BMI - Brain Hippocampus
sheng Qian
2021-2-6
Last updated: 2022-02-21
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bbf6737. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/AF/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/BMI_Brain_Hippocampus.Rmd) and HTML (docs/BMI_Brain_Hippocampus.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bbf6737 | sq-96 | 2022-02-21 | update |
| html | 91f38fa | sq-96 | 2022-02-13 | Build site. |
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 10973#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1076 769 643 427 529 614 510 402 415 437 663 585 220 377 370 516
17 18 19 20 21 22
664 166 865 330 117 278 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 8798#proportion of imputed weights without missing variants
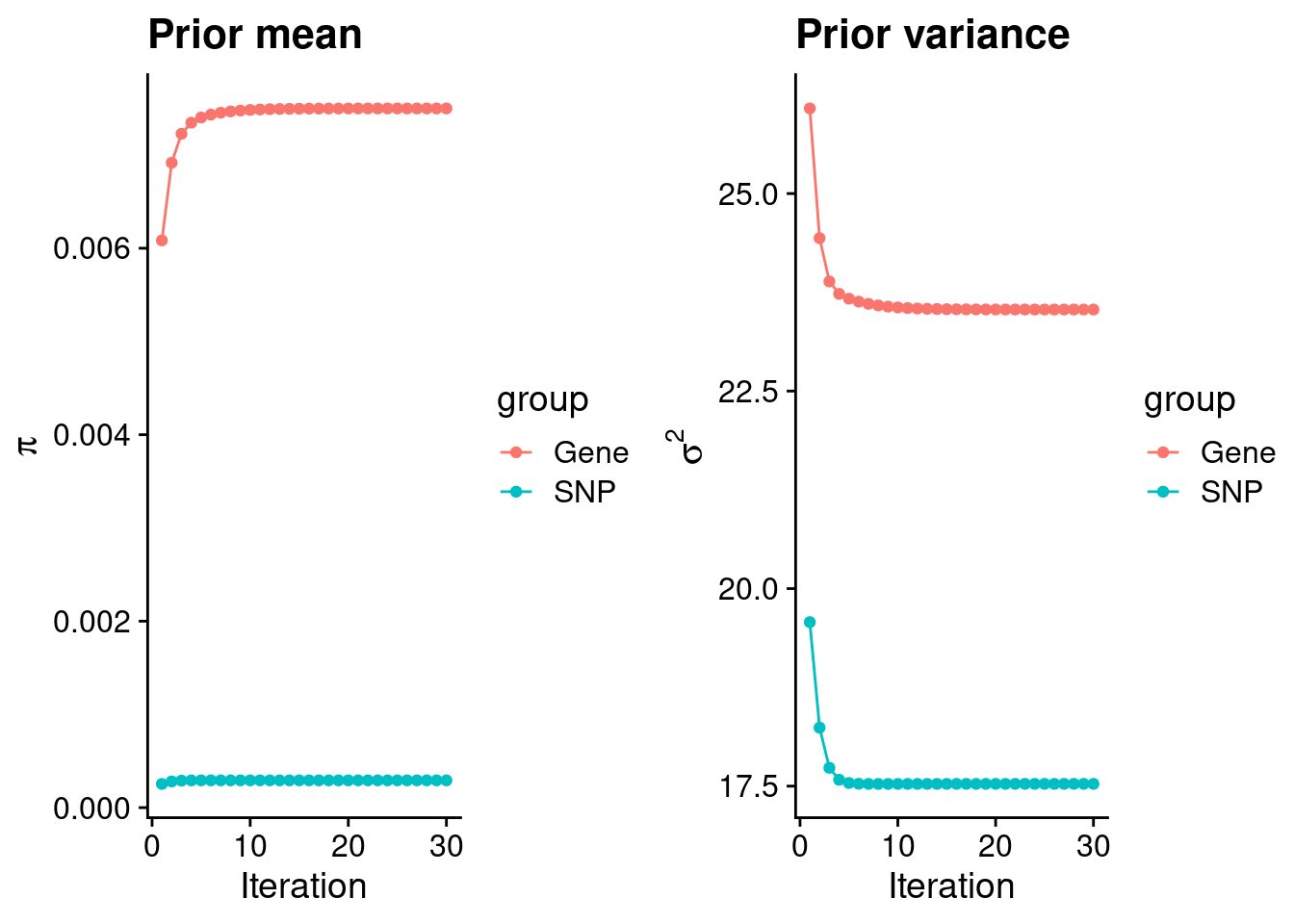
mean(qclist_all$nmiss==0)[1] 0.8018Check convergence of parameters
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0074990 0.0002925 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
23.53 17.53 #report sample size
print(sample_size)[1] 336107#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 10973 7535010#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.005761 0.114953 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.2223 18.6719Genes with highest PIPs
