Process data for Buenrostro et al (2018) scATAC-seq dataset
Kaixuan Luo
Last updated: 2022-02-24
Checks: 7 0
Knit directory: scATACseq-topics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200729) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0be72fd. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: output/clustering-Cusanovich2018.rds
Untracked: paper/
Unstaged changes:
Modified: analysis/assess_fits_Buenrostro2018_Chen2019pipeline.Rmd
Modified: analysis/clusters_Cusanovich2018_k13.Rmd
Modified: analysis/gene_analysis_Buenrostro2018_Chen2019pipeline.Rmd
Modified: analysis/gene_analysis_Cusanovich2018.Rmd
Modified: analysis/motif_analysis_Buenrostro2018_Chen2019pipeline.Rmd
Modified: analysis/motif_analysis_Cusanovich2018.Rmd
Modified: analysis/plots_Cusanovich2018.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/process_data_Buenrostro2018.Rmd) and HTML (docs/process_data_Buenrostro2018.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0be72fd | kevinlkx | 2022-02-24 | output data processing messages |
| html | 95a1145 | kevinlkx | 2022-02-24 | Build site. |
| html | 478d922 | kevinlkx | 2021-01-06 | Build site. |
| Rmd | 3890d38 | kevinlkx | 2021-01-06 | wflow_publish("analysis/process_data_Buenrostro2018.Rmd") |
| html | 17fc08a | kevinlkx | 2021-01-06 | Build site. |
| Rmd | 0c32e6a | kevinlkx | 2021-01-06 | wflow_publish("analysis/process_data_Buenrostro2018.Rmd") |
| html | 04cb767 | kevinlkx | 2021-01-06 | Build site. |
| Rmd | 97c421d | kevinlkx | 2021-01-06 | put all four data processing options together |
| html | 694fd3b | kevinlkx | 2020-11-19 | Build site. |
| Rmd | 4b7d5fa | kevinlkx | 2020-11-19 | process data with peaks in less than 1% samples filtered out |
| html | d5b411d | kevinlkx | 2020-11-18 | Build site. |
| Rmd | ee608fd | kevinlkx | 2020-11-18 | process counts for aggregated single-cell peaks using chromVAR |
| html | 594fd86 | kevinlkx | 2020-11-10 | Build site. |
| Rmd | 614814b | kevinlkx | 2020-11-10 | process Buenrostro2018 data using Downloaded scATAC-seq processed data from GEO |
| Rmd | c113844 | kevinlkx | 2020-11-10 | wflow_rename("analysis/process_data_Buenrostro2018.Rmd", "analysis/process_data_Buenrostro2018_Chen2019pipeline.Rmd") |
| html | c113844 | kevinlkx | 2020-11-10 | wflow_rename("analysis/process_data_Buenrostro2018.Rmd", "analysis/process_data_Buenrostro2018_Chen2019pipeline.Rmd") |
| html | 1905c58 | kevinlkx | 2020-11-05 | Build site. |
| Rmd | 1da32f8 | kevinlkx | 2020-11-05 | set binarized counts as sparse matrix |
| html | 89b45be | kevinlkx | 2020-11-04 | Build site. |
| Rmd | 857e1e8 | kevinlkx | 2020-11-04 | process data from Buenrostro 2018 paper |
| html | 907fa65 | kevinlkx | 2020-11-04 | Build site. |
| Rmd | 12bf4b3 | kevinlkx | 2020-11-04 | process data from Buenrostro 2018 paper |
About Buenrostro 2018 dataset
Reference: Buenrostro, J. D. et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 173, 1535–1548.e16 (2018).
Data were downloaded from GEO: GSE96772
More details about the samples: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2540299
RCC directory: /project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/
Prepare data for topic modeling
Here we tried four different options to process the scATAC-seq data.
In option 1, we used the processed counts and peaks from Buenrostro et al (2018) paper. However, the ATAC-seq peaks were defined using the bulk ATAC-seq samples. This may not be ideal as this approach may miss cell-type specific peaks that do not pass the peak calling threshold using all the bulk ATAC-seq samples. Thus, we did not use this option in our topic modeling of the dataset.
