T2D - Pancreas
sheng Qian
2021-2-6
Last updated: 2022-02-26
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0e6a2f2. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/AF/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Unstaged changes:
Modified: analysis/BMI_Brain_Amygdala_S.Rmd
Modified: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Modified: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Modified: analysis/BMI_Brain_Cerebellum_S.Rmd
Modified: analysis/BMI_Brain_Cortex.Rmd
Modified: analysis/BMI_Brain_Cortex_S.Rmd
Modified: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Modified: analysis/BMI_Brain_Hippocampus_S.Rmd
Modified: analysis/BMI_Brain_Hypothalamus_S.Rmd
Modified: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Modified: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Modified: analysis/LDL_Liver_S.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/T2D_Pancreas.Rmd) and HTML (docs/T2D_Pancreas.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0e6a2f2 | sq-96 | 2022-02-26 | update |
| html | 91f38fa | sq-96 | 2022-02-13 | Build site. |
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
[1] 7605
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
748 581 480 310 401 436 387 270 287 327 513 475 161 273 282 292 407 119 367 228
21 22
92 169 [1] 4452[1] 0.5854043Load ctwas results
Check convergence of parameters
********************************************************Note: As of version 1.0.0, cowplot does not change the default ggplot2 theme anymore. To recover the previous behavior, execute:
theme_set(theme_cowplot())********************************************************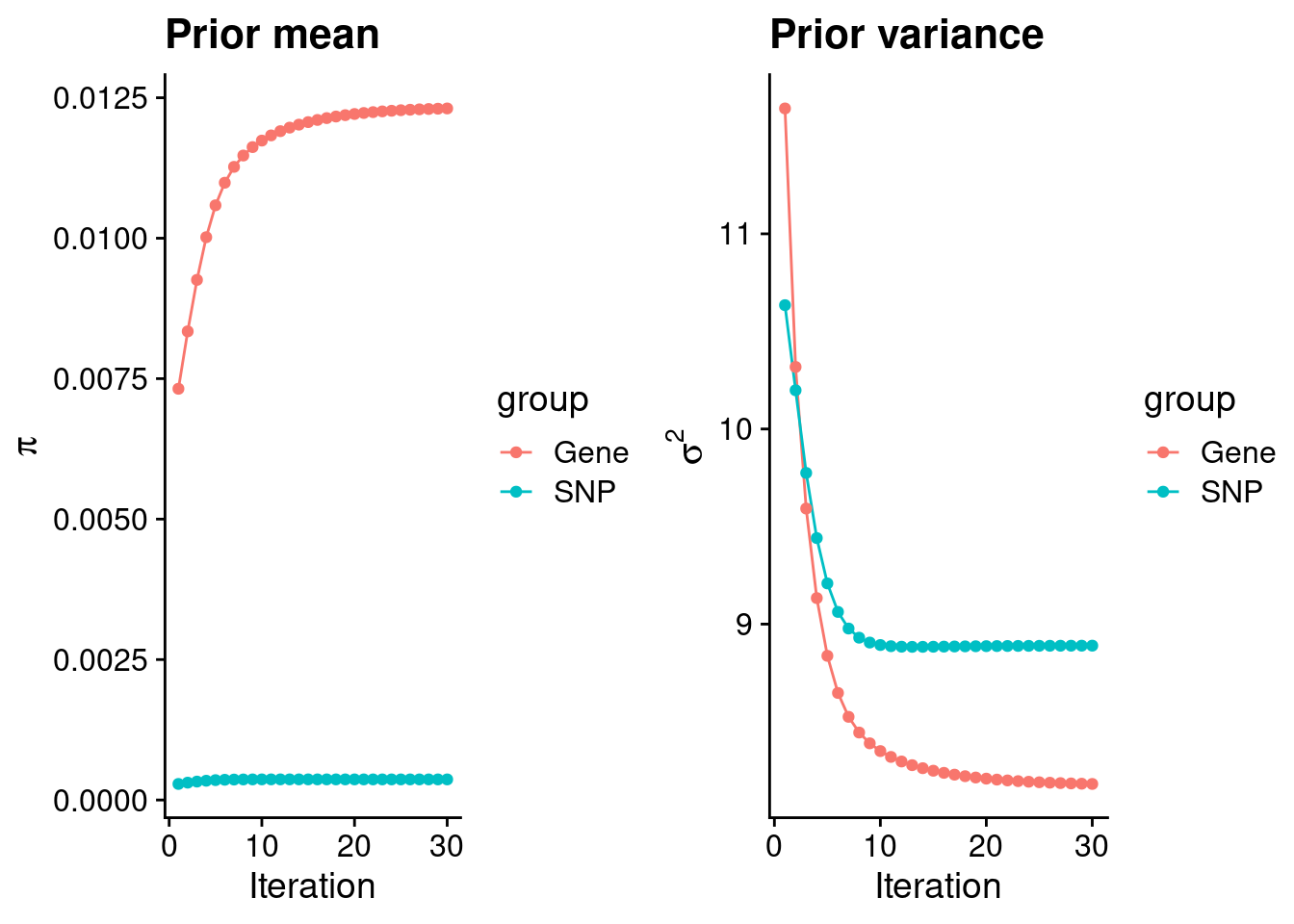
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
gene snp
0.0123097006 0.0003667405 gene snp
8.180928 8.889625 [1] 62892[1] 7605 5017190 gene snp
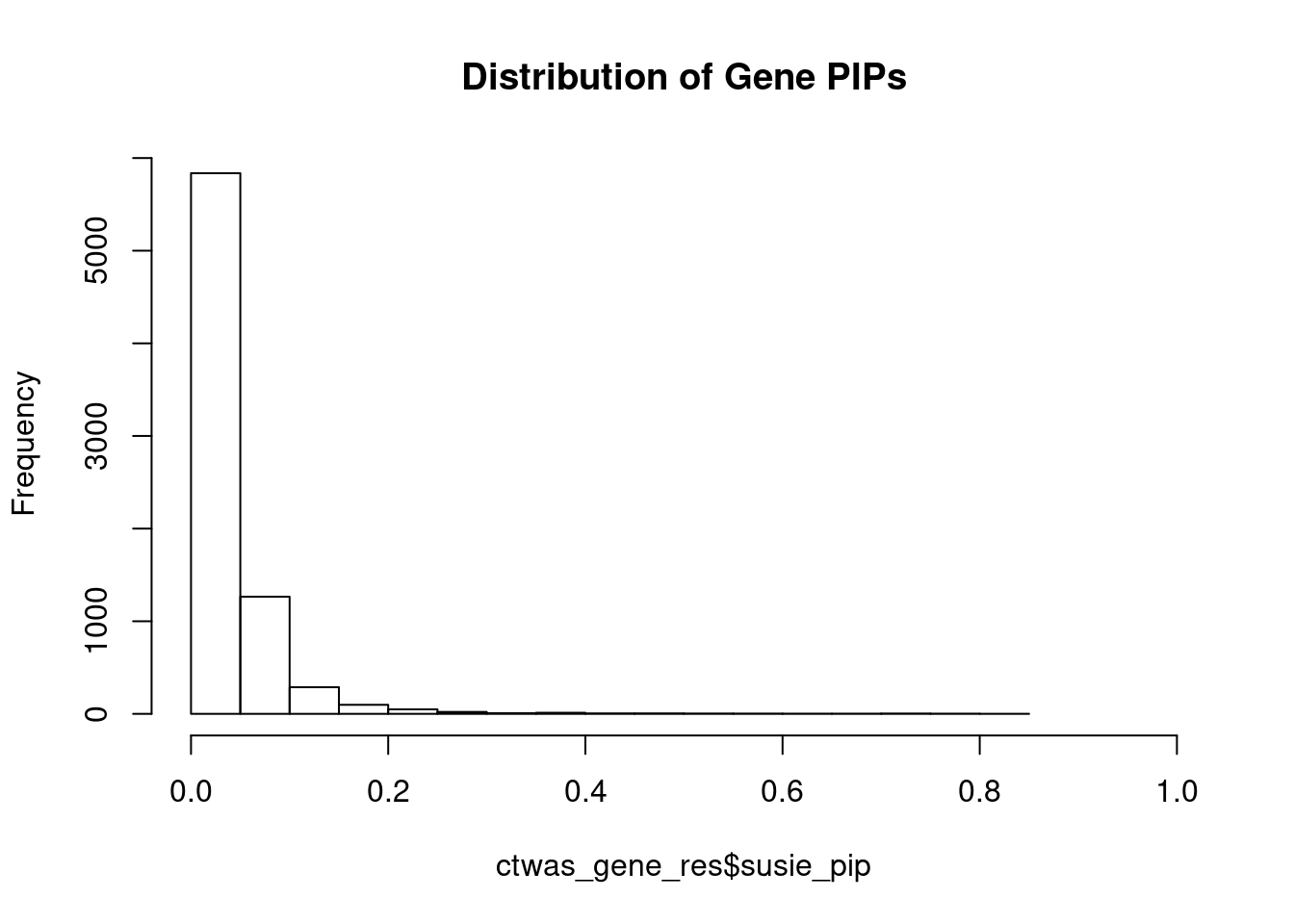
0.01217738 0.26008027 [1] 0.06992339 1.44416811Genes with highest PIPs
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
genename region_tag susie_pip mu2 PVE z num_eqtl
5708 SCRN2 17_28 0.8106840 23.09802 0.0002977357 -4.970331 2
3452 ARG1 6_87 0.7825383 28.70545 0.0003571696 -5.418037 2
8741 ONECUT1 15_22 0.7596047 27.19347 0.0003284406 5.078652 1
10062 ARL6IP4 12_75 0.7433361 36.01403 0.0004256587 5.620253 1
11158 PARVA 11_9 0.7137719 21.89032 0.0002484369 -3.861836 2
2227 DNASE2 19_10 0.7083131 19.08862 0.0002149831 -3.744186 1
4461 ZNF236 18_45 0.7058864 20.69653 0.0002322934 -4.378049 1
7604 CFAP221 2_69 0.6764235 20.29448 0.0002182736 -4.049666 2
10527 GSAP 7_49 0.6504951 24.56257 0.0002540519 -4.185185 1
183 GIPR 19_32 0.6326493 36.31838 0.0003653374 -6.632184 1
