Differential celltype composition between groups
Sarah Williams
Last updated: 2024-04-18
Checks: 7 0
Knit directory: spatialsnippets/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231017) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1dc6730. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rproj.user/
Ignored: renv/library/
Ignored: renv/staging/
Unstaged changes:
Modified: analysis/e_DEPseudobulk.Rmd
Modified: renv.lock
Modified: renv/activate.R
Modified: spatialsnippets.Rproj
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/e_CompositionChange.Rmd)
and HTML (docs/e_CompositionChange.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1dc6730 | swbioinf | 2024-04-18 | wflow_publish(c("analysis/index.Rmd", "analysis/e_CompositionChange.Rmd", |
| html | 8423279 | swbioinf | 2024-04-17 | Build site. |
| Rmd | 048c672 | swbioinf | 2024-04-17 | wflow_publish("analysis/e_CompositionChange.Rmd") |
| html | 456dd2f | swbioinf | 2024-04-10 | Build site. |
| Rmd | 8ef9cd9 | swbioinf | 2024-04-10 | wflow_publish("analysis/") |
| html | 30da140 | Sarah Williams | 2024-03-22 | Build site. |
| Rmd | 89c3371 | Sarah Williams | 2024-03-22 | wflow_publish(c("analysis/index_data.Rmd", "analysis/index.Rmd", |
Is there a difference in the celltype composition between individuals with Ulcerative colitis or Crohn’s disease, and Healthy controls?
Load libraries and data object
library(Seurat)
library(speckle)
library(tidyverse)
library(DT)data_dir <- file.path("~/projects/spatialsnippets/datasets/GSE234713_IBDcosmx_GarridoTrigo2023/processed_data")
seurat_file_01_loaded <- file.path(data_dir, "GSE234713_CosMx_IBD_seurat_01_loaded.RDS")so <- readRDS(seurat_file_01_loaded)Looking at the data
There are three individuals per condition (one tissue sample from each individual). With multiple fovs on each physical tissue sample.
sample_table <- select(as_tibble(so@meta.data), condition, individual_code, fov_name) %>%
unique() %>%
group_by(condition, individual_code) %>%
summarise(n_fovs= n(), item = str_c(fov_name, collapse = ", "))
DT::datatable(sample_table)Full worked example
Count how many cells of each type in your data