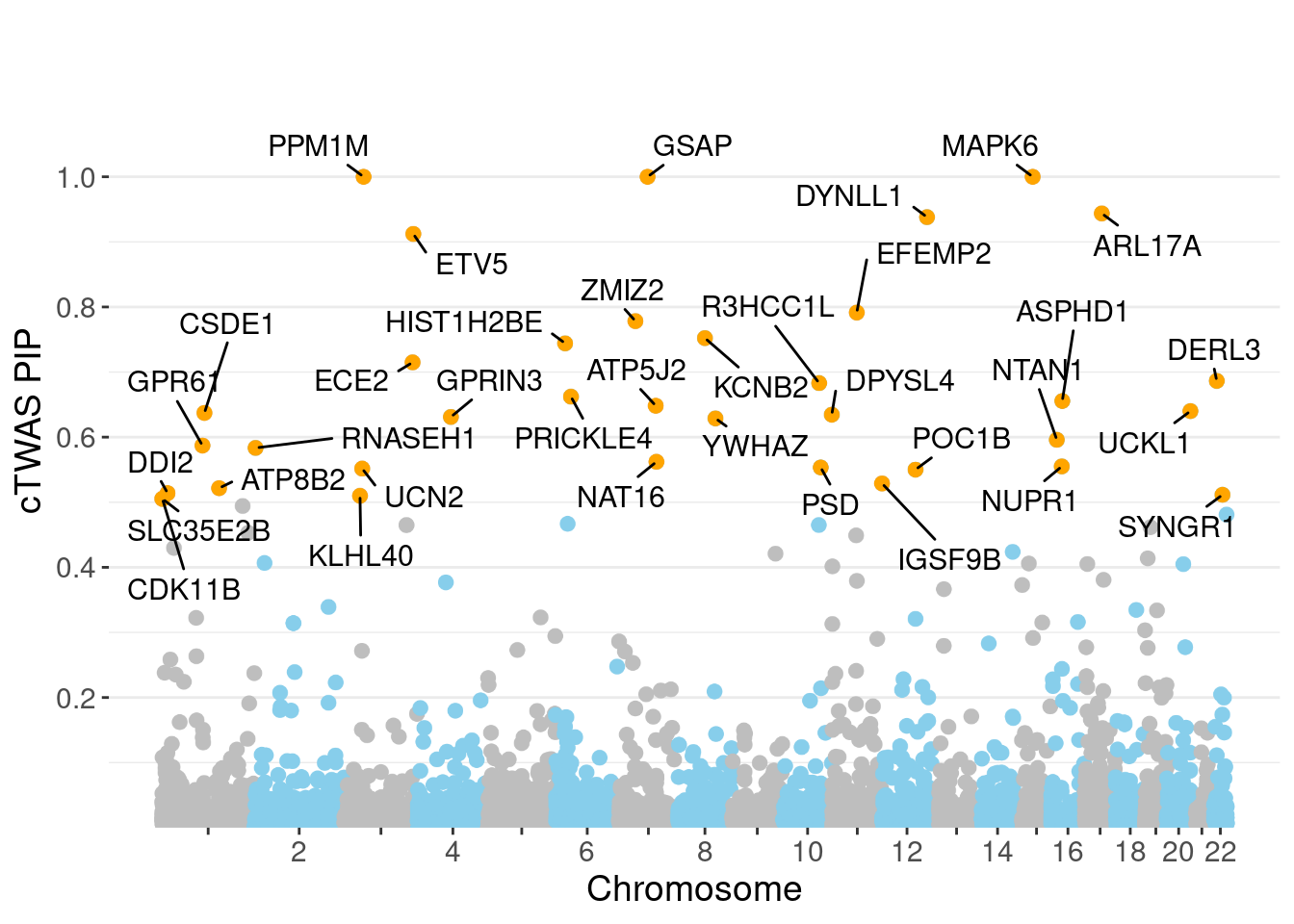
genename region_tag susie_pip mu2 PVE z num_eqtl
717 MAPK6 15_21 1.0000 34111.66 1.015e-01 -4.600 1
10029 GSAP 7_49 1.0000 32344.92 9.623e-02 5.260 1
7429 PPM1M 3_36 1.0000 241.22 7.177e-04 4.468 2
9989 ARL17A 17_27 0.9437 32.41 9.100e-05 5.325 1
1199 DYNLL1 12_74 0.9380 59.08 1.649e-04 -5.806 1
12053 ETV5 3_114 0.9122 96.75 2.626e-04 9.862 1
8671 EFEMP2 11_36 0.7914 56.03 1.319e-04 -8.201 1
3564 ZMIZ2 7_33 0.7783 66.49 1.540e-04 -8.105 1
9621 KCNB2 8_53 0.7522 66.19 1.481e-04 -8.226 1
13243 HIST1H2BE 6_20 0.7441 31.10 6.885e-05 -6.515 1
5796 ECE2 3_113 0.7149 29.87 6.354e-05 -5.305 1
1445 DERL3 22_6 0.6864 22.98 4.694e-05 4.037 1
7736 R3HCC1L 10_62 0.6830 40.52 8.234e-05 7.439 1
13421 PRICKLE4 6_32 0.6622 24.65 4.858e-05 -4.797 1
8923 ASPHD1 16_24 0.6556 578.64 1.129e-03 -11.938 1
11969 ATP5J2 7_61 0.6482 53.45 1.031e-04 -7.117 1
10750 UCKL1 20_38 0.6400 25.31 4.819e-05 3.573 1
151 CSDE1 1_71 0.6372 22.68 4.300e-05 -4.745 1
6243 DPYSL4 10_83 0.6345 43.79 8.266e-05 -6.801 1
9938 GPRIN3 4_60 0.6310 25.08 4.709e-05 -3.769 2Genes with largest effect sizes
genename region_tag susie_pip mu2 PVE z num_eqtl
10 SEMA3F 3_35 0.000e+00 73861 0.000e+00 7.681 1
