Test susieI using chr17 to 22
Last updated: 2020-08-20
Checks: 6 1
Knit directory: causal-TWAS/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191103) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 86ac96e. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/workflow/.ipynb_checkpoints/
Ignored: data/
Unstaged changes:
Modified: analysis/simulation-susieI-ukbchr17to22-gtex.adipose.Rmd
Modified: code/run_test_mr.ash2s.R
Modified: code/run_test_susieI.R
Modified: code/workflow/workflow-ashtest4.ipynb
Modified: code/workflow/workflow-susieI-20200813.ipynb
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/simulation-susieI-ukbchr17to22-gtex.adipose.Rmd) and HTML (docs/simulation-susieI-ukbchr17to22-gtex.adipose.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 86ac96e | simingz | 2020-08-19 | more iterations |
| html | 86ac96e | simingz | 2020-08-19 | more iterations |
| Rmd | bd402eb | simingz | 2020-08-19 | susieI bug fix |
| html | bd402eb | simingz | 2020-08-19 | susieI bug fix |
| Rmd | f125882 | simingz | 2020-08-19 | susieI |
| html | f125882 | simingz | 2020-08-19 | susieI |
| Rmd | d299f60 | simingz | 2020-08-19 | susieI |
| html | d299f60 | simingz | 2020-08-19 | susieI |
| Rmd | 846fb96 | simingz | 2020-08-14 | susieI |
| html | 846fb96 | simingz | 2020-08-14 | susieI |
| Rmd | fd909a1 | simingz | 2020-08-14 | susieI |
Test susieI for ukb chr 17 to chr 22 combined. SNPs are downsampled to 1/10, eQTLs defined by FUSION-TWAS (Adipose/GTEx) lasso effect size > 0 were kept, p= 86k, n = 20k.
susieI is an iterative version of susie, learning parameters (prior weights) iteratively using the top region from mr.ash for genes only.
library(mr.ash.alpha)
library(data.table)
suppressMessages({library(plotly)})
library(tidyr)
library(plyr)
library(stringr)
library(kableExtra)
source("analysis/summarize_twas_plots.R")simdatadir <- "~/causalTWAS/simulations/simulation_ashtest_20200721/"
outputdir <- "~/causalTWAS/simulations/simulation_susieI_20200813/"
susiedir <- "~/causalTWAS/simulations/simulation_susieI_20200813/"
mrashdir <- "~/causalTWAS/simulations/simulation_ashtest_20200721/"
tag <- '20200813-1-1'
Niter <- 30MR.ASH pip distribution
- Use mr.ash for gene only:
rpipf <- paste0(outputdir, tag, "-mr.ash.rPIP.txt")
a <- read.table(rpipf, header = T)
a.c <- a[a$nCausal > 0, ]
ax <- pretty(0:max(a$rPIP), n = 30)
par(mfrow=c(2,1))
h1 <- hist(a$rPIP, breaks = 100, xlab = "PIP", main = "PIP distribution-all", col = "grey"); grid()
h2 <- hist(a.c$rPIP, breaks = h1$breaks, xlab = "PIP", main = "PIP distribution-causal", col = "salmon");grid()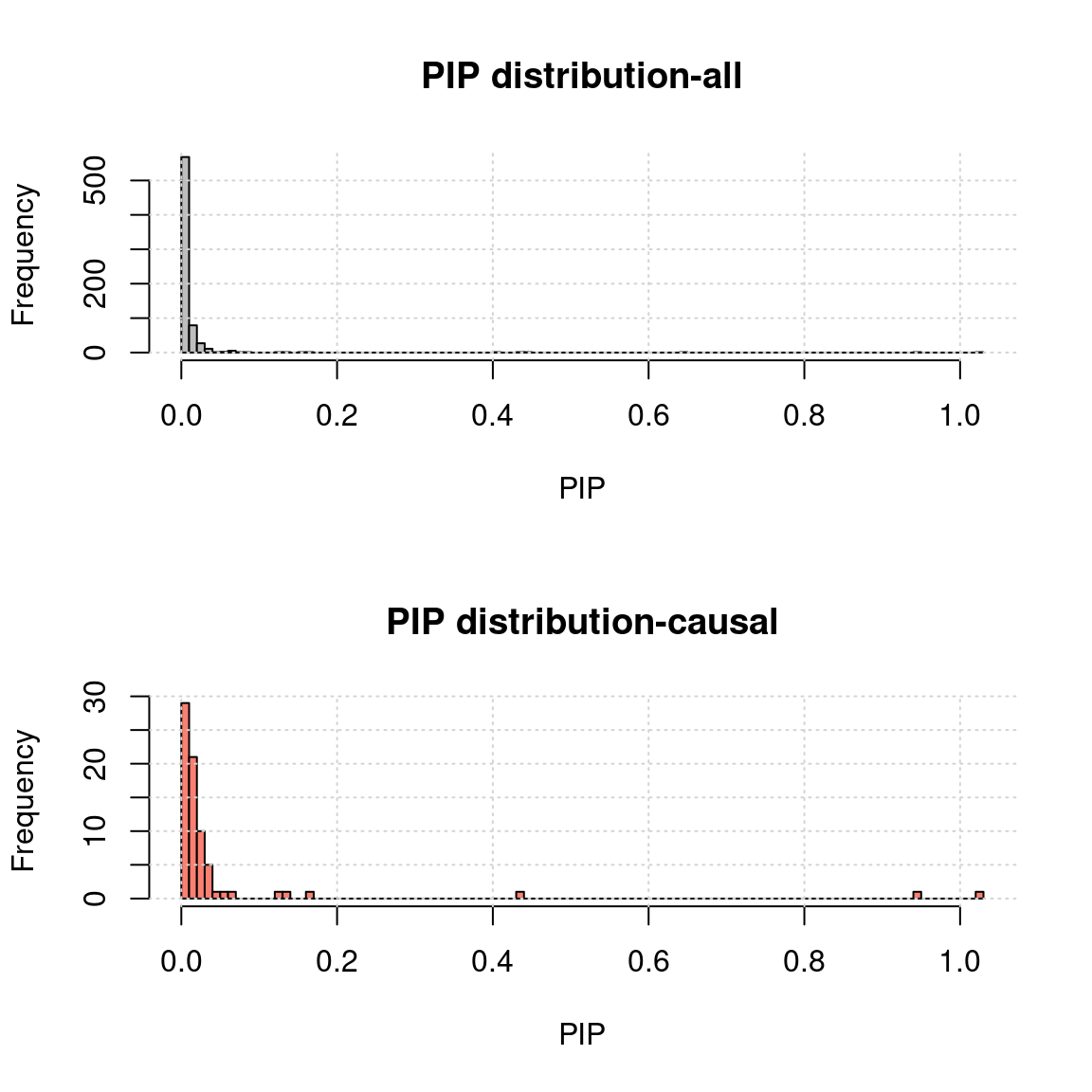
- MR.ash2s results for the same simulated data
rpipf <- "~/causalTWAS/simulations/simulation_ashtest_20200721/20200721-1-1-mr.ash2s.lassoes-es.rPIP.txt"
