GO_after_comments
ERM
2023-12-19
Last updated: 2023-12-19
Checks: 6 1
Knit directory: Cardiotoxicity/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230109) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| C:/Program Files/R_WD/Cardiotoxicity/data/DEG-GO/DAspgostres3.RDS | data/DEG-GO/DAspgostres3.RDS |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e993745. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/figure/
Ignored: analysis/variance_values by gene.png
Ignored: data/41588_2018_171_MOESM3_ESMeQTL_ST2_for paper.csv
Ignored: data/Arr_GWAS.txt
Ignored: data/Arr_geneset.RDS
Ignored: data/BC_cell_lines.csv
Ignored: data/BurridgeDOXTOX.RDS
Ignored: data/CADGWASgene_table.csv
Ignored: data/CAD_geneset.RDS
Ignored: data/CALIMA_Data/
Ignored: data/CMD04_75DRCviability.csv
Ignored: data/CMD04_87DRCviability.csv
Ignored: data/CMD05_75DRCviability.csv
Ignored: data/CMD05_87DRCviability.csv
Ignored: data/Clamp_Summary.csv
Ignored: data/Cormotif_24_k1-5_raw.RDS
Ignored: data/Counts_RNA_ERMatthews.txt
Ignored: data/DAgostres24.RDS
Ignored: data/DAtable1.csv
Ignored: data/DDEMresp_list.csv
Ignored: data/DDE_reQTL.txt
Ignored: data/DDEresp_list.csv
Ignored: data/DEG-GO/
Ignored: data/DEG_cormotif.RDS
Ignored: data/DF_Plate_Peak.csv
Ignored: data/DRC48hoursdata.csv
Ignored: data/Da24counts.txt
Ignored: data/Dx24counts.txt
Ignored: data/Dx_reQTL_specific.txt
Ignored: data/EPIstorelist24.RDS
Ignored: data/Ep24counts.txt
Ignored: data/FC_necela.RDS
Ignored: data/FC_necela_names.RDS
Ignored: data/Full_LD_rep.csv
Ignored: data/GOIsig.csv
Ignored: data/GOplots.R
Ignored: data/GTEX_setsimple.csv
Ignored: data/GTEX_sig24.RDS
Ignored: data/GTEx_gene_list.csv
Ignored: data/HFGWASgene_table.csv
Ignored: data/HF_geneset.RDS
Ignored: data/Heart_Left_Ventricle.v8.egenes.txt
Ignored: data/Heatmap_mat.RDS
Ignored: data/Heatmap_sig.RDS
Ignored: data/Hf_GWAS.txt
Ignored: data/K_cluster
Ignored: data/K_cluster_kisthree.csv
Ignored: data/K_cluster_kistwo.csv
Ignored: data/Knowles_log2cpm_real.RDS
Ignored: data/Knowles_variation_data.RDS
Ignored: data/Knowlesvarlist.RDS
Ignored: data/LD50_05via.csv
Ignored: data/LDH48hoursdata.csv
Ignored: data/Mt24counts.txt
Ignored: data/NoRespDEG_final.csv
Ignored: data/RINsamplelist.txt
Ignored: data/RNA_seq_trial.RDS
Ignored: data/Schneider_GWAS.txt
Ignored: data/Seonane2019supp1.txt
Ignored: data/TMMnormed_x.RDS
Ignored: data/TOP2Bi-24hoursGO_analysis.csv
Ignored: data/TR24counts.txt
Ignored: data/TableS10.csv
Ignored: data/TableS11.csv
Ignored: data/TableS9.csv
Ignored: data/Top2_expression.RDS
Ignored: data/Top2biresp_cluster24h.csv
Ignored: data/Var_test_list.RDS
Ignored: data/Var_test_list24.RDS
Ignored: data/Var_test_list24alt.RDS
Ignored: data/Var_test_list3.RDS
Ignored: data/Vargenes.RDS
Ignored: data/Viabilitylistfull.csv
Ignored: data/allexpressedgenes.txt
Ignored: data/allfinal3hour.RDS
Ignored: data/allgenes.txt
Ignored: data/allmatrix.RDS
Ignored: data/allmymatrix.RDS
Ignored: data/annotation_data_frame.RDS
Ignored: data/averageviabilitytable.RDS
Ignored: data/averageviabilitytable.csv
Ignored: data/avgLD50.RDS
Ignored: data/avg_LD50.RDS
Ignored: data/backGL.txt
Ignored: data/burr_genes.RDS
Ignored: data/calcium_data.RDS
Ignored: data/clamp_summary.RDS
Ignored: data/cormotif_3hk1-8.RDS
Ignored: data/cormotif_initalK5.RDS
Ignored: data/cormotif_initialK5.RDS
Ignored: data/cormotif_initialall.RDS
Ignored: data/cormotifprobs.csv
Ignored: data/counts24hours.RDS
Ignored: data/cpmcount.RDS
Ignored: data/cpmnorm_counts.csv
Ignored: data/crispr_genes.csv
Ignored: data/ctnnt_results.txt
Ignored: data/cvd_GWAS.txt
Ignored: data/dat_cpm.RDS
Ignored: data/data_outline.txt
Ignored: data/drug_noveh1.csv
Ignored: data/efit2.RDS
Ignored: data/efit2_final.RDS
Ignored: data/efit2results.RDS
Ignored: data/ensembl_backup.RDS
Ignored: data/ensgtotal.txt
Ignored: data/filcpm_counts.RDS
Ignored: data/filenameonly.txt
Ignored: data/filtered_cpm_counts.csv
Ignored: data/filtered_raw_counts.csv
Ignored: data/filtermatrix_x.RDS
Ignored: data/folder_05top/
Ignored: data/framefun24.RDS
Ignored: data/geneDoxonlyQTL.csv
Ignored: data/gene_corr_df.RDS
Ignored: data/gene_corr_frame.RDS
Ignored: data/gene_prob_tran3h.RDS
Ignored: data/gene_probabilityk5.RDS
Ignored: data/geneset_24.RDS
Ignored: data/gostresTop2bi_ER.RDS
Ignored: data/gostresTop2bi_LR
Ignored: data/gostresTop2bi_LR.RDS
Ignored: data/gostresTop2bi_TI.RDS
Ignored: data/gostrescoNR
Ignored: data/gtex/
Ignored: data/heartgenes.csv
Ignored: data/highly_var_genelist.RDS
Ignored: data/hsa_kegg_anno.RDS
Ignored: data/individualDRCfile.RDS
Ignored: data/individual_DRC48.RDS
Ignored: data/individual_LDH48.RDS
Ignored: data/indv_noveh1.csv
Ignored: data/kegglistDEG.RDS
Ignored: data/kegglistDEG24.RDS
Ignored: data/kegglistDEG3.RDS
Ignored: data/knowfig4.csv
Ignored: data/knowfig5.csv
Ignored: data/label_list.RDS
Ignored: data/ld50_table.csv
Ignored: data/mean_vardrug1.csv
Ignored: data/mean_varframe.csv
Ignored: data/mymatrix.RDS
Ignored: data/new_ld50avg.RDS
Ignored: data/nonresponse_cluster24h.csv
Ignored: data/norm_LDH.csv
Ignored: data/norm_counts.csv
Ignored: data/old_sets/
Ignored: data/organized_drugframe.csv
Ignored: data/plan2plot.png
Ignored: data/plot_intv_list.RDS
Ignored: data/plot_list_DRC.RDS
Ignored: data/qval24hr.RDS
Ignored: data/qval3hr.RDS
Ignored: data/qvalueEPItemp.RDS
Ignored: data/raw_counts.csv
Ignored: data/response_cluster24h.csv
Ignored: data/sampsettrz.RDS
Ignored: data/schneider_closest_output.RDS
Ignored: data/sigVDA24.txt
Ignored: data/sigVDA3.txt
Ignored: data/sigVDX24.txt
Ignored: data/sigVDX3.txt
Ignored: data/sigVEP24.txt
Ignored: data/sigVEP3.txt
Ignored: data/sigVMT24.txt
Ignored: data/sigVMT3.txt
Ignored: data/sigVTR24.txt
Ignored: data/sigVTR3.txt
Ignored: data/siglist.RDS
Ignored: data/siglist_final.RDS
Ignored: data/siglist_old.RDS
Ignored: data/slope_table.csv
Ignored: data/supp10_24hlist.RDS
Ignored: data/supp10_3hlist.RDS
Ignored: data/supp_normLDH48.RDS
Ignored: data/supp_pca_all_anno.RDS
Ignored: data/table3a.omar
Ignored: data/test_run_sample_list.txt
Ignored: data/testlist.txt
Ignored: data/toplistall.RDS
Ignored: data/trtonly_24h_genes.RDS
Ignored: data/trtonly_3h_genes.RDS
Ignored: data/tvl24hour.txt
Ignored: data/tvl24hourw.txt
Ignored: data/venn_code.R
Ignored: data/viability.RDS
Untracked files:
Untracked: .RDataTmp
Untracked: .RDataTmp1
Untracked: .RDataTmp2
Untracked: .RDataTmp3
Untracked: 3hr all.pdf
Untracked: Code_files_list.csv
Untracked: Data_files_list.csv
Untracked: Doxorubicin_vehicle_3_24.csv
Untracked: Doxtoplist.csv
Untracked: EPIqvalue_analysis.Rmd
Untracked: GWAS_list_of_interest.xlsx
Untracked: KEGGpathwaylist.R
Untracked: OmicNavigator_learn.R
Untracked: SNP_egenes_allfiles.RDS
Untracked: SigDoxtoplist.csv
Untracked: analysis/DRC_viability_check.Rmd
Untracked: analysis/New_code_dec-23.R
Untracked: analysis/cellcycle_kegg_genes.R
Untracked: analysis/ciFIT.R
Untracked: analysis/export_to_excel.R
Untracked: analysis/featureCountsPLAY.R
Untracked: cleanupfiles_script.R
Untracked: code/biomart_gene_names.R
Untracked: code/constantcode.R
Untracked: code/corMotifcustom.R
Untracked: code/cpm_boxplot.R
Untracked: code/extracting_ggplot_data.R
Untracked: code/movingfilesto_ppl.R
Untracked: code/pearson_extract_func.R
Untracked: code/pearson_tox_extract.R
Untracked: code/plot1C.fun.R
Untracked: code/spearman_extract_func.R
Untracked: code/venndiagramcolor_control.R
Untracked: cormotif_p.post.list_4.csv
Untracked: figS1024h.pdf
Untracked: individual-legenddark2.png
Untracked: installed_old.rda
Untracked: listoftranscripts
Untracked: motif_ER.txt
Untracked: motif_LR.txt
Untracked: motif_NR.txt
Untracked: motif_TI.txt
Untracked: output/ABHD8_dif_values.RDS
Untracked: output/C3orf18_dif_values.RDS
Untracked: output/Cardiotox_dif_values.RDS
Untracked: output/DNR_DEGlist.csv
Untracked: output/DNRvenn.RDS
Untracked: output/DOX_DEGlist.csv
Untracked: output/DOX_de_goi.csv
Untracked: output/DOXvenn.RDS
Untracked: output/EEF1B2_dif_values.RDS
Untracked: output/EEIG1_dif_values.RDS
Untracked: output/EPI_DEGlist.csv
Untracked: output/EPIvenn.RDS
Untracked: output/ESGN_rds.RDS
Untracked: output/FC_necela.RDS
Untracked: output/FC_necela_names.RDS
Untracked: output/FRS2_dif_values.RDS
Untracked: output/Figures/
Untracked: output/GTEXv8_gene_median_tpm.RDS
Untracked: output/GTEXv8_gene_tpm_heart_left_ventricle.RDS
Untracked: output/HDDC2_dif_values.RDS
Untracked: output/HER2_gene.RDS
Untracked: output/KEGGcellcyclegenes.RDS
Untracked: output/Knowles_S13.csv
Untracked: output/Knowles_log2cpm.csv
Untracked: output/Knowles_supp13.csv
Untracked: output/MTX_DEGlist.csv
Untracked: output/MTXvenn.RDS
Untracked: output/PEX16_dif_values.RDS
Untracked: output/RASIP1_dif_values.RDS
Untracked: output/RMI1_dif_values.RDS
Untracked: output/RSID_QTL_list_full.txt
Untracked: output/SETA_analysis_reyes.RDS
Untracked: output/SGWAS_top50_order.csv
Untracked: output/SLC27A1_dif_values.RDS
Untracked: output/SLC28A3_dif_values.RDS
Untracked: output/SNP_egenes_allfiles.RDS
Untracked: output/SNP_list_ID.RDS
Untracked: output/SNP_list_full.txt
Untracked: output/SNP_supp.RDS
Untracked: output/TGFBR3L_dif_values.RDS
Untracked: output/TNS2_dif_values.RDS
Untracked: output/TOP_50SNPreffile.csv
Untracked: output/TRZ_DEGlist.csv
Untracked: output/TableS8.csv
Untracked: output/Volcanoplot_10
Untracked: output/Volcanoplot_10.RDS
Untracked: output/ZNF740_dif_values.RDS
Untracked: output/allfinal_sup10.RDS
Untracked: output/counts_v8_heart_left_ventricle_gct.RDS
Untracked: output/crisprfoldchange.RDS
Untracked: output/endocytosisgenes.csv
Untracked: output/expre7k.csv
Untracked: output/gene_corr_fig9.RDS
Untracked: output/genes.RDS
Untracked: output/legend_b.RDS
Untracked: output/motif_ERrep.RDS
Untracked: output/motif_LRrep.RDS
Untracked: output/motif_NRrep.RDS
Untracked: output/motif_TI_rep.RDS
Untracked: output/near_genes_SNP1.RDS
Untracked: output/necela_list_test.RDS
Untracked: output/necela_val_genes.RDS
Untracked: output/output-old/
Untracked: output/rank24genes.csv
Untracked: output/rank3genes.csv
Untracked: output/reneem@ls6.tacc.utexas.edu
Untracked: output/sequencinginformationforsupp.csv
Untracked: output/sequencinginformationforsupp.prn
Untracked: output/sigVDA24.txt
Untracked: output/sigVDA3.txt
Untracked: output/sigVDX24.txt
Untracked: output/sigVDX3.txt
Untracked: output/sigVEP24.txt
Untracked: output/sigVEP3.txt
Untracked: output/sigVMT24.txt
Untracked: output/sigVMT3.txt
Untracked: output/sigVTR24.txt
Untracked: output/sigVTR3.txt
Untracked: output/supplementary_motif_list_GO.RDS
Untracked: output/test_biomart_run.RDS
Untracked: output/toptablebydrug.RDS
Untracked: output/trop_knowles_fun.csv
Untracked: output/tvl24hour.txt
Untracked: output/x_counts.RDS
Untracked: reneebasecode.R
Unstaged changes:
Modified: analysis/DRC_analysis.Rmd
Modified: analysis/GOI_plots.Rmd
Modified: analysis/GTEx_genes.Rmd
Modified: analysis/Knowels_trop_analysis.Rmd
Modified: analysis/Knowles2019.Rmd
Modified: analysis/Var_genes.Rmd
Modified: analysis/after_comments.Rmd
Modified: analysis/diff_tox.Rmd
Modified: analysis/variance_scrip.Rmd
Modified: output/daplot.RDS
Modified: output/dxplot.RDS
Modified: output/epplot.RDS
Modified: output/mtplot.RDS
Modified: output/plan2plot.png
Modified: output/trplot.RDS
Modified: output/veplot.RDS
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/GO_after_comments.Rmd) and
HTML (docs/GO_after_comments.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | e993745 | reneeisnowhere | 2023-12-19 | Adding pathway enrichment plots |
| html | b442590 | reneeisnowhere | 2023-11-21 | Build site. |
| Rmd | b464f89 | reneeisnowhere | 2023-11-21 | updates to links again |
| html | fd6d33c | reneeisnowhere | 2023-11-21 | Build site. |
| Rmd | cbefbd1 | reneeisnowhere | 2023-11-21 | adding links |
library(tidyverse)
library(ggsignif)
library(cowplot)
library(ggpubr)
library(scales)
library(sjmisc)
library(kableExtra)
library(broom)
library(biomaRt)
library(RColorBrewer)
library(gprofiler2)
library(qvalue)FC_mine <- readRDS("data/toplistall.RDS")
TRZ_list <- FC_mine %>%
filter(id=="TRZ",P.Value < 0.05) #%>%
backGL <- read_csv("data/backGL.txt",
col_types = cols(...1 = col_skip()))
TRZ_list3h <- FC_mine %>%
filter(id=="TRZ",P.Value < 0.05, time == "3_hours")
TRZ_list24h <- FC_mine %>%
filter(id=="TRZ",P.Value < 0.05, time == "24_hours")
TRZ_listqvalue <- FC_mine %>%
filter(id=="TRZ", time=="24_hours") %>%
dplyr::select(ENTREZID,P.Value)
qobj <- qvalue(TRZ_listqvalue$P.Value)
hist(qobj)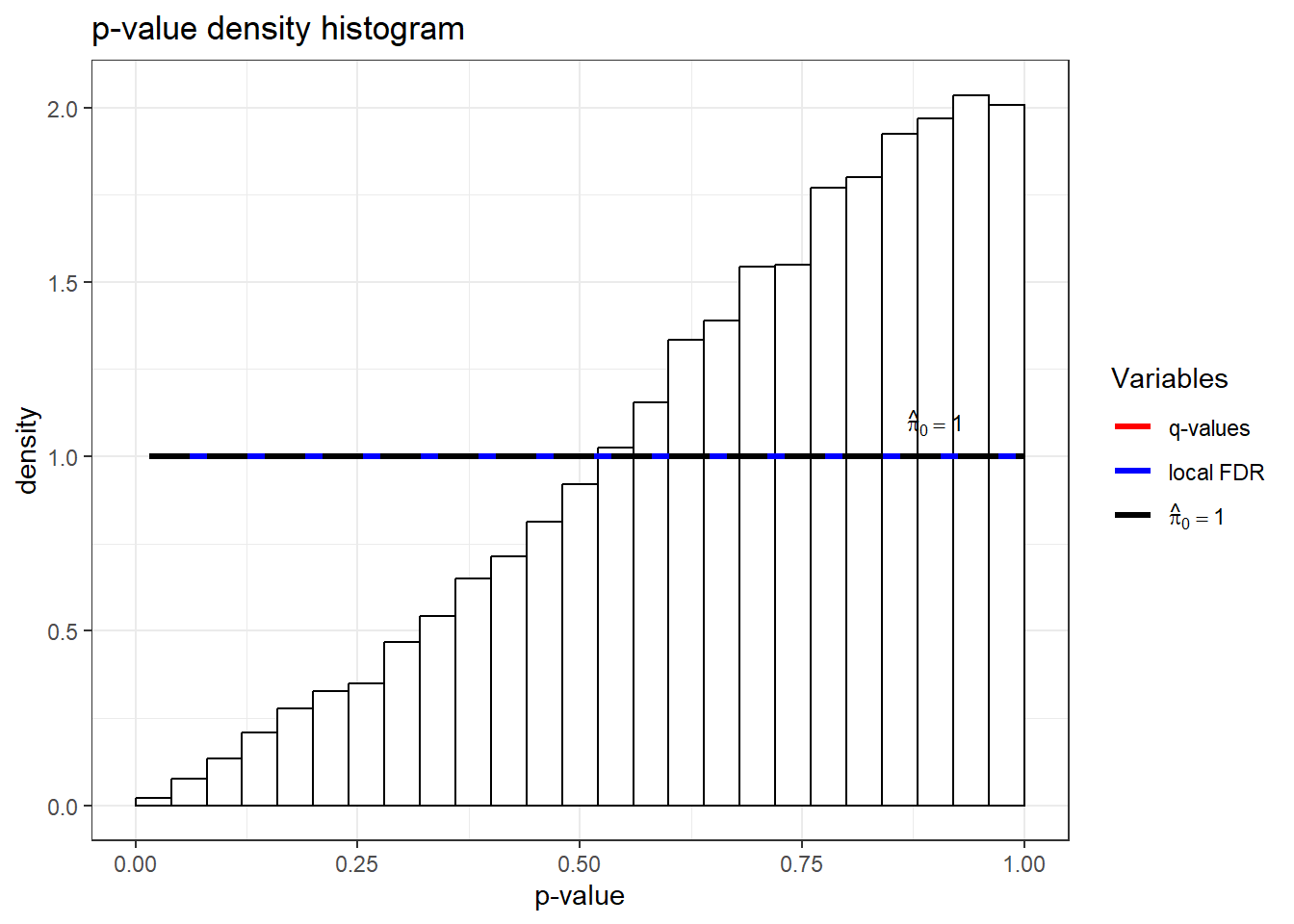
plot(qobj)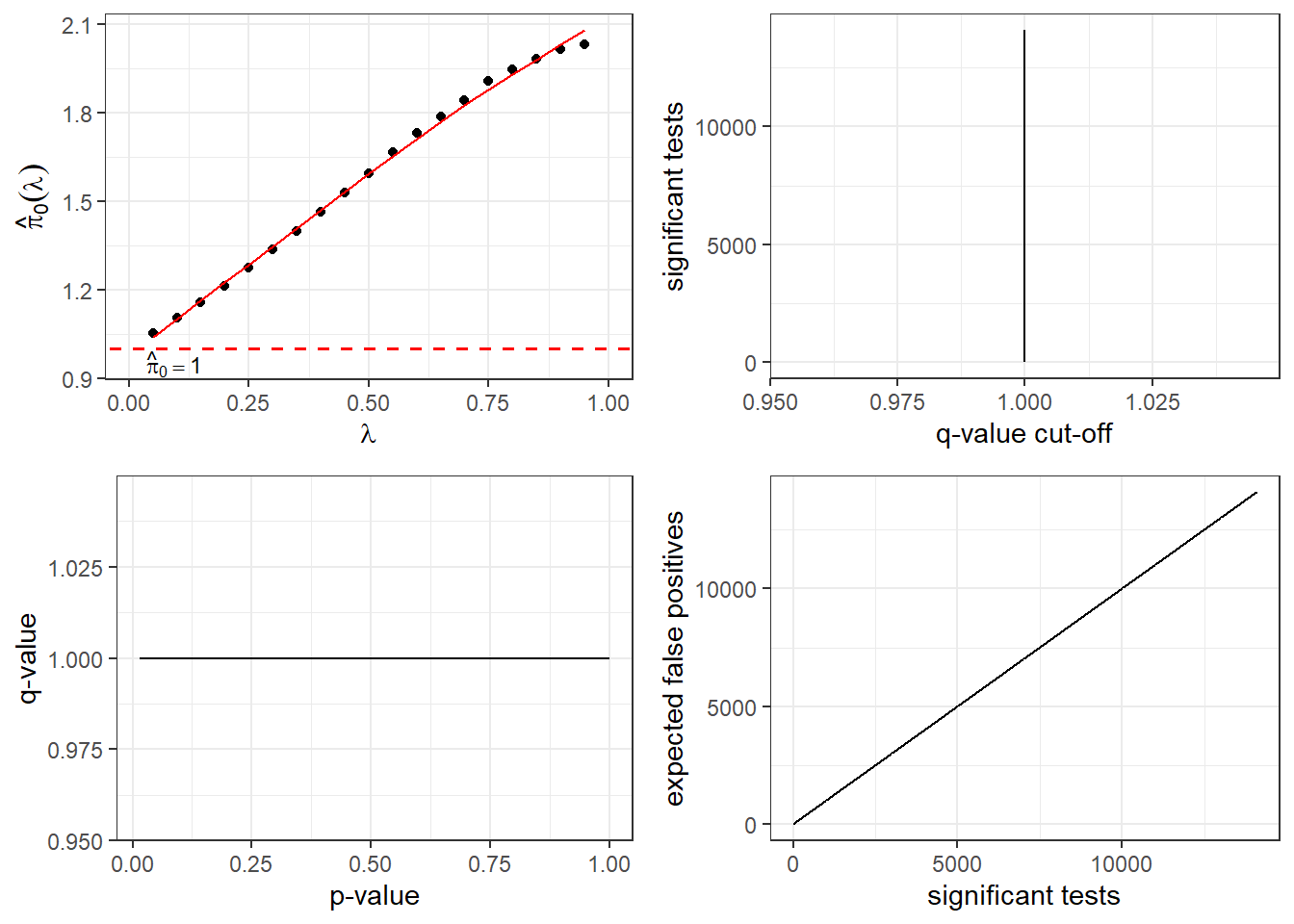
TRZ nom de (3 & 24 hour)
# TRZresgenes <- gost(query = TRZ_list$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(TRZresgenes,"data/DEG-GO/TRZresgenes.RDS")
TRZresgenes <- readRDS("data/DEG-GO/TRZresgenes.RDS")
TRZnom <- gostplot(TRZresgenes, capped = FALSE, interactive = TRUE)
TRZnomTRZnomtable <- TRZresgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
TRZnomtable %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:1902746 | regulation of lens fiber cell differentiation | 2 | 8 | 6.14e-02 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 1 | 2 | 6.14e-02 |
| GO:BP | GO:0042078 | germ-line stem cell division | 1 | 2 | 6.14e-02 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 1 | 2 | 6.14e-02 |
| GO:BP | GO:1902747 | negative regulation of lens fiber cell differentiation | 2 | 5 | 6.14e-02 |
| GO:BP | GO:0048133 | male germ-line stem cell asymmetric division | 1 | 2 | 6.14e-02 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 2 | 10 | 6.14e-02 |
| GO:BP | GO:0044849 | estrous cycle | 2 | 14 | 7.09e-02 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 2 | 15 | 8.54e-02 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 2 | 22 | 8.66e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 3 | 218 | 8.66e-02 |
| GO:BP | GO:1901386 | negative regulation of voltage-gated calcium channel activity | 2 | 14 | 8.66e-02 |
| GO:BP | GO:0035809 | regulation of urine volume | 2 | 14 | 8.66e-02 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 2 | 31 | 9.28e-02 |
| GO:BP | GO:0098708 | glucose import across plasma membrane | 1 | 5 | 9.28e-02 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 3 | 72 | 9.28e-02 |
| GO:BP | GO:0098739 | import across plasma membrane | 3 | 130 | 9.28e-02 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 2 | 28 | 9.28e-02 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 2 | 72 | 9.28e-02 |
| GO:BP | GO:0045210 | FasL biosynthetic process | 1 | 1 | 9.28e-02 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 2 | 32 | 9.28e-02 |
| GO:BP | GO:0098704 | carbohydrate import across plasma membrane | 1 | 5 | 9.28e-02 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 2 | 29 | 9.28e-02 |
| GO:BP | GO:0019249 | lactate biosynthetic process | 1 | 3 | 9.28e-02 |
| GO:BP | GO:0018345 | protein palmitoylation | 1 | 30 | 9.28e-02 |
| GO:BP | GO:0140271 | hexose import across plasma membrane | 1 | 5 | 9.28e-02 |
| GO:BP | GO:0097167 | circadian regulation of translation | 1 | 4 | 9.28e-02 |
| GO:BP | GO:0051764 | actin crosslink formation | 1 | 9 | 9.28e-02 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 2 | 30 | 9.28e-02 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 2 | 17 | 9.28e-02 |
| GO:BP | GO:0007281 | germ cell development | 2 | 194 | 9.28e-02 |
| GO:BP | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 1 | 7 | 9.28e-02 |
| GO:BP | GO:0042918 | alkanesulfonate transport | 1 | 6 | 9.28e-02 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 4 | 205 | 9.28e-02 |
| GO:BP | GO:0038066 | p38MAPK cascade | 2 | 42 | 9.28e-02 |
| GO:BP | GO:0008356 | asymmetric cell division | 1 | 14 | 9.28e-02 |
| GO:BP | GO:0036159 | inner dynein arm assembly | 1 | 8 | 9.28e-02 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 2 | 38 | 9.28e-02 |
| GO:BP | GO:0051939 | gamma-aminobutyric acid import | 1 | 3 | 9.28e-02 |
| GO:BP | GO:0051946 | regulation of glutamate uptake involved in transmission of nerve impulse | 1 | 3 | 9.28e-02 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 1 | 26 | 9.28e-02 |
| GO:BP | GO:0060252 | positive regulation of glial cell proliferation | 1 | 14 | 9.28e-02 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 2 | 29 | 9.28e-02 |
| GO:BP | GO:0006979 | response to oxidative stress | 3 | 328 | 9.28e-02 |
| GO:BP | GO:0060485 | mesenchyme development | 4 | 250 | 9.28e-02 |
| GO:BP | GO:0061643 | chemorepulsion of axon | 1 | 6 | 9.28e-02 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 3 | 269 | 9.28e-02 |
| GO:BP | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission | 1 | 3 | 9.28e-02 |
| GO:BP | GO:0018231 | peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine | 1 | 21 | 9.28e-02 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 2 | 255 | 9.28e-02 |
| GO:BP | GO:1904059 | regulation of locomotor rhythm | 1 | 3 | 9.28e-02 |
| GO:BP | GO:0010232 | vascular transport | 2 | 72 | 9.28e-02 |
| GO:BP | GO:1990708 | conditioned place preference | 1 | 2 | 9.28e-02 |
| GO:BP | GO:0015734 | taurine transport | 1 | 6 | 9.28e-02 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 9.28e-02 |
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 2 | 31 | 9.28e-02 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 1 | 44 | 9.28e-02 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 3 | 139 | 9.28e-02 |
| GO:BP | GO:0014009 | glial cell proliferation | 1 | 40 | 9.28e-02 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 9.28e-02 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 2 | 35 | 9.28e-02 |
| GO:BP | GO:0015757 | galactose transmembrane transport | 1 | 4 | 9.28e-02 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 9.28e-02 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 10 | 9.28e-02 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 10 | 9.28e-02 |
| GO:BP | GO:0017145 | stem cell division | 1 | 25 | 9.28e-02 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 12 | 9.28e-02 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 3 | 198 | 9.28e-02 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 2 | 35 | 9.28e-02 |
| GO:BP | GO:0015800 | acidic amino acid transport | 2 | 39 | 9.28e-02 |
| GO:BP | GO:0018230 | peptidyl-L-cysteine S-palmitoylation | 1 | 21 | 9.28e-02 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 9.28e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 3 | 115 | 9.49e-02 |
| GO:BP | GO:0045226 | extracellular polysaccharide biosynthetic process | 1 | 2 | 9.49e-02 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 1 | 14 | 9.49e-02 |
| GO:BP | GO:1900625 | positive regulation of monocyte aggregation | 1 | 3 | 9.49e-02 |
| GO:BP | GO:0048232 | male gamete generation | 2 | 346 | 9.49e-02 |
| GO:BP | GO:0046379 | extracellular polysaccharide metabolic process | 1 | 2 | 9.49e-02 |
| GO:BP | GO:0007283 | spermatogenesis | 2 | 333 | 9.49e-02 |
| GO:BP | GO:0007288 | sperm axoneme assembly | 1 | 20 | 9.49e-02 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 1 | 15 | 9.49e-02 |
| GO:BP | GO:1900623 | regulation of monocyte aggregation | 1 | 3 | 9.49e-02 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 3 | 116 | 9.54e-02 |
| GO:BP | GO:0070345 | negative regulation of fat cell proliferation | 1 | 4 | 9.57e-02 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 1 | 63 | 9.57e-02 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 1 | 41 | 9.57e-02 |
| GO:BP | GO:0000379 | tRNA-type intron splice site recognition and cleavage | 1 | 3 | 9.96e-02 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 1 | 66 | 9.96e-02 |
| GO:BP | GO:0003013 | circulatory system process | 4 | 445 | 1.04e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 1 | 16 | 1.04e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 2 | 54 | 1.04e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 2 | 35 | 1.04e-01 |
| GO:BP | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway | 1 | 3 | 1.04e-01 |
| GO:BP | GO:1900127 | positive regulation of hyaluronan biosynthetic process | 1 | 4 | 1.04e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 3 | 127 | 1.04e-01 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 2 | 70 | 1.04e-01 |
| GO:BP | GO:0015812 | gamma-aminobutyric acid transport | 1 | 11 | 1.04e-01 |
| GO:BP | GO:0120316 | sperm flagellum assembly | 1 | 27 | 1.04e-01 |
| GO:BP | GO:0070286 | axonemal dynein complex assembly | 1 | 21 | 1.04e-01 |
| GO:BP | GO:0051935 | glutamate reuptake | 1 | 4 | 1.04e-01 |
| GO:BP | GO:0070837 | dehydroascorbic acid transport | 1 | 7 | 1.04e-01 |
| GO:BP | GO:0090500 | endocardial cushion to mesenchymal transition | 1 | 4 | 1.04e-01 |
| GO:BP | GO:0032328 | alanine transport | 1 | 13 | 1.04e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 1.04e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 2 | 45 | 1.05e-01 |
| GO:BP | GO:0007276 | gamete generation | 2 | 465 | 1.05e-01 |
| GO:BP | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | 1 | 30 | 1.05e-01 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 2 | 58 | 1.05e-01 |
| GO:BP | GO:0034089 | establishment of meiotic sister chromatid cohesion | 1 | 4 | 1.05e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 1.07e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 2 | 71 | 1.07e-01 |
| GO:BP | GO:0006858 | extracellular transport | 1 | 33 | 1.07e-01 |
| GO:BP | GO:0019852 | L-ascorbic acid metabolic process | 1 | 10 | 1.07e-01 |
| GO:BP | GO:1901334 | lactone metabolic process | 1 | 10 | 1.07e-01 |
| GO:BP | GO:0060437 | lung growth | 1 | 4 | 1.07e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 2 | 93 | 1.07e-01 |
| GO:BP | GO:0010887 | negative regulation of cholesterol storage | 1 | 8 | 1.07e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 1 | 19 | 1.08e-01 |
| GO:BP | GO:0070487 | monocyte aggregation | 1 | 4 | 1.08e-01 |
| GO:BP | GO:0002036 | regulation of L-glutamate import across plasma membrane | 1 | 6 | 1.08e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 2 | 54 | 1.08e-01 |
| GO:BP | GO:1901842 | negative regulation of high voltage-gated calcium channel activity | 1 | 4 | 1.09e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 3 | 135 | 1.09e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 1 | 28 | 1.16e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 2 | 52 | 1.17e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 2 | 538 | 1.17e-01 |
| GO:BP | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 8 | 1.20e-01 |
| GO:BP | GO:0070295 | renal water absorption | 1 | 6 | 1.20e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 2 | 76 | 1.22e-01 |
| GO:BP | GO:0051933 | amino acid neurotransmitter reuptake | 1 | 5 | 1.24e-01 |
| GO:BP | GO:0070344 | regulation of fat cell proliferation | 1 | 8 | 1.24e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 2 | 558 | 1.25e-01 |
| GO:BP | GO:0006497 | protein lipidation | 1 | 89 | 1.25e-01 |
| GO:BP | GO:0038003 | G protein-coupled opioid receptor signaling pathway | 1 | 5 | 1.25e-01 |
| GO:BP | GO:0044458 | motile cilium assembly | 1 | 46 | 1.25e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 4 | 396 | 1.26e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 3 | 302 | 1.26e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 1 | 92 | 1.26e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 2 | 59 | 1.26e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 2 | 603 | 1.26e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 1 | 137 | 1.26e-01 |
| GO:BP | GO:1900125 | regulation of hyaluronan biosynthetic process | 1 | 6 | 1.26e-01 |
| GO:BP | GO:0060449 | bud elongation involved in lung branching | 1 | 7 | 1.26e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 2 | 57 | 1.27e-01 |
| GO:BP | GO:0043951 | negative regulation of cAMP-mediated signaling | 1 | 10 | 1.33e-01 |
| GO:BP | GO:0051382 | kinetochore assembly | 1 | 17 | 1.33e-01 |
| GO:BP | GO:0042754 | negative regulation of circadian rhythm | 1 | 8 | 1.33e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 1 | 190 | 1.33e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 2 | 633 | 1.33e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 2 | 64 | 1.33e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 2 | 85 | 1.33e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 2 | 95 | 1.33e-01 |
| GO:BP | GO:0106070 | regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 11 | 1.33e-01 |
| GO:BP | GO:0006865 | amino acid transport | 2 | 103 | 1.33e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 2 | 86 | 1.33e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 2 | 97 | 1.35e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 1 | 34 | 1.36e-01 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 1.36e-01 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 1 | 13 | 1.36e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 2 | 87 | 1.36e-01 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 2 | 111 | 1.36e-01 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 2 | 112 | 1.37e-01 |
| GO:BP | GO:0003097 | renal water transport | 1 | 7 | 1.39e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 5 | 471 | 1.39e-01 |
| GO:BP | GO:0070341 | fat cell proliferation | 1 | 11 | 1.39e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 2 | 70 | 1.39e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 1 | 13 | 1.39e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 2 | 81 | 1.39e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 2 | 52 | 1.39e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 1 | 116 | 1.40e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 2 | 70 | 1.40e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 1 | 222 | 1.41e-01 |
| GO:BP | GO:0051383 | kinetochore organization | 1 | 21 | 1.41e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 2 | 549 | 1.41e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 2 | 113 | 1.41e-01 |
| GO:BP | GO:0015711 | organic anion transport | 3 | 270 | 1.41e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 1 | 20 | 1.41e-01 |
| GO:BP | GO:0035082 | axoneme assembly | 1 | 54 | 1.41e-01 |
| GO:BP | GO:0010958 | regulation of amino acid import across plasma membrane | 1 | 14 | 1.42e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 4 | 357 | 1.42e-01 |
| GO:BP | GO:1903789 | regulation of amino acid transmembrane transport | 1 | 14 | 1.42e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 2 | 96 | 1.42e-01 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 2 | 98 | 1.45e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 1 | 48 | 1.45e-01 |
| GO:BP | GO:0010888 | negative regulation of lipid storage | 1 | 15 | 1.47e-01 |
| GO:BP | GO:0098712 | L-glutamate import across plasma membrane | 1 | 11 | 1.47e-01 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 1.47e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 1 | 10 | 1.49e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 2 | 74 | 1.50e-01 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 1 | 26 | 1.50e-01 |