10261 SLC38A3 3_35 0.000e+00 69034 0.000e+00 6.726 1
7591 CCDC171 9_13 0.000e+00 44932 0.000e+00 7.405 2
8624 NEGR1 1_46 0.000e+00 43597 0.000e+00 -10.375 2
40 RBM6 3_35 0.000e+00 41746 0.000e+00 12.536 1
6640 ZNF689 16_24 0.000e+00 39994 0.000e+00 -6.014 1
7425 MST1R 3_35 0.000e+00 35624 0.000e+00 -12.635 2
717 MAPK6 15_21 1.000e+00 34112 1.015e-01 -4.600 1
10029 GSAP 7_49 1.000e+00 32345 9.623e-02 5.260 1
9293 STX19 3_59 0.000e+00 31600 0.000e+00 -5.060 1
9289 DHFR2 3_59 0.000e+00 25976 0.000e+00 4.031 2
5274 MFAP1 15_16 1.202e-06 24147 8.636e-08 4.303 1
7420 RNF123 3_35 0.000e+00 23601 0.000e+00 -10.959 1
12024 NAT6 3_35 0.000e+00 23005 0.000e+00 -6.362 1
10512 C6orf106 6_29 0.000e+00 22878 0.000e+00 2.962 1
11433 CKMT1A 15_16 0.000e+00 21625 0.000e+00 4.130 1
1326 WDR76 15_16 0.000e+00 21190 0.000e+00 4.963 2
1785 ZNF629 16_24 0.000e+00 20375 0.000e+00 4.335 1
10290 DPYD 1_60 0.000e+00 19961 0.000e+00 -3.213 1
10101 HYAL3 3_35 0.000e+00 18111 0.000e+00 6.243 2Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
717 MAPK6 15_21 1.00000 34111.66 1.015e-01 -4.600 1
10029 GSAP 7_49 1.00000 32344.92 9.623e-02 5.260 1
821 SDHA 5_1 0.22996 12142.57 8.308e-03 2.907 1
8923 ASPHD1 16_24 0.65557 578.64 1.129e-03 -11.938 1
7429 PPM1M 3_36 1.00000 241.22 7.177e-04 4.468 2
