ld_analysis
sq-96
2021-07-05
Last updated: 2021-07-26
Checks: 7 0
Knit directory: LD_Analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210705) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 51b7a14. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/.ipynb_checkpoints/
Untracked files:
Untracked: analysis/.Rmd
Untracked: analysis/.ipynb_checkpoints/
Untracked: data/EUR_subpop1.txt
Untracked: data/EUR_subpop2.txt
Untracked: data/EUR_subpop3.txt
Untracked: data/chr10_18681005_18770105_EUR/
Untracked: data/chr15_91416560_91429040_EUR/
Untracked: data/chr8_4177794_4192544_EUR/
Untracked: plink.log
Untracked: plink.nosex
Unstaged changes:
Deleted: analysis/first-analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ld_analysis.Rmd) and HTML (docs/ld_analysis.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 51b7a14 | sq-96 | 2021-07-26 | Publish the initial files for myproject |
| html | f5ec7d8 | sq-96 | 2021-07-26 | Build site. |
| Rmd | f697973 | sq-96 | 2021-07-26 | Publish the initial files for myproject |
| html | 317626c | sq-96 | 2021-07-07 | Build site. |
| Rmd | 87b9948 | sq-96 | 2021-07-07 | Publish the initial files for myproject |
| html | c0164e0 | sq-96 | 2021-07-07 | Build site. |
| Rmd | 822ad33 | sq-96 | 2021-07-07 | Publish the initial files for myproject |
| html | 42306c5 | sq-96 | 2021-07-07 | Build site. |
| Rmd | 3ffae79 | sq-96 | 2021-07-07 | Publish the initial files for myproject |
| html | d0e6281 | sq-96 | 2021-07-07 | Build site. |
| Rmd | f7c4b47 | sq-96 | 2021-07-07 | Publish the initial files for myproject |
| html | 1569358 | sq-96 | 2021-07-05 | Build site. |
| Rmd | 64f9844 | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | d4588ec | sq-96 | 2021-07-05 | Build site. |
| Rmd | cdf47fa | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | 9745cc6 | sq-96 | 2021-07-05 | Build site. |
| Rmd | 859c344 | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | b5b98f1 | sq-96 | 2021-07-05 | Build site. |
| Rmd | 24a94c7 | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | 2fd6c2f | sq-96 | 2021-07-05 | Build site. |
| Rmd | 6272671 | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | 457808c | sq-96 | 2021-07-05 | Build site. |
| Rmd | 82cfe8c | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | a7420d8 | sq-96 | 2021-07-05 | Build site. |
| Rmd | d817a3f | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | e77076d | sq-96 | 2021-07-05 | Build site. |
| Rmd | 8de5405 | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | 18a0f2f | sq-96 | 2021-07-05 | Build site. |
| Rmd | 643b482 | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | db2ad35 | sq-96 | 2021-07-05 | Build site. |
| html | f0c6cb5 | sq-96 | 2021-07-05 | Build site. |
| html | 76ce23b | sq-96 | 2021-07-05 | Build site. |
| Rmd | fd1293a | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | d5bf4c4 | sq-96 | 2021-07-05 | Build site. |
| Rmd | f66f509 | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | b538793 | sq-96 | 2021-07-05 | Build site. |
| Rmd | ec874a3 | sq-96 | 2021-07-05 | Publish the initial files for myproject |
| html | a5a1ae8 | sq-96 | 2021-07-05 | Build site. |
| html | ee7390b | sq-96 | 2021-07-05 | Build site. |
| html | 81812fd | sq-96 | 2021-07-05 | Build site. |
| Rmd | 3e38b1c | sq-96 | 2021-07-05 | Publish the initial files for myproject |
Objective
Comparing eigenvalues and eigenvectors of LD matrices calcualted with different subsamples.