| GO:BP | GO:0042063 | gliogenesis | 1 | 244 | 1.51e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 1 | 15 | 1.51e-01 |
| GO:BP | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway | 1 | 11 | 1.52e-01 |
| GO:BP | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway | 1 | 8 | 1.52e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 8 | 1122 | 1.52e-01 |
| GO:BP | GO:0010878 | cholesterol storage | 1 | 18 | 1.52e-01 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 1.53e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 2 | 103 | 1.53e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 1.53e-01 |
| GO:BP | GO:0051549 | positive regulation of keratinocyte migration | 1 | 11 | 1.53e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 1 | 36 | 1.54e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 33 | 1.57e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 3 | 265 | 1.57e-01 |
| GO:BP | GO:0006089 | lactate metabolic process | 1 | 16 | 1.57e-01 |
| GO:BP | GO:0048511 | rhythmic process | 3 | 233 | 1.57e-01 |
| GO:BP | GO:0001578 | microtubule bundle formation | 1 | 79 | 1.57e-01 |
| GO:BP | GO:0070486 | leukocyte aggregation | 1 | 9 | 1.58e-01 |
| GO:BP | GO:0030213 | hyaluronan biosynthetic process | 1 | 10 | 1.58e-01 |
| GO:BP | GO:0036302 | atrioventricular canal development | 1 | 12 | 1.58e-01 |
| GO:BP | GO:1901841 | regulation of high voltage-gated calcium channel activity | 1 | 12 | 1.62e-01 |
| GO:BP | GO:0003014 | renal system process | 2 | 76 | 1.62e-01 |
| GO:BP | GO:0015804 | neutral amino acid transport | 1 | 43 | 1.62e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 1 | 31 | 1.62e-01 |
| GO:BP | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 1 | 10 | 1.62e-01 |
| GO:BP | GO:0043543 | protein acylation | 1 | 193 | 1.62e-01 |
| GO:BP | GO:0010838 | positive regulation of keratinocyte proliferation | 1 | 9 | 1.62e-01 |
| GO:BP | GO:0097722 | sperm motility | 1 | 62 | 1.62e-01 |
| GO:BP | GO:0034508 | centromere complex assembly | 1 | 29 | 1.62e-01 |
| GO:BP | GO:0051547 | regulation of keratinocyte migration | 1 | 13 | 1.62e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 1 | 62 | 1.62e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 4 | 394 | 1.63e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 30 | 1.65e-01 |
| GO:BP | GO:0007254 | JNK cascade | 2 | 137 | 1.65e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 2 | 141 | 1.65e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 1 | 38 | 1.65e-01 |
| GO:BP | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation | 1 | 13 | 1.65e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 1.65e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 1 | 318 | 1.65e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 2 | 160 | 1.65e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 1 | 69 | 1.65e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 1 | 72 | 1.66e-01 |
| GO:BP | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation | 1 | 14 | 1.66e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 2 | 126 | 1.66e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 1 | 23 | 1.66e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 4 | 403 | 1.66e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 1 | 35 | 1.66e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 3 | 290 | 1.66e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 1 | 35 | 1.66e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 1 | 11 | 1.66e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 1 | 15 | 1.66e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 2 | 96 | 1.66e-01 |
| GO:BP | GO:0021591 | ventricular system development | 1 | 29 | 1.66e-01 |
| GO:BP | GO:0022414 | reproductive process | 2 | 924 | 1.66e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 1 | 15 | 1.66e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 2 | 88 | 1.66e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 1 | 72 | 1.66e-01 |
| GO:BP | GO:0000003 | reproduction | 2 | 933 | 1.66e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 2 | 127 | 1.68e-01 |
| GO:BP | GO:0009648 | photoperiodism | 1 | 24 | 1.73e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 16 | 1.73e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 122 | 1.73e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 2 | 153 | 1.73e-01 |
| GO:BP | GO:0001822 | kidney development | 3 | 246 | 1.74e-01 |
| GO:BP | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 1 | 16 | 1.74e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 2 | 157 | 1.78e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 1 | 44 | 1.78e-01 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 1 | 27 | 1.79e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 1 | 27 | 1.79e-01 |
| GO:BP | GO:0015813 | L-glutamate transmembrane transport | 1 | 18 | 1.79e-01 |
| GO:BP | GO:0050919 | negative chemotaxis | 1 | 42 | 1.82e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 1 | 41 | 1.82e-01 |
| GO:BP | GO:0072001 | renal system development | 3 | 253 | 1.82e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 2 | 100 | 1.83e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 2 | 100 | 1.83e-01 |
| GO:BP | GO:0060602 | branch elongation of an epithelium | 1 | 15 | 1.84e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 2 | 757 | 1.84e-01 |
| GO:BP | GO:0051546 | keratinocyte migration | 1 | 15 | 1.86e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 1 | 19 | 1.86e-01 |
| GO:BP | GO:0043950 | positive regulation of cAMP-mediated signaling | 1 | 5 | 1.86e-01 |
| GO:BP | GO:0003341 | cilium movement | 1 | 97 | 1.86e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 1 | 31 | 1.86e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 1 | 14 | 1.88e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 2 | 135 | 1.88e-01 |
| GO:BP | GO:0007286 | spermatid development | 1 | 108 | 1.88e-01 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 1 | 43 | 1.91e-01 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 1 | 43 | 1.91e-01 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 1 | 20 | 1.91e-01 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 1 | 43 | 1.91e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 1 | 114 | 1.91e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 1 | 192 | 1.92e-01 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 1 | 45 | 1.92e-01 |
| GO:BP | GO:0009988 | cell-cell recognition | 1 | 31 | 1.92e-01 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 1 | 18 | 1.92e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 1 | 21 | 1.92e-01 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 1 | 44 | 1.92e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 1 | 34 | 1.92e-01 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 1 | 45 | 1.92e-01 |
| GO:BP | GO:0051180 | vitamin transport | 1 | 29 | 1.92e-01 |
| GO:BP | GO:0042755 | eating behavior | 1 | 16 | 1.92e-01 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 1 | 45 | 1.92e-01 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 1 | 45 | 1.92e-01 |
| GO:BP | GO:0051301 | cell division | 1 | 569 | 1.93e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 1.93e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 1 | 24 | 1.94e-01 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 1 | 46 | 1.94e-01 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 1 | 46 | 1.94e-01 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 1 | 46 | 1.94e-01 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 1 | 18 | 1.94e-01 |
| GO:BP | GO:0007517 | muscle organ development | 2 | 279 | 1.94e-01 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 1 | 22 | 1.94e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 3 | 335 | 1.94e-01 |
| GO:BP | GO:0021756 | striatum development | 1 | 18 | 1.94e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 2 | 212 | 1.94e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 3 | 334 | 1.94e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 2 | 145 | 1.94e-01 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 1 | 23 | 1.94e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 1 | 49 | 1.94e-01 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 31 | 1.94e-01 |
| GO:BP | GO:0006612 | protein targeting to membrane | 1 | 120 | 1.94e-01 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 1 | 20 | 1.94e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 2 | 71 | 1.94e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 1 | 14 | 1.94e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 4 | 473 | 1.94e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 1 | 51 | 1.95e-01 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 1 | 51 | 1.97e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 2 | 115 | 1.97e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 1 | 44 | 1.97e-01 |
| GO:BP | GO:0035265 | organ growth | 2 | 128 | 1.98e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 1.98e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 5 | 799 | 1.98e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 2 | 188 | 1.99e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 1 | 50 | 1.99e-01 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 1 | 53 | 2.00e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 1 | 38 | 2.02e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 2.03e-01 |
| GO:BP | GO:0007130 | synaptonemal complex assembly | 1 | 10 | 2.03e-01 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 1 | 18 | 2.04e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 1 | 59 | 2.05e-01 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 1 | 56 | 2.06e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 1 | 60 | 2.06e-01 |
| GO:BP | GO:0051955 | regulation of amino acid transport | 1 | 28 | 2.06e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 2 | 194 | 2.06e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 2 | 154 | 2.10e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 1 | 25 | 2.10e-01 |
| GO:BP | GO:0006833 | water transport | 1 | 12 | 2.11e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 1 | 29 | 2.11e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 1 | 40 | 2.11e-01 |
| GO:BP | GO:0060441 | epithelial tube branching involved in lung morphogenesis | 1 | 21 | 2.11e-01 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 1 | 56 | 2.11e-01 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 1 | 59 | 2.11e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 1 | 28 | 2.11e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 1 | 40 | 2.11e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 1 | 593 | 2.11e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 1 | 30 | 2.11e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 5 | 654 | 2.11e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 1 | 618 | 2.11e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 5 | 730 | 2.12e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 22 | 2.12e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 2 | 120 | 2.12e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 1 | 41 | 2.12e-01 |
| GO:BP | GO:0070193 | synaptonemal complex organization | 1 | 13 | 2.12e-01 |
| GO:BP | GO:0021544 | subpallium development | 1 | 19 | 2.12e-01 |
| GO:BP | GO:0003401 | axis elongation | 1 | 25 | 2.12e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 2.12e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 1 | 30 | 2.13e-01 |
| GO:BP | GO:0050890 | cognition | 2 | 226 | 2.13e-01 |
| GO:BP | GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | 1 | 36 | 2.14e-01 |
| GO:BP | GO:0070293 | renal absorption | 1 | 24 | 2.15e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 44 | 2.15e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 2 | 173 | 2.15e-01 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 1 | 25 | 2.15e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 55 | 2.18e-01 |
| GO:BP | GO:0001654 | eye development | 3 | 273 | 2.19e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 1 | 23 | 2.20e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 2.20e-01 |
| GO:BP | GO:0030212 | hyaluronan metabolic process | 1 | 21 | 2.20e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 2 | 177 | 2.20e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 1 | 70 | 2.20e-01 |
| GO:BP | GO:0150063 | visual system development | 3 | 274 | 2.20e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 1 | 45 | 2.20e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 1 | 45 | 2.20e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 1 | 43 | 2.20e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 1 | 17 | 2.20e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 23 | 2.20e-01 |
| GO:BP | GO:0048880 | sensory system development | 3 | 276 | 2.24e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 1 | 258 | 2.24e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 1 | 744 | 2.24e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 1 | 31 | 2.24e-01 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 1 | 69 | 2.24e-01 |
| GO:BP | GO:0048864 | stem cell development | 1 | 75 | 2.29e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 1 | 27 | 2.29e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 2.29e-01 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 1 | 32 | 2.32e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 1 | 32 | 2.32e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 2 | 194 | 2.32e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 1 | 25 | 2.33e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 2 | 160 | 2.33e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 4 | 553 | 2.34e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 36 | 2.35e-01 |
| GO:BP | GO:0006605 | protein targeting | 1 | 310 | 2.36e-01 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 1 | 36 | 2.36e-01 |
| GO:BP | GO:0002931 | response to ischemia | 1 | 48 | 2.36e-01 |
| GO:BP | GO:0060271 | cilium assembly | 1 | 296 | 2.36e-01 |
| GO:BP | GO:0042044 | fluid transport | 1 | 19 | 2.36e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 1 | 79 | 2.36e-01 |
| GO:BP | GO:0051304 | chromosome separation | 1 | 75 | 2.36e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 1 | 25 | 2.36e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 1 | 82 | 2.36e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 5 | 1150 | 2.36e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 2 | 191 | 2.36e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 3 | 316 | 2.36e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 1 | 28 | 2.36e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 21 | 2.36e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 1 | 19 | 2.36e-01 |
| GO:BP | GO:0003008 | system process | 6 | 1194 | 2.38e-01 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 1 | 32 | 2.38e-01 |
| GO:BP | GO:0046942 | carboxylic acid transport | 2 | 213 | 2.39e-01 |
| GO:BP | GO:0140013 | meiotic nuclear division | 2 | 117 | 2.39e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 2 | 490 | 2.39e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 2 | 490 | 2.39e-01 |
| GO:BP | GO:0015849 | organic acid transport | 2 | 214 | 2.39e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 2.40e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 2 | 494 | 2.41e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 1 | 90 | 2.43e-01 |
| GO:BP | GO:0044782 | cilium organization | 1 | 317 | 2.43e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 2 | 197 | 2.43e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 9 | 1832 | 2.45e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 1 | 35 | 2.45e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 1 | 36 | 2.45e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 1 | 35 | 2.45e-01 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 1 | 37 | 2.45e-01 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 1 | 87 | 2.45e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 1 | 37 | 2.45e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 1 | 44 | 2.46e-01 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 1 | 112 | 2.46e-01 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 1 | 37 | 2.46e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 2 | 512 | 2.46e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 1 | 300 | 2.46e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 2 | 1186 | 2.48e-01 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 1 | 87 | 2.48e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 918 | 2.48e-01 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 1 | 88 | 2.48e-01 |
| GO:BP | GO:1902475 | L-alpha-amino acid transmembrane transport | 1 | 51 | 2.48e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 1 | 94 | 2.48e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 1 | 886 | 2.48e-01 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 1 | 91 | 2.48e-01 |
| GO:BP | GO:0060348 | bone development | 2 | 157 | 2.48e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 5 | 1813 | 2.48e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 1 | 96 | 2.48e-01 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 1 | 89 | 2.48e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 1 | 63 | 2.48e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 1 | 41 | 2.50e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 2.50e-01 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 1 | 92 | 2.51e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 1 | 1005 | 2.52e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 1 | 957 | 2.52e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 1 | 185 | 2.52e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 1 | 185 | 2.52e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 2 | 207 | 2.52e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 1 | 46 | 2.53e-01 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 1 | 58 | 2.53e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 1 | 65 | 2.53e-01 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 1 | 37 | 2.53e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 3 | 413 | 2.53e-01 |
| GO:BP | GO:0008037 | cell recognition | 1 | 82 | 2.53e-01 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 211 | 2.53e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 5 | 707 | 2.53e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 1 | 1014 | 2.53e-01 |
| GO:BP | GO:0055092 | sterol homeostasis | 1 | 59 | 2.53e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 1 | 64 | 2.53e-01 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 2 | 132 | 2.53e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 68 | 2.53e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 1 | 39 | 2.53e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 4 | 941 | 2.53e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 3 | 445 | 2.53e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 1 | 35 | 2.53e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 2 | 232 | 2.53e-01 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 1 | 94 | 2.53e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 1 | 75 | 2.53e-01 |
| GO:BP | GO:0000280 | nuclear division | 3 | 343 | 2.53e-01 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 1 | 99 | 2.53e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 1 | 997 | 2.53e-01 |
| GO:BP | GO:0070085 | glycosylation | 1 | 197 | 2.53e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 1 | 84 | 2.53e-01 |
| GO:BP | GO:0032890 | regulation of organic acid transport | 1 | 48 | 2.53e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 1 | 1058 | 2.53e-01 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 1 | 93 | 2.54e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 2 | 155 | 2.54e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 2 | 222 | 2.54e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 2 | 225 | 2.54e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 1 | 1112 | 2.54e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 1 | 87 | 2.54e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 102 | 2.54e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 1 | 68 | 2.54e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 2 | 251 | 2.54e-01 |
| GO:BP | GO:0048675 | axon extension | 1 | 108 | 2.54e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 39 | 2.57e-01 |
| GO:BP | GO:0048468 | cell development | 2 | 1950 | 2.57e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 1 | 88 | 2.57e-01 |
| GO:BP | GO:0008306 | associative learning | 1 | 65 | 2.57e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 2 | 227 | 2.58e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 1 | 51 | 2.58e-01 |
| GO:BP | GO:0048699 | generation of neurons | 1 | 1120 | 2.59e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 1 | 105 | 2.61e-01 |
| GO:BP | GO:0071702 | organic substance transport | 7 | 2133 | 2.61e-01 |
| GO:BP | GO:0050000 | chromosome localization | 1 | 101 | 2.62e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 2 | 214 | 2.63e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 3 | 408 | 2.63e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 1 | 62 | 2.63e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 1 | 39 | 2.65e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 1 | 1203 | 2.66e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 1 | 52 | 2.66e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 1 | 70 | 2.66e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 1 | 72 | 2.66e-01 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 1 | 99 | 2.66e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 2 | 167 | 2.66e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 1 | 117 | 2.69e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 1 | 50 | 2.69e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 1 | 36 | 2.69e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 1 | 95 | 2.69e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 1 | 530 | 2.69e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 1 | 47 | 2.70e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 1 | 73 | 2.70e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 39 | 2.70e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 1 | 1103 | 2.72e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 1 | 158 | 2.73e-01 |
| GO:BP | GO:0051293 | establishment of spindle localization | 1 | 49 | 2.73e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 1 | 81 | 2.73e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 7 | 1662 | 2.73e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 1 | 463 | 2.73e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 2 | 189 | 2.73e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 1 | 61 | 2.73e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 1 | 113 | 2.73e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 2 | 113 | 2.74e-01 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 1 | 24 | 2.74e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 1 | 1298 | 2.76e-01 |
| GO:BP | GO:0022008 | neurogenesis | 1 | 1297 | 2.76e-01 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 1 | 121 | 2.76e-01 |
| GO:BP | GO:0007416 | synapse assembly | 1 | 153 | 2.76e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 1 | 473 | 2.76e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 1 | 71 | 2.76e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 1 | 54 | 2.76e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 3 | 421 | 2.76e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 1 | 54 | 2.76e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 1 | 1305 | 2.76e-01 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 56 | 2.77e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 1 | 119 | 2.78e-01 |
| GO:BP | GO:0048285 | organelle fission | 3 | 387 | 2.78e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 3 | 439 | 2.80e-01 |
| GO:BP | GO:0051668 | localization within membrane | 1 | 611 | 2.80e-01 |
| GO:BP | GO:0051653 | spindle localization | 1 | 53 | 2.80e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 2.80e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 1 | 82 | 2.80e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 2 | 200 | 2.80e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 1 | 52 | 2.80e-01 |
| GO:BP | GO:0007631 | feeding behavior | 1 | 51 | 2.80e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 3 | 429 | 2.82e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 2 | 192 | 2.82e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 2 | 249 | 2.83e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 1 | 130 | 2.83e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 1 | 47 | 2.84e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 1 | 1425 | 2.86e-01 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 1 | 36 | 2.86e-01 |
| GO:BP | GO:0015918 | sterol transport | 1 | 93 | 2.86e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 1 | 57 | 2.86e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 7 | 1318 | 2.87e-01 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 1 | 137 | 2.87e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 1 | 114 | 2.88e-01 |
| GO:BP | GO:0098657 | import into cell | 3 | 664 | 2.92e-01 |
| GO:BP | GO:0015837 | amine transport | 1 | 59 | 2.92e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 79 | 2.92e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 3 | 585 | 2.92e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 2 | 2340 | 2.94e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 1 | 287 | 2.94e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 2 | 261 | 2.94e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 4 | 643 | 2.97e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 1 | 70 | 2.97e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 2 | 212 | 2.97e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 4 | 707 | 2.98e-01 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 1 | 59 | 2.98e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 2 | 179 | 2.98e-01 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 1 | 39 | 2.98e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 1 | 91 | 2.98e-01 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 1 | 32 | 2.98e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 1 | 145 | 2.99e-01 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 1 | 99 | 2.99e-01 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 1 | 35 | 3.00e-01 |
