T2D - Liver
sheng Qian
2021-2-6
Last updated: 2022-02-26
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5c37a5d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/AF/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Unstaged changes:
Modified: analysis/BMI_Brain_Amygdala_S.Rmd
Modified: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Modified: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Modified: analysis/BMI_Brain_Cerebellum_S.Rmd
Modified: analysis/BMI_Brain_Cortex.Rmd
Modified: analysis/BMI_Brain_Cortex_S.Rmd
Modified: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Modified: analysis/BMI_Brain_Hippocampus_S.Rmd
Modified: analysis/BMI_Brain_Hypothalamus_S.Rmd
Modified: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Modified: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Modified: analysis/LDL_Liver_S.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/T2D_Liver.Rmd) and HTML (docs/T2D_Liver.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5c37a5d | sq-96 | 2022-02-26 | update |
| html | 3fa3a64 | sq-96 | 2022-02-26 | Build site. |
| Rmd | 0e6a2f2 | sq-96 | 2022-02-26 | update |
| html | 0974c69 | sq-96 | 2022-02-14 | Build site. |
| Rmd | cf57e2f | sq-96 | 2022-02-14 | update |
| Rmd | 721f7e8 | sq-96 | 2022-02-14 | update |
| html | 91f38fa | sq-96 | 2022-02-13 | Build site. |