1451 CWF19L1 10_64 0.6279579 33.14247 0.0003309177 -5.802916 2
12493 LINC01184 5_78 0.5774450 20.30140 0.0001863979 3.793478 1
4519 TUBG1 17_25 0.5615797 23.49127 0.0002097599 5.267913 2
6236 CRIP3 6_33 0.5552431 21.65792 0.0001912073 4.544995 2
2221 MIER2 19_1 0.5414438 23.95810 0.0002062578 3.683544 1
6019 MRPS5 2_57 0.5399310 21.72457 0.0001865065 -3.736842 1
11243 ZNF251 8_94 0.5343093 24.42065 0.0002074696 -4.886076 1
1411 TYRO3 15_15 0.5095761 23.08603 0.0001870522 4.865854 1
3830 KBTBD4 11_29 0.4950396 25.44472 0.0002002821 -5.097561 1
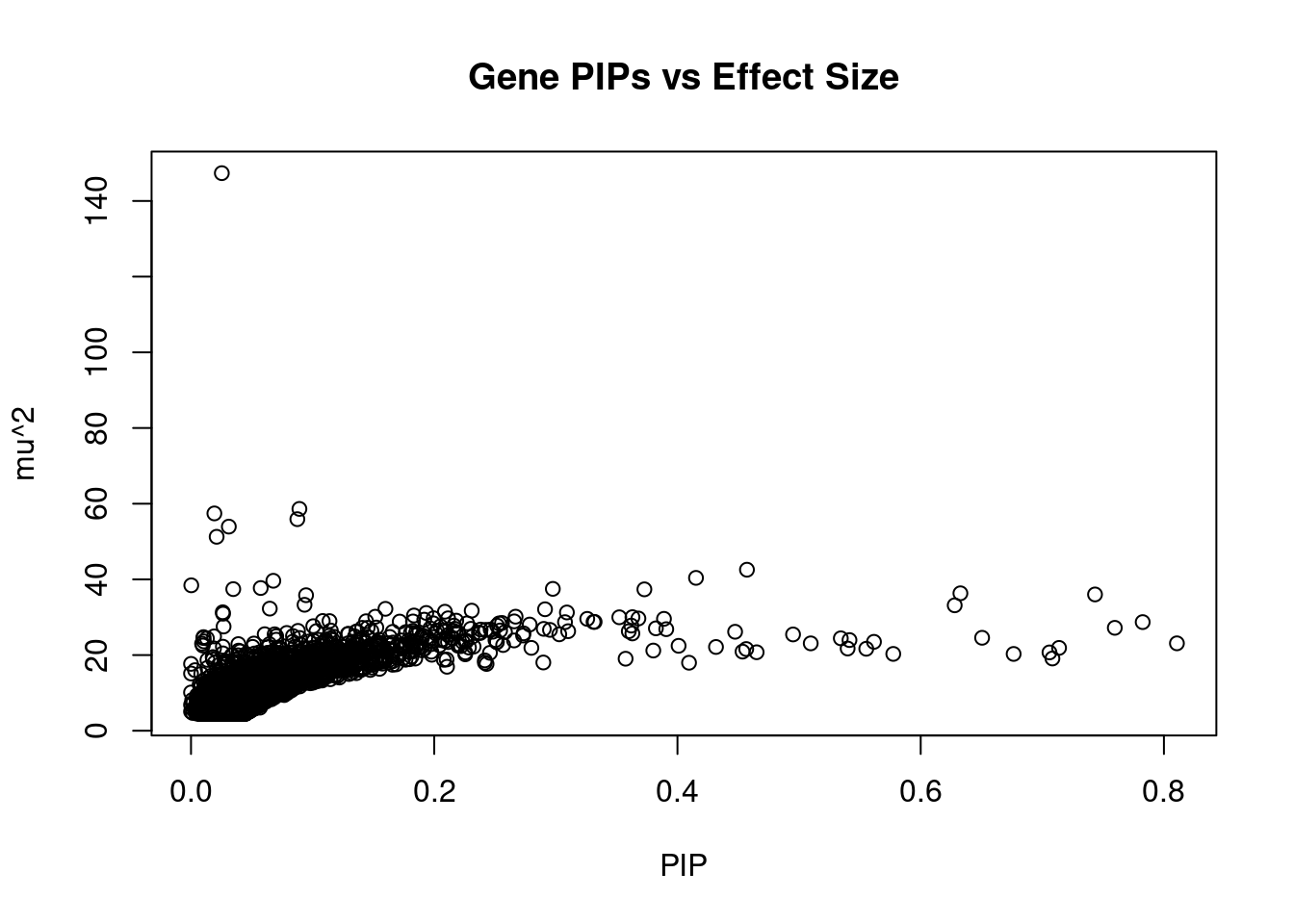
12664 LINC01933 5_89 0.4650992 20.73843 0.0001533649 3.780488 1Genes with largest effect sizes
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
genename region_tag susie_pip mu2 PVE z
6795 JAZF1 7_23 0.0253350265 147.33975 5.935344e-05 -13.081610
474 BCAR1 16_40 0.0891403843 58.61868 8.308357e-05 -7.720000
10584 ARAP1 11_41 0.0192135642 57.42519 1.754345e-05 7.885714
6345 CDKN2B 9_16 0.0874432759 55.90606 7.773022e-05 -8.191781
13639 LINC01126 2_27 0.0310929522 53.93953 2.666697e-05 -8.376923
10742 NCR3LG1 11_12 0.0210811758 51.25289 1.717979e-05 -7.369863
7991 NKX6-3 8_36 0.4571378966 42.54881 3.092710e-04 -6.780976
9232 PEAK1 15_36 0.4152425579 40.39351 2.666970e-04 6.885057
7192 ATP5G1 17_28 0.0676819144 39.62674 4.264475e-05 6.400000
