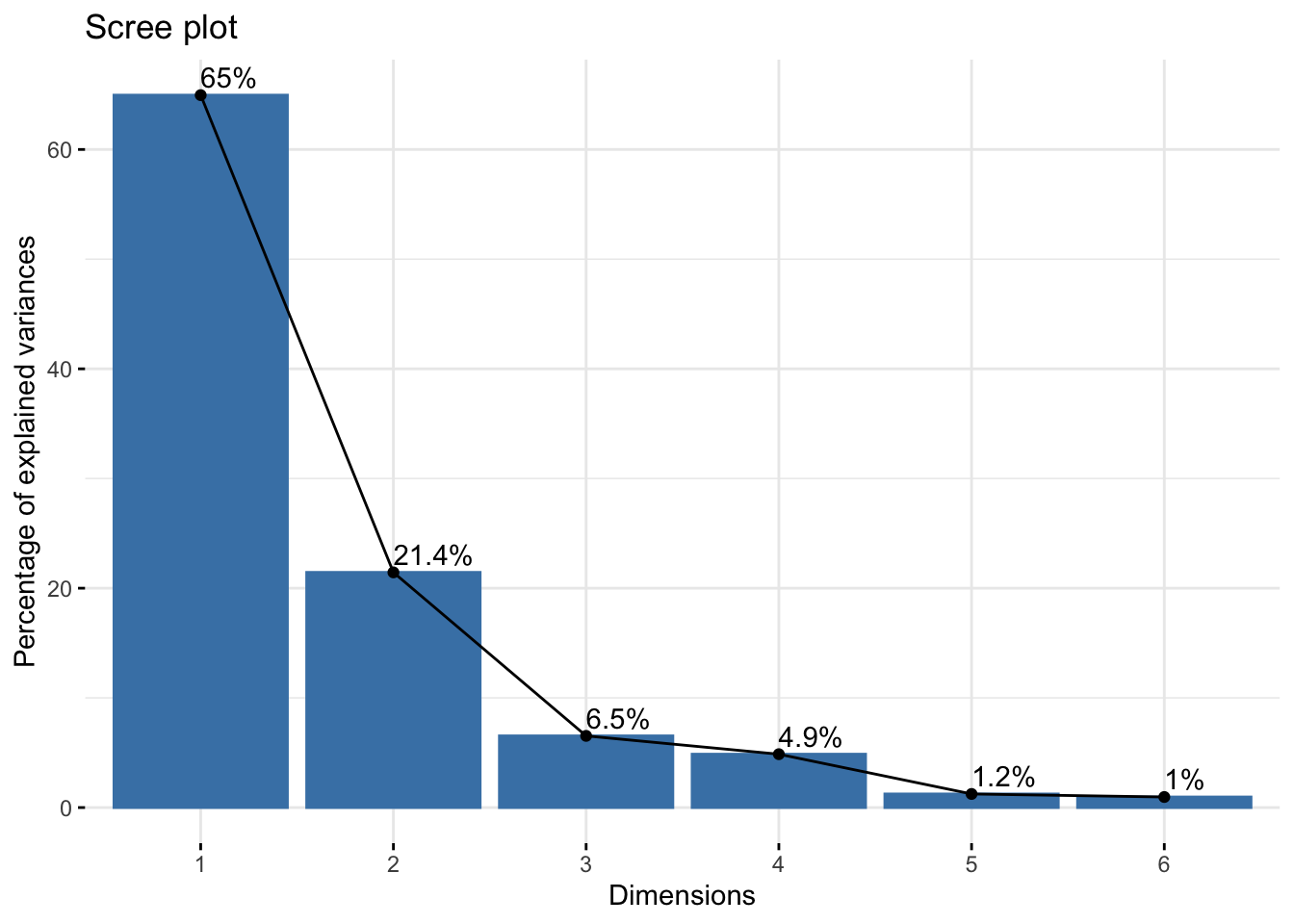
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_pca_var(six_cie_pca, col.var = "black")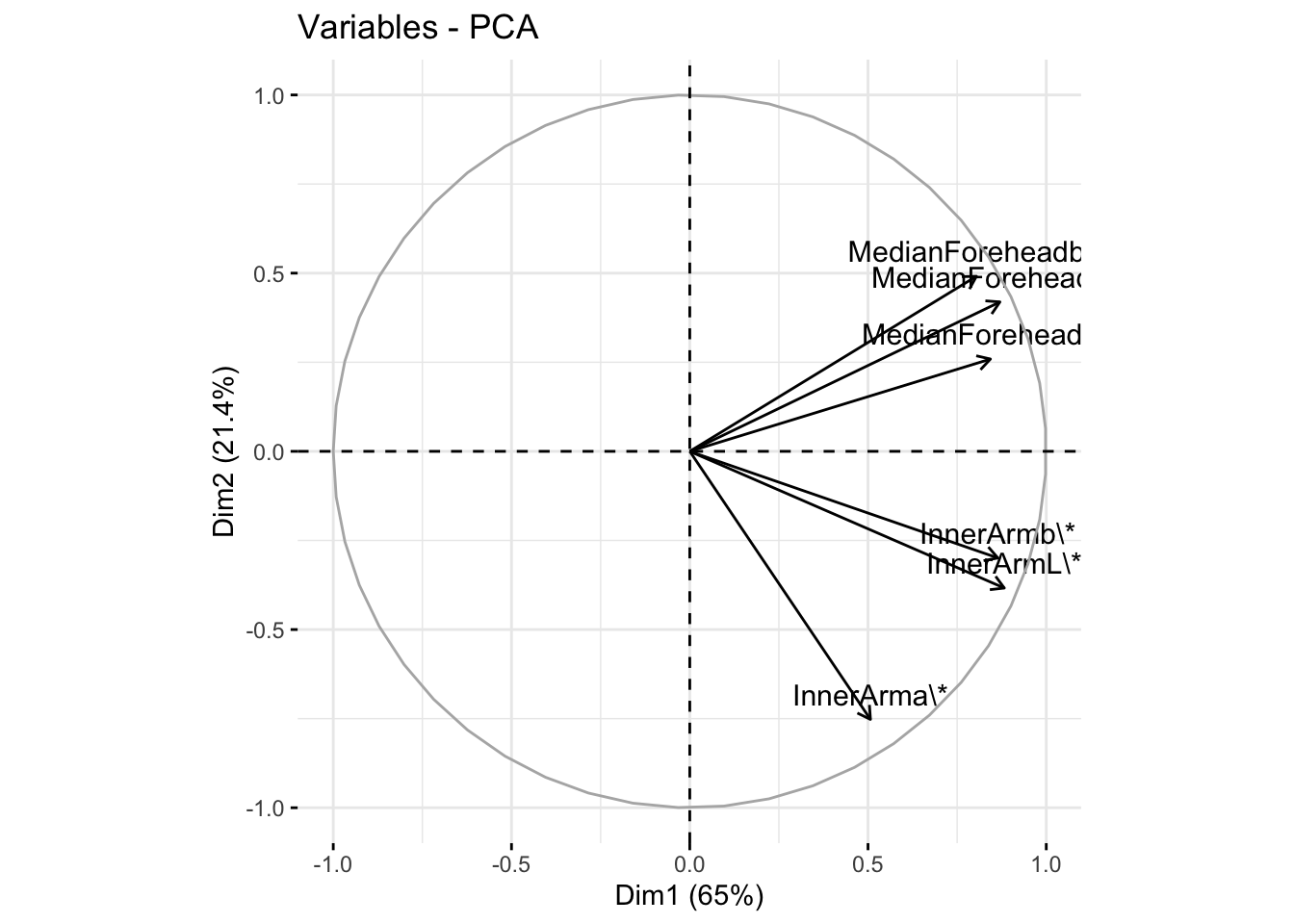
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_cos2(six_cie_pca, choice = "var", axes = 1:2)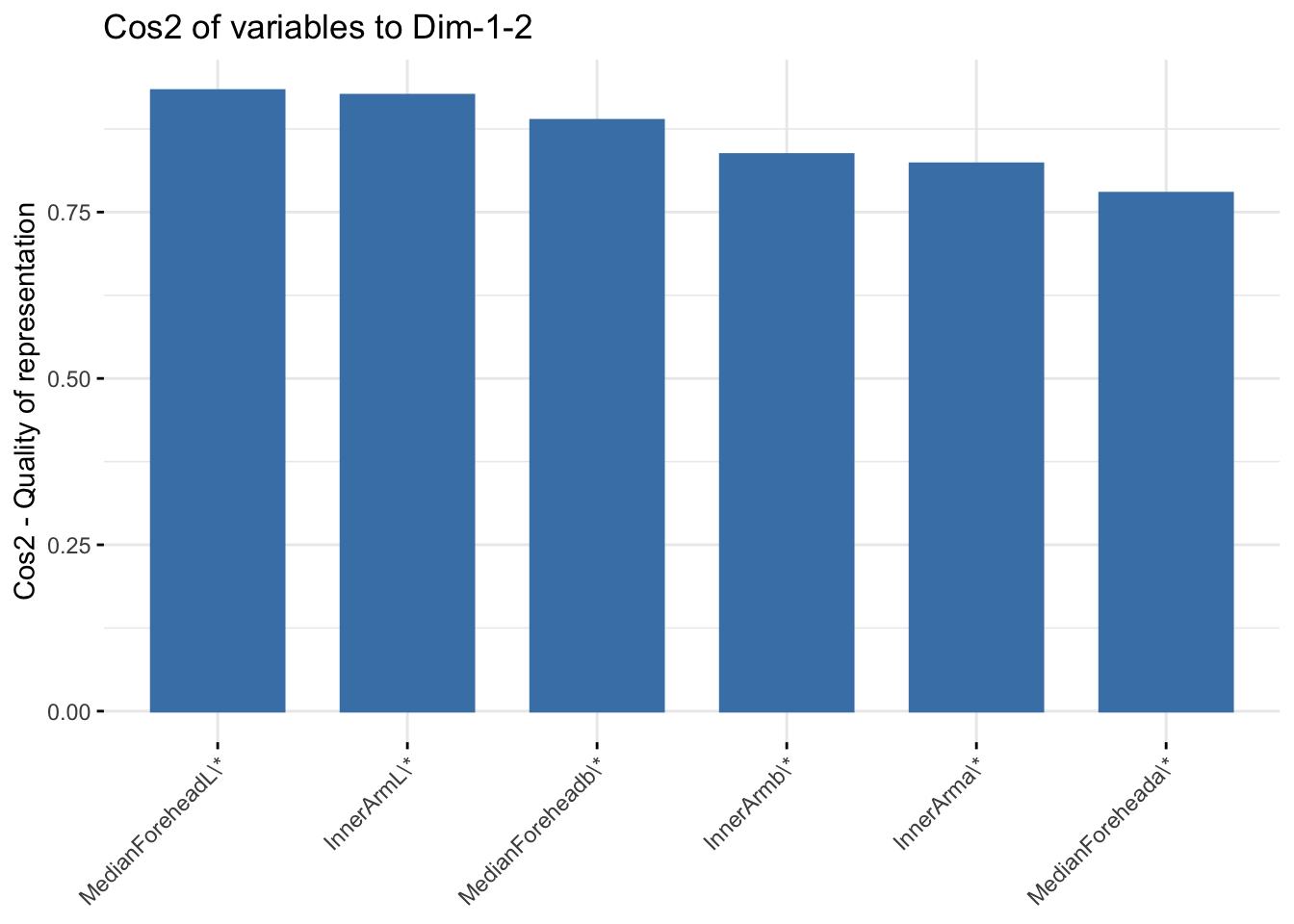
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
six_cie_comps <- as.data.frame(six_cie_pca$x)
six_cie_new <- cbind(six_week_data,six_cie_comps[,c(1,2)])
ggplot(six_rgb_new, aes(x=PC1, y=PC2, col = Ethnicity, fill = Ethnicity)) +
stat_ellipse(geom = "polygon", col= "black", alpha =0.5)+
geom_point(shape=21, col="black") +
labs(title = "Six Week CIELAB PCA")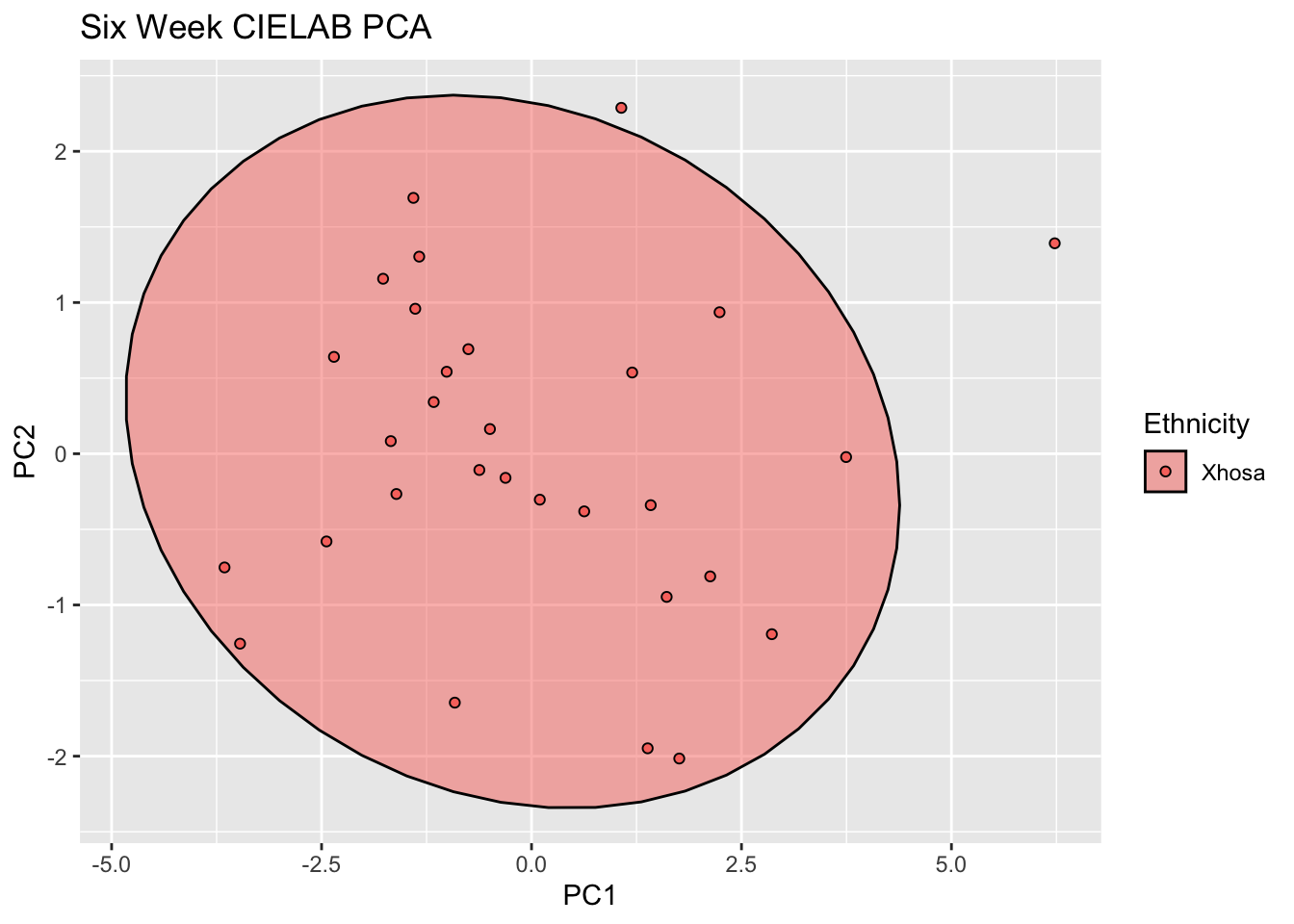
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(six_cie_pca, choice = "var", axes = 1, top = 10)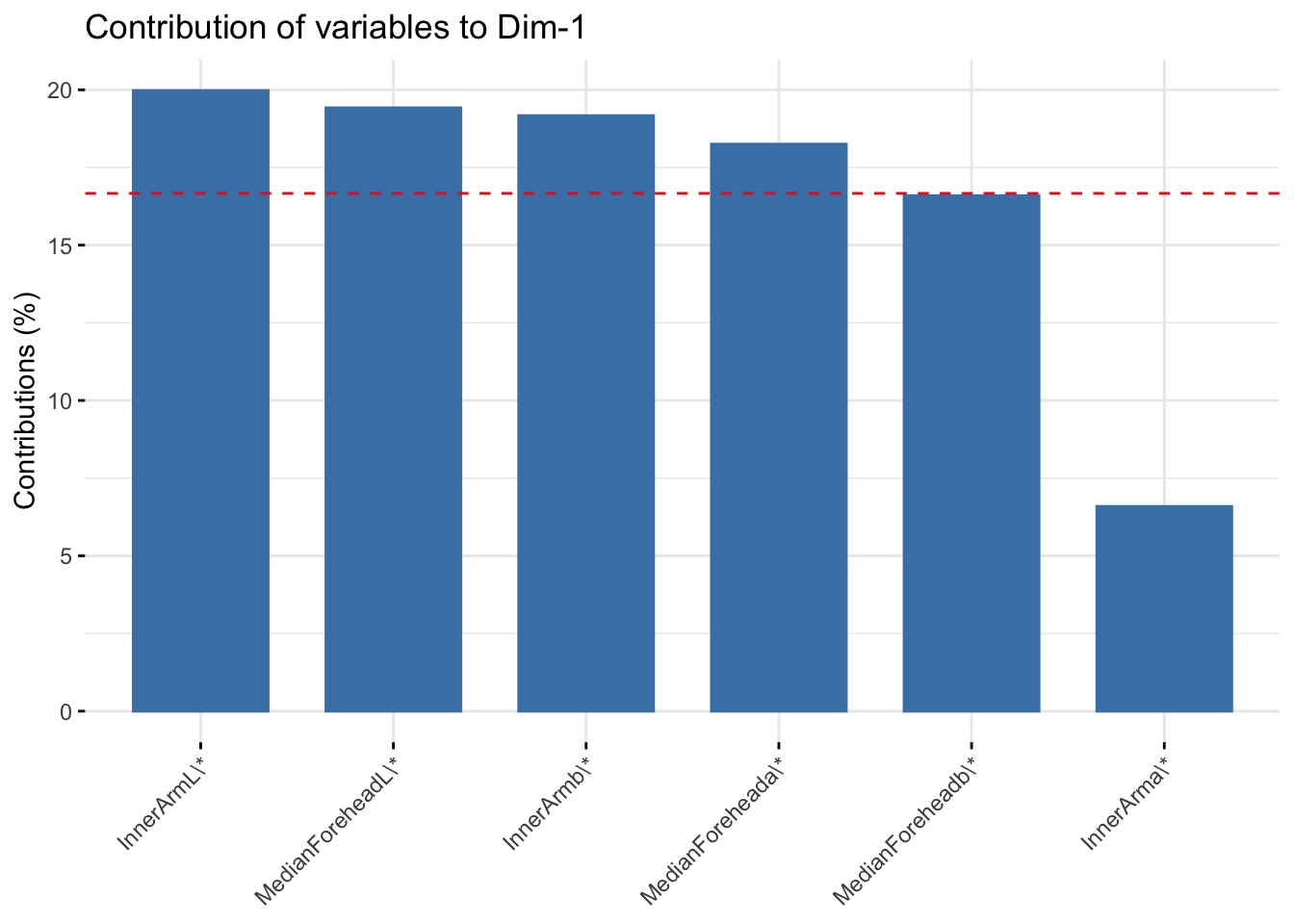
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
fviz_contrib(six_cie_pca, choice = "var", axes = 2, top = 10)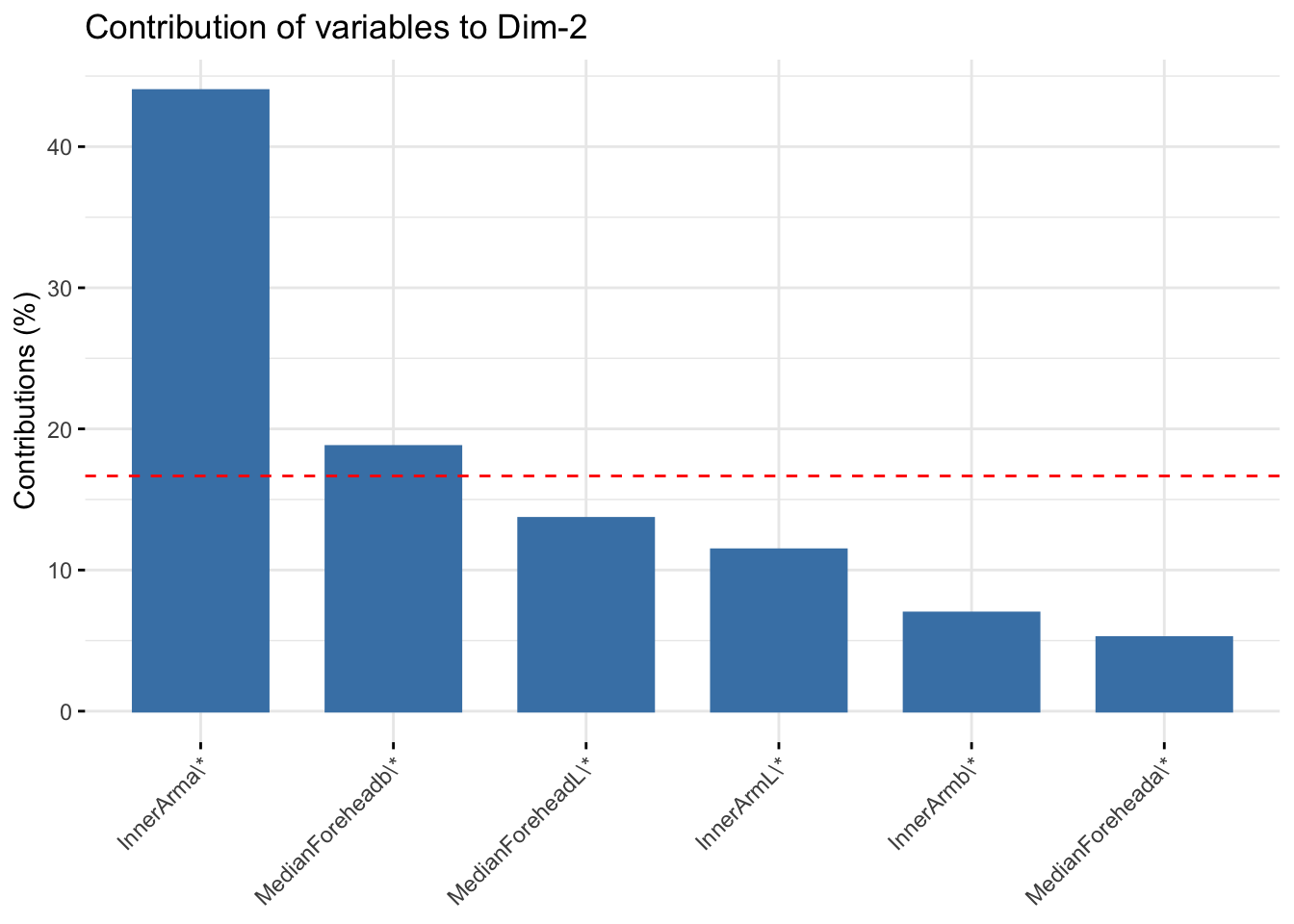
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
