Main simulation study
Scenario 1
Bla bla.
library("KODAMA")Loading required package: minervaLoading required package: RtsneLoading required package: umap
Attaching package: 'KODAMA'The following object is masked from 'package:umap':
umap.defaultslibrary("KODAMAextra")Loading required package: parallelLoading required package: doParallelLoading required package: foreachLoading required package: iteratorsLoading required package: e1071x1=runif(100,min=-1,max=0)
x2=runif(100,min=0,max=1)
y1=runif(100)
y2=runif(100)
x=c(x1,x2)
y=c(y1,y2)
xy=cbind(x,y)
labels=rep(c(TRUE,FALSE),each=100)
ss=sample(100,5)
labels[ss]=!labels[ss]
labels=labels
region=rep(1:0,each=100)
data=cbind(rnorm(200,mean=labels,sd=0.1),
rnorm(200,mean=labels,sd=0.1))
color.code=c("#1d79d0aa","#53ca3eaa")
kk <- KODAMA.matrix.parallel(data,spatial=xy,spatial.resolution=0.1,M=100,
FUN="PLS",
landmarks = 100000,
splitting = 100,
f.par.pls = 2,
n.cores=4)socket cluster with 4 nodes on host 'localhost'
================================================================================[1] "Finished parallel computation"
[1] "Calculation of dissimilarity matrix..."
================================================================================vis=RunKODAMAvisualization(kk,method="UMAP")
u=umap(data)$layout
par(mfrow=c(1,3))
plot(x,y,bg=color.code[region+1],pch=21+2*!labels,cex=1+1.5*region,axes=FALSE,main="Position")
axis(1)
axis(2,las=2)
box()
plot(vis,type="n",axes=FALSE,main="KODAMA")
axis(1)
axis(2,las=2)
box()
points(vis[!labels,],bg=color.code[region+1][!labels],pch=21+2,cex=1+1.5*region[!labels])
points(vis[labels,],bg=color.code[region+1][labels],pch=21+2*0,cex=1+1.5*region[labels])
plot(u,type="n",axes=FALSE,xlab="Dimension 1",ylab="Dimension 2",main="UMAP")
axis(1)
axis(2,las=2)
box()
points(u[!labels,],bg=color.code[region+1][!labels],pch=21+2,cex=1+1.5*region[!labels])
points(u[labels,],bg=color.code[region+1][labels],pch=21+2*0,cex=1+1.5*region[labels])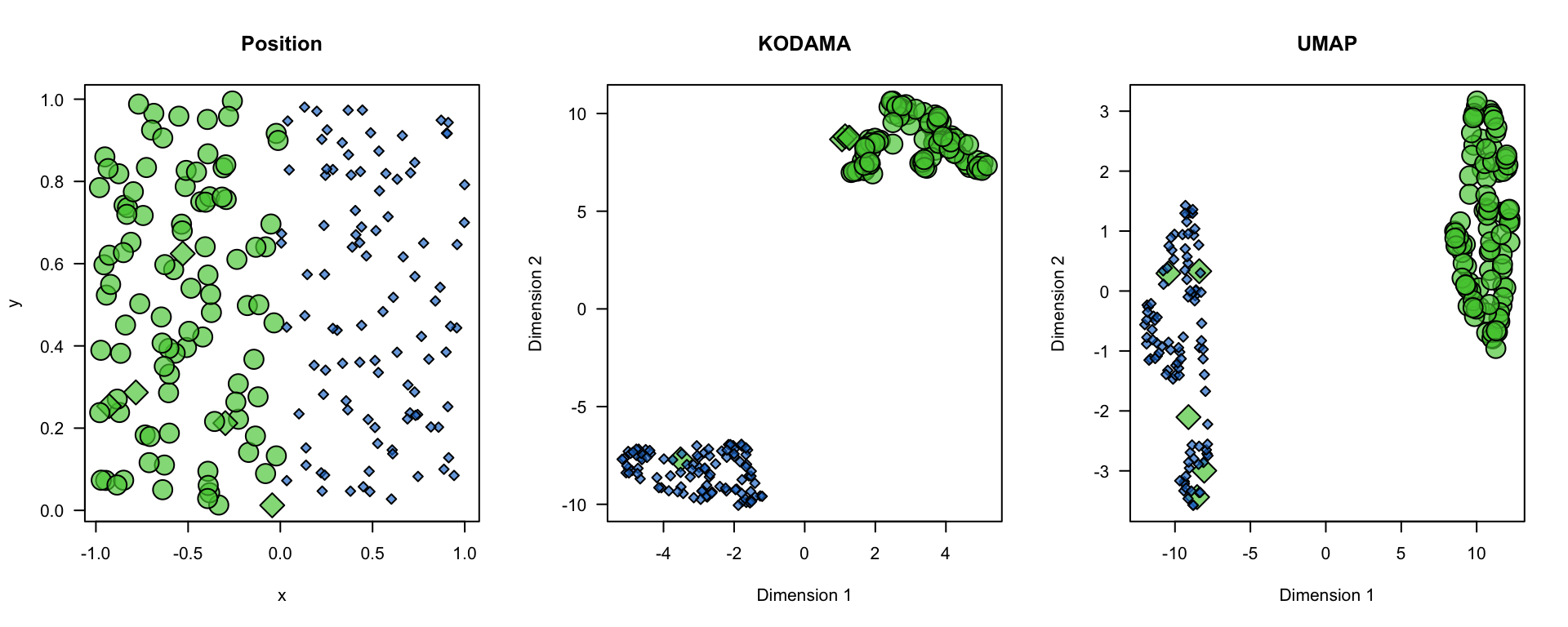
| Version | Author | Date |
|---|---|---|
| 4eae145 | Stefano Cacciatore | 2024-06-18 |
| f1649ac | Stefano Cacciatore | 2024-06-18 |
Scenario 2
Bla bla.
