Node summary
Robert Schlegel
2019-07-09
Last updated: 2019-07-09
workflowr checks: (Click a bullet for more information)-
✖ R Markdown file: uncommitted changes
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can runwflow_publishto commit the R Markdown file and build the HTML. -
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20190513)The command
set.seed(20190513)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 2edffcb
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: data/NAPA_clim_U.Rda Ignored: data/NAPA_clim_V.Rda Ignored: data/NAPA_clim_W.Rda Ignored: data/NAPA_clim_emp_ice.Rda Ignored: data/NAPA_clim_emp_oce.Rda Ignored: data/NAPA_clim_fmmflx.Rda Ignored: data/NAPA_clim_mldkz5.Rda Ignored: data/NAPA_clim_mldr10_1.Rda Ignored: data/NAPA_clim_qemp_oce.Rda Ignored: data/NAPA_clim_qla_oce.Rda Ignored: data/NAPA_clim_qns.Rda Ignored: data/NAPA_clim_qsb_oce.Rda Ignored: data/NAPA_clim_qt.Rda Ignored: data/NAPA_clim_runoffs.Rda Ignored: data/NAPA_clim_ssh.Rda Ignored: data/NAPA_clim_sss.Rda Ignored: data/NAPA_clim_sst.Rda Ignored: data/NAPA_clim_taum.Rda Ignored: data/NAPA_clim_vars.Rda Ignored: data/NAPA_clim_vecs.Rda Ignored: data/node_mean_all_anom.Rda Ignored: data/packet_all_anom.Rda Ignored: data/som_all_anom.Rda Ignored: data/synoptic_states.Rda Ignored: data/synoptic_vec_states.Rda Untracked files: Untracked: docs/assets/ Unstaged changes: Modified: analysis/node-summary.Rmd
Expand here to see past versions:
Introduction
This vignette will show the summary figures for the 12 nodes. The code used to create these summary figures may be found in ‘code/functions.R’. The nodes are listed below by their number. Use the table of contents on the left of the screen to move qucikly between nodes of interest as desired.
Figure caption key
The captions for the figures below have a lot of acronyms in them but they are consistently used and I will provide a list of what they are here. Whenever one sees an acronym in lower case letters it is refering to one of the regions of the study area as seen in the following figure.
knitr::include_graphics("assets/study_regions.png")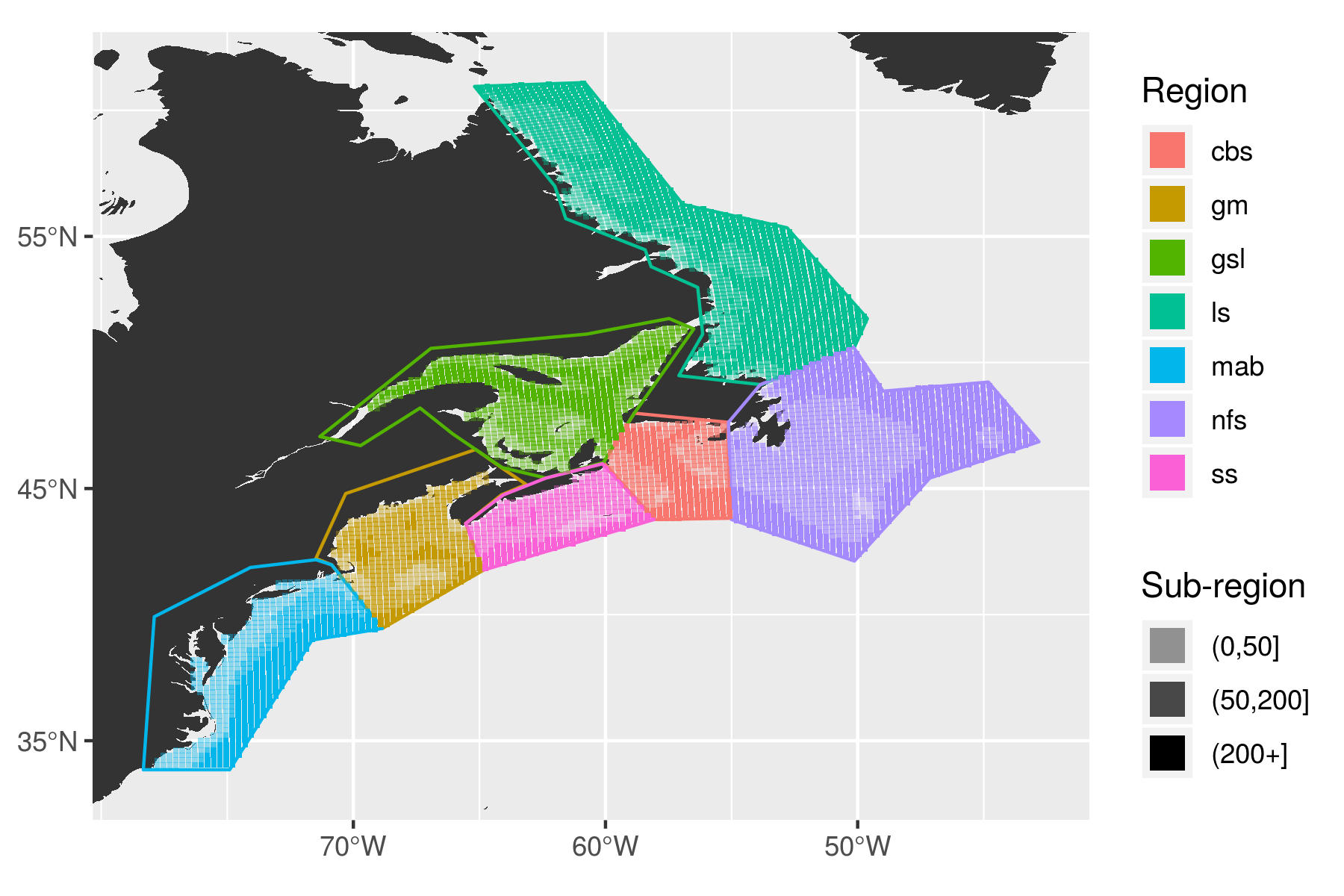
The regions of the coast were devided up by their temperature and salinity regimes based on work by Richaud et al. (2016). The region abbreviations are: gm = Gulf of Maine, gls = Gulf of St. Lawrence, ls = Labrador Shelf, mab = Mid-Atlantic Bight, nfs = Newfoundland Shelf, ss = Scotian Shelf. The regions were furthered divided into sub-regions based on their depth.
