statsplot
Last updated: 2025-02-04
Checks: 6 1
Knit directory: sapphire/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240923) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/sapphire/data/serum_vit_D_study_with_lab_results.xlsx | data/serum_vit_D_study_with_lab_results.xlsx |
| ~/sapphire/data/sun_expos_data/sun_expos_long.csv | data/sun_expos_data/sun_expos_long.csv |
| ~/sapphire/data/summer_winter.csv | data/summer_winter.csv |
| ~/sapphire/SAfrADMIX/long.csv | SAfrADMIX/long.csv |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5636f2f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Untracked files:
Untracked: .lock_gPLINK
Untracked: .metafile_gPLINK
Untracked: SAfrADMIX/
Untracked: analysis/genotype.R
Untracked: analysis/lilyanalysis.Rmd
Untracked: data/dist_pops.csv
Untracked: data/summer_winter.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/statsplot.Rmd) and HTML
(docs/statsplot.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d9fb6da | Lily Heald | 2025-02-04 | wflow |
| html | d9fb6da | Lily Heald | 2025-02-04 | wflow |
| Rmd | ce202c9 | Lily Heald | 2025-02-04 | create statsplot.Rmd |
file_path <- "~/sapphire/data/serum_vit_D_study_with_lab_results.xlsx"
data_summer <- read_excel(file_path, sheet = "ScreeningDataCollectionSummer")
data_winter <- read_excel(file_path, sheet = "ScreeningDataCollectionWinter")
data_6weeks <- read_excel(file_path, sheet = "ScreeningDataCollection6Weeks")
sun_expos <- read.csv("~/sapphire/data/sun_expos_data/sun_expos_long.csv")
sun_expos_summer <- sun_expos[sun_expos$collection_period == 'Summer', ]
sun_expos_winter <- sun_expos[sun_expos$collection_period == 'Winter', ]
sun_expos_6Weeks <- sun_expos[sun_expos$collection_period == '6Weeks', ]
summer_winter <- read.csv("~/sapphire/data/summer_winter.csv")long_data <- summer_winter %>%
pivot_longer(
cols = -ParticipantCentreID,
names_to = c(".value", "Season"), # Extract variable name and season
names_sep = "_" # Separate at "_"
)
numpairs <- long_data %>%
filter(!is.na(MedianForeheadM)) %>%
group_by(ParticipantCentreID) %>%
filter(all(c('summer', 'winter') %in% Season)) %>%
summarise(count = n_distinct(ParticipantCentreID))
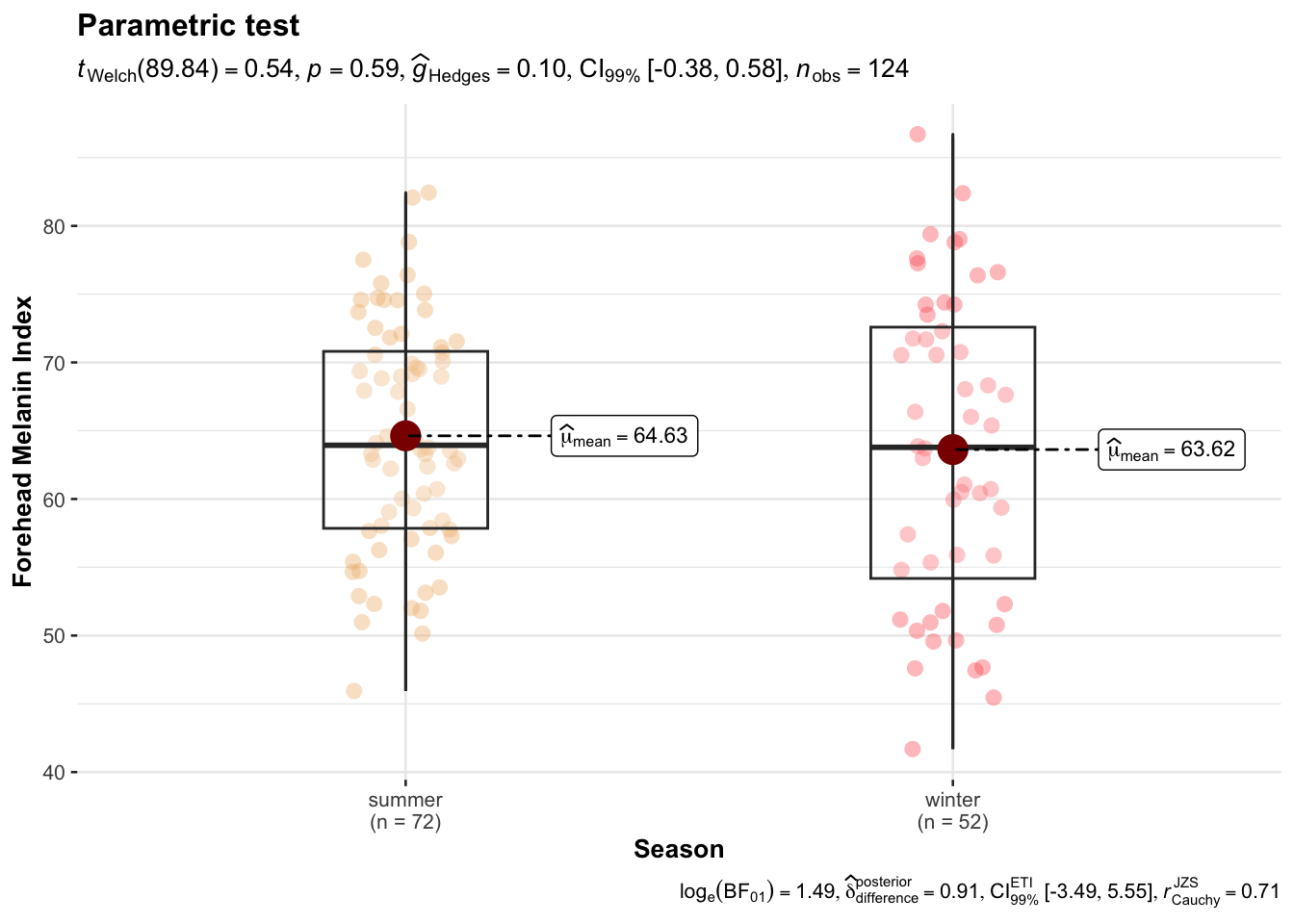
write.csv(long_data, "~/sapphire/SAfrADMIX/long.csv", row.names = F)ggbetweenstats(
data = long_data,
x = Season,
y = MedianForeheadM,
xlab ="Season",
ylab = "Forehead Melanin Index",
violin.args = list(width = 0),
type = "p",
conf.level = 0.99,
title = "Parametric test",
package = "wesanderson",
palette = "GrandBudapest1"
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
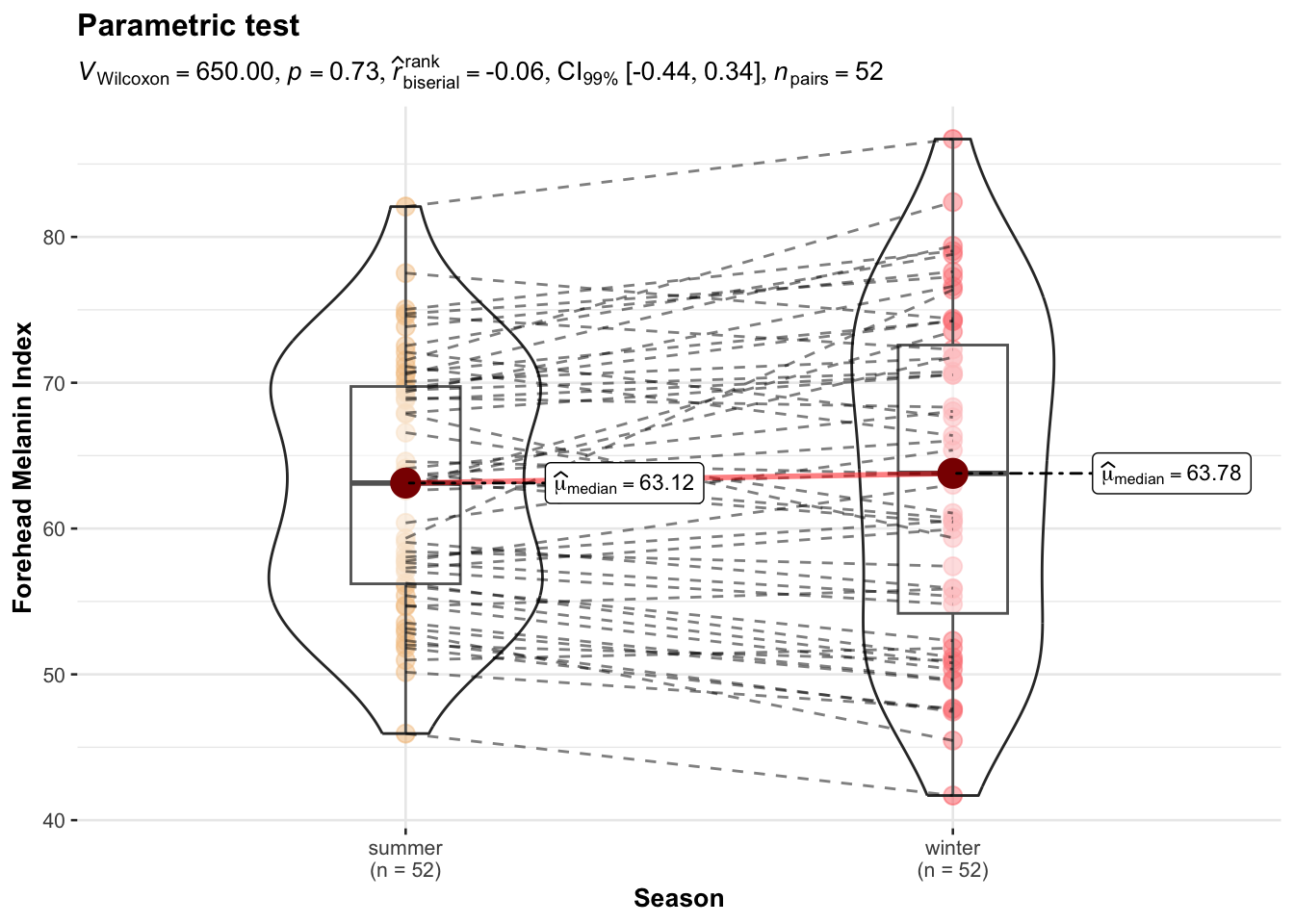
ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadM,
xlab ="Season",
ylab = "Forehead Melanin Index",
type = "np",
conf.level = 0.99,
title = "Parametric test",
package = "wesanderson",
palette = "GrandBudapest1"
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
Goal: winter versus summer t test across all colors
# p1 <- ggwithinstats(
# data = long_data,
# x = Season,
# y = MedianForeheadM,
# type = "p",
# effsize.type = "d",
# conf.level = 0.99,
# title = "Parametric test",
# package = "ggsci",
# palette = "nrc_npg"
# )
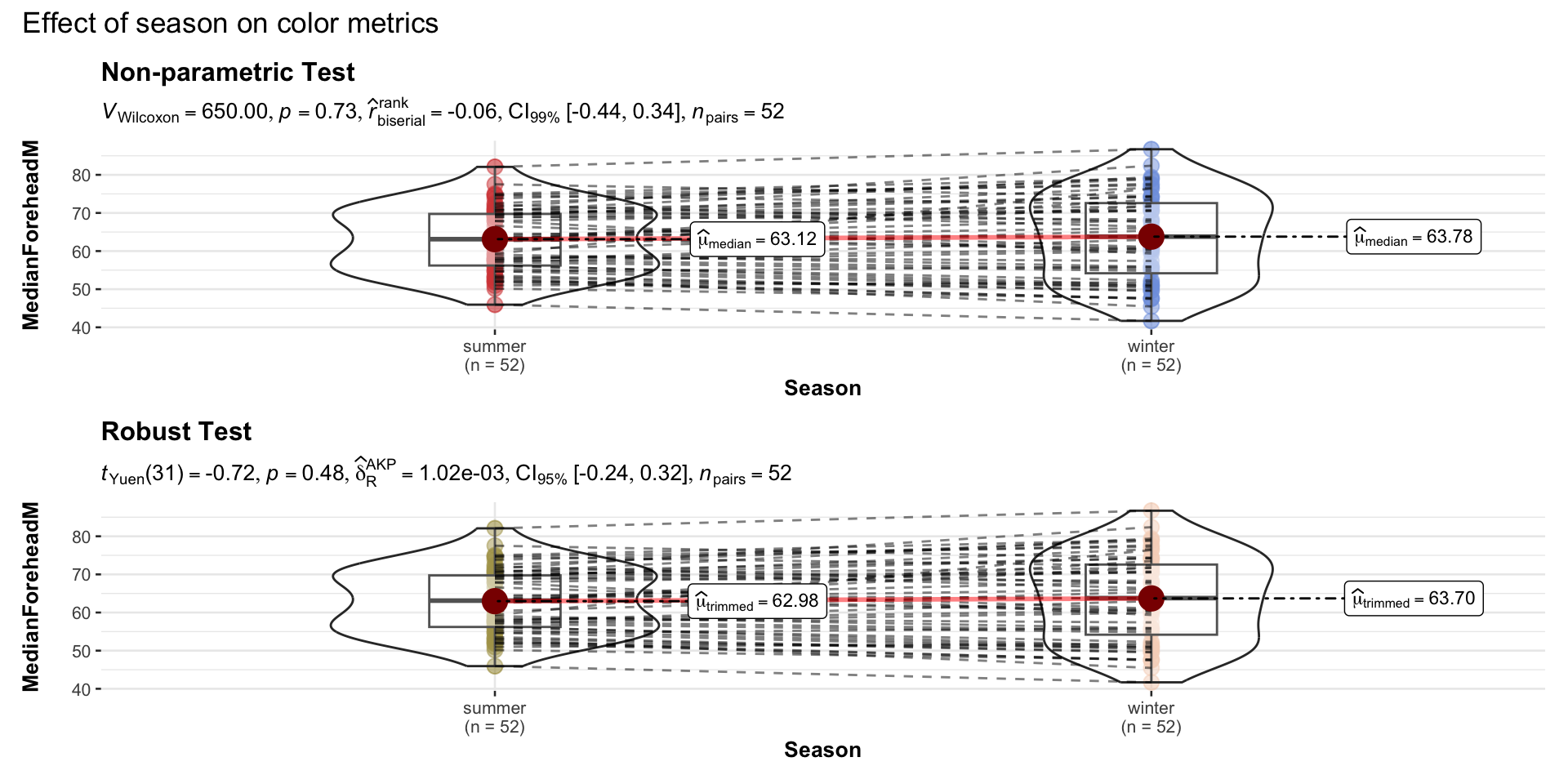
## Mann-Whitney U test (nonparametric test)
p2 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadM,
xlab = "Season",
ylab = "MedianForeheadM
",
type = "np",
conf.level = 0.99,
title = "Non-parametric Test",
package = "ggsci",
palette = "uniform_startrek"
)
## robust t-test
p3 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadM,
xlab = "Season",
