BMI - Brain Nucleus accumbens basal ganglia
sheng Qian
2021-2-6
Last updated: 2022-02-13
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version eb13ecf. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/AF/
Untracked: data/BMI/
Untracked: data/T2D/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia.Rmd) and HTML (docs/BMI_Brain_Nucleus_accumbens_basal_ganglia.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
[1] 11487
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1134 801 689 451 561 646 551 431 414 460 695 611 220 384 375 529
17 18 19 20 21 22
672 184 903 357 131 288 [1] 9066[1] 0.78924Load ctwas results
Check convergence of parameters
********************************************************Note: As of version 1.0.0, cowplot does not change the default ggplot2 theme anymore. To recover the previous behavior, execute:
theme_set(theme_cowplot())********************************************************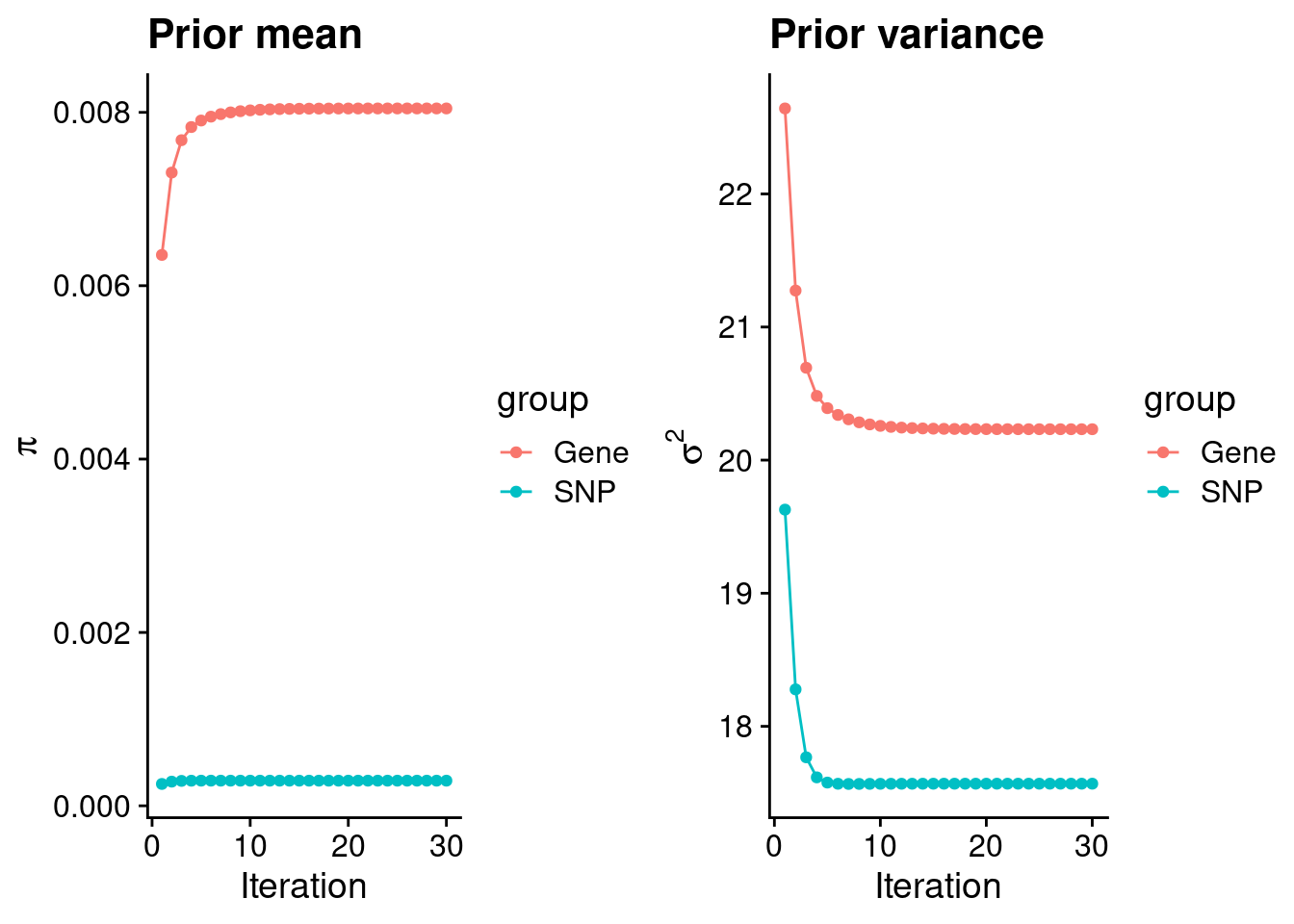
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
gene snp
0.0080450365 0.0002899309 gene snp
20.23213 17.56895 [1] 336107[1] 11487 7535010 gene snp
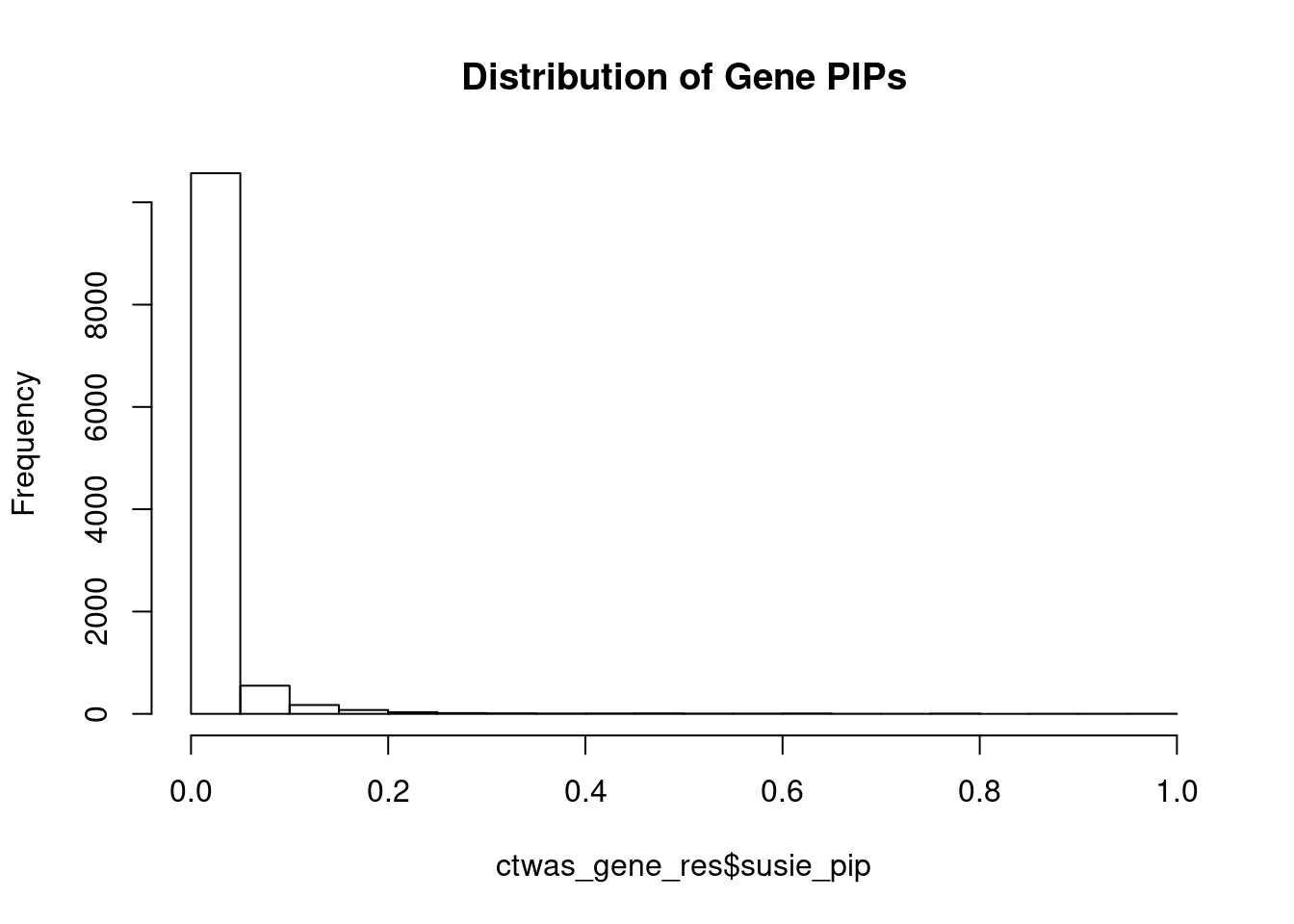
0.005562868 0.114194849 [1] 0.05015945 17.88795499Genes with highest PIPs
