T2D - Adipose Subcutaneous
sheng Qian
2021-2-6
Last updated: 2022-02-26
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0e6a2f2. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/AF/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Unstaged changes:
Modified: analysis/BMI_Brain_Amygdala_S.Rmd
Modified: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Modified: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Modified: analysis/BMI_Brain_Cerebellum_S.Rmd
Modified: analysis/BMI_Brain_Cortex.Rmd
Modified: analysis/BMI_Brain_Cortex_S.Rmd
Modified: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Modified: analysis/BMI_Brain_Hippocampus_S.Rmd
Modified: analysis/BMI_Brain_Hypothalamus_S.Rmd
Modified: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Modified: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Modified: analysis/LDL_Liver_S.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/T2D_Adipose_Subcutaneous.Rmd) and HTML (docs/T2D_Adipose_Subcutaneous.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 91f38fa | sq-96 | 2022-02-13 | Build site. |
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
[1] 8591
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
874 679 536 364 458 498 421 323 333 362 534 523 182 297 322 342 434 142 429 246
21 22
102 190 [1] 4545[1] 0.529042Load ctwas results
Check convergence of parameters
********************************************************Note: As of version 1.0.0, cowplot does not change the default ggplot2 theme anymore. To recover the previous behavior, execute:
theme_set(theme_cowplot())********************************************************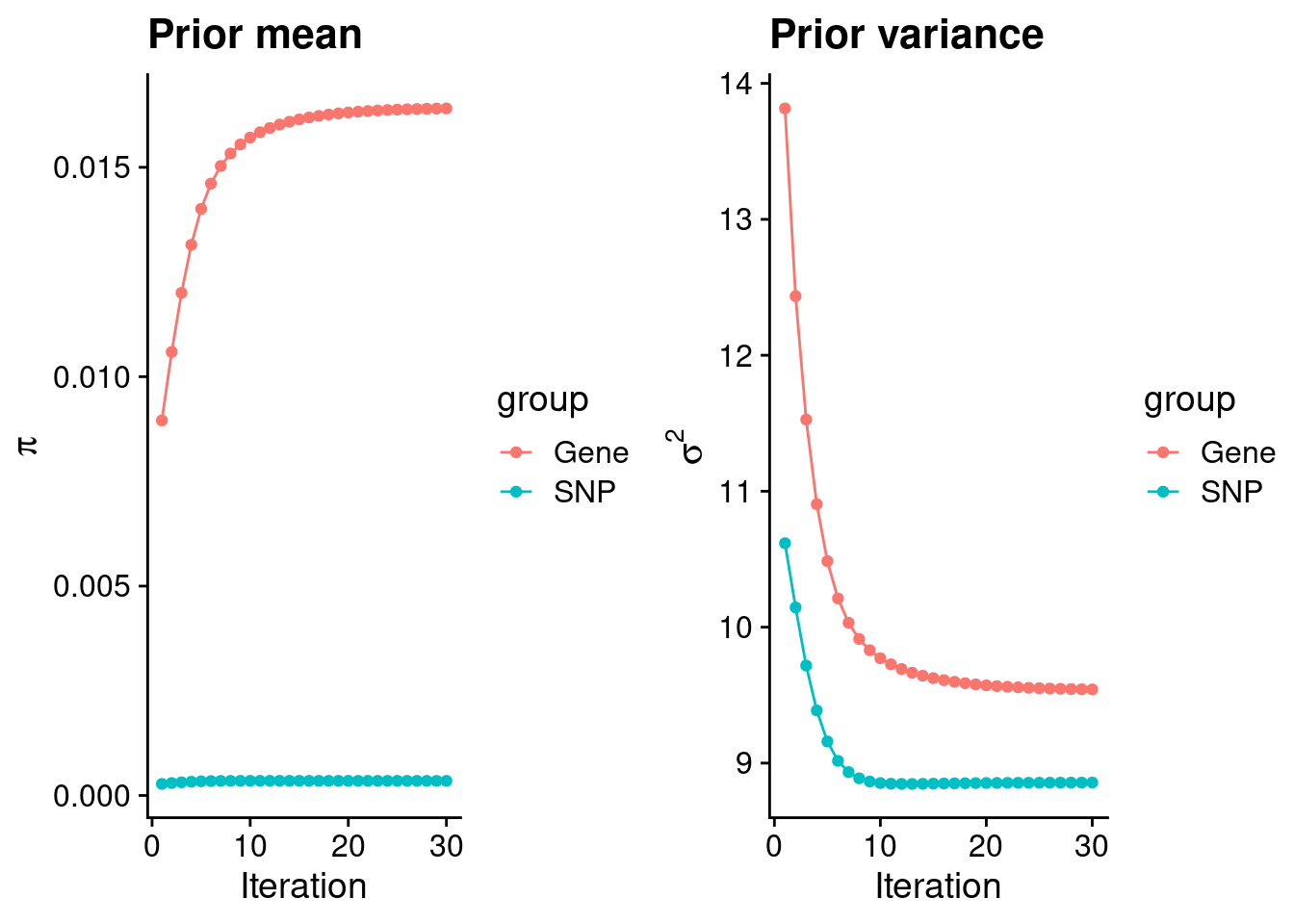
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
gene snp
0.0164052744 0.0003491226 gene snp
9.541881 8.856665 [1] 62892[1] 8591 5017200 gene snp
0.02138286 0.24666880 [1] 0.097563 1.394008Genes with highest PIPs
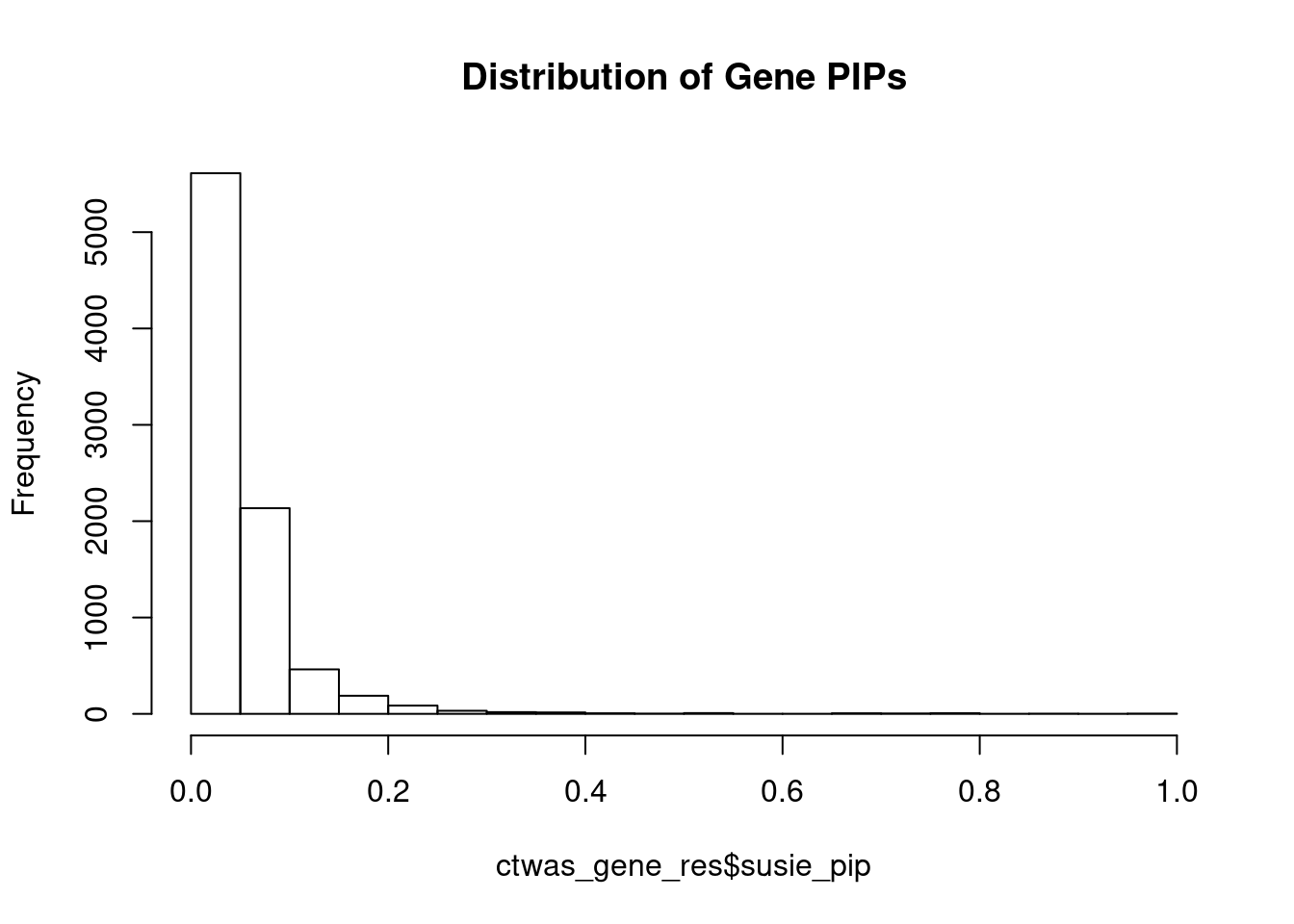
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
genename region_tag susie_pip mu2 PVE z num_eqtl
7109 LONRF1 8_15 0.9929090 29.21253 0.0004611935 -5.424976 2
5112 P2RX4 12_74 0.9614949 25.94139 0.0003965927 5.081395 1
250 ANGEL1 14_36 0.9577010 22.67702 0.0003453190 4.529353 2
7272 KCNS2 8_68 0.9084550 26.21428 0.0003786570 -5.148148 1
7654 PTH1R 3_33 0.8979756 30.11423 0.0004299728 -5.646341 1
6918 ARL14EP 11_21 0.8714865 23.28173 0.0003226120 -4.877816 2
13239 N4BP2L2 13_11 0.8441484 24.33744 0.0003266617 -4.430380 1
10030 FAM89B 11_36 0.7961326 24.33365 0.0003080330 5.011364 1
8836 FAM234A 16_1 0.7887942 31.18972 0.0003911829 5.621053 1
11712 PARVA 11_9 0.7846449 20.35377 0.0002539351 -3.862069 1