ylab = "MedianForeheadM",
type = "r",
conf.level = 0.99,
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
## Bayes Factor for parametric t-test
# p4 <- ggwithinstats(
# data = long_data,
# x = Season,
# y = MedianForeheadM,
# xlab = "Season",
# ylab = "MedianForeheadM",
# type = "bayes",
# title = "Bayesian Test",
# package = "ggsci",
# palette = "nrc_npg"
# )
# combining the individual plots into a single plot
combine_plots(
plotlist = list(p2, p3),
plotgrid.args = list(nrow = 2),
annotation.args = list(
title = "Effect of season on color metrics"),
height = 10, # Increase the height (adjust as needed)
width = 12
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
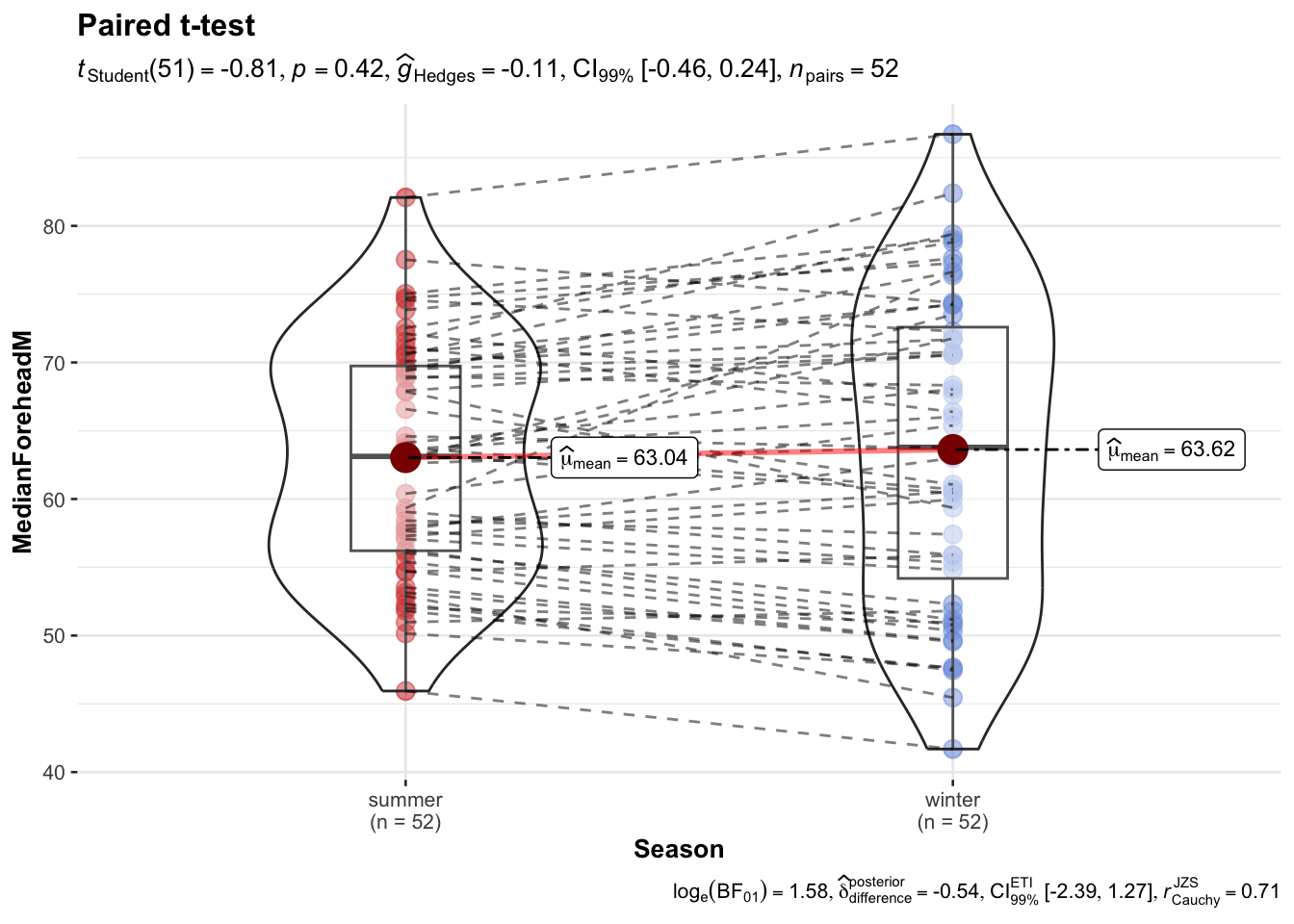
library(ggstatsplot)
ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadM,
paired = TRUE, # must be TRUE for repeated measures
id = "ParticipantCentreID", # ID column to match pairs
type = "parametric", # paired t-test
conf.level = 0.99,
title = "Paired t-test",
xlab = "Season",
ylab = "MedianForeheadM",
package = "ggsci",
palette = "uniform_startrek"
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
p1 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadM,
xlab = "Season",
ylab = "MedianForeheadM",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2",
centrality.label.args = list(size = 5)
)
p2 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadE,
xlab = "Season",
ylab = "MedianForeheadE",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2",
centrality.label.args = list(size = 5)
)
p3 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadL,
xlab = "Season",
ylab = "MedianForeheadL",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2",
centrality.label.args = list(size = 5)
)
p4 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheada,
xlab = "Season",
ylab = "MedianForeheada",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2",
centrality.label.args = list(size = 5)
)
p5 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadb,
xlab = "Season",
ylab = "MedianForeheadb",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2",
centrality.label.args = list(size = 5)
)
p6 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadR,
xlab = "Season",
ylab = "MedianForeheadR",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2",
centrality.label.args = list(size = 5)
)
p7 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadG,
xlab = "Season",
ylab = "MedianForeheadG",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2",
centrality.label.args = list(size = 5)
)
p8 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadB,
xlab = "Season",
ylab = "MedianForeheadB",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2",
centrality.label.args = list(size = 5)
)
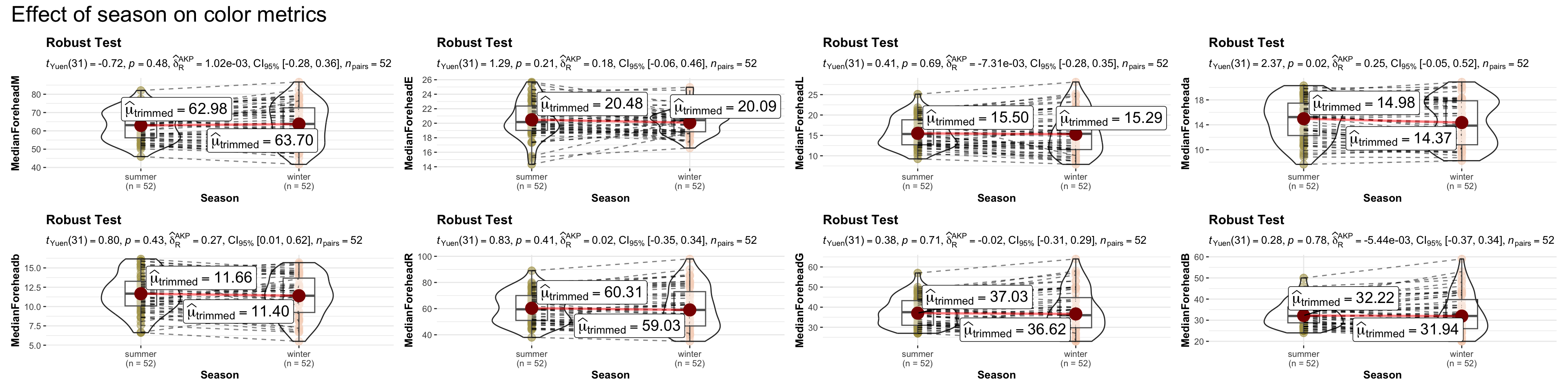
# combining the individual plots into a single plot
combine_plots(
plotlist = list(p1, p2, p3, p4, p5, p6, p7, p8),
plotgrid.args = list(nrow = 2),
annotation.args = list(
title = "Effect of season on color metrics",
theme = ggplot2::theme(
plot.title = element_text(size = 20)
)
)
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
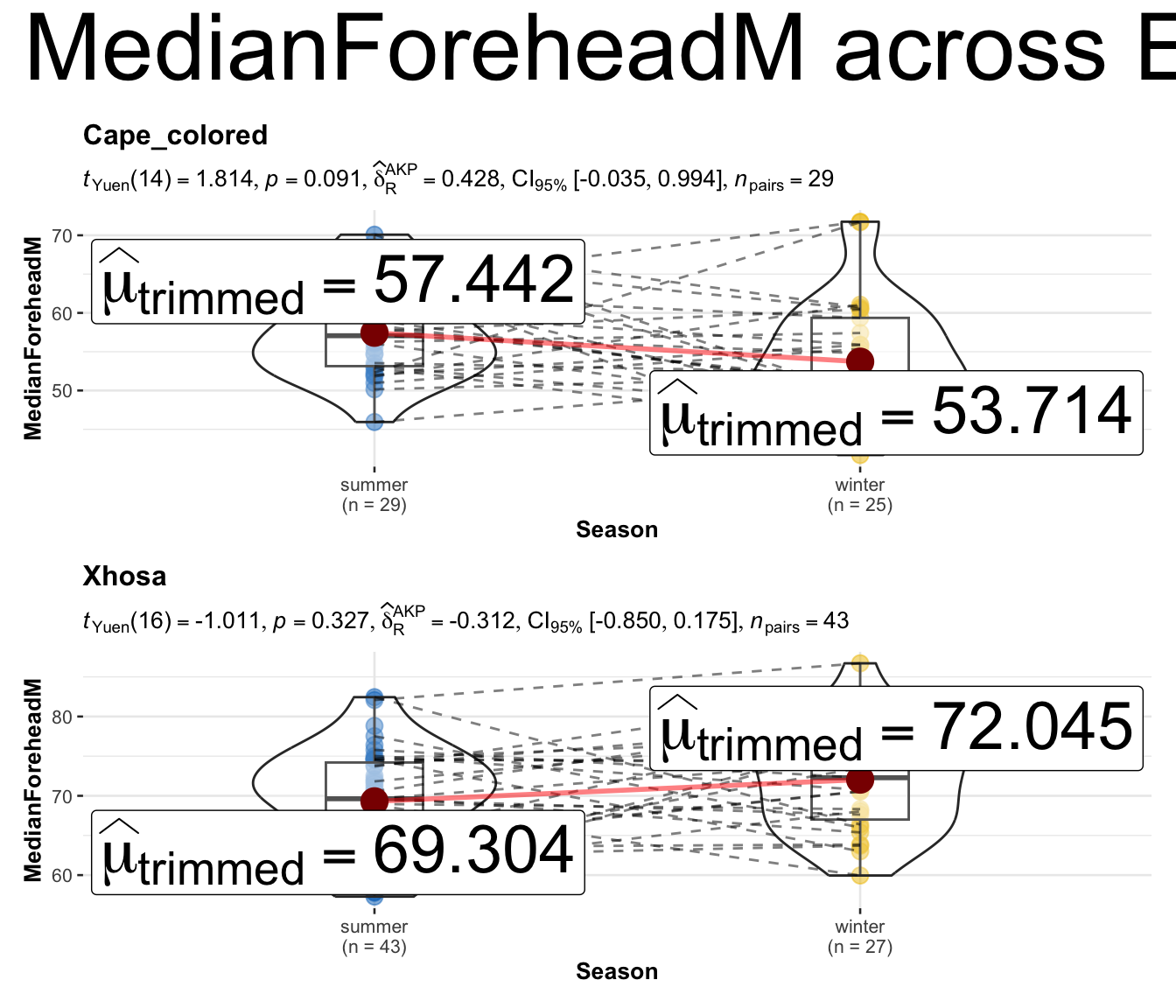
grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = MedianForeheadM,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "MedianForeheadM",
type = "robust", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
centrality.label.args = list(size = 10),
## arguments relevant for combine_plots
annotation.args = list(title = "MedianForeheadM across Ethnicity",
theme = ggplot2::theme(plot.title = element_text(size = 40))),
