T2D - Adipose Visceral Omentum
sheng Qian
2021-2-6
Last updated: 2022-02-26
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5c37a5d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/AF/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Unstaged changes:
Modified: analysis/BMI_Brain_Amygdala_S.Rmd
Modified: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Modified: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Modified: analysis/BMI_Brain_Cerebellum_S.Rmd
Modified: analysis/BMI_Brain_Cortex.Rmd
Modified: analysis/BMI_Brain_Cortex_S.Rmd
Modified: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Modified: analysis/BMI_Brain_Hippocampus_S.Rmd
Modified: analysis/BMI_Brain_Hypothalamus_S.Rmd
Modified: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Modified: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Modified: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Modified: analysis/LDL_Liver_S.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/T2D_Adipose_Visceral_Omentum.Rmd) and HTML (docs/T2D_Adipose_Visceral_Omentum.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5c37a5d | sq-96 | 2022-02-26 | update |
| html | 3fa3a64 | sq-96 | 2022-02-26 | Build site. |
| html | 91f38fa | sq-96 | 2022-02-13 | Build site. |
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 8279#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
860 632 533 357 434 491 417 287 316 367 525 508 179 281 284 320 451 139 400 239
21 22
83 176 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 4625#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.5586Check convergence of parameters
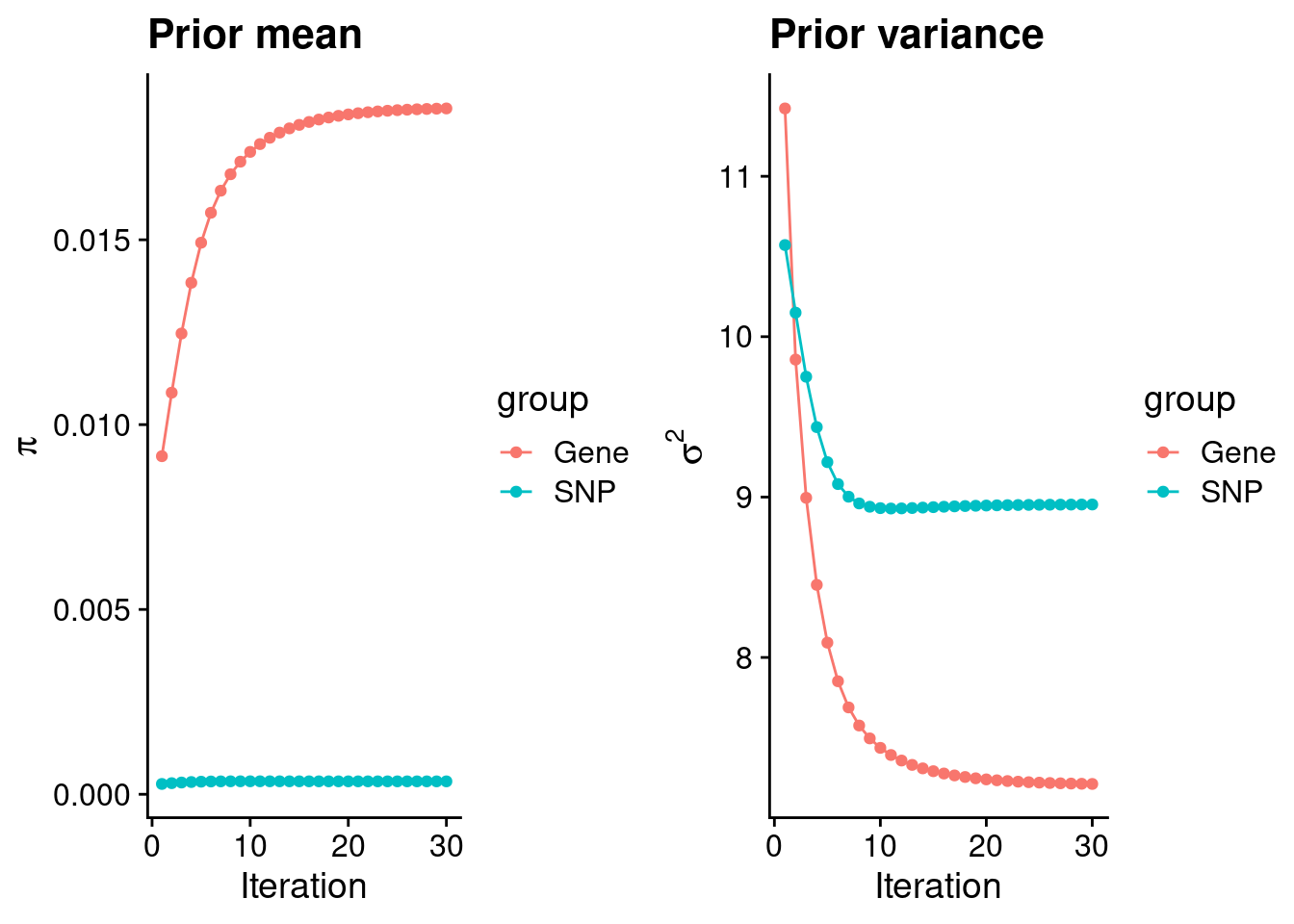
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0185530 0.0003497 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
7.211 8.954 #report sample size
print(sample_size)[1] 62892#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 8279 5017190#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.01761 0.24979 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.1023 1.4004Genes with highest PIPs
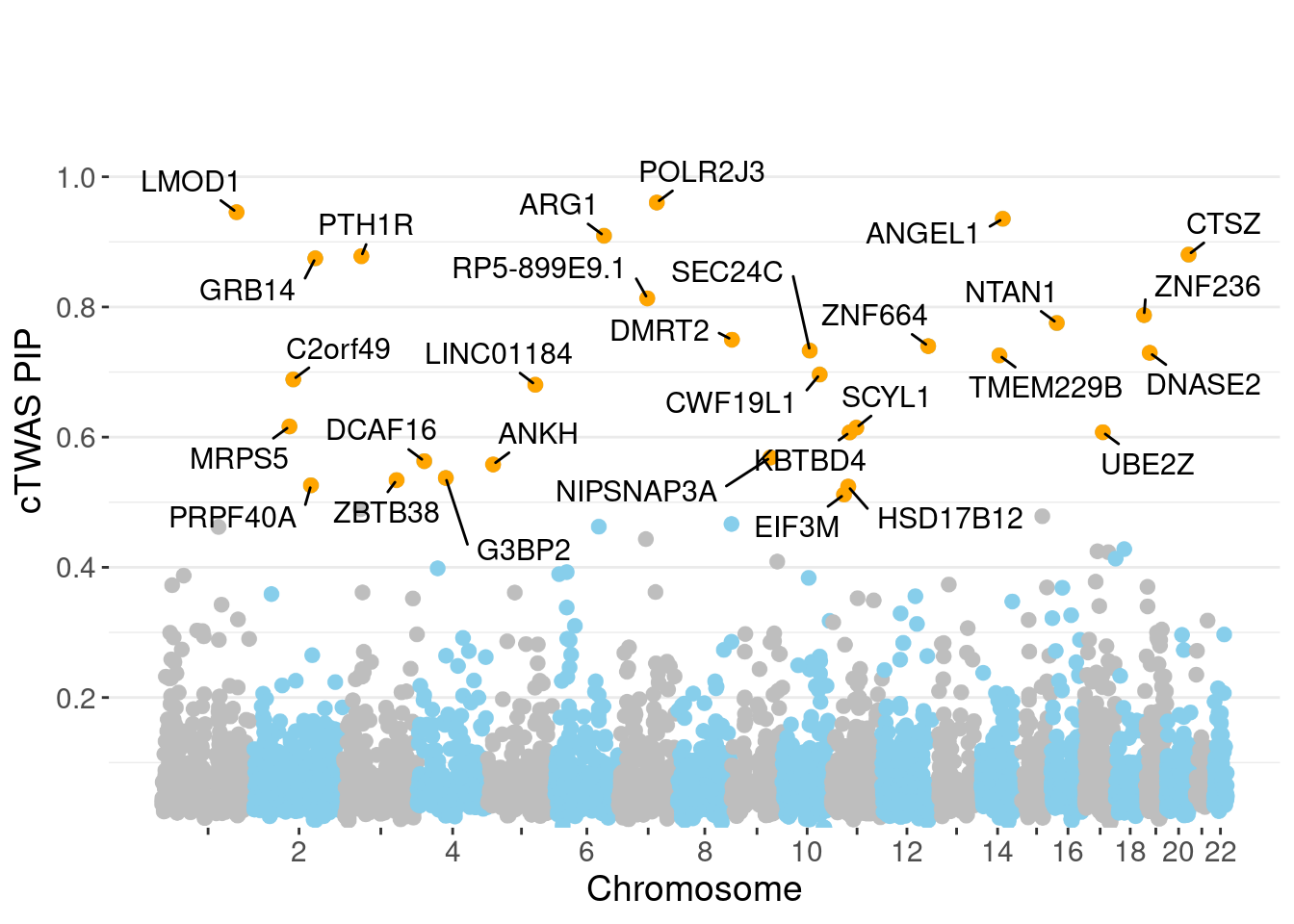
genename region_tag susie_pip mu2 PVE z num_eqtl
8882 POLR2J3 7_63 0.9604 25.74 0.0003930 4.987 1
7980 LMOD1 1_102 0.9456 24.43 0.0003672 -4.907 1
249 ANGEL1 14_36 0.9355 21.80 0.0003243 4.558 2
3617 ARG1 6_87 0.9094 29.87 0.0004319 -5.606 1
1848 CTSZ 20_34 0.8805 20.18 0.0002824 -3.896 1
7655 PTH1R 3_33 0.8780 28.47 0.0003974 -5.646 1
3290 GRB14 2_100 0.8747 25.00 0.0003478 5.163 1
14287 RP5-899E9.1 7_49 0.8132 19.91 0.0002575 -4.333 1
4668 ZNF236 18_45 0.7874 20.14 0.0002522 -4.378 1
7322 NTAN1 16_15 0.7754 19.37 0.0002388 4.192 1
9579 DMRT2 9_1 0.7497 19.40 0.0002313 -4.318 1
10241 ZNF664 12_75 0.7398 41.11 0.0004836 -6.452 1
9990 SEC24C 10_49 0.7330 26.90 0.0003135 -4.862 1
2306 DNASE2 19_10 0.7296 18.47 0.0002143 -3.744 1
11753 TMEM229B 14_31 0.7256 18.31 0.0002112 -3.658 1
1508 CWF19L1 10_64 0.6962 33.33 0.0003690 -5.810 1
5258 C2orf49 2_62 0.6887 26.88 0.0002943 5.235 1
13192 LINC01184 5_78 0.6805 18.64 0.0002016 3.793 1
6264 MRPS5 2_57 0.6164 20.20 0.0001980 -3.737 1
6029 SCYL1 11_36 0.6145 22.23 0.0002172 -4.814 1Genes with largest effect sizes