7267 GDF6 8_67 0.7821456 21.36036 0.0002656444 -4.371795 1
3924 SEPT7 7_26 0.7750777 20.71351 0.0002552722 -4.166667 1
3988 ZC3H13 13_19 0.7732369 20.55708 0.0002527427 4.310345 1
2323 DNASE2 19_10 0.7718480 18.79133 0.0002306184 -3.744186 1
4009 KBTBD4 11_29 0.7473759 26.46156 0.0003144555 -5.097561 1
3282 GRB14 2_100 0.7261964 21.31644 0.0002461349 4.744898 1
11149 SLIT1 10_62 0.7054051 24.44621 0.0002741919 4.797347 2
7490 UBE2Z 17_28 0.7027425 47.07794 0.0005260394 -7.392405 1
6245 MRPS5 2_57 0.6981455 19.67432 0.0002183988 -3.809979 2
10469 TMEM132E 17_20 0.6953548 21.32974 0.0002358287 -3.692999 2Genes with largest effect sizes
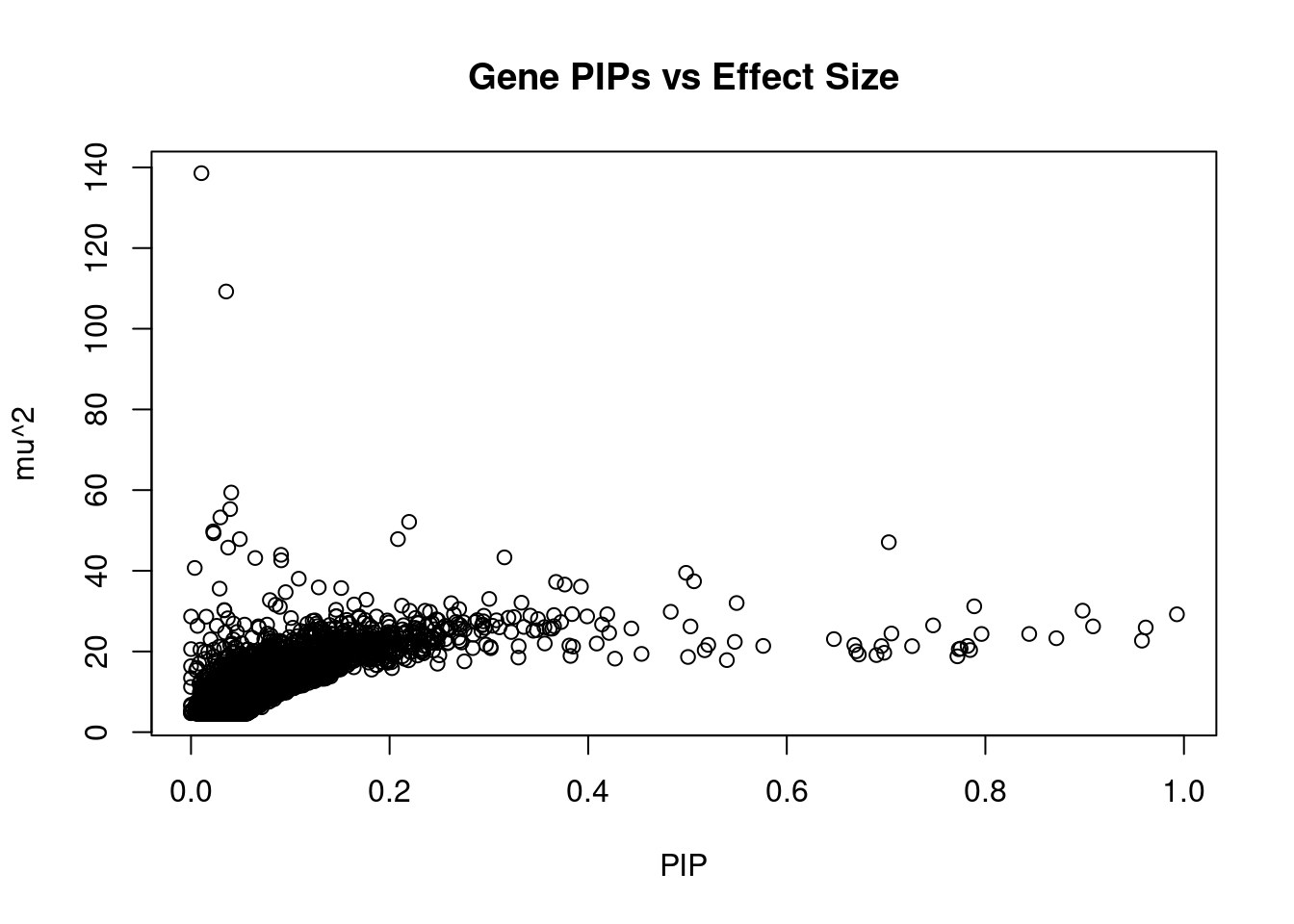
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
genename region_tag susie_pip mu2 PVE z
7048 JAZF1 7_23 0.010489657 138.57315 2.311240e-05 -12.730769
14285 RP11-395N3.2 2_133 0.035388941 109.23628 6.146658e-05 -11.370370
356 ANK1 8_36 0.040525872 59.39273 3.827104e-05 8.666667
6954 ZFP36L2 2_27 0.039417323 55.29285 3.465458e-05 8.376923
2723 WFS1 4_7 0.029567029 53.22895 2.502420e-05 10.641975
8537 C2 6_26 0.219640555 52.12414 1.820355e-04 7.148936
11174 KCNJ11 11_12 0.022352569 49.76935 1.768862e-05 7.230769
11270 NCR3LG1 11_12 0.022725263 49.30958 1.781742e-05 -7.168932
14659 LINC01126 2_27 0.049133590 47.84997 3.738219e-05 -7.520384