11440 RBM20 10_70 0.0002240545 38.42672 1.368962e-07 -3.000000
7194 SNF8 17_28 0.0573205865 37.71067 3.437000e-05 6.300000
1291 P2RX7 12_74 0.2975091723 37.48946 1.773430e-04 5.483748
10224 BMP8A 1_24 0.0347386872 37.42298 2.067076e-05 6.296296
7087 AP3S2 15_41 0.3727948286 37.36237 2.214669e-04 6.356322
183 GIPR 19_32 0.6326492659 36.31838 3.653374e-04 -6.632184
10062 ARL6IP4 12_75 0.7433361169 36.01403 4.256587e-04 5.620253
10674 HAPLN4 19_15 0.0946101030 35.80928 5.386884e-05 5.347368
11533 MICB 6_25 0.0933548640 33.28052 4.940054e-05 5.545585
1451 CWF19L1 10_64 0.6279578506 33.14247 3.309177e-04 -5.802916
8540 TAP1 6_27 0.0647763566 32.26518 3.323190e-05 -5.575000
num_eqtl
6795 2
474 1
10584 1
6345 1
13639 1
10742 1
7991 2
9232 1
7192 1
11440 1
7194 1
1291 2
10224 1
7087 1
183 1
10062 1
10674 1
11533 2
1451 2
8540 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
10062 ARL6IP4 12_75 0.7433361 36.01403 0.0004256587 5.620253 1
183 GIPR 19_32 0.6326493 36.31838 0.0003653374 -6.632184 1
3452 ARG1 6_87 0.7825383 28.70545 0.0003571696 -5.418037 2
1451 CWF19L1 10_64 0.6279579 33.14247 0.0003309177 -5.802916 2
8741 ONECUT1 15_22 0.7596047 27.19347 0.0003284406 5.078652 1
7991 NKX6-3 8_36 0.4571379 42.54881 0.0003092710 -6.780976 2
5708 SCRN2 17_28 0.8106840 23.09802 0.0002977357 -4.970331 2
9232 PEAK1 15_36 0.4152426 40.39351 0.0002666970 6.885057 1