a <- read.table(rpipf, header = T)
a.c <- a[a$nCausal > 0 ,]
ax <- pretty(0:max(a$rPIP), n = 30) # Make a neat vector for the breakpoints
par(mfrow=c(2,1))
h1 <- hist(a$rPIP, breaks = 100, xlab = "PIP", main = "PIP distribution-all", col = "grey"); grid()
h2 <- hist(a.c$rPIP, breaks = h1$breaks, xlab = "PIP", main = "PIP distribution-causal", col = "salmon");grid()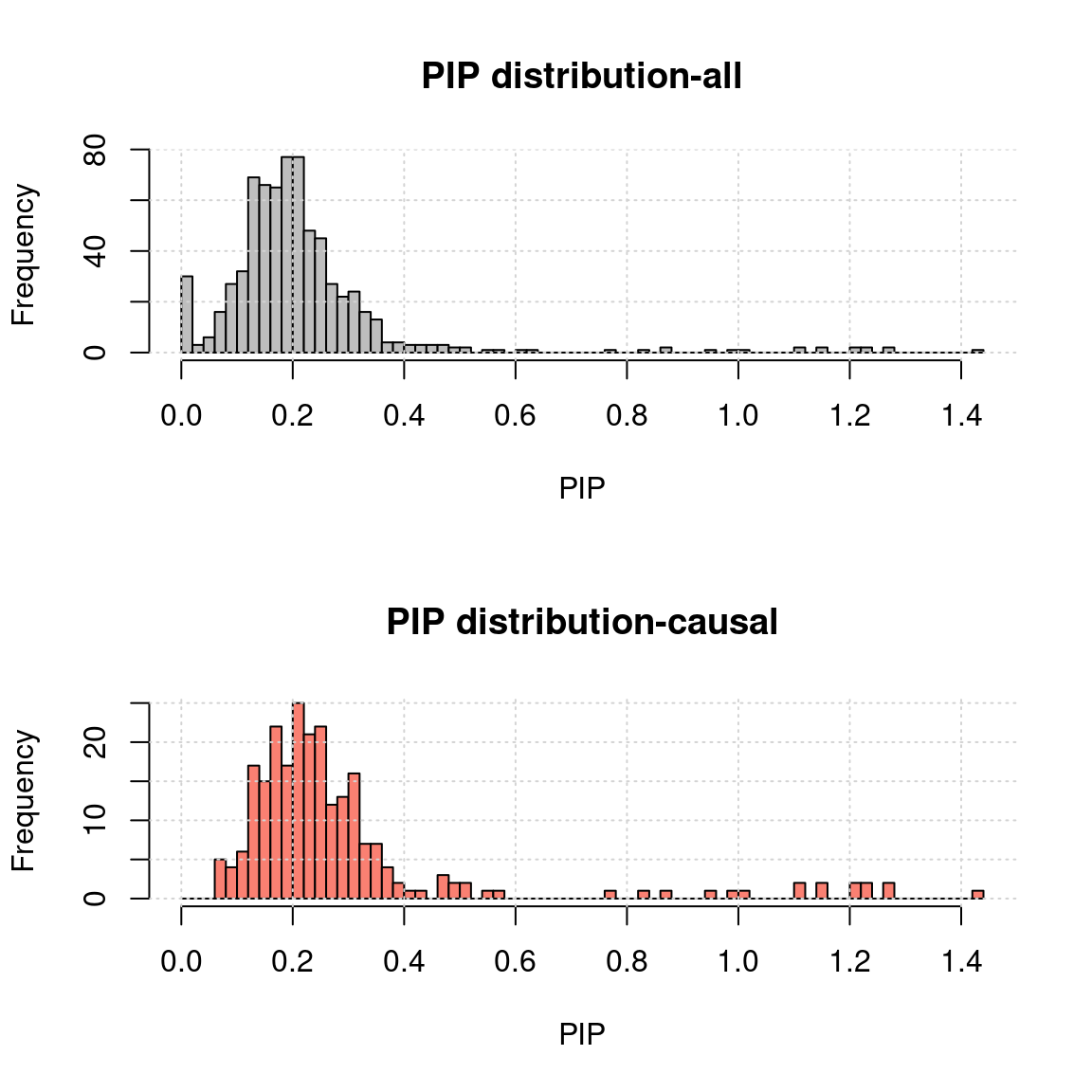
susieI genes
Regions: We use regional PIP of both SNP and gene from mr.ash2s > 0.3 , and also require at least one causal gene or SNP is present, also, at least one gene.
One example
Prior change for each iteration:
load(paste0(outputdir, tag, ".susieIres.Rd"))
df <- cbind(prior.gene_rec, prior.SNP_rec)
rownames(df) <- paste("Iteration", 1:Niter)
colnames(df) <- c("Prior.gene", "Prior.SNP")
df Prior.gene Prior.SNP
Iteration 1 0.00853819 0.006018864
Iteration 2 0.01049847 0.005956966
Iteration 3 0.01202068 0.005908900
Iteration 4 0.01318421 0.005872160
Iteration 5 0.01405232 0.005844749
Iteration 6 0.01469379 0.005824494
Iteration 7 0.01516465 0.005809626
Iteration 8 0.01550865 0.005798763
Iteration 9 0.01575915 0.005790854
Iteration 10 0.01594113 0.005785108
Iteration 11 0.01607310 0.005780940
Iteration 12 0.01616870 0.005777922
Iteration 13 0.01623788 0.005775737
Iteration 14 0.01628792 0.005774157
Iteration 15 0.01632410 0.005773015
Iteration 16 0.01635024 0.005772189
Iteration 17 0.01636913 0.005771593
Iteration 18 0.01638277 0.005771162
Iteration 19 0.01639263 0.005770851
Iteration 20 0.01639975 0.005770626
Iteration 21 0.01640489 0.005770464
Iteration 22 0.01640860 0.005770346
Iteration 23 0.01641129 0.005770262
Iteration 24 0.01641322 0.005770201
Iteration 25 0.01641462 0.005770157
Iteration 26 0.01641563 0.005770125
Iteration 27 0.01641636 0.005770102
Iteration 28 0.01641688 0.005770085
Iteration 29 0.01641726 0.005770073
Iteration 30 0.01641754 0.005770064Results for each region:
regionlist <- readRDS(paste0(outputdir, tag, "_regionlist.rds"))
ng <- unlist(lapply(lapply(regionlist, '[[', "anno"), function(x) dim(x[x$type == "gene", , drop = F ])[1]))
ns <- unlist(lapply(lapply(regionlist, '[[', "anno"), function(x) dim(x[x$type == "SNP", , drop = F ])[1]))
ng.c <- unlist(lapply(lapply(regionlist, '[[', "anno"), function(x) dim(x[x$type == "gene" & x$ifcausal == 1, , drop = F])[1]))
ns.c <- unlist(lapply(lapply(regionlist, '[[', "anno"), function(x) dim(x[x$type == "SNP" & x$ifcausal == 1, , drop = F])[1]))
ngdf <- data.frame("ngene" = ng, "nsnp" = ns, "ncausalgene" = ng.c, "ncausalSNP" = ns.c)
cat("causal gene fraction:", sum(ngdf$ncausalgene)/sum(ngdf$ngene) )causal gene fraction: 0.06355932cat("causal snp fraction:", sum(ngdf$ncausalSNP)/sum(ngdf$ns))causal snp fraction: 0.006288467for (i in seq(1, Niter, 10)){
resf <- paste0(outputdir, tag, ".", i, ".susieIres.Rd")