part 2: visualizations
joined_data <- joined_data %>%
select(matches("Participant|Ethnicity_|InnerArm|MedianForehead|VitD|Diff"))
long_data <- joined_data %>%
pivot_longer(
cols = -ParticipantCentreID,
names_to = c(".value", "Season"), # Split column names into two parts
names_sep = "_" # Separate at _
)ggplot(long_data, aes(x = MedianForeheadM, fill = Ethnicity)) +
geom_histogram(alpha = 0.5, position = "identity") `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 68 rows containing non-finite outside the scale range
(`stat_bin()`).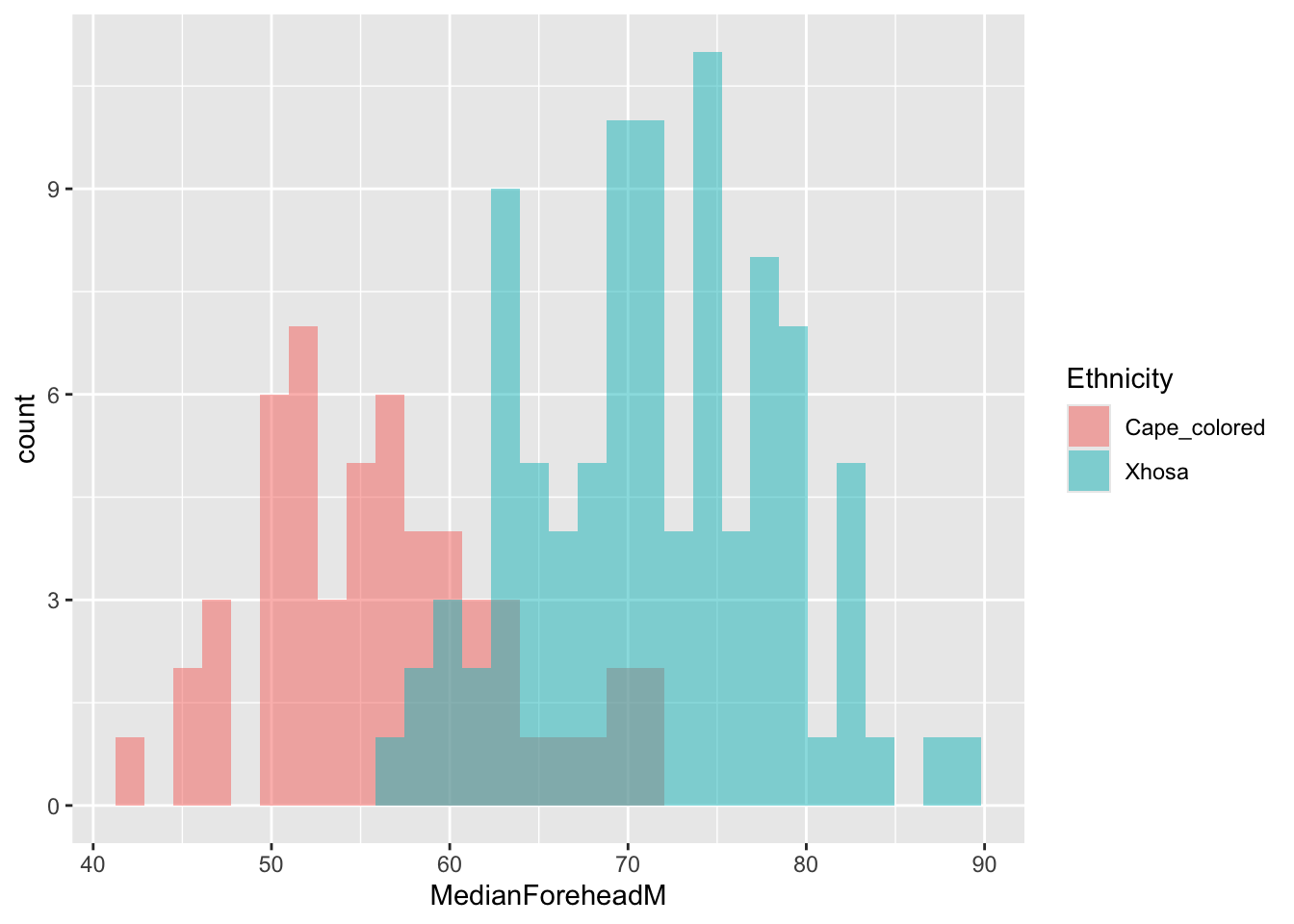
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
cape_colored <- long_data[long_data$Ethnicity == "Cape_colored",]
ggplot(cape_colored, aes(x = MedianForeheadM, fill = ..x..)) +
geom_histogram() +
scale_fill_gradient(low="#D2B48C", high="#1B0000") +
theme_minimal() +
labs(
title = "Distribution of Forehead Melanin Index by Ethnicity ~ Cape_colored",
x = "Forehead Melanin Index",
y = "Count"
)Warning: The dot-dot notation (`..x..`) was deprecated in ggplot2 3.4.0.
