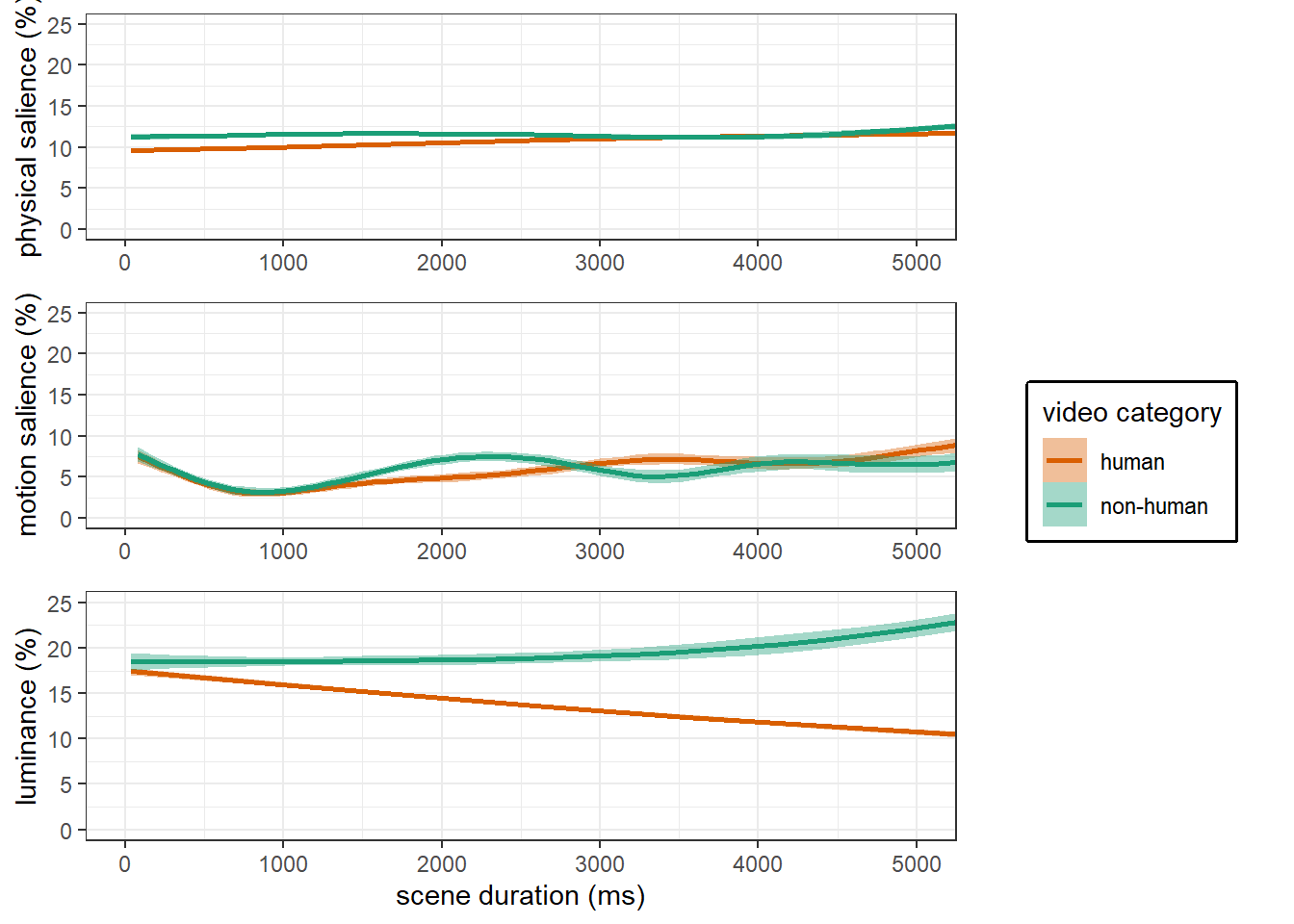
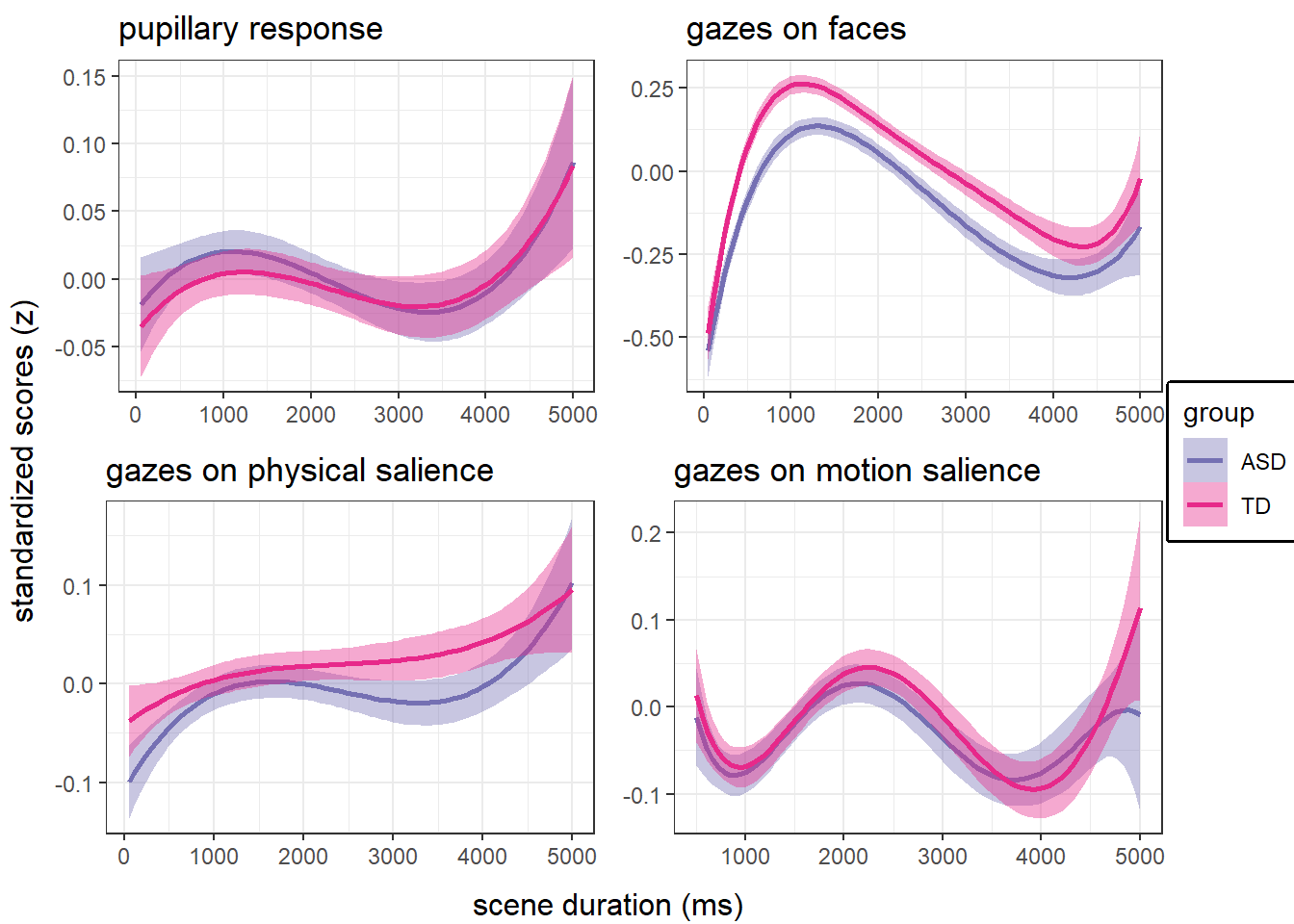
main analyses
sensory salience
2.5 % 97.5 %
vid_socialTRUE -0.32 -0.46 -0.18 2.5 % 97.5 %
rpd_z -0.03 -0.05 -0.01 contrast estimate SE df asymp.LCL asymp.UCL
FALSE - TRUE 0.303 0.0706 Inf 0.164 0.441
Results are averaged over the levels of: t1_diagnosis, t1_sex
Note: contrasts are still on the scale scale
Degrees-of-freedom method: asymptotic
Confidence level used: 0.95 vid_social rpd_z.trend SE df asymp.LCL asymp.UCL
FALSE -0.0368 0.00818 Inf -0.0529 -0.02081
TRUE -0.0115 0.00929 Inf -0.0298 0.00667
Results are averaged over the levels of: t1_diagnosis, t1_sex
Degrees-of-freedom method: asymptotic
Confidence level used: 0.95 | Sum Sq | df1 | df2 | F | p | p_adj | |
|---|---|---|---|---|---|---|
| group | 2.452 | 1 | 255.985 | 2.892 | 0.090 | 0.270 |
| pupillary response (PR) | 19.264 | 1 | 50077.514 | 22.718 | 0.000 | 0.000 |
| video category (VC) | 16.627 | 1 | 74.058 | 19.609 | 0.000 | 0.000 |
| time^1 | 6.380 | 1 | 60383.221 | 7.524 | 0.006 | 0.018 |
| time^2 | 3.466 | 1 | 60402.842 | 4.088 | 0.043 | 0.129 |
| time^3 | 2.010 | 1 | 60409.761 | 2.370 | 0.124 | 0.372 |
| sex | 0.034 | 1 | 233.071 | 0.040 | 0.842 | 1.000 |
| age | 0.633 | 1 | 260.651 | 0.746 | 0.388 | 1.000 |
| perceptual IQ | 0.858 | 1 | 255.764 | 1.012 | 0.315 | 0.945 |
| ADHD inattention | 0.729 | 1 | 260.368 | 0.859 | 0.355 | 1.000 |
| anxiety | 1.503 | 1 | 200.375 | 1.772 | 0.185 | 0.555 |
| depression | 0.572 | 1 | 214.709 | 0.674 | 0.412 | 1.000 |
| accuracy | 0.631 | 1 | 290.413 | 0.745 | 0.389 | 1.000 |
| precision | 0.107 | 1 | 355.764 | 0.127 | 0.722 | 1.000 |