load(resf)
ngdf[, paste("Prior.gene", i)] <- unlist(lapply(gene.rpiplist, "mean"))
ngdf[, paste("Prior.SNP", i)] <- unlist(lapply(snp.rpiplist, "mean"))
}
ngdf ngene nsnp ncausalgene ncausalSNP Prior.gene 1 Prior.SNP 1
1 1 177 1 2 1.121460e-03 0.0056433816
2 1 131 0 1 1.399491e-02 0.0075267564
3 7 116 1 0 1.149406e-02 0.0079270825
4 4 112 0 2 1.446824e-03 0.0088768991
5 7 99 0 1 2.187797e-03 0.0099463174
6 10 154 1 1 8.959135e-02 0.0006758864
7 4 89 0 1 3.618684e-04 0.0112196913
8 1 89 0 1 1.064940e-04 0.0112347585
10 6 145 1 0 1.057093e-02 0.0064591341
11 11 155 2 1 3.212361e-03 0.0062236389
12 2 173 0 2 6.221626e-06 0.0057802749
13 12 166 2 0 6.603292e-03 0.0055467500
14 7 154 0 2 3.895577e-04 0.0064757993
18 2 97 1 0 3.556363e-02 0.0095760076
19 2 158 1 0 7.285274e-03 0.0062368953
22 1 209 0 1 4.761905e-03 0.0047619048
24 6 191 0 1 4.803413e-03 0.0050847096
25 4 178 1 0 5.987876e-03 0.0054834185
26 4 108 0 1 1.366526e-07 0.0092592542
27 2 160 0 1 1.455019e-02 0.0060681226
28 4 216 0 1 4.545455e-03 0.0045454545
29 4 100 0 1 2.693962e-03 0.0098922415
30 3 274 0 1 3.491355e-03 0.0036114085
31 4 205 1 0 4.784689e-03 0.0047846890
32 7 192 1 0 1.149907e-02 0.0047890964
33 2 125 0 2 1.612216e-02 0.0077420454
34 9 133 0 1 7.297093e-08 0.0075187921
35 8 217 0 1 4.444444e-03 0.0044444444
36 6 236 0 1 9.325782e-06 0.0042370510
37 11 181 0 1 5.208333e-03 0.0052083333
39 1 90 0 1 7.830050e-05 0.0111102411
43 1 139 0 2 2.392592e-03 0.0071770317
44 3 159 0 1 1.331606e-03 0.0062641835
46 4 137 0 1 6.858883e-12 0.0072992701
47 4 182 1 0 7.264653e-03 0.0053348428
53 5 169 0 3 8.244458e-04 0.0058927679
54 3 119 0 1 8.196721e-03 0.0081967213
55 6 177 0 1 1.818031e-03 0.0055880894
56 2 138 0 1 5.244884e-03 0.0071703640
57 10 231 0 2 4.586726e-03 0.0041304448
58 5 215 0 1 4.545455e-03 0.0045454545
59 14 163 0 1 7.228010e-03 0.0055141587
60 4 161 0 1 6.060606e-03 0.0060606061
61 4 192 0 1 1.626941e-03 0.0051744387
62 3 123 0 2 7.342575e-04 0.0081121726
63 3 193 0 1 3.341177e-03 0.0051294118
64 12 146 1 0 7.098813e-03 0.0062658510
Prior.gene 11 Prior.SNP 11 Prior.gene 21 Prior.SNP 21
1 3.084535e-03 0.0056322908 3.180950e-03 0.0056317460
2 3.764638e-02 0.0073462109 3.878124e-02 0.0073375478
3 2.775258e-02 0.0069459650 2.844939e-02 0.0069039159
4 3.963215e-03 0.0087870280 4.086279e-03 0.0087826329
5 5.881132e-03 0.0096851725 6.058213e-03 0.0096726516
6 9.596150e-02 0.0002622402 9.607958e-02 0.0002545729
7 9.954318e-04 0.0111912165 1.026553e-03 0.0111898178
8 2.934224e-04 0.0112326582 3.026202e-04 0.0112325548
10 2.621497e-02 0.0058117943 2.690438e-02 0.0057832670
11 8.483562e-03 0.0058495536 8.731592e-03 0.0058319515
12 1.714378e-05 0.0057801486 1.768125e-05 0.0057801424
13 1.609830e-02 0.0048603637 1.650836e-02 0.0048307212