ℹ Please use `after_stat(x)` instead.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 68 rows containing non-finite outside the scale range
(`stat_bin()`).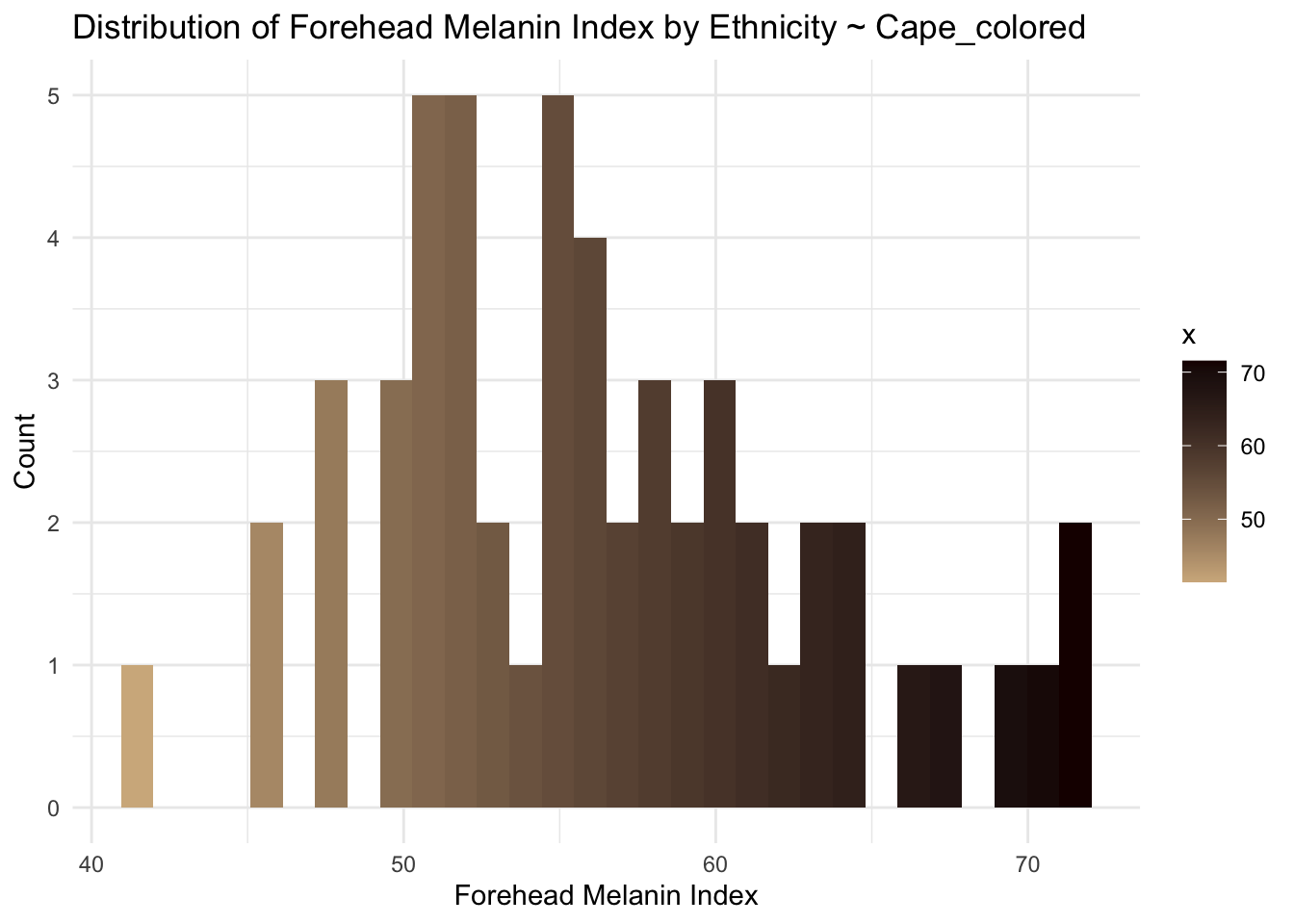
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
xhosa <- long_data[long_data$Ethnicity == "Xhosa",]
ggplot(xhosa, aes(x = MedianForeheadM, fill = ..x..)) +
geom_histogram() +
scale_fill_gradient(low="#D2B48C", high="#1B0000") +
theme_minimal() +
labs(
title = "Distribution of Forehead Melanin Index by Ethnicity ~ Xhosa",
x = "Forehead Melanin Index",
y = "Count"
)`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 68 rows containing non-finite outside the scale range
(`stat_bin()`).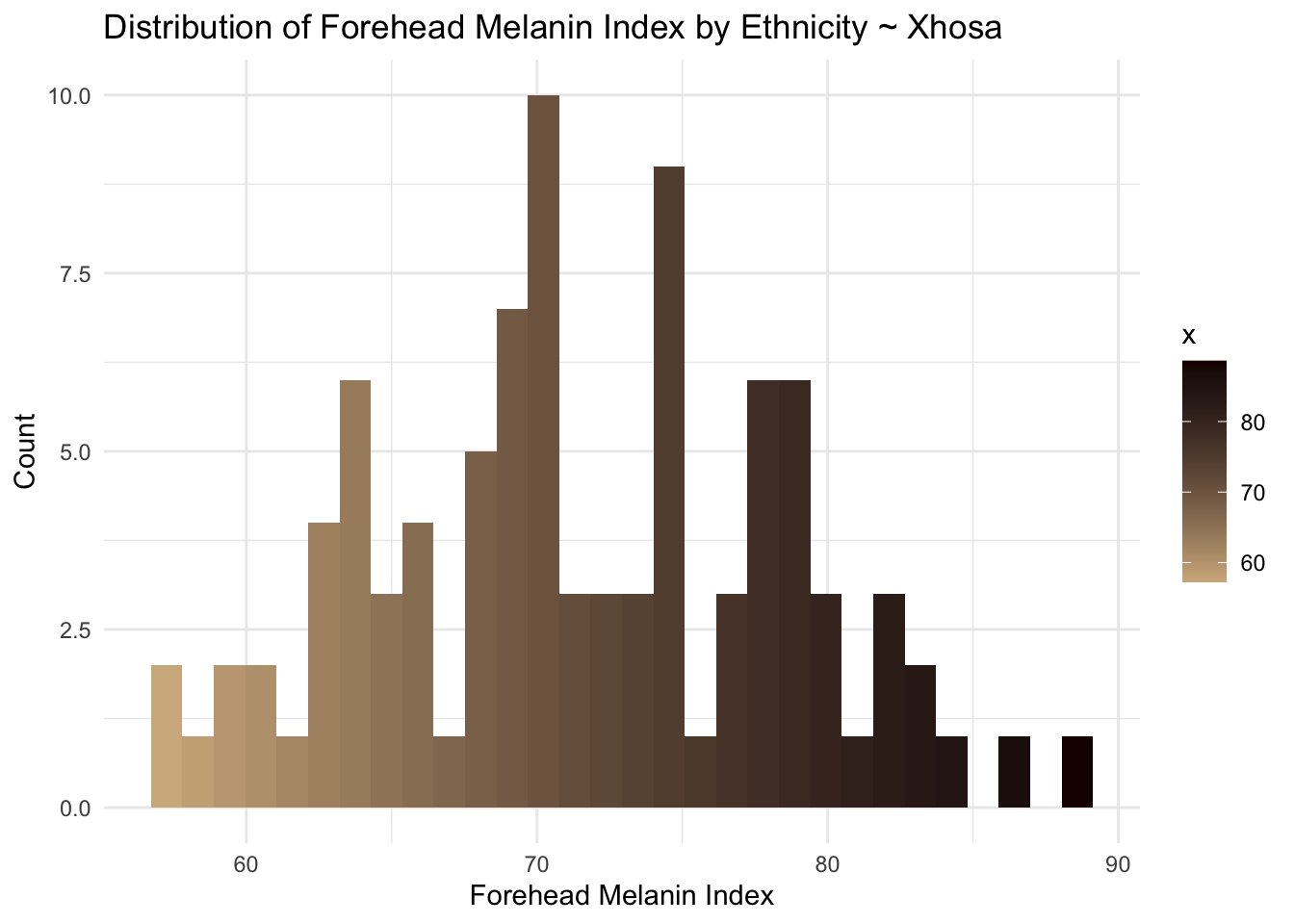
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
filtered_data <- long_data %>%
filter(!is.na(Ethnicity))
ggplot(filtered_data, aes(x = MedianForeheadM, fill = ..x..)) +
geom_histogram() +
facet_grid(Ethnicity ~ .) +
scale_fill_gradient(
low = "#D2B48C", # Light tan
high = "#1B0000" # Dark brown
) +
theme_minimal() +
labs(
title = "Distribution of Forehead Melanin Index by Ethnicity",
x = "Forehead Melanin Index",
y = "Count",
fill = "Melanin Index"
)`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.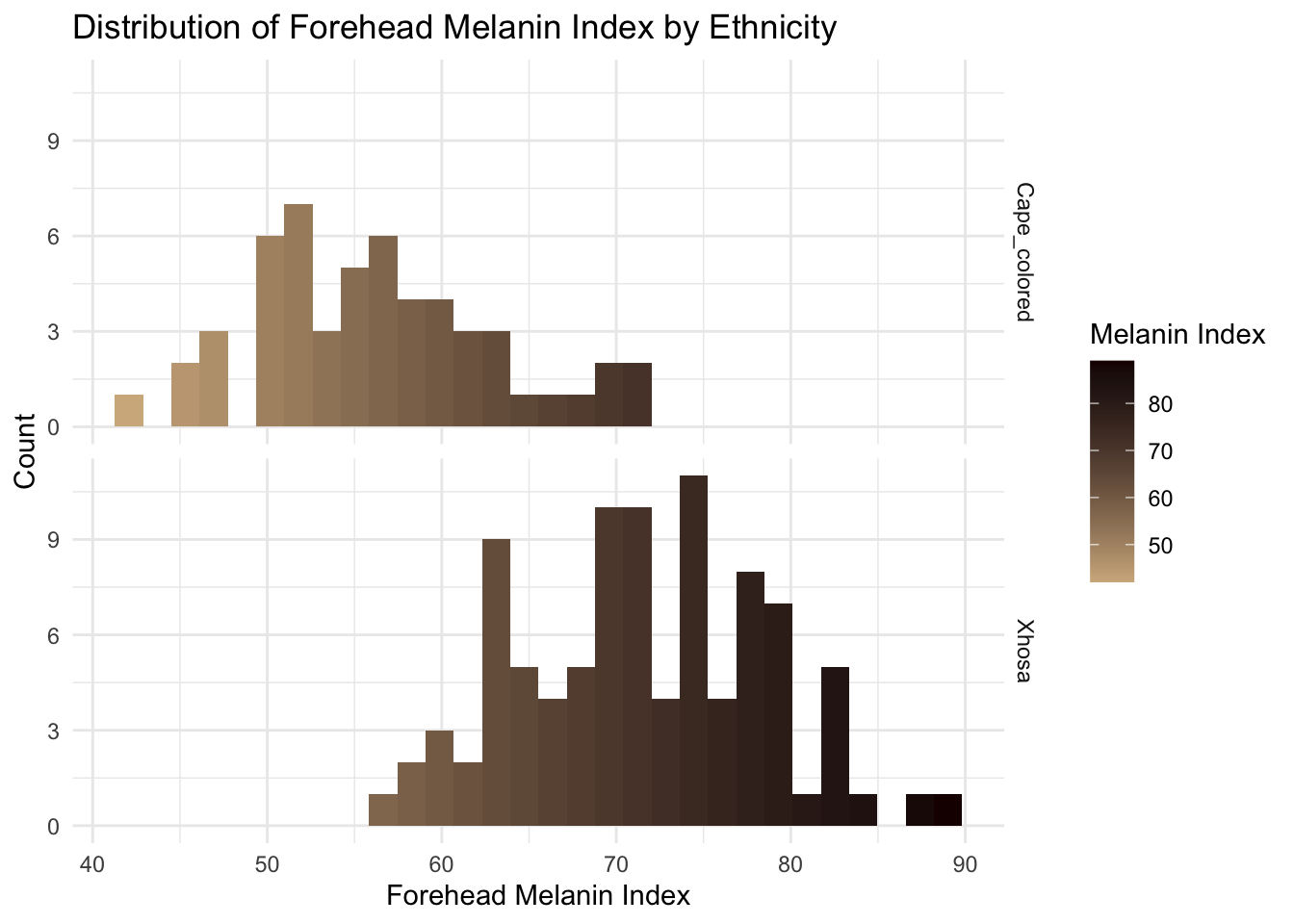
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
filtered_data <- long_data %>%
filter(!is.na(Ethnicity))
ggplot(filtered_data, aes(x = MedianForeheadM, fill = ..x..)) +
geom_histogram() +
facet_grid(Ethnicity ~ Season) +
scale_fill_gradient(
low = "#D2B48C", # Light tan
high = "#1B0000" # Dark brown
) +
theme_minimal() +
labs(
title = "Distribution of Forehead Melanin Index by Ethnicity",
x = "Forehead Melanin Index",
y = "Count",
fill = "Melanin Index"
)`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.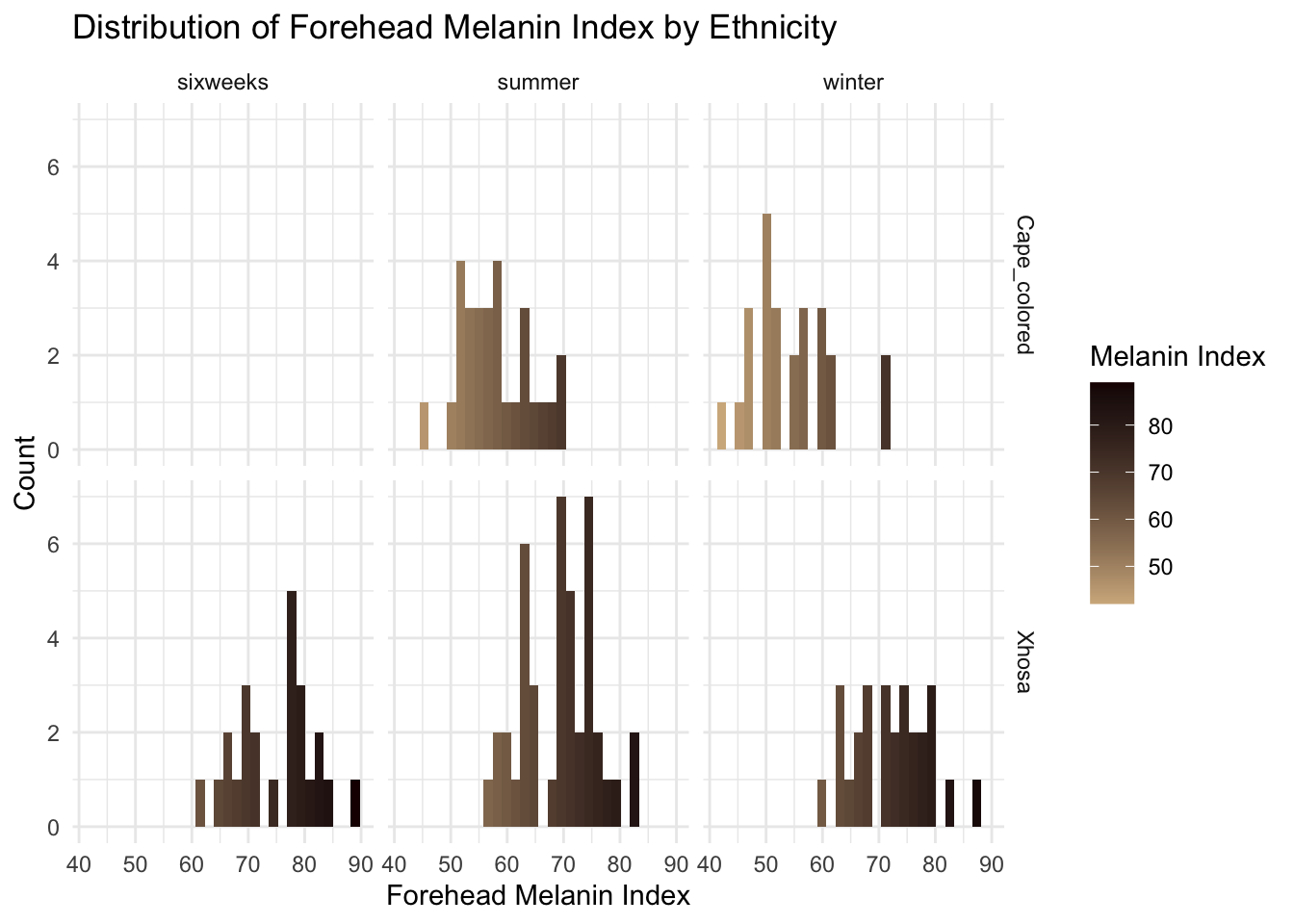