| center deviation | 601.857 | 1 | 55270.456 | 709.762 | 0.000 | 0.000 |
| luminance | 6.081 | 1 | 5226.350 | 7.171 | 0.007 | 0.021 |
| group x PR | 1.422 | 1 | 52203.539 | 1.676 | 0.195 | 0.585 |
| group x VC | 0.863 | 1 | 45266.169 | 1.017 | 0.313 | 0.939 |
| PR x VC | 7.781 | 1 | 23092.014 | 9.177 | 0.002 | 0.006 |
| group x time^1 | 0.045 | 1 | 60332.853 | 0.053 | 0.818 | 1.000 |
| group x time^2 | 0.020 | 1 | 60344.828 | 0.024 | 0.878 | 1.000 |
| group x time^3 | 0.012 | 1 | 60347.178 | 0.015 | 0.904 | 1.000 |
| PR x time^1 | 0.089 | 1 | 60361.277 | 0.104 | 0.747 | 1.000 |
| PR x time^2 | 0.001 | 1 | 60384.879 | 0.001 | 0.979 | 1.000 |
| PR x time^3 | 0.018 | 1 | 60391.595 | 0.021 | 0.883 | 1.000 |
| VC x time^1 | 0.705 | 1 | 60396.575 | 0.831 | 0.362 | 1.000 |
| VC x time^2 | 2.031 | 1 | 60412.980 | 2.395 | 0.122 | 0.366 |
| VC x time^3 | 2.045 | 1 | 60422.044 | 2.412 | 0.120 | 0.360 |
| group x PR x VC | 0.025 | 1 | 25032.049 | 0.030 | 0.863 | 1.000 |
| group x PR x time^1 | 0.801 | 1 | 60355.653 | 0.944 | 0.331 | 0.993 |
| group x PR x time^2 | 0.335 | 1 | 60378.604 | 0.395 | 0.530 | 1.000 |
| group x PR x time^3 | 0.100 | 1 | 60383.294 | 0.118 | 0.731 | 1.000 |
| group x VC x time^1 | 0.061 | 1 | 60349.925 | 0.072 | 0.789 | 1.000 |
| group x VC x time^2 | 0.026 | 1 | 60363.250 | 0.031 | 0.860 | 1.000 |
| group x VC x time^3 | 0.013 | 1 | 60362.491 | 0.016 | 0.900 | 1.000 |
| PR x VC x time^1 | 0.758 | 1 | 60368.465 | 0.893 | 0.345 | 1.000 |
| PR x VC x time^2 | 0.625 | 1 | 60384.248 | 0.737 | 0.391 | 1.000 |
| PR x VC x time^3 | 0.881 | 1 | 60385.331 | 1.039 | 0.308 | 0.924 |
| group x PR x VC x time^1 | 1.980 | 1 | 60352.732 | 2.335 | 0.127 | 0.381 |