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
genename region_tag susie_pip mu2 PVE z
8160 SMAD3 15_31 0.9999995 8015.87282 2.384916e-02 -2.852677
9866 TMIE 3_33 0.9989779 34.99719 1.040187e-04 -7.091659
3378 CCND2 12_4 0.9648998 28.86368 8.286217e-05 -5.119990
9479 NIM1K 5_28 0.9385399 3262.53413 9.110249e-03 5.550554
7777 ZNF12 7_9 0.9363982 27.17655 7.571420e-05 5.127453
13756 NOL12 22_15 0.8990493 61.44780 1.643661e-04 -4.510837
7320 TAL1 1_29 0.7874439 49.34191 1.156001e-04 -6.865849
1678 NINL 20_19 0.7830803 32.87526 7.659455e-05 -5.599931
12851 CYP2A6 19_28 0.7799696 21.54230 4.999105e-05 -3.989619
13574 RP11-108M9.6 1_12 0.7771895 28.35156 6.555809e-05 -4.914590
7010 NBL1 1_13 0.7555923 34.04073 7.652597e-05 -5.638117
4404 TUBG1 17_25 0.7230289 30.27122 6.511904e-05 -5.660250
13759 RP11-823E8.3 12_54 0.7000492 103.65878 2.159022e-04 -6.438012
4137 PODXL 7_80 0.6950352 26.19268 5.416382e-05 4.018009
9422 SOX11 2_4 0.6665243 26.52941 5.260971e-05 4.517012
7829 YWHAZ 8_69 0.6475680 22.77961 4.388884e-05 4.235328
5519 CDH13 16_46 0.6397455 23.81304 4.532570e-05 -4.826363
1132 RRN3 16_15 0.6239131 22.13907 4.109661e-05 4.374275
12949 RP11-566J3.2 14_52 0.6215001 34.21108 6.326019e-05 -5.489252
2821 SENP6 6_52 0.6090897 25.38256 4.599801e-05 -4.618392