plotgrid.args = list(ncol = 1)
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
c1 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = MedianForeheadL,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "MedianForeheadL",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "MedianForeheadL across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
c2 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = MedianForeheada,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "MedianForeheada",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "MedianForeheada across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
c3 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = MedianForeheadb,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "MedianForeheadb",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "MedianForeheadb across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
c4 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = InnerArmL,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "InnerArmL",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "InnerArmL across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
c5 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = InnerArma,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "InnerArma",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "InnerArma across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
c6 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = InnerArmb,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "InnerArmb",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "InnerArmb across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
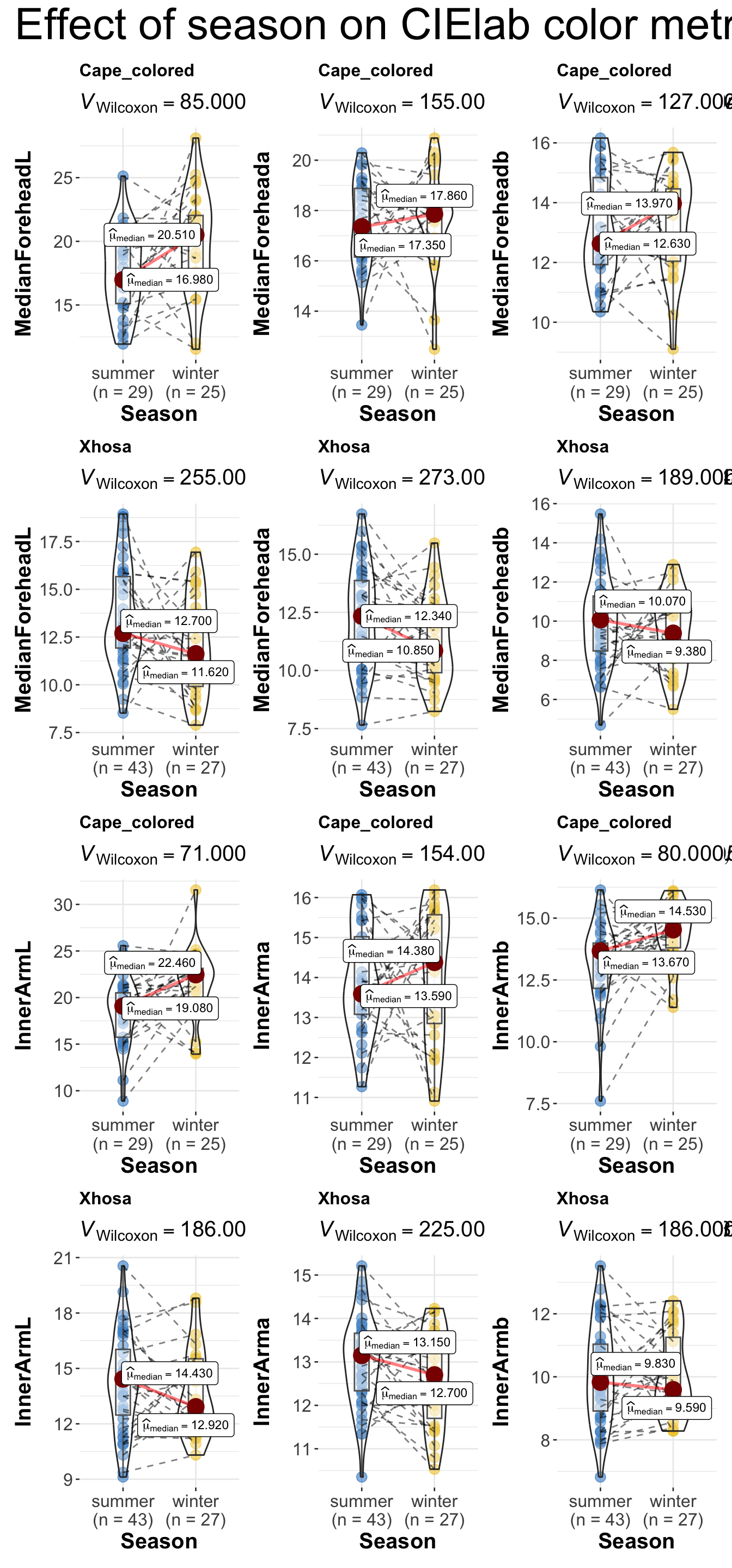
combine_plots(
plotlist = list(c1, c2, c3, c4, c5, c6),
plotgrid.args = list(nrow = 2),
annotation.args = list(
title = "Effect of season on CIElab color metrics across ethnicity",
theme = ggplot2::theme(plot.title = element_text(size = 30)))
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
r1 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = MedianForeheadR,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "MedianForeheadR",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "MedianForeheadR across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
r2 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = MedianForeheadG,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "MedianForeheadG",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "MedianForeheadG across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
r3 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = MedianForeheadB,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "MedianForeheadB",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "MedianForeheadB across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
r4 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = InnerArmR,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "InnerArmR",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "InnerArmR across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
r5 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = InnerArmG,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "InnerArmG",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "InnerArmG across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
r6 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = InnerArmB,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "InnerArmB",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "InnerArmB across Ethnicity"),