In option 2-4, we obtained the processed counts using the BAM reads and peaks from aggregated single cell ATAC-seq data. The processed peaks and BAM files were downloaded from scATAC-benchmarking website from Chen et al
Reference: Chen, H. et al. Assessment of computational methods for the analysis of single-cell ATAC-seq data. Genome Biol. 20, 241 (2019).
In option 2, we used the read count processing pipeline from Chen et al (2019), which uses bedtools to count reads in peaks. But it seems they did not consider the paired-end reads from the ATAC-seq data, i.e. they treated the paired-end reads as independent reads. This would bias the actual read counts, but should not affect the binarized counts (which we used in our topic modeling). The count processing pipeline from Chen et al (2019) also filtered peaks by requring peaks to be accessbile in at least 1% samples.
In option 3 and 4, we used chromVAR to count the reads in peaks, which considers the paired-end reads. In option 4, we further filtered the peaks by requring peaks to be accessbile in at least 1% samples. This filtering was also done using chormVAR.
Option 1. Using peaks called from bulk ATAC-seq samples
The Buenrostro et al (2018) paper called ATAC-seq peaks from the bulk ATAC-seq samples.
Using the previously described approach (Corces et al., 2016), we defined a peak list using all bulk hematopoietic data analyzed here, resulting in 491,437 500bp non-overlapping peaks which we use for the remainder of this study.
Downloaded scATAC-seq processed data from GEO: GSE96769
- Peaks called from bulk ATAC-seq samples
GSE96769_PeakFile_20160207.bed.gz - scATAC-seq sparse count matrix
GSE96769_scATACseq_counts.txt.gz
library(Matrix)
library(tools)
library(readr)
library(data.table)Load the fragment counts as a 2,953 x 491,437 sparse matrix.
# The first row has the sample names
file_counts <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/GEO_data/GSE96769_scATACseq_counts.txt.gz"
sample_names <- readLines(file_counts,n = 1)
sample_names <- unlist(strsplit(sample_names,"\t",fixed = TRUE))
sample_names <- unlist(strsplit(sample_names,";",fixed = TRUE))
sample_names <- sample_names[-1]
# Load the fragment counts as sparse matrix.
dat <- fread(file_counts,sep = "\t",skip = 1)
class(dat) <- "data.frame"
names(dat) <- c("i","j","x")
counts <- sparseMatrix(i = dat$i,j = dat$j,x = dat$x)
counts <- t(counts)
rownames(counts) <- sample_names
dim(counts)# [1] 2953 491437peaks (from bulk ATAC-seq)
peaks <- fread("/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/GEO_data/GSE96769_PeakFile_20160207.bed.gz")
peak_names <- paste(peaks$V1,peaks$V2,peaks$V3,sep = "_")
rownames(peaks) <- peak_names
cat(sprintf("Number of peaks: %d\n",nrow(peaks)))
colnames(counts) <- peak_names# Number of peaks: 491437The supplementary Table S1 provides more details about these samples. https://ars.els-cdn.com/content/image/1-s2.0-S009286741830446X-mmc1.xlsx
Select the 2,034 samples (cells) passing quality filtering
Single-cell profiles were of consistent high-quality with 2,034 cells passing stringent quality filtering, yielding a median of 8,268 fragments per cell with 76% of those fragments mapping to peaks, resulting in a median of 6,442 fragments in peaks per cell (figure 1E).
Single-cells were filtered for quality requiring at least 60% of fragments in peaks and requiring greater than 1,000 fragments passing quality filters, quality filters are previously described (Corces et al., 2016) which includes removal of mitochondrial reads and low alignment quality (Q30).
The 2,034 samples (pass filter) were included in the metadata.tsv file on the scATAC-benchmarking website from Chen et al.