genename region_tag susie_pip mu2 PVE z num_eqtl
7057 JAZF1 7_23 0.277121 130.42 5.747e-04 -12.662 1
2722 WFS1 4_7 0.158490 64.77 1.632e-04 11.434 1
3379 THADA 2_27 0.063978 59.34 6.037e-05 8.667 1
14565 LINC01126 2_27 0.050344 52.18 4.177e-05 -8.377 1
10525 UBE2E2 3_17 0.084142 48.51 6.491e-05 7.166 2
11136 KCNJ11 11_12 0.028462 45.08 2.040e-05 7.075 2
11227 NCR3LG1 11_12 0.035887 44.08 2.515e-05 -6.854 2
3273 NRBP1 2_16 0.057481 43.86 4.008e-05 -6.625 1
7493 UBE2Z 17_28 0.607571 43.82 4.234e-04 -7.392 1
3656 CCDC92 12_75 0.135611 42.88 9.246e-05 -5.887 2
1401 PABPC4 1_24 0.103952 42.78 7.072e-05 -6.817 1
7491 ATP5G1 17_28 0.090095 41.35 5.923e-05 6.400 1
10241 ZNF664 12_75 0.739843 41.11 4.836e-04 -6.452 1
1158 COBLL1 2_100 0.072551 39.99 4.613e-05 -5.375 1
12780 CYP21A2 6_26 0.055455 39.91 3.519e-05 6.453 1
7494 SNF8 17_28 0.077698 39.44 4.873e-05 6.300 1
9611 PEAK1 15_36 0.478500 39.02 2.969e-04 -6.885 1
6467 CDKAL1 6_15 0.004169 37.81 2.506e-06 -8.192 1
7378 AP3S2 15_41 0.369116 36.18 2.124e-04 6.356 1
5126 P2RX4 12_74 0.263755 35.42 1.485e-04 4.096 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
7057 JAZF1 7_23 0.2771 130.42 0.0005747 -12.662 1
10241 ZNF664 12_75 0.7398 41.11 0.0004836 -6.452 1
3617 ARG1 6_87 0.9094 29.87 0.0004319 -5.606 1
7493 UBE2Z 17_28 0.6076 43.82 0.0004234 -7.392 1
7655 PTH1R 3_33 0.8780 28.47 0.0003974 -5.646 1
8882 POLR2J3 7_63 0.9604 25.74 0.0003930 4.987 1
1508 CWF19L1 10_64 0.6962 33.33 0.0003690 -5.810 1
7980 LMOD1 1_102 0.9456 24.43 0.0003672 -4.907 1
3290 GRB14 2_100 0.8747 25.00 0.0003478 5.163 1