x1=runif(100,min=-1,max=0)
x2=runif(100,min=0,max=1)
y1=runif(100)
y2=runif(100)
x=c(x1,x2)
y=c(y1,y2)
xy=cbind(x,y)
labels=rep(c(TRUE,FALSE),each=100)
region=rep(1:0,each=100)
data=cbind(rnorm(200,mean=1+labels,sd=0.1),
rnorm(200,mean=1+labels,sd=0.1),
rnorm(200,mean=1+labels,sd=0.1),
rnorm(200,mean=1+labels,sd=0.1))
ll=length(data)
ss=sample(ll,ll*0.5)
data[ss]=0
color.code=c("#1d79d0aa","#53ca3eaa")
sel=apply(data,1,function(x) sum(x>0))>2
data=data[sel,]
region=region[sel]
labels=labels[sel]
xy=xy[sel,]
labels=data>0
labels=labels[,1]+
labels[,2]*2+
labels[,3]*4+
labels[,4]*8
labels=as.numeric(as.factor(labels))+1
pca=prcomp(scale(data))$x
kk <- KODAMA.matrix.parallel(pca,spatial=xy,spatial.resolution=0.1,M=100,
FUN="PLS",
landmarks = 100000,
splitting = 100,
f.par.pls = 10,
n.cores=4)The number of components selected for PLS-DA is too high and it will be automatically reduced to 4socket cluster with 4 nodes on host 'localhost'
================================================================================[1] "Finished parallel computation"
[1] "Calculation of dissimilarity matrix..."
================================================================================config=umap.defaults
config$n_neighbors=15
vis=RunKODAMAvisualization(kk,method="UMAP",config=config)
u=umap(pca)$layout
old.par = par(mfrow=c(1,3))
plot(xy,bg=labels,pch=21+2*(region),cex=2+1*region,axes=FALSE,main="Position")
axis(1)
axis(2,las=2)
box()
plot(vis,bg=labels,pch=21+2*(region),cex=1+1.5*region,axes=FALSE,main="KODAMA")
axis(1)
axis(2,las=2)
box()
plot(u,bg=labels,pch=21+2*(region),cex=1+1.5*region,axes=FALSE,main="UMAP")