samples_filtered <- read.table('/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/input_Chen_2019/metadata.tsv', header = TRUE, stringsAsFactors=FALSE, quote="",row.names=1)
samples_filtered$name <- rownames(samples_filtered)
idx_samples_filtered <- match(samples_filtered$name, sample_names)
sample_names_filtered <- sample_names[idx_samples_filtered]
counts <- counts[idx_samples_filtered,]
samples <- samples_filtered[,c("name", "label")]
dim(counts)# [1] 2034 491437Remove peaks not exist in any of the cells
j <- which(colSums(counts > 0) >= 1)
peaks <- peaks[j,]
counts <- counts[,j]
cat(length(j), "peaks after filtering. \n")# 465536 peaks after filtering.Binarize counts
binarized_counts <- as.matrix((counts > 0) + 0)
binarized_counts <- Matrix(binarized_counts, sparse = TRUE)
dim(binarized_counts)# [1] 2034 465536Save processed data: /project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data/
data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data/"
dir.create(data.dir, showWarnings = FALSE, recursive = TRUE)
saveRDS(counts, file.path(data.dir, "counts_Buenrostro_2018.rds"))
saveRDS(binarized_counts, file.path(data.dir, "binarized_counts_Buenrostro_2018.rds"))
save(list = c("samples", "peaks", "counts"),
file = file.path(data.dir, "Buenrostro_2018.RData"))
counts <- binarized_counts
save(list = c("samples", "peaks", "counts"),
file = file.path(data.dir, "Buenrostro_2018_binarized.RData"))data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data/"
load(file.path(data.dir, "Buenrostro_2018.RData"))
cat(sprintf("Loaded %d x %d counts matrix.\n",nrow(counts),ncol(counts)))
cat(sprintf("Number of samples (cells): %d\n",nrow(counts)))
cat(sprintf("Number of peaks: %d\n",ncol(counts)))
cat(sprintf("Proportion of counts that are non-zero: %0.1f%%.\n",
100*mean(counts > 0)))# Loaded 2034 x 465536 counts matrix.
# Number of samples (cells): 2034
# Number of peaks: 465536
# Proportion of counts that are non-zero: 1.5%.Option 2. Using the single-cell ATAC-seq data processing pipeline adapted from Chen et al (2019)
Processed peaks (from aggregated single cell ATAC-seq data) and BAM files were downloaded from the scATAC-benchmarking website (Chen et al 2019) (https://github.com/pinellolab/scATAC-benchmarking/)
Process counts using Chen et al pipeline
library(Matrix)
library(tools)
library(readr)
library(data.table)count scATAC-seq reads in peaks
sbatch ~/projects/scATACseq-topics/scripts/count_reads_peaks_Buenrostro_2018.sbatchcombine counts from all samples
dir_readcount_output <- '/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/count_reads_peaks_output/'
files <- list.files(dir_readcount_output, pattern = "\\.txt$")
sample_names <- sapply(strsplit(files,'\\.'),'[', 1)
cat(length(sample_names), "samples. \n")# 2034 samples.datalist <- lapply(files, function(x)fread(file.path(dir_readcount_output,x))$V4)
counts <- do.call("cbind", datalist)data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data_Chen2019pipeline/"