249 ANGEL1 14_36 0.9355 21.80 0.0003243 4.558 2
9990 SEC24C 10_49 0.7330 26.90 0.0003135 -4.862 1
9611 PEAK1 15_36 0.4785 39.02 0.0002969 -6.885 1
5258 C2orf49 2_62 0.6887 26.88 0.0002943 5.235 1
1848 CTSZ 20_34 0.8805 20.18 0.0002824 -3.896 1
14287 RP5-899E9.1 7_49 0.8132 19.91 0.0002575 -4.333 1
4668 ZNF236 18_45 0.7874 20.14 0.0002522 -4.378 1
7322 NTAN1 16_15 0.7754 19.37 0.0002388 4.192 1
4021 KBTBD4 11_29 0.6073 24.30 0.0002346 -5.098 1
9579 DMRT2 9_1 0.7497 19.40 0.0002313 -4.318 1
6029 SCYL1 11_36 0.6145 22.23 0.0002172 -4.814 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
7057 JAZF1 7_23 0.277121 130.42 5.747e-04 -12.662 1
2722 WFS1 4_7 0.158490 64.77 1.632e-04 11.434 1
3379 THADA 2_27 0.063978 59.34 6.037e-05 8.667 1
14565 LINC01126 2_27 0.050344 52.18 4.177e-05 -8.377 1
6467 CDKAL1 6_15 0.004169 37.81 2.506e-06 -8.192 1
7493 UBE2Z 17_28 0.607571 43.82 4.234e-04 -7.392 1
10525 UBE2E2 3_17 0.084142 48.51 6.491e-05 7.166 2
11136 KCNJ11 11_12 0.028462 45.08 2.040e-05 7.075 2
9611 PEAK1 15_36 0.478500 39.02 2.969e-04 -6.885 1
11227 NCR3LG1 11_12 0.035887 44.08 2.515e-05 -6.854 2
1401 PABPC4 1_24 0.103952 42.78 7.072e-05 -6.817 1
3273 NRBP1 2_16 0.057481 43.86 4.008e-05 -6.625 1
12062 MICB 6_25 0.392658 34.75 2.170e-04 6.462 2
12780 CYP21A2 6_26 0.055455 39.91 3.519e-05 6.453 1
10241 ZNF664 12_75 0.739843 41.11 4.836e-04 -6.452 1
7491 ATP5G1 17_28 0.090095 41.35 5.923e-05 6.400 1
7378 AP3S2 15_41 0.369116 36.18 2.124e-04 6.356 1
7494 SNF8 17_28 0.077698 39.44 4.873e-05 6.300 1
13095 ARPIN 15_41 0.224871 34.65 1.239e-04 6.250 1
