SCZ 2018 - Brain_Caudate_basal_ganglia
sheng Qian
2021-2-6
Last updated: 2022-05-19
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bcaadf3. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: G_list.RData
Untracked: Rplot.png
Untracked: SCZ_annotation.xlsx
Untracked: analysis/.ipynb_checkpoints/
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/Autism_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/SCZ_2014_EUR_out/
Untracked: code/SCZ_2018_S_out/
Untracked: code/SCZ_2018_out/
Untracked: code/SCZ_2020_Single_out/
Untracked: code/SCZ_2020_out/
Untracked: code/SCZ_S_out/
Untracked: code/SCZ_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/process_scz_2018_snps.R
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_Autism_analysis.sbatch
Untracked: code/run_Autism_analysis.sh
Untracked: code/run_Autism_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2014_EUR_analysis.sbatch
Untracked: code/run_SCZ_2014_EUR_analysis.sh
Untracked: code/run_SCZ_2014_EUR_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2018_analysis.sbatch
Untracked: code/run_SCZ_2018_analysis.sh
Untracked: code/run_SCZ_2018_analysis_S.sbatch
Untracked: code/run_SCZ_2018_analysis_S.sh
Untracked: code/run_SCZ_2018_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2018_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2020_Single_analysis.sbatch
Untracked: code/run_SCZ_2020_Single_analysis.sh
Untracked: code/run_SCZ_2020_Single_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2020_analysis.sbatch