The upper case acronyms used in the following figure captions are as follows:
- SST = Sea surface temperature
- SOM = Self-organising map(s)
- GS = Gulf Stream
- AO = Atlantic Ocean
- LS = Labrador Sea
- LC = Labrador Current
- NS = Nova Scotia
If I have missed any please don’t hesitate to send me a message.
SST overview
The figure below shows the SST anomaly per node as a means of providing a very general overview of what each node looks like so as to make navigating between them more convenient.

The SST anomaly per node of the SOM. Note that the SST patterns are calculated in conjunction with the several other variables and are not the only variable taken into account. They are shown here by themselves as I think they provide the most convenient single snap shot of the results.
Node 1
knitr::include_graphics("~/MHWNWA/output/node_1_panels.png")
Warm pulse of GS near the NS coast. Shallowing mixed layer, low wind stress, and strong negative heat flux. Mostly gm and ss, almost no nfs. Almost entirely summer and autumn from 2013 - 2016. Mostly smaller evets but a few are massive.
Node 2
knitr::include_graphics("~/MHWNWA/output/node_2_panels.png")
Cold GS with warm LC caused by positive heat flux, low wind stress, and shallow mixed layer. Mostly cbs with some gsl and no mab. Occurred in only one year in two pulses in spring and summer. Normal intensity but short duration.
Node 3
knitr::include_graphics("~/MHWNWA/output/node_3_panels.png")
Calm sea state with some positive heatflux into the LC causing events. Shallower mixed layer everywhere. Mostly nfs with progressively fewer events in regions down the coast. Almost none in ls. Smaller events with a couple of large ones. All seasons from 1999 - 2014.
Node 4
knitr::include_graphics("~/MHWNWA/output/node_4_panels.png")
Extremely shallow mixed layer with a strong positive heatflux and low wind stress. Mostly nfs with progressively fewer events further away. Smaller events. Autumn, Winter, and Spring from 1999 - 2014.
Node 5
knitr::include_graphics("~/MHWNWA/output/node_5_panels.png")
Slightly shallow slightly fast push of the GS into the coast becoming slightly deeper near WHOI before coming back away from the coast and chilling out. The core of the pulse has negative heatflux but the surrounding GS has a strong positive heatflux and snall wind stress. Almost exclusively occurs in mab with only a bit everywhere else. Smallish events with a few massive ones. All seasons from 2003 - 2015.
