alkati_cell_line_tae684_response
Haider Inam
3/6/2019
Last updated: 2019-06-03
Checks: 5 1
Knit directory: pair_con_select/
This reproducible R Markdown analysis was created with workflowr (version 1.3.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190211) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: alk_luad_synonymous.csv
Untracked: analysis/alk_luad_mutation_bias.Rmd
Untracked: analysis/mutation_bias_gist_brca.Rmd
Untracked: bias.brca.csv
Untracked: bias.gist.csv
Untracked: code/alldata_compiler.R
Untracked: code/contab_maker.R
Untracked: code/mut_excl_genes_datapoints.R
Untracked: code/mut_excl_genes_generator.R
Untracked: code/quadratic_solver.R
Untracked: code/simresults_generator.R
Untracked: data/ALKATI_ccle.csv
Untracked: data/All_Data_V2.csv
Untracked: data/CCLE_NP24.2009_Drug_data_2015.02.24.csv
Untracked: data/alkati_growthcurvedata.csv
Untracked: data/alkati_growthcurvedata_popdoublings.csv
Untracked: data/alkati_melanoma_vemurafenib_figure_data.csv
Untracked: data/alkati_simulations_compiled_1000_12319.csv
Untracked: data/all_data.csv
Untracked: data/gist_expression/
Untracked: data/tcga_brca_expression/
Untracked: data/tcga_luad_expression/
Untracked: data/tcga_skcm_expression/
Untracked: docs/figure/Filteranalysis.Rmd/
Untracked: mutation_bias_brca_n.pdf
Untracked: mutation_bias_brca_probabilities.pdf
Untracked: mutation_bias_brca_probabilities2.pdf
Untracked: mutation_bias_gist.pdf
Untracked: mutation_bias_gist_n.pdf
Untracked: mutation_bias_gist_probabilities.pdf
Untracked: mutation_bias_gist_probabilities2.pdf
Untracked: mutation_bias_luad.pdf
Untracked: mutation_bias_luad_deviations.pdf
Untracked: mutation_bias_luad_diagnostic.pdf
Untracked: mutation_probability.csv
Untracked: output/ alkati_subsamplesize_orval_fig1c.pdf
Untracked: output/alkati_ccle_tae684_plot.pdf
Untracked: output/alkati_filtercutoff_allfilters.csv
Untracked: output/alkati_luad_exonimbalance.pdf
Untracked: output/alkati_mtn_pval_fig2B.pdf
Untracked: output/alkati_skcm_exonimbalance.pdf
Untracked: output/alkati_subsamplesize_pval_fig.pdf
Untracked: output/alkati_subsamplesize_pval_fig1c.pdf
Untracked: output/all_data_luad.csv
Untracked: output/all_data_luad_egfr.csv
Untracked: output/all_data_skcm.csv
Untracked: output/baf3_alkati_figure_deltaadjusted_doublings.pdf
Untracked: output/baf3_barplot.pdf
Untracked: output/baf3_elisa_barplot.pdf
Untracked: output/egfr_luad_exonimbalance.pdf
Untracked: output/fig1c_3719_4.pdf
Untracked: output/fig1c_52219.pdf
Untracked: output/fig2b2_filtercutoff_atinras_totalalk.pdf
Untracked: output/fig2b_filtercutoff_atibraf.pdf
Untracked: output/fig2b_filtercutoff_atinras.pdf
Untracked: output/luad_alk_exon_expression.csv
Untracked: output/luad_egfr_exon_expression.csv
Untracked: output/melanoma_vemurafenib_fig.pdf
Untracked: output/skcm_alk_exon_expression.csv
Untracked: output/suppfig1..pdf
Untracked: output/suppfig1_52219.pdf
Untracked: suppfig1..pdf
Unstaged changes:
Modified: analysis/ALKATI_Filter_Cutoff_Analysis.Rmd
Modified: analysis/ALK_ExonImbalance_SKCM_Analysis.Rmd
Modified: analysis/TCGA_luad_data_parser.Rmd
Modified: analysis/alkati_cell_line_tae684_response.Rmd
Modified: analysis/alkati_subsampling_simulations.Rmd
Modified: analysis/baf3_alkati_transformations.Rmd
Modified: analysis/index.Rmd
Modified: analysis/pairwise_comparisons_conditional_selection_simulated_cohorts.Rmd
Modified: analysis/practice.Rmd
Modified: analysis/tcga_luad_data_parser_egfr.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | cd475bf | haiderinam | 2019-03-06 | Build site. |
| Rmd | 6f57396 | haiderinam | 2019-03-06 | Added CCLE analyses on cancer cell line TAE684 response. This includes a logistic |
Here, we combine data from 3 sources: TCGA SKCM ALK expressiossion, CCLE TAE684 IC50, and ALK expression for some of the cell lines on CCLE
The inputs here are 3 .csv files.