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 6749#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
693 529 449 280 351 381 339 268 267 299 405 423 131 247 240 250 343 112 336 180
21 22
67 159 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 4358#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.6457Check convergence of parameters
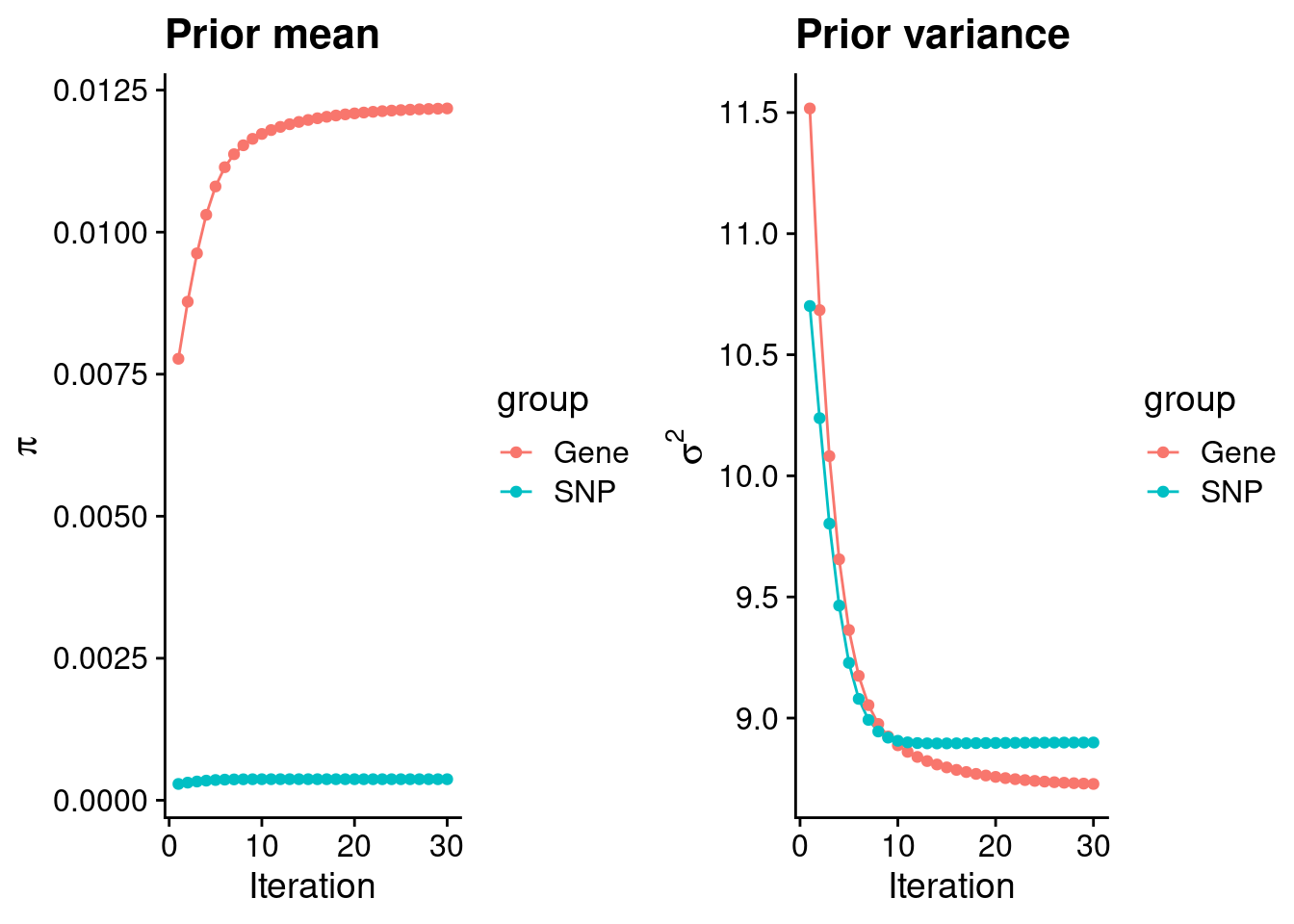
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0121777 0.0003704 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
8.728 8.899 #report sample size
print(sample_size)[1] 62892#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 6749 5017190#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.01141 0.26295 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.06078 1.45686Genes with highest PIPs
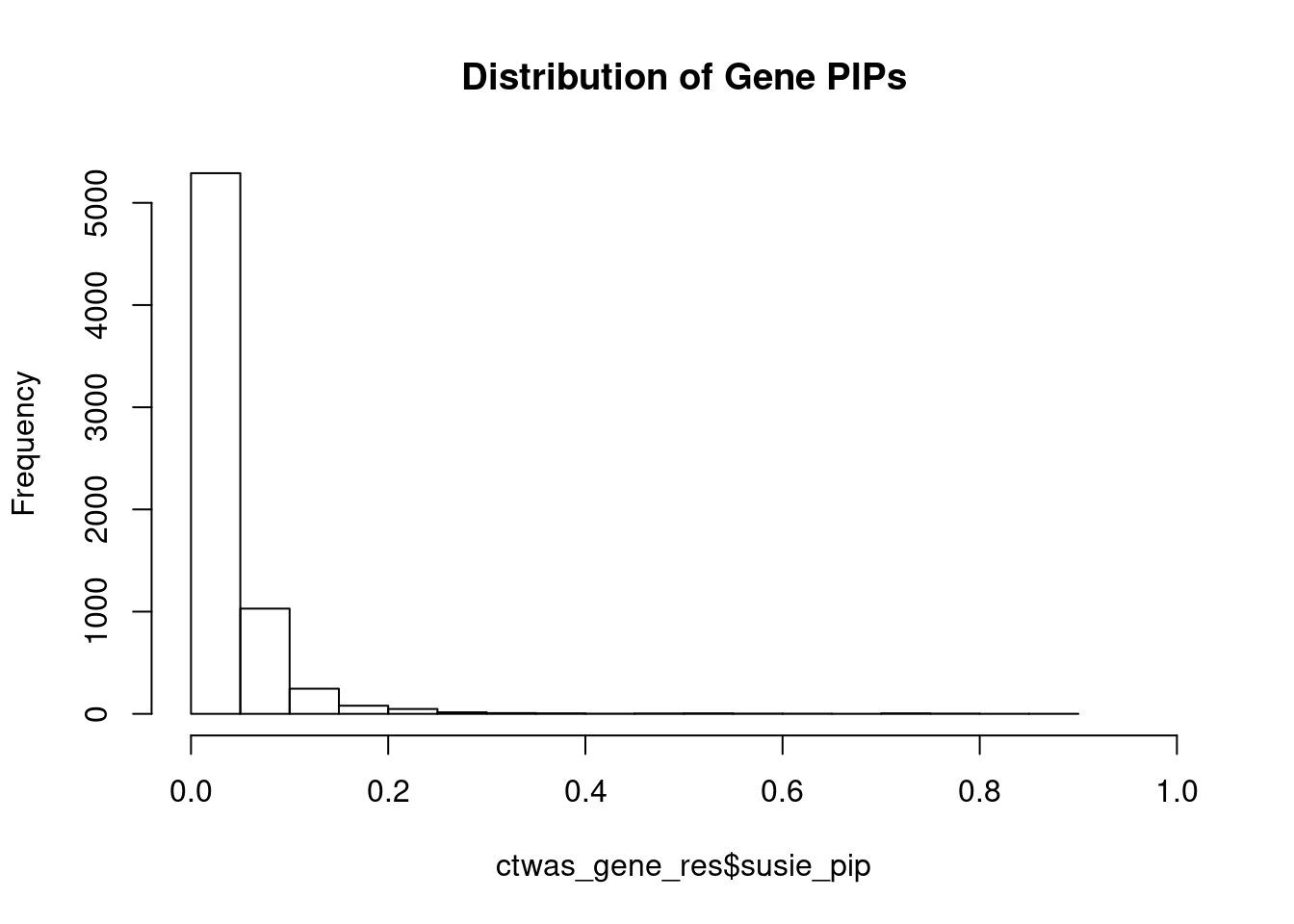
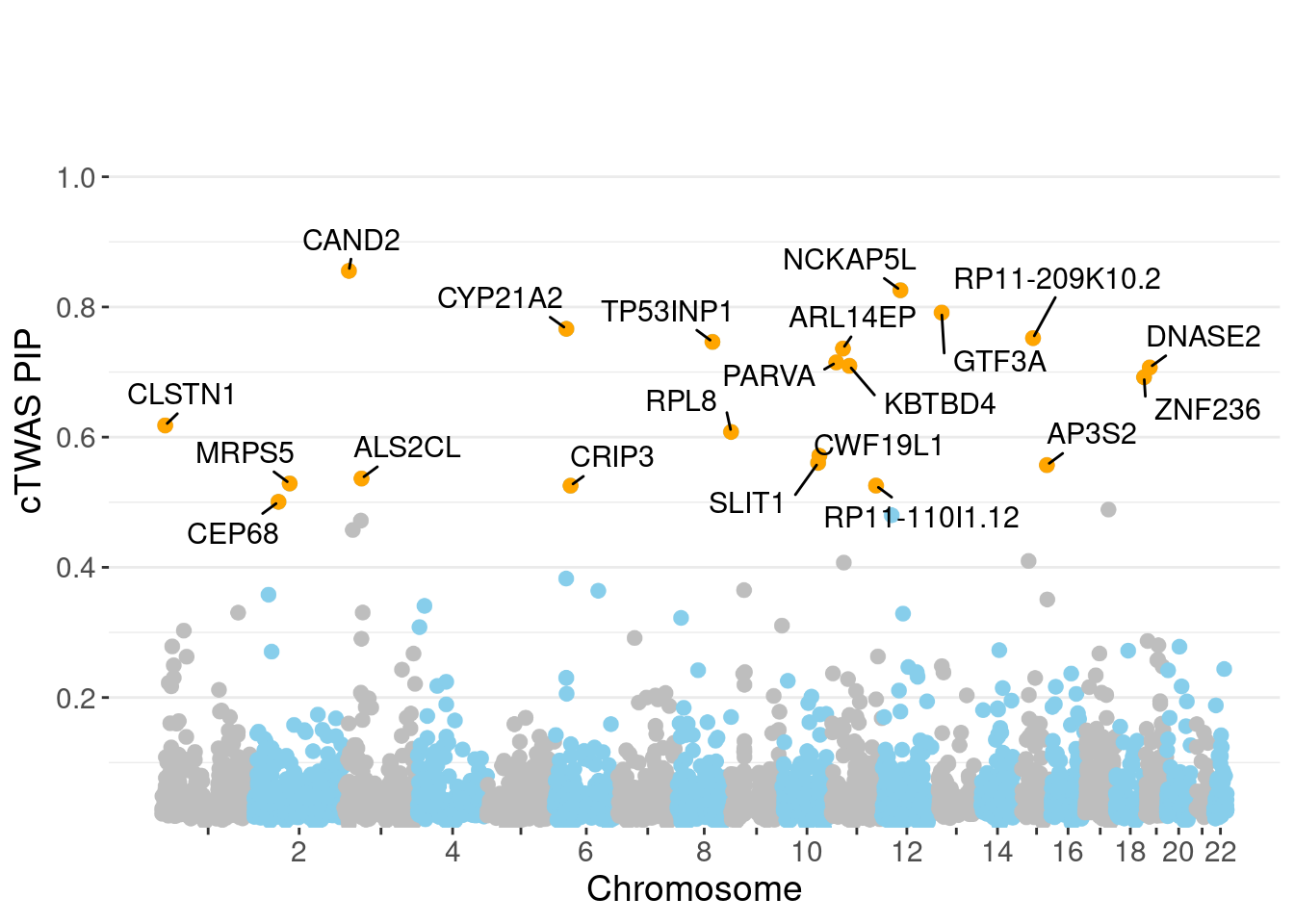
genename region_tag susie_pip mu2 PVE z num_eqtl
5632 CAND2 3_9 0.8555 22.92 0.0003117 -4.854 1
7788 NCKAP5L 12_31 0.8257 27.05 0.0003551 5.000 1
3444 GTF3A 13_7 0.7913 23.01 0.0002896 -4.478 2
11216 CYP21A2 6_26 0.7664 38.72 0.0004718 7.835 1
11883 RP11-209K10.2 15_22 0.7522 27.46 0.0003284 -5.056 1
7394 TP53INP1 8_66 0.7463 25.33 0.0003006 -5.474 1
6171 ARL14EP 11_21 0.7361 22.13 0.0002590 -4.512 3
10272 PARVA 11_9 0.7148 21.99 0.0002500 3.862 1
3551 KBTBD4 11_29 0.7095 26.51 0.0002990 -5.098 1