3656 CCDC92 12_75 0.135611 42.88 9.246e-05 -5.887 2Comparing z scores and PIPs
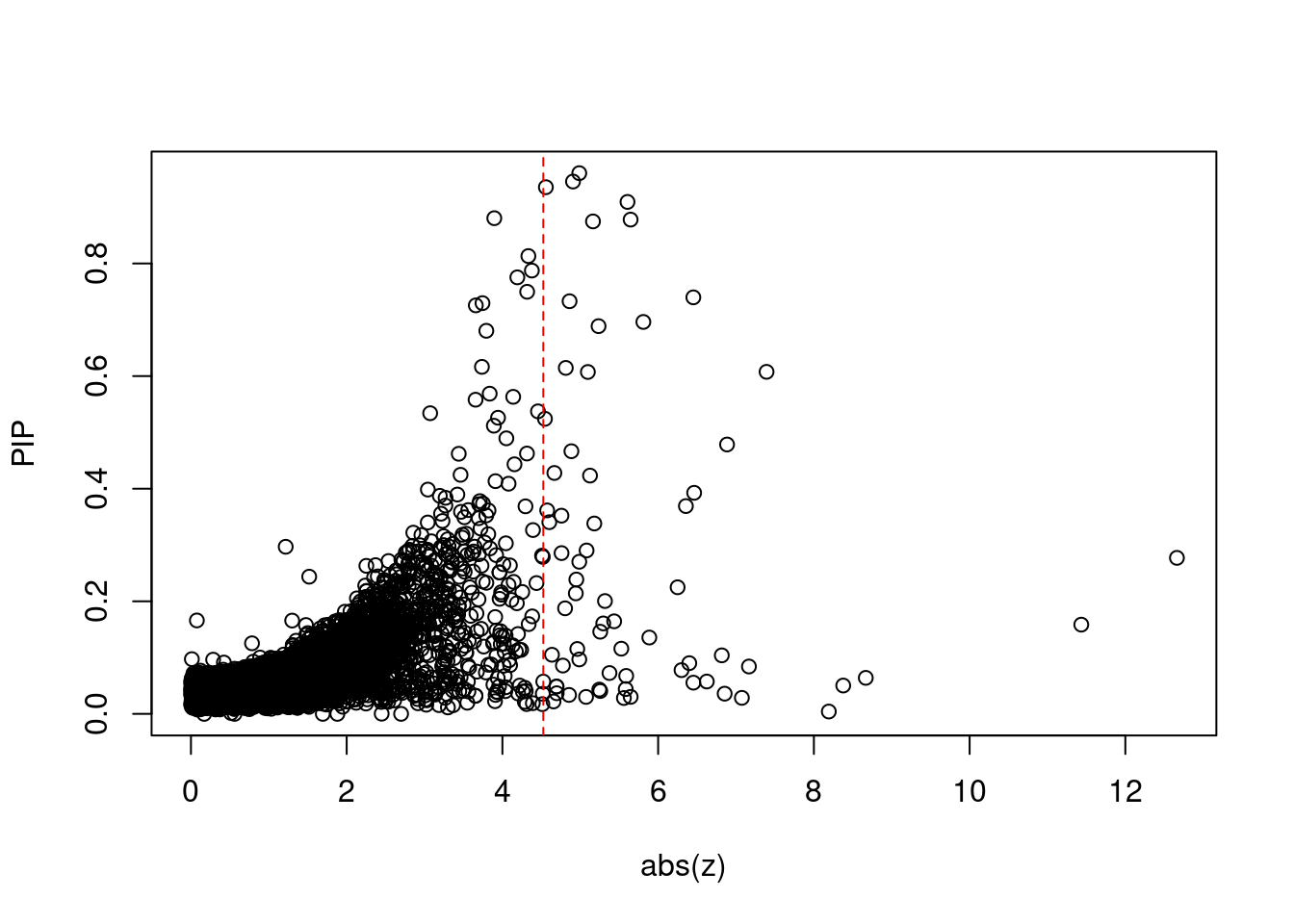
[1] 0.008093 genename region_tag susie_pip mu2 PVE z num_eqtl
7057 JAZF1 7_23 0.277121 130.42 5.747e-04 -12.662 1
2722 WFS1 4_7 0.158490 64.77 1.632e-04 11.434 1
3379 THADA 2_27 0.063978 59.34 6.037e-05 8.667 1
14565 LINC01126 2_27 0.050344 52.18 4.177e-05 -8.377 1
6467 CDKAL1 6_15 0.004169 37.81 2.506e-06 -8.192 1
7493 UBE2Z 17_28 0.607571 43.82 4.234e-04 -7.392 1
10525 UBE2E2 3_17 0.084142 48.51 6.491e-05 7.166 2
11136 KCNJ11 11_12 0.028462 45.08 2.040e-05 7.075 2
9611 PEAK1 15_36 0.478500 39.02 2.969e-04 -6.885 1
11227 NCR3LG1 11_12 0.035887 44.08 2.515e-05 -6.854 2
1401 PABPC4 1_24 0.103952 42.78 7.072e-05 -6.817 1
3273 NRBP1 2_16 0.057481 43.86 4.008e-05 -6.625 1
12062 MICB 6_25 0.392658 34.75 2.170e-04 6.462 2
12780 CYP21A2 6_26 0.055455 39.91 3.519e-05 6.453 1
10241 ZNF664 12_75 0.739843 41.11 4.836e-04 -6.452 1
7491 ATP5G1 17_28 0.090095 41.35 5.923e-05 6.400 1
7378 AP3S2 15_41 0.369116 36.18 2.124e-04 6.356 1
7494 SNF8 17_28 0.077698 39.44 4.873e-05 6.300 1
13095 ARPIN 15_41 0.224871 34.65 1.239e-04 6.250 1
3656 CCDC92 12_75 0.135611 42.88 9.246e-05 -5.887 2GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 30Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio BgRatio
30 Ataxia, Spinocerebellar 0.006521 2/13 34/9703
35 Jansen type metaphyseal chondrodysplasia 0.006521 1/13 1/9703
45 Spinocerebellar Ataxia Type 1 0.006521 2/13 34/9703
46 Spinocerebellar Ataxia Type 2 0.006521 2/13 34/9703
47 Spinocerebellar Ataxia Type 4 0.006521 2/13 35/9703