12053 ETV5 3_114 0.91224 96.75 2.626e-04 9.862 1
6834 ADPGK 15_34 0.05907 1201.55 2.112e-04 5.872 3
1199 DYNLL1 12_74 0.93799 59.08 1.649e-04 -5.806 1
3564 ZMIZ2 7_33 0.77828 66.49 1.540e-04 -8.105 1
9621 KCNB2 8_53 0.75221 66.19 1.481e-04 -8.226 1
6587 GPR61 1_67 0.58712 79.87 1.395e-04 8.755 1
5143 USO1 4_51 0.37687 123.87 1.389e-04 -2.134 1
8671 EFEMP2 11_36 0.79142 56.03 1.319e-04 -8.201 1
9035 NUPR1 16_23 0.55509 69.67 1.151e-04 -10.643 2
11969 ATP5J2 7_61 0.64819 53.45 1.031e-04 -7.117 1
10366 SLC35E2B 1_1 0.50528 63.53 9.550e-05 -7.654 1
12120 CDK11B 1_1 0.50528 63.53 9.550e-05 -7.654 1
9989 ARL17A 17_27 0.94369 32.41 9.100e-05 5.325 1
6243 DPYSL4 10_83 0.63454 43.79 8.266e-05 -6.801 1
7736 R3HCC1L 10_62 0.68300 40.52 8.234e-05 7.439 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
7425 MST1R 3_35 0.000e+00 35623.61 0.000e+00 -12.635 2
40 RBM6 3_35 0.000e+00 41746.10 0.000e+00 12.536 1
8923 ASPHD1 16_24 6.556e-01 578.64 1.129e-03 -11.938 1
8924 KCTD13 16_24 4.364e-03 498.27 6.469e-06 11.491 1
8275 INO80E 16_24 1.638e-04 1631.75 7.953e-07 11.077 1
7420 RNF123 3_35 0.000e+00 23601.01 0.000e+00 -10.959 1
6146 TAOK2 16_24 4.676e-06 1891.49 2.632e-08 10.738 1
9035 NUPR1 16_23 5.551e-01 69.67 1.151e-04 -10.643 2