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
xhosa <- xhosa %>%
filter(!is.na(Ethnicity)) %>%
filter(!is.na(Season))
xhosa %>%
group_by(Season) %>%
shapiro_test(InnerArmM)# A tibble: 3 × 4
Season variable statistic p
<chr> <chr> <dbl> <dbl>
1 sixweeks InnerArmM 0.954 0.337
2 summer InnerArmM 0.977 0.520
3 winter InnerArmM 0.952 0.239# xhosa is normally distributed in each season (p>0.05)
cape_colored <- cape_colored %>%
filter(!is.na(Ethnicity)) %>%
filter(!is.na(Season))
cape_colored %>%
group_by(Season) %>%
shapiro_test(InnerArmM)# A tibble: 2 × 4
Season variable statistic p
<chr> <chr> <dbl> <dbl>
1 summer InnerArmM 0.895 0.00727
2 winter InnerArmM 0.873 0.00509# cape_colored is not normally distributed in each season (p<0.05)
ggqqplot(cape_colored, "InnerArmM", facet.by = "Season")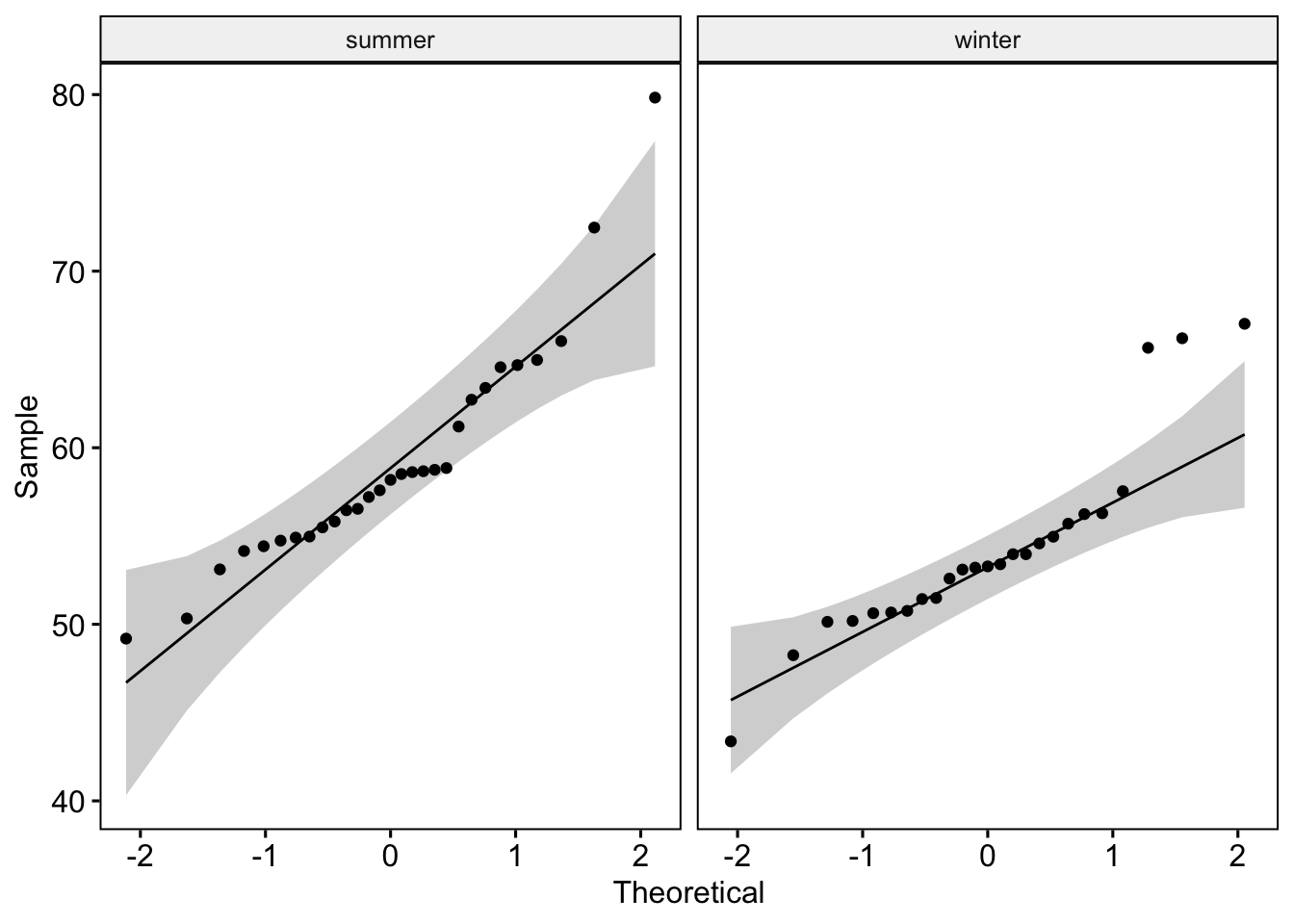
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
ggqqplot(xhosa, "InnerArmM", facet.by = "Season")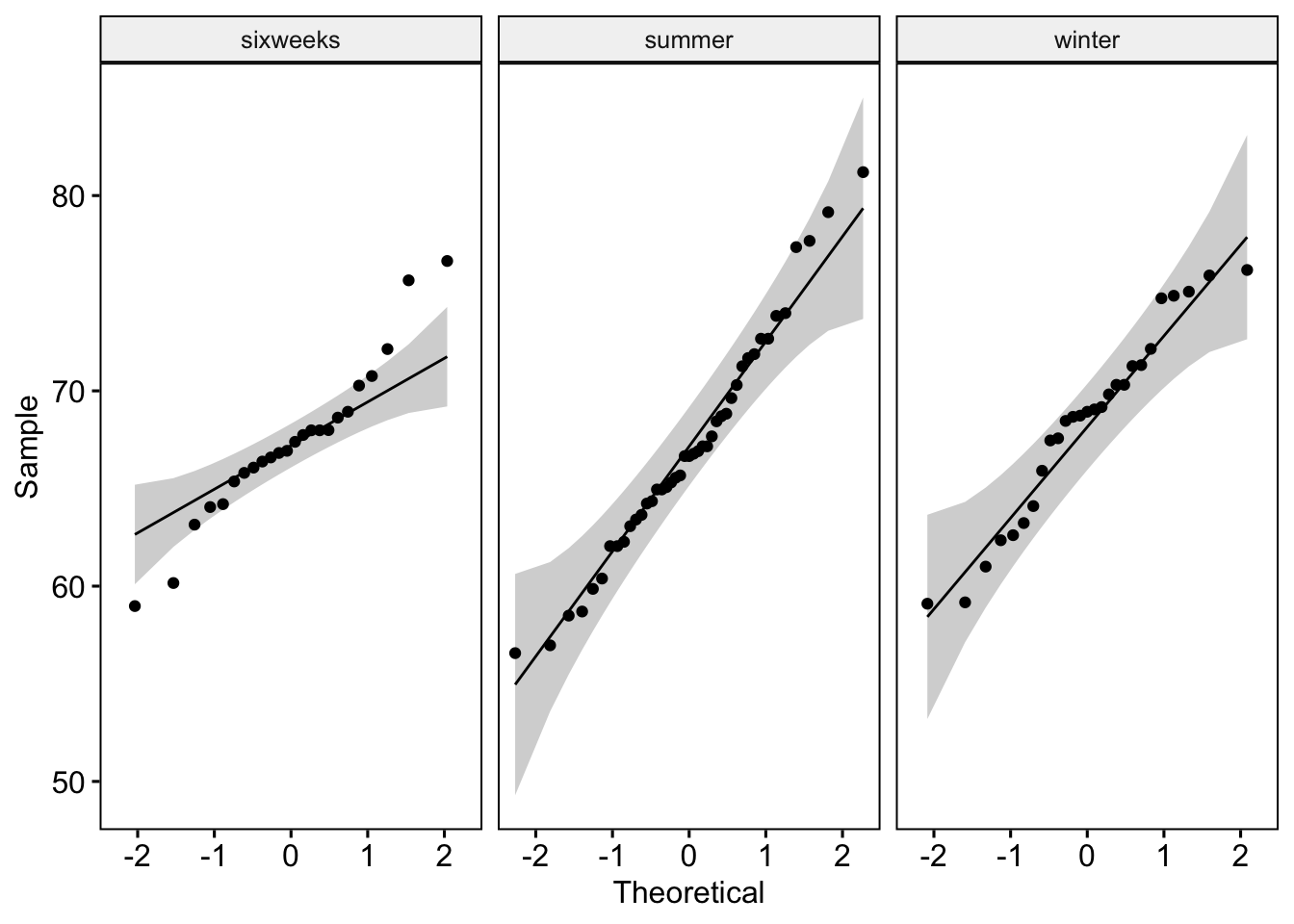
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
aov_result_x <- aov(InnerArmM ~ Season, data = xhosa)
broom::tidy(aov_result_x)# A tibble: 2 × 6
term df sumsq meansq statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Season 2 29.8 14.9 0.560 0.573
2 Residuals 91 2420. 26.6 NA NA # filtered_data <- long_data %>%
# filter(!is.na(Ethnicity))
# ggplot(filtered_data, aes(x = MDifference)) +
# geom_histogram() +
# facet_grid(Ethnicity ~ Season) +
# theme_minimal() +
# labs(
# title = "Difference in Forehead and Inner Arm Melanin Index by Ethnicity and Season",
# x = "Difference in Forehead MI and Inner Arm MI",
# y = "Count",
# )
#
# filtered_data <- long_data %>%
# filter(!is.na(Ethnicity))
# ggplot(filtered_data, aes(x = Ethnicity, y = MDifference)) +
# geom_boxplot() +
# facet_grid(.