48 Spinocerebellar Ataxia Type 5 0.006521 2/13 34/9703
49 Spinocerebellar Ataxia Type 6 (disorder) 0.006521 2/13 34/9703
50 Spinocerebellar Ataxia Type 7 0.006521 2/13 34/9703
52 Calcium pyrophosphate arthropathy 0.006521 1/13 1/9703
58 Eiken Skeletal Dysplasia 0.006521 1/13 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 72#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 23#significance threshold for TWAS
print(sig_thresh)[1] 4.525#number of ctwas genes
length(ctwas_genes)[1] 30#number of TWAS genes
length(twas_genes)[1] 67#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
6264 MRPS5 2_57 0.6164 20.20 0.0001980 -3.737 1
11469 PRPF40A 2_91 0.5260 19.92 0.0001666 -3.944 2
10022 ZBTB38 3_86 0.5340 17.12 0.0001454 -3.072 1
7953 DCAF16 4_17 0.5632 20.45 0.0001831 -4.139 3
5664 G3BP2 4_51 0.5373 20.37 0.0001740 -4.457 1
7088 ANKH 5_12 0.5580 21.14 0.0001876 3.655 1
13192 LINC01184 5_78 0.6805 18.64 0.0002016 3.793 1
5358 NIPSNAP3A 9_52 0.5688 19.29 0.0001745 -3.837 2
6679 EIF3M 11_22 0.5118 17.81 0.0001449 -3.890 1
11753 TMEM229B 14_31 0.7256 18.31 0.0002112 -3.658 1
7322 NTAN1 16_15 0.7754 19.37 0.0002388 4.192 1
4668 ZNF236 18_45 0.7874 20.14 0.0002522 -4.378 1
2306 DNASE2 19_10 0.7296 18.47 0.0002143 -3.744 1
14287 RP5-899E9.1 7_49 0.8132 19.91 0.0002575 -4.333 1
9579 DMRT2 9_1 0.7497 19.40 0.0002313 -4.318 1
1848 CTSZ 20_34 0.8805 20.18 0.0002824 -3.896 1#sensitivity / recall
print(sensitivity) ctwas TWAS
0.01389 0.04167 #specificity
print(specificity) ctwas TWAS
0.9965 0.9922 #precision / PPV
print(precision) ctwas TWAS
0.03333 0.04478