dir.create(data.dir, showWarnings = FALSE, recursive = TRUE)
# saveRDS(counts, file.path(data.dir, "raw_counts_Buenrostro_2018.rds"))
counts <- readRDS(file.path(data.dir, "raw_counts_Buenrostro_2018.rds"))Peaks
peaks <- read.csv("/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/input_Chen_2019/combined.sorted.merged.bed",
sep = '\t',header=FALSE,stringsAsFactors=FALSE)
peaknames <- paste(peaks$V1,peaks$V2,peaks$V3,sep = "_")
rownames(peaks) <- peaknames
cat(sprintf("Number of peaks: %d\n",nrow(peaks)))
head(peaknames)
colnames(counts) <- sample_names
rownames(counts) <- peaknames
dim(counts)# Number of peaks: 237450
# [1] "chr1_10413_10625" "chr1_13380_13624" "chr1_16145_16354"
# [4] "chr1_96388_96812" "chr1_115650_115812" "chr1_237625_237888"
# [1] 237450 2034Samples metadata
samples <- read.table('/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/input_Chen_2019/metadata.tsv', header = TRUE, stringsAsFactors=FALSE, quote="",row.names=1)
head(samples)# label
# BM1077-CLP-Frozen-160106-13 CLP
# BM1077-CLP-Frozen-160106-14 CLP
# BM1077-CLP-Frozen-160106-2 CLP
# BM1077-CLP-Frozen-160106-21 CLP
# BM1077-CLP-Frozen-160106-27 CLP
# BM1077-CLP-Frozen-160106-3 CLPFilter peaks (filter out peaks with counts in < 1% samples)
# adapted from Chen et al. Genome Biology 2019 paper https://github.com/pinellolab/scATAC-benchmarking/blob/master/Real_Data/Buenrostro_2018/run_methods/Control/Control_buenrostro2018.ipynb
# filter peaks with counts in at least 1% samples
filter_peaks <- function (datafr,cutoff = 0.01){
binary_mat = as.matrix((datafr > 0) + 0)
binary_mat = Matrix(binary_mat, sparse = TRUE)
num_cells_ncounted = Matrix::rowSums(binary_mat)
ncounts = binary_mat[num_cells_ncounted >= dim(binary_mat)[2]*cutoff,]
ncounts = ncounts[rowSums(ncounts) > 0,]
options(repr.plot.width=4, repr.plot.height=4)
hist(log10(num_cells_ncounted),main="No. of Cells Each Site is Observed In",breaks=50)
abline(v=log10(min(num_cells_ncounted[num_cells_ncounted >= dim(binary_mat)[2]*cutoff])),lwd=2,col="indianred")
# hist(log10(new_counts),main="Number of Sites Each Cell Uses",breaks=50)
peaks_selected = rownames(ncounts)
return(peaks_selected)
}peaks_selected <- filter_peaks(counts)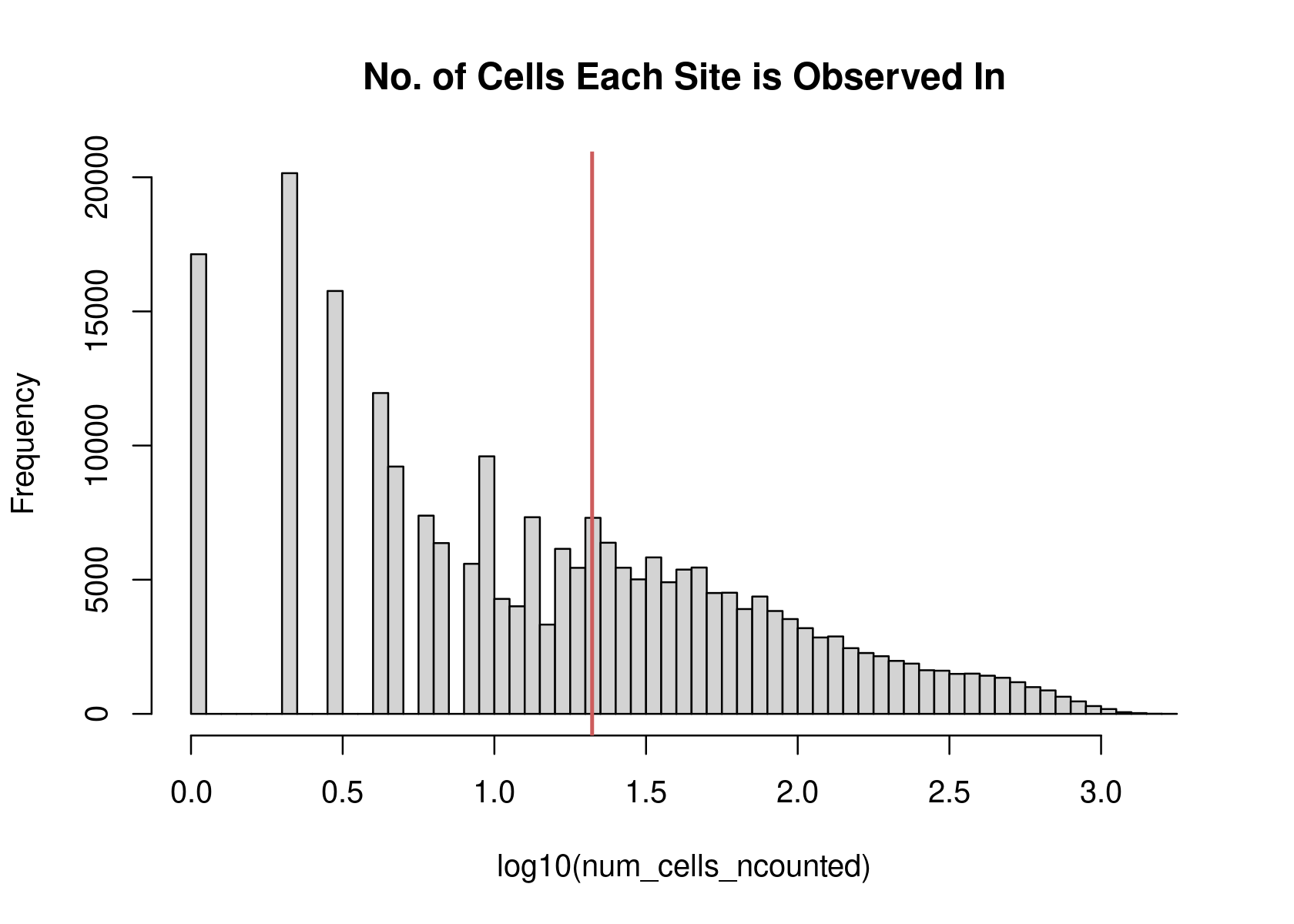
counts <- counts[peaks_selected,]
peaks <- peaks[peaks_selected, ]
counts <- t(counts)
counts <- Matrix(counts, sparse = TRUE)
dim(counts)# [1] 2034 101172Binarize counts
binarized_counts <- as.matrix((counts > 0) + 0)
binarized_counts <- Matrix(binarized_counts, sparse = TRUE)
dim(binarized_counts)# [1] 2034 101172Save processed data: /project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data_Chen2019pipeline/
saveRDS(counts, file.path(data.dir, "counts_Buenrostro_2018.rds"))
saveRDS(binarized_counts, file.path(data.dir, "binarized_counts_Buenrostro_2018.rds"))
save(list = c("samples", "peaks", "counts"),
file = file.path(data.dir, "Buenrostro_2018_counts.RData"))
counts <- binarized_counts
save(list = c("samples", "peaks", "counts"),