2050 DNASE2 19_10 0.7071 19.19 0.0002157 -3.744 1
4127 ZNF236 18_45 0.6921 20.89 0.0002298 -4.378 1
8335 CLSTN1 1_7 0.6180 20.20 0.0001985 3.978 1
6831 RPL8 8_94 0.6080 26.54 0.0002566 -5.063 1
1320 CWF19L1 10_64 0.5714 32.84 0.0002984 -5.742 2
9797 SLIT1 10_62 0.5606 23.74 0.0002116 4.762 1
6558 AP3S2 15_41 0.5570 39.32 0.0003483 6.483 1
8968 ALS2CL 3_33 0.5364 22.70 0.0001936 3.405 1
5574 MRPS5 2_57 0.5288 22.15 0.0001862 -3.737 1
11765 RP11-110I1.12 11_71 0.5256 18.71 0.0001563 3.747 1
5773 CRIP3 6_33 0.5254 21.27 0.0001777 4.511 2Genes with largest effect sizes
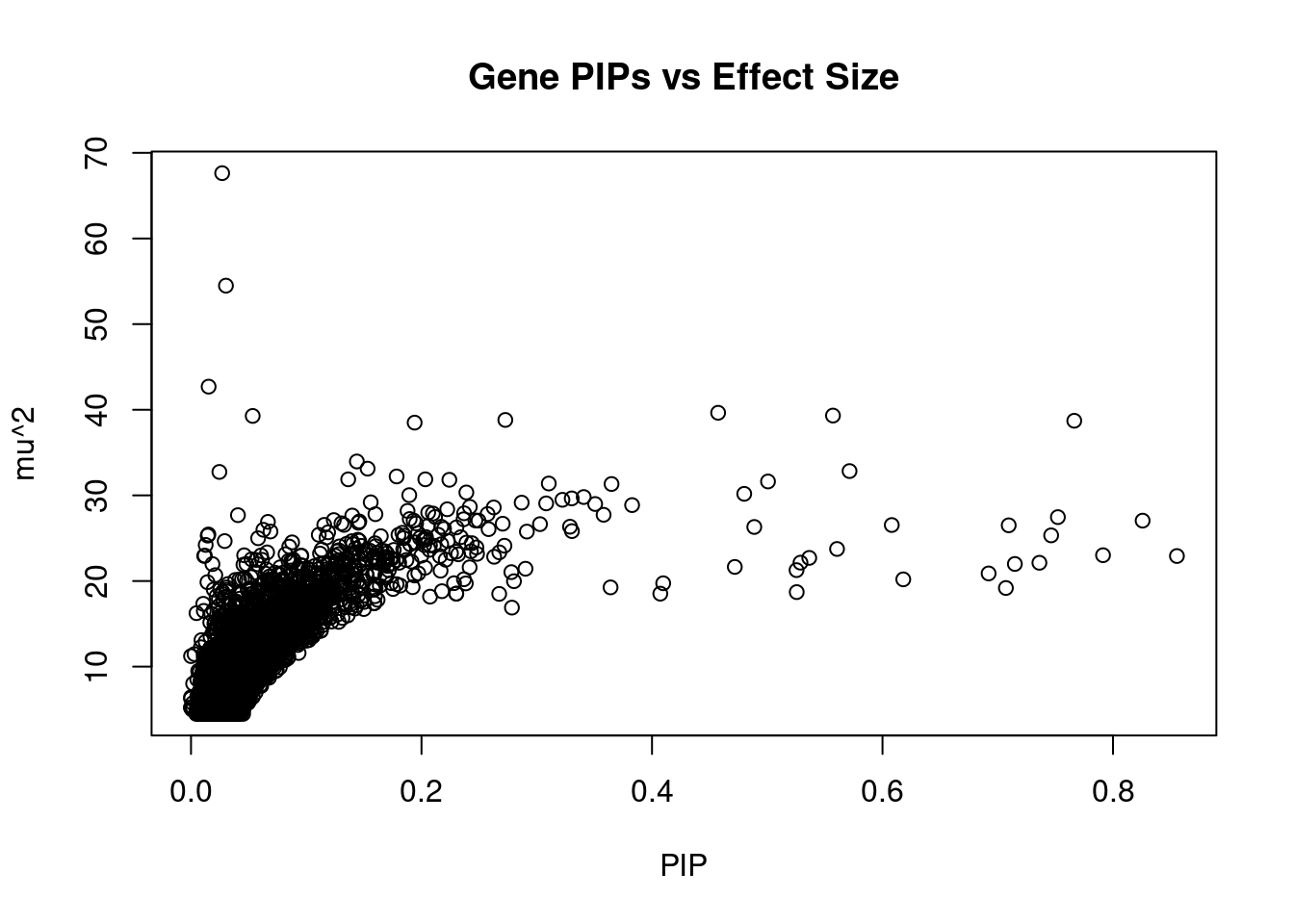
genename region_tag susie_pip mu2 PVE z num_eqtl
9887 NCR3LG1 11_12 0.02703 67.63 2.907e-05 -8.447 2
12661 LINC01126 2_27 0.03031 54.49 2.626e-05 -8.377 1
6291 JAZF1 7_23 0.01529 42.70 1.038e-05 -6.628 1
9311 UBE2E2 3_17 0.45738 39.65 2.883e-04 6.058 2
6558 AP3S2 15_41 0.55702 39.32 3.483e-04 6.483 1
6667 UBE2Z 17_28 0.05352 39.28 3.343e-05 -6.797 1
10351 TMEM229B 14_32 0.27272 38.81 1.683e-04 -3.685 2
11216 CYP21A2 6_26 0.76639 38.72 4.718e-04 7.835 1
4550 P2RX4 12_74 0.19404 38.50 1.188e-04 4.087 1
2084 RASA4 7_63 0.14390 33.96 7.771e-05 -4.470 1
10830 SYNJ2BP 14_32 0.15324 33.12 8.070e-05 -3.228 1
1320 CWF19L1 10_64 0.57140 32.84 2.984e-04 -5.742 2
2887 NRBP1 2_16 0.02462 32.74 1.281e-05 -5.595 1
6867 FMNL3 12_31 0.17840 32.21 9.137e-05 3.719 1
