SNP & TAD
Renee Matthews
2025-06-10
Last updated: 2025-07-29
Checks: 7 0
Knit directory: ATAC_learning/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231016) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version ad110ea. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/H3K27ac_integration_noM.Rmd
Ignored: data/ACresp_SNP_table.csv
Ignored: data/ARR_SNP_table.csv
Ignored: data/All_merged_peaks.tsv
Ignored: data/CAD_gwas_dataframe.RDS
Ignored: data/CTX_SNP_table.csv
Ignored: data/Collapsed_expressed_NG_peak_table.csv
Ignored: data/DEG_toplist_sep_n45.RDS
Ignored: data/FRiP_first_run.txt
Ignored: data/Final_four_data/
Ignored: data/Frip_1_reads.csv
Ignored: data/Frip_2_reads.csv
Ignored: data/Frip_3_reads.csv
Ignored: data/Frip_4_reads.csv
Ignored: data/Frip_5_reads.csv
Ignored: data/Frip_6_reads.csv
Ignored: data/GO_KEGG_analysis/
Ignored: data/HF_SNP_table.csv
Ignored: data/Ind1_75DA24h_dedup_peaks.csv
Ignored: data/Ind1_TSS_peaks.RDS
Ignored: data/Ind1_firstfragment_files.txt
Ignored: data/Ind1_fragment_files.txt
Ignored: data/Ind1_peaks_list.RDS
Ignored: data/Ind1_summary.txt
Ignored: data/Ind2_TSS_peaks.RDS
Ignored: data/Ind2_fragment_files.txt
Ignored: data/Ind2_peaks_list.RDS
Ignored: data/Ind2_summary.txt
Ignored: data/Ind3_TSS_peaks.RDS
Ignored: data/Ind3_fragment_files.txt
Ignored: data/Ind3_peaks_list.RDS
Ignored: data/Ind3_summary.txt
Ignored: data/Ind4_79B24h_dedup_peaks.csv
Ignored: data/Ind4_TSS_peaks.RDS
Ignored: data/Ind4_V24h_fraglength.txt
Ignored: data/Ind4_fragment_files.txt
Ignored: data/Ind4_fragment_filesN.txt
Ignored: data/Ind4_peaks_list.RDS
Ignored: data/Ind4_summary.txt
Ignored: data/Ind5_TSS_peaks.RDS
Ignored: data/Ind5_fragment_files.txt
Ignored: data/Ind5_fragment_filesN.txt
Ignored: data/Ind5_peaks_list.RDS
Ignored: data/Ind5_summary.txt
Ignored: data/Ind6_TSS_peaks.RDS
Ignored: data/Ind6_fragment_files.txt
Ignored: data/Ind6_peaks_list.RDS
Ignored: data/Ind6_summary.txt
Ignored: data/Knowles_4.RDS
Ignored: data/Knowles_5.RDS
Ignored: data/Knowles_6.RDS
Ignored: data/LiSiLTDNRe_TE_df.RDS
Ignored: data/MI_gwas.RDS
Ignored: data/SNP_GWAS_PEAK_MRC_id
Ignored: data/SNP_GWAS_PEAK_MRC_id.csv
Ignored: data/SNP_gene_cat_list.tsv
Ignored: data/SNP_supp_schneider.RDS
Ignored: data/TE_info/
Ignored: data/TFmapnames.RDS
Ignored: data/all_TSSE_scores.RDS
Ignored: data/all_four_filtered_counts.txt
Ignored: data/aln_run1_results.txt
Ignored: data/anno_ind1_DA24h.RDS
Ignored: data/anno_ind4_V24h.RDS
Ignored: data/annotated_gwas_SNPS.csv
Ignored: data/background_n45_he_peaks.RDS
Ignored: data/cardiac_muscle_FRIP.csv
Ignored: data/cardiomyocyte_FRIP.csv
Ignored: data/col_ng_peak.csv
Ignored: data/cormotif_full_4_run.RDS
Ignored: data/cormotif_full_4_run_he.RDS
Ignored: data/cormotif_full_6_run.RDS
Ignored: data/cormotif_full_6_run_he.RDS
Ignored: data/cormotif_probability_45_list.csv
Ignored: data/cormotif_probability_45_list_he.csv
Ignored: data/cormotif_probability_all_6_list.csv
Ignored: data/cormotif_probability_all_6_list_he.csv
Ignored: data/datasave.RDS
Ignored: data/embryo_heart_FRIP.csv
Ignored: data/enhancer_list_ENCFF126UHK.bed
Ignored: data/enhancerdata/
Ignored: data/filt_Peaks_efit2.RDS
Ignored: data/filt_Peaks_efit2_bl.RDS
Ignored: data/filt_Peaks_efit2_n45.RDS
Ignored: data/first_Peaksummarycounts.csv
Ignored: data/first_run_frag_counts.txt
Ignored: data/full_bedfiles/
Ignored: data/gene_ref.csv
Ignored: data/gwas_1_dataframe.RDS
Ignored: data/gwas_2_dataframe.RDS
Ignored: data/gwas_3_dataframe.RDS
Ignored: data/gwas_4_dataframe.RDS
Ignored: data/gwas_5_dataframe.RDS
Ignored: data/high_conf_peak_counts.csv
Ignored: data/high_conf_peak_counts.txt
Ignored: data/high_conf_peaks_bl_counts.txt
Ignored: data/high_conf_peaks_counts.txt
Ignored: data/hits_files/
Ignored: data/hyper_files/
Ignored: data/hypo_files/
Ignored: data/ind1_DA24hpeaks.RDS
Ignored: data/ind1_TSSE.RDS
Ignored: data/ind2_TSSE.RDS
Ignored: data/ind3_TSSE.RDS
Ignored: data/ind4_TSSE.RDS
Ignored: data/ind4_V24hpeaks.RDS
Ignored: data/ind5_TSSE.RDS
Ignored: data/ind6_TSSE.RDS
Ignored: data/initial_complete_stats_run1.txt
Ignored: data/left_ventricle_FRIP.csv
Ignored: data/median_24_lfc.RDS
Ignored: data/median_3_lfc.RDS
Ignored: data/mergedPeads.gff
Ignored: data/mergedPeaks.gff
Ignored: data/motif_list_full
Ignored: data/motif_list_n45
Ignored: data/motif_list_n45.RDS
Ignored: data/multiqc_fastqc_run1.txt
Ignored: data/multiqc_fastqc_run2.txt
Ignored: data/multiqc_genestat_run1.txt
Ignored: data/multiqc_genestat_run2.txt
Ignored: data/my_hc_filt_counts.RDS
Ignored: data/my_hc_filt_counts_n45.RDS
Ignored: data/n45_bedfiles/
Ignored: data/n45_files
Ignored: data/other_papers/
Ignored: data/peakAnnoList_1.RDS
Ignored: data/peakAnnoList_2.RDS
Ignored: data/peakAnnoList_24_full.RDS
Ignored: data/peakAnnoList_24_n45.RDS
Ignored: data/peakAnnoList_3.RDS
Ignored: data/peakAnnoList_3_full.RDS
Ignored: data/peakAnnoList_3_n45.RDS
Ignored: data/peakAnnoList_4.RDS
Ignored: data/peakAnnoList_5.RDS
Ignored: data/peakAnnoList_6.RDS
Ignored: data/peakAnnoList_Eight.RDS
Ignored: data/peakAnnoList_full_motif.RDS
Ignored: data/peakAnnoList_n45_motif.RDS
Ignored: data/siglist_full.RDS
Ignored: data/siglist_n45.RDS
Ignored: data/summarized_peaks_dataframe.txt
Ignored: data/summary_peakIDandReHeat.csv
Ignored: data/test.list.RDS
Ignored: data/testnames.txt
Ignored: data/toplist_6.RDS
Ignored: data/toplist_full.RDS
Ignored: data/toplist_full_DAR_6.RDS
Ignored: data/toplist_n45.RDS
Ignored: data/trimmed_seq_length.csv
Ignored: data/unclassified_full_set_peaks.RDS
Ignored: data/unclassified_n45_set_peaks.RDS
Ignored: data/xstreme/
Untracked files:
Untracked: RNA_seq_integration.Rmd
Untracked: Rplot.pdf
Untracked: Sig_meta
Untracked: analysis/.gitignore
Untracked: analysis/Cormotif_analysis_testing diff.Rmd
Untracked: analysis/Diagnosis-tmm.Rmd
Untracked: analysis/Expressed_RNA_associations.Rmd
Untracked: analysis/LFC_corr.Rmd
Untracked: analysis/SVA.Rmd
Untracked: analysis/Tan2020.Rmd
Untracked: analysis/making_master_peaks_list.Rmd
Untracked: analysis/my_hc_filt_counts.csv
Untracked: code/Concatenations_for_export.R
Untracked: code/IGV_snapshot_code.R
Untracked: code/LongDARlist.R
Untracked: code/just_for_Fun.R
Untracked: my_plot.pdf
Untracked: my_plot.png
Untracked: output/cormotif_probability_45_list.csv
Untracked: output/cormotif_probability_all_6_list.csv
Untracked: setup.RData
Unstaged changes:
Modified: ATAC_learning.Rproj
Modified: analysis/AC_shared_analysis.Rmd
Modified: analysis/AF_HF_SNP_DAR.Rmd
Modified: analysis/AF_HF_SNPs.Rmd
Modified: analysis/Cardiotox_SNPs.Rmd
Modified: analysis/Cormotif_analysis.Rmd
Modified: analysis/DEG_analysis.Rmd
Modified: analysis/H3K27ac_initial_QC.Rmd
Modified: analysis/H3K27ac_integration.Rmd
Modified: analysis/Jaspar_motif.Rmd
Modified: analysis/Jaspar_motif_ff.Rmd
Modified: analysis/TE_analysis_norm.Rmd
Modified: analysis/Top2B_analysis.Rmd
Modified: analysis/final_four_analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/SNP_TAD_peaks.Rmd) and
HTML (docs/SNP_TAD_peaks.html) files. If you’ve configured
a remote Git repository (see ?wflow_git_remote), click on
the hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | ad110ea | reneeisnowhere | 2025-07-29 | updates |
| html | 1429820 | reneeisnowhere | 2025-07-21 | Build site. |
| Rmd | b838697 | reneeisnowhere | 2025-07-21 | adding in SNP overlap heatmap |
| html | 1a9df02 | reneeisnowhere | 2025-07-09 | Build site. |
| Rmd | 0fa75c3 | reneeisnowhere | 2025-07-09 | wflow_publish("analysis/SNP_TAD_peaks.Rmd") |
library(tidyverse)
library(kableExtra)
library(broom)
library(RColorBrewer)
library(ChIPseeker)
library(ChIPpeakAnno)
library("TxDb.Hsapiens.UCSC.hg38.knownGene")
library("org.Hs.eg.db")
library(rtracklayer)
library(edgeR)
library(ggfortify)
library(limma)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(VennDiagram)
library(scales)
library(BiocParallel)
library(ggpubr)
library(devtools)
library(biomaRt)
library(eulerr)
library(smplot2)
library(genomation)
library(ggsignif)
library(plyranges)
library(ggrepel)
library(epitools)
library(circlize)
library(readxl)
library(ComplexHeatmap)
library(gwascat)
library(liftOver)Loading data frames
### Pulling the all regions granges list from the motif list of lists
Motif_list_gr <- readRDS("data/Final_four_data/re_analysis/Motif_list_granges.RDS")
### no change motif_list_gr names so they do not overwrite the dataframes
names(Motif_list_gr) <- paste0(names(Motif_list_gr), "_gr")
list2env(Motif_list_gr[10],envir= .GlobalEnv)<environment: R_GlobalEnv>annotated_DARs<- readRDS("data/Final_four_data/re_analysis/DOX_DAR_annotated_peaks_chipannno.RDS")
Left_ventricle_TAD <- import(con = "C://Users/renee/Downloads/hg38.TADs/hg38/VentricleLeft_STL003_Leung_2015-raw_TADs.txt", format = "bed",genome="hg38")
mcols(Left_ventricle_TAD)$TAD_id <- paste0("TAD_", seq_along(Left_ventricle_TAD))
# mcols(Left_ventricle_TAD)$name <- Left_ventricle_TAD$TAD_id
##exporting the LEFt ventricle tad info for IGV visualization
# export(Left_ventricle_TAD, con = "data/Final_four_data/re_analysis/Other_bed_files/TAD_regions.bed", format="bed")
Schneider_all_SNPS <- read_delim("data/other_papers/Schneider_all_SNPS.txt",
delim = "\t", escape_double = FALSE,
trim_ws = TRUE)
Schneider_all_SNPS_df <- Schneider_all_SNPS %>%
dplyr::rename("RSID"="#Uploaded_variation") %>%
dplyr::select(RSID,Location,SYMBOL,Gene, SOURCE) %>%
distinct(RSID,Location,SYMBOL,.keep_all = TRUE) %>%
dplyr::rename("Close_SYMBOL"="SYMBOL") %>%
dplyr::filter(!str_starts(Location, "H")) %>%
separate_wider_delim(Location,delim=":",names=c("Chr","Coords")) %>%
separate_wider_delim(Coords,delim= "-", names= c("Start","End")) %>%
mutate(Chr=paste0("chr",Chr)) %>%
group_by(RSID) %>%
reframe(Chr=unique(Chr),
Start=unique(Start),
End=unique(End),
Close_SYMBOL=paste(unique(Close_SYMBOL),collapse=";"),
Gene=paste(Gene,collapse=";"),
SOURCE=paste(SOURCE,collapse=";")
) %>%
GRanges() %>% as.data.frame
schneider_gr <-Schneider_all_SNPS_df%>%
dplyr::select(seqnames,start,end,RSID:SOURCE) %>%
distinct() %>%
GRanges()
# export(schneider_gr, con = "data/Final_four_data/re_analysis/Other_bed_files/CardiotoxSNPs.bed", format="bed")
toptable_results <- readRDS("data/Final_four_data/re_analysis/Toptable_results.RDS")
all_results <- toptable_results %>%
imap(~ .x %>% tibble::rownames_to_column(var = "rowname") %>%
mutate(source = .y)) %>%
bind_rows()
all_results_pivot <- all_results %>%
dplyr::select(genes,logFC,source) %>%
pivot_wider(., id_cols = genes, names_from = source, values_from = logFC) %>%
dplyr::select(genes,DOX_3,EPI_3,DNR_3,MTX_3,TRZ_3,DOX_24,EPI_24,DNR_24,MTX_24,TRZ_24)
toplistall_RNA <- readRDS("data/other_papers/toplistall_RNA.RDS") %>%
mutate(logFC = logFC*(-1))
Assigned_genes_toPeak <- annotated_DARs$DOX_24 %>% as.data.frame() %>%
dplyr::select(mcols.genes,annotation, geneId, distanceToTSS) %>%
dplyr::rename("Peakid"=mcols.genes)
RNA_results <-
toplistall_RNA %>%
dplyr::select(time:logFC) %>%
tidyr::unite("sample",time, id) %>%
pivot_wider(., id_cols = c(ENTREZID,SYMBOL),names_from = sample, values_from = logFC) %>%
rename_with(~ str_replace(., "hours", "RNA"))
Peak_gene_RNA_LFC <- Assigned_genes_toPeak %>%
left_join(., RNA_results, by =c("geneId"="ENTREZID"))
entrez_ids <- Assigned_genes_toPeak$geneId
gene_info <- AnnotationDbi::select(
org.Hs.eg.db,
keys = entrez_ids,
columns = c("SYMBOL"),
keytype = "ENTREZID"
)
gene_info_collapsed <- gene_info %>%
group_by(ENTREZID) %>%
summarise(SYMBOL = paste(unique(SYMBOL), collapse = ","), .groups = "drop")
DOX_DAR_24hr_table <- annotated_DARs$DOX_24 %>%
as.data.frame()
Top2b_peaks <- import(con="data/other_papers/ChIP3_TOP2B_CM_87-1.bed",format = "bed",genome="hg38")
# for_export <- DOX_24_DAR%>%
# # as.data.frame() %>%
# ### mark significance with color
# mutate(sig_24=if_else(mcols.adj.P.Val<0.05,"TRUE","FALSE")) %>%
# dplyr::select(seqnames:mcols.genes,sig_24) %>%
# GRanges
#
# ### add to the granges thingy for exporting
# mcols(for_export)$itemRgb <- ifelse(mcols(for_export)$sig_24,
# "255,0,0", # red for significant
# "190,190,190") # gray for not significant
# mcols(for_export)$sig_24 <- as.logical(mcols(for_export)$sig_24)
# mcols(for_export)$itemRgb <- as.character(mcols(for_export)$itemRgb)
#
# bed_df <- data.frame(
# seqnames = seqnames(for_export),
# start = start(for_export) - 1, # BED is 0-based
# end = end(for_export),
# strand = "*",
# thickStart = start(for_export) - 1,
# thickEnd = end(for_export),
# itemRgb = ifelse(mcols(for_export)$sig_24, "255,0,0", "190,190,190"),
# stringsAsFactors = FALSE)
# # )
#
# write.table(bed_df,
# file = "data/Final_four_data/re_analysis/Other_bed_files/DOX_24hour_sig_notsig_regions.bed",
# quote = FALSE, sep = "\t", row.names = FALSE, col.names = FALSE)Enrichment test of sig DAR and non-sig DAR of DOX within SNP-containing TADS
test_ol <- join_overlap_intersect(Left_ventricle_TAD, schneider_gr)
df <- as.data.frame(test_ol, row.names = NULL)
TAD_SNP_ol <- test_ol %>% as.data.frame() %>%
distinct(TAD_id, RSID)
peak_ol <- join_overlap_intersect(all_regions_gr, Left_ventricle_TAD)
TAD_SNP_Peak_ol <- peak_ol %>%
as.data.frame() %>%
dplyr::filter(TAD_id %in% TAD_SNP_ol$TAD_id)
snp_ol <- join_overlap_inner(schneider_gr, Left_ventricle_TAD)
TAD_peak_ol <- peak_ol %>%
as.data.frame() %>%
distinct(Peakid,.keep_all = TRUE)
left_ventricle_ol <- join_overlap_inner(all_regions_gr ,Left_ventricle_TAD) %>%
as.data.frame() %>%
distinct(Peakid,.keep_all = TRUE) %>%
dplyr::filter(TAD_id %in% TAD_SNP_ol$TAD_id)
peak_df <- as.data.frame(left_ventricle_ol)
SNP_df <- as.data.frame(snp_ol)
peak_snp_pairs <- inner_join(peak_df, SNP_df, by = "TAD_id", suffix = c(".peak", ".snp")) %>%
mutate(
peak_center = (start.peak + end.peak) / 2,
distance = abs(peak_center - start.snp) # or any metric you prefer
)reds <- colorRampPalette(brewer.pal(9, "Reds")[3:9])(12)
greens <- colorRampPalette(brewer.pal(9, "Greens")[3:9])(12)
blues <- colorRampPalette(brewer.pal(9, "Blues")[3:9])(12)
purples <- colorRampPalette(brewer.pal(9, "Purples")[3:9])(12)
oranges <- colorRampPalette(brewer.pal(9, "Oranges")[3:9])(12)
tads <- unique(peak_snp_pairs$TAD_id)
num_tads <- length(tads)
color_spectrum <- c(reds, greens, blues, purples, oranges)[1:num_tads]
if (num_tads > length(color_spectrum)) {
stop("Not enough colors for TADs. Add more palettes.")
}
tad_colors <- color_spectrum[1:num_tads]
names(tad_colors) <- tads # Assign color names to TAD IDs
#ha <- HeatmapAnnotation(TAD = df$TAD_id, col = list(TAD = tad_colors))Top2b_overlap_regions <-join_overlap_inner(all_regions_gr ,Top2b_peaks) %>%
as.data.frame() %>%
distinct(Peakid,.keep_all = TRUE) DOX_24_DAR <- as.data.frame(annotated_DARs$DOX_24)
EPI_24_DAR <- as.data.frame(annotated_DARs$EPI_24)
DNR_24_DAR <- as.data.frame(annotated_DARs$DNR_24)
MTX_24_DAR <- as.data.frame(annotated_DARs$MTX_24)
DOX_3_DAR <- as.data.frame(annotated_DARs$DOX_3)
EPI_3_DAR <- as.data.frame(annotated_DARs$EPI_3)
DNR_3_DAR <- as.data.frame(annotated_DARs$DNR_3)
MTX_3_DAR <- as.data.frame(annotated_DARs$MTX_3)
TAD_count_df <- DOX_24_DAR %>%
dplyr::select(mcols.genes, mcols.adj.P.Val,annotation:distanceToTSS) %>%
mutate(sig_24=if_else(mcols.adj.P.Val<0.05,"sig","not_sig")) %>%
mutate(sig_24=factor(sig_24, levels = c("sig","not_sig"))) %>%
mutate(TAD_all_status=if_else(mcols.genes %in% peak_ol$Peakid,"TAD_peak","not_TAD_peak")) %>%
mutate(SNP_TAD_status= if_else(mcols.genes %in% TAD_SNP_Peak_ol$Peakid,"SNP_TAD","not_SNP_TAD")) %>%
mutate(Top2b_peak= if_else(mcols.genes %in% Top2b_overlap_regions$Peakid, "TOP2B_peak","not_TOP2B_peak"))
TAD_count_df %>% #dplyr::filter(TAD_all_status=="TAD_peak") %>%
group_by(sig_24,SNP_TAD_status,TAD_all_status) %>%
tally #%>% # A tibble: 6 × 4
# Groups: sig_24, SNP_TAD_status [4]
sig_24 SNP_TAD_status TAD_all_status n
<fct> <chr> <chr> <int>
1 sig SNP_TAD TAD_peak 2047
2 sig not_SNP_TAD TAD_peak 56865
3 sig not_SNP_TAD not_TAD_peak 5908
4 not_sig SNP_TAD TAD_peak 3111
5 not_sig not_SNP_TAD TAD_peak 78627
6 not_sig not_SNP_TAD not_TAD_peak 8999 # pivot_wider(., id_cols = sig_24, names_from = SNP_TAD_status, values_from = n)
# print("Odds ratio of SNP_TADs across DOXall regions, regardless of TAD_status")
# TAD_count_df %>% #dplyr::filter(TAD_all_status=="TAD_peak") %>%
# group_by(sig_24,SNP_TAD_status) %>%
# tally %>%
# pivot_wider(., id_cols = sig_24, names_from = SNP_TAD_status, values_from = n) %>%
# column_to_rownames( "sig_24") %>% as.matrix() %>%
# # chisq.test()
# epitools::oddsratio()
print("Odds ratio testing proportion SNP-containing TADs of sig-DOX DARs vs non-sig DARs at 24 hours")[1] "Odds ratio testing proportion SNP-containing TADs of sig-DOX DARs vs non-sig DARs at 24 hours"TAD_count_df %>% dplyr::filter(TAD_all_status=="TAD_peak") %>%
group_by(sig_24,SNP_TAD_status) %>%
tally %>%
pivot_wider(., id_cols = sig_24, names_from = SNP_TAD_status, values_from = n) %>%
column_to_rownames( "sig_24") %>% as.matrix() %>%
# chisq.test()
epitools::oddsratio(method = "wald")$data
SNP_TAD not_SNP_TAD Total
sig 2047 56865 58912
not_sig 3111 78627 81738
Total 5158 135492 140650
$measure
NA
odds ratio with 95% C.I. estimate lower upper
sig 1.000000 NA NA
not_sig 0.909797 0.8595486 0.9629828
$p.value
NA
two-sided midp.exact fisher.exact chi.square
sig NA NA NA
not_sig 0.00107761 0.001098901 0.001105101
$correction
[1] FALSE
attr(,"method")
[1] "Unconditional MLE & normal approximation (Wald) CI"TAD_count_df %>%
dplyr::filter(TAD_all_status=="TAD_peak") %>%
group_by(sig_24,SNP_TAD_status) %>%
tally ()%>%
mutate(sig_24=factor(sig_24, levels = c("sig","not_sig"))) %>%
ggplot(.,aes(x=sig_24, y= n,fill=SNP_TAD_status))+
geom_col(position="fill")+
theme_bw()+
ggtitle("Proportion of significant regions by 24 hours")+
ylab("proportion")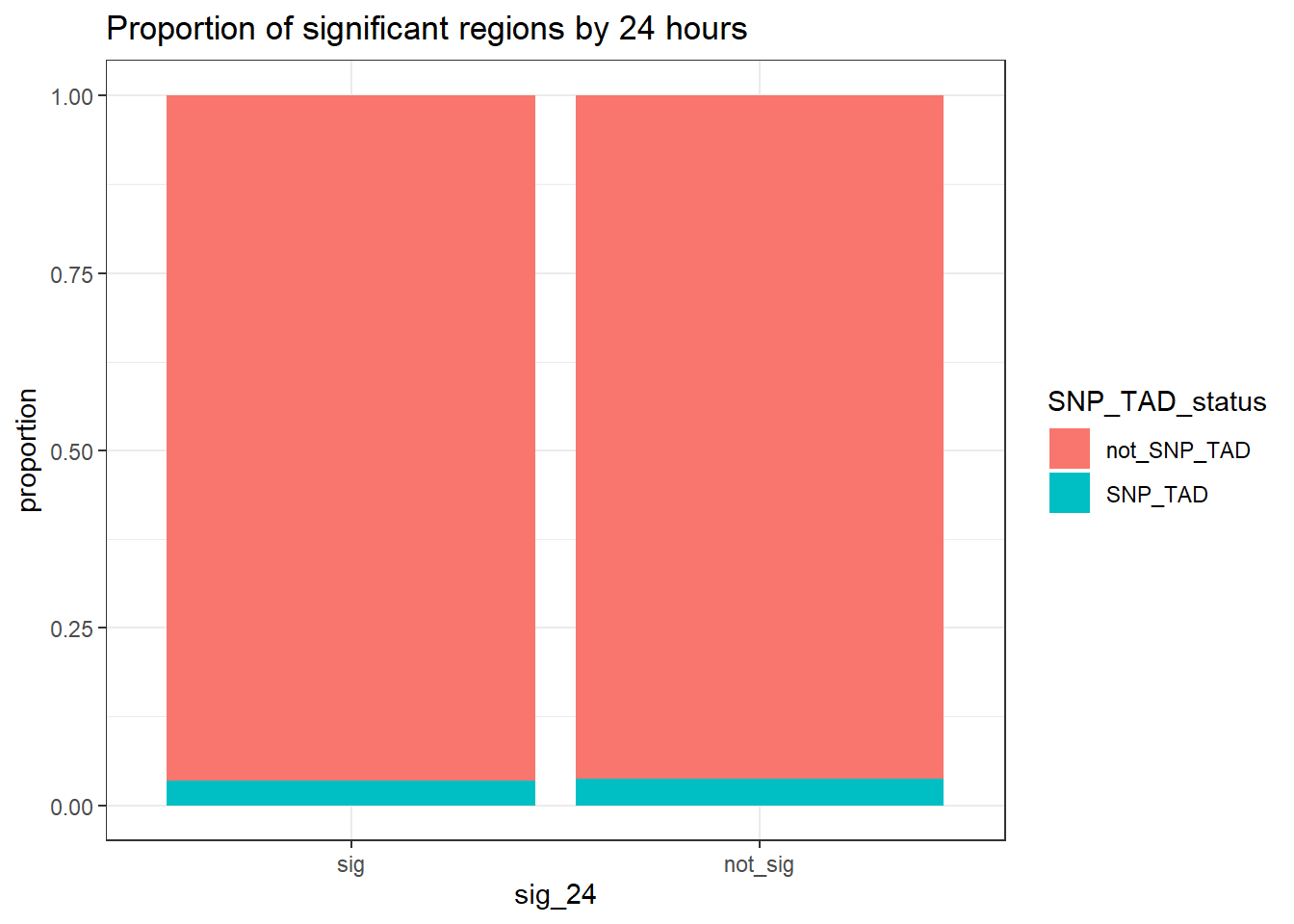
# TAD_count_df %>%
# dplyr::filter(TAD_all_status=="TAD_peak") %>%
#
# group_by(sig_24,SNP_TAD_status) %>%
# tally ()%>%
# mutate(sig_24=factor(sig_24, levels = c("sig","not_sig"))) %>%
# ggplot(.,aes(x=SNP_TAD_status, y= n,fill=sig_24))+
# geom_col(position="fill")+
# theme_bw()+
# ggtitle("Proportion of significant regions by 24 hours")+
# ylab("proportion")Proportion of DARs that overlap TOP2B peaks in a TAD
TAD_count_df %>%
dplyr::filter((TAD_all_status=="TAD_peak")) %>%
dplyr::filter(SNP_TAD_status=="SNP_TAD") %>%
group_by(SNP_TAD_status, Top2b_peak, sig_24) %>%
tally() %>%
pivot_wider(., id_cols=sig_24, names_from = Top2b_peak, values_from = n) %>%
print() %>%
column_to_rownames("sig_24") %>%
fisher.test()# A tibble: 2 × 3
sig_24 TOP2B_peak not_TOP2B_peak
<fct> <int> <int>
1 sig 32 2015
2 not_sig 121 2990
Fisher's Exact Test for Count Data
data: .