14 1.068784e-03 0.0064449254 1.102056e-03 0.0064434130
18 8.738874e-02 0.0085074486 8.964968e-02 0.0084608313
19 1.959184e-02 0.0060811160 2.018215e-02 0.0060736437
22 1.301286e-02 0.0047224265 1.341536e-02 0.0047205007
24 1.327761e-02 0.0048185044 1.365976e-02 0.0048064996
25 1.583544e-02 0.0052621248 1.629960e-02 0.0052516943
26 3.765524e-07 0.0092592453 3.883578e-07 0.0092592449
27 3.826661e-02 0.0057716673 3.937740e-02 0.0057577825
28 1.213778e-02 0.0044048559 1.249929e-02 0.0043981612
29 7.306144e-03 0.0097077542 7.529344e-03 0.0096988262
30 9.449546e-03 0.0035461729 9.737281e-03 0.0035430225
31 1.275585e-02 0.0046291542 1.313475e-02 0.0046217610
32 2.776620e-02 0.0041960238 2.846341e-02 0.0041706049
33 4.210671e-02 0.0073262926 4.331543e-02 0.0073069532
34 2.010746e-07 0.0075187834 2.073785e-07 0.0075187830
35 1.208195e-02 0.0041628772 1.242524e-02 0.0041502217
36 2.569632e-05 0.0042366348 2.650187e-05 0.0042366144
37 1.304024e-02 0.0047323612 1.338886e-02 0.0047111745
39 2.157407e-04 0.0111087140 2.225034e-04 0.0111086389
43 6.573432e-03 0.0071469537 6.778533e-03 0.0071454782
44 3.648856e-03 0.0062204618 3.762221e-03 0.0062183229
46 1.889997e-11 0.0072992701 1.949251e-11 0.0072992701
47 1.934185e-02 0.0050694100 1.991257e-02 0.0050568666
53 2.260495e-03 0.0058502812 2.330793e-03 0.0058482014
54 2.165175e-02 0.0078575188 2.228518e-02 0.0078415500
55 4.944899e-03 0.0054820938 5.096684e-03 0.0054769486
56 1.423868e-02 0.0070400191 1.467438e-02 0.0070337046
57 1.170211e-02 0.0038224195 1.202510e-02 0.0038084371
58 1.204464e-02 0.0043710548 1.239885e-02 0.0043628175
59 1.694286e-02 0.0046797543 1.734628e-02 0.0046451045
60 1.601855e-02 0.0058132037 1.648763e-02 0.0058015495
61 4.465785e-03 0.0051152961 4.604931e-03 0.0051123973
62 2.017586e-03 0.0080808719 2.080552e-03 0.0080793361
63 9.125601e-03 0.0050394984 9.407651e-03 0.0050351142
64 1.701870e-02 0.0054505177 1.744068e-02 0.0054158341more examples
Each row represents one simulation run. Runs labeled 1-x have the same simulation parameters, as you can tell from the true parameters. (and so are runs labled 2-x and 3-x)
show_param <- function(tags){
mr.ashfs <- paste0(mrashdir, "20200721-", tags, "-mr.ash2s.lassoes-es.param.txt" )
param <- do.call(rbind, lapply(mr.ashfs, function(x) t(read.table(x))[2:1,]))
pars <- cbind(param[seq(1,nrow(param), 2), 1], param[seq(2,nrow(param), 2), 1], param[seq(1,nrow(param), 2), 3], param[seq(2,nrow(param), 2), 3])
colnames(pars) <- paste(rep(c("pi1.gene_", "pi1.SNP_"), each = 2), c("truth", "mr.ash2"), sep = "")
susieIfs <- paste0(outputdir, "20200813-", tags, ".susieIres.Rd" )
param.s <- do.call(rbind, lapply(susieIfs, function(x) {load(x); c(tail(prior.gene_rec, 1), tail(prior.SNP_rec,1))}))
susieIfs <- paste0(outputdir, "20200813-", tags, ".1.txt")
