Corrmotif overlap
Last updated: 2025-03-04
Checks: 6 1
Knit directory: CX5461_Project/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20250129) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 56e1527. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Unstaged changes:
Modified: analysis/Corrmotif_overlap.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Corrmotif_overlap.Rmd) and
HTML (docs/Corrmotif_overlap.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 56e1527 | sayanpaul01 | 2025-03-04 | Commit |
| html | 56e1527 | sayanpaul01 | 2025-03-04 | Commit |
| Rmd | 63ae929 | sayanpaul01 | 2025-03-04 | Commit |
| html | 63ae929 | sayanpaul01 | 2025-03-04 | Commit |
📌 Overlap of GO functions between Corrmotif all and corrmotif Conc.
library(UpSetR)
library(dplyr)
library(tools)
library(GO.db)
# Set the folder path
folder_path <- "data/all_GO"
# Get a list of all CSV files in the folder
csv_files <- list.files(folder_path, pattern = "\\.csv$", full.names = TRUE)
# Loop through each file and assign it as a variable in the global environment
for (file in csv_files) {
# Generate a valid R variable name from the file name (remove extension and replace spaces)
file_name <- tools::file_path_sans_ext(basename(file))
file_name <- gsub(" ", "_", file_name) # Replace spaces with underscores
file_name <- make.names(file_name) # Ensure the name is valid in R
# Assign the CSV file as a variable in the environment
assign(file_name, read.csv(file, stringsAsFactors = FALSE))
}
# Define datasets (lists of Entrez Gene IDs)
sets <- list(
"Non response all" = prob_all_1$ID,
"CX_DOX shared late response all" = prob_all_2$ID,
"Dox specific response all" = prob_all_3$ID,
"Late high dose DOX specific response all" = prob_all_4$ID,
"Non response (0.1)" = prob_1_0.1$ID,
"DOX only mid-late (0.1)" = prob_2_0.1$ID,
"CX_DOX mid-late (0.1)" = prob_3_0.1$ID,
"Non response (0.5)" = prob_1_0.5$ID,
"DOX only early-mid (0.5)" = prob_2_0.5$ID,
"DOX only mid-late (0.5)" = prob_3_0.5$ID,
"CX only mid-late (0.5)" = prob_4_0.5$ID,
"CX_DOX mid-late (0.5)" = prob_5_0.5$ID
)
# Create a binary matrix for UpSet plot
all_genes <- unique(unlist(sets)) # Get all unique Entrez Gene IDs
binary_matrix <- data.frame(Gene_ID = all_genes) # Initialize DataFrame
# Convert gene lists into a presence/absence matrix (1 = present, 0 = absent)
for (set_name in names(sets)) {
binary_matrix[[set_name]] <- as.integer(all_genes %in% sets[[set_name]])
}
# Remove Gene_ID column as UpSetR only needs the binary matrix
binary_matrix <- binary_matrix[, -1]
upset(binary_matrix,
sets = names(sets),
order.by = "freq",
sets.bar.color = "#56B4E9", # Blue bars for set sizes
mainbar.y.label = "Number of Shared Functions",
sets.x.label = "GO terms per set",
text.scale = 1.2,
nintersects = 30)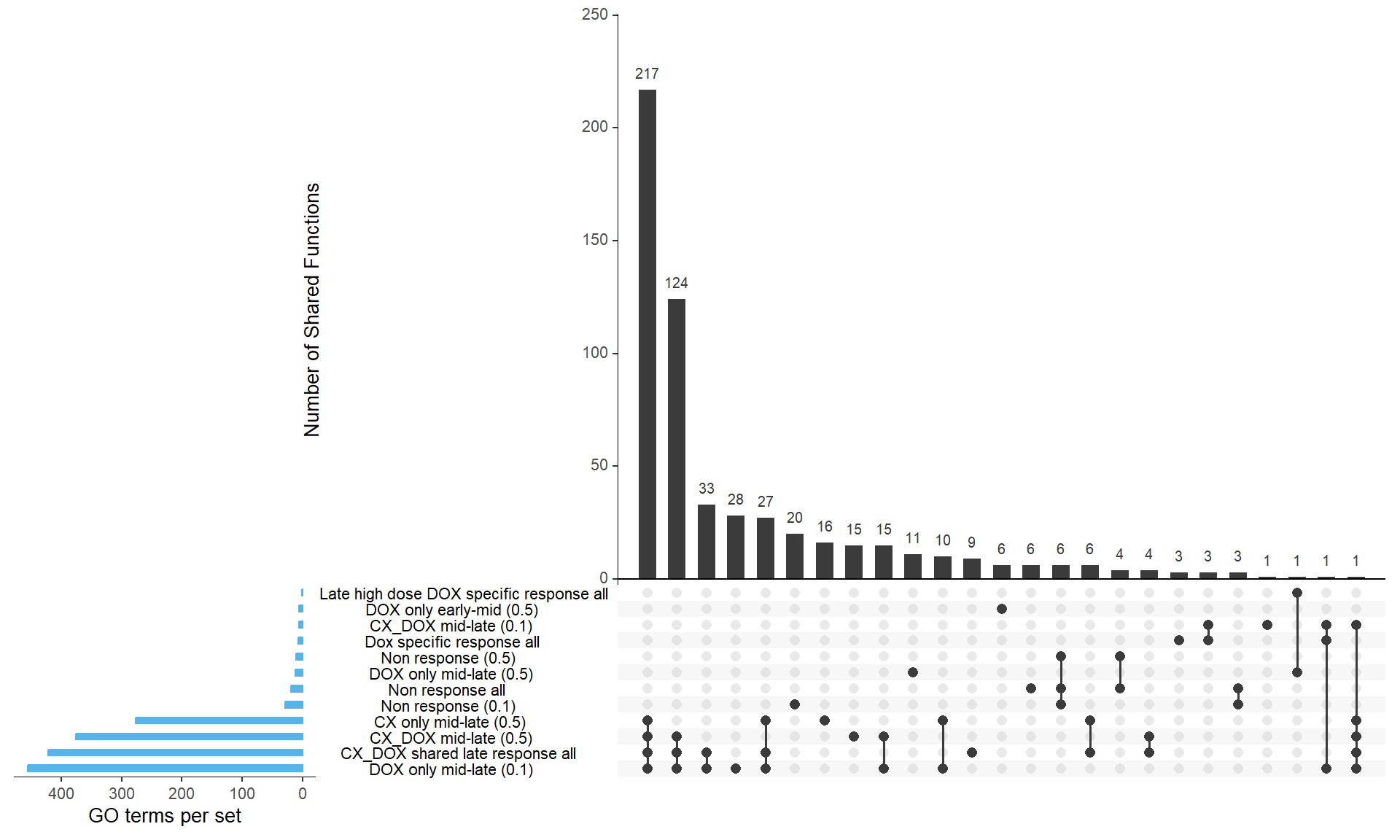
| Version | Author | Date |
|---|---|---|
| 63ae929 | sayanpaul01 | 2025-03-04 |
📌 Identify Unique GO Terms for Each Response Group
# Create a list to store unique GO terms per category
unique_go_terms <- list()