| GO:BP | GO:0061061 | muscle structure development | 2 | 552 | 3.00e-01 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 1 | 61 | 3.00e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 2 | 211 | 3.00e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 149 | 3.00e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 64 | 3.00e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 1 | 1589 | 3.00e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 1 | 328 | 3.00e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 3 | 392 | 3.00e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 2 | 315 | 3.01e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 1 | 48 | 3.01e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 1 | 45 | 3.02e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 1 | 866 | 3.02e-01 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 2 | 302 | 3.04e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 1 | 81 | 3.04e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 2 | 186 | 3.04e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 1 | 803 | 3.04e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 4 | 570 | 3.04e-01 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 1 | 106 | 3.04e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 1 | 161 | 3.04e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 1 | 160 | 3.04e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 1 | 59 | 3.04e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 1 | 102 | 3.04e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 1 | 53 | 3.04e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 1 | 61 | 3.04e-01 |
| GO:BP | GO:0007015 | actin filament organization | 1 | 372 | 3.04e-01 |
| GO:BP | GO:0008015 | blood circulation | 2 | 372 | 3.04e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 2 | 395 | 3.04e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 1 | 114 | 3.04e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 1 | 1568 | 3.09e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 1 | 49 | 3.09e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 2 | 183 | 3.11e-01 |
| GO:BP | GO:0048870 | cell motility | 2 | 1169 | 3.12e-01 |
| GO:BP | GO:0016125 | sterol metabolic process | 1 | 108 | 3.12e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 2 | 592 | 3.13e-01 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 1 | 93 | 3.14e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 1 | 67 | 3.22e-01 |
| GO:BP | GO:0007423 | sensory organ development | 3 | 395 | 3.22e-01 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 1 | 165 | 3.22e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 2 | 182 | 3.23e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 4 | 741 | 3.23e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 165 | 3.23e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 4 | 1014 | 3.23e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 2 | 237 | 3.23e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 1 | 116 | 3.25e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 1 | 110 | 3.25e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 1 | 75 | 3.25e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 2 | 264 | 3.28e-01 |
| GO:BP | GO:0035601 | protein deacylation | 1 | 91 | 3.28e-01 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 1 | 178 | 3.28e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 2 | 232 | 3.28e-01 |
| GO:BP | GO:0007049 | cell cycle | 7 | 1515 | 3.29e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 1 | 1764 | 3.29e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 1 | 92 | 3.29e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 3 | 544 | 3.29e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 2 | 288 | 3.31e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 1 | 98 | 3.33e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 1 | 98 | 3.33e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 1 | 143 | 3.33e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 184 | 3.33e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 1 | 98 | 3.34e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 1 | 1852 | 3.36e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 2 | 375 | 3.36e-01 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 1 | 174 | 3.36e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 2 | 1496 | 3.36e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 13 | 2943 | 3.36e-01 |
| GO:BP | GO:0007399 | nervous system development | 1 | 1886 | 3.36e-01 |
| GO:BP | GO:0030154 | cell differentiation | 13 | 2942 | 3.36e-01 |
| GO:BP | GO:0007612 | learning | 1 | 114 | 3.36e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 194 | 3.37e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 298 | 3.38e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 3 | 549 | 3.38e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 1 | 58 | 3.38e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 4 | 762 | 3.39e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 177 | 3.43e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1 | 1896 | 3.43e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 1 | 177 | 3.43e-01 |
| GO:BP | GO:1903510 | mucopolysaccharide metabolic process | 1 | 75 | 3.43e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 1 | 140 | 3.43e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 2 | 237 | 3.43e-01 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 1 | 51 | 3.43e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 1 | 392 | 3.44e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 1 | 71 | 3.44e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 2 | 221 | 3.44e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 1 | 102 | 3.45e-01 |
| GO:BP | GO:0048589 | developmental growth | 3 | 522 | 3.46e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1 | 1972 | 3.46e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 1 | 196 | 3.46e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 80 | 3.46e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 79 | 3.46e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 1 | 1857 | 3.46e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 1 | 204 | 3.46e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 1 | 72 | 3.46e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 1 | 72 | 3.46e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 1 | 1958 | 3.46e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 1 | 90 | 3.46e-01 |
| GO:BP | GO:0070925 | organelle assembly | 1 | 828 | 3.46e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 1 | 86 | 3.46e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 1 | 133 | 3.46e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 1 | 87 | 3.48e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 1 | 1669 | 3.48e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1 | 68 | 3.52e-01 |
| GO:BP | GO:0070848 | response to growth factor | 3 | 571 | 3.52e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 1 | 81 | 3.52e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 1 | 1708 | 3.52e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 1 | 59 | 3.53e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 1 | 96 | 3.53e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 1 | 1721 | 3.53e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 1 | 64 | 3.57e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 1 | 75 | 3.58e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 209 | 3.58e-01 |
| GO:BP | GO:0000819 | sister chromatid segregation | 1 | 217 | 3.59e-01 |
| GO:BP | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 1 | 78 | 3.59e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 1 | 76 | 3.59e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 1 | 76 | 3.59e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 1 | 119 | 3.61e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 9 | 2036 | 3.65e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 1 | 190 | 3.66e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 1 | 114 | 3.66e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1 | 2052 | 3.66e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 1 | 85 | 3.66e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 1 | 171 | 3.67e-01 |
| GO:BP | GO:0001823 | mesonephros development | 1 | 77 | 3.67e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 1 | 566 | 3.76e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 1 | 2367 | 3.76e-01 |
| GO:BP | GO:0030203 | glycosaminoglycan metabolic process | 1 | 87 | 3.77e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 1 | 343 | 3.79e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 1 | 344 | 3.79e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 1 | 132 | 3.80e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 1 | 235 | 3.80e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 2 | 271 | 3.82e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 1 | 153 | 3.84e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 234 | 3.86e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 3 | 684 | 3.90e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 1 | 2069 | 3.91e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 1 | 108 | 3.91e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 1 | 99 | 3.94e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 1 | 89 | 3.94e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 3 | 439 | 3.94e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 3.94e-01 |
| GO:BP | GO:0006022 | aminoglycan metabolic process | 1 | 92 | 3.95e-01 |
| GO:BP | GO:0006936 | muscle contraction | 1 | 266 | 3.95e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 2 | 358 | 3.97e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1 | 2131 | 3.97e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 1 | 139 | 3.97e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 1 | 84 | 3.97e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 1 | 254 | 3.99e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 1 | 80 | 3.99e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 1 | 135 | 3.99e-01 |
| GO:BP | GO:0015031 | protein transport | 1 | 1366 | 4.00e-01 |
| GO:BP | GO:0046907 | intracellular transport | 1 | 1440 | 4.00e-01 |
| GO:BP | GO:0030073 | insulin secretion | 1 | 144 | 4.01e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 1 | 643 | 4.01e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 1 | 137 | 4.01e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 1 | 2518 | 4.01e-01 |
| GO:BP | GO:0140014 | mitotic nuclear division | 1 | 251 | 4.01e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 4.01e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 4.01e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 1 | 139 | 4.01e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 1 | 2560 | 4.01e-01 |
| GO:BP | GO:0034502 | protein localization to chromosome | 1 | 101 | 4.02e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 1 | 82 | 4.02e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 2 | 347 | 4.02e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 1 | 2577 | 4.02e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1 | 2607 | 4.04e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 2 | 317 | 4.05e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 4 | 744 | 4.05e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 1 | 97 | 4.05e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 5 | 1092 | 4.05e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 1 | 262 | 4.05e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 1 | 347 | 4.06e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 1 | 146 | 4.07e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 211 | 4.07e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 1 | 666 | 4.07e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1 | 2297 | 4.07e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 1 | 2669 | 4.07e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 1 | 1468 | 4.08e-01 |
| GO:BP | GO:0051656 | establishment of organelle localization | 2 | 397 | 4.08e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 1 | 2699 | 4.10e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 2 | 360 | 4.10e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 5 | 1267 | 4.12e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 271 | 4.12e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 1 | 214 | 4.15e-01 |
| GO:BP | GO:0007127 | meiosis I | 1 | 73 | 4.15e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 2 | 275 | 4.18e-01 |
| GO:BP | GO:0048565 | digestive tract development | 1 | 92 | 4.19e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 1 | 2824 | 4.22e-01 |
| GO:BP | GO:0006810 | transport | 2 | 3294 | 4.24e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 1 | 187 | 4.24e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 373 | 4.24e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 5 | 1124 | 4.24e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1 | 2846 | 4.25e-01 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 1 | 77 | 4.26e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 1 | 132 | 4.27e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 1 | 187 | 4.27e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 1 | 306 | 4.27e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 4 | 1017 | 4.28e-01 |
| GO:BP | GO:0040011 | locomotion | 4 | 854 | 4.28e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 3 | 759 | 4.28e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 1 | 121 | 4.28e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 9 | 2560 | 4.30e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 1 | 181 | 4.30e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 1 | 174 | 4.30e-01 |
| GO:BP | GO:0006950 | response to stress | 1 | 2763 | 4.30e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 2 | 296 | 4.31e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 9 | 2567 | 4.33e-01 |
| GO:BP | GO:0055123 | digestive system development | 1 | 100 | 4.33e-01 |
| GO:BP | GO:0048731 | system development | 1 | 2886 | 4.33e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 2 | 314 | 4.33e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 1 | 116 | 4.33e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 1 | 305 | 4.33e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 1 | 116 | 4.33e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 1 | 169 | 4.33e-01 |
| GO:BP | GO:0008033 | tRNA processing | 1 | 131 | 4.33e-01 |
| GO:BP | GO:0042552 | myelination | 1 | 114 | 4.33e-01 |
| GO:BP | GO:0042221 | response to chemical | 1 | 2456 | 4.33e-01 |
| GO:BP | GO:0003012 | muscle system process | 1 | 334 | 4.33e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 97 | 4.33e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 1 | 134 | 4.34e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 1 | 3054 | 4.34e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1 | 3059 | 4.35e-01 |
| GO:BP | GO:0051234 | establishment of localization | 2 | 3453 | 4.35e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 1 | 197 | 4.35e-01 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 1 | 123 | 4.35e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 1 | 113 | 4.35e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 1 | 113 | 4.35e-01 |
| GO:BP | GO:0002790 | peptide secretion | 1 | 173 | 4.35e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 1 | 3115 | 4.38e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 1 | 163 | 4.38e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 1 | 3116 | 4.38e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 1 | 113 | 4.38e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1 | 1203 | 4.38e-01 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 1 | 157 | 4.39e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 1 | 171 | 4.40e-01 |
| GO:BP | GO:0051179 | localization | 4 | 3983 | 4.40e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 1 | 124 | 4.42e-01 |
| GO:BP | GO:0007611 | learning or memory | 1 | 201 | 4.42e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 1 | 1689 | 4.43e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 3 | 537 | 4.44e-01 |
| GO:BP | GO:0030030 | cell projection organization | 1 | 1236 | 4.44e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 1 | 310 | 4.44e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 1 | 186 | 4.46e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 1 | 3221 | 4.46e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 203 | 4.49e-01 |
| GO:BP | GO:0015833 | peptide transport | 1 | 182 | 4.50e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 1 | 352 | 4.50e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 1 | 254 | 4.53e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 291 | 4.53e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 1 | 248 | 4.53e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1842 | 4.53e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 1 | 180 | 4.54e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 1 | 141 | 4.57e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 3 | 554 | 4.59e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 111 | 4.61e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 1 | 156 | 4.61e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 374 | 4.61e-01 |
| GO:BP | GO:0009888 | tissue development | 6 | 1425 | 4.61e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 1 | 122 | 4.61e-01 |
| GO:BP | GO:0007409 | axonogenesis | 1 | 350 | 4.61e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 1 | 114 | 4.61e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 1 | 337 | 4.61e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 1 | 719 | 4.61e-01 |
| GO:BP | GO:0030324 | lung development | 1 | 144 | 4.61e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 1 | 143 | 4.61e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 1 | 148 | 4.66e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 1 | 147 | 4.66e-01 |
| GO:BP | GO:0030001 | metal ion transport | 2 | 568 | 4.66e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 1 | 3355 | 4.66e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 1 | 251 | 4.66e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 4 | 1088 | 4.66e-01 |
| GO:BP | GO:0051235 | maintenance of location | 1 | 263 | 4.66e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 1 | 274 | 4.66e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 4 | 1090 | 4.66e-01 |
| GO:BP | GO:0006935 | chemotaxis | 1 | 264 | 4.66e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 1 | 364 | 4.66e-01 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 1 | 144 | 4.66e-01 |
| GO:BP | GO:0042330 | taxis | 1 | 264 | 4.66e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 1 | 781 | 4.66e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 4 | 1096 | 4.67e-01 |
| GO:BP | GO:0032879 | regulation of localization | 4 | 1570 | 4.70e-01 |
| GO:BP | GO:0008104 | protein localization | 2 | 2128 | 4.72e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2 | 2137 | 4.73e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 5 | 1116 | 4.73e-01 |
| GO:BP | GO:0001501 | skeletal system development | 2 | 361 | 4.76e-01 |
| GO:BP | GO:0048839 | inner ear development | 1 | 129 | 4.76e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 1 | 155 | 4.79e-01 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 2 | 711 | 4.79e-01 |
| GO:BP | GO:0048736 | appendage development | 1 | 141 | 4.83e-01 |
| GO:BP | GO:0060173 | limb development | 1 | 141 | 4.83e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 1 | 3743 | 4.83e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 1 | 263 | 4.83e-01 |
| GO:BP | GO:0046879 | hormone secretion | 1 | 212 | 4.83e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 1 | 241 | 4.84e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 1 | 275 | 4.84e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 2 | 582 | 4.84e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 1 | 387 | 4.85e-01 |
| GO:BP | GO:0016049 | cell growth | 1 | 416 | 4.85e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 1 | 805 | 4.85e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 1 | 120 | 4.85e-01 |
| GO:BP | GO:0060541 | respiratory system development | 1 | 161 | 4.86e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 1 | 223 | 4.87e-01 |
| GO:BP | GO:0061564 | axon development | 1 | 388 | 4.89e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 152 | 4.89e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 1 | 224 | 4.89e-01 |
| GO:BP | GO:0009914 | hormone transport | 1 | 217 | 4.89e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 3 | 758 | 4.89e-01 |
| GO:BP | GO:0051640 | organelle localization | 2 | 521 | 4.90e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 1 | 142 | 4.90e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 1 | 883 | 4.90e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 3 | 870 | 4.90e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 1 | 411 | 4.91e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 2 | 473 | 4.91e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 9 | 4807 | 4.91e-01 |
| GO:BP | GO:0040007 | growth | 3 | 737 | 4.94e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 2 | 456 | 4.95e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 1 | 288 | 4.96e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 1 | 171 | 4.96e-01 |
| GO:BP | GO:0016477 | cell migration | 2 | 1065 | 4.96e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 1 | 906 | 4.97e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 1 | 841 | 5.00e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 2 | 684 | 5.02e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 2 | 369 | 5.02e-01 |
| GO:BP | GO:0043583 | ear development | 1 | 150 | 5.06e-01 |
| GO:BP | GO:0051216 | cartilage development | 1 | 149 | 5.06e-01 |
| GO:BP | GO:0006996 | organelle organization | 2 | 2974 | 5.09e-01 |
| GO:BP | GO:0042886 | amide transport | 1 | 237 | 5.12e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 1 | 189 | 5.12e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 2 | 4100 | 5.12e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 1 | 195 | 5.12e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 2 | 699 | 5.12e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 1 | 156 | 5.12e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 1 | 159 | 5.13e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 2 | 461 | 5.13e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 2 | 703 | 5.14e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 1 | 3587 | 5.15e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 1 | 451 | 5.15e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 586 | 5.15e-01 |
| GO:BP | GO:0009306 | protein secretion | 1 | 262 | 5.15e-01 |
| GO:BP | GO:0051098 | regulation of binding | 1 | 314 | 5.15e-01 |
| GO:BP | GO:0006869 | lipid transport | 1 | 271 | 5.15e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 1 | 438 | 5.15e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 1 | 262 | 5.15e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1 | 3773 | 5.15e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 2 | 542 | 5.20e-01 |
| GO:BP | GO:0007507 | heart development | 2 | 512 | 5.20e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 1 | 4243 | 5.20e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 1 | 266 | 5.20e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 2 | 650 | 5.21e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 1 | 126 | 5.21e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1 | 3702 | 5.22e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 1 | 2448 | 5.24e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 1 | 202 | 5.25e-01 |
| GO:BP | GO:0022402 | cell cycle process | 4 | 1078 | 5.25e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1 | 4197 | 5.27e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 1 | 860 | 5.27e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 2 | 677 | 5.27e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 1 | 3794 | 5.30e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 1 | 4371 | 5.30e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1 | 3820 | 5.32e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 1 | 1003 | 5.33e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 1 | 176 | 5.33e-01 |
| GO:BP | GO:0040008 | regulation of growth | 1 | 493 | 5.33e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 2 | 438 | 5.33e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 1 | 276 | 5.34e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 3 | 841 | 5.34e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1 | 4026 | 5.39e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 1 | 379 | 5.42e-01 |
| GO:BP | GO:0036211 | protein modification process | 1 | 2751 | 5.42e-01 |
| GO:BP | GO:0010876 | lipid localization | 1 | 309 | 5.42e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 1 | 500 | 5.42e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 1 | 496 | 5.42e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 1 | 380 | 5.42e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 1 | 529 | 5.42e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 1 | 217 | 5.42e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 2 | 548 | 5.47e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 1 | 515 | 5.47e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1 | 157 | 5.49e-01 |
| GO:BP | GO:0006417 | regulation of translation | 1 | 373 | 5.49e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 1 | 519 | 5.49e-01 |
| GO:BP | GO:0042592 | homeostatic process | 5 | 1189 | 5.51e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 4 | 1097 | 5.53e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 3 | 725 | 5.53e-01 |
| GO:BP | GO:0009314 | response to radiation | 1 | 339 | 5.53e-01 |
| GO:BP | GO:0051049 | regulation of transport | 3 | 1268 | 5.53e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 7 | 2271 | 5.53e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 1 | 200 | 5.53e-01 |
| GO:BP | GO:0032502 | developmental process | 2 | 4445 | 5.55e-01 |
| GO:BP | GO:0051276 | chromosome organization | 2 | 533 | 5.55e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 1 | 539 | 5.55e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 1 | 202 | 5.55e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1 | 4566 | 5.55e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 5 | 1249 | 5.57e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 5 | 1252 | 5.57e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 1 | 2948 | 5.57e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 1 | 157 | 5.57e-01 |