| group x PR x VC x time^2 | 2.143 | 1 | 60369.720 | 2.527 | 0.112 | 0.336 |
| group x PR x VC x time^3 | 2.078 | 1 | 60372.326 | 2.450 | 0.117 | 0.351 |
| Sum Sq | df1 | df2 | F | p | p_adj | |
|---|---|---|---|---|---|---|
| group | 0.011 | 1 | 266.263 | 0.013 | 0.910 | 1.000 |
| pupillary response (PR) | 0.521 | 1 | 37749.348 | 0.576 | 0.448 | 1.000 |
| video category (VC) | 3.307 | 1 | 77.178 | 3.659 | 0.059 | 0.177 |
| time^1 | 162.528 | 1 | 60460.016 | 179.856 | 0.000 | 0.000 |
| time^2 | 156.434 | 1 | 60472.791 | 173.112 | 0.000 | 0.000 |
| time^3 | 135.227 | 1 | 60477.847 | 149.644 | 0.000 | 0.000 |
| sex | 0.007 | 1 | 224.167 | 0.008 | 0.928 | 1.000 |
| age | 0.277 | 1 | 257.570 | 0.307 | 0.580 | 1.000 |
| perceptual IQ | 0.153 | 1 | 237.975 | 0.169 | 0.681 | 1.000 |
| ADHD inattention | 0.534 | 1 | 268.147 | 0.591 | 0.443 | 1.000 |
| anxiety | 0.021 | 1 | 183.443 | 0.023 | 0.878 | 1.000 |
| depression | 0.049 | 1 | 208.174 | 0.054 | 0.817 | 1.000 |
| accuracy | 0.789 | 1 | 347.071 | 0.873 | 0.351 | 1.000 |
| precision | 1.227 | 1 | 457.477 | 1.358 | 0.245 | 0.735 |
| center deviation | 72.929 | 1 | 45083.562 | 80.704 | 0.000 | 0.000 |
| luminance | 2.870 | 1 | 8189.613 | 3.176 | 0.075 | 0.225 |
| group x PR | 1.570 | 1 | 40994.681 | 1.738 | 0.187 | 0.561 |
| group x VC | 0.465 | 1 | 38807.474 | 0.514 | 0.473 | 1.000 |
| PR x VC | 0.116 | 1 | 10454.605 | 0.128 | 0.721 | 1.000 |
| group x time^1 | 1.110 | 1 | 60420.469 | 1.228 | 0.268 | 0.804 |
| group x time^2 | 0.505 | 1 | 60426.347 | 0.559 | 0.455 | 1.000 |
| group x time^3 | 0.267 | 1 | 60426.832 | 0.296 | 0.587 | 1.000 |