# Loop through each set to find unique GO terms
for (set_name in names(sets)) {
# Get the GO terms for the current set
current_go_terms <- sets[[set_name]]
# Find GO terms that appear **only** in this set and not in others
unique_terms <- current_go_terms[!(current_go_terms %in% unlist(sets[names(sets) != set_name]))]
# Store in the list if there are any unique terms
if (length(unique_terms) > 0) {
unique_go_terms[[set_name]] <- unique_terms
}
}
# Display unique GO terms for each category
unique_go_terms$`Non response all`
[1] "GO:0071339" "GO:1990204" "GO:0101031" "GO:0070469" "GO:0070971"
[6] "GO:0044665"
$`CX_DOX shared late response all`
[1] "GO:0009132" "GO:0097421" "GO:0140719" "GO:0140299" "GO:0035064"
[6] "GO:0140034" "GO:1990498" "GO:0005682" "GO:0030532"
$`Dox specific response all`
[1] "GO:0045177" "GO:0016323" "GO:0009925"
$`Non response (0.1)`
[1] "GO:0034470" "GO:0042254" "GO:0022613" "GO:0140053" "GO:0006413"
[6] "GO:0006364" "GO:0032543" "GO:0048193" "GO:0033108" "GO:0016072"
[11] "GO:0008135" "GO:0090079" "GO:0030684" "GO:0005759" "GO:0098800"
[16] "GO:0098803" "GO:0010494" "GO:0000313" "GO:0005761" "GO:0035770"
$`DOX only mid-late (0.1)`
[1] "GO:0009162" "GO:0009130" "GO:0006999" "GO:0006221" "GO:0007096"
[6] "GO:0045859" "GO:0006978" "GO:0009129" "GO:0046785" "GO:0002562"
[11] "GO:0016444" "GO:0042772" "GO:0010458" "GO:0071900" "GO:0051292"
[16] "GO:0006289" "GO:0008584" "GO:0008406" "GO:0046546" "GO:0045739"
[21] "GO:0045137" "GO:0043549" "GO:0009124" "GO:0048144" "GO:0008301"
[26] "GO:0017056" "GO:0000803" "GO:0043240"
$`CX_DOX mid-late (0.1)`
[1] "GO:0005402"
$`DOX only early-mid (0.5)`
[1] "GO:0140297" "GO:0061629" "GO:0090575" "GO:0005667" "GO:0097550"
[6] "GO:0005669"
$`DOX only mid-late (0.5)`
[1] "GO:0007186" "GO:0048738" "GO:0014706" "GO:0099084" "GO:0099173"
[6] "GO:0045598" "GO:0046486" "GO:0003013" "GO:0045444" "GO:0046620"
[11] "GO:0016236"
$`CX only mid-late (0.5)`
[1] "GO:0048599" "GO:0090305" "GO:0009994" "GO:2000243" "GO:0018105"
[6] "GO:0018209" "GO:0035561" "GO:0090657" "GO:0048477" "GO:0071732"
[11] "GO:0006264" "GO:0000722" "GO:0071731" "GO:1902170" "GO:0004518"
[16] "GO:0019205"
$`CX_DOX mid-late (0.5)`
[1] "GO:0048146" "GO:0030865" "GO:0051493" "GO:0051782" "GO:0090399"
[6] "GO:2000279" "GO:0030010" "GO:1901875" "GO:0032147" "GO:0031398"
[11] "GO:0007163" "GO:0051972" "GO:0000235" "GO:0005818" "GO:0101019"📌 Map GO IDs to Function Names Using GO.db
map_go_terms_local <- function(go_ids) {
go_names <- unlist(mget(go_ids, GOTERM, ifnotfound = NA)) # Retrieve function names
go_descriptions <- sapply(go_names, function(x) if (!is.na(x)) Term(x) else NA)
# Create a dataframe
go_mapping <- data.frame(GO_ID = go_ids, Function = go_descriptions, stringsAsFactors = FALSE)
# Remove NAs (unrecognized GO IDs)
go_mapping <- go_mapping[!is.na(go_mapping$Function), ]
return(go_mapping)
}
mapped_unique_go_terms <- list()
for (set_name in names(unique_go_terms)) {
if (length(unique_go_terms[[set_name]]) > 0) {
