Enrichment Overview
Last updated: 2024-01-30
Checks: 7 0
Knit directory: dgrp-starve/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221101) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bc296df. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .snakemake/
Ignored: code/methodComp/bglr/err-bglr-f.5381.err
Ignored: code/methodComp/bglr/err-bglr-m.5382.err
Ignored: code/methodComp/m/meth-m.4676.err
Ignored: code/methodComp/m/meth-m.4685.err
Ignored: code/methodComp/method-f.4751.out
Ignored: data/fb/
Ignored: data/snake/
Ignored: snake/.snakemake/
Ignored: snake/GOfile.yaml
Ignored: snake/ReadMe.md
Ignored: snake/code/bayesC/
Ignored: snake/code/misc/
Ignored: snake/customL.Rds
Ignored: snake/data/
Ignored: snake/datafile.yaml
Ignored: snake/dgrp.yaml
Ignored: snake/fig/
Ignored: snake/fullGO.yaml
Ignored: snake/gofig.yaml
Ignored: snake/guide/
Ignored: snake/logs/
Ignored: snake/pbs_smake.pbs
Ignored: snake/pbsmake.sbatch
Ignored: snake/slurm/
Ignored: snake/smake.sbatch
Ignored: snake/snubnose.sbatch
Ignored: snake/zz_lost/
Ignored: zz_lost/
Untracked files:
Untracked: analysis/allotter.R
Untracked: analysis/old_index.Rmd.Rmd
Untracked: analysis/sparseComp.Rmd
Untracked: forester.R
Untracked: malegofind.R
Untracked: moarnotes.R
Untracked: pippinRMDbackup.R
Untracked: snake/code/binner.R
Untracked: snake/code/combine_GO.R
Untracked: snake/code/dataFinGO.R
Untracked: snake/code/datafile.yaml
Untracked: snake/code/filterNcombine_GO.R
Untracked: snake/code/filter_GO.R
Untracked: snake/code/go/
Untracked: snake/code/idTEMP.R
Untracked: snake/code/imstuff.R
Untracked: snake/code/method/bayesHome.R
Untracked: snake/code/method/multiplotGO.Rmd
Untracked: snake/code/scheming.R
Untracked: snake/code/scripts/
Untracked: snake/code/srfile.yaml
Untracked: teno.R
Unstaged changes:
Modified: analysis/Method/BayesC.Rmd
Modified: analysis/bigGO.Rmd
Modified: snake/code/method/bayesGO.R
Modified: snake/code/method/goFish.R
Modified: snake/code/method/varbvs.R
Modified: snake/temp.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/enrich.Rmd) and HTML (docs/enrich.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bc296df | nklimko | 2024-01-30 | wflow_publish(“analysis/enrich.Rmd”) |
| html | a9c9555 | nklimko | 2024-01-30 | Build site. |
| Rmd | e3dd8b2 | nklimko | 2024-01-30 | wflow_publish(“analysis/enrich.Rmd”) |
| html | 65745c3 | nklimko | 2024-01-30 | Build site. |
| Rmd | d373edf | nklimko | 2024-01-30 | wflow_publish(“analysis/enrich.Rmd”) |
| html | 8e39493 | nklimko | 2024-01-23 | Build site. |
| Rmd | 2683f74 | nklimko | 2024-01-23 | wflow_publish(“analysis/enrich.Rmd”) |
| html | 991d04c | nklimko | 2024-01-23 | Build site. |
| Rmd | 7c20291 | nklimko | 2024-01-23 | wflow_publish(“analysis/enrich.Rmd”) |
| html | 02fdd7e | nklimko | 2024-01-23 | Build site. |
| Rmd | c5835d5 | nklimko | 2024-01-23 | wflow_publish(“analysis/enrich.Rmd”) |
| html | 8f35a08 | nklimko | 2024-01-23 | Build site. |
| Rmd | dd3c95c | nklimko | 2024-01-23 | wflow_publish(“analysis/enrich.Rmd”) |
Dive back in:
We left off looking to modify the selection cutoff for top terms. While various ideas were floated(standard devs, percentiles, flat cutoff), there was not a consistently most useful method for looking at both male and female data. As such, I opted to run a series of flat cutoffs(100, 50, 25) for both complete methods per sex.
I first looked at the top terms ordered to look for patterns in the data. While I’m unsure of the significance, the clustering of correlations for top terms shares a shape within sexes across methods. Notably, female data had a sharper increase(negative slope) for top results
load('snake/data/go/50_tables/enrichment.Rdata')
topGrid <- function(data, sex, psize, custom.title, custom.Xlab, custom.Ylab){
plothole <- ggplot(data, aes(x=index, y=cor, label=term))+
geom_point(color=viridis(1, begin=0.5), size=psize)+
theme_minimal() +
labs(x=custom.Xlab, y=custom.Ylab, tag=sex, title=custom.title) +
theme(text=element_text(size=10), plot.tag = element_text(size=15))
return(plothole)
}
gg[[1]] <- topGrid(blupF, 'F', 1, 'Top Female Results: TBLUP', 'Rank', 'Correlation')
gg[[2]] <- topGrid(blupM, 'M', 1, 'Top Male Results: TBLUP', 'Rank', 'Correlation')
gg[[3]] <- topGrid(bayesF, 'F', 1, 'Top Female Results: BayesC', 'Rank', 'Correlation')
gg[[4]] <- topGrid(bayesM, 'M', 1, 'Top Male Results: BayesC', 'Rank', 'Correlation')
plot_grid(gg[[1]], gg[[2]], gg[[3]], gg[[4]], ncol=2)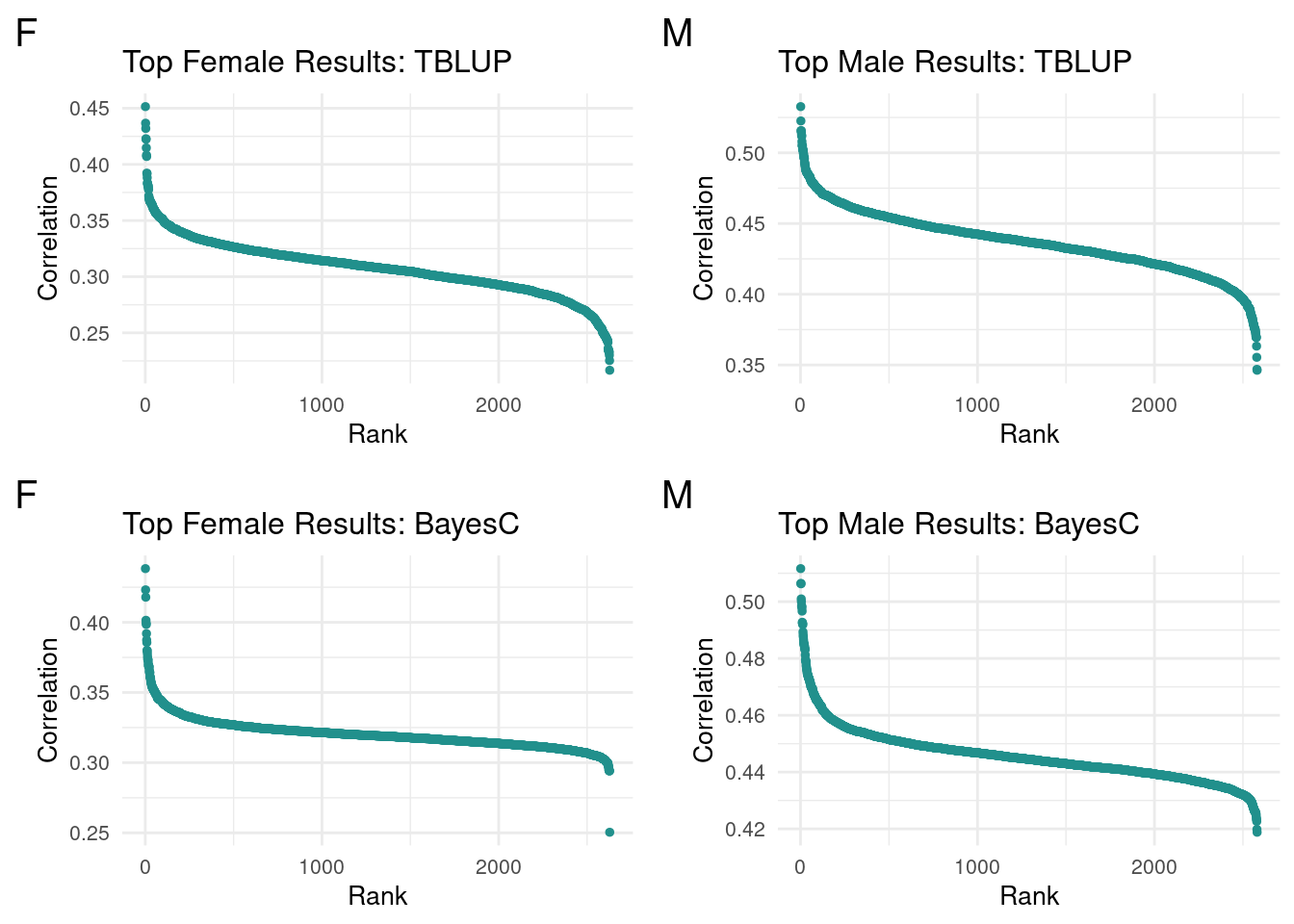
| Version | Author | Date |
|---|---|---|
| 8f35a08 | nklimko | 2024-01-23 |
After this, I found the correlation between the two methods to see how similar generated results are.