In this dataset there are two different levels of categorisation. We’d like to look at the celltype proportions at a more granular resolution - but we need to ensure there are enough cells of each type for meaningful statistics.
- Celltype_subset; Some broad groupings of cell types.
- celltype_SingleR2: Much more specific groupings, predicted with the singleR method.
In the ‘Celltype_subset’ column, there are 5 broad categories;
celltype_summary_table <- so@meta.data %>%
group_by(condition, individual_code, fov_name, celltype_subset) %>%
summarise(cells=n(), .groups = 'drop')
DT::datatable(celltype_summary_table)Here T cells are rare, but there are still a decent distribution of them with 10-100+ cells in a FOV. We should be able to see changes in these.
ggplot(celltype_summary_table, aes(x=cells, col=celltype_subset)) +
geom_density() +
geom_rug(alpha=0.2) +
scale_x_log10() +
theme_bw() +
ggtitle("Cells per FOV by celltype")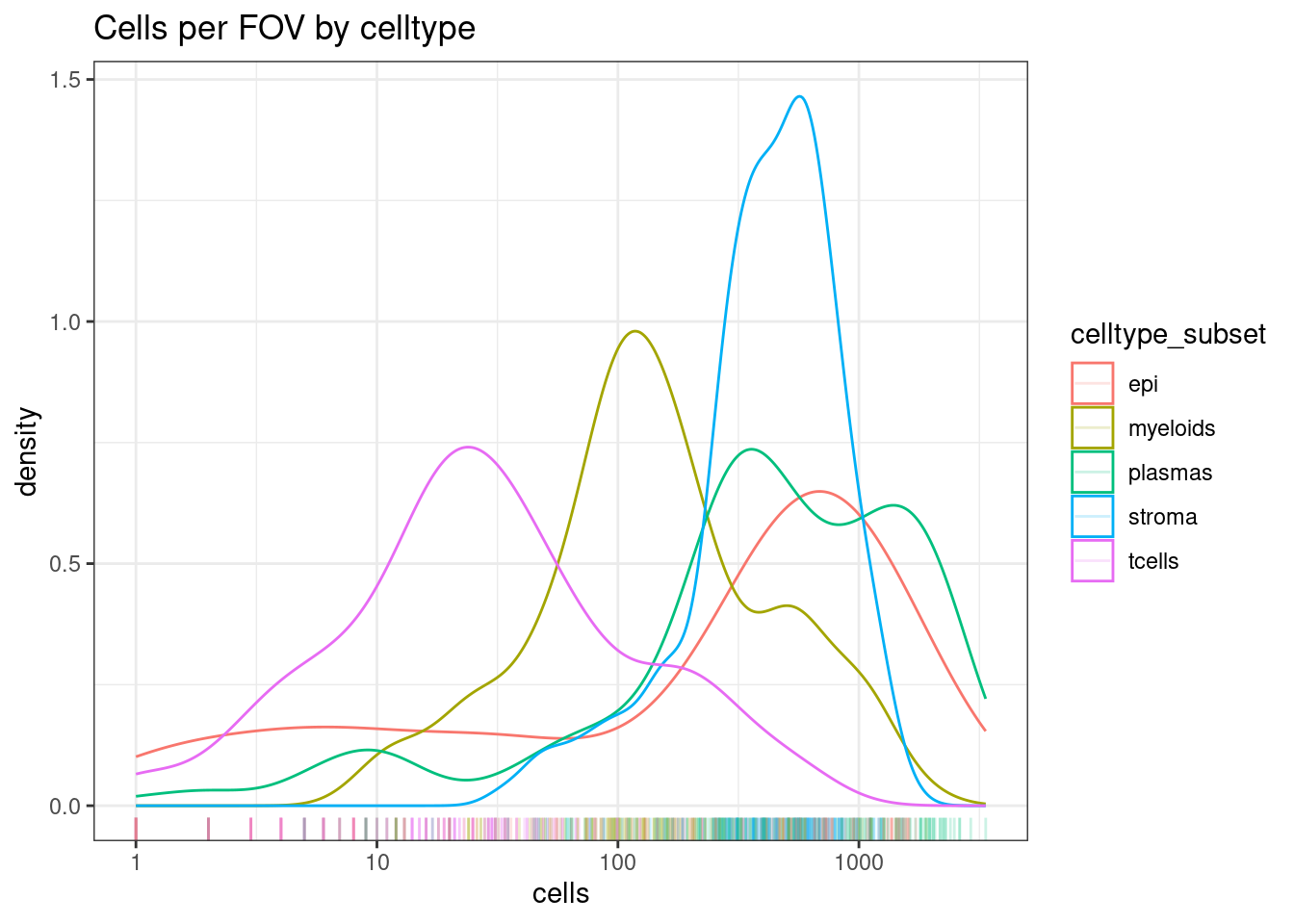
On the other hand, could we use the more fine-grained categoriesation in the ‘celltype_SingleR2’ grouping?
There are many different types here - which is typical if you’re using celltype assignment with a detailed reference.
In this case, there are just too many cell type categories, and we should stick with the broad categorisation.
Approaches to handle this when you want to keep the specific types:
- Drop ‘nonsense’ cell types: When cell typing with a broad refernece, you might get a handful of irrelevant cell types called (e.g. 4 hepatocytes on a non-liver sample).
- Pool subtypes: Some celltypes are simply rare. Rather then dropping them entirely, you can pool transcriptionally similar cells (e.g. T cell subtypes).
The more cell types you have, the more agressive your FDR multiple hypothesis correction will need to be. Its best to remove cell types that can never reach statistical significance.
celltype_summary_table.SingleR <- so@meta.data %>%
group_by(condition, individual_code, fov_name, celltype_SingleR2) %>%
summarise(cells=n(), .groups = 'drop')
DT::datatable(celltype_summary_table.SingleR)ggplot(celltype_summary_table.SingleR, aes(x=cells, col=celltype_SingleR2)) +
geom_density() +
geom_rug(alpha=0.2) +
scale_x_log10() +
theme_bw() +
ggtitle("Cells per FOV by celltype")
Look at your samples.