Node 6
knitr::include_graphics("~/MHWNWA/output/node_6_panels.png")
Slightly warmer LS and LC with cooler GS. Minor poitive heat flux into LS and large positive heat flux into GS. Normal mixed layer with low wind stress over the LS and high over the GS. Mostly in the ls with a bit in the mab with almost none elsewhere. Occurred over 1999 - 2010 in spring and summer. Smaller events that have not been increasing over time.
Node 7
knitr::include_graphics("~/MHWNWA/output/node_7_panels.png")
Warm waters from LS to LC to GSL and a cold GS. Strong downward heat flux over northern waters and negative flux over GS. Shallow northern waters with low wind stress while high stress over GS. Equally high in ls and nfs. A bit in cbs but almost none elsewhere. Spring - Autumn from 2000 - 2014. 2006 was a particularly strong year. Events are overall not particularly large.
Node 8
knitr::include_graphics("~/MHWNWA/output/node_8_panels.png")
Warm northern waters with a cold GS. Strong positive flux over LS with weaker positive flux over GSL and negative over GS. Very shallow LS and very deep GS. Affects all northern waters but highest in gsl and ls. No events in mab and almost none in gm. Almost always Autumn and Winter from 2006 - 2013. Some more intense events later on with 2010/11 being a larger year.
Node 9
knitr::include_graphics("~/MHWNWA/output/node_9_panels.png")
Similar to node 5. Strong nearshore GS pulse. Strong negative flux over LS and GS but positive over the rest of the Atlantic. Very strong wind stress over LS and eastern part of Atlantic, weak over the warm heat flux area of the Atlantic. Extremely deep LS and shallow GS. Occurred over 2002 - 2016 for winter and spring, events began occurring in Autumn from 2013. Evens becoming rather intense as time progresses with some massive ones. Increasing in intensity in most regions.
Node 10
knitr::include_graphics("~/MHWNWA/output/node_10_panels.png")
Very unstable mostly cold GS with warm GM and SS waters. Negative heat flux into shelf waters and positive into GS. High wind stress over LS and low over shelf waters. Deep GS and GM waters but shallow over SS. Spread out over most regions with fewest events in mab and nfs. A few tiny events from 2009 - 2011 but really got going from 2012 - 2013. Spring of 2013 was small while Autumn/WInter of 2012/13 was noteworthy.
Node 11
knitr::include_graphics("~/MHWNWA/output/node_11_panels.png")
Energetic but normal temperature GS with warm inshore waters and slightly warm LS. Positive heat flux into GS and LS but negative into inshore waters. High wind stress above LS and a bit over central AO, but negative everywhere else. Very deep mixed layer next to coast in MAB but relatively normal everywhere else. Relatively equivalent occurrence in all regions. Occurred only from July - October, 2012. A few decent sized events. Mean max intensity is decent.
Node 12
knitr::include_graphics("~/MHWNWA/output/node_12_panels.png")
Warm inshore and LS waters with cold GS and AO. GS is moving fast and consistent. Negative heatflux into GS and inshore waters, slightly positive into LS and AO. High wind stress over GS and AO, negative over inshore waters and LS. Very deep mixed layer along coast in mab and very shallow along coast in ls. Mostly events occurring in gsl, but also in other northern areas. Occurred every even year from 2008 to 2014 from ~June - September. Relatively small (short) events but with decent max intensities.
Richaud, B., Kwon, Y.-O., Joyce, T. M., Fratantoni, P. S., and Lentz, S. J. (2016). Surface and bottom temperature and salinity climatology along the continental shelf off the canadian and us east coasts. Continental Shelf Research 124, 165–181.
Session information
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 16.04.5 LTS
Matrix products: default
BLAS: /usr/lib/openblas-base/libblas.so.3
LAPACK: /usr/lib/libopenblasp-r0.2.18.so
locale:
[1] LC_CTYPE=en_CA.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_CA.UTF-8 LC_COLLATE=en_CA.UTF-8
[5] LC_MONETARY=en_CA.UTF-8 LC_MESSAGES=en_CA.UTF-8
[7] LC_PAPER=en_CA.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_CA.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] workflowr_1.1.1 Rcpp_0.12.18 digest_0.6.16
[4] rprojroot_1.3-2 R.methodsS3_1.7.1 backports_1.1.2
[7] git2r_0.23.0 magrittr_1.5 evaluate_0.11
[10] highr_0.7 stringi_1.2.4 whisker_0.3-2
[13] R.oo_1.22.0 R.utils_2.7.0 rmarkdown_1.10
[16] tools_3.6.1 stringr_1.3.1 yaml_2.2.0
[19] compiler_3.6.1 htmltools_0.3.6 knitr_1.20 This reproducible R Markdown analysis was created with workflowr 1.1.1