Log2021
Last updated: 2021-01-04
Checks: 7 0
Knit directory: GradLog/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20201014) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 361c43e. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: code/.DS_Store
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Unstaged changes:
Deleted: analysis/Log.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Log2021.Rmd) and HTML (docs/Log2021.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 23d78aa | Lili Wang | 2021-01-04 | wflow_git_commit(c("analysis/_site.yml“,”analysis/index.Rmd", |
Jan 05
eQTLGen
eQTLGen description
This full dataset includes 19942 genes that showed expression in blood tested and 10317 SNPs that are trait-associated SNPs based on GWAS Catalog.
After gene filter steps described in
Dec 10, there are 4963 genes left. Applying the same filtering to 13634 DGN genes, there are 3695 genes left, among which 3642 are also included in eQTLGen. So, I will use these genes to do the downstream analysis, e.g. constructing co-expressed gene modules. These 3642 genes result in 19 gene modules.Replication of DGN signals in eQTLGen
The table below gives the signals found in eQTLGen and DGN.
The first two rows give results based on eQTLGen zscores, with row 1 using
qvaluefor FDR correction (threshold \(0.05\)) and row 2 using \(\frac{0.05}{\#DGN signals}\) as significance threshold. The third row is based on the same gene modules and SNPs but tensorQTL zscores using DGN expression data. The FDR correction uses the empirical distribution of pvalues from the combined chr’s and modules (10 permutations).
| Dataset | FDR | minp | (QTL, module) | unique QTL | independent QTL |
|---|---|---|---|---|---|
| eQTLGen | qvalue | 5.82e-04 | 2195 | 909 | 348 |
| eQTLGen | 0.05/#DGN signals | 1.18e-04 | 1707 | 762 | 286 |
| eQTLGen_DGN | combined chr+module #10-perms | 1.00e-07 | 420 | 374 | 28 |
Among 374 eQTLGen_DGN signals, 16 are replicated in 762 eQTLGen signals. These 16 replicated signals consists of 6 independent SNPs, including (based on GRCh37),
- rs12485738 : 3:56865776, intron variant of ARHGEF3.
