Run_all_analysis
ERM
20230414
Last updated: 2023-05-26
Checks: 7 0
Knit directory: Cardiotoxicity/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230109) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f99a753. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/41588_2018_171_MOESM3_ESMeQTL_ST2_for paper.csv
Ignored: data/Arr_GWAS.txt
Ignored: data/BC_cell_lines.csv
Ignored: data/CADGWASgene_table.csv
Ignored: data/Clamp_Summary.csv
Ignored: data/Cormotif_24_k1-5_raw.RDS
Ignored: data/DAgostres24.RDS
Ignored: data/DAtable1.csv
Ignored: data/DDEMresp_list.csv
Ignored: data/DDE_reQTL.txt
Ignored: data/DDEresp_list.csv
Ignored: data/DEG-GO/
Ignored: data/DEG_cormotif.RDS
Ignored: data/DF_Plate_Peak.csv
Ignored: data/Da24counts.txt
Ignored: data/Dx24counts.txt
Ignored: data/Dx_reQTL_specific.txt
Ignored: data/Ep24counts.txt
Ignored: data/GOplots.R
Ignored: data/GTEX_setsimple.csv
Ignored: data/HFGWASgene_table.csv
Ignored: data/Heart_Left_Ventricle.v8.egenes.txt
Ignored: data/Hf_GWAS.txt
Ignored: data/K_cluster
Ignored: data/K_cluster_kisthree.csv
Ignored: data/K_cluster_kistwo.csv
Ignored: data/LDH48hoursdata.csv
Ignored: data/Mt24counts.txt
Ignored: data/RINsamplelist.txt
Ignored: data/Seonane2019supp1.txt
Ignored: data/TOP2Bi-24hoursGO_analysis.csv
Ignored: data/TR24counts.txt
Ignored: data/Top2biresp_cluster24h.csv
Ignored: data/Viabilitylistfull.csv
Ignored: data/allexpressedgenes.txt
Ignored: data/allgenes.txt
Ignored: data/allmatrix.RDS
Ignored: data/avgLD50.RDS
Ignored: data/backGL.txt
Ignored: data/cormotif_3hk1-8.RDS
Ignored: data/cormotif_initalK5.RDS
Ignored: data/cormotif_initialK5.RDS
Ignored: data/cormotif_initialall.RDS
Ignored: data/counts24hours.RDS