(add spekcle example)
ggplot(celltype_summary_table, aes(x=fov_name, y=cells, fill=celltype_subset)) +
geom_bar(position="fill", stat="identity") +
theme_bw() +
coord_flip() +
theme(legend.position = "bottom") +
facet_wrap(~condition, ncol=3, scales = 'free_y') +
scale_y_continuous(expand = c(0,0)) 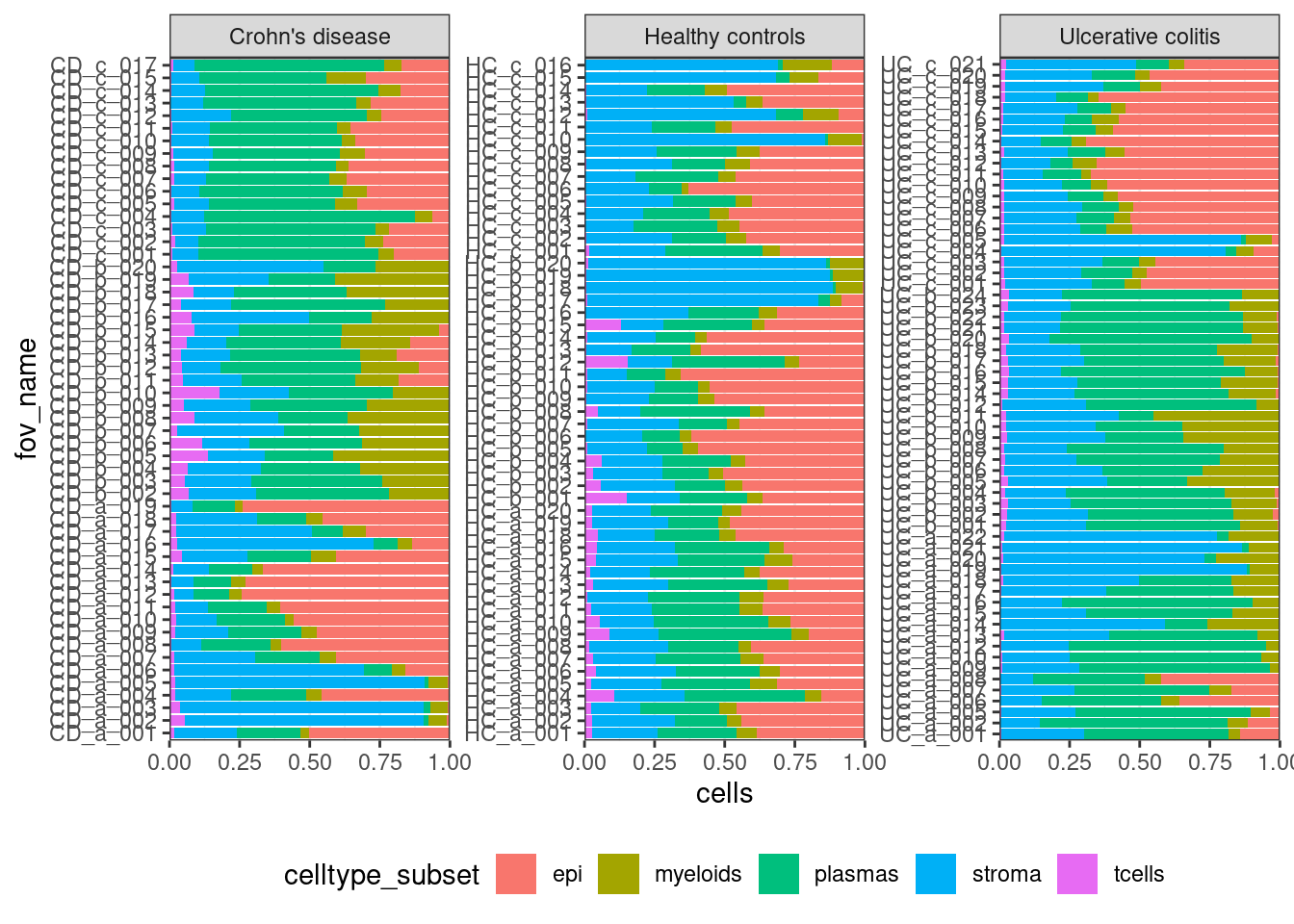
Calculate stats.