1417 PABPC4 1_24 0.208389179 47.84681 1.585378e-04 -7.054348
7490 UBE2Z 17_28 0.702742531 47.07794 5.260394e-04 -7.392405
3368 THADA 2_27 0.037435740 45.74582 2.722967e-05 7.450450
943 ZZEF1 17_4 0.090686616 43.96105 6.338928e-05 6.916667
10282 ZNF664 12_75 0.315660986 43.32163 2.174354e-04 -6.452055
7491 SNF8 17_28 0.064661984 43.16571 4.438053e-05 6.300000
10726 BMP8A 1_24 0.090840980 42.58988 6.151667e-05 6.868132
6449 CDKAL1 6_15 0.003751902 40.70165 2.428108e-06 -8.191860
10555 UBE2E2 3_17 0.498580721 39.49123 3.130695e-04 6.080882
3647 CCDC92 12_75 0.108517987 38.03243 6.562365e-05 -5.498713
13467 RP11-419C23.1 8_33 0.506566550 37.39164 3.011727e-04 -6.316832
num_eqtl
7048 1
14285 1
356 1
6954 1
2723 1
8537 1
11174 1
11270 2
14659 2
1417 1
7490 1
3368 1
943 1
10282 1
7491 1
10726 1
6449 1
10555 1
3647 5
13467 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z
7490 UBE2Z 17_28 0.7027425 47.07794 0.0005260394 -7.392405
7109 LONRF1 8_15 0.9929090 29.21253 0.0004611935 -5.424976
7654 PTH1R 3_33 0.8979756 30.11423 0.0004299728 -5.646341
5112 P2RX4 12_74 0.9614949 25.94139 0.0003965927 5.081395
8836 FAM234A 16_1 0.7887942 31.18972 0.0003911829 5.621053
7272 KCNS2 8_68 0.9084550 26.21428 0.0003786570 -5.148148
250 ANGEL1 14_36 0.9577010 22.67702 0.0003453190 4.529353