Ignored: data/cpmnorm_counts.csv
Ignored: data/cvd_GWAS.txt
Ignored: data/dat_cpm.RDS
Ignored: data/data_outline.txt
Ignored: data/efit2.RDS
Ignored: data/efit2results.RDS
Ignored: data/ensembl_backup.RDS
Ignored: data/ensgtotal.txt
Ignored: data/filenameonly.txt
Ignored: data/filtered_cpm_counts.csv
Ignored: data/filtered_raw_counts.csv
Ignored: data/filtermatrix_x.RDS
Ignored: data/folder_05top/
Ignored: data/gene_prob_tran3h.RDS
Ignored: data/gene_probabilityk5.RDS
Ignored: data/gostresTop2bi_ER.RDS
Ignored: data/gostresTop2bi_LR
Ignored: data/gostresTop2bi_LR.RDS
Ignored: data/gostresTop2bi_TI.RDS
Ignored: data/gostrescoNR
Ignored: data/gtex/
Ignored: data/heartgenes.csv
Ignored: data/individualDRCfile.RDS
Ignored: data/individual_LDH48.RDS
Ignored: data/knowfig4.csv
Ignored: data/knowfig5.csv
Ignored: data/knowles56.GMT
Ignored: data/knowlesGMT.GMT
Ignored: data/mymatrix.RDS
Ignored: data/nonresponse_cluster24h.csv
Ignored: data/norm_LDH.csv
Ignored: data/norm_counts.csv
Ignored: data/old_sets/
Ignored: data/plan2plot.png
Ignored: data/raw_counts.csv
Ignored: data/response_cluster24h.csv
Ignored: data/sigVDA24.txt
Ignored: data/sigVDA3.txt
Ignored: data/sigVDX24.txt
Ignored: data/sigVDX3.txt
Ignored: data/sigVEP24.txt
Ignored: data/sigVEP3.txt
Ignored: data/sigVMT24.txt
Ignored: data/sigVMT3.txt
Ignored: data/sigVTR24.txt
Ignored: data/sigVTR3.txt
Ignored: data/siglist.RDS
Ignored: data/table3a.omar
Ignored: data/toplistall.RDS
Ignored: data/tvl24hour.txt
Ignored: data/tvl24hourw.txt
Ignored: data/venn_code.R
Untracked files:
Untracked: .RDataTmp
Untracked: .RDataTmp1
Untracked: .RDataTmp2
Untracked: cormotif_probability_genelist.csv
Untracked: individual-legenddark2.png