| PR x time^1 | 0.490 | 1 | 60435.585 | 0.542 | 0.462 | 1.000 |
| PR x time^2 | 1.334 | 1 | 60443.048 | 1.476 | 0.224 | 0.672 |
| PR x time^3 | 1.879 | 1 | 60443.185 | 2.079 | 0.149 | 0.447 |
| VC x time^1 | 3.570 | 1 | 60466.539 | 3.950 | 0.047 | 0.141 |
| VC x time^2 | 5.010 | 1 | 60475.764 | 5.544 | 0.019 | 0.057 |
| VC x time^3 | 7.066 | 1 | 60481.539 | 7.820 | 0.005 | 0.015 |
| group x PR x VC | 0.046 | 1 | 12426.176 | 0.051 | 0.821 | 1.000 |
| group x PR x time^1 | 0.058 | 1 | 60430.323 | 0.064 | 0.800 | 1.000 |
| group x PR x time^2 | 0.012 | 1 | 60433.107 | 0.013 | 0.908 | 1.000 |
| group x PR x time^3 | 0.119 | 1 | 60432.031 | 0.131 | 0.717 | 1.000 |
| group x VC x time^1 | 0.195 | 1 | 60426.810 | 0.215 | 0.643 | 1.000 |
| group x VC x time^2 | 0.401 | 1 | 60431.023 | 0.444 | 0.505 | 1.000 |
| group x VC x time^3 | 0.657 | 1 | 60430.932 | 0.727 | 0.394 | 1.000 |
| PR x VC x time^1 | 21.441 | 1 | 60439.836 | 23.726 | 0.000 | 0.000 |
| PR x VC x time^2 | 28.245 | 1 | 60443.100 | 31.256 | 0.000 | 0.000 |
| PR x VC x time^3 | 30.210 | 1 | 60442.944 | 33.431 | 0.000 | 0.000 |
| group x PR x VC x time^1 | 0.116 | 1 | 60430.735 | 0.128 | 0.720 | 1.000 |
| group x PR x VC x time^2 | 0.002 | 1 | 60433.282 | 0.003 | 0.958 | 1.000 |
| group x PR x VC x time^3 | 0.026 | 1 | 60432.961 | 0.029 | 0.865 | 1.000 |
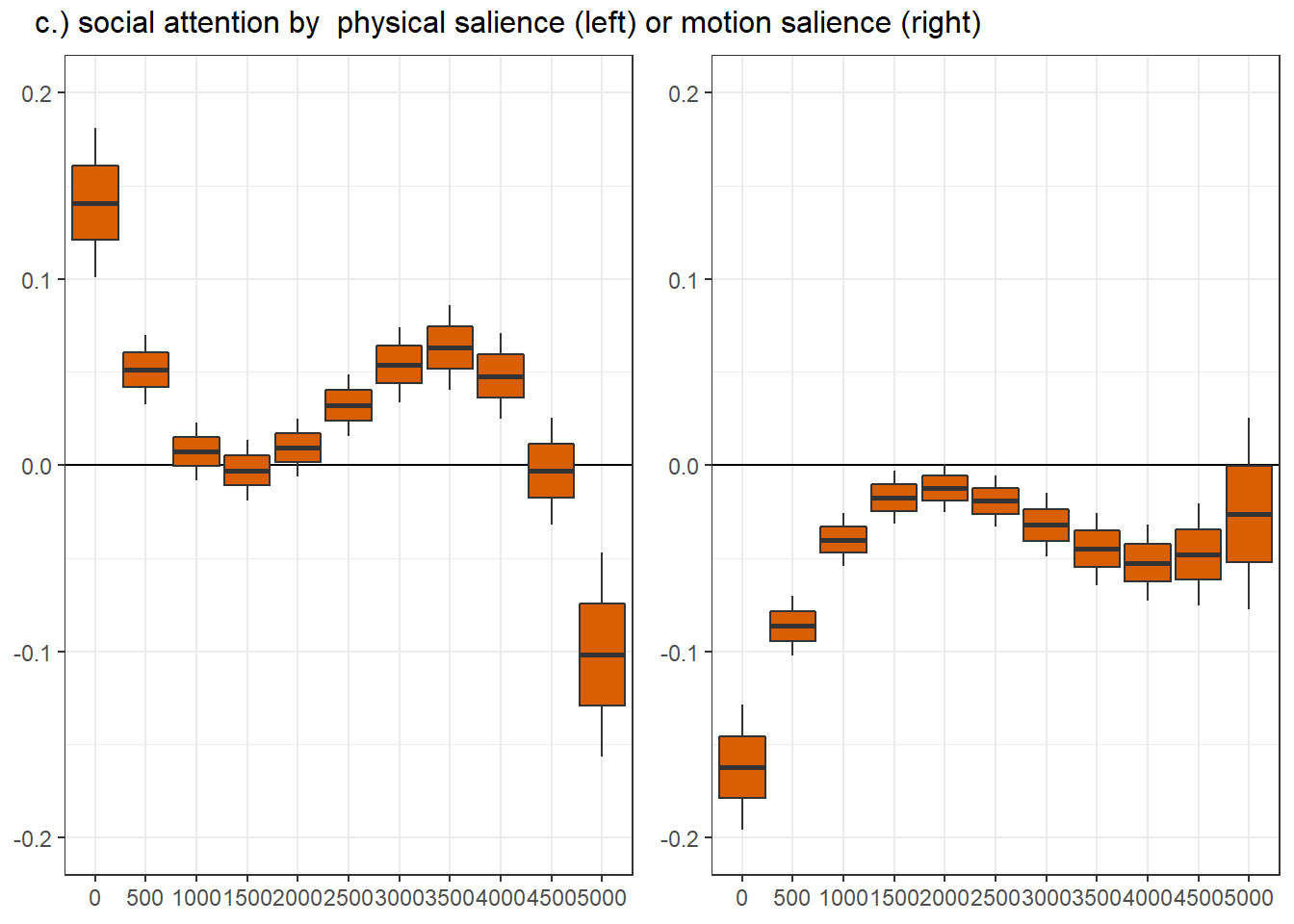
vid_social ts.scene rpd_z.trend SE df asymp.LCL asymp.UCL
FALSE 1000 -0.0376 0.00849 Inf -0.05427 -0.020979
TRUE 1000 0.0154 0.00962 Inf -0.00342 0.034283
FALSE 3000 0.0455 0.01160 Inf 0.02281 0.068263
TRUE 3000 -0.0237 0.01189 Inf -0.04700 -0.000379
Results are averaged over the levels of: t1_diagnosis, t1_sex