p-value = 8.347e-07
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
0.2560764 0.5863054
sample estimates:
odds ratio
0.3924942 Calculating Distance to TAD-SNP from peak
DOX 24 hours
DOX_DAR_sig <- DOX_24_DAR %>%
dplyr::filter(mcols.adj.P.Val<0.05) %>%
distinct (mcols.genes) %>%
dplyr::rename("Peakid"="mcols.genes")
DOX_DAR_sig_3 <- DOX_3_DAR %>%
dplyr::filter(mcols.adj.P.Val<0.05) %>%
distinct (mcols.genes) %>%
dplyr::rename("Peakid"="mcols.genes")
EPI_DAR_sig <- EPI_24_DAR %>%
dplyr::filter(mcols.adj.P.Val<0.05) %>%
distinct (mcols.genes) %>%
dplyr::rename("Peakid"="mcols.genes")
EPI_DAR_sig_3 <- EPI_3_DAR %>%
dplyr::filter(mcols.adj.P.Val<0.05) %>%
distinct (mcols.genes) %>%
dplyr::rename("Peakid"="mcols.genes")
DNR_DAR_sig <- DNR_24_DAR %>%
dplyr::filter(mcols.adj.P.Val<0.05) %>%
distinct (mcols.genes) %>%
dplyr::rename("Peakid"="mcols.genes")
DNR_DAR_sig_3 <- DNR_3_DAR %>%
dplyr::filter(mcols.adj.P.Val<0.05) %>%
distinct (mcols.genes) %>%
dplyr::rename("Peakid"="mcols.genes")
MTX_DAR_sig <- MTX_24_DAR %>%
dplyr::filter(mcols.adj.P.Val<0.05) %>%
distinct (mcols.genes) %>%
dplyr::rename("Peakid"="mcols.genes")
MTX_DAR_sig_3 <- MTX_3_DAR %>%
dplyr::filter(mcols.adj.P.Val<0.05) %>%
distinct (mcols.genes) %>%
dplyr::rename("Peakid"="mcols.genes")
MCF7_DAR_snp_pairs_dist <- readRDS("data/Final_four_data/re_analysis/MCF7_DAR_snp_pairs_dist.RDS") %>%
dplyr::rename("Peakid"=names) %>%
mutate(sig_24="MCF7_DAR")
snp_tad_df <-
join_overlap_inner(schneider_gr, Left_ventricle_TAD) %>%
as_tibble() %>%
dplyr::select(RSID, snp_start = start, snp_chr = seqnames, TAD_id)
peak_tad_df <-
join_overlap_inner(all_regions_gr, Left_ventricle_TAD) %>%
as_tibble() %>%
dplyr::select(Peakid, peak_start = start, peak_chr = seqnames, TAD_id)
peak_snp_pairs <- peak_tad_df %>%
inner_join(snp_tad_df, by = "TAD_id")
peak_snp_pairs_dist <- peak_snp_pairs %>%
mutate(distance = abs(peak_start - snp_start)) %>%
mutate(sig_24= if_else(Peakid %in% DOX_DAR_sig$Peakid, "sig","not_sig"))
peak_snp_pairs_dist %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig"))) %>%
ggplot(., aes(x= sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
map_signif_level = FALSE, test = "wilcox.test")+
ggtitle("DOX 24 hour distances of DAR-SNP pairs and non-DAR-SNP pairs")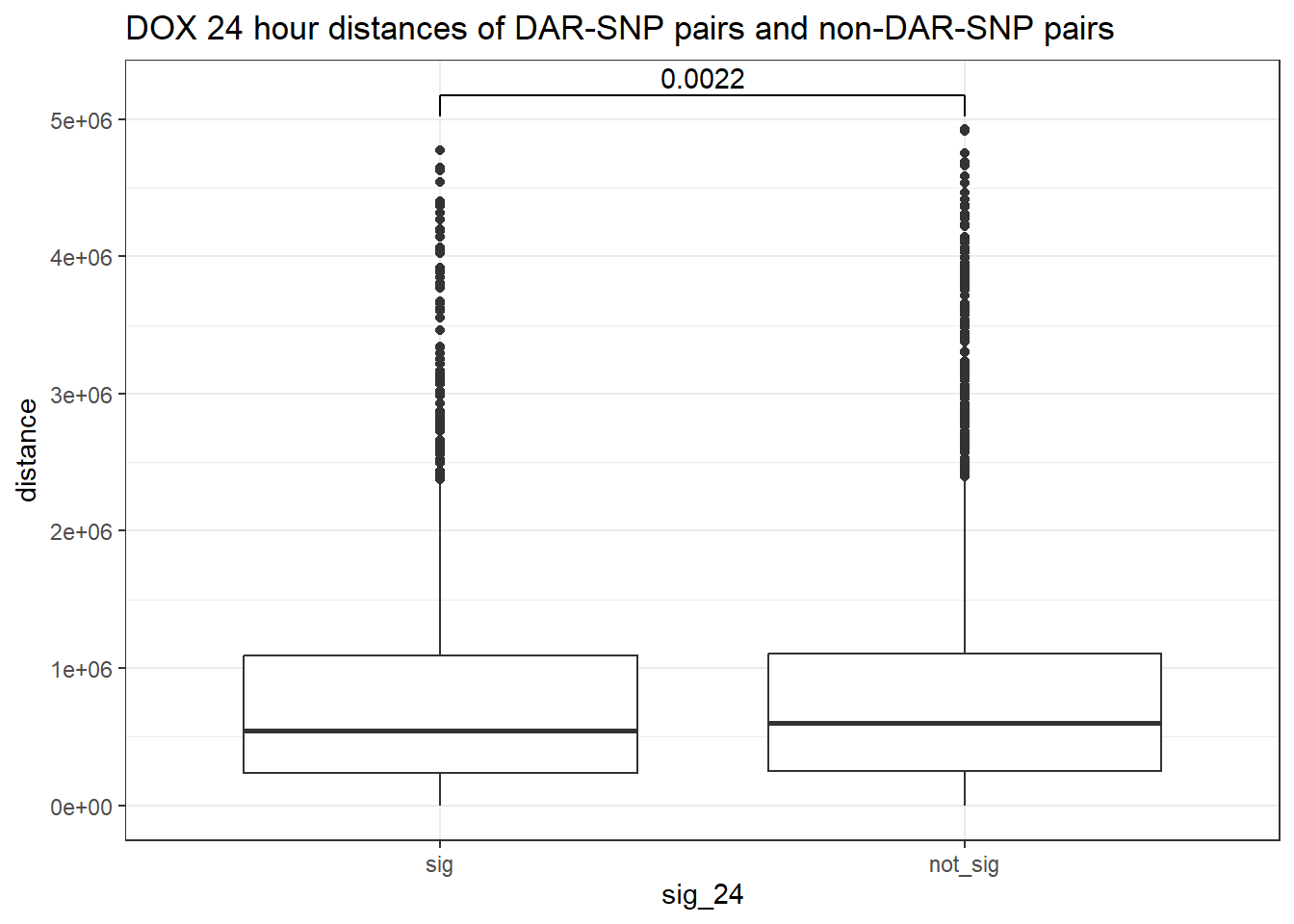
wilcox.test(distance ~ sig_24, data = peak_snp_pairs_dist)
Wilcoxon rank sum test with continuity correction
data: distance by sig_24
W = 9463083, p-value = 0.002185
alternative hypothesis: true location shift is not equal to 0Cardiotox_gwas_df <- peak_snp_pairs_dist %>%
dplyr::filter(sig_24=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)
Cardiotox_gwas_collaped_df <-
peak_snp_pairs_dist %>%
dplyr::filter(sig_24=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
group_by(Peakid, peak_chr, peak_start, TAD_id, sig_24) %>%
summarise(
min_distance = min(distance),
mean_distance = mean(distance),
snp_list = paste(unique(RSID), collapse = ","),
.groups = "drop"
) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
left_join(., Peak_gene_RNA_LFC, by=c("Peakid"="Peakid")) %>%
left_join(.,gene_info_collapsed, by=c("geneId"="ENTREZID")) %>%
mutate(SYMBOL=if_else(is.na(SYMBOL.x),SYMBOL.y,if_else(SYMBOL.x==SYMBOL.y, SYMBOL.x,paste0(SYMBOL.x,"_",SYMBOL.y)))) %>%
tidyr::unite(., name,Peakid,SYMBOL,snp_list) %>%
mutate(snp_dist=case_when(min_distance <2000 ~"2kb",
min_distance > 2000 & min_distance<20000 ~ "20kb",
min_distance >20000 ~">20kb"))peak_snp_pairs_dist_DOX_3 <- peak_snp_pairs %>%
mutate(distance = abs(peak_start - snp_start)) %>%
mutate(sig_3= if_else(Peakid %in% DOX_DAR_sig_3$Peakid, "sig","not_sig"))
peak_snp_pairs_dist_DOX_3 %>%
mutate(sig_3=factor(sig_3, levels= c("sig","not_sig"))) %>%
ggplot(., aes(x= sig_3, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
map_signif_level = FALSE, test = "wilcox.test")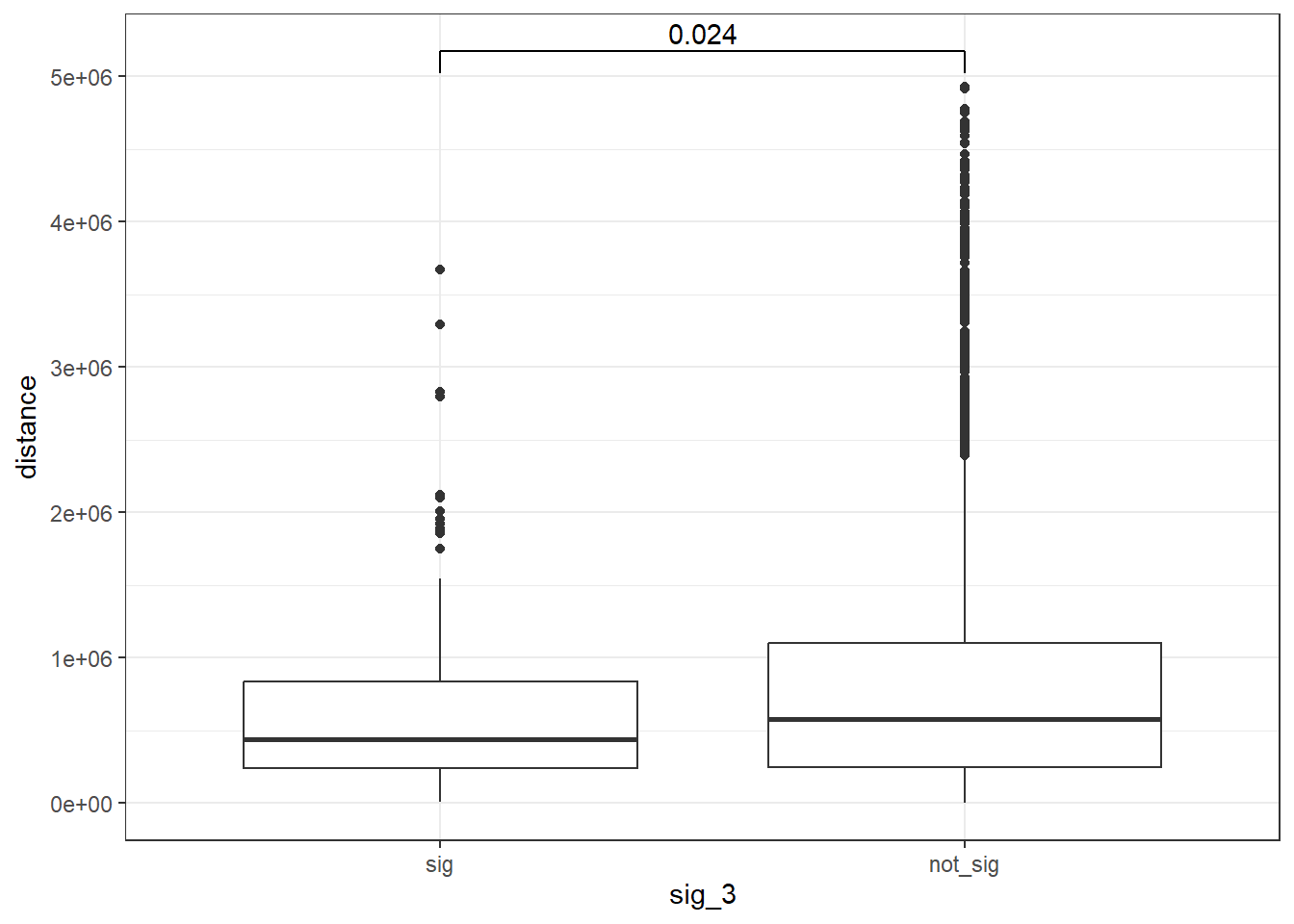
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
wilcox.test(distance ~ sig_3, data = peak_snp_pairs_dist_DOX_3)
Wilcoxon rank sum test with continuity correction
data: distance by sig_3
W = 837367, p-value = 0.0241
alternative hypothesis: true location shift is not equal to 0Cardiotox_gwas_EPI <- peak_snp_pairs_dist_DOX_3 %>%
dplyr::filter(sig_3=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)Looking at SNPs that directly overlap DARs
snp_peak_ol <- join_overlap_inner(all_regions_gr,schneider_gr)
SNP_DAR_overlap_direct <- snp_peak_ol %>%
as.data.frame() %>%
mutate(Dox_24=if_else(Peakid %in% DOX_DAR_sig$Peakid,"yes","no")) %>%
mutate(Epi_24=if_else(Peakid %in% EPI_DAR_sig$Peakid,"yes","no")) %>%
mutate(Dnr_24=if_else(Peakid %in% DNR_DAR_sig$Peakid,"yes","no")) %>%
mutate(MTx_24=if_else(Peakid %in% MTX_DAR_sig$Peakid,"yes","no")) %>%
mutate(Dox_3=if_else(Peakid %in% DOX_DAR_sig_3$Peakid,"yes","no")) %>%
mutate(Epi_3=if_else(Peakid %in% EPI_DAR_sig_3$Peakid,"yes","no")) %>%