file = file.path(data.dir, "Buenrostro_2018_binarized_counts.RData"))data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data/"
load(file.path(data.dir, "Buenrostro_2018_counts.RData"))
cat(sprintf("Loaded %d x %d counts matrix.\n",nrow(counts),ncol(counts)))
cat(sprintf("Number of samples (cells): %d\n",nrow(counts)))
cat(sprintf("Number of peaks: %d\n",ncol(counts)))
cat(sprintf("Proportion of counts that are non-zero: %0.1f%%.\n",
100*mean(counts > 0)))# Loaded 2953 x 491437 counts matrix.
# Number of samples (cells): 2953
# Number of peaks: 491437
# Proportion of counts that are non-zero: 1.3%.Option 3. Using BAM reads and peaks called from aggregated single-cell ATAC-seq data processed by Chen et al (2019)
Processed peaks (from aggregated scATAC-seq) and BAM files were downloaded from the scATAC-benchmarking website (Chen et al 2019) (https://github.com/pinellolab/scATAC-benchmarking/)
Count paired-end scATAC-seq reads in peaks using
chromVAR, following the pipeline
Load chromVAR and related packages
library(chromVAR)
library(motifmatchr)
library(Matrix)
library(SummarizedExperiment)
library(BiocParallel)
library('JASPAR2016')
library(BSgenome.Hsapiens.UCSC.hg19)
register(MulticoreParam(10))
data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data_Chen2019pipeline/chromVAR/"
dir.create(data.dir, showWarnings = FALSE, recursive = TRUE)samples metadata
samples <- read.table('/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/input_Chen_2019/metadata.tsv', header = TRUE, stringsAsFactors=FALSE, quote="",row.names=1)peaks (from aggregated scATAC-seq)
peakfile <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/input_Chen_2019/combined.sorted.merged.bed"
peaks <- getPeaks(peakfile, sort_peaks = TRUE)# Warning in getPeaks(peakfile, sort_peaks = TRUE): Peaks are not equal width!Use
# resize(peaks, width = x, fix = "center") to make peaks equal in size, where x is
# the desired size of the peaks)# Peaks sortedpeaks <- resize(peaks, width = 500, fix = "center")
cat(length(peaks), "peaks \n")# 237450 peaksBAM files of scATAC-seq reads
dir_bamfiles <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/input_Chen_2019/sc-bams_nodup/"
bamfiles <- list.files(path = dir_bamfiles, pattern = "\\.bam$")
cellnames <- sapply(strsplit(bamfiles,'.',fixed = TRUE), "[[", 1)
sum(cellnames == rownames(samples))# [1] 2034count scATAC-seq paired-end fragments in the peaks using chromVAR
fragment_counts <- getCounts(file.path(dir_bamfiles,bamfile),
peaks,
paired = TRUE,
by_rg = TRUE,
format = "bam",
colData = data.frame(celltype = cellnames))
saveRDS(fragment_counts, file.path(data.dir, "fragment_counts_scPeaks_chromVAR_Buenrostro_2018.rds"))fragment_counts <- readRDS(file.path(data.dir, "fragment_counts_scPeaks_chromVAR_Buenrostro_2018.rds"))
counts <- assay(fragment_counts)
counts <- t(counts)
peaks <- as.data.frame(peaks)[,1:3]
colnames(peaks) <- c("chr", "start", "end")
peak_names <- paste(peaks$chr, peaks$start, peaks$end, sep = "_")