| GO:BP | GO:0051641 | cellular localization | 1 | 2822 | 5.59e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 1 | 412 | 5.60e-01 |
| GO:BP | GO:0001568 | blood vessel development | 2 | 537 | 5.60e-01 |
| GO:BP | GO:0006915 | apoptotic process | 5 | 1452 | 5.68e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 1 | 599 | 5.68e-01 |
| GO:BP | GO:0035295 | tube development | 3 | 822 | 5.68e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1151 | 5.68e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 1 | 1215 | 5.75e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 1 | 242 | 5.78e-01 |
| GO:BP | GO:0021537 | telencephalon development | 1 | 208 | 5.78e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 3 | 689 | 5.78e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 2 | 689 | 5.78e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 3 | 1386 | 5.78e-01 |
| GO:BP | GO:0001944 | vasculature development | 2 | 562 | 5.79e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 1 | 206 | 5.79e-01 |
| GO:BP | GO:0061448 | connective tissue development | 1 | 212 | 5.83e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 1 | 253 | 5.83e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 1 | 426 | 5.83e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 1 | 239 | 5.83e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 1 | 4563 | 5.83e-01 |
| GO:BP | GO:0090132 | epithelium migration | 1 | 253 | 5.84e-01 |
| GO:BP | GO:0072359 | circulatory system development | 3 | 888 | 5.85e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 1 | 4593 | 5.85e-01 |
| GO:BP | GO:0023061 | signal release | 1 | 329 | 5.85e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 1 | 788 | 5.88e-01 |
| GO:BP | GO:0090130 | tissue migration | 1 | 257 | 5.89e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 628 | 5.89e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 1 | 188 | 5.89e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 1 | 254 | 5.90e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 2 | 5148 | 5.92e-01 |
| GO:BP | GO:0012501 | programmed cell death | 5 | 1498 | 5.93e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 1 | 627 | 5.93e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 1 | 3573 | 5.93e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 1 | 233 | 5.94e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 3 | 949 | 5.94e-01 |
| GO:BP | GO:0008219 | cell death | 5 | 1504 | 5.94e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 1 | 234 | 5.94e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 3 | 950 | 5.94e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 1 | 234 | 5.94e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 1 | 276 | 5.97e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 1 | 287 | 5.97e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 1 | 404 | 5.98e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 1 | 423 | 5.98e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 3 | 1102 | 5.99e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 6 | 1905 | 6.07e-01 |
| GO:BP | GO:0007420 | brain development | 2 | 546 | 6.08e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 1 | 274 | 6.10e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 1 | 622 | 6.12e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 1 | 428 | 6.13e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 1 | 4998 | 6.14e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 2 | 664 | 6.15e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 1 | 337 | 6.17e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 1 | 271 | 6.18e-01 |
| GO:BP | GO:0044057 | regulation of system process | 1 | 398 | 6.20e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4929 | 6.20e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 1 | 680 | 6.21e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1 | 4018 | 6.25e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 2 | 544 | 6.25e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 3 | 1074 | 6.25e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 1 | 382 | 6.26e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 1 | 208 | 6.27e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 1 | 256 | 6.29e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 1 | 210 | 6.29e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 268 | 6.30e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 1 | 1320 | 6.32e-01 |
| GO:BP | GO:0060322 | head development | 2 | 588 | 6.35e-01 |
| GO:BP | GO:0060429 | epithelium development | 3 | 849 | 6.35e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 1 | 432 | 6.35e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 2 | 663 | 6.49e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 4 | 1260 | 6.49e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 1 | 221 | 6.51e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 740 | 6.53e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 2 | 815 | 6.55e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 1 | 413 | 6.55e-01 |
| GO:BP | GO:0007610 | behavior | 1 | 439 | 6.55e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1 | 4423 | 6.55e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 1 | 434 | 6.59e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 1 | 233 | 6.61e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 5 | 1895 | 6.62e-01 |
| GO:BP | GO:0031175 | neuron projection development | 1 | 774 | 6.63e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 2 | 989 | 6.65e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 1 | 4537 | 6.66e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 2 | 5813 | 6.67e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 1 | 4561 | 6.69e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 1 | 329 | 6.69e-01 |
| GO:BP | GO:0048513 | animal organ development | 7 | 2156 | 6.70e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 2 | 680 | 6.71e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 1 | 520 | 6.71e-01 |
| GO:BP | GO:0034097 | response to cytokine | 2 | 609 | 6.71e-01 |
| GO:BP | GO:0007600 | sensory perception | 2 | 259 | 6.71e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 2 | 5875 | 6.73e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 2 | 741 | 6.78e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 2 | 5912 | 6.78e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 1 | 4713 | 6.82e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 690 | 6.83e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 8 | 2877 | 6.84e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 318 | 6.86e-01 |
| GO:BP | GO:0030900 | forebrain development | 1 | 290 | 6.88e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 3 | 1130 | 6.89e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 1 | 4202 | 6.91e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 1 | 4913 | 6.91e-01 |
| GO:BP | GO:0023052 | signaling | 10 | 4134 | 6.97e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 1 | 1731 | 7.05e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 1 | 1964 | 7.07e-01 |
| GO:BP | GO:0007155 | cell adhesion | 1 | 996 | 7.07e-01 |
| GO:BP | GO:0048666 | neuron development | 1 | 873 | 7.07e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 1 | 292 | 7.08e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 1 | 319 | 7.08e-01 |
| GO:BP | GO:0050896 | response to stimulus | 1 | 5721 | 7.12e-01 |
| GO:BP | GO:0006412 | translation | 1 | 657 | 7.18e-01 |
| GO:BP | GO:0007154 | cell communication | 10 | 4210 | 7.19e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 1 | 397 | 7.21e-01 |
| GO:BP | GO:0007417 | central nervous system development | 1 | 750 | 7.22e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 1 | 886 | 7.23e-01 |
| GO:BP | GO:0010467 | gene expression | 1 | 4733 | 7.26e-01 |
| GO:BP | GO:0006325 | chromatin organization | 1 | 631 | 7.29e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 1 | 677 | 7.29e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 1 | 415 | 7.30e-01 |
| GO:BP | GO:0001525 | angiogenesis | 1 | 389 | 7.33e-01 |
| GO:BP | GO:0032940 | secretion by cell | 1 | 573 | 7.43e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 5 | 1557 | 7.43e-01 |
| GO:BP | GO:0033993 | response to lipid | 1 | 594 | 7.60e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 1 | 1001 | 7.60e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 1 | 634 | 7.60e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2459 | 7.61e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 1 | 815 | 7.61e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 1 | 4919 | 7.62e-01 |
| GO:BP | GO:0140352 | export from cell | 1 | 617 | 7.67e-01 |
| GO:BP | GO:0009056 | catabolic process | 1 | 2108 | 7.67e-01 |
| GO:BP | GO:0016310 | phosphorylation | 3 | 1338 | 7.79e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 1 | 785 | 7.82e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 470 | 7.82e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2722 | 7.83e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 2 | 1981 | 7.83e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 1 | 364 | 7.84e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 1 | 355 | 7.88e-01 |
| GO:BP | GO:0046903 | secretion | 1 | 647 | 7.88e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 1 | 369 | 7.88e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 1 | 501 | 7.88e-01 |
| GO:BP | GO:0009790 | embryo development | 2 | 859 | 7.91e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 3 | 1349 | 7.92e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 1 | 736 | 8.03e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 1 | 449 | 8.03e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 1 | 465 | 8.03e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 1 | 410 | 8.26e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 2 | 7431 | 8.29e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 1 | 475 | 8.29e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 1 | 416 | 8.29e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 1 | 561 | 8.30e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 1 | 511 | 8.31e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 1 | 848 | 8.33e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 3 | 2085 | 8.40e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 2 | 7084 | 8.54e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 1 | 386 | 8.54e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 1 | 559 | 8.56e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 1 | 535 | 8.56e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 1 | 559 | 8.56e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 1 | 1132 | 8.56e-01 |
| GO:BP | GO:0006974 | DNA damage response | 1 | 795 | 8.67e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 1 | 498 | 8.74e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 1 | 662 | 8.84e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 1 | 664 | 8.86e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 1 | 7632 | 8.90e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 1 | 685 | 8.90e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 2 | 7788 | 8.93e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 3 | 1390 | 8.93e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 4 | 1889 | 8.95e-01 |
| GO:BP | GO:0009725 | response to hormone | 1 | 624 | 8.98e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 1 | 583 | 9.06e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 1 | 802 | 9.10e-01 |
| GO:BP | GO:0007165 | signal transduction | 11 | 3803 | 9.12e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 4 | 1952 | 9.20e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 2 | 7562 | 9.21e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 1 | 8017 | 9.25e-01 |
| GO:BP | GO:0016043 | cellular component organization | 2 | 5106 | 9.27e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 2 | 1635 | 9.30e-01 |
| GO:BP | GO:0010033 | response to organic substance | 4 | 1942 | 9.31e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1 | 1219 | 9.34e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 2 | 5307 | 9.35e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 1 | 868 | 9.40e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 795 | 9.41e-01 |
| GO:BP | GO:0065007 | biological regulation | 1 | 8282 | 9.50e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 3 | 2096 | 9.56e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 3 | 2118 | 9.58e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 288 | 9.63e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 288 | 9.63e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 292 | 9.63e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 1 | 1067 | 9.63e-01 |
| GO:BP | GO:0008380 | RNA splicing | 2 | 426 | 9.64e-01 |
| GO:BP | GO:0006397 | mRNA processing | 2 | 451 | 9.67e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 2 | 671 | 9.84e-01 |
| GO:BP | GO:0009987 | cellular process | 1 | 11410 | 1.00e+00 |
| GO:BP | GO:0006396 | RNA processing | 1 | 928 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12027 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8522 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8176 | 1.00e+00 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 4 | 128 | 8.80e-03 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 3 | 67 | 1.20e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 3 | 87 | 1.53e-02 |
| KEGG | KEGG:05216 | Thyroid cancer | 2 | 35 | 1.53e-02 |
| KEGG | KEGG:05226 | Gastric cancer | 3 | 117 | 2.09e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 2 | 75 | 2.09e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 2 | 57 | 2.09e-02 |
| KEGG | KEGG:05214 | Glioma | 2 | 67 | 2.09e-02 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 2.09e-02 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 2 | 49 | 2.09e-02 |
| KEGG | KEGG:04115 | p53 signaling pathway | 2 | 65 | 2.09e-02 |
| KEGG | KEGG:05218 | Melanoma | 2 | 58 | 2.09e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 2 | 75 | 2.09e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 2 | 84 | 2.29e-02 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 2 | 71 | 2.96e-02 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 2 | 109 | 4.44e-02 |
| KEGG | KEGG:04210 | Apoptosis | 2 | 118 | 4.44e-02 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 4.62e-02 |
| KEGG | KEGG:05224 | Breast cancer | 2 | 117 | 4.66e-02 |
| KEGG | KEGG:04110 | Cell cycle | 2 | 151 | 4.77e-02 |
| KEGG | KEGG:04218 | Cellular senescence | 2 | 145 | 4.77e-02 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 2 | 145 | 5.04e-02 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 2 | 153 | 6.66e-02 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 7.37e-02 |
| KEGG | KEGG:05016 | Huntington disease | 1 | 256 | 1.00e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 1 | 303 | 1.14e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 2 | 240 | 1.14e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 1 | 56 | 1.22e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 1 | 385 | 1.32e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 69 | 1.56e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 3 | 409 | 1.56e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 1.77e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 1 | 108 | 3.11e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 1 | 108 | 4.34e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 5.30e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 5.41e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 1 | 415 | 6.12e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5548 | 6.12e-01 |
TRZnomtable %>%
filter(source=="KEGG") %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "Enrichment of KEGG terms using nominal pvalue TRZ genes in my data") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 4 | 128 | 8.80e-03 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 3 | 67 | 1.20e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 3 | 87 | 1.53e-02 |
| KEGG | KEGG:05216 | Thyroid cancer | 2 | 35 | 1.53e-02 |
| KEGG | KEGG:05226 | Gastric cancer | 3 | 117 | 2.09e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 2 | 75 | 2.09e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 2 | 57 | 2.09e-02 |
| KEGG | KEGG:05214 | Glioma | 2 | 67 | 2.09e-02 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 2.09e-02 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 2 | 49 | 2.09e-02 |
| KEGG | KEGG:04115 | p53 signaling pathway | 2 | 65 | 2.09e-02 |
| KEGG | KEGG:05218 | Melanoma | 2 | 58 | 2.09e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 2 | 75 | 2.09e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 2 | 84 | 2.29e-02 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 2 | 71 | 2.96e-02 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 2 | 109 | 4.44e-02 |
| KEGG | KEGG:04210 | Apoptosis | 2 | 118 | 4.44e-02 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 4.62e-02 |
| KEGG | KEGG:05224 | Breast cancer | 2 | 117 | 4.66e-02 |
| KEGG | KEGG:04110 | Cell cycle | 2 | 151 | 4.77e-02 |
| KEGG | KEGG:04218 | Cellular senescence | 2 | 145 | 4.77e-02 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 2 | 145 | 5.04e-02 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 2 | 153 | 6.66e-02 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 7.37e-02 |
| KEGG | KEGG:05016 | Huntington disease | 1 | 256 | 1.00e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 1 | 303 | 1.14e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 2 | 240 | 1.14e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 1 | 56 | 1.22e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 1 | 385 | 1.32e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 69 | 1.56e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 3 | 409 | 1.56e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 1.77e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 1 | 108 | 3.11e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 1 | 108 | 4.34e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 5.30e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 5.41e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 1 | 415 | 6.12e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5548 | 6.12e-01 |
TRZ 3 hour nom de
# TRZresgenes3h <- gost(query = TRZ_list3h$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(TRZresgenes3h,"data/DEG-GO/TRZresgenes3h.RDS")
TRZresgenes3h <- readRDS("data/DEG-GO/TRZresgenes3h.RDS")
TRZnom3h <- gostplot(TRZresgenes3h, capped = FALSE, interactive = TRUE)
TRZnom3hTRZnomtable3h <- TRZresgenes3h$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
TRZnomtable3h %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 2 | 22 | 6.71e-03 |
| GO:BP | GO:1902746 | regulation of lens fiber cell differentiation | 2 | 8 | 6.71e-03 |
| GO:BP | GO:1902747 | negative regulation of lens fiber cell differentiation | 2 | 5 | 6.71e-03 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 2 | 72 | 8.98e-03 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 2 | 32 | 8.98e-03 |
| GO:BP | GO:0038066 | p38MAPK cascade | 2 | 42 | 1.06e-02 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 2 | 15 | 1.06e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 2 | 115 | 1.12e-02 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 2 | 116 | 1.12e-02 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 2 | 127 | 1.25e-02 |
| GO:BP | GO:0001659 | temperature homeostasis | 2 | 135 | 1.35e-02 |
| GO:BP | GO:0097167 | circadian regulation of translation | 1 | 4 | 1.51e-02 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 2 | 29 | 1.51e-02 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 4 | 396 | 1.51e-02 |
| GO:BP | GO:0019249 | lactate biosynthetic process | 1 | 3 | 1.51e-02 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 3 | 139 | 1.51e-02 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 2 | 71 | 1.51e-02 |
| GO:BP | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission | 1 | 3 | 1.51e-02 |
| GO:BP | GO:0051946 | regulation of glutamate uptake involved in transmission of nerve impulse | 1 | 3 | 1.51e-02 |
| GO:BP | GO:0070345 | negative regulation of fat cell proliferation | 1 | 4 | 1.51e-02 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 2 | 30 | 1.61e-02 |
| GO:BP | GO:0051935 | glutamate reuptake | 1 | 4 | 1.65e-02 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 2 | 97 | 1.74e-02 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 2 | 95 | 1.74e-02 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 2 | 35 | 1.74e-02 |
| GO:BP | GO:0002036 | regulation of L-glutamate import across plasma membrane | 1 | 6 | 1.86e-02 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 2 | 113 | 1.90e-02 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 2 | 31 | 2.01e-02 |
| GO:BP | GO:0051933 | amino acid neurotransmitter reuptake | 1 | 5 | 2.01e-02 |
| GO:BP | GO:0070344 | regulation of fat cell proliferation | 1 | 8 | 2.01e-02 |
| GO:BP | GO:0042754 | negative regulation of circadian rhythm | 1 | 8 | 2.27e-02 |
| GO:BP | GO:0045210 | FasL biosynthetic process | 1 | 1 | 2.27e-02 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 3 | 394 | 2.35e-02 |
| GO:BP | GO:0007254 | JNK cascade | 2 | 137 | 2.35e-02 |
| GO:BP | GO:1903789 | regulation of amino acid transmembrane transport | 1 | 14 | 2.39e-02 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 3 | 205 | 2.39e-02 |
| GO:BP | GO:0070341 | fat cell proliferation | 1 | 11 | 2.39e-02 |
| GO:BP | GO:0010958 | regulation of amino acid import across plasma membrane | 1 | 14 | 2.39e-02 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 2 | 157 | 2.39e-02 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 3 | 403 | 2.39e-02 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 2 | 153 | 2.39e-02 |
| GO:BP | GO:0098712 | L-glutamate import across plasma membrane | 1 | 11 | 2.44e-02 |
| GO:BP | GO:0050872 | white fat cell differentiation | 1 | 15 | 2.44e-02 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 2 | 58 | 2.44e-02 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 2 | 54 | 2.44e-02 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 2.44e-02 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 2.44e-02 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 4 | 473 | 2.47e-02 |
| GO:BP | GO:0006089 | lactate metabolic process | 1 | 16 | 2.47e-02 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 3 | 265 | 2.73e-02 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 1 | 23 | 2.79e-02 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 2 | 188 | 2.79e-02 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 2 | 194 | 2.89e-02 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 2 | 76 | 2.97e-02 |
| GO:BP | GO:0009648 | photoperiodism | 1 | 24 | 2.97e-02 |
| GO:BP | GO:0015813 | L-glutamate transmembrane transport | 1 | 18 | 2.99e-02 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 1 | 27 | 2.99e-02 |
| GO:BP | GO:0051726 | regulation of cell cycle | 3 | 941 | 2.99e-02 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 3 | 290 | 2.99e-02 |
| GO:BP | GO:0060485 | mesenchyme development | 3 | 250 | 3.09e-02 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 2 | 57 | 3.09e-02 |
| GO:BP | GO:1900623 | regulation of monocyte aggregation | 1 | 3 | 3.11e-02 |
| GO:BP | GO:0045226 | extracellular polysaccharide biosynthetic process | 1 | 2 | 3.11e-02 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 1 | 21 | 3.11e-02 |
| GO:BP | GO:0046379 | extracellular polysaccharide metabolic process | 1 | 2 | 3.11e-02 |
| GO:BP | GO:0051938 | L-glutamate import | 1 | 24 | 3.11e-02 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 2 | 85 | 3.11e-02 |
| GO:BP | GO:1900625 | positive regulation of monocyte aggregation | 1 | 3 | 3.11e-02 |
| GO:BP | GO:0000165 | MAPK cascade | 4 | 553 | 3.11e-02 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 2 | 86 | 3.11e-02 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 2 | 87 | 3.20e-02 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 3 | 335 | 3.45e-02 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 1 | 40 | 3.45e-02 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 2 | 96 | 3.45e-02 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 3 | 334 | 3.45e-02 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 1 | 29 | 3.45e-02 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 1 | 40 | 3.45e-02 |
| GO:BP | GO:0051955 | regulation of amino acid transport | 1 | 28 | 3.45e-02 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 2 | 81 | 3.45e-02 |
| GO:BP | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway | 1 | 3 | 3.45e-02 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 2 | 98 | 3.45e-02 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 2 | 570 | 3.48e-02 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 44 | 3.48e-02 |
| GO:BP | GO:1900127 | positive regulation of hyaluronan biosynthetic process | 1 | 4 | 3.48e-02 |
| GO:BP | GO:0090500 | endocardial cushion to mesenchymal transition | 1 | 4 | 3.48e-02 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 1 | 38 | 3.51e-02 |
| GO:BP | GO:0000379 | tRNA-type intron splice site recognition and cleavage | 1 | 3 | 3.54e-02 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 1 | 43 | 3.57e-02 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 2 | 103 | 3.61e-02 |
| GO:BP | GO:0060437 | lung growth | 1 | 4 | 3.69e-02 |
| GO:BP | GO:0002931 | response to ischemia | 1 | 48 | 3.92e-02 |
| GO:BP | GO:0070487 | monocyte aggregation | 1 | 4 | 3.93e-02 |
| GO:BP | GO:0015800 | acidic amino acid transport | 1 | 39 | 4.03e-02 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 4.05e-02 |
| GO:BP | GO:0034089 | establishment of meiotic sister chromatid cohesion | 1 | 4 | 4.06e-02 |
| GO:BP | GO:1901842 | negative regulation of high voltage-gated calcium channel activity | 1 | 4 | 4.08e-02 |
| GO:BP | GO:1902475 | L-alpha-amino acid transmembrane transport | 1 | 51 | 4.30e-02 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 1 | 63 | 4.32e-02 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 2 | 126 | 4.34e-02 |
| GO:BP | GO:0070295 | renal water absorption | 1 | 6 | 4.34e-02 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 2 | 127 | 4.35e-02 |
| GO:BP | GO:0005977 | glycogen metabolic process | 1 | 64 | 4.35e-02 |
| GO:BP | GO:0060449 | bud elongation involved in lung branching | 1 | 7 | 4.35e-02 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 122 | 4.35e-02 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 4 | 707 | 4.35e-02 |
| GO:BP | GO:0060349 | bone morphogenesis | 2 | 74 | 4.35e-02 |