mutate(Dnr_3=if_else(Peakid %in% DNR_DAR_sig_3$Peakid,"yes","no")) %>%
mutate(Mtx_3=if_else(Peakid %in% MTX_DAR_sig_3$Peakid,"yes","no")) %>%
dplyr::select(Peakid,RSID,Dox_24:Mtx_3)
SNP_DAR_overlap_direct Peakid RSID Dox_24 Epi_24 Dnr_24 MTx_24 Dox_3 Epi_3
1 chr18.58336709.58336869 rs12051934 yes yes yes no no no
2 chr2.176086015.176086512 rs6752623 no no no no no no
3 chr22.46249819.46250733 rs7291763 no no yes no no no
4 chr7.134834642.134835353 rs7777356 no yes no no no yes
Dnr_3 Mtx_3
1 no no
2 no no
3 no no
4 yes noDOX_MCF7 added
bind_rows(MCF7_DAR_snp_pairs_dist,peak_snp_pairs_dist) %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig", "MCF7_DAR"))) %>% ggplot(., aes(x= sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig"),
c("sig","MCF7_DAR"),
c("not_sig","MCF7_DAR")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("DOX 24 hour distances of DAR-SNP pairs\n and non-DAR-SNP pairs with MCF7 DARs")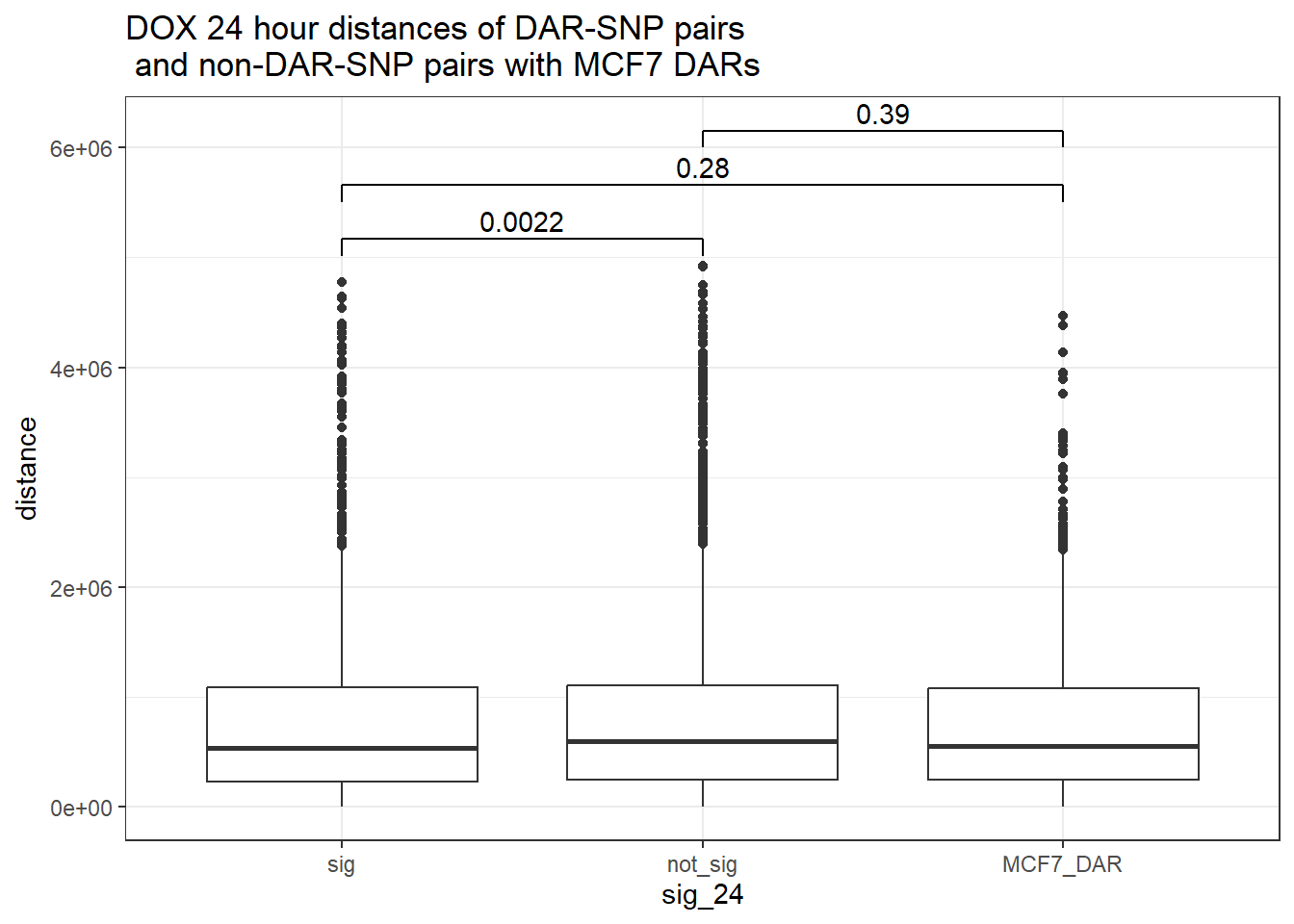
EPI 24 hours
peak_snp_pairs_dist_EPI <- peak_snp_pairs %>%
mutate(distance = abs(peak_start - snp_start)) %>%
mutate(sig_24= if_else(Peakid %in% EPI_DAR_sig$Peakid, "sig","not_sig"))
peak_snp_pairs_dist_EPI %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig"))) %>%
ggplot(., aes(x= sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
map_signif_level = FALSE, test = "wilcox.test")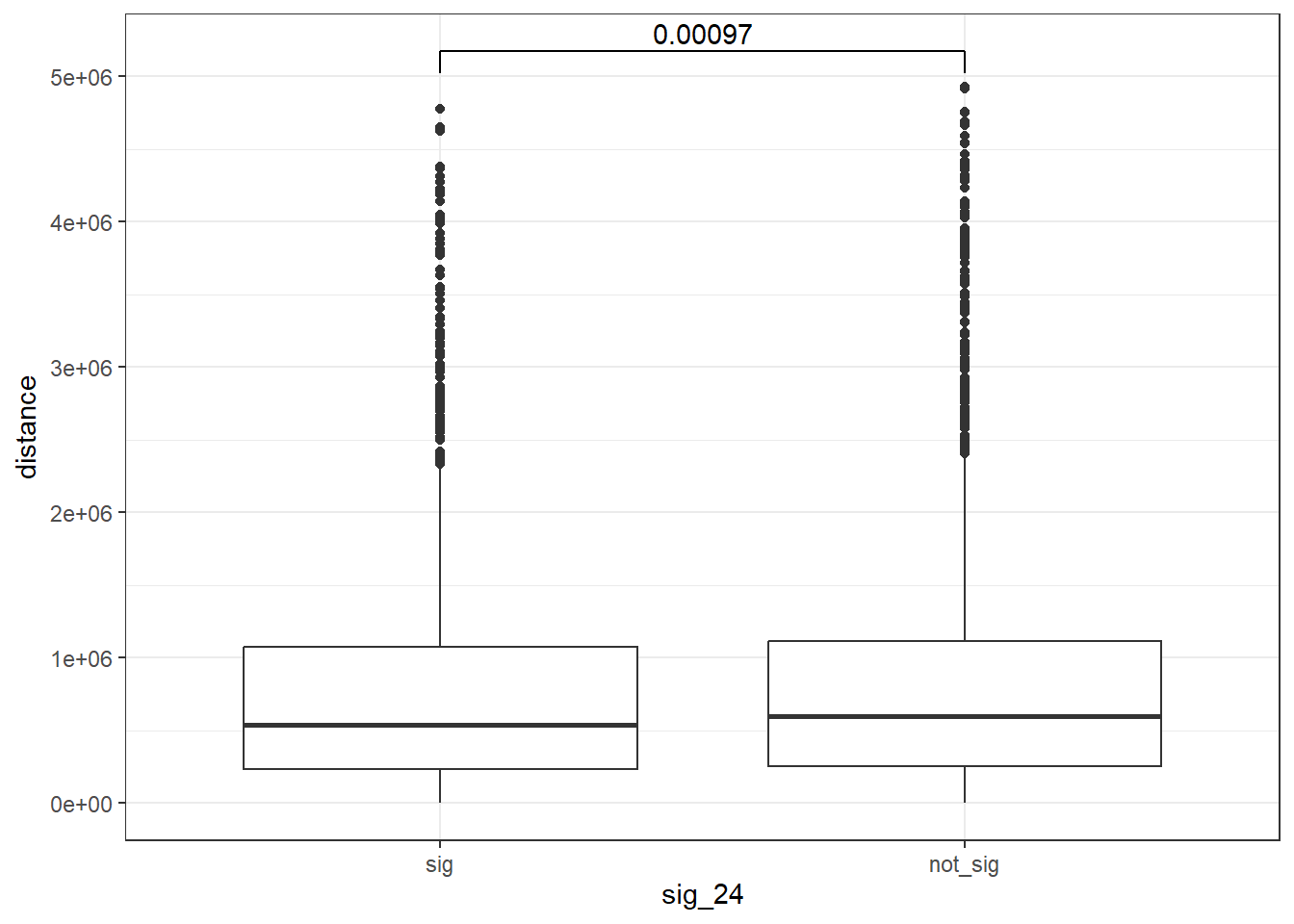
| Version | Author | Date |
|---|---|---|
| 1a9df02 | reneeisnowhere | 2025-07-09 |
wilcox.test(distance ~ sig_24, data = peak_snp_pairs_dist)
Wilcoxon rank sum test with continuity correction
data: distance by sig_24
W = 9463083, p-value = 0.002185
alternative hypothesis: true location shift is not equal to 0Cardiotox_gwas_EPI <- peak_snp_pairs_dist_EPI %>%
dplyr::filter(sig_24=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)EPI_MCF7 added
bind_rows(MCF7_DAR_snp_pairs_dist,peak_snp_pairs_dist_EPI) %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig", "MCF7_DAR"))) %>% ggplot(., aes(x= sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig"),
c("sig","MCF7_DAR"),
c("not_sig","MCF7_DAR")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("EPI 24 hour distances of DAR-SNP pairs\n and non-DAR-SNP pairs with MCF7 DARs")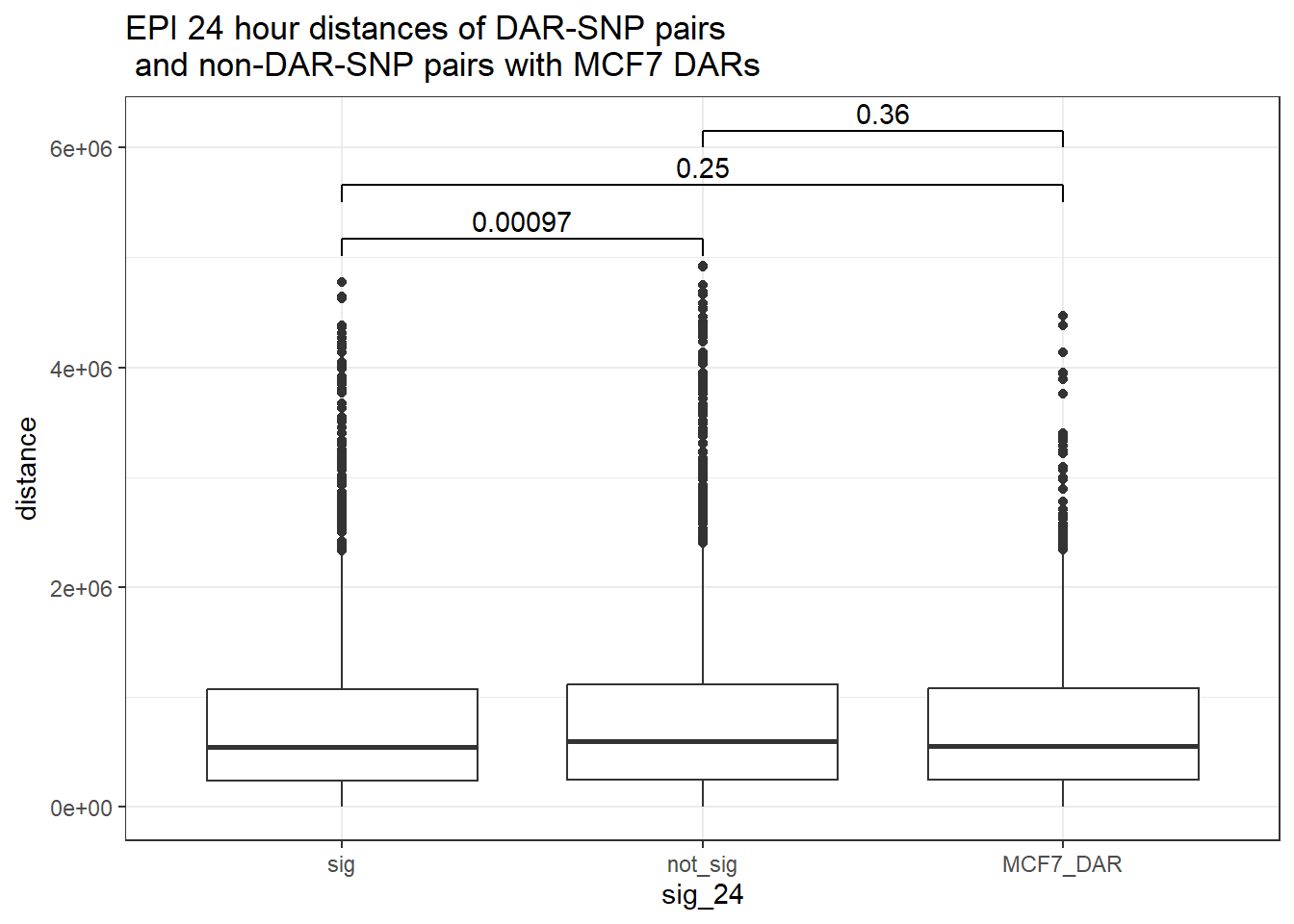
peak_snp_pairs_dist_EPI_3 <- peak_snp_pairs %>%
mutate(distance = abs(peak_start - snp_start)) %>%
mutate(sig_3= if_else(Peakid %in% EPI_DAR_sig_3$Peakid, "sig","not_sig"))
peak_snp_pairs_dist_EPI_3 %>%
mutate(sig_3=factor(sig_3, levels= c("sig","not_sig"))) %>%
ggplot(., aes(x= sig_3, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
map_signif_level = FALSE, test = "wilcox.test")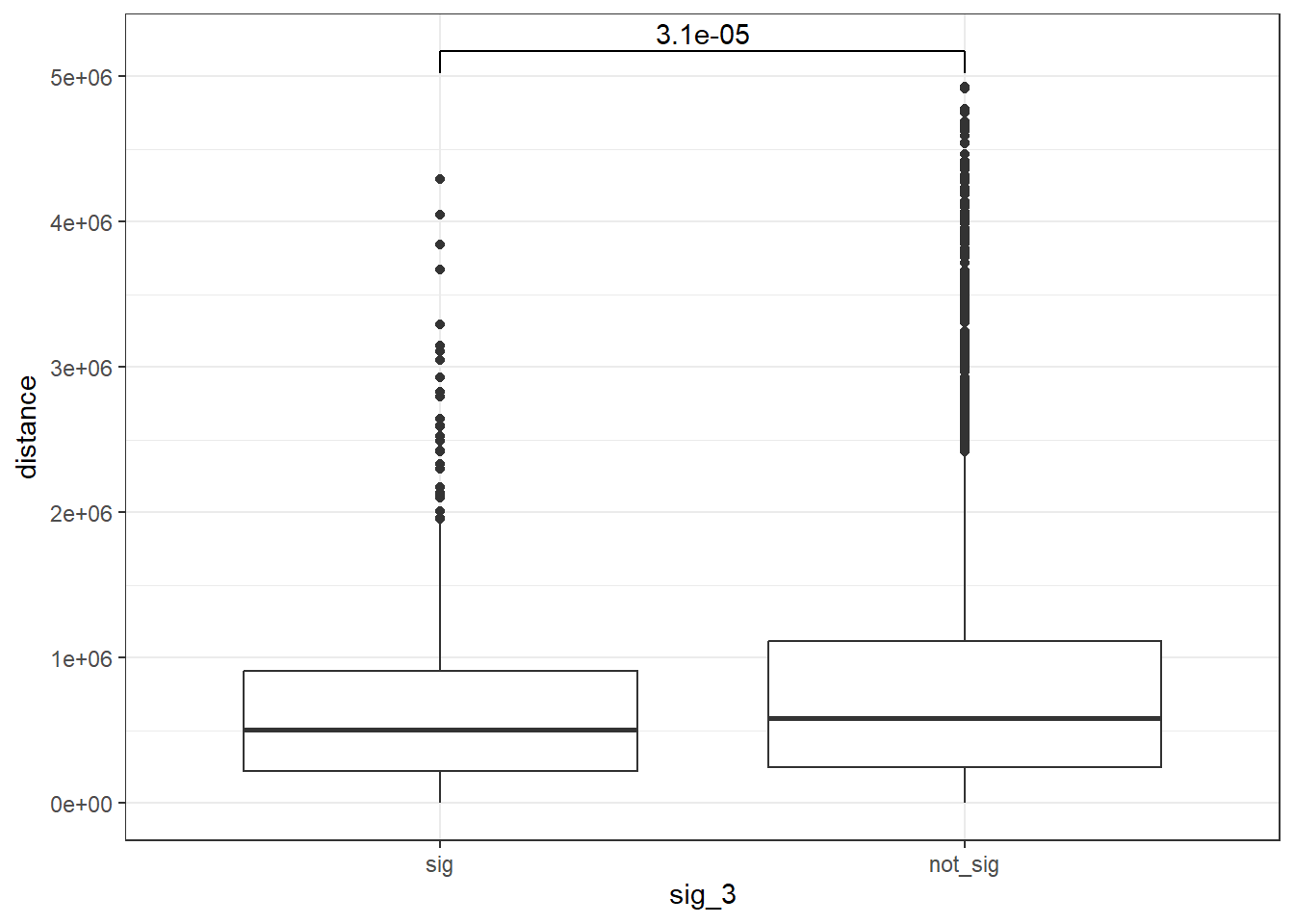
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
wilcox.test(distance ~ sig_3, data = peak_snp_pairs_dist_EPI_3)
Wilcoxon rank sum test with continuity correction
data: distance by sig_3
W = 3249493, p-value = 3.114e-05
alternative hypothesis: true location shift is not equal to 0Cardiotox_gwas_EPI_3 <- peak_snp_pairs_dist_EPI_3 %>%
dplyr::filter(sig_3=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)DNR 24 hours
peak_snp_pairs_dist_DNR <- peak_snp_pairs %>%
mutate(distance = abs(peak_start - snp_start)) %>%
mutate(sig_24= if_else(Peakid %in% DNR_DAR_sig$Peakid, "sig","not_sig"))
peak_snp_pairs_dist_DNR %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig"))) %>%
ggplot(., aes(x= sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
map_signif_level = FALSE, test = "wilcox.test")+
ggtitle("DNR 24 hour distances of DAR-SNP pairs and non-DAR-SNP pairs")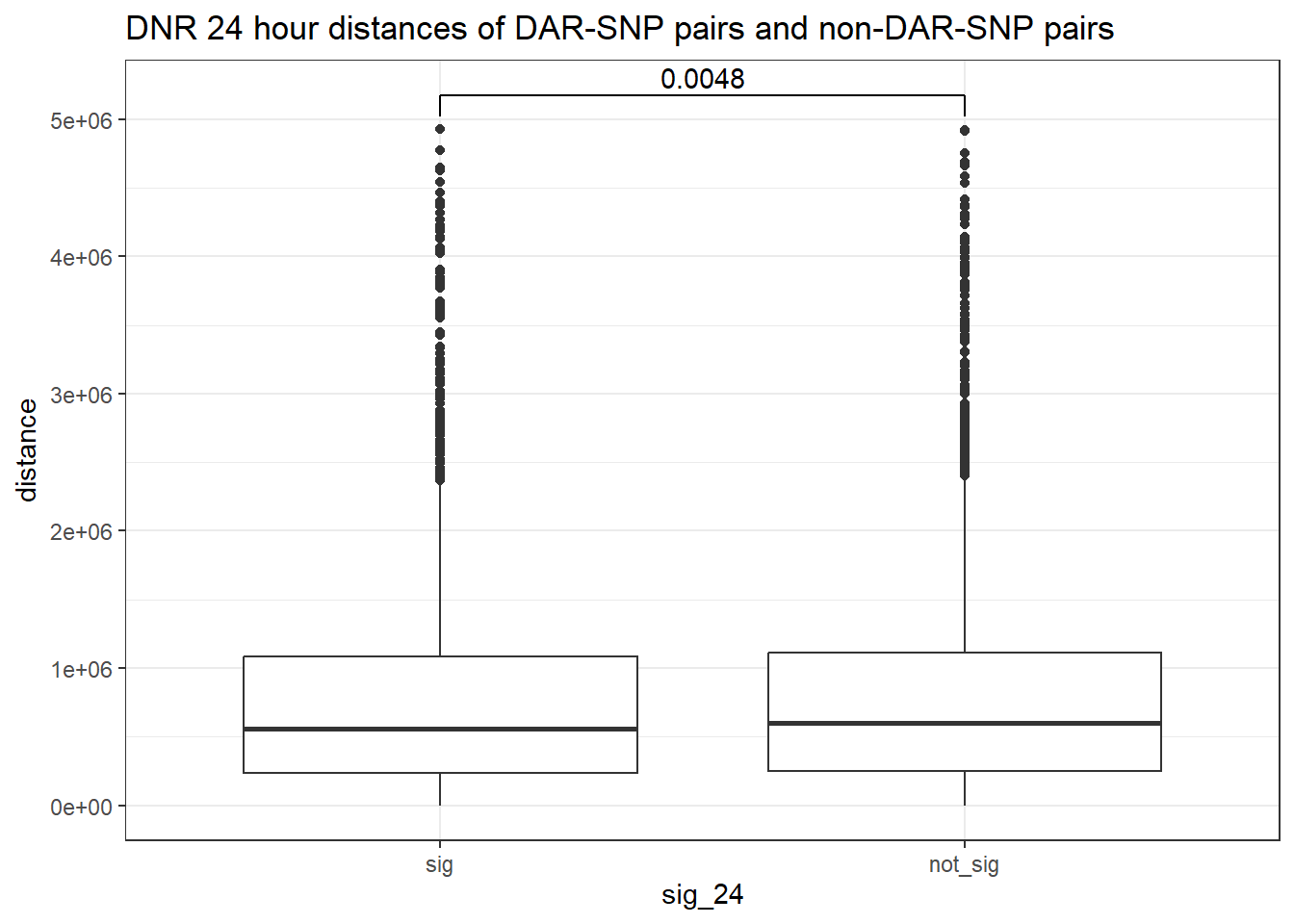
| Version | Author | Date |
|---|---|---|
| 1a9df02 | reneeisnowhere | 2025-07-09 |
wilcox.test(distance ~ sig_24, data = peak_snp_pairs_dist)
Wilcoxon rank sum test with continuity correction
data: distance by sig_24
W = 9463083, p-value = 0.002185
alternative hypothesis: true location shift is not equal to 0Cardiotox_gwas_DNR <- peak_snp_pairs_dist_DNR %>%
dplyr::filter(sig_24=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)DNR_MCF7 added
bind_rows(MCF7_DAR_snp_pairs_dist,peak_snp_pairs_dist_DNR) %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig", "MCF7_DAR"))) %>% ggplot(., aes(x= sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig"),
c("sig","MCF7_DAR"),
c("not_sig","MCF7_DAR")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("DNR 24 hour distances of DAR-SNP pairs\n and non-DAR-SNP pairs with MCF7 DARs")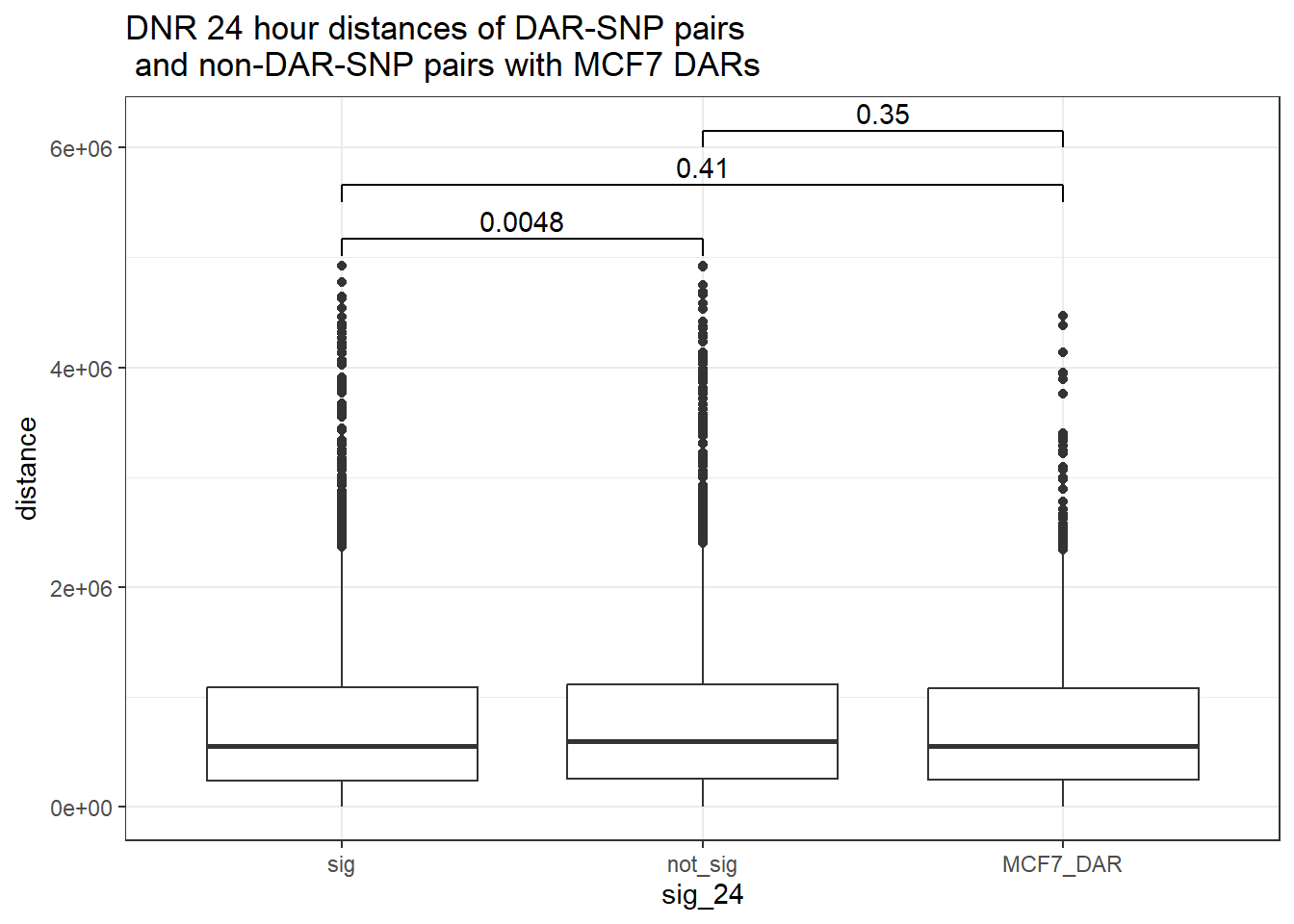
peak_snp_pairs_dist_DNR_3 <- peak_snp_pairs %>%
mutate(distance = abs(peak_start - snp_start)) %>%
mutate(sig_3= if_else(Peakid %in% DNR_DAR_sig_3$Peakid, "sig","not_sig"))
peak_snp_pairs_dist_DNR_3 %>%
mutate(sig_3=factor(sig_3, levels= c("sig","not_sig"))) %>%
ggplot(., aes(x= sig_3, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
map_signif_level = FALSE, test = "wilcox.test")+