~Season) +
# theme_minimal() +
# labs(
# title = "Difference in Forehead and Inner Arm Melanin Index by Ethnicity and Season",
# x = "Difference in Forehead MI and Inner Arm MI",
# y = "Count"
# )summer_data$Ethnicity <- as.factor (summer_data$Ethnicity)
names(summer_data) <- gsub("\\\\", "", names(summer_data))
names(summer_data) <- gsub("\\*", "", names(summer_data))
#Xhosa is value 2
winter_data$Ethnicity <- as.factor (winter_data$Ethnicity)
names(winter_data) <- gsub("\\\\", "", names(winter_data))
names(winter_data) <- gsub("\\*", "", names(winter_data))
# cielab colorspaces different between groups
lstarresult <- t.test(MedianForeheadL ~ Ethnicity, data = summer_data)
lstarp_value <- lstarresult$p.value
ggplot(summer_data, aes(x = Ethnicity, y = MedianForeheadL, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(summer_data$MedianForeheadL),
label = paste("p =", signif(lstarp_value, digits = 3)),
size = 5) +
labs(title = "summer L star")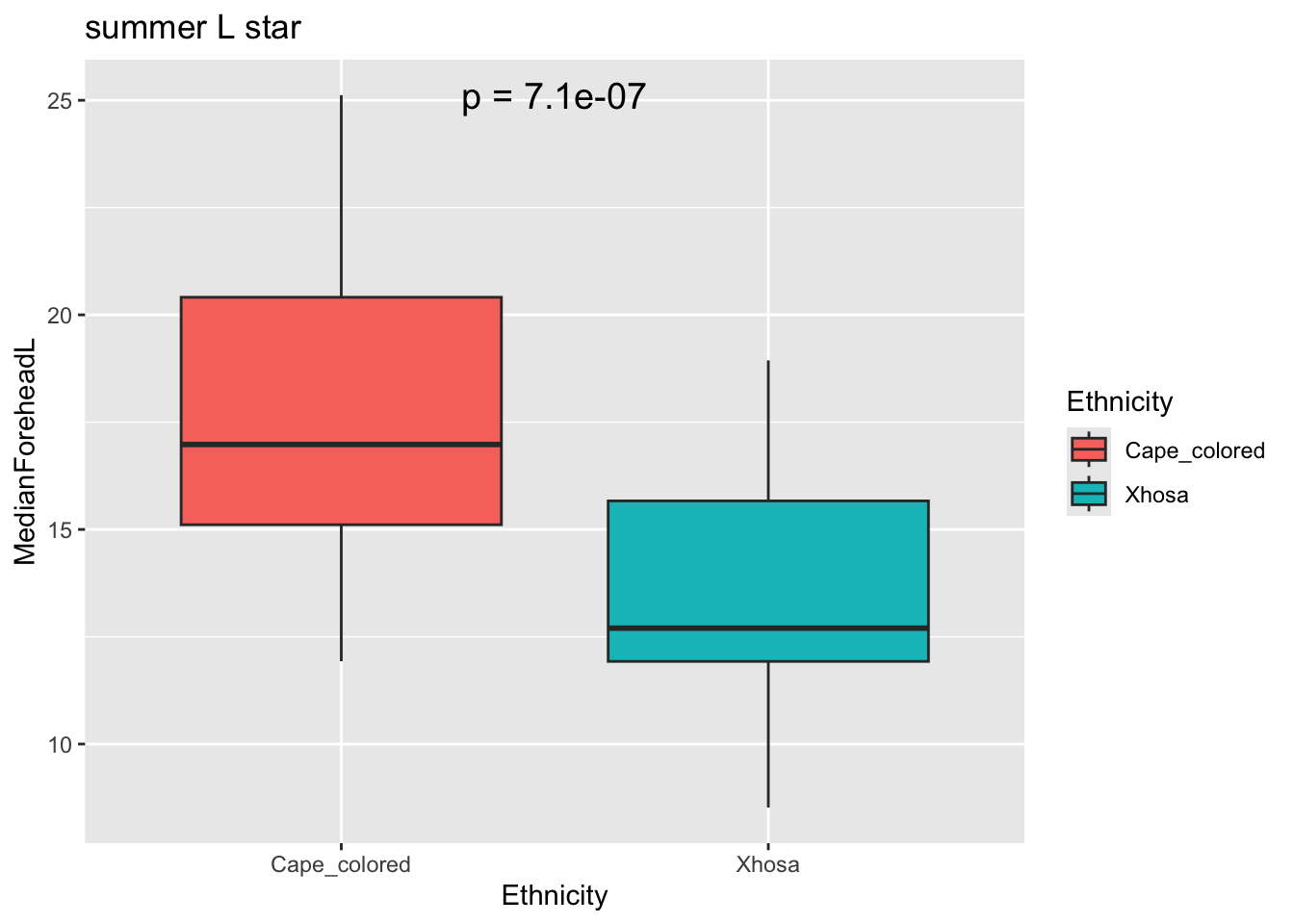
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
astarresult <- t.test(MedianForeheada ~ Ethnicity, data = summer_data)
astarp_value <- astarresult$p.value
ggplot(summer_data, aes(x = Ethnicity, y = MedianForeheada, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(summer_data$MedianForeheada),
label = paste("p =", signif(astarp_value, digits = 3)),
size = 5) +
labs(title = "summer a star")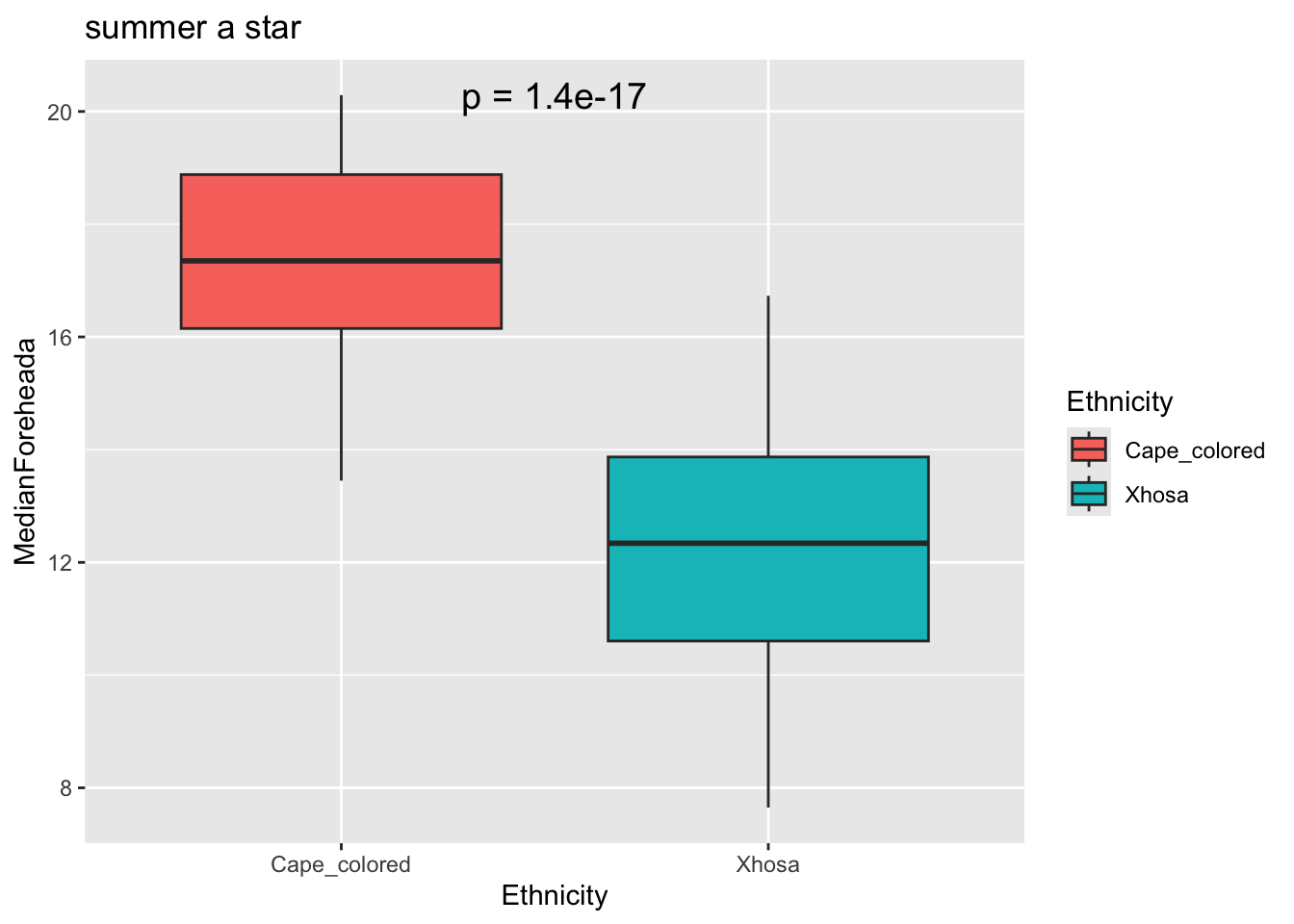
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
bstarresult <- t.test(MedianForeheadb ~ Ethnicity, data = summer_data)
bstarp_value <- bstarresult$p.value
ggplot(summer_data, aes(x = Ethnicity, y = MedianForeheadb, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(summer_data$MedianForeheadb),
label = paste("p =", signif(bstarp_value, digits = 3)),
size = 5) +