num_eqtl
8160 2
9866 2
3378 1
9479 2
7777 2
13756 2
7320 1
1678 2
12851 1
13574 1
7010 1
4404 1
13759 1
4137 1
9422 1
7829 1
5519 1
1132 1
12949 1
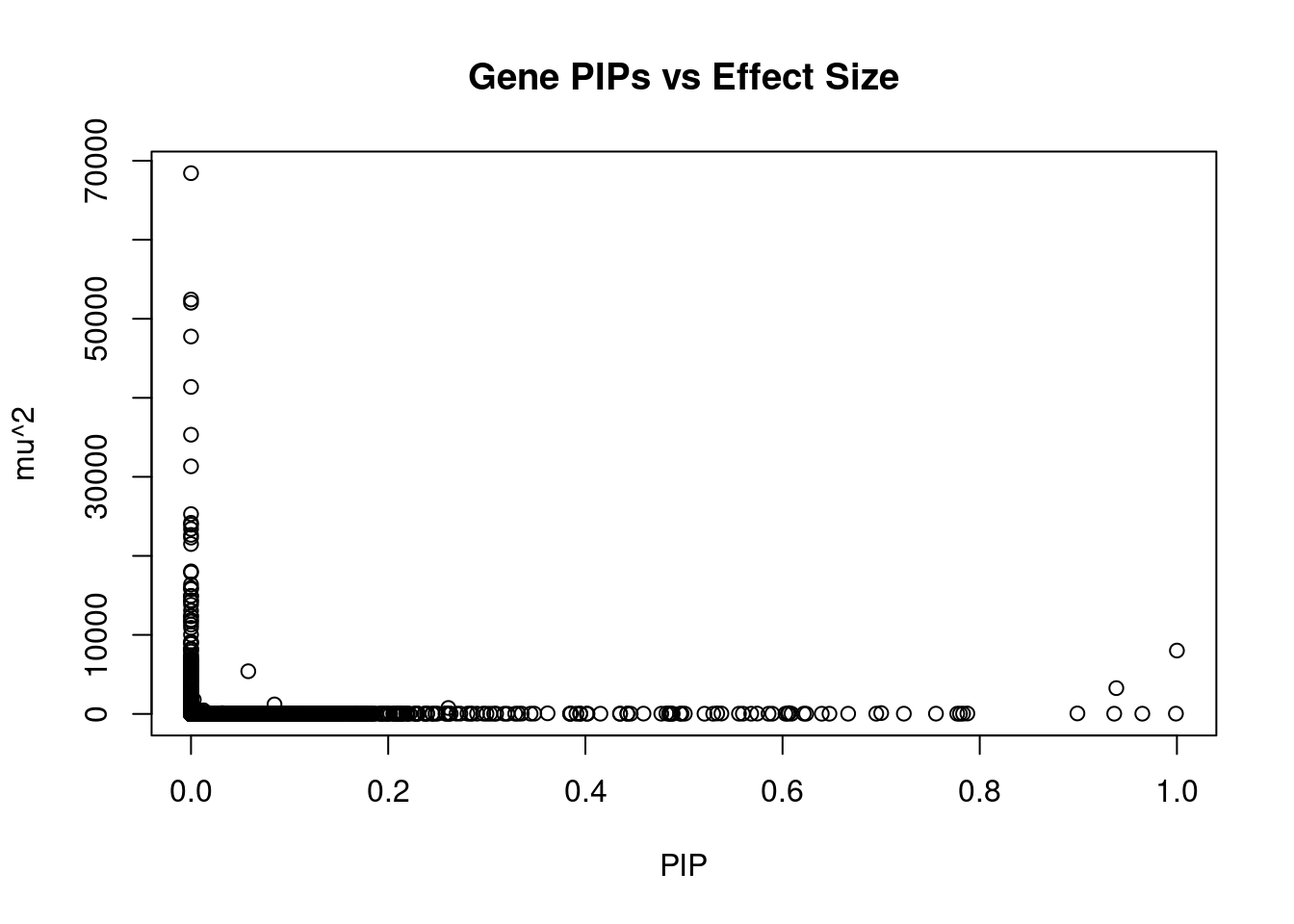
2821 2Genes with largest effect sizes
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
genename region_tag susie_pip mu2 PVE z
10604 SLC38A3 3_35 0.000000e+00 68425.51 0.000000e+00 6.725828
7846 CCDC171 9_13 0.000000e+00 52440.50 0.000000e+00 7.972850
7671 CAMKV 3_35 0.000000e+00 52028.02 0.000000e+00 -9.574047
2153 PIK3R2 19_14 0.000000e+00 47754.62 0.000000e+00 -7.140312
36 RBM6 3_35 0.000000e+00 41375.93 0.000000e+00 12.536042
7673 MST1R 3_35 0.000000e+00 35329.04 0.000000e+00 -12.622779
9620 STX19 3_59 0.000000e+00 31328.70 0.000000e+00 -5.059656
5542 NOB1 16_37 0.000000e+00 25284.54 0.000000e+00 2.762572
10375 GSAP 7_49 0.000000e+00 24182.56 0.000000e+00 5.119800
5442 MFAP1 15_16 4.268919e-12 23938.26 3.040415e-13 4.302998
7668 RNF123 3_35 0.000000e+00 23402.76 0.000000e+00 -10.957103
5264 TMOD3 15_21 0.000000e+00 22653.47 0.000000e+00 -4.222248
5446 LYSMD2 15_21 0.000000e+00 22304.53 0.000000e+00 -4.402599
1353 WDR76 15_16 0.000000e+00 21497.96 0.000000e+00 4.808984
901 MCM6 2_80 0.000000e+00 18037.55 0.000000e+00 -3.886179