print(cor(blupF$cor, bayesF$cor))[1] 0.9490421Female Overall
print(cor(blupF[1:200,cor], bayesF[1:200, cor]))[1] 0.9937145Female Top 200
print(cor(blupM$cor, bayesM$cor))[1] 0.9598015Male Overall
print(cor(blupM[1:200,cor], bayesM[1:200, cor]))[1] 0.9967979Male Top 100
Moving past this, I wanted to assess the effect of term count on enrichment——000000——000000——000000——000000——000000
load("snake/data/go/50_tables/enrich/kables.Rdata")
percentModder <- function(dataKable){
dataKable[,5] <- dataKable[,5]*2
dataKable[,8] <- dataKable[,8]*2
colnames(dataKable) <- rep(c('Flybase Gene', 'Percent', 'Gene'), 3)
return(dataKable)
}
bayesF_KableMod <- percentModder(bayesF_Kable)
bayesM_KableMod <- percentModder(bayesM_Kable)
blupF_KableMod <- percentModder(blupF_Kable )
blupM_KableMod <- percentModder(blupM_Kable )
kable(bayesF_KableMod, caption="Female BayesC", "simple") %>%
add_header_above(c('Top 100 GO Terms' = 3, 'Top 50 GO Terms' = 3, 'Top 25 GO Terms' = 3))Warning in add_header_above(., c(`Top 100 GO Terms` = 3, `Top 50 GO Terms` = 3,
: Please specify format in kable. kableExtra can customize either HTML or LaTeX
outputs. See https://haozhu233.github.io/kableExtra/ for details.| Flybase Gene | Percent | Gene | Flybase Gene | Percent | Gene | Flybase Gene | Percent | Gene |
|---|---|---|---|---|---|---|---|---|
| FBgn0262738 | 15 | norpA | FBgn0025595 | 18 | AkhR | FBgn0025595 | 16 | AkhR |
| FBgn0003731 | 13 | Egfr | FBgn0003731 | 18 | Egfr | FBgn0000575 | 12 | emc |
| FBgn0004635 | 13 | rho | FBgn0003205 | 18 | Ras85D | FBgn0004552 | 8 | Akh |
| FBgn0003205 | 10 | Ras85D | FBgn0004635 | 18 | rho | FBgn0283499 | 8 | InR |
| FBgn0025595 | 9 | AkhR | FBgn0000575 | 14 | emc | FBgn0000490 | 8 | dpp |
| FBgn0003310 | 8 | NULL | FBgn0262738 | 14 | norpA | FBgn0010303 | 6 | hep |
| FBgn0000575 | 7 | emc | FBgn0004552 | 10 | Akh | FBgn0015279 | 6 | Pi3K92E |
| FBgn0015795 | 7 | Rab7 | FBgn0015279 | 10 | Pi3K92E | FBgn0033799 | 6 | GLaz |
| FBgn0283499 | 6 | InR | FBgn0283499 | 10 | InR | FBgn0036449 | 6 | bmm |
| FBgn0039114 | 6 | Lsd-1 | FBgn0003310 | 10 | NULL | FBgn0003731 | 6 | Egfr |
| FBgn0004552 | 5 | Akh | FBgn0035586 | 8 | CG10671 | FBgn0003205 | 6 | Ras85D |
| FBgn0015279 | 5 | Pi3K92E | FBgn0026252 | 8 | msk | FBgn0003463 | 6 | sog |
| FBgn0035586 | 5 | CG10671 | FBgn0000490 | 8 | dpp | FBgn0003719 | 6 | tld |
| FBgn0003463 | 5 | sog | FBgn0261648 | 8 | salm | FBgn0262738 | 6 | norpA |
| FBgn0003719 | 5 | tld | FBgn0010303 | 6 | hep | |||
| FBgn0036545 | 5 | GXIVsPLA2 | FBgn0036046 | 6 | Ilp2 | |||
| FBgn0039655 | 5 | CG14507 | FBgn0024248 | 6 | chico | |||
| FBgn0003118 | 5 | pnt | FBgn0033799 | 6 | GLaz | |||
| FBgn0005672 | 5 | spi | FBgn0036449 | 6 | bmm | |||
| FBgn0036046 | 4 | Ilp2 | FBgn0036260 | 6 | Rh7 | |||
| FBgn0003651 | 4 | svp | FBgn0003463 | 6 | sog | |||
| FBgn0010379 | 4 | Akt1 | FBgn0003719 | 6 | tld | |||
| FBgn0024248 | 4 | chico | FBgn0003984 | 6 | vn | |||
| FBgn0036449 | 4 | bmm | FBgn0038197 | 6 | foxo | |||
| FBgn0026252 | 4 | msk | FBgn0039114 | 6 | Lsd-1 | |||
| FBgn0036260 | 4 | Rh7 | FBgn0003218 | 6 | rdgB | |||
| FBgn0000490 | 4 | dpp | FBgn0026207 | 6 | mbo | |||
| FBgn0003984 | 4 | vn | FBgn0027537 | 6 | Nup93-1 | |||
| FBgn0261648 | 4 | salm | FBgn0031078 | 6 | Nup205 | |||
| FBgn0029720 | 4 | CG3009 | FBgn0033737 | 6 | Nup54 | |||
| FBgn0030013 | 4 | GIIIspla2 | FBgn0033766 | 6 | CG8771 | |||
| FBgn0033170 | 4 | sPLA2 | FBgn0038274 | 6 | Nup93-2 | |||
| FBgn0050503 | 4 | CG30503 | FBgn0061200 | 6 | Nup153 | |||
| FBgn0250862 | 4 | CG42237 | FBgn0003256 | 6 | rl | |||
| FBgn0003218 | 4 | rdgB | FBgn0034140 | 6 | Lst | |||
| FBgn0030608 | 4 | Lsd-2 | FBgn0015795 | 6 | Rab7 | |||
| FBgn0035206 | 4 | CG9186 | FBgn0002940 | 6 | ninaE | |||
| FBgn0040336 | 4 | Seipin | FBgn0003295 | 6 | ru | |||
| FBgn0026207 | 4 | mbo | ||||||
| FBgn0004435 | 4 | Galphaq | ||||||
| FBgn0000257 | 4 | car | ||||||
| FBgn0000482 | 4 | dor | ||||||
| FBgn0003256 | 4 | rl | ||||||
| FBgn0035871 | 4 | BI-1 | ||||||
| FBgn0052350 | 4 | Vps11 | ||||||
| FBgn0285911 | 4 | NULL | ||||||
| FBgn0015754 | 4 | Lis-1 | ||||||
| FBgn0004647 | 4 | NULL | ||||||
| FBgn0002940 | 4 | ninaE | ||||||
| FBgn0003295 | 4 | ru | ||||||
| FBgn0020386 | 4 | Pdk1 | ||||||
| FBgn0262451 | 4 | ban | ||||||
| FBgn0004784 | 4 | inaC | ||||||
| FBgn0003169 | 3 | put | ||||||
| FBgn0010303 | 3 | hep | ||||||
| FBgn0028717 | 3 | Lnk | ||||||
| FBgn0010051 | 3 | Itp-r83A | ||||||
| FBgn0030607 | 3 | dob | ||||||
| FBgn0033226 | 3 | CG1882 | ||||||
| FBgn0033799 | 3 | GLaz | ||||||
| FBgn0262103 | 3 | Sik3 | ||||||
| FBgn0002576 | 3 | lz | ||||||
| FBgn0004885 | 3 | tok | ||||||
| FBgn0050418 | 3 | nord | ||||||
| FBgn0039152 | 3 | Root | ||||||
| FBgn0038197 | 3 | foxo | ||||||
| FBgn0028741 | 3 | fab1 | ||||||
| FBgn0029870 | 3 | Marf | ||||||
| FBgn0052703 | 3 | Erk7 | ||||||
| FBgn0027537 | 3 | Nup93-1 | ||||||
| FBgn0031078 | 3 | Nup205 | ||||||
| FBgn0033737 | 3 | Nup54 | ||||||
| FBgn0033766 | 3 | CG8771 | ||||||
| FBgn0038274 | 3 | Nup93-2 | ||||||
| FBgn0061200 | 3 | Nup153 | ||||||
| FBgn0261549 | 3 | rdgA | ||||||
| FBgn0002566 | 3 | lt | ||||||
| FBgn0034140 | 3 | Lst | ||||||
| FBgn0038659 | 3 | EndoA | ||||||
| FBgn0086676 | 3 | spin | ||||||
| FBgn0015721 | 3 | ktub | ||||||
| FBgn0086687 | 3 | Desat1 | ||||||
| FBgn0014010 | 3 | Rab5 | ||||||
| FBgn0025680 | 3 | cry | ||||||
| FBgn0038167 | 3 | Lkb1 | ||||||
| FBgn0004611 | 3 | Plc21C | ||||||
| FBgn0001263 | 3 | inaD | ||||||
| FBgn0052699 | 3 | LPCAT | ||||||
| FBgn0265048 | 3 | cv-d |
kable(bayesM_KableMod, caption="Male BayesC", "simple") %>%
add_header_above(c('Top 100 GO Terms' = 3, 'Top 50 GO Terms' = 3, 'Top 25 GO Terms' = 3))Warning in add_header_above(., c(`Top 100 GO Terms` = 3, `Top 50 GO Terms` = 3,
: Please specify format in kable. kableExtra can customize either HTML or LaTeX
outputs. See https://haozhu233.github.io/kableExtra/ for details.| Flybase Gene | Percent | Gene | Flybase Gene | Percent | Gene | Flybase Gene | Percent | Gene |
|---|---|---|---|---|---|---|---|---|
| FBgn0265778 | 13 | PDZ-GEF | FBgn0261873 | 22 | sdt | FBgn0261873 | 16 | sdt |
| FBgn0261873 | 11 | sdt | FBgn0265778 | 20 | PDZ-GEF | FBgn0261854 | 12 | aPKC |
| FBgn0283499 | 10 | InR | FBgn0283499 | 16 | InR | FBgn0265778 | 10 | PDZ-GEF |
| FBgn0015279 | 9 | Pi3K92E | FBgn0261854 | 16 | aPKC | FBgn0025595 | 10 | AkhR |
| FBgn0036046 | 9 | Ilp2 | FBgn0025595 | 14 | AkhR | FBgn0036046 | 10 | Ilp2 |
| FBgn0261854 | 9 | aPKC | FBgn0036046 | 14 | Ilp2 | FBgn0283499 | 10 | InR |