13239 N4BP2L2 13_11 0.8441484 24.33744 0.0003266617 -4.430380
6918 ARL14EP 11_21 0.8714865 23.28173 0.0003226120 -4.877816
4009 KBTBD4 11_29 0.7473759 26.46156 0.0003144555 -5.097561
10555 UBE2E2 3_17 0.4985807 39.49123 0.0003130695 6.080882
10030 FAM89B 11_36 0.7961326 24.33365 0.0003080330 5.011364
13467 RP11-419C23.1 8_33 0.5065666 37.39164 0.0003011727 -6.316832
6645 CAMK2G 10_49 0.5494966 32.00592 0.0002796404 5.012658
11149 SLIT1 10_62 0.7054051 24.44621 0.0002741919 4.797347
7267 GDF6 8_67 0.7821456 21.36036 0.0002656444 -4.371795
3924 SEPT7 7_26 0.7750777 20.71351 0.0002552722 -4.166667
11712 PARVA 11_9 0.7846449 20.35377 0.0002539351 -3.862069
3988 ZC3H13 13_19 0.7732369 20.55708 0.0002527427 4.310345
3282 GRB14 2_100 0.7261964 21.31644 0.0002461349 4.744898
num_eqtl
7490 1
7109 2
7654 1
5112 1
8836 1
7272 1
250 2
13239 1
6918 2
4009 1
10555 1
10030 1
13467 1
6645 1
11149 2
7267 1
3924 1
11712 1
3988 1
3282 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z
7048 JAZF1 7_23 0.010489657 138.57315 2.311240e-05 -12.730769
14285 RP11-395N3.2 2_133 0.035388941 109.23628 6.146658e-05 -11.370370
2723 WFS1 4_7 0.029567029 53.22895 2.502420e-05 10.641975
356 ANK1 8_36 0.040525872 59.39273 3.827104e-05 8.666667
6954 ZFP36L2 2_27 0.039417323 55.29285 3.465458e-05 8.376923
6449 CDKAL1 6_15 0.003751902 40.70165 2.428108e-06 -8.191860