2155 PDE4C 19_14 0.000000e+00 17890.71 0.000000e+00 6.667721
5158 TUBGCP4 15_16 0.000000e+00 16376.38 0.000000e+00 3.554938
1434 MAST3 19_14 0.000000e+00 15938.64 0.000000e+00 -2.208055
9613 NSUN3 3_59 0.000000e+00 15791.55 0.000000e+00 4.755360
8457 LCMT2 15_16 0.000000e+00 14934.90 0.000000e+00 -2.861302
num_eqtl
10604 1
7846 2
7671 2
2153 1
36 1
7673 2
9620 1
5542 1
10375 3
5442 1
7668 1
5264 1
5446 1
1353 2
901 1
2155 1
5158 2
1434 1
9613 1
8457 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z
8160 SMAD3 15_31 0.99999954 8015.87282 2.384916e-02 -2.852677
9479 NIM1K 5_28 0.93853991 3262.53413 9.110249e-03 5.550554
13498 CTC-498M16.4 5_52 0.05807462 5398.39878 9.327683e-04 7.882989
10849 TTC30B 2_107 0.26101401 752.00020 5.839884e-04 -3.137443
7062 ADPGK 15_34 0.08467030 1200.49261 3.024217e-04 5.925192
13759 RP11-823E8.3 12_54 0.70004918 103.65878 2.159022e-04 -6.438012
10660 SKOR1 15_31 0.60657759 97.95638 1.767834e-04 -9.879755
13756 NOL12 22_15 0.89904930 61.44780 1.643661e-04 -4.510837
7320 TAL1 1_29 0.78744386 49.34191 1.156001e-04 -6.865849
9866 TMIE 3_33 0.99897790 34.99719 1.040187e-04 -7.091659
13962 DHRS11 17_22 0.53374351 61.96358 9.839920e-05 -8.128326
8419 ATXN2L 16_23 0.36166789 77.22963 8.310294e-05 -10.702325
3378 CCND2 12_4 0.96489977 28.86368 8.286217e-05 -5.119990