Untracked: code/run_SCZ_2020_analysis.sh
Untracked: code/run_SCZ_2020_ctwas_rss_LDR.R
Untracked: code/run_SCZ_analysis.sbatch
Untracked: code/run_SCZ_analysis.sh
Untracked: code/run_SCZ_analysis_S.sbatch
Untracked: code/run_SCZ_analysis_S.sh
Untracked: code/run_SCZ_ctwas_rss_LDR.R
Untracked: code/run_SCZ_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: code/wflow_build.R
Untracked: code/wflow_build.sbatch
Untracked: data/.ipynb_checkpoints/
Untracked: data/GO_Terms/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/SCZ/
Untracked: data/SCZ_2014_EUR/
Untracked: data/SCZ_2018/
Untracked: data/SCZ_2018_S/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_S/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/T2D/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Untracked: top_genes_32.txt
Untracked: top_genes_37.txt
Untracked: top_genes_43.txt
Untracked: top_genes_54.txt
Untracked: top_genes_81.txt
Untracked: z_snp_pos_SCZ.RData
Untracked: z_snp_pos_SCZ_2014_EUR.RData
Untracked: z_snp_pos_SCZ_2018.RData
Untracked: z_snp_pos_SCZ_2020.RData
Unstaged changes:
Deleted: analysis/BMI_S_results.Rmd
Modified: analysis/SCZ_2018_Brain_Amygdala_S.Rmd
Modified: analysis/SCZ_2018_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Modified: analysis/SCZ_2018_Brain_Caudate_basal_ganglia_S.Rmd
Modified: analysis/SCZ_2018_Brain_Cerebellar_Hemisphere_S.Rmd
Modified: analysis/SCZ_2018_Brain_Cerebellum_S.Rmd
Modified: analysis/SCZ_2018_Brain_Cortex_S.Rmd
Modified: analysis/SCZ_2018_Brain_Frontal_Cortex_BA9_S.Rmd