colnames(counts) <- peak_namesFilter peaks (accessible in at least 1 sample) using filterPeaks in chromVAR
idx_peaks_filtered <- filterPeaks(fragment_counts, min_fragments_per_peak = 1, non_overlapping = TRUE, ix_return = TRUE)
peaks <- peaks[idx_peaks_filtered,]
counts <- counts[,idx_peaks_filtered]
cat(length(idx_peaks_filtered), "peaks after filtering. \n")# 228965 peaks after filtering.Binarize counts
binarized_counts <- as.matrix((counts > 0) + 0)
binarized_counts <- Matrix(binarized_counts, sparse = TRUE)
dim(binarized_counts)# [1] 2034 228965Save processed data: /project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data_Chen2019pipeline/chromVAR/
saveRDS(counts, file.path(data.dir, "counts_scPeaks_Buenrostro_2018.rds"))
saveRDS(binarized_counts, file.path(data.dir, "binarized_counts_scPeaks_Buenrostro_2018.rds"))
save(list = c("samples", "peaks", "counts"),
file = file.path(data.dir, "Buenrostro_2018_scPeaks.RData"))
counts <- binarized_counts
save(list = c("samples", "peaks", "counts"),
file = file.path(data.dir, "Buenrostro_2018_binarized_scPeaks.RData"))data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data_Chen2019pipeline/chromVAR/"
load(file.path(data.dir, "Buenrostro_2018_binarized_scPeaks.RData"))
cat(sprintf("Loaded %d x %d counts matrix.\n",nrow(counts),ncol(counts)))
cat(sprintf("Number of samples (cells): %d\n",nrow(counts)))
cat(sprintf("Number of peaks: %d\n",ncol(counts)))
cat(sprintf("Proportion of counts that are non-zero: %0.1f%%.\n",
100*mean(counts > 0)))# Loaded 2034 x 228965 counts matrix.
# Number of samples (cells): 2034
# Number of peaks: 228965
# Proportion of counts that are non-zero: 2.6%.Option 4. Using BAM reads and peaks called from aggregated single-cell ATAC-seq data processed by Chen et al (2019) and filter peaks that are accessible in less than 1% samples
Processed peaks (from aggregated scATAC-seq) and BAM files were downloaded from the scATAC-benchmarking website (Chen et al 2019) (https://github.com/pinellolab/scATAC-benchmarking/)
Count paired-end scATAC-seq reads in peaks using
chromVAR, following the pipeline
Load chromVAR and related packages
library(chromVAR)
library(motifmatchr)
library(Matrix)
library(SummarizedExperiment)
library(BiocParallel)
library('JASPAR2016')
library(BSgenome.Hsapiens.UCSC.hg19)
register(MulticoreParam(10))
data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data_Chen2019pipeline/chromVAR/"
dir.create(data.dir, showWarnings = FALSE, recursive = TRUE)samples metadata
samples <- read.table('/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/input_Chen_2019/metadata.tsv', header = TRUE, stringsAsFactors=FALSE, quote="",row.names=1)peaks (from aggregated scATAC-seq)
peakfile <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/input_Chen_2019/combined.sorted.merged.bed"
peaks <- getPeaks(peakfile, sort_peaks = TRUE)# Warning in getPeaks(peakfile, sort_peaks = TRUE): Peaks are not equal width!Use