plotgrid.args = list(ncol = 1),
ggplot.component = list(theme(text = element_text(size = 15)))
)
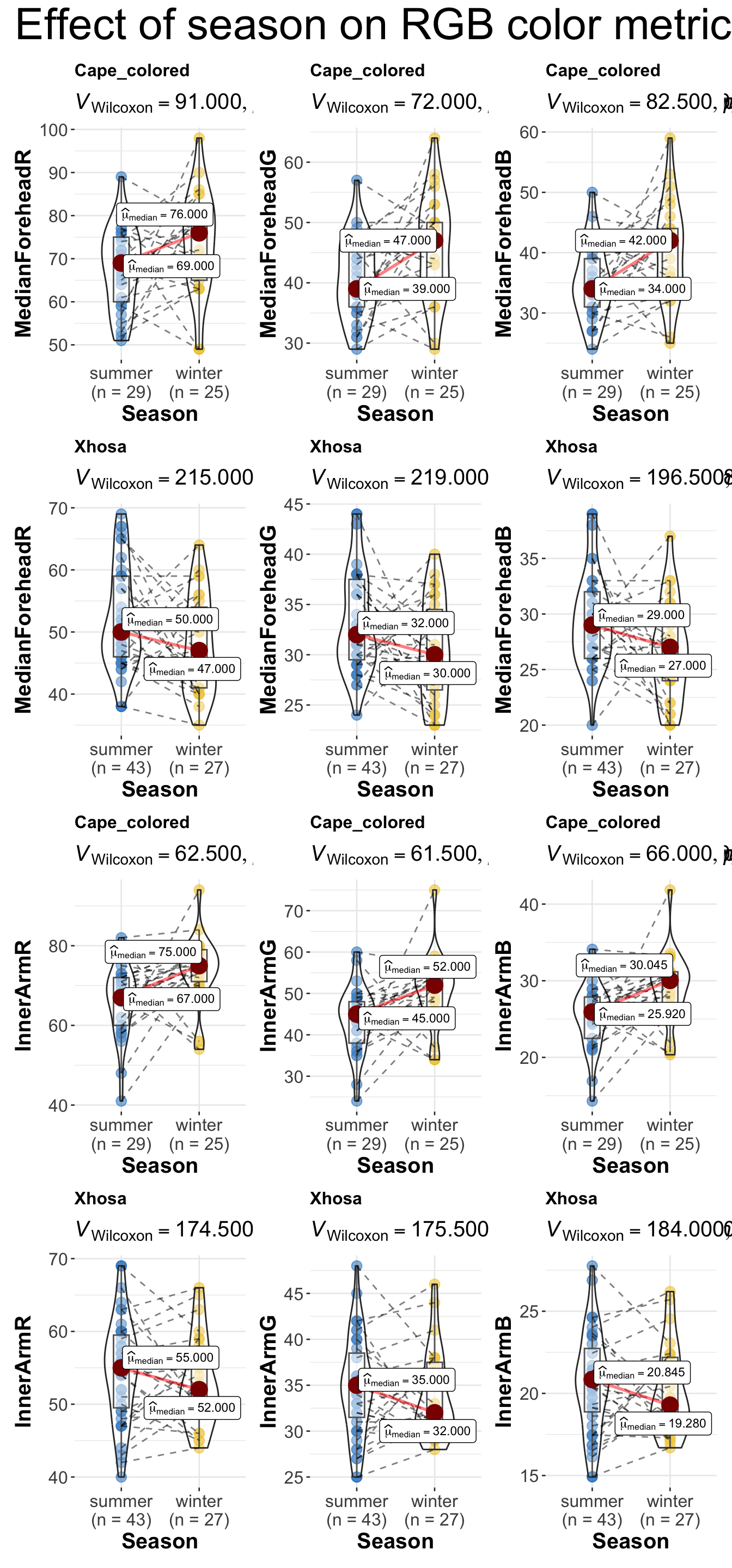
combine_plots(
plotlist = list(r1, r2, r3, r4, r5, r6),
plotgrid.args = list(nrow = 2),
annotation.args = list(
title = "Effect of season on RGB color metrics across ethnicity",
theme = ggplot2::theme(plot.title = element_text(size = 30)))
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
m1 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = MedianForeheadE,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "MedianForeheadE",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "MedianForeheadE across Ethnicity"),
plotgrid.args = list(ncol = 1)
)
m3 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = InnerArmE,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "InnerArmE",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "InnerArmE across Ethnicity"),
plotgrid.args = list(ncol = 1)
)
m2 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = MedianForeheadM,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "MedianForheadM",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "MedianForheadM across Ethnicity"),
plotgrid.args = list(ncol = 1)
)
m4 <- grouped_ggwithinstats(
## arguments relevant for ggwithinstats
data = long_data,
x = Season,
y = InnerArmM,
grouping.var = Ethnicity,
xlab = "Season",
ylab = "InnerArmM",
type = "nonparametric", ## type of test
pairwise.display = "all", ## display only all pairwise comparisons
p.adjust.method = "BH", ## adjust p-values for multiple tests using this method
# ggtheme = ggthemes::theme_tufte(),
package = "ggsci",
palette = "default_jco",
digits = 3,
## arguments relevant for combine_plots
annotation.args = list(title = "InnerArmM across Ethnicity"),
plotgrid.args = list(ncol = 1)
)
combine_plots(
plotlist = list(m1, m2, m3, m4),
plotgrid.args = list(nrow = 2),
annotation.args = list(
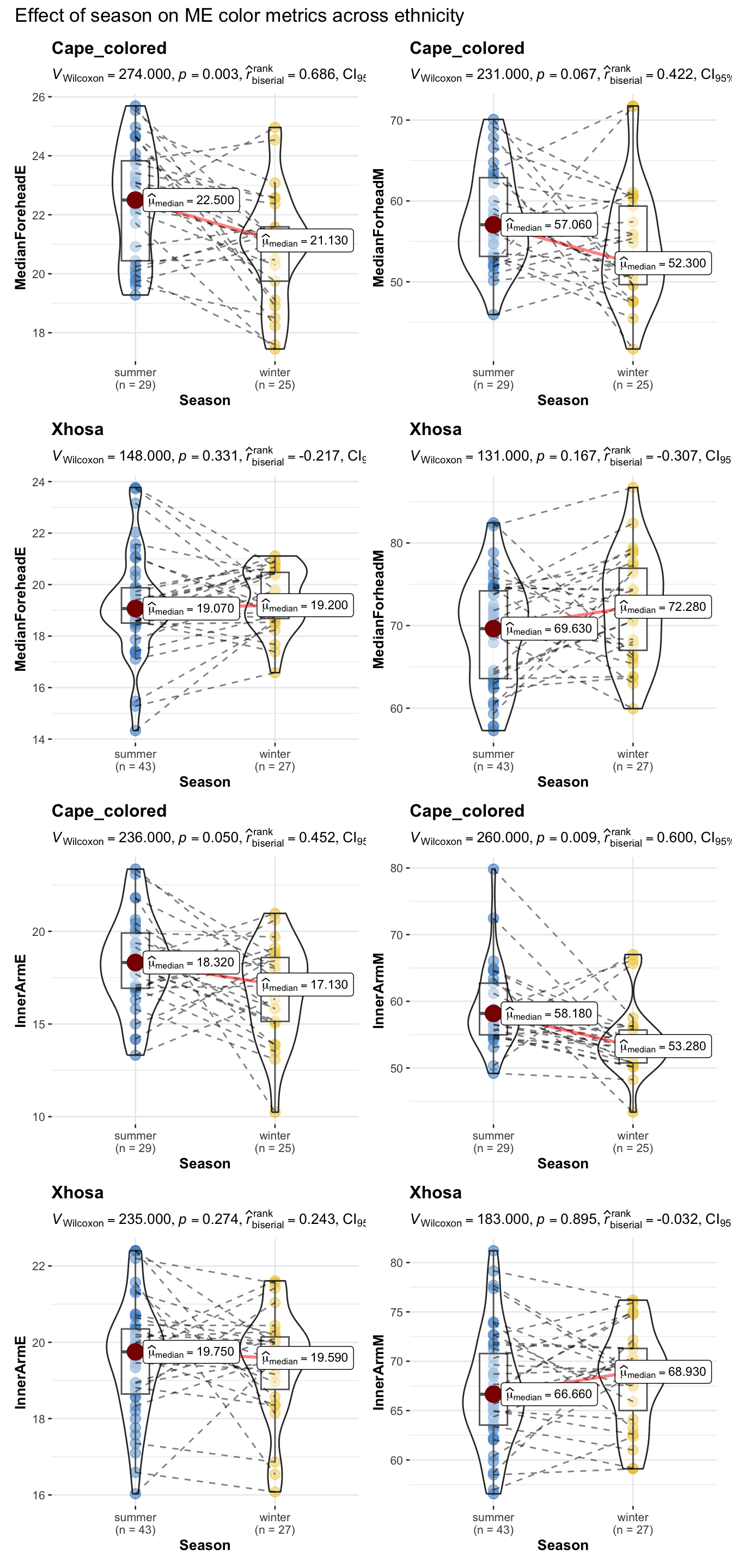
title = "Effect of season on ME color metrics across ethnicity")
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
p1 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadM,
type = "p",
effsize.type = "d",
conf.level = 0.99,
title = "Parametric test",
package = "ggsci",
palette = "nrc_npg"
)
## Mann-Whitney U test (nonparametric test)
p2 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadM,
xlab = "Season",
ylab = "MedianForeheadM
",
type = "np",
conf.level = 0.99,
title = "Non-parametric Test",
package = "ggsci",
palette = "uniform_startrek"
)
## robust t-test
p3 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadM,
xlab = "Season",
ylab = "MedianForeheadM",
type = "r",
conf.level = 0.99,
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
## Bayes Factor for parametric t-test
p4 <- ggwithinstats(
data = long_data,
x = Season,
y = MedianForeheadM,
xlab = "Season",
ylab = "MedianForeheadM",
type = "bayes",
title = "Bayesian Test",
package = "ggsci",
palette = "nrc_npg"
)
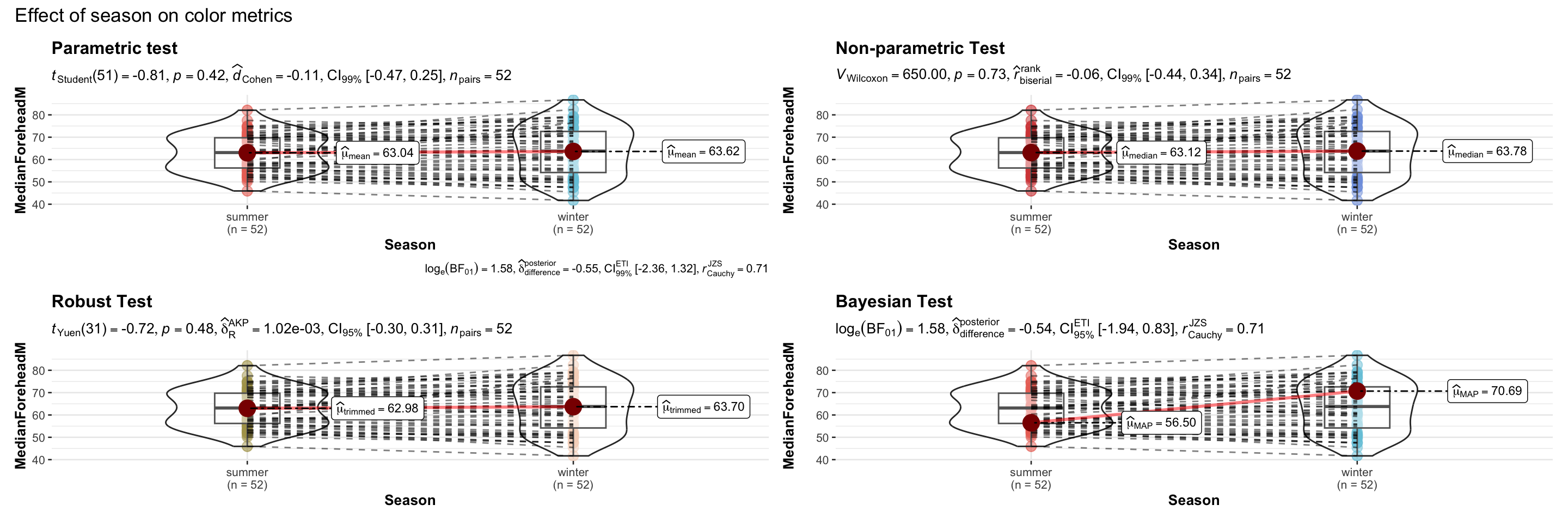
# combining the individual plots into a single plot
combine_plots(
plotlist = list(p1, p2, p3, p4),
plotgrid.args = list(nrow = 2),
annotation.args = list(
title = "Effect of season on color metrics"),
height = 10, # Increase the height (adjust as needed)
width = 12
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
e1 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = MedianForeheadM_summer,
xlab = "Ethnicity",
ylab = "MedianForeheadM",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e2 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = MedianForeheadE_summer,
xlab = "Ethnicity",