labs(title = "summer b star")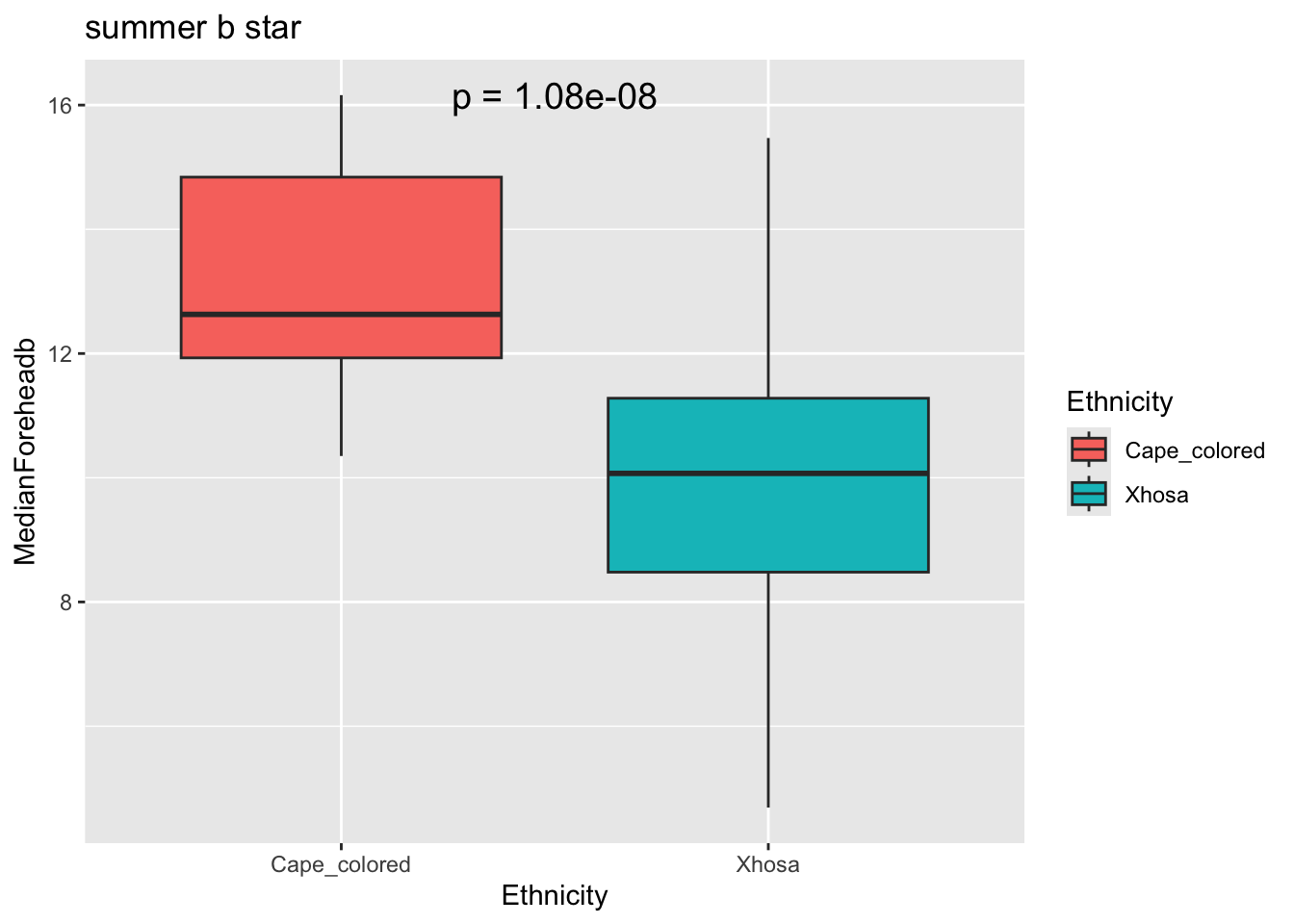
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
winter versus summer t test across all colors
# ME colorspaces different between groups
Mresult <- t.test(MedianForeheadM ~ Ethnicity, data = summer_data)
Mp_value <- Mresult$p.value
ggplot(summer_data, aes(x = Ethnicity, y = MedianForeheadM, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(summer_data$MedianForeheadM),
label = paste("p =", signif(Mp_value, digits = 3)),
size = 5) +
labs(title = "Summer Melanin")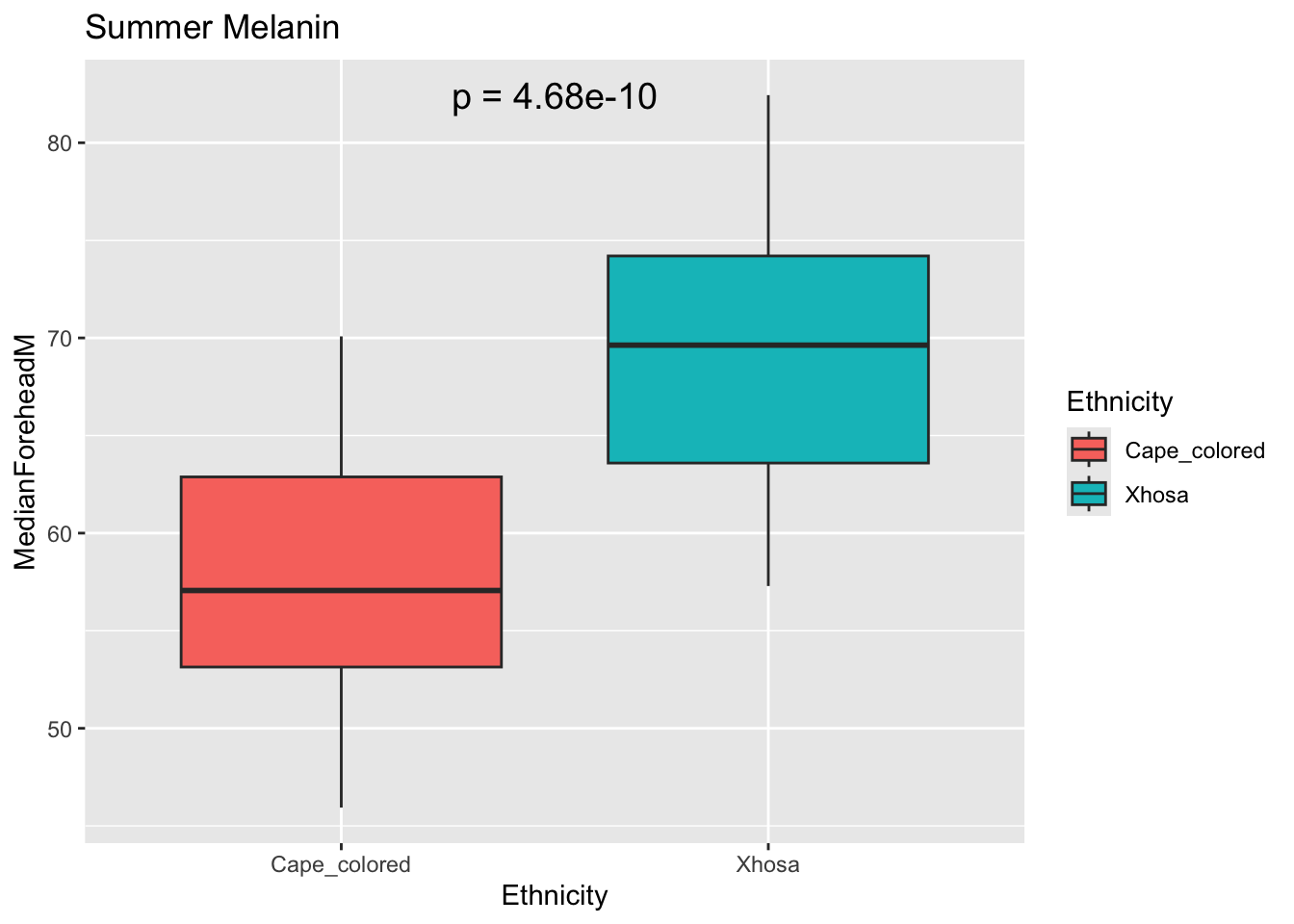
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
Eresult <- t.test(MedianForeheadE ~ Ethnicity, data = summer_data)
Ep_value <- Eresult$p.value
ggplot(summer_data, aes(x = Ethnicity, y = MedianForeheadE, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(summer_data$MedianForeheadE),
label = paste("p =", signif(Ep_value, digits = 3)),
size = 5) +
labs(title = "Summer Erythema")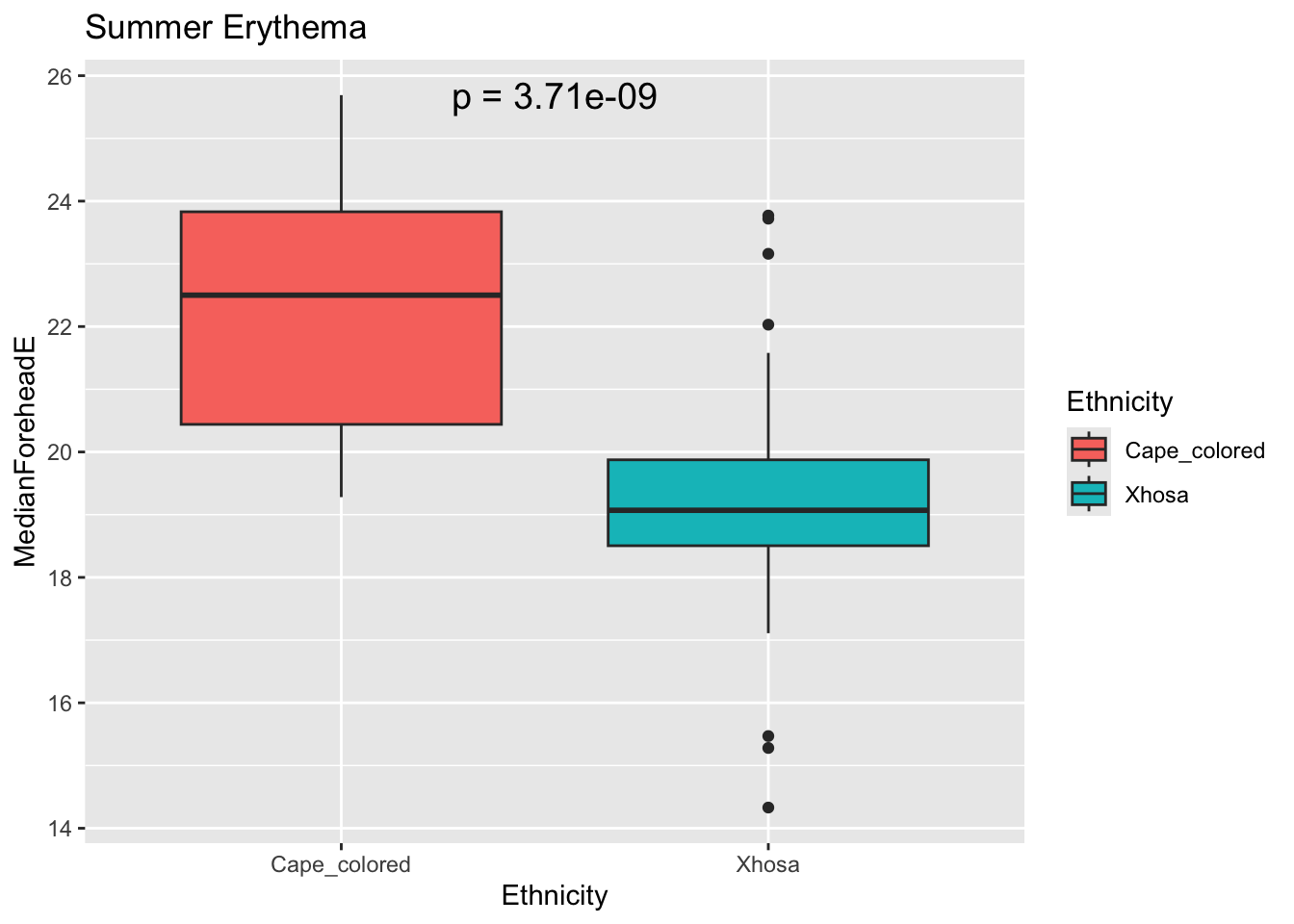
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
Mresult <- t.test(MedianForeheadM ~ Ethnicity, data = winter_data)
Mp_value <- Mresult$p.value
ggplot(winter_data, aes(x = Ethnicity, y = MedianForeheadM, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(winter_data$MedianForeheadM),
label = paste("p =", signif(Mp_value, digits = 3)),
size = 5) +
labs(title = "Winter Melanin")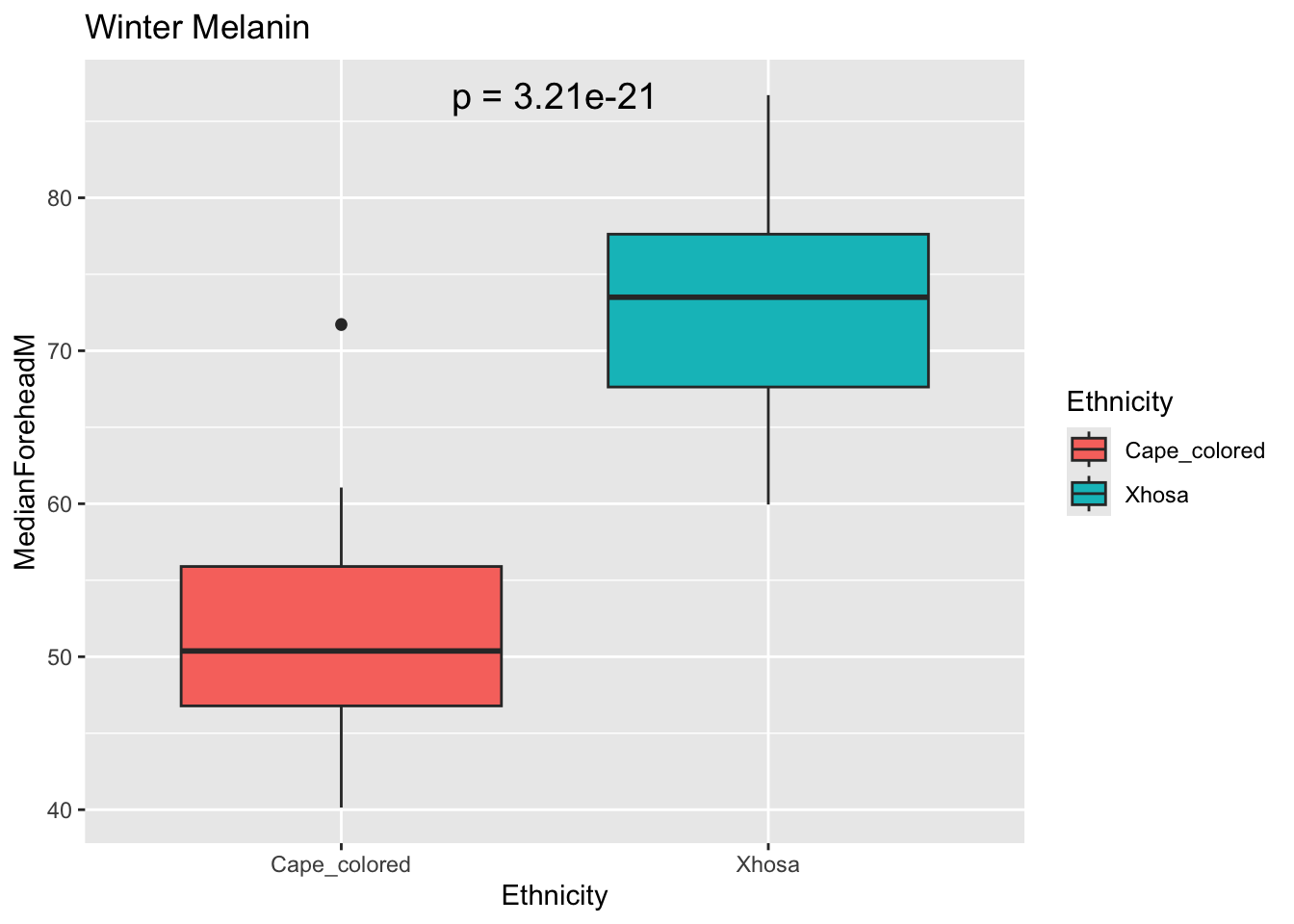
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
Eresult <- t.test(MedianForeheadE ~ Ethnicity, data = winter_data)
Ep_value <- Eresult$p.value
ggplot(winter_data, aes(x = Ethnicity, y = MedianForeheadE, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(winter_data$MedianForeheadE),
label = paste("p =", signif(Ep_value, digits = 3)),
size = 5) +