Modified: analysis/SCZ_2018_Brain_Hippocampus_S.Rmd
Modified: analysis/SCZ_2018_Brain_Hypothalamus_S.Rmd
Modified: analysis/SCZ_2018_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Modified: analysis/SCZ_2018_Brain_Putamen_basal_ganglia_S.Rmd
Modified: analysis/SCZ_2018_Brain_Spinal_cord_cervical_c-1_S.Rmd
Modified: analysis/SCZ_2018_Brain_Substantia_nigra_S.Rmd
Modified: analysis/ttt.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/SCZ_2018_Brain_Caudate_basal_ganglia_S.Rmd) and HTML (docs/SCZ_2018_Brain_Caudate_basal_ganglia_S.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bcaadf3 | sq-96 | 2022-05-19 | update |
| html | bcaadf3 | sq-96 | 2022-05-19 | update |
| Rmd | be614ed | sq-96 | 2022-05-19 | update |
| html | be614ed | sq-96 | 2022-05-19 | update |
| Rmd | 7d08c9b | sq-96 | 2022-05-18 | update |
| html | 7d08c9b | sq-96 | 2022-05-18 | update |
| Rmd | 2749be9 | sq-96 | 2022-05-12 | update |
| html | 2749be9 | sq-96 | 2022-05-12 | update |
| html | 011327d | sq-96 | 2022-05-12 | update |
| Rmd | 6c6abbd | sq-96 | 2022-05-12 | update |
library(reticulate)
use_python("/scratch/midway2/shengqian/miniconda3/envs/PythonForR/bin/python",required=T)Weight QC
#number of imputed weights
nrow(qclist_all)[1] 21263#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1943 1490 1296 830 843 1056 1229 736 851 971 1284 1164 399 806 759 863
17 18 19 20 21 22
1510 310 1515 698 42 668 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 18726#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.8807INFO:numexpr.utils:Note: NumExpr detected 56 cores but "NUMEXPR_MAX_THREADS" not set, so enforcing safe limit of 8.finish