6223 GPR180 13_47 0.20330 31.88 1.030e-04 -3.353 1
8847 CCDC121 2_16 0.13639 31.87 6.912e-05 3.505 1
6456 ART3 4_51 0.22417 31.82 1.134e-04 -3.686 2
191 CEP68 2_42 0.50066 31.63 2.518e-04 6.229 2
7489 SDCCAG3 9_73 0.31043 31.39 1.549e-04 -3.739 2
9802 RP11-195F19.5 9_27 0.36489 31.33 1.818e-04 -3.408 2Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
11216 CYP21A2 6_26 0.7664 38.72 0.0004718 7.835 1
7788 NCKAP5L 12_31 0.8257 27.05 0.0003551 5.000 1
6558 AP3S2 15_41 0.5570 39.32 0.0003483 6.483 1
11883 RP11-209K10.2 15_22 0.7522 27.46 0.0003284 -5.056 1
5632 CAND2 3_9 0.8555 22.92 0.0003117 -4.854 1
7394 TP53INP1 8_66 0.7463 25.33 0.0003006 -5.474 1
3551 KBTBD4 11_29 0.7095 26.51 0.0002990 -5.098 1
1320 CWF19L1 10_64 0.5714 32.84 0.0002984 -5.742 2
3444 GTF3A 13_7 0.7913 23.01 0.0002896 -4.478 2
9311 UBE2E2 3_17 0.4574 39.65 0.0002883 6.058 2
6171 ARL14EP 11_21 0.7361 22.13 0.0002590 -4.512 3
6831 RPL8 8_94 0.6080 26.54 0.0002566 -5.063 1
191 CEP68 2_42 0.5007 31.63 0.0002518 6.229 2
10272 PARVA 11_9 0.7148 21.99 0.0002500 3.862 1
3522 BHLHE41 12_18 0.4800 30.18 0.0002304 5.640 1
4127 ZNF236 18_45 0.6921 20.89 0.0002298 -4.378 1
2050 DNASE2 19_10 0.7071 19.19 0.0002157 -3.744 1
9797 SLIT1 10_62 0.5606 23.74 0.0002116 4.762 1
10501 MAP3K3 17_37 0.4887 26.31 0.0002045 -5.170 1
8335 CLSTN1 1_7 0.6180 20.20 0.0001985 3.978 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