labs(title = "Winter Erythema")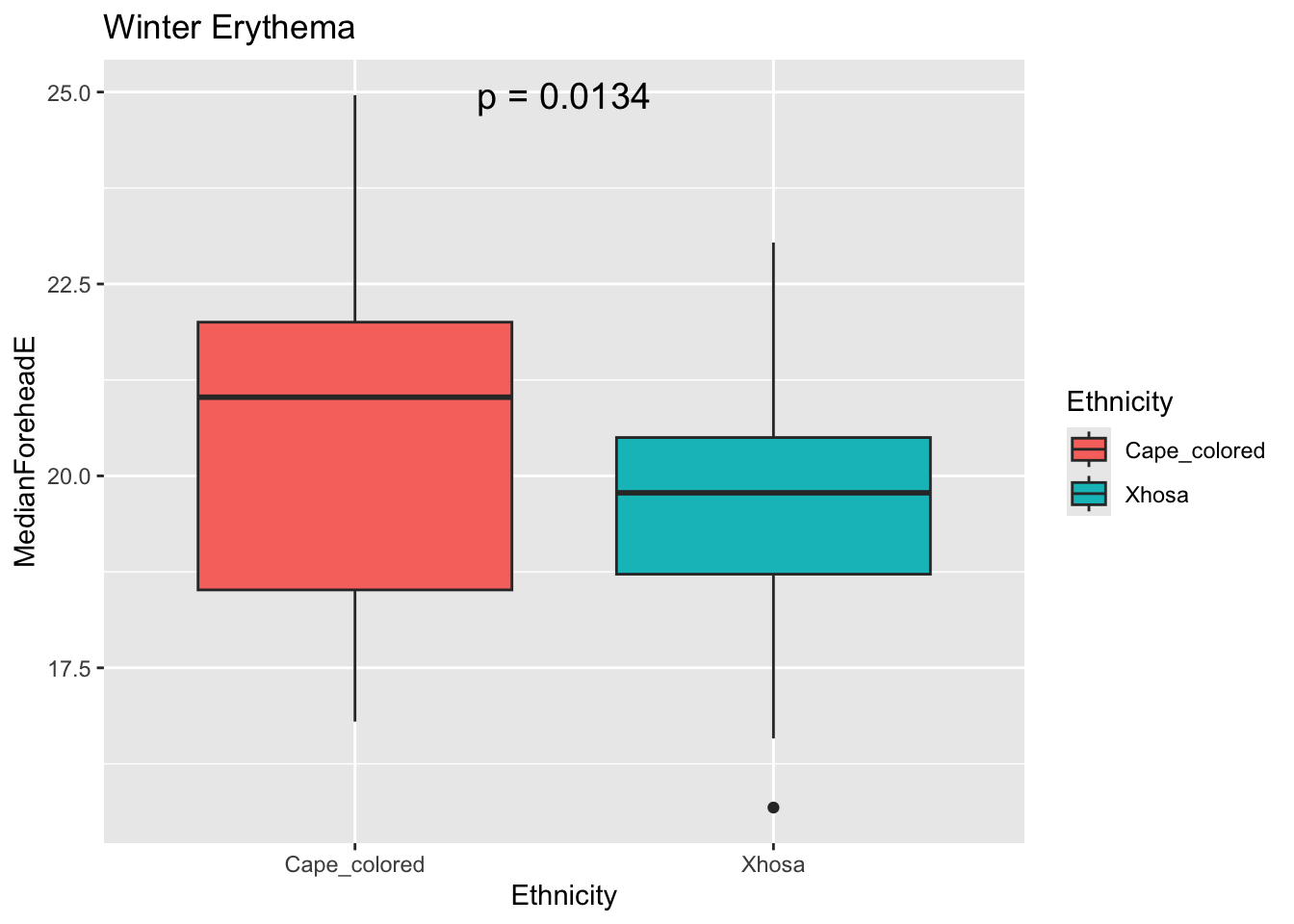
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
# RGB colorspaces different between groups
Rresult <- t.test(MedianForeheadR ~ Ethnicity, data = winter_data)
Rp_value <- Rresult$p.value
ggplot(winter_data, aes(x = Ethnicity, y = MedianForeheadR, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(winter_data$MedianForeheadR),
label = paste("p =", signif(Rp_value, digits = 3)),
size = 5) +
labs(title = "Winter R")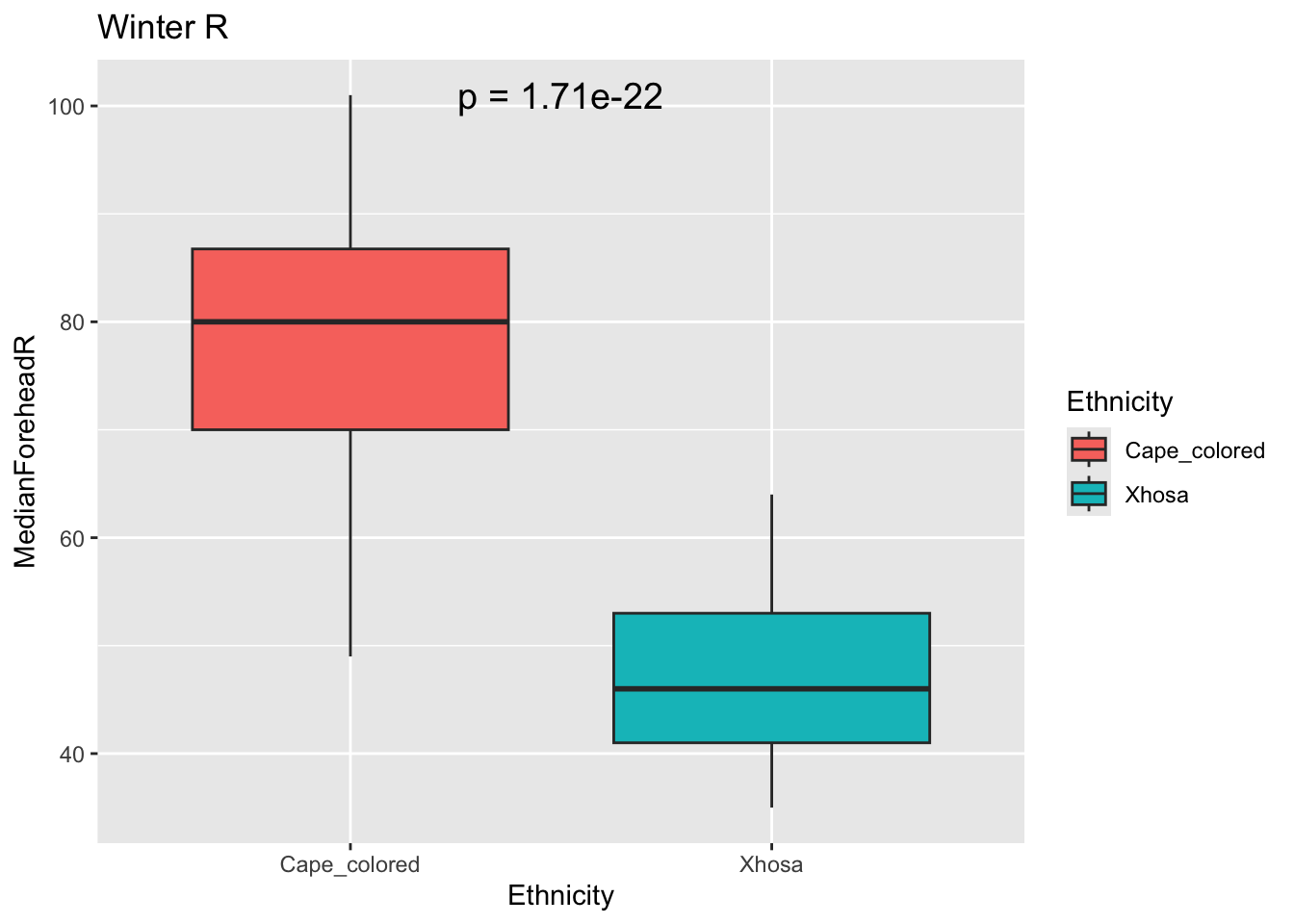
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
winterB <- t.test(MedianForeheadB ~ Ethnicity, data = winter_data)
winterBp <- winterB$p.value
ggplot(winter_data, aes(x = Ethnicity, y = MedianForeheadB, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(winter_data$MedianForeheadB),
label = paste("p =", signif(winterBp, digits = 3)),
size = 5) +
labs(title = "Winter B")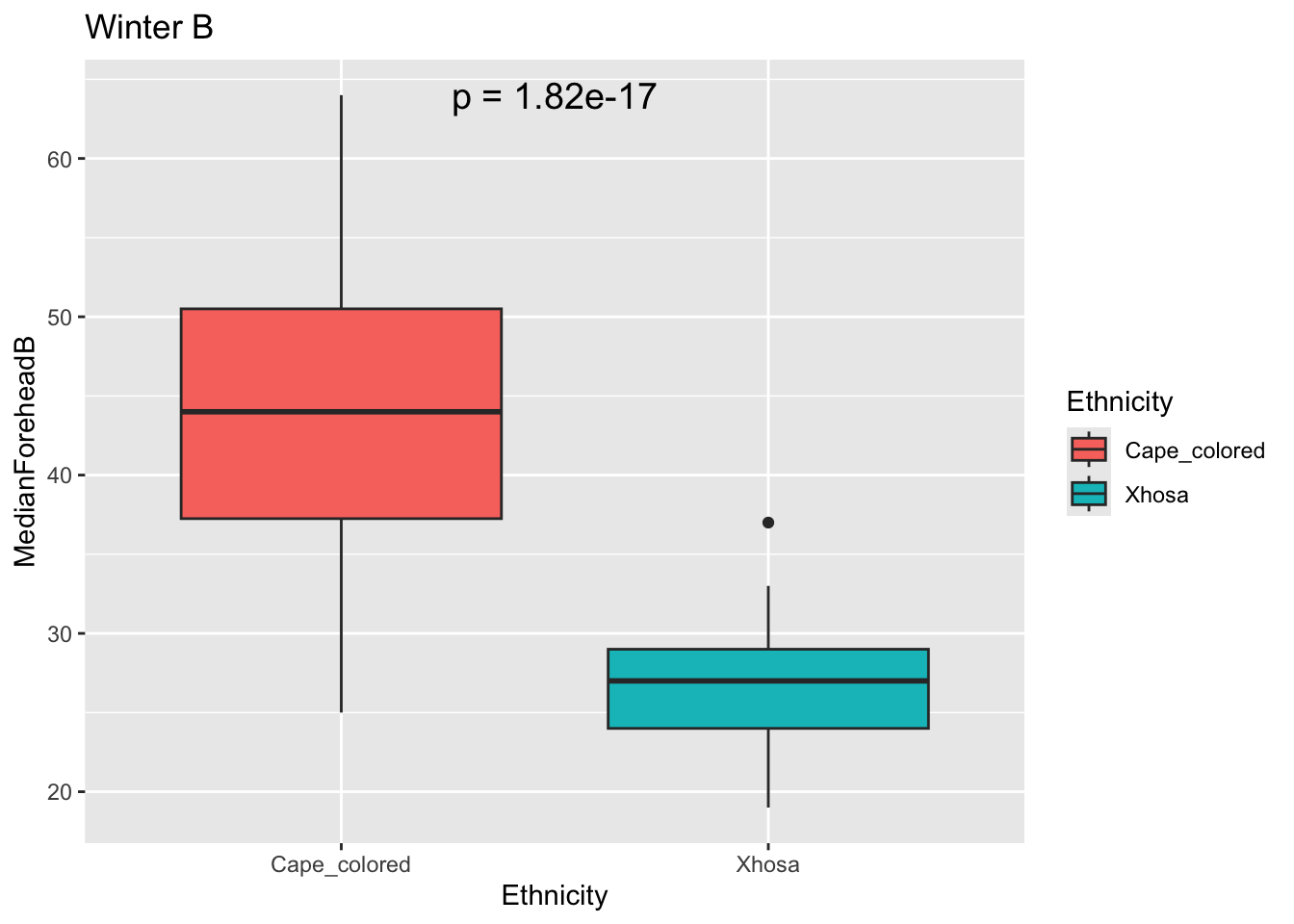
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
winterG <- t.test(MedianForeheadG ~ Ethnicity, data = winter_data)
winterGp <- winterG$p.value
ggplot(winter_data, aes(x = Ethnicity, y = MedianForeheadG, fill = Ethnicity)) +
geom_boxplot() +
annotate("text", x = 1.5, y = max(winter_data$MedianForeheadG),
label = paste("p =", signif(winterGp, digits = 3)),
size = 5) +
labs(title = "Winter G")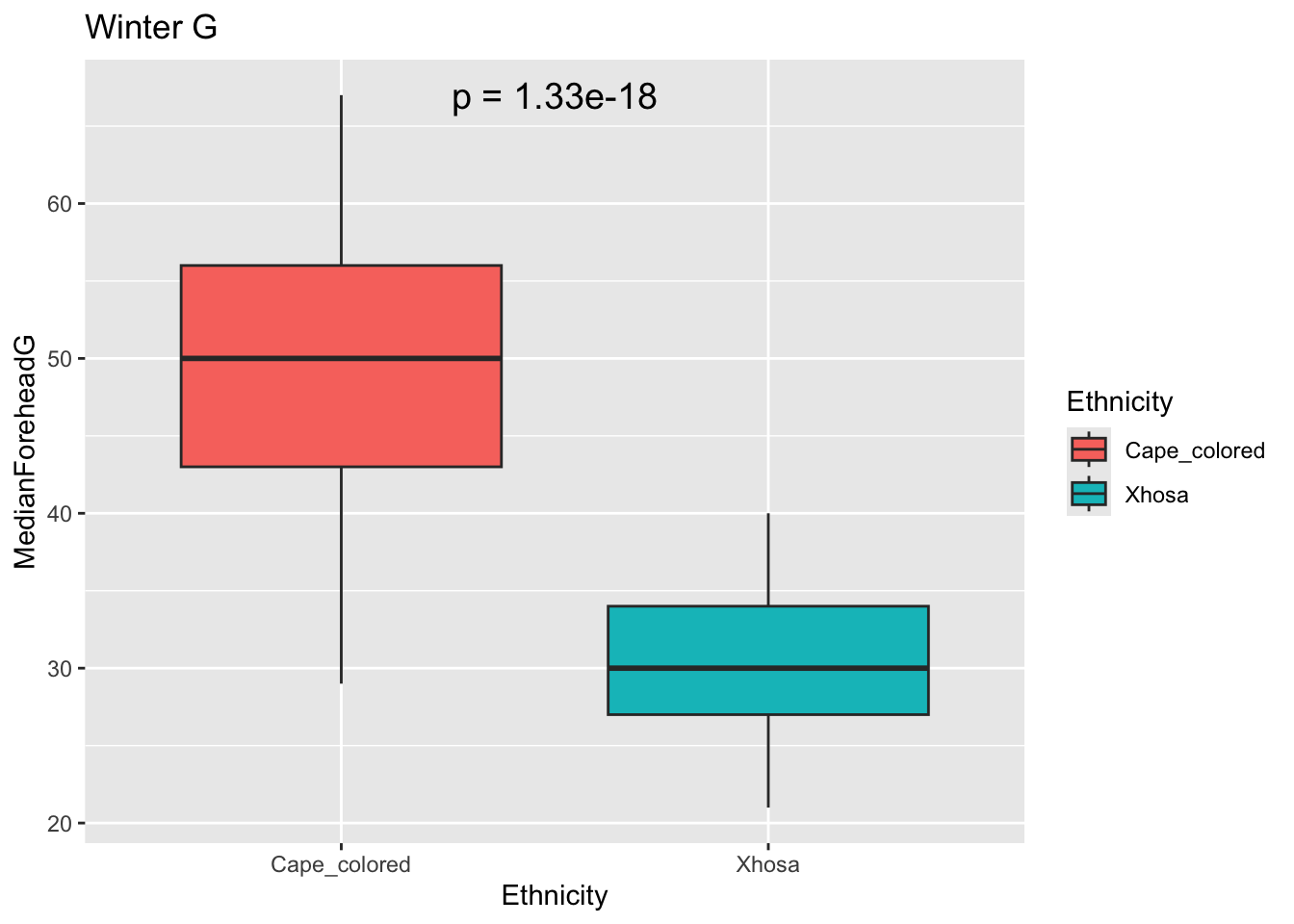
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
sessionInfo()R version 4.4.2 (2024-10-31)
Platform: aarch64-apple-darwin20
Running under: macOS Monterey 12.5.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/Detroit
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] lme4_1.1-36 Matrix_1.7-1 rstatix_0.7.2 ggpubr_0.6.0
[5] ggfortify_0.4.17 wesanderson_0.3.7 missMDA_1.19 FactoMineR_2.11
[9] factoextra_1.0.7 ggcorrplot_0.1.4.1 corrr_0.4.4 readxl_1.4.3