10527 GSAP 7_49 0.6504951 24.56257 0.0002540519 -4.185185 1
11158 PARVA 11_9 0.7137719 21.89032 0.0002484369 -3.861836 2
4461 ZNF236 18_45 0.7058864 20.69653 0.0002322934 -4.378049 1
7087 AP3S2 15_41 0.3727948 37.36237 0.0002214669 6.356322 1
7604 CFAP221 2_69 0.6764235 20.29448 0.0002182736 -4.049666 2
2227 DNASE2 19_10 0.7083131 19.08862 0.0002149831 -3.744186 1
4519 TUBG1 17_25 0.5615797 23.49127 0.0002097599 5.267913 2
11243 ZNF251 8_94 0.5343093 24.42065 0.0002074696 -4.886076 1
2221 MIER2 19_1 0.5414438 23.95810 0.0002062578 3.683544 1
3830 KBTBD4 11_29 0.4950396 25.44472 0.0002002821 -5.097561 1
6236 CRIP3 6_33 0.5552431 21.65792 0.0001912073 4.544995 2
1411 TYRO3 15_15 0.5095761 23.08603 0.0001870522 4.865854 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z
6795 JAZF1 7_23 0.02533503 147.33975 5.935344e-05 -13.081610
13639 LINC01126 2_27 0.03109295 53.93953 2.666697e-05 -8.376923
6345 CDKN2B 9_16 0.08744328 55.90606 7.773022e-05 -8.191781
10584 ARAP1 11_41 0.01921356 57.42519 1.754345e-05 7.885714
474 BCAR1 16_40 0.08914038 58.61868 8.308357e-05 -7.720000
10742 NCR3LG1 11_12 0.02108118 51.25289 1.717979e-05 -7.369863
9232 PEAK1 15_36 0.41524256 40.39351 2.666970e-04 6.885057
7991 NKX6-3 8_36 0.45713790 42.54881 3.092710e-04 -6.780976
183 GIPR 19_32 0.63264927 36.31838 3.653374e-04 -6.632184