# resize(peaks, width = x, fix = "center") to make peaks equal in size, where x is
# the desired size of the peaks)# Peaks sortedpeaks <- resize(peaks, width = 500, fix = "center")
cat(length(peaks), "peaks \n")# 237450 peaksBAM reads
dir_bamfiles <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/data/input_Chen_2019/sc-bams_nodup/"
bamfiles <- list.files(path = dir_bamfiles, pattern = "\\.bam$")
cellnames <- sapply(strsplit(bamfiles,'.',fixed = TRUE), "[[", 1)
sum(cellnames == rownames(samples))# [1] 2034count scATAC-seq paired-end fragments in the peaks
fragment_counts <- getCounts(file.path(dir_bamfiles,bamfile),
peaks,
paired = TRUE,
by_rg = TRUE,
format = "bam",
colData = data.frame(celltype = cellnames))
saveRDS(fragment_counts, file.path(data.dir, "fragment_counts_scPeaks_chromVAR_Buenrostro_2018.rds"))fragment_counts <- readRDS(file.path(data.dir, "fragment_counts_scPeaks_chromVAR_Buenrostro_2018.rds"))
fragment_counts# class: RangedSummarizedExperiment
# dim: 237450 2034
# metadata(0):
# assays(1): counts
# rownames: NULL
# rowData names(0):
# colnames(2034): BM1077-CLP-Frozen-160106-13 BM1077-CLP-Frozen-160106-14
# ... singles-PB1022-mono-160128-95 singles-PB1022-mono-160128-96
# colData names(2): celltype depthcounts <- assay(fragment_counts)
counts <- t(counts)
peaks <- as.data.frame(peaks)[,1:3]
colnames(peaks) <- c("chr", "start", "end")
peak_names <- paste(peaks$chr, peaks$start, peaks$end, sep = "_")
colnames(counts) <- peak_namesFilter peaks (filter out peaks with fragments in < 1% samples)
fragment_binarized_counts <- fragment_counts
assay(fragment_binarized_counts) <- Matrix(as.matrix((assay(fragment_binarized_counts) > 0) + 0), sparse = TRUE)
idx_peaks_filtered <- filterPeaks(fragment_binarized_counts, min_fragments_per_peak = floor(nrow(samples)*0.01), non_overlapping = TRUE, ix_return = TRUE)
cat(length(idx_peaks_filtered), "peaks selected after filtering. \n")
peaks <- peaks[idx_peaks_filtered,]
counts <- counts[,idx_peaks_filtered]# 104502 peaks selected after filtering.Binarize counts
binarized_counts <- as.matrix((counts > 0) + 0)
binarized_counts <- Matrix(binarized_counts, sparse = TRUE)
dim(binarized_counts)# [1] 2034 104502Save processed data: /project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data_Chen2019pipeline/chromVAR/
save(list = c("samples", "peaks", "counts"),
file = file.path(data.dir, "Buenrostro_2018_scPeaks_filtered.RData"))
counts <- binarized_counts
save(list = c("samples", "peaks", "counts"),
file = file.path(data.dir, "Buenrostro_2018_binarized_scPeaks_filtered.RData"))data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data_Chen2019pipeline/chromVAR/"
load(file.path(data.dir, "Buenrostro_2018_binarized_scPeaks_filtered.RData"))
cat(sprintf("Loaded %d x %d counts matrix.\n",nrow(counts),ncol(counts)))
cat(sprintf("Number of samples (cells): %d\n",nrow(counts)))
cat(sprintf("Number of peaks: %d\n",ncol(counts)))
cat(sprintf("Proportion of counts that are non-zero: %0.1f%%.\n",
100*mean(counts > 0)))# Loaded 2034 x 104502 counts matrix.