Degrees-of-freedom method: asymptotic
Confidence level used: 0.95 pupillary response
| Sum Sq | df1 | df2 | F | p | p_adj | |
|---|---|---|---|---|---|---|
| group | 0.002 | 1 | 301.764 | 0.003 | 0.954 | 1.000 |
| video category (VC) | 15.431 | 1 | 78.138 | 21.252 | 0.000 | 0.000 |
| time^1 | 1.281 | 1 | 60190.477 | 1.765 | 0.184 | 0.552 |
| time^2 | 1.222 | 1 | 60195.699 | 1.683 | 0.195 | 0.585 |
| time^3 | 0.913 | 1 | 60198.634 | 1.257 | 0.262 | 0.786 |
| sex | 0.416 | 1 | 303.843 | 0.573 | 0.449 | 1.000 |
| age | 0.250 | 1 | 314.546 | 0.344 | 0.558 | 1.000 |
| perceptual IQ | 1.132 | 1 | 313.407 | 1.559 | 0.213 | 0.639 |
| ADHD inattention | 0.160 | 1 | 309.328 | 0.220 | 0.639 | 1.000 |
| anxiety | 2.931 | 1 | 294.514 | 4.037 | 0.045 | 0.135 |
| depression | 0.978 | 1 | 297.002 | 1.347 | 0.247 | 0.741 |
| accuracy | 0.000 | 1 | 312.085 | 0.000 | 0.984 | 1.000 |
| precision | 0.003 | 1 | 331.139 | 0.004 | 0.951 | 1.000 |
| center deviation | 333.439 | 1 | 60448.581 | 459.205 | 0.000 | 0.000 |
| luminance | 0.017 | 1 | 25590.455 | 0.023 | 0.880 | 1.000 |
| group x VC | 159.276 | 1 | 60337.877 | 219.352 | 0.000 | 0.000 |
| group x time^1 | 0.365 | 1 | 60169.957 | 0.503 | 0.478 | 1.000 |
| group x time^2 | 0.335 | 1 | 60173.710 | 0.461 | 0.497 | 1.000 |
| group x time^3 | 0.194 | 1 | 60174.954 | 0.268 | 0.605 | 1.000 |
| VC x time^1 | 1.192 | 1 | 60200.641 | 1.642 | 0.200 | 0.600 |
| VC x time^2 | 1.149 | 1 | 60204.672 | 1.582 | 0.208 | 0.624 |
| VC x time^3 | 1.641 | 1 | 60208.101 | 2.261 | 0.133 | 0.399 |
| group x VC x time^1 | 5.974 | 1 | 60180.107 | 8.228 | 0.004 | 0.012 |
| group x VC x time^2 | 5.904 | 1 | 60184.079 | 8.130 | 0.004 | 0.012 |