7192 ATP5G1 17_28 0.06768191 39.62674 4.264475e-05 6.400000
7087 AP3S2 15_41 0.37279483 37.36237 2.214669e-04 6.356322
7194 SNF8 17_28 0.05732059 37.71067 3.437000e-05 6.300000
10224 BMP8A 1_24 0.03473869 37.42298 2.067076e-05 6.296296
1451 CWF19L1 10_64 0.62795785 33.14247 3.309177e-04 -5.802916
10062 ARL6IP4 12_75 0.74333612 36.01403 4.256587e-04 5.620253
3128 NRBP1 2_16 0.02608681 31.30510 1.298496e-05 -5.594937
8540 TAP1 6_27 0.06477636 32.26518 3.323190e-05 -5.575000
11533 MICB 6_25 0.09335486 33.28052 4.940054e-05 5.545585
3134 PPM1G 2_16 0.02636672 30.92622 1.296545e-05 -5.544304
1291 P2RX7 12_74 0.29750917 37.48946 1.773430e-04 5.483748
num_eqtl
6795 2
13639 1
6345 1
10584 1
474 1
10742 1
9232 1
7991 2
183 1
7192 1
7087 1
7194 1
10224 1
1451 2
10062 1
3128 1
8540 1
11533 2
3134 1
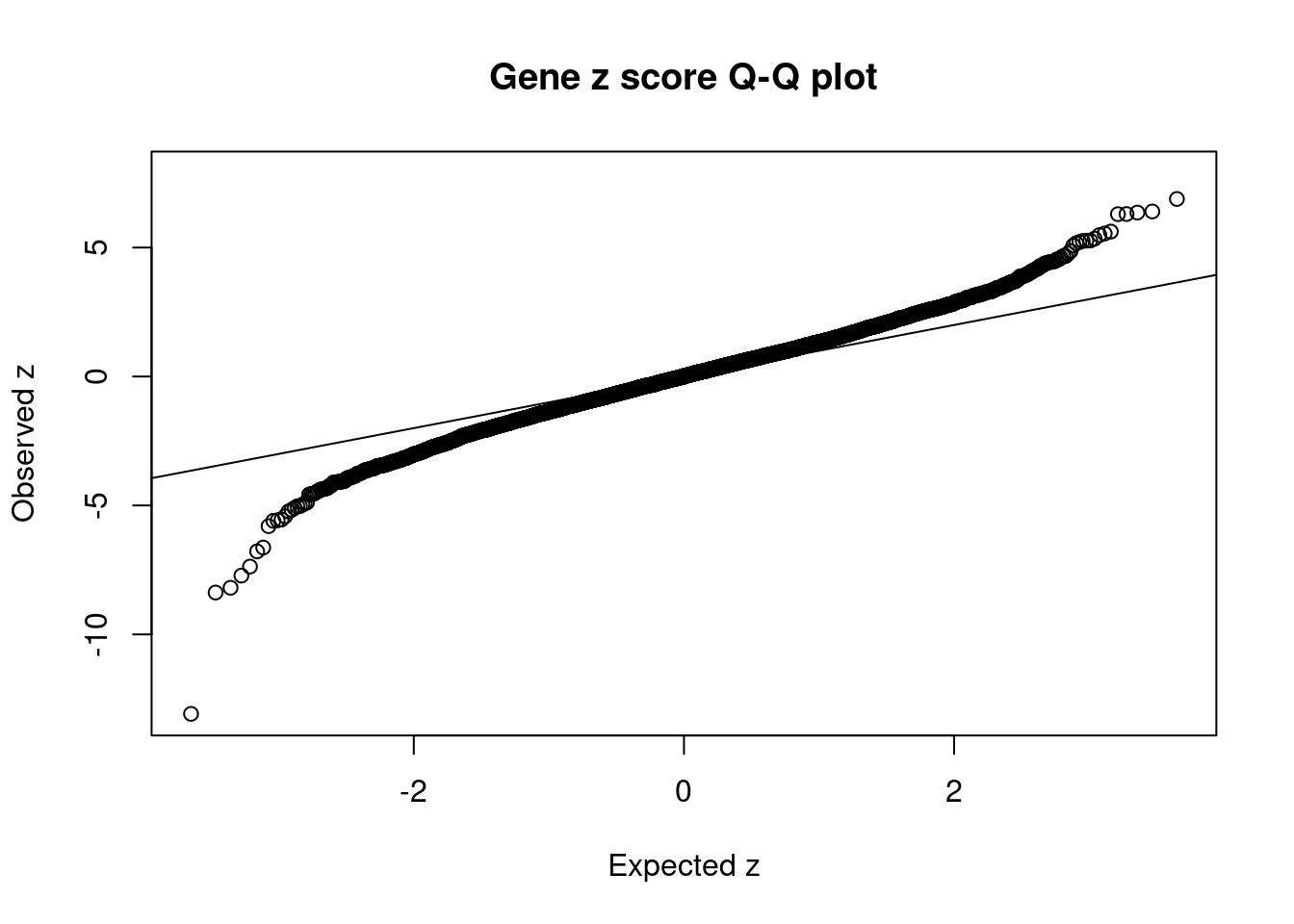
1291 2Comparing z scores and PIPs
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
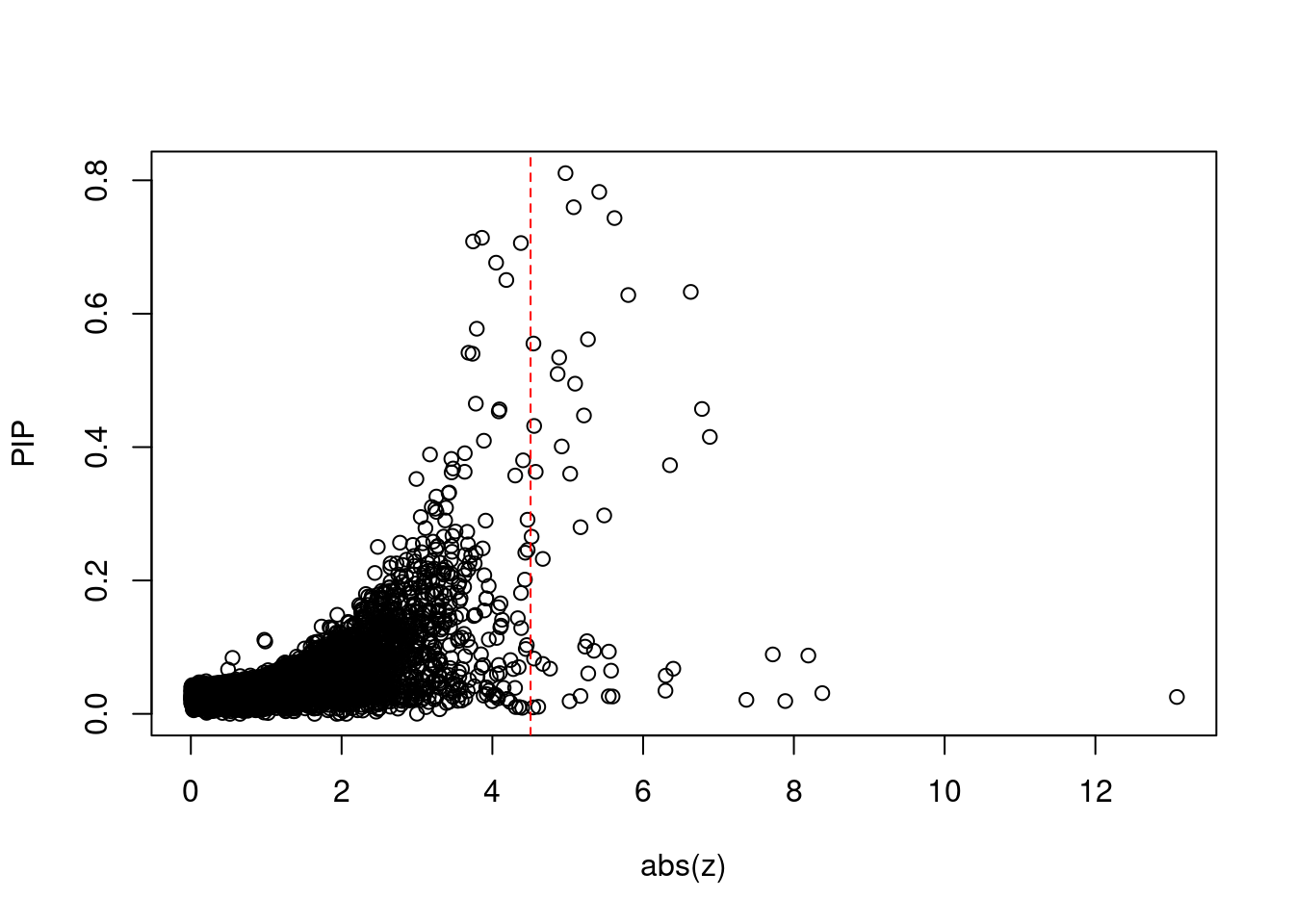
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
[1] 0.006180145 genename region_tag susie_pip mu2 PVE z
6795 JAZF1 7_23 0.02533503 147.33975 5.935344e-05 -13.081610
13639 LINC01126 2_27 0.03109295 53.93953 2.666697e-05 -8.376923
6345 CDKN2B 9_16 0.08744328 55.90606 7.773022e-05 -8.191781
10584 ARAP1 11_41 0.01921356 57.42519 1.754345e-05 7.885714
474 BCAR1 16_40 0.08914038 58.61868 8.308357e-05 -7.720000