Untracked: installed_old.rda
Untracked: motif_ER.txt
Untracked: motif_LR.txt
Untracked: motif_NR.txt
Untracked: motif_TI.txt
Untracked: output/figure_1.Rmd
Untracked: output/output-old/
Untracked: output/plan2plot.png
Untracked: output/plan48ldh.png
Untracked: reneebasecode.R
Unstaged changes:
Modified: analysis/DEG-GO_analysis.Rmd
Modified: code/Compare_tnni_ldh.R
Modified: code/sequencing_info_collection.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/run_all_analysis.Rmd) and
HTML (docs/run_all_analysis.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f99a753 | reneeisnowhere | 2023-05-26 | volcano plot limits added |
| Rmd | 8d87a52 | reneeisnowhere | 2023-05-17 | end of day |
| html | bdbf1c2 | reneeisnowhere | 2023-04-21 | Build site. |
| Rmd | 5a99763 | reneeisnowhere | 2023-04-21 | correct graph to log 2! |
| html | 1f4237c | reneeisnowhere | 2023-04-20 | Build site. |
| Rmd | 13e7b3d | reneeisnowhere | 2023-04-20 | updatedsequences graph with bar at 20,000,000 |
| html | 5ef62c9 | reneeisnowhere | 2023-04-17 | Build site. |
| Rmd | 8a5a1e1 | reneeisnowhere | 2023-04-17 | update html page |
| html | 7f62d67 | reneeisnowhere | 2023-04-17 | Build site. |
| Rmd | 363ddad | reneeisnowhere | 2023-04-17 | update with GO links |
| html | 21fd945 | reneeisnowhere | 2023-04-17 | Build site. |
| Rmd | e1c63d9 | reneeisnowhere | 2023-04-17 | wflow_publish("analysis/run_all_analysis.Rmd") |
| html | 8221ec3 | reneeisnowhere | 2023-04-16 | Build site. |
| Rmd | 9e88c22 | reneeisnowhere | 2023-04-16 | updated run data |
| Rmd | 6d925a2 | reneeisnowhere | 2023-04-16 | updating cormotif with updated RNAseq counts |
| html | 8d08bd2 | reneeisnowhere | 2023-04-11 | Build site. |