ylab = "MedianForeheadE",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e3 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = MedianForeheadL_summer,
xlab = "Ethnicity",
ylab = "MedianForeheadL",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e4 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = MedianForeheada_summer,
xlab = "Ethnicity",
ylab = "MedianForeheada",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e5 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = MedianForeheadb_summer,
xlab = "Ethnicity",
ylab = "MedianForeheadb",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e6 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = MedianForeheadR_summer,
xlab = "Ethnicity",
ylab = "MedianForeheadR",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e7 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = MedianForeheadG_summer,
xlab = "Ethnicity",
ylab = "MedianForeheadG",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e8 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = MedianForeheadB_summer,
xlab = "Ethnicity",
ylab = "MedianForeheadB",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
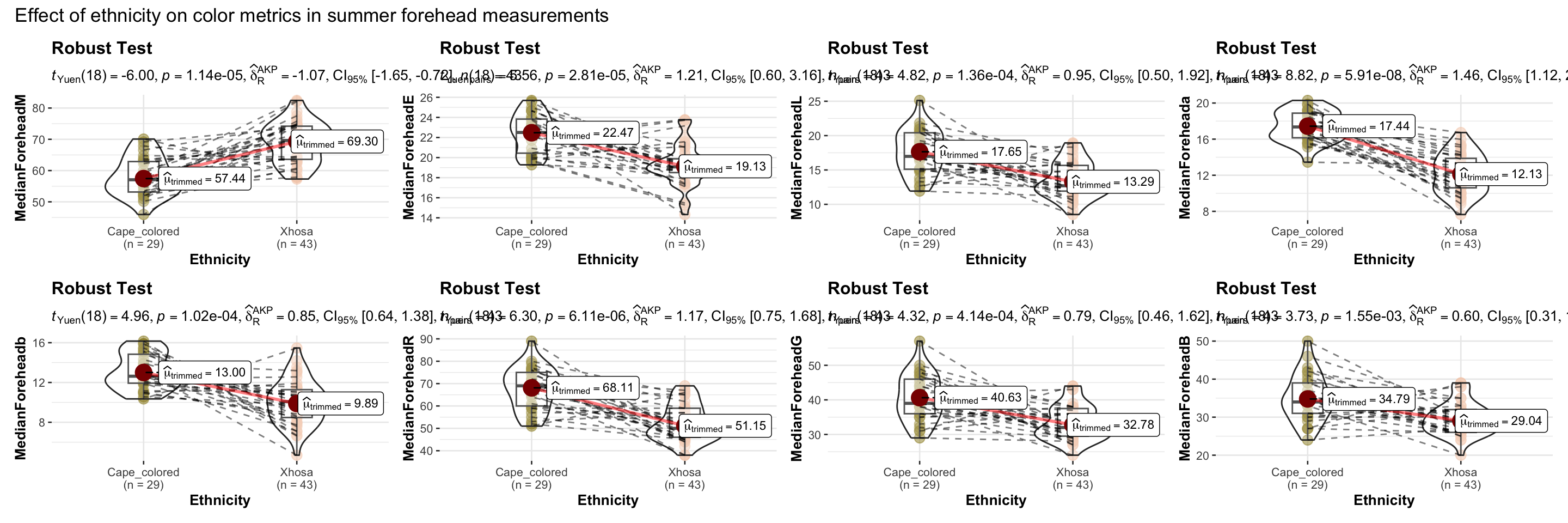
# combining the individual plots into a single plot
combine_plots(
plotlist = list(e1, e2, e3, e4, e5, e6, e7, e8),
plotgrid.args = list(nrow = 2),
annotation.args = list(
title = "Effect of ethnicity on color metrics in summer forehead measurements"),
height = 10, # Increase the height (adjust as needed)
width = 12
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
e1 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = MedianForeheadM_winter,
xlab = "Ethnicity",
ylab = "MedianForeheadM",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e2 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = MedianForeheadE_winter,
xlab = "Ethnicity",
ylab = "MedianForeheadE",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e3 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = MedianForeheadL_winter,
xlab = "Ethnicity",
ylab = "MedianForeheadL",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e4 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = MedianForeheada_winter,
xlab = "Ethnicity",
ylab = "MedianForeheada",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e5 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = MedianForeheadb_winter,
xlab = "Ethnicity",
ylab = "MedianForeheadb",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e6 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = MedianForeheadR_winter,
xlab = "Ethnicity",
ylab = "MedianForeheadR",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e7 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = MedianForeheadG_winter,
xlab = "Ethnicity",
ylab = "MedianForeheadG",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e8 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = MedianForeheadB_winter,
xlab = "Ethnicity",
ylab = "MedianForeheadB",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
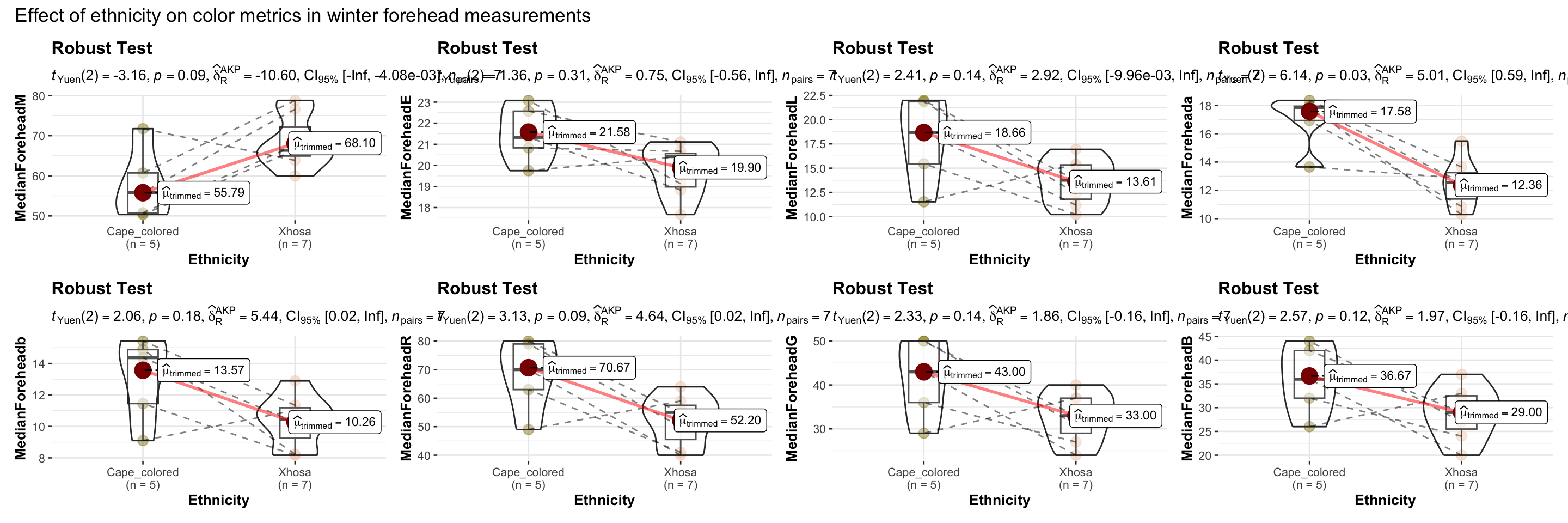
# combining the individual plots into a single plot
combine_plots(
plotlist = list(e1, e2, e3, e4, e5, e6, e7, e8),
plotgrid.args = list(nrow = 2),
annotation.args = list(
title = "Effect of ethnicity on color metrics in winter forehead measurements"),
height = 10, # Increase the height (adjust as needed)
width = 12
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
e1 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = InnerArmM_summer,
xlab = "Ethnicity",