- alldata.csv contains SKCM mutation data for BRAF,NRAS and expression data for ALK
- CCLE_NP24.2009_Drug_data_2015.02.24.csv contains SKCM dose responses for different drugs for many cell lines. Of these drugs, only TAE684 targets ALK. There are 504 cell lines with dose repsonses for drug. We had expression data for only 54 of these 504 cell lines.
- ALKATI_ccle.csv contains ALK exon expression RPKMs for 54 cell lines. Obtained from crownbio
ccle_drug=read.csv("data/CCLE_NP24.2009_Drug_data_2015.02.24.csv",sep=",",header=T,stringsAsFactors=F)
sort(unique(ccle_drug$Compound)) [1] "17-AAG" "AEW541" "AZD0530" "AZD6244"
[5] "Erlotinib" "Irinotecan" "L-685458" "Lapatinib"
[9] "LBW242" "Nilotinib" "Nutlin-3" "Paclitaxel"
[13] "Panobinostat" "PD-0325901" "PD-0332991" "PF2341066"
[17] "PHA-665752" "PLX4720" "RAF265" "Sorafenib"
[21] "TAE684" "TKI258" "Topotecan" "ZD-6474" ccle_alk=ccle_drug[ccle_drug$Target=="ALK",]
rm(ccle_drug)
ccle_rpkm=read.csv("data/ALKATI_ccle.csv", sep=",",header=T, stringsAsFactors=F)
ccleRpkmT=t(ccle_rpkm)
#Extracting our desired cell lines
data_mat=data.frame(ccleRpkmT[5:57,])
colnames(data_mat)[1:28]=ccleRpkmT[4,2:29]
data_mat_rpkm=data.frame(cbind(rownames(data_mat),data_mat))
rownames(data_mat_rpkm)=NULL
data_mat_rpkm$rownames.data_mat.=sub("^G[0-9]{5}.([A-Za-z0-9._]*).[0-9].bam","\\1",data_mat_rpkm$rownames.data_mat.)
############################################################################################################
ccle_alk$CCLE.Cell.Line.Name=sub("^([0-9A-Z]*)_[A-Za-z0-9_]*","\\1",ccle_alk$CCLE.Cell.Line.Name)
data_mat_rpkm$rownames.data_mat.=gsub("[_]","",data_mat_rpkm$rownames.data_mat.)
data_mat_rpkm$rownames.data_mat.=gsub("[.]","",data_mat_rpkm$rownames.data_mat.)
data_mat_rpkm$rownames.data_mat.=toupper(data_mat_rpkm$rownames.data_mat.)