| Rmd | 0aaa63d | reneeisnowhere | 2023-04-11 | cormotif analysis update |
| Rmd | 575fd81 | reneeisnowhere | 2023-04-11 | updating cormotif |
| html | 4cd8ac4 | reneeisnowhere | 2023-04-11 | Build site. |
| html | 08936e7 | reneeisnowhere | 2023-04-10 | Build site. |
| Rmd | fa2cbeb | reneeisnowhere | 2023-04-10 | monday end |
| html | 85526c5 | reneeisnowhere | 2023-04-10 | Build site. |
| Rmd | 1444a85 | reneeisnowhere | 2023-04-10 | update push of new data |
| html | b266b76 | reneeisnowhere | 2023-04-10 | Build site. |
| Rmd | d3f8cf7 | reneeisnowhere | 2023-04-10 | update push of new data |
| html | f0a75e1 | reneeisnowhere | 2023-04-10 | Build site. |
| Rmd | 8ca4c7e | reneeisnowhere | 2023-04-10 | first rmd commit |
| Rmd | 2e69969 | reneeisnowhere | 2023-04-10 | adding data |
| Rmd | 0f1f1da | reneeisnowhere | 2023-04-10 | final run analysis |
This starts the documentation of the RNA-seq cardiotoxicity analysis for my manuscript
library(edgeR)#
library(limma)#
library(RColorBrewer)
library(mixOmics)
library(gridExtra)#
library(reshape2)#
library(data.table)
library(AnnotationHub)
library(tidyverse)
library(scales)
library(biomaRt)#
library(Homo.sapiens)
library(cowplot)#
library(ggrepel)#
library(corrplot)
library(Hmisc)
library(ggpubr) ###now we add genenames to the geneid###
geneid <- rownames(mymatrix) ### pulls the names we have in the counts file
genes <- select(Homo.sapiens, keys=geneid, columns=c("SYMBOL"),
keytype="ENTREZID")
genes <- genes[!duplicated(genes$ENTREZID),]
mymatrix$genes <- genes
#saveRDS(mymatrix, "data/allmatrix.RDS")
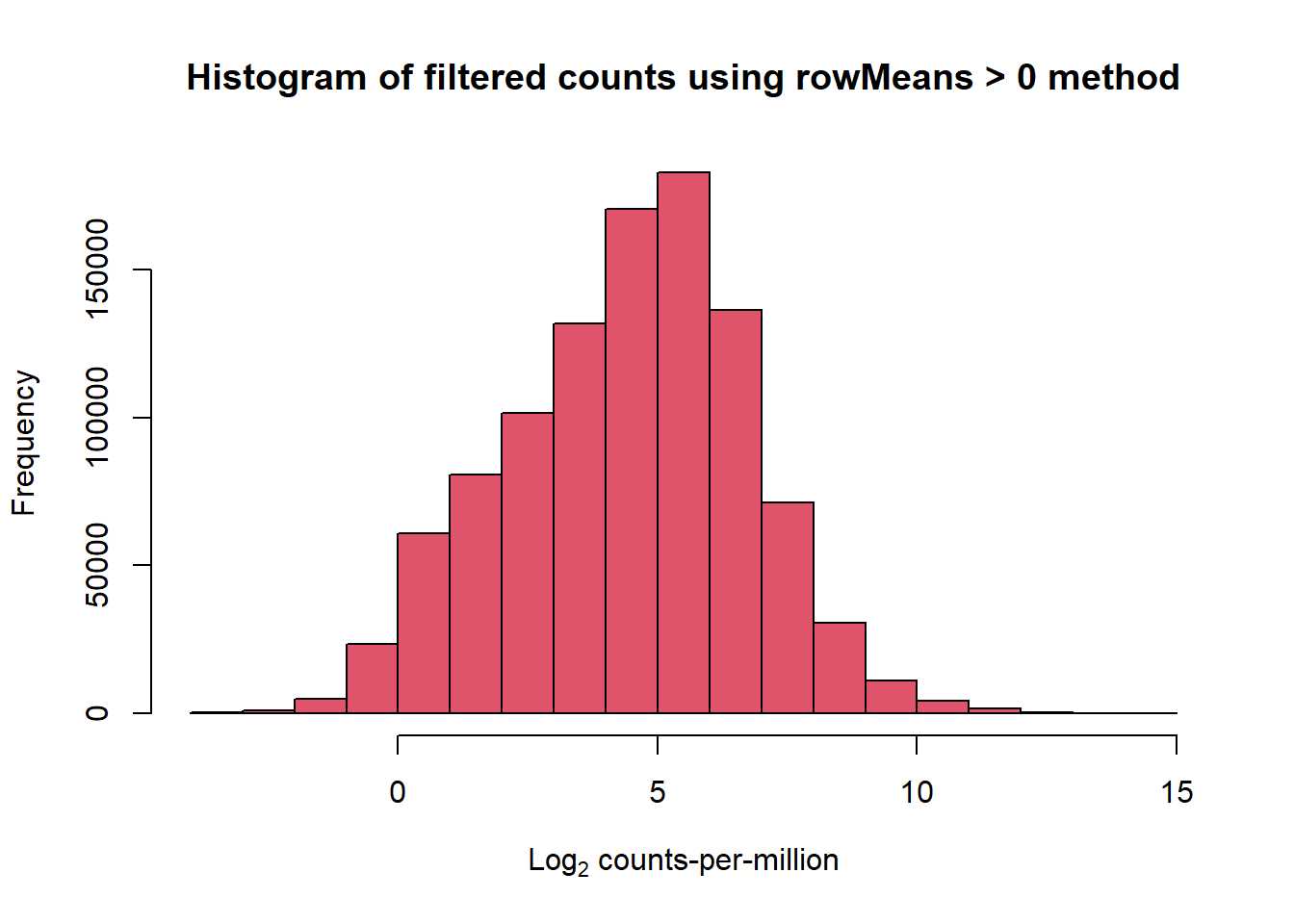
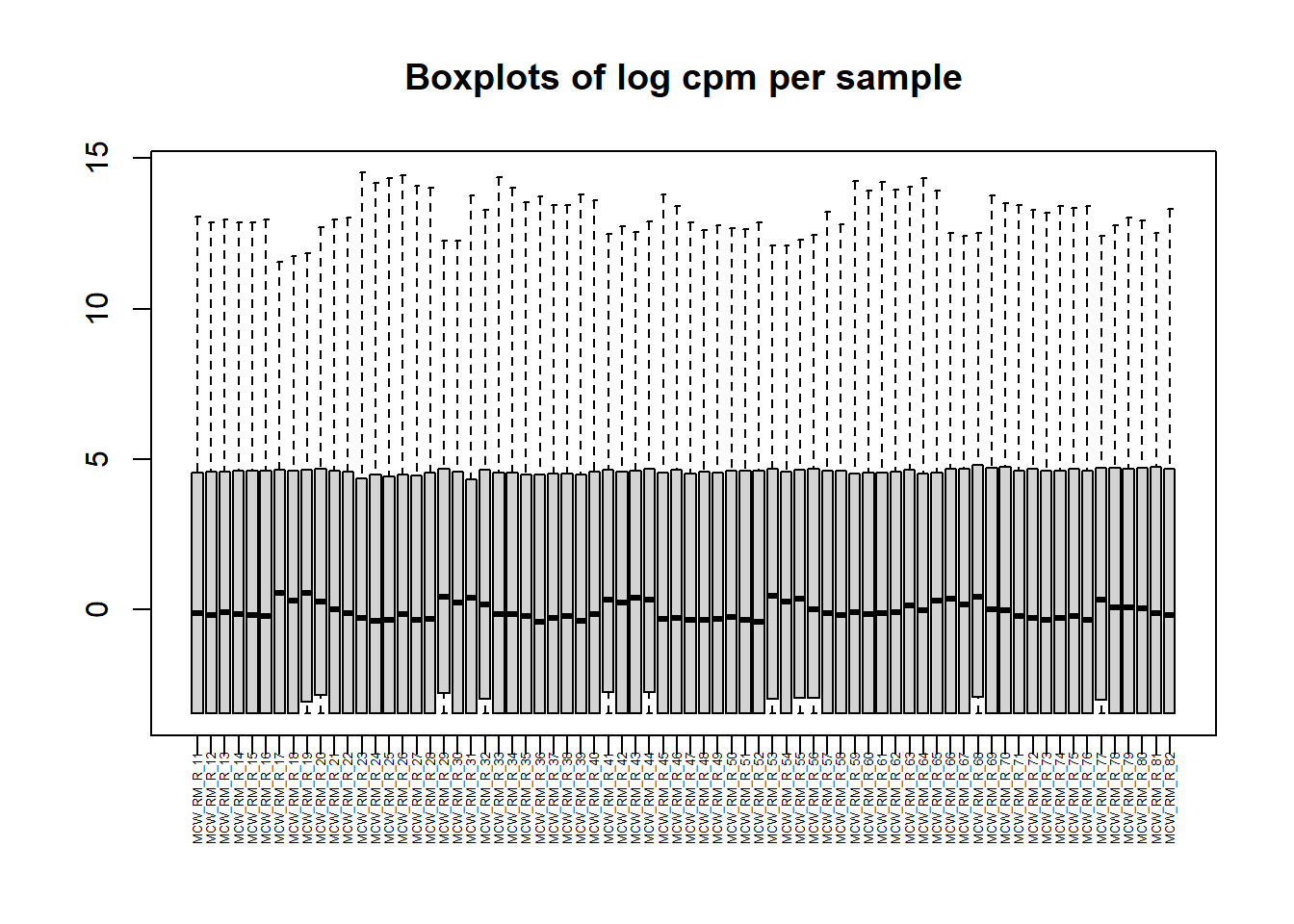
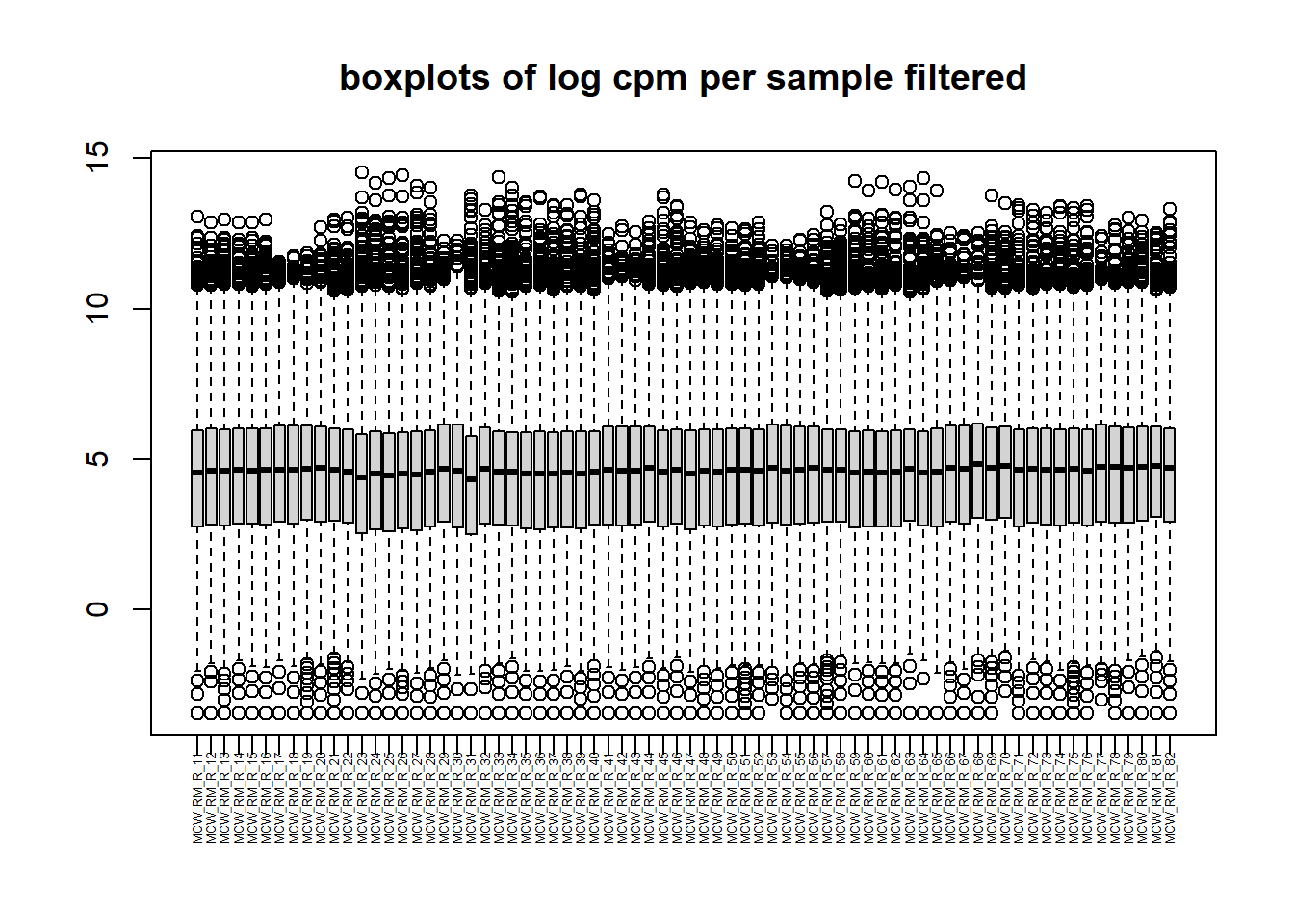