| FBgn0011661 | 8 | Moe | FBgn0067864 | 14 | Patj | FBgn0067864 | 10 | Patj |
| FBgn0020386 | 8 | Pdk1 | FBgn0026192 | 14 | par-6 | FBgn0026192 | 10 | par-6 |
| FBgn0025595 | 7 | AkhR | FBgn0015279 | 12 | Pi3K92E | FBgn0259685 | 8 | crb |
| FBgn0003205 | 7 | Ras85D | FBgn0011661 | 12 | Moe | FBgn0263289 | 8 | scrib |
| FBgn0041191 | 7 | Rheb | FBgn0020386 | 12 | Pdk1 | FBgn0003391 | 8 | shg |
| FBgn0067864 | 7 | Patj | FBgn0024248 | 12 | chico | FBgn0086687 | 8 | Desat1 |
| FBgn0026192 | 7 | par-6 | FBgn0003205 | 10 | Ras85D | FBgn0036449 | 6 | bmm |
| FBgn0010379 | 7 | Akt1 | FBgn0041191 | 10 | Rheb | FBgn0004552 | 6 | Akh |
| FBgn0024248 | 7 | chico | FBgn0259685 | 10 | crb | FBgn0003205 | 6 | Ras85D |
| FBgn0263289 | 6 | scrib | FBgn0263289 | 10 | scrib | FBgn0015279 | 6 | Pi3K92E |
| FBgn0000163 | 6 | baz | FBgn0000163 | 10 | baz | FBgn0041191 | 6 | Rheb |
| FBgn0086687 | 6 | Desat1 | FBgn0010379 | 10 | Akt1 | FBgn0002121 | 6 | l(2)gl |
| FBgn0262656 | 6 | Myc | FBgn0262656 | 10 | Myc | FBgn0000163 | 6 | baz |
| FBgn0025741 | 6 | PlexA | FBgn0036449 | 8 | bmm | FBgn0024248 | 6 | chico |
| FBgn0014020 | 6 | Rho1 | FBgn0004552 | 8 | Akh | |||
| FBgn0023172 | 6 | RhoGEF2 | FBgn0003079 | 8 | Raf | |||
| FBgn0259685 | 5 | crb | FBgn0010269 | 8 | Dsor1 | |||
| FBgn0003391 | 5 | shg | FBgn0003391 | 8 | shg | |||
| FBgn0004636 | 5 | Rap1 | FBgn0086687 | 8 | Desat1 | |||
| FBgn0000547 | 5 | ed | FBgn0004636 | 8 | Rap1 | |||
| FBgn0003256 | 5 | rl | FBgn0004638 | 8 | drk | |||
| FBgn0051217 | 4 | modSP | FBgn0015402 | 8 | ksr | |||
| FBgn0036449 | 4 | bmm | FBgn0003256 | 8 | rl | |||
| FBgn0004552 | 4 | Akh | FBgn0051217 | 6 | modSP | |||
| FBgn0025800 | 4 | Smox | FBgn0033205 | 6 | CG2064 | |||
| FBgn0003079 | 4 | Raf | FBgn0030607 | 6 | dob | |||
| FBgn0003118 | 4 | pnt | FBgn0033226 | 6 | CG1882 | |||
| FBgn0010269 | 4 | Dsor1 | FBgn0028717 | 6 | Lnk | |||
| FBgn0028717 | 4 | Lnk | FBgn0001624 | 6 | dlg1 | |||
| FBgn0027108 | 4 | Inx2 | FBgn0002121 | 6 | l(2)gl | |||
| FBgn0266756 | 4 | btsz | FBgn0004167 | 6 | kst | |||
| FBgn0003514 | 4 | sqh | FBgn0000117 | 6 | arm | |||
| FBgn0004647 | 4 | NULL | FBgn0024846 | 6 | p38b | |||
| FBgn0001965 | 4 | Sos | FBgn0037874 | 6 | Tctp | |||
| FBgn0004638 | 4 | drk | FBgn0266756 | 6 | btsz | |||
| FBgn0015402 | 4 | ksr | FBgn0000206 | 6 | boss | |||
| FBgn0283472 | 4 | S6k | FBgn0038167 | 6 | Lkb1 | |||
| FBgn0003731 | 4 | Egfr | FBgn0024836 | 6 | stan | |||
| FBgn0026379 | 4 | Pten | FBgn0000547 | 6 | ed | |||
| FBgn0010333 | 4 | Rac1 | FBgn0003285 | 6 | rst | |||
| FBgn0010389 | 4 | htl | FBgn0034577 | 6 | cpa | |||
| FBgn0011259 | 4 | Sema1a | FBgn0262614 | 6 | pyd | |||
| FBgn0030926 | 3 | psh | FBgn0287864 | 6 | NULL | |||
| FBgn0030608 | 3 | Lsd-2 | FBgn0001965 | 6 | Sos | |||
| FBgn0033205 | 3 | CG2064 | FBgn0021796 | 6 | Tor | |||
| FBgn0035586 | 3 | CG10671 | FBgn0283472 | 6 | S6k | |||
| FBgn0040336 | 3 | Seipin | FBgn0023172 | 6 | RhoGEF2 | |||
| FBgn0030607 | 3 | dob | FBgn0262103 | 6 | Sik3 | |||
| FBgn0033226 | 3 | CG1882 | ||||||
| FBgn0011300 | 3 | babo | ||||||
| FBgn0024913 | 3 | Actbeta | ||||||
| FBgn0000382 | 3 | csw | ||||||
| FBgn0044048 | 3 | Ilp5 | ||||||
| FBgn0001624 | 3 | dlg1 | ||||||
| FBgn0002121 | 3 | l(2)gl | ||||||
| FBgn0004167 | 3 | kst | ||||||
| FBgn0000117 | 3 | arm | ||||||
| FBgn0027538 | 3 | beta4GalNAcTA | ||||||
| FBgn0039625 | 3 | beta4GalNAcTB | ||||||
| FBgn0005558 | 3 | ey | ||||||
| FBgn0024846 | 3 | p38b | ||||||
| FBgn0037874 | 3 | Tctp | ||||||
| FBgn0044047 | 3 | Ilp6 | ||||||
| FBgn0000206 | 3 | boss | ||||||
| FBgn0015721 | 3 | ktub | ||||||
| FBgn0032940 | 3 | Mondo | ||||||
| FBgn0038167 | 3 | Lkb1 | ||||||
| FBgn0020388 | 3 | Gcn5 | ||||||
| FBgn0028387 | 3 | chm | ||||||
| FBgn0037376 | 3 | Hat1 | ||||||
| FBgn0024836 | 3 | stan | ||||||
| FBgn0027509 | 3 | TBCD | ||||||
| FBgn0003285 | 3 | rst | ||||||
| FBgn0004143 | 3 | nullo | ||||||
| FBgn0034577 | 3 | cpa | ||||||
| FBgn0086384 | 3 | Mer | ||||||
| FBgn0259212 | 3 | cno | ||||||
| FBgn0262614 | 3 | pyd | ||||||
| FBgn0287864 | 3 | NULL | ||||||
| FBgn0021796 | 3 | Tor | ||||||
| FBgn0029840 | 3 | raptor | ||||||
| FBgn0085407 | 3 | Pvf3 | ||||||
| FBgn0266136 | 3 | Gyc76C | ||||||
| FBgn0010909 | 3 | msn | ||||||
| FBgn0004456 | 3 | mew | ||||||
| FBgn0262103 | 3 | Sik3 | ||||||
| FBgn0004956 | 3 | upd1 | ||||||
| FBgn0000721 | 3 | for | ||||||
| FBgn0261574 | 3 | kug | ||||||
| FBgn0003254 | 3 | rib | ||||||
| FBgn0004657 | 3 | mys | ||||||
| FBgn0025740 | 3 | PlexB | ||||||
| FBgn0004644 | 3 | hh | ||||||
| FBgn0051935 | 3 | CG31935 |
kable(blupF_KableMod, caption="Female TBLUP", "simple") %>%
add_header_above(c('Top 100 GO Terms' = 3, 'Top 50 GO Terms' = 3, 'Top 25 GO Terms' = 3))Warning in add_header_above(., c(`Top 100 GO Terms` = 3, `Top 50 GO Terms` = 3,
: Please specify format in kable. kableExtra can customize either HTML or LaTeX
outputs. See https://haozhu233.github.io/kableExtra/ for details.| Flybase Gene | Percent | Gene | Flybase Gene | Percent | Gene | Flybase Gene | Percent | Gene |
|---|---|---|---|---|---|---|---|---|
| FBgn0003731 | 14 | Egfr | FBgn0003731 | 22 | Egfr | FBgn0003731 | 14 | Egfr |
| FBgn0003205 | 11 | Ras85D | FBgn0283499 | 16 | InR | FBgn0010379 | 12 | Akt1 |
| FBgn0003256 | 11 | rl | FBgn0025595 | 14 | AkhR | FBgn0283499 | 12 | InR |
| FBgn0262738 | 11 | norpA | FBgn0000575 | 14 | emc | FBgn0262103 | 10 | Sik3 |
| FBgn0283499 | 10 | InR | FBgn0010303 | 14 | hep | FBgn0020386 | 10 | Pdk1 |
| FBgn0010379 | 9 | Akt1 | FBgn0010379 | 12 | Akt1 | FBgn0028484 | 10 | Ack |
| FBgn0025595 | 9 | AkhR | FBgn0004635 | 12 | rho | FBgn0003093 | 10 | Pkc98E |
| FBgn0000575 | 9 | emc | FBgn0262103 | 10 | Sik3 | FBgn0003256 | 10 | rl |
| FBgn0004635 | 9 | rho | FBgn0283472 | 10 | S6k | FBgn0052703 | 10 | Erk7 |
| FBgn0010303 | 9 | hep | FBgn0003205 | 10 | Ras85D | FBgn0025595 | 8 | AkhR |
| FBgn0003079 | 8 | Raf | FBgn0003169 | 10 | put | FBgn0035039 | 8 | Adck |
| FBgn0283472 | 7 | S6k | FBgn0011300 | 10 | babo | FBgn0261984 | 8 | Ire1 |
| FBgn0003169 | 7 | put | FBgn0260945 | 10 | Atg1 | FBgn0283472 | 8 | S6k |
| FBgn0011300 | 7 | babo | FBgn0020386 | 10 | Pdk1 | FBgn0000575 | 8 | emc |
| FBgn0260945 | 7 | Atg1 | FBgn0028484 | 10 | Ack | FBgn0004635 | 8 | rho |
| FBgn0004784 | 7 | inaC | FBgn0003093 | 10 | Pkc98E | FBgn0035142 | 8 | Hipk |
| FBgn0052703 | 7 | Erk7 | FBgn0003256 | 10 | rl | FBgn0003169 | 8 | put |
| FBgn0263395 | 7 | hppy | FBgn0003744 | 10 | trc | FBgn0011300 | 8 | babo |