Method
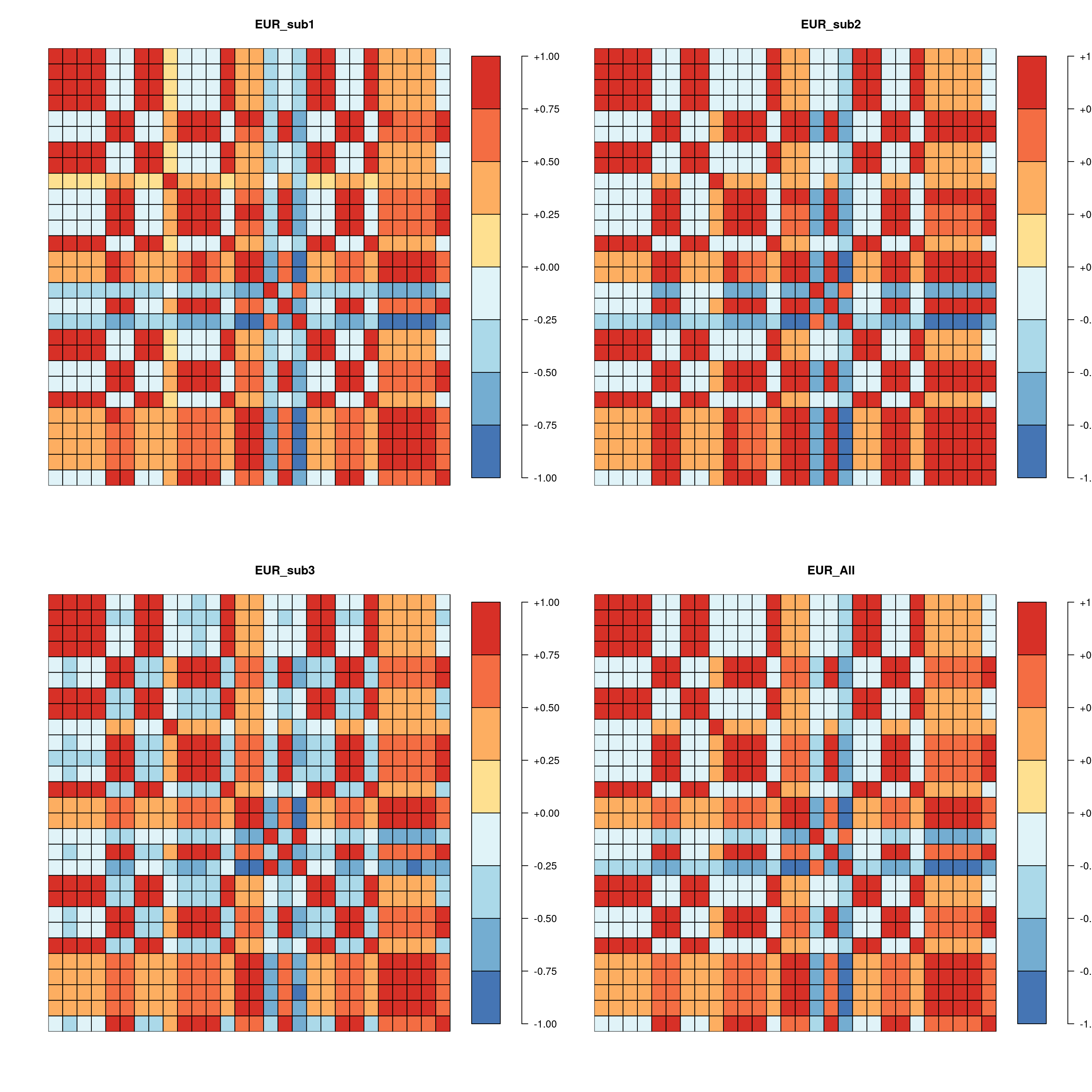
Data is from 1000 Genome EUR ancestry (~500 individuals). Samples are randomly splitted into 3 subsets, each has 160 individuals. Using PLINL to alculate LD matrices with variants (MAF > 0.05) in three SCZ loci.