8624 NEGR1 1_46 0.000e+00 43597.35 0.000e+00 -10.375 2
11727 RP11-196G11.6 16_24 4.163e-08 7371.39 9.131e-10 10.011 1
8623 C1QTNF4 11_29 2.297e-02 95.38 6.518e-06 9.951 2
12053 ETV5 3_114 9.122e-01 96.75 2.626e-04 9.862 1
5469 SAE1 19_33 4.553e-03 100.30 1.359e-06 9.849 1
461 PRSS8 16_24 2.775e-09 6922.87 5.715e-11 -9.765 1
7720 RAPSN 11_29 1.189e-02 88.21 3.121e-06 9.729 1
11241 LAT 16_23 2.439e-01 56.17 4.076e-05 -9.553 1
2491 MTCH2 11_29 9.481e-03 83.10 2.344e-06 -9.514 1
10579 IL27 16_23 6.176e-03 50.94 9.360e-07 9.140 1
3527 POLK 5_44 1.242e-02 53.54 1.978e-06 8.884 1
7718 SLC39A13 11_29 9.708e-03 72.48 2.093e-06 -8.831 1Comparing z scores and PIPs
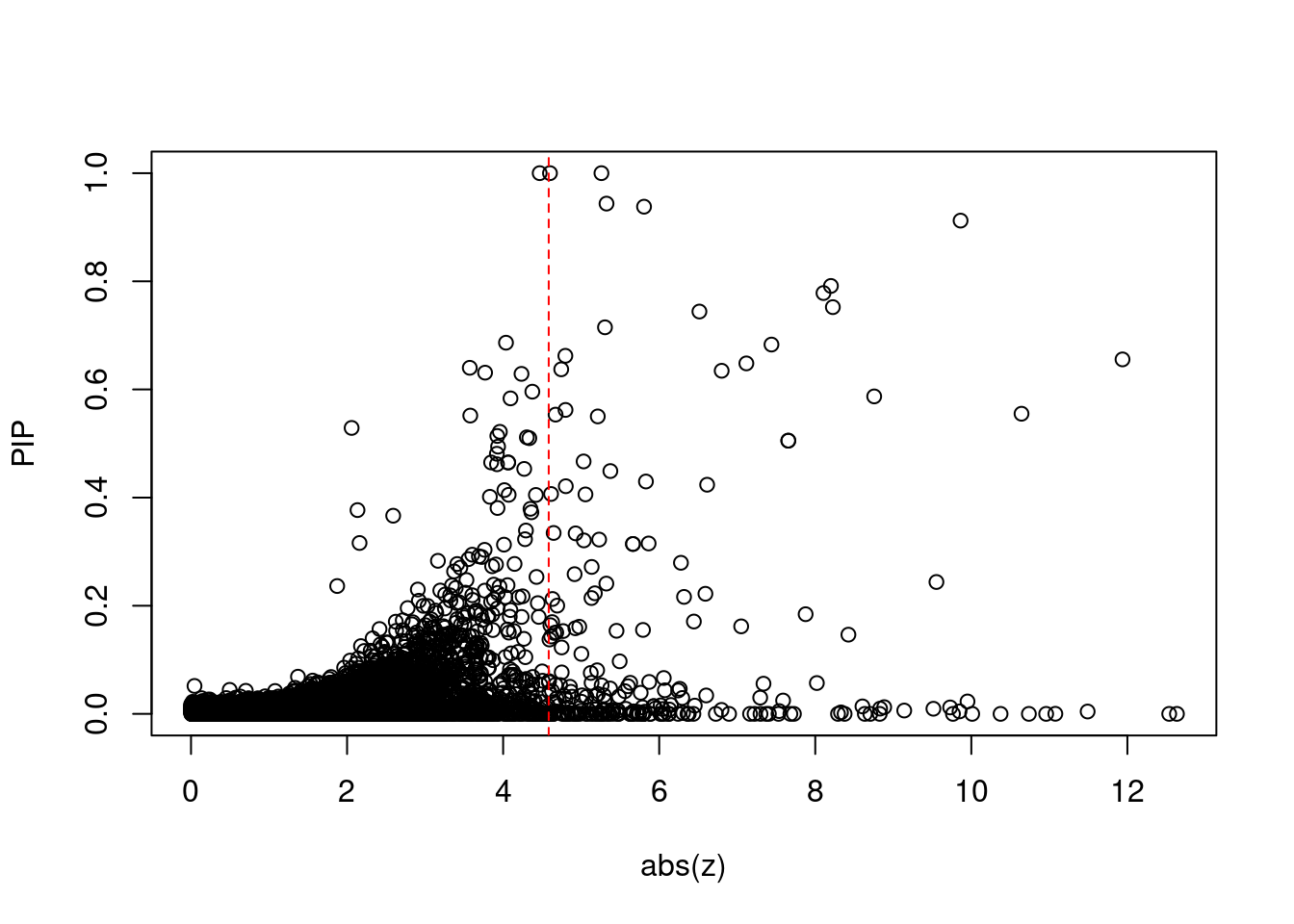
[1] 0.0216 genename region_tag susie_pip mu2 PVE z num_eqtl
7425 MST1R 3_35 0.000e+00 35623.61 0.000e+00 -12.635 2
40 RBM6 3_35 0.000e+00 41746.10 0.000e+00 12.536 1
8923 ASPHD1 16_24 6.556e-01 578.64 1.129e-03 -11.938 1
8924 KCTD13 16_24 4.364e-03 498.27 6.469e-06 11.491 1
8275 INO80E 16_24 1.638e-04 1631.75 7.953e-07 11.077 1
7420 RNF123 3_35 0.000e+00 23601.01 0.000e+00 -10.959 1
6146 TAOK2 16_24 4.676e-06 1891.49 2.632e-08 10.738 1
9035 NUPR1 16_23 5.551e-01 69.67 1.151e-04 -10.643 2
8624 NEGR1 1_46 0.000e+00 43597.35 0.000e+00 -10.375 2