| FBgn0000229 | 7 | bsk | FBgn0024846 | 10 | p38b | FBgn0260945 | 8 | Atg1 |
| FBgn0013759 | 7 | CASK | FBgn0025743 | 10 | mbt | FBgn0002413 | 8 | dco |
| FBgn0086657 | 7 | IKKepsilon | FBgn0034568 | 10 | CG3216 | FBgn0003091 | 8 | Pkc53E |
| FBgn0015279 | 6 | Pi3K92E | FBgn0051183 | 10 | CG31183 | FBgn0003502 | 8 | Btk29A |
| FBgn0261984 | 6 | Ire1 | FBgn0052703 | 10 | Erk7 | FBgn0003744 | 8 | trc |
| FBgn0262103 | 6 | Sik3 | FBgn0261456 | 10 | hpo | FBgn0004784 | 8 | inaC |
| FBgn0002413 | 6 | dco | FBgn0262738 | 10 | norpA | FBgn0004864 | 8 | hop |
| FBgn0020386 | 6 | Pdk1 | FBgn0263395 | 10 | hppy | FBgn0010197 | 8 | Gyc32E |
| FBgn0021796 | 6 | Tor | FBgn0010389 | 10 | htl | FBgn0010441 | 8 | pll |
| FBgn0003093 | 6 | Pkc98E | FBgn0035039 | 8 | Adck | FBgn0011817 | 8 | nmo |
| FBgn0003744 | 6 | trc | FBgn0036449 | 8 | bmm | FBgn0013987 | 8 | MAPk-Ak2 |
| FBgn0023169 | 6 | AMPKalpha | FBgn0261984 | 8 | Ire1 | FBgn0015765 | 8 | p38a |
| FBgn0024846 | 6 | p38b | FBgn0035142 | 8 | Hipk | FBgn0017581 | 8 | Lk6 |
| FBgn0025743 | 6 | mbt | FBgn0002413 | 8 | dco | FBgn0020621 | 8 | Pkn |
| FBgn0038167 | 6 | Lkb1 | FBgn0023076 | 8 | Clk | FBgn0023169 | 8 | AMPKalpha |
| FBgn0261456 | 6 | hpo | FBgn0003091 | 8 | Pkc53E | FBgn0024846 | 8 | p38b |
| FBgn0261854 | 6 | aPKC | FBgn0003502 | 8 | Btk29A | FBgn0025625 | 8 | Sik2 |
| FBgn0262866 | 6 | S6kII | FBgn0004784 | 8 | inaC | FBgn0025743 | 8 | mbt |
| FBgn0267390 | 6 | dop | FBgn0004864 | 8 | hop | FBgn0026063 | 8 | KP78b |
| FBgn0003317 | 6 | sax | FBgn0010197 | 8 | Gyc32E | FBgn0026064 | 8 | KP78a |
| FBgn0003716 | 6 | tkv | FBgn0010441 | 8 | pll | FBgn0031299 | 8 | CG4629 |
| FBgn0003733 | 6 | tor | FBgn0011817 | 8 | nmo | FBgn0031784 | 8 | CG9222 |
| FBgn0015402 | 6 | ksr | FBgn0013987 | 8 | MAPk-Ak2 | FBgn0033915 | 8 | CG8485 |
| FBgn0031030 | 6 | Tao | FBgn0015765 | 8 | p38a | FBgn0034568 | 8 | CG3216 |
| FBgn0038816 | 6 | Lrrk | FBgn0017581 | 8 | Lk6 | FBgn0034950 | 8 | Pask |
| FBgn0014020 | 6 | Rho1 | FBgn0020621 | 8 | Pkn | FBgn0036368 | 8 | CG10738 |
| FBgn0000463 | 6 | Dl | FBgn0023169 | 8 | AMPKalpha | FBgn0036544 | 8 | sff |
| FBgn0002940 | 6 | ninaE | FBgn0025625 | 8 | Sik2 | FBgn0037098 | 8 | Wnk |
| FBgn0004647 | 6 | NULL | FBgn0026063 | 8 | KP78b | FBgn0038167 | 8 | Lkb1 |
| FBgn0263111 | 6 | cac | FBgn0026064 | 8 | KP78a | FBgn0038630 | 8 | CG14305 |
| FBgn0010051 | 5 | Itp-r83A | FBgn0031299 | 8 | CG4629 | FBgn0039083 | 8 | CG10177 |
| FBgn0035142 | 5 | Hipk | FBgn0031784 | 8 | CG9222 | FBgn0040056 | 8 | CG17698 |
| FBgn0004170 | 5 | sc | FBgn0033915 | 8 | CG8485 | FBgn0044826 | 8 | Pak3 |
| FBgn0005672 | 5 | spi | FBgn0034950 | 8 | Pask | FBgn0046706 | 8 | Haspin |
| FBgn0028484 | 5 | Ack | FBgn0036368 | 8 | CG10738 | FBgn0051183 | 8 | CG31183 |
| FBgn0032006 | 5 | Pvr | FBgn0036544 | 8 | sff | FBgn0052666 | 8 | Drak |
| FBgn0003091 | 5 | Pkc53E | FBgn0037098 | 8 | Wnk | FBgn0052944 | 8 | CG32944 |
| FBgn0003502 | 5 | Btk29A | FBgn0038167 | 8 | Lkb1 | FBgn0085386 | 8 | CG34357 |
| FBgn0010441 | 5 | pll | FBgn0038630 | 8 | CG14305 | FBgn0259712 | 8 | CG42366 |
| FBgn0011817 | 5 | nmo | FBgn0039083 | 8 | CG10177 | FBgn0260399 | 8 | gwl |
| FBgn0015765 | 5 | p38a | FBgn0040056 | 8 | CG17698 | FBgn0260934 | 8 | par-1 |
| FBgn0017581 | 5 | Lk6 | FBgn0044826 | 8 | Pak3 | FBgn0261278 | 8 | grp |
| FBgn0020621 | 5 | Pkn | FBgn0046706 | 8 | Haspin | FBgn0261360 | 8 | CG42637 |
| FBgn0025625 | 5 | Sik2 | FBgn0052666 | 8 | Drak | FBgn0261387 | 8 | CG17528 |
| FBgn0026063 | 5 | KP78b | FBgn0052944 | 8 | CG32944 | FBgn0261456 | 8 | hpo |
| FBgn0026064 | 5 | KP78a | FBgn0085386 | 8 | CG34357 | FBgn0261854 | 8 | aPKC |
| FBgn0028717 | 5 | Lnk | FBgn0259712 | 8 | CG42366 | FBgn0262866 | 8 | S6kII |
| FBgn0031299 | 5 | CG4629 | FBgn0260399 | 8 | gwl | FBgn0263395 | 8 | hppy |
| FBgn0031784 | 5 | CG9222 | FBgn0260934 | 8 | par-1 | FBgn0266136 | 8 | Gyc76C |
| FBgn0033915 | 5 | CG8485 | FBgn0261278 | 8 | grp | FBgn0267339 | 8 | p38c |
| FBgn0034568 | 5 | CG3216 | FBgn0261360 | 8 | CG42637 | FBgn0267390 | 8 | dop |
| FBgn0034950 | 5 | Pask | FBgn0261387 | 8 | CG17528 | FBgn0267698 | 8 | Pak |
| FBgn0036368 | 5 | CG10738 | FBgn0261854 | 8 | aPKC | FBgn0283657 | 8 | Tlk |
| FBgn0036544 | 5 | sff | FBgn0262866 | 8 | S6kII | FBgn0002466 | 8 | sti |
| FBgn0037098 | 5 | Wnk | FBgn0266136 | 8 | Gyc76C | FBgn0010303 | 8 | hep |
| FBgn0038630 | 5 | CG14305 | FBgn0267339 | 8 | p38c | FBgn0024227 | 8 | aurB |
| FBgn0039083 | 5 | CG10177 | FBgn0267390 | 8 | dop | FBgn0026181 | 8 | Rok |
| FBgn0040056 | 5 | CG17698 | FBgn0267698 | 8 | Pak | FBgn0285896 | 8 | btl |
| FBgn0044826 | 5 | Pak3 | FBgn0283657 | 8 | Tlk | FBgn0004552 | 6 | Akh |
| FBgn0051183 | 5 | CG31183 | FBgn0000147 | 8 | aurA | FBgn0026252 | 6 | msk |
| FBgn0052666 | 5 | Drak | FBgn0000229 | 8 | bsk | FBgn0021796 | 6 | Tor |
| FBgn0052944 | 5 | CG32944 | FBgn0000274 | 8 | Pka-C2 | FBgn0032006 | 6 | Pvr |
| FBgn0259712 | 5 | CG42366 | FBgn0002466 | 8 | sti | FBgn0027497 | 6 | Madm |
| FBgn0260399 | 5 | gwl | FBgn0003317 | 8 | sax | FBgn0028741 | 6 | fab1 |
| FBgn0260934 | 5 | par-1 | FBgn0003366 | 8 | sev | FBgn0262738 | 6 | norpA |
| FBgn0261278 | 5 | grp | FBgn0003716 | 8 | tkv | FBgn0000017 | 6 | Abl |
| FBgn0261387 | 5 | CG17528 | FBgn0003733 | 8 | tor | FBgn0000063 | 6 | Mps1 |
| FBgn0267339 | 5 | p38c | FBgn0004839 | 8 | otk | FBgn0000147 | 6 | aurA |
| FBgn0267698 | 5 | Pak | FBgn0005666 | 8 | bt | FBgn0000229 | 6 | bsk |
| FBgn0283657 | 5 | Tlk | FBgn0010407 | 8 | Ror | FBgn0000273 | 6 | Pka-C1 |
| FBgn0000147 | 5 | aurA | FBgn0013759 | 8 | CASK | FBgn0000274 | 6 | Pka-C2 |
| FBgn0000274 | 5 | Pka-C2 | FBgn0013762 | 8 | Cdk5 | FBgn0000442 | 6 | Pkg21D |
| FBgn0001079 | 5 | fu | FBgn0015618 | 8 | Cdk8 | FBgn0000489 | 6 | Pka-C3 |
| FBgn0002466 | 5 | sti | FBgn0022800 | 8 | Cad96Ca | FBgn0000721 | 6 | for |
| FBgn0002938 | 5 | ninaC | FBgn0024179 | 8 | wit | FBgn0000723 | 6 | FER |
| FBgn0003371 | 5 | sgg | FBgn0024222 | 8 | IKKbeta | FBgn0000826 | 6 | png |
| FBgn0005640 | 5 | Eip63E | FBgn0024227 | 8 | aurB | FBgn0001079 | 6 | fu |
| FBgn0005666 | 5 | bt | FBgn0025936 | 8 | Eph | FBgn0002938 | 6 | ninaC |
| FBgn0010269 | 5 | Dsor1 | FBgn0026181 | 8 | Rok | FBgn0003079 | 6 | Raf |
| FBgn0010389 | 5 | htl | FBgn0031030 | 8 | Tao | FBgn0003124 | 6 | polo |
| FBgn0013762 | 5 | Cdk5 | FBgn0037327 | 8 | PEK | FBgn0003317 | 6 | sax |
| FBgn0015024 | 5 | CkIalpha | FBgn0038816 | 8 | Lrrk | FBgn0003366 | 6 | sev |
| FBgn0015618 | 5 | Cdk8 | FBgn0040298 | 8 | Myt1 | FBgn0003371 | 6 | sgg |