param.s.truth <- do.call(rbind, lapply(susieIfs, function(x) {
a <- read.table(x, header = T);
c(nrow(a[a$ifcausal == 1 & a$type == "gene", , drop = F])/ nrow(a[a$type == "gene", , drop = F]),
nrow(a[a$ifcausal == 1 & a$type == "SNP", , drop = F])/ nrow(a[a$type == "SNP", , drop = F]))
}))
pars.s <- cbind(param.s.truth, param.s)[, c(1,3,2,4)]
colnames(pars.s) <- paste(rep(c("pi1.gene_", "pi1.SNP_"), each = 2), c("susietruth", "susieI"), sep = "")
df <- cbind(pars, pars.s)
rownames(df) <- tags
df %>%
kable(format = "html", col.names = colnames(df)) %>%
kable_styling() %>%
kableExtra::scroll_box(width = "100%")
}
tags <- paste(rep(1:3, each = 5), 1:5, sep ="-")
show_param(tags)| pi1.gene_truth | pi1.gene_mr.ash2 | pi1.SNP_truth | pi1.SNP_mr.ash2 | pi1.gene_susietruth | pi1.gene_susieI | pi1.SNP_susietruth | pi1.SNP_susieI | |
|---|---|---|---|---|---|---|---|---|
| 1-1 | 0.0502117 | 0.0031626 | 0.0024981 | 0.0017442 | 0.0635593 | 0.0164175 | 0.0062885 | 0.0057701 |
| 1-2 | 0.0502117 | 0.0023143 | 0.0024981 | 0.0019384 | 0.0898876 | 0.0272836 | 0.0068851 | 0.0055209 |
| 1-3 | 0.0502117 | 0.0237669 | 0.0024981 | 0.0022848 | 0.0961003 | 0.0514446 | 0.0054195 | 0.0050422 |
| 1-4 | 0.0502117 | 0.0159417 | 0.0024981 | 0.0025405 | 0.0989957 | 0.0425986 | 0.0053842 | 0.0053504 |
| 1-5 | 0.0502117 | 0.0588623 | 0.0024981 | 0.0015320 | 0.1041968 | 0.0761682 | 0.0054099 | 0.0042034 |
| 2-1 | 0.0998185 | 0.0105981 | 0.0024981 | 0.0022373 | 0.1428571 | 0.0000000 | 0.0062785 | 0.0062785 |
| 2-2 | 0.0998185 | 0.0220761 | 0.0024981 | 0.0021912 | 0.1366906 | 0.0329071 | 0.0090887 | 0.0052631 |
| 2-3 | 0.0998185 | 0.0352456 | 0.0024981 | 0.0015849 | 0.0930233 | 0.0727722 | 0.0123698 | 0.0057753 |
| 2-4 | 0.0998185 | 0.0190760 | 0.0024981 | 0.0016220 | 0.1379310 | 0.0409841 | 0.0090259 | 0.0058046 |
| 2-5 | 0.0998185 | 0.0111937 | 0.0024981 | 0.0021376 | 0.1881188 | 0.0370117 | 0.0081091 | 0.0063626 |
| 3-1 | 0.0199637 | 0.0132199 | 0.0024981 | 0.0031756 | 0.0782609 | 0.0658229 | 0.0056657 | 0.0042274 |
| 3-2 | 0.0199637 | 0.0187323 | 0.0024981 | 0.0026544 | 0.0858896 | 0.0508288 | 0.0089696 | 0.0045318 |
| 3-3 | 0.0199637 | 0.0093039 | 0.0024981 | 0.0018025 | 0.0666667 | 0.0439207 | 0.0068643 | 0.0056309 |
| 3-4 | 0.0199637 | 0.0252941 | 0.0024981 | 0.0024017 | 0.0833333 | 0.1054679 | 0.0064229 | 0.0030996 |
| 3-5 | 0.0199637 | 0.0128022 | 0.0024981 | 0.0024385 | 0.0495050 | 0.0395866 | 0.0084349 | 0.0056238 |
susieI shuffle genes
one example
Shuffle genes to different regions.
Prior change for each iteration:
tag <- '20200813-1-1-shuffleg'
load(paste0(outputdir, tag, ".susieIres.Rd"))
df <- cbind(prior.gene_rec, prior.SNP_rec)
rownames(df) <- paste("Iteration", 1:nrow(df))
colnames(df) <- c("Prior.gene", "Prior.SNP")
df Prior.gene Prior.SNP
Iteration 1 0.01786469 0.005724369