| group x VC x time^3 | 5.101 | 1 | 60183.608 | 7.025 | 0.008 | 0.024 |
2.5 % 97.5 %
vid_socialTRUE 0.4 0.19 0.62vid_social = FALSE:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control 0.0978 0.0342 Inf 0.0308 0.165
vid_social = TRUE:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control -0.0775 0.0339 Inf -0.1439 -0.011
Results are averaged over the levels of: t1_sex
Degrees-of-freedom method: asymptotic
Confidence level used: 0.95 vid_social = FALSE, ts.scene = 0:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control 0.1560 0.0475 Inf 0.0628 0.2491
vid_social = TRUE, ts.scene = 0:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control -0.1716 0.0476 Inf -0.2649 -0.0783
vid_social = FALSE, ts.scene = 1000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control 0.0975 0.0342 Inf 0.0305 0.1646
vid_social = TRUE, ts.scene = 1000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control -0.0744 0.0340 Inf -0.1410 -0.0077
vid_social = FALSE, ts.scene = 2000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control 0.1004 0.0341 Inf 0.0335 0.1673
vid_social = TRUE, ts.scene = 2000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control -0.0828 0.0338 Inf -0.1490 -0.0165
vid_social = FALSE, ts.scene = 3000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control 0.1218 0.0368 Inf 0.0498 0.1939
vid_social = TRUE, ts.scene = 3000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control -0.1334 0.0360 Inf -0.2039 -0.0628
vid_social = FALSE, ts.scene = 4000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control 0.1192 0.0389 Inf 0.0430 0.1953