| FBgn0024179 | 5 | wit | FBgn0040505 | 8 | Alk | FBgn0003716 | 6 | tkv |
| FBgn0024222 | 5 | IKKbeta | FBgn0053531 | 8 | Ddr | FBgn0003733 | 6 | tor |
| FBgn0024227 | 5 | aurB | FBgn0086657 | 8 | IKKepsilon | FBgn0004106 | 6 | Cdk1 |
| FBgn0026181 | 5 | Rok | FBgn0265998 | 8 | Doa | FBgn0004107 | 6 | Cdk2 |
| FBgn0040298 | 5 | Myt1 | FBgn0285896 | 8 | btl | FBgn0004839 | 6 | otk |
| FBgn0264607 | 5 | CaMKII | FBgn0286813 | 8 | NULL | FBgn0004876 | 6 | cdi |
| FBgn0265998 | 5 | Doa | FBgn0004552 | 6 | Akh | FBgn0005640 | 6 | Eip63E |
| FBgn0286813 | 5 | NULL | FBgn0030607 | 6 | dob | FBgn0005666 | 6 | bt |
| FBgn0013343 | 5 | Syx1A | FBgn0035586 | 6 | CG10671 | FBgn0010269 | 6 | Dsor1 |
| FBgn0003310 | 5 | NULL | FBgn0041191 | 6 | Rheb | FBgn0010389 | 6 | htl |
| FBgn0003218 | 5 | rdgB | FBgn0000212 | 6 | brm | FBgn0010407 | 6 | Ror |
| FBgn0000022 | 5 | ac | FBgn0026252 | 6 | msk | FBgn0010909 | 6 | msn |
| FBgn0000206 | 4 | boss | FBgn0036741 | 6 | CG7510 | FBgn0011737 | 6 | Wee1 |
| FBgn0003651 | 4 | svp | FBgn0261648 | 6 | salm | FBgn0011739 | 6 | wts |
| FBgn0004552 | 4 | Akh | FBgn0261983 | 6 | l(2)gd1 | FBgn0011754 | 6 | PhKgamma |
| FBgn0035039 | 4 | Adck | FBgn0004170 | 6 | sc | FBgn0011829 | 6 | Ret |
| FBgn0035586 | 4 | CG10671 | FBgn0021796 | 6 | Tor | FBgn0013435 | 6 | Cdc2rk |
| FBgn0036449 | 4 | bmm | FBgn0032006 | 6 | Pvr | FBgn0013759 | 6 | CASK |
| FBgn0041191 | 4 | Rheb | FBgn0027497 | 6 | Madm | FBgn0013762 | 6 | Cdk5 |
| FBgn0000317 | 4 | ck | FBgn0028717 | 6 | Lnk | FBgn0014006 | 6 | Ask1 |
| FBgn0000490 | 4 | dpp | FBgn0028741 | 6 | fab1 | FBgn0014073 | 6 | Tie |
| FBgn0003463 | 4 | sog | FBgn0000017 | 6 | Abl | FBgn0015024 | 6 | CkIalpha |
| FBgn0003984 | 4 | vn | FBgn0000063 | 6 | Mps1 | FBgn0015295 | 6 | Shark |
| FBgn0036741 | 4 | CG7510 | FBgn0000241 | 6 | bw | FBgn0015380 | 6 | drl |
| FBgn0261648 | 4 | salm | FBgn0000273 | 6 | Pka-C1 | FBgn0015402 | 6 | ksr |
| FBgn0261983 | 4 | l(2)gd1 | FBgn0000442 | 6 | Pkg21D | FBgn0015618 | 6 | Cdk8 |
| FBgn0023076 | 4 | Clk | FBgn0000489 | 6 | Pka-C3 | FBgn0015772 | 6 | Nak |
| FBgn0004864 | 4 | hop | FBgn0000721 | 6 | for | FBgn0016126 | 6 | CaMKI |
| FBgn0010197 | 4 | Gyc32E | FBgn0000723 | 6 | FER | FBgn0016131 | 6 | Cdk4 |
| FBgn0013987 | 4 | MAPk-Ak2 | FBgn0000826 | 6 | png | FBgn0016696 | 6 | Pitslre |
| FBgn0015296 | 4 | Shc | FBgn0001079 | 6 | fu | FBgn0016930 | 6 | Dyrk2 |
| FBgn0027497 | 4 | Madm | FBgn0002921 | 6 | Atpalpha | FBgn0019686 | 6 | lok |
| FBgn0046706 | 4 | Haspin | FBgn0002938 | 6 | ninaC | FBgn0019949 | 6 | Cdk9 |
| FBgn0085386 | 4 | CG34357 | FBgn0003079 | 6 | Raf | FBgn0019990 | 6 | Gcn2 |
| FBgn0261360 | 4 | CG42637 | FBgn0003124 | 6 | polo | FBgn0020391 | 6 | Nrk |
| FBgn0266136 | 4 | Gyc76C | FBgn0003371 | 6 | sgg | FBgn0020412 | 6 | JIL-1 |
| FBgn0000063 | 4 | Mps1 | FBgn0003515 | 6 | st | FBgn0020440 | 6 | Fak |
| FBgn0000241 | 4 | bw | FBgn0003996 | 6 | NULL | FBgn0022800 | 6 | Cad96Ca |
| FBgn0000273 | 4 | Pka-C1 | FBgn0004106 | 6 | Cdk1 | FBgn0023081 | 6 | gek |
| FBgn0000442 | 4 | Pkg21D | FBgn0004107 | 6 | Cdk2 | FBgn0023083 | 6 | fray |
| FBgn0000489 | 4 | Pka-C3 | FBgn0004512 | 6 | Mdr49 | FBgn0024179 | 6 | wit |
| FBgn0000721 | 4 | for | FBgn0004513 | 6 | Mdr65 | FBgn0024222 | 6 | IKKbeta |
| FBgn0000826 | 4 | png | FBgn0004876 | 6 | cdi | FBgn0024245 | 6 | dnt |
| FBgn0003124 | 4 | polo | FBgn0005640 | 6 | Eip63E | FBgn0024326 | 6 | Mkk4 |
| FBgn0003366 | 4 | sev | FBgn0010241 | 6 | Mdr50 | FBgn0024329 | 6 | Mekk1 |
| FBgn0003515 | 4 | st | FBgn0010269 | 6 | Dsor1 | FBgn0025702 | 6 | Srpk79D |
| FBgn0003996 | 4 | NULL | FBgn0010549 | 6 | l(2)03659 | FBgn0025936 | 6 | Eph |
| FBgn0004106 | 4 | Cdk1 | FBgn0010909 | 6 | msn | FBgn0026323 | 6 | Tak1 |
| FBgn0004512 | 4 | Mdr49 | FBgn0011737 | 6 | Wee1 | FBgn0026371 | 6 | SAK |
| FBgn0004513 | 4 | Mdr65 | FBgn0011739 | 6 | wts | FBgn0027101 | 6 | Dyrk3 |
| FBgn0004839 | 4 | otk | FBgn0011754 | 6 | PhKgamma | FBgn0027504 | 6 | CG8878 |
| FBgn0004876 | 4 | cdi | FBgn0011829 | 6 | Ret | FBgn0027587 | 6 | CG7028 |
| FBgn0010241 | 4 | Mdr50 | FBgn0013435 | 6 | Cdc2rk | FBgn0027889 | 6 | ball |
| FBgn0010407 | 4 | Ror | FBgn0014006 | 6 | Ask1 | FBgn0028360 | 6 | Cdc7 |
| FBgn0010549 | 4 | l(2)03659 | FBgn0014073 | 6 | Tie | FBgn0028410 | 6 | Pk34A |
| FBgn0010909 | 4 | msn | FBgn0015024 | 6 | CkIalpha | FBgn0028427 | 6 | Ilk |
| FBgn0011739 | 4 | wts | FBgn0015295 | 6 | Shark | FBgn0028978 | 6 | trbl |
| FBgn0011754 | 4 | PhKgamma | FBgn0015380 | 6 | drl | FBgn0029736 | 6 | CG4041 |
| FBgn0013435 | 4 | Cdc2rk | FBgn0015402 | 6 | ksr | FBgn0029891 | 6 | Pink1 |
| FBgn0015772 | 4 | Nak | FBgn0015772 | 6 | Nak | FBgn0029970 | 6 | Nek2 |
| FBgn0016131 | 4 | Cdk4 | FBgn0016126 | 6 | CaMKI | FBgn0030018 | 6 | slpr |
| FBgn0016696 | 4 | Pitslre | FBgn0016131 | 6 | Cdk4 | FBgn0030384 | 6 | CG2577 |
| FBgn0016930 | 4 | Dyrk2 | FBgn0016696 | 6 | Pitslre | FBgn0030697 | 6 | CG8565 |
| FBgn0019686 | 4 | lok | FBgn0016930 | 6 | Dyrk2 | FBgn0030864 | 6 | CG8173 |
| FBgn0020412 | 4 | JIL-1 | FBgn0019686 | 6 | lok | FBgn0031030 | 6 | Tao |
| FBgn0020445 | 4 | E23 | FBgn0019949 | 6 | Cdk9 | FBgn0031441 | 6 | CG9962 |
| FBgn0020762 | 4 | Atet | FBgn0019990 | 6 | Gcn2 | FBgn0031518 | 6 | CG3277 |
| FBgn0022800 | 4 | Cad96Ca | FBgn0020391 | 6 | Nrk | FBgn0031643 | 6 | CG3008 |
| FBgn0023081 | 4 | gek | FBgn0020412 | 6 | JIL-1 | FBgn0031696 | 6 | Bub1 |
| FBgn0023083 | 4 | fray | FBgn0020440 | 6 | Fak | FBgn0031730 | 6 | CG7236 |
| FBgn0023536 | 4 | CG3156 | FBgn0020445 | 6 | E23 | FBgn0031855 | 6 | CG11221 |
| FBgn0025702 | 4 | Srpk79D | FBgn0020762 | 6 | Atet | FBgn0032187 | 6 | CG4839 |
| FBgn0025936 | 4 | Eph | FBgn0023081 | 6 | gek | FBgn0032650 | 6 | CG7094 |
| FBgn0026323 | 4 | Tak1 | FBgn0023083 | 6 | fray | FBgn0032677 | 6 | CG5790 |
| FBgn0026371 | 4 | SAK | FBgn0023536 | 6 | CG3156 | FBgn0033244 | 6 | CG8726 |
| FBgn0027101 | 4 | Dyrk3 | FBgn0024245 | 6 | dnt | FBgn0033773 | 6 | Mos |
| FBgn0027504 | 4 | CG8878 | FBgn0024326 | 6 | Mkk4 | FBgn0034137 | 6 | CG4945 |
| FBgn0027587 | 4 | CG7028 | FBgn0024329 | 6 | Mekk1 | FBgn0035001 | 6 | Slik |
| FBgn0027889 | 4 | ball | FBgn0025702 | 6 | Srpk79D | FBgn0035590 | 6 | Prpk |
| FBgn0028360 | 4 | Cdc7 | FBgn0026323 | 6 | Tak1 | FBgn0036187 | 6 | RIOK1 |
| FBgn0028410 | 4 | Pk34A | FBgn0026371 | 6 | SAK | FBgn0036896 | 6 | wnd |
| FBgn0028539 | 4 | CG31731 | FBgn0027101 | 6 | Dyrk3 | FBgn0037093 | 6 | Cdk12 |
| FBgn0028675 | 4 | Sur | FBgn0027504 | 6 | CG8878 | FBgn0037218 | 6 | aux |