| GO:BP | GO:0032890 | regulation of organic acid transport | 1 | 48 | 4.35e-02 |
| GO:BP | GO:0044042 | glucan metabolic process | 1 | 65 | 4.35e-02 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 3 | 316 | 4.41e-02 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 4 | 799 | 4.41e-02 |
| GO:BP | GO:0042310 | vasoconstriction | 1 | 51 | 4.46e-02 |
| GO:BP | GO:1900125 | regulation of hyaluronan biosynthetic process | 1 | 6 | 4.52e-02 |
| GO:BP | GO:0015807 | L-amino acid transport | 1 | 62 | 4.53e-02 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 2 | 100 | 4.53e-02 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 1 | 72 | 4.53e-02 |
| GO:BP | GO:0006094 | gluconeogenesis | 1 | 70 | 4.53e-02 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 1 | 73 | 4.65e-02 |
| GO:BP | GO:0030010 | establishment of cell polarity | 2 | 135 | 4.72e-02 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 1 | 61 | 4.72e-02 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 1 | 71 | 4.83e-02 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 56 | 4.84e-02 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 1 | 82 | 4.90e-02 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 1 | 70 | 4.91e-02 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 3 | 585 | 5.05e-02 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 2 | 145 | 5.14e-02 |
| GO:BP | GO:0003097 | renal water transport | 1 | 7 | 5.14e-02 |
| GO:BP | GO:0015837 | amine transport | 1 | 59 | 5.14e-02 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 1 | 93 | 5.33e-02 |
| GO:BP | GO:0007049 | cell cycle | 3 | 1515 | 5.42e-02 |
| GO:BP | GO:0051101 | regulation of DNA binding | 1 | 102 | 5.53e-02 |
| GO:BP | GO:0001654 | eye development | 3 | 273 | 5.66e-02 |
| GO:BP | GO:0150063 | visual system development | 3 | 274 | 5.77e-02 |
| GO:BP | GO:0048880 | sensory system development | 3 | 276 | 5.88e-02 |
| GO:BP | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway | 1 | 11 | 5.88e-02 |
| GO:BP | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway | 1 | 8 | 5.88e-02 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 4 | 730 | 6.19e-02 |
| GO:BP | GO:0035296 | regulation of tube diameter | 1 | 98 | 6.19e-02 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 1 | 98 | 6.19e-02 |
| GO:BP | GO:0051549 | positive regulation of keratinocyte migration | 1 | 11 | 6.19e-02 |
| GO:BP | GO:0035150 | regulation of tube size | 1 | 98 | 6.19e-02 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 2 | 173 | 6.21e-02 |
| GO:BP | GO:0006865 | amino acid transport | 1 | 103 | 6.21e-02 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 1 | 111 | 6.25e-02 |
| GO:BP | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 1 | 10 | 6.25e-02 |
| GO:BP | GO:0030213 | hyaluronan biosynthetic process | 1 | 10 | 6.25e-02 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 6.25e-02 |
| GO:BP | GO:0036302 | atrioventricular canal development | 1 | 12 | 6.25e-02 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 2 | 177 | 6.25e-02 |
| GO:BP | GO:0070486 | leukocyte aggregation | 1 | 9 | 6.25e-02 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 1 | 112 | 6.25e-02 |
| GO:BP | GO:0051100 | negative regulation of binding | 1 | 133 | 6.32e-02 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 1 | 10 | 6.33e-02 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 1 | 119 | 6.54e-02 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 16 | 6.54e-02 |
| GO:BP | GO:0010838 | positive regulation of keratinocyte proliferation | 1 | 9 | 6.54e-02 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 2 | 194 | 6.54e-02 |
| GO:BP | GO:1901841 | regulation of high voltage-gated calcium channel activity | 1 | 12 | 6.54e-02 |
| GO:BP | GO:0051547 | regulation of keratinocyte migration | 1 | 13 | 6.54e-02 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 2 | 191 | 6.67e-02 |
| GO:BP | GO:0035265 | organ growth | 2 | 128 | 6.71e-02 |
| GO:BP | GO:0044849 | estrous cycle | 1 | 14 | 6.72e-02 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 1 | 132 | 6.90e-02 |
| GO:BP | GO:0006006 | glucose metabolic process | 1 | 153 | 6.97e-02 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 2 | 160 | 6.99e-02 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 1 | 15 | 6.99e-02 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 2 | 197 | 7.06e-02 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 2 | 1103 | 7.06e-02 |
| GO:BP | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 1 | 16 | 7.08e-02 |
| GO:BP | GO:0098739 | import across plasma membrane | 1 | 130 | 7.12e-02 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 1 | 135 | 7.12e-02 |
| GO:BP | GO:0006836 | neurotransmitter transport | 1 | 141 | 7.12e-02 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 1 | 137 | 7.12e-02 |
| GO:BP | GO:0030073 | insulin secretion | 1 | 144 | 7.12e-02 |
| GO:BP | GO:0007623 | circadian rhythm | 1 | 160 | 7.12e-02 |
| GO:BP | GO:0090087 | regulation of peptide transport | 1 | 139 | 7.15e-02 |
| GO:BP | GO:0042592 | homeostatic process | 2 | 1189 | 7.33e-02 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 1 | 146 | 7.34e-02 |
| GO:BP | GO:0060602 | branch elongation of an epithelium | 1 | 15 | 7.41e-02 |
| GO:BP | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation | 1 | 13 | 7.41e-02 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 211 | 7.42e-02 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 1 | 15 | 7.42e-02 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 1 | 11 | 7.42e-02 |
| GO:BP | GO:0045444 | fat cell differentiation | 1 | 187 | 7.54e-02 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 2 | 222 | 7.54e-02 |
| GO:BP | GO:0019318 | hexose metabolic process | 1 | 187 | 7.54e-02 |
| GO:BP | GO:0051546 | keratinocyte migration | 1 | 15 | 7.54e-02 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 1 | 19 | 7.54e-02 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 5 | 1092 | 7.54e-02 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 2 | 225 | 7.54e-02 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 1 | 174 | 7.54e-02 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 2 | 232 | 7.54e-02 |
| GO:BP | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation | 1 | 14 | 7.54e-02 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 2 | 227 | 7.61e-02 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 1 | 18 | 7.61e-02 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 1 | 20 | 7.61e-02 |
| GO:BP | GO:0030072 | peptide hormone secretion | 1 | 169 | 7.61e-02 |
| GO:BP | GO:1903522 | regulation of blood circulation | 1 | 197 | 7.61e-02 |
| GO:BP | GO:0002790 | peptide secretion | 1 | 173 | 7.61e-02 |
| GO:BP | GO:0035809 | regulation of urine volume | 1 | 14 | 7.61e-02 |
| GO:BP | GO:1901386 | negative regulation of voltage-gated calcium channel activity | 1 | 14 | 7.61e-02 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 1 | 171 | 7.76e-02 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 1 | 212 | 7.77e-02 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 3 | 549 | 7.77e-02 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 1 | 198 | 7.77e-02 |
| GO:BP | GO:0050708 | regulation of protein secretion | 1 | 186 | 7.77e-02 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 5 | 1124 | 7.77e-02 |
| GO:BP | GO:0015833 | peptide transport | 1 | 182 | 7.81e-02 |
| GO:BP | GO:0031399 | regulation of protein modification process | 4 | 1096 | 7.81e-02 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 1 | 20 | 7.83e-02 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 1 | 23 | 7.83e-02 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 1 | 22 | 7.83e-02 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 1 | 30 | 7.91e-02 |
| GO:BP | GO:0003417 | growth plate cartilage development | 1 | 14 | 8.01e-02 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 1 | 28 | 8.02e-02 |
| GO:BP | GO:0003091 | renal water homeostasis | 1 | 14 | 8.02e-02 |
| GO:BP | GO:0070848 | response to growth factor | 3 | 571 | 8.31e-02 |
| GO:BP | GO:0048511 | rhythmic process | 1 | 233 | 8.33e-02 |
| GO:BP | GO:0043950 | positive regulation of cAMP-mediated signaling | 1 | 5 | 8.40e-02 |
| GO:BP | GO:0009416 | response to light stimulus | 1 | 241 | 8.56e-02 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 2 | 192 | 8.56e-02 |
| GO:BP | GO:0016567 | protein ubiquitination | 3 | 684 | 8.56e-02 |
| GO:BP | GO:0046879 | hormone secretion | 1 | 212 | 8.56e-02 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 1 | 25 | 8.56e-02 |
| GO:BP | GO:0060348 | bone development | 2 | 157 | 8.58e-02 |
| GO:BP | GO:0060441 | epithelial tube branching involved in lung morphogenesis | 1 | 21 | 8.58e-02 |
| GO:BP | GO:0021756 | striatum development | 1 | 18 | 8.58e-02 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 1 | 224 | 8.58e-02 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 1 | 223 | 8.58e-02 |
| GO:BP | GO:0009914 | hormone transport | 1 | 217 | 8.58e-02 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 1 | 18 | 8.58e-02 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 1 | 302 | 8.66e-02 |
| GO:BP | GO:0003401 | axis elongation | 1 | 25 | 8.75e-02 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 1 | 288 | 8.75e-02 |
| GO:BP | GO:0140013 | meiotic nuclear division | 2 | 117 | 8.80e-02 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 2 | 261 | 8.80e-02 |
| GO:BP | GO:0006833 | water transport | 1 | 12 | 8.80e-02 |
| GO:BP | GO:0015849 | organic acid transport | 1 | 214 | 8.80e-02 |
| GO:BP | GO:0046942 | carboxylic acid transport | 1 | 213 | 8.80e-02 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 1 | 25 | 8.80e-02 |
| GO:BP | GO:0009306 | protein secretion | 1 | 262 | 8.92e-02 |
| GO:BP | GO:0051098 | regulation of binding | 1 | 314 | 8.92e-02 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 2 | 155 | 8.92e-02 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 4 | 1116 | 8.92e-02 |
| GO:BP | GO:0042886 | amide transport | 1 | 237 | 8.92e-02 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 1 | 262 | 8.92e-02 |
| GO:BP | GO:0007423 | sensory organ development | 3 | 395 | 8.92e-02 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 36 | 8.92e-02 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 2 | 211 | 8.92e-02 |
| GO:BP | GO:0070293 | renal absorption | 1 | 24 | 8.92e-02 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 1 | 18 | 8.95e-02 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 1 | 266 | 8.98e-02 |
| GO:BP | GO:0007130 | synaptonemal complex assembly | 1 | 10 | 9.09e-02 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 302 | 9.15e-02 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 1 | 17 | 9.23e-02 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 9.23e-02 |
| GO:BP | GO:0030212 | hyaluronan metabolic process | 1 | 21 | 9.23e-02 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 1 | 276 | 9.31e-02 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 3 | 759 | 9.36e-02 |
| GO:BP | GO:0034405 | response to fluid shear stress | 1 | 31 | 9.36e-02 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 1 | 32 | 9.36e-02 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 2 | 132 | 9.48e-02 |
| GO:BP | GO:0021544 | subpallium development | 1 | 19 | 9.48e-02 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 9.48e-02 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 22 | 9.48e-02 |
| GO:BP | GO:0006417 | regulation of translation | 1 | 373 | 9.51e-02 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 1 | 36 | 9.51e-02 |
| GO:BP | GO:0070193 | synaptonemal complex organization | 1 | 13 | 9.51e-02 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 1 | 44 | 9.51e-02 |
| GO:BP | GO:0043010 | camera-type eye development | 2 | 237 | 9.51e-02 |
| GO:BP | GO:0009314 | response to radiation | 1 | 339 | 9.51e-02 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 1 | 32 | 9.51e-02 |
| GO:BP | GO:0015711 | organic anion transport | 1 | 270 | 9.66e-02 |
| GO:BP | GO:0042044 | fluid transport | 1 | 19 | 9.94e-02 |
| GO:BP | GO:0098868 | bone growth | 1 | 23 | 1.01e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 1 | 23 | 1.01e-01 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 1 | 32 | 1.01e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 1 | 357 | 1.01e-01 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 1 | 37 | 1.01e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 1 | 35 | 1.01e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 298 | 1.02e-01 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 1 | 37 | 1.02e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 1 | 426 | 1.02e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 1.03e-01 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 1 | 28 | 1.03e-01 |
| GO:BP | GO:0023061 | signal release | 1 | 329 | 1.03e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 3 | 471 | 1.03e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 1 | 27 | 1.03e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 1 | 35 | 1.03e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 1 | 36 | 1.03e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 1 | 37 | 1.03e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 1 | 25 | 1.04e-01 |
| GO:BP | GO:0008015 | blood circulation | 1 | 372 | 1.04e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 1 | 395 | 1.05e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 1 | 404 | 1.05e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 1 | 423 | 1.05e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 1 | 25 | 1.05e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 4 | 1318 | 1.06e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 1 | 28 | 1.06e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 21 | 1.06e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 1.06e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 2 | 179 | 1.06e-01 |
| GO:BP | GO:0006915 | apoptotic process | 3 | 1452 | 1.08e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 1 | 46 | 1.08e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 1 | 428 | 1.08e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 1 | 337 | 1.08e-01 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 1 | 29 | 1.08e-01 |
| GO:BP | GO:0044057 | regulation of system process | 1 | 398 | 1.08e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 4 | 1252 | 1.09e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 4 | 1249 | 1.09e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 1 | 375 | 1.09e-01 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 1 | 17 | 1.09e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 1 | 382 | 1.09e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 39 | 1.10e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 1 | 439 | 1.10e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 2 | 186 | 1.10e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 3 | 439 | 1.10e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 1 | 19 | 1.11e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 1 | 432 | 1.11e-01 |
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 1 | 31 | 1.11e-01 |
| GO:BP | GO:0003013 | circulatory system process | 1 | 445 | 1.11e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 3 | 758 | 1.11e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 1 | 392 | 1.11e-01 |
| GO:BP | GO:0012501 | programmed cell death | 3 | 1498 | 1.12e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 3 | 870 | 1.12e-01 |
| GO:BP | GO:0008219 | cell death | 3 | 1504 | 1.12e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 1 | 52 | 1.14e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 1 | 413 | 1.14e-01 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 1 | 35 | 1.14e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 1 | 434 | 1.14e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 1 | 50 | 1.14e-01 |
| GO:BP | GO:0001822 | kidney development | 2 | 246 | 1.16e-01 |
| GO:BP | GO:0051293 | establishment of spindle localization | 1 | 49 | 1.18e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 2 | 2052 | 1.18e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 1 | 520 | 1.18e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 1 | 35 | 1.19e-01 |
| GO:BP | GO:0072001 | renal system development | 2 | 253 | 1.20e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 1 | 47 | 1.20e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 39 | 1.20e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 1 | 54 | 1.21e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 1 | 70 | 1.22e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 3 | 2560 | 1.22e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 3 | 2567 | 1.23e-01 |
| GO:BP | GO:0051653 | spindle localization | 1 | 53 | 1.23e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 2 | 183 | 1.24e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 1 | 54 | 1.25e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 1 | 39 | 1.30e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 1 | 81 | 1.31e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 3 | 841 | 1.31e-01 |
| GO:BP | GO:0006412 | translation | 1 | 657 | 1.32e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 1 | 57 | 1.33e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 1 | 36 | 1.33e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 2 | 1074 | 1.34e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 1 | 677 | 1.35e-01 |
| GO:BP | GO:0006325 | chromatin organization | 1 | 631 | 1.35e-01 |
| GO:BP | GO:0032940 | secretion by cell | 1 | 573 | 1.39e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 4 | 997 | 1.39e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 1.39e-01 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 1 | 59 | 1.40e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 1 | 593 | 1.41e-01 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 1 | 24 | 1.41e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 1 | 54 | 1.41e-01 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 1 | 61 | 1.41e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 64 | 1.41e-01 |
| GO:BP | GO:0035601 | protein deacylation | 1 | 91 | 1.41e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 1 | 59 | 1.42e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 1 | 92 | 1.42e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 1 | 48 | 1.42e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 1 | 643 | 1.42e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 1 | 47 | 1.43e-01 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 1 | 711 | 1.43e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 2 | 445 | 1.43e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 1 | 719 | 1.43e-01 |
| GO:BP | GO:0140352 | export from cell | 1 | 617 | 1.43e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 1 | 52 | 1.43e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 1 | 61 | 1.44e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 1 | 781 | 1.47e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 1 | 785 | 1.49e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 4 | 1097 | 1.49e-01 |
| GO:BP | GO:0098657 | import into cell | 1 | 664 | 1.50e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 1 | 52 | 1.50e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 1 | 684 | 1.50e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 1 | 102 | 1.50e-01 |
| GO:BP | GO:0046903 | secretion | 1 | 647 | 1.51e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 1 | 703 | 1.52e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 1 | 699 | 1.52e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 1 | 67 | 1.52e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 1 | 744 | 1.52e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 1 | 35 | 1.52e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 1 | 805 | 1.53e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 1 | 75 | 1.55e-01 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 1 | 32 | 1.55e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 1 | 736 | 1.55e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 3 | 950 | 1.55e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 3 | 949 | 1.55e-01 |
| GO:BP | GO:0006950 | response to stress | 3 | 2763 | 1.55e-01 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 1 | 35 | 1.55e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 1 | 59 | 1.58e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 1 | 53 | 1.58e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 3 | 1102 | 1.58e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 3 | 654 | 1.58e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 4 | 1662 | 1.59e-01 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 1 | 39 | 1.59e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 2 | 2846 | 1.59e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 1 | 71 | 1.59e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 2 | 413 | 1.59e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 1 | 49 | 1.59e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 2 | 408 | 1.59e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 1 | 72 | 1.60e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 80 | 1.60e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 1 | 72 | 1.60e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 1 | 475 | 1.60e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 1 | 848 | 1.61e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 1 | 45 | 1.61e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 1 | 64 | 1.61e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 1 | 87 | 1.61e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 1 | 86 | 1.61e-01 |
| GO:BP | GO:1903510 | mucopolysaccharide metabolic process | 1 | 75 | 1.61e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 1 | 96 | 1.63e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 2 | 473 | 1.63e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 1 | 81 | 1.63e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1 | 68 | 1.63e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 1 | 59 | 1.63e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 79 | 1.63e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 4 | 1122 | 1.64e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 2 | 421 | 1.64e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 1 | 90 | 1.64e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 2 | 456 | 1.64e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 5 | 2036 | 1.65e-01 |
| GO:BP | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 1 | 78 | 1.65e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 1 | 70 | 1.67e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 1 | 85 | 1.69e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 1 | 1003 | 1.69e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 1 | 70 | 1.69e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 2 | 429 | 1.69e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 4 | 1557 | 1.69e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 2 | 314 | 1.69e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 1 | 64 | 1.69e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 1 | 58 | 1.74e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 1 | 1005 | 1.80e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 3 | 2943 | 1.81e-01 |
| GO:BP | GO:0030154 | cell differentiation | 3 | 2942 | 1.81e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 1 | 1014 | 1.81e-01 |
| GO:BP | GO:0030203 | glycosaminoglycan metabolic process | 1 | 87 | 1.81e-01 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 1 | 51 | 1.83e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 1.84e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 2 | 461 | 1.86e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 5 | 1832 | 1.88e-01 |
| GO:BP | GO:0003014 | renal system process | 1 | 76 | 1.89e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 1 | 989 | 1.89e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 3 | 725 | 1.90e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 1 | 99 | 1.90e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 1 | 75 | 1.90e-01 |
| GO:BP | GO:0006022 | aminoglycan metabolic process | 1 | 92 | 1.90e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 1 | 76 | 1.91e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 1 | 1112 | 1.91e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 1 | 76 | 1.91e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 4 | 1260 | 1.92e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 1 | 97 | 1.92e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 2 | 548 | 1.92e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 1 | 52 | 1.92e-01 |
| GO:BP | GO:0001823 | mesonephros development | 1 | 77 | 1.96e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 1 | 82 | 1.97e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 4 | 1895 | 1.98e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 3 | 1130 | 1.98e-01 |
| GO:BP | GO:0035295 | tube development | 3 | 822 | 1.98e-01 |
| GO:BP | GO:0001501 | skeletal system development | 2 | 361 | 1.98e-01 |
| GO:BP | GO:0000280 | nuclear division | 2 | 343 | 1.98e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 2 | 544 | 1.98e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 1 | 163 | 1.98e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 1 | 1203 | 2.00e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 1 | 88 | 2.00e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 1 | 96 | 2.00e-01 |
| GO:BP | GO:0036211 | protein modification process | 5 | 2751 | 2.01e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 3 | 744 | 2.01e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 1 | 1014 | 2.01e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 1 | 121 | 2.06e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 1 | 108 | 2.06e-01 |
| GO:BP | GO:0072359 | circulatory system development | 3 | 888 | 2.07e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 1 | 180 | 2.08e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 1 | 89 | 2.08e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 1 | 84 | 2.11e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 97 | 2.11e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 1 | 132 | 2.11e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 1 | 1150 | 2.11e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 1 | 80 | 2.11e-01 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 1 | 123 | 2.11e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 1 | 1298 | 2.11e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 1 | 1305 | 2.11e-01 |
| GO:BP | GO:0022008 | neurogenesis | 1 | 1297 | 2.11e-01 |