ylab = "InnerArmM",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e2 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = InnerArmE_summer,
xlab = "Ethnicity",
ylab = "InnerArmE",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e3 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = InnerArmL_summer,
xlab = "Ethnicity",
ylab = "InnerArmL",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e4 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = InnerArma_summer,
xlab = "Ethnicity",
ylab = "InnerArma",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e5 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = InnerArmb_summer,
xlab = "Ethnicity",
ylab = "InnerArmb",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e6 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = InnerArmR_summer,
xlab = "Ethnicity",
ylab = "InnerArmR",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e7 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = InnerArmG_summer,
xlab = "Ethnicity",
ylab = "InnerArmG",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e8 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_summer,
y = InnerArmB_summer,
xlab = "Ethnicity",
ylab = "InnerArmB",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
# combining the individual plots into a single plot
combine_plots(
plotlist = list(e1, e2, e3, e4, e5, e6, e7, e8),
plotgrid.args = list(nrow = 2),
annotation.args = list(
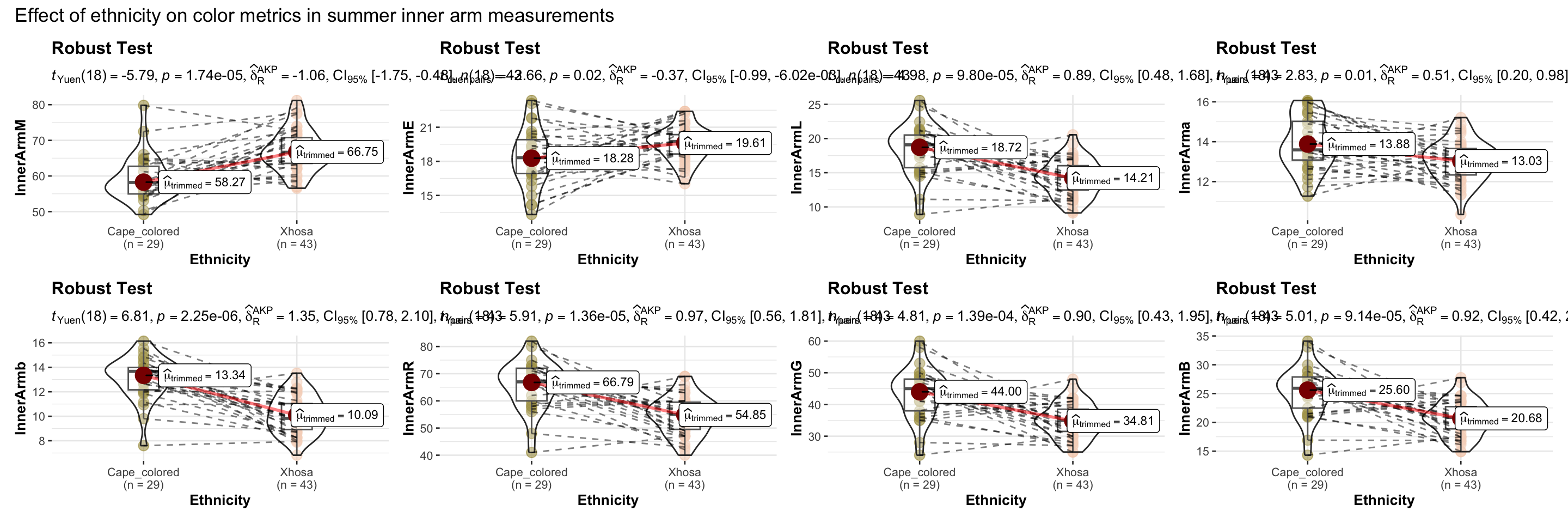
title = "Effect of ethnicity on color metrics in summer inner arm measurements"),
height = 10, # Increase the height (adjust as needed)
width = 12
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
e1 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = InnerArmM_winter,
xlab = "Ethnicity",
ylab = "InnerArmM",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e2 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = InnerArmE_winter,
xlab = "Ethnicity",
ylab = "InnerArmE",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e3 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = InnerArmL_winter,
xlab = "Ethnicity",
ylab = "InnerArmL",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e4 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = InnerArma_winter,
xlab = "Ethnicity",
ylab = "InnerArma",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e5 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = InnerArmb_winter,
xlab = "Ethnicity",
ylab = "InnerArmb",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e6 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = InnerArmR_winter,
xlab = "Ethnicity",
ylab = "InnerArmR",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e7 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = InnerArmG_winter,
xlab = "Ethnicity",
ylab = "InnerArmG",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
e8 <- ggwithinstats(
data = summer_winter,
x = Ethnicity_winter,
y = InnerArmB_winter,
xlab = "Ethnicity",
ylab = "InnerArmB",
type = "r",
title = "Robust Test",
package = "wesanderson",
palette = "Royal2"
)
# combining the individual plots into a single plot
combine_plots(
plotlist = list(e1, e2, e3, e4, e5, e6, e7, e8),
plotgrid.args = list(nrow = 2),
annotation.args = list(
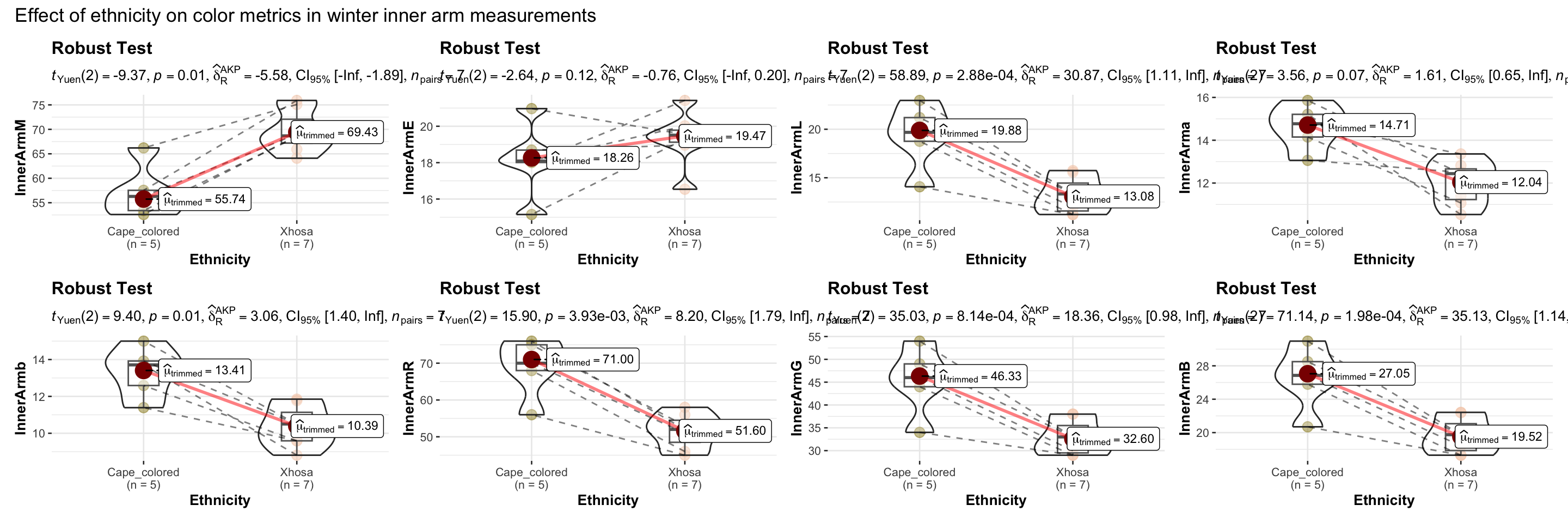
title = "Effect of ethnicity on color metrics in winter inner arm measurements"),
height = 10, # Increase the height (adjust as needed)
width = 12
)
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
linear mixed effects model
fixed effects: we expect will have an effect on the response variable
random effects: trying to control - season
Y_{ijk} = _0 + 1 + (1|) + {ijk}