Result
chr15 91416560-91429040
The first ten eigenvalues
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
EUR_sub1 14.507 10.845 0.885 0.744 0.458 0.190 0.135 0.061 0.054 0.037
EUR_sub2 14.503 11.196 0.862 0.605 0.419 0.173 0.082 0.054 0.048 0.015
EUR_sub3 13.488 11.461 1.059 0.815 0.572 0.187 0.130 0.084 0.073 0.058
EUR_All 14.084 11.226 0.874 0.792 0.482 0.188 0.088 0.067 0.055 0.036Pairwise correlation of the ith eigenvectors
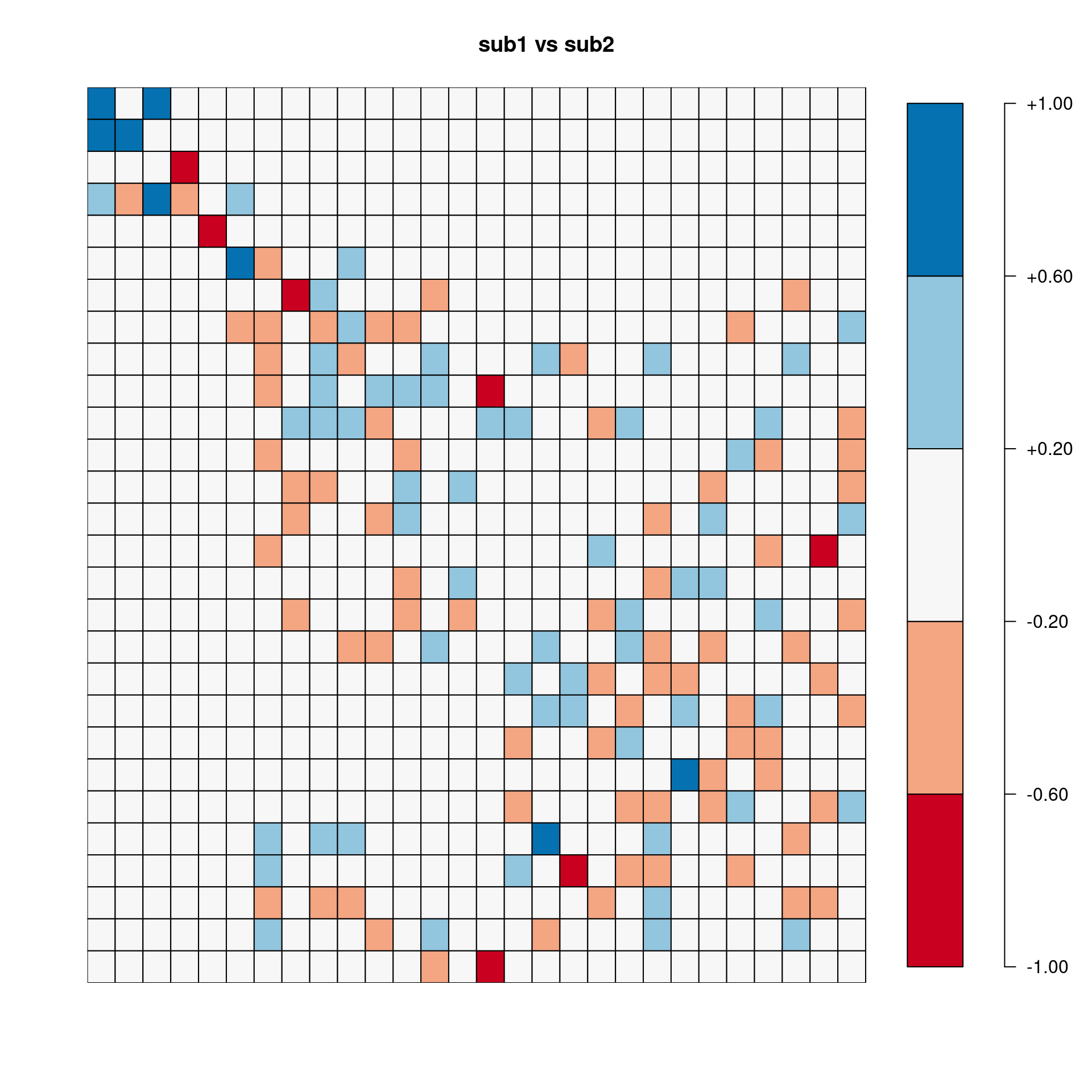
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
sub1.vs.sub2 0.990 0.998 0.965 0.961 -0.940 0.947 -0.528 0.379 -0.365 -0.021
sub1.vs.sub3 0.864 0.956 -0.051 -0.216 -0.972 0.868 -0.106 0.137 0.347 0.161
sub2.vs.sub3 0.925 0.972 -0.167 -0.395 0.910 0.912 0.085 -0.503 -0.575 0.004
sub1.vs.All 0.980 0.996 -0.937 -0.905 0.991 0.969 0.904 -0.769 -0.687 0.219
sub2.vs.All 0.998 0.999 -0.958 -0.955 -0.968 0.980 -0.797 -0.698 0.637 -0.131
sub3.vs.All 0.948 0.979 0.389 0.590 -0.977 0.953 -0.206 -0.019 -0.576 0.536chr8 4177794-4192544