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
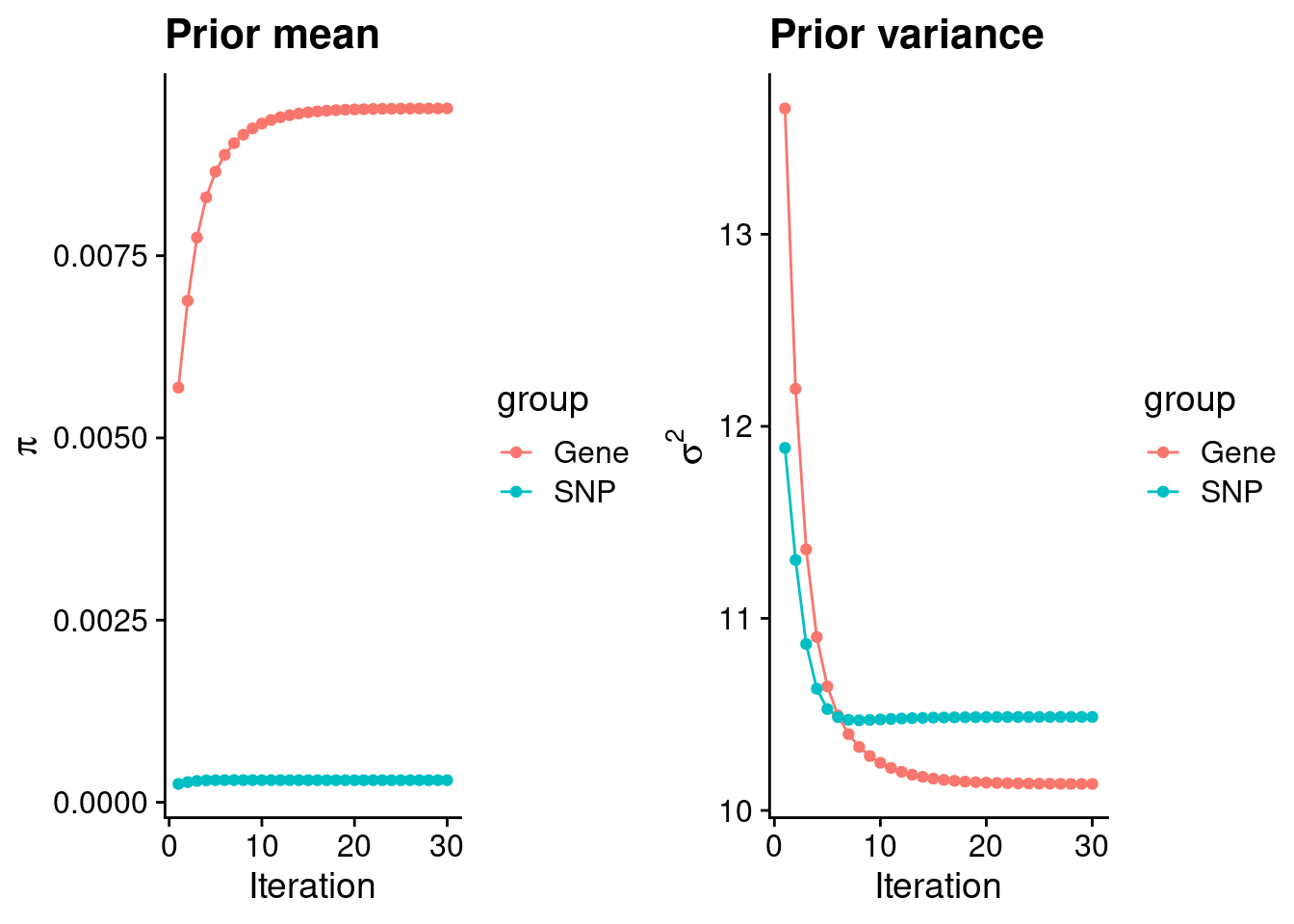
intersect, setdiff, setequal, unionCheck convergence of parameters
| Version | Author | Date |
|---|---|---|
| 2749be9 | sq-96 | 2022-05-12 |
gene snp
0.0095212 0.0003029 gene snp
10.14 10.49 [1] 105318[1] 7589 6309950 gene snp
0.006955 0.190306 [1] 0.02263 1.04773Genes with highest PIPs
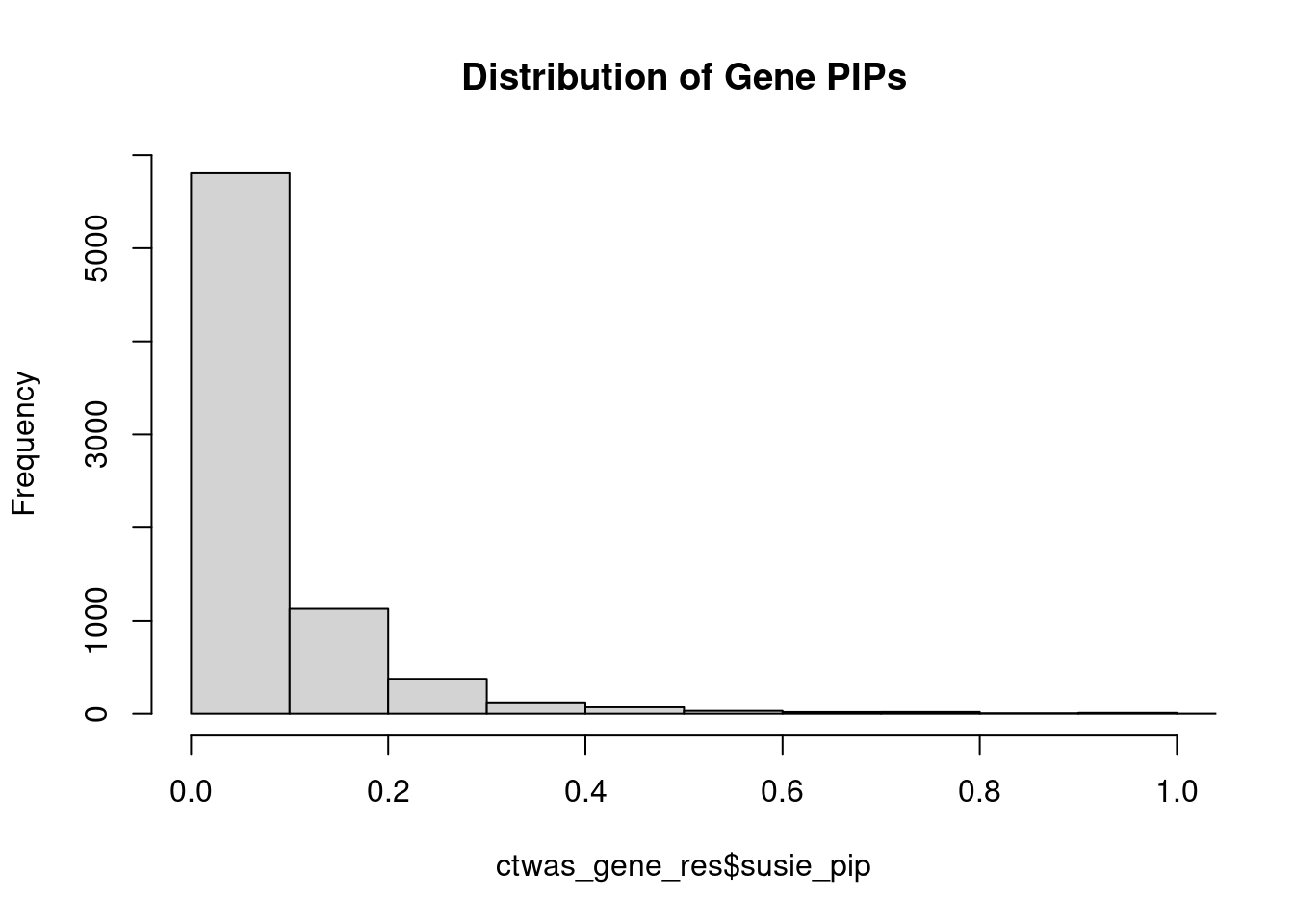
genename region_tag susie_pip mu2 PVE z num_intron num_sqtl
2339 FAM177A1 14_9 1.1961 23.35 2.552e-04 4.820 15 16
3586 LRP8 1_33 1.1731 31.79 3.182e-04 4.820 6 6
5170 R3HDM2 12_36 1.0831 42.25 4.412e-04 6.634 5 6
3407 LAMA5 20_36 0.9784 25.18 2.057e-04 -4.341 14 18
5118 PTPA 9_66 0.9768 22.61 1.987e-04 4.650 8 10
3475 LINC00320 21_6 0.9730 28.66 2.418e-04 5.336 4 4
7343 ZDHHC20 13_2 0.9550 24.33 2.005e-04 -4.784 4 5
293 AKT3 1_128 0.9385 34.39 2.613e-04 -6.291 7 7
2459 FEZF1 7_74 0.9335 24.01 1.987e-04 -4.812 1 1
818 BUB1B-PAK6 15_14 0.9306 29.32 2.343e-04 -5.588 3 3
454 APOPT1 14_54 0.9107 42.67 3.249e-04 7.429 7 9
785 BRCA1 17_25 0.9021 30.88 1.315e-04 -3.794 21 23
6150 SNRPA1 15_50 0.8892 22.00 1.516e-04 -3.925 6 7
673 B3GAT1 11_84 0.8677 22.71 1.434e-04 4.348 8 13
7215 WDR27 6_111 0.8638 14.16 6.448e-05 -2.146 20 27
5161 PYROXD2 10_62 0.8241 21.34 1.255e-04 3.755 11 12
2651 GIGYF1 7_62 0.8160 27.02 1.650e-04 -5.266 3 3
1201 CD46 1_105 0.7757 19.49 1.006e-04 3.804 10 10