##note-not filtered!Initial RNA-seq quality checks
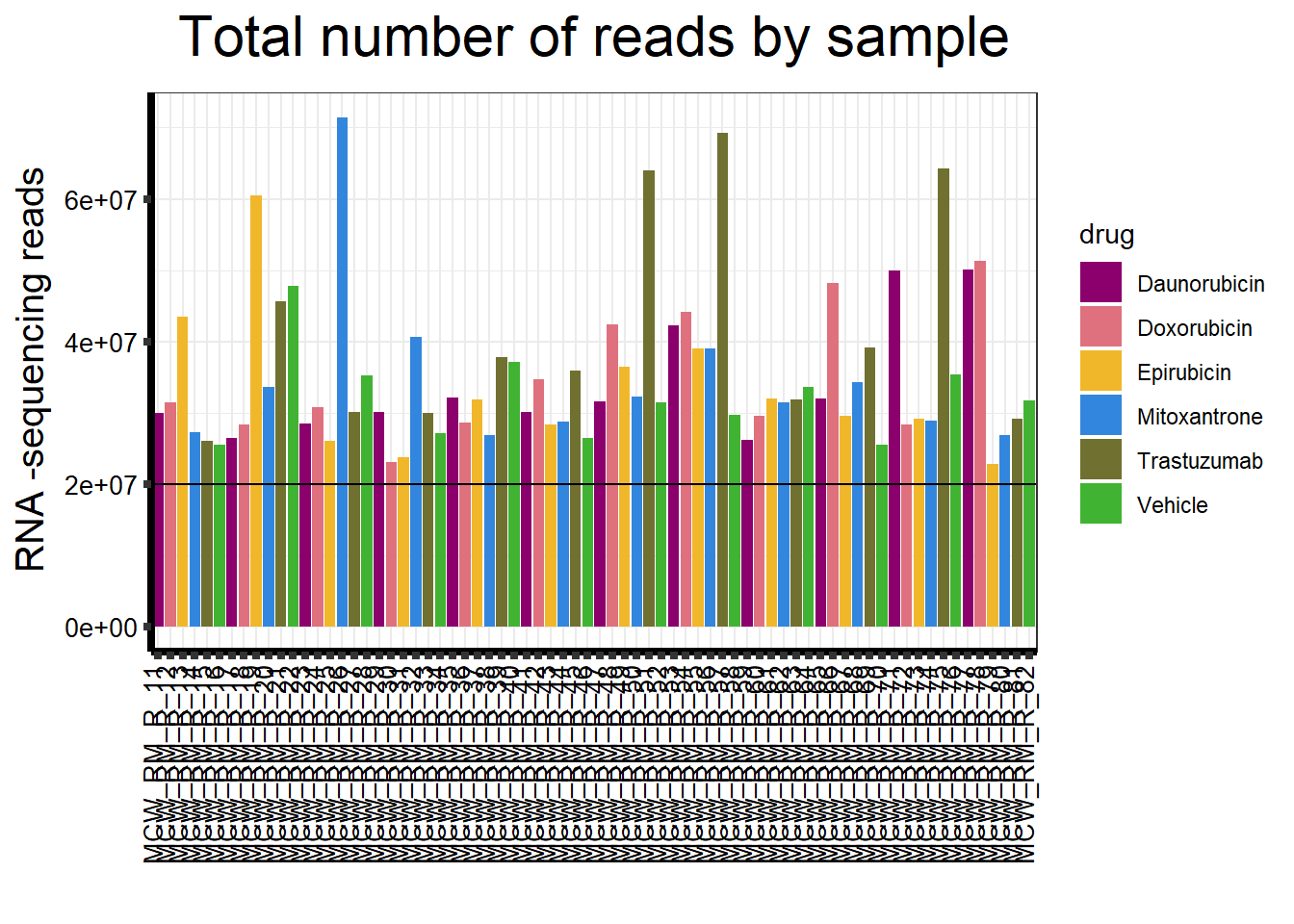
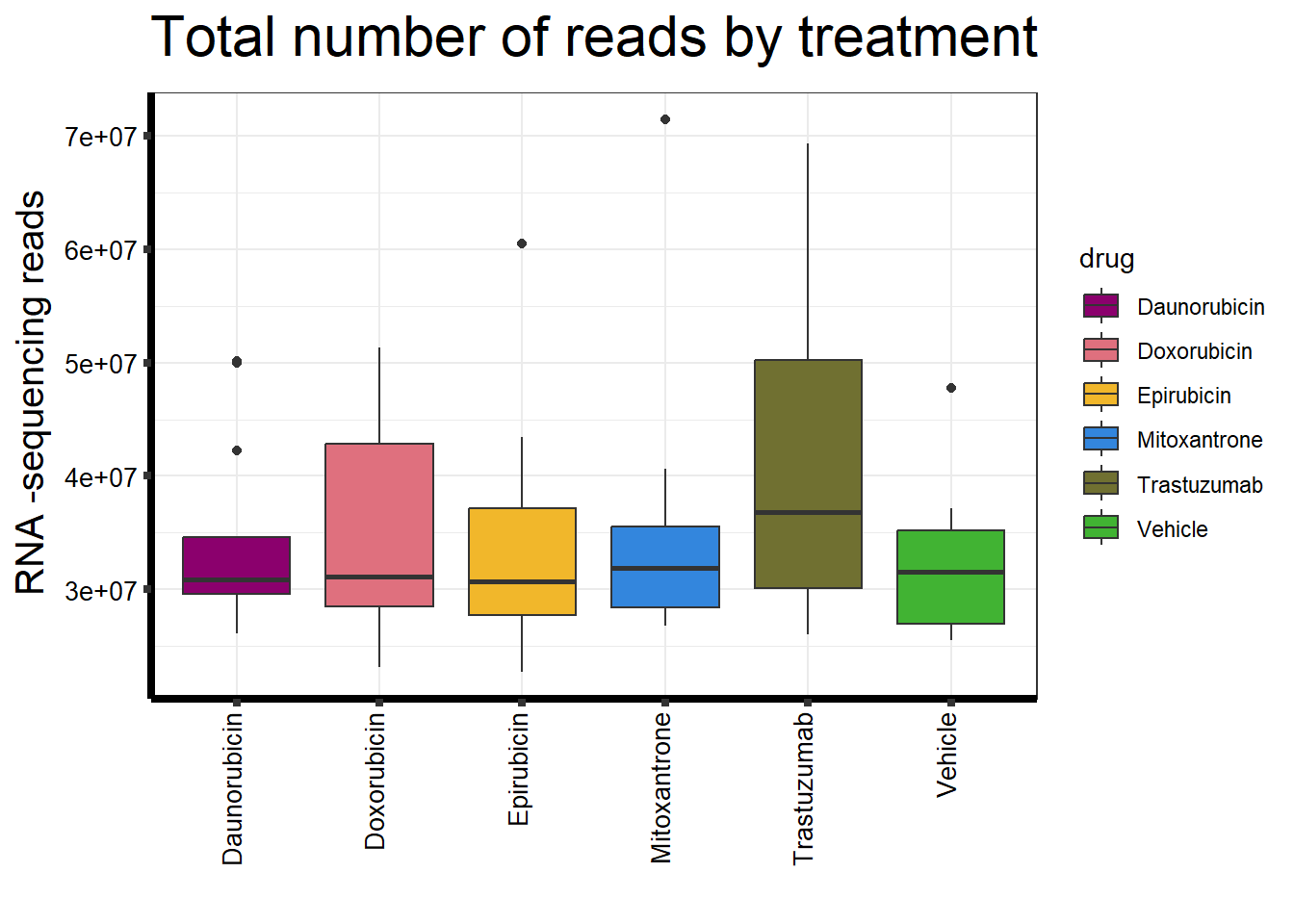
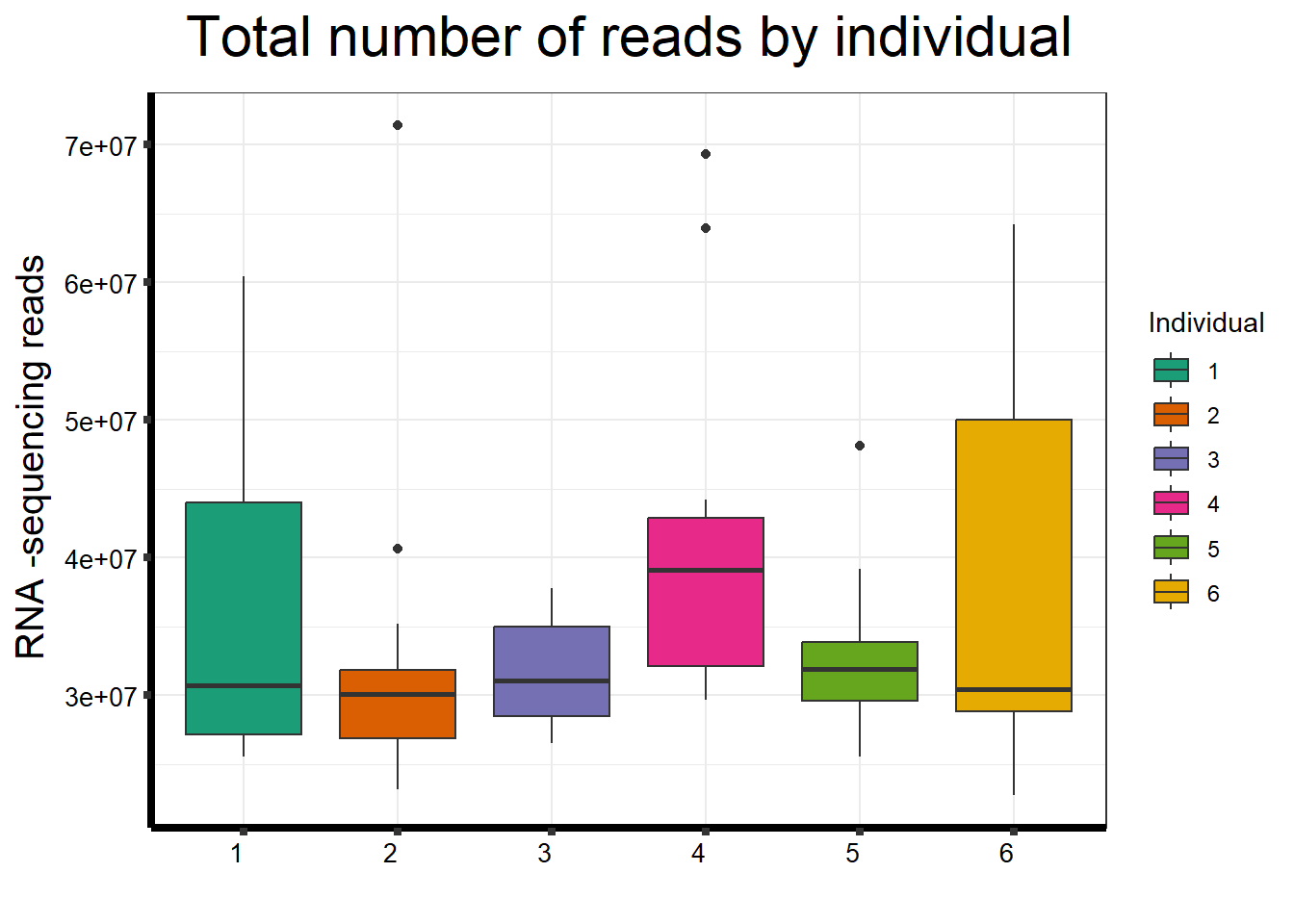
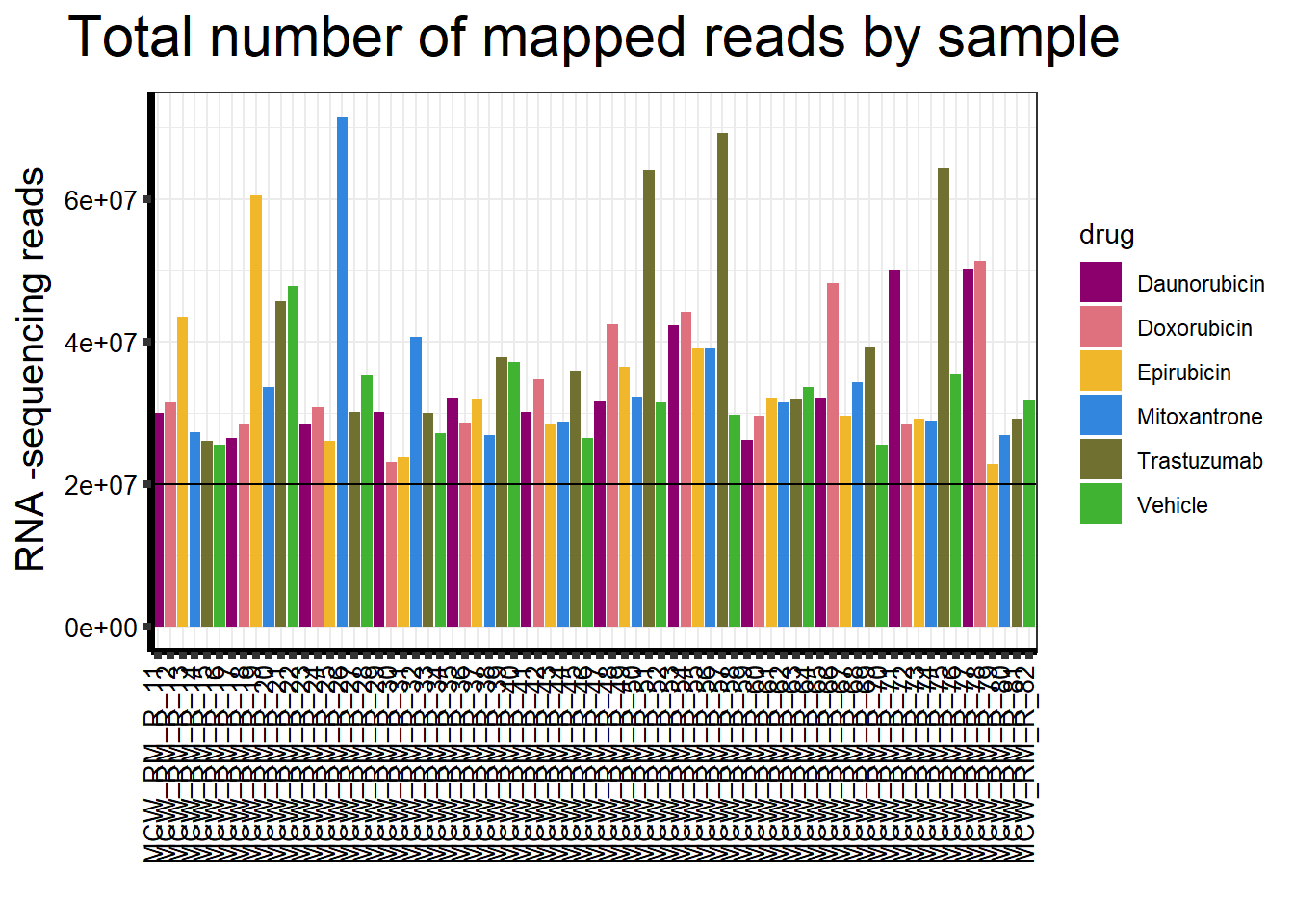
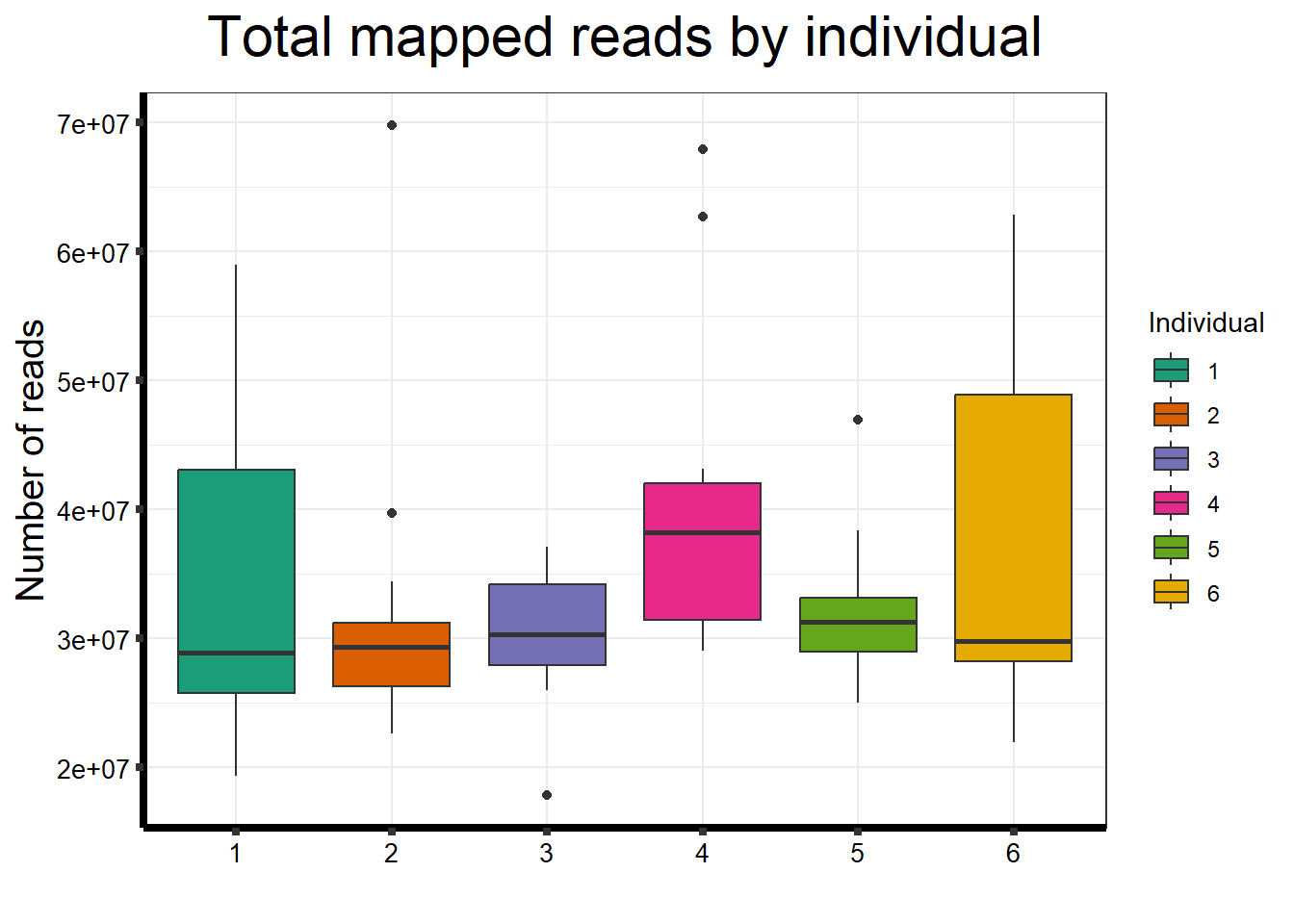
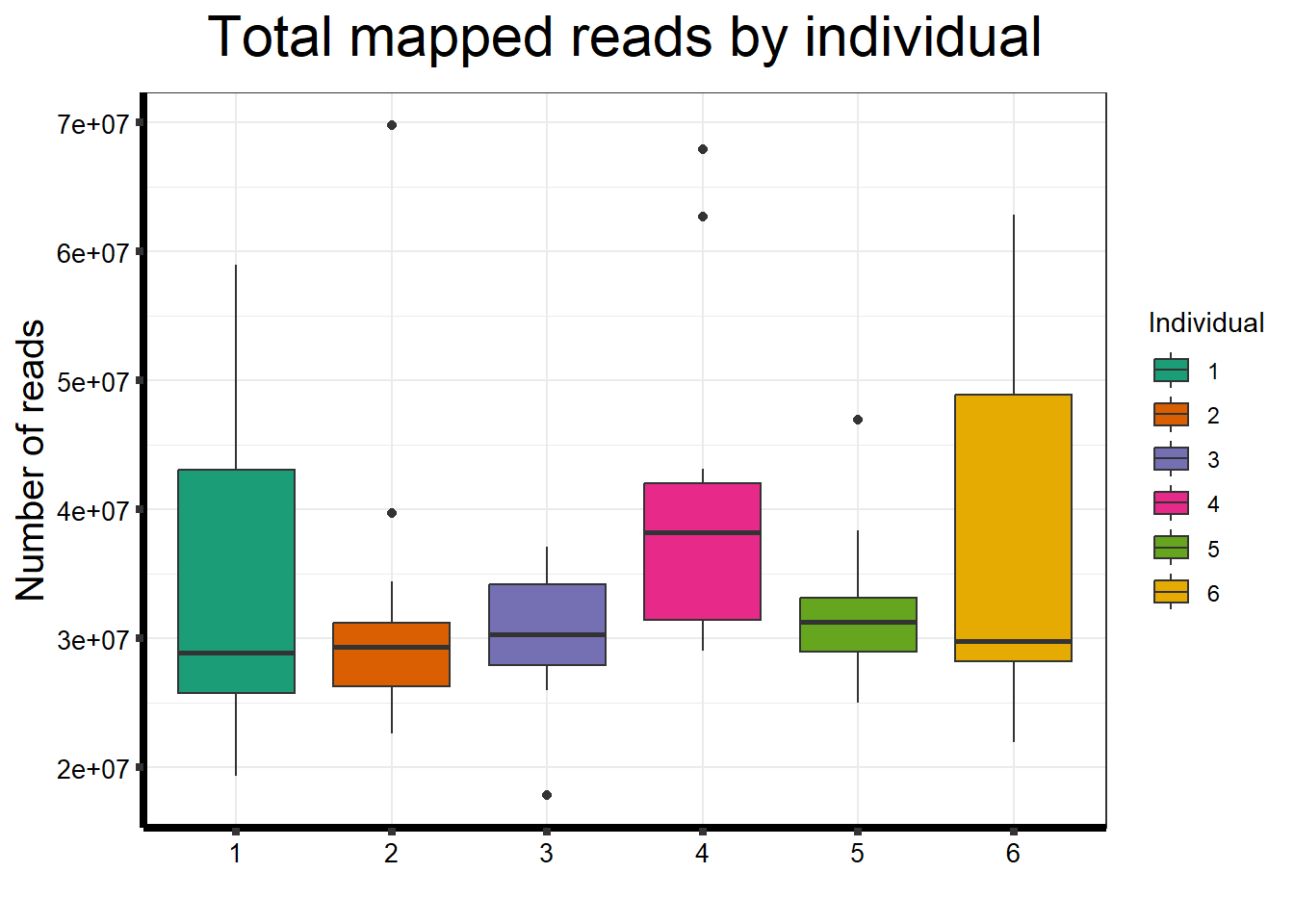
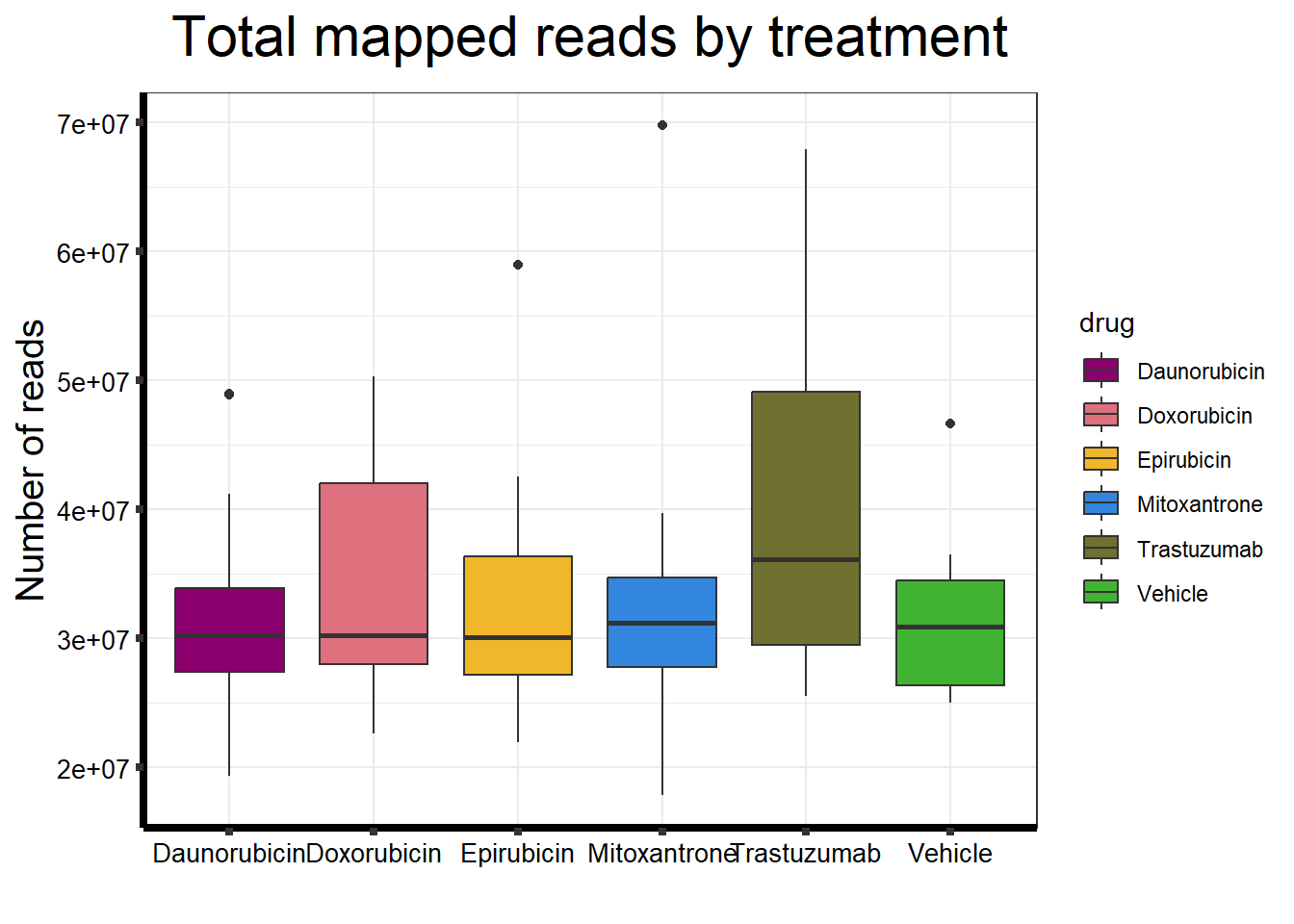
Initial histograms from count matrix
[1] 28395 72[1] 14084 72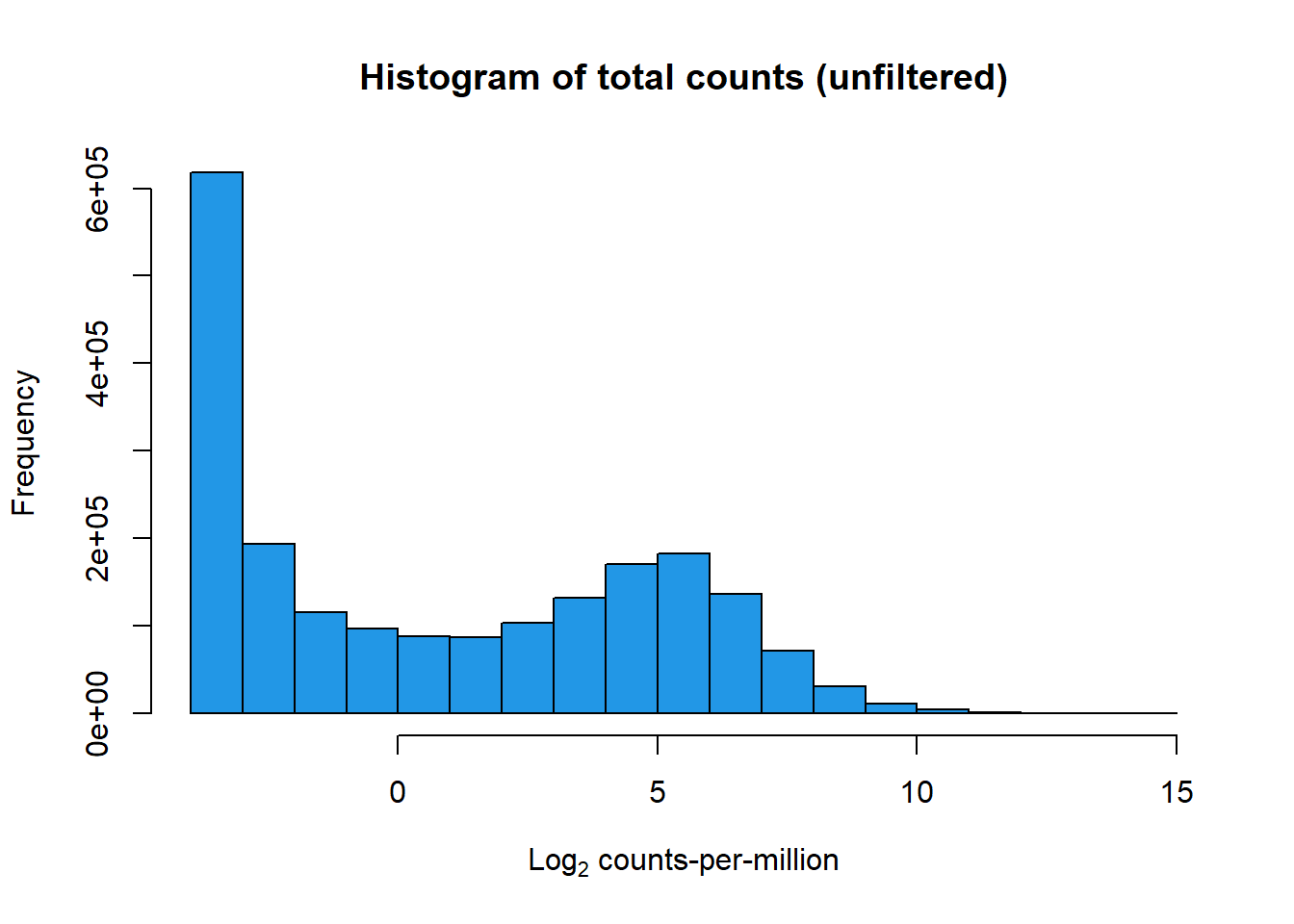
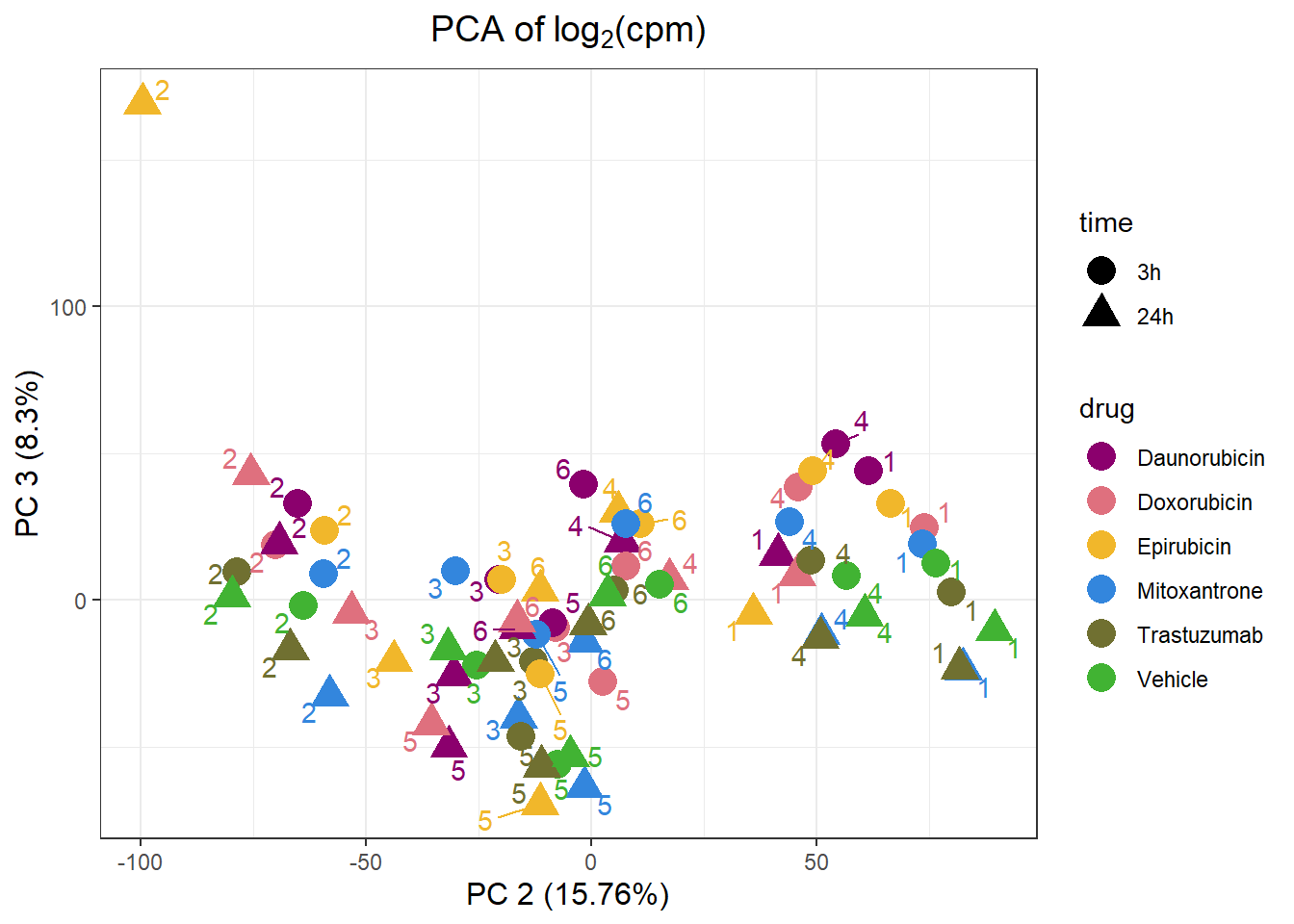
PCA by treatment and as a whole
### Vehicle 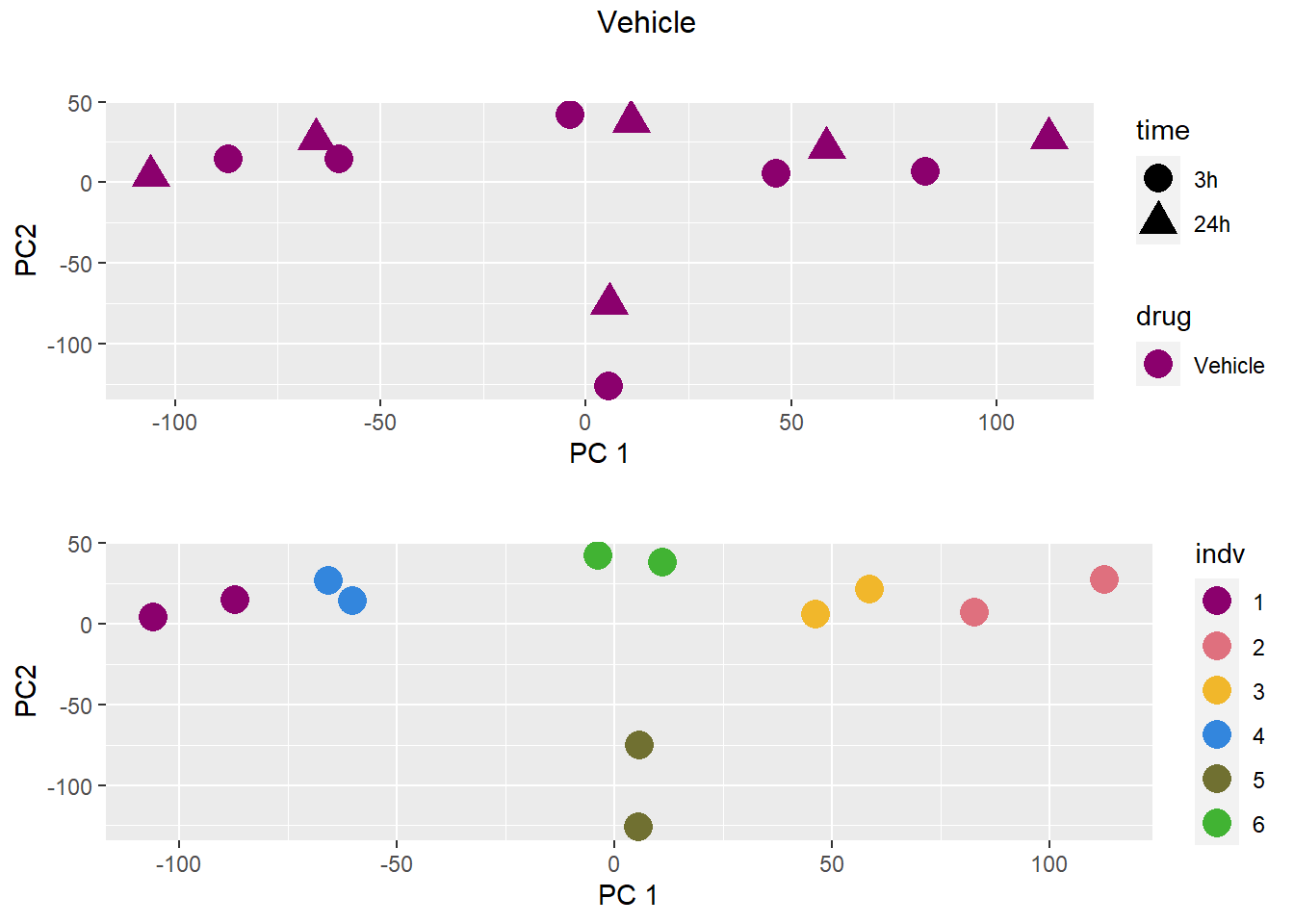
### Daunorubicin 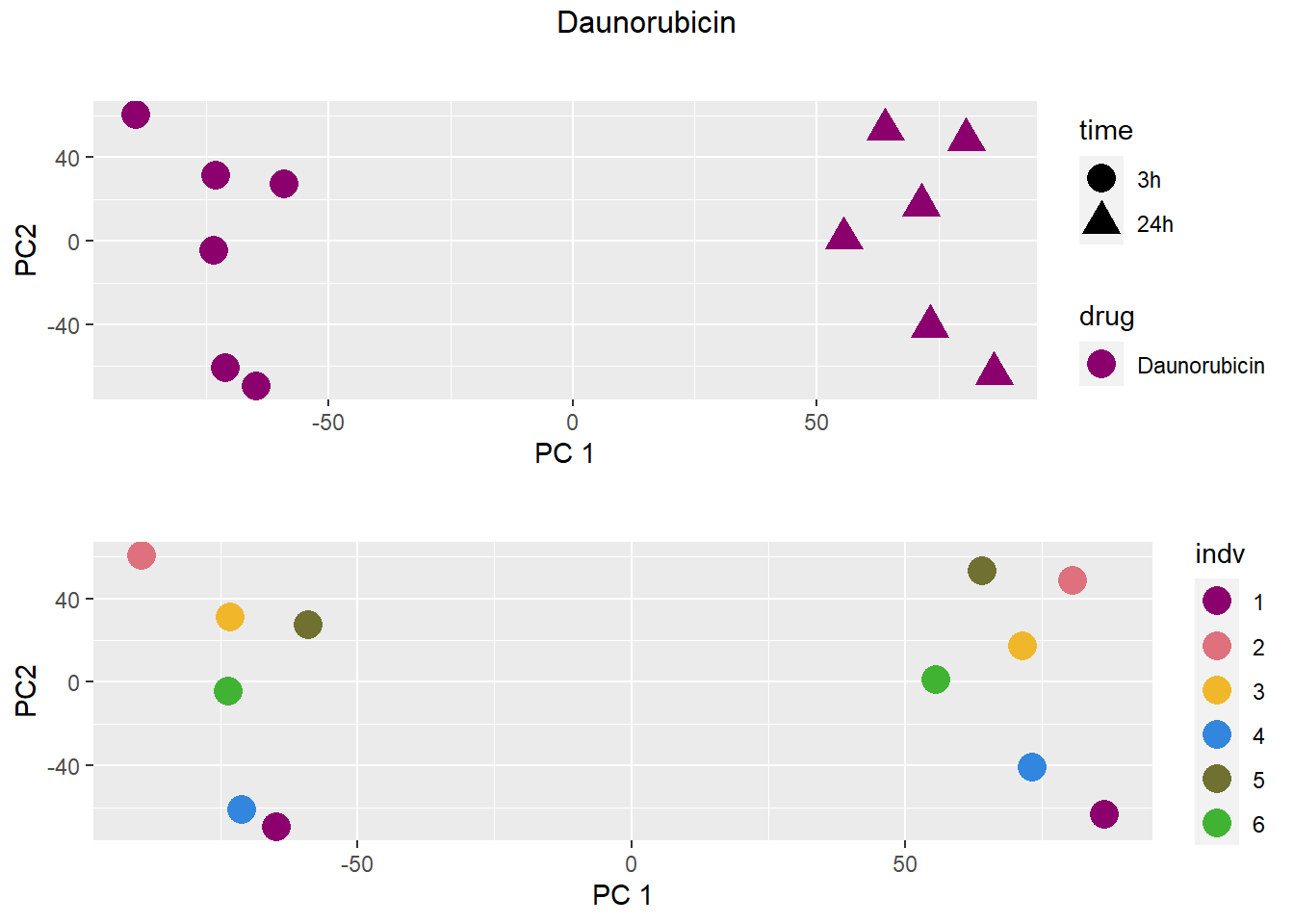
### Doxorubicin 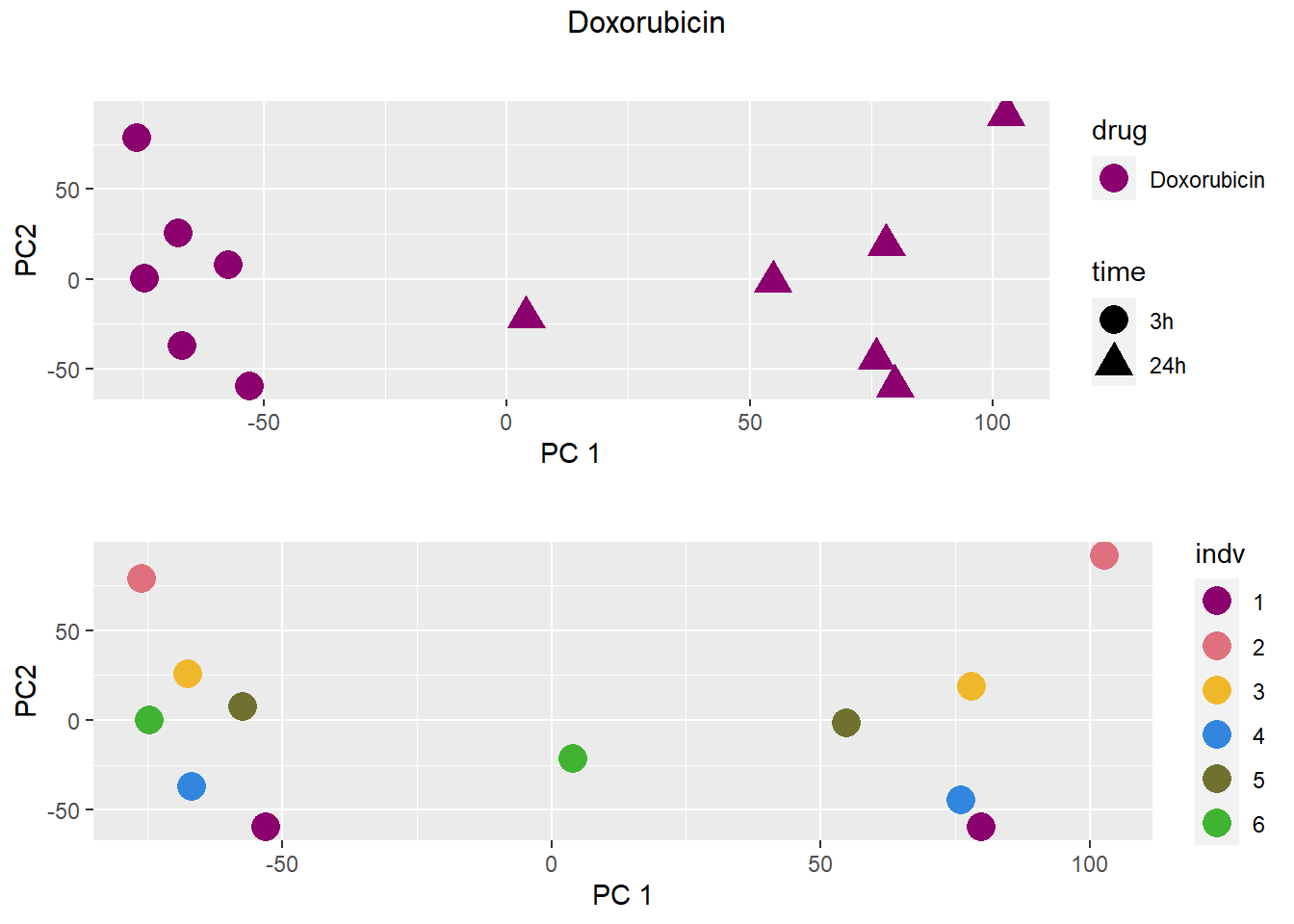
### Epirubicin 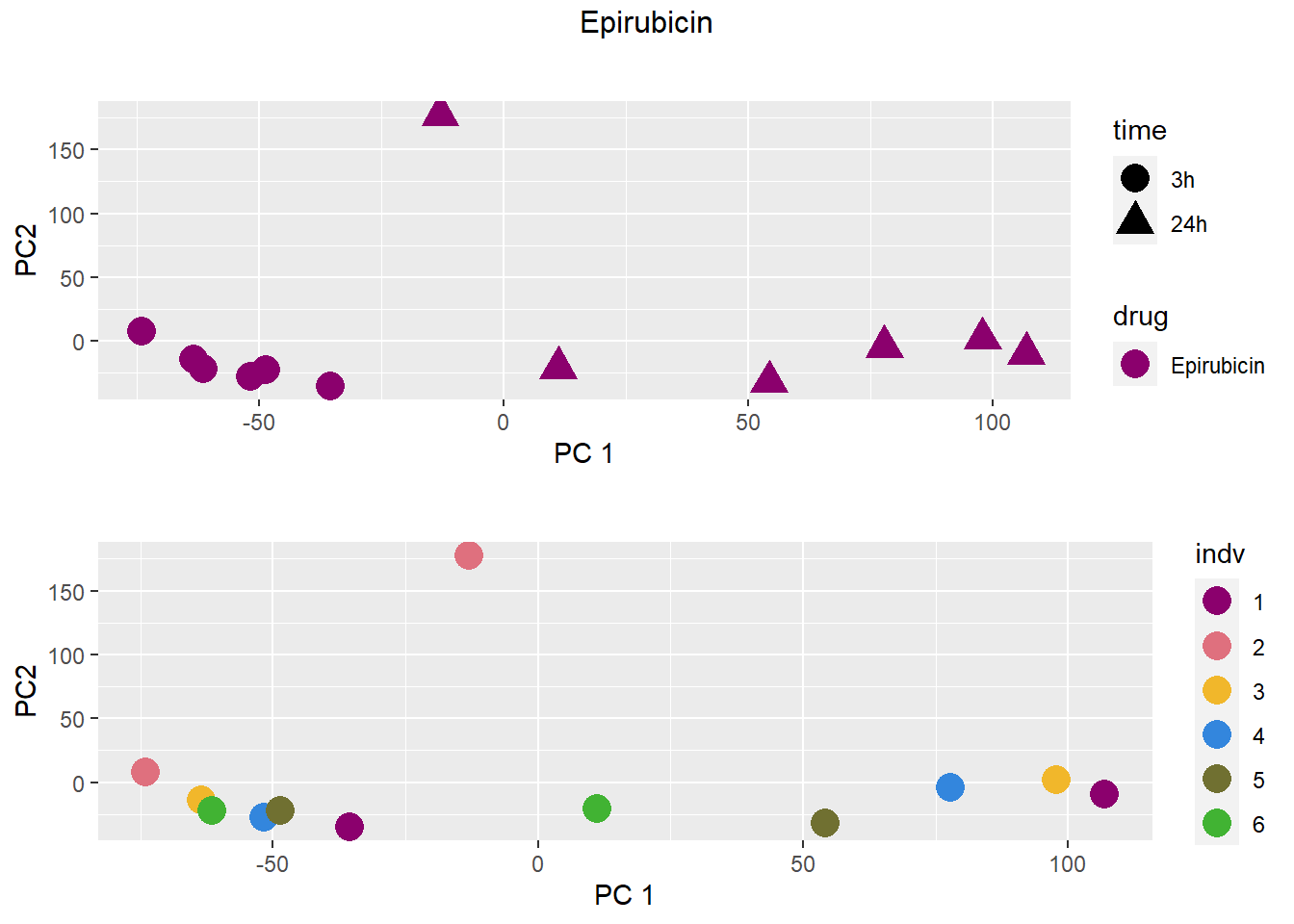
### Mitoxantrone 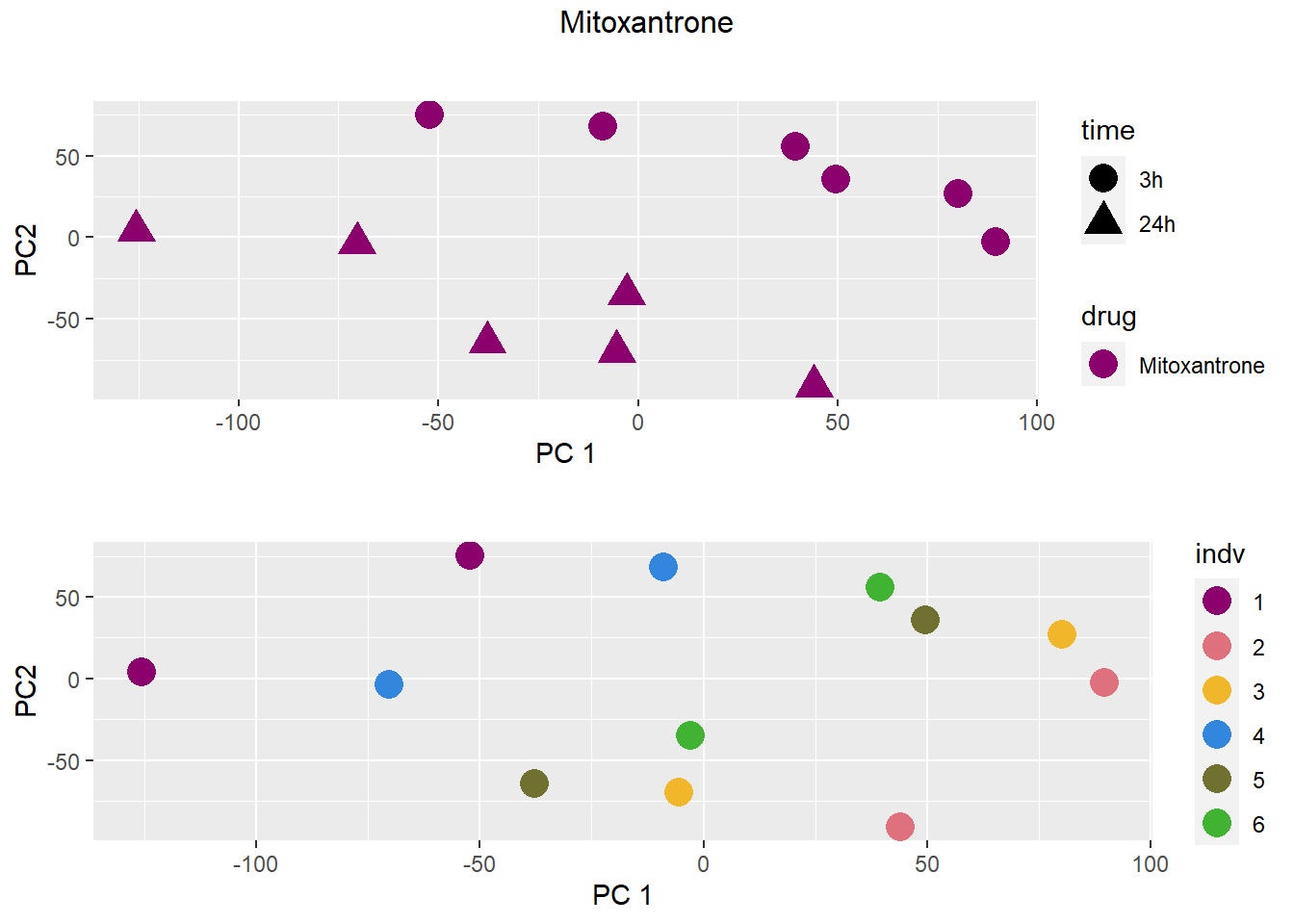
### Trastuzumab 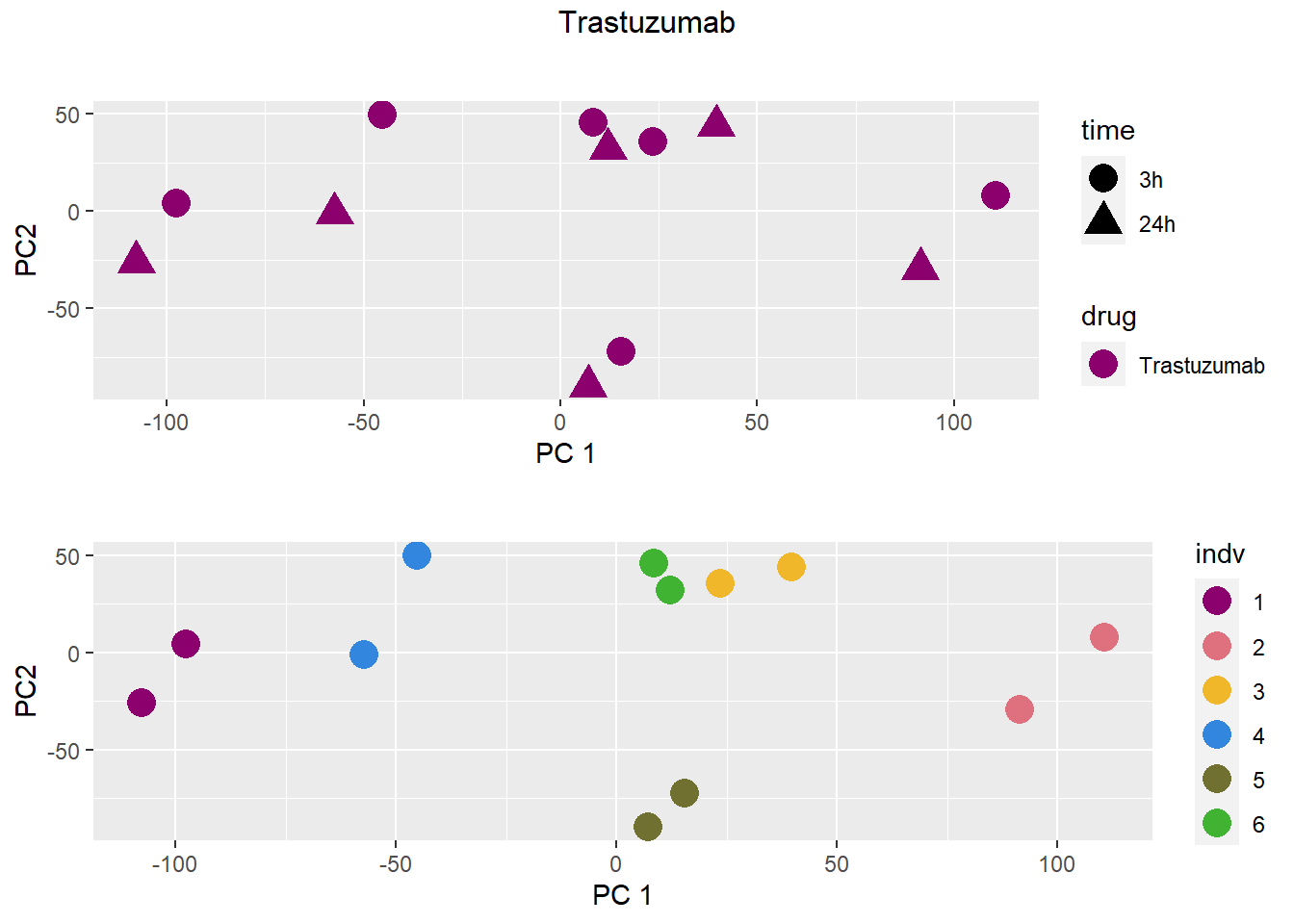
samplenames indv drug time RIN group PC1 PC2
Da.1.3h MCW_RM_R_11 1 Daunorubicin 3h 9.3 1 -18.33154 61.71013
Do.1.3h MCW_RM_R_12 1 Doxorubicin 3h 9.8 2 -12.36280 73.97678
Ep.1.3h MCW_RM_R_13 1 Epirubicin 3h 9.8 3 -11.16205 66.48794
Mi.1.3h MCW_RM_R_14 1 Mitoxantrone 3h 10 4 -10.19948 73.48343
Tr.1.3h MCW_RM_R_15 1 Trastuzumab 3h 9.6 5 -12.17619 80.01454
Ve.1.3h MCW_RM_R_16 1 Vehicle 3h 9.9 6 -14.98226 76.62199
PC3 PC4 PC5 PC6
Da.1.3h 44.039139 -4.547031 24.642107 -35.03245
Do.1.3h 24.576395 -8.626528 -19.908580 -18.97447
Ep.1.3h 33.025628 -9.349549 18.083569 -43.06551
Mi.1.3h 19.016766 -14.639651 -9.065324 -24.29908
Tr.1.3h 2.640624 -17.019296 -34.253925 -11.77881
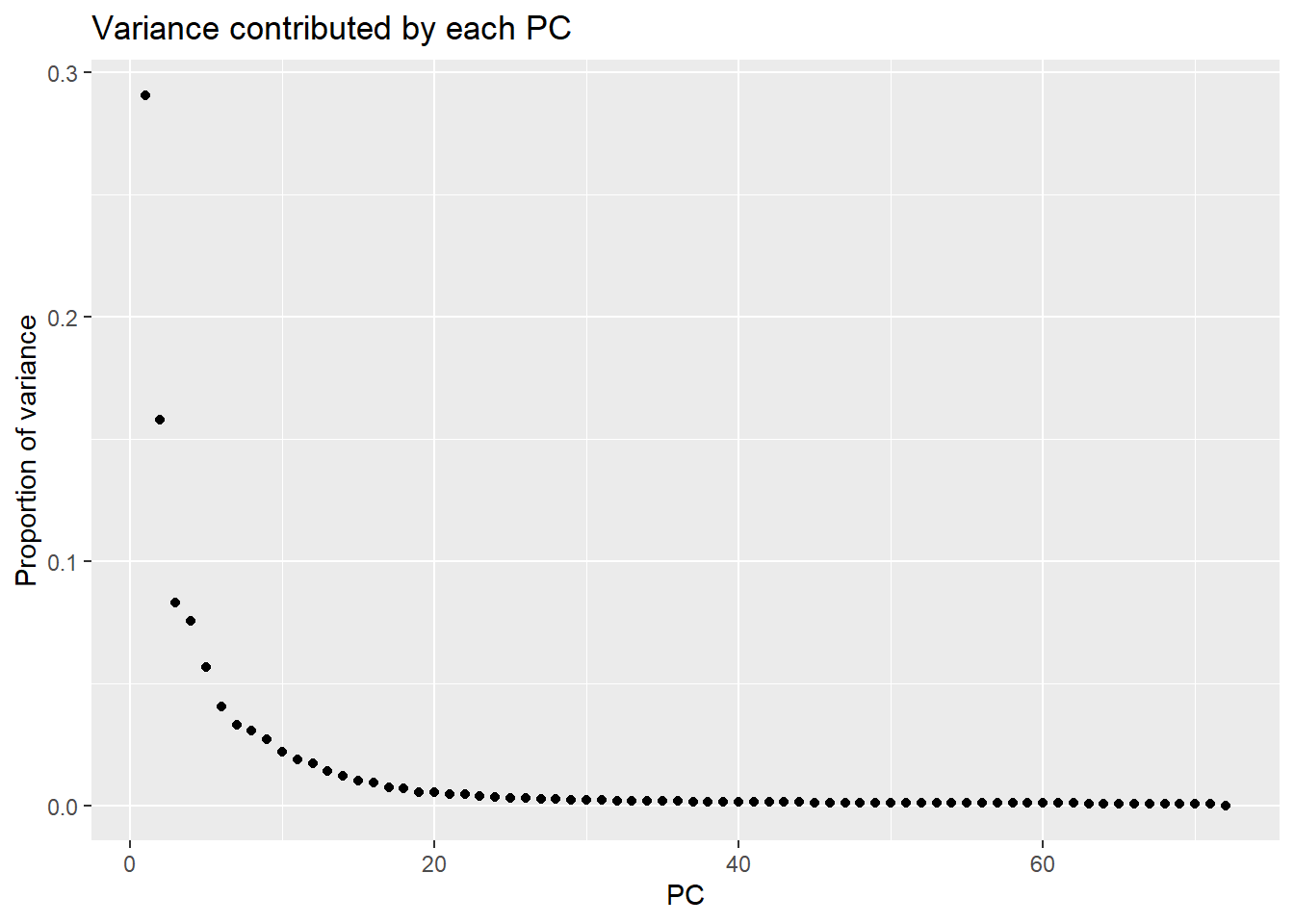
Ve.1.3h 12.706808 -4.173412 -39.846595 -17.16213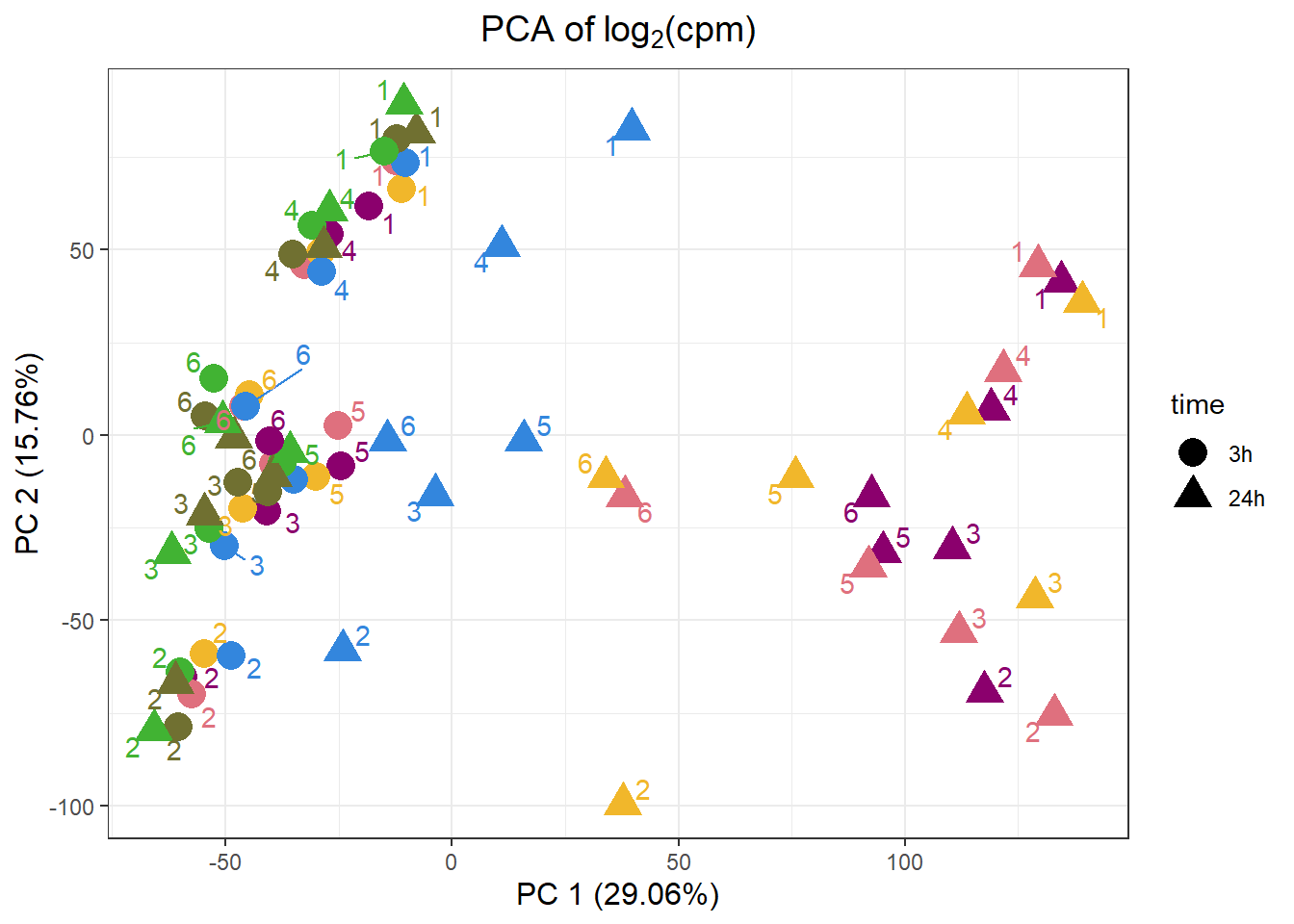
PC1 PC2 PC3 PC4 PC5 PC6