go_data <- map_go_terms_local(unique_go_terms[[set_name]])
mapped_unique_go_terms[[set_name]] <- go_data
}
}Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'
Warning in is.na(x): is.na() applied to non-(list or vector) of type 'S4'# Display mapped GO terms
mapped_unique_go_terms$`Non response all`
GO_ID Function
GO:0071339 GO:0071339 MLL1 complex
GO:1990204 GO:1990204 oxidoreductase complex
GO:0101031 GO:0101031 protein folding chaperone complex
GO:0070469 GO:0070469 respirasome
GO:0070971 GO:0070971 endoplasmic reticulum exit site
GO:0044665 GO:0044665 MLL1/2 complex
$`CX_DOX shared late response all`
GO_ID Function
GO:0009132 GO:0009132 nucleoside diphosphate metabolic process
GO:0097421 GO:0097421 liver regeneration
GO:0140719 GO:0140719 constitutive heterochromatin formation
GO:0140299 GO:0140299 small molecule sensor activity
GO:0035064 GO:0035064 methylated histone binding
GO:0140034 GO:0140034 methylation-dependent protein binding
GO:1990498 GO:1990498 mitotic spindle microtubule
GO:0005682 GO:0005682 U5 snRNP
GO:0030532 GO:0030532 small nuclear ribonucleoprotein complex
$`Dox specific response all`
GO_ID Function
GO:0045177 GO:0045177 apical part of cell
GO:0016323 GO:0016323 basolateral plasma membrane
GO:0009925 GO:0009925 basal plasma membrane
$`Non response (0.1)`
GO_ID Function
GO:0034470 GO:0034470 ncRNA processing
GO:0042254 GO:0042254 ribosome biogenesis
GO:0022613 GO:0022613 ribonucleoprotein complex biogenesis
GO:0140053 GO:0140053 mitochondrial gene expression
GO:0006413 GO:0006413 translational initiation
GO:0006364 GO:0006364 rRNA processing
GO:0032543 GO:0032543 mitochondrial translation
GO:0048193 GO:0048193 Golgi vesicle transport
GO:0033108 GO:0033108 mitochondrial respiratory chain complex assembly
GO:0016072 GO:0016072 rRNA metabolic process
GO:0008135 GO:0008135 translation factor activity, RNA binding
GO:0090079 GO:0090079 translation regulator activity, nucleic acid binding
GO:0030684 GO:0030684 preribosome
GO:0005759 GO:0005759 mitochondrial matrix
GO:0098800 GO:0098800 inner mitochondrial membrane protein complex
GO:0098803 GO:0098803 respiratory chain complex
GO:0010494 GO:0010494 cytoplasmic stress granule
GO:0000313 GO:0000313 organellar ribosome
GO:0005761 GO:0005761 mitochondrial ribosome
GO:0035770 GO:0035770 ribonucleoprotein granule
$`DOX only mid-late (0.1)`
GO_ID
GO:0009162 GO:0009162
GO:0009130 GO:0009130
GO:0006999 GO:0006999
GO:0006221 GO:0006221
GO:0007096 GO:0007096
GO:0045859 GO:0045859
GO:0006978 GO:0006978
GO:0009129 GO:0009129
GO:0046785 GO:0046785
GO:0002562 GO:0002562
GO:0016444 GO:0016444
GO:0042772 GO:0042772
GO:0010458 GO:0010458
GO:0071900 GO:0071900
GO:0051292 GO:0051292
GO:0006289 GO:0006289
GO:0008584 GO:0008584
GO:0008406 GO:0008406
GO:0046546 GO:0046546
GO:0045739 GO:0045739
GO:0045137 GO:0045137
GO:0043549 GO:0043549
GO:0009124 GO:0009124
GO:0048144 GO:0048144
GO:0008301 GO:0008301
GO:0017056 GO:0017056
GO:0000803 GO:0000803
GO:0043240 GO:0043240
Function