681 B9D1 17_16 0.7748 27.45 1.554e-04 5.282 3 3
5869 SGCE 7_58 0.7698 20.54 1.084e-04 4.413 6 8Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_intron num_sqtl
5170 R3HDM2 12_36 1.0831 42.25 0.0004412 6.634 5 6
454 APOPT1 14_54 0.9107 42.67 0.0003249 7.429 7 9
3586 LRP8 1_33 1.1731 31.79 0.0003182 4.820 6 6
293 AKT3 1_128 0.9385 34.39 0.0002613 -6.291 7 7
2339 FAM177A1 14_9 1.1961 23.35 0.0002552 4.820 15 16
3475 LINC00320 21_6 0.9730 28.66 0.0002418 5.336 4 4
818 BUB1B-PAK6 15_14 0.9306 29.32 0.0002343 -5.588 3 3
5855 SF3B1 2_117 0.7434 44.53 0.0002219 7.053 3 3
6695 TMEM219 16_24 0.6954 46.18 0.0002114 -7.020 2 2
3407 LAMA5 20_36 0.9784 25.18 0.0002057 -4.341 14 18
7343 ZDHHC20 13_2 0.9550 24.33 0.0002005 -4.784 4 5
5118 PTPA 9_66 0.9768 22.61 0.0001987 4.650 8 10
2459 FEZF1 7_74 0.9335 24.01 0.0001987 -4.812 1 1
4405 NT5C2 10_66 0.6860 46.35 0.0001860 8.475 11 15
2651 GIGYF1 7_62 0.8160 27.02 0.0001650 -5.266 3 3
681 B9D1 17_16 0.7748 27.45 0.0001554 5.282 3 3
6150 SNRPA1 15_50 0.8892 22.00 0.0001516 -3.925 6 7
673 B3GAT1 11_84 0.8677 22.71 0.0001434 4.348 8 13
7349 ZDHHC8 22_4 0.7294 35.56 0.0001336 -4.861 5 5
785 BRCA1 17_25 0.9021 30.88 0.0001315 -3.794 21 23Comparing z scores and PIPs
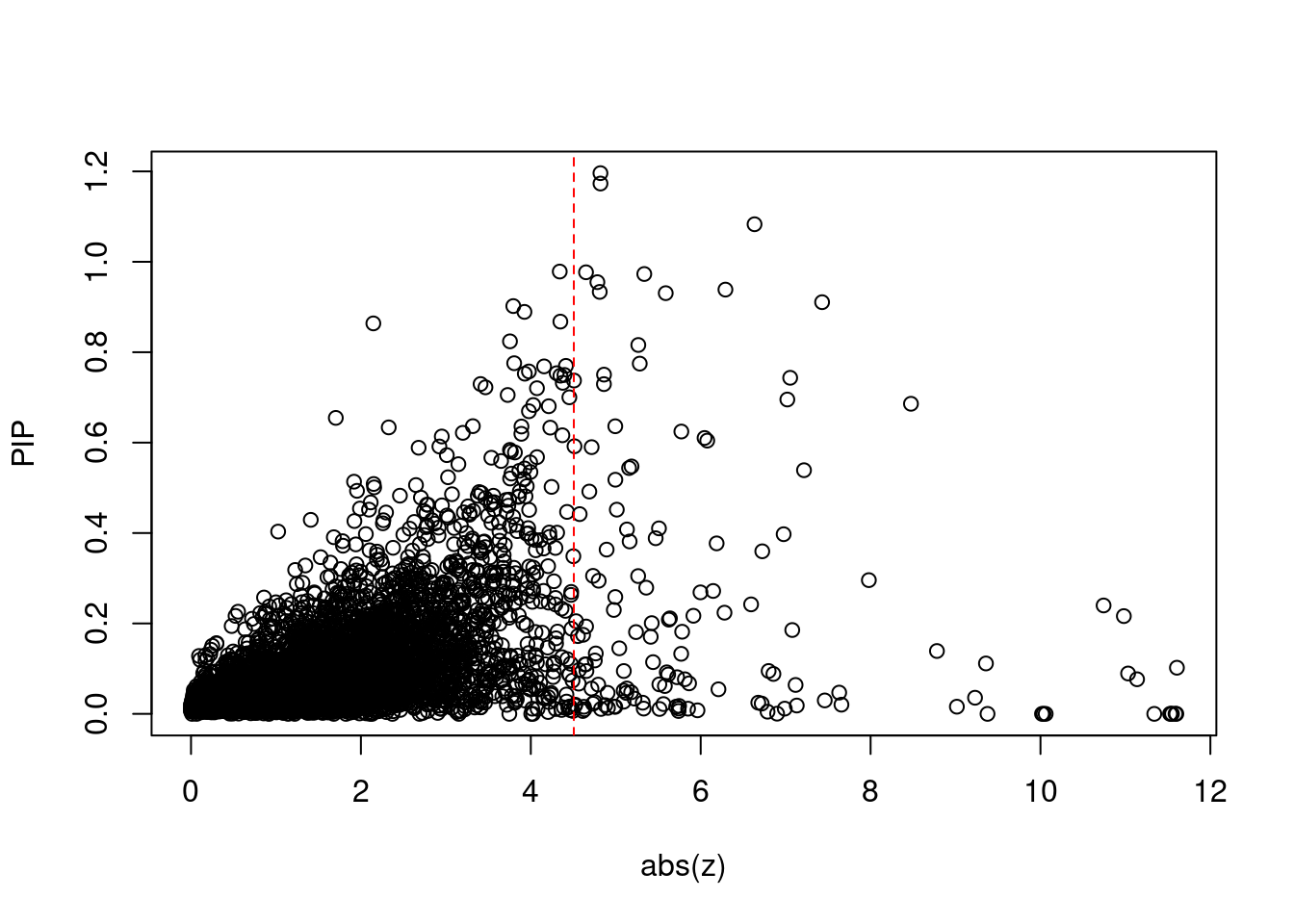
[1] 0.01884 genename region_tag susie_pip mu2 PVE z num_intron num_sqtl
811 BTN2A1 6_20 1.021e-01 107.10 4.502e-06 -11.606 5 5
3622 LSM2 6_26 9.237e-05 214.43 1.737e-11 -11.599 1 1
453 APOM 6_26 1.969e-04 214.01 7.828e-11 11.590 3 3
7130 VARS 6_26 8.850e-05 212.38 1.580e-11 -11.548 1 1