9887 NCR3LG1 11_12 0.02703 67.63 2.907e-05 -8.447 2
12661 LINC01126 2_27 0.03031 54.49 2.626e-05 -8.377 1
11216 CYP21A2 6_26 0.76639 38.72 4.718e-04 7.835 1
6667 UBE2Z 17_28 0.05352 39.28 3.343e-05 -6.797 1
6291 JAZF1 7_23 0.01529 42.70 1.038e-05 -6.628 1
6558 AP3S2 15_41 0.55702 39.32 3.483e-04 6.483 1
191 CEP68 2_42 0.50066 31.63 2.518e-04 6.229 2
9311 UBE2E2 3_17 0.45738 39.65 2.883e-04 6.058 2
10639 MICB 6_25 0.38273 28.87 1.757e-04 5.917 1
1320 CWF19L1 10_64 0.57140 32.84 2.984e-04 -5.742 2
3522 BHLHE41 12_18 0.48002 30.18 2.304e-04 5.640 1
2887 NRBP1 2_16 0.02462 32.74 1.281e-05 -5.595 1
11110 LTA 6_25 0.04071 27.69 1.793e-05 5.500 1
7394 TP53INP1 8_66 0.74634 25.33 3.006e-04 -5.474 1
326 ATP6V0A1 17_25 0.11561 26.56 4.882e-05 5.188 2
10501 MAP3K3 17_37 0.48875 26.31 2.045e-04 -5.170 1
3848 TSPAN8 12_44 0.24678 27.07 1.062e-04 5.137 1
3551 KBTBD4 11_29 0.70951 26.51 2.990e-04 -5.098 1
10594 PSMB8 6_27 0.20581 27.99 9.160e-05 5.081 1
6831 RPL8 8_94 0.60805 26.54 2.566e-04 -5.063 1Comparing z scores and PIPs
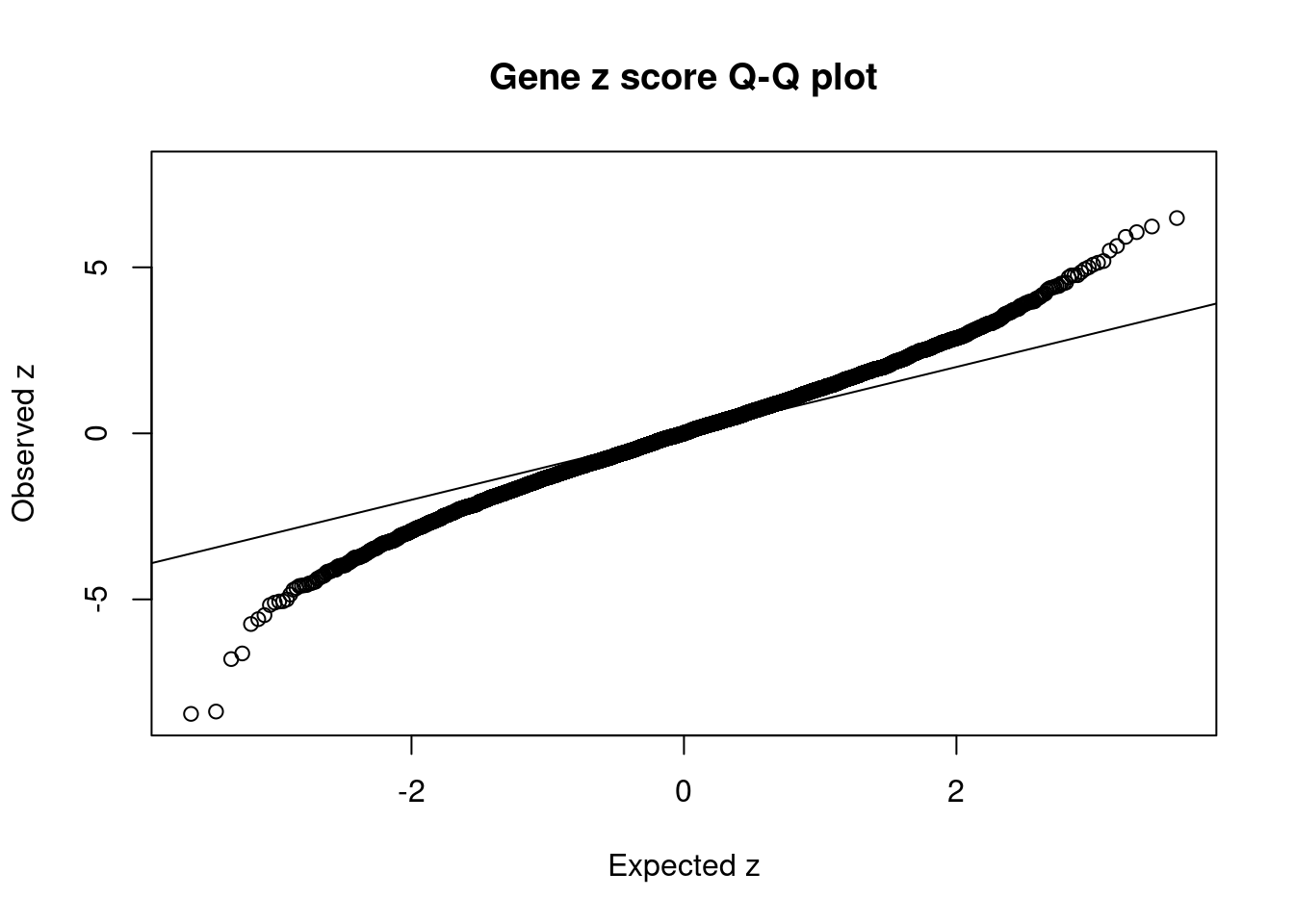
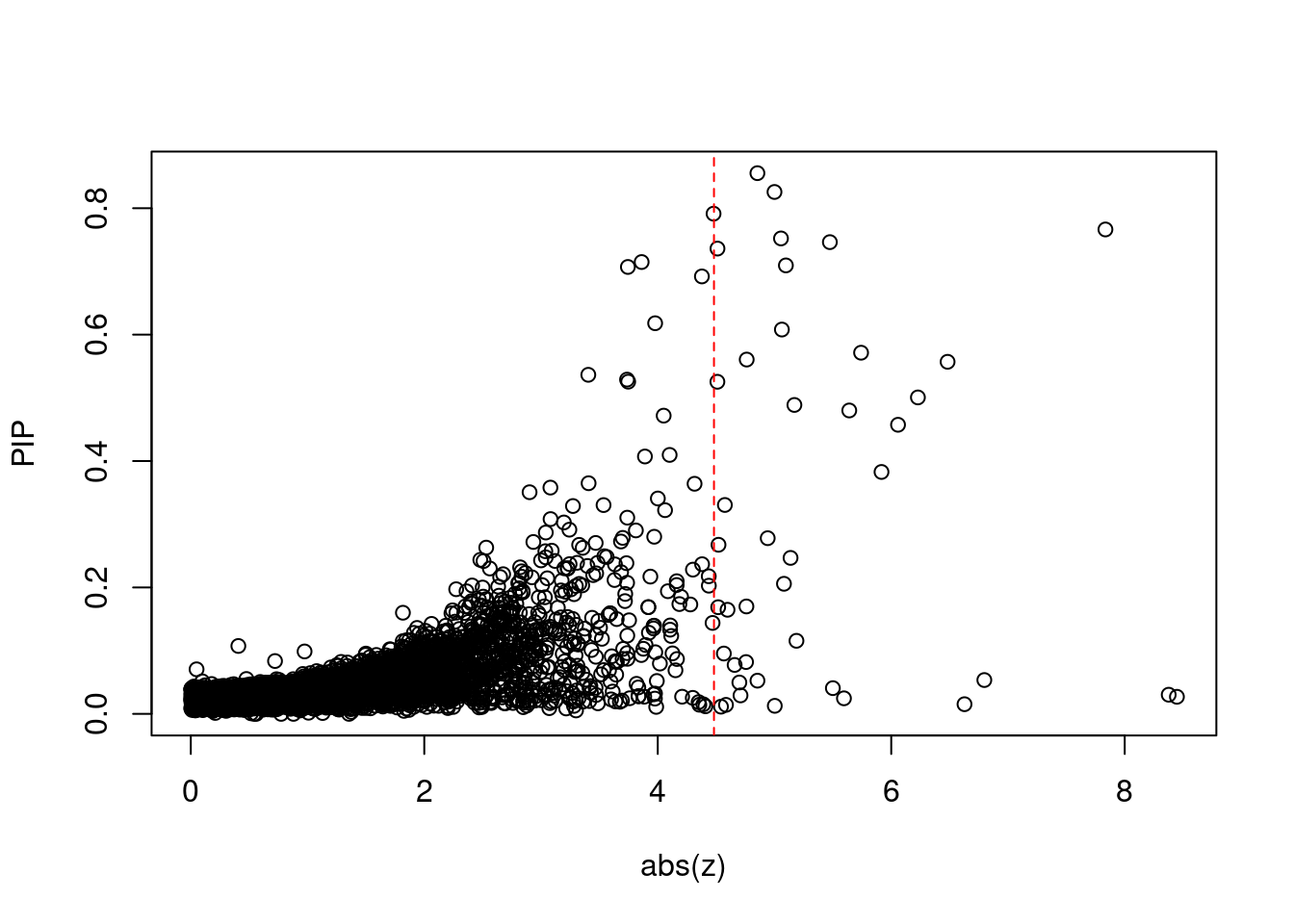
[1] 0.006075 genename region_tag susie_pip mu2 PVE z num_eqtl
9887 NCR3LG1 11_12 0.02703 67.63 2.907e-05 -8.447 2
12661 LINC01126 2_27 0.03031 54.49 2.626e-05 -8.377 1
11216 CYP21A2 6_26 0.76639 38.72 4.718e-04 7.835 1