10742 NCR3LG1 11_12 0.02108118 51.25289 1.717979e-05 -7.369863
9232 PEAK1 15_36 0.41524256 40.39351 2.666970e-04 6.885057
7991 NKX6-3 8_36 0.45713790 42.54881 3.092710e-04 -6.780976
183 GIPR 19_32 0.63264927 36.31838 3.653374e-04 -6.632184
7192 ATP5G1 17_28 0.06768191 39.62674 4.264475e-05 6.400000
7087 AP3S2 15_41 0.37279483 37.36237 2.214669e-04 6.356322
7194 SNF8 17_28 0.05732059 37.71067 3.437000e-05 6.300000
10224 BMP8A 1_24 0.03473869 37.42298 2.067076e-05 6.296296
1451 CWF19L1 10_64 0.62795785 33.14247 3.309177e-04 -5.802916
10062 ARL6IP4 12_75 0.74333612 36.01403 4.256587e-04 5.620253
3128 NRBP1 2_16 0.02608681 31.30510 1.298496e-05 -5.594937
8540 TAP1 6_27 0.06477636 32.26518 3.323190e-05 -5.575000
11533 MICB 6_25 0.09335486 33.28052 4.940054e-05 5.545585
3134 PPM1G 2_16 0.02636672 30.92622 1.296545e-05 -5.544304
1291 P2RX7 12_74 0.29750917 37.48946 1.773430e-04 5.483748
num_eqtl
6795 2
13639 1
6345 1
10584 1
474 1
10742 1
9232 1
7991 2
183 1
7192 1
7087 1
7194 1
10224 1
1451 2
10062 1
3128 1
8540 1
11533 2
3134 1
1291 2Sensitivity, specificity and precision for silver standard genes
[1] 72[1] 27[1] 4.507014[1] 18[1] 47 genename region_tag susie_pip mu2 PVE z num_eqtl
6019 MRPS5 2_57 0.5399310 21.72457 0.0001865065 -3.736842 1
12493 LINC01184 5_78 0.5774450 20.30140 0.0001863979 3.793478 1