4031 MSH5 6_26 1.671e-04 212.05 5.316e-11 11.538 5 5
690 BAG6 6_26 1.071e-04 208.16 2.197e-11 -11.525 5 7
1742 CYP21A2 6_26 1.973e-05 205.68 7.605e-13 -11.340 1 1
7131 VARS2 6_25 7.654e-02 101.41 5.642e-06 -11.137 1 1
950 C6orf136 6_24 8.935e-02 78.68 5.963e-06 11.031 2 2
2500 FLOT1 6_24 2.163e-01 77.34 3.429e-05 10.981 6 6
814 BTN3A2 6_20 2.398e-01 91.77 2.183e-05 -10.743 6 6
4989 PPT2 6_26 3.405e-05 147.91 1.534e-12 -10.061 5 5
2093 EGFL8 6_26 3.178e-05 147.12 1.305e-12 10.036 6 6
5049 PRRT1 6_26 2.733e-05 146.36 1.038e-12 -10.018 1 1
2773 GPSM3 6_26 2.302e-06 119.97 6.034e-15 9.377 1 1
1157 CCHCR1 6_25 1.115e-01 63.28 2.757e-06 -9.358 11 14
7382 ZKSCAN3 6_22 3.568e-02 54.53 4.183e-07 -9.230 3 3
1805 DDR1 6_25 1.590e-02 67.83 1.627e-07 9.016 1 1
2944 HLA-DMA 6_27 1.391e-01 66.33 4.945e-06 8.781 6 10
4405 NT5C2 10_66 6.860e-01 46.35 1.860e-04 8.475 11 15GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 85Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"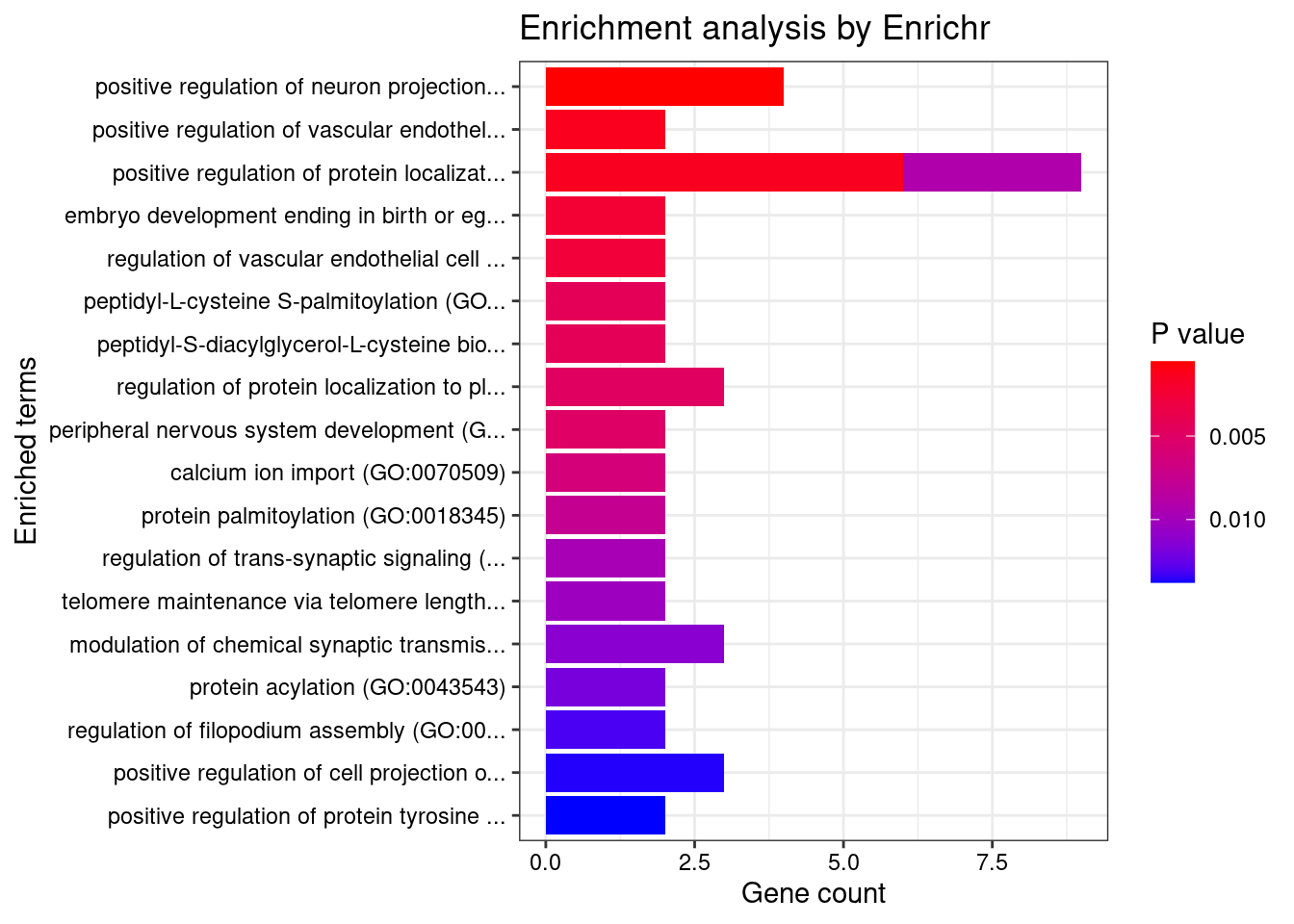
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"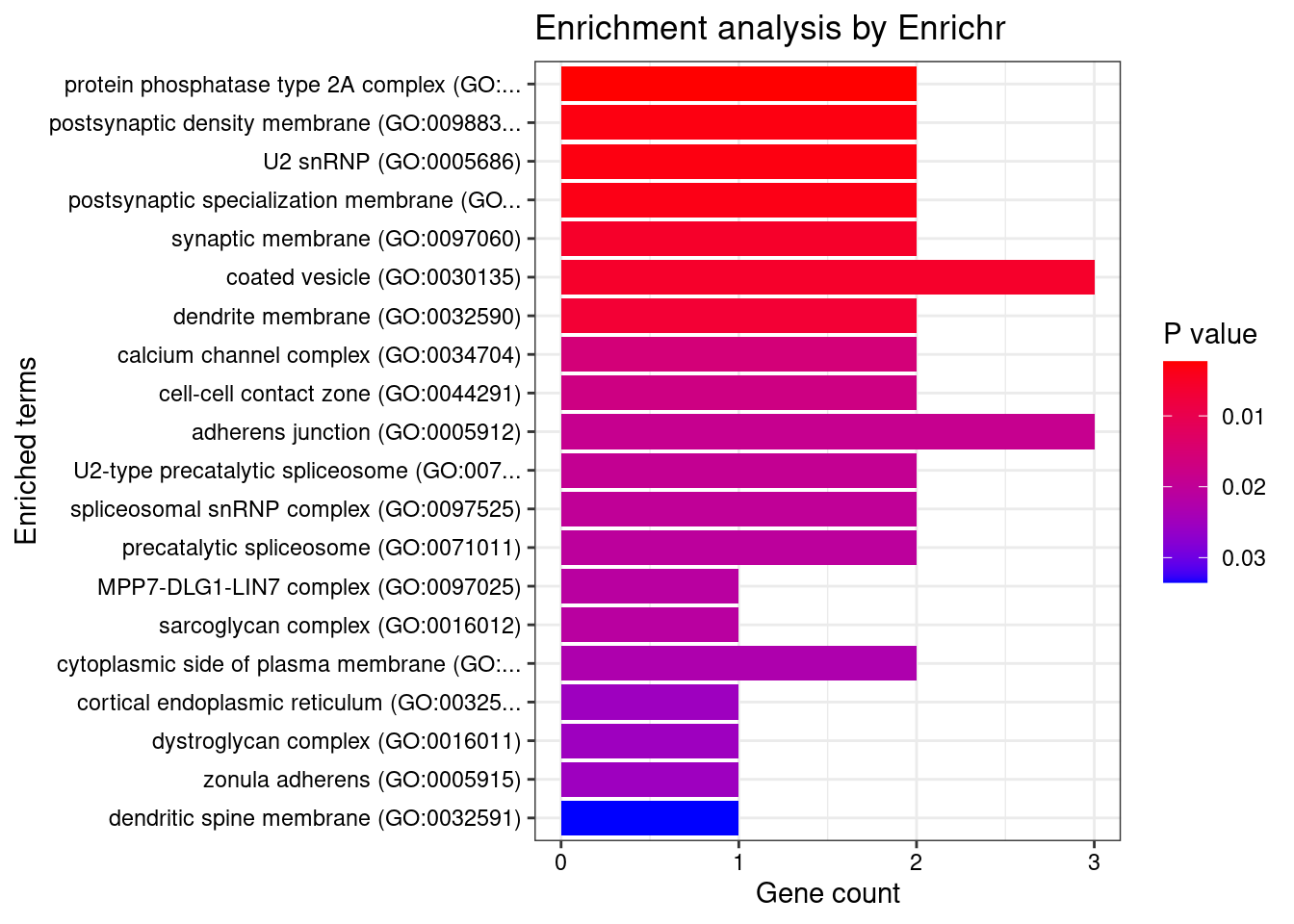
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"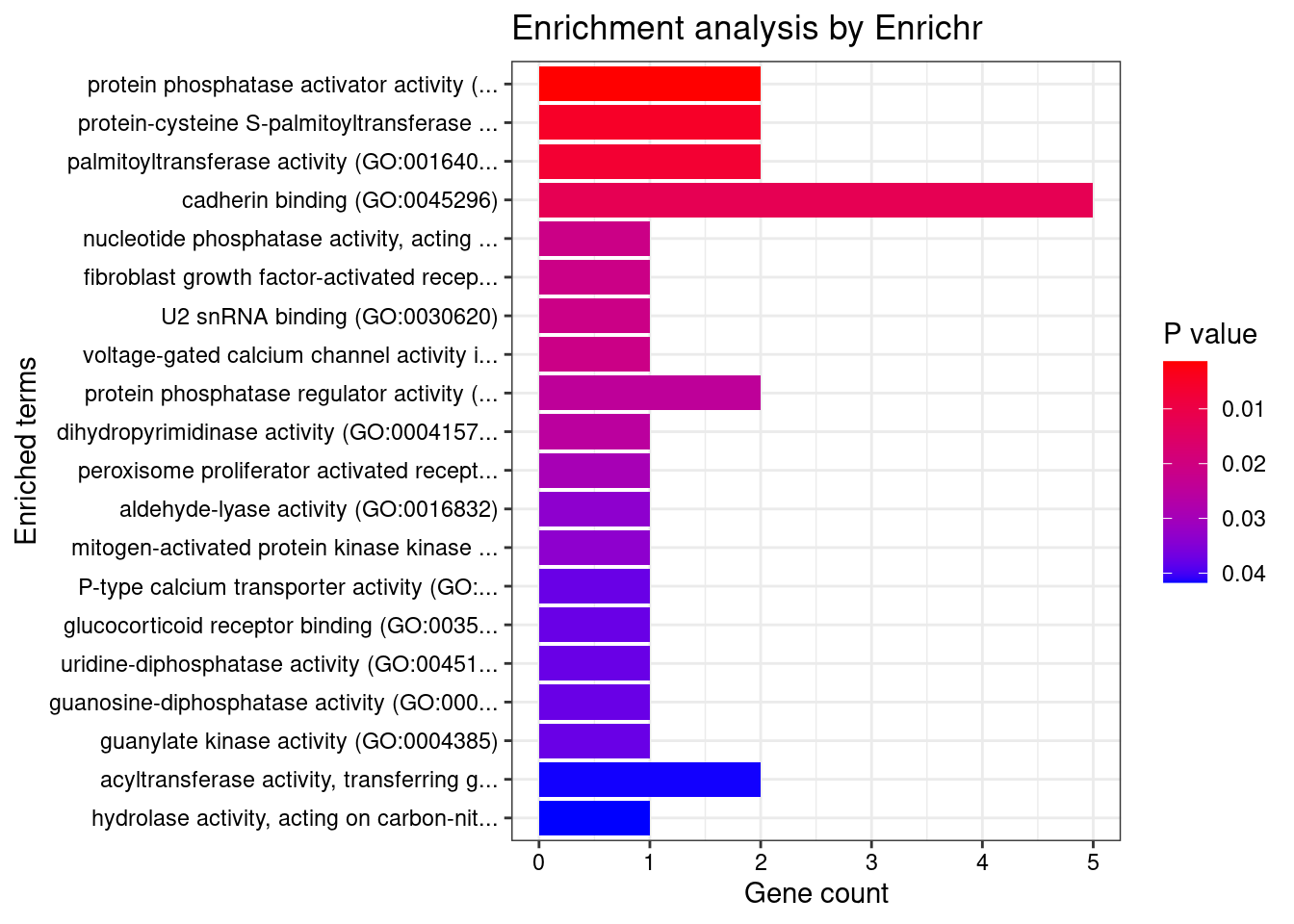
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio BgRatio
10 Balo's Concentric Sclerosis 0.04698 1/46 1/9703
19 Malignant Neoplasms 0.04698 4/46 128/9703
28 Diffuse Cerebral Sclerosis of Schilder 0.04698 1/46 1/9703
73 Measles 0.04698 1/46 1/9703
104 Schizophrenia 0.04698 12/46 883/9703
130 Electroencephalogram abnormal 0.04698 1/46 1/9703