Standard deviation 63.98853 47.11608 34.21502 32.58775 28.22245 23.90977
Proportion of Variance 0.29072 0.15762 0.08312 0.07540 0.05655 0.04059
Cumulative Proportion 0.29072 0.44834 0.53146 0.60687 0.66342 0.70401
PC7
Standard deviation 21.56133
Proportion of Variance 0.03301
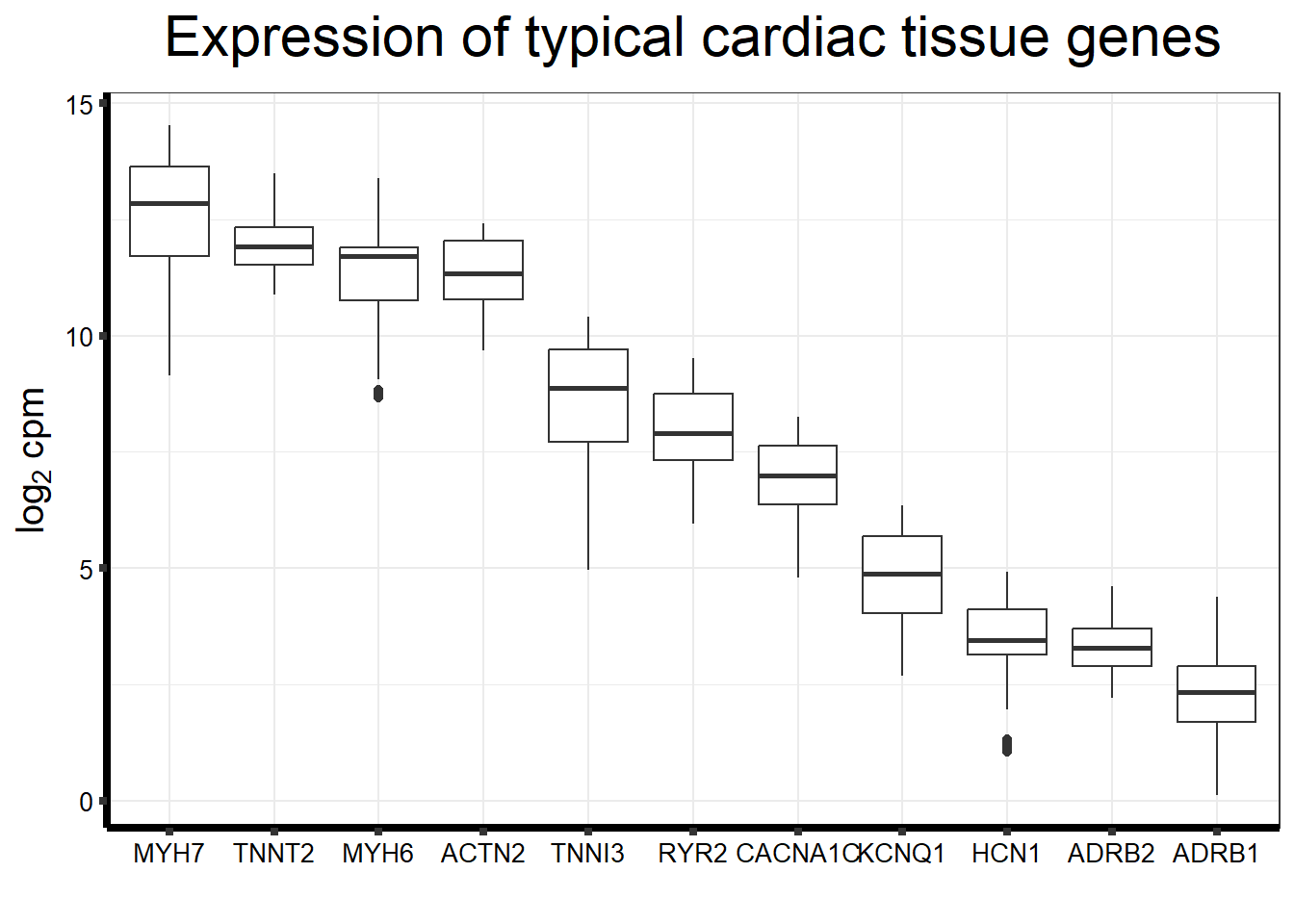
Cumulative Proportion 0.73702Typical genes expressed in iPSC-CMS
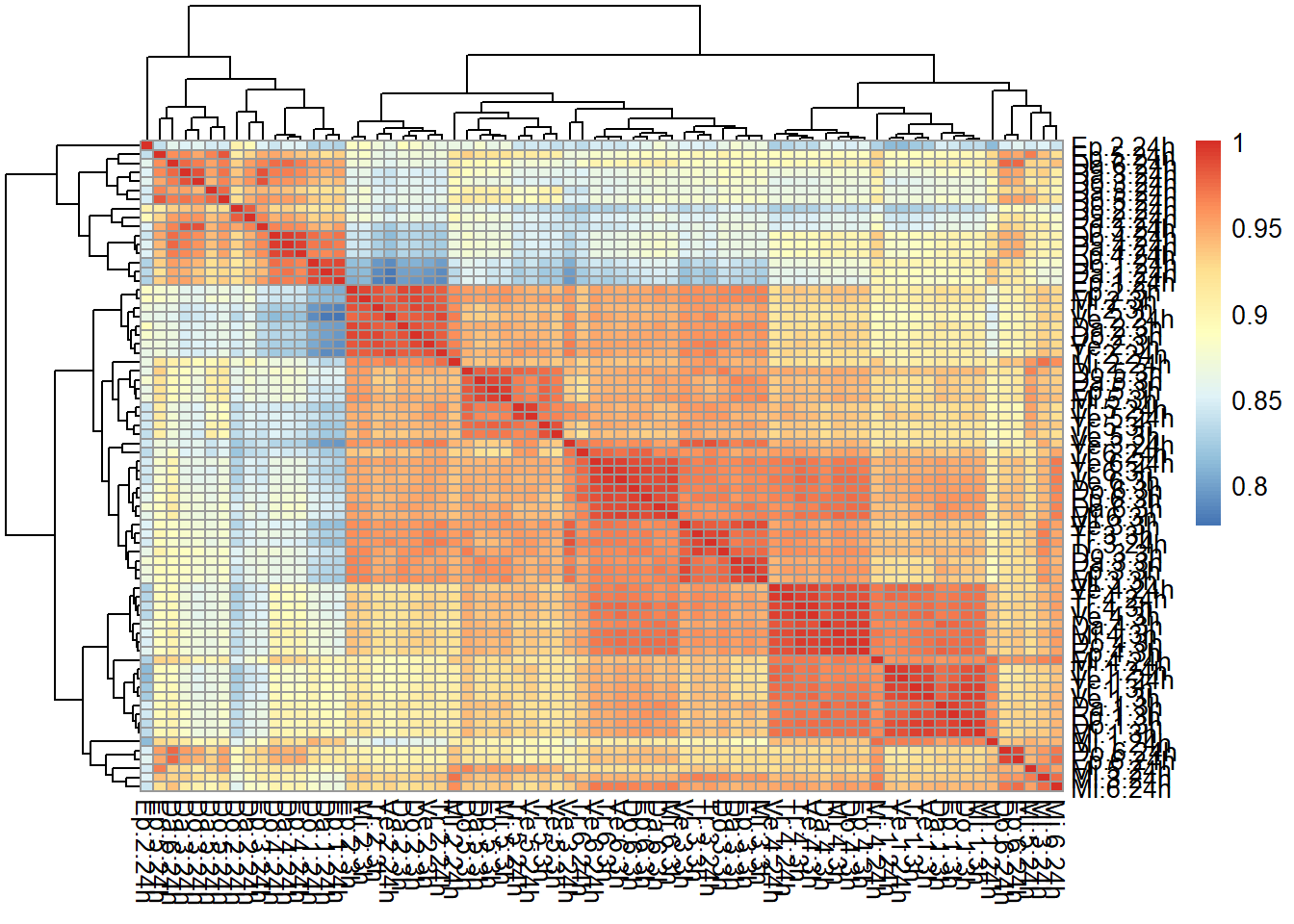
correlation heatmap of counts matrix
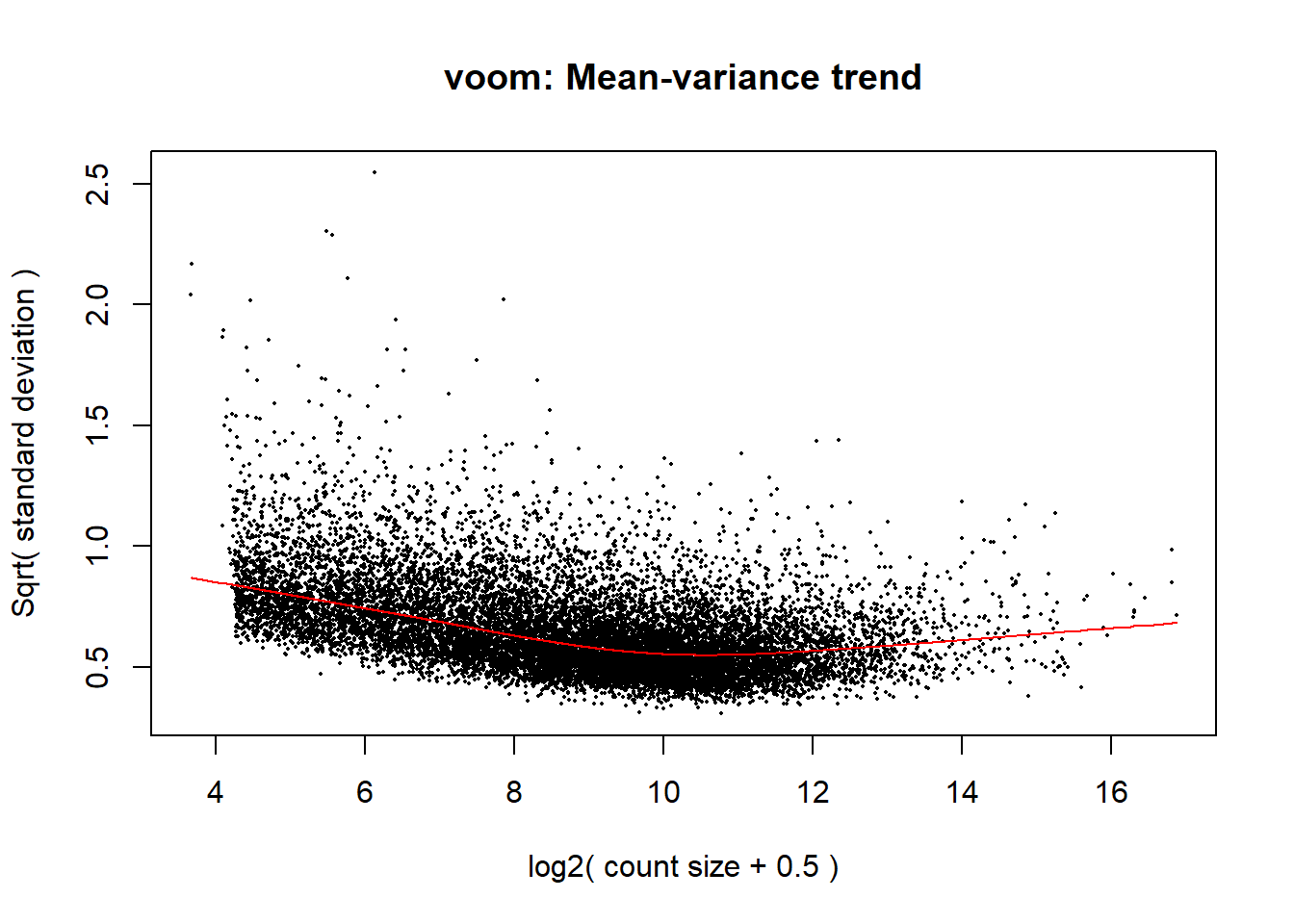
now to get the counts set for DEG!!
DEG analysis
summary
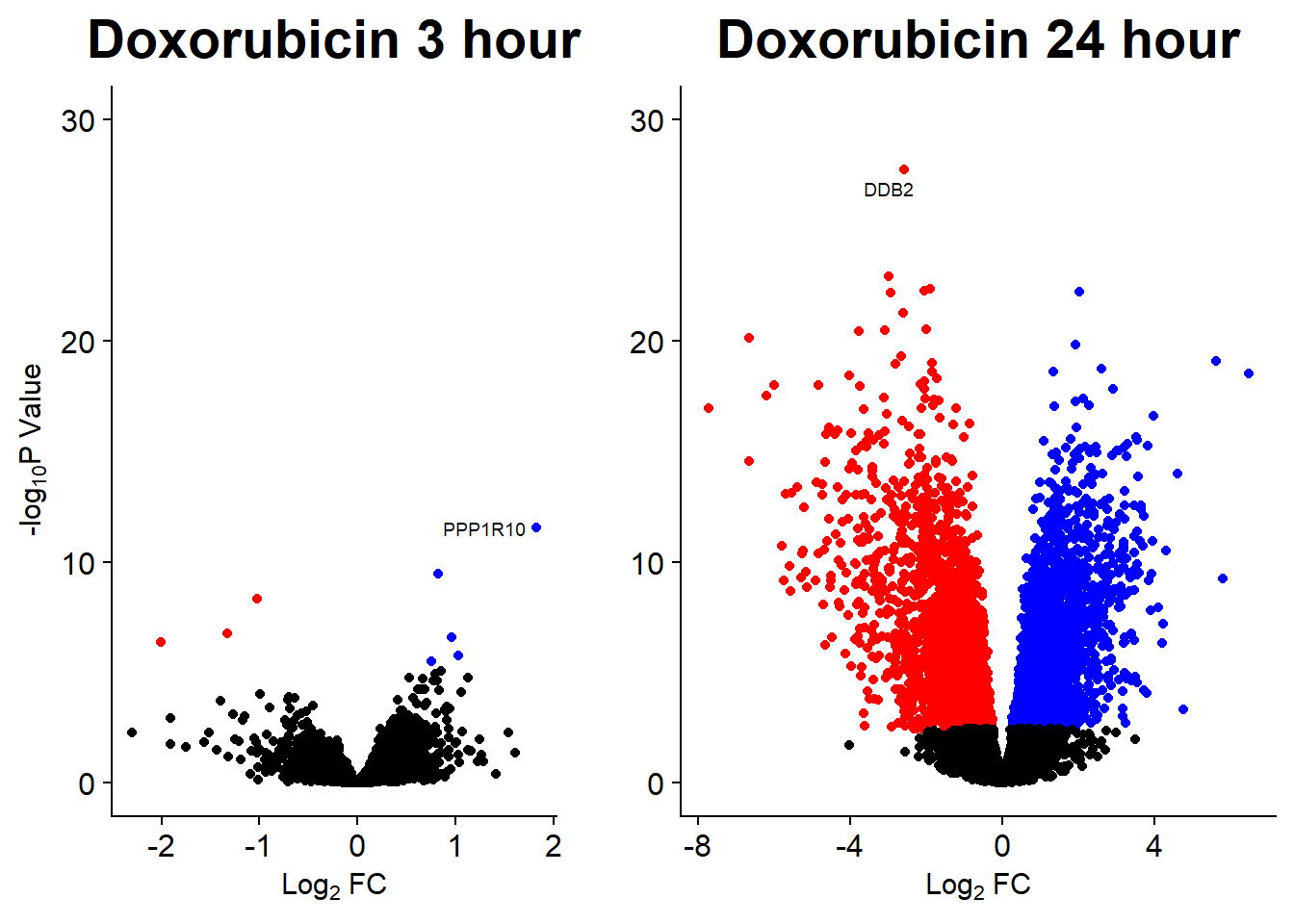
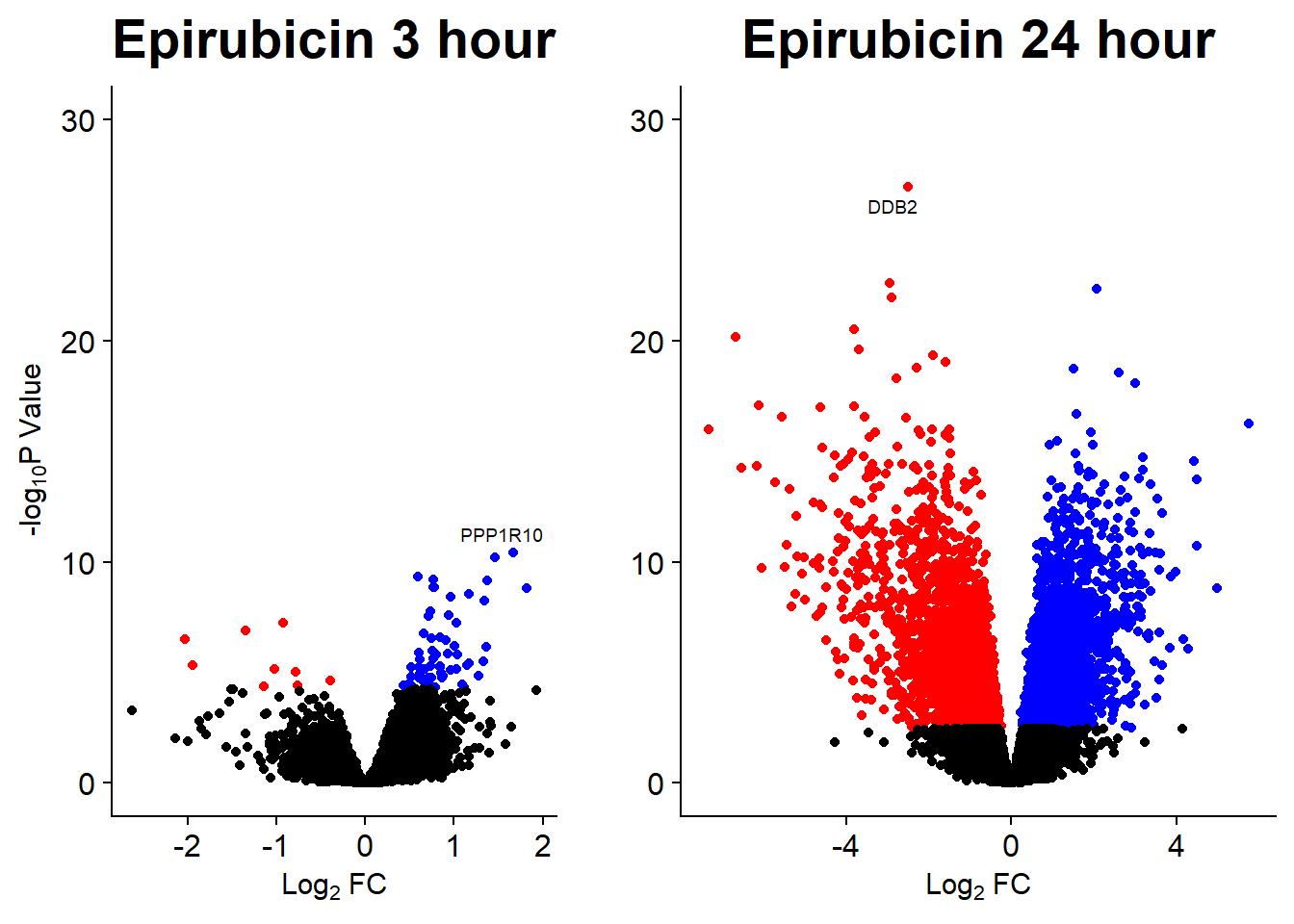
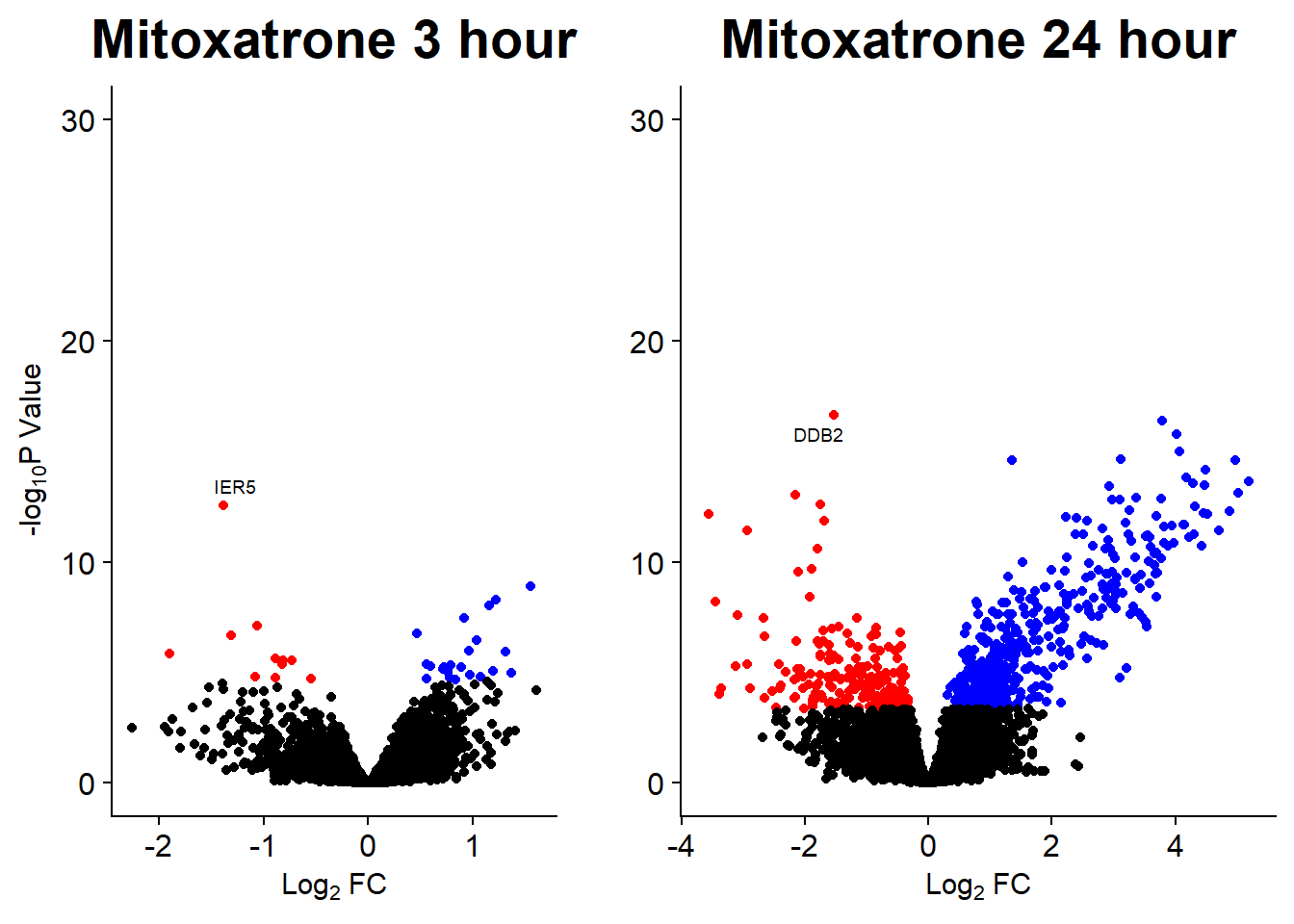
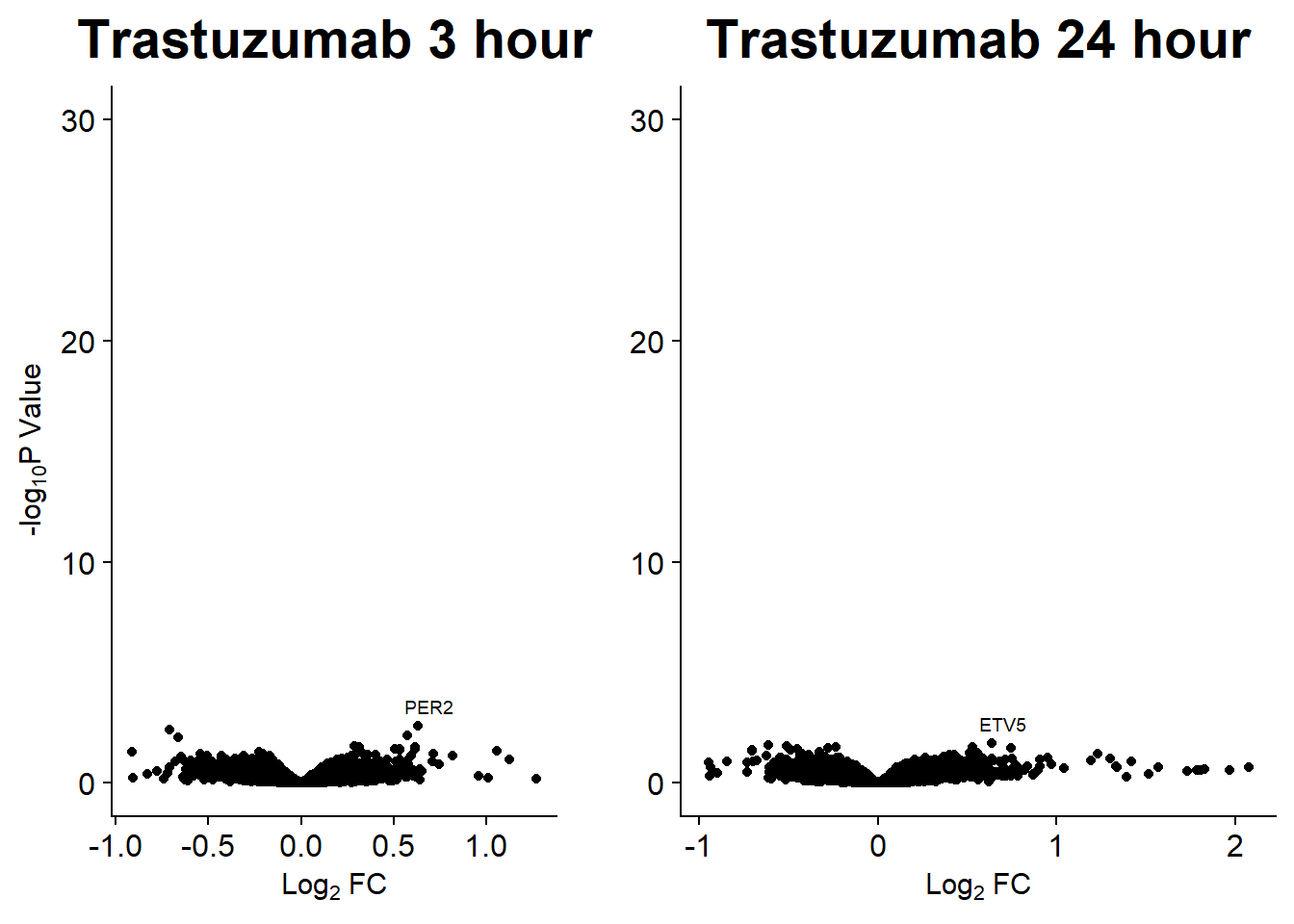
V.DA V.DX V.EP V.MT V.TR V.DA24 V.DX24 V.EP24 V.MT24 V.TR24
Down 89 4 23 19 0 3722 3386 2953 849 0
NotSig 13529 14068 13864 14026 14084 7220 7568 7882 12757 14084
Up 466 12 197 39 0 3142 3130 3249 478 0cormotif analysis More In-depth
written summary so far: Response sets look similar to previous results. This data is based on the filtered count matrix (using rowmeans>0 of cpm(log=true)). Classification of patterns appear to be:
motif 1- No Response set: 7504 (gene list made by filtering likelihood of gene belonging to cluster 1 <0.5)
motif 2- Time-independent Top2\(\beta\)i response cluster: 528 (gene list made by filtering likelihood of gene belonging to cluster 2 <0.5)
motif 3- Early Top2\(\beta\)i response cluster: 444 (gene list made by filtering likelihood of gene belonging to cluster 3 <0.5)
motif 4- Late Top2\(\beta\)i response cluster: 5545 (gene list made by filtering likelihood of gene belonging to cluster 4 <0.5)
NOTE: these are based on the most recent counts (motif numbers have changed a little)