| FBgn0028978 | 4 | trbl | FBgn0027587 | 6 | CG7028 | FBgn0037325 | 6 | CG12147 |
| FBgn0029891 | 4 | Pink1 | FBgn0027889 | 6 | ball | FBgn0037327 | 6 | PEK |
| FBgn0029970 | 4 | Nek2 | FBgn0028360 | 6 | Cdc7 | FBgn0037491 | 6 | CG1227 |
| FBgn0030018 | 4 | slpr | FBgn0028410 | 6 | Pk34A | FBgn0037679 | 6 | CG8866 |
| FBgn0030384 | 4 | CG2577 | FBgn0028427 | 6 | Ilk | FBgn0038588 | 6 | CG7156 |
| FBgn0030403 | 4 | CG1824 | FBgn0028539 | 6 | CG31731 | FBgn0038603 | 6 | PKD |
| FBgn0030697 | 4 | CG8565 | FBgn0028675 | 6 | Sur | FBgn0038816 | 6 | Lrrk |
| FBgn0030864 | 4 | CG8173 | FBgn0028978 | 6 | trbl | FBgn0038902 | 6 | CG6800 |
| FBgn0031069 | 4 | Pmp70 | FBgn0029736 | 6 | CG4041 | FBgn0039015 | 6 | Takl2 |
| FBgn0031169 | 4 | CG1494 | FBgn0029891 | 6 | Pink1 | FBgn0039306 | 6 | RIOK2 |
| FBgn0031170 | 4 | CG1718 | FBgn0029970 | 6 | Nek2 | FBgn0039796 | 6 | CG12069 |
| FBgn0031171 | 4 | CG1801 | FBgn0030018 | 6 | slpr | FBgn0039908 | 6 | Asator |
| FBgn0031220 | 4 | CG4822 | FBgn0030384 | 6 | CG2577 | FBgn0040298 | 6 | Myt1 |
| FBgn0031441 | 4 | CG9962 | FBgn0030403 | 6 | CG1824 | FBgn0040505 | 6 | Alk |
| FBgn0031449 | 4 | CG31689 | FBgn0030697 | 6 | CG8565 | FBgn0045980 | 6 | niki |
| FBgn0031515 | 4 | CG9664 | FBgn0030864 | 6 | CG8173 | FBgn0046332 | 6 | gskt |
| FBgn0031516 | 4 | CG9663 | FBgn0031069 | 6 | Pmp70 | FBgn0046685 | 6 | Wsck |
| FBgn0031643 | 4 | CG3008 | FBgn0031169 | 6 | CG1494 | FBgn0046689 | 6 | Takl1 |
| FBgn0031730 | 4 | CG7236 | FBgn0031170 | 6 | CG1718 | FBgn0046692 | 6 | Stlk |
| FBgn0031855 | 4 | CG11221 | FBgn0031171 | 6 | CG1801 | FBgn0053519 | 6 | Unc-89 |
| FBgn0032018 | 4 | CG7806 | FBgn0031220 | 6 | CG4822 | FBgn0053531 | 6 | Ddr |
| FBgn0032026 | 4 | CG7627 | FBgn0031441 | 6 | CG9962 | FBgn0085385 | 6 | bma |
| FBgn0032167 | 4 | CG5853 | FBgn0031449 | 6 | CG31689 | FBgn0086657 | 6 | IKKepsilon |
| FBgn0032187 | 4 | CG4839 | FBgn0031515 | 6 | CG9664 | FBgn0250823 | 6 | gish |
| FBgn0032456 | 4 | MRP | FBgn0031516 | 6 | CG9663 | FBgn0259168 | 6 | mnb |
| FBgn0032650 | 4 | CG7094 | FBgn0031518 | 6 | CG3277 | FBgn0259678 | 6 | sqa |
| FBgn0032677 | 4 | CG5790 | FBgn0031643 | 6 | CG3008 | FBgn0260798 | 6 | Gprk1 |
| FBgn0032908 | 4 | CG9270 | FBgn0031696 | 6 | Bub1 | FBgn0260935 | 6 | Vps15 |
| FBgn0034137 | 4 | CG4945 | FBgn0031730 | 6 | CG7236 | FBgn0260990 | 6 | yata |
| FBgn0034493 | 4 | CG8908 | FBgn0031734 | 6 | CG11147 | FBgn0261524 | 6 | lic |
| FBgn0034612 | 4 | CG10505 | FBgn0031855 | 6 | CG11221 | FBgn0261988 | 6 | Gprk2 |
| FBgn0035001 | 4 | Slik | FBgn0032018 | 6 | CG7806 | FBgn0262081 | 6 | Csk |
| FBgn0035244 | 4 | ABCB7 | FBgn0032026 | 6 | CG7627 | FBgn0262617 | 6 | CG43143 |
| FBgn0035590 | 4 | Prpk | FBgn0032167 | 6 | CG5853 | FBgn0262733 | 6 | Src64B |
| FBgn0035695 | 4 | CG10226 | FBgn0032187 | 6 | CG4839 | FBgn0263237 | 6 | Cdk7 |
| FBgn0036187 | 4 | RIOK1 | FBgn0032456 | 6 | MRP | FBgn0263855 | 6 | BubR1 |
| FBgn0036747 | 4 | CG6052 | FBgn0032650 | 6 | CG7094 | FBgn0263998 | 6 | Ack-like |
| FBgn0037218 | 4 | aux | FBgn0032677 | 6 | CG5790 | FBgn0264492 | 6 | CkIIalpha |
| FBgn0037325 | 4 | CG12147 | FBgn0032908 | 6 | CG9270 | FBgn0264607 | 6 | CaMKII |
| FBgn0037327 | 4 | PEK | FBgn0033244 | 6 | CG8726 | FBgn0264959 | 6 | Src42A |
| FBgn0037491 | 4 | CG1227 | FBgn0033773 | 6 | Mos | FBgn0265045 | 6 | Strn-Mlck |
| FBgn0037679 | 4 | CG8866 | FBgn0034137 | 6 | CG4945 | FBgn0265998 | 6 | Doa |
| FBgn0038588 | 4 | CG7156 | FBgn0034493 | 6 | CG8908 | FBgn0266465 | 6 | GckIII |
| FBgn0038603 | 4 | PKD | FBgn0034612 | 6 | CG10505 | FBgn0283473 | 6 | S6KL |
| FBgn0038740 | 4 | CG4562 | FBgn0035001 | 6 | Slik | FBgn0283712 | 6 | LIMK1 |
| FBgn0038902 | 4 | CG6800 | FBgn0035244 | 6 | ABCB7 | FBgn0286813 | 6 | NULL |
| FBgn0039207 | 4 | CG5789 | FBgn0035590 | 6 | Prpk | FBgn0287828 | 6 | NULL |
| FBgn0039306 | 4 | RIOK2 | FBgn0035632 | 6 | Ppat-Dpck | FBgn0014020 | 6 | Rho1 |
| FBgn0039644 | 4 | CG11897 | FBgn0035695 | 6 | CG10226 | |||
| FBgn0039645 | 4 | CG11898 | FBgn0036187 | 6 | RIOK1 | |||
| FBgn0039796 | 4 | CG12069 | FBgn0036747 | 6 | CG6052 | |||
| FBgn0039890 | 4 | CG2316 | FBgn0036896 | 6 | wnd | |||
| FBgn0039908 | 4 | Asator | FBgn0037093 | 6 | Cdk12 | |||
| FBgn0040505 | 4 | Alk | FBgn0037218 | 6 | aux | |||
| FBgn0045980 | 4 | niki | FBgn0037325 | 6 | CG12147 | |||
| FBgn0046332 | 4 | gskt | FBgn0037491 | 6 | CG1227 | |||
| FBgn0051121 | 4 | CG31121 | FBgn0037679 | 6 | CG8866 | |||
| FBgn0051213 | 4 | CG31213 | FBgn0038279 | 6 | Sdr | |||
| FBgn0051792 | 4 | CG31792 | FBgn0038376 | 6 | Hmt-1 | |||
| FBgn0051793 | 4 | CG31793 | FBgn0038588 | 6 | CG7156 | |||
| FBgn0052091 | 4 | CG32091 | FBgn0038603 | 6 | PKD | |||
| FBgn0052451 | 4 | SPoCk | FBgn0038740 | 6 | CG4562 | |||
| FBgn0053531 | 4 | Ddr | FBgn0038902 | 6 | CG6800 | |||
| FBgn0083956 | 4 | CG34120 | FBgn0039015 | 6 | Takl2 | |||
| FBgn0250823 | 4 | gish | FBgn0039207 | 6 | CG5789 | |||
| FBgn0259168 | 4 | mnb | FBgn0039306 | 6 | RIOK2 | |||
| FBgn0259678 | 4 | sqa | FBgn0039594 | 6 | CG9990 | |||
| FBgn0260798 | 4 | Gprk1 | FBgn0039644 | 6 | CG11897 | |||
| FBgn0260935 | 4 | Vps15 | FBgn0039645 | 6 | CG11898 | |||
| FBgn0261524 | 4 | lic | FBgn0039796 | 6 | CG12069 | |||
| FBgn0261988 | 4 | Gprk2 | FBgn0039890 | 6 | CG2316 | |||
| FBgn0261998 | 4 | CG42816 | FBgn0039908 | 6 | Asator | |||
| FBgn0262617 | 4 | CG43143 | FBgn0045980 | 6 | niki | |||
| FBgn0262733 | 4 | Src64B | FBgn0046332 | 6 | gskt | |||
| FBgn0263237 | 4 | Cdk7 | FBgn0046685 | 6 | Wsck | |||
| FBgn0263316 | 4 | Mrp4 | FBgn0046689 | 6 | Takl1 | |||
| FBgn0263747 | 4 | CG43672 | FBgn0046692 | 6 | Stlk | |||
| FBgn0264492 | 4 | CkIIalpha | FBgn0051121 | 6 | CG31121 | |||
| FBgn0264494 | 4 | CG17646 | FBgn0051213 | 6 | CG31213 | |||
| FBgn0265045 | 4 | Strn-Mlck | FBgn0051792 | 6 | CG31792 | |||
| FBgn0266465 | 4 | GckIII | FBgn0051793 | 6 | CG31793 | |||
| FBgn0283473 | 4 | S6KL | FBgn0052091 | 6 | CG32091 | |||
| FBgn0283712 | 4 | LIMK1 | FBgn0053519 | 6 | Unc-89 | |||
| FBgn0285896 | 4 | btl | FBgn0053970 | 6 | CG33970 | |||
| FBgn0287828 | 4 | NULL | FBgn0083956 | 6 | CG34120 | |||
| FBgn0288229 | 4 | NULL | FBgn0085385 | 6 | bma | |||
| FBgn0036046 | 4 | Ilp2 | FBgn0250823 | 6 | gish | |||
| FBgn0037163 | 4 | laza | FBgn0259168 | 6 | mnb | |||
| FBgn0037166 | 4 | CG11426 | FBgn0259678 | 6 | sqa | |||
| FBgn0003392 | 4 | shi | FBgn0260798 | 6 | Gprk1 | |||
| FBgn0014010 | 4 | Rab5 | FBgn0260935 | 6 | Vps15 | |||
| FBgn0004367 | 4 | mei-41 | FBgn0260990 | 6 | yata | |||
| FBgn0000137 | 4 | ase | FBgn0261524 | 6 | lic | |||