The first ten eigenvalues
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
EUR_sub1 38.418 29.334 17.410 3.603 2.218 1.479 1.220 1.083 0.939 0.712
EUR_sub2 41.208 27.896 16.586 3.776 2.082 1.514 1.252 1.136 0.987 0.587
EUR_sub3 43.011 28.383 15.097 3.686 2.217 1.173 1.091 0.962 0.817 0.706
EUR_All 40.807 28.743 16.199 3.586 2.177 1.282 1.074 1.032 0.996 0.687Pairwise correlation of the ith eigenvectors
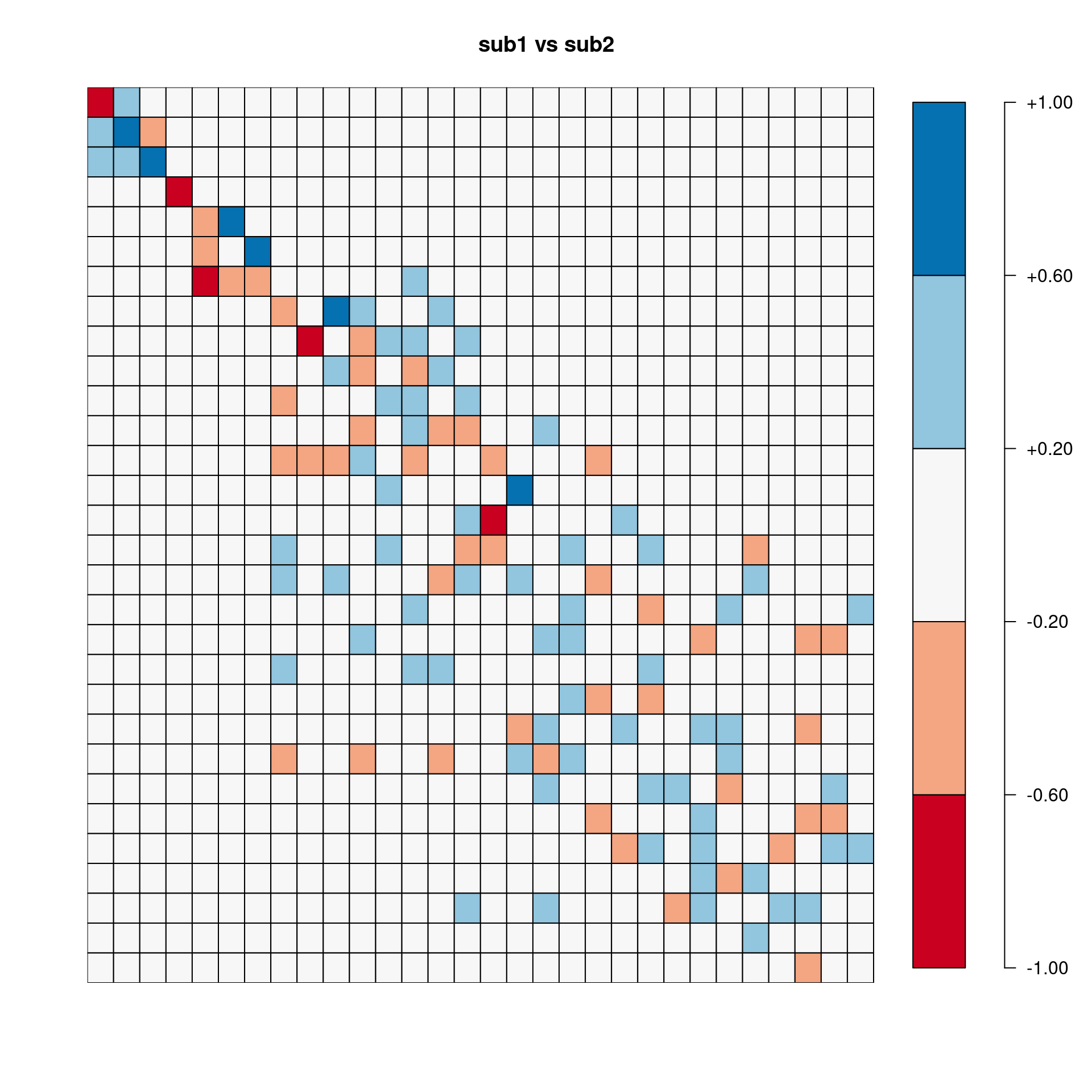
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
sub1.vs.sub2 0.481 -0.815 0.860 -0.952 0.823 0.087 0.021 -0.543 0.517 -0.325
sub1.vs.sub3 0.996 -0.989 0.984 0.920 -0.872 0.477 0.308 -0.690 0.012 0.482
sub2.vs.sub3 0.518 0.769 0.890 -0.835 -0.778 0.735 0.226 0.197 -0.378 -0.321
sub1.vs.All 0.997 -0.996 0.995 -0.992 0.953 0.515 -0.159 0.089 0.298 0.760
sub2.vs.All 0.450 0.842 0.884 0.968 0.927 0.784 0.344 0.491 0.627 -0.101
sub3.vs.All 0.991 0.990 0.995 -0.933 -0.928 0.901 0.718 -0.378 -0.571 0.457Heatmap of LD matrix