GO:0009162 deoxyribonucleoside monophosphate metabolic process
GO:0009130 pyrimidine nucleoside monophosphate biosynthetic process
GO:0006999 nuclear pore organization
GO:0006221 pyrimidine nucleotide biosynthetic process
GO:0007096 regulation of exit from mitosis
GO:0045859 regulation of protein kinase activity
GO:0006978 DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator
GO:0009129 pyrimidine nucleoside monophosphate metabolic process
GO:0046785 microtubule polymerization
GO:0002562 somatic diversification of immune receptors via germline recombination within a single locus
GO:0016444 somatic cell DNA recombination
GO:0042772 DNA damage response, signal transduction resulting in transcription
GO:0010458 exit from mitosis
GO:0071900 regulation of protein serine/threonine kinase activity
GO:0051292 nuclear pore complex assembly
GO:0006289 nucleotide-excision repair
GO:0008584 male gonad development
GO:0008406 gonad development
GO:0046546 development of primary male sexual characteristics
GO:0045739 positive regulation of DNA repair
GO:0045137 development of primary sexual characteristics
GO:0043549 regulation of kinase activity
GO:0009124 nucleoside monophosphate biosynthetic process
GO:0048144 fibroblast proliferation
GO:0008301 DNA binding, bending
GO:0017056 structural constituent of nuclear pore
GO:0000803 sex chromosome
GO:0043240 Fanconi anaemia nuclear complex
$`CX_DOX mid-late (0.1)`
GO_ID Function
GO:0005402 GO:0005402 carbohydrate:monoatomic cation symporter activity
$`DOX only early-mid (0.5)`
GO_ID
GO:0140297 GO:0140297
GO:0061629 GO:0061629
GO:0090575 GO:0090575
GO:0005667 GO:0005667
GO:0097550 GO:0097550
GO:0005669 GO:0005669
Function
GO:0140297 DNA-binding transcription factor binding
GO:0061629 RNA polymerase II-specific DNA-binding transcription factor binding
GO:0090575 RNA polymerase II transcription regulator complex
GO:0005667 transcription regulator complex
GO:0097550 transcription preinitiation complex
GO:0005669 transcription factor TFIID complex
$`DOX only mid-late (0.5)`
GO_ID Function
GO:0007186 GO:0007186 G protein-coupled receptor signaling pathway
GO:0048738 GO:0048738 cardiac muscle tissue development
GO:0014706 GO:0014706 striated muscle tissue development
GO:0099084 GO:0099084 postsynaptic specialization organization
GO:0099173 GO:0099173 postsynapse organization
GO:0045598 GO:0045598 regulation of fat cell differentiation
GO:0046486 GO:0046486 glycerolipid metabolic process
GO:0003013 GO:0003013 circulatory system process
GO:0045444 GO:0045444 fat cell differentiation
GO:0046620 GO:0046620 regulation of organ growth
GO:0016236 GO:0016236 macroautophagy
$`CX only mid-late (0.5)`
GO_ID Function
GO:0048599 GO:0048599 oocyte development
GO:0090305 GO:0090305 nucleic acid phosphodiester bond hydrolysis
GO:0009994 GO:0009994 oocyte differentiation
GO:2000243 GO:2000243 positive regulation of reproductive process
GO:0018105 GO:0018105 peptidyl-serine phosphorylation
GO:0018209 GO:0018209 peptidyl-serine modification
GO:0035561 GO:0035561 regulation of chromatin binding