More analysis on corMotif (aka Baysian gene anaylsis can be found on this page:) CorMotif
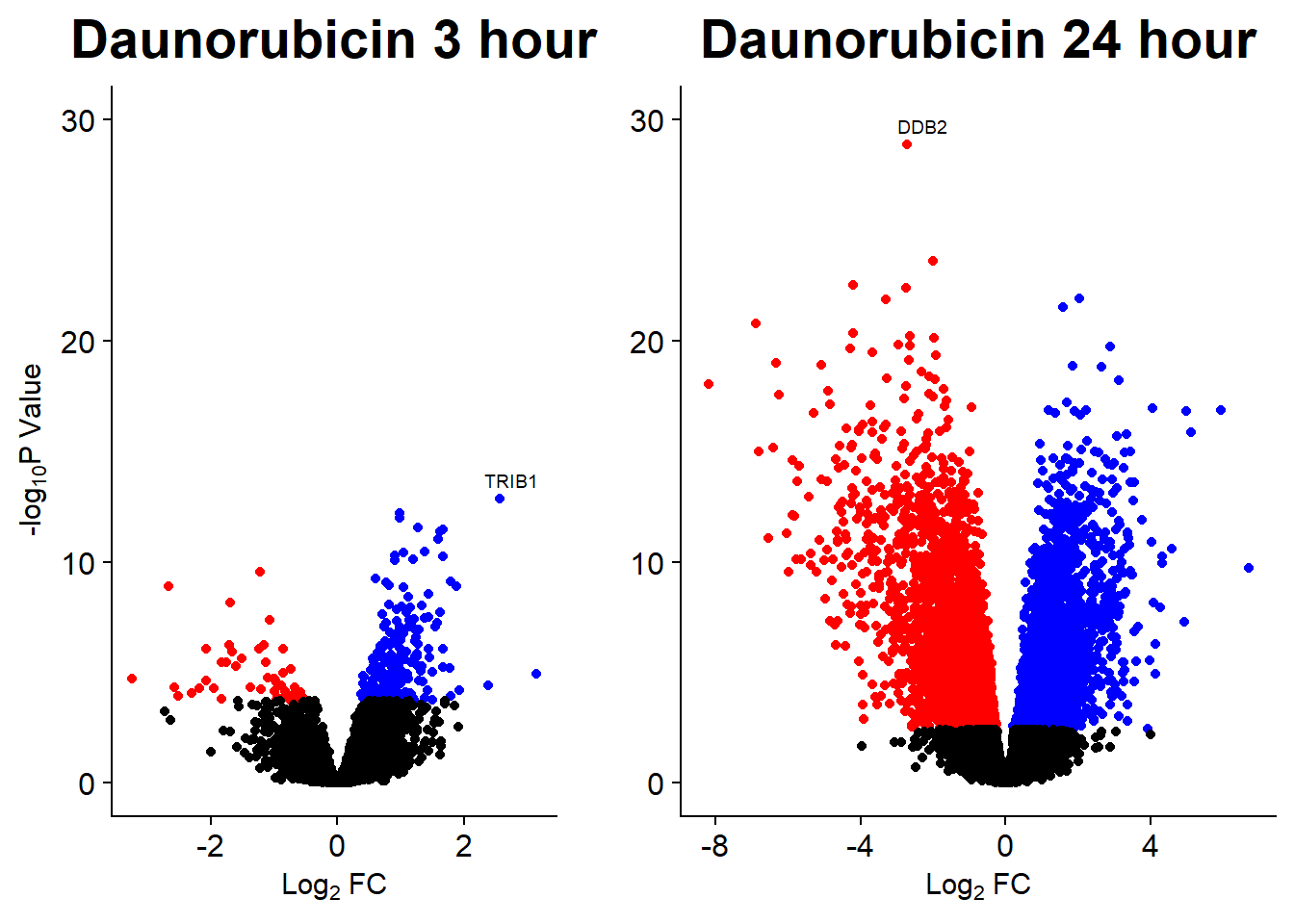
Volcano plots from pairwise gene analysis
R version 4.2.2 (2022-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] ggpubr_0.6.0
[2] Hmisc_4.8-0
[3] Formula_1.2-5
[4] survival_3.5-3
[5] corrplot_0.92
[6] ggrepel_0.9.3
[7] cowplot_1.1.1
[8] Homo.sapiens_1.3.1
[9] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[10] org.Hs.eg.db_3.15.0
[11] GO.db_3.15.0
[12] OrganismDbi_1.38.1
[13] GenomicFeatures_1.48.4
[14] GenomicRanges_1.48.0
[15] GenomeInfoDb_1.32.4
[16] AnnotationDbi_1.58.0
[17] IRanges_2.30.1
[18] S4Vectors_0.34.0
[19] Biobase_2.56.0
[20] biomaRt_2.52.0
[21] scales_1.2.1
[22] lubridate_1.9.2
[23] forcats_1.0.0
[24] stringr_1.5.0
[25] dplyr_1.1.0
[26] purrr_1.0.1
[27] readr_2.1.4
[28] tidyr_1.3.0
[29] tibble_3.1.8
[30] tidyverse_2.0.0
[31] AnnotationHub_3.4.0
[32] BiocFileCache_2.4.0
[33] dbplyr_2.3.1
[34] BiocGenerics_0.42.0
[35] data.table_1.14.8
[36] reshape2_1.4.4
[37] gridExtra_2.3
[38] mixOmics_6.20.0
[39] ggplot2_3.4.1
[40] lattice_0.20-45
[41] MASS_7.3-58.2
[42] RColorBrewer_1.1-3
[43] edgeR_3.38.4
[44] limma_3.52.4
[45] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] utf8_1.2.3 tidyselect_1.2.0
[3] RSQLite_2.3.0 htmlwidgets_1.6.1
[5] grid_4.2.2 BiocParallel_1.30.4
[7] munsell_0.5.0 codetools_0.2-19
[9] statmod_1.5.0 interp_1.1-3
[11] withr_2.5.0 colorspace_2.1-0
[13] filelock_1.0.2 highr_0.10
[15] knitr_1.42 rstudioapi_0.14
[17] ggsignif_0.6.4 MatrixGenerics_1.8.1
[19] labeling_0.4.2 git2r_0.31.0
[21] GenomeInfoDbData_1.2.8 pheatmap_1.0.12
[23] farver_2.1.1 bit64_4.0.5
[25] rprojroot_2.0.3 vctrs_0.5.2