chr10 18681005-18770105
The first ten eigenvalues
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
EUR_sub1 85.111 63.778 53.515 26.972 11.070 8.549 7.872 3.639 3.398 3.001
EUR_sub2 80.116 68.940 47.786 29.973 11.169 10.196 7.929 4.504 3.271 2.721
EUR_sub3 85.548 69.447 53.013 28.336 8.967 8.474 5.279 3.999 3.307 2.724
EUR_All 81.628 67.213 52.643 28.061 10.654 9.106 6.961 3.648 3.072 3.037Pairwise correlation of the ith eigenvectors
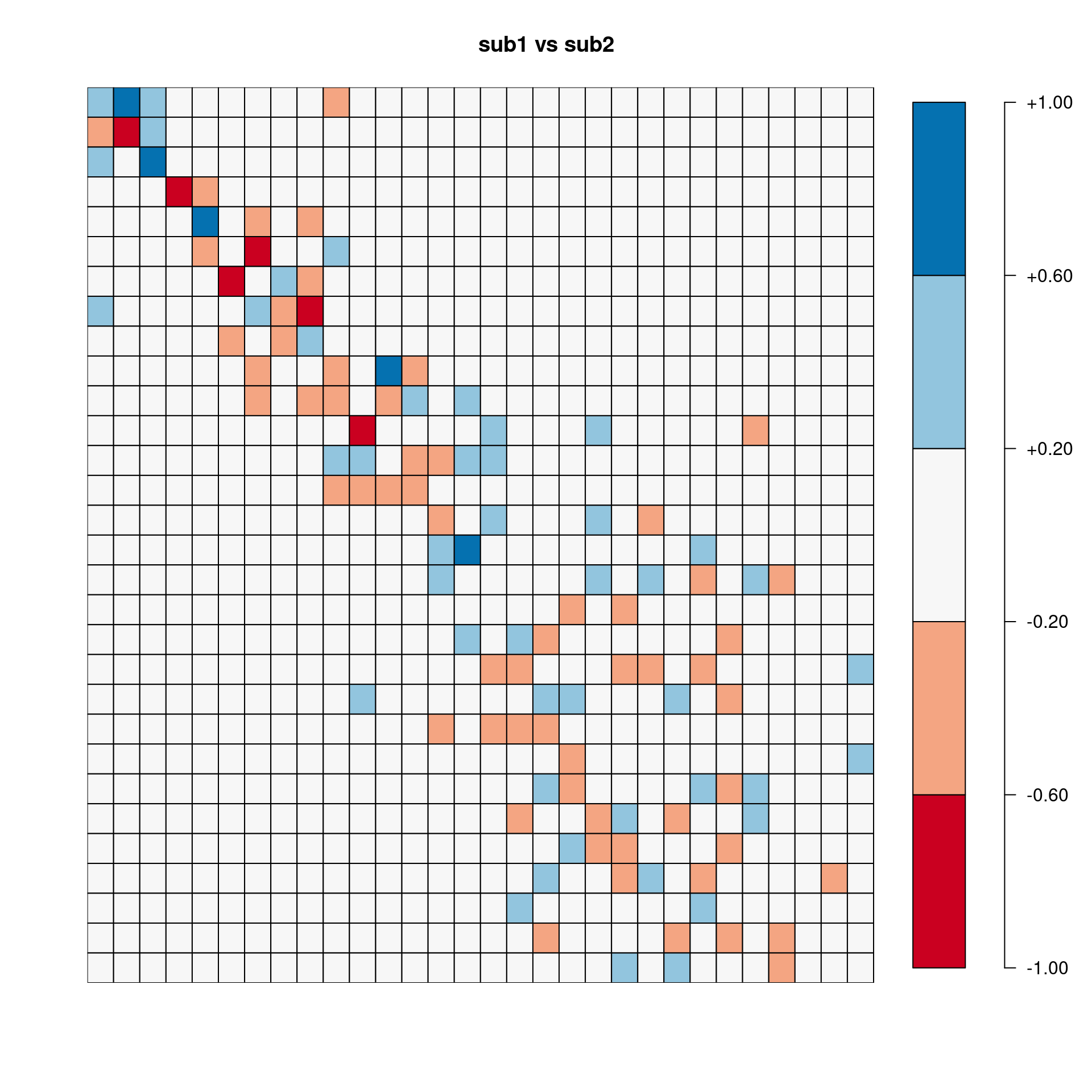
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9]
sub1.vs.sub2 -0.833 0.618 0.819 -0.934 -0.266 -0.054 -0.424 -0.384 -0.717
sub1.vs.sub3 0.912 0.865 0.917 0.921 0.837 -0.557 -0.256 0.150 -0.652
sub2.vs.sub3 -0.736 0.708 0.844 -0.871 -0.057 0.185 0.875 0.498 0.496
sub1.vs.All 0.970 0.868 0.932 0.972 0.970 -0.636 -0.494 0.496 0.479
sub2.vs.All -0.860 0.836 0.953 -0.965 -0.216 0.214 0.980 -0.869 -0.663
sub3.vs.All 0.956 0.929 0.954 0.953 0.902 0.906 0.871 -0.281 -0.023