Iteration 2 0.02870735 0.005382000
Iteration 3 0.03553571 0.005166386
Iteration 4 0.03936470 0.005045482
Iteration 5 0.04142174 0.004980528
Iteration 6 0.04250774 0.004946237
Iteration 7 0.04307662 0.004928274
Iteration 8 0.04337351 0.004918899
Iteration 9 0.04352816 0.004914016
Iteration 10 0.04360865 0.004911474
Iteration 11 0.04365052 0.004910152
Iteration 12 0.04367229 0.004909465
Iteration 13 0.04368361 0.004909107
Iteration 14 0.04368950 0.004908921
Iteration 15 0.04369256 0.004908825
Iteration 16 0.04369415 0.004908775
Iteration 17 0.04369498 0.004908748
Iteration 18 0.04369541 0.004908735
Iteration 19 0.04369563 0.004908728
Iteration 20 0.04369575 0.004908724
Iteration 21 0.04369581 0.004908722
Iteration 22 0.04369584 0.004908721
Iteration 23 0.04369585 0.004908721
Iteration 24 0.04369586 0.004908720
Iteration 25 0.04369587 0.004908720
Iteration 26 0.04369587 0.004908720
Iteration 27 0.04369587 0.004908720
Iteration 28 0.04369587 0.004908720
Iteration 29 0.04369587 0.004908720
Iteration 30 0.04369587 0.004908720
Iteration 31 0.04369587 0.004908720
Iteration 32 0.04369587 0.004908720
Iteration 33 0.04369587 0.004908720
Iteration 34 0.04369587 0.004908720
Iteration 35 0.04369587 0.004908720
Iteration 36 0.04369587 0.004908720
Iteration 37 0.04369587 0.004908720
Iteration 38 0.04369587 0.004908720
Iteration 39 0.04369587 0.004908720
Iteration 40 0.04369587 0.004908720
Iteration 41 0.04369587 0.004908720
Iteration 42 0.04369587 0.004908720
Iteration 43 0.04369587 0.004908720
Iteration 44 0.04369587 0.004908720
Iteration 45 0.04369587 0.004908720
Iteration 46 0.04369587 0.004908720
Iteration 47 0.04369587 0.004908720
Iteration 48 0.04369587 0.004908720
Iteration 49 0.04369587 0.004908720
Iteration 50 0.04369587 0.004908720Results for each region:
regionlist <- readRDS(paste0(outputdir, tag, "_regionlist.rds"))
ng <- unlist(lapply(lapply(regionlist, '[[', "anno"), function(x) dim(x[x$type == "gene", , drop = F ])[1]))
ns <- unlist(lapply(lapply(regionlist, '[[', "anno"), function(x) dim(x[x$type == "SNP", , drop = F ])[1]))
ng.c <- unlist(lapply(lapply(regionlist, '[[', "anno"), function(x) dim(x[x$type == "gene" & x$ifcausal == 1, , drop = F])[1]))
ns.c <- unlist(lapply(lapply(regionlist, '[[', "anno"), function(x) dim(x[x$type == "SNP" & x$ifcausal == 1, , drop = F])[1]))
ngdf <- data.frame("ngene" = ng, "nsnp" = ns, "ncausalgene" = ng.c, "ncausalSNP" = ns.c)
cat("causal gene fraction:", sum(ngdf$ncausalgene)/sum(ngdf$ngene))causal gene fraction: 0.06355932cat("causal snp fraction:", sum(ngdf$ncausalSNP)/sum(ngdf$ns))causal snp fraction: 0.006288467for (i in seq(1, Niter, 10)){
resf <- paste0(outputdir, tag, ".", i, ".susieIres.Rd")
load(resf)
ngdf[, paste("Prior.gene", i)] <- unlist(lapply(gene.rpiplist, "mean"))
ngdf[, paste("Prior.SNP", i)] <- unlist(lapply(snp.rpiplist, "mean"))