[13] lubridate_1.9.4 forcats_1.0.0 stringr_1.5.1 purrr_1.0.2
[17] tibble_3.2.1 ggplot2_3.5.1 tidyverse_2.0.0 tidyr_1.3.1
[21] dplyr_1.1.4 readr_2.1.5 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] Rdpack_2.6.2 gridExtra_2.3 sandwich_3.1-1
[4] rlang_1.1.4 magrittr_2.0.3 git2r_0.35.0
[7] multcomp_1.4-28 compiler_4.4.2 getPass_0.2-4
[10] callr_3.7.6 vctrs_0.6.5 crayon_1.5.3
[13] pkgconfig_2.0.3 shape_1.4.6.1 fastmap_1.2.0
[16] backports_1.5.0 labeling_0.4.3 utf8_1.2.4
[19] promises_1.3.2 rmarkdown_2.29 tzdb_0.4.0
[22] nloptr_2.1.1 ps_1.8.1 xfun_0.50
[25] glmnet_4.1-8 jomo_2.7-6 cachem_1.1.0
[28] jsonlite_1.8.9 flashClust_1.01-2 later_1.4.1
[31] pan_1.9 broom_1.0.7 parallel_4.4.2
[34] cluster_2.1.8 R6_2.5.1 bslib_0.8.0
[37] stringi_1.8.4 car_3.1-3 rpart_4.1.24
[40] boot_1.3-31 jquerylib_0.1.4 cellranger_1.1.0
[43] estimability_1.5.1 Rcpp_1.0.14 iterators_1.0.14
[46] knitr_1.49 zoo_1.8-12 nnet_7.3-20
[49] httpuv_1.6.15 splines_4.4.2 timechange_0.3.0
[52] tidyselect_1.2.1 abind_1.4-8 rstudioapi_0.17.1
[55] yaml_2.3.10 doParallel_1.0.17 codetools_0.2-20
[58] processx_3.8.5 lattice_0.22-6 withr_3.0.2
[61] coda_0.19-4.1 evaluate_1.0.3 survival_3.8-3
[64] pillar_1.10.1 carData_3.0-5 mice_3.17.0
[67] whisker_0.4.1 DT_0.33 foreach_1.5.2
[70] reformulas_0.4.0 generics_0.1.3 rprojroot_2.0.4
[73] hms_1.1.3 munsell_0.5.1 scales_1.3.0
[76] minqa_1.2.8 xtable_1.8-4 leaps_3.2
[79] glue_1.8.0 emmeans_1.10.6 scatterplot3d_0.3-44
[82] tools_4.4.2 ggsignif_0.6.4 fs_1.6.5
[85] mvtnorm_1.3-3 grid_4.4.2 rbibutils_2.3
[88] colorspace_2.1-1 nlme_3.1-166 Formula_1.2-5
[91] cli_3.6.3 gtable_0.3.6 sass_0.4.9
[94] digest_0.6.37 ggrepel_0.9.6 TH.data_1.1-3
[97] farver_2.1.2 htmlwidgets_1.6.4 htmltools_0.5.8.1
[100] lifecycle_1.0.4 httr_1.4.7 multcompView_0.1-10
[103] mitml_0.4-5 MASS_7.3-64