6667 UBE2Z 17_28 0.05352 39.28 3.343e-05 -6.797 1
6291 JAZF1 7_23 0.01529 42.70 1.038e-05 -6.628 1
6558 AP3S2 15_41 0.55702 39.32 3.483e-04 6.483 1
191 CEP68 2_42 0.50066 31.63 2.518e-04 6.229 2
9311 UBE2E2 3_17 0.45738 39.65 2.883e-04 6.058 2
10639 MICB 6_25 0.38273 28.87 1.757e-04 5.917 1
1320 CWF19L1 10_64 0.57140 32.84 2.984e-04 -5.742 2
3522 BHLHE41 12_18 0.48002 30.18 2.304e-04 5.640 1
2887 NRBP1 2_16 0.02462 32.74 1.281e-05 -5.595 1
11110 LTA 6_25 0.04071 27.69 1.793e-05 5.500 1
7394 TP53INP1 8_66 0.74634 25.33 3.006e-04 -5.474 1
326 ATP6V0A1 17_25 0.11561 26.56 4.882e-05 5.188 2
10501 MAP3K3 17_37 0.48875 26.31 2.045e-04 -5.170 1
3848 TSPAN8 12_44 0.24678 27.07 1.062e-04 5.137 1
3551 KBTBD4 11_29 0.70951 26.51 2.990e-04 -5.098 1
10594 PSMB8 6_27 0.20581 27.99 9.160e-05 5.081 1
6831 RPL8 8_94 0.60805 26.54 2.566e-04 -5.063 1GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 21Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR
22 Late onset congenital adrenal hyperplasia 0.01134
32 Hyperandrogenism, Nonclassic Type, due to 21-Hydroxylase Deficiency 0.01134
34 Congenital adrenal hyperplasia due to 21 hydroxylase deficiency 0.01134
38 SPINOCEREBELLAR ATAXIA, AUTOSOMAL RECESSIVE 17 0.01134
1 Congenital adrenal hyperplasia 0.05394
2 Atrial Fibrillation 0.05394
8 Glomerulonephritis, Membranoproliferative 0.05394