5575 C18orf8 18_12 0.48218671 57.70858 8.279004e-05 7.477424
1678 NINL 20_19 0.78308033 32.87526 7.659455e-05 -5.599931
7010 NBL1 1_13 0.75559234 34.04073 7.652597e-05 -5.638117
7777 ZNF12 7_9 0.93639821 27.17655 7.571420e-05 5.127453
5380 SUOX 12_35 0.53798979 47.10209 7.539398e-05 -5.190773
385 PHLPP2 16_38 0.57430624 41.84005 7.149212e-05 4.618775
13574 RP11-108M9.6 1_12 0.77718948 28.35156 6.555809e-05 -4.914590
num_eqtl
8160 2
9479 2
13498 1
10849 1
7062 3
13759 1
10660 2
13756 2
7320 1
9866 2
13962 1
8419 1
3378 1
5575 2
1678 2
7010 1
7777 2
5380 1
385 1
13574 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z
7673 MST1R 3_35 0.0000000000 35329.04340 0.000000e+00 -12.622779
36 RBM6 3_35 0.0000000000 41375.93494 0.000000e+00 12.536042
7668 RNF123 3_35 0.0000000000 23402.76132 0.000000e+00 -10.957103
8554 INO80E 16_24 0.0316645156 99.46356 9.370425e-06 10.886319
8419 ATXN2L 16_23 0.3616678947 77.22963 8.310294e-05 -10.702325
10629 CLN3 16_23 0.0417000087 75.23465 9.334187e-06 10.363250
10875 C6orf106 6_28 0.0000256066 120.40834 9.173411e-09 -10.263559
12178 NPIPB7 16_23 0.0366258145 74.39248 8.106601e-06 10.037986
8218 ZNF668 16_24 0.1121179514 77.18576 2.574748e-05 10.000364
10660 SKOR1 15_31 0.6065775943 97.95638 1.767834e-04 -9.879755