14659 LINC01126 2_27 0.049133590 47.84997 3.738219e-05 -7.520384
3368 THADA 2_27 0.037435740 45.74582 2.722967e-05 7.450450
7490 UBE2Z 17_28 0.702742531 47.07794 5.260394e-04 -7.392405
11174 KCNJ11 11_12 0.022352569 49.76935 1.768862e-05 7.230769
11270 NCR3LG1 11_12 0.022725263 49.30958 1.781742e-05 -7.168932
8537 C2 6_26 0.219640555 52.12414 1.820355e-04 7.148936
1417 PABPC4 1_24 0.208389179 47.84681 1.585378e-04 -7.054348
943 ZZEF1 17_4 0.090686616 43.96105 6.338928e-05 6.916667
10726 BMP8A 1_24 0.090840980 42.58988 6.151667e-05 6.868132
10282 ZNF664 12_75 0.315660986 43.32163 2.174354e-04 -6.452055
12119 MICB 6_25 0.483137672 29.83545 2.291966e-04 6.443315
13467 RP11-419C23.1 8_33 0.506566550 37.39164 3.011727e-04 -6.316832
7491 SNF8 17_28 0.064661984 43.16571 4.438053e-05 6.300000
7368 AP3S2 15_41 0.392663862 36.07772 2.252499e-04 6.272710
num_eqtl
7048 1
14285 1
2723 1
356 1
6954 1
6449 1
14659 2
3368 1
7490 1
11174 1
11270 2
8537 1
1417 1
943 1
10726 1
10282 1
12119 3
13467 1
7491 1
7368 2Comparing z scores and PIPs
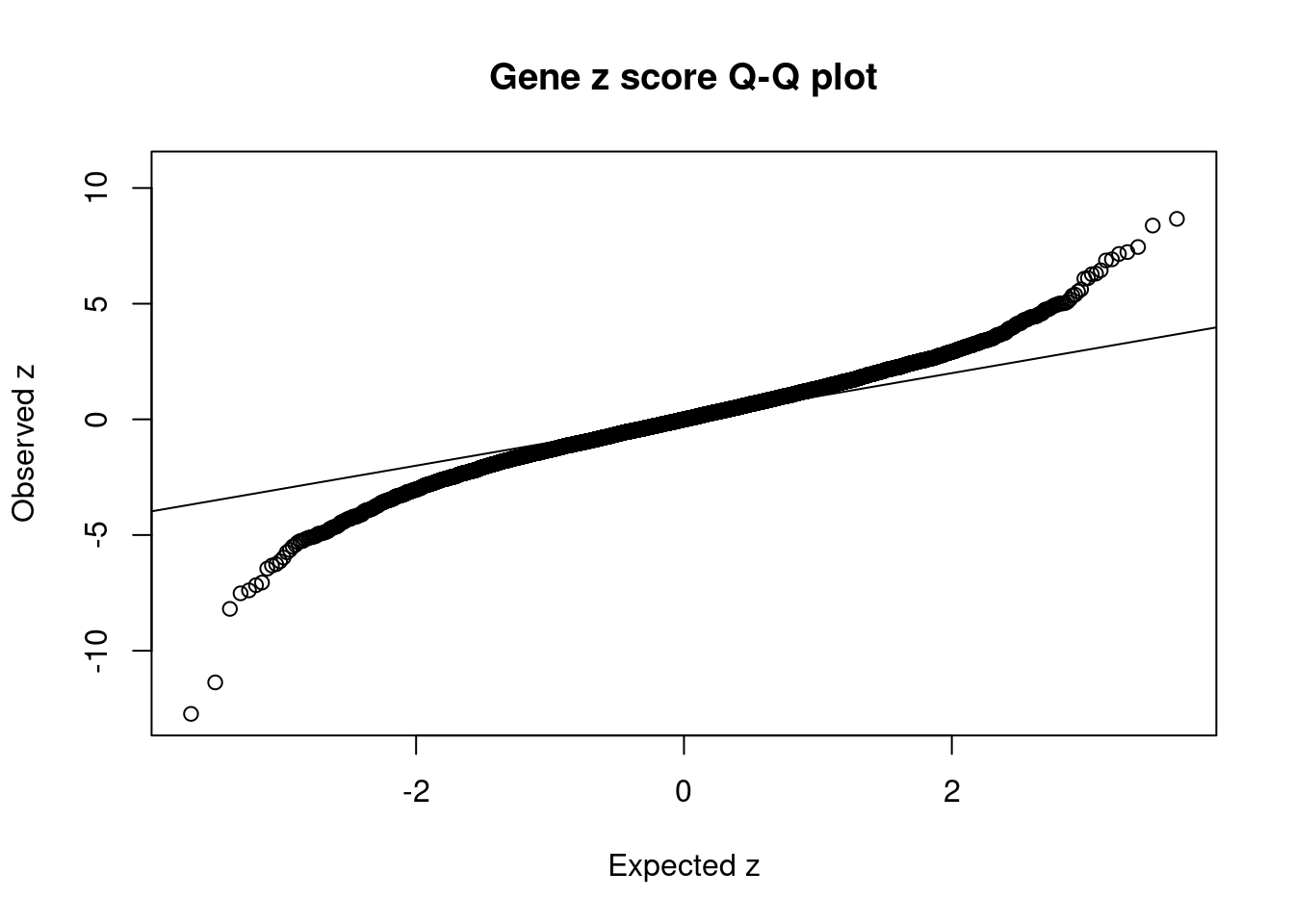
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
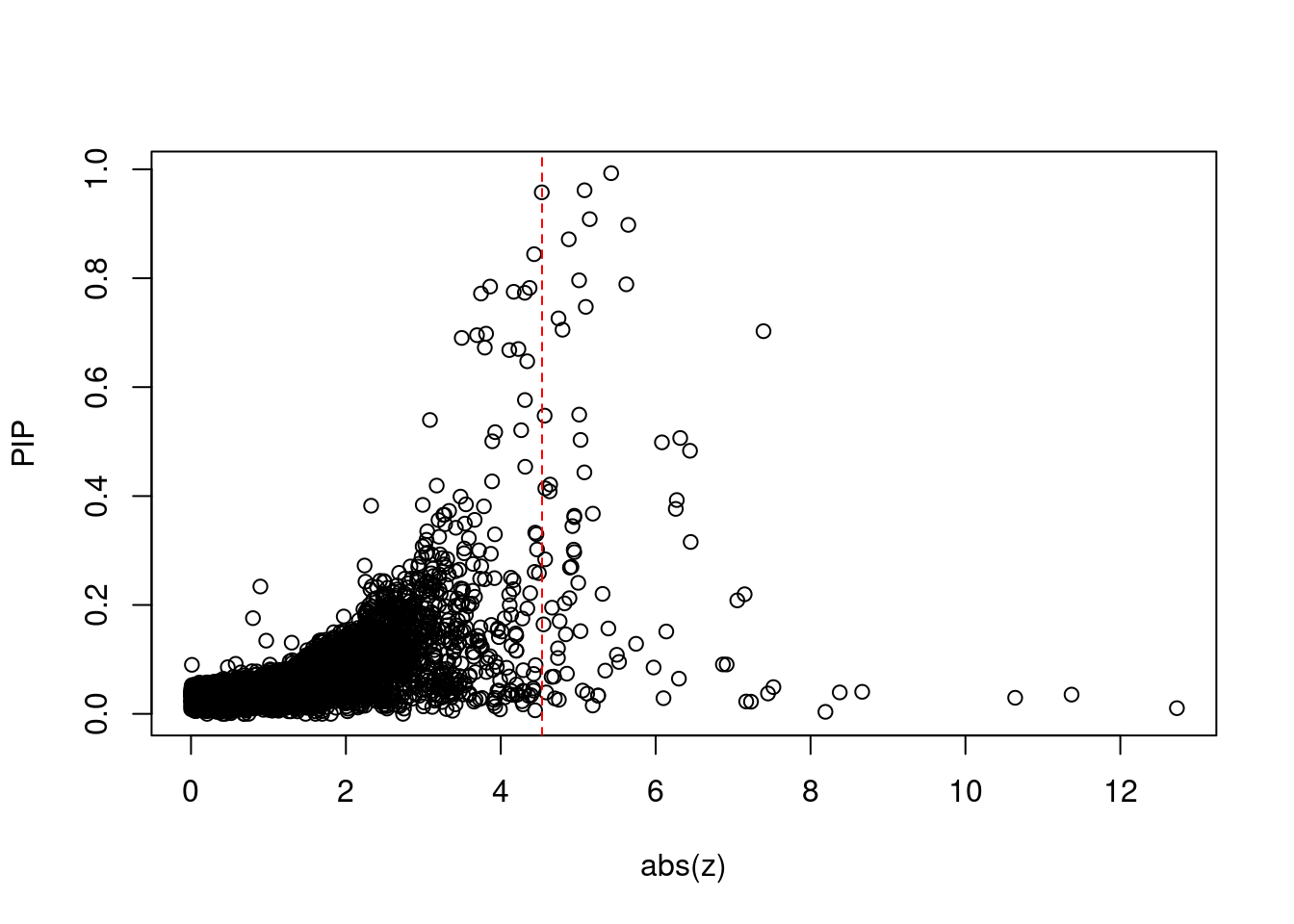
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
[1] 0.009079269 genename region_tag susie_pip mu2 PVE z