axis(1)
axis(2,las=2)
box()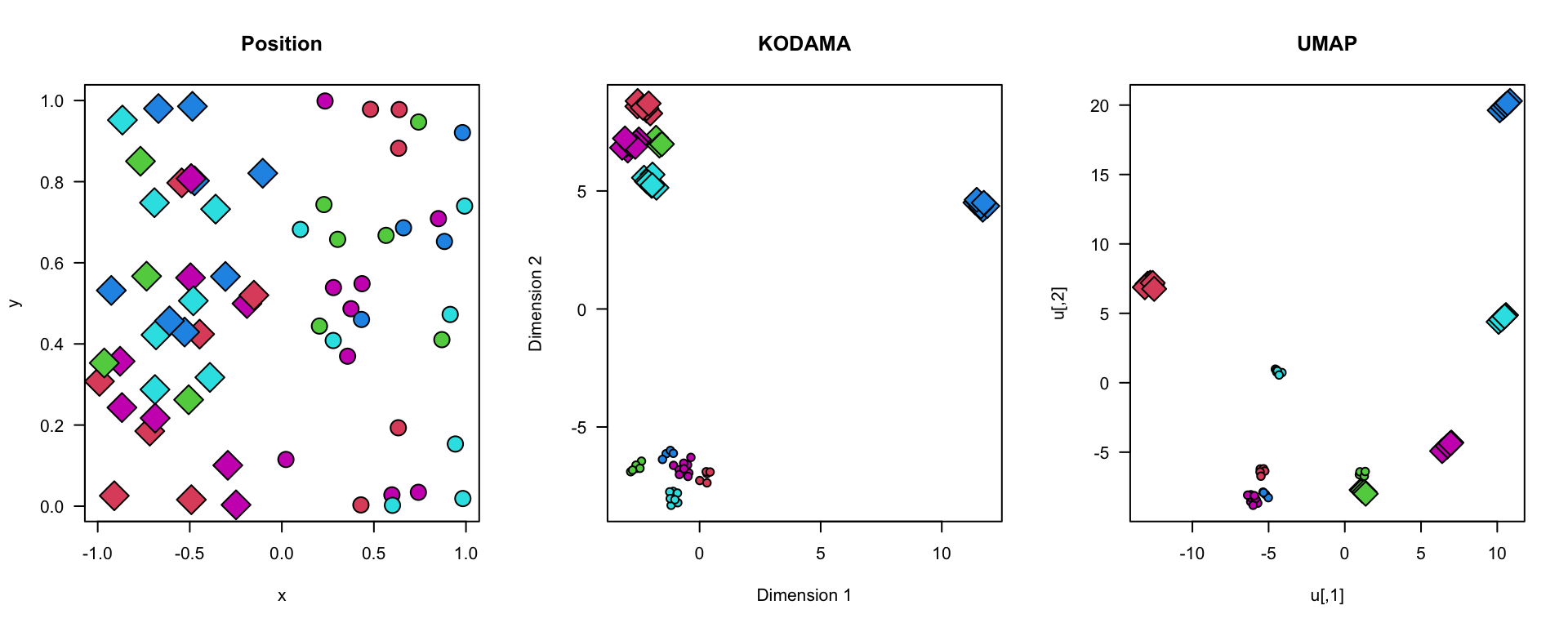
| Version | Author | Date |
|---|---|---|
| fe6339e | Stefano Cacciatore | 2024-06-18 |
| 299901b | Stefano Cacciatore | 2024-06-18 |
| 4d57ea9 | Stefano Cacciatore | 2024-06-18 |
| a94ae9e | Stefano Cacciatore | 2024-06-18 |
| f1649ac | Stefano Cacciatore | 2024-06-18 |
par(old.par)
sessionInfo()R version 4.3.3 (2024-02-29)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS Sonoma 14.5
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/Sao_Paulo
tzcode source: internal
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] KODAMAextra_1.0 e1071_1.7-14 doParallel_1.0.17 iterators_1.0.14
[5] foreach_1.5.2 KODAMA_3.0 umap_0.2.10.0 Rtsne_0.17
[9] minerva_1.5.10 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] sass_0.4.9 utf8_1.2.4 class_7.3-22 stringi_1.8.3
[5] lattice_0.22-6 digest_0.6.35 magrittr_2.0.3 evaluate_0.23
[9] grid_4.3.3 fastmap_1.1.1 rprojroot_2.0.4 jsonlite_1.8.8
[13] Matrix_1.6-5 processx_3.8.4 whisker_0.4.1 RSpectra_0.16-1
[17] doSNOW_1.0.20 ps_1.7.6 promises_1.3.0 httr_1.4.7
[21] fansi_1.0.6 codetools_0.2-20 jquerylib_0.1.4 cli_3.6.2
[25] rlang_1.1.3 cachem_1.0.8 yaml_2.3.8 tools_4.3.3
[29] httpuv_1.6.15 reticulate_1.36.0 vctrs_0.6.5 R6_2.5.1
[33] png_0.1-8 proxy_0.4-27 lifecycle_1.0.4 git2r_0.33.0
[37] stringr_1.5.1 fs_1.6.3 pkgconfig_2.0.3 callr_3.7.6
[41] pillar_1.9.0 bslib_0.7.0 later_1.3.2 glue_1.7.0
[45] Rcpp_1.0.12 highr_0.10 xfun_0.43 tibble_3.2.1
[49] rstudioapi_0.16.0 knitr_1.45 snow_0.4-4 htmltools_0.5.8.1
[53] rmarkdown_2.26 compiler_4.3.3 getPass_0.2-4 askpass_1.2.0
[57] openssl_2.1.1