[,10]
sub1.vs.sub2 0.404
sub1.vs.sub3 -0.153
sub2.vs.sub3 0.379
sub1.vs.All -0.114
sub2.vs.All 0.378
sub3.vs.All -0.082Pairwise correlation between two sets of eigenvectors
| Version | Author | Date |
|---|---|---|
| f5ec7d8 | sq-96 | 2021-07-26 |
sessionInfo()R version 4.0.4 (2021-02-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] RColorBrewer_1.1-2 plot.matrix_1.6 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 whisker_0.4 knitr_1.33 magrittr_2.0.1
[5] R6_2.5.0 rlang_0.4.11 fansi_0.5.0 highr_0.9
[9] stringr_1.4.0 tools_4.0.4 xfun_0.24 utf8_1.2.1
[13] git2r_0.28.0 jquerylib_0.1.4 htmltools_0.5.1.1 ellipsis_0.3.2
[17] rprojroot_2.0.2 yaml_2.2.1 digest_0.6.27 tibble_3.1.2
[21] lifecycle_1.0.0 crayon_1.4.1 later_1.2.0 sass_0.4.0
[25] vctrs_0.3.8 promises_1.2.0.1 fs_1.5.0 glue_1.4.2
[29] evaluate_0.14 rmarkdown_2.9 stringi_1.6.2 bslib_0.2.5.1
[33] compiler_4.0.4 pillar_1.6.1 jsonlite_1.7.2 httpuv_1.6.1
[37] pkgconfig_2.0.3