vid_social = TRUE, ts.scene = 4000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control -0.1627 0.0381 Inf -0.2374 -0.0880
vid_social = FALSE, ts.scene = 5000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control 0.0497 0.0682 Inf -0.0841 0.1834
vid_social = TRUE, ts.scene = 5000:
contrast estimate SE df asymp.LCL asymp.UCL
ASD - Control -0.1074 0.0656 Inf -0.2359 0.0211
Results are averaged over the levels of: t1_sex
Degrees-of-freedom method: asymptotic
Confidence level used: 0.95 panel figure - temporal effects
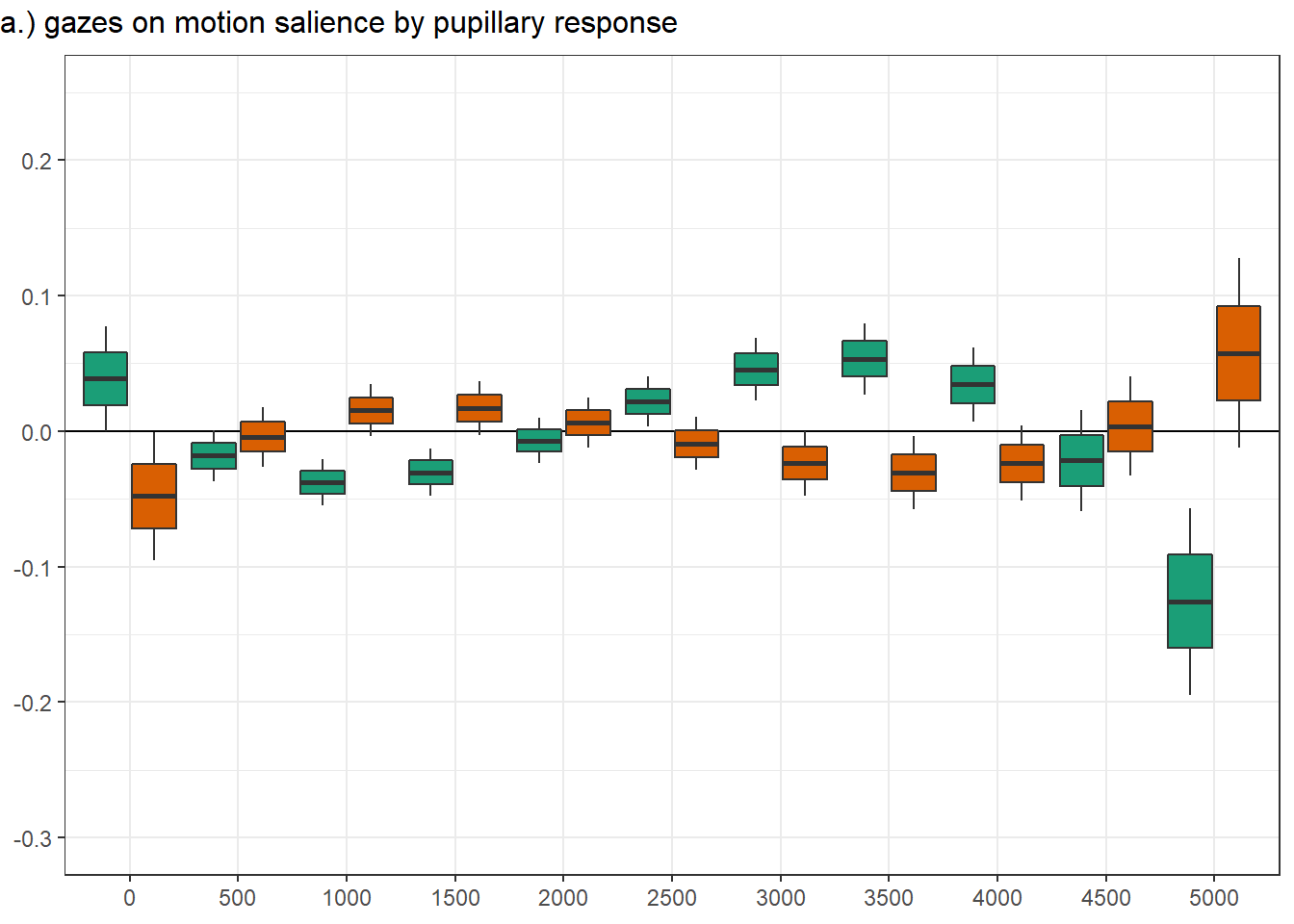
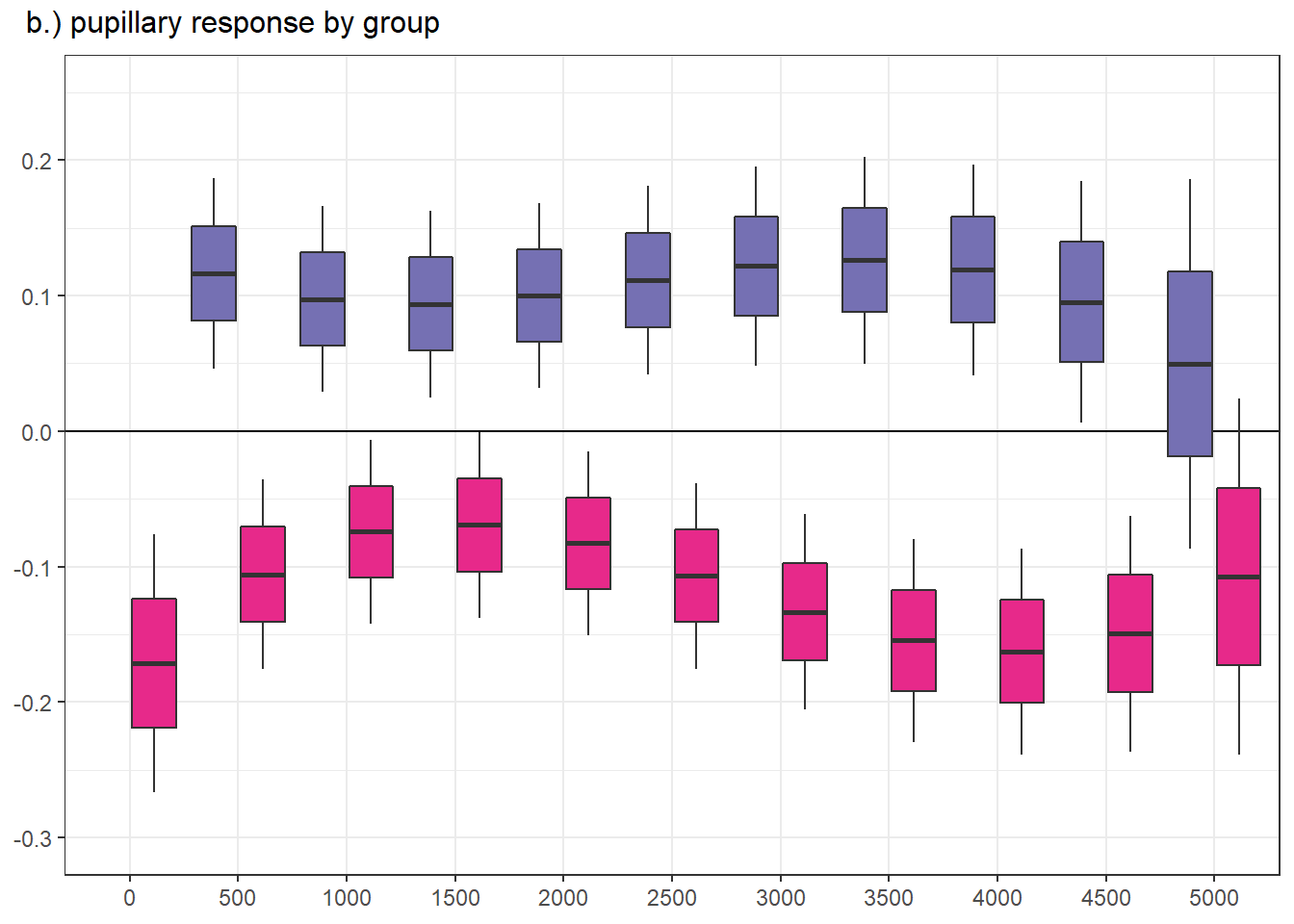
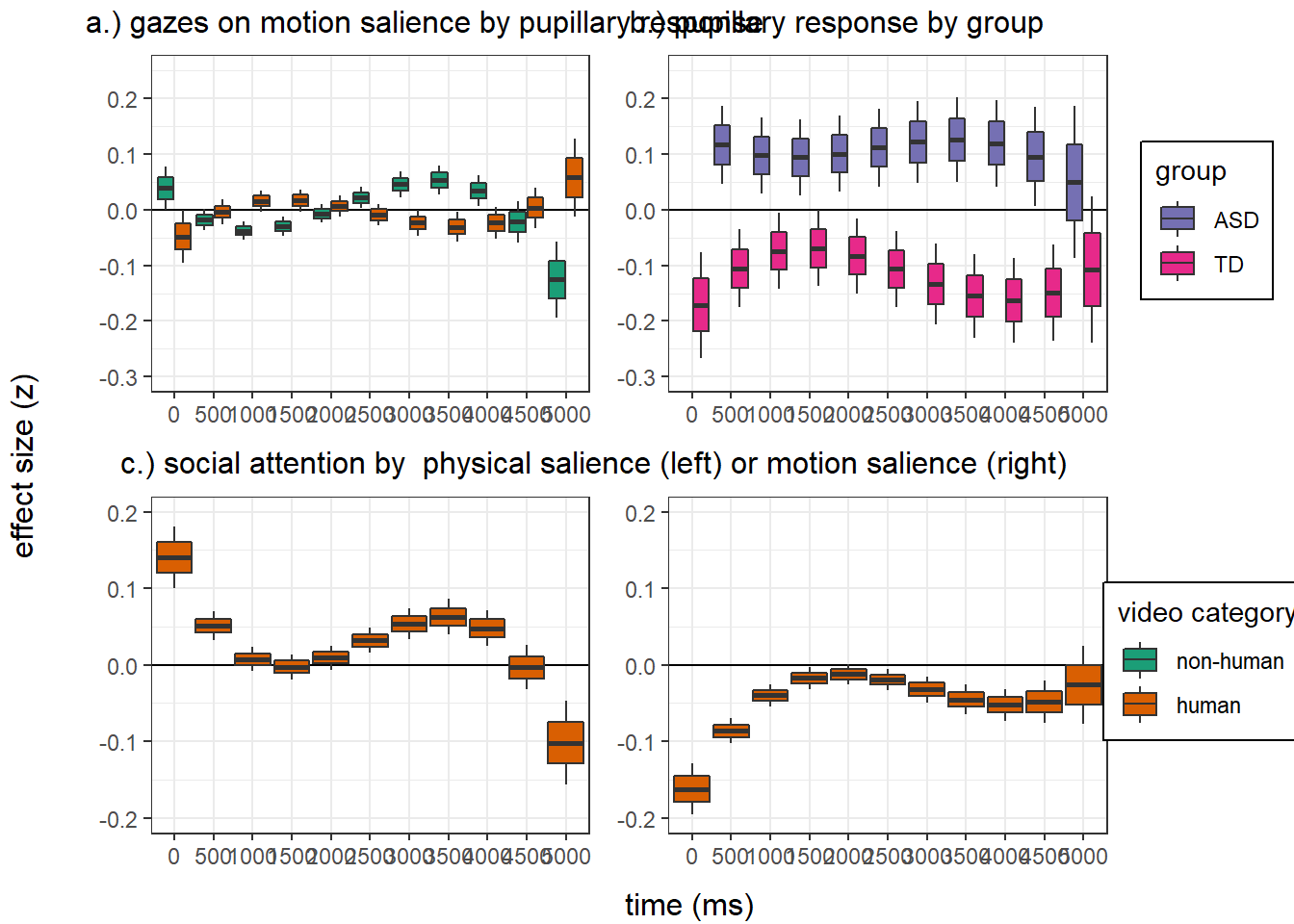
png
2
R version 3.6.1 (2019-07-05)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19043)
Matrix products: default
locale:
[1] LC_COLLATE=German_Germany.1252 LC_CTYPE=German_Germany.1252
[3] LC_MONETARY=German_Germany.1252 LC_NUMERIC=C
[5] LC_TIME=German_Germany.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] simr_1.0.5 missMDA_1.18 psych_1.9.12.31 MuMIn_1.43.17
[5] emmeans_1.4.3.01 lmerTest_3.1-2 lme4_1.1-23 Matrix_1.2-18
[9] kableExtra_1.3.1 gridExtra_2.3 sjmisc_2.8.5 sjPlot_2.8.5
[13] ggplot2_3.2.1 reshape2_1.4.3 R.matlab_3.6.2 MatchIt_3.0.2
[17] Gmisc_1.11.0 htmlTable_2.1.0 Rcpp_1.0.3 mice_3.7.0
[21] lattice_0.20-38 zoo_1.8-6 readxl_1.3.1
loaded via a namespace (and not attached):
[1] backports_1.1.5 Hmisc_4.3-0 workflowr_1.6.2
[4] plyr_1.8.5 lazyeval_0.2.2 splines_3.6.1
[7] RLRsim_3.1-3 digest_0.6.23 foreach_1.5.1
[10] htmltools_0.4.0 magrittr_1.5 checkmate_1.9.4
[13] cluster_2.1.0 doParallel_1.0.16 openxlsx_4.1.4
[16] modelr_0.1.5 R.utils_2.9.2 jpeg_0.1-8.1