| GO:BP | GO:0001568 | blood vessel development | 2 | 537 | 2.12e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 2.13e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 1 | 100 | 2.13e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 2.13e-01 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 1 | 157 | 2.14e-01 |
| GO:BP | GO:0051049 | regulation of transport | 1 | 1268 | 2.15e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 1 | 251 | 2.17e-01 |
| GO:BP | GO:0034502 | protein localization to chromosome | 1 | 101 | 2.19e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 1 | 1496 | 2.20e-01 |
| GO:BP | GO:0048285 | organelle fission | 2 | 387 | 2.20e-01 |
| GO:BP | GO:0015031 | protein transport | 1 | 1366 | 2.20e-01 |
| GO:BP | GO:0001944 | vasculature development | 2 | 562 | 2.21e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 1 | 1386 | 2.22e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 1 | 1425 | 2.22e-01 |
| GO:BP | GO:0007507 | heart development | 2 | 512 | 2.24e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 1 | 141 | 2.24e-01 |
| GO:BP | GO:0048565 | digestive tract development | 1 | 92 | 2.26e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 111 | 2.26e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 1 | 156 | 2.26e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 1 | 1468 | 2.27e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 1 | 154 | 2.27e-01 |
| GO:BP | GO:0030324 | lung development | 1 | 144 | 2.27e-01 |
| GO:BP | GO:0007127 | meiosis I | 1 | 73 | 2.27e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 1 | 143 | 2.27e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 5 | 2948 | 2.29e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 2 | 664 | 2.29e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 1 | 148 | 2.29e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 2 | 438 | 2.31e-01 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 1 | 144 | 2.32e-01 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 1 | 77 | 2.34e-01 |
| GO:BP | GO:0042552 | myelination | 1 | 114 | 2.35e-01 |
| GO:BP | GO:0055123 | digestive system development | 1 | 100 | 2.36e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 1 | 1589 | 2.36e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 1 | 116 | 2.36e-01 |
| GO:BP | GO:0060429 | epithelium development | 3 | 849 | 2.36e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 1 | 116 | 2.36e-01 |
| GO:BP | GO:0048839 | inner ear development | 1 | 129 | 2.36e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 1 | 147 | 2.36e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 1 | 134 | 2.36e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 1 | 155 | 2.37e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 1 | 113 | 2.37e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 1 | 113 | 2.37e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 1 | 113 | 2.40e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1570 | 2.41e-01 |
| GO:BP | GO:0016310 | phosphorylation | 3 | 1338 | 2.41e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 1 | 120 | 2.42e-01 |
| GO:BP | GO:0060541 | respiratory system development | 1 | 161 | 2.42e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 1 | 124 | 2.42e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 2 | 1905 | 2.43e-01 |
| GO:BP | GO:0008033 | tRNA processing | 1 | 131 | 2.43e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 1 | 120 | 2.43e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 1 | 315 | 2.44e-01 |
| GO:BP | GO:0048589 | developmental growth | 2 | 522 | 2.44e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 1 | 1568 | 2.44e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 2 | 544 | 2.45e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1194 | 2.51e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 152 | 2.52e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 1 | 1689 | 2.53e-01 |
| GO:BP | GO:0009888 | tissue development | 4 | 1425 | 2.54e-01 |
| GO:BP | GO:0043583 | ear development | 1 | 150 | 2.55e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 1 | 122 | 2.55e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 1 | 114 | 2.55e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 71 | 2.56e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 1 | 171 | 2.58e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 1 | 115 | 2.60e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1842 | 2.60e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 2 | 663 | 2.60e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2 | 4197 | 2.60e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 1 | 1764 | 2.61e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 2 | 741 | 2.64e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 1 | 1813 | 2.66e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 207 | 2.68e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 1 | 195 | 2.68e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 1 | 159 | 2.69e-01 |
| GO:BP | GO:0007399 | nervous system development | 1 | 1886 | 2.69e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 1 | 1852 | 2.70e-01 |
| GO:BP | GO:0048736 | appendage development | 1 | 141 | 2.72e-01 |
| GO:BP | GO:0060173 | limb development | 1 | 141 | 2.72e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1 | 1896 | 2.76e-01 |
| GO:BP | GO:0034097 | response to cytokine | 2 | 609 | 2.77e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 1 | 142 | 2.82e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 2 | 707 | 2.82e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 1 | 1958 | 2.83e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 1 | 1635 | 2.83e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1 | 1972 | 2.85e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 1 | 176 | 2.85e-01 |
| GO:BP | GO:0008104 | protein localization | 1 | 2128 | 2.86e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1 | 2137 | 2.87e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 1 | 1669 | 2.87e-01 |
| GO:BP | GO:0048468 | cell development | 1 | 1950 | 2.88e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 4 | 2271 | 2.90e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 1 | 1708 | 2.91e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 1 | 200 | 2.91e-01 |
| GO:BP | GO:0051216 | cartilage development | 1 | 149 | 2.91e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 1 | 1721 | 2.91e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 1 | 202 | 2.92e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 1 | 217 | 2.92e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 1 | 157 | 2.94e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 1 | 156 | 2.95e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 2 | 4566 | 2.95e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1 | 157 | 2.95e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 2 | 866 | 2.98e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 1 | 2085 | 2.98e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 2 | 741 | 2.98e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 1 | 249 | 3.00e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 1 | 167 | 3.02e-01 |
| GO:BP | GO:0023052 | signaling | 2 | 4134 | 3.05e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 1 | 242 | 3.06e-01 |
| GO:BP | GO:0032502 | developmental process | 2 | 4445 | 3.06e-01 |
| GO:BP | GO:0071702 | organic substance transport | 1 | 2133 | 3.06e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 1 | 202 | 3.06e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 1 | 189 | 3.06e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 1 | 206 | 3.07e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 1 | 189 | 3.07e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 1 | 2367 | 3.07e-01 |
| GO:BP | GO:0007154 | cell communication | 2 | 4210 | 3.10e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 2 | 762 | 3.10e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 1 | 254 | 3.17e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 1 | 200 | 3.17e-01 |
| GO:BP | GO:0022402 | cell cycle process | 3 | 1078 | 3.17e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 1 | 214 | 3.17e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 1 | 2069 | 3.17e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 6 | 4202 | 3.17e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 2 | 841 | 3.17e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 1 | 2340 | 3.17e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 1 | 2448 | 3.17e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 1 | 253 | 3.17e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 1 | 126 | 3.19e-01 |
| GO:BP | GO:0090132 | epithelium migration | 1 | 253 | 3.19e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 1 | 287 | 3.20e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 1 | 276 | 3.20e-01 |
| GO:BP | GO:0090130 | tissue migration | 1 | 257 | 3.23e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 1 | 188 | 3.23e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1 | 2131 | 3.24e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 1 | 212 | 3.26e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 1 | 233 | 3.27e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 1 | 234 | 3.27e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 1 | 274 | 3.27e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 1 | 234 | 3.27e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 1 | 2518 | 3.27e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 1 | 2560 | 3.30e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 1 | 2577 | 3.31e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1 | 2607 | 3.33e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 1 | 288 | 3.33e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1 | 2297 | 3.39e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 1 | 2669 | 3.39e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 1 | 2699 | 3.42e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 1 | 271 | 3.45e-01 |
| GO:BP | GO:0021537 | telencephalon development | 1 | 208 | 3.45e-01 |
| GO:BP | GO:0051641 | cellular localization | 1 | 2822 | 3.45e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 1 | 113 | 3.47e-01 |
| GO:BP | GO:0061448 | connective tissue development | 1 | 212 | 3.49e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 2 | 1017 | 3.50e-01 |
| GO:BP | GO:0040007 | growth | 2 | 737 | 3.50e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 1 | 239 | 3.50e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 1 | 232 | 3.51e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 1 | 2824 | 3.51e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 268 | 3.53e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 1 | 210 | 3.53e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 1 | 237 | 3.53e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 2 | 4998 | 3.64e-01 |
| GO:BP | GO:0048731 | system development | 1 | 2886 | 3.67e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 1 | 221 | 3.68e-01 |
| GO:BP | GO:0040011 | locomotion | 2 | 854 | 3.72e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 1 | 1964 | 3.72e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 1 | 3054 | 3.72e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1 | 3059 | 3.72e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 1 | 3115 | 3.77e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 1 | 3116 | 3.77e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 1 | 233 | 3.77e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 7 | 4919 | 3.79e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 2 | 1088 | 3.83e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 2 | 1090 | 3.83e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 1 | 264 | 3.86e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 1 | 3221 | 3.87e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 1 | 256 | 3.91e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 2 | 4807 | 3.91e-01 |
| GO:BP | GO:0051656 | establishment of organelle localization | 1 | 397 | 3.93e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 1 | 358 | 3.93e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 182 | 3.94e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 1 | 271 | 3.95e-01 |
| GO:BP | GO:0022414 | reproductive process | 3 | 924 | 3.99e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 2 | 2877 | 3.99e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 318 | 3.99e-01 |
| GO:BP | GO:0000003 | reproduction | 3 | 933 | 4.02e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 1 | 3355 | 4.13e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 1 | 208 | 4.13e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 1 | 317 | 4.16e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 1 | 292 | 4.19e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 1 | 319 | 4.19e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3294 | 4.19e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 1 | 415 | 4.24e-01 |
| GO:BP | GO:0001525 | angiogenesis | 1 | 389 | 4.24e-01 |
| GO:BP | GO:0009790 | embryo development | 2 | 859 | 4.24e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 1 | 329 | 4.24e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 1 | 275 | 4.24e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 1 | 3743 | 4.28e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 1 | 221 | 4.31e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3453 | 4.31e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 6 | 4243 | 4.35e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 1 | 296 | 4.35e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 1 | 347 | 4.41e-01 |
| GO:BP | GO:0030900 | forebrain development | 1 | 290 | 4.42e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 3 | 2096 | 4.46e-01 |
| GO:BP | GO:0016477 | cell migration | 2 | 1065 | 4.50e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 3 | 2118 | 4.52e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 3 | 1390 | 4.53e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 2 | 1267 | 4.58e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 2 | 689 | 4.58e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 1 | 360 | 4.58e-01 |
| GO:BP | GO:0051640 | organelle localization | 1 | 521 | 4.61e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 470 | 4.67e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 6 | 4371 | 4.69e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 1 | 3587 | 4.73e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1 | 3773 | 4.73e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 1 | 501 | 4.75e-01 |
| GO:BP | GO:0051179 | localization | 1 | 3983 | 4.75e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1 | 3702 | 4.81e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 1 | 542 | 4.85e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 1 | 549 | 4.85e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 2 | 633 | 4.85e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 1 | 465 | 4.89e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 1 | 3794 | 4.89e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 1 | 397 | 4.89e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1 | 3820 | 4.90e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 1 | 4100 | 4.95e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 1 | 1349 | 4.95e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 1 | 369 | 4.97e-01 |
| GO:BP | GO:0050896 | response to stimulus | 2 | 5721 | 4.97e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1 | 4026 | 4.97e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 1 | 592 | 5.08e-01 |
| GO:BP | GO:0048870 | cell motility | 2 | 1169 | 5.08e-01 |
| GO:BP | GO:0048513 | animal organ development | 4 | 2156 | 5.11e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 1 | 511 | 5.16e-01 |
| GO:BP | GO:0006974 | DNA damage response | 1 | 795 | 5.18e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 1 | 689 | 5.30e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 1 | 559 | 5.50e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 1 | 559 | 5.50e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 288 | 5.55e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 288 | 5.55e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 292 | 5.55e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 1 | 449 | 5.55e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 1 | 4563 | 5.55e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 1 | 4593 | 5.58e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 1 | 364 | 5.62e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 1 | 535 | 5.64e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 1 | 355 | 5.66e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 1 | 369 | 5.67e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 1 | 3573 | 5.68e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 1 | 680 | 5.73e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 1 | 662 | 5.79e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 426 | 5.79e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 1 | 664 | 5.81e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 1 | 815 | 5.86e-01 |
| GO:BP | GO:0061061 | muscle structure development | 1 | 552 | 5.86e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 1 | 685 | 5.86e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 451 | 5.86e-01 |
| GO:BP | GO:0051276 | chromosome organization | 1 | 533 | 5.87e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 259 | 6.00e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 1 | 561 | 6.00e-01 |
| GO:BP | GO:0051301 | cell division | 1 | 569 | 6.00e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 1 | 757 | 6.00e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 1 | 680 | 6.00e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1 | 4018 | 6.00e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 1 | 410 | 6.01e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 1 | 416 | 6.06e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 1 | 538 | 6.11e-01 |
| GO:BP | GO:0007420 | brain development | 1 | 546 | 6.19e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 1 | 537 | 6.19e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 1 | 599 | 6.24e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 1 | 671 | 6.26e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 1 | 554 | 6.28e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 1 | 558 | 6.28e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 1 | 802 | 6.30e-01 |
| GO:BP | GO:0007165 | signal transduction | 2 | 3803 | 6.30e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1 | 4423 | 6.31e-01 |
| GO:BP | GO:0030001 | metal ion transport | 1 | 568 | 6.34e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 3 | 1952 | 6.34e-01 |
| GO:BP | GO:0060322 | head development | 1 | 588 | 6.35e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 3 | 1857 | 6.35e-01 |
| GO:BP | GO:0016043 | cellular component organization | 1 | 5106 | 6.35e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 1 | 4537 | 6.40e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 1 | 4561 | 6.43e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 1 | 582 | 6.48e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1 | 5307 | 6.49e-01 |
| GO:BP | GO:0010033 | response to organic substance | 3 | 1942 | 6.54e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 1 | 498 | 6.55e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 1 | 4713 | 6.57e-01 |
| GO:BP | GO:0009725 | response to hormone | 1 | 624 | 6.59e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 1 | 622 | 6.59e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 1 | 868 | 6.62e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 795 | 6.65e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 1 | 4913 | 6.66e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 3 | 1981 | 6.77e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 1 | 650 | 6.81e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 1 | 677 | 6.86e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 1 | 583 | 7.01e-01 |
| GO:BP | GO:0010467 | gene expression | 1 | 4733 | 7.05e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 1 | 886 | 7.14e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 1 | 1067 | 7.19e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 2 | 1889 | 7.19e-01 |
| GO:BP | GO:0007417 | central nervous system development | 1 | 750 | 7.22e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 1 | 5148 | 7.39e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 1 | 1186 | 7.39e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 628 | 7.47e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 1 | 788 | 7.50e-01 |
| GO:BP | GO:0030030 | cell projection organization | 1 | 1236 | 7.67e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 2 | 7632 | 7.71e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2 | 4929 | 7.73e-01 |
| GO:BP | GO:0007155 | cell adhesion | 1 | 996 | 7.76e-01 |
| GO:BP | GO:0006396 | RNA processing | 1 | 928 | 7.79e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 918 | 7.83e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 1 | 860 | 7.85e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 1 | 5813 | 7.93e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 1 | 5875 | 7.99e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 1 | 5912 | 8.02e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 1 | 1001 | 8.14e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 2 | 8017 | 8.37e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 1 | 386 | 8.37e-01 |
| GO:BP | GO:0042221 | response to chemical | 3 | 2456 | 8.73e-01 |
| GO:BP | GO:0065007 | biological regulation | 2 | 8282 | 8.93e-01 |
| GO:BP | GO:0006996 | organelle organization | 3 | 2974 | 8.99e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 1 | 7431 | 9.01e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 1 | 7084 | 9.16e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 1 | 7788 | 9.40e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 1 | 7562 | 9.57e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 2 | 2459 | 9.95e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 2 | 2722 | 9.97e-01 |
| GO:BP | GO:0009987 | cellular process | 1 | 11410 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8522 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12027 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8176 | 1.00e+00 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 3 | 128 | 3.30e-04 |
| KEGG | KEGG:04115 | p53 signaling pathway | 2 | 65 | 5.02e-04 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 3 | 67 | 5.02e-04 |
| KEGG | KEGG:05212 | Pancreatic cancer | 2 | 75 | 5.02e-04 |
| KEGG | KEGG:05222 | Small cell lung cancer | 3 | 87 | 5.02e-04 |
| KEGG | KEGG:05213 | Endometrial cancer | 2 | 57 | 5.02e-04 |
| KEGG | KEGG:05214 | Glioma | 2 | 67 | 5.02e-04 |
| KEGG | KEGG:05216 | Thyroid cancer | 2 | 35 | 5.02e-04 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 2 | 75 | 5.02e-04 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 2 | 49 | 5.02e-04 |
| KEGG | KEGG:05218 | Melanoma | 2 | 58 | 5.02e-04 |
| KEGG | KEGG:05210 | Colorectal cancer | 2 | 84 | 5.89e-04 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 2 | 71 | 7.65e-04 |
| KEGG | KEGG:05226 | Gastric cancer | 3 | 117 | 1.09e-03 |
| KEGG | KEGG:04210 | Apoptosis | 2 | 118 | 1.09e-03 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 2 | 109 | 1.09e-03 |
| KEGG | KEGG:05224 | Breast cancer | 2 | 117 | 1.21e-03 |
| KEGG | KEGG:04218 | Cellular senescence | 2 | 145 | 1.24e-03 |
| KEGG | KEGG:04110 | Cell cycle | 2 | 151 | 1.24e-03 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 2 | 145 | 1.31e-03 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 2 | 153 | 1.78e-03 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 3.41e-03 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 2 | 240 | 3.55e-03 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 1 | 56 | 6.16e-03 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 69 | 8.57e-03 |
| KEGG | KEGG:05200 | Pathways in cancer | 2 | 409 | 9.94e-03 |
| KEGG | KEGG:04114 | Oocyte meiosis | 1 | 108 | 1.57e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 2.40e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 1 | 415 | 2.41e-01 |
| KEGG | KEGG:00000 | KEGG root term | 2 | 5548 | 3.61e-01 |
TRZnomtable3h %>%
filter(source=="KEGG") %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "Enrichment of KEGG terms using nominal pvalue TRZ 3hour genes in my data") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 3 | 128 | 3.30e-04 |
| KEGG | KEGG:04115 | p53 signaling pathway | 2 | 65 | 5.02e-04 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 3 | 67 | 5.02e-04 |
| KEGG | KEGG:05212 | Pancreatic cancer | 2 | 75 | 5.02e-04 |
| KEGG | KEGG:05222 | Small cell lung cancer | 3 | 87 | 5.02e-04 |
| KEGG | KEGG:05213 | Endometrial cancer | 2 | 57 | 5.02e-04 |
| KEGG | KEGG:05214 | Glioma | 2 | 67 | 5.02e-04 |
| KEGG | KEGG:05216 | Thyroid cancer | 2 | 35 | 5.02e-04 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 2 | 75 | 5.02e-04 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 2 | 49 | 5.02e-04 |
| KEGG | KEGG:05218 | Melanoma | 2 | 58 | 5.02e-04 |
| KEGG | KEGG:05210 | Colorectal cancer | 2 | 84 | 5.89e-04 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 2 | 71 | 7.65e-04 |
| KEGG | KEGG:05226 | Gastric cancer | 3 | 117 | 1.09e-03 |
| KEGG | KEGG:04210 | Apoptosis | 2 | 118 | 1.09e-03 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 2 | 109 | 1.09e-03 |
| KEGG | KEGG:05224 | Breast cancer | 2 | 117 | 1.21e-03 |
| KEGG | KEGG:04218 | Cellular senescence | 2 | 145 | 1.24e-03 |
| KEGG | KEGG:04110 | Cell cycle | 2 | 151 | 1.24e-03 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 2 | 145 | 1.31e-03 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 2 | 153 | 1.78e-03 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 3.41e-03 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 2 | 240 | 3.55e-03 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 1 | 56 | 6.16e-03 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 69 | 8.57e-03 |
| KEGG | KEGG:05200 | Pathways in cancer | 2 | 409 | 9.94e-03 |
| KEGG | KEGG:04114 | Oocyte meiosis | 1 | 108 | 1.57e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 2.40e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 1 | 415 | 2.41e-01 |
| KEGG | KEGG:00000 | KEGG root term | 2 | 5548 | 3.61e-01 |
TRZ 24 hour nom de
# TRZresgenes24h <- gost(query = TRZ_list24h$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(TRZresgenes24h,"data/DEG-GO/TRZresgenes24h.RDS")
TRZresgenes24h <- readRDS("data/DEG-GO/TRZresgenes24h.RDS")