messyjoin <- left_join(data_summer, data_winter,
by = "ParticipantCentreID",
suffix = c("_summer", "_winter"))
messyjoin <- messyjoin %>%
select(matches("SkinReflectance|ParticipantCentreID|VitD"))
names(messyjoin) <- gsub("\\\\", "", names(messyjoin))
names(messyjoin) <- gsub("\\*", "", names(messyjoin))
df_long <- messyjoin %>%
pivot_longer(
cols = -ParticipantCentreID,
names_to = c("Site", "Metric", "Replicate", "Season"),
names_pattern = "SkinReflectance(.*)([EMRGBLab])(\\d+)_(.*)",
values_to = "Value"
)
df_long <- df_long %>%
mutate(Ethnicity = case_when(
grepl("^VDTG", ParticipantCentreID) ~ "CM",
grepl("^VDKH", ParticipantCentreID) ~ "XH",
TRUE ~ NA))df_long$Season <- as.factor (df_long$Season)
df_long$Ethnicity <- as.factor (df_long$Ethnicity)
head(df_long)# A tibble: 6 × 7
ParticipantCentreID Site Metric Replicate Season Value Ethnicity
<chr> <chr> <chr> <chr> <fct> <dbl> <fct>
1 VDKH001 Forehead E 1 summer 17.0 XH
2 VDKH001 Forehead E 2 summer 18.7 XH
3 VDKH001 Forehead E 3 summer 18.6 XH
4 VDKH001 Forehead M 1 summer 56.0 XH
5 VDKH001 Forehead M 2 summer 70.6 XH
6 VDKH001 Forehead M 3 summer 76.3 XH melanin <- df_long[df_long$Metric == "M",]
erythema <- df_long[df_long$Metric == "E",]
library(lme4)
melanin.mixed.lmer <- lmer(Value ~ Ethnicity + Season + (1|ParticipantCentreID/Site) + (1|Replicate), data = melanin)boundary (singular) fit: see help('isSingular')melanin.reduced.lmer <- lmer(Value ~ 1 + (1|Season) + (1|ParticipantCentreID/Site) + (1|Replicate), data = melanin)boundary (singular) fit: see help('isSingular')a1 <- anova(melanin.reduced.lmer, melanin.mixed.lmer)refitting model(s) with ML (instead of REML)erythema.mixed.lmer <- lmer(Value ~ Ethnicity + Season + (1|ParticipantCentreID/Site) + (1|Replicate), data = erythema)boundary (singular) fit: see help('isSingular')erythema.reduced.lmer <- lmer(Value ~ 1 + (1|Season) + (1|ParticipantCentreID/Site) + (1|Replicate), data = erythema)boundary (singular) fit: see help('isSingular')a2 <- anova(erythema.reduced.lmer, erythema.mixed.lmer)refitting model(s) with ML (instead of REML)a1_new <- data.frame(a1)
a2_new <- data.frame(a2)
anova_results <- data.frame(cbind(c("Ethnicity", "Residuals", "Ethnicity", "Residuals"),
rbind(a1_new, a2_new)))
colnames(anova_results) <- c("","npar","AIC","BIC","logLik","deviance","Chisq","Df","Pr(>Chisq)" )
row.names(anova_results) <- NULL
library(kableExtra)
Attaching package: 'kableExtra'The following object is masked from 'package:dplyr':
group_rowsanova_results %>% kable("html", digits=2) %>%
kable_styling(bootstrap_options = "striped", full_width = F) %>%
pack_rows(., "Melanin", 1, 2) %>% # groups rows with label
pack_rows(., "Erythema", 3, 4) # groups rows with label| npar | AIC | BIC | logLik | deviance | Chisq | Df | Pr(>Chisq) | |
|---|---|---|---|---|---|---|---|---|
| Melanin | ||||||||
| Ethnicity | 6 | 14911.18 | 14943.75 | -7449.59 | 14899.18 | NA | NA | NA |
| Residuals | 7 | 14896.94 | 14934.94 | -7441.47 | 14882.94 | 16.24 | 1 | 0 |
| Erythema | ||||||||
| Ethnicity | 6 | 8148.99 | 8181.56 | -4068.49 | 8136.99 | NA | NA | NA |
| Residuals | 7 | 8124.75 | 8162.74 | -4055.37 | 8110.75 | 26.24 | 1 | 0 |
melanin1 <- stats::lm(formula = Value ~ Ethnicity + Season, data = melanin)
combine_plots(
plotlist = list(
ggcoefstats(melanin1) +
ggplot2::labs(x = parse(text = "'regression coefficient' ~italic(beta)")),
ggcoefstats(melanin.mixed.lmer) +
ggplot2::labs(
x = parse(text = "'regression coefficient' ~italic(beta)"),
y = "fixed effects"
)
),
plotgrid.args = list(nrow = 2),