ggtitle("3 hour DNR")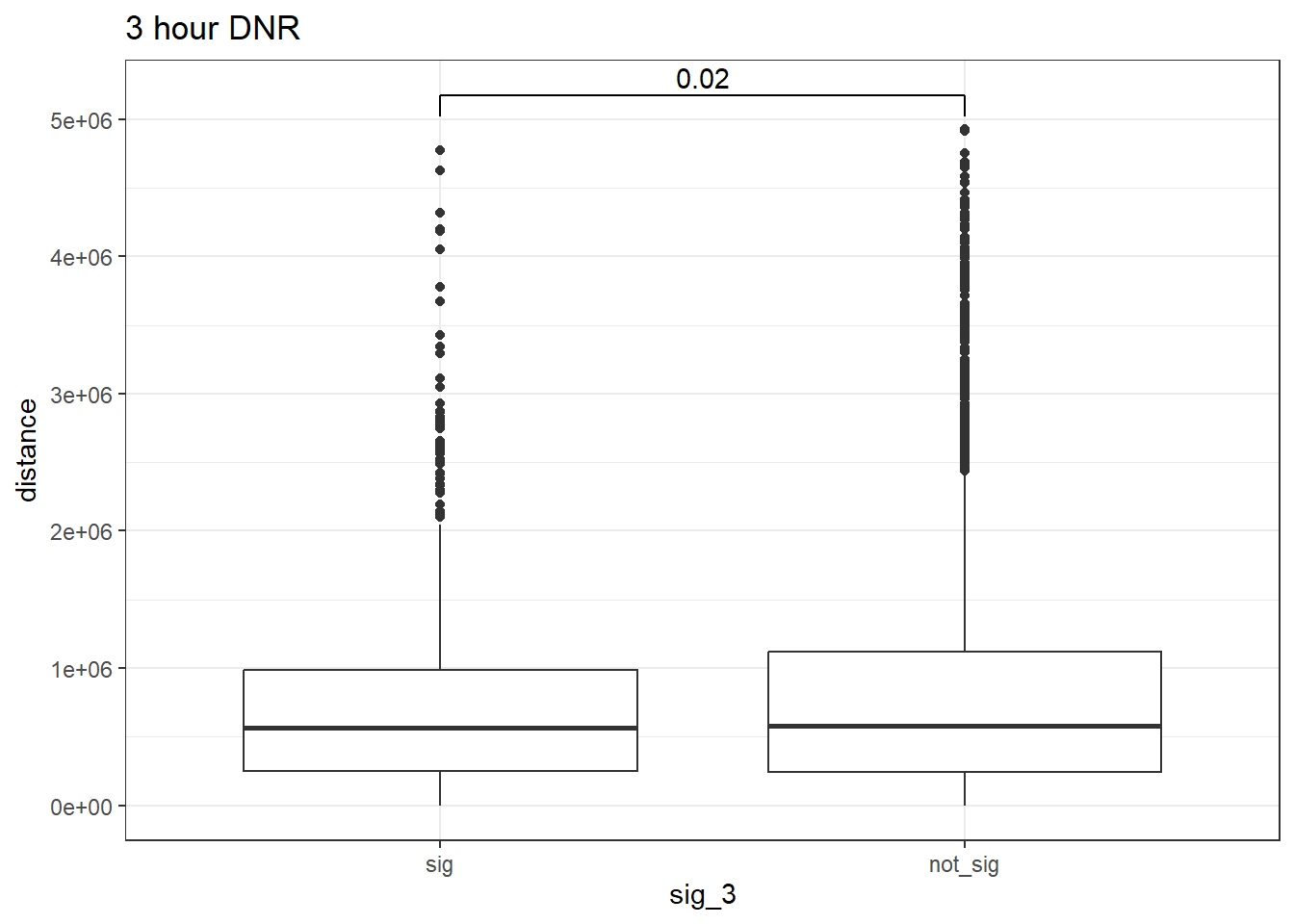
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
wilcox.test(distance ~ sig_3, data = peak_snp_pairs_dist_DNR_3)
Wilcoxon rank sum test with continuity correction
data: distance by sig_3
W = 4576878, p-value = 0.02023
alternative hypothesis: true location shift is not equal to 0Cardiotox_gwas_DNR_3 <- peak_snp_pairs_dist_DNR_3 %>%
dplyr::filter(sig_3=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)MTX 24 hours
peak_snp_pairs_dist_MTX <- peak_snp_pairs %>%
mutate(distance = abs(peak_start - snp_start)) %>%
mutate(sig_24= if_else(Peakid %in% MTX_DAR_sig$Peakid, "sig","not_sig"))
peak_snp_pairs_dist_MTX %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig"))) %>%
ggplot(., aes(x= sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
map_signif_level = FALSE, test = "wilcox.test")+
ggtitle("MTX 24 hour distances of DAR-SNP pairs and non-DAR-SNP pairs")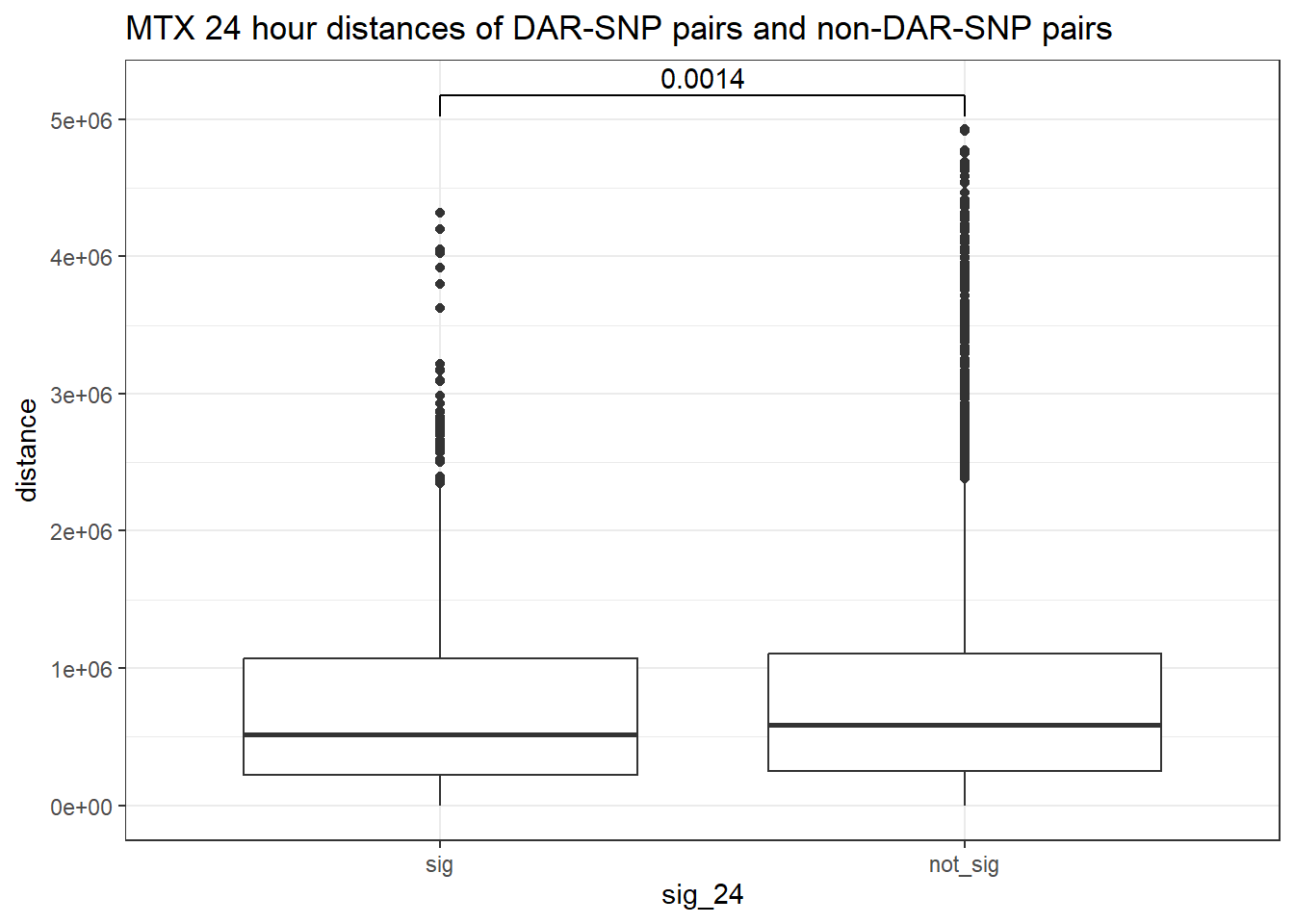
| Version | Author | Date |
|---|---|---|
| 1a9df02 | reneeisnowhere | 2025-07-09 |
wilcox.test(distance ~ sig_24, data = peak_snp_pairs_dist)
Wilcoxon rank sum test with continuity correction
data: distance by sig_24
W = 9463083, p-value = 0.002185
alternative hypothesis: true location shift is not equal to 0Cardiotox_gwas_MTX <- peak_snp_pairs_dist_MTX %>%
dplyr::filter(sig_24=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)MTX_MCF7 added
bind_rows(MCF7_DAR_snp_pairs_dist,peak_snp_pairs_dist_MTX) %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig", "MCF7_DAR"))) %>% ggplot(., aes(x= sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig"),
c("sig","MCF7_DAR"),
c("not_sig","MCF7_DAR")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("MTX 24 hour distances of DAR-SNP pairs\n and non-DAR-SNP pairs with MCF7 DARs")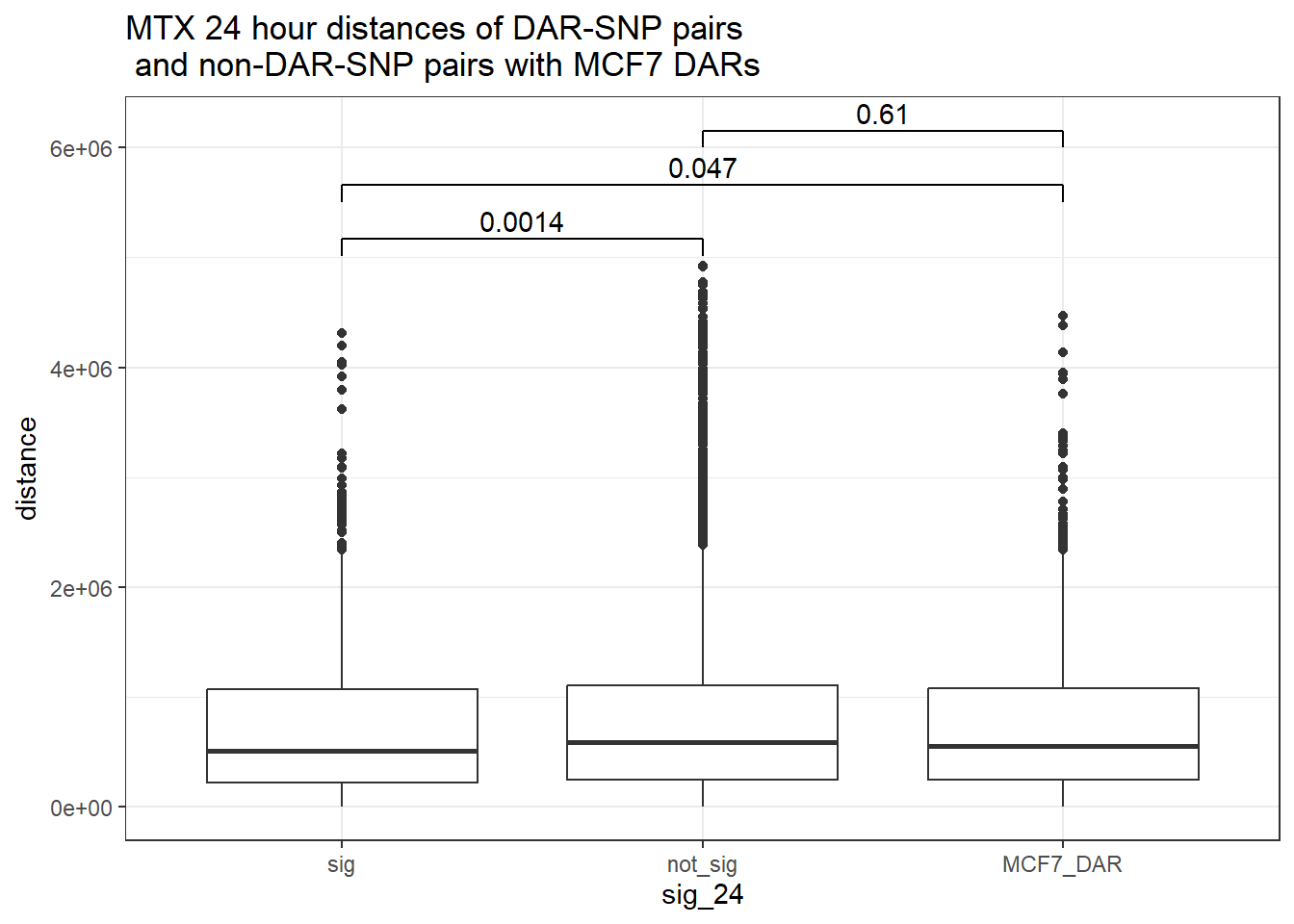
peak_snp_pairs_dist_MTX_3 <- peak_snp_pairs %>%
mutate(distance = abs(peak_start - snp_start)) %>%
mutate(sig_3= if_else(Peakid %in% MTX_DAR_sig_3$Peakid, "sig","not_sig"))
peak_snp_pairs_dist_MTX_3 %>%
mutate(sig_3=factor(sig_3, levels= c("sig","not_sig"))) %>%
ggplot(., aes(x= sig_3, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
map_signif_level = FALSE, test = "wilcox.test")+
ggtitle("3 hour MTX")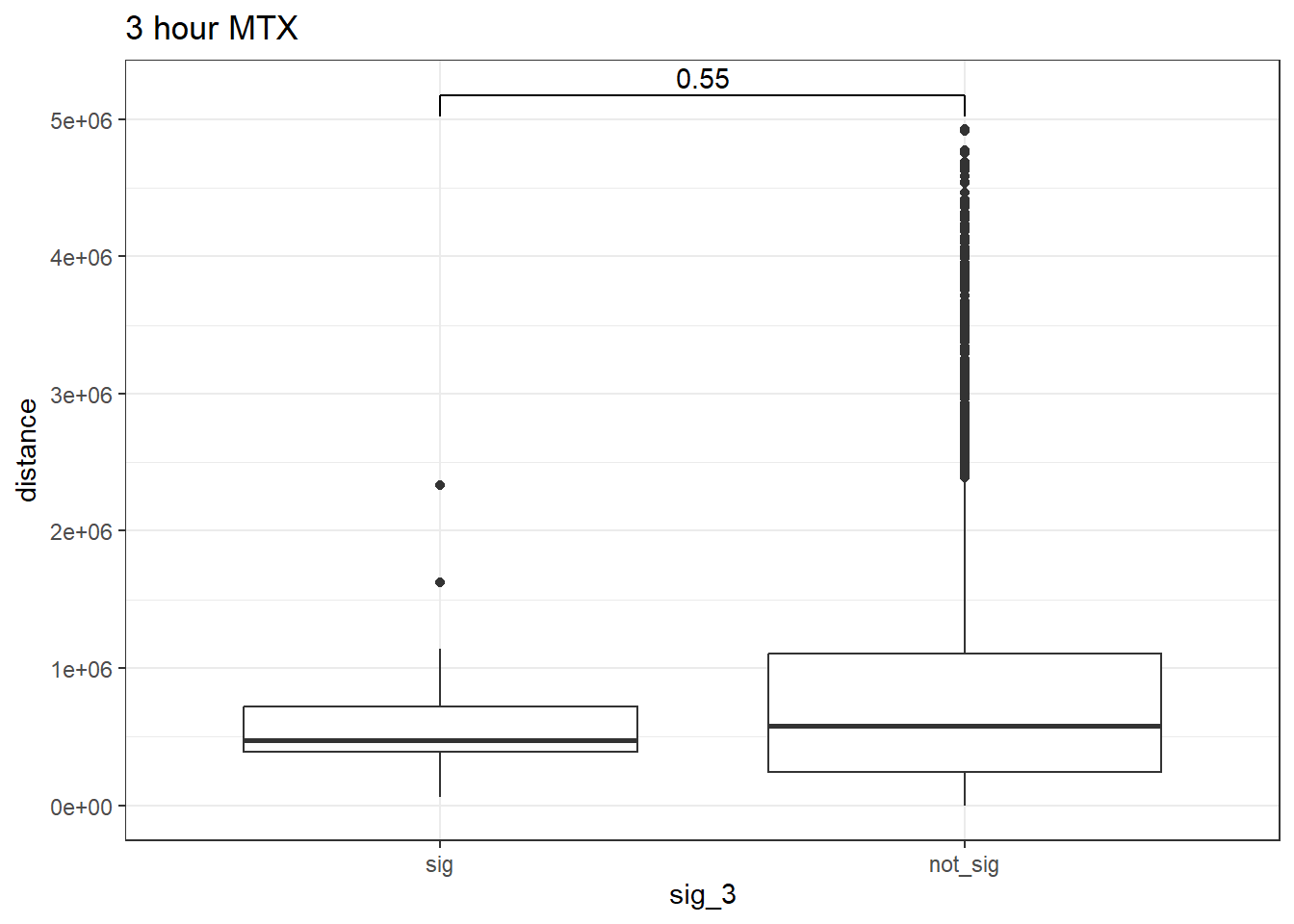
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
wilcox.test(distance ~ sig_3, data = peak_snp_pairs_dist_MTX_3)
Wilcoxon rank sum test with continuity correction
data: distance by sig_3
W = 219052, p-value = 0.5511
alternative hypothesis: true location shift is not equal to 0Cardiotox_gwas_MTX_3 <- peak_snp_pairs_dist_MTX_3 %>%
dplyr::filter(sig_3=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)Creating SNP_TAD distance DF
For combining the above 24 hour trt-distance to SNP data frames for box-plots
drug_pal <- c("#8B006D","#DF707E","#F1B72B", "#3386DD","#707031","#41B333")
SNP_TAD_dist_DF <- bind_rows((peak_snp_pairs_dist_MTX %>%
mutate(trt="MTX")),
(peak_snp_pairs_dist %>%
mutate(trt="DOX"))) %>%
bind_rows(.,(peak_snp_pairs_dist_EPI %>%
mutate(trt="EPI"))) %>%
bind_rows(.,(peak_snp_pairs_dist_DNR %>%
mutate(trt="DNR"))) %>%
mutate(trt=factor(trt,levels=c("DOX","EPI","DNR","MTX"))) %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig")))
SNP_TAD_dist_DF%>%
ggplot(., aes(x= interaction(sig_24,trt), y=distance))+
geom_boxplot(aes(fill=trt))+
theme_bw()+
geom_signif(comparisons = list(c("sig.DOX", "not_sig.DOX"),
c("sig.EPI","not_sig.EPI"),
c("sig.DNR", "not_sig.DNR"),
c("sig.MTX", "not_sig.MTX")),
# step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("ALL dist 24 hours")+
scale_fill_manual(values=drug_pal)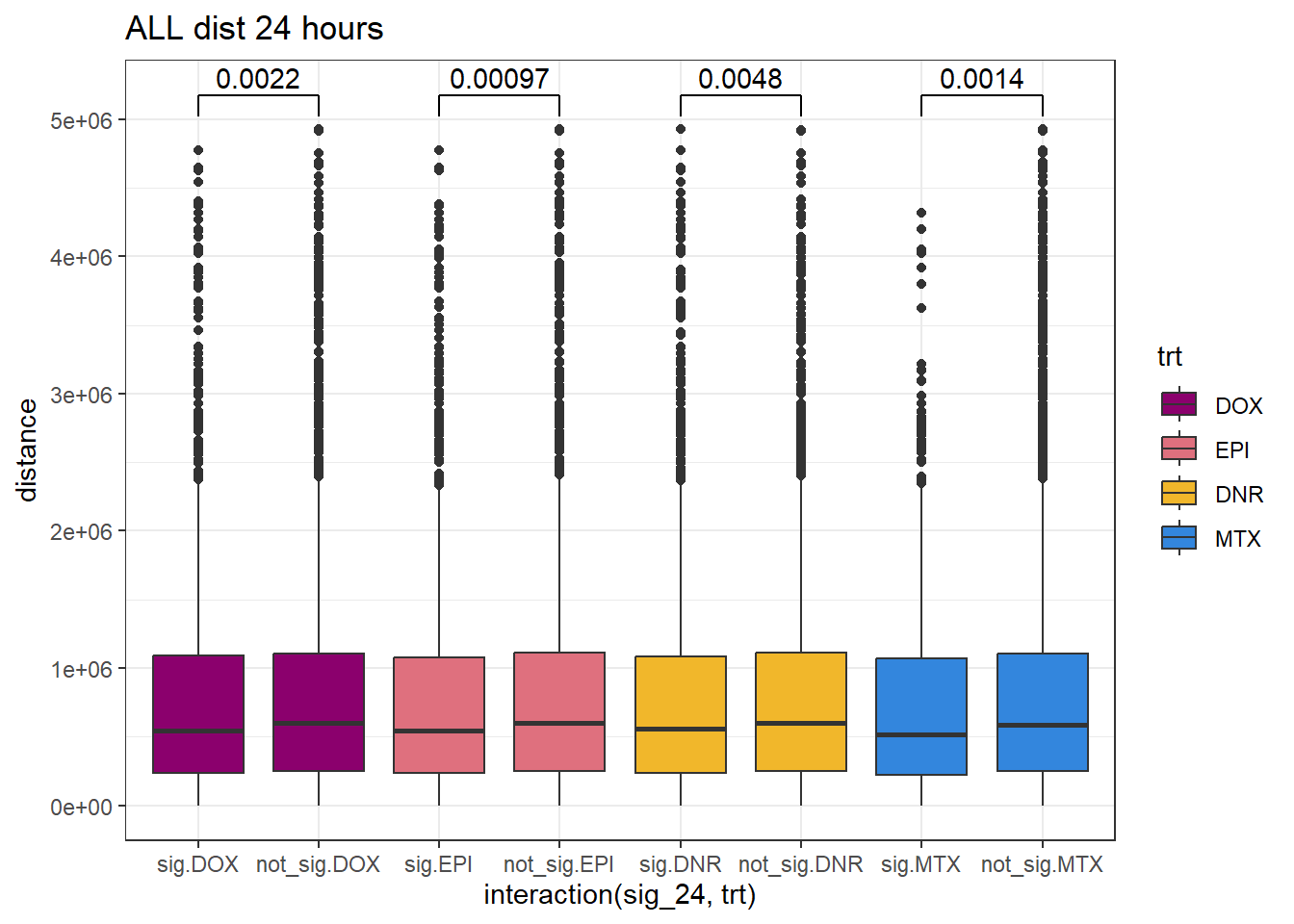
SNP_TAD_dist_DF_3 <- bind_rows((peak_snp_pairs_dist_MTX_3 %>%
mutate(trt="MTX")),
(peak_snp_pairs_dist_DOX_3 %>%
mutate(trt="DOX"))) %>%
bind_rows(.,(peak_snp_pairs_dist_EPI_3 %>%
mutate(trt="EPI"))) %>%
bind_rows(.,(peak_snp_pairs_dist_DNR_3 %>%
mutate(trt="DNR"))) %>%
mutate(trt=factor(trt,levels=c("DOX","EPI","DNR","MTX"))) %>%
mutate(sig_3=factor(sig_3, levels= c("sig","not_sig")))
SNP_TAD_dist_DF_3%>%
ggplot(., aes(x= interaction(sig_3,trt), y=distance))+
geom_boxplot(aes(fill=trt))+
theme_bw()+
geom_signif(comparisons = list(c("sig.DOX", "not_sig.DOX"),
c("sig.EPI","not_sig.EPI"),
c("sig.DNR", "not_sig.DNR"),
c("sig.MTX", "not_sig.MTX")),
# step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("ALL dist 3 hours")+
scale_fill_manual(values=drug_pal)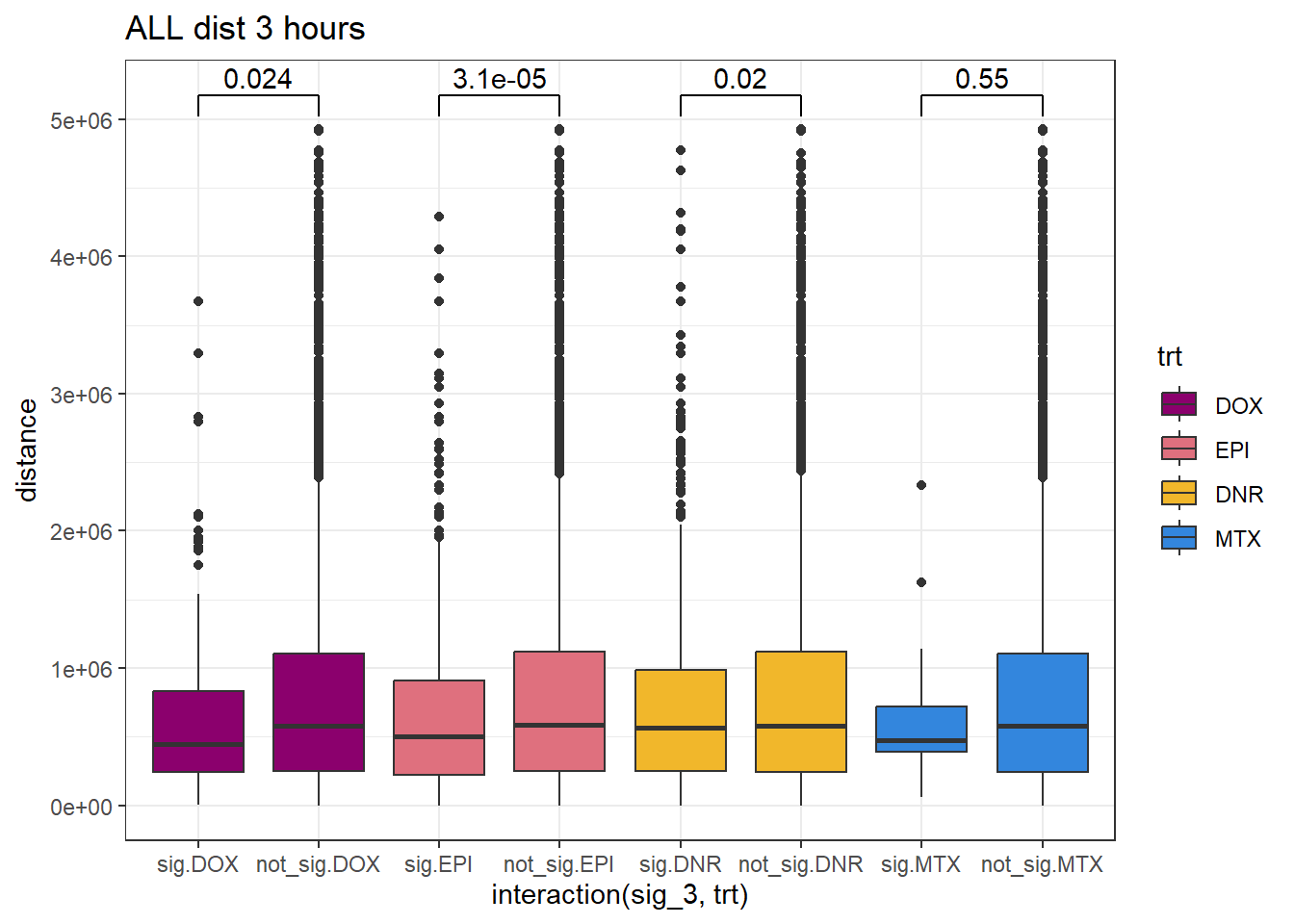
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
MCF7 tissue specific, non-tissue specific.
Here I am overlapping my data with the MCF7 DAR data. This will create DARS for each treatment that overlap MCF7 DARS (tissue-shared regions) and DARS that do not overlap MCF7 DARS (tissue-specific regions). I will then calculate the distance between the SNPs in the shared vs specific for each treatment.