}
ngdf ngene nsnp ncausalgene ncausalSNP Prior.gene 1 Prior.SNP 1
1 3 177 0 2 1.717934e-07 5.649715e-03
2 4 131 0 1 2.270815e-03 7.564250e-03
3 7 116 0 0 2.096937e-02 7.355296e-03
4 3 112 0 2 1.389850e-03 8.891343e-03
5 4 99 0 1 1.034140e-04 1.009683e-02
6 6 154 0 1 3.226609e-07 6.493494e-03
7 10 89 0 1 4.790372e-04 1.118213e-02
8 6 89 1 1 1.058470e-03 1.116460e-02
10 7 145 1 0 2.449904e-02 5.713839e-03
11 11 155 0 1 3.414922e-03 6.209264e-03
12 2 173 0 2 1.727517e-05 5.780147e-03
13 4 166 1 0 7.619547e-03 5.840493e-03
14 5 154 0 2 3.616045e-06 6.493389e-03
18 1 97 1 0 6.775795e-01 3.323923e-03
19 1 158 0 0 1.372879e-03 6.320425e-03
22 1 209 0 1 4.761905e-03 4.761905e-03
24 1 191 0 1 1.279136e-02 5.168632e-03
25 4 178 1 0 5.987876e-03 5.483419e-03
26 2 108 0 1 1.309371e-06 9.259235e-03
27 2 160 0 1 2.954673e-01 2.556658e-03
28 6 216 0 1 4.504505e-03 4.504505e-03
29 8 100 0 1 1.052669e-03 9.915786e-03
30 2 274 0 1 2.025748e-03 3.634849e-03
31 2 205 1 0 2.102774e-01 2.826562e-03
32 2 192 1 0 3.028851e-03 5.176783e-03
33 3 125 0 2 4.055900e-05 7.999027e-03
34 4 133 0 1 2.141720e-09 7.518797e-03
35 12 217 2 1 5.544644e-03 4.301679e-03
36 1 236 0 1 2.261070e-05 4.237192e-03
37 3 181 0 1 5.434783e-03 5.434783e-03
39 6 90 0 1 1.641715e-05 1.111002e-02
43 7 139 1 2 6.858337e-03 6.848861e-03
44 1 159 0 1 7.556348e-04 6.284556e-03
46 5 137 0 1 1.552294e-11 7.299270e-03
47 4 182 1 0 4.251523e-03 5.401065e-03
53 11 169 2 3 5.785313e-04 5.879504e-03
54 12 119 1 1 3.079473e-03 8.092826e-03
55 14 177 0 1 4.639140e-03 5.282780e-03
56 4 138 0 1 1.679274e-03 7.197702e-03
57 10 231 1 2 9.999684e-02 1.368013e-07
58 4 215 0 1 4.566210e-03 4.566210e-03
59 4 163 0 1 2.626825e-03 6.070507e-03
60 3 161 0 1 8.139826e-03 6.059506e-03
61 4 192 0 1 6.556847e-03 5.071732e-03
62 9 123 0 2 6.773697e-03 7.634445e-03
63 7 193 0 1 8.651600e-02 2.043461e-03
64 4 146 0 0 4.394473e-03 6.728919e-03
Prior.gene 11 Prior.SNP 11 Prior.gene 21 Prior.SNP 21
1 1.525338e-06 5.649692e-03 1.529241e-06 5.649692e-03
2 1.937611e-02 7.041951e-03 1.942352e-02 7.040503e-03
3 8.865712e-02 3.270691e-03 8.874525e-02 3.265373e-03
4 1.211209e-02 8.604141e-03 1.214243e-02 8.603328e-03
5 9.167296e-04 1.006397e-02 9.190709e-04 1.006388e-02
6 2.864862e-06 6.493395e-03 2.872192e-06 6.493395e-03
7 4.135651e-03 1.077128e-02 4.145901e-03 1.077012e-02
8 9.048265e-03 1.062596e-02 9.070440e-03 1.062446e-02
10 9.462254e-02 2.328567e-03 9.470595e-02 2.324540e-03
11 2.476081e-02 4.694394e-03 2.481072e-02 4.690852e-03
12 1.533590e-04 5.778574e-03 1.537513e-04 5.778569e-03
13 5.530764e-02 4.691382e-03 5.541932e-02 4.688691e-03
14 3.210410e-05 6.492464e-03 3.218624e-05 6.492461e-03
18 9.492147e-01 5.235593e-04 9.493378e-01 5.222905e-04
19 1.213501e-02 6.252310e-03 1.216591e-02 6.252115e-03