- rs643381 : 6:139839423. (rs590856, 6:139844429).
- rs149007767 7:50370254: , intron variant of IKZF1.
- rs12718597 : 7:50428428, intron variant of IKZF1.
- rs35979828 : 12:54685880, 500B downstream variant of NFE2.
- rs7210990 : 17:16170764, intron variant of PIGL.
GTEx
Results
The table below summarizes all results I have so far. (The dataset “DGN_new” represents DGN through the standard filtering (see “Gene filter” on Dec 10).)
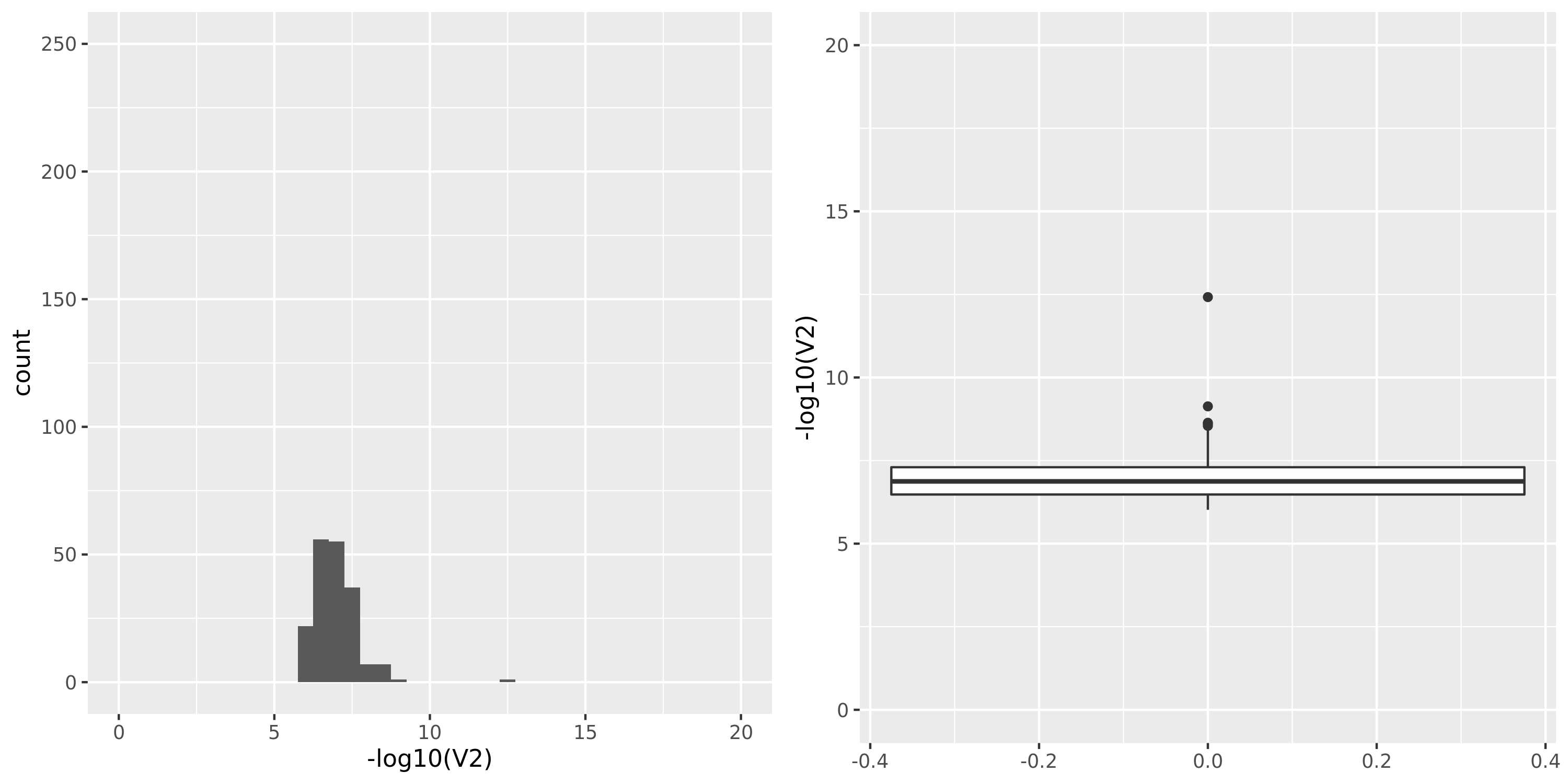
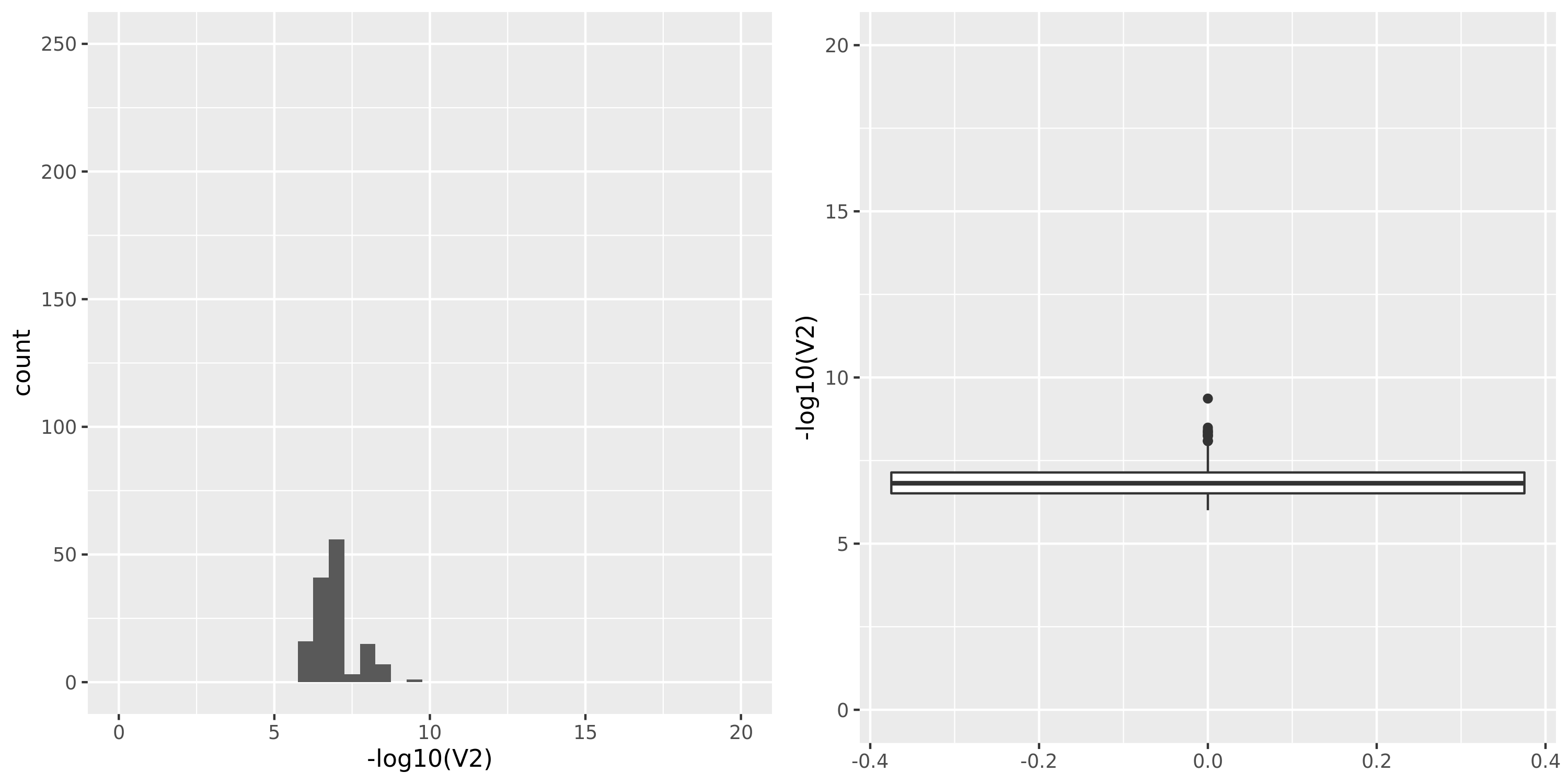
The following figures give the distributions of pvalues in various datasets.
| Dataset | Nsample | All | Annotated | Filtered | Filtered_info | Final | Nmodule | FDR | Nperm | QTL_Module | unique_QTL | independent_QTL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DGN | 913 | 13634 | 11979 | 8120 | 0;165;1663;8073 | 3859 | 21 | combined chr+module | 20 | 659 | 623 | 40 |
| DGN_new | 913 | 13634 | 12585 | 9939 | 1;453;1798;8705 | 3695 | 19 | combined chr+module | 10 | 331 | 275 | 26 |
| TCGA | 788 | 17656 | 15994 | 12694 | 336;3504;10697;598 | 4962 | ||||||
| GTEx_v8.Whole_Blood | 670 | 20315 | 20315 | 15245 | 619;4544;3677;13096 | 5070 | 22 | combined chr+module | 10 | 4 | 4 | 2 |
| GTEx_v8.Muscle_Skeletal | 706 | 21031 | 21031 | 15601 | 675;4447;3631;13490 | 5430 | 18 | combined chr+module | 10 | 38 | 38 | 3 |
| GTEx_v8.Skin_Sun_Exposed_Lower_leg | 605 | 25196 | 25196 | 18801 | 807;6458;4557;15650 | 6395 | 30 | combined chr+module | 10 | 1 | 1 | 1 |
| GTEx_v8.Artery_Tibial | 584 | 23304 | 23304 | 17390 | 795;5588;4202;14730 | 5914 | 20 | combined chr+module | 10 | 0 | 0 | 0 |
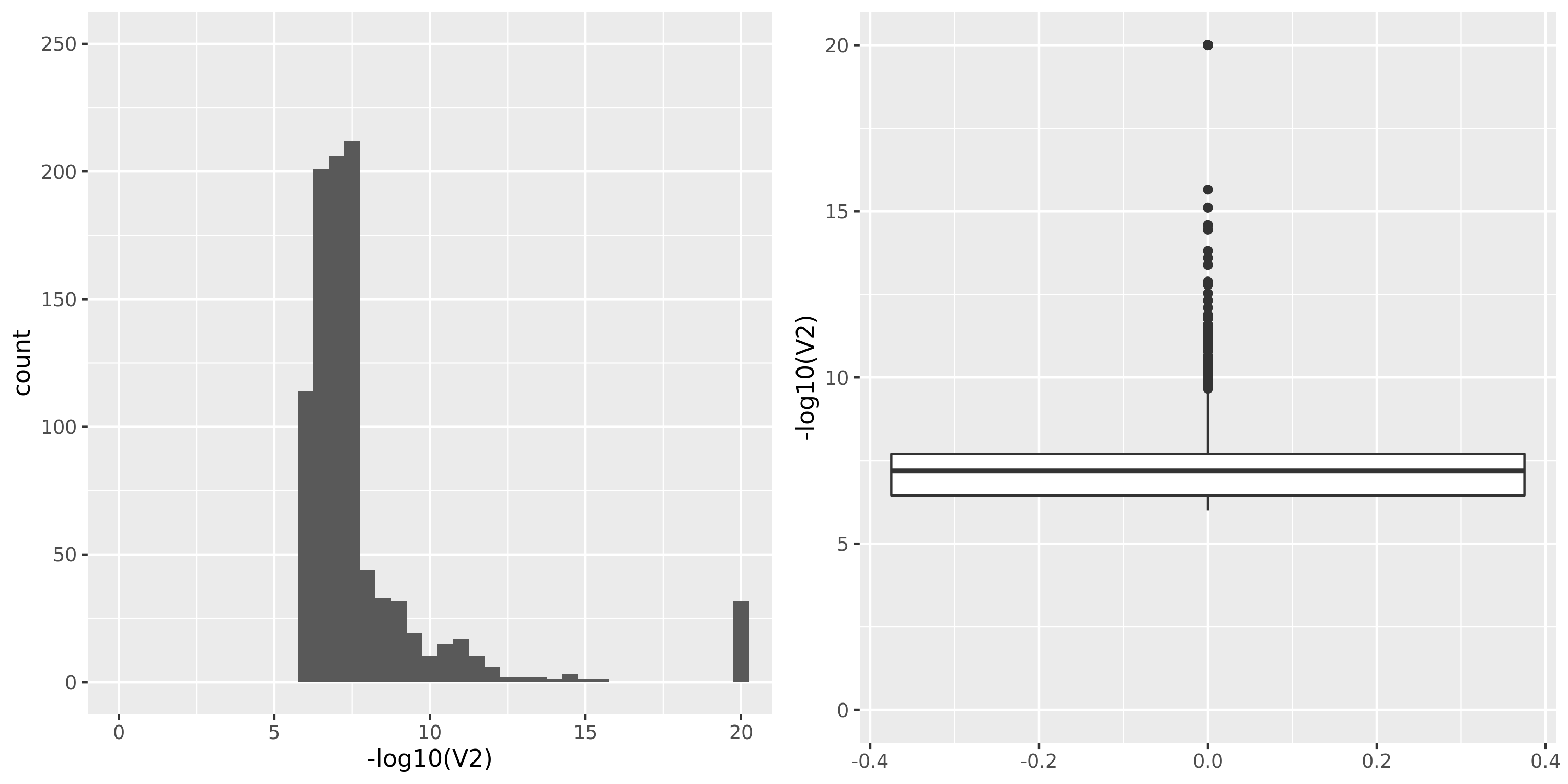
DGN
| Version | Author | Date |
|---|---|---|
| b110975 | Lili Wang | 2020-12-17 |
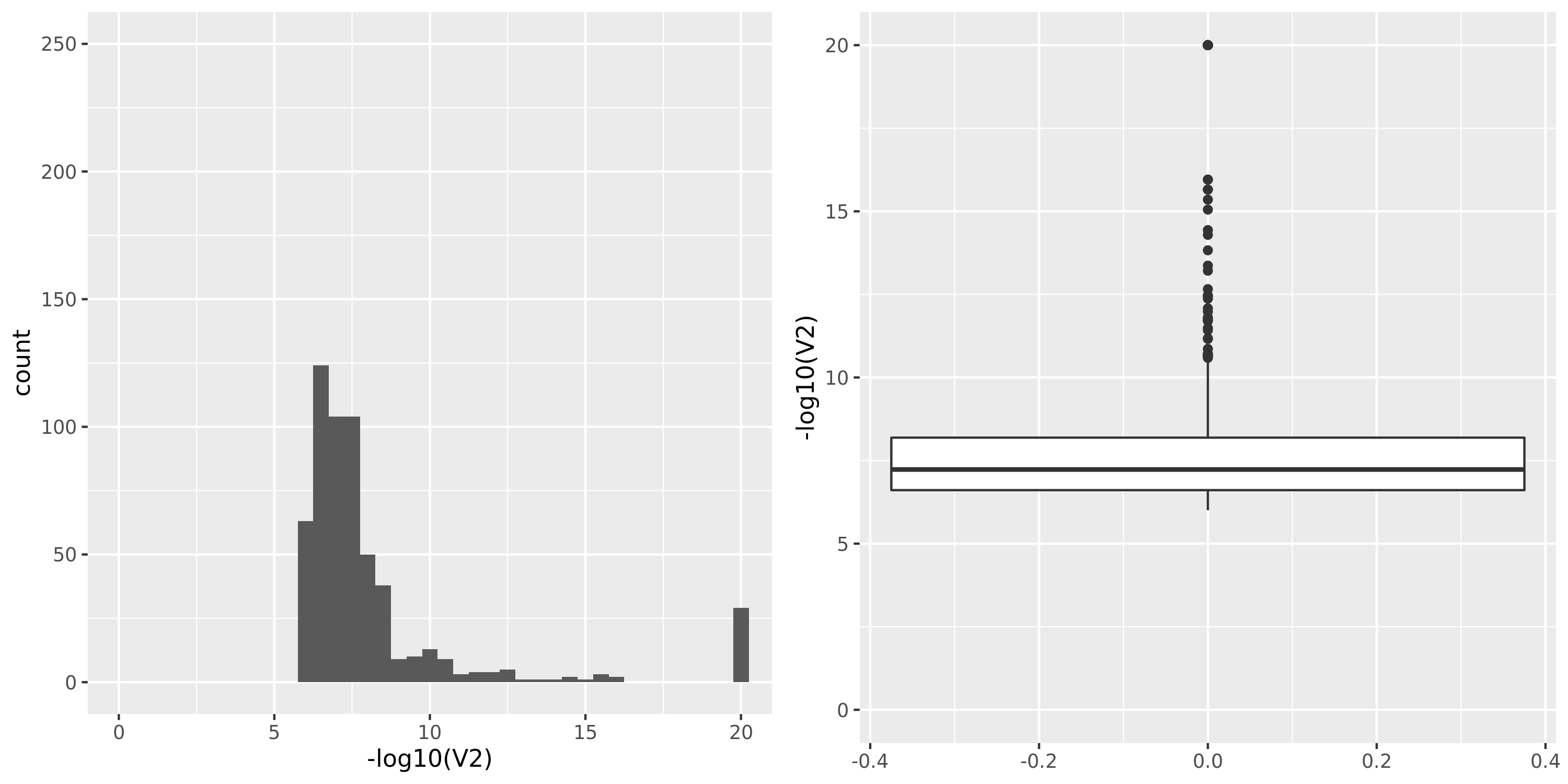
DGN_new
| Version | Author | Date |
|---|---|---|
| 361c43e | Lili Wang | 2021-01-04 |
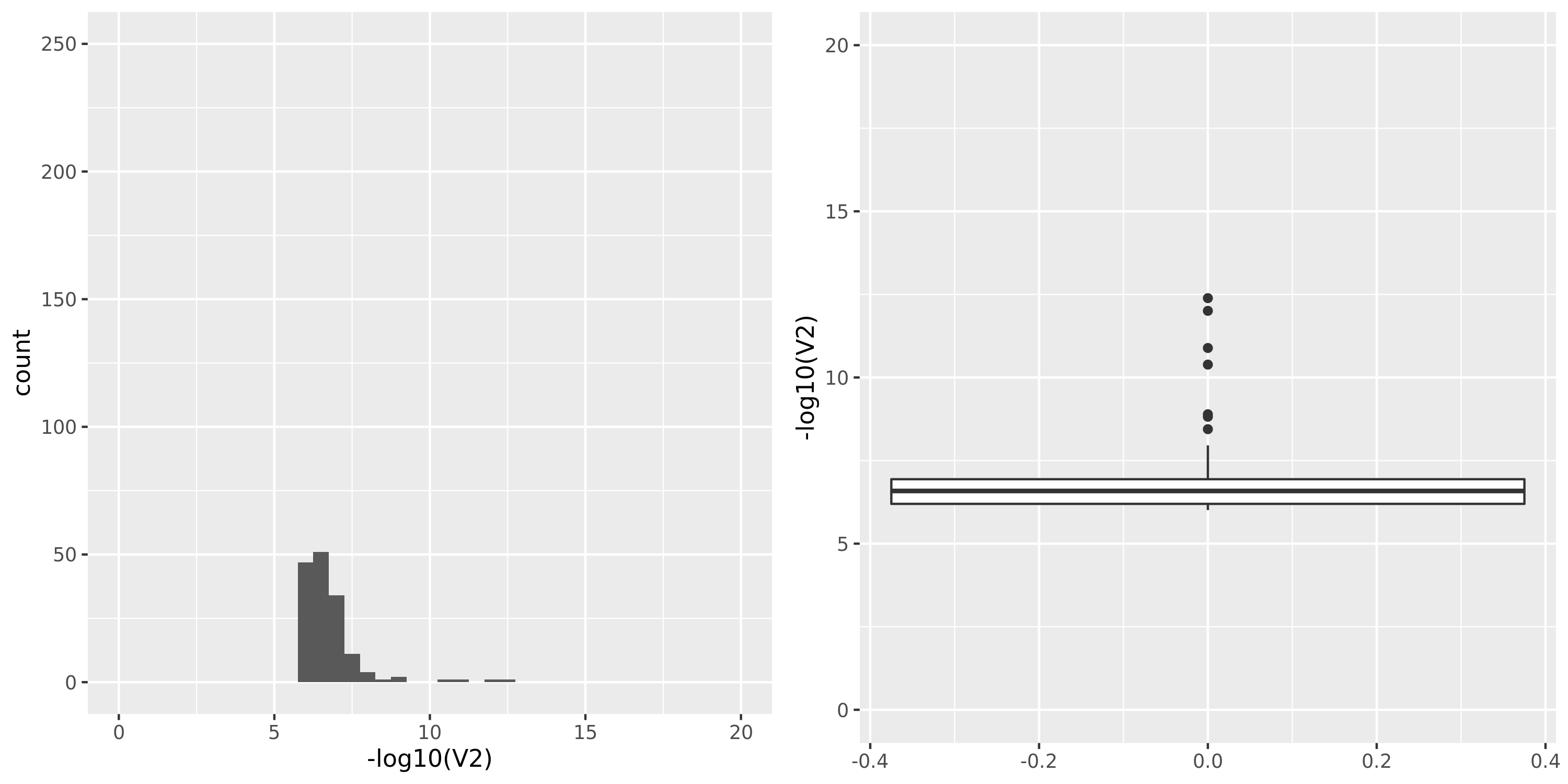
Whole_Blood
| Version | Author | Date |
|---|---|---|
| 361c43e | Lili Wang | 2021-01-04 |
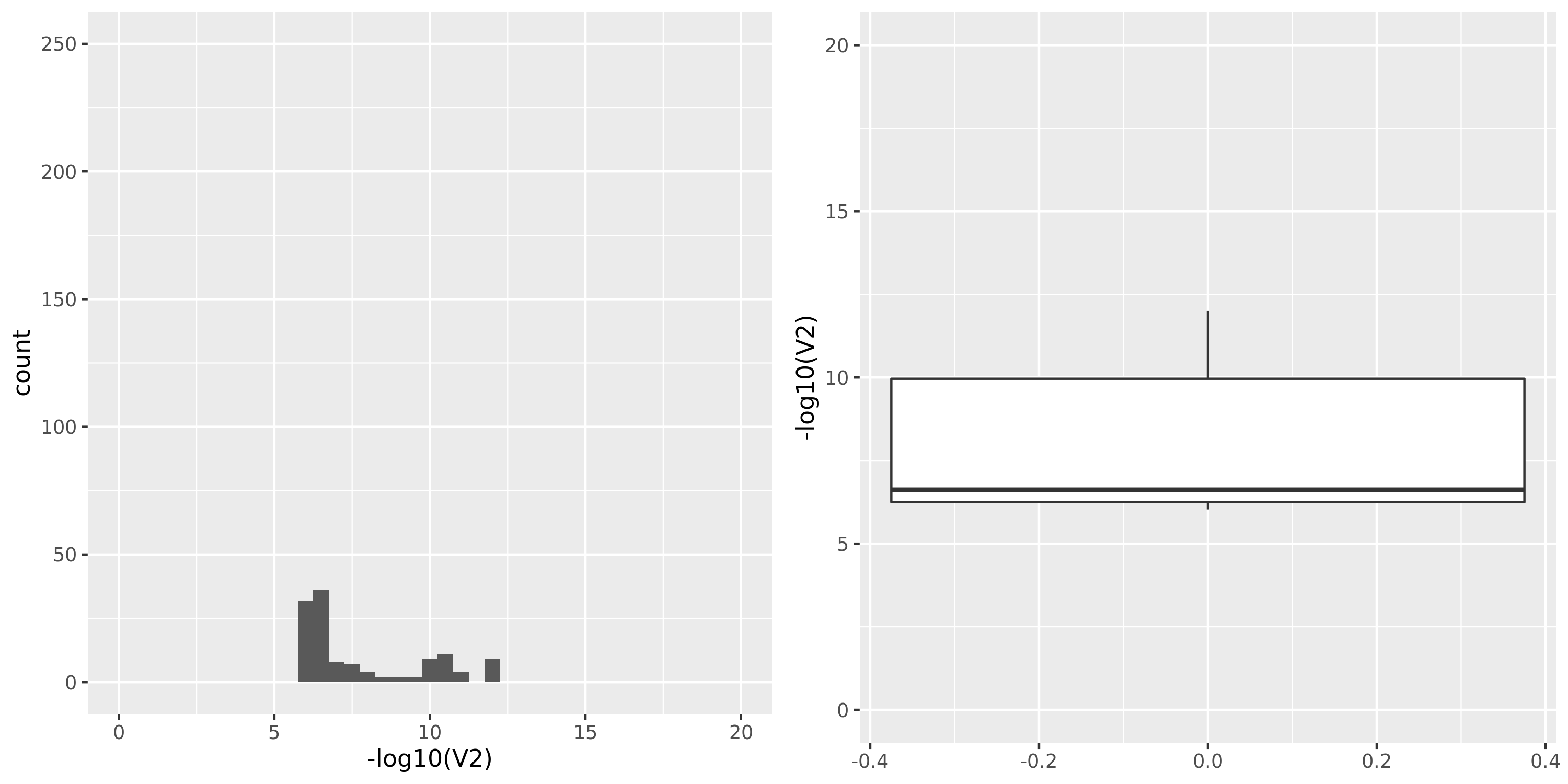
Muscle_Skeletal
| Version | Author | Date |
|---|---|---|
| 361c43e | Lili Wang | 2021-01-04 |
Skin_Sun_Exposed_Lower_leg
| Version | Author | Date |
|---|---|---|
| 361c43e | Lili Wang | 2021-01-04 |
Artery_Tibial
| Version | Author | Date |
|---|---|---|
| 361c43e | Lili Wang | 2021-01-04 |
Remarks
- GTEx datasets generally have few significant signals and relatively large pvalues (compared to DGN).
New TCGA by the standard filtering
R version 3.6.3 (2020-02-29)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.4 rprojroot_1.3-2 digest_0.6.25 later_1.0.0
[5] R6_2.4.1 backports_1.1.5 git2r_0.27.1 magrittr_1.5
[9] evaluate_0.14 highr_0.8 stringi_1.4.6 rlang_0.4.7
[13] fs_1.3.2 promises_1.1.0 whisker_0.4 rmarkdown_2.1
[17] tools_3.6.3 stringr_1.4.0 glue_1.3.2 httpuv_1.5.2
[21] xfun_0.12 yaml_2.2.1 compiler_3.6.3 htmltools_0.4.0
[25] knitr_1.28