MCF7_DARs_hyper <- read_excel("C:/Users/renee/Downloads/MCF7-doxATAC/Table 4.XLSX",
sheet = "hyper") %>% GRanges()
MCF7_DARs_hypo <- read_excel("C:/Users/renee/Downloads/MCF7-doxATAC/Table 4.XLSX",
sheet = "hypo") %>% GRanges()
# MCF7_ARsmcf7_1 <- read_excel("C:/Users/renee/Downloads/MCF7-doxATAC/Table 3.XLSX") %>%
# GRanges()
MCF7_DARs_hyper$names <- paste0("hyper_", seq_along(seqnames(MCF7_DARs_hyper)))
MCF7_DARs_hypo$names <- paste0("hypo_", seq_along(seqnames(MCF7_DARs_hypo)))
MCF7_DAR_all <- c(MCF7_DARs_hyper,MCF7_DARs_hypo)
ch = import.chain("C:/Users/renee/ATAC_folder/liftOver_genome/hg19ToHg38.over.chain")
# MCF7_ARsmcf7_1_LO <- as.data.frame(liftOver(MCF7_ARsmcf7_1,ch)) %>%
# GRanges()
MCF7_DARs_hyper_LO <- as.data.frame(liftOver(MCF7_DARs_hyper,ch)) %>%
GRanges()
MCF7_DARs_hypo_LO <- as.data.frame(liftOver(MCF7_DARs_hypo,ch)) %>%
GRanges()
MCF7_DAR_all_LO <- c(MCF7_DARs_hyper_LO,MCF7_DARs_hypo_LO)
MCF7DAR_AR_ol <- join_overlap_intersect(all_regions_gr, MCF7_DAR_all_LO)
DAR_AR_overlap_df <-MCF7DAR_AR_ol %>%
as.data.frame() %>%
distinct(Peakid)Attempting with DOX
peak_snp_pairs_dist %>%
dplyr::filter(sig_24 =="sig") %>%
mutate(specific=if_else(Peakid %in%DAR_AR_overlap_df$Peakid,"specific","not_specific")) %>%
mutate(specific=factor(specific,levels= c("specific","not_specific"))) %>%
ggplot(.,aes(x=specific, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("specific", "not_specific")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("DOX 24 hour distances of DAR-SNP pairs\n that are specific for iPSC-CMs or not-specific")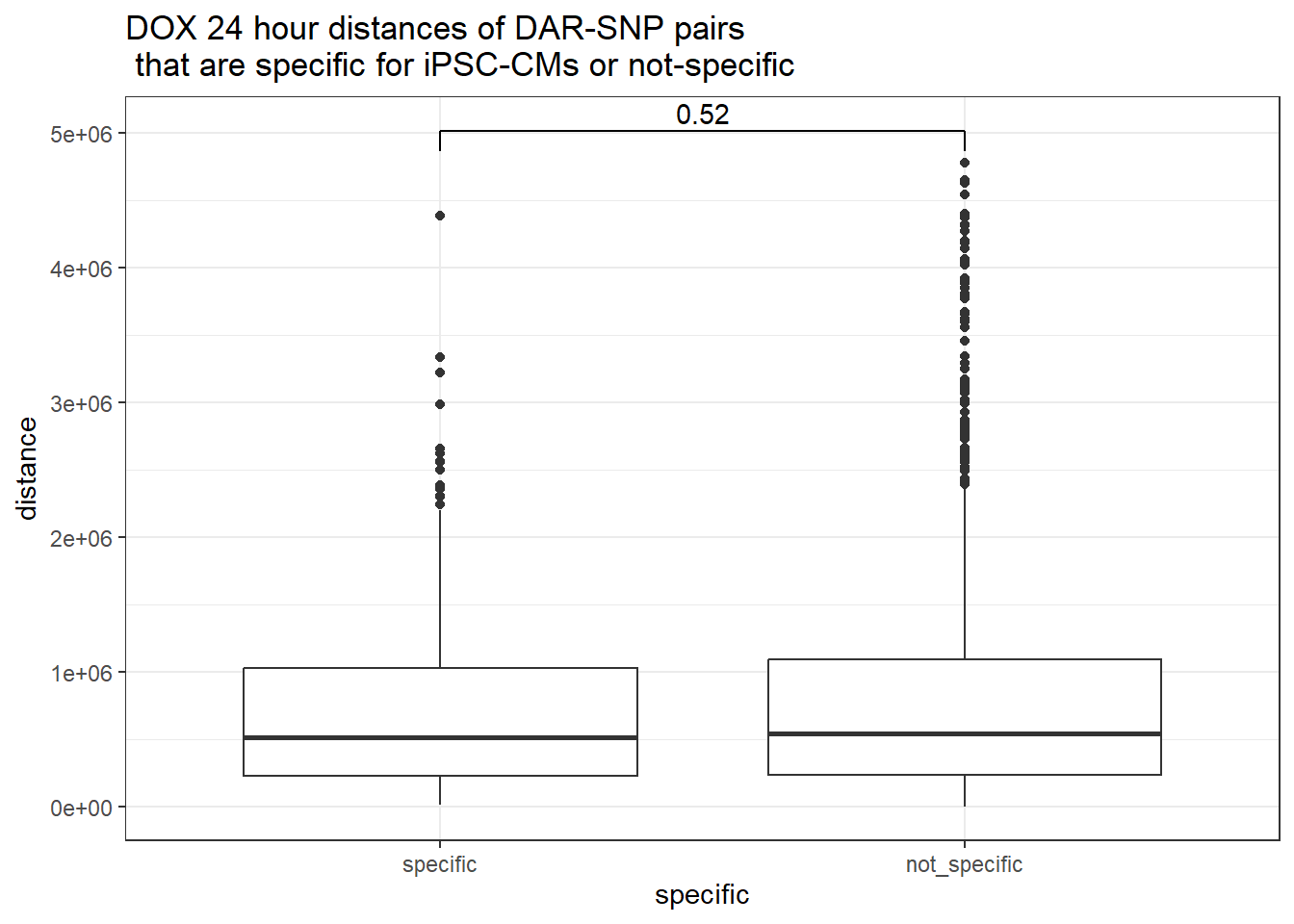
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
peak_snp_pairs_dist_EPI %>%
dplyr::filter(sig_24 =="sig") %>%
mutate(specific=if_else(Peakid %in%DAR_AR_overlap_df$Peakid,"specific","not_specific")) %>%
mutate(specific=factor(specific,levels= c("specific","not_specific"))) %>%
ggplot(.,aes(x=specific, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("specific", "not_specific")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("EPI 24 hour distances of DAR-SNP pairs\n that are specific for iPSC-CMs or not-specific")
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
peak_snp_pairs_dist_DNR %>%
dplyr::filter(sig_24 =="sig") %>%
mutate(specific=if_else(Peakid %in%DAR_AR_overlap_df$Peakid,"specific","not_specific")) %>%
mutate(specific=factor(specific,levels= c("specific","not_specific"))) %>%
ggplot(.,aes(x=specific, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("specific", "not_specific")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("DNR 24 hour distances of DAR-SNP pairs\n that are specific for iPSC-CMs or not-specific")
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
peak_snp_pairs_dist_MTX %>%
dplyr::filter(sig_24 =="sig") %>%
mutate(specific=if_else(Peakid %in%DAR_AR_overlap_df$Peakid,"specific","not_specific")) %>%
mutate(specific=factor(specific,levels= c("specific","not_specific"))) %>%
ggplot(.,aes(x=specific, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("specific", "not_specific")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("MTX 24 hour distances of DAR-SNP pairs\n that are specific for iPSC-CMs or not-specific")
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
peak_snp_pairs_dist %>%
dplyr::filter(!Peakid %in% DAR_AR_overlap_df$Peakid) %>%
mutate(sig_24=factor(sig_24,levels = c ("sig","not_sig"))) %>%
ggplot(.,aes(x=sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("DOX 24 hour distances of DAR-SNP pairs\n that are specific for iPSC-CMs")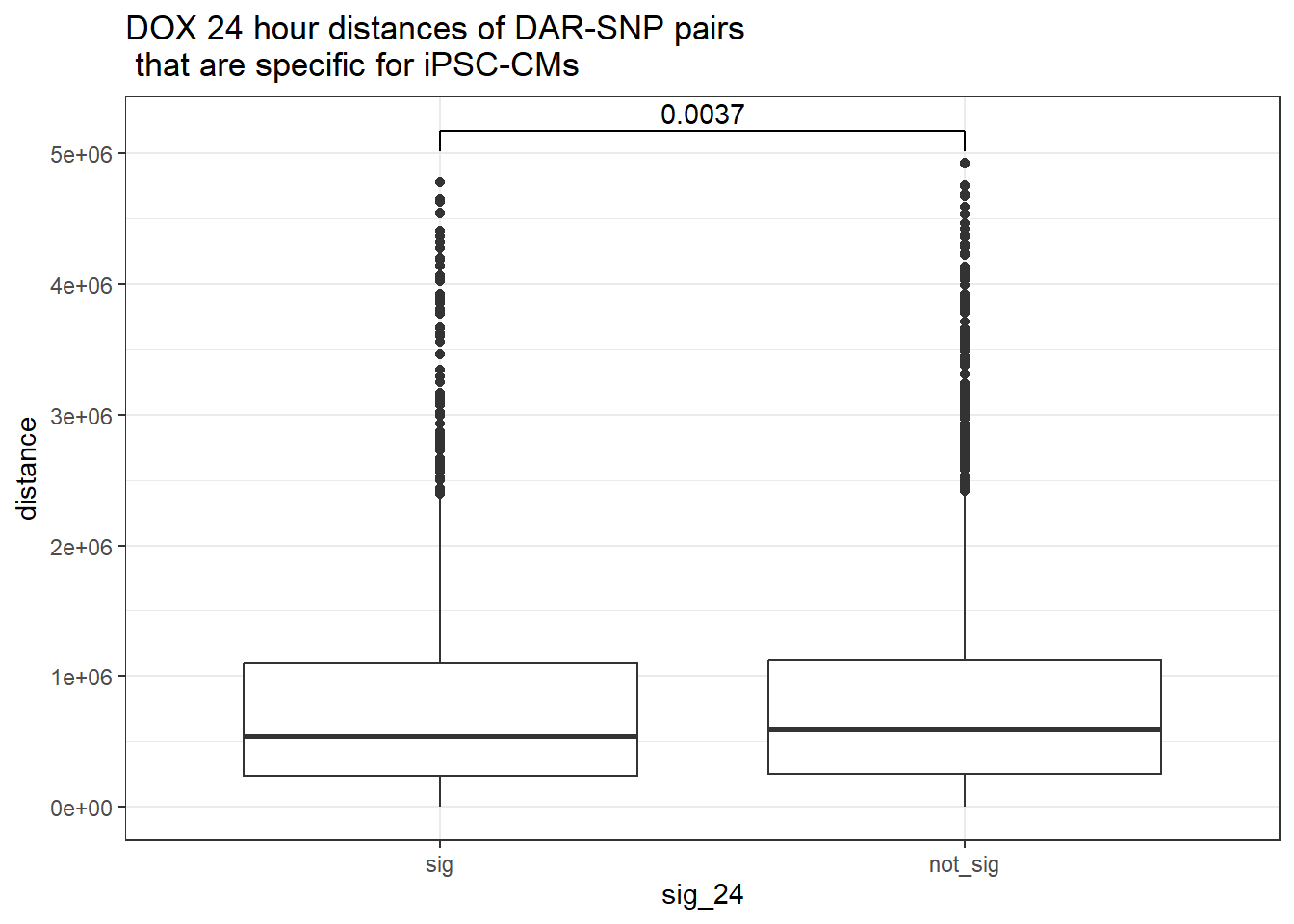
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
peak_snp_pairs_dist_EPI %>%
dplyr::filter(!Peakid %in% DAR_AR_overlap_df$Peakid) %>%
mutate(sig_24=factor(sig_24,levels = c ("sig","not_sig"))) %>%
ggplot(.,aes(x=sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("EPI 24 hour distances of DAR-SNP pairs\n that are specific for iPSC-CMs")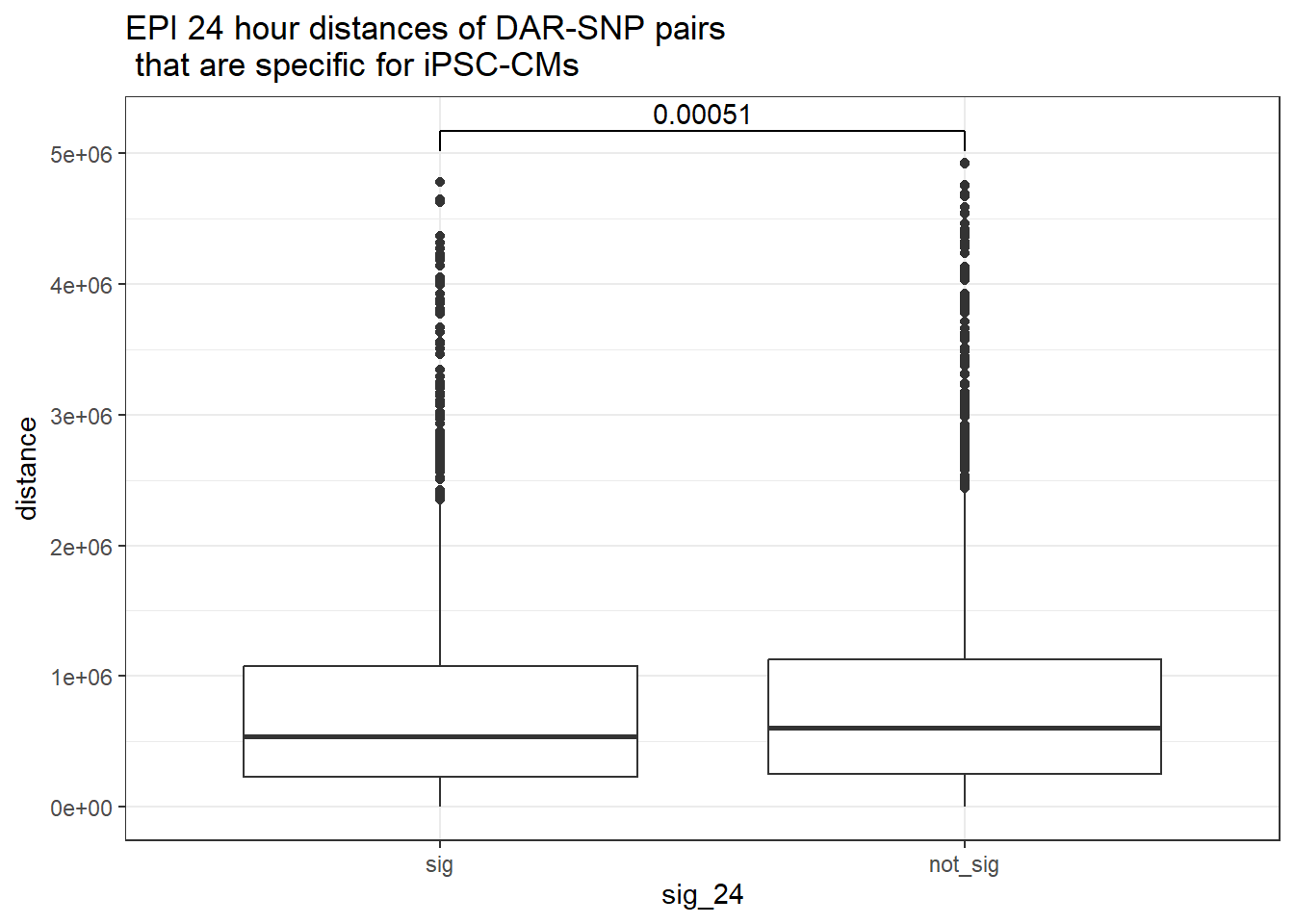
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
peak_snp_pairs_dist_DNR %>%
dplyr::filter(!Peakid %in% DAR_AR_overlap_df$Peakid) %>%
mutate(sig_24=factor(sig_24,levels = c ("sig","not_sig"))) %>%
ggplot(.,aes(x=sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("DNR 24 hour distances of DAR-SNP pairs\n that are specific for iPSC-CMs")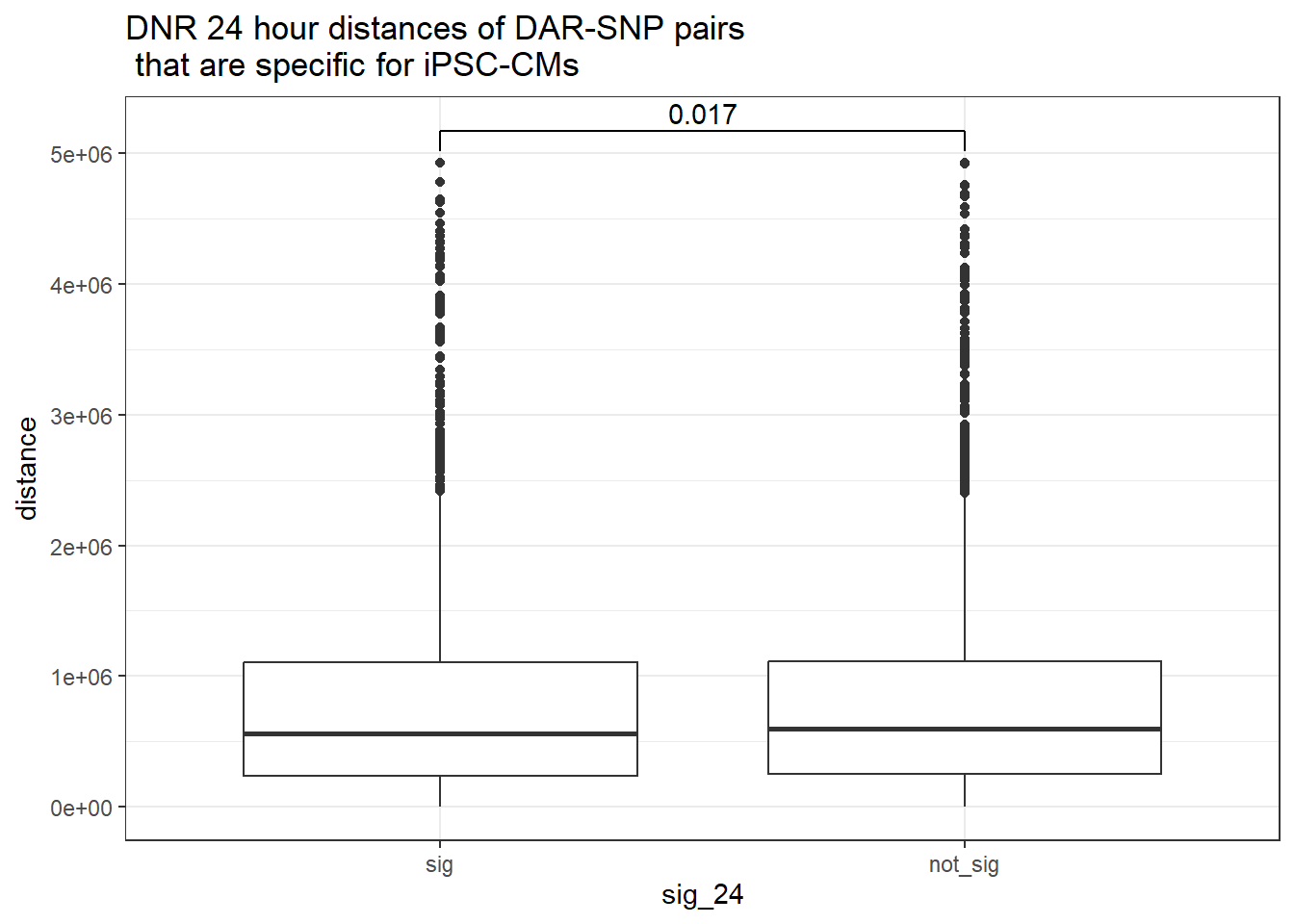
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
peak_snp_pairs_dist_MTX %>%
dplyr::filter(!Peakid %in% DAR_AR_overlap_df$Peakid) %>%
mutate(sig_24=factor(sig_24,levels = c ("sig","not_sig"))) %>%
ggplot(.,aes(x=sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
step_increase = 0.1,
map_signif_level = FALSE,
test = "wilcox.test")+
ggtitle("MTX 24 hour distances of DAR-SNP pairs\n that are specific for iPSC-CMs")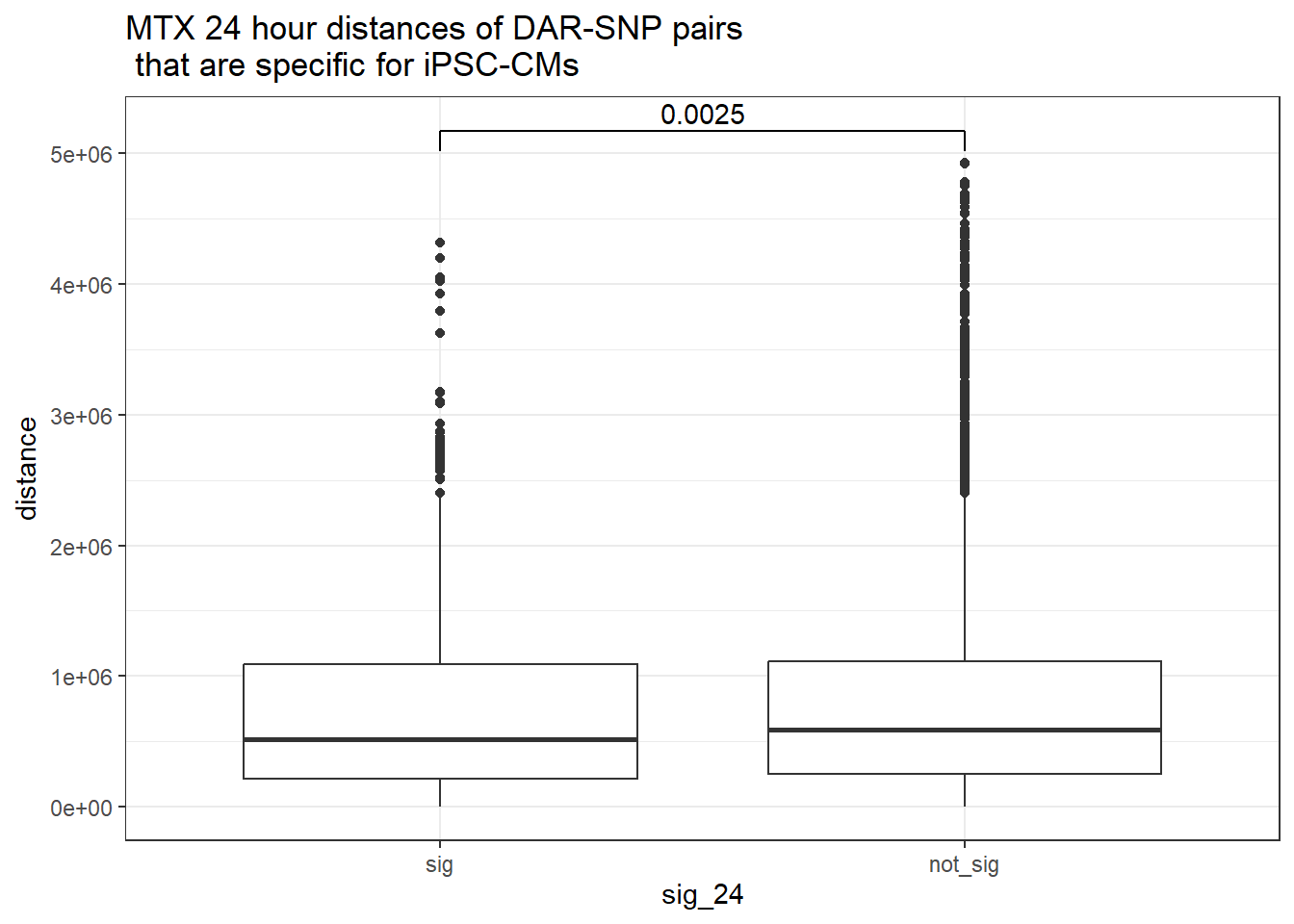
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
Attempt at heatmap with RNA expression, decided not to use or plot here.
Cardotox_mat <- Cardiotox_gwas_collaped_df %>%
dplyr::select(name,DOX_3:TRZ_24,`3_RNA_DOX`,`3_RNA_EPI`,`3_RNA_DNR`,`3_RNA_MTX`,`3_RNA_TRZ`,`24_RNA_DOX`,`24_RNA_EPI`,`24_RNA_DNR`,`24_RNA_MTX`,`24_RNA_TRZ`) %>%
column_to_rownames("name") %>%
as.matrix()
annot_map_df <- Cardiotox_gwas_collaped_df %>%
dplyr::select(name,snp_dist) %>%
column_to_rownames("name")
annot_map <-
rowAnnotation(
snp_dist=Cardiotox_gwas_collaped_df$snp_dist,
TAD_id=Cardiotox_gwas_collaped_df$TAD_id,
col= list(snp_dist=c("2kb"="goldenrod4",
"20kb"="pink",
">20kb"="tan2"),
TAD_id=tad_colors))
simply_map_lfc <- ComplexHeatmap::Heatmap(Cardotox_mat,
# col = col_fun,
left_annotation = annot_map,
show_row_names = TRUE,
row_names_max_width= ComplexHeatmap::max_text_width(rownames(Cardotox_mat), gp=gpar(fontsize=14)),
heatmap_legend_param = list(direction = "horizontal"),
show_column_names = TRUE,
cluster_rows = FALSE,
cluster_columns = FALSE)
ComplexHeatmap::draw(simply_map_lfc,
merge_legend = TRUE,
heatmap_legend_side = "left",
annotation_legend_side = "left")Alternative of final graph
ATAC_all_adj.pvals <- all_results%>%
dplyr::select(source,genes,adj.P.Val) %>%
pivot_wider(id_cols=genes, values_from = adj.P.Val, names_from = source)
# saveRDS(ATAC_all_adj.pvals,"data/Final_four_data/re_analysis/ATAC_all_adj_pvals.RDS")
sig_mat_cardiotox <- ATAC_all_adj.pvals %>%
dplyr::filter(genes %in% peak_snp_pairs_dist$Peakid) %>%
left_join(peak_snp_pairs_dist, by=c("genes"="Peakid")) %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
group_by(genes, peak_chr, peak_start, TAD_id, sig_24) %>%
summarise(
min_distance = min(distance),
mean_distance = mean(distance),
snp_list = paste(unique(RSID), collapse = ","),
.groups = "drop"
) %>%
left_join(ATAC_all_adj.pvals) %>%
tidyr::unite(., name,genes,snp_list) %>%
dplyr::select(name, DNR_3:TRZ_24) %>%
column_to_rownames("name") %>%
as.matrix()
AR_Cardiotox_gwas_collaped_df <-
peak_snp_pairs_dist %>%
# dplyr::filter(sig_24=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
group_by(Peakid, peak_chr, peak_start, TAD_id, sig_24) %>%
summarise(
min_distance = min(distance),
mean_distance = mean(distance),
snp_list = paste(unique(RSID), collapse = ","),
.groups = "drop"
) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
# left_join(., Peak_gene_RNA_LFC, by=c("Peakid"="Peakid")) %>%
# left_join(.,gene_info_collapsed, by=c("geneId"="ENTREZID")) %>%
# mutate(SYMBOL=if_else(is.na(SYMBOL.x),SYMBOL.y,if_else(SYMBOL.x==SYMBOL.y, SYMBOL.x,paste0(SYMBOL.x,"_",SYMBOL.y)))) %>%
tidyr::unite(., name,Peakid,snp_list) %>%
mutate(snp_dist=case_when(min_distance <2000 ~"2kb",
min_distance > 2000 & min_distance<20000 ~ "20kb",
min_distance >20000 ~">20kb"))
Cardotox_mat_2 <- AR_Cardiotox_gwas_collaped_df %>%
dplyr::select(name,DOX_3:TRZ_24) %>%
column_to_rownames("name") %>%
as.matrix()
annot_map_df_2 <- AR_Cardiotox_gwas_collaped_df %>%
dplyr::select(name,snp_dist,sig_24) %>%
column_to_rownames("name")
annot_map_2 <-
ComplexHeatmap::rowAnnotation(
snp_dist=AR_Cardiotox_gwas_collaped_df$snp_dist,
TAD_id=AR_Cardiotox_gwas_collaped_df$TAD_id,
DOX_24hr_DAR=AR_Cardiotox_gwas_collaped_df$sig_24,
col= list(snp_dist=c("2kb"="goldenrod4",
"20kb"="pink",
">20kb"="tan2"),
TAD_id=tad_colors))
simply_map_lfc_2 <- ComplexHeatmap::Heatmap(Cardotox_mat_2,
left_annotation = annot_map_2,
show_row_names = TRUE,
row_names_max_width= ComplexHeatmap::max_text_width(rownames(Cardotox_mat_2), gp=gpar(fontsize=14)),
heatmap_legend_param = list(direction = "horizontal"),
show_column_names = TRUE,
cluster_rows = FALSE,
cluster_columns = FALSE,
cell_fun = function(j, i, x, y, width, height, fill) {
if (!is.na(sig_mat_cardiotox[i, j]) && sig_mat_cardiotox[i, j] <0.05) {
grid.text("*", x, y, gp = gpar(fontsize = 20)) # Add star if significant
} })
ComplexHeatmap::draw(simply_map_lfc_2,
merge_legend = TRUE,
heatmap_legend_side = "left",
annotation_legend_side = "left")
ATAC_all_adj.pvals %>%
dplyr::filter(genes %in% peak_snp_pairs_dist$Peakid) %>%
left_join(peak_snp_pairs_dist, by=c("genes"="Peakid")) %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
group_by(genes, peak_chr, peak_start, TAD_id, sig_24) %>%
summarise(
min_distance = min(distance),
mean_distance = mean(distance),
snp_list = paste(unique(RSID), collapse = ","),
.groups = "drop"
) %>%
left_join(ATAC_all_adj.pvals) %>%
tidyr::unite(., name,genes,snp_list) %>%
dplyr::select(name, DNR_3:TRZ_24) %>%
column_to_rownames("name") %>%
as.matrix() DNR_3
chr1.108829511.108829805_rs10857972,rs6704266 0.2604386136
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 0.9941823649
chr1.19640454.19641134_rs7523079 0.2751816852
chr1.19644587.19645224_rs7522389 0.1110929630
chr10.16792579.16793007_rs7068265,rs7068881 0.1754416544
chr10.76209088.76209420_rs7094302 0.6062032046
chr11.124863016.124863456_rs7111513 0.9095139649
chr11.124865334.124865878_rs5017048 0.8068193757
chr11.133816825.133817041_rs10894749 0.9576490254
chr11.36409327.36409838_rs10836550 0.4507242896
chr11.36420818.36421386_rs1365120 0.1671941818
chr12.120317069.120317911_rs16950058 0.8463250455
chr12.120325389.120326108_rs5634 0.5597350198
chr12.120367731.120368309_rs2238161 0.0496726775
chr12.19222646.19223206_rs7399293 0.7530113056
chr13.100722024.100722184_rs12863645 0.1369620985
chr13.46774697.46774984_rs1745836 0.1103175482
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 0.3060666798
chr13.91724572.91724918_rs342715,rs171060 0.0506322122
chr13.99908163.99908652_rs9557321 0.1382557035
chr14.55721341.55721700_rs2184559 0.9748799763
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 0.0975418891
chr15.93645160.93645583_rs4238475 0.9900179537
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 0.2887664071
chr17.51461539.51461982_rs10514983 0.5052014759
chr18.32469230.32469379_rs1458866 0.7599187815
chr2.104528125.104528417_rs11123997 0.8552557266
chr2.12759282.12759669_rs6722342,rs7596623 0.4098477471
chr2.12766229.12766820_rs13413220,rs13397389 0.4019189553
chr2.176086015.176086512_rs6752623 0.7542466264
chr2.41038390.41038934_rs6746423 0.3221761584
chr2.77457604.77458071_rs12614202,rs17014128 0.1955860057
chr20.22217873.22218175_rs76464104 0.7146552246
chr20.53735535.53736570_rs12480103 0.0039717853
chr22.46225398.46226192_rs4253763 0.0323062960
chr22.46233211.46233644_rs11090819 0.0061385813
chr22.46249819.46250733_rs7291763 0.2194815027
chr3.107240266.107241585_rs11710776 0.0039425887
chr3.118380545.118381165_rs779206,rs13087409 0.9685916399
chr3.193461783.193462089_rs9879227 0.3898544491
chr3.20682627.20683020_rs11715178,rs10510504 0.9624468773
chr4.137238844.137239180_rs13148112 0.2635250836
chr4.176615030.176615396_rs6553973 0.5016560972
chr4.32226261.32226503_rs13125385 0.7169684281
chr4.40731697.40732054_rs11727143 0.8934529766
chr4.40749770.40751218_rs10012367 0.4517700481
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 0.2945038500
chr5.102865289.102866656_rs32680 0.0543776655
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 0.2877953785
chr5.164619344.164619596_rs250594 0.0304643100
chr5.7415226.7416045_rs7710510 0.1140516622
chr5.7464212.7464571_rs6883259,rs766643 0.6488216182
chr6.160899655.160900401_rs2489944,rs2465874 0.8175996745
chr6.27026322.27026687_rs36022097 0.9851482375
chr6.32521585.32522598_rs115541994 0.5746006297
chr6.71712533.71713167_rs1204328 0.7516231731
chr7.10239630.10240386_rs17162825 0.0275441848
chr7.116499279.116500903_rs17138749 0.0004354869
chr7.134834642.134835353_rs7777356 0.0468197649
chr7.26147128.26148142_rs10248605 0.1920101330
chr8.141160879.141162083_rs428300 0.4304731801
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 0.0209345846
chr8.20115130.20115538_rs12548222 0.4403856042
chr8.69455933.69456585_rs12545665 0.5785563175
chr8.701154.701537_rs13273765 0.0847512055
chr8.81096615.81097088_rs16908579 0.3593169617
chr9.18632808.18633319_rs776777 0.4978362823
chr9.19720338.19720665_rs7869360,rs7855757 0.9888222941
DOX_3
chr1.108829511.108829805_rs10857972,rs6704266 0.28996020
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 0.79324477
chr1.19640454.19641134_rs7523079 0.10235976
chr1.19644587.19645224_rs7522389 0.76107014
chr10.16792579.16793007_rs7068265,rs7068881 0.52334599
chr10.76209088.76209420_rs7094302 0.87870169
chr11.124863016.124863456_rs7111513 0.87999911
chr11.124865334.124865878_rs5017048 0.94606145
chr11.133816825.133817041_rs10894749 0.69326462
chr11.36409327.36409838_rs10836550 0.87860628
chr11.36420818.36421386_rs1365120 0.22233972
chr12.120317069.120317911_rs16950058 0.72036630
chr12.120325389.120326108_rs5634 0.94148931
chr12.120367731.120368309_rs2238161 0.86619208
chr12.19222646.19223206_rs7399293 0.80126526
chr13.100722024.100722184_rs12863645 0.13362272
chr13.46774697.46774984_rs1745836 0.10024827
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 0.31772047
chr13.91724572.91724918_rs342715,rs171060 0.30032478
chr13.99908163.99908652_rs9557321 0.33019997
chr14.55721341.55721700_rs2184559 0.81016947
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 0.07429395
chr15.93645160.93645583_rs4238475 0.93216164
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 0.42769913
chr17.51461539.51461982_rs10514983 0.49879726
chr18.32469230.32469379_rs1458866 0.73139860
chr2.104528125.104528417_rs11123997 0.46759370
chr2.12759282.12759669_rs6722342,rs7596623 0.58364196
chr2.12766229.12766820_rs13413220,rs13397389 0.54471722
chr2.176086015.176086512_rs6752623 0.95595571
chr2.41038390.41038934_rs6746423 0.95043912
chr2.77457604.77458071_rs12614202,rs17014128 0.75361165
chr20.22217873.22218175_rs76464104 0.42265673
chr20.53735535.53736570_rs12480103 0.08343468