TRZnom24h <- gostplot(TRZresgenes24h, capped = FALSE, interactive = TRUE)
TRZnom24hTRZnomtable24h <- TRZresgenes24h$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
TRZnomtable24h %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 1 | 2 | 6.00e-02 |
| GO:BP | GO:0048133 | male germ-line stem cell asymmetric division | 1 | 2 | 6.00e-02 |
| GO:BP | GO:0042078 | germ-line stem cell division | 1 | 2 | 6.00e-02 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 1 | 2 | 6.00e-02 |
| GO:BP | GO:0015757 | galactose transmembrane transport | 1 | 4 | 9.34e-02 |
| GO:BP | GO:0098708 | glucose import across plasma membrane | 1 | 5 | 9.34e-02 |
| GO:BP | GO:0098704 | carbohydrate import across plasma membrane | 1 | 5 | 9.34e-02 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 3 | 269 | 9.34e-02 |
| GO:BP | GO:0061643 | chemorepulsion of axon | 1 | 6 | 9.34e-02 |
| GO:BP | GO:0060252 | positive regulation of glial cell proliferation | 1 | 14 | 9.34e-02 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 1 | 26 | 9.34e-02 |
| GO:BP | GO:0051939 | gamma-aminobutyric acid import | 1 | 3 | 9.34e-02 |
| GO:BP | GO:0051764 | actin crosslink formation | 1 | 9 | 9.34e-02 |
| GO:BP | GO:0006979 | response to oxidative stress | 3 | 328 | 9.34e-02 |
| GO:BP | GO:0048232 | male gamete generation | 2 | 346 | 9.34e-02 |
| GO:BP | GO:0014009 | glial cell proliferation | 1 | 40 | 9.34e-02 |
| GO:BP | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 1 | 7 | 9.34e-02 |
| GO:BP | GO:0042918 | alkanesulfonate transport | 1 | 6 | 9.34e-02 |
| GO:BP | GO:0008356 | asymmetric cell division | 1 | 14 | 9.34e-02 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 1 | 44 | 9.34e-02 |
| GO:BP | GO:0007283 | spermatogenesis | 2 | 333 | 9.34e-02 |
| GO:BP | GO:0007288 | sperm axoneme assembly | 1 | 20 | 9.34e-02 |
| GO:BP | GO:0036159 | inner dynein arm assembly | 1 | 8 | 9.34e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 3 | 218 | 9.34e-02 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 2 | 255 | 9.34e-02 |
| GO:BP | GO:0018345 | protein palmitoylation | 1 | 30 | 9.34e-02 |
| GO:BP | GO:0018231 | peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine | 1 | 21 | 9.34e-02 |
| GO:BP | GO:0018230 | peptidyl-L-cysteine S-palmitoylation | 1 | 21 | 9.34e-02 |
| GO:BP | GO:0017145 | stem cell division | 1 | 25 | 9.34e-02 |
| GO:BP | GO:0015734 | taurine transport | 1 | 6 | 9.34e-02 |
| GO:BP | GO:0007281 | germ cell development | 2 | 194 | 9.34e-02 |
| GO:BP | GO:0010232 | vascular transport | 2 | 72 | 9.34e-02 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 10 | 9.34e-02 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 10 | 9.34e-02 |
| GO:BP | GO:1904059 | regulation of locomotor rhythm | 1 | 3 | 9.34e-02 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 9.34e-02 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 9.34e-02 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 9.34e-02 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 9.34e-02 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 1 | 14 | 9.34e-02 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 2 | 72 | 9.34e-02 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 12 | 9.34e-02 |
| GO:BP | GO:1990708 | conditioned place preference | 1 | 2 | 9.34e-02 |
| GO:BP | GO:0140271 | hexose import across plasma membrane | 1 | 5 | 9.34e-02 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 1 | 15 | 9.57e-02 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 1 | 41 | 9.78e-02 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 1 | 63 | 9.78e-02 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 1 | 66 | 1.02e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 1 | 16 | 1.05e-01 |
| GO:BP | GO:0070286 | axonemal dynein complex assembly | 1 | 21 | 1.10e-01 |
| GO:BP | GO:0015812 | gamma-aminobutyric acid transport | 1 | 11 | 1.10e-01 |
| GO:BP | GO:0120316 | sperm flagellum assembly | 1 | 27 | 1.10e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 1.10e-01 |
| GO:BP | GO:0070837 | dehydroascorbic acid transport | 1 | 7 | 1.10e-01 |
| GO:BP | GO:0032328 | alanine transport | 1 | 13 | 1.10e-01 |
| GO:BP | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | 1 | 30 | 1.10e-01 |
| GO:BP | GO:1901334 | lactone metabolic process | 1 | 10 | 1.11e-01 |
| GO:BP | GO:0006858 | extracellular transport | 1 | 33 | 1.11e-01 |
| GO:BP | GO:0010887 | negative regulation of cholesterol storage | 1 | 8 | 1.11e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 1.11e-01 |
| GO:BP | GO:0007276 | gamete generation | 2 | 465 | 1.11e-01 |
| GO:BP | GO:0019852 | L-ascorbic acid metabolic process | 1 | 10 | 1.11e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 1 | 19 | 1.14e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 1 | 28 | 1.25e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 2 | 538 | 1.27e-01 |
| GO:BP | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 8 | 1.30e-01 |
| GO:BP | GO:0044458 | motile cilium assembly | 1 | 46 | 1.34e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 1 | 92 | 1.34e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 1 | 137 | 1.34e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 2 | 130 | 1.34e-01 |
| GO:BP | GO:0038003 | G protein-coupled opioid receptor signaling pathway | 1 | 5 | 1.34e-01 |
| GO:BP | GO:0006497 | protein lipidation | 1 | 89 | 1.34e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 2 | 558 | 1.34e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 2 | 603 | 1.34e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 2 | 633 | 1.43e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 1.43e-01 |
| GO:BP | GO:0106070 | regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 11 | 1.43e-01 |
| GO:BP | GO:0051382 | kinetochore assembly | 1 | 17 | 1.43e-01 |
| GO:BP | GO:0043951 | negative regulation of cAMP-mediated signaling | 1 | 10 | 1.43e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 1 | 190 | 1.43e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 1 | 34 | 1.46e-01 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 1.46e-01 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 1 | 13 | 1.47e-01 |
| GO:BP | GO:0044849 | estrous cycle | 1 | 14 | 1.51e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 1 | 13 | 1.51e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 1 | 116 | 1.54e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 1 | 20 | 1.54e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 1 | 222 | 1.54e-01 |
| GO:BP | GO:0035082 | axoneme assembly | 1 | 54 | 1.54e-01 |
| GO:BP | GO:0051383 | kinetochore organization | 1 | 21 | 1.54e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 2 | 549 | 1.54e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 1 | 48 | 1.61e-01 |
| GO:BP | GO:0010888 | negative regulation of lipid storage | 1 | 15 | 1.63e-01 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 1.63e-01 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 1 | 38 | 1.67e-01 |
| GO:BP | GO:0010878 | cholesterol storage | 1 | 18 | 1.67e-01 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 1 | 26 | 1.67e-01 |
| GO:BP | GO:0042063 | gliogenesis | 1 | 244 | 1.67e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 2 | 198 | 1.67e-01 |
| GO:BP | GO:1901386 | negative regulation of voltage-gated calcium channel activity | 1 | 14 | 1.67e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 1 | 36 | 1.71e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 33 | 1.74e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 1 | 14 | 1.74e-01 |
| GO:BP | GO:0001578 | microtubule bundle formation | 1 | 79 | 1.74e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 302 | 1.75e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 1 | 62 | 1.78e-01 |
| GO:BP | GO:0043543 | protein acylation | 1 | 193 | 1.78e-01 |
| GO:BP | GO:0034508 | centromere complex assembly | 1 | 29 | 1.78e-01 |
| GO:BP | GO:0015804 | neutral amino acid transport | 1 | 43 | 1.78e-01 |
| GO:BP | GO:0097722 | sperm motility | 1 | 62 | 1.78e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 1 | 31 | 1.78e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 1.81e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 1 | 38 | 1.81e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 1 | 35 | 1.81e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 1 | 35 | 1.81e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 1 | 39 | 1.81e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 30 | 1.81e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 1 | 69 | 1.81e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 1 | 318 | 1.81e-01 |
| GO:BP | GO:0021591 | ventricular system development | 1 | 29 | 1.82e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 1 | 357 | 1.82e-01 |
| GO:BP | GO:0022414 | reproductive process | 2 | 924 | 1.82e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 1 | 72 | 1.82e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 1 | 72 | 1.82e-01 |
| GO:BP | GO:0000003 | reproduction | 2 | 933 | 1.83e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 1 | 44 | 2.02e-01 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 1 | 27 | 2.02e-01 |
| GO:BP | GO:0003013 | circulatory system process | 3 | 445 | 2.03e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 1 | 41 | 2.03e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 4 | 1122 | 2.03e-01 |
| GO:BP | GO:0050919 | negative chemotaxis | 1 | 42 | 2.03e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 2 | 757 | 2.05e-01 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 1 | 17 | 2.05e-01 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 1 | 45 | 2.06e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 1 | 34 | 2.06e-01 |
| GO:BP | GO:0003341 | cilium movement | 1 | 97 | 2.06e-01 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 1 | 28 | 2.06e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 1 | 192 | 2.06e-01 |
| GO:BP | GO:0051301 | cell division | 1 | 569 | 2.06e-01 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 1 | 46 | 2.06e-01 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 1 | 45 | 2.06e-01 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 1 | 43 | 2.06e-01 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 1 | 43 | 2.06e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 1 | 31 | 2.06e-01 |
| GO:BP | GO:0051180 | vitamin transport | 1 | 29 | 2.06e-01 |
| GO:BP | GO:0042755 | eating behavior | 1 | 16 | 2.06e-01 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 1 | 46 | 2.06e-01 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 1 | 45 | 2.06e-01 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 1 | 45 | 2.06e-01 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 1 | 43 | 2.06e-01 |
| GO:BP | GO:0009988 | cell-cell recognition | 1 | 31 | 2.06e-01 |
| GO:BP | GO:0007286 | spermatid development | 1 | 108 | 2.06e-01 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 1 | 46 | 2.06e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 2.06e-01 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 1 | 44 | 2.06e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 1 | 114 | 2.06e-01 |
| GO:BP | GO:0007517 | muscle organ development | 2 | 279 | 2.08e-01 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 1 | 70 | 2.08e-01 |
| GO:BP | GO:0006612 | protein targeting to membrane | 1 | 120 | 2.08e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 1 | 49 | 2.08e-01 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 31 | 2.09e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 1 | 51 | 2.12e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 1 | 44 | 2.12e-01 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 1 | 51 | 2.12e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 2.14e-01 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 1 | 29 | 2.15e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 1 | 50 | 2.15e-01 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 1 | 53 | 2.15e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 1 | 38 | 2.17e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 2.18e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 1 | 59 | 2.20e-01 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 1 | 56 | 2.20e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 1 | 60 | 2.20e-01 |
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 1 | 31 | 2.22e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 1 | 593 | 2.27e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 1 | 30 | 2.27e-01 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 1 | 56 | 2.27e-01 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 1 | 59 | 2.27e-01 |
| GO:BP | GO:0015711 | organic anion transport | 2 | 270 | 2.28e-01 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 1 | 35 | 2.28e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 1 | 618 | 2.28e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 1 | 41 | 2.29e-01 |
| GO:BP | GO:0050890 | cognition | 2 | 226 | 2.31e-01 |
| GO:BP | GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | 1 | 36 | 2.32e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 55 | 2.38e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 1 | 70 | 2.45e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 1 | 45 | 2.45e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 1 | 45 | 2.45e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 1 | 744 | 2.46e-01 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 1 | 69 | 2.46e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 1 | 35 | 2.46e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 1 | 258 | 2.46e-01 |
| GO:BP | GO:0006865 | amino acid transport | 1 | 103 | 2.46e-01 |
| GO:BP | GO:0048864 | stem cell development | 1 | 75 | 2.52e-01 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 1 | 111 | 2.53e-01 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 1 | 112 | 2.54e-01 |
| GO:BP | GO:0060271 | cilium assembly | 1 | 296 | 2.62e-01 |
| GO:BP | GO:0006605 | protein targeting | 1 | 310 | 2.62e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 1 | 79 | 2.62e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 1 | 82 | 2.62e-01 |
| GO:BP | GO:0051304 | chromosome separation | 1 | 75 | 2.62e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 2.67e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 2 | 490 | 2.69e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 2 | 490 | 2.69e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 2 | 494 | 2.71e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 1 | 90 | 2.73e-01 |
| GO:BP | GO:0044782 | cilium organization | 1 | 317 | 2.73e-01 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 1 | 37 | 2.74e-01 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 1 | 58 | 2.74e-01 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 1 | 87 | 2.74e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 918 | 2.74e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 1 | 185 | 2.74e-01 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 1 | 112 | 2.74e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 1 | 1005 | 2.74e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 1 | 300 | 2.74e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 1 | 94 | 2.74e-01 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 1 | 92 | 2.74e-01 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 1 | 91 | 2.74e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 2 | 1186 | 2.74e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 1 | 52 | 2.74e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 1 | 54 | 2.74e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 1 | 96 | 2.74e-01 |
| GO:BP | GO:0008037 | cell recognition | 1 | 82 | 2.74e-01 |
| GO:BP | GO:0055092 | sterol homeostasis | 1 | 59 | 2.74e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 1 | 75 | 2.74e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 1 | 886 | 2.74e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 1 | 185 | 2.74e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 2 | 512 | 2.74e-01 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 1 | 87 | 2.74e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 1 | 957 | 2.74e-01 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 1 | 88 | 2.74e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 1 | 1014 | 2.74e-01 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 1 | 94 | 2.74e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 1 | 41 | 2.74e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 5 | 1813 | 2.74e-01 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 1 | 89 | 2.74e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 68 | 2.74e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 1 | 141 | 2.74e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 1 | 59 | 2.75e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 1 | 87 | 2.75e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 1 | 84 | 2.75e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 102 | 2.75e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 1 | 997 | 2.75e-01 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 1 | 99 | 2.75e-01 |
| GO:BP | GO:0070085 | glycosylation | 1 | 197 | 2.75e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 1 | 68 | 2.75e-01 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 1 | 93 | 2.75e-01 |
| GO:BP | GO:0048675 | axon extension | 1 | 108 | 2.75e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 1 | 39 | 2.75e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 1 | 1112 | 2.75e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 1 | 1058 | 2.75e-01 |
| GO:BP | GO:0048468 | cell development | 2 | 1950 | 2.76e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 1 | 52 | 2.76e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 1 | 88 | 2.76e-01 |
| GO:BP | GO:0008306 | associative learning | 1 | 65 | 2.76e-01 |
| GO:BP | GO:0048699 | generation of neurons | 1 | 1120 | 2.78e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 1 | 105 | 2.80e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 1 | 64 | 2.81e-01 |
| GO:BP | GO:0050000 | chromosome localization | 1 | 101 | 2.81e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 1 | 1203 | 2.86e-01 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 1 | 99 | 2.86e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 1 | 117 | 2.89e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 1 | 95 | 2.89e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 2 | 471 | 2.89e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 1 | 530 | 2.89e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 1 | 70 | 2.91e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 1 | 158 | 2.92e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 1 | 1103 | 2.92e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 1 | 70 | 2.92e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 1 | 81 | 2.92e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 1 | 463 | 2.93e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 1 | 113 | 2.93e-01 |
| GO:BP | GO:0007416 | synapse assembly | 1 | 153 | 2.95e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 1 | 1305 | 2.95e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 1 | 473 | 2.95e-01 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 1 | 121 | 2.95e-01 |
| GO:BP | GO:0022008 | neurogenesis | 1 | 1297 | 2.95e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 1 | 1298 | 2.95e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 1 | 119 | 2.99e-01 |
| GO:BP | GO:0051668 | localization within membrane | 1 | 611 | 3.03e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 3.03e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 1 | 251 | 3.03e-01 |
| GO:BP | GO:0007631 | feeding behavior | 1 | 51 | 3.03e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 1 | 130 | 3.06e-01 |
| GO:BP | GO:0015918 | sterol transport | 1 | 93 | 3.09e-01 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 1 | 36 | 3.09e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 1 | 1425 | 3.09e-01 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 1 | 137 | 3.10e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 1 | 114 | 3.11e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 1 | 93 | 3.13e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 1 | 115 | 3.13e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 79 | 3.14e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 1 | 1662 | 3.14e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 2 | 2340 | 3.14e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 1 | 287 | 3.14e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 1 | 91 | 3.23e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 1 | 145 | 3.23e-01 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 1 | 99 | 3.23e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 1 | 1589 | 3.24e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 149 | 3.24e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 1 | 328 | 3.24e-01 |
| GO:BP | GO:0061061 | muscle structure development | 2 | 552 | 3.24e-01 |
| GO:BP | GO:0003014 | renal system process | 1 | 76 | 3.25e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 4 | 1150 | 3.26e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 1 | 866 | 3.26e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 1 | 96 | 3.26e-01 |
| GO:BP | GO:0007015 | actin filament organization | 1 | 372 | 3.26e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 1 | 160 | 3.26e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 1 | 114 | 3.26e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 1 | 154 | 3.26e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 1 | 88 | 3.26e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 1 | 803 | 3.26e-01 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 1 | 106 | 3.28e-01 |
| GO:BP | GO:0003008 | system process | 4 | 1194 | 3.28e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 1 | 161 | 3.28e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 1 | 1568 | 3.31e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 1 | 315 | 3.31e-01 |
| GO:BP | GO:0048870 | cell motility | 2 | 1169 | 3.33e-01 |
| GO:BP | GO:0016125 | sterol metabolic process | 1 | 108 | 3.33e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 2 | 592 | 3.34e-01 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 1 | 93 | 3.34e-01 |
| GO:BP | GO:0046942 | carboxylic acid transport | 1 | 213 | 3.34e-01 |
| GO:BP | GO:0015849 | organic acid transport | 1 | 214 | 3.34e-01 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 1 | 165 | 3.41e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 165 | 3.43e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 1 | 1832 | 3.43e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 71 | 3.43e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 1 | 100 | 3.43e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 1 | 110 | 3.43e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 1 | 116 | 3.43e-01 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 1 | 178 | 3.48e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 1 | 1764 | 3.48e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 1 | 143 | 3.55e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 184 | 3.55e-01 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 1 | 174 | 3.57e-01 |
| GO:BP | GO:0007612 | learning | 1 | 114 | 3.57e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 1 | 1852 | 3.57e-01 |
| GO:BP | GO:0007399 | nervous system development | 1 | 1886 | 3.57e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 2 | 1496 | 3.57e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 194 | 3.58e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1 | 1896 | 3.65e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 1 | 140 | 3.65e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 1 | 177 | 3.65e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 177 | 3.65e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 1 | 1857 | 3.65e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 1 | 1958 | 3.65e-01 |
| GO:BP | GO:0071702 | organic substance transport | 6 | 2133 | 3.65e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 2 | 654 | 3.65e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 1 | 204 | 3.65e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 207 | 3.65e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 1 | 392 | 3.65e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 1 | 205 | 3.65e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 2 | 707 | 3.65e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 1 | 196 | 3.65e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1 | 1972 | 3.65e-01 |
| GO:BP | GO:0070925 | organelle assembly | 1 | 828 | 3.66e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 1 | 1669 | 3.68e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 1 | 1708 | 3.75e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 1 | 1721 | 3.75e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 209 | 3.82e-01 |
| GO:BP | GO:0000819 | sister chromatid segregation | 1 | 217 | 3.83e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 1 | 120 | 3.87e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 2 | 741 | 3.87e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1 | 2052 | 3.90e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 1 | 171 | 3.90e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 1 | 114 | 3.90e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 1 | 190 | 3.90e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 1 | 212 | 3.94e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 1 | 214 | 3.96e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 1 | 2367 | 3.96e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 1 | 566 | 3.96e-01 |
| GO:BP | GO:0098657 | import into cell | 2 | 664 | 3.96e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 1 | 343 | 3.98e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 1 | 344 | 3.98e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 2 | 762 | 3.98e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 1 | 235 | 3.99e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 234 | 4.06e-01 |
| GO:BP | GO:0030154 | cell differentiation | 2 | 2942 | 4.08e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 2 | 2943 | 4.08e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 1 | 160 | 4.08e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 1 | 2069 | 4.08e-01 |
| GO:BP | GO:0006936 | muscle contraction | 1 | 266 | 4.14e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 1 | 250 | 4.14e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 1 | 139 | 4.16e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1 | 2131 | 4.16e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 1 | 254 | 4.18e-01 |