#they were character because of auto import
for (i in 2:ncol(data_mat_rpkm)){
data_mat_rpkm[,i]=as.numeric(as.character(data_mat_rpkm[,i]))
}
for (i in 2:nrow(data_mat_rpkm)){
data_mat_rpkm[i,31]=sum(data_mat_rpkm[i,2:29])
data_mat_rpkm[i,32]=sum(data_mat_rpkm[i,2:19])/19
data_mat_rpkm[i,33]=sum(data_mat_rpkm[i,20:29])/10
}
colnames(data_mat_rpkm)[31]="SumRPKM"
colnames(data_mat_rpkm)[32]="Avg1_19RPKM"
colnames(data_mat_rpkm)[33]="Avg20_29RPKM"
alldata=merge(data_mat_rpkm,ccle_alk, by.x="rownames.data_mat.", by.y="CCLE.Cell.Line.Name")
alldata=data.frame(cbind(alldata,alldata$Avg20_29RPKM/alldata$Avg1_19RPKM))
data_mat_rpkm=data.frame(cbind(data_mat_rpkm,data_mat_rpkm$Avg20_29RPKM/data_mat_rpkm$Avg1_19RPKM))
#####Makingderivative columns for RPKM ratio and Sum RPKM#########
skcm=read.csv("data/all_data.csv",sep=",",header=T,stringsAsFactors=F)#Downloaded from firehose 02-xx-2016,compiled with mutation data, 340 patients with RNAseq and Muts
skcm_comp=data.frame(cbind(skcm,skcm$mean_RPKM_20.29/skcm$mean_RPKM_1.19))
dim(skcm_comp)[1] 351 11skcm_comp[12]=skcm_comp[5]
skcm_comp[13]=data.frame(20*(skcm_comp$mean_RPKM_1.19)+10*(skcm_comp$mean_RPKM_20.29))
colnames(skcm_comp)[13]="Total_RPKM"
colnames(skcm_comp)[12]="Ratio_ALK_Exons20_29vsExons1_19"
skcm_comp[14]=data.frame(skcm_comp$BRAF!=NaN)
colnames(skcm_comp)[14]="is.BRAF"
skcm_comp[15]=data.frame(grepl("V600",skcm_comp$BRAF),"BRAF")Warning in `[<-.data.frame`(`*tmp*`, 15, value =
structure(list(grepl..V600...skcm_comp.BRAF. = c(TRUE, : provided 2
variables to replace 1 variablescolnames(skcm_comp)[15]="is.V600"
skcm_comp[16]=data.frame(skcm_comp$NRAS!=NaN)
colnames(skcm_comp)[16]="is.NRAS"
skcm_comp[17]=data.frame(grepl("Q61",skcm_comp$NRAS),"NRAS")Warning in `[<-.data.frame`(`*tmp*`, 17, value =
structure(list(grepl..Q61...skcm_comp.NRAS. = c(FALSE, : provided 2
variables to replace 1 variablescolnames(skcm_comp)[17]="is.Q61"
skcm_comp[skcm_comp$BRAF==NaN & skcm_comp$NRAS==NaN, 18]= "DoubleNegative"
colnames(skcm_comp)[18]="is.neitherBRAForNRAS"
skcm_comp[skcm_comp$BRAF!=NaN, 19]= "BRAF Mutant"