22 4.075168e-02 4.589705e-03 4.085169e-02 4.589226e-03
24 1.279402e-01 4.565758e-03 1.282585e-01 4.564092e-03
25 4.473792e-02 4.612631e-03 4.483189e-02 4.610519e-03
26 1.162566e-05 9.259044e-03 1.165541e-05 9.259043e-03
27 4.678708e-01 4.016154e-04 4.679488e-01 4.006403e-04
28 3.323960e-02 3.706308e-03 3.330814e-02 3.704404e-03
29 8.968987e-03 9.282481e-03 8.990889e-03 9.280729e-03
30 1.759843e-02 3.521179e-03 1.764236e-02 3.520859e-03
31 4.389208e-01 5.958942e-04 4.390601e-01 5.945354e-04
32 2.581906e-02 4.939385e-03 2.588213e-02 4.938728e-03
33 3.599408e-04 7.991361e-03 3.608612e-04 7.991339e-03
34 1.901619e-08 7.518796e-03 1.906484e-08 7.518796e-03
35 3.383258e-02 2.737369e-03 3.388516e-02 2.734461e-03
36 2.007378e-04 4.236438e-03 2.012513e-04 4.236435e-03
37 3.815143e-02 4.892518e-03 3.824009e-02 4.891048e-03
39 1.457049e-04 1.110140e-02 1.460775e-04 1.110137e-02
43 4.475378e-02 4.940457e-03 4.483365e-02 4.936435e-03
44 6.680545e-03 6.247292e-03 6.697556e-03 6.247185e-03
46 1.378271e-10 7.299270e-03 1.381798e-10 7.299270e-03
47 3.707958e-02 4.679570e-03 3.717470e-02 4.677479e-03
53 4.986926e-03 5.592567e-03 4.999267e-03 5.591764e-03
54 2.282058e-02 6.102126e-03 2.286797e-02 6.097348e-03
55 2.766829e-02 3.461265e-03 2.771250e-02 3.457768e-03
56 1.456335e-02 6.824251e-03 1.459966e-02 6.823198e-03
57 9.999964e-02 1.540769e-08 9.999964e-02 1.536836e-08
58 3.544261e-02 3.991765e-03 3.552041e-02 3.990318e-03
59 2.213809e-02 5.591703e-03 2.219149e-02 5.590393e-03
60 6.177075e-02 5.060172e-03 6.190255e-02 5.057716e-03
61 4.834652e-02 4.201114e-03 4.844647e-02 4.199032e-03
62 4.071366e-02 5.151033e-03 4.077977e-02 5.146196e-03
63 1.339318e-01 3.237160e-04 1.339535e-01 3.229313e-04
64 3.813653e-02 5.804479e-03 3.823352e-02 5.801821e-03
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] kableExtra_1.1.0 stringr_1.4.0 plyr_1.8.6
[4] tidyr_0.8.3 plotly_4.9.2.9000 ggplot2_3.3.1
[7] data.table_1.12.7 mr.ash.alpha_0.1-34
loaded via a namespace (and not attached):
[1] tidyselect_1.1.0 purrr_0.3.4 lattice_0.20-38
[4] colorspace_1.3-2 vctrs_0.3.1 generics_0.0.2
[7] htmltools_0.3.6 viridisLite_0.3.0 yaml_2.2.0
[10] rlang_0.4.6 later_0.7.5 pillar_1.4.4
[13] glue_1.4.1 withr_2.1.2 lifecycle_0.2.0
[16] munsell_0.5.0 gtable_0.2.0 workflowr_1.6.2
[19] rvest_0.3.2 htmlwidgets_1.3 evaluate_0.12
[22] knitr_1.20 httpuv_1.4.5 highr_0.7
[25] Rcpp_1.0.4.6 readr_1.3.1 promises_1.0.1
[28] scales_1.0.0 backports_1.1.2 webshot_0.5.1
[31] jsonlite_1.6.1 fs_1.3.1 hms_0.4.2
[34] digest_0.6.25 stringi_1.3.1 dplyr_1.0.0
[37] grid_3.5.1 rprojroot_1.3-2 tools_3.5.1
[40] magrittr_1.5 lazyeval_0.2.1 tibble_3.0.1
[43] crayon_1.3.4 whisker_0.3-2 pkgconfig_2.0.2
[46] ellipsis_0.3.1 Matrix_1.2-15 xml2_1.2.0
[49] rmarkdown_1.10 httr_1.4.1 rstudioapi_0.11
[52] R6_2.3.0 git2r_0.26.1 compiler_3.5.1