9435 NFATC2IP 16_23 0.0457742783 75.97829 1.034745e-05 -9.863387
4330 ZC3H4 19_33 0.0052482859 98.99347 1.545776e-06 9.848747
1906 KAT8 16_24 0.0170220382 72.06602 3.649762e-06 -9.785239
1905 BCKDK 16_24 0.0124311403 68.64839 2.539006e-06 9.637985
11636 NDUFS3 11_29 0.0131019911 84.88855 3.309092e-06 -9.629039
7671 CAMKV 3_35 0.0000000000 52028.02173 0.000000e+00 -9.574047
8924 C1QTNF4 11_29 0.0113720906 83.81941 2.836007e-06 9.563515
11640 LAT 16_23 0.0724465661 77.05500 1.660891e-05 -9.552834
10852 FAM180B 11_29 0.0108339097 82.28414 2.652307e-06 -9.489956
2550 MTCH2 11_29 0.0105269672 81.10908 2.540359e-06 -9.432202
num_eqtl
7673 2
36 1
7668 1
8554 2
8419 1
10629 2
10875 1
12178 1
8218 1
10660 2
9435 1
4330 1
1906 1
1905 1
11636 2
7671 2
8924 1
11640 1
10852 1
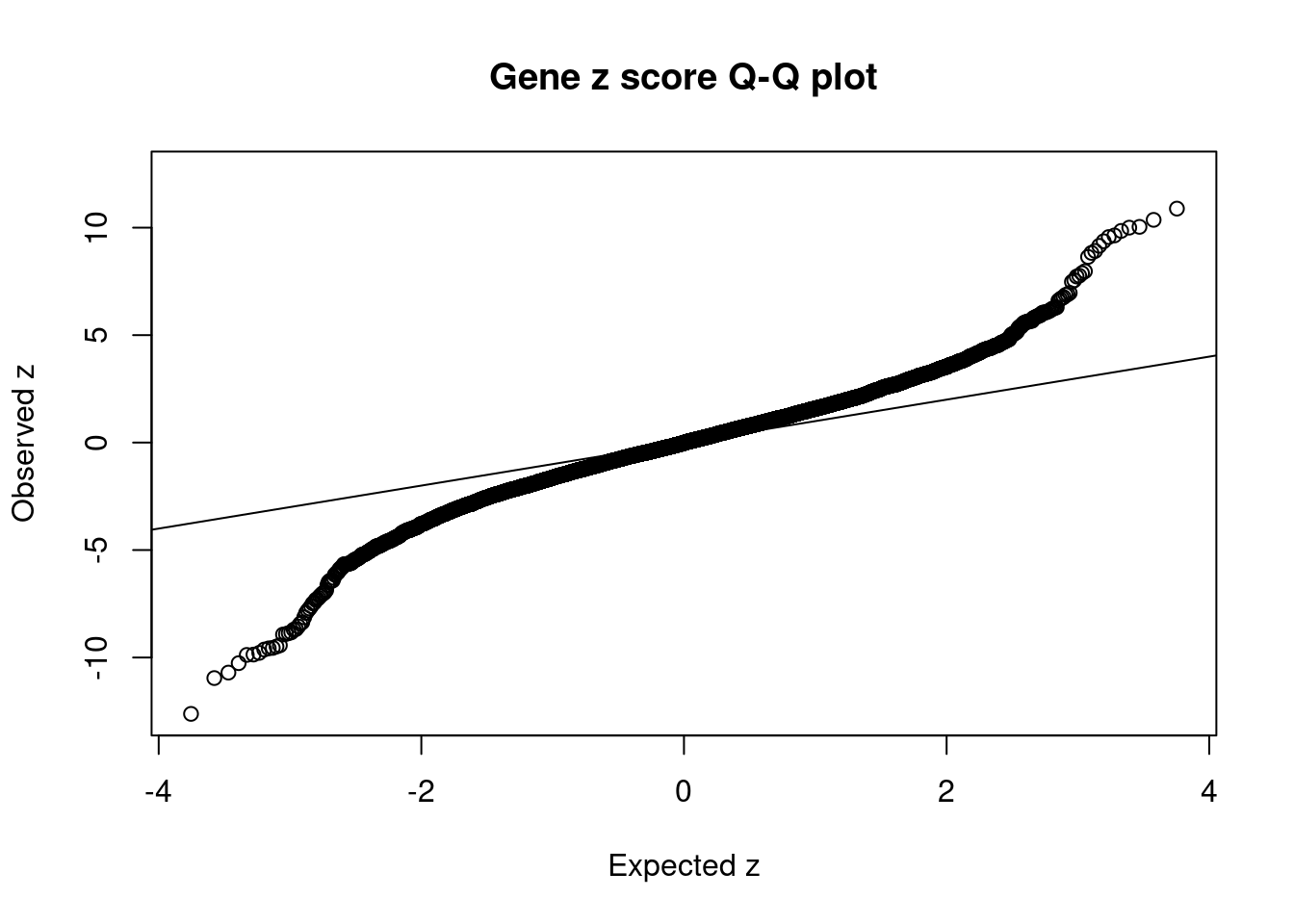
2550 1Comparing z scores and PIPs
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
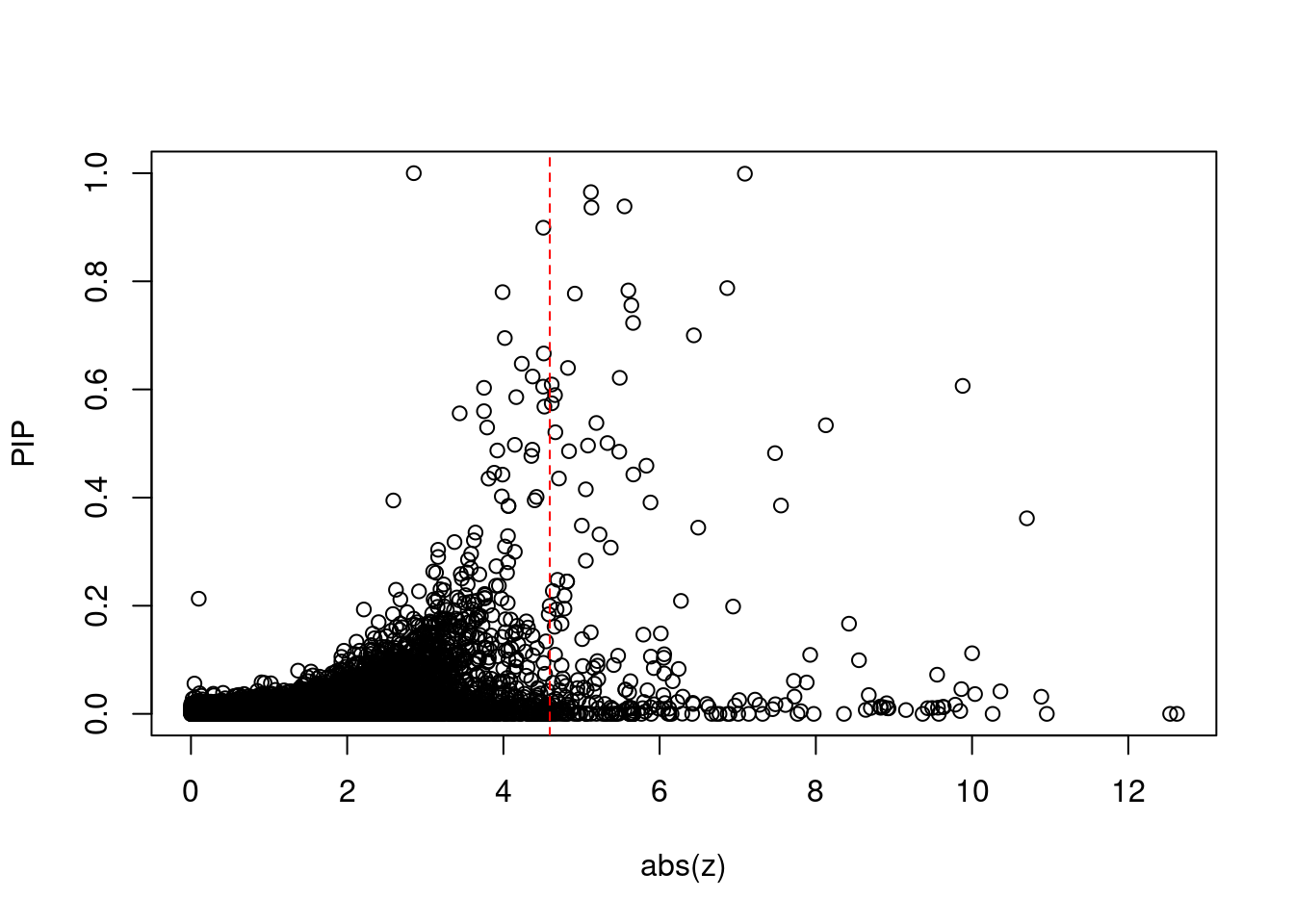
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
[1] 0.02002263 genename region_tag susie_pip mu2 PVE z
7673 MST1R 3_35 0.0000000000 35329.04340 0.000000e+00 -12.622779
36 RBM6 3_35 0.0000000000 41375.93494 0.000000e+00 12.536042
7668 RNF123 3_35 0.0000000000 23402.76132 0.000000e+00 -10.957103