skcm_comp[skcm_comp$NRAS!=NaN, 19]= "NRAS Mutant"
skcm_comp[skcm_comp$BRAF==NaN & skcm_comp$NRAS==NaN, 19]= "DoubleNegative"
colnames(skcm_comp)[19]="is.mutant"
######Graphing########
ggplot()+geom_point(data=skcm_comp,aes(x=Ratio_ALK_Exons20_29vsExons1_19, y=Total_RPKM,color=is.mutant))+geom_point(data=alldata,aes(x=alldata.Avg20_29RPKM.alldata.Avg1_19RPKM,y=SumRPKM,size=IC50..uM.))+scale_x_log10()+scale_y_log10()Warning: Transformation introduced infinite values in continuous y-axisWarning: Removed 2 rows containing missing values (geom_point).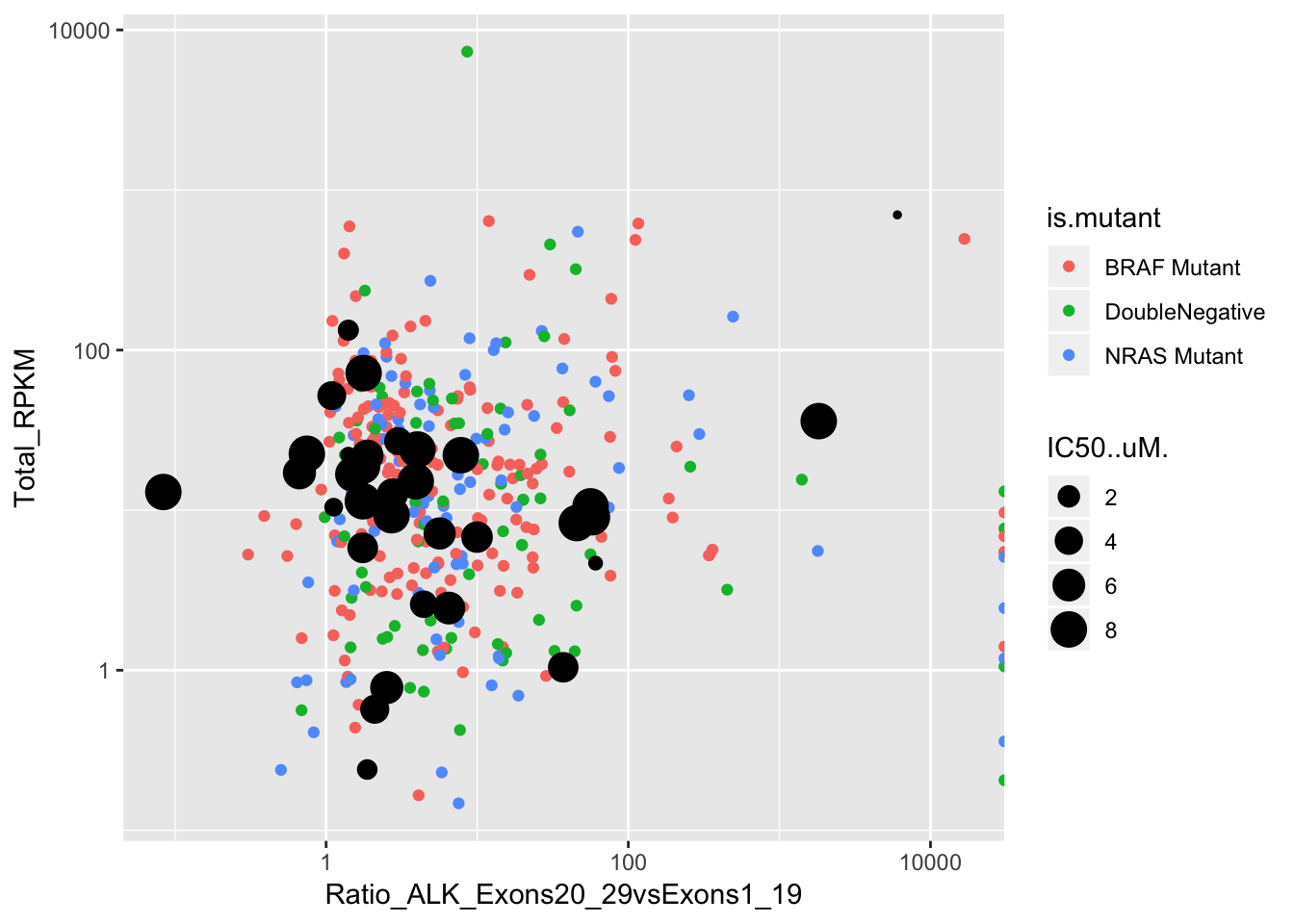
| Version | Author | Date |
|---|---|---|
| cd475bf | haiderinam | 2019-03-06 |
# ggsave("cellline,IC50.png",width=10,length=10)
# ggplot()+geom_point(data=skcm_comp,aes(x=Ratio_ALK_Exons20_29vsExons1_19, y=Total_RPKM,color=is.mutant))+geom_point(data=data_mat_rpkm,aes(x=alldata.Avg20_29RPKM.alldata.Avg1_19RPKM,y=SumRPKM,size=IC50..uM.))+scale_x_log10()+scale_y_log10()
# ggsave("cellline_clinical_all.png",width=10,length=10)
ggplot()+geom_point(data=skcm_comp,aes(x=Ratio_ALK_Exons20_29vsExons1_19, y=Total_RPKM,color=factor(skcm_comp$is.mutant,levels = c("DoubleNegative","BRAF Mutant","NRAS Mutant"))))+
geom_point(data=alldata,aes(x=alldata.Avg20_29RPKM.alldata.Avg1_19RPKM,y=SumRPKM,size=IC50..uM.))+
annotate("rect", xmin = 10, xmax = Inf, ymin = 10, ymax = Inf,fill="#66C2A5",alpha = .4)+
scale_x_log10()+
scale_y_log10()+
cleanup+
# scale_color_brewer(palette="Set2",name="Mutation")+
scale_color_manual(values =c("#FFD92F","#E78AC3","#8DA0CB"),name="Mutation")+
scale_size_continuous("Cell Line \nIC50 (uM)",range = (c(.1,2.5)))+
xlab("Ratio ALK Ex20-29 to Ex1-19")+
ylab("Total RPKM")+
theme(plot.title = element_text(hjust=.5),
text = element_text(size=10,face="bold"),
axis.title = element_text(face="bold",size="10",color="black"),
axis.text=element_text(face="bold",size="10",color="black"),
legend.position = "bottom")Warning: Transformation introduced infinite values in continuous y-axis
Warning: Removed 2 rows containing missing values (geom_point).