[19] colorspace_1.4-1 rvest_0.3.5 ggrepel_0.8.1
[22] haven_2.2.0 pan_1.6 xfun_0.11
[25] dplyr_1.0.2 crayon_1.3.4 survival_3.1-8
[28] iterators_1.0.12 glue_1.4.2 gtable_0.3.0
[31] webshot_0.5.2 sjstats_0.18.0 car_3.0-6
[34] jomo_2.6-10 abind_1.4-5 scales_1.1.0
[37] mvtnorm_1.0-11 ggeffects_0.16.0 plotrix_3.7-7
[40] viridisLite_0.3.0 xtable_1.8-4 performance_0.5.0
[43] flashClust_1.01-2 foreign_0.8-74 Formula_1.2-3
[46] stats4_3.6.1 DT_0.11 htmlwidgets_1.5.1
[49] httr_1.4.1 RColorBrewer_1.1-2 acepack_1.4.1
[52] ellipsis_0.3.0 pkgconfig_2.0.3 XML_3.99-0.3
[55] R.methodsS3_1.7.1 binom_1.1-1 nnet_7.3-12
[58] tidyselect_1.1.0 rlang_0.4.8 later_1.0.0
[61] effectsize_0.0.1 munsell_0.5.0 cellranger_1.1.0
[64] tools_3.6.1 generics_0.0.2 sjlabelled_1.1.6
[67] broom_0.7.2 evaluate_0.14 stringr_1.4.0
[70] yaml_2.2.0 knitr_1.26 fs_1.3.1
[73] zip_2.0.4 forestplot_1.10 purrr_0.3.3
[76] mitml_0.3-7 nlme_3.1-143 R.oo_1.23.0
[79] leaps_3.1 xml2_1.2.2 pbkrtest_0.4-7
[82] compiler_3.6.1 rstudioapi_0.10 curl_4.3
[85] png_0.1-7 tibble_3.0.4 statmod_1.4.34
[88] stringi_1.4.5 highr_0.8 parameters_0.8.5
[91] forcats_0.4.0 nloptr_1.2.1 vctrs_0.3.4
[94] pillar_1.4.3 lifecycle_0.2.0 estimability_1.3
[97] data.table_1.12.8 insight_0.9.6 httpuv_1.5.2
[100] R6_2.4.1 latticeExtra_0.6-29 promises_1.1.0
[103] rio_0.5.16 codetools_0.2-16 boot_1.3-24
[106] MASS_7.3-51.5 rprojroot_1.3-2 withr_2.1.2
[109] mnormt_1.5-5 hms_0.5.3 mgcv_1.8-31
[112] bayestestR_0.7.2 parallel_3.6.1 grid_3.6.1
[115] rpart_4.1-15 tidyr_1.0.0 coda_0.19-4
[118] minqa_1.2.4 rmarkdown_2.0 carData_3.0-3
[121] git2r_0.26.1 numDeriv_2016.8-1.1 scatterplot3d_0.3-41
[124] lubridate_1.7.9 base64enc_0.1-3 FactoMineR_2.4