results.anova <- propeller(clusters= so$celltype_subset,
sample = so$individual_code,
group = so$condition)
results.anova BaselineProp PropMean.Crohn.s.disease PropMean.Healthy.controls
epi 0.2336112 0.25884773 0.40528423
myeloids 0.1242437 0.12933965 0.06414522
stroma 0.2397435 0.21120914 0.24761081
plasmas 0.3692386 0.36680895 0.25076104
tcells 0.0331629 0.03379454 0.03219870
PropMean.Ulcerative.colitis Fstatistic P.Value FDR
epi 0.21803174 1.2861724 0.3216825 0.7534123
myeloids 0.10678699 0.4468305 0.6528237 0.7534123
stroma 0.27412907 0.4228438 0.6672889 0.7534123
plasmas 0.38515297 0.3440366 0.7176420 0.7534123
tcells 0.01589923 0.2920340 0.7534123 0.7534123# If a column is preferred over rownames
results.anova.table <- rownames_to_column( results.anova, var="celltype_subset")so.UCvsHC <- so[,so$condition %in% c("Healthy controls", "Ulcerative colitis")]
results.pair <- propeller( clusters= so.UCvsHC$celltype_subset,
sample = so.UCvsHC$individual_code,
group = so.UCvsHC$condition)Code snippet
library(speckle)
# seurat object so
results_table <- propeller(clusters = so$cluster,
sample = so$sample,
group = so$condition)Results
Paired
results.pair BaselineProp.clusters BaselineProp.Freq PropMean.Healthy.controls
epi epi 0.27031229 0.40528423
myeloids myeloids 0.09419929 0.06414522
plasmas plasmas 0.35035166 0.25076104
tcells tcells 0.02610896 0.03219870
stroma stroma 0.25902780 0.24761081
PropMean.Ulcerative.colitis PropRatio Tstatistic P.Value FDR
epi 0.21803174 1.8588314 1.5285142 0.1807322 0.6416075
myeloids 0.10678699 0.6006839 -1.2428579 0.2634330 0.6416075
plasmas 0.38515297 0.6510687 -0.7489044 0.4841729 0.6416075
tcells 0.01589923 2.0251739 0.5451820 0.6066415 0.6416075
stroma 0.27412907 0.9032636 -0.4916983 0.6416075 0.6416075- rownames : The tested cell types
- BaselineProp : The overall proportion of the cell type across the dataset. (Overall, not a average of PropMean per group).
- PropMean.X : The proportion of cell type in each group. Here that’s PropMean.Healthy.controls, PropMean.Ulcerative.colitis)
- PropRatio : Ratio of proprotions of first to second group.
- Tstatistic : The T statistic.
- P.Value : P.value
- FDR : A multiple-hypothesis corrected p-value
Anova
results.anova BaselineProp PropMean.Crohn.s.disease PropMean.Healthy.controls
epi 0.2336112 0.25884773 0.40528423
myeloids 0.1242437 0.12933965 0.06414522
stroma 0.2397435 0.21120914 0.24761081
plasmas 0.3692386 0.36680895 0.25076104
tcells 0.0331629 0.03379454 0.03219870
PropMean.Ulcerative.colitis Fstatistic P.Value FDR
epi 0.21803174 1.2861724 0.3216825 0.7534123
myeloids 0.10678699 0.4468305 0.6528237 0.7534123
stroma 0.27412907 0.4228438 0.6672889 0.7534123
plasmas 0.38515297 0.3440366 0.7176420 0.7534123
tcells 0.01589923 0.2920340 0.7534123 0.7534123- rownames : The tested cell types
- BaselineProp : The overall proportion of the cell type across the dataset. (Overall, not a average of PropMean per group).
- PropMean.X : The proportion of cell type in each group. Here that’s PropMean.Chrones.s.disease, PropMean.Healthy.controls, PropMean.Ulcerative.colitis)
- Fstatistic : The Fstatistic calculated.
- P.Value : P.value
- FDR : A multiple-hypothesis corrected p-value
More information
- Speckle Vignette (https://www.bioconductor.org/packages/release/bioc/vignettes/speckle/inst/doc/speckle.html) : Comphrehensive details of how to use different tests for speckle.