10527 GSAP 7_49 0.6504951 24.56257 0.0002540519 -4.185185 1
11158 PARVA 11_9 0.7137719 21.89032 0.0002484369 -3.861836 2
4461 ZNF236 18_45 0.7058864 20.69653 0.0002322934 -4.378049 1
2221 MIER2 19_1 0.5414438 23.95810 0.0002062578 3.683544 1
2227 DNASE2 19_10 0.7083131 19.08862 0.0002149831 -3.744186 1
7604 CFAP221 2_69 0.6764235 20.29448 0.0002182736 -4.049666 2 ctwas TWAS
0.01388889 0.01388889 ctwas TWAS
0.9977567 0.9939298 ctwas TWAS
0.05555556 0.02127660
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] tidyselect_1.1.1 xfun_0.29 purrr_0.3.4 colorspace_2.0-2
[5] vctrs_0.3.8 generics_0.1.1 htmltools_0.5.2 yaml_2.2.1
[9] utf8_1.2.2 blob_1.2.2 rlang_1.0.1 jquerylib_0.1.4
[13] later_0.8.0 pillar_1.6.4 glue_1.6.2 withr_2.4.3
[17] DBI_1.1.2 bit64_4.0.5 lifecycle_1.0.1 stringr_1.4.0
[21] cellranger_1.1.0 munsell_0.5.0 gtable_0.3.0 evaluate_0.14
[25] memoise_2.0.1 labeling_0.4.2 knitr_1.36 fastmap_1.1.0
[29] httpuv_1.5.1 fansi_1.0.2 highr_0.9 Rcpp_1.0.8
[33] promises_1.0.1 scales_1.1.1 cachem_1.0.6 farver_2.1.0
[37] fs_1.5.2 bit_4.0.4 digest_0.6.29 stringi_1.7.6
[41] dplyr_1.0.7 rprojroot_2.0.2 grid_3.6.1 cli_3.1.0
[45] tools_3.6.1 magrittr_2.0.2 tibble_3.1.6 RSQLite_2.2.8
[49] crayon_1.5.0 whisker_0.3-2 pkgconfig_2.0.3 ellipsis_0.3.2
[53] data.table_1.14.2 assertthat_0.2.1 rmarkdown_2.11 R6_2.5.1
[57] git2r_0.26.1 compiler_3.6.1