| FBgn0002561 | 4 | l(1)sc | FBgn0261988 | 6 | Gprk2 | |||
| FBgn0032940 | 4 | Mondo | FBgn0261998 | 6 | CG42816 | |||
| FBgn0267821 | 4 | da | FBgn0262081 | 6 | Csk | |||
| FBgn0028734 | 4 | Fmr1 | FBgn0262617 | 6 | CG43143 | |||
| FBgn0036260 | 4 | Rh7 | FBgn0262733 | 6 | Src64B | |||
| FBgn0037671 | 4 | VhaM8.9 | FBgn0263237 | 6 | Cdk7 | |||
| FBgn0266671 | 4 | Sec6 | FBgn0263316 | 6 | Mrp4 | |||
| FBgn0266672 | 4 | Sec8 | FBgn0263747 | 6 | CG43672 | |||
| FBgn0261617 | 4 | nej | FBgn0263855 | 6 | BubR1 | |||
| FBgn0025680 | 4 | cry | FBgn0263998 | 6 | Ack-like | |||
| FBgn0284084 | 4 | wg | FBgn0264492 | 6 | CkIIalpha | |||
| FBgn0266670 | 4 | Sec5 | FBgn0264494 | 6 | CG17646 | |||
| FBgn0004638 | 4 | drk | FBgn0264607 | 6 | CaMKII | |||
| FBgn0003892 | 3 | ptc | FBgn0264959 | 6 | Src42A | |||
| FBgn0024248 | 3 | chico | FBgn0265045 | 6 | Strn-Mlck | |||
| FBgn0026379 | 3 | Pten | FBgn0266465 | 6 | GckIII | |||
| FBgn0027572 | 3 | CG5009 | FBgn0283473 | 6 | S6KL | |||
| FBgn0029924 | 3 | CG4586 | FBgn0283712 | 6 | LIMK1 | |||
| FBgn0030607 | 3 | dob | FBgn0287828 | 6 | NULL | |||
| FBgn0031813 | 3 | CG9527 | FBgn0288229 | 6 | NULL | |||
| FBgn0032775 | 3 | CG17544 | FBgn0014020 | 6 | Rho1 | |||
| FBgn0033799 | 3 | GLaz | FBgn0003310 | 6 | NULL | |||
| FBgn0034628 | 3 | Acox57D-p | FBgn0037163 | 6 | laza | |||
| FBgn0034629 | 3 | Acox57D-d | FBgn0037165 | 6 | CG11437 | |||
| FBgn0086712 | 3 | Egm | FBgn0037166 | 6 | CG11426 | |||
| FBgn0263593 | 3 | Lpin | FBgn0037167 | 6 | CG11425 | |||
| FBgn0000212 | 3 | brm | FBgn0037341 | 6 | CG12746 | |||
| FBgn0024234 | 3 | gbb | FBgn0261794 | 6 | kcc | |||
| FBgn0026252 | 3 | msk | FBgn0029870 | 6 | Marf | |||
| FBgn0037657 | 3 | hyx | FBgn0023094 | 6 | cyc | |||
| FBgn0050418 | 3 | nord | FBgn0032940 | 6 | Mondo | |||
| FBgn0025800 | 3 | Smox | FBgn0262139 | 6 | trh | |||
| FBgn0002573 | 3 | sens | FBgn0266411 | 6 | sima | |||
| FBgn0034368 | 3 | CG5482 | FBgn0002778 | 6 | mnd | |||
| FBgn0035871 | 3 | BI-1 | FBgn0002940 | 6 | ninaE | |||
| FBgn0262451 | 3 | ban | FBgn0031064 | 6 | CG12531 | |||
| FBgn0263107 | 3 | bft | FBgn0036984 | 6 | CG13248 | |||
| FBgn0284421 | 3 | Psn | FBgn0261617 | 6 | nej | |||
| FBgn0027537 | 3 | Nup93-1 | ||||||
| FBgn0038274 | 3 | Nup93-2 | ||||||
| FBgn0039004 | 3 | Nup133 | ||||||
| FBgn0002576 | 3 | lz | ||||||
| FBgn0028741 | 3 | fab1 | ||||||
| FBgn0030869 | 3 | Socs16D | ||||||
| FBgn0261549 | 3 | rdgA | ||||||
| FBgn0033247 | 3 | Nup44A | ||||||
| FBgn0000017 | 3 | Abl | ||||||
| FBgn0000723 | 3 | FER | ||||||
| FBgn0001218 | 3 | Hsc70-3 | ||||||
| FBgn0002921 | 3 | Atpalpha | ||||||
| FBgn0004107 | 3 | Cdk2 | ||||||
| FBgn0011737 | 3 | Wee1 | ||||||
| FBgn0011829 | 3 | Ret | ||||||
| FBgn0014006 | 3 | Ask1 | ||||||
| FBgn0014073 | 3 | Tie | ||||||
| FBgn0015295 | 3 | Shark | ||||||
| FBgn0015380 | 3 | drl | ||||||
| FBgn0016126 | 3 | CaMKI | ||||||
| FBgn0019949 | 3 | Cdk9 | ||||||
| FBgn0019990 | 3 | Gcn2 | ||||||
| FBgn0020391 | 3 | Nrk | ||||||
| FBgn0020440 | 3 | Fak | ||||||
| FBgn0024245 | 3 | dnt | ||||||
| FBgn0024326 | 3 | Mkk4 | ||||||
| FBgn0024329 | 3 | Mekk1 | ||||||
| FBgn0027582 | 3 | CG6230 | ||||||
| FBgn0028427 | 3 | Ilk | ||||||
| FBgn0029736 | 3 | CG4041 | ||||||
| FBgn0030343 | 3 | ATP7 | ||||||
| FBgn0031518 | 3 | CG3277 | ||||||
| FBgn0031696 | 3 | Bub1 | ||||||
| FBgn0031734 | 3 | CG11147 | ||||||
| FBgn0033244 | 3 | CG8726 | ||||||
| FBgn0033773 | 3 | Mos | ||||||
| FBgn0035632 | 3 | Ppat-Dpck | ||||||
| FBgn0036896 | 3 | wnd | ||||||
| FBgn0037093 | 3 | Cdk12 | ||||||
| FBgn0038279 | 3 | Sdr | ||||||
| FBgn0038376 | 3 | Hmt-1 | ||||||
| FBgn0039015 | 3 | Takl2 | ||||||
| FBgn0039594 | 3 | CG9990 | ||||||
| FBgn0046685 | 3 | Wsck | ||||||
| FBgn0046689 | 3 | Takl1 | ||||||
| FBgn0046692 | 3 | Stlk | ||||||
| FBgn0053519 | 3 | Unc-89 | ||||||
| FBgn0053970 | 3 | CG33970 | ||||||
| FBgn0085385 | 3 | bma | ||||||
| FBgn0260990 | 3 | yata | ||||||
| FBgn0262081 | 3 | Csk | ||||||
| FBgn0263006 | 3 | SERCA | ||||||
| FBgn0263855 | 3 | BubR1 | ||||||
| FBgn0263864 | 3 | Dark | ||||||
| FBgn0263998 | 3 | Ack-like | ||||||
| FBgn0264959 | 3 | Src42A | ||||||
| FBgn0283469 | 3 | Vps4 | ||||||
| FBgn0031090 | 3 | Rab35 | ||||||
| FBgn0250791 | 3 | alphaSnap | ||||||
| FBgn0261385 | 3 | scra | ||||||
| FBgn0285937 | 3 | NULL | ||||||
| FBgn0037165 | 3 | CG11437 | ||||||
| FBgn0037167 | 3 | CG11425 | ||||||
| FBgn0037341 | 3 | CG12746 | ||||||
| FBgn0051145 | 3 | CG31145 | ||||||
| FBgn0263112 | 3 | Mitf | ||||||
| FBgn0029687 | 3 | Vap33 | ||||||
| FBgn0283500 | 3 | Sac1 | ||||||
| FBgn0015795 | 3 | Rab7 | ||||||
| FBgn0010355 | 3 | Taf1 | ||||||
| FBgn0035934 | 3 | TrpA1 | ||||||
| FBgn0261794 | 3 | kcc | ||||||
| FBgn0029870 | 3 | Marf | ||||||
| FBgn0004666 | 3 | sim | ||||||
| FBgn0023094 | 3 | cyc | ||||||
| FBgn0262139 | 3 | trh | ||||||
| FBgn0266411 | 3 | sima | ||||||
| FBgn0035187 | 3 | Trh | ||||||
| FBgn0038197 | 3 | foxo | ||||||
| FBgn0000257 | 3 | car | ||||||
| FBgn0002778 | 3 | mnd | ||||||
| FBgn0003249 | 3 | Rh3 | ||||||
| FBgn0003250 | 3 | Rh4 | ||||||
| FBgn0003295 | 3 | ru | ||||||
| FBgn0004197 | 3 | Ser | ||||||
| FBgn0010350 | 3 | CdsA | ||||||
| FBgn0011708 | 3 | Syx5 | ||||||
| FBgn0014019 | 3 | Rh5 | ||||||
| FBgn0019940 | 3 | Rh6 | ||||||
| FBgn0025742 | 3 | mtm | ||||||
| FBgn0030670 | 3 | Pis | ||||||
| FBgn0030964 | 3 | Pvf1 | ||||||
| FBgn0031064 | 3 | CG12531 | ||||||
| FBgn0031106 | 3 | Syx16 | ||||||
| FBgn0033483 | 3 | egr | ||||||
| FBgn0035540 | 3 | Syx17 | ||||||
| FBgn0036984 | 3 | CG13248 | ||||||
| FBgn0038659 | 3 | EndoA | ||||||
| FBgn0038984 | 3 | AdipoR | ||||||
| FBgn0045495 | 3 | Gr28b | ||||||
| FBgn0062413 | 3 | Ctr1A | ||||||
| FBgn0086676 | 3 | spin | ||||||
| FBgn0086687 | 3 | Desat1 | ||||||
| FBgn0004435 | 3 | Galphaq | ||||||
| FBgn0034140 | 3 | Lst | ||||||
| FBgn0000482 | 3 | dor | ||||||
| FBgn0052350 | 3 | Vps11 | ||||||
| FBgn0263968 | 3 | nonC | ||||||
| FBgn0015790 | 3 | Rab11 | ||||||
| FBgn0015791 | 3 | Rab14 | ||||||
| FBgn0266668 | 3 | Exo84 | ||||||
| FBgn0003118 | 3 | pnt | ||||||
| FBgn0026404 | 3 | Dronc | ||||||
| FBgn0266673 | 3 | Sec10 |
kable(blupM_KableMod, caption="Male TBLUP", "simple") %>%
add_header_above(c('Top 100 GO Terms' = 3, 'Top 50 GO Terms' = 3, 'Top 25 GO Terms' = 3))Warning in add_header_above(., c(`Top 100 GO Terms` = 3, `Top 50 GO Terms` = 3,
: Please specify format in kable. kableExtra can customize either HTML or LaTeX
outputs. See https://haozhu233.github.io/kableExtra/ for details.| Flybase Gene | Percent | Gene | Flybase Gene | Percent | Gene | Flybase Gene | Percent | Gene |