| Version | Author | Date |
|---|---|---|
| cd475bf | haiderinam | 2019-03-06 |
# ggsave("output/alkati_ccle_tae684_plot.pdf",height=3,width=4.5, useDingbats=FALSE)Logistic regression to see if IC50 can predict ALKATI or Overexpression
Didn’t end up using logistic regression. Used linear regression.
Linear regression to see if IC50 can predict ALKATI or Overexpression
Linear regression of total RPKM and 20-29/1-19 ratio predicting IC50 yielded a p-value of .06 when
alldata=alldata%>%filter(rownames.data_mat.!="SUPM2")
alkati_lm=lm(IC50..uM.~alldata.Avg20_29RPKM.alldata.Avg1_19RPKM+SumRPKM,data = alldata)
summary(alkati_lm)
Call:
lm(formula = IC50..uM. ~ alldata.Avg20_29RPKM.alldata.Avg1_19RPKM +
SumRPKM, data = alldata)
Residuals:
Min 1Q Median 3Q Max
-5.4559 -1.1478 0.1536 2.2514 3.0534
Coefficients:
Estimate Std. Error t value
(Intercept) 5.871911 0.564334 10.405
alldata.Avg20_29RPKM.alldata.Avg1_19RPKM 0.001421 0.001422 0.999
SumRPKM -0.012923 0.017739 -0.729
Pr(>|t|)
(Intercept) 2.66e-11 ***
alldata.Avg20_29RPKM.alldata.Avg1_19RPKM 0.326
SumRPKM 0.472
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.525 on 29 degrees of freedom
(2 observations deleted due to missingness)
Multiple R-squared: 0.04576, Adjusted R-squared: -0.02005
F-statistic: 0.6954 on 2 and 29 DF, p-value: 0.507
sessionInfo()R version 3.5.2 (2018-12-20)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14.5
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel grid stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] ggsignif_0.5.0 usethis_1.5.0 devtools_2.0.2
[4] RColorBrewer_1.1-2 reshape2_1.4.3 ggplot2_3.1.1
[7] doParallel_1.0.14 iterators_1.0.10 foreach_1.4.4
[10] dplyr_0.8.1 VennDiagram_1.6.20 futile.logger_1.4.3
[13] workflowr_1.3.0 tictoc_1.0 knitr_1.23
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 xfun_0.7 remotes_2.0.4
[4] purrr_0.3.2 colorspace_1.4-1 htmltools_0.3.6
[7] yaml_2.2.0 rlang_0.3.4 pkgbuild_1.0.3
[10] pillar_1.4.1 glue_1.3.1 withr_2.1.2
[13] lambda.r_1.2.3 sessioninfo_1.1.1 plyr_1.8.4
[16] stringr_1.4.0 munsell_0.5.0 gtable_0.3.0
[19] codetools_0.2-16 evaluate_0.14 memoise_1.1.0
[22] labeling_0.3 callr_3.2.0 ps_1.3.0
[25] Rcpp_1.0.1 scales_1.0.0 backports_1.1.4
[28] formatR_1.6 desc_1.2.0 pkgload_1.0.2
[31] fs_1.3.1 digest_0.6.19 stringi_1.4.3
[34] processx_3.3.1 rprojroot_1.3-2 cli_1.1.0
[37] tools_3.5.2 magrittr_1.5 lazyeval_0.2.2
[40] tibble_2.1.2 futile.options_1.0.1 crayon_1.3.4
[43] whisker_0.3-2 pkgconfig_2.0.2 prettyunits_1.0.2
[46] assertthat_0.2.1 rmarkdown_1.13 R6_2.4.0
[49] git2r_0.25.2 compiler_3.5.2