8554 INO80E 16_24 0.0316645156 99.46356 9.370425e-06 10.886319
8419 ATXN2L 16_23 0.3616678947 77.22963 8.310294e-05 -10.702325
10629 CLN3 16_23 0.0417000087 75.23465 9.334187e-06 10.363250
10875 C6orf106 6_28 0.0000256066 120.40834 9.173411e-09 -10.263559
12178 NPIPB7 16_23 0.0366258145 74.39248 8.106601e-06 10.037986
8218 ZNF668 16_24 0.1121179514 77.18576 2.574748e-05 10.000364
10660 SKOR1 15_31 0.6065775943 97.95638 1.767834e-04 -9.879755
9435 NFATC2IP 16_23 0.0457742783 75.97829 1.034745e-05 -9.863387
4330 ZC3H4 19_33 0.0052482859 98.99347 1.545776e-06 9.848747
1906 KAT8 16_24 0.0170220382 72.06602 3.649762e-06 -9.785239
1905 BCKDK 16_24 0.0124311403 68.64839 2.539006e-06 9.637985
11636 NDUFS3 11_29 0.0131019911 84.88855 3.309092e-06 -9.629039
7671 CAMKV 3_35 0.0000000000 52028.02173 0.000000e+00 -9.574047
8924 C1QTNF4 11_29 0.0113720906 83.81941 2.836007e-06 9.563515
11640 LAT 16_23 0.0724465661 77.05500 1.660891e-05 -9.552834
10852 FAM180B 11_29 0.0108339097 82.28414 2.652307e-06 -9.489956
2550 MTCH2 11_29 0.0105269672 81.10908 2.540359e-06 -9.432202
num_eqtl
7673 2
36 1
7668 1
8554 2
8419 1
10629 2
10875 1
12178 1
8218 1
10660 2
9435 1
4330 1
1906 1
1905 1
11636 2
7671 2
8924 1
11640 1
10852 1
2550 1Sensitivity, specificity and precision for silver standard genes