[27] generics_0.1.3 xfun_0.37
[29] timechange_0.2.0 R6_2.5.1
[31] locfit_1.5-9.7 bitops_1.0-7
[33] cachem_1.0.7 DelayedArray_0.22.0
[35] promises_1.2.0.1 BiocIO_1.6.0
[37] nnet_7.3-18 gtable_0.3.1
[39] processx_3.8.0 rlang_1.0.6
[41] splines_4.2.2 rtracklayer_1.56.1
[43] rstatix_0.7.2 broom_1.0.3
[45] checkmate_2.1.0 BiocManager_1.30.20
[47] yaml_2.3.7 abind_1.4-5
[49] backports_1.4.1 httpuv_1.6.9
[51] RBGL_1.72.0 tools_4.2.2
[53] ellipsis_0.3.2 jquerylib_0.1.4
[55] Rcpp_1.0.10 plyr_1.8.8
[57] base64enc_0.1-3 progress_1.2.2
[59] zlibbioc_1.42.0 RCurl_1.98-1.10
[61] ps_1.7.2 prettyunits_1.1.1
[63] rpart_4.1.19 deldir_1.0-6
[65] SummarizedExperiment_1.26.1 cluster_2.1.4
[67] fs_1.6.1 magrittr_2.0.3
[69] RSpectra_0.16-1 whisker_0.4.1
[71] matrixStats_0.63.0 hms_1.1.2
[73] mime_0.12 evaluate_0.20
[75] xtable_1.8-4 XML_3.99-0.13
[77] jpeg_0.1-10 compiler_4.2.2
[79] ellipse_0.4.3 crayon_1.5.2
[81] htmltools_0.5.4 corpcor_1.6.10
[83] later_1.3.0 tzdb_0.3.0
[85] DBI_1.1.3 rappdirs_0.3.3
[87] Matrix_1.5-3 car_3.1-1
[89] cli_3.6.0 parallel_4.2.2
[91] igraph_1.4.1 pkgconfig_2.0.3
[93] getPass_0.2-2 GenomicAlignments_1.32.1
[95] foreign_0.8-84 xml2_1.3.3
[97] rARPACK_0.11-0 bslib_0.4.2
[99] XVector_0.36.0 callr_3.7.3
[101] digest_0.6.31 graph_1.74.0
[103] Biostrings_2.64.1 rmarkdown_2.20
[105] htmlTable_2.4.1 restfulr_0.0.15
[107] curl_5.0.0 shiny_1.7.4
[109] Rsamtools_2.12.0 rjson_0.2.21
[111] lifecycle_1.0.3 jsonlite_1.8.4
[113] carData_3.0-5 fansi_1.0.4
[115] pillar_1.8.1 KEGGREST_1.36.3
[117] fastmap_1.1.1 httr_1.4.5
[119] interactiveDisplayBase_1.34.0 glue_1.6.2
[121] png_0.1-8 BiocVersion_3.15.2
[123] bit_4.0.5 stringi_1.7.12
[125] sass_0.4.5 blob_1.2.3
[127] latticeExtra_0.6-30 memoise_2.0.1