11727 RP11-196G11.6 16_24 4.163e-08 7371.39 9.131e-10 10.011 1
8623 C1QTNF4 11_29 2.297e-02 95.38 6.518e-06 9.951 2
12053 ETV5 3_114 9.122e-01 96.75 2.626e-04 9.862 1
5469 SAE1 19_33 4.553e-03 100.30 1.359e-06 9.849 1
461 PRSS8 16_24 2.775e-09 6922.87 5.715e-11 -9.765 1
7720 RAPSN 11_29 1.189e-02 88.21 3.121e-06 9.729 1
11241 LAT 16_23 2.439e-01 56.17 4.076e-05 -9.553 1
2491 MTCH2 11_29 9.481e-03 83.10 2.344e-06 -9.514 1
10579 IL27 16_23 6.176e-03 50.94 9.360e-07 9.140 1
3527 POLK 5_44 1.242e-02 53.54 1.978e-06 8.884 1
7718 SLC39A13 11_29 9.708e-03 72.48 2.093e-06 -8.831 1GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 36Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"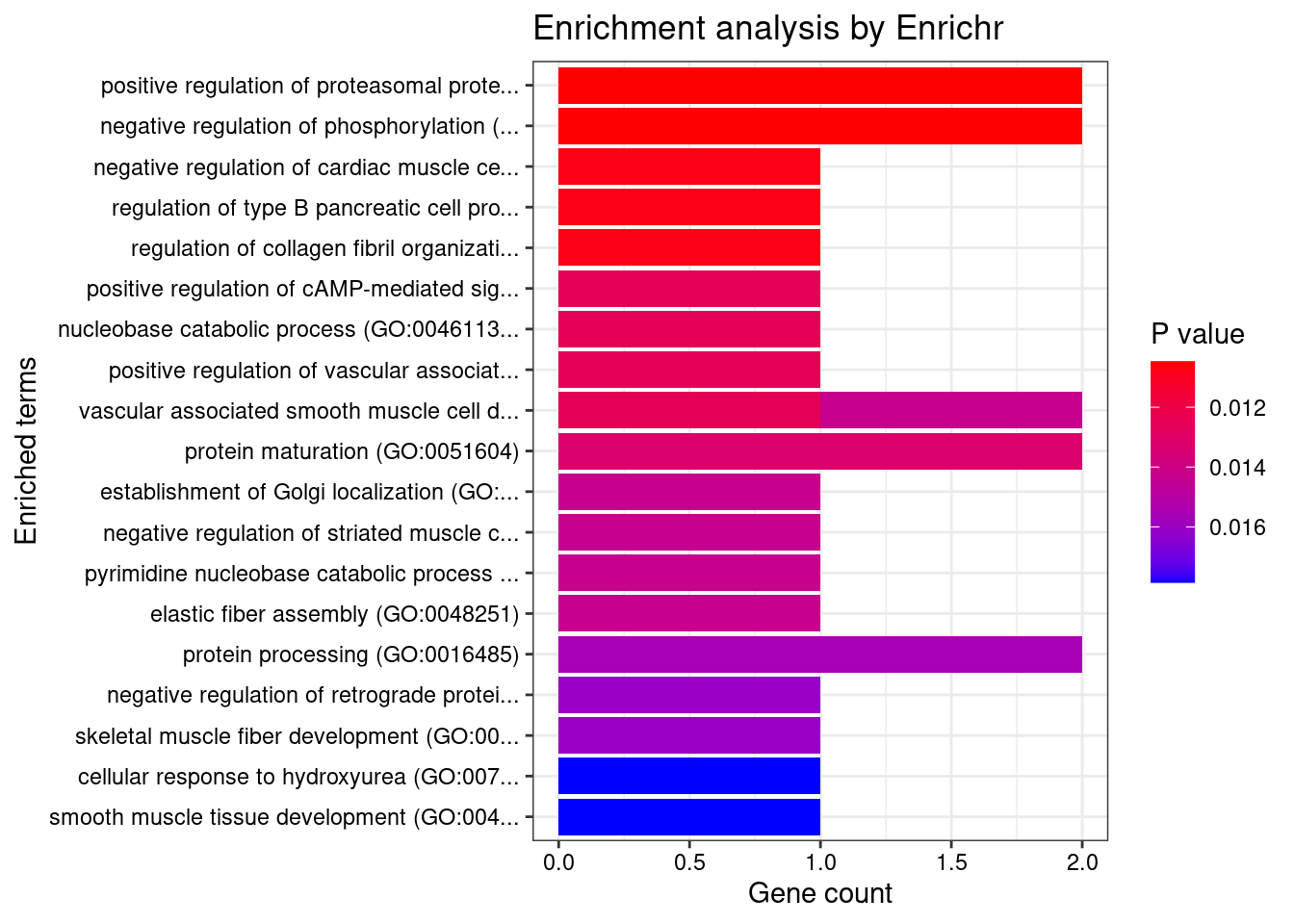
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"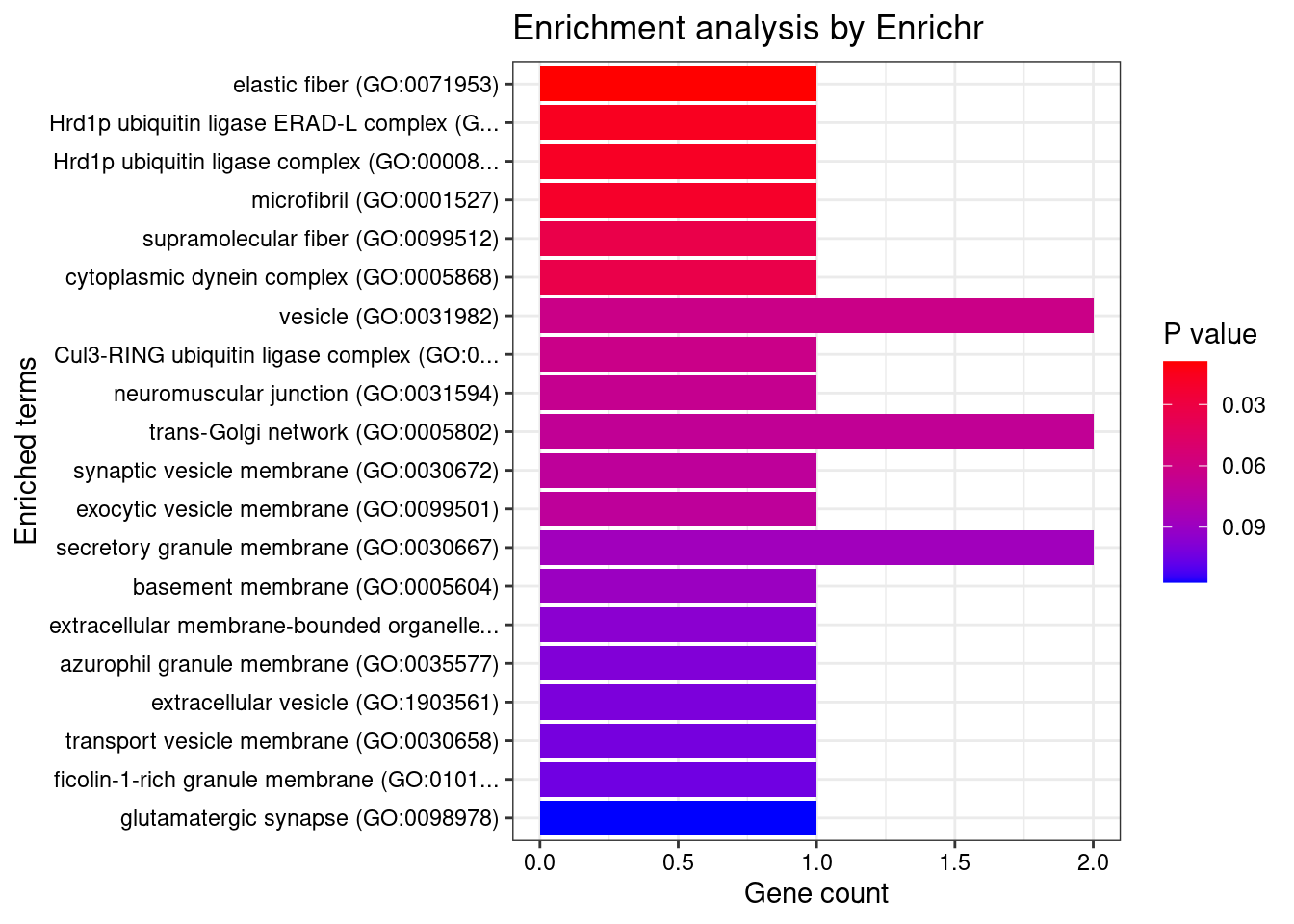
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"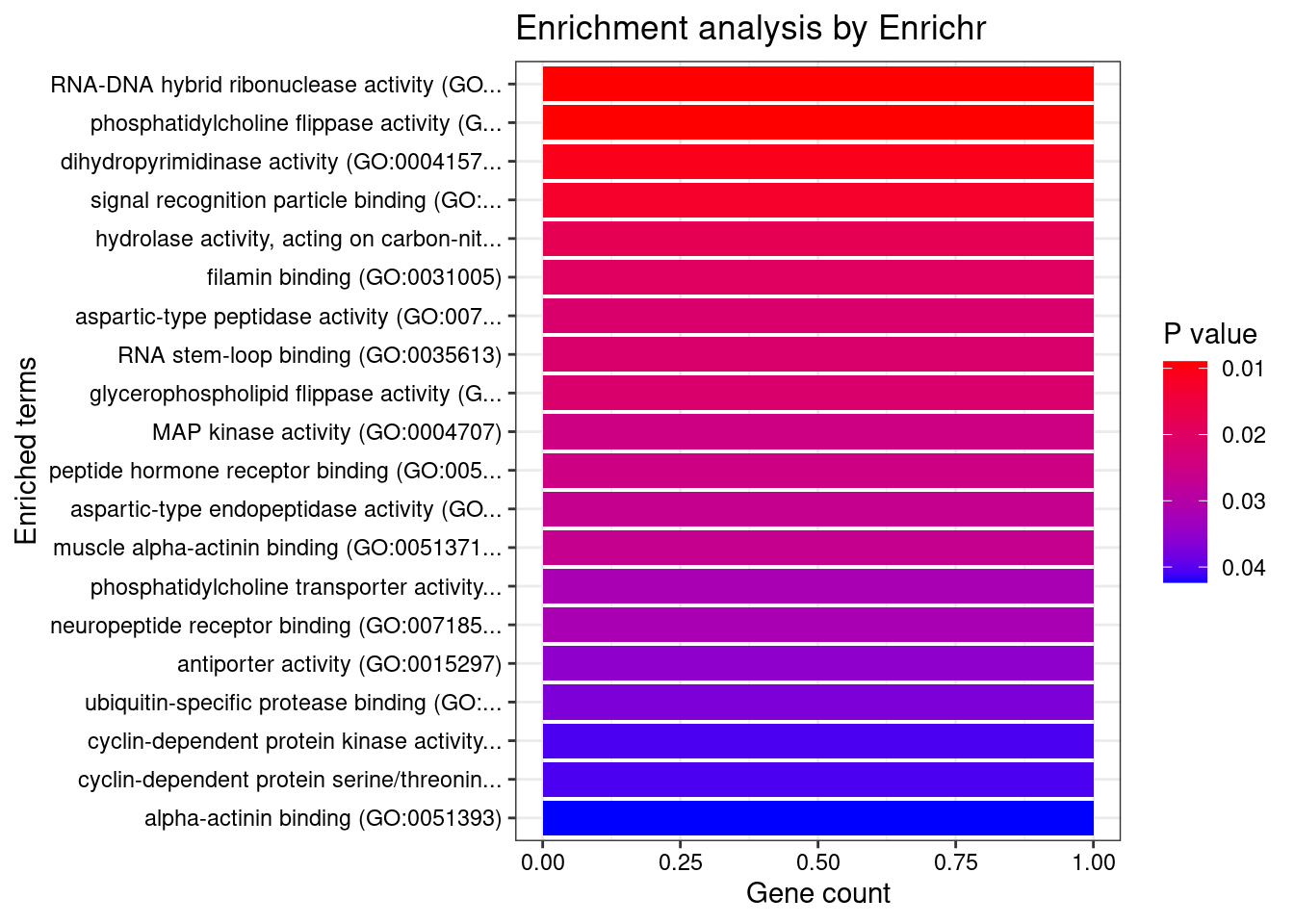
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description
44 CUTIS LAXA, AUTOSOMAL RECESSIVE, TYPE IB
47 NEMALINE MYOPATHY 8
48 CONE-ROD DYSTROPHY 20
49 PROGRESSIVE EXTERNAL OPHTHALMOPLEGIA WITH MITOCHONDRIAL DNA DELETIONS, AUTOSOMAL RECESSIVE 2
32 Cutis Laxa, Autosomal Recessive, Type I
36 Cutis laxa, recessive, type I