[1] 41[1] 24[1] 4.593787[1] 6[1] 230 genename region_tag susie_pip mu2 PVE z num_eqtl
8160 SMAD3 15_31 0.9999995 8015.8728 0.0238491585 -2.852677 2
13756 NOL12 22_15 0.8990493 61.4478 0.0001643661 -4.510837 2 ctwas TWAS
0.00000000 0.04878049 ctwas TWAS
0.9994766 0.9801099 ctwas TWAS
0.000000000 0.008695652 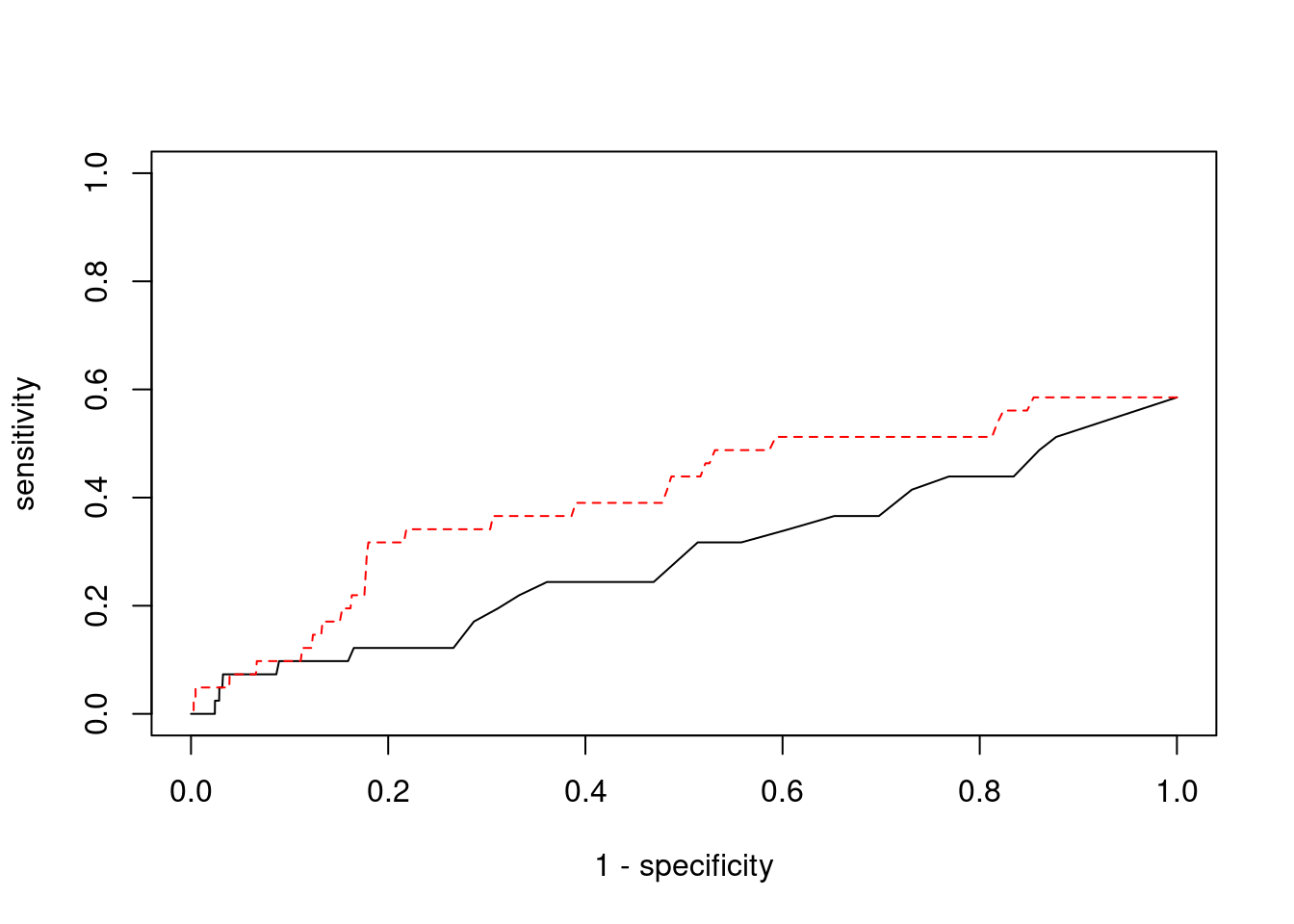
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] tidyselect_1.1.1 xfun_0.29 purrr_0.3.4 colorspace_2.0-2
[5] vctrs_0.3.8 generics_0.1.1 htmltools_0.5.2 yaml_2.2.1
[9] utf8_1.2.2 blob_1.2.2 rlang_0.4.12 jquerylib_0.1.4
[13] later_0.8.0 pillar_1.6.4 glue_1.5.1 withr_2.4.3
[17] DBI_1.1.1 bit64_4.0.5 lifecycle_1.0.1 stringr_1.4.0
[21] cellranger_1.1.0 munsell_0.5.0 gtable_0.3.0 evaluate_0.14
[25] memoise_2.0.1 labeling_0.4.2 knitr_1.36 fastmap_1.1.0
[29] httpuv_1.5.1 fansi_0.5.0 highr_0.9 Rcpp_1.0.7
[33] promises_1.0.1 scales_1.1.1 cachem_1.0.6 farver_2.1.0
[37] fs_1.5.2 bit_4.0.4 digest_0.6.29 stringi_1.7.6
[41] dplyr_1.0.7 rprojroot_2.0.2 grid_3.6.1 tools_3.6.1
[45] magrittr_2.0.1 tibble_3.1.6 RSQLite_2.2.8 crayon_1.4.2
[49] whisker_0.3-2 pkgconfig_2.0.3 ellipsis_0.3.2 data.table_1.14.2
[53] assertthat_0.2.1 rmarkdown_2.11 R6_2.5.1 git2r_0.26.1
[57] compiler_3.6.1