|---|---|---|---|---|---|---|---|---|
| FBgn0261854 | 11 | aPKC | FBgn0261854 | 14 | aPKC | FBgn0261873 | 10 | sdt |
| FBgn0261873 | 11 | sdt | FBgn0265778 | 14 | PDZ-GEF | FBgn0036046 | 8 | Ilp2 |
| FBgn0026192 | 9 | par-6 | FBgn0283499 | 12 | InR | FBgn0086687 | 8 | Desat1 |
| FBgn0036046 | 8 | Ilp2 | FBgn0261873 | 12 | sdt | FBgn0283499 | 8 | InR |
| FBgn0283499 | 8 | InR | FBgn0003205 | 12 | Ras85D | FBgn0261854 | 8 | aPKC |
| FBgn0265778 | 8 | PDZ-GEF | FBgn0036046 | 10 | Ilp2 | FBgn0020386 | 8 | Pdk1 |
| FBgn0020386 | 8 | Pdk1 | FBgn0086687 | 10 | Desat1 | FBgn0010333 | 8 | Rac1 |
| FBgn0003205 | 8 | Ras85D | FBgn0026192 | 10 | par-6 | FBgn0024248 | 6 | chico |
| FBgn0011661 | 8 | Moe | FBgn0020386 | 10 | Pdk1 | FBgn0024836 | 6 | stan |
| FBgn0067864 | 7 | Patj | FBgn0041191 | 10 | Rheb | FBgn0026192 | 6 | par-6 |
| FBgn0041191 | 7 | Rheb | FBgn0024836 | 8 | stan | FBgn0265778 | 6 | PDZ-GEF |
| FBgn0263289 | 6 | scrib | FBgn0067864 | 8 | Patj | FBgn0067864 | 6 | Patj |
| FBgn0259685 | 6 | crb | FBgn0003079 | 8 | Raf | FBgn0037874 | 6 | Tctp |
| FBgn0003256 | 6 | rl | FBgn0010333 | 8 | Rac1 | FBgn0011661 | 6 | Moe |
| FBgn0014020 | 6 | Rho1 | FBgn0015402 | 8 | ksr | |||
| FBgn0015279 | 5 | Pi3K92E | FBgn0001965 | 8 | Sos | |||
| FBgn0024248 | 5 | chico | FBgn0003256 | 8 | rl | |||
| FBgn0086687 | 5 | Desat1 | FBgn0011661 | 8 | Moe | |||
| FBgn0001624 | 5 | dlg1 | FBgn0004636 | 8 | Rap1 | |||
| FBgn0010379 | 5 | Akt1 | FBgn0014020 | 8 | Rho1 | |||
| FBgn0262656 | 5 | Myc | FBgn0005558 | 6 | ey | |||
| FBgn0003079 | 5 | Raf | FBgn0015279 | 6 | Pi3K92E | |||
| FBgn0010269 | 5 | Dsor1 | FBgn0024248 | 6 | chico | |||
| FBgn0010333 | 5 | Rac1 | FBgn0025595 | 6 | AkhR | |||
| FBgn0023172 | 5 | RhoGEF2 | FBgn0014135 | 6 | bnl | |||
| FBgn0003514 | 5 | sqh | FBgn0051217 | 6 | modSP | |||
| FBgn0000163 | 5 | baz | FBgn0259685 | 6 | crb | |||
| FBgn0011300 | 4 | babo | FBgn0010379 | 6 | Akt1 | |||
| FBgn0025595 | 4 | AkhR | FBgn0037874 | 6 | Tctp | |||
| FBgn0025800 | 4 | Smox | FBgn0262656 | 6 | Myc | |||
| FBgn0014135 | 4 | bnl | FBgn0266756 | 6 | btsz | |||
| FBgn0051217 | 4 | modSP | FBgn0010269 | 6 | Dsor1 | |||
| FBgn0003391 | 4 | shg | FBgn0033205 | 6 | CG2064 | |||
| FBgn0024836 | 4 | stan | FBgn0266136 | 6 | Gyc76C | |||
| FBgn0033205 | 4 | CG2064 | FBgn0004638 | 6 | drk | |||
| FBgn0015402 | 4 | ksr | FBgn0020224 | 6 | Cbl | |||
| FBgn0001965 | 4 | Sos | FBgn0025741 | 6 | PlexA | |||
| FBgn0004638 | 4 | drk | FBgn0020388 | 6 | Gcn5 | |||
| FBgn0021796 | 4 | Tor | FBgn0028387 | 6 | chm | |||
| FBgn0000017 | 4 | Abl | FBgn0037376 | 6 | Hat1 | |||
| FBgn0013733 | 4 | shot | FBgn0023172 | 6 | RhoGEF2 | |||
| FBgn0026181 | 4 | Rok | FBgn0003514 | 6 | sqh | |||
| FBgn0025741 | 4 | PlexA | ||||||
| FBgn0000547 | 4 | ed | ||||||
| FBgn0020388 | 4 | Gcn5 | ||||||
| FBgn0028387 | 4 | chm | ||||||
| FBgn0034975 | 4 | enok | ||||||
| FBgn0004636 | 4 | Rap1 | ||||||
| FBgn0011202 | 4 | dia | ||||||
| FBgn0011674 | 4 | insc | ||||||
| FBgn0003169 | 3 | put | ||||||
| FBgn0005558 | 3 | ey | ||||||
| FBgn0024913 | 3 | Actbeta | ||||||
| FBgn0038167 | 3 | Lkb1 | ||||||
| FBgn0030926 | 3 | psh | ||||||
| FBgn0040321 | 3 | GNBP3 | ||||||
| FBgn0285896 | 3 | btl | ||||||
| FBgn0000504 | 3 | dsx | ||||||
| FBgn0002121 | 3 | l(2)gl | ||||||
| FBgn0004167 | 3 | kst | ||||||
| FBgn0024846 | 3 | p38b | ||||||
| FBgn0037874 | 3 | Tctp | ||||||
| FBgn0266756 | 3 | btsz | ||||||
| FBgn0003118 | 3 | pnt | ||||||
| FBgn0040336 | 3 | Seipin | ||||||
| FBgn0023388 | 3 | Dap160 | ||||||
| FBgn0040323 | 3 | GNBP1 | ||||||
| FBgn0266136 | 3 | Gyc76C | ||||||
| FBgn0028717 | 3 | Lnk | ||||||
| FBgn0036449 | 3 | bmm | ||||||
| FBgn0029840 | 3 | raptor | ||||||
| FBgn0283472 | 3 | S6k | ||||||
| FBgn0020224 | 3 | Cbl | ||||||
| FBgn0002543 | 3 | robo2 | ||||||
| FBgn0010389 | 3 | htl | ||||||
| FBgn0021776 | 3 | mira | ||||||
| FBgn0023081 | 3 | gek | ||||||
| FBgn0027596 | 3 | Kank | ||||||
| FBgn0085408 | 3 | Shroom | ||||||
| FBgn0086384 | 3 | Mer | ||||||
| FBgn0027598 | 3 | cindr | ||||||
| FBgn0011642 | 3 | Zyx | ||||||
| FBgn0014340 | 3 | mof | ||||||
| FBgn0026080 | 3 | Tip60 | ||||||
| FBgn0032691 | 3 | Atac2 | ||||||
| FBgn0036039 | 3 | Naa60 | ||||||
| FBgn0037376 | 3 | Hat1 | ||||||
| FBgn0039585 | 3 | CG1894 | ||||||
| FBgn0261617 | 3 | nej | ||||||
| FBgn0002577 | 3 | NULL | ||||||
| FBgn0004956 | 3 | upd1 | ||||||
| FBgn0010909 | 3 | msn | ||||||
| FBgn0036985 | 3 | zye | ||||||
| FBgn0066365 | 3 | dyl | ||||||
| FBgn0284435 | 3 | tyn | ||||||
| FBgn0004595 | 3 | pros | ||||||
| FBgn0028734 | 3 | Fmr1 | ||||||
| FBgn0029866 | 3 | CG3842 | ||||||
| FBgn0033203 | 3 | CG2070 | ||||||
| FBgn0033204 | 3 | CG2065 | ||||||
| FBgn0050491 | 3 | CG30491 | ||||||
| FBgn0050495 | 3 | CG30495 | ||||||
| FBgn0000117 | 3 | arm | ||||||
| FBgn0003520 | 3 | stau | ||||||
| FBgn0003731 | 3 | Egfr | ||||||
| FBgn0035142 | 3 | Hipk |
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Rocky Linux 8.5 (Green Obsidian)
Matrix products: default
BLAS/LAPACK: /opt/ohpc/pub/libs/gnu9/openblas/0.3.7/lib/libopenblasp-r0.3.7.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] DT_0.31 kableExtra_1.3.4 knitr_1.43 reshape2_1.4.4
[5] melt_1.10.0 ggcorrplot_0.1.4.1 lubridate_1.9.3 forcats_1.0.0
[9] stringr_1.5.0 purrr_1.0.1 readr_2.1.4 tidyr_1.3.0
[13] tibble_3.2.1 tidyverse_2.0.0 scales_1.2.1 viridis_0.6.4
[17] viridisLite_0.4.2 qqman_0.1.9 cowplot_1.1.1 ggplot2_3.4.4
[21] data.table_1.14.8 dplyr_1.1.3 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] httr_1.4.7 sass_0.4.7 jsonlite_1.8.7 bslib_0.5.0
[5] getPass_0.2-2 highr_0.10 yaml_2.3.7 pillar_1.9.0
[9] glue_1.6.2 digest_0.6.33 promises_1.2.0.1 rvest_1.0.3
[13] colorspace_2.1-0 htmltools_0.5.5 httpuv_1.6.12 plyr_1.8.9
[17] pkgconfig_2.0.3 calibrate_1.7.7 webshot_0.5.5 processx_3.8.2
[21] svglite_2.1.2 whisker_0.4.1 later_1.3.1 tzdb_0.4.0
[25] timechange_0.2.0 git2r_0.32.0 generics_0.1.3 farver_2.1.1
[29] cachem_1.0.8 withr_2.5.0 cli_3.6.1 magrittr_2.0.3
[33] evaluate_0.21 ps_1.7.5 fs_1.6.3 fansi_1.0.4
[37] MASS_7.3-60 xml2_1.3.3 tools_4.1.2 hms_1.1.3
[41] lifecycle_1.0.3 munsell_0.5.0 callr_3.7.3 compiler_4.1.2
[45] jquerylib_0.1.4 systemfonts_1.0.5 rlang_1.1.1 grid_4.1.2
[49] rstudioapi_0.15.0 htmlwidgets_1.6.2 labeling_0.4.3 rmarkdown_2.23
[53] gtable_0.3.4 R6_2.5.1 gridExtra_2.3 fastmap_1.1.1
[57] utf8_1.2.3 rprojroot_2.0.3 stringi_1.7.12 Rcpp_1.0.11
[61] vctrs_0.6.4 tidyselect_1.2.0 xfun_0.39