149 gliosarcoma 0.04698 2/46 16/9703
175 Dyskeratosis Congenita 0.04698 2/46 16/9703
177 Gastric Antral Vascular Ectasia 0.04698 1/46 1/9703
207 Idiopathic hypogonadotropic hypogonadism 0.04698 2/46 18/9703WebGestalt enrichment analysis for genes with PIP>0.5
Warning: replacing previous import 'lifecycle::last_warnings' by
'rlang::last_warnings' when loading 'hms'Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Warning: ggrepel: 45 unlabeled data points (too many overlaps). Consider
increasing max.overlaps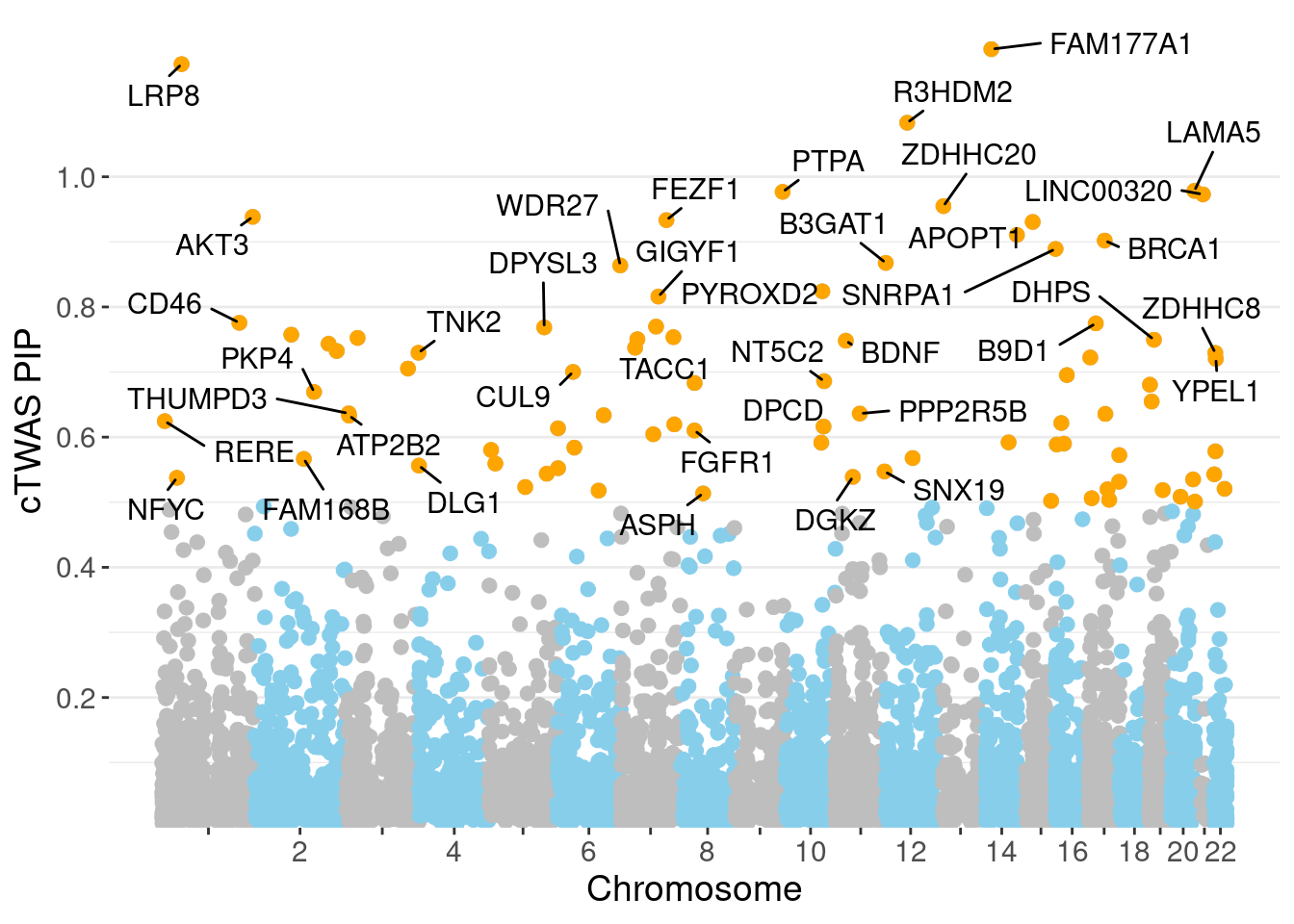
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 130#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 50#significance threshold for TWAS
print(sig_thresh)[1] 4.507#number of ctwas genes
length(ctwas_genes)[1] 17#number of TWAS genes
length(twas_genes)[1] 143#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_intron num_sqtl
673 B3GAT1 11_84 0.8677 22.71 1.434e-04 4.348 8 13
785 BRCA1 17_25 0.9021 30.88 1.315e-04 -3.794 21 23
3407 LAMA5 20_36 0.9784 25.18 2.057e-04 -4.341 14 18
5161 PYROXD2 10_62 0.8241 21.34 1.255e-04 3.755 11 12
6150 SNRPA1 15_50 0.8892 22.00 1.516e-04 -3.925 6 7
7215 WDR27 6_111 0.8638 14.16 6.448e-05 -2.146 20 27#sensitivity / recall
print(sensitivity) ctwas TWAS
0.02308 0.09231 #specificity
print(specificity) ctwas TWAS
0.9981 0.9826 #precision / PPV
print(precision) ctwas TWAS
0.17647 0.08392
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.4.0 forcats_0.5.1 stringr_1.4.0 purrr_0.3.4
[5] readr_1.4.0 tidyr_1.1.3 tidyverse_1.3.1 tibble_3.1.7