7048 JAZF1 7_23 0.010489657 138.57315 2.311240e-05 -12.730769
14285 RP11-395N3.2 2_133 0.035388941 109.23628 6.146658e-05 -11.370370
2723 WFS1 4_7 0.029567029 53.22895 2.502420e-05 10.641975
356 ANK1 8_36 0.040525872 59.39273 3.827104e-05 8.666667
6954 ZFP36L2 2_27 0.039417323 55.29285 3.465458e-05 8.376923
6449 CDKAL1 6_15 0.003751902 40.70165 2.428108e-06 -8.191860
14659 LINC01126 2_27 0.049133590 47.84997 3.738219e-05 -7.520384
3368 THADA 2_27 0.037435740 45.74582 2.722967e-05 7.450450
7490 UBE2Z 17_28 0.702742531 47.07794 5.260394e-04 -7.392405
11174 KCNJ11 11_12 0.022352569 49.76935 1.768862e-05 7.230769
11270 NCR3LG1 11_12 0.022725263 49.30958 1.781742e-05 -7.168932
8537 C2 6_26 0.219640555 52.12414 1.820355e-04 7.148936
1417 PABPC4 1_24 0.208389179 47.84681 1.585378e-04 -7.054348
943 ZZEF1 17_4 0.090686616 43.96105 6.338928e-05 6.916667
10726 BMP8A 1_24 0.090840980 42.58988 6.151667e-05 6.868132
10282 ZNF664 12_75 0.315660986 43.32163 2.174354e-04 -6.452055
12119 MICB 6_25 0.483137672 29.83545 2.291966e-04 6.443315
13467 RP11-419C23.1 8_33 0.506566550 37.39164 3.011727e-04 -6.316832
7491 SNF8 17_28 0.064661984 43.16571 4.438053e-05 6.300000
7368 AP3S2 15_41 0.392663862 36.07772 2.252499e-04 6.272710
num_eqtl
7048 1
14285 1
2723 1
356 1
6954 1
6449 1
14659 2
3368 1
7490 1
11174 1
11270 2
8537 1
1417 1
943 1
10726 1
10282 1
12119 3