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[5] purrr_0.3.4 readr_2.1.1 tidyr_1.1.4 tidyverse_1.3.1
[9] tibble_3.1.6 WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[13] cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.2 lubridate_1.8.0 bit64_4.0.5 doParallel_1.0.17
[5] httr_1.4.2 rprojroot_2.0.2 tools_3.6.1 backports_1.4.1
[9] doRNG_1.8.2 utf8_1.2.2 R6_2.5.1 vipor_0.4.5
[13] DBI_1.1.2 colorspace_2.0-2 withr_2.4.3 ggrastr_1.0.1
[17] tidyselect_1.1.1 bit_4.0.4 curl_4.3.2 compiler_3.6.1
[21] git2r_0.26.1 rvest_1.0.2 cli_3.1.0 Cairo_1.5-12.2
[25] xml2_1.3.3 labeling_0.4.2 scales_1.1.1 apcluster_1.4.8
[29] digest_0.6.29 rmarkdown_2.11 svglite_1.2.2 pkgconfig_2.0.3
[33] htmltools_0.5.2 dbplyr_2.1.1 fastmap_1.1.0 highr_0.9
[37] rlang_1.0.1 rstudioapi_0.13 RSQLite_2.2.8 jquerylib_0.1.4
[41] farver_2.1.0 generics_0.1.1 jsonlite_1.7.2 vroom_1.5.7
[45] magrittr_2.0.2 Matrix_1.2-18 ggbeeswarm_0.6.0 Rcpp_1.0.8
[49] munsell_0.5.0 fansi_1.0.2 gdtools_0.1.9 lifecycle_1.0.1
[53] stringi_1.7.6 whisker_0.3-2 yaml_2.2.1 plyr_1.8.6
[57] grid_3.6.1 blob_1.2.2 ggrepel_0.9.1 parallel_3.6.1
[61] promises_1.0.1 crayon_1.5.0 lattice_0.20-38 haven_2.4.3
[65] hms_1.1.1 knitr_1.36 pillar_1.6.4 igraph_1.2.10
[69] rjson_0.2.20 rngtools_1.5.2 reshape2_1.4.4 codetools_0.2-16
[73] reprex_2.0.1 glue_1.6.2 evaluate_0.14 data.table_1.14.2
[77] modelr_0.1.8 vctrs_0.3.8 tzdb_0.2.0 httpuv_1.5.1
[81] foreach_1.5.2 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] cachem_1.0.6 xfun_0.29 broom_0.7.10 later_0.8.0
[89] iterators_1.0.14 beeswarm_0.2.3 memoise_2.0.1 ellipsis_0.3.2