[9] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0 cowplot_1.1.1
[13] ggplot2_3.3.5 dplyr_1.0.7 reticulate_1.25 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] fs_1.5.0 lubridate_1.7.10 doParallel_1.0.16 httr_1.4.2
[5] rprojroot_2.0.2 tools_4.1.0 backports_1.2.1 doRNG_1.8.2
[9] bslib_0.2.5.1 utf8_1.2.1 R6_2.5.0 vipor_0.4.5
[13] DBI_1.1.1 colorspace_2.0-2 withr_2.4.2 ggrastr_1.0.1
[17] tidyselect_1.1.1 processx_3.5.2 curl_4.3.2 compiler_4.1.0
[21] git2r_0.28.0 rvest_1.0.0 cli_3.0.0 Cairo_1.5-15
[25] xml2_1.3.2 labeling_0.4.2 sass_0.4.0 scales_1.1.1
[29] callr_3.7.0 systemfonts_1.0.4 apcluster_1.4.9 digest_0.6.27
[33] rmarkdown_2.9 svglite_2.0.0 pkgconfig_2.0.3 htmltools_0.5.1.1
[37] dbplyr_2.1.1 highr_0.9 rlang_1.0.2 rstudioapi_0.13
[41] jquerylib_0.1.4 farver_2.1.0 generics_0.1.0 jsonlite_1.7.2
[45] magrittr_2.0.1 Matrix_1.3-3 ggbeeswarm_0.6.0 Rcpp_1.0.7
[49] munsell_0.5.0 fansi_0.5.0 lifecycle_1.0.0 stringi_1.6.2
[53] whisker_0.4 yaml_2.2.1 plyr_1.8.6 grid_4.1.0
[57] ggrepel_0.9.1 parallel_4.1.0 promises_1.2.0.1 crayon_1.4.1
[61] lattice_0.20-44 haven_2.4.1 hms_1.1.0 knitr_1.33
[65] ps_1.6.0 pillar_1.7.0 igraph_1.2.6 rjson_0.2.20
[69] rngtools_1.5 reshape2_1.4.4 codetools_0.2-18 reprex_2.0.0
[73] glue_1.4.2 evaluate_0.14 getPass_0.2-2 modelr_0.1.8
[77] data.table_1.14.0 png_0.1-7 vctrs_0.3.8 httpuv_1.6.1
[81] foreach_1.5.1 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] xfun_0.24 broom_0.7.8 later_1.2.0 iterators_1.0.13
[89] beeswarm_0.4.0 ellipsis_0.3.2 here_1.0.1