50 Adult-onset chronic progressive external ophthalmoplegia with mitochondrial myopathy
8 Cutis Laxa
2 Aortic Aneurysm
25 Paranoid Schizophrenia
FDR Ratio BgRatio
44 0.01840 1/14 1/9703
47 0.01840 1/14 1/9703
48 0.01840 1/14 1/9703
49 0.01840 1/14 1/9703
32 0.02101 1/14 2/9703
36 0.02101 1/14 2/9703
50 0.02101 1/14 2/9703
8 0.05501 1/14 6/9703
2 0.05701 1/14 7/9703
25 0.08628 1/14 13/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 41#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 25#significance threshold for TWAS
print(sig_thresh)[1] 4.584#number of ctwas genes
length(ctwas_genes)[1] 6#number of TWAS genes
length(twas_genes)[1] 237#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
7429 PPM1M 3_36 1 241.2 0.0007177 4.468 2#sensitivity / recall
print(sensitivity) ctwas TWAS
0.00000 0.09756 #specificity
print(specificity) ctwas TWAS
0.9995 0.9787 #precision / PPV
print(precision) ctwas TWAS
0.00000 0.01688 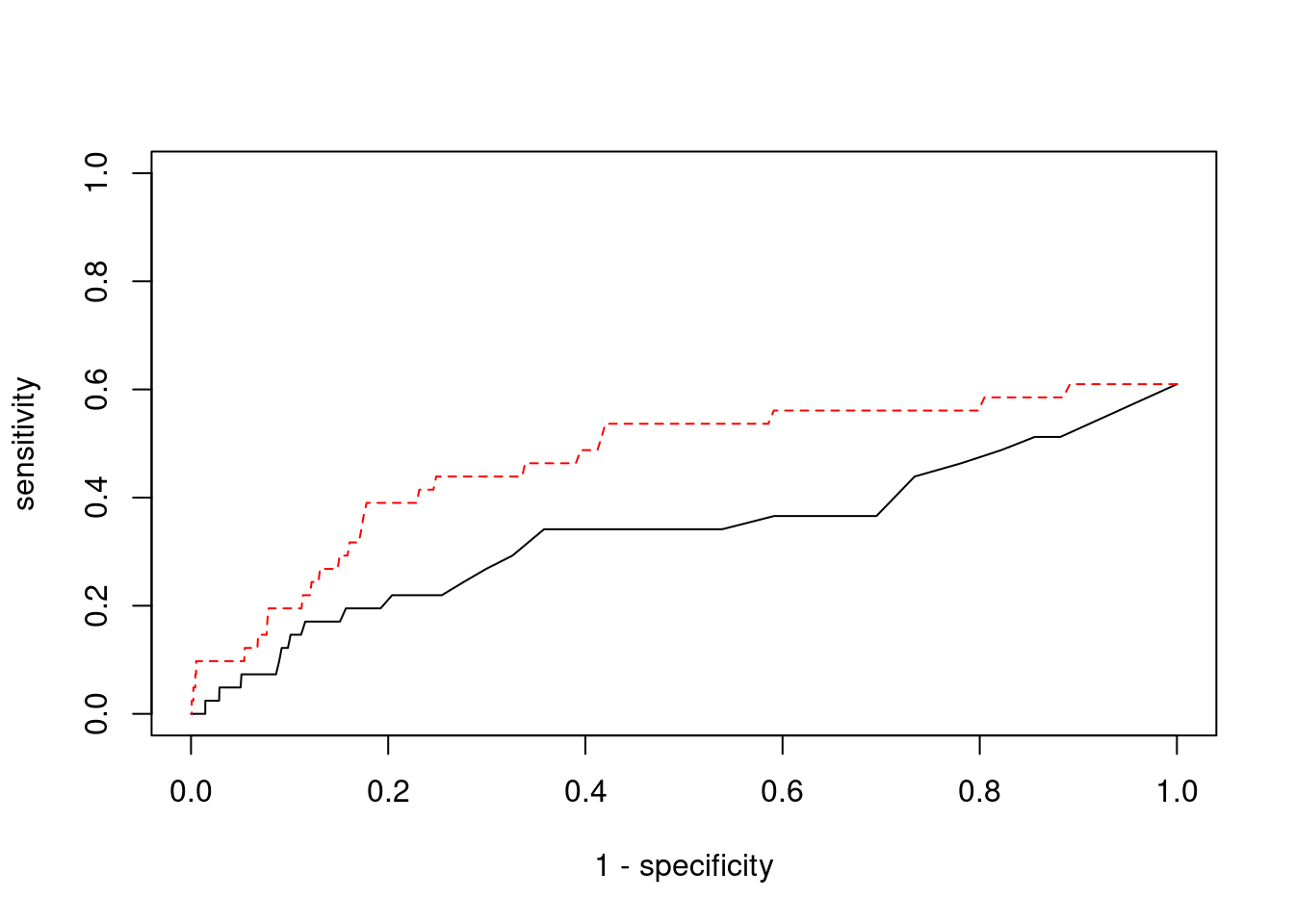
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[5] purrr_0.3.4 readr_2.1.1 tidyr_1.1.4 tidyverse_1.3.1
[9] tibble_3.1.6 WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[13] cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.2 lubridate_1.8.0 bit64_4.0.5 doParallel_1.0.16
[5] httr_1.4.2 rprojroot_2.0.2 tools_3.6.1 backports_1.4.1