13467 1
7491 1
7368 2Sensitivity, specificity and precision for silver standard genes
[1] 72[1] 25[1] 4.532824[1] 7[1] 78 genename region_tag susie_pip mu2 PVE z num_eqtl
13239 N4BP2L2 13_11 0.8441484 24.33744 0.0003266617 -4.430380 1
250 ANGEL1 14_36 0.9577010 22.67702 0.0003453190 4.529353 2 ctwas TWAS
0.00000000 0.02777778 ctwas TWAS
0.9991828 0.9911277 ctwas TWAS
0.00000000 0.02564103 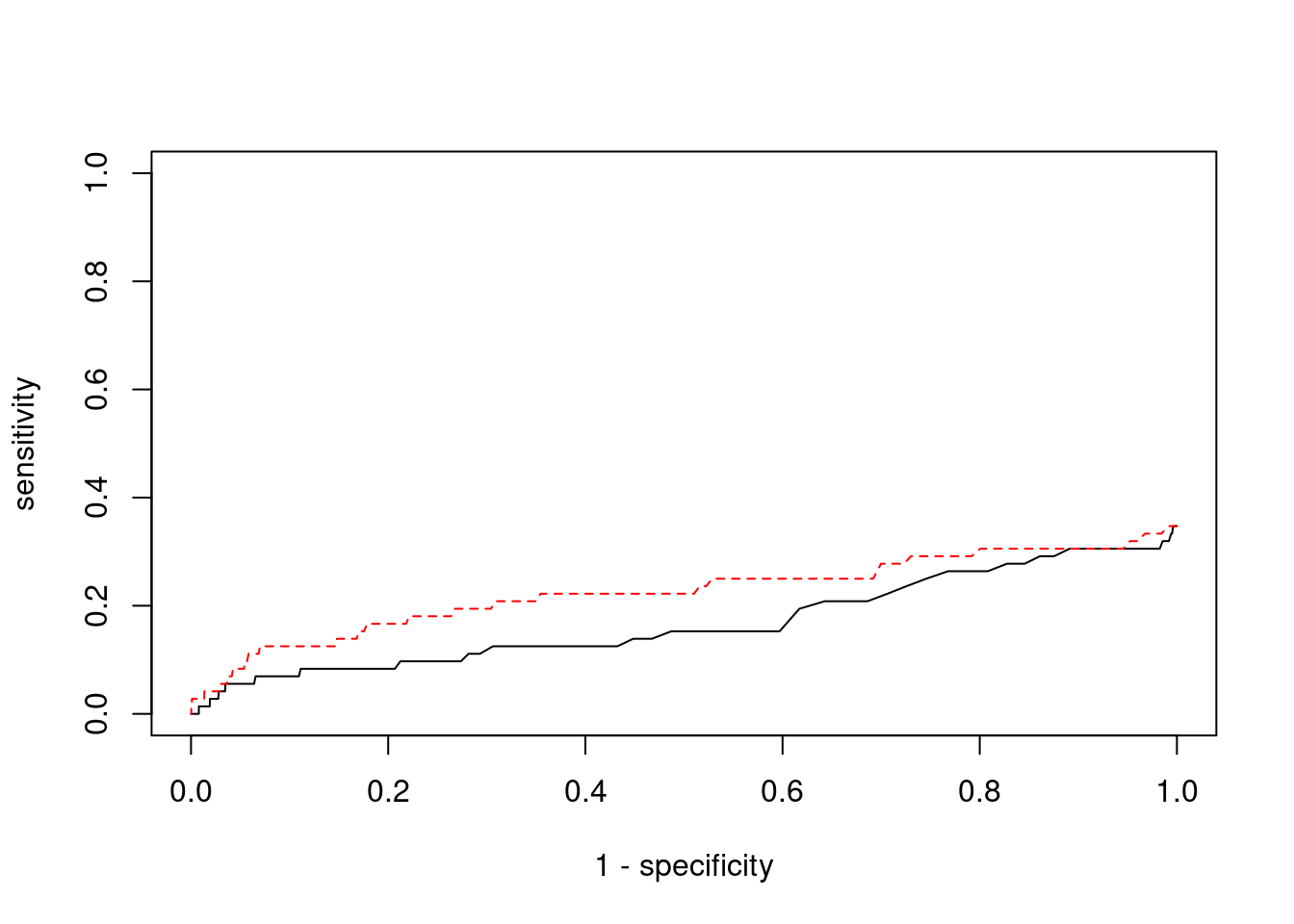
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] tidyselect_1.1.1 xfun_0.29 purrr_0.3.4 colorspace_2.0-2
[5] vctrs_0.3.8 generics_0.1.1 htmltools_0.5.2 yaml_2.2.1
[9] utf8_1.2.2 blob_1.2.2 rlang_1.0.1 jquerylib_0.1.4
[13] later_0.8.0 pillar_1.6.4 glue_1.6.2 withr_2.4.3
[17] DBI_1.1.2 bit64_4.0.5 lifecycle_1.0.1 stringr_1.4.0
[21] cellranger_1.1.0 munsell_0.5.0 gtable_0.3.0 evaluate_0.14
[25] memoise_2.0.1 labeling_0.4.2 knitr_1.36 fastmap_1.1.0
[29] httpuv_1.5.1 fansi_1.0.2 highr_0.9 Rcpp_1.0.8
[33] promises_1.0.1 scales_1.1.1 cachem_1.0.6 farver_2.1.0
[37] fs_1.5.2 bit_4.0.4 digest_0.6.29 stringi_1.7.6
[41] dplyr_1.0.7 rprojroot_2.0.2 grid_3.6.1 cli_3.1.0
[45] tools_3.6.1 magrittr_2.0.2 tibble_3.1.6 RSQLite_2.2.8
[49] crayon_1.5.0 whisker_0.3-2 pkgconfig_2.0.3 ellipsis_0.3.2
[53] data.table_1.14.2 assertthat_0.2.1 rmarkdown_2.11 R6_2.5.1
[57] git2r_0.26.1 compiler_3.6.1