# Number of samples (cells): 2034
# Number of peaks: 104502
# Proportion of counts that are non-zero: 5.3%.
sessionInfo()# R version 4.0.4 (2021-02-15)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Scientific Linux 7.4 (Nitrogen)
#
# Matrix products: default
# BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] parallel stats4 tools stats graphics grDevices utils
# [8] datasets methods base
#
# other attached packages:
# [1] BSgenome.Hsapiens.UCSC.hg19_1.4.3 BSgenome_1.58.0
# [3] rtracklayer_1.50.0 Biostrings_2.58.0
# [5] XVector_0.30.0 JASPAR2016_1.18.0
# [7] BiocParallel_1.24.1 SummarizedExperiment_1.20.0
# [9] Biobase_2.50.0 GenomicRanges_1.42.0
# [11] GenomeInfoDb_1.26.7 IRanges_2.24.1
# [13] S4Vectors_0.28.1 BiocGenerics_0.36.1
# [15] MatrixGenerics_1.2.1 matrixStats_0.61.0
# [17] motifmatchr_1.12.0 chromVAR_1.12.0
# [19] data.table_1.14.2 readr_2.1.1
# [21] Matrix_1.4-0 workflowr_1.7.0
#
# loaded via a namespace (and not attached):
# [1] colorspace_2.0-3 ellipsis_0.3.2
# [3] rprojroot_2.0.2 fs_1.5.2
# [5] rstudioapi_0.13 DT_0.20
# [7] bit64_4.0.5 AnnotationDbi_1.52.0
# [9] fansi_1.0.2 R.methodsS3_1.8.1
# [11] cachem_1.0.6 knitr_1.37
# [13] jsonlite_1.7.3 Rsamtools_2.6.0
# [15] seqLogo_1.56.0 annotate_1.68.0
# [17] GO.db_3.12.1 png_0.1-7
# [19] R.oo_1.24.0 shiny_1.7.1
# [21] compiler_4.0.4 httr_1.4.2
# [23] lazyeval_0.2.2 assertthat_0.2.1
# [25] fastmap_1.1.0 cli_3.2.0
# [27] later_1.3.0 htmltools_0.5.2
# [29] gtable_0.3.0 glue_1.6.2
# [31] TFMPvalue_0.0.8 GenomeInfoDbData_1.2.4
# [33] reshape2_1.4.4 dplyr_1.0.8
# [35] Rcpp_1.0.8 jquerylib_0.1.4
# [37] vctrs_0.3.8 xfun_0.29
# [39] CNEr_1.26.0 stringr_1.4.0
# [41] ps_1.6.0 mime_0.12
# [43] miniUI_0.1.1.1 lifecycle_1.0.1
# [45] poweRlaw_0.70.6 gtools_3.9.2
# [47] XML_3.99-0.8 getPass_0.2-2
# [49] zlibbioc_1.36.0 scales_1.1.1
# [51] vroom_1.5.7 hms_1.1.1
# [53] promises_1.2.0.1 yaml_2.2.2
# [55] memoise_2.0.1 ggplot2_3.3.5
# [57] sass_0.4.0 stringi_1.7.6
# [59] RSQLite_2.2.9 highr_0.9
# [61] caTools_1.18.2 rlang_1.0.1
# [63] pkgconfig_2.0.3 bitops_1.0-7
# [65] pracma_2.3.6 evaluate_0.14
# [67] lattice_0.20-45 purrr_0.3.4
# [69] GenomicAlignments_1.26.0 htmlwidgets_1.5.4
# [71] bit_4.0.4 processx_3.5.2
# [73] tidyselect_1.1.2 plyr_1.8.6
# [75] magrittr_2.0.2 R6_2.5.1
# [77] generics_0.1.2 DelayedArray_0.16.3
# [79] DBI_1.1.2 pillar_1.7.0
# [81] whisker_0.4 KEGGREST_1.30.1
# [83] RCurl_1.98-1.5 tibble_3.1.6
# [85] crayon_1.5.0 utf8_1.2.2
# [87] plotly_4.10.0 tzdb_0.2.0
# [89] rmarkdown_2.11 TFBSTools_1.28.0
# [91] grid_4.0.4 blob_1.2.2
# [93] callr_3.7.0 git2r_0.29.0
# [95] digest_0.6.29 xtable_1.8-4
# [97] tidyr_1.1.4 httpuv_1.6.5
# [99] R.utils_2.11.0 munsell_0.5.0
# [101] DirichletMultinomial_1.32.0 viridisLite_0.4.0
# [103] bslib_0.3.1