[9] doRNG_1.8.2 utf8_1.2.2 R6_2.5.1 vipor_0.4.5
[13] DBI_1.1.1 colorspace_2.0-2 withr_2.4.3 ggrastr_1.0.1
[17] tidyselect_1.1.1 bit_4.0.4 curl_4.3.2 compiler_3.6.1
[21] git2r_0.26.1 cli_3.1.0 rvest_1.0.2 Cairo_1.5-12.2
[25] xml2_1.3.3 labeling_0.4.2 scales_1.1.1 apcluster_1.4.8
[29] digest_0.6.29 rmarkdown_2.11 svglite_1.2.2 pkgconfig_2.0.3
[33] htmltools_0.5.2 dbplyr_2.1.1 fastmap_1.1.0 highr_0.9
[37] rlang_0.4.12 rstudioapi_0.13 RSQLite_2.2.8 jquerylib_0.1.4
[41] farver_2.1.0 generics_0.1.1 jsonlite_1.7.2 vroom_1.5.7
[45] magrittr_2.0.1 Matrix_1.2-18 ggbeeswarm_0.6.0 Rcpp_1.0.7
[49] munsell_0.5.0 fansi_0.5.0 gdtools_0.1.9 lifecycle_1.0.1
[53] stringi_1.7.6 whisker_0.3-2 yaml_2.2.1 plyr_1.8.6
[57] grid_3.6.1 blob_1.2.2 ggrepel_0.9.1 parallel_3.6.1
[61] promises_1.0.1 crayon_1.4.2 lattice_0.20-38 haven_2.4.3
[65] hms_1.1.1 knitr_1.36 pillar_1.6.4 igraph_1.2.10
[69] rjson_0.2.20 rngtools_1.5.2 reshape2_1.4.4 codetools_0.2-16
[73] reprex_2.0.1 glue_1.5.1 evaluate_0.14 data.table_1.14.2
[77] modelr_0.1.8 vctrs_0.3.8 tzdb_0.2.0 httpuv_1.5.1
[81] foreach_1.5.1 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] cachem_1.0.6 xfun_0.29 broom_0.7.10 later_0.8.0
[89] iterators_1.0.13 beeswarm_0.2.3 memoise_2.0.1 ellipsis_0.3.2