chr22.46225398.46226192_rs4253763 0.39046192
chr22.46233211.46233644_rs11090819 0.11013515
chr22.46249819.46250733_rs7291763 0.41625015
chr3.107240266.107241585_rs11710776 0.12136474
chr3.118380545.118381165_rs779206,rs13087409 0.66214382
chr3.193461783.193462089_rs9879227 0.42860660
chr3.20682627.20683020_rs11715178,rs10510504 0.99221246
chr4.137238844.137239180_rs13148112 0.91587514
chr4.176615030.176615396_rs6553973 0.97276867
chr4.32226261.32226503_rs13125385 0.78012067
chr4.40731697.40732054_rs11727143 0.99026586
chr4.40749770.40751218_rs10012367 0.91621966
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 0.21644958
chr5.102865289.102866656_rs32680 0.39419001
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 0.53263972
chr5.164619344.164619596_rs250594 0.37800156
chr5.7415226.7416045_rs7710510 0.59452990
chr5.7464212.7464571_rs6883259,rs766643 0.96323882
chr6.160899655.160900401_rs2489944,rs2465874 0.87685451
chr6.27026322.27026687_rs36022097 0.96215537
chr6.32521585.32522598_rs115541994 0.86310747
chr6.71712533.71713167_rs1204328 0.77580074
chr7.10239630.10240386_rs17162825 0.06055191
chr7.116499279.116500903_rs17138749 0.03946688
chr7.134834642.134835353_rs7777356 0.06100145
chr7.26147128.26148142_rs10248605 0.98340441
chr8.141160879.141162083_rs428300 0.66713952
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 0.20823369
chr8.20115130.20115538_rs12548222 0.60645494
chr8.69455933.69456585_rs12545665 0.58621301
chr8.701154.701537_rs13273765 0.14027293
chr8.81096615.81097088_rs16908579 0.54794256
chr9.18632808.18633319_rs776777 0.38921211
chr9.19720338.19720665_rs7869360,rs7855757 0.89538037
EPI_3
chr1.108829511.108829805_rs10857972,rs6704266 0.627792912
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 0.709914257
chr1.19640454.19641134_rs7523079 0.415719858
chr1.19644587.19645224_rs7522389 0.601889717
chr10.16792579.16793007_rs7068265,rs7068881 0.130614071
chr10.76209088.76209420_rs7094302 0.728255472
chr11.124863016.124863456_rs7111513 0.698068653
chr11.124865334.124865878_rs5017048 0.658177438
chr11.133816825.133817041_rs10894749 0.519814821
chr11.36409327.36409838_rs10836550 0.204486043
chr11.36420818.36421386_rs1365120 0.126493056
chr12.120317069.120317911_rs16950058 0.896505610
chr12.120325389.120326108_rs5634 0.972895082
chr12.120367731.120368309_rs2238161 0.332993063
chr12.19222646.19223206_rs7399293 0.755566564
chr13.100722024.100722184_rs12863645 0.097501269
chr13.46774697.46774984_rs1745836 0.041216934
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 0.478988902
chr13.91724572.91724918_rs342715,rs171060 0.266646540
chr13.99908163.99908652_rs9557321 0.512417777
chr14.55721341.55721700_rs2184559 0.628238659
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 0.002865416
chr15.93645160.93645583_rs4238475 0.436573116
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 0.031459171
chr17.51461539.51461982_rs10514983 0.399674713
chr18.32469230.32469379_rs1458866 0.679191345
chr2.104528125.104528417_rs11123997 0.898534678
chr2.12759282.12759669_rs6722342,rs7596623 0.536775555
chr2.12766229.12766820_rs13413220,rs13397389 0.748199265
chr2.176086015.176086512_rs6752623 0.995945463
chr2.41038390.41038934_rs6746423 0.907599244
chr2.77457604.77458071_rs12614202,rs17014128 0.277297264
chr20.22217873.22218175_rs76464104 0.615438543
chr20.53735535.53736570_rs12480103 0.060097328
chr22.46225398.46226192_rs4253763 0.046303317
chr22.46233211.46233644_rs11090819 0.079390213
chr22.46249819.46250733_rs7291763 0.632752727
chr3.107240266.107241585_rs11710776 0.009720301
chr3.118380545.118381165_rs779206,rs13087409 0.851331297
chr3.193461783.193462089_rs9879227 0.213935344
chr3.20682627.20683020_rs11715178,rs10510504 0.628774319
chr4.137238844.137239180_rs13148112 0.771367568
chr4.176615030.176615396_rs6553973 0.316307474
chr4.32226261.32226503_rs13125385 0.515065253
chr4.40731697.40732054_rs11727143 0.771939694
chr4.40749770.40751218_rs10012367 0.640910831
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 0.595412025
chr5.102865289.102866656_rs32680 0.231037136
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 0.392516335
chr5.164619344.164619596_rs250594 0.232154081
chr5.7415226.7416045_rs7710510 0.089960026
chr5.7464212.7464571_rs6883259,rs766643 0.564584818
chr6.160899655.160900401_rs2489944,rs2465874 0.852939108
chr6.27026322.27026687_rs36022097 0.849269257
chr6.32521585.32522598_rs115541994 0.835105641
chr6.71712533.71713167_rs1204328 0.398006470
chr7.10239630.10240386_rs17162825 0.003879779
chr7.116499279.116500903_rs17138749 0.005247476
chr7.134834642.134835353_rs7777356 0.006546935
chr7.26147128.26148142_rs10248605 0.063537276
chr8.141160879.141162083_rs428300 0.297732616
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 0.011869205
chr8.20115130.20115538_rs12548222 0.168385512
chr8.69455933.69456585_rs12545665 0.978951269
chr8.701154.701537_rs13273765 0.032741476
chr8.81096615.81097088_rs16908579 0.503472900
chr9.18632808.18633319_rs776777 0.635914438
chr9.19720338.19720665_rs7869360,rs7855757 0.871034405
MTX_3
chr1.108829511.108829805_rs10857972,rs6704266 0.94362679
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 0.99802465
chr1.19640454.19641134_rs7523079 0.48437917
chr1.19644587.19645224_rs7522389 0.11226483
chr10.16792579.16793007_rs7068265,rs7068881 0.56212854
chr10.76209088.76209420_rs7094302 0.73886646
chr11.124863016.124863456_rs7111513 0.48404517
chr11.124865334.124865878_rs5017048 0.80172074
chr11.133816825.133817041_rs10894749 0.83460367
chr11.36409327.36409838_rs10836550 0.61712982
chr11.36420818.36421386_rs1365120 0.06195368
chr12.120317069.120317911_rs16950058 0.83007788
chr12.120325389.120326108_rs5634 0.33879927
chr12.120367731.120368309_rs2238161 0.99849458
chr12.19222646.19223206_rs7399293 0.77288115
chr13.100722024.100722184_rs12863645 0.40520448
chr13.46774697.46774984_rs1745836 0.47094774
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 0.98094184
chr13.91724572.91724918_rs342715,rs171060 0.64047737
chr13.99908163.99908652_rs9557321 0.90367908
chr14.55721341.55721700_rs2184559 0.93540948
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 0.06195368
chr15.93645160.93645583_rs4238475 0.91785720
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 0.64181703
chr17.51461539.51461982_rs10514983 0.88083022
chr18.32469230.32469379_rs1458866 0.62913275
chr2.104528125.104528417_rs11123997 0.82707716
chr2.12759282.12759669_rs6722342,rs7596623 0.95995037
chr2.12766229.12766820_rs13413220,rs13397389 0.75742139
chr2.176086015.176086512_rs6752623 0.66588181
chr2.41038390.41038934_rs6746423 0.88641352
chr2.77457604.77458071_rs12614202,rs17014128 0.24139438
chr20.22217873.22218175_rs76464104 0.63615304
chr20.53735535.53736570_rs12480103 0.65896538
chr22.46225398.46226192_rs4253763 0.91036202
chr22.46233211.46233644_rs11090819 0.73870849
chr22.46249819.46250733_rs7291763 0.10613245
chr3.107240266.107241585_rs11710776 0.33473496
chr3.118380545.118381165_rs779206,rs13087409 0.39468863
chr3.193461783.193462089_rs9879227 0.74709279
chr3.20682627.20683020_rs11715178,rs10510504 0.83552292
chr4.137238844.137239180_rs13148112 0.53961070
chr4.176615030.176615396_rs6553973 0.32815975
chr4.32226261.32226503_rs13125385 0.57101791
chr4.40731697.40732054_rs11727143 0.88665099
chr4.40749770.40751218_rs10012367 0.57024703
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 0.30425568
chr5.102865289.102866656_rs32680 0.45625089
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 0.44068330
chr5.164619344.164619596_rs250594 0.98347369
chr5.7415226.7416045_rs7710510 0.96134937
chr5.7464212.7464571_rs6883259,rs766643 0.74209321
chr6.160899655.160900401_rs2489944,rs2465874 0.97225243
chr6.27026322.27026687_rs36022097 0.72606878
chr6.32521585.32522598_rs115541994 0.80282001
chr6.71712533.71713167_rs1204328 0.98124991
chr7.10239630.10240386_rs17162825 0.29934433
chr7.116499279.116500903_rs17138749 0.08841947
chr7.134834642.134835353_rs7777356 0.18102442
chr7.26147128.26148142_rs10248605 0.64095699
chr8.141160879.141162083_rs428300 0.68455466
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 0.11269613
chr8.20115130.20115538_rs12548222 0.43458342
chr8.69455933.69456585_rs12545665 0.97431132
chr8.701154.701537_rs13273765 0.57807309
chr8.81096615.81097088_rs16908579 0.80238348
chr9.18632808.18633319_rs776777 0.42825715
chr9.19720338.19720665_rs7869360,rs7855757 0.67064494
TRZ_3
chr1.108829511.108829805_rs10857972,rs6704266 0.9999782
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 0.9999782
chr1.19640454.19641134_rs7523079 0.9999782
chr1.19644587.19645224_rs7522389 0.9999782
chr10.16792579.16793007_rs7068265,rs7068881 0.9999782
chr10.76209088.76209420_rs7094302 0.9999782
chr11.124863016.124863456_rs7111513 0.9999782
chr11.124865334.124865878_rs5017048 0.9999782
chr11.133816825.133817041_rs10894749 0.9999782
chr11.36409327.36409838_rs10836550 0.9999782
chr11.36420818.36421386_rs1365120 0.9999782
chr12.120317069.120317911_rs16950058 0.9999782
chr12.120325389.120326108_rs5634 0.9999782
chr12.120367731.120368309_rs2238161 0.9999782
chr12.19222646.19223206_rs7399293 0.9999782
chr13.100722024.100722184_rs12863645 0.9999782
chr13.46774697.46774984_rs1745836 0.9999782
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 0.9999782
chr13.91724572.91724918_rs342715,rs171060 0.9999782
chr13.99908163.99908652_rs9557321 0.9999782
chr14.55721341.55721700_rs2184559 0.9999782
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 0.9999782
chr15.93645160.93645583_rs4238475 0.9999782
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 0.9999782
chr17.51461539.51461982_rs10514983 0.9999782
chr18.32469230.32469379_rs1458866 0.9999782
chr2.104528125.104528417_rs11123997 0.9999782
chr2.12759282.12759669_rs6722342,rs7596623 0.9999782
chr2.12766229.12766820_rs13413220,rs13397389 0.9999782
chr2.176086015.176086512_rs6752623 0.9999782
chr2.41038390.41038934_rs6746423 0.9999782
chr2.77457604.77458071_rs12614202,rs17014128 0.9999782
chr20.22217873.22218175_rs76464104 0.9999782
chr20.53735535.53736570_rs12480103 0.9999782
chr22.46225398.46226192_rs4253763 0.9999782
chr22.46233211.46233644_rs11090819 0.9999782
chr22.46249819.46250733_rs7291763 0.9999782
chr3.107240266.107241585_rs11710776 0.9999782
chr3.118380545.118381165_rs779206,rs13087409 0.9999782
chr3.193461783.193462089_rs9879227 0.9999782
chr3.20682627.20683020_rs11715178,rs10510504 0.9999782
chr4.137238844.137239180_rs13148112 0.9999782
chr4.176615030.176615396_rs6553973 0.9999782
chr4.32226261.32226503_rs13125385 0.9999782
chr4.40731697.40732054_rs11727143 0.9999782
chr4.40749770.40751218_rs10012367 0.9999782
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 0.9999782
chr5.102865289.102866656_rs32680 0.9999782
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 0.9999782
chr5.164619344.164619596_rs250594 0.9999782
chr5.7415226.7416045_rs7710510 0.9999782
chr5.7464212.7464571_rs6883259,rs766643 0.9999782
chr6.160899655.160900401_rs2489944,rs2465874 0.9999782
chr6.27026322.27026687_rs36022097 0.9999782
chr6.32521585.32522598_rs115541994 0.9999782
chr6.71712533.71713167_rs1204328 0.9999782
chr7.10239630.10240386_rs17162825 0.9999782
chr7.116499279.116500903_rs17138749 0.9999782
chr7.134834642.134835353_rs7777356 0.9999782
chr7.26147128.26148142_rs10248605 0.9999782
chr8.141160879.141162083_rs428300 0.9999782
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 0.9999782
chr8.20115130.20115538_rs12548222 0.9999782
chr8.69455933.69456585_rs12545665 0.9999782
chr8.701154.701537_rs13273765 0.9999782
chr8.81096615.81097088_rs16908579 0.9999782
chr9.18632808.18633319_rs776777 0.9999782
chr9.19720338.19720665_rs7869360,rs7855757 0.9999782
DNR_24
chr1.108829511.108829805_rs10857972,rs6704266 3.653165e-01
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 3.414595e-04
chr1.19640454.19641134_rs7523079 2.258959e-02
chr1.19644587.19645224_rs7522389 1.284788e-01
chr10.16792579.16793007_rs7068265,rs7068881 2.915057e-07
chr10.76209088.76209420_rs7094302 6.989744e-05
chr11.124863016.124863456_rs7111513 7.043568e-01
chr11.124865334.124865878_rs5017048 5.194175e-01
chr11.133816825.133817041_rs10894749 2.019903e-01
chr11.36409327.36409838_rs10836550 9.991173e-01
chr11.36420818.36421386_rs1365120 6.204942e-04
chr12.120317069.120317911_rs16950058 3.122067e-01
chr12.120325389.120326108_rs5634 9.454181e-01
chr12.120367731.120368309_rs2238161 7.491181e-02
chr12.19222646.19223206_rs7399293 4.739298e-01
chr13.100722024.100722184_rs12863645 7.478253e-01
chr13.46774697.46774984_rs1745836 7.803478e-01
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 3.708060e-01
chr13.91724572.91724918_rs342715,rs171060 1.630170e-01
chr13.99908163.99908652_rs9557321 3.824181e-02
chr14.55721341.55721700_rs2184559 6.334068e-03
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 3.962670e-05
chr15.93645160.93645583_rs4238475 1.777890e-02
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 2.727988e-03
chr17.51461539.51461982_rs10514983 2.881889e-01
chr18.32469230.32469379_rs1458866 5.533204e-01
chr2.104528125.104528417_rs11123997 2.379898e-02
chr2.12759282.12759669_rs6722342,rs7596623 2.178708e-02
chr2.12766229.12766820_rs13413220,rs13397389 2.030141e-04
chr2.176086015.176086512_rs6752623 6.773493e-01
chr2.41038390.41038934_rs6746423 1.109757e-05
chr2.77457604.77458071_rs12614202,rs17014128 9.910401e-01
chr20.22217873.22218175_rs76464104 2.370751e-02
chr20.53735535.53736570_rs12480103 2.754945e-01
chr22.46225398.46226192_rs4253763 7.221211e-05
chr22.46233211.46233644_rs11090819 5.820372e-06
chr22.46249819.46250733_rs7291763 1.285664e-04
chr3.107240266.107241585_rs11710776 6.211429e-01
chr3.118380545.118381165_rs779206,rs13087409 8.252985e-03
chr3.193461783.193462089_rs9879227 8.472228e-01
chr3.20682627.20683020_rs11715178,rs10510504 3.903876e-01
chr4.137238844.137239180_rs13148112 8.523507e-03
chr4.176615030.176615396_rs6553973 6.898894e-02
chr4.32226261.32226503_rs13125385 8.301549e-01
chr4.40731697.40732054_rs11727143 7.963452e-02
chr4.40749770.40751218_rs10012367 3.622968e-01
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 3.565007e-02
chr5.102865289.102866656_rs32680 1.018215e-01
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 2.602205e-06
chr5.164619344.164619596_rs250594 5.145805e-05
chr5.7415226.7416045_rs7710510 1.723460e-02
chr5.7464212.7464571_rs6883259,rs766643 3.098879e-03
chr6.160899655.160900401_rs2489944,rs2465874 4.020312e-02
chr6.27026322.27026687_rs36022097 3.769879e-03
chr6.32521585.32522598_rs115541994 4.042327e-02
chr6.71712533.71713167_rs1204328 2.657015e-03
chr7.10239630.10240386_rs17162825 3.560149e-04
chr7.116499279.116500903_rs17138749 1.597110e-02