annotation.args = list(title = "Relationship between MI value and ethnicity")
)Random effect variances not available. Returned R2 does not account for random effects.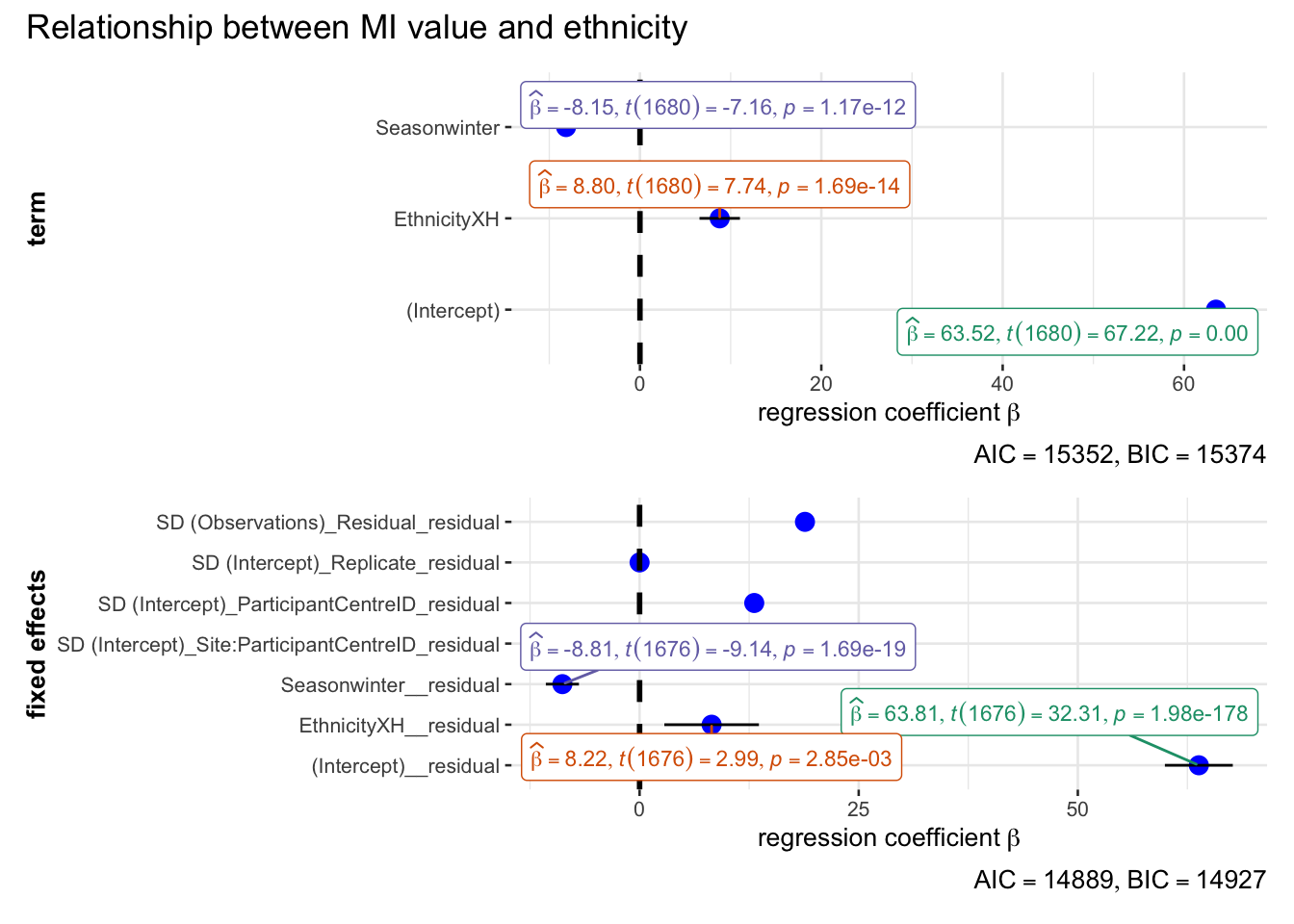
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
erythema1 <- stats::lm(formula = Value ~ Ethnicity + Season, data = erythema)
combine_plots(
plotlist = list(
ggcoefstats(erythema1) +
ggplot2::labs(x = parse(text = "'regression coefficient' ~italic(beta)")),
ggcoefstats(erythema.mixed.lmer) +
ggplot2::labs(
x = parse(text = "'regression coefficient' ~italic(beta)"),
y = "fixed effects"
)
),
plotgrid.args = list(nrow = 2),
annotation.args = list(title = "Relationship between EI value and ethnicity")
)Random effect variances not available. Returned R2 does not account for random effects.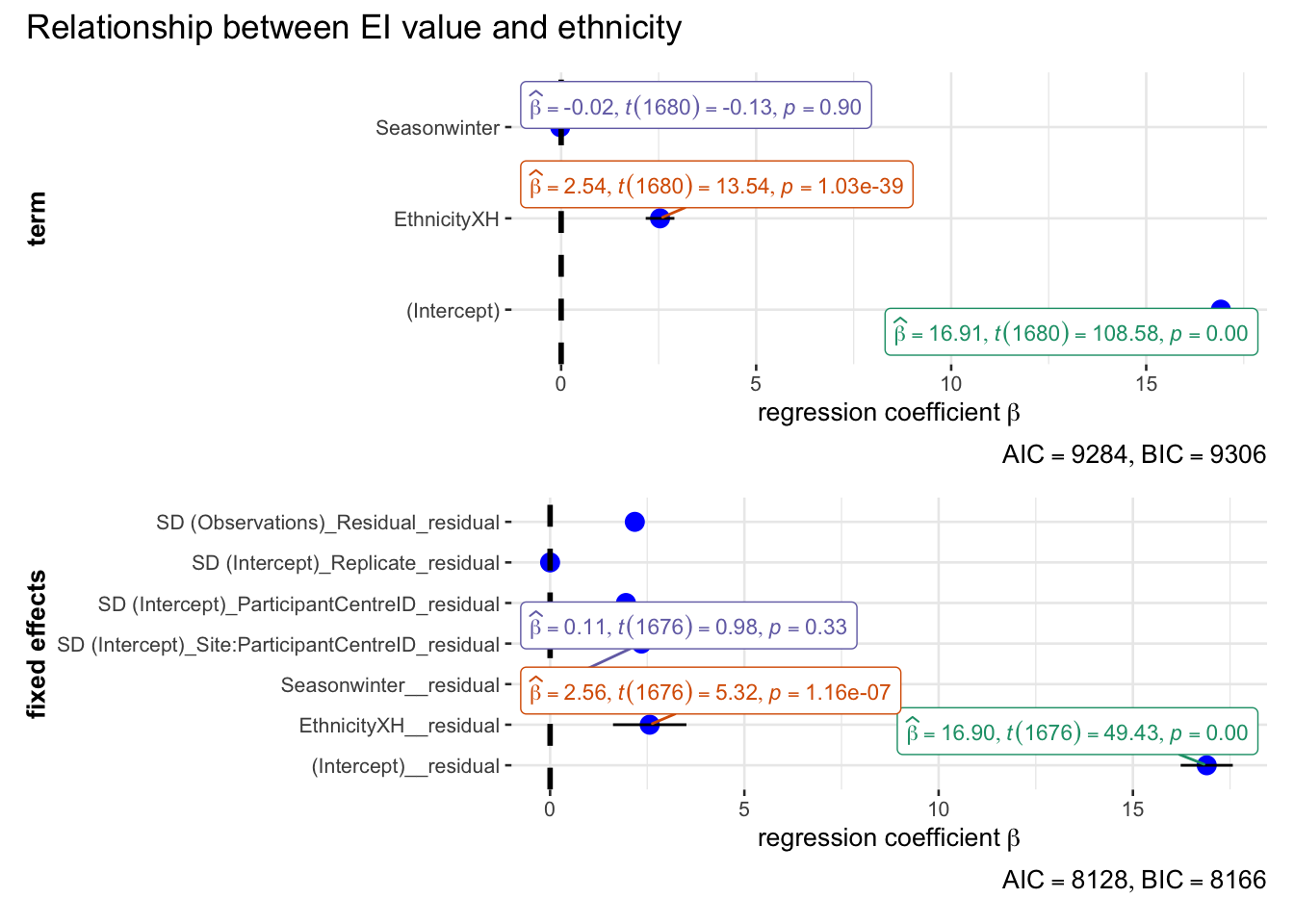
| Version | Author | Date |
|---|---|---|
| d9fb6da | Lily Heald | 2025-02-04 |
sessionInfo()R version 4.4.2 (2024-10-31)
Platform: aarch64-apple-darwin20
Running under: macOS Monterey 12.5.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/Detroit
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] kableExtra_1.4.0 ggstatsplot_0.13.0 lme4_1.1-36 Matrix_1.7-1
[5] rstatix_0.7.2 ggpubr_0.6.0 ggfortify_0.4.17 wesanderson_0.3.7
[9] missMDA_1.19 FactoMineR_2.11 factoextra_1.0.7 ggcorrplot_0.1.4.1
[13] corrr_0.4.4 readxl_1.4.3 lubridate_1.9.4 forcats_1.0.0
[17] stringr_1.5.1 purrr_1.0.2 tibble_3.2.1 ggplot2_3.5.1
[21] tidyverse_2.0.0 tidyr_1.3.1 dplyr_1.1.4 readr_2.1.5
[25] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] rstudioapi_0.17.1 jsonlite_1.8.9 shape_1.4.6.1
[4] correlation_0.8.6 datawizard_1.0.0 magrittr_2.0.3
[7] TH.data_1.1-3 estimability_1.5.1 jomo_2.7-6
[10] farver_2.1.2 nloptr_2.1.1 rmarkdown_2.29
[13] fs_1.6.5 vctrs_0.6.5 minqa_1.2.8
[16] paletteer_1.6.0 effectsize_1.0.0 htmltools_0.5.8.1
[19] broom_1.0.7 cellranger_1.1.0 Formula_1.2-5
[22] mitml_0.4-5 sass_0.4.9 bslib_0.8.0
[25] htmlwidgets_1.6.4 plyr_1.8.9 sandwich_3.1-1
[28] emmeans_1.10.6 zoo_1.8-12 cachem_1.1.0
[31] whisker_0.4.1 lifecycle_1.0.4 iterators_1.0.14
[34] pkgconfig_2.0.3 R6_2.5.1 fastmap_1.2.0
[37] rbibutils_2.3 BayesFactor_0.9.12-4.7 digest_0.6.37
[40] reshape_0.8.9 colorspace_2.1-1 rematch2_2.1.2
[43] patchwork_1.3.0 ps_1.8.1 rprojroot_2.0.4
[46] prismatic_1.1.2 labeling_0.4.3 WRS2_1.1-6
[49] timechange_0.3.0 httr_1.4.7 abind_1.4-8
[52] compiler_4.4.2 withr_3.0.2 doParallel_1.0.17
[55] backports_1.5.0 carData_3.0-5 performance_0.13.0
[58] ggsignif_0.6.4 pan_1.9 MASS_7.3-64
[61] scatterplot3d_0.3-44 flashClust_1.01-2 tools_4.4.2
[64] httpuv_1.6.15 statsExpressions_1.6.2 nnet_7.3-20
[67] glue_1.8.0 callr_3.7.6 nlme_3.1-166
[70] promises_1.3.2 grid_4.4.2 getPass_0.2-4
[73] cluster_2.1.8 generics_0.1.3 gtable_0.3.6
[76] tzdb_0.4.0 hms_1.1.3 xml2_1.3.6
[79] utf8_1.2.4 car_3.1-3 ggrepel_0.9.6
[82] foreach_1.5.2 pillar_1.10.1 later_1.4.1
[85] splines_4.4.2 lattice_0.22-6 survival_3.8-3
[88] tidyselect_1.2.1 pbapply_1.7-2 knitr_1.49
[91] git2r_0.35.0 reformulas_0.4.0 gridExtra_2.3
[94] svglite_2.1.3 xfun_0.50 DT_0.33
[97] stringi_1.8.4 yaml_2.3.10 boot_1.3-31
[100] evaluate_1.0.3 codetools_0.2-20 multcompView_0.1-10
[103] cli_3.6.3 RcppParallel_5.1.10 rpart_4.1.24
[106] systemfonts_1.2.0 xtable_1.8-4 parameters_0.24.1
[109] Rdpack_2.6.2 munsell_0.5.1 processx_3.8.5
[112] jquerylib_0.1.4 Rcpp_1.0.14 zeallot_0.1.0
[115] coda_0.19-4.1 parallel_4.4.2 MatrixModels_0.5-3
[118] rstantools_2.4.0 leaps_3.2 bayestestR_0.15.0
[121] glmnet_4.1-8 viridisLite_0.4.2 mvtnorm_1.3-3
[124] scales_1.3.0 insight_1.0.1 rlang_1.1.4
[127] multcomp_1.4-28 mice_3.17.0