19 Paroxysmal atrial fibrillation 0.05394
33 Persistent atrial fibrillation 0.05394
35 familial atrial fibrillation 0.05394
Ratio BgRatio
22 1/11 1/9703
32 1/11 1/9703
34 1/11 1/9703
38 1/11 1/9703
1 1/11 9/9703
2 2/11 160/9703
8 1/11 7/9703
19 2/11 156/9703
33 2/11 156/9703
35 2/11 156/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 72#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 20#significance threshold for TWAS
print(sig_thresh)[1] 4.482#number of ctwas genes
length(ctwas_genes)[1] 21#number of TWAS genes
length(twas_genes)[1] 41#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
8335 CLSTN1 1_7 0.6180 20.20 0.0001985 3.978 1
5574 MRPS5 2_57 0.5288 22.15 0.0001862 -3.737 1
8968 ALS2CL 3_33 0.5364 22.70 0.0001936 3.405 1
10272 PARVA 11_9 0.7148 21.99 0.0002500 3.862 1
11765 RP11-110I1.12 11_71 0.5256 18.71 0.0001563 3.747 1
4127 ZNF236 18_45 0.6921 20.89 0.0002298 -4.378 1
2050 DNASE2 19_10 0.7071 19.19 0.0002157 -3.744 1
3444 GTF3A 13_7 0.7913 23.01 0.0002896 -4.478 2#sensitivity / recall
print(sensitivity)ctwas TWAS
0 0 #specificity
print(specificity) ctwas TWAS
0.9969 0.9939 #precision / PPV
print(precision)ctwas TWAS
0 0
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[5] purrr_0.3.4 readr_2.1.1 tidyr_1.1.4 tidyverse_1.3.1
[9] tibble_3.1.6 WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[13] cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.2 lubridate_1.8.0 bit64_4.0.5 doParallel_1.0.17