GO:0090657 GO:0090657 telomeric loop disassembly
GO:0048477 GO:0048477 oogenesis
GO:0071732 GO:0071732 cellular response to nitric oxide
GO:0006264 GO:0006264 mitochondrial DNA replication
GO:0000722 GO:0000722 telomere maintenance via recombination
GO:0071731 GO:0071731 response to nitric oxide
GO:1902170 GO:1902170 cellular response to reactive nitrogen species
GO:0004518 GO:0004518 nuclease activity
GO:0019205 GO:0019205 nucleobase-containing compound kinase activity
$`CX_DOX mid-late (0.5)`
GO_ID
GO:0048146 GO:0048146
GO:0030865 GO:0030865
GO:0051493 GO:0051493
GO:0051782 GO:0051782
GO:0090399 GO:0090399
GO:2000279 GO:2000279
GO:0030010 GO:0030010
GO:1901875 GO:1901875
GO:0032147 GO:0032147
GO:0031398 GO:0031398
GO:0007163 GO:0007163
GO:0051972 GO:0051972
GO:0000235 GO:0000235
GO:0005818 GO:0005818
GO:0101019 GO:0101019
Function
GO:0048146 positive regulation of fibroblast proliferation
GO:0030865 cortical cytoskeleton organization
GO:0051493 regulation of cytoskeleton organization
GO:0051782 negative regulation of cell division
GO:0090399 replicative senescence
GO:2000279 negative regulation of DNA biosynthetic process
GO:0030010 establishment of cell polarity
GO:1901875 positive regulation of post-translational protein modification
GO:0032147 activation of protein kinase activity
GO:0031398 positive regulation of protein ubiquitination
GO:0007163 establishment or maintenance of cell polarity
GO:0051972 regulation of telomerase activity
GO:0000235 astral microtubule
GO:0005818 aster
GO:0101019 nucleolar exosome (RNase complex)
sessionInfo()R version 4.3.0 (2023-04-21 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 11 x64 (build 22631)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats4 tools stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] GO.db_3.18.0 AnnotationDbi_1.64.1 IRanges_2.36.0
[4] S4Vectors_0.40.1 Biobase_2.62.0 BiocGenerics_0.48.1
[7] dplyr_1.1.4 UpSetR_1.4.0
loaded via a namespace (and not attached):
[1] KEGGREST_1.42.0 gtable_0.3.6 xfun_0.50
[4] bslib_0.8.0 ggplot2_3.5.1 vctrs_0.6.5
[7] bitops_1.0-7 generics_0.1.3 tibble_3.2.1
[10] RSQLite_2.3.3 blob_1.2.4 pkgconfig_2.0.3
[13] lifecycle_1.0.4 GenomeInfoDbData_1.2.11 farver_2.1.2
[16] compiler_4.3.0 stringr_1.5.1 git2r_0.35.0
[19] Biostrings_2.70.1 munsell_0.5.1 httpuv_1.6.15
[22] GenomeInfoDb_1.38.8 htmltools_0.5.8.1 sass_0.4.9
[25] RCurl_1.98-1.13 yaml_2.3.10 later_1.3.2
[28] pillar_1.10.1 crayon_1.5.3 jquerylib_0.1.4
[31] whisker_0.4.1 cachem_1.0.8 tidyselect_1.2.1
[34] digest_0.6.34 stringi_1.8.3 labeling_0.4.3
[37] rprojroot_2.0.4 fastmap_1.1.1 grid_4.3.0
[40] colorspace_2.1-0 cli_3.6.1 magrittr_2.0.3
[43] withr_3.0.2 scales_1.3.0 promises_1.3.0
[46] bit64_4.0.5 rmarkdown_2.29 XVector_0.42.0
[49] httr_1.4.7 bit_4.0.5 gridExtra_2.3
[52] workflowr_1.7.1 png_0.1-8 memoise_2.0.1
[55] evaluate_1.0.3 knitr_1.49 rlang_1.1.3
[58] Rcpp_1.0.12 glue_1.7.0 DBI_1.2.3
[61] rstudioapi_0.17.1 jsonlite_1.8.9 R6_2.5.1
[64] plyr_1.8.9 fs_1.6.3 zlibbioc_1.48.0