chr7.134834642.134835353_rs7777356 6.953804e-02
chr7.26147128.26148142_rs10248605 7.987717e-01
chr8.141160879.141162083_rs428300 3.799798e-01
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 3.324496e-07
chr8.20115130.20115538_rs12548222 1.006294e-01
chr8.69455933.69456585_rs12545665 5.878684e-05
chr8.701154.701537_rs13273765 3.284827e-03
chr8.81096615.81097088_rs16908579 2.772015e-01
chr9.18632808.18633319_rs776777 4.468125e-03
chr9.19720338.19720665_rs7869360,rs7855757 1.200819e-02
DOX_24
chr1.108829511.108829805_rs10857972,rs6704266 5.414089e-01
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 5.101642e-04
chr1.19640454.19641134_rs7523079 5.044156e-02
chr1.19644587.19645224_rs7522389 9.963477e-02
chr10.16792579.16793007_rs7068265,rs7068881 7.171811e-07
chr10.76209088.76209420_rs7094302 4.306295e-05
chr11.124863016.124863456_rs7111513 2.556428e-02
chr11.124865334.124865878_rs5017048 4.975003e-01
chr11.133816825.133817041_rs10894749 1.878919e-01
chr11.36409327.36409838_rs10836550 5.445648e-01
chr11.36420818.36421386_rs1365120 1.860554e-02
chr12.120317069.120317911_rs16950058 6.946001e-02
chr12.120325389.120326108_rs5634 5.175766e-01
chr12.120367731.120368309_rs2238161 2.843598e-01
chr12.19222646.19223206_rs7399293 2.917495e-01
chr13.100722024.100722184_rs12863645 2.093724e-01
chr13.46774697.46774984_rs1745836 8.325236e-01
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 7.745944e-02
chr13.91724572.91724918_rs342715,rs171060 1.779978e-01
chr13.99908163.99908652_rs9557321 3.726412e-02
chr14.55721341.55721700_rs2184559 2.531649e-01
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 4.650302e-03
chr15.93645160.93645583_rs4238475 8.588334e-02
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 2.812001e-02
chr17.51461539.51461982_rs10514983 7.835901e-01
chr18.32469230.32469379_rs1458866 9.620150e-01
chr2.104528125.104528417_rs11123997 7.692048e-02
chr2.12759282.12759669_rs6722342,rs7596623 6.708164e-02
chr2.12766229.12766820_rs13413220,rs13397389 5.432938e-03
chr2.176086015.176086512_rs6752623 8.114451e-02
chr2.41038390.41038934_rs6746423 6.858211e-03
chr2.77457604.77458071_rs12614202,rs17014128 6.968894e-01
chr20.22217873.22218175_rs76464104 9.972604e-02
chr20.53735535.53736570_rs12480103 1.540196e-01
chr22.46225398.46226192_rs4253763 3.655236e-01
chr22.46233211.46233644_rs11090819 4.213655e-06
chr22.46249819.46250733_rs7291763 7.254337e-02
chr3.107240266.107241585_rs11710776 5.299319e-01
chr3.118380545.118381165_rs779206,rs13087409 1.816010e-02
chr3.193461783.193462089_rs9879227 7.037356e-01
chr3.20682627.20683020_rs11715178,rs10510504 2.512577e-01
chr4.137238844.137239180_rs13148112 6.937904e-02
chr4.176615030.176615396_rs6553973 9.848660e-01
chr4.32226261.32226503_rs13125385 6.958245e-01
chr4.40731697.40732054_rs11727143 8.199057e-01
chr4.40749770.40751218_rs10012367 9.311694e-01
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 6.523537e-01
chr5.102865289.102866656_rs32680 2.317915e-01
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 3.395487e-04
chr5.164619344.164619596_rs250594 2.588807e-03
chr5.7415226.7416045_rs7710510 1.421706e-01
chr5.7464212.7464571_rs6883259,rs766643 8.472525e-04
chr6.160899655.160900401_rs2489944,rs2465874 1.472753e-01
chr6.27026322.27026687_rs36022097 1.942271e-02
chr6.32521585.32522598_rs115541994 9.139523e-03
chr6.71712533.71713167_rs1204328 2.869743e-02
chr7.10239630.10240386_rs17162825 7.585279e-02
chr7.116499279.116500903_rs17138749 1.217002e-02
chr7.134834642.134835353_rs7777356 1.883007e-01
chr7.26147128.26148142_rs10248605 9.003688e-01
chr8.141160879.141162083_rs428300 5.742705e-01
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 4.234505e-05
chr8.20115130.20115538_rs12548222 3.558188e-01
chr8.69455933.69456585_rs12545665 1.755253e-04
chr8.701154.701537_rs13273765 3.824064e-01
chr8.81096615.81097088_rs16908579 3.685826e-01
chr9.18632808.18633319_rs776777 1.446595e-03
chr9.19720338.19720665_rs7869360,rs7855757 6.070919e-01
EPI_24
chr1.108829511.108829805_rs10857972,rs6704266 3.146685e-01
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 4.083676e-04
chr1.19640454.19641134_rs7523079 1.258701e-02
chr1.19644587.19645224_rs7522389 2.094945e-02
chr10.16792579.16793007_rs7068265,rs7068881 1.814685e-06
chr10.76209088.76209420_rs7094302 6.278692e-05
chr11.124863016.124863456_rs7111513 2.672714e-01
chr11.124865334.124865878_rs5017048 6.841295e-01
chr11.133816825.133817041_rs10894749 2.454422e-02
chr11.36409327.36409838_rs10836550 5.990061e-01
chr11.36420818.36421386_rs1365120 2.180274e-04
chr12.120317069.120317911_rs16950058 3.078297e-01
chr12.120325389.120326108_rs5634 6.709758e-01
chr12.120367731.120368309_rs2238161 4.294827e-02
chr12.19222646.19223206_rs7399293 9.681589e-01
chr13.100722024.100722184_rs12863645 7.117580e-01
chr13.46774697.46774984_rs1745836 5.460103e-01
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 2.059572e-01
chr13.91724572.91724918_rs342715,rs171060 5.553094e-01
chr13.99908163.99908652_rs9557321 2.811388e-01
chr14.55721341.55721700_rs2184559 1.582115e-02
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 7.433648e-02
chr15.93645160.93645583_rs4238475 2.793523e-01
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 2.119829e-03
chr17.51461539.51461982_rs10514983 9.947625e-01
chr18.32469230.32469379_rs1458866 8.123747e-01
chr2.104528125.104528417_rs11123997 4.754784e-03
chr2.12759282.12759669_rs6722342,rs7596623 2.560375e-02
chr2.12766229.12766820_rs13413220,rs13397389 1.466140e-04
chr2.176086015.176086512_rs6752623 2.730192e-01
chr2.41038390.41038934_rs6746423 2.705464e-03
chr2.77457604.77458071_rs12614202,rs17014128 1.341606e-02
chr20.22217873.22218175_rs76464104 2.532489e-03
chr20.53735535.53736570_rs12480103 8.756351e-01
chr22.46225398.46226192_rs4253763 8.978233e-02
chr22.46233211.46233644_rs11090819 5.119405e-06
chr22.46249819.46250733_rs7291763 7.275134e-02
chr3.107240266.107241585_rs11710776 2.995115e-01
chr3.118380545.118381165_rs779206,rs13087409 2.956848e-02
chr3.193461783.193462089_rs9879227 5.332866e-01
chr3.20682627.20683020_rs11715178,rs10510504 1.687950e-01
chr4.137238844.137239180_rs13148112 2.262285e-03
chr4.176615030.176615396_rs6553973 2.155857e-02
chr4.32226261.32226503_rs13125385 3.487420e-01
chr4.40731697.40732054_rs11727143 6.510506e-01
chr4.40749770.40751218_rs10012367 3.703610e-01
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 4.313594e-01
chr5.102865289.102866656_rs32680 5.569582e-01
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 5.021422e-04
chr5.164619344.164619596_rs250594 6.012673e-02
chr5.7415226.7416045_rs7710510 5.959757e-02
chr5.7464212.7464571_rs6883259,rs766643 1.565716e-03
chr6.160899655.160900401_rs2489944,rs2465874 5.664559e-01
chr6.27026322.27026687_rs36022097 1.768075e-01
chr6.32521585.32522598_rs115541994 2.489476e-01
chr6.71712533.71713167_rs1204328 2.896963e-01
chr7.10239630.10240386_rs17162825 4.425513e-03
chr7.116499279.116500903_rs17138749 2.101737e-02
chr7.134834642.134835353_rs7777356 1.173219e-02
chr7.26147128.26148142_rs10248605 1.566356e-01
chr8.141160879.141162083_rs428300 2.443513e-01
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 2.261059e-04
chr8.20115130.20115538_rs12548222 6.742704e-02
chr8.69455933.69456585_rs12545665 3.088158e-03
chr8.701154.701537_rs13273765 5.405245e-02
chr8.81096615.81097088_rs16908579 1.451063e-01
chr9.18632808.18633319_rs776777 3.813429e-02
chr9.19720338.19720665_rs7869360,rs7855757 4.255916e-01
MTX_24
chr1.108829511.108829805_rs10857972,rs6704266 0.143612204
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 0.166985401
chr1.19640454.19641134_rs7523079 0.386560760
chr1.19644587.19645224_rs7522389 0.083459693
chr10.16792579.16793007_rs7068265,rs7068881 0.007940749
chr10.76209088.76209420_rs7094302 0.000924940
chr11.124863016.124863456_rs7111513 0.120123879
chr11.124865334.124865878_rs5017048 0.307342372
chr11.133816825.133817041_rs10894749 0.368842737
chr11.36409327.36409838_rs10836550 0.231801783
chr11.36420818.36421386_rs1365120 0.027092796
chr12.120317069.120317911_rs16950058 0.172473549
chr12.120325389.120326108_rs5634 0.646604873
chr12.120367731.120368309_rs2238161 0.535934368
chr12.19222646.19223206_rs7399293 0.616081509
chr13.100722024.100722184_rs12863645 0.465107643
chr13.46774697.46774984_rs1745836 0.939257304
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 0.532181117
chr13.91724572.91724918_rs342715,rs171060 0.837246328
chr13.99908163.99908652_rs9557321 0.761667808
chr14.55721341.55721700_rs2184559 0.162819249
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 0.217838852
chr15.93645160.93645583_rs4238475 0.476711994
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 0.308763031
chr17.51461539.51461982_rs10514983 0.640685868
chr18.32469230.32469379_rs1458866 0.512354849
chr2.104528125.104528417_rs11123997 0.225281685
chr2.12759282.12759669_rs6722342,rs7596623 0.510320433
chr2.12766229.12766820_rs13413220,rs13397389 0.022657513
chr2.176086015.176086512_rs6752623 0.967536271
chr2.41038390.41038934_rs6746423 0.140763597
chr2.77457604.77458071_rs12614202,rs17014128 0.762871185
chr20.22217873.22218175_rs76464104 0.050031470
chr20.53735535.53736570_rs12480103 0.445159074
chr22.46225398.46226192_rs4253763 0.012437681
chr22.46233211.46233644_rs11090819 0.016770628
chr22.46249819.46250733_rs7291763 0.183926449
chr3.107240266.107241585_rs11710776 0.231162659
chr3.118380545.118381165_rs779206,rs13087409 0.494393320
chr3.193461783.193462089_rs9879227 0.515773248
chr3.20682627.20683020_rs11715178,rs10510504 0.309606866
chr4.137238844.137239180_rs13148112 0.577743705
chr4.176615030.176615396_rs6553973 0.897856151
chr4.32226261.32226503_rs13125385 0.993026830
chr4.40731697.40732054_rs11727143 0.961489010
chr4.40749770.40751218_rs10012367 0.675404619
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 0.848771811
chr5.102865289.102866656_rs32680 0.157419963
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 0.002836755
chr5.164619344.164619596_rs250594 0.286580523
chr5.7415226.7416045_rs7710510 0.636407880
chr5.7464212.7464571_rs6883259,rs766643 0.135326952
chr6.160899655.160900401_rs2489944,rs2465874 0.583409984
chr6.27026322.27026687_rs36022097 0.099562150
chr6.32521585.32522598_rs115541994 0.273102359
chr6.71712533.71713167_rs1204328 0.915814676
chr7.10239630.10240386_rs17162825 0.579189402
chr7.116499279.116500903_rs17138749 0.389947603
chr7.134834642.134835353_rs7777356 0.894533553
chr7.26147128.26148142_rs10248605 0.647506619
chr8.141160879.141162083_rs428300 0.883193455
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 0.005202160
chr8.20115130.20115538_rs12548222 0.047002399
chr8.69455933.69456585_rs12545665 0.004285771
chr8.701154.701537_rs13273765 0.318646385
chr8.81096615.81097088_rs16908579 0.635859465
chr9.18632808.18633319_rs776777 0.158385280
chr9.19720338.19720665_rs7869360,rs7855757 0.143104689
TRZ_24
chr1.108829511.108829805_rs10857972,rs6704266 0.9999654
chr1.173473597.173473889_rs4916358,rs10798282,rs10753081 0.9999654
chr1.19640454.19641134_rs7523079 0.9999654
chr1.19644587.19645224_rs7522389 0.9999654
chr10.16792579.16793007_rs7068265,rs7068881 0.9999654
chr10.76209088.76209420_rs7094302 0.9999654
chr11.124863016.124863456_rs7111513 0.9999654
chr11.124865334.124865878_rs5017048 0.9999654
chr11.133816825.133817041_rs10894749 0.9999654
chr11.36409327.36409838_rs10836550 0.9999654
chr11.36420818.36421386_rs1365120 0.9999654
chr12.120317069.120317911_rs16950058 0.9999654
chr12.120325389.120326108_rs5634 0.9999654
chr12.120367731.120368309_rs2238161 0.9999654
chr12.19222646.19223206_rs7399293 0.9999654
chr13.100722024.100722184_rs12863645 0.9999654
chr13.46774697.46774984_rs1745836 0.9999654
chr13.64945176.64945505_rs925558,rs1513001,rs7994896 0.9999654
chr13.91724572.91724918_rs342715,rs171060 0.9999654
chr13.99908163.99908652_rs9557321 0.9999654
chr14.55721341.55721700_rs2184559 0.9999654
chr15.23483710.23484436_rs28684656,rs28714259,rs1875910 0.9999654
chr15.93645160.93645583_rs4238475 0.9999654
chr16.13378005.13378513_rs2188738,rs1018891,rs886218,rs1468171 0.9999654
chr17.51461539.51461982_rs10514983 0.9999654
chr18.32469230.32469379_rs1458866 0.9999654
chr2.104528125.104528417_rs11123997 0.9999654
chr2.12759282.12759669_rs6722342,rs7596623 0.9999654
chr2.12766229.12766820_rs13413220,rs13397389 0.9999654
chr2.176086015.176086512_rs6752623 0.9999654
chr2.41038390.41038934_rs6746423 0.9999654
chr2.77457604.77458071_rs12614202,rs17014128 0.9999654
chr20.22217873.22218175_rs76464104 0.9999654
chr20.53735535.53736570_rs12480103 0.9999654
chr22.46225398.46226192_rs4253763 0.9999654
chr22.46233211.46233644_rs11090819 0.9999654
chr22.46249819.46250733_rs7291763 0.9999654
chr3.107240266.107241585_rs11710776 0.9999654
chr3.118380545.118381165_rs779206,rs13087409 0.9999654
chr3.193461783.193462089_rs9879227 0.9999654
chr3.20682627.20683020_rs11715178,rs10510504 0.9999654
chr4.137238844.137239180_rs13148112 0.9999654
chr4.176615030.176615396_rs6553973 0.9999654
chr4.32226261.32226503_rs13125385 0.9999654
chr4.40731697.40732054_rs11727143 0.9999654
chr4.40749770.40751218_rs10012367 0.9999654
chr4.9880847.9881265_rs4697900,rs9684729,rs6449090 0.9999654
chr5.102865289.102866656_rs32680 0.9999654
chr5.102947036.102947471_rs6871111,rs6860588,rs3776874,rs11952361 0.9999654
chr5.164619344.164619596_rs250594 0.9999654
chr5.7415226.7416045_rs7710510 0.9999654
chr5.7464212.7464571_rs6883259,rs766643 0.9999654
chr6.160899655.160900401_rs2489944,rs2465874 0.9999654
chr6.27026322.27026687_rs36022097 0.9999654
chr6.32521585.32522598_rs115541994 0.9999654
chr6.71712533.71713167_rs1204328 0.9999654
chr7.10239630.10240386_rs17162825 0.9999654
chr7.116499279.116500903_rs17138749 0.9999654
chr7.134834642.134835353_rs7777356 0.9999654
chr7.26147128.26148142_rs10248605 0.9999654
chr8.141160879.141162083_rs428300 0.9999654
chr8.15830942.15831184_rs11203734,rs7016422,rs12549742,rs2410329 0.9999654
chr8.20115130.20115538_rs12548222 0.9999654
chr8.69455933.69456585_rs12545665 0.9999654
chr8.701154.701537_rs13273765 0.9999654
chr8.81096615.81097088_rs16908579 0.9999654
chr9.18632808.18633319_rs776777 0.9999654
chr9.19720338.19720665_rs7869360,rs7855757 0.9999654AR_Cardiotox_gwas_collaped_df# A tibble: 68 × 18
name peak_chr peak_start TAD_id sig_24 min_distance mean_distance DOX_3
<chr> <fct> <int> <chr> <chr> <int> <dbl> <dbl>
1 chr1.10… chr1 108829511 TAD_71 not_s… 12303 13172 -1.27
2 chr1.17… chr1 173473597 TAD_1… sig 795 8481. 0.195
3 chr1.19… chr1 19640454 TAD_13 not_s… 4074 4074 -0.989
4 chr1.19… chr1 19644587 TAD_13 not_s… 2277 2277 0.241
5 chr10.1… chr10 16792579 TAD_1… sig 3839 4064. -0.310