[5] httr_1.4.2 rprojroot_2.0.2 tools_3.6.1 backports_1.4.1
[9] doRNG_1.8.2 utf8_1.2.2 R6_2.5.1 vipor_0.4.5
[13] DBI_1.1.2 colorspace_2.0-2 withr_2.4.3 ggrastr_1.0.1
[17] tidyselect_1.1.1 bit_4.0.4 curl_4.3.2 compiler_3.6.1
[21] git2r_0.26.1 rvest_1.0.2 cli_3.1.0 Cairo_1.5-12.2
[25] xml2_1.3.3 labeling_0.4.2 scales_1.1.1 apcluster_1.4.8
[29] digest_0.6.29 rmarkdown_2.11 svglite_1.2.2 pkgconfig_2.0.3
[33] htmltools_0.5.2 dbplyr_2.1.1 fastmap_1.1.0 highr_0.9
[37] rlang_1.0.1 rstudioapi_0.13 RSQLite_2.2.8 jquerylib_0.1.4
[41] farver_2.1.0 generics_0.1.1 jsonlite_1.7.2 vroom_1.5.7
[45] magrittr_2.0.2 Matrix_1.2-18 ggbeeswarm_0.6.0 Rcpp_1.0.8
[49] munsell_0.5.0 fansi_1.0.2 gdtools_0.1.9 lifecycle_1.0.1
[53] stringi_1.7.6 whisker_0.3-2 yaml_2.2.1 plyr_1.8.6
[57] grid_3.6.1 blob_1.2.2 ggrepel_0.9.1 parallel_3.6.1
[61] promises_1.0.1 crayon_1.5.0 lattice_0.20-38 haven_2.4.3
[65] hms_1.1.1 knitr_1.36 pillar_1.6.4 igraph_1.2.10
[69] rjson_0.2.20 rngtools_1.5.2 reshape2_1.4.4 codetools_0.2-16
[73] reprex_2.0.1 glue_1.6.2 evaluate_0.14 data.table_1.14.2
[77] modelr_0.1.8 vctrs_0.3.8 tzdb_0.2.0 httpuv_1.5.1
[81] foreach_1.5.2 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] cachem_1.0.6 xfun_0.29 broom_0.7.10 later_0.8.0
[89] iterators_1.0.14 beeswarm_0.2.3 memoise_2.0.1 ellipsis_0.3.2