- Propeller paper: (https://academic.oup.com/bioinformatics/article/38/20/4720/6675456)
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices datasets utils methods base
other attached packages:
[1] DT_0.33 lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1
[5] dplyr_1.1.4 purrr_1.0.2 readr_2.1.5 tidyr_1.3.1
[9] tibble_3.2.1 ggplot2_3.5.0 tidyverse_2.0.0 speckle_1.2.0
[13] Seurat_5.0.3 SeuratObject_5.0.1 sp_2.1-3 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] RcppAnnoy_0.0.22 splines_4.3.2
[3] later_1.3.2 bitops_1.0-7
[5] polyclip_1.10-6 fastDummies_1.7.3
[7] lifecycle_1.0.4 edgeR_4.0.16
[9] rprojroot_2.0.4 globals_0.16.3
[11] processx_3.8.4 lattice_0.22-6
[13] MASS_7.3-60.0.1 crosstalk_1.2.1
[15] magrittr_2.0.3 limma_3.58.1
[17] plotly_4.10.4 sass_0.4.9
[19] rmarkdown_2.26 jquerylib_0.1.4
[21] yaml_2.3.8 httpuv_1.6.15
[23] sctransform_0.4.1 spam_2.10-0
[25] spatstat.sparse_3.0-3 reticulate_1.35.0
[27] cowplot_1.1.3 pbapply_1.7-2
[29] RColorBrewer_1.1-3 abind_1.4-5
[31] zlibbioc_1.48.2 Rtsne_0.17
[33] GenomicRanges_1.54.1 BiocGenerics_0.48.1
[35] RCurl_1.98-1.14 git2r_0.33.0
[37] GenomeInfoDbData_1.2.11 IRanges_2.36.0
[39] S4Vectors_0.40.2 ggrepel_0.9.5
[41] irlba_2.3.5.1 listenv_0.9.1
[43] spatstat.utils_3.0-4 goftest_1.2-3
[45] RSpectra_0.16-1 spatstat.random_3.2-3
[47] fitdistrplus_1.1-11 parallelly_1.37.1
[49] leiden_0.4.3.1 codetools_0.2-20
[51] DelayedArray_0.28.0 tidyselect_1.2.1
[53] farver_2.1.1 matrixStats_1.2.0
[55] stats4_4.3.2 spatstat.explore_3.2-7
[57] jsonlite_1.8.8 progressr_0.14.0
[59] ggridges_0.5.6 survival_3.5-8
[61] tools_4.3.2 ica_1.0-3
[63] Rcpp_1.0.12 glue_1.7.0
[65] gridExtra_2.3 SparseArray_1.2.4
[67] xfun_0.43 MatrixGenerics_1.14.0
[69] GenomeInfoDb_1.38.8 withr_3.0.0
[71] BiocManager_1.30.22 fastmap_1.1.1
[73] fansi_1.0.6 callr_3.7.6
[75] digest_0.6.35 timechange_0.3.0
[77] R6_2.5.1 mime_0.12
[79] colorspace_2.1-0 scattermore_1.2
[81] tensor_1.5 spatstat.data_3.0-4
[83] utf8_1.2.4 generics_0.1.3
[85] renv_1.0.5 data.table_1.15.4
[87] httr_1.4.7 htmlwidgets_1.6.4
[89] S4Arrays_1.2.1 whisker_0.4.1
[91] uwot_0.1.16 pkgconfig_2.0.3
[93] gtable_0.3.4 lmtest_0.9-40
[95] SingleCellExperiment_1.24.0 XVector_0.42.0
[97] htmltools_0.5.8 dotCall64_1.1-1
[99] scales_1.3.0 Biobase_2.62.0
[101] png_0.1-8 knitr_1.45
[103] rstudioapi_0.16.0 tzdb_0.4.0
[105] reshape2_1.4.4 nlme_3.1-164
[107] cachem_1.0.8 zoo_1.8-12
[109] KernSmooth_2.23-22 parallel_4.3.2
[111] miniUI_0.1.1.1 pillar_1.9.0
[113] grid_4.3.2 vctrs_0.6.5
[115] RANN_2.6.1 promises_1.2.1
[117] xtable_1.8-4 cluster_2.1.6
[119] evaluate_0.23 cli_3.6.2
[121] locfit_1.5-9.9 compiler_4.3.2
[123] rlang_1.1.3 crayon_1.5.2
[125] future.apply_1.11.2 labeling_0.4.3
[127] ps_1.7.6 getPass_0.2-4
[129] plyr_1.8.9 fs_1.6.3
[131] stringi_1.8.3 viridisLite_0.4.2
[133] deldir_2.0-4 munsell_0.5.1
[135] lazyeval_0.2.2 spatstat.geom_3.2-9
[137] Matrix_1.6-5 RcppHNSW_0.6.0
[139] hms_1.1.3 patchwork_1.2.0
[141] future_1.33.2 statmod_1.5.0
[143] shiny_1.8.1.1 highr_0.10
[145] SummarizedExperiment_1.32.0 ROCR_1.0-11
[147] igraph_2.0.3 bslib_0.7.0