6 chr10.7… chr10 76209088 TAD_1… sig 2753 2753 0.311
7 chr11.1… chr11 124863016 TAD_1… sig 465 465 0.188
8 chr11.1… chr11 124865334 TAD_1… not_s… 759 759 0.0823
9 chr11.1… chr11 133816825 TAD_1… not_s… 4468 4468 -0.585
10 chr11.3… chr11 36409327 TAD_1… not_s… 2984 2984 -0.171
# ℹ 58 more rows
# ℹ 10 more variables: EPI_3 <dbl>, DNR_3 <dbl>, MTX_3 <dbl>, TRZ_3 <dbl>,
# DOX_24 <dbl>, EPI_24 <dbl>, DNR_24 <dbl>, MTX_24 <dbl>, TRZ_24 <dbl>,
# snp_dist <chr>adding in Park data:
ParkSNPs <- readRDS("data/other_papers/ParkSNPs_pull_VEF.RDS")
ParkSNP_table <-
ParkSNPs %>%
dplyr::select(1:2) %>%
distinct() %>%
separate_wider_delim(.,Location,delim=":",names=c("chr","position"), cols_remove=FALSE) %>%
separate_wider_delim(.,position,delim="-",names=c("begin","term")) %>%
mutate(chr=paste0("chr",chr))
ParkSNP_gr <- ParkSNP_table %>%
mutate("start" = begin, "end"=term) %>%
GRanges()
Park_snp_tad_df <- join_overlap_inner(ParkSNP_gr, Left_ventricle_TAD) %>%
as_tibble() %>%
dplyr::rename("RSID"=X.Uploaded_variation) %>%
dplyr::select(RSID, snp_start = start, snp_chr = seqnames, TAD_id)
Park_snp_pairs <- peak_tad_df %>%
inner_join(Park_snp_tad_df, by = "TAD_id")
Park_snp_pairs %>%
distinct(RSID)
Park_snp_pairs_dist <- Park_snp_pairs %>%
mutate(distance = abs(peak_start - snp_start)) %>%
mutate(sig_24= if_else(Peakid %in% DOX_DAR_sig$Peakid, "sig","not_sig"))
Park_snp_pairs_dist %>%
mutate(sig_24=factor(sig_24, levels= c("sig","not_sig"))) %>%
ggplot(., aes(x= sig_24, y=distance))+
geom_boxplot()+
theme_bw()+
geom_signif(comparisons = list(c("sig", "not_sig")),
map_signif_level = FALSE, test = "wilcox.test")
wilcox.test(distance ~ sig_24, data = Park_snp_pairs_dist)
Park_Cardiotox_gwas_df <- Park_snp_pairs_dist %>%
dplyr::filter(sig_24=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)Park_Cardiotox_gwas_collaped_df <-
Park_snp_pairs_dist %>%
# dplyr::filter(sig_24=="sig") %>%
group_by(TAD_id,RSID) %>%
slice_min(order_by = distance, with_ties = FALSE) %>%
ungroup() %>%
arrange(snp_chr,snp_start) %>%
group_by(Peakid, peak_chr, peak_start, TAD_id, sig_24) %>%
summarise(
min_distance = min(distance),
mean_distance = mean(distance),
snp_list = paste(unique(RSID), collapse = ","),
.groups = "drop"
) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
# left_join(., Peak_gene_RNA_LFC, by=c("Peakid"="Peakid")) %>%
# left_join(.,gene_info_collapsed, by=c("geneId"="ENTREZID")) %>%
# mutate(SYMBOL=if_else(is.na(SYMBOL.x),SYMBOL.y,if_else(SYMBOL.x==SYMBOL.y, SYMBOL.x,paste0(SYMBOL.x,"_",SYMBOL.y)))) %>%
tidyr::unite(., name,Peakid,snp_list) %>%
mutate(snp_dist=case_when(min_distance <2000 ~"2kb",
min_distance > 2000 & min_distance<20000 ~ "20kb",
min_distance >20000 ~">20kb"))
Cardotox_mat_park <- Park_Cardiotox_gwas_collaped_df %>%
dplyr::select(name,DOX_3:TRZ_24) %>%
column_to_rownames("name") %>%
as.matrix()
annot_map_df_park <- Park_Cardiotox_gwas_collaped_df %>%
dplyr::select(name,snp_dist,sig_24) %>%
column_to_rownames("name")
annot_map_park <-
ComplexHeatmap::rowAnnotation(
snp_dist=Park_Cardiotox_gwas_collaped_df$snp_dist,
TAD_id=Park_Cardiotox_gwas_collaped_df$TAD_id,
DOX_24hr_DAR=Park_Cardiotox_gwas_collaped_df$sig_24,
col= list(snp_dist=c("2kb"="goldenrod4",
"20kb"="pink",
">20kb"="tan2")))
simply_map_lfc_park <- ComplexHeatmap::Heatmap(Cardotox_mat_park,
# col = col_fun,
left_annotation = annot_map_park,
show_row_names = TRUE,
row_names_max_width= ComplexHeatmap::max_text_width(rownames(Cardotox_mat_park), gp=gpar(fontsize=14)),
heatmap_legend_param = list(direction = "horizontal"),
show_column_names = TRUE,
cluster_rows = FALSE,
cluster_columns = FALSE)
ComplexHeatmap::draw(simply_map_lfc_park,
merge_legend = TRUE,
heatmap_legend_side = "left",
annotation_legend_side = "left")Accessibility changes of SNP- directly overlapping DARs
drug_pal <- c("#8B006D","#DF707E","#F1B72B", "#3386DD","#707031","#41B333")
raw_counts <- read_delim("data/Final_four_data/re_analysis/Raw_unfiltered_counts.tsv",delim="\t") %>%
column_to_rownames("Peakid") %>%
as.matrix()
lcpm <- cpm(raw_counts, log= TRUE)
### for determining the basic cutoffs
filt_raw_counts <- raw_counts[rowMeans(lcpm)> 0,]
filt_raw_counts_noY <- filt_raw_counts[!grepl("chrY",rownames(filt_raw_counts)),]
ATAC_adj.pvals <-all_results %>%
dplyr::select(source,genes,adj.P.Val) %>%
dplyr::filter(genes %in% SNP_DAR_overlap_direct$Peakid) %>%
separate(source, into = c("trt", "time")) %>%
mutate(
time = paste0(time, "h"), # convert "3" → "3h"
trt = factor(trt, levels = c("DOX", "EPI", "DNR", "MTX", "TRZ")),
group=paste0(trt,"_",time)) %>%
mutate(group=factor(group,levels = c("DOX_3h", "EPI_3h", "DNR_3h", "MTX_3h", "TRZ_3h", "VEH_3h",
"DOX_24h", "EPI_24h", "DNR_24h", "MTX_24h", "TRZ_24h", "VEH_24h"))) %>%
dplyr::rename("Peakid"=genes)ATAC_counts_lcpm <- filt_raw_counts_noY %>%
cpm(., log = TRUE) %>%
as.data.frame() %>%
rownames_to_column("Peakid")for (peak in SNP_DAR_overlap_direct$Peakid) {
PEAK <- SNP_DAR_overlap_direct$Peakid[SNP_DAR_overlap_direct$Peakid == peak]
# Prep expression data
peak_expr <- ATAC_counts_lcpm %>%
filter(Peakid == peak) %>%
pivot_longer(cols = !Peakid, names_to = "sample", values_to = "lcpm") %>%
separate(sample, into = c("ind", "trt", "time")) %>%
mutate(
time = paste0(time), # if already "3h"/"24h"
group = paste0(trt, "_", time),
group = factor(group, levels = c(
"DOX_3h", "EPI_3h", "DNR_3h", "MTX_3h", "TRZ_3h", "VEH_3h",
"DOX_24h", "EPI_24h", "DNR_24h", "MTX_24h", "TRZ_24h", "VEH_24h"
))
)
# Get peak-specific p-values
peak_pvals <- ATAC_adj.pvals %>%
filter(Peakid==peak)
# Merge in p-values by group
peak_plot_data <- left_join(peak_expr, peak_pvals, by = c("Peakid", "group", "time"))
# Create label position below box
label_positions <- peak_plot_data %>%
group_by(group) %>%
summarise(y = min(lcpm, na.rm = TRUE) - 0.5, .groups = "drop")
peak_plot_data <- left_join(peak_plot_data, label_positions, by = "group")
peak_plot_data <- peak_plot_data %>%
separate(group, into = c("trt", "time"), sep = "_", remove = FALSE)
# Plot
peak_plot <- ggplot(peak_plot_data, aes(x = group, y = lcpm)) +
geom_boxplot(aes(fill = trt)) +
geom_text(
aes(y = y,
label = ifelse(trt != "VEH" & !is.na(adj.P.Val),
paste0("", signif(adj.P.Val, 2)),
"")),
size = 3,
vjust = 1.2
) +
scale_fill_manual(values = drug_pal) +
theme_bw() +
ggtitle(paste0("ATAC Log2cpm of ", PEAK)) +
ylab("log2 cpm ATAC") +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
plot(peak_plot)
}
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
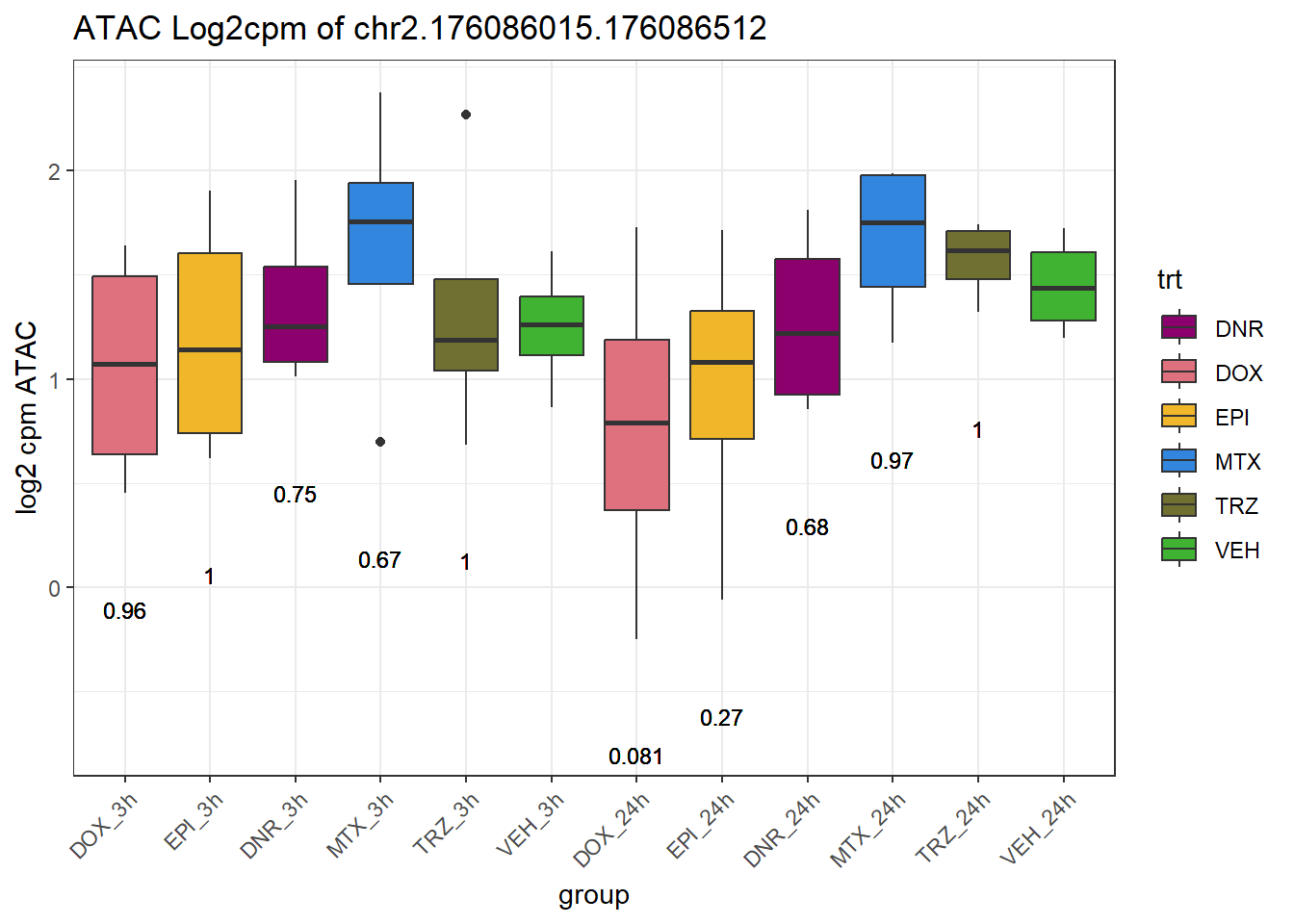
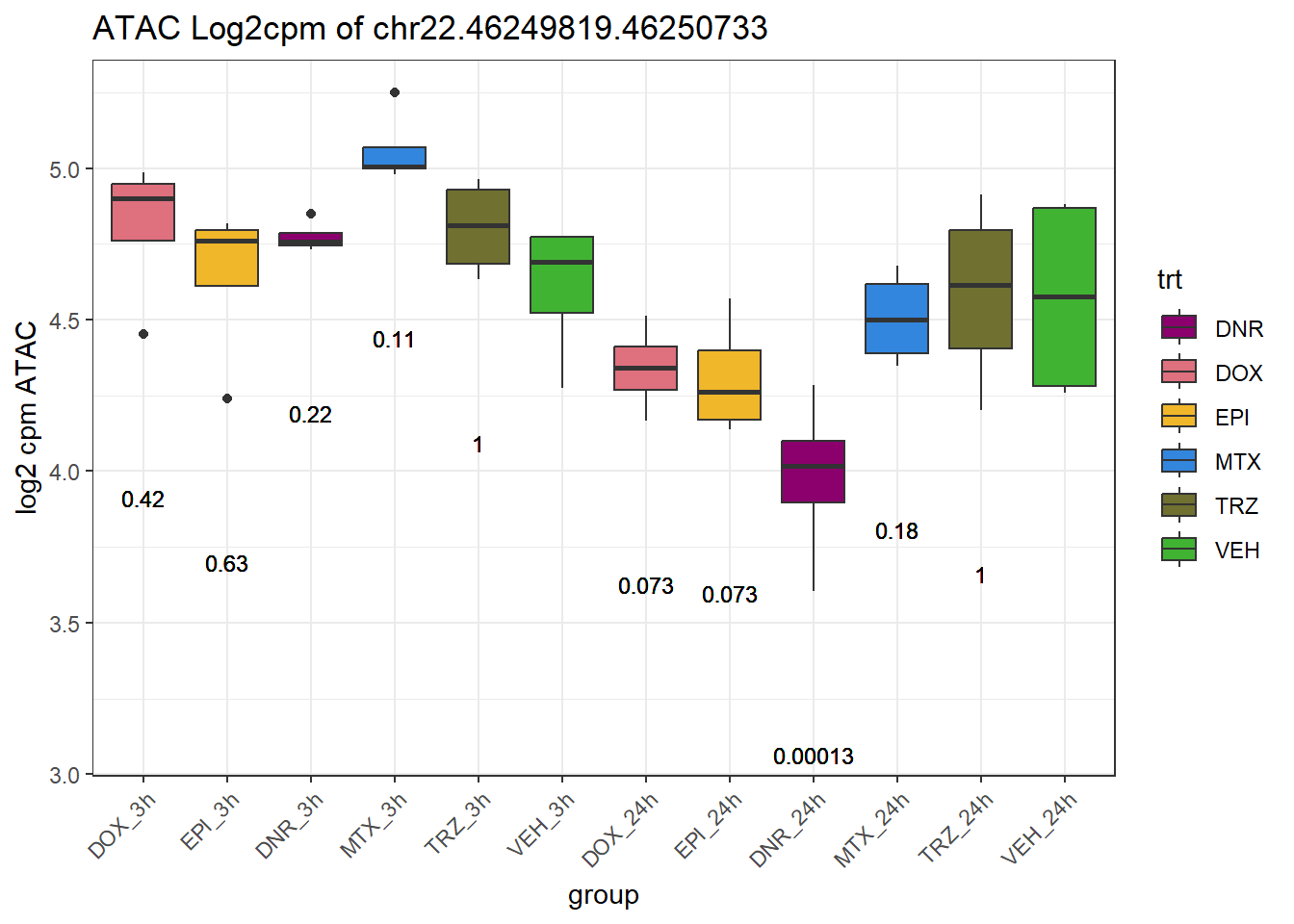
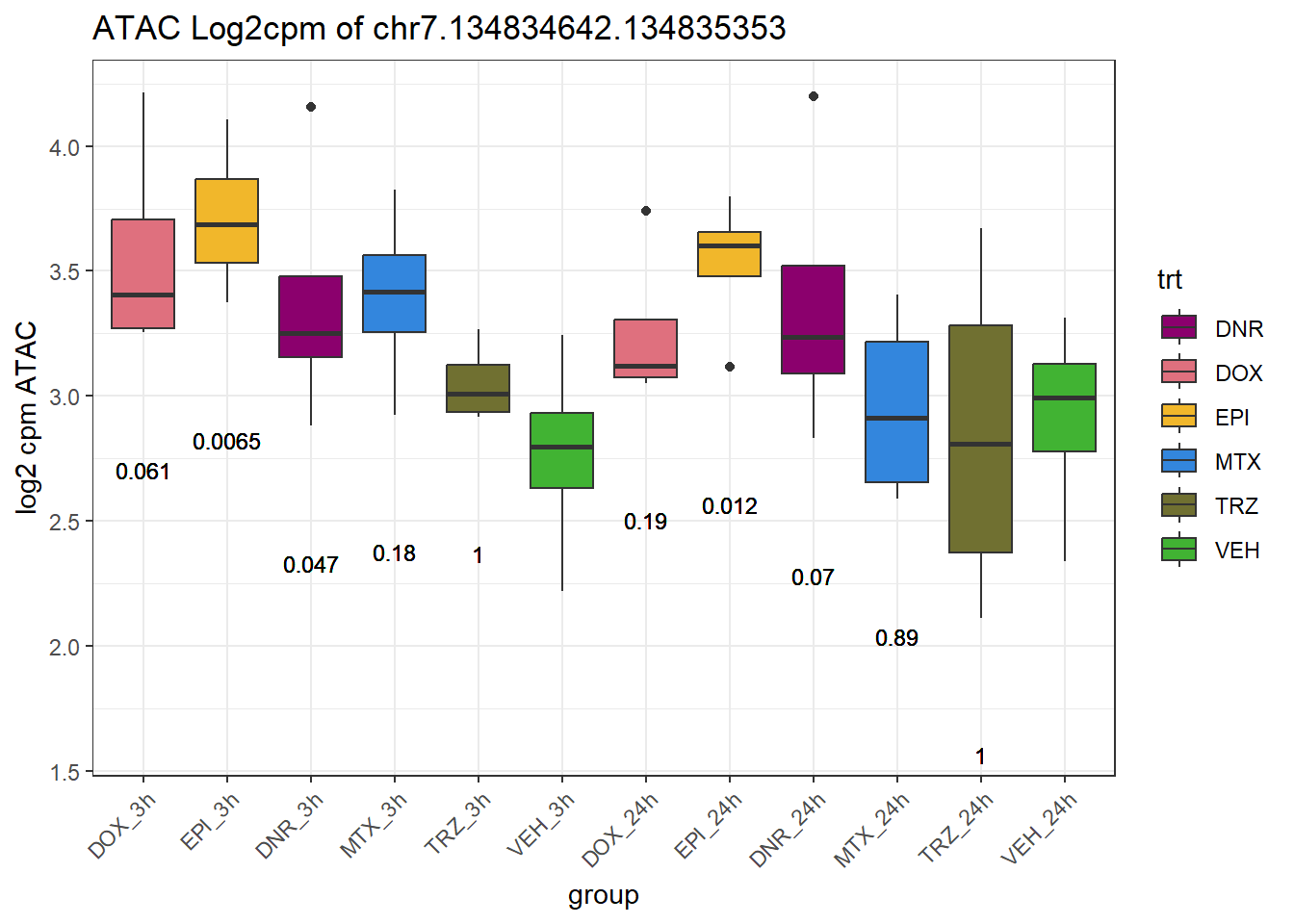
# for (peak in SNP_DAR_overlap_direct$Peakid) {
# PEAK <- SNP_DAR_overlap_direct$Peakid[SNP_DAR_overlap_direct$Peakid == peak]
#
#
# # Filter and plot
# gene_plot <- ATAC_counts_lcpm %>%
# filter(Peakid == peak) %>%
# pivot_longer(cols = !Peakid, names_to = "sample", values_to = "lcpm") %>%
# separate(sample, into = c("trt", "ind", "time")) %>%
# mutate(
# time = factor(time, levels = c("3h", "24h")),
# trt = factor(trt, levels = c("DOX", "EPI", "DNR", "MTX", "TRZ", "VEH"))
# ) %>%
# ggplot(aes(x = time, y = counts)) +
# geom_boxplot(aes(fill = trt)) +
# scale_fill_manual(values = drug_pal) +
# theme_bw() +
# ylab("log2 cpm RNA") +
# ggtitle(paste0(" Log2cpm of ", PEAK))
#
# plot(gene_plot)
# }
#
filt_raw_counts_noY %>%
cpm(., log = TRUE) %>%
as.data.frame() %>%
rownames_to_column("Peakid") %>%
dplyr::filter(Peakid %in% SNP_DAR_overlap_direct$Peakid) %>%
pivot_longer(., cols= !Peakid, names_to = "sample",values_to = "log2cpm") %>%
separate_wider_delim(, cols=sample, names =c("ind","trt","time"),delim="_",cols_remove = FALSE) %>%
mutate(
time = factor(time, levels = c("3h", "24h")),
trt = factor(trt, levels = c("DOX", "EPI", "DNR", "MTX", "TRZ", "VEH"))
) %>%
ggplot(aes(x = time, y = log2cpm)) +
geom_boxplot(aes(fill = trt)) +
scale_fill_manual(values = drug_pal) +
theme_bw() +
facet_wrap(~Peakid, scales="free_y")+
ylab("log2 cpm ATAC regions") 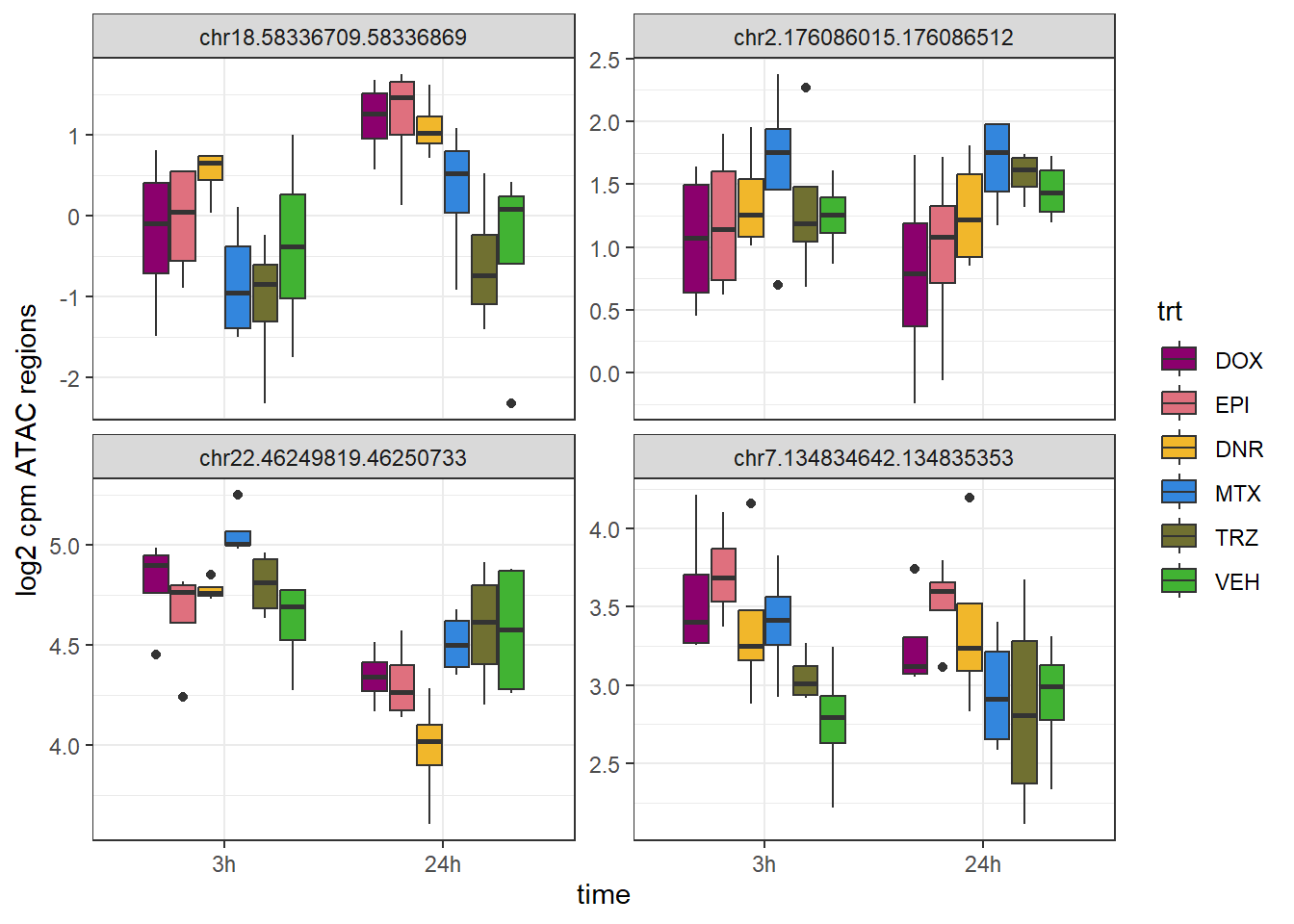
| Version | Author | Date |
|---|---|---|
| 1429820 | reneeisnowhere | 2025-07-21 |
SNP DAR direct overlap heatmap
SNP_DAR_overlap_mat <-
SNP_DAR_overlap_direct %>%
dplyr::select(Peakid,RSID) %>%
left_join(., snp_tad_df,by= c("RSID"="RSID")) %>%
dplyr::select(Peakid, TAD_id, RSID) %>%
left_join(., all_results_pivot, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID)
SNP_DAR_sig_mat <- SNP_DAR_overlap_direct %>%
dplyr::select(Peakid,RSID) %>%
left_join(., snp_tad_df,by= c("RSID"="RSID")) %>%
dplyr::select(Peakid, TAD_id, RSID) %>%
left_join(., ATAC_all_adj.pvals, by=c("Peakid"="genes")) %>%
tidyr::unite(., name,Peakid,RSID) %>%
column_to_rownames("name") %>%
as.matrix()
Cardotox_mat_3 <- SNP_DAR_overlap_mat %>%
dplyr::select(name,DOX_3:TRZ_24) %>%
column_to_rownames("name") %>%
as.matrix()
annot_map_df_3 <- SNP_DAR_overlap_mat %>%
dplyr::select(name,TAD_id) %>%
column_to_rownames("name")
annot_map_3 <-
ComplexHeatmap::rowAnnotation(TAD_id=SNP_DAR_overlap_mat$TAD_id)
simply_map_lfc_3 <- ComplexHeatmap::Heatmap(Cardotox_mat_3,
# col = col_fun,
left_annotation = annot_map_3,
column_title="Cardiotox SNP direct overlaps",
show_row_names = TRUE,
row_names_max_width= ComplexHeatmap::max_text_width(rownames(Cardotox_mat_3), gp=gpar(fontsize=14)),
heatmap_legend_param = list(direction = "horizontal"),
show_column_names = TRUE,
cluster_rows = FALSE,
cluster_columns = FALSE,
cell_fun = function(j, i, x, y, width, height, fill) {
if (!is.na(SNP_DAR_sig_mat[i, j]) && SNP_DAR_sig_mat[i, j] <0.05) {
grid.text("*", x, y, gp = gpar(fontsize = 20)) # Add star if significant
} })
ComplexHeatmap::draw(simply_map_lfc_3,
merge_legend = TRUE,
heatmap_legend_side = "left",
annotation_legend_side = "left")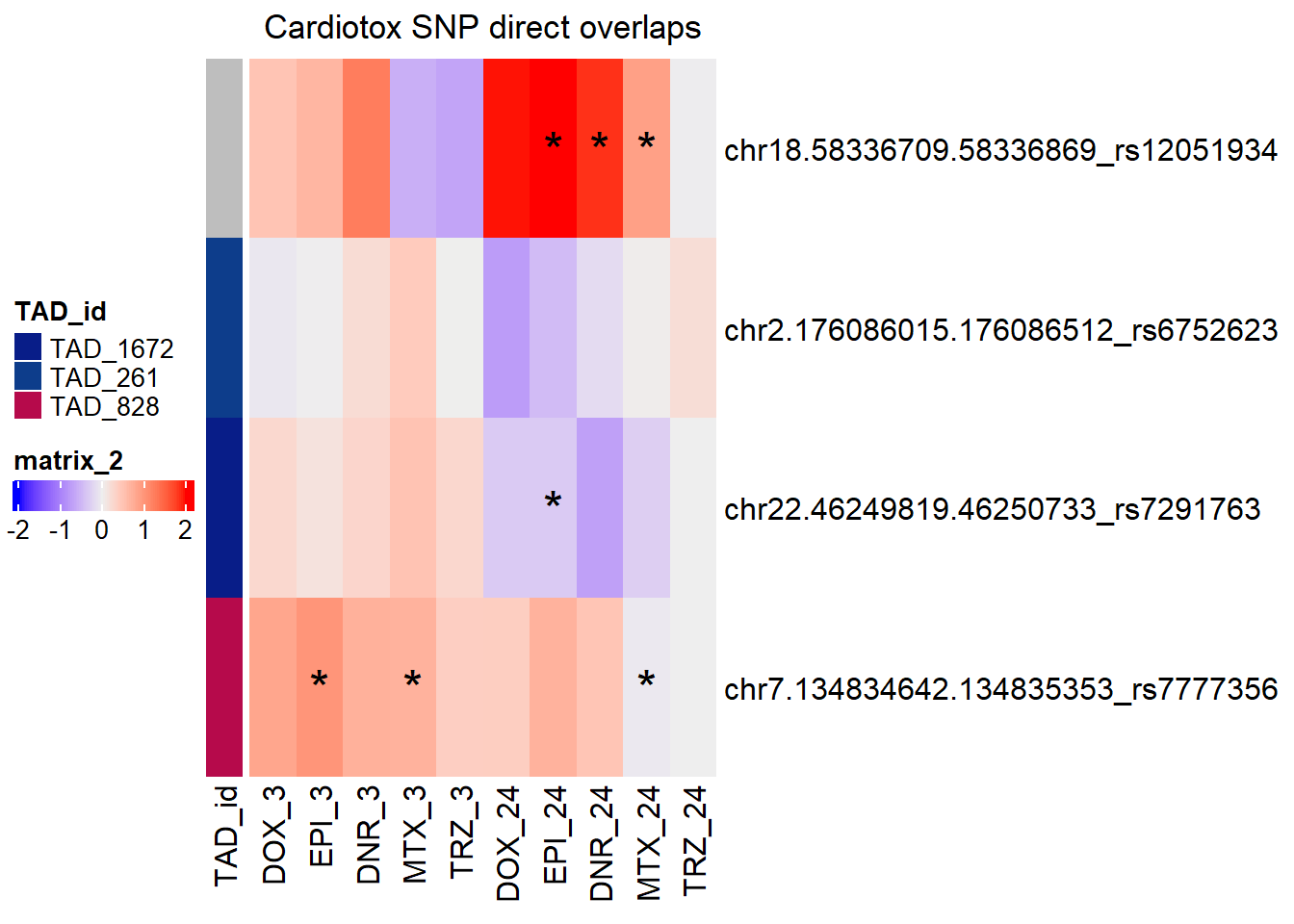
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 26100)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] BSgenome.Hsapiens.UCSC.hg38_1.4.5
[2] BSgenome_1.74.0
[3] BiocIO_1.16.0
[4] Biostrings_2.74.1
[5] XVector_0.46.0
[6] liftOver_1.30.0
[7] Homo.sapiens_1.3.1
[8] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[9] GO.db_3.20.0
[10] OrganismDbi_1.48.0
[11] gwascat_2.38.0
[12] ComplexHeatmap_2.22.0
[13] readxl_1.4.5
[14] circlize_0.4.16
[15] epitools_0.5-10.1
[16] ggrepel_0.9.6
[17] plyranges_1.26.0
[18] ggsignif_0.6.4
[19] genomation_1.38.0
[20] smplot2_0.2.5
[21] eulerr_7.0.2
[22] biomaRt_2.62.1
[23] devtools_2.4.5
[24] usethis_3.1.0
[25] ggpubr_0.6.1
[26] BiocParallel_1.40.2
[27] scales_1.4.0
[28] VennDiagram_1.7.3
[29] futile.logger_1.4.3
[30] gridExtra_2.3
[31] ggfortify_0.4.18
[32] edgeR_4.4.2
[33] limma_3.62.2
[34] rtracklayer_1.66.0
[35] org.Hs.eg.db_3.20.0
[36] TxDb.Hsapiens.UCSC.hg38.knownGene_3.20.0
[37] GenomicFeatures_1.58.0
[38] AnnotationDbi_1.68.0
[39] Biobase_2.66.0
[40] ChIPpeakAnno_3.40.0
[41] GenomicRanges_1.58.0
[42] GenomeInfoDb_1.42.3
[43] IRanges_2.40.1
[44] S4Vectors_0.44.0
[45] BiocGenerics_0.52.0
[46] ChIPseeker_1.42.1
[47] RColorBrewer_1.1-3
[48] broom_1.0.8
[49] kableExtra_1.4.0
[50] lubridate_1.9.4
[51] forcats_1.0.0
[52] stringr_1.5.1
[53] dplyr_1.1.4
[54] purrr_1.0.4
[55] readr_2.1.5
[56] tidyr_1.3.1
[57] tibble_3.3.0
[58] ggplot2_3.5.2
[59] tidyverse_2.0.0
[60] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] R.methodsS3_1.8.2 dichromat_2.0-0.1
[3] vroom_1.6.5 progress_1.2.3
[5] urlchecker_1.0.1 nnet_7.3-20
[7] vctrs_0.6.5 ggtangle_0.0.7
[9] digest_0.6.37 png_0.1-8
[11] shape_1.4.6.1 git2r_0.36.2
[13] magick_2.8.7 MASS_7.3-65
[15] reshape2_1.4.4 foreach_1.5.2
[17] httpuv_1.6.16 qvalue_2.38.0
[19] withr_3.0.2 xfun_0.52
[21] ggfun_0.1.9 ellipsis_0.3.2
[23] survival_3.8-3 memoise_2.0.1
[25] profvis_0.4.0 systemfonts_1.2.3
[27] tidytree_0.4.6 zoo_1.8-14
[29] GlobalOptions_0.1.2 gtools_3.9.5
[31] R.oo_1.27.1 Formula_1.2-5
[33] prettyunits_1.2.0 KEGGREST_1.46.0
[35] promises_1.3.3 httr_1.4.7
[37] rstatix_0.7.2 restfulr_0.0.16
[39] ps_1.9.1 rstudioapi_0.17.1
[41] UCSC.utils_1.2.0 miniUI_0.1.2
[43] generics_0.1.4 DOSE_4.0.1
[45] base64enc_0.1-3 processx_3.8.6
[47] curl_6.4.0 zlibbioc_1.52.0
[49] GenomeInfoDbData_1.2.13 SparseArray_1.6.2
[51] RBGL_1.82.0 xtable_1.8-4
[53] doParallel_1.0.17 evaluate_1.0.4
[55] S4Arrays_1.6.0 BiocFileCache_2.14.0
[57] hms_1.1.3 colorspace_2.1-1
[59] filelock_1.0.3 magrittr_2.0.3
[61] later_1.4.2 ggtree_3.14.0
[63] lattice_0.22-7 getPass_0.2-4
[65] XML_3.99-0.18 cowplot_1.1.3
[67] matrixStats_1.5.0 Hmisc_5.2-3
[69] pillar_1.11.0 nlme_3.1-168
[71] iterators_1.0.14 pwalign_1.2.0
[73] gridBase_0.4-7 caTools_1.18.3
[75] compiler_4.4.2 stringi_1.8.7
[77] SummarizedExperiment_1.36.0 GenomicAlignments_1.42.0
[79] plyr_1.8.9 crayon_1.5.3
[81] abind_1.4-8 gridGraphics_0.5-1
[83] locfit_1.5-9.12 bit_4.6.0
[85] fastmatch_1.1-6 whisker_0.4.1
[87] codetools_0.2-20 textshaping_1.0.1
[89] bslib_0.9.0 GetoptLong_1.0.5
[91] multtest_2.62.0 mime_0.13
[93] splines_4.4.2 Rcpp_1.1.0
[95] dbplyr_2.5.0 cellranger_1.1.0
[97] utf8_1.2.6 knitr_1.50
[99] blob_1.2.4 clue_0.3-66
[101] AnnotationFilter_1.30.0 fs_1.6.6
[103] checkmate_2.3.2 pkgbuild_1.4.8
[105] ggplotify_0.1.2 Matrix_1.7-3
[107] callr_3.7.6 statmod_1.5.0
[109] tzdb_0.5.0 svglite_2.2.1
[111] pkgconfig_2.0.3 tools_4.4.2
[113] cachem_1.1.0 RSQLite_2.4.1
[115] viridisLite_0.4.2 DBI_1.2.3
[117] impute_1.80.0 fastmap_1.2.0
[119] rmarkdown_2.29 Rsamtools_2.22.0
[121] sass_0.4.10 patchwork_1.3.1
[123] BiocManager_1.30.26 VariantAnnotation_1.52.0
[125] graph_1.84.1 carData_3.0-5
[127] rpart_4.1.24 farver_2.1.2
[129] yaml_2.3.10 MatrixGenerics_1.18.1
[131] foreign_0.8-90 cli_3.6.5
[133] txdbmaker_1.2.1 lifecycle_1.0.4
[135] lambda.r_1.2.4 sessioninfo_1.2.3
[137] backports_1.5.0 timechange_0.3.0
[139] gtable_0.3.6 rjson_0.2.23
[141] parallel_4.4.2 ape_5.8-1
[143] jsonlite_2.0.0 bitops_1.0-9
[145] bit64_4.6.0-1 pwr_1.3-0
[147] yulab.utils_0.2.0 futile.options_1.0.1
[149] jquerylib_0.1.4 GOSemSim_2.32.0
[151] R.utils_2.13.0 snpStats_1.56.0
[153] lazyeval_0.2.2 shiny_1.11.1
[155] htmltools_0.5.8.1 enrichplot_1.26.6
[157] rappdirs_0.3.3 formatR_1.14
[159] ensembldb_2.30.0 glue_1.8.0
[161] httr2_1.1.2 RCurl_1.98-1.17
[163] InteractionSet_1.34.0 rprojroot_2.0.4
[165] treeio_1.30.0 boot_1.3-31
[167] universalmotif_1.24.2 igraph_2.1.4
[169] R6_2.6.1 gplots_3.2.0
[171] labeling_0.4.3 cluster_2.1.8.1
[173] pkgload_1.4.0 regioneR_1.38.0
[175] aplot_0.2.8 DelayedArray_0.32.0
[177] tidyselect_1.2.1 plotrix_3.8-4
[179] ProtGenerics_1.38.0 htmlTable_2.4.3
[181] xml2_1.3.8 car_3.1-3
[183] seqPattern_1.38.0 KernSmooth_2.23-26
[185] data.table_1.17.6 htmlwidgets_1.6.4
[187] fgsea_1.32.4 rlang_1.1.6
[189] remotes_2.5.0 Cairo_1.6-2