| GO:BP | GO:0015031 | protein transport | 1 | 1366 | 4.19e-01 |
| GO:BP | GO:0046907 | intracellular transport | 1 | 1440 | 4.19e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 1 | 643 | 4.20e-01 |
| GO:BP | GO:0140014 | mitotic nuclear division | 1 | 251 | 4.20e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 1 | 2518 | 4.20e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 1 | 2560 | 4.21e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 1 | 2577 | 4.22e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1 | 2607 | 4.24e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 211 | 4.26e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 1 | 262 | 4.26e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 1 | 544 | 4.26e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 1 | 666 | 4.26e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 1 | 2669 | 4.26e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 1 | 347 | 4.26e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1 | 2297 | 4.26e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 1 | 1468 | 4.27e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 1 | 2699 | 4.29e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 271 | 4.32e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 1 | 214 | 4.35e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 1 | 113 | 4.35e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 1 | 2824 | 4.42e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 373 | 4.43e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 1 | 264 | 4.43e-01 |
| GO:BP | GO:0006810 | transport | 2 | 3294 | 4.43e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1 | 2846 | 4.43e-01 |
| GO:BP | GO:0001822 | kidney development | 1 | 246 | 4.46e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 1 | 306 | 4.46e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 1 | 249 | 4.47e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 1 | 181 | 4.51e-01 |
| GO:BP | GO:0006950 | response to stress | 1 | 2763 | 4.51e-01 |
| GO:BP | GO:0072001 | renal system development | 1 | 253 | 4.52e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 1 | 167 | 4.52e-01 |
| GO:BP | GO:0048731 | system development | 1 | 2886 | 4.52e-01 |
| GO:BP | GO:0042221 | response to chemical | 1 | 2456 | 4.53e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 1 | 305 | 4.53e-01 |
| GO:BP | GO:0003012 | muscle system process | 1 | 334 | 4.53e-01 |
| GO:BP | GO:0051234 | establishment of localization | 2 | 3453 | 4.55e-01 |
| GO:BP | GO:0040011 | locomotion | 2 | 854 | 4.55e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1 | 3059 | 4.55e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 1 | 3054 | 4.55e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 1 | 189 | 4.56e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 1 | 3115 | 4.60e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1 | 1203 | 4.60e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 1 | 3116 | 4.60e-01 |
| GO:BP | GO:0051179 | localization | 4 | 3983 | 4.62e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 1 | 200 | 4.64e-01 |
| GO:BP | GO:0007611 | learning or memory | 1 | 201 | 4.64e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 1 | 1689 | 4.64e-01 |
| GO:BP | GO:0030030 | cell projection organization | 1 | 1236 | 4.64e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 1 | 310 | 4.64e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 1 | 3221 | 4.67e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 2 | 537 | 4.69e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 203 | 4.69e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 1 | 248 | 4.69e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 1 | 352 | 4.69e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 1 | 254 | 4.69e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 182 | 4.69e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 1 | 358 | 4.69e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1842 | 4.69e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 291 | 4.69e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 1 | 212 | 4.75e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 2 | 554 | 4.77e-01 |
| GO:BP | GO:0048511 | rhythmic process | 1 | 233 | 4.77e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 374 | 4.77e-01 |
| GO:BP | GO:0007409 | axonogenesis | 1 | 350 | 4.77e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 1 | 719 | 4.77e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 1 | 337 | 4.77e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 1 | 288 | 4.77e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 1 | 274 | 4.80e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 1 | 364 | 4.80e-01 |
| GO:BP | GO:0006935 | chemotaxis | 1 | 264 | 4.80e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 1 | 251 | 4.80e-01 |
| GO:BP | GO:0030001 | metal ion transport | 2 | 568 | 4.80e-01 |
| GO:BP | GO:0051235 | maintenance of location | 1 | 263 | 4.80e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 1 | 781 | 4.80e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 1 | 3355 | 4.80e-01 |
| GO:BP | GO:0042330 | taxis | 1 | 264 | 4.80e-01 |
| GO:BP | GO:0008104 | protein localization | 2 | 2128 | 4.87e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2 | 2137 | 4.88e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 3 | 1014 | 4.89e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 1 | 360 | 4.90e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 1 | 347 | 4.90e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 1 | 263 | 4.96e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 1 | 3743 | 4.96e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 1 | 805 | 4.97e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 1 | 275 | 4.97e-01 |
| GO:BP | GO:0016049 | cell growth | 1 | 416 | 4.97e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 1 | 799 | 4.97e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 1 | 387 | 4.97e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 2 | 582 | 4.97e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 1 | 232 | 4.97e-01 |
| GO:BP | GO:0061564 | axon development | 1 | 388 | 4.99e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 1 | 395 | 4.99e-01 |
| GO:BP | GO:0000280 | nuclear division | 1 | 343 | 4.99e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 3 | 1267 | 5.04e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 1 | 883 | 5.04e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 9 | 4807 | 5.04e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 1 | 411 | 5.04e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 1 | 221 | 5.10e-01 |
| GO:BP | GO:0016477 | cell migration | 2 | 1065 | 5.10e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 1 | 906 | 5.10e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 1 | 408 | 5.10e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 1 | 841 | 5.12e-01 |
| GO:BP | GO:0051656 | establishment of organelle localization | 1 | 397 | 5.15e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 1 | 237 | 5.15e-01 |
| GO:BP | GO:0006996 | organelle organization | 2 | 2974 | 5.21e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 1 | 421 | 5.25e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 2 | 4100 | 5.25e-01 |
| GO:BP | GO:0048285 | organelle fission | 1 | 387 | 5.26e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 586 | 5.27e-01 |
| GO:BP | GO:0006869 | lipid transport | 1 | 271 | 5.27e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 1 | 3587 | 5.27e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 1 | 429 | 5.27e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 1 | 451 | 5.27e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 1 | 438 | 5.27e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1 | 3773 | 5.27e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 1 | 4243 | 5.33e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 2 | 650 | 5.34e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1 | 3702 | 5.35e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 1 | 271 | 5.35e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 1 | 2448 | 5.35e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 1 | 860 | 5.38e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 2 | 677 | 5.38e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1 | 4197 | 5.38e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 1 | 4371 | 5.40e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 1 | 3794 | 5.40e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1 | 3820 | 5.41e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 1 | 413 | 5.41e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 1 | 439 | 5.41e-01 |
| GO:BP | GO:0040008 | regulation of growth | 1 | 493 | 5.41e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 1 | 1003 | 5.41e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1 | 4026 | 5.47e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 1 | 380 | 5.47e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 1 | 529 | 5.47e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 1 | 317 | 5.47e-01 |
| GO:BP | GO:0010876 | lipid localization | 1 | 309 | 5.47e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 1 | 496 | 5.47e-01 |
| GO:BP | GO:0036211 | protein modification process | 1 | 2751 | 5.47e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 1 | 500 | 5.47e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 1 | 379 | 5.47e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 1 | 515 | 5.51e-01 |
| GO:BP | GO:0048589 | developmental growth | 1 | 522 | 5.51e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 1 | 542 | 5.51e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 1 | 519 | 5.52e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 1 | 275 | 5.57e-01 |
| GO:BP | GO:0032502 | developmental process | 2 | 4445 | 5.61e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 1 | 539 | 5.62e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1 | 4566 | 5.62e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 1 | 2948 | 5.64e-01 |
| GO:BP | GO:0051641 | cellular localization | 1 | 2822 | 5.66e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 1 | 296 | 5.66e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 1 | 412 | 5.66e-01 |
| GO:BP | GO:0051640 | organelle localization | 1 | 521 | 5.69e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 1 | 599 | 5.73e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1151 | 5.73e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 1 | 1215 | 5.80e-01 |
| GO:BP | GO:0051276 | chromosome organization | 1 | 533 | 5.85e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 1 | 375 | 5.90e-01 |
| GO:BP | GO:0008015 | blood circulation | 1 | 372 | 5.90e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 1 | 4563 | 5.91e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 1 | 4593 | 5.94e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 1 | 788 | 5.97e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 628 | 5.98e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 2 | 5148 | 6.01e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 1 | 445 | 6.01e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 2 | 1017 | 6.01e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 1 | 627 | 6.01e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 1 | 3573 | 6.01e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 1 | 622 | 6.27e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 1 | 4998 | 6.30e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 1 | 369 | 6.35e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4929 | 6.35e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 1 | 643 | 6.35e-01 |
| GO:BP | GO:0007420 | brain development | 1 | 546 | 6.35e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 1 | 680 | 6.35e-01 |
| GO:BP | GO:0032879 | regulation of localization | 3 | 1570 | 6.37e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 1 | 689 | 6.37e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1 | 4018 | 6.37e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 1 | 392 | 6.37e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 2 | 1090 | 6.42e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 2 | 1088 | 6.42e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 1 | 1320 | 6.44e-01 |
| GO:BP | GO:0060322 | head development | 1 | 588 | 6.52e-01 |
| GO:BP | GO:0040007 | growth | 1 | 737 | 6.56e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 740 | 6.67e-01 |
| GO:BP | GO:0007610 | behavior | 1 | 439 | 6.69e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1 | 4423 | 6.69e-01 |
| GO:BP | GO:0031175 | neuron projection development | 1 | 774 | 6.78e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 1 | 4537 | 6.81e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 2 | 5813 | 6.82e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 1 | 4561 | 6.83e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 1 | 684 | 6.88e-01 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 1 | 711 | 6.88e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 2 | 5875 | 6.88e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 1 | 699 | 6.93e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 2 | 5912 | 6.93e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 1 | 703 | 6.93e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 1 | 815 | 6.93e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 1 | 4713 | 6.93e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 690 | 6.94e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 1 | 4202 | 7.03e-01 |
| GO:BP | GO:0009888 | tissue development | 2 | 1425 | 7.03e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 1 | 4913 | 7.03e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 744 | 7.05e-01 |
| GO:BP | GO:0048513 | animal organ development | 3 | 2156 | 7.10e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 1 | 1731 | 7.14e-01 |
| GO:BP | GO:0007155 | cell adhesion | 1 | 996 | 7.15e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 1 | 1964 | 7.15e-01 |
| GO:BP | GO:0048666 | neuron development | 1 | 873 | 7.15e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 1 | 680 | 7.20e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 2 | 1386 | 7.20e-01 |
| GO:BP | GO:0050896 | response to stimulus | 1 | 5721 | 7.20e-01 |
| GO:BP | GO:0007417 | central nervous system development | 1 | 750 | 7.30e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 1 | 886 | 7.30e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 2 | 1318 | 7.32e-01 |
| GO:BP | GO:0010467 | gene expression | 1 | 4733 | 7.32e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 1 | 1895 | 7.42e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 4 | 2560 | 7.49e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 1 | 941 | 7.50e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 4 | 2567 | 7.50e-01 |
| GO:BP | GO:0051049 | regulation of transport | 2 | 1268 | 7.50e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 3 | 2036 | 7.51e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 1 | 689 | 7.59e-01 |
| GO:BP | GO:0033993 | response to lipid | 1 | 594 | 7.60e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 1 | 1001 | 7.60e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2459 | 7.60e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 1 | 634 | 7.60e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 1 | 815 | 7.60e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 1 | 4919 | 7.60e-01 |
| GO:BP | GO:0009056 | catabolic process | 1 | 2108 | 7.65e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 259 | 7.70e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 1 | 730 | 7.72e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2722 | 7.79e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 2 | 1981 | 7.79e-01 |
| GO:BP | GO:0022402 | cell cycle process | 1 | 1078 | 7.85e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 3 | 2271 | 8.26e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 2 | 7431 | 8.32e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 4 | 2877 | 8.43e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 2 | 7084 | 8.57e-01 |
| GO:BP | GO:0023052 | signaling | 7 | 4134 | 8.57e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 1 | 386 | 8.57e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 1 | 1132 | 8.57e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 1 | 989 | 8.57e-01 |
| GO:BP | GO:0007154 | cell communication | 7 | 4210 | 8.69e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 1 | 7632 | 8.93e-01 |
| GO:BP | GO:0007049 | cell cycle | 1 | 1515 | 8.93e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 2 | 7788 | 8.95e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 2 | 1905 | 8.99e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 1 | 1349 | 9.03e-01 |
| GO:BP | GO:0042592 | homeostatic process | 1 | 1189 | 9.05e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 2 | 1889 | 9.12e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 2 | 7562 | 9.22e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 1 | 8017 | 9.26e-01 |
| GO:BP | GO:0016043 | cellular component organization | 2 | 5106 | 9.27e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1 | 1219 | 9.35e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 2 | 5307 | 9.36e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 2 | 2085 | 9.37e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1 | 1952 | 9.39e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 1 | 1635 | 9.39e-01 |
| GO:BP | GO:0065007 | biological regulation | 1 | 8282 | 9.47e-01 |
| GO:BP | GO:0007165 | signal transduction | 5 | 3803 | 9.55e-01 |
| GO:BP | GO:0010033 | response to organic substance | 1 | 1942 | 9.90e-01 |
| GO:BP | GO:0008150 | biological_process | 1 | 12027 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8176 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8522 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11410 | 1.00e+00 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 1 | 128 | 7.07e-02 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 7.07e-02 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 7.29e-02 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 1 | 303 | 1.56e-01 |
| KEGG | KEGG:05016 | Huntington disease | 1 | 256 | 1.56e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 1 | 385 | 1.68e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 2.12e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 1 | 108 | 4.86e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 5.69e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5548 | 6.01e-01 |
TRZnomtable24h %>%
filter(source=="KEGG") %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "Enrichment of KEGG terms using nominal pvalue TRZ 24hour genes in my data") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 1 | 128 | 7.07e-02 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 7.07e-02 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 7.29e-02 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 1 | 303 | 1.56e-01 |
| KEGG | KEGG:05016 | Huntington disease | 1 | 256 | 1.56e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 1 | 385 | 1.68e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 2.12e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 1 | 108 | 4.86e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 5.69e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5548 | 6.01e-01 |
# DAgostres3 <- readRDS("data/DEG-GO/DAgostres3.RDS")
# DAgostres24 <- readRDS("data/DAgostres24.RDS")
# DXgostres <- readRDS("data/DEG-GO/DXgostres.RDS")
# DXgostres24 <- readRDS("data/DEG-GO/DXgostres24.RDS")
# EPgostres <- readRDS("data/DEG-GO/EPgostres.RDS")
# EPgostres24 <- readRDS("data/DEG-GO/EPgostres24.RDS")
# MTgostres <- readRDS("data/DEG-GO/MTgostres.RDS")
# MTgostres24 <- readRDS("data/DEG-GO/MTgostres24.RDS")
# DDEgostres24only <- readRDS("data/DEG-GO/DDEgostres24only.RDS")
# DDEgostres3only <- readRDS("data/DEG-GO/DDEgostres3only.RDS")
# DDEMgostres3 <- readRDS("data/DEG-GO/DDEMgostres3.RDS")
# DDEMgostres24 <- readRDS("data/DEG-GO/DDEMgostres24.RDS")
# TRZresgenes24h <- readRDS("data/DEG-GO/TRZresgenes24h.RDS")
# TRZresgenes3h <- readRDS("data/DEG-GO/TRZresgenes3h.RDS")
# saveRDS(list3hr,"data/DEG-GO/list3hr.RDS")
# saveRDS(list24hr,"data/DEG-GO/list24hr.RDS")
# list3hr <- list(DAgostres3=DAgostres3,DXgostres=DXgostres,EPgostres=EPgostres,MTgostres=MTgostres,TRZresgenes3h=TRZresgenes3h,DDEgostres3only=DDEgostres3only,DDEMgostres3=DDEMgostres3)
# list24hr <- list(DAgostres24=DAgostres24,DXgostres24=DXgostres24,EPgostres24=EPgostres24,MTgostres24=MTgostres24,TRZresgenes243h=TRZresgenes24h,DDEgostres24only=DDEgostres24only,DDEMgostres24=DDEMgostres24)
list3hr <- readRDS("data/DEG-GO/list3hr.RDS")
list24hr <- readRDS("data/DEG-GO/list24hr.RDS")
KEGGtrz3 <- c("KEGG:05202", "KEGG:04115", "KEGG:05223", "KEGG:05212","KEGG:05222")
KEGGtrz24 <- c("KEGG:05202", "KEGG:05215", "KEGG:00510")
# testframe <- lapply(list3hr, `[`,'result')
hr3_results <- list3hr |>
map(~ .x$result |> dplyr::select(p_value,term_id,term_name,intersection_size))
hr24_results <- list24hr |>
map(~ .x$result |> dplyr::select(p_value,term_id,term_name,,intersection_size))
table_3hr <- list_rbind(hr3_results,names_to = "set")%>%
mutate(log_val = -log10(p_value)) %>%
dplyr::filter(term_id %in% KEGGtrz3) %>%
mutate(treatment = case_when(set =="DAgostres3"~"DNR",set=="DXgostres" ~ "DOX",set=="EPgostres"~"EPI",set=="MTgostres"~"MTX",set=="TRZresgenes3h"~"TRZ",set=="DDEgostres23only"~"AC",set=="DDEMgostres3"~"TOP2i"))
table_24hr <- list_rbind(hr24_results,names_to = "set") %>%
mutate(log_val = -log10(p_value))%>%
dplyr::filter(term_id %in% KEGGtrz24) %>%
mutate(treatment = case_when(set =="DAgostres24"~"DNR",set=="DXgostres24" ~ "DOX",set=="EPgostres24"~"EPI",set=="MTgostres24"~"MTX",set=="TRZresgenes243h"~"TRZ",set=="DDEgostres24only"~"AC",set=="DDEMgostres24"~"TOP2i"))
hr3_results[["TRZresgenes3h"]] %>%
mutate(log_val = -log10(p_value)) %>%
dplyr::filter(str_detect(term_id,"^KEGG:")) %>%
slice_min(., n=5,order_by=p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value),col=intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = scales::label_wrap(30))+
geom_vline(xintercept = (-log10(0.05)), col="red")+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('TRZ 3 hour nominal DE\n KEGG pathway enrichment') +
xlab(expression(" -"~log[10]~("nominal p-value")))+
ylab("KEGG")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))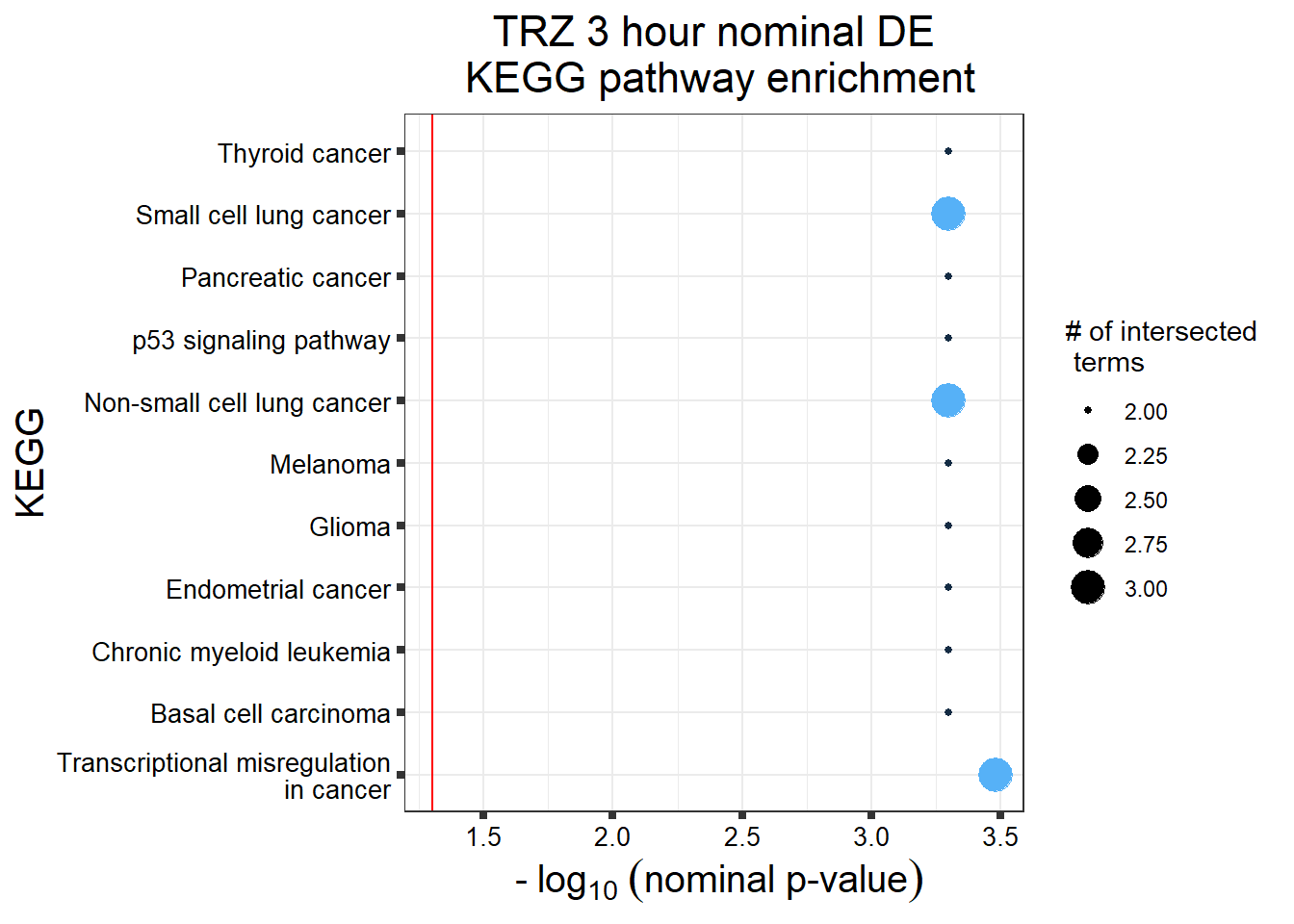
hr24_results[["TRZresgenes243h"]] %>%
mutate(log_val = -log10(p_value)) %>%
dplyr::filter(str_detect(term_id,"^KEGG:")) %>%
slice_min(., n=5,order_by=p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col=intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = scales::label_wrap(30))+
geom_vline(xintercept = (-log10(0.05)), col="red")+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('TRZ 24 hour nominal DE\n KEGG pathway enrichment') +
xlab(expression(" -"~log[10]~("nominal p-value")))+
ylab("KEGG")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))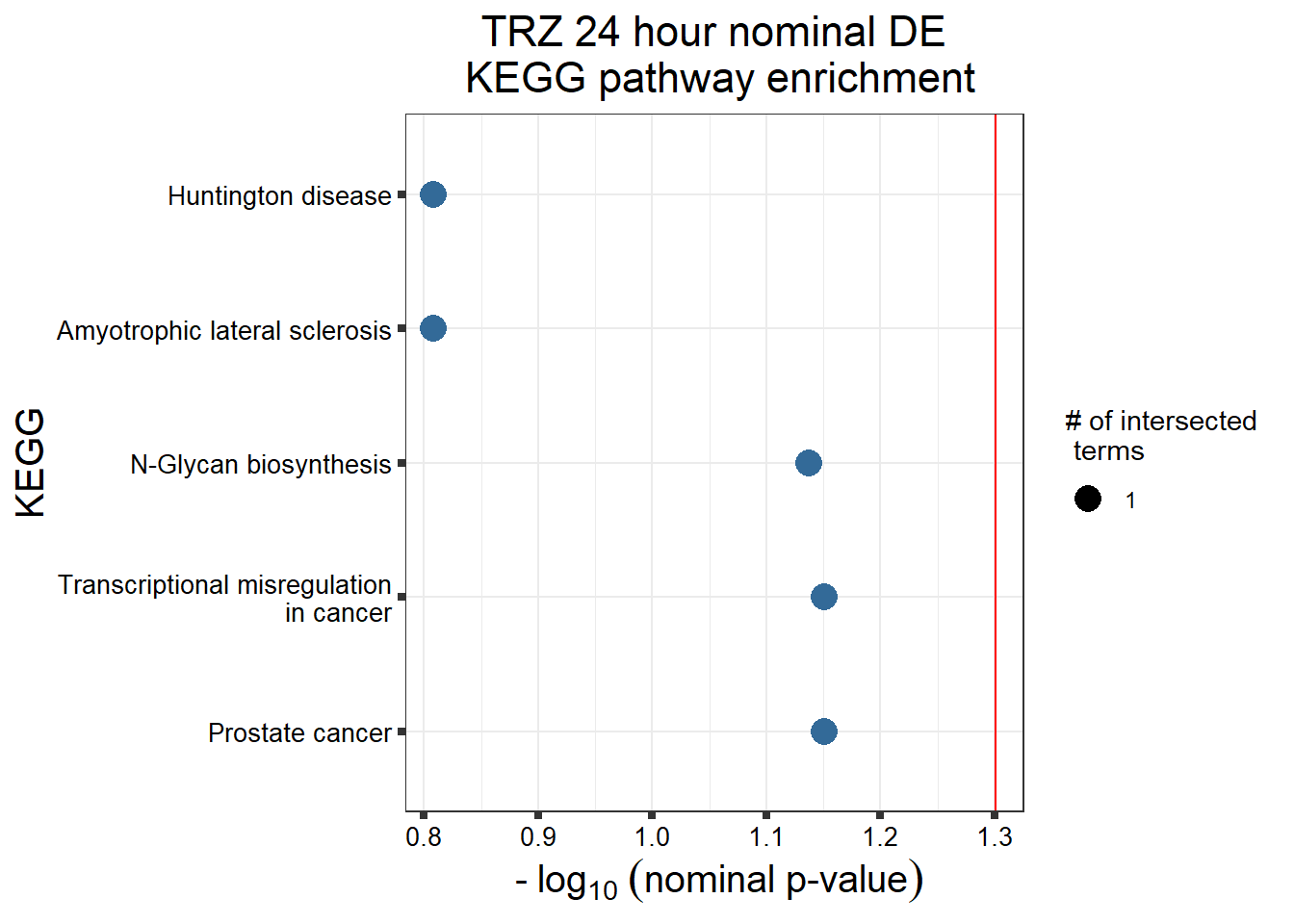
hr3_results[["TRZresgenes3h"]] %>%
mutate(log_val = -log10(p_value)) %>%
dplyr::filter(str_detect(term_id,"^GO:")) %>%
slice_min(., n=5,order_by=p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value),col=intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = scales::label_wrap(30))+
geom_vline(xintercept = (-log10(0.05)), col="red")+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('TRZ 3 hour nominal DE\n GO:BP enrichment') +
xlab(expression(" -"~log[10]~("nominal p-value")))+
ylab("KEGG")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))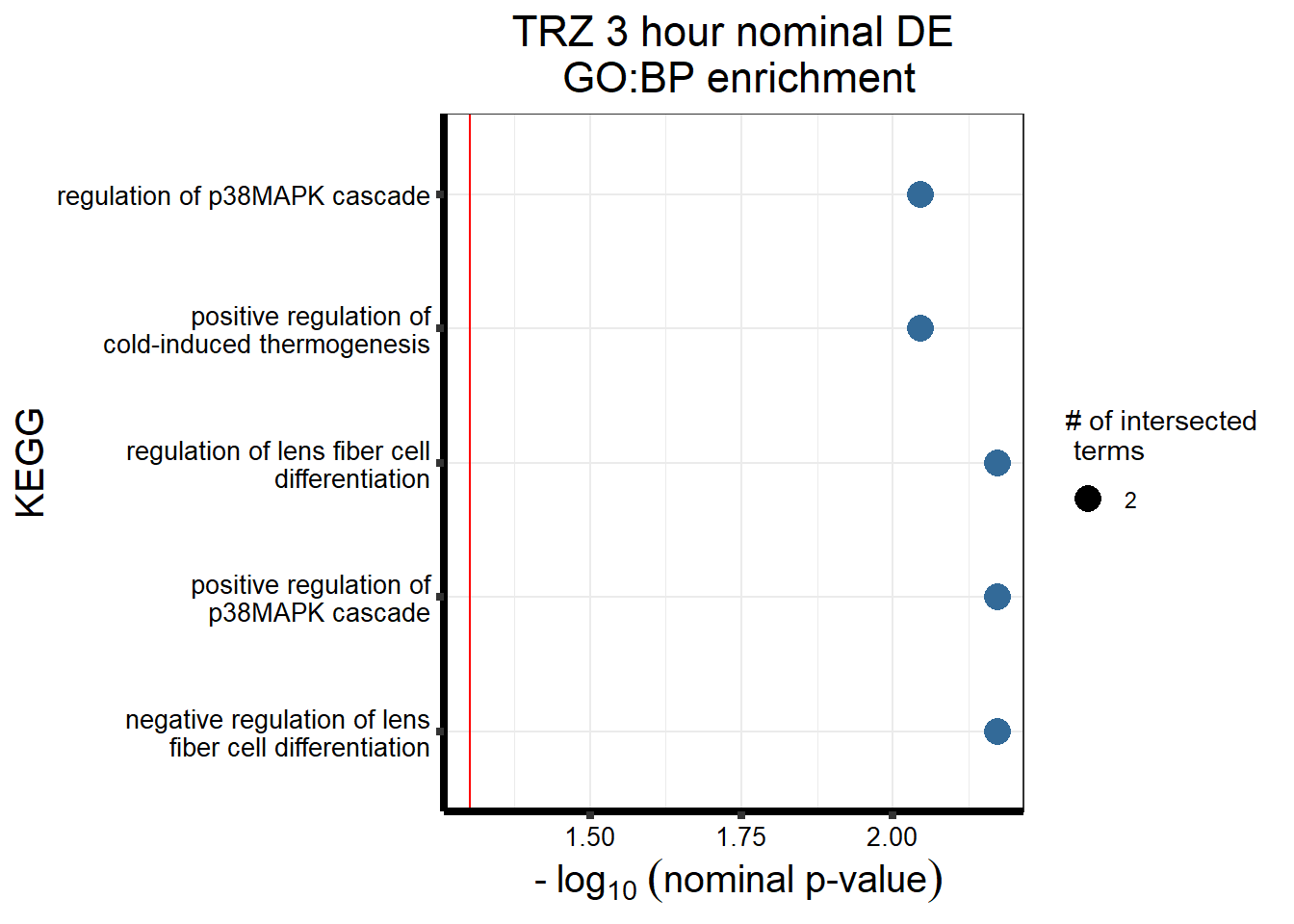
hr24_results[["TRZresgenes243h"]] %>%
mutate(log_val = -log10(p_value)) %>%
dplyr::filter(str_detect(term_id,"^GO:")) %>%
slice_min(., n=4,order_by=p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col=intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = scales::label_wrap(30))+
geom_vline(xintercept = (-log10(0.05)), col="red")+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('TRZ 24 hour nominal DE\n GO:BP enrichment') +
xlab(expression(" -"~log[10]~("nominal p-value")))+
ylab("KEGG")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))
Link to other pages here:
Other analysis aka after_comments data here
sessionInfo()R version 4.3.1 (2023-06-16 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] qvalue_2.32.0 gprofiler2_0.2.2 RColorBrewer_1.1-3 biomaRt_2.56.1
[5] broom_1.0.5 kableExtra_1.3.4 sjmisc_2.8.9 scales_1.2.1
[9] ggpubr_0.6.0 cowplot_1.1.1 ggsignif_0.6.4 lubridate_1.9.3
[13] forcats_1.0.0 stringr_1.5.0 dplyr_1.1.3 purrr_1.0.2
[17] readr_2.1.4 tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.4
[21] tidyverse_2.0.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] rstudioapi_0.15.0 jsonlite_1.8.7 magrittr_2.0.3
[4] farver_2.1.1 rmarkdown_2.25 fs_1.6.3
[7] zlibbioc_1.46.0 vctrs_0.6.4 memoise_2.0.1
[10] RCurl_1.98-1.13 rstatix_0.7.2 webshot_0.5.5
[13] htmltools_0.5.7 progress_1.2.2 curl_5.1.0
[16] sass_0.4.7 bslib_0.5.1 htmlwidgets_1.6.2
[19] plyr_1.8.9 plotly_4.10.3 cachem_1.0.8
[22] whisker_0.4.1 mime_0.12 lifecycle_1.0.4
[25] pkgconfig_2.0.3 sjlabelled_1.2.0 R6_2.5.1
[28] fastmap_1.1.1 shiny_1.7.5.1 GenomeInfoDbData_1.2.10
[31] digest_0.6.33 colorspace_2.1-0 AnnotationDbi_1.62.2
[34] S4Vectors_0.38.2 ps_1.7.5 rprojroot_2.0.4
[37] crosstalk_1.2.0 RSQLite_2.3.3 filelock_1.0.2
[40] labeling_0.4.3 fansi_1.0.5 timechange_0.2.0
[43] httr_1.4.7 abind_1.4-5 compiler_4.3.1
[46] bit64_4.0.5 withr_2.5.2 backports_1.4.1
[49] carData_3.0-5 DBI_1.1.3 highr_0.10
[52] rappdirs_0.3.3 tools_4.3.1 httpuv_1.6.12
[55] glue_1.6.2 callr_3.7.3 promises_1.2.1
[58] grid_4.3.1 getPass_0.2-2 reshape2_1.4.4
[61] generics_0.1.3 gtable_0.3.4 tzdb_0.4.0
[64] data.table_1.14.8 hms_1.1.3 xml2_1.3.5
[67] car_3.1-2 utf8_1.2.4 XVector_0.40.0
[70] BiocGenerics_0.46.0 pillar_1.9.0 vroom_1.6.4
[73] later_1.3.1 splines_4.3.1 BiocFileCache_2.8.0
[76] bit_4.0.5 tidyselect_1.2.0 Biostrings_2.68.1
[79] knitr_1.45 git2r_0.32.0 IRanges_2.34.1
[82] svglite_2.1.2 stats4_4.3.1 xfun_0.41
[85] Biobase_2.60.0 stringi_1.7.12 lazyeval_0.2.2
[88] yaml_2.3.7 evaluate_0.23 cli_3.6.1
[91] xtable_1.8-4 systemfonts_1.0.5 munsell_0.5.0
[94] processx_3.8.2 jquerylib_0.1.4 Rcpp_1.0.11
[97] GenomeInfoDb_1.36.4 dbplyr_2.3.4 png_0.1-8
[100] XML_3.99-0.15 parallel_4.3.1 ellipsis_0.3.2
[103] blob_1.2.4 prettyunits_1.2.0 bitops_1.0-7
[106] viridisLite_0.4.2 insight_0.19.6 crayon_1.5.2
[109] rlang_1.1.2 